code
stringlengths 38
801k
| repo_path
stringlengths 6
263
|
|---|---|
# +
"""
17. How to compute the mean squared error on a truth and predicted series?
"""
"""
Difficulty Level: L2
"""
"""
Compute the mean squared error of truth and pred series.
"""
"""
Input
"""
"""
truth = pd.Series(range(10))
pred = pd.Series(range(10)) + np.random.random(10)
"""
# Input
truth = pd.Series(range(10))
pred = pd.Series(range(10)) + np.random.random(10)
# Solution
np.mean((truth-pred)**2)
| pset_pandas_ext/101problems/solutions/nb/p17.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ---
# layout: post
# title: "Entropy와 Gini계수"
# author: "<NAME>"
# categories: Data분석
# tags: [DecisionTree, 의사결정나무, 불순도, Entropy와, Gini, 엔트로피, 지니계수, InformationGain, information]
# image: 03_entropy_gini.png
# ---
# ## **목적**
# - 지난번 포스팅에 ensemble 모델에 관하여 이야기하면서 약한 모형으로 의사결정나무를 많이 사용하는 것을 알 수 있었습니다. 이번에는 의사결정 나무를 만들기 위하여 사용되는 Entropy와 gini index에 대해서 알아보도록 하겠습니다.
# <br/>
# <br/>
#
# ### **트리 구축의 원칙**
# 
# > 출처 : https://m.blog.naver.com/PostView.naver?isHttpsRedirect=true&blogId=ehdrndd&logNo=221158124011
# - 결정 트리를 구축할 때는 Occamm의 면도날처럼 데이터의 특성을 가장 잘 반영하는 간단한 가설을 먼저 채택하도록 되어있습니다. 어떻게 간단하고 합리적인 트리를 만들 수 있을 지 알아보겠습니다.
# <br>
# <br>
# ---
#
# ### **1. 결정 트리**
# 의사결정나무를 효율적으로 만들기 위해서는 변수의 기준에 따라 불순도/불확실성을 낮추는 방식으로 선택하여 만들게 됩니다.<br>
# 이에 불순도(Impurity) / 불확실성(Uncertainty)를 감소하는 것을 Information gain이라고 하며 이것을 최소화시키기 위하여 Gini Index와 Entropy라는 개념을 사용하게 되고 의사결정 나무의 종류에 따라 다르게 쓰입니다.<br>
# sklearn에서 default로 쓰이는 건 gini계수이며 이는 CART(Classificatioin And Regression Tree)에 쓰입니다.<br>
# ID3 그리고 이것을 개선한 C4.5, C5.0에서는 Entropy를 계산한다고 합니다. <br>
# CART tree는 항상 2진 분류를 하는 방식으로 나타나며, Entropy 혹은 Entropy 기반으로 계산되는 Information gain으로 계산되며 다중 분리가 됩니다. <br>
#
# - Gini계수와 Entropy 모두 높을수록 불순도가 높아져 분류를 하기 어렵습니다. <br>
# 
#
# |비 고|ID3|C4.5, C5|CART|
# |:---:|:---:|:---:|:---:|
# |평가지수|Entropy|Information gain|Gini Index(범주), 분산의 차이(수치)|
# |분리방식|다지분리|다지분리(범주) 및 이진분리(수치)|항상2진 분리|
# |비고|수치형 데이터 못 다룸|||
#
# <br>
# <br>
# > 출처/참고자료 : https://ko.wikipedia.org/wiki/%EA%B2%B0%EC%A0%95_%ED%8A%B8%EB%A6%AC_%ED%95%99%EC%8A%B5%EB%B2%95 <br>
# > 출처/참고자료 : https://m.blog.naver.com/PostView.naver?isHttpsRedirect=true&blogId=trashx&logNo=60099037740 <br>
# > 출처/참고자료 : https://ratsgo.github.io/machine%20learning/2017/03/26/tree/
# ---
#
# ### **1. Gini Index**
# 일단 sklearn의 DecisionTreeClassifier의 default 값인 Gini 계수에 대해서 먼저 설명하겠습니다. <br>
# 우선 Gini index의 공식입니다. <br>
#
# - 영역의 데이터 비율을 제곱하여 더한 값을 1에서 빼주게 된다.<br>
# 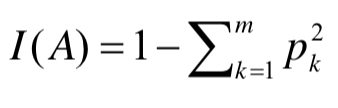 <br>
# <br>
# - 두개 영역 이상이 되면 비율의 제곱의 비율을 곱하여 1에서 빼주게 된다.<br>
# 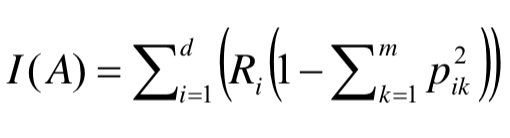
# > 출처 : https://soobarkbar.tistory.com/17
#
# <br>
#
# - 최대값을 보게되면 1 - ( (1/2)^2 + (1/2)^2 ) = 0.5
# - 최소값을 보게되면 1 - ( 1^2 + 0^2 ) = 0
# +
import os
import sys
import warnings
import math
import random
import numpy as np
import pandas as pd
import scipy
from sklearn import tree
from sklearn.tree import DecisionTreeClassifier
import matplotlib as mpl
from matplotlib import pyplot as plt
from plotnine import *
import graphviz
from sklearn.preprocessing import OneHotEncoder
# %matplotlib inline
warnings.filterwarnings("ignore")
# -
tennis = pd.read_csv("data/tennis.csv", index_col = "Day")
tennis
# - 위와 같은 데이터가 있다고 할 때, 우리는 어떤 요인이 가장 확실한(불확실성이 적은) 변수일지 생각을 하고 트리를 만들어야합니다.
# <br>
# <br>
#
# 아무것도 나누지 않았을 때 gini계수를 구하는 함수를 만든 후 얼마인지 출력해보겠습니다
def get_unique_dict(df) :
return {x : list(df[x].unique()) for x in ["Outlook", "Temperature", "Humidity", "Wind"]}
def get_gini(df, y_col) :
Ys = df[y_col].value_counts()
total_row = len(df)
return 1 - np.sum([np.square(len(df[df[y_col] == y]) / total_row) for y in Ys.index])
def gini_split(df, y_col, col, feature) :
r1 = len(df[df[col] == feature])
Y1 = dict(df[df[col] == feature][y_col].value_counts())
r2 = len(df[df[col] != feature])
Y2 = dict(df[df[col] != feature][y_col].value_counts())
ratio = r1 / (r1 + r2)
gi1 = 1 - np.sum([np.square(len(df[(df[col] == feature) & (df[y_col] == x)]) / r1) for x, y in Y1.items()])
gi2 = 1 - np.sum([np.square(len(df[(df[col] != feature) & (df[y_col] == x)]) / r2) for x, y in Y2.items()])
return (ratio * gi1) + ((1-ratio) * gi2)
# 어떤 기준으로 나누었을 때 gini계수를 구하는 함수를 만들어 예시로 Outlook이 Sunny일 때 gini 계수를 구해보겠습니다.
get_gini(tennis, "PlayTennis")
# 아무것도 나누지 않았을 때보다, Sunny로 나누었을 때 gini계수가 줄어드는 것을 볼 수 있습니다.<br>
# 이 때 이 차이값을 Information gain(정보획득)이라고 합니다. 그리고 정보획득량이 많은 쪽을 선택하여 트리의 구조를 만들기 시작합니다.
split_point = ["Outlook", "Sunny"]
print("{}, {} 기준 split 후 gini 계수 : {}".format(*split_point, gini_split(tennis, "PlayTennis", *split_point)))
print("information gain : {}".format(get_gini(tennis, "PlayTennis") - gini_split(tennis, "PlayTennis", *split_point)))
# - 이제 모든 변수에 대해서 각각의 gini계수를 구하여 정보획득량이 많은, 즉 gini계수가 적은 변수를 선정하여 트리를 만들어갑니다.
y_col = "PlayTennis"
unique_dict = get_unique_dict(tennis)
unique_dict
[f"col : {idx}, split_feature : {v} : gini_index = {gini_split(tennis, y_col, idx, v)}" for idx, val in unique_dict.items() for v in val]
gini_df = pd.DataFrame([[idx, v, gini_split(tennis, y_col, idx, v)] for idx, val in unique_dict.items() for v in val], columns = ["cat1", "cat2", "gini"])
print(gini_df.iloc[gini_df["gini"].argmax()])
print(gini_df.iloc[gini_df["gini"].argmin()])
# ---
# 임의로 x, y좌표를 생성하여 정보들이 얼마나 흩어져있는지 확인해보겠습니다.
def generate_xy(df, split_col = None, split_value = None) :
if split_col == None :
return df.assign(x = [random.random() for _ in range(len(df))], y = [random.random() for _ in range(len(df))])
else :
tmp_ = df[df[split_col] == split_value]
tmp__ = df[df[split_col] != split_value]
return pd.concat([tmp_.assign(x = [random.random() / 2 for _ in range(len(tmp_))], y = [random.random() for _ in range(len(tmp_))]),
tmp__.assign(x = [(random.random() / 2) + 0.5 for _ in range(len(tmp__))], y = [random.random() for _ in range(len(tmp__))])] )
# - 아무런 기준을 두지 않았을 때는 정보를 구분할 수 있는 정보가 없습니다.
p = (
ggplot(data = generate_xy(tennis), mapping = aes(x = "x", y = "y", color = y_col)) +
geom_point() +
theme_bw()
)
p.save(filename = "../assets/img/2021-06-01-Entropy/1.jpg")
# 
# - Outlook이 Overcast로 나누었을 때, Yes 4개가 확실히 구분되는 것을 볼 수 있습니다.
split_list = ["Outlook", "Overcast"]
p = (
ggplot(data = generate_xy(tennis, *split_list), mapping = aes(x = "x", y = "y", color = y_col)) +
geom_point() +
geom_vline(xintercept = 0.5, color = "red", alpha = 0.7) +
theme_bw()
)
p.save(filename = "../assets/img/2021-06-01-Entropy/2.jpg")
# 
# - 정보획득량이 가장 큰 Temperature가 Mild로 나누었을 때입니다.
split_list = ["Temperature", "Mild"]
p = (
ggplot(data = generate_xy(tennis, *split_list), mapping = aes(x = "x", y = "y", color = y_col)) +
geom_point() +
geom_vline(xintercept = 0.5, color = "red", alpha = 0.7) +
theme_bw()
)
p.save(filename = "../assets/img/2021-06-01-Entropy/3.jpg")
# 
# - Outlook이 Sunny, Rain으로 각각 나누었을 때입니다.
split_list = ["Outlook", "Sunny"]
p = (
ggplot(data = generate_xy(tennis, *split_list), mapping = aes(x = "x", y = "y", color = y_col)) +
geom_point() +
geom_vline(xintercept = 0.5, color = "red", alpha = 0.7) +
theme_bw()
)
p.save(filename = "../assets/img/2021-06-01-Entropy/4.jpg")
# 
split_list = ["Outlook", "Rain"]
p = (
ggplot(data = generate_xy(tennis, *split_list), mapping = aes(x = "x", y = "y", color = y_col)) +
geom_point() +
geom_vline(xintercept = 0.5, color = "red", alpha = 0.7) +
theme_bw()
)
p.save("../assets/img/2021-06-01-Entropy/5.jpg")
# 
# #### **실제 tree 모델과 비교하기 위하여 OneHotEncoding 후 트리모형을 돌려보도록 하겠습니다.
cols = ["Outlook", "Temperature", "Humidity", "Wind"]
oe = OneHotEncoder()
Xs = pd.get_dummies(tennis[cols])
Ys = tennis[y_col]
dt_gini = DecisionTreeClassifier(criterion="gini")
dt_gini.fit(Xs, Ys)
def save_graphviz(grp, grp_num) :
p = graphviz.Source(grp)
p.save(filename = f"../assets/img/2021-06-01-Entropy/{grp_num}")
p.render(filename = f"../assets/img/2021-06-01-Entropy/{grp_num}", format = "jpg")
grp = tree.export_graphviz(dt_gini, out_file = None, feature_names=Xs.columns,
class_names=Ys.unique(),
filled=True)
save_graphviz(grp, 6)
# 
# #### **실제로 이 순서가 맞는지 확인해보겠습니다**
get_gini(tennis, "PlayTennis")
gini_df.iloc[gini_df["gini"].argmin()]
tennis_node1 = tennis[tennis["Outlook"] != "Overcast"]
[print(f"col : {idx}, split_feature : {v} : gini_index = {gini_split(tennis_node1, y_col, idx, v)}") for idx, val in get_unique_dict(tennis_node1).items() for v in val]
gini_df = pd.DataFrame([[idx, v, gini_split(tennis_node1, y_col, idx, v)] for idx, val in get_unique_dict(tennis_node1).items() for v in val], columns = ["cat1", "cat2", "gini"])
print("")
print("gini index : {}".format(get_gini(tennis_node1, y_col)))
print(gini_df.iloc[gini_df["gini"].argmin()])
tennis_node2 = tennis[(tennis["Outlook"] != "Overcast") & (tennis["Humidity"] == "High")]
[print(f"col : {idx}, split_feature : {v} : gini_index = {gini_split(tennis_node2, y_col, idx, v)}") for idx, val in get_unique_dict(tennis_node2).items() for v in val]
gini_df = pd.DataFrame([[idx, v, gini_split(tennis_node2, y_col, idx, v)] for idx, val in get_unique_dict(tennis_node2).items() for v in val], columns = ["cat1", "cat2", "gini"])
print("")
print("gini index : {}".format(get_gini(tennis_node2, y_col)))
gini_df.iloc[gini_df["gini"].argmin()]
# #### - gini계수가 0이면 가장 끝쪽에 있는 terminal node가 됩니다.(데이터가 많으면 overfitting을 막기위하여 가지치기 컨셉이 활용됩니다)
tennis_ter1 = tennis[tennis["Outlook"] == "Overcast"]
[print(f"col : {idx}, split_feature : {v} : gini_index = {gini_split(tennis_ter1, y_col, idx, v)}") for idx, val in get_unique_dict(tennis_ter1).items() for v in val]
gini_df = pd.DataFrame([[idx, v, gini_split(tennis_ter1, y_col, idx, v)] for idx, val in get_unique_dict(tennis_ter1).items() for v in val], columns = ["cat1", "cat2", "gini"])
gini_df.iloc[gini_df["gini"].argmin()]
# ---
#
# ### **2. Entropy**
# 다음은 ID3, C4.5 등 트리에서 정보획득량을 측정하기 위해 쓰이는 Entropy입니다.<br>
# 우선 Entropy의 공식입니다. <br>
#
# - 영역의 데이터 비율을 제곱하여 더한 값을 1에서 빼주게 된다.<br>
# 
# <br>
max_entropy = (-1 * ((0.5*np.log2(0.5)) + (0.5*np.log2(0.5))))
min_entropy = (-1 * ((1*np.log2(1))))
print(f"Entropy의 최대값 : {max_entropy}")
print(f"Entropy의 최대값 : {min_entropy}")
tennis
def get_entropy(df, y_col) :
Ys = df[y_col].value_counts()
total_row = len(df)
(-1 * ((0.5*np.log2(0.5)) + (0.5*np.log2(0.5))))
return -1 * np.sum([(len(df[df[y_col] == y]) / total_row) * np.log2(len(df[df[y_col] == y]) / total_row) for y in Ys.index])
get_entropy(tennis, y_col)
def entropy_split(df, y_col, col, feature) :
r1 = len(df[df[col] == feature])
Y1 = dict(df[df[col] == feature][y_col].value_counts())
r2 = len(df[df[col] != feature])
Y2 = dict(df[df[col] != feature][y_col].value_counts())
ratio = r1 / (r1 + r2)
ent1 = np.sum([(len(df[(df[col] == feature) & (df[y_col] == x)]) / r1) * np.log2(len(df[(df[col] == feature) & (df[y_col] == x)]) / r1) for x, y in Y1.items()])
ent2 = np.sum([(len(df[(df[col] != feature) & (df[y_col] == x)]) / r2) * np.log2(len(df[(df[col] != feature) & (df[y_col] == x)]) / r2) for x, y in Y2.items()])
return -1 * ((ratio * ent1) + ((1-ratio) * ent2))
entropy_split(tennis, "PlayTennis", "Outlook", "Sunny")
# Entropy 역시 gini index와 똑같은 개념으로 아무것도 나누지 않았을 때보다, Sunny로 나누었을 때 줄어드는 것을 볼 수 있습니다.<br>
# 이 때 차이값(Information gain)을 이용하여 트리를 만들면 ID3, C4.5 등의 트리 구조를 만들게 됩니다.
[f"col : {idx}, split_feature : {v} : Entropy = {entropy_split(tennis, y_col, idx, v)}" for idx, val in get_unique_dict(tennis).items() for v in val]
entropy_df = pd.DataFrame([[idx, v, entropy_split(tennis, y_col, idx, v)] for idx, val in unique_dict.items() for v in val], columns = ["cat1", "cat2", "entropy"])
print(entropy_df.iloc[entropy_df["entropy"].argmin()])
print(entropy_df.iloc[gini_df["gini"].argmax()])
# #### **실제 tree 모델과 비교하기 위하여 OneHotEncoding 후 트리모형을 돌려보도록 하겠습니다.**
dt_entropy = DecisionTreeClassifier(criterion="entropy")
dt_entropy.fit(Xs, Ys)
grp = tree.export_graphviz(dt_entropy, out_file = None, feature_names=Xs.columns,
class_names=Ys.unique(),
filled=True)
save_graphviz(grp, 7)
# 
# #### **실제로 이 순서가 맞는지 확인해보겠습니다**
get_entropy(tennis, "PlayTennis")
entropy_df.iloc[entropy_df["entropy"].argmin()]
tennis_ter1 = tennis[tennis["Outlook"] == "Overcast"]
[print(f"col : {idx}, split_feature : {v} : entropy = {entropy_split(tennis_ter1, y_col, idx, v)}") for idx, val in get_unique_dict(tennis_ter1).items() for v in val]
entropy_df = pd.DataFrame([[idx, v, entropy_split(tennis, y_col, idx, v)] for idx, val in get_unique_dict(tennis_ter1).items() for v in val], columns = ["cat1", "cat2", "entropy"])
entropy_df.iloc[entropy_df["entropy"].argmin()]
tennis_ter1 = tennis[tennis["Outlook"] != "Overcast"]
[print(f"col : {idx}, split_feature : {v} : entropy = {entropy_split(tennis_ter1, y_col, idx, v)}") for idx, val in get_unique_dict(tennis_ter1).items() for v in val]
entropy_df = pd.DataFrame([[idx, v, entropy_split(tennis, y_col, idx, v)] for idx, val in get_unique_dict(tennis_ter1).items() for v in val], columns = ["cat1", "cat2", "entropy"])
entropy_df.iloc[entropy_df["entropy"].argmin()]
# ---
# ### **마지막으로 gini index와 entropy를 활용한 tree가 어떻게 노드가 나뉘었는지 보고 포스팅 마치겠습니다.**
#  
# <br>
# <br>
#
# ---
#
# <br>
#
# - code : [https://github.com/Chanjun-kim/Chanjun-kim.github.io/blob/main/_ipynb/2021-06-01-Entropy.ipynb](https://github.com/Chanjun-kim/Chanjun-kim.github.io/blob/main/_ipynb/2021-06-01-Entropy.ipynb) <br>
# - 참고 자료 : [https://m.blog.naver.com/PostView.naver?isHttpsRedirect=true&blogId=ehdrndd&logNo=221158124011](https://m.blog.naver.com/PostView.naver?isHttpsRedirect=true&blogId=ehdrndd&logNo=221158124011)
| _ipynb/2021-06-01-Entropy.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="KWRAbsYfeuQn" colab_type="code" outputId="67dc3c10-c34c-4fc5-fe46-d89eb7499296" colab={"base_uri": "https://localhost:8080/", "height": 73}
import torch
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# + id="6tkM3eZGfObT" colab_type="code" outputId="ae73e9d8-8b1f-4844-b16d-35f31b4db3f7" colab={"base_uri": "https://localhost:8080/", "height": 237}
data = pd.read_csv('Admission_Predict.csv', index_col=0)
data.head()
# + id="9Yb8b2rFfhNj" colab_type="code" outputId="5023441a-5dee-442c-fc91-03f535e49a1a" colab={"base_uri": "https://localhost:8080/", "height": 35}
data.shape
# + id="hv3oA4QkfwAk" colab_type="code" outputId="03e59d28-63c8-4c00-9338-d87f7bea74b9" colab={"base_uri": "https://localhost:8080/", "height": 300}
data.describe()
# + id="dWnd5ID-ixMb" colab_type="code" outputId="b34221c5-c503-4bfc-b705-1cb9b5c94a3a" colab={"base_uri": "https://localhost:8080/", "height": 128}
# !pip3 install --upgrade pandas
# + id="UiA1Bb3afx3w" colab_type="code" outputId="349e166f-187c-43df-de1e-43763e0686e3" colab={"base_uri": "https://localhost:8080/", "height": 206}
data = data.rename(index=str, columns={"Chance of Admit " : "admit_probability"})
data = data.reset_index()
data = data[['GRE Score', 'TOEFL Score', 'University Rating', 'LOR', 'SOP', 'CGPA', 'Research', 'admit_probability']]
data.head()
# + id="GSWnEB3egrRy" colab_type="code" outputId="7e758766-bc73-4486-8276-7d79ed8d31c1" colab={"base_uri": "https://localhost:8080/", "height": 513}
plt.figure(figsize = (8,8))
fig = sns.regplot(x='GRE Score', y='TOEFL Score', data=data)
plt.title("GRE v/s TOEFL Score")
plt.show(fig)
# + id="YKnNpLs9kkuP" colab_type="code" outputId="c77d5569-a383-4ca1-cfe1-1d056930dc4f" colab={"base_uri": "https://localhost:8080/", "height": 513}
plt.figure(figsize = (8,8))
fig = sns.regplot(x='GRE Score', y='CGPA', data=data)
plt.title("GRE Score v/s CGPA")
plt.show(fig)
# + id="hTtP96Tzlg1s" colab_type="code" outputId="95ed8581-b575-4a87-a435-61aedaf8ca7e" colab={"base_uri": "https://localhost:8080/", "height": 514}
plt.figure(figsize = (8,8))
fig = sns.scatterplot(x='admit_probability', y='CGPA', data=data, hue='Research')
plt.title("Scatterplot")
plt.show(fig)
# + id="NwPubRvXlqsK" colab_type="code" outputId="97c6450a-cf14-42c4-db90-88e1db861a15" colab={"base_uri": "https://localhost:8080/", "height": 684}
plt.figure(figsize = (10,10))
fig = sns.heatmap(data.corr(), annot=True, linewidths=0.05, fmt='.2f')
plt.title("Heatmap")
plt.show(fig)
# + [markdown] id="XiONHwAVp7c2" colab_type="text"
# #Data Preprocessing
# + id="1PyGC_ZsopzU" colab_type="code" colab={}
from sklearn import preprocessing
# + id="AEqJXE3xqClI" colab_type="code" colab={}
data[['GRE Score', 'TOEFL Score', 'LOR', 'SOP', 'CGPA']] = \
preprocessing.scale(data[['GRE Score', 'TOEFL Score', 'LOR', 'SOP', 'CGPA']])
# + id="hWL4ba0gqdYn" colab_type="code" outputId="bc091656-a260-464d-a7a6-3bb29539853f" colab={"base_uri": "https://localhost:8080/", "height": 363}
data.sample(10)
# + id="_3HZX3PXqgQf" colab_type="code" colab={}
col = ['GRE Score', 'TOEFL Score', 'LOR', 'SOP', 'CGPA', 'Research']
features = data[col]
# + id="MvScGNqTq0Dj" colab_type="code" outputId="74e1a5d4-00d8-4459-b07f-a88df61ef6de" colab={"base_uri": "https://localhost:8080/", "height": 206}
features.head()
# + id="O1YiruP9q5uQ" colab_type="code" colab={}
target = data[['admit_probability']]
# + id="MGv2czmHrB_N" colab_type="code" colab={}
y = target.copy()
# + id="1U62J-6OrG76" colab_type="code" outputId="e0e0f410-cb4e-4120-8ee9-9ea23386fe2c" colab={"base_uri": "https://localhost:8080/", "height": 424}
y.replace(to_replace = target[target >= 0.85], value = int(2), inplace=True)
y.replace(to_replace = target[target >= 0.65], value = int(1), inplace=True)
y.replace(to_replace = target[target < 0.65], value = int(0), inplace=False)
# + id="GFLAEjF_rkhs" colab_type="code" colab={}
target = y
# + id="CGlU8xvYrn3x" colab_type="code" outputId="c8295a98-7c46-4679-d9dd-341f649238fc" colab={"base_uri": "https://localhost:8080/", "height": 235}
target['admit_probability'].unique
# + id="T8PCIEUzrutl" colab_type="code" colab={}
from sklearn.model_selection import train_test_split
# + id="O5JRZmqEr74q" colab_type="code" colab={}
X_train, x_test, Y_train, y_test = train_test_split(features, target, test_size=0.2)
# + id="wDoseJgYsIK0" colab_type="code" colab={}
xtrain = torch.from_numpy(X_train.values).float()
xtest = torch.from_numpy(x_test.values).float()
# + id="P8xHDxl5sphm" colab_type="code" outputId="d50a6855-8f67-4cc7-8552-d92777f7309b" colab={"base_uri": "https://localhost:8080/", "height": 35}
xtrain.shape
# + id="-5BnkQJBstPT" colab_type="code" outputId="b61fefa0-3510-488f-fb61-0a81fd4b7a1b" colab={"base_uri": "https://localhost:8080/", "height": 35}
xtrain.dtype
# + id="4dWDyiuHsuxe" colab_type="code" colab={}
ytrain = torch.from_numpy(Y_train.values).view(-1, 1)[0].long()
ytest = torch.from_numpy(y_test.values).view(-1, 1)[0].long()
# + [markdown] id="b84js1JBt9RO" colab_type="text"
# #Making the model
# + id="JGOSf7qYuYpZ" colab_type="code" colab={}
import torch.nn as nn
import torch.nn.functional as F
# + id="c28rnvpvtcil" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 35} outputId="885be287-62e5-431a-b461-2ffc16986a59"
input_size = xtrain.shape[0]
output_size = len(target['admit_probability'].unique())
# xtrain = xtrain.unsqueeze_(2)
input_size
# + id="Uet491eLuSRI" colab_type="code" colab={}
class Net(nn.Module):
def __init__(self, hidden_size, activation_fn = 'relu', apply_dropout=False):
super(Net, self).__init__()
self.fc1 = nn.Linear(input_size, hidden_size)
self.fc2 = nn.Linear(hidden_size, hidden_size)
self.fc3 = nn.Linear(hidden_size, output_size)
self.hidden_size = hidden_size
self.activation_fn = activation_fn
self.dropout = None
if apply_dropout:
self.dropout = nn.dropout(0.2)
def forward(self, x):
activation_fn = None
if self.activation_fn == 'sigmoid':
activation_fn = F.torch.sigmoid
elif self.activation_fn == 'tanh':
activation_fn = F.torch.tanh
elif self.activation_fn == 'relu':
activation_fn = F.relu
x = activation_fn(self.fc1(x))
x = activation_fn(self.fc2(x))
if self.dropout != None:
x = self.dropout(x)
x = self.fc3(x)
return F.log_softmax(x, dim=-1)
# + id="iZA6zASk0cT5" colab_type="code" colab={}
import torch.optim as optim
# + id="s8FTknbH05yl" colab_type="code" colab={}
def train_and_evaluate_model(model, learn_rate=0.001):
epoch_data = []
epochs = 1001
optimizer = optim.Adam(model.parameters(), lr=learn_rate)
loss_fn = nn.NLLLoss()
test_accuracy = 0.0
for epoch in range(1, epochs):
optimizer.zero_grad()
model.train()
ypred = model(xtrain)
loss = loss_fn(ypred, ytrain)
loss.backward()
optimizer.step()
model.eval()
ypred_test = model(xtest)
loss_test = loss_fn(ypred_test, ytest)
_, pred = ypred_test.data.max(1)
test_accuracy = pred.eq(ytest.data).sum().item() / y_test.values.size
epoch_data.append([epoch, loss.data.item(), loss_test.data.item(), test_accuracy])
if epoch % 100 == 0:
print('epoch - %d (%d%%) train loss - %.2f test loss - %.2f test accuracy - %.4f'\
% (epoch, epoch/150 * 10, loss.data.item(), loss_test.data.item(),
test_accuracy))
return {
'model' : model,
'epoch_data' : epoch_data,
'num_epochs' : epochs,
'optimizer' : optimizer,
'loss_fn' : loss_fn,
'test_accuracy' : test_accuracy,
'_, pred' : ypred_test.data.max(1),
'actual_test_label' : ytest,
}
# + [markdown] id="zuocRRPP5hGB" colab_type="text"
# #Training the model
# + id="5u4n9Dtl5PJe" colab_type="code" outputId="3ee678ae-3279-4361-a5ba-68c46d111523" colab={"base_uri": "https://localhost:8080/", "height": 108}
signet = Net(hidden_size=50, activation_fn = 'sigmoid')
signet
# + id="4fg8bGhx8AGO" colab_type="code" colab={}
result_signet = train_and_evaluate_model(signet)
# + id="CQj6BnxK8IgA" colab_type="code" colab={}
| AdmissionPredictorNN.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="Agebac83jwLd"
import pandas as pd
import numpy as np
import io
import matplotlib.pyplot as plt
from google.colab import files
# + colab={"resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "<KEY>", "ok": true, "headers": [["content-type", "application/javascript"]], "status": 200, "status_text": ""}}, "base_uri": "https://localhost:8080/", "height": 72} id="Ze5DNKHyl2-U" outputId="2b3e03d8-4389-44da-aa5f-3cf994b101f7"
uploaded = files.upload()
# + colab={"base_uri": "https://localhost:8080/", "height": 224} id="IGVoGF6jmdGD" outputId="c5d0a36a-a812-42df-8e40-d06b6dad2235"
df_original = pd.read_csv(io.BytesIO(uploaded['property-tax-report.csv']))
df = df_original
df.head()
# + id="VYWKuHKNpc-O" colab={"base_uri": "https://localhost:8080/", "height": 224} outputId="5972ddcb-6e10-4d55-f995-70a0ed2ee612"
df['2021'] = 0
df['2020'] = 0
df['2019'] = 0
df.head()
# + id="7CaYdSq5qOh0"
df.dropna(inplace=True)
indexes_to_drop = []
for index, row in df.iterrows():
if row['TAX_ASSESSMENT_YEAR'] == 2021:
df.at[index, '2021'] = row['CURRENT_LAND_VALUE']
df.at[index, '2020'] = row['PREVIOUS_LAND_VALUE']
elif row['TAX_ASSESSMENT_YEAR'] == 2020:
df.at[index, '2020'] = row['CURRENT_LAND_VALUE']
df.at[index, '2019'] = row['PREVIOUS_LAND_VALUE']
else:
continue
df.head()
# + id="PpLhG80aGCey"
count_2019_one = 0
count_2020_one = 0
count_2021_one = 0
sum_2019_one = 0
sum_2020_one = 0
sum_2021_one = 0
count_2019_two = 0
count_2020_two = 0
count_2021_two = 0
sum_2019_two = 0
sum_2020_two = 0
sum_2021_two = 0
count_2019_multi = 0
count_2020_multi = 0
count_2021_multi = 0
sum_2019_multi = 0
sum_2020_multi = 0
sum_2021_multi = 0
for index, row in df.iterrows():
if row['ZONING_CLASSIFICATION'] == 'One-Family Dwelling':
if(row['2019']):
count_2019_one +=1
sum_2019_one += row['2019']
if(row['2020']):
count_2020_one +=1
sum_2020_one += row['2020']
if(row['2021']):
count_2021_one +=1
sum_2021_one += row['2021']
elif row['ZONING_CLASSIFICATION'] == 'Two-Family Dwelling':
if(row['2019']):
count_2019_two +=1
sum_2019_two += row['2019']
if(row['2020']):
count_2020_two +=1
sum_2020_two += row['2020']
if(row['2021']):
count_2021_two +=1
sum_2021_two += row['2021']
elif row['ZONING_CLASSIFICATION'] == 'Multiple Dwelling':
if(row['2019']):
count_2019_multi +=1
sum_2019_multi += row['2019']
if(row['2020']):
count_2020_multi +=1
sum_2020_multi += row['2020']
if(row['2021']):
count_2021_multi +=1
sum_2021_multi += row['2021']
avg_2019_one = sum_2019_one/count_2019_one
avg_2020_one = sum_2020_one/count_2020_one
avg_2021_one = sum_2021_one/count_2021_one
avg_2019_two = sum_2019_two/count_2019_two
avg_2020_two = sum_2020_two/count_2020_two
avg_2021_two = sum_2021_two/count_2021_two
avg_2019_multi = sum_2019_multi/count_2019_multi
avg_2020_multi = sum_2020_multi/count_2020_multi
avg_2021_multi = sum_2021_multi/count_2021_multi
data = {'Zone_Classification':['One-Family Dwelling', 'Two-Family Dwelling', 'Multiple Dwelling'],
'2019':[avg_2019_one, avg_2019_two, avg_2019_multi], '2020':[avg_2020_one, avg_2020_two, avg_2020_multi], '2021':[avg_2021_one, avg_2021_two, avg_2021_multi]}
# + id="c6S2Knn4qObz" colab={"base_uri": "https://localhost:8080/", "height": 142} outputId="48bea29f-6251-4308-8e4d-66fe43b6b6d0"
results = pd.DataFrame(data)
results
# + id="87f20zVdqOT0" colab={"base_uri": "https://localhost:8080/", "height": 293} outputId="9d841cbc-063d-4f5a-f0c8-cf32420afcf4"
x = np.arange(3)
y1 = [avg_2019_one, avg_2020_one, avg_2021_one]
y2 = [avg_2019_two, avg_2020_two, avg_2021_two]
y3 = [avg_2019_multi, avg_2020_multi, avg_2021_multi]
width = 0.20
plt.bar(x-0.2, y1, width)
plt.bar(x, y2, width)
plt.bar(x+0.2, y3, width)
plt.xticks(x, ['One-Family', 'Two-Family', 'Multiple'])
plt.legend(["2019", "2020", "2021"])
plt.ylabel("Land Values (xe6)")
# + id="UjuqT2EYqODg"
| notebooks/LandCostAveragePerYear.ipynb |
# +
# Copyright 2010-2018 Google LLC
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# [START program]
"""Simple Vehicles Routing Problem."""
# [START import]
from __future__ import print_function
from ortools.constraint_solver import routing_enums_pb2
from ortools.constraint_solver import pywrapcp
# [END import]
# [START data_model]
def create_data_model():
"""Stores the data for the problem."""
data = {}
data['distance_matrix'] = [
[
0, 548, 776, 696, 582, 274, 502, 194, 308, 194, 536, 502, 388, 354,
468, 776, 662
],
[
548, 0, 684, 308, 194, 502, 730, 354, 696, 742, 1084, 594, 480, 674,
1016, 868, 1210
],
[
776, 684, 0, 992, 878, 502, 274, 810, 468, 742, 400, 1278, 1164,
1130, 788, 1552, 754
],
[
696, 308, 992, 0, 114, 650, 878, 502, 844, 890, 1232, 514, 628, 822,
1164, 560, 1358
],
[
582, 194, 878, 114, 0, 536, 764, 388, 730, 776, 1118, 400, 514, 708,
1050, 674, 1244
],
[
274, 502, 502, 650, 536, 0, 228, 308, 194, 240, 582, 776, 662, 628,
514, 1050, 708
],
[
502, 730, 274, 878, 764, 228, 0, 536, 194, 468, 354, 1004, 890, 856,
514, 1278, 480
],
[
194, 354, 810, 502, 388, 308, 536, 0, 342, 388, 730, 468, 354, 320,
662, 742, 856
],
[
308, 696, 468, 844, 730, 194, 194, 342, 0, 274, 388, 810, 696, 662,
320, 1084, 514
],
[
194, 742, 742, 890, 776, 240, 468, 388, 274, 0, 342, 536, 422, 388,
274, 810, 468
],
[
536, 1084, 400, 1232, 1118, 582, 354, 730, 388, 342, 0, 878, 764,
730, 388, 1152, 354
],
[
502, 594, 1278, 514, 400, 776, 1004, 468, 810, 536, 878, 0, 114,
308, 650, 274, 844
],
[
388, 480, 1164, 628, 514, 662, 890, 354, 696, 422, 764, 114, 0, 194,
536, 388, 730
],
[
354, 674, 1130, 822, 708, 628, 856, 320, 662, 388, 730, 308, 194, 0,
342, 422, 536
],
[
468, 1016, 788, 1164, 1050, 514, 514, 662, 320, 274, 388, 650, 536,
342, 0, 764, 194
],
[
776, 868, 1552, 560, 674, 1050, 1278, 742, 1084, 810, 1152, 274,
388, 422, 764, 0, 798
],
[
662, 1210, 754, 1358, 1244, 708, 480, 856, 514, 468, 354, 844, 730,
536, 194, 798, 0
],
]
data['num_vehicles'] = 4
# [START starts_ends]
data['starts'] = [1, 2, 15, 16]
data['ends'] = [0, 0, 0, 0]
# [END starts_ends]
return data
# [END data_model]
# [START solution_printer]
def print_solution(data, manager, routing, solution):
"""Prints solution on console."""
max_route_distance = 0
for vehicle_id in range(data['num_vehicles']):
index = routing.Start(vehicle_id)
plan_output = 'Route for vehicle {}:\n'.format(vehicle_id)
route_distance = 0
while not routing.IsEnd(index):
plan_output += ' {} -> '.format(manager.IndexToNode(index))
previous_index = index
index = solution.Value(routing.NextVar(index))
route_distance += routing.GetArcCostForVehicle(
previous_index, index, vehicle_id)
plan_output += '{}\n'.format(manager.IndexToNode(index))
plan_output += 'Distance of the route: {}m\n'.format(route_distance)
print(plan_output)
max_route_distance = max(route_distance, max_route_distance)
print('Maximum of the route distances: {}m'.format(max_route_distance))
# [END solution_printer]
"""Entry point of the program."""
# Instantiate the data problem.
# [START data]
data = create_data_model()
# [END data]
# Create the routing index manager.
# [START index_manager]
manager = pywrapcp.RoutingIndexManager(len(data['distance_matrix']),
data['num_vehicles'], data['starts'],
data['ends'])
# [END index_manager]
# Create Routing Model.
# [START routing_model]
routing = pywrapcp.RoutingModel(manager)
# [END routing_model]
# Create and register a transit callback.
# [START transit_callback]
def distance_callback(from_index, to_index):
"""Returns the distance between the two nodes."""
# Convert from routing variable Index to distance matrix NodeIndex.
from_node = manager.IndexToNode(from_index)
to_node = manager.IndexToNode(to_index)
return data['distance_matrix'][from_node][to_node]
transit_callback_index = routing.RegisterTransitCallback(distance_callback)
# [END transit_callback]
# Define cost of each arc.
# [START arc_cost]
routing.SetArcCostEvaluatorOfAllVehicles(transit_callback_index)
# [END arc_cost]
# Add Distance constraint.
# [START distance_constraint]
dimension_name = 'Distance'
routing.AddDimension(
transit_callback_index,
0, # no slack
2000, # vehicle maximum travel distance
True, # start cumul to zero
dimension_name)
distance_dimension = routing.GetDimensionOrDie(dimension_name)
distance_dimension.SetGlobalSpanCostCoefficient(100)
# [END distance_constraint]
# Setting first solution heuristic.
# [START parameters]
search_parameters = pywrapcp.DefaultRoutingSearchParameters()
search_parameters.first_solution_strategy = (
routing_enums_pb2.FirstSolutionStrategy.PATH_CHEAPEST_ARC)
# [END parameters]
# Solve the problem.
# [START solve]
solution = routing.SolveWithParameters(search_parameters)
# [END solve]
# Print solution on console.
# [START print_solution]
if solution:
print_solution(data, manager, routing, solution)
# [END print_solution]
| examples/notebook/constraint_solver/vrp_starts_ends.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] deletable=true editable=true
# # Numpy 介紹
# + deletable=true editable=true
# 起手式
import numpy as np
# + [markdown] deletable=true editable=true
# ### 建立 ndarray
# + deletable=true editable=true
np.array([1,2,3,4])
# + deletable=true editable=true
x = _
# + deletable=true editable=true
y = np.array([[1.,2,3],[4,5,6]])
y
# + [markdown] deletable=true editable=true
# 看 ndarray 的第一件事情: shape , dtype
# + deletable=true editable=true
x.shape
# + deletable=true editable=true
y.shape
# + deletable=true editable=true
x.dtype
# + deletable=true editable=true
y.dtype
# + [markdown] deletable=true editable=true
# ### 有時候,可以看圖
# + deletable=true editable=true
# import matplotlib
# %matplotlib inline
import matplotlib.pyplot as plt
# 畫圖
plt.plot(x, 'x');
# + [markdown] deletable=true editable=true
# ### 有很多其他建立的方式
# + deletable=true editable=true
# 建立 0 array
np.zeros_like(y)
# + deletable=true editable=true
np.zeros((10,10))
# + deletable=true editable=true
# 跟 range 差不多
x = np.arange(0, 10, 0.1)
# 亂數
y = np.random.uniform(-1,1, size=x.shape)
plt.plot(x, y)
# + [markdown] deletable=true editable=true
# 這是一堆資料
# * 資料有什麼資訊?
# * 資料有什麼限制?
# * 這些限制有什麼意義?好處?
# * 以前碰過什麼類似的東西?
# * 可以套用在哪些東西上面?
# * 可以怎麼用(運算)?
# + [markdown] deletable=true editable=true
# 最簡單的計算是 **逐項計算**
# see also `np.vectorize`
# + deletable=true editable=true
x = np.linspace(0, 2* np.pi, 1000)
plt.plot(x, np.sin(x))
# + [markdown] deletable=true editable=true
# ## Q0:
# 畫出 $y=x^2+1$ 或其他函數的圖形
# 用
# ```python
# # plt.plot?
# ```
# 看看 plot 還有什麼參數可以玩
# + deletable=true editable=true
#可以用 %run -i 跑參考範例
# %run -i q0.py
# + deletable=true editable=true
# 或者看看參考範例
# #%load q0.py
# + [markdown] deletable=true editable=true
# ## Q1:
# 試試看圖片。
# 使用
# ```python
# from PIL import Image
# # 讀入 PIL Image (這張圖是從 openclipart 來的 cc0)
# img = Image.open('img/Green-Rolling-Hills-Landscape-800px.png')
# # 圖片轉成 ndarray
# img_array = np.array(img)
# # ndarray 轉成 PIL Image
# Image.fromarray(img_array)
# ```
# 看看這個圖片的內容, dtype 和 shape
# + deletable=true editable=true
# 參考答案
# #%load q1.py
# + [markdown] deletable=true editable=true
# ### Indexing
# 可以用類似 list 的 indexing
# + deletable=true editable=true
a = np.arange(30)
a
# + deletable=true editable=true
a[5]
# + deletable=true editable=true
a[3:7]
# + deletable=true editable=true
# 列出所有奇數項
a[1::2]
# + deletable=true editable=true
# 還可以用來設定值
a[1::2] = -1
a
# + deletable=true editable=true
# 或是
a[1::2] = -a[::2]-1
a
# + [markdown] deletable=true editable=true
# ## Q2
# 給定
# ```python
# x = np.arange(30)
# a = np.arange(30)
# a[1::2] = -a[1::2]
# ```
# 畫出下面的圖
# + deletable=true editable=true
# %run -i q2.py
# #%load q2.py
# + [markdown] deletable=true editable=true
# ### ndarray 也可以
# + deletable=true editable=true
b = np.array([[1,2,3], [4,5,6], [7,8,9]])
b
# + deletable=true editable=true
b[1][2]
# + deletable=true editable=true
b[1,2]
# + deletable=true editable=true
b[1]
# + [markdown] deletable=true editable=true
# ## Q3
# 動手試試看各種情況
# 比方
# ```python
# b = np.random.randint(0,99, size=(10,10))
# b[::2, 2]
# ```
# + [markdown] deletable=true editable=true
# ### Fancy indexing
# + deletable=true editable=true
b = np.random.randint(0,99, size=(5,10))
b
# + [markdown] deletable=true editable=true
# 試試看下面的結果
#
# 想一下是怎麼一回事(numpy 在想什麼?)
# + deletable=true editable=true
b[[1,3]]
# + deletable=true editable=true
b[(1,3)]
# + deletable=true editable=true
b[[1,2], [3,4]]
# + deletable=true editable=true
b[[(1,2),(3,4)]]
# + deletable=true editable=true
b[[True, False, False, True, False]]
# + [markdown] deletable=true editable=true
# ## Q4
# 把 `b` 中的偶數都變成 `-1`
# + deletable=true editable=true
#參考範例
# %run -i q4.py
# + [markdown] deletable=true editable=true
# # 用圖形來練習
# + deletable=true editable=true
# 還記得剛才的
from PIL import Image
img = Image.open('img/Green-Rolling-Hills-Landscape-800px.png')
img_array = np.array(img)
Image.fromarray(img_array)
# + deletable=true editable=true
# 用來顯示圖片的函數
from IPython.display import display
def show(img_array):
display(Image.fromarray(img_array))
# + [markdown] deletable=true editable=true
# ## Q
# * 將圖片縮小成一半
# * 擷取中間一小塊
# * 圖片上下顛倒
# * 左右鏡射
# * 去掉綠色
# * 將圖片放大兩倍
# * 貼另外一張圖到大圖中
# ```python
# from urllib.request import urlopen
# url = "https://raw.githubusercontent.com/playcanvas/engine/master/examples/images/animation.png"
# simg = Image.open(urlopen(url))
# ```
# * 紅綠交換
# * 團片變成黑白 參考 `Y=0.299R+0.587G+0.114B`
# * 會碰到什麼困難? 要如何解決
# + deletable=true editable=true
# 將圖片縮小成一半
# %run -i q_half.py
# + deletable=true editable=true
# 將圖片放大
# %run -i q_scale2.py
# + deletable=true editable=true
# 圖片上下顛倒
show(img_array[::-1])
# + deletable=true editable=true
# %run -i q_paste.py
# + deletable=true editable=true
# %run -i q_grayscale.py
# + [markdown] deletable=true editable=true
# ## Q
# * 挖掉個圓圈? (300,300)中心,半徑 100
# * 旋轉九十度? x,y 互換?
# + deletable=true editable=true
# 用迴圈畫圓
# %run -i q_slow_circle.py
# + deletable=true editable=true
# 用 fancy index 畫圓
# %run -i q_fast_circle.py
# + [markdown] deletable=true editable=true
# ### indexing 的其他用法
# + deletable=true editable=true
# 還可以做模糊化
a = img_array.astype(float)
for i in range(10):
a[1:,1:] = (a[1:,1:]+a[:-1,1:]+a[1:,:-1]+a[:-1,:-1])/4
show(a.astype('uint8'))
# + deletable=true editable=true
# 求邊界
a = img_array.astype(float)
a = a @ [0.299, 0.587, 0.114, 0]
a = np.abs((a[1:]-a[:-1]))*2
show(a.astype('uint8'))
# + [markdown] deletable=true editable=true
# ## Reshaping
# `.flatten` 拉平看看資料在電腦中如何儲存?
#
#
#
# 查看
# `.reshape, .T, np.rot00, .swapaxes .rollaxis`
# 然後再做一下上面的事情
# + deletable=true editable=true
# reshaping 的應用
R,G,B,A = img_array.reshape(-1,4).T
plt.hist((R,G,B,A), color="rgby");
# + [markdown] deletable=true editable=true
# ## 堆疊在一起
# 查看 `np.vstack` `np.hstack` `np.concatenate` 然後試試看
# + deletable=true editable=true
# 例子
show(np.hstack([img_array, img_array2]))
# + deletable=true editable=true
# 例子
np.concatenate([img_array, img_array2], axis=2).shape
# + [markdown] deletable=true editable=true
# ## 作用在整個 array/axis 的函數
# + deletable=true editable=true
np.max([1,2,3,4])
# + deletable=true editable=true
np.sum([1,2,3,4])
# + deletable=true editable=true
np.mean([1,2,3,4])
# + deletable=true editable=true
np.min([1,2,3,4])
# + [markdown] deletable=true editable=true
# 多重意義的運用, 水平平均,整合垂直平均
# + deletable=true editable=true
x_mean = img_array.astype(float).min(axis=0, keepdims=True)
print(x_mean.dtype, x_mean.shape)
y_mean = img_array.astype(float).min(axis=1, keepdims=True)
print(y_mean.dtype, y_mean.shape)
# 自動 broadcast
xy_combined = ((x_mean+y_mean)/2).astype('uint8')
show(xy_combined)
# + [markdown] deletable=true editable=true
# ## Tensor 乘法
# + [markdown] deletable=true editable=true
# 先從點積開始
# + deletable=true editable=true
# = 1*4 + 2*5 + 4*6
np.dot([1,2,3], [4,5,6])
# + deletable=true editable=true
u=np.array([1,2,3])
v=np.array([4,5,6])
print( u@v )
print( (u*v).sum() )
# + [markdown] deletable=true editable=true
# ### 矩陣乘法
# 如果忘記矩陣乘法是什麼了, 參考這裡 http://matrixmultiplication.xyz/
# 或者 http://eli.thegreenplace.net/2015/visualizing-matrix-multiplication-as-a-linear-combination/
#
#
# 矩陣乘法可以看成是:
# * 所有組合(其他軸)的內積(共有軸)
# * 多個行向量線性組合
# * 代入線性方程式 [A1-矩陣與基本列運算.ipynb](A1-矩陣與基本列運算.ipynb)
# * 用 numpy 來理解
# ```python
# np.sum(a[:,:, np.newaxis] * b[np.newaxis, : , :], axis=1)
# dot(a, b)[i,k] = sum(a[i,:] * b[:, k])
# ```
#
# ### 高維度
# 要如何推廣?
# * tensordot, tensor contraction, a.shape=(3,4,5), b.shape=(4,5,6), axis = 2 時等價於
# ```python
# np.sum(a[..., np.newaxis] * b[np.newaxis, ...], axis=(1, 2))
# tensordot(a,b)[i,k]=sum(a[i, ...]* b[..., k])
# ```
# https://en.wikipedia.org/wiki/Tensor_contraction
#
# * dot
# ```python
# dot(a, b)[i,j,k,m] = sum(a[i,j,:] * b[k,:,m])
# np.tensordot(a,b, axes=(-1,-2))
# ```
# * matmul 最後兩個 index 當成 matrix
# ```python
# a=np.random.random(size=(3,4,5))
# b=np.random.random(size=(3,5,7))
# (a @ b).shape
# np.sum(a[..., np.newaxis] * np.moveaxis(b[..., np.newaxis], -1,-3), axis=-2)
# ```
# * einsum https://en.wikipedia.org/wiki/Einstein_notation
# ```python
# np.einsum('ii', a) # trace(a)
# np.einsum('ii->i', a) #diag(a)
# np.einsum('ijk,jkl', a, b) # tensordot(a,b)
# np.einsum('ijk,ikl->ijl', a,b ) # matmul(a,b)
# ```
# + deletable=true editable=true
A=np.random.randint(0,10, size=(5,3))
A
# + deletable=true editable=true
B=np.random.randint(0,10, size=(3,7))
B
# + deletable=true editable=true
A.dot(B)
# + [markdown] deletable=true editable=true
# ## Q
# * 手動算算看 A,B 的 dot
# * 試試看其他的乘法
# + [markdown] deletable=true editable=true
# ### 小結
# + [markdown] deletable=true editable=true
# numpy 以 ndarray 為中心
# * 最基本的運算是逐項運算(用 `np.vectorize`把一般的函數變成逐項運算 )
# * indexing 很好用
# * reshaping
# * 整合的操作與計算
| Week01/01-Numpy.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import pandas as pd
import numpy as np
import math
import tensorflow as tf
import matplotlib.pyplot as plt
print(pd.__version__)
import progressbar
from tensorflow import keras
# ### Load of the test data
from process import loaddata
class_data = loaddata("../data/classifier/100-high-ene.csv")
np.random.shuffle(class_data)
y = class_data[:,-7:-4]
x = class_data[:,1:7]
# ### Model Load
model = keras.models.load_model('../models/classificationandregression/large_mse250.h5')
# ### Test of the Classification&Regression NN
model.fit(x, y)
# ### Test spectrum
#
# A quick way of saying how well the network is doing. We reproduce the electrons final spectrum using the Neural Network's prediction and we compare it to the real "spectrum", the one obtained from OSIRIS.
def energy_spectrum(energy_array, bins):
energy_array = np.array(energy_array)
plt.hist(energy_array, bins, histtype=u'step')
plt.yscale("log")
plt.show()
final_e = []
for y_ in y:
final_e.append(np.linalg.norm(y_))
energy_spectrum(final_e, 75)
prediction = model.predict(x)
from tensorflow import keras
final_e_nn = []
bar = progressbar.ProgressBar(maxval=len(prediction),
widgets=[progressbar.Bar('=', '[', ']'), ' ',
progressbar.Percentage(),
" of {0}".format(len(prediction))])
bar.start()
for i, pred in enumerate(prediction):
final_e_nn.append(np.linalg.norm(pred))
bar.update(i+1)
bar.finish()
plt.hist(final_e, bins=100, alpha = 0.5, color = 'mediumslateblue', label='Electrons spectrum', density = True)
plt.legend(loc='upper right')
plt.yscale('log')
plt.savefig('../plots/onenetwork/highene/electronspectrum.png')
plt.savefig('../plots/onenetwork/highene/electronspectrum.pdf')
plt.show()
plt.hist(final_e_nn, bins=100, alpha = 0.5, color = 'indianred', label='NN prediction', density = True)
plt.legend(loc='upper right')
plt.yscale('log')
plt.savefig('../plots/onenetwork/highene/NNprediction.png')
plt.savefig('../plots/onenetwork/highene/NNprediction.pdf')
plt.show()
plt.hist(final_e_nn, bins=100, alpha = 0.5, color = 'mediumslateblue', label='NN prediction', density = True)
plt.hist(final_e, bins=100, alpha = 0.5, color = 'indianred', label='Electron Momentum from simulations', density = True)
plt.legend(loc = 'upper right')
plt.yscale('log')
plt.savefig('../plots/onenetwork/highene/comparison.png')
plt.savefig('../plots/onenetwork/highene/comparison.pdf')
plt.show()
| notebooks/Cool ideas...but not working/Test_NeuralNetwork-Highenergy_RegressionSpectrum.ipynb |
-- -*- coding: utf-8 -*-
-- ---
-- jupyter:
-- jupytext:
-- text_representation:
-- extension: .hs
-- format_name: light
-- format_version: '1.5'
-- jupytext_version: 1.14.4
-- kernelspec:
-- display_name: Haskell
-- language: haskell
-- name: haskell
-- ---
-- # 5. List comprehensions
-- ### 5.1 Basic concepts
-- $\displaystyle \big\{x2 \mid x\in\{1 . . 5\} \big\}$
--
-- $\{1,4,9,16,25\}$
[x^2 | x <- [1..5]]
-- 표현식 `x <- [1..5]` 에서 식 `[1..5]`가 제너레이터(generator)에 해당됨.
[(x,y) | x <- [1,2,3], y <- [4,5]]
[(x,y) | y <- [4,5], x <- [1,2,3]]
[(x,y) | x <- [1,2,3], y <- [1,2,3]]
:type concat
concat :: [[a]] -> [a]
concat x2 = [x | x1 <- x2, x <- x1]
-- concat xss = [x | xs <- xss, x <- xs]
concat [[1,2],[3,4,5],[6,7,8,9]]
firsts :: [(a,b)] -> [a]
firsts ps = [x | (x, _) <- ps]
firsts [(1,'a'), (2,'b'), (3,'c')] -- [1,2,3]
length :: [a] -> Int
length xs = sum [1 | _ <- xs]
length [1,2,3,4,5] -- 5
xs = [1,2,3,4,5]
[1 | _ <- xs]
sum [1, 2, 3, 4, 5]
-- ### 5.2 Guards (조건)
[x | x <- [1..10], even x]
[x | x <- [1..10], odd x]
-- 표현식 `even x` 와 `odd x` 는 가드라고 부른다.
factors :: Int -> [Int]
factors n = [x | x <- [1..n], n `mod` x == 0]
factors 15 -- [1,3,5,15]
factors 7 -- [1,7]
prime :: Int -> Bool
prime n = factors n == [1, n]
prime 15
prime 7
primes' :: Int -> [Int]
primes' t = [x | x <- [2..t], prime x]
primes' 40
find :: Eq a => a -> [(a, b)] -> [b]
find k t = [v | (k',v) <- t, k == k']
find 'a' [('a',1), ('b',2), ('c',3), ('b',4)] -- [1]
find 'b' [('a',1), ('b',2), ('c',3), ('b',4)] -- [2,4]
find 'c' [('a',1), ('b',2), ('c',3), ('b',4)] -- [3]
find 'd' [('a',1), ('b',2), ('c',3), ('b',4)] -- []
find' :: Eq b => b -> [(a, b)] -> [a]
find' t k = [v | (v, t') <- k, t == t']
find' 1 [('a',1), ('b',2), ('c',3), ('b',4)] -- ['a']
find' 2 [('a',1), ('b',2), ('c',3), ('b',4)] -- ['b']
find' 3 [('a',1), ('b',2), ('c',3), ('b',4)] -- ['c']
find' 4 [('a',1), ('b',2), ('c',3), ('b',4)] -- ['d']
--
find''
find'' :: Eq a => a -> [(a,(b,c))] -> [(b,c)]
find'' k t = [v | (k',(v)) <- t, k == k']
find'' 'a' [('a',(1,2)), ('b',(3,4)), ('c',(5,6)), ('b',(7,8))] -- ['a']
-- 결론은
-- #### [처음 식 리스트 중 하나를 이용한 값(표현식) | 처음 식 리스트 중 하나 <- 처음 식, 조건] 구조로 이루어짐.
-- ### 5.3 The `zip` function
zip ['a','b','c'] [1,2,3,4]
:type zip
pairs :: [a] -> [(a, a)]
pairs ns = zip ns (tail ns)
xs = [1,2,3,4]
tail xs
zip [1,2,3] [2,3,4]
pairs [1,2,3,4] -- [(1,2),(2,3),(3,4)]
-- * `[1,2,3,4,5,6,7,8]` 을 인접한 수끼리 묶은 튜플 리스트를 만들고 싶다면
-- `zip [1,2,3,4,5,6,7] [2,3,4,5,6,7,8]`
zip [1,2,3,4,5,6,7] [2,3,4,5,6,7,8]
sorted :: Ord a => [a] -> Bool
sorted xs = and [x <= y | (x,y) <- pairs xs]
sorted [1,2,3,4] -- True
sorted [1,3,2,4] -- False
:type and
and [True, False]
sorted :: Ord a => [a] -> [a]
sorted xs = [if x > y then x else y | (x,y) <- pairs xs]
sorted [1,2,3,4] -- True
sorted [1,3,2,4] -- False
positions :: Eq a => a -> [a] -> [Int] -- zip a b 사용
positions x xs = [i | (x', i) <- zip xs [0..], x == x']
zip [True, False, True, False] [0..]
positions False [True, False, True, False] -- [1,3]
-- ### 5.4 String comprehensions
("abc" :: String) == (['a','b','c'] :: [Char])
"abcde" !! 2
take 3 "abcde"
length "abcde"
-- +
import Data.Char (isLower)
lowers :: String -> Int
lowers cs = length [c | c <- cs, isLower c]
-- -
lowers "Haskell" -- 6
lowers "LaTeX" -- 2
count :: Char -> String -> Int
count c cs = sum [1 | c' <- cs, c == c']
count' c cs = length [c' | c' <- cs, c == c']
count 's' "Mississippi"
count' 's' "Mississippi"
-- ### 5.5 The Caesar cipher
| PiHchap05.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:pg]
# language: python
# name: conda-env-pg-py
# ---
# + [markdown] toc=true
# <h1>Inhaltsverzeichnis<span class="tocSkip"></span></h1>
# <div class="toc"><ul class="toc-item"></ul></div>
# -
import pygimli as pg
import matplotlib.pyplot as plt
import pygimli.meshtools as mt
import os
from os import system
import numpy as np
def callTriangle(filename,
quality=33,
triangle='triangle',
verbose=True):
filebody = filename.replace('.poly', '')
syscal = triangle + ' -pq' + str(quality)
syscal += 'Aa ' + filebody + '.poly'
if verbose:
print(syscal)
system(syscal)
world = mt.createWorld(start=[-2e5, -2e5],
end=[2e5, 2e5],
layers=[0, 75e3],
area=[0, 1e7, 0],
marker=[1, 2, 3],
worldMarker=False)
blockleft = mt.createRectangle(start=[-22e3, 7e3],
end=[-4e3, 9e3],
marker=4,
markerPosition=[-18e3, 8e3],
area=1e4, boundaryMarker=10)
blockright = mt.createRectangle(start=[10e3, 7e3],
end=[22e3, 9e3],
marker=5,
markerPosition=[18e3, 8e3],
area=1e4, boundaryMarker=12)
geom = world + blockleft + blockright
ax, _ = pg.show(geom,
showNodes=False,
boundaryMarker=False)
ax.set_ylim(ax.get_ylim()[::-1]);
mt.exportPLC(geom,
'../meshes/commemi2d2.poly',
float_format='.8e')
callTriangle('../meshes/commemi2d2.poly',
quality=34.2,
verbose=False)
mesh = mt.createMesh(geom, quality=34.2)
ax, _ = pg.show(mesh)
ax.set_ylim(ax.get_ylim()[::-1]);
| Notebooks/COMMEMI_2D_2.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
from load_data_df import *
# # Data Paths
aes_data_dir = '../aes'
uart_data_dir = '../uart'
or1200_data_dir = '../or1200'
picorv32_data_dir = '../picorv32'
# # Plot Settings
# +
# Plot Settings
FIG_WIDTH = 12
FIG_HEIGHT = 6
HIST_SAVE_AS_PDF = True
AES_FP_SAVE_AS_PDF = False
UART_FP_SAVE_AS_PDF = False
OR1200_FP_SAVE_AS_PDF = False
# Plot PDF Filenames
HIST_PDF_FILENAME = 'cntr_sizes_histogram.pdf'
AES_FP_PDF_FILENAME = 'aes_fps_vs_time.pdf'
UART_FP_PDF_FILENAME = 'uart_fps_vs_time.pdf'
OR1200_FP_PDF_FILENAME = 'or1200_fps_vs_time.pdf'
# -
# # Plot Counter Size Histogram
# +
def plt_histogram(data, ax, title, color_index):
bins = [0,8,16,32,64,128,256] # your bins
# Create Histogram
hist, bin_edges = np.histogram(data,bins) # make the histogram
# Make x-tick labels list
xtick_labels = ['{}'.format(bins[i]) for i,j in enumerate(hist)]
xtick_labels[0] = 1
# Plot Histogram
ax.bar(\
range(len(hist)),\
hist,\
width = 1,\
align = 'center',\
tick_label = xtick_labels, \
color = current_palette[color_index])
# Format Histogram
ax.set_title(title, fontweight='bold', fontsize=14)
ax.set_xlabel('Register Size (# bits)', fontsize=12)
ax.set_ylabel('# Registers', fontsize=12)
ax.grid(axis='y', alpha=0.5)
ax.set_ylim(0, 400)
# Text on the top of each barplot
for i in range(len(hist)):
ax.text(x = i , y = hist[i] + 10, s = hist[i], size = 12, horizontalalignment='center')
# Load Data
aes_counter_sizes = load_counter_sizes(aes_data_dir)
uart_counter_sizes = load_counter_sizes(uart_data_dir)
or1200_counter_sizes = load_counter_sizes(or1200_data_dir)
picorv32_counter_sizes = load_counter_sizes(picorv32_data_dir)
# Create Figure
sns.set()
current_palette = sns.color_palette()
fig, axes = plt.subplots(1, 4, figsize=(14, 3))
plt_histogram(aes_counter_sizes['Coalesced Sizes'], axes[0], 'AES', 0)
plt_histogram(uart_counter_sizes['Coalesced Sizes'], axes[1], 'UART', 1)
plt_histogram(or1200_counter_sizes['Coalesced Sizes'], axes[2], 'OR1200', 2)
plt_histogram(picorv32_counter_sizes['Coalesced Sizes'], axes[3], 'RISC-V', 3)
# Show
plt.tight_layout(h_pad=1)
plt.show()
# +
def plt_histogram_grouped(data_1, data_2, data_3, data_4, ax):
bins = [0,8,16,32,64,128,256] # your bins
# Create Histograms
hist_1, bin_edges_1 = np.histogram(data_1, bins) # make the histogram
hist_2, bin_edges_2 = np.histogram(data_2, bins) # make the histogram
hist_3, bin_edges_3 = np.histogram(data_3, bins) # make the histogram
hist_4, bin_edges_4 = np.histogram(data_4, bins) # make the histogram
# Make x-tick labels list
xtick_labels = ['{}'.format(bins[i]) for i,j in enumerate(hist_1)]
xtick_labels[0] = 1
# Set bar widths
bar_width = 0.2
r_1 = range(len(hist_1))
r_2 = [x + bar_width for x in r_1]
r_2end = [x + bar_width + 0.1 for x in r_1]
r_3 = [x + bar_width for x in r_2]
r_4 = [x + bar_width for x in r_3]
# Plot Histogram
ax.bar(r_1, hist_1, color=current_palette[0], width=bar_width, edgecolor='white', label='AES')
ax.bar(r_2, hist_2, color=current_palette[1], width=bar_width, edgecolor='white', label='UART')
ax.bar(r_3, hist_3, color=current_palette[2], width=bar_width, edgecolor='white', label='RISC-V')
ax.bar(r_4, hist_4, color='#FAC000', width=bar_width, edgecolor='white', label='OR1200')
# Format Histogram
plt.xticks(r_2end, xtick_labels, fontsize=14)
plt.yticks(range(0, 401, 100), range(0, 401, 100), fontsize=14)
plt.legend(fontsize=14)
ax.set_xlabel('Register Size (# bits)', fontsize=14)
ax.set_ylabel('# Registers', fontsize=14)
ax.grid(axis='y', alpha=0.5)
ax.set_ylim(0, 400)
# Text on the top of each barplot
for i in range(len(hist_1)):
ax.text(x = i , y = hist_1[i] + 10, s = hist_1[i], size = 12, horizontalalignment='center')
ax.text(x = i + bar_width, y = hist_2[i] + 10, s = hist_2[i], size = 12, horizontalalignment='center')
ax.text(x = i + (2*bar_width), y = hist_3[i] + 10, s = hist_3[i], size = 12, horizontalalignment='center')
ax.text(x = i + (3*bar_width), y = hist_4[i] + 10, s = hist_4[i], size = 12, horizontalalignment='center')
# Load Data
aes_counter_sizes = load_counter_sizes(aes_data_dir)
uart_counter_sizes = load_counter_sizes(uart_data_dir)
picorv32_counter_sizes = load_counter_sizes(picorv32_data_dir)
or1200_counter_sizes = load_counter_sizes(or1200_data_dir)
# Create Figure
sns.set_style(style="whitegrid")
current_palette = sns.color_palette()
fig, axes = plt.subplots(1, 1, figsize=(8, 4))
plt_histogram_grouped(\
aes_counter_sizes['Coalesced Sizes'],\
uart_counter_sizes['Coalesced Sizes'],\
picorv32_counter_sizes['Coalesced Sizes'],\
or1200_counter_sizes['Coalesced Sizes'],\
axes)
# Save Histogram to PDF
plt.tight_layout(h_pad=1)
if HIST_SAVE_AS_PDF:
plt.savefig(HIST_PDF_FILENAME, format='pdf')
plt.show()
| circuits/plots/exp_001_design_complexities.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics import mean_squared_error
from scipy import stats
plt.rcParams['figure.figsize'] = [12,6]
import warnings
warnings.filterwarnings('ignore')
# + [markdown] heading_collapsed=true
# # Trend
# + hidden=true
df = pd.read_excel('India_Exchange_Rate_Dataset.xls', index_col=0, parse_dates=True)
df.head()
# + hidden=true
df.plot()
plt.show()
# + [markdown] heading_collapsed=true hidden=true
# ## Detecting Trend using Hodrick-Prescott Filter
# + hidden=true
from statsmodels.tsa.filters.hp_filter import hpfilter
# + hidden=true
cycle, trend = hpfilter(df.EXINUS, lamb=129600)
# + hidden=true
trend.plot()
plt.title('Trend Plot')
plt.show()
# + hidden=true
cycle.plot()
plt.title('Cyclic plot')
plt.show()
# + [markdown] heading_collapsed=true hidden=true
# ## Detrending Time Series
# + [markdown] hidden=true
# 1. Pandas Differencing
# 2. SciPy Signal
# 3. Hp filter
# + [markdown] heading_collapsed=true
# # Seasonality
# + [markdown] hidden=true
# ## Seasonal Decomposition
# + hidden=true
from statsmodels.tsa.seasonal import seasonal_decompose
result = seasonal_decompose(df.EXINUS, model='multiplicative', extrapolate_trend='freq')
# + hidden=true
result.plot();
# + hidden=true
deseason = df.EXINUS - result.seasonal
deseason.plot()
plt.title("Deseason Data")
plt.show()
# + [markdown] heading_collapsed=true
# # Smoothing Methods
# + [markdown] heading_collapsed=true hidden=true
# ## Simple Exponential Smoothing
# + hidden=true
facebook = pd.read_csv('https://raw.githubusercontent.com/Apress/hands-on-time-series-analylsis-python/master/Data/FB.csv',
parse_dates=True, index_col=0)
facebook.head()
# + hidden=true
X = facebook['Close']
train = X.iloc[:-30]
test = X.iloc[-30:]
# + hidden=true
from statsmodels.tsa.api import SimpleExpSmoothing
# + hidden=true
ses = SimpleExpSmoothing(X).fit(smoothing_level=0.9)
# + hidden=true
ses.summary()
# + hidden=true
preds = ses.forecast(30)
rmse = np.sqrt(mean_squared_error(test, preds))
print('RMSE:', rmse)
# + [markdown] heading_collapsed=true hidden=true
# ## Double Exponential Smoothing
# + hidden=true
from statsmodels.tsa.api import ExponentialSmoothing, Holt
# + hidden=true
model1 = Holt(train, damped_trend=False).fit(smoothing_level=0.9, smoothing_trend=0.6,
damping_trend=0.1, optimized=False)
model1.summary()
# + hidden=true
preds = model1.forecast(30)
rmse = np.sqrt(mean_squared_error(test, preds))
print('RMSE:', rmse)
# + hidden=true
model_auto = Holt(train).fit(optimized=True, use_brute=True)
model_auto.summary()
# + hidden=true
preds = model_auto.forecast(30)
rmse = np.sqrt(mean_squared_error(test, preds))
print('RMSE:', rmse)
# + [markdown] hidden=true
# ## Triple Exponential Smoothing
# + hidden=true
model2 = ExponentialSmoothing(train, trend='mul',
damped_trend=False,
seasonal_periods=3).fit(smoothing_level=0.9,
smoothing_trend=0.6,
damping_trend=0.6,
use_boxcox=False,
use_basinhopping=True,
optimized=False)
# + hidden=true
model2.summary()
# -
# # Regression Extension Techniques for Time Series Data
from pmdarima import auto_arima
from statsmodels.tsa.stattools import adfuller
adfuller(facebook['Close'])
# - Fail to reject the null hypothesis. It means data is non-stationary
| Time Series Analysis.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Self-Driving Car Engineer Nanodegree
#
#
# ## Project: **Finding Lane Lines on the Road**
# ***
# In this project, you will use the tools you learned about in the lesson to identify lane lines on the road. You can develop your pipeline on a series of individual images, and later apply the result to a video stream (really just a series of images). Check out the video clip "raw-lines-example.mp4" (also contained in this repository) to see what the output should look like after using the helper functions below.
#
# Once you have a result that looks roughly like "raw-lines-example.mp4", you'll need to get creative and try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video "P1_example.mp4". Ultimately, you would like to draw just one line for the left side of the lane, and one for the right.
#
# In addition to implementing code, there is a brief writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) that can be used to guide the writing process. Completing both the code in the Ipython notebook and the writeup template will cover all of the [rubric points](https://review.udacity.com/#!/rubrics/322/view) for this project.
#
# ---
# Let's have a look at our first image called 'test_images/solidWhiteRight.jpg'. Run the 2 cells below (hit Shift-Enter or the "play" button above) to display the image.
#
# **Note: If, at any point, you encounter frozen display windows or other confounding issues, you can always start again with a clean slate by going to the "Kernel" menu above and selecting "Restart & Clear Output".**
#
# ---
# **The tools you have are color selection, region of interest selection, grayscaling, Gaussian smoothing, Canny Edge Detection and Hough Tranform line detection. You are also free to explore and try other techniques that were not presented in the lesson. Your goal is piece together a pipeline to detect the line segments in the image, then average/extrapolate them and draw them onto the image for display (as below). Once you have a working pipeline, try it out on the video stream below.**
#
# ---
#
# <figure>
# <img src="examples/line-segments-example.jpg" width="380" alt="Combined Image" />
# <figcaption>
# <p></p>
# <p style="text-align: center;"> Your output should look something like this (above) after detecting line segments using the helper functions below </p>
# </figcaption>
# </figure>
# <p></p>
# <figure>
# <img src="examples/laneLines_thirdPass.jpg" width="380" alt="Combined Image" />
# <figcaption>
# <p></p>
# <p style="text-align: center;"> Your goal is to connect/average/extrapolate line segments to get output like this</p>
# </figcaption>
# </figure>
# **Run the cell below to import some packages. If you get an `import error` for a package you've already installed, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.**
# ## Import Packages
#importing some useful packages
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
from ipywidgets import widgets
import numpy as np
import os
import cv2
# %matplotlib inline
# ## Read in an Image
# +
#reading in an image
image = mpimg.imread('test_images/solidWhiteRight.jpg')
#printing out some stats and plotting
print('This image is:', type(image), 'with dimensions:', image.shape)
plt.imshow(image) # if you wanted to show a single color channel image called 'gray', for example, call as plt.imshow(gray, cmap='gray')
# -
# ## Ideas for Lane Detection Pipeline
# **Some OpenCV functions (beyond those introduced in the lesson) that might be useful for this project are:**
#
# `cv2.inRange()` for color selection
# `cv2.fillPoly()` for regions selection
# `cv2.line()` to draw lines on an image given endpoints
# `cv2.addWeighted()` to coadd / overlay two images
# `cv2.cvtColor()` to grayscale or change color
# `cv2.imwrite()` to output images to file
# `cv2.bitwise_and()` to apply a mask to an image
#
# **Check out the OpenCV documentation to learn about these and discover even more awesome functionality!**
# ## Helper Functions
# Below are some helper functions to help get you started. They should look familiar from the lesson!
# +
import math
class FindLanes(object):
def __init__(self):
left_lane = [0,0,0,0]
right_lane = [0,0,0,0]
def grayscale(self, img):
"""Applies the Grayscale transform
This will return an image with only one color channel
but NOTE: to see the returned image as grayscale
(assuming your grayscaled image is called 'gray')
you should call plt.imshow(gray, cmap='gray')"""
return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
# Or use BGR2GRAY if you read an image with cv2.imread()
# return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
def canny(self, img, low_threshold, high_threshold):
"""Applies the Canny transform"""
return cv2.Canny(img, low_threshold, high_threshold)
def gaussian_blur(self, img, kernel_size):
"""Applies a Gaussian Noise kernel"""
return cv2.GaussianBlur(img, (kernel_size, kernel_size), 0)
def region_of_interest(self, img, vertices):
"""
Applies an image mask.
Only keeps the region of the image defined by the polygon
formed from `vertices`. The rest of the image is set to black.
`vertices` should be a numpy array of integer points.
"""
#defining a blank mask to start with
mask = np.zeros_like(img)
#defining a 3 channel or 1 channel color to fill the mask with depending on the input image
if len(img.shape) > 2:
channel_count = img.shape[2] # i.e. 3 or 4 depending on your image
ignore_mask_color = (255,) * channel_count
else:
ignore_mask_color = 255
#filling pixels inside the polygon defined by "vertices" with the fill color
cv2.fillPoly(mask, vertices, ignore_mask_color)
#returning the image only where mask pixels are nonzero
masked_image = cv2.bitwise_and(img, mask)
return masked_image
def draw_lines(self, img, lines, color=[255, 0, 0], thickness=2, interp_tol=10):
"""
NOTE: this is the function you might want to use as a starting point once you want to
average/extrapolate the line segments you detect to map out the full
extent of the lane (going from the result shown in raw-lines-example.mp4
to that shown in P1_example.mp4).
Think about things like separating line segments by their
slope ((y2-y1)/(x2-x1)) to decide which segments are part of the left
line vs. the right line. Then, you can average the position of each of
the lines and extrapolate to the top and bottom of the lane.
This function draws `lines` with `color` and `thickness`.
Lines are drawn on the image inplace (mutates the image).
If you want to make the lines semi-transparent, think about combining
this function with the weighted_img() function below
"""
"""
Left Lane: > 0.0
Right Lane: < 0.0
"""
right_slopes = []
left_slopes = []
right_intercepts = []
left_intercepts = []
x_min_interp = 0
y_max = img.shape[0]
y_min_left = img.shape[0] + 1
y_min_right = img.shape[0] + 1
y_min_interp = 320 + 15 #top of ROI plus some offset for aesthetics
for line in lines:
for x1,y1,x2,y2 in line:
current_slope = (y2-y1)/(x2-x1)
# For each detected line, seperate lines into left and right lanes.
# Calculate the current slope and intercept and keep a history for averaging.
if current_slope < 0.0 and current_slope > -math.inf:
right_slopes.append(current_slope)
right_intercepts.append(y1 - current_slope*x1)
y_min_right = min(y_min_right, y1, y2)
if current_slope > 0.0 and current_slope < math.inf:
left_slopes.append(current_slope)
left_intercepts.append(y1 - current_slope*x1)
y_min_left = min(y_min_left, y1, y2)
# Calculate the average of the slopes, intercepts, x_min and x_max
# Interpolate the average line to the end of the region of interest (using equation of slopes)
if len(left_slopes) > 0:
ave_left_slope = sum(left_slopes) / len(left_slopes)
ave_intercept = sum(left_intercepts) / len(left_intercepts)
x_min=int((y_min_left - ave_intercept)/ ave_left_slope)
x_max = int((y_max - ave_intercept)/ ave_left_slope)
x_min_interp = int(((x_min*y_min_interp) - (x_min*y_max) - (x_max*y_min_interp) + (x_max*y_min_left))/(y_min_left - y_max))
self.left_lane = [x_min_interp, y_min_interp, x_max, y_max]
# Draw the left lane line
cv2.line(img, (self.left_lane[0], self.left_lane[1]), (self.left_lane[2], self.left_lane[3]), [255, 0, 0], 12)
if len(right_slopes) > 0:
ave_right_slope = sum(right_slopes) / len(right_slopes)
ave_intercept = sum(right_intercepts) / len(right_intercepts)
x_min = int((y_min_right - ave_intercept)/ ave_right_slope)
x_max = int((y_max - ave_intercept)/ ave_right_slope)
x_min_interp = int(((x_min*y_min_interp) - (x_min*y_max) - (x_max*y_min_interp) + (x_max*y_min_right))/(y_min_right - y_max))
self.right_lane = [x_min_interp, y_min_interp, x_max, y_max]
# Draw the right lane line
cv2.line(img, (self.right_lane[0], self.right_lane[1]), (self.right_lane[2], self.right_lane[3]), [255, 0, 0], 12)
def hough_lines(self, img, rho, theta, threshold, min_line_len, max_line_gap):
"""
`img` should be the output of a Canny transform.
Returns an image with hough lines drawn.
"""
lines = cv2.HoughLinesP(img, rho, theta, threshold, np.array([]), minLineLength=min_line_len, maxLineGap=max_line_gap)
line_img = np.zeros((img.shape[0], img.shape[1], 3), dtype=np.uint8)
self.draw_lines(line_img, lines)
return line_img
# Python 3 has support for cool math symbols.
def weighted_img(self, img, initial_img, α=0.8, β=1., γ=0.):
"""
`img` is the output of the hough_lines(), An image with lines drawn on it.
Should be a blank image (all black) with lines drawn on it.
`initial_img` should be the image before any processing.
The result image is computed as follows:
initial_img * α + img * β + γ
NOTE: initial_img and img must be the same shape!
"""
return cv2.addWeighted(initial_img, α, img, β, γ)
# -
# ## Test Images
#
# Build your pipeline to work on the images in the directory "test_images"
# **You should make sure your pipeline works well on these images before you try the videos.**
import os
os.listdir("test_images/")
# ## Build a Lane Finding Pipeline
#
#
# Build the pipeline and run your solution on all test_images. Make copies into the `test_images_output` directory, and you can use the images in your writeup report.
#
# Try tuning the various parameters, especially the low and high Canny thresholds as well as the Hough lines parameters.
# +
# TODO: Build your pipeline that will draw lane lines on the test_images
# then save them to the test_images_output directory.
# Load test images
dir = "test_images/"
out_dir = "test_images_output/"
input = os.listdir(dir)
# Define globals
kernel_size = 3
canny_thresh = [75,150]
rho = 2
theta = np.pi/180
threshold = 90
min_line_length = 20
max_line_gap = 20
fl = FindLanes()
# Setup pipeline
def process_image(image):
# Preprocess image
gray = fl.grayscale(image)
plt.imsave(out_dir + "gray.jpg", gray, cmap="gray")
blur = fl.gaussian_blur(gray, kernel_size)
plt.imsave(out_dir + "blur.jpg", blur, cmap="gray")
# Find edges
edges = fl.canny(blur, canny_thresh[0], canny_thresh[1])
plt.imsave(out_dir + "edges.jpg", edges, cmap="gray")
# Region of interest
verts = np.array([[(100,image.shape[0]),(450, 320), (500, 320), (image.shape[1],image.shape[0])]], dtype=np.int32)
masked_edges = fl.region_of_interest(edges, verts)
plt.imsave(out_dir + "masked_edges.jpg", masked_edges, cmap="gray")
# Draw hough lines
lines_image = fl.hough_lines(masked_edges, rho, theta, threshold, min_line_length, max_line_gap)
plt.imsave(out_dir + "lines.jpg", lines_image, cmap="gray")
result = fl.weighted_img(lines_image, image)
plt.imsave(out_dir + "final.jpg", result, cmap="gray")
return result
# Jupyter notebook validation error -----
# widgets.interact(process_image, canny_thresh=widgets.IntRangeSlider(min=0, max=255, step=1, value=canny_thresh))
# -----
for test_image in input:
image = mpimg.imread(dir + test_image)
processed = process_image(image)
plt.imshow(processed)
plt.show()
# -
# ## Test on Videos
#
# You know what's cooler than drawing lanes over images? Drawing lanes over video!
#
# We can test our solution on two provided videos:
#
# `solidWhiteRight.mp4`
#
# `solidYellowLeft.mp4`
#
# **Note: if you get an import error when you run the next cell, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.**
#
# **If you get an error that looks like this:**
# ```
# NeedDownloadError: Need ffmpeg exe.
# You can download it by calling:
# imageio.plugins.ffmpeg.download()
# ```
# **Follow the instructions in the error message and check out [this forum post](https://discussions.udacity.com/t/project-error-of-test-on-videos/274082) for more troubleshooting tips across operating systems.**
# Import everything needed to edit/save/watch video clips
from moviepy.editor import VideoFileClip
from IPython.display import HTML
# +
# Define globals
# def process_image(image):
# # NOTE: The output you return should be a color image (3 channel) for processing video below
# # TODO: put your pipeline here,
# # you should return the final output (image where lines are drawn on lanes)
# -
# Let's try the one with the solid white lane on the right first ...
white_output = 'test_videos_output/solidWhiteRight.mp4'
## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video
## To do so add .subclip(start_second,end_second) to the end of the line below
## Where start_second and end_second are integer values representing the start and end of the subclip
## You may also uncomment the following line for a subclip of the first 5 seconds
##clip1 = VideoFileClip("test_videos/solidWhiteRight.mp4").subclip(0,5)
clip1 = VideoFileClip("test_videos/solidWhiteRight.mp4")
white_clip = clip1.fl_image(process_image) #NOTE: this function expects color images!!
# %time white_clip.write_videofile(white_output, audio=False)
# Play the video inline, or if you prefer find the video in your filesystem (should be in the same directory) and play it in your video player of choice.
HTML("""
<video width="960" height="540" controls>
<source src="{0}">
</video>
""".format(white_output))
# ## Improve the draw_lines() function
#
# **At this point, if you were successful with making the pipeline and tuning parameters, you probably have the Hough line segments drawn onto the road, but what about identifying the full extent of the lane and marking it clearly as in the example video (P1_example.mp4)? Think about defining a line to run the full length of the visible lane based on the line segments you identified with the Hough Transform. As mentioned previously, try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video "P1_example.mp4".**
#
# **Go back and modify your draw_lines function accordingly and try re-running your pipeline. The new output should draw a single, solid line over the left lane line and a single, solid line over the right lane line. The lines should start from the bottom of the image and extend out to the top of the region of interest.**
# Now for the one with the solid yellow lane on the left. This one's more tricky!
yellow_output = 'test_videos_output/solidYellowLeft.mp4'
## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video
## To do so add .subclip(start_second,end_second) to the end of the line below
## Where start_second and end_second are integer values representing the start and end of the subclip
## You may also uncomment the following line for a subclip of the first 5 seconds
##clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4').subclip(0,5)
clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4')
yellow_clip = clip2.fl_image(process_image)
# %time yellow_clip.write_videofile(yellow_output, audio=False)
HTML("""
<video width="960" height="540" controls>
<source src="{0}">
</video>
""".format(yellow_output))
# ## Writeup and Submission
#
# If you're satisfied with your video outputs, it's time to make the report writeup in a pdf or markdown file. Once you have this Ipython notebook ready along with the writeup, it's time to submit for review! Here is a [link](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) to the writeup template file.
#
# ## Optional Challenge
#
# Try your lane finding pipeline on the video below. Does it still work? Can you figure out a way to make it more robust? If you're up for the challenge, modify your pipeline so it works with this video and submit it along with the rest of your project!
challenge_output = 'test_videos_output/challenge.mp4'
## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video
## To do so add .subclip(start_second,end_second) to the end of the line below
## Where start_second and end_second are integer values representing the start and end of the subclip
## You may also uncomment the following line for a subclip of the first 5 seconds
##clip3 = VideoFileClip('test_videos/challenge.mp4').subclip(0,5)
clip3 = VideoFileClip('test_videos/challenge.mp4')
challenge_clip = clip3.fl_image(process_image)
# %time challenge_clip.write_videofile(challenge_output, audio=False)
HTML("""
<video width="960" height="540" controls>
<source src="{0}">
</video>
""".format(challenge_output))
| P1.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import logging
import pandas as pd
from pandas import Series
from iso3166 import countries
from datetime import datetime
from csv import QUOTE_NONNUMERIC
from unicef_schools_attribute_cleaning.pandas.dataframe_cleaner import dataframe_cleaner
# make INFO logs visible
logging.basicConfig(level=logging.INFO)
# -
src_df = pd.read_csv('../../data/UNICE_schools_raw_2020_Jun/2019_ASC_schools.csv', low_memory=False)
src_df
# +
# ^ Problem 1: there are 3764 columns in the source data. This makes it difficult to correctly map columns onto the unicef schema.
# The fuzzy matching in dataframe_cleaner will collect some of them.
# -
# Problem 2: the 'date' values are not an ISO format, and also have NaN float values- so cannot be automatically parsed without formatting.
src_df['date']
def fix_date(value):
if not isinstance(value, str):
return None
return datetime.strptime(value, '%m/%d/%Y %H:%M')
preprocess_df = src_df
preprocess_df['date'] = preprocess_df['date'].apply(fix_date)
preprocess_df['date']
# run the dataframe_cleaner
country = countries.get('SL')
with open('sierra_leone_columns_report.txt', mode='w', encoding='utf-8') as filehandle:
df = dataframe_cleaner(
dataframe=preprocess_df,
country=country,
removed_columns_report=filehandle,
is_private=True,
provider="ASC",
provider_is_private=True
)
df
df.to_csv('sierra_leone_cleaned.csv', quoting=QUOTE_NONNUMERIC, index=False)
# open in LibreOffice, Excel, other
# !open sierra_leone_cleaned.csv
| unicef_schools_attribute_cleaning/notebooks/sierra_leone_demo.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda root]
# language: python
# name: conda-root-py
# ---
import pandas as pd, numpy as np, matplotlib.pyplot as plt, requests, urllib2
from bs4 import BeautifulSoup
# %matplotlib inline
# Import CNC
cnc_path='../../universal/countries/'
cnc=pd.read_excel(cnc_path+'cnc.xlsx').set_index('Name')
cnc
# Capture airport lists
L={}
F=[]
baseurl='https://www.airportia.com/'
for k in range(len(cnc.columns)):
c=cnc.columns[k]
if c not in L and F:
#capture token
url=baseurl+c.lower().replace(' ','-')
s = requests.Session()
cookiesopen = s.get(url)
cookies=str(s.cookies)
fcookies=[[k[:k.find('=')],k[k.find('=')+1:k.find(' for ')]] for k in cookies[cookies.find('Cookie '):].split('Cookie ')[1:]]
#push token
opener = urllib2.build_opener()
for k in fcookies:
opener.addheaders.append(('Cookie', k[0]+'='+k[1]))
#read html
content=s.get(url).content
soup = BeautifulSoup(content, "lxml")
if len(soup.findAll(attrs={'class':'textlist'}))>0:
links=soup.findAll(attrs={'class':'textlist'})[0].findAll('a')
L[c]=[str(i)[str(i).find('href')+6:str(i).find('title')-2] for i in links]
print 'Success',url
else:
F.append(c)
print 'Fail',url
# Fix failures
lnc={
"Brunei Darussalam":"brunei",
"Cent African Rep":"central-african-republic",
"Cote d'Ivoire":"ivory-coast",
"People's Republic of Korea":"north-korea",
"Dem. Rep. of Congo":"congo",
"Guinea-Bissau":"guinea_bissau",
"Holy See":"Holy See",
"Lao People's Dem. Rep.":"laos",
"Libyan Arab Jamahiriya":"libya",
"Liechtenstein":"Liechtenstein",
"Macao, China":"Macao, China",
"Micronesia (Federated States of)":"micronesia",
"Myanmar (Burma)":"burma",
"Netherlands Antilles":"Netherlands Antilles",
"Palestinian Territories":"palestinian-territory",
"Peru":u"perú",
"Rep. of Korea":"south-korea",
"Rep. of Moldova":"moldova",
"Russian Federation":"russia",
"San Marino":"San Marino",
"Syrian Arab Republic":"syria",
"FYR of Macedonia":"macedonia",
"Timor-Leste":"timor_leste",
"Tokelau":"Tokelau",
"Zanzibar":"Zanzibar",
"Svalbard and Jan Mayen":"Svalbard and Jan Mayen",
"US Virgin Islands":"US Virgin Islands"
}
F2=[]
for k in range(len(cnc.columns)):
c=cnc.columns[k]
if c not in L and F2:
#capture token
url=baseurl+lnc[c]
s = requests.Session()
cookiesopen = s.get(url)
cookies=str(s.cookies)
fcookies=[[k[:k.find('=')],k[k.find('=')+1:k.find(' for ')]] for k in cookies[cookies.find('Cookie '):].split('Cookie ')[1:]]
#push token
opener = urllib2.build_opener()
for k in fcookies:
opener.addheaders.append(('Cookie', k[0]+'='+k[1]))
#read html
content=s.get(url).content
soup = BeautifulSoup(content, "lxml")
if len(soup.findAll(attrs={'class':'textlist'}))>0:
links=soup.findAll(attrs={'class':'textlist'})[0].findAll('a')
L[c]=[str(i)[str(i).find('href')+6:str(i).find('title')-2] for i in links]
print 'Success',url
else:
F2.append(c)
print 'Fail',url
#absolute failures
F2
# Update CNC
g=[]
for k in range(len(cnc.columns)):
c=cnc.columns[k]
if c in F2:
g.append(np.nan)
elif c in F:
g.append(lnc[c])
else:
g.append(c.lower().replace(' ','-'))
dnc=cnc.T
dnc['Airportia']=g
cnc=dnc.T
cnc
cnc.to_excel(cnc_path+'cnc1.xlsx')
# Save links
import json
file('../json/L.json','w').write(json.dumps(L))
# Create folder structure
import os.path
for i in cnc.loc['ISO2']:
if str(i).lower()!='nan':
directory='../countries/'+i.lower()
if not os.path.exists(directory) :
os.makedirs(directory)
for j in ['code','d3','json','map']:
if not os.path.exists(directory+'/'+j):
os.makedirs(directory+'/'+j)
| code/.ipynb_checkpoints/countryparser-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] colab_type="text" id="rX8mhOLljYeM"
# ##### Copyright 2019 The TensorFlow Authors.
# + cellView="form" colab={} colab_type="code" id="BZSlp3DAjdYf"
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# + [markdown] colab_type="text" id="3wF5wszaj97Y"
# # <div dir="rtl"> تمهيد سريع للمبتدئين حول Tensorflow-2</div>
#
# + [markdown] colab_type="text" id="DUNzJc4jTj6G"
# <table class="tfo-notebook-buttons" align="right" dir="rtl">
# <td>
# <a target="_blank" href="https://www.tensorflow.org/tutorials/quickstart/beginner?hl=ar"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />إفتح المحتوى على موقع TensorFlow.org</a>
# </td>
# <td>
# <a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/ar/tutorials/quickstart/beginner.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />تفاعل مع المحتوى على Google Colab</a>
# </td>
# <td>
# <a target="_blank" href="https://github.com/tensorflow/docs-l10n/blob/master/site/ar/tutorials/quickstart/beginner.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />اعرض المصدر على Github</a>
# </td>
# <td>
# <a href="https://storage.googleapis.com/tensorflow_docs/docs-l10n/site/ar/tutorials/quickstart/beginner.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />نزّل الدّفتر</a>
# </td>
# </table>
# + [markdown] colab_type="text" id="04QgGZc9bF5D"
# <div dir="rtl">
#
# يستخدم هذا التمهيد القصير
# [Keras](https://www.tensorflow.org/guide/keras/overview)
# من أجل:
#
# 1. بناء شبكة عصبية تصنّف الصور.
# 2. تدريب هذه الشبكة العصبية.
# 3. وأخيرًا ، تقييم دقة النموذج.
#
# </div>
#
# + [markdown] colab_type="text" id="hiH7AC-NTniF"
# <div dir="rtl">
#
# هذا الملفّ هو دفتر
# [Google Colaboratory](https://colab.research.google.com/notebooks/welcome.ipynb).
# بواسطته ، يمكنك تشغيل برامج Python مباشرة في المتصفّح -
# و هي طريقة رائعة لتعلّم و إستخدام Tensorflow.
#
# لمتابعة هذا البرنامج التعليمي ، قم بتشغيل الدفتر التّفاعلي في
# Google Colab
# بالنقر فوق الزر، ذي نفس التسمية ، الموجود في أعلى هذه الصفحة.
#
# 1. في Colab ، اتصل بمحرّك تشغيل Python بالطريقة التّالية: إذهب إلى قائمة الإختيارات في أعلى يسار الدفتر ، ثمّ إضغط على *CONNECT*.
# 2. شغّل كافة خلايا الدفتر التّفاعلي بإختيار *Runtime* ثمّ الضغط على *Run all*.
#
# </div>
# + [markdown] colab_type="text" id="nnrWf3PCEzXL"
# <div dir="rtl">
#
# قم بتنزيل وتثبيت TensorFlow 2. ثمّ قم بإستيراد حزمة TensorFlow في برنامجك:
#
# Note:
# قم بتحديث
# pip
# لتتمكّن من تثبيت حزمة
# Tensorflow 2.
# أنظر في
# [دليل التثبيت](https://www.tensorflow.org/install)
# للحصول على المزيد من التفاصيل.
#
# </div>
#
# + colab={} colab_type="code" id="0trJmd6DjqBZ"
from __future__ import absolute_import, division, print_function, unicode_literals
try:
# # %tensorflow_version only exists in Colab.
# %tensorflow_version 2.x
except Exception:
pass
import tensorflow as tf
# + [markdown] colab_type="text" id="7NAbSZiaoJ4z"
# <div dir="rtl">
#
# قم بتحميل و إعداد
# [قاعدة بيانات MNIST](http://yann.lecun.com/exdb/mnist/)
# و تحويل العيّنات من أعداد صحيحة إلى أعداد الفاصلة العائمة
# (floating-point numbers):
#
# </div>
# + colab={} colab_type="code" id="7FP5258xjs-v"
mnist = tf.keras.datasets.mnist
(x_train, y_train), (x_test, y_test) = mnist.load_data()
x_train, x_test = x_train / 255.0, x_test / 255.0
# + [markdown] colab_type="text" id="BPZ68wASog_I"
# <div dir="rtl">
#
# قم ببناء نموذج
# `tf.keras.Sequential`
# عن طريق تكديس الطبقات.
# لتدريب النموذج قم بإختيار خوارزميّة تحسين
# (optimizer)
# و دالّة خسارة
# (loss function).
#
# </div>
# + colab={} colab_type="code" id="h3IKyzTCDNGo"
model = tf.keras.models.Sequential([
tf.keras.layers.Flatten(input_shape=(28, 28)),
tf.keras.layers.Dense(128, activation='relu'),
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Dense(10)
])
# + [markdown] colab_type="text" id="l2hiez2eIUz8"
# <div dir="rtl">
#
# لكلّ مثال في قاعدة التّدريب، يعرض النّموذج متجّها
# (vector)
# متكوّنا من نتائج تمثّل
# ["logits"](https://developers.google.com/machine-learning/glossary#logits)
# أو
# ["log-odds"](https://developers.google.com/machine-learning/glossary#log-odds)
# كلُّ قيمة في هذا المتّجه تمثّل واحدة من الأقسام التّي نريد تصنيف الصّور حسبها.
#
# </div>
#
# + colab={} colab_type="code" id="OeOrNdnkEEcR"
predictions = model(x_train[:1]).numpy()
predictions
# + [markdown] colab_type="text" id="tgjhDQGcIniO"
# <div dir="rtl">
#
# تحول الدّالة
# `tf.nn.softmax`
# هذه النتائج ، المتمثلة في أعداد حقيقية ، إلى "احتمالات"
# لكل فئة ، حيث تكون قيمة كُلّ واحدة من هذه الإحتمالات بين 0 و 1
# و يساوي مجموع كُلّ القيم 1:
# </div>
# + colab={} colab_type="code" id="zWSRnQ0WI5eq"
tf.nn.softmax(predictions).numpy()
# + [markdown] colab_type="text" id="he5u_okAYS4a"
# <div dir="rtl">
#
# ملاحظة: يمكن جعل الدّالة
# `tf.nn.softmax`
# جزءًا من الشبكة العصبيّة التي بنيناها سابقا، حيث يمكن إعتبارها دالّة تنشيط
# (activation function)
# للطبقة الأخيرة من الشبكة العصبيّة.
# بهذه الطريقة ستصير نتيجة النموذج أكثر قابليّة للتفسير بشكل مباشر من دون معالجات إضافيّة.
# إلاّ أنّ هذه الطريقة غير منصوح بها ، لأنّه من المستحيل توفير حساب خسارة دقيق و مستقرّ رقميًّا لجميع النماذج عند إستخدام نتائج
# `softmax`.
#
# </div>
# + [markdown] colab_type="text" id="hQyugpgRIyrA"
# <div dir="rtl">
#
# تأخذ دالّة الخسار
# `losses.SparseCategoricalCrossentropy`
# متجّها متكوّنا من
# logits
# و مؤشرًا عن الفئة الصحيحة لكلّ مثال ، ثمّ تقوم بإنتاج عدد حقيقي يمثلّ قيمة الخسارة لكلّ مثال.
#
# </div>
# + colab={} colab_type="code" id="RSkzdv8MD0tT"
loss_fn = tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True)
# + [markdown] colab_type="text" id="SfR4MsSDU880"
# <div dir="rtl">
#
# تساوي قيمة هذه الخسارة القيمة السلبية اللوغاريتمية للإحتمال
# (negative log probability)
# الذّي أعطاه النموذج للفئة الصحيحة للمثال:
# تكون هذه القيمة قريبة من الصفر إذا كان النموذج متأكّدا من تصنيفه للمثال حسب الفئة الصحيحة.
#
# يعطي النموذج الحالي ، غير المدرّب ، إحتمالات عشوائيّة تقارب 1/10
# لكلّ فئة، لذا فإنّ قيمة الخسارة الأوليّة لهذا النموذج ستكون تقريبا
# :
# `tf.log(1/10) ~= 2.3`
#
# </div>
# + colab={} colab_type="code" id="NJWqEVrrJ7ZB"
loss_fn(y_train[:1], predictions).numpy()
# + colab={} colab_type="code" id="9foNKHzTD2Vo"
model.compile(optimizer='adam',
loss=loss_fn,
metrics=['accuracy'])
# + [markdown] colab_type="text" id="ix4mEL65on-w"
# <div dir="rtl">
#
# تقوم الدّالة
# `Model.fit`
# بضبط معلما
# (parameters)
# النموذج لتقليل الخسارة:
#
# </div>
# + colab={} colab_type="code" id="y7suUbJXVLqP"
model.fit(x_train, y_train, epochs=5)
# + [markdown] colab_type="text" id="4mDAAPFqVVgn"
# <div dir="rtl">
#
# تقوم الدّالة
# `Model.evaluate`
# بالتحقّق من أداء النّموذج على مجموعة بيانات معزولة عن تلك المستخدمة في التّدريب و تسمّى مجموعة التحقّق
# ([Validation-set](https://developers.google.com/machine-learning/glossary#validation-set)).
#
# </div>
# + colab={} colab_type="code" id="F7dTAzgHDUh7"
model.evaluate(x_test, y_test, verbose=2)
# + [markdown] colab_type="text" id="T4JfEh7kvx6m"
# <div dir="rtl">
#
# الآن ، و بعد إستخدام الدّالة
# `Model.fit`
# ، تمّ تدريب مصنّف الصور إلى دقة 98% تقريبًا على مجموعة البيانات هذه.
#
# لتعلّم المزيد ، اقرأ
# [الدروس التعليمية](https://www.tensorflow.org/tutorials/)
# الأخرى على موقع
# TensorFlow.
#
# </div>
# + [markdown] colab_type="text" id="Aj8NrlzlJqDG"
# <div dir="rtl">
#
# إذا أردت أن تكون نتائج النموذج المدرّب في شكل إحتمالات ، فيمكنك لفّه باستعمال الدّالة
# `tf.keras.Sequential`
# و إرفاقه بدالّة
# softmax:
#
# </div>
# + colab={} colab_type="code" id="rYb6DrEH0GMv"
probability_model = tf.keras.Sequential([
model,
tf.keras.layers.Softmax()
])
# + colab={} colab_type="code" id="cnqOZtUp1YR_"
probability_model(x_test[:5])
| site/ar/tutorials/quickstart/beginner.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import torch
import torchvision.datasets as datasets
from matplotlib import pyplot as plt
# Dataset! Yay that torchvision make everything easy
train_dataset = datasets.MNIST("mnist", train=True, download=True)
test_dataset = datasets.MNIST("mnist", train=False, download=True)
# +
# Sanity check that we have the right number of data points and that they look legit
assert len(train_dataset) == 60000
assert len(test_dataset) == 10000
plt.title("Label: %i" % train_dataset[0][1].item())
plt.imshow(train_dataset[0][0])
plt.show()
| MNIST Dataset.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ----------------
# # Spiegazione
# <img src="fig/heatmap.PNG">
#
# **LEGEND**
#
# Il grafico mette a confronto i tentativi utilizzati da _knuth_ con i tentativi utilizzati dalla rete in esame.
#
# In particolare, nell'asse delle y abbiamo i tentativi che ha impiegato _knuth_ per risolvere il match, mentre nell'asse delle x il numero di tentativi del player.
#
# - E.g. Scrorrendo la riga 1 abbiamo il numero di partite vinte da knuth con 1 tentativo, e in corrispondenza della colonna 4 abbiamo un +1, Questo vuol dire che knuth ha risolto 1a partita con 1 solo tentativo, mentre per la stessa partita il player ha impiegato 4 tentativi.
#
# - E.g. Scorrendo la riga 3 possiamo vedere come Knuth abbia vinto 3+8+16+23+2+2+2+5 partite in 3 tentativi. Mentre il player di queste partite non ne ha concluse 5 (colonna 10+), e nella maggior parte dei casi (23volte) ha impiegato 6 tentativi
#
# -------------------------------
# !ls ./matchesPlayed/
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
sns.set()
# --------------
# **PARAMS**
# +
KNUTH_PATH = "../2_database/dbPlayers/knuth/knuth_optimal.csv"
PLAYER_PATH = "./matchesPlayed/model_20191026_1947_play.csv"
# -
# -------------------
# +
df_list = []
features_name = ['Guess 1', 'Guess 2', 'Guess 3', 'Guess 4', 'Guess 5',
'Guess 6', 'Guess 7', 'Guess 8', 'Guess 9', 'Guess 10']
target_name = 'PASSWORD'
# Optimize the df load
feature_type = {}
cuts_type = {}
feature_type[target_name] = str
for feature in features_name:
feature_type[feature] = str
# +
df_knuth = pd.read_csv(KNUTH_PATH, delimiter=',',encoding='utf-8', skip_blank_lines=True)
df_player = pd.read_csv(PLAYER_PATH, delimiter=',',encoding='utf-8', skip_blank_lines=True)
assert len(df_knuth) == len(df_player), "DF must be the same lenght of 1296 matches"
assert all(df_knuth.iloc[:][target_name] == df_player.iloc[:][target_name]), "DF must be ordered in the same way"
# -
# *The PASSWORD field must be ordered*
df_knuth.head()
df_player.head()
def calculate_match_len(row):
match_len = 0
for guess in row:
if guess != '<pad>':
match_len += 1
if guess == 'XXXX':
match_len = 11
return match_len
matrix_analysis = np.zeros((5,11))
for match_knuth, match_player in zip(df_knuth[features_name].values, df_player[features_name].values):
knuth_len = calculate_match_len(match_knuth)
player_len = calculate_match_len(match_player)
matrix_analysis[(knuth_len - 1), (player_len - 1)] += 1
matrix_analysis
plt.figure(figsize=(16, 6))
plt.xlabel('Player')
plt.ylabel('Knuth')
plt.title('Tentativi del player (axis-x) vs tentativi knuth(axis-y)')
xticklabels = [str(i) for i in range(1,11)]
xticklabels.append('10+')
yticklabels = [str(i) for i in range(1,6)]
sns.heatmap(matrix_analysis, annot=True, cmap="YlGnBu", fmt="g",
xticklabels=xticklabels, yticklabels=yticklabels)
| 5_analysis/matrix_analysis.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Contrasts Overview
import numpy as np
import statsmodels.api as sm
# This document is based heavily on this excellent resource from UCLA http://www.ats.ucla.edu/stat/r/library/contrast_coding.htm
# A categorical variable of K categories, or levels, usually enters a regression as a sequence of K-1 dummy variables. This amounts to a linear hypothesis on the level means. That is, each test statistic for these variables amounts to testing whether the mean for that level is statistically significantly different from the mean of the base category. This dummy coding is called Treatment coding in R parlance, and we will follow this convention. There are, however, different coding methods that amount to different sets of linear hypotheses.
#
# In fact, the dummy coding is not technically a contrast coding. This is because the dummy variables add to one and are not functionally independent of the model's intercept. On the other hand, a set of *contrasts* for a categorical variable with `k` levels is a set of `k-1` functionally independent linear combinations of the factor level means that are also independent of the sum of the dummy variables. The dummy coding is not wrong *per se*. It captures all of the coefficients, but it complicates matters when the model assumes independence of the coefficients such as in ANOVA. Linear regression models do not assume independence of the coefficients and thus dummy coding is often the only coding that is taught in this context.
#
# To have a look at the contrast matrices in Patsy, we will use data from UCLA ATS. First let's load the data.
# #### Example Data
import pandas as pd
url = 'https://stats.idre.ucla.edu/stat/data/hsb2.csv'
hsb2 = pd.read_table(url, delimiter=",")
hsb2.head(10)
# It will be instructive to look at the mean of the dependent variable, write, for each level of race ((1 = Hispanic, 2 = Asian, 3 = African American and 4 = Caucasian)).
hsb2.groupby('race')['write'].mean()
# #### Treatment (Dummy) Coding
# Dummy coding is likely the most well known coding scheme. It compares each level of the categorical variable to a base reference level. The base reference level is the value of the intercept. It is the default contrast in Patsy for unordered categorical factors. The Treatment contrast matrix for race would be
from patsy.contrasts import Treatment
levels = [1,2,3,4]
contrast = Treatment(reference=0).code_without_intercept(levels)
print(contrast.matrix)
# Here we used `reference=0`, which implies that the first level, Hispanic, is the reference category against which the other level effects are measured. As mentioned above, the columns do not sum to zero and are thus not independent of the intercept. To be explicit, let's look at how this would encode the `race` variable.
hsb2.race.head(10)
print(contrast.matrix[hsb2.race-1, :][:20])
sm.categorical(hsb2.race.values)
# This is a bit of a trick, as the `race` category conveniently maps to zero-based indices. If it does not, this conversion happens under the hood, so this will not work in general but nonetheless is a useful exercise to fix ideas. The below illustrates the output using the three contrasts above
from statsmodels.formula.api import ols
mod = ols("write ~ C(race, Treatment)", data=hsb2)
res = mod.fit()
print(res.summary())
# We explicitly gave the contrast for race; however, since Treatment is the default, we could have omitted this.
# ### Simple Coding
# Like Treatment Coding, Simple Coding compares each level to a fixed reference level. However, with simple coding, the intercept is the grand mean of all the levels of the factors. Patsy does not have the Simple contrast included, but you can easily define your own contrasts. To do so, write a class that contains a code_with_intercept and a code_without_intercept method that returns a patsy.contrast.ContrastMatrix instance
# +
from patsy.contrasts import ContrastMatrix
def _name_levels(prefix, levels):
return ["[%s%s]" % (prefix, level) for level in levels]
class Simple(object):
def _simple_contrast(self, levels):
nlevels = len(levels)
contr = -1./nlevels * np.ones((nlevels, nlevels-1))
contr[1:][np.diag_indices(nlevels-1)] = (nlevels-1.)/nlevels
return contr
def code_with_intercept(self, levels):
contrast = np.column_stack((np.ones(len(levels)),
self._simple_contrast(levels)))
return ContrastMatrix(contrast, _name_levels("Simp.", levels))
def code_without_intercept(self, levels):
contrast = self._simple_contrast(levels)
return ContrastMatrix(contrast, _name_levels("Simp.", levels[:-1]))
# -
hsb2.groupby('race')['write'].mean().mean()
contrast = Simple().code_without_intercept(levels)
print(contrast.matrix)
mod = ols("write ~ C(race, Simple)", data=hsb2)
res = mod.fit()
print(res.summary())
# ### Sum (Deviation) Coding
# Sum coding compares the mean of the dependent variable for a given level to the overall mean of the dependent variable over all the levels. That is, it uses contrasts between each of the first k-1 levels and level k In this example, level 1 is compared to all the others, level 2 to all the others, and level 3 to all the others.
from patsy.contrasts import Sum
contrast = Sum().code_without_intercept(levels)
print(contrast.matrix)
mod = ols("write ~ C(race, Sum)", data=hsb2)
res = mod.fit()
print(res.summary())
# This corresponds to a parameterization that forces all the coefficients to sum to zero. Notice that the intercept here is the grand mean where the grand mean is the mean of means of the dependent variable by each level.
hsb2.groupby('race')['write'].mean().mean()
# ### Backward Difference Coding
# In backward difference coding, the mean of the dependent variable for a level is compared with the mean of the dependent variable for the prior level. This type of coding may be useful for a nominal or an ordinal variable.
from patsy.contrasts import Diff
contrast = Diff().code_without_intercept(levels)
print(contrast.matrix)
mod = ols("write ~ C(race, Diff)", data=hsb2)
res = mod.fit()
print(res.summary())
# For example, here the coefficient on level 1 is the mean of `write` at level 2 compared with the mean at level 1. Ie.,
res.params["C(race, Diff)[D.1]"]
hsb2.groupby('race').mean()["write"][2] - \
hsb2.groupby('race').mean()["write"][1]
# ### Helmert Coding
# Our version of Helmert coding is sometimes referred to as Reverse Helmert Coding. The mean of the dependent variable for a level is compared to the mean of the dependent variable over all previous levels. Hence, the name 'reverse' being sometimes applied to differentiate from forward Helmert coding. This comparison does not make much sense for a nominal variable such as race, but we would use the Helmert contrast like so:
from patsy.contrasts import Helmert
contrast = Helmert().code_without_intercept(levels)
print(contrast.matrix)
mod = ols("write ~ C(race, Helmert)", data=hsb2)
res = mod.fit()
print(res.summary())
# To illustrate, the comparison on level 4 is the mean of the dependent variable at the previous three levels taken from the mean at level 4
grouped = hsb2.groupby('race')
grouped.mean()["write"][4] - grouped.mean()["write"][:3].mean()
# As you can see, these are only equal up to a constant. Other versions of the Helmert contrast give the actual difference in means. Regardless, the hypothesis tests are the same.
k = 4
1./k * (grouped.mean()["write"][k] - grouped.mean()["write"][:k-1].mean())
k = 3
1./k * (grouped.mean()["write"][k] - grouped.mean()["write"][:k-1].mean())
# ### Orthogonal Polynomial Coding
# The coefficients taken on by polynomial coding for `k=4` levels are the linear, quadratic, and cubic trends in the categorical variable. The categorical variable here is assumed to be represented by an underlying, equally spaced numeric variable. Therefore, this type of encoding is used only for ordered categorical variables with equal spacing. In general, the polynomial contrast produces polynomials of order `k-1`. Since `race` is not an ordered factor variable let's use `read` as an example. First we need to create an ordered categorical from `read`.
hsb2['readcat'] = np.asarray(pd.cut(hsb2.read, bins=3))
hsb2.groupby('readcat').mean()['write']
from patsy.contrasts import Poly
levels = hsb2.readcat.unique().tolist()
contrast = Poly().code_without_intercept(levels)
print(contrast.matrix)
mod = ols("write ~ C(readcat, Poly)", data=hsb2)
res = mod.fit()
print(res.summary())
# As you can see, readcat has a significant linear effect on the dependent variable `write` but not a significant quadratic or cubic effect.
| examples/notebooks/contrasts.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# <small><i>This notebook was prepared by [<NAME>](http://donnemartin.com). Source and license info is on [GitHub](https://github.com/donnemartin/interactive-coding-challenges).</i></small>
# # Challenge Notebook
# ## Problem: Implement a hash table with set, get, and remove methods.
#
# * [Constraints](#Constraints)
# * [Test Cases](#Test-Cases)
# * [Algorithm](#Algorithm)
# * [Code](#Code)
# * [Unit Test](#Unit-Test)
# * [Solution Notebook](#Solution-Notebook)
# ## Constraints
#
# * For simplicity, are the keys integers only?
# * Yes
# * For collision resolution, can we use chaining?
# * Yes
# * Do we have to worry about load factors?
# * No
# ## Test Cases
#
# * get on an empty hash table index
# * set on an empty hash table index
# * set on a non empty hash table index
# * set on a key that already exists
# * remove on a key with an entry
# * remove on a key without an entry
# ## Algorithm
#
# Refer to the [Solution Notebook](http://nbviewer.ipython.org/github/donnemartin/interactive-coding-challenges/blob/master/arrays_strings/hash_map/hash_map_solution.ipynb). If you are stuck and need a hint, the solution notebook's algorithm discussion might be a good place to start.
# ## Code
# +
class Item(object):
def __init__(self, key, value):
# TODO: Implement me
pass
class HashTable(object):
def __init__(self, size):
# TODO: Implement me
pass
def hash_function(self, key):
# TODO: Implement me
pass
def set(self, key, value):
# TODO: Implement me
pass
def get(self, key):
# TODO: Implement me
pass
def remove(self, key):
# TODO: Implement me
pass
# -
# ## Unit Test
#
#
# **The following unit test is expected to fail until you solve the challenge.**
# +
# # %load test_hash_map.py
from nose.tools import assert_equal
class TestHashMap(object):
# TODO: It would be better if we had unit tests for each
# method in addition to the following end-to-end test
def test_end_to_end(self):
hash_table = HashTable(10)
print("Test: get on an empty hash table index")
assert_equal(hash_table.get(0), None)
print("Test: set on an empty hash table index")
hash_table.set(0, 'foo')
assert_equal(hash_table.get(0), 'foo')
hash_table.set(1, 'bar')
assert_equal(hash_table.get(1), 'bar')
print("Test: set on a non empty hash table index")
hash_table.set(10, 'foo2')
assert_equal(hash_table.get(0), 'foo')
assert_equal(hash_table.get(10), 'foo2')
print("Test: set on a key that already exists")
hash_table.set(10, 'foo3')
assert_equal(hash_table.get(0), 'foo')
assert_equal(hash_table.get(10), 'foo3')
print("Test: remove on a key that already exists")
hash_table.remove(10)
assert_equal(hash_table.get(0), 'foo')
assert_equal(hash_table.get(10), None)
print("Test: remove on a key that doesn't exist")
hash_table.remove(-1)
print('Success: test_end_to_end')
def main():
test = TestHashMap()
test.test_end_to_end()
if __name__ == '__main__':
main()
# -
# ## Solution Notebook
#
# Review the [Solution Notebook](http://nbviewer.ipython.org/github/donnemartin/interactive-coding-challenges/blob/master/arrays_strings/hash_map/hash_map_solution.ipynb) for a discussion on algorithms and code solutions.
| arrays_strings/hash_map/hash_map_challenge.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: 'Python 3.6.9 64-bit (''elfi36'': conda)'
# language: python
# name: python36964bitelfi36conda8bec451552304612bd355a97e3742bbb
# ---
# # Adaptive distance
#
# [ABC](https://elfi.readthedocs.io/en/latest/usage/tutorial.html#approximate-bayesian-computation) provides means to sample an approximate posterior distribution over unknown parameters based on comparison between observed and simulated data.
# This comparison is often based on distance between features that summarise the data and are informative about the parameter values.
#
# Here we assume that the summaries calculated based on observed and simulated data are compared based on weighted distance with weight $w_i=1/\sigma_i$ calculated based on their standard deviation $\sigma_i$.
# This ensures that the selected summaries to have an equal contribution in the distance between observed and simulated data.
#
# This notebook studies [adaptive distance](https://projecteuclid.org/euclid.ba/1460641065) [SMC-ABC](https://elfi.readthedocs.io/en/latest/usage/tutorial.html#sequential-monte-carlo-abc) where $\sigma_i$ and $w_i$ are recalculated between SMC iterations as proposed in [[1](#Reference)].
import numpy as np
import scipy.stats as ss
import matplotlib.pyplot as plt
# %matplotlib inline
import elfi
# ## Example 1:
#
# Assume we have an unknown parameter with prior distribution $\theta\sim U(0,50)$ and two simulator outputs $S_1\sim N(\theta, 1)$ and $S_2\sim N(\theta, 100)$ whose observed values are 20.
def simulator(mu, batch_size=1, random_state=None):
batches_mu = np.asarray(mu).reshape((-1,1))
obs_1 = ss.norm.rvs(loc=batches_mu, scale=1, random_state=random_state).reshape((-1,1))
obs_2 = ss.norm.rvs(loc=batches_mu, scale=100, random_state=random_state).reshape((-1,1))
return np.hstack((obs_1, obs_2))
observed_data = np.array([20,20])[None,:]
# Here the simulator outputs are both informative about the unknown model parameter, but $S_2$ has more observation noise than $S_1$. We do not calculate separate summaries in this example, but compare observed and simulated data based on these two variables.
#
# Euclidean distance between observed and simulated outputs or summaries can be used to find parameter values that could produce the observed data. Here we describe dependencies between the unknown parameter value and observed distances as an ELFI model `m` and sample the approximate posterior distribution with the [rejection sampler](https://elfi.readthedocs.io/en/latest/usage/tutorial.html#inference-with-rejection-sampling).
m = elfi.new_model()
theta = elfi.Prior(ss.uniform, 0, 50, model=m)
sim = elfi.Simulator(simulator, theta, observed=observed_data)
d = elfi.Distance('euclidean', sim)
rej = elfi.Rejection(d, batch_size=10000, seed=123)
# Let us sample 100 parameters with `quantile=0.01`. This means that we sample 10000 candidate parameters from the prior distribution and take the 100 parameters that produce simulated data closest to the observed data.
sample = rej.sample(100, quantile=0.01)
sample
plt.hist(sample.samples_array,range=(0,50),bins=20)
plt.xlabel('theta');
# The approximate posterior sample is concentrated around $\theta=20$ as expected in this example. However the sample distribution is much wider than we would observe in case the sample was selected based on $S_1$ alone.
#
# Now let us test adaptive distance in the same example.
#
# First we switch the distance node `d` to an adaptive distance node and initialise adaptive distance SMC-ABC. Initialisation is identical to the rejection sampler, and here we use the same batch size and seed as earlier, so that the methods are presented with the exact same candidate parameters.
d.become(elfi.AdaptiveDistance(sim))
ada_smc = elfi.AdaptiveDistanceSMC(d, batch_size=10000, seed=123)
# Since this is an iterative method, we must decide both sample size (`n_samples`) and how many populations are sampled (`rounds`). In addition we can decide the $\alpha$ quantile (`quantile`) used in estimation.
#
# Each population with `n_samples` parameter values is sampled as follows: 1. `n_samples/quantile` parameters are sampled from the current proposal distribution with acceptance threshold determined based on the previous population and 2. the distance measure is updated based on the observed sample and `n_samples` with the smallest distance are selected as the new population. The first population is sampled from the prior distribution and all samples are accepted in step 1.
#
# Here we sample one population with `quantile=0.01`. This means that the total simulation count will be the same as with the rejection sampler, but now the distance function is updated based on the 10000 simulated observations, and the 100 parameters included in the posterior sample are selected based on the new distance measure.
sample_ada = ada_smc.sample(100, 1, quantile=0.01)
sample_ada
plt.hist(sample_ada.samples_array,range=(0,50),bins=20)
plt.xlabel('theta');
# We see that the posterior distribution over unknown parameter values is narrower than in the previous example. This is because the simulator outputs are now normalised based on their estimated standard deviation.
#
# We can see $w_1$ and $w_2$:
sample_ada.adaptive_distance_w
# ## Example 2:
#
# This is the normal distribution example presented in [[1](#Reference)].
#
# Here we have an unknown parameter with prior distribution $\theta\sim N(0,100)$ and two simulator outputs $S_1\sim N(\theta, 0.1)$ and $S_2\sim N(1, 1)$ whose observed values are 0.
def simulator(mu, batch_size=1, random_state=None):
batches_mu = np.asarray(mu).reshape((-1,1))
obs_1 = ss.norm.rvs(loc=batches_mu, scale=0.1, random_state=random_state).reshape((-1,1))
obs_2 = ss.norm.rvs(loc=1, scale=1, size=batch_size, random_state=random_state).reshape((-1,1))
return np.hstack((obs_1, obs_2))
observed_data = np.array([0,0])[None,:]
# $S_1$ is now informative and $S_2$ uninformative about the unknown parameter value, and we note that between the two output variables, $S_1$ has larger variance under the prior predictive distribution. This means that normalisation estimated based on output data observed in the initial round or based on a separate sample would not work well in this example.
#
# Let us define a new model and initialise adaptive distance SMC-ABC.
m = elfi.new_model()
theta = elfi.Prior(ss.norm, 0, 100, model=m)
sim = elfi.Simulator(simulator, theta, observed=observed_data)
d = elfi.AdaptiveDistance(sim)
ada_smc = elfi.AdaptiveDistanceSMC(d, batch_size=2000, seed=123)
# Next we sample 1000 parameter values in 5 rounds with the default `quantile=0.5` which is recommended in sequential estimation [[1](#Reference)]:
sample_ada = ada_smc.sample(1000, 5)
sample_ada
plt.hist(sample_ada.samples_array, range=(-25,25), bins=20)
plt.xlabel(theta);
# The sample distribution is concentrated around $\theta=0$ but wider than could be expected. However we can continue the iterative estimation process. Here we sample two more populations:
sample_ada = ada_smc.sample(1000, 2)
sample_ada
plt.hist(sample_ada.samples_array, range=(-25,25), bins=20)
plt.xlabel('theta');
# We observe that the sample mean is now closer to zero and the sample distribution is narrower.
#
# Let us examine $w_1$ and $w_2$:
sample_ada.adaptive_distance_w
# We can see that $w_2$ (second column) is constant across iterations whereas $w_1$ increases as the method learns more about possible parameter values and the proposal distribution becomes more concentrated around $\theta=0$.
#
# ## Notes
#
# The adaptive distance SMC-ABC method demonstrated in this notebook normalises simulator outputs or summaries calculated based on simulator output based on their estimated standard deviation under the proposal distribution in each iteration. This ensures that all outputs or summaries have an equal contribution to the distance between simulated and observed data in all iterations.
#
# It is important to note that the method does not evaluate whether outputs or summaries are needed or informative. In both examples studied in this notebook, results would improve if inference was carried out based on $S_1$ alone. Hence one should choose the summaries used in adaptive distance SMC-ABC with the usual care. ELFI tools that aid in the selection process are discussed in the diagnostics notebook available [here](https://github.com/elfi-dev/notebooks/tree/master).
# ## Reference
# [1] <NAME> (2017). Adapting the ABC Distance Function. Bayesian Analysis 12(1): 289-309, 2017. https://projecteuclid.org/euclid.ba/1460641065
| adaptive_distance.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <img style="float: left; margin: 30px 15px 15px 15px;" src="https://pngimage.net/wp-content/uploads/2018/06/logo-iteso-png-5.png" width="300" height="500" />
#
#
# ### <font color='navy'> Simulación de procesos financieros.
#
# **Nombres:** <NAME> y <NAME>
#
# **Fecha:** 22 de Febrero del 2021
#
# **Expediente** : if721470
# **Profesor:** <NAME>.
#
# ### LINK DE GITHUB:
#
# # Tarea 4: Clase 7. Ejemplos Simulación
# ## Enunciado de tarea
# > # 1
# Como ejemplo simple de una simulación de Monte Carlo, considere calcular la probabilidad de una suma particular del lanzamiento de tres dados (cada dado tiene valores del uno al seis). Además cada dado tiene las siguientes carácterísticas: el primer dado no está cargado (distribución uniforme todos son equiprobables); el segundo y tercer dado están cargados basados en una distribución binomial con parámetros (`n=5, p=0.5` y `n=5, p=0.2`). Calcule la probabilidad de que la suma resultante sea 7, 14 o 18.
#
#
# > # 2 Ejercicio de aplicación- Cafetería Central
#
# Premisas para la simulación:
# - Negocio de alimentos que vende bebidas y alimentos.
# - Negocio dentro del ITESO.
# - Negocio en cafetería central.
# - Tipo de clientes (hombres y mujeres).
# - Rentabilidad del 60%.
#
# ## Objetivo
# Realizar una simulación estimado el tiempo medio que se tardaran los clientes en ser atendidos entre el horario de 6:30 a 1 pm. Además saber el consumo.
# **Analizar supuestos y limitantes**
#
# ## Supuestos en simulación
# Clasificación de clientes:
# - Mujer = 1 $\longrightarrow$ aleatorio < 0.5
# - Hombre = 0 $\longrightarrow$ aleatorio $\geq$ 0.5.
#
# Condiciones iniciales:
# - Todas las distrubuciones de probabilidad se supondrán uniformes.
# - Tiempo de simulación: 6:30 am - 1:30pm $\longrightarrow$ T = 7 horas = 25200 seg.
# - Tiempo de llegada hasta ser atendido: Min=5seg, Max=30seg.
# - Tiempo que tardan los clientes en ser atendidos:
# - Mujer: Min = 1 min= 60seg, Max = 5 min = 300 seg
# - Hombre: Min = 40 seg, Max = 2 min= 120 seg
# - Consumo según el tipo de cliente:
# - Mujer: Min = 30 pesos, Max = 100 pesos
# - Hombre: Min = 20 pesos, Max = 80 pesos
#
# Responder las siguientes preguntas basados en los datos del problema:
# 1. ¿Cuáles fueron los gastos de los hombres y las mujeres en 5 días de trabajo?.
# 2. ¿Cuál fue el consumo promedio de los hombres y mujeres?
# 3. ¿Cuál fue el número de personas atendidas por día?
# 4. ¿Cuál fue el tiempo de atención promedio?
# 5. ¿Cuánto fue la ganancia promedio de la cafetería en 5 días de trabajo y su respectiva rentabilidad?
# ### Ejercicio 1:
# Como ejemplo simple de una simulación de Monte Carlo, considere calcular la probabilidad de una suma particular del lanzamiento de tres dados (cada dado tiene valores del uno al seis). Además cada dado tiene las siguientes carácterísticas: el primer dado no está cargado (distribución uniforme todos son equiprobables); el segundo y tercer dado están cargados basados en una distribución binomial con parámetros (`n=5, p=0.5` y `n=5, p=0.2`). Calcule la probabilidad de que la suma resultante sea 7, 14 o 18.
#
#
# +
# SOLUCION CRISTINA
# -
# Código de solución
.
.
.
.
# +
# SOLUCION DAYANA
# +
# Código de solución
import numpy as np
import scipy.stats as st
def suma1():
d1_no_caragado = np.random.randint(1,7)
d2_cargado = st.binom(n = 5, p=0.5).rvs(size = 1)
d3_cargado = st.binom(n = 5, p=0.2).rvs(size = 1)
suma_dados = d1_no_caragado + d2_cargado + d3_cargado
if suma_dados == 7:
return True
else:
return False
def suma2():
d1_no_caragado = np.random.randint(1,7)
d2_cargado = st.binom(n = 5, p=0.5).rvs(size = 1)
d3_cargado = st.binom(n = 5, p=0.2).rvs(size = 1)
suma_dados = d1_no_caragado + d2_cargado + d3_cargado
if suma_dados == 14:
return True
else:
return False
def suma3():
d1_no_caragado = np.random.randint(1,7)
d2_cargado = st.binom(n = 5, p=0.5).rvs(size = 1)
d3_cargado = st.binom(n = 5, p=0.2).rvs(size = 1)
suma_dados = d1_no_caragado + d2_cargado + d3_cargado
if suma_dados == 18:
return True
else:
return False
N=100
dado_1=[suma1( ) for i in range(N)]
dado_2=[suma2( ) for i in range(N)]
dado_3=[suma3( ) for i in range(N)]
dado_1.count(True)/N, dado_2.count(True)/N, dado_3.count(True)/N
# -
# ### Ejercicio 2 de aplicación- Cafetería Central
#
# Premisas para la simulación:
# - Negocio de alimentos que vende bebidas y alimentos.
# - Negocio dentro del ITESO.
# - Negocio en cafetería central.
# - Tipo de clientes (hombres y mujeres).
# - Rentabilidad del 60%.
#
# #### Objetivo
# Realizar una simulación estimado el tiempo medio que se tardaran los clientes en ser atendidos entre el horario de 6:30 a 1 pm. Además saber el consumo.
# **Analizar supuestos y limitantes**
#
# #### Supuestos en simulación
# Clasificación de clientes:
# - Mujer = 1 $\longrightarrow$ aleatorio < 0.5
# - Hombre = 0 $\longrightarrow$ aleatorio $\geq$ 0.5.
#
# Condiciones iniciales:
# - Todas las distrubuciones de probabilidad se supondrán uniformes.
# - Tiempo de simulación: 6:30 am - 1:30pm $\longrightarrow$ T = 7 horas = 25200 seg.
# - Tiempo de llegada hasta ser atendido: Min=5seg, Max=30seg.
# - Tiempo que tardan los clientes en ser atendidos:
# - Mujer: Min = 1 min= 60seg, Max = 5 min = 300 seg
# - Hombre: Min = 40 seg, Max = 2 min= 120 seg
# - Consumo según el tipo de cliente:
# - Mujer: Min = 30 pesos, Max = 100 pesos
# - Hombre: Min = 20 pesos, Max = 80 pesos
#
# Responder las siguientes preguntas basados en los datos del problema:
# 1. ¿Cuáles fueron los gastos de los hombres y las mujeres en 5 días de trabajo?.
# 2. ¿Cuál fue el consumo promedio de los hombres y mujeres?
# 3. ¿Cuál fue el número de personas atendidas por día?
# 4. ¿Cuál fue el tiempo de atención promedio?
# 5. ¿Cuánto fue la ganancia promedio de la cafetería en 5 días de trabajo y su respectiva rentabilidad?
################## Datos del problema
d = 5
T =25200
T_at_min = 5; T_at_max = 30
T_mujer_min =60; T_mujer_max = 300
T_hombre_min = 40; T_hombre_max = 120
C_mujer_min = 30; C_mujer_max = 100
C_hombre_min = 20; C_hombre_max = 80
# +
# SOLUCION CRISTINA
# -
# Código de solución
.
.
.
.
# +
# SOLUCION DAYANA
# -
# Código de solución
.
.
.
.
| TAREA_4. VazquezVargas_Cristina_NavarroValencia_Dayana.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] raw_mimetype="text/restructuredtext"
# .. _nb_interface_minimize:
# -
# ## Minimize
#
#
# The `minimize` function provides the external interface for any kind of optimization to be performed. The minimize method arguments and options look as follows:
# ```python
# def minimize(problem,
# algorithm,
# termination=None,
# seed=None,
# verbose=False,
# display=None,
# callback=None,
# return_least_infeasible=False,
# save_history=False
# )
# ```
# - `problem`: A [Problem](../problems/index.ipynb) object that contains the problem to be solved.
# - `algorithm`: An [Algorithm](../algorithms/index.ipynb) objective which represents the algorithm to be used.
# - `termination`: A [Termination](termination.ipynb) object or a tuple defining when the algorithm has terminated. If not provided, a default termination criterion will be used. Purposefully, we list the *termination* as a parameter and not an option. Specific algorithms might need some refinement of the termination to work reliably.
# - `seed`: Most algorithms underly some randomness. Setting the *seed* to a positive integer value ensures reproducible results. If not provided, a random seed will be set automatically, and the used integer will be stored in the [Result](result.ipynb) object.
# - `verbose`: Boolean value defining whether the output should be printed during the run or not.
# - `display`: You can overwrite what output is supposed to be printed in each iteration. Therefore, a custom [Display](display.ipynb) object can be used for customization purposes.
# - `save_history`: A boolean value representing whether a snapshot of the algorithm should be stored in each iteration. If enabled, the [Result](result.ipynb) object contains the history.
# - `return_least_infeasible`: Whether if the algorithm can not find a feasible solution, the least infeasible solution should be returned. By default `False`.
# Note, the `minimize` function creates a **deep copy** of the algorithm object before the run.
# This ensures that two independent runs with the same algorithm and same random seed have the same results without any side effects. However, to access the algorithm's internals, you can access the object being used by `res.algorithm` where `res` is an instance of the [Result](result.ipynb) object.
# ### API
# + raw_mimetype="text/restructuredtext" active=""
# .. autofunction:: pymoo.optimize.minimize
| source/interface/minimize.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import os
import cudf
from cuml import neighbors
from blazingsql import BlazingContext
bc = BlazingContext()
# identify path to data
path = os.getcwd()
path = path + '/data/iris.csv'
path
# +
# create table
bc.create_table('iris', path, header=0)
# pull x & y training data
y = bc.sql('select target from iris')
x = bc.sql('select sepal_length, sepal_width, petal_length, petal_width from iris')
# set knn model
knn = neighbors.KNeighborsClassifier(n_neighbors=3)
# train model
knn.fit(x, y)
# make simple test case
t = cudf.DataFrame({'sepal_length':5.1, 'sepal_width':3.5, 'petal_length':1.4, 'petal_width':.2})
# predict flower type
knn.predict(t)
| bsql_iris.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="qn8CLGQ1U_sw"
#
# Assignment 1 Part 1: Graph Class
# + id="00cUFtx6Sixf" colab={"base_uri": "https://localhost:8080/", "height": 50} outputId="be253fd7-1337-4755-e4f3-f66b042c2b79"
graph_elements = { "Apple" : ["Banana","Cherry"],
"Banana" : ["Apple", "Durian"],
"Cherry" : ["Apple", "Durian"],
"Durian" : ["Orange"],
"Orange" : ["Durian"]
}
class graph:
def __init__(self, value=None):
self.value = value
def getVertices(self):
return list(self.value.keys())
def getEdges(self):
return self.edges()
def edges(self):
edgelist = []
for vertex in self.value:
for neighbor in self.value[vertex]:
if {neighbor, vertex} not in edgelist:
edgelist.append({vertex, neighbor})
return edgelist
g = graph(graph_elements)
print(g.getVertices())
print(g.getEdges())
# + [markdown] id="QzmgYXn5VVk1"
# Depth First Traversal of a graph
# + id="4wh5icuWSkt8" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="37d45899-4560-4535-807f-0f8713b8cec1"
def depth_first_search(graph, start, visited=None):
if visited is None:
visited = set()
visited.add(start)
#print(start)
for next in graph[start] - visited:
depth_first_search(graph, next, visited)
return visited
graph = {'Apple': set(['Banana', 'Cherry']),
'Banana': set(['Apple', 'Durian', 'Orange']),
'Cherry': set(['Apple']),
'Durian': set(['Banana', 'Orange']),
'Orange': set(['Cherry', 'Durian'])
}
depth_first_search(graph, 'Banana')
# + [markdown] id="90iwo9W1Wanw"
# Breadth-First Traversal of a Graph
# + id="_QEoUEm-SkhT" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="c283fe39-4004-4770-a3bc-3b80a1102dbb"
import collections
def breadth_first_search(graph, root):
visited, queue = set(), collections.deque([root])
visited.add(root)
while queue:
vertex = queue.popleft()
print(str(vertex) + " ", end="")
for neighbour in graph[vertex]:
if neighbour not in visited:
visited.add(neighbour)
queue.append(neighbour)
if __name__ == '__main__':
graph = {0: [1, 2],
1: [0, 2],
2: [3],
3: [1, 2]
}
breadth_first_search(graph, 1)
# + id="nHtTTpI5SkHs"
# + [markdown] id="_DAwWMYGWwcW"
# Assignment 1 Part 2: Election Data Search
# + id="XytUX-nemkr0"
import pandas as pd
# + id="k0V_IGLnl7P7" colab={"base_uri": "https://localhost:8080/", "height": 84} outputId="e6919695-6c1f-4631-c315-2967cd1b2d02"
from google.colab import drive
drive.mount('/data/')
data_dir = '/data/My Drive/Colab Notebooks/FEC dataset'
# !ls '/data/My Drive/Colab Notebooks/FEC dataset'
# + [markdown] id="pl-0E8cE-hGy"
# Search on 'CN20.zip' folder
# + id="zw1KZ0NmmWQw" colab={"base_uri": "https://localhost:8080/", "height": 400} outputId="cfb24d9f-b184-42d8-9b4d-5c5ad20b32b7"
from zipfile import ZipFile
header = pd.read_csv(data_dir+'/cn_header_file.csv')
with ZipFile(data_dir+'/cn20.zip') as zip:
candidates = pd.read_csv(zip.open('cn.txt'), sep='|', names=header.columns)
candidates.head()
# + id="CZ2y9RTAnTzU" colab={"base_uri": "https://localhost:8080/", "height": 333} outputId="d698ba69-e8bf-4db5-b657-c79efd645bdf"
candidates[candidates['CAND_NAME'].str.contains('WALKER')].head()
# + id="tdUayozq-9Cb" colab={"base_uri": "https://localhost:8080/", "height": 417} outputId="a26fa3cc-48cc-48aa-aef1-3195d29110a7"
tg = candidates[(candidates['CAND_ELECTION_YR'] == 2020) & (candidates['CAND_OFFICE_ST'] == 'FL')]
tg.head()
# + id="ytvXx2wHCvs1" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="f379bdaf-43ee-4446-a7e5-1c4302a9ec9c"
len(tg.index)
# + id="mymZWrEypYla" colab={"base_uri": "https://localhost:8080/", "height": 130} outputId="6f97a11e-f339-4022-fd05-136583e1ab0f"
candidates[candidates['CAND_NAME'].str.contains('TRUMP, DONALD')]
# + [markdown] id="AoFXVna2--1J"
# Search on 'Pas220.zip' folder
# + id="wIKz_EIYiHLj" colab={"base_uri": "https://localhost:8080/", "height": 400} outputId="1bb2e9a1-c58c-460c-e64d-2f73e34c291f"
header = pd.read_csv(data_dir+'/pas2_header_file.csv')
with ZipFile(data_dir+'/pas220.zip') as zip:
#print(zip.namelist())
spending = pd.read_csv(zip.open('itpas2.txt'), sep='|', names=header.columns)
spending.head()
# + id="cJReRZAi7RSR" colab={"base_uri": "https://localhost:8080/", "height": 195} outputId="8f41dac7-2e34-4db1-b661-2b46ab2debab"
house_spending_FL = spending[(spending['CAND_ID'].str.startswith('H')==True) & (spending['STATE']=='FL')]
df_house = house_spending_FL[['CMTE_ID', 'NAME', 'STATE', 'TRANSACTION_AMT', 'CAND_ID']]
df_house.head()
# + [markdown] id="L-O7kKyr_OkU"
# Search on 'CM20.zip' folder
# + id="UX-S1tm7nOo2" colab={"base_uri": "https://localhost:8080/", "height": 518} outputId="145959fb-bb59-4e7a-c6e1-31f3d64148c5"
header = pd.read_csv(data_dir+'/cm_header_file.csv')
with ZipFile(data_dir+'/cm20.zip') as zip:
#print(zip.namelist())
df = pd.read_csv(zip.open('cm.txt'), sep='|', names=header.columns)
df.head()
# + id="Gz5FKlS11ZGQ" colab={"base_uri": "https://localhost:8080/", "height": 195} outputId="caef84a1-84e2-4340-d070-4103622b12fa"
df_name = df[['CMTE_ID', 'CMTE_NM']]
df_name.head()
# + id="MpE777-JE3Gt" colab={"base_uri": "https://localhost:8080/", "height": 618} outputId="4d62b2f1-8c4e-4678-df9b-b028eeda0ea3"
Society_Organization = df[df['CONNECTED_ORG_NM'].str.contains('SOCIETY', na=False)]
Society_Organization.head()
# + id="GErXTSsJGOV-" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="f2899955-e0ec-47b2-9310-24ba8fa7def4"
len(Society_Organization.index)
# + [markdown] id="drRP2x2Z_lZS"
# Search on 'CCL20.zip' folder
# + id="ombc-hjIn0KR" colab={"base_uri": "https://localhost:8080/", "height": 195} outputId="20885340-ea71-466c-e66d-ba3684884989"
header = pd.read_csv(data_dir+'/ccl_header_file.csv')
with ZipFile(data_dir+'/ccl20.zip') as zip:
#print(zip.namelist())
linkage = pd.read_csv(zip.open('ccl.txt'), sep='|', names=header.columns)
linkage.head()
# + id="vb8dIlKKE_4m"
df1 = pd.concat([candidates, linkage, df], axis=1, sort=False).reset_index()
# + id="yEgZmnNHT2xw" colab={"base_uri": "https://localhost:8080/", "height": 534} outputId="d25d1e93-80a7-4ebd-a382-97460212b7ae"
df1.head()
# + id="d_oy0b5jT8eY" colab={"base_uri": "https://localhost:8080/", "height": 333} outputId="6650d9ab-983e-4277-843d-157107978f50"
df2 = pd.merge(candidates, linkage, on='CAND_ID')
df2.head()
# + id="5pJTEGPm0OXR"
df_merge = pd.merge(df2, df_name, on='CMTE_ID')
#df_merge.head()
df_sort = df_merge[['CAND_ID', 'CAND_NAME', 'CAND_ST', 'CMTE_NM']]
# + id="jAIGmy572dYD" colab={"base_uri": "https://localhost:8080/", "height": 343} outputId="bbeade85-9d03-4b76-88d7-ba6ecf5ec6f0"
df_sort[df_sort['CAND_ST']=='FL'].head(10)
# + id="geYa-uW_34Aq" colab={"base_uri": "https://localhost:8080/", "height": 195} outputId="8ec71abb-0aa5-42cf-a6f4-41716b07891e"
df_sort_house = pd.merge(df_sort, df_house, on='CAND_ID')
df_sort_house.head()
# + id="hRa7Rywa-Rby" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="71f63e33-c35c-413b-c882-c3863e073ba0"
df_sort_house['TRANSACTION_AMT'].sum()
# + id="lJ51BCl5YHBM"
| assets/EMSE6574/Week2_Assignment.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Initialization
#
# Test notebook for the C-MAPPS benchmark. Test different MLP architectures.
#
# First we import the necessary packages and create the global variables.
# +
import math
import numpy as np
import csv
import copy
from scipy import stats
from sklearn.preprocessing import StandardScaler, MinMaxScaler
import sys
sys.path.append('/Users/davidlaredorazo/Documents/University_of_California/Research/Projects')
#sys.path.append('/media/controlslab/DATA/Projects')
from ann_framework.data_handlers.data_handler_CMAPSS import CMAPSSDataHandler
from ann_framework.tunable_model.tunable_model import SequenceTunableModelRegression
from ann_framework import aux_functions
#import custom_scores
from keras.models import Sequential, Model
from keras.layers import Dense, Input, Dropout, Reshape, Conv2D, Flatten, MaxPooling2D
from keras.optimizers import Adam
from keras.callbacks import LearningRateScheduler
from keras import backend as K
from keras import regularizers
from keras.layers import LSTM, CuDNNLSTM
# -
# # Define architectures
#
# Define each one of the different architectures to be tested.
# +
K.clear_session() #Clear the previous tensorflow graph
l2_lambda_regularization = 0.20
l1_lambda_regularization = 0.20
def RULmodel_LSTM(input_shape):
"""Define the RNN model"""
#Create a sequential model
model = Sequential()
#model.add(Masking(mask_value=0, imput))
#model.add(LSTM(input_shape=input_shape, units=100, return_sequences=True, name='lstm1')))
model.add(CuDNNLSTM(input_shape=input_shape, units=20, return_sequences=False, name='lstm2'))
model.add(Dense(10, input_dim=input_shape, activation='relu', kernel_initializer='glorot_normal',
kernel_regularizer=regularizers.l2(l2_lambda_regularization), name='fc1'))
model.add(Dense(1, activation='linear', name='out'))
return model
def RULmodel_SN_5(input_shape):
#Create a sequential model
model = Sequential()
#Add the layers for the model
model.add(Dense(20, input_dim=input_shape, activation='relu', kernel_initializer='glorot_normal',
kernel_regularizer=regularizers.l2(l2_lambda_regularization),
name='fc1'))
model.add(Dense(20, input_dim=input_shape, activation='relu', kernel_initializer='glorot_normal',
kernel_regularizer=regularizers.l2(l2_lambda_regularization),
name='fc2'))
model.add(Dense(1, activation='linear', name='out'))
return model
# -
def get_compiled_model(model_def, shape, model_type='lstm'):
#Shared parameters for the models
optimizer = Adam(lr=0, beta_1=0.5)
lossFunction = "mean_squared_error"
metrics = ["mse"]
model = None
#Create and compile the models
if model_type=='ann':
model = model_def(shape)
model.compile(optimizer = optimizer, loss = lossFunction, metrics = metrics)
elif model_type=='lstm':
model = RULmodel_LSTM(shape)
model.compile(optimizer = optimizer, loss = lossFunction, metrics = metrics)
else:
pass
return model
# +
#Define the usable models for this notebook
#models = {'shallow-20':RULmodel_SN_5,'rnn-20-10':RULmodel_LSTM}
models = {'shallow-20':RULmodel_SN_5}
# -
# # Process Data
# +
features = ['T2', 'T24', 'T30', 'T50', 'P2', 'P15', 'P30', 'Nf', 'Nc', 'epr', 'Ps30', 'phi', 'NRf', 'NRc',
'BPR', 'farB', 'htBleed', 'Nf_dmd', 'PCNfR_dmd', 'W31', 'W32']
selected_indices = np.array([2, 3, 4, 7, 8, 9, 11, 12, 13, 14, 15, 17, 20, 21])
selected_features = list(features[i] for i in selected_indices-1)
data_folder = '../CMAPSSData'
window_size = 30
window_stride = 1
max_rul = 128
min_max_scaler = MinMaxScaler(feature_range=(-1, 1))
dHandler_cmaps = CMAPSSDataHandler(data_folder, 1, selected_features, max_rul,
window_size, window_stride)
# -
# # Build the model
# +
optimizer = Adam(lr=0, beta_1=0.5)
lossFunction = "mean_squared_error"
metrics = ["mse"]
#Create and compile the models
nFeatures = len(selected_features)
shapeSN = nFeatures*window_size
shapeLSTM = (window_size,nFeatures)
model = get_compiled_model(models['shallow-20'], shapeSN, model_type='ann')
tModel = SequenceTunableModelRegression('mlpnn', model, lib_type='keras', data_handler=dHandler_cmaps)
# -
# # Load Data
# +
#For LSTM
tModel.data_handler.data_scaler = min_max_scaler
tModel.data_scaler = None
#For ANN
#tModel.data_handler.data_scaler = min_max_scaler
#tModel.data_scaler = min_max_scaler
tModel.data_handler.sequence_length = 30
#tModel.data_handler.sequence_length = maxWindowSize[datasetNumber]
tModel.data_handler.sequence_stride = 1
tModel.data_handler.max_rul = 128
tModel.load_data(unroll=False, verbose=1, cross_validation_ratio=0)
tModel.print_data()
# -
# # Test on dataset 1
# +
iterations = 10
tModel.epochs = 100
lrate = LearningRateScheduler(aux_functions.step_decay)
num_features = len(selected_features)
windowSize = 30
windowStride = 1
constRul = 140
file = open("results/MLP/ResultsDatasets_1_test.csv", "w")
csvfile = csv.writer(file, lineterminator='\n')
for key, model_def in models.items():
print("For model "+str(key))
#file.write("For model "+str(key)+'\n\n')
for i in range(1,2):
dataset = i
print("Computing for dataset "+str(i))
#file.write("Computing for dataset "+str(i)+'\n\n')
tempScoresRMSE = np.zeros((iterations,1))
tempScoresRHS = np.zeros((iterations,1))
tempTime = np.zeros((iterations,1))
input_shape = windowSize*num_features #For simple ANN
tModel.data_handler.change_dataset(i)
tModel.data_handler.sequence_length = windowSize
tModel.data_handler.sequence_stride = windowStride
tModel.data_handler.max_rul = constRul
tModel.load_data(unroll=True, verbose=0, cross_validation_ratio=0)
#tModel.print_data()
for j in range(iterations):
#Model needs to be recompiled everytime since they are different runs so weights should be reinit
model = get_compiled_model(model_def, input_shape, model_type='ann')
tModel.change_model(key, model, 'keras')
tModel.train_model(learningRate_scheduler=lrate, verbose=0)
tModel.evaluate_model(['rhs', 'rmse'], round=2)
#print("scores")
#print(j)
cScores = tModel.scores
rmse = math.sqrt(cScores['score_1'])
rmse2 = cScores['rmse']
rhs = cScores['rhs']
time = tModel.train_time
tempScoresRMSE[j] = rmse2
tempScoresRHS[j] = rhs
tempTime[j] = time
print("Results for model " + key)
print(stats.describe(tempScoresRMSE))
print(stats.describe(tempScoresRHS))
print(stats.describe(tempTime))
tempScoresRMSE = np.reshape(tempScoresRMSE, (iterations,))
tempScoresRHS = np.reshape(tempScoresRHS, (iterations,))
tempTime = np.reshape(tempTime, (iterations,))
csvfile.writerow(tempScoresRMSE)
csvfile.writerow(tempScoresRHS)
csvfile.writerow(tempTime)
file.close()
# -
# # Test on all Datasets
# +
datasets = [1,2,3,4]
iterations = 2
tModel.epochs = 150
lrate = LearningRateScheduler(aux_functions.step_decay)
scores ={1:[], 2:[], 3:[], 4:[]}
window_sizes = {1:30,2:20,3:30,4:18}
strides = {1:1,2:2,3:1,4:2}
max_ruls = {1:140, 2:134, 3:128, 4:134}
num_features = len(selected_features)
input_shape = None
#For each model
for key, model_def in models.items():
file = open("results/MLP/ResultsDatasets_1_test"+key+".csv", "w")
csvfile = csv.writer(file, lineterminator='\n')
print(model.summary())
print("Generating statistics for model " + key)
#For each dataset
for i in range(1,2):
print("Working on dataset " + str(i))
tempScoresRMSE = np.zeros((iterations,1))
tempScoresRHS = np.zeros((iterations,1))
tempTime = np.zeros((iterations,1))
input_shape = window_sizes[i]*num_features #For simple ANN
#input_shape = (window_sizes[i],num_features) #For RNN
print(input_shape)
tModel.data_handler.change_dataset(i)
tModel.data_handler.sequence_length = window_sizes[i]
tModel.data_handler.sequence_stride = strides[i]
tModel.data_handler.max_rul = max_ruls[i]
tModel.load_data(unroll=True, verbose=0, cross_validation_ratio=0)
#tModel.print_data()
#tModel.print_data()
for j in range(iterations):
#Model needs to be recompiled everytime since they are different runs so weights should be reinit
model = get_compiled_model(model_def, input_shape, model_type='ann')
tModel.change_model(key, model, 'keras')
tModel.train_model(learningRate_scheduler=lrate, verbose=0)
tModel.evaluate_model(['rhs', 'rmse'], round=2)
#print("scores")
#print(j)
cScores = tModel.scores
rmse = math.sqrt(cScores['score_1'])
rmse2 = cScores['rmse']
rhs = cScores['rhs']
time = tModel.train_time
tempScoresRMSE[j] = rmse2
tempScoresRHS[j] = rhs
tempTime[j] = time
print("Results for model " + key)
print(stats.describe(tempScoresRMSE))
print(stats.describe(tempScoresRHS))
print(stats.describe(tempTime))
tempScoresRMSE = np.reshape(tempScoresRMSE, (iterations,))
tempScoresRHS = np.reshape(tempScoresRHS, (iterations,))
tempTime = np.reshape(tempTime, (iterations,))
csvfile.writerow(tempScoresRMSE)
csvfile.writerow(tempScoresRHS)
csvfile.writerow(tempTime)
file.close()
# -
| code/TestArchitectures.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import json
import yaml
import pandas as pd
from SPARQLWrapper import SPARQLWrapper, JSON
# + pycharm={"name": "#%%\n"}
with open('../ontos.txt') as fp:
ontologies = [l.strip() for l in fp.readlines()]
# All ontologies wildcard
ontologies.append('ontos')
endpoints = dict()
for ontology in ontologies:
e = SPARQLWrapper(f'http://127.0.0.1:9999/blazegraph/namespace/obo-{ontology}/sparql')
e.setRequestMethod('postdirectly')
e.setMethod('POST')
e.setReturnFormat(JSON)
endpoints[ontology] = e
# + pycharm={"name": "#%%\n"}
with open('props_o_types.rq') as fp:
otypes_query = fp.read()
ont_results = dict()
for ontology, e in list(endpoints.items()):
e.setQuery(otypes_query)
data = e.query().convert()['results']['bindings']
ont_results[ontology] = [
{
'prop': r['p']['value'],
'count': int(r['count']['value']),
'uris': int(r['uris']['value']),
'blanks': int(r['blanks']['value']),
'literals': int(r['literals']['value']),
'range': r['range1']['value'] if 'range1' in r else '',
'type': r['type1']['value'] if 'type1' in r else '',
}
for r in data
]
print(ontology)
# + pycharm={"name": "#%%\n"}
with open('results/otypes.json', 'w') as fp:
json.dump(ont_results, fp, indent=4)
# + pycharm={"name": "#%%\n"}
ont_dfs = {ontology: pd.DataFrame.from_records(data).set_index('prop') for ontology, data in ont_results.items()}
def calc_errors(r):
return min(r['uris'] + r['blanks'], r['literals']), max(r['uris'] + r['blanks'], r['literals'])
for df in ont_dfs.values():
df['errors_min'], df['errors_max'] = zip(*df.apply(calc_errors, axis=1))
# + pycharm={"name": "#%%\n"}
df_all = ont_dfs['ontos'].copy()
df_all['prop'] = df_all.index
for ont, df in ont_dfs.items():
if ont == 'ontos':
continue
df_all[[f'{ont}_uris', f'{ont}_blanks', f'{ont}_literals']] = df[['uris', 'blanks', 'literals']]
# + pycharm={"name": "#%%\n"}
with open('results/otypes_review.yaml') as fp:
review = yaml.safe_load(fp)
df_review = pd.DataFrame.from_records(review)
df_review = df_review.join(df_all, on='prop', lsuffix='1')
# + pycharm={"name": "#%%\n"}
df_review.to_json('results/otypes_invalid.json', indent=4, orient='index')
| obo/2_prop_values/otypes_get.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import pandas as pd
dota2_matches = pd.read_csv('C:/Users/ashis/Documents/Data/new_match_list.csv')
radiant_win = dota2_matches['r_win']
X = dota2_matches.iloc[:, 0:10].values
labels = radiant_win.values.astype(int)
# +
# Importing Modules
from sklearn.manifold import TSNE
import matplotlib.pyplot as plt
# Loading dataset
dataset = X
# Defining Model
model = TSNE(learning_rate = 100)
# save the model to disk
#ilename = 'Dota2_TSNE.sav'
#ickle.dump(model, open(filename, 'wb'))
# train a logistic regression model on the training set
#mport pickle
# load the model from disk
#filename = 'Dota2_unsupervised.sav'
#sne_model = pickle.load(open(filename, 'rb'))
# Fitting Model
transformed = model.fit_transform(dataset.data)
# Plotting 2d t-Sne
x_axis = transformed[:, 0]
y_axis = transformed[:, 1]
plt.scatter(x_axis, y_axis, c=labels)
plt.show()
# -
| jupyter notebook/Unsupervised-Clustering visualization.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: ra
# language: python
# name: ra
# ---
import iris
from iris.experimental.equalise_cubes import equalise_attributes
import numpy as np
import glob
import matplotlib.pyplot as plt
from cf_units import Unit
import warnings
import datetime
def get_concatenated_cube(path):
files = glob.glob(path, recursive=True)
assert files
files.sort()
cube_list = iris.load(files)
equalise_attributes(cube_list)
out_cube = cube_list.concatenate_cube()
return out_cube
def cube_multiply(big_cube, small_cube):
"""Returns the multiplication of two input cubes accounting for
missing coordinates in the smaller cube."""
dim_diff = big_cube.ndim - small_cube.ndim
assert big_cube.shape[dim_diff:] == small_cube.shape
temp_cube = small_cube.copy() #as to not make changes to small_cube
#add aux coords to smaller cube to enable multiply
for coord in big_cube.coords():
if coord not in temp_cube.coords():
temp_cube.add_aux_coord(coord[0])
out_cube = big_cube * temp_cube
return out_cube
def date_BOM(date):
"""Return beginning of month of given date."""
return date.replace(day=1, hour=0, minute=0, second=0, microsecond=0)
def time_constraint(cube):
"""Returns iris constraint to extract all time coordinates in the same month."""
cube_coord = cube.coord('time')
cube_coord_points = cube_coord.units.num2date(cube_coord.points)
constraint = iris.Constraint(time = lambda cell: date_BOM(cell.point) \
in [date_BOM(date) for date in cube_coord_points])
return constraint
model_path = {'ACCESS-CM2': '/g/data/fs38/publications/CMIP6/FAFMIP/CSIRO-ARCCSS/ACCESS-CM2/',
'CAS-ESM2-0': '/g/data/oi10/replicas/CMIP6/FAFMIP/CAS/CAS-ESM2-0/',
'FGOALS-g3': '/g/data/oi10/replicas/CMIP6/FAFMIP/CAS/FGOALS-g3/',
'CANESM5': '/g/data/oi10/replicas/CMIP6/FAFMIP/CCCma/CanESM5/',
'MIROC6': '/g/data/oi10/replicas/CMIP6/FAFMIP/MIROC/MIROC6/',
'MPI-ESM1-2-HR': '/g/data/oi10/replicas/CMIP6/FAFMIP/MPI-M/MPI-ESM1-2-HR/',
'MRI-ESM2-0': '/g/data/oi10/replicas/CMIP6/FAFMIP/MRI/MRI-ESM2-0/',
'CESM2': '/g/data/oi10/replicas/CMIP6/FAFMIP/NCAR/CESM2/',
'GFDL-ESM2M': '/g/data/oi10/replicas/CMIP6/FAFMIP/NOAA-GFDL/GFDL-ESM2M/'
}
experiment_path = {'faf-heat': 'faf-heat/r1i1p1f1/',
'faf-passiveheat': 'faf-passiveheat/r1i1p1f1/',
'faf-water': 'faf-water/r1i1p1f1/',
'faf-stress': 'faf-stress/r1i1p1f1/',
'faf-all': 'faf-all/r1i1p1f1/'
}
variable_path = {'areacello': 'Ofx/areacello/gn/v*/**/*.nc',
'volcello': 'Ofx/volcello/gn/v*/**/*.nc',
'thetao': 'Omon/thetao/gn/v*/**/*.nc',
'bigthetao': 'Omon/bigthetao/gn/v*/**/*.nc',
'so': 'Omon/so/gn/v*/**/*.nc',
'hfds': 'Omon/hfds/gn/v*/**/*.nc',
'wfo': 'Omon/wfo/gn/v*/**/*.nc',
'ocontemptend': 'Oyr/ocontemptend/gn/v*/**/*.nc',
'opottemptend': 'Oyr/opottemptend/gn/v*/**/*.nc',
'ocontempdiff': 'Oyr/ocontempdiff/gn/v*/**/*.nc',
'ocontemppmdiff': 'Oyr/ocontemppmdiff/gn/v*/**/*.nc',
'ocontemppadvect': 'Oyr/ocontemppadvect/gn/v*/**/*.nc',
'ocontemppsmadvect': 'Oyr/ocontemppsmadvect/gn/v*/**/*.nc',
'ocontemprmadvect': 'Oyr/ocontemprmadvect/gn/v*/**/*.nc',
'osaltdiff': 'Oyr/osaltdiff/gn/v*/**/*.nc',
'osaltpadvect': 'Oyr/osaltpadvect/gn/v*/**/*.nc',
'osaltpmdiff': 'Oyr/osaltpmdiff/gn/v*/**/*.nc',
'osaltpsmadvect': 'Oyr/osaltpsmadvect/gn/v*/**/*.nc',
'osaltrmadvect': 'Oyr/osaltrmadvect/gn/v*/**/*.nc',
'osalttend': 'Oyr/osalttend/gn/v*/**/*.nc',
'rsdoabsorb': 'Oyr/rsdoabsorb/gn/v*/**/*.nc'
}
# ### Parameters
model = 'ACCESS-CM2'
experiment = 'faf-heat'
nbins = 100
bin_edges = np.linspace(-5,40,nbins)
# + jupyter={"outputs_hidden": true}
#Loading cubes
cube_list = {}
for var, path in variable_path.items():
try:
cube_list[var] = get_concatenated_cube(model_path[model]+experiment_path[experiment]+path)
except AssertionError:
print(f'No files in {path}')
# -
hist={}
for var,cube in cube_list.items():
if cube.units == Unit('W m-2'):
w_cube = cube_multiply(cube,cube_list['areacello'])
temp_cube=cube_list['bigthetao'].extract(time_constraint(w_cube))
if 'depth' not in [coord.standard_name for coord in w_cube.coords()]:
temp_cube = temp_cube.extract(iris.Constraint(depth=0))
assert w_cube.shape == temp_cube.shape
hist[var] = [np.histogram(temp_cube.data.compressed(),bins=bin_edges, weights=w_cube.data.compressed()),len(w_cube.coord('time').points)]
fig, ax = plt.subplots(figsize=(10,5))
for var,[[dist,edges],tlen] in hist.items():
if var != 'rsdoabsorb':
ax.plot(edges[1:],np.cumsum((dist/tlen)[::-1])[::-1],label = var)
ax.legend()
ax.set_xlabel(r'Conservative Temperature $\Theta$ ($^\circ$ C)')
ax.set_ylabel('Flux (W)')
ax.set_title('Annually averaged global heat budget of water volume greater than $\Theta$')
fig.savefig('ACCESS-CM2_budget_temp',dpi=1000)
# ### Other exploratory stuff
fig,ax = plt.subplots(figsize=(10,5))
for exp in hfds_plot:
ax.plot(hfds_plot[exp][1][1:],np.cumsum(hfds_plot[exp][0][::-1]/840.)[::-1],'--',label=exp)
ax.legend()
ax.set_title(r'ACCESS-CM2 Surface fluxes binned by temperature into waters warmer than $\Theta$')
ax.set_ylabel('Surface downward heat flux (W)')
ax.set_xlabel(r'Potential tempeature $\Theta$ ($^\circ$C)')
fig.savefig('../results/ACCESS-CM2_surface_fluxes_temp',dpi=1000)
[var for var,cube in cube_list.items() if cube.units == Unit('W m-2')]
hfds_cube = cube_multiply(cube_list['hfds'].extract(time_constraint(cube_list['rsdoabsorb'])),cube_list['areacello'])
rsdo_cube=cube_multiply(cube_list['rsdoabsorb'].collapsed('depth',iris.analysis.SUM),cube_list['areacello'])
temp_cube = cube_list['bigthetao'].extract(time_constraint(cube_list['rsdoabsorb']) & iris.Constraint(depth=0))
dist,edges = np.histogram(temp_cube.data.compressed(),bins=bin_edges,weights=hfds_cube.data.compressed())
hist['modhfds'] = [np.histogram(temp_cube.data.compressed(),bins=bin_edges,weights=hfds_cube.data.compressed()-rsdo_cube.data.compressed()),70]
fig,axs = plt.subplots(3,1,figsize=(10,15))
axs[0].set_title('hfds less rsdoabsorb')
axs[0].plot(hist['modhfds'][0][1][1:],np.cumsum((hist['modhfds'][0][0]/70)[::-1])[::-1])
axs[1].set_title('rsdoabsorb')
axs[1].plot(hist['rsdoabsorb'][0][1][1:],np.cumsum((hist['rsdoabsorb'][0][0]/70)[::-1])[::-1])
axs[2].set_title('sum of the two')
axs[2].plot(hist['modhfds'][0][1][1:],np.cumsum(((hist['rsdoabsorb'][0][0]+hist['modhfds'][0][0])/70)[::-1])[::-1])
fig.savefig('../results/hfds_rsdoabsorb_modification')
| bin/flux_binning.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# %aiida
# +
import numpy as np
import scipy.constants as const
import ipywidgets as ipw
from IPython.display import display, clear_output, HTML
import re
import gzip
from collections import OrderedDict
import urllib.parse
import io
import zipfile
from apps.scanning_probe import common
from apps.scanning_probe import series_plotter
# + language="javascript"
# IPython.OutputArea.prototype._should_scroll = function(lines) {
# return false;
# }
# +
local_ref_index = None
cp2k_calc = None
orb_calc = None
def load_pk(b):
global cp2k_calc, orb_calc
global local_ref_index
try:
workcalc = load_node(pk=pk_select.value)
cp2k_calc = common.get_calc_by_label(workcalc, 'scf_diag')
orb_calc = common.get_calc_by_label(workcalc, 'orb')
except:
print("Incorrect pk.")
return
geom_info.value = common.get_slab_calc_info(workcalc.inputs.structure)
n_homo_inttext.value = max([int(workcalc.inputs.stm_params['--n_homo']) - 2, 1])
n_lumo_inttext.value = max([int(workcalc.inputs.stm_params['--n_lumo']) - 2, 1])
### ----------------------------------------------------
### Information about the calculation
with misc_info:
clear_output()
dft_inp_params = dict(workcalc.inputs['dft_params'])
dft_out_params = dict(cp2k_calc.outputs.output_parameters)
with misc_info:
if dft_inp_params['uks']:
print("UKS multiplicity %d" % dft_inp_params['multiplicity'])
else:
print("RKS")
if 'charge' in dft_inp_params:
print('Charge %d' % dft_inp_params['charge'])
else:
print('Charge 0')
if 'init_nel_spin1' in dft_out_params:
print("Number of alpha (s0) electrons: %d" % dft_out_params['init_nel_spin1'])
print("Number of beta (s1) electrons: %d" % dft_out_params['init_nel_spin2'])
print("Energy [au]: %.6f" % (dft_out_params['energy']))
print("Energy [eV]: %.6f" % (dft_out_params['energy'] * 27.211386245988))
if '--p_tip_ratios' in dict(workcalc.inputs.stm_params):
p_tip_ratio = workcalc.inputs.stm_params['--p_tip_ratios']
print("Tip p-wave contrib: %.2f" % p_tip_ratio)
### Ionization potential, if it's there
with orb_calc.outputs.retrieved.open('_scheduler-stdout.txt') as std_out_file:
std_out = std_out_file.read()
matches = re.findall("IONIZATION POTENIAL \(eV\): ([\d\.\d]+)", std_out)
if len(matches) > 0:
with misc_info:
print("Ionization potential: %.4f eV" % float(matches[0]))
### ----------------------------------------------------
### Load data
with orb_calc.outputs.retrieved.open('orb.npz') as npz_handle:
loaded_data = np.load(npz_handle.name, allow_pickle=True)
s0_orb_general_info = loaded_data['s0_orb_general_info'][()]
s0_orb_series_info = loaded_data['s0_orb_series_info']
s0_orb_series_data = loaded_data['s0_orb_series_data']
series_plotter_inst.add_series_collection(s0_orb_general_info, s0_orb_series_info, s0_orb_series_data)
ref_index = s0_orb_general_info['homo']
if 's1_orb_general_info' in loaded_data.files:
s1_orb_general_info = loaded_data['s1_orb_general_info'][()]
s1_orb_series_info = loaded_data['s1_orb_series_info']
s1_orb_series_data = loaded_data['s1_orb_series_data']
series_plotter_inst.add_series_collection(s1_orb_general_info, s1_orb_series_info, s1_orb_series_data)
ref_index = int(0.5 * (ref_index + s1_orb_general_info['homo']))
series_plotter_inst.setup_added_collections(workcalc.pk)
wfn_kit_button.disabled = False
local_ref_index = np.where(s0_orb_general_info['orb_indexes'] == ref_index)
local_ref_index = local_ref_index[0][0]
style = {'description_width': '50px'}
layout = {'width': '70%'}
pk_select = ipw.IntText(value=0, description='pk', style=style, layout=layout)
load_pk_btn = ipw.Button(description='Load pk', style=style, layout=layout)
load_pk_btn.on_click(load_pk)
geom_info = ipw.HTML()
display(ipw.HBox([ipw.VBox([pk_select, load_pk_btn]), geom_info]))
misc_info = ipw.Output()
display(misc_info)
# -
# # Orbital images
def selected_orbital_indexes():
n_homo = n_homo_inttext.value
n_lumo = n_lumo_inttext.value
i_start = local_ref_index - n_homo + 1
i_start = 0 if i_start < 0 else i_start
i_end = local_ref_index + n_lumo + 1
i_end = 0 if i_end < 0 else i_end
#i_start_ = np.where(np.logical_and(orb_indexes[0] <= 1, orb_indexes[0] > -n_homo))[0]
#i_start = i_start_[0] if len(i_start_) != 0 else 1
#
#i_end_ = np.where(np.logical_and(orb_indexes[0] > 0, orb_indexes[0] < n_lumo+2))[0]
#i_end = i_end_[-1] if len(i_end_) != 0 else len(orb_indexes[0])
return np.arange(i_start, i_end)
# +
style = {'description_width': '80px'}
layout = {'width': '40%'}
series_plotter_inst = series_plotter.SeriesPlotter(
select_indexes_function = selected_orbital_indexes,
zip_prepend = 'orbs'
)
### -----------------------------------------------
### Plot selector
n_homo_inttext = ipw.IntText(
description='num HOMO',
min=0,
max=100,
value=10,
style=style, layout=layout)
n_lumo_inttext = ipw.IntText(
description='num LUMO',
min=0,
max=100,
value=10,
style=style, layout=layout)
n_orb_select = ipw.HBox([n_homo_inttext, n_lumo_inttext],
style=style, layout={'width': '60%'})
### -----------------------------------------------
display(series_plotter_inst.selector_widget, n_orb_select,
series_plotter_inst.plot_btn, series_plotter_inst.clear_btn, series_plotter_inst.plot_output)
# -
# # Export
# **Image zip** exports the currently selected orbital images in png, txt and IGOR pro formats.
#
# **Cube creation kit** creates an archive containing all necessary ingredients to generate the Kohn-Sham orbital cube files with the `cube_from_wfn.py` script available from https://github.com/nanotech-empa/cp2k-spm-tools.
display(ipw.HBox([series_plotter_inst.zip_btn, series_plotter_inst.zip_progress]), series_plotter_inst.link_out)
# +
def create_wfn_zip(b):
wfn_kit_button.disabled=True
# ! mkdir -p tmp
label = "cube-kit-pk%d" % int(pk_select.value)
cube_kit_name = label + ".zip"
zipf = zipfile.ZipFile('tmp/%s'%cube_kit_name, 'w', zipfile.ZIP_DEFLATED)
fd = cp2k_calc.outputs['retrieved']
for fn in ['BASIS_MOLOPT', 'aiida.inp', 'aiida.out', 'geom.xyz', 'aiida-RESTART.wfn']:
zipf.write(fd.open(fn).name, arcname=label + '/' + fn)
run_script_path = "/home/aiida/apps/scanning_probe/orb/misc/run_cube_from_wfn.sh"
zipf.write(run_script_path, arcname=label + '/' +"run_cube_from_wfn.sh")
zipf.close()
with wfn_kit_output:
display(HTML('<a href="tmp/%s" target="_blank">download zip</a>' %cube_kit_name))
wfn_kit_button = ipw.Button(description='Cube creation kit', disabled=True)
wfn_kit_button.on_click(create_wfn_zip)
wfn_kit_output = ipw.Output()
display(wfn_kit_button, wfn_kit_output)
# +
def clear_tmp(b):
# ! rm -rf tmp && mkdir tmp
with series_plotter_inst.link_out:
clear_output()
series_plotter_inst.zip_progress.value = 0.0
with wfn_kit_output:
clear_output()
if series_plotter_inst.series is not None:
series_plotter_inst.zip_btn.disabled = False
wfn_kit_button.disabled = False
clear_tmp_btn = ipw.Button(description='clear tmp')
clear_tmp_btn.on_click(clear_tmp)
display(clear_tmp_btn)
# -
### Load the URL after everything is set up ###
try:
url = urllib.parse.urlsplit(jupyter_notebook_url)
pk_select.value = urllib.parse.parse_qs(url.query)['pk'][0]
load_pk(0)
except:
pass
| orb/view_orb.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
from charistools import hypsometry, modis_info
from netCDF4 import Dataset
import numpy as np
from osgeo import gdal
import matplotlib.pyplot as plt
modis_info.area_per_500m_pixel_km2
help(hypsometry)
hyps = hypsometry.Hypsometry(comments=['some comments'])
hyps.comments
hyps.data
# %cd /Users/brodzik/projects/CHARIS/snow_cover/modice.v0.4/min05yr_nc
# %ls
file = Dataset('MODICE.v0.4.h23v05.1strike.min05yr.mask.nc', mode='r', format='NETCDF4')
file
modice = file.variables["modice_min_year_mask"][:]
np.shape(modice)
# %cd /Users/brodzik/projects/CHARIS/elevation_data/SRTMGL3
# %ls
elevation_file = 'SRTMGL3.v0.1.h23v05.tif'
dataset = gdal.Open(elevation_file, gdal.GA_ReadOnly)
dataset.RasterCount
band = dataset.GetRasterBand(1)
elevation = band.ReadAsArray()
np.shape(elevation)
np.amin(elevation)
np.amax(elevation)
hyps
hyps.comments
hyps.append(elevation,modice,min_contour_m=1400,verbose=True)
hyps.data
hypsh23v05 = hyps
elevation_file_h24 = 'SRTMGL3.v0.1.h24v05.tif'
dataset24 = gdal.Open(elevation_file_h24, gdal.GA_ReadOnly)
band24 = dataset24.GetRasterBand(1)
elevation24 = band24.ReadAsArray()
np.amin(elevation24)
np.amax(elevation24)
# %cd /Users/brodzik/projects/CHARIS/snow_cover/modice.v0.4/min05yr_nc
# %ls
file24 = Dataset('MODICE.v0.4.h24v05.1strike.min05yr.mask.nc', mode='r', format='NETCDF4')
modice24 = file24.variables["modice_min_year_mask"][:]
print(np.amin(modice24),np.amax(modice24))
hypsh24v05 = hypsometry.Hypsometry(comments=['h24v05'])
hypsh24v05.append(elevation24,modice24,min_contour_m=1400,verbose=True)
hypsh23v05.data
hypsh24v05.data
all = hypsh23v05.data + hypsh24v05.data
all
orig = hypsometry.Hypsometry(filename='/Users/brodzik/projects/CHARIS/snow_cover/modice.v0.4/IN_Hunza_at_Danyour.0100m.modicev04_1strike_area_by_elev.txt')
orig.data
| charis/Playing with charistools.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import pandas as pd
import numpy as np
import pyreadstat
import matplotlib.pyplot as plt
from scipy import stats
import seaborn as sns
# %matplotlib inline
# -
w68spssord, metapassord = pyreadstat.read_sav('~/Desktop/cap1/pew_w68_media_literacy_insights/data/W68.sav')
w68spssord
df68ord = pd.DataFrame(w68spssord)
w68spss, metaspss = pyreadstat.read_sav('~/Desktop/cap1/pew_w68_media_literacy_insights/data/W68.sav'
, apply_value_formats = True
, formats_as_category = True )
fields = ['QKEY', 'COVIDFOL_W68', 'COVIDCOVER1_W68', 'COVIDFACTS_b_W68', 'COVIDNEWSCHNG_a_W68', 'COVIDNEWSCHNG_c_W68', 'COVIDNEWSCHNG_e_W68', 'COVIDNEWSCHNG_d_W68', 'COVIDINFODIFF_W68', 'COVIDLOCINFO_W68', 'COVIDPLANHRD_W68', 'COVIDPLANTRUE_W68', 'COVIDPLANWATCH_W68', 'F_METRO', 'F_EDUCCAT', 'F_PARTY_FINAL']
df68 = pd.DataFrame(w68spss)
df68 = pd.read_spss('~/Desktop/cap1/pew_w68_media_literacy_insights/data/W68.sav', usecols=fields)
df68.head()
df68.columns = [x.lower() for x in df68.columns]
df68.tail()
planbd = df68.covidplantrue_w68.value_counts(sort=True)
planbd
# +
tot = 0
for ct in planbd:
tot += ct
print(tot)
# -
plansee = df68.covidplanwatch_w68.value_counts(sort=True)
plansee
planhear = df68.covidplanhrd_w68.value_counts(sort=True)
planhear
# +
tot = 0
for ct in planhear:
tot += ct
print(tot)
# -
# matplotlib.pyplot.figure(num=None, figsize=None, dpi=None, facecolor=None, edgecolor=None, frameon=True, FigureClass=<class 'matplotlib.figure.Figure'>, clear=False, **kwargs)
# +
import matplotlib.pyplot as plt
plantrue = df68['covidplanhrd_w68'].value_counts()
sns.set(style="darkgrid")
sns.barplot(x = plantrue.index, y = plantrue.values, alpha=0.9)
plt.title('How much have you heard of the "Plandemic" conspiracy theory?')
plt.ylabel('Number of Respondents', fontsize=16)
plt.xticks(rotation = 65)
plt.xlabel('Response', fontsize=16)
plt.show()
# -
# Of all 9654 surveyed, 1432 had heard "a lot" about the theory portrayed in an online video entitled "Plandemic". That theory states that powerful people intentionally planned the coronavirus outbreak. I am curious about why certain people had hear a lot about this video and, as a follow-up to that question, of the people who had heard of this video, why might some of them have been more prone to believe the theory it proposed.
# +
import matplotlib.pyplot as plt
planhrd = df68['covidplantrue_w68'].value_counts()
sns.set(style="darkgrid")
sns.barplot(x = planhrd.index, y = planhrd.values, alpha=0.9)
plt.title('Do You Think that "Plandemic" is true?')
plt.ylabel('Number of Respondents', fontsize=16)
plt.xticks(rotation = 65)
plt.xlabel('Response', fontsize=16)
plt.show()
# -
# This question was not posed to all those surveyed. In total, 6818 people answered this question. (Note to self, shouild I re-order the columns to Def NOT, Prob NOT, Prob true, Def true, not sure, refused?) Again, I notice that relatively few people believed this theory to be true, but a surprising amount of people thought that it could "probably" be true. What factors are involved in this belief? I am looking at not only demographics, but also other indicators as outlined in the survey questions. I am especially interested in the amound of news media consumed. Interestingly, this dataset includes several questions from previous waves that might be helpful in addressing these questions. They ask about news formats most often consumed, specific news outlets, and party alignment of the news that those surveyed most trust.
# Ref for catplot video https://www.youtube.com/watch?v=vUmpqpb-FvA
# This chart does not do what I had hoped it would, but it does give insights into the patterns of refusal to answer questions. There were no respondents who refused both of these questions. It appears that the older the respondent was, the more likely they were to refuse to answer this question. (Note to self, how can I change this ugly title location?)
party_follow = sns.catplot('covidfol_w68', 'f_party_final', data=df68)
party_follow.fig.suptitle("How closely have you followed pandemic news?")
party_follow.set_xticklabels(rotation=30)
party_plan_true = sns.catplot('covidplantrue_w68', 'f_party_final', data=df68)
party_plan_true.fig.suptitle("Do you think it's true that powerful people planned the pandemic?")
party_plan_true.set_xticklabels(rotation=30)
plan_party = sns.catplot(y= 'covidplantrue_w68',hue='f_party_final', kind="count",
palette="deep", edgecolor=".6",
data=df68)
coverage_quality = sns.catplot(x='covidcover1_w68', kind="count", palette="deep", data = df68)
coverage_quality.set_xticklabels(rotation=30)
follow_party = sns.catplot(y= 'covidfol_w68',hue='f_party_final', kind="count",
palette="deep", edgecolor=".6",
data=df68)
difficulty_party = sns.catplot(y= 'f_party_final',hue='covidinfodiff_w68', kind="count",
palette="deep", edgecolor=".6",
data=df68)
edu_follow = pd.crosstab([df68.f_educcat], [df68.covidfol_w68])
(chi2, p, dof, _) = stats.chi2_contingency([edu_follow.iloc[0].values,edu_follow.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
edu_cover = pd.crosstab([df68.f_educcat], [df68.covidcover1_w68])
(chi2, p, dof, _) = stats.chi2_contingency([edu_cover.iloc[0].values,edu_cover.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
edu_facts = pd.crosstab([df68.f_educcat], [df68.covidfacts_b_w68])
(chi2, p, dof, _) = stats.chi2_contingency([edu_facts.iloc[0].values,edu_facts.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
edu_newsa = pd.crosstab([df68.f_educcat], [df68.covidnewschng_a_w68])
(chi2, p, dof, _) = stats.chi2_contingency([edu_newsa.iloc[0].values,edu_newsa.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
edu_newsc = pd.crosstab([df68.f_educcat], [df68.covidnewschng_c_w68])
(chi2, p, dof, _) = stats.chi2_contingency([edu_newsc.iloc[0].values,edu_newsc.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
edu_newsd = pd.crosstab([df68.f_educcat], [df68.covidnewschng_d_w68])
(chi2, p, dof, _) = stats.chi2_contingency([edu_newsd.iloc[0].values,edu_newsd.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
edu_newse = pd.crosstab([df68.f_educcat], [df68.covidnewschng_e_w68])
(chi2, p, dof, _) = stats.chi2_contingency([edu_newse.iloc[0].values,edu_newse.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
edu_diff = pd.crosstab([df68.f_educcat], [df68.covidinfodiff_w68])
(chi2, p, dof, _) = stats.chi2_contingency([edu_diff.iloc[0].values,edu_diff.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
edu_loc = pd.crosstab([df68.f_educcat], [df68.covidlocinfo_w68],)
(chi2, p, dof, _) = stats.chi2_contingency([edu_loc.iloc[0].values,edu_loc.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
edu_hrd = pd.crosstab([df68.f_educcat], [df68.covidplanhrd_w68])
(chi2, p, dof, _) = stats.chi2_contingency([edu_hrd.iloc[0].values,edu_hrd.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
edu_true = pd.crosstab([df68.f_educcat], [df68.covidplantrue_w68])
(chi2, p, dof, _) = stats.chi2_contingency([edu_true.iloc[0].values,edu_true.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
edu_watch = pd.crosstab([df68.f_educcat], [df68.covidplanwatch_w68])
(chi2, p, dof, _) = stats.chi2_contingency([edu_watch.iloc[0].values,edu_watch.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
party_fol = pd.crosstab([df68.f_party_final], [df68.covidfol_w68])
(chi2, p, dof, _) = stats.chi2_contingency([party_fol.iloc[0].values,party_fol.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
party_cover = pd.crosstab([df68.f_party_final], [df68.covidcover1_w68])
(chi2, p, dof, _) = stats.chi2_contingency([party_cover.iloc[0].values,party_cover.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
party_facts = pd.crosstab([df68.f_party_final], [df68.covidfacts_b_w68])
(chi2, p, dof, _) = stats.chi2_contingency([party_facts.iloc[0].values,party_facts.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
party_newsa = pd.crosstab([df68.f_party_final], [df68.covidnewschng_a_w68])
(chi2, p, dof, _) = stats.chi2_contingency([party_newsa.iloc[0].values,party_newsa.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
party_newsc = pd.crosstab([df68.f_party_final], [df68.covidnewschng_c_w68])
(chi2, p, dof, _) = stats.chi2_contingency([party_newsc.iloc[0].values,party_newsc.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
party_newsd = pd.crosstab([df68.f_party_final], [df68.covidnewschng_d_w68])
(chi2, p, dof, _) = stats.chi2_contingency([party_newsd.iloc[0].values,party_newsd.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
party_newse = pd.crosstab([df68.f_party_final], [df68.covidnewschng_e_w68])
(chi2, p, dof, _) = stats.chi2_contingency([party_newse.iloc[0].values,party_newse.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
party_diff = pd.crosstab([df68.f_party_final], [df68.covidinfodiff_w68])
(chi2, p, dof, _) = stats.chi2_contingency([party_diff.iloc[0].values,party_diff.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
party_loc = pd.crosstab([df68.f_party_final], [df68.covidlocinfo_w68])
(chi2, p, dof, _) = stats.chi2_contingency([party_loc.iloc[0].values,party_loc.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
party_hrd = pd.crosstab([df68.f_party_final], [df68.covidplanhrd_w68])
(chi2, p, dof, _) = stats.chi2_contingency([party_hrd.iloc[0].values,party_hrd.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
party_true = pd.crosstab([df68.f_party_final], [df68.covidplantrue_w68])
(chi2, p, dof, _) = stats.chi2_contingency([party_true.iloc[0].values,party_true.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
party_watch = pd.crosstab([df68.f_party_final], [df68.covidplanwatch_w68])
(chi2, p, dof, _) = stats.chi2_contingency([party_watch.iloc[0].values,party_watch.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
metro_fol = pd.crosstab([df68.f_metro], [df68.covidfol_w68])
(chi2, p, dof, _) = stats.chi2_contingency([metro_fol.iloc[0].values,metro_fol.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
metro_cov = pd.crosstab([df68.f_metro], [df68.covidcover1_w68])
(chi2, p, dof, _) = stats.chi2_contingency([metro_cov.iloc[0].values,metro_cov.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
metro_facts = pd.crosstab([df68.f_metro], [df68.covidfacts_b_w68])
(chi2, p, dof, _) = stats.chi2_contingency([metro_facts.iloc[0].values,metro_facts.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
metro_newsa = pd.crosstab([df68.f_metro], [df68.covidnewschng_a_w68])
(chi2, p, dof, _) = stats.chi2_contingency([metro_newsa.iloc[0].values,metro_newsa.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
metro_newsc = pd.crosstab([df68.f_metro], [df68.covidnewschng_c_w68])
(chi2, p, dof, _) = stats.chi2_contingency([metro_newsc.iloc[0].values,metro_newsc.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
metro_newsd = pd.crosstab([df68.f_metro], [df68.covidnewschng_d_w68])
(chi2, p, dof, _) = stats.chi2_contingency([metro_newsd.iloc[0].values,metro_newsd.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
metro_newse = pd.crosstab([df68.f_metro], [df68.covidnewschng_e_w68])
(chi2, p, dof, _) = stats.chi2_contingency([metro_newse.iloc[0].values,metro_newse.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
metro_diff = pd.crosstab([df68.f_metro], [df68.covidinfodiff_w68])
(chi2, p, dof, _) = stats.chi2_contingency([metro_diff.iloc[0].values,metro_diff.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
metro_loc = pd.crosstab([df68.f_metro], [df68.covidlocinfo_w68])
(chi2, p, dof, _) = stats.chi2_contingency([metro_loc.iloc[0].values,metro_loc.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
metro_hrd = pd.crosstab([df68.f_metro], [df68.covidplanhrd_w68])
(chi2, p, dof, _) = stats.chi2_contingency([metro_hrd.iloc[0].values,metro_hrd.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
metro_true = pd.crosstab([df68.f_metro], [df68.covidplantrue_w68])
(chi2, p, dof, _) = stats.chi2_contingency([metro_true.iloc[0].values,metro_true.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
metro_watch = pd.crosstab([df68.f_metro], [df68.covidplanwatch_w68])
(chi2, p, dof, _) = stats.chi2_contingency([metro_watch.iloc[0].values,metro_watch.iloc[1].values])
print('chi2: ', chi2)
print('p-value: ', p)
print('Degrees of Freedom: ', dof)
# reference video on cross-tabulation or contingency tables https://www.youtube.com/watch?v=I_kUj-MfYys <br>
# also: reference video on chi squared https://www.youtube.com/watch?v=H9AULpvRxgM
edu_plan_true = pd.crosstab([df68.f_educcat], [df68.covidplantrue_w68], normalize='index')
edu_plan_true
edu_plan_heard = pd.crosstab([df68.f_educcat], [df68.covidplanhrd_w68], normalize='index')
edu_plan_heard
diff_party = pd.crosstab([df68.f_party_final], [df68.covidinfodiff_w68], normalize = 'index')
diff_party
diff_metroy = pd.crosstab([df68.f_metro], [df68.covidinfodiff_w68], normalize = 'index')
diff_metroy
# And interesting takeaway is that when using cross-tabulation, the x and y axis you choose makes a huge difference. I suspect that it's because the responses are more likely to be evenly distributed than the demographic profiles. The table above would look very skewed if the x and y were swapped because the population of this survey is mastly more metropolitan.
diff_metron = pd.crosstab([df68.covidinfodiff_w68], [df68.f_metro], normalize = 'index')
diff_metron
plan_party = pd.crosstab([df68.covidplantrue_w68], [df68.f_party_final])
plan_party
plan_party_norm = pd.crosstab([df68.covidplantrue_w68], [df68.f_party_final], normalize = 'index')
plan_party_norm
| notebooks/eda.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Regression
# ### a. Importing Libraries
# +
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from sklearn.linear_model import LinearRegression, LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix, plot_confusion_matrix, f1_score
from sklearn.metrics import mean_squared_error, r2_score
# -
# ### b. Importing Dataset
dataset = pd.read_csv("winequality-white.csv",sep=";")
dataset.head(5)
train_set, test_set = train_test_split(dataset, test_size=0.2)
print(train_set.shape, test_set.shape)
# ### c. Creating Regression Dataset
acidity_X_train = train_set[["quality"]].values
acidity_y_train = train_set[["residual sugar"]].values
acidity_X_test = test_set[["quality"]].values
acidity_y_test = test_set[["residual sugar"]].values
acidity_X_train.ravel().shape
acidity_y_train.shape
# ## 1. Linear Regression
#
# <u>1. Declaring the model</u>
model = LinearRegression().fit(acidity_X_train, acidity_y_train.ravel())
# <u>2. Fitting the model</u>
model.fit(acidity_X_train, acidity_y_train)
# <u>3. Testing the model</u>
pred = model.predict(acidity_X_test)
print('Mean squared error: %.2f' % mean_squared_error(acidity_y_test, pred))
print('Coefficient of determination: %.2f'% r2_score(acidity_y_test, pred))
# +
plt.scatter(acidity_X_test, acidity_y_test, color='black')
plt.plot(acidity_X_test, pred, color='blue', linewidth=3)
plt.show()
# -
# ## 2. Logistic Regression
#
# <u>1. Declaring the model</u>
model = LogisticRegression(max_iter=300,solver="liblinear")
# <u>2. Fitting the model</u>
model.fit(acidity_X_train, acidity_y_train)
# <u>3. Testing the model</u>
# +
pred = model.predict(acidity_X_test)
disp = plot_confusion_matrix(model, acidity_X_test, acidity_y_test,
cmap=plt.cm.Blues,
normalize=None)
disp.ax_.set_title('Confusion matrix')
print("F1 score : ", f1_score(pred,acidity_y_test,average='micro'))
| linear_regression/linear_regression.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Daten, wir brauchen Daten!
#
# Das Presseportal veröffentlicht unter dieser [Seite](https://www.presseportal.de/blaulicht/r/Baden-Baden) Polizeimeldungen für Baden-Baden (unter ähnlichen Links finden sich Meldungen für andere Städte).
#
# Die Seite bietet auch einen [RSS Feed](https://www.presseportal.de/rss/polizei/r/Baden-Baden.rss2), also einen maschinenlesbaren Nachrichtenticker, welcher von anderen Programmen abonniert werden kann. Dieser Feed wäre eine vorzügliche Quelle für unser kleines Projekt, schauen wir uns diesen Feed einmal an.
# +
import requests
feed_url = "https://www.presseportal.de/rss/polizei/r/Baden-Baden.rss2"
feed = requests.get(feed_url)
print(f"Status Code der Anfrage: {feed.status_code}")
print()
print(f"Erste Zeichen der Antwort: {feed.text[:256]}")
# -
# Der Feed antwortet und das Ergebnis sieht, wie erwartet, nach XML aus, dieses XML können wir parsen.
# +
from bs4 import BeautifulSoup
feed_soup = BeautifulSoup(feed.text, "html.parser")
feed_items = feed_soup.find_all("item")
print(f"{len(feed_items)} Items im Feed:")
for item in feed_items:
print(f"\t{item.title.get_text()}")
# -
# Auf der einen Seite funktioniert das Parsing und wir können den Feed auslesen, auf der anderen liefert der Feed aber nur die 15 aktuellsten Meldungen (welche auch anscheinend nicht alle direkt Baden-Baden betreffen), das ist für unsere Zwecke etwas dürftig. Daher verwerfen wir die Idee des RSS Feeds.
#
# Wir müssen uns unsere Daten wohl auf anderem Weg beschaffen. Schauen wir uns die Seite [https://www.presseportal.de/blaulicht/r/Baden-Baden](https://www.presseportal.de/blaulicht/r/Baden-Baden) noch einmal an. Auf dieser Seite werden 27 Meldungen gelistet, außerdem gibt es eine Pagination für ältere Meldungen, die zweite Seite hat die Adresse [https://www.presseportal.de/blaulicht/r/Baden-Baden/27](https://www.presseportal.de/blaulicht/r/Baden-Baden/27), die dritte Seite [https://www.presseportal.de/blaulicht/r/Baden-Baden/54](https://www.presseportal.de/blaulicht/r/Baden-Baden/54).
#
# Es scheint, ab der zweiten Seite, ein Muster zu geben. Die Zahl in der Adresse wird pro Seite immer um 27 erhöht (und jede Seite liefert 27 Meldungen). Wenn wir manuell diese Zahlen ändern und diese Adressen aufrufen, erkennen wir dass diese Zahl der ersten Meldung auf der Seite entspricht, wenn alle Meldungen **null-basiert** hochgezählt werden. Seite 1 zeigt also die die Meldungen 0-26, Seite 2 zeigt 27-53 usw. Folglich müsste die erste Seite auch unter [https://www.presseportal.de/blaulicht/r/Baden-Baden/0](https://www.presseportal.de/blaulicht/r/Baden-Baden/0) erreichbar sein.
#
# Wir probieren es aus und Bingo! Folglich können wir uns die Adressen für alle Seiten recht einfach generieren:
for i in range(5):
print(f"https://www.presseportal.de/blaulicht/r/Baden-Baden/{i * 27}")
# Jetzt wissen wir, wo wir unsere Information herbekommen, Zeit sich um das Wie zu kümmern. Daher schauen wir eine solche Seite mal genauer an.
url = "https://www.presseportal.de/blaulicht/r/Baden-Baden/0"
r = requests.get(url)
r.status_code
# Wenn wir im Browser die Entwicklertools öffnen, können wir feststellen, dass jede Einzelmeldung innerhalb eines `<article>` Tag mit der Klasse `news` erscheint. Die eigentliche Schlagzeile befindet sich in einem `<h3>` Tag mit der Klasse `news-headline-clamp`. Somit können wir die 27 Schlagzeilen aus der Seite extrahieren:
# +
soup = BeautifulSoup(r.text, "html.parser")
for i, article in enumerate(soup.find_all("article", class_="news")):
print(f"{i} -> {article.find('h3', class_='news-headline-clamp').get_text()}")
# -
# Jeder `<article>` Tag enthält in einem `data-url` Attribut die URL-Slug zum eigentlichen Artikel. Damit haben wir das Rüstzeug, um uns eine größere Anzahl von Artikeln zu generieren:
# +
articles = []
for i in range(5):
soup = BeautifulSoup(
requests.get(f"https://www.presseportal.de/blaulicht/r/Baden-Baden/{i * 27}").text,
"html.parser"
)
for article in soup.find_all("article", class_="news"):
articles.append({
"url": article["data-url"],
"headline": article.find("h3", class_="news-headline-clamp").get_text()
})
len(articles)
# -
# In der obigen Aufzählung sehen wir auch Artikel, welche nicht direkt Baden-Baden betreffen. In diesen Artikleln ist meist die Staatsanwaltschaft Baden-Baden involviert oder es wurden Verletzte in das Klinikum Baden-Baden gebracht. Diese Meldungen interessieren uns nicht.
#
# Der Anfang einer jeden Schlagzeile ist die Dienststelle, gefolgt von einem Doppeltpunkt. Uns interessieren nur die Meldungen des Polizeipräsidiums Offenburg (POL-OG), diese scheinen auch die Mehrheit auszumachen.
# +
from collections import Counter
Counter(article.get("headline").split(":")[0] for article in articles)
# -
# Von 135 Meldungen bleiben noch 120, da es darunter auch indirekte Nachrichten zu geben scheint, wollen wir zusätzlich auf das Vorhandensein von "Baden-Baden" innerhalb der Schlagzeile f
# filtern.
# +
pol_og_articles = [article for article in articles if article.get("headline").startswith("POL-OG:")]
pol_og_bad_articles = [article for article in pol_og_articles if "Baden-Baden" in article.get("headline")]
len(pol_og_bad_articles)
# -
# Jetzt basteln wir aus allem eine Funktion.
# +
def get_list_of_articles(number_of_pages):
articles = []
for i in range(number_of_pages):
soup = BeautifulSoup(
requests.get(f"https://www.presseportal.de/blaulicht/r/Baden-Baden/{i * 27}").text,
"html.parser"
)
for article in soup.find_all("article", class_="news"):
headline = article.find("h3", class_="news-headline-clamp").get_text()
if (headline.startswith("POL-OG:") and "Baden-Baden" in headline):
articles.append({
"url": article["data-url"],
"headline": headline
})
return articles
len(get_list_of_articles(5))
# -
# Nun lagern wir diese Funktion in eine Python Datei aus, damit wir sie auch in anderen Notebooks einfach verweden können.
# +
from scraping import get_list_of_articles as gloa
len(gloa(5))
# -
# Et voilà !
| code/01_daten_wir_brauchen_daten.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# Importing Required Python Packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings('ignore')
pd.set_option('display.max_columns',None)
# Loading Reduced feature Training set
X_train_red = pd.read_csv('X_train_final.csv')
y_train = pd.read_csv('y_train.final.csv')
# Loading Reduced feature Test set
X_test_red = pd.read_csv('X_test_final.csv')
y_test = pd.read_csv('y_test.final.csv')
# ## Model_16: Stacking Classifier
# #### Loading the best Voting Classifier model & Neural Network (with Equal Nodes in all the Layers)
# Importing Joblib module
import joblib
import tensorflow
from tensorflow import keras
# Importing best Voting Classifier
voting_clf = joblib.load('Voting_Red.joblib')
# ### Preparing the Training Set for Stacking Classifier
# Instantiating an empty DataFrame to store training set for Stacking Classifier
df_train_stack = pd.DataFrame(columns=['Voting','Neural','True'])
# Initializing the 10 Fold object
from sklearn.model_selection import StratifiedKFold
cv_strat = StratifiedKFold(10,random_state=42)
# #### Getting predictions of the Voting & Neural Net classifier for each of 10 folds of Training set.
# Importing the Sklearn's roc_auc_score module
from sklearn.metrics import roc_auc_score
# Importing cross val score from sklearn
from sklearn.model_selection import cross_val_score
# Importing train test split from Sklearn to produce validation set
from sklearn.model_selection import train_test_split
# Defining the exponential decay learning rate.
def exponential_decay_fn(epoch):
return 0.01 * 0.1**(epoch /4 )
def Neural_Pred(clf, X_tr, y_tr, X_tst):
'''
This function returns the predictions of Neural Classifier(clf) for the test folds
Parameters:
X_tr : Training set Features
y_tr : Training set Labels
X_tst : Test set Features
'''
# Splitting the Training set further into training & validation set.
X_tr_r, X_val, y_tr_r, y_val = train_test_split(X_tr, y_tr, test_size=0.1, random_state=42, stratify=y_tr)
# Compiling & Training the Neural Net
clf.compile(loss="binary_crossentropy", optimizer=keras.optimizers.Nadam(beta_1=0.9, beta_2=0.999), metrics=["accuracy"])
# defining Checkpoints
checkpoint_cb = keras.callbacks.ModelCheckpoint('best_model.h5',save_best_only=True) # 1st Callback
early_stopping_cb = keras.callbacks.EarlyStopping(patience=10)# 2nd Callback
lr_scheduler_cb = keras.callbacks.LearningRateScheduler(exponential_decay_fn)# 3rd Callback
# Fitting The model
clf.fit(X_tr_r, y_tr_r, epochs=50, validation_data=(X_val,y_val),batch_size=32,
class_weight={0: 1.0, 1: 10.0},callbacks=[checkpoint_cb,early_stopping_cb,lr_scheduler_cb])
# Loading the best Neural model after training & before making predictions
clf_best = keras.models.load_model('best_model.h5')
# Saving the predictions for every fold in a numpy array
return (clf_best.predict_proba(X_tst).flatten())
# Computing Voting Classifier's & Neural Net 10 fold conditional prob predictions on the training set for Stacking classifier
for train_index, test_index in cv_strat.split(X_train_red, y_train):
# Creating Folds
X_tr, X_tst = X_train_red.loc[train_index], X_train_red.loc[test_index]
y_tr, y_tst = y_train.loc[train_index], y_train.loc[test_index]
# Fitting the best Voting Classifier on the Training Folds
voting_clf.fit(X_tr, y_tr)
# Making Predictions on the testing Fold.
y_pred_voting = voting_clf.predict_proba(X_tst)[:,1]
# Loading the fresh best Neural net classifier
neural = keras.models.load_model('Best_model_Selu_eq_Learn.h5')
# Fitting the best Neural Net on Training Folds & obtaining predictions
y_pred_neural = Neural_Pred(neural, X_tr, y_tr, X_tst)
# Storing the predictions in a dataframe
df_temp = pd.DataFrame(columns=['Voting','Neural','True'])
df_temp['Voting'] = y_pred_voting
df_temp['Neural'] = y_pred_neural
df_temp['True'] = y_tst.values.flatten() # As y_tst is a dataframe we first convert it into a 2d numpy vector
# array, which is is then converted to 1d array using .flatten()
# Appending the df_temp to df_train_stack
df_train_stack = df_train_stack.append(df_temp,ignore_index=True)
# Getting the info of the Training dataframe of the Stacking Classifier
df_train_stack.info()
# Converting the True Column to the int type
df_train_stack['True'] = df_train_stack['True'].astype('int8')
# Re-checking the info of the df_train_stack
df_train_stack.info()
# Printing the top 5 rows of the df_train_stack
df_train_stack.head()
# ### Selecting Random forest as the Stacking classifier as it has one of highest test set roc_auc as well as R_R ratio for this dataset.
# ### Tuning the hyperparameters of the Random Forest on the Stacked Training set using Optuna
# Defining the class weights
cl_weight = [None,'balanced',{0:1.0,1:9.0},{0:1.0,1:10},{0:1.0,1:11},{0:1.0,1:12},{0:1.0,1:13},{0:1.0,1:14},{0:1.0,1:15}]
# Defining the appropriate objective function for the Random Forest classifier
def objective_wrappper_rf(X_tr, y_tr, cls=None, cv_strat=None):
'''
Optimizes Random Forest parameters on the given training set X_tr,y_tr
using cv_strat cross-validation object
'''
def objective(trial):
params = {
'max_depth': trial.suggest_categorical('max_depth',list(range(2,50))+ [None]),
'n_estimators':trial.suggest_int('n_estimators',100,2000,10),
'class_weight':trial.suggest_categorical('class_weight',cl_weight),
'min_samples_leaf':trial.suggest_loguniform('min_samples_leaf',.00001,.1)
}
cls.set_params(**params)#Initializing the model with the parameters
return np.mean(cross_val_score(cls, X_tr, y_tr, cv=cv_strat, n_jobs=5, scoring='roc_auc'))
return objective
# Importing hyperparamater tuning optimizer optuna
import optuna
# Defining the evaluation function for study's best parameters
def train_test_roc_auc(X_tr, y_tr, cls, obj_func, cv_strat, n_trials=100):
''' Computes the best hyper parameters of the classsifier and returns
Optuna's study's best score & clasifier parameters'''
study = optuna.create_study(direction='maximize')
study.optimize(obj_func(X_tr, y_tr, cls, cv_strat), n_trials)
best_score = study.best_value
best_params = study.best_params
return (best_score,best_params)
# Importing Random Forest Classifier from Sklearn
from sklearn.ensemble import RandomForestClassifier
# Instantiating the Random forest classifier
rf_s = RandomForestClassifier(n_jobs=5, random_state=42)
# Segregating the Features and class labels from the Training Dataset
X_train_stacked = df_train_stack[['Voting','Neural']]
y_train_stacked = df_train_stack['True']
# Extracting the best model parameters and best study score
best_study_score,best_study_params = train_test_roc_auc(X_train_stacked, y_train_stacked, rf_s, objective_wrappper_rf, cv_strat, n_trials=150)
print('The best roc_auc_score for the study is: ', best_study_score)
print('The best study parameters for the classifier are: ', best_study_params)
# Obtaining the best Stacking Random Forest model by setting best study parameters.
rf_stack = rf_s.set_params(**best_study_params)
# fitting the best Stacking Random Forest model on the whole training set
rf_stack.fit(X_train_stacked, y_train_stacked)
# ### Preparing the Test set observations for the Stacking Classifier.
# Training the Voting Classifier on the whole Training set
voting_clf.fit(X_train_red,y_train)
# Computing the predictions of the voting classifier on the test set.
y_pred_test_voting = voting_clf.predict_proba(X_test_red)[:,1]
# Reloading the best Neural Classifier
neural = keras.models.load_model('Best_model_Selu_eq_Learn.h5')
# Computing the predictions of the Neural Classifier on the test set.
y_pred_test_neural = neural.predict_proba(X_test_red).flatten()
# Preparing the Test feature set for the Stacking Classifier
X_test_stacked = pd.DataFrame({'Voting':y_pred_test_voting,'Neural':y_pred_test_neural})
# Checking the info of the Stacking Test Features
X_test_stacked.info()
# Getting the Stacking Classifier Predictions for the Test set
y_pred_stacked = rf_stack.predict_proba(X_test_stacked)[:,1]
# Getting the stacking Clasiifier roc_auc score for the Test Set.
print('The test set roc_auc score for the Stacking Classifier is: ',roc_auc_score(y_test,y_pred_stacked))
# ### Calculating R_R ratio for Stacking Classifier.
# Computing the CV scores using sklearn's cross_val_score
score_Stacking = cross_val_score(rf_stack, X_train_stacked, y_train_stacked, cv=cv_strat, n_jobs=5, scoring='roc_auc')
print('The reward associated with the tuned Stacking Classifier using roc_auc metric is: ',np.mean(score_Stacking))
print('The risk associated with the tuned Stacking Classifier using roc_auc metric is: ',np.std(score_Stacking))
R_R_Ratio_Stacking = np.mean(score_Stacking)/np.std(score_Stacking)
print('The reward risk ratio for the tuned Stacking Classifier using roc_auc metric is: ',R_R_Ratio_Stacking)
# ## Observations:
# ### 1) The test set roc_auc score for the Stacking classifier is more than that of Neural Net classifier , but less than that of the Voting classifier , both of which were used to create the training as well as test set for the stacking classifier.
# ### 2) Similarly the R_R ratio of the Stacking classifier is approx. equal (although more) to that of the Voting Classifier, but much less than that of the Neural Net. Thus, even with added complexity, the Stacking Classifier still hasn't been able to beat the tuned Random Forest Classifier on this dataset.
#
#
#
# ### The R_R Ratio for the tuned Stacking Classifier using roc_auc metric is: 48.351649337461005
| 13.1_a_Modeling.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
from project_functions_Mete import load_and_process_file1
from project_functions_Mete import load_and_process_file2
from project_functions_Mete import merge_and_process_dataframes
btc= load_and_process_file2("/Users/metec/school/project-group08-project/data/processed/Bitcoin Historical Data cleaned.csv")
btc
eth= load_and_process_file1("/Users/metec/school/project-group08-project/data/processed/etherium cleaned.csv")
eth
merge1= merge_and_process_dataframes(btc,eth)
merge1
| notebooks/ungraded/mete_cil_wfunctions.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Sense and Move
#
# In this notebook, let's put all of what we've learned together and see what happens to an initial probability distribution as a robot goes trough cycles of sensing then moving then sensing then moving, and so on! Recall that each time a robot senses (in this case a red or green color)it gains information about its environment, and everytime it moves, it loses some information due to motion uncertainty.
#
#
# <img src='images/sense_move.png' width=50% height=50% />
#
# First let's include our usual resource imports and display function.
# importing resources
import matplotlib.pyplot as plt
import numpy as np
# A helper function for visualizing a distribution.
def display_map(grid, bar_width=1):
if(len(grid) > 0):
x_labels = range(len(grid))
plt.bar(x_labels, height=grid, width=bar_width, color='b')
plt.xlabel('Grid Cell')
plt.ylabel('Probability')
plt.ylim(0, 1) # range of 0-1 for probability values
plt.title('Probability of the robot being at each cell in the grid')
plt.xticks(np.arange(min(x_labels), max(x_labels)+1, 1))
plt.show()
else:
print('Grid is empty')
# ### QUIZ: Given the list motions=[1,1], compute the posterior distribution if the robot first senses red, then moves right one, then senses green, then moves right again, starting with a uniform prior distribution, `p`.
#
# `motions=[1,1]` mean that the robot moves right one cell and then right again. You are given the initial variables and the complete `sense` and `move` function, below.
# +
# given initial variables
p=[0.2, 0.2, 0.2, 0.2, 0.2]
# the color of each grid cell in the 1D world
world=['green', 'red', 'red', 'green', 'green']
# Z, the sensor reading ('red' or 'green')
measurements = ['red', 'green']
pHit = 0.6
pMiss = 0.2
motions = [1,1]
pExact = 0.8
pOvershoot = 0.1
pUndershoot = 0.1
# You are given the complete sense function
def sense(p, Z):
''' Takes in a current probability distribution, p, and a sensor reading, Z.
Returns a *normalized* distribution after the sensor measurement has been made, q.
This should be accurate whether Z is 'red' or 'green'. '''
q=[]
# loop through all grid cells
for i in range(len(p)):
# check if the sensor reading is equal to the color of the grid cell
# if so, hit = 1
# if not, hit = 0
hit = (Z == world[i])
q.append(p[i] * (hit * pHit + (1-hit) * pMiss))
# sum up all the components
s = sum(q)
# divide all elements of q by the sum to normalize
for i in range(len(p)):
q[i] = q[i] / s
return q
# The complete move function
def move(p, U):
q=[]
# iterate through all values in p
for i in range(len(p)):
# use the modulo operator to find the new location for a p value
# this finds an index that is shifted by the correct amount
index = (i-U) % len(p)
nextIndex = (index+1) % len(p)
prevIndex = (index-1) % len(p)
s = pExact * p[index]
s = s + pOvershoot * p[nextIndex]
s = s + pUndershoot * p[prevIndex]
# append the correct, modified value of p to q
q.append(s)
return q
## TODO: Compute the posterior distribution if the robot first senses red, then moves
## right one, then senses green, then moves right again, starting with a uniform prior distribution.
for idx in range(len(motions)):
p = sense(p, measurements[idx])
p = move(p, motions[idx])
## print/display that distribution
print(p)
display_map(p)
# -
# ### Clarification about Entropy
#
# The video mentions that entropy will go down after the update step and that entropy will go up after the measurement step.
#
# In general, **entropy measures the amount of uncertainty**. Since the update step increases uncertainty, then entropy should increase. The measurement step decreases uncertainty, so entropy should decrease.
#
# Let's look at our current example where the robot could be at five different positions. The maximum uncertainty occurs when all positions have equal probabilities $[0.2, 0.2, 0.2, 0.2, 0.2]$
#
# Following the formula $$\text{Entropy} = \Sigma (-p \times log(p))$$we get $$-5 \times (.2)\times log(0.2) = 0.699$$
#
# Taking a measurement should decrease uncertainty and thus decrease entropy. Let's say after taking a measurement, the probabilities become <span class="mathquill">[0.05, 0.05, 0.05, 0.8, 0.05]</span>. Now the entropy decreased to 0.338. Hence a measurement step should decrease entropy whereas an update step should increase entropy.
| 4_2_Robot_Localization/9_1. Sense and Move, exercise.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Fairness part of the Workshop
# Analyze fairness of a dataset with different techniques
# Imports
import pandas as pd
from IPython.display import display
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
# # Loading data
#
# We are first going to work with data from the Lending Club Dataset, a dataset of around 800k lending club users. This dataset doesn't have information about race or gender, so we will artificially create a "race" column to test our fairness metrics. Let's create a heavily unfair dataset: 80% of the users that were given a loan will be of race "1", and only 10% of the users who were not given a loan will be of race "1".
# +
# Data management
df = pd.read_csv('../data/loans_data.csv')
# Subset
df = df.sample(frac=0.1)
display(df)
# Random vector to add noise to protected class
p_nochange=0.9
random_vec = np.random.choice([0,1], size=len(df), p=[p_nochange,1-p_nochange])
print(len(df), sum(random_vec))
# Add protected class
df['protected_class'] = df['loan_status']^random_vec
print(df[['protected_class','loan_status']])
# +
quant_cols = ['loan_amnt', 'int_rate', 'annual_inc', 'dti', 'delinq_2yrs', 'fico_range_low',\
'inq_last_6mths', 'mths_since_last_delinq', 'mths_since_last_record', 'open_acc', 'pub_rec',\
'revol_bal', 'revol_util', 'total_acc', 'acc_now_delinq', 'tot_coll_amt',\
'tot_cur_bal', 'tax_liens', 'total_bal_ex_mort', 'total_bc_limit', 'total_il_high_credit_limit',\
'age_of_cr_line', 'installment','protected_class']
cat_cols = ['grade','emp_length', 'home_ownership','verification_status', 'term', 'initial_list_status',\
'disbursement_method', 'application_type']
other_cols = ['zip_code']
response_col = 'loan_status'
# Get train and test
df_x = pd.get_dummies(df[quant_cols+cat_cols], drop_first=False, columns=cat_cols)
df_y = df[response_col]
x_train, x_test, y_train, y_test = train_test_split(df_x, df_y, test_size=0.3, random_state=42)
# -
# ## Fitting our model
#
# Let's fit a Random Forest to our data. Because we artificially added a biased "protected class" column, our classifier will not be fair.
# +
# Fit model
model = RandomForestClassifier(n_estimators=25, max_depth=None, #class_weight='balanced_subsample', \
random_state=42).fit(x_train, y_train)
# -
# ## Getting predictions
# +
# Get predictions
preds_test = model.predict(x_test)
acc_train = model.score(x_train, y_train)
acc_test = model.score(x_test, y_test)
print(acc_train)
print(acc_test)
# -
# # Statistical Parity
#
# We will first test our model's predictions with statistical parity, a simple fairness measure that is easy to compute.
#
# ## What is statistical parity?
#
# This metric measures the difference between the probability of positive decisions for the protected group and the probability of positive decisions for ghe unprotected group. Mathematically:
# $$Sp = P(d=1|G=0) - P(d=1|G=1)$$
#
# This can be easily approximated with our data by calculating the proportion of positive decisions amongst people from race "0" and substracting the proportion of positive decisions amongst people from race "1":
#
# $$Sp = \frac{ \text{# people with positive decision and race 0}} { \text{ # people from race 0} } - \frac{ \text{# people with positive decision and race 1}} { \text{ # people from race 1}}$$
# Let's code a simple function that will calculate this for our dataset. In the next cell, complete the function `evaluate_statistical_parity` to perform the calculation above. The function definition and docstring will guide you.
# Statistical parity function
def evaluate_statistical_parity(predictions, protected_class_array):
"""Function to calculate statistical parity.
Parameters
----------
predictions (numpy array): binary decision labels outputted by our trained model.
protected_class_array (numpy array): boolean mask where protected rows are marked True
Returns
-------
bias (float): statistical parity bias
"""
# --------------
# --------------
# Your code here
# --------------
# --------------
prob_g = np.sum(predictions & protected_class_array) / np.sum(protected_class_array)
prob_not_g = np.sum(predictions & ~protected_class_array) / np.sum(~protected_class_array)
bias = np.abs(prob_g - prob_not_g)
return bias
# # Conditional Parity
#
# Statistical parity is a simple measure, and it gives a fast overview on our model's fairness. However, it disregards important aspects of our dataset, such as the values of the features of each row. We could have a situation where the statistical parity measure tells us that we are giving loans to 20% of people from race 0 and 20% of people from race 1, which would be fair, but those 20% from race 0 are random, while the 20% from race 1 are people from developed countries. Our model would be hiding another layer of unfairness: we are not giving loans equally to people from race 1.
#
# We can use conditional parity to detect these types of imbalances. Conditional parity allows us to test for unfairness in a similar way as Statistical Parity, but conditioning on another feature (for example, country of origin). The equation is:
#
# $$Cp = P(d=1|G=0, L=l) - P(d=1|G=1, L=l)$$
#
# Again, this can be easily calculated by counting the number of positive outcome cases in por both protected groups, but this time only looking at the people that fulfill our conditional constraint (L=l)
# Conditional parity function
def evaluate_conditional_parity(predictions, protected_class_array, condition_array):
"""Function to calculate Conditional statistical parity.
Parameters
----------
predictions (numpy array): binary (decision) labels for X
protected_class_array (numpy array): boolean array where protected rows are marked True
condition_array (numpy array): boolean array that indicates conditional status
Returns
-------
bias (float): conditional parity bias
"""
# --------------
# --------------
# Your code here
# --------------
# --------------
prob_g = np.sum(predictions & condition_array & protected_class_array) / np.sum(predictions & protected_class_array)
prob_not_g = np.sum(predictions & condition_array & ~protected_class_array) / np.sum(predictions & ~protected_class_array)
bias = np.abs(prob_g - prob_not_g)
return bias
# Evaluate statistical and conditional parity
stat_parity = evaluate_statistical_parity([bool(x) for x in preds_test], ~x_test['protected_class'].apply(lambda x: bool(x)))
cond_parity = evaluate_conditional_parity([bool(x) for x in preds_test], ~x_test['protected_class'].apply(lambda x: bool(x)), x_test['loan_amnt']>10000)
print(stat_parity)
print(cond_parity)
# # False Positive (Negative) Error Rate Balance
#
# The previous measures don't take into account the real labels of each observation; they only consider the predictions. The measure of fairness proposed here controls for equal poportions of false positives/false negatives in protected and unprotected classes. This measure is ideal in cases where committing mistakes disproportionately for different protected groups can bring negative outcomes.
#
# We will again code these measures as they are rather easy to understand. The function definition below will guide you through the process.
# +
# False positive and false negative rates
def evaluate_false_negative_rate(predictions, protected, y):
"""evaluate fnr
Parameters
----------
predictions (numpy array): binary (decision) labels for X predicted by our model
protected (numpy array): boolean mask where protected rows are marked True or 1
y (numpy array): boolean array that marks ground truth
Note:
FNR: FN / CP where FN=(predictions==0) & (y==1) CN = (y==1)
Returns
-------
bias (float)
"""
# --------------
# --------------
# Your code here
# --------------
# --------------
cond_pos_protected = np.sum((y==1) & protected)
cond_pos_not_protected = np.sum((y==1) & ~protected)
if cond_pos_protected == 0:
return 'No Condition Positive in Protected'
if cond_pos_not_protected == 0:
return 'No Condition Positive in Not Protected'
false_neg_protected = np.sum((y==1) & (predictions==0) & protected)
false_neg_not_protected = np.sum((y==1) & (predictions==0) & ~protected)
fnr_g = false_neg_protected / cond_pos_protected
fnr_not_g = false_neg_not_protected / cond_pos_not_protected
bias = np.abs(fnr_g - fnr_not_g)
return bias
def evaluate_false_positive_rate(predictions, protected, y):
"""evaluate fpr
Parameters
----------
predictions (numpy array): binary (decision) labels for X predicted by our model
protected (numpy array): boolean mask where protected rows are marked True or 1
y (numpy array): boolean array that marks ground truth
Note:
FPR: FP / CN where FP=(predictions==1) & (y==0) CN = (y==0)
Returns
-------
bias (float)
"""
# --------------
# --------------
# Your code here
# --------------
# --------------
cond_neg_protected = np.sum((y==0) & protected)
cond_neg_not_protected = np.sum((y==0) & ~protected)
if cond_neg_protected == 0:
return 'No Condition Negative in Protected'
if cond_neg_not_protected == 0:
return 'No Condition Negative in Not Protected'
false_pos_protected = np.sum((y==0) & predictions & protected)
false_pos_not_protected = np.sum((y==0) & predictions & ~protected)
fpr_g = false_pos_protected / cond_neg_protected
fpr_not_g = false_pos_not_protected / cond_neg_not_protected
bias = np.abs(fpr_g - fpr_not_g)
return bias
# +
# Test FPR and FNR on this dataset
fnr = evaluate_false_negative_rate(x_test, preds_test, ~x_test['protected_class'], y_test)
fpr = evaluate_false_positive_rate(x_test, preds_test, ~x_test['protected_class'], y_test)
print(fpr)
print(fnr)
# -
# As we can see, the values of FPR and FNR are significantly higher than expected, showing that our dataset is clearly unfair.
# ## Other Fairness metrics
#
# We have coded and tested some basic Fairness metrics, but there are multiple other metrics that can be used, depending on the situation. Some of them are:
#
# **Predictive parity:**
# The fraction of correct positive predictions should be the same for protected and unprotected groups.
# $$P(Y=1|d=1, G=m) = P(Y=1|d=1, G=f)$$
#
#
# **Equalized odds:**
# Applicants with a good actual credit scope and applicants with a bad actual credit
# score should have a similar classification, regardless of the value of the protected class.
# $$P(d=1|Y=i, G=m) = P(d=1|Y=i, G=f), i\in \{0,1\}$$
#
#
# **Overall accuracy equality:**
# Both protected and unprotected groups have equal prediction accuracy.
# $$P(d=Y, G=m) = P(d=Y, G=f)$$
#
#
# **Treatment Equality:**
# Looks at ratio of errors a classifier makes instead of its accuracy. Satisfied if both protected and unprotected groups have equal ratio of false negatives and false positives.
#
# ## Fairness concepts
# - **Fairness through unawareness:**
# No sensitive attributes used in the decision making process.
# - **Fairness through awareness:**
# Similar individuals should have similar classification.
# - **Disparate impact:**
# Exists when decision outcomes disproportionately benefits or hurts individuals of a certain group.
# - **Disparate treatment:**
# Decision changes when protected feature changes.
# - **Disparate mistreatment:**
# Missclassification rates are different for people of different protected groups
#
# We refer the reader to http://fairware.cs.umass.edu/papers/Verma.pdf for more information.
# # Creating a Fair Model
#
# Once we have characterized and measured the fairness of the model, we might want to build a model that avoids discrimination given a protected class. As there are multiple ways to define fairness, there are also multiple ways to build a fair classifier, depending on what notion we want to emphasize.
#
# Some options are:
# - Preprocessing the data to remove biases, and training normal classifiers on that data
# - Training the classifier and post-processing the predictions to accomodate our measures of fairness
# - Training a modified classifier with clear constraints that enforce fairness
#
# We will exemplify the Optimized Preprocessing technique, published by our very own Flavio Calmon.
#
# ![../optimized.PNG]
# ### Census Income dataset
#
# The previous dataset was thorough and complex enough to demonstrate interpretability techniques, but as it is an anonymized dataset, it has little to no information on sensitive features. We will switch to another dataset for this part that is more suited to analyzing fairness techniques, as it possesses information on gender and race.
#
# This dataset is called the **Census Income dataset**, and it associates features of working adults to **whether or not they make more than $50k/yr**. It is extracted from the 1994 Census database, and contains **48842 observations** with a mix of continuous and categorical features (14 in total).
#
# List of features:
# - **age:** continuous.
# - **workclass:** categorical.
# - **education:** categorical.
# - **education-num:** continuous.
# - **marital-status:** categorical.
# - **relationship:** categorical.
# - **race:** categorical.
# - **sex:** categorical.
# - **capital-gain:** continuous.
# - **capital-loss:** continuous.
# - **hours-per-week:** continuous.
# - **fnlwgt:** (final weight) continuous.
# - **native-country:** categorical.
#
# Response: binary, corresponding to >50K (1) or <=50K (0).
#
#
# #### Reference:
# <NAME>, "Scaling Up the Accuracy of Naive-Bayes Classifiers: a Decision-Tree Hybrid", Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, 1996
#
# +
# Imports
from aif360.algorithms.preprocessing import OptimPreproc
from sklearn.preprocessing import StandardScaler
from aif360.algorithms.preprocessing.optim_preproc_helpers.data_preproc_functions import load_preproc_data_german
from aif360.algorithms.preprocessing.optim_preproc_helpers.distortion_functions import get_distortion_german
from aif360.algorithms.preprocessing.optim_preproc_helpers.opt_tools import OptTools
from aif360.datasets import BinaryLabelDataset
from aif360.datasets import AdultDataset, GermanDataset, CompasDataset
from aif360.metrics import BinaryLabelDatasetMetric
from aif360.metrics import ClassificationMetric
from aif360.metrics.utils import compute_boolean_conditioning_vector
from aif360.algorithms.preprocessing.optim_preproc import OptimPreproc
from aif360.algorithms.preprocessing.optim_preproc_helpers.data_preproc_functions\
import load_preproc_data_adult, load_preproc_data_german, load_preproc_data_compas
from aif360.algorithms.preprocessing.optim_preproc_helpers.distortion_functions\
import get_distortion_adult, get_distortion_german, get_distortion_compas
from aif360.algorithms.preprocessing.optim_preproc_helpers.opt_tools import OptTools
from sklearn.linear_model import LogisticRegression
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import accuracy_score
from IPython.display import Markdown, display
import matplotlib.pyplot as plt
# +
# Load German dataset with 'sex' as protected attribute
# privileged_groups = [{'sex': 1}]
# unprivileged_groups = [{'sex': 0}]
# dataset_orig = load_preproc_data_german(['sex'])
# optim_options = {
# "distortion_fun": get_distortion_german,
# "epsilon": 0.05,
# "clist": [0.99, 1.99, 2.99],
# "dlist": [.1, 0.05, 0]
# }
privileged_groups = [{'sex': 1}]
unprivileged_groups = [{'sex': 0}]
dataset_orig = load_preproc_data_adult(['sex'])
optim_options = {
"distortion_fun": get_distortion_adult,
"epsilon": 0.05,
"clist": [0.99, 1.99, 2.99],
"dlist": [.1, 0.05, 0]
}
# Split into train and test
dataset_orig_train, dataset_orig_test = dataset_orig.split([0.7], shuffle=True)
# -
print('Training Dataset shape:',dataset_orig_train.features.shape)
# print('Favorable and unfavorable labels:',dataset_orig_train.favorable_label, dataset_orig_train.unfavorable_label)
print('Protected attribute names:',dataset_orig_train.protected_attribute_names)
print('Privileged and unprivileged protected attribute values:' ,dataset_orig_train.privileged_protected_attributes,
dataset_orig_train.unprivileged_protected_attributes)
print('Dataset feature names:',dataset_orig_train.feature_names)
# +
# Extract data from AIF360 Data object
# We define a scaler to normalize our data
scale_orig = StandardScaler()
# We get our training numpy arrays
x_train = scale_orig.fit_transform(dataset_orig_train.features) #This fit_transform scales our data feature-wise.
y_train = (dataset_orig_train.labels.ravel()-2)*-1
y_train = dataset_orig_train.labels.ravel()
# And our testing arrays
x_test = scale_orig.transform(dataset_orig_test.features) # Here, we only transform, as we can't use the testing set to define the scaling factors.
y_test = (dataset_orig_test.labels.ravel()-2)*-1
y_test = dataset_orig_test.labels.ravel()
# +
# Train classifier on original data
rf_model = RandomForestClassifier(n_estimators=25,
max_depth=None,
random_state=42).fit(x_train, y_train)
rf_model.fit(x_train, y_train)
# +
# Getting accuracy and fairness metrics on test
acc_orig = rf_model.score(x_test, y_test)
print('Accuracy on test with original data:', acc_orig)
print(dataset_orig_test.feature_names)
predictions = rf_model.predict(x_test)>0.5
protected_class_array = dataset_orig_test.features[:,1]==1 # Here, we're taking the column corresponding to 'sex' and we are transforming it into a boolean array
statistical_parity_orig = evaluate_statistical_parity(predictions, protected_class_array)
fpr_orig = evaluate_false_positive_rate(predictions, protected_class_array, y_test)
fnr_orig = evaluate_false_negative_rate(predictions, protected_class_array, y_test)
print(statistical_parity_orig, fpr_orig, fnr_orig)
# metric_test_bef = compute_metrics(dataset_transf_test, dataset_transf_test_pred,
# unprivileged_groups, privileged_groups, disp=disp)
# bal_acc_arr_transf.append(metric_test_bef["Balanced accuracy"])
# avg_odds_diff_arr_transf.append(metric_test_bef["Average odds difference"])
# disp_imp_arr_transf.append(metric_test_bef["Disparate impact"])
# -
# ### Now, let's apply a dataset transformation to increase fairness !
# +
# Instantiate OptimizedDataPreprocessing module from AIF360
OP = OptimPreproc(OptTools, optim_options,
unprivileged_groups = unprivileged_groups,
privileged_groups = privileged_groups)
# Fit the module to the training data, effectively creating the mapping from original data to transformed, fair data
OP = OP.fit(dataset_orig_train)
# +
# Transform training data and align features
dataset_transf_train = OP.transform(dataset_orig_train, transform_Y=True)
dataset_transf_train = dataset_orig_train.align_datasets(dataset_transf_train)
# Same with test data
dataset_transf_test = OP.transform(dataset_orig_test, transform_Y = True)
dataset_transf_test = dataset_orig_test.align_datasets(dataset_transf_test)
# +
# Again, we have to get our training numpy arrays, this time on the TRANSFORMED training data
x_train_transf = scale_orig.fit_transform(dataset_transf_train.features)
y_train_transf = (dataset_transf_train.labels.ravel()-2)*-1
y_train_transf = dataset_transf_train.labels.ravel()
# And our testing arrays, on the TRANSFORMED test data
x_test_transf = scale_orig.transform(dataset_transf_test.features) # Here, we only transform, as we can't use the testing set to define the scaling factors.
y_test_transf = (dataset_transf_test.labels.ravel()-2)*-1
y_test_transf = dataset_transf_test.labels.ravel()
# -
# Train same classifier on TRANSFORMED data
rf_model_transf = RandomForestClassifier(n_estimators=25,
max_depth=None,
random_state=42).fit(x_train_transf, y_train_transf)
rf_model_transf.fit(x_train_transf, y_train_transf)
# +
# Getting accuracy and fairness metrics on TRANSFORMED test set
acc_transf = rf_model_transf.score(x_test_transf, y_test_transf)
print('Accuracy on test with original data (we should expect a bit less than before):', acc_transf)
predictions_transf = rf_model_transf.predict(x_test_transf)>0.5
protected_class_array_transf = dataset_orig_test.features[:,1]==1
statistical_parity_transf = evaluate_statistical_parity(predictions_transf, protected_class_array_transf)
fpr_transf = evaluate_false_positive_rate(predictions_transf, protected_class_array_transf, y_test_transf)
fnr_transf = evaluate_false_negative_rate(predictions_transf, protected_class_array_transf, y_test_transf)
print(statistical_parity_transf, fpr_transf, fnr_transf)
# +
# Compare the Results
# +
# +
# Predictions and fairness metrics on transformed test set
# -
dataset_transf_test_pred = dataset_transf_test.copy(deepcopy=True)
X_test = scale_transf.transform(dataset_transf_test_pred.features)
y_test = dataset_transf_test_pred.labels
dataset_transf_test_pred.scores = lmod.predict_proba(X_test)[:,pos_ind].reshape(-1,1)
# +
# Results
# -
Disparate impact
Average odds difference
Balanced accuracy
# # Conclusion
#
# We have analyzed particular fairness metrics and observed their behavior on an artificial dataset. It is important to remember that Fairness has multiple definitions, each one approriate for analyzing a specific situation. Statistical notions of fairness as described above are easy to measure. However, it is important to keep in mind that statistical definitions are insufficient in some cases (for example, when similarity has to be taken into account). Moreover, most valuable statistical metrics assume availability of actual, verified outcomes. While such outcomes are available for the training data, it is unclear whether the real classified data always conforms to the same distribution.
# # Appendix: extra resources
#
# ## Interesting Fairness analysis tools
# - Pymetrics audit-ai (https://github.com/pymetrics/audit-ai)
# - fairness metrics github (https://github.com/megantosh/fairness_measures_code)
# - fairness-comparison github (https://github.com/algofairness/fairness-comparison)
# - IBM AIF360 (https://github.com/IBM/AIF360, https://arxiv.org/pdf/1810.01943.pdf)
# - Themis ML (https://themis-ml.readthedocs.io/en/latest/)
# - FairML (https://github.com/adebayoj/fairml)
# - BlackBoxAuditing (https://github.com/algofairness/BlackBoxAuditing)
#
# ## Interesting papers
# - Learning Fair Representations (seminal paper) http://proceedings.mlr.press/v28/zemel13.pdf
# - Optimized Data Pre-Processing for Discrimination Prevention (by <NAME>) https://arxiv.org/pdf/1704.03354.pdf
# - Fairness Definitions Explained http://fairware.cs.umass.edu/papers/Verma.pdf
# - From parity to Preference-based notions of fairness https://arxiv.org/abs/1707.00010
# - Certifying and removing disparate impact https://arxiv.org/pdf/1412.3756.pdf
# - Learning Classification without Disparate Mistreatment https://arxiv.org/pdf/1610.08452.pdf
# - Fairness Constraints: Mechanisms for Fair Classification https://arxiv.org/abs/1507.05259
# - Fairness GAN https://arxiv.org/pdf/1805.09910.pdf
# - Adversarial Debiasing https://arxiv.org/pdf/1801.07593.pdf
# - Classification with Fairness Constraints: A Meta-Algorithm with Provable Guarantees https://arxiv.org/pdf/1806.06055.pdf
#
#
#
#
# +
# add IBM AIF360 examples if time
# +
# German Loan Dataset
import aif360
# +
from aif360.algorithms.preprocessing import DisparateImpactRemover
from aif360.datasets import AdultDataset
from aif360.metrics import BinaryLabelDatasetMetric
protected = 'sex'
# ad = AdultDataset(protected_attribute_names=[protected],
# privileged_classes=[['Male']], categorical_features=[],
# features_to_keep=['age', 'education-num', 'capital-gain', 'capital-loss', 'hours-per-week'])
data = GermanDataset()
print(data.feature_names)
aif360.algorithms.preprocessing.OptimPreproc(optimizer, optim_options, unprivileged_groups, privileged_groups, verbose=False, seed=None)
['age', 'sex', 'credit_history=Delay', 'credit_history=None/Paid', 'credit_history=Other', 'savings=500+', 'savings=<500', 'savings=Unknown/None', 'employment=1-4 years', 'employment=4+ years', 'employment=Unemployed']
# +
import urllib.request
url1 = 'https://archive.ics.uci.edu/ml/machine-learning-databases/statlog/german/german.data'
url2 = 'https://archive.ics.uci.edu/ml/machine-learning-databases/statlog/german/german.doc'
urllib.request.urlretrieve(url1,'C:/Users/Camilo/Anaconda3/lib/site-packages/aif360/data/raw/german/german.data')
urllib.request.urlretrieve(url2,'C:/Users/Camilo/Anaconda3/lib/site-packages/aif360/data/raw/german/german.doc')
# -
| notebooks/cf_fairness.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + colab={} colab_type="code" id="OG2JlAXx3VoJ"
#Declare a string and store it in a variable.
s = 'I am a student of Learnbay'
#Check the type and print the id of the same.
print(type(s))
print(id(s))
# + colab={} colab_type="code" id="wPxm3G8W3dqN"
#Which are valid/invalid strings
1. 'This is Python class'
valid/invalid #Valid
2. "This is Python class"
valid/invalid #Valid
3. '''This is Python class''' #Valid
valid/invalid
4. """This is Python class""" #Valid
valid/invalid
5. 'This is Python's class'
valid/invalid #Invalid
6. "Learnbay provides "Java", "Python" classes"
valid/invalid #Invalid
7. "Learnbay provides 'Java', 'Python' classes"
valid/invalid #Valid
8. "This is Python's class"
valid/invalid #Valid
9. """Learnbay provides "Java", "Python" classes"""
valid/invalid #Valid
10. '''Learnbay provides "Java", "Python" classes'''
valid/invalid #Valid
11. '''Learnbay provides
"Java", "Python"
classes'''
valid/invalid #Valid
12. 'This is
Python
class'
valid/invalid #Invalid
# + colab={} colab_type="code" id="Drx8fhT23g7a"
#Write the code to get the output mentioned below print statement
my_str = "Although that way may not be obvious at first unless you're Dutch."
my_str1 = "Although that way may not be obvious at first unless you're Dutch."
print('The length of my_str is',len(my_str))
#output:- The length of my_str is 66
print('id of my_str and my_str1 is same? -',id(my_str) == id(my_str1))
#output:- id of my_str and my_str1 is same? - True #False. For true, we've to do my_str1 = my_str
# print(id(my_str))
# print(id(my_str1))
print('Type of my_str is:',type(my_str))
#output:- Type of my_str is: str
# + colab={} colab_type="code" id="403e-I3A3lCO"
#Indexing
my_str = "Although 8 that way may not be obvious at first unless you're Dutch"
#Write the code to get the output,instructions are mentioned below print statement. use indexing
print('The first character in my_str is:',my_str[0])
#output:- The first character in my_str is: A
#Note:- Use positive indexing
print('The first character in my_str is:',my_str[len(my_str)-1])
#output:- The first character in my_str is: h
#Note:- Use len() function.
print('The character at index 10 in my_str is:',my_str[10])
#output:- The character at index 10 in my_str is: c #c is not at index 10
#Note:- Use positive indexing
print('The last character in my_str is:',my_str[-1])
#output:- The last character in my_str is: h
#Note:- Use negative indexing.
print('The last character in my_str is:',my_str[len(my_str)-1])
#output:- The last character in my_str is: h
#Note:- Use len() function.
print('The character in my_str is:',my_str[9])
#output:- The character in my_str is: 8
#Note:- Use positive index
# + colab={} colab_type="code" id="NK_QdtsM3luu"
#Slicing
my_str = "Although that way may not be obvious at first unless you're Dutch."
#Write the code to get the output,instructions are mentioned below print statement. use slicing
print(my_str[::])
#output:- You have sliced: Although that way may not be obvious at first unless you're Dutch.Without begin, end and step
print(my_str[0:len(my_str):])
#output:- You have sliced: Although that way may not be obvious at first unless you're Dutch.with begin as 0 end using len and without step
print(my_str[::1])
#output:- You have sliced: Although that way may not be obvious at first unless you're Dutch.without begin and end but using step
print(my_str[0:len(my_str):1])
#output:- You have sliced: Although that way may not be obvious at first unless you're Dutch.With begin, end and step
print(my_str[0:1:-1])
#output:- You have sliced: .with using begin and end using postive values and step as negative values.
#Slicing command should print empty string.
print(my_str[0:len(my_str):2])
#output:- You have sliced: Atog htwymyntb biu tfrtuls o'eDth
print(my_str[0:len(my_str):3])
#output:- You have sliced: Ahgttam tebo r lsorDc
print(my_str[::-1])
#output:- You have sliced: .hctuD er'uoy sselnu tsrif ta suoivbo eb ton yam yaw taht hguohtlA. Use only step
print(my_str[-1:-len(my_str)-1:-1])
#output:- You have sliced: .hctuD er'uoy sselnu tsrif ta suoivbo eb ton yam yaw taht hguohtlA. Use begin end and step.
print(my_str[::-2])
#output:- You have sliced: .cu ruysen si asovoe o a a athuhl. use only step
print(my_str[-1:-len(my_str)-1:-2])
#output:- You have sliced: .cu ruysen si asovoe o a a athuhl. use begin, end and step.
print(my_str[10:17:-1])
#What will be the output? # Nothing as the beginning index should be greater than ending index while the step is negative
print(my_str[16:10:-1])
#output:- You have sliced: yaw ta, Using begin, end and step.
print(my_str[-17:-10:1])
#output:- You have sliced: ess you. Using begin, end and step.
# + colab={} colab_type="code" id="qUSYa5x-3n5j"
#Basic operation on string
str1 = 'Learnbay'
str2 = 'Python'
#Write the code to get the output,instructions are mentioned below.
#Output is: Learnbay Python
print(str1+' '+str2)
#Error: TypeError: can only concatenate str (not "int") to str
# print(str+1)
#Error: TypeError: can only concatenate str (not "float") to str
# print(str+1.1)
#Find below Output
#Output is: LearnbayLearnbayLearnbay
print(str1*3)
#Error: TypeError: can't multiply sequence by non-int of type 'float'
# print(str1*3.0)
#Error: TypeError: can't multiply sequence by non-int of type 'str'
# print(str1*str2)
# + colab={} colab_type="code" id="gREffmnr3s-p"
#Find below Output
str1 = 'Python'
str2 = 'Python'
str3 = 'Python$'
str4 = 'Python$'
#print True by using identity operator between str1 and str2
print(str1 is str2)
#print False by using identity operator between str1 and str3
print(str1 is str3)
#print False by using identity operator between str4 and str3
print(str4 is str2)
#Check if P is available in str1 and print True by using membership operator
print('P' in str1)
#Check if $ is available in str3 and print True by using membership operator
print('$' in str3)
#Check if N is available in str3 and print False by using membership operator
print('N' in str3)
# + colab={} colab_type="code" id="QNFjxDr73u2H"
#Complete the below code
str1 = 'This is Python class'
#write the code to replace 'Python' with 'Java' and you should get below error.
#TypeError: 'str' object does not support item assignment.
str1[8] = 'J'
# + colab={} colab_type="code" id="-JgFbPmn3w3D"
str1 = 'A'
str2 = 'A'
#Compare str1 and str2 and print True using comparison operator
print(str1 >= str2)
#Compare str1 and str2 and print True using equality operator
print(str1 == str2)
#Compare str1 and str2 and print False using equality operator
print(str1 != str2)
#Compare str1 and str2 and print False using comparison operator
print(str1 > str2)
# + colab={} colab_type="code" id="fJ46_L-53yhW"
str1 = 'A'
str2 = 'a'
#Compare str1 and str2 and print True using comparison operator
print(str1 < str2)
#Compare str1 and str2 and print True using equality operator
print(str1!=str2)
#Compare str1 and str2 and print False using equality operator
print(str1 == str2)
#Compare str1 and str2 and print False using comparison operator
print(str1 > str2)
# + colab={} colab_type="code" id="e-Lr9va330gi"
str1 = 'A'
str2 = '65'
#Compare str1 and str2 using comparison operator and it should give below error.
# print(str1 >= int(str2))
#Error: TypeError: '>=' not supported between instances of 'str' and 'int'
#Compare str1 and str2 and print True using equality operator
print(str1 != str2)
#Compare str1 and str2 and print False using equality operator
print(str1 == str2)
# + colab={} colab_type="code" id="JO04jmpN32Im"
str1 = 'Python'
str2 = 'Python'
#Compare str1 and str2 and print True using comparison operator
print(str1 >= str2)
#Compare str1 and str2 and print True using equality operator
print(str1 == str2)
#Compare str1 and str2 and print False using equality operator
print(str1 != str2)
#Compare str1 and str2 and print False using comparison operator
print(str1 > str2)
# + colab={} colab_type="code" id="7ulv5ith37OJ"
a = 'Python'
b = ''
#Apply logical opereators (and, or & not) on above string values and observe the output.
print(a and b)
print(a or b)
print(not a)
print(not b)
# print(a not b)
# + colab={} colab_type="code" id="Yg_gsZBL383n"
a = ''
b = ''
#Apply logical opereators (and, or & not) on above string values and observe the output.
print(a and b)
print(a or b)
print(not a)
# + colab={} colab_type="code" id="cIWkP9Hf3-q5"
a = 'Python'
b = 'learnbay'
#Apply logical opereators (and, or & not) on above string values and observe the output.
print(a and b)
print(a or b)
print(not a)
# + colab={} colab_type="code" id="Y0Ot_E704AX5"
my_str = "Although 8 that way may not be obvious at first unless you're Dutch"
#Write the code to get the total count of 't' in above string. Use find() and index() method.
print(my_str.find('t'))
print(my_str.index('t'))
#Write the code to get the index of '8' in my_str. Use find() and index() method.
print(my_str.find('8'))
print(my_str.index('8'))
#What will be the output of below code?
# print(my_str.find('the')) ------------ -1
# print(my_str.index('the')) ----------- Error
# print(my_str.find('t', 9, 15)) ------- 11
# print(my_str.rfind('u')) ------------- 63
# print(my_str.rindex('u')) ------------ 63
# + colab={} colab_type="code" id="Gommq59Q4CMi"
#W A P which applies strip() method if any string, which will be taken from user, starts and ends with space, or applies
#rrstrip() method if that string only ends with space or applies lstrip() method if that string only starts with a space.
#For example:-
#input:- ' Python '
#output:- 'Python'
s = ' Python '
print(s.strip())
#input:- ' Python'
#output:- 'Python'
s = ' Python '
print(s.strip())
#input:- 'Python '
#output:- 'Python'
s = ' Python '
print(s.strip())
# + colab={} colab_type="code" id="9IUy-cpS4Dtt"
my_str = "Although 8 that way may not be obvious at first unless you're Dutch"
#Write the code to convert all alphabets in my_str into upper case.
print(my_str.upper())
#Write the code to convert all alphabets in my_str into lower case.
print(my_str.lower())
#Write the code to swap the cases of all alphabets in my_str.(lower to upper and upper to lower)
print(my_str.swapcase())
# + colab={} colab_type="code" id="-BjS-z674HKW"
#Write the code which takes one string from user and if it starts with small case letter then convert it to corresponding
#capital letter otherwise if starts with capital letters then convert first character of every word in that string into capital.
str1 = "this is a python class"
if str1[0] >= 'a' and str1[0] <= 'z':
str1 = str1.capitalize()
else:
str1 = str1.title()
print(str1)
str2 = "This is a python class"
if str2[0] >= 'a' and str2[0] <= 'z':
str2 = str2.capitalize()
else:
str2 = str2.title()
print(str2)
# + colab={} colab_type="code" id="-MCuDy5J4JQS"
#Take a string from user and check if it is:-
# 1. alphanumeric
# 2. alphabets
# 3. digit
# 4. all letters are in lower case
# 5. all letters are in upper case
# 6. in title case
# 7. a space character
# 8. numeric
# 9. all number elements in string are decimal
str1 = 'adsn54asd'
print(str1.isalnum())
print(str1.isalpha())
print(str1.isdigit())
print(str1.islower())
print(str1.isupper())
print(str1.istitle())
print(str1.isspace())
print(str1.isnumeric())
print(str1.isdecimal())
# + colab={} colab_type="code" id="tiTN-4ik4KAG"
#W A P which takes a string as an input and prints True if the string is valid identifier else returns False.
#Sample Input:- 'abc', 'abc1', 'ab1c', '1abc', 'abc$', '_abc', 'if'
str1 = 'abc'
str2 = 'abc1'
str3 = 'ab1c'
str4 = '1abc'
str5 = 'abc$'
str6 = '_abc'
str7 = 'if'
print(str1.isidentifier())
print(str2.isidentifier())
print(str3.isidentifier())
print(str4.isidentifier())
print(str5.isidentifier())
print(str6.isidentifier())
print(str7.isidentifier())
# + colab={} colab_type="code" id="tAMkSIN84MPe"
#What will be output of below code?
s = chr(65) + chr(97) # A + a
print(s.isprintable())
s = chr(27) + chr(97) # non-alphabet + a
print(s.isprintable())
s = '\n'
print(s.isprintable()) # \n is an Enter, not a character
s = ''
print(s.isprintable()) # '' is an empty string
# + colab={} colab_type="code" id="pU9Os0mD4Pk1"
#What will be output of below code?
my_string = ' '
print(my_string.isascii()) # ascii contains only english characters
my_string = 'Studytonight'
print(my_string.isascii())
my_string = 'Study tonight'
print(my_string.isascii())
my_string = 'Studytonight@123'
print(my_string.isascii())
my_string = '°'
print(my_string.isascii())
my_string = 'ö'
print(my_string.isascii())
# + colab={} colab_type="code" id="VsLV8FF74QTB"
#What will be the output of below code?
firstString = "der Fluß"
secondString = "der Fluss"
if firstString.casefold() == secondString.casefold(): #Casefolds contain more characters
print('The strings are equal.')
else:
print('The strings are not equal.')
# + colab={} colab_type="code" id="1m0U7jLa4Wjf"
#Write the code to get below output
#O/P 1:- python** (using ljust method)
s = 'python'
print(s.ljust(8,'*'))
#Write the code to get below output
#O/P 1:- **python (using rjust method)
print(s.rjust(8,'*'))
#Write the code to get below output
#O/P 1:- **python** (using rjust method)
print('python**'.rjust(10,'*'))
# print('python'.center(10,'*'))
# + colab={} colab_type="code" id="cRVkvW6s4YKK"
#Write a Python program to find the length of the my_str:-
#Input:- 'Write a Python program to find the length of the my_str'
#Output:- 55
s = 'Write a Python program to find the length of the my_str'
print(len(s))
# + colab={} colab_type="code" id="SjABWI284Zuz"
#Write a Python program to find the total number of times letter 'p' is appeared in the below string:-
#Input:- '<NAME> picked a peck of pickled peppers.'
#Output:- 9
s = '<NAME> picked a peck of pickled peppers.'
print(s.count('p'))
# + colab={} colab_type="code" id="-iZtqbEo4bQp"
#Write a Python Program, to print all the indexes of all occurences of letter 'p' appeared in the string:-
#Input:- '<NAME> picked a peck of pickled peppers.'
#Output:-
# 0
# 6
# 8
# 12
# 21
# 29
# 37
# 39
# 40
s = '<NAME> picked a peck of pickled peppers.'
for i in range(len(s)):
if s[i] == 'p':
print(i)
# + colab={} colab_type="code" id="_A8Eu8f84daM"
#Write a python program to find below output:-
#Input:- '<NAME> picked a peck of pickled peppers.'
#Output:- ['peter', 'piper', 'picked', 'a', 'peck', 'of', 'pickled', 'peppers']
s = '<NAME> picked a peck of pickled peppers.'
lst = s.split()
print(lst)
# + colab={} colab_type="code" id="juh7tQuL4fNc"
#Write a python program to find below output:-
#Input:- '<NAME> picked a peck of pickled peppers.'
#Output:- 'peppers pickled of peck a picked piper peter'
s = '<NAME> picked a peck of pickled peppers.'
s = s.replace('.','')
lst = s.split()[::-1]
print(lst)
# + colab={} colab_type="code" id="aaHQYz194fW7"
#Write a python program to find below output:-
#Input:- '<NAME> picked a peck of pickled peppers.'
#Output:- 'sreppep delkcip fo kcep a dekcip repip retep'
s = 'pe<NAME> picked a peck of pickled peppers.'
s = s.replace('.','')[::-1]
print(s)
# + colab={} colab_type="code" id="LSgj9glC4feI"
#Write a python program to find below output:-
#Input:- '<NAME> picked a peck of pickled peppers.'
#Output:- 'retep repip dekcip a kcep fo delkcip sreppep'
s = 'pe<NAME> picked a peck of pickled peppers.'.replace('.','')[::-1]
lst = s.split(' ')[::-1]
s = ' '.join(lst)
print(s)
# + colab={} colab_type="code" id="WUxPAkY94foo"
#Write a python program to find below output:-
#Input:- '<NAME> picked a peck of pickled peppers.'
#Output:- '<NAME> Picked A Peck Of Pickled Peppers'
s = '<NAME> picked a peck of pickled peppers.'.replace('.','')
print(s.title())
# + colab={} colab_type="code" id="IT1NSPxA4mww"
#Write a python program to find below output:-
#Input:- '<NAME> Picked A Peck Of Pickled Peppers.'
#Output:- '<NAME> picked a peck of pickled peppers'
s = '<NAME>per picked a peck of pickled peppers.'.replace('.','')
print(s.capitalize())
# + colab={} colab_type="code" id="3Gg65vSc4m3C"
#Write a python program to implement index method. If sub_str is found in my_str then it will print the index
# of first occurrence of first character of matching string in my_str:-
#Input:- my_str = '<NAME> Picked A Peck Of Pickled Peppers.', sub_str = 'Pickl'
#Output:- 29
# my_str = '<NAME> picked a peck of pickled peppers.'.replace('.','')
# sub_str = 'Pickl'
# my_str.index(sub_str)
my_str = '<NAME> picked a peck of pickled peppers.'.replace('.','')
sub_str = 'pickle'
my_str.index(sub_str)
# + colab={} colab_type="code" id="iFRhp_JI4m6k"
#Write a python program to implement replace method. If sub_str is found in my_str then it will replace the first
#occurrence of sub_str with new_str else it will will print sub_str not found:-
#Input:- my_str = '<NAME> Picked A Peck Of Pickled Peppers.', sub_str = 'Peck', new_str = 'Pack'
#Output:- '<NAME> Picked A Pack Of Pickled Peppers.'
my_str = '<NAME> Picked A Peck Of Pickled Peppers.'
sub_str = 'Peck'
new_str = 'Pack'
my_str.replace(sub_str,new_str,1)
# + colab={} colab_type="code" id="dQ9YhBBc4m9V"
#Write a python program to find below output (implements rjust and ljust):-
#Input:- '<NAME> Picked A Peck Of Pickled Peppers.', sub_str = 'Peck',
#Output:- '*********************Peck********************'
my_str = '<NAME> Picked A Peck Of Pickled Peppers.'
sub_str = 'Peck'
print(sub_str.center(len(my_str),'*'))
# + colab={} colab_type="code" id="w37BRLdt4tYT"
#Write a python program to find below output (implement partition and rpartition):-
#Input:- 'This is Python class', sep = 'is',
#Output:- ['This', 'is', 'Python class']
s = 'This is Python class'
sep = 'is'
list(s.partition(' is'))
# + colab={} colab_type="code" id="W64gCpeu4vaF"
#Write a python program which takes one input string from user and encode it in below format:-
# 1. #Input:- 'Python'
# #Output:- 'R{vjqp'
s = 'Python'
print(s)
for x in s:
a = ord(x)
b = chr(a+2)
s = s.replace(x,b)
print(s)
# 2. #Input:- 'Python'
# #Output:- 'Rwvfql'
s = 'Python'
print(s)
for i in range(len(s)):
if i%2 != 0:
a = ord(s[i])
b = chr(a-2)
s = s.replace(s[i],b)
else:
a = ord(s[i])
b = chr(a+2)
s = s.replace(s[i],b)
print(s)
# 3. #Input:- 'Python'
# #Output:- 'R{vfml'
s = 'Python'
print(s)
for i in range(len(s)):
if i <= len(s)/2-1:
a = ord(s[i])
b = chr(a+2)
s = s.replace(s[i],b)
else:
a = ord(s[i])
b = chr(a-2)
s = s.replace(s[i],b)
print(s)
# -
| Amit/Amit_String_Assignment.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# Will solve:
#
# https://github.com/CONABIO/kube_sipecam_playground/issues/14
# # Set up minikube and usage of docker image for MAD-Mex + kale in AWS
# Will follow:
#
# * For minikube: [minikube_sipecam/setup](https://github.com/CONABIO/kube_sipecam/tree/master/minikube_sipecam/setup#aws)
#
# * docker image for MAD-Mex: [kube_sipecam/dockerfiles/MAD_Mex/odc_kale](https://github.com/CONABIO/kube_sipecam/tree/master/dockerfiles/MAD_Mex/odc_kale) and [minikube_sipecam/deployments/MAD_Mex](https://github.com/CONABIO/kube_sipecam/tree/master/minikube_sipecam/deployments/MAD_Mex/)
#
# * Reference for this nbook:
#
# [1_issue_5_basic_setup_in_AWS_for_MAD_Mex_classif_pipeline](https://github.com/CONABIO/kube_sipecam_playground/blob/master/MAD_Mex/notebooks/1_issue_5_basic_setup_in_AWS_for_MAD_Mex_classif_pipeline.ipynb)
#
# [1_issue_10_basic_setup_in_AWS_for_MAD_Mex_classif_pipeline](https://github.com/CONABIO/kube_sipecam_playground/blob/master/MAD_Mex/notebooks/2_issues_and_nbooks/1_issue_10_basic_setup_in_AWS_for_MAD_Mex_classif_pipeline.ipynb.ipynb)
# Will use [minikube_sipecam/deployments/MAD_Mex/hostpath_pv](https://github.com/CONABIO/kube_sipecam/tree/master/minikube_sipecam/deployments/MAD_Mex/hostpath_pv)
# ## Instance
#
# In AWS account we can select ami: `minikube-sipecam` which has next description:
#
# *Based in k8s-1.16-debian-buster-amd64-hvm-ebs-2020-04-27 - ami-0ab39819e336a3f3f Contains kubectl 1.19.1 minikube 1.13.0 kubeflow 1.0.2*
#
# and instance `m5.2xlarge` with `100` gb of disk.
#
# Use next bash script for user data:
#
# ```
# # #!/bin/bash
# ##variables:
# region=us-west-2
# name_instance=minikube-10-09-2020
# ##System update
# apt-get update -yq
# ##Tag instance
# INSTANCE_ID=$(curl -s http://instance-data/latest/meta-data/instance-id)
# PUBLIC_IP=$(curl -s http://instance-data/latest/meta-data/public-ipv4)
# aws ec2 create-tags --resources $INSTANCE_ID --tag Key=Name,Value=$name_instance-$PUBLIC_IP --region=$region
# ```
# **Ssh to instance, all commands will be executed as root**
#
# `sudo su`
#
# **Next will start minikube and kubeflow pods:**
# ```
# # cd /root && minikube start --driver=none
#
# # cd /opt/kf-test && /root/kfctl apply -V -f kfctl_k8s_istio.v1.0.2.yaml
# ```
#
# Check pods and status with:
#
# ```
# minikube status
#
# minikube
# type: Control Plane
# host: Running
# kubelet: Running
# apiserver: Running
# kubeconfig: Configured
# ```
#
# ```
# kubectl get pods -n kubeflow
#
# #all running except:
# spark-operatorcrd-cleanup-2p7x2 0/2 Completed 0 7m6s
# ```
#
#
# **To access kubeflow UI set:**
#
# ```
# export INGRESS_HOST=$(minikube ip)
# export INGRESS_PORT=$(kubectl -n istio-system get service istio-ingressgateway -o jsonpath='{.spec.ports[?(@.name=="http2")].nodePort}')
# # echo $INGRESS_PORT
# ```
#
# **And go to:**
#
# ```
# http://<ipv4 of ec2 instance>:$INGRESS_PORT
# ```
#
#
# ## Deployments and services
#
# **Set:**
# ```
# MAD_MEX_LOAD_BALANCER_SERVICE=loadbalancer-mad-mex-0.1.0_1.7.0_0.5.0-hostpath-pv
# MAD_MEX_PV=hostpath-pv
# MAD_MEX_PVC=hostpath-pvc
# MAD_MEX_JUPYTERLAB_SERVICE=jupyterlab-mad-mex-0.1.0_1.7.0_0.5.0-hostpath-pv
# MAD_MEX_URL=https://raw.githubusercontent.com/CONABIO/kube_sipecam/master/minikube_sipecam/deployments/MAD_Mex/
# ```
# **Create storage:**
#
# ```
# kubectl create -f $MAD_MEX_URL/hostpath_pv/$MAD_MEX_PV.yaml
# kubectl create -f $MAD_MEX_URL/hostpath_pv/$MAD_MEX_PVC.yaml
# ```
# **Create service:**
# ```
# kubectl create -f $MAD_MEX_URL/hostpath_pv/$MAD_MEX_LOAD_BALANCER_SERVICE.yaml
# ```
# **Create deployment:**
# ```
# kubectl create -f $MAD_MEX_URL/hostpath_pv/$MAD_MEX_JUPYTERLAB_SERVICE.yaml
# ```
# **And go to:**
#
# ```
# http://<ipv4 of ec2 instance>:30001/madmexurl
# ```
#
# # Set up postgresql instance in AWS
# Will follow:
#
# https://github.com/CONABIO/antares3-docker/tree/master/postgresql/local_deployment
# **Clone, init DB**
# ```
# # cd /shared_volume
# dir=/shared_volume/postgresql_volume_docker
# # mkdir $dir
#
# git clone https://github.com/CONABIO/antares3-docker.git $dir/antares3-docker
#
# # mkdir -p $dir/etc/postgresql
# # mkdir -p $dir/var/log/postgresql
# # mkdir -p $dir/var/lib/postgresql
#
# docker run -v $dir/etc/postgresql:/etc/postgresql \
# -v $dir/var/log/postgresql:/var/log/postgresql \
# -v $dir/var/lib/postgresql:/var/lib/postgresql \
# -v $dir/antares3-docker/postgresql/local_deployment/conf/:/home/postgres/conf/ \
# -w /home/postgres \
# -p 2225:22 -p 2345:5432 --name postgresql-madmex-odc --hostname postgresql-madmex \
# -dit madmex/postgresql-madmex-local:v8 /bin/bash
#
# docker exec -it postgresql-madmex-odc /usr/local/bin/entrypoint.sh
# docker exec -u=postgres -it postgresql-madmex-odc /home/postgres/conf/setup.sh
# ```
#
# # Create `/shared_volume/.geonode_conabio`:
# ```
# HOST_NAME="<ipv4 DNS of ec2>"
# USER_GEOSERVER="super"
# PASSWORD_GEOSERVER="<PASSWORD>"
# PASSWORD_DB_GEONODE_DATA="<PASSWORD>"
# ```
# ## Init files for antares3 and ODC
# **Next commands in jupyterlab**
# `~/.datacube.conf`
#
# ```
# [user]
# default_environment: datacube
# #default_environment: s3aio_env
#
# [datacube]
# db_hostname: 172.17.0.1
# db_database: antares_datacube
# db_username: postgres
# db_password: <PASSWORD>
# db_port: 2345
#
#
# execution_engine.use_s3: False
#
# [s3aio_env]
# db_hostname: 172.17.0.1
# db_database: antares_datacube
# db_username: postgres
# db_password: <PASSWORD>
# db_port: 2345
#
# #index_driver: s3aio_index
#
# execution_engine.use_s3: False
# ```
# `~/.antares`
#
# ```
# # Django settings
# SECRET_KEY=<key>
# DEBUG=True
# DJANGO_LOG_LEVEL=DEBUG
# ALLOWED_HOSTS=
# # Database
# DATABASE_NAME=antares_datacube
# DATABASE_USER=postgres
# DATABASE_PASSWORD=<PASSWORD>
# DATABASE_HOST=172.17.0.1
# DATABASE_PORT=2345
# # Datacube
# SERIALIZED_OBJECTS_DIR=/shared_volume/datacube_ingest/serialized_objects/
# INGESTION_PATH=/shared_volume/datacube_ingest
# #DRIVER=s3aio
# DRIVER='NetCDF CF'
# #INGESTION_BUCKET=datacube-s2-jalisco-test
# # Query and download
# USGS_USER=<username>
# USGS_PASSWORD=<password>
# SCIHUB_USER=
# SCIHUB_PASSWORD=
# # Misc
# BIS_LICENSE=<license>
# TEMP_DIR=/shared_volume/temp
# SEGMENTATION_DIR=/shared_volume/segmentation/
# #SEGMENTATION_BUCKET=<name of bucket>
#
# ```
# **Create dir for segmentation if will hold results of that process:**
#
# `mkdir /shared_volume/segmentation/`
# **Upgrade antares with no deps:**
#
# `pip3 install --user git+https://github.com/CONABIO/antares3.git@develop --upgrade --no-deps`
# **Init antares and datacube:**
#
# ```
# ~/.local/bin/antares init
# datacube -v system init
# ```
#
# **Check:**
#
# `datacube -v system check`
# **Create spatial indexes:**
#
# ```
# apt-get install -y postgresql-client
# psql -h 172.17.0.1 -d antares_datacube -U postgres -p 2345
# #password postgres
# CREATE INDEX madmex_predictobject_gix ON public.madmex_predictobject USING GIST (the_geom);
# CREATE INDEX madmex_trainobject_gix ON public.madmex_trainobject USING GIST (the_geom);
# ```
# **There are some notes that could be followed [Notes](https://github.com/CONABIO/antares3-docker/tree/master/postgresql/local_deployment#note) for docker container of postgresql**
#
# # Register and ingest LANDSAT 8 data into ODC
# S3 bucket that has data: `landsat-images-kube-sipecam-mad-mex`
# **Prepare metadata:**
# ```
# ~/.local/bin/antares prepare_metadata --path "/" --bucket landsat-images-kube-sipecam-mad-mex --dataset_name landsat_espa --outfile /shared_volume/metadata_mex_l8.yaml --multi 2
# ```
# **Datacube ingestion:**
# ```
# datacube -v product add ~/.config/madmex/indexing/ls8_espa_scenes.yaml
# datacube -v dataset add /shared_volume/metadata_mex_l8.yaml
# datacube -v ingest -c ~/.config/madmex/ingestion/ls8_espa_mexico.yaml --executor multiproc 6
# ```
# # Register and ingest SRTM data into ODC
# Using https://conabio.github.io/antares3/example_s2_land_cover.html#prepare-terrain-metrics
# From http://dwtkns.com/srtm/ will download srtm data for Chiapas:
#
# ```
# # cd /shared_volume
# wget http://srtm.csi.cgiar.org/wp-content/uploads/files/srtm_5x5/tiff/srtm_18_09.zip
# apt-get install -y unzip
# unzip srtm_18_09.zip -d /shared_volume/srtm_18_09
# # mkdir /shared_volume/srtm_mosaic
# # cp /shared_volume/srtm_18_09/srtm_18_09.tif /shared_volume/srtm_mosaic/srtm_mosaic.tif
# gdaldem slope /shared_volume/srtm_mosaic/srtm_mosaic.tif /shared_volume/srtm_mosaic/slope_mosaic.tif -s 111120
# gdaldem aspect /shared_volume/srtm_mosaic/srtm_mosaic.tif /shared_volume/srtm_mosaic/aspect_mosaic.tif
# ```
# ## Create product and Index mosaic
#
# `datacube -v product add ~/.config/madmex/indexing/srtm_cgiar.yaml`
#
# ```
# ~/.local/bin/antares prepare_metadata --path /shared_volume/srtm_mosaic --dataset_name srtm_cgiar --outfile /shared_volume/metadata_srtm.yaml
#
# datacube -v dataset add /shared_volume/metadata_srtm.yaml
# datacube -v ingest -c ~/.config/madmex/ingestion/srtm_cgiar_mexico.yaml --executor multiproc 6
# ```
# # Ingest Mexico's shapefile to antares-datacube DB
# `~/.local/bin/antares init -c 'MEX'`
# # Ingest training data in antares-datacube DB
# **Training data is in bucket `training-data-kube-sipecam-mad-mex`**
#
# ```
# Chiapas_31.shp
# Chiapas_31.shx
# Chiapas_31.prj
# Chiapas_31.dbf
# ```
# ```
# ~/.local/bin/antares ingest_training_from_vector /shared_volume/training_data/Chiapas_31.shp --scheme madmex --year 2015 --name train_chiapas_dummy --field class
# ```
# # Deploy geonode
# **Being in EC2 instance as root**
# Following: https://github.com/CONABIO/geonode/tree/master/deployment_using_spcgeonode
# Being root `sudo su`
# Install docker-compose:
# ```
# # cd ~
# curl -L "https://github.com/docker/compose/releases/download/1.26.2/docker-compose-$(uname -s)-$(uname -m)" -o /usr/local/bin/docker-compose
# chmod +x /usr/local/bin/docker-compose
# ```
# Deploy geonode using https://github.com/CONABIO/geonode/tree/master/deployment_using_spcgeonode instructions
# When cloning repo of geonode in `/shared_volume` change `/shared_volume/geonode/scripts/spcgeonode/nginx/nginx.conf.envsubst` to `server_names_hash_bucket_size 128;` and use in `/shared_volume/geonode/scripts/spcgeonode/.env` `ipv4 dns of ec2 instance`
#
#
#
# And add rule in security groups for `80` port
# ## Deployments and services
#
# **Set:**
# ```
# GEONODE_CONABIO_LOAD_BALANCER_SERVICE=loadbalancer-geonode-conabio-0.1_0.5.0-hostpath-pv
# GEONODE_CONABIO_PV=hostpath-pv
# GEONODE_CONABIO_PVC=hostpath-pvc
# GEONODE_CONABIO_JUPYTERLAB_SERVICE_HOSTPATH_PV=jupyterlab-geonode-conabio-0.1_0.5.0-hostpath-pv
# GEONODE_CONABIO_URL=https://raw.githubusercontent.com/CONABIO/kube_sipecam/master/minikube_sipecam/deployments/geonode_conabio/
#
# ```
# **Create storage:**
#
# ```
# kubectl create -f $GEONODE_CONABIO_URL/hostpath_pv/$GEONODE_CONABIO_PV.yaml
# kubectl create -f $GEONODE_CONABIO_URL/hostpath_pv/$GEONODE_CONABIO_PVC.yaml
# ```
# **Create service:**
# ```
# kubectl create -f $GEONODE_CONABIO_URL/hostpath_pv/$GEONODE_CONABIO_LOAD_BALANCER_SERVICE.yaml
# ```
# **Create deployment:**
# ```
# kubectl create -f $GEONODE_CONABIO_URL/hostpath_pv/$GEONODE_CONABIO_JUPYTERLAB_SERVICE_HOSTPATH_PV.yaml
# ```
# **And go to:**
#
# ```
# http://<ipv4 of ec2 instance>:30002/geonodeurl
# ```
#
# # Note:
#
# If disk is full which could happen if a kubeflow pipeline will be uploaded from kale:
#
# ```
# HTTP response headers: HTTPHeaderDict({'Date': 'Tue, 01 Sep 2020 18:12:22 GMT', 'Content-Length': '487', 'Content-Type': 'text/plain; charset=utf-8'})
# HTTP response body: {"error_message":"Error creating pipeline: Create pipeline failed: InternalServerError: Failed to store b2fa5a70-cab4-4c89-8784-9c0cb118d1b4: Storage backend has reached its minimum free disk threshold. Please delete a few objects to proceed.","error_details":"Error creating pipeline: Create pipeline failed: InternalServerError: Failed to store b2fa5a70-cab4-4c89-8784-9c0cb118d1b4: Storage backend has reached its minimum free disk threshold. Please delete a few objects to proceed."}
# ```
#
# Delete kubeflow (MAD-Mex and geonode deployments)
#
# To free space:
#
# ```
# minikube stop
# minikube delete
# ```
#
# Check:
#
# ```
# docker system df
# docker system prune --all --volumes
# # rm -r /root/.minikube/*
# # rm -r /root/.kube/*
# # rm -r /opt/kf-test
# ```
#
# Start again (being in root dir):
#
# ```
# CONFIG_URI="https://raw.githubusercontent.com/kubeflow/manifests/v1.0-branch/kfdef/kfctl_k8s_istio.v1.0.2.yaml"
# source ~/.profile
# chmod gou+wrx -R /opt/
# # mkdir -p ${KF_DIR}
# #minikube start
# # cd /root && minikube start --driver=none
# #kubeflow start
# # cd ${KF_DIR}
#
# wget $CONFIG_URI
# wget https://codeload.github.com/kubeflow/manifests/tar.gz/v1.0.2 -O v1.0.2.tar.gz
#
# ```
#
# change kfctl_k8s_istio.v1.0.2.yaml at the end uri:
#
# ```
# #this section:
# repos:
# - name: manifests
# uri: https://github.com/kubeflow/manifests/archive/v1.0.2.tar.gz
# #for:
# repos:
# - name: manifests
# uri: file:///opt/kf-test/v1.0.2.tar.gz
# ```
#
# Then:
#
# ```
# kfctl apply -V -f kfctl_k8s_istio.v1.0.2.yaml
# ```
#
#
#
# ref: https://github.com/aws-samples/eks-workshop/issues/639
#
# If there's problems with geonode (because stack of docker-compose was deleted, clone again repo and deploy geonode)
| MAD_Mex/notebooks/3_issues_and_nbooks/1_issue_14_basic_setup_in_AWS_for_MAD_Mex_classif_pipeline.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Occupation
# ### Introduction:
#
# Special thanks to: https://github.com/justmarkham for sharing the dataset and materials.
#
# ### Step 1. Import the necessary libraries
# + jupyter={"outputs_hidden": false}
# -
# ### Step 2. Import the dataset from this [address](https://raw.githubusercontent.com/justmarkham/DAT8/master/data/u.user).
# ### Step 3. Assign it to a variable called users.
# + jupyter={"outputs_hidden": false}
# -
# ### Step 4. Discover what is the mean age per occupation
# + jupyter={"outputs_hidden": false}
# -
# ### Step 5. Discover the Male ratio per occupation and sort it from the most to the least
# + jupyter={"outputs_hidden": false}
# -
# ### Step 6. For each occupation, calculate the minimum and maximum ages
# + jupyter={"outputs_hidden": false}
# -
# ### Step 7. For each combination of occupation and gender, calculate the mean age
# + jupyter={"outputs_hidden": false}
# -
# ### Step 8. For each occupation present the percentage of women and men
# + jupyter={"outputs_hidden": false}
| 03_Grouping/Occupation/Solutions.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# Full netCDF4 module documentation http://unidata.github.io/netcdf4-python/
# Example code taken from http://pyhogs.github.io/intro_netcdf4.html
from netCDF4 import Dataset
import numpy as np
import netCDF4 as nc4
import arrow
region_data = ["CH", "LI"]
tech_data = ["WPP1"]
timestep_data = ["2010-01-01 01:00","2010-01-01 02:00"]
supply_data = [np.matrix([[1, 2],[0, 0]])]
demand_data = np.matrix([[0.5, 1.7],[2, 2]])
f = nc4.Dataset('D:/netcdf_datapackage.nc','w', format='NETCDF4') #'w' stands for write
# Global dimensions will be added to the Dataset
f.createDimension('region', len(region_data))
f.createDimension('timestep', len(timestep_data))
f.createDimension('tech', len(tech_data))
region = f.createVariable('Region', 'S4', 'region')
timestep = f.createVariable('Timestep', 'S12', 'timestep')
tech = f.createVariable('Technology', 'S8', 'tech')
for i, ireg in enumerate(region_data):
region[i] = ireg
for i, itime in enumerate(timestep_data):
timestep[i] = itime
for i, itech in enumerate(tech_data):
tech[i] = itech
# Scenario data is added to the scenario group
# groups represent the hirarchical structure in netcdf4 files
tempgrp = f.createGroup('Scenario_1')
supply = tempgrp.createVariable('Temperature', 'f4', ('timestep', 'region', 'tech'))
demand = tempgrp.createVariable('Demand', 'f4', ('timestep', 'region'))
supply[:,:,:] = supply_data
demand[:,:] = demand_data
#Add local attributes to variable instances
supply.units = 'MWh'
demand.units = 'MWh'
#Add global attributes
f.description = "NetCDF4 example data for 8th OpenMod"
f.history = "Created {}".format(arrow.utcnow().format('YYYY-MM-DD HH:mm:ss ZZ'))
f.close()
# -
# Open the netcdf file and navigate the Scenario_1 group
with nc4.Dataset('D:/netcdf_datapackage.nc','r') as f:
print(f['Scenario_1'].variables)
| examples/netcdf4_datapackage/NetCDF4_writer.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
from pyspark.sql import SparkSession
spark = SparkSession.builder.appName('Basics').getOrCreate()
df = spark.read.json('Spark_DataFrames/people.json')
df.show()
df.printSchema()
df.columns
df.describe()
df.describe().show()
from pyspark.sql.types import (StructField, StructType,
StringType, IntegerType)
data_schema = [StructField('age', IntegerType(), True),
StructField('name', StringType(), True)]
final_schema = StructType(fields=data_schema)
final_schema
df = spark.read.json('Spark_DataFrames/people.json', schema=final_schema)
df.printSchema()
type(df.select('age'))
type(df.head(2)[0])
df.withColumn('newage', df['age']+2).show()
df.createOrReplaceTempView('people')
results = spark.sql("SELECT * FROM people")
results.show()
new_results = spark.sql("SELECT * FROM people WHERE age >=30")
new_results.show()
| src/pySpark/pyspark_01.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# +
import json
from pprint import pprint
import requests
from requests.packages.urllib3.exceptions import InsecureRequestWarning
requests.packages.urllib3.disable_warnings(InsecureRequestWarning)
service_load_balancer = 'http://localhost:8000'
# -
# *We are going to take the time to clear out any existing reminders*
response = requests.get(service_load_balancer, verify=False).json()
pprint(response)
for reminder in response['reminders']:
requests.delete((service_load_balancer + '/{}'.format(reminder['reminder_id'])),
verify=False)
response = requests.get(service_load_balancer, verify=False).json()
pprint(response)
demo_reminder = {'contact_number': 'my_did_goes_here', # fill me in
'appointment_time': '2016-08-08T16:30-0500',
'notify_window': 1,
'location': 'Lucerne',
'particpant': 'ClueCon'}
demo_post_response = requests.post(service_load_balancer,
json=demo_reminder, verify=False)
demo_reminder_id = demo_post_response.json()['reminder_id']
pprint(demo_post_response.text)
demo_get_response = requests.get((service_load_balancer + '/{}'.format(demo_reminder_id)), verify=False)
pprint(demo_get_response.json())
resp = requests.delete((service_load_balancer + '/{}'.format(demo_reminder_id)),
verify=False)
print(resp.text)
pizza_party_reminder = {'contact_number': 'my_did_goes_here', # fill me in
'appointment_time': '2016-08-08T19:00-0500',
'notify_window': 3,
'location': "Giovanni's",
'participant': '<NAME>'}
pizza_party_post_response = requests.post(service_load_balancer,
json=pizza_party_reminder, verify=False)
pizza_party_reminder_id = pizza_party_post_response.json()['reminder_id']
pprint(pizza_party_post_response.text)
party_reminder_get_response = requests.get(
(service_load_balancer + '/{}'.format(pizza_party_reminder_id)),
verify=False)
pprint(party_reminder_get_response.json())
| SMS-Reminder.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: dl
# language: python
# name: dl
# ---
# # Skip-gram Word2Vec
#
# In this notebook, I'll lead you through using PyTorch to implement the [Word2Vec algorithm](https://en.wikipedia.org/wiki/Word2vec) using the skip-gram architecture. By implementing this, you'll learn about embedding words for use in natural language processing. This will come in handy when dealing with things like machine translation.
#
# ## Readings
#
# Here are the resources I used to build this notebook. I suggest reading these either beforehand or while you're working on this material.
#
# * A really good [conceptual overview](http://mccormickml.com/2016/04/19/word2vec-tutorial-the-skip-gram-model/) of Word2Vec from <NAME>
# * [First Word2Vec paper](https://arxiv.org/pdf/1301.3781.pdf) from Mikolov et al.
# * [Neural Information Processing Systems, paper](http://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf) with improvements for Word2Vec also from Mikolov et al.
#
# ---
# ## Word embeddings
#
# When you're dealing with words in text, you end up with tens of thousands of word classes to analyze; one for each word in a vocabulary. Trying to one-hot encode these words is massively inefficient because most values in a one-hot vector will be set to zero. So, the matrix multiplication that happens in between a one-hot input vector and a first, hidden layer will result in mostly zero-valued hidden outputs.
#
# To solve this problem and greatly increase the efficiency of our networks, we use what are called **embeddings**. Embeddings are just a fully connected layer like you've seen before. We call this layer the embedding layer and the weights are embedding weights. We skip the multiplication into the embedding layer by instead directly grabbing the hidden layer values from the weight matrix. We can do this because the multiplication of a one-hot encoded vector with a matrix returns the row of the matrix corresponding the index of the "on" input unit.
#
# <img src='assets/lookup_matrix.png' width=50%>
#
# Instead of doing the matrix multiplication, we use the weight matrix as a lookup table. We encode the words as integers, for example "heart" is encoded as 958, "mind" as 18094. Then to get hidden layer values for "heart", you just take the 958th row of the embedding matrix. This process is called an **embedding lookup** and the number of hidden units is the **embedding dimension**.
#
# There is nothing magical going on here. The embedding lookup table is just a weight matrix. The embedding layer is just a hidden layer. The lookup is just a shortcut for the matrix multiplication. The lookup table is trained just like any weight matrix.
#
# Embeddings aren't only used for words of course. You can use them for any model where you have a massive number of classes. A particular type of model called **Word2Vec** uses the embedding layer to find vector representations of words that contain semantic meaning.
# ---
# ## Word2Vec
#
# The Word2Vec algorithm finds much more efficient representations by finding vectors that represent the words. These vectors also contain semantic information about the words.
#
# <img src="assets/context_drink.png" width=40%>
#
# Words that show up in similar **contexts**, such as "coffee", "tea", and "water" will have vectors near each other. Different words will be further away from one another, and relationships can be represented by distance in vector space.
#
#
# There are two architectures for implementing Word2Vec:
# >* CBOW (Continuous Bag-Of-Words) and
# * Skip-gram
#
# <img src="assets/word2vec_architectures.png" width=60%>
#
# In this implementation, we'll be using the **skip-gram architecture** with **negative sampling** because it performs better than CBOW and trains faster with negative sampling. Here, we pass in a word and try to predict the words surrounding it in the text. In this way, we can train the network to learn representations for words that show up in similar contexts.
# ---
# ## Loading Data
#
# Next, we'll ask you to load in data and place it in the `data` directory
#
# 1. Load the [text8 dataset](https://s3.amazonaws.com/video.udacity-data.com/topher/2018/October/5bbe6499_text8/text8.zip); a file of cleaned up *Wikipedia article text* from <NAME>.
# 2. Place that data in the `data` folder in the home directory.
# 3. Then you can extract it and delete the archive, zip file to save storage space.
#
# After following these steps, you should have one file in your data directory: `data/text8`.
# +
# read in the extracted text file
with open('data/text8') as f:
text = f.read()
# print out the first 100 characters
print(text[:100])
# -
# ## Pre-processing
#
# Here I'm fixing up the text to make training easier. This comes from the `utils.py` file. The `preprocess` function does a few things:
# >* It converts any punctuation into tokens, so a period is changed to ` <PERIOD> `. In this data set, there aren't any periods, but it will help in other NLP problems.
# * It removes all words that show up five or *fewer* times in the dataset. This will greatly reduce issues due to noise in the data and improve the quality of the vector representations.
# * It returns a list of words in the text.
#
# This may take a few seconds to run, since our text file is quite large. If you want to write your own functions for this stuff, go for it!
# +
import utils
# get list of words
words = utils.preprocess(text)
print(words[:30])
# -
# print some stats about this word data
print("Total words in text: {}".format(len(words)))
print("Unique words: {}".format(len(set(words)))) # `set` removes any duplicate words
# ### Dictionaries
#
# Next, I'm creating two dictionaries to convert words to integers and back again (integers to words). This is again done with a function in the `utils.py` file. `create_lookup_tables` takes in a list of words in a text and returns two dictionaries.
# >* The integers are assigned in descending frequency order, so the most frequent word ("the") is given the integer 0 and the next most frequent is 1, and so on.
#
# Once we have our dictionaries, the words are converted to integers and stored in the list `int_words`.
# +
vocab_to_int, int_to_vocab = utils.create_lookup_tables(words)
int_words = [vocab_to_int[word] for word in words]
print(int_words[:30])
# -
# ## Subsampling
#
# Words that show up often such as "the", "of", and "for" don't provide much context to the nearby words. If we discard some of them, we can remove some of the noise from our data and in return get faster training and better representations. This process is called subsampling by Mikolov. For each word $w_i$ in the training set, we'll discard it with probability given by
#
# $$ P(w_i) = 1 - \sqrt{\frac{t}{f(w_i)}} $$
#
# where $t$ is a threshold parameter and $f(w_i)$ is the frequency of word $w_i$ in the total dataset.
#
# > Implement subsampling for the words in `int_words`. That is, go through `int_words` and discard each word given the probablility $P(w_i)$ shown above. Note that $P(w_i)$ is the probability that a word is discarded. Assign the subsampled data to `train_words`.
# +
from collections import Counter
import random
import numpy as np
threshold = 1e-5
word_counts = Counter(int_words)
#print(list(word_counts.items())[0]) # dictionary of int_words, how many times they appear
total_count = len(int_words)
freqs = {word: count/total_count for word, count in word_counts.items()}
p_drop = {word: 1 - np.sqrt(threshold/freqs[word]) for word in word_counts}
# discard some frequent words, according to the subsampling equation
# create a new list of words for training
train_words = [word for word in int_words if random.random() < (1 - p_drop[word])]
print(train_words[:30])
# -
# ## Making batches
# Now that our data is in good shape, we need to get it into the proper form to pass it into our network. With the skip-gram architecture, for each word in the text, we want to define a surrounding _context_ and grab all the words in a window around that word, with size $C$.
#
# From [Mikolov et al.](https://arxiv.org/pdf/1301.3781.pdf):
#
# "Since the more distant words are usually less related to the current word than those close to it, we give less weight to the distant words by sampling less from those words in our training examples... If we choose $C = 5$, for each training word we will select randomly a number $R$ in range $[ 1: C ]$, and then use $R$ words from history and $R$ words from the future of the current word as correct labels."
#
# > **Exercise:** Implement a function `get_target` that receives a list of words, an index, and a window size, then returns a list of words in the window around the index. Make sure to use the algorithm described above, where you chose a random number of words to from the window.
#
# Say, we have an input and we're interested in the idx=2 token, `741`:
# ```
# [5233, 58, 741, 10571, 27349, 0, 15067, 58112, 3580, 58, 10712]
# ```
#
# For `R=2`, `get_target` should return a list of four values:
# ```
# [5233, 58, 10571, 27349]
# ```
def get_target(words, idx, window_size=5):
''' Get a list of words in a window around an index. '''
R = np.random.randint(1, window_size+1)
start = idx - R if (idx - R) > 0 else 0
stop = idx + R
target_words = words[start:idx] + words[idx+1:stop+1]
return list(target_words)
# +
# test your code!
# run this cell multiple times to check for random window selection
int_text = [i for i in range(10)]
print('Input: ', int_text)
idx=5 # word index of interest
target = get_target(int_text, idx=idx, window_size=5)
print('Target: ', target) # you should get some indices around the idx
# -
# ### Generating Batches
#
# Here's a generator function that returns batches of input and target data for our model, using the `get_target` function from above. The idea is that it grabs `batch_size` words from a words list. Then for each of those batches, it gets the target words in a window.
def get_batches(words, batch_size, window_size=5):
''' Create a generator of word batches as a tuple (inputs, targets) '''
n_batches = len(words)//batch_size
# only full batches
words = words[:n_batches*batch_size]
for idx in range(0, len(words), batch_size):
x, y = [], []
batch = words[idx:idx+batch_size]
for ii in range(len(batch)):
batch_x = batch[ii]
batch_y = get_target(batch, ii, window_size)
y.extend(batch_y)
x.extend([batch_x]*len(batch_y))
yield x, y
# +
int_text = [i for i in range(20)]
x,y = next(get_batches(int_text, batch_size=4, window_size=5))
print('x\n', x)
print('y\n', y)
# -
# ---
# ## Validation
#
# Here, I'm creating a function that will help us observe our model as it learns. We're going to choose a few common words and few uncommon words. Then, we'll print out the closest words to them using the cosine similarity:
#
# <img src="assets/two_vectors.png" width=30%>
#
# $$
# \mathrm{similarity} = \cos(\theta) = \frac{\vec{a} \cdot \vec{b}}{|\vec{a}||\vec{b}|}
# $$
#
#
# We can encode the validation words as vectors $\vec{a}$ using the embedding table, then calculate the similarity with each word vector $\vec{b}$ in the embedding table. With the similarities, we can print out the validation words and words in our embedding table semantically similar to those words. It's a nice way to check that our embedding table is grouping together words with similar semantic meanings.
def cosine_similarity(embedding, valid_size=16, valid_window=100, device='cpu'):
""" Returns the cosine similarity of validation words with words in the embedding matrix.
Here, embedding should be a PyTorch embedding module.
"""
# Here we're calculating the cosine similarity between some random words and
# our embedding vectors. With the similarities, we can look at what words are
# close to our random words.
# sim = (a . b) / |a||b|
embed_vectors = embedding.weight
# magnitude of embedding vectors, |b|
magnitudes = embed_vectors.pow(2).sum(dim=1).sqrt().unsqueeze(0)
# pick N words from our ranges (0,window) and (1000,1000+window). lower id implies more frequent
valid_examples = np.array(random.sample(range(valid_window), valid_size//2))
valid_examples = np.append(valid_examples,
random.sample(range(1000,1000+valid_window), valid_size//2))
valid_examples = torch.LongTensor(valid_examples).to(device)
valid_vectors = embedding(valid_examples)
similarities = torch.mm(valid_vectors, embed_vectors.t())/magnitudes
return valid_examples, similarities
# ---
# # SkipGram model
#
# Define and train the SkipGram model.
# > You'll need to define an [embedding layer](https://pytorch.org/docs/stable/nn.html#embedding) and a final, softmax output layer.
#
# An Embedding layer takes in a number of inputs, importantly:
# * **num_embeddings** – the size of the dictionary of embeddings, or how many rows you'll want in the embedding weight matrix
# * **embedding_dim** – the size of each embedding vector; the embedding dimension
#
# Below is an approximate diagram of the general structure of our network.
# <img src="assets/skip_gram_arch.png" width=60%>
#
# >* The input words are passed in as batches of input word tokens.
# * This will go into a hidden layer of linear units (our embedding layer).
# * Then, finally into a softmax output layer.
#
# We'll use the softmax layer to make a prediction about the context words by sampling, as usual.
# ---
# ## Negative Sampling
#
# For every example we give the network, we train it using the output from the softmax layer. That means for each input, we're making very small changes to millions of weights even though we only have one true example. This makes training the network very inefficient. We can approximate the loss from the softmax layer by only updating a small subset of all the weights at once. We'll update the weights for the correct example, but only a small number of incorrect, or noise, examples. This is called ["negative sampling"](http://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf).
#
# There are two modifications we need to make. First, since we're not taking the softmax output over all the words, we're really only concerned with one output word at a time. Similar to how we use an embedding table to map the input word to the hidden layer, we can now use another embedding table to map the hidden layer to the output word. Now we have two embedding layers, one for input words and one for output words. Secondly, we use a modified loss function where we only care about the true example and a small subset of noise examples.
#
# $$
# - \large \log{\sigma\left(u_{w_O}\hspace{0.001em}^\top v_{w_I}\right)} -
# \sum_i^N \mathbb{E}_{w_i \sim P_n(w)}\log{\sigma\left(-u_{w_i}\hspace{0.001em}^\top v_{w_I}\right)}
# $$
#
# This is a little complicated so I'll go through it bit by bit. $u_{w_O}\hspace{0.001em}^\top$ is the embedding vector for our "output" target word (transposed, that's the $^\top$ symbol) and $v_{w_I}$ is the embedding vector for the "input" word. Then the first term
#
# $$\large \log{\sigma\left(u_{w_O}\hspace{0.001em}^\top v_{w_I}\right)}$$
#
# says we take the log-sigmoid of the inner product of the output word vector and the input word vector. Now the second term, let's first look at
#
# $$\large \sum_i^N \mathbb{E}_{w_i \sim P_n(w)}$$
#
# This means we're going to take a sum over words $w_i$ drawn from a noise distribution $w_i \sim P_n(w)$. The noise distribution is basically our vocabulary of words that aren't in the context of our input word. In effect, we can randomly sample words from our vocabulary to get these words. $P_n(w)$ is an arbitrary probability distribution though, which means we get to decide how to weight the words that we're sampling. This could be a uniform distribution, where we sample all words with equal probability. Or it could be according to the frequency that each word shows up in our text corpus, the unigram distribution $U(w)$. The authors found the best distribution to be $U(w)^{3/4}$, empirically.
#
# Finally, in
#
# $$\large \log{\sigma\left(-u_{w_i}\hspace{0.001em}^\top v_{w_I}\right)},$$
#
# we take the log-sigmoid of the negated inner product of a noise vector with the input vector.
#
# <img src="assets/neg_sampling_loss.png" width=50%>
#
# To give you an intuition for what we're doing here, remember that the sigmoid function returns a probability between 0 and 1. The first term in the loss pushes the probability that our network will predict the correct word $w_O$ towards 1. In the second term, since we are negating the sigmoid input, we're pushing the probabilities of the noise words towards 0.
import torch
from torch import nn
import torch.optim as optim
class SkipGramNeg(nn.Module):
def __init__(self, n_vocab, n_embed, noise_dist=None):
super().__init__()
self.n_vocab = n_vocab
self.n_embed = n_embed
self.noise_dist = noise_dist
# define embedding layers for input and output words
self.in_embed = nn.Embedding(n_vocab, n_embed)
self.out_embed = nn.Embedding(n_vocab, n_embed)
# Initialize both embedding tables with uniform distribution
self.in_embed.weight.data.uniform_(-1, 1)
self.out_embed.weight.data.uniform_(-1, 1)
def forward_input(self, input_words):
# return input vector embeddings
input_vectors = self.in_embed(input_words)
return input_vectors
def forward_output(self, output_words):
# return output vector embeddings
output_vectors = self.out_embed(output_words)
return output_vectors
def forward_noise(self, batch_size, n_samples):
""" Generate noise vectors with shape (batch_size, n_samples, n_embed)"""
if self.noise_dist is None:
# Sample words uniformly
noise_dist = torch.ones(self.n_vocab)
else:
noise_dist = self.noise_dist
# Sample words from our noise distribution
noise_words = torch.multinomial(noise_dist,
batch_size * n_samples,
replacement=True)
device = "cuda" if model.out_embed.weight.is_cuda else "cpu"
noise_words = noise_words.to(device)
## TODO: get the noise embeddings
# reshape the embeddings so that they have dims (batch_size, n_samples, n_embed)
noise_vectors = self.out_embed(noise_words).view(batch_size, n_samples, self.n_embed)
return noise_vectors
class NegativeSamplingLoss(nn.Module):
def __init__(self):
super().__init__()
def forward(self, input_vectors, output_vectors, noise_vectors):
batch_size, embed_size = input_vectors.shape
# Input vectors should be a batch of column vectors
input_vectors = input_vectors.view(batch_size, embed_size, 1)
# Output vectors should be a batch of row vectors
output_vectors = output_vectors.view(batch_size, 1, embed_size)
# bmm = batch matrix multiplication
# correct log-sigmoid loss
out_loss = torch.bmm(output_vectors, input_vectors).sigmoid().log()
out_loss = out_loss.squeeze()
# incorrect log-sigmoid loss
noise_loss = torch.bmm(noise_vectors.neg(), input_vectors).sigmoid().log()
noise_loss = noise_loss.squeeze().sum(1) # sum the losses over the sample of noise vectors
# negate and sum correct and noisy log-sigmoid losses
# return average batch loss
return -(out_loss + noise_loss).mean()
# ### Training
#
# Below is our training loop, and I recommend that you train on GPU, if available.
# +
device = 'cuda' if torch.cuda.is_available() else 'cpu'
# Get our noise distribution
# Using word frequencies calculated earlier in the notebook
word_freqs = np.array(sorted(freqs.values(), reverse=True))
unigram_dist = word_freqs/word_freqs.sum()
noise_dist = torch.from_numpy(unigram_dist**(0.75)/np.sum(unigram_dist**(0.75)))
# instantiating the model
embedding_dim = 300
model = SkipGramNeg(len(vocab_to_int), embedding_dim, noise_dist=noise_dist).to(device)
# using the loss that we defined
criterion = NegativeSamplingLoss()
optimizer = optim.Adam(model.parameters(), lr=0.003)
print_every = 1500
steps = 0
epochs = 5
# train for some number of epochs
for e in range(epochs):
# get our input, target batches
for input_words, target_words in get_batches(train_words, 512):
steps += 1
inputs, targets = torch.LongTensor(input_words), torch.LongTensor(target_words)
inputs, targets = inputs.to(device), targets.to(device)
# input, outpt, and noise vectors
input_vectors = model.forward_input(inputs)
output_vectors = model.forward_output(targets)
noise_vectors = model.forward_noise(inputs.shape[0], 5)
# negative sampling loss
loss = criterion(input_vectors, output_vectors, noise_vectors)
optimizer.zero_grad()
loss.backward()
optimizer.step()
# loss stats
if steps % print_every == 0:
print("Epoch: {}/{}".format(e+1, epochs))
print("Loss: ", loss.item()) # avg batch loss at this point in training
valid_examples, valid_similarities = cosine_similarity(model.in_embed, device=device)
_, closest_idxs = valid_similarities.topk(6)
valid_examples, closest_idxs = valid_examples.to('cpu'), closest_idxs.to('cpu')
for ii, valid_idx in enumerate(valid_examples):
closest_words = [int_to_vocab[idx.item()] for idx in closest_idxs[ii]][1:]
print(int_to_vocab[valid_idx.item()] + " | " + ', '.join(closest_words))
print("...\n")
# -
# ## Visualizing the word vectors
#
# Below we'll use T-SNE to visualize how our high-dimensional word vectors cluster together. T-SNE is used to project these vectors into two dimensions while preserving local stucture. Check out [this post from <NAME>](http://colah.github.io/posts/2014-10-Visualizing-MNIST/) to learn more about T-SNE and other ways to visualize high-dimensional data.
# +
# %matplotlib inline
# %config InlineBackend.figure_format = 'retina'
import matplotlib.pyplot as plt
from sklearn.manifold import TSNE
# -
# getting embeddings from the embedding layer of our model, by name
embeddings = model.in_embed.weight.to('cpu').data.numpy()
viz_words = 380
tsne = TSNE()
embed_tsne = tsne.fit_transform(embeddings[:viz_words, :])
fig, ax = plt.subplots(figsize=(16, 16))
for idx in range(viz_words):
plt.scatter(*embed_tsne[idx, :], color='steelblue')
plt.annotate(int_to_vocab[idx], (embed_tsne[idx, 0], embed_tsne[idx, 1]), alpha=0.7)
| word2vec-embeddings/Negative_Sampling_Exercise.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# In this tutorial we'll use pandas and seaborn. If you haven't already, you can install them with:
#
# ```shell
# pip install pandas
# pip install seaborn
# ```
# +
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import torch
import os
# Used to make paths work both in Windows and Unix
path = lambda path: os.path.join(*path.split('/'))
# -
# # Download and preprocess dataset
# !mkdir datasets
# !mkdir datasets/concrete
# !wget -cO - \
# https://archive.ics.uci.edu/ml/machine-learning-databases/concrete/compressive/Concrete_Data.xls \
# > datasets/concrete/data.xls
# +
# import some data to play with
df = pd.read_excel('datasets/concrete/data.xls')
df.columns = [
'cement',
'blast_furnace_slag',
'fly_ash',
'water',
'superplasticizer',
'coarse_aggregate',
'fine_aggregate',
'age',
'concrete_compressive_strength'
]
df.head()
# -
df.dtypes
# We remove age because it's a discrete, ordinal variable.
# These can still be modelled with flows, but they tend to be troublesome.
# For the purposes of this tutorial, we'll skip them.
df = df.drop('age', 1)
# +
# Plot histograms
_, axes = plt.subplots(2, 4, figsize=(6 * 4, 4 * 2))
axes = axes.flatten()
for i, ax in enumerate(axes):
ax.hist(df.iloc[:, i])
ax.set_title(df.columns[i])
# -
# Look at variable correlations
sns.heatmap(df.corr(), vmin=-1, vmax=1, cmap='twilight')
# +
# The biggest correlations are:
# water - superplasticizer
# cement - concrete-compressive-strength
_, axes = plt.subplots(1, 2, figsize=(6 * 2, 4))
df.plot.scatter('water', 'superplasticizer', ax=axes[0], alpha=.1)
df.plot.scatter('cement', 'concrete_compressive_strength', ax=axes[1], alpha=.1)
# -
# # Prepare torch dataset
# Let's transform our dataset into a torch Tensor, and divide into train, validation and test.
#
# We'll then create a simple flow that learns the dataset distribution.
# +
X = torch.Tensor(df.values)
np.random.seed(123)
torch.random.manual_seed(123)
split = np.random.choice(range(3), replace=True, size=len(X), p=[.75, .15, .1]) # train/val/test
split = torch.Tensor(split) # to tensor
trainX = X[split == 0]
valX = X[split == 1]
testX = X[split == 2]
# -
# Our flow will be a MADE - DSF combination.
#
# But, before that, it is important to normalize the original distribution.
# That's why we use BatchNorm first.
#
# To concatenate several flows together, use the Sequential flow.
# Its syntax is equivalent to torch.nn.Sequential.
#
# Finally, we will train with the included train function, that trains flows with early stopping.
from flow.flow import Sequential
from flow.conditioner import MADE
from flow.transformer import DSF
from flow.modules import BatchNorm
from flow.training import train, plot_losses, test_nll, get_device
# +
device = get_device() # returns cuda if available, cpu otherwise
dim = trainX.size(1) # dimension of the flow, 4 in this case
flow = Sequential(
BatchNorm(dim=dim), # to normalize distribution
MADE(DSF(dim=dim)) # the actual transformation
).to(device)
# +
train_losses, val_losses = train(flow, trainX, valX, patience=100)
plot_losses(train_losses, val_losses)
# -
# Now that the flow is trained, we can sample and compute log-likelihoods with it.
test_nll(flow, testX) # average negative log-likelihood (nll) of the test set
# +
# Want to compute the nll of any sample? Use .nll(sample)
with torch.no_grad(): # no need for gradients now
nll = flow.nll(testX.to(device)) # remember to move the tensor to the flow's device
nll[:5] # show the first 5 nlls of the test set
# +
# Want to generate new samples from the learned distribution? Use .sample(N)
with torch.no_grad():
sample = flow.sample(1000)
# This returns a tensor in the flow's device.
# Let's move it to numpy and transform it to a DataFrame
# to plot its histograms and its correlation heatmap
sample = pd.DataFrame(
sample.cpu().numpy(),
columns=df.columns
)
# +
# Plot histograms
_, axes = plt.subplots(2, 4, figsize=(6 * 4, 4 * 2))
axes = axes.flatten()
for i, ax in enumerate(axes):
ax.hist(df.iloc[:, i], label='real', alpha=.75)
ax.hist(sample.iloc[:, i], label='fake', alpha=.75)
ax.set_title(df.columns[i])
ax.legend()
# +
# Look at variable correlations
_, axes = plt.subplots(1, 2, figsize=(6 * 2, 4))
sns.heatmap(df.corr(), vmin=-1, vmax=1, cmap='twilight', ax=axes[0])
axes[0].set_title('real')
sns.heatmap(sample.corr(), vmin=-1, vmax=1, cmap='twilight', ax=axes[1], yticklabels=False)
axes[1].set_title('fake');
# +
# Check original and sample scatterplots
_, axes = plt.subplots(2, 2, figsize=(6 * 2, 4 * 2))
df.plot.scatter('water', 'superplasticizer', ax=axes[0, 0], alpha=.1, title='real')
sample.plot.scatter('water', 'superplasticizer', ax=axes[0, 1], alpha=.1, title='fake')
df.plot.scatter('cement', 'concrete_compressive_strength', ax=axes[1, 0], alpha=.1)
sample.plot.scatter('cement', 'concrete_compressive_strength', ax=axes[1, 1], alpha=.1)
| tutorials/2 - Train flows - concrete.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "b6359e23-7c62-445c-b681-477321cbb9a3", "showTitle": false, "title": ""}
# # XGBoost training
# This is an auto-generated notebook. To reproduce these results, attach this notebook to the **10-3-ML-Cluster** cluster and rerun it.
# - Compare trials in the [MLflow experiment](#mlflow/experiments/406583024052808/s?orderByKey=metrics.%60val_f1_score%60&orderByAsc=false)
# - Navigate to the parent notebook [here](#notebook/406583024052798) (If you launched the AutoML experiment using the Experiments UI, this link isn't very useful.)
# - Clone this notebook into your project folder by selecting **File > Clone** in the notebook toolbar.
#
# Runtime Version: _10.3.x-cpu-ml-scala2.12_
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "f489ff0e-8577-4808-9059-d2c0eba3c133", "showTitle": false, "title": ""}
import mlflow
import databricks.automl_runtime
# Use MLflow to track experiments
mlflow.set_experiment("/Users/<EMAIL>/databricks_automl/label_news_articles_csv-2022_03_12-15_38")
target_col = "label"
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "3d0c6d31-5159-49b8-b20f-1f648688f328", "showTitle": false, "title": ""}
# ## Load Data
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "7ad90423-7c24-42a0-aff0-62b6a24167a7", "showTitle": false, "title": ""}
from mlflow.tracking import MlflowClient
import os
import uuid
import shutil
import pandas as pd
# Create temp directory to download input data from MLflow
input_temp_dir = os.path.join(os.environ["SPARK_LOCAL_DIRS"], "tmp", str(uuid.uuid4())[:8])
os.makedirs(input_temp_dir)
# Download the artifact and read it into a pandas DataFrame
input_client = MlflowClient()
input_data_path = input_client.download_artifacts("c2dfe80b419d4a8dbc88a90e3274369a", "data", input_temp_dir)
df_loaded = pd.read_parquet(os.path.join(input_data_path, "training_data"))
# Delete the temp data
shutil.rmtree(input_temp_dir)
# Preview data
df_loaded.head(5)
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "1a014d7b-a090-4985-b6aa-44fd5481145d", "showTitle": false, "title": ""}
df_loaded.head(1).to_dict()
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "075f6d94-3911-4fb4-8ab8-5876d10fc4b4", "showTitle": false, "title": ""}
# ### Select supported columns
# Select only the columns that are supported. This allows us to train a model that can predict on a dataset that has extra columns that are not used in training.
# `[]` are dropped in the pipelines. See the Alerts tab of the AutoML Experiment page for details on why these columns are dropped.
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "46c0a724-febf-4e58-b34a-f527bf66c85a", "showTitle": false, "title": ""}
from databricks.automl_runtime.sklearn.column_selector import ColumnSelector
supported_cols = ["text_without_stopwords", "published", "language", "main_img_url", "site_url", "hasImage", "title_without_stopwords", "text", "title", "type", "author"]
col_selector = ColumnSelector(supported_cols)
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "db576856-ca24-46bb-b144-db7181b1a741", "showTitle": false, "title": ""}
# ## Preprocessors
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "9d47028f-66ca-4236-909f-5b6363b3b879", "showTitle": false, "title": ""}
transformers = []
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "ffec277f-4679-4e97-a6e1-5cf011b277c3", "showTitle": false, "title": ""}
# ### Categorical columns
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "e5318b8e-8cb5-42ec-99df-60b036f447a0", "showTitle": false, "title": ""}
# #### Low-cardinality categoricals
# Convert each low-cardinality categorical column into multiple binary columns through one-hot encoding.
# For each input categorical column (string or numeric), the number of output columns is equal to the number of unique values in the input column.
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "9b213020-c4a9-4f9a-a662-21e359d8b417", "showTitle": false, "title": ""}
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import OneHotEncoder
one_hot_encoder = OneHotEncoder(handle_unknown="ignore")
transformers.append(("onehot", one_hot_encoder, ["published", "language", "site_url", "hasImage", "title", "title_without_stopwords", "text_without_stopwords"]))
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "000b220c-9aa5-48ca-81c0-e4cbda653b70", "showTitle": false, "title": ""}
# #### Medium-cardinality categoricals
# Convert each medium-cardinality categorical column into a numerical representation.
# Each string column is hashed to 1024 float columns.
# Each numeric column is imputed with zeros.
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "40c3ca0d-e7ef-4d3f-84c6-0c29983daed0", "showTitle": false, "title": ""}
from sklearn.feature_extraction import FeatureHasher
from sklearn.impute import SimpleImputer
from sklearn.pipeline import Pipeline
for feature in ["text", "main_img_url"]:
hash_transformer = Pipeline(steps=[
("imputer", SimpleImputer(missing_values=None, strategy="constant", fill_value="")),
(f"{feature}_hasher", FeatureHasher(n_features=1024, input_type="string"))])
transformers.append((f"{feature}_hasher", hash_transformer, [feature]))
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "1d3d483a-464e-4ddf-ae19-61b7402c9eec", "showTitle": false, "title": ""}
# ### Text features
# Convert each feature to a fixed-length vector using TF-IDF vectorization. The length of the output
# vector is equal to 1024. Each column corresponds to one of the top word n-grams
# where n is in the range [1, 2].
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "941e66c0-dbc2-4aaa-a42c-68a276eb1b5b", "showTitle": false, "title": ""}
import numpy as np
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.impute import SimpleImputer
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import FunctionTransformer
for col in {'type', 'author'}:
vectorizer = Pipeline(steps=[
("imputer", SimpleImputer(missing_values=None, strategy="constant", fill_value="")),
# Reshape to 1D since SimpleImputer changes the shape of the input to 2D
("reshape", FunctionTransformer(np.reshape, kw_args={"newshape":-1})),
("tfidf", TfidfVectorizer(decode_error="ignore", ngram_range = (1, 2), max_features=1024))])
transformers.append((f"text_{col}", vectorizer, [col]))
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "1829b323-689b-46de-b4d7-89a84dcd3089", "showTitle": false, "title": ""}
from sklearn.compose import ColumnTransformer
preprocessor = ColumnTransformer(transformers, remainder="passthrough", sparse_threshold=0)
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "9642d315-192d-4198-8f4e-de25e6e84c73", "showTitle": false, "title": ""}
# ### Feature standardization
# Scale all feature columns to be centered around zero with unit variance.
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "81209c7e-04f3-4b2e-bb82-cc527635bb97", "showTitle": false, "title": ""}
from sklearn.preprocessing import StandardScaler
standardizer = StandardScaler()
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "132672f8-5bfa-46a1-b20a-d53c05674e8d", "showTitle": false, "title": ""}
# ## Train - Validation - Test Split
# Split the input data into 3 sets:
# - Train (60% of the dataset used to train the model)
# - Validation (20% of the dataset used to tune the hyperparameters of the model)
# - Test (20% of the dataset used to report the true performance of the model on an unseen dataset)
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "ec74ca8c-55da-4cc0-9b6a-8bf697d41d03", "showTitle": false, "title": ""}
df_loaded.columns
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "038c10bf-b26e-4f03-ad32-35d24b246f9b", "showTitle": false, "title": ""}
from sklearn.model_selection import train_test_split
split_X = df_loaded.drop([target_col], axis=1)
split_y = df_loaded[target_col]
# Split out train data
X_train, split_X_rem, y_train, split_y_rem = train_test_split(split_X, split_y, train_size=0.6, random_state=799811440, stratify=split_y)
# Split remaining data equally for validation and test
X_val, X_test, y_val, y_test = train_test_split(split_X_rem, split_y_rem, test_size=0.5, random_state=799811440, stratify=split_y_rem)
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "70fb1e86-67b4-44cd-b9f3-d20cd4f29708", "showTitle": false, "title": ""}
# ## Train classification model
# - Log relevant metrics to MLflow to track runs
# - All the runs are logged under [this MLflow experiment](#mlflow/experiments/406583024052808/s?orderByKey=metrics.%60val_f1_score%60&orderByAsc=false)
# - Change the model parameters and re-run the training cell to log a different trial to the MLflow experiment
# - To view the full list of tunable hyperparameters, check the output of the cell below
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "413f6481-158c-4185-8343-b909c37724ff", "showTitle": false, "title": ""}
from xgboost import XGBClassifier
help(XGBClassifier)
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "23c1fc84-759b-4421-a5b5-65a01e735b45", "showTitle": false, "title": ""}
import mlflow
import sklearn
from sklearn import set_config
from sklearn.pipeline import Pipeline
set_config(display="diagram")
xgbc_classifier = XGBClassifier(
colsample_bytree=0.7324555878929649,
learning_rate=0.007636627530856404,
max_depth=7,
min_child_weight=6,
n_estimators=106,
n_jobs=100,
subsample=0.6972187716458148,
verbosity=0,
random_state=799811440,
)
model = Pipeline([
("column_selector", col_selector),
("preprocessor", preprocessor),
("standardizer", standardizer),
("classifier", xgbc_classifier),
])
# Create a separate pipeline to transform the validation dataset. This is used for early stopping.
pipeline = Pipeline([
("column_selector", col_selector),
("preprocessor", preprocessor),
("standardizer", standardizer),
])
mlflow.sklearn.autolog(disable=True)
X_val_processed = pipeline.fit_transform(X_val, y_val)
model
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "db0342f7-973d-4d5b-afa4-4c6cfa11c0d7", "showTitle": false, "title": ""}
# Enable automatic logging of input samples, metrics, parameters, and models
mlflow.sklearn.autolog(log_input_examples=True, silent=True)
with mlflow.start_run(run_name="xgboost") as mlflow_run:
model.fit(X_train, y_train, classifier__early_stopping_rounds=5, classifier__eval_set=[(X_val_processed,y_val)], classifier__verbose=False)
# Training metrics are logged by MLflow autologging
# Log metrics for the validation set
xgbc_val_metrics = mlflow.sklearn.eval_and_log_metrics(model, X_val, y_val, prefix="val_")
# Log metrics for the test set
xgbc_test_metrics = mlflow.sklearn.eval_and_log_metrics(model, X_test, y_test, prefix="test_")
# Display the logged metrics
xgbc_val_metrics = {k.replace("val_", ""): v for k, v in xgbc_val_metrics.items()}
xgbc_test_metrics = {k.replace("test_", ""): v for k, v in xgbc_test_metrics.items()}
display(pd.DataFrame([xgbc_val_metrics, xgbc_test_metrics], index=["validation", "test"]))
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "a0a7ceed-f6f6-4b0c-b996-19ab6875d09b", "showTitle": false, "title": ""}
# Patch requisite packages to the model environment YAML for model serving
import os
import shutil
import uuid
import yaml
None
import xgboost
from mlflow.tracking import MlflowClient
xgbc_temp_dir = os.path.join(os.environ["SPARK_LOCAL_DIRS"], str(uuid.uuid4())[:8])
os.makedirs(xgbc_temp_dir)
xgbc_client = MlflowClient()
xgbc_model_env_path = xgbc_client.download_artifacts(mlflow_run.info.run_id, "model/conda.yaml", xgbc_temp_dir)
xgbc_model_env_str = open(xgbc_model_env_path)
xgbc_parsed_model_env_str = yaml.load(xgbc_model_env_str, Loader=yaml.FullLoader)
xgbc_parsed_model_env_str["dependencies"][-1]["pip"].append(f"xgboost=={xgboost.__version__}")
with open(xgbc_model_env_path, "w") as f:
f.write(yaml.dump(xgbc_parsed_model_env_str))
xgbc_client.log_artifact(run_id=mlflow_run.info.run_id, local_path=xgbc_model_env_path, artifact_path="model")
shutil.rmtree(xgbc_temp_dir)
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "9e667aaf-af28-4692-9304-948d3c7ec856", "showTitle": false, "title": ""}
# ## Feature importance
#
# SHAP is a game-theoretic approach to explain machine learning models, providing a summary plot
# of the relationship between features and model output. Features are ranked in descending order of
# importance, and impact/color describe the correlation between the feature and the target variable.
# - Generating SHAP feature importance is a very memory intensive operation, so to ensure that AutoML can run trials without
# running out of memory, we disable SHAP by default.<br />
# You can set the flag defined below to `shap_enabled = True` and re-run this notebook to see the SHAP plots.
# - To reduce the computational overhead of each trial, a single example is sampled from the validation set to explain.<br />
# For more thorough results, increase the sample size of explanations, or provide your own examples to explain.
# - SHAP cannot explain models using data with nulls; if your dataset has any, both the background data and
# examples to explain will be imputed using the mode (most frequent values). This affects the computed
# SHAP values, as the imputed samples may not match the actual data distribution.
#
# For more information on how to read Shapley values, see the [SHAP documentation](https://shap.readthedocs.io/en/latest/example_notebooks/overviews/An%20introduction%20to%20explainable%20AI%20with%20Shapley%20values.html).
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "20b1ac9f-31de-4cae-a97a-73c8754560ca", "showTitle": false, "title": ""}
# Set this flag to True and re-run the notebook to see the SHAP plots
shap_enabled = True
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "70419482-fdef-45d9-b490-76e1d08f613b", "showTitle": false, "title": ""}
if shap_enabled:
from shap import KernelExplainer, summary_plot
# SHAP cannot explain models using data with nulls.
# To enable SHAP to succeed, both the background data and examples to explain are imputed with the mode (most frequent values).
mode = X_train.mode().iloc[0]
# Sample background data for SHAP Explainer. Increase the sample size to reduce variance.
train_sample = X_train.sample(n=min(100, len(X_train.index))).fillna(mode)
# Sample a single example from the validation set to explain. Increase the sample size and rerun for more thorough results.
example = X_val.sample(n=1).fillna(mode)
# Use Kernel SHAP to explain feature importance on the example from the validation set.
predict = lambda x: model.predict_proba(pd.DataFrame(x, columns=X_train.columns))
explainer = KernelExplainer(predict, train_sample, link="logit")
shap_values = explainer.shap_values(example, l1_reg=False)
summary_plot(shap_values, example, class_names=model.classes_)
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "b7ba68ec-1277-4de3-bd8b-b0e0f7f44a45", "showTitle": false, "title": ""}
# ## Inference
# [The MLflow Model Registry](https://docs.databricks.com/applications/mlflow/model-registry.html) is a collaborative hub where teams can share ML models, work together from experimentation to online testing and production, integrate with approval and governance workflows, and monitor ML deployments and their performance. The snippets below show how to add the model trained in this notebook to the model registry and to retrieve it later for inference.
#
# > **NOTE:** The `model_uri` for the model already trained in this notebook can be found in the cell below
#
# ### Register to Model Registry
# ```
# model_name = "Example"
#
# model_uri = f"runs:/{ mlflow_run.info.run_id }/model"
# registered_model_version = mlflow.register_model(model_uri, model_name)
# ```
#
# ### Load from Model Registry
# ```
# model_name = "Example"
# model_version = registered_model_version.version
#
# model = mlflow.pyfunc.load_model(model_uri=f"models:/{model_name}/{model_version}")
# model.predict(input_X)
# ```
#
# ### Load model without registering
# ```
# model_uri = f"runs:/{ mlflow_run.info.run_id }/model"
#
# model = mlflow.pyfunc.load_model(model_uri)
# model.predict(input_X)
# ```
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "b73abc77-5b61-4b2c-8640-937e2f133edd", "showTitle": false, "title": ""}
# model_uri for the generated model
print(f"runs:/{ mlflow_run.info.run_id }/model")
# + [markdown] application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "6de9cdf2-b461-47a3-ac27-873f53f797eb", "showTitle": false, "title": ""}
# ### Loading model to make prediction
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "001cea05-9cc0-469b-b204-1c151985e4f8", "showTitle": false, "title": ""}
model_uri = f"runs:/51c0348482e042ea8e4b7983ab6bff99/model"
model = mlflow.pyfunc.load_model(model_uri)
#model.predict(input_X)
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "7c79178a-e4d9-4688-9a72-5e80ef722f54", "showTitle": false, "title": ""}
import pandas as pd
data = {'author': {0: 'bigjim.com'},
'published': {0: '2016-10-27T18:05:26.351+03:00'},
'title': {0: 'aliens are coming to invade earth'},
'text': {0: 'aliens are coming to invade earth'},
'language': {0: 'english'},
'site_url': {0: 'cnn.com'},
'main_img_url': {0: 'https://2.bp.blogspot.com/-0mdp0nZiwMI/UYwYvexmW2I/AAAAAAAAVQM/7C_X5WRE_mQ/w1200-h630-p-nu/Edison-Stock-Ticker.jpg'},
'type': {0: 'bs'},
'title_without_stopwords': {0: 'aliens are coming to invade earth'},
'text_without_stopwords': {0: 'aliens are coming to invade earth'},
'hasImage': {0: 1.0}}
df = pd.DataFrame(data=data)
df.head()
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "43bfd651-4f88-4f8b-bea8-c457e735a645", "showTitle": false, "title": ""}
model.predict(df)
# + application/vnd.databricks.v1+cell={"inputWidgets": {}, "nuid": "dcfd4fd1-f90f-46d8-87e6-4b9192f27828", "showTitle": false, "title": ""}
| XGBoost-fake-news-automl.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # TV Script Generation
# In this project, you'll generate your own [Simpsons](https://en.wikipedia.org/wiki/The_Simpsons) TV scripts using RNNs. You'll be using part of the [Simpsons dataset](https://www.kaggle.com/wcukierski/the-simpsons-by-the-data) of scripts from 27 seasons. The Neural Network you'll build will generate a new TV script for a scene at [Moe's Tavern](https://simpsonswiki.com/wiki/Moe's_Tavern).
# ## Get the Data
# The data is already provided for you. You'll be using a subset of the original dataset. It consists of only the scenes in Moe's Tavern. This doesn't include other versions of the tavern, like "Moe's Cavern", "Flaming Moe's", "Uncle Moe's Family Feed-Bag", etc..
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import helper
data_dir = './data/simpsons/moes_tavern_lines.txt'
text = helper.load_data(data_dir)
# Ignore notice, since we don't use it for analysing the data
text = text[81:]
# -
# ## Explore the Data
# Play around with `view_sentence_range` to view different parts of the data.
# +
view_sentence_range = (0, 10)
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import numpy as np
print('Dataset Stats')
print('Roughly the number of unique words: {}'.format(len({word: None for word in text.split()})))
scenes = text.split('\n\n')
print('Number of scenes: {}'.format(len(scenes)))
sentence_count_scene = [scene.count('\n') for scene in scenes]
print('Average number of sentences in each scene: {}'.format(np.average(sentence_count_scene)))
sentences = [sentence for scene in scenes for sentence in scene.split('\n')]
print('Number of lines: {}'.format(len(sentences)))
word_count_sentence = [len(sentence.split()) for sentence in sentences]
print('Average number of words in each line: {}'.format(np.average(word_count_sentence)))
print()
print('The sentences {} to {}:'.format(*view_sentence_range))
print('\n'.join(text.split('\n')[view_sentence_range[0]:view_sentence_range[1]]))
# -
# ## Implement Preprocessing Functions
# The first thing to do to any dataset is preprocessing. Implement the following preprocessing functions below:
# - Lookup Table
# - Tokenize Punctuation
#
# ### Lookup Table
# To create a word embedding, you first need to transform the words to ids. In this function, create two dictionaries:
# - Dictionary to go from the words to an id, we'll call `vocab_to_int`
# - Dictionary to go from the id to word, we'll call `int_to_vocab`
#
# Return these dictionaries in the following tuple `(vocab_to_int, int_to_vocab)`
# +
import numpy as np
import problem_unittests as tests
def create_lookup_tables(text):
"""
Create lookup tables for vocabulary
:param text: The text of tv scripts split into words
:return: A tuple of dicts (vocab_to_int, int_to_vocab)
"""
# TODO: Implement Function
vocab = set(text)
vocab_to_int = {word: i for i,word in enumerate(vocab)}
int_to_vocab = dict(enumerate(vocab))
print(vocab_to_int)
print("\n")
print(int_to_vocab)
return vocab_to_int, int_to_vocab
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_create_lookup_tables(create_lookup_tables)
# -
# ### Tokenize Punctuation
# We'll be splitting the script into a word array using spaces as delimiters. However, punctuations like periods and exclamation marks make it hard for the neural network to distinguish between the word "bye" and "bye!".
#
# Implement the function `token_lookup` to return a dict that will be used to tokenize symbols like "!" into "||Exclamation_Mark||". Create a dictionary for the following symbols where the symbol is the key and value is the token:
# - Period ( . )
# - Comma ( , )
# - Quotation Mark ( " )
# - Semicolon ( ; )
# - Exclamation mark ( ! )
# - Question mark ( ? )
# - Left Parentheses ( ( )
# - Right Parentheses ( ) )
# - Dash ( -- )
# - Return ( \n )
#
# This dictionary will be used to token the symbols and add the delimiter (space) around it. This separates the symbols as it's own word, making it easier for the neural network to predict on the next word. Make sure you don't use a token that could be confused as a word. Instead of using the token "dash", try using something like "||dash||".
# +
def token_lookup():
"""
Generate a dict to turn punctuation into a token.
:return: Tokenize dictionary where the key is the punctuation and the value is the token
"""
# TODO: Implement Function
punc = {'.': '||period||',
',': '||comma||',
'"': '||quotation_mark||',
';': '||semicolon||',
'!': '||exclamation_mark||',
'?': '||question_mark||',
'(': '||left_parentheses',
')': '||right_parentheses',
'--': '||dash||',
'\n': '||return||'}
return punc
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_tokenize(token_lookup)
# -
# ## Preprocess all the data and save it
# Running the code cell below will preprocess all the data and save it to file.
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
# Preprocess Training, Validation, and Testing Data
helper.preprocess_and_save_data(data_dir, token_lookup, create_lookup_tables)
# # Check Point
# This is your first checkpoint. If you ever decide to come back to this notebook or have to restart the notebook, you can start from here. The preprocessed data has been saved to disk.
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import helper
import numpy as np
import problem_unittests as tests
int_text, vocab_to_int, int_to_vocab, token_dict = helper.load_preprocess()
# -
# ## Build the Neural Network
# You'll build the components necessary to build a RNN by implementing the following functions below:
# - get_inputs
# - get_init_cell
# - get_embed
# - build_rnn
# - build_nn
# - get_batches
#
# ### Check the Version of TensorFlow and Access to GPU
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
from distutils.version import LooseVersion
import warnings
import tensorflow as tf
# Check TensorFlow Version
assert LooseVersion(tf.__version__) >= LooseVersion('1.3'), 'Please use TensorFlow version 1.3 or newer'
print('TensorFlow Version: {}'.format(tf.__version__))
# Check for a GPU
if not tf.test.gpu_device_name():
warnings.warn('No GPU found. Please use a GPU to train your neural network.')
else:
print('Default GPU Device: {}'.format(tf.test.gpu_device_name()))
# -
# ### Input
# Implement the `get_inputs()` function to create TF Placeholders for the Neural Network. It should create the following placeholders:
# - Input text placeholder named "input" using the [TF Placeholder](https://www.tensorflow.org/api_docs/python/tf/placeholder) `name` parameter.
# - Targets placeholder
# - Learning Rate placeholder
#
# Return the placeholders in the following tuple `(Input, Targets, LearningRate)`
# +
def get_inputs():
"""
Create TF Placeholders for input, targets, and learning rate.
:return: Tuple (input, targets, learning rate)
"""
# TODO: Implement Function
inputs = tf.placeholder(tf.int32, [None, None], name="input")
targets = tf.placeholder(tf.int32, [None, None], name="targets")
learning_rate = tf.placeholder(tf.float32, name="learning_rate")
return inputs, targets, learning_rate
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_get_inputs(get_inputs)
# -
# ### Build RNN Cell and Initialize
# Stack one or more [`BasicLSTMCells`](https://www.tensorflow.org/api_docs/python/tf/contrib/rnn/BasicLSTMCell) in a [`MultiRNNCell`](https://www.tensorflow.org/api_docs/python/tf/contrib/rnn/MultiRNNCell).
# - The Rnn size should be set using `rnn_size`
# - Initalize Cell State using the MultiRNNCell's [`zero_state()`](https://www.tensorflow.org/api_docs/python/tf/contrib/rnn/MultiRNNCell#zero_state) function
# - Apply the name "initial_state" to the initial state using [`tf.identity()`](https://www.tensorflow.org/api_docs/python/tf/identity)
#
# Return the cell and initial state in the following tuple `(Cell, InitialState)`
# +
def get_init_cell(batch_size, rnn_size):
"""
Create an RNN Cell and initialize it.
:param batch_size: Size of batches
:param rnn_size: Size of RNNs
:return: Tuple (cell, initialize state)
"""
# TODO: Implement Function
layers = 2
cells = []
for i in range(layers):
lstm = tf.contrib.rnn.BasicLSTMCell(rnn_size)
drop = tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=0.7)
cells.append(drop)
cell = tf.contrib.rnn.MultiRNNCell(cells)
initial_state = tf.identity(cell.zero_state(batch_size, tf.float32), "initial_state")
return cell, initial_state
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_get_init_cell(get_init_cell)
# -
# ### Word Embedding
# Apply embedding to `input_data` using TensorFlow. Return the embedded sequence.
# +
def get_embed(input_data, vocab_size, embed_dim):
"""
Create embedding for <input_data>.
:param input_data: TF placeholder for text input.
:param vocab_size: Number of words in vocabulary.
:param embed_dim: Number of embedding dimensions
:return: Embedded input.
"""
# TODO: Implement Function
return tf.contrib.layers.embed_sequence(input_data, vocab_size, embed_dim)
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_get_embed(get_embed)
# -
# ### Build RNN
# You created a RNN Cell in the `get_init_cell()` function. Time to use the cell to create a RNN.
# - Build the RNN using the [`tf.nn.dynamic_rnn()`](https://www.tensorflow.org/api_docs/python/tf/nn/dynamic_rnn)
# - Apply the name "final_state" to the final state using [`tf.identity()`](https://www.tensorflow.org/api_docs/python/tf/identity)
#
# Return the outputs and final_state state in the following tuple `(Outputs, FinalState)`
# +
def build_rnn(cell, inputs):
"""
Create a RNN using a RNN Cell
:param cell: RNN Cell
:param inputs: Input text data
:return: Tuple (Outputs, Final State)
"""
# TODO: Implement Function
outputs, final_state = tf.nn.dynamic_rnn(cell, inputs, dtype=tf.float32)
final_state = tf.identity(final_state, "final_state")
return outputs, final_state
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_build_rnn(build_rnn)
# -
# ### Build the Neural Network
# Apply the functions you implemented above to:
# - Apply embedding to `input_data` using your `get_embed(input_data, vocab_size, embed_dim)` function.
# - Build RNN using `cell` and your `build_rnn(cell, inputs)` function.
# - Apply a fully connected layer with a linear activation and `vocab_size` as the number of outputs.
#
# Return the logits and final state in the following tuple (Logits, FinalState)
# +
def build_nn(cell, rnn_size, input_data, vocab_size, embed_dim):
"""
Build part of the neural network
:param cell: RNN cell
:param rnn_size: Size of rnns
:param input_data: Input data
:param vocab_size: Vocabulary size
:param embed_dim: Number of embedding dimensions
:return: Tuple (Logits, FinalState)
"""
# TODO: Implement Function
embed = get_embed(input_data, vocab_size, embed_dim)
outputs, final_state = build_rnn(cell, embed)
logits = tf.contrib.layers.fully_connected(outputs, vocab_size, activation_fn=None)
return logits, final_state
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_build_nn(build_nn)
# -
# ### Batches
# Implement `get_batches` to create batches of input and targets using `int_text`. The batches should be a Numpy array with the shape `(number of batches, 2, batch size, sequence length)`. Each batch contains two elements:
# - The first element is a single batch of **input** with the shape `[batch size, sequence length]`
# - The second element is a single batch of **targets** with the shape `[batch size, sequence length]`
#
# If you can't fill the last batch with enough data, drop the last batch.
#
# For example, `get_batches([1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20], 3, 2)` would return a Numpy array of the following:
# ```
# [
# # First Batch
# [
# # Batch of Input
# [[ 1 2], [ 7 8], [13 14]]
# # Batch of targets
# [[ 2 3], [ 8 9], [14 15]]
# ]
#
# # Second Batch
# [
# # Batch of Input
# [[ 3 4], [ 9 10], [15 16]]
# # Batch of targets
# [[ 4 5], [10 11], [16 17]]
# ]
#
# # Third Batch
# [
# # Batch of Input
# [[ 5 6], [11 12], [17 18]]
# # Batch of targets
# [[ 6 7], [12 13], [18 1]]
# ]
# ]
# ```
#
# Notice that the last target value in the last batch is the first input value of the first batch. In this case, `1`. This is a common technique used when creating sequence batches, although it is rather unintuitive.
# +
def get_batches(int_text, batch_size, seq_length):
"""
Return batches of input and target
:param int_text: Text with the words replaced by their ids
:param batch_size: The size of batch
:param seq_length: The length of sequence
:return: Batches as a Numpy array
"""
# TODO: Implement Function
chars_per_batch = batch_size * seq_length
num_of_batches = len(int_text) // chars_per_batch
inputs = np.array(int_text[:num_of_batches*chars_per_batch])
targets = np.array(int_text[1:num_of_batches*chars_per_batch] + [int_text[0]])
inputs = inputs.reshape(batch_size, -1)
targets = targets.reshape(batch_size, -1)
inputs = np.split(inputs, num_of_batches, axis=1)
targets = np.split(targets, num_of_batches, axis=1)
batches = np.array(list(zip(inputs, targets)))
batches.reshape(num_of_batches, 2, batch_size, seq_length)
return batches
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_get_batches(get_batches)
# -
# ## Neural Network Training
# ### Hyperparameters
# Tune the following parameters:
#
# - Set `num_epochs` to the number of epochs.
# - Set `batch_size` to the batch size.
# - Set `rnn_size` to the size of the RNNs.
# - Set `embed_dim` to the size of the embedding.
# - Set `seq_length` to the length of sequence.
# - Set `learning_rate` to the learning rate.
# - Set `show_every_n_batches` to the number of batches the neural network should print progress.
# +
# Number of Epochs
num_epochs = 70
# Batch Size
batch_size = 64
# RNN Size
rnn_size = 1000
# Embedding Dimension Size
embed_dim = 500
# Sequence Length
seq_length = 14
# Learning Rate
learning_rate = 0.001
# Show stats for every n number of batches
show_every_n_batches = 15
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
save_dir = './save'
# -
# ### Build the Graph
# Build the graph using the neural network you implemented.
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
from tensorflow.contrib import seq2seq
train_graph = tf.Graph()
with train_graph.as_default():
vocab_size = len(int_to_vocab)
input_text, targets, lr = get_inputs()
input_data_shape = tf.shape(input_text)
cell, initial_state = get_init_cell(input_data_shape[0], rnn_size)
logits, final_state = build_nn(cell, rnn_size, input_text, vocab_size, embed_dim)
# Probabilities for generating words
probs = tf.nn.softmax(logits, name='probs')
# Loss function
cost = seq2seq.sequence_loss(
logits,
targets,
tf.ones([input_data_shape[0], input_data_shape[1]]))
# Optimizer
optimizer = tf.train.AdamOptimizer(lr)
# Gradient Clipping
gradients = optimizer.compute_gradients(cost)
capped_gradients = [(tf.clip_by_value(grad, -1., 1.), var) for grad, var in gradients if grad is not None]
train_op = optimizer.apply_gradients(capped_gradients)
# -
# ## Train
# Train the neural network on the preprocessed data. If you have a hard time getting a good loss, check the [forums](https://discussions.udacity.com/) to see if anyone is having the same problem.
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
batches = get_batches(int_text, batch_size, seq_length)
with tf.Session(graph=train_graph) as sess:
sess.run(tf.global_variables_initializer())
for epoch_i in range(num_epochs):
state = sess.run(initial_state, {input_text: batches[0][0]})
for batch_i, (x, y) in enumerate(batches):
feed = {
input_text: x,
targets: y,
initial_state: state,
lr: learning_rate}
train_loss, state, _ = sess.run([cost, final_state, train_op], feed)
# Show every <show_every_n_batches> batches
if (epoch_i * len(batches) + batch_i) % show_every_n_batches == 0:
print('Epoch {:>3} Batch {:>4}/{} train_loss = {:.3f}'.format(
epoch_i,
batch_i,
len(batches),
train_loss))
# Save Model
saver = tf.train.Saver()
saver.save(sess, save_dir)
print('Model Trained and Saved')
# -
# ## Save Parameters
# Save `seq_length` and `save_dir` for generating a new TV script.
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
# Save parameters for checkpoint
helper.save_params((seq_length, save_dir))
# # Checkpoint
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import tensorflow as tf
import numpy as np
import helper
import problem_unittests as tests
_, vocab_to_int, int_to_vocab, token_dict = helper.load_preprocess()
seq_length, load_dir = helper.load_params()
# -
# ## Implement Generate Functions
# ### Get Tensors
# Get tensors from `loaded_graph` using the function [`get_tensor_by_name()`](https://www.tensorflow.org/api_docs/python/tf/Graph#get_tensor_by_name). Get the tensors using the following names:
# - "input:0"
# - "initial_state:0"
# - "final_state:0"
# - "probs:0"
#
# Return the tensors in the following tuple `(InputTensor, InitialStateTensor, FinalStateTensor, ProbsTensor)`
# +
def get_tensors(loaded_graph):
"""
Get input, initial state, final state, and probabilities tensor from <loaded_graph>
:param loaded_graph: TensorFlow graph loaded from file
:return: Tuple (InputTensor, InitialStateTensor, FinalStateTensor, ProbsTensor)
"""
# TODO: Implement Function
InputTensor = loaded_graph.get_tensor_by_name("input:0")
InitialStateTensor = loaded_graph.get_tensor_by_name("initial_state:0")
FinalStateTensor = loaded_graph.get_tensor_by_name("final_state:0")
ProbsTensor = loaded_graph.get_tensor_by_name("probs:0")
return InputTensor, InitialStateTensor, FinalStateTensor, ProbsTensor
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_get_tensors(get_tensors)
# -
# ### Choose Word
# Implement the `pick_word()` function to select the next word using `probabilities`.
# +
def pick_word(probabilities, int_to_vocab):
"""
Pick the next word in the generated text
:param probabilities: Probabilites of the next word
:param int_to_vocab: Dictionary of word ids as the keys and words as the values
:return: String of the predicted word
"""
# TODO: Implement Function
predict = np.random.choice(range(0, len(int_to_vocab)), size=1, p=probabilities)
return int_to_vocab[predict[0]]
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_pick_word(pick_word)
# -
# ## Generate TV Script
# This will generate the TV script for you. Set `gen_length` to the length of TV script you want to generate.
# +
gen_length = 200
# homer_simpson, moe_szyslak, or Barney_Gumble
prime_word = 'moe_szyslak'
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
loaded_graph = tf.Graph()
with tf.Session(graph=loaded_graph) as sess:
# Load saved model
loader = tf.train.import_meta_graph(load_dir + '.meta')
loader.restore(sess, load_dir)
# Get Tensors from loaded model
input_text, initial_state, final_state, probs = get_tensors(loaded_graph)
# Sentences generation setup
gen_sentences = [prime_word + ':']
prev_state = sess.run(initial_state, {input_text: np.array([[1]])})
# Generate sentences
for n in range(gen_length):
# Dynamic Input
dyn_input = [[vocab_to_int[word] for word in gen_sentences[-seq_length:]]]
dyn_seq_length = len(dyn_input[0])
# Get Prediction
probabilities, prev_state = sess.run(
[probs, final_state],
{input_text: dyn_input, initial_state: prev_state})
pred_word = pick_word(probabilities[0][dyn_seq_length-1], int_to_vocab)
gen_sentences.append(pred_word)
# Remove tokens
tv_script = ' '.join(gen_sentences)
for key, token in token_dict.items():
ending = ' ' if key in ['\n', '(', '"'] else ''
tv_script = tv_script.replace(' ' + token.lower(), key)
tv_script = tv_script.replace('\n ', '\n')
tv_script = tv_script.replace('( ', '(')
print(tv_script)
# -
# # The TV Script is Nonsensical
# It's ok if the TV script doesn't make any sense. We trained on less than a megabyte of text. In order to get good results, you'll have to use a smaller vocabulary or get more data. Luckily there's more data! As we mentioned in the beggining of this project, this is a subset of [another dataset](https://www.kaggle.com/wcukierski/the-simpsons-by-the-data). We didn't have you train on all the data, because that would take too long. However, you are free to train your neural network on all the data. After you complete the project, of course.
# # Submitting This Project
# When submitting this project, make sure to run all the cells before saving the notebook. Save the notebook file as "dlnd_tv_script_generation.ipynb" and save it as a HTML file under "File" -> "Download as". Include the "helper.py" and "problem_unittests.py" files in your submission.
| dlnd_tv_script_generation.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] _cell_guid="455c5288-0893-4f5c-be8f-b70a3ba51231" _uuid="978e24e319343f241547ac0ae94868a678c7f2cc" endofcell="--"
# *This tutorial is part of the [Learn Machine Learning](https://www.kaggle.com/learn/machine-learning/) series. In this step, you will learn what data leakage is and how to prevent it.*
#
#
# # What is Data Leakage
# Data leakage is one of the most important issues for a data scientist to understand. If you don't know how to prevent it, leakage will come up frequently, and it will ruin your models in the most subtle and dangerous ways. Specifically, leakage causes a model to look accurate until you start making decisions with the model, and then the model becomes very inaccurate. This tutorial will show you what leakage is and how to avoid it.
#
# There are two main types of leakage: **Leaky Predictors** and a **Leaky Validation Strategies.**
#
# ## Leaky Predictors
# This occurs when your predictors include data that will not be available at the time you make predictions.
#
# For example, imagine you want to predict who will get sick with pneumonia. The top few rows of your raw data might look like this:
#
# | got_pneumonia | age | weight | male | took_antibiotic_medicine | ... |
# |:-------------:|:---:|:------:|:-----:|:------------------------:|-----|
# | False | 65 | 100 | False | False | ... |
# | False | 72 | 130 | True | False | ... |
# | True | 58 | 100 | False | True | ... |
# -
#
#
# People take antibiotic medicines after getting pneumonia in order to recover. So the raw data shows a strong relationship between those columns. But *took_antibiotic_medicine* is frequently changed **after** the value for *got_pneumonia* is determined. This is target leakage.
#
# The model would see that anyone who has a value of `False` for `took_antibiotic_medicine` didn't have pneumonia. Validation data comes from the same source, so the pattern will repeat itself in validation, and the model will have great validation (or cross-validation) scores. But the model will be very inaccurate when subsequently deployed in the real world.
#
# To prevent this type of data leakage, any variable updated (or created) after the target value is realized should be excluded. Because when we use this model to make new predictions, that data won't be available to the model.
#
# 
# --
# + [markdown] _cell_guid="cca14623-d55f-49ba-8907-501e9ac2acca" _uuid="e5a3cf9b1bd44f7d2c8e8672b9e8594150d30ad6"
# ## Leaky Validation Strategy
#
# A much different type of leak occurs when you aren't careful distinguishing training data from validation data. For example, this happens if you run preprocessing (like fitting the Imputer for missing values) before calling train_test_split. Validation is meant to be a measure of how the model does on data it hasn't considered before. You can corrupt this process in subtle ways if the validation data affects the preprocessing behavoir.. The end result? Your model will get very good validation scores, giving you great confidence in it, but perform poorly when you deploy it to make decisions.
#
#
# ## Preventing Leaky Predictors
# There is no single solution that universally prevents leaky predictors. It requires knowledge about your data, case-specific inspection and common sense.
#
# However, leaky predictors frequently have high statistical correlations to the target. So two tactics to keep in mind:
# * To screen for possible leaky predictors, look for columns that are statistically correlated to your target.
# * If you build a model and find it extremely accurate, you likely have a leakage problem.
#
# ## Preventing Leaky Validation Strategies
#
# If your validation is based on a simple train-test split, exclude the validation data from any type of *fitting*, including the fitting of preprocessing steps. This is easier if you use [scikit-learn Pipelines](https://www.kaggle.com/dansbecker/pipelines). When using cross-validation, it's even more critical that you use pipelines and do your preprocessing inside the pipeline.
#
# # Example
# We will use a small dataset about credit card applications, and we will build a model predicting which applications were accepted (stored in a variable called *card*). Here is a look at the data:
# + _cell_guid="29c264f4-3836-4b48-b8c7-828e7bec45a0" _uuid="b95201cc2da5de79c022ab8c7cdfe38c16723907"
import pandas as pd
data = pd.read_csv('../input/AER_credit_card_data.csv',
true_values = ['yes'],
false_values = ['no'])
print(data.head())
# + [markdown] _cell_guid="12d34c22-ad00-4e6c-9d0b-443ab54caf35" _uuid="60436a2d7d8e7b87ce891639a2727b77761ff08d"
# We can see with `data.shape` that this is a small dataset (1312 rows), so we should use cross-validation to ensure accurate measures of model quality
# + _cell_guid="29a27f97-44b1-408f-95fb-00a6f01ea93f" _uuid="46cad269244b866c179b00f7f9048b9dc29e9de9"
data.shape
# + _cell_guid="2e15597f-171e-4da0-8c52-96c42f073a36" _uuid="90ea96255857648ffd6e74f33bf7e23c1c3da467"
from sklearn.pipeline import make_pipeline
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_val_score
y = data.card
X = data.drop(['card'], axis=1)
# Since there was no preprocessing, we didn't need a pipeline here. Used anyway as best practice
modeling_pipeline = make_pipeline(RandomForestClassifier())
cv_scores = cross_val_score(modeling_pipeline, X, y, scoring='accuracy')
print("Cross-val accuracy: %f" %cv_scores.mean())
# + [markdown] _cell_guid="722ccd83-3d23-4d4e-989e-a9f3a719b308" _uuid="d7faed322b2b993b56f18922dac556950bfa8190"
# With experience, you'll find that it's very rare to find models that are accurate 98% of the time. It happens, but it's rare enough that we should inspect the data more closely to see if it is target leakage.
#
# Here is a summary of the data, which you can also find under the data tab:
#
# - **card:** Dummy variable, 1 if application for credit card accepted, 0 if not
# - **reports:** Number of major derogatory reports
# - **age:** Age n years plus twelfths of a year
# - **income:** Yearly income (divided by 10,000)
# - **share:** Ratio of monthly credit card expenditure to yearly income
# - **expenditure:** Average monthly credit card expenditure
# - **owner:** 1 if owns their home, 0 if rent
# - **selfempl:** 1 if self employed, 0 if not.
# - **dependents:** 1 + number of dependents
# - **months:** Months living at current address
# - **majorcards:** Number of major credit cards held
# - **active:** Number of active credit accounts
#
# A few variables look suspicious. For example, does **expenditure** mean expenditure on this card or on cards used before appying?
#
# At this point, basic data comparisons can be very helpful:
# + _cell_guid="1778dd97-db7e-47e6-a8bd-bb48e6da6327" _uuid="f6e587b54c565c9ca7990da8ff74ec4252c4ae49"
expenditures_cardholders = data.expenditure[data.card]
expenditures_noncardholders = data.expenditure[~data.card]
print('Fraction of those who received a card with no expenditures: %.2f' \
%(( expenditures_cardholders == 0).mean()))
print('Fraction of those who received a card with no expenditures: %.2f' \
%((expenditures_noncardholders == 0).mean()))
# + [markdown] _cell_guid="6ce5490e-f5eb-4ab8-b6cd-9309c1b3d832" _uuid="a5120e2851e400a9d70018496ceb4523a680ca06"
# Everyone with `card == False` had no expenditures, while only 2% of those with `card == True` had no expenditures. It's not surprising that our model appeared to have a high accuracy. But this seems a data leak, where expenditures probably means *expenditures on the card they applied for.**.
#
# Since **share** is partially determined by **expenditure**, it should be excluded too. The variables **active**, **majorcards** are a little less clear, but from the description, they sound concerning. In most situations, it's better to be safe than sorry if you can't track down the people who created the data to find out more.
#
# We would run a model without leakage as follows:
# + _cell_guid="6364e8d5-bb75-4f57-b635-f43627b72d62" _uuid="85f3ba9371ddb581005e096825c603faeacb9ffd"
potential_leaks = ['expenditure', 'share', 'active', 'majorcards']
X2 = X.drop(potential_leaks, axis=1)
cv_scores = cross_val_score(modeling_pipeline, X2, y, scoring='accuracy')
print("Cross-val accuracy: %f" %cv_scores.mean())
# + [markdown] _cell_guid="2b471192-b646-4b66-87db-3e977424f090" _uuid="f52399825ef93cc40a46d28c931cb3238c4bc613"
# This accuracy is quite a bit lower, which on the one hand is disappointing. However, we can expect it to be right about 80% of the time when used on new applications, whereas the leaky model would likely do much worse then that (even in spite of it's higher apparent score in cross-validation.).
#
# # Conclusion
# Data leakage can be multi-million dollar mistake in many data science applications. Careful separation of training and validation data is a first step, and pipelines can help implement this separation. Leaking predictors are a more frequent issue, and leaking predictors are harder to track down. A combination of caution, common sense and data exploration can help identify leaking predictors so you remove them from your model.
#
# # Exercise
# Review the data in your ongoing project. Are there any predictors that may cause leakage? As a hint, most datasets from Kaggle competitions don't have these variables. Once you get past those carefully curated datasets, this becomes a common issue.
#
# Click **[here](https://www.kaggle.com/learn/machine-learning)** to return the main page for *Learning Machine Learning.*
| ML_Learning/kernel (4).ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # Sketch Classifier for "How Do Humans Sketch Objects?"
# A sketch classifier using the dataset from the paper <a href='http://cybertron.cg.tu-berlin.de/eitz/projects/classifysketch/'>How Do Humans Sketch Objects?</a> where the authors collected 20,000 unique sketches evenly distributed over 250 object categories - we will use a CNN (using Keras) to classify a sketch.
# <img src='http://cybertron.cg.tu-berlin.de/eitz/projects/classifysketch/teaser_siggraph.jpg'/>
from __future__ import print_function
import matplotlib.pyplot as plt
import numpy as np
from scipy.misc import imresize
import os
import random
# +
# %matplotlib inline
# %config InlineBackend.figure_format = 'retina'
plt.style.use('ggplot')
# -
import keras
keras.__version__
# +
from keras import layers
from keras import models
from keras import optimizers
from keras import callbacks
from keras import Input
from keras.utils import plot_model
from keras import backend as K
from keras import preprocessing
from keras.preprocessing import image
# -
# ## Trained on Floydhub
DEST_SKETCH_DIR = '/Users/Joshua.Newnham/Dropbox/Method - Personal/Machine Learning with CoreML/TrainingData/Chapter8/sketches/'
TARGET_SIZE = (256,256)
CATEGORIES_COUNT = 205
TRAINING_SAMPLES = 12736
VALIDATION_SAMPLES = 3184
# ## Create model
# More data always proves to be useful; exposing the network to more samples means that the model has a better chance of identifying and extracting the features that best *describe* a category. Images provide has an advantage over text in that we can easily modify it to expose the network to more samples, such as shifting the image around, rotating and flipping. This task is called *data augmentation* and Keras makes it easy, as shown below. For more information, check out the official Keras <a href='https://keras.io/preprocessing/image/'>documentation</a>.
# Inutitive we can see that, unlike photos, sketches don't have mine details but are better described by their strokes. This is how I had previously tackled the problem; using <a href='https://en.wikipedia.org/wiki/Histogram_of_oriented_gradients'>Histogram of oriented gradients (HOG)</a> filters, I built a bag of visual words for each image. These 'visual words' were dominate gradients of each patch. We can replicate something similar using a ConvNet, but unlike my previous attempt, the feature engineering will be defined and refined during training using the lower layers of the network.
def create_model(input_shape=(256,256,1), classes=CATEGORIES_COUNT, is_training=True):
"""
Create a CNN model
"""
model = models.Sequential()
model.add(layers.Conv2D(16, kernel_size=(7,7), strides=(3,3),
padding='same', activation='relu', input_shape=input_shape))
model.add(layers.MaxPooling2D(2,2))
model.add(layers.Conv2D(32, kernel_size=(5,5), padding='same', activation='relu'))
model.add(layers.MaxPooling2D(2,2))
model.add(layers.Conv2D(64, (5,5), padding='same', activation='relu'))
model.add(layers.MaxPooling2D(2,2))
if is_training:
model.add(layers.Dropout(0.125))
model.add(layers.Conv2D(128, (5,5), padding='same', activation='relu'))
model.add(layers.MaxPooling2D(2,2))
model.add(layers.Flatten())
model.add(layers.Dense(512, activation='relu', name='dense_2_512'))
if is_training:
model.add(layers.Dropout(0.5))
model.add(layers.Dense(classes, activation='softmax', name='output'))
return model
model = create_model()
model.summary()
# ---
# ## Load (pretrained) model
model = create_model()
model.load_weights("output/cnn_sketch_weights_2.h5")
model.summary()
def get_validation_metadata():
"""
Walk the root directory and for each subdirectory, obtain the
list of .png image files creating (and returning) a list for each category label and
associated filepath
"""
image_file_paths = []
categories = []
labels = []
validation_dir = os.path.join(DEST_SKETCH_DIR, 'validation')
validation_directories = sorted(os.listdir(validation_dir))
for d in validation_directories:
if not os.path.isdir(os.path.join(validation_dir, d)):
continue
labels.append(d)
label = d
for f in os.listdir(os.path.join(validation_dir, d)):
full_path = os.path.join(os.path.join(validation_dir, d), f)
if os.path.isfile(full_path) and ".png" in full_path.lower():
categories.append(label)
image_file_paths.append(full_path)
return labels, image_file_paths, categories
def make_predictions(model, categories, filepaths, labels, sample_idx, target_dimension=256.0):
filename = filepaths[sample_idx]
img = plt.imread(filename) # load image
scale = 1.0
# resize if size doesn't match the target dimension (assuming image is square)
if img.shape[0] != target_dimension:
scale = target_dimension / img.shape[0]
img = imresize(img, scale)
img = img[:,:,0] # take the single channel
img = img.reshape(1, int(target_dimension), int(target_dimension), 1)
probs = model.predict(img)[0]
predicted_index = np.argmax(probs)
plt.imshow(imresize(plt.imread(filename), scale),
cmap='gray',
interpolation='nearest')
plt.title("Pred: {} ({}%), Actual: {}".format(
categories[predicted_index],
int(probs[predicted_index]*100),
labels[sample_idx]))
plt.tight_layout()
plt.show()
return predicted_index
categories, validation_filepaths, labels = get_validation_metadata()
categories[0]
_ = make_predictions(model,
categories,
validation_filepaths,
labels,
random.randint(0,len(validation_filepaths) - 1))
_ = make_predictions(model,
categories,
validation_filepaths,
labels,
0)
_ = make_predictions(model,
categories,
validation_filepaths,
labels,
10)
_ = make_predictions(model,
categories,
validation_filepaths,
labels,
801)
# ### Confusion matrix
#
# One way to evaluate the quality of your classifier (especially for multi-class) is visualising a confusion matrix. The diagonal elements represent the number of point for which the predicted label is equal to the true label, while the off-diagonal elements are those that are mislabeled by the classifier. The higher the diagonal values of the confusion matrix, the better the model is performing i.e. we should see a dark line diagonally along the plot.
def plot_confusion_matrix(model, categories, filepaths, labels,
target_dimension=256.0, normalize=False, cmap=plt.cm.Blues):
from sklearn.metrics import confusion_matrix
y_true = []
y_pred = []
category_2_idx = {category:idx for idx, category in enumerate(categories)}
idx_2_category = {idx:category for idx, category in enumerate(categories)}
for idx, filename in enumerate(filepaths):
img = plt.imread(filename) # load image
scale = 1.0
# resize if size doesn't match the target dimension (assuming image is square)
if img.shape[0] != target_dimension:
scale = target_dimension / img.shape[0]
img = imresize(img, scale)
img = img[:,:,0] # take the single channel
img = img.reshape(1, int(target_dimension), int(target_dimension), 1)
probs = model.predict(img)[0]
predicted_index = np.argmax(probs)
actual_index = category_2_idx[labels[idx]]
y_true.append(labels[idx])
y_pred.append(idx_2_category[predicted_index])
y_true = np.array(y_true)
y_pred = np.array(y_pred)
cnf_matrix = confusion_matrix(y_true=y_true, y_pred=y_pred, labels=categories)
if normalize:
cnf_matrix = cnf_matrix.astype('float') / cnf_matrix.sum(axis=1)[:, np.newaxis]
plt.figure(figsize=(12, 12))
plt.imshow(cnf_matrix, interpolation='nearest', cmap=cmap)
plt.title('Confusion Matrix')
plt.colorbar()
#tick_marks = np.arange(len(categories))
#plt.xticks(tick_marks, categories, rotation=45)
#plt.yticks(tick_marks, categories)
#plt.tight_layout()
plt.ylabel('True')
plt.xlabel('Predicted')
plt.show()
plot_confusion_matrix(model,
categories,
validation_filepaths,
labels)
# ---
# # Visualise the activations
#
#
def get_activations(model, categories, filepaths, labels, sample_idx, target_dimension=256.0):
filename = filepaths[sample_idx]
img = plt.imread(filename) # load image
scale = 1.0
# resize if size doesn't match the target dimension (assuming image is square)
if img.shape[0] != target_dimension:
scale = target_dimension / img.shape[0]
img = imresize(img, scale)
img = img[:,:,0] # take the single channel
img = img.reshape(1, int(target_dimension), int(target_dimension), 1)
return model.predict(img)
model.summary()
model.summary()
# extract the first
layer_outputs = [layer.output for layer in model.layers[:10]]
activation_model = models.Model(inputs=model.input, outputs=layer_outputs)
activation_model.summary()
# ## Visualise activations
activations = get_activations(activation_model,
categories,
validation_filepaths,
labels,
801)
first_layer_activation = activations[0]
print(first_layer_activation.shape)
plt.matshow(first_layer_activation[0, :, :, 3], cmap='viridis')
plt.matshow(first_layer_activation[0, :, :, 10], cmap='viridis')
# ## Visualise Feature Maps
def deprocess_image(x):
x -= x.mean()
x /= (x.std() + 1e-5)
x *= 0.1
x += 0.5
x = np.clip(x, 0, 1)
x *= 255
x = np.clip(x, 0, 255).astype('uint8')
return x
def generate_pattern(layer_name, filter_index, size=256):
layer_output = model.get_layer(layer_name).output
loss = K.mean(layer_output[:, :, :, filter_index])
grads = K.gradients(loss, model.input)[0]
grads /= (K.sqrt(K.mean(K.square(grads))) + 1e-5)
iterate = K.function([model.input], [loss, grads])
input_img_data = np.random.random((1, size, size, 1)) * 20 + 128.
step = 1.
for i in range(40):
loss_value, grads_value = iterate([input_img_data])
input_img_data += grads_value * step
img = input_img_data[0]
return deprocess_image(img)
def visualise_feature_maps_for_layer(model, layer_name):
size = 256
margin = 5
results = np.zeros((8 * size + 7 * margin, 8 * size + 7 * margin, 3))
for i in range(8):
for j in range(8):
filter_img = generate_pattern(layer_name, i + (j * 8), size=size)
filter_img = filter_img
horizontal_start = i * size + i * margin
horizontal_end = horizontal_start + size
vertical_start = j * size + j * margin
vertical_end = vertical_start + size
results[horizontal_start: horizontal_end,
vertical_start: vertical_end, :] = filter_img
plt.figure(figsize=(20, 20))
plt.imshow(results.resize(size, size), cmap='gray',
interpolation='nearest')
def visualise_feature_maps(model, layer_names=None):
if layer_names is None:
layer_names = []
for layer in model.layers[:8]:
if "conv" in layer.name:
layer_names.append(layer.name)
for layer_name in layer_names:
visualise_feature_maps_for_layer(model, layer_name)
return
visualise_feature_maps(model, layer_names=['conv2d_21'])
# ---
def plot_feature_maps_for_layer(model, layer_name, size=256):
filter_index = 0
layer_output = model.get_layer(layer_name).output
loss = K.mean(layer_output[:, :, :, filter_index])
grads = K.gradients(loss, model.input)[0]
grads /= (K.sqrt(K.mean(K.square(grads))) + 1e-5)
iterate = K.function([model.input], [loss, grads])
loss_value, grads_value = iterate([np.zeros((1, size, size, 1))])
input_img_data = np.random.random((1, size, size, 1)) * 20 + 128.
step = 1.
for i in range(40):
loss_value, grads_value = iterate([input_img_data])
input_img_data += grads_value * step
img = input_img_data[0]
img = deprocess_image(img)
plt.imshow(img.reshape(size, size),
cmap='gray',
interpolation='nearest')
plot_feature_maps_for_layer(model, layer_name='conv2d_21')
plot_feature_maps_for_layer(model, layer_name='conv2d_22')
plot_feature_maps_for_layer(model, layer_name='conv2d_23')
| Chapter07/Notebooks/Training/CNNSketchClassifier_3_Visualise.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### Import Libraries
# import the necessary packages
from tensorflow.keras.applications.mobilenet_v2 import preprocess_input
from tensorflow.keras.preprocessing.image import img_to_array
from tensorflow.keras.models import load_model
from imutils.video import VideoStream
import numpy as np
import argparse
import imutils
import time
import cv2
import os
# ### Upload Alarm Sound
from pygame import mixer
mixer.init()
sound = mixer.Sound('mixkit-security-facility-breach-alarm-994.wav')
# ### Image Pre-Processing
def mask_detection_prediction(frame, faceNet, maskNet):
# find the dimension of frame and construct a blob
(h, w) = frame.shape[:2]
blob = cv2.dnn.blobFromImage(frame, 1.0, (224, 224),(104.0, 177.0, 123.0))
# pass the blob through the network and obtain the face detections
faceNet.setInput(blob)
detections = faceNet.forward()
# create a empty list which'll store list of faces,face location and prediction
faces = []
locs = []
preds = []
# loop over the detections
for i in range(0, detections.shape[2]):
# find the confidence or probability associated with the detection
confidence = detections[0, 0, i, 2]
# filter the strong detection [confidence > min confidence(let 0.5)]
if confidence > 0.5:
# find starting and ending coordinates of boundry box
box = detections[0, 0, i, 3:7] * np.array([w, h, w, h])
(startX, startY, endX, endY) = box.astype("int")
# make sure bounding boxes fall within the dimensions of the frame
(startX, startY) = (max(0, startX), max(0, startY))
(endX, endY) = (min(w - 1, endX), min(h - 1, endY))
# extract the face ROI, convert it from BGR to RGB channel
# ordering, resize it to 224x224, and preprocess it
face = frame[startY:endY, startX:endX]
face = cv2.cvtColor(face, cv2.COLOR_BGR2RGB)
face = cv2.resize(face, (224, 224))
face = img_to_array(face)
face = preprocess_input(face)
# append the face and bounding boxes to their respective lists
faces.append(face)
locs.append((startX, startY, endX, endY))
# only make a predictions if at least one face was detected
if len(faces) > 0:
# for faster inference we'll make batch predictions on *all*
# faces at the same time rather than one-by-one predictions
# in the above `for` loop
faces = np.array(faces, dtype="float32")
preds = maskNet.predict(faces, batch_size=32)
# return a 2-tuple of the face locations and their corresponding prediction
return (locs, preds)
# ### Load Caffe Model
# Caffe (Convolutional Architecture for Fast Feature Embedding) is a deep learning framework that allows users to create image classification and image segmentation models. It is a Caffe model which is based on the Single Shot-Multibox Detector (SSD) and uses ResNet-10 architecture as its backbone. It was introduced post OpenCV 3.3 in its deep neural network module.
# +
# load our serialized face detector model from disk
from os.path import dirname, join
prototxtPath = join("face_detector", "deploy.prototxt")
weightsPath = join("face_detector", "res10_300x300_ssd_iter_140000.caffemodel")
faceNet = cv2.dnn.readNet(prototxtPath, weightsPath)
# load the face mask detector model from disk
maskNet = load_model("fmd_model.h5")
# -
# ### Face Detection on Live Camera
# +
# initialize the video stream
print("[INFO] starting video stream...")
vs = VideoStream(src=0).start()
# loop over the frames from the video stream
while True:
# grab the frame from the threaded video stream and resize it
# to have a maximum width of 400 pixels
frame = vs.read()
frame = imutils.resize(frame, width=400)
# detect faces in the frame and determine if they are wearing a
# face mask or not
(locs, preds) = mask_detection_prediction(frame, faceNet, maskNet)
# loop over the detected face locations and their corresponding
# locations
for (box, pred) in zip(locs, preds):
# unpack the bounding box and predictions
(startX, startY, endX, endY) = box
(mask, withoutMask) = pred
if mask>withoutMask:
label = "Mask"
color = (0, 255, 0)
print("Normal")
else:
label = "No Mask"
color = (0, 0, 255)
sound.play()
print("Alert!!!")
# include the probability in the label
label = "{}: {:.2f}%".format(label, max(mask, withoutMask) * 100)
# display the label and bounding box rectangle on the output frame
cv2.putText(frame, label, (startX, startY - 10),cv2.FONT_HERSHEY_SIMPLEX, 0.45, color, 2)
cv2.rectangle(frame, (startX, startY), (endX, endY), color, 2)
# show the output frame
cv2.imshow("Frame", frame)
key = cv2.waitKey(1) & 0xFF
# if the `q` key was pressed, break from the loop
if key == ord("q"):
break
# do a bit of cleanup
cv2.destroyAllWindows()
vs.stop()
| detect_mask_live.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import torch
import torchvision
import torchvision.transforms as transforms
import os
import glob
# +
"""
Importing the required libraries.''
"""
import matplotlib.pyplot as plt
from PIL import Image
from tqdm import tqdm
import numpy as np
import glob
"""
Displaying the sample sketch and color images.
"""
for file in glob.glob('../input/anime-sketch-colorization-pair/data/train/*')[:5]:
f, a = plt.subplots(1,2, figsize=(10,5))
a = a.flatten()
img = Image.open(file).convert('RGB')
print(img)
a[0].imshow(img.crop((0, 0, 512,512))); a[0].axis('off');
a[1].imshow(img.crop((512, 0, 1024, 512))); a[1].axis('off');
plt.show()
print(file)
# + _cell_guid="b1076dfc-b9ad-4769-8c92-a6c4dae69d19" _uuid="8f2839f25d086af736a60e9eeb907d3b93b6e0e5"
'''
"""
Creating a directory for training data.
"""
get_ipython().system('rm -rf trainData_Image/Imagess')
get_ipython().system('rm -rf trainData_Sketches/Sketches')
get_ipython().system('rm -rf trainData_Images')
get_ipython().system('rm -rf trainData_Sketches')
get_ipython().system('mkdir trainData_Images')
get_ipython().system('mkdir trainData_Sketches')
get_ipython().system('mkdir trainData_Images/Images')
get_ipython().system('mkdir trainData_Sketches/Sketches')
"""
Preprocessing and saving the training data to corresponding directory.
"""
for idx, file in tqdm(enumerate(glob.glob('../input/anime-sketch-colorization-pair/data/train/*.png'))):
img = Image.open(file).convert('RGB')
img.crop((0, 0, 512,512)).save('./trainData_Images/Images/{}.png'.format(idx))
img.crop((512, 0, 1024, 512)).save('./trainData_Sketches/Sketches/{}.png'.format(idx))
'''
# +
"""
Creating a directory for validation/test data.
"""
get_ipython().system('rm -rf testData_Images')
get_ipython().system('rm -rf testData_Sketches')
get_ipython().system('rm -rf testData_Images/Images')
get_ipython().system('rm -rf testData_Sketches/Sketches')
get_ipython().system('mkdir testData_Images')
get_ipython().system('mkdir testData_Sketches')
get_ipython().system('mkdir testData_Images/Images')
get_ipython().system('mkdir testData_Sketches/Sketches')
print('yo')
"""
Preprocessing and saving the validation/test data to corresponding directory.
"""
for idx, file in tqdm(enumerate(glob.glob('../input/anime-sketch-colorization-pair/data/val/*.png'))):
if (idx ==1):
break
img = Image.open(file).convert('RGB')
img.crop((0, 0, 512,512)).save('./testData_Images/Images/{}.png'.format(idx))
img.crop((512, 0, 1024, 512)).save('./testData_Sketches/Sketches/{}.png'.format(idx))
# -
# # **import datas**
# ! ls testData_Sketches/Sketches
# +
import matplotlib.pyplot as plt
import numpy as np
import cv2
i=0
for file in enumerate(glob.glob('./testData_Images/Images/*.png')):
i+=1
file=glob.glob('./testData_Images/Images/*.png')
op=[]
print(i)
for i in range (0,1):
op.append(np.array(Image.open(file[i]).convert('RGB')))
#print(op)
# + _cell_guid="79c7e3d0-c299-4dcb-8224-4455121ee9b0" _uuid="d629ff2d2480ee46fbb7e2d37f6b5fab8052498a"
import torch
import torchvision
import torchvision.transforms as transforms
import os
from torch.utils.data import random_split
import glob
##### import CIFAR-10 dataset into trainset, testset# color dataset
transform_color = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.5), (0.5))
])
batchSize = 1
testset_color = torchvision.datasets.ImageFolder(root='./testData_Images/',
transform=transform_color)
testloader_color = torch.utils.data.DataLoader(testset_color, batch_size=batchSize,
shuffle=False, num_workers=2)
testset_sketch = torchvision.datasets.ImageFolder(root='./testData_Sketches/',
transform=transform_color)
testloader_sketch = torch.utils.data.DataLoader(testset_sketch, batch_size=batchSize,
shuffle=False, num_workers=2)
# Number of GPUs available. Use 0 for CPU mode.
ngpu = 1
# Decide which device we want to run on
device = torch.device("cuda:0" if (torch.cuda.is_available() and ngpu > 0) else "cpu")
print(device)
# +
import matplotlib.pyplot as plt
import numpy as np
import cv2
# functions to show an image
def imshow(img):
img = img / 2 + 0.5 # unnormalize
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1, 2, 0)))
plt.show()
# get some random training images
dataiter = iter(testloader_color)
images_color, labels = dataiter.next()
# show images
imshow(torchvision.utils.make_grid(images_color))
# show dataiter shape
print(images_color.shape)
# +
import matplotlib.pyplot as plt
import numpy as np
import cv2
# functions to show an image
def imshow(img):
img = img / 2 + 0.5 # unnormalize
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1, 2, 0)))
plt.show()
# get some random training images
dataiter = iter(testloader_sketch)
images_gray, labels = dataiter.next()
# show images
imshow(torchvision.utils.make_grid(images_gray))
# show dataiter shape
print(images_gray.shape)
print("images_gray shape: ", images_gray.shape)
# -
# ### Define the MLP network
# +
import torch
import torchvision
import torchvision.transforms as transforms
import os
import glob
import torch.nn as nn
import torch.nn.functional as F
class MLP_net(nn.Module):
def __init__(self):
# Input layer is 786432, since gray_image is 3x512x512
# Output layer is 786432, since color_image is 32x32x3
super(MLP_net, self).__init__()
self.layer1 = nn.Linear(786432,1024).cuda() #TODO changer ça
#self.bn1 = nn.BatchNorm1d(1024).cuda()
self.layer2 = nn.Linear(1024,786432).cuda()
def forward(self, x):
# convert tensor
x = x.view(x.size(0), -1)
x = self.layer1(x)
#x = self.bn1(x)
x = self.layer2(x)
return x
print("yo")
ngpu = 1
device = torch.device("cuda:0" if (torch.cuda.is_available() and ngpu > 0) else "cpu")
print(device)
mlp_net = MLP_net().to(device)
print("is cude: ", next(mlp_net.parameters()).is_cuda)
# -
# ### Define the loss function and optimizer
# +
import torch.optim as optim
criterion = nn.MSELoss()
optimizer = optim.SGD(mlp_net.parameters(), lr=0.001, momentum=0.9)
# -
# ### Tensorboard to save all training output logs
# +
from torch.utils.tensorboard import SummaryWriter
# Writer will output to ./runs/ directory by default
writer = SummaryWriter("./runs_mlp/")
# -
# ### Train the network
history_loss=[]
history_val_loss=[]
for epoch in range(50): # loop over the dataset multiple times
running_loss = 0.0
num_training = 0
for i, data_color in enumerate(testloader_color, 0):
for j, data_gray in enumerate(testloader_color, 0):
if (i==j):
# get the inputs; data is a list of [inputs, labels]
images_color, labels_color = data_color
images_gray, labels_color = data_gray
# put data in gpu/cpu
images_color = images_color.to(device)
images_gray = images_gray.to(device)
# zero the parameter gradients
optimizer.zero_grad()
# forward + backward + optimize
# input: images_gray
images_gray = images_gray.view(images_gray.shape[0],-1)
print("images_gray: ", images_gray.shape)
outputs = mlp_net(images_gray)
# change the shape of images_color: [batchSize,3,32,32] -> [batchSize,3072]
images_color = images_color.reshape(images_color.shape[0], -1)
loss = criterion(outputs, images_color)
loss.backward()
optimizer.step()
# print output statistics
running_loss += loss.item()
j = 200 # print every 200 mini-batches
if i % j == (j-1): # print every 200 mini-batches
print("trainingloss:",'[%d, %5d] loss: %.3f' %
(epoch + 1, i + 1, running_loss / j))
writer.add_scalar('Loss/train', running_loss/j, len(trainset_color)*epoch + i)
history_loss.append(running_loss / j)
running_loss = 0.0
break
'''
running_val_loss = 0.0
for i, data_val_color in enumerate(valloader_color, 0):
# get the inputs; data is a list of [inputs, labels]
images_val_color, labels_val_color = data_val_color
images_val_gray = rgb2gray_batch(images_val_color)
# put data in gpu/cpu
images_val_color = images_val_color.to(device)
images_val_gray = images_val_gray.to(device)
# forward + backward + optimize
# input: images_val_gray
images_val_gray = images_val_gray.view(images_val_gray.shape[0],-1)
# print("images_val_gray: ", images_val_gray.shape)
outputs_val = mlp_net(images_val_gray)
# change the shape of images_val_color: [batchSize,3,32,32] -> [batchSize,3072]
images_val_color = images_val_color.reshape(images_val_color.shape[0], -1)
loss_val = criterion(outputs_val, images_val_color)
# print output statistics
running_val_loss += loss_val.item()
j = 10 # print every 200 mini-batches
if i % j == (j-1): # print every 200 mini-batches
print("validation",'[%d, %5d] loss: %.3f' %
(epoch + 1, i + 1, running_val_loss / j))
writer.add_scalar('Loss/val', running_val_loss/j, len(valset_color)*epoch + i)
history_val_loss.append(running_val_loss / j)
running_val_loss = 0.0
'''
writer.flush()
writer.close()
print('Finished Training')
# ### Loss graphic evolution displayed (on training set and validation set)
# +
# Defining Figure
f = plt.figure(figsize=(10,7))
f.add_subplot()
#Adding Subplot
plt.plot(range(len(history_loss)), history_loss, label = "loss") # Loss curve for training set
plt.plot(range(len(history_val_loss)), history_val_loss, label = "val_loss") # Loss curve for validation set
plt.title("Loss Curve",fontsize=18)
plt.xlabel("Epochs",fontsize=15)
plt.ylabel("Loss",fontsize=15)
plt.grid(alpha=0.3)
plt.legend()
plt.savefig("Loss_curve.png")
plt.show()
# -
# ### Inference on train dataset
# +
# get some random training images
dataiter = iter(trainloader_color)
images_color, labels_color = dataiter.next()
print("images_color: ", images_color.shape)
# show images
images_color_show = images_color.reshape(batchSize,3,32,32)
imshow(torchvision.utils.make_grid(images_color_show.detach()))
images_gray = rgb2gray_batch(images_color)
print("images_gray: ", images_gray.shape)
# show images
images_gray = images_gray.reshape(batchSize,1,32,32)
imshow(torchvision.utils.make_grid(images_gray.detach()))
# run inference on the network
# oututs [4,3072]
images_gray = images_gray.to(device)
outputs = mlp_net(images_gray)
print(outputs.shape)
images_color = outputs.reshape(batchSize,3,32,32)
print(images_color.shape)
images_color = images_color.to("cpu")
imshow(torchvision.utils.make_grid(images_color.detach()))
# -
# ### Inference on validation dataset
# +
# get some random training images
dataiter = iter(valloader_color)
images_color, labels_color = dataiter.next()
print("images_color: ", images_color.shape)
# show images
images_color_show = images_color.reshape(batchSize,3,32,32)
imshow(torchvision.utils.make_grid(images_color_show.detach()))
images_gray = rgb2gray_batch(images_color)
print("images_gray: ", images_gray.shape)
# show images
images_gray = images_gray.reshape(batchSize,1,32,32)
imshow(torchvision.utils.make_grid(images_gray.detach()))
# run inference on the network
# oututs [4,3072]
images_gray = images_gray.to(device)
outputs = mlp_net(images_gray)
print(outputs.shape)
images_color = outputs.reshape(batchSize,3,32,32)
print(images_color.shape)
images_color = images_color.to("cpu")
imshow(torchvision.utils.make_grid(images_color.detach()))
# -
# ### Inference on test dataset
# +
# get some random training images
dataiter = iter(testloader_color)
images_color, labels_color = dataiter.next()
print("images_color: ", images_color.shape)
# show images
images_color_show = images_color.reshape(batchSize,3,32,32)
imshow(torchvision.utils.make_grid(images_color_show.detach()))
images_gray = rgb2gray_batch(images_color)
print("images_gray: ", images_gray.shape)
# show images
images_gray = images_gray.reshape(batchSize,1,32,32)
imshow(torchvision.utils.make_grid(images_gray.detach()))
# run inference on the network
# oututs [batchSize,3072]
images_gray = images_gray.to(device)
outputs = mlp_net(images_gray)
print(outputs.shape)
images_color = outputs.reshape(batchSize,3,32,32)
print(images_color.shape)
images_color = images_color.to("cpu")
imshow(torchvision.utils.make_grid(images_color.detach()))
images_gray = images_gray.to("cpu")
images_gray = torch.cat((images_gray, images_gray, images_gray), 1)
final_result_display = torch.cat((images_color_show, images_gray, images_color), 0)
imshow(torchvision.utils.make_grid(final_result_display.detach()))
| src_Kaggle/anime-colorization-mlp-pytorch.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Troubleshooting node id's
#
# <NAME>, July 2018
#
# The problem is that a large portion of households aren't being matched to walk network aggregations, which might be affecting the regression results.
import numpy as np
import pandas as pd
import sys
print(sys.version)
# ### Check if saved data tables exhibit the problem
p = pd.read_csv('../data/parcels_with_nodes.csv').set_index('primary_id')
len(p)
p.columns
nw = pd.read_csv('../data/nodeswalk_vars.csv').set_index('osmid')
len(nw)
nw.head(3)
len(p.loc[~p.node_id_walk.isin(nw.index)])
len(p.loc[~p.node_id_walk.isin(nw.index)])/len(p)
# Yes, 29% of the parcels have node id's that aren't in the aggregation table
# +
# How many nodes is that?
len(p.loc[~p.node_id_walk.isin(nw.index)].node_id_walk.unique())
# -
len(p.loc[~p.node_id_walk.isin(nw.index)].node_id_walk.unique())/len(p.node_id_walk.unique())
# ### What does the network file look like?
nodes = pd.read_csv('../data/bayarea_walk_nodes.csv').set_index('osmid')
len(nodes)
nodes.head(3)
len(p.loc[~p.node_id_walk.isin(nodes.index)])
# ### Any nodes in the aggregation table that aren't in the units table?
len(nw.loc[~nw.index.isin(p.node_id_walk)])
# Yes, meaning that it might not be a pandana problem..
# ### Create some fresh aggregations
import orca
import os; os.chdir('..')
import pandana as pdna
from urbansim.utils import misc, networks
import warnings;warnings.simplefilter('ignore')
os.getcwd()
pdna.__version__
d = 'data/'
# +
@orca.table(cache=True)
def parcels():
df = pd.read_csv(
d + 'mtc_data_platform_format_7-6-18/' + 'parcel_attr.csv',
# d + 'parcels_with_nodes.csv',
index_col='primary_id', dtype={'primary_id': int, 'block_id':str})
return df
@orca.table(cache=True)
def buildings():
df = pd.read_csv(
d + 'mtc_data_platform_format_7-6-18/' + 'buildings_v2.csv',
index_col='building_id', dtype={'building_id': int, 'parcel_id': int})
df['res_sqft_per_unit'] = df['residential_sqft'] / df['residential_units']
df['res_sqft_per_unit'][df['res_sqft_per_unit'] == np.inf] = 0
return df
@orca.table(cache=True)
def units():
df = pd.read_csv(
d + 'mtc_data_platform_format_7-6-18/' + 'units_v2.csv',
index_col='unit_id', dtype={'unit_id': int, 'building_id': int})
return df
@orca.table(cache=True)
def households():
df = pd.read_csv(
d + 'mtc_data_platform_format_7-6-18/' + 'households_v2.csv',
index_col='household_id', dtype={
'househould_id': int, 'block_group_id': str, 'state': str,
'county': str, 'tract': str, 'block_group': str,
'building_id': int, 'unit_id': int})
return df
# -
@orca.step()
def initialize_network_walk():
@orca.injectable('netwalk', cache=True)
def build_networkwalk():
nodeswalk = pd.read_csv(d + 'bayarea_walk_nodes.csv') \
.set_index('osmid')
edgeswalk = pd.read_csv(d + 'bayarea_walk_edges.csv')
netwalk = pdna.Network(nodeswalk.x, nodeswalk.y, edgeswalk.u, \
edgeswalk.v, edgeswalk[['length']], twoway=True)
netwalk.precompute(2500)
return netwalk
parcels = orca.get_table('parcels').to_frame(columns=['x', 'y'])
idswalk_parcel = orca.get_injectable('netwalk').get_node_ids(parcels.x, parcels.y)
orca.add_column('parcels', 'node_id_walk', idswalk_parcel, cache=False)
orca.broadcast('nodeswalk', 'parcels', cast_index=True, onto_on='node_id_walk')
@orca.column('buildings', 'node_id_walk')
def node_id(parcels, buildings):
return misc.reindex(parcels.node_id_walk, buildings.parcel_id)
@orca.column('units', 'node_id_walk')
def node_id(buildings, units):
return misc.reindex(buildings.node_id_walk, units.building_id)
@orca.column('households', 'node_id_walk')
def node_id(units, households):
return misc.reindex(units.node_id_walk, households.unit_id)
@orca.step()
def network_aggregations_walk_test(netwalk):
nodeswalk = networks.from_yaml(netwalk, 'network_aggregations_walk_test.yaml')
nodeswalk = nodeswalk.fillna(0)
print(nodeswalk.describe())
orca.add_table('nodeswalk', nodeswalk)
orca.run(["initialize_network_walk"])
# ### Aggregations are of households and buildings - what do those id's look like?
h = orca.get_table('households').to_frame()
len(h.loc[(~h.node_id_walk.isnull()) & (~h.node_id_walk.isin(nw.index))])
b = orca.get_table('buildings').to_frame()
len(b.loc[~b.node_id_walk.isin(nw.index)])
# Those tables also have some ids missing from the saved aggregation table
# ### Ok, do the aggregations
orca.run(["network_aggregations_walk_test"])
nw2 = orca.get_table('nodeswalk').to_frame()
len(h.loc[(h.building_id>-1) & (~h.node_id_walk.isin(nw2.index))])
len(p.loc[~p.node_id_walk.isin(nw2.index)])
ids = orca.get_injectable('netwalk').node_ids
len(ids)
len(nw2)
len(nw2.loc[~nw2.index.isin(p.node_id_walk)])
# All the nodes are included in this aggregation table, including nodes that are in the network but not matched to any parcels. So it's not a pandana or core urbansim bug.
nw2.to_csv('data/nodeswalk_test.csv')
# ### Code to run an aggregation with no external file dependencies
with open('configs/net_test.yaml', 'w') as t:
t.write('''
name: network_aggregations
desc: Network aggregations
model_type: networks
node_col: node_id_walk
variable_definitions:
- name: pop_500_walk
dataframe: households
varname: persons
radius: 500
decay: flat
''')
@orca.step()
def net_test(netwalk):
nodestest = networks.from_yaml(netwalk, 'net_test.yaml')
nodestest = nodestest.fillna(0)
print(nodestest.describe())
orca.add_table('nodestest', nodestest)
orca.run(["net_test"])
nt = orca.get_table('nodestest').to_frame()
len(h.loc[(h.building_id>-1) & (~h.node_id_walk.isin(nt.index))])
| summer-2018-model/notebooks-sam/Node-id-troubleshooting.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Pre-processing and Training Data Development
# - Goal is to create a cleaned development dataset you can use to complete the modeling step of this project.
# +
# Import libraries
import pandas as pd
pd.options.mode.chained_assignment = None # default='warn'
import os
from sklearn.preprocessing import MaxAbsScaler
import matplotlib.pyplot as plt
#suppress future warnings
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
warnings.simplefilter(action='ignore', category=UserWarning)
# Show plots inline
# %matplotlib inline
# -
os.getcwd()
df = pd.read_csv("data/Quora_cleaned.csv")
df.shape
df.head(5)
df['question_stemmed']= df['question_stemmed'].str.replace('”', '')
df['question_stemmed']= df['question_stemmed'].str.replace('“', '')
df.info()
# Drop index columns
df.drop('Unnamed: 0',axis=1, inplace=True)
df.head(10)
# Rename question_stemmed column
df = df.rename(columns = {"question_stemmed":"question_final"})
df.head()
# Print the data type of each column
print(df.dtypes)
# ### Standardize the magnitude of numeric features using a scaler
# +
# Find the average length of word
df['avg_word_length'] = df['question_length'] / df['total_words']
# Print the first 5 rows of these columns
print(df[['question_final', 'question_length', 'total_words', 'avg_word_length']].head(5))
# -
# Create subset of only the numeric columns
numeric_df = df.select_dtypes(include=['int64','float'])
print(numeric_df.columns)
# Create subset of only the categorical variable columns
categorical_df = df.select_dtypes(include=['object'])
print(categorical_df.columns)
numeric_df['avg_word_length'].describe()
# +
# Instantiate StandardScaler to generate the absolute values are mapped in the range [0, 1].
# This is optional step for our dataset since there are not many outliers, however, this condenses data even better
scaler = MaxAbsScaler()
# Fit SS_scaler to the data
scaler.fit(numeric_df[['avg_word_length']])
# Transform the data using the fitted scaler
numeric_df['avg_word_length_scaled'] = scaler.transform(numeric_df[['avg_word_length']])
# Compare the origional and transformed column
print(numeric_df[['avg_word_length_scaled', 'avg_word_length']].head(5))
# +
# Create a histogram for the 2
plt.rcParams["figure.figsize"] = (10,5)
numeric_df.hist(['avg_word_length','avg_word_length_scaled'])
plt.show()
# -
# Drop non scaled column
numeric_df.drop(columns='avg_word_length', inplace=True)
numeric_df.head(5)
numeric_df['avg_word_length_scaled'].describe()
df_numeric_scaled = pd.concat([categorical_df, numeric_df], axis=1, sort=False)
df_numeric_scaled.head(5)
df_numeric_scaled.info()
# ### Perform Count vectorization (OR TFIDF) on categorical feature to fit and transform data
# +
from sklearn.feature_extraction.text import ENGLISH_STOP_WORDS
#'quantum' in ENGLISH_STOP_WORDS
#'scammers' in ENGLISH_STOP_WORDS
'ask stupid question' in ENGLISH_STOP_WORDS
# +
# Import CountVectorizer
#from sklearn.feature_extraction.text import CountVectorizer
#from nltk.tokenize import TreebankWordTokenizer
#import re
# Instantiate a trigram vectorizer
# In data Wrangling step, stop words were removed using nltk. However at this step, we will remove stop_words using sklearn library too
#cv_bigram_vec = CountVectorizer(stop_words='english',
# max_features=500,
# tokenizer=TreebankWordTokenizer().tokenize,
# ngram_range=(1,1))
# Fit and apply trigram vectorizer
#cv_bigram = cv_bigram_vec.fit_transform(df_numeric_scaled['question_final'])
# Print the trigram features
#print(cv_bigram_vec.get_feature_names())
# Create a DataFrame of the features
#cv_bi_df = pd.DataFrame(cv_bigram.toarray(),
# columns=cv_bigram_vec.get_feature_names())
# +
#print(cv_bi_df.sum().sort_values(ascending=False).head(5))
# +
#TFIDF representation (ignoring common words)
# Import TfidfVectorizer
from sklearn.feature_extraction.text import TfidfVectorizer
# Instantiate TfidfVectorizer
tv = TfidfVectorizer(max_features=200, ngram_range=(1, 1), stop_words = 'english')
# Fit the vectroizer and transform the data
tv_transformed = tv.fit_transform(df['question_final'])
# Create a DataFrame with these features
tv_df = pd.DataFrame(tv_transformed.toarray(),
columns=tv.get_feature_names())
print(tv_df.head())
# +
# Isolate the row to be examined
sample_row = tv_df.iloc[0]
# Print the top 5 words of the sorted output
print(sample_row.sort_values(ascending=False).head(5))
# -
df_final_tfidf = pd.concat([df_numeric_scaled, tv_df], axis=1, sort=False)
df_final_tfidf.head(5)
# +
#df_final = pd.concat([df_numeric_scaled, cv_bi_df], axis=1, sort=False)
#df_final.head(5)
# -
# ### Split into testing and training datasets
# +
#from sklearn.model_selection import train_test_split
#train_df, test_df = train_test_split(df_final, test_size=0.3)
# +
from sklearn.model_selection import train_test_split
train_df_tfidf, test_df_tfidf = train_test_split(df_final_tfidf, test_size=0.3)
# -
train_df_tfidf.shape
test_df_tfidf.shape
# +
#train_df.shape
# +
#test_df.shape
# +
#test_df.head(5)
# +
#test_df.to_csv("data/FeatureEngineering_Test.csv")
# +
#train_df.to_csv("data/FeatureEngineering_Train.csv")
# -
test_df_tfidf.to_csv("data/FeatureEngineering_tfidf_Test.csv")
train_df_tfidf.to_csv("data/FeatureEngineering_tfidf_Train.csv")
| notebooks/3. Preprocessing and Training Data Development.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .jl
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Julia 0.5.0-rc4
# language: julia
# name: julia-0.5
# ---
# ### Integral BL code for cylinder
include("bl_cyl.jl")
t, x, del, E = IBLm_cyl()
using PyPlot
plot(x[2:end], diff(del))
# +
### Express an airfoil as a Fourier series.
using PyPlot
using ForwardDiff
function thick(x)
num = 5
b1 = 0.2969
b2 = -0.1260
b3 = -0.3516
b4 = 0.2843
b5 = -0.1015
th = 5.*num*(b1*sqrt(x) + b2*x + b3*x^2 + b4*x^3 + b5*x^4)
end
# +
# Consider a NACA 0012 airfoil.
ncell = 64
num = 0.12
x = zeros(ncell + 2)
th = zeros(ncell + 2)
thdot = zeros(ncell + 2)
dx = 1./(ncell + 1.)
for ic = 1:ncell+2
x[ic] = real(ic-1)*dx
end
for ic = 1:ncell+2
th[ic] = thick(x[ic])
end
for ic = 1:ncell+2
thdot[ic] = ForwardDiff.derivative(thick,x[ic])
end
# -
thdot
thdot[10] = ForwardDiff.derivative(thick,1.1)
thdot
| Notebooks/BL_cyl.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### Part 1
# #### Code that downloads the page to your local file system (for hotels.com)
# #### Code that stores URLs from tripadvisor into a text file
# Please note we considered Hotel Listings between 15th to 16th March,2020
#
# +
import os
#Create a directory for final project
try:
if not os.path.exists('Final_DDR_Assignment'):
os.makedirs('Final_DDR_Assignment')
print('Final_DDR_Assignment:Created')
except OSError:
print ('Error: Creating directory. ' + 'Final_DDR_Assignment')
#os.chdir(new_path) --to change the directory
# -
#getting the current directory
os.getcwd()
#Importing libraries
import requests
import re
from requests import get
from bs4 import BeautifulSoup
import time
# #### Source 1: TripAdvisor
# #### Creating a list of URLs for scraping
# +
with open("hotel_links_tripadvisor.txt", "w") as file:
for i in range(0,270,30):
url = "https://www.tripadvisor.com/Hotels-g60713-oa"+str(i)+"-San_Francisco_California-Hotels"
user_agent = {'User-agent': 'Mozilla/5.0'}
# Requesting the tripadvisor website access to the server and storing the response
res = get(url,user_agent)
# Parsing the response and storing the resulting html
data = BeautifulSoup(res.text, 'html.parser')
items = data.select('div.listing_title')
#print(items)
for item in items:
url_b=item.find('a')['href']
url= "https://www.tripadvisor.com"+url_b
print(url)
file.write(url+"\n")
file.close()
# -
# #### Source 2: Hotels.com
# #### Getting hotel pages from Hotels.com for scraping
base_url = '''https://www.hotels.com/search.do?q-destination=San%20Francisco&
q-check-in=2020-03-15&q-check-out=2020-03-16&q-rooms=1&q-room-0-adults=2&q-room-0-children=0&pn='''
headers = {'User-Agent': 'Mozilla/5.0'}
a=list(range(1,100))
for number in a:
user_agent='Mozilla/5.0'
url=base_url+str(number)
response= requests.get(url, headers)
filename = 'hotels_pg'+str(number)+'.htm'
print(filename)
with open(filename,'w') as file:
file.write(str(response.content))
# ### Part 2
# #### Web-scraping
# - Connecting to MySql
# - Creating structures for the database tables
# - Scraping and simultaneously inserting
#
# Also, We used INSERT IGNORE to avoid duplicate entries in our table as it ignores whenever a duplicate row tries to insert. Since we made Hotel Name an unique column. It worked properly
# +
#Connecting to MySQL using Python connector
import mysql.connector
#establishing the connection
#Adding an exception block to know connection status
try:
conn = conn = mysql.connector.connect(
user='root', password='', host='localhost'
)
except mysql.connector.Error as err:
if err.errno == errorcode.ER_ACCESS_DENIED_ERROR:
print("Something is wrong with your user name or password")
elif err.errno == errorcode.ER_BAD_DB_ERROR:
print("Database does not exist")
else:
print(err)
else:
if (conn.is_connected()):
print("Connected")
# -
#Creating a cursor object using the cursor() method
cursor = conn.cursor()
#Executing cursor with execute method and pass SQL query
#cursor.execute("CREATE DATABASE Hotel_data")
#Get list of all databases to check
cursor.execute("SHOW DATABASES")
#print all databases
for db in cursor:
print(db)
# #### Creating Table structure
# ##### For TRIPADVISOR
# +
#Dropping tripadvisor table if already exists.
#cursor = conn.cursor()
cursor.execute("USE Hotel_data")
cursor.execute("DROP TABLE IF EXISTS Hotel_data.tripadvisor_sf_hotel")
#Creating table as per requirement
sql ='''CREATE TABLE tripadvisor_sf_hotel(
HOTEL_ID INT(30) PRIMARY KEY AUTO_INCREMENT NOT NULL ,
Hotel_Name VARCHAR(120) UNIQUE NOT NULL,
Neighborhood VARCHAR(120),
Hotel_Class FLOAT,
Total_Reviews INT(7),
Lang_spoken VARCHAR(200),
Amenities VARCHAR(500),
Deal_Price INT(4),
Special_Offer CHAR,
Offer_Desc VARCHAR(150),
Walking_Score INT(3),
Num_Restaurants INT(3),
Num_Attractions INT(3),
SFO_dist_mi FLOAT,
OAK_dist_mi FLOAT,
Contact_No VARCHAR(20),
Contact_Address VARCHAR(200)
)'''
cursor.execute(sql)
# -
# ##### Hotels.com details
# +
#Dropping OMDB_test table if already exists.
#cursor = conn.cursor()
cursor.execute("USE Hotel_data")
cursor.execute("DROP TABLE IF EXISTS Hotel_data.hotelscom_sf_hotel")
#Creating table as per requirement
sql ='''CREATE TABLE hotelscom_sf_hotel(
HOTEL_ID INT(30) PRIMARY KEY AUTO_INCREMENT NOT NULL ,
hotel_name VARCHAR(150) UNIQUE,
hotel_addr VARCHAR(500),
hotel_location_1 VARCHAR(150),
hotel_location_2 VARCHAR(150),
hotel_rewards VARCHAR(150),
hotel_amenities VARCHAR(400),
hotel_ratings FLOAT,
hotel_reviews INT(6),
hotel_specialoffer CHAR,
hotel_specialdeal VARCHAR(50),
hotel_specialdeal_details VARCHAR(100),
hotel_price INT(4),
fully_booked VARCHAR(100),
hotel_sponsored VARCHAR(25),
hotel_deals VARCHAR(20)
)'''
cursor.execute(sql)
#Get database table'
cursor.execute("SHOW TABLES")
for table in cursor:
print(table)
# +
# importing required libraries
import requests, json, random
from bs4 import BeautifulSoup
import time, re
import pandas as pd
# -
# #### Inserting scraped data from Tripadvisor to corresponding Table
# +
import numpy as np
#Opening the file in the directory
with open("hotel_links_tripadv.txt", "r") as f:
#for each link in the text file
for x in f:
#open each file in read bytes mode
trial_url=x.rstrip('\n')
headers = {'user-agent': 'Mozilla/5.0'}
response = requests.get(trial_url, headers = headers)
#parse it through Beautiful soup
data = BeautifulSoup(response.text, 'html.parser')
#Get Hotel-name
Hotel_name=data.find('h1',id='HEADING').string
#Get total number of reviews
try:
tot_reviews=data.find('a', href='#REVIEWS').find_next(string=True).strip('reviews').astype(int)
except AttributeError:
tot_reviews=None
#Hotel Class
h_class=data.find('div',class_='ssr-init-26f').find('span').string
#Neighborhood Name
try:
neigh_name=data.find('div',class_=re.compile('.*Neighborhood__name.*')).get_text()
except AttributeError:
neigh_name='Not Available'
#Deal Price Offered
try:
deal_price=data.find('div',class_=re.compile('hotels-hotel-offers-DetailChevronOffer__price--py2LH.*')).get_text().lstrip('$')
except AttributeError:
deal_price=None
try:
for i in data.find_all('div',class_='hotels-hr-about-layout-TextItem__textitem--2JToc'):
if i.previous_sibling.string=='HOTEL STYLE':
h_style=i.get_text()
elif i.previous_sibling.string=='Languages Spoken':
lang_spoken=i.get_text()
elif i.previous_sibling.string=='NUMBER OF ROOMS':
num_of_rooms=i.get_text()
else: pass
except AttributeError:
h_style=None
lang_spoken=None
num_rooms=None
#distance from nearby airports
distance=data.select('span.hotels-hotel-review-location-NearbyTransport__distance--24tmE > span >span.number')
SFO_dist=distance[0].text #SFO internation Airport
Oak_dist=distance[1].text #Oakland Intl Airport
#Hotel Address
address= data.find('span',class_='public-business-listing-ContactInfo__ui_link--1_7Zp public-business-listing-ContactInfo__level_4--3JgmI').get_text()
#Hotel Phone number
try:
contact_no= data.find('span',class_=re.compile('public-business-listing-ContactInfo__nonWebLinkText--nGymU public-business-listing.*')).get_text().strip(' ')
except AttributeError:
contact_no=None
#List of amenities available
a= data.find_all('div', attrs={"data-test-target": "amenity_text"})
amenities_list=[]
for i in a:
amenities_list.append(i.get_text())
#flag for special offer
special_offer= True if data.find('div',class_=re.compile('.*SpecialOffer,*')) is not None else False
#Capture the specific specialoffer
offer_text= 'None' if special_offer is False else data.find('div',class_=re.compile('.*SpecialOffer,*')).find_next(string=True)
#Walking Score --grabbing the number after 'Grade: '
try:
walk_score=data.find('div',class_=re.compile('.*Infographic__walkscoreSubtitle.*')).find_next(string=True)[7:10]
#please note planning to use Regex here instead to extract number from the string
except AttributeError:
walk_score=None
#Number of restaurants closeby
try:
num_rest=data.find('span',class_=re.compile('.*Highlight__orange--1N.*')).string
except AttributeError:
num_rest=None
#Number of attractions closeby
try:
num_attrac=data.find('span',class_=re.compile('.*location-layout-Highlight__blue.*')).string
except AttributeError:
num_attrac=None
#Closing File
#f.close()
#inserting into the table omdb
sql_query = '''INSERT IGNORE INTO tripadvisor_sf_hotel(Hotel_Name,Neighborhood,Hotel_Class,Total_Reviews,Lang_spoken,Amenities,Deal_Price,Special_Offer,Offer_Desc,Walking_Score,Num_Restaurants,Num_Attractions,SFO_dist_mi,OAK_dist_mi,Contact_No,Contact_Address
) VALUES(%s, %s, %s, %s, %s, %s,%s, %s, %s, %s, %s, %s, %s, %s, %s, %s)'''
record=(Hotel_name, neigh_name,h_class,tot_reviews,lang_spoken,json.dumps(amenities_list[0:10]),deal_price,special_offer,offer_text,walk_score,num_rest,num_attrac, SFO_dist, Oak_dist,contact_no,address)
cursor.execute(sql_query,record)
print("inserted")
print("\n")
# -
# #### QC:
# check the first 100 rows in the tripadvisor table
# +
conn.commit()
#validating whether records were inserted
cursor.execute("SELECT * FROM tripadvisor_sf_hotel LIMIT 100")
records = cursor.fetchall()
print("First 100 rows in tripadvisor dataset are: ")
for record in records:
print(record)
# -
# ##### Inserting scraped data from hotels.com to the corresponding table
# +
import string
import re
a=list(range(1,100))
for number in a:
filename = 'hotels_pg'+str(number)+'.htm'
with open(filename, 'r', encoding='utf-8') as f:
text = f.read()
soup = BeautifulSoup(text, 'html.parser')
all_hotels=soup.find_all('article')
for hotel in all_hotels:
#name
hotel_name=hotel.find('h3').get_text()
#addr
hotel_addr=hotel.find('address').get_text()
#location
if hotel.find('a', attrs={'class':'map-link xs-welcome-rewards'}) is None:
hotel_location_1=None
else:
hotel_location_1=hotel.find('a', attrs={'class':'map-link xs-welcome-rewards'}).get_text()
hotel_location_2=hotel.find('ul', attrs={'class':'property-landmarks'}).get_text()
#rewards
hotel_rewards_check=hotel.find('div', attrs={'class':'welcome-rewards widget-tooltip widget-tooltip-tr'})
if hotel_rewards_check is None:
hotel_rewards=None
else:
hotel_rewards=hotel.find('div', attrs={'class':'welcome-rewards widget-tooltip widget-tooltip-tr'}).get_text()
if hotel.find('ul', attrs={'class':'hmvt8258-amenities'}) is None:
hotel_amenities=None
else:
hotel_amenities=hotel.find('ul', attrs={'class':'hmvt8258-amenities'}).get_text()
#ratings
try:
hotel_ratings=hotel.find('strong').get_text()
hotel_ratings=float(re.sub('[A-Z|a-z]+', '',hotel_ratings).replace(' ',''))
except (AttributeError, ValueError):
hotel_ratings=None
#Reviews
try:
hotel_reviews=hotel.find('span', attrs={'class':'small-view'}).get_text().replace(',', '')
NumRegex = re.compile(r'([0-9])+')
number = NumRegex.search(hotel_reviews)
hotel_reviews=int(number.group())
except AttributeError:
hotel_reviews=None
#special_offer
if hotel.find('span', attrs={'class':'deal-text'}) is None:
hotel_specialoffer=None
else:
hotel_specialoffer=hotel.find('span', attrs={'class':'deal-text'}).get_text()
#special_deal
if hotel.find('span', attrs={'class':'special-deal-badge'}) is None:
hotel_specialdeal='N'
hotel_specialdeal_details=None
else:
hotel_specialdeal='Y'
hotel_specialdeal1=hotel.find('span', attrs={'class':'special-deal-badge'})
hotel_specialdeal_details=hotel_specialdeal1.find_next('span').get_text()
if hotel.find('ins') is None:
hotel_price=None
if hotel.find('p', attrs={'class':'sold-out-message'}) is not None:
fully_booked=hotel.find('p', attrs={'class':'sold-out-message'}).get_text()
else:
fully_booked=None
else:
hotel_price=hotel.find('ins').get_text().lstrip('$')
fully_booked=None
if hotel.find('div', attrs={'class':'sponsored-label'}) is None:
hotel_sponsored="Not sponsored"
else:
hotel_sponsored=hotel.find('div', attrs={'class':'sponsored-label'}).get_text()
#other deals _free cancellation & pay at hotel
if hotel.find('ul', attrs={'class':'deals'}) is None:
hotel_deals=None
else:
hotel_deals=(hotel.find('ul', attrs={'class':'deals'}).get_text()).strip()
sql_query = '''INSERT IGNORE INTO hotelscom_sf_hotel(hotel_name,hotel_addr,hotel_location_1,hotel_location_2,
hotel_rewards,hotel_amenities,hotel_ratings,hotel_reviews,hotel_specialoffer,hotel_specialdeal,hotel_specialdeal_details,
hotel_price,fully_booked,hotel_sponsored,hotel_deals)
VALUES(%s, %s, %s, %s, %s, %s,%s, %s, %s, %s, %s, %s, %s, %s, %s)'''
record=(hotel_name,hotel_addr,hotel_location_1,hotel_location_2,hotel_rewards,hotel_amenities,hotel_ratings,hotel_reviews,hotel_specialoffer,hotel_specialdeal,hotel_specialdeal_details,hotel_price,fully_booked,hotel_sponsored,hotel_deals)
cursor.execute(sql_query,record)
print("inserted")
print("\n")
# -
# #### QC:
# Check first 100 rows for hotels.com
# +
conn.commit()
#validating whether records were inserted
cursor.execute("SELECT * FROM hotelscom_sf_hotel LIMIT 100")
records = cursor.fetchall()
print("First 100 rows in Hotels.com dataset are: ")
for record in records:
print(record)
# -
# #### Checking how many hotels are common across the two sources
# +
sql ='''SELECT a.hotel_name , b.hotel_name
FROM hotelscom_sf_hotel a
JOIN tripadvisor_sf_hotel b
ON a. Hotel_name=b.Hotel_name ;)
)'''
cursor.execute(sql)
records = cursor.fetchall()
#Viewing the results in tabular format
results_table=pd.DataFrame(records)
results_table.columns=("TripAdvisor","Hotels.com")
results_table
# -
# #### Closing all the connections
cursor.close()
conn.close()
| Scraped Hotel Listings.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: 'Python 3.7.3 64-bit (''base'': conda)'
# language: python
# name: python37364bitbaseconda65b5f305a1974c36abb2297a98801d43
# ---
# # ```Linear_Support_Vector_Machine``` Example
# Import the modules. We are also going to import `make_blobs()` function just to create the dataset.
import matplotlib.pyplot as plt
from Ardi.ml import Linear_Support_Vector_Machine_2D
from sklearn.datasets import make_blobs
# Create the dataset.
X, y = make_blobs(n_samples=50, n_features=2, centers=2, cluster_std=1.05, random_state=80)
# Display the data using scatter plot, where the data with label of 0 are displayed in blue, and the rest are of labels 1.
plt.scatter(X[:,0], X[:,1], c=y, cmap='winter')
# Initialize the Support Vector Machine model with linear kernel. Refer to the documentation to see the available parameters.
svm = Linear_Support_Vector_Machine_2D(iterations=800)
# Load the data that we just created to the `svm` model.
svm.take_data_raw(X, y)
# If you want to load the data from a csv file instead, you can use the following code.
# +
#svm.take_data_csv('dataset_test/sepal_petal_length.csv')
# -
# The following code is used just to ensure that our features and labels have been loaded properly.
svm.X[:5]
svm.y[:5]
# Display the first 5 data. Keep in mind that the `svm` model automatically converts label 0 to -1 because that's just how an SVM works.
plt.scatter(X[:5,0], X[:5,1], c=y[:5], cmap='winter')
# How to train the model.
svm.train()
# The `bias` and `weights` term before and after training. Note that both `bias` and `weights` are initially just a random number.
print('svm.bias\t\t:', svm.bias)
print('svm.updated_bias\t:', svm.updated_bias)
print()
print('svm.weights\t\t:', svm.weights)
print('svm.updated_weights\t:', svm.updated_weights)
# Perdicting multiple samples.
svm.predict_multiple_samples(svm.X)
# How the error decrease looks like. Note that these error values are the sum of hinge loss and its regularization term.
svm.plot_errors(print_details=True)
# How the decision boundary looks like before training.
svm.visualize_before()
# How the decision boundary looks like after training.
svm.visualize_after()
| Documentation/Linear_Support_Vector_Machine_2D Example.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: .venv_dev
# language: python
# name: .venv_dev
# ---
# ## Task-01 Clue-02: Fetching S3 Bucket with dataset
# +
import boto3
s3_resource = boto3.resource('s3')
all_buckets = s3_resource.buckets.all()
for bucket in all_buckets:
if (bucket.name.startswith('sagemaker')):
BUCKET = bucket.name
print ("SageMaker Default Bucket for Training Data: ", BUCKET)
jam_bucket = s3_resource.Bucket(BUCKET)
# -
# ## Task-01 Clue-03: Extracting Zipped Dataset into S3
# +
import boto3
import zipfile
from io import BytesIO
ZIPPED_DATA = 'radiography_train_data.zip'
TRAIN_DATA_PREFIX = 'radiography_train_data'
print ("Unzipping ", ZIPPED_DATA)
zip_obj = s3_resource.Object(bucket_name=BUCKET, key=ZIPPED_DATA)
buffer = BytesIO(zip_obj.get()["Body"].read())
z = zipfile.ZipFile(buffer)
for filename in z.namelist():
file_info = z.getinfo(filename)
s3_resource.meta.client.upload_fileobj(
z.open(filename),
Bucket=BUCKET,
Key=TRAIN_DATA_PREFIX+'/'+f'{filename}'
)
print ("Completed Unzipping Training Data")
# -
# removing hidden files after unzipping, if any
for obj in jam_bucket.objects.filter(Prefix=TRAIN_DATA_PREFIX+'/'):
if '/.' in obj.key:
s3_resource.Object(jam_bucket.name, obj.key).delete()
# ## Task-01 Clue-04: Extracting Zipped Dataset into S3
# +
import boto3
def get_size(bucket, path):
s3 = boto3.resource('s3')
my_bucket = s3.Bucket(bucket)
total_size = 0
image_count = 0
for obj in my_bucket.objects.filter(Prefix=path):
if ".png" in obj.key:
total_size = total_size + obj.size
image_count = image_count + 1
return total_size, image_count
folder_size, image_count = get_size(BUCKET, TRAIN_DATA_PREFIX+'/train')
average_image_size = folder_size / image_count
print (average_image_size)
| get-started-task-1-all-clues-q0dMD.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
from enmspring.k_b0_plot import CorrelationPlot
import matplotlib.pyplot as plt
rootfolder = '/home/yizaochen/codes/dna_rna/fluctmatch_sequence'
cutoff = 4.7
# -
# ### Part 0: Initialize
# +
corre_type = 'mean' # 'mean' or 'median'
plot_agent = CorrelationPlot(rootfolder, cutoff, corre_type)
# -
plot_agent.d_df['a_tract_21mer']
# ### Part 1: Single Correlation Plot
# +
pairtype_x = 'PP0'
pairtype_y = 'hb'
fig, ax = plt.subplots(figsize=(8, 3))
coefs = plot_agent.plot_single_one_curve(ax, pairtype_x, pairtype_y)
#plt.savefig(f'{pairtype_x}_{pairtype_y}.eps')
plt.show()
# -
# ### Part 2: Write Heatmap array
heatmap_array = plot_agent.write_heatmap_array()
# ### Part 3: Draw Heatmap
# +
fig, ax = plt.subplots(figsize=(12, 12))
plot_agent.heatmap_pearson_coefficient(ax, textcolor='black')
#plt.savefig(f'heatmap_{corre_type}.svg')
#plt.savefig(f'heatmap_{corre_type}.eps')
plt.show()
# -
| notebooks/.ipynb_checkpoints/correlation_plot_sequence-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # ETL Pipeline
#
# ### 1. Import libraries and load datasets
# - Import Python libraries
# - Load `messages.csv` into a dataframe and inspect the first few lines.
# - Load `categories.csv` into a dataframe and inspect the first few lines.
import numpy as np
import pandas as pd
from sqlalchemy import create_engine
messages = pd.read_csv("data/messages.csv")
messages.head()
categories = pd.read_csv("data/categories.csv")
categories.head()
# ### 2. Merge datasets
# - Merge the messages and categories datasets using the common id
# - Assign this combined dataset to `df`, which will be cleaned in the following steps
df = pd.merge(messages, categories, on = "id")
df.head()
# ### 3. Split `categories` into separate category columns
# - Split the values in the `categories` column on the `;` character so that each value becomes a separate column. You'll find [this method](https://pandas.pydata.org/pandas-docs/version/0.23/generated/pandas.Series.str.split.html) very helpful! Make sure to set `expand=True`.
# - Use the first row of categories dataframe to create column names for the categories data.
# - Rename columns of `categories` with new column names.
categories = df["categories"].str.split(";", expand = True)
categories.columns = categories.iloc[0,:].str.replace("-0","").str.replace("-1","").values
categories.head()
# ### 4. Convert category values to just numbers 0 or 1
# - Iterate through the category columns in df to keep only the last character of each string (the 1 or 0). For example, `related-0` becomes `0`, `related-1` becomes `1`. Convert the string to a numeric value.
#
# +
for col in categories.columns:
categories[col] = categories[col].str[-1].astype(float)
categories.head()
# -
categories["related"][categories["related"] == 2] = 1
((categories != 0) & (categories != 1)).sum()
# ### 5. Replace `categories` column in `df` with new category columns
# - Drop the categories column from the df dataframe since it is no longer needed.
# - Concatenate df and categories data frames.
df = pd.concat([df, categories], axis = 1).drop(["categories"], axis = 1)
df[:2]
# ### 6. Remove duplicates.
# - Check how many duplicates are in this dataset.
# - Drop the duplicates.
# - Confirm duplicates were removed.
df.duplicated().sum()
df = df.drop_duplicates()
df.duplicated().sum()
# ### 7. Save the clean dataset into an sqlite database
# - Save the data to a database
# - Test the connection
#
engine = create_engine('sqlite:///Data/Database.db')
df.to_sql('TheDRTable', engine, index=False)
connection = engine.connect()
df = pd.read_sql_query('''SELECT * FROM TheDRTable''', connection)
connection.close()
df[:3]
| dev/ETL Pipeline Dev.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# + [markdown] slideshow={"slide_type": "slide"}
# ## Differential expression anlaysis of the TCGA breast cancer set
#
# This notebook can be run locally or on a remote cloud computer by clicking the badge below:
#
# [](https://mybinder.org/v2/gh/statisticalbiotechnology/cb2030/master?filepath=nb%2Ftesting%2Ftesting.ipynb)
#
# First we retrieve the breast cancer RNAseq data as well as the clinical classification of the sets from cbioportal.org.
#
# The gene expresion data is stored in the [DataFrame](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.html) `brca`, and the adherent clinical information of the cancers and their patients is stored in the [DataFrame](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.html) `brca_clin`. It can be woth exploring these data structures.
#
# + slideshow={"slide_type": "fragment"}
import pandas as pd
import seaborn as sns
import numpy as np
import tarfile
import gzip
from scipy.stats import ttest_ind
import sys
sys.path.append("..") # Read loacal modules for tcga access and qvalue calculations
import tcga_read as tcga
brca = tcga.get_expression_data("../../data/brca.tsv.gz", 'http://download.cbioportal.org/brca_tcga_pub2015.tar.gz',"data_RNA_Seq_v2_expression_median.txt")
brca_clin = tcga.get_clinical_data("../../data/brca_clin.tsv.gz", 'http://download.cbioportal.org/brca_tcga_pub2015.tar.gz',"data_clinical_sample.txt")
# + [markdown] slideshow={"slide_type": "slide"}
# Before any further analysis we clean our data. This includes removal of genes where no transcripts were found for any of the samples , i.e. their values are either [NaN](https://en.wikipedia.org/wiki/NaN) or zero.
#
# The data is also log transformed. It is generally assumed that expression values follow a log-normal distribution, and hence the log transformation implies that the new values follow a nomal distribution.
# + slideshow={"slide_type": "fragment"}
brca.dropna(axis=0, how='any', inplace=True)
brca = brca.loc[~(brca<=0.0).any(axis=1)]
brca = pd.DataFrame(data=np.log2(brca),index=brca.index,columns=brca.columns)
# + [markdown] slideshow={"slide_type": "slide"}
# We can get an overview of the expression data:
# + slideshow={"slide_type": "fragment"}
brca
# + [markdown] slideshow={"slide_type": "slide"}
# and the clinical data:
# + slideshow={"slide_type": "fragment"}
brca_clin
# + [markdown] slideshow={"slide_type": "slide"}
# ### Differential expression analysis
#
# The goal of the excercise is to determine which genes that are differentially expressed in so called tripple negative cancers as compared to other cancers. A breast cancer is triple negative when it does not express either [Progesterone receptors](https://en.wikipedia.org/wiki/Progesterone_receptor), [Estrogen receptors](https://en.wikipedia.org/wiki/Estrogen_receptor) or [Epidermal growth factor receptor 2](https://en.wikipedia.org/wiki/HER2/neu). Such cancers are known to behave different than other cancers, and are not amendable to regular [hormonal theraphies](https://en.wikipedia.org/wiki/Hormonal_therapy_(oncology)).
#
# We first create a vector of booleans, that track which cancers that are tripple negative. This will be needed as an input for subsequent significance estimation.
# + slideshow={"slide_type": "fragment"}
brca_clin.loc["3N"]= (brca_clin.loc["PR status by ihc"]=="Negative") & (brca_clin.loc["ER Status By IHC"]=="Negative") & (brca_clin.loc["IHC-HER2"]=="Negative")
tripple_negative_bool = (brca_clin.loc["3N"] == True)
# + [markdown] slideshow={"slide_type": "slide"}
# Next, for each transcript that has been measured, we calculate (1) log of the average Fold Change difference between tripple negative and other cancers, and (2) the significance of the difference between tripple negative and other cancers.
#
# An easy way to do so is by defining a separate function, `get_significance_two_groups(row)`, that can do such calculations for any row of the `brca` DataFrame, and subsequently we use the function `apply` for the function to execute on each row of the DataFrame. For the significance test we use a $t$ test, which is provided by the function [`ttest_ind`.](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.ttest_ind.html)
#
# This results in a new table with gene names and their $p$ values of differential concentration, and their fold changes.
# + slideshow={"slide_type": "fragment"}
def get_significance_two_groups(row):
log_fold_change = row[tripple_negative_bool].mean() - row[~tripple_negative_bool].mean() # Calculate the log Fold Change
p = ttest_ind(row[tripple_negative_bool],row[~tripple_negative_bool],equal_var=False)[1] # Calculate the significance
return [p,-np.log10(p),log_fold_change]
pvalues = brca.apply(get_significance_two_groups,axis=1,result_type="expand")
pvalues.rename(columns = {list(pvalues)[0]: 'p', list(pvalues)[1]: '-log_p', list(pvalues)[2]: 'log_FC'}, inplace = True)
# + [markdown] slideshow={"slide_type": "slide"}
# The resulting list can be further investigated.
# + slideshow={"slide_type": "fragment"}
pvalues
# + [markdown] slideshow={"slide_type": "slide"}
# A common way to illustrate the diffrential expression values are by plotting the negative log of the $p$ values, as a function of the mean [fold change](https://en.wikipedia.org/wiki/Fold_change) of each transcript. This is known as a [Volcano plot](https://en.wikipedia.org/wiki/Volcano_plot_(statistics)).
# + slideshow={"slide_type": "fragment"}
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("white")
sns.set_context("talk")
ax = sns.relplot(data=pvalues,x="log_FC",y="-log_p",aspect=1.5,height=6)
ax.set(xlabel="$log_2(TN/not TN)$", ylabel="$-log_{10}(p)$");
# + [markdown] slideshow={"slide_type": "fragment"}
# The regular interpretation of a Volcano plot is that the ges in the top left and the top right corner are the most interesting ones, as the have a large fold change between the conditions as well as being very significant.
# -
| nb/testing/testing.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# 
#
# ## 一. Density Estimation 密度估计
#
#
# 假如要更为正式定义异常检测问题,首先我们有一组从 $x^{(1)}$ 到 $x^{(m)}$ m个样本,且这些样本均为正常的。我们将这些样本数据建立一个模型 p(x) , p(x) 表示为 x 的分布概率。
#
#
# 
#
#
# 那么假如我们的测试集 $x_{test}$ 概率 p 低于阈值 $\varepsilon$ ,那么则将其标记为异常。
#
#
# 异常检测的核心就在于找到一个概率模型,帮助我们知道一个样本落入正常样本中的概率,从而帮助我们区分正常和异常样本。高斯分布(Gaussian Distribution)模型就是异常检测算法最常使用的概率分布模型。
#
# ### 1. 高斯分布
#
#
# 假如 x 服从高斯分布,那么我们将表示为: $x\sim N(\mu,\sigma^2)$ 。其分布概率为:
#
# $$p(x;\mu,\sigma^2)=\frac{1}{\sqrt{2\pi}\sigma}exp(-\frac{(x-\mu)^2}{2\sigma^2})$$
#
# 其中 $\mu$ 为期望值(均值), $\sigma^2$ 为方差。
#
# 其中,期望值 $\mu$ 决定了其轴的位置,标准差 $\sigma$ 决定了分布的幅度宽窄。当 $\mu=0,\sigma=1$ 时的正态分布是标准正态分布。
#
# 
#
#
# 期望值:$$\mu=\frac{1}{m}\sum_{i=1}^{m}{x^{(i)}}$$
#
# 方差: $$\sigma^2=\frac{1}{m}\sum_{i=1}^{m}{(x^{(i)}-\mu)}^2$$
#
#
# 假如我们有一组 m 个无标签训练集,其中每个训练数据又有 n 个特征,那么这个训练集应该是 m 个 n 维向量构成的样本矩阵。
#
#
# 在概率论中,对有限个样本进行参数估计
#
# $$\mu_j = \frac{1}{m} \sum_{i=1}^{m}x_j^{(i)}\;\;\;,\;\;\; \delta^2_j = \frac{1}{m} \sum_{i=1}^{m}(x_j^{(i)}-\mu_j)^2$$
#
# 这里对参数 $\mu$ 和参数 $\delta^2$ 的估计就是二者的极大似然估计。
#
# 假定每一个特征 $x_{1}$ 到 $x_{n}$ 均服从正态分布,则其模型的概率为:
#
# $$
# \begin{align*}
# p(x)&=p(x_1;\mu_1,\sigma_1^2)p(x_2;\mu_2,\sigma_2^2) \cdots p(x_n;\mu_n,\sigma_n^2)\\
# &=\prod_{j=1}^{n}p(x_j;\mu_j,\sigma_j^2)\\
# &=\prod_{j=1}^{n} \frac{1}{\sqrt{2\pi}\sigma_{j}}exp(-\frac{(x_{j}-\mu_{j})^2}{2\sigma_{j}^2})
# \end{align*}
# $$
#
#
# 当 $p(x)<\varepsilon$时,$x$ 为异常样本。
#
# ### 2. 举例
#
# 假定我们有两个特征 $x_1$ 、 $x_2$ ,它们都服从于高斯分布,并且通过参数估计,我们知道了分布参数:
#
# 
#
# 则模型 $p(x)$ 能由如下的热力图反映,热力图越热的地方,是正常样本的概率越高,参数 $\varepsilon$ 描述了一个截断高度,当概率落到了截断高度以下(下图紫色区域所示),则为异常样本:
#
# 
#
# 将 $p(x)$ 投影到特征 $x_1$ 、$x_2$ 所在平面,下图紫色曲线就反映了 $\varepsilon$ 的投影,它是一条截断曲线,落在截断曲线以外的样本,都会被认为是异常样本:
#
# 
#
#
# ### 3. 算法评估
#
# 由于异常样本是非常少的,所以整个数据集是非常偏斜的,我们不能单纯的用预测准确率来评估算法优劣,所以用我们之前的查准率(Precision)和召回率(Recall)计算出 F 值进行衡量异常检测算法了。
#
# - 真阳性、假阳性、真阴性、假阴性
# - 查准率(Precision)与 召回率(Recall)
# - F1 Score
#
# 我们还有一个参数 $\varepsilon$ ,这个 $\varepsilon$ 是我们用来决定什么时候把一个样本当做是异常样本的阈值。我们应该试用多个不同的 $\varepsilon$ 值,选取一个使得 F 值最大的那个 $\varepsilon$ 。
#
#
#
#
# ----------------------------------------------------------------------------------------------------------------
#
#
#
# ## 二. Building an Anomaly Detection System
#
#
#
# ### 1. 有监督学习与异常检测
#
#
#
#
# |有监督学习| 异常检测|
# | :----------: | :---: |
# |数据分布均匀 |数据非常偏斜,异常样本数目远小于正常样本数目
# |可以根据对正样本的拟合来知道正样本的形态,从而预测新来的样本是否是正样本 |异常的类型不一,很难根据对现有的异常样本(即正样本)的拟合来判断出异常样本的形态|
#
#
# 下面的表格则展示了二者的一些应用场景:
#
# |有监督学习| 异常检测|
# | :----------: | :---: |
# |垃圾邮件检测| 故障检测|
# |天气预测(预测雨天、晴天、或是多云天气)| 某数据中心对于机器设备的监控|
# |癌症的分类| 制造业判断一个零部件是否异常|
#
# 
#
# 假如我们的数据看起来不是很服从高斯分布,可以通过对数、指数、幂等数学变换让其接近于高斯分布。
#
#
# ----------------------------------------------------------------------------------------------------------------
#
#
#
# ## 三. Multivariate Gaussian Distribution (Optional)
#
#
#
# ### 1. 多元高斯分布模型
#
#
# 
#
#
# 我们以数据中心的监控计算机为例子。 $x_1$ 是CPU的负载,$x_2$ 是内存的使用量。其正常样本如左图红色点所示。假如我们有一个异常的样本(图中左上角绿色点),在图中看很明显它并不是正常样本所在的范围。但是在计算概率 $p(x)$ 的时候,因为它在 $x_1$ 和 $x_2$ 的高斯分布都属于正常范围,所以该点并不会被判断为异常点。
#
# 这是因为在高斯分布中,它并不能察觉在蓝色椭圆处才是正常样本概率高的范围,其概率是通过圆圈逐渐向外减小。所以在同一个圆圈内,虽然在计算中概率是一样的,但是在实际上却往往有很大偏差。
#
# 所以我们开发了一种改良版的异常检测算法:多元高斯分布。
#
#
#
# 我们不将每一个特征值都分开进行高斯分布的计算,而是作为整个模型进行高斯分布的拟合。
#
# 其概率模型为: $$p(x;\mu,\Sigma)=\frac{1}{(2\pi)^{\frac{n}{2}}|\Sigma|^{\frac{1}{2}}}exp(-\frac{1}{2}(x-\mu)^T\Sigma^{-1}(x-\mu))$$ (其中 $|\Sigma|$ 是 $\Sigma$ 的行列式,$\mu$ 表示样本均值,$\Sigma$ 表示样本协方差矩阵。)。
#
# 多元高斯分布模型的热力图如下:
#
#
# 
#
#
# $\Sigma$ 是一个协方差矩阵,所以它衡量的是方差。减小 $\Sigma$ 其宽度也随之减少,增大反之。
#
#
# 
#
#
# $\Sigma$ 中第一个数字是衡量 $x_1$ 的,假如减少第一个数字,则可从图中观察到 $x_1$ 的范围也随之被压缩,变成了一个椭圆。
#
#
# 
#
#
# 多元高斯分布还可以给数据的相关性建立模型。假如我们在非主对角线上改变数据(如图中间那副),则其图像会根据 $y=x$ 这条直线上进行高斯分布。
#
#
# 
#
#
# 反之亦然。
#
#
# 
#
#
# 改变 $\mu$ 的值则是改变其中心点的位置。
#
#
# ### 2. 参数估计
#
#
# 多元高斯分布模型的参数估计如下:
#
#
#
# $$\mu=\frac{1}{m}\sum_{i=1}^{m}{x^{(i)}}$$
#
# $$\Sigma=\frac{1}{m}\sum_{i=1}^{m}{(x^{(i)}-\mu)(x^{(i)}-\mu)^T}$$
#
#
#
# ### 3. 算法流程
#
#
# 采用了多元高斯分布的异常检测算法流程如下:
#
# 1. 选择一些足够反映异常样本的特征 $x_j$ 。
# 2. 对各个样本进行参数估计:
# $$\mu=\frac{1}{m}\sum_{i=1}^{m}{x^{(i)}}$$
# $$\Sigma=\frac{1}{m}\sum_{i=1}^{m}{(x^{(i)}-\mu)(x^{(i)}-\mu)^T}$$
# 3. 当新的样本 x 到来时,计算 $p(x)$ :
#
# $$p(x)=\frac{1}{(2\pi)^{\frac{n}{2}}|\Sigma|^{\frac{1}{2}}}exp(-\frac{1}{2}(x-\mu)^T\Sigma^{-1}(x-\mu))$$
#
# 如果 $p(x)<\varepsilon $ ,则认为样本 x 是异常样本。
#
#
#
# ### 4. 多元高斯分布模型与一般高斯分布模型的差异
#
# 一般的高斯分布模型只是多元高斯分布模型的一个约束,它将多元高斯分布的等高线约束到了如下所示同轴分布(概率密度的等高线是沿着轴向的):
#
# 
#
#
# 当: $\Sigma=\left[ \begin{array}{ccc}\sigma_1^2 \\ & \sigma_2^2 \\ &&…\\&&&\sigma_n^2\end{array} \right]$ 的时候,此时的多元高斯分布即是原来的多元高斯分布。(因为只有主对角线方差,并没有其它斜率的变化)
#
#
# 对比
#
# ### 模型定义
#
# 一般高斯模型:
#
# $$
# \begin{align*}
# p(x)&=p(x_1;\mu_1,\sigma_1^2)p(x_2;\mu_2,\sigma_2^2) \cdots p(x_n;\mu_n,\sigma_n^2)\\
# &=\prod_{j=1}^{n}p(x_j;\mu_j,\sigma_j^2)\\
# &=\prod_{j=1}^{n} \frac{1}{\sqrt{2\pi}\sigma_{j}}exp(-\frac{(x_{j}-\mu_{j})^2}{2\sigma_{j}^2})
# \end{align*}
# $$
#
# 多元高斯模型:
#
#
# $$p(x)=\frac{1}{(2\pi)^{\frac{n}{2}}|\Sigma|^{\frac{1}{2}}}exp(-\frac{1}{2}(x-\mu)^T\Sigma^{-1}(x-\mu))$$
#
#
# ### 相关性
#
# 一般高斯模型:
#
# 需要手动创建一些特征来描述某些特征的相关性
#
# 多元高斯模型:
#
# 利用协方差矩阵$\Sigma$获得了各个特征相关性
#
#
# ### 复杂度
#
# 一般高斯模型:
#
# 计算复杂度低,适用于高维特征
#
# 多元高斯模型:
#
# 计算复杂
#
# ### 效果
#
#
# 一般高斯模型:
#
# 在样本数目 m 较小时也工作良好
#
# 多元高斯模型:
#
# 需要 $\Sigma$ 可逆,亦即需要 $m>n$ ,且各个特征不能线性相关,如不能存在 $x_2=3x_1$ 或者 $x_3=x_1+2x_2$
#
#
#
# 结论:**基于多元高斯分布模型的异常检测应用十分有限**。
#
# ----------------------------------------------------------------------------------------------------------------
#
#
# ## 四. Anomaly Detection 测试
#
#
# ### 1. Question 1
#
#
# For which of the following problems would anomaly detection be a suitable algorithm?
#
# A. Given a dataset of credit card transactions, identify unusual transactions to flag them as possibly fraudulent.
#
# B. Given data from credit card transactions, classify each transaction according to type of purchase (for example: food, transportation, clothing).
#
# C. Given an image of a face, determine whether or not it is the face of a particular famous individual.
#
# D. From a large set of primary care patient records, identify individuals who might have unusual health conditions.
#
# 解答:A、D
#
# A、D 才适合异常检测算法。
#
#
# ### 2. Question 2
#
# Suppose you have trained an anomaly detection system for fraud detection, and your system that flags anomalies when $p(x)$ is less than ε, and you find on the cross-validation set that it is missing many fradulent transactions (i.e., failing to flag them as anomalies). What should you do?
#
#
# A. Decrease $\varepsilon$
#
# B. Increase $\varepsilon$
#
# 解答:B
#
#
#
# ### 3. Question 3
#
# Suppose you are developing an anomaly detection system to catch manufacturing defects in airplane engines. You model uses
#
# $$p(x) = \prod_{j=1}^{n}p(x_{j};\mu_{j},\sigma_{j}^{2})$$
#
# You have two features $x_1$ = vibration intensity, and $x_2$ = heat generated. Both $x_1$ and $x_2$ take on values between 0 and 1 (and are strictly greater than 0), and for most "normal" engines you expect that $x_1 \approx x_2$. One of the suspected anomalies is that a flawed engine may vibrate very intensely even without generating much heat (large $x_1$, small $x_2$), even though the particular values of $x_1$ and $x_2$ may not fall outside their typical ranges of values. What additional feature $x_3$ should you create to capture these types of anomalies:
#
#
# A. $x_3 = \frac{x_1}{x_2}$
#
# B. $x_3 = x_1^2\times x_2^2$
#
# C. $x_3 = (x_1 + x_2)^2$
#
# D. $x_3 = x_1 \times x_2^2$
#
#
# 解答:A
#
# 假如特征量 $x_1$ 和 $x_2$ ,可建立特征量 $x_3=\frac{x_1}{x_2}$ 结合两者。
#
# ### 4. Question 4
#
# Which of the following are true? Check all that apply.
#
#
# A. When evaluating an anomaly detection algorithm on the cross validation set (containing some positive and some negative examples), classification accuracy is usually a good evaluation metric to use.
#
# B. When developing an anomaly detection system, it is often useful to select an appropriate numerical performance metric to evaluate the effectiveness of the learning algorithm.
#
# C. In a typical anomaly detection setting, we have a large number of anomalous examples, and a relatively small number of normal/non-anomalous examples.
#
# D. In anomaly detection, we fit a model p(x) to a set of negative (y=0) examples, without using any positive examples we may have collected of previously observed anomalies.
#
# 解答:B、D
#
#
# ### 5. Question 5
#
# You have a 1-D dataset $\begin{Bmatrix}
# x^{(i)},\cdots,x^{(m)}
# \end{Bmatrix}$ and you want to detect outliers in the dataset. You first plot the dataset and it looks like this:
#
# 
#
# Suppose you fit the gaussian distribution parameters $\mu_1$ and $\sigma_1^2$ to this dataset. Which of the following values for $\mu_1$ and $\sigma_1^2$ might you get?
#
# A. $\mu = -3$,$\sigma_1^2 = 4$
#
# B. $\mu = -6$,$\sigma_1^2 = 4$
#
# C. $\mu = -3$,$\sigma_1^2 = 2$
#
# D. $\mu = -6$,$\sigma_1^2 = 2$
#
#
# 解答:A
#
# 中心点在-3,在-3周围即(-4,-2)周围仍比较密集,所以 $\sigma_1=2$ 。
#
#
# ----------------------------------------------------------------------------------------------------------------
# > GitHub Repo:[Halfrost-Field](https://github.com/halfrost/Halfrost-Field)
# >
# > Follow: [halfrost · GitHub](https://github.com/halfrost)
# >
# > Source: [https://github.com/halfrost/Halfrost-Field/blob/master/contents/Machine\_Learning/Anomaly\_Detection.ipynb](https://github.com/halfrost/Halfrost-Field/blob/master/contents/Machine_Learning/Anomaly_Detection.ipynb)
| contents/Machine_Learning/Anomaly_Detection.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: SageMath 9.0
# language: sage
# name: sagemath
# ---
# # Euclidean Algorithm
#
# Here we explore one implementation of the Euclidean Algorithm. Although this may not be the most efficient impelmentation of the Euclidean Algorithm, it is the one I wrote. Finding the GCD is the easy part of this Algorithm, it is the back substitution that needs some creativity here. In this document, we will go through the process of developing an algorithm that implements the both sides of the Euclidean Algorithm.
#
# Here we look at an implementation of the Euclidean Algorithm with no back substitution, that is, it only returns the greatest common divisor of two numbers. Here we override the gcd function that is provided to us by SageMath, and create our own. Let's take a look.
# define the gcd function that will take in two parameters which must be integers.
def gcd(a,b):
# set the variable atemp to the maximum of the two numbers.
# Note that we take the absolute value of each of the numbers,
# as this does not change the gcd.
atemp = max(abs(a),abs(b))
# similar with btemp, but the minimum.
btemp = min(abs(a),abs(b))
# let a be the maximum of the two, and b be the minimum of the two,
# where both are now positive, if they were not before
a = atemp
b = btemp
# while b is non zero...
while b > 0:
# obtain the quotient of a/b...
quotient = floor(a/b)
# as well as the remainder of a/b...
remainder = a % b
# then, for the next step in the Euclidean Algorithm, set
# a to be the current value of b, and set b to be the remainder
# obtained from the previous step, continue this process untill
# the remainder is zero.
a = b
b = remainder
# return the quotient, as this will be the greatest common divisor.
return a
# Here we give this function a test. We test a few different numbers, as well as some large ones to demonstrate the speed and efficiency of the Euclidean Algorithm.
print(f"gcd(5,12)={gcd(5,12)}")
result = gcd(5^23,23^5)
print(f"gcd(5^23,23^5)={result}")
# Now we turn our attention to the problem of finding some linear combination of the two numbers that equals their gcd. To see how this is done, consider the following general Euclidean Algorithm:
# \begin{align*}
# a = q_1 b+r_1 \\
# b = q_2 r_1 + r_2 \\
# \vdots \\
# r_{n-2}=q_n r_{n-1}+r_n \\
# r_{n-1}=q_{n+1}r_n
# \end{align*}
# Where we know that $\gcd(a,b)=r_n$. We can then work our way back up from the second to last line, we can write
#
# $$ r_n=r_{n-2}-q_{n}r_{n-1} $$
# But, since
#
# $$r_{n-1}=r_{n-3}-q_{n-1}r_{n-2} $$
#
# We can write
#
# $$r_n=r_{n-2}-q_n(r_{n-3}-q_{n-1}r_{n-2}) =(1+q_nq_{n-1})r_{n-2}-q_nr_{n-3}$$
#
# And so on through until we reach the top level, and have some linear combination of $a$ and $b$. To implement this in an algorithm, we consider the following steps a computer might take.
#
# > first, let $x=1$ and $y=-q_n$
# > then $r_n=xr_{n-2}+yr_{n-1}$
# > then write $r_n=xr_{n-2}+y(r_{n-3}-q_{n-1}r_{n-2})=(x-q_{n-1}y)r_{n-2}+yr_{n-3}$
# > set $x=x-q_{n-2}y$ and $y=y$
# > then $r_n=xr_{n-2}+yr_{n-3}$
# > then write $r_n=x(r_{n-4}-q_{n-2}r_{n-3})+yr_{n-3}=xr_{n-4}+(y-q_{n-2}x)r_{n-3}$
# > set $x=x$ and $y=y-q_{n-2}x)$
# > then $r_n=xr_{n-4}+yr_{n-3}$
# > continue until we reach the top equation, then we will have a
# > combination of $a$ and $b$ that equals $r_n=gcd(a,b)$
# redefine our gcd function, similar as before, must have
# two intengers as input.
def gcd_full(a: int,b: int):
# store the original values of a and b into the
# variables aOrig and bOrig, to be used at the end
aOrig = a
bOrig = b
# let atemp be the maximum of the absolute values of a and b
# let btemp be the minimum
atemp = max(abs(a),abs(b))
btemp = min(abs(a),abs(b))
# reassign the max value to be a and the minimum value to be b
a = atemp
b = btemp
# create a list to hold all of the quotients, there is no need to
# save all the remainders, as they are not used in the pseudocode
# above
quotients = []
# same as before, go through and perform the steps of the Euclidean
# algorithm, only this time, save all of the quotients into the list
# that we just defined.
while b > 0:
quotient = floor(a/b)
remainder = a % b
a = b
b = remainder
quotients.append(quotient)
# let d, be the value of the gcd
d = a
# throw the very last equation (the one with no remainder)
# away, since we do not use it in the pseudocode above
quotients.pop()
# set x and y to their initial values
x = 1
y = -quotients.pop()
# set the count equal to one, this will be used to alternate which one of x and y
# we change in each step
count = 0
# follow the pseudocode above until there are no longer any quotients left
while len(quotients)!=0:
if count % 2 == 0:
x = x-quotients.pop()*y
if count % 2 == 1:
y = y-quotients.pop()*x
count = count + 1
# this part is a little messy (sorry about that), but test to see which linear
# combination of the original values of a and b give us the gcd, then
# return those values
if x*aOrig+y*bOrig==d:
return [d,x,y]
elif (-x)*aOrig+y*bOrig==d:
return [d,-x,y]
elif x*aOrig+(-y)*bOrig==d:
return [d,x,-y]
elif (-x)*aOrig+(-y)*bOrig==d:
return [d,-x,-y]
elif y*aOrig+x*bOrig==d:
return [d,y,x]
elif (-y)*aOrig+x*bOrig==d:
return [d,-y,x]
elif y*aOrig+(-x)*bOrig==d:
return [d,y,-x]
elif (-y)*aOrig+(-x)*bOrig==d:
return [d,-y,-x]
# Here we go through an example of both of the above methods.
print(gcd(-339348,5423493))
print(gcd_full(-339348,5423493))
# The output of the above cell tells us that
#
# $$ \gcd(-339348,5423493)=3 $$
#
# and that
#
# $$ -339348(-146412)+5423493(-9161)=3 $$
| Misc/Euclid.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (system-wide)
# language: python
# metadata:
# cocalc:
# description: Python 3 programming language
# priority: 100
# url: https://www.python.org/
# name: python3
# ---
# # Laboratory 14
# ## Full name:
# ## R#:
# ## HEX:
# ## Title of the notebook
# ## Date:
# ### Important Terminology:
# __Plotting Position:__ An empirical distribution, based on a random sample from a (possibly unknown) probability distribution, obtained by plotting the exceedance (or cumulative) probability of the sample distribution against the sample value. <br>
# The exceedance probability for a particular sample value is a function of sample size and the rank of the particular sample. For exceedance probabilities, the sample values are ranked from largest to smallest. The general expression in common use for plotting position is
#
# $$ P = \frac{m - b}{N + 1 -2b}\ $$
#
# where m is the ordered rank of a sample value, N is the sample size, and b is a constant between 0 and 1, depending on the plotting method.<br>
#
# 
#
# __*From:__<br>
# __*https://glossary.ametsoc.org/wiki/*__<br>
#
# __Let's work on example. First, import the necessary packages:__
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# __Read the "lab14_E1data.csv" file as a dataset:__
data = pd.read_csv("lab14_E1data.csv")
data
# __The dataset contains two sets of values: "Set1" and "Set2". Use descriptive functions to learn more the sets.__
# Let's check out set1 and set2
set1 = data['Set1']
set2 = data['Set2']
print(set1)
print(set2)
set1.describe()
set2.describe()
# __Remember the Weibull Plotting Position formula from last session. Use Weibull Plotting Position formula to plot set1 and set2 quantiles on the same graph.__<br>
# __Do they look different? How?__
def weibull_pp(sample): # Weibull plotting position function
# returns a list of plotting positions; sample must be a numeric list
weibull_pp = [] # null list to return after fill
sample.sort() # sort the sample list in place
for i in range(0,len(sample),1):
weibull_pp.append((i+1)/(len(sample)+1)) #values from the gringorten formula
return weibull_pp
#Convert to numpy arrays
set1 = np.array(set1)
set2 = np.array(set2)
#Apply the weibull pp function
set1_wei = weibull_pp(set1)
set2_wei = weibull_pp(set2)
myfigure = matplotlib.pyplot.figure(figsize = (4,8)) # generate a object from the figure class, set aspect ratio
matplotlib.pyplot.scatter(set1_wei, set1 ,color ='blue')
matplotlib.pyplot.scatter(set2_wei, set2 ,color ='orange')
matplotlib.pyplot.xlabel("Density or Quantile Value")
matplotlib.pyplot.ylabel("Value")
matplotlib.pyplot.title("Quantile Plot for Set1 and Set2 based on Weibull Plotting Function")
matplotlib.pyplot.show()
# __Do they look different? How?__
# __Define functions for Gringorten, Cunnane, California, and Hazen Plotting Position Formulas. Overlay and Plot them all for set 1 and set2 on two different graphs.__<br>
def gringorten_pp(sample): # plotting position function
# returns a list of plotting positions; sample must be a numeric list
gringorten_pp = [] # null list to return after fill
sample.sort() # sort the sample list in place
for i in range(0,len(sample),1):
gringorten_pp.append((i+1-0.44)/(len(sample)+0.12)) #values from the gringorten formula
return gringorten_pp
set1_grin = gringorten_pp(set1)
set2_grin = gringorten_pp(set2)
def cunnane_pp(sample): # plotting position function
# returns a list of plotting positions; sample must be a numeric list
cunnane_pp = [] # null list to return after fill
sample.sort() # sort the sample list in place
for i in range(0,len(sample),1):
cunnane_pp.append((i+1-0.40)/(len(sample)+0.2)) #values from the cunnane formula
return cunnane_pp
set1_cun = cunnane_pp(set1)
set2_cun = cunnane_pp(set2)
def california_pp(sample): # plotting position function
# returns a list of plotting positions; sample must be a numeric list
california_pp = [] # null list to return after fill
sample.sort() # sort the sample list in place
for i in range(0,len(sample),1):
california_pp.append((i+1)/(len(sample))) #values from the cunnane formula
return california_pp
set1_cal = california_pp(set1)
set2_cal = california_pp(set2)
def hazen_pp(sample): # plotting position function
# returns a list of plotting positions; sample must be a numeric list
hazen_pp = [] # null list to return after fill
sample.sort() # sort the sample list in place
for i in range(0,len(sample),1):
hazen_pp.append((i+1-0.5)/(len(sample))) #values from the cunnane formula
return hazen_pp
set1_haz = hazen_pp(set1)
set2_haz = hazen_pp(set2)
myfigure = matplotlib.pyplot.figure(figsize = (12,8)) # generate a object from the figure class, set aspect ratio
matplotlib.pyplot.scatter(set1_wei, set1 ,color ='blue',
marker ="^",
s = 50)
matplotlib.pyplot.scatter(set1_grin, set1 ,color ='red',
marker ="o",
s = 20)
matplotlib.pyplot.scatter(set1_cun, set1 ,color ='green',
marker ="s",
s = 20)
matplotlib.pyplot.scatter(set1_cal, set1 ,color ='yellow',
marker ="p",
s = 20)
matplotlib.pyplot.scatter(set1_haz, set1 ,color ='black',
marker ="*",
s = 20)
matplotlib.pyplot.xlabel("Density or Quantile Value")
matplotlib.pyplot.ylabel("Value")
matplotlib.pyplot.title("Quantile Plot for Set1 based on Weibull, Gringorton, Cunnane, California, and Hazen Plotting Functions")
matplotlib.pyplot.show()
myfigure = matplotlib.pyplot.figure(figsize = (12,8)) # generate a object from the figure class, set aspect ratio
matplotlib.pyplot.scatter(set2_wei, set2 ,color ='blue',
marker ="^",
s = 50)
matplotlib.pyplot.scatter(set2_grin, set2 ,color ='red',
marker ="o",
s = 20)
matplotlib.pyplot.scatter(set2_cun, set2 ,color ='green',
marker ="s",
s = 20)
matplotlib.pyplot.scatter(set2_cal, set2 ,color ='yellow',
marker ="p",
s = 20)
matplotlib.pyplot.scatter(set2_haz, set2 ,color ='black',
marker ="*",
s = 20)
matplotlib.pyplot.xlabel("Density or Quantile Value")
matplotlib.pyplot.ylabel("Value")
matplotlib.pyplot.title("Quantile Plot for Set2 based on Weibull, Gringorton, Cunnane, California, and Hazen Plotting Functions")
matplotlib.pyplot.show()
# __Plot a histogram of Set1 with 10 bins.__<br>
# +
import matplotlib.pyplot as plt
myfigure = matplotlib.pyplot.figure(figsize = (10,5)) # generate a object from the figure class, set aspect ratio
set1 = data['Set1']
set1.plot.hist(grid=False, bins=10, rwidth=1,
color='navy')
plt.title('Histogram of Set1')
plt.xlabel('Value')
plt.ylabel('Counts')
plt.grid(axis='y',color='yellow', alpha=1)
# -
# __Plot a histogram of Set2 with 10 bins.__<br>
set2 = data['Set2']
set2.plot.hist(grid=False, bins=10, rwidth=1,
color='darkorange')
plt.title('Histogram of Set2')
plt.xlabel('Value')
plt.ylabel('Counts')
plt.grid(axis='y',color='yellow', alpha=1)
# __Plot a histogram of both Set1 and Set2 and discuss the differences.__<br>
fig, ax = plt.subplots()
data.plot.hist(density=False, ax=ax, title='Histogram: Set1 vs. Set2', bins=40)
ax.set_ylabel('Count')
ax.grid(axis='y')
# __The cool 'seaborn' package: Another way for plotting histograms and more!__<br>
#
import seaborn as sns
sns.displot(set1,color='navy', rug=True)
sns.displot(set2,color='darkorange', rug=True)
# ### Important Terminology:
# __Kernel Density Estimation (KDE):__ a non-parametric way to estimate the probability density function of a random variable. Kernel density estimation is a fundamental data smoothing problem where inferences about the population are made, based on a finite data sample. This can be useful if you want to visualize just the “shape” of some data, as a kind of continuous replacement for the discrete histogram.<br>
#
# __*From:__<br>
# __*https://en.wikipedia.org/wiki/Kernel_density_estimation*__<br>
# __*https://mathisonian.github.io/kde/* >> A SUPERCOOL Blog!__<br>
# __*https://www.youtube.com/watch?v=fJoR3QsfXa0* >> A Nice Intro to distplot in seaborn | Note that displot is pretty much the same thing!__<br>
#
#
#
sns.displot(set1,color='navy',kind='kde',rug=True)
sns.displot(set1,color='navy',kde=True)
sns.displot(set2,color='orange',kde=True)
# ### Important Terminology:
# __Empirical Cumulative Distribution Function (ECDF):__ the distribution function associated with the empirical measure of a sample. This cumulative distribution function is a step function that jumps up by 1/n at each of the n data points. Its value at any specified value of the measured variable is the fraction of observations of the measured variable that are less than or equal to the specified value. <br>
#
# __*From:__<br>
# __*https://en.wikipedia.org/wiki/Empirical_distribution_function*__<br>
sns.displot(set1,color='navy',kind='ecdf')
# __Fit a Normal distribution data model to both Set1 and Set2. Plot them seperately. Describe the fit.__<br>
# +
set1 = data['Set1']
set2 = data['Set2']
set1 = np.array(set1)
set2 = np.array(set2)
set1_wei = weibull_pp(set1)
set2_wei = weibull_pp(set2)
# Normal Quantile Function
import math
def normdist(x,mu,sigma):
argument = (x - mu)/(math.sqrt(2.0)*sigma)
normdist = (1.0 + math.erf(argument))/2.0
return normdist
# For set1
mu = set1.mean() # Fitted Model
sigma = set1.std()
x = []; ycdf = []
xlow = 0; xhigh = 1.2*max(set1) ; howMany = 100
xstep = (xhigh - xlow)/howMany
for i in range(0,howMany+1,1):
x.append(xlow + i*xstep)
yvalue = normdist(xlow + i*xstep,mu,sigma)
ycdf.append(yvalue)
# Fitting Data to Normal Data Model
# Now plot the sample values and plotting position
myfigure = matplotlib.pyplot.figure(figsize = (7,9)) # generate a object from the figure class, set aspect ratio
matplotlib.pyplot.scatter(set1_wei, set1 ,color ='navy')
matplotlib.pyplot.plot(ycdf, x, color ='gold',linewidth=3)
matplotlib.pyplot.xlabel("Quantile Value")
matplotlib.pyplot.ylabel("Set1 Value")
mytitle = "Normal Distribution Data Model sample mean = : " + str(mu)+ " sample variance =:" + str(sigma**2)
matplotlib.pyplot.title(mytitle)
matplotlib.pyplot.show()
# -
# For set2
mu = set2.mean() # Fitted Model
sigma = set2.std()
x = []; ycdf = []
xlow = 0; xhigh = 1.2*max(set2) ; howMany = 100
xstep = (xhigh - xlow)/howMany
for i in range(0,howMany+1,1):
x.append(xlow + i*xstep)
yvalue = normdist(xlow + i*xstep,mu,sigma)
ycdf.append(yvalue)
# Fitting Data to Normal Data Model
# Now plot the sample values and plotting position
myfigure = matplotlib.pyplot.figure(figsize = (7,9)) # generate a object from the figure class, set aspect ratio
matplotlib.pyplot.scatter(set2_wei, set2 ,color ='orange')
matplotlib.pyplot.plot(ycdf, x, color ='purple',linewidth=3)
matplotlib.pyplot.xlabel("Quantile Value")
matplotlib.pyplot.ylabel("Set2 Value")
mytitle = "Normal Distribution Data Model sample mean = : " + str(mu)+ " sample variance =:" + str(sigma**2)
matplotlib.pyplot.title(mytitle)
matplotlib.pyplot.show()
# __Since it was an appropriate fit, we can use the normal distrubation to generate another sample randomly from the same population. Use a histogram with the new generated sets and compare them visually.__<br>
mu1 = set1.mean()
sd1 = set1.std()
mu2 = set2.mean()
sd2 = set2.std()
set1_s = np.random.normal(mu1, sd1, 100)
set2_s = np.random.normal(mu2, sd2, 100)
# +
data_d = pd.DataFrame({'Set1s':set1_s,'Set2s':set2_s})
fig, ax = plt.subplots()
data_d.plot.hist(density=False, ax=ax, title='Histogram: Set1 samples vs. Set2 samples', bins=40)
ax.set_ylabel('Count')
ax.grid(axis='y')
# +
fig, ax = plt.subplots()
data_d.plot.hist(density=False, ax=ax, title='Histogram: Set1 and Set1 samples vs. Set2 and Set2 samples', bins=40)
data.plot.hist(density=False, ax=ax, bins=40)
ax.set_ylabel('Count')
ax.grid(axis='y')
# -
# __Use boxplots to compare the four sets. Discuss their differences.__<br>
fig = plt.figure(figsize =(10, 7))
plt.boxplot ([set1, set1_s, set2, set2_s],1, '')
plt.show()
# __The first pair and the second pair look similar while the two pairs look differnet, right? The question is how can we KNOW if two sets are truly (significantly) different or not?__<br>
# ### Exercise 1:
# - __Step1:Read the "lab14_E2data.csv" file as a dataset.__<br>
# - __Step2:Describe the dataset numerically (using descriptive functions) and in your own words.__<br>
# - __Step3:Plot histograms and compare the sets in the dataset. What do you infer from the histograms?__<br>
# - __Step3*: This is a bonus step | Use "seaborn" to plot histograms with KDE and rugs!__<br>
# - __Step4:Write appropriate functions for the Beard, Tukey, and Adamowski Plotting Position Formulas.__<br>
# - __Step5:Apply your functions for the Beard, Tukey, and Adamowski Plotting Position Formulas on both sets and make quantile plots.__<br>
# - __Step6:Use the Tukey Plotting Position Formula and fit a Normal and a LogNormal distribution data model. Plot them and visually assess which one provides a better fit for each set__<br>
# - __Step7:Use the best distribution data model and a create two sample sets (one for each set) with 100 values.__<br>
# - __Step8:Use boxplots and illustrate the differences and similarities between the sets. What do you infer from the boxplots?__<br>
# Step1:
#Step2:
# Step3:
#Step3*: Bonus Step
#Step4: Functions for the Beard, Tukey, and Adamowski Plotting Position Formulas
#Step 5:
#Step6:
#Step7:
#Step8:
| 1-Lessons/Lesson14/Lab14/.ipynb_checkpoints/Lab14_Class-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Pre-process Allen Brain morphology data
# +
# %matplotlib inline
import os
import sys
notebook_path = os.path.abspath('')
sources_path = os.path.abspath(os.path.join(notebook_path, 'sources'))
sys.path.insert(0, sources_path)
import numpy as np
import matplotlib.pylab as plt
from allensdk.core.cell_types_cache import CellTypesCache
from allen_data import ProcessedAllenNeuronMorphology
# -
optosynth_data_path = '/home/jupyter/mb-ml-data-disk/Optosynth'
morphology_output_path = os.path.join(optosynth_data_path, 'processed_morphology_new')
if not os.path.exists(morphology_output_path):
os.mkdir(morphology_output_path)
ctc = CellTypesCache(manifest_file=os.path.join(optosynth_data_path, 'allen_data', 'manifest.json'))
cell_ids = np.load(os.path.join(optosynth_data_path, 'allen_data', 'good_cell_ids.npy'))
for idx, cell_id in enumerate(cell_ids):
print(f'Processing cell #{idx + 1} ({cell_id}) ...')
morph = ctc.get_reconstruction(cell_id)
neuron = ProcessedAllenNeuronMorphology.from_morphology(
cell_id, morph,
soma_scale_factor=1.0,
dendrite_scale_factor=5.0,
soma_relative_wiggle_size = 0.5,
soma_wiggle_n_components = 8)
neuron.save(morphology_output_path)
cell_id = cell_ids[400]
neuron = ProcessedAllenNeuronMorphology.from_file(
os.path.join(morphology_output_path, f'{cell_id}_processed_morphology.pkl'))
fig = plt.figure(figsize=(5, 5))
ax = plt.gca()
ax.set_aspect('equal')
ax.imshow(neuron.mask)
| process_allen_morphology.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: conda_pytorch_p36
# language: python
# name: conda_pytorch_p36
# ---
# # Predicting Boston Housing Prices
#
# ## Using XGBoost in SageMaker (Deploy)
#
# _Deep Learning Nanodegree Program | Deployment_
#
# ---
#
# As an introduction to using SageMaker's Low Level Python API we will look at a relatively simple problem. Namely, we will use the [Boston Housing Dataset](https://www.cs.toronto.edu/~delve/data/boston/bostonDetail.html) to predict the median value of a home in the area of Boston Mass.
#
# The documentation reference for the API used in this notebook is the [SageMaker Developer's Guide](https://docs.aws.amazon.com/sagemaker/latest/dg/)
#
# ## General Outline
#
# Typically, when using a notebook instance with SageMaker, you will proceed through the following steps. Of course, not every step will need to be done with each project. Also, there is quite a lot of room for variation in many of the steps, as you will see throughout these lessons.
#
# 1. Download or otherwise retrieve the data.
# 2. Process / Prepare the data.
# 3. Upload the processed data to S3.
# 4. Train a chosen model.
# 5. Test the trained model (typically using a batch transform job).
# 6. Deploy the trained model.
# 7. Use the deployed model.
#
# In this notebook we will be skipping step 5, testing the model. We will still test the model but we will do so by first deploying it and then sending the test data to the deployed model.
# ## Step 0: Setting up the notebook
#
# We begin by setting up all of the necessary bits required to run our notebook. To start that means loading all of the Python modules we will need.
# +
# %matplotlib inline
import os
import time
from time import gmtime, strftime
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import load_boston
import sklearn.model_selection
# -
# In addition to the modules above, we need to import the various bits of SageMaker that we will be using.
# +
import sagemaker
from sagemaker import get_execution_role
from sagemaker.amazon.amazon_estimator import get_image_uri
# This is an object that represents the SageMaker session that we are currently operating in. This
# object contains some useful information that we will need to access later such as our region.
session = sagemaker.Session()
# This is an object that represents the IAM role that we are currently assigned. When we construct
# and launch the training job later we will need to tell it what IAM role it should have. Since our
# use case is relatively simple we will simply assign the training job the role we currently have.
role = get_execution_role()
# -
# ## Step 1: Downloading the data
#
# Fortunately, this dataset can be retrieved using sklearn and so this step is relatively straightforward.
boston = load_boston()
# ## Step 2: Preparing and splitting the data
#
# Given that this is clean tabular data, we don't need to do any processing. However, we do need to split the rows in the dataset up into train, test and validation sets.
# +
# First we package up the input data and the target variable (the median value) as pandas dataframes. This
# will make saving the data to a file a little easier later on.
X_bos_pd = pd.DataFrame(boston.data, columns=boston.feature_names)
Y_bos_pd = pd.DataFrame(boston.target)
# We split the dataset into 2/3 training and 1/3 testing sets.
X_train, X_test, Y_train, Y_test = sklearn.model_selection.train_test_split(X_bos_pd, Y_bos_pd, test_size=0.33)
# Then we split the training set further into 2/3 training and 1/3 validation sets.
X_train, X_val, Y_train, Y_val = sklearn.model_selection.train_test_split(X_train, Y_train, test_size=0.33)
# -
# ## Step 3: Uploading the training and validation files to S3
#
# When a training job is constructed using SageMaker, a container is executed which performs the training operation. This container is given access to data that is stored in S3. This means that we need to upload the data we want to use for training to S3. We can use the SageMaker API to do this and hide some of the details.
#
# ### Save the data locally
#
# First we need to create the train and validation csv files which we will then upload to S3.
# This is our local data directory. We need to make sure that it exists.
data_dir = '../data/boston'
if not os.path.exists(data_dir):
os.makedirs(data_dir)
# +
# We use pandas to save our train and validation data to csv files. Note that we make sure not to include header
# information or an index as this is required by the built in algorithms provided by Amazon. Also, it is assumed
# that the first entry in each row is the target variable.
pd.concat([Y_val, X_val], axis=1).to_csv(os.path.join(data_dir, 'validation.csv'), header=False, index=False)
pd.concat([Y_train, X_train], axis=1).to_csv(os.path.join(data_dir, 'train.csv'), header=False, index=False)
# -
# ### Upload to S3
#
# Since we are currently running inside of a SageMaker session, we can use the object which represents this session to upload our data to the 'default' S3 bucket. Note that it is good practice to provide a custom prefix (essentially an S3 folder) to make sure that you don't accidentally interfere with data uploaded from some other notebook or project.
# +
prefix = 'boston-xgboost-deploy-ll'
val_location = session.upload_data(os.path.join(data_dir, 'validation.csv'), key_prefix=prefix)
train_location = session.upload_data(os.path.join(data_dir, 'train.csv'), key_prefix=prefix)
# -
# ## Step 4: Train and construct the XGBoost model
#
# Now that we have the training and validation data uploaded to S3, we can construct a training job for our XGBoost model and build the model itself.
#
# ### Set up the training job
#
# First, we will set up and execute a training job for our model. To do this we need to specify some information that SageMaker will use to set up and properly execute the computation. For additional documentation on constructing a training job, see the [CreateTrainingJob API](https://docs.aws.amazon.com/sagemaker/latest/dg/API_CreateTrainingJob.html) reference.
# +
# We will need to know the name of the container that we want to use for training. SageMaker provides
# a nice utility method to construct this for us.
container = get_image_uri(session.boto_region_name, 'xgboost')
# We now specify the parameters we wish to use for our training job
training_params = {}
# We need to specify the permissions that this training job will have. For our purposes we can use
# the same permissions that our current SageMaker session has.
training_params['RoleArn'] = role
# Here we describe the algorithm we wish to use. The most important part is the container which
# contains the training code.
training_params['AlgorithmSpecification'] = {
"TrainingImage": container,
"TrainingInputMode": "File"
}
# We also need to say where we would like the resulting model artifacst stored.
training_params['OutputDataConfig'] = {
"S3OutputPath": "s3://" + session.default_bucket() + "/" + prefix + "/output"
}
# We also need to set some parameters for the training job itself. Namely we need to describe what sort of
# compute instance we wish to use along with a stopping condition to handle the case that there is
# some sort of error and the training script doesn't terminate.
training_params['ResourceConfig'] = {
"InstanceCount": 1,
"InstanceType": "ml.m4.xlarge",
"VolumeSizeInGB": 5
}
training_params['StoppingCondition'] = {
"MaxRuntimeInSeconds": 86400
}
# Next we set the algorithm specific hyperparameters. You may wish to change these to see what effect
# there is on the resulting model.
training_params['HyperParameters'] = {
"max_depth": "5",
"eta": "0.2",
"gamma": "4",
"min_child_weight": "6",
"subsample": "0.8",
"objective": "reg:linear",
"early_stopping_rounds": "10",
"num_round": "200"
}
# Now we need to tell SageMaker where the data should be retrieved from.
training_params['InputDataConfig'] = [
{
"ChannelName": "train",
"DataSource": {
"S3DataSource": {
"S3DataType": "S3Prefix",
"S3Uri": train_location,
"S3DataDistributionType": "FullyReplicated"
}
},
"ContentType": "csv",
"CompressionType": "None"
},
{
"ChannelName": "validation",
"DataSource": {
"S3DataSource": {
"S3DataType": "S3Prefix",
"S3Uri": val_location,
"S3DataDistributionType": "FullyReplicated"
}
},
"ContentType": "csv",
"CompressionType": "None"
}
]
# -
# ### Execute the training job
#
# Now that we've built the dict containing the training job parameters, we can ask SageMaker to execute the job.
# +
# First we need to choose a training job name. This is useful for if we want to recall information about our
# training job at a later date. Note that SageMaker requires a training job name and that the name needs to
# be unique, which we accomplish by appending the current timestamp.
training_job_name = "boston-xgboost-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
training_params['TrainingJobName'] = training_job_name
# And now we ask SageMaker to create (and execute) the training job
training_job = session.sagemaker_client.create_training_job(**training_params)
# -
# The training job has now been created by SageMaker and is currently running. Since we need the output of the training job, we may wish to wait until it has finished. We can do so by asking SageMaker to output the logs generated by the training job and continue doing so until the training job terminates.
session.logs_for_job(training_job_name, wait=True)
# ### Build the model
#
# Now that the training job has completed, we have some model artifacts which we can use to build a model. Note that here we mean SageMaker's definition of a model, which is a collection of information about a specific algorithm along with the artifacts which result from a training job.
# +
# We begin by asking SageMaker to describe for us the results of the training job. The data structure
# returned contains a lot more information than we currently need, try checking it out yourself in
# more detail.
training_job_info = session.sagemaker_client.describe_training_job(TrainingJobName=training_job_name)
model_artifacts = training_job_info['ModelArtifacts']['S3ModelArtifacts']
# +
# Just like when we created a training job, the model name must be unique
model_name = training_job_name + "-model"
# We also need to tell SageMaker which container should be used for inference and where it should
# retrieve the model artifacts from. In our case, the xgboost container that we used for training
# can also be used for inference.
primary_container = {
"Image": container,
"ModelDataUrl": model_artifacts
}
# And lastly we construct the SageMaker model
model_info = session.sagemaker_client.create_model(
ModelName = model_name,
ExecutionRoleArn = role,
PrimaryContainer = primary_container)
# -
# ## Step 5: Test the trained model
#
# We will be skipping this step for now. We will still test our trained model but we are going to do it by using the deployed model, rather than setting up a batch transform job.
#
# ## Step 6: Create and deploy the endpoint
#
# Now that we have trained and constructed a model it is time to build the associated endpoint and deploy it. As in the earlier steps, we first need to construct the appropriate configuration.
# +
# As before, we need to give our endpoint configuration a name which should be unique
endpoint_config_name = "boston-xgboost-endpoint-config-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we ask SageMaker to construct the endpoint configuration
endpoint_config_info = session.sagemaker_client.create_endpoint_config(
EndpointConfigName = endpoint_config_name,
ProductionVariants = [{
"InstanceType": "ml.m4.xlarge",
"InitialVariantWeight": 1,
"InitialInstanceCount": 1,
"ModelName": model_name,
"VariantName": "AllTraffic"
}])
# -
# And now that the endpoint configuration has been created we can deploy the endpoint itself.
#
# **NOTE:** When deploying a model you are asking SageMaker to launch an compute instance that will wait for data to be sent to it. As a result, this compute instance will continue to run until *you* shut it down. This is important to know since the cost of a deployed endpoint depends on how long it has been running for.
#
# In other words **If you are no longer using a deployed endpoint, shut it down!**
# +
# Again, we need a unique name for our endpoint
endpoint_name = "boston-xgboost-endpoint-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we can deploy our endpoint
endpoint_info = session.sagemaker_client.create_endpoint(
EndpointName = endpoint_name,
EndpointConfigName = endpoint_config_name)
# -
# Just like when we created a training job, SageMaker is now requisitioning and launching our endpoint. Since we can't do much until the endpoint has been completely deployed we can wait for it to finish.
endpoint_dec = session.wait_for_endpoint(endpoint_name)
# ## Step 7: Use the model
#
# Now that our model is trained and deployed we can send test data to it and evaluate the results. Here, because our test data is so small, we can send it all using a single call to our endpoint. If our test dataset was larger we would need to split it up and send the data in chunks, making sure to accumulate the results.
# First we need to serialize the input data. In this case we want to send the test data as a csv and
# so we manually do this. Of course, there are many other ways to do this.
payload = [[str(entry) for entry in row] for row in X_test.values]
payload = '\n'.join([','.join(row) for row in payload])
# +
# This time we use the sagemaker runtime client rather than the sagemaker client so that we can invoke
# the endpoint that we created.
response = session.sagemaker_runtime_client.invoke_endpoint(
EndpointName = endpoint_name,
ContentType = 'text/csv',
Body = payload)
# We need to make sure that we deserialize the result of our endpoint call.
result = response['Body'].read().decode("utf-8")
Y_pred = np.fromstring(result, sep=',')
# -
# To see how well our model works we can create a simple scatter plot between the predicted and actual values. If the model was completely accurate the resulting scatter plot would look like the line $x=y$. As we can see, our model seems to have done okay but there is room for improvement.
plt.scatter(Y_test, Y_pred)
plt.xlabel("Median Price")
plt.ylabel("Predicted Price")
plt.title("Median Price vs Predicted Price")
# ## Delete the endpoint
#
# Since we are no longer using the deployed model we need to make sure to shut it down. Remember that you have to pay for the length of time that your endpoint is deployed so the longer it is left running, the more it costs.
session.sagemaker_client.delete_endpoint(EndpointName = endpoint_name)
# ## Optional: Clean up
#
# The default notebook instance on SageMaker doesn't have a lot of excess disk space available. As you continue to complete and execute notebooks you will eventually fill up this disk space, leading to errors which can be difficult to diagnose. Once you are completely finished using a notebook it is a good idea to remove the files that you created along the way. Of course, you can do this from the terminal or from the notebook hub if you would like. The cell below contains some commands to clean up the created files from within the notebook.
# +
# First we will remove all of the files contained in the data_dir directory
# !rm $data_dir/*
# And then we delete the directory itself
# !rmdir $data_dir
# -
| sagemaker_deployment/Tutorials/Boston Housing - XGBoost (Deploy) - Low Level.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# header files
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import torchvision
from PIL import Image
import slideio
from matplotlib import pyplot as plt
import cv2
import os
os.environ['KMP_DUPLICATE_LIB_OK'] = 'True'
print("Header files loaded!")
# -
# get patches from the image
slide = slideio.open_slide("../data/TCGA-23-1123.svs", 'SVS')
scene = slide.get_scene(0)
image = scene.read_block((74001, 50001, 2000, 2000))
cv2.imwrite("../data/TCGA-23-1123_5.png", cv2.cvtColor(image, cv2.COLOR_RGB2BGR))
plt.imshow(image)
| notebooks/generate_patches_from_image.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Codebook
# **Authors:** <NAME>
# Documenting existing data files of DaanMatch with information about location, owner, "version", source etc.
import boto3
import numpy as np
import pandas as pd
pd.plotting.register_matplotlib_converters()
import matplotlib.pyplot as plt
# %matplotlib inline
from collections import Counter
import statistics
client = boto3.client('s3')
resource = boto3.resource('s3')
my_bucket = resource.Bucket('daanmatchdatafiles')
# # Expenditure_Gov_India_2017-18_2019-20.csv
#
# ## TOC:
# * [About this dataset](#1)
# * [What's in this dataset](#2)
# * [Codebook](#3)
# * [Missing values](#3.1)
# * [Summary statistics](#3.2)
# * [Columns](#4)
# * [Sl. No.](#4.1)
# * [Category](#4.2)
# * [Sub Head](#4.3)
# * [2017-2018 - Actuals](#4.4)
# * [2018-2019 - Budget Estimates](#4.5)
# * [2018-2019 - Revised Estimates](#4.6)
# * [2019-2020 - Budget Estimates](#4.7)
# **About this dataset** <a class="anchor" id="1"></a>
# Data provided by: Unknown.
# Source: https://daanmatchdatafiles.s3.us-west-1.amazonaws.com/Expenditure_Gov_India_2017-18_2019-20.csv
# Type: csv
# Last Modified: June 14, 2021, 21:47:22 (UTC-07:00)
# Size: 786.0 B
path = "s3://daanmatchdatafiles/Expenditure_Gov_India_2017-18_2019-20.csv"
expenditure = pd.read_csv(path)
expenditure
# **What's in this dataset?** <a class="anchor" id="2"></a>
print("Shape:", expenditure.shape)
print("Rows:", expenditure.shape[0])
print("Columns:", expenditure.shape[1])
print("Each row is a department of expenditures.")
# **Codebook** <a class="anchor" id="3"></a>
# +
expenditure_columns = [column for column in expenditure.columns]
expenditure_description = ["Serial number.",
"Expenditure category.",
"Sub-category of the category for expenditure.",
"Actual amount spent in expenditures in 2017-2018.",
"Predicted expenditures for 2018-2019.",
"Updated and revised estimated expenditures for 2018-2019.",
"Predicted expenditures for 2019-2020."]
expenditure_dtypes = [dtype for dtype in expenditure.dtypes]
data = {"Column Name": expenditure_columns, "Description": expenditure_description, "Type": expenditure_dtypes}
expenditure_codebook = pd.DataFrame(data)
expenditure_codebook.style.set_properties(subset=['Description'], **{'width': '600px'})
# -
# **Missing values** <a class="anchor" id="3.1"></a>
expenditure.isnull().sum()
# **Summary statistics** <a class="anchor" id="3.2"></a>
expenditure.describe()
# ## Columns
# <a class="anchor" id="4"></a>
# ### Sl. No.
# <a class="anchor" id="4.1"></a>
# Serial number.
column = expenditure["Sl. No."]
column
print("No. of unique values:", len(column.unique()))
counter = dict(Counter(column))
duplicates = {key:value for key, value in counter.items() if value > 1}
print("Duplicates:", duplicates)
expenditure[expenditure["Sl. No."].isin(duplicates)]
# Duplicates in ```Sl. No.``` are not duplicates in rows.
# ### Category
# <a class="anchor" id="4.2"></a>
# Expenditure category.
column = expenditure["Category"]
column
print("No. of unique values:", len(column.unique()))
counter = dict(Counter(column))
duplicates = {key:value for key, value in counter.items() if value > 1}
print("Duplicates:", duplicates)
if len(duplicates) > 0:
print("No. of duplicates:", len(duplicates))
expenditure[expenditure["Category"].isin(duplicates)].sort_values('Category')
# Duplicates in ```Category``` are not duplicates in rows.
# ### Sub Head
# <a class="anchor" id="4.3"></a>
# Sub-category of the category for expenditure.
column = expenditure["Sub Head"]
column
print("No. of unique values:", len(column.unique()))
counter = dict(Counter(column))
duplicates = {key:value for key, value in counter.items() if value > 1}
print("Duplicates:", duplicates)
if len(duplicates) > 0:
print("No. of duplicates:", len(duplicates))
# ### 2017-2018 - Actuals
# <a class="anchor" id="4.4"></a>
# Actual amount spent in expenditures in 2017-2018.
column = expenditure["2017-2018 - Actuals"]
column
actual_17_18 = column[7]
sum(column[:7]) == column[7]
print('Sum of first 7 rows:', sum(column[:7]))
print('Value in the total row:', column[7])
print('Difference:', sum(column[:7]) - column[7])
# ### 2018-2019 - Budget Estimates
# <a class="anchor" id="4.5"></a>
# Predicted expenditures for 2018-2019.
column = expenditure["2018-2019 - Budget Estimates"]
column
sum(column[:7]) == column[7]
print('Sum of first 7 rows:', sum(column[:7]))
print('Value in the total row:', column[7])
print('Difference:', sum(column[:7]) - column[7])
# ### 2018-2019 - Revised Estimates
# <a class="anchor" id="4.6"></a>
# Updated and revised estimated expenditures for 2018-2019.
column = expenditure["2018-2019 - Revised Estimates"]
column
estimate_18_19 = column[7]
sum(column[:7]) == column[7]
print('Sum of first 7 rows:', sum(column[:7]))
print('Value in the total row:', column[7])
print('Difference:', sum(column[:7]) - column[7])
# ### 2019-2020 - Budget Estimates
# <a class="anchor" id="4.7"></a>
# Predicted expenditures for 2019-2020.
column = expenditure["2019-2020 - Budget Estimates"]
column
estimate_19_20 = column[7]
sum(column[:7]) == column[7]
print('Sum of first 7 rows:', sum(column[:7]))
print('Value in the total row:', column[7])
print('Difference:', sum(column[:7]) - column[7])
# #### Visualization
amounts = [actual_17_18, estimate_18_19, estimate_19_20]
years = ['Actual 2017-18', 'Estimate 2018-19', 'Estimate 2019-20']
expenditure_df = pd.DataFrame(data = {'Year' : years, 'Amounts' : amounts})
expenditure_df
plt.figure(figsize = (8, 5))
plt.bar(expenditure_df["Year"], expenditure_df["Amounts"])
plt.title("Total Expenditure per Year")
plt.xlabel("Year")
plt.ylabel("Expenditure Amount")
plt.show()
| Expenditure_Gov_India_2017-18_2019-20/Expenditure_Gov_India_2017-18_2019-20.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:.conda-dpm360]
# language: python
# name: conda-env-.conda-dpm360-py
# ---
# + [markdown] tags=[]
# # IHM Example Using LSTM
#
# This notebook shows an example of using `LSTM` to model In-hospital mortality from MIMIC-III dataset.
#
# Data is presumed to have been already extracted from cohort and defined via a yaml configuration as below:
# + [markdown] tags=[]
# ## Pre-amble
#
# The following code cell imports the required libraries and sets up the notebook
# +
# Jupyter notebook specific imports
# %matplotlib inline
import warnings
warnings.filterwarnings('ignore')
# Imports injecting into namespace
from tqdm.auto import tqdm
tqdm.pandas()
# General imports
import os
import json
import pickle
from pathlib import Path
import pandas as pd
import numpy as np
from getpass import getpass
import argparse
from sklearn.preprocessing import StandardScaler
from sklearn.exceptions import NotFittedError
import torch as T
from torch import nn
from pytorch_lightning import Trainer
from lightsaber import constants as C
import lightsaber.data_utils.utils as du
from lightsaber.data_utils import pt_dataset as ptd
from lightsaber.trainers import pt_trainer as ptr
from lightsaber.model_lib.pt_sota_models import rnn
# -
import logging
log = logging.getLogger()
# +
data_dir = Path(os.environ.get('LS_DATA_PATH', './data'))
assert data_dir.is_dir()
conf_path = os.environ.get('LS_CONF_PATH', os.path.abspath('./ihm_expt_config.yml'))
expt_conf = du.yaml.load(open(conf_path).read().format(DATA_DIR=data_dir),
Loader=du._Loader)
# + [markdown] tags=[]
# ## IHM Model Training
#
# In general, user need to follow the following steps to train a `HistGBT` for IHM model.
#
# * _Data Ingestion_: The first step involves setting up the pre-processors to train an IHM model. In this example, we will use a `StandardScaler` from `scikit-learn` using filters defined within lightsaber.
#
# - We would next read the train, test, and validation dataset. In some cases, users may also want to define a calibration dataset
#
# * _Model Definition_: We would next need to define a base model for classification. In this example, we will use a pre-packaged `LSTM` model from `lightsaber`
#
# * _Model Training_: Once the models are defined, we can use `lightsaber` to train the model via the pre-packaged `PyModel` and the corresponding trainer code. This step will also generate the relevant `metrics` for this problem.
# -
# ### Data Ingestion
#
# We firs start by reading extracted cohort data and use a `StandardScaler` demonstrating the proper usage of a pre-processor
preprocessor = StandardScaler()
train_filter = [ptd.filter_preprocessor(cols=expt_conf['numerical'],
preprocessor=preprocessor,
refit=True),
ptd.filter_fillna(fill_value=expt_conf['normal_values'],
time_order_col=expt_conf['time_order_col'])
]
transform = ptd.transform_drop_cols(cols_to_drop=expt_conf['time_order_col'])
# + tags=[]
train_dataset = ptd.BaseDataset(tgt_file=expt_conf['train']['tgt_file'],
feat_file=expt_conf['train']['feat_file'],
idx_col=expt_conf['idx_cols'],
tgt_col=expt_conf['tgt_col'],
feat_columns=expt_conf['feat_cols'],
time_order_col=expt_conf['time_order_col'],
category_map=expt_conf['category_map'],
transform=transform,
filter=train_filter,
)
# print(train_dataset.data.head())
print(train_dataset.shape, len(train_dataset))
# +
# For other datasets use fitted preprocessors
fitted_filter = [ptd.filter_preprocessor(cols=expt_conf['numerical'],
preprocessor=preprocessor, refit=False),
ptd.filter_fillna(fill_value=expt_conf['normal_values'],
time_order_col=expt_conf['time_order_col'])
]
val_dataset = ptd.BaseDataset(tgt_file=expt_conf['val']['tgt_file'],
feat_file=expt_conf['val']['feat_file'],
idx_col=expt_conf['idx_cols'],
tgt_col=expt_conf['tgt_col'],
feat_columns=expt_conf['feat_cols'],
time_order_col=expt_conf['time_order_col'],
category_map=expt_conf['category_map'],
transform=transform,
filter=fitted_filter,
)
test_dataset = ptd.BaseDataset(tgt_file=expt_conf['test']['tgt_file'],
feat_file=expt_conf['test']['feat_file'],
idx_col=expt_conf['idx_cols'],
tgt_col=expt_conf['tgt_col'],
feat_columns=expt_conf['feat_cols'],
time_order_col=expt_conf['time_order_col'],
category_map=expt_conf['category_map'],
transform=transform,
filter=fitted_filter,
)
print(val_dataset.shape, len(val_dataset))
print(test_dataset.shape, len(test_dataset))
# +
# Handling imbala
input_dim, target_dim = train_dataset.shape
output_dim = 2
weight_labels = train_dataset.target.iloc[:, 0].value_counts()
weight_labels = (weight_labels.max() / ((weight_labels + 0.0000001) ** (1)))
weight_labels.sort_index(inplace=True)
weights = T.FloatTensor(weight_labels.values).to(train_dataset.device)
print(weights)
# -
# ## Single Run
# +
# For most models you need to change only this part
hparams = argparse.Namespace(gpus=[0],
lr=0.01,
max_epochs=50,
batch_size=32,
hidden_dim=32,
rnn_class='LSTM',
n_layers=2,
dropout=0.1,
recurrent_dropout=0.1,
bidirectional=False,
)
hparams.rnn_class = C.PYTORCH_CLASS_DICT[hparams.rnn_class]
base_model = rnn.RNNClassifier(input_dim, output_dim,
hidden_dim=hparams.hidden_dim,
rnn_class=hparams.rnn_class,
n_layers=hparams.n_layers,
dropout=hparams.dropout,
recurrent_dropout=hparams.recurrent_dropout,
bidirectional=hparams.bidirectional
)
criterion = nn.CrossEntropyLoss(weight=weights)
# optimizer = T.optim.Adam(base_model.parameters(),
# lr=hparams.lr,
# weight_decay=1e-5 # standard value)
# )
# scheduler = T.optim.lr_scheduler.ReduceLROnPlateau(optimizer, 'min')
# Creating the wrapped model
wrapped_model = ptr.PyModel(hparams, base_model,
train_dataset=train_dataset,
val_dataset=val_dataset, # None
test_dataset=None, #test_dataset, # test_dataset
#optimizer=optimizer,
loss_func=criterion,
#scheduler=scheduler,
collate_fn=ptd.collate_fn
)
# +
# Training
overfit_pct, fast_dev_run, terminate_on_nan, auto_lr_find = 0, False, False, False
trainer = Trainer(max_epochs=hparams.max_epochs,
gpus=hparams.gpus,
default_root_dir=os.path.join('./out/', 'classifier_ihm'),
terminate_on_nan=terminate_on_nan,
auto_lr_find=auto_lr_find,
overfit_pct=overfit_pct,
fast_dev_run=fast_dev_run #True if devugging
)
# + tags=[]
mlflow_conf = dict(experiment_name=f'classifier_ihm')
artifacts = dict(preprocessor=preprocessor,
weight_labels=weight_labels,
)
experiment_tags = dict(model='RNNClassifier',
input_dim=input_dim,
output_dim=output_dim
)
(run_id, metrics,
val_y, val_yhat, val_pred_proba,
test_y, test_yhat, test_pred_proba) = ptr.run_training_with_mlflow(mlflow_conf,
trainer,
wrapped_model,
overfit_pct=overfit_pct,
artifacts=artifacts,
**experiment_tags)
print(f"MLFlow Experiment: {mlflow_conf['experiment_name']} \t | Run ID: {run_id}")
print(metrics)
# + [markdown] tags=[]
# ## IHM Model Registration
#
# This block shows how to register a model for subsequent steps. Given a `run_id` this block can be run independtly of other aspects
#
# Internally, the following steps happen:
#
# - a saved model (along with hyper-params and weights) is retrieved using `run_id`
# - model is initialized using the weights
# - model is logged to mlflow under registered model name
# -
print(f"Registering model for run: {run_id}")
# +
# Reading things from mlflow
# Model coders can create functions to repeat this - part of model init
import torch
from lightsaber.trainers import helper
data_dir = Path(os.environ.get('LS_DATA_PATH', './data'))
assert data_dir.is_dir()
conf_path = os.environ.get('LS_CONF_PATH', os.path.abspath('./ihm_expt_config.yml'))
expt_conf = du.yaml.load(open(conf_path).read().format(DATA_DIR=data_dir),
Loader=du._Loader)
mlflow_conf = dict(experiment_name=f'classifier_ihm')
registered_model_name = 'classifier_ihm_rnn_v0'
## Loading model attributes from mlflow
mlflow_setup = helper.setup_mlflow(**mlflow_conf)
run_data = helper.fetch_mlflow_run(run_id,
mlflow_uri=mlflow_setup['mlflow_uri'],
artifacts_prefix=['artifact/weight_labels'],
parse_params=True
)
hparams = run_data['params']
hparams = argparse.Namespace(**hparams)
hparams.rnn_class = helper.import_model_class(hparams.rnn_class.split("'")[1::2][0])
weight_labels = pickle.load(open(helper.get_artifact_path(run_data['artifact_paths'][0],
artifact_uri=run_data['info'].artifact_uri), 'rb'))
weights = T.FloatTensor(weight_labels.values)
## Setting model weights
base_model = rnn.RNNClassifier(input_dim=input_dim,
output_dim=output_dim,
hidden_dim=hparams.hidden_dim,
rnn_class=hparams.rnn_class,
n_layers=hparams.n_layers,
dropout=hparams.dropout,
recurrent_dropout=hparams.recurrent_dropout,
bidirectional=hparams.bidirectional
)
criterion = nn.CrossEntropyLoss(weight=weights)
wrapped_model = ptr.PyModel(hparams, base_model,
train_dataset=None,
val_dataset=None, # None
test_dataset=None, # test_dataset
#optimizer=optimizer,
loss_func=criterion,
#scheduler=scheduler,
collate_fn=ptd.collate_fn
)
# +
# Recreate models
base_model = rnn.RNNClassifier(input_dim=int(run_data['tags']['input_dim']),
output_dim=int(run_data['tags']['output_dim']),
hidden_dim=hparams.hidden_dim,
rnn_class=hparams.rnn_class,
n_layers=hparams.n_layers,
dropout=hparams.dropout,
recurrent_dropout=hparams.recurrent_dropout,
bidirectional=hparams.bidirectional
)
criterion = nn.CrossEntropyLoss(weight=weights)
# Creating the wrapped model
wrapped_model = ptr.PyModel(hparams, base_model,
train_dataset=None,
val_dataset=None, # None
test_dataset=None, # test_dataset
cal_dataset=None,
loss_func=criterion,
collate_fn=ptd.collate_fn
)
print('model ready for logging')
# + tags=[]
# Register model
ptr.register_model_with_mlflow(run_id, mlflow_conf, wrapped_model,
registered_model_name=registered_model_name,
test_feat_file=expt_conf['test']['feat_file'],
test_tgt_file=expt_conf['test']['tgt_file'],
config=os.path.abspath('./ihm_expt_config.yml'),
model_path='model_checkpoint'
)
# + [markdown] tags=[]
# ## IHM Model Inference
#
# `Lightsaber` also natively supports conducting inferences on new patients using the registered model. The key steps involve:
#
# * loading the registerd model from mlflow
# * Ingest the new test data using `BaseDataset` in inference mode (setting `tgt_file` to `None`)
# * Use the `PyModel.predict_patient` method to generate inference for the patient of interest
#
# It is to be noted, for the first step, users may need to perform additional setup as show below
# -
print(f"Inference using model for run: {run_id}")
# +
# Reading things from mlflow
# Model coders can create functions to repeat this - part of model init
import torch
from lightsaber.trainers import helper
data_dir = Path(os.environ.get('LS_DATA_PATH', './data'))
assert data_dir.is_dir()
conf_path = os.environ.get('LS_CONF_PATH', os.path.abspath('./ihm_expt_config.yml'))
expt_conf = du.yaml.load(open(conf_path).read().format(DATA_DIR=data_dir),
Loader=du._Loader)
mlflow_conf = dict(experiment_name=f'classifier_ihm')
registered_model_name = 'classifier_ihm_rnn_v0'
## Loading model attributes from mlflow
mlflow_setup = helper.setup_mlflow(**mlflow_conf)
run_data = helper.fetch_mlflow_run(run_id,
mlflow_uri=mlflow_setup['mlflow_uri'],
artifacts_prefix=['artifact/weight_labels'],
parse_params=True
)
hparams = run_data['params']
hparams = argparse.Namespace(**hparams)
hparams.rnn_class = helper.import_model_class(hparams.rnn_class.split("'")[1::2][0])
weight_labels = pickle.load(open(helper.get_artifact_path(run_data['artifact_paths'][0],
artifact_uri=run_data['info'].artifact_uri), 'rb'))
weights = T.FloatTensor(weight_labels.values)
## Setting model weights
base_model = rnn.RNNClassifier(input_dim=input_dim,
output_dim=output_dim,
hidden_dim=hparams.hidden_dim,
rnn_class=hparams.rnn_class,
n_layers=hparams.n_layers,
dropout=hparams.dropout,
recurrent_dropout=hparams.recurrent_dropout,
bidirectional=hparams.bidirectional
)
criterion = nn.CrossEntropyLoss(weight=weights)
wrapped_model = ptr.PyModel(hparams, base_model,
train_dataset=None,
val_dataset=None, # None
test_dataset=None, # test_dataset
#optimizer=optimizer,
loss_func=criterion,
#scheduler=scheduler,
collate_fn=ptd.collate_fn
)
# -
# Loading saved model from mlflow
wrapped_model = ptr.load_model_from_mlflow(run_id, mlflow_conf, wrapped_model)
# +
inference_dataloader = ptd.BaseDataset(tgt_file=None,
feat_file=expt_conf['test']['feat_file'],
idx_col=expt_conf['idx_cols'],
tgt_col=expt_conf['tgt_col'],
feat_columns=expt_conf['feat_cols'],
time_order_col=expt_conf['time_order_col'],
category_map=expt_conf['category_map'],
transform=transform,
filter=fitted_filter,
)
print(inference_dataloader.shape, len(inference_dataloader))
# + tags=[]
patient_id = inference_dataloader.sample_idx.index[0]
print(f"Inference for patient: {patient_id}")
# patient_id = '10011_episode1_timeseries.csv'
wrapped_model.predict_patient(patient_id, inference_dataloader)
| lightsaber/examples/in_hosptial_mortality/Exp_LSTM.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# The pupose of this notebook is to illustrate the use of a webscraper in Python 3 to gather text based data.
#
# The tutorial will be based on the below link and will be extended to other websites to gather information of interest.
#
# https://www.dataquest.io/blog/web-scraping-tutorial-python/
# +
# -*- coding: utf-8 -*-
"""
Created on Tue Oct 30 13:12:50 2018
@author: <NAME>
"""
#import libraries
import requests
from bs4 import BeautifulSoup
# -
# A GET request fetches the whole HTML page.
#Send GET HTML request to get webpage content
page = requests.get("http://dataquestio.github.io/web-scraping-pages/simple.html")
#check request status
page
#check the content of the webpage
page.content
# Beautifulsoup helps to parse and analyse the data.
# +
#Parse/structure the webpage content using Beautifulsoup
soup = BeautifulSoup(page.content, 'html.parser')
#check object strucutre
print(soup.prettify())
# +
#move through the object one level at a time
#Note that children returns a list generator, so we need to call the list function on it
list(soup.children)
# +
#check the items present in the list
[type(item) for item in list(soup.children)]
# -
# It is noted that there are three objects from the above:
#
# * The first is a Doctype object, which contains information about the type of the document.
# * The second is a NavigableString, which represents text found in the HTML document.
# * The final item is a Tag object, which contains other nested tags.
#
# The Tag object allows us to navigate through an HTML document, and extract other tags and text.
html = list(soup.children)[2]
print(html)
# There are two tags above, head, and body.
#
# The part of interest is the "p" tag.
#
# +
#Find all instances of the "p" tag
soup = BeautifulSoup(page.content, 'html.parser')
soup.find_all('p')
# +
#Since the above command retuns a list, use get_text to extract the text object
soup.find_all('p')[0].get_text()
# -
# These techniques will now be extended to construct a pandas dataframe from a weather forecast page
#Download the whole weather page
weatherpage = requests.get("https://forecast.weather.gov/MapClick.php?lat=37.7772&lon=-122.4168")
weatherpage
# +
#Get appears to be successful
#check the content of the webpage
#weatherpage.content
#There is too much output, web inspector (ctrl+shift+c) in firefox will be used to identify the element of choice
# +
#Based on indentation a tombstone container is used
#First parse the webpage
weathersoup = BeautifulSoup(weatherpage.content, 'html.parser')
#Find the seven day forecast
seven_day = weathersoup.find(id="seven-day-forecast")
#Extract all the instances of the tombstone container from the seven_day forecast
forecast_items = seven_day.find_all(class_="tombstone-container")
#Show tonight's forecast
tonight = forecast_items[0]
print(tonight.prettify())
# -
# From the above, there are cetain elements that can be accessed:
#
# * Short description : Sunny
# * temp : temp-high
# * Forecast item/date : Tonight
#
# +
#From the tonight object find the related text for each of the above.
period = tonight.find(class_="period-name").get_text()
short_desc = tonight.find(class_="short-desc").get_text()
temp = tonight.find(class_="temp").get_text()
print(period)
print(short_desc)
print(temp)
# +
#Extract the full description from the image attribute
img = tonight.find("img")
desc = img['title']
print(desc)
# +
#Select all the days in the forecast by selecting the whole tombstone container
period_tags = seven_day.select(".tombstone-container .period-name")
periods = [pt.get_text() for pt in period_tags]
periods
# +
#Get all the items for weather descriptions from tombstone container
short_descs = [sd.get_text() for sd in seven_day.select(".tombstone-container .short-desc")]
temps = [t.get_text() for t in seven_day.select(".tombstone-container .temp")]
descs = [d["title"] for d in seven_day.select(".tombstone-container img")]
print(short_descs)
print(temps)
print(descs)
# +
#Combine in pandas dataframe for future analysis
import pandas as pd
weather = pd.DataFrame({
"period": periods,
"short_desc": short_descs,
"temp": temps,
"desc":descs
})
weather
| Webcrawler_Tutorial.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# +
import os
import glob
import shutil
from pathlib import Path
import random
import numpy
import tensorflow as tf
from model_builder import model_builder, relabel, class_merger, balancer
import tools_keras
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.applications import resnet_v2, vgg19, efficientnet
random.seed(42)
numpy.random.seed(42)
tf.random.set_seed(42)
# -
specs = {
'chips': "../../chips_gb/32_shuffled/",
'chips_combined': "../../chips_gb/32_shuffled_combined_12_named/",
'chips_balanced': "../../chips_gb/32_shuffled_sample_12_named/",
'folder': "../../urbangrammar_samba/spatial_signatures/ai/gb_32_sample/",
}
# +
group_mapping = [
['9_0', '9_1', '9_2', '9_4', '9_5'],
['2_0'],
['2_1'],
['2_2'],
['1_0'],
['3_0'],
['5_0'],
['6_0'],
['8_0'],
['0_0'],
['4_0'],
['7_0']
]
group_naming = [
"Urbanity",
"Dense residential neighbourhoods",
"Connected residential neighbourhoods",
"Dense urban neighbourhoods",
"Accessible suburbia",
"Open sprawl",
"Warehouse_Park land",
"Gridded residential quarters",
"Disconnected suburbia",
"Countryside agriculture",
"Wild countryside",
"Urban buffer"
]
# +
# for subset in ["train", "validation", "secret"]:
# total = 3500 if subset == "train" else 500
# os.makedirs(specs['chips_balanced'] + subset, exist_ok=True)
# for folder in glob.glob(specs["chips_combined"] + f"{subset}/*"):
# os.makedirs(specs['chips_balanced'] + subset + "/" + Path(folder).name, exist_ok=True)
# files = glob.glob(folder + "/*")
# random.shuffle(files)
# for f in files[:total]:
# f = Path(f)
# shutil.copy(f, specs['chips_balanced'] + subset + "/" + Path(folder).name + "/" + f.name)
# -
model_specs = {
'meta_class_map': group_mapping,
'meta_class_names': group_naming,
'meta_chip_size': 32,
}
model = model_builder(
model_name="efficientnet",
bridge="pooling",
top_layer_neurons=256,
n_labels=12,
input_shape=(224, 224, 3),
metrics=["accuracy"]
)
h = tools_keras.fit_phase(
model,
specs['chips_balanced'] + 'train',
specs['chips_balanced'] + 'validation',
specs['chips_balanced'] + 'secret',
log_folder=specs["folder"] + "logs",
pred_folder=specs["folder"] + "pred",
model_folder=specs["folder"] + "model",
json_folder=specs["folder"] + "json",
specs=model_specs,
epochs=2,
patience=5,
batch_size=32,
verbose=True,
)
# ## verify results
datagen = keras.preprocessing.image.ImageDataGenerator()
generator = datagen.flow_from_directory(
"../../chips_gb/32_shuffled_sample_12_named/train/",
target_size=(224, 224),
batch_size=32,
class_mode='sparse',
shuffle=False)
def accuracy(y, y_pred):
a = tf.keras.metrics.Accuracy()
a.update_state(y, y_pred)
return a.result().numpy()
oy_pred_probs = model.predict(generator)
oy_pred = numpy.argmax(oy_pred_probs, axis=1)
y = generator.labels
accuracy(y, oy_pred)
generator.class_indices
model.save("model_folder", save_format="tf")
model.save("model.h5", save_format="h5")
model_folder = keras.models.load_model("model_folder")
y_pred_probs = model_folder.predict(generator)
y_pred = numpy.argmax(y_pred_probs, axis=1)
accuracy(y, y_pred)
model_h5 = keras.models.load_model("model.h5")
y_pred_probs = model_h5.predict(generator)
y_pred = numpy.argmax(y_pred_probs, axis=1)
accuracy(y, y_pred)
| ai_experiments/_debug_save_load.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
from __future__ import division
from sympy import *
init_session(quiet=True)
# +
T,delta,rho_r,b_m,c_m,a_m,R_u = symbols('T,delta,rho_r,b_m,c_m,a_m,R_u')
W = symbols('W', cls=Function)(delta)
alphar = -log(1-delta*rho_r*(b_m-c_m)) - sqrt(2)*a_m/(4*R_u*T*b_m)*log(W);
display(alphar)
for ndelta in range(1,5):
ss = simplify(diff(alphar, delta, ndelta))
display(ss)
W =(1+delta*rho_r*(b_m*(1+sqrt(2)+c_m))) / (1+delta*rho_r*(b_m*(1-sqrt(2)+c_m)))
for ndelta in range(1,5):
display(diff(W,delta,ndelta))
# -
| doc/notebooks/Sympy derivatives of alphar from VTPR.ipynb |
# -*- coding: utf-8 -*-
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .jl
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Julia 1.1.0
# language: julia
# name: julia-1.1
# ---
#
# <a id='mccall'></a>
# <div id="qe-notebook-header" style="text-align:right;">
# <a href="https://quantecon.org/" title="quantecon.org">
# <img style="width:250px;display:inline;" src="https://assets.quantecon.org/img/qe-menubar-logo.svg" alt="QuantEcon">
# </a>
# </div>
# # Job Search I: The McCall Search Model
# ## Contents
#
# - [Job Search I: The McCall Search Model](#Job-Search-I:-The-McCall-Search-Model)
# - [Overview](#Overview)
# - [The McCall Model](#The-McCall-Model)
# - [Computing the Optimal Policy: Take 1](#Computing-the-Optimal-Policy:-Take-1)
# - [Computing the Optimal Policy: Take 2](#Computing-the-Optimal-Policy:-Take-2)
# - [Exercises](#Exercises)
# - [Solutions](#Solutions)
# > “Questioning a McCall worker is like having a conversation with an out-of-work friend:
# > ‘Maybe you are setting your sights too high’, or ‘Why did you quit your old job before you
# > had a new one lined up?’ This is real social science: an attempt to model, to understand,
# > human behavior by visualizing the situation people find themselves in, the options they face
# > and the pros and cons as they themselves see them.” – <NAME>, Jr.
# ## Overview
#
# The McCall search model [[McC70]](zreferences.html#mccall1970) helped transform economists’ way of thinking about labor markets
#
# To clarify vague notions such as “involuntary” unemployment, McCall modeled the decision problem of unemployed agents directly, in terms of factors such as
#
# - current and likely future wages
# - impatience
# - unemployment compensation
#
#
# To solve the decision problem he used dynamic programming
#
# Here we set up McCall’s model and adopt the same solution method
#
# As we’ll see, McCall’s model is not only interesting in its own right but also an excellent vehicle for learning dynamic programming
# ## The McCall Model
#
#
# <a id='index-0'></a>
# An unemployed worker receives in each period a job offer at wage $ W_t $
#
# At time $ t $, our worker has two choices:
#
# 1. Accept the offer and work permanently at constant wage $ W_t $
# 1. Reject the offer, receive unemployment compensation $ c $, and reconsider next period
#
#
# The wage sequence $ \{W_t\} $ is assumed to be iid with probability mass function $ p_1, \ldots, p_n $
#
# Here $ p_i $ is the probability of observing wage offer $ W_t = w_i $ in the set $ w_1, \ldots, w_n $
#
# The worker is infinitely lived and aims to maximize the expected discounted sum of earnings
#
# $$
# \mathbb{E} \sum_{t=0}^{\infty} \beta^t Y_t
# $$
#
# The constant $ \beta $ lies in $ (0, 1) $ and is called a **discount factor**
#
# The smaller is $ \beta $, the more the worker discounts future utility relative to current utility
#
# The variable $ Y_t $ is income, equal to
#
# - his wage $ W_t $ when employed
# - unemployment compensation $ c $ when unemployed
# ### A Trade Off
#
# The worker faces a trade-off:
#
# - Waiting too long for a good offer is costly, since the future is discounted
# - Accepting too early is costly, since better offers might arrive in the future
#
#
# To decide optimally in the face of this trade off, we use dynamic programming
#
# Dynamic programming can be thought of as a two step procedure that
#
# 1. first assigns values to “states” and
# 1. then deduces optimal actions given those values
#
#
# We’ll go through these steps in turn
# ### The Value Function
#
# In order to optimally trade off current and future rewards, we need to think about two things:
#
# 1. the current payoffs we get from different choices
# 1. the different states that those choices will lead to next period (in this case, either employment or unemployment)
#
#
# To weigh these two aspects of the decision problem, we need to assign *values* to states
#
# To this end, let $ V(w) $ be the total lifetime *value* accruing to an unemployed worker who enters the current period unemployed but with wage offer $ w $ in hand
#
# More precisely, $ V(w) $ denotes the value of the objective function [(1)](mccall_model_with_separation.html#equation-objective) when an agent in this situation makes *optimal* decisions now and at all future points in time
#
# Of course $ V(w) $ is not trivial to calculate because we don’t yet know what decisions are optimal and what aren’t!
#
# But think of $ V $ as a function that assigns to each possible wage $ w $ the maximal lifetime value that can be obtained with that offer in hand
#
# A crucial observation is that this function $ V $ must satisfy the recursion
#
#
# <a id='equation-odu-pv'></a>
# $$
# V(w)
# = \max \left\{
# \frac{w}{1 - \beta}, \, c + \beta \sum_{i=1}^n V(w_i) p_i
# \right\} \tag{1}
# $$
#
# for every possible $ w_i $ in $ w_1, \ldots, w_n $
#
# This important equation is a version of the **Bellman equation**, which is
# ubiquitous in economic dynamics and other fields involving planning over time
#
# The intuition behind it is as follows:
#
# - the first term inside the max operation is the lifetime payoff from accepting current offer $ w $, since
#
#
# $$
# w + \beta w + \beta^2 w + \cdots = \frac{w}{1 - \beta}
# $$
#
# - the second term inside the max operation is the **continuation value**, which is the lifetime payoff from rejecting the current offer and then behaving optimally in all subsequent periods
#
#
# If we optimize and pick the best of these two options, we obtain maximal lifetime value from today, given current offer $ w $
#
# But this is precisely $ V(w) $, which is the l.h.s. of [(1)](#equation-odu-pv)
# ### The Optimal Policy
#
# Suppose for now that we are able to solve [(1)](#equation-odu-pv) for the unknown
# function $ V $
#
# Once we have this function in hand we can behave optimally (i.e., make the
# right choice between accept and reject)
#
# All we have to do is select the maximal choice on the r.h.s. of [(1)](#equation-odu-pv)
#
# The optimal action is best thought of as a **policy**, which is, in general, a map from
# states to actions
#
# In our case, the state is the current wage offer $ w $
#
# Given *any* $ w $, we can read off the corresponding best choice (accept or
# reject) by picking the max on the r.h.s. of [(1)](#equation-odu-pv)
#
# Thus, we have a map from $ \mathbb{R} $ to $ \{0, 1\} $, with 1 meaning accept and zero meaning reject
#
# We can write the policy as follows
#
# $$
# \sigma(w) := \mathbf{1}
# \left\{
# \frac{w}{1 - \beta} \geq c + \beta \sum_{i=1}^n V(w_i) p_i
# \right\}
# $$
#
# Here $ \mathbf{1}\{ P \} = 1 $ if statement $ P $ is true and equals zero otherwise
#
# We can also write this as
#
# $$
# \sigma(w) := \mathbf{1} \{ w \geq \bar w \}
# $$
#
# where
#
#
# <a id='equation-odu-barw'></a>
# $$
# \bar w := (1 - \beta) \left\{ c + \beta \sum_{i=1}^n V(w_i) p_i \right\} \tag{2}
# $$
#
# Here $ \bar w $ is a constant depending on $ \beta, c $ and the wage distribution, called the *reservation wage*
#
# The agent should accept if and only if the current wage offer exceeds the reservation wage
#
# Clearly, we can compute this reservation wage if we can compute the value function
# ## Computing the Optimal Policy: Take 1
#
# To put the above ideas into action, we need to compute the value function at
# points $ w_1, \ldots, w_n $
#
# In doing so, we can identify these values with the vector $ v = (v_i) $ where $ v_i := V(w_i) $
#
# In view of [(1)](#equation-odu-pv), this vector satisfies the nonlinear system of equations
#
#
# <a id='equation-odu-pv2'></a>
# $$
# v_i
# = \max \left\{
# \frac{w_i}{1 - \beta}, \, c + \beta \sum_{i=1}^n v_i p_i
# \right\}
# \quad
# \text{for } i = 1, \ldots, n \tag{3}
# $$
#
# It turns out that there is exactly one vector $ v := (v_i)_{i=1}^n $ in
# $ \mathbb R^n $ that satisfies this equation
# ### The Algorithm
#
# To compute this vector, we proceed as follows:
#
# Step 1: pick an arbitrary initial guess $ v \in \mathbb R^n $
#
# Step 2: compute a new vector $ v' \in \mathbb R^n $ via
#
#
# <a id='equation-odu-pv2p'></a>
# $$
# v'_i
# = \max \left\{
# \frac{w_i}{1 - \beta}, \, c + \beta \sum_{i=1}^n v_i p_i
# \right\}
# \quad
# \text{for } i = 1, \ldots, n \tag{4}
# $$
#
# Step 3: calculate a measure of the deviation between $ v $ and $ v' $, such as $ \max_i |v_i - v_i'| $
#
# Step 4: if the deviation is larger than some fixed tolerance, set $ v = v' $ and go to step 2, else continue
#
# Step 5: return $ v $
#
# This algorithm returns an arbitrarily good approximation to the true solution
# to [(3)](#equation-odu-pv2), which represents the value function
#
# (Arbitrarily good means here that the approximation converges to the true
# solution as the tolerance goes to zero)
# ### The Fixed Point Theory
#
# What’s the math behind these ideas?
#
# First, one defines a mapping $ T $ from $ \mathbb R^n $ to
# itself via
#
#
# <a id='equation-odu-pv3'></a>
# $$
# Tv_i
# = \max \left\{
# \frac{w_i}{1 - \beta}, \, c + \beta \sum_{i=1}^n v_i p_i
# \right\}
# \quad
# \text{for } i = 1, \ldots, n \tag{5}
# $$
#
# (A new vector $ Tv $ is obtained from given vector $ v $ by evaluating
# the r.h.s. at each $ i $)
#
# One can show that the conditions of the Banach contraction mapping theorem are
# satisfied by $ T $ as a self-mapping on $ \mathbb{R}^n $
#
# One implication is that $ T $ has a unique fixed point in $ \mathbb R^n $
#
# Moreover, it’s immediate from the definition of $ T $ that this fixed
# point is precisely the value function
#
# The iterative algorithm presented above corresponds to iterating with
# $ T $ from some initial guess $ v $
#
# The Banach contraction mapping theorem tells us that this iterative process
# generates a sequence that converges to the fixed point
# ### Implementation
# ### Setup
# + hide-output=false
using InstantiateFromURL
activate_github("QuantEcon/QuantEconLecturePackages", tag = "v0.9.8");
# + hide-output=true
using LinearAlgebra, Statistics, Compat
using Distributions, Expectations, NLsolve, Roots, Random, Plots, Parameters
# + hide-output=false
gr(fmt = :png);;
# -
# Here’s the distribution of wage offers we’ll work with
# + hide-output=false
n = 50
dist = BetaBinomial(n, 200, 100) # probability distribution
@show support(dist)
w = range(10.0, 60.0, length = n+1) # linearly space wages
using StatsPlots
plt = plot(w, dist, xlabel = "wages", ylabel = "probabilities", legend = false)
# -
# We can explore taking expectations over this distribution
# + hide-output=false
E = expectation(dist) # expectation operator
# exploring the properties of the operator
wage(i) = w[i+1] # +1 to map from support of 0
E_w = E(wage)
E_w_2 = E(i -> wage(i)^2) - E_w^2 # variance
@show E_w, E_w_2
# use operator with left-multiply
@show E * w # the `w` are values assigned for the discrete states
@show dot(pdf.(dist, support(dist)), w); # identical calculation
# -
# To implement our algorithm, let’s have a look at the sequence of approximate value functions that
# this fixed point algorithm generates
#
# Default parameter values are embedded in the function
#
# Our initial guess $ v $ is the value of accepting at every given wage
# + hide-output=false
# parameters and constant objects
c = 25
β = 0.99
num_plots = 6
# Operator
T(v) = max.(w/(1 - β), c + β * E*v) # (5) broadcasts over the w, fixes the v
# alternatively, T(v) = [max(wval/(1 - β), c + β * E*v) for wval in w]
# fill in matrix of vs
vs = zeros(n + 1, 6) # data to fill
vs[:, 1] .= w / (1-β) # initial guess of "accept all"
# manually applying operator
for col in 2:num_plots
v_last = vs[:, col - 1]
vs[:, col] .= T(v_last) # apply operator
end
plot(vs)
# -
# One approach to solving the model is to directly implement this sort of iteration, and continues until measured deviation
# between successive iterates is below tol
# + hide-output=false
function compute_reservation_wage_direct(params; v_iv = collect(w ./(1-β)), max_iter = 500,
tol = 1e-6)
@unpack c, β, w = params
# create a closure for the T operator
T(v) = max.(w/(1 - β), c + β * E*v) # (5) fixing the parameter values
v = copy(v_iv) # start at initial value. copy to prevent v_iv modification
v_next = similar(v)
i = 0
error = Inf
while i < max_iter && error > tol
v_next .= T(v) # (4)
error = norm(v_next - v)
i += 1
v .= v_next # copy contents into v. Also could have used v[:] = v_next
end
# now compute the reservation wage
return (1 - β) * (c + β * E*v) # (2)
end
# -
# In the above, we use `v = copy(v_iv)` rather than just `v_iv = v`
#
# To understand why, first recall that `v_iv` is a function argument – either defaulting to the given value, or passed into the function
#
# > - If we had gone `v = v_iv` instead, then it would have simply created a new name `v` which binds to whatever is located at `v_iv`
# - Since we later use `v .= v_next` later in the algorithm, the values in it would be modified
# - Hence, we would be modifying the `v_iv` vector we were passed in, which may not be what the caller of the function wanted
# - The big issue this creates are “side-effects” where you can call a function and strange things can happen outside of the function that you didn’t expect
# - If you intended for the modification to potentially occur, then the Julia style guide says that we should call the function `compute_reservation_wage_direct!` to make the possible side-effects clear
#
#
#
# As usual, we are better off using a package, which may give a better algorithm and is likely to less error prone
#
# In this case, we can use the `fixedpoint` algorithm discussed in [our Julia by Example lecture](getting_started_julia/julia_by_example.html) to find the fixed point of the $ T $ operator
# + hide-output=false
function compute_reservation_wage(params; v_iv = collect(w ./(1-β)), iterations = 500,
ftol = 1e-6, m = 6)
@unpack c, β, w = params
T(v) = max.(w/(1 - β), c + β * E*v) # (5) fixing the parameter values
v_star = fixedpoint(T, v_iv, iterations = iterations, ftol = ftol,
m = 6).zero # (5)
return (1 - β) * (c + β * E*v_star) # (3)
end
# -
# Let’s compute the reservation wage at the default parameters
# + hide-output=false
mcm = @with_kw (c=25.0, β=0.99, w=w) # named tuples
compute_reservation_wage(mcm()) # call with default parameters
# -
# ### Comparative Statics
#
# Now we know how to compute the reservation wage, let’s see how it varies with
# parameters
#
# In particular, let’s look at what happens when we change $ \beta $ and
# $ c $
# + hide-output=false
grid_size = 25
R = rand(grid_size, grid_size)
c_vals = range(10.0, 30.0, length = grid_size)
β_vals = range(0.9, 0.99, length = grid_size)
for (i, c) in enumerate(c_vals)
for (j, β) in enumerate(β_vals)
R[i, j] = compute_reservation_wage(mcm(c=c, β=β)) # change from defaults
end
end
# + hide-output=false
contour(c_vals, β_vals, R',
title = "Reservation Wage",
xlabel = "c",
ylabel = "beta",
fill = true)
# -
# As expected, the reservation wage increases both with patience and with
# unemployment compensation
# ## Computing the Optimal Policy: Take 2
#
# The approach to dynamic programming just described is very standard and
# broadly applicable
#
# For this particular problem, there’s also an easier way, which circumvents the
# need to compute the value function
#
# Let $ \psi $ denote the value of not accepting a job in this period but
# then behaving optimally in all subsequent periods
#
# That is,
#
#
# <a id='equation-j1'></a>
# $$
# \psi
# = c + \beta
# \sum_{i=1}^n V(w_i) p_i \tag{6}
# $$
#
# where $ V $ is the value function
#
# By the Bellman equation, we then have
#
# $$
# V(w_i)
# = \max \left\{ \frac{w_i}{1 - \beta}, \, \psi \right\}
# $$
#
# Substituting this last equation into [(6)](#equation-j1) gives
#
#
# <a id='equation-j2'></a>
# $$
# \psi
# = c + \beta
# \sum_{i=1}^n
# \max \left\{
# \frac{w_i}{1 - \beta}, \psi
# \right\} p_i \tag{7}
# $$
#
# Which we could also write as $ \psi = T_{\psi}(\psi) $ for the appropriate operator
#
# This is a nonlinear equation that we can solve for $ \psi $
#
# One solution method for this kind of nonlinear equation is iterative
#
# That is,
#
# Step 1: pick an initial guess $ \psi $
#
# Step 2: compute the update $ \psi' $ via
#
#
# <a id='equation-j3'></a>
# $$
# \psi'
# = c + \beta
# \sum_{i=1}^n
# \max \left\{
# \frac{w_i}{1 - \beta}, \psi
# \right\} p_i \tag{8}
# $$
#
# Step 3: calculate the deviation $ |\psi - \psi'| $
#
# Step 4: if the deviation is larger than some fixed tolerance, set $ \psi = \psi' $ and go to step 2, else continue
#
# Step 5: return $ \psi $
#
# Once again, one can use the Banach contraction mapping theorem to show that this process always converges
#
# The big difference here, however, is that we’re iterating on a single number, rather than an $ n $-vector
#
# Here’s an implementation:
# + hide-output=false
function compute_reservation_wage_ψ(c, β; ψ_iv = E * w ./ (1 - β), max_iter = 500,
tol = 1e-5)
T_ψ(ψ) = [c + β * E*max.((w ./ (1 - β)), ψ[1])] # (7)
# using vectors since fixedpoint doesn't support scalar
ψ_star = fixedpoint(T_ψ, [ψ_iv]).zero[1]
return (1 - β) * (c + β * ψ_star) # (2)
end
compute_reservation_wage_ψ(c, β)
# -
# You can use this code to solve the exercise below
#
# Another option is to solve for the root of the $ T_{\psi}(\psi) - \psi $ equation
# + hide-output=false
function compute_reservation_wage_ψ2(c, β; ψ_iv = E * w ./ (1 - β), max_iter = 500,
tol = 1e-5)
root_ψ(ψ) = c + β * E*max.((w ./ (1 - β)), ψ) - ψ # (7)
ψ_star = find_zero(root_ψ, ψ_iv)
return (1 - β) * (c + β * ψ_star) # (2)
end
compute_reservation_wage_ψ2(c, β)
# -
# ## Exercises
# ### Exercise 1
#
# Compute the average duration of unemployment when $ \beta=0.99 $ and
# $ c $ takes the following values
#
# > `c_vals = range(10, 40, length = 25)`
#
#
# That is, start the agent off as unemployed, computed their reservation wage
# given the parameters, and then simulate to see how long it takes to accept
#
# Repeat a large number of times and take the average
#
# Plot mean unemployment duration as a function of $ c $ in `c_vals`
# ## Solutions
# ### Exercise 1
#
# Here’s one solution
# + hide-output=false
function compute_stopping_time(w̄; seed=1234)
Random.seed!(seed)
stopping_time = 0
t = 1
# make sure the constraint is sometimes binding
@assert length(w) - 1 ∈ support(dist) && w̄ <= w[end]
while true
# Generate a wage draw
w_val = w[rand(dist)] # the wage dist set up earlier
if w_val ≥ w̄
stopping_time = t
break
else
t += 1
end
end
return stopping_time
end
compute_mean_stopping_time(w̄, num_reps=10000) = mean(i ->
compute_stopping_time(w̄,
seed = i), 1:num_reps)
c_vals = range(10, 40, length = 25)
stop_times = similar(c_vals)
beta = 0.99
for (i, c) in enumerate(c_vals)
w̄ = compute_reservation_wage_ψ(c, beta)
stop_times[i] = compute_mean_stopping_time(w̄)
end
plot(c_vals, stop_times, label = "mean unemployment duration",
xlabel = "unemployment compensation", ylabel = "months")
| dynamic_programming/mccall_model.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# !python -m spacy download en_core_web_sm
import spacy
nlp = spacy.load("en_core_web_sm")
example_message = """
Dr <NAME>, one of the founding 'fathers' of the atomic bomb, has left a deadly legacy to humanity. For he is the inventor of ice-nine, a lethal chemical capable of freezing the entire planet. Writer Jonah's search for his whereabouts leads him to Hoenikker's three eccentric children, to an island republic in the Caribbean where the absurd religion of Bokononism is practised, to love and to insanity. Told with deadpan humour and bitter irony, <NAME>'s cult tale of global destruction is a frightening and funny satire on the end of the world and the madness of mankind.
"""
doc = nlp(example_message)
nlp.pipeline
doc.ents
# # Add new step to the pipeline
import redis
r = redis.Redis()
sub = r.pubsub(ignore_subscribe_messages=True)
sub.subscribe(['processed'])
import mq
for message in sub.listen():
sentence = mq.read_message_data(message)
| step3_nlp/NER.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:ml-laboratory] *
# language: python
# name: conda-env-ml-laboratory-py
# ---
# +
from keras.layers import Input, Dense, Flatten, Reshape, Conv2D, MaxPooling2D, UpSampling2D
from keras.models import Model, Sequential
from keras.datasets import mnist
from keras.callbacks import Callback
from keras.models import load_model
import numpy as np
import matplotlib.pyplot as plt
# %matplotlib inline
# Workaround for Error #15: Initializing libiomp5.dylib, but found libiomp5.dylib already initialized
# https://stackoverflow.com/questions/53014306/error-15-initializing-libiomp5-dylib-but-found-libiomp5-dylib-already-initial
# https://github.com/dmlc/xgboost/issues/1715
# export KMP_DUPLICATE_LIB_OK=True
import os
os.environ['KMP_DUPLICATE_LIB_OK']='True'
# -----
def add_noise(img, noise_factor = 0.5):
img_noisy = img + noise_factor * np.random.normal(loc=0.0, scale=1.0, size=img.shape)
img_noisy = np.clip(img_noisy, 0., 1.)
return img_noisy
def plot_images(images, title=""):
figure = plt.figure()
i = 0
for img in images:
ax = figure.add_subplot(len(images)/8 + 1, 8, i + 1, xticks=[], yticks=[])
ax.imshow(np.squeeze(img), cmap = 'gray')
i += 1
figure.suptitle(title, fontsize=25)
# -
# we don't care about the label in this scenario
(x_train, _), (x_test, _) = mnist.load_data()
# normalized the data
x_train = x_train.astype('float32') / 255.
x_test = x_test.astype('float32') / 255.
# visualize some test images
plot_images(x_train[:31,:], 'Some training images')
plot_images(x_test[:31,:], 'Some test images')
# First version
model = Sequential()
model.add(Flatten(input_shape = (28, 28)))
model.add(Dense(encoding_dim, activation='relu'))
model.add(Dense(28*28, activation='sigmoid'))
model.add(Reshape((28,28)))
model.compile(optimizer='adam', loss='mse')
model.summary()
encoding_dim = 32
epochs = 10
model.fit(x_train, x_train, epochs=epochs, validation_data=(x_test, x_test))
model.save('auto.e10.h5')
model = load_model('auto.e10.h5')
# +
# Run the auto encoder model on another input image
def show_encoding(model, test_set, sample_size=8, noise=0.5)
test_images = test_set[np.random.choice(len(test_set),
sample_size=8,
replace=False)]
test_images = [add_noise(img, noise) for img in test_images]
plot_images(test_images, title='Original images')
generated = [model.predict(img.reshape(1,28,28))[0].reshape(28,28,1) for img in test_images]
plot_images(np.array(generated), title='Generated images')
show_encoding(model, x_test, sample_size=8, noise=0.2)
# -
encoding_dim = 32
epochs = 50
model.fit(x_train, x_train, epochs=epochs, validation_data=(x_test, x_test))
model.save('auto.e50.h5')
# Run the auto encoder model on another input image
model = load_model('auto.e50.h5')
show_encoding(model, x_test, sample_size=8, noise=0.2)
# +
# test_images = x_test[np.random.choice(len(test_images), size=8, replace=False)]
# test_images = [add_noise(img, 0.25) for img in test_images]
# plot_images(test_images, title='Original images')
# generated = [model.predict(img.reshape(1,28,28))[0].reshape(28,28,1) for img in test_images]
# plot_images(np.array(generated), title='Generated images')
# +
# 2nd version: CNN
cnn = Sequential()
cnn.add(Reshape((28, 28, 1), input_shape=(28, 28)))
cnn.add(Conv2D(32, (3, 3), activation='relu', padding='same'))
cnn.add(MaxPooling2D(2, 2))
cnn.add(Conv2D(32, (3, 3), activation='relu', padding='same'))
cnn.add(UpSampling2D((2, 2)))
cnn.add(Conv2D(1, (3, 3), activation='relu', padding='same'))
cnn.add(Reshape((28, 28)))
cnn.compile(optimizer='adam', loss='mse')
cnn.summary()
# -
encoding_dim = 32
epochs = 10
cnn.fit(x_train, x_train, epochs=epochs, validation_data=(x_test, x_test))
cnn.save('auto.cnn.e10.h5')
model = load_model('auto.cnn.e50.h5')
show_encoding(model, x_test, sample_size=8, noise=0.2)
| tensorflow/notebooks/keras/4_Auto-encoders-with-keras.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# #### 1. Write a Python program to print the following string in a specific format (see the output)
#
# Sample String : "Twinkle, twinkle, little star, How I wonder what you are! Up above the world so high, Like a diamond in the sky. Twinkle, twinkle, little star, How I wonder what you are" Output :
#
# Twinkle, twinkle, little star,
# How I wonder what you are!
# Up above the world so high,
# Like a diamond in the sky.
# Twinkle, twinkle, little star,
# How I wonder what you are
simple_string = "Twinkle, twinkle, little star, How I wonder what you are! Up above the world so high,\nLike a diamond in the sky. Twinkle, twinkle, little star, How I wonder what you are"
print(simple_string)
# #### 2. Write a Python program to get the Python version you are using.
import sys
print("Python verison:", sys.version)
print("Version info:", sys.version_info)
# #### 3. Write a Python program to display the current date and time.
# Sample Output :
# Current date and time : 2014-07-05 14:34:14
# +
import datetime
print(f"Current date and time:", datetime.datetime.now().strftime("%Y-%m-%d %H:%M:%S"))
# -
# #### 4. Write a Python program which accepts the radius of a circle from the user and compute the area.
# Sample Output :
# r = 1.1
# Area = 3.8013271108436504
import math
# Close for all runs
# r = float(input("Input the radius of the circle: "))
r = 2.2
print(f"r = {r} Area = {math.pi * r**2}")
# #### 5. Write a Python program which accepts the user's first and last name and print them in reverse order with a space between them.
# first_name = input("Input your first name: ")
# last_name = input("In your last name: ")
first_name = "Hieu"
last_name = "<NAME>"
print(f"Wellcome! {last_name} {first_name}")
# #### 6. Write a Python program which accepts a sequence of comma-separated numbers from user and generate a list and a tuple with those numbers.
#
# Sample data : 3, 5, 7, 23
# Output:
# List : ['3', ' 5', ' 7', ' 23']
# Tuple : ('3', ' 5', ' 7', ' 23')
# sample_input_values = input("Input some comma-separated numbers: ")
sample_input_values = '4, 6, 5, 3, 6, 4, 12'
list_input_values = list(sample_input_values.split(','))
tuple_input_values = tuple(list_input_values)
print("List:", list_input_values)
print("Tuple:", tuple_input_values)
# #### 7. Write a Python program to accept a filename from the user and print the extension of that.
#
# Sample filename : abc.java
# Output : java
# sample_input_filename = input("Input a filename: ")
sample_input_filename = "Working.py"
print("The file name is:", sample_input_filename.split('.')[-1])
# #### 8. Write a Python program to display the first and last colors from the following list.
#
# color_list = ["Red","Green","White" ,"Black"]
color_list = ["Red","Green","White" ,"Black"]
print(f"The first color of the list is {color_list[0]}, last color is {color_list[-1]}")
# #### 9. Write a Python program to display the examination schedule. (extract the date from exam_st_date).
#
# exam_st_date = (11, 12, 2014)
import datetime
exam_st_date = (11, 12, 2014)
exam_st_date_parse = datetime.date(*exam_st_date[::-1])
print(exam_st_date_parse)
# #### 10. Write a Python program that accepts an integer (n) and computes the value of n+nn+nnn.
#
# Sample value of n is 5
# Expected Result : 615
# simple_n = input("Input a integer number: ")
simple_n = '5'
print("result of n+nn+nnn:", int(simple_n)+int(simple_n+simple_n)+int(simple_n+simple_n+simple_n))
# #### 11. Write a Python program to print the documents (syntax, description etc.) of Python built-in function(s).
#
# Sample function : abs()
# Expected Result :
# abs(number) -> number
# Return the absolute value of the argument.
print(help(abs))
print(abs.__doc__)
# #### 12. Write a Python program to print the calendar of a given month and year.
# Note : Use 'calendar' module.
import calendar
print(calendar.month(2021, 11, w=0, l=0))
# #### 13. Write a Python program to print the following 'here document'. Go to the editor
# Sample string :
# a string that you "don't" have to escape
# This
# is a ....... multi-line
# heredoc string --------> example
print("""
a string that you "don't" have to escape
This
is a ....... multi-line
heredoc string --------> example""")
# #### 14. Write a Python program to calculate number of days between two dates.
# Sample dates : (2014, 7, 2), (2014, 7, 11)
# Expected output : 9 days
from datetime import date
Date_1 = date(2014, 7, 2)
Date_2 = date(2014, 7, 11)
days_diff= Date_2 - Date_1
print(days_diff.days)
# #### 15. Write a Python program to get the volume of a sphere with radius 6.
# Click me to see the sample solution
import math
r = 6
print("Volume of a sphere with radius 6:", 4/3*r*math.pi)
# #### 16. Write a Python program to get the difference between a given number and 17, if the number is greater than 17 return double the absolute difference.
# a_number = int(input("Input an integer number: "))
a_number = 66
if a_number > 17:
print(2*abs(a_number-17))
# #### 17. Write a Python program to test whether a number is within 100 of 1000 or 2000.
def check_number(x):
if (x >=100 and x <=1000) or (x>=1900 and x <= 2000):
return True
else:
return False
print(check_number(555))
print(check_number(44))
print(check_number(905))
print(check_number(1975))
# #### 18. Write a Python program to calculate the sum of three given numbers, if the values are equal then return three times of their sum.
# #### 19. Write a Python program to get a new string from a given string where "Is" has been added to the front. If the given string already begins with "Is" then return the string unchanged.
# #### 20. Write a Python program to get a string which is n (non-negative integer) copies of a given string.
| Python/Basic - Part-I/Python basic Part - I .ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + colab={"base_uri": "https://localhost:8080/"} id="DIhNg2zIIn43" outputId="93b2d516-f73b-4cd3-dcc8-d69c4b9fed99"
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import tensorflow as tf
# !pip install tensorflow-addons
import tensorflow_addons as tfa
from sklearn.model_selection import train_test_split
# + colab={"base_uri": "https://localhost:8080/"} id="mzatsb06In49" outputId="3d949007-5174-4f3b-fa2d-72235ebcc7d8"
from google.colab import drive
drive.mount('/content/gdrive')
# + colab={"base_uri": "https://localhost:8080/"} id="HLgJ9pNFIn4-" outputId="9fab9e84-35bd-4df8-8209-7e2ce1dc4b8b"
# !git clone https://github.com/naufalhisyam/TurbidityPrediction-thesis.git
os.chdir('/content/TurbidityPrediction-thesis')
# + [markdown] id="V4-H6ee_OtAo"
# LOAD MODEL
# + id="tqJKFEs_Op2v"
model_path = r'/content/gdrive/MyDrive/MODEL BERHASIL/0deg/ResNet_0deg_none_lr1e-3_decay1e-6_bs8_huber/resnet-epoch100-loss9.602346420288086'
name = 'ResNet50_0deg_noTL'
if not os.path.exists(f'plots/{name}'):
os.makedirs(f'plots/{name}')
model = tf.keras.models.load_model(model_path) #loads saved model
# + id="IHVhzXKjIn4_"
images = pd.read_csv(r'./Datasets/0degree/0degInfo.csv') #load dataset info
train_df, test_df = train_test_split(images, train_size=0.9, shuffle=True, random_state=1) #Split into train and test set
train_generator = tf.keras.preprocessing.image.ImageDataGenerator(
horizontal_flip=True
)
test_generator = tf.keras.preprocessing.image.ImageDataGenerator(
horizontal_flip=True
)
# + colab={"base_uri": "https://localhost:8080/"} id="RBZ6WlHXIn5A" outputId="c81c804c-d911-4585-868c-60ddcf242da8"
train_images = train_generator.flow_from_dataframe(
dataframe=train_df,
x_col='Filepath',
y_col='Turbidity',
target_size=(224, 224),
color_mode='rgb',
class_mode='raw',
batch_size=32,
shuffle=False,
seed=42,
)
test_images = test_generator.flow_from_dataframe(
dataframe=test_df,
x_col='Filepath',
y_col='Turbidity',
target_size=(224, 224),
color_mode='rgb',
class_mode='raw',
batch_size=32,
shuffle=False
)
# + [markdown] id="RbjEOfwkIn5B"
# **Plotting Model Graphs**
# + colab={"base_uri": "https://localhost:8080/"} id="-EoG1gcTIn5J" outputId="bb53c81f-f84d-4ab5-f6ef-aa0f8a766a2c"
test_pred = np.squeeze(model.predict(test_images))
test_true = test_images.labels
test_residuals = test_true - test_pred
train_pred = np.squeeze(model.predict(train_images))
train_true = train_images.labels
train_residuals = train_true - train_pred
train_score = model.evaluate(train_images)
test_score = model.evaluate(test_images)
print('test ',test_score)
print('train ', train_score)
# + [markdown] id="9SNaYE58In5N"
# Residual Plot
# + colab={"base_uri": "https://localhost:8080/", "height": 405} id="arOYaNzoIn5O" outputId="8ee86293-80d8-47c2-9fa1-4f2d827b7767"
f, axs = plt.subplots(1, 2, figsize=(8,6), gridspec_kw={'width_ratios': [4, 1]})
f.suptitle(f'Residual Plot - {name}', fontsize=13, fontweight='bold', y=0.92)
axs[0].scatter(train_pred,train_residuals, label='Train Set', alpha=0.75, color='tab:blue')
axs[0].scatter(test_pred,test_residuals, label='Test Set', alpha=0.75, color='tab:orange')
axs[0].set_ylabel('Residual (NTU)')
axs[0].set_xlabel('Predicted Turbidity (NTU)')
axs[0].axhline(0, color='black')
axs[0].legend()
axs[0].grid()
axs[1].hist(train_residuals, bins=50, orientation="horizontal", density=True, alpha=0.9, color='tab:blue')
axs[1].hist(test_residuals, bins=50, orientation="horizontal", density=True, alpha=0.75, color='tab:orange')
axs[1].axhline(0, color='black')
axs[1].set_xlabel('Distribution')
axs[1].yaxis.tick_right()
axs[1].grid(axis='y')
plt.subplots_adjust(wspace=0.05)
plt.savefig(f'plots/{name}/residualPlot_{name}.png', dpi=150)
plt.show()
# + [markdown] id="nH7eDNfiIn5P"
# Measured vs Predicted Plot
# + colab={"base_uri": "https://localhost:8080/", "height": 423} id="C9HK-G42In5Q" outputId="bef4c0d6-b958-43bb-fcd9-18c64ea65dc4"
fig, ax = plt.subplots(1,2,figsize=(13,6))
fig.suptitle(f'Nilai Prediksi vs Observasi - {name}', fontsize=13, fontweight='bold', y=0.96)
ax[0].scatter(test_true,test_pred, label=f'$Test\ R^2=${round(test_score[3],3)}',color='tab:orange', alpha=0.75)
theta = np.polyfit(test_true, test_pred, 1)
y_line = theta[1] + theta[0] * test_true
ax[0].plot([test_true.min(), test_true.max()], [y_line.min(), y_line.max()],'k--', lw=2,label='best fit')
ax[0].plot([test_true.min(), test_true.max()], [test_true.min(), test_true.max()], 'k--', lw=2, label='identity',color='dimgray')
ax[0].set_xlabel('Measured Turbidity (NTU)')
ax[0].set_ylabel('Predicted Turbidity (NTU)')
ax[0].set_title(f'Test Set', fontsize=10, fontweight='bold')
ax[0].set_xlim([0, 200])
ax[0].set_ylim([0, 200])
ax[0].grid()
ax[0].legend()
ax[1].scatter(train_true,train_pred, label=f'$Train\ R^2=${round(train_score[3],3)}', color='tab:blue', alpha=0.75)
theta2 = np.polyfit(train_true, train_pred, 1)
y_line2 = theta2[1] + theta2[0] * train_true
ax[1].plot([train_true.min(), train_true.max()], [y_line2.min(), y_line2.max()],'k--', lw=2,label='best fit')
ax[1].plot([train_true.min(), train_true.max()], [train_true.min(),train_true.max()], 'k--', lw=2, label='identity',color='dimgray')
ax[1].set_xlabel('Measured Turbidity (NTU)')
ax[1].set_ylabel('Predicted Turbidity (NTU)')
ax[1].set_title(f'Train Set', fontsize=10, fontweight='bold')
ax[1].set_xlim([0, 200])
ax[1].set_ylim([0, 200])
ax[1].grid()
ax[1].legend()
plt.savefig(f'plots/{name}/predErrorPlot_{name}.png', dpi=150)
plt.show()
# + [markdown] id="VQ-4qDSVIn5R"
# Training History
# + id="Tr86rxM8In5R"
history = pd.read_csv(f'{model_path}/history.csv')
loss = history['loss']
val_loss = history['val_loss']
ma_error = history['mae']
val_ma_error = history['val_mae']
r2 = history['R2']
val_r2 = history['val_R2']
ms_error = history['mse']
val_ms_error = history['val_mse']
# + colab={"base_uri": "https://localhost:8080/", "height": 749} id="xNFBLEQ4In5T" outputId="b01a6dbd-6ee8-4fab-962d-5d5d9a06324a"
epochs = range(1, len(loss) + 1)
f, axs = plt.subplots(2, 2, figsize=(12,12))
axs[0][0].plot(epochs, loss, 'tab:blue', label='train_loss (huber)')
axs[0][0].plot(epochs, val_loss, 'tab:red', label='val_loss (huber)')
axs[0][0].set_title('Loss selama Training', fontsize=10, fontweight='bold')
axs[0][0].set(yscale="log")
axs[0][0].set_xlabel('Epoch')
axs[0][0].set_ylabel('Loss')
axs[0][0].legend(facecolor='white')
axs[0][0].grid()
axs[0][1].plot(epochs, ms_error, 'tab:blue', label='train_mse')
axs[0][1].plot(epochs, val_ms_error, 'tab:red', label='val_mse')
axs[0][1].set_title('MSE selama Training ', fontsize=10, fontweight='bold')
axs[0][1].set_xlabel('Epoch')
axs[0][1].set_ylabel('MSE')
axs[0][1].set(yscale="log")
axs[0][1].legend(facecolor='white')
axs[0][1].grid()
axs[1][0].plot(epochs, ma_error, 'tab:blue', label='train_mae')
axs[1][0].plot(epochs, val_ma_error, 'tab:red', label='val_mae')
axs[1][0].set_title('MAE selama Training', fontsize=10, fontweight='bold')
axs[1][0].set_xlabel('Epoch')
axs[1][0].set_ylabel('MAE')
axs[1][0].set(yscale="log")
axs[1][0].legend(facecolor='white')
axs[1][0].grid()
axs[1][1].plot(epochs, r2, 'tab:blue', label='train_R2')
axs[1][1].plot(epochs, val_r2, 'tab:red', label='val_R2')
axs[1][1].set_title(r'$R^2$ selama Training', fontsize=10, fontweight='bold')
axs[1][1].set_xlabel('Epoch')
axs[1][1].set_ylabel(r'$R^2$')
axs[1][1].set(yscale="log")
axs[1][1].legend(facecolor='white')
axs[1][1].grid()
#plt.tight_layout()
f.suptitle(f'Grafik Training - {name}', fontsize=13, fontweight='bold', y=0.92)
plt.savefig(f'plots/{name}/trainPlot_{name}.png', dpi=150)
plt.show()
# + id="z6NY5wfxKHLA" outputId="efc5e6cc-4f86-4fcb-b292-692b6b35aee5" colab={"base_uri": "https://localhost:8080/"}
save_path = f"/content/gdrive/MyDrive/MODEL BERHASIL/Plots"
if not os.path.exists(save_path):
os.makedirs(save_path)
oripath = "plots/."
# !cp -a "{oripath}" "{save_path}" # copies files to google drive
print("Done!")
| Utils/evaluate_model.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/sigvehaug/Introduction-to-Python-for-Medical-Researchers/blob/master/07-Exercise-2.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="rzrzHykrUoFO"
# Introduction to Python Programming for Medical Researchers, University of Bern, <NAME>
# + [markdown] id="cpigRldCUqre"
# # Exercise 2 - (60 min) - Homework for next lecture
#
# Skin cancer is the most common cancer globally, with melanoma being the most deadly form. [ISIC](https://challenge2019.isic-archive.com/) arranges machine learning challenges on automatic classification based on training with many thousand labeled skin lesian images. In this course we don't do machine learning, however some basic image management and data analysis with Python.
#
# In the [github repository](https://github.com/sigvehaug/Introduction-to-Python-for-Medical-Researchers/tree/master/Data) of the course there are about 100 images and an excel (csv) file with the image labeling indicating the type of cancer.
#
# Use course examples 1 and 2 to fullfil the following tasks in a Jupyter notebook.
#
# ### Tasks
#
# - What is the relative weight (%) of each cancer type in the dataset? For this task, load the excel file into a datafame and extract the numbers and plot them with a histogram.
# - Use the dataframe to identify 10 images from each class and plot them from the images available in the github repository.
#
# ### Delivery and Deadline
#
# You will be assigned to another course colleague and you solve the exercise togehter.
# - Upload your notebook to the e-learningplatform of the course with the following file name convention: EX2-NamePerson1-NamePerson2.ipynb
# - Deadline for upload is September 15 (evening).
#
# Many thanks and have fun !
# ---
#
# + [markdown] id="1b7ZWKZgeln0"
# ---
#
# ## Mandatory Feedback (one question)
#
# Please fill this [feedback form](https://forms.gle/J5AwyCXcR5sXcJsv6). Many thanks !
| Exercises/Exercise-2.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 ana-1.2.9-py3
# language: python
# name: ana-1.2.9-py3
# ---
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
from lmfit import Model
#matplotlib.rcParams.keys()
plt.style.use('dark_background')
mpl.rcParams['figure.figsize'] = (11, 7)
mpl.rc('font', size=14)
length = 8
width = 1.5
mpl.rcParams['xtick.major.size'] = length
mpl.rcParams['ytick.major.size'] = length
mpl.rcParams['xtick.major.width'] = width
mpl.rcParams['ytick.major.width'] = width
""" CONSTANTS """
e_0 = 8.85E-12 # [s/(Ohm*m)]
c = 299792458 # [m/s]
h = 4.135667516e-15 # eV*s
# # XPCS spot size
# +
def speckle_size(lam, footprint, L):
return lam*L/footprint
display = 'plot'
E = 1200 # eV
lam = h*c/E*1e6
# lam = 1E-3 # [um]
iangle = np.deg2rad(90)
spot_size = np.arange(45,260,10) # [um]
footprint = spot_size/np.sin(iangle)
L = 3.5E6 # [um]
S = speckle_size(lam, footprint, L)
if display=='print':
if S.ndim!=1:
S = np.asarray([S])
st = 'Speckle size: ' + ', '.join('{:0.2f}'.format(k) for k in S) + ' [um]'
print(st)
elif display=='plot':
x = np.argmax([np.array([lam]).size, np.array([footprint]).size, np.array([L]).size])
if x==0:
x = lam*1000
xlabel = 'Wavelength [nm]'
elif x==1:
x = footprint
xlabel = 'Footprint size [um]'
elif x==2:
x = L*10**-6
xlabel = 'Samplt - det distance [m]'
fig, ax = plt.subplots()
ax.plot(x,S,'-o')
ax.set_xlabel(xlabel)
ax.set_ylabel('Speckel size [um]')
ax.grid()
plt.show()
# -
# # Optical properties
# +
""" MATERIAL AND LIGHT DATA """
sigma = 500 # [1/(Ohm*cm)]
sigma = sigma*100 # [1/(Ohm*m)]
e_r = 0.5 # [] real part of epsilon
wvl = 800E-9 # [m] wavelength
f = c/wvl # [1/s]
omega = 2*np.pi*f # [1/s]
"""#########################################################################"""
n = np.sqrt(e_r+1j*sigma/e_0/omega)
alpha = 2*n.imag*omega/c
str = '\nPenetration depth 1/alpha = {:0.2f} [nm]'.format((1/alpha)*10**9)
print(str)
""" REFLECTIVITY """
nr = n.real;
ni = n.imag;
thetai = np.deg2rad(np.arange(0,91,1))
thetat = np.arcsin( np.sin(thetai)/n )
r = (np.cos(thetai)-n*np.cos(thetat)) / (np.cos(thetai)+n*np.cos(thetat))
R = np.absolute( r )**2 # reflectivity at incident angle theta1 (s-pol)
R_N = ( 1-n / (1+n) )**2 # normal incidence reflectivity
""" p-polarized light """
num = -( (nr**2-ni**2+1j*(2*nr*ni)) )*np.cos(thetai) + np.sqrt( (nr**2-ni**2-np.sin(thetai)**2+1j*2*nr*ni) )
den = ( (nr**2-ni**2+1j*(2*nr*ni)) )*np.cos(thetai) + np.sqrt( (nr**2-ni**2-np.sin(thetai)**2+1j*2*nr*ni) )
R_p = np.absolute(num / den)**2
""" s-polarized light """
num = np.cos(thetai) - np.sqrt( (nr**2-ni**2-np.sin(thetai)**2+1j*2*nr*ni) )
den = np.cos(thetai) + np.sqrt( (nr**2-ni**2-np.sin(thetai)**2+1j*2*nr*ni) )
R_s = np.absolute(num / den)**2
thetai = np.rad2deg(thetai)
thetat = np.rad2deg(thetat.real)
plt.figure('Reflectivity')
plt.title('Reflectivity')
plt.plot(thetai, R_p, label='p-polarized')
plt.xlabel('incident angle')
plt.ylabel('reflectance')
plt.plot(thetai, R_s, label='s-polarized')
plt.xlabel('incident angle')
plt.ylabel('reflectance')
plt.legend(loc='upper left')
plt.grid()
plt.figure('Refraction')
plt.title('Refraction')
plt.plot(thetai, thetat)
plt.xlabel('theta_i')
plt.ylabel('theta_t')
plt.grid()
plt.show()
# -
# # Fluence
# +
""" material properties """
""" low T """
z0 = 52 # effective penetration depth [nm]
T = 0.8 # transmission
Cp = 5 # heat capacity [J/K(/mol)]
""" high T """
z0 = 64
T = 0.82
Cp = 10
""" laser properties """
power = 5 # [mW]
rep_rate = 1000 # [Hz]
spotx = 0.046 # FWHM [cm]
spoty = 0.052 # FWHM [cm]
err_spot = 0.001 # [cm]
""" fluence """
fluence = power/rep_rate/spotx/spoty # [mJ/cm^2]
fluence_max = power/rep_rate/(spotx-err_spot)/(spoty-err_spot)
fluence_min = power/rep_rate/(spotx+err_spot)/(spoty+err_spot)
print('\nfluence = {:0.2f} ({:1.2f}, {:2.2f}) mJ/cm^2\n'.format(fluence, fluence_min-fluence, fluence_max-fluence))
""" energy density """
dlayer = 1 # [nm]
flu_top = fluence*T*np.exp(-0*dlayer/z0)
flu_bottom = fluence*T*np.exp(-1*dlayer/z0)
n0 = (flu_top-flu_bottom)/(dlayer*1E-7)/1000
flu_top = fluence_max*T*np.exp(-0*dlayer/z0)
flu_bottom = fluence_max*T*np.exp(-1*dlayer/z0)
n0_max = (flu_top-flu_bottom)/(dlayer*1E-7)/1000
flu_top = fluence_min*T*np.exp(-0*dlayer/z0)
flu_bottom = fluence_min*T*np.exp(-1*dlayer/z0)
n0_min = (flu_top-flu_bottom)/(dlayer*1E-7)/1000
print('n = {:0.2f} ({:1.2f}, {:2.2f}) mJ/cm^2\n'.format(n0, n0_min-n0, n0_max-n0))
""" Average heating """
heating = power/rep_rate/Cp*T *1000 #[mK]
print('heating = {:0.3f} K'.format(heating))
# -
| utilities.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import sqlite3
import pandas as pd
import matplotlib.dates as dates
from datetime import datetime as dt
import warnings
warnings.filterwarnings('ignore')
import matplotlib.pyplot as plt
import numpy as np
from matplotlib.ticker import FuncFormatter
import Cdf
# -
params = {'axes.labelsize' : 14, 'axes.titlesize' : 14,
'font.size' : 14, 'legend.fontsize' : 14,
'xtick.labelsize' : 14, 'ytick.labelsize' : 14}
plt.rcParams.update(params)
# +
conn = sqlite3.connect('../data/netflix-data-aggregated.db')
netflix = pd.read_sql_query('select * from netflix_meta',
con=conn, parse_dates=['dtime'])
traceroute = pd.read_sql_query('select * from traceroute_meta',
con=conn, parse_dates=['dtime'])
conn.close()
# -
netflix['delta_connect_time'] = netflix['delta_connect_time']/1000.0
for df in [netflix, traceroute]:
df['v4_cache'] = df['src_asn_v4'] == df['dst_asn_v4']
df['v6_cache'] = df['src_asn_v6'] == df['dst_asn_v6']
netflix_v4_cache = netflix[netflix['v4_cache'] & ~netflix['v6_cache']]
netflix_v6_cache = netflix[~netflix['v4_cache'] & netflix['v6_cache']]
netflix_neither_caches = netflix[~netflix['v4_cache'] & ~netflix['v6_cache']]
netflix_both_caches = netflix[netflix['v4_cache'] & netflix['v6_cache']]
netflix_v4_cache
netflix_v6_cache
netflix_neither_caches
netflix_both_caches
traceroute_v4_cache = traceroute[traceroute['v4_cache'] & ~traceroute['v6_cache']]
traceroute_v6_cache = traceroute[~traceroute['v4_cache'] & traceroute['v6_cache']]
traceroute_neither_caches = traceroute[~traceroute['v4_cache'] & ~traceroute['v6_cache']]
traceroute_both_caches = traceroute[traceroute['v4_cache'] & traceroute['v6_cache']]
traceroute_v4_cache
traceroute_v6_cache
traceroute_neither_caches
traceroute_both_caches
ttl_cache_v4 = Cdf.MakeCdfFromList(traceroute_v4_cache['delta_ttl'])
ttl_cache_v6 = Cdf.MakeCdfFromList(traceroute_v6_cache['delta_ttl'])
ttl_cache_both = Cdf.MakeCdfFromList(traceroute_both_caches['delta_ttl'])
ttl_cache_neither = Cdf.MakeCdfFromList(traceroute_neither_caches['delta_ttl'])
# +
cdf_fig, cdf_ax = plt.subplots(figsize = (7, 2))
cdf_ax.plot(ttl_cache_v4.xs, ttl_cache_v4.ps, marker = '^', linewidth = 0.3, markersize = 5, fillstyle = 'none', color = 'blue')
cdf_ax.plot(ttl_cache_v6.xs, ttl_cache_v6.ps, marker = 'v', linewidth = 0.3, markersize = 5, fillstyle = 'none', color = 'red')
cdf_ax.plot(ttl_cache_both.xs, ttl_cache_both.ps, marker = 'd', linewidth = 0.3, markersize = 5, fillstyle = 'none', color = 'purple')
cdf_ax.plot(ttl_cache_neither.xs, ttl_cache_neither.ps, marker = 'o', linewidth = 0.3, markersize = 5, fillstyle = 'none', color = 'green')
cdf_ax.grid(False)
yticks = np.arange(0,1.1,0.2)
cdf_ax.set_yticks(yticks)
cdf_ax.set_ylim([-0.05,1.05])
cdf_ax.set_xlim([-25,25])
cdf_ax.set_xscale('linear')
cdf_ax.set_xlabel('Delta')
cdf_ax.set_ylabel('CDF')
cdf_ax.legend(["IPv4 Only", "IPv6 Only", "Both", "Neither"], fontsize = 'small', loc = 'lower right')
cdf_ax.spines['right'].set_color('none')
cdf_ax.spines['top'].set_color('none')
cdf_ax.yaxis.set_ticks_position('left')
cdf_ax.xaxis.set_ticks_position('bottom')
cdf_ax.spines['bottom'].set_position(('axes', -0.03))
cdf_ax.spines['left'].set_position(('axes', -0.03))
cdf_ax.axvline(x = 0, linewidth = 1.0, ymax = 0.95, ymin = 0, color = 'black', ls = 'dotted')
cdf_ax.annotate('', xy = (0.4, 1.1), xycoords = 'axes fraction', xytext = (0.0, 1.1),
arrowprops = dict(arrowstyle = '<-'))
cdf_ax.annotate('', xy = (1.0, 1.1), xycoords = 'axes fraction', xytext = (0.6, 1.1),
arrowprops = dict(arrowstyle = '->'))
cdf_ax.text(-22.5, 1.2, 'IPv6 paths longer')
cdf_ax.text(7, 1.2, 'IPv6 paths shorter')
cdf_ax.set_title('TTL', y = 1.30)
cdf_fig.savefig('../plots/traceroute-ttl-deltas-cache-pairs.pdf', bbox_inches='tight')
plt.show()
# -
print('------ TTL V4 CACHE ------')
print('m_delta_ttl; cdf')
print('--------------------------')
for x, y in zip(ttl_cache_v4.xs, ttl_cache_v4.ps):
print('%.2f; %.5f' % (x, y))
print('------ TTL V6 CACHE ------')
print('m_delta_ttl; cdf')
print('--------------------------')
for x, y in zip(ttl_cache_v6.xs, ttl_cache_v6.ps):
print('%.2f; %.5f' % (x, y))
print('------ TTL CACHE BOTH ------')
print('m_delta_ttl; cdf')
print('----------------------------')
for x, y in zip(ttl_cache_both.xs, ttl_cache_both.ps):
print('%.2f; %.5f' % (x, y))
print('------ TTL CACHE NEITHER ------')
print('m_delta_ttl; cdf')
print('-------------------------------')
for x, y in zip(ttl_cache_neither.xs, ttl_cache_neither.ps):
print('%.2f; %.5f' % (x, y))
tcp_conn_cache_v4 = Cdf.MakeCdfFromList(netflix_v4_cache['delta_connect_time'])
tcp_conn_cache_v6 = Cdf.MakeCdfFromList(netflix_v6_cache['delta_connect_time'])
tcp_conn_cache_both = Cdf.MakeCdfFromList(netflix_both_caches['delta_connect_time'])
tcp_conn_cache_neither = Cdf.MakeCdfFromList(netflix_neither_caches['delta_connect_time'])
# +
cdf_fig, cdf_ax = plt.subplots(figsize = (7, 2))
cdf_ax.plot(tcp_conn_cache_v4.xs, tcp_conn_cache_v4.ps, marker = '^', linewidth = 0.3, markersize = 5, fillstyle = 'none', color = 'blue', markevery = 25)
cdf_ax.plot(tcp_conn_cache_v6.xs, tcp_conn_cache_v6.ps, marker = 'v', linewidth = 0.3, markersize = 5, fillstyle = 'none', color = 'red', markevery = 25)
cdf_ax.plot(tcp_conn_cache_both.xs, tcp_conn_cache_both.ps, marker = 'd', linewidth = 0.3, markersize = 5, fillstyle = 'none', color = 'purple', markevery = 25)
cdf_ax.plot(tcp_conn_cache_neither.xs, tcp_conn_cache_neither.ps, marker = 'o', linewidth = 0.3, markersize = 5, fillstyle = 'none', color = 'green', markevery = 25)
cdf_ax.grid(False)
yticks = np.arange(0,1.1,0.2)
cdf_ax.set_yticks(yticks)
cdf_ax.set_ylim([-0.05,1.05])
cdf_ax.set_xscale('symlog')
cdf_ax.set_xlabel('Delta (ms)')
cdf_ax.set_ylabel('CDF')
cdf_ax.legend(["IPv4 Only", "IPv6 Only", "Both", "Neither"], fontsize = 'small', loc = 'upper left')
cdf_ax.spines['right'].set_color('none')
cdf_ax.spines['top'].set_color('none')
cdf_ax.yaxis.set_ticks_position('left')
cdf_ax.xaxis.set_ticks_position('bottom')
cdf_ax.spines['bottom'].set_position(('axes', -0.03))
cdf_ax.spines['left'].set_position(('axes', -0.03))
cdf_ax.axvline(x = 0, linewidth = 1.0, ymax = 0.95, ymin = 0, color='black', ls = 'dotted')
cdf_ax.annotate('', xy = (0.4, 1.1), xycoords = 'axes fraction', xytext = (0.1, 1.1),
arrowprops = dict(arrowstyle = '<-'))
cdf_ax.annotate('', xy = (0.9, 1.1), xycoords = 'axes fraction', xytext = (0.6, 1.1),
arrowprops = dict(arrowstyle = '->'))
cdf_ax.text(-450, 1.2, 'IPv6 slower')
cdf_ax.text(7.5, 1.2, 'IPv6 faster')
cdf_ax.set_title('TCP Connect Time', y = 1.30)
cdf_fig.savefig('../plots/tcp-conn-deltas-cache-pairs.pdf', bbox_inches = 'tight')
plt.show()
# -
print('------ TCP CONN V4 CACHE ------')
print('m_delta; cdf')
print('-------------------------------')
for x, y in list(zip(tcp_conn_cache_v4.xs, tcp_conn_cache_v4.ps))[0::100]:
print('%.2f; %.5f' % (x, y))
print('------ TCP CONN V6 CACHE ------')
print('m_delta; cdf')
print('-------------------------------')
for x, y in list(zip(tcp_conn_cache_v6.xs, tcp_conn_cache_v6.ps))[0::50]:
print('%.2f; %.5f' % (x, y))
print('------ TCP CONN CACHE BOTH ------')
print('m_delta; cdf')
print('---------------------------------')
for x, y in list(zip(tcp_conn_cache_both.xs, tcp_conn_cache_both.ps))[0::150]:
print('%.2f; %.5f' % (x, y))
print('------ TCP CONN CACHE NEITHER ------')
print('m_delta; cdf')
print('------------------------------------')
for x, y in list(zip(tcp_conn_cache_neither.xs, tcp_conn_cache_neither.ps))[0::500]:
print('%.2f; %.5f' % (x, y))
| notebooks/fig-13-cdn-vs-cache-four-split-different-AF.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: 'Python 3.9.5 64-bit (''portfolioConstr'': conda)'
# name: python395jvsc74a57bd043b94212bc69ae27b3b77c14012133cb38ae29affcd91d98ac2dfc98bb0b5c6c
# ---
# # Measuring Max Drawdown
# ## Motivation: Another measure of risk other than volatility
#
# - *Perspective*: Volatility is just deviation from the mean. Risk is the possibility of losing money.
# - *Maximum Drawdown*: One another way of looking at risk.
#
# > **Maximum Drawdown** : Maximum loss that you could have experienced if you bought the asset at its peak and sold it at the bottom
# - Worst return of peak to trough that you could have experienced over the given time series
#
# ### How to take a return series and convert to a Drawdown
# 1. Take return series and convert to a wealth index.
# > **Wealth Index**: What would have happened if you'd bought an asset and kept it over time.
#
# > **Drawdown**: Decrease from peak to the trough at any given point in time.
# 2. Compute the previous peaks.
# - At any point in time, keep track of what is the highest value experienced since inception.
# - When wealth rises, so does the peak. But when wealth falls, peak remains the same.
# 3. Compute drawdowns.
# - Drawdown: distance between previous peak and current position.
# - Distance is how much you feel you've lost.
# - In line with known behavioural effects/ biases.
# - Calculated as wealth value as a percentage of previous peak.
# - We can plot drawdowns to see how long it takes to recover from drawdowns.
#
# > **Calmar Ratio**: Ratio of annualized return over the trailing 36 months to the _maximum drawdown_ over those trailing 36 months.
#
# ### Problems with drawdowns
# 1. Defined by two points, and so sensitive to outliers
# 2. Depend on frequency of observations => Daily/weekly drawdown that's very deep might almost completely disappear based on monthly data.
# 3. Other measures of extreme risk exist that are more robust. (e.g. VAR, CVAR)
#
#
#
def compound_int (principal, rate, n, t):
return principal * (1+(rate/n))**(n*t)
# ## Computing Drawdowns
# +
import pandas as pd
import numpy as np
me_m = pd.read_csv(".\data\Portfolios_Formed_on_ME_monthly_EW.csv", header=0, index_col=0, parse_dates=True, na_values=-99.99)
rets = me_m[['Lo 10', 'Hi 10']]
rets.columns = ['SmallCap', 'LargeCap']
rets = rets/100
rets.head()
rets.plot.line()
# -
# Changing the index to time-series
rets.index = pd.to_datetime(rets.index, format="%Y%m")
rets.index
rets.head()
# Index date is first of a month. The data pertains to the entire _period_. Can change it thus:
rets.index = rets.index.to_period("M")
rets.head()
# Neat. Filtering is even neater.
rets["1982"]
# Even better! Can filter rows by years or months now.
rets["1982-01": "1983-06"]
nov_mask = rets.index.map(lambda x: x.month) == 11
rets[nov_mask]
# Nifty! Back to our scheduled programming now.
#
# ### Computing Drawdowns.
# 1. Compute wealth index.
# 2. Compute previous peaks.
# 3. Compute drawdowns: wealth value as a percentage of previous peak.
#
# #### 1. Compute Wealth Index.
wealth_index = 1000*(1+rets["LargeCap"]).cumprod()
wealth_index.head()
wealth_index.plot.line()
# #### 2. Compute previous peaks.
previous_peaks = wealth_index.cummax()
previous_peaks.plot.line()
# #### 3. Compute drawdowns.
drawdown = (wealth_index - previous_peaks)/previous_peaks
drawdown.plot()
# drawdown.tail()
# - The decimation in 1929 was 80% - the largest ever.
# - Will be interesting to see the figures in 2020. (This data goes only till 2018)
# - You can use min() to find out the precise figure in 1929.
#
drawdown.min()
# Or slice and then min. The following gives the minimum drawdown since 1975.
drawdown["1975":].min()
# Or return the index of the min(), i.e., _when_ did the minimum drawdown occur.
print(drawdown.idxmin())
print(drawdown["1950":].idxmin())
# Ah! The least drawdown was in _May 1932_, and not 1929. Also, the least drawdown since 1950 was in Feb 2009.
#
# We can generalize this as a function.
def drawdown(return_series: pd.Series):
"""
Takes a time-series of asset returns.
Computes and returns a DataFrame that contains:
1. The wealth index
2. The previous peaks
3. Percentage drawdowns
"""
wealth_index = 1000*(1+return_series).cumprod()
previous_peaks = wealth_index.cummax()
drawdowns = (wealth_index - previous_peaks)/previous_peaks
return pd.DataFrame({
"Wealth": wealth_index,
"Peaks": previous_peaks,
"Drawdown": drawdowns
})
drawdown(rets["LargeCap"]).head()
# This works! Let's do a graph.
drawdown(rets["LargeCap"])[["Wealth", "Peaks"]].plot()
print((9300*13+29800+1000)*1.05)
drawdown(rets[:"1950"]["LargeCap"])[["Wealth", "Peaks"]].plot()
drawdown(rets["LargeCap"])["Drawdown"].min()
drawdown(rets["SmallCap"])["Drawdown"].min()
# Drawdowns in large and small caps are roughly the same amount. When did they happen? Let's use idxmin()
drawdown(rets["SmallCap"])["Drawdown"].idxmin()
drawdown(rets["LargeCap"])["Drawdown"].idxmin()
# Yup, May 1932, when the Great Crash happened.
#
# What about the period after that? What is the min and when did it happen?
print("-----Small Caps-----")
print("Least drawdown since 1940: {minDrawDown:.2%}".format(minDrawDown = drawdown(rets["1940":]["SmallCap"])["Drawdown"].min()))
print("The year-month this happened : {minDrawDown:}".format(minDrawDown = drawdown(rets["1940":]["SmallCap"])["Drawdown"].idxmin()))
print("Least drawdown since 1940: {minDrawDown:.2%}".format(minDrawDown = drawdown(rets["1975":]["SmallCap"])["Drawdown"].min()))
print("The year-month this happened : {minDrawDown:}".format(minDrawDown = drawdown(rets["1975":]["SmallCap"])["Drawdown"].idxmin()))
print("-----Large Caps-----")
print("Least drawdown since 1940: {minDrawDown:.2%}".format(minDrawDown = drawdown(rets["1940":]["LargeCap"])["Drawdown"].min()))
print("The year-month this happened : {minDrawDown:}".format(minDrawDown = drawdown(rets["1940":]["LargeCap"])["Drawdown"].idxmin()))
print("Least drawdown since 1940: {minDrawDown:.2%}".format(minDrawDown = drawdown(rets["1975":]["LargeCap"])["Drawdown"].min()))
print("The year-month this happened : {minDrawDown:}".format(minDrawDown = drawdown(rets["1975":]["LargeCap"])["Drawdown"].idxmin()))
# Abstracting this out as functions.
def min_drawdown_amt(rets: pd.Series, cap: str, start_year: str):
return drawdown(rets[start_year:][cap])["Drawdown"].min()
def min_drawdown_year(rets: pd.Series, cap: str, start_year: str):
return drawdown(rets[start_year:][cap])["Drawdown"].idxmin()
# +
start_year = "1940"
print("-----Small Caps-----")
caps = "SmallCap"
print("Least drawdown since 1940: {minDrawDown:.2%}".format(minDrawDown = min_drawdown_amt(rets,caps, start_year)))
print("Least drawdown since 1940: {minDrawDown:}".format(minDrawDown = min_drawdown_year(rets,caps, start_year)))
print("-----Large Caps-----")
caps = "LargeCap"
print("Least drawdown since 1940: {minDrawDown:.2%}".format(minDrawDown = min_drawdown_amt(rets,caps, start_year)))
print("Least drawdown since 1940: {minDrawDown:}".format(minDrawDown = min_drawdown_year(rets,caps, start_year)))
# print(min_drawdown_year(rets,caps,start_year))
# -
| Week 1- Measuring Max Drawdown.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Generate and visualize toy data sets
# +
import zfit
import numpy as np
from scipy.stats import norm, expon
from matplotlib import pyplot as plt
zfit.settings.set_seed(10) # fix seed
bounds = (0, 10)
obs = zfit.Space('x', limits=bounds)
# true parameters for signal and background
truth_n_sig = 1000
Nsig = zfit.Parameter("Nsig", truth_n_sig)
mean_sig = zfit.Parameter("mean_sig", 5.0)
sigma_sig = zfit.Parameter("sigma_sig", 0.5)
sig_pdf = zfit.pdf.Gauss(obs=obs, mu=mean_sig, sigma=sigma_sig).create_extended(Nsig)
truth_n_bkg = 10000
Nbkg = zfit.Parameter("Nbkg", truth_n_bkg)
lambda_bkg = zfit.Parameter("lambda_bkg", -1/4.0)
bkg_pdf = zfit.pdf.Exponential(obs=obs, lambda_=lambda_bkg).create_extended(Nbkg)
truth_sig_t = (1.0,)
truth_bkg_t = (2.5, 2.0)
# make a data set
m_sig = sig_pdf.sample(truth_n_sig).numpy()
m_bkg = bkg_pdf.sample(truth_n_bkg).numpy()
m = np.concatenate([m_sig, m_bkg]).flatten()
# fill t variables
t_sig = expon(0, *truth_sig_t).rvs(truth_n_sig)
t_bkg = norm(*truth_bkg_t).rvs(truth_n_bkg)
t = np.concatenate([t_sig, t_bkg])
# cut out range (0, 10) in m, t
ma = (bounds[0] < t) & (t < bounds[1])
m = m[ma]
t = t[ma]
fig, ax = plt.subplots(1, 3, figsize=(16, 4.5))
ax[0].hist2d(m, t, bins=(50, 50))
ax[0].set_xlabel("m")
ax[0].set_ylabel("t")
ax[1].hist([m_bkg, m_sig], bins=50, stacked=True, label=("background", "signal"))
ax[1].set_xlabel("m")
ax[1].legend()
ax[2].hist((t[truth_n_sig:], t[:truth_n_sig]), bins=50, stacked=True, label=("background", "signal"))
ax[2].set_xlabel("t")
ax[2].legend();
sorter = np.argsort(m)
m = m[sorter]
t = t[sorter]
# -
# # Fit toy data set
# +
from zfit.loss import ExtendedUnbinnedNLL
from zfit.minimize import Minuit
tot_pdf = zfit.pdf.SumPDF([sig_pdf, bkg_pdf])
loss = ExtendedUnbinnedNLL(model=tot_pdf, data=zfit.data.Data.from_numpy(obs=obs, array=m))
minimizer = Minuit()
minimum = minimizer.minimize(loss=loss)
minimum.hesse()
print(minimum)
# -
# ## Visualize fitted model
# +
from utils import pltdist, plotfitresult
fig = plt.figure(figsize=(8, 5.5))
nbins = 80
pltdist(m, nbins, bounds)
plotfitresult(tot_pdf, bounds, nbins, label="total model", color="crimson")
plotfitresult(bkg_pdf, bounds, nbins, label="background", color="forestgreen")
plotfitresult(sig_pdf, bounds, nbins, label="signal", color="orange")
plt.xlabel("m")
plt.ylabel("number of events")
plt.legend();
# -
# ## Compute sWeights
# +
from hepstats.splot import compute_sweights
weights = compute_sweights(tot_pdf, m)
print("Sum of signal sWeights: ", np.sum(weights[Nsig]))
# +
fig, ax = plt.subplots(1, 2, figsize=(16, 4.5))
plt.sca(ax[0])
nbins = 40
plt.plot(m, weights[Nsig], label="$w_\\mathrm{sig}$")
plt.plot(m, weights[Nbkg], label="$w_\\mathrm{bkg}$")
plt.plot(m, weights[Nsig] + weights[Nbkg], "-k")
plt.axhline(0, color="0.5")
plt.legend()
plt.sca(ax[1])
plt.hist(t, bins=nbins, range=bounds, weights=weights[Nsig], label="weighted histogram")
plt.hist(t_sig, bins=nbins, range=bounds, histtype="step", label="true histogram")
t1 = np.linspace(*bounds, nbins)
tcdf = expon(0, 1).pdf(t1) * np.sum(weights[Nsig]) * (bounds[1] - bounds[0])/nbins
plt.plot(t1, tcdf, label="model with $\lambda_\\mathrm{sig}$")
plt.xlabel("t")
plt.legend();
# -
np.average(t, weights=weights[Nsig])
np.average(t_sig)
| notebooks/splots/splot_example_2.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="VYfg3I_fSt6e"
# # WebScraping with Selenium
#
# This notebook shows how use selenium to scrape data from an Pagine Gialle web site.
# The scope is only to understand the capabilities of web scraping and prepare a dataset for academic purporse.
#
# Let’s begin writing our scraper!
#
# We will first install important modules and packages for our Notebook
#
#
# * Selenium
# * Chromium-chromedriver
#
#
# + id="6E11q1711nmO" colab={"base_uri": "https://localhost:8080/"} outputId="0f98d052-995f-4374-87b2-47c37d6fcef6"
# !pip install selenium
# !apt-get update
# !apt install chromium-chromedriver
# !cp /usr/lib/chromium-browser/chromedriver /usr/bin
# + [markdown] id="kuhZcDh4UHYp"
# And now we will import some modules on our Notebook
#
#
# 1. sys: to setup the path of chrome driver
# 2. selenium: to emulate the user behaviour
# 3. pandas: to work with data
# 4. tqdm: to show a progress bar in our notebook
# 5. json: to work with json format
#
#
#
#
# + id="YRt5yEXxA-jW"
import sys
sys.path.insert(0,'/usr/lib/chromium-browser/chromedriver')
from selenium import webdriver
from tqdm import tqdm_notebook as tqdm
import pandas
import json
import pprint
# + [markdown] id="ksyefP35VQc2"
# First we need to set the options for the ghost browser.
# The most important is `--headless` because we are in a "cloud" notebook. In our local notebook we can remove the `--headless` option.
#
# + id="DTfvdy_gBCBU"
chrome_options = webdriver.ChromeOptions()
chrome_options.add_argument('--headless')
chrome_options.add_argument('--no-sandbox')
chrome_options.add_argument('--disable-dev-shm-usage')
chrome_options.add_argument("window-size=1900,800")
chrome_options.add_argument("--enable-javascript")
chrome_options.add_argument("user-agent=Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/88.0.4324.150 Safari/537.36")
# + [markdown] id="0DpumOn7VoFw"
# `webdriver` is the most importa object in Selenium.
# With `webdriver` we can start the phantom browser, emulate the user navigation and scrape our data.
#
# Let’s now create a new instance of google chrome.
#
#
# We will navitage to page by a `get` request. With http we can do a get or a post (or others but they are not important for us): https://www.w3schools.com/tags/ref_httpmethods.asp
#
#
#
# ```
# wd.get("https://www.paginegialle.it/ricerca/ceramica")
# ```
#
#
# + id="D6MLvqs9BIzI" colab={"base_uri": "https://localhost:8080/"} outputId="f15d9d9b-1d88-4ec0-f256-579d27a4a95c"
wd = webdriver.Chrome('chromedriver',chrome_options=chrome_options)
wd.get("https://www.paginegialle.it/ricerca/ceramica")
# + [markdown] id="p3Sbfud-WbV-"
# Take a look to the screenshot from our phantom browser.
#
# Note:
# - la language and the locale: It is en_US!
# - the screen ration: is it the same of our notebook?
# + id="iBCZSC4dWfKW" colab={"base_uri": "https://localhost:8080/", "height": 210} outputId="075a9025-df2b-45f2-9c55-2acea95ba3dd"
wd.save_screenshot('screenshot.png')
# %pylab inline
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
img=mpimg.imread('/content/screenshot.png')
imgplot = plt.imshow(img)
plt.show()
# + [markdown] id="QV7mMMKn8kbx"
# Click on cookie accept button
# + id="0yURJOQW8kDU"
wd.find_elements_by_css_selector("button.iubenda-cs-customize-btn")[0].click()
# + [markdown] id="kRr2IAQsa1s_"
# Let's try to extract some information from the page
#
#
#
# `title` contains the title of our page.
#
#
#
# + id="X033nUzra6uq" colab={"base_uri": "https://localhost:8080/"} outputId="1280b9be-c049-42e8-c6a9-173f83d035e4"
print(wd.title)
# + [markdown] id="zK7Cf_mradiL"
# ### How do we extract the values inside a page with CSS Selector?
#
# Selenium has a method called `find_elements_by_css_selector`.
#
# We will pass our CSS Selector into this method and get a list of selenium elements. Once we have the element, we can extract the text inside it using the `text` function.
# + id="WFuRVKIdbWN3" colab={"base_uri": "https://localhost:8080/"} outputId="d383e7fd-6d50-47eb-a61f-23848b107cf6"
list_companies = wd.find_elements_by_css_selector("h2.itemTitle")
print(len(list_companies))
# + id="cRPP5xUrbwcE" colab={"base_uri": "https://localhost:8080/"} outputId="4ab06b6c-9f47-4c67-e5f9-815b5ec3d886"
print(list_companies[0].text)
# + [markdown] id="A1ETBqlPcQhl"
# # Extract the companies list
#
# Let's begin to download the list of companies.
#
# The list of companies is defined by the css selection rule
# `section.vcard`
#
#
# + id="_V1hNO9ZcJy6" colab={"base_uri": "https://localhost:8080/"} outputId="0e583a25-aeda-40fb-bc39-9bb48eacbd0d"
list_companies = wd.find_elements_by_css_selector("section.vcard")
print(len(list_companies))
# + [markdown] id="k_fSFWX0cktI"
# Gread! We find 10 items!
#
# Now, we will extract the attribute from each single element.
# The scope is to create one dict for each item in the list with the attribute:
#
#
# 1. company name
# 2. link to the detail page
# 3. number of employees
# 4. vat code
# 5. economic sector
# 6. tags
#
#
# + id="1J2xAL6qcj9-" colab={"base_uri": "https://localhost:8080/"} outputId="1ea1d105-2160-4b7e-e7b8-1db3f99d4291"
import pprint
import time
items = []
for item in list_companies:
company_name = item.find_elements_by_css_selector("h2.itemTitle")[0].text
url = item.find_elements_by_css_selector("a.icn-vetrina")[0].get_attribute("href")
items.append({'company_name': company_name,
'url': url})
pprint.pprint(items[0:5])
# + [markdown] id="NgvnPlxqBpP4"
# Get the list of the first 100 companies
# + colab={"base_uri": "https://localhost:8080/"} id="OkoyTU9TI3QW" outputId="2fb8df21-bc08-4fcc-d092-8d7200839469"
import math
wd.get("https://www.paginegialle.it/ricerca/ceramica/Italia/p-1")
total_items = wd.find_elements_by_css_selector('span.searchResNum span')[0].text
page_total = math.ceil(int(total_items) / 20)
print(page_total)
# + id="TXEFPcS0_1du"
items = []
for page in range(1, 16):
wd.get(f"https://www.paginegialle.it/ricerca/ceramica/Italia/p-{page}")
time.sleep(1)
list_companies = wd.find_elements_by_css_selector("section.vcard")
for item in list_companies:
company_name = item.find_elements_by_css_selector("h2.itemTitle")[0].text
url = item.find_elements_by_css_selector("a.icn-vetrina")[0].get_attribute("href")
items.append({'company_name': company_name,
'url': url})
# + [markdown] id="w6o9zLQBBwRP"
# Check the list returned by our scraper:
# + id="d0-ystf3VAY0" colab={"base_uri": "https://localhost:8080/"} outputId="97f33a25-2e86-4a23-ef4e-c4e144ed7400"
len(items)
# + colab={"base_uri": "https://localhost:8080/"} id="pduDvl8BA0e1" outputId="b7bd87e1-ef42-4192-c48d-b1cd3d6ff86e"
pprint.pprint(items[90:95])
# + [markdown] id="0oakp01EBza0"
# Navigate to each detail page and get the details
# + colab={"base_uri": "https://localhost:8080/", "height": 116, "referenced_widgets": ["007167e7bb33479aa4d9f56bb356157f", "d70fe51185e4491ea62ccf92b730b441", "<KEY>", "f11ba03dea854fa4ad8f8ba336c4e02f", "da31e6b42234430d8752643d2522f1ae", "b5010fa144c24452bb5a700547d28cc0", "564bb5cb57124693a094e6a4a0f2aa5e", "f5dca2a7c0ec43a7a0d87d50f664dd4f"]} id="552PDNctBDWr" outputId="ba72cf76-1c86-45f6-ae7b-59c7df83d077"
details = []
for item in tqdm(items):
wd.get(item['url'])
# print(item['url'])
time.sleep(1)
description = wd.find_elements_by_css_selector("#descrizioneAzienda")[0].text
abstract = ""
try:
abstract = wd.find_elements_by_css_selector("h2.dsabstract")[0].text
except:
abstract = ""
wd.find_elements_by_css_selector('.btn-show-phone')[0].click()
phone = wd.find_elements_by_css_selector('span.tel')[0].text
url = ""
try:
url = wd.find_elements_by_css_selector("a.icn-sitoWeb")[0].get_attribute("href")
except:
url = item['url']
address = wd.find_elements_by_css_selector('div.street-address')[0].text
altre_informazioni = wd.find_elements_by_css_selector("section.altre-info-azienda div")
vat_code = ""
for box in altre_informazioni:
if 'Partita IVA' in box.text:
vat_code = box.find_elements_by_css_selector("span")[0].text
company_name = item['company_name']
details.append({'company_name': company_name,
'address': address,
'phone': phone,
'description': description,
'vat_code': vat_code,
'abstract': abstract,
'url': url})
# + colab={"base_uri": "https://localhost:8080/"} id="ZYKijANaDZX_" outputId="3fda5d22-f2c9-4801-f6ff-de4639756832"
pprint.pprint(details[0:5])
# + [markdown] id="TqxsicNNovfy"
# # Pandas and data processing
#
# **Well!**
# We're starting to see another amazing library for working with data!
#
# It's **pandas**: *Python library for data analysis*.
#
# Basically, with pandas we can manipulate a data set or a historical series in Python.
#
# For now, we start to give a couple of concepts.
#
# Pandas is based on two types of data: **Series** and **DataFrame**:
# - `Series` represents a list of data
# - `DataFrame` represent a data set in tabular format
#
# Each column of a `DataFrame` is a `Series`.
#
# We can create a `DataFrame` using the `pd.DataFrame` method by passing our dictionary as an input parameter.
#
# A `DataFrame` pandas can be easily exported in *CSV*, *Excel*, ...
#
# For more information about `Pandas` see
#
# https://pandas.pydata.org/getting_started.html
#
# + id="MumFgyNypoBo" colab={"base_uri": "https://localhost:8080/", "height": 363} outputId="f8a14ab3-d1dd-41b0-9fa6-763b89af2861"
import pandas as pd
ds_items = pd.DataFrame(details)
ds_items.set_index("vat_code")
ds_items.head()
# + id="Z--9BRv6Jo0W"
ds_items.info()
# + id="3lUw4-_BrQDh"
ds_items.to_csv('ds_items.csv')
| 1_Pagine_gialle.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
from nltk.tokenize import word_tokenize
# # Tokenization
# * Tokens are the building blocks of Natural Language.
# * Tokenization is a way of separating a piece of text into smaller units called tokens.
# * Here, tokens can be either words, characters, or subwords. Hence,
# * Tokenization can be broadly classified into 3 types –
# * Word,
# * Character, or Sentence
# * Subword (n-gram characters) tokenization.
# ## Word Tokenization
# 
Text = "The Cate Sat on the mat."
word_tokenize(Text)
File = "E:\NLP\NLP_v01-class2\exercise\Data\cnus.txt"
text = ''
with open('Data/cnus.txt','r') as f:
text = " ".join([l.strip() for l in f.readlines()])
text
li = word_tokenize(text) # Out is in the form list
li.count('THE')
# ## Tokenization: Sentences
from nltk.tokenize import sent_tokenize
S_LI = sent_tokenize(Text)
S_LI
s_li = sent_tokenize(text)
s_li
my_text = "Hi Mr. Smith! I’m going to buy some vegetables (tomatoes and cucumbers) from the store. Should I pick up some black-eyed peas as well?"
sent_tokenize(my_text)
# ## Tokenization (N-Grams) or SubWord
#
from nltk.util import ngrams
my_words = word_tokenize(my_text) # This is will give you the list of all words
twograms = list(ngrams(my_words,2)) # This Will do for two-word combos, but can pick any n
twograms
threegrams = list(ngrams(my_words,3)) # This Will do for three-word combos, but can pick any n
threegrams
# ## Tokenization (Regular Expressions)
#
# Let’s say you want to tokenize by some other type of grouping or pattern.
#
# * Regular expressions (regex) allows you to do so.
#
# * Some examples of regular expressions:
# * Find white spaces: \s+
# * Find words starting with capital letters: [A-Z]['\w]+
#
from nltk.tokenize import RegexpTokenizer
# i want to match all Capitalized word
cap_tokenize = RegexpTokenizer("[A-Z]['\w]+")
cap_tokenize.tokenize(my_text)
cap_tokenize_atoz = RegexpTokenizer("[a-z]['\w]+")
cap_tokenize_atoz.tokenize(my_text)
| Tokenization/Tokenization.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # "Non-Intrusive Load Monitoring - NILM"
# > "A source separation problem that can enable a better, smarter electric grid"
#
# - toc:true
# - branch: master
# - badges: false
# - comments: true
# - author: <NAME>
# - categories: [projects]
# ## Context
#
# I spent just over three years at the research arm of Tata Consultancy Services (then called the Innovation Labs) from 2011 to 2014 (before I took a sabbatical for my doctoral studies and eventually moved out). The experience of working on futuristic technology problems in an industrial environment was interesting. I had the opportunity to witness an idea grow from its budding stage to eventual proof of concept and further adoption.
#
# One research problem that occupied most of my time during this stint was *Source Separation*. The idea underlying the problem stems from the need to analyse the multiple sources that give rise to a multitude of effects on a single (or a small number of) measurements. One application problem that came out of it (which was in its infancy during that time, and is at various stages of implementation today): [Non-intrusive load monitoring (NILM)](https://en.wikipedia.org/wiki/Nonintrusive_load_monitoring).
#
# In this article, I will discuss my experiences working on this problem.
# ## An intro
#
# A couple of years or so back, EDF, the French electric giant, with whom I have a contract for my house's electricity, updated their app/interface with a possible option to follow our daily consumption profile. This followed the installation of the smart electric meters, known as `Linky`, in our apartment. While this did not provide me with insights that I did not already have, there was something more on offer. The application also allowed me to dive further into the categories of my consumption based on different groupings:
# 1. Refrigeration
# 1. Water heating
# 1. Electric heater
# 1. Cooking
# 1. Infotainment and Computers.
#
# The recent screen capture of this looks something like this (I redacted the amount and my address):
#
# <img src="./../images/nilm/nilm_edf.jpg" alt="NILM - EDF" width="300"/>
#
# The refrigerator consumed 9% of the total electricity consumption. And one can get a vague idea already about how much the other categories consumed. Given the furore over data privacy, I had to *unlock* these features by deliberately giving EDF the rights handle my data.
#
# By now, one would have got the idea of what NILM means:
# > Note: To disaggregate or distinguish between different electrical appliances and their power consumption using a single power meter data.
# Or, one could view NILM algorithms acting like a prism
#
# <img src="./../images/nilm/nilm_prism.png" alt="NILM - Prism" width="500"/>
#
# When I first started to work on the problem of NILM, the academic papers were all interested in a sampling rate of once in 1 second. And right in line, the open datasets were also developed to the same tune. For example, the [REDD](http://redd.csail.mit.edu/) dataset from (the then) MIT team with [<NAME>](http://zicokolter.com/) at the helm provided data in that sampling rate range. Subsequent open data sets followed suit with similar ones. And some, like the popular dataset from Ubicomp Lab at the University of Washington on [Kaggle](https://www.kaggle.com/c/belkin-energy-disaggregation-competition) (that featured in the Belkin competition), had an even higher frequency of operation.
#
# However, the *Linky* smart meters installed in my apartment collects data once in 15 minutes and maps more closely to my ventures just before I started my PhD. In this work, we explored the use of AMI type data for NILM. AMI stands for Advanced Metering Infrastructure, the type of metering companies were hoping to install in households (sampling the cumulated power consumption every 15 minutes or so). This is in contrast with the high-frequency data over which most of the academic research work were based on. While challenging, that seemed unrealistic and so we wrote this paper summarizing our then-ongoing efforts.
#
# [Springer Link behind paywall](https://link.springer.com/chapter/10.1007/978-3-319-04960-1_8) or perhaps more useful would be the [link to Pre-print](https://github.com/krishnans14/feedback-control/tree/master/files/sirs_14_final.pdf)
#
#
# ## Client Project
#
# In our initial work, we focused on using the open datasets (for lack of data from our side). Things changed due to a pilot project to implement NILM in the Netherlands for a startup client. The unfortunate thing was we started without any data to work with and limited assumptions. The IT team built a data handling infrastructure awaiting installation of sensors, but developing a machine learning algorithm without data was a cruel joke (we weren't even sure on what would be the sampling rate of data because our client was still discussing with their potential clients about it).
#
# So we decided to do what today is terms as [*Transfer Learning*](https://en.wikipedia.org/wiki/Transfer_learning) which (as per Wikipedia)
# > focuses on storing knowledge gained while solving one problem and applying it to a different but related problem.
#
# without actually knowing what the term *transfer learning*. We collected some statistics/pattern about the characteristics of various appliances from the open datasets available. Then we created a pseudo database for different appliances that could then be used for training when the data arrived.
#
# An extra problem that plagued the initial efforts were in obtaining appliance level signatures. The pilot was in households where the wiring of the appliances was well integrated into the walls and it was difficult to put plugs to tap them. This led to an extra aspect of *gamification* introduced to label data. The process was as follows:
#
#
# * Using the transfer learning-based database, our NILM algorithm will provide detection of appliances.
# * The inhabitants of the households will get notifications at the end of the day on these detections through a mobile App.
# * The user labels the detection (correct or wrong) based on their own knowledge.
# * The NILM algorithm trains a model specific to each household based on this labelling.
#
# Several aspects of the above process were in flux. For instance, we were exploring different algorithms that can perform NILM (note that back in 2013, this area was fresh and had limited success) or the *gamification* aspects were not clear (how much to trust the feedback, etc.). But with all the limitations, a pilot went forward and looked good. But everything also came to an abrupt end due to financial constraints at the startup.
#
# During the same period, several other startups, notably in the US were working on the NILM problem. Some of them are still active: [Bidgely](https://www.bidgely.com/), [Opower](https://en.wikipedia.org/wiki/Opower), and more (check this [2012 entry on Oliver Parson's blog](https://blog.oliverparson.co.uk/2012/05/nialm-in-industry.html) on companies working on NILM). Several other companies came in and disappeared as it always happens. I worked with one of them.
#
# ## NILM Techniques Explored
#
# It is not surprising that we tried several techniques to realize NILM. Further, unlike academic freedom, we had to work with a limited set of assumptions and hence the need to use customized techniques (which were of course not published). But here are some techniques that were published:
# * Bayesian Inference [IEEE link behind paywall](https://ieeexplore.ieee.org/abstract/document/6603710) or [Preprint](https://github.com/krishnans14/feedback-control/tree/master/files/esiot13_final.pdf)
# * Factor Graphs [IEEE link behind paywall](https://ieeexplore.ieee.org/abstract/document/6637447/) or [Preprint](https://github.com/krishnans14/feedback-control/tree/master/files/icacci13_final.pdf)
#
# Apart from the different techniques, we also presented a paper on the approach to use a mix of transfer learning and simulation to generate labelled data over which NILM algorithms could be tested: [ACM link behind paywall](https://dl.acm.org/doi/abs/10.1145/2559627.2559630) or [Preprint](https://github.com/krishnans14/feedback-control/tree/master/files/es4cps_final.pdf)
#
# I will discuss these techniques and those we explored in more details sometime in the future.
# ## Beyond household disaggregation
#
# Our explorations for the application of NILM went beyond the household energy disaggregation pilot project with the Dutch startup. The following were the other problems that were at explored:
# * Cost-savings for a large building
# * A large office building of an enterprise also contained a cafeteria serving hundreds of diners. The power to these were supplied from a single transformer. This means that the electricity tarrif paid by the enterprise was corresponding to a commercial establishment (the cafeteria) and not the workplace. The latter was much cheaper. We gave a proposal of how one could attempt to use NILM and obtain an estimation of the two entities and save cost in the electricity bill paid.
# * Condition monitoring of appliances or industrial equipments
# * Today, the central aspect of Industry 4.0 is the condition monitoring of equipment to perform predictive maintenance, so much so that even [Amazon is into it](https://krishnans14.github.io/feedback-control/musings/industry-watch/2020/12/17/Monitron-and-Predictive-Maintenance.html). A couple of proposals were floated in that direction back in 2013-14.
# * Disaggregation of load versus generation (with rooftop solar installation)
# * When the rooftop solar installations became popular, different countries took different approaches to their integration with the grid. In some countries, there were no restrictions on how an individual household decides to integrate solar panels with their own usage or to connect back to the grid. We floated ideas on how to use NILM on the smart meter data to estimate generation capacity in a household.
# * A related application was whether we can use a single smart meter to identify defects in a host of solar PV panels (say on rooftops or a farm).
# * Activity monitoring
# * A more contentious application of NILM was on activity monitoring. A [patent](https://patents.google.com/patent/WO2015124972A1/en) was filed in this application towards the fag end of my stay in the lab.
#
# These are the applications that I remember from the top of my head.
# ## A few words before the end
#
# Recently, I bumped into the [NILM workshop for 2020](http://nilmworkshop.org/2020/) organized online and came across their papers and the YouTube live videos.
#
# I would try to spend some more time into these papers and posters and synthesize some thoughts for a future post where I would also like to discuss the [NILMTK python module](https://github.com/nilmtk/nilmtk), which I had been itching to try.
#
# It would be disingenious not to acknowledge the contributions of co-workers/supervisors in the above endeavours, though one can see the presence of [<NAME>](https://in.linkedin.com/in/mgirishchandra), [<NAME>](https://ca.linkedin.com/in/goutam-yelluru-gopal-93549428) prominently in all the publications.
| _notebooks/2020-12-30-NILM.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
#default_exp crawler
# -
#hide
from nbdev.showdoc import *
# # Crawler
# > Defines methods to crawl all web pages in a specific domain, extract contents from them and store them in a DataFrame
# +
#hide
#export
from search_engine.scrapper import parse_webpage
from urllib.parse import urljoin, urlparse
from collections import deque
import pandas as pd
import re
# -
#hide
#export
def link_filter(link, domain, base_url):
"""
Filters out links if they... \n
1. Are not from specified domain \n
2. Contain extensions - pdf|jpg|jpeg|doc|docx|ppt|pptx|png|txt|exe|ps|psb \n
3. Contain an `@` \n
4. Have already been visited
"""
is_valid = lambda url: not bool(re.search('pdf|jpg|jpeg|doc|docx|ppt|pptx|png|txt|exe|ps|psb|@',
url))
return link is not None and is_valid(link) and (link.startswith('/') or domain in link) \
and urljoin(base_url, link) != base_url
show_doc(link_filter)
#hide
#export
def link_modifier(url, base_url):
"""
Converts `relative` urls to absolute ones.
"""
url = urljoin(base_url, url)
if url[-1]=='/':
url= url[:-1]
if 'https' not in url:
url = url.replace("http", "https")
return url
show_doc(link_modifier)
#hide
#export
def crawl(domain='uic.edu',
url='https://cs.uic.edu',
num_pages=5):
"""
Starts crawling the specified url and linked urls in a breadth-first fashion,
extracts content and puts them in a DataFrame that will be returned
"""
# Queue links to crawl
crawl_q = deque([])
# Already crawled links
crawled_links = set([])
# Redundant crawl_q
crawl_q_set = set(crawl_q)
# Number of links crawled
crawl_count = 0
pages = pd.DataFrame(columns=['id', 'url', 'content', 'graph'])
crawl_q.append(url)
while len(crawl_q) > 0 and crawl_count < num_pages:
try:
crawl_q_set = set(crawl_q)
url = crawl_q.popleft()
if url in crawled_links:
continue
crawled_links.add(url)
content, links = parse_webpage(url)
# Remove invalid links
links = list(filter(lambda link: link_filter(link, domain, url), links))
# Modify relative urls to absolute
links = list(map(lambda link: link_modifier(link, url), links))
# Remove duplicates within the links
links = list(set(links))
pages = pages.append({'id': crawl_count, 'url': url, 'content': content, 'outgoing_links': links}, ignore_index=True)
print(f'Crawled {url}')
# Add links to crawl_q if they are not in crawled links or not already in crawl_q
crawl_q.extend(list(filter(lambda l: l not in crawl_q_set and l not in crawled_links, links)))
crawl_count += 1
except:
print('Error')
# Clear all lists and queues
crawl_q.clear()
crawled_links = set([])
crawl_q_set = set(crawl_q)
return pages
show_doc(crawl)
| nbs/03_crawler.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import pandas as pd
import numpy as np
from sklearn.feature_extraction.text import CountVectorizer,TfidfVectorizer
# %matplotlib inline
import matplotlib.pyplot as plt
# +
#train = pd.read_csv('data/set',header=None,error_bad_lines=False,encoding='gbk2312', chunksize=4)
# train = pd.read_csv('data/set',header=None,error_bad_lines=False,encoding='gbk')
# y = train[0]
# -
#set2是set经过排序后删除重复项
d = []
f =open('data/set2','r')
for line in f.readlines():
d.append(line.replace('\n','').split(','))
f.close()
#能力小于等于1的删除
data=[]
label=[]
for i in d:
if len(i)>2:
label.append(i[0])
data.append(i[1:])
# +
#保存为datanew即可以理解为x 标签为lable可以理解为y
file = open('data/datanew','a')
for i in range(len(data)):
s = str(data[i]).replace('[','').replace(']','')#去除[],这两行按数据不同,可以选择
s = s.replace("'",'').replace(',','') +'\n' #去除单引号,逗号,每行末尾追加换行符
file.write(s)
file.close()
file = open('data/lable','a')
for i in range(len(label)):
s = str(label[i]).replace('[','').replace(']','')#去除[],这两行按数据不同,可以选择
s = s.replace("'",'').replace(',','') +'\n' #去除单引号,逗号,每行末尾追加换行符
file.write(s)
file.close()
| src/Prediction/position/datawashposition.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="PkQNSCoX38Ba"
# ### **This notebook is used to generate segmentation results for input size of 64 X 64**
# + colab={"base_uri": "https://localhost:8080/"} id="W3QVwm5gIScE" outputId="fb2dcf80-3b70-4ce3-ce44-476e24c7d479"
from google.colab import drive
drive.mount('/content/drive')
# + id="UbvE2cHVF74G" colab={"base_uri": "https://localhost:8080/"} outputId="4089579a-a53c-4f62-980e-fb1265d7dfa9"
pip install nilearn
# + id="n1IQDSXYFmRa" colab={"base_uri": "https://localhost:8080/"} outputId="1e2b9a18-5148-4b5e-dda0-4008a6d0c6e4"
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import tensorflow as tf
from sklearn.model_selection import train_test_split
import keras
from keras.models import Model, load_model
from keras.layers import Input ,BatchNormalization , Activation
from keras.layers.convolutional import Conv2D, UpSampling2D
from keras.layers.pooling import MaxPooling2D
from keras.layers.merge import concatenate
from keras.callbacks import EarlyStopping, ModelCheckpoint
from keras import optimizers
from sklearn.model_selection import train_test_split
import os
import nibabel as nib
import cv2 as cv
import matplotlib.pyplot as plt
from keras import backend as K
import glob
import skimage.io as io
import skimage.color as color
import random as r
import math
from nilearn import plotting
import pickle
import skimage.transform as skTrans
from nilearn import image
from nilearn.image import resample_img
import nibabel.processing
import warnings
# + id="X8qqXfe6GBmn"
for dirname, _, filenames in os.walk('/content/drive/MyDrive/MRI Data'):
for filename in filenames:
print(os.path.join(dirname, filename))
# + id="tW44p9imgYik"
"""
rescale_Nii(nifti_file):
This function takes a .nii files as an input and rescales it according to the
values of voxel_dims by creating a new affine transform. The new affine transform
is used in resample_img() function from nibable library which will transform the images
accordinly inputs for resample_img() are tagert image, target affine transform and target shape
Input:
nifti_file: A .nii file which we want to rescale
Output:
rescaled .nii file with dimensions as defined in target_shape
"""
def rescale_Nii(nifti_file):
warnings.filterwarnings("ignore")
img=nifti_file
voxel_dims=[3.8, 3.8,1]
#voxel_dims=[1.60, 1.60,1]
# downl sample to 128*128*155
target_shape=(64,64,130)
#target_shape=(128,128,155)
# Initialize target_affine
target_affine = img.affine.copy()
# Calculate the translation part of the affine
spatial_dimensions = (img.header['dim'] * img.header['pixdim'])[1:4]
# Calculate the translation affine as a proportion of the real world
# spatial dimensions
image_center_as_prop = img.affine[0:3,3] / spatial_dimensions
# Calculate the equivalent center coordinates in the target image
dimensions_of_target_image = (np.array(voxel_dims) * np.array(target_shape))
target_center_coords = dimensions_of_target_image * image_center_as_prop
# Decompose the image affine to allow scaling
u,s,v = np.linalg.svd(target_affine[:3,:3],full_matrices=False)
# Rescale the image to the appropriate voxel dimensions
s = voxel_dims
# Reconstruct the affine
target_affine[:3,:3] = u @ np.diag(s) @ v
# Set the translation component of the affine computed from the input
target_affine[:3,3] = target_center_coords
#target_affine = rescale_affine(target_affine,voxel_dims,target_center_coords)
resampled_img = resample_img(img, target_affine=target_affine,target_shape=target_shape)
resampled_img.header.set_zooms((np.absolute(voxel_dims)))
return resampled_img
# + id="36R35K1oGtdg"
"""
Data_Concatenate(Input_Data):
Converts a numpy array with fused images ( eg: (20, 130, 128, 128, 2))
into an array with two partitions(eg: (2, 2600, 128, 128, 1)).
Input:
Input_Data= Array which contains information of all the modalities
Output:
Fused array which will used for making the training and testing split
"""
def Data_Concatenate(Input_Data):
counter=0
Output= []
for i in range(5):
print('$')
c=0
counter=0
for ii in range(len(Input_Data)):
if (counter < len(Input_Data)-1):
a= Input_Data[counter][:,:,:,i]
#print('a={}'.format(a.shape))
b= Input_Data[counter+1][:,:,:,i]
#print('b={}'.format(b.shape))
if (counter==0):
c= np.concatenate((a, b), axis=0)
print('c1={}'.format(c.shape))
counter= counter+2
else:
c1= np.concatenate((a, b), axis=0)
c= np.concatenate((c, c1), axis=0)
print('c2={}'.format(c.shape))
counter= counter+2
if (counter == len(Input_Data)-1):
a= Input_Data[counter][:,:,:,i]
c= np.concatenate((c, a), axis=0)
print('c2={}'.format(c.shape))
counter=counter+2
c= c[:,:,:,np.newaxis]
Output.append(c)
return Output
# + [markdown] id="0-X2CDif7jbb"
# ### **Data Preprocessing**
# + id="J1YiOfDWGQFc"
Path= '/content/drive/MyDrive/MRI Data/BraTS2020_TrainingData/MICCAI_BraTS2020_TrainingData'
p=os.listdir(Path)
Input_Data= []
def Data_Preprocessing(modalities_dir):
all_modalities = []
for modality in modalities_dir:
nifti_file = nib.load(modality)
nifti_file= rescale_Nii(nifti_file)
brain_numpy = np.asarray(nifti_file.dataobj)
all_modalities.append(brain_numpy)
brain_affine = nifti_file.affine
all_modalities = np.array(all_modalities)
all_modalities = np.rint(all_modalities).astype(np.int16)
all_modalities = all_modalities[:, :, :, :]
all_modalities = np.transpose(all_modalities)
return all_modalities
for i in p[:369]:
brain_dir = os.path.normpath(Path+'/'+i)
flair = glob.glob(os.path.join(brain_dir, '*_flair*.nii'))
t1 = glob.glob(os.path.join(brain_dir, '*_t1*.nii'))
t1ce = glob.glob(os.path.join(brain_dir, '*_t1ce*.nii'))
t2 = glob.glob(os.path.join(brain_dir, '*_t2*.nii'))
gt = glob.glob( os.path.join(brain_dir, '*_seg*.nii'))
modalities_dir = [flair[0], t1[0], t1ce[0], t2[0], gt[0]]
P_Data = Data_Preprocessing(modalities_dir)
Input_Data.append(P_Data)
# + id="2lG9yQiYJgoS"
# save the input data for future use
with open("/content/drive/MyDrive/MRI Data/TrainingData NumPy/1_369.txt", "wb") as fp: # pickling
pickle.dump(Input_Data,fp)
# + id="lSeUv2EYbM7c"
with open("/content/drive/MyDrive/MRI Data/TrainingData NumPy/1_369_(64*64*130).txt", "rb") as fp: # un-pickling
Input_Data= pickle.load(fp)
# + id="cDWEI4AIGvk5" colab={"base_uri": "https://localhost:8080/"} outputId="00888390-9c6c-4c44-fc95-87f48d11664e"
InData= Data_Concatenate(Input_Data)
# + id="TuSgSGBFGyIT"
AIO= concatenate(InData, axis=3)
# + id="AuvqaAulqjgh"
AIO=np.array(AIO,dtype='float32')
TR=np.array(AIO[:,:,:,1],dtype='float32')
TRL=np.array(AIO[:,:,:,4],dtype='float32')#segmentation
X_train , X_test, Y_train, Y_test = train_test_split(TR, TRL, test_size=0.15, random_state=32)
# + [markdown] id="-lNvMFnn7oZx"
# ### **Model Training**
# + id="ve0aDOG4G2lN"
def Convolution(input_tensor,filters):
x = Conv2D(filters=filters,kernel_size=(3, 3),padding = 'same',strides=(1, 1))(input_tensor)
x = BatchNormalization()(x)
x = Activation('relu')(x)
return x
def model(input_shape):
inputs = Input((input_shape))
conv_1 = Convolution(inputs,32)
maxp_1 = MaxPooling2D(pool_size = (2, 2), strides = (2, 2), padding = 'same') (conv_1)
conv_2 = Convolution(maxp_1,64)
maxp_2 = MaxPooling2D(pool_size = (2, 2), strides = (2, 2), padding = 'same') (conv_2)
conv_3 = Convolution(maxp_2,128)
maxp_3 = MaxPooling2D(pool_size = (2, 2), strides = (2, 2), padding = 'same') (conv_3)
conv_4 = Convolution(maxp_3,256)
maxp_4 = MaxPooling2D(pool_size = (2, 2), strides = (2, 2), padding = 'same') (conv_4)
conv_5 = Convolution(maxp_4,512)
upsample_6 = UpSampling2D((2, 2)) (conv_5)
conv_6 = Convolution(upsample_6,256)
upsample_7 = UpSampling2D((2, 2)) (conv_6)
upsample_7 = concatenate([upsample_7, conv_3])
conv_7 = Convolution(upsample_7,128)
upsample_8 = UpSampling2D((2, 2)) (conv_7)
conv_8 = Convolution(upsample_8,64)
upsample_9 = UpSampling2D((2, 2)) (conv_8)
upsample_9 = concatenate([upsample_9, conv_1])
conv_9 = Convolution(upsample_9,32)
outputs = Conv2D(1, (1, 1), activation='sigmoid') (conv_9)
model = Model(inputs=[inputs], outputs=[outputs])
return model
# + id="htbRT_xFG6Ix" colab={"base_uri": "https://localhost:8080/"} outputId="a91187c7-b68f-42e0-a3a7-45868487ece5"
# Loding the Light weighted CNN
model = model(input_shape = (64,64,1))
model.summary()
# + id="pdLM2pWkG9Ut"
# Computing Dice_Coefficient
def dice_coef(y_true, y_pred, smooth=1.0):
y_true_f = K.flatten(y_true)
y_pred_f = K.flatten(y_pred)
intersection = K.sum(y_true_f * y_pred_f)
return (2 * intersection + smooth) / (K.sum(y_true_f) + K.sum(y_pred_f) + smooth)/100
# Computing Precision
def precision(y_true, y_pred):
true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
predicted_positives = K.sum(K.round(K.clip(y_pred, 0, 1)))
precision = true_positives / (predicted_positives + K.epsilon())
return precision
# Computing Sensitivity
def sensitivity(y_true, y_pred):
true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
possible_positives = K.sum(K.round(K.clip(y_true, 0, 1)))
return true_positives / (possible_positives + K.epsilon())
# Computing Specificity
def specificity(y_true, y_pred):
true_negatives = K.sum(K.round(K.clip((1-y_true) * (1-y_pred), 0, 1)))
possible_negatives = K.sum(K.round(K.clip(1-y_true, 0, 1)))
return true_negatives / (possible_negatives + K.epsilon())
# + id="eBhMf0GiHAPs"
# Compiling the model
Adam=optimizers.Adam(lr=0.001)
model.compile(optimizer=Adam, loss='binary_crossentropy', metrics=['accuracy',dice_coef,precision,sensitivity,specificity])
# + id="g0IR8Rp_HChL" colab={"base_uri": "https://localhost:8080/"} outputId="2388cfd6-3ad4-48a5-877f-201b3055979b"
# Fitting the model over the data
history = model.fit(X_train,Y_train,batch_size=32,epochs=40,validation_split=0.20,verbose=1,initial_epoch=0)
# + id="L8Hs2JQWHI3X"
# Evaluating the model on the training and testing data
model.evaluate(x=X_test, y=Y_test, batch_size=32, verbose=1, sample_weight=None, steps=None)
# + id="oDetgIfJHJzv"
def Accuracy_Graph(history):
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
#plt.title('Model accuracy')
plt.ylabel('Accuracy')
plt.xlabel('Epoch')
plt.legend(['Train', 'Validation'], loc='upper left')
plt.subplots_adjust(top=1.00, bottom=0.0, left=0.0, right=0.95, hspace=0.25,
wspace=0.35)
plt.show()
# Dice Similarity Coefficient vs Epoch
def Dice_coefficient_Graph(history):
plt.plot(history.history['dice_coef'])
plt.plot(history.history['val_dice_coef'])
#plt.title('Dice_Coefficient')
plt.ylabel('Dice_Coefficient')
plt.xlabel('Epoch')
plt.legend(['Train', 'Validation'], loc='upper left')
plt.subplots_adjust(top=1.00, bottom=0.0, left=0.0, right=0.95, hspace=0.25,
wspace=0.35)
plt.show()
# Loss vs Epoch
def Loss_Graph(history):
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
#plt.title('Model loss')
plt.ylabel('Loss')
plt.xlabel('Epoch')
plt.legend(['Train', 'Validation'], loc='upper left')
plt.subplots_adjust(top=1.00, bottom=0.0, left=0.0, right=0.95, hspace=0.25,
wspace=0.35)
plt.show()
# + id="Pm_Yz2yfHOFh"
# Plotting the Graphs of Accuracy, Dice_coefficient, Loss at each epoch on Training and Testing data
Accuracy_Graph(history)
Dice_coefficient_Graph(history)
Loss_Graph(history)
# + id="Ex4TBCdOHPyP"
# save model for future use
model.save('/content/drive/MyDrive/MRI Data/TrainingData NumPy/64*64.h5')
# + [markdown] id="FQ-DfqbY7tQa"
# ### **Predicting Tumor**
# + id="acoh3vFtHRcS"
#load weights
model.load_weights('/content/drive/MyDrive/MRI Data/TrainingData NumPy/64*64.h5')
# + id="vnWg9QWqHUi_" colab={"base_uri": "https://localhost:8080/", "height": 322} outputId="6f2ca009-4a1a-4da9-aeac-3383fe3c5049"
fig = plt.figure(figsize=(5,5))
immmg = TR[205,:,:]
imgplot = plt.imshow(immmg)
plt.show()
# + id="gPCyRSNbHWZQ"
pref_Tumor = model.predict(TR)
# + id="x1juVnaiHYft" colab={"base_uri": "https://localhost:8080/", "height": 322} outputId="c1d813fa-a4fe-4511-993f-f71bba543698"
fig = plt.figure(figsize=(5,5))
immmg = pref_Tumor[205,:,:,0]
imgplot = plt.imshow(immmg)
plt.show()
| Python Files/UNet_64_64.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import functools
input = open('inputs/day10.txt', 'r').read().splitlines()
nums = [int(l) for l in input]
dev = max(nums)+3
nums.extend([0, dev])
nums.sort()
diffs = [b-a for (a,b) in zip(nums, nums[1:])]
diffs.count(1) * diffs.count(3)
# +
# %%time
@functools.lru_cache()
def count(n):
if n == dev:
return 1
valid = [x for x in nums if x > n and x <= n + 3]
return sum(map(count, valid))
count(0)
| aoc_2020/day10.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="qKIsKr1RJl94" executionInfo={"status": "ok", "timestamp": 1630826180768, "user_tz": -540, "elapsed": 632, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}}
# !mkdir -p ~/.kaggle
# !cp kaggle.json ~/.kaggle
# !chmod 600 ~/.kaggle/kaggle.json
# + colab={"base_uri": "https://localhost:8080/"} id="htpWdxXLJ2BD" executionInfo={"status": "ok", "timestamp": 1630826180148, "user_tz": -540, "elapsed": 4381, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="f0a1162a-8192-422c-a089-54d54f6120ee"
# 타이타닉 데이터 다운로드
# !kaggle competitions download -c titanic
# + id="ZmtmGb7eJ-lD" executionInfo={"status": "ok", "timestamp": 1630826180148, "user_tz": -540, "elapsed": 13, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}}
## 데이터 분석 관련
import pandas as pd
from pandas import Series, DataFrame
import numpy as np
## 데이터 시각화 관련
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style('whitegrid') # matplotlib의 스타일에 관련한 함
## 그래프 출력에 필요한 IPython 명령어
# %matplotlib inline
# + colab={"base_uri": "https://localhost:8080/", "height": 204} id="LjFQNUyKKQ-h" executionInfo={"status": "ok", "timestamp": 1630826180768, "user_tz": -540, "elapsed": 17, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="3f3d6327-edcf-40be-c050-b577a78047f1"
train_df = pd.read_csv("train.csv")
test_df = pd.read_csv("test.csv")
gender_submission = pd.read_csv("gender_submission.csv")
train_df.head()
# + id="k0DSjVQoKaGo" colab={"base_uri": "https://localhost:8080/"} executionInfo={"status": "ok", "timestamp": 1630826180768, "user_tz": -540, "elapsed": 16, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="bb993487-5640-4865-feff-92d0d0376df5"
train_df.info()
print('-'*28)
test_df.info()
# train의 attribute는 12개 test의 attribute는 11개인 이유는 train은 Survied의 여부를 알고 있기 때문이다.
# + [markdown] id="IEHEBEj_kfL8"
# ###**여기서 주의 깊게 봐야할 부분은 다음과 같다.**
# # + 각 데이터는 빈 부분이 있는가?
# + 빈 부분이 있다면, drop할 것인가 아니면 default값으로 채워 넣을 것인가.
# + cabin, Age, Embarked 세 항목에 주의
# # + 데이터는 float64로 변환할 수 있는가.
# + 아니라면 범주형 데이터로 만들 수 있는가.
# + id="pnRDQcfejz6t" executionInfo={"status": "ok", "timestamp": 1630826180769, "user_tz": -540, "elapsed": 15, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}}
# 이름과 티켓에서 가져올 수 있는 데이터는 없기 때문에 PassengerID와 이름, 티켓을 지운다.
# 하지만 이 문제에서 결과물은 'PassengerId', 'Survived' 요소가 필요하므로 훈련데이터에서만 삭제한다.
train_df = train_df.drop(['PassengerId', 'Ticket'], axis=1) # axis = 1은 열을 지운다.
test_df = test_df.drop(['Ticket'], axis=1) # 결과물은 test에서 나온다. 즉, PassengerId를 지우면 안된다.
# + [markdown] id="lOqcP7wllwdj"
# ###**데이터 하나하나 처리하기**
# 이제 남은 데이터 종류는 다음과 같다.
# 1. Pclass
# 2. Sex
# 3. SibSp
# 4. Parch
# 5. Fare
# 6. Cabin
# 7. Embarked
# 8. Name
# 9. Age(추가)
# + id="dPRYi_bxljnl" executionInfo={"status": "ok", "timestamp": 1630826180769, "user_tz": -540, "elapsed": 15, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}}
# # 1. Pclass
# # 서수형 데이터이다. 1등석, 2등석, 3등석과 같은 정보. 처음에 확인시에 데이터가 비어있지 않은 것을 확인할 수 있었다.
# # 데이터에 대한 확인과 데이터를 변환해보도록 하겠다. 우선 각 unique한 value에 대한 카운팅은 value_counts() 메서드로 확인할 수 있다.
# train_df['Pclass'].value_counts()
# # 1, 2, 3은 정수이니, 그냥 실수로만 바꾸면 되지않을까 생각할 수 있다. 하지만 1, 2, 3 등급은 경우에 따라 다를 수 있지만 연속적인 정보가 아니며, 각 차이 또한 균등하지 않다.
# # 그렇기에 범주형(카테고리) 데이터로 인식하고 인코딩해야한다.(비슷한 예시로 영화 별점 등이 있다.)
# # 이 데이터는 범주형 데이터이므로 one-hot-encoding을 pd.get_dummies() 메서드로 인코딩한다.
# pclass_train_dummies = pd.get_dummies(train_df['Pclass'])
# pclass_test_dummies = pd.get_dummies(test_df['Pclass'])
# train_df.drop(['Pclass'], axis=1, inplace=True)
# test_df.drop(['Pclass'], axis=1, inplace=True)
# train_df = train_df.join(pclass_train_dummies)
# test_df = test_df.join(pclass_test_dummies)
# + colab={"base_uri": "https://localhost:8080/", "height": 204} id="nJLYp9c3lrHH" executionInfo={"status": "ok", "timestamp": 1630826180769, "user_tz": -540, "elapsed": 15, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="031e475c-0d23-4c40-d6e2-25499522d155"
train_df.head() # 원래는 columns의 이름을 설정하고, 넣어줘야하는데 실수로 넣지 않아 1, 2, 3 이라는 컬럼으로 데이터가 들어갔다.
# + id="ldd-zZK7pFqp" colab={"base_uri": "https://localhost:8080/", "height": 204} executionInfo={"status": "ok", "timestamp": 1630826180769, "user_tz": -540, "elapsed": 15, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="541a37d9-ef62-40c3-f661-792330bea7ef"
# 2. Sex
# 성별이라는 뜻으로 남과 여로 나뉘므로 이 또한 one-hot-encoding을 진행
# # [[[ dummie 방식 ]]]
# sex_train_dummies = pd.get_dummies(train_df['Sex'])
# sex_test_dummies = pd.get_dummies(test_df['Sex'])
# train_df.drop(['Sex'], axis=1, inplace=True)
# test_df.drop(['Sex'], axis=1, inplace=True)
# train_df = train_df.join(sex_train_dummies)
# test_df = test_df.join(sex_test_dummies)
# category .cat.codes방식 numeric
train_df['Sex'] = train_df['Sex'].astype('category').cat.codes
test_df['Sex'] = test_df['Sex'].astype('category').cat.codes
train_df.head()
# + id="T-Xt00Lkq34v" colab={"base_uri": "https://localhost:8080/", "height": 204} executionInfo={"status": "ok", "timestamp": 1630826180769, "user_tz": -540, "elapsed": 14, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="771cfceb-9a04-432f-8b00-01d9e10cccf6"
# 3,4. SibSp & Parch
# 형제 자매와 부모님은 가족으로 함께 처리할 수 있다. 하지만 굳이 바꿀필요는 없다.
train_df['Family'] = 1 + train_df['SibSp'] + train_df['Parch']
test_df['Family'] = 1 + train_df['SibSp'] + test_df['Parch']
train_df = train_df.drop(['SibSp', 'Parch'], axis=1)
test_df = test_df.drop(['SibSp', 'Parch'], axis=1)
# # + Solo : 내가 혼자 탔는지 다른 가족과 탔는지 여부를 구분해주는 데이터 추가
train_df['Solo'] = (train_df['Family'] == 1)
test_df['Solo'] = (test_df['Family'] == 1)
# 5. Fare
# 탑승료이다. 신기하게 test 데이터셋에 1개의 데이터가 비어있다. 아마 디카프리오인듯 하다. 우선 빈 부분을 fillna 메서드로 채운다.
# 데이터 누락이 아닌 무단 탑승이라 생각하고 0으로 입력
test_df['Fare'].fillna(0, inplace=True)
train_df.head()
# + id="G0FFULCiwdTU" executionInfo={"status": "ok", "timestamp": 1630826180770, "user_tz": -540, "elapsed": 14, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}}
# 6. Cabin
# 객실이라는 뜻이다. 대부분이 NaN이므로 버린다.
train_df = train_df.drop(['Cabin'],axis=1)
test_df = test_df.drop(['Cabin'], axis=1)
# + colab={"base_uri": "https://localhost:8080/"} id="4q35OL45rUuF" executionInfo={"status": "ok", "timestamp": 1630826180771, "user_tz": -540, "elapsed": 15, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="b873a88e-796f-4710-f42a-011a3b397e50"
# 7. Embarked
# 탑승 항구를 의미, 우선 데이터를 확인
train_df['Embarked'].value_counts()
# + colab={"base_uri": "https://localhost:8080/"} id="dvSqwZo6sABg" executionInfo={"status": "ok", "timestamp": 1630826180771, "user_tz": -540, "elapsed": 14, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="475f2fe0-c6da-4f13-c4ef-1f6246a0d321"
test_df['Embarked'].value_counts()
# + id="M1KGaqPgsOcs" colab={"base_uri": "https://localhost:8080/", "height": 204} executionInfo={"status": "ok", "timestamp": 1630826180771, "user_tz": -540, "elapsed": 13, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="c0ea5fc0-9191-4e72-8668-a20ec9920f8f"
# S가 대다수이고 일부 데이터가 비어있는 것을 알 수 있다. 빈 부분은 S로 우선 채운다(.info로 확인했을 때 빈 부분이 있는 줄 몰랐다)
train_df["Embarked"].fillna('S', inplace=True)
# Embarked 컬럼 역시 numeric 한 데이터로 변경
train_df['Embarked'] = train_df['Embarked'].astype('category').cat.codes
test_df['Embarked'] = test_df['Embarked'].astype('category').cat.codes
train_df.head()
# + colab={"base_uri": "https://localhost:8080/"} id="g82WkzAekyPg" executionInfo={"status": "ok", "timestamp": 1630826180771, "user_tz": -540, "elapsed": 12, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="1b20c5e4-1b13-4790-e014-238bb47f582d"
# 8. Name
# Title은 'Name' 칼럼에서 ~씨와 같은 t itle을 추출하여 새롭게 생성해주는 컬럼,
# 단 주의해야할 점은 TItle을 추출하여 카테고리와 해주면, 데이터의 총량 비하여 너무 복잡도가 올라가는 경향이있다.
# 그렇기에 모수가 적은 Mlle, Mme, Ms는 단일화 시켜주어야 한다.
train_df['Title'] = train_df['Name'].str.extract(' ([A-Za-z]+)\.', expand=False)
test_df['Title'] = test_df['Name'].str.extract(' ([A-Za-z]+)\.', expand=False)
train_df['Title'] = train_df['Title'].replace(['Lady', 'Countess', 'Capt', 'Col', 'Don', 'Dr', 'Major', 'Rev', 'Sir', 'Jonkheer', 'Dona'], 'Other')
test_df['Title'] = test_df['Title'].replace(['Lady', 'Countess', 'Capt', 'Col', 'Don', 'Dr', 'Major', 'Rev', 'Sir', 'Jonkheer', 'Dona'], 'Other')
train_df = train_df.drop(['Name'], axis=1)
test_df = test_df.drop(['Name'], axis=1)
train_df['Title'].value_counts()
# + colab={"base_uri": "https://localhost:8080/", "height": 204} id="sEU5zYHOoVFg" executionInfo={"status": "ok", "timestamp": 1630826180772, "user_tz": -540, "elapsed": 12, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="172f2c9f-3a03-41e2-8214-185886c0d842"
# 위에서 세어보니 Mlle, Mme, Ms의 수가 적어서 단일화 해주는 작업을 해야한다.
train_df['Title'] = train_df['Title'].replace(['Mlle', 'Ms'], 'Miss')
train_df['Title'] = train_df['Title'].replace('Mme', 'Mrs')
test_df['Title'] = test_df['Title'].replace(['Mlle', 'Ms'], 'Miss')
test_df['Title'] = test_df['Title'].replace('Mme', 'Mrs')
train_df['Title'] = train_df['Title'].astype('category').cat.codes
test_df['Title'] = test_df['Title'].astype('category').cat.codes
train_df.head()
# + colab={"base_uri": "https://localhost:8080/", "height": 204} id="d7kk7eZcuEsw" executionInfo={"status": "ok", "timestamp": 1630826180772, "user_tz": -540, "elapsed": 12, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="5951fd6f-865c-4037-f54b-de2ce98b986b"
# 9. Age
# 나이는 연속형 데이터이므로, 큰 처리가 필요없다. (카테고리화를 하여 일부 알고리즘에 더 유용한 결과를 만들 수 있다.)
# 하지만 일부 NaN 데이터가 있으니 이를 채울 수 있는 방법에 대해서 생각해보자
# 1. 랜덤(random), 2. 평균값(mean), 3. 중간값(median), 4. 데이터 버리기(drop)
# groupyby 함수를 이용해 "Title" 컬럼의 그룹을 나누어(1, 2, 3) 해당 그룹의 "Age"칼럼의 median을 fillna에 대입
train_df["Age"].fillna(train_df.groupby("Title")["Age"].transform("median"), inplace=True)
test_df["Age"].fillna(test_df.groupby("Title")["Age"].transform("median"), inplace=True)
train_df.head()
# + colab={"base_uri": "https://localhost:8080/", "height": 204} id="Ki-72F68uonI" executionInfo={"status": "ok", "timestamp": 1630826181287, "user_tz": -540, "elapsed": 526, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="eb580247-0082-494d-f918-c0f989422bfd"
# Age를 구간화 (Binning), 5세 단위로 자르고 50대는 10세단위 그리고 60세 이상은 모두 묶어서 Binning해줌
# Train
train_df.loc[ train_df['Age'] <= 10, 'Age'] = 0
train_df.loc[(train_df['Age'] > 10) & (train_df['Age'] <= 16), 'Age'] = 1
train_df.loc[(train_df['Age'] > 16) & (train_df['Age'] <= 20), 'Age'] = 2
train_df.loc[(train_df['Age'] > 20) & (train_df['Age'] <= 26), 'Age'] = 3
train_df.loc[(train_df['Age'] > 26) & (train_df['Age'] <= 30), 'Age'] = 4
train_df.loc[(train_df['Age'] > 30) & (train_df['Age'] <= 36), 'Age'] = 5
train_df.loc[(train_df['Age'] > 36) & (train_df['Age'] <= 40), 'Age'] = 6
train_df.loc[(train_df['Age'] > 40) & (train_df['Age'] <= 46), 'Age'] = 7
train_df.loc[(train_df['Age'] > 46) & (train_df['Age'] <= 50), 'Age'] = 8
train_df.loc[(train_df['Age'] > 50) & (train_df['Age'] <= 60), 'Age'] = 9
train_df.loc[ train_df['Age'] > 60, 'Age'] = 10
# Test
test_df.loc[ test_df['Age'] <= 10, 'Age'] = 0
test_df.loc[(test_df['Age'] > 10) & (test_df['Age'] <= 16), 'Age'] = 1
test_df.loc[(test_df['Age'] > 16) & (test_df['Age'] <= 20), 'Age'] = 2
test_df.loc[(test_df['Age'] > 20) & (test_df['Age'] <= 26), 'Age'] = 3
test_df.loc[(test_df['Age'] > 26) & (test_df['Age'] <= 30), 'Age'] = 4
test_df.loc[(test_df['Age'] > 30) & (test_df['Age'] <= 36), 'Age'] = 5
test_df.loc[(test_df['Age'] > 36) & (test_df['Age'] <= 40), 'Age'] = 6
test_df.loc[(test_df['Age'] > 40) & (test_df['Age'] <= 46), 'Age'] = 7
test_df.loc[(test_df['Age'] > 46) & (test_df['Age'] <= 50), 'Age'] = 8
test_df.loc[(test_df['Age'] > 50) & (test_df['Age'] <= 60), 'Age'] = 9
test_df.loc[ test_df['Age'] > 60, 'Age'] = 10
train_df.head()
# + [markdown] id="RrlQyHHKwYuG"
# ### Feature와 Label을 정의하기
# + id="Lgr1m06SwUvH" executionInfo={"status": "ok", "timestamp": 1630826181287, "user_tz": -540, "elapsed": 4, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}}
feature = [
'Pclass',
'Sex',
'Age',
'Fare',
'Embarked',
'Family',
'Solo',
"Title",
]
label = [
'Survived',
]
# + [markdown] id="30eR99cPyKMY"
# ### HyperParameter
# 여러 가지 모델링을 해본 결과, 이 블로그에서 진행항 pre-processing 데이터셋에는 RnadomForestClassifier가 가장 좋은 결과를 가져다 주었다.
#
# ---
#
# 우선, 이번 Titanic 생존자 예측 대회에서는 dataset의 복잡도가 크지 않고, 사이즈도 매우 적기 때문에 n_estimator 값은 최대한 줄이는 전략을 취했다.
# 또한 max_depth도 제한을 두어 너무 깊어지지 않도록 했으며, 다른 parameter는 별도로 건들이지 않았다.
# + id="3CGTTkTruuW7" colab={"base_uri": "https://localhost:8080/"} executionInfo={"status": "ok", "timestamp": 1630826352211, "user_tz": -540, "elapsed": 2173, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="ab10cee7-d3ba-44e5-f6fa-accdb81b3f89"
from sklearn.model_selection import KFold, cross_val_score
from sklearn.ensemble import RandomForestClassifier
data = train_df[feature]
target = train_df[label]
k_fold = KFold(n_splits=10, shuffle=True, random_state=0)
clf = RandomForestClassifier(n_estimators=50, max_depth=6, random_state=0)
cross_val_score(clf, data, target, cv=k_fold, scoring='accuracy', ).mean()
# + colab={"base_uri": "https://localhost:8080/"} id="mPblffmvzjgX" executionInfo={"status": "ok", "timestamp": 1630826341427, "user_tz": -540, "elapsed": 402, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="dfcf9ddf-2f89-490e-c174-2974af00eed1"
train_x = train_df[feature]
train_y = train_df[label]
test_x = test_df[feature]
clf = RandomForestClassifier(n_estimators=100, max_depth=6, random_state=0)
clf.fit(train_x, train_y)
gender_submission['Survived'] = clf.predict(test_x)
gender_submission.to_csv('titanic-submission.csv',index=False)
# + [markdown] id="pzE5dTDHwMAZ"
# ###**여러 머신러닝 알고리즘 적용 해보기**
# + colab={"base_uri": "https://localhost:8080/"} id="E7pewssKvLvf" executionInfo={"status": "ok", "timestamp": 1630826531293, "user_tz": -540, "elapsed": 540, "user": {"displayName": "\uc190\uc720\uc131", "photoUrl": "", "userId": "04792335917307913817"}} outputId="0f8a7155-0e19-4f7b-8d53-f8596a7fbdeb"
## Scikit-Learn의 다양한 머신러닝 모듈을 불러옵니다.
## 분류 알고리즘 중에서 선형회귀, 서포트벡터머신, 랜덤포레스트, K-최근접이웃 알고리즘을 사용해보려고 한다.
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC, LinearSVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
# Logistic Regression
logreg = LogisticRegression(max_iter=1000) # max_iter을 이용하여 오류해결
logreg.fit(train_x, train_y)
pred_y = logreg.predict(test_x)
logreg.score(train_x, train_y)
# Support Vector Machines
svc = SVC()
svc.fit(train_x, train_y)
pred_y = svc.predict(test_x)
svc.score(train_x, train_y)
# Random Forests
random_forest = RandomForestClassifier(n_estimators=100)
random_forest.fit(train_x, train_y)
pred_y = random_forest.predict(test_x)
random_forest.score(train_x, train_y)
# K-Neigbor
knn = KNeighborsClassifier(n_neighbors = 3)
knn.fit(train_x, train_y)
pred_y = knn.predict(test_x)
knn.score(train_x, train_y)
# Random Forests
random_forest = RandomForestClassifier(n_estimators=1)
random_forest.fit(train_x, train_y)
pred_y = random_forest.predict(test_x)
random_forest.score(train_x, train_y)
| Titanic/Titanic.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Now You Code 2: Is That An Email Address?
#
# Let's use Python's built-in string functions to write our own function to detect if a string is an email address.
#
# The function `isEmail(text)` should return `True` when `text` is an email address, `False` otherwise.
#
# For simplicity's sake we will define an email address to be any string with just ONE `@` symbol in it, where the `@` is not at the beginning or end of the string. So `a@b` is considered an email (even though it really isn't).
#
# The program should detect emails until you enter quit.
#
# Sample run:
# ```
# Email address detector. Type quit to exit.
# Email: <EMAIL>
# <EMAIL> ==> email
# Email: mafudge@
# mafudge@ ==> NOT EMAIL
# Email: mafudge
# mafudge ==> NOT EMAIL
# Email: @syr.edu
# @syr.edu ==> NOT EMAIL
# Email: @
# @ ==> NOT EMAIL
# Email: <EMAIL>
# <EMAIL> ==> NOT EMAIL
# Email: <EMAIL>
# <EMAIL> ==> NOT EMAIL
# ```
#
# Once again we will use the problem simplification technique to writing this program.
#
# First we will write the `isEmail(text)` function, then we will write the main program.
#
# ## Step 1: Problem Analysis for isEmail function only
#
# Inputs (function arguments): an email address
#
# Outputs (what is returns): true/false
#
# Algorithm (Steps in Function): enter email, check validity, print real or not
#
#
#
help(str)
# +
## Step 2: Todo write the function definition for isEmail functiuon
def isEmail(text):
if text.count("@") ==1 and text.find ("@") >0 and text.find("@") <= len(text)-3: #makes sure there is 1 @ at a position greater than the first and not the last.
return True
else:
return False
email=input("Enter an email address: ")
isEmail(email)
# +
## Step 3: Write some tests, to ensure the function works, for example
## Make sure to test all cases!
print("WHEN text=<EMAIL> We EXPECT isEmail(text) to return True", "ACTUAL", isEmail("<EMAIL>") )
print("WHEN text=mike@ We EXPECT isEmail(text) to return False", "ACTUAL", isEmail("mike@") )
# -
# ## Step 4: Problem Analysis for full Program
#
# Inputs: enter an email address
#
# Outputs: if email real or not
#
# Algorithm (Steps in Program): enter email, call def, check true/false, print if real or not
#
#
# +
## Step 5: todo write code for full problem, using the isEmail function to help you solve the problem
email=input("Enter your email address: ")
if isEmail(email)==True:
print("That's a real email")
else:
print("Invalid email")
# -
# ## Step 6: Questions
#
# 1. How many test cases should you have in step 3 to ensure you've tested all the cases?
# 2. What should kind of logic should we add to make our `isEmail` function even better, so that is detects emails more accurately?
#
# +
#1. A few, one a real email, one that doesn't meet one of the conditions,
#2. besides the @ finder, we should add to check if there is a period at least a few spaces after the @ and before the last spot. we could eventually check if the email domain is real.
# -
# ## Reminder of Evaluation Criteria
#
# 1. What the problem attempted (analysis, code, and answered questions) ?
# 2. What the problem analysis thought out? (does the program match the plan?)
# 3. Does the code execute without syntax error?
# 4. Does the code solve the intended problem?
# 5. Is the code well written? (easy to understand, modular, and self-documenting, handles errors)
#
# +
#1. to check if the inputted email is a real email or not
#2. the thought out might not be thoroughly thoought out but it makes sense for me to follow
#3. there are none
#4. yes
#5. yes
| content/lessons/07/Now-You-Code/NYC2-Email-Address.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:ds-twitter]
# language: python
# name: conda-env-ds-twitter-py
# ---
# ## Word clouds
# This NB contains the code required to generate the word clouds in [this article](https://www.datacamp.com/community/blog/metoo-twitter-analysis). You will need to get the tweets and, to do so, check out the README.md in [this repository](https://github.com/datacamp/datacamp-metoo-analysis). Find out more about <NAME>'s word cloud generator [here](https://github.com/amueller/word_cloud).
# +
# Imports
import pandas as pd
import matplotlib.pyplot as plt
# %matplotlib inline
import seaborn as sns
from wordcloud import WordCloud, STOPWORDS
import json
sns.set()
# Get datasets into DataFrames
df = pd.read_csv('data/tweets1.csv')
df2 = pd.read_csv('data/tweets2.csv')
# +
# Get English language tweets, split into words and remove stopwords
text = df[df.lang == 'en'].text.str.cat(sep = ' ').lower()
stopwords = set(STOPWORDS)
stopwords.update(['https', 'RT', 'co'])
# Generate wordcloud and save to file
wordcloud = WordCloud(width=1000, height=500, max_font_size=90, collocations=False, stopwords=stopwords).generate(text)
wordcloud.to_file("wc1.png")
# Display the generated image:
# the matplotlib way:
#import matplotlib.pyplot as plt
plt.imshow(wordcloud, interpolation='bilinear')
#plt.figure(figsize=(8, 6))
plt.axis("off")
# +
# Get English language tweets, split into words and remove stopwords
text = df2[df2.lang == 'en'].text.str.cat(sep = ' ').lower()
stopwords = set(STOPWORDS)
stopwords.update(['https', 'RT', 'co'])
# Generate wordcloud and save to file
wordcloud = WordCloud(width=1000, height=500, max_font_size=90, collocations=False, stopwords=stopwords).generate(text)
wordcloud.to_file("wc2.png")
# Display the generated image:
# the matplotlib way:
#import matplotlib.pyplot as plt
plt.imshow(wordcloud, interpolation='bilinear')
#plt.figure(figsize=(8, 6))
plt.axis("off")
| word_clouds.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Python Documentation
#
# * Main Python Site: www.python.org
# * Python 3 Documentation: https://docs.python.org/3/
# * Tutorial - takes about 3-4 hours to complete. Contains many examples of the language and its usage: https://docs.python.org/3/tutorial/index.html
# * Standard Library: https://docs.python.org/3/library/index.html
# * Glossary: https://docs.python.org/3/glossary.html
#
#
# * Problems - w3resource.com/python-exercises/
# # Sequence Types
#
#
# ## List
#
# ### Definition
#
# A list is a sequence of values where the values can be of the same or different types. A list is defined by writing a series of comma-separated values enclosed by brackets.
#
# ### Nomenclature
#
# l = [1, 2, 3, 4] # List of four integers
# l = ['a', 'b', 'c', 'd'] # list of four characters
# l = [] # empty list
# l = list() # empty list
#
# ### Operations
#
# * append(x)
# * clear()
# * copy()
# * extend(t)
# * insert(x)
# * pop([i])
# * remove(x)
# * reverse()
#
# ## Tuple
#
# ### Definition
#
# Tuples are immutable sequences, typically used to store collections of heterogeneous data. A tuple is defined by writing a series of comma-separated values encluded by parenthesis.
#
# **Note: As an immutable type, tuple is hashable and therefore can be used as a key in a dictionary and stored in a set.**
#
# ### Nomenclature
#
# t = (1, 2, 3) # Tuple of three integers
# t = (1,) # Tuple of single integer
# t = ('a', 'b', 'c') # Tuple of three characters
# t = 'this', 'that', 'these', 'those' # Tuple of four strings
# t = tuple() # Empty tuple
# t = () # Empty tuple
#
# ### Operations
#
# See Common Sequence Operations
#
# ## Range
#
# ### Definition
#
# The range type represents an immutable sequence of numbers and is commonly used for looping a specific number of times in for loops.
#
# ### Nomenclature
#
#
# ### Operations
#
#
# ## Set
#
# ### Definition
#
# A set object is a mutable unordered collection of distinct hashable objects. Common uses include membership testing, removing duplicates from a sequence, and computing mathematical operations such as intersection, union, difference, and symmetric difference.
#
# ### Nomenclature
#
# s = set([1, 2, 3, 4, 5]) # Set of five integers from list
# s = set(('a', 'b', 'c', 'd')) # Set of characters from tuple
# s = set('asdf') # Set of four characters from string
# s = set() # Empty set
#
# ### Operations
#
# * isdisjoint
# * issubset or set <= other
# * set < other
# * issuperset or set >= other
# * set > other
# * union or set | other | ...
# * intersection or set & other & ...
# * difference or set - other - ...
# * symmetric_difference or set ^ other
# * copy
# * update or set |= other
# * intersection_update or set &= other
# * difference_update or set -= other
# * symmetric_difference_update or set ^= other
# * add(x)
# * remove(x)
# * discard(x)
# * pop()
# * clear()
#
# ## Bytes
#
# ### Definitions
#
# Bytes objects are immutable sequences of single bytes.
#
# ### Nomenclature
#
# b = b'12345656' # Bytes object of length 8
# b = bytes(10) # Zero-filled bytes object of length 10
# b = bytes(range(10)) # Bytes object of length 10 filled with values 0 through 9
#
# ### Operations
#
# * hex() - Converts ASCII to hex values
# * fromhex(x) - Converts hex string to ASCII bytes
#
# **Note: Contains many ASCII-based (string-type) operations. Care must be taken when using these operations with non-ASCII data.
#
#
# ## ByteArray
#
# Immutable counterpart to Bytes
#
# ## Common Sequence Operations
#
# * x in s
# * x not in s
# * s + t (concatenate)
# * s * n or n * s (repeat)
# * s[i]
# * s[start:stop]
# * s[start:stop:step]
# * len(s)
# * min(s)
# * max(s)
# * s.index(x)
# * s.count(x)
#
#
# ## Exercises
#
# ### List
#
# ### Tuple
#
# ### Set
#
# ### Bytes/ByteArray
#
#
# # Mapping
#
# ## Dictionary
#
# ### Keys
#
# ### Values
#
# ### Items
#
# ## Exercise
#
# Rock, Paper, Scissors
#
#
# # Files
#
#
# # Functions
#
#
#
# * Problem
# * Count the number of words in a file
#
#
# Now that we have learned some basics of programming, input, processing and output, its time to learn new ways to represent the information that is stored within a program. The mechanisms we use to store information are called data structures. Python has several useful builtin data structures and also allows a programmer to create her own.
# **List**
#
#
#
# **Note: While a list can hold values of different types, typically, a list contains values of a single type.**
[1, 2, 3, 4, 5]
['a', 'b', 'c', 'd', 'e']
[1, 'a', 2, 'b', 3, 'c']
# Python also provides a **list** function that will create a list from an iterable. An iterable is defined as:
#
# Iterable: An object capable of returning its members one at a time.
#
# Iteration is a fundamental property used by many data structures throughout Python.
list('abcde')
list('12345')
list(range(10))
# **List Operations**
#
# append
# index
# slice
# insert
# pop
#
# * Append
#
# l = []
# l.append('a')
# l.append('b')
# l.append('c')
# l.append('d')
#
# * Index
# l = [1, 2, 3, 4, 5]
# l.index[3]
# l.index[0]
# l.index[-1]
#
# * Slice
# l = [1, 2, 3, 4, 5]
# l[0:5]
# l[:]
# l[3:5]
# l[-1:-3]
#
# * Push
# l = []
# l.push('a')
# l.push('b')
# l.push('c')
#
# * Pop
# l = [1, 2, 3, 4, 5]
# l.pop(0)
# **List Exercises**
# 1. Write a Python program to sum all the items in a list.
l = [1, 2, 3, 4, 5]
sum(l)
# 2. Write a Python program to multiplies all the items in a list.
# +
def mul(iterable):
x = 1
for i in iterable:
x*=i
return x
l = [1, 2, 3, 4, 5]
mul(l)
# -
# 3. Write a Python program to get the largest number from a list.
l = [1, 2, 3, 4, 5]
max(l)
# 4. Write a Python program to get the smallest number from a list.
l = [1, 2, 3, 4, 5]
min(l)
# 5. Write a Python program to count the number of strings where the string length is 2 or more and the first and last character are same from a given list of strings.
# +
s = ['this', 'that', 'these', 'those', 'it', 'what', 'wish']
for i in s:
if len(i) >= 2 and i[0] == i[-1]:
print(i)
# -
# 6. Write a Python program to get a list, sorted in increasing order by the last element in each tuple from a given list of non-empty tuples.
l = [(1, 2), (3, 2), (2, 3), (5, 3), (7,2)]
sorted(l, key=lambda i: i[1], reverse=True)
# 7. Write a Python program to remove duplicates from a list.
l = [1, 2, 3, 4, 1, 5, 6, 5, 7, 6]
list(set(l))
# 8. Write a Python program to check a list is empty or not.
# +
def is_empty(l):
return l is None
l = [1, 2, 3, 4, 5]
print(is_empty(l))
l = None
print(is_empty(l))
# -
# 9. Write a Python program to clone or copy a list.
# +
l = [1, 2, 3, 4, 5]
print(id(l))
m = l.copy()
print(id(m))
# -
# 10. Write a Python program to find the list of words that are longer than n from a given list of words.
l = ['this', 'that', 'these', 'those', 'which', 'what', 'who', 'where']
n = 4
m = [i for i in l if len(i) > n]
print(m)
# 11. Write a Python function that takes two lists and returns True if they have at least one common member.
# +
l = [1, 2, 3, 4, 5, 6, 7, 8, 9]
m = [9, 5, 3, 1, 3, 8]
True if True in [
i == j
for i in m
for j in l] else False
# -
# 12. Write a Python program to print a specified list after removing the 0th, 4th and 5th elements.
# +
l = [1, 3, 2, 6, 5, 7, 8, 9, 4]
r = [0, 4, 5]
m = [v for i,v in enumerate(l) if i not in r]
print(m)
# -
# 13. Write a Python program to generate a 3*4*6 3D array whose each element is *.
# +
def make_3d_array(i, j, k):
return [[[ '*' for _i in range(i)] for _j in range(j)] for _k in range(k)]
make_3d_array(3, 4, 6)
# -
# 14. Write a Python program to print the numbers of a specified list after removing even numbers from it.
l = [1, 4, 3, 2, 5, 7 ,4, 5, 8]
[i for i in l if i % 2 == 0]
# 15. Write a Python program to shuffle and print a specified list.
# +
import random
l = [1, 2, 3, 4, 5, 6]
random.shuffle(l)
print(l)
# -
# 16. Write a Python program to generate and print a list of first and last 5 elements where the values are square of numbers between 1 and 30 (both included).
# +
import random
import math
l = [random.randint(0, 1000) for _ in range(100)]
m = [i for i in l if 1 < math.sqrt(i) < 30]
print(m[:5] + m[-5:])
# -
# 17. Write a Python program to generate and print a list except for the first 5 elements, where the values are square of numbers between 1 and 30 (both included).
# +
import random
l = [random.randint(0, 1000) for _ in range(100)]
m = [i for i in l if 1 < i < 30*30]
print(m[5:])
# -
# 18. Write a Python program to generate all permutations of a list in Python.
# +
import random
import itertools
n = 3
l = [random.randint(0,n) for i in range(n)]
[i for i in itertools.permutations(l)]
# -
# 19. Write a Python program to get the difference between the two lists.
# +
l = [1, 2, 3]
m = [5, 8, 3, 2, 1]
print(list(set(m)-set(l)))
# -
# 20. Write a Python program access the index of a list.
# +
l = [1, 2, 3, 4, 5]
for k, v in enumerate(l):
print(k, v)
# -
# 21. Write a Python program to convert a list of characters into a string.
# +
l = ['a', 'b', 'c', 'd', 'e', 'f', 'g', 'h']
''.join(l)
# -
# 22. Write a Python program to find the index of an item in a specified list.
# +
l = ['a', 'b', 'c', 'd', 'e', 'f', 'g']
print(l.index('a'))
print(l.index('e'))
# -
# 23. Write a Python program to flatten a shallow list.
# +
import itertools
l = [[2,4,3],[1,5,6], [9], [7,9,0]]
m = list(itertools.chain(*l))
print(m)
# -
# 24. Write a Python program to append a list to the second list.
# +
l = [1, 2, 3, 4, 5]
m = ['a', 'b', 'c', 'd']
print(m + l)
m.extend(l)
print(m)
# -
# 25. Write a Python program to select an item randomly from a list.
# +
import random
l = [1, 2, 3, 4, 5, 6]
l[random.randint(0, len(l))]
# -
# 26. Write a python program to check whether two lists are circularly identical.
# +
list1 = [10, 10, 0, 0, 10]
list2 = [10, 10, 10, 0, 0]
list3 = [1, 10, 10, 0, 0]
def rotate(l, n=1):
t = l.pop(0)
l.append(t)
def linearly_circular(l1, l2):
for i in range(len(l1)):
rotate(l1)
if l1 == l2:
return True
return False
linearly_circular(list2, list3)
# +
list1 = [10, 10, 0, 0, 10]
list2 = [10, 10, 10, 0, 0]
list3 = [1, 10, 10, 0, 0]
def linearly_circular(l1, l2):
return ''.join(map(str, l1)) in ''.join(map(str, l2*2))
linearly_circular(list1, list2)
# -
# 27. Write a Python program to find the second smallest number in a list.
# +
import random
l = [random.randint(0, 100) for _ in range(100)]
l.sort()
print(l[1])
# -
# 28. Write a Python program to find the second largest number in a list.
# +
import random
l = [random.randint(0, 100) for _ in range(100)]
l.sort()
print(l[-2])
# -
# 29. Write a Python program to get unique values from a list.
# +
import random
l = [random.randint(0, 100) for _ in range(100)]
print(set(l))
# -
# 30. Write a Python program to get the frequency of the elements in a list.
# +
import random
import collections
l = [random.randint(0, 100) for _ in range(100)]
collections.Counter(l)
# -
# 31. Write a Python program to count the number of elements in a list within a specified range.
# +
import random
import collections
l = [random.randint(0, 100) for _ in range(100)]
def count_elements_in_range(l, min, max):
return collections.Counter([i for i in l if min < i < max])
count_elements_in_range(l, 40, 60)
# -
# 32. Write a Python program to check whether a list contains a sublist.
# +
l = [1, 2, 3, 4, 5]
s = [1, 2, 3]
print(set(s).issubset(set(l)) is not None)
print(set(s) <= set(l))
# -
# **Dictionary**
# http://www.w3resource.com/python-exercises/
#
| Beginning Python - Sequences, Mapping, Files, and Functions.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### Import all the relevant libraries
import numpy as np
import matplotlib.pyplot as plt
# ### load model
# +
from keras.models import load_model
from keras.applications.inception_v3 import preprocess_input
model = load_model('filename.model')
# -
# ### precict() function for prediction of class
def predict(image_path, model):
img = image.load_img(image_path, target_size = (299,299))
x = image.img_to_array(img)
x = np.expand_dims(x, axis = 0)
x = preprocess_input(x)
preds = model.predict(x)
if preds[0][0]<0.5:
return(str("PNEUMONIA"))
else:
return(str("NORMAL"))
# ### Predict the image using predict function
# +
from PIL import Image
from keras.preprocessing import image
img = image.load_img('person1946_bacteria_4875.jpeg', target_size = (299,299))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
x = preprocess_input(x)
preds = model.predict(x)
# -
preds
p = predict('person1946_bacteria_4875.jpeg', model)
p
| predict_class.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Problem 1
# With this code, we first open and read the lines from both archives. The status 'alligned' will be true unless the AGI locus codes from both archives don't match each row.
germplasm_data = open('Germplasm.tsv', 'r').readlines()
locusgene_data = open('LocusGene.tsv', 'r').readlines()
alligned = True
for line in range(len(germplasm_data)):
if germplasm_data[line].split('\t')[0] != locusgene_data[line].split('\t')[0]:
print('AGI locus code not alligned at line ', line)
alligned = False
if alligned:
print('All AGI locus codes are alligned')
# ## Problem 2
# Generate the database exam_2, containing the tables germplasm and locus_gene.
# Visualization
for line in germplasm_data[:4]:
print(line)
print('-'*60)
for line in locusgene_data[:4]:
print(line)
# %load_ext sql
# %config SqlMagic.autocommit=False
# %sql mysql+pymysql://root:root@127.0.0.1:3306/mysql
# #%sql drop database exam_2; #In case of restart
# %sql create database exam_2;
# %sql use exam_2
# %sql CREATE TABLE germplasm(locus VARCHAR(15) NOT NULL PRIMARY KEY, germplasm VARCHAR(50), phenotype VARCHAR(500), pubmed INTEGER);
# %sql CREATE TABLE locus_gene(locus VARCHAR(15) NOT NULL PRIMARY KEY, gene VARCHAR(50), protein_length INTEGER);
# %sql describe germplasm
# %sql describe locus_gene
# ## Problem 3
# Fill the databases. For each table, we insert the rows using a for loop.
# +
import pymysql.cursors
germplasm_data = open('Germplasm.tsv', 'r').readlines()
locusgene_data = open('LocusGene.tsv', 'r').readlines()
# Connect to the database
connection = pymysql.connect(host='localhost',
user='root',
password='<PASSWORD>',
db='exam_2',
charset='utf8mb4',
cursorclass=pymysql.cursors.DictCursor,
autocommit = True)
# Fill germplasm
try:
with connection.cursor() as cursor:
for line in germplasm_data[1:]:
line = line.strip()
locus, germplasm, phenotype, pubmed = line.split('\t')
sql = f'''INSERT INTO germplasm (locus, germplasm, phenotype, pubmed) VALUES ('{locus}', '{germplasm}', '{phenotype}', '{pubmed}');'''
cursor.execute(sql)
for line in locusgene_data[1:]:
line = line.strip()
locus, gene, protein_length = line.split('\t')
sql = f'''INSERT INTO locus_gene (locus, gene, protein_length) VALUES ('{locus}', '{gene}', '{protein_length}');'''
cursor.execute(sql)
finally:
print("Done")
connection.close()
# -
# Comprobation
# %sql select * from germplasm, locus_gene where germplasm.locus = locus_gene.locus;
# ## Problem 4
# 1. Create a report that shows the full, joined, content of the two database tables (including a header line).
# First, we create a connection to the mysql server. Then, we select both tables, joined by the locus code. Finally, using the DictWriter object, we store the data into a tsv file.
# +
import csv, io, pymysql
# Connect to the database
connection = pymysql.connect(host='localhost',
user='root',
password='<PASSWORD>',
db='exam_2',
charset='utf8mb4',
cursorclass=pymysql.cursors.DictCursor,
autocommit = True)
# Execute query and obtain the data
try:
with connection.cursor() as cursor:
sql = 'select * from germplasm, locus_gene where germplasm.locus = locus_gene.locus'
cursor.execute(sql)
results = cursor.fetchall()
except:
print("Extraction error")
# Write the report
report_file = open('report_exam2.tsv', 'w')
report_file.write('Problem 4.1\n')
writer = csv.DictWriter(report_file, delimiter = '\t', quotechar = '"', fieldnames = results[0].keys())
writer.writeheader()
writer.writerows(results)
report_file.close()
# -
# 2. Create a joined report that only includes the Genes SKOR and MAA3
# +
# Execute query and obtain the data (connexion is open from problem 4.1)
try:
with connection.cursor() as cursor:
sql = '''select * from germplasm, locus_gene where germplasm.locus = locus_gene.locus and (locus_gene.gene = "SKOR" or locus_gene.gene = "MAA3")'''
cursor.execute(sql)
results = cursor.fetchall()
except:
print("Extraction error")
# Write the report
report_file = open('report_exam2.tsv', 'a')
report_file.write('\nProblem 4.2\n')
writer = csv.DictWriter(report_file, delimiter = '\t', quotechar = '"', fieldnames = results[0].keys())
writer.writeheader()
writer.writerows(results)
report_file.close()
# -
# 3. Create a report that counts the number of entries for each Chromosome (AT1Gxxxxxx to AT5Gxxxxxxx)
# +
# Execute query and obtain the data
try:
with connection.cursor() as cursor:
sql = '''select locus from germplasm'''
cursor.execute(sql)
results = cursor.fetchall() # Here we have the loci from all entries at the germplasm table
except:
print("Extraction error")
report_file = open('report_exam2.tsv', 'a')
report_file.write('\nProblem 4.3\n')
chromo_list = list() #List of all chromosome numbers
for item in results:
chromo_list.append(int(item['locus'][2])) # Get the third character from all loci (chromosome number)
for number in range(1, 6):
report_file.write(f'Chromosome {number}: {chromo_list.count(number)} genes in the database\n')
report_file.close()
# -
# 4. Create a report that shows the average protein length for the genes on each Chromosome (AT1Gxxxxxx to AT5Gxxxxxxx)
# Execute query and obtain the data
report_file = open('report_exam2.tsv', 'a')
report_file.write('\nProblem 4.4\n')
try:
with connection.cursor() as cursor:
for number in range(1,6):
sql = f'''select avg(protein_length) from locus_gene where locus regexp '^..{number}' '''
cursor.execute(sql)
results = cursor.fetchall()
report_file.write(f'Average protein length in chromosome {number}: {list(results[0].values())[0]}\n')
except:
print("Extraction error")
connection.close()
report_file.close()
| Exams/.ipynb_checkpoints/Exam_2_Answers-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# This is hamoye Data science track introduction to python for machine learning code snippets
#
# # Lesson 1 no code snippet
# # Lesson 2 Numpy array and vectorization
#
#
# convention for importing numpy
import numpy as np
# +
arr = [6, 7, 8, 9]
print(type(arr)) # prints <class 'list'>
a = np.array(arr)
print(type(a)) # prints <class 'numpy.ndarray'>
print(a.shape) # prints (4, ) - a is a 1d array with 4 items
print(a.dtype) # prints int64
# get the dimension of a with ndim
print(a.ndim) # print's 1
b = np.array([
[3, 4, 5], [4, 5, 8]
])
print(b) # prints [[3 4 5] [4 5 8]]
print(b.ndim) # prints 2 - b is a 2d array
print(b.shape) # prints (2, 3) - b a 2d array with 2 rows and 3 columns
# -
# There are also inbuilt functions that can be used to initialize numpy which include empty(), zeros(), ones(), full(), random.random()
# +
# a 2x3 array with random values
print(np.random.random((2, 3))) # = array([[0.60793904, 0.02881965, 0.73022145], [0.34183628, 0.63274067, 0.07945224]])
# a 2x3 array of zeros
print(np.zeros((2, 3))) # = array([[0., 0., 0.], [0., 0., 0.]])
# a 2x3 array of ones
print(np.ones((2, 3))) # = array([[1., 1., 1.], [1., 1., 1]])
# a 3x3 identity matrix
print(np.zeros(3))# = array([[1., 0., 0.], [0., 0., 1.]])
# +
# Intra operability of arrays and scalars
c = np.array([[9.0, 8.0, 7.0], [1.0, 2.0, 3.0]])
d = np.array([[4.0, 5.0, 6.0], [9.0, 8.0, 7.0]])
print(c + d) # prints [[13. 13. 13.] [10. 10. 10.]]
print(5/d) # prints [[1.25 1. 0.83333333] [0.55555556 0.625 0.71428571]]
print(c ** 2) # prints [[81. 64. 49.] [ 1. 4. 9.]]
# -
# Indexing with arrays and using arrays for data processing
# +
print(d[1, 0:2]) # prints [9. 8.]
e = np.array([[10, 11, 12], [13, 14, 15], [16, 17, 18], [19, 20, 21]])
#print(e.shape, e.ndim)
# list slicing
print(e[:3, :2]) # prints 3 rows and 2 columns
# -
# There are other advanced methods of indexing which are shown below
# +
# Integer indexing
print(e[[2, 0, 3, 1], [2, 1, 0, 2]]) # prints [18 11 19 15]
# boolean indexing meeting a specified condition
print(e[e>15]) # prints [16 17 18 19 20 21]
# -
# # Lesson 3
# # Pandas - so much more than a cute animal
# it is a series of one dimensional array
#
# convention for importing pandas
import pandas as pd
# +
days = pd.Series(['Monday', 'Tuesday', 'Wodnesday', 'Thursday', 'Friday'])
days
# +
# using numpy array
list_days = np.array(['Monday', 'Tuesday', 'Wodnesday', 'Thursday', 'Friday'])
numpy_days = pd.Series(list_days)
numpy_days
# -
# using strings as index
d = pd.Series(['Monday', 'Tuesday', 'Wodnesday', 'Thursday', 'Friday'], ['a', 'b', 'c', 'd', 'e'])
d
# +
# create series from dictionary
d1 = pd.Series({'a':'Monday', 'b':'Tuesday', 'c':'Wednesday', 'd':'Thursday', 'e':'Friday'})
d1
# -
# Series can be accessed using specified index
d1[0]
d1[1:]
d1['c']
# # Pandas DataFrame
#
pd.DataFrame() # prints an empty dataframe
# +
# Create a dataframe from a dictionary
df_dict ={"Country": ['Ethiopia', 'Kenya', 'Nigeria', 'Ghana', 'Uganda'],
"Capital": ['Addis Ababa', 'Nirobi', 'Abuja', 'Accra', 'Kampala'],
"Population": [100000, 80500, 150000, 40000, 50000],
"Age": [60, 80, 70, 67, 90]}
df = pd.DataFrame(df_dict, index = [2, 4, 6, 8, 10])
df
# +
# create a dataframe from a list
df_list = [["Ethiopia", "<NAME>", 100000, 60],
["Kenya", "Nirobi", 805000, 80],
['Nigeria', 'Abuja', 150000, 70],
['Ghana', 'Accra', 40000, 67],
['Uganda', 'Kampala', 50000, 90]]
df1 = pd.DataFrame(df_list, columns=['Country', 'Capital', 'Population', 'Age'], index=[i+1 for i in range(len(df_list))])
df1
# -
# # at, iat, iloc, loc are accessors used to retrieve data in DataFrame
# Select the row at the index 0
df.iloc[0]
# select the Capital column
df['Capital']
# Select row with index label 6
df.loc[6]
# select single value with at label 6
df.at[6, 'Country']
# select single value using iat
df.iat[3, 0]
# # Statistical analysis
# find the sum of population
df['Population'].sum()
df.mean()
df.std()
df.median()
df.describe()
df.info()
# # missing value
# +
df_dict2 = {'Name':['Dejene', 'Asibeh', 'Tenager', np.nan],
'Profession':['Researcher', 'Software Engineer', 'Doctor', 'Data Scientist'],
'Experience':[7, np.nan, 8, 10],
'Height': [np.nan, 175, 180, 150]}
new_df = pd.DataFrame(df_dict2)
new_df
# -
# check for cells with missing values as True
new_df.isnull()
# remove rows with missing values
new_df.dropna()
# # Data types and Data wrangling
#
# - Working with different types of data: text files, CSV, JSON objects, HTML and databases
#
# Pandas can connect to databases, get data with queries and save in a dataframe
# impoting pandas library
import pandas as pd
# +
url = url='https://github.com/WalePhenomenon/climate_change/blob/master/fuel_ferc1.csv?raw=true'
fuel_df = pd.read_csv(url, error_bad_lines=False)
fuel_df.to_csv('fuel_data.csv', index=False)
# -
fuel_data = pd.read_csv('fuel_data.csv')
fuel_data.head()
fuel_data.describe(include='all')
# shows the skewness of the fuel data in two decimal points
round(fuel_data.skew(), 2)
# shows the kurtios of the fuel data in two decimal points
round(fuel_data.kurt(), 2)
# the correlation of the fuel data
fuel_data.corr()
# # Check for missing values
fuel_data.isnull().sum()
# Use groupby to count the sum of each unique value in the fuel unit column
fuel_data['fuel_count']= fuel_data.groupby('fuel_unit')['fuel_unit'].count()
fuel_count
fuel_data[['fuel_unit']] = fuel_data[['fuel_unit']].fillna(value='mcf')
# Check if missing values have been filled
fuel_data.isnull().sum()
# Count the number of report year
fuel_data.groupby('report_year')['report_year'].count()
# The average fuel_cost_per_unit_delivered in each year
fuel_data.groupby('report_year')['fuel_cost_per_unit_delivered'].mean()
# Merging in Pandas can be likened to join operations in relational databases like SQL.
# Group by the fuel type code and print the first entries in all the groups formed
fuel_data.groupby('fuel_type_code_pudl').first()
# Split the fuel data into two groups and merge using different methods
fuel_df1 = fuel_data.iloc[0:19000].reset_index(drop=True)
fuel_df2 = fuel_data.iloc[19000:].reset_index(drop=True)
# check that the length of both dataframes sum to the expexted length
assert len(fuel_data) == (len(fuel_df1) + len(fuel_df2))
# an inner merge will lose rows that do not match in both dataframes
pd.merge(fuel_df1, fuel_df2, how='inner')
# outer merge returns all rows in both dataframes
pd.merge(fuel_df1, fuel_df2, how='outer')
# removes rows from the right dataframe that do not have a match with the left and keeps all rows from the left
pd.merge(fuel_df1, fuel_df2, how='left')
# Concatenation is performed with the concat() function
data_to_concat = pd.DataFrame(np.zeros(fuel_data.shape))
pd.concat([fuel_data, data_to_concat]).reset_index(drop=True)
# Duplicates are a common occurrence in datasets which alter the results of analysis
# check for duplicate rows
fuel_data.duplicated().any()
# # Data Visualization and Representation in Python
# - The Anscombe Quartet and the importance of visualizing data
# +
# Import plotting library
import matplotlib.pyplot as plt
import seaborn as sns
plt.figure(figsize=(7, 4))
plt.xticks(rotation=90)
fuel_unit = pd.DataFrame({'unit': ['BBL', 'GAL', 'GRAMSU', 'KGU', 'MCF', 'MMBTU', 'MWDTH', 'MWHTH', 'TON'],
'count': [7998, 84, 464, 110, 11354, 180, 95, 100, 8958]})
sns.barplot(data=fuel_unit, x='unit', y='count')
plt.xlabel('Fuel Unit')
# -
# Because of the extreme range of the values for the fuel unit, we can plot the barchart by taking the logarithm of the y-axis as follows:
g = sns.barplot(data=fuel_unit, x='unit', y='count')
plt.xticks(rotation=90)
g.set_yscale('log')
g.set_ylim(1, 12000)
plt.xlabel('Fuel Unit')
# Select a sample of the dataset
sample_df = fuel_data.sample(n=50, random_state=4)
sns.regplot(x=sample_df['utility_id_ferc1'], y=sample_df['fuel_cost_per_mmbtu'], fit_reg=False)
# - Advanced plotting: Kerbel Density Estimate plots, box plots and violin plots
# box plot
sns.boxplot(x='fuel_type_code_pudl', y='utility_id_ferc1', palette=['m','g'], data=fuel_data)
# KDE plot
sns.kdeplot(sample_df['fuel_cost_per_unit_burned'], shade=True, color='b')
# A heatmap is a representation of data that uses a spectrum of colours to indicate different values. It gives quick summaries and identifies patterns especially in large datasets. Alternatively, heatmaps can be described as table visualisations where the colour of each cell relates the values. The image below is an example of a heatmap
sns.heatmap(sample_df.corr())
| SectionA.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
import matplotlib.pyplot as plt
npdizi = np.linspace(0,10,8)
npdizi2 = npdizi**3
# +
figur2 = plt.figure() #iç içe grafik oluşturma figure eksen döndürmez.
eksen1 = figur2.add_axes([0.1,0.1,0.9,0.9])
eksen2 = figur2.add_axes([0.3,0.6,0.3,0.3])
eksen1.set_xlabel("X EKSENİ")
eksen1.set_ylabel("Y EKSENİ")
eksen1.set_title("DIŞ GRAFİK")
eksen1.plot(npdizi,npdizi2,"g*-")
eksen2.set_xlabel("X Line")
eksen2.set_ylabel("Y Line")
eksen2.set_title("İÇ GRAFİK")
eksen2.plot(npdizi2,npdizi,"r--")
# -
eksen = plt.subplot() #burada eksen de gelir bize tuple döndürür
eksen.plot(npdizi,npdizi2,"b*-")
(eksen1,eksen2) = plt.subplots(1,2) #nrows satır sayısı, ncols = colon sayısı
for eksen in eksen2:
eksen.set_xlabel("x ekseni")
eksen.set_ylabel("y tarafı")
eksen.plot(npdizi,npdizi2,"g*-")
plt.tight_layout() #grafiklerin birbinee girmesini engeller
| Matplotlib 3.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
from copy import deepcopy
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import gym
import matplotlib.pyplot as plt
from tqdm.auto import tqdm
import pandas as pd
class ReplayBuffer():
def __init__(self, mem_size, batch_size, input_dims):
self.mem_size = mem_size
self.mem_centr = 0
self.batch_size = batch_size
self.state_memory = np.zeros(
(self.mem_size, *input_dims), dtype=np.float32)
self.new_state_memory = np.zeros(
(self.mem_size, *input_dims), dtype=np.float32)
self.action_memory = np.zeros(self.mem_size, dtype=np.int32)
self.reward_memory = np.zeros(self.mem_size, dtype=np.float32)
self.terminal_memory = np.zeros(self.mem_size, dtype=np.int32)
def store_transitions(self, state, action, reward, new_state, done):
index = self.mem_centr % self.mem_size
self.state_memory[index] = state
self.new_state_memory[index] = new_state
self.action_memory[index] = action
self.reward_memory[index] = reward
self.terminal_memory[index] = 1 - int(done)
self.mem_centr = self.mem_centr + 1
def is_sampleable(self):
if self.mem_centr >= self.batch_size:
return True
else:
return False
def sample_buffer(self):
if not(self.is_sampleable()):
return []
max_mem = min(self.mem_size, self.mem_centr)
batch = np.random.choice(max_mem, self.batch_size, replace=False)
states = self.state_memory[batch]
new_states = self.new_state_memory[batch]
actions = self.action_memory[batch]
rewards = self.reward_memory[batch]
terminals = self.terminal_memory[batch]
return states, new_states, actions, rewards, terminals
class NeuralNetwork(nn.Module):
def __init__(self, input_dims, n_actions):
super(NeuralNetwork, self).__init__()
self.layer1 = nn.Linear(input_dims[0], 100)
self.layer2 = nn.Linear(100, 100)
self.layer3 = nn.Linear(100, n_actions)
def forward(self, x):
l1 = self.layer1(x)
l1 = F.relu(l1)
l2 = self.layer2(l1)
l2 = F.relu(l2)
l3 = self.layer3(l2)
output = l3
return output
# +
device = 'cuda' if torch.cuda.is_available() else 'cpu'
## Force Use a Device
#device = 'cuda' #for GPU
#device = 'cpu' #for CPU
print(f'Using {device} device')
# -
class Agent():
def __init__(self, n_actions, input_dims,
lr=1e-4, gamma=0.9, mem_size=128, batch_size=64,
epsilon_decay=0.995):
self.n_actions = n_actions
self.input_dims = input_dims
self.gamma = gamma
self.epsilon_decay = epsilon_decay
self.batch_size = batch_size
self.policy_network = NeuralNetwork(input_dims=input_dims, n_actions=n_actions).to(device)
self.loss_function = nn.MSELoss()
self.optimizer = torch.optim.Adam(self.policy_network.parameters(), lr = lr)
self.replay_mem = ReplayBuffer(
mem_size=mem_size, batch_size=batch_size, input_dims=input_dims)
self.epsilon = 1
def choose_action(self, obs):
if np.random.random() < self.epsilon:
action = np.random.randint(self.n_actions)
else:
obs_T = torch.tensor(obs, device=device).float()
with torch.no_grad():
policy_values = self.policy_network(obs_T).cpu().detach().numpy()
action = np.argmax(policy_values)
return action
def store_memory(self, state, action, reward, new_state, done):
self.replay_mem.store_transitions(state, action, reward, new_state, done)
def train(self):
if not(self.replay_mem.is_sampleable()):
return 0
states, new_states, actions, rewards, dones = self.replay_mem.sample_buffer()
states_T = torch.tensor(states, device=device).float()
new_states_T = torch.tensor(new_states, device=device).float()
rewards_T = torch.tensor(rewards, device=device).float()
dones_T = torch.tensor(dones, device=device).float()
actions_T = torch.tensor(actions, device=device).type(torch.int64).unsqueeze(1)
q_eval = self.policy_network(states_T).gather(1, actions_T).squeeze(1)
with torch.no_grad():
q_next = self.policy_network(new_states_T).max(1)[0].detach()
q_target = rewards_T + ( self.gamma * q_next ) * dones_T
loss = self.loss_function(q_eval, q_target)
self.optimizer.zero_grad()
loss.backward()
self.optimizer.step()
self.epsilon = max(self.epsilon * self.epsilon_decay, 0.1)
return loss.item()
def save_model(self, file_path='./model/torch_dqn_model.model'):
torch.save(self.policy_network.state_dict(), file_path)
def load_model(self, file_path='./model/torch_dqn_model.model'):
self.policy_network.load_state_dict(torch.load(file_path))
# +
lr = 3e-4
gamma = 0.99
epsilon_decay = 1 - (2e-5)
episodes = 600
# -
mem_size = 1024
batch_size = 32
env = gym.make('LunarLander-v2')
agent = Agent(n_actions=env.action_space.n, input_dims=env.observation_space.shape,
lr=lr, gamma=gamma, mem_size=mem_size, batch_size=batch_size,
epsilon_decay=epsilon_decay)
scores = []
eps = []
losses = []
# +
pbar = tqdm(range(episodes))
for i in pbar:
done = False
score = 0
obs = env.reset()
ep_loss = []
while not(done):
action = agent.choose_action(obs)
new_obs, reward, done, _ = env.step(action)
#env.render()
score = score + reward
agent.store_memory(state=obs, action=action, reward=reward, new_state=new_obs, done=done)
obs = deepcopy(new_obs)
loss = agent.train()
ep_loss.append(loss)
scores.append(score)
eps.append(agent.epsilon)
losses.append(ep_loss)
pbar.set_description("Current_score = %s" % score)
# -
agent.save_model()
env.close()
plt.plot(eps, label="epsilon")
plt.legend()
plt.savefig('./plots/torch/dqn/epsilon.png')
plt.show()
losses_array = []
for x in losses:
losses_array.append(np.mean(np.array(x)))
plt.plot(losses_array, label="loss")
plt.legend()
plt.savefig('./plots/torch/dqn/losses.png')
plt.show()
# +
resolution = 50
cumsum_losses = np.array(pd.Series(np.array(losses_array)).rolling(window=resolution).mean() )
plt.plot(cumsum_losses, label="loss")
plt.legend()
plt.savefig('./plots/torch/dqn/losses_trend.png')
plt.show()
# -
plt.plot(scores, label="rewards")
plt.legend()
plt.savefig('./plots/torch/dqn/rewards.png')
plt.show()
# +
resolution = 50
cumsum_rewards = np.array(pd.Series(np.array(scores)).rolling(window=resolution).mean() )
plt.plot(cumsum_rewards, label="rewards")
plt.legend()
plt.savefig('./plots/torch/dqn/rewards_trend.png')
plt.show()
# +
test_env = gym.make('LunarLander-v2')
test_agent = Agent(n_actions=test_env.action_space.n, input_dims=test_env.observation_space.shape)
test_agent.epsilon = 0.0
test_agent.load_model()
# +
test_episodes = 10
pbar = tqdm(range(test_episodes))
for i in pbar:
done = False
score = 0
obs = test_env.reset()
test_env.render()
while not(done):
action = test_agent.choose_action(obs)
new_obs, reward, done, _ = test_env.step(action)
test_env.render()
score = score + reward
obs = deepcopy(new_obs)
pbar.set_description("Current_score = %s" % score)
print("score in episode ", (i+1) ," : ",score)
test_env.close()
| Week5/3_dqn_torch_lunar_lander.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [default]
# language: python
# name: python3
# ---
# + [markdown] id="wkNujasdKO2d"
# # Setup Google Colab
# + [markdown] id="c0RICCQHKvGI"
# https://colab.research.google.com/drive/1j7wXb-4bg4jU0uYeAoDGhguN9osRuage
# + id="ZzopSER3KN6N"
run_on_colab = True
# + [markdown] id="ZMn9CISXKM8V"
# # Download & Setup datasets
# + [markdown] id="55siaUvJKM8Y"
# Download Datasets
# > wget
# > -nc: to prevent redownload
# + colab={"base_uri": "https://localhost:8080/"} id="XRTL6ts4KM8Z" outputId="bde710c6-f5af-449e-bd37-91fdfc5ea5c5"
# !wget -nc "https://s3-us-west-1.amazonaws.com/udacity-aind/dog-project/lfw.zip"
# + colab={"base_uri": "https://localhost:8080/"} id="c0986GR9KM8a" outputId="af9f077e-109e-4b98-b475-5b679f9eda75"
# !wget -nc "https://s3-us-west-1.amazonaws.com/udacity-aind/dog-project/dogImages.zip"
# + [markdown] id="H-Oy49FuKM8a"
# unzip datasets
# > -n to prevent overwriting
# + colab={"base_uri": "https://localhost:8080/"} id="VbvN08r3KM8b" outputId="935de07c-b72d-4932-f16b-c2b919f9bd5b"
# !unzip -n "lfw.zip"
# + colab={"base_uri": "https://localhost:8080/"} id="28c2En6uKM8b" outputId="a78aa0d7-45a4-4050-bdb0-ec3efd081a28"
# !unzip -n "dogImages.zip"
# + [markdown] id="iExKjPIBKM8b"
# > remove unneeded folder
# + id="eT4aH3_NKM8c"
# !rm -r "./__MACOSX"
# + [markdown] id="LXmT4ja5pTNw"
# # Imports
# + id="gwLMaZwOpSh7"
import os
from glob import glob
from tqdm import tqdm
import numpy as np
import cv2
import matplotlib.pyplot as plt
import torch
import torch.optim as optim
import torch.nn as nn
from torchvision import datasets
import torchvision.models as models
import torchvision.transforms as transforms
from PIL import Image
from PIL import ImageFile
ImageFile.LOAD_TRUNCATED_IMAGES = True # Set PIL to be tolerant of image files that are truncated.
# + [markdown] id="tE9D0jDXKM8c"
# # Convolutional Neural Networks
#
# ## Project: Write an Algorithm for a Dog Identification App
#
# ---
#
# In this notebook, some template code has already been provided for you, and you will need to implement additional functionality to successfully complete this project. You will not need to modify the included code beyond what is requested. Sections that begin with **'(IMPLEMENTATION)'** in the header indicate that the following block of code will require additional functionality which you must provide. Instructions will be provided for each section, and the specifics of the implementation are marked in the code block with a 'TODO' statement. Please be sure to read the instructions carefully!
#
# > **Note**: Once you have completed all of the code implementations, you need to finalize your work by exporting the Jupyter Notebook as an HTML document. Before exporting the notebook to html, all of the code cells need to have been run so that reviewers can see the final implementation and output. You can then export the notebook by using the menu above and navigating to **File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission.
#
# In addition to implementing code, there will be questions that you must answer which relate to the project and your implementation. Each section where you will answer a question is preceded by a **'Question X'** header. Carefully read each question and provide thorough answers in the following text boxes that begin with **'Answer:'**. Your project submission will be evaluated based on your answers to each of the questions and the implementation you provide.
#
# >**Note:** Code and Markdown cells can be executed using the **Shift + Enter** keyboard shortcut. Markdown cells can be edited by double-clicking the cell to enter edit mode.
#
# The rubric contains _optional_ "Stand Out Suggestions" for enhancing the project beyond the minimum requirements. If you decide to pursue the "Stand Out Suggestions", you should include the code in this Jupyter notebook.
#
#
#
# ---
# ### Why We're Here
#
# In this notebook, you will make the first steps towards developing an algorithm that could be used as part of a mobile or web app. At the end of this project, your code will accept any user-supplied image as input. If a dog is detected in the image, it will provide an estimate of the dog's breed. If a human is detected, it will provide an estimate of the dog breed that is most resembling. The image below displays potential sample output of your finished project (... but we expect that each student's algorithm will behave differently!).
#
# 
#
# In this real-world setting, you will need to piece together a series of models to perform different tasks; for instance, the algorithm that detects humans in an image will be different from the CNN that infers dog breed. There are many points of possible failure, and no perfect algorithm exists. Your imperfect solution will nonetheless create a fun user experience!
#
# ### The Road Ahead
#
# We break the notebook into separate steps. Feel free to use the links below to navigate the notebook.
#
# * [Step 0](#step0): Import Datasets
# * [Step 1](#step1): Detect Humans
# * [Step 2](#step2): Detect Dogs
# * [Step 3](#step3): Create a CNN to Classify Dog Breeds (from Scratch)
# * [Step 4](#step4): Create a CNN to Classify Dog Breeds (using Transfer Learning)
# * [Step 5](#step5): Write your Algorithm
# * [Step 6](#step6): Test Your Algorithm
#
# ---
# <a id='step0'></a>
# ## Step 0: Import Datasets
#
# Make sure that you've downloaded the required human and dog datasets:
# * Download the [dog dataset](https://s3-us-west-1.amazonaws.com/udacity-aind/dog-project/dogImages.zip). Unzip the folder and place it in this project's home directory, at the location `/dogImages`.
#
# * Download the [human dataset](https://s3-us-west-1.amazonaws.com/udacity-aind/dog-project/lfw.zip). Unzip the folder and place it in the home directory, at location `/lfw`.
#
# *Note: If you are using a Windows machine, you are encouraged to use [7zip](http://www.7-zip.org/) to extract the folder.*
#
# In the code cell below, we save the file paths for both the human (LFW) dataset and dog dataset in the numpy arrays `human_files` and `dog_files`.
# + colab={"base_uri": "https://localhost:8080/"} id="-fObLuOtKM8d" outputId="8b0495fe-e8e1-49e2-f3e0-ba47a71a8fa1"
import numpy as np
from glob import glob
# load filenames for human and dog images
human_files = np.array(glob("./lfw/*/*"))
dog_files = np.array(glob("./dogImages/*/*/*"))
# print number of images in each dataset
print('There are %d total human images.' % len(human_files))
print('There are %d total dog images.' % len(dog_files))
# + id="Nl5oDCEVaYyl"
# check if CUDA is available
use_cuda = torch.cuda.is_available()
# + [markdown] id="khpLs6IQKM8e"
# <a id='step1'></a>
# ## Step 1: Detect Humans
#
# In this section, we use OpenCV's implementation of [Haar feature-based cascade classifiers](http://docs.opencv.org/trunk/d7/d8b/tutorial_py_face_detection.html) to detect human faces in images.
#
# OpenCV provides many pre-trained face detectors, stored as XML files on [github](https://github.com/opencv/opencv/tree/master/data/haarcascades). We have downloaded one of these detectors and stored it in the `haarcascades` directory. In the next code cell, we demonstrate how to use this detector to find human faces in a sample image.
# + colab={"base_uri": "https://localhost:8080/", "height": 286} id="du6Whtc-KM8e" outputId="4ace9c7d-61a1-462e-bd24-98604059d47c"
import cv2
import matplotlib.pyplot as plt
# %matplotlib inline
show_human_or_dog = True
# extract pre-trained face detector
if run_on_colab:
face_cascade = cv2.CascadeClassifier('haarcascade_frontalface_alt.xml')
else:
face_cascade = cv2.CascadeClassifier('haarcascades/haarcascade_frontalface_alt.xml')
# load color (BGR) image
if show_human_or_dog:
img = cv2.imread(human_files[0])
else:
img = cv2.imread(dog_files[0])
# convert BGR image to grayscale
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
# find faces in image
faces = face_cascade.detectMultiScale(gray)
# print number of faces detected in the image
print('Number of faces detected:', len(faces))
# get bounding box for each detected face
for (x,y,w,h) in faces:
# add bounding box to color image
cv2.rectangle(img,(x,y),(x+w,y+h),(255,0,0),2)
# convert BGR image to RGB for plotting
cv_rgb = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
# display the image, along with bounding box
plt.imshow(cv_rgb)
plt.show()
# + [markdown] id="M9SUIgbzKM8f"
# Before using any of the face detectors, it is standard procedure to convert the images to grayscale. The `detectMultiScale` function executes the classifier stored in `face_cascade` and takes the grayscale image as a parameter.
#
# In the above code, `faces` is a numpy array of detected faces, where each row corresponds to a detected face. Each detected face is a 1D array with four entries that specifies the bounding box of the detected face. The first two entries in the array (extracted in the above code as `x` and `y`) specify the horizontal and vertical positions of the top left corner of the bounding box. The last two entries in the array (extracted here as `w` and `h`) specify the width and height of the box.
#
# ### Write a Human Face Detector
#
# We can use this procedure to write a function that returns `True` if a human face is detected in an image and `False` otherwise. This function, aptly named `face_detector`, takes a string-valued file path to an image as input and appears in the code block below.
# + id="-J9xUNAoKM8f"
# returns "True" if face is detected in image stored at img_path
def face_detector(img_path):
img = cv2.imread(img_path)
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
faces = face_cascade.detectMultiScale(gray)
return len(faces) > 0
# + [markdown] id="nyDawVR3KM8f"
# ### (IMPLEMENTATION) Assess the Human Face Detector
#
# __Question 1:__ Use the code cell below to test the performance of the `face_detector` function.
# - What percentage of the first 100 images in `human_files` have a detected human face?
# - What percentage of the first 100 images in `dog_files` have a detected human face?
#
# Ideally, we would like 100% of human images with a detected face and 0% of dog images with a detected face. You will see that our algorithm falls short of this goal, but still gives acceptable performance. We extract the file paths for the first 100 images from each of the datasets and store them in the numpy arrays `human_files_short` and `dog_files_short`.
# + [markdown] id="dTZ1LF4JKM8f"
# __Answer:__
# (You can print out your results and/or write your percentages in this cell)
# + id="xH0q29oTKM8f"
from tqdm import tqdm
human_files_short = human_files[:100]
dog_files_short = dog_files[:100]
#-#-# Do NOT modify the code above this line. #-#-#
## TODO: Test the performance of the face_detector algorithm
## on the images in human_files_short and dog_files_short.
n_detected_humans_in_human_files = 0.0
n_detected_humans_in_dog_files = 0.0
for h in human_files_short:
n_detected_humans_in_human_files += face_detector(h)
for d in dog_files_short:
n_detected_humans_in_dog_files += face_detector(d)
human_accuracy = 100*n_detected_humans_in_human_files/len(human_files_short)
dog_accuracy = 100*n_detected_humans_in_dog_files/len(dog_files_short)
print(f"Human dataset : {human_accuracy:.2f} % humans detected")
print(f"Dog dataset : {dog_accuracy:.2f} % humans detected")
# + [markdown] id="8dFaFsBBKM8g"
# We suggest the face detector from OpenCV as a potential way to detect human images in your algorithm, but you are free to explore other approaches, especially approaches that make use of deep learning :). Please use the code cell below to design and test your own face detection algorithm. If you decide to pursue this _optional_ task, report performance on `human_files_short` and `dog_files_short`.
# + id="Dtt8YYB4KM8g"
### (Optional)
### TODO: Test performance of another face detection algorithm.
### Feel free to use as many code cells as needed.
# + [markdown] id="Wn9iksJ8KM8g"
# ---
# <a id='step2'></a>
# ## Step 2: Detect Dogs
#
# In this section, we use a [pre-trained model](http://pytorch.org/docs/master/torchvision/models.html) to detect dogs in images.
#
# ### Obtain Pre-trained VGG-16 Model
#
# The code cell below downloads the VGG-16 model, along with weights that have been trained on [ImageNet](http://www.image-net.org/), a very large, very popular dataset used for image classification and other vision tasks. ImageNet contains over 10 million URLs, each linking to an image containing an object from one of [1000 categories](https://gist.github.com/yrevar/942d3a0ac09ec9e5eb3a).
# + id="V4F-ePDzKM8g"
import torch
import torchvision.models as models
# define VGG16 model
VGG16 = models.vgg16(pretrained=True)
# check if CUDA is available
use_cuda = torch.cuda.is_available()
# move model to GPU if CUDA is available
if use_cuda:
print("Using CUDA")
VGG16 = VGG16.cuda()
else:
print("Using CPU")
# + [markdown] id="DqnGA2SUKM8h"
# Given an image, this pre-trained VGG-16 model returns a prediction (derived from the 1000 possible categories in ImageNet) for the object that is contained in the image.
# + [markdown] id="ubibrLSoKM8h"
# ### (IMPLEMENTATION) Making Predictions with a Pre-trained Model
#
# In the next code cell, you will write a function that accepts a path to an image (such as `'dogImages/train/001.Affenpinscher/Affenpinscher_00001.jpg'`) as input and returns the index corresponding to the ImageNet class that is predicted by the pre-trained VGG-16 model. The output should always be an integer between 0 and 999, inclusive.
#
# Before writing the function, make sure that you take the time to learn how to appropriately pre-process tensors for pre-trained models in the [PyTorch documentation](https://pytorch.org/vision/stable/models.html).
# + id="LniGcVIiKM8h"
from PIL import Image
import torchvision.transforms as transforms
# Set PIL to be tolerant of image files that are truncated.
from PIL import ImageFile
ImageFile.LOAD_TRUNCATED_IMAGES = True
def VGG16_predict(img_path):
'''
Use pre-trained VGG-16 model to obtain index corresponding to
predicted ImageNet class for image at specified path
Args:
img_path: path to an image
Returns:
Index corresponding to VGG-16 model's prediction
'''
## TODO: Complete the function.
## Load and pre-process an image from the given img_path
img = Image.open(img_path).convert('RGB')
in_transform = transforms.Compose([
transforms.Resize(224),
transforms.ToTensor(),
transforms.Normalize((0.485, 0.456, 0.406), (0.229, 0.224, 0.225)) # normalize according to pytorch documentation
])
# discard the transparent, alpha channel (that's the :3) and add the batch dimension
img = in_transform(img)[:3,:,:].unsqueeze(0)
img = img.cuda()
## Return the *index* of the predicted class for that image
pred = torch.argmax(VGG16(img).cpu()).item()
return pred
# + [markdown] id="PZhDQOclKM8h"
# ### (IMPLEMENTATION) Write a Dog Detector
#
# While looking at the [dictionary](https://gist.github.com/yrevar/942d3a0ac09ec9e5eb3a), you will notice that the categories corresponding to dogs appear in an uninterrupted sequence and correspond to dictionary keys 151-268, inclusive, to include all categories from `'Chihuahua'` to `'Mexican hairless'`. Thus, in order to check to see if an image is predicted to contain a dog by the pre-trained VGG-16 model, we need only check if the pre-trained model predicts an index between 151 and 268 (inclusive).
#
# Use these ideas to complete the `dog_detector` function below, which returns `True` if a dog is detected in an image (and `False` if not).
# + id="RRyWzUqXKM8h"
### returns "True" if a dog is detected in the image stored at img_path
def dog_detector(img_path):
## TODO: Complete the function.
pred = VGG16_predict(img_path)
return 151 <= pred <= 268 # true/false
# + id="AAZDqtrvKM8i"
# @AVTsoof: test VGG16_predict and dog_detector
with open("imagenet1000_clsidx_to_labels.txt") as f:
idx2label = eval(f.read())
dog_file = dog_files[100]
pred = VGG16_predict(dog_file)
is_dog = dog_detector(dog_file)
print(f"{idx2label[pred]} - is a dog? {is_dog}")
# + [markdown] id="sG8hfIKiKM8i"
# ### (IMPLEMENTATION) Assess the Dog Detector
#
# __Question 2:__ Use the code cell below to test the performance of your `dog_detector` function.
# - What percentage of the images in `human_files_short` have a detected dog?
# - What percentage of the images in `dog_files_short` have a detected dog?
# + [markdown] id="pXGEiXL6KM8i"
# __Answer:__
#
# + id="xQ9JJOvdKM8i" colab={"base_uri": "https://localhost:8080/"} outputId="dbfdd7a9-c148-4cf2-98ba-f7f08bf65162"
### TODO: Test the performance of the dog_detector function
### on the images in human_files_short and dog_files_short.
n_dogs_detected_in_human_files = 0.0
n_dogs_detected_in_dog_files = 0.0
for h in human_files_short:
n_dogs_detected_in_human_files += dog_detector(h)
for d in dog_files_short:
n_dogs_detected_in_dog_files += dog_detector(d)
human_accuracy = 100*n_dogs_detected_in_human_files/len(human_files_short)
dog_accuracy = 100*n_dogs_detected_in_dog_files/len(dog_files_short)
print(f"Human dataset : {human_accuracy:.2f} % dogs detected")
print(f"Dog dataset : {dog_accuracy:.2f} % dogs detected")
# + [markdown] id="I5GWkwfKKM8i"
# We suggest VGG-16 as a potential network to detect dog images in your algorithm, but you are free to explore other pre-trained networks (such as [Inception-v3](http://pytorch.org/docs/master/torchvision/models.html#inception-v3), [ResNet-50](http://pytorch.org/docs/master/torchvision/models.html#id3), etc). Please use the code cell below to test other pre-trained PyTorch models. If you decide to pursue this _optional_ task, report performance on `human_files_short` and `dog_files_short`.
# + id="APmJOJlEKM8i"
### (Optional)
### TODO: Report the performance of another pre-trained network.
### Feel free to use as many code cells as needed.
# + [markdown] id="hgopkem0KM8j"
# ---
# <a id='step3'></a>
# ## Step 3: Create a CNN to Classify Dog Breeds (from Scratch)
#
# Now that we have functions for detecting humans and dogs in images, we need a way to predict breed from images. In this step, you will create a CNN that classifies dog breeds. You must create your CNN _from scratch_ (so, you can't use transfer learning _yet_!), and you must attain a test accuracy of at least 10%. In Step 4 of this notebook, you will have the opportunity to use transfer learning to create a CNN that attains greatly improved accuracy.
#
# We mention that the task of assigning breed to dogs from images is considered exceptionally challenging. To see why, consider that *even a human* would have trouble distinguishing between a Brittany and a Welsh Springer Spaniel.
#
# Brittany | Welsh Springer Spaniel
# - | -
# <img src="images/Brittany_02625.jpg" width="100"> | <img src="images/Welsh_springer_spaniel_08203.jpg" width="200">
#
# It is not difficult to find other dog breed pairs with minimal inter-class variation (for instance, Curly-Coated Retrievers and American Water Spaniels).
#
# Curly-Coated Retriever | American Water Spaniel
# - | -
# <img src="images/Curly-coated_retriever_03896.jpg" width="200"> | <img src="images/American_water_spaniel_00648.jpg" width="200">
#
#
# Likewise, recall that labradors come in yellow, chocolate, and black. Your vision-based algorithm will have to conquer this high intra-class variation to determine how to classify all of these different shades as the same breed.
#
# Yellow Labrador | Chocolate Labrador | Black Labrador
# - | -
# <img src="images/Labrador_retriever_06457.jpg" width="150"> | <img src="images/Labrador_retriever_06455.jpg" width="240"> | <img src="images/Labrador_retriever_06449.jpg" width="220">
#
# We also mention that random chance presents an exceptionally low bar: setting aside the fact that the classes are slightly imabalanced, a random guess will provide a correct answer roughly 1 in 133 times, which corresponds to an accuracy of less than 1%.
#
# Remember that the practice is far ahead of the theory in deep learning. Experiment with many different architectures, and trust your intuition. And, of course, have fun!
#
# ### (IMPLEMENTATION) Specify Data Loaders for the Dog Dataset
#
# Use the code cell below to write three separate [data loaders](http://pytorch.org/docs/stable/data.html#torch.utils.data.DataLoader) for the training, validation, and test datasets of dog images (located at `dogImages/train`, `dogImages/valid`, and `dogImages/test`, respectively). You may find [this documentation on custom datasets](http://pytorch.org/docs/stable/torchvision/datasets.html) to be a useful resource. If you are interested in augmenting your training and/or validation data, check out the wide variety of [transforms](http://pytorch.org/docs/stable/torchvision/transforms.html?highlight=transform)!
# + id="crQLvAqcKM8j"
import os
from torchvision import datasets
import torchvision.transforms as transforms
### TODO: Write data loaders for training, validation, and test sets
## Specify appropriate transforms, and batch_sizes
# prepare data loaders (combine dataset and sampler)
batch_size = 40
train_transform = transforms.Compose([
transforms.Resize(256),
transforms.CenterCrop(224),
transforms.RandomHorizontalFlip(),
transforms.RandomRotation(10),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))
])
valid_transform = transforms.Compose([
transforms.Resize(256),
transforms.CenterCrop(224),
transforms.RandomHorizontalFlip(),
transforms.RandomRotation(10),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))
])
test_transform = transforms.Compose([
transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
train_data = datasets.ImageFolder("dogImages/train", transform=train_transform)
valid_data = datasets.ImageFolder("dogImages/valid", transform=valid_transform)
test_data = datasets.ImageFolder("dogImages/test", transform=test_transform)
train_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size, shuffle=True)
valid_loader = torch.utils.data.DataLoader(valid_data, batch_size=batch_size, shuffle=True)
test_loader = torch.utils.data.DataLoader(test_data, batch_size=batch_size, shuffle=True)
loaders_scratch = {"train": train_loader, "valid": valid_loader, "test": test_loader}
# + [markdown] id="wVA8Y1SuKM8j"
# **Question 3:** Describe your chosen procedure for preprocessing the data.
# - How does your code resize the images (by cropping, stretching, etc)? What size did you pick for the input tensor, and why?
# - Did you decide to augment the dataset? If so, how (through translations, flips, rotations, etc)? If not, why not?
#
# + [markdown] id="SXHSCG8FKM8j"
# **Answer**:
# + [markdown] id="jum-XotUKM8j"
# ### (IMPLEMENTATION) Model Architecture
#
# Create a CNN to classify dog breed. Use the template in the code cell below.
# + id="IsewwAOpKM8k"
import torch.nn as nn
import torch.nn.functional as F
# define the CNN architecture
class Net(nn.Module):
### TODO: choose an architecture, and complete the class
def __init__(self, imsize=224, n_classes=133):
super(Net, self).__init__()
## Define layers of a CNN
# note: based on VGG16 arch
self.layers = nn.Sequential(
nn.Conv2d(3, 64, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.Conv2d(64, 64, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.Conv2d(64, 128, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.MaxPool2d(2, 2), # imsize=112
nn.Conv2d(128, 128, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.Conv2d(128, 256, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.MaxPool2d(2, 2), # imsize=56
nn.Conv2d(256, 256, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.Conv2d(256, 256, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.Conv2d(256, 512, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.MaxPool2d(2, 2), # imsize=28
nn.Conv2d(512, 512, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.Conv2d(512, 512, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.Conv2d(512, 512, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.MaxPool2d(2, 2), # imsize=14
nn.Conv2d(512, 512, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.Conv2d(512, 512, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.Conv2d(512, 512, kernel_size=3, padding=1),
nn.LeakyReLU(),
nn.MaxPool2d(2, 2), # imsize=7
nn.Flatten(),
nn.Linear(7 * 7 * 512, 4096),
nn.LeakyReLU(),
nn.Dropout(p=0.2),
nn.Linear(4096, 4096),
nn.LeakyReLU(),
nn.Dropout(p=0.2),
nn.Linear(4096, n_classes),
nn.Softmax(dim=1) # dim=1 for summing accross classes per example (dim 0 holds examples, dim 1 holds classes)
)
def forward(self, x):
## Define forward behavior
for l in self.layers:
x = l(x)
return x
#-#-# You do NOT have to modify the code below this line. #-#-#
# instantiate the CNN
try:
model_scratch = None
del model_scratch
except:
pass
model_scratch = Net()
# + id="I-oQzjACMgLY"
# Test model
TEST_SCRATCH_MODEL = False
if TEST_SCRATCH_MODEL:
data, target = next(iter(train_loader))
y = model_scratch(data)
print(data.size(), target.size(), y.size())
y = None
del y
# + [markdown] id="t17Jf7ZbKM8k"
# __Question 4:__ Outline the steps you took to get to your final CNN architecture and your reasoning at each step.
# + [markdown] id="wxQT35ClKM8k"
# __Answer:__
# Based on the VGG16 architecture, but with fewer layer for less parameters.
# + [markdown] id="euvf0IovKM8k"
# ### (IMPLEMENTATION) Specify Loss Function and Optimizer
#
# Use the next code cell to specify a [loss function](http://pytorch.org/docs/stable/nn.html#loss-functions) and [optimizer](http://pytorch.org/docs/stable/optim.html). Save the chosen loss function as `criterion_scratch`, and the optimizer as `optimizer_scratch` below.
# + id="eiBl29kzKM8k"
import torch.optim as optim
### TODO: select loss function
criterion_scratch = nn.CrossEntropyLoss()
### TODO: select optimizer
optimizer_scratch = optim.Adam(params=model_scratch.parameters(), lr=0.1)
# + [markdown] id="cezQnwTPKM8k"
# ### (IMPLEMENTATION) Train and Validate the Model
#
# Train and validate your model in the code cell below. [Save the final model parameters](http://pytorch.org/docs/master/notes/serialization.html) at filepath `'model_scratch.pt'`.
# + id="WBsoIIBNKM8k"
# the following import is required for training to be robust to truncated images
from PIL import ImageFile
ImageFile.LOAD_TRUNCATED_IMAGES = True
def train(n_epochs, loaders, model, optimizer, criterion, use_cuda, save_path):
"""returns trained model"""
# initialize tracker for minimum validation loss
valid_loss_min = np.Inf
for epoch in range(1, n_epochs+1):
# initialize variables to monitor training and validation loss
train_loss = 0.0
valid_loss = 0.0
correct = 0.0
total = 0.0
###################
# train the model #
###################
print() # newline
n_train_batches = len(loaders['train'])
model.train()
for batch_idx, (data, target) in enumerate(loaders['train']):
# move to GPU
if use_cuda:
data, target = data.cuda(), target.cuda()
## find the loss and update the model parameters accordingly
optimizer.zero_grad()
logits = model(data)
loss = criterion(logits, target)
loss.backward()
optimizer.step()
## record the average training loss
train_loss = train_loss + ((1 / (batch_idx + 1)) * (loss.data - train_loss))
print('\rEpoch: {}/{} \tTraining Batch: {}/{} \tTraining Loss: {:.6f} '.format(
epoch, n_epochs,
batch_idx+1, n_train_batches,
train_loss
), end='')
######################
# validate the model #
######################
print() # newline
n_valid_batches = len(loaders['valid'])
model.eval()
with torch.no_grad():
for batch_idx, (data, target) in enumerate(loaders['valid']):
# move to GPU
if use_cuda:
data, target = data.cuda(), target.cuda()
## update the average validation loss
logits = model(data)
loss = criterion(logits, target)
valid_loss = valid_loss + ((1 / (batch_idx + 1)) * (loss.data - valid_loss))
# convert output probabilities to predicted class
pred = logits.data.max(1, keepdim=True)[1]
# compare predictions to true label
correct += np.sum(np.squeeze(pred.eq(target.data.view_as(pred))).cpu().numpy())
total += data.size(0)
print('\rEpoch: {}/{} \tValidation Batch: {}/{} \tValidation Loss: {:.6f} '.format(
epoch, n_epochs,
batch_idx+1, n_valid_batches,
valid_loss
), end='')
# print training/validation statistics
print() # newline
print('# Epoch: {}/{} \tTraining Loss: {:.6f} \tValidation Loss: {:.6f} \tTest Accuracy: {:.2f}% ({}/{})'.format(
epoch, n_epochs,
train_loss,
valid_loss,
100. * correct / total, int(correct), int(total),
))
## TODO: save the model if validation loss has decreased
if valid_loss < valid_loss_min:
valid_loss_min = valid_loss
torch.save(model.state_dict(), save_path)
print(f"Valid Loss decreased - Saved model checkpoint to: {save_path}")
# return trained model
return model
# + colab={"base_uri": "https://localhost:8080/", "height": 234} id="ctlkC4AaWpEP" outputId="0dd0610c-4cca-422c-f6c6-e8bd9b247310"
# move tensors to GPU if CUDA is available
if use_cuda:
torch.cuda.empty_cache()
model_scratch.cuda()
# train the model
model_scratch = train(5, loaders_scratch, model_scratch, optimizer_scratch,
criterion_scratch, use_cuda, 'model_scratch.pt')
# load the model that got the best validation accuracy
model_scratch.load_state_dict(torch.load('model_scratch.pt'))
# + [markdown] id="CiMzoT7vKM8l"
# ### (IMPLEMENTATION) Test the Model
#
# Try out your model on the test dataset of dog images. Use the code cell below to calculate and print the test loss and accuracy. Ensure that your test accuracy is greater than 10%.
# + id="h6u0AvqBKM8l"
def test(loaders, model, criterion, use_cuda):
# monitor test loss and accuracy
test_loss = 0.
correct = 0.
total = 0.
model.eval()
for batch_idx, (data, target) in enumerate(loaders['test']):
# move to GPU
if use_cuda:
data, target = data.cuda(), target.cuda()
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the loss
loss = criterion(output, target)
# update average test loss
test_loss = test_loss + ((1 / (batch_idx + 1)) * (loss.data - test_loss))
# convert output probabilities to predicted class
pred = output.data.max(1, keepdim=True)[1]
# compare predictions to true label
correct += np.sum(np.squeeze(pred.eq(target.data.view_as(pred))).cpu().numpy())
total += data.size(0)
print('Test Loss: {:.6f}\n'.format(test_loss))
print('\nTest Accuracy: %2d%% (%2d/%2d)' % (
100. * correct / total, correct, total))
# + id="zPcRobmOWquo"
# call test function
test(loaders_scratch, model_scratch, criterion_scratch, use_cuda)
# + id="WLIOag22Tch9"
del model_scratch
torch.cuda.empty_cache()
# + [markdown] id="qeZFIVBwKM8l"
# ---
# <a id='step4'></a>
# ## Step 4: Create a CNN to Classify Dog Breeds (using Transfer Learning)
#
# You will now use transfer learning to create a CNN that can identify dog breed from images. Your CNN must attain at least 60% accuracy on the test set.
#
# ### (IMPLEMENTATION) Specify Data Loaders for the Dog Dataset
#
# Use the code cell below to write three separate [data loaders](http://pytorch.org/docs/master/data.html#torch.utils.data.DataLoader) for the training, validation, and test datasets of dog images (located at `dogImages/train`, `dogImages/valid`, and `dogImages/test`, respectively).
#
# If you like, **you are welcome to use the same data loaders from the previous step**, when you created a CNN from scratch.
# + id="uyn2wa-qKM8l"
import os
from torchvision import datasets
import torchvision.transforms as transforms
batch_size = 40
train_transform = transforms.Compose([
transforms.Resize(256),
transforms.CenterCrop(224),
transforms.RandomHorizontalFlip(),
transforms.RandomRotation(10),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])
])
valid_transform = transforms.Compose([
transforms.Resize(256),
transforms.CenterCrop(224),
transforms.RandomHorizontalFlip(),
transforms.RandomRotation(10),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])
])
test_transform = transforms.Compose([
transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])
])
train_data = datasets.ImageFolder("dogImages/train", transform=train_transform)
valid_data = datasets.ImageFolder("dogImages/valid", transform=valid_transform)
test_data = datasets.ImageFolder("dogImages/test", transform=test_transform)
train_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size, shuffle=True)
valid_loader = torch.utils.data.DataLoader(valid_data, batch_size=batch_size, shuffle=True)
test_loader = torch.utils.data.DataLoader(test_data, batch_size=batch_size, shuffle=True)
loaders_transfer = {"train": train_loader, "valid": valid_loader, "test": test_loader}
# + [markdown] id="gZo25lmuKM8l"
# ### (IMPLEMENTATION) Model Architecture
#
# Use transfer learning to create a CNN to classify dog breed. Use the code cell below, and save your initialized model as the variable `model_transfer`.
# + id="WoFQ1L6XKM8l" colab={"base_uri": "https://localhost:8080/"} outputId="574b91d7-284d-4757-e0c2-3eba1710f942"
import torchvision.models as models
import torch.nn as nn
## TODO: Specify model architecture
model_transfer = torch.hub.load('pytorch/vision:v0.10.0', 'resnet50', pretrained=True)
# + id="yZDlqWmtUEeQ"
model_transfer
# + id="YILbxG5kUDQZ"
# freeze features
for param in model_transfer.parameters():
param.requires_grad = False
# + id="_nPr8A_Ve_c5" colab={"base_uri": "https://localhost:8080/"} outputId="eecabe24-3d29-43ac-e6ac-cb49d06eb0ee"
model_transfer.fc
# + id="YLzTY7RZe6gB"
# replace classifier output to 133 classes (number of dog breeds in the dataset)
n_classes = 133
model_transfer.fc = nn.Linear(in_features=model_transfer.fc.in_features, out_features=n_classes)
# move to GPU
if use_cuda:
model_transfer = model_transfer.cuda()
# + [markdown] id="RrUDwc-4KM8l"
# __Question 5:__ Outline the steps you took to get to your final CNN architecture and your reasoning at each step. Describe why you think the architecture is suitable for the current problem.
# + [markdown] id="Jubn5tCBKM8l"
# __Answer:__
#
# + [markdown] id="hIlAsBeAKM8l"
# ### (IMPLEMENTATION) Specify Loss Function and Optimizer
#
# Use the next code cell to specify a [loss function](http://pytorch.org/docs/master/nn.html#loss-functions) and [optimizer](http://pytorch.org/docs/master/optim.html). Save the chosen loss function as `criterion_transfer`, and the optimizer as `optimizer_transfer` below.
# + id="RkFiWa1hKM8l"
import torch.optim as optim
criterion_transfer = nn.CrossEntropyLoss()
optimizer_transfer = optim.Adam(params=model_transfer.fc.parameters(), lr=0.003)
# + [markdown] id="aVMc9oxGKM8m"
# ### (IMPLEMENTATION) Train and Validate the Model
#
# Train and validate your model in the code cell below. [Save the final model parameters](http://pytorch.org/docs/master/notes/serialization.html) at filepath `'model_transfer.pt'`.
# + id="IoNPFl4FKM8m" colab={"base_uri": "https://localhost:8080/"} outputId="fe4e064d-de8e-4d0c-9134-8d1abf583fc3"
# train the model
n_epochs = 5
model_transfer = train(n_epochs, loaders_transfer, model_transfer, optimizer_transfer, criterion_transfer, use_cuda, 'model_transfer.pt')
# + [markdown] id="xRvcP4ChKM8m"
# ### (IMPLEMENTATION) Test the Model
#
# Try out your model on the test dataset of dog images. Use the code cell below to calculate and print the test loss and accuracy. Ensure that your test accuracy is greater than 60%.
# + colab={"base_uri": "https://localhost:8080/"} id="8uT6vu__bls8" outputId="967d7133-eba1-49f2-abc6-2e8bbec1825a"
# load the model that got the best validation accuracy (uncomment the line below)
model_transfer.load_state_dict(torch.load('model_transfer.pt'))
# + id="7Fq-aD_HKM8m" colab={"base_uri": "https://localhost:8080/"} outputId="1f26dc4d-c32e-4a46-9834-d68b2db162f0"
test(loaders_transfer, model_transfer, criterion_transfer, use_cuda)
# + [markdown] id="e8dUolwJKM8m"
# ### (IMPLEMENTATION) Predict Dog Breed with the Model
#
# Write a function that takes an image path as input and returns the dog breed (`Affenpinscher`, `Afghan hound`, etc) that is predicted by your model.
# + colab={"base_uri": "https://localhost:8080/"} id="lyAgqgH8kRAR" outputId="3f805ed9-b31d-461d-fb46-93dd167ddea1"
# example to extract class names and number
dir_idx = 0
print(glob("dogImages/train/*"))
print(glob("dogImages/train/*")[dir_idx])
print(glob("dogImages/train/*")[dir_idx].split("/"))
print(glob("dogImages/train/*")[dir_idx].split("/")[-1])
print(glob("dogImages/train/*")[dir_idx].split("/")[-1].split("."))
c = glob("dogImages/train/*")[dir_idx].split("/")[-1].split(".")
c_dict = {int(c[0]): c[1]}
print(c_dict)
# + id="lYWrRZsVl3bO"
# create class names dict
class_dirs = glob("dogImages/train/*")
class_names = [c.split("/")[-1].split(".") for c in class_dirs]
class_names = {int(c[0]): c[1] for c in class_names}
class_names
# + id="mWZoFFCAKM8m"
### TODO: Write a function that takes a path to an image as input
### and returns the dog breed that is predicted by the model.
# list of class names by index, i.e. a name can be accessed like class_names[0]
# class_names = [item[4:].replace("_", " ") for item in data_transfer['train'].classes]
def predict_breed_transfer(img_path):
# load the image and return the predicted breed
img = Image.open(img_path).convert('RGB')
in_transform = transforms.Compose([
transforms.Resize(225),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize((0.485, 0.456, 0.406), (0.229, 0.224, 0.225)) # normalize according to pytorch documentation
])
# discard the transparent, alpha channel (that's the :3) and add the batch dimension
img = in_transform(img)[:3,:,:].unsqueeze(0)
if use_cuda:
img = img.cuda()
## Return the *index* of the predicted class for that image
with torch.no_grad():
pred = torch.argmax(model_transfer(img).cpu()).item()
return class_names[pred+1]
# + colab={"base_uri": "https://localhost:8080/", "height": 35} id="6T40iok7m9Id" outputId="017db65a-20f1-432f-a51f-277b2974b372"
# test predict_breed_transfer() function
predict_breed_transfer("dogImages/test/001.Affenpinscher/Affenpinscher_00003.jpg")
# + [markdown] id="BXWxm6sLKM8m"
# ---
# <a id='step5'></a>
# ## Step 5: Write your Algorithm
#
# Write an algorithm that accepts a file path to an image and first determines whether the image contains a human, dog, or neither. Then,
# - if a __dog__ is detected in the image, return the predicted breed.
# - if a __human__ is detected in the image, return the resembling dog breed.
# - if __neither__ is detected in the image, provide output that indicates an error.
#
# You are welcome to write your own functions for detecting humans and dogs in images, but feel free to use the `face_detector` and `dog_detector` functions developed above. You are __required__ to use your CNN from Step 4 to predict dog breed.
#
# Some sample output for our algorithm is provided below, but feel free to design your own user experience!
#
# 
#
#
# ### (IMPLEMENTATION) Write your Algorithm
# + id="qkyulycVKM8m"
### TODO: Write your algorithm.
### Feel free to use as many code cells as needed.
def run_app(img_path):
img_contains = None
breed = None
## handle cases for a human face, dog, and neither
if face_detector(img_path):
img_contains = "human"
elif dog_detector(img_path):
img_contains = "dog"
breed = predict_breed_transfer(img_path)
if img_contains == "human":
print(f"This human looks like a '{breed}'")
elif img_contains == "dog":
print(f"This dog is a '{breed}'")
else:
print("APP ERROR: No dog or human detected!")
# + [markdown] id="pF0v9CwPKM8m"
# ---
# <a id='step6'></a>
# ## Step 6: Test Your Algorithm
#
# In this section, you will take your new algorithm for a spin! What kind of dog does the algorithm think that _you_ look like? If you have a dog, does it predict your dog's breed accurately? If you have a cat, does it mistakenly think that your cat is a dog?
#
# ### (IMPLEMENTATION) Test Your Algorithm on Sample Images!
#
# Test your algorithm at least six images on your computer. Feel free to use any images you like. Use at least two human and two dog images.
#
# __Question 6:__ Is the output better than you expected :) ? Or worse :( ? Provide at least three possible points of improvement for your algorithm.
# + [markdown] id="AetqaHLTKM8n"
# __Answer:__ (Three possible points for improvement)
# + colab={"base_uri": "https://localhost:8080/", "height": 182} id="v_MN-SbitI8O" outputId="cd89f4ce-2683-462b-eb33-0bed696c19ca"
n_min_files = min(len(dog_files), len(human_files))
np.random.randint(0, n_min_files-1, (6,1))
# + id="GJ5C4_AKKM8n" colab={"base_uri": "https://localhost:8080/"} outputId="1e6c7ec0-d6fc-4b8f-a694-27a0011fd1a0"
## TODO: Execute your algorithm from Step 6 on
## at least 6 images on your computer.
## Feel free to use as many code cells as needed.
## suggested code, below
for file in np.hstack((human_files[:3], dog_files[:3])):
run_app(file)
# + id="NodS9qYvscdV"
| project-dog-classification/dog_app.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# + [markdown] heading_collapsed=true
# # PA005: High Value Customer Identification ( Insiders )
# + [markdown] hidden=true
# ## 0.0 Planejamento da Solução( IOT )
# + [markdown] heading_collapsed=true hidden=true
# ### Input - Entrada
# + [markdown] hidden=true
# 1. Problema de Negócio
# - Selecionar os clientes mais valiosos para integrar um programa de Fidelização.
#
# 2. Conjunto de Dados
# - Vendas de um e-commerce online, durante o período de um ano.
# + [markdown] heading_collapsed=true hidden=true
# ### Outpout - Saída
# + [markdown] hidden=true
# 1. indicação das pessoas que farão parte do programa de Insiders
# - Lista: client_id|is_insider|
#
# 10323 | yes/1
# 32413 | no/1
#
# 2. Relatório com as respostas das perguntas de negócio.
#
# - Quem são as pessoas elegíveis para participar do programa de
# Insiders ?
# - Quantos clientes farão parte do grupo?
# - Quais as principais características desses clientes ?
# - Qual a porcentagem de contribuição do faturamento, vinda do
# Insiders ?
# - Qual a expectativa de faturamento desse grupo para os próximos
# meses ?
# - Quais as condições para uma pessoa ser elegível ao Insiders ?
# - Quais as condições para uma pessoa ser removida do Insiders ?
# - Qual a garantia que o programa Insiders é melhor que o restante da
# base ?
# - Quais ações o time de marketing pode realizar para aumentar o
# # faturamento?
# + [markdown] hidden=true
# ### Tasks - Tarefas
# + [markdown] hidden=true
# 1. Quem são as pessoas elegíveis para participar do programa de
# Insiders ?
# - O que é ser elegível ? O que são clientes de maior "valor"
# - Faturamento:
# - Alto Ticket médio.
# - Alto LTV(receita média por cliente durante o relacionamento com a empresa.)
# - Baixa Recência(tempo desde a ultima compra)
# - Alto bascket size( Tamanho da sexta de compra)
# - Baixa probabilidade de churn(Cliente para de comprar).
#
#
# - Custo:
# - Baixa Taxa de devolução.
#
#
# - Experiência de Compra:
# - Média alta das avaliação.
#
#
# 2. Quantos clientes farão parte do grupo?
# - Número total de clientes.
# - % do grupo Insiders
#
# 3. Quais as principais características desses clientes ?
# - Escrever características do cliente:
# - Idade
# - Localização.
#
# - Escrever características do consumo.
# - Atributos da clusterização.
#
# 4. Qual a porcentagem de contribuição do faturamento, vinda do Insiders ?
# - Faturamento total do ano.
# - Faturamento do grupo de Insiders
#
# 5. Qual a expectativa de faturamento desse grupo para os próximos meses ?
# - LTV do grupo Insiders
# - Análise de cohort()
#
# 6. Quais as condições para uma pessoa ser elegível ao Insiders ?
# - Definir a periodicidade( 1 mês, 3 mêses )
# - A pessoa precisa ser similar ou parecido com uma pessoa grupo.
#
# 7. Quais as condições para uma pessoa ser removida do Insiders ?
# - Definir a periodicidade( 1 mês, 3 mêses )
# - A pessoa precisa ser desimilar ou não-parecido com uma pessoa grupo.
#
# 8. Qual a garantia que o programa Insiders é melhor que o restante da base ?
# - Teste A/B
# - Teste A/B Baysiano
# - Teste de hipótese
#
# 9. Quais ações o time de marketing pode realizar para aumentar o faturamento?
# - Desconto
# - Preferencia de compra
# - Frente
# - Visita a empresa
# -
# # 0.0 Imports
# ## 0.1. Libries
# +
import pandas as pd
import numpy as np
import seaborn as sns
import umap.umap_ as umap
from matplotlib import pyplot as plt
from sklearn import cluster as c
from sklearn.cluster import KMeans
from yellowbrick.cluster import KElbowVisualizer, SilhouetteVisualizer
from sklearn import metrics as m
from plotly import express as px
import warnings
warnings.filterwarnings('ignore')
# -
# ## 0.2. Helper Function
# + [markdown] heading_collapsed=true
# ## 0.3. Load Dataset
# + hidden=true
#Load data
df_raw = pd.read_csv('data/Ecommerce.csv', sep=',', encoding = "ISO-8859-1")
#Drop extra column
df_raw = df_raw.drop( columns = ['Unnamed: 8'], axis = 1 )
# + hidden=true
df_raw.head()
# -
# # 1.0. Descrição dos Dados
df1 = df_raw.copy()
# ## 1.1 Rename Columuns
df1.columns
cols_new= ['invoice_no', 'stock_code', 'description', 'quantity', 'invoice_date', 'unit_price', 'customer_id', 'country']
df1.columns = cols_new
df1.columns
# ## 1.2. Data Dimensions
print( 'Number of Rows: {}'.format( df1.shape[0] ) )
print( 'Number of cols: {}'.format( df1.shape[1] ) )
# ## 1.3. Data Types
df1.dtypes
# + [markdown] heading_collapsed=true
# ## 1.4. Check NA
# + hidden=true
df1.isna().sum()
# + [markdown] heading_collapsed=true
# ## 1.5 Replace NA
# + hidden=true
df1 = df1.dropna( subset = ['description', 'customer_id'])
print( 'Foram removidos: {:.2f}% dos dados'.format( 1- (df1.shape[0]/df_raw.shape[0])))
# + hidden=true
df1.isna().sum()
# + [markdown] heading_collapsed=true
# ## 1.6 Change Types
# + hidden=true
#invoice date
df1['invoice_date'] = pd.to_datetime( df1['invoice_date'], format = '%d-%b-%y')
#customer id
df1['customer_id'] = df1['customer_id'].astype( int )
df1.head(1)
# + hidden=true
df1.dtypes
# + [markdown] heading_collapsed=true
# ## 1.7. Desctiptive Statistical
# + [markdown] hidden=true
#
# -
# # 2.0. Feature Engeneering
df2 = df1.copy()
# ## 2.1. Feature Creation
df2.head()
#data reference
df_ref = df2.drop(['invoice_no', 'stock_code', 'description','quantity','unit_price','invoice_date','country'], axis = 1).drop_duplicates(ignore_index = True )
# +
# Gross Revenue ( Faturamento ) quanty + price
df2['gross_revenue'] = df2[ 'quantity' ] * df2[ 'unit_price' ]
# Monetary
df_monetary = df2[['customer_id', 'gross_revenue']].groupby( 'customer_id').sum().reset_index()
df_ref = pd.merge( df_ref, df_monetary, on = 'customer_id', how= 'left')
# Recency - Last day purchase
df_recency = df2[['customer_id', 'invoice_date']].groupby( 'customer_id').max().reset_index()
df_recency['recency_days'] = (df2['invoice_date'].max() - df_recency['invoice_date'] ).dt.days
df_recency = df_recency[['customer_id', 'recency_days']].copy()
df_ref = pd.merge(df_ref, df_recency, on = 'customer_id', how = 'left' )
#Frequency
df_freq = df2[['customer_id', 'invoice_no']].drop_duplicates().groupby( 'customer_id').count().reset_index()
df_ref = pd.merge(df_ref, df_freq, on = 'customer_id', how = 'left' )
#Avg Ticket
df_avg_ticket = df2[['customer_id', 'gross_revenue']].groupby( 'customer_id' ).mean().reset_index().rename( columns = {'gross_revenue' : 'avg_ticket'})
df_ref = pd.merge( df_ref, df_avg_ticket, on='customer_id', how = 'left')
df_ref.head(2)
# -
# # 3.0. Data Filtering
df3 = df_ref.copy()
# # 4.0. EDA(Exploratory Data Analysis)
df4 = df3.copy()
# # 5.0. Data Preparation
df5 = df4.copy()
# # 6.0. Feature Selection
df6 = df5.copy()
# + [markdown] heading_collapsed=true
# # 7.0. Hyperparameter Fine-Tunning
# + hidden=true
x = df6.drop( columns = ['customer_id'])
# + hidden=true
clusters = [2, 3, 4, 5, 6, 7 ]
# + [markdown] heading_collapsed=true hidden=true
# ## 7.1 Within-Cluster Sum of Square( WSS )
# + hidden=true
#wss = []
#for k in clusters:
#model definition
# kmeans = c.KMeans( init = 'random', n_clusters = k, n_init = 10, max_iter = 300, random_state = 42 )
#model training
# kmeans.fit( x )
#validation
# wss.append( kmeans.inertia_)
#plot wss - elbow methods
#plt.plot( clusters, wss, linestyle = '--', marker = 'o', color = 'b')
#plt.xlabel( 'k' )
#plt.ylabel( 'Within-Cluster Sum of Square' );
#plt.title( 'WSS vc K')
# + hidden=true
kmeans = KElbowVisualizer( c.KMeans(), k = clusters, timining = False )
kmeans.fit( x )
kmeans.show();
# + [markdown] hidden=true
# > A métrica wss leva em consideração o tamanho dos cluster, usando como parâmetro para os calculos a distância intra clusters.
#
# > Analisando os resultados, queremos encontrar o chamado "joelho", que é a quantidade de cluster que há uma diferença visual grande no gráfico, no caso de nosso plot este número é o k=3
# + [markdown] heading_collapsed=true hidden=true
# ## 7.2 Silhouette Score( SS )
# + hidden=true
kmeans = KElbowVisualizer( c.KMeans(), k = clusters, metric='silhouette', timining = False )
kmeans.fit( x )
kmeans.show();
# + [markdown] hidden=true
# > A métrica SS, leva em consideração para o calculo tanto a distância intra cluster como a distânci entre os clusters.
#
# > Analisando os resultados, queremos encontrar o número de K, que teja um valor de SS mais próximo de 1. No nosso caso o melhor S encontrado foi em K=2, porém vale ressaltar euq o k=3, ainda esta acima de 0,95 o que é muito bom também.
#
# > Como o wSS o k=3 teve melhor desempenho e no SS os desempenhos de k=2 e k=3 ficaram semelhantes, vamos definir de inicio o valor de k=3.
# + [markdown] heading_collapsed=true hidden=true
# ## 7.3 Silhouette Analysis
# + hidden=true
fig, ax = plt.subplots(3, 2, figsize = (25, 18 ) )
for k in clusters:
km = c.KMeans( n_clusters = k, init= 'random', n_init = 10, max_iter = 100, random_state = 42 )
q, mod = divmod( k, 2 )
visualizer = SilhouetteVisualizer( km, colors = 'yellowbrick', ax=ax[q-1][mod] )
visualizer.fit( x );
visualizer.finalize( );
# + [markdown] heading_collapsed=true
# # 8.0. Model Training
# + [markdown] heading_collapsed=true hidden=true
# ## 8.1 K-Means
# + [markdown] hidden=true
# #### Model Definition
# + hidden=true
# model definition
k=4
kmeans = c.KMeans( init = 'random', n_clusters=k, n_init=10, max_iter=300 )
#model training
kmeans.fit( x )
#clustering
labels = kmeans.labels_
# + [markdown] hidden=true
# #### Cluster Validation
# + hidden=true
## wss( within-cluster sum of square )
print( 'wss values: {}'.format( kmeans.inertia_ ))
## SS( Silhouette Score)
print('SS Values:{}'.format( m.silhouette_score( x, labels)))
# + [markdown] heading_collapsed=true
# # 9. Cluster Analysis
# + hidden=true
df9 = df6.copy()
df9['cluster'] = labels
df9.head()
# + [markdown] heading_collapsed=true hidden=true
# ## 9.1. Visualizatoin Inspection
# + hidden=true
#fig = px.scatter_3d( df8, x= 'recency_days', y = 'invoice_no', z='gross_revenue', color= 'cluster' );
#fig.show()
visualizer = SilhouetteVisualizer( kmeans, colors = 'yellowbrick')
visualizer.fit(x)
visualizer.finalize();
# + [markdown] heading_collapsed=true hidden=true
# ## 9.2. 2d plot
# + hidden=true
# 2d plot
df_viz = df9.drop( columns = 'customer_id', axis = 1 )
sns.pairplot( df_viz, hue = 'cluster' )
# + [markdown] heading_collapsed=true hidden=true
# ## 9.3. UMAP
# + hidden=true
#UMAP
reducer = umap.UMAP( n_neighbors = 20, random_state=42 )
embedding = reducer.fit_transform( x )
#embedding
df_viz['embedding_x'] = embedding[:, 0]
df_viz['embedding_y'] = embedding[:, 1]
#plot UMAP
sns.scatterplot( x= 'embedding_x', y = 'embedding_y',
hue = 'cluster',
palette = sns.color_palette( 'hls', n_colors = len(df_viz['cluster'].unique() )),
data= df_viz )
# + [markdown] hidden=true
# > O UMAP projeta uma visualização de mais alta dimensionalidade. Assim conseguimos analisar o quão misturados estão os clusters.
#
# > Os pontos próximos são classificados com a mesma cor, da para ver uma clara dominação do cluster 3(roxo), um dos motivos pode ser de não ter sido feito uma reescala dos cluster ainda.
#
# > Alterando o parâmetro n_neighbors, é possível visualizar estruturas mais globais, quando diminui, ele mostra visualização com menos agrupação, pois diminui a quantidade de visinhos.
# + [markdown] heading_collapsed=true hidden=true
# ## 9.4. Cluster Profile
# + hidden=true
# Number of customer
df_cluster = df9[['customer_id', 'cluster']].groupby( 'cluster' ).count().reset_index()
df_cluster['perc_customer'] = 100*( df_cluster['customer_id'] / df_cluster[ 'customer_id' ].sum() )
# Avg Gross revenue
df_avg_gross_revenue = df9[['gross_revenue', 'cluster']].groupby( 'cluster' ).mean().reset_index()
df_cluster = pd.merge ( df_cluster, df_avg_gross_revenue, how = 'inner', on = 'cluster')
#Avg recency days
df_avg_recency_days = df9[['recency_days', 'cluster']].groupby ( 'cluster' ).mean().reset_index()
df_cluster = pd.merge( df_cluster, df_avg_recency_days, how = 'inner', on='cluster' )
#Avg invoice_no
df_invoice_no = df9[['invoice_no', 'cluster']].groupby ( 'cluster' ).mean().reset_index()
df_cluster = pd.merge( df_cluster, df_invoice_no, how = 'inner', on='cluster' )
#AVG TIcket
df_ticket = df9[['avg_ticket', 'cluster']].groupby( 'cluster' ).mean().reset_index()
df_cluster = pd.merge(df_cluster, df_ticket, how='inner', on='cluster' )
df_cluster
# + [markdown] hidden=true
# Cluster 01: ( Candidato à Insider )
# - Número de customers: 6 (0.14% do customers )
# - Recência em média: 7 dias
# - Compras em média: 89 compras
# - Receita em média: $182.182,00 dólares
#
# Cluster 02:
# - Número de customers: 31 (0.71 do customers )
# - Recência em média: 14 dias
# - Compras em média: 53 compras
# - Receita em média: $40.543,52 dólares
#
# Cluster 03:
# - Número de customers: 4.335 (99% do customers )
# - Recência em média: 92 dias
# - Compras em média: 5 compras
# - Receita em média: $1.372,57 dólares
# -
# # 10.0. Deploy To Production
| c02_end-to-end_clustering.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# +
from selenium import webdriver
from selenium.webdriver.common.keys import Keys
from selenium.webdriver.chrome.options import Options
import time
# -
chromeOption = Options()
driver = webdriver.Chrome(options= chromeOption)
for x in range(5000):
time.sleep(7)
driver.execute_script('''<button class="vjs-big-play-button" type="button" title="Play Video" aria-disabled="false"><span aria-hidden="true" class="vjs-icon-placeholder"></span><span class="vjs-control-text" aria-live="polite">Play Video</span></button>
''')
Video = driver.find_element_by_class_name('vjs-poster').click()
| Scrapping/VideoRobot.ipynb |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.