Commit
•
5d45dc7
1
Parent(s):
217ed5f
Upload 361 files
Browse filesThis view is limited to 50 files because it contains too many changes.
See raw diff
- Titanic/Data/gender_submission.csv +419 -0
- Titanic/Data/test.csv +419 -0
- Titanic/Data/train.csv +892 -0
- Titanic/Kernels/AdaBoost/.ipynb_checkpoints/0-introduction-to-ensembling-stacking-in-python-checkpoint.ipynb +0 -0
- Titanic/Kernels/AdaBoost/.ipynb_checkpoints/1-a-data-science-framework-to-achieve-99-accuracy-checkpoint.ipynb +0 -0
- Titanic/Kernels/AdaBoost/.ipynb_checkpoints/10-a-comprehensive-guide-to-titanic-machine-learning-checkpoint.ipynb +0 -0
- Titanic/Kernels/AdaBoost/.ipynb_checkpoints/2-titanic-top-4-with-ensemble-modeling-checkpoint.ipynb +0 -0
- Titanic/Kernels/AdaBoost/.ipynb_checkpoints/3-eda-to-prediction-dietanic-checkpoint.ipynb +0 -0
- Titanic/Kernels/AdaBoost/.ipynb_checkpoints/4-a-statistical-analysis-ml-workflow-of-titanic-checkpoint.ipynb +0 -0
- Titanic/Kernels/AdaBoost/.ipynb_checkpoints/6-titanic-best-working-classifier-checkpoint.ipynb +1504 -0
- Titanic/Kernels/AdaBoost/.ipynb_checkpoints/7-titanic-survival-prediction-end-to-end-ml-pipeline-checkpoint.ipynb +0 -0
- Titanic/Kernels/AdaBoost/.ipynb_checkpoints/9-titanic-top-solution-checkpoint.ipynb +0 -0
- Titanic/Kernels/AdaBoost/10-a-comprehensive-guide-to-titanic-machine-learning.ipynb +0 -0
- Titanic/Kernels/AdaBoost/10-a-comprehensive-guide-to-titanic-machine-learning.py +0 -0
- Titanic/Kernels/AdaBoost/2-titanic-top-4-with-ensemble-modeling.ipynb +0 -0
- Titanic/Kernels/AdaBoost/2-titanic-top-4-with-ensemble-modeling.py +1110 -0
- Titanic/Kernels/AdaBoost/3-eda-to-prediction-dietanic.ipynb +0 -0
- Titanic/Kernels/AdaBoost/3-eda-to-prediction-dietanic.py +1152 -0
- Titanic/Kernels/AdaBoost/4-a-statistical-analysis-ml-workflow-of-titanic.ipynb +0 -0
- Titanic/Kernels/AdaBoost/4-a-statistical-analysis-ml-workflow-of-titanic.py +0 -0
- Titanic/Kernels/AdaBoost/6-titanic-best-working-classifier.ipynb +1504 -0
- Titanic/Kernels/AdaBoost/6-titanic-best-working-classifier.py +269 -0
- Titanic/Kernels/AdaBoost/7-titanic-survival-prediction-end-to-end-ml-pipeline.ipynb +0 -0
- Titanic/Kernels/AdaBoost/7-titanic-survival-prediction-end-to-end-ml-pipeline.py +919 -0
- Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/0-introduction-to-ensembling-stacking-in-python-checkpoint.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/10-ensemble-learning-techniques-tutorial-checkpoint.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/11-titanic-a-step-by-step-intro-to-machine-learning-checkpoint.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/2-titanic-top-4-with-ensemble-modeling-checkpoint.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/3-a-statistical-analysis-ml-workflow-of-titanic-checkpoint.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/4-applied-machine-learning-checkpoint.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/5-titanic-the-only-notebook-you-need-to-see-checkpoint.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/6-titanic-top-solution-checkpoint.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/7-titanic-eda-model-pipeline-keras-nn-checkpoint.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/8-a-comprehensive-guide-to-titanic-machine-learning-checkpoint.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/9-top-3-efficient-ensembling-in-few-lines-of-code-checkpoint.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/0-introduction-to-ensembling-stacking-in-python.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/0-introduction-to-ensembling-stacking-in-python.py +779 -0
- Titanic/Kernels/ExtraTrees/11-titanic-a-step-by-step-intro-to-machine-learning.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/11-titanic-a-step-by-step-intro-to-machine-learning.py +1445 -0
- Titanic/Kernels/ExtraTrees/2-titanic-top-4-with-ensemble-modeling.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/2-titanic-top-4-with-ensemble-modeling.py +1110 -0
- Titanic/Kernels/ExtraTrees/3-a-statistical-analysis-ml-workflow-of-titanic.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/3-a-statistical-analysis-ml-workflow-of-titanic.py +0 -0
- Titanic/Kernels/ExtraTrees/8-a-comprehensive-guide-to-titanic-machine-learning.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/8-a-comprehensive-guide-to-titanic-machine-learning.py +0 -0
- Titanic/Kernels/ExtraTrees/9-top-3-efficient-ensembling-in-few-lines-of-code.ipynb +0 -0
- Titanic/Kernels/ExtraTrees/9-top-3-efficient-ensembling-in-few-lines-of-code.py +944 -0
- Titanic/Kernels/GBC/.ipynb_checkpoints/0-introduction-to-ensembling-stacking-in-python-checkpoint.ipynb +0 -0
- Titanic/Kernels/GBC/.ipynb_checkpoints/1-a-data-science-framework-to-achieve-99-accuracy-checkpoint.ipynb +0 -0
- Titanic/Kernels/GBC/.ipynb_checkpoints/10-titanic-survival-prediction-end-to-end-ml-pipeline-checkpoint.ipynb +0 -0
Titanic/Data/gender_submission.csv
ADDED
|
@@ -0,0 +1,419 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
PassengerId,Survived
|
| 2 |
+
892,0
|
| 3 |
+
893,1
|
| 4 |
+
894,0
|
| 5 |
+
895,0
|
| 6 |
+
896,1
|
| 7 |
+
897,0
|
| 8 |
+
898,1
|
| 9 |
+
899,0
|
| 10 |
+
900,1
|
| 11 |
+
901,0
|
| 12 |
+
902,0
|
| 13 |
+
903,0
|
| 14 |
+
904,1
|
| 15 |
+
905,0
|
| 16 |
+
906,1
|
| 17 |
+
907,1
|
| 18 |
+
908,0
|
| 19 |
+
909,0
|
| 20 |
+
910,1
|
| 21 |
+
911,1
|
| 22 |
+
912,0
|
| 23 |
+
913,0
|
| 24 |
+
914,1
|
| 25 |
+
915,0
|
| 26 |
+
916,1
|
| 27 |
+
917,0
|
| 28 |
+
918,1
|
| 29 |
+
919,0
|
| 30 |
+
920,0
|
| 31 |
+
921,0
|
| 32 |
+
922,0
|
| 33 |
+
923,0
|
| 34 |
+
924,1
|
| 35 |
+
925,1
|
| 36 |
+
926,0
|
| 37 |
+
927,0
|
| 38 |
+
928,1
|
| 39 |
+
929,1
|
| 40 |
+
930,0
|
| 41 |
+
931,0
|
| 42 |
+
932,0
|
| 43 |
+
933,0
|
| 44 |
+
934,0
|
| 45 |
+
935,1
|
| 46 |
+
936,1
|
| 47 |
+
937,0
|
| 48 |
+
938,0
|
| 49 |
+
939,0
|
| 50 |
+
940,1
|
| 51 |
+
941,1
|
| 52 |
+
942,0
|
| 53 |
+
943,0
|
| 54 |
+
944,1
|
| 55 |
+
945,1
|
| 56 |
+
946,0
|
| 57 |
+
947,0
|
| 58 |
+
948,0
|
| 59 |
+
949,0
|
| 60 |
+
950,0
|
| 61 |
+
951,1
|
| 62 |
+
952,0
|
| 63 |
+
953,0
|
| 64 |
+
954,0
|
| 65 |
+
955,1
|
| 66 |
+
956,0
|
| 67 |
+
957,1
|
| 68 |
+
958,1
|
| 69 |
+
959,0
|
| 70 |
+
960,0
|
| 71 |
+
961,1
|
| 72 |
+
962,1
|
| 73 |
+
963,0
|
| 74 |
+
964,1
|
| 75 |
+
965,0
|
| 76 |
+
966,1
|
| 77 |
+
967,0
|
| 78 |
+
968,0
|
| 79 |
+
969,1
|
| 80 |
+
970,0
|
| 81 |
+
971,1
|
| 82 |
+
972,0
|
| 83 |
+
973,0
|
| 84 |
+
974,0
|
| 85 |
+
975,0
|
| 86 |
+
976,0
|
| 87 |
+
977,0
|
| 88 |
+
978,1
|
| 89 |
+
979,1
|
| 90 |
+
980,1
|
| 91 |
+
981,0
|
| 92 |
+
982,1
|
| 93 |
+
983,0
|
| 94 |
+
984,1
|
| 95 |
+
985,0
|
| 96 |
+
986,0
|
| 97 |
+
987,0
|
| 98 |
+
988,1
|
| 99 |
+
989,0
|
| 100 |
+
990,1
|
| 101 |
+
991,0
|
| 102 |
+
992,1
|
| 103 |
+
993,0
|
| 104 |
+
994,0
|
| 105 |
+
995,0
|
| 106 |
+
996,1
|
| 107 |
+
997,0
|
| 108 |
+
998,0
|
| 109 |
+
999,0
|
| 110 |
+
1000,0
|
| 111 |
+
1001,0
|
| 112 |
+
1002,0
|
| 113 |
+
1003,1
|
| 114 |
+
1004,1
|
| 115 |
+
1005,1
|
| 116 |
+
1006,1
|
| 117 |
+
1007,0
|
| 118 |
+
1008,0
|
| 119 |
+
1009,1
|
| 120 |
+
1010,0
|
| 121 |
+
1011,1
|
| 122 |
+
1012,1
|
| 123 |
+
1013,0
|
| 124 |
+
1014,1
|
| 125 |
+
1015,0
|
| 126 |
+
1016,0
|
| 127 |
+
1017,1
|
| 128 |
+
1018,0
|
| 129 |
+
1019,1
|
| 130 |
+
1020,0
|
| 131 |
+
1021,0
|
| 132 |
+
1022,0
|
| 133 |
+
1023,0
|
| 134 |
+
1024,1
|
| 135 |
+
1025,0
|
| 136 |
+
1026,0
|
| 137 |
+
1027,0
|
| 138 |
+
1028,0
|
| 139 |
+
1029,0
|
| 140 |
+
1030,1
|
| 141 |
+
1031,0
|
| 142 |
+
1032,1
|
| 143 |
+
1033,1
|
| 144 |
+
1034,0
|
| 145 |
+
1035,0
|
| 146 |
+
1036,0
|
| 147 |
+
1037,0
|
| 148 |
+
1038,0
|
| 149 |
+
1039,0
|
| 150 |
+
1040,0
|
| 151 |
+
1041,0
|
| 152 |
+
1042,1
|
| 153 |
+
1043,0
|
| 154 |
+
1044,0
|
| 155 |
+
1045,1
|
| 156 |
+
1046,0
|
| 157 |
+
1047,0
|
| 158 |
+
1048,1
|
| 159 |
+
1049,1
|
| 160 |
+
1050,0
|
| 161 |
+
1051,1
|
| 162 |
+
1052,1
|
| 163 |
+
1053,0
|
| 164 |
+
1054,1
|
| 165 |
+
1055,0
|
| 166 |
+
1056,0
|
| 167 |
+
1057,1
|
| 168 |
+
1058,0
|
| 169 |
+
1059,0
|
| 170 |
+
1060,1
|
| 171 |
+
1061,1
|
| 172 |
+
1062,0
|
| 173 |
+
1063,0
|
| 174 |
+
1064,0
|
| 175 |
+
1065,0
|
| 176 |
+
1066,0
|
| 177 |
+
1067,1
|
| 178 |
+
1068,1
|
| 179 |
+
1069,0
|
| 180 |
+
1070,1
|
| 181 |
+
1071,1
|
| 182 |
+
1072,0
|
| 183 |
+
1073,0
|
| 184 |
+
1074,1
|
| 185 |
+
1075,0
|
| 186 |
+
1076,1
|
| 187 |
+
1077,0
|
| 188 |
+
1078,1
|
| 189 |
+
1079,0
|
| 190 |
+
1080,1
|
| 191 |
+
1081,0
|
| 192 |
+
1082,0
|
| 193 |
+
1083,0
|
| 194 |
+
1084,0
|
| 195 |
+
1085,0
|
| 196 |
+
1086,0
|
| 197 |
+
1087,0
|
| 198 |
+
1088,0
|
| 199 |
+
1089,1
|
| 200 |
+
1090,0
|
| 201 |
+
1091,1
|
| 202 |
+
1092,1
|
| 203 |
+
1093,0
|
| 204 |
+
1094,0
|
| 205 |
+
1095,1
|
| 206 |
+
1096,0
|
| 207 |
+
1097,0
|
| 208 |
+
1098,1
|
| 209 |
+
1099,0
|
| 210 |
+
1100,1
|
| 211 |
+
1101,0
|
| 212 |
+
1102,0
|
| 213 |
+
1103,0
|
| 214 |
+
1104,0
|
| 215 |
+
1105,1
|
| 216 |
+
1106,1
|
| 217 |
+
1107,0
|
| 218 |
+
1108,1
|
| 219 |
+
1109,0
|
| 220 |
+
1110,1
|
| 221 |
+
1111,0
|
| 222 |
+
1112,1
|
| 223 |
+
1113,0
|
| 224 |
+
1114,1
|
| 225 |
+
1115,0
|
| 226 |
+
1116,1
|
| 227 |
+
1117,1
|
| 228 |
+
1118,0
|
| 229 |
+
1119,1
|
| 230 |
+
1120,0
|
| 231 |
+
1121,0
|
| 232 |
+
1122,0
|
| 233 |
+
1123,1
|
| 234 |
+
1124,0
|
| 235 |
+
1125,0
|
| 236 |
+
1126,0
|
| 237 |
+
1127,0
|
| 238 |
+
1128,0
|
| 239 |
+
1129,0
|
| 240 |
+
1130,1
|
| 241 |
+
1131,1
|
| 242 |
+
1132,1
|
| 243 |
+
1133,1
|
| 244 |
+
1134,0
|
| 245 |
+
1135,0
|
| 246 |
+
1136,0
|
| 247 |
+
1137,0
|
| 248 |
+
1138,1
|
| 249 |
+
1139,0
|
| 250 |
+
1140,1
|
| 251 |
+
1141,1
|
| 252 |
+
1142,1
|
| 253 |
+
1143,0
|
| 254 |
+
1144,0
|
| 255 |
+
1145,0
|
| 256 |
+
1146,0
|
| 257 |
+
1147,0
|
| 258 |
+
1148,0
|
| 259 |
+
1149,0
|
| 260 |
+
1150,1
|
| 261 |
+
1151,0
|
| 262 |
+
1152,0
|
| 263 |
+
1153,0
|
| 264 |
+
1154,1
|
| 265 |
+
1155,1
|
| 266 |
+
1156,0
|
| 267 |
+
1157,0
|
| 268 |
+
1158,0
|
| 269 |
+
1159,0
|
| 270 |
+
1160,1
|
| 271 |
+
1161,0
|
| 272 |
+
1162,0
|
| 273 |
+
1163,0
|
| 274 |
+
1164,1
|
| 275 |
+
1165,1
|
| 276 |
+
1166,0
|
| 277 |
+
1167,1
|
| 278 |
+
1168,0
|
| 279 |
+
1169,0
|
| 280 |
+
1170,0
|
| 281 |
+
1171,0
|
| 282 |
+
1172,1
|
| 283 |
+
1173,0
|
| 284 |
+
1174,1
|
| 285 |
+
1175,1
|
| 286 |
+
1176,1
|
| 287 |
+
1177,0
|
| 288 |
+
1178,0
|
| 289 |
+
1179,0
|
| 290 |
+
1180,0
|
| 291 |
+
1181,0
|
| 292 |
+
1182,0
|
| 293 |
+
1183,1
|
| 294 |
+
1184,0
|
| 295 |
+
1185,0
|
| 296 |
+
1186,0
|
| 297 |
+
1187,0
|
| 298 |
+
1188,1
|
| 299 |
+
1189,0
|
| 300 |
+
1190,0
|
| 301 |
+
1191,0
|
| 302 |
+
1192,0
|
| 303 |
+
1193,0
|
| 304 |
+
1194,0
|
| 305 |
+
1195,0
|
| 306 |
+
1196,1
|
| 307 |
+
1197,1
|
| 308 |
+
1198,0
|
| 309 |
+
1199,0
|
| 310 |
+
1200,0
|
| 311 |
+
1201,1
|
| 312 |
+
1202,0
|
| 313 |
+
1203,0
|
| 314 |
+
1204,0
|
| 315 |
+
1205,1
|
| 316 |
+
1206,1
|
| 317 |
+
1207,1
|
| 318 |
+
1208,0
|
| 319 |
+
1209,0
|
| 320 |
+
1210,0
|
| 321 |
+
1211,0
|
| 322 |
+
1212,0
|
| 323 |
+
1213,0
|
| 324 |
+
1214,0
|
| 325 |
+
1215,0
|
| 326 |
+
1216,1
|
| 327 |
+
1217,0
|
| 328 |
+
1218,1
|
| 329 |
+
1219,0
|
| 330 |
+
1220,0
|
| 331 |
+
1221,0
|
| 332 |
+
1222,1
|
| 333 |
+
1223,0
|
| 334 |
+
1224,0
|
| 335 |
+
1225,1
|
| 336 |
+
1226,0
|
| 337 |
+
1227,0
|
| 338 |
+
1228,0
|
| 339 |
+
1229,0
|
| 340 |
+
1230,0
|
| 341 |
+
1231,0
|
| 342 |
+
1232,0
|
| 343 |
+
1233,0
|
| 344 |
+
1234,0
|
| 345 |
+
1235,1
|
| 346 |
+
1236,0
|
| 347 |
+
1237,1
|
| 348 |
+
1238,0
|
| 349 |
+
1239,1
|
| 350 |
+
1240,0
|
| 351 |
+
1241,1
|
| 352 |
+
1242,1
|
| 353 |
+
1243,0
|
| 354 |
+
1244,0
|
| 355 |
+
1245,0
|
| 356 |
+
1246,1
|
| 357 |
+
1247,0
|
| 358 |
+
1248,1
|
| 359 |
+
1249,0
|
| 360 |
+
1250,0
|
| 361 |
+
1251,1
|
| 362 |
+
1252,0
|
| 363 |
+
1253,1
|
| 364 |
+
1254,1
|
| 365 |
+
1255,0
|
| 366 |
+
1256,1
|
| 367 |
+
1257,1
|
| 368 |
+
1258,0
|
| 369 |
+
1259,1
|
| 370 |
+
1260,1
|
| 371 |
+
1261,0
|
| 372 |
+
1262,0
|
| 373 |
+
1263,1
|
| 374 |
+
1264,0
|
| 375 |
+
1265,0
|
| 376 |
+
1266,1
|
| 377 |
+
1267,1
|
| 378 |
+
1268,1
|
| 379 |
+
1269,0
|
| 380 |
+
1270,0
|
| 381 |
+
1271,0
|
| 382 |
+
1272,0
|
| 383 |
+
1273,0
|
| 384 |
+
1274,1
|
| 385 |
+
1275,1
|
| 386 |
+
1276,0
|
| 387 |
+
1277,1
|
| 388 |
+
1278,0
|
| 389 |
+
1279,0
|
| 390 |
+
1280,0
|
| 391 |
+
1281,0
|
| 392 |
+
1282,0
|
| 393 |
+
1283,1
|
| 394 |
+
1284,0
|
| 395 |
+
1285,0
|
| 396 |
+
1286,0
|
| 397 |
+
1287,1
|
| 398 |
+
1288,0
|
| 399 |
+
1289,1
|
| 400 |
+
1290,0
|
| 401 |
+
1291,0
|
| 402 |
+
1292,1
|
| 403 |
+
1293,0
|
| 404 |
+
1294,1
|
| 405 |
+
1295,0
|
| 406 |
+
1296,0
|
| 407 |
+
1297,0
|
| 408 |
+
1298,0
|
| 409 |
+
1299,0
|
| 410 |
+
1300,1
|
| 411 |
+
1301,1
|
| 412 |
+
1302,1
|
| 413 |
+
1303,1
|
| 414 |
+
1304,1
|
| 415 |
+
1305,0
|
| 416 |
+
1306,1
|
| 417 |
+
1307,0
|
| 418 |
+
1308,0
|
| 419 |
+
1309,0
|
Titanic/Data/test.csv
ADDED
|
@@ -0,0 +1,419 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
PassengerId,Pclass,Name,Sex,Age,SibSp,Parch,Ticket,Fare,Cabin,Embarked
|
| 2 |
+
892,3,"Kelly, Mr. James",male,34.5,0,0,330911,7.8292,,Q
|
| 3 |
+
893,3,"Wilkes, Mrs. James (Ellen Needs)",female,47,1,0,363272,7,,S
|
| 4 |
+
894,2,"Myles, Mr. Thomas Francis",male,62,0,0,240276,9.6875,,Q
|
| 5 |
+
895,3,"Wirz, Mr. Albert",male,27,0,0,315154,8.6625,,S
|
| 6 |
+
896,3,"Hirvonen, Mrs. Alexander (Helga E Lindqvist)",female,22,1,1,3101298,12.2875,,S
|
| 7 |
+
897,3,"Svensson, Mr. Johan Cervin",male,14,0,0,7538,9.225,,S
|
| 8 |
+
898,3,"Connolly, Miss. Kate",female,30,0,0,330972,7.6292,,Q
|
| 9 |
+
899,2,"Caldwell, Mr. Albert Francis",male,26,1,1,248738,29,,S
|
| 10 |
+
900,3,"Abrahim, Mrs. Joseph (Sophie Halaut Easu)",female,18,0,0,2657,7.2292,,C
|
| 11 |
+
901,3,"Davies, Mr. John Samuel",male,21,2,0,A/4 48871,24.15,,S
|
| 12 |
+
902,3,"Ilieff, Mr. Ylio",male,,0,0,349220,7.8958,,S
|
| 13 |
+
903,1,"Jones, Mr. Charles Cresson",male,46,0,0,694,26,,S
|
| 14 |
+
904,1,"Snyder, Mrs. John Pillsbury (Nelle Stevenson)",female,23,1,0,21228,82.2667,B45,S
|
| 15 |
+
905,2,"Howard, Mr. Benjamin",male,63,1,0,24065,26,,S
|
| 16 |
+
906,1,"Chaffee, Mrs. Herbert Fuller (Carrie Constance Toogood)",female,47,1,0,W.E.P. 5734,61.175,E31,S
|
| 17 |
+
907,2,"del Carlo, Mrs. Sebastiano (Argenia Genovesi)",female,24,1,0,SC/PARIS 2167,27.7208,,C
|
| 18 |
+
908,2,"Keane, Mr. Daniel",male,35,0,0,233734,12.35,,Q
|
| 19 |
+
909,3,"Assaf, Mr. Gerios",male,21,0,0,2692,7.225,,C
|
| 20 |
+
910,3,"Ilmakangas, Miss. Ida Livija",female,27,1,0,STON/O2. 3101270,7.925,,S
|
| 21 |
+
911,3,"Assaf Khalil, Mrs. Mariana (Miriam"")""",female,45,0,0,2696,7.225,,C
|
| 22 |
+
912,1,"Rothschild, Mr. Martin",male,55,1,0,PC 17603,59.4,,C
|
| 23 |
+
913,3,"Olsen, Master. Artur Karl",male,9,0,1,C 17368,3.1708,,S
|
| 24 |
+
914,1,"Flegenheim, Mrs. Alfred (Antoinette)",female,,0,0,PC 17598,31.6833,,S
|
| 25 |
+
915,1,"Williams, Mr. Richard Norris II",male,21,0,1,PC 17597,61.3792,,C
|
| 26 |
+
916,1,"Ryerson, Mrs. Arthur Larned (Emily Maria Borie)",female,48,1,3,PC 17608,262.375,B57 B59 B63 B66,C
|
| 27 |
+
917,3,"Robins, Mr. Alexander A",male,50,1,0,A/5. 3337,14.5,,S
|
| 28 |
+
918,1,"Ostby, Miss. Helene Ragnhild",female,22,0,1,113509,61.9792,B36,C
|
| 29 |
+
919,3,"Daher, Mr. Shedid",male,22.5,0,0,2698,7.225,,C
|
| 30 |
+
920,1,"Brady, Mr. John Bertram",male,41,0,0,113054,30.5,A21,S
|
| 31 |
+
921,3,"Samaan, Mr. Elias",male,,2,0,2662,21.6792,,C
|
| 32 |
+
922,2,"Louch, Mr. Charles Alexander",male,50,1,0,SC/AH 3085,26,,S
|
| 33 |
+
923,2,"Jefferys, Mr. Clifford Thomas",male,24,2,0,C.A. 31029,31.5,,S
|
| 34 |
+
924,3,"Dean, Mrs. Bertram (Eva Georgetta Light)",female,33,1,2,C.A. 2315,20.575,,S
|
| 35 |
+
925,3,"Johnston, Mrs. Andrew G (Elizabeth Lily"" Watson)""",female,,1,2,W./C. 6607,23.45,,S
|
| 36 |
+
926,1,"Mock, Mr. Philipp Edmund",male,30,1,0,13236,57.75,C78,C
|
| 37 |
+
927,3,"Katavelas, Mr. Vassilios (Catavelas Vassilios"")""",male,18.5,0,0,2682,7.2292,,C
|
| 38 |
+
928,3,"Roth, Miss. Sarah A",female,,0,0,342712,8.05,,S
|
| 39 |
+
929,3,"Cacic, Miss. Manda",female,21,0,0,315087,8.6625,,S
|
| 40 |
+
930,3,"Sap, Mr. Julius",male,25,0,0,345768,9.5,,S
|
| 41 |
+
931,3,"Hee, Mr. Ling",male,,0,0,1601,56.4958,,S
|
| 42 |
+
932,3,"Karun, Mr. Franz",male,39,0,1,349256,13.4167,,C
|
| 43 |
+
933,1,"Franklin, Mr. Thomas Parham",male,,0,0,113778,26.55,D34,S
|
| 44 |
+
934,3,"Goldsmith, Mr. Nathan",male,41,0,0,SOTON/O.Q. 3101263,7.85,,S
|
| 45 |
+
935,2,"Corbett, Mrs. Walter H (Irene Colvin)",female,30,0,0,237249,13,,S
|
| 46 |
+
936,1,"Kimball, Mrs. Edwin Nelson Jr (Gertrude Parsons)",female,45,1,0,11753,52.5542,D19,S
|
| 47 |
+
937,3,"Peltomaki, Mr. Nikolai Johannes",male,25,0,0,STON/O 2. 3101291,7.925,,S
|
| 48 |
+
938,1,"Chevre, Mr. Paul Romaine",male,45,0,0,PC 17594,29.7,A9,C
|
| 49 |
+
939,3,"Shaughnessy, Mr. Patrick",male,,0,0,370374,7.75,,Q
|
| 50 |
+
940,1,"Bucknell, Mrs. William Robert (Emma Eliza Ward)",female,60,0,0,11813,76.2917,D15,C
|
| 51 |
+
941,3,"Coutts, Mrs. William (Winnie Minnie"" Treanor)""",female,36,0,2,C.A. 37671,15.9,,S
|
| 52 |
+
942,1,"Smith, Mr. Lucien Philip",male,24,1,0,13695,60,C31,S
|
| 53 |
+
943,2,"Pulbaum, Mr. Franz",male,27,0,0,SC/PARIS 2168,15.0333,,C
|
| 54 |
+
944,2,"Hocking, Miss. Ellen Nellie""""",female,20,2,1,29105,23,,S
|
| 55 |
+
945,1,"Fortune, Miss. Ethel Flora",female,28,3,2,19950,263,C23 C25 C27,S
|
| 56 |
+
946,2,"Mangiavacchi, Mr. Serafino Emilio",male,,0,0,SC/A.3 2861,15.5792,,C
|
| 57 |
+
947,3,"Rice, Master. Albert",male,10,4,1,382652,29.125,,Q
|
| 58 |
+
948,3,"Cor, Mr. Bartol",male,35,0,0,349230,7.8958,,S
|
| 59 |
+
949,3,"Abelseth, Mr. Olaus Jorgensen",male,25,0,0,348122,7.65,F G63,S
|
| 60 |
+
950,3,"Davison, Mr. Thomas Henry",male,,1,0,386525,16.1,,S
|
| 61 |
+
951,1,"Chaudanson, Miss. Victorine",female,36,0,0,PC 17608,262.375,B61,C
|
| 62 |
+
952,3,"Dika, Mr. Mirko",male,17,0,0,349232,7.8958,,S
|
| 63 |
+
953,2,"McCrae, Mr. Arthur Gordon",male,32,0,0,237216,13.5,,S
|
| 64 |
+
954,3,"Bjorklund, Mr. Ernst Herbert",male,18,0,0,347090,7.75,,S
|
| 65 |
+
955,3,"Bradley, Miss. Bridget Delia",female,22,0,0,334914,7.725,,Q
|
| 66 |
+
956,1,"Ryerson, Master. John Borie",male,13,2,2,PC 17608,262.375,B57 B59 B63 B66,C
|
| 67 |
+
957,2,"Corey, Mrs. Percy C (Mary Phyllis Elizabeth Miller)",female,,0,0,F.C.C. 13534,21,,S
|
| 68 |
+
958,3,"Burns, Miss. Mary Delia",female,18,0,0,330963,7.8792,,Q
|
| 69 |
+
959,1,"Moore, Mr. Clarence Bloomfield",male,47,0,0,113796,42.4,,S
|
| 70 |
+
960,1,"Tucker, Mr. Gilbert Milligan Jr",male,31,0,0,2543,28.5375,C53,C
|
| 71 |
+
961,1,"Fortune, Mrs. Mark (Mary McDougald)",female,60,1,4,19950,263,C23 C25 C27,S
|
| 72 |
+
962,3,"Mulvihill, Miss. Bertha E",female,24,0,0,382653,7.75,,Q
|
| 73 |
+
963,3,"Minkoff, Mr. Lazar",male,21,0,0,349211,7.8958,,S
|
| 74 |
+
964,3,"Nieminen, Miss. Manta Josefina",female,29,0,0,3101297,7.925,,S
|
| 75 |
+
965,1,"Ovies y Rodriguez, Mr. Servando",male,28.5,0,0,PC 17562,27.7208,D43,C
|
| 76 |
+
966,1,"Geiger, Miss. Amalie",female,35,0,0,113503,211.5,C130,C
|
| 77 |
+
967,1,"Keeping, Mr. Edwin",male,32.5,0,0,113503,211.5,C132,C
|
| 78 |
+
968,3,"Miles, Mr. Frank",male,,0,0,359306,8.05,,S
|
| 79 |
+
969,1,"Cornell, Mrs. Robert Clifford (Malvina Helen Lamson)",female,55,2,0,11770,25.7,C101,S
|
| 80 |
+
970,2,"Aldworth, Mr. Charles Augustus",male,30,0,0,248744,13,,S
|
| 81 |
+
971,3,"Doyle, Miss. Elizabeth",female,24,0,0,368702,7.75,,Q
|
| 82 |
+
972,3,"Boulos, Master. Akar",male,6,1,1,2678,15.2458,,C
|
| 83 |
+
973,1,"Straus, Mr. Isidor",male,67,1,0,PC 17483,221.7792,C55 C57,S
|
| 84 |
+
974,1,"Case, Mr. Howard Brown",male,49,0,0,19924,26,,S
|
| 85 |
+
975,3,"Demetri, Mr. Marinko",male,,0,0,349238,7.8958,,S
|
| 86 |
+
976,2,"Lamb, Mr. John Joseph",male,,0,0,240261,10.7083,,Q
|
| 87 |
+
977,3,"Khalil, Mr. Betros",male,,1,0,2660,14.4542,,C
|
| 88 |
+
978,3,"Barry, Miss. Julia",female,27,0,0,330844,7.8792,,Q
|
| 89 |
+
979,3,"Badman, Miss. Emily Louisa",female,18,0,0,A/4 31416,8.05,,S
|
| 90 |
+
980,3,"O'Donoghue, Ms. Bridget",female,,0,0,364856,7.75,,Q
|
| 91 |
+
981,2,"Wells, Master. Ralph Lester",male,2,1,1,29103,23,,S
|
| 92 |
+
982,3,"Dyker, Mrs. Adolf Fredrik (Anna Elisabeth Judith Andersson)",female,22,1,0,347072,13.9,,S
|
| 93 |
+
983,3,"Pedersen, Mr. Olaf",male,,0,0,345498,7.775,,S
|
| 94 |
+
984,1,"Davidson, Mrs. Thornton (Orian Hays)",female,27,1,2,F.C. 12750,52,B71,S
|
| 95 |
+
985,3,"Guest, Mr. Robert",male,,0,0,376563,8.05,,S
|
| 96 |
+
986,1,"Birnbaum, Mr. Jakob",male,25,0,0,13905,26,,C
|
| 97 |
+
987,3,"Tenglin, Mr. Gunnar Isidor",male,25,0,0,350033,7.7958,,S
|
| 98 |
+
988,1,"Cavendish, Mrs. Tyrell William (Julia Florence Siegel)",female,76,1,0,19877,78.85,C46,S
|
| 99 |
+
989,3,"Makinen, Mr. Kalle Edvard",male,29,0,0,STON/O 2. 3101268,7.925,,S
|
| 100 |
+
990,3,"Braf, Miss. Elin Ester Maria",female,20,0,0,347471,7.8542,,S
|
| 101 |
+
991,3,"Nancarrow, Mr. William Henry",male,33,0,0,A./5. 3338,8.05,,S
|
| 102 |
+
992,1,"Stengel, Mrs. Charles Emil Henry (Annie May Morris)",female,43,1,0,11778,55.4417,C116,C
|
| 103 |
+
993,2,"Weisz, Mr. Leopold",male,27,1,0,228414,26,,S
|
| 104 |
+
994,3,"Foley, Mr. William",male,,0,0,365235,7.75,,Q
|
| 105 |
+
995,3,"Johansson Palmquist, Mr. Oskar Leander",male,26,0,0,347070,7.775,,S
|
| 106 |
+
996,3,"Thomas, Mrs. Alexander (Thamine Thelma"")""",female,16,1,1,2625,8.5167,,C
|
| 107 |
+
997,3,"Holthen, Mr. Johan Martin",male,28,0,0,C 4001,22.525,,S
|
| 108 |
+
998,3,"Buckley, Mr. Daniel",male,21,0,0,330920,7.8208,,Q
|
| 109 |
+
999,3,"Ryan, Mr. Edward",male,,0,0,383162,7.75,,Q
|
| 110 |
+
1000,3,"Willer, Mr. Aaron (Abi Weller"")""",male,,0,0,3410,8.7125,,S
|
| 111 |
+
1001,2,"Swane, Mr. George",male,18.5,0,0,248734,13,F,S
|
| 112 |
+
1002,2,"Stanton, Mr. Samuel Ward",male,41,0,0,237734,15.0458,,C
|
| 113 |
+
1003,3,"Shine, Miss. Ellen Natalia",female,,0,0,330968,7.7792,,Q
|
| 114 |
+
1004,1,"Evans, Miss. Edith Corse",female,36,0,0,PC 17531,31.6792,A29,C
|
| 115 |
+
1005,3,"Buckley, Miss. Katherine",female,18.5,0,0,329944,7.2833,,Q
|
| 116 |
+
1006,1,"Straus, Mrs. Isidor (Rosalie Ida Blun)",female,63,1,0,PC 17483,221.7792,C55 C57,S
|
| 117 |
+
1007,3,"Chronopoulos, Mr. Demetrios",male,18,1,0,2680,14.4542,,C
|
| 118 |
+
1008,3,"Thomas, Mr. John",male,,0,0,2681,6.4375,,C
|
| 119 |
+
1009,3,"Sandstrom, Miss. Beatrice Irene",female,1,1,1,PP 9549,16.7,G6,S
|
| 120 |
+
1010,1,"Beattie, Mr. Thomson",male,36,0,0,13050,75.2417,C6,C
|
| 121 |
+
1011,2,"Chapman, Mrs. John Henry (Sara Elizabeth Lawry)",female,29,1,0,SC/AH 29037,26,,S
|
| 122 |
+
1012,2,"Watt, Miss. Bertha J",female,12,0,0,C.A. 33595,15.75,,S
|
| 123 |
+
1013,3,"Kiernan, Mr. John",male,,1,0,367227,7.75,,Q
|
| 124 |
+
1014,1,"Schabert, Mrs. Paul (Emma Mock)",female,35,1,0,13236,57.75,C28,C
|
| 125 |
+
1015,3,"Carver, Mr. Alfred John",male,28,0,0,392095,7.25,,S
|
| 126 |
+
1016,3,"Kennedy, Mr. John",male,,0,0,368783,7.75,,Q
|
| 127 |
+
1017,3,"Cribb, Miss. Laura Alice",female,17,0,1,371362,16.1,,S
|
| 128 |
+
1018,3,"Brobeck, Mr. Karl Rudolf",male,22,0,0,350045,7.7958,,S
|
| 129 |
+
1019,3,"McCoy, Miss. Alicia",female,,2,0,367226,23.25,,Q
|
| 130 |
+
1020,2,"Bowenur, Mr. Solomon",male,42,0,0,211535,13,,S
|
| 131 |
+
1021,3,"Petersen, Mr. Marius",male,24,0,0,342441,8.05,,S
|
| 132 |
+
1022,3,"Spinner, Mr. Henry John",male,32,0,0,STON/OQ. 369943,8.05,,S
|
| 133 |
+
1023,1,"Gracie, Col. Archibald IV",male,53,0,0,113780,28.5,C51,C
|
| 134 |
+
1024,3,"Lefebre, Mrs. Frank (Frances)",female,,0,4,4133,25.4667,,S
|
| 135 |
+
1025,3,"Thomas, Mr. Charles P",male,,1,0,2621,6.4375,,C
|
| 136 |
+
1026,3,"Dintcheff, Mr. Valtcho",male,43,0,0,349226,7.8958,,S
|
| 137 |
+
1027,3,"Carlsson, Mr. Carl Robert",male,24,0,0,350409,7.8542,,S
|
| 138 |
+
1028,3,"Zakarian, Mr. Mapriededer",male,26.5,0,0,2656,7.225,,C
|
| 139 |
+
1029,2,"Schmidt, Mr. August",male,26,0,0,248659,13,,S
|
| 140 |
+
1030,3,"Drapkin, Miss. Jennie",female,23,0,0,SOTON/OQ 392083,8.05,,S
|
| 141 |
+
1031,3,"Goodwin, Mr. Charles Frederick",male,40,1,6,CA 2144,46.9,,S
|
| 142 |
+
1032,3,"Goodwin, Miss. Jessie Allis",female,10,5,2,CA 2144,46.9,,S
|
| 143 |
+
1033,1,"Daniels, Miss. Sarah",female,33,0,0,113781,151.55,,S
|
| 144 |
+
1034,1,"Ryerson, Mr. Arthur Larned",male,61,1,3,PC 17608,262.375,B57 B59 B63 B66,C
|
| 145 |
+
1035,2,"Beauchamp, Mr. Henry James",male,28,0,0,244358,26,,S
|
| 146 |
+
1036,1,"Lindeberg-Lind, Mr. Erik Gustaf (Mr Edward Lingrey"")""",male,42,0,0,17475,26.55,,S
|
| 147 |
+
1037,3,"Vander Planke, Mr. Julius",male,31,3,0,345763,18,,S
|
| 148 |
+
1038,1,"Hilliard, Mr. Herbert Henry",male,,0,0,17463,51.8625,E46,S
|
| 149 |
+
1039,3,"Davies, Mr. Evan",male,22,0,0,SC/A4 23568,8.05,,S
|
| 150 |
+
1040,1,"Crafton, Mr. John Bertram",male,,0,0,113791,26.55,,S
|
| 151 |
+
1041,2,"Lahtinen, Rev. William",male,30,1,1,250651,26,,S
|
| 152 |
+
1042,1,"Earnshaw, Mrs. Boulton (Olive Potter)",female,23,0,1,11767,83.1583,C54,C
|
| 153 |
+
1043,3,"Matinoff, Mr. Nicola",male,,0,0,349255,7.8958,,C
|
| 154 |
+
1044,3,"Storey, Mr. Thomas",male,60.5,0,0,3701,,,S
|
| 155 |
+
1045,3,"Klasen, Mrs. (Hulda Kristina Eugenia Lofqvist)",female,36,0,2,350405,12.1833,,S
|
| 156 |
+
1046,3,"Asplund, Master. Filip Oscar",male,13,4,2,347077,31.3875,,S
|
| 157 |
+
1047,3,"Duquemin, Mr. Joseph",male,24,0,0,S.O./P.P. 752,7.55,,S
|
| 158 |
+
1048,1,"Bird, Miss. Ellen",female,29,0,0,PC 17483,221.7792,C97,S
|
| 159 |
+
1049,3,"Lundin, Miss. Olga Elida",female,23,0,0,347469,7.8542,,S
|
| 160 |
+
1050,1,"Borebank, Mr. John James",male,42,0,0,110489,26.55,D22,S
|
| 161 |
+
1051,3,"Peacock, Mrs. Benjamin (Edith Nile)",female,26,0,2,SOTON/O.Q. 3101315,13.775,,S
|
| 162 |
+
1052,3,"Smyth, Miss. Julia",female,,0,0,335432,7.7333,,Q
|
| 163 |
+
1053,3,"Touma, Master. Georges Youssef",male,7,1,1,2650,15.2458,,C
|
| 164 |
+
1054,2,"Wright, Miss. Marion",female,26,0,0,220844,13.5,,S
|
| 165 |
+
1055,3,"Pearce, Mr. Ernest",male,,0,0,343271,7,,S
|
| 166 |
+
1056,2,"Peruschitz, Rev. Joseph Maria",male,41,0,0,237393,13,,S
|
| 167 |
+
1057,3,"Kink-Heilmann, Mrs. Anton (Luise Heilmann)",female,26,1,1,315153,22.025,,S
|
| 168 |
+
1058,1,"Brandeis, Mr. Emil",male,48,0,0,PC 17591,50.4958,B10,C
|
| 169 |
+
1059,3,"Ford, Mr. Edward Watson",male,18,2,2,W./C. 6608,34.375,,S
|
| 170 |
+
1060,1,"Cassebeer, Mrs. Henry Arthur Jr (Eleanor Genevieve Fosdick)",female,,0,0,17770,27.7208,,C
|
| 171 |
+
1061,3,"Hellstrom, Miss. Hilda Maria",female,22,0,0,7548,8.9625,,S
|
| 172 |
+
1062,3,"Lithman, Mr. Simon",male,,0,0,S.O./P.P. 251,7.55,,S
|
| 173 |
+
1063,3,"Zakarian, Mr. Ortin",male,27,0,0,2670,7.225,,C
|
| 174 |
+
1064,3,"Dyker, Mr. Adolf Fredrik",male,23,1,0,347072,13.9,,S
|
| 175 |
+
1065,3,"Torfa, Mr. Assad",male,,0,0,2673,7.2292,,C
|
| 176 |
+
1066,3,"Asplund, Mr. Carl Oscar Vilhelm Gustafsson",male,40,1,5,347077,31.3875,,S
|
| 177 |
+
1067,2,"Brown, Miss. Edith Eileen",female,15,0,2,29750,39,,S
|
| 178 |
+
1068,2,"Sincock, Miss. Maude",female,20,0,0,C.A. 33112,36.75,,S
|
| 179 |
+
1069,1,"Stengel, Mr. Charles Emil Henry",male,54,1,0,11778,55.4417,C116,C
|
| 180 |
+
1070,2,"Becker, Mrs. Allen Oliver (Nellie E Baumgardner)",female,36,0,3,230136,39,F4,S
|
| 181 |
+
1071,1,"Compton, Mrs. Alexander Taylor (Mary Eliza Ingersoll)",female,64,0,2,PC 17756,83.1583,E45,C
|
| 182 |
+
1072,2,"McCrie, Mr. James Matthew",male,30,0,0,233478,13,,S
|
| 183 |
+
1073,1,"Compton, Mr. Alexander Taylor Jr",male,37,1,1,PC 17756,83.1583,E52,C
|
| 184 |
+
1074,1,"Marvin, Mrs. Daniel Warner (Mary Graham Carmichael Farquarson)",female,18,1,0,113773,53.1,D30,S
|
| 185 |
+
1075,3,"Lane, Mr. Patrick",male,,0,0,7935,7.75,,Q
|
| 186 |
+
1076,1,"Douglas, Mrs. Frederick Charles (Mary Helene Baxter)",female,27,1,1,PC 17558,247.5208,B58 B60,C
|
| 187 |
+
1077,2,"Maybery, Mr. Frank Hubert",male,40,0,0,239059,16,,S
|
| 188 |
+
1078,2,"Phillips, Miss. Alice Frances Louisa",female,21,0,1,S.O./P.P. 2,21,,S
|
| 189 |
+
1079,3,"Davies, Mr. Joseph",male,17,2,0,A/4 48873,8.05,,S
|
| 190 |
+
1080,3,"Sage, Miss. Ada",female,,8,2,CA. 2343,69.55,,S
|
| 191 |
+
1081,2,"Veal, Mr. James",male,40,0,0,28221,13,,S
|
| 192 |
+
1082,2,"Angle, Mr. William A",male,34,1,0,226875,26,,S
|
| 193 |
+
1083,1,"Salomon, Mr. Abraham L",male,,0,0,111163,26,,S
|
| 194 |
+
1084,3,"van Billiard, Master. Walter John",male,11.5,1,1,A/5. 851,14.5,,S
|
| 195 |
+
1085,2,"Lingane, Mr. John",male,61,0,0,235509,12.35,,Q
|
| 196 |
+
1086,2,"Drew, Master. Marshall Brines",male,8,0,2,28220,32.5,,S
|
| 197 |
+
1087,3,"Karlsson, Mr. Julius Konrad Eugen",male,33,0,0,347465,7.8542,,S
|
| 198 |
+
1088,1,"Spedden, Master. Robert Douglas",male,6,0,2,16966,134.5,E34,C
|
| 199 |
+
1089,3,"Nilsson, Miss. Berta Olivia",female,18,0,0,347066,7.775,,S
|
| 200 |
+
1090,2,"Baimbrigge, Mr. Charles Robert",male,23,0,0,C.A. 31030,10.5,,S
|
| 201 |
+
1091,3,"Rasmussen, Mrs. (Lena Jacobsen Solvang)",female,,0,0,65305,8.1125,,S
|
| 202 |
+
1092,3,"Murphy, Miss. Nora",female,,0,0,36568,15.5,,Q
|
| 203 |
+
1093,3,"Danbom, Master. Gilbert Sigvard Emanuel",male,0.33,0,2,347080,14.4,,S
|
| 204 |
+
1094,1,"Astor, Col. John Jacob",male,47,1,0,PC 17757,227.525,C62 C64,C
|
| 205 |
+
1095,2,"Quick, Miss. Winifred Vera",female,8,1,1,26360,26,,S
|
| 206 |
+
1096,2,"Andrew, Mr. Frank Thomas",male,25,0,0,C.A. 34050,10.5,,S
|
| 207 |
+
1097,1,"Omont, Mr. Alfred Fernand",male,,0,0,F.C. 12998,25.7417,,C
|
| 208 |
+
1098,3,"McGowan, Miss. Katherine",female,35,0,0,9232,7.75,,Q
|
| 209 |
+
1099,2,"Collett, Mr. Sidney C Stuart",male,24,0,0,28034,10.5,,S
|
| 210 |
+
1100,1,"Rosenbaum, Miss. Edith Louise",female,33,0,0,PC 17613,27.7208,A11,C
|
| 211 |
+
1101,3,"Delalic, Mr. Redjo",male,25,0,0,349250,7.8958,,S
|
| 212 |
+
1102,3,"Andersen, Mr. Albert Karvin",male,32,0,0,C 4001,22.525,,S
|
| 213 |
+
1103,3,"Finoli, Mr. Luigi",male,,0,0,SOTON/O.Q. 3101308,7.05,,S
|
| 214 |
+
1104,2,"Deacon, Mr. Percy William",male,17,0,0,S.O.C. 14879,73.5,,S
|
| 215 |
+
1105,2,"Howard, Mrs. Benjamin (Ellen Truelove Arman)",female,60,1,0,24065,26,,S
|
| 216 |
+
1106,3,"Andersson, Miss. Ida Augusta Margareta",female,38,4,2,347091,7.775,,S
|
| 217 |
+
1107,1,"Head, Mr. Christopher",male,42,0,0,113038,42.5,B11,S
|
| 218 |
+
1108,3,"Mahon, Miss. Bridget Delia",female,,0,0,330924,7.8792,,Q
|
| 219 |
+
1109,1,"Wick, Mr. George Dennick",male,57,1,1,36928,164.8667,,S
|
| 220 |
+
1110,1,"Widener, Mrs. George Dunton (Eleanor Elkins)",female,50,1,1,113503,211.5,C80,C
|
| 221 |
+
1111,3,"Thomson, Mr. Alexander Morrison",male,,0,0,32302,8.05,,S
|
| 222 |
+
1112,2,"Duran y More, Miss. Florentina",female,30,1,0,SC/PARIS 2148,13.8583,,C
|
| 223 |
+
1113,3,"Reynolds, Mr. Harold J",male,21,0,0,342684,8.05,,S
|
| 224 |
+
1114,2,"Cook, Mrs. (Selena Rogers)",female,22,0,0,W./C. 14266,10.5,F33,S
|
| 225 |
+
1115,3,"Karlsson, Mr. Einar Gervasius",male,21,0,0,350053,7.7958,,S
|
| 226 |
+
1116,1,"Candee, Mrs. Edward (Helen Churchill Hungerford)",female,53,0,0,PC 17606,27.4458,,C
|
| 227 |
+
1117,3,"Moubarek, Mrs. George (Omine Amenia"" Alexander)""",female,,0,2,2661,15.2458,,C
|
| 228 |
+
1118,3,"Asplund, Mr. Johan Charles",male,23,0,0,350054,7.7958,,S
|
| 229 |
+
1119,3,"McNeill, Miss. Bridget",female,,0,0,370368,7.75,,Q
|
| 230 |
+
1120,3,"Everett, Mr. Thomas James",male,40.5,0,0,C.A. 6212,15.1,,S
|
| 231 |
+
1121,2,"Hocking, Mr. Samuel James Metcalfe",male,36,0,0,242963,13,,S
|
| 232 |
+
1122,2,"Sweet, Mr. George Frederick",male,14,0,0,220845,65,,S
|
| 233 |
+
1123,1,"Willard, Miss. Constance",female,21,0,0,113795,26.55,,S
|
| 234 |
+
1124,3,"Wiklund, Mr. Karl Johan",male,21,1,0,3101266,6.4958,,S
|
| 235 |
+
1125,3,"Linehan, Mr. Michael",male,,0,0,330971,7.8792,,Q
|
| 236 |
+
1126,1,"Cumings, Mr. John Bradley",male,39,1,0,PC 17599,71.2833,C85,C
|
| 237 |
+
1127,3,"Vendel, Mr. Olof Edvin",male,20,0,0,350416,7.8542,,S
|
| 238 |
+
1128,1,"Warren, Mr. Frank Manley",male,64,1,0,110813,75.25,D37,C
|
| 239 |
+
1129,3,"Baccos, Mr. Raffull",male,20,0,0,2679,7.225,,C
|
| 240 |
+
1130,2,"Hiltunen, Miss. Marta",female,18,1,1,250650,13,,S
|
| 241 |
+
1131,1,"Douglas, Mrs. Walter Donald (Mahala Dutton)",female,48,1,0,PC 17761,106.425,C86,C
|
| 242 |
+
1132,1,"Lindstrom, Mrs. Carl Johan (Sigrid Posse)",female,55,0,0,112377,27.7208,,C
|
| 243 |
+
1133,2,"Christy, Mrs. (Alice Frances)",female,45,0,2,237789,30,,S
|
| 244 |
+
1134,1,"Spedden, Mr. Frederic Oakley",male,45,1,1,16966,134.5,E34,C
|
| 245 |
+
1135,3,"Hyman, Mr. Abraham",male,,0,0,3470,7.8875,,S
|
| 246 |
+
1136,3,"Johnston, Master. William Arthur Willie""""",male,,1,2,W./C. 6607,23.45,,S
|
| 247 |
+
1137,1,"Kenyon, Mr. Frederick R",male,41,1,0,17464,51.8625,D21,S
|
| 248 |
+
1138,2,"Karnes, Mrs. J Frank (Claire Bennett)",female,22,0,0,F.C.C. 13534,21,,S
|
| 249 |
+
1139,2,"Drew, Mr. James Vivian",male,42,1,1,28220,32.5,,S
|
| 250 |
+
1140,2,"Hold, Mrs. Stephen (Annie Margaret Hill)",female,29,1,0,26707,26,,S
|
| 251 |
+
1141,3,"Khalil, Mrs. Betros (Zahie Maria"" Elias)""",female,,1,0,2660,14.4542,,C
|
| 252 |
+
1142,2,"West, Miss. Barbara J",female,0.92,1,2,C.A. 34651,27.75,,S
|
| 253 |
+
1143,3,"Abrahamsson, Mr. Abraham August Johannes",male,20,0,0,SOTON/O2 3101284,7.925,,S
|
| 254 |
+
1144,1,"Clark, Mr. Walter Miller",male,27,1,0,13508,136.7792,C89,C
|
| 255 |
+
1145,3,"Salander, Mr. Karl Johan",male,24,0,0,7266,9.325,,S
|
| 256 |
+
1146,3,"Wenzel, Mr. Linhart",male,32.5,0,0,345775,9.5,,S
|
| 257 |
+
1147,3,"MacKay, Mr. George William",male,,0,0,C.A. 42795,7.55,,S
|
| 258 |
+
1148,3,"Mahon, Mr. John",male,,0,0,AQ/4 3130,7.75,,Q
|
| 259 |
+
1149,3,"Niklasson, Mr. Samuel",male,28,0,0,363611,8.05,,S
|
| 260 |
+
1150,2,"Bentham, Miss. Lilian W",female,19,0,0,28404,13,,S
|
| 261 |
+
1151,3,"Midtsjo, Mr. Karl Albert",male,21,0,0,345501,7.775,,S
|
| 262 |
+
1152,3,"de Messemaeker, Mr. Guillaume Joseph",male,36.5,1,0,345572,17.4,,S
|
| 263 |
+
1153,3,"Nilsson, Mr. August Ferdinand",male,21,0,0,350410,7.8542,,S
|
| 264 |
+
1154,2,"Wells, Mrs. Arthur Henry (Addie"" Dart Trevaskis)""",female,29,0,2,29103,23,,S
|
| 265 |
+
1155,3,"Klasen, Miss. Gertrud Emilia",female,1,1,1,350405,12.1833,,S
|
| 266 |
+
1156,2,"Portaluppi, Mr. Emilio Ilario Giuseppe",male,30,0,0,C.A. 34644,12.7375,,C
|
| 267 |
+
1157,3,"Lyntakoff, Mr. Stanko",male,,0,0,349235,7.8958,,S
|
| 268 |
+
1158,1,"Chisholm, Mr. Roderick Robert Crispin",male,,0,0,112051,0,,S
|
| 269 |
+
1159,3,"Warren, Mr. Charles William",male,,0,0,C.A. 49867,7.55,,S
|
| 270 |
+
1160,3,"Howard, Miss. May Elizabeth",female,,0,0,A. 2. 39186,8.05,,S
|
| 271 |
+
1161,3,"Pokrnic, Mr. Mate",male,17,0,0,315095,8.6625,,S
|
| 272 |
+
1162,1,"McCaffry, Mr. Thomas Francis",male,46,0,0,13050,75.2417,C6,C
|
| 273 |
+
1163,3,"Fox, Mr. Patrick",male,,0,0,368573,7.75,,Q
|
| 274 |
+
1164,1,"Clark, Mrs. Walter Miller (Virginia McDowell)",female,26,1,0,13508,136.7792,C89,C
|
| 275 |
+
1165,3,"Lennon, Miss. Mary",female,,1,0,370371,15.5,,Q
|
| 276 |
+
1166,3,"Saade, Mr. Jean Nassr",male,,0,0,2676,7.225,,C
|
| 277 |
+
1167,2,"Bryhl, Miss. Dagmar Jenny Ingeborg ",female,20,1,0,236853,26,,S
|
| 278 |
+
1168,2,"Parker, Mr. Clifford Richard",male,28,0,0,SC 14888,10.5,,S
|
| 279 |
+
1169,2,"Faunthorpe, Mr. Harry",male,40,1,0,2926,26,,S
|
| 280 |
+
1170,2,"Ware, Mr. John James",male,30,1,0,CA 31352,21,,S
|
| 281 |
+
1171,2,"Oxenham, Mr. Percy Thomas",male,22,0,0,W./C. 14260,10.5,,S
|
| 282 |
+
1172,3,"Oreskovic, Miss. Jelka",female,23,0,0,315085,8.6625,,S
|
| 283 |
+
1173,3,"Peacock, Master. Alfred Edward",male,0.75,1,1,SOTON/O.Q. 3101315,13.775,,S
|
| 284 |
+
1174,3,"Fleming, Miss. Honora",female,,0,0,364859,7.75,,Q
|
| 285 |
+
1175,3,"Touma, Miss. Maria Youssef",female,9,1,1,2650,15.2458,,C
|
| 286 |
+
1176,3,"Rosblom, Miss. Salli Helena",female,2,1,1,370129,20.2125,,S
|
| 287 |
+
1177,3,"Dennis, Mr. William",male,36,0,0,A/5 21175,7.25,,S
|
| 288 |
+
1178,3,"Franklin, Mr. Charles (Charles Fardon)",male,,0,0,SOTON/O.Q. 3101314,7.25,,S
|
| 289 |
+
1179,1,"Snyder, Mr. John Pillsbury",male,24,1,0,21228,82.2667,B45,S
|
| 290 |
+
1180,3,"Mardirosian, Mr. Sarkis",male,,0,0,2655,7.2292,F E46,C
|
| 291 |
+
1181,3,"Ford, Mr. Arthur",male,,0,0,A/5 1478,8.05,,S
|
| 292 |
+
1182,1,"Rheims, Mr. George Alexander Lucien",male,,0,0,PC 17607,39.6,,S
|
| 293 |
+
1183,3,"Daly, Miss. Margaret Marcella Maggie""""",female,30,0,0,382650,6.95,,Q
|
| 294 |
+
1184,3,"Nasr, Mr. Mustafa",male,,0,0,2652,7.2292,,C
|
| 295 |
+
1185,1,"Dodge, Dr. Washington",male,53,1,1,33638,81.8583,A34,S
|
| 296 |
+
1186,3,"Wittevrongel, Mr. Camille",male,36,0,0,345771,9.5,,S
|
| 297 |
+
1187,3,"Angheloff, Mr. Minko",male,26,0,0,349202,7.8958,,S
|
| 298 |
+
1188,2,"Laroche, Miss. Louise",female,1,1,2,SC/Paris 2123,41.5792,,C
|
| 299 |
+
1189,3,"Samaan, Mr. Hanna",male,,2,0,2662,21.6792,,C
|
| 300 |
+
1190,1,"Loring, Mr. Joseph Holland",male,30,0,0,113801,45.5,,S
|
| 301 |
+
1191,3,"Johansson, Mr. Nils",male,29,0,0,347467,7.8542,,S
|
| 302 |
+
1192,3,"Olsson, Mr. Oscar Wilhelm",male,32,0,0,347079,7.775,,S
|
| 303 |
+
1193,2,"Malachard, Mr. Noel",male,,0,0,237735,15.0458,D,C
|
| 304 |
+
1194,2,"Phillips, Mr. Escott Robert",male,43,0,1,S.O./P.P. 2,21,,S
|
| 305 |
+
1195,3,"Pokrnic, Mr. Tome",male,24,0,0,315092,8.6625,,S
|
| 306 |
+
1196,3,"McCarthy, Miss. Catherine Katie""""",female,,0,0,383123,7.75,,Q
|
| 307 |
+
1197,1,"Crosby, Mrs. Edward Gifford (Catherine Elizabeth Halstead)",female,64,1,1,112901,26.55,B26,S
|
| 308 |
+
1198,1,"Allison, Mr. Hudson Joshua Creighton",male,30,1,2,113781,151.55,C22 C26,S
|
| 309 |
+
1199,3,"Aks, Master. Philip Frank",male,0.83,0,1,392091,9.35,,S
|
| 310 |
+
1200,1,"Hays, Mr. Charles Melville",male,55,1,1,12749,93.5,B69,S
|
| 311 |
+
1201,3,"Hansen, Mrs. Claus Peter (Jennie L Howard)",female,45,1,0,350026,14.1083,,S
|
| 312 |
+
1202,3,"Cacic, Mr. Jego Grga",male,18,0,0,315091,8.6625,,S
|
| 313 |
+
1203,3,"Vartanian, Mr. David",male,22,0,0,2658,7.225,,C
|
| 314 |
+
1204,3,"Sadowitz, Mr. Harry",male,,0,0,LP 1588,7.575,,S
|
| 315 |
+
1205,3,"Carr, Miss. Jeannie",female,37,0,0,368364,7.75,,Q
|
| 316 |
+
1206,1,"White, Mrs. John Stuart (Ella Holmes)",female,55,0,0,PC 17760,135.6333,C32,C
|
| 317 |
+
1207,3,"Hagardon, Miss. Kate",female,17,0,0,AQ/3. 30631,7.7333,,Q
|
| 318 |
+
1208,1,"Spencer, Mr. William Augustus",male,57,1,0,PC 17569,146.5208,B78,C
|
| 319 |
+
1209,2,"Rogers, Mr. Reginald Harry",male,19,0,0,28004,10.5,,S
|
| 320 |
+
1210,3,"Jonsson, Mr. Nils Hilding",male,27,0,0,350408,7.8542,,S
|
| 321 |
+
1211,2,"Jefferys, Mr. Ernest Wilfred",male,22,2,0,C.A. 31029,31.5,,S
|
| 322 |
+
1212,3,"Andersson, Mr. Johan Samuel",male,26,0,0,347075,7.775,,S
|
| 323 |
+
1213,3,"Krekorian, Mr. Neshan",male,25,0,0,2654,7.2292,F E57,C
|
| 324 |
+
1214,2,"Nesson, Mr. Israel",male,26,0,0,244368,13,F2,S
|
| 325 |
+
1215,1,"Rowe, Mr. Alfred G",male,33,0,0,113790,26.55,,S
|
| 326 |
+
1216,1,"Kreuchen, Miss. Emilie",female,39,0,0,24160,211.3375,,S
|
| 327 |
+
1217,3,"Assam, Mr. Ali",male,23,0,0,SOTON/O.Q. 3101309,7.05,,S
|
| 328 |
+
1218,2,"Becker, Miss. Ruth Elizabeth",female,12,2,1,230136,39,F4,S
|
| 329 |
+
1219,1,"Rosenshine, Mr. George (Mr George Thorne"")""",male,46,0,0,PC 17585,79.2,,C
|
| 330 |
+
1220,2,"Clarke, Mr. Charles Valentine",male,29,1,0,2003,26,,S
|
| 331 |
+
1221,2,"Enander, Mr. Ingvar",male,21,0,0,236854,13,,S
|
| 332 |
+
1222,2,"Davies, Mrs. John Morgan (Elizabeth Agnes Mary White) ",female,48,0,2,C.A. 33112,36.75,,S
|
| 333 |
+
1223,1,"Dulles, Mr. William Crothers",male,39,0,0,PC 17580,29.7,A18,C
|
| 334 |
+
1224,3,"Thomas, Mr. Tannous",male,,0,0,2684,7.225,,C
|
| 335 |
+
1225,3,"Nakid, Mrs. Said (Waika Mary"" Mowad)""",female,19,1,1,2653,15.7417,,C
|
| 336 |
+
1226,3,"Cor, Mr. Ivan",male,27,0,0,349229,7.8958,,S
|
| 337 |
+
1227,1,"Maguire, Mr. John Edward",male,30,0,0,110469,26,C106,S
|
| 338 |
+
1228,2,"de Brito, Mr. Jose Joaquim",male,32,0,0,244360,13,,S
|
| 339 |
+
1229,3,"Elias, Mr. Joseph",male,39,0,2,2675,7.2292,,C
|
| 340 |
+
1230,2,"Denbury, Mr. Herbert",male,25,0,0,C.A. 31029,31.5,,S
|
| 341 |
+
1231,3,"Betros, Master. Seman",male,,0,0,2622,7.2292,,C
|
| 342 |
+
1232,2,"Fillbrook, Mr. Joseph Charles",male,18,0,0,C.A. 15185,10.5,,S
|
| 343 |
+
1233,3,"Lundstrom, Mr. Thure Edvin",male,32,0,0,350403,7.5792,,S
|
| 344 |
+
1234,3,"Sage, Mr. John George",male,,1,9,CA. 2343,69.55,,S
|
| 345 |
+
1235,1,"Cardeza, Mrs. James Warburton Martinez (Charlotte Wardle Drake)",female,58,0,1,PC 17755,512.3292,B51 B53 B55,C
|
| 346 |
+
1236,3,"van Billiard, Master. James William",male,,1,1,A/5. 851,14.5,,S
|
| 347 |
+
1237,3,"Abelseth, Miss. Karen Marie",female,16,0,0,348125,7.65,,S
|
| 348 |
+
1238,2,"Botsford, Mr. William Hull",male,26,0,0,237670,13,,S
|
| 349 |
+
1239,3,"Whabee, Mrs. George Joseph (Shawneene Abi-Saab)",female,38,0,0,2688,7.2292,,C
|
| 350 |
+
1240,2,"Giles, Mr. Ralph",male,24,0,0,248726,13.5,,S
|
| 351 |
+
1241,2,"Walcroft, Miss. Nellie",female,31,0,0,F.C.C. 13528,21,,S
|
| 352 |
+
1242,1,"Greenfield, Mrs. Leo David (Blanche Strouse)",female,45,0,1,PC 17759,63.3583,D10 D12,C
|
| 353 |
+
1243,2,"Stokes, Mr. Philip Joseph",male,25,0,0,F.C.C. 13540,10.5,,S
|
| 354 |
+
1244,2,"Dibden, Mr. William",male,18,0,0,S.O.C. 14879,73.5,,S
|
| 355 |
+
1245,2,"Herman, Mr. Samuel",male,49,1,2,220845,65,,S
|
| 356 |
+
1246,3,"Dean, Miss. Elizabeth Gladys Millvina""""",female,0.17,1,2,C.A. 2315,20.575,,S
|
| 357 |
+
1247,1,"Julian, Mr. Henry Forbes",male,50,0,0,113044,26,E60,S
|
| 358 |
+
1248,1,"Brown, Mrs. John Murray (Caroline Lane Lamson)",female,59,2,0,11769,51.4792,C101,S
|
| 359 |
+
1249,3,"Lockyer, Mr. Edward",male,,0,0,1222,7.8792,,S
|
| 360 |
+
1250,3,"O'Keefe, Mr. Patrick",male,,0,0,368402,7.75,,Q
|
| 361 |
+
1251,3,"Lindell, Mrs. Edvard Bengtsson (Elin Gerda Persson)",female,30,1,0,349910,15.55,,S
|
| 362 |
+
1252,3,"Sage, Master. William Henry",male,14.5,8,2,CA. 2343,69.55,,S
|
| 363 |
+
1253,2,"Mallet, Mrs. Albert (Antoinette Magnin)",female,24,1,1,S.C./PARIS 2079,37.0042,,C
|
| 364 |
+
1254,2,"Ware, Mrs. John James (Florence Louise Long)",female,31,0,0,CA 31352,21,,S
|
| 365 |
+
1255,3,"Strilic, Mr. Ivan",male,27,0,0,315083,8.6625,,S
|
| 366 |
+
1256,1,"Harder, Mrs. George Achilles (Dorothy Annan)",female,25,1,0,11765,55.4417,E50,C
|
| 367 |
+
1257,3,"Sage, Mrs. John (Annie Bullen)",female,,1,9,CA. 2343,69.55,,S
|
| 368 |
+
1258,3,"Caram, Mr. Joseph",male,,1,0,2689,14.4583,,C
|
| 369 |
+
1259,3,"Riihivouri, Miss. Susanna Juhantytar Sanni""""",female,22,0,0,3101295,39.6875,,S
|
| 370 |
+
1260,1,"Gibson, Mrs. Leonard (Pauline C Boeson)",female,45,0,1,112378,59.4,,C
|
| 371 |
+
1261,2,"Pallas y Castello, Mr. Emilio",male,29,0,0,SC/PARIS 2147,13.8583,,C
|
| 372 |
+
1262,2,"Giles, Mr. Edgar",male,21,1,0,28133,11.5,,S
|
| 373 |
+
1263,1,"Wilson, Miss. Helen Alice",female,31,0,0,16966,134.5,E39 E41,C
|
| 374 |
+
1264,1,"Ismay, Mr. Joseph Bruce",male,49,0,0,112058,0,B52 B54 B56,S
|
| 375 |
+
1265,2,"Harbeck, Mr. William H",male,44,0,0,248746,13,,S
|
| 376 |
+
1266,1,"Dodge, Mrs. Washington (Ruth Vidaver)",female,54,1,1,33638,81.8583,A34,S
|
| 377 |
+
1267,1,"Bowen, Miss. Grace Scott",female,45,0,0,PC 17608,262.375,,C
|
| 378 |
+
1268,3,"Kink, Miss. Maria",female,22,2,0,315152,8.6625,,S
|
| 379 |
+
1269,2,"Cotterill, Mr. Henry Harry""""",male,21,0,0,29107,11.5,,S
|
| 380 |
+
1270,1,"Hipkins, Mr. William Edward",male,55,0,0,680,50,C39,S
|
| 381 |
+
1271,3,"Asplund, Master. Carl Edgar",male,5,4,2,347077,31.3875,,S
|
| 382 |
+
1272,3,"O'Connor, Mr. Patrick",male,,0,0,366713,7.75,,Q
|
| 383 |
+
1273,3,"Foley, Mr. Joseph",male,26,0,0,330910,7.8792,,Q
|
| 384 |
+
1274,3,"Risien, Mrs. Samuel (Emma)",female,,0,0,364498,14.5,,S
|
| 385 |
+
1275,3,"McNamee, Mrs. Neal (Eileen O'Leary)",female,19,1,0,376566,16.1,,S
|
| 386 |
+
1276,2,"Wheeler, Mr. Edwin Frederick""""",male,,0,0,SC/PARIS 2159,12.875,,S
|
| 387 |
+
1277,2,"Herman, Miss. Kate",female,24,1,2,220845,65,,S
|
| 388 |
+
1278,3,"Aronsson, Mr. Ernst Axel Algot",male,24,0,0,349911,7.775,,S
|
| 389 |
+
1279,2,"Ashby, Mr. John",male,57,0,0,244346,13,,S
|
| 390 |
+
1280,3,"Canavan, Mr. Patrick",male,21,0,0,364858,7.75,,Q
|
| 391 |
+
1281,3,"Palsson, Master. Paul Folke",male,6,3,1,349909,21.075,,S
|
| 392 |
+
1282,1,"Payne, Mr. Vivian Ponsonby",male,23,0,0,12749,93.5,B24,S
|
| 393 |
+
1283,1,"Lines, Mrs. Ernest H (Elizabeth Lindsey James)",female,51,0,1,PC 17592,39.4,D28,S
|
| 394 |
+
1284,3,"Abbott, Master. Eugene Joseph",male,13,0,2,C.A. 2673,20.25,,S
|
| 395 |
+
1285,2,"Gilbert, Mr. William",male,47,0,0,C.A. 30769,10.5,,S
|
| 396 |
+
1286,3,"Kink-Heilmann, Mr. Anton",male,29,3,1,315153,22.025,,S
|
| 397 |
+
1287,1,"Smith, Mrs. Lucien Philip (Mary Eloise Hughes)",female,18,1,0,13695,60,C31,S
|
| 398 |
+
1288,3,"Colbert, Mr. Patrick",male,24,0,0,371109,7.25,,Q
|
| 399 |
+
1289,1,"Frolicher-Stehli, Mrs. Maxmillian (Margaretha Emerentia Stehli)",female,48,1,1,13567,79.2,B41,C
|
| 400 |
+
1290,3,"Larsson-Rondberg, Mr. Edvard A",male,22,0,0,347065,7.775,,S
|
| 401 |
+
1291,3,"Conlon, Mr. Thomas Henry",male,31,0,0,21332,7.7333,,Q
|
| 402 |
+
1292,1,"Bonnell, Miss. Caroline",female,30,0,0,36928,164.8667,C7,S
|
| 403 |
+
1293,2,"Gale, Mr. Harry",male,38,1,0,28664,21,,S
|
| 404 |
+
1294,1,"Gibson, Miss. Dorothy Winifred",female,22,0,1,112378,59.4,,C
|
| 405 |
+
1295,1,"Carrau, Mr. Jose Pedro",male,17,0,0,113059,47.1,,S
|
| 406 |
+
1296,1,"Frauenthal, Mr. Isaac Gerald",male,43,1,0,17765,27.7208,D40,C
|
| 407 |
+
1297,2,"Nourney, Mr. Alfred (Baron von Drachstedt"")""",male,20,0,0,SC/PARIS 2166,13.8625,D38,C
|
| 408 |
+
1298,2,"Ware, Mr. William Jeffery",male,23,1,0,28666,10.5,,S
|
| 409 |
+
1299,1,"Widener, Mr. George Dunton",male,50,1,1,113503,211.5,C80,C
|
| 410 |
+
1300,3,"Riordan, Miss. Johanna Hannah""""",female,,0,0,334915,7.7208,,Q
|
| 411 |
+
1301,3,"Peacock, Miss. Treasteall",female,3,1,1,SOTON/O.Q. 3101315,13.775,,S
|
| 412 |
+
1302,3,"Naughton, Miss. Hannah",female,,0,0,365237,7.75,,Q
|
| 413 |
+
1303,1,"Minahan, Mrs. William Edward (Lillian E Thorpe)",female,37,1,0,19928,90,C78,Q
|
| 414 |
+
1304,3,"Henriksson, Miss. Jenny Lovisa",female,28,0,0,347086,7.775,,S
|
| 415 |
+
1305,3,"Spector, Mr. Woolf",male,,0,0,A.5. 3236,8.05,,S
|
| 416 |
+
1306,1,"Oliva y Ocana, Dona. Fermina",female,39,0,0,PC 17758,108.9,C105,C
|
| 417 |
+
1307,3,"Saether, Mr. Simon Sivertsen",male,38.5,0,0,SOTON/O.Q. 3101262,7.25,,S
|
| 418 |
+
1308,3,"Ware, Mr. Frederick",male,,0,0,359309,8.05,,S
|
| 419 |
+
1309,3,"Peter, Master. Michael J",male,,1,1,2668,22.3583,,C
|
Titanic/Data/train.csv
ADDED
|
@@ -0,0 +1,892 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
PassengerId,Survived,Pclass,Name,Sex,Age,SibSp,Parch,Ticket,Fare,Cabin,Embarked
|
| 2 |
+
1,0,3,"Braund, Mr. Owen Harris",male,22,1,0,A/5 21171,7.25,,S
|
| 3 |
+
2,1,1,"Cumings, Mrs. John Bradley (Florence Briggs Thayer)",female,38,1,0,PC 17599,71.2833,C85,C
|
| 4 |
+
3,1,3,"Heikkinen, Miss. Laina",female,26,0,0,STON/O2. 3101282,7.925,,S
|
| 5 |
+
4,1,1,"Futrelle, Mrs. Jacques Heath (Lily May Peel)",female,35,1,0,113803,53.1,C123,S
|
| 6 |
+
5,0,3,"Allen, Mr. William Henry",male,35,0,0,373450,8.05,,S
|
| 7 |
+
6,0,3,"Moran, Mr. James",male,,0,0,330877,8.4583,,Q
|
| 8 |
+
7,0,1,"McCarthy, Mr. Timothy J",male,54,0,0,17463,51.8625,E46,S
|
| 9 |
+
8,0,3,"Palsson, Master. Gosta Leonard",male,2,3,1,349909,21.075,,S
|
| 10 |
+
9,1,3,"Johnson, Mrs. Oscar W (Elisabeth Vilhelmina Berg)",female,27,0,2,347742,11.1333,,S
|
| 11 |
+
10,1,2,"Nasser, Mrs. Nicholas (Adele Achem)",female,14,1,0,237736,30.0708,,C
|
| 12 |
+
11,1,3,"Sandstrom, Miss. Marguerite Rut",female,4,1,1,PP 9549,16.7,G6,S
|
| 13 |
+
12,1,1,"Bonnell, Miss. Elizabeth",female,58,0,0,113783,26.55,C103,S
|
| 14 |
+
13,0,3,"Saundercock, Mr. William Henry",male,20,0,0,A/5. 2151,8.05,,S
|
| 15 |
+
14,0,3,"Andersson, Mr. Anders Johan",male,39,1,5,347082,31.275,,S
|
| 16 |
+
15,0,3,"Vestrom, Miss. Hulda Amanda Adolfina",female,14,0,0,350406,7.8542,,S
|
| 17 |
+
16,1,2,"Hewlett, Mrs. (Mary D Kingcome) ",female,55,0,0,248706,16,,S
|
| 18 |
+
17,0,3,"Rice, Master. Eugene",male,2,4,1,382652,29.125,,Q
|
| 19 |
+
18,1,2,"Williams, Mr. Charles Eugene",male,,0,0,244373,13,,S
|
| 20 |
+
19,0,3,"Vander Planke, Mrs. Julius (Emelia Maria Vandemoortele)",female,31,1,0,345763,18,,S
|
| 21 |
+
20,1,3,"Masselmani, Mrs. Fatima",female,,0,0,2649,7.225,,C
|
| 22 |
+
21,0,2,"Fynney, Mr. Joseph J",male,35,0,0,239865,26,,S
|
| 23 |
+
22,1,2,"Beesley, Mr. Lawrence",male,34,0,0,248698,13,D56,S
|
| 24 |
+
23,1,3,"McGowan, Miss. Anna ""Annie""",female,15,0,0,330923,8.0292,,Q
|
| 25 |
+
24,1,1,"Sloper, Mr. William Thompson",male,28,0,0,113788,35.5,A6,S
|
| 26 |
+
25,0,3,"Palsson, Miss. Torborg Danira",female,8,3,1,349909,21.075,,S
|
| 27 |
+
26,1,3,"Asplund, Mrs. Carl Oscar (Selma Augusta Emilia Johansson)",female,38,1,5,347077,31.3875,,S
|
| 28 |
+
27,0,3,"Emir, Mr. Farred Chehab",male,,0,0,2631,7.225,,C
|
| 29 |
+
28,0,1,"Fortune, Mr. Charles Alexander",male,19,3,2,19950,263,C23 C25 C27,S
|
| 30 |
+
29,1,3,"O'Dwyer, Miss. Ellen ""Nellie""",female,,0,0,330959,7.8792,,Q
|
| 31 |
+
30,0,3,"Todoroff, Mr. Lalio",male,,0,0,349216,7.8958,,S
|
| 32 |
+
31,0,1,"Uruchurtu, Don. Manuel E",male,40,0,0,PC 17601,27.7208,,C
|
| 33 |
+
32,1,1,"Spencer, Mrs. William Augustus (Marie Eugenie)",female,,1,0,PC 17569,146.5208,B78,C
|
| 34 |
+
33,1,3,"Glynn, Miss. Mary Agatha",female,,0,0,335677,7.75,,Q
|
| 35 |
+
34,0,2,"Wheadon, Mr. Edward H",male,66,0,0,C.A. 24579,10.5,,S
|
| 36 |
+
35,0,1,"Meyer, Mr. Edgar Joseph",male,28,1,0,PC 17604,82.1708,,C
|
| 37 |
+
36,0,1,"Holverson, Mr. Alexander Oskar",male,42,1,0,113789,52,,S
|
| 38 |
+
37,1,3,"Mamee, Mr. Hanna",male,,0,0,2677,7.2292,,C
|
| 39 |
+
38,0,3,"Cann, Mr. Ernest Charles",male,21,0,0,A./5. 2152,8.05,,S
|
| 40 |
+
39,0,3,"Vander Planke, Miss. Augusta Maria",female,18,2,0,345764,18,,S
|
| 41 |
+
40,1,3,"Nicola-Yarred, Miss. Jamila",female,14,1,0,2651,11.2417,,C
|
| 42 |
+
41,0,3,"Ahlin, Mrs. Johan (Johanna Persdotter Larsson)",female,40,1,0,7546,9.475,,S
|
| 43 |
+
42,0,2,"Turpin, Mrs. William John Robert (Dorothy Ann Wonnacott)",female,27,1,0,11668,21,,S
|
| 44 |
+
43,0,3,"Kraeff, Mr. Theodor",male,,0,0,349253,7.8958,,C
|
| 45 |
+
44,1,2,"Laroche, Miss. Simonne Marie Anne Andree",female,3,1,2,SC/Paris 2123,41.5792,,C
|
| 46 |
+
45,1,3,"Devaney, Miss. Margaret Delia",female,19,0,0,330958,7.8792,,Q
|
| 47 |
+
46,0,3,"Rogers, Mr. William John",male,,0,0,S.C./A.4. 23567,8.05,,S
|
| 48 |
+
47,0,3,"Lennon, Mr. Denis",male,,1,0,370371,15.5,,Q
|
| 49 |
+
48,1,3,"O'Driscoll, Miss. Bridget",female,,0,0,14311,7.75,,Q
|
| 50 |
+
49,0,3,"Samaan, Mr. Youssef",male,,2,0,2662,21.6792,,C
|
| 51 |
+
50,0,3,"Arnold-Franchi, Mrs. Josef (Josefine Franchi)",female,18,1,0,349237,17.8,,S
|
| 52 |
+
51,0,3,"Panula, Master. Juha Niilo",male,7,4,1,3101295,39.6875,,S
|
| 53 |
+
52,0,3,"Nosworthy, Mr. Richard Cater",male,21,0,0,A/4. 39886,7.8,,S
|
| 54 |
+
53,1,1,"Harper, Mrs. Henry Sleeper (Myna Haxtun)",female,49,1,0,PC 17572,76.7292,D33,C
|
| 55 |
+
54,1,2,"Faunthorpe, Mrs. Lizzie (Elizabeth Anne Wilkinson)",female,29,1,0,2926,26,,S
|
| 56 |
+
55,0,1,"Ostby, Mr. Engelhart Cornelius",male,65,0,1,113509,61.9792,B30,C
|
| 57 |
+
56,1,1,"Woolner, Mr. Hugh",male,,0,0,19947,35.5,C52,S
|
| 58 |
+
57,1,2,"Rugg, Miss. Emily",female,21,0,0,C.A. 31026,10.5,,S
|
| 59 |
+
58,0,3,"Novel, Mr. Mansouer",male,28.5,0,0,2697,7.2292,,C
|
| 60 |
+
59,1,2,"West, Miss. Constance Mirium",female,5,1,2,C.A. 34651,27.75,,S
|
| 61 |
+
60,0,3,"Goodwin, Master. William Frederick",male,11,5,2,CA 2144,46.9,,S
|
| 62 |
+
61,0,3,"Sirayanian, Mr. Orsen",male,22,0,0,2669,7.2292,,C
|
| 63 |
+
62,1,1,"Icard, Miss. Amelie",female,38,0,0,113572,80,B28,
|
| 64 |
+
63,0,1,"Harris, Mr. Henry Birkhardt",male,45,1,0,36973,83.475,C83,S
|
| 65 |
+
64,0,3,"Skoog, Master. Harald",male,4,3,2,347088,27.9,,S
|
| 66 |
+
65,0,1,"Stewart, Mr. Albert A",male,,0,0,PC 17605,27.7208,,C
|
| 67 |
+
66,1,3,"Moubarek, Master. Gerios",male,,1,1,2661,15.2458,,C
|
| 68 |
+
67,1,2,"Nye, Mrs. (Elizabeth Ramell)",female,29,0,0,C.A. 29395,10.5,F33,S
|
| 69 |
+
68,0,3,"Crease, Mr. Ernest James",male,19,0,0,S.P. 3464,8.1583,,S
|
| 70 |
+
69,1,3,"Andersson, Miss. Erna Alexandra",female,17,4,2,3101281,7.925,,S
|
| 71 |
+
70,0,3,"Kink, Mr. Vincenz",male,26,2,0,315151,8.6625,,S
|
| 72 |
+
71,0,2,"Jenkin, Mr. Stephen Curnow",male,32,0,0,C.A. 33111,10.5,,S
|
| 73 |
+
72,0,3,"Goodwin, Miss. Lillian Amy",female,16,5,2,CA 2144,46.9,,S
|
| 74 |
+
73,0,2,"Hood, Mr. Ambrose Jr",male,21,0,0,S.O.C. 14879,73.5,,S
|
| 75 |
+
74,0,3,"Chronopoulos, Mr. Apostolos",male,26,1,0,2680,14.4542,,C
|
| 76 |
+
75,1,3,"Bing, Mr. Lee",male,32,0,0,1601,56.4958,,S
|
| 77 |
+
76,0,3,"Moen, Mr. Sigurd Hansen",male,25,0,0,348123,7.65,F G73,S
|
| 78 |
+
77,0,3,"Staneff, Mr. Ivan",male,,0,0,349208,7.8958,,S
|
| 79 |
+
78,0,3,"Moutal, Mr. Rahamin Haim",male,,0,0,374746,8.05,,S
|
| 80 |
+
79,1,2,"Caldwell, Master. Alden Gates",male,0.83,0,2,248738,29,,S
|
| 81 |
+
80,1,3,"Dowdell, Miss. Elizabeth",female,30,0,0,364516,12.475,,S
|
| 82 |
+
81,0,3,"Waelens, Mr. Achille",male,22,0,0,345767,9,,S
|
| 83 |
+
82,1,3,"Sheerlinck, Mr. Jan Baptist",male,29,0,0,345779,9.5,,S
|
| 84 |
+
83,1,3,"McDermott, Miss. Brigdet Delia",female,,0,0,330932,7.7875,,Q
|
| 85 |
+
84,0,1,"Carrau, Mr. Francisco M",male,28,0,0,113059,47.1,,S
|
| 86 |
+
85,1,2,"Ilett, Miss. Bertha",female,17,0,0,SO/C 14885,10.5,,S
|
| 87 |
+
86,1,3,"Backstrom, Mrs. Karl Alfred (Maria Mathilda Gustafsson)",female,33,3,0,3101278,15.85,,S
|
| 88 |
+
87,0,3,"Ford, Mr. William Neal",male,16,1,3,W./C. 6608,34.375,,S
|
| 89 |
+
88,0,3,"Slocovski, Mr. Selman Francis",male,,0,0,SOTON/OQ 392086,8.05,,S
|
| 90 |
+
89,1,1,"Fortune, Miss. Mabel Helen",female,23,3,2,19950,263,C23 C25 C27,S
|
| 91 |
+
90,0,3,"Celotti, Mr. Francesco",male,24,0,0,343275,8.05,,S
|
| 92 |
+
91,0,3,"Christmann, Mr. Emil",male,29,0,0,343276,8.05,,S
|
| 93 |
+
92,0,3,"Andreasson, Mr. Paul Edvin",male,20,0,0,347466,7.8542,,S
|
| 94 |
+
93,0,1,"Chaffee, Mr. Herbert Fuller",male,46,1,0,W.E.P. 5734,61.175,E31,S
|
| 95 |
+
94,0,3,"Dean, Mr. Bertram Frank",male,26,1,2,C.A. 2315,20.575,,S
|
| 96 |
+
95,0,3,"Coxon, Mr. Daniel",male,59,0,0,364500,7.25,,S
|
| 97 |
+
96,0,3,"Shorney, Mr. Charles Joseph",male,,0,0,374910,8.05,,S
|
| 98 |
+
97,0,1,"Goldschmidt, Mr. George B",male,71,0,0,PC 17754,34.6542,A5,C
|
| 99 |
+
98,1,1,"Greenfield, Mr. William Bertram",male,23,0,1,PC 17759,63.3583,D10 D12,C
|
| 100 |
+
99,1,2,"Doling, Mrs. John T (Ada Julia Bone)",female,34,0,1,231919,23,,S
|
| 101 |
+
100,0,2,"Kantor, Mr. Sinai",male,34,1,0,244367,26,,S
|
| 102 |
+
101,0,3,"Petranec, Miss. Matilda",female,28,0,0,349245,7.8958,,S
|
| 103 |
+
102,0,3,"Petroff, Mr. Pastcho (""Pentcho"")",male,,0,0,349215,7.8958,,S
|
| 104 |
+
103,0,1,"White, Mr. Richard Frasar",male,21,0,1,35281,77.2875,D26,S
|
| 105 |
+
104,0,3,"Johansson, Mr. Gustaf Joel",male,33,0,0,7540,8.6542,,S
|
| 106 |
+
105,0,3,"Gustafsson, Mr. Anders Vilhelm",male,37,2,0,3101276,7.925,,S
|
| 107 |
+
106,0,3,"Mionoff, Mr. Stoytcho",male,28,0,0,349207,7.8958,,S
|
| 108 |
+
107,1,3,"Salkjelsvik, Miss. Anna Kristine",female,21,0,0,343120,7.65,,S
|
| 109 |
+
108,1,3,"Moss, Mr. Albert Johan",male,,0,0,312991,7.775,,S
|
| 110 |
+
109,0,3,"Rekic, Mr. Tido",male,38,0,0,349249,7.8958,,S
|
| 111 |
+
110,1,3,"Moran, Miss. Bertha",female,,1,0,371110,24.15,,Q
|
| 112 |
+
111,0,1,"Porter, Mr. Walter Chamberlain",male,47,0,0,110465,52,C110,S
|
| 113 |
+
112,0,3,"Zabour, Miss. Hileni",female,14.5,1,0,2665,14.4542,,C
|
| 114 |
+
113,0,3,"Barton, Mr. David John",male,22,0,0,324669,8.05,,S
|
| 115 |
+
114,0,3,"Jussila, Miss. Katriina",female,20,1,0,4136,9.825,,S
|
| 116 |
+
115,0,3,"Attalah, Miss. Malake",female,17,0,0,2627,14.4583,,C
|
| 117 |
+
116,0,3,"Pekoniemi, Mr. Edvard",male,21,0,0,STON/O 2. 3101294,7.925,,S
|
| 118 |
+
117,0,3,"Connors, Mr. Patrick",male,70.5,0,0,370369,7.75,,Q
|
| 119 |
+
118,0,2,"Turpin, Mr. William John Robert",male,29,1,0,11668,21,,S
|
| 120 |
+
119,0,1,"Baxter, Mr. Quigg Edmond",male,24,0,1,PC 17558,247.5208,B58 B60,C
|
| 121 |
+
120,0,3,"Andersson, Miss. Ellis Anna Maria",female,2,4,2,347082,31.275,,S
|
| 122 |
+
121,0,2,"Hickman, Mr. Stanley George",male,21,2,0,S.O.C. 14879,73.5,,S
|
| 123 |
+
122,0,3,"Moore, Mr. Leonard Charles",male,,0,0,A4. 54510,8.05,,S
|
| 124 |
+
123,0,2,"Nasser, Mr. Nicholas",male,32.5,1,0,237736,30.0708,,C
|
| 125 |
+
124,1,2,"Webber, Miss. Susan",female,32.5,0,0,27267,13,E101,S
|
| 126 |
+
125,0,1,"White, Mr. Percival Wayland",male,54,0,1,35281,77.2875,D26,S
|
| 127 |
+
126,1,3,"Nicola-Yarred, Master. Elias",male,12,1,0,2651,11.2417,,C
|
| 128 |
+
127,0,3,"McMahon, Mr. Martin",male,,0,0,370372,7.75,,Q
|
| 129 |
+
128,1,3,"Madsen, Mr. Fridtjof Arne",male,24,0,0,C 17369,7.1417,,S
|
| 130 |
+
129,1,3,"Peter, Miss. Anna",female,,1,1,2668,22.3583,F E69,C
|
| 131 |
+
130,0,3,"Ekstrom, Mr. Johan",male,45,0,0,347061,6.975,,S
|
| 132 |
+
131,0,3,"Drazenoic, Mr. Jozef",male,33,0,0,349241,7.8958,,C
|
| 133 |
+
132,0,3,"Coelho, Mr. Domingos Fernandeo",male,20,0,0,SOTON/O.Q. 3101307,7.05,,S
|
| 134 |
+
133,0,3,"Robins, Mrs. Alexander A (Grace Charity Laury)",female,47,1,0,A/5. 3337,14.5,,S
|
| 135 |
+
134,1,2,"Weisz, Mrs. Leopold (Mathilde Francoise Pede)",female,29,1,0,228414,26,,S
|
| 136 |
+
135,0,2,"Sobey, Mr. Samuel James Hayden",male,25,0,0,C.A. 29178,13,,S
|
| 137 |
+
136,0,2,"Richard, Mr. Emile",male,23,0,0,SC/PARIS 2133,15.0458,,C
|
| 138 |
+
137,1,1,"Newsom, Miss. Helen Monypeny",female,19,0,2,11752,26.2833,D47,S
|
| 139 |
+
138,0,1,"Futrelle, Mr. Jacques Heath",male,37,1,0,113803,53.1,C123,S
|
| 140 |
+
139,0,3,"Osen, Mr. Olaf Elon",male,16,0,0,7534,9.2167,,S
|
| 141 |
+
140,0,1,"Giglio, Mr. Victor",male,24,0,0,PC 17593,79.2,B86,C
|
| 142 |
+
141,0,3,"Boulos, Mrs. Joseph (Sultana)",female,,0,2,2678,15.2458,,C
|
| 143 |
+
142,1,3,"Nysten, Miss. Anna Sofia",female,22,0,0,347081,7.75,,S
|
| 144 |
+
143,1,3,"Hakkarainen, Mrs. Pekka Pietari (Elin Matilda Dolck)",female,24,1,0,STON/O2. 3101279,15.85,,S
|
| 145 |
+
144,0,3,"Burke, Mr. Jeremiah",male,19,0,0,365222,6.75,,Q
|
| 146 |
+
145,0,2,"Andrew, Mr. Edgardo Samuel",male,18,0,0,231945,11.5,,S
|
| 147 |
+
146,0,2,"Nicholls, Mr. Joseph Charles",male,19,1,1,C.A. 33112,36.75,,S
|
| 148 |
+
147,1,3,"Andersson, Mr. August Edvard (""Wennerstrom"")",male,27,0,0,350043,7.7958,,S
|
| 149 |
+
148,0,3,"Ford, Miss. Robina Maggie ""Ruby""",female,9,2,2,W./C. 6608,34.375,,S
|
| 150 |
+
149,0,2,"Navratil, Mr. Michel (""Louis M Hoffman"")",male,36.5,0,2,230080,26,F2,S
|
| 151 |
+
150,0,2,"Byles, Rev. Thomas Roussel Davids",male,42,0,0,244310,13,,S
|
| 152 |
+
151,0,2,"Bateman, Rev. Robert James",male,51,0,0,S.O.P. 1166,12.525,,S
|
| 153 |
+
152,1,1,"Pears, Mrs. Thomas (Edith Wearne)",female,22,1,0,113776,66.6,C2,S
|
| 154 |
+
153,0,3,"Meo, Mr. Alfonzo",male,55.5,0,0,A.5. 11206,8.05,,S
|
| 155 |
+
154,0,3,"van Billiard, Mr. Austin Blyler",male,40.5,0,2,A/5. 851,14.5,,S
|
| 156 |
+
155,0,3,"Olsen, Mr. Ole Martin",male,,0,0,Fa 265302,7.3125,,S
|
| 157 |
+
156,0,1,"Williams, Mr. Charles Duane",male,51,0,1,PC 17597,61.3792,,C
|
| 158 |
+
157,1,3,"Gilnagh, Miss. Katherine ""Katie""",female,16,0,0,35851,7.7333,,Q
|
| 159 |
+
158,0,3,"Corn, Mr. Harry",male,30,0,0,SOTON/OQ 392090,8.05,,S
|
| 160 |
+
159,0,3,"Smiljanic, Mr. Mile",male,,0,0,315037,8.6625,,S
|
| 161 |
+
160,0,3,"Sage, Master. Thomas Henry",male,,8,2,CA. 2343,69.55,,S
|
| 162 |
+
161,0,3,"Cribb, Mr. John Hatfield",male,44,0,1,371362,16.1,,S
|
| 163 |
+
162,1,2,"Watt, Mrs. James (Elizabeth ""Bessie"" Inglis Milne)",female,40,0,0,C.A. 33595,15.75,,S
|
| 164 |
+
163,0,3,"Bengtsson, Mr. John Viktor",male,26,0,0,347068,7.775,,S
|
| 165 |
+
164,0,3,"Calic, Mr. Jovo",male,17,0,0,315093,8.6625,,S
|
| 166 |
+
165,0,3,"Panula, Master. Eino Viljami",male,1,4,1,3101295,39.6875,,S
|
| 167 |
+
166,1,3,"Goldsmith, Master. Frank John William ""Frankie""",male,9,0,2,363291,20.525,,S
|
| 168 |
+
167,1,1,"Chibnall, Mrs. (Edith Martha Bowerman)",female,,0,1,113505,55,E33,S
|
| 169 |
+
168,0,3,"Skoog, Mrs. William (Anna Bernhardina Karlsson)",female,45,1,4,347088,27.9,,S
|
| 170 |
+
169,0,1,"Baumann, Mr. John D",male,,0,0,PC 17318,25.925,,S
|
| 171 |
+
170,0,3,"Ling, Mr. Lee",male,28,0,0,1601,56.4958,,S
|
| 172 |
+
171,0,1,"Van der hoef, Mr. Wyckoff",male,61,0,0,111240,33.5,B19,S
|
| 173 |
+
172,0,3,"Rice, Master. Arthur",male,4,4,1,382652,29.125,,Q
|
| 174 |
+
173,1,3,"Johnson, Miss. Eleanor Ileen",female,1,1,1,347742,11.1333,,S
|
| 175 |
+
174,0,3,"Sivola, Mr. Antti Wilhelm",male,21,0,0,STON/O 2. 3101280,7.925,,S
|
| 176 |
+
175,0,1,"Smith, Mr. James Clinch",male,56,0,0,17764,30.6958,A7,C
|
| 177 |
+
176,0,3,"Klasen, Mr. Klas Albin",male,18,1,1,350404,7.8542,,S
|
| 178 |
+
177,0,3,"Lefebre, Master. Henry Forbes",male,,3,1,4133,25.4667,,S
|
| 179 |
+
178,0,1,"Isham, Miss. Ann Elizabeth",female,50,0,0,PC 17595,28.7125,C49,C
|
| 180 |
+
179,0,2,"Hale, Mr. Reginald",male,30,0,0,250653,13,,S
|
| 181 |
+
180,0,3,"Leonard, Mr. Lionel",male,36,0,0,LINE,0,,S
|
| 182 |
+
181,0,3,"Sage, Miss. Constance Gladys",female,,8,2,CA. 2343,69.55,,S
|
| 183 |
+
182,0,2,"Pernot, Mr. Rene",male,,0,0,SC/PARIS 2131,15.05,,C
|
| 184 |
+
183,0,3,"Asplund, Master. Clarence Gustaf Hugo",male,9,4,2,347077,31.3875,,S
|
| 185 |
+
184,1,2,"Becker, Master. Richard F",male,1,2,1,230136,39,F4,S
|
| 186 |
+
185,1,3,"Kink-Heilmann, Miss. Luise Gretchen",female,4,0,2,315153,22.025,,S
|
| 187 |
+
186,0,1,"Rood, Mr. Hugh Roscoe",male,,0,0,113767,50,A32,S
|
| 188 |
+
187,1,3,"O'Brien, Mrs. Thomas (Johanna ""Hannah"" Godfrey)",female,,1,0,370365,15.5,,Q
|
| 189 |
+
188,1,1,"Romaine, Mr. Charles Hallace (""Mr C Rolmane"")",male,45,0,0,111428,26.55,,S
|
| 190 |
+
189,0,3,"Bourke, Mr. John",male,40,1,1,364849,15.5,,Q
|
| 191 |
+
190,0,3,"Turcin, Mr. Stjepan",male,36,0,0,349247,7.8958,,S
|
| 192 |
+
191,1,2,"Pinsky, Mrs. (Rosa)",female,32,0,0,234604,13,,S
|
| 193 |
+
192,0,2,"Carbines, Mr. William",male,19,0,0,28424,13,,S
|
| 194 |
+
193,1,3,"Andersen-Jensen, Miss. Carla Christine Nielsine",female,19,1,0,350046,7.8542,,S
|
| 195 |
+
194,1,2,"Navratil, Master. Michel M",male,3,1,1,230080,26,F2,S
|
| 196 |
+
195,1,1,"Brown, Mrs. James Joseph (Margaret Tobin)",female,44,0,0,PC 17610,27.7208,B4,C
|
| 197 |
+
196,1,1,"Lurette, Miss. Elise",female,58,0,0,PC 17569,146.5208,B80,C
|
| 198 |
+
197,0,3,"Mernagh, Mr. Robert",male,,0,0,368703,7.75,,Q
|
| 199 |
+
198,0,3,"Olsen, Mr. Karl Siegwart Andreas",male,42,0,1,4579,8.4042,,S
|
| 200 |
+
199,1,3,"Madigan, Miss. Margaret ""Maggie""",female,,0,0,370370,7.75,,Q
|
| 201 |
+
200,0,2,"Yrois, Miss. Henriette (""Mrs Harbeck"")",female,24,0,0,248747,13,,S
|
| 202 |
+
201,0,3,"Vande Walle, Mr. Nestor Cyriel",male,28,0,0,345770,9.5,,S
|
| 203 |
+
202,0,3,"Sage, Mr. Frederick",male,,8,2,CA. 2343,69.55,,S
|
| 204 |
+
203,0,3,"Johanson, Mr. Jakob Alfred",male,34,0,0,3101264,6.4958,,S
|
| 205 |
+
204,0,3,"Youseff, Mr. Gerious",male,45.5,0,0,2628,7.225,,C
|
| 206 |
+
205,1,3,"Cohen, Mr. Gurshon ""Gus""",male,18,0,0,A/5 3540,8.05,,S
|
| 207 |
+
206,0,3,"Strom, Miss. Telma Matilda",female,2,0,1,347054,10.4625,G6,S
|
| 208 |
+
207,0,3,"Backstrom, Mr. Karl Alfred",male,32,1,0,3101278,15.85,,S
|
| 209 |
+
208,1,3,"Albimona, Mr. Nassef Cassem",male,26,0,0,2699,18.7875,,C
|
| 210 |
+
209,1,3,"Carr, Miss. Helen ""Ellen""",female,16,0,0,367231,7.75,,Q
|
| 211 |
+
210,1,1,"Blank, Mr. Henry",male,40,0,0,112277,31,A31,C
|
| 212 |
+
211,0,3,"Ali, Mr. Ahmed",male,24,0,0,SOTON/O.Q. 3101311,7.05,,S
|
| 213 |
+
212,1,2,"Cameron, Miss. Clear Annie",female,35,0,0,F.C.C. 13528,21,,S
|
| 214 |
+
213,0,3,"Perkin, Mr. John Henry",male,22,0,0,A/5 21174,7.25,,S
|
| 215 |
+
214,0,2,"Givard, Mr. Hans Kristensen",male,30,0,0,250646,13,,S
|
| 216 |
+
215,0,3,"Kiernan, Mr. Philip",male,,1,0,367229,7.75,,Q
|
| 217 |
+
216,1,1,"Newell, Miss. Madeleine",female,31,1,0,35273,113.275,D36,C
|
| 218 |
+
217,1,3,"Honkanen, Miss. Eliina",female,27,0,0,STON/O2. 3101283,7.925,,S
|
| 219 |
+
218,0,2,"Jacobsohn, Mr. Sidney Samuel",male,42,1,0,243847,27,,S
|
| 220 |
+
219,1,1,"Bazzani, Miss. Albina",female,32,0,0,11813,76.2917,D15,C
|
| 221 |
+
220,0,2,"Harris, Mr. Walter",male,30,0,0,W/C 14208,10.5,,S
|
| 222 |
+
221,1,3,"Sunderland, Mr. Victor Francis",male,16,0,0,SOTON/OQ 392089,8.05,,S
|
| 223 |
+
222,0,2,"Bracken, Mr. James H",male,27,0,0,220367,13,,S
|
| 224 |
+
223,0,3,"Green, Mr. George Henry",male,51,0,0,21440,8.05,,S
|
| 225 |
+
224,0,3,"Nenkoff, Mr. Christo",male,,0,0,349234,7.8958,,S
|
| 226 |
+
225,1,1,"Hoyt, Mr. Frederick Maxfield",male,38,1,0,19943,90,C93,S
|
| 227 |
+
226,0,3,"Berglund, Mr. Karl Ivar Sven",male,22,0,0,PP 4348,9.35,,S
|
| 228 |
+
227,1,2,"Mellors, Mr. William John",male,19,0,0,SW/PP 751,10.5,,S
|
| 229 |
+
228,0,3,"Lovell, Mr. John Hall (""Henry"")",male,20.5,0,0,A/5 21173,7.25,,S
|
| 230 |
+
229,0,2,"Fahlstrom, Mr. Arne Jonas",male,18,0,0,236171,13,,S
|
| 231 |
+
230,0,3,"Lefebre, Miss. Mathilde",female,,3,1,4133,25.4667,,S
|
| 232 |
+
231,1,1,"Harris, Mrs. Henry Birkhardt (Irene Wallach)",female,35,1,0,36973,83.475,C83,S
|
| 233 |
+
232,0,3,"Larsson, Mr. Bengt Edvin",male,29,0,0,347067,7.775,,S
|
| 234 |
+
233,0,2,"Sjostedt, Mr. Ernst Adolf",male,59,0,0,237442,13.5,,S
|
| 235 |
+
234,1,3,"Asplund, Miss. Lillian Gertrud",female,5,4,2,347077,31.3875,,S
|
| 236 |
+
235,0,2,"Leyson, Mr. Robert William Norman",male,24,0,0,C.A. 29566,10.5,,S
|
| 237 |
+
236,0,3,"Harknett, Miss. Alice Phoebe",female,,0,0,W./C. 6609,7.55,,S
|
| 238 |
+
237,0,2,"Hold, Mr. Stephen",male,44,1,0,26707,26,,S
|
| 239 |
+
238,1,2,"Collyer, Miss. Marjorie ""Lottie""",female,8,0,2,C.A. 31921,26.25,,S
|
| 240 |
+
239,0,2,"Pengelly, Mr. Frederick William",male,19,0,0,28665,10.5,,S
|
| 241 |
+
240,0,2,"Hunt, Mr. George Henry",male,33,0,0,SCO/W 1585,12.275,,S
|
| 242 |
+
241,0,3,"Zabour, Miss. Thamine",female,,1,0,2665,14.4542,,C
|
| 243 |
+
242,1,3,"Murphy, Miss. Katherine ""Kate""",female,,1,0,367230,15.5,,Q
|
| 244 |
+
243,0,2,"Coleridge, Mr. Reginald Charles",male,29,0,0,W./C. 14263,10.5,,S
|
| 245 |
+
244,0,3,"Maenpaa, Mr. Matti Alexanteri",male,22,0,0,STON/O 2. 3101275,7.125,,S
|
| 246 |
+
245,0,3,"Attalah, Mr. Sleiman",male,30,0,0,2694,7.225,,C
|
| 247 |
+
246,0,1,"Minahan, Dr. William Edward",male,44,2,0,19928,90,C78,Q
|
| 248 |
+
247,0,3,"Lindahl, Miss. Agda Thorilda Viktoria",female,25,0,0,347071,7.775,,S
|
| 249 |
+
248,1,2,"Hamalainen, Mrs. William (Anna)",female,24,0,2,250649,14.5,,S
|
| 250 |
+
249,1,1,"Beckwith, Mr. Richard Leonard",male,37,1,1,11751,52.5542,D35,S
|
| 251 |
+
250,0,2,"Carter, Rev. Ernest Courtenay",male,54,1,0,244252,26,,S
|
| 252 |
+
251,0,3,"Reed, Mr. James George",male,,0,0,362316,7.25,,S
|
| 253 |
+
252,0,3,"Strom, Mrs. Wilhelm (Elna Matilda Persson)",female,29,1,1,347054,10.4625,G6,S
|
| 254 |
+
253,0,1,"Stead, Mr. William Thomas",male,62,0,0,113514,26.55,C87,S
|
| 255 |
+
254,0,3,"Lobb, Mr. William Arthur",male,30,1,0,A/5. 3336,16.1,,S
|
| 256 |
+
255,0,3,"Rosblom, Mrs. Viktor (Helena Wilhelmina)",female,41,0,2,370129,20.2125,,S
|
| 257 |
+
256,1,3,"Touma, Mrs. Darwis (Hanne Youssef Razi)",female,29,0,2,2650,15.2458,,C
|
| 258 |
+
257,1,1,"Thorne, Mrs. Gertrude Maybelle",female,,0,0,PC 17585,79.2,,C
|
| 259 |
+
258,1,1,"Cherry, Miss. Gladys",female,30,0,0,110152,86.5,B77,S
|
| 260 |
+
259,1,1,"Ward, Miss. Anna",female,35,0,0,PC 17755,512.3292,,C
|
| 261 |
+
260,1,2,"Parrish, Mrs. (Lutie Davis)",female,50,0,1,230433,26,,S
|
| 262 |
+
261,0,3,"Smith, Mr. Thomas",male,,0,0,384461,7.75,,Q
|
| 263 |
+
262,1,3,"Asplund, Master. Edvin Rojj Felix",male,3,4,2,347077,31.3875,,S
|
| 264 |
+
263,0,1,"Taussig, Mr. Emil",male,52,1,1,110413,79.65,E67,S
|
| 265 |
+
264,0,1,"Harrison, Mr. William",male,40,0,0,112059,0,B94,S
|
| 266 |
+
265,0,3,"Henry, Miss. Delia",female,,0,0,382649,7.75,,Q
|
| 267 |
+
266,0,2,"Reeves, Mr. David",male,36,0,0,C.A. 17248,10.5,,S
|
| 268 |
+
267,0,3,"Panula, Mr. Ernesti Arvid",male,16,4,1,3101295,39.6875,,S
|
| 269 |
+
268,1,3,"Persson, Mr. Ernst Ulrik",male,25,1,0,347083,7.775,,S
|
| 270 |
+
269,1,1,"Graham, Mrs. William Thompson (Edith Junkins)",female,58,0,1,PC 17582,153.4625,C125,S
|
| 271 |
+
270,1,1,"Bissette, Miss. Amelia",female,35,0,0,PC 17760,135.6333,C99,S
|
| 272 |
+
271,0,1,"Cairns, Mr. Alexander",male,,0,0,113798,31,,S
|
| 273 |
+
272,1,3,"Tornquist, Mr. William Henry",male,25,0,0,LINE,0,,S
|
| 274 |
+
273,1,2,"Mellinger, Mrs. (Elizabeth Anne Maidment)",female,41,0,1,250644,19.5,,S
|
| 275 |
+
274,0,1,"Natsch, Mr. Charles H",male,37,0,1,PC 17596,29.7,C118,C
|
| 276 |
+
275,1,3,"Healy, Miss. Hanora ""Nora""",female,,0,0,370375,7.75,,Q
|
| 277 |
+
276,1,1,"Andrews, Miss. Kornelia Theodosia",female,63,1,0,13502,77.9583,D7,S
|
| 278 |
+
277,0,3,"Lindblom, Miss. Augusta Charlotta",female,45,0,0,347073,7.75,,S
|
| 279 |
+
278,0,2,"Parkes, Mr. Francis ""Frank""",male,,0,0,239853,0,,S
|
| 280 |
+
279,0,3,"Rice, Master. Eric",male,7,4,1,382652,29.125,,Q
|
| 281 |
+
280,1,3,"Abbott, Mrs. Stanton (Rosa Hunt)",female,35,1,1,C.A. 2673,20.25,,S
|
| 282 |
+
281,0,3,"Duane, Mr. Frank",male,65,0,0,336439,7.75,,Q
|
| 283 |
+
282,0,3,"Olsson, Mr. Nils Johan Goransson",male,28,0,0,347464,7.8542,,S
|
| 284 |
+
283,0,3,"de Pelsmaeker, Mr. Alfons",male,16,0,0,345778,9.5,,S
|
| 285 |
+
284,1,3,"Dorking, Mr. Edward Arthur",male,19,0,0,A/5. 10482,8.05,,S
|
| 286 |
+
285,0,1,"Smith, Mr. Richard William",male,,0,0,113056,26,A19,S
|
| 287 |
+
286,0,3,"Stankovic, Mr. Ivan",male,33,0,0,349239,8.6625,,C
|
| 288 |
+
287,1,3,"de Mulder, Mr. Theodore",male,30,0,0,345774,9.5,,S
|
| 289 |
+
288,0,3,"Naidenoff, Mr. Penko",male,22,0,0,349206,7.8958,,S
|
| 290 |
+
289,1,2,"Hosono, Mr. Masabumi",male,42,0,0,237798,13,,S
|
| 291 |
+
290,1,3,"Connolly, Miss. Kate",female,22,0,0,370373,7.75,,Q
|
| 292 |
+
291,1,1,"Barber, Miss. Ellen ""Nellie""",female,26,0,0,19877,78.85,,S
|
| 293 |
+
292,1,1,"Bishop, Mrs. Dickinson H (Helen Walton)",female,19,1,0,11967,91.0792,B49,C
|
| 294 |
+
293,0,2,"Levy, Mr. Rene Jacques",male,36,0,0,SC/Paris 2163,12.875,D,C
|
| 295 |
+
294,0,3,"Haas, Miss. Aloisia",female,24,0,0,349236,8.85,,S
|
| 296 |
+
295,0,3,"Mineff, Mr. Ivan",male,24,0,0,349233,7.8958,,S
|
| 297 |
+
296,0,1,"Lewy, Mr. Ervin G",male,,0,0,PC 17612,27.7208,,C
|
| 298 |
+
297,0,3,"Hanna, Mr. Mansour",male,23.5,0,0,2693,7.2292,,C
|
| 299 |
+
298,0,1,"Allison, Miss. Helen Loraine",female,2,1,2,113781,151.55,C22 C26,S
|
| 300 |
+
299,1,1,"Saalfeld, Mr. Adolphe",male,,0,0,19988,30.5,C106,S
|
| 301 |
+
300,1,1,"Baxter, Mrs. James (Helene DeLaudeniere Chaput)",female,50,0,1,PC 17558,247.5208,B58 B60,C
|
| 302 |
+
301,1,3,"Kelly, Miss. Anna Katherine ""Annie Kate""",female,,0,0,9234,7.75,,Q
|
| 303 |
+
302,1,3,"McCoy, Mr. Bernard",male,,2,0,367226,23.25,,Q
|
| 304 |
+
303,0,3,"Johnson, Mr. William Cahoone Jr",male,19,0,0,LINE,0,,S
|
| 305 |
+
304,1,2,"Keane, Miss. Nora A",female,,0,0,226593,12.35,E101,Q
|
| 306 |
+
305,0,3,"Williams, Mr. Howard Hugh ""Harry""",male,,0,0,A/5 2466,8.05,,S
|
| 307 |
+
306,1,1,"Allison, Master. Hudson Trevor",male,0.92,1,2,113781,151.55,C22 C26,S
|
| 308 |
+
307,1,1,"Fleming, Miss. Margaret",female,,0,0,17421,110.8833,,C
|
| 309 |
+
308,1,1,"Penasco y Castellana, Mrs. Victor de Satode (Maria Josefa Perez de Soto y Vallejo)",female,17,1,0,PC 17758,108.9,C65,C
|
| 310 |
+
309,0,2,"Abelson, Mr. Samuel",male,30,1,0,P/PP 3381,24,,C
|
| 311 |
+
310,1,1,"Francatelli, Miss. Laura Mabel",female,30,0,0,PC 17485,56.9292,E36,C
|
| 312 |
+
311,1,1,"Hays, Miss. Margaret Bechstein",female,24,0,0,11767,83.1583,C54,C
|
| 313 |
+
312,1,1,"Ryerson, Miss. Emily Borie",female,18,2,2,PC 17608,262.375,B57 B59 B63 B66,C
|
| 314 |
+
313,0,2,"Lahtinen, Mrs. William (Anna Sylfven)",female,26,1,1,250651,26,,S
|
| 315 |
+
314,0,3,"Hendekovic, Mr. Ignjac",male,28,0,0,349243,7.8958,,S
|
| 316 |
+
315,0,2,"Hart, Mr. Benjamin",male,43,1,1,F.C.C. 13529,26.25,,S
|
| 317 |
+
316,1,3,"Nilsson, Miss. Helmina Josefina",female,26,0,0,347470,7.8542,,S
|
| 318 |
+
317,1,2,"Kantor, Mrs. Sinai (Miriam Sternin)",female,24,1,0,244367,26,,S
|
| 319 |
+
318,0,2,"Moraweck, Dr. Ernest",male,54,0,0,29011,14,,S
|
| 320 |
+
319,1,1,"Wick, Miss. Mary Natalie",female,31,0,2,36928,164.8667,C7,S
|
| 321 |
+
320,1,1,"Spedden, Mrs. Frederic Oakley (Margaretta Corning Stone)",female,40,1,1,16966,134.5,E34,C
|
| 322 |
+
321,0,3,"Dennis, Mr. Samuel",male,22,0,0,A/5 21172,7.25,,S
|
| 323 |
+
322,0,3,"Danoff, Mr. Yoto",male,27,0,0,349219,7.8958,,S
|
| 324 |
+
323,1,2,"Slayter, Miss. Hilda Mary",female,30,0,0,234818,12.35,,Q
|
| 325 |
+
324,1,2,"Caldwell, Mrs. Albert Francis (Sylvia Mae Harbaugh)",female,22,1,1,248738,29,,S
|
| 326 |
+
325,0,3,"Sage, Mr. George John Jr",male,,8,2,CA. 2343,69.55,,S
|
| 327 |
+
326,1,1,"Young, Miss. Marie Grice",female,36,0,0,PC 17760,135.6333,C32,C
|
| 328 |
+
327,0,3,"Nysveen, Mr. Johan Hansen",male,61,0,0,345364,6.2375,,S
|
| 329 |
+
328,1,2,"Ball, Mrs. (Ada E Hall)",female,36,0,0,28551,13,D,S
|
| 330 |
+
329,1,3,"Goldsmith, Mrs. Frank John (Emily Alice Brown)",female,31,1,1,363291,20.525,,S
|
| 331 |
+
330,1,1,"Hippach, Miss. Jean Gertrude",female,16,0,1,111361,57.9792,B18,C
|
| 332 |
+
331,1,3,"McCoy, Miss. Agnes",female,,2,0,367226,23.25,,Q
|
| 333 |
+
332,0,1,"Partner, Mr. Austen",male,45.5,0,0,113043,28.5,C124,S
|
| 334 |
+
333,0,1,"Graham, Mr. George Edward",male,38,0,1,PC 17582,153.4625,C91,S
|
| 335 |
+
334,0,3,"Vander Planke, Mr. Leo Edmondus",male,16,2,0,345764,18,,S
|
| 336 |
+
335,1,1,"Frauenthal, Mrs. Henry William (Clara Heinsheimer)",female,,1,0,PC 17611,133.65,,S
|
| 337 |
+
336,0,3,"Denkoff, Mr. Mitto",male,,0,0,349225,7.8958,,S
|
| 338 |
+
337,0,1,"Pears, Mr. Thomas Clinton",male,29,1,0,113776,66.6,C2,S
|
| 339 |
+
338,1,1,"Burns, Miss. Elizabeth Margaret",female,41,0,0,16966,134.5,E40,C
|
| 340 |
+
339,1,3,"Dahl, Mr. Karl Edwart",male,45,0,0,7598,8.05,,S
|
| 341 |
+
340,0,1,"Blackwell, Mr. Stephen Weart",male,45,0,0,113784,35.5,T,S
|
| 342 |
+
341,1,2,"Navratil, Master. Edmond Roger",male,2,1,1,230080,26,F2,S
|
| 343 |
+
342,1,1,"Fortune, Miss. Alice Elizabeth",female,24,3,2,19950,263,C23 C25 C27,S
|
| 344 |
+
343,0,2,"Collander, Mr. Erik Gustaf",male,28,0,0,248740,13,,S
|
| 345 |
+
344,0,2,"Sedgwick, Mr. Charles Frederick Waddington",male,25,0,0,244361,13,,S
|
| 346 |
+
345,0,2,"Fox, Mr. Stanley Hubert",male,36,0,0,229236,13,,S
|
| 347 |
+
346,1,2,"Brown, Miss. Amelia ""Mildred""",female,24,0,0,248733,13,F33,S
|
| 348 |
+
347,1,2,"Smith, Miss. Marion Elsie",female,40,0,0,31418,13,,S
|
| 349 |
+
348,1,3,"Davison, Mrs. Thomas Henry (Mary E Finck)",female,,1,0,386525,16.1,,S
|
| 350 |
+
349,1,3,"Coutts, Master. William Loch ""William""",male,3,1,1,C.A. 37671,15.9,,S
|
| 351 |
+
350,0,3,"Dimic, Mr. Jovan",male,42,0,0,315088,8.6625,,S
|
| 352 |
+
351,0,3,"Odahl, Mr. Nils Martin",male,23,0,0,7267,9.225,,S
|
| 353 |
+
352,0,1,"Williams-Lambert, Mr. Fletcher Fellows",male,,0,0,113510,35,C128,S
|
| 354 |
+
353,0,3,"Elias, Mr. Tannous",male,15,1,1,2695,7.2292,,C
|
| 355 |
+
354,0,3,"Arnold-Franchi, Mr. Josef",male,25,1,0,349237,17.8,,S
|
| 356 |
+
355,0,3,"Yousif, Mr. Wazli",male,,0,0,2647,7.225,,C
|
| 357 |
+
356,0,3,"Vanden Steen, Mr. Leo Peter",male,28,0,0,345783,9.5,,S
|
| 358 |
+
357,1,1,"Bowerman, Miss. Elsie Edith",female,22,0,1,113505,55,E33,S
|
| 359 |
+
358,0,2,"Funk, Miss. Annie Clemmer",female,38,0,0,237671,13,,S
|
| 360 |
+
359,1,3,"McGovern, Miss. Mary",female,,0,0,330931,7.8792,,Q
|
| 361 |
+
360,1,3,"Mockler, Miss. Helen Mary ""Ellie""",female,,0,0,330980,7.8792,,Q
|
| 362 |
+
361,0,3,"Skoog, Mr. Wilhelm",male,40,1,4,347088,27.9,,S
|
| 363 |
+
362,0,2,"del Carlo, Mr. Sebastiano",male,29,1,0,SC/PARIS 2167,27.7208,,C
|
| 364 |
+
363,0,3,"Barbara, Mrs. (Catherine David)",female,45,0,1,2691,14.4542,,C
|
| 365 |
+
364,0,3,"Asim, Mr. Adola",male,35,0,0,SOTON/O.Q. 3101310,7.05,,S
|
| 366 |
+
365,0,3,"O'Brien, Mr. Thomas",male,,1,0,370365,15.5,,Q
|
| 367 |
+
366,0,3,"Adahl, Mr. Mauritz Nils Martin",male,30,0,0,C 7076,7.25,,S
|
| 368 |
+
367,1,1,"Warren, Mrs. Frank Manley (Anna Sophia Atkinson)",female,60,1,0,110813,75.25,D37,C
|
| 369 |
+
368,1,3,"Moussa, Mrs. (Mantoura Boulos)",female,,0,0,2626,7.2292,,C
|
| 370 |
+
369,1,3,"Jermyn, Miss. Annie",female,,0,0,14313,7.75,,Q
|
| 371 |
+
370,1,1,"Aubart, Mme. Leontine Pauline",female,24,0,0,PC 17477,69.3,B35,C
|
| 372 |
+
371,1,1,"Harder, Mr. George Achilles",male,25,1,0,11765,55.4417,E50,C
|
| 373 |
+
372,0,3,"Wiklund, Mr. Jakob Alfred",male,18,1,0,3101267,6.4958,,S
|
| 374 |
+
373,0,3,"Beavan, Mr. William Thomas",male,19,0,0,323951,8.05,,S
|
| 375 |
+
374,0,1,"Ringhini, Mr. Sante",male,22,0,0,PC 17760,135.6333,,C
|
| 376 |
+
375,0,3,"Palsson, Miss. Stina Viola",female,3,3,1,349909,21.075,,S
|
| 377 |
+
376,1,1,"Meyer, Mrs. Edgar Joseph (Leila Saks)",female,,1,0,PC 17604,82.1708,,C
|
| 378 |
+
377,1,3,"Landergren, Miss. Aurora Adelia",female,22,0,0,C 7077,7.25,,S
|
| 379 |
+
378,0,1,"Widener, Mr. Harry Elkins",male,27,0,2,113503,211.5,C82,C
|
| 380 |
+
379,0,3,"Betros, Mr. Tannous",male,20,0,0,2648,4.0125,,C
|
| 381 |
+
380,0,3,"Gustafsson, Mr. Karl Gideon",male,19,0,0,347069,7.775,,S
|
| 382 |
+
381,1,1,"Bidois, Miss. Rosalie",female,42,0,0,PC 17757,227.525,,C
|
| 383 |
+
382,1,3,"Nakid, Miss. Maria (""Mary"")",female,1,0,2,2653,15.7417,,C
|
| 384 |
+
383,0,3,"Tikkanen, Mr. Juho",male,32,0,0,STON/O 2. 3101293,7.925,,S
|
| 385 |
+
384,1,1,"Holverson, Mrs. Alexander Oskar (Mary Aline Towner)",female,35,1,0,113789,52,,S
|
| 386 |
+
385,0,3,"Plotcharsky, Mr. Vasil",male,,0,0,349227,7.8958,,S
|
| 387 |
+
386,0,2,"Davies, Mr. Charles Henry",male,18,0,0,S.O.C. 14879,73.5,,S
|
| 388 |
+
387,0,3,"Goodwin, Master. Sidney Leonard",male,1,5,2,CA 2144,46.9,,S
|
| 389 |
+
388,1,2,"Buss, Miss. Kate",female,36,0,0,27849,13,,S
|
| 390 |
+
389,0,3,"Sadlier, Mr. Matthew",male,,0,0,367655,7.7292,,Q
|
| 391 |
+
390,1,2,"Lehmann, Miss. Bertha",female,17,0,0,SC 1748,12,,C
|
| 392 |
+
391,1,1,"Carter, Mr. William Ernest",male,36,1,2,113760,120,B96 B98,S
|
| 393 |
+
392,1,3,"Jansson, Mr. Carl Olof",male,21,0,0,350034,7.7958,,S
|
| 394 |
+
393,0,3,"Gustafsson, Mr. Johan Birger",male,28,2,0,3101277,7.925,,S
|
| 395 |
+
394,1,1,"Newell, Miss. Marjorie",female,23,1,0,35273,113.275,D36,C
|
| 396 |
+
395,1,3,"Sandstrom, Mrs. Hjalmar (Agnes Charlotta Bengtsson)",female,24,0,2,PP 9549,16.7,G6,S
|
| 397 |
+
396,0,3,"Johansson, Mr. Erik",male,22,0,0,350052,7.7958,,S
|
| 398 |
+
397,0,3,"Olsson, Miss. Elina",female,31,0,0,350407,7.8542,,S
|
| 399 |
+
398,0,2,"McKane, Mr. Peter David",male,46,0,0,28403,26,,S
|
| 400 |
+
399,0,2,"Pain, Dr. Alfred",male,23,0,0,244278,10.5,,S
|
| 401 |
+
400,1,2,"Trout, Mrs. William H (Jessie L)",female,28,0,0,240929,12.65,,S
|
| 402 |
+
401,1,3,"Niskanen, Mr. Juha",male,39,0,0,STON/O 2. 3101289,7.925,,S
|
| 403 |
+
402,0,3,"Adams, Mr. John",male,26,0,0,341826,8.05,,S
|
| 404 |
+
403,0,3,"Jussila, Miss. Mari Aina",female,21,1,0,4137,9.825,,S
|
| 405 |
+
404,0,3,"Hakkarainen, Mr. Pekka Pietari",male,28,1,0,STON/O2. 3101279,15.85,,S
|
| 406 |
+
405,0,3,"Oreskovic, Miss. Marija",female,20,0,0,315096,8.6625,,S
|
| 407 |
+
406,0,2,"Gale, Mr. Shadrach",male,34,1,0,28664,21,,S
|
| 408 |
+
407,0,3,"Widegren, Mr. Carl/Charles Peter",male,51,0,0,347064,7.75,,S
|
| 409 |
+
408,1,2,"Richards, Master. William Rowe",male,3,1,1,29106,18.75,,S
|
| 410 |
+
409,0,3,"Birkeland, Mr. Hans Martin Monsen",male,21,0,0,312992,7.775,,S
|
| 411 |
+
410,0,3,"Lefebre, Miss. Ida",female,,3,1,4133,25.4667,,S
|
| 412 |
+
411,0,3,"Sdycoff, Mr. Todor",male,,0,0,349222,7.8958,,S
|
| 413 |
+
412,0,3,"Hart, Mr. Henry",male,,0,0,394140,6.8583,,Q
|
| 414 |
+
413,1,1,"Minahan, Miss. Daisy E",female,33,1,0,19928,90,C78,Q
|
| 415 |
+
414,0,2,"Cunningham, Mr. Alfred Fleming",male,,0,0,239853,0,,S
|
| 416 |
+
415,1,3,"Sundman, Mr. Johan Julian",male,44,0,0,STON/O 2. 3101269,7.925,,S
|
| 417 |
+
416,0,3,"Meek, Mrs. Thomas (Annie Louise Rowley)",female,,0,0,343095,8.05,,S
|
| 418 |
+
417,1,2,"Drew, Mrs. James Vivian (Lulu Thorne Christian)",female,34,1,1,28220,32.5,,S
|
| 419 |
+
418,1,2,"Silven, Miss. Lyyli Karoliina",female,18,0,2,250652,13,,S
|
| 420 |
+
419,0,2,"Matthews, Mr. William John",male,30,0,0,28228,13,,S
|
| 421 |
+
420,0,3,"Van Impe, Miss. Catharina",female,10,0,2,345773,24.15,,S
|
| 422 |
+
421,0,3,"Gheorgheff, Mr. Stanio",male,,0,0,349254,7.8958,,C
|
| 423 |
+
422,0,3,"Charters, Mr. David",male,21,0,0,A/5. 13032,7.7333,,Q
|
| 424 |
+
423,0,3,"Zimmerman, Mr. Leo",male,29,0,0,315082,7.875,,S
|
| 425 |
+
424,0,3,"Danbom, Mrs. Ernst Gilbert (Anna Sigrid Maria Brogren)",female,28,1,1,347080,14.4,,S
|
| 426 |
+
425,0,3,"Rosblom, Mr. Viktor Richard",male,18,1,1,370129,20.2125,,S
|
| 427 |
+
426,0,3,"Wiseman, Mr. Phillippe",male,,0,0,A/4. 34244,7.25,,S
|
| 428 |
+
427,1,2,"Clarke, Mrs. Charles V (Ada Maria Winfield)",female,28,1,0,2003,26,,S
|
| 429 |
+
428,1,2,"Phillips, Miss. Kate Florence (""Mrs Kate Louise Phillips Marshall"")",female,19,0,0,250655,26,,S
|
| 430 |
+
429,0,3,"Flynn, Mr. James",male,,0,0,364851,7.75,,Q
|
| 431 |
+
430,1,3,"Pickard, Mr. Berk (Berk Trembisky)",male,32,0,0,SOTON/O.Q. 392078,8.05,E10,S
|
| 432 |
+
431,1,1,"Bjornstrom-Steffansson, Mr. Mauritz Hakan",male,28,0,0,110564,26.55,C52,S
|
| 433 |
+
432,1,3,"Thorneycroft, Mrs. Percival (Florence Kate White)",female,,1,0,376564,16.1,,S
|
| 434 |
+
433,1,2,"Louch, Mrs. Charles Alexander (Alice Adelaide Slow)",female,42,1,0,SC/AH 3085,26,,S
|
| 435 |
+
434,0,3,"Kallio, Mr. Nikolai Erland",male,17,0,0,STON/O 2. 3101274,7.125,,S
|
| 436 |
+
435,0,1,"Silvey, Mr. William Baird",male,50,1,0,13507,55.9,E44,S
|
| 437 |
+
436,1,1,"Carter, Miss. Lucile Polk",female,14,1,2,113760,120,B96 B98,S
|
| 438 |
+
437,0,3,"Ford, Miss. Doolina Margaret ""Daisy""",female,21,2,2,W./C. 6608,34.375,,S
|
| 439 |
+
438,1,2,"Richards, Mrs. Sidney (Emily Hocking)",female,24,2,3,29106,18.75,,S
|
| 440 |
+
439,0,1,"Fortune, Mr. Mark",male,64,1,4,19950,263,C23 C25 C27,S
|
| 441 |
+
440,0,2,"Kvillner, Mr. Johan Henrik Johannesson",male,31,0,0,C.A. 18723,10.5,,S
|
| 442 |
+
441,1,2,"Hart, Mrs. Benjamin (Esther Ada Bloomfield)",female,45,1,1,F.C.C. 13529,26.25,,S
|
| 443 |
+
442,0,3,"Hampe, Mr. Leon",male,20,0,0,345769,9.5,,S
|
| 444 |
+
443,0,3,"Petterson, Mr. Johan Emil",male,25,1,0,347076,7.775,,S
|
| 445 |
+
444,1,2,"Reynaldo, Ms. Encarnacion",female,28,0,0,230434,13,,S
|
| 446 |
+
445,1,3,"Johannesen-Bratthammer, Mr. Bernt",male,,0,0,65306,8.1125,,S
|
| 447 |
+
446,1,1,"Dodge, Master. Washington",male,4,0,2,33638,81.8583,A34,S
|
| 448 |
+
447,1,2,"Mellinger, Miss. Madeleine Violet",female,13,0,1,250644,19.5,,S
|
| 449 |
+
448,1,1,"Seward, Mr. Frederic Kimber",male,34,0,0,113794,26.55,,S
|
| 450 |
+
449,1,3,"Baclini, Miss. Marie Catherine",female,5,2,1,2666,19.2583,,C
|
| 451 |
+
450,1,1,"Peuchen, Major. Arthur Godfrey",male,52,0,0,113786,30.5,C104,S
|
| 452 |
+
451,0,2,"West, Mr. Edwy Arthur",male,36,1,2,C.A. 34651,27.75,,S
|
| 453 |
+
452,0,3,"Hagland, Mr. Ingvald Olai Olsen",male,,1,0,65303,19.9667,,S
|
| 454 |
+
453,0,1,"Foreman, Mr. Benjamin Laventall",male,30,0,0,113051,27.75,C111,C
|
| 455 |
+
454,1,1,"Goldenberg, Mr. Samuel L",male,49,1,0,17453,89.1042,C92,C
|
| 456 |
+
455,0,3,"Peduzzi, Mr. Joseph",male,,0,0,A/5 2817,8.05,,S
|
| 457 |
+
456,1,3,"Jalsevac, Mr. Ivan",male,29,0,0,349240,7.8958,,C
|
| 458 |
+
457,0,1,"Millet, Mr. Francis Davis",male,65,0,0,13509,26.55,E38,S
|
| 459 |
+
458,1,1,"Kenyon, Mrs. Frederick R (Marion)",female,,1,0,17464,51.8625,D21,S
|
| 460 |
+
459,1,2,"Toomey, Miss. Ellen",female,50,0,0,F.C.C. 13531,10.5,,S
|
| 461 |
+
460,0,3,"O'Connor, Mr. Maurice",male,,0,0,371060,7.75,,Q
|
| 462 |
+
461,1,1,"Anderson, Mr. Harry",male,48,0,0,19952,26.55,E12,S
|
| 463 |
+
462,0,3,"Morley, Mr. William",male,34,0,0,364506,8.05,,S
|
| 464 |
+
463,0,1,"Gee, Mr. Arthur H",male,47,0,0,111320,38.5,E63,S
|
| 465 |
+
464,0,2,"Milling, Mr. Jacob Christian",male,48,0,0,234360,13,,S
|
| 466 |
+
465,0,3,"Maisner, Mr. Simon",male,,0,0,A/S 2816,8.05,,S
|
| 467 |
+
466,0,3,"Goncalves, Mr. Manuel Estanslas",male,38,0,0,SOTON/O.Q. 3101306,7.05,,S
|
| 468 |
+
467,0,2,"Campbell, Mr. William",male,,0,0,239853,0,,S
|
| 469 |
+
468,0,1,"Smart, Mr. John Montgomery",male,56,0,0,113792,26.55,,S
|
| 470 |
+
469,0,3,"Scanlan, Mr. James",male,,0,0,36209,7.725,,Q
|
| 471 |
+
470,1,3,"Baclini, Miss. Helene Barbara",female,0.75,2,1,2666,19.2583,,C
|
| 472 |
+
471,0,3,"Keefe, Mr. Arthur",male,,0,0,323592,7.25,,S
|
| 473 |
+
472,0,3,"Cacic, Mr. Luka",male,38,0,0,315089,8.6625,,S
|
| 474 |
+
473,1,2,"West, Mrs. Edwy Arthur (Ada Mary Worth)",female,33,1,2,C.A. 34651,27.75,,S
|
| 475 |
+
474,1,2,"Jerwan, Mrs. Amin S (Marie Marthe Thuillard)",female,23,0,0,SC/AH Basle 541,13.7917,D,C
|
| 476 |
+
475,0,3,"Strandberg, Miss. Ida Sofia",female,22,0,0,7553,9.8375,,S
|
| 477 |
+
476,0,1,"Clifford, Mr. George Quincy",male,,0,0,110465,52,A14,S
|
| 478 |
+
477,0,2,"Renouf, Mr. Peter Henry",male,34,1,0,31027,21,,S
|
| 479 |
+
478,0,3,"Braund, Mr. Lewis Richard",male,29,1,0,3460,7.0458,,S
|
| 480 |
+
479,0,3,"Karlsson, Mr. Nils August",male,22,0,0,350060,7.5208,,S
|
| 481 |
+
480,1,3,"Hirvonen, Miss. Hildur E",female,2,0,1,3101298,12.2875,,S
|
| 482 |
+
481,0,3,"Goodwin, Master. Harold Victor",male,9,5,2,CA 2144,46.9,,S
|
| 483 |
+
482,0,2,"Frost, Mr. Anthony Wood ""Archie""",male,,0,0,239854,0,,S
|
| 484 |
+
483,0,3,"Rouse, Mr. Richard Henry",male,50,0,0,A/5 3594,8.05,,S
|
| 485 |
+
484,1,3,"Turkula, Mrs. (Hedwig)",female,63,0,0,4134,9.5875,,S
|
| 486 |
+
485,1,1,"Bishop, Mr. Dickinson H",male,25,1,0,11967,91.0792,B49,C
|
| 487 |
+
486,0,3,"Lefebre, Miss. Jeannie",female,,3,1,4133,25.4667,,S
|
| 488 |
+
487,1,1,"Hoyt, Mrs. Frederick Maxfield (Jane Anne Forby)",female,35,1,0,19943,90,C93,S
|
| 489 |
+
488,0,1,"Kent, Mr. Edward Austin",male,58,0,0,11771,29.7,B37,C
|
| 490 |
+
489,0,3,"Somerton, Mr. Francis William",male,30,0,0,A.5. 18509,8.05,,S
|
| 491 |
+
490,1,3,"Coutts, Master. Eden Leslie ""Neville""",male,9,1,1,C.A. 37671,15.9,,S
|
| 492 |
+
491,0,3,"Hagland, Mr. Konrad Mathias Reiersen",male,,1,0,65304,19.9667,,S
|
| 493 |
+
492,0,3,"Windelov, Mr. Einar",male,21,0,0,SOTON/OQ 3101317,7.25,,S
|
| 494 |
+
493,0,1,"Molson, Mr. Harry Markland",male,55,0,0,113787,30.5,C30,S
|
| 495 |
+
494,0,1,"Artagaveytia, Mr. Ramon",male,71,0,0,PC 17609,49.5042,,C
|
| 496 |
+
495,0,3,"Stanley, Mr. Edward Roland",male,21,0,0,A/4 45380,8.05,,S
|
| 497 |
+
496,0,3,"Yousseff, Mr. Gerious",male,,0,0,2627,14.4583,,C
|
| 498 |
+
497,1,1,"Eustis, Miss. Elizabeth Mussey",female,54,1,0,36947,78.2667,D20,C
|
| 499 |
+
498,0,3,"Shellard, Mr. Frederick William",male,,0,0,C.A. 6212,15.1,,S
|
| 500 |
+
499,0,1,"Allison, Mrs. Hudson J C (Bessie Waldo Daniels)",female,25,1,2,113781,151.55,C22 C26,S
|
| 501 |
+
500,0,3,"Svensson, Mr. Olof",male,24,0,0,350035,7.7958,,S
|
| 502 |
+
501,0,3,"Calic, Mr. Petar",male,17,0,0,315086,8.6625,,S
|
| 503 |
+
502,0,3,"Canavan, Miss. Mary",female,21,0,0,364846,7.75,,Q
|
| 504 |
+
503,0,3,"O'Sullivan, Miss. Bridget Mary",female,,0,0,330909,7.6292,,Q
|
| 505 |
+
504,0,3,"Laitinen, Miss. Kristina Sofia",female,37,0,0,4135,9.5875,,S
|
| 506 |
+
505,1,1,"Maioni, Miss. Roberta",female,16,0,0,110152,86.5,B79,S
|
| 507 |
+
506,0,1,"Penasco y Castellana, Mr. Victor de Satode",male,18,1,0,PC 17758,108.9,C65,C
|
| 508 |
+
507,1,2,"Quick, Mrs. Frederick Charles (Jane Richards)",female,33,0,2,26360,26,,S
|
| 509 |
+
508,1,1,"Bradley, Mr. George (""George Arthur Brayton"")",male,,0,0,111427,26.55,,S
|
| 510 |
+
509,0,3,"Olsen, Mr. Henry Margido",male,28,0,0,C 4001,22.525,,S
|
| 511 |
+
510,1,3,"Lang, Mr. Fang",male,26,0,0,1601,56.4958,,S
|
| 512 |
+
511,1,3,"Daly, Mr. Eugene Patrick",male,29,0,0,382651,7.75,,Q
|
| 513 |
+
512,0,3,"Webber, Mr. James",male,,0,0,SOTON/OQ 3101316,8.05,,S
|
| 514 |
+
513,1,1,"McGough, Mr. James Robert",male,36,0,0,PC 17473,26.2875,E25,S
|
| 515 |
+
514,1,1,"Rothschild, Mrs. Martin (Elizabeth L. Barrett)",female,54,1,0,PC 17603,59.4,,C
|
| 516 |
+
515,0,3,"Coleff, Mr. Satio",male,24,0,0,349209,7.4958,,S
|
| 517 |
+
516,0,1,"Walker, Mr. William Anderson",male,47,0,0,36967,34.0208,D46,S
|
| 518 |
+
517,1,2,"Lemore, Mrs. (Amelia Milley)",female,34,0,0,C.A. 34260,10.5,F33,S
|
| 519 |
+
518,0,3,"Ryan, Mr. Patrick",male,,0,0,371110,24.15,,Q
|
| 520 |
+
519,1,2,"Angle, Mrs. William A (Florence ""Mary"" Agnes Hughes)",female,36,1,0,226875,26,,S
|
| 521 |
+
520,0,3,"Pavlovic, Mr. Stefo",male,32,0,0,349242,7.8958,,S
|
| 522 |
+
521,1,1,"Perreault, Miss. Anne",female,30,0,0,12749,93.5,B73,S
|
| 523 |
+
522,0,3,"Vovk, Mr. Janko",male,22,0,0,349252,7.8958,,S
|
| 524 |
+
523,0,3,"Lahoud, Mr. Sarkis",male,,0,0,2624,7.225,,C
|
| 525 |
+
524,1,1,"Hippach, Mrs. Louis Albert (Ida Sophia Fischer)",female,44,0,1,111361,57.9792,B18,C
|
| 526 |
+
525,0,3,"Kassem, Mr. Fared",male,,0,0,2700,7.2292,,C
|
| 527 |
+
526,0,3,"Farrell, Mr. James",male,40.5,0,0,367232,7.75,,Q
|
| 528 |
+
527,1,2,"Ridsdale, Miss. Lucy",female,50,0,0,W./C. 14258,10.5,,S
|
| 529 |
+
528,0,1,"Farthing, Mr. John",male,,0,0,PC 17483,221.7792,C95,S
|
| 530 |
+
529,0,3,"Salonen, Mr. Johan Werner",male,39,0,0,3101296,7.925,,S
|
| 531 |
+
530,0,2,"Hocking, Mr. Richard George",male,23,2,1,29104,11.5,,S
|
| 532 |
+
531,1,2,"Quick, Miss. Phyllis May",female,2,1,1,26360,26,,S
|
| 533 |
+
532,0,3,"Toufik, Mr. Nakli",male,,0,0,2641,7.2292,,C
|
| 534 |
+
533,0,3,"Elias, Mr. Joseph Jr",male,17,1,1,2690,7.2292,,C
|
| 535 |
+
534,1,3,"Peter, Mrs. Catherine (Catherine Rizk)",female,,0,2,2668,22.3583,,C
|
| 536 |
+
535,0,3,"Cacic, Miss. Marija",female,30,0,0,315084,8.6625,,S
|
| 537 |
+
536,1,2,"Hart, Miss. Eva Miriam",female,7,0,2,F.C.C. 13529,26.25,,S
|
| 538 |
+
537,0,1,"Butt, Major. Archibald Willingham",male,45,0,0,113050,26.55,B38,S
|
| 539 |
+
538,1,1,"LeRoy, Miss. Bertha",female,30,0,0,PC 17761,106.425,,C
|
| 540 |
+
539,0,3,"Risien, Mr. Samuel Beard",male,,0,0,364498,14.5,,S
|
| 541 |
+
540,1,1,"Frolicher, Miss. Hedwig Margaritha",female,22,0,2,13568,49.5,B39,C
|
| 542 |
+
541,1,1,"Crosby, Miss. Harriet R",female,36,0,2,WE/P 5735,71,B22,S
|
| 543 |
+
542,0,3,"Andersson, Miss. Ingeborg Constanzia",female,9,4,2,347082,31.275,,S
|
| 544 |
+
543,0,3,"Andersson, Miss. Sigrid Elisabeth",female,11,4,2,347082,31.275,,S
|
| 545 |
+
544,1,2,"Beane, Mr. Edward",male,32,1,0,2908,26,,S
|
| 546 |
+
545,0,1,"Douglas, Mr. Walter Donald",male,50,1,0,PC 17761,106.425,C86,C
|
| 547 |
+
546,0,1,"Nicholson, Mr. Arthur Ernest",male,64,0,0,693,26,,S
|
| 548 |
+
547,1,2,"Beane, Mrs. Edward (Ethel Clarke)",female,19,1,0,2908,26,,S
|
| 549 |
+
548,1,2,"Padro y Manent, Mr. Julian",male,,0,0,SC/PARIS 2146,13.8625,,C
|
| 550 |
+
549,0,3,"Goldsmith, Mr. Frank John",male,33,1,1,363291,20.525,,S
|
| 551 |
+
550,1,2,"Davies, Master. John Morgan Jr",male,8,1,1,C.A. 33112,36.75,,S
|
| 552 |
+
551,1,1,"Thayer, Mr. John Borland Jr",male,17,0,2,17421,110.8833,C70,C
|
| 553 |
+
552,0,2,"Sharp, Mr. Percival James R",male,27,0,0,244358,26,,S
|
| 554 |
+
553,0,3,"O'Brien, Mr. Timothy",male,,0,0,330979,7.8292,,Q
|
| 555 |
+
554,1,3,"Leeni, Mr. Fahim (""Philip Zenni"")",male,22,0,0,2620,7.225,,C
|
| 556 |
+
555,1,3,"Ohman, Miss. Velin",female,22,0,0,347085,7.775,,S
|
| 557 |
+
556,0,1,"Wright, Mr. George",male,62,0,0,113807,26.55,,S
|
| 558 |
+
557,1,1,"Duff Gordon, Lady. (Lucille Christiana Sutherland) (""Mrs Morgan"")",female,48,1,0,11755,39.6,A16,C
|
| 559 |
+
558,0,1,"Robbins, Mr. Victor",male,,0,0,PC 17757,227.525,,C
|
| 560 |
+
559,1,1,"Taussig, Mrs. Emil (Tillie Mandelbaum)",female,39,1,1,110413,79.65,E67,S
|
| 561 |
+
560,1,3,"de Messemaeker, Mrs. Guillaume Joseph (Emma)",female,36,1,0,345572,17.4,,S
|
| 562 |
+
561,0,3,"Morrow, Mr. Thomas Rowan",male,,0,0,372622,7.75,,Q
|
| 563 |
+
562,0,3,"Sivic, Mr. Husein",male,40,0,0,349251,7.8958,,S
|
| 564 |
+
563,0,2,"Norman, Mr. Robert Douglas",male,28,0,0,218629,13.5,,S
|
| 565 |
+
564,0,3,"Simmons, Mr. John",male,,0,0,SOTON/OQ 392082,8.05,,S
|
| 566 |
+
565,0,3,"Meanwell, Miss. (Marion Ogden)",female,,0,0,SOTON/O.Q. 392087,8.05,,S
|
| 567 |
+
566,0,3,"Davies, Mr. Alfred J",male,24,2,0,A/4 48871,24.15,,S
|
| 568 |
+
567,0,3,"Stoytcheff, Mr. Ilia",male,19,0,0,349205,7.8958,,S
|
| 569 |
+
568,0,3,"Palsson, Mrs. Nils (Alma Cornelia Berglund)",female,29,0,4,349909,21.075,,S
|
| 570 |
+
569,0,3,"Doharr, Mr. Tannous",male,,0,0,2686,7.2292,,C
|
| 571 |
+
570,1,3,"Jonsson, Mr. Carl",male,32,0,0,350417,7.8542,,S
|
| 572 |
+
571,1,2,"Harris, Mr. George",male,62,0,0,S.W./PP 752,10.5,,S
|
| 573 |
+
572,1,1,"Appleton, Mrs. Edward Dale (Charlotte Lamson)",female,53,2,0,11769,51.4792,C101,S
|
| 574 |
+
573,1,1,"Flynn, Mr. John Irwin (""Irving"")",male,36,0,0,PC 17474,26.3875,E25,S
|
| 575 |
+
574,1,3,"Kelly, Miss. Mary",female,,0,0,14312,7.75,,Q
|
| 576 |
+
575,0,3,"Rush, Mr. Alfred George John",male,16,0,0,A/4. 20589,8.05,,S
|
| 577 |
+
576,0,3,"Patchett, Mr. George",male,19,0,0,358585,14.5,,S
|
| 578 |
+
577,1,2,"Garside, Miss. Ethel",female,34,0,0,243880,13,,S
|
| 579 |
+
578,1,1,"Silvey, Mrs. William Baird (Alice Munger)",female,39,1,0,13507,55.9,E44,S
|
| 580 |
+
579,0,3,"Caram, Mrs. Joseph (Maria Elias)",female,,1,0,2689,14.4583,,C
|
| 581 |
+
580,1,3,"Jussila, Mr. Eiriik",male,32,0,0,STON/O 2. 3101286,7.925,,S
|
| 582 |
+
581,1,2,"Christy, Miss. Julie Rachel",female,25,1,1,237789,30,,S
|
| 583 |
+
582,1,1,"Thayer, Mrs. John Borland (Marian Longstreth Morris)",female,39,1,1,17421,110.8833,C68,C
|
| 584 |
+
583,0,2,"Downton, Mr. William James",male,54,0,0,28403,26,,S
|
| 585 |
+
584,0,1,"Ross, Mr. John Hugo",male,36,0,0,13049,40.125,A10,C
|
| 586 |
+
585,0,3,"Paulner, Mr. Uscher",male,,0,0,3411,8.7125,,C
|
| 587 |
+
586,1,1,"Taussig, Miss. Ruth",female,18,0,2,110413,79.65,E68,S
|
| 588 |
+
587,0,2,"Jarvis, Mr. John Denzil",male,47,0,0,237565,15,,S
|
| 589 |
+
588,1,1,"Frolicher-Stehli, Mr. Maxmillian",male,60,1,1,13567,79.2,B41,C
|
| 590 |
+
589,0,3,"Gilinski, Mr. Eliezer",male,22,0,0,14973,8.05,,S
|
| 591 |
+
590,0,3,"Murdlin, Mr. Joseph",male,,0,0,A./5. 3235,8.05,,S
|
| 592 |
+
591,0,3,"Rintamaki, Mr. Matti",male,35,0,0,STON/O 2. 3101273,7.125,,S
|
| 593 |
+
592,1,1,"Stephenson, Mrs. Walter Bertram (Martha Eustis)",female,52,1,0,36947,78.2667,D20,C
|
| 594 |
+
593,0,3,"Elsbury, Mr. William James",male,47,0,0,A/5 3902,7.25,,S
|
| 595 |
+
594,0,3,"Bourke, Miss. Mary",female,,0,2,364848,7.75,,Q
|
| 596 |
+
595,0,2,"Chapman, Mr. John Henry",male,37,1,0,SC/AH 29037,26,,S
|
| 597 |
+
596,0,3,"Van Impe, Mr. Jean Baptiste",male,36,1,1,345773,24.15,,S
|
| 598 |
+
597,1,2,"Leitch, Miss. Jessie Wills",female,,0,0,248727,33,,S
|
| 599 |
+
598,0,3,"Johnson, Mr. Alfred",male,49,0,0,LINE,0,,S
|
| 600 |
+
599,0,3,"Boulos, Mr. Hanna",male,,0,0,2664,7.225,,C
|
| 601 |
+
600,1,1,"Duff Gordon, Sir. Cosmo Edmund (""Mr Morgan"")",male,49,1,0,PC 17485,56.9292,A20,C
|
| 602 |
+
601,1,2,"Jacobsohn, Mrs. Sidney Samuel (Amy Frances Christy)",female,24,2,1,243847,27,,S
|
| 603 |
+
602,0,3,"Slabenoff, Mr. Petco",male,,0,0,349214,7.8958,,S
|
| 604 |
+
603,0,1,"Harrington, Mr. Charles H",male,,0,0,113796,42.4,,S
|
| 605 |
+
604,0,3,"Torber, Mr. Ernst William",male,44,0,0,364511,8.05,,S
|
| 606 |
+
605,1,1,"Homer, Mr. Harry (""Mr E Haven"")",male,35,0,0,111426,26.55,,C
|
| 607 |
+
606,0,3,"Lindell, Mr. Edvard Bengtsson",male,36,1,0,349910,15.55,,S
|
| 608 |
+
607,0,3,"Karaic, Mr. Milan",male,30,0,0,349246,7.8958,,S
|
| 609 |
+
608,1,1,"Daniel, Mr. Robert Williams",male,27,0,0,113804,30.5,,S
|
| 610 |
+
609,1,2,"Laroche, Mrs. Joseph (Juliette Marie Louise Lafargue)",female,22,1,2,SC/Paris 2123,41.5792,,C
|
| 611 |
+
610,1,1,"Shutes, Miss. Elizabeth W",female,40,0,0,PC 17582,153.4625,C125,S
|
| 612 |
+
611,0,3,"Andersson, Mrs. Anders Johan (Alfrida Konstantia Brogren)",female,39,1,5,347082,31.275,,S
|
| 613 |
+
612,0,3,"Jardin, Mr. Jose Neto",male,,0,0,SOTON/O.Q. 3101305,7.05,,S
|
| 614 |
+
613,1,3,"Murphy, Miss. Margaret Jane",female,,1,0,367230,15.5,,Q
|
| 615 |
+
614,0,3,"Horgan, Mr. John",male,,0,0,370377,7.75,,Q
|
| 616 |
+
615,0,3,"Brocklebank, Mr. William Alfred",male,35,0,0,364512,8.05,,S
|
| 617 |
+
616,1,2,"Herman, Miss. Alice",female,24,1,2,220845,65,,S
|
| 618 |
+
617,0,3,"Danbom, Mr. Ernst Gilbert",male,34,1,1,347080,14.4,,S
|
| 619 |
+
618,0,3,"Lobb, Mrs. William Arthur (Cordelia K Stanlick)",female,26,1,0,A/5. 3336,16.1,,S
|
| 620 |
+
619,1,2,"Becker, Miss. Marion Louise",female,4,2,1,230136,39,F4,S
|
| 621 |
+
620,0,2,"Gavey, Mr. Lawrence",male,26,0,0,31028,10.5,,S
|
| 622 |
+
621,0,3,"Yasbeck, Mr. Antoni",male,27,1,0,2659,14.4542,,C
|
| 623 |
+
622,1,1,"Kimball, Mr. Edwin Nelson Jr",male,42,1,0,11753,52.5542,D19,S
|
| 624 |
+
623,1,3,"Nakid, Mr. Sahid",male,20,1,1,2653,15.7417,,C
|
| 625 |
+
624,0,3,"Hansen, Mr. Henry Damsgaard",male,21,0,0,350029,7.8542,,S
|
| 626 |
+
625,0,3,"Bowen, Mr. David John ""Dai""",male,21,0,0,54636,16.1,,S
|
| 627 |
+
626,0,1,"Sutton, Mr. Frederick",male,61,0,0,36963,32.3208,D50,S
|
| 628 |
+
627,0,2,"Kirkland, Rev. Charles Leonard",male,57,0,0,219533,12.35,,Q
|
| 629 |
+
628,1,1,"Longley, Miss. Gretchen Fiske",female,21,0,0,13502,77.9583,D9,S
|
| 630 |
+
629,0,3,"Bostandyeff, Mr. Guentcho",male,26,0,0,349224,7.8958,,S
|
| 631 |
+
630,0,3,"O'Connell, Mr. Patrick D",male,,0,0,334912,7.7333,,Q
|
| 632 |
+
631,1,1,"Barkworth, Mr. Algernon Henry Wilson",male,80,0,0,27042,30,A23,S
|
| 633 |
+
632,0,3,"Lundahl, Mr. Johan Svensson",male,51,0,0,347743,7.0542,,S
|
| 634 |
+
633,1,1,"Stahelin-Maeglin, Dr. Max",male,32,0,0,13214,30.5,B50,C
|
| 635 |
+
634,0,1,"Parr, Mr. William Henry Marsh",male,,0,0,112052,0,,S
|
| 636 |
+
635,0,3,"Skoog, Miss. Mabel",female,9,3,2,347088,27.9,,S
|
| 637 |
+
636,1,2,"Davis, Miss. Mary",female,28,0,0,237668,13,,S
|
| 638 |
+
637,0,3,"Leinonen, Mr. Antti Gustaf",male,32,0,0,STON/O 2. 3101292,7.925,,S
|
| 639 |
+
638,0,2,"Collyer, Mr. Harvey",male,31,1,1,C.A. 31921,26.25,,S
|
| 640 |
+
639,0,3,"Panula, Mrs. Juha (Maria Emilia Ojala)",female,41,0,5,3101295,39.6875,,S
|
| 641 |
+
640,0,3,"Thorneycroft, Mr. Percival",male,,1,0,376564,16.1,,S
|
| 642 |
+
641,0,3,"Jensen, Mr. Hans Peder",male,20,0,0,350050,7.8542,,S
|
| 643 |
+
642,1,1,"Sagesser, Mlle. Emma",female,24,0,0,PC 17477,69.3,B35,C
|
| 644 |
+
643,0,3,"Skoog, Miss. Margit Elizabeth",female,2,3,2,347088,27.9,,S
|
| 645 |
+
644,1,3,"Foo, Mr. Choong",male,,0,0,1601,56.4958,,S
|
| 646 |
+
645,1,3,"Baclini, Miss. Eugenie",female,0.75,2,1,2666,19.2583,,C
|
| 647 |
+
646,1,1,"Harper, Mr. Henry Sleeper",male,48,1,0,PC 17572,76.7292,D33,C
|
| 648 |
+
647,0,3,"Cor, Mr. Liudevit",male,19,0,0,349231,7.8958,,S
|
| 649 |
+
648,1,1,"Simonius-Blumer, Col. Oberst Alfons",male,56,0,0,13213,35.5,A26,C
|
| 650 |
+
649,0,3,"Willey, Mr. Edward",male,,0,0,S.O./P.P. 751,7.55,,S
|
| 651 |
+
650,1,3,"Stanley, Miss. Amy Zillah Elsie",female,23,0,0,CA. 2314,7.55,,S
|
| 652 |
+
651,0,3,"Mitkoff, Mr. Mito",male,,0,0,349221,7.8958,,S
|
| 653 |
+
652,1,2,"Doling, Miss. Elsie",female,18,0,1,231919,23,,S
|
| 654 |
+
653,0,3,"Kalvik, Mr. Johannes Halvorsen",male,21,0,0,8475,8.4333,,S
|
| 655 |
+
654,1,3,"O'Leary, Miss. Hanora ""Norah""",female,,0,0,330919,7.8292,,Q
|
| 656 |
+
655,0,3,"Hegarty, Miss. Hanora ""Nora""",female,18,0,0,365226,6.75,,Q
|
| 657 |
+
656,0,2,"Hickman, Mr. Leonard Mark",male,24,2,0,S.O.C. 14879,73.5,,S
|
| 658 |
+
657,0,3,"Radeff, Mr. Alexander",male,,0,0,349223,7.8958,,S
|
| 659 |
+
658,0,3,"Bourke, Mrs. John (Catherine)",female,32,1,1,364849,15.5,,Q
|
| 660 |
+
659,0,2,"Eitemiller, Mr. George Floyd",male,23,0,0,29751,13,,S
|
| 661 |
+
660,0,1,"Newell, Mr. Arthur Webster",male,58,0,2,35273,113.275,D48,C
|
| 662 |
+
661,1,1,"Frauenthal, Dr. Henry William",male,50,2,0,PC 17611,133.65,,S
|
| 663 |
+
662,0,3,"Badt, Mr. Mohamed",male,40,0,0,2623,7.225,,C
|
| 664 |
+
663,0,1,"Colley, Mr. Edward Pomeroy",male,47,0,0,5727,25.5875,E58,S
|
| 665 |
+
664,0,3,"Coleff, Mr. Peju",male,36,0,0,349210,7.4958,,S
|
| 666 |
+
665,1,3,"Lindqvist, Mr. Eino William",male,20,1,0,STON/O 2. 3101285,7.925,,S
|
| 667 |
+
666,0,2,"Hickman, Mr. Lewis",male,32,2,0,S.O.C. 14879,73.5,,S
|
| 668 |
+
667,0,2,"Butler, Mr. Reginald Fenton",male,25,0,0,234686,13,,S
|
| 669 |
+
668,0,3,"Rommetvedt, Mr. Knud Paust",male,,0,0,312993,7.775,,S
|
| 670 |
+
669,0,3,"Cook, Mr. Jacob",male,43,0,0,A/5 3536,8.05,,S
|
| 671 |
+
670,1,1,"Taylor, Mrs. Elmer Zebley (Juliet Cummins Wright)",female,,1,0,19996,52,C126,S
|
| 672 |
+
671,1,2,"Brown, Mrs. Thomas William Solomon (Elizabeth Catherine Ford)",female,40,1,1,29750,39,,S
|
| 673 |
+
672,0,1,"Davidson, Mr. Thornton",male,31,1,0,F.C. 12750,52,B71,S
|
| 674 |
+
673,0,2,"Mitchell, Mr. Henry Michael",male,70,0,0,C.A. 24580,10.5,,S
|
| 675 |
+
674,1,2,"Wilhelms, Mr. Charles",male,31,0,0,244270,13,,S
|
| 676 |
+
675,0,2,"Watson, Mr. Ennis Hastings",male,,0,0,239856,0,,S
|
| 677 |
+
676,0,3,"Edvardsson, Mr. Gustaf Hjalmar",male,18,0,0,349912,7.775,,S
|
| 678 |
+
677,0,3,"Sawyer, Mr. Frederick Charles",male,24.5,0,0,342826,8.05,,S
|
| 679 |
+
678,1,3,"Turja, Miss. Anna Sofia",female,18,0,0,4138,9.8417,,S
|
| 680 |
+
679,0,3,"Goodwin, Mrs. Frederick (Augusta Tyler)",female,43,1,6,CA 2144,46.9,,S
|
| 681 |
+
680,1,1,"Cardeza, Mr. Thomas Drake Martinez",male,36,0,1,PC 17755,512.3292,B51 B53 B55,C
|
| 682 |
+
681,0,3,"Peters, Miss. Katie",female,,0,0,330935,8.1375,,Q
|
| 683 |
+
682,1,1,"Hassab, Mr. Hammad",male,27,0,0,PC 17572,76.7292,D49,C
|
| 684 |
+
683,0,3,"Olsvigen, Mr. Thor Anderson",male,20,0,0,6563,9.225,,S
|
| 685 |
+
684,0,3,"Goodwin, Mr. Charles Edward",male,14,5,2,CA 2144,46.9,,S
|
| 686 |
+
685,0,2,"Brown, Mr. Thomas William Solomon",male,60,1,1,29750,39,,S
|
| 687 |
+
686,0,2,"Laroche, Mr. Joseph Philippe Lemercier",male,25,1,2,SC/Paris 2123,41.5792,,C
|
| 688 |
+
687,0,3,"Panula, Mr. Jaako Arnold",male,14,4,1,3101295,39.6875,,S
|
| 689 |
+
688,0,3,"Dakic, Mr. Branko",male,19,0,0,349228,10.1708,,S
|
| 690 |
+
689,0,3,"Fischer, Mr. Eberhard Thelander",male,18,0,0,350036,7.7958,,S
|
| 691 |
+
690,1,1,"Madill, Miss. Georgette Alexandra",female,15,0,1,24160,211.3375,B5,S
|
| 692 |
+
691,1,1,"Dick, Mr. Albert Adrian",male,31,1,0,17474,57,B20,S
|
| 693 |
+
692,1,3,"Karun, Miss. Manca",female,4,0,1,349256,13.4167,,C
|
| 694 |
+
693,1,3,"Lam, Mr. Ali",male,,0,0,1601,56.4958,,S
|
| 695 |
+
694,0,3,"Saad, Mr. Khalil",male,25,0,0,2672,7.225,,C
|
| 696 |
+
695,0,1,"Weir, Col. John",male,60,0,0,113800,26.55,,S
|
| 697 |
+
696,0,2,"Chapman, Mr. Charles Henry",male,52,0,0,248731,13.5,,S
|
| 698 |
+
697,0,3,"Kelly, Mr. James",male,44,0,0,363592,8.05,,S
|
| 699 |
+
698,1,3,"Mullens, Miss. Katherine ""Katie""",female,,0,0,35852,7.7333,,Q
|
| 700 |
+
699,0,1,"Thayer, Mr. John Borland",male,49,1,1,17421,110.8833,C68,C
|
| 701 |
+
700,0,3,"Humblen, Mr. Adolf Mathias Nicolai Olsen",male,42,0,0,348121,7.65,F G63,S
|
| 702 |
+
701,1,1,"Astor, Mrs. John Jacob (Madeleine Talmadge Force)",female,18,1,0,PC 17757,227.525,C62 C64,C
|
| 703 |
+
702,1,1,"Silverthorne, Mr. Spencer Victor",male,35,0,0,PC 17475,26.2875,E24,S
|
| 704 |
+
703,0,3,"Barbara, Miss. Saiide",female,18,0,1,2691,14.4542,,C
|
| 705 |
+
704,0,3,"Gallagher, Mr. Martin",male,25,0,0,36864,7.7417,,Q
|
| 706 |
+
705,0,3,"Hansen, Mr. Henrik Juul",male,26,1,0,350025,7.8542,,S
|
| 707 |
+
706,0,2,"Morley, Mr. Henry Samuel (""Mr Henry Marshall"")",male,39,0,0,250655,26,,S
|
| 708 |
+
707,1,2,"Kelly, Mrs. Florence ""Fannie""",female,45,0,0,223596,13.5,,S
|
| 709 |
+
708,1,1,"Calderhead, Mr. Edward Pennington",male,42,0,0,PC 17476,26.2875,E24,S
|
| 710 |
+
709,1,1,"Cleaver, Miss. Alice",female,22,0,0,113781,151.55,,S
|
| 711 |
+
710,1,3,"Moubarek, Master. Halim Gonios (""William George"")",male,,1,1,2661,15.2458,,C
|
| 712 |
+
711,1,1,"Mayne, Mlle. Berthe Antonine (""Mrs de Villiers"")",female,24,0,0,PC 17482,49.5042,C90,C
|
| 713 |
+
712,0,1,"Klaber, Mr. Herman",male,,0,0,113028,26.55,C124,S
|
| 714 |
+
713,1,1,"Taylor, Mr. Elmer Zebley",male,48,1,0,19996,52,C126,S
|
| 715 |
+
714,0,3,"Larsson, Mr. August Viktor",male,29,0,0,7545,9.4833,,S
|
| 716 |
+
715,0,2,"Greenberg, Mr. Samuel",male,52,0,0,250647,13,,S
|
| 717 |
+
716,0,3,"Soholt, Mr. Peter Andreas Lauritz Andersen",male,19,0,0,348124,7.65,F G73,S
|
| 718 |
+
717,1,1,"Endres, Miss. Caroline Louise",female,38,0,0,PC 17757,227.525,C45,C
|
| 719 |
+
718,1,2,"Troutt, Miss. Edwina Celia ""Winnie""",female,27,0,0,34218,10.5,E101,S
|
| 720 |
+
719,0,3,"McEvoy, Mr. Michael",male,,0,0,36568,15.5,,Q
|
| 721 |
+
720,0,3,"Johnson, Mr. Malkolm Joackim",male,33,0,0,347062,7.775,,S
|
| 722 |
+
721,1,2,"Harper, Miss. Annie Jessie ""Nina""",female,6,0,1,248727,33,,S
|
| 723 |
+
722,0,3,"Jensen, Mr. Svend Lauritz",male,17,1,0,350048,7.0542,,S
|
| 724 |
+
723,0,2,"Gillespie, Mr. William Henry",male,34,0,0,12233,13,,S
|
| 725 |
+
724,0,2,"Hodges, Mr. Henry Price",male,50,0,0,250643,13,,S
|
| 726 |
+
725,1,1,"Chambers, Mr. Norman Campbell",male,27,1,0,113806,53.1,E8,S
|
| 727 |
+
726,0,3,"Oreskovic, Mr. Luka",male,20,0,0,315094,8.6625,,S
|
| 728 |
+
727,1,2,"Renouf, Mrs. Peter Henry (Lillian Jefferys)",female,30,3,0,31027,21,,S
|
| 729 |
+
728,1,3,"Mannion, Miss. Margareth",female,,0,0,36866,7.7375,,Q
|
| 730 |
+
729,0,2,"Bryhl, Mr. Kurt Arnold Gottfrid",male,25,1,0,236853,26,,S
|
| 731 |
+
730,0,3,"Ilmakangas, Miss. Pieta Sofia",female,25,1,0,STON/O2. 3101271,7.925,,S
|
| 732 |
+
731,1,1,"Allen, Miss. Elisabeth Walton",female,29,0,0,24160,211.3375,B5,S
|
| 733 |
+
732,0,3,"Hassan, Mr. Houssein G N",male,11,0,0,2699,18.7875,,C
|
| 734 |
+
733,0,2,"Knight, Mr. Robert J",male,,0,0,239855,0,,S
|
| 735 |
+
734,0,2,"Berriman, Mr. William John",male,23,0,0,28425,13,,S
|
| 736 |
+
735,0,2,"Troupiansky, Mr. Moses Aaron",male,23,0,0,233639,13,,S
|
| 737 |
+
736,0,3,"Williams, Mr. Leslie",male,28.5,0,0,54636,16.1,,S
|
| 738 |
+
737,0,3,"Ford, Mrs. Edward (Margaret Ann Watson)",female,48,1,3,W./C. 6608,34.375,,S
|
| 739 |
+
738,1,1,"Lesurer, Mr. Gustave J",male,35,0,0,PC 17755,512.3292,B101,C
|
| 740 |
+
739,0,3,"Ivanoff, Mr. Kanio",male,,0,0,349201,7.8958,,S
|
| 741 |
+
740,0,3,"Nankoff, Mr. Minko",male,,0,0,349218,7.8958,,S
|
| 742 |
+
741,1,1,"Hawksford, Mr. Walter James",male,,0,0,16988,30,D45,S
|
| 743 |
+
742,0,1,"Cavendish, Mr. Tyrell William",male,36,1,0,19877,78.85,C46,S
|
| 744 |
+
743,1,1,"Ryerson, Miss. Susan Parker ""Suzette""",female,21,2,2,PC 17608,262.375,B57 B59 B63 B66,C
|
| 745 |
+
744,0,3,"McNamee, Mr. Neal",male,24,1,0,376566,16.1,,S
|
| 746 |
+
745,1,3,"Stranden, Mr. Juho",male,31,0,0,STON/O 2. 3101288,7.925,,S
|
| 747 |
+
746,0,1,"Crosby, Capt. Edward Gifford",male,70,1,1,WE/P 5735,71,B22,S
|
| 748 |
+
747,0,3,"Abbott, Mr. Rossmore Edward",male,16,1,1,C.A. 2673,20.25,,S
|
| 749 |
+
748,1,2,"Sinkkonen, Miss. Anna",female,30,0,0,250648,13,,S
|
| 750 |
+
749,0,1,"Marvin, Mr. Daniel Warner",male,19,1,0,113773,53.1,D30,S
|
| 751 |
+
750,0,3,"Connaghton, Mr. Michael",male,31,0,0,335097,7.75,,Q
|
| 752 |
+
751,1,2,"Wells, Miss. Joan",female,4,1,1,29103,23,,S
|
| 753 |
+
752,1,3,"Moor, Master. Meier",male,6,0,1,392096,12.475,E121,S
|
| 754 |
+
753,0,3,"Vande Velde, Mr. Johannes Joseph",male,33,0,0,345780,9.5,,S
|
| 755 |
+
754,0,3,"Jonkoff, Mr. Lalio",male,23,0,0,349204,7.8958,,S
|
| 756 |
+
755,1,2,"Herman, Mrs. Samuel (Jane Laver)",female,48,1,2,220845,65,,S
|
| 757 |
+
756,1,2,"Hamalainen, Master. Viljo",male,0.67,1,1,250649,14.5,,S
|
| 758 |
+
757,0,3,"Carlsson, Mr. August Sigfrid",male,28,0,0,350042,7.7958,,S
|
| 759 |
+
758,0,2,"Bailey, Mr. Percy Andrew",male,18,0,0,29108,11.5,,S
|
| 760 |
+
759,0,3,"Theobald, Mr. Thomas Leonard",male,34,0,0,363294,8.05,,S
|
| 761 |
+
760,1,1,"Rothes, the Countess. of (Lucy Noel Martha Dyer-Edwards)",female,33,0,0,110152,86.5,B77,S
|
| 762 |
+
761,0,3,"Garfirth, Mr. John",male,,0,0,358585,14.5,,S
|
| 763 |
+
762,0,3,"Nirva, Mr. Iisakki Antino Aijo",male,41,0,0,SOTON/O2 3101272,7.125,,S
|
| 764 |
+
763,1,3,"Barah, Mr. Hanna Assi",male,20,0,0,2663,7.2292,,C
|
| 765 |
+
764,1,1,"Carter, Mrs. William Ernest (Lucile Polk)",female,36,1,2,113760,120,B96 B98,S
|
| 766 |
+
765,0,3,"Eklund, Mr. Hans Linus",male,16,0,0,347074,7.775,,S
|
| 767 |
+
766,1,1,"Hogeboom, Mrs. John C (Anna Andrews)",female,51,1,0,13502,77.9583,D11,S
|
| 768 |
+
767,0,1,"Brewe, Dr. Arthur Jackson",male,,0,0,112379,39.6,,C
|
| 769 |
+
768,0,3,"Mangan, Miss. Mary",female,30.5,0,0,364850,7.75,,Q
|
| 770 |
+
769,0,3,"Moran, Mr. Daniel J",male,,1,0,371110,24.15,,Q
|
| 771 |
+
770,0,3,"Gronnestad, Mr. Daniel Danielsen",male,32,0,0,8471,8.3625,,S
|
| 772 |
+
771,0,3,"Lievens, Mr. Rene Aime",male,24,0,0,345781,9.5,,S
|
| 773 |
+
772,0,3,"Jensen, Mr. Niels Peder",male,48,0,0,350047,7.8542,,S
|
| 774 |
+
773,0,2,"Mack, Mrs. (Mary)",female,57,0,0,S.O./P.P. 3,10.5,E77,S
|
| 775 |
+
774,0,3,"Elias, Mr. Dibo",male,,0,0,2674,7.225,,C
|
| 776 |
+
775,1,2,"Hocking, Mrs. Elizabeth (Eliza Needs)",female,54,1,3,29105,23,,S
|
| 777 |
+
776,0,3,"Myhrman, Mr. Pehr Fabian Oliver Malkolm",male,18,0,0,347078,7.75,,S
|
| 778 |
+
777,0,3,"Tobin, Mr. Roger",male,,0,0,383121,7.75,F38,Q
|
| 779 |
+
778,1,3,"Emanuel, Miss. Virginia Ethel",female,5,0,0,364516,12.475,,S
|
| 780 |
+
779,0,3,"Kilgannon, Mr. Thomas J",male,,0,0,36865,7.7375,,Q
|
| 781 |
+
780,1,1,"Robert, Mrs. Edward Scott (Elisabeth Walton McMillan)",female,43,0,1,24160,211.3375,B3,S
|
| 782 |
+
781,1,3,"Ayoub, Miss. Banoura",female,13,0,0,2687,7.2292,,C
|
| 783 |
+
782,1,1,"Dick, Mrs. Albert Adrian (Vera Gillespie)",female,17,1,0,17474,57,B20,S
|
| 784 |
+
783,0,1,"Long, Mr. Milton Clyde",male,29,0,0,113501,30,D6,S
|
| 785 |
+
784,0,3,"Johnston, Mr. Andrew G",male,,1,2,W./C. 6607,23.45,,S
|
| 786 |
+
785,0,3,"Ali, Mr. William",male,25,0,0,SOTON/O.Q. 3101312,7.05,,S
|
| 787 |
+
786,0,3,"Harmer, Mr. Abraham (David Lishin)",male,25,0,0,374887,7.25,,S
|
| 788 |
+
787,1,3,"Sjoblom, Miss. Anna Sofia",female,18,0,0,3101265,7.4958,,S
|
| 789 |
+
788,0,3,"Rice, Master. George Hugh",male,8,4,1,382652,29.125,,Q
|
| 790 |
+
789,1,3,"Dean, Master. Bertram Vere",male,1,1,2,C.A. 2315,20.575,,S
|
| 791 |
+
790,0,1,"Guggenheim, Mr. Benjamin",male,46,0,0,PC 17593,79.2,B82 B84,C
|
| 792 |
+
791,0,3,"Keane, Mr. Andrew ""Andy""",male,,0,0,12460,7.75,,Q
|
| 793 |
+
792,0,2,"Gaskell, Mr. Alfred",male,16,0,0,239865,26,,S
|
| 794 |
+
793,0,3,"Sage, Miss. Stella Anna",female,,8,2,CA. 2343,69.55,,S
|
| 795 |
+
794,0,1,"Hoyt, Mr. William Fisher",male,,0,0,PC 17600,30.6958,,C
|
| 796 |
+
795,0,3,"Dantcheff, Mr. Ristiu",male,25,0,0,349203,7.8958,,S
|
| 797 |
+
796,0,2,"Otter, Mr. Richard",male,39,0,0,28213,13,,S
|
| 798 |
+
797,1,1,"Leader, Dr. Alice (Farnham)",female,49,0,0,17465,25.9292,D17,S
|
| 799 |
+
798,1,3,"Osman, Mrs. Mara",female,31,0,0,349244,8.6833,,S
|
| 800 |
+
799,0,3,"Ibrahim Shawah, Mr. Yousseff",male,30,0,0,2685,7.2292,,C
|
| 801 |
+
800,0,3,"Van Impe, Mrs. Jean Baptiste (Rosalie Paula Govaert)",female,30,1,1,345773,24.15,,S
|
| 802 |
+
801,0,2,"Ponesell, Mr. Martin",male,34,0,0,250647,13,,S
|
| 803 |
+
802,1,2,"Collyer, Mrs. Harvey (Charlotte Annie Tate)",female,31,1,1,C.A. 31921,26.25,,S
|
| 804 |
+
803,1,1,"Carter, Master. William Thornton II",male,11,1,2,113760,120,B96 B98,S
|
| 805 |
+
804,1,3,"Thomas, Master. Assad Alexander",male,0.42,0,1,2625,8.5167,,C
|
| 806 |
+
805,1,3,"Hedman, Mr. Oskar Arvid",male,27,0,0,347089,6.975,,S
|
| 807 |
+
806,0,3,"Johansson, Mr. Karl Johan",male,31,0,0,347063,7.775,,S
|
| 808 |
+
807,0,1,"Andrews, Mr. Thomas Jr",male,39,0,0,112050,0,A36,S
|
| 809 |
+
808,0,3,"Pettersson, Miss. Ellen Natalia",female,18,0,0,347087,7.775,,S
|
| 810 |
+
809,0,2,"Meyer, Mr. August",male,39,0,0,248723,13,,S
|
| 811 |
+
810,1,1,"Chambers, Mrs. Norman Campbell (Bertha Griggs)",female,33,1,0,113806,53.1,E8,S
|
| 812 |
+
811,0,3,"Alexander, Mr. William",male,26,0,0,3474,7.8875,,S
|
| 813 |
+
812,0,3,"Lester, Mr. James",male,39,0,0,A/4 48871,24.15,,S
|
| 814 |
+
813,0,2,"Slemen, Mr. Richard James",male,35,0,0,28206,10.5,,S
|
| 815 |
+
814,0,3,"Andersson, Miss. Ebba Iris Alfrida",female,6,4,2,347082,31.275,,S
|
| 816 |
+
815,0,3,"Tomlin, Mr. Ernest Portage",male,30.5,0,0,364499,8.05,,S
|
| 817 |
+
816,0,1,"Fry, Mr. Richard",male,,0,0,112058,0,B102,S
|
| 818 |
+
817,0,3,"Heininen, Miss. Wendla Maria",female,23,0,0,STON/O2. 3101290,7.925,,S
|
| 819 |
+
818,0,2,"Mallet, Mr. Albert",male,31,1,1,S.C./PARIS 2079,37.0042,,C
|
| 820 |
+
819,0,3,"Holm, Mr. John Fredrik Alexander",male,43,0,0,C 7075,6.45,,S
|
| 821 |
+
820,0,3,"Skoog, Master. Karl Thorsten",male,10,3,2,347088,27.9,,S
|
| 822 |
+
821,1,1,"Hays, Mrs. Charles Melville (Clara Jennings Gregg)",female,52,1,1,12749,93.5,B69,S
|
| 823 |
+
822,1,3,"Lulic, Mr. Nikola",male,27,0,0,315098,8.6625,,S
|
| 824 |
+
823,0,1,"Reuchlin, Jonkheer. John George",male,38,0,0,19972,0,,S
|
| 825 |
+
824,1,3,"Moor, Mrs. (Beila)",female,27,0,1,392096,12.475,E121,S
|
| 826 |
+
825,0,3,"Panula, Master. Urho Abraham",male,2,4,1,3101295,39.6875,,S
|
| 827 |
+
826,0,3,"Flynn, Mr. John",male,,0,0,368323,6.95,,Q
|
| 828 |
+
827,0,3,"Lam, Mr. Len",male,,0,0,1601,56.4958,,S
|
| 829 |
+
828,1,2,"Mallet, Master. Andre",male,1,0,2,S.C./PARIS 2079,37.0042,,C
|
| 830 |
+
829,1,3,"McCormack, Mr. Thomas Joseph",male,,0,0,367228,7.75,,Q
|
| 831 |
+
830,1,1,"Stone, Mrs. George Nelson (Martha Evelyn)",female,62,0,0,113572,80,B28,
|
| 832 |
+
831,1,3,"Yasbeck, Mrs. Antoni (Selini Alexander)",female,15,1,0,2659,14.4542,,C
|
| 833 |
+
832,1,2,"Richards, Master. George Sibley",male,0.83,1,1,29106,18.75,,S
|
| 834 |
+
833,0,3,"Saad, Mr. Amin",male,,0,0,2671,7.2292,,C
|
| 835 |
+
834,0,3,"Augustsson, Mr. Albert",male,23,0,0,347468,7.8542,,S
|
| 836 |
+
835,0,3,"Allum, Mr. Owen George",male,18,0,0,2223,8.3,,S
|
| 837 |
+
836,1,1,"Compton, Miss. Sara Rebecca",female,39,1,1,PC 17756,83.1583,E49,C
|
| 838 |
+
837,0,3,"Pasic, Mr. Jakob",male,21,0,0,315097,8.6625,,S
|
| 839 |
+
838,0,3,"Sirota, Mr. Maurice",male,,0,0,392092,8.05,,S
|
| 840 |
+
839,1,3,"Chip, Mr. Chang",male,32,0,0,1601,56.4958,,S
|
| 841 |
+
840,1,1,"Marechal, Mr. Pierre",male,,0,0,11774,29.7,C47,C
|
| 842 |
+
841,0,3,"Alhomaki, Mr. Ilmari Rudolf",male,20,0,0,SOTON/O2 3101287,7.925,,S
|
| 843 |
+
842,0,2,"Mudd, Mr. Thomas Charles",male,16,0,0,S.O./P.P. 3,10.5,,S
|
| 844 |
+
843,1,1,"Serepeca, Miss. Augusta",female,30,0,0,113798,31,,C
|
| 845 |
+
844,0,3,"Lemberopolous, Mr. Peter L",male,34.5,0,0,2683,6.4375,,C
|
| 846 |
+
845,0,3,"Culumovic, Mr. Jeso",male,17,0,0,315090,8.6625,,S
|
| 847 |
+
846,0,3,"Abbing, Mr. Anthony",male,42,0,0,C.A. 5547,7.55,,S
|
| 848 |
+
847,0,3,"Sage, Mr. Douglas Bullen",male,,8,2,CA. 2343,69.55,,S
|
| 849 |
+
848,0,3,"Markoff, Mr. Marin",male,35,0,0,349213,7.8958,,C
|
| 850 |
+
849,0,2,"Harper, Rev. John",male,28,0,1,248727,33,,S
|
| 851 |
+
850,1,1,"Goldenberg, Mrs. Samuel L (Edwiga Grabowska)",female,,1,0,17453,89.1042,C92,C
|
| 852 |
+
851,0,3,"Andersson, Master. Sigvard Harald Elias",male,4,4,2,347082,31.275,,S
|
| 853 |
+
852,0,3,"Svensson, Mr. Johan",male,74,0,0,347060,7.775,,S
|
| 854 |
+
853,0,3,"Boulos, Miss. Nourelain",female,9,1,1,2678,15.2458,,C
|
| 855 |
+
854,1,1,"Lines, Miss. Mary Conover",female,16,0,1,PC 17592,39.4,D28,S
|
| 856 |
+
855,0,2,"Carter, Mrs. Ernest Courtenay (Lilian Hughes)",female,44,1,0,244252,26,,S
|
| 857 |
+
856,1,3,"Aks, Mrs. Sam (Leah Rosen)",female,18,0,1,392091,9.35,,S
|
| 858 |
+
857,1,1,"Wick, Mrs. George Dennick (Mary Hitchcock)",female,45,1,1,36928,164.8667,,S
|
| 859 |
+
858,1,1,"Daly, Mr. Peter Denis ",male,51,0,0,113055,26.55,E17,S
|
| 860 |
+
859,1,3,"Baclini, Mrs. Solomon (Latifa Qurban)",female,24,0,3,2666,19.2583,,C
|
| 861 |
+
860,0,3,"Razi, Mr. Raihed",male,,0,0,2629,7.2292,,C
|
| 862 |
+
861,0,3,"Hansen, Mr. Claus Peter",male,41,2,0,350026,14.1083,,S
|
| 863 |
+
862,0,2,"Giles, Mr. Frederick Edward",male,21,1,0,28134,11.5,,S
|
| 864 |
+
863,1,1,"Swift, Mrs. Frederick Joel (Margaret Welles Barron)",female,48,0,0,17466,25.9292,D17,S
|
| 865 |
+
864,0,3,"Sage, Miss. Dorothy Edith ""Dolly""",female,,8,2,CA. 2343,69.55,,S
|
| 866 |
+
865,0,2,"Gill, Mr. John William",male,24,0,0,233866,13,,S
|
| 867 |
+
866,1,2,"Bystrom, Mrs. (Karolina)",female,42,0,0,236852,13,,S
|
| 868 |
+
867,1,2,"Duran y More, Miss. Asuncion",female,27,1,0,SC/PARIS 2149,13.8583,,C
|
| 869 |
+
868,0,1,"Roebling, Mr. Washington Augustus II",male,31,0,0,PC 17590,50.4958,A24,S
|
| 870 |
+
869,0,3,"van Melkebeke, Mr. Philemon",male,,0,0,345777,9.5,,S
|
| 871 |
+
870,1,3,"Johnson, Master. Harold Theodor",male,4,1,1,347742,11.1333,,S
|
| 872 |
+
871,0,3,"Balkic, Mr. Cerin",male,26,0,0,349248,7.8958,,S
|
| 873 |
+
872,1,1,"Beckwith, Mrs. Richard Leonard (Sallie Monypeny)",female,47,1,1,11751,52.5542,D35,S
|
| 874 |
+
873,0,1,"Carlsson, Mr. Frans Olof",male,33,0,0,695,5,B51 B53 B55,S
|
| 875 |
+
874,0,3,"Vander Cruyssen, Mr. Victor",male,47,0,0,345765,9,,S
|
| 876 |
+
875,1,2,"Abelson, Mrs. Samuel (Hannah Wizosky)",female,28,1,0,P/PP 3381,24,,C
|
| 877 |
+
876,1,3,"Najib, Miss. Adele Kiamie ""Jane""",female,15,0,0,2667,7.225,,C
|
| 878 |
+
877,0,3,"Gustafsson, Mr. Alfred Ossian",male,20,0,0,7534,9.8458,,S
|
| 879 |
+
878,0,3,"Petroff, Mr. Nedelio",male,19,0,0,349212,7.8958,,S
|
| 880 |
+
879,0,3,"Laleff, Mr. Kristo",male,,0,0,349217,7.8958,,S
|
| 881 |
+
880,1,1,"Potter, Mrs. Thomas Jr (Lily Alexenia Wilson)",female,56,0,1,11767,83.1583,C50,C
|
| 882 |
+
881,1,2,"Shelley, Mrs. William (Imanita Parrish Hall)",female,25,0,1,230433,26,,S
|
| 883 |
+
882,0,3,"Markun, Mr. Johann",male,33,0,0,349257,7.8958,,S
|
| 884 |
+
883,0,3,"Dahlberg, Miss. Gerda Ulrika",female,22,0,0,7552,10.5167,,S
|
| 885 |
+
884,0,2,"Banfield, Mr. Frederick James",male,28,0,0,C.A./SOTON 34068,10.5,,S
|
| 886 |
+
885,0,3,"Sutehall, Mr. Henry Jr",male,25,0,0,SOTON/OQ 392076,7.05,,S
|
| 887 |
+
886,0,3,"Rice, Mrs. William (Margaret Norton)",female,39,0,5,382652,29.125,,Q
|
| 888 |
+
887,0,2,"Montvila, Rev. Juozas",male,27,0,0,211536,13,,S
|
| 889 |
+
888,1,1,"Graham, Miss. Margaret Edith",female,19,0,0,112053,30,B42,S
|
| 890 |
+
889,0,3,"Johnston, Miss. Catherine Helen ""Carrie""",female,,1,2,W./C. 6607,23.45,,S
|
| 891 |
+
890,1,1,"Behr, Mr. Karl Howell",male,26,0,0,111369,30,C148,C
|
| 892 |
+
891,0,3,"Dooley, Mr. Patrick",male,32,0,0,370376,7.75,,Q
|
Titanic/Kernels/AdaBoost/.ipynb_checkpoints/0-introduction-to-ensembling-stacking-in-python-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/.ipynb_checkpoints/1-a-data-science-framework-to-achieve-99-accuracy-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/.ipynb_checkpoints/10-a-comprehensive-guide-to-titanic-machine-learning-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/.ipynb_checkpoints/2-titanic-top-4-with-ensemble-modeling-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/.ipynb_checkpoints/3-eda-to-prediction-dietanic-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/.ipynb_checkpoints/4-a-statistical-analysis-ml-workflow-of-titanic-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/.ipynb_checkpoints/6-titanic-best-working-classifier-checkpoint.ipynb
ADDED
|
@@ -0,0 +1,1504 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "markdown",
|
| 5 |
+
"metadata": {
|
| 6 |
+
"_cell_guid": "25b1e1db-8bc5-7029-f719-91da523bd121"
|
| 7 |
+
},
|
| 8 |
+
"source": [
|
| 9 |
+
"## Introduction ##\n",
|
| 10 |
+
"\n",
|
| 11 |
+
"This is my first work of machine learning. the notebook is written in python and has inspired from [\"Exploring Survival on Titanic\" by Megan Risdal, a Kernel in R on Kaggle][1].\n",
|
| 12 |
+
"\n",
|
| 13 |
+
"\n",
|
| 14 |
+
" [1]: https://www.kaggle.com/mrisdal/titanic/exploring-survival-on-the-titanic"
|
| 15 |
+
]
|
| 16 |
+
},
|
| 17 |
+
{
|
| 18 |
+
"cell_type": "code",
|
| 19 |
+
"execution_count": 1,
|
| 20 |
+
"metadata": {
|
| 21 |
+
"_cell_guid": "2ce68358-02ec-556d-ba88-e773a50bc18b"
|
| 22 |
+
},
|
| 23 |
+
"outputs": [
|
| 24 |
+
{
|
| 25 |
+
"name": "stdout",
|
| 26 |
+
"output_type": "stream",
|
| 27 |
+
"text": [
|
| 28 |
+
"<class 'pandas.core.frame.DataFrame'>\n",
|
| 29 |
+
"RangeIndex: 891 entries, 0 to 890\n",
|
| 30 |
+
"Data columns (total 12 columns):\n",
|
| 31 |
+
" # Column Non-Null Count Dtype \n",
|
| 32 |
+
"--- ------ -------------- ----- \n",
|
| 33 |
+
" 0 PassengerId 891 non-null int64 \n",
|
| 34 |
+
" 1 Survived 891 non-null int64 \n",
|
| 35 |
+
" 2 Pclass 891 non-null int64 \n",
|
| 36 |
+
" 3 Name 891 non-null object \n",
|
| 37 |
+
" 4 Sex 891 non-null object \n",
|
| 38 |
+
" 5 Age 714 non-null float64\n",
|
| 39 |
+
" 6 SibSp 891 non-null int64 \n",
|
| 40 |
+
" 7 Parch 891 non-null int64 \n",
|
| 41 |
+
" 8 Ticket 891 non-null object \n",
|
| 42 |
+
" 9 Fare 891 non-null float64\n",
|
| 43 |
+
" 10 Cabin 204 non-null object \n",
|
| 44 |
+
" 11 Embarked 889 non-null object \n",
|
| 45 |
+
"dtypes: float64(2), int64(5), object(5)\n",
|
| 46 |
+
"memory usage: 83.7+ KB\n",
|
| 47 |
+
"None\n"
|
| 48 |
+
]
|
| 49 |
+
}
|
| 50 |
+
],
|
| 51 |
+
"source": [
|
| 52 |
+
"%matplotlib inline\n",
|
| 53 |
+
"import numpy as np\n",
|
| 54 |
+
"import pandas as pd\n",
|
| 55 |
+
"import re as re\n",
|
| 56 |
+
"\n",
|
| 57 |
+
"train = pd.read_csv('../../Data/train.csv', header = 0, dtype={'Age': np.float64})\n",
|
| 58 |
+
"test = pd.read_csv('../../Data/test.csv' , header = 0, dtype={'Age': np.float64})\n",
|
| 59 |
+
"full_data = [train, test]\n",
|
| 60 |
+
"\n",
|
| 61 |
+
"print (train.info())"
|
| 62 |
+
]
|
| 63 |
+
},
|
| 64 |
+
{
|
| 65 |
+
"cell_type": "code",
|
| 66 |
+
"execution_count": 2,
|
| 67 |
+
"metadata": {},
|
| 68 |
+
"outputs": [],
|
| 69 |
+
"source": [
|
| 70 |
+
"from aif360.datasets import StandardDataset\n",
|
| 71 |
+
"from aif360.metrics import BinaryLabelDatasetMetric, ClassificationMetric\n",
|
| 72 |
+
"import matplotlib.patches as patches\n",
|
| 73 |
+
"from aif360.algorithms.preprocessing import Reweighing\n",
|
| 74 |
+
"#from packages import *\n",
|
| 75 |
+
"#from ml_fairness import *\n",
|
| 76 |
+
"import matplotlib.pyplot as plt\n",
|
| 77 |
+
"import seaborn as sns\n",
|
| 78 |
+
"\n",
|
| 79 |
+
"\n",
|
| 80 |
+
"\n",
|
| 81 |
+
"from IPython.display import Markdown, display"
|
| 82 |
+
]
|
| 83 |
+
},
|
| 84 |
+
{
|
| 85 |
+
"cell_type": "markdown",
|
| 86 |
+
"metadata": {
|
| 87 |
+
"_cell_guid": "f9595646-65c9-6fc4-395f-0befc4d122ce"
|
| 88 |
+
},
|
| 89 |
+
"source": [
|
| 90 |
+
"# Feature Engineering #"
|
| 91 |
+
]
|
| 92 |
+
},
|
| 93 |
+
{
|
| 94 |
+
"cell_type": "markdown",
|
| 95 |
+
"metadata": {
|
| 96 |
+
"_cell_guid": "9b4c278b-aaca-e92c-ba77-b9b48379d1f1"
|
| 97 |
+
},
|
| 98 |
+
"source": [
|
| 99 |
+
"## 1. Pclass ##\n",
|
| 100 |
+
"there is no missing value on this feature and already a numerical value. so let's check it's impact on our train set."
|
| 101 |
+
]
|
| 102 |
+
},
|
| 103 |
+
{
|
| 104 |
+
"cell_type": "code",
|
| 105 |
+
"execution_count": 3,
|
| 106 |
+
"metadata": {
|
| 107 |
+
"_cell_guid": "4680d950-cf7d-a6ae-e813-535e2247d88e"
|
| 108 |
+
},
|
| 109 |
+
"outputs": [
|
| 110 |
+
{
|
| 111 |
+
"name": "stdout",
|
| 112 |
+
"output_type": "stream",
|
| 113 |
+
"text": [
|
| 114 |
+
" Pclass Survived\n",
|
| 115 |
+
"0 1 0.629630\n",
|
| 116 |
+
"1 2 0.472826\n",
|
| 117 |
+
"2 3 0.242363\n"
|
| 118 |
+
]
|
| 119 |
+
}
|
| 120 |
+
],
|
| 121 |
+
"source": [
|
| 122 |
+
"print (train[['Pclass', 'Survived']].groupby(['Pclass'], as_index=False).mean())"
|
| 123 |
+
]
|
| 124 |
+
},
|
| 125 |
+
{
|
| 126 |
+
"cell_type": "markdown",
|
| 127 |
+
"metadata": {
|
| 128 |
+
"_cell_guid": "5e70f81c-d4e2-1823-f0ba-a7c9b46984ff"
|
| 129 |
+
},
|
| 130 |
+
"source": [
|
| 131 |
+
"## 2. Sex ##"
|
| 132 |
+
]
|
| 133 |
+
},
|
| 134 |
+
{
|
| 135 |
+
"cell_type": "code",
|
| 136 |
+
"execution_count": 4,
|
| 137 |
+
"metadata": {
|
| 138 |
+
"_cell_guid": "6729681d-7915-1631-78d2-ddf3c35a424c"
|
| 139 |
+
},
|
| 140 |
+
"outputs": [
|
| 141 |
+
{
|
| 142 |
+
"name": "stdout",
|
| 143 |
+
"output_type": "stream",
|
| 144 |
+
"text": [
|
| 145 |
+
" Sex Survived\n",
|
| 146 |
+
"0 female 0.742038\n",
|
| 147 |
+
"1 male 0.188908\n"
|
| 148 |
+
]
|
| 149 |
+
}
|
| 150 |
+
],
|
| 151 |
+
"source": [
|
| 152 |
+
"print (train[[\"Sex\", \"Survived\"]].groupby(['Sex'], as_index=False).mean())"
|
| 153 |
+
]
|
| 154 |
+
},
|
| 155 |
+
{
|
| 156 |
+
"cell_type": "markdown",
|
| 157 |
+
"metadata": {
|
| 158 |
+
"_cell_guid": "7c58b7ee-d6a1-0cc9-2346-81c47846a54a"
|
| 159 |
+
},
|
| 160 |
+
"source": [
|
| 161 |
+
"## 3. SibSp and Parch ##\n",
|
| 162 |
+
"With the number of siblings/spouse and the number of children/parents we can create new feature called Family Size."
|
| 163 |
+
]
|
| 164 |
+
},
|
| 165 |
+
{
|
| 166 |
+
"cell_type": "code",
|
| 167 |
+
"execution_count": 5,
|
| 168 |
+
"metadata": {
|
| 169 |
+
"_cell_guid": "1a537f10-7cec-d0b7-8a34-fa9975655190"
|
| 170 |
+
},
|
| 171 |
+
"outputs": [
|
| 172 |
+
{
|
| 173 |
+
"name": "stdout",
|
| 174 |
+
"output_type": "stream",
|
| 175 |
+
"text": [
|
| 176 |
+
" FamilySize Survived\n",
|
| 177 |
+
"0 1 0.303538\n",
|
| 178 |
+
"1 2 0.552795\n",
|
| 179 |
+
"2 3 0.578431\n",
|
| 180 |
+
"3 4 0.724138\n",
|
| 181 |
+
"4 5 0.200000\n",
|
| 182 |
+
"5 6 0.136364\n",
|
| 183 |
+
"6 7 0.333333\n",
|
| 184 |
+
"7 8 0.000000\n",
|
| 185 |
+
"8 11 0.000000\n"
|
| 186 |
+
]
|
| 187 |
+
}
|
| 188 |
+
],
|
| 189 |
+
"source": [
|
| 190 |
+
"for dataset in full_data:\n",
|
| 191 |
+
" dataset['FamilySize'] = dataset['SibSp'] + dataset['Parch'] + 1\n",
|
| 192 |
+
"print (train[['FamilySize', 'Survived']].groupby(['FamilySize'], as_index=False).mean())"
|
| 193 |
+
]
|
| 194 |
+
},
|
| 195 |
+
{
|
| 196 |
+
"cell_type": "markdown",
|
| 197 |
+
"metadata": {
|
| 198 |
+
"_cell_guid": "e4861d3e-10db-1a23-8728-44e4d5251844"
|
| 199 |
+
},
|
| 200 |
+
"source": [
|
| 201 |
+
"it seems has a good effect on our prediction but let's go further and categorize people to check whether they are alone in this ship or not."
|
| 202 |
+
]
|
| 203 |
+
},
|
| 204 |
+
{
|
| 205 |
+
"cell_type": "code",
|
| 206 |
+
"execution_count": 6,
|
| 207 |
+
"metadata": {
|
| 208 |
+
"_cell_guid": "8c35e945-c928-e3bc-bd9c-d6ddb287e4c9"
|
| 209 |
+
},
|
| 210 |
+
"outputs": [
|
| 211 |
+
{
|
| 212 |
+
"name": "stdout",
|
| 213 |
+
"output_type": "stream",
|
| 214 |
+
"text": [
|
| 215 |
+
" IsAlone Survived\n",
|
| 216 |
+
"0 0 0.505650\n",
|
| 217 |
+
"1 1 0.303538\n"
|
| 218 |
+
]
|
| 219 |
+
}
|
| 220 |
+
],
|
| 221 |
+
"source": [
|
| 222 |
+
"for dataset in full_data:\n",
|
| 223 |
+
" dataset['IsAlone'] = 0\n",
|
| 224 |
+
" dataset.loc[dataset['FamilySize'] == 1, 'IsAlone'] = 1\n",
|
| 225 |
+
"print (train[['IsAlone', 'Survived']].groupby(['IsAlone'], as_index=False).mean())"
|
| 226 |
+
]
|
| 227 |
+
},
|
| 228 |
+
{
|
| 229 |
+
"cell_type": "markdown",
|
| 230 |
+
"metadata": {
|
| 231 |
+
"_cell_guid": "2780ca4e-7923-b845-0b6b-5f68a45f6b93"
|
| 232 |
+
},
|
| 233 |
+
"source": [
|
| 234 |
+
"good! the impact is considerable."
|
| 235 |
+
]
|
| 236 |
+
},
|
| 237 |
+
{
|
| 238 |
+
"cell_type": "markdown",
|
| 239 |
+
"metadata": {
|
| 240 |
+
"_cell_guid": "8aa419c0-6614-7efc-7797-97f4a5158b19"
|
| 241 |
+
},
|
| 242 |
+
"source": [
|
| 243 |
+
"## 4. Embarked ##\n",
|
| 244 |
+
"the embarked feature has some missing value. and we try to fill those with the most occurred value ( 'S' )."
|
| 245 |
+
]
|
| 246 |
+
},
|
| 247 |
+
{
|
| 248 |
+
"cell_type": "code",
|
| 249 |
+
"execution_count": 7,
|
| 250 |
+
"metadata": {
|
| 251 |
+
"_cell_guid": "0e70e9af-d7cc-8c40-b7d4-2643889c376d"
|
| 252 |
+
},
|
| 253 |
+
"outputs": [
|
| 254 |
+
{
|
| 255 |
+
"name": "stdout",
|
| 256 |
+
"output_type": "stream",
|
| 257 |
+
"text": [
|
| 258 |
+
" Embarked Survived\n",
|
| 259 |
+
"0 C 0.553571\n",
|
| 260 |
+
"1 Q 0.389610\n",
|
| 261 |
+
"2 S 0.339009\n"
|
| 262 |
+
]
|
| 263 |
+
}
|
| 264 |
+
],
|
| 265 |
+
"source": [
|
| 266 |
+
"for dataset in full_data:\n",
|
| 267 |
+
" dataset['Embarked'] = dataset['Embarked'].fillna('S')\n",
|
| 268 |
+
"print (train[['Embarked', 'Survived']].groupby(['Embarked'], as_index=False).mean())"
|
| 269 |
+
]
|
| 270 |
+
},
|
| 271 |
+
{
|
| 272 |
+
"cell_type": "markdown",
|
| 273 |
+
"metadata": {
|
| 274 |
+
"_cell_guid": "e08c9ee8-d6d1-99b7-38bd-f0042c18a5d9"
|
| 275 |
+
},
|
| 276 |
+
"source": [
|
| 277 |
+
"## 5. Fare ##\n",
|
| 278 |
+
"Fare also has some missing value and we will replace it with the median. then we categorize it into 4 ranges."
|
| 279 |
+
]
|
| 280 |
+
},
|
| 281 |
+
{
|
| 282 |
+
"cell_type": "code",
|
| 283 |
+
"execution_count": 8,
|
| 284 |
+
"metadata": {
|
| 285 |
+
"_cell_guid": "a21335bd-4e8d-66e8-e6a5-5d2173b72d3b"
|
| 286 |
+
},
|
| 287 |
+
"outputs": [
|
| 288 |
+
{
|
| 289 |
+
"name": "stdout",
|
| 290 |
+
"output_type": "stream",
|
| 291 |
+
"text": [
|
| 292 |
+
" CategoricalFare Survived\n",
|
| 293 |
+
"0 (-0.001, 7.91] 0.197309\n",
|
| 294 |
+
"1 (7.91, 14.454] 0.303571\n",
|
| 295 |
+
"2 (14.454, 31.0] 0.454955\n",
|
| 296 |
+
"3 (31.0, 512.329] 0.581081\n"
|
| 297 |
+
]
|
| 298 |
+
}
|
| 299 |
+
],
|
| 300 |
+
"source": [
|
| 301 |
+
"for dataset in full_data:\n",
|
| 302 |
+
" dataset['Fare'] = dataset['Fare'].fillna(train['Fare'].median())\n",
|
| 303 |
+
"train['CategoricalFare'] = pd.qcut(train['Fare'], 4)\n",
|
| 304 |
+
"print (train[['CategoricalFare', 'Survived']].groupby(['CategoricalFare'], as_index=False).mean())"
|
| 305 |
+
]
|
| 306 |
+
},
|
| 307 |
+
{
|
| 308 |
+
"cell_type": "markdown",
|
| 309 |
+
"metadata": {
|
| 310 |
+
"_cell_guid": "ec8d1b22-a95f-9f16-77ab-7b60d2103852"
|
| 311 |
+
},
|
| 312 |
+
"source": [
|
| 313 |
+
"## 6. Age ##\n",
|
| 314 |
+
"we have plenty of missing values in this feature. # generate random numbers between (mean - std) and (mean + std).\n",
|
| 315 |
+
"then we categorize age into 5 range."
|
| 316 |
+
]
|
| 317 |
+
},
|
| 318 |
+
{
|
| 319 |
+
"cell_type": "code",
|
| 320 |
+
"execution_count": 9,
|
| 321 |
+
"metadata": {
|
| 322 |
+
"_cell_guid": "b90c2870-ce5d-ae0e-a33d-59e35445500e"
|
| 323 |
+
},
|
| 324 |
+
"outputs": [
|
| 325 |
+
{
|
| 326 |
+
"name": "stdout",
|
| 327 |
+
"output_type": "stream",
|
| 328 |
+
"text": [
|
| 329 |
+
" CategoricalAge Survived\n",
|
| 330 |
+
"0 (-0.08, 16.0] 0.530973\n",
|
| 331 |
+
"1 (16.0, 32.0] 0.353741\n",
|
| 332 |
+
"2 (32.0, 48.0] 0.369650\n",
|
| 333 |
+
"3 (48.0, 64.0] 0.434783\n",
|
| 334 |
+
"4 (64.0, 80.0] 0.090909\n"
|
| 335 |
+
]
|
| 336 |
+
},
|
| 337 |
+
{
|
| 338 |
+
"name": "stderr",
|
| 339 |
+
"output_type": "stream",
|
| 340 |
+
"text": [
|
| 341 |
+
"\n",
|
| 342 |
+
"A value is trying to be set on a copy of a slice from a DataFrame\n",
|
| 343 |
+
"\n",
|
| 344 |
+
"See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n"
|
| 345 |
+
]
|
| 346 |
+
}
|
| 347 |
+
],
|
| 348 |
+
"source": [
|
| 349 |
+
"for dataset in full_data:\n",
|
| 350 |
+
" age_avg \t = dataset['Age'].mean()\n",
|
| 351 |
+
" age_std \t = dataset['Age'].std()\n",
|
| 352 |
+
" age_null_count = dataset['Age'].isnull().sum()\n",
|
| 353 |
+
" \n",
|
| 354 |
+
" age_null_random_list = np.random.randint(age_avg - age_std, age_avg + age_std, size=age_null_count)\n",
|
| 355 |
+
" dataset['Age'][np.isnan(dataset['Age'])] = age_null_random_list\n",
|
| 356 |
+
" dataset['Age'] = dataset['Age'].astype(int)\n",
|
| 357 |
+
" \n",
|
| 358 |
+
"train['CategoricalAge'] = pd.cut(train['Age'], 5)\n",
|
| 359 |
+
"\n",
|
| 360 |
+
"print (train[['CategoricalAge', 'Survived']].groupby(['CategoricalAge'], as_index=False).mean())"
|
| 361 |
+
]
|
| 362 |
+
},
|
| 363 |
+
{
|
| 364 |
+
"cell_type": "markdown",
|
| 365 |
+
"metadata": {
|
| 366 |
+
"_cell_guid": "bd25ec3f-b601-c1cc-d701-991fac1621f9"
|
| 367 |
+
},
|
| 368 |
+
"source": [
|
| 369 |
+
"## 7. Name ##\n",
|
| 370 |
+
"inside this feature we can find the title of people."
|
| 371 |
+
]
|
| 372 |
+
},
|
| 373 |
+
{
|
| 374 |
+
"cell_type": "code",
|
| 375 |
+
"execution_count": 10,
|
| 376 |
+
"metadata": {
|
| 377 |
+
"_cell_guid": "ad042f43-bfe0-ded0-4171-379d8caaa749"
|
| 378 |
+
},
|
| 379 |
+
"outputs": [
|
| 380 |
+
{
|
| 381 |
+
"name": "stdout",
|
| 382 |
+
"output_type": "stream",
|
| 383 |
+
"text": [
|
| 384 |
+
"Sex female male\n",
|
| 385 |
+
"Title \n",
|
| 386 |
+
"Capt 0 1\n",
|
| 387 |
+
"Col 0 2\n",
|
| 388 |
+
"Countess 1 0\n",
|
| 389 |
+
"Don 0 1\n",
|
| 390 |
+
"Dr 1 6\n",
|
| 391 |
+
"Jonkheer 0 1\n",
|
| 392 |
+
"Lady 1 0\n",
|
| 393 |
+
"Major 0 2\n",
|
| 394 |
+
"Master 0 40\n",
|
| 395 |
+
"Miss 182 0\n",
|
| 396 |
+
"Mlle 2 0\n",
|
| 397 |
+
"Mme 1 0\n",
|
| 398 |
+
"Mr 0 517\n",
|
| 399 |
+
"Mrs 125 0\n",
|
| 400 |
+
"Ms 1 0\n",
|
| 401 |
+
"Rev 0 6\n",
|
| 402 |
+
"Sir 0 1\n"
|
| 403 |
+
]
|
| 404 |
+
}
|
| 405 |
+
],
|
| 406 |
+
"source": [
|
| 407 |
+
"def get_title(name):\n",
|
| 408 |
+
"\ttitle_search = re.search(' ([A-Za-z]+)\\.', name)\n",
|
| 409 |
+
"\t# If the title exists, extract and return it.\n",
|
| 410 |
+
"\tif title_search:\n",
|
| 411 |
+
"\t\treturn title_search.group(1)\n",
|
| 412 |
+
"\treturn \"\"\n",
|
| 413 |
+
"\n",
|
| 414 |
+
"for dataset in full_data:\n",
|
| 415 |
+
" dataset['Title'] = dataset['Name'].apply(get_title)\n",
|
| 416 |
+
"\n",
|
| 417 |
+
"print(pd.crosstab(train['Title'], train['Sex']))"
|
| 418 |
+
]
|
| 419 |
+
},
|
| 420 |
+
{
|
| 421 |
+
"cell_type": "markdown",
|
| 422 |
+
"metadata": {
|
| 423 |
+
"_cell_guid": "ca5fff8c-7a0d-6c18-2173-b8df6293c50a"
|
| 424 |
+
},
|
| 425 |
+
"source": [
|
| 426 |
+
" so we have titles. let's categorize it and check the title impact on survival rate."
|
| 427 |
+
]
|
| 428 |
+
},
|
| 429 |
+
{
|
| 430 |
+
"cell_type": "code",
|
| 431 |
+
"execution_count": 11,
|
| 432 |
+
"metadata": {
|
| 433 |
+
"_cell_guid": "8357238b-98fe-632a-acd5-33674a6132ce"
|
| 434 |
+
},
|
| 435 |
+
"outputs": [
|
| 436 |
+
{
|
| 437 |
+
"name": "stdout",
|
| 438 |
+
"output_type": "stream",
|
| 439 |
+
"text": [
|
| 440 |
+
" Title Survived\n",
|
| 441 |
+
"0 Master 0.575000\n",
|
| 442 |
+
"1 Miss 0.702703\n",
|
| 443 |
+
"2 Mr 0.156673\n",
|
| 444 |
+
"3 Mrs 0.793651\n",
|
| 445 |
+
"4 Rare 0.347826\n"
|
| 446 |
+
]
|
| 447 |
+
}
|
| 448 |
+
],
|
| 449 |
+
"source": [
|
| 450 |
+
"for dataset in full_data:\n",
|
| 451 |
+
" dataset['Title'] = dataset['Title'].replace(['Lady', 'Countess','Capt', 'Col',\\\n",
|
| 452 |
+
" \t'Don', 'Dr', 'Major', 'Rev', 'Sir', 'Jonkheer', 'Dona'], 'Rare')\n",
|
| 453 |
+
"\n",
|
| 454 |
+
" dataset['Title'] = dataset['Title'].replace('Mlle', 'Miss')\n",
|
| 455 |
+
" dataset['Title'] = dataset['Title'].replace('Ms', 'Miss')\n",
|
| 456 |
+
" dataset['Title'] = dataset['Title'].replace('Mme', 'Mrs')\n",
|
| 457 |
+
"\n",
|
| 458 |
+
"print (train[['Title', 'Survived']].groupby(['Title'], as_index=False).mean())"
|
| 459 |
+
]
|
| 460 |
+
},
|
| 461 |
+
{
|
| 462 |
+
"cell_type": "markdown",
|
| 463 |
+
"metadata": {
|
| 464 |
+
"_cell_guid": "68fa2057-e27a-e252-0d1b-869c00a303ba"
|
| 465 |
+
},
|
| 466 |
+
"source": [
|
| 467 |
+
"# Data Cleaning #\n",
|
| 468 |
+
"great! now let's clean our data and map our features into numerical values."
|
| 469 |
+
]
|
| 470 |
+
},
|
| 471 |
+
{
|
| 472 |
+
"cell_type": "code",
|
| 473 |
+
"execution_count": 12,
|
| 474 |
+
"metadata": {
|
| 475 |
+
"_cell_guid": "2502bb70-ce6f-2497-7331-7d1f80521470"
|
| 476 |
+
},
|
| 477 |
+
"outputs": [
|
| 478 |
+
{
|
| 479 |
+
"name": "stdout",
|
| 480 |
+
"output_type": "stream",
|
| 481 |
+
"text": [
|
| 482 |
+
" Survived Pclass Sex Age Fare Embarked IsAlone Title\n",
|
| 483 |
+
"0 0 3 0 1 0 0 0 1\n",
|
| 484 |
+
"1 1 1 1 2 3 1 0 3\n",
|
| 485 |
+
"2 1 3 1 1 1 0 1 2\n",
|
| 486 |
+
"3 1 1 1 2 3 0 0 3\n",
|
| 487 |
+
"4 0 3 0 2 1 0 1 1\n",
|
| 488 |
+
"5 0 3 0 2 1 2 1 1\n",
|
| 489 |
+
"6 0 1 0 3 3 0 1 1\n",
|
| 490 |
+
"7 0 3 0 0 2 0 0 4\n",
|
| 491 |
+
"8 1 3 1 1 1 0 0 3\n",
|
| 492 |
+
"9 1 2 1 0 2 1 0 3\n"
|
| 493 |
+
]
|
| 494 |
+
}
|
| 495 |
+
],
|
| 496 |
+
"source": [
|
| 497 |
+
"for dataset in full_data:\n",
|
| 498 |
+
" # Mapping Sex\n",
|
| 499 |
+
" dataset['Sex'] = dataset['Sex'].map( {'female': 1, 'male': 0} ).astype(int)\n",
|
| 500 |
+
" \n",
|
| 501 |
+
" # Mapping titles\n",
|
| 502 |
+
" title_mapping = {\"Mr\": 1, \"Miss\": 2, \"Mrs\": 3, \"Master\": 4, \"Rare\": 5}\n",
|
| 503 |
+
" dataset['Title'] = dataset['Title'].map(title_mapping)\n",
|
| 504 |
+
" dataset['Title'] = dataset['Title'].fillna(0)\n",
|
| 505 |
+
" \n",
|
| 506 |
+
" # Mapping Embarked\n",
|
| 507 |
+
" dataset['Embarked'] = dataset['Embarked'].map( {'S': 0, 'C': 1, 'Q': 2} ).astype(int)\n",
|
| 508 |
+
" \n",
|
| 509 |
+
" # Mapping Fare\n",
|
| 510 |
+
" dataset.loc[ dataset['Fare'] <= 7.91, 'Fare'] \t\t\t\t\t\t = 0\n",
|
| 511 |
+
" dataset.loc[(dataset['Fare'] > 7.91) & (dataset['Fare'] <= 14.454), 'Fare'] = 1\n",
|
| 512 |
+
" dataset.loc[(dataset['Fare'] > 14.454) & (dataset['Fare'] <= 31), 'Fare'] = 2\n",
|
| 513 |
+
" dataset.loc[ dataset['Fare'] > 31, 'Fare'] \t\t\t\t\t\t\t = 3\n",
|
| 514 |
+
" dataset['Fare'] = dataset['Fare'].astype(int)\n",
|
| 515 |
+
" \n",
|
| 516 |
+
" # Mapping Age\n",
|
| 517 |
+
" dataset.loc[ dataset['Age'] <= 16, 'Age'] \t\t\t\t\t = 0\n",
|
| 518 |
+
" dataset.loc[(dataset['Age'] > 16) & (dataset['Age'] <= 32), 'Age'] = 1\n",
|
| 519 |
+
" dataset.loc[(dataset['Age'] > 32) & (dataset['Age'] <= 48), 'Age'] = 2\n",
|
| 520 |
+
" dataset.loc[(dataset['Age'] > 48) & (dataset['Age'] <= 64), 'Age'] = 3\n",
|
| 521 |
+
" dataset.loc[ dataset['Age'] > 64, 'Age'] = 4\n",
|
| 522 |
+
"\n",
|
| 523 |
+
"# Feature Selection\n",
|
| 524 |
+
"drop_elements = ['PassengerId', 'Name', 'Ticket', 'Cabin', 'SibSp',\\\n",
|
| 525 |
+
" 'Parch', 'FamilySize']\n",
|
| 526 |
+
"train = train.drop(drop_elements, axis = 1)\n",
|
| 527 |
+
"train = train.drop(['CategoricalAge', 'CategoricalFare'], axis = 1)\n",
|
| 528 |
+
"\n",
|
| 529 |
+
"test = test.drop(drop_elements, axis = 1)\n",
|
| 530 |
+
"\n",
|
| 531 |
+
"print (train.head(10))\n",
|
| 532 |
+
"train_df = train\n",
|
| 533 |
+
"train = train.values\n",
|
| 534 |
+
"test = test.values"
|
| 535 |
+
]
|
| 536 |
+
},
|
| 537 |
+
{
|
| 538 |
+
"cell_type": "markdown",
|
| 539 |
+
"metadata": {
|
| 540 |
+
"_cell_guid": "8aaaf2bc-e282-79cc-008a-e2e801b51b07"
|
| 541 |
+
},
|
| 542 |
+
"source": [
|
| 543 |
+
"good! now we have a clean dataset and ready to predict. let's find which classifier works better on this dataset. "
|
| 544 |
+
]
|
| 545 |
+
},
|
| 546 |
+
{
|
| 547 |
+
"cell_type": "markdown",
|
| 548 |
+
"metadata": {
|
| 549 |
+
"_cell_guid": "23b55b45-572b-7276-32e7-8f7a0dcfd25e"
|
| 550 |
+
},
|
| 551 |
+
"source": [
|
| 552 |
+
"# Classifier Comparison #"
|
| 553 |
+
]
|
| 554 |
+
},
|
| 555 |
+
{
|
| 556 |
+
"cell_type": "code",
|
| 557 |
+
"execution_count": 13,
|
| 558 |
+
"metadata": {
|
| 559 |
+
"_cell_guid": "31ded30a-8de4-6507-e7f7-5805a0f1eaf1"
|
| 560 |
+
},
|
| 561 |
+
"outputs": [
|
| 562 |
+
{
|
| 563 |
+
"data": {
|
| 564 |
+
"text/plain": [
|
| 565 |
+
"<AxesSubplot:title={'center':'Classifier Accuracy'}, xlabel='Accuracy', ylabel='Classifier'>"
|
| 566 |
+
]
|
| 567 |
+
},
|
| 568 |
+
"execution_count": 13,
|
| 569 |
+
"metadata": {},
|
| 570 |
+
"output_type": "execute_result"
|
| 571 |
+
},
|
| 572 |
+
{
|
| 573 |
+
"data": {
|
| 574 |
+
"image/png": "iVBORw0KGgoAAAANSUhEUgAAAgwAAAEWCAYAAAAKI89vAAAAOXRFWHRTb2Z0d2FyZQBNYXRwbG90bGliIHZlcnNpb24zLjMuMiwgaHR0cHM6Ly9tYXRwbG90bGliLm9yZy8vihELAAAACXBIWXMAAAsTAAALEwEAmpwYAAA98klEQVR4nO3dd7xcVb3//9ebIiEkBAnIJYgEEEEIECCgKEhVFBFQ6QgGQYw/FQG5XxtGxAZy0Su9augiVdqlCITekpBOUwJSFJASSqjh/ftjryOTYc6ZOclpCe/n43EeZ2bttdf67H0C+7PXWjNbtomIiIjoyEK9HUBERET0fUkYIiIioqkkDBEREdFUEoaIiIhoKglDRERENJWEISIiIppKwhAR8x1Jh0k6uxvbnyZp8/Jakv4o6XlJd0vaVNID3dV3RF+VhCEi+iRJe0gaJ+llSf+U9H+SNumJvm2vZXtsebsJ8Gngg7Y3sn2L7dW7us+SBFnSx7q67YiukIQhIvocSQcD/wv8ClgO+BBwArBDL4SzEvCI7VfmtSFJi7RTLmBv4Lnyu8e0F1NEvSQMEdGnSBoEHA58y/bFtl+x/abty23/dzv7XCDpX5JmSrpZ0lo127aVNF3SS5KekHRIKV9G0hWSXpD0nKRbJC1Utj0iaWtJ+wKnARuXkY6fSdpc0uM17Q+RdJGkZyTNkHRAzbbDJF0o6WxJLwIj2znsTYHlgQOA3SS9r6aNxSUdLenRcny3Slq8bNtE0u3lGB6TNLKUj5W0X00bIyXdWvPekr4l6SHgoVL2+9LGi5LGS9q0pv7Ckn4k6e/lPI6XtKKk4yUdXfe3uEzSQe0cZ8zHkjBERF+zMdAPuKQT+/wfsBrwAWACcE7NttOBb9geCAwDbijl3wMeB5alGsX4ETDHd+XbPh0YBdxhe4Dtn9ZuLwnG5cAkYAVgK+BASdvUVNsBuBBYqi6uWl8t7fy5vP9Czbb/ATYAPgEsDfw/4G1JK5XjPrYcw3BgYjvtN7Ij8DFgzfL+ntLG0sC5wAWS+pVtBwO7A9sCSwJfA2YBZwC71yRaywBbl/1jAZOEISL6msHAv22/1eoOtv9g+yXbrwOHAeuWkQqAN4E1JS1p+3nbE2rKlwdWKiMYt7jzD9fZEFjW9uG237D9MHAqsFtNnTtsX2r7bduv1jcgqT+wM3Cu7Tepkou9y7aFqC7O37X9hO3Ztm8vx7kH8Ffb55X4n7U9sROx/9r2c20x2T67tPGW7aOBxYC2tRr7AYfafsCVSaXu3cBMqkSJctxjbT/ViThiPpGEISL6mmeBZVqdWy/D5UeU4fIXgUfKpmXK7y9T3Rk/KukmSRuX8qOAvwHXSnpY0g/mItaVgCFlSuAFSS9QjVQsV1PnsSZtfBF4C7iqvD8H+JykZcsx9AP+3mC/Fdspb9UccUk6RNJ9ZdrjBWAQ75zDjvo6A/hKef0V4Kx5iCn6sCQMEdHX3AG8TjVk3oo9qIb9t6a6yA0t5QKwfY/tHaimKy6lDPuXEYnv2V4F2B44WNJWdM5jwAzbS9X8DLS9bU2dZqMWXwUGAP+Q9C/gAmDRclz/Bl4DVm2n70blAK8A/Wve/1eDOv+Jq6xX+H/ALsD7bS9FNXKgFvo6G9hB0rrAR6nOcSyAkjBERJ9ieyYwGjhe0o6S+ktaVNLnJP2mwS4DqRKMZ6kukr9q2yDpfZL2lDSoDPe/CLxdtm0n6cPlEwozgdlt2zrhbuAlSd8vixMXljRM0oat7Cypbd3DdlTrB4YD6wJHAnvbfhv4A/DbsrhyYUkbS1qMaiRia0m7SFpE0mBJw0vTE4EvlXP3YWDfJqEMpBrleAZYRNJoqrUKbU4Dfi5pNVXWkTQYwPbjVOsfzgIuajTtEguGJAwR0eeUOfSDgUOpLmKPAd+m8d3rmcCjwBPAdODOuu17AY+U6YpRwJ6lfDXgr8DLVKMaJ9i+sZNxzuadi/0MqhGB06hGOlqxFzDR9rW2/9X2AxwDrCNpGHAIMIXqovwcVTKxkO1/UE21fK+UT6RKNgB+B7wBPEU1ZdDeYss21wBXAw9SncvXmHPK4rdUIzPXUiVdpwOL12w/A1ibTEcs0NT5NT4RERHvkPQpqqmJleZi4WjMJzLCEBERc03SosB3gdOSLCzYkjBERMRckfRR4AWqj6f+b68GE90uUxIRERHRVEYYIiIioqk8dCQWSMsss4yHDh3a22FERMxXxo8f/2/byzbaloQhFkhDhw5l3LhxvR1GRMR8RdKj7W3LlEREREQ0lRGGWCDNePIl9hg9trfDiIjotHMP37y3Q2goIwwRERHRVBKGiIiIaCoJQ0RERDSVhCEiIiKaSsLQAkkv17zeVtKDklaSdJikWZI+0KhuB+1dJWmpJnXGShrRoHykpOM6eQgtkXSIpPslTZR0j6S9O4plLvsYIemY8noxSX8t/e0q6TRJa3ZFPxER0bXyKYlOkLQV1WNnt7H9qCSoHmf7PeD7rbZje9vuibBjqgKW7bcbbBsFfBrYyPaLkpYEvtjVMdgeB7R9QcJ6pWx4eX9+Z9qStHB5vHBERHSzjDC0qDy+9VRgO9t/r9n0B2BXSUs32Ocrku4ud9AnS1q4lD8iaZny+ieSHpB0q6TzJB1S08TOZf8HJW1aU75iuet/SNJPa/o7WNLU8nNgKRta2j8TmFr2HVPqTJF0UNn9R8A3bb8IYPtF22c0OKYTJY2TNE3Sz2rKj5A0XdJkSf9TynYu/UySdHMp21zSFWVU5mxgw3J+Vq0dyZD0GUl3SJog6QJJA2rO3ZGSJgA7N/u7RURE18gIQ2sWAy4FNrd9f922l6mShu8CtRfvjwK7Ap+0/aakE4A9gTNr6mwIfBlYF1gUmACMr2l7EdsbSdq2tL11Kd8IGAbMAu6RdCVgYB/gY4CAuyTdBDwPrAZ81fadkjYAVrA9rMSwVBlNGGj74RbOxY9tP1eSn+slrQM8QTUasYZt10y3jKYajXmifgrG9tOS9gMOsb1diaXtvCwDHApsbfsVSd8HDgYOL7s/a3v9+sAk7Q/sD9B/0HItHEpERLQqIwyteRO4Hdi3ne3HAF+VNLCmbCtgA6oL+sTyfpW6/T4J/MX2a7ZfAi6v235x+T0eGFpTfp3tZ22/WupsUn4usf2K7ZdLeduoxKO27yyvHwZWkXSspM8CL3Z86O+yS7m7vxdYC1gTmAm8Bpwu6UtUiQzAbcAYSV8HFu5EHx8v7d5Wzt1XgZVqtjecurB9iu0Rtkf06z+oE91FREQzSRha8zawC7CRpB/Vb7T9AnAu8K2aYgFn2B5efla3fVgn+329/J7NnKNB9c8kb/aM8ldqYn2eakRjLDAKOK1MQ7wsqT6hmYOklYFDgK1srwNcCfSz/RbVqMeFwHbA1aWvUVQjBSsC4yUNbhLnf7qiSorazt2atmuTtVfa2zEiIrpHEoYW2Z4FfB7YU1KjkYbfAt/gnQv79cBObZ+gkLS0pJXq9rkN+IKkfmWOfrsWw/l0aW9xYMfSzi3AjpL6S1qCaorglvody3D/QrYvorqYtw3t/xo4vkxPIGlA26ckaixJdbGeKWk54HNtdYFBtq8CDqJKSJC0qu27bI8GnqFKHFpxJ/BJSR8u7Swh6SMt7hsREd0gaxg6oczdfxa4WdIzddv+LekSqgsmtqdLOhS4VtJCVNMa3wIerdnnHkmXAZOBp4ApVMP7zdwNXAR8EDi7fPIASWPKNqhGDu6VNLRu3xWAP5aYAH5Yfp8IDKCaQnmzxHt03TFOknQvcD/wGFWiAjAQ+IukflSjAweX8qMkrVbKrgcmAZs1Ozjbz0gaCZwnabFSfCjwYLN9IyKie8huNpod3UnSANsvS+oP3Azsb3tCb8c1vxs8ZHVvs9/JvR1GRESn9ebDpySNt93we3cywtD7TlH1ZUX9qNY8JFmIiIg+JwlDL7O9R2/HEBER0UwWPUZERERTGWGIBdLKQwb26jxgRMSCJiMMERER0VQShoiIiGgqCUNEREQ0lTUMsUCa8eRL7DF6bG+HERHRo7pz7VZGGCIiIqKpJAwRERHRVBKGiIiIaCoJQ0RERDSVhCEiIiKaSsIQPU7SjyVNkzRZ0kRJP5X067o6wyXdV14PkHSypL9LGi9prKSP9U70ERHvTflYZfQoSRsD2wHr235d0jLAmsAY4Ic1VXcDziuvTwNmAKvZflvSymWfiIjoIUkYoqctD/zb9usAtv8N3CzpeUkfs31XqbcLsI2kVYGPAXvafrvsM4MqgYiIiB6SKYnoadcCK0p6UNIJkjYr5edRjSog6ePAc7YfAtYCJtqe3axhSftLGidp3GuzZnZX/BER70lJGKJH2X4Z2ADYH3gGOF/SSOB8YCdJCzHndERn2j7F9gjbI/r1H9SFUUdERKYkoseV0YKxwFhJU4Cv2h4jaQawGfBlYONSfRqwrqSFWxlliIiI7pERhuhRklaXtFpN0XDg0fL6POB3wMO2Hwew/XdgHPAzSSptDJX0+Z6LOiIikjBETxsAnCFpuqTJVJ92OKxsu4BqzUL9dMR+wHLA3yRNpfpExdM9Em1ERACZkogeZns88Il2tv0bWLRB+YvA17s5tIiI6EBGGCIiIqKpJAwRERHRVBKGiIiIaCprGGKBtPKQgZx7+Oa9HUZExAIjIwwRERHRVBKGiIiIaCoJQ0RERDSVNQyxQJrx5EvsMXpsb4cREdEtemONVkYYIiIioqkkDBEREdFUEoaIiIhoKglDRERENJWEISIiIppKwlBD0mxJEyVNkzRJ0vckzdU5knS4pK072D5K0t5z0e42JcaJkl6W9EB5febcxFnX9iGS7i/t3dMWn6SxkkbMa/ulrRGSjimvF5P019LfrpJOk7RmV/QTERFdKx+rnNOrtocDSPoAcC6wJPDTzjZke3ST7SfNTYC2rwGuKTGOBQ6xPa62jqSFbc/uTLuSRgGfBjay/aKkJYEvzk2MHSmxtsW7XikbXt6f35m25uY4IyJi7mSEoR22nwb2B76tysKSjip33pMlfaOtrqTvS5pSRiWOKGVjJO1UXh8haXrZ739K2WGSDimvh0u6s2y/RNL7S/lYSUdKulvSg5I2bS9eSY+UuhOAnSV9RtIdkiZIukDSgFJvA0k3SRov6RpJy5cmfgR80/aL5fhftH1Gg35OlDSujML8rKa80THuLGlqOS83l7LNJV1RErKzgQ3LCMOqtSMZHcQ/x3F29u8aERFzJyMMHbD9sKSFgQ8AOwAzbW8oaTHgNknXAmuUbR+zPUvS0rVtSBpMdae+hm1LWqpBV2cC37F9k6TDqUY0DizbFrG9kaRtS3m70xzAs7bXl7QMcDGwte1XJH0fOFjSr4FjgR1sPyNpV+CXkg4EBtp+uIXT8mPbz5Xzcr2kdYAn2jnG0cA2tp+oP27bT0vaj2qEZLtyrtrO2TLAofXxA4fXHmd9YJL2p0ry6D9ouRYOJSIiWpWEoXWfAdZpGzUABgGrUV3A/2h7FoDt5+r2mwm8Bpwu6QrgitqNkgYBS9m+qRSdAVxQU+Xi8ns8MLRJjG1D+h8H1qRKagDeB9wBrA4MA64r5QsD/2zSZr1dyoV5EWD50s90Gh/jbcAYSX+uOY5WtBd/m4ZTF7ZPAU4BGDxkdXeiv4iIaCIJQwckrQLMBp4GRDUKcE1dnW06asP2W5I2ArYCdgK+DWzZiTBeL79n0/zv9UpbWMB1tnevi3VtYJrtjet3VLWAcpWORhkkrQwcAmxo+3lJY4B+7R2j7VGSPgZ8HhgvaYNmB9tR/A2OMyIiekjWMLRD0rLAScBxtk210PCbkhYt2z8iaQngOmAfSf1Lef2UxABgkO2rgIOAdWu3254JPF+zPmEv4CbmzZ3AJyV9uMSwhKSPAA8Ay0rauJQvKmmtss+vgeNVLXZE0gC9+1McS1JdrGdKWg74XEfHKGlV23eVBaDPACvOY/wREdFLMsIwp8UlTQQWBd4CzgJ+W7adRjUlMEHVOPkzwI62r5Y0HBgn6Q3gKqoFhG0GAn+R1I/qzvngBv1+FTipJB0PA/vMy0GU9QkjgfPKeguAQ20/WKZUjilTIYsA/wtMA04EBgD3SHoTeBM4uq7dSZLuBe4HHqOacujoGI+StFopux6YBGw2t/EDD3bqRERERJdRdfMcsWAZPGR1b7Pfyb0dRkREt+iup1VKGm+74ffuZEoiIiIimkrCEBEREU1lDUMskFYeMrDbhuwiIt6LMsIQERERTSVhiIiIiKaSMERERERTSRgiIiKiqSx6jAXSjCdfYo/RY3s7jIiIbtWTi7szwhARERFNJWGIiIiIppIwRERERFNJGCIiIqKp91TCIGm2pImSpkq6XNJSXdTuSEnHdVFbj0iaUuKcKOkTXdFug36GS9q2ruxzksZJmi7pXklHl/LDJB3ShX3fXvP6KEnTyu9RDR6pHRERfcB77VMSr9oeDiDpDOBbwC97NaLGtrD9787sIGkR2291YpfhwAiqx3EjaRhwHPB52/dLWhjYvzMxtMp2bRK0P7C07dmdbWcujjkiIubSe2qEoc4dwAoAkjaSdEe5q75d0uqlfKSkiyVdLekhSb9p21nSPpIelHQ38Mma8qGSbpA0WdL1kj5UysdIOlHSnZIelrS5pD9Iuk/SmI4CbdLmSZLuAn4jadUS63hJt0hao9TbuYyqTJJ0s6T3AYcDu5ZRjF2B/wf80vb9ALZn2z6xQSxfl3RPaesiSf0b9VHK1pJ0d+ljsqTVSvnL5fdlwABgvKRda0cyOjiWOY65E3/viIiYB+/JhKHcPW8FXFaK7gc2tb0eMBr4VU314cCuwNpUF9gVJS0P/IwqUdgEWLOm/rHAGbbXAc4BjqnZ9n5gY+Cg0vfvgLWAtSUNr6l3Y7nI3tVCmx8EPmH7YOAU4Du2NwAOAU4odUYD29heF9je9hul7Hzbw22fDwwDxjc9eXCx7Q1LW/cB+zbqo5SNAn5fRnVGAI/XNmR7e8qoT4mhVnvHUn/M/yFp/zKlMu61WTNbOJSIiGjVe21KYnFJE6lGFu4Drivlg4Azyh2wgUVr9rne9kwASdOBlYBlgLG2nynl5wMfKfU3Br5UXp/FnHfBl9u2pCnAU7anlP2nAUOBiaVe/ZRER21eYHu2pAHAJ4ALJLVtW6z8vg0YI+nPwMUdnaAWDJP0C2ApqtGBazro4w7gx5I+SJVoPNRKB02OBcox1+9n+xSqRIPBQ1Z3Zw4qIiI69l4bYWhbw7ASIKo1DAA/B260PQz4AtCvZp/Xa17PZt6SrLa23q5r9+15aPeV8nsh4IVyt97281EA26OAQ4EVqYb/BzdoZxqwQQv9jQG+bXttqlGWfu31YftcqtGGV4GrJG3Z4jG1eyx1xxwRET3kvZYwAGB7FnAA8D1Ji1CNMDxRNo9soYm7gM0kDZa0KLBzzbbbgd3K6z2BW7og5KZt2n4RmCFpZwBV1i2vV7V9l+3RwDNUF/WXgIE1TRwF/EjSR8o+C0ka1SCWgcA/y3Hv2VbYqA9JqwAP2z4G+AuwTisH29GxRERE73hPJgwAtu8FJgO7Uw3x/1rSvbRwp2/7n8BhVEPut1FNb7T5DrCPpMnAXsB3uyDcVtvcE9hX0iSqEYMdSvlRqj6qOZUq+ZgE3Ais2bbo0fZk4EDgPEn3AVOBVRr08ROqhOk2qrUfbRr1sQswtUwDDQPO7MQxt3csERHRC2RnqjcWPIOHrO5t9ju5t8OIiOhWXf3wKUnjbY9otO09O8IQERERrUvCEBEREU0lYYiIiIim3mvfwxDvESsPGdjlc3sREe9lGWGIiIiIppIwRERERFNJGCIiIqKppgmDpIUl3d+sXkRERCy4WvlWw9mSHpD0Idv/6ImgIubVjCdfYo/RY3s7jIiILtWbi7lb/ZTE+4Fpku6m5sE/5fHEERERsYBrNWH4SbdGEREREX1aSwmD7ZskrQSsZvuvkvoDC3dvaBEREdFXtPQpCUlfBy4E2p7mswJwaTfFFBEREX1Mqx+r/BbwSeBFANsPAR/orqDmZ5J2lGRJa7Szfaykhk8Cq6vzQHn09H2S9u/iGEdKGlLzflFJR0h6SNIESXdI+lzZ9oikZbqo3+0l/aC8XlbSXZLulbSppKskLdUV/URERNdrNWF43fYbbW8kLQLkudiN7Q7cWn7Piz1tD6dK1I6U9L55DazGSGBIzfufA8sDw2yvD+wIDOzC/gCwfZntI8rbrYApttezfYvtbW2/0GpbkjIlFhHRg1pNGG6S9CNgcUmfBi4ALu++sOZPkgYAmwD7AruVssUl/amMFFwCLF5T/0RJ4yRNk/SzdpodQPXJlNlln90lTZE0VdKRNW29q7x8h8aYUjZF0kGSdgJGAOeUEYwlgK8D37H9OoDtp2z/ucHxXSppfIl3//b6KOUHSJouabKkP5WykZKOkzQc+A2wQ4lh8dqRDElfkXR32XZyW3Ig6WVJR0uaBGw8F3+iiIiYS61+SuIHVBfBKcA3gKuA07orqPnYDsDVth+U9KykDYDNgFm2PyppHWBCTf0f236uXBCvl7SO7cll2zmSXgdWAw4s34cxBDgS2AB4HrhW0o7A3e2UPwasYHsYgKSlbL8g6dvAIbbHlZj+YfvFFo7vayXexYF7JF0EDK3vo9T9AbCy7dfrpxpsT5Q0Ghhh+9tlP8rvjwK7Ap+0/aakE4A9gTOBJYC7bH+vUXAlidkfoP+g5Vo4nIiIaFWrn5J4Gzi1/ET7dgd+X17/qbz/MHAMgO3JkibX1N+lXOQWoZoSWBNo275nuaAvC9wu6WpgODDW9jMAks4BPkU1PdSo/OfAKpKOBa4Erp3H4ztA0hfL6xWpkpkH2uljMlXScymdWyC7FVXic09JIhYHni7bZgMXtbej7VOAUwAGD1k9U2YREV2ow4RB0p9t7yJpCg3WLNhep9sim89IWhrYElhbkqk+dmrg3nbqrwwcAmxo+3lJY4B+9fVsPyNpAvAx4PXOxFTaXRfYBhgF7AJ8ra7a34APSVqyo1EGSZsDWwMb254laSzQr4M+Pk+VtHwB+LGktVsMW8AZtn/YYNtrtme32E5ERHShZmsYDiy/t6P6H3/9T7xjJ+As2yvZHmp7RWAGMB7YA0DSMKAtyVqSam3CTEnLAZ9r1Kiq77xYD/g71dTDZpKWKdMYuwM3tVde1gQsZPsi4FBg/dLsS5RFjbZnAacDv29bWFk+wbBzXSiDgOdLsrAG8PFS9119SFoIWNH2jcD3y74DWjyP1wM7SfpAaX9pVd8BEhERvajZlMQVVBeZX9jeqwfimZ/tTrWOoNZFVBf7xSXdB9xHlUBge5Kke4H7qdYa3Fa37zmSXgUWA8bYHg+g6mOJN1LdiV9p+y/tlZc7/z+WCzhA2137GOCk0v7GVBf6XwDTJb1GlciMrovnamBUOY4HgDtL+QoN+lgYOFvSoBLPMWXtRLNziO3pkg6lWoexEPAm1cd6H226c0REdBvZ7U/1SpoK/IpqLvy/67fbvrj7QouYe4OHrO5t9ju5ecWIiPlIdz98StJ42w2/K6jZCMMoqhXqS/HuKQgDSRgiIiLeAzpMGGzfCtwqaZzt03sopoiIiOhjmn1KYkvbNwDPS/pS/fZMSURERLw3NJuS2Ay4gcafiMiURPRZKw8Z2O1zfRER7yXNpiR+Wn7v0zPhRERERF/U6uOtvytpSVVOU/VEw890d3ARERHRN7T68KmvlW8B/AwwGNgLOKLjXSIiImJB0erDp9q+cWdb4Ezb09TKt/BE9JIZT77EHqPH9nYYERHdqifXarU6wjBe0rVUCcM1kgYCb3dfWBEREdGXtDrCsC/VkxIfLs8SWBrIQsiIiIj3iFZHGDYGHijPA/gK1bMHZnZfWBEREdGXtJownAjMKg8z+h7VkxPP7LaoIiIiok9pNWF4y9VTqnYAjrN9POXxyBEREbHgazVheEnSD4GvAFeWxw4v2n1htU/ScpLOlfSwpPGS7pD0xXlo7zBJh5TXh0vaei7bGS5p25r3IyU9I2mipGmSLpTUf27jbKG/7csjrue2vUUlHSHpofI9G3dI+lzZ9oikZboo7v/EKWlZSXdJulfSppKukrRUV/QTERFdq9WEYVfgdWBf2/8CPggc1W1RtaN8lPNS4Gbbq9jeANitxFNbr9XFnHOwPdr2X+cyvOFUnyKpdb7t4bbXAt6gOo9dZY7+bF9me16+G+PnwPLAMNvrAzvSDaNIdXFuBUyxvZ7tW2xva/uFVtuStHBXxxcREY21lDDY/pft39q+pbz/h+3eWMOwJfCG7ZNqYnvU9rHljv4ySTcA10saIOn6crc8RdIObftI+rGkByXdCqxeUz5G0k7l9QaSbiqjGNdIWr6Uj5V0pKS7SxubSnofcDiwaxlRmCMxKAnMEsDz5f1QSTdImlxi/FCT8p0lTZU0SdLNjforx39czXEcI+n2MhLTdkwLSTpB0v2Srit39DuVkY+vA9+x/Xo5r0/Z/nP9H0DSpeWcTJO0fylbuPQ5tZzrg0r5AZKml+P5UykbKek4ScOB3wA7lGNYvHYkQ9JXyjmeKOnktuRA0suSjpY0iWoxbkRE9IBWvxr645LuKf+zfkPSbEm98SmJtYAJHWxfH9jJ9mbAa8AXy93yFsDRqrSNSgynukPfsL4RSYsCx5a2NgD+APyypsoitjcCDgR+avsNYDTvjCicX+rtKmki8ASwNHB5KT8WOMP2OsA5wDFNykcD29heF9i+g/5qLQ9sAmzHO9/K+SVgKLAm1bd1tl1wPwz8o3ybZzNfK+dkBHCApMFU53IF28Nsrw38sdT9AbBeOZ5RtY3Ynlh3DK+2bZP0UarRmE/aHg7MBvYsm5cA7rK9bnn8OjX77S9pnKRxr83Kh3giIrpSq1MSxwG7Aw8BiwP7ASd0V1CtknR8ueu+pxRdZ/u5ts3AryRNBv4KrAAsB2wKXGJ7VrlAXtag6dWBYcB15YJ/KHNOe7Q9pXM81QW4PeeXC95/AVOA/y7lGwPnltdnUV3YOyq/DRgj6etAq8Pwl9p+2/Z0quOmtHdBKf8XcGOLbdU6oNzd3wmsCKwGPAysIulYSZ8F2hKPycA5qj6K+1Yn+tgK2AC4p5z/rYBVyrbZwEWNdrJ9iu0Rtkf06z+ok4cVEREdaTVhwPbfgIVtz7b9R+Cz3RdWu6ZRjSK0xfQtqovJsqXolZq6e5byDcpF+ymgX4v9CJhW7nyH217bdu3Dtl4vv2fTwpdflU+YXA58qsX+6/cfRZW0rEj1rZuDW9jt9ZrXzb7G+2/AhyQt2VElSZsDWwMbl9GOe4F+tp8H1gXGUo0knFZ2+TxwPNXf7J5OrC0R1UhL2/lf3fZhZdtrtme32E5ERHSRVhOGWWXefKKk35Q56paTjS50A9BP0jdrytr75MEg4Gnbb0raAliplN8M7FjmzAcCX2iw7wPAspI2hv98gmCtJrG9RMeLBDeh+v4KgNuppkWgSmxu6ahc0qq277I9GniGKnFo1l8jtwFfLmsZlgM2B7A9Czgd+H35O7d9gmHnuv0HAc+Xb/tcA/h4qbsMsJDti6gSm/VVfZJmRds3At8v+w5oMc7rgZ0kfaC0v7SklZrsExER3ajVi/5eVEPh36a6i18R+HJ3BdWecqe+I7CZpBmS7gbOoLog1TsHGCFpCrA3cH9pYwJwPjAJ+D/gnvodyxqBnYAjy/D7ROATTcK7EVizbtFj26LEycB6VJ9EAPgOsE8p3wv4bpPyo8piwqlUScWkdvpr5iLgcWA6cDbVepC2yf5DqZKR6aWfK3hnaqHN1cAiku6jWhdxZylfARhbpg/OBn5I9e/l7HL+7wWOafUTEGUa5VDg2nIurqNakxEREb1E1TU43iskDbD9cpnWuJtqYeG/ejuurjZ4yOreZr+TezuMiIhu1dVPq5Q03vaIRts6nFMud4ftZhRl9XvMX65Q9eVI7wN+viAmCxER0fWaLUL7EtUK+8fqylcEcqGZD9nevLdjiIiI+U+zNQy/A2aWL0f6zw/VvPfvuj+8iIiI6AuajTAsZ3tKfaHtKZKGdk9IEfNu5SEDu3xuLyLivazZCMNSHWxbvAvjiIiIiD6sWcIwrny74Bwk7Uf1LYcRERHxHtBsSuJA4BJJe/JOgjCCaoX9XD9SOiIiIuYvHSYMtp8CPlG+KXFYKb7S9g3dHlnEPJjx5EvsMXpsb4cREdGS+WHNVUvf7V++3nduHlQUERERC4DeeB5EREREzGeSMERERERTSRgiIiKiqSQMERER0VQShgWMpOUknSvpYUnjJd0hqVs/AitphKRj5mH/RyRdVPN+J0ljyuuRkp4pj/GeJulCSf27IOyIiOiEJAwLEEkCLgVutr2K7Q2A3YAPdme/tsfZPmAem9lA0prtbDvf9nDbawFvALvOY18REdFJSRgWLFsCb9g+qa2gPDDsWElDJd0iaUL5+QSApM0lXdFWX9JxkkaW10dImi5psqT/KWU7S5oqaZKkm+vbkLRRGdW4V9LtklYv5SMlXSzpakkPSfpNXexHAz/u6OAkLQIsATw/b6cpIiI6q6XvYYj5xlrAhHa2PQ182vZrklYDzqP61s6GJA2m+jbPNWxb0lJl02hgG9tP1JTVuh/Y1PZbkrYGfgV8uWwbDqwHvA48IOlY222PTv8z8P9J+nCDNneVtAmwPPAgcHk7Me8P7A/Qf9By7R1aRETMhYwwLMAkHV9GAu4BFgVOlTQFuABob/i/zUzgNeB0SV8CZpXy24Ax5RkjCzfYbxBwgaSpVI9AX6tm2/W2Z9p+DZgOrFSzbTZwFPDDBm2eb3s48F/AFOC/GwVs+xTbI2yP6Nd/UJPDi4iIzkjCsGCZBqzf9sb2t4CtgGWBg4CngHV553kgAG8x57+DfmXft4CNgAuB7YCrS/ko4FBgRWB8GYmo9XPgRtvDgC+0tVe8XvN6Nu8e4ToL+FRp+11sm2p04VONtkdERPdJwrBguQHoJ+mbNWVtnygYBPzT9tvAXrwzOvAosKakxcoUw1YAkgYAg2xfRZVsrFvKV7V9l+3RwDO8++I+CHiivB7ZmeBtv0k1KnFQB9U2Af7emXYjImLeJWFYgJQ78B2BzSTNkHQ3cAbwfeAE4KuSJgFrAK+UfR6jWj8wtfy+tzQ3ELhC0mTgVuDgUn6UpCllyuF2YFJdGL8Bfi3pXuZujczpDfbbtXyscjLVGoifz0W7ERExD1RdYyIWLIOHrO5t9ju5t8OIiGhJX3lapaTxthsuiM8IQ0RERDSVhCEiIiKayvcwxAJp5SED+8wQX0TEgiAjDBEREdFUEoaIiIhoKglDRERENJWEISIiIprKosdYIM148iX2GD22t8OIiJgnfWnxdkYYIiIioqkkDBEREdFUEoaIiIhoKglDRERENNXnEwZJLzcoGyVp7x7o+5HyZMYpkqZL+oWkfmXbEEkXdkEf20v6QSf3uao8irrLSBoqaY8G5f8r6QlJ8/RvpZzLZeZivy4/1oiI6Lw+nzA0Yvsk22d2V/uqtJ2bLWyvDWwErAKcXGJ40vZO89jPIrYvs31EZ/azva3tF+al7waGAnMkDOUcfBF4DNisi/trSTcda0REdNJ8mTBIOkzSIeX1WElHSrpb0oOSNi3lC0s6StI9kiZL+kYpHyDpekkTysjBDqV8qKQHJJ0JTAVWrO3T9svAKGBHSUuX+lPLvmuV/ieWvlYr5XuX95MknVXKxkg6SdJdwG8kjZR0XM22EyXdKelhSZtL+oOk+ySNqTn+RyQtU2K4T9KpkqZJulbS4qXO18uxT5J0kaT+NX0cI+n20kdb0nMEsGk5hoNK2ebANOBEYPe68/+Hcu4flnRAzbZLJY0v8ezf4G93uKQDa97/UtJ3JS0v6ebS/9Sav2PbsS4h6cpyPFMl7drav5aIiOgK82XC0MAitjcCDgR+Wsr2BWba3hDYEPi6pJWB14Av2l4f2AI4WpLKPqsBJ9hey/aj9Z3YfhGYUerVGgX83vZwYATwuKS1gEOBLW2vC3y3pv4HgU/YPrjBsbwf2Bg4CLgM+B2wFrC2pOEN6q8GHG97LeAF4Mul/GLbG5a+7yvno83ywCbAdlSJAsAPgFtsD7f9u1K2O3AecAnweUmL1rSxBrAN1cjLT2u2fc32BuU8HCBpcF28fwD2hv+MYOwGnE01unFNOYfrAhPr9vss8KTtdW0PA66uPxGS9pc0TtK412bNbHCqIiJibi0oCcPF5fd4qqF1gM8Ae0uaCNwFDKa6uAr4laTJwF+BFYDlyj6P2r6zSV9qUHYH8CNJ3wdWsv0qsCVwge1/A9h+rqb+BbZnt9P+5bYNTAGesj3F9ttUd/pDG9SfYXtieV17/MMk3SJpCrAnVdLR5lLbb9uezjvHPudBSu8Dti11X6Q6h9vUVLnS9uvl+J6uaecASZOAO6lGaeZIrmw/AjwraT2qv9G9tp8F7gH2kXQYsLbtl+pCmgJ8uowmbWr7XRmB7VNsj7A9ol//QY0OKyIi5tKCkjC8Xn7P5p1vrxTwnXLHPNz2yravpbp4LgtsUO5mnwL6lX1e6agTSQOpLsgP1pbbPhfYHngVuErSlk3i7aiftmN5u+Z12/tG38xZW6f2+McA3y7rL37GO8dYv0+jBAiq5GApYIqkR6hGJHav2f6ufiVtDmwNbFxGNu6t67fNacBIYB+qEQds3wx8CngCGKO6Ra22HwTWp0ocfiFpdDtxR0REN1hQEoZGrgG+2TZULukjkpYABgFP235T0hbASq00JmkAcALVHffzddtWAR62fQzwF2Ad4AZg57YheUlLd9FxtWog8M9y/Hu2UP+lsk+b3YH9bA+1PRRYmeoOv38HbQwCnrc9S9IawMfbqXcJ1RTDhlR/JyStRDWicipVQrF+7Q6ShgCzbJ8NHFW/PSIiutf88CyJ/pIer3n/2xb3O41qNGBCWaPwDLAjcA5weRmqHwfc36SdG8v+C1Fd6H7eoM4uwF6S3gT+BfzK9nOSfgncJGk21d32yBZj7wo/oZpGeKb8HthxdSYDs8t0wp+pLuij2jbafkXSrcAXOmjjamCUpPuAB6imJd7F9huSbgReqJma2Rz473IOX6asc6ixNnCUpLeBN4FvNjmeiIjoQqqmyyN6TlnsOAHY2fZD3dHH4CGre5v9Tu6OpiMiekxPP3xK0njbIxptW5CnJKIPkrQm8Dfg+u5KFiIiouvND1MSsQApn8xYpbfjiIiIzskIQ0RERDSVEYZYIK08ZGCPz/1FRCzIMsIQERERTSVhiIiIiKaSMERERERTWcMQC6QZT77EHqPH9nYYERFdoi+sycoIQ0RERDSVhCEiIiKaSsIQERERTSVhiIiIiKaSMERERERT3ZowSPqgpL9IekjSw5KOk7RYF7S7uaQrOrnPUEl71LwfIemYJvs8ImlK+Zku6ReS+pVtQyRdOHdHMEcf20v6QSf3uUrSUvPad12bc5yfmvL/lfREecLkvLT/iKRl5mK/Lj/WiIjovG5LGCQJuBi41PZqwGrA4sBvurHPjj4mOhT4zwXR9jjbB7TQ7Ba21wY2onpo0sll/ydt7zQP4SJpEduX2T6iM/vZ3tb2C/PSdwNDqTk/8J/HUH8ReAzYrIv7a0k3HWtERHRSd44wbAm8ZvuPALZnAwcBe0v6tqTj2ipKukLS5uX1iZLGSZom6Wc1dT4r6X5JE4Av1ZQfJuksSbcBZ5U75VskTSg/nyhVjwA2lTRR0kG1oxSSBkj6YxlJmCzpy/UHY/tlYBSwo6SlSz9Ty/5rSbq7tD1Z0mqlfO/yfpKks0rZGEknSboL+I2kkW3nomw7UdKdZURmc0l/kHSfpDE1x/yIpGVKDPdJOrWcr2slLV7qfF3SPaXviyT1r+njGEm3lz7akp45zk8p2xyYBpwI7F53zv8gaWxp44CabZdKGl/i2b/+PEo6XNKBNe9/Kem7kpaXdHPpf6qkTeuOdQlJV5bjmSpp1/q2IyKi+3RnwrAWML62wPaLwCN0/IVRP7Y9AlgH2EzSOmUa4FTgC8AGwH/V7bMmsLXt3YGngU/bXh/YFWibdvgBcIvt4bZ/V7f/T4CZtte2vQ5wQ6PASvwzqEZLao0Cfm97ODACeFzSWsChwJa21wW+W1P/g8AnbB/coJv3AxtTJVeXAb+jOpdrSxreoP5qwPG21wJeANqSnYttb1j6vg/Yt2af5YFNgO2oEgVofH52B84DLgE+L2nRmjbWALahGnn5ac22r9neoJyHAyQNrov3D8De8J8RjN2As6lGN64p53BdYGLdfp8FnrS9ru1hwNX1J0LS/iXZHPfarJkNTlVERMytvrjocZcyinAv1YVyTaqL0wzbD9k21QWm1mW2Xy2vFwVOlTQFuKDs38zWwPFtb2w/30FdNSi7A/iRpO8DK5VYtgQusP3v0uZzNfUvKCMujVxejnEK8JTtKbbfprrTH9qg/gzbE8vr8TV1hpWRlinAnlTnss2ltt+2PR1YruFBSu8Dti11XwTuokoQ2lxp+/VyfE/XtHOApEnAncCK1CVXth8BnpW0HvAZ4F7bzwL3APtIOgxY2/ZLdSFNAT4t6UhJm9p+V0Zg+xTbI2yP6Nd/UKPDioiIudSdCcN0qtGA/5C0JNXowLN1fbctJFwZOATYqtzpX9m2rYlXal4fBDxFdZc6AnjfXMb/LpIGUl2QH6wtt30usD3wKnCVpC07EW+918vvt2tet71vNDJTW2d2TZ0xwLfL+oufMed5rN2nUQIEVXKwFDBF0iNUIxK712x/V79lWmlrYOMysnEvjf9+pwEjgX2oRhywfTPwKeAJYIykvWt3sP0gsD5V4vALSaPbiTsiIrpBdyYM1wP92/7HL2lh4GjgOKph/eGSFpK0ItWwNsCSVBfTmZKWAz5Xyu8HhkpatbyvvXDVGwT8s9yV7wUsXMpfAga2s891wLfa3kh6f30FSQOAE6juuJ+v27YK8LDtY4C/UE2n3ADs3DYkL2npDmLuDgOBf5apgj1bqF9/fnYH9rM91PZQYGWqO/z+HbQxCHje9ixJawAfb6feJVRTDBsC1wBIWolqROVUqoRi/dodJA0BZtk+GziqfntERHSvbksYyrD6F4GdJD1ENarwtu1fArdRJQ3TqdYYTCj7TKK6K70fOLfUw/ZrwP7AlWW64ukOuj4B+GoZFl+Dd+7mJwOzy6K5g+r2+QXw/rKYbhKwRc22G8vixruBfwDfaNDnLsBUSROBYcCZtqcBvwRuKm3+toOYu8NPqKYRbqM6n83Unp8fU13Qr2zbaPsV4FaqdSTtuZpqpOE+qrURdzaqZPsN4EbgzzVTM5sDkyTdS7X25Pd1u60N3F3O8U+p/mYREdFDVF3Xe6Cj6tMK5wFftD2hRzqNPqksdpwA7Gz7oe7oY/CQ1b3Nfid3R9MRET2up55WKWl8+eDBu/TY461t3w6s1FP9Rd8kaU3gCuCS7koWIiKi6/VYwhABUD6ZsUpvxxEREZ3TFz9WGREREX1MRhhigbTykIE9NucXEfFekBGGiIiIaCoJQ0RERDSVhCEiIiKayhqGWCDNePIl9hg9trfDiIiYZ31lPVZGGCIiIqKpJAwRERHRVBKGiIiIaCoJQ0RERDSVhCEiIiKaSsLQxSS93AVtjJB0TAfbh0rao9X6pc4jkqZImizpJkl95kFgkkZJ2ru344iIiPYlYeiDbI+zfUAHVYYC/0kYWqjfZgvb6wBjgUPnKUhAlXn+N2T7JNtnzms7ERHRfZIw9ABJwyXdWe7uL5H0/lK+YSmbKOkoSVNL+eaSriivNyvbJ0q6V9JA4Ahg01J2UF39AZL+WDOa8OUGId0BrFDqLyvpIkn3lJ9P1pRfJ2mapNMkPSppmTK68YCkM4GpwIqS/rvsO1nSz8r+S0i6UtIkSVMl7VrKj5A0vdT9n1J2mKRDmpyrsZKOlHS3pAclbdo9f62IiGgkCUPPOBP4frm7nwL8tJT/EfiG7eHA7Hb2PQT4VqmzKfAq8APgFtvDbf+urv5PgJm21y793dCgzc8Cl5bXvwd+Z3tD4MvAaaX8p8ANttcCLgQ+VLP/asAJZdvq5f1GwHBgA0mfKn08aXtd28OAqyUNBr4IrFVi+0UnzhXAIrY3Ag6sKwdA0v6Sxkka99qsmQ2ajoiIuZWEoZtJGgQsZfumUnQG8ClJSwEDbd9Rys9tp4nbgN9KOqC081aTLrcGjm97Y/v5mm03SnoC+BxwXk394yRNBC4DlpQ0ANgE+FNp42qgtp1Hbd9ZXn+m/NwLTADWoEogpgCfLqMCm9qeCcwEXgNOl/QlYFZt4O2dq5oqF5ff46mmZeZg+xTbI2yP6Nd/UPtnKCIiOi0JQx9n+whgP2Bx4DZJa8xDc1sAKwETgZ+VsoWAj5fRiuG2V7DdbOHmKzWvBfy6Zv8P2z7d9oPA+lSJwy8kjS7JzkZUIxbbAVd3Mv7Xy+/Z5GvNIyJ6VBKGblburJ+vmXPfC7jJ9gvAS5I+Vsp3a7S/pFVtT7F9JHAP1R38S8DAdrq8DvhWzf7vr4vnLaoh/b0lLQ1cC3ynpv7w8vI2YJdS9hlgjnZqXAN8rYxKIGkFSR+QNASYZfts4Chg/VJnkO2rgIOAdetia3iu2uk3IiJ6UO7Sul5/SY/XvP8t8FXgJEn9gYeBfcq2fYFTJb1NdWFsNPF+oKQtgLeBacD/ldezJU0CxlBNB7T5BXB8WUA5m2ok4eLaBm3/U9J5VInFAaX+ZKp/DzcDo8p+50nai2qR5L+oEpUBdW1dK+mjwB2SAF4GvgJ8GDiqHNubwDepkpy/SOpHNTJxcIPjbe9cRUREL5Lt3o7hPUvSgLbhf0k/AJa3/d1eDgsASYsBs22/JWlj4MSy8HK+MHjI6t5mv5N7O4yIiHnWk0+rlDTe9ohG2zLC0Ls+L+mHVH+HR4GRvRvOHD4E/Ll8z8IbwNd7OZ6IiOhFSRh6ke3zgfN7O45GbD8ErNfbcURERN+QRY8RERHRVEYYYoG08pCBPTrvFxGxoMsIQ0RERDSVT0nEAknSS8ADvR1HO5YB/t3bQbQjsc2dvhpbX40LEtvc6u7YVrK9bKMNmZKIBdUD7X00qLdJGpfYOi+xdV5fjQsS29zqzdgyJRERERFNJWGIiIiIppIwxILqlN4OoAOJbe4kts7rq3FBYptbvRZbFj1GREREUxlhiIiIiKaSMERERERTSRhivibps5IekPS38sTP+u2LSTq/bL9L0tA+FNunJE2Q9JaknXoqrhZjO1jSdEmTJV0vaaU+FNsoSVMkTZR0q6Q1+0JcNfW+LMmSeuyjby2cs5GSninnbKKk/fpKbKXOLuXf2zRJ5/aV2CT9ruacPSjphT4S14ck3Sjp3vLf6LY9ERe285Of+fIHWBj4O7AK8D5gErBmXZ3/DzipvN4NOL8PxTYUWAc4E9ipj523LYD+5fU3+9h5W7Lm9fbA1X0hrlJvIHAzcCcwog+ds5HAcT31b6yTsa0G3Au8v7z/QF+Jra7+d4A/9IW4qBY+frO8XhN4pCfOWUYYYn62EfA32w/bfgP4E7BDXZ0dgDPK6wuBrSSpL8Rm+xHbk4G3eyCezsZ2o+1Z5e2dwAf7UGwv1rxdAuiJldut/FsD+DlwJPBaD8TU2dh6QyuxfR043vbzALaf7kOx1dodOK+PxGVgyfJ6EPBkD8SVhCHmaysAj9W8f7yUNaxj+y1gJjC4j8TWWzob277A/3VrRO9oKTZJ35L0d+A3wAF9IS5J6wMr2r6yB+Kp1erf88tl+PpCSSv2TGgtxfYR4COSbpN0p6TP9qHYAChTcisDN/SRuA4DviLpceAqqtGPbpeEISLaJekrwAjgqN6OpZbt422vCnwfOLS345G0EPBb4Hu9HUs7LgeG2l4HuI53Rt36gkWopiU2p7qLP1XSUr0ZUAO7ARfant3bgRS7A2NsfxDYFjir/BvsVkkYYn72BFB7p/TBUtawjqRFqIbvnu0jsfWWlmKTtDXwY2B726/3pdhq/AnYsTsDKprFNRAYBoyV9AjwceCyHlr42PSc2X625m94GrBBD8TVUmxUd9CX2X7T9gzgQaoEoi/E1mY3emY6AlqLa1/gzwC27wD6UT2UqlslYYj52T3AapJWlvQ+qv+oL6urcxnw1fJ6J+AGl5VCfSC23tI0NknrASdTJQs9Nafcamy1F5PPAw/1dly2Z9pexvZQ20Op1n1sb3tcb8cGIGn5mrfbA/f1QFwtxQZcSjW6gKRlqKYoHu4jsSFpDeD9wB09EFOrcf0D2KrE91GqhOGZbo+sJ1ZW5ic/3fVDNRz3INWq4h+XssOp/mdN+Q/pAuBvwN3AKn0otg2p7q5eoRr1mNaHYvsr8BQwsfxc1odi+z0wrcR1I7BWX4irru5YeuhTEi2es1+XczapnLM1+lBsoprOmQ5MAXbrK7GV94cBR/RUTC2eszWB28rfcyLwmZ6IK18NHREREU1lSiIiIiKaSsIQERERTSVhiIiIiKaSMERERERTSRgiIiKiqSQMEREdkLRjefrkGr0dS0RvSsIQEdGx3YFby+9uIWnh7mo7oqskYYiIaIekAcAmVF/Fu1spW1jS/0iaWh7m9J1SvqGk2yVNknS3pIGSRko6rqa9KyRtXl6/LOloSZOAjSWNlnRPafeUtqeqSvqwpL+WdidIWlXSmZJ2rGn3HEl95QmVsYBKwhAR0b4dgKttPwg8K2kDYH9gKDDc1cOczilf4Xs+8F3b6wJbA682aXsJ4C7b69q+FTjO9oa2hwGLA9uVeudQPf55XeATwD+B04GRAJIGlfKefkpmvMckYYiIaN/uVA+4ovzenSoZONnV49Kx/RywOvBP2/eUshfbtndgNnBRzfstJN0laQqwJbCWpIHACrYvKe2+ZnuW7ZuonjewbInpohb6i5gni/R2ABERfZGkpaku3GtLMrAwYKqHA7XqLea8MetX8/o1l8clS+oHnED1/InHJB1WV7eRM4GvUE2V7NOJmCLmSkYYIiIa2wk4y/ZKrp5CuSIwg+qBP98oj0tvSyweAJaXtGEpG1i2PwIMl7SQpBWBjdrpqy05+HdZN7ETgO2XgMfb1itIWkxS/1J3DHBgqTe9y446oh1JGCIiGtsduKSu7CJgearHC08uCxb3sP0GsCtwbCm7jioJuI0qyZgOHANMaNSR7ReAU4GpwDXMOYqxF3CApMnA7cB/lX2eonpM9R/n9UAjWpGnVUZEzIfKSMMUYH3bM3s7nljwZYQhImI+I2lrqtGFY5MsRE/JCENEREQ0lRGGiIiIaCoJQ0RERDSVhCEiIiKaSsIQERERTSVhiIiIiKb+f/AEJGNVFut0AAAAAElFTkSuQmCC\n",
|
| 575 |
+
"text/plain": [
|
| 576 |
+
"<Figure size 432x288 with 1 Axes>"
|
| 577 |
+
]
|
| 578 |
+
},
|
| 579 |
+
"metadata": {
|
| 580 |
+
"needs_background": "light"
|
| 581 |
+
},
|
| 582 |
+
"output_type": "display_data"
|
| 583 |
+
}
|
| 584 |
+
],
|
| 585 |
+
"source": [
|
| 586 |
+
"import matplotlib.pyplot as plt\n",
|
| 587 |
+
"import seaborn as sns\n",
|
| 588 |
+
"\n",
|
| 589 |
+
"from sklearn.model_selection import StratifiedShuffleSplit\n",
|
| 590 |
+
"from sklearn.metrics import accuracy_score, log_loss\n",
|
| 591 |
+
"from sklearn.neighbors import KNeighborsClassifier\n",
|
| 592 |
+
"from sklearn.svm import SVC\n",
|
| 593 |
+
"from sklearn.tree import DecisionTreeClassifier\n",
|
| 594 |
+
"from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, GradientBoostingClassifier\n",
|
| 595 |
+
"from sklearn.naive_bayes import GaussianNB\n",
|
| 596 |
+
"from sklearn.discriminant_analysis import LinearDiscriminantAnalysis, QuadraticDiscriminantAnalysis\n",
|
| 597 |
+
"from sklearn.linear_model import LogisticRegression\n",
|
| 598 |
+
"\n",
|
| 599 |
+
"classifiers = [\n",
|
| 600 |
+
" KNeighborsClassifier(3),\n",
|
| 601 |
+
" SVC(probability=True),\n",
|
| 602 |
+
" DecisionTreeClassifier(),\n",
|
| 603 |
+
" RandomForestClassifier(),\n",
|
| 604 |
+
"\tAdaBoostClassifier(),\n",
|
| 605 |
+
" GradientBoostingClassifier(),\n",
|
| 606 |
+
" GaussianNB(),\n",
|
| 607 |
+
" LinearDiscriminantAnalysis(),\n",
|
| 608 |
+
" QuadraticDiscriminantAnalysis(),\n",
|
| 609 |
+
" LogisticRegression()]\n",
|
| 610 |
+
"\n",
|
| 611 |
+
"log_cols = [\"Classifier\", \"Accuracy\"]\n",
|
| 612 |
+
"log \t = pd.DataFrame(columns=log_cols)\n",
|
| 613 |
+
"\n",
|
| 614 |
+
"sss = StratifiedShuffleSplit(n_splits=10, test_size=0.1, random_state=0)\n",
|
| 615 |
+
"\n",
|
| 616 |
+
"X = train[0::, 1::]\n",
|
| 617 |
+
"y = train[0::, 0]\n",
|
| 618 |
+
"\n",
|
| 619 |
+
"acc_dict = {}\n",
|
| 620 |
+
"\n",
|
| 621 |
+
"for train_index, test_index in sss.split(X, y):\n",
|
| 622 |
+
"\tX_train, X_test = X[train_index], X[test_index]\n",
|
| 623 |
+
"\ty_train, y_test = y[train_index], y[test_index]\n",
|
| 624 |
+
"\t\n",
|
| 625 |
+
"\tfor clf in classifiers:\n",
|
| 626 |
+
"\t\tname = clf.__class__.__name__\n",
|
| 627 |
+
"\t\tclf.fit(X_train, y_train)\n",
|
| 628 |
+
"\t\ttrain_predictions = clf.predict(X_test)\n",
|
| 629 |
+
"\t\tacc = accuracy_score(y_test, train_predictions)\n",
|
| 630 |
+
"\t\tif name in acc_dict:\n",
|
| 631 |
+
"\t\t\tacc_dict[name] += acc\n",
|
| 632 |
+
"\t\telse:\n",
|
| 633 |
+
"\t\t\tacc_dict[name] = acc\n",
|
| 634 |
+
"\n",
|
| 635 |
+
"for clf in acc_dict:\n",
|
| 636 |
+
"\tacc_dict[clf] = acc_dict[clf] / 10.0\n",
|
| 637 |
+
"\tlog_entry = pd.DataFrame([[clf, acc_dict[clf]]], columns=log_cols)\n",
|
| 638 |
+
"\tlog = log.append(log_entry)\n",
|
| 639 |
+
"\n",
|
| 640 |
+
"plt.xlabel('Accuracy')\n",
|
| 641 |
+
"plt.title('Classifier Accuracy')\n",
|
| 642 |
+
"\n",
|
| 643 |
+
"sns.set_color_codes(\"muted\")\n",
|
| 644 |
+
"sns.barplot(x='Accuracy', y='Classifier', data=log, color=\"b\")"
|
| 645 |
+
]
|
| 646 |
+
},
|
| 647 |
+
{
|
| 648 |
+
"cell_type": "markdown",
|
| 649 |
+
"metadata": {
|
| 650 |
+
"_cell_guid": "438585cf-b7ad-73ba-49aa-87688ff21233"
|
| 651 |
+
},
|
| 652 |
+
"source": [
|
| 653 |
+
"# Prediction #\n",
|
| 654 |
+
"now we can use SVC classifier to predict our data."
|
| 655 |
+
]
|
| 656 |
+
},
|
| 657 |
+
{
|
| 658 |
+
"cell_type": "code",
|
| 659 |
+
"execution_count": 13,
|
| 660 |
+
"metadata": {
|
| 661 |
+
"_cell_guid": "24967b57-732b-7180-bfd5-005beff75974"
|
| 662 |
+
},
|
| 663 |
+
"outputs": [],
|
| 664 |
+
"source": [
|
| 665 |
+
"candidate_classifier = SVC()\n",
|
| 666 |
+
"candidate_classifier.fit(train[0::, 1::], train[0::, 0])\n",
|
| 667 |
+
"result = candidate_classifier.predict(test)"
|
| 668 |
+
]
|
| 669 |
+
},
|
| 670 |
+
{
|
| 671 |
+
"cell_type": "markdown",
|
| 672 |
+
"metadata": {},
|
| 673 |
+
"source": [
|
| 674 |
+
"## Fairness"
|
| 675 |
+
]
|
| 676 |
+
},
|
| 677 |
+
{
|
| 678 |
+
"cell_type": "code",
|
| 679 |
+
"execution_count": 14,
|
| 680 |
+
"metadata": {},
|
| 681 |
+
"outputs": [],
|
| 682 |
+
"source": [
|
| 683 |
+
"# This DataFrame is created to stock differents models and fair metrics that we produce in this notebook\n",
|
| 684 |
+
"algo_metrics = pd.DataFrame(columns=['model', 'fair_metrics', 'prediction', 'probs'])\n",
|
| 685 |
+
"\n",
|
| 686 |
+
"def add_to_df_algo_metrics(algo_metrics, model, fair_metrics, preds, probs, name):\n",
|
| 687 |
+
" return algo_metrics.append(pd.DataFrame(data=[[model, fair_metrics, preds, probs]], columns=['model', 'fair_metrics', 'prediction', 'probs'], index=[name]))"
|
| 688 |
+
]
|
| 689 |
+
},
|
| 690 |
+
{
|
| 691 |
+
"cell_type": "code",
|
| 692 |
+
"execution_count": 15,
|
| 693 |
+
"metadata": {},
|
| 694 |
+
"outputs": [],
|
| 695 |
+
"source": [
|
| 696 |
+
"def fair_metrics(dataset, pred, pred_is_dataset=False):\n",
|
| 697 |
+
" if pred_is_dataset:\n",
|
| 698 |
+
" dataset_pred = pred\n",
|
| 699 |
+
" else:\n",
|
| 700 |
+
" dataset_pred = dataset.copy()\n",
|
| 701 |
+
" dataset_pred.labels = pred\n",
|
| 702 |
+
" \n",
|
| 703 |
+
" cols = ['statistical_parity_difference', 'equal_opportunity_difference', 'average_abs_odds_difference', 'disparate_impact', 'theil_index']\n",
|
| 704 |
+
" obj_fairness = [[0,0,0,1,0]]\n",
|
| 705 |
+
" \n",
|
| 706 |
+
" fair_metrics = pd.DataFrame(data=obj_fairness, index=['objective'], columns=cols)\n",
|
| 707 |
+
" \n",
|
| 708 |
+
" for attr in dataset_pred.protected_attribute_names:\n",
|
| 709 |
+
" idx = dataset_pred.protected_attribute_names.index(attr)\n",
|
| 710 |
+
" privileged_groups = [{attr:dataset_pred.privileged_protected_attributes[idx][0]}] \n",
|
| 711 |
+
" unprivileged_groups = [{attr:dataset_pred.unprivileged_protected_attributes[idx][0]}] \n",
|
| 712 |
+
" \n",
|
| 713 |
+
" classified_metric = ClassificationMetric(dataset, \n",
|
| 714 |
+
" dataset_pred,\n",
|
| 715 |
+
" unprivileged_groups=unprivileged_groups,\n",
|
| 716 |
+
" privileged_groups=privileged_groups)\n",
|
| 717 |
+
"\n",
|
| 718 |
+
" metric_pred = BinaryLabelDatasetMetric(dataset_pred,\n",
|
| 719 |
+
" unprivileged_groups=unprivileged_groups,\n",
|
| 720 |
+
" privileged_groups=privileged_groups)\n",
|
| 721 |
+
"\n",
|
| 722 |
+
" acc = classified_metric.accuracy()\n",
|
| 723 |
+
"\n",
|
| 724 |
+
" row = pd.DataFrame([[metric_pred.mean_difference(),\n",
|
| 725 |
+
" classified_metric.equal_opportunity_difference(),\n",
|
| 726 |
+
" classified_metric.average_abs_odds_difference(),\n",
|
| 727 |
+
" metric_pred.disparate_impact(),\n",
|
| 728 |
+
" classified_metric.theil_index()]],\n",
|
| 729 |
+
" columns = cols,\n",
|
| 730 |
+
" index = [attr]\n",
|
| 731 |
+
" )\n",
|
| 732 |
+
" fair_metrics = fair_metrics.append(row) \n",
|
| 733 |
+
" \n",
|
| 734 |
+
" fair_metrics = fair_metrics.replace([-np.inf, np.inf], 2)\n",
|
| 735 |
+
" \n",
|
| 736 |
+
" return fair_metrics\n",
|
| 737 |
+
"\n",
|
| 738 |
+
"def plot_fair_metrics(fair_metrics):\n",
|
| 739 |
+
" fig, ax = plt.subplots(figsize=(20,4), ncols=5, nrows=1)\n",
|
| 740 |
+
"\n",
|
| 741 |
+
" plt.subplots_adjust(\n",
|
| 742 |
+
" left = 0.125, \n",
|
| 743 |
+
" bottom = 0.1, \n",
|
| 744 |
+
" right = 0.9, \n",
|
| 745 |
+
" top = 0.9, \n",
|
| 746 |
+
" wspace = .5, \n",
|
| 747 |
+
" hspace = 1.1\n",
|
| 748 |
+
" )\n",
|
| 749 |
+
"\n",
|
| 750 |
+
" y_title_margin = 1.2\n",
|
| 751 |
+
"\n",
|
| 752 |
+
" plt.suptitle(\"Fairness metrics\", y = 1.09, fontsize=20)\n",
|
| 753 |
+
" sns.set(style=\"dark\")\n",
|
| 754 |
+
"\n",
|
| 755 |
+
" cols = fair_metrics.columns.values\n",
|
| 756 |
+
" obj = fair_metrics.loc['objective']\n",
|
| 757 |
+
" size_rect = [0.2,0.2,0.2,0.4,0.25]\n",
|
| 758 |
+
" rect = [-0.1,-0.1,-0.1,0.8,0]\n",
|
| 759 |
+
" bottom = [-1,-1,-1,0,0]\n",
|
| 760 |
+
" top = [1,1,1,2,1]\n",
|
| 761 |
+
" bound = [[-0.1,0.1],[-0.1,0.1],[-0.1,0.1],[0.8,1.2],[0,0.25]]\n",
|
| 762 |
+
"\n",
|
| 763 |
+
" display(Markdown(\"### Check bias metrics :\"))\n",
|
| 764 |
+
" display(Markdown(\"A model can be considered bias if just one of these five metrics show that this model is biased.\"))\n",
|
| 765 |
+
" for attr in fair_metrics.index[1:len(fair_metrics)].values:\n",
|
| 766 |
+
" display(Markdown(\"#### For the %s attribute :\"%attr))\n",
|
| 767 |
+
" check = [bound[i][0] < fair_metrics.loc[attr][i] < bound[i][1] for i in range(0,5)]\n",
|
| 768 |
+
" display(Markdown(\"With default thresholds, bias against unprivileged group detected in **%d** out of 5 metrics\"%(5 - sum(check))))\n",
|
| 769 |
+
"\n",
|
| 770 |
+
" for i in range(0,5):\n",
|
| 771 |
+
" plt.subplot(1, 5, i+1)\n",
|
| 772 |
+
" ax = sns.barplot(x=fair_metrics.index[1:len(fair_metrics)], y=fair_metrics.iloc[1:len(fair_metrics)][cols[i]])\n",
|
| 773 |
+
" \n",
|
| 774 |
+
" for j in range(0,len(fair_metrics)-1):\n",
|
| 775 |
+
" a, val = ax.patches[j], fair_metrics.iloc[j+1][cols[i]]\n",
|
| 776 |
+
" marg = -0.2 if val < 0 else 0.1\n",
|
| 777 |
+
" ax.text(a.get_x()+a.get_width()/5, a.get_y()+a.get_height()+marg, round(val, 3), fontsize=15,color='black')\n",
|
| 778 |
+
"\n",
|
| 779 |
+
" plt.ylim(bottom[i], top[i])\n",
|
| 780 |
+
" plt.setp(ax.patches, linewidth=0)\n",
|
| 781 |
+
" ax.add_patch(patches.Rectangle((-5,rect[i]), 10, size_rect[i], alpha=0.3, facecolor=\"green\", linewidth=1, linestyle='solid'))\n",
|
| 782 |
+
" plt.axhline(obj[i], color='black', alpha=0.3)\n",
|
| 783 |
+
" plt.title(cols[i])\n",
|
| 784 |
+
" ax.set_ylabel('') \n",
|
| 785 |
+
" ax.set_xlabel('')"
|
| 786 |
+
]
|
| 787 |
+
},
|
| 788 |
+
{
|
| 789 |
+
"cell_type": "code",
|
| 790 |
+
"execution_count": 16,
|
| 791 |
+
"metadata": {},
|
| 792 |
+
"outputs": [],
|
| 793 |
+
"source": [
|
| 794 |
+
"def get_fair_metrics_and_plot(data, model, plot=False, model_aif=False):\n",
|
| 795 |
+
" pred = model.predict(data).labels if model_aif else model.predict(data.features)\n",
|
| 796 |
+
" # fair_metrics function available in the metrics.py file\n",
|
| 797 |
+
" fair = fair_metrics(data, pred)\n",
|
| 798 |
+
"\n",
|
| 799 |
+
" if plot:\n",
|
| 800 |
+
" # plot_fair_metrics function available in the visualisations.py file\n",
|
| 801 |
+
" # The visualisation of this function is inspired by the dashboard on the demo of IBM aif360 \n",
|
| 802 |
+
" plot_fair_metrics(fair)\n",
|
| 803 |
+
" display(fair)\n",
|
| 804 |
+
" \n",
|
| 805 |
+
" return fair"
|
| 806 |
+
]
|
| 807 |
+
},
|
| 808 |
+
{
|
| 809 |
+
"cell_type": "code",
|
| 810 |
+
"execution_count": 17,
|
| 811 |
+
"metadata": {},
|
| 812 |
+
"outputs": [
|
| 813 |
+
{
|
| 814 |
+
"data": {
|
| 815 |
+
"text/html": [
|
| 816 |
+
"<div>\n",
|
| 817 |
+
"<style scoped>\n",
|
| 818 |
+
" .dataframe tbody tr th:only-of-type {\n",
|
| 819 |
+
" vertical-align: middle;\n",
|
| 820 |
+
" }\n",
|
| 821 |
+
"\n",
|
| 822 |
+
" .dataframe tbody tr th {\n",
|
| 823 |
+
" vertical-align: top;\n",
|
| 824 |
+
" }\n",
|
| 825 |
+
"\n",
|
| 826 |
+
" .dataframe thead th {\n",
|
| 827 |
+
" text-align: right;\n",
|
| 828 |
+
" }\n",
|
| 829 |
+
"</style>\n",
|
| 830 |
+
"<table border=\"1\" class=\"dataframe\">\n",
|
| 831 |
+
" <thead>\n",
|
| 832 |
+
" <tr style=\"text-align: right;\">\n",
|
| 833 |
+
" <th></th>\n",
|
| 834 |
+
" <th>Survived</th>\n",
|
| 835 |
+
" <th>Pclass</th>\n",
|
| 836 |
+
" <th>Sex</th>\n",
|
| 837 |
+
" <th>Age</th>\n",
|
| 838 |
+
" <th>Fare</th>\n",
|
| 839 |
+
" <th>Embarked</th>\n",
|
| 840 |
+
" <th>IsAlone</th>\n",
|
| 841 |
+
" <th>Title</th>\n",
|
| 842 |
+
" </tr>\n",
|
| 843 |
+
" </thead>\n",
|
| 844 |
+
" <tbody>\n",
|
| 845 |
+
" <tr>\n",
|
| 846 |
+
" <th>0</th>\n",
|
| 847 |
+
" <td>0</td>\n",
|
| 848 |
+
" <td>3</td>\n",
|
| 849 |
+
" <td>0</td>\n",
|
| 850 |
+
" <td>1</td>\n",
|
| 851 |
+
" <td>0</td>\n",
|
| 852 |
+
" <td>0</td>\n",
|
| 853 |
+
" <td>0</td>\n",
|
| 854 |
+
" <td>1</td>\n",
|
| 855 |
+
" </tr>\n",
|
| 856 |
+
" <tr>\n",
|
| 857 |
+
" <th>1</th>\n",
|
| 858 |
+
" <td>1</td>\n",
|
| 859 |
+
" <td>1</td>\n",
|
| 860 |
+
" <td>1</td>\n",
|
| 861 |
+
" <td>2</td>\n",
|
| 862 |
+
" <td>3</td>\n",
|
| 863 |
+
" <td>1</td>\n",
|
| 864 |
+
" <td>0</td>\n",
|
| 865 |
+
" <td>3</td>\n",
|
| 866 |
+
" </tr>\n",
|
| 867 |
+
" <tr>\n",
|
| 868 |
+
" <th>2</th>\n",
|
| 869 |
+
" <td>1</td>\n",
|
| 870 |
+
" <td>3</td>\n",
|
| 871 |
+
" <td>1</td>\n",
|
| 872 |
+
" <td>1</td>\n",
|
| 873 |
+
" <td>1</td>\n",
|
| 874 |
+
" <td>0</td>\n",
|
| 875 |
+
" <td>1</td>\n",
|
| 876 |
+
" <td>2</td>\n",
|
| 877 |
+
" </tr>\n",
|
| 878 |
+
" <tr>\n",
|
| 879 |
+
" <th>3</th>\n",
|
| 880 |
+
" <td>1</td>\n",
|
| 881 |
+
" <td>1</td>\n",
|
| 882 |
+
" <td>1</td>\n",
|
| 883 |
+
" <td>2</td>\n",
|
| 884 |
+
" <td>3</td>\n",
|
| 885 |
+
" <td>0</td>\n",
|
| 886 |
+
" <td>0</td>\n",
|
| 887 |
+
" <td>3</td>\n",
|
| 888 |
+
" </tr>\n",
|
| 889 |
+
" <tr>\n",
|
| 890 |
+
" <th>4</th>\n",
|
| 891 |
+
" <td>0</td>\n",
|
| 892 |
+
" <td>3</td>\n",
|
| 893 |
+
" <td>0</td>\n",
|
| 894 |
+
" <td>2</td>\n",
|
| 895 |
+
" <td>1</td>\n",
|
| 896 |
+
" <td>0</td>\n",
|
| 897 |
+
" <td>1</td>\n",
|
| 898 |
+
" <td>1</td>\n",
|
| 899 |
+
" </tr>\n",
|
| 900 |
+
" <tr>\n",
|
| 901 |
+
" <th>...</th>\n",
|
| 902 |
+
" <td>...</td>\n",
|
| 903 |
+
" <td>...</td>\n",
|
| 904 |
+
" <td>...</td>\n",
|
| 905 |
+
" <td>...</td>\n",
|
| 906 |
+
" <td>...</td>\n",
|
| 907 |
+
" <td>...</td>\n",
|
| 908 |
+
" <td>...</td>\n",
|
| 909 |
+
" <td>...</td>\n",
|
| 910 |
+
" </tr>\n",
|
| 911 |
+
" <tr>\n",
|
| 912 |
+
" <th>886</th>\n",
|
| 913 |
+
" <td>0</td>\n",
|
| 914 |
+
" <td>2</td>\n",
|
| 915 |
+
" <td>0</td>\n",
|
| 916 |
+
" <td>1</td>\n",
|
| 917 |
+
" <td>1</td>\n",
|
| 918 |
+
" <td>0</td>\n",
|
| 919 |
+
" <td>1</td>\n",
|
| 920 |
+
" <td>5</td>\n",
|
| 921 |
+
" </tr>\n",
|
| 922 |
+
" <tr>\n",
|
| 923 |
+
" <th>887</th>\n",
|
| 924 |
+
" <td>1</td>\n",
|
| 925 |
+
" <td>1</td>\n",
|
| 926 |
+
" <td>1</td>\n",
|
| 927 |
+
" <td>1</td>\n",
|
| 928 |
+
" <td>2</td>\n",
|
| 929 |
+
" <td>0</td>\n",
|
| 930 |
+
" <td>1</td>\n",
|
| 931 |
+
" <td>2</td>\n",
|
| 932 |
+
" </tr>\n",
|
| 933 |
+
" <tr>\n",
|
| 934 |
+
" <th>888</th>\n",
|
| 935 |
+
" <td>0</td>\n",
|
| 936 |
+
" <td>3</td>\n",
|
| 937 |
+
" <td>1</td>\n",
|
| 938 |
+
" <td>0</td>\n",
|
| 939 |
+
" <td>2</td>\n",
|
| 940 |
+
" <td>0</td>\n",
|
| 941 |
+
" <td>0</td>\n",
|
| 942 |
+
" <td>2</td>\n",
|
| 943 |
+
" </tr>\n",
|
| 944 |
+
" <tr>\n",
|
| 945 |
+
" <th>889</th>\n",
|
| 946 |
+
" <td>1</td>\n",
|
| 947 |
+
" <td>1</td>\n",
|
| 948 |
+
" <td>0</td>\n",
|
| 949 |
+
" <td>1</td>\n",
|
| 950 |
+
" <td>2</td>\n",
|
| 951 |
+
" <td>1</td>\n",
|
| 952 |
+
" <td>1</td>\n",
|
| 953 |
+
" <td>1</td>\n",
|
| 954 |
+
" </tr>\n",
|
| 955 |
+
" <tr>\n",
|
| 956 |
+
" <th>890</th>\n",
|
| 957 |
+
" <td>0</td>\n",
|
| 958 |
+
" <td>3</td>\n",
|
| 959 |
+
" <td>0</td>\n",
|
| 960 |
+
" <td>1</td>\n",
|
| 961 |
+
" <td>0</td>\n",
|
| 962 |
+
" <td>2</td>\n",
|
| 963 |
+
" <td>1</td>\n",
|
| 964 |
+
" <td>1</td>\n",
|
| 965 |
+
" </tr>\n",
|
| 966 |
+
" </tbody>\n",
|
| 967 |
+
"</table>\n",
|
| 968 |
+
"<p>891 rows × 8 columns</p>\n",
|
| 969 |
+
"</div>"
|
| 970 |
+
],
|
| 971 |
+
"text/plain": [
|
| 972 |
+
" Survived Pclass Sex Age Fare Embarked IsAlone Title\n",
|
| 973 |
+
"0 0 3 0 1 0 0 0 1\n",
|
| 974 |
+
"1 1 1 1 2 3 1 0 3\n",
|
| 975 |
+
"2 1 3 1 1 1 0 1 2\n",
|
| 976 |
+
"3 1 1 1 2 3 0 0 3\n",
|
| 977 |
+
"4 0 3 0 2 1 0 1 1\n",
|
| 978 |
+
".. ... ... ... ... ... ... ... ...\n",
|
| 979 |
+
"886 0 2 0 1 1 0 1 5\n",
|
| 980 |
+
"887 1 1 1 1 2 0 1 2\n",
|
| 981 |
+
"888 0 3 1 0 2 0 0 2\n",
|
| 982 |
+
"889 1 1 0 1 2 1 1 1\n",
|
| 983 |
+
"890 0 3 0 1 0 2 1 1\n",
|
| 984 |
+
"\n",
|
| 985 |
+
"[891 rows x 8 columns]"
|
| 986 |
+
]
|
| 987 |
+
},
|
| 988 |
+
"execution_count": 17,
|
| 989 |
+
"metadata": {},
|
| 990 |
+
"output_type": "execute_result"
|
| 991 |
+
}
|
| 992 |
+
],
|
| 993 |
+
"source": [
|
| 994 |
+
"##train['Sex'] = train['Sex'].map( {'female': 1, 'male': 0} ).astype(int)\n",
|
| 995 |
+
"train_df\n",
|
| 996 |
+
"\n",
|
| 997 |
+
"#features = [\"Pclass\", \"Sex\", \"SibSp\", \"Parch\", \"Survived\"]\n",
|
| 998 |
+
"#X = pd.get_dummies(train_data[features])"
|
| 999 |
+
]
|
| 1000 |
+
},
|
| 1001 |
+
{
|
| 1002 |
+
"cell_type": "code",
|
| 1003 |
+
"execution_count": 18,
|
| 1004 |
+
"metadata": {},
|
| 1005 |
+
"outputs": [],
|
| 1006 |
+
"source": [
|
| 1007 |
+
"privileged_groups = [{'Sex': 1}]\n",
|
| 1008 |
+
"unprivileged_groups = [{'Sex': 0}]\n",
|
| 1009 |
+
"dataset_orig = StandardDataset(train_df,\n",
|
| 1010 |
+
" label_name='Survived',\n",
|
| 1011 |
+
" protected_attribute_names=['Sex'],\n",
|
| 1012 |
+
" favorable_classes=[1],\n",
|
| 1013 |
+
" privileged_classes=[[1]])\n",
|
| 1014 |
+
"\n"
|
| 1015 |
+
]
|
| 1016 |
+
},
|
| 1017 |
+
{
|
| 1018 |
+
"cell_type": "code",
|
| 1019 |
+
"execution_count": 19,
|
| 1020 |
+
"metadata": {},
|
| 1021 |
+
"outputs": [
|
| 1022 |
+
{
|
| 1023 |
+
"data": {
|
| 1024 |
+
"text/markdown": [
|
| 1025 |
+
"#### Original training dataset"
|
| 1026 |
+
],
|
| 1027 |
+
"text/plain": [
|
| 1028 |
+
"<IPython.core.display.Markdown object>"
|
| 1029 |
+
]
|
| 1030 |
+
},
|
| 1031 |
+
"metadata": {},
|
| 1032 |
+
"output_type": "display_data"
|
| 1033 |
+
},
|
| 1034 |
+
{
|
| 1035 |
+
"name": "stdout",
|
| 1036 |
+
"output_type": "stream",
|
| 1037 |
+
"text": [
|
| 1038 |
+
"Difference in mean outcomes between unprivileged and privileged groups = -0.553130\n"
|
| 1039 |
+
]
|
| 1040 |
+
}
|
| 1041 |
+
],
|
| 1042 |
+
"source": [
|
| 1043 |
+
"metric_orig_train = BinaryLabelDatasetMetric(dataset_orig, \n",
|
| 1044 |
+
" unprivileged_groups=unprivileged_groups,\n",
|
| 1045 |
+
" privileged_groups=privileged_groups)\n",
|
| 1046 |
+
"display(Markdown(\"#### Original training dataset\"))\n",
|
| 1047 |
+
"print(\"Difference in mean outcomes between unprivileged and privileged groups = %f\" % metric_orig_train.mean_difference())"
|
| 1048 |
+
]
|
| 1049 |
+
},
|
| 1050 |
+
{
|
| 1051 |
+
"cell_type": "code",
|
| 1052 |
+
"execution_count": 41,
|
| 1053 |
+
"metadata": {},
|
| 1054 |
+
"outputs": [],
|
| 1055 |
+
"source": [
|
| 1056 |
+
"import ipynbname\n",
|
| 1057 |
+
"nb_fname = ipynbname.name()\n",
|
| 1058 |
+
"nb_path = ipynbname.path()\n",
|
| 1059 |
+
"\n",
|
| 1060 |
+
"from sklearn.ensemble import AdaBoostClassifier\n",
|
| 1061 |
+
"import pickle\n",
|
| 1062 |
+
"\n",
|
| 1063 |
+
"data_orig_train, data_orig_test = dataset_orig.split([0.7], shuffle=True)\n",
|
| 1064 |
+
"X_train = data_orig_train.features\n",
|
| 1065 |
+
"y_train = data_orig_train.labels.ravel()\n",
|
| 1066 |
+
"\n",
|
| 1067 |
+
"X_test = data_orig_test.features\n",
|
| 1068 |
+
"y_test = data_orig_test.labels.ravel()\n",
|
| 1069 |
+
"num_estimators = 100\n",
|
| 1070 |
+
"\n",
|
| 1071 |
+
"model = AdaBoostClassifier(n_estimators=1)\n",
|
| 1072 |
+
"\n",
|
| 1073 |
+
"mdl = model.fit(X_train, y_train)\n",
|
| 1074 |
+
"with open('../../Results/AdaBoost/' + nb_fname + '.pkl', 'wb') as f:\n",
|
| 1075 |
+
" pickle.dump(mdl, f)\n",
|
| 1076 |
+
"\n",
|
| 1077 |
+
"with open('../../Results/AdaBoost/' + nb_fname + '_Train' + '.pkl', 'wb') as f:\n",
|
| 1078 |
+
" pickle.dump(data_orig_train, f) \n",
|
| 1079 |
+
" \n",
|
| 1080 |
+
"with open('../../Results/AdaBoost/' + nb_fname + '_Test' + '.pkl', 'wb') as f:\n",
|
| 1081 |
+
" pickle.dump(data_orig_test, f) "
|
| 1082 |
+
]
|
| 1083 |
+
},
|
| 1084 |
+
{
|
| 1085 |
+
"cell_type": "code",
|
| 1086 |
+
"execution_count": 22,
|
| 1087 |
+
"metadata": {},
|
| 1088 |
+
"outputs": [
|
| 1089 |
+
{
|
| 1090 |
+
"name": "stdout",
|
| 1091 |
+
"output_type": "stream",
|
| 1092 |
+
"text": [
|
| 1093 |
+
"0\n",
|
| 1094 |
+
"1\n",
|
| 1095 |
+
"2\n",
|
| 1096 |
+
"3\n",
|
| 1097 |
+
"4\n",
|
| 1098 |
+
"5\n",
|
| 1099 |
+
"6\n",
|
| 1100 |
+
"7\n",
|
| 1101 |
+
"8\n",
|
| 1102 |
+
"9\n",
|
| 1103 |
+
"STD [3.02765035 0.06749158 0.08874808 0.09476216 0.03541161 0.01255178]\n",
|
| 1104 |
+
"[4.5, -0.6693072547146794, -0.581259725046272, 0.49612085216852686, -2.1276205667545494, 0.1590111172017386]\n",
|
| 1105 |
+
"-2.7230555771452356\n",
|
| 1106 |
+
"0.8093283582089552\n",
|
| 1107 |
+
"0.7401892992453725\n"
|
| 1108 |
+
]
|
| 1109 |
+
}
|
| 1110 |
+
],
|
| 1111 |
+
"source": [
|
| 1112 |
+
"final_metrics = []\n",
|
| 1113 |
+
"accuracy = []\n",
|
| 1114 |
+
"f1= []\n",
|
| 1115 |
+
"from statistics import mean\n",
|
| 1116 |
+
"from sklearn.metrics import accuracy_score, f1_score\n",
|
| 1117 |
+
"from sklearn.ensemble import AdaBoostClassifier\n",
|
| 1118 |
+
"\n",
|
| 1119 |
+
"\n",
|
| 1120 |
+
"for i in range(0,10):\n",
|
| 1121 |
+
" \n",
|
| 1122 |
+
" data_orig_train, data_orig_test = dataset_orig.split([0.7], shuffle=True)\n",
|
| 1123 |
+
" print(i)\n",
|
| 1124 |
+
" X_train = data_orig_train.features\n",
|
| 1125 |
+
" y_train = data_orig_train.labels.ravel()\n",
|
| 1126 |
+
"\n",
|
| 1127 |
+
" X_test = data_orig_test.features\n",
|
| 1128 |
+
" y_test = data_orig_test.labels.ravel()\n",
|
| 1129 |
+
" model = GradientBoostingClassifier(n_estimators = 200)\n",
|
| 1130 |
+
" \n",
|
| 1131 |
+
" mdl = model.fit(X_train, y_train)\n",
|
| 1132 |
+
" yy = mdl.predict(X_test)\n",
|
| 1133 |
+
" accuracy.append(accuracy_score(y_test, yy))\n",
|
| 1134 |
+
" f1.append(f1_score(y_test, yy))\n",
|
| 1135 |
+
" fair = get_fair_metrics_and_plot(data_orig_test, mdl) \n",
|
| 1136 |
+
" fair_list = fair.iloc[1].tolist()\n",
|
| 1137 |
+
" fair_list.insert(0, i)\n",
|
| 1138 |
+
" final_metrics.append(fair_list)\n",
|
| 1139 |
+
"\n",
|
| 1140 |
+
" \n",
|
| 1141 |
+
"element_wise_std = np.std(final_metrics, 0, ddof=1)\n",
|
| 1142 |
+
"print(\"STD \" + str(element_wise_std))\n",
|
| 1143 |
+
"final_metrics = list(map(mean, zip(*final_metrics)))\n",
|
| 1144 |
+
"accuracy = mean(accuracy)\n",
|
| 1145 |
+
"f1 = mean(f1)\n",
|
| 1146 |
+
"final_metrics[4] = np.log(final_metrics[4])\n",
|
| 1147 |
+
"print(final_metrics)\n",
|
| 1148 |
+
"print(sum(final_metrics[1:]))\n",
|
| 1149 |
+
"print(accuracy)\n",
|
| 1150 |
+
"print(f1)"
|
| 1151 |
+
]
|
| 1152 |
+
},
|
| 1153 |
+
{
|
| 1154 |
+
"cell_type": "code",
|
| 1155 |
+
"execution_count": 42,
|
| 1156 |
+
"metadata": {},
|
| 1157 |
+
"outputs": [],
|
| 1158 |
+
"source": [
|
| 1159 |
+
"from csv import writer\n",
|
| 1160 |
+
"from sklearn.metrics import accuracy_score, f1_score\n",
|
| 1161 |
+
"\n",
|
| 1162 |
+
"final_metrics = []\n",
|
| 1163 |
+
"accuracy = []\n",
|
| 1164 |
+
"f1= []\n",
|
| 1165 |
+
"\n",
|
| 1166 |
+
"for i in range(1,num_estimators+1):\n",
|
| 1167 |
+
" \n",
|
| 1168 |
+
" model = AdaBoostClassifier(n_estimators=i)\n",
|
| 1169 |
+
" \n",
|
| 1170 |
+
" mdl = model.fit(X_train, y_train)\n",
|
| 1171 |
+
" yy = mdl.predict(X_test)\n",
|
| 1172 |
+
" accuracy.append(accuracy_score(y_test, yy))\n",
|
| 1173 |
+
" f1.append(f1_score(y_test, yy))\n",
|
| 1174 |
+
" fair = get_fair_metrics_and_plot(data_orig_test, mdl) \n",
|
| 1175 |
+
" fair_list = fair.iloc[1].tolist()\n",
|
| 1176 |
+
" fair_list.insert(0, i)\n",
|
| 1177 |
+
" final_metrics.append(fair_list)\n"
|
| 1178 |
+
]
|
| 1179 |
+
},
|
| 1180 |
+
{
|
| 1181 |
+
"cell_type": "code",
|
| 1182 |
+
"execution_count": 43,
|
| 1183 |
+
"metadata": {},
|
| 1184 |
+
"outputs": [
|
| 1185 |
+
{
|
| 1186 |
+
"data": {
|
| 1187 |
+
"text/html": [
|
| 1188 |
+
"<div>\n",
|
| 1189 |
+
"<style scoped>\n",
|
| 1190 |
+
" .dataframe tbody tr th:only-of-type {\n",
|
| 1191 |
+
" vertical-align: middle;\n",
|
| 1192 |
+
" }\n",
|
| 1193 |
+
"\n",
|
| 1194 |
+
" .dataframe tbody tr th {\n",
|
| 1195 |
+
" vertical-align: top;\n",
|
| 1196 |
+
" }\n",
|
| 1197 |
+
"\n",
|
| 1198 |
+
" .dataframe thead th {\n",
|
| 1199 |
+
" text-align: right;\n",
|
| 1200 |
+
" }\n",
|
| 1201 |
+
"</style>\n",
|
| 1202 |
+
"<table border=\"1\" class=\"dataframe\">\n",
|
| 1203 |
+
" <thead>\n",
|
| 1204 |
+
" <tr style=\"text-align: right;\">\n",
|
| 1205 |
+
" <th></th>\n",
|
| 1206 |
+
" <th>classifier</th>\n",
|
| 1207 |
+
" <th>T0</th>\n",
|
| 1208 |
+
" <th>T1</th>\n",
|
| 1209 |
+
" <th>T2</th>\n",
|
| 1210 |
+
" <th>T3</th>\n",
|
| 1211 |
+
" <th>T4</th>\n",
|
| 1212 |
+
" <th>T5</th>\n",
|
| 1213 |
+
" <th>T6</th>\n",
|
| 1214 |
+
" <th>T7</th>\n",
|
| 1215 |
+
" <th>T8</th>\n",
|
| 1216 |
+
" <th>...</th>\n",
|
| 1217 |
+
" <th>T90</th>\n",
|
| 1218 |
+
" <th>T91</th>\n",
|
| 1219 |
+
" <th>T92</th>\n",
|
| 1220 |
+
" <th>T93</th>\n",
|
| 1221 |
+
" <th>T94</th>\n",
|
| 1222 |
+
" <th>T95</th>\n",
|
| 1223 |
+
" <th>T96</th>\n",
|
| 1224 |
+
" <th>T97</th>\n",
|
| 1225 |
+
" <th>T98</th>\n",
|
| 1226 |
+
" <th>T99</th>\n",
|
| 1227 |
+
" </tr>\n",
|
| 1228 |
+
" </thead>\n",
|
| 1229 |
+
" <tbody>\n",
|
| 1230 |
+
" <tr>\n",
|
| 1231 |
+
" <th>accuracy</th>\n",
|
| 1232 |
+
" <td>0.787313</td>\n",
|
| 1233 |
+
" <td>0.764925</td>\n",
|
| 1234 |
+
" <td>0.764925</td>\n",
|
| 1235 |
+
" <td>0.779851</td>\n",
|
| 1236 |
+
" <td>0.750000</td>\n",
|
| 1237 |
+
" <td>0.783582</td>\n",
|
| 1238 |
+
" <td>0.779851</td>\n",
|
| 1239 |
+
" <td>0.783582</td>\n",
|
| 1240 |
+
" <td>0.791045</td>\n",
|
| 1241 |
+
" <td>0.787313</td>\n",
|
| 1242 |
+
" <td>...</td>\n",
|
| 1243 |
+
" <td>0.787313</td>\n",
|
| 1244 |
+
" <td>0.787313</td>\n",
|
| 1245 |
+
" <td>0.787313</td>\n",
|
| 1246 |
+
" <td>0.787313</td>\n",
|
| 1247 |
+
" <td>0.787313</td>\n",
|
| 1248 |
+
" <td>0.787313</td>\n",
|
| 1249 |
+
" <td>0.787313</td>\n",
|
| 1250 |
+
" <td>0.787313</td>\n",
|
| 1251 |
+
" <td>0.787313</td>\n",
|
| 1252 |
+
" <td>0.787313</td>\n",
|
| 1253 |
+
" </tr>\n",
|
| 1254 |
+
" <tr>\n",
|
| 1255 |
+
" <th>f1</th>\n",
|
| 1256 |
+
" <td>0.729858</td>\n",
|
| 1257 |
+
" <td>0.729614</td>\n",
|
| 1258 |
+
" <td>0.729614</td>\n",
|
| 1259 |
+
" <td>0.735426</td>\n",
|
| 1260 |
+
" <td>0.621469</td>\n",
|
| 1261 |
+
" <td>0.715686</td>\n",
|
| 1262 |
+
" <td>0.730594</td>\n",
|
| 1263 |
+
" <td>0.715686</td>\n",
|
| 1264 |
+
" <td>0.730769</td>\n",
|
| 1265 |
+
" <td>0.727273</td>\n",
|
| 1266 |
+
" <td>...</td>\n",
|
| 1267 |
+
" <td>0.729858</td>\n",
|
| 1268 |
+
" <td>0.729858</td>\n",
|
| 1269 |
+
" <td>0.729858</td>\n",
|
| 1270 |
+
" <td>0.727273</td>\n",
|
| 1271 |
+
" <td>0.729858</td>\n",
|
| 1272 |
+
" <td>0.729858</td>\n",
|
| 1273 |
+
" <td>0.727273</td>\n",
|
| 1274 |
+
" <td>0.729858</td>\n",
|
| 1275 |
+
" <td>0.727273</td>\n",
|
| 1276 |
+
" <td>0.729858</td>\n",
|
| 1277 |
+
" </tr>\n",
|
| 1278 |
+
" <tr>\n",
|
| 1279 |
+
" <th>statistical_parity_difference</th>\n",
|
| 1280 |
+
" <td>-0.814846</td>\n",
|
| 1281 |
+
" <td>-0.867052</td>\n",
|
| 1282 |
+
" <td>-0.867052</td>\n",
|
| 1283 |
+
" <td>-0.908549</td>\n",
|
| 1284 |
+
" <td>-0.489565</td>\n",
|
| 1285 |
+
" <td>-0.578096</td>\n",
|
| 1286 |
+
" <td>-0.947977</td>\n",
|
| 1287 |
+
" <td>-0.708549</td>\n",
|
| 1288 |
+
" <td>-0.799574</td>\n",
|
| 1289 |
+
" <td>-0.793794</td>\n",
|
| 1290 |
+
" <td>...</td>\n",
|
| 1291 |
+
" <td>-0.814846</td>\n",
|
| 1292 |
+
" <td>-0.814846</td>\n",
|
| 1293 |
+
" <td>-0.814846</td>\n",
|
| 1294 |
+
" <td>-0.793794</td>\n",
|
| 1295 |
+
" <td>-0.814846</td>\n",
|
| 1296 |
+
" <td>-0.814846</td>\n",
|
| 1297 |
+
" <td>-0.793794</td>\n",
|
| 1298 |
+
" <td>-0.814846</td>\n",
|
| 1299 |
+
" <td>-0.793794</td>\n",
|
| 1300 |
+
" <td>-0.814846</td>\n",
|
| 1301 |
+
" </tr>\n",
|
| 1302 |
+
" <tr>\n",
|
| 1303 |
+
" <th>equal_opportunity_difference</th>\n",
|
| 1304 |
+
" <td>-0.775214</td>\n",
|
| 1305 |
+
" <td>-0.731707</td>\n",
|
| 1306 |
+
" <td>-0.731707</td>\n",
|
| 1307 |
+
" <td>-0.766974</td>\n",
|
| 1308 |
+
" <td>-0.477917</td>\n",
|
| 1309 |
+
" <td>-0.531641</td>\n",
|
| 1310 |
+
" <td>-0.853659</td>\n",
|
| 1311 |
+
" <td>-0.759064</td>\n",
|
| 1312 |
+
" <td>-0.761701</td>\n",
|
| 1313 |
+
" <td>-0.761701</td>\n",
|
| 1314 |
+
" <td>...</td>\n",
|
| 1315 |
+
" <td>-0.775214</td>\n",
|
| 1316 |
+
" <td>-0.775214</td>\n",
|
| 1317 |
+
" <td>-0.775214</td>\n",
|
| 1318 |
+
" <td>-0.761701</td>\n",
|
| 1319 |
+
" <td>-0.775214</td>\n",
|
| 1320 |
+
" <td>-0.775214</td>\n",
|
| 1321 |
+
" <td>-0.761701</td>\n",
|
| 1322 |
+
" <td>-0.775214</td>\n",
|
| 1323 |
+
" <td>-0.761701</td>\n",
|
| 1324 |
+
" <td>-0.775214</td>\n",
|
| 1325 |
+
" </tr>\n",
|
| 1326 |
+
" <tr>\n",
|
| 1327 |
+
" <th>average_abs_odds_difference</th>\n",
|
| 1328 |
+
" <td>0.702001</td>\n",
|
| 1329 |
+
" <td>0.820399</td>\n",
|
| 1330 |
+
" <td>0.820399</td>\n",
|
| 1331 |
+
" <td>0.864548</td>\n",
|
| 1332 |
+
" <td>0.322833</td>\n",
|
| 1333 |
+
" <td>0.370799</td>\n",
|
| 1334 |
+
" <td>0.915466</td>\n",
|
| 1335 |
+
" <td>0.539705</td>\n",
|
| 1336 |
+
" <td>0.675223</td>\n",
|
| 1337 |
+
" <td>0.671435</td>\n",
|
| 1338 |
+
" <td>...</td>\n",
|
| 1339 |
+
" <td>0.702001</td>\n",
|
| 1340 |
+
" <td>0.702001</td>\n",
|
| 1341 |
+
" <td>0.702001</td>\n",
|
| 1342 |
+
" <td>0.671435</td>\n",
|
| 1343 |
+
" <td>0.702001</td>\n",
|
| 1344 |
+
" <td>0.702001</td>\n",
|
| 1345 |
+
" <td>0.671435</td>\n",
|
| 1346 |
+
" <td>0.702001</td>\n",
|
| 1347 |
+
" <td>0.671435</td>\n",
|
| 1348 |
+
" <td>0.702001</td>\n",
|
| 1349 |
+
" </tr>\n",
|
| 1350 |
+
" <tr>\n",
|
| 1351 |
+
" <th>disparate_impact</th>\n",
|
| 1352 |
+
" <td>-2.545325</td>\n",
|
| 1353 |
+
" <td>-2.017797</td>\n",
|
| 1354 |
+
" <td>-2.017797</td>\n",
|
| 1355 |
+
" <td>-2.503652</td>\n",
|
| 1356 |
+
" <td>-2.248073</td>\n",
|
| 1357 |
+
" <td>-1.713065</td>\n",
|
| 1358 |
+
" <td>-2.956067</td>\n",
|
| 1359 |
+
" <td>-2.277845</td>\n",
|
| 1360 |
+
" <td>-2.608239</td>\n",
|
| 1361 |
+
" <td>-2.521227</td>\n",
|
| 1362 |
+
" <td>...</td>\n",
|
| 1363 |
+
" <td>-2.545325</td>\n",
|
| 1364 |
+
" <td>-2.545325</td>\n",
|
| 1365 |
+
" <td>-2.545325</td>\n",
|
| 1366 |
+
" <td>-2.521227</td>\n",
|
| 1367 |
+
" <td>-2.545325</td>\n",
|
| 1368 |
+
" <td>-2.545325</td>\n",
|
| 1369 |
+
" <td>-2.521227</td>\n",
|
| 1370 |
+
" <td>-2.545325</td>\n",
|
| 1371 |
+
" <td>-2.521227</td>\n",
|
| 1372 |
+
" <td>-2.545325</td>\n",
|
| 1373 |
+
" </tr>\n",
|
| 1374 |
+
" <tr>\n",
|
| 1375 |
+
" <th>theil_index</th>\n",
|
| 1376 |
+
" <td>0.179316</td>\n",
|
| 1377 |
+
" <td>0.157679</td>\n",
|
| 1378 |
+
" <td>0.157679</td>\n",
|
| 1379 |
+
" <td>0.164565</td>\n",
|
| 1380 |
+
" <td>0.265484</td>\n",
|
| 1381 |
+
" <td>0.193705</td>\n",
|
| 1382 |
+
" <td>0.171370</td>\n",
|
| 1383 |
+
" <td>0.193705</td>\n",
|
| 1384 |
+
" <td>0.181456</td>\n",
|
| 1385 |
+
" <td>0.182624</td>\n",
|
| 1386 |
+
" <td>...</td>\n",
|
| 1387 |
+
" <td>0.179316</td>\n",
|
| 1388 |
+
" <td>0.179316</td>\n",
|
| 1389 |
+
" <td>0.179316</td>\n",
|
| 1390 |
+
" <td>0.182624</td>\n",
|
| 1391 |
+
" <td>0.179316</td>\n",
|
| 1392 |
+
" <td>0.179316</td>\n",
|
| 1393 |
+
" <td>0.182624</td>\n",
|
| 1394 |
+
" <td>0.179316</td>\n",
|
| 1395 |
+
" <td>0.182624</td>\n",
|
| 1396 |
+
" <td>0.179316</td>\n",
|
| 1397 |
+
" </tr>\n",
|
| 1398 |
+
" </tbody>\n",
|
| 1399 |
+
"</table>\n",
|
| 1400 |
+
"<p>7 rows × 101 columns</p>\n",
|
| 1401 |
+
"</div>"
|
| 1402 |
+
],
|
| 1403 |
+
"text/plain": [
|
| 1404 |
+
" classifier T0 T1 T2 \\\n",
|
| 1405 |
+
"accuracy 0.787313 0.764925 0.764925 0.779851 \n",
|
| 1406 |
+
"f1 0.729858 0.729614 0.729614 0.735426 \n",
|
| 1407 |
+
"statistical_parity_difference -0.814846 -0.867052 -0.867052 -0.908549 \n",
|
| 1408 |
+
"equal_opportunity_difference -0.775214 -0.731707 -0.731707 -0.766974 \n",
|
| 1409 |
+
"average_abs_odds_difference 0.702001 0.820399 0.820399 0.864548 \n",
|
| 1410 |
+
"disparate_impact -2.545325 -2.017797 -2.017797 -2.503652 \n",
|
| 1411 |
+
"theil_index 0.179316 0.157679 0.157679 0.164565 \n",
|
| 1412 |
+
"\n",
|
| 1413 |
+
" T3 T4 T5 T6 \\\n",
|
| 1414 |
+
"accuracy 0.750000 0.783582 0.779851 0.783582 \n",
|
| 1415 |
+
"f1 0.621469 0.715686 0.730594 0.715686 \n",
|
| 1416 |
+
"statistical_parity_difference -0.489565 -0.578096 -0.947977 -0.708549 \n",
|
| 1417 |
+
"equal_opportunity_difference -0.477917 -0.531641 -0.853659 -0.759064 \n",
|
| 1418 |
+
"average_abs_odds_difference 0.322833 0.370799 0.915466 0.539705 \n",
|
| 1419 |
+
"disparate_impact -2.248073 -1.713065 -2.956067 -2.277845 \n",
|
| 1420 |
+
"theil_index 0.265484 0.193705 0.171370 0.193705 \n",
|
| 1421 |
+
"\n",
|
| 1422 |
+
" T7 T8 ... T90 T91 \\\n",
|
| 1423 |
+
"accuracy 0.791045 0.787313 ... 0.787313 0.787313 \n",
|
| 1424 |
+
"f1 0.730769 0.727273 ... 0.729858 0.729858 \n",
|
| 1425 |
+
"statistical_parity_difference -0.799574 -0.793794 ... -0.814846 -0.814846 \n",
|
| 1426 |
+
"equal_opportunity_difference -0.761701 -0.761701 ... -0.775214 -0.775214 \n",
|
| 1427 |
+
"average_abs_odds_difference 0.675223 0.671435 ... 0.702001 0.702001 \n",
|
| 1428 |
+
"disparate_impact -2.608239 -2.521227 ... -2.545325 -2.545325 \n",
|
| 1429 |
+
"theil_index 0.181456 0.182624 ... 0.179316 0.179316 \n",
|
| 1430 |
+
"\n",
|
| 1431 |
+
" T92 T93 T94 T95 \\\n",
|
| 1432 |
+
"accuracy 0.787313 0.787313 0.787313 0.787313 \n",
|
| 1433 |
+
"f1 0.729858 0.727273 0.729858 0.729858 \n",
|
| 1434 |
+
"statistical_parity_difference -0.814846 -0.793794 -0.814846 -0.814846 \n",
|
| 1435 |
+
"equal_opportunity_difference -0.775214 -0.761701 -0.775214 -0.775214 \n",
|
| 1436 |
+
"average_abs_odds_difference 0.702001 0.671435 0.702001 0.702001 \n",
|
| 1437 |
+
"disparate_impact -2.545325 -2.521227 -2.545325 -2.545325 \n",
|
| 1438 |
+
"theil_index 0.179316 0.182624 0.179316 0.179316 \n",
|
| 1439 |
+
"\n",
|
| 1440 |
+
" T96 T97 T98 T99 \n",
|
| 1441 |
+
"accuracy 0.787313 0.787313 0.787313 0.787313 \n",
|
| 1442 |
+
"f1 0.727273 0.729858 0.727273 0.729858 \n",
|
| 1443 |
+
"statistical_parity_difference -0.793794 -0.814846 -0.793794 -0.814846 \n",
|
| 1444 |
+
"equal_opportunity_difference -0.761701 -0.775214 -0.761701 -0.775214 \n",
|
| 1445 |
+
"average_abs_odds_difference 0.671435 0.702001 0.671435 0.702001 \n",
|
| 1446 |
+
"disparate_impact -2.521227 -2.545325 -2.521227 -2.545325 \n",
|
| 1447 |
+
"theil_index 0.182624 0.179316 0.182624 0.179316 \n",
|
| 1448 |
+
"\n",
|
| 1449 |
+
"[7 rows x 101 columns]"
|
| 1450 |
+
]
|
| 1451 |
+
},
|
| 1452 |
+
"execution_count": 43,
|
| 1453 |
+
"metadata": {},
|
| 1454 |
+
"output_type": "execute_result"
|
| 1455 |
+
}
|
| 1456 |
+
],
|
| 1457 |
+
"source": [
|
| 1458 |
+
"import numpy as np\n",
|
| 1459 |
+
"final_result = pd.DataFrame(final_metrics)\n",
|
| 1460 |
+
"final_result[4] = np.log(final_result[4])\n",
|
| 1461 |
+
"final_result = final_result.transpose()\n",
|
| 1462 |
+
"final_result.loc[0] = f1 # add f1 and acc to df\n",
|
| 1463 |
+
"acc = pd.DataFrame(accuracy).transpose()\n",
|
| 1464 |
+
"acc = acc.rename(index={0: 'accuracy'})\n",
|
| 1465 |
+
"final_result = pd.concat([acc,final_result])\n",
|
| 1466 |
+
"final_result = final_result.rename(index={0: 'f1', 1: 'statistical_parity_difference', 2: 'equal_opportunity_difference', 3: 'average_abs_odds_difference', 4: 'disparate_impact', 5: 'theil_index'})\n",
|
| 1467 |
+
"final_result.columns = ['T' + str(col) for col in final_result.columns]\n",
|
| 1468 |
+
"final_result.insert(0, \"classifier\", final_result['T' + str(num_estimators - 1)]) ##Add final metrics add the beginning of the df\n",
|
| 1469 |
+
"final_result.to_csv('../../Results/AdaBoost/' + nb_fname + '.csv')\n",
|
| 1470 |
+
"final_result"
|
| 1471 |
+
]
|
| 1472 |
+
},
|
| 1473 |
+
{
|
| 1474 |
+
"cell_type": "code",
|
| 1475 |
+
"execution_count": null,
|
| 1476 |
+
"metadata": {},
|
| 1477 |
+
"outputs": [],
|
| 1478 |
+
"source": []
|
| 1479 |
+
}
|
| 1480 |
+
],
|
| 1481 |
+
"metadata": {
|
| 1482 |
+
"_change_revision": 2,
|
| 1483 |
+
"_is_fork": false,
|
| 1484 |
+
"kernelspec": {
|
| 1485 |
+
"display_name": "Python 3",
|
| 1486 |
+
"language": "python",
|
| 1487 |
+
"name": "python3"
|
| 1488 |
+
},
|
| 1489 |
+
"language_info": {
|
| 1490 |
+
"codemirror_mode": {
|
| 1491 |
+
"name": "ipython",
|
| 1492 |
+
"version": 3
|
| 1493 |
+
},
|
| 1494 |
+
"file_extension": ".py",
|
| 1495 |
+
"mimetype": "text/x-python",
|
| 1496 |
+
"name": "python",
|
| 1497 |
+
"nbconvert_exporter": "python",
|
| 1498 |
+
"pygments_lexer": "ipython3",
|
| 1499 |
+
"version": "3.8.5"
|
| 1500 |
+
}
|
| 1501 |
+
},
|
| 1502 |
+
"nbformat": 4,
|
| 1503 |
+
"nbformat_minor": 1
|
| 1504 |
+
}
|
Titanic/Kernels/AdaBoost/.ipynb_checkpoints/7-titanic-survival-prediction-end-to-end-ml-pipeline-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/.ipynb_checkpoints/9-titanic-top-solution-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/10-a-comprehensive-guide-to-titanic-machine-learning.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/10-a-comprehensive-guide-to-titanic-machine-learning.py
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/2-titanic-top-4-with-ensemble-modeling.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/2-titanic-top-4-with-ensemble-modeling.py
ADDED
|
@@ -0,0 +1,1110 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python
|
| 2 |
+
# coding: utf-8
|
| 3 |
+
|
| 4 |
+
# # Titanic Top 4% with ensemble modeling
|
| 5 |
+
# ### **Yassine Ghouzam, PhD**
|
| 6 |
+
# #### 13/07/2017
|
| 7 |
+
#
|
| 8 |
+
# * **1 Introduction**
|
| 9 |
+
# * **2 Load and check data**
|
| 10 |
+
# * 2.1 load data
|
| 11 |
+
# * 2.2 Outlier detection
|
| 12 |
+
# * 2.3 joining train and test set
|
| 13 |
+
# * 2.4 check for null and missing values
|
| 14 |
+
# * **3 Feature analysis**
|
| 15 |
+
# * 3.1 Numerical values
|
| 16 |
+
# * 3.2 Categorical values
|
| 17 |
+
# * **4 Filling missing Values**
|
| 18 |
+
# * 4.1 Age
|
| 19 |
+
# * **5 Feature engineering**
|
| 20 |
+
# * 5.1 Name/Title
|
| 21 |
+
# * 5.2 Family Size
|
| 22 |
+
# * 5.3 Cabin
|
| 23 |
+
# * 5.4 Ticket
|
| 24 |
+
# * **6 Modeling**
|
| 25 |
+
# * 6.1 Simple modeling
|
| 26 |
+
# * 6.1.1 Cross validate models
|
| 27 |
+
# * 6.1.2 Hyperparamater tunning for best models
|
| 28 |
+
# * 6.1.3 Plot learning curves
|
| 29 |
+
# * 6.1.4 Feature importance of the tree based classifiers
|
| 30 |
+
# * 6.2 Ensemble modeling
|
| 31 |
+
# * 6.2.1 Combining models
|
| 32 |
+
# * 6.3 Prediction
|
| 33 |
+
# * 6.3.1 Predict and Submit results
|
| 34 |
+
#
|
| 35 |
+
|
| 36 |
+
# ## 1. Introduction
|
| 37 |
+
#
|
| 38 |
+
# This is my first kernel at Kaggle. I choosed the Titanic competition which is a good way to introduce feature engineering and ensemble modeling. Firstly, I will display some feature analyses then ill focus on the feature engineering. Last part concerns modeling and predicting the survival on the Titanic using an voting procedure.
|
| 39 |
+
#
|
| 40 |
+
# This script follows three main parts:
|
| 41 |
+
#
|
| 42 |
+
# * **Feature analysis**
|
| 43 |
+
# * **Feature engineering**
|
| 44 |
+
# * **Modeling**
|
| 45 |
+
|
| 46 |
+
# In[1]:
|
| 47 |
+
|
| 48 |
+
|
| 49 |
+
import pandas as pd
|
| 50 |
+
import numpy as np
|
| 51 |
+
import matplotlib.pyplot as plt
|
| 52 |
+
import seaborn as sns
|
| 53 |
+
get_ipython().run_line_magic('matplotlib', 'inline')
|
| 54 |
+
|
| 55 |
+
from collections import Counter
|
| 56 |
+
|
| 57 |
+
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, GradientBoostingClassifier, ExtraTreesClassifier, VotingClassifier
|
| 58 |
+
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
|
| 59 |
+
from sklearn.linear_model import LogisticRegression
|
| 60 |
+
from sklearn.neighbors import KNeighborsClassifier
|
| 61 |
+
from sklearn.tree import DecisionTreeClassifier
|
| 62 |
+
from sklearn.neural_network import MLPClassifier
|
| 63 |
+
from sklearn.svm import SVC
|
| 64 |
+
from sklearn.model_selection import GridSearchCV, cross_val_score, StratifiedKFold, learning_curve
|
| 65 |
+
|
| 66 |
+
sns.set(style='white', context='notebook', palette='deep')
|
| 67 |
+
|
| 68 |
+
|
| 69 |
+
# ## 2. Load and check data
|
| 70 |
+
# ### 2.1 Load data
|
| 71 |
+
|
| 72 |
+
# In[2]:
|
| 73 |
+
|
| 74 |
+
|
| 75 |
+
# Load data
|
| 76 |
+
##### Load train and Test set
|
| 77 |
+
|
| 78 |
+
train = pd.read_csv("../input/train.csv")
|
| 79 |
+
test = pd.read_csv("../input/test.csv")
|
| 80 |
+
IDtest = test["PassengerId"]
|
| 81 |
+
|
| 82 |
+
|
| 83 |
+
# ### 2.2 Outlier detection
|
| 84 |
+
|
| 85 |
+
# In[3]:
|
| 86 |
+
|
| 87 |
+
|
| 88 |
+
# Outlier detection
|
| 89 |
+
|
| 90 |
+
def detect_outliers(df,n,features):
|
| 91 |
+
"""
|
| 92 |
+
Takes a dataframe df of features and returns a list of the indices
|
| 93 |
+
corresponding to the observations containing more than n outliers according
|
| 94 |
+
to the Tukey method.
|
| 95 |
+
"""
|
| 96 |
+
outlier_indices = []
|
| 97 |
+
|
| 98 |
+
# iterate over features(columns)
|
| 99 |
+
for col in features:
|
| 100 |
+
# 1st quartile (25%)
|
| 101 |
+
Q1 = np.percentile(df[col], 25)
|
| 102 |
+
# 3rd quartile (75%)
|
| 103 |
+
Q3 = np.percentile(df[col],75)
|
| 104 |
+
# Interquartile range (IQR)
|
| 105 |
+
IQR = Q3 - Q1
|
| 106 |
+
|
| 107 |
+
# outlier step
|
| 108 |
+
outlier_step = 1.5 * IQR
|
| 109 |
+
|
| 110 |
+
# Determine a list of indices of outliers for feature col
|
| 111 |
+
outlier_list_col = df[(df[col] < Q1 - outlier_step) | (df[col] > Q3 + outlier_step )].index
|
| 112 |
+
|
| 113 |
+
# append the found outlier indices for col to the list of outlier indices
|
| 114 |
+
outlier_indices.extend(outlier_list_col)
|
| 115 |
+
|
| 116 |
+
# select observations containing more than 2 outliers
|
| 117 |
+
outlier_indices = Counter(outlier_indices)
|
| 118 |
+
multiple_outliers = list( k for k, v in outlier_indices.items() if v > n )
|
| 119 |
+
|
| 120 |
+
return multiple_outliers
|
| 121 |
+
|
| 122 |
+
# detect outliers from Age, SibSp , Parch and Fare
|
| 123 |
+
Outliers_to_drop = detect_outliers(train,2,["Age","SibSp","Parch","Fare"])
|
| 124 |
+
|
| 125 |
+
|
| 126 |
+
# Since outliers can have a dramatic effect on the prediction (espacially for regression problems), i choosed to manage them.
|
| 127 |
+
#
|
| 128 |
+
# I used the Tukey method (Tukey JW., 1977) to detect ouliers which defines an interquartile range comprised between the 1st and 3rd quartile of the distribution values (IQR). An outlier is a row that have a feature value outside the (IQR +- an outlier step).
|
| 129 |
+
#
|
| 130 |
+
#
|
| 131 |
+
# I decided to detect outliers from the numerical values features (Age, SibSp, Sarch and Fare). Then, i considered outliers as rows that have at least two outlied numerical values.
|
| 132 |
+
|
| 133 |
+
# In[4]:
|
| 134 |
+
|
| 135 |
+
|
| 136 |
+
train.loc[Outliers_to_drop] # Show the outliers rows
|
| 137 |
+
|
| 138 |
+
|
| 139 |
+
# We detect 10 outliers. The 28, 89 and 342 passenger have an high Ticket Fare
|
| 140 |
+
#
|
| 141 |
+
# The 7 others have very high values of SibSP.
|
| 142 |
+
|
| 143 |
+
# In[5]:
|
| 144 |
+
|
| 145 |
+
|
| 146 |
+
# Drop outliers
|
| 147 |
+
train = train.drop(Outliers_to_drop, axis = 0).reset_index(drop=True)
|
| 148 |
+
|
| 149 |
+
|
| 150 |
+
# ### 2.3 joining train and test set
|
| 151 |
+
|
| 152 |
+
# In[6]:
|
| 153 |
+
|
| 154 |
+
|
| 155 |
+
## Join train and test datasets in order to obtain the same number of features during categorical conversion
|
| 156 |
+
train_len = len(train)
|
| 157 |
+
dataset = pd.concat(objs=[train, test], axis=0).reset_index(drop=True)
|
| 158 |
+
|
| 159 |
+
|
| 160 |
+
# I join train and test datasets to obtain the same number of features during categorical conversion (See feature engineering).
|
| 161 |
+
|
| 162 |
+
# ### 2.4 check for null and missing values
|
| 163 |
+
|
| 164 |
+
# In[7]:
|
| 165 |
+
|
| 166 |
+
|
| 167 |
+
# Fill empty and NaNs values with NaN
|
| 168 |
+
dataset = dataset.fillna(np.nan)
|
| 169 |
+
|
| 170 |
+
# Check for Null values
|
| 171 |
+
dataset.isnull().sum()
|
| 172 |
+
|
| 173 |
+
|
| 174 |
+
# Age and Cabin features have an important part of missing values.
|
| 175 |
+
#
|
| 176 |
+
# **Survived missing values correspond to the join testing dataset (Survived column doesn't exist in test set and has been replace by NaN values when concatenating the train and test set)**
|
| 177 |
+
|
| 178 |
+
# In[8]:
|
| 179 |
+
|
| 180 |
+
|
| 181 |
+
# Infos
|
| 182 |
+
train.info()
|
| 183 |
+
train.isnull().sum()
|
| 184 |
+
|
| 185 |
+
|
| 186 |
+
# In[9]:
|
| 187 |
+
|
| 188 |
+
|
| 189 |
+
train.head()
|
| 190 |
+
|
| 191 |
+
|
| 192 |
+
# In[10]:
|
| 193 |
+
|
| 194 |
+
|
| 195 |
+
train.dtypes
|
| 196 |
+
|
| 197 |
+
|
| 198 |
+
# In[11]:
|
| 199 |
+
|
| 200 |
+
|
| 201 |
+
### Summarize data
|
| 202 |
+
# Summarie and statistics
|
| 203 |
+
train.describe()
|
| 204 |
+
|
| 205 |
+
|
| 206 |
+
# ## 3. Feature analysis
|
| 207 |
+
# ### 3.1 Numerical values
|
| 208 |
+
|
| 209 |
+
# In[12]:
|
| 210 |
+
|
| 211 |
+
|
| 212 |
+
# Correlation matrix between numerical values (SibSp Parch Age and Fare values) and Survived
|
| 213 |
+
g = sns.heatmap(train[["Survived","SibSp","Parch","Age","Fare"]].corr(),annot=True, fmt = ".2f", cmap = "coolwarm")
|
| 214 |
+
|
| 215 |
+
|
| 216 |
+
# Only Fare feature seems to have a significative correlation with the survival probability.
|
| 217 |
+
#
|
| 218 |
+
# It doesn't mean that the other features are not usefull. Subpopulations in these features can be correlated with the survival. To determine this, we need to explore in detail these features
|
| 219 |
+
|
| 220 |
+
# #### SibSP
|
| 221 |
+
|
| 222 |
+
# In[13]:
|
| 223 |
+
|
| 224 |
+
|
| 225 |
+
# Explore SibSp feature vs Survived
|
| 226 |
+
g = sns.factorplot(x="SibSp",y="Survived",data=train,kind="bar", size = 6 ,
|
| 227 |
+
palette = "muted")
|
| 228 |
+
g.despine(left=True)
|
| 229 |
+
g = g.set_ylabels("survival probability")
|
| 230 |
+
|
| 231 |
+
|
| 232 |
+
# It seems that passengers having a lot of siblings/spouses have less chance to survive
|
| 233 |
+
#
|
| 234 |
+
# Single passengers (0 SibSP) or with two other persons (SibSP 1 or 2) have more chance to survive
|
| 235 |
+
#
|
| 236 |
+
# This observation is quite interesting, we can consider a new feature describing these categories (See feature engineering)
|
| 237 |
+
|
| 238 |
+
# #### Parch
|
| 239 |
+
|
| 240 |
+
# In[14]:
|
| 241 |
+
|
| 242 |
+
|
| 243 |
+
# Explore Parch feature vs Survived
|
| 244 |
+
g = sns.factorplot(x="Parch",y="Survived",data=train,kind="bar", size = 6 ,
|
| 245 |
+
palette = "muted")
|
| 246 |
+
g.despine(left=True)
|
| 247 |
+
g = g.set_ylabels("survival probability")
|
| 248 |
+
|
| 249 |
+
|
| 250 |
+
# Small families have more chance to survive, more than single (Parch 0), medium (Parch 3,4) and large families (Parch 5,6 ).
|
| 251 |
+
#
|
| 252 |
+
# Be carefull there is an important standard deviation in the survival of passengers with 3 parents/children
|
| 253 |
+
|
| 254 |
+
# #### Age
|
| 255 |
+
|
| 256 |
+
# In[15]:
|
| 257 |
+
|
| 258 |
+
|
| 259 |
+
# Explore Age vs Survived
|
| 260 |
+
g = sns.FacetGrid(train, col='Survived')
|
| 261 |
+
g = g.map(sns.distplot, "Age")
|
| 262 |
+
|
| 263 |
+
|
| 264 |
+
# Age distribution seems to be a tailed distribution, maybe a gaussian distribution.
|
| 265 |
+
#
|
| 266 |
+
# We notice that age distributions are not the same in the survived and not survived subpopulations. Indeed, there is a peak corresponding to young passengers, that have survived. We also see that passengers between 60-80 have less survived.
|
| 267 |
+
#
|
| 268 |
+
# So, even if "Age" is not correlated with "Survived", we can see that there is age categories of passengers that of have more or less chance to survive.
|
| 269 |
+
#
|
| 270 |
+
# It seems that very young passengers have more chance to survive.
|
| 271 |
+
|
| 272 |
+
# In[16]:
|
| 273 |
+
|
| 274 |
+
|
| 275 |
+
# Explore Age distibution
|
| 276 |
+
g = sns.kdeplot(train["Age"][(train["Survived"] == 0) & (train["Age"].notnull())], color="Red", shade = True)
|
| 277 |
+
g = sns.kdeplot(train["Age"][(train["Survived"] == 1) & (train["Age"].notnull())], ax =g, color="Blue", shade= True)
|
| 278 |
+
g.set_xlabel("Age")
|
| 279 |
+
g.set_ylabel("Frequency")
|
| 280 |
+
g = g.legend(["Not Survived","Survived"])
|
| 281 |
+
|
| 282 |
+
|
| 283 |
+
# When we superimpose the two densities , we cleary see a peak correponsing (between 0 and 5) to babies and very young childrens.
|
| 284 |
+
|
| 285 |
+
# #### Fare
|
| 286 |
+
|
| 287 |
+
# In[17]:
|
| 288 |
+
|
| 289 |
+
|
| 290 |
+
dataset["Fare"].isnull().sum()
|
| 291 |
+
|
| 292 |
+
|
| 293 |
+
# In[18]:
|
| 294 |
+
|
| 295 |
+
|
| 296 |
+
#Fill Fare missing values with the median value
|
| 297 |
+
dataset["Fare"] = dataset["Fare"].fillna(dataset["Fare"].median())
|
| 298 |
+
|
| 299 |
+
|
| 300 |
+
# Since we have one missing value , i decided to fill it with the median value which will not have an important effect on the prediction.
|
| 301 |
+
|
| 302 |
+
# In[19]:
|
| 303 |
+
|
| 304 |
+
|
| 305 |
+
# Explore Fare distribution
|
| 306 |
+
g = sns.distplot(dataset["Fare"], color="m", label="Skewness : %.2f"%(dataset["Fare"].skew()))
|
| 307 |
+
g = g.legend(loc="best")
|
| 308 |
+
|
| 309 |
+
|
| 310 |
+
# As we can see, Fare distribution is very skewed. This can lead to overweigth very high values in the model, even if it is scaled.
|
| 311 |
+
#
|
| 312 |
+
# In this case, it is better to transform it with the log function to reduce this skew.
|
| 313 |
+
|
| 314 |
+
# In[20]:
|
| 315 |
+
|
| 316 |
+
|
| 317 |
+
# Apply log to Fare to reduce skewness distribution
|
| 318 |
+
dataset["Fare"] = dataset["Fare"].map(lambda i: np.log(i) if i > 0 else 0)
|
| 319 |
+
|
| 320 |
+
|
| 321 |
+
# In[21]:
|
| 322 |
+
|
| 323 |
+
|
| 324 |
+
g = sns.distplot(dataset["Fare"], color="b", label="Skewness : %.2f"%(dataset["Fare"].skew()))
|
| 325 |
+
g = g.legend(loc="best")
|
| 326 |
+
|
| 327 |
+
|
| 328 |
+
# Skewness is clearly reduced after the log transformation
|
| 329 |
+
|
| 330 |
+
# ### 3.2 Categorical values
|
| 331 |
+
# #### Sex
|
| 332 |
+
|
| 333 |
+
# In[22]:
|
| 334 |
+
|
| 335 |
+
|
| 336 |
+
g = sns.barplot(x="Sex",y="Survived",data=train)
|
| 337 |
+
g = g.set_ylabel("Survival Probability")
|
| 338 |
+
|
| 339 |
+
|
| 340 |
+
# In[23]:
|
| 341 |
+
|
| 342 |
+
|
| 343 |
+
train[["Sex","Survived"]].groupby('Sex').mean()
|
| 344 |
+
|
| 345 |
+
|
| 346 |
+
# It is clearly obvious that Male have less chance to survive than Female.
|
| 347 |
+
#
|
| 348 |
+
# So Sex, might play an important role in the prediction of the survival.
|
| 349 |
+
#
|
| 350 |
+
# For those who have seen the Titanic movie (1997), I am sure, we all remember this sentence during the evacuation : "Women and children first".
|
| 351 |
+
|
| 352 |
+
# #### Pclass
|
| 353 |
+
|
| 354 |
+
# In[24]:
|
| 355 |
+
|
| 356 |
+
|
| 357 |
+
# Explore Pclass vs Survived
|
| 358 |
+
g = sns.factorplot(x="Pclass",y="Survived",data=train,kind="bar", size = 6 ,
|
| 359 |
+
palette = "muted")
|
| 360 |
+
g.despine(left=True)
|
| 361 |
+
g = g.set_ylabels("survival probability")
|
| 362 |
+
|
| 363 |
+
|
| 364 |
+
# In[25]:
|
| 365 |
+
|
| 366 |
+
|
| 367 |
+
# Explore Pclass vs Survived by Sex
|
| 368 |
+
g = sns.factorplot(x="Pclass", y="Survived", hue="Sex", data=train,
|
| 369 |
+
size=6, kind="bar", palette="muted")
|
| 370 |
+
g.despine(left=True)
|
| 371 |
+
g = g.set_ylabels("survival probability")
|
| 372 |
+
|
| 373 |
+
|
| 374 |
+
# The passenger survival is not the same in the 3 classes. First class passengers have more chance to survive than second class and third class passengers.
|
| 375 |
+
#
|
| 376 |
+
# This trend is conserved when we look at both male and female passengers.
|
| 377 |
+
|
| 378 |
+
# #### Embarked
|
| 379 |
+
|
| 380 |
+
# In[26]:
|
| 381 |
+
|
| 382 |
+
|
| 383 |
+
dataset["Embarked"].isnull().sum()
|
| 384 |
+
|
| 385 |
+
|
| 386 |
+
# In[27]:
|
| 387 |
+
|
| 388 |
+
|
| 389 |
+
#Fill Embarked nan values of dataset set with 'S' most frequent value
|
| 390 |
+
dataset["Embarked"] = dataset["Embarked"].fillna("S")
|
| 391 |
+
|
| 392 |
+
|
| 393 |
+
# Since we have two missing values , i decided to fill them with the most fequent value of "Embarked" (S).
|
| 394 |
+
|
| 395 |
+
# In[28]:
|
| 396 |
+
|
| 397 |
+
|
| 398 |
+
# Explore Embarked vs Survived
|
| 399 |
+
g = sns.factorplot(x="Embarked", y="Survived", data=train,
|
| 400 |
+
size=6, kind="bar", palette="muted")
|
| 401 |
+
g.despine(left=True)
|
| 402 |
+
g = g.set_ylabels("survival probability")
|
| 403 |
+
|
| 404 |
+
|
| 405 |
+
# It seems that passenger coming from Cherbourg (C) have more chance to survive.
|
| 406 |
+
#
|
| 407 |
+
# My hypothesis is that the proportion of first class passengers is higher for those who came from Cherbourg than Queenstown (Q), Southampton (S).
|
| 408 |
+
#
|
| 409 |
+
# Let's see the Pclass distribution vs Embarked
|
| 410 |
+
|
| 411 |
+
# In[29]:
|
| 412 |
+
|
| 413 |
+
|
| 414 |
+
# Explore Pclass vs Embarked
|
| 415 |
+
g = sns.factorplot("Pclass", col="Embarked", data=train,
|
| 416 |
+
size=6, kind="count", palette="muted")
|
| 417 |
+
g.despine(left=True)
|
| 418 |
+
g = g.set_ylabels("Count")
|
| 419 |
+
|
| 420 |
+
|
| 421 |
+
# Indeed, the third class is the most frequent for passenger coming from Southampton (S) and Queenstown (Q), whereas Cherbourg passengers are mostly in first class which have the highest survival rate.
|
| 422 |
+
#
|
| 423 |
+
# At this point, i can't explain why first class has an higher survival rate. My hypothesis is that first class passengers were prioritised during the evacuation due to their influence.
|
| 424 |
+
|
| 425 |
+
# ## 4. Filling missing Values
|
| 426 |
+
# ### 4.1 Age
|
| 427 |
+
#
|
| 428 |
+
# As we see, Age column contains 256 missing values in the whole dataset.
|
| 429 |
+
#
|
| 430 |
+
# Since there is subpopulations that have more chance to survive (children for example), it is preferable to keep the age feature and to impute the missing values.
|
| 431 |
+
#
|
| 432 |
+
# To adress this problem, i looked at the most correlated features with Age (Sex, Parch , Pclass and SibSP).
|
| 433 |
+
|
| 434 |
+
# In[30]:
|
| 435 |
+
|
| 436 |
+
|
| 437 |
+
# Explore Age vs Sex, Parch , Pclass and SibSP
|
| 438 |
+
g = sns.factorplot(y="Age",x="Sex",data=dataset,kind="box")
|
| 439 |
+
g = sns.factorplot(y="Age",x="Sex",hue="Pclass", data=dataset,kind="box")
|
| 440 |
+
g = sns.factorplot(y="Age",x="Parch", data=dataset,kind="box")
|
| 441 |
+
g = sns.factorplot(y="Age",x="SibSp", data=dataset,kind="box")
|
| 442 |
+
|
| 443 |
+
|
| 444 |
+
# Age distribution seems to be the same in Male and Female subpopulations, so Sex is not informative to predict Age.
|
| 445 |
+
#
|
| 446 |
+
# However, 1rst class passengers are older than 2nd class passengers who are also older than 3rd class passengers.
|
| 447 |
+
#
|
| 448 |
+
# Moreover, the more a passenger has parents/children the older he is and the more a passenger has siblings/spouses the younger he is.
|
| 449 |
+
|
| 450 |
+
# In[31]:
|
| 451 |
+
|
| 452 |
+
|
| 453 |
+
# convert Sex into categorical value 0 for male and 1 for female
|
| 454 |
+
dataset["Sex"] = dataset["Sex"].map({"male": 0, "female":1})
|
| 455 |
+
|
| 456 |
+
|
| 457 |
+
# In[32]:
|
| 458 |
+
|
| 459 |
+
|
| 460 |
+
g = sns.heatmap(dataset[["Age","Sex","SibSp","Parch","Pclass"]].corr(),cmap="BrBG",annot=True)
|
| 461 |
+
|
| 462 |
+
|
| 463 |
+
# The correlation map confirms the factorplots observations except for Parch. Age is not correlated with Sex, but is negatively correlated with Pclass, Parch and SibSp.
|
| 464 |
+
#
|
| 465 |
+
# In the plot of Age in function of Parch, Age is growing with the number of parents / children. But the general correlation is negative.
|
| 466 |
+
#
|
| 467 |
+
# So, i decided to use SibSP, Parch and Pclass in order to impute the missing ages.
|
| 468 |
+
#
|
| 469 |
+
# The strategy is to fill Age with the median age of similar rows according to Pclass, Parch and SibSp.
|
| 470 |
+
|
| 471 |
+
# In[33]:
|
| 472 |
+
|
| 473 |
+
|
| 474 |
+
# Filling missing value of Age
|
| 475 |
+
|
| 476 |
+
## Fill Age with the median age of similar rows according to Pclass, Parch and SibSp
|
| 477 |
+
# Index of NaN age rows
|
| 478 |
+
index_NaN_age = list(dataset["Age"][dataset["Age"].isnull()].index)
|
| 479 |
+
|
| 480 |
+
for i in index_NaN_age :
|
| 481 |
+
age_med = dataset["Age"].median()
|
| 482 |
+
age_pred = dataset["Age"][((dataset['SibSp'] == dataset.iloc[i]["SibSp"]) & (dataset['Parch'] == dataset.iloc[i]["Parch"]) & (dataset['Pclass'] == dataset.iloc[i]["Pclass"]))].median()
|
| 483 |
+
if not np.isnan(age_pred) :
|
| 484 |
+
dataset['Age'].iloc[i] = age_pred
|
| 485 |
+
else :
|
| 486 |
+
dataset['Age'].iloc[i] = age_med
|
| 487 |
+
|
| 488 |
+
|
| 489 |
+
# In[34]:
|
| 490 |
+
|
| 491 |
+
|
| 492 |
+
g = sns.factorplot(x="Survived", y = "Age",data = train, kind="box")
|
| 493 |
+
g = sns.factorplot(x="Survived", y = "Age",data = train, kind="violin")
|
| 494 |
+
|
| 495 |
+
|
| 496 |
+
# No difference between median value of age in survived and not survived subpopulation.
|
| 497 |
+
#
|
| 498 |
+
# But in the violin plot of survived passengers, we still notice that very young passengers have higher survival rate.
|
| 499 |
+
|
| 500 |
+
# ## 5. Feature engineering
|
| 501 |
+
# ### 5.1 Name/Title
|
| 502 |
+
|
| 503 |
+
# In[35]:
|
| 504 |
+
|
| 505 |
+
|
| 506 |
+
dataset["Name"].head()
|
| 507 |
+
|
| 508 |
+
|
| 509 |
+
# The Name feature contains information on passenger's title.
|
| 510 |
+
#
|
| 511 |
+
# Since some passenger with distingused title may be preferred during the evacuation, it is interesting to add them to the model.
|
| 512 |
+
|
| 513 |
+
# In[36]:
|
| 514 |
+
|
| 515 |
+
|
| 516 |
+
# Get Title from Name
|
| 517 |
+
dataset_title = [i.split(",")[1].split(".")[0].strip() for i in dataset["Name"]]
|
| 518 |
+
dataset["Title"] = pd.Series(dataset_title)
|
| 519 |
+
dataset["Title"].head()
|
| 520 |
+
|
| 521 |
+
|
| 522 |
+
# In[37]:
|
| 523 |
+
|
| 524 |
+
|
| 525 |
+
g = sns.countplot(x="Title",data=dataset)
|
| 526 |
+
g = plt.setp(g.get_xticklabels(), rotation=45)
|
| 527 |
+
|
| 528 |
+
|
| 529 |
+
# There is 17 titles in the dataset, most of them are very rare and we can group them in 4 categories.
|
| 530 |
+
|
| 531 |
+
# In[38]:
|
| 532 |
+
|
| 533 |
+
|
| 534 |
+
# Convert to categorical values Title
|
| 535 |
+
dataset["Title"] = dataset["Title"].replace(['Lady', 'the Countess','Countess','Capt', 'Col','Don', 'Dr', 'Major', 'Rev', 'Sir', 'Jonkheer', 'Dona'], 'Rare')
|
| 536 |
+
dataset["Title"] = dataset["Title"].map({"Master":0, "Miss":1, "Ms" : 1 , "Mme":1, "Mlle":1, "Mrs":1, "Mr":2, "Rare":3})
|
| 537 |
+
dataset["Title"] = dataset["Title"].astype(int)
|
| 538 |
+
|
| 539 |
+
|
| 540 |
+
# In[39]:
|
| 541 |
+
|
| 542 |
+
|
| 543 |
+
g = sns.countplot(dataset["Title"])
|
| 544 |
+
g = g.set_xticklabels(["Master","Miss/Ms/Mme/Mlle/Mrs","Mr","Rare"])
|
| 545 |
+
|
| 546 |
+
|
| 547 |
+
# In[40]:
|
| 548 |
+
|
| 549 |
+
|
| 550 |
+
g = sns.factorplot(x="Title",y="Survived",data=dataset,kind="bar")
|
| 551 |
+
g = g.set_xticklabels(["Master","Miss-Mrs","Mr","Rare"])
|
| 552 |
+
g = g.set_ylabels("survival probability")
|
| 553 |
+
|
| 554 |
+
|
| 555 |
+
# "Women and children first"
|
| 556 |
+
#
|
| 557 |
+
# It is interesting to note that passengers with rare title have more chance to survive.
|
| 558 |
+
|
| 559 |
+
# In[41]:
|
| 560 |
+
|
| 561 |
+
|
| 562 |
+
# Drop Name variable
|
| 563 |
+
dataset.drop(labels = ["Name"], axis = 1, inplace = True)
|
| 564 |
+
|
| 565 |
+
|
| 566 |
+
# ### 5.2 Family size
|
| 567 |
+
#
|
| 568 |
+
# We can imagine that large families will have more difficulties to evacuate, looking for theirs sisters/brothers/parents during the evacuation. So, i choosed to create a "Fize" (family size) feature which is the sum of SibSp , Parch and 1 (including the passenger).
|
| 569 |
+
|
| 570 |
+
# In[42]:
|
| 571 |
+
|
| 572 |
+
|
| 573 |
+
# Create a family size descriptor from SibSp and Parch
|
| 574 |
+
dataset["Fsize"] = dataset["SibSp"] + dataset["Parch"] + 1
|
| 575 |
+
|
| 576 |
+
|
| 577 |
+
# In[43]:
|
| 578 |
+
|
| 579 |
+
|
| 580 |
+
g = sns.factorplot(x="Fsize",y="Survived",data = dataset)
|
| 581 |
+
g = g.set_ylabels("Survival Probability")
|
| 582 |
+
|
| 583 |
+
|
| 584 |
+
# The family size seems to play an important role, survival probability is worst for large families.
|
| 585 |
+
#
|
| 586 |
+
# Additionally, i decided to created 4 categories of family size.
|
| 587 |
+
|
| 588 |
+
# In[44]:
|
| 589 |
+
|
| 590 |
+
|
| 591 |
+
# Create new feature of family size
|
| 592 |
+
dataset['Single'] = dataset['Fsize'].map(lambda s: 1 if s == 1 else 0)
|
| 593 |
+
dataset['SmallF'] = dataset['Fsize'].map(lambda s: 1 if s == 2 else 0)
|
| 594 |
+
dataset['MedF'] = dataset['Fsize'].map(lambda s: 1 if 3 <= s <= 4 else 0)
|
| 595 |
+
dataset['LargeF'] = dataset['Fsize'].map(lambda s: 1 if s >= 5 else 0)
|
| 596 |
+
|
| 597 |
+
|
| 598 |
+
# In[45]:
|
| 599 |
+
|
| 600 |
+
|
| 601 |
+
g = sns.factorplot(x="Single",y="Survived",data=dataset,kind="bar")
|
| 602 |
+
g = g.set_ylabels("Survival Probability")
|
| 603 |
+
g = sns.factorplot(x="SmallF",y="Survived",data=dataset,kind="bar")
|
| 604 |
+
g = g.set_ylabels("Survival Probability")
|
| 605 |
+
g = sns.factorplot(x="MedF",y="Survived",data=dataset,kind="bar")
|
| 606 |
+
g = g.set_ylabels("Survival Probability")
|
| 607 |
+
g = sns.factorplot(x="LargeF",y="Survived",data=dataset,kind="bar")
|
| 608 |
+
g = g.set_ylabels("Survival Probability")
|
| 609 |
+
|
| 610 |
+
|
| 611 |
+
# Factorplots of family size categories show that Small and Medium families have more chance to survive than single passenger and large families.
|
| 612 |
+
|
| 613 |
+
# In[46]:
|
| 614 |
+
|
| 615 |
+
|
| 616 |
+
# convert to indicator values Title and Embarked
|
| 617 |
+
dataset = pd.get_dummies(dataset, columns = ["Title"])
|
| 618 |
+
dataset = pd.get_dummies(dataset, columns = ["Embarked"], prefix="Em")
|
| 619 |
+
|
| 620 |
+
|
| 621 |
+
# In[47]:
|
| 622 |
+
|
| 623 |
+
|
| 624 |
+
dataset.head()
|
| 625 |
+
|
| 626 |
+
|
| 627 |
+
# At this stage, we have 22 features.
|
| 628 |
+
|
| 629 |
+
# ### 5.3 Cabin
|
| 630 |
+
|
| 631 |
+
# In[48]:
|
| 632 |
+
|
| 633 |
+
|
| 634 |
+
dataset["Cabin"].head()
|
| 635 |
+
|
| 636 |
+
|
| 637 |
+
# In[49]:
|
| 638 |
+
|
| 639 |
+
|
| 640 |
+
dataset["Cabin"].describe()
|
| 641 |
+
|
| 642 |
+
|
| 643 |
+
# In[50]:
|
| 644 |
+
|
| 645 |
+
|
| 646 |
+
dataset["Cabin"].isnull().sum()
|
| 647 |
+
|
| 648 |
+
|
| 649 |
+
# The Cabin feature column contains 292 values and 1007 missing values.
|
| 650 |
+
#
|
| 651 |
+
# I supposed that passengers without a cabin have a missing value displayed instead of the cabin number.
|
| 652 |
+
|
| 653 |
+
# In[51]:
|
| 654 |
+
|
| 655 |
+
|
| 656 |
+
dataset["Cabin"][dataset["Cabin"].notnull()].head()
|
| 657 |
+
|
| 658 |
+
|
| 659 |
+
# In[52]:
|
| 660 |
+
|
| 661 |
+
|
| 662 |
+
# Replace the Cabin number by the type of cabin 'X' if not
|
| 663 |
+
dataset["Cabin"] = pd.Series([i[0] if not pd.isnull(i) else 'X' for i in dataset['Cabin'] ])
|
| 664 |
+
|
| 665 |
+
|
| 666 |
+
# The first letter of the cabin indicates the Desk, i choosed to keep this information only, since it indicates the probable location of the passenger in the Titanic.
|
| 667 |
+
|
| 668 |
+
# In[53]:
|
| 669 |
+
|
| 670 |
+
|
| 671 |
+
g = sns.countplot(dataset["Cabin"],order=['A','B','C','D','E','F','G','T','X'])
|
| 672 |
+
|
| 673 |
+
|
| 674 |
+
# In[54]:
|
| 675 |
+
|
| 676 |
+
|
| 677 |
+
g = sns.factorplot(y="Survived",x="Cabin",data=dataset,kind="bar",order=['A','B','C','D','E','F','G','T','X'])
|
| 678 |
+
g = g.set_ylabels("Survival Probability")
|
| 679 |
+
|
| 680 |
+
|
| 681 |
+
# Because of the low number of passenger that have a cabin, survival probabilities have an important standard deviation and we can't distinguish between survival probability of passengers in the different desks.
|
| 682 |
+
#
|
| 683 |
+
# But we can see that passengers with a cabin have generally more chance to survive than passengers without (X).
|
| 684 |
+
#
|
| 685 |
+
# It is particularly true for cabin B, C, D, E and F.
|
| 686 |
+
|
| 687 |
+
# In[55]:
|
| 688 |
+
|
| 689 |
+
|
| 690 |
+
dataset = pd.get_dummies(dataset, columns = ["Cabin"],prefix="Cabin")
|
| 691 |
+
|
| 692 |
+
|
| 693 |
+
# ### 5.4 Ticket
|
| 694 |
+
|
| 695 |
+
# In[56]:
|
| 696 |
+
|
| 697 |
+
|
| 698 |
+
dataset["Ticket"].head()
|
| 699 |
+
|
| 700 |
+
|
| 701 |
+
# It could mean that tickets sharing the same prefixes could be booked for cabins placed together. It could therefore lead to the actual placement of the cabins within the ship.
|
| 702 |
+
#
|
| 703 |
+
# Tickets with same prefixes may have a similar class and survival.
|
| 704 |
+
#
|
| 705 |
+
# So i decided to replace the Ticket feature column by the ticket prefixe. Which may be more informative.
|
| 706 |
+
|
| 707 |
+
# In[57]:
|
| 708 |
+
|
| 709 |
+
|
| 710 |
+
## Treat Ticket by extracting the ticket prefix. When there is no prefix it returns X.
|
| 711 |
+
|
| 712 |
+
Ticket = []
|
| 713 |
+
for i in list(dataset.Ticket):
|
| 714 |
+
if not i.isdigit() :
|
| 715 |
+
Ticket.append(i.replace(".","").replace("/","").strip().split(' ')[0]) #Take prefix
|
| 716 |
+
else:
|
| 717 |
+
Ticket.append("X")
|
| 718 |
+
|
| 719 |
+
dataset["Ticket"] = Ticket
|
| 720 |
+
dataset["Ticket"].head()
|
| 721 |
+
|
| 722 |
+
|
| 723 |
+
# In[58]:
|
| 724 |
+
|
| 725 |
+
|
| 726 |
+
dataset = pd.get_dummies(dataset, columns = ["Ticket"], prefix="T")
|
| 727 |
+
|
| 728 |
+
|
| 729 |
+
# In[59]:
|
| 730 |
+
|
| 731 |
+
|
| 732 |
+
# Create categorical values for Pclass
|
| 733 |
+
dataset["Pclass"] = dataset["Pclass"].astype("category")
|
| 734 |
+
dataset = pd.get_dummies(dataset, columns = ["Pclass"],prefix="Pc")
|
| 735 |
+
|
| 736 |
+
|
| 737 |
+
# In[60]:
|
| 738 |
+
|
| 739 |
+
|
| 740 |
+
# Drop useless variables
|
| 741 |
+
dataset.drop(labels = ["PassengerId"], axis = 1, inplace = True)
|
| 742 |
+
|
| 743 |
+
|
| 744 |
+
# In[61]:
|
| 745 |
+
|
| 746 |
+
|
| 747 |
+
dataset.head()
|
| 748 |
+
|
| 749 |
+
|
| 750 |
+
# ## 6. MODELING
|
| 751 |
+
|
| 752 |
+
# In[62]:
|
| 753 |
+
|
| 754 |
+
|
| 755 |
+
## Separate train dataset and test dataset
|
| 756 |
+
|
| 757 |
+
train = dataset[:train_len]
|
| 758 |
+
test = dataset[train_len:]
|
| 759 |
+
test.drop(labels=["Survived"],axis = 1,inplace=True)
|
| 760 |
+
|
| 761 |
+
|
| 762 |
+
# In[63]:
|
| 763 |
+
|
| 764 |
+
|
| 765 |
+
## Separate train features and label
|
| 766 |
+
|
| 767 |
+
train["Survived"] = train["Survived"].astype(int)
|
| 768 |
+
|
| 769 |
+
Y_train = train["Survived"]
|
| 770 |
+
|
| 771 |
+
X_train = train.drop(labels = ["Survived"],axis = 1)
|
| 772 |
+
|
| 773 |
+
|
| 774 |
+
# ### 6.1 Simple modeling
|
| 775 |
+
# #### 6.1.1 Cross validate models
|
| 776 |
+
#
|
| 777 |
+
# I compared 10 popular classifiers and evaluate the mean accuracy of each of them by a stratified kfold cross validation procedure.
|
| 778 |
+
#
|
| 779 |
+
# * SVC
|
| 780 |
+
# * Decision Tree
|
| 781 |
+
# * AdaBoost
|
| 782 |
+
# * Random Forest
|
| 783 |
+
# * Extra Trees
|
| 784 |
+
# * Gradient Boosting
|
| 785 |
+
# * Multiple layer perceprton (neural network)
|
| 786 |
+
# * KNN
|
| 787 |
+
# * Logistic regression
|
| 788 |
+
# * Linear Discriminant Analysis
|
| 789 |
+
|
| 790 |
+
# In[64]:
|
| 791 |
+
|
| 792 |
+
|
| 793 |
+
# Cross validate model with Kfold stratified cross val
|
| 794 |
+
kfold = StratifiedKFold(n_splits=10)
|
| 795 |
+
|
| 796 |
+
|
| 797 |
+
# In[65]:
|
| 798 |
+
|
| 799 |
+
|
| 800 |
+
# Modeling step Test differents algorithms
|
| 801 |
+
random_state = 2
|
| 802 |
+
classifiers = []
|
| 803 |
+
classifiers.append(SVC(random_state=random_state))
|
| 804 |
+
classifiers.append(DecisionTreeClassifier(random_state=random_state))
|
| 805 |
+
classifiers.append(AdaBoostClassifier(DecisionTreeClassifier(random_state=random_state),random_state=random_state,learning_rate=0.1))
|
| 806 |
+
classifiers.append(RandomForestClassifier(random_state=random_state))
|
| 807 |
+
classifiers.append(ExtraTreesClassifier(random_state=random_state))
|
| 808 |
+
classifiers.append(GradientBoostingClassifier(random_state=random_state))
|
| 809 |
+
classifiers.append(MLPClassifier(random_state=random_state))
|
| 810 |
+
classifiers.append(KNeighborsClassifier())
|
| 811 |
+
classifiers.append(LogisticRegression(random_state = random_state))
|
| 812 |
+
classifiers.append(LinearDiscriminantAnalysis())
|
| 813 |
+
|
| 814 |
+
cv_results = []
|
| 815 |
+
for classifier in classifiers :
|
| 816 |
+
cv_results.append(cross_val_score(classifier, X_train, y = Y_train, scoring = "accuracy", cv = kfold, n_jobs=4))
|
| 817 |
+
|
| 818 |
+
cv_means = []
|
| 819 |
+
cv_std = []
|
| 820 |
+
for cv_result in cv_results:
|
| 821 |
+
cv_means.append(cv_result.mean())
|
| 822 |
+
cv_std.append(cv_result.std())
|
| 823 |
+
|
| 824 |
+
cv_res = pd.DataFrame({"CrossValMeans":cv_means,"CrossValerrors": cv_std,"Algorithm":["SVC","DecisionTree","AdaBoost",
|
| 825 |
+
"RandomForest","ExtraTrees","GradientBoosting","MultipleLayerPerceptron","KNeighboors","LogisticRegression","LinearDiscriminantAnalysis"]})
|
| 826 |
+
|
| 827 |
+
g = sns.barplot("CrossValMeans","Algorithm",data = cv_res, palette="Set3",orient = "h",**{'xerr':cv_std})
|
| 828 |
+
g.set_xlabel("Mean Accuracy")
|
| 829 |
+
g = g.set_title("Cross validation scores")
|
| 830 |
+
|
| 831 |
+
|
| 832 |
+
# I decided to choose the SVC, AdaBoost, RandomForest , ExtraTrees and the GradientBoosting classifiers for the ensemble modeling.
|
| 833 |
+
|
| 834 |
+
# #### 6.1.2 Hyperparameter tunning for best models
|
| 835 |
+
#
|
| 836 |
+
# I performed a grid search optimization for AdaBoost, ExtraTrees , RandomForest, GradientBoosting and SVC classifiers.
|
| 837 |
+
#
|
| 838 |
+
# I set the "n_jobs" parameter to 4 since i have 4 cpu . The computation time is clearly reduced.
|
| 839 |
+
#
|
| 840 |
+
# But be carefull, this step can take a long time, i took me 15 min in total on 4 cpu.
|
| 841 |
+
|
| 842 |
+
# In[66]:
|
| 843 |
+
|
| 844 |
+
|
| 845 |
+
### META MODELING WITH ADABOOST, RF, EXTRATREES and GRADIENTBOOSTING
|
| 846 |
+
|
| 847 |
+
# Adaboost
|
| 848 |
+
DTC = DecisionTreeClassifier()
|
| 849 |
+
|
| 850 |
+
adaDTC = AdaBoostClassifier(DTC, random_state=7)
|
| 851 |
+
|
| 852 |
+
ada_param_grid = {"base_estimator__criterion" : ["gini", "entropy"],
|
| 853 |
+
"base_estimator__splitter" : ["best", "random"],
|
| 854 |
+
"algorithm" : ["SAMME","SAMME.R"],
|
| 855 |
+
"n_estimators" :[1,2],
|
| 856 |
+
"learning_rate": [0.0001, 0.001, 0.01, 0.1, 0.2, 0.3,1.5]}
|
| 857 |
+
|
| 858 |
+
gsadaDTC = GridSearchCV(adaDTC,param_grid = ada_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 859 |
+
|
| 860 |
+
gsadaDTC.fit(X_train,Y_train)
|
| 861 |
+
|
| 862 |
+
ada_best = gsadaDTC.best_estimator_
|
| 863 |
+
|
| 864 |
+
|
| 865 |
+
# In[67]:
|
| 866 |
+
|
| 867 |
+
|
| 868 |
+
gsadaDTC.best_score_
|
| 869 |
+
|
| 870 |
+
|
| 871 |
+
# In[68]:
|
| 872 |
+
|
| 873 |
+
|
| 874 |
+
#ExtraTrees
|
| 875 |
+
ExtC = ExtraTreesClassifier()
|
| 876 |
+
|
| 877 |
+
|
| 878 |
+
## Search grid for optimal parameters
|
| 879 |
+
ex_param_grid = {"max_depth": [None],
|
| 880 |
+
"max_features": [1, 3, 10],
|
| 881 |
+
"min_samples_split": [2, 3, 10],
|
| 882 |
+
"min_samples_leaf": [1, 3, 10],
|
| 883 |
+
"bootstrap": [False],
|
| 884 |
+
"n_estimators" :[100,300],
|
| 885 |
+
"criterion": ["gini"]}
|
| 886 |
+
|
| 887 |
+
|
| 888 |
+
gsExtC = GridSearchCV(ExtC,param_grid = ex_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 889 |
+
|
| 890 |
+
gsExtC.fit(X_train,Y_train)
|
| 891 |
+
|
| 892 |
+
ExtC_best = gsExtC.best_estimator_
|
| 893 |
+
|
| 894 |
+
# Best score
|
| 895 |
+
gsExtC.best_score_
|
| 896 |
+
|
| 897 |
+
|
| 898 |
+
# In[69]:
|
| 899 |
+
|
| 900 |
+
|
| 901 |
+
# RFC Parameters tunning
|
| 902 |
+
RFC = RandomForestClassifier()
|
| 903 |
+
|
| 904 |
+
|
| 905 |
+
## Search grid for optimal parameters
|
| 906 |
+
rf_param_grid = {"max_depth": [None],
|
| 907 |
+
"max_features": [1, 3, 10],
|
| 908 |
+
"min_samples_split": [2, 3, 10],
|
| 909 |
+
"min_samples_leaf": [1, 3, 10],
|
| 910 |
+
"bootstrap": [False],
|
| 911 |
+
"n_estimators" :[100,300],
|
| 912 |
+
"criterion": ["gini"]}
|
| 913 |
+
|
| 914 |
+
|
| 915 |
+
gsRFC = GridSearchCV(RFC,param_grid = rf_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 916 |
+
|
| 917 |
+
gsRFC.fit(X_train,Y_train)
|
| 918 |
+
|
| 919 |
+
RFC_best = gsRFC.best_estimator_
|
| 920 |
+
|
| 921 |
+
# Best score
|
| 922 |
+
gsRFC.best_score_
|
| 923 |
+
|
| 924 |
+
|
| 925 |
+
# In[70]:
|
| 926 |
+
|
| 927 |
+
|
| 928 |
+
# Gradient boosting tunning
|
| 929 |
+
|
| 930 |
+
GBC = GradientBoostingClassifier()
|
| 931 |
+
gb_param_grid = {'loss' : ["deviance"],
|
| 932 |
+
'n_estimators' : [100,200,300],
|
| 933 |
+
'learning_rate': [0.1, 0.05, 0.01],
|
| 934 |
+
'max_depth': [4, 8],
|
| 935 |
+
'min_samples_leaf': [100,150],
|
| 936 |
+
'max_features': [0.3, 0.1]
|
| 937 |
+
}
|
| 938 |
+
|
| 939 |
+
gsGBC = GridSearchCV(GBC,param_grid = gb_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 940 |
+
|
| 941 |
+
gsGBC.fit(X_train,Y_train)
|
| 942 |
+
|
| 943 |
+
GBC_best = gsGBC.best_estimator_
|
| 944 |
+
|
| 945 |
+
# Best score
|
| 946 |
+
gsGBC.best_score_
|
| 947 |
+
|
| 948 |
+
|
| 949 |
+
# In[71]:
|
| 950 |
+
|
| 951 |
+
|
| 952 |
+
### SVC classifier
|
| 953 |
+
SVMC = SVC(probability=True)
|
| 954 |
+
svc_param_grid = {'kernel': ['rbf'],
|
| 955 |
+
'gamma': [ 0.001, 0.01, 0.1, 1],
|
| 956 |
+
'C': [1, 10, 50, 100,200,300, 1000]}
|
| 957 |
+
|
| 958 |
+
gsSVMC = GridSearchCV(SVMC,param_grid = svc_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 959 |
+
|
| 960 |
+
gsSVMC.fit(X_train,Y_train)
|
| 961 |
+
|
| 962 |
+
SVMC_best = gsSVMC.best_estimator_
|
| 963 |
+
|
| 964 |
+
# Best score
|
| 965 |
+
gsSVMC.best_score_
|
| 966 |
+
|
| 967 |
+
|
| 968 |
+
# #### 6.1.3 Plot learning curves
|
| 969 |
+
#
|
| 970 |
+
# Learning curves are a good way to see the overfitting effect on the training set and the effect of the training size on the accuracy.
|
| 971 |
+
|
| 972 |
+
# In[72]:
|
| 973 |
+
|
| 974 |
+
|
| 975 |
+
def plot_learning_curve(estimator, title, X, y, ylim=None, cv=None,
|
| 976 |
+
n_jobs=-1, train_sizes=np.linspace(.1, 1.0, 5)):
|
| 977 |
+
"""Generate a simple plot of the test and training learning curve"""
|
| 978 |
+
plt.figure()
|
| 979 |
+
plt.title(title)
|
| 980 |
+
if ylim is not None:
|
| 981 |
+
plt.ylim(*ylim)
|
| 982 |
+
plt.xlabel("Training examples")
|
| 983 |
+
plt.ylabel("Score")
|
| 984 |
+
train_sizes, train_scores, test_scores = learning_curve(
|
| 985 |
+
estimator, X, y, cv=cv, n_jobs=n_jobs, train_sizes=train_sizes)
|
| 986 |
+
train_scores_mean = np.mean(train_scores, axis=1)
|
| 987 |
+
train_scores_std = np.std(train_scores, axis=1)
|
| 988 |
+
test_scores_mean = np.mean(test_scores, axis=1)
|
| 989 |
+
test_scores_std = np.std(test_scores, axis=1)
|
| 990 |
+
plt.grid()
|
| 991 |
+
|
| 992 |
+
plt.fill_between(train_sizes, train_scores_mean - train_scores_std,
|
| 993 |
+
train_scores_mean + train_scores_std, alpha=0.1,
|
| 994 |
+
color="r")
|
| 995 |
+
plt.fill_between(train_sizes, test_scores_mean - test_scores_std,
|
| 996 |
+
test_scores_mean + test_scores_std, alpha=0.1, color="g")
|
| 997 |
+
plt.plot(train_sizes, train_scores_mean, 'o-', color="r",
|
| 998 |
+
label="Training score")
|
| 999 |
+
plt.plot(train_sizes, test_scores_mean, 'o-', color="g",
|
| 1000 |
+
label="Cross-validation score")
|
| 1001 |
+
|
| 1002 |
+
plt.legend(loc="best")
|
| 1003 |
+
return plt
|
| 1004 |
+
|
| 1005 |
+
g = plot_learning_curve(gsRFC.best_estimator_,"RF mearning curves",X_train,Y_train,cv=kfold)
|
| 1006 |
+
g = plot_learning_curve(gsExtC.best_estimator_,"ExtraTrees learning curves",X_train,Y_train,cv=kfold)
|
| 1007 |
+
g = plot_learning_curve(gsSVMC.best_estimator_,"SVC learning curves",X_train,Y_train,cv=kfold)
|
| 1008 |
+
g = plot_learning_curve(gsadaDTC.best_estimator_,"AdaBoost learning curves",X_train,Y_train,cv=kfold)
|
| 1009 |
+
g = plot_learning_curve(gsGBC.best_estimator_,"GradientBoosting learning curves",X_train,Y_train,cv=kfold)
|
| 1010 |
+
|
| 1011 |
+
|
| 1012 |
+
# GradientBoosting and Adaboost classifiers tend to overfit the training set. According to the growing cross-validation curves GradientBoosting and Adaboost could perform better with more training examples.
|
| 1013 |
+
#
|
| 1014 |
+
# SVC and ExtraTrees classifiers seem to better generalize the prediction since the training and cross-validation curves are close together.
|
| 1015 |
+
|
| 1016 |
+
# #### 6.1.4 Feature importance of tree based classifiers
|
| 1017 |
+
#
|
| 1018 |
+
# In order to see the most informative features for the prediction of passengers survival, i displayed the feature importance for the 4 tree based classifiers.
|
| 1019 |
+
|
| 1020 |
+
# In[73]:
|
| 1021 |
+
|
| 1022 |
+
|
| 1023 |
+
nrows = ncols = 2
|
| 1024 |
+
fig, axes = plt.subplots(nrows = nrows, ncols = ncols, sharex="all", figsize=(15,15))
|
| 1025 |
+
|
| 1026 |
+
names_classifiers = [("AdaBoosting", ada_best),("ExtraTrees",ExtC_best),("RandomForest",RFC_best),("GradientBoosting",GBC_best)]
|
| 1027 |
+
|
| 1028 |
+
nclassifier = 0
|
| 1029 |
+
for row in range(nrows):
|
| 1030 |
+
for col in range(ncols):
|
| 1031 |
+
name = names_classifiers[nclassifier][0]
|
| 1032 |
+
classifier = names_classifiers[nclassifier][1]
|
| 1033 |
+
indices = np.argsort(classifier.feature_importances_)[::-1][:40]
|
| 1034 |
+
g = sns.barplot(y=X_train.columns[indices][:40],x = classifier.feature_importances_[indices][:40] , orient='h',ax=axes[row][col])
|
| 1035 |
+
g.set_xlabel("Relative importance",fontsize=12)
|
| 1036 |
+
g.set_ylabel("Features",fontsize=12)
|
| 1037 |
+
g.tick_params(labelsize=9)
|
| 1038 |
+
g.set_title(name + " feature importance")
|
| 1039 |
+
nclassifier += 1
|
| 1040 |
+
|
| 1041 |
+
|
| 1042 |
+
# I plot the feature importance for the 4 tree based classifiers (Adaboost, ExtraTrees, RandomForest and GradientBoosting).
|
| 1043 |
+
#
|
| 1044 |
+
# We note that the four classifiers have different top features according to the relative importance. It means that their predictions are not based on the same features. Nevertheless, they share some common important features for the classification , for example 'Fare', 'Title_2', 'Age' and 'Sex'.
|
| 1045 |
+
#
|
| 1046 |
+
# Title_2 which indicates the Mrs/Mlle/Mme/Miss/Ms category is highly correlated with Sex.
|
| 1047 |
+
#
|
| 1048 |
+
# We can say that:
|
| 1049 |
+
#
|
| 1050 |
+
# - Pc_1, Pc_2, Pc_3 and Fare refer to the general social standing of passengers.
|
| 1051 |
+
#
|
| 1052 |
+
# - Sex and Title_2 (Mrs/Mlle/Mme/Miss/Ms) and Title_3 (Mr) refer to the gender.
|
| 1053 |
+
#
|
| 1054 |
+
# - Age and Title_1 (Master) refer to the age of passengers.
|
| 1055 |
+
#
|
| 1056 |
+
# - Fsize, LargeF, MedF, Single refer to the size of the passenger family.
|
| 1057 |
+
#
|
| 1058 |
+
# **According to the feature importance of this 4 classifiers, the prediction of the survival seems to be more associated with the Age, the Sex, the family size and the social standing of the passengers more than the location in the boat.**
|
| 1059 |
+
|
| 1060 |
+
# In[74]:
|
| 1061 |
+
|
| 1062 |
+
|
| 1063 |
+
test_Survived_RFC = pd.Series(RFC_best.predict(test), name="RFC")
|
| 1064 |
+
test_Survived_ExtC = pd.Series(ExtC_best.predict(test), name="ExtC")
|
| 1065 |
+
test_Survived_SVMC = pd.Series(SVMC_best.predict(test), name="SVC")
|
| 1066 |
+
test_Survived_AdaC = pd.Series(ada_best.predict(test), name="Ada")
|
| 1067 |
+
test_Survived_GBC = pd.Series(GBC_best.predict(test), name="GBC")
|
| 1068 |
+
|
| 1069 |
+
|
| 1070 |
+
# Concatenate all classifier results
|
| 1071 |
+
ensemble_results = pd.concat([test_Survived_RFC,test_Survived_ExtC,test_Survived_AdaC,test_Survived_GBC, test_Survived_SVMC],axis=1)
|
| 1072 |
+
|
| 1073 |
+
|
| 1074 |
+
g= sns.heatmap(ensemble_results.corr(),annot=True)
|
| 1075 |
+
|
| 1076 |
+
|
| 1077 |
+
# The prediction seems to be quite similar for the 5 classifiers except when Adaboost is compared to the others classifiers.
|
| 1078 |
+
#
|
| 1079 |
+
# The 5 classifiers give more or less the same prediction but there is some differences. Theses differences between the 5 classifier predictions are sufficient to consider an ensembling vote.
|
| 1080 |
+
|
| 1081 |
+
# ### 6.2 Ensemble modeling
|
| 1082 |
+
# #### 6.2.1 Combining models
|
| 1083 |
+
#
|
| 1084 |
+
# I choosed a voting classifier to combine the predictions coming from the 5 classifiers.
|
| 1085 |
+
#
|
| 1086 |
+
# I preferred to pass the argument "soft" to the voting parameter to take into account the probability of each vote.
|
| 1087 |
+
|
| 1088 |
+
# In[75]:
|
| 1089 |
+
|
| 1090 |
+
|
| 1091 |
+
votingC = VotingClassifier(estimators=[('rfc', RFC_best), ('extc', ExtC_best),
|
| 1092 |
+
('svc', SVMC_best), ('adac',ada_best),('gbc',GBC_best)], voting='soft', n_jobs=4)
|
| 1093 |
+
|
| 1094 |
+
votingC = votingC.fit(X_train, Y_train)
|
| 1095 |
+
|
| 1096 |
+
|
| 1097 |
+
# ### 6.3 Prediction
|
| 1098 |
+
# #### 6.3.1 Predict and Submit results
|
| 1099 |
+
|
| 1100 |
+
# In[76]:
|
| 1101 |
+
|
| 1102 |
+
|
| 1103 |
+
test_Survived = pd.Series(votingC.predict(test), name="Survived")
|
| 1104 |
+
|
| 1105 |
+
results = pd.concat([IDtest,test_Survived],axis=1)
|
| 1106 |
+
|
| 1107 |
+
results.to_csv("ensemble_python_voting.csv",index=False)
|
| 1108 |
+
|
| 1109 |
+
|
| 1110 |
+
# If you found this notebook helpful or you just liked it , some upvotes would be very much appreciated - That will keep me motivated :)
|
Titanic/Kernels/AdaBoost/3-eda-to-prediction-dietanic.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/3-eda-to-prediction-dietanic.py
ADDED
|
@@ -0,0 +1,1152 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python
|
| 2 |
+
# coding: utf-8
|
| 3 |
+
|
| 4 |
+
# # EDA To Prediction (DieTanic)
|
| 5 |
+
#
|
| 6 |
+
|
| 7 |
+
# ### *Sometimes life has a cruel sense of humor, giving you the thing you always wanted at the worst time possible.*
|
| 8 |
+
# -Lisa Kleypas
|
| 9 |
+
#
|
| 10 |
+
#
|
| 11 |
+
|
| 12 |
+
# The sinking of the Titanic is one of the most infamous shipwrecks in history. On April 15, 1912, during her maiden voyage, the Titanic sank after colliding with an iceberg, killing 1502 out of 2224 passengers and crew. That's why the name **DieTanic**. This is a very unforgetable disaster that no one in the world can forget.
|
| 13 |
+
#
|
| 14 |
+
# It took about $7.5 million to build the Titanic and it sunk under the ocean due to collision. The Titanic Dataset is a very good dataset for begineers to start a journey in data science and participate in competitions in Kaggle.
|
| 15 |
+
#
|
| 16 |
+
# The Objective of this notebook is to give an **idea how is the workflow in any predictive modeling problem**. How do we check features, how do we add new features and some Machine Learning Concepts. I have tried to keep the notebook as basic as possible so that even newbies can understand every phase of it.
|
| 17 |
+
#
|
| 18 |
+
# If You Like the notebook and think that it helped you..**PLEASE UPVOTE**. It will keep me motivated.
|
| 19 |
+
|
| 20 |
+
# ## Contents of the Notebook:
|
| 21 |
+
#
|
| 22 |
+
# #### Part1: Exploratory Data Analysis(EDA):
|
| 23 |
+
# 1)Analysis of the features.
|
| 24 |
+
#
|
| 25 |
+
# 2)Finding any relations or trends considering multiple features.
|
| 26 |
+
# #### Part2: Feature Engineering and Data Cleaning:
|
| 27 |
+
# 1)Adding any few features.
|
| 28 |
+
#
|
| 29 |
+
# 2)Removing redundant features.
|
| 30 |
+
#
|
| 31 |
+
# 3)Converting features into suitable form for modeling.
|
| 32 |
+
# #### Part3: Predictive Modeling
|
| 33 |
+
# 1)Running Basic Algorithms.
|
| 34 |
+
#
|
| 35 |
+
# 2)Cross Validation.
|
| 36 |
+
#
|
| 37 |
+
# 3)Ensembling.
|
| 38 |
+
#
|
| 39 |
+
# 4)Important Features Extraction.
|
| 40 |
+
|
| 41 |
+
# ## Part1: Exploratory Data Analysis(EDA)
|
| 42 |
+
|
| 43 |
+
# In[ ]:
|
| 44 |
+
|
| 45 |
+
|
| 46 |
+
import numpy as np
|
| 47 |
+
import pandas as pd
|
| 48 |
+
import matplotlib.pyplot as plt
|
| 49 |
+
import seaborn as sns
|
| 50 |
+
plt.style.use('fivethirtyeight')
|
| 51 |
+
import warnings
|
| 52 |
+
warnings.filterwarnings('ignore')
|
| 53 |
+
get_ipython().run_line_magic('matplotlib', 'inline')
|
| 54 |
+
|
| 55 |
+
|
| 56 |
+
# In[ ]:
|
| 57 |
+
|
| 58 |
+
|
| 59 |
+
data=pd.read_csv('../input/train.csv')
|
| 60 |
+
|
| 61 |
+
|
| 62 |
+
# In[ ]:
|
| 63 |
+
|
| 64 |
+
|
| 65 |
+
data.head()
|
| 66 |
+
|
| 67 |
+
|
| 68 |
+
# In[ ]:
|
| 69 |
+
|
| 70 |
+
|
| 71 |
+
data.isnull().sum() #checking for total null values
|
| 72 |
+
|
| 73 |
+
|
| 74 |
+
# The **Age, Cabin and Embarked** have null values. I will try to fix them.
|
| 75 |
+
|
| 76 |
+
# ### How many Survived??
|
| 77 |
+
|
| 78 |
+
# In[ ]:
|
| 79 |
+
|
| 80 |
+
|
| 81 |
+
f,ax=plt.subplots(1,2,figsize=(18,8))
|
| 82 |
+
data['Survived'].value_counts().plot.pie(explode=[0,0.1],autopct='%1.1f%%',ax=ax[0],shadow=True)
|
| 83 |
+
ax[0].set_title('Survived')
|
| 84 |
+
ax[0].set_ylabel('')
|
| 85 |
+
sns.countplot('Survived',data=data,ax=ax[1])
|
| 86 |
+
ax[1].set_title('Survived')
|
| 87 |
+
plt.show()
|
| 88 |
+
|
| 89 |
+
|
| 90 |
+
# It is evident that not many passengers survived the accident.
|
| 91 |
+
#
|
| 92 |
+
# Out of 891 passengers in training set, only around 350 survived i.e Only **38.4%** of the total training set survived the crash. We need to dig down more to get better insights from the data and see which categories of the passengers did survive and who didn't.
|
| 93 |
+
#
|
| 94 |
+
# We will try to check the survival rate by using the different features of the dataset. Some of the features being Sex, Port Of Embarcation, Age,etc.
|
| 95 |
+
#
|
| 96 |
+
# First let us understand the different types of features.
|
| 97 |
+
|
| 98 |
+
# ## Types Of Features
|
| 99 |
+
#
|
| 100 |
+
# ### Categorical Features:
|
| 101 |
+
# A categorical variable is one that has two or more categories and each value in that feature can be categorised by them.For example, gender is a categorical variable having two categories (male and female). Now we cannot sort or give any ordering to such variables. They are also known as **Nominal Variables**.
|
| 102 |
+
#
|
| 103 |
+
# **Categorical Features in the dataset: Sex,Embarked.**
|
| 104 |
+
#
|
| 105 |
+
# ### Ordinal Features:
|
| 106 |
+
# An ordinal variable is similar to categorical values, but the difference between them is that we can have relative ordering or sorting between the values. For eg: If we have a feature like **Height** with values **Tall, Medium, Short**, then Height is a ordinal variable. Here we can have a relative sort in the variable.
|
| 107 |
+
#
|
| 108 |
+
# **Ordinal Features in the dataset: PClass**
|
| 109 |
+
#
|
| 110 |
+
# ### Continous Feature:
|
| 111 |
+
# A feature is said to be continous if it can take values between any two points or between the minimum or maximum values in the features column.
|
| 112 |
+
#
|
| 113 |
+
# **Continous Features in the dataset: Age**
|
| 114 |
+
|
| 115 |
+
# ## Analysing The Features
|
| 116 |
+
|
| 117 |
+
# ## Sex--> Categorical Feature
|
| 118 |
+
|
| 119 |
+
# In[ ]:
|
| 120 |
+
|
| 121 |
+
|
| 122 |
+
data.groupby(['Sex','Survived'])['Survived'].count()
|
| 123 |
+
|
| 124 |
+
|
| 125 |
+
# In[ ]:
|
| 126 |
+
|
| 127 |
+
|
| 128 |
+
f,ax=plt.subplots(1,2,figsize=(18,8))
|
| 129 |
+
data[['Sex','Survived']].groupby(['Sex']).mean().plot.bar(ax=ax[0])
|
| 130 |
+
ax[0].set_title('Survived vs Sex')
|
| 131 |
+
sns.countplot('Sex',hue='Survived',data=data,ax=ax[1])
|
| 132 |
+
ax[1].set_title('Sex:Survived vs Dead')
|
| 133 |
+
plt.show()
|
| 134 |
+
|
| 135 |
+
|
| 136 |
+
# This looks interesting. The number of men on the ship is lot more than the number of women. Still the number of women saved is almost twice the number of males saved. The survival rates for a **women on the ship is around 75% while that for men in around 18-19%.**
|
| 137 |
+
#
|
| 138 |
+
# This looks to be a **very important** feature for modeling. But is it the best?? Lets check other features.
|
| 139 |
+
|
| 140 |
+
# ## Pclass --> Ordinal Feature
|
| 141 |
+
|
| 142 |
+
# In[ ]:
|
| 143 |
+
|
| 144 |
+
|
| 145 |
+
pd.crosstab(data.Pclass,data.Survived,margins=True).style.background_gradient(cmap='summer_r')
|
| 146 |
+
|
| 147 |
+
|
| 148 |
+
# In[ ]:
|
| 149 |
+
|
| 150 |
+
|
| 151 |
+
f,ax=plt.subplots(1,2,figsize=(18,8))
|
| 152 |
+
data['Pclass'].value_counts().plot.bar(color=['#CD7F32','#FFDF00','#D3D3D3'],ax=ax[0])
|
| 153 |
+
ax[0].set_title('Number Of Passengers By Pclass')
|
| 154 |
+
ax[0].set_ylabel('Count')
|
| 155 |
+
sns.countplot('Pclass',hue='Survived',data=data,ax=ax[1])
|
| 156 |
+
ax[1].set_title('Pclass:Survived vs Dead')
|
| 157 |
+
plt.show()
|
| 158 |
+
|
| 159 |
+
|
| 160 |
+
# People say **Money Can't Buy Everything**. But we can clearly see that Passenegers Of Pclass 1 were given a very high priority while rescue. Even though the the number of Passengers in Pclass 3 were a lot higher, still the number of survival from them is very low, somewhere around **25%**.
|
| 161 |
+
#
|
| 162 |
+
# For Pclass 1 %survived is around **63%** while for Pclass2 is around **48%**. So money and status matters. Such a materialistic world.
|
| 163 |
+
#
|
| 164 |
+
# Lets Dive in little bit more and check for other interesting observations. Lets check survival rate with **Sex and Pclass** Together.
|
| 165 |
+
|
| 166 |
+
# In[ ]:
|
| 167 |
+
|
| 168 |
+
|
| 169 |
+
pd.crosstab([data.Sex,data.Survived],data.Pclass,margins=True).style.background_gradient(cmap='summer_r')
|
| 170 |
+
|
| 171 |
+
|
| 172 |
+
# In[ ]:
|
| 173 |
+
|
| 174 |
+
|
| 175 |
+
sns.factorplot('Pclass','Survived',hue='Sex',data=data)
|
| 176 |
+
plt.show()
|
| 177 |
+
|
| 178 |
+
|
| 179 |
+
# We use **FactorPlot** in this case, because they make the seperation of categorical values easy.
|
| 180 |
+
#
|
| 181 |
+
# Looking at the **CrossTab** and the **FactorPlot**, we can easily infer that survival for **Women from Pclass1** is about **95-96%**, as only 3 out of 94 Women from Pclass1 died.
|
| 182 |
+
#
|
| 183 |
+
# It is evident that irrespective of Pclass, Women were given first priority while rescue. Even Men from Pclass1 have a very low survival rate.
|
| 184 |
+
#
|
| 185 |
+
# Looks like Pclass is also an important feature. Lets analyse other features.
|
| 186 |
+
|
| 187 |
+
# ## Age--> Continous Feature
|
| 188 |
+
#
|
| 189 |
+
|
| 190 |
+
# In[ ]:
|
| 191 |
+
|
| 192 |
+
|
| 193 |
+
print('Oldest Passenger was of:',data['Age'].max(),'Years')
|
| 194 |
+
print('Youngest Passenger was of:',data['Age'].min(),'Years')
|
| 195 |
+
print('Average Age on the ship:',data['Age'].mean(),'Years')
|
| 196 |
+
|
| 197 |
+
|
| 198 |
+
# In[ ]:
|
| 199 |
+
|
| 200 |
+
|
| 201 |
+
f,ax=plt.subplots(1,2,figsize=(18,8))
|
| 202 |
+
sns.violinplot("Pclass","Age", hue="Survived", data=data,split=True,ax=ax[0])
|
| 203 |
+
ax[0].set_title('Pclass and Age vs Survived')
|
| 204 |
+
ax[0].set_yticks(range(0,110,10))
|
| 205 |
+
sns.violinplot("Sex","Age", hue="Survived", data=data,split=True,ax=ax[1])
|
| 206 |
+
ax[1].set_title('Sex and Age vs Survived')
|
| 207 |
+
ax[1].set_yticks(range(0,110,10))
|
| 208 |
+
plt.show()
|
| 209 |
+
|
| 210 |
+
|
| 211 |
+
# #### Observations:
|
| 212 |
+
#
|
| 213 |
+
# 1)The number of children increases with Pclass and the survival rate for passenegers below Age 10(i.e children) looks to be good irrespective of the Pclass.
|
| 214 |
+
#
|
| 215 |
+
# 2)Survival chances for Passenegers aged 20-50 from Pclass1 is high and is even better for Women.
|
| 216 |
+
#
|
| 217 |
+
# 3)For males, the survival chances decreases with an increase in age.
|
| 218 |
+
|
| 219 |
+
# As we had seen earlier, the Age feature has **177** null values. To replace these NaN values, we can assign them the mean age of the dataset.
|
| 220 |
+
#
|
| 221 |
+
# But the problem is, there were many people with many different ages. We just cant assign a 4 year kid with the mean age that is 29 years. Is there any way to find out what age-band does the passenger lie??
|
| 222 |
+
#
|
| 223 |
+
# **Bingo!!!!**, we can check the **Name** feature. Looking upon the feature, we can see that the names have a salutation like Mr or Mrs. Thus we can assign the mean values of Mr and Mrs to the respective groups.
|
| 224 |
+
#
|
| 225 |
+
# **''What's In A Name??''**---> **Feature** :p
|
| 226 |
+
|
| 227 |
+
# In[ ]:
|
| 228 |
+
|
| 229 |
+
|
| 230 |
+
data['Initial']=0
|
| 231 |
+
for i in data:
|
| 232 |
+
data['Initial']=data.Name.str.extract('([A-Za-z]+)\.') #lets extract the Salutations
|
| 233 |
+
|
| 234 |
+
|
| 235 |
+
# Okay so here we are using the Regex: **[A-Za-z]+)\.**. So what it does is, it looks for strings which lie between **A-Z or a-z** and followed by a **.(dot)**. So we successfully extract the Initials from the Name.
|
| 236 |
+
|
| 237 |
+
# In[ ]:
|
| 238 |
+
|
| 239 |
+
|
| 240 |
+
pd.crosstab(data.Initial,data.Sex).T.style.background_gradient(cmap='summer_r') #Checking the Initials with the Sex
|
| 241 |
+
|
| 242 |
+
|
| 243 |
+
# Okay so there are some misspelled Initials like Mlle or Mme that stand for Miss. I will replace them with Miss and same thing for other values.
|
| 244 |
+
|
| 245 |
+
# In[ ]:
|
| 246 |
+
|
| 247 |
+
|
| 248 |
+
data['Initial'].replace(['Mlle','Mme','Ms','Dr','Major','Lady','Countess','Jonkheer','Col','Rev','Capt','Sir','Don'],['Miss','Miss','Miss','Mr','Mr','Mrs','Mrs','Other','Other','Other','Mr','Mr','Mr'],inplace=True)
|
| 249 |
+
|
| 250 |
+
|
| 251 |
+
# In[ ]:
|
| 252 |
+
|
| 253 |
+
|
| 254 |
+
data.groupby('Initial')['Age'].mean() #lets check the average age by Initials
|
| 255 |
+
|
| 256 |
+
|
| 257 |
+
# ### Filling NaN Ages
|
| 258 |
+
|
| 259 |
+
# In[ ]:
|
| 260 |
+
|
| 261 |
+
|
| 262 |
+
## Assigning the NaN Values with the Ceil values of the mean ages
|
| 263 |
+
data.loc[(data.Age.isnull())&(data.Initial=='Mr'),'Age']=33
|
| 264 |
+
data.loc[(data.Age.isnull())&(data.Initial=='Mrs'),'Age']=36
|
| 265 |
+
data.loc[(data.Age.isnull())&(data.Initial=='Master'),'Age']=5
|
| 266 |
+
data.loc[(data.Age.isnull())&(data.Initial=='Miss'),'Age']=22
|
| 267 |
+
data.loc[(data.Age.isnull())&(data.Initial=='Other'),'Age']=46
|
| 268 |
+
|
| 269 |
+
|
| 270 |
+
# In[ ]:
|
| 271 |
+
|
| 272 |
+
|
| 273 |
+
data.Age.isnull().any() #So no null values left finally
|
| 274 |
+
|
| 275 |
+
|
| 276 |
+
# In[ ]:
|
| 277 |
+
|
| 278 |
+
|
| 279 |
+
f,ax=plt.subplots(1,2,figsize=(20,10))
|
| 280 |
+
data[data['Survived']==0].Age.plot.hist(ax=ax[0],bins=20,edgecolor='black',color='red')
|
| 281 |
+
ax[0].set_title('Survived= 0')
|
| 282 |
+
x1=list(range(0,85,5))
|
| 283 |
+
ax[0].set_xticks(x1)
|
| 284 |
+
data[data['Survived']==1].Age.plot.hist(ax=ax[1],color='green',bins=20,edgecolor='black')
|
| 285 |
+
ax[1].set_title('Survived= 1')
|
| 286 |
+
x2=list(range(0,85,5))
|
| 287 |
+
ax[1].set_xticks(x2)
|
| 288 |
+
plt.show()
|
| 289 |
+
|
| 290 |
+
|
| 291 |
+
# ### Observations:
|
| 292 |
+
# 1)The Toddlers(age<5) were saved in large numbers(The Women and Child First Policy).
|
| 293 |
+
#
|
| 294 |
+
# 2)The oldest Passenger was saved(80 years).
|
| 295 |
+
#
|
| 296 |
+
# 3)Maximum number of deaths were in the age group of 30-40.
|
| 297 |
+
|
| 298 |
+
# In[ ]:
|
| 299 |
+
|
| 300 |
+
|
| 301 |
+
sns.factorplot('Pclass','Survived',col='Initial',data=data)
|
| 302 |
+
plt.show()
|
| 303 |
+
|
| 304 |
+
|
| 305 |
+
# The Women and Child first policy thus holds true irrespective of the class.
|
| 306 |
+
|
| 307 |
+
# ## Embarked--> Categorical Value
|
| 308 |
+
|
| 309 |
+
# In[ ]:
|
| 310 |
+
|
| 311 |
+
|
| 312 |
+
pd.crosstab([data.Embarked,data.Pclass],[data.Sex,data.Survived],margins=True).style.background_gradient(cmap='summer_r')
|
| 313 |
+
|
| 314 |
+
|
| 315 |
+
# ### Chances for Survival by Port Of Embarkation
|
| 316 |
+
|
| 317 |
+
# In[ ]:
|
| 318 |
+
|
| 319 |
+
|
| 320 |
+
sns.factorplot('Embarked','Survived',data=data)
|
| 321 |
+
fig=plt.gcf()
|
| 322 |
+
fig.set_size_inches(5,3)
|
| 323 |
+
plt.show()
|
| 324 |
+
|
| 325 |
+
|
| 326 |
+
# The chances for survival for Port C is highest around 0.55 while it is lowest for S.
|
| 327 |
+
|
| 328 |
+
# In[ ]:
|
| 329 |
+
|
| 330 |
+
|
| 331 |
+
f,ax=plt.subplots(2,2,figsize=(20,15))
|
| 332 |
+
sns.countplot('Embarked',data=data,ax=ax[0,0])
|
| 333 |
+
ax[0,0].set_title('No. Of Passengers Boarded')
|
| 334 |
+
sns.countplot('Embarked',hue='Sex',data=data,ax=ax[0,1])
|
| 335 |
+
ax[0,1].set_title('Male-Female Split for Embarked')
|
| 336 |
+
sns.countplot('Embarked',hue='Survived',data=data,ax=ax[1,0])
|
| 337 |
+
ax[1,0].set_title('Embarked vs Survived')
|
| 338 |
+
sns.countplot('Embarked',hue='Pclass',data=data,ax=ax[1,1])
|
| 339 |
+
ax[1,1].set_title('Embarked vs Pclass')
|
| 340 |
+
plt.subplots_adjust(wspace=0.2,hspace=0.5)
|
| 341 |
+
plt.show()
|
| 342 |
+
|
| 343 |
+
|
| 344 |
+
# ### Observations:
|
| 345 |
+
# 1)Maximum passenegers boarded from S. Majority of them being from Pclass3.
|
| 346 |
+
#
|
| 347 |
+
# 2)The Passengers from C look to be lucky as a good proportion of them survived. The reason for this maybe the rescue of all the Pclass1 and Pclass2 Passengers.
|
| 348 |
+
#
|
| 349 |
+
# 3)The Embark S looks to the port from where majority of the rich people boarded. Still the chances for survival is low here, that is because many passengers from Pclass3 around **81%** didn't survive.
|
| 350 |
+
#
|
| 351 |
+
# 4)Port Q had almost 95% of the passengers were from Pclass3.
|
| 352 |
+
|
| 353 |
+
# In[ ]:
|
| 354 |
+
|
| 355 |
+
|
| 356 |
+
sns.factorplot('Pclass','Survived',hue='Sex',col='Embarked',data=data)
|
| 357 |
+
plt.show()
|
| 358 |
+
|
| 359 |
+
|
| 360 |
+
# ### Observations:
|
| 361 |
+
#
|
| 362 |
+
# 1)The survival chances are almost 1 for women for Pclass1 and Pclass2 irrespective of the Pclass.
|
| 363 |
+
#
|
| 364 |
+
# 2)Port S looks to be very unlucky for Pclass3 Passenegers as the survival rate for both men and women is very low.**(Money Matters)**
|
| 365 |
+
#
|
| 366 |
+
# 3)Port Q looks looks to be unlukiest for Men, as almost all were from Pclass 3.
|
| 367 |
+
#
|
| 368 |
+
|
| 369 |
+
# ### Filling Embarked NaN
|
| 370 |
+
#
|
| 371 |
+
# As we saw that maximum passengers boarded from Port S, we replace NaN with S.
|
| 372 |
+
|
| 373 |
+
# In[ ]:
|
| 374 |
+
|
| 375 |
+
|
| 376 |
+
data['Embarked'].fillna('S',inplace=True)
|
| 377 |
+
|
| 378 |
+
|
| 379 |
+
# In[ ]:
|
| 380 |
+
|
| 381 |
+
|
| 382 |
+
data.Embarked.isnull().any()# Finally No NaN values
|
| 383 |
+
|
| 384 |
+
|
| 385 |
+
# ## SibSip-->Discrete Feature
|
| 386 |
+
# This feature represents whether a person is alone or with his family members.
|
| 387 |
+
#
|
| 388 |
+
# Sibling = brother, sister, stepbrother, stepsister
|
| 389 |
+
#
|
| 390 |
+
# Spouse = husband, wife
|
| 391 |
+
|
| 392 |
+
# In[ ]:
|
| 393 |
+
|
| 394 |
+
|
| 395 |
+
pd.crosstab([data.SibSp],data.Survived).style.background_gradient(cmap='summer_r')
|
| 396 |
+
|
| 397 |
+
|
| 398 |
+
# In[ ]:
|
| 399 |
+
|
| 400 |
+
|
| 401 |
+
f,ax=plt.subplots(1,2,figsize=(20,8))
|
| 402 |
+
sns.barplot('SibSp','Survived',data=data,ax=ax[0])
|
| 403 |
+
ax[0].set_title('SibSp vs Survived')
|
| 404 |
+
sns.factorplot('SibSp','Survived',data=data,ax=ax[1])
|
| 405 |
+
ax[1].set_title('SibSp vs Survived')
|
| 406 |
+
plt.close(2)
|
| 407 |
+
plt.show()
|
| 408 |
+
|
| 409 |
+
|
| 410 |
+
# In[ ]:
|
| 411 |
+
|
| 412 |
+
|
| 413 |
+
pd.crosstab(data.SibSp,data.Pclass).style.background_gradient(cmap='summer_r')
|
| 414 |
+
|
| 415 |
+
|
| 416 |
+
# ### Observations:
|
| 417 |
+
#
|
| 418 |
+
#
|
| 419 |
+
# The barplot and factorplot shows that if a passenger is alone onboard with no siblings, he have 34.5% survival rate. The graph roughly decreases if the number of siblings increase. This makes sense. That is, if I have a family on board, I will try to save them instead of saving myself first. Surprisingly the survival for families with 5-8 members is **0%**. The reason may be Pclass??
|
| 420 |
+
#
|
| 421 |
+
# The reason is **Pclass**. The crosstab shows that Person with SibSp>3 were all in Pclass3. It is imminent that all the large families in Pclass3(>3) died.
|
| 422 |
+
|
| 423 |
+
# ## Parch
|
| 424 |
+
|
| 425 |
+
# In[ ]:
|
| 426 |
+
|
| 427 |
+
|
| 428 |
+
pd.crosstab(data.Parch,data.Pclass).style.background_gradient(cmap='summer_r')
|
| 429 |
+
|
| 430 |
+
|
| 431 |
+
# The crosstab again shows that larger families were in Pclass3.
|
| 432 |
+
|
| 433 |
+
# In[ ]:
|
| 434 |
+
|
| 435 |
+
|
| 436 |
+
f,ax=plt.subplots(1,2,figsize=(20,8))
|
| 437 |
+
sns.barplot('Parch','Survived',data=data,ax=ax[0])
|
| 438 |
+
ax[0].set_title('Parch vs Survived')
|
| 439 |
+
sns.factorplot('Parch','Survived',data=data,ax=ax[1])
|
| 440 |
+
ax[1].set_title('Parch vs Survived')
|
| 441 |
+
plt.close(2)
|
| 442 |
+
plt.show()
|
| 443 |
+
|
| 444 |
+
|
| 445 |
+
# ### Observations:
|
| 446 |
+
#
|
| 447 |
+
# Here too the results are quite similar. Passengers with their parents onboard have greater chance of survival. It however reduces as the number goes up.
|
| 448 |
+
#
|
| 449 |
+
# The chances of survival is good for somebody who has 1-3 parents on the ship. Being alone also proves to be fatal and the chances for survival decreases when somebody has >4 parents on the ship.
|
| 450 |
+
|
| 451 |
+
# ## Fare--> Continous Feature
|
| 452 |
+
|
| 453 |
+
# In[ ]:
|
| 454 |
+
|
| 455 |
+
|
| 456 |
+
print('Highest Fare was:',data['Fare'].max())
|
| 457 |
+
print('Lowest Fare was:',data['Fare'].min())
|
| 458 |
+
print('Average Fare was:',data['Fare'].mean())
|
| 459 |
+
|
| 460 |
+
|
| 461 |
+
# The lowest fare is **0.0**. Wow!! a free luxorious ride.
|
| 462 |
+
|
| 463 |
+
# In[ ]:
|
| 464 |
+
|
| 465 |
+
|
| 466 |
+
f,ax=plt.subplots(1,3,figsize=(20,8))
|
| 467 |
+
sns.distplot(data[data['Pclass']==1].Fare,ax=ax[0])
|
| 468 |
+
ax[0].set_title('Fares in Pclass 1')
|
| 469 |
+
sns.distplot(data[data['Pclass']==2].Fare,ax=ax[1])
|
| 470 |
+
ax[1].set_title('Fares in Pclass 2')
|
| 471 |
+
sns.distplot(data[data['Pclass']==3].Fare,ax=ax[2])
|
| 472 |
+
ax[2].set_title('Fares in Pclass 3')
|
| 473 |
+
plt.show()
|
| 474 |
+
|
| 475 |
+
|
| 476 |
+
# There looks to be a large distribution in the fares of Passengers in Pclass1 and this distribution goes on decreasing as the standards reduces. As this is also continous, we can convert into discrete values by using binning.
|
| 477 |
+
|
| 478 |
+
# ## Observations in a Nutshell for all features:
|
| 479 |
+
# **Sex:** The chance of survival for women is high as compared to men.
|
| 480 |
+
#
|
| 481 |
+
# **Pclass:**There is a visible trend that being a **1st class passenger** gives you better chances of survival. The survival rate for **Pclass3 is very low**. For **women**, the chance of survival from **Pclass1** is almost 1 and is high too for those from **Pclass2**. **Money Wins!!!**.
|
| 482 |
+
#
|
| 483 |
+
# **Age:** Children less than 5-10 years do have a high chance of survival. Passengers between age group 15 to 35 died a lot.
|
| 484 |
+
#
|
| 485 |
+
# **Embarked:** This is a very interesting feature. **The chances of survival at C looks to be better than even though the majority of Pclass1 passengers got up at S.** Passengers at Q were all from **Pclass3**.
|
| 486 |
+
#
|
| 487 |
+
# **Parch+SibSp:** Having 1-2 siblings,spouse on board or 1-3 Parents shows a greater chance of probablity rather than being alone or having a large family travelling with you.
|
| 488 |
+
|
| 489 |
+
# ## Correlation Between The Features
|
| 490 |
+
|
| 491 |
+
# In[ ]:
|
| 492 |
+
|
| 493 |
+
|
| 494 |
+
sns.heatmap(data.corr(),annot=True,cmap='RdYlGn',linewidths=0.2) #data.corr()-->correlation matrix
|
| 495 |
+
fig=plt.gcf()
|
| 496 |
+
fig.set_size_inches(10,8)
|
| 497 |
+
plt.show()
|
| 498 |
+
|
| 499 |
+
|
| 500 |
+
# ### Interpreting The Heatmap
|
| 501 |
+
#
|
| 502 |
+
# The first thing to note is that only the numeric features are compared as it is obvious that we cannot correlate between alphabets or strings. Before understanding the plot, let us see what exactly correlation is.
|
| 503 |
+
#
|
| 504 |
+
# **POSITIVE CORRELATION:** If an **increase in feature A leads to increase in feature B, then they are positively correlated**. A value **1 means perfect positive correlation**.
|
| 505 |
+
#
|
| 506 |
+
# **NEGATIVE CORRELATION:** If an **increase in feature A leads to decrease in feature B, then they are negatively correlated**. A value **-1 means perfect negative correlation**.
|
| 507 |
+
#
|
| 508 |
+
# Now lets say that two features are highly or perfectly correlated, so the increase in one leads to increase in the other. This means that both the features are containing highly similar information and there is very little or no variance in information. This is known as **MultiColinearity** as both of them contains almost the same information.
|
| 509 |
+
#
|
| 510 |
+
# So do you think we should use both of them as **one of them is redundant**. While making or training models, we should try to eliminate redundant features as it reduces training time and many such advantages.
|
| 511 |
+
#
|
| 512 |
+
# Now from the above heatmap,we can see that the features are not much correlated. The highest correlation is between **SibSp and Parch i.e 0.41**. So we can carry on with all features.
|
| 513 |
+
|
| 514 |
+
# ## Part2: Feature Engineering and Data Cleaning
|
| 515 |
+
#
|
| 516 |
+
# Now what is Feature Engineering?
|
| 517 |
+
#
|
| 518 |
+
# Whenever we are given a dataset with features, it is not necessary that all the features will be important. There maybe be many redundant features which should be eliminated. Also we can get or add new features by observing or extracting information from other features.
|
| 519 |
+
#
|
| 520 |
+
# An example would be getting the Initals feature using the Name Feature. Lets see if we can get any new features and eliminate a few. Also we will tranform the existing relevant features to suitable form for Predictive Modeling.
|
| 521 |
+
|
| 522 |
+
# ## Age_band
|
| 523 |
+
#
|
| 524 |
+
# #### Problem With Age Feature:
|
| 525 |
+
# As I have mentioned earlier that **Age is a continous feature**, there is a problem with Continous Variables in Machine Learning Models.
|
| 526 |
+
#
|
| 527 |
+
# **Eg:**If I say to group or arrange Sports Person by **Sex**, We can easily segregate them by Male and Female.
|
| 528 |
+
#
|
| 529 |
+
# Now if I say to group them by their **Age**, then how would you do it? If there are 30 Persons, there may be 30 age values. Now this is problematic.
|
| 530 |
+
#
|
| 531 |
+
# We need to convert these **continous values into categorical values** by either Binning or Normalisation. I will be using binning i.e group a range of ages into a single bin or assign them a single value.
|
| 532 |
+
#
|
| 533 |
+
# Okay so the maximum age of a passenger was 80. So lets divide the range from 0-80 into 5 bins. So 80/5=16.
|
| 534 |
+
# So bins of size 16.
|
| 535 |
+
|
| 536 |
+
# In[ ]:
|
| 537 |
+
|
| 538 |
+
|
| 539 |
+
data['Age_band']=0
|
| 540 |
+
data.loc[data['Age']<=16,'Age_band']=0
|
| 541 |
+
data.loc[(data['Age']>16)&(data['Age']<=32),'Age_band']=1
|
| 542 |
+
data.loc[(data['Age']>32)&(data['Age']<=48),'Age_band']=2
|
| 543 |
+
data.loc[(data['Age']>48)&(data['Age']<=64),'Age_band']=3
|
| 544 |
+
data.loc[data['Age']>64,'Age_band']=4
|
| 545 |
+
data.head(2)
|
| 546 |
+
|
| 547 |
+
|
| 548 |
+
# In[ ]:
|
| 549 |
+
|
| 550 |
+
|
| 551 |
+
data['Age_band'].value_counts().to_frame().style.background_gradient(cmap='summer')#checking the number of passenegers in each band
|
| 552 |
+
|
| 553 |
+
|
| 554 |
+
# In[ ]:
|
| 555 |
+
|
| 556 |
+
|
| 557 |
+
sns.factorplot('Age_band','Survived',data=data,col='Pclass')
|
| 558 |
+
plt.show()
|
| 559 |
+
|
| 560 |
+
|
| 561 |
+
# True that..the survival rate decreases as the age increases irrespective of the Pclass.
|
| 562 |
+
#
|
| 563 |
+
# ## Family_Size and Alone
|
| 564 |
+
# At this point, we can create a new feature called "Family_size" and "Alone" and analyse it. This feature is the summation of Parch and SibSp. It gives us a combined data so that we can check if survival rate have anything to do with family size of the passengers. Alone will denote whether a passenger is alone or not.
|
| 565 |
+
|
| 566 |
+
# In[ ]:
|
| 567 |
+
|
| 568 |
+
|
| 569 |
+
data['Family_Size']=0
|
| 570 |
+
data['Family_Size']=data['Parch']+data['SibSp']#family size
|
| 571 |
+
data['Alone']=0
|
| 572 |
+
data.loc[data.Family_Size==0,'Alone']=1#Alone
|
| 573 |
+
|
| 574 |
+
f,ax=plt.subplots(1,2,figsize=(18,6))
|
| 575 |
+
sns.factorplot('Family_Size','Survived',data=data,ax=ax[0])
|
| 576 |
+
ax[0].set_title('Family_Size vs Survived')
|
| 577 |
+
sns.factorplot('Alone','Survived',data=data,ax=ax[1])
|
| 578 |
+
ax[1].set_title('Alone vs Survived')
|
| 579 |
+
plt.close(2)
|
| 580 |
+
plt.close(3)
|
| 581 |
+
plt.show()
|
| 582 |
+
|
| 583 |
+
|
| 584 |
+
# **Family_Size=0 means that the passeneger is alone.** Clearly, if you are alone or family_size=0,then chances for survival is very low. For family size > 4,the chances decrease too. This also looks to be an important feature for the model. Lets examine this further.
|
| 585 |
+
|
| 586 |
+
# In[ ]:
|
| 587 |
+
|
| 588 |
+
|
| 589 |
+
sns.factorplot('Alone','Survived',data=data,hue='Sex',col='Pclass')
|
| 590 |
+
plt.show()
|
| 591 |
+
|
| 592 |
+
|
| 593 |
+
# It is visible that being alone is harmful irrespective of Sex or Pclass except for Pclass3 where the chances of females who are alone is high than those with family.
|
| 594 |
+
#
|
| 595 |
+
# ## Fare_Range
|
| 596 |
+
#
|
| 597 |
+
# Since fare is also a continous feature, we need to convert it into ordinal value. For this we will use **pandas.qcut**.
|
| 598 |
+
#
|
| 599 |
+
# So what **qcut** does is it splits or arranges the values according the number of bins we have passed. So if we pass for 5 bins, it will arrange the values equally spaced into 5 seperate bins or value ranges.
|
| 600 |
+
|
| 601 |
+
# In[ ]:
|
| 602 |
+
|
| 603 |
+
|
| 604 |
+
data['Fare_Range']=pd.qcut(data['Fare'],4)
|
| 605 |
+
data.groupby(['Fare_Range'])['Survived'].mean().to_frame().style.background_gradient(cmap='summer_r')
|
| 606 |
+
|
| 607 |
+
|
| 608 |
+
# As discussed above, we can clearly see that as the **fare_range increases, the chances of survival increases.**
|
| 609 |
+
#
|
| 610 |
+
# Now we cannot pass the Fare_Range values as it is. We should convert it into singleton values same as we did in **Age_Band**
|
| 611 |
+
|
| 612 |
+
# In[ ]:
|
| 613 |
+
|
| 614 |
+
|
| 615 |
+
data['Fare_cat']=0
|
| 616 |
+
data.loc[data['Fare']<=7.91,'Fare_cat']=0
|
| 617 |
+
data.loc[(data['Fare']>7.91)&(data['Fare']<=14.454),'Fare_cat']=1
|
| 618 |
+
data.loc[(data['Fare']>14.454)&(data['Fare']<=31),'Fare_cat']=2
|
| 619 |
+
data.loc[(data['Fare']>31)&(data['Fare']<=513),'Fare_cat']=3
|
| 620 |
+
|
| 621 |
+
|
| 622 |
+
# In[ ]:
|
| 623 |
+
|
| 624 |
+
|
| 625 |
+
sns.factorplot('Fare_cat','Survived',data=data,hue='Sex')
|
| 626 |
+
plt.show()
|
| 627 |
+
|
| 628 |
+
|
| 629 |
+
# Clearly, as the Fare_cat increases, the survival chances increases. This feature may become an important feature during modeling along with the Sex.
|
| 630 |
+
#
|
| 631 |
+
# ## Converting String Values into Numeric
|
| 632 |
+
#
|
| 633 |
+
# Since we cannot pass strings to a machine learning model, we need to convert features loke Sex, Embarked, etc into numeric values.
|
| 634 |
+
|
| 635 |
+
# In[ ]:
|
| 636 |
+
|
| 637 |
+
|
| 638 |
+
data['Sex'].replace(['male','female'],[0,1],inplace=True)
|
| 639 |
+
data['Embarked'].replace(['S','C','Q'],[0,1,2],inplace=True)
|
| 640 |
+
data['Initial'].replace(['Mr','Mrs','Miss','Master','Other'],[0,1,2,3,4],inplace=True)
|
| 641 |
+
|
| 642 |
+
|
| 643 |
+
# ### Dropping UnNeeded Features
|
| 644 |
+
#
|
| 645 |
+
# **Name**--> We don't need name feature as it cannot be converted into any categorical value.
|
| 646 |
+
#
|
| 647 |
+
# **Age**--> We have the Age_band feature, so no need of this.
|
| 648 |
+
#
|
| 649 |
+
# **Ticket**--> It is any random string that cannot be categorised.
|
| 650 |
+
#
|
| 651 |
+
# **Fare**--> We have the Fare_cat feature, so unneeded
|
| 652 |
+
#
|
| 653 |
+
# **Cabin**--> A lot of NaN values and also many passengers have multiple cabins. So this is a useless feature.
|
| 654 |
+
#
|
| 655 |
+
# **Fare_Range**--> We have the fare_cat feature.
|
| 656 |
+
#
|
| 657 |
+
# **PassengerId**--> Cannot be categorised.
|
| 658 |
+
|
| 659 |
+
# In[ ]:
|
| 660 |
+
|
| 661 |
+
|
| 662 |
+
data.drop(['Name','Age','Ticket','Fare','Cabin','Fare_Range','PassengerId'],axis=1,inplace=True)
|
| 663 |
+
sns.heatmap(data.corr(),annot=True,cmap='RdYlGn',linewidths=0.2,annot_kws={'size':20})
|
| 664 |
+
fig=plt.gcf()
|
| 665 |
+
fig.set_size_inches(18,15)
|
| 666 |
+
plt.xticks(fontsize=14)
|
| 667 |
+
plt.yticks(fontsize=14)
|
| 668 |
+
plt.show()
|
| 669 |
+
|
| 670 |
+
|
| 671 |
+
# Now the above correlation plot, we can see some positively related features. Some of them being **SibSp andd Family_Size** and **Parch and Family_Size** and some negative ones like **Alone and Family_Size.**
|
| 672 |
+
|
| 673 |
+
# # Part3: Predictive Modeling
|
| 674 |
+
#
|
| 675 |
+
# We have gained some insights from the EDA part. But with that, we cannot accurately predict or tell whether a passenger will survive or die. So now we will predict the whether the Passenger will survive or not using some great Classification Algorithms.Following are the algorithms I will use to make the model:
|
| 676 |
+
#
|
| 677 |
+
# 1)Logistic Regression
|
| 678 |
+
#
|
| 679 |
+
# 2)Support Vector Machines(Linear and radial)
|
| 680 |
+
#
|
| 681 |
+
# 3)Random Forest
|
| 682 |
+
#
|
| 683 |
+
# 4)K-Nearest Neighbours
|
| 684 |
+
#
|
| 685 |
+
# 5)Naive Bayes
|
| 686 |
+
#
|
| 687 |
+
# 6)Decision Tree
|
| 688 |
+
#
|
| 689 |
+
# 7)Logistic Regression
|
| 690 |
+
|
| 691 |
+
# In[ ]:
|
| 692 |
+
|
| 693 |
+
|
| 694 |
+
#importing all the required ML packages
|
| 695 |
+
from sklearn.linear_model import LogisticRegression #logistic regression
|
| 696 |
+
from sklearn import svm #support vector Machine
|
| 697 |
+
from sklearn.ensemble import RandomForestClassifier #Random Forest
|
| 698 |
+
from sklearn.neighbors import KNeighborsClassifier #KNN
|
| 699 |
+
from sklearn.naive_bayes import GaussianNB #Naive bayes
|
| 700 |
+
from sklearn.tree import DecisionTreeClassifier #Decision Tree
|
| 701 |
+
from sklearn.model_selection import train_test_split #training and testing data split
|
| 702 |
+
from sklearn import metrics #accuracy measure
|
| 703 |
+
from sklearn.metrics import confusion_matrix #for confusion matrix
|
| 704 |
+
|
| 705 |
+
|
| 706 |
+
# In[ ]:
|
| 707 |
+
|
| 708 |
+
|
| 709 |
+
train,test=train_test_split(data,test_size=0.3,random_state=0,stratify=data['Survived'])
|
| 710 |
+
train_X=train[train.columns[1:]]
|
| 711 |
+
train_Y=train[train.columns[:1]]
|
| 712 |
+
test_X=test[test.columns[1:]]
|
| 713 |
+
test_Y=test[test.columns[:1]]
|
| 714 |
+
X=data[data.columns[1:]]
|
| 715 |
+
Y=data['Survived']
|
| 716 |
+
|
| 717 |
+
|
| 718 |
+
# ### Radial Support Vector Machines(rbf-SVM)
|
| 719 |
+
|
| 720 |
+
# In[ ]:
|
| 721 |
+
|
| 722 |
+
|
| 723 |
+
model=svm.SVC(kernel='rbf',C=1,gamma=0.1)
|
| 724 |
+
model.fit(train_X,train_Y)
|
| 725 |
+
prediction1=model.predict(test_X)
|
| 726 |
+
print('Accuracy for rbf SVM is ',metrics.accuracy_score(prediction1,test_Y))
|
| 727 |
+
|
| 728 |
+
|
| 729 |
+
# ### Linear Support Vector Machine(linear-SVM)
|
| 730 |
+
|
| 731 |
+
# In[ ]:
|
| 732 |
+
|
| 733 |
+
|
| 734 |
+
model=svm.SVC(kernel='linear',C=0.1,gamma=0.1)
|
| 735 |
+
model.fit(train_X,train_Y)
|
| 736 |
+
prediction2=model.predict(test_X)
|
| 737 |
+
print('Accuracy for linear SVM is',metrics.accuracy_score(prediction2,test_Y))
|
| 738 |
+
|
| 739 |
+
|
| 740 |
+
# ### Logistic Regression
|
| 741 |
+
|
| 742 |
+
# In[ ]:
|
| 743 |
+
|
| 744 |
+
|
| 745 |
+
model = LogisticRegression()
|
| 746 |
+
model.fit(train_X,train_Y)
|
| 747 |
+
prediction3=model.predict(test_X)
|
| 748 |
+
print('The accuracy of the Logistic Regression is',metrics.accuracy_score(prediction3,test_Y))
|
| 749 |
+
|
| 750 |
+
|
| 751 |
+
# ### Decision Tree
|
| 752 |
+
|
| 753 |
+
# In[ ]:
|
| 754 |
+
|
| 755 |
+
|
| 756 |
+
model=DecisionTreeClassifier()
|
| 757 |
+
model.fit(train_X,train_Y)
|
| 758 |
+
prediction4=model.predict(test_X)
|
| 759 |
+
print('The accuracy of the Decision Tree is',metrics.accuracy_score(prediction4,test_Y))
|
| 760 |
+
|
| 761 |
+
|
| 762 |
+
# ### K-Nearest Neighbours(KNN)
|
| 763 |
+
|
| 764 |
+
# In[ ]:
|
| 765 |
+
|
| 766 |
+
|
| 767 |
+
model=KNeighborsClassifier()
|
| 768 |
+
model.fit(train_X,train_Y)
|
| 769 |
+
prediction5=model.predict(test_X)
|
| 770 |
+
print('The accuracy of the KNN is',metrics.accuracy_score(prediction5,test_Y))
|
| 771 |
+
|
| 772 |
+
|
| 773 |
+
# Now the accuracy for the KNN model changes as we change the values for **n_neighbours** attribute. The default value is **5**. Lets check the accuracies over various values of n_neighbours.
|
| 774 |
+
|
| 775 |
+
# In[ ]:
|
| 776 |
+
|
| 777 |
+
|
| 778 |
+
a_index=list(range(1,11))
|
| 779 |
+
a=pd.Series()
|
| 780 |
+
x=[0,1,2,3,4,5,6,7,8,9,10]
|
| 781 |
+
for i in list(range(1,11)):
|
| 782 |
+
model=KNeighborsClassifier(n_neighbors=i)
|
| 783 |
+
model.fit(train_X,train_Y)
|
| 784 |
+
prediction=model.predict(test_X)
|
| 785 |
+
a=a.append(pd.Series(metrics.accuracy_score(prediction,test_Y)))
|
| 786 |
+
plt.plot(a_index, a)
|
| 787 |
+
plt.xticks(x)
|
| 788 |
+
fig=plt.gcf()
|
| 789 |
+
fig.set_size_inches(12,6)
|
| 790 |
+
plt.show()
|
| 791 |
+
print('Accuracies for different values of n are:',a.values,'with the max value as ',a.values.max())
|
| 792 |
+
|
| 793 |
+
|
| 794 |
+
# ### Gaussian Naive Bayes
|
| 795 |
+
|
| 796 |
+
# In[ ]:
|
| 797 |
+
|
| 798 |
+
|
| 799 |
+
model=GaussianNB()
|
| 800 |
+
model.fit(train_X,train_Y)
|
| 801 |
+
prediction6=model.predict(test_X)
|
| 802 |
+
print('The accuracy of the NaiveBayes is',metrics.accuracy_score(prediction6,test_Y))
|
| 803 |
+
|
| 804 |
+
|
| 805 |
+
# ### Random Forests
|
| 806 |
+
|
| 807 |
+
# In[ ]:
|
| 808 |
+
|
| 809 |
+
|
| 810 |
+
model=RandomForestClassifier(n_estimators=100)
|
| 811 |
+
model.fit(train_X,train_Y)
|
| 812 |
+
prediction7=model.predict(test_X)
|
| 813 |
+
print('The accuracy of the Random Forests is',metrics.accuracy_score(prediction7,test_Y))
|
| 814 |
+
|
| 815 |
+
|
| 816 |
+
# The accuracy of a model is not the only factor that determines the robustness of the classifier. Let's say that a classifier is trained over a training data and tested over the test data and it scores an accuracy of 90%.
|
| 817 |
+
#
|
| 818 |
+
# Now this seems to be very good accuracy for a classifier, but can we confirm that it will be 90% for all the new test sets that come over??. The answer is **No**, because we can't determine which all instances will the classifier will use to train itself. As the training and testing data changes, the accuracy will also change. It may increase or decrease. This is known as **model variance**.
|
| 819 |
+
#
|
| 820 |
+
# To overcome this and get a generalized model,we use **Cross Validation**.
|
| 821 |
+
#
|
| 822 |
+
#
|
| 823 |
+
# # Cross Validation
|
| 824 |
+
#
|
| 825 |
+
# Many a times, the data is imbalanced, i.e there may be a high number of class1 instances but less number of other class instances. Thus we should train and test our algorithm on each and every instance of the dataset. Then we can take an average of all the noted accuracies over the dataset.
|
| 826 |
+
#
|
| 827 |
+
# 1)The K-Fold Cross Validation works by first dividing the dataset into k-subsets.
|
| 828 |
+
#
|
| 829 |
+
# 2)Let's say we divide the dataset into (k=5) parts. We reserve 1 part for testing and train the algorithm over the 4 parts.
|
| 830 |
+
#
|
| 831 |
+
# 3)We continue the process by changing the testing part in each iteration and training the algorithm over the other parts. The accuracies and errors are then averaged to get a average accuracy of the algorithm.
|
| 832 |
+
#
|
| 833 |
+
# This is called K-Fold Cross Validation.
|
| 834 |
+
#
|
| 835 |
+
# 4)An algorithm may underfit over a dataset for some training data and sometimes also overfit the data for other training set. Thus with cross-validation, we can achieve a generalised model.
|
| 836 |
+
|
| 837 |
+
# In[ ]:
|
| 838 |
+
|
| 839 |
+
|
| 840 |
+
from sklearn.model_selection import KFold #for K-fold cross validation
|
| 841 |
+
from sklearn.model_selection import cross_val_score #score evaluation
|
| 842 |
+
from sklearn.model_selection import cross_val_predict #prediction
|
| 843 |
+
kfold = KFold(n_splits=10, random_state=22) # k=10, split the data into 10 equal parts
|
| 844 |
+
xyz=[]
|
| 845 |
+
accuracy=[]
|
| 846 |
+
std=[]
|
| 847 |
+
classifiers=['Linear Svm','Radial Svm','Logistic Regression','KNN','Decision Tree','Naive Bayes','Random Forest']
|
| 848 |
+
models=[svm.SVC(kernel='linear'),svm.SVC(kernel='rbf'),LogisticRegression(),KNeighborsClassifier(n_neighbors=9),DecisionTreeClassifier(),GaussianNB(),RandomForestClassifier(n_estimators=100)]
|
| 849 |
+
for i in models:
|
| 850 |
+
model = i
|
| 851 |
+
cv_result = cross_val_score(model,X,Y, cv = kfold,scoring = "accuracy")
|
| 852 |
+
cv_result=cv_result
|
| 853 |
+
xyz.append(cv_result.mean())
|
| 854 |
+
std.append(cv_result.std())
|
| 855 |
+
accuracy.append(cv_result)
|
| 856 |
+
new_models_dataframe2=pd.DataFrame({'CV Mean':xyz,'Std':std},index=classifiers)
|
| 857 |
+
new_models_dataframe2
|
| 858 |
+
|
| 859 |
+
|
| 860 |
+
# In[ ]:
|
| 861 |
+
|
| 862 |
+
|
| 863 |
+
plt.subplots(figsize=(12,6))
|
| 864 |
+
box=pd.DataFrame(accuracy,index=[classifiers])
|
| 865 |
+
box.T.boxplot()
|
| 866 |
+
|
| 867 |
+
|
| 868 |
+
# In[ ]:
|
| 869 |
+
|
| 870 |
+
|
| 871 |
+
new_models_dataframe2['CV Mean'].plot.barh(width=0.8)
|
| 872 |
+
plt.title('Average CV Mean Accuracy')
|
| 873 |
+
fig=plt.gcf()
|
| 874 |
+
fig.set_size_inches(8,5)
|
| 875 |
+
plt.show()
|
| 876 |
+
|
| 877 |
+
|
| 878 |
+
# The classification accuracy can be sometimes misleading due to imbalance. We can get a summarized result with the help of confusion matrix, which shows where did the model go wrong, or which class did the model predict wrong.
|
| 879 |
+
#
|
| 880 |
+
# ## Confusion Matrix
|
| 881 |
+
#
|
| 882 |
+
# It gives the number of correct and incorrect classifications made by the classifier.
|
| 883 |
+
|
| 884 |
+
# In[ ]:
|
| 885 |
+
|
| 886 |
+
|
| 887 |
+
f,ax=plt.subplots(3,3,figsize=(12,10))
|
| 888 |
+
y_pred = cross_val_predict(svm.SVC(kernel='rbf'),X,Y,cv=10)
|
| 889 |
+
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[0,0],annot=True,fmt='2.0f')
|
| 890 |
+
ax[0,0].set_title('Matrix for rbf-SVM')
|
| 891 |
+
y_pred = cross_val_predict(svm.SVC(kernel='linear'),X,Y,cv=10)
|
| 892 |
+
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[0,1],annot=True,fmt='2.0f')
|
| 893 |
+
ax[0,1].set_title('Matrix for Linear-SVM')
|
| 894 |
+
y_pred = cross_val_predict(KNeighborsClassifier(n_neighbors=9),X,Y,cv=10)
|
| 895 |
+
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[0,2],annot=True,fmt='2.0f')
|
| 896 |
+
ax[0,2].set_title('Matrix for KNN')
|
| 897 |
+
y_pred = cross_val_predict(RandomForestClassifier(n_estimators=100),X,Y,cv=10)
|
| 898 |
+
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[1,0],annot=True,fmt='2.0f')
|
| 899 |
+
ax[1,0].set_title('Matrix for Random-Forests')
|
| 900 |
+
y_pred = cross_val_predict(LogisticRegression(),X,Y,cv=10)
|
| 901 |
+
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[1,1],annot=True,fmt='2.0f')
|
| 902 |
+
ax[1,1].set_title('Matrix for Logistic Regression')
|
| 903 |
+
y_pred = cross_val_predict(DecisionTreeClassifier(),X,Y,cv=10)
|
| 904 |
+
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[1,2],annot=True,fmt='2.0f')
|
| 905 |
+
ax[1,2].set_title('Matrix for Decision Tree')
|
| 906 |
+
y_pred = cross_val_predict(GaussianNB(),X,Y,cv=10)
|
| 907 |
+
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[2,0],annot=True,fmt='2.0f')
|
| 908 |
+
ax[2,0].set_title('Matrix for Naive Bayes')
|
| 909 |
+
plt.subplots_adjust(hspace=0.2,wspace=0.2)
|
| 910 |
+
plt.show()
|
| 911 |
+
|
| 912 |
+
|
| 913 |
+
# ### Interpreting Confusion Matrix
|
| 914 |
+
#
|
| 915 |
+
# The left diagonal shows the number of correct predictions made for each class while the right diagonal shows the number of wrong prredictions made. Lets consider the first plot for rbf-SVM:
|
| 916 |
+
#
|
| 917 |
+
# 1)The no. of correct predictions are **491(for dead) + 247(for survived)** with the mean CV accuracy being **(491+247)/891 = 82.8%** which we did get earlier.
|
| 918 |
+
#
|
| 919 |
+
# 2)**Errors**--> Wrongly Classified 58 dead people as survived and 95 survived as dead. Thus it has made more mistakes by predicting dead as survived.
|
| 920 |
+
#
|
| 921 |
+
# By looking at all the matrices, we can say that rbf-SVM has a higher chance in correctly predicting dead passengers but NaiveBayes has a higher chance in correctly predicting passengers who survived.
|
| 922 |
+
|
| 923 |
+
# ### Hyper-Parameters Tuning
|
| 924 |
+
#
|
| 925 |
+
# The machine learning models are like a Black-Box. There are some default parameter values for this Black-Box, which we can tune or change to get a better model. Like the C and gamma in the SVM model and similarly different parameters for different classifiers, are called the hyper-parameters, which we can tune to change the learning rate of the algorithm and get a better model. This is known as Hyper-Parameter Tuning.
|
| 926 |
+
#
|
| 927 |
+
# We will tune the hyper-parameters for the 2 best classifiers i.e the SVM and RandomForests.
|
| 928 |
+
#
|
| 929 |
+
# #### SVM
|
| 930 |
+
|
| 931 |
+
# In[ ]:
|
| 932 |
+
|
| 933 |
+
|
| 934 |
+
from sklearn.model_selection import GridSearchCV
|
| 935 |
+
C=[0.05,0.1,0.2,0.3,0.25,0.4,0.5,0.6,0.7,0.8,0.9,1]
|
| 936 |
+
gamma=[0.1,0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,1.0]
|
| 937 |
+
kernel=['rbf','linear']
|
| 938 |
+
hyper={'kernel':kernel,'C':C,'gamma':gamma}
|
| 939 |
+
gd=GridSearchCV(estimator=svm.SVC(),param_grid=hyper,verbose=True)
|
| 940 |
+
gd.fit(X,Y)
|
| 941 |
+
print(gd.best_score_)
|
| 942 |
+
print(gd.best_estimator_)
|
| 943 |
+
|
| 944 |
+
|
| 945 |
+
# #### Random Forests
|
| 946 |
+
|
| 947 |
+
# In[ ]:
|
| 948 |
+
|
| 949 |
+
|
| 950 |
+
n_estimators=range(100,1000,100)
|
| 951 |
+
hyper={'n_estimators':n_estimators}
|
| 952 |
+
gd=GridSearchCV(estimator=RandomForestClassifier(random_state=0),param_grid=hyper,verbose=True)
|
| 953 |
+
gd.fit(X,Y)
|
| 954 |
+
print(gd.best_score_)
|
| 955 |
+
print(gd.best_estimator_)
|
| 956 |
+
|
| 957 |
+
|
| 958 |
+
# The best score for Rbf-Svm is **82.82% with C=0.05 and gamma=0.1**.
|
| 959 |
+
# For RandomForest, score is abt **81.8% with n_estimators=900**.
|
| 960 |
+
|
| 961 |
+
# # Ensembling
|
| 962 |
+
#
|
| 963 |
+
# Ensembling is a good way to increase the accuracy or performance of a model. In simple words, it is the combination of various simple models to create a single powerful model.
|
| 964 |
+
#
|
| 965 |
+
# Lets say we want to buy a phone and ask many people about it based on various parameters. So then we can make a strong judgement about a single product after analysing all different parameters. This is **Ensembling**, which improves the stability of the model. Ensembling can be done in ways like:
|
| 966 |
+
#
|
| 967 |
+
# 1)Voting Classifier
|
| 968 |
+
#
|
| 969 |
+
# 2)Bagging
|
| 970 |
+
#
|
| 971 |
+
# 3)Boosting.
|
| 972 |
+
|
| 973 |
+
# ## Voting Classifier
|
| 974 |
+
#
|
| 975 |
+
# It is the simplest way of combining predictions from many different simple machine learning models. It gives an average prediction result based on the prediction of all the submodels. The submodels or the basemodels are all of diiferent types.
|
| 976 |
+
|
| 977 |
+
# In[ ]:
|
| 978 |
+
|
| 979 |
+
|
| 980 |
+
from sklearn.ensemble import VotingClassifier
|
| 981 |
+
ensemble_lin_rbf=VotingClassifier(estimators=[('KNN',KNeighborsClassifier(n_neighbors=10)),
|
| 982 |
+
('RBF',svm.SVC(probability=True,kernel='rbf',C=0.5,gamma=0.1)),
|
| 983 |
+
('RFor',RandomForestClassifier(n_estimators=500,random_state=0)),
|
| 984 |
+
('LR',LogisticRegression(C=0.05)),
|
| 985 |
+
('DT',DecisionTreeClassifier(random_state=0)),
|
| 986 |
+
('NB',GaussianNB()),
|
| 987 |
+
('svm',svm.SVC(kernel='linear',probability=True))
|
| 988 |
+
],
|
| 989 |
+
voting='soft').fit(train_X,train_Y)
|
| 990 |
+
print('The accuracy for ensembled model is:',ensemble_lin_rbf.score(test_X,test_Y))
|
| 991 |
+
cross=cross_val_score(ensemble_lin_rbf,X,Y, cv = 10,scoring = "accuracy")
|
| 992 |
+
print('The cross validated score is',cross.mean())
|
| 993 |
+
|
| 994 |
+
|
| 995 |
+
# ## Bagging
|
| 996 |
+
#
|
| 997 |
+
# Bagging is a general ensemble method. It works by applying similar classifiers on small partitions of the dataset and then taking the average of all the predictions. Due to the averaging,there is reduction in variance. Unlike Voting Classifier, Bagging makes use of similar classifiers.
|
| 998 |
+
#
|
| 999 |
+
# #### Bagged KNN
|
| 1000 |
+
#
|
| 1001 |
+
# Bagging works best with models with high variance. An example for this can be Decision Tree or Random Forests. We can use KNN with small value of **n_neighbours**, as small value of n_neighbours.
|
| 1002 |
+
|
| 1003 |
+
# In[ ]:
|
| 1004 |
+
|
| 1005 |
+
|
| 1006 |
+
from sklearn.ensemble import BaggingClassifier
|
| 1007 |
+
model=BaggingClassifier(base_estimator=KNeighborsClassifier(n_neighbors=3),random_state=0,n_estimators=700)
|
| 1008 |
+
model.fit(train_X,train_Y)
|
| 1009 |
+
prediction=model.predict(test_X)
|
| 1010 |
+
print('The accuracy for bagged KNN is:',metrics.accuracy_score(prediction,test_Y))
|
| 1011 |
+
result=cross_val_score(model,X,Y,cv=10,scoring='accuracy')
|
| 1012 |
+
print('The cross validated score for bagged KNN is:',result.mean())
|
| 1013 |
+
|
| 1014 |
+
|
| 1015 |
+
# #### Bagged DecisionTree
|
| 1016 |
+
#
|
| 1017 |
+
|
| 1018 |
+
# In[ ]:
|
| 1019 |
+
|
| 1020 |
+
|
| 1021 |
+
model=BaggingClassifier(base_estimator=DecisionTreeClassifier(),random_state=0,n_estimators=100)
|
| 1022 |
+
model.fit(train_X,train_Y)
|
| 1023 |
+
prediction=model.predict(test_X)
|
| 1024 |
+
print('The accuracy for bagged Decision Tree is:',metrics.accuracy_score(prediction,test_Y))
|
| 1025 |
+
result=cross_val_score(model,X,Y,cv=10,scoring='accuracy')
|
| 1026 |
+
print('The cross validated score for bagged Decision Tree is:',result.mean())
|
| 1027 |
+
|
| 1028 |
+
|
| 1029 |
+
# ## Boosting
|
| 1030 |
+
#
|
| 1031 |
+
# Boosting is an ensembling technique which uses sequential learning of classifiers. It is a step by step enhancement of a weak model.Boosting works as follows:
|
| 1032 |
+
#
|
| 1033 |
+
# A model is first trained on the complete dataset. Now the model will get some instances right while some wrong. Now in the next iteration, the learner will focus more on the wrongly predicted instances or give more weight to it. Thus it will try to predict the wrong instance correctly. Now this iterative process continous, and new classifers are added to the model until the limit is reached on the accuracy.
|
| 1034 |
+
|
| 1035 |
+
# #### AdaBoost(Adaptive Boosting)
|
| 1036 |
+
#
|
| 1037 |
+
# The weak learner or estimator in this case is a Decsion Tree. But we can change the dafault base_estimator to any algorithm of our choice.
|
| 1038 |
+
|
| 1039 |
+
# In[ ]:
|
| 1040 |
+
|
| 1041 |
+
|
| 1042 |
+
from sklearn.ensemble import AdaBoostClassifier
|
| 1043 |
+
ada=AdaBoostClassifier(n_estimators=200,random_state=0,learning_rate=0.1)
|
| 1044 |
+
result=cross_val_score(ada,X,Y,cv=10,scoring='accuracy')
|
| 1045 |
+
print('The cross validated score for AdaBoost is:',result.mean())
|
| 1046 |
+
|
| 1047 |
+
|
| 1048 |
+
# #### Stochastic Gradient Boosting
|
| 1049 |
+
#
|
| 1050 |
+
# Here too the weak learner is a Decision Tree.
|
| 1051 |
+
|
| 1052 |
+
# In[ ]:
|
| 1053 |
+
|
| 1054 |
+
|
| 1055 |
+
from sklearn.ensemble import GradientBoostingClassifier
|
| 1056 |
+
grad=GradientBoostingClassifier(n_estimators=500,random_state=0,learning_rate=0.1)
|
| 1057 |
+
result=cross_val_score(grad,X,Y,cv=10,scoring='accuracy')
|
| 1058 |
+
print('The cross validated score for Gradient Boosting is:',result.mean())
|
| 1059 |
+
|
| 1060 |
+
|
| 1061 |
+
# #### XGBoost
|
| 1062 |
+
|
| 1063 |
+
# In[ ]:
|
| 1064 |
+
|
| 1065 |
+
|
| 1066 |
+
import xgboost as xg
|
| 1067 |
+
xgboost=xg.XGBClassifier(n_estimators=900,learning_rate=0.1)
|
| 1068 |
+
result=cross_val_score(xgboost,X,Y,cv=10,scoring='accuracy')
|
| 1069 |
+
print('The cross validated score for XGBoost is:',result.mean())
|
| 1070 |
+
|
| 1071 |
+
|
| 1072 |
+
# We got the highest accuracy for AdaBoost. We will try to increase it with Hyper-Parameter Tuning
|
| 1073 |
+
#
|
| 1074 |
+
# #### Hyper-Parameter Tuning for AdaBoost
|
| 1075 |
+
|
| 1076 |
+
# In[ ]:
|
| 1077 |
+
|
| 1078 |
+
|
| 1079 |
+
n_estimators=list(range(100,1100,100))
|
| 1080 |
+
learn_rate=[0.05,0.1,0.2,0.3,0.25,0.4,0.5,0.6,0.7,0.8,0.9,1]
|
| 1081 |
+
hyper={'n_estimators':n_estimators,'learning_rate':learn_rate}
|
| 1082 |
+
gd=GridSearchCV(estimator=AdaBoostClassifier(),param_grid=hyper,verbose=True)
|
| 1083 |
+
gd.fit(X,Y)
|
| 1084 |
+
print(gd.best_score_)
|
| 1085 |
+
print(gd.best_estimator_)
|
| 1086 |
+
|
| 1087 |
+
|
| 1088 |
+
# The maximum accuracy we can get with AdaBoost is **83.16% with n_estimators=200 and learning_rate=0.05**
|
| 1089 |
+
|
| 1090 |
+
# ### Confusion Matrix for the Best Model
|
| 1091 |
+
|
| 1092 |
+
# In[ ]:
|
| 1093 |
+
|
| 1094 |
+
|
| 1095 |
+
ada=AdaBoostClassifier(n_estimators=200,random_state=0,learning_rate=0.05)
|
| 1096 |
+
result=cross_val_predict(ada,X,Y,cv=10)
|
| 1097 |
+
sns.heatmap(confusion_matrix(Y,result),cmap='winter',annot=True,fmt='2.0f')
|
| 1098 |
+
plt.show()
|
| 1099 |
+
|
| 1100 |
+
|
| 1101 |
+
# ## Feature Importance
|
| 1102 |
+
|
| 1103 |
+
# In[ ]:
|
| 1104 |
+
|
| 1105 |
+
|
| 1106 |
+
f,ax=plt.subplots(2,2,figsize=(15,12))
|
| 1107 |
+
model=RandomForestClassifier(n_estimators=500,random_state=0)
|
| 1108 |
+
model.fit(X,Y)
|
| 1109 |
+
pd.Series(model.feature_importances_,X.columns).sort_values(ascending=True).plot.barh(width=0.8,ax=ax[0,0])
|
| 1110 |
+
ax[0,0].set_title('Feature Importance in Random Forests')
|
| 1111 |
+
model=AdaBoostClassifier(n_estimators=200,learning_rate=0.05,random_state=0)
|
| 1112 |
+
model.fit(X,Y)
|
| 1113 |
+
pd.Series(model.feature_importances_,X.columns).sort_values(ascending=True).plot.barh(width=0.8,ax=ax[0,1],color='#ddff11')
|
| 1114 |
+
ax[0,1].set_title('Feature Importance in AdaBoost')
|
| 1115 |
+
model=GradientBoostingClassifier(n_estimators=500,learning_rate=0.1,random_state=0)
|
| 1116 |
+
model.fit(X,Y)
|
| 1117 |
+
pd.Series(model.feature_importances_,X.columns).sort_values(ascending=True).plot.barh(width=0.8,ax=ax[1,0],cmap='RdYlGn_r')
|
| 1118 |
+
ax[1,0].set_title('Feature Importance in Gradient Boosting')
|
| 1119 |
+
model=xg.XGBClassifier(n_estimators=900,learning_rate=0.1)
|
| 1120 |
+
model.fit(X,Y)
|
| 1121 |
+
pd.Series(model.feature_importances_,X.columns).sort_values(ascending=True).plot.barh(width=0.8,ax=ax[1,1],color='#FD0F00')
|
| 1122 |
+
ax[1,1].set_title('Feature Importance in XgBoost')
|
| 1123 |
+
plt.show()
|
| 1124 |
+
|
| 1125 |
+
|
| 1126 |
+
# We can see the important features for various classifiers like RandomForests, AdaBoost,etc.
|
| 1127 |
+
#
|
| 1128 |
+
# #### Observations:
|
| 1129 |
+
#
|
| 1130 |
+
# 1)Some of the common important features are Initial,Fare_cat,Pclass,Family_Size.
|
| 1131 |
+
#
|
| 1132 |
+
# 2)The Sex feature doesn't seem to give any importance, which is shocking as we had seen earlier that Sex combined with Pclass was giving a very good differentiating factor. Sex looks to be important only in RandomForests.
|
| 1133 |
+
#
|
| 1134 |
+
# However, we can see the feature Initial, which is at the top in many classifiers.We had already seen the positive correlation between Sex and Initial, so they both refer to the gender.
|
| 1135 |
+
#
|
| 1136 |
+
# 3)Similarly the Pclass and Fare_cat refer to the status of the passengers and Family_Size with Alone,Parch and SibSp.
|
| 1137 |
+
|
| 1138 |
+
# I hope all of you did gain some insights to Machine Learning. Some other great notebooks for Machine Learning are:
|
| 1139 |
+
# 1) For R:[Divide and Conquer by Oscar Takeshita](https://www.kaggle.com/pliptor/divide-and-conquer-0-82297/notebook)
|
| 1140 |
+
#
|
| 1141 |
+
# 2)For Python:[Pytanic by Heads and Tails](https://www.kaggle.com/headsortails/pytanic)
|
| 1142 |
+
#
|
| 1143 |
+
# 3)For Python:[Introduction to Ensembling/Stacking by Anisotropic](https://www.kaggle.com/arthurtok/introduction-to-ensembling-stacking-in-python)
|
| 1144 |
+
#
|
| 1145 |
+
# ### Thanks a lot for having a look at this notebook. If you found this notebook useful, **Do Upvote**.
|
| 1146 |
+
#
|
| 1147 |
+
|
| 1148 |
+
# In[ ]:
|
| 1149 |
+
|
| 1150 |
+
|
| 1151 |
+
|
| 1152 |
+
|
Titanic/Kernels/AdaBoost/4-a-statistical-analysis-ml-workflow-of-titanic.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/4-a-statistical-analysis-ml-workflow-of-titanic.py
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/6-titanic-best-working-classifier.ipynb
ADDED
|
@@ -0,0 +1,1504 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "markdown",
|
| 5 |
+
"metadata": {
|
| 6 |
+
"_cell_guid": "25b1e1db-8bc5-7029-f719-91da523bd121"
|
| 7 |
+
},
|
| 8 |
+
"source": [
|
| 9 |
+
"## Introduction ##\n",
|
| 10 |
+
"\n",
|
| 11 |
+
"This is my first work of machine learning. the notebook is written in python and has inspired from [\"Exploring Survival on Titanic\" by Megan Risdal, a Kernel in R on Kaggle][1].\n",
|
| 12 |
+
"\n",
|
| 13 |
+
"\n",
|
| 14 |
+
" [1]: https://www.kaggle.com/mrisdal/titanic/exploring-survival-on-the-titanic"
|
| 15 |
+
]
|
| 16 |
+
},
|
| 17 |
+
{
|
| 18 |
+
"cell_type": "code",
|
| 19 |
+
"execution_count": 1,
|
| 20 |
+
"metadata": {
|
| 21 |
+
"_cell_guid": "2ce68358-02ec-556d-ba88-e773a50bc18b"
|
| 22 |
+
},
|
| 23 |
+
"outputs": [
|
| 24 |
+
{
|
| 25 |
+
"name": "stdout",
|
| 26 |
+
"output_type": "stream",
|
| 27 |
+
"text": [
|
| 28 |
+
"<class 'pandas.core.frame.DataFrame'>\n",
|
| 29 |
+
"RangeIndex: 891 entries, 0 to 890\n",
|
| 30 |
+
"Data columns (total 12 columns):\n",
|
| 31 |
+
" # Column Non-Null Count Dtype \n",
|
| 32 |
+
"--- ------ -------------- ----- \n",
|
| 33 |
+
" 0 PassengerId 891 non-null int64 \n",
|
| 34 |
+
" 1 Survived 891 non-null int64 \n",
|
| 35 |
+
" 2 Pclass 891 non-null int64 \n",
|
| 36 |
+
" 3 Name 891 non-null object \n",
|
| 37 |
+
" 4 Sex 891 non-null object \n",
|
| 38 |
+
" 5 Age 714 non-null float64\n",
|
| 39 |
+
" 6 SibSp 891 non-null int64 \n",
|
| 40 |
+
" 7 Parch 891 non-null int64 \n",
|
| 41 |
+
" 8 Ticket 891 non-null object \n",
|
| 42 |
+
" 9 Fare 891 non-null float64\n",
|
| 43 |
+
" 10 Cabin 204 non-null object \n",
|
| 44 |
+
" 11 Embarked 889 non-null object \n",
|
| 45 |
+
"dtypes: float64(2), int64(5), object(5)\n",
|
| 46 |
+
"memory usage: 83.7+ KB\n",
|
| 47 |
+
"None\n"
|
| 48 |
+
]
|
| 49 |
+
}
|
| 50 |
+
],
|
| 51 |
+
"source": [
|
| 52 |
+
"%matplotlib inline\n",
|
| 53 |
+
"import numpy as np\n",
|
| 54 |
+
"import pandas as pd\n",
|
| 55 |
+
"import re as re\n",
|
| 56 |
+
"\n",
|
| 57 |
+
"train = pd.read_csv('../../Data/train.csv', header = 0, dtype={'Age': np.float64})\n",
|
| 58 |
+
"test = pd.read_csv('../../Data/test.csv' , header = 0, dtype={'Age': np.float64})\n",
|
| 59 |
+
"full_data = [train, test]\n",
|
| 60 |
+
"\n",
|
| 61 |
+
"print (train.info())"
|
| 62 |
+
]
|
| 63 |
+
},
|
| 64 |
+
{
|
| 65 |
+
"cell_type": "code",
|
| 66 |
+
"execution_count": 2,
|
| 67 |
+
"metadata": {},
|
| 68 |
+
"outputs": [],
|
| 69 |
+
"source": [
|
| 70 |
+
"from aif360.datasets import StandardDataset\n",
|
| 71 |
+
"from aif360.metrics import BinaryLabelDatasetMetric, ClassificationMetric\n",
|
| 72 |
+
"import matplotlib.patches as patches\n",
|
| 73 |
+
"from aif360.algorithms.preprocessing import Reweighing\n",
|
| 74 |
+
"#from packages import *\n",
|
| 75 |
+
"#from ml_fairness import *\n",
|
| 76 |
+
"import matplotlib.pyplot as plt\n",
|
| 77 |
+
"import seaborn as sns\n",
|
| 78 |
+
"\n",
|
| 79 |
+
"\n",
|
| 80 |
+
"\n",
|
| 81 |
+
"from IPython.display import Markdown, display"
|
| 82 |
+
]
|
| 83 |
+
},
|
| 84 |
+
{
|
| 85 |
+
"cell_type": "markdown",
|
| 86 |
+
"metadata": {
|
| 87 |
+
"_cell_guid": "f9595646-65c9-6fc4-395f-0befc4d122ce"
|
| 88 |
+
},
|
| 89 |
+
"source": [
|
| 90 |
+
"# Feature Engineering #"
|
| 91 |
+
]
|
| 92 |
+
},
|
| 93 |
+
{
|
| 94 |
+
"cell_type": "markdown",
|
| 95 |
+
"metadata": {
|
| 96 |
+
"_cell_guid": "9b4c278b-aaca-e92c-ba77-b9b48379d1f1"
|
| 97 |
+
},
|
| 98 |
+
"source": [
|
| 99 |
+
"## 1. Pclass ##\n",
|
| 100 |
+
"there is no missing value on this feature and already a numerical value. so let's check it's impact on our train set."
|
| 101 |
+
]
|
| 102 |
+
},
|
| 103 |
+
{
|
| 104 |
+
"cell_type": "code",
|
| 105 |
+
"execution_count": 3,
|
| 106 |
+
"metadata": {
|
| 107 |
+
"_cell_guid": "4680d950-cf7d-a6ae-e813-535e2247d88e"
|
| 108 |
+
},
|
| 109 |
+
"outputs": [
|
| 110 |
+
{
|
| 111 |
+
"name": "stdout",
|
| 112 |
+
"output_type": "stream",
|
| 113 |
+
"text": [
|
| 114 |
+
" Pclass Survived\n",
|
| 115 |
+
"0 1 0.629630\n",
|
| 116 |
+
"1 2 0.472826\n",
|
| 117 |
+
"2 3 0.242363\n"
|
| 118 |
+
]
|
| 119 |
+
}
|
| 120 |
+
],
|
| 121 |
+
"source": [
|
| 122 |
+
"print (train[['Pclass', 'Survived']].groupby(['Pclass'], as_index=False).mean())"
|
| 123 |
+
]
|
| 124 |
+
},
|
| 125 |
+
{
|
| 126 |
+
"cell_type": "markdown",
|
| 127 |
+
"metadata": {
|
| 128 |
+
"_cell_guid": "5e70f81c-d4e2-1823-f0ba-a7c9b46984ff"
|
| 129 |
+
},
|
| 130 |
+
"source": [
|
| 131 |
+
"## 2. Sex ##"
|
| 132 |
+
]
|
| 133 |
+
},
|
| 134 |
+
{
|
| 135 |
+
"cell_type": "code",
|
| 136 |
+
"execution_count": 4,
|
| 137 |
+
"metadata": {
|
| 138 |
+
"_cell_guid": "6729681d-7915-1631-78d2-ddf3c35a424c"
|
| 139 |
+
},
|
| 140 |
+
"outputs": [
|
| 141 |
+
{
|
| 142 |
+
"name": "stdout",
|
| 143 |
+
"output_type": "stream",
|
| 144 |
+
"text": [
|
| 145 |
+
" Sex Survived\n",
|
| 146 |
+
"0 female 0.742038\n",
|
| 147 |
+
"1 male 0.188908\n"
|
| 148 |
+
]
|
| 149 |
+
}
|
| 150 |
+
],
|
| 151 |
+
"source": [
|
| 152 |
+
"print (train[[\"Sex\", \"Survived\"]].groupby(['Sex'], as_index=False).mean())"
|
| 153 |
+
]
|
| 154 |
+
},
|
| 155 |
+
{
|
| 156 |
+
"cell_type": "markdown",
|
| 157 |
+
"metadata": {
|
| 158 |
+
"_cell_guid": "7c58b7ee-d6a1-0cc9-2346-81c47846a54a"
|
| 159 |
+
},
|
| 160 |
+
"source": [
|
| 161 |
+
"## 3. SibSp and Parch ##\n",
|
| 162 |
+
"With the number of siblings/spouse and the number of children/parents we can create new feature called Family Size."
|
| 163 |
+
]
|
| 164 |
+
},
|
| 165 |
+
{
|
| 166 |
+
"cell_type": "code",
|
| 167 |
+
"execution_count": 5,
|
| 168 |
+
"metadata": {
|
| 169 |
+
"_cell_guid": "1a537f10-7cec-d0b7-8a34-fa9975655190"
|
| 170 |
+
},
|
| 171 |
+
"outputs": [
|
| 172 |
+
{
|
| 173 |
+
"name": "stdout",
|
| 174 |
+
"output_type": "stream",
|
| 175 |
+
"text": [
|
| 176 |
+
" FamilySize Survived\n",
|
| 177 |
+
"0 1 0.303538\n",
|
| 178 |
+
"1 2 0.552795\n",
|
| 179 |
+
"2 3 0.578431\n",
|
| 180 |
+
"3 4 0.724138\n",
|
| 181 |
+
"4 5 0.200000\n",
|
| 182 |
+
"5 6 0.136364\n",
|
| 183 |
+
"6 7 0.333333\n",
|
| 184 |
+
"7 8 0.000000\n",
|
| 185 |
+
"8 11 0.000000\n"
|
| 186 |
+
]
|
| 187 |
+
}
|
| 188 |
+
],
|
| 189 |
+
"source": [
|
| 190 |
+
"for dataset in full_data:\n",
|
| 191 |
+
" dataset['FamilySize'] = dataset['SibSp'] + dataset['Parch'] + 1\n",
|
| 192 |
+
"print (train[['FamilySize', 'Survived']].groupby(['FamilySize'], as_index=False).mean())"
|
| 193 |
+
]
|
| 194 |
+
},
|
| 195 |
+
{
|
| 196 |
+
"cell_type": "markdown",
|
| 197 |
+
"metadata": {
|
| 198 |
+
"_cell_guid": "e4861d3e-10db-1a23-8728-44e4d5251844"
|
| 199 |
+
},
|
| 200 |
+
"source": [
|
| 201 |
+
"it seems has a good effect on our prediction but let's go further and categorize people to check whether they are alone in this ship or not."
|
| 202 |
+
]
|
| 203 |
+
},
|
| 204 |
+
{
|
| 205 |
+
"cell_type": "code",
|
| 206 |
+
"execution_count": 6,
|
| 207 |
+
"metadata": {
|
| 208 |
+
"_cell_guid": "8c35e945-c928-e3bc-bd9c-d6ddb287e4c9"
|
| 209 |
+
},
|
| 210 |
+
"outputs": [
|
| 211 |
+
{
|
| 212 |
+
"name": "stdout",
|
| 213 |
+
"output_type": "stream",
|
| 214 |
+
"text": [
|
| 215 |
+
" IsAlone Survived\n",
|
| 216 |
+
"0 0 0.505650\n",
|
| 217 |
+
"1 1 0.303538\n"
|
| 218 |
+
]
|
| 219 |
+
}
|
| 220 |
+
],
|
| 221 |
+
"source": [
|
| 222 |
+
"for dataset in full_data:\n",
|
| 223 |
+
" dataset['IsAlone'] = 0\n",
|
| 224 |
+
" dataset.loc[dataset['FamilySize'] == 1, 'IsAlone'] = 1\n",
|
| 225 |
+
"print (train[['IsAlone', 'Survived']].groupby(['IsAlone'], as_index=False).mean())"
|
| 226 |
+
]
|
| 227 |
+
},
|
| 228 |
+
{
|
| 229 |
+
"cell_type": "markdown",
|
| 230 |
+
"metadata": {
|
| 231 |
+
"_cell_guid": "2780ca4e-7923-b845-0b6b-5f68a45f6b93"
|
| 232 |
+
},
|
| 233 |
+
"source": [
|
| 234 |
+
"good! the impact is considerable."
|
| 235 |
+
]
|
| 236 |
+
},
|
| 237 |
+
{
|
| 238 |
+
"cell_type": "markdown",
|
| 239 |
+
"metadata": {
|
| 240 |
+
"_cell_guid": "8aa419c0-6614-7efc-7797-97f4a5158b19"
|
| 241 |
+
},
|
| 242 |
+
"source": [
|
| 243 |
+
"## 4. Embarked ##\n",
|
| 244 |
+
"the embarked feature has some missing value. and we try to fill those with the most occurred value ( 'S' )."
|
| 245 |
+
]
|
| 246 |
+
},
|
| 247 |
+
{
|
| 248 |
+
"cell_type": "code",
|
| 249 |
+
"execution_count": 7,
|
| 250 |
+
"metadata": {
|
| 251 |
+
"_cell_guid": "0e70e9af-d7cc-8c40-b7d4-2643889c376d"
|
| 252 |
+
},
|
| 253 |
+
"outputs": [
|
| 254 |
+
{
|
| 255 |
+
"name": "stdout",
|
| 256 |
+
"output_type": "stream",
|
| 257 |
+
"text": [
|
| 258 |
+
" Embarked Survived\n",
|
| 259 |
+
"0 C 0.553571\n",
|
| 260 |
+
"1 Q 0.389610\n",
|
| 261 |
+
"2 S 0.339009\n"
|
| 262 |
+
]
|
| 263 |
+
}
|
| 264 |
+
],
|
| 265 |
+
"source": [
|
| 266 |
+
"for dataset in full_data:\n",
|
| 267 |
+
" dataset['Embarked'] = dataset['Embarked'].fillna('S')\n",
|
| 268 |
+
"print (train[['Embarked', 'Survived']].groupby(['Embarked'], as_index=False).mean())"
|
| 269 |
+
]
|
| 270 |
+
},
|
| 271 |
+
{
|
| 272 |
+
"cell_type": "markdown",
|
| 273 |
+
"metadata": {
|
| 274 |
+
"_cell_guid": "e08c9ee8-d6d1-99b7-38bd-f0042c18a5d9"
|
| 275 |
+
},
|
| 276 |
+
"source": [
|
| 277 |
+
"## 5. Fare ##\n",
|
| 278 |
+
"Fare also has some missing value and we will replace it with the median. then we categorize it into 4 ranges."
|
| 279 |
+
]
|
| 280 |
+
},
|
| 281 |
+
{
|
| 282 |
+
"cell_type": "code",
|
| 283 |
+
"execution_count": 8,
|
| 284 |
+
"metadata": {
|
| 285 |
+
"_cell_guid": "a21335bd-4e8d-66e8-e6a5-5d2173b72d3b"
|
| 286 |
+
},
|
| 287 |
+
"outputs": [
|
| 288 |
+
{
|
| 289 |
+
"name": "stdout",
|
| 290 |
+
"output_type": "stream",
|
| 291 |
+
"text": [
|
| 292 |
+
" CategoricalFare Survived\n",
|
| 293 |
+
"0 (-0.001, 7.91] 0.197309\n",
|
| 294 |
+
"1 (7.91, 14.454] 0.303571\n",
|
| 295 |
+
"2 (14.454, 31.0] 0.454955\n",
|
| 296 |
+
"3 (31.0, 512.329] 0.581081\n"
|
| 297 |
+
]
|
| 298 |
+
}
|
| 299 |
+
],
|
| 300 |
+
"source": [
|
| 301 |
+
"for dataset in full_data:\n",
|
| 302 |
+
" dataset['Fare'] = dataset['Fare'].fillna(train['Fare'].median())\n",
|
| 303 |
+
"train['CategoricalFare'] = pd.qcut(train['Fare'], 4)\n",
|
| 304 |
+
"print (train[['CategoricalFare', 'Survived']].groupby(['CategoricalFare'], as_index=False).mean())"
|
| 305 |
+
]
|
| 306 |
+
},
|
| 307 |
+
{
|
| 308 |
+
"cell_type": "markdown",
|
| 309 |
+
"metadata": {
|
| 310 |
+
"_cell_guid": "ec8d1b22-a95f-9f16-77ab-7b60d2103852"
|
| 311 |
+
},
|
| 312 |
+
"source": [
|
| 313 |
+
"## 6. Age ##\n",
|
| 314 |
+
"we have plenty of missing values in this feature. # generate random numbers between (mean - std) and (mean + std).\n",
|
| 315 |
+
"then we categorize age into 5 range."
|
| 316 |
+
]
|
| 317 |
+
},
|
| 318 |
+
{
|
| 319 |
+
"cell_type": "code",
|
| 320 |
+
"execution_count": 9,
|
| 321 |
+
"metadata": {
|
| 322 |
+
"_cell_guid": "b90c2870-ce5d-ae0e-a33d-59e35445500e"
|
| 323 |
+
},
|
| 324 |
+
"outputs": [
|
| 325 |
+
{
|
| 326 |
+
"name": "stdout",
|
| 327 |
+
"output_type": "stream",
|
| 328 |
+
"text": [
|
| 329 |
+
" CategoricalAge Survived\n",
|
| 330 |
+
"0 (-0.08, 16.0] 0.530973\n",
|
| 331 |
+
"1 (16.0, 32.0] 0.353741\n",
|
| 332 |
+
"2 (32.0, 48.0] 0.369650\n",
|
| 333 |
+
"3 (48.0, 64.0] 0.434783\n",
|
| 334 |
+
"4 (64.0, 80.0] 0.090909\n"
|
| 335 |
+
]
|
| 336 |
+
},
|
| 337 |
+
{
|
| 338 |
+
"name": "stderr",
|
| 339 |
+
"output_type": "stream",
|
| 340 |
+
"text": [
|
| 341 |
+
"\n",
|
| 342 |
+
"A value is trying to be set on a copy of a slice from a DataFrame\n",
|
| 343 |
+
"\n",
|
| 344 |
+
"See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n"
|
| 345 |
+
]
|
| 346 |
+
}
|
| 347 |
+
],
|
| 348 |
+
"source": [
|
| 349 |
+
"for dataset in full_data:\n",
|
| 350 |
+
" age_avg \t = dataset['Age'].mean()\n",
|
| 351 |
+
" age_std \t = dataset['Age'].std()\n",
|
| 352 |
+
" age_null_count = dataset['Age'].isnull().sum()\n",
|
| 353 |
+
" \n",
|
| 354 |
+
" age_null_random_list = np.random.randint(age_avg - age_std, age_avg + age_std, size=age_null_count)\n",
|
| 355 |
+
" dataset['Age'][np.isnan(dataset['Age'])] = age_null_random_list\n",
|
| 356 |
+
" dataset['Age'] = dataset['Age'].astype(int)\n",
|
| 357 |
+
" \n",
|
| 358 |
+
"train['CategoricalAge'] = pd.cut(train['Age'], 5)\n",
|
| 359 |
+
"\n",
|
| 360 |
+
"print (train[['CategoricalAge', 'Survived']].groupby(['CategoricalAge'], as_index=False).mean())"
|
| 361 |
+
]
|
| 362 |
+
},
|
| 363 |
+
{
|
| 364 |
+
"cell_type": "markdown",
|
| 365 |
+
"metadata": {
|
| 366 |
+
"_cell_guid": "bd25ec3f-b601-c1cc-d701-991fac1621f9"
|
| 367 |
+
},
|
| 368 |
+
"source": [
|
| 369 |
+
"## 7. Name ##\n",
|
| 370 |
+
"inside this feature we can find the title of people."
|
| 371 |
+
]
|
| 372 |
+
},
|
| 373 |
+
{
|
| 374 |
+
"cell_type": "code",
|
| 375 |
+
"execution_count": 10,
|
| 376 |
+
"metadata": {
|
| 377 |
+
"_cell_guid": "ad042f43-bfe0-ded0-4171-379d8caaa749"
|
| 378 |
+
},
|
| 379 |
+
"outputs": [
|
| 380 |
+
{
|
| 381 |
+
"name": "stdout",
|
| 382 |
+
"output_type": "stream",
|
| 383 |
+
"text": [
|
| 384 |
+
"Sex female male\n",
|
| 385 |
+
"Title \n",
|
| 386 |
+
"Capt 0 1\n",
|
| 387 |
+
"Col 0 2\n",
|
| 388 |
+
"Countess 1 0\n",
|
| 389 |
+
"Don 0 1\n",
|
| 390 |
+
"Dr 1 6\n",
|
| 391 |
+
"Jonkheer 0 1\n",
|
| 392 |
+
"Lady 1 0\n",
|
| 393 |
+
"Major 0 2\n",
|
| 394 |
+
"Master 0 40\n",
|
| 395 |
+
"Miss 182 0\n",
|
| 396 |
+
"Mlle 2 0\n",
|
| 397 |
+
"Mme 1 0\n",
|
| 398 |
+
"Mr 0 517\n",
|
| 399 |
+
"Mrs 125 0\n",
|
| 400 |
+
"Ms 1 0\n",
|
| 401 |
+
"Rev 0 6\n",
|
| 402 |
+
"Sir 0 1\n"
|
| 403 |
+
]
|
| 404 |
+
}
|
| 405 |
+
],
|
| 406 |
+
"source": [
|
| 407 |
+
"def get_title(name):\n",
|
| 408 |
+
"\ttitle_search = re.search(' ([A-Za-z]+)\\.', name)\n",
|
| 409 |
+
"\t# If the title exists, extract and return it.\n",
|
| 410 |
+
"\tif title_search:\n",
|
| 411 |
+
"\t\treturn title_search.group(1)\n",
|
| 412 |
+
"\treturn \"\"\n",
|
| 413 |
+
"\n",
|
| 414 |
+
"for dataset in full_data:\n",
|
| 415 |
+
" dataset['Title'] = dataset['Name'].apply(get_title)\n",
|
| 416 |
+
"\n",
|
| 417 |
+
"print(pd.crosstab(train['Title'], train['Sex']))"
|
| 418 |
+
]
|
| 419 |
+
},
|
| 420 |
+
{
|
| 421 |
+
"cell_type": "markdown",
|
| 422 |
+
"metadata": {
|
| 423 |
+
"_cell_guid": "ca5fff8c-7a0d-6c18-2173-b8df6293c50a"
|
| 424 |
+
},
|
| 425 |
+
"source": [
|
| 426 |
+
" so we have titles. let's categorize it and check the title impact on survival rate."
|
| 427 |
+
]
|
| 428 |
+
},
|
| 429 |
+
{
|
| 430 |
+
"cell_type": "code",
|
| 431 |
+
"execution_count": 11,
|
| 432 |
+
"metadata": {
|
| 433 |
+
"_cell_guid": "8357238b-98fe-632a-acd5-33674a6132ce"
|
| 434 |
+
},
|
| 435 |
+
"outputs": [
|
| 436 |
+
{
|
| 437 |
+
"name": "stdout",
|
| 438 |
+
"output_type": "stream",
|
| 439 |
+
"text": [
|
| 440 |
+
" Title Survived\n",
|
| 441 |
+
"0 Master 0.575000\n",
|
| 442 |
+
"1 Miss 0.702703\n",
|
| 443 |
+
"2 Mr 0.156673\n",
|
| 444 |
+
"3 Mrs 0.793651\n",
|
| 445 |
+
"4 Rare 0.347826\n"
|
| 446 |
+
]
|
| 447 |
+
}
|
| 448 |
+
],
|
| 449 |
+
"source": [
|
| 450 |
+
"for dataset in full_data:\n",
|
| 451 |
+
" dataset['Title'] = dataset['Title'].replace(['Lady', 'Countess','Capt', 'Col',\\\n",
|
| 452 |
+
" \t'Don', 'Dr', 'Major', 'Rev', 'Sir', 'Jonkheer', 'Dona'], 'Rare')\n",
|
| 453 |
+
"\n",
|
| 454 |
+
" dataset['Title'] = dataset['Title'].replace('Mlle', 'Miss')\n",
|
| 455 |
+
" dataset['Title'] = dataset['Title'].replace('Ms', 'Miss')\n",
|
| 456 |
+
" dataset['Title'] = dataset['Title'].replace('Mme', 'Mrs')\n",
|
| 457 |
+
"\n",
|
| 458 |
+
"print (train[['Title', 'Survived']].groupby(['Title'], as_index=False).mean())"
|
| 459 |
+
]
|
| 460 |
+
},
|
| 461 |
+
{
|
| 462 |
+
"cell_type": "markdown",
|
| 463 |
+
"metadata": {
|
| 464 |
+
"_cell_guid": "68fa2057-e27a-e252-0d1b-869c00a303ba"
|
| 465 |
+
},
|
| 466 |
+
"source": [
|
| 467 |
+
"# Data Cleaning #\n",
|
| 468 |
+
"great! now let's clean our data and map our features into numerical values."
|
| 469 |
+
]
|
| 470 |
+
},
|
| 471 |
+
{
|
| 472 |
+
"cell_type": "code",
|
| 473 |
+
"execution_count": 12,
|
| 474 |
+
"metadata": {
|
| 475 |
+
"_cell_guid": "2502bb70-ce6f-2497-7331-7d1f80521470"
|
| 476 |
+
},
|
| 477 |
+
"outputs": [
|
| 478 |
+
{
|
| 479 |
+
"name": "stdout",
|
| 480 |
+
"output_type": "stream",
|
| 481 |
+
"text": [
|
| 482 |
+
" Survived Pclass Sex Age Fare Embarked IsAlone Title\n",
|
| 483 |
+
"0 0 3 0 1 0 0 0 1\n",
|
| 484 |
+
"1 1 1 1 2 3 1 0 3\n",
|
| 485 |
+
"2 1 3 1 1 1 0 1 2\n",
|
| 486 |
+
"3 1 1 1 2 3 0 0 3\n",
|
| 487 |
+
"4 0 3 0 2 1 0 1 1\n",
|
| 488 |
+
"5 0 3 0 2 1 2 1 1\n",
|
| 489 |
+
"6 0 1 0 3 3 0 1 1\n",
|
| 490 |
+
"7 0 3 0 0 2 0 0 4\n",
|
| 491 |
+
"8 1 3 1 1 1 0 0 3\n",
|
| 492 |
+
"9 1 2 1 0 2 1 0 3\n"
|
| 493 |
+
]
|
| 494 |
+
}
|
| 495 |
+
],
|
| 496 |
+
"source": [
|
| 497 |
+
"for dataset in full_data:\n",
|
| 498 |
+
" # Mapping Sex\n",
|
| 499 |
+
" dataset['Sex'] = dataset['Sex'].map( {'female': 1, 'male': 0} ).astype(int)\n",
|
| 500 |
+
" \n",
|
| 501 |
+
" # Mapping titles\n",
|
| 502 |
+
" title_mapping = {\"Mr\": 1, \"Miss\": 2, \"Mrs\": 3, \"Master\": 4, \"Rare\": 5}\n",
|
| 503 |
+
" dataset['Title'] = dataset['Title'].map(title_mapping)\n",
|
| 504 |
+
" dataset['Title'] = dataset['Title'].fillna(0)\n",
|
| 505 |
+
" \n",
|
| 506 |
+
" # Mapping Embarked\n",
|
| 507 |
+
" dataset['Embarked'] = dataset['Embarked'].map( {'S': 0, 'C': 1, 'Q': 2} ).astype(int)\n",
|
| 508 |
+
" \n",
|
| 509 |
+
" # Mapping Fare\n",
|
| 510 |
+
" dataset.loc[ dataset['Fare'] <= 7.91, 'Fare'] \t\t\t\t\t\t = 0\n",
|
| 511 |
+
" dataset.loc[(dataset['Fare'] > 7.91) & (dataset['Fare'] <= 14.454), 'Fare'] = 1\n",
|
| 512 |
+
" dataset.loc[(dataset['Fare'] > 14.454) & (dataset['Fare'] <= 31), 'Fare'] = 2\n",
|
| 513 |
+
" dataset.loc[ dataset['Fare'] > 31, 'Fare'] \t\t\t\t\t\t\t = 3\n",
|
| 514 |
+
" dataset['Fare'] = dataset['Fare'].astype(int)\n",
|
| 515 |
+
" \n",
|
| 516 |
+
" # Mapping Age\n",
|
| 517 |
+
" dataset.loc[ dataset['Age'] <= 16, 'Age'] \t\t\t\t\t = 0\n",
|
| 518 |
+
" dataset.loc[(dataset['Age'] > 16) & (dataset['Age'] <= 32), 'Age'] = 1\n",
|
| 519 |
+
" dataset.loc[(dataset['Age'] > 32) & (dataset['Age'] <= 48), 'Age'] = 2\n",
|
| 520 |
+
" dataset.loc[(dataset['Age'] > 48) & (dataset['Age'] <= 64), 'Age'] = 3\n",
|
| 521 |
+
" dataset.loc[ dataset['Age'] > 64, 'Age'] = 4\n",
|
| 522 |
+
"\n",
|
| 523 |
+
"# Feature Selection\n",
|
| 524 |
+
"drop_elements = ['PassengerId', 'Name', 'Ticket', 'Cabin', 'SibSp',\\\n",
|
| 525 |
+
" 'Parch', 'FamilySize']\n",
|
| 526 |
+
"train = train.drop(drop_elements, axis = 1)\n",
|
| 527 |
+
"train = train.drop(['CategoricalAge', 'CategoricalFare'], axis = 1)\n",
|
| 528 |
+
"\n",
|
| 529 |
+
"test = test.drop(drop_elements, axis = 1)\n",
|
| 530 |
+
"\n",
|
| 531 |
+
"print (train.head(10))\n",
|
| 532 |
+
"train_df = train\n",
|
| 533 |
+
"train = train.values\n",
|
| 534 |
+
"test = test.values"
|
| 535 |
+
]
|
| 536 |
+
},
|
| 537 |
+
{
|
| 538 |
+
"cell_type": "markdown",
|
| 539 |
+
"metadata": {
|
| 540 |
+
"_cell_guid": "8aaaf2bc-e282-79cc-008a-e2e801b51b07"
|
| 541 |
+
},
|
| 542 |
+
"source": [
|
| 543 |
+
"good! now we have a clean dataset and ready to predict. let's find which classifier works better on this dataset. "
|
| 544 |
+
]
|
| 545 |
+
},
|
| 546 |
+
{
|
| 547 |
+
"cell_type": "markdown",
|
| 548 |
+
"metadata": {
|
| 549 |
+
"_cell_guid": "23b55b45-572b-7276-32e7-8f7a0dcfd25e"
|
| 550 |
+
},
|
| 551 |
+
"source": [
|
| 552 |
+
"# Classifier Comparison #"
|
| 553 |
+
]
|
| 554 |
+
},
|
| 555 |
+
{
|
| 556 |
+
"cell_type": "code",
|
| 557 |
+
"execution_count": 13,
|
| 558 |
+
"metadata": {
|
| 559 |
+
"_cell_guid": "31ded30a-8de4-6507-e7f7-5805a0f1eaf1"
|
| 560 |
+
},
|
| 561 |
+
"outputs": [
|
| 562 |
+
{
|
| 563 |
+
"data": {
|
| 564 |
+
"text/plain": [
|
| 565 |
+
"<AxesSubplot:title={'center':'Classifier Accuracy'}, xlabel='Accuracy', ylabel='Classifier'>"
|
| 566 |
+
]
|
| 567 |
+
},
|
| 568 |
+
"execution_count": 13,
|
| 569 |
+
"metadata": {},
|
| 570 |
+
"output_type": "execute_result"
|
| 571 |
+
},
|
| 572 |
+
{
|
| 573 |
+
"data": {
|
| 574 |
+
"image/png": "iVBORw0KGgoAAAANSUhEUgAAAgwAAAEWCAYAAAAKI89vAAAAOXRFWHRTb2Z0d2FyZQBNYXRwbG90bGliIHZlcnNpb24zLjMuMiwgaHR0cHM6Ly9tYXRwbG90bGliLm9yZy8vihELAAAACXBIWXMAAAsTAAALEwEAmpwYAAA98klEQVR4nO3dd7xcVb3//9ebIiEkBAnIJYgEEEEIECCgKEhVFBFQ6QgGQYw/FQG5XxtGxAZy0Su9augiVdqlCITekpBOUwJSFJASSqjh/ftjryOTYc6ZOclpCe/n43EeZ2bttdf67H0C+7PXWjNbtomIiIjoyEK9HUBERET0fUkYIiIioqkkDBEREdFUEoaIiIhoKglDRERENJWEISIiIppKwhAR8x1Jh0k6uxvbnyZp8/Jakv4o6XlJd0vaVNID3dV3RF+VhCEi+iRJe0gaJ+llSf+U9H+SNumJvm2vZXtsebsJ8Gngg7Y3sn2L7dW7us+SBFnSx7q67YiukIQhIvocSQcD/wv8ClgO+BBwArBDL4SzEvCI7VfmtSFJi7RTLmBv4Lnyu8e0F1NEvSQMEdGnSBoEHA58y/bFtl+x/abty23/dzv7XCDpX5JmSrpZ0lo127aVNF3SS5KekHRIKV9G0hWSXpD0nKRbJC1Utj0iaWtJ+wKnARuXkY6fSdpc0uM17Q+RdJGkZyTNkHRAzbbDJF0o6WxJLwIj2znsTYHlgQOA3SS9r6aNxSUdLenRcny3Slq8bNtE0u3lGB6TNLKUj5W0X00bIyXdWvPekr4l6SHgoVL2+9LGi5LGS9q0pv7Ckn4k6e/lPI6XtKKk4yUdXfe3uEzSQe0cZ8zHkjBERF+zMdAPuKQT+/wfsBrwAWACcE7NttOBb9geCAwDbijl3wMeB5alGsX4ETDHd+XbPh0YBdxhe4Dtn9ZuLwnG5cAkYAVgK+BASdvUVNsBuBBYqi6uWl8t7fy5vP9Czbb/ATYAPgEsDfw/4G1JK5XjPrYcw3BgYjvtN7Ij8DFgzfL+ntLG0sC5wAWS+pVtBwO7A9sCSwJfA2YBZwC71yRaywBbl/1jAZOEISL6msHAv22/1eoOtv9g+yXbrwOHAeuWkQqAN4E1JS1p+3nbE2rKlwdWKiMYt7jzD9fZEFjW9uG237D9MHAqsFtNnTtsX2r7bduv1jcgqT+wM3Cu7Tepkou9y7aFqC7O37X9hO3Ztm8vx7kH8Ffb55X4n7U9sROx/9r2c20x2T67tPGW7aOBxYC2tRr7AYfafsCVSaXu3cBMqkSJctxjbT/ViThiPpGEISL6mmeBZVqdWy/D5UeU4fIXgUfKpmXK7y9T3Rk/KukmSRuX8qOAvwHXSnpY0g/mItaVgCFlSuAFSS9QjVQsV1PnsSZtfBF4C7iqvD8H+JykZcsx9AP+3mC/Fdspb9UccUk6RNJ9ZdrjBWAQ75zDjvo6A/hKef0V4Kx5iCn6sCQMEdHX3AG8TjVk3oo9qIb9t6a6yA0t5QKwfY/tHaimKy6lDPuXEYnv2V4F2B44WNJWdM5jwAzbS9X8DLS9bU2dZqMWXwUGAP+Q9C/gAmDRclz/Bl4DVm2n70blAK8A/Wve/1eDOv+Jq6xX+H/ALsD7bS9FNXKgFvo6G9hB0rrAR6nOcSyAkjBERJ9ieyYwGjhe0o6S+ktaVNLnJP2mwS4DqRKMZ6kukr9q2yDpfZL2lDSoDPe/CLxdtm0n6cPlEwozgdlt2zrhbuAlSd8vixMXljRM0oat7Cypbd3DdlTrB4YD6wJHAnvbfhv4A/DbsrhyYUkbS1qMaiRia0m7SFpE0mBJw0vTE4EvlXP3YWDfJqEMpBrleAZYRNJoqrUKbU4Dfi5pNVXWkTQYwPbjVOsfzgIuajTtEguGJAwR0eeUOfSDgUOpLmKPAd+m8d3rmcCjwBPAdODOuu17AY+U6YpRwJ6lfDXgr8DLVKMaJ9i+sZNxzuadi/0MqhGB06hGOlqxFzDR9rW2/9X2AxwDrCNpGHAIMIXqovwcVTKxkO1/UE21fK+UT6RKNgB+B7wBPEU1ZdDeYss21wBXAw9SncvXmHPK4rdUIzPXUiVdpwOL12w/A1ibTEcs0NT5NT4RERHvkPQpqqmJleZi4WjMJzLCEBERc03SosB3gdOSLCzYkjBERMRckfRR4AWqj6f+b68GE90uUxIRERHRVEYYIiIioqk8dCQWSMsss4yHDh3a22FERMxXxo8f/2/byzbaloQhFkhDhw5l3LhxvR1GRMR8RdKj7W3LlEREREQ0lRGGWCDNePIl9hg9trfDiIjotHMP37y3Q2goIwwRERHRVBKGiIiIaCoJQ0RERDSVhCEiIiKaSsLQAkkv17zeVtKDklaSdJikWZI+0KhuB+1dJWmpJnXGShrRoHykpOM6eQgtkXSIpPslTZR0j6S9O4plLvsYIemY8noxSX8t/e0q6TRJa3ZFPxER0bXyKYlOkLQV1WNnt7H9qCSoHmf7PeD7rbZje9vuibBjqgKW7bcbbBsFfBrYyPaLkpYEvtjVMdgeB7R9QcJ6pWx4eX9+Z9qStHB5vHBERHSzjDC0qDy+9VRgO9t/r9n0B2BXSUs32Ocrku4ud9AnS1q4lD8iaZny+ieSHpB0q6TzJB1S08TOZf8HJW1aU75iuet/SNJPa/o7WNLU8nNgKRta2j8TmFr2HVPqTJF0UNn9R8A3bb8IYPtF22c0OKYTJY2TNE3Sz2rKj5A0XdJkSf9TynYu/UySdHMp21zSFWVU5mxgw3J+Vq0dyZD0GUl3SJog6QJJA2rO3ZGSJgA7N/u7RURE18gIQ2sWAy4FNrd9f922l6mShu8CtRfvjwK7Ap+0/aakE4A9gTNr6mwIfBlYF1gUmACMr2l7EdsbSdq2tL11Kd8IGAbMAu6RdCVgYB/gY4CAuyTdBDwPrAZ81fadkjYAVrA9rMSwVBlNGGj74RbOxY9tP1eSn+slrQM8QTUasYZt10y3jKYajXmifgrG9tOS9gMOsb1diaXtvCwDHApsbfsVSd8HDgYOL7s/a3v9+sAk7Q/sD9B/0HItHEpERLQqIwyteRO4Hdi3ne3HAF+VNLCmbCtgA6oL+sTyfpW6/T4J/MX2a7ZfAi6v235x+T0eGFpTfp3tZ22/WupsUn4usf2K7ZdLeduoxKO27yyvHwZWkXSspM8CL3Z86O+yS7m7vxdYC1gTmAm8Bpwu6UtUiQzAbcAYSV8HFu5EHx8v7d5Wzt1XgZVqtjecurB9iu0Rtkf06z+oE91FREQzSRha8zawC7CRpB/Vb7T9AnAu8K2aYgFn2B5efla3fVgn+329/J7NnKNB9c8kb/aM8ldqYn2eakRjLDAKOK1MQ7wsqT6hmYOklYFDgK1srwNcCfSz/RbVqMeFwHbA1aWvUVQjBSsC4yUNbhLnf7qiSorazt2atmuTtVfa2zEiIrpHEoYW2Z4FfB7YU1KjkYbfAt/gnQv79cBObZ+gkLS0pJXq9rkN+IKkfmWOfrsWw/l0aW9xYMfSzi3AjpL6S1qCaorglvody3D/QrYvorqYtw3t/xo4vkxPIGlA26ckaixJdbGeKWk54HNtdYFBtq8CDqJKSJC0qu27bI8GnqFKHFpxJ/BJSR8u7Swh6SMt7hsREd0gaxg6oczdfxa4WdIzddv+LekSqgsmtqdLOhS4VtJCVNMa3wIerdnnHkmXAZOBp4ApVMP7zdwNXAR8EDi7fPIASWPKNqhGDu6VNLRu3xWAP5aYAH5Yfp8IDKCaQnmzxHt03TFOknQvcD/wGFWiAjAQ+IukflSjAweX8qMkrVbKrgcmAZs1Ozjbz0gaCZwnabFSfCjwYLN9IyKie8huNpod3UnSANsvS+oP3Azsb3tCb8c1vxs8ZHVvs9/JvR1GRESn9ebDpySNt93we3cywtD7TlH1ZUX9qNY8JFmIiIg+JwlDL7O9R2/HEBER0UwWPUZERERTGWGIBdLKQwb26jxgRMSCJiMMERER0VQShoiIiGgqCUNEREQ0lTUMsUCa8eRL7DF6bG+HERHRo7pz7VZGGCIiIqKpJAwRERHRVBKGiIiIaCoJQ0RERDSVhCEiIiKaSsIQPU7SjyVNkzRZ0kRJP5X067o6wyXdV14PkHSypL9LGi9prKSP9U70ERHvTflYZfQoSRsD2wHr235d0jLAmsAY4Ic1VXcDziuvTwNmAKvZflvSymWfiIjoIUkYoqctD/zb9usAtv8N3CzpeUkfs31XqbcLsI2kVYGPAXvafrvsM4MqgYiIiB6SKYnoadcCK0p6UNIJkjYr5edRjSog6ePAc7YfAtYCJtqe3axhSftLGidp3GuzZnZX/BER70lJGKJH2X4Z2ADYH3gGOF/SSOB8YCdJCzHndERn2j7F9gjbI/r1H9SFUUdERKYkoseV0YKxwFhJU4Cv2h4jaQawGfBlYONSfRqwrqSFWxlliIiI7pERhuhRklaXtFpN0XDg0fL6POB3wMO2Hwew/XdgHPAzSSptDJX0+Z6LOiIikjBETxsAnCFpuqTJVJ92OKxsu4BqzUL9dMR+wHLA3yRNpfpExdM9Em1ERACZkogeZns88Il2tv0bWLRB+YvA17s5tIiI6EBGGCIiIqKpJAwRERHRVBKGiIiIaCprGGKBtPKQgZx7+Oa9HUZExAIjIwwRERHRVBKGiIiIaCoJQ0RERDSVNQyxQJrx5EvsMXpsb4cREdEtemONVkYYIiIioqkkDBEREdFUEoaIiIhoKglDRERENJWEISIiIppKwlBD0mxJEyVNkzRJ0vckzdU5knS4pK072D5K0t5z0e42JcaJkl6W9EB5febcxFnX9iGS7i/t3dMWn6SxkkbMa/ulrRGSjimvF5P019LfrpJOk7RmV/QTERFdKx+rnNOrtocDSPoAcC6wJPDTzjZke3ST7SfNTYC2rwGuKTGOBQ6xPa62jqSFbc/uTLuSRgGfBjay/aKkJYEvzk2MHSmxtsW7XikbXt6f35m25uY4IyJi7mSEoR22nwb2B76tysKSjip33pMlfaOtrqTvS5pSRiWOKGVjJO1UXh8haXrZ739K2WGSDimvh0u6s2y/RNL7S/lYSUdKulvSg5I2bS9eSY+UuhOAnSV9RtIdkiZIukDSgFJvA0k3SRov6RpJy5cmfgR80/aL5fhftH1Gg35OlDSujML8rKa80THuLGlqOS83l7LNJV1RErKzgQ3LCMOqtSMZHcQ/x3F29u8aERFzJyMMHbD9sKSFgQ8AOwAzbW8oaTHgNknXAmuUbR+zPUvS0rVtSBpMdae+hm1LWqpBV2cC37F9k6TDqUY0DizbFrG9kaRtS3m70xzAs7bXl7QMcDGwte1XJH0fOFjSr4FjgR1sPyNpV+CXkg4EBtp+uIXT8mPbz5Xzcr2kdYAn2jnG0cA2tp+oP27bT0vaj2qEZLtyrtrO2TLAofXxA4fXHmd9YJL2p0ry6D9ouRYOJSIiWpWEoXWfAdZpGzUABgGrUV3A/2h7FoDt5+r2mwm8Bpwu6QrgitqNkgYBS9m+qRSdAVxQU+Xi8ns8MLRJjG1D+h8H1qRKagDeB9wBrA4MA64r5QsD/2zSZr1dyoV5EWD50s90Gh/jbcAYSX+uOY5WtBd/m4ZTF7ZPAU4BGDxkdXeiv4iIaCIJQwckrQLMBp4GRDUKcE1dnW06asP2W5I2ArYCdgK+DWzZiTBeL79n0/zv9UpbWMB1tnevi3VtYJrtjet3VLWAcpWORhkkrQwcAmxo+3lJY4B+7R2j7VGSPgZ8HhgvaYNmB9tR/A2OMyIiekjWMLRD0rLAScBxtk210PCbkhYt2z8iaQngOmAfSf1Lef2UxABgkO2rgIOAdWu3254JPF+zPmEv4CbmzZ3AJyV9uMSwhKSPAA8Ay0rauJQvKmmtss+vgeNVLXZE0gC9+1McS1JdrGdKWg74XEfHKGlV23eVBaDPACvOY/wREdFLMsIwp8UlTQQWBd4CzgJ+W7adRjUlMEHVOPkzwI62r5Y0HBgn6Q3gKqoFhG0GAn+R1I/qzvngBv1+FTipJB0PA/vMy0GU9QkjgfPKeguAQ20/WKZUjilTIYsA/wtMA04EBgD3SHoTeBM4uq7dSZLuBe4HHqOacujoGI+StFopux6YBGw2t/EDD3bqRERERJdRdfMcsWAZPGR1b7Pfyb0dRkREt+iup1VKGm+74ffuZEoiIiIimkrCEBEREU1lDUMskFYeMrDbhuwiIt6LMsIQERERTSVhiIiIiKaSMERERERTSRgiIiKiqSx6jAXSjCdfYo/RY3s7jIiIbtWTi7szwhARERFNJWGIiIiIppIwRERERFNJGCIiIqKp91TCIGm2pImSpkq6XNJSXdTuSEnHdVFbj0iaUuKcKOkTXdFug36GS9q2ruxzksZJmi7pXklHl/LDJB3ShX3fXvP6KEnTyu9RDR6pHRERfcB77VMSr9oeDiDpDOBbwC97NaLGtrD9787sIGkR2291YpfhwAiqx3EjaRhwHPB52/dLWhjYvzMxtMp2bRK0P7C07dmdbWcujjkiIubSe2qEoc4dwAoAkjaSdEe5q75d0uqlfKSkiyVdLekhSb9p21nSPpIelHQ38Mma8qGSbpA0WdL1kj5UysdIOlHSnZIelrS5pD9Iuk/SmI4CbdLmSZLuAn4jadUS63hJt0hao9TbuYyqTJJ0s6T3AYcDu5ZRjF2B/wf80vb9ALZn2z6xQSxfl3RPaesiSf0b9VHK1pJ0d+ljsqTVSvnL5fdlwABgvKRda0cyOjiWOY65E3/viIiYB+/JhKHcPW8FXFaK7gc2tb0eMBr4VU314cCuwNpUF9gVJS0P/IwqUdgEWLOm/rHAGbbXAc4BjqnZ9n5gY+Cg0vfvgLWAtSUNr6l3Y7nI3tVCmx8EPmH7YOAU4Du2NwAOAU4odUYD29heF9je9hul7Hzbw22fDwwDxjc9eXCx7Q1LW/cB+zbqo5SNAn5fRnVGAI/XNmR7e8qoT4mhVnvHUn/M/yFp/zKlMu61WTNbOJSIiGjVe21KYnFJE6lGFu4Drivlg4Azyh2wgUVr9rne9kwASdOBlYBlgLG2nynl5wMfKfU3Br5UXp/FnHfBl9u2pCnAU7anlP2nAUOBiaVe/ZRER21eYHu2pAHAJ4ALJLVtW6z8vg0YI+nPwMUdnaAWDJP0C2ApqtGBazro4w7gx5I+SJVoPNRKB02OBcox1+9n+xSqRIPBQ1Z3Zw4qIiI69l4bYWhbw7ASIKo1DAA/B260PQz4AtCvZp/Xa17PZt6SrLa23q5r9+15aPeV8nsh4IVyt97281EA26OAQ4EVqYb/BzdoZxqwQQv9jQG+bXttqlGWfu31YftcqtGGV4GrJG3Z4jG1eyx1xxwRET3kvZYwAGB7FnAA8D1Ji1CNMDxRNo9soYm7gM0kDZa0KLBzzbbbgd3K6z2BW7og5KZt2n4RmCFpZwBV1i2vV7V9l+3RwDNUF/WXgIE1TRwF/EjSR8o+C0ka1SCWgcA/y3Hv2VbYqA9JqwAP2z4G+AuwTisH29GxRERE73hPJgwAtu8FJgO7Uw3x/1rSvbRwp2/7n8BhVEPut1FNb7T5DrCPpMnAXsB3uyDcVtvcE9hX0iSqEYMdSvlRqj6qOZUq+ZgE3Ais2bbo0fZk4EDgPEn3AVOBVRr08ROqhOk2qrUfbRr1sQswtUwDDQPO7MQxt3csERHRC2RnqjcWPIOHrO5t9ju5t8OIiOhWXf3wKUnjbY9otO09O8IQERERrUvCEBEREU0lYYiIiIim3mvfwxDvESsPGdjlc3sREe9lGWGIiIiIppIwRERERFNJGCIiIqKppgmDpIUl3d+sXkRERCy4WvlWw9mSHpD0Idv/6ImgIubVjCdfYo/RY3s7jIiILtWbi7lb/ZTE+4Fpku6m5sE/5fHEERERsYBrNWH4SbdGEREREX1aSwmD7ZskrQSsZvuvkvoDC3dvaBEREdFXtPQpCUlfBy4E2p7mswJwaTfFFBEREX1Mqx+r/BbwSeBFANsPAR/orqDmZ5J2lGRJa7Szfaykhk8Cq6vzQHn09H2S9u/iGEdKGlLzflFJR0h6SNIESXdI+lzZ9oikZbqo3+0l/aC8XlbSXZLulbSppKskLdUV/URERNdrNWF43fYbbW8kLQLkudiN7Q7cWn7Piz1tD6dK1I6U9L55DazGSGBIzfufA8sDw2yvD+wIDOzC/gCwfZntI8rbrYApttezfYvtbW2/0GpbkjIlFhHRg1pNGG6S9CNgcUmfBi4ALu++sOZPkgYAmwD7AruVssUl/amMFFwCLF5T/0RJ4yRNk/SzdpodQPXJlNlln90lTZE0VdKRNW29q7x8h8aYUjZF0kGSdgJGAOeUEYwlgK8D37H9OoDtp2z/ucHxXSppfIl3//b6KOUHSJouabKkP5WykZKOkzQc+A2wQ4lh8dqRDElfkXR32XZyW3Ig6WVJR0uaBGw8F3+iiIiYS61+SuIHVBfBKcA3gKuA07orqPnYDsDVth+U9KykDYDNgFm2PyppHWBCTf0f236uXBCvl7SO7cll2zmSXgdWAw4s34cxBDgS2AB4HrhW0o7A3e2UPwasYHsYgKSlbL8g6dvAIbbHlZj+YfvFFo7vayXexYF7JF0EDK3vo9T9AbCy7dfrpxpsT5Q0Ghhh+9tlP8rvjwK7Ap+0/aakE4A9gTOBJYC7bH+vUXAlidkfoP+g5Vo4nIiIaFWrn5J4Gzi1/ET7dgd+X17/qbz/MHAMgO3JkibX1N+lXOQWoZoSWBNo275nuaAvC9wu6WpgODDW9jMAks4BPkU1PdSo/OfAKpKOBa4Erp3H4ztA0hfL6xWpkpkH2uljMlXScymdWyC7FVXic09JIhYHni7bZgMXtbej7VOAUwAGD1k9U2YREV2ow4RB0p9t7yJpCg3WLNhep9sim89IWhrYElhbkqk+dmrg3nbqrwwcAmxo+3lJY4B+9fVsPyNpAvAx4PXOxFTaXRfYBhgF7AJ8ra7a34APSVqyo1EGSZsDWwMb254laSzQr4M+Pk+VtHwB+LGktVsMW8AZtn/YYNtrtme32E5ERHShZmsYDiy/t6P6H3/9T7xjJ+As2yvZHmp7RWAGMB7YA0DSMKAtyVqSam3CTEnLAZ9r1Kiq77xYD/g71dTDZpKWKdMYuwM3tVde1gQsZPsi4FBg/dLsS5RFjbZnAacDv29bWFk+wbBzXSiDgOdLsrAG8PFS9119SFoIWNH2jcD3y74DWjyP1wM7SfpAaX9pVd8BEhERvajZlMQVVBeZX9jeqwfimZ/tTrWOoNZFVBf7xSXdB9xHlUBge5Kke4H7qdYa3Fa37zmSXgUWA8bYHg+g6mOJN1LdiV9p+y/tlZc7/z+WCzhA2137GOCk0v7GVBf6XwDTJb1GlciMrovnamBUOY4HgDtL+QoN+lgYOFvSoBLPMWXtRLNziO3pkg6lWoexEPAm1cd6H226c0REdBvZ7U/1SpoK/IpqLvy/67fbvrj7QouYe4OHrO5t9ju5ecWIiPlIdz98StJ42w2/K6jZCMMoqhXqS/HuKQgDSRgiIiLeAzpMGGzfCtwqaZzt03sopoiIiOhjmn1KYkvbNwDPS/pS/fZMSURERLw3NJuS2Ay4gcafiMiURPRZKw8Z2O1zfRER7yXNpiR+Wn7v0zPhRERERF/U6uOtvytpSVVOU/VEw890d3ARERHRN7T68KmvlW8B/AwwGNgLOKLjXSIiImJB0erDp9q+cWdb4Ezb09TKt/BE9JIZT77EHqPH9nYYERHdqifXarU6wjBe0rVUCcM1kgYCb3dfWBEREdGXtDrCsC/VkxIfLs8SWBrIQsiIiIj3iFZHGDYGHijPA/gK1bMHZnZfWBEREdGXtJownAjMKg8z+h7VkxPP7LaoIiIiok9pNWF4y9VTqnYAjrN9POXxyBEREbHgazVheEnSD4GvAFeWxw4v2n1htU/ScpLOlfSwpPGS7pD0xXlo7zBJh5TXh0vaei7bGS5p25r3IyU9I2mipGmSLpTUf27jbKG/7csjrue2vUUlHSHpofI9G3dI+lzZ9oikZboo7v/EKWlZSXdJulfSppKukrRUV/QTERFdq9WEYVfgdWBf2/8CPggc1W1RtaN8lPNS4Gbbq9jeANitxFNbr9XFnHOwPdr2X+cyvOFUnyKpdb7t4bbXAt6gOo9dZY7+bF9me16+G+PnwPLAMNvrAzvSDaNIdXFuBUyxvZ7tW2xva/uFVtuStHBXxxcREY21lDDY/pft39q+pbz/h+3eWMOwJfCG7ZNqYnvU9rHljv4ySTcA10saIOn6crc8RdIObftI+rGkByXdCqxeUz5G0k7l9QaSbiqjGNdIWr6Uj5V0pKS7SxubSnofcDiwaxlRmCMxKAnMEsDz5f1QSTdImlxi/FCT8p0lTZU0SdLNjforx39czXEcI+n2MhLTdkwLSTpB0v2Srit39DuVkY+vA9+x/Xo5r0/Z/nP9H0DSpeWcTJO0fylbuPQ5tZzrg0r5AZKml+P5UykbKek4ScOB3wA7lGNYvHYkQ9JXyjmeKOnktuRA0suSjpY0iWoxbkRE9IBWvxr645LuKf+zfkPSbEm98SmJtYAJHWxfH9jJ9mbAa8AXy93yFsDRqrSNSgynukPfsL4RSYsCx5a2NgD+APyypsoitjcCDgR+avsNYDTvjCicX+rtKmki8ASwNHB5KT8WOMP2OsA5wDFNykcD29heF9i+g/5qLQ9sAmzHO9/K+SVgKLAm1bd1tl1wPwz8o3ybZzNfK+dkBHCApMFU53IF28Nsrw38sdT9AbBeOZ5RtY3Ynlh3DK+2bZP0UarRmE/aHg7MBvYsm5cA7rK9bnn8OjX77S9pnKRxr83Kh3giIrpSq1MSxwG7Aw8BiwP7ASd0V1CtknR8ueu+pxRdZ/u5ts3AryRNBv4KrAAsB2wKXGJ7VrlAXtag6dWBYcB15YJ/KHNOe7Q9pXM81QW4PeeXC95/AVOA/y7lGwPnltdnUV3YOyq/DRgj6etAq8Pwl9p+2/Z0quOmtHdBKf8XcGOLbdU6oNzd3wmsCKwGPAysIulYSZ8F2hKPycA5qj6K+1Yn+tgK2AC4p5z/rYBVyrbZwEWNdrJ9iu0Rtkf06z+ok4cVEREdaTVhwPbfgIVtz7b9R+Cz3RdWu6ZRjSK0xfQtqovJsqXolZq6e5byDcpF+ymgX4v9CJhW7nyH217bdu3Dtl4vv2fTwpdflU+YXA58qsX+6/cfRZW0rEj1rZuDW9jt9ZrXzb7G+2/AhyQt2VElSZsDWwMbl9GOe4F+tp8H1gXGUo0knFZ2+TxwPNXf7J5OrC0R1UhL2/lf3fZhZdtrtme32E5ERHSRVhOGWWXefKKk35Q56paTjS50A9BP0jdrytr75MEg4Gnbb0raAliplN8M7FjmzAcCX2iw7wPAspI2hv98gmCtJrG9RMeLBDeh+v4KgNuppkWgSmxu6ahc0qq277I9GniGKnFo1l8jtwFfLmsZlgM2B7A9Czgd+H35O7d9gmHnuv0HAc+Xb/tcA/h4qbsMsJDti6gSm/VVfZJmRds3At8v+w5oMc7rgZ0kfaC0v7SklZrsExER3ajVi/5eVEPh36a6i18R+HJ3BdWecqe+I7CZpBmS7gbOoLog1TsHGCFpCrA3cH9pYwJwPjAJ+D/gnvodyxqBnYAjy/D7ROATTcK7EVizbtFj26LEycB6VJ9EAPgOsE8p3wv4bpPyo8piwqlUScWkdvpr5iLgcWA6cDbVepC2yf5DqZKR6aWfK3hnaqHN1cAiku6jWhdxZylfARhbpg/OBn5I9e/l7HL+7wWOafUTEGUa5VDg2nIurqNakxEREb1E1TU43iskDbD9cpnWuJtqYeG/ejuurjZ4yOreZr+TezuMiIhu1dVPq5Q03vaIRts6nFMud4ftZhRl9XvMX65Q9eVI7wN+viAmCxER0fWaLUL7EtUK+8fqylcEcqGZD9nevLdjiIiI+U+zNQy/A2aWL0f6zw/VvPfvuj+8iIiI6AuajTAsZ3tKfaHtKZKGdk9IEfNu5SEDu3xuLyLivazZCMNSHWxbvAvjiIiIiD6sWcIwrny74Bwk7Uf1LYcRERHxHtBsSuJA4BJJe/JOgjCCaoX9XD9SOiIiIuYvHSYMtp8CPlG+KXFYKb7S9g3dHlnEPJjx5EvsMXpsb4cREdGS+WHNVUvf7V++3nduHlQUERERC4DeeB5EREREzGeSMERERERTSRgiIiKiqSQMERER0VQShgWMpOUknSvpYUnjJd0hqVs/AitphKRj5mH/RyRdVPN+J0ljyuuRkp4pj/GeJulCSf27IOyIiOiEJAwLEEkCLgVutr2K7Q2A3YAPdme/tsfZPmAem9lA0prtbDvf9nDbawFvALvOY18REdFJSRgWLFsCb9g+qa2gPDDsWElDJd0iaUL5+QSApM0lXdFWX9JxkkaW10dImi5psqT/KWU7S5oqaZKkm+vbkLRRGdW4V9LtklYv5SMlXSzpakkPSfpNXexHAz/u6OAkLQIsATw/b6cpIiI6q6XvYYj5xlrAhHa2PQ182vZrklYDzqP61s6GJA2m+jbPNWxb0lJl02hgG9tP1JTVuh/Y1PZbkrYGfgV8uWwbDqwHvA48IOlY222PTv8z8P9J+nCDNneVtAmwPPAgcHk7Me8P7A/Qf9By7R1aRETMhYwwLMAkHV9GAu4BFgVOlTQFuABob/i/zUzgNeB0SV8CZpXy24Ax5RkjCzfYbxBwgaSpVI9AX6tm2/W2Z9p+DZgOrFSzbTZwFPDDBm2eb3s48F/AFOC/GwVs+xTbI2yP6Nd/UJPDi4iIzkjCsGCZBqzf9sb2t4CtgGWBg4CngHV553kgAG8x57+DfmXft4CNgAuB7YCrS/ko4FBgRWB8GYmo9XPgRtvDgC+0tVe8XvN6Nu8e4ToL+FRp+11sm2p04VONtkdERPdJwrBguQHoJ+mbNWVtnygYBPzT9tvAXrwzOvAosKakxcoUw1YAkgYAg2xfRZVsrFvKV7V9l+3RwDO8++I+CHiivB7ZmeBtv0k1KnFQB9U2Af7emXYjImLeJWFYgJQ78B2BzSTNkHQ3cAbwfeAE4KuSJgFrAK+UfR6jWj8wtfy+tzQ3ELhC0mTgVuDgUn6UpCllyuF2YFJdGL8Bfi3pXuZujczpDfbbtXyscjLVGoifz0W7ERExD1RdYyIWLIOHrO5t9ju5t8OIiGhJX3lapaTxthsuiM8IQ0RERDSVhCEiIiKayvcwxAJp5SED+8wQX0TEgiAjDBEREdFUEoaIiIhoKglDRERENJWEISIiIprKosdYIM148iX2GD22t8OIiJgnfWnxdkYYIiIioqkkDBEREdFUEoaIiIhoKglDRERENNXnEwZJLzcoGyVp7x7o+5HyZMYpkqZL+oWkfmXbEEkXdkEf20v6QSf3uao8irrLSBoqaY8G5f8r6QlJ8/RvpZzLZeZivy4/1oiI6Lw+nzA0Yvsk22d2V/uqtJ2bLWyvDWwErAKcXGJ40vZO89jPIrYvs31EZ/azva3tF+al7waGAnMkDOUcfBF4DNisi/trSTcda0REdNJ8mTBIOkzSIeX1WElHSrpb0oOSNi3lC0s6StI9kiZL+kYpHyDpekkTysjBDqV8qKQHJJ0JTAVWrO3T9svAKGBHSUuX+lPLvmuV/ieWvlYr5XuX95MknVXKxkg6SdJdwG8kjZR0XM22EyXdKelhSZtL+oOk+ySNqTn+RyQtU2K4T9KpkqZJulbS4qXO18uxT5J0kaT+NX0cI+n20kdb0nMEsGk5hoNK2ebANOBEYPe68/+Hcu4flnRAzbZLJY0v8ezf4G93uKQDa97/UtJ3JS0v6ebS/9Sav2PbsS4h6cpyPFMl7drav5aIiOgK82XC0MAitjcCDgR+Wsr2BWba3hDYEPi6pJWB14Av2l4f2AI4WpLKPqsBJ9hey/aj9Z3YfhGYUerVGgX83vZwYATwuKS1gEOBLW2vC3y3pv4HgU/YPrjBsbwf2Bg4CLgM+B2wFrC2pOEN6q8GHG97LeAF4Mul/GLbG5a+7yvno83ywCbAdlSJAsAPgFtsD7f9u1K2O3AecAnweUmL1rSxBrAN1cjLT2u2fc32BuU8HCBpcF28fwD2hv+MYOwGnE01unFNOYfrAhPr9vss8KTtdW0PA66uPxGS9pc0TtK412bNbHCqIiJibi0oCcPF5fd4qqF1gM8Ae0uaCNwFDKa6uAr4laTJwF+BFYDlyj6P2r6zSV9qUHYH8CNJ3wdWsv0qsCVwge1/A9h+rqb+BbZnt9P+5bYNTAGesj3F9ttUd/pDG9SfYXtieV17/MMk3SJpCrAnVdLR5lLbb9uezjvHPudBSu8Dti11X6Q6h9vUVLnS9uvl+J6uaecASZOAO6lGaeZIrmw/AjwraT2qv9G9tp8F7gH2kXQYsLbtl+pCmgJ8uowmbWr7XRmB7VNsj7A9ol//QY0OKyIi5tKCkjC8Xn7P5p1vrxTwnXLHPNz2yravpbp4LgtsUO5mnwL6lX1e6agTSQOpLsgP1pbbPhfYHngVuErSlk3i7aiftmN5u+Z12/tG38xZW6f2+McA3y7rL37GO8dYv0+jBAiq5GApYIqkR6hGJHav2f6ufiVtDmwNbFxGNu6t67fNacBIYB+qEQds3wx8CngCGKO6Ra22HwTWp0ocfiFpdDtxR0REN1hQEoZGrgG+2TZULukjkpYABgFP235T0hbASq00JmkAcALVHffzddtWAR62fQzwF2Ad4AZg57YheUlLd9FxtWog8M9y/Hu2UP+lsk+b3YH9bA+1PRRYmeoOv38HbQwCnrc9S9IawMfbqXcJ1RTDhlR/JyStRDWicipVQrF+7Q6ShgCzbJ8NHFW/PSIiutf88CyJ/pIer3n/2xb3O41qNGBCWaPwDLAjcA5weRmqHwfc36SdG8v+C1Fd6H7eoM4uwF6S3gT+BfzK9nOSfgncJGk21d32yBZj7wo/oZpGeKb8HthxdSYDs8t0wp+pLuij2jbafkXSrcAXOmjjamCUpPuAB6imJd7F9huSbgReqJma2Rz473IOX6asc6ixNnCUpLeBN4FvNjmeiIjoQqqmyyN6TlnsOAHY2fZD3dHH4CGre5v9Tu6OpiMiekxPP3xK0njbIxptW5CnJKIPkrQm8Dfg+u5KFiIiouvND1MSsQApn8xYpbfjiIiIzskIQ0RERDSVEYZYIK08ZGCPz/1FRCzIMsIQERERTSVhiIiIiKaSMERERERTWcMQC6QZT77EHqPH9nYYERFdoi+sycoIQ0RERDSVhCEiIiKaSsIQERERTSVhiIiIiKaSMERERERT3ZowSPqgpL9IekjSw5KOk7RYF7S7uaQrOrnPUEl71LwfIemYJvs8ImlK+Zku6ReS+pVtQyRdOHdHMEcf20v6QSf3uUrSUvPad12bc5yfmvL/lfREecLkvLT/iKRl5mK/Lj/WiIjovG5LGCQJuBi41PZqwGrA4sBvurHPjj4mOhT4zwXR9jjbB7TQ7Ba21wY2onpo0sll/ydt7zQP4SJpEduX2T6iM/vZ3tb2C/PSdwNDqTk/8J/HUH8ReAzYrIv7a0k3HWtERHRSd44wbAm8ZvuPALZnAwcBe0v6tqTj2ipKukLS5uX1iZLGSZom6Wc1dT4r6X5JE4Av1ZQfJuksSbcBZ5U75VskTSg/nyhVjwA2lTRR0kG1oxSSBkj6YxlJmCzpy/UHY/tlYBSwo6SlSz9Ty/5rSbq7tD1Z0mqlfO/yfpKks0rZGEknSboL+I2kkW3nomw7UdKdZURmc0l/kHSfpDE1x/yIpGVKDPdJOrWcr2slLV7qfF3SPaXviyT1r+njGEm3lz7akp45zk8p2xyYBpwI7F53zv8gaWxp44CabZdKGl/i2b/+PEo6XNKBNe9/Kem7kpaXdHPpf6qkTeuOdQlJV5bjmSpp1/q2IyKi+3RnwrAWML62wPaLwCN0/IVRP7Y9AlgH2EzSOmUa4FTgC8AGwH/V7bMmsLXt3YGngU/bXh/YFWibdvgBcIvt4bZ/V7f/T4CZtte2vQ5wQ6PASvwzqEZLao0Cfm97ODACeFzSWsChwJa21wW+W1P/g8AnbB/coJv3AxtTJVeXAb+jOpdrSxreoP5qwPG21wJeANqSnYttb1j6vg/Yt2af5YFNgO2oEgVofH52B84DLgE+L2nRmjbWALahGnn5ac22r9neoJyHAyQNrov3D8De8J8RjN2As6lGN64p53BdYGLdfp8FnrS9ru1hwNX1J0LS/iXZHPfarJkNTlVERMytvrjocZcyinAv1YVyTaqL0wzbD9k21QWm1mW2Xy2vFwVOlTQFuKDs38zWwPFtb2w/30FdNSi7A/iRpO8DK5VYtgQusP3v0uZzNfUvKCMujVxejnEK8JTtKbbfprrTH9qg/gzbE8vr8TV1hpWRlinAnlTnss2ltt+2PR1YruFBSu8Dti11XwTuokoQ2lxp+/VyfE/XtHOApEnAncCK1CVXth8BnpW0HvAZ4F7bzwL3APtIOgxY2/ZLdSFNAT4t6UhJm9p+V0Zg+xTbI2yP6Nd/UKPDioiIudSdCcN0qtGA/5C0JNXowLN1fbctJFwZOATYqtzpX9m2rYlXal4fBDxFdZc6AnjfXMb/LpIGUl2QH6wtt30usD3wKnCVpC07EW+918vvt2tet71vNDJTW2d2TZ0xwLfL+oufMed5rN2nUQIEVXKwFDBF0iNUIxK712x/V79lWmlrYOMysnEvjf9+pwEjgX2oRhywfTPwKeAJYIykvWt3sP0gsD5V4vALSaPbiTsiIrpBdyYM1wP92/7HL2lh4GjgOKph/eGSFpK0ItWwNsCSVBfTmZKWAz5Xyu8HhkpatbyvvXDVGwT8s9yV7wUsXMpfAga2s891wLfa3kh6f30FSQOAE6juuJ+v27YK8LDtY4C/UE2n3ADs3DYkL2npDmLuDgOBf5apgj1bqF9/fnYH9rM91PZQYGWqO/z+HbQxCHje9ixJawAfb6feJVRTDBsC1wBIWolqROVUqoRi/dodJA0BZtk+GziqfntERHSvbksYyrD6F4GdJD1ENarwtu1fArdRJQ3TqdYYTCj7TKK6K70fOLfUw/ZrwP7AlWW64ukOuj4B+GoZFl+Dd+7mJwOzy6K5g+r2+QXw/rKYbhKwRc22G8vixruBfwDfaNDnLsBUSROBYcCZtqcBvwRuKm3+toOYu8NPqKYRbqM6n83Unp8fU13Qr2zbaPsV4FaqdSTtuZpqpOE+qrURdzaqZPsN4EbgzzVTM5sDkyTdS7X25Pd1u60N3F3O8U+p/mYREdFDVF3Xe6Cj6tMK5wFftD2hRzqNPqksdpwA7Gz7oe7oY/CQ1b3Nfid3R9MRET2up55WKWl8+eDBu/TY461t3w6s1FP9Rd8kaU3gCuCS7koWIiKi6/VYwhABUD6ZsUpvxxEREZ3TFz9WGREREX1MRhhigbTykIE9NucXEfFekBGGiIiIaCoJQ0RERDSVhCEiIiKayhqGWCDNePIl9hg9trfDiIiYZ31lPVZGGCIiIqKpJAwRERHRVBKGiIiIaCoJQ0RERDSVhCEiIiKaSsLQxSS93AVtjJB0TAfbh0rao9X6pc4jkqZImizpJkl95kFgkkZJ2ru344iIiPYlYeiDbI+zfUAHVYYC/0kYWqjfZgvb6wBjgUPnKUhAlXn+N2T7JNtnzms7ERHRfZIw9ABJwyXdWe7uL5H0/lK+YSmbKOkoSVNL+eaSriivNyvbJ0q6V9JA4Ahg01J2UF39AZL+WDOa8OUGId0BrFDqLyvpIkn3lJ9P1pRfJ2mapNMkPSppmTK68YCkM4GpwIqS/rvsO1nSz8r+S0i6UtIkSVMl7VrKj5A0vdT9n1J2mKRDmpyrsZKOlHS3pAclbdo9f62IiGgkCUPPOBP4frm7nwL8tJT/EfiG7eHA7Hb2PQT4VqmzKfAq8APgFtvDbf+urv5PgJm21y793dCgzc8Cl5bXvwd+Z3tD4MvAaaX8p8ANttcCLgQ+VLP/asAJZdvq5f1GwHBgA0mfKn08aXtd28OAqyUNBr4IrFVi+0UnzhXAIrY3Ag6sKwdA0v6Sxkka99qsmQ2ajoiIuZWEoZtJGgQsZfumUnQG8ClJSwEDbd9Rys9tp4nbgN9KOqC081aTLrcGjm97Y/v5mm03SnoC+BxwXk394yRNBC4DlpQ0ANgE+FNp42qgtp1Hbd9ZXn+m/NwLTADWoEogpgCfLqMCm9qeCcwEXgNOl/QlYFZt4O2dq5oqF5ff46mmZeZg+xTbI2yP6Nd/UPtnKCIiOi0JQx9n+whgP2Bx4DZJa8xDc1sAKwETgZ+VsoWAj5fRiuG2V7DdbOHmKzWvBfy6Zv8P2z7d9oPA+lSJwy8kjS7JzkZUIxbbAVd3Mv7Xy+/Z5GvNIyJ6VBKGblburJ+vmXPfC7jJ9gvAS5I+Vsp3a7S/pFVtT7F9JHAP1R38S8DAdrq8DvhWzf7vr4vnLaoh/b0lLQ1cC3ynpv7w8vI2YJdS9hlgjnZqXAN8rYxKIGkFSR+QNASYZfts4Chg/VJnkO2rgIOAdetia3iu2uk3IiJ6UO7Sul5/SY/XvP8t8FXgJEn9gYeBfcq2fYFTJb1NdWFsNPF+oKQtgLeBacD/ldezJU0CxlBNB7T5BXB8WUA5m2ok4eLaBm3/U9J5VInFAaX+ZKp/DzcDo8p+50nai2qR5L+oEpUBdW1dK+mjwB2SAF4GvgJ8GDiqHNubwDepkpy/SOpHNTJxcIPjbe9cRUREL5Lt3o7hPUvSgLbhf0k/AJa3/d1eDgsASYsBs22/JWlj4MSy8HK+MHjI6t5mv5N7O4yIiHnWk0+rlDTe9ohG2zLC0Ls+L+mHVH+HR4GRvRvOHD4E/Ll8z8IbwNd7OZ6IiOhFSRh6ke3zgfN7O45GbD8ErNfbcURERN+QRY8RERHRVEYYYoG08pCBPTrvFxGxoMsIQ0RERDSVT0nEAknSS8ADvR1HO5YB/t3bQbQjsc2dvhpbX40LEtvc6u7YVrK9bKMNmZKIBdUD7X00qLdJGpfYOi+xdV5fjQsS29zqzdgyJRERERFNJWGIiIiIppIwxILqlN4OoAOJbe4kts7rq3FBYptbvRZbFj1GREREUxlhiIiIiKaSMERERERTSRhivibps5IekPS38sTP+u2LSTq/bL9L0tA+FNunJE2Q9JaknXoqrhZjO1jSdEmTJV0vaaU+FNsoSVMkTZR0q6Q1+0JcNfW+LMmSeuyjby2cs5GSninnbKKk/fpKbKXOLuXf2zRJ5/aV2CT9ruacPSjphT4S14ck3Sjp3vLf6LY9ERe285Of+fIHWBj4O7AK8D5gErBmXZ3/DzipvN4NOL8PxTYUWAc4E9ipj523LYD+5fU3+9h5W7Lm9fbA1X0hrlJvIHAzcCcwog+ds5HAcT31b6yTsa0G3Au8v7z/QF+Jra7+d4A/9IW4qBY+frO8XhN4pCfOWUYYYn62EfA32w/bfgP4E7BDXZ0dgDPK6wuBrSSpL8Rm+xHbk4G3eyCezsZ2o+1Z5e2dwAf7UGwv1rxdAuiJldut/FsD+DlwJPBaD8TU2dh6QyuxfR043vbzALaf7kOx1dodOK+PxGVgyfJ6EPBkD8SVhCHmaysAj9W8f7yUNaxj+y1gJjC4j8TWWzob277A/3VrRO9oKTZJ35L0d+A3wAF9IS5J6wMr2r6yB+Kp1erf88tl+PpCSSv2TGgtxfYR4COSbpN0p6TP9qHYAChTcisDN/SRuA4DviLpceAqqtGPbpeEISLaJekrwAjgqN6OpZbt422vCnwfOLS345G0EPBb4Hu9HUs7LgeG2l4HuI53Rt36gkWopiU2p7qLP1XSUr0ZUAO7ARfant3bgRS7A2NsfxDYFjir/BvsVkkYYn72BFB7p/TBUtawjqRFqIbvnu0jsfWWlmKTtDXwY2B726/3pdhq/AnYsTsDKprFNRAYBoyV9AjwceCyHlr42PSc2X625m94GrBBD8TVUmxUd9CX2X7T9gzgQaoEoi/E1mY3emY6AlqLa1/gzwC27wD6UT2UqlslYYj52T3AapJWlvQ+qv+oL6urcxnw1fJ6J+AGl5VCfSC23tI0NknrASdTJQs9Nafcamy1F5PPAw/1dly2Z9pexvZQ20Op1n1sb3tcb8cGIGn5mrfbA/f1QFwtxQZcSjW6gKRlqKYoHu4jsSFpDeD9wB09EFOrcf0D2KrE91GqhOGZbo+sJ1ZW5ic/3fVDNRz3INWq4h+XssOp/mdN+Q/pAuBvwN3AKn0otg2p7q5eoRr1mNaHYvsr8BQwsfxc1odi+z0wrcR1I7BWX4irru5YeuhTEi2es1+XczapnLM1+lBsoprOmQ5MAXbrK7GV94cBR/RUTC2eszWB28rfcyLwmZ6IK18NHREREU1lSiIiIiKaSsIQERERTSVhiIiIiKaSMERERERTSRgiIiKiqSQMEREdkLRjefrkGr0dS0RvSsIQEdGx3YFby+9uIWnh7mo7oqskYYiIaIekAcAmVF/Fu1spW1jS/0iaWh7m9J1SvqGk2yVNknS3pIGSRko6rqa9KyRtXl6/LOloSZOAjSWNlnRPafeUtqeqSvqwpL+WdidIWlXSmZJ2rGn3HEl95QmVsYBKwhAR0b4dgKttPwg8K2kDYH9gKDDc1cOczilf4Xs+8F3b6wJbA682aXsJ4C7b69q+FTjO9oa2hwGLA9uVeudQPf55XeATwD+B04GRAJIGlfKefkpmvMckYYiIaN/uVA+4ovzenSoZONnV49Kx/RywOvBP2/eUshfbtndgNnBRzfstJN0laQqwJbCWpIHACrYvKe2+ZnuW7ZuonjewbInpohb6i5gni/R2ABERfZGkpaku3GtLMrAwYKqHA7XqLea8MetX8/o1l8clS+oHnED1/InHJB1WV7eRM4GvUE2V7NOJmCLmSkYYIiIa2wk4y/ZKrp5CuSIwg+qBP98oj0tvSyweAJaXtGEpG1i2PwIMl7SQpBWBjdrpqy05+HdZN7ETgO2XgMfb1itIWkxS/1J3DHBgqTe9y446oh1JGCIiGtsduKSu7CJgearHC08uCxb3sP0GsCtwbCm7jioJuI0qyZgOHANMaNSR7ReAU4GpwDXMOYqxF3CApMnA7cB/lX2eonpM9R/n9UAjWpGnVUZEzIfKSMMUYH3bM3s7nljwZYQhImI+I2lrqtGFY5MsRE/JCENEREQ0lRGGiIiIaCoJQ0RERDSVhCEiIiKaSsIQERERTSVhiIiIiKb+f/AEJGNVFut0AAAAAElFTkSuQmCC\n",
|
| 575 |
+
"text/plain": [
|
| 576 |
+
"<Figure size 432x288 with 1 Axes>"
|
| 577 |
+
]
|
| 578 |
+
},
|
| 579 |
+
"metadata": {
|
| 580 |
+
"needs_background": "light"
|
| 581 |
+
},
|
| 582 |
+
"output_type": "display_data"
|
| 583 |
+
}
|
| 584 |
+
],
|
| 585 |
+
"source": [
|
| 586 |
+
"import matplotlib.pyplot as plt\n",
|
| 587 |
+
"import seaborn as sns\n",
|
| 588 |
+
"\n",
|
| 589 |
+
"from sklearn.model_selection import StratifiedShuffleSplit\n",
|
| 590 |
+
"from sklearn.metrics import accuracy_score, log_loss\n",
|
| 591 |
+
"from sklearn.neighbors import KNeighborsClassifier\n",
|
| 592 |
+
"from sklearn.svm import SVC\n",
|
| 593 |
+
"from sklearn.tree import DecisionTreeClassifier\n",
|
| 594 |
+
"from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, GradientBoostingClassifier\n",
|
| 595 |
+
"from sklearn.naive_bayes import GaussianNB\n",
|
| 596 |
+
"from sklearn.discriminant_analysis import LinearDiscriminantAnalysis, QuadraticDiscriminantAnalysis\n",
|
| 597 |
+
"from sklearn.linear_model import LogisticRegression\n",
|
| 598 |
+
"\n",
|
| 599 |
+
"classifiers = [\n",
|
| 600 |
+
" KNeighborsClassifier(3),\n",
|
| 601 |
+
" SVC(probability=True),\n",
|
| 602 |
+
" DecisionTreeClassifier(),\n",
|
| 603 |
+
" RandomForestClassifier(),\n",
|
| 604 |
+
"\tAdaBoostClassifier(),\n",
|
| 605 |
+
" GradientBoostingClassifier(),\n",
|
| 606 |
+
" GaussianNB(),\n",
|
| 607 |
+
" LinearDiscriminantAnalysis(),\n",
|
| 608 |
+
" QuadraticDiscriminantAnalysis(),\n",
|
| 609 |
+
" LogisticRegression()]\n",
|
| 610 |
+
"\n",
|
| 611 |
+
"log_cols = [\"Classifier\", \"Accuracy\"]\n",
|
| 612 |
+
"log \t = pd.DataFrame(columns=log_cols)\n",
|
| 613 |
+
"\n",
|
| 614 |
+
"sss = StratifiedShuffleSplit(n_splits=10, test_size=0.1, random_state=0)\n",
|
| 615 |
+
"\n",
|
| 616 |
+
"X = train[0::, 1::]\n",
|
| 617 |
+
"y = train[0::, 0]\n",
|
| 618 |
+
"\n",
|
| 619 |
+
"acc_dict = {}\n",
|
| 620 |
+
"\n",
|
| 621 |
+
"for train_index, test_index in sss.split(X, y):\n",
|
| 622 |
+
"\tX_train, X_test = X[train_index], X[test_index]\n",
|
| 623 |
+
"\ty_train, y_test = y[train_index], y[test_index]\n",
|
| 624 |
+
"\t\n",
|
| 625 |
+
"\tfor clf in classifiers:\n",
|
| 626 |
+
"\t\tname = clf.__class__.__name__\n",
|
| 627 |
+
"\t\tclf.fit(X_train, y_train)\n",
|
| 628 |
+
"\t\ttrain_predictions = clf.predict(X_test)\n",
|
| 629 |
+
"\t\tacc = accuracy_score(y_test, train_predictions)\n",
|
| 630 |
+
"\t\tif name in acc_dict:\n",
|
| 631 |
+
"\t\t\tacc_dict[name] += acc\n",
|
| 632 |
+
"\t\telse:\n",
|
| 633 |
+
"\t\t\tacc_dict[name] = acc\n",
|
| 634 |
+
"\n",
|
| 635 |
+
"for clf in acc_dict:\n",
|
| 636 |
+
"\tacc_dict[clf] = acc_dict[clf] / 10.0\n",
|
| 637 |
+
"\tlog_entry = pd.DataFrame([[clf, acc_dict[clf]]], columns=log_cols)\n",
|
| 638 |
+
"\tlog = log.append(log_entry)\n",
|
| 639 |
+
"\n",
|
| 640 |
+
"plt.xlabel('Accuracy')\n",
|
| 641 |
+
"plt.title('Classifier Accuracy')\n",
|
| 642 |
+
"\n",
|
| 643 |
+
"sns.set_color_codes(\"muted\")\n",
|
| 644 |
+
"sns.barplot(x='Accuracy', y='Classifier', data=log, color=\"b\")"
|
| 645 |
+
]
|
| 646 |
+
},
|
| 647 |
+
{
|
| 648 |
+
"cell_type": "markdown",
|
| 649 |
+
"metadata": {
|
| 650 |
+
"_cell_guid": "438585cf-b7ad-73ba-49aa-87688ff21233"
|
| 651 |
+
},
|
| 652 |
+
"source": [
|
| 653 |
+
"# Prediction #\n",
|
| 654 |
+
"now we can use SVC classifier to predict our data."
|
| 655 |
+
]
|
| 656 |
+
},
|
| 657 |
+
{
|
| 658 |
+
"cell_type": "code",
|
| 659 |
+
"execution_count": 13,
|
| 660 |
+
"metadata": {
|
| 661 |
+
"_cell_guid": "24967b57-732b-7180-bfd5-005beff75974"
|
| 662 |
+
},
|
| 663 |
+
"outputs": [],
|
| 664 |
+
"source": [
|
| 665 |
+
"candidate_classifier = SVC()\n",
|
| 666 |
+
"candidate_classifier.fit(train[0::, 1::], train[0::, 0])\n",
|
| 667 |
+
"result = candidate_classifier.predict(test)"
|
| 668 |
+
]
|
| 669 |
+
},
|
| 670 |
+
{
|
| 671 |
+
"cell_type": "markdown",
|
| 672 |
+
"metadata": {},
|
| 673 |
+
"source": [
|
| 674 |
+
"## Fairness"
|
| 675 |
+
]
|
| 676 |
+
},
|
| 677 |
+
{
|
| 678 |
+
"cell_type": "code",
|
| 679 |
+
"execution_count": 14,
|
| 680 |
+
"metadata": {},
|
| 681 |
+
"outputs": [],
|
| 682 |
+
"source": [
|
| 683 |
+
"# This DataFrame is created to stock differents models and fair metrics that we produce in this notebook\n",
|
| 684 |
+
"algo_metrics = pd.DataFrame(columns=['model', 'fair_metrics', 'prediction', 'probs'])\n",
|
| 685 |
+
"\n",
|
| 686 |
+
"def add_to_df_algo_metrics(algo_metrics, model, fair_metrics, preds, probs, name):\n",
|
| 687 |
+
" return algo_metrics.append(pd.DataFrame(data=[[model, fair_metrics, preds, probs]], columns=['model', 'fair_metrics', 'prediction', 'probs'], index=[name]))"
|
| 688 |
+
]
|
| 689 |
+
},
|
| 690 |
+
{
|
| 691 |
+
"cell_type": "code",
|
| 692 |
+
"execution_count": 15,
|
| 693 |
+
"metadata": {},
|
| 694 |
+
"outputs": [],
|
| 695 |
+
"source": [
|
| 696 |
+
"def fair_metrics(dataset, pred, pred_is_dataset=False):\n",
|
| 697 |
+
" if pred_is_dataset:\n",
|
| 698 |
+
" dataset_pred = pred\n",
|
| 699 |
+
" else:\n",
|
| 700 |
+
" dataset_pred = dataset.copy()\n",
|
| 701 |
+
" dataset_pred.labels = pred\n",
|
| 702 |
+
" \n",
|
| 703 |
+
" cols = ['statistical_parity_difference', 'equal_opportunity_difference', 'average_abs_odds_difference', 'disparate_impact', 'theil_index']\n",
|
| 704 |
+
" obj_fairness = [[0,0,0,1,0]]\n",
|
| 705 |
+
" \n",
|
| 706 |
+
" fair_metrics = pd.DataFrame(data=obj_fairness, index=['objective'], columns=cols)\n",
|
| 707 |
+
" \n",
|
| 708 |
+
" for attr in dataset_pred.protected_attribute_names:\n",
|
| 709 |
+
" idx = dataset_pred.protected_attribute_names.index(attr)\n",
|
| 710 |
+
" privileged_groups = [{attr:dataset_pred.privileged_protected_attributes[idx][0]}] \n",
|
| 711 |
+
" unprivileged_groups = [{attr:dataset_pred.unprivileged_protected_attributes[idx][0]}] \n",
|
| 712 |
+
" \n",
|
| 713 |
+
" classified_metric = ClassificationMetric(dataset, \n",
|
| 714 |
+
" dataset_pred,\n",
|
| 715 |
+
" unprivileged_groups=unprivileged_groups,\n",
|
| 716 |
+
" privileged_groups=privileged_groups)\n",
|
| 717 |
+
"\n",
|
| 718 |
+
" metric_pred = BinaryLabelDatasetMetric(dataset_pred,\n",
|
| 719 |
+
" unprivileged_groups=unprivileged_groups,\n",
|
| 720 |
+
" privileged_groups=privileged_groups)\n",
|
| 721 |
+
"\n",
|
| 722 |
+
" acc = classified_metric.accuracy()\n",
|
| 723 |
+
"\n",
|
| 724 |
+
" row = pd.DataFrame([[metric_pred.mean_difference(),\n",
|
| 725 |
+
" classified_metric.equal_opportunity_difference(),\n",
|
| 726 |
+
" classified_metric.average_abs_odds_difference(),\n",
|
| 727 |
+
" metric_pred.disparate_impact(),\n",
|
| 728 |
+
" classified_metric.theil_index()]],\n",
|
| 729 |
+
" columns = cols,\n",
|
| 730 |
+
" index = [attr]\n",
|
| 731 |
+
" )\n",
|
| 732 |
+
" fair_metrics = fair_metrics.append(row) \n",
|
| 733 |
+
" \n",
|
| 734 |
+
" fair_metrics = fair_metrics.replace([-np.inf, np.inf], 2)\n",
|
| 735 |
+
" \n",
|
| 736 |
+
" return fair_metrics\n",
|
| 737 |
+
"\n",
|
| 738 |
+
"def plot_fair_metrics(fair_metrics):\n",
|
| 739 |
+
" fig, ax = plt.subplots(figsize=(20,4), ncols=5, nrows=1)\n",
|
| 740 |
+
"\n",
|
| 741 |
+
" plt.subplots_adjust(\n",
|
| 742 |
+
" left = 0.125, \n",
|
| 743 |
+
" bottom = 0.1, \n",
|
| 744 |
+
" right = 0.9, \n",
|
| 745 |
+
" top = 0.9, \n",
|
| 746 |
+
" wspace = .5, \n",
|
| 747 |
+
" hspace = 1.1\n",
|
| 748 |
+
" )\n",
|
| 749 |
+
"\n",
|
| 750 |
+
" y_title_margin = 1.2\n",
|
| 751 |
+
"\n",
|
| 752 |
+
" plt.suptitle(\"Fairness metrics\", y = 1.09, fontsize=20)\n",
|
| 753 |
+
" sns.set(style=\"dark\")\n",
|
| 754 |
+
"\n",
|
| 755 |
+
" cols = fair_metrics.columns.values\n",
|
| 756 |
+
" obj = fair_metrics.loc['objective']\n",
|
| 757 |
+
" size_rect = [0.2,0.2,0.2,0.4,0.25]\n",
|
| 758 |
+
" rect = [-0.1,-0.1,-0.1,0.8,0]\n",
|
| 759 |
+
" bottom = [-1,-1,-1,0,0]\n",
|
| 760 |
+
" top = [1,1,1,2,1]\n",
|
| 761 |
+
" bound = [[-0.1,0.1],[-0.1,0.1],[-0.1,0.1],[0.8,1.2],[0,0.25]]\n",
|
| 762 |
+
"\n",
|
| 763 |
+
" display(Markdown(\"### Check bias metrics :\"))\n",
|
| 764 |
+
" display(Markdown(\"A model can be considered bias if just one of these five metrics show that this model is biased.\"))\n",
|
| 765 |
+
" for attr in fair_metrics.index[1:len(fair_metrics)].values:\n",
|
| 766 |
+
" display(Markdown(\"#### For the %s attribute :\"%attr))\n",
|
| 767 |
+
" check = [bound[i][0] < fair_metrics.loc[attr][i] < bound[i][1] for i in range(0,5)]\n",
|
| 768 |
+
" display(Markdown(\"With default thresholds, bias against unprivileged group detected in **%d** out of 5 metrics\"%(5 - sum(check))))\n",
|
| 769 |
+
"\n",
|
| 770 |
+
" for i in range(0,5):\n",
|
| 771 |
+
" plt.subplot(1, 5, i+1)\n",
|
| 772 |
+
" ax = sns.barplot(x=fair_metrics.index[1:len(fair_metrics)], y=fair_metrics.iloc[1:len(fair_metrics)][cols[i]])\n",
|
| 773 |
+
" \n",
|
| 774 |
+
" for j in range(0,len(fair_metrics)-1):\n",
|
| 775 |
+
" a, val = ax.patches[j], fair_metrics.iloc[j+1][cols[i]]\n",
|
| 776 |
+
" marg = -0.2 if val < 0 else 0.1\n",
|
| 777 |
+
" ax.text(a.get_x()+a.get_width()/5, a.get_y()+a.get_height()+marg, round(val, 3), fontsize=15,color='black')\n",
|
| 778 |
+
"\n",
|
| 779 |
+
" plt.ylim(bottom[i], top[i])\n",
|
| 780 |
+
" plt.setp(ax.patches, linewidth=0)\n",
|
| 781 |
+
" ax.add_patch(patches.Rectangle((-5,rect[i]), 10, size_rect[i], alpha=0.3, facecolor=\"green\", linewidth=1, linestyle='solid'))\n",
|
| 782 |
+
" plt.axhline(obj[i], color='black', alpha=0.3)\n",
|
| 783 |
+
" plt.title(cols[i])\n",
|
| 784 |
+
" ax.set_ylabel('') \n",
|
| 785 |
+
" ax.set_xlabel('')"
|
| 786 |
+
]
|
| 787 |
+
},
|
| 788 |
+
{
|
| 789 |
+
"cell_type": "code",
|
| 790 |
+
"execution_count": 16,
|
| 791 |
+
"metadata": {},
|
| 792 |
+
"outputs": [],
|
| 793 |
+
"source": [
|
| 794 |
+
"def get_fair_metrics_and_plot(data, model, plot=False, model_aif=False):\n",
|
| 795 |
+
" pred = model.predict(data).labels if model_aif else model.predict(data.features)\n",
|
| 796 |
+
" # fair_metrics function available in the metrics.py file\n",
|
| 797 |
+
" fair = fair_metrics(data, pred)\n",
|
| 798 |
+
"\n",
|
| 799 |
+
" if plot:\n",
|
| 800 |
+
" # plot_fair_metrics function available in the visualisations.py file\n",
|
| 801 |
+
" # The visualisation of this function is inspired by the dashboard on the demo of IBM aif360 \n",
|
| 802 |
+
" plot_fair_metrics(fair)\n",
|
| 803 |
+
" display(fair)\n",
|
| 804 |
+
" \n",
|
| 805 |
+
" return fair"
|
| 806 |
+
]
|
| 807 |
+
},
|
| 808 |
+
{
|
| 809 |
+
"cell_type": "code",
|
| 810 |
+
"execution_count": 17,
|
| 811 |
+
"metadata": {},
|
| 812 |
+
"outputs": [
|
| 813 |
+
{
|
| 814 |
+
"data": {
|
| 815 |
+
"text/html": [
|
| 816 |
+
"<div>\n",
|
| 817 |
+
"<style scoped>\n",
|
| 818 |
+
" .dataframe tbody tr th:only-of-type {\n",
|
| 819 |
+
" vertical-align: middle;\n",
|
| 820 |
+
" }\n",
|
| 821 |
+
"\n",
|
| 822 |
+
" .dataframe tbody tr th {\n",
|
| 823 |
+
" vertical-align: top;\n",
|
| 824 |
+
" }\n",
|
| 825 |
+
"\n",
|
| 826 |
+
" .dataframe thead th {\n",
|
| 827 |
+
" text-align: right;\n",
|
| 828 |
+
" }\n",
|
| 829 |
+
"</style>\n",
|
| 830 |
+
"<table border=\"1\" class=\"dataframe\">\n",
|
| 831 |
+
" <thead>\n",
|
| 832 |
+
" <tr style=\"text-align: right;\">\n",
|
| 833 |
+
" <th></th>\n",
|
| 834 |
+
" <th>Survived</th>\n",
|
| 835 |
+
" <th>Pclass</th>\n",
|
| 836 |
+
" <th>Sex</th>\n",
|
| 837 |
+
" <th>Age</th>\n",
|
| 838 |
+
" <th>Fare</th>\n",
|
| 839 |
+
" <th>Embarked</th>\n",
|
| 840 |
+
" <th>IsAlone</th>\n",
|
| 841 |
+
" <th>Title</th>\n",
|
| 842 |
+
" </tr>\n",
|
| 843 |
+
" </thead>\n",
|
| 844 |
+
" <tbody>\n",
|
| 845 |
+
" <tr>\n",
|
| 846 |
+
" <th>0</th>\n",
|
| 847 |
+
" <td>0</td>\n",
|
| 848 |
+
" <td>3</td>\n",
|
| 849 |
+
" <td>0</td>\n",
|
| 850 |
+
" <td>1</td>\n",
|
| 851 |
+
" <td>0</td>\n",
|
| 852 |
+
" <td>0</td>\n",
|
| 853 |
+
" <td>0</td>\n",
|
| 854 |
+
" <td>1</td>\n",
|
| 855 |
+
" </tr>\n",
|
| 856 |
+
" <tr>\n",
|
| 857 |
+
" <th>1</th>\n",
|
| 858 |
+
" <td>1</td>\n",
|
| 859 |
+
" <td>1</td>\n",
|
| 860 |
+
" <td>1</td>\n",
|
| 861 |
+
" <td>2</td>\n",
|
| 862 |
+
" <td>3</td>\n",
|
| 863 |
+
" <td>1</td>\n",
|
| 864 |
+
" <td>0</td>\n",
|
| 865 |
+
" <td>3</td>\n",
|
| 866 |
+
" </tr>\n",
|
| 867 |
+
" <tr>\n",
|
| 868 |
+
" <th>2</th>\n",
|
| 869 |
+
" <td>1</td>\n",
|
| 870 |
+
" <td>3</td>\n",
|
| 871 |
+
" <td>1</td>\n",
|
| 872 |
+
" <td>1</td>\n",
|
| 873 |
+
" <td>1</td>\n",
|
| 874 |
+
" <td>0</td>\n",
|
| 875 |
+
" <td>1</td>\n",
|
| 876 |
+
" <td>2</td>\n",
|
| 877 |
+
" </tr>\n",
|
| 878 |
+
" <tr>\n",
|
| 879 |
+
" <th>3</th>\n",
|
| 880 |
+
" <td>1</td>\n",
|
| 881 |
+
" <td>1</td>\n",
|
| 882 |
+
" <td>1</td>\n",
|
| 883 |
+
" <td>2</td>\n",
|
| 884 |
+
" <td>3</td>\n",
|
| 885 |
+
" <td>0</td>\n",
|
| 886 |
+
" <td>0</td>\n",
|
| 887 |
+
" <td>3</td>\n",
|
| 888 |
+
" </tr>\n",
|
| 889 |
+
" <tr>\n",
|
| 890 |
+
" <th>4</th>\n",
|
| 891 |
+
" <td>0</td>\n",
|
| 892 |
+
" <td>3</td>\n",
|
| 893 |
+
" <td>0</td>\n",
|
| 894 |
+
" <td>2</td>\n",
|
| 895 |
+
" <td>1</td>\n",
|
| 896 |
+
" <td>0</td>\n",
|
| 897 |
+
" <td>1</td>\n",
|
| 898 |
+
" <td>1</td>\n",
|
| 899 |
+
" </tr>\n",
|
| 900 |
+
" <tr>\n",
|
| 901 |
+
" <th>...</th>\n",
|
| 902 |
+
" <td>...</td>\n",
|
| 903 |
+
" <td>...</td>\n",
|
| 904 |
+
" <td>...</td>\n",
|
| 905 |
+
" <td>...</td>\n",
|
| 906 |
+
" <td>...</td>\n",
|
| 907 |
+
" <td>...</td>\n",
|
| 908 |
+
" <td>...</td>\n",
|
| 909 |
+
" <td>...</td>\n",
|
| 910 |
+
" </tr>\n",
|
| 911 |
+
" <tr>\n",
|
| 912 |
+
" <th>886</th>\n",
|
| 913 |
+
" <td>0</td>\n",
|
| 914 |
+
" <td>2</td>\n",
|
| 915 |
+
" <td>0</td>\n",
|
| 916 |
+
" <td>1</td>\n",
|
| 917 |
+
" <td>1</td>\n",
|
| 918 |
+
" <td>0</td>\n",
|
| 919 |
+
" <td>1</td>\n",
|
| 920 |
+
" <td>5</td>\n",
|
| 921 |
+
" </tr>\n",
|
| 922 |
+
" <tr>\n",
|
| 923 |
+
" <th>887</th>\n",
|
| 924 |
+
" <td>1</td>\n",
|
| 925 |
+
" <td>1</td>\n",
|
| 926 |
+
" <td>1</td>\n",
|
| 927 |
+
" <td>1</td>\n",
|
| 928 |
+
" <td>2</td>\n",
|
| 929 |
+
" <td>0</td>\n",
|
| 930 |
+
" <td>1</td>\n",
|
| 931 |
+
" <td>2</td>\n",
|
| 932 |
+
" </tr>\n",
|
| 933 |
+
" <tr>\n",
|
| 934 |
+
" <th>888</th>\n",
|
| 935 |
+
" <td>0</td>\n",
|
| 936 |
+
" <td>3</td>\n",
|
| 937 |
+
" <td>1</td>\n",
|
| 938 |
+
" <td>0</td>\n",
|
| 939 |
+
" <td>2</td>\n",
|
| 940 |
+
" <td>0</td>\n",
|
| 941 |
+
" <td>0</td>\n",
|
| 942 |
+
" <td>2</td>\n",
|
| 943 |
+
" </tr>\n",
|
| 944 |
+
" <tr>\n",
|
| 945 |
+
" <th>889</th>\n",
|
| 946 |
+
" <td>1</td>\n",
|
| 947 |
+
" <td>1</td>\n",
|
| 948 |
+
" <td>0</td>\n",
|
| 949 |
+
" <td>1</td>\n",
|
| 950 |
+
" <td>2</td>\n",
|
| 951 |
+
" <td>1</td>\n",
|
| 952 |
+
" <td>1</td>\n",
|
| 953 |
+
" <td>1</td>\n",
|
| 954 |
+
" </tr>\n",
|
| 955 |
+
" <tr>\n",
|
| 956 |
+
" <th>890</th>\n",
|
| 957 |
+
" <td>0</td>\n",
|
| 958 |
+
" <td>3</td>\n",
|
| 959 |
+
" <td>0</td>\n",
|
| 960 |
+
" <td>1</td>\n",
|
| 961 |
+
" <td>0</td>\n",
|
| 962 |
+
" <td>2</td>\n",
|
| 963 |
+
" <td>1</td>\n",
|
| 964 |
+
" <td>1</td>\n",
|
| 965 |
+
" </tr>\n",
|
| 966 |
+
" </tbody>\n",
|
| 967 |
+
"</table>\n",
|
| 968 |
+
"<p>891 rows × 8 columns</p>\n",
|
| 969 |
+
"</div>"
|
| 970 |
+
],
|
| 971 |
+
"text/plain": [
|
| 972 |
+
" Survived Pclass Sex Age Fare Embarked IsAlone Title\n",
|
| 973 |
+
"0 0 3 0 1 0 0 0 1\n",
|
| 974 |
+
"1 1 1 1 2 3 1 0 3\n",
|
| 975 |
+
"2 1 3 1 1 1 0 1 2\n",
|
| 976 |
+
"3 1 1 1 2 3 0 0 3\n",
|
| 977 |
+
"4 0 3 0 2 1 0 1 1\n",
|
| 978 |
+
".. ... ... ... ... ... ... ... ...\n",
|
| 979 |
+
"886 0 2 0 1 1 0 1 5\n",
|
| 980 |
+
"887 1 1 1 1 2 0 1 2\n",
|
| 981 |
+
"888 0 3 1 0 2 0 0 2\n",
|
| 982 |
+
"889 1 1 0 1 2 1 1 1\n",
|
| 983 |
+
"890 0 3 0 1 0 2 1 1\n",
|
| 984 |
+
"\n",
|
| 985 |
+
"[891 rows x 8 columns]"
|
| 986 |
+
]
|
| 987 |
+
},
|
| 988 |
+
"execution_count": 17,
|
| 989 |
+
"metadata": {},
|
| 990 |
+
"output_type": "execute_result"
|
| 991 |
+
}
|
| 992 |
+
],
|
| 993 |
+
"source": [
|
| 994 |
+
"##train['Sex'] = train['Sex'].map( {'female': 1, 'male': 0} ).astype(int)\n",
|
| 995 |
+
"train_df\n",
|
| 996 |
+
"\n",
|
| 997 |
+
"#features = [\"Pclass\", \"Sex\", \"SibSp\", \"Parch\", \"Survived\"]\n",
|
| 998 |
+
"#X = pd.get_dummies(train_data[features])"
|
| 999 |
+
]
|
| 1000 |
+
},
|
| 1001 |
+
{
|
| 1002 |
+
"cell_type": "code",
|
| 1003 |
+
"execution_count": 18,
|
| 1004 |
+
"metadata": {},
|
| 1005 |
+
"outputs": [],
|
| 1006 |
+
"source": [
|
| 1007 |
+
"privileged_groups = [{'Sex': 1}]\n",
|
| 1008 |
+
"unprivileged_groups = [{'Sex': 0}]\n",
|
| 1009 |
+
"dataset_orig = StandardDataset(train_df,\n",
|
| 1010 |
+
" label_name='Survived',\n",
|
| 1011 |
+
" protected_attribute_names=['Sex'],\n",
|
| 1012 |
+
" favorable_classes=[1],\n",
|
| 1013 |
+
" privileged_classes=[[1]])\n",
|
| 1014 |
+
"\n"
|
| 1015 |
+
]
|
| 1016 |
+
},
|
| 1017 |
+
{
|
| 1018 |
+
"cell_type": "code",
|
| 1019 |
+
"execution_count": 19,
|
| 1020 |
+
"metadata": {},
|
| 1021 |
+
"outputs": [
|
| 1022 |
+
{
|
| 1023 |
+
"data": {
|
| 1024 |
+
"text/markdown": [
|
| 1025 |
+
"#### Original training dataset"
|
| 1026 |
+
],
|
| 1027 |
+
"text/plain": [
|
| 1028 |
+
"<IPython.core.display.Markdown object>"
|
| 1029 |
+
]
|
| 1030 |
+
},
|
| 1031 |
+
"metadata": {},
|
| 1032 |
+
"output_type": "display_data"
|
| 1033 |
+
},
|
| 1034 |
+
{
|
| 1035 |
+
"name": "stdout",
|
| 1036 |
+
"output_type": "stream",
|
| 1037 |
+
"text": [
|
| 1038 |
+
"Difference in mean outcomes between unprivileged and privileged groups = -0.553130\n"
|
| 1039 |
+
]
|
| 1040 |
+
}
|
| 1041 |
+
],
|
| 1042 |
+
"source": [
|
| 1043 |
+
"metric_orig_train = BinaryLabelDatasetMetric(dataset_orig, \n",
|
| 1044 |
+
" unprivileged_groups=unprivileged_groups,\n",
|
| 1045 |
+
" privileged_groups=privileged_groups)\n",
|
| 1046 |
+
"display(Markdown(\"#### Original training dataset\"))\n",
|
| 1047 |
+
"print(\"Difference in mean outcomes between unprivileged and privileged groups = %f\" % metric_orig_train.mean_difference())"
|
| 1048 |
+
]
|
| 1049 |
+
},
|
| 1050 |
+
{
|
| 1051 |
+
"cell_type": "code",
|
| 1052 |
+
"execution_count": 41,
|
| 1053 |
+
"metadata": {},
|
| 1054 |
+
"outputs": [],
|
| 1055 |
+
"source": [
|
| 1056 |
+
"import ipynbname\n",
|
| 1057 |
+
"nb_fname = ipynbname.name()\n",
|
| 1058 |
+
"nb_path = ipynbname.path()\n",
|
| 1059 |
+
"\n",
|
| 1060 |
+
"from sklearn.ensemble import AdaBoostClassifier\n",
|
| 1061 |
+
"import pickle\n",
|
| 1062 |
+
"\n",
|
| 1063 |
+
"data_orig_train, data_orig_test = dataset_orig.split([0.7], shuffle=True)\n",
|
| 1064 |
+
"X_train = data_orig_train.features\n",
|
| 1065 |
+
"y_train = data_orig_train.labels.ravel()\n",
|
| 1066 |
+
"\n",
|
| 1067 |
+
"X_test = data_orig_test.features\n",
|
| 1068 |
+
"y_test = data_orig_test.labels.ravel()\n",
|
| 1069 |
+
"num_estimators = 100\n",
|
| 1070 |
+
"\n",
|
| 1071 |
+
"model = AdaBoostClassifier(n_estimators=1)\n",
|
| 1072 |
+
"\n",
|
| 1073 |
+
"mdl = model.fit(X_train, y_train)\n",
|
| 1074 |
+
"with open('../../Results/AdaBoost/' + nb_fname + '.pkl', 'wb') as f:\n",
|
| 1075 |
+
" pickle.dump(mdl, f)\n",
|
| 1076 |
+
"\n",
|
| 1077 |
+
"with open('../../Results/AdaBoost/' + nb_fname + '_Train' + '.pkl', 'wb') as f:\n",
|
| 1078 |
+
" pickle.dump(data_orig_train, f) \n",
|
| 1079 |
+
" \n",
|
| 1080 |
+
"with open('../../Results/AdaBoost/' + nb_fname + '_Test' + '.pkl', 'wb') as f:\n",
|
| 1081 |
+
" pickle.dump(data_orig_test, f) "
|
| 1082 |
+
]
|
| 1083 |
+
},
|
| 1084 |
+
{
|
| 1085 |
+
"cell_type": "code",
|
| 1086 |
+
"execution_count": 22,
|
| 1087 |
+
"metadata": {},
|
| 1088 |
+
"outputs": [
|
| 1089 |
+
{
|
| 1090 |
+
"name": "stdout",
|
| 1091 |
+
"output_type": "stream",
|
| 1092 |
+
"text": [
|
| 1093 |
+
"0\n",
|
| 1094 |
+
"1\n",
|
| 1095 |
+
"2\n",
|
| 1096 |
+
"3\n",
|
| 1097 |
+
"4\n",
|
| 1098 |
+
"5\n",
|
| 1099 |
+
"6\n",
|
| 1100 |
+
"7\n",
|
| 1101 |
+
"8\n",
|
| 1102 |
+
"9\n",
|
| 1103 |
+
"STD [3.02765035 0.06749158 0.08874808 0.09476216 0.03541161 0.01255178]\n",
|
| 1104 |
+
"[4.5, -0.6693072547146794, -0.581259725046272, 0.49612085216852686, -2.1276205667545494, 0.1590111172017386]\n",
|
| 1105 |
+
"-2.7230555771452356\n",
|
| 1106 |
+
"0.8093283582089552\n",
|
| 1107 |
+
"0.7401892992453725\n"
|
| 1108 |
+
]
|
| 1109 |
+
}
|
| 1110 |
+
],
|
| 1111 |
+
"source": [
|
| 1112 |
+
"final_metrics = []\n",
|
| 1113 |
+
"accuracy = []\n",
|
| 1114 |
+
"f1= []\n",
|
| 1115 |
+
"from statistics import mean\n",
|
| 1116 |
+
"from sklearn.metrics import accuracy_score, f1_score\n",
|
| 1117 |
+
"from sklearn.ensemble import AdaBoostClassifier\n",
|
| 1118 |
+
"\n",
|
| 1119 |
+
"\n",
|
| 1120 |
+
"for i in range(0,10):\n",
|
| 1121 |
+
" \n",
|
| 1122 |
+
" data_orig_train, data_orig_test = dataset_orig.split([0.7], shuffle=True)\n",
|
| 1123 |
+
" print(i)\n",
|
| 1124 |
+
" X_train = data_orig_train.features\n",
|
| 1125 |
+
" y_train = data_orig_train.labels.ravel()\n",
|
| 1126 |
+
"\n",
|
| 1127 |
+
" X_test = data_orig_test.features\n",
|
| 1128 |
+
" y_test = data_orig_test.labels.ravel()\n",
|
| 1129 |
+
" model = GradientBoostingClassifier(n_estimators = 200)\n",
|
| 1130 |
+
" \n",
|
| 1131 |
+
" mdl = model.fit(X_train, y_train)\n",
|
| 1132 |
+
" yy = mdl.predict(X_test)\n",
|
| 1133 |
+
" accuracy.append(accuracy_score(y_test, yy))\n",
|
| 1134 |
+
" f1.append(f1_score(y_test, yy))\n",
|
| 1135 |
+
" fair = get_fair_metrics_and_plot(data_orig_test, mdl) \n",
|
| 1136 |
+
" fair_list = fair.iloc[1].tolist()\n",
|
| 1137 |
+
" fair_list.insert(0, i)\n",
|
| 1138 |
+
" final_metrics.append(fair_list)\n",
|
| 1139 |
+
"\n",
|
| 1140 |
+
" \n",
|
| 1141 |
+
"element_wise_std = np.std(final_metrics, 0, ddof=1)\n",
|
| 1142 |
+
"print(\"STD \" + str(element_wise_std))\n",
|
| 1143 |
+
"final_metrics = list(map(mean, zip(*final_metrics)))\n",
|
| 1144 |
+
"accuracy = mean(accuracy)\n",
|
| 1145 |
+
"f1 = mean(f1)\n",
|
| 1146 |
+
"final_metrics[4] = np.log(final_metrics[4])\n",
|
| 1147 |
+
"print(final_metrics)\n",
|
| 1148 |
+
"print(sum(final_metrics[1:]))\n",
|
| 1149 |
+
"print(accuracy)\n",
|
| 1150 |
+
"print(f1)"
|
| 1151 |
+
]
|
| 1152 |
+
},
|
| 1153 |
+
{
|
| 1154 |
+
"cell_type": "code",
|
| 1155 |
+
"execution_count": 42,
|
| 1156 |
+
"metadata": {},
|
| 1157 |
+
"outputs": [],
|
| 1158 |
+
"source": [
|
| 1159 |
+
"from csv import writer\n",
|
| 1160 |
+
"from sklearn.metrics import accuracy_score, f1_score\n",
|
| 1161 |
+
"\n",
|
| 1162 |
+
"final_metrics = []\n",
|
| 1163 |
+
"accuracy = []\n",
|
| 1164 |
+
"f1= []\n",
|
| 1165 |
+
"\n",
|
| 1166 |
+
"for i in range(1,num_estimators+1):\n",
|
| 1167 |
+
" \n",
|
| 1168 |
+
" model = AdaBoostClassifier(n_estimators=i)\n",
|
| 1169 |
+
" \n",
|
| 1170 |
+
" mdl = model.fit(X_train, y_train)\n",
|
| 1171 |
+
" yy = mdl.predict(X_test)\n",
|
| 1172 |
+
" accuracy.append(accuracy_score(y_test, yy))\n",
|
| 1173 |
+
" f1.append(f1_score(y_test, yy))\n",
|
| 1174 |
+
" fair = get_fair_metrics_and_plot(data_orig_test, mdl) \n",
|
| 1175 |
+
" fair_list = fair.iloc[1].tolist()\n",
|
| 1176 |
+
" fair_list.insert(0, i)\n",
|
| 1177 |
+
" final_metrics.append(fair_list)\n"
|
| 1178 |
+
]
|
| 1179 |
+
},
|
| 1180 |
+
{
|
| 1181 |
+
"cell_type": "code",
|
| 1182 |
+
"execution_count": 43,
|
| 1183 |
+
"metadata": {},
|
| 1184 |
+
"outputs": [
|
| 1185 |
+
{
|
| 1186 |
+
"data": {
|
| 1187 |
+
"text/html": [
|
| 1188 |
+
"<div>\n",
|
| 1189 |
+
"<style scoped>\n",
|
| 1190 |
+
" .dataframe tbody tr th:only-of-type {\n",
|
| 1191 |
+
" vertical-align: middle;\n",
|
| 1192 |
+
" }\n",
|
| 1193 |
+
"\n",
|
| 1194 |
+
" .dataframe tbody tr th {\n",
|
| 1195 |
+
" vertical-align: top;\n",
|
| 1196 |
+
" }\n",
|
| 1197 |
+
"\n",
|
| 1198 |
+
" .dataframe thead th {\n",
|
| 1199 |
+
" text-align: right;\n",
|
| 1200 |
+
" }\n",
|
| 1201 |
+
"</style>\n",
|
| 1202 |
+
"<table border=\"1\" class=\"dataframe\">\n",
|
| 1203 |
+
" <thead>\n",
|
| 1204 |
+
" <tr style=\"text-align: right;\">\n",
|
| 1205 |
+
" <th></th>\n",
|
| 1206 |
+
" <th>classifier</th>\n",
|
| 1207 |
+
" <th>T0</th>\n",
|
| 1208 |
+
" <th>T1</th>\n",
|
| 1209 |
+
" <th>T2</th>\n",
|
| 1210 |
+
" <th>T3</th>\n",
|
| 1211 |
+
" <th>T4</th>\n",
|
| 1212 |
+
" <th>T5</th>\n",
|
| 1213 |
+
" <th>T6</th>\n",
|
| 1214 |
+
" <th>T7</th>\n",
|
| 1215 |
+
" <th>T8</th>\n",
|
| 1216 |
+
" <th>...</th>\n",
|
| 1217 |
+
" <th>T90</th>\n",
|
| 1218 |
+
" <th>T91</th>\n",
|
| 1219 |
+
" <th>T92</th>\n",
|
| 1220 |
+
" <th>T93</th>\n",
|
| 1221 |
+
" <th>T94</th>\n",
|
| 1222 |
+
" <th>T95</th>\n",
|
| 1223 |
+
" <th>T96</th>\n",
|
| 1224 |
+
" <th>T97</th>\n",
|
| 1225 |
+
" <th>T98</th>\n",
|
| 1226 |
+
" <th>T99</th>\n",
|
| 1227 |
+
" </tr>\n",
|
| 1228 |
+
" </thead>\n",
|
| 1229 |
+
" <tbody>\n",
|
| 1230 |
+
" <tr>\n",
|
| 1231 |
+
" <th>accuracy</th>\n",
|
| 1232 |
+
" <td>0.787313</td>\n",
|
| 1233 |
+
" <td>0.764925</td>\n",
|
| 1234 |
+
" <td>0.764925</td>\n",
|
| 1235 |
+
" <td>0.779851</td>\n",
|
| 1236 |
+
" <td>0.750000</td>\n",
|
| 1237 |
+
" <td>0.783582</td>\n",
|
| 1238 |
+
" <td>0.779851</td>\n",
|
| 1239 |
+
" <td>0.783582</td>\n",
|
| 1240 |
+
" <td>0.791045</td>\n",
|
| 1241 |
+
" <td>0.787313</td>\n",
|
| 1242 |
+
" <td>...</td>\n",
|
| 1243 |
+
" <td>0.787313</td>\n",
|
| 1244 |
+
" <td>0.787313</td>\n",
|
| 1245 |
+
" <td>0.787313</td>\n",
|
| 1246 |
+
" <td>0.787313</td>\n",
|
| 1247 |
+
" <td>0.787313</td>\n",
|
| 1248 |
+
" <td>0.787313</td>\n",
|
| 1249 |
+
" <td>0.787313</td>\n",
|
| 1250 |
+
" <td>0.787313</td>\n",
|
| 1251 |
+
" <td>0.787313</td>\n",
|
| 1252 |
+
" <td>0.787313</td>\n",
|
| 1253 |
+
" </tr>\n",
|
| 1254 |
+
" <tr>\n",
|
| 1255 |
+
" <th>f1</th>\n",
|
| 1256 |
+
" <td>0.729858</td>\n",
|
| 1257 |
+
" <td>0.729614</td>\n",
|
| 1258 |
+
" <td>0.729614</td>\n",
|
| 1259 |
+
" <td>0.735426</td>\n",
|
| 1260 |
+
" <td>0.621469</td>\n",
|
| 1261 |
+
" <td>0.715686</td>\n",
|
| 1262 |
+
" <td>0.730594</td>\n",
|
| 1263 |
+
" <td>0.715686</td>\n",
|
| 1264 |
+
" <td>0.730769</td>\n",
|
| 1265 |
+
" <td>0.727273</td>\n",
|
| 1266 |
+
" <td>...</td>\n",
|
| 1267 |
+
" <td>0.729858</td>\n",
|
| 1268 |
+
" <td>0.729858</td>\n",
|
| 1269 |
+
" <td>0.729858</td>\n",
|
| 1270 |
+
" <td>0.727273</td>\n",
|
| 1271 |
+
" <td>0.729858</td>\n",
|
| 1272 |
+
" <td>0.729858</td>\n",
|
| 1273 |
+
" <td>0.727273</td>\n",
|
| 1274 |
+
" <td>0.729858</td>\n",
|
| 1275 |
+
" <td>0.727273</td>\n",
|
| 1276 |
+
" <td>0.729858</td>\n",
|
| 1277 |
+
" </tr>\n",
|
| 1278 |
+
" <tr>\n",
|
| 1279 |
+
" <th>statistical_parity_difference</th>\n",
|
| 1280 |
+
" <td>-0.814846</td>\n",
|
| 1281 |
+
" <td>-0.867052</td>\n",
|
| 1282 |
+
" <td>-0.867052</td>\n",
|
| 1283 |
+
" <td>-0.908549</td>\n",
|
| 1284 |
+
" <td>-0.489565</td>\n",
|
| 1285 |
+
" <td>-0.578096</td>\n",
|
| 1286 |
+
" <td>-0.947977</td>\n",
|
| 1287 |
+
" <td>-0.708549</td>\n",
|
| 1288 |
+
" <td>-0.799574</td>\n",
|
| 1289 |
+
" <td>-0.793794</td>\n",
|
| 1290 |
+
" <td>...</td>\n",
|
| 1291 |
+
" <td>-0.814846</td>\n",
|
| 1292 |
+
" <td>-0.814846</td>\n",
|
| 1293 |
+
" <td>-0.814846</td>\n",
|
| 1294 |
+
" <td>-0.793794</td>\n",
|
| 1295 |
+
" <td>-0.814846</td>\n",
|
| 1296 |
+
" <td>-0.814846</td>\n",
|
| 1297 |
+
" <td>-0.793794</td>\n",
|
| 1298 |
+
" <td>-0.814846</td>\n",
|
| 1299 |
+
" <td>-0.793794</td>\n",
|
| 1300 |
+
" <td>-0.814846</td>\n",
|
| 1301 |
+
" </tr>\n",
|
| 1302 |
+
" <tr>\n",
|
| 1303 |
+
" <th>equal_opportunity_difference</th>\n",
|
| 1304 |
+
" <td>-0.775214</td>\n",
|
| 1305 |
+
" <td>-0.731707</td>\n",
|
| 1306 |
+
" <td>-0.731707</td>\n",
|
| 1307 |
+
" <td>-0.766974</td>\n",
|
| 1308 |
+
" <td>-0.477917</td>\n",
|
| 1309 |
+
" <td>-0.531641</td>\n",
|
| 1310 |
+
" <td>-0.853659</td>\n",
|
| 1311 |
+
" <td>-0.759064</td>\n",
|
| 1312 |
+
" <td>-0.761701</td>\n",
|
| 1313 |
+
" <td>-0.761701</td>\n",
|
| 1314 |
+
" <td>...</td>\n",
|
| 1315 |
+
" <td>-0.775214</td>\n",
|
| 1316 |
+
" <td>-0.775214</td>\n",
|
| 1317 |
+
" <td>-0.775214</td>\n",
|
| 1318 |
+
" <td>-0.761701</td>\n",
|
| 1319 |
+
" <td>-0.775214</td>\n",
|
| 1320 |
+
" <td>-0.775214</td>\n",
|
| 1321 |
+
" <td>-0.761701</td>\n",
|
| 1322 |
+
" <td>-0.775214</td>\n",
|
| 1323 |
+
" <td>-0.761701</td>\n",
|
| 1324 |
+
" <td>-0.775214</td>\n",
|
| 1325 |
+
" </tr>\n",
|
| 1326 |
+
" <tr>\n",
|
| 1327 |
+
" <th>average_abs_odds_difference</th>\n",
|
| 1328 |
+
" <td>0.702001</td>\n",
|
| 1329 |
+
" <td>0.820399</td>\n",
|
| 1330 |
+
" <td>0.820399</td>\n",
|
| 1331 |
+
" <td>0.864548</td>\n",
|
| 1332 |
+
" <td>0.322833</td>\n",
|
| 1333 |
+
" <td>0.370799</td>\n",
|
| 1334 |
+
" <td>0.915466</td>\n",
|
| 1335 |
+
" <td>0.539705</td>\n",
|
| 1336 |
+
" <td>0.675223</td>\n",
|
| 1337 |
+
" <td>0.671435</td>\n",
|
| 1338 |
+
" <td>...</td>\n",
|
| 1339 |
+
" <td>0.702001</td>\n",
|
| 1340 |
+
" <td>0.702001</td>\n",
|
| 1341 |
+
" <td>0.702001</td>\n",
|
| 1342 |
+
" <td>0.671435</td>\n",
|
| 1343 |
+
" <td>0.702001</td>\n",
|
| 1344 |
+
" <td>0.702001</td>\n",
|
| 1345 |
+
" <td>0.671435</td>\n",
|
| 1346 |
+
" <td>0.702001</td>\n",
|
| 1347 |
+
" <td>0.671435</td>\n",
|
| 1348 |
+
" <td>0.702001</td>\n",
|
| 1349 |
+
" </tr>\n",
|
| 1350 |
+
" <tr>\n",
|
| 1351 |
+
" <th>disparate_impact</th>\n",
|
| 1352 |
+
" <td>-2.545325</td>\n",
|
| 1353 |
+
" <td>-2.017797</td>\n",
|
| 1354 |
+
" <td>-2.017797</td>\n",
|
| 1355 |
+
" <td>-2.503652</td>\n",
|
| 1356 |
+
" <td>-2.248073</td>\n",
|
| 1357 |
+
" <td>-1.713065</td>\n",
|
| 1358 |
+
" <td>-2.956067</td>\n",
|
| 1359 |
+
" <td>-2.277845</td>\n",
|
| 1360 |
+
" <td>-2.608239</td>\n",
|
| 1361 |
+
" <td>-2.521227</td>\n",
|
| 1362 |
+
" <td>...</td>\n",
|
| 1363 |
+
" <td>-2.545325</td>\n",
|
| 1364 |
+
" <td>-2.545325</td>\n",
|
| 1365 |
+
" <td>-2.545325</td>\n",
|
| 1366 |
+
" <td>-2.521227</td>\n",
|
| 1367 |
+
" <td>-2.545325</td>\n",
|
| 1368 |
+
" <td>-2.545325</td>\n",
|
| 1369 |
+
" <td>-2.521227</td>\n",
|
| 1370 |
+
" <td>-2.545325</td>\n",
|
| 1371 |
+
" <td>-2.521227</td>\n",
|
| 1372 |
+
" <td>-2.545325</td>\n",
|
| 1373 |
+
" </tr>\n",
|
| 1374 |
+
" <tr>\n",
|
| 1375 |
+
" <th>theil_index</th>\n",
|
| 1376 |
+
" <td>0.179316</td>\n",
|
| 1377 |
+
" <td>0.157679</td>\n",
|
| 1378 |
+
" <td>0.157679</td>\n",
|
| 1379 |
+
" <td>0.164565</td>\n",
|
| 1380 |
+
" <td>0.265484</td>\n",
|
| 1381 |
+
" <td>0.193705</td>\n",
|
| 1382 |
+
" <td>0.171370</td>\n",
|
| 1383 |
+
" <td>0.193705</td>\n",
|
| 1384 |
+
" <td>0.181456</td>\n",
|
| 1385 |
+
" <td>0.182624</td>\n",
|
| 1386 |
+
" <td>...</td>\n",
|
| 1387 |
+
" <td>0.179316</td>\n",
|
| 1388 |
+
" <td>0.179316</td>\n",
|
| 1389 |
+
" <td>0.179316</td>\n",
|
| 1390 |
+
" <td>0.182624</td>\n",
|
| 1391 |
+
" <td>0.179316</td>\n",
|
| 1392 |
+
" <td>0.179316</td>\n",
|
| 1393 |
+
" <td>0.182624</td>\n",
|
| 1394 |
+
" <td>0.179316</td>\n",
|
| 1395 |
+
" <td>0.182624</td>\n",
|
| 1396 |
+
" <td>0.179316</td>\n",
|
| 1397 |
+
" </tr>\n",
|
| 1398 |
+
" </tbody>\n",
|
| 1399 |
+
"</table>\n",
|
| 1400 |
+
"<p>7 rows × 101 columns</p>\n",
|
| 1401 |
+
"</div>"
|
| 1402 |
+
],
|
| 1403 |
+
"text/plain": [
|
| 1404 |
+
" classifier T0 T1 T2 \\\n",
|
| 1405 |
+
"accuracy 0.787313 0.764925 0.764925 0.779851 \n",
|
| 1406 |
+
"f1 0.729858 0.729614 0.729614 0.735426 \n",
|
| 1407 |
+
"statistical_parity_difference -0.814846 -0.867052 -0.867052 -0.908549 \n",
|
| 1408 |
+
"equal_opportunity_difference -0.775214 -0.731707 -0.731707 -0.766974 \n",
|
| 1409 |
+
"average_abs_odds_difference 0.702001 0.820399 0.820399 0.864548 \n",
|
| 1410 |
+
"disparate_impact -2.545325 -2.017797 -2.017797 -2.503652 \n",
|
| 1411 |
+
"theil_index 0.179316 0.157679 0.157679 0.164565 \n",
|
| 1412 |
+
"\n",
|
| 1413 |
+
" T3 T4 T5 T6 \\\n",
|
| 1414 |
+
"accuracy 0.750000 0.783582 0.779851 0.783582 \n",
|
| 1415 |
+
"f1 0.621469 0.715686 0.730594 0.715686 \n",
|
| 1416 |
+
"statistical_parity_difference -0.489565 -0.578096 -0.947977 -0.708549 \n",
|
| 1417 |
+
"equal_opportunity_difference -0.477917 -0.531641 -0.853659 -0.759064 \n",
|
| 1418 |
+
"average_abs_odds_difference 0.322833 0.370799 0.915466 0.539705 \n",
|
| 1419 |
+
"disparate_impact -2.248073 -1.713065 -2.956067 -2.277845 \n",
|
| 1420 |
+
"theil_index 0.265484 0.193705 0.171370 0.193705 \n",
|
| 1421 |
+
"\n",
|
| 1422 |
+
" T7 T8 ... T90 T91 \\\n",
|
| 1423 |
+
"accuracy 0.791045 0.787313 ... 0.787313 0.787313 \n",
|
| 1424 |
+
"f1 0.730769 0.727273 ... 0.729858 0.729858 \n",
|
| 1425 |
+
"statistical_parity_difference -0.799574 -0.793794 ... -0.814846 -0.814846 \n",
|
| 1426 |
+
"equal_opportunity_difference -0.761701 -0.761701 ... -0.775214 -0.775214 \n",
|
| 1427 |
+
"average_abs_odds_difference 0.675223 0.671435 ... 0.702001 0.702001 \n",
|
| 1428 |
+
"disparate_impact -2.608239 -2.521227 ... -2.545325 -2.545325 \n",
|
| 1429 |
+
"theil_index 0.181456 0.182624 ... 0.179316 0.179316 \n",
|
| 1430 |
+
"\n",
|
| 1431 |
+
" T92 T93 T94 T95 \\\n",
|
| 1432 |
+
"accuracy 0.787313 0.787313 0.787313 0.787313 \n",
|
| 1433 |
+
"f1 0.729858 0.727273 0.729858 0.729858 \n",
|
| 1434 |
+
"statistical_parity_difference -0.814846 -0.793794 -0.814846 -0.814846 \n",
|
| 1435 |
+
"equal_opportunity_difference -0.775214 -0.761701 -0.775214 -0.775214 \n",
|
| 1436 |
+
"average_abs_odds_difference 0.702001 0.671435 0.702001 0.702001 \n",
|
| 1437 |
+
"disparate_impact -2.545325 -2.521227 -2.545325 -2.545325 \n",
|
| 1438 |
+
"theil_index 0.179316 0.182624 0.179316 0.179316 \n",
|
| 1439 |
+
"\n",
|
| 1440 |
+
" T96 T97 T98 T99 \n",
|
| 1441 |
+
"accuracy 0.787313 0.787313 0.787313 0.787313 \n",
|
| 1442 |
+
"f1 0.727273 0.729858 0.727273 0.729858 \n",
|
| 1443 |
+
"statistical_parity_difference -0.793794 -0.814846 -0.793794 -0.814846 \n",
|
| 1444 |
+
"equal_opportunity_difference -0.761701 -0.775214 -0.761701 -0.775214 \n",
|
| 1445 |
+
"average_abs_odds_difference 0.671435 0.702001 0.671435 0.702001 \n",
|
| 1446 |
+
"disparate_impact -2.521227 -2.545325 -2.521227 -2.545325 \n",
|
| 1447 |
+
"theil_index 0.182624 0.179316 0.182624 0.179316 \n",
|
| 1448 |
+
"\n",
|
| 1449 |
+
"[7 rows x 101 columns]"
|
| 1450 |
+
]
|
| 1451 |
+
},
|
| 1452 |
+
"execution_count": 43,
|
| 1453 |
+
"metadata": {},
|
| 1454 |
+
"output_type": "execute_result"
|
| 1455 |
+
}
|
| 1456 |
+
],
|
| 1457 |
+
"source": [
|
| 1458 |
+
"import numpy as np\n",
|
| 1459 |
+
"final_result = pd.DataFrame(final_metrics)\n",
|
| 1460 |
+
"final_result[4] = np.log(final_result[4])\n",
|
| 1461 |
+
"final_result = final_result.transpose()\n",
|
| 1462 |
+
"final_result.loc[0] = f1 # add f1 and acc to df\n",
|
| 1463 |
+
"acc = pd.DataFrame(accuracy).transpose()\n",
|
| 1464 |
+
"acc = acc.rename(index={0: 'accuracy'})\n",
|
| 1465 |
+
"final_result = pd.concat([acc,final_result])\n",
|
| 1466 |
+
"final_result = final_result.rename(index={0: 'f1', 1: 'statistical_parity_difference', 2: 'equal_opportunity_difference', 3: 'average_abs_odds_difference', 4: 'disparate_impact', 5: 'theil_index'})\n",
|
| 1467 |
+
"final_result.columns = ['T' + str(col) for col in final_result.columns]\n",
|
| 1468 |
+
"final_result.insert(0, \"classifier\", final_result['T' + str(num_estimators - 1)]) ##Add final metrics add the beginning of the df\n",
|
| 1469 |
+
"final_result.to_csv('../../Results/AdaBoost/' + nb_fname + '.csv')\n",
|
| 1470 |
+
"final_result"
|
| 1471 |
+
]
|
| 1472 |
+
},
|
| 1473 |
+
{
|
| 1474 |
+
"cell_type": "code",
|
| 1475 |
+
"execution_count": null,
|
| 1476 |
+
"metadata": {},
|
| 1477 |
+
"outputs": [],
|
| 1478 |
+
"source": []
|
| 1479 |
+
}
|
| 1480 |
+
],
|
| 1481 |
+
"metadata": {
|
| 1482 |
+
"_change_revision": 2,
|
| 1483 |
+
"_is_fork": false,
|
| 1484 |
+
"kernelspec": {
|
| 1485 |
+
"display_name": "Python 3",
|
| 1486 |
+
"language": "python",
|
| 1487 |
+
"name": "python3"
|
| 1488 |
+
},
|
| 1489 |
+
"language_info": {
|
| 1490 |
+
"codemirror_mode": {
|
| 1491 |
+
"name": "ipython",
|
| 1492 |
+
"version": 3
|
| 1493 |
+
},
|
| 1494 |
+
"file_extension": ".py",
|
| 1495 |
+
"mimetype": "text/x-python",
|
| 1496 |
+
"name": "python",
|
| 1497 |
+
"nbconvert_exporter": "python",
|
| 1498 |
+
"pygments_lexer": "ipython3",
|
| 1499 |
+
"version": "3.8.5"
|
| 1500 |
+
}
|
| 1501 |
+
},
|
| 1502 |
+
"nbformat": 4,
|
| 1503 |
+
"nbformat_minor": 1
|
| 1504 |
+
}
|
Titanic/Kernels/AdaBoost/6-titanic-best-working-classifier.py
ADDED
|
@@ -0,0 +1,269 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python
|
| 2 |
+
# coding: utf-8
|
| 3 |
+
|
| 4 |
+
# ## Introduction ##
|
| 5 |
+
#
|
| 6 |
+
# This is my first work of machine learning. the notebook is written in python and has inspired from ["Exploring Survival on Titanic" by Megan Risdal, a Kernel in R on Kaggle][1].
|
| 7 |
+
#
|
| 8 |
+
#
|
| 9 |
+
# [1]: https://www.kaggle.com/mrisdal/titanic/exploring-survival-on-the-titanic
|
| 10 |
+
|
| 11 |
+
# In[1]:
|
| 12 |
+
|
| 13 |
+
|
| 14 |
+
get_ipython().run_line_magic('matplotlib', 'inline')
|
| 15 |
+
import numpy as np
|
| 16 |
+
import pandas as pd
|
| 17 |
+
import re as re
|
| 18 |
+
|
| 19 |
+
train = pd.read_csv('../input/train.csv', header = 0, dtype={'Age': np.float64})
|
| 20 |
+
test = pd.read_csv('../input/test.csv' , header = 0, dtype={'Age': np.float64})
|
| 21 |
+
full_data = [train, test]
|
| 22 |
+
|
| 23 |
+
print (train.info())
|
| 24 |
+
|
| 25 |
+
|
| 26 |
+
# # Feature Engineering #
|
| 27 |
+
|
| 28 |
+
# ## 1. Pclass ##
|
| 29 |
+
# there is no missing value on this feature and already a numerical value. so let's check it's impact on our train set.
|
| 30 |
+
|
| 31 |
+
# In[2]:
|
| 32 |
+
|
| 33 |
+
|
| 34 |
+
print (train[['Pclass', 'Survived']].groupby(['Pclass'], as_index=False).mean())
|
| 35 |
+
|
| 36 |
+
|
| 37 |
+
# ## 2. Sex ##
|
| 38 |
+
|
| 39 |
+
# In[3]:
|
| 40 |
+
|
| 41 |
+
|
| 42 |
+
print (train[["Sex", "Survived"]].groupby(['Sex'], as_index=False).mean())
|
| 43 |
+
|
| 44 |
+
|
| 45 |
+
# ## 3. SibSp and Parch ##
|
| 46 |
+
# With the number of siblings/spouse and the number of children/parents we can create new feature called Family Size.
|
| 47 |
+
|
| 48 |
+
# In[4]:
|
| 49 |
+
|
| 50 |
+
|
| 51 |
+
for dataset in full_data:
|
| 52 |
+
dataset['FamilySize'] = dataset['SibSp'] + dataset['Parch'] + 1
|
| 53 |
+
print (train[['FamilySize', 'Survived']].groupby(['FamilySize'], as_index=False).mean())
|
| 54 |
+
|
| 55 |
+
|
| 56 |
+
# it seems has a good effect on our prediction but let's go further and categorize people to check whether they are alone in this ship or not.
|
| 57 |
+
|
| 58 |
+
# In[5]:
|
| 59 |
+
|
| 60 |
+
|
| 61 |
+
for dataset in full_data:
|
| 62 |
+
dataset['IsAlone'] = 0
|
| 63 |
+
dataset.loc[dataset['FamilySize'] == 1, 'IsAlone'] = 1
|
| 64 |
+
print (train[['IsAlone', 'Survived']].groupby(['IsAlone'], as_index=False).mean())
|
| 65 |
+
|
| 66 |
+
|
| 67 |
+
# good! the impact is considerable.
|
| 68 |
+
|
| 69 |
+
# ## 4. Embarked ##
|
| 70 |
+
# the embarked feature has some missing value. and we try to fill those with the most occurred value ( 'S' ).
|
| 71 |
+
|
| 72 |
+
# In[6]:
|
| 73 |
+
|
| 74 |
+
|
| 75 |
+
for dataset in full_data:
|
| 76 |
+
dataset['Embarked'] = dataset['Embarked'].fillna('S')
|
| 77 |
+
print (train[['Embarked', 'Survived']].groupby(['Embarked'], as_index=False).mean())
|
| 78 |
+
|
| 79 |
+
|
| 80 |
+
# ## 5. Fare ##
|
| 81 |
+
# Fare also has some missing value and we will replace it with the median. then we categorize it into 4 ranges.
|
| 82 |
+
|
| 83 |
+
# In[7]:
|
| 84 |
+
|
| 85 |
+
|
| 86 |
+
for dataset in full_data:
|
| 87 |
+
dataset['Fare'] = dataset['Fare'].fillna(train['Fare'].median())
|
| 88 |
+
train['CategoricalFare'] = pd.qcut(train['Fare'], 4)
|
| 89 |
+
print (train[['CategoricalFare', 'Survived']].groupby(['CategoricalFare'], as_index=False).mean())
|
| 90 |
+
|
| 91 |
+
|
| 92 |
+
# ## 6. Age ##
|
| 93 |
+
# we have plenty of missing values in this feature. # generate random numbers between (mean - std) and (mean + std).
|
| 94 |
+
# then we categorize age into 5 range.
|
| 95 |
+
|
| 96 |
+
# In[8]:
|
| 97 |
+
|
| 98 |
+
|
| 99 |
+
for dataset in full_data:
|
| 100 |
+
age_avg = dataset['Age'].mean()
|
| 101 |
+
age_std = dataset['Age'].std()
|
| 102 |
+
age_null_count = dataset['Age'].isnull().sum()
|
| 103 |
+
|
| 104 |
+
age_null_random_list = np.random.randint(age_avg - age_std, age_avg + age_std, size=age_null_count)
|
| 105 |
+
dataset['Age'][np.isnan(dataset['Age'])] = age_null_random_list
|
| 106 |
+
dataset['Age'] = dataset['Age'].astype(int)
|
| 107 |
+
|
| 108 |
+
train['CategoricalAge'] = pd.cut(train['Age'], 5)
|
| 109 |
+
|
| 110 |
+
print (train[['CategoricalAge', 'Survived']].groupby(['CategoricalAge'], as_index=False).mean())
|
| 111 |
+
|
| 112 |
+
|
| 113 |
+
# ## 7. Name ##
|
| 114 |
+
# inside this feature we can find the title of people.
|
| 115 |
+
|
| 116 |
+
# In[9]:
|
| 117 |
+
|
| 118 |
+
|
| 119 |
+
def get_title(name):
|
| 120 |
+
title_search = re.search(' ([A-Za-z]+)\.', name)
|
| 121 |
+
# If the title exists, extract and return it.
|
| 122 |
+
if title_search:
|
| 123 |
+
return title_search.group(1)
|
| 124 |
+
return ""
|
| 125 |
+
|
| 126 |
+
for dataset in full_data:
|
| 127 |
+
dataset['Title'] = dataset['Name'].apply(get_title)
|
| 128 |
+
|
| 129 |
+
print(pd.crosstab(train['Title'], train['Sex']))
|
| 130 |
+
|
| 131 |
+
|
| 132 |
+
# so we have titles. let's categorize it and check the title impact on survival rate.
|
| 133 |
+
|
| 134 |
+
# In[10]:
|
| 135 |
+
|
| 136 |
+
|
| 137 |
+
for dataset in full_data:
|
| 138 |
+
dataset['Title'] = dataset['Title'].replace(['Lady', 'Countess','Capt', 'Col', 'Don', 'Dr', 'Major', 'Rev', 'Sir', 'Jonkheer', 'Dona'], 'Rare')
|
| 139 |
+
|
| 140 |
+
dataset['Title'] = dataset['Title'].replace('Mlle', 'Miss')
|
| 141 |
+
dataset['Title'] = dataset['Title'].replace('Ms', 'Miss')
|
| 142 |
+
dataset['Title'] = dataset['Title'].replace('Mme', 'Mrs')
|
| 143 |
+
|
| 144 |
+
print (train[['Title', 'Survived']].groupby(['Title'], as_index=False).mean())
|
| 145 |
+
|
| 146 |
+
|
| 147 |
+
# # Data Cleaning #
|
| 148 |
+
# great! now let's clean our data and map our features into numerical values.
|
| 149 |
+
|
| 150 |
+
# In[11]:
|
| 151 |
+
|
| 152 |
+
|
| 153 |
+
for dataset in full_data:
|
| 154 |
+
# Mapping Sex
|
| 155 |
+
dataset['Sex'] = dataset['Sex'].map( {'female': 0, 'male': 1} ).astype(int)
|
| 156 |
+
|
| 157 |
+
# Mapping titles
|
| 158 |
+
title_mapping = {"Mr": 1, "Miss": 2, "Mrs": 3, "Master": 4, "Rare": 5}
|
| 159 |
+
dataset['Title'] = dataset['Title'].map(title_mapping)
|
| 160 |
+
dataset['Title'] = dataset['Title'].fillna(0)
|
| 161 |
+
|
| 162 |
+
# Mapping Embarked
|
| 163 |
+
dataset['Embarked'] = dataset['Embarked'].map( {'S': 0, 'C': 1, 'Q': 2} ).astype(int)
|
| 164 |
+
|
| 165 |
+
# Mapping Fare
|
| 166 |
+
dataset.loc[ dataset['Fare'] <= 7.91, 'Fare'] = 0
|
| 167 |
+
dataset.loc[(dataset['Fare'] > 7.91) & (dataset['Fare'] <= 14.454), 'Fare'] = 1
|
| 168 |
+
dataset.loc[(dataset['Fare'] > 14.454) & (dataset['Fare'] <= 31), 'Fare'] = 2
|
| 169 |
+
dataset.loc[ dataset['Fare'] > 31, 'Fare'] = 3
|
| 170 |
+
dataset['Fare'] = dataset['Fare'].astype(int)
|
| 171 |
+
|
| 172 |
+
# Mapping Age
|
| 173 |
+
dataset.loc[ dataset['Age'] <= 16, 'Age'] = 0
|
| 174 |
+
dataset.loc[(dataset['Age'] > 16) & (dataset['Age'] <= 32), 'Age'] = 1
|
| 175 |
+
dataset.loc[(dataset['Age'] > 32) & (dataset['Age'] <= 48), 'Age'] = 2
|
| 176 |
+
dataset.loc[(dataset['Age'] > 48) & (dataset['Age'] <= 64), 'Age'] = 3
|
| 177 |
+
dataset.loc[ dataset['Age'] > 64, 'Age'] = 4
|
| 178 |
+
|
| 179 |
+
# Feature Selection
|
| 180 |
+
drop_elements = ['PassengerId', 'Name', 'Ticket', 'Cabin', 'SibSp', 'Parch', 'FamilySize']
|
| 181 |
+
train = train.drop(drop_elements, axis = 1)
|
| 182 |
+
train = train.drop(['CategoricalAge', 'CategoricalFare'], axis = 1)
|
| 183 |
+
|
| 184 |
+
test = test.drop(drop_elements, axis = 1)
|
| 185 |
+
|
| 186 |
+
print (train.head(10))
|
| 187 |
+
|
| 188 |
+
train = train.values
|
| 189 |
+
test = test.values
|
| 190 |
+
|
| 191 |
+
|
| 192 |
+
# good! now we have a clean dataset and ready to predict. let's find which classifier works better on this dataset.
|
| 193 |
+
|
| 194 |
+
# # Classifier Comparison #
|
| 195 |
+
|
| 196 |
+
# In[12]:
|
| 197 |
+
|
| 198 |
+
|
| 199 |
+
import matplotlib.pyplot as plt
|
| 200 |
+
import seaborn as sns
|
| 201 |
+
|
| 202 |
+
from sklearn.model_selection import StratifiedShuffleSplit
|
| 203 |
+
from sklearn.metrics import accuracy_score, log_loss
|
| 204 |
+
from sklearn.neighbors import KNeighborsClassifier
|
| 205 |
+
from sklearn.svm import SVC
|
| 206 |
+
from sklearn.tree import DecisionTreeClassifier
|
| 207 |
+
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, GradientBoostingClassifier
|
| 208 |
+
from sklearn.naive_bayes import GaussianNB
|
| 209 |
+
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis, QuadraticDiscriminantAnalysis
|
| 210 |
+
from sklearn.linear_model import LogisticRegression
|
| 211 |
+
|
| 212 |
+
classifiers = [
|
| 213 |
+
KNeighborsClassifier(3),
|
| 214 |
+
SVC(probability=True),
|
| 215 |
+
DecisionTreeClassifier(),
|
| 216 |
+
RandomForestClassifier(),
|
| 217 |
+
AdaBoostClassifier(),
|
| 218 |
+
GradientBoostingClassifier(),
|
| 219 |
+
GaussianNB(),
|
| 220 |
+
LinearDiscriminantAnalysis(),
|
| 221 |
+
QuadraticDiscriminantAnalysis(),
|
| 222 |
+
LogisticRegression()]
|
| 223 |
+
|
| 224 |
+
log_cols = ["Classifier", "Accuracy"]
|
| 225 |
+
log = pd.DataFrame(columns=log_cols)
|
| 226 |
+
|
| 227 |
+
sss = StratifiedShuffleSplit(n_splits=10, test_size=0.1, random_state=0)
|
| 228 |
+
|
| 229 |
+
X = train[0::, 1::]
|
| 230 |
+
y = train[0::, 0]
|
| 231 |
+
|
| 232 |
+
acc_dict = {}
|
| 233 |
+
|
| 234 |
+
for train_index, test_index in sss.split(X, y):
|
| 235 |
+
X_train, X_test = X[train_index], X[test_index]
|
| 236 |
+
y_train, y_test = y[train_index], y[test_index]
|
| 237 |
+
|
| 238 |
+
for clf in classifiers:
|
| 239 |
+
name = clf.__class__.__name__
|
| 240 |
+
clf.fit(X_train, y_train)
|
| 241 |
+
train_predictions = clf.predict(X_test)
|
| 242 |
+
acc = accuracy_score(y_test, train_predictions)
|
| 243 |
+
if name in acc_dict:
|
| 244 |
+
acc_dict[name] += acc
|
| 245 |
+
else:
|
| 246 |
+
acc_dict[name] = acc
|
| 247 |
+
|
| 248 |
+
for clf in acc_dict:
|
| 249 |
+
acc_dict[clf] = acc_dict[clf] / 10.0
|
| 250 |
+
log_entry = pd.DataFrame([[clf, acc_dict[clf]]], columns=log_cols)
|
| 251 |
+
log = log.append(log_entry)
|
| 252 |
+
|
| 253 |
+
plt.xlabel('Accuracy')
|
| 254 |
+
plt.title('Classifier Accuracy')
|
| 255 |
+
|
| 256 |
+
sns.set_color_codes("muted")
|
| 257 |
+
sns.barplot(x='Accuracy', y='Classifier', data=log, color="b")
|
| 258 |
+
|
| 259 |
+
|
| 260 |
+
# # Prediction #
|
| 261 |
+
# now we can use SVC classifier to predict our data.
|
| 262 |
+
|
| 263 |
+
# In[13]:
|
| 264 |
+
|
| 265 |
+
|
| 266 |
+
candidate_classifier = SVC()
|
| 267 |
+
candidate_classifier.fit(train[0::, 1::], train[0::, 0])
|
| 268 |
+
result = candidate_classifier.predict(test)
|
| 269 |
+
|
Titanic/Kernels/AdaBoost/7-titanic-survival-prediction-end-to-end-ml-pipeline.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/AdaBoost/7-titanic-survival-prediction-end-to-end-ml-pipeline.py
ADDED
|
@@ -0,0 +1,919 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python
|
| 2 |
+
# coding: utf-8
|
| 3 |
+
|
| 4 |
+
# ## Introduction
|
| 5 |
+
#
|
| 6 |
+
# **_Poonam Ligade_**
|
| 7 |
+
#
|
| 8 |
+
# *27th Dec 2016*
|
| 9 |
+
#
|
| 10 |
+
# I am are trying to find out how many people on titanic survived from disaster.
|
| 11 |
+
#
|
| 12 |
+
# Here goes Titanic Survival Prediction End to End ML Pipeline
|
| 13 |
+
#
|
| 14 |
+
# 1) **Introduction**
|
| 15 |
+
#
|
| 16 |
+
# 1. Import Libraries
|
| 17 |
+
# 2. Load data
|
| 18 |
+
# 3. Run Statistical summeries
|
| 19 |
+
# 4. Figure out missing value columns
|
| 20 |
+
#
|
| 21 |
+
#
|
| 22 |
+
#
|
| 23 |
+
# 2) **Visualizations**
|
| 24 |
+
#
|
| 25 |
+
# 1. Correlation with target variable
|
| 26 |
+
#
|
| 27 |
+
#
|
| 28 |
+
# 3) **Missing values imputation**
|
| 29 |
+
#
|
| 30 |
+
# 1. train data Missing columns- Embarked,Age,Cabin
|
| 31 |
+
# 2. test data Missing columns- Age and Fare
|
| 32 |
+
#
|
| 33 |
+
#
|
| 34 |
+
# 4) **Feature Engineering**
|
| 35 |
+
#
|
| 36 |
+
# 1. Calculate total family size
|
| 37 |
+
# 2. Get title from name
|
| 38 |
+
# 3. Find out which deck passenger belonged to
|
| 39 |
+
# 4. Dealing with Categorical Variables
|
| 40 |
+
# * Label encoding
|
| 41 |
+
# 5. Feature Scaling
|
| 42 |
+
#
|
| 43 |
+
#
|
| 44 |
+
# 5) **Prediction**
|
| 45 |
+
#
|
| 46 |
+
# 1. Split into training & test sets
|
| 47 |
+
# 2. Build the model
|
| 48 |
+
# 3. Feature importance
|
| 49 |
+
# 4. Predictions
|
| 50 |
+
# 5. Ensemble : Majority voting
|
| 51 |
+
#
|
| 52 |
+
# 6) **Submission**
|
| 53 |
+
|
| 54 |
+
# Import libraries
|
| 55 |
+
# ================
|
| 56 |
+
|
| 57 |
+
# In[1]:
|
| 58 |
+
|
| 59 |
+
|
| 60 |
+
# We can use the pandas library in python to read in the csv file.
|
| 61 |
+
import pandas as pd
|
| 62 |
+
#for numerical computaions we can use numpy library
|
| 63 |
+
import numpy as np
|
| 64 |
+
|
| 65 |
+
|
| 66 |
+
# Load train & test data
|
| 67 |
+
# ======================
|
| 68 |
+
|
| 69 |
+
# In[2]:
|
| 70 |
+
|
| 71 |
+
|
| 72 |
+
# This creates a pandas dataframe and assigns it to the titanic variable.
|
| 73 |
+
titanic = pd.read_csv("../input/train.csv")
|
| 74 |
+
# Print the first 5 rows of the dataframe.
|
| 75 |
+
titanic.head()
|
| 76 |
+
|
| 77 |
+
|
| 78 |
+
# In[3]:
|
| 79 |
+
|
| 80 |
+
|
| 81 |
+
titanic_test = pd.read_csv("../input/test.csv")
|
| 82 |
+
#transpose
|
| 83 |
+
titanic_test.head().T
|
| 84 |
+
#note their is no Survived column here which is our target varible we are trying to predict
|
| 85 |
+
|
| 86 |
+
|
| 87 |
+
# In[4]:
|
| 88 |
+
|
| 89 |
+
|
| 90 |
+
#shape command will give number of rows/samples/examples and number of columns/features/predictors in dataset
|
| 91 |
+
#(rows,columns)
|
| 92 |
+
titanic.shape
|
| 93 |
+
|
| 94 |
+
|
| 95 |
+
# In[5]:
|
| 96 |
+
|
| 97 |
+
|
| 98 |
+
#Describe gives statistical information about numerical columns in the dataset
|
| 99 |
+
titanic.describe()
|
| 100 |
+
#you can check from count if there are missing vales in columns, here age has got missing values
|
| 101 |
+
|
| 102 |
+
|
| 103 |
+
# In[6]:
|
| 104 |
+
|
| 105 |
+
|
| 106 |
+
#info method provides information about dataset like
|
| 107 |
+
#total values in each column, null/not null, datatype, memory occupied etc
|
| 108 |
+
titanic.info()
|
| 109 |
+
|
| 110 |
+
|
| 111 |
+
# In[7]:
|
| 112 |
+
|
| 113 |
+
|
| 114 |
+
#lets see if there are any more columns with missing values
|
| 115 |
+
null_columns=titanic.columns[titanic.isnull().any()]
|
| 116 |
+
titanic.isnull().sum()
|
| 117 |
+
|
| 118 |
+
|
| 119 |
+
# **yes even Embarked and cabin has missing values.**
|
| 120 |
+
|
| 121 |
+
# In[8]:
|
| 122 |
+
|
| 123 |
+
|
| 124 |
+
#how about test set??
|
| 125 |
+
titanic_test.isnull().sum()
|
| 126 |
+
|
| 127 |
+
|
| 128 |
+
# **Age, Fare and cabin has missing values.
|
| 129 |
+
# we will see how to fill missing values next.**
|
| 130 |
+
|
| 131 |
+
# In[9]:
|
| 132 |
+
|
| 133 |
+
|
| 134 |
+
get_ipython().run_line_magic('matplotlib', 'inline')
|
| 135 |
+
import matplotlib.pyplot as plt
|
| 136 |
+
import seaborn as sns
|
| 137 |
+
sns.set(font_scale=1)
|
| 138 |
+
|
| 139 |
+
pd.options.display.mpl_style = 'default'
|
| 140 |
+
labels = []
|
| 141 |
+
values = []
|
| 142 |
+
for col in null_columns:
|
| 143 |
+
labels.append(col)
|
| 144 |
+
values.append(titanic[col].isnull().sum())
|
| 145 |
+
ind = np.arange(len(labels))
|
| 146 |
+
width=0.6
|
| 147 |
+
fig, ax = plt.subplots(figsize=(6,5))
|
| 148 |
+
rects = ax.barh(ind, np.array(values), color='purple')
|
| 149 |
+
ax.set_yticks(ind+((width)/2.))
|
| 150 |
+
ax.set_yticklabels(labels, rotation='horizontal')
|
| 151 |
+
ax.set_xlabel("Count of missing values")
|
| 152 |
+
ax.set_ylabel("Column Names")
|
| 153 |
+
ax.set_title("Variables with missing values");
|
| 154 |
+
|
| 155 |
+
|
| 156 |
+
# **Visualizations**
|
| 157 |
+
# ==============
|
| 158 |
+
|
| 159 |
+
# In[10]:
|
| 160 |
+
|
| 161 |
+
|
| 162 |
+
titanic.hist(bins=10,figsize=(9,7),grid=False);
|
| 163 |
+
|
| 164 |
+
|
| 165 |
+
# **we can see that Age and Fare are measured on very different scaling. So we need to do feature scaling before predictions.**
|
| 166 |
+
|
| 167 |
+
# In[11]:
|
| 168 |
+
|
| 169 |
+
|
| 170 |
+
g = sns.FacetGrid(titanic, col="Sex", row="Survived", margin_titles=True)
|
| 171 |
+
g.map(plt.hist, "Age",color="purple");
|
| 172 |
+
|
| 173 |
+
|
| 174 |
+
# In[12]:
|
| 175 |
+
|
| 176 |
+
|
| 177 |
+
g = sns.FacetGrid(titanic, hue="Survived", col="Pclass", margin_titles=True,
|
| 178 |
+
palette={1:"seagreen", 0:"gray"})
|
| 179 |
+
g=g.map(plt.scatter, "Fare", "Age",edgecolor="w").add_legend();
|
| 180 |
+
|
| 181 |
+
|
| 182 |
+
# In[13]:
|
| 183 |
+
|
| 184 |
+
|
| 185 |
+
g = sns.FacetGrid(titanic, hue="Survived", col="Sex", margin_titles=True,
|
| 186 |
+
palette="Set1",hue_kws=dict(marker=["^", "v"]))
|
| 187 |
+
g.map(plt.scatter, "Fare", "Age",edgecolor="w").add_legend()
|
| 188 |
+
plt.subplots_adjust(top=0.8)
|
| 189 |
+
g.fig.suptitle('Survival by Gender , Age and Fare');
|
| 190 |
+
|
| 191 |
+
|
| 192 |
+
# In[14]:
|
| 193 |
+
|
| 194 |
+
|
| 195 |
+
titanic.Embarked.value_counts().plot(kind='bar', alpha=0.55)
|
| 196 |
+
plt.title("Passengers per boarding location");
|
| 197 |
+
|
| 198 |
+
|
| 199 |
+
# In[15]:
|
| 200 |
+
|
| 201 |
+
|
| 202 |
+
sns.factorplot(x = 'Embarked',y="Survived", data = titanic,color="r");
|
| 203 |
+
|
| 204 |
+
|
| 205 |
+
# In[16]:
|
| 206 |
+
|
| 207 |
+
|
| 208 |
+
sns.set(font_scale=1)
|
| 209 |
+
g = sns.factorplot(x="Sex", y="Survived", col="Pclass",
|
| 210 |
+
data=titanic, saturation=.5,
|
| 211 |
+
kind="bar", ci=None, aspect=.6)
|
| 212 |
+
(g.set_axis_labels("", "Survival Rate")
|
| 213 |
+
.set_xticklabels(["Men", "Women"])
|
| 214 |
+
.set_titles("{col_name} {col_var}")
|
| 215 |
+
.set(ylim=(0, 1))
|
| 216 |
+
.despine(left=True))
|
| 217 |
+
plt.subplots_adjust(top=0.8)
|
| 218 |
+
g.fig.suptitle('How many Men and Women Survived by Passenger Class');
|
| 219 |
+
|
| 220 |
+
|
| 221 |
+
# In[17]:
|
| 222 |
+
|
| 223 |
+
|
| 224 |
+
ax = sns.boxplot(x="Survived", y="Age",
|
| 225 |
+
data=titanic)
|
| 226 |
+
ax = sns.stripplot(x="Survived", y="Age",
|
| 227 |
+
data=titanic, jitter=True,
|
| 228 |
+
edgecolor="gray")
|
| 229 |
+
sns.plt.title("Survival by Age",fontsize=12);
|
| 230 |
+
|
| 231 |
+
|
| 232 |
+
# In[18]:
|
| 233 |
+
|
| 234 |
+
|
| 235 |
+
titanic.Age[titanic.Pclass == 1].plot(kind='kde')
|
| 236 |
+
titanic.Age[titanic.Pclass == 2].plot(kind='kde')
|
| 237 |
+
titanic.Age[titanic.Pclass == 3].plot(kind='kde')
|
| 238 |
+
# plots an axis lable
|
| 239 |
+
plt.xlabel("Age")
|
| 240 |
+
plt.title("Age Distribution within classes")
|
| 241 |
+
# sets our legend for our graph.
|
| 242 |
+
plt.legend(('1st Class', '2nd Class','3rd Class'),loc='best') ;
|
| 243 |
+
|
| 244 |
+
|
| 245 |
+
# In[19]:
|
| 246 |
+
|
| 247 |
+
|
| 248 |
+
corr=titanic.corr()#["Survived"]
|
| 249 |
+
plt.figure(figsize=(10, 10))
|
| 250 |
+
|
| 251 |
+
sns.heatmap(corr, vmax=.8, linewidths=0.01,
|
| 252 |
+
square=True,annot=True,cmap='YlGnBu',linecolor="white")
|
| 253 |
+
plt.title('Correlation between features');
|
| 254 |
+
|
| 255 |
+
|
| 256 |
+
# In[20]:
|
| 257 |
+
|
| 258 |
+
|
| 259 |
+
#correlation of features with target variable
|
| 260 |
+
titanic.corr()["Survived"]
|
| 261 |
+
|
| 262 |
+
|
| 263 |
+
# **Looks like Pclass has got highest negative correlation with "Survived" followed by Fare, Parch and Age**
|
| 264 |
+
|
| 265 |
+
# In[21]:
|
| 266 |
+
|
| 267 |
+
|
| 268 |
+
g = sns.factorplot(x="Age", y="Embarked",
|
| 269 |
+
hue="Sex", row="Pclass",
|
| 270 |
+
data=titanic[titanic.Embarked.notnull()],
|
| 271 |
+
orient="h", size=2, aspect=3.5,
|
| 272 |
+
palette={'male':"purple", 'female':"blue"},
|
| 273 |
+
kind="violin", split=True, cut=0, bw=.2);
|
| 274 |
+
|
| 275 |
+
|
| 276 |
+
# Missing Value Imputation
|
| 277 |
+
# ========================
|
| 278 |
+
#
|
| 279 |
+
# **Its important to fill missing values, because some machine learning algorithms can't accept them eg SVM.**
|
| 280 |
+
#
|
| 281 |
+
# *But filling missing values with mean/median/mode is also a prediction which may not be 100% accurate, instead you can use models like Decision Trees and Random Forest which handle missing values very well.*
|
| 282 |
+
|
| 283 |
+
# **Embarked Column**
|
| 284 |
+
|
| 285 |
+
# In[22]:
|
| 286 |
+
|
| 287 |
+
|
| 288 |
+
#Lets check which rows have null Embarked column
|
| 289 |
+
titanic[titanic['Embarked'].isnull()]
|
| 290 |
+
|
| 291 |
+
|
| 292 |
+
# **PassengerId 62 and 830** have missing embarked values
|
| 293 |
+
#
|
| 294 |
+
# Both have ***Passenger class 1*** and ***fare $80.***
|
| 295 |
+
#
|
| 296 |
+
# Lets plot a graph to visualize and try to guess from where they embarked
|
| 297 |
+
|
| 298 |
+
# In[23]:
|
| 299 |
+
|
| 300 |
+
|
| 301 |
+
sns.boxplot(x="Embarked", y="Fare", hue="Pclass", data=titanic);
|
| 302 |
+
|
| 303 |
+
|
| 304 |
+
# In[24]:
|
| 305 |
+
|
| 306 |
+
|
| 307 |
+
titanic["Embarked"] = titanic["Embarked"].fillna('C')
|
| 308 |
+
|
| 309 |
+
|
| 310 |
+
# We can see that for ***1st class*** median line is coming around ***fare $80*** for ***embarked*** value ***'C'***.
|
| 311 |
+
# So we can replace NA values in Embarked column with 'C'
|
| 312 |
+
|
| 313 |
+
# In[25]:
|
| 314 |
+
|
| 315 |
+
|
| 316 |
+
#there is an empty fare column in test set
|
| 317 |
+
titanic_test.describe()
|
| 318 |
+
|
| 319 |
+
|
| 320 |
+
# ***Fare Column***
|
| 321 |
+
|
| 322 |
+
# In[26]:
|
| 323 |
+
|
| 324 |
+
|
| 325 |
+
titanic_test[titanic_test['Fare'].isnull()]
|
| 326 |
+
|
| 327 |
+
|
| 328 |
+
# In[27]:
|
| 329 |
+
|
| 330 |
+
|
| 331 |
+
#we can replace missing value in fare by taking median of all fares of those passengers
|
| 332 |
+
#who share 3rd Passenger class and Embarked from 'S'
|
| 333 |
+
def fill_missing_fare(df):
|
| 334 |
+
median_fare=df[(df['Pclass'] == 3) & (df['Embarked'] == 'S')]['Fare'].median()
|
| 335 |
+
#'S'
|
| 336 |
+
#print(median_fare)
|
| 337 |
+
df["Fare"] = df["Fare"].fillna(median_fare)
|
| 338 |
+
return df
|
| 339 |
+
|
| 340 |
+
titanic_test=fill_missing_fare(titanic_test)
|
| 341 |
+
|
| 342 |
+
|
| 343 |
+
# Feature Engineering
|
| 344 |
+
# ===================
|
| 345 |
+
|
| 346 |
+
# ***Deck- Where exactly were passenger on the ship?***
|
| 347 |
+
|
| 348 |
+
# In[28]:
|
| 349 |
+
|
| 350 |
+
|
| 351 |
+
titanic["Deck"]=titanic.Cabin.str[0]
|
| 352 |
+
titanic_test["Deck"]=titanic_test.Cabin.str[0]
|
| 353 |
+
titanic["Deck"].unique() # 0 is for null values
|
| 354 |
+
|
| 355 |
+
|
| 356 |
+
# In[29]:
|
| 357 |
+
|
| 358 |
+
|
| 359 |
+
g = sns.factorplot("Survived", col="Deck", col_wrap=4,
|
| 360 |
+
data=titanic[titanic.Deck.notnull()],
|
| 361 |
+
kind="count", size=2.5, aspect=.8);
|
| 362 |
+
|
| 363 |
+
|
| 364 |
+
# In[30]:
|
| 365 |
+
|
| 366 |
+
|
| 367 |
+
titanic = titanic.assign(Deck=titanic.Deck.astype(object)).sort("Deck")
|
| 368 |
+
g = sns.FacetGrid(titanic, col="Pclass", sharex=False,
|
| 369 |
+
gridspec_kws={"width_ratios": [5, 3, 3]})
|
| 370 |
+
g.map(sns.boxplot, "Deck", "Age");
|
| 371 |
+
|
| 372 |
+
|
| 373 |
+
# In[31]:
|
| 374 |
+
|
| 375 |
+
|
| 376 |
+
titanic.Deck.fillna('Z', inplace=True)
|
| 377 |
+
titanic_test.Deck.fillna('Z', inplace=True)
|
| 378 |
+
titanic["Deck"].unique() # Z is for null values
|
| 379 |
+
|
| 380 |
+
|
| 381 |
+
# ***How Big is your family?***
|
| 382 |
+
|
| 383 |
+
# In[32]:
|
| 384 |
+
|
| 385 |
+
|
| 386 |
+
# Create a family size variable including the passenger themselves
|
| 387 |
+
titanic["FamilySize"] = titanic["SibSp"] + titanic["Parch"]+1
|
| 388 |
+
titanic_test["FamilySize"] = titanic_test["SibSp"] + titanic_test["Parch"]+1
|
| 389 |
+
print(titanic["FamilySize"].value_counts())
|
| 390 |
+
|
| 391 |
+
|
| 392 |
+
# In[33]:
|
| 393 |
+
|
| 394 |
+
|
| 395 |
+
# Discretize family size
|
| 396 |
+
titanic.loc[titanic["FamilySize"] == 1, "FsizeD"] = 'singleton'
|
| 397 |
+
titanic.loc[(titanic["FamilySize"] > 1) & (titanic["FamilySize"] < 5) , "FsizeD"] = 'small'
|
| 398 |
+
titanic.loc[titanic["FamilySize"] >4, "FsizeD"] = 'large'
|
| 399 |
+
|
| 400 |
+
titanic_test.loc[titanic_test["FamilySize"] == 1, "FsizeD"] = 'singleton'
|
| 401 |
+
titanic_test.loc[(titanic_test["FamilySize"] >1) & (titanic_test["FamilySize"] <5) , "FsizeD"] = 'small'
|
| 402 |
+
titanic_test.loc[titanic_test["FamilySize"] >4, "FsizeD"] = 'large'
|
| 403 |
+
print(titanic["FsizeD"].unique())
|
| 404 |
+
print(titanic["FsizeD"].value_counts())
|
| 405 |
+
|
| 406 |
+
|
| 407 |
+
# In[34]:
|
| 408 |
+
|
| 409 |
+
|
| 410 |
+
sns.factorplot(x="FsizeD", y="Survived", data=titanic);
|
| 411 |
+
|
| 412 |
+
|
| 413 |
+
# ***Do you have longer names?***
|
| 414 |
+
|
| 415 |
+
# In[35]:
|
| 416 |
+
|
| 417 |
+
|
| 418 |
+
#Create feture for length of name
|
| 419 |
+
# The .apply method generates a new series
|
| 420 |
+
titanic["NameLength"] = titanic["Name"].apply(lambda x: len(x))
|
| 421 |
+
|
| 422 |
+
titanic_test["NameLength"] = titanic_test["Name"].apply(lambda x: len(x))
|
| 423 |
+
#print(titanic["NameLength"].value_counts())
|
| 424 |
+
|
| 425 |
+
bins = [0, 20, 40, 57, 85]
|
| 426 |
+
group_names = ['short', 'okay', 'good', 'long']
|
| 427 |
+
titanic['NlengthD'] = pd.cut(titanic['NameLength'], bins, labels=group_names)
|
| 428 |
+
titanic_test['NlengthD'] = pd.cut(titanic_test['NameLength'], bins, labels=group_names)
|
| 429 |
+
|
| 430 |
+
sns.factorplot(x="NlengthD", y="Survived", data=titanic)
|
| 431 |
+
print(titanic["NlengthD"].unique())
|
| 432 |
+
|
| 433 |
+
|
| 434 |
+
# ***Whats in the name?***
|
| 435 |
+
|
| 436 |
+
# In[36]:
|
| 437 |
+
|
| 438 |
+
|
| 439 |
+
import re
|
| 440 |
+
|
| 441 |
+
#A function to get the title from a name.
|
| 442 |
+
def get_title(name):
|
| 443 |
+
# Use a regular expression to search for a title. Titles always consist of capital and lowercase letters, and end with a period.
|
| 444 |
+
title_search = re.search(' ([A-Za-z]+)\.', name)
|
| 445 |
+
#If the title exists, extract and return it.
|
| 446 |
+
if title_search:
|
| 447 |
+
return title_search.group(1)
|
| 448 |
+
return ""
|
| 449 |
+
|
| 450 |
+
#Get all the titles and print how often each one occurs.
|
| 451 |
+
titles = titanic["Name"].apply(get_title)
|
| 452 |
+
print(pd.value_counts(titles))
|
| 453 |
+
|
| 454 |
+
|
| 455 |
+
#Add in the title column.
|
| 456 |
+
titanic["Title"] = titles
|
| 457 |
+
|
| 458 |
+
# Titles with very low cell counts to be combined to "rare" level
|
| 459 |
+
rare_title = ['Dona', 'Lady', 'Countess','Capt', 'Col', 'Don',
|
| 460 |
+
'Dr', 'Major', 'Rev', 'Sir', 'Jonkheer']
|
| 461 |
+
|
| 462 |
+
# Also reassign mlle, ms, and mme accordingly
|
| 463 |
+
titanic.loc[titanic["Title"] == "Mlle", "Title"] = 'Miss'
|
| 464 |
+
titanic.loc[titanic["Title"] == "Ms", "Title"] = 'Miss'
|
| 465 |
+
titanic.loc[titanic["Title"] == "Mme", "Title"] = 'Mrs'
|
| 466 |
+
titanic.loc[titanic["Title"] == "Dona", "Title"] = 'Rare Title'
|
| 467 |
+
titanic.loc[titanic["Title"] == "Lady", "Title"] = 'Rare Title'
|
| 468 |
+
titanic.loc[titanic["Title"] == "Countess", "Title"] = 'Rare Title'
|
| 469 |
+
titanic.loc[titanic["Title"] == "Capt", "Title"] = 'Rare Title'
|
| 470 |
+
titanic.loc[titanic["Title"] == "Col", "Title"] = 'Rare Title'
|
| 471 |
+
titanic.loc[titanic["Title"] == "Don", "Title"] = 'Rare Title'
|
| 472 |
+
titanic.loc[titanic["Title"] == "Major", "Title"] = 'Rare Title'
|
| 473 |
+
titanic.loc[titanic["Title"] == "Rev", "Title"] = 'Rare Title'
|
| 474 |
+
titanic.loc[titanic["Title"] == "Sir", "Title"] = 'Rare Title'
|
| 475 |
+
titanic.loc[titanic["Title"] == "Jonkheer", "Title"] = 'Rare Title'
|
| 476 |
+
titanic.loc[titanic["Title"] == "Dr", "Title"] = 'Rare Title'
|
| 477 |
+
|
| 478 |
+
#titanic.loc[titanic["Title"].isin(['Dona', 'Lady', 'Countess','Capt', 'Col', 'Don',
|
| 479 |
+
# 'Dr', 'Major', 'Rev', 'Sir', 'Jonkheer']), "Title"] = 'Rare Title'
|
| 480 |
+
|
| 481 |
+
#titanic[titanic['Title'].isin(['Dona', 'Lady', 'Countess'])]
|
| 482 |
+
#titanic.query("Title in ('Dona', 'Lady', 'Countess')")
|
| 483 |
+
|
| 484 |
+
titanic["Title"].value_counts()
|
| 485 |
+
|
| 486 |
+
|
| 487 |
+
titles = titanic_test["Name"].apply(get_title)
|
| 488 |
+
print(pd.value_counts(titles))
|
| 489 |
+
|
| 490 |
+
#Add in the title column.
|
| 491 |
+
titanic_test["Title"] = titles
|
| 492 |
+
|
| 493 |
+
# Titles with very low cell counts to be combined to "rare" level
|
| 494 |
+
rare_title = ['Dona', 'Lady', 'Countess','Capt', 'Col', 'Don',
|
| 495 |
+
'Dr', 'Major', 'Rev', 'Sir', 'Jonkheer']
|
| 496 |
+
|
| 497 |
+
# Also reassign mlle, ms, and mme accordingly
|
| 498 |
+
titanic_test.loc[titanic_test["Title"] == "Mlle", "Title"] = 'Miss'
|
| 499 |
+
titanic_test.loc[titanic_test["Title"] == "Ms", "Title"] = 'Miss'
|
| 500 |
+
titanic_test.loc[titanic_test["Title"] == "Mme", "Title"] = 'Mrs'
|
| 501 |
+
titanic_test.loc[titanic_test["Title"] == "Dona", "Title"] = 'Rare Title'
|
| 502 |
+
titanic_test.loc[titanic_test["Title"] == "Lady", "Title"] = 'Rare Title'
|
| 503 |
+
titanic_test.loc[titanic_test["Title"] == "Countess", "Title"] = 'Rare Title'
|
| 504 |
+
titanic_test.loc[titanic_test["Title"] == "Capt", "Title"] = 'Rare Title'
|
| 505 |
+
titanic_test.loc[titanic_test["Title"] == "Col", "Title"] = 'Rare Title'
|
| 506 |
+
titanic_test.loc[titanic_test["Title"] == "Don", "Title"] = 'Rare Title'
|
| 507 |
+
titanic_test.loc[titanic_test["Title"] == "Major", "Title"] = 'Rare Title'
|
| 508 |
+
titanic_test.loc[titanic_test["Title"] == "Rev", "Title"] = 'Rare Title'
|
| 509 |
+
titanic_test.loc[titanic_test["Title"] == "Sir", "Title"] = 'Rare Title'
|
| 510 |
+
titanic_test.loc[titanic_test["Title"] == "Jonkheer", "Title"] = 'Rare Title'
|
| 511 |
+
titanic_test.loc[titanic_test["Title"] == "Dr", "Title"] = 'Rare Title'
|
| 512 |
+
|
| 513 |
+
titanic_test["Title"].value_counts()
|
| 514 |
+
|
| 515 |
+
|
| 516 |
+
# ***Ticket column***
|
| 517 |
+
|
| 518 |
+
# In[37]:
|
| 519 |
+
|
| 520 |
+
|
| 521 |
+
titanic["Ticket"].tail()
|
| 522 |
+
|
| 523 |
+
|
| 524 |
+
# In[38]:
|
| 525 |
+
|
| 526 |
+
|
| 527 |
+
titanic["TicketNumber"] = titanic["Ticket"].str.extract('(\d{2,})', expand=True)
|
| 528 |
+
titanic["TicketNumber"] = titanic["TicketNumber"].apply(pd.to_numeric)
|
| 529 |
+
|
| 530 |
+
|
| 531 |
+
titanic_test["TicketNumber"] = titanic_test["Ticket"].str.extract('(\d{2,})', expand=True)
|
| 532 |
+
titanic_test["TicketNumber"] = titanic_test["TicketNumber"].apply(pd.to_numeric)
|
| 533 |
+
|
| 534 |
+
|
| 535 |
+
# In[39]:
|
| 536 |
+
|
| 537 |
+
|
| 538 |
+
#some rows in ticket column dont have numeric value so we got NaN there
|
| 539 |
+
titanic[titanic["TicketNumber"].isnull()]
|
| 540 |
+
|
| 541 |
+
|
| 542 |
+
# In[40]:
|
| 543 |
+
|
| 544 |
+
|
| 545 |
+
titanic.TicketNumber.fillna(titanic["TicketNumber"].median(), inplace=True)
|
| 546 |
+
titanic_test.TicketNumber.fillna(titanic_test["TicketNumber"].median(), inplace=True)
|
| 547 |
+
|
| 548 |
+
|
| 549 |
+
# Convert Categorical variables into Numerical ones
|
| 550 |
+
# =================================================
|
| 551 |
+
|
| 552 |
+
# In[41]:
|
| 553 |
+
|
| 554 |
+
|
| 555 |
+
from sklearn.preprocessing import LabelEncoder,OneHotEncoder
|
| 556 |
+
|
| 557 |
+
labelEnc=LabelEncoder()
|
| 558 |
+
|
| 559 |
+
cat_vars=['Embarked','Sex',"Title","FsizeD","NlengthD",'Deck']
|
| 560 |
+
for col in cat_vars:
|
| 561 |
+
titanic[col]=labelEnc.fit_transform(titanic[col])
|
| 562 |
+
titanic_test[col]=labelEnc.fit_transform(titanic_test[col])
|
| 563 |
+
|
| 564 |
+
titanic.head()
|
| 565 |
+
|
| 566 |
+
|
| 567 |
+
# ***Age Column***
|
| 568 |
+
#
|
| 569 |
+
# Age seems to be promising feature.
|
| 570 |
+
# So it doesnt make sense to simply fill null values out with median/mean/mode.
|
| 571 |
+
#
|
| 572 |
+
# We will use ***Random Forest*** algorithm to predict ages.
|
| 573 |
+
|
| 574 |
+
# In[42]:
|
| 575 |
+
|
| 576 |
+
|
| 577 |
+
with sns.plotting_context("notebook",font_scale=1.5):
|
| 578 |
+
sns.set_style("whitegrid")
|
| 579 |
+
sns.distplot(titanic["Age"].dropna(),
|
| 580 |
+
bins=80,
|
| 581 |
+
kde=False,
|
| 582 |
+
color="red")
|
| 583 |
+
sns.plt.title("Age Distribution")
|
| 584 |
+
plt.ylabel("Count");
|
| 585 |
+
|
| 586 |
+
|
| 587 |
+
# In[43]:
|
| 588 |
+
|
| 589 |
+
|
| 590 |
+
from sklearn.ensemble import RandomForestRegressor
|
| 591 |
+
#predicting missing values in age using Random Forest
|
| 592 |
+
def fill_missing_age(df):
|
| 593 |
+
|
| 594 |
+
#Feature set
|
| 595 |
+
age_df = df[['Age','Embarked','Fare', 'Parch', 'SibSp',
|
| 596 |
+
'TicketNumber', 'Title','Pclass','FamilySize',
|
| 597 |
+
'FsizeD','NameLength',"NlengthD",'Deck']]
|
| 598 |
+
# Split sets into train and test
|
| 599 |
+
train = age_df.loc[ (df.Age.notnull()) ]# known Age values
|
| 600 |
+
test = age_df.loc[ (df.Age.isnull()) ]# null Ages
|
| 601 |
+
|
| 602 |
+
# All age values are stored in a target array
|
| 603 |
+
y = train.values[:, 0]
|
| 604 |
+
|
| 605 |
+
# All the other values are stored in the feature array
|
| 606 |
+
X = train.values[:, 1::]
|
| 607 |
+
|
| 608 |
+
# Create and fit a model
|
| 609 |
+
rtr = RandomForestRegressor(n_estimators=2000, n_jobs=-1)
|
| 610 |
+
rtr.fit(X, y)
|
| 611 |
+
|
| 612 |
+
# Use the fitted model to predict the missing values
|
| 613 |
+
predictedAges = rtr.predict(test.values[:, 1::])
|
| 614 |
+
|
| 615 |
+
# Assign those predictions to the full data set
|
| 616 |
+
df.loc[ (df.Age.isnull()), 'Age' ] = predictedAges
|
| 617 |
+
|
| 618 |
+
return df
|
| 619 |
+
|
| 620 |
+
|
| 621 |
+
# In[44]:
|
| 622 |
+
|
| 623 |
+
|
| 624 |
+
titanic=fill_missing_age(titanic)
|
| 625 |
+
titanic_test=fill_missing_age(titanic_test)
|
| 626 |
+
|
| 627 |
+
|
| 628 |
+
# In[45]:
|
| 629 |
+
|
| 630 |
+
|
| 631 |
+
with sns.plotting_context("notebook",font_scale=1.5):
|
| 632 |
+
sns.set_style("whitegrid")
|
| 633 |
+
sns.distplot(titanic["Age"].dropna(),
|
| 634 |
+
bins=80,
|
| 635 |
+
kde=False,
|
| 636 |
+
color="tomato")
|
| 637 |
+
sns.plt.title("Age Distribution")
|
| 638 |
+
plt.ylabel("Count")
|
| 639 |
+
plt.xlim((15,100));
|
| 640 |
+
|
| 641 |
+
|
| 642 |
+
# **Feature Scaling**
|
| 643 |
+
# ===============
|
| 644 |
+
#
|
| 645 |
+
# We can see that Age, Fare are measured on different scales, so we need to do Feature Scaling first before we proceed with predictions.
|
| 646 |
+
|
| 647 |
+
# In[46]:
|
| 648 |
+
|
| 649 |
+
|
| 650 |
+
from sklearn import preprocessing
|
| 651 |
+
|
| 652 |
+
std_scale = preprocessing.StandardScaler().fit(titanic[['Age', 'Fare']])
|
| 653 |
+
titanic[['Age', 'Fare']] = std_scale.transform(titanic[['Age', 'Fare']])
|
| 654 |
+
|
| 655 |
+
|
| 656 |
+
std_scale = preprocessing.StandardScaler().fit(titanic_test[['Age', 'Fare']])
|
| 657 |
+
titanic_test[['Age', 'Fare']] = std_scale.transform(titanic_test[['Age', 'Fare']])
|
| 658 |
+
|
| 659 |
+
|
| 660 |
+
# Correlation of features with target
|
| 661 |
+
# =======================
|
| 662 |
+
|
| 663 |
+
# In[47]:
|
| 664 |
+
|
| 665 |
+
|
| 666 |
+
titanic.corr()["Survived"]
|
| 667 |
+
|
| 668 |
+
|
| 669 |
+
# Predict Survival
|
| 670 |
+
# ================
|
| 671 |
+
|
| 672 |
+
# *Linear Regression*
|
| 673 |
+
# -------------------
|
| 674 |
+
|
| 675 |
+
# In[48]:
|
| 676 |
+
|
| 677 |
+
|
| 678 |
+
# Import the linear regression class
|
| 679 |
+
from sklearn.linear_model import LinearRegression
|
| 680 |
+
# Sklearn also has a helper that makes it easy to do cross validation
|
| 681 |
+
from sklearn.cross_validation import KFold
|
| 682 |
+
|
| 683 |
+
# The columns we'll use to predict the target
|
| 684 |
+
predictors = ["Pclass", "Sex", "Age","SibSp", "Parch", "Fare",
|
| 685 |
+
"Embarked","NlengthD", "FsizeD", "Title","Deck"]
|
| 686 |
+
target="Survived"
|
| 687 |
+
# Initialize our algorithm class
|
| 688 |
+
alg = LinearRegression()
|
| 689 |
+
|
| 690 |
+
# Generate cross validation folds for the titanic dataset. It return the row indices corresponding to train and test.
|
| 691 |
+
# We set random_state to ensure we get the same splits every time we run this.
|
| 692 |
+
kf = KFold(titanic.shape[0], n_folds=3, random_state=1)
|
| 693 |
+
|
| 694 |
+
predictions = []
|
| 695 |
+
|
| 696 |
+
|
| 697 |
+
# In[49]:
|
| 698 |
+
|
| 699 |
+
|
| 700 |
+
for train, test in kf:
|
| 701 |
+
# The predictors we're using the train the algorithm. Note how we only take the rows in the train folds.
|
| 702 |
+
train_predictors = (titanic[predictors].iloc[train,:])
|
| 703 |
+
# The target we're using to train the algorithm.
|
| 704 |
+
train_target = titanic[target].iloc[train]
|
| 705 |
+
# Training the algorithm using the predictors and target.
|
| 706 |
+
alg.fit(train_predictors, train_target)
|
| 707 |
+
# We can now make predictions on the test fold
|
| 708 |
+
test_predictions = alg.predict(titanic[predictors].iloc[test,:])
|
| 709 |
+
predictions.append(test_predictions)
|
| 710 |
+
|
| 711 |
+
|
| 712 |
+
# In[50]:
|
| 713 |
+
|
| 714 |
+
|
| 715 |
+
predictions = np.concatenate(predictions, axis=0)
|
| 716 |
+
# Map predictions to outcomes (only possible outcomes are 1 and 0)
|
| 717 |
+
predictions[predictions > .5] = 1
|
| 718 |
+
predictions[predictions <=.5] = 0
|
| 719 |
+
|
| 720 |
+
|
| 721 |
+
accuracy=sum(titanic["Survived"]==predictions)/len(titanic["Survived"])
|
| 722 |
+
accuracy
|
| 723 |
+
|
| 724 |
+
|
| 725 |
+
# *Logistic Regression*
|
| 726 |
+
# -------------------
|
| 727 |
+
|
| 728 |
+
# In[51]:
|
| 729 |
+
|
| 730 |
+
|
| 731 |
+
from sklearn import cross_validation
|
| 732 |
+
from sklearn.linear_model import LogisticRegression
|
| 733 |
+
from sklearn.model_selection import cross_val_score
|
| 734 |
+
from sklearn.model_selection import ShuffleSplit
|
| 735 |
+
|
| 736 |
+
predictors = ["Pclass", "Sex", "Fare", "Embarked","Deck","Age",
|
| 737 |
+
"FsizeD", "NlengthD","Title","Parch"]
|
| 738 |
+
|
| 739 |
+
# Initialize our algorithm
|
| 740 |
+
lr = LogisticRegression(random_state=1)
|
| 741 |
+
# Compute the accuracy score for all the cross validation folds.
|
| 742 |
+
cv = ShuffleSplit(n_splits=10, test_size=0.3, random_state=50)
|
| 743 |
+
|
| 744 |
+
scores = cross_val_score(lr, titanic[predictors],
|
| 745 |
+
titanic["Survived"],scoring='f1', cv=cv)
|
| 746 |
+
# Take the mean of the scores (because we have one for each fold)
|
| 747 |
+
print(scores.mean())
|
| 748 |
+
|
| 749 |
+
|
| 750 |
+
# *Random Forest *
|
| 751 |
+
# -------------------
|
| 752 |
+
|
| 753 |
+
# In[52]:
|
| 754 |
+
|
| 755 |
+
|
| 756 |
+
from sklearn import cross_validation
|
| 757 |
+
from sklearn.ensemble import RandomForestClassifier
|
| 758 |
+
from sklearn.cross_validation import KFold
|
| 759 |
+
from sklearn.model_selection import cross_val_predict
|
| 760 |
+
|
| 761 |
+
import numpy as np
|
| 762 |
+
predictors = ["Pclass", "Sex", "Age",
|
| 763 |
+
"Fare","NlengthD","NameLength", "FsizeD", "Title","Deck"]
|
| 764 |
+
|
| 765 |
+
# Initialize our algorithm with the default paramters
|
| 766 |
+
# n_estimators is the number of trees we want to make
|
| 767 |
+
# min_samples_split is the minimum number of rows we need to make a split
|
| 768 |
+
# min_samples_leaf is the minimum number of samples we can have at the place where a tree branch ends (the bottom points of the tree)
|
| 769 |
+
rf = RandomForestClassifier(random_state=1, n_estimators=10, min_samples_split=2,
|
| 770 |
+
min_samples_leaf=1)
|
| 771 |
+
kf = KFold(titanic.shape[0], n_folds=5, random_state=1)
|
| 772 |
+
cv = ShuffleSplit(n_splits=10, test_size=0.3, random_state=50)
|
| 773 |
+
|
| 774 |
+
predictions = cross_validation.cross_val_predict(rf, titanic[predictors],titanic["Survived"],cv=kf)
|
| 775 |
+
predictions = pd.Series(predictions)
|
| 776 |
+
scores = cross_val_score(rf, titanic[predictors], titanic["Survived"],
|
| 777 |
+
scoring='f1', cv=kf)
|
| 778 |
+
# Take the mean of the scores (because we have one for each fold)
|
| 779 |
+
print(scores.mean())
|
| 780 |
+
|
| 781 |
+
|
| 782 |
+
# In[53]:
|
| 783 |
+
|
| 784 |
+
|
| 785 |
+
predictors = ["Pclass", "Sex", "Age",
|
| 786 |
+
"Fare","NlengthD","NameLength", "FsizeD", "Title","Deck","TicketNumber"]
|
| 787 |
+
rf = RandomForestClassifier(random_state=1, n_estimators=50, max_depth=9,min_samples_split=6, min_samples_leaf=4)
|
| 788 |
+
rf.fit(titanic[predictors],titanic["Survived"])
|
| 789 |
+
kf = KFold(titanic.shape[0], n_folds=5, random_state=1)
|
| 790 |
+
predictions = cross_validation.cross_val_predict(rf, titanic[predictors],titanic["Survived"],cv=kf)
|
| 791 |
+
predictions = pd.Series(predictions)
|
| 792 |
+
scores = cross_val_score(rf, titanic[predictors], titanic["Survived"],scoring='f1', cv=kf)
|
| 793 |
+
# Take the mean of the scores (because we have one for each fold)
|
| 794 |
+
print(scores.mean())
|
| 795 |
+
|
| 796 |
+
|
| 797 |
+
# Important features
|
| 798 |
+
# ==================
|
| 799 |
+
|
| 800 |
+
# In[54]:
|
| 801 |
+
|
| 802 |
+
|
| 803 |
+
importances=rf.feature_importances_
|
| 804 |
+
std = np.std([rf.feature_importances_ for tree in rf.estimators_],
|
| 805 |
+
axis=0)
|
| 806 |
+
indices = np.argsort(importances)[::-1]
|
| 807 |
+
sorted_important_features=[]
|
| 808 |
+
for i in indices:
|
| 809 |
+
sorted_important_features.append(predictors[i])
|
| 810 |
+
#predictors=titanic.columns
|
| 811 |
+
plt.figure()
|
| 812 |
+
plt.title("Feature Importances By Random Forest Model")
|
| 813 |
+
plt.bar(range(np.size(predictors)), importances[indices],
|
| 814 |
+
color="r", yerr=std[indices], align="center")
|
| 815 |
+
plt.xticks(range(np.size(predictors)), sorted_important_features, rotation='vertical')
|
| 816 |
+
|
| 817 |
+
plt.xlim([-1, np.size(predictors)]);
|
| 818 |
+
|
| 819 |
+
|
| 820 |
+
# *Gradient Boosting*
|
| 821 |
+
# -------------------
|
| 822 |
+
|
| 823 |
+
# In[55]:
|
| 824 |
+
|
| 825 |
+
|
| 826 |
+
import numpy as np
|
| 827 |
+
from sklearn.ensemble import GradientBoostingClassifier
|
| 828 |
+
|
| 829 |
+
from sklearn.feature_selection import SelectKBest, f_classif
|
| 830 |
+
from sklearn.cross_validation import KFold
|
| 831 |
+
get_ipython().run_line_magic('matplotlib', 'inline')
|
| 832 |
+
import matplotlib.pyplot as plt
|
| 833 |
+
#predictors = ["Pclass", "Sex", "Age", "Fare",
|
| 834 |
+
# "FsizeD", "Embarked", "NlengthD","Deck","TicketNumber"]
|
| 835 |
+
predictors = ["Pclass", "Sex", "Age",
|
| 836 |
+
"Fare","NlengthD", "FsizeD","NameLength","Deck","Embarked"]
|
| 837 |
+
# Perform feature selection
|
| 838 |
+
selector = SelectKBest(f_classif, k=5)
|
| 839 |
+
selector.fit(titanic[predictors], titanic["Survived"])
|
| 840 |
+
|
| 841 |
+
# Get the raw p-values for each feature, and transform from p-values into scores
|
| 842 |
+
scores = -np.log10(selector.pvalues_)
|
| 843 |
+
|
| 844 |
+
indices = np.argsort(scores)[::-1]
|
| 845 |
+
|
| 846 |
+
sorted_important_features=[]
|
| 847 |
+
for i in indices:
|
| 848 |
+
sorted_important_features.append(predictors[i])
|
| 849 |
+
|
| 850 |
+
plt.figure()
|
| 851 |
+
plt.title("Feature Importances By SelectKBest")
|
| 852 |
+
plt.bar(range(np.size(predictors)), scores[indices],
|
| 853 |
+
color="seagreen", yerr=std[indices], align="center")
|
| 854 |
+
plt.xticks(range(np.size(predictors)), sorted_important_features, rotation='vertical')
|
| 855 |
+
|
| 856 |
+
plt.xlim([-1, np.size(predictors)]);
|
| 857 |
+
|
| 858 |
+
|
| 859 |
+
# In[56]:
|
| 860 |
+
|
| 861 |
+
|
| 862 |
+
from sklearn import cross_validation
|
| 863 |
+
from sklearn.linear_model import LogisticRegression
|
| 864 |
+
predictors = ["Pclass", "Sex", "Age", "Fare", "Embarked","NlengthD",
|
| 865 |
+
"FsizeD", "Title","Deck"]
|
| 866 |
+
|
| 867 |
+
# Initialize our algorithm
|
| 868 |
+
lr = LogisticRegression(random_state=1)
|
| 869 |
+
# Compute the accuracy score for all the cross validation folds.
|
| 870 |
+
cv = ShuffleSplit(n_splits=10, test_size=0.3, random_state=50)
|
| 871 |
+
scores = cross_val_score(lr, titanic[predictors], titanic["Survived"], scoring='f1',cv=cv)
|
| 872 |
+
print(scores.mean())
|
| 873 |
+
|
| 874 |
+
|
| 875 |
+
# *AdaBoost *
|
| 876 |
+
# --------------------
|
| 877 |
+
|
| 878 |
+
# In[57]:
|
| 879 |
+
|
| 880 |
+
|
| 881 |
+
from sklearn.ensemble import AdaBoostClassifier
|
| 882 |
+
predictors = ["Pclass", "Sex", "Age", "Fare", "Embarked","NlengthD",
|
| 883 |
+
"FsizeD", "Title","Deck","TicketNumber"]
|
| 884 |
+
adb=AdaBoostClassifier()
|
| 885 |
+
adb.fit(titanic[predictors],titanic["Survived"])
|
| 886 |
+
cv = ShuffleSplit(n_splits=10, test_size=0.3, random_state=50)
|
| 887 |
+
scores = cross_val_score(adb, titanic[predictors], titanic["Survived"], scoring='f1',cv=cv)
|
| 888 |
+
print(scores.mean())
|
| 889 |
+
|
| 890 |
+
|
| 891 |
+
# Maximum Voting ensemble and Submission
|
| 892 |
+
# =======
|
| 893 |
+
|
| 894 |
+
# In[58]:
|
| 895 |
+
|
| 896 |
+
|
| 897 |
+
predictions=["Pclass", "Sex", "Age", "Fare", "Embarked","NlengthD",
|
| 898 |
+
"FsizeD", "Title","Deck","NameLength","TicketNumber"]
|
| 899 |
+
from sklearn.ensemble import VotingClassifier
|
| 900 |
+
eclf1 = VotingClassifier(estimators=[
|
| 901 |
+
('lr', lr), ('rf', rf), ('adb', adb)], voting='soft')
|
| 902 |
+
eclf1 = eclf1.fit(titanic[predictors], titanic["Survived"])
|
| 903 |
+
predictions=eclf1.predict(titanic[predictors])
|
| 904 |
+
predictions
|
| 905 |
+
|
| 906 |
+
test_predictions=eclf1.predict(titanic_test[predictors])
|
| 907 |
+
|
| 908 |
+
test_predictions=test_predictions.astype(int)
|
| 909 |
+
submission = pd.DataFrame({
|
| 910 |
+
"PassengerId": titanic_test["PassengerId"],
|
| 911 |
+
"Survived": test_predictions
|
| 912 |
+
})
|
| 913 |
+
|
| 914 |
+
submission.to_csv("titanic_submission.csv", index=False)
|
| 915 |
+
|
| 916 |
+
|
| 917 |
+
# ***To do: stacking!. Watch this space…***
|
| 918 |
+
|
| 919 |
+
# ***Hope you find it useful. :)please upvote***
|
Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/0-introduction-to-ensembling-stacking-in-python-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/10-ensemble-learning-techniques-tutorial-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/11-titanic-a-step-by-step-intro-to-machine-learning-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/2-titanic-top-4-with-ensemble-modeling-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/3-a-statistical-analysis-ml-workflow-of-titanic-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/4-applied-machine-learning-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/5-titanic-the-only-notebook-you-need-to-see-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/6-titanic-top-solution-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/7-titanic-eda-model-pipeline-keras-nn-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/8-a-comprehensive-guide-to-titanic-machine-learning-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/.ipynb_checkpoints/9-top-3-efficient-ensembling-in-few-lines-of-code-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/0-introduction-to-ensembling-stacking-in-python.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/0-introduction-to-ensembling-stacking-in-python.py
ADDED
|
@@ -0,0 +1,779 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python
|
| 2 |
+
# coding: utf-8
|
| 3 |
+
|
| 4 |
+
# # Introduction
|
| 5 |
+
#
|
| 6 |
+
# This notebook is a very basic and simple introductory primer to the method of ensembling (combining) base learning models, in particular the variant of ensembling known as Stacking. In a nutshell stacking uses as a first-level (base), the predictions of a few basic classifiers and then uses another model at the second-level to predict the output from the earlier first-level predictions.
|
| 7 |
+
#
|
| 8 |
+
# The Titanic dataset is a prime candidate for introducing this concept as many newcomers to Kaggle start out here. Furthermore even though stacking has been responsible for many a team winning Kaggle competitions there seems to be a dearth of kernels on this topic so I hope this notebook can fill somewhat of that void.
|
| 9 |
+
#
|
| 10 |
+
# I myself am quite a newcomer to the Kaggle scene as well and the first proper ensembling/stacking script that I managed to chance upon and study was one written in the AllState Severity Claims competition by the great Faron. The material in this notebook borrows heavily from Faron's script although ported to factor in ensembles of classifiers whilst his was ensembles of regressors. Anyway please check out his script here:
|
| 11 |
+
#
|
| 12 |
+
# [Stacking Starter][1] : by Faron
|
| 13 |
+
#
|
| 14 |
+
#
|
| 15 |
+
# Now onto the notebook at hand and I hope that it manages to do justice and convey the concept of ensembling in an intuitive and concise manner. My other standalone Kaggle [script][2] which implements exactly the same ensembling steps (albeit with different parameters) discussed below gives a Public LB score of 0.808 which is good enough to get to the top 9% and runs just under 4 minutes. Therefore I am pretty sure there is a lot of room to improve and add on to that script. Anyways please feel free to leave me any comments with regards to how I can improve
|
| 16 |
+
#
|
| 17 |
+
#
|
| 18 |
+
# [1]: https://www.kaggle.com/mmueller/allstate-claims-severity/stacking-starter/run/390867
|
| 19 |
+
# [2]: https://www.kaggle.com/arthurtok/titanic/simple-stacking-with-xgboost-0-808
|
| 20 |
+
|
| 21 |
+
# In[1]:
|
| 22 |
+
|
| 23 |
+
|
| 24 |
+
# Load in our libraries
|
| 25 |
+
import pandas as pd
|
| 26 |
+
import numpy as np
|
| 27 |
+
import re
|
| 28 |
+
import sklearn
|
| 29 |
+
import xgboost as xgb
|
| 30 |
+
import seaborn as sns
|
| 31 |
+
import matplotlib.pyplot as plt
|
| 32 |
+
get_ipython().run_line_magic('matplotlib', 'inline')
|
| 33 |
+
|
| 34 |
+
import plotly.offline as py
|
| 35 |
+
py.init_notebook_mode(connected=True)
|
| 36 |
+
import plotly.graph_objs as go
|
| 37 |
+
import plotly.tools as tls
|
| 38 |
+
|
| 39 |
+
import warnings
|
| 40 |
+
warnings.filterwarnings('ignore')
|
| 41 |
+
|
| 42 |
+
# Going to use these 5 base models for the stacking
|
| 43 |
+
from sklearn.ensemble import (RandomForestClassifier, AdaBoostClassifier,
|
| 44 |
+
GradientBoostingClassifier, ExtraTreesClassifier)
|
| 45 |
+
from sklearn.svm import SVC
|
| 46 |
+
from sklearn.cross_validation import KFold
|
| 47 |
+
|
| 48 |
+
|
| 49 |
+
# # Feature Exploration, Engineering and Cleaning
|
| 50 |
+
#
|
| 51 |
+
# Now we will proceed much like how most kernels in general are structured, and that is to first explore the data on hand, identify possible feature engineering opportunities as well as numerically encode any categorical features.
|
| 52 |
+
|
| 53 |
+
# In[2]:
|
| 54 |
+
|
| 55 |
+
|
| 56 |
+
# Load in the train and test datasets
|
| 57 |
+
train = pd.read_csv('../input/train.csv')
|
| 58 |
+
test = pd.read_csv('../input/test.csv')
|
| 59 |
+
|
| 60 |
+
# Store our passenger ID for easy access
|
| 61 |
+
PassengerId = test['PassengerId']
|
| 62 |
+
|
| 63 |
+
train.head(3)
|
| 64 |
+
|
| 65 |
+
|
| 66 |
+
# Well it is no surprise that our task is to somehow extract the information out of the categorical variables
|
| 67 |
+
#
|
| 68 |
+
# **Feature Engineering**
|
| 69 |
+
#
|
| 70 |
+
# Here, credit must be extended to Sina's very comprehensive and well-thought out notebook for the feature engineering ideas so please check out his work
|
| 71 |
+
#
|
| 72 |
+
# [Titanic Best Working Classfier][1] : by Sina
|
| 73 |
+
#
|
| 74 |
+
#
|
| 75 |
+
# [1]: https://www.kaggle.com/sinakhorami/titanic/titanic-best-working-classifier
|
| 76 |
+
|
| 77 |
+
# In[3]:
|
| 78 |
+
|
| 79 |
+
|
| 80 |
+
full_data = [train, test]
|
| 81 |
+
|
| 82 |
+
# Some features of my own that I have added in
|
| 83 |
+
# Gives the length of the name
|
| 84 |
+
train['Name_length'] = train['Name'].apply(len)
|
| 85 |
+
test['Name_length'] = test['Name'].apply(len)
|
| 86 |
+
# Feature that tells whether a passenger had a cabin on the Titanic
|
| 87 |
+
train['Has_Cabin'] = train["Cabin"].apply(lambda x: 0 if type(x) == float else 1)
|
| 88 |
+
test['Has_Cabin'] = test["Cabin"].apply(lambda x: 0 if type(x) == float else 1)
|
| 89 |
+
|
| 90 |
+
# Feature engineering steps taken from Sina
|
| 91 |
+
# Create new feature FamilySize as a combination of SibSp and Parch
|
| 92 |
+
for dataset in full_data:
|
| 93 |
+
dataset['FamilySize'] = dataset['SibSp'] + dataset['Parch'] + 1
|
| 94 |
+
# Create new feature IsAlone from FamilySize
|
| 95 |
+
for dataset in full_data:
|
| 96 |
+
dataset['IsAlone'] = 0
|
| 97 |
+
dataset.loc[dataset['FamilySize'] == 1, 'IsAlone'] = 1
|
| 98 |
+
# Remove all NULLS in the Embarked column
|
| 99 |
+
for dataset in full_data:
|
| 100 |
+
dataset['Embarked'] = dataset['Embarked'].fillna('S')
|
| 101 |
+
# Remove all NULLS in the Fare column and create a new feature CategoricalFare
|
| 102 |
+
for dataset in full_data:
|
| 103 |
+
dataset['Fare'] = dataset['Fare'].fillna(train['Fare'].median())
|
| 104 |
+
train['CategoricalFare'] = pd.qcut(train['Fare'], 4)
|
| 105 |
+
# Create a New feature CategoricalAge
|
| 106 |
+
for dataset in full_data:
|
| 107 |
+
age_avg = dataset['Age'].mean()
|
| 108 |
+
age_std = dataset['Age'].std()
|
| 109 |
+
age_null_count = dataset['Age'].isnull().sum()
|
| 110 |
+
age_null_random_list = np.random.randint(age_avg - age_std, age_avg + age_std, size=age_null_count)
|
| 111 |
+
dataset['Age'][np.isnan(dataset['Age'])] = age_null_random_list
|
| 112 |
+
dataset['Age'] = dataset['Age'].astype(int)
|
| 113 |
+
train['CategoricalAge'] = pd.cut(train['Age'], 5)
|
| 114 |
+
# Define function to extract titles from passenger names
|
| 115 |
+
def get_title(name):
|
| 116 |
+
title_search = re.search(' ([A-Za-z]+)\.', name)
|
| 117 |
+
# If the title exists, extract and return it.
|
| 118 |
+
if title_search:
|
| 119 |
+
return title_search.group(1)
|
| 120 |
+
return ""
|
| 121 |
+
# Create a new feature Title, containing the titles of passenger names
|
| 122 |
+
for dataset in full_data:
|
| 123 |
+
dataset['Title'] = dataset['Name'].apply(get_title)
|
| 124 |
+
# Group all non-common titles into one single grouping "Rare"
|
| 125 |
+
for dataset in full_data:
|
| 126 |
+
dataset['Title'] = dataset['Title'].replace(['Lady', 'Countess','Capt', 'Col','Don', 'Dr', 'Major', 'Rev', 'Sir', 'Jonkheer', 'Dona'], 'Rare')
|
| 127 |
+
|
| 128 |
+
dataset['Title'] = dataset['Title'].replace('Mlle', 'Miss')
|
| 129 |
+
dataset['Title'] = dataset['Title'].replace('Ms', 'Miss')
|
| 130 |
+
dataset['Title'] = dataset['Title'].replace('Mme', 'Mrs')
|
| 131 |
+
|
| 132 |
+
for dataset in full_data:
|
| 133 |
+
# Mapping Sex
|
| 134 |
+
dataset['Sex'] = dataset['Sex'].map( {'female': 0, 'male': 1} ).astype(int)
|
| 135 |
+
|
| 136 |
+
# Mapping titles
|
| 137 |
+
title_mapping = {"Mr": 1, "Miss": 2, "Mrs": 3, "Master": 4, "Rare": 5}
|
| 138 |
+
dataset['Title'] = dataset['Title'].map(title_mapping)
|
| 139 |
+
dataset['Title'] = dataset['Title'].fillna(0)
|
| 140 |
+
|
| 141 |
+
# Mapping Embarked
|
| 142 |
+
dataset['Embarked'] = dataset['Embarked'].map( {'S': 0, 'C': 1, 'Q': 2} ).astype(int)
|
| 143 |
+
|
| 144 |
+
# Mapping Fare
|
| 145 |
+
dataset.loc[ dataset['Fare'] <= 7.91, 'Fare'] = 0
|
| 146 |
+
dataset.loc[(dataset['Fare'] > 7.91) & (dataset['Fare'] <= 14.454), 'Fare'] = 1
|
| 147 |
+
dataset.loc[(dataset['Fare'] > 14.454) & (dataset['Fare'] <= 31), 'Fare'] = 2
|
| 148 |
+
dataset.loc[ dataset['Fare'] > 31, 'Fare'] = 3
|
| 149 |
+
dataset['Fare'] = dataset['Fare'].astype(int)
|
| 150 |
+
|
| 151 |
+
# Mapping Age
|
| 152 |
+
dataset.loc[ dataset['Age'] <= 16, 'Age'] = 0
|
| 153 |
+
dataset.loc[(dataset['Age'] > 16) & (dataset['Age'] <= 32), 'Age'] = 1
|
| 154 |
+
dataset.loc[(dataset['Age'] > 32) & (dataset['Age'] <= 48), 'Age'] = 2
|
| 155 |
+
dataset.loc[(dataset['Age'] > 48) & (dataset['Age'] <= 64), 'Age'] = 3
|
| 156 |
+
dataset.loc[ dataset['Age'] > 64, 'Age'] = 4 ;
|
| 157 |
+
|
| 158 |
+
|
| 159 |
+
# In[4]:
|
| 160 |
+
|
| 161 |
+
|
| 162 |
+
# Feature selection
|
| 163 |
+
drop_elements = ['PassengerId', 'Name', 'Ticket', 'Cabin', 'SibSp']
|
| 164 |
+
train = train.drop(drop_elements, axis = 1)
|
| 165 |
+
train = train.drop(['CategoricalAge', 'CategoricalFare'], axis = 1)
|
| 166 |
+
test = test.drop(drop_elements, axis = 1)
|
| 167 |
+
|
| 168 |
+
|
| 169 |
+
# All right so now having cleaned the features and extracted relevant information and dropped the categorical columns our features should now all be numeric, a format suitable to feed into our Machine Learning models. However before we proceed let us generate some simple correlation and distribution plots of our transformed dataset to observe ho
|
| 170 |
+
#
|
| 171 |
+
# ## Visualisations
|
| 172 |
+
|
| 173 |
+
# In[5]:
|
| 174 |
+
|
| 175 |
+
|
| 176 |
+
train.head(3)
|
| 177 |
+
|
| 178 |
+
|
| 179 |
+
# **Pearson Correlation Heatmap**
|
| 180 |
+
#
|
| 181 |
+
# let us generate some correlation plots of the features to see how related one feature is to the next. To do so, we will utilise the Seaborn plotting package which allows us to plot heatmaps very conveniently as follows
|
| 182 |
+
|
| 183 |
+
# In[6]:
|
| 184 |
+
|
| 185 |
+
|
| 186 |
+
colormap = plt.cm.RdBu
|
| 187 |
+
plt.figure(figsize=(14,12))
|
| 188 |
+
plt.title('Pearson Correlation of Features', y=1.05, size=15)
|
| 189 |
+
sns.heatmap(train.astype(float).corr(),linewidths=0.1,vmax=1.0,
|
| 190 |
+
square=True, cmap=colormap, linecolor='white', annot=True)
|
| 191 |
+
|
| 192 |
+
|
| 193 |
+
# **Takeaway from the Plots**
|
| 194 |
+
#
|
| 195 |
+
# One thing that that the Pearson Correlation plot can tell us is that there are not too many features strongly correlated with one another. This is good from a point of view of feeding these features into your learning model because this means that there isn't much redundant or superfluous data in our training set and we are happy that each feature carries with it some unique information. Here are two most correlated features are that of Family size and Parch (Parents and Children). I'll still leave both features in for the purposes of this exercise.
|
| 196 |
+
#
|
| 197 |
+
# **Pairplots**
|
| 198 |
+
#
|
| 199 |
+
# Finally let us generate some pairplots to observe the distribution of data from one feature to the other. Once again we use Seaborn to help us.
|
| 200 |
+
|
| 201 |
+
# In[7]:
|
| 202 |
+
|
| 203 |
+
|
| 204 |
+
g = sns.pairplot(train[[u'Survived', u'Pclass', u'Sex', u'Age', u'Parch', u'Fare', u'Embarked',
|
| 205 |
+
u'FamilySize', u'Title']], hue='Survived', palette = 'seismic',size=1.2,diag_kind = 'kde',diag_kws=dict(shade=True),plot_kws=dict(s=10) )
|
| 206 |
+
g.set(xticklabels=[])
|
| 207 |
+
|
| 208 |
+
|
| 209 |
+
# # Ensembling & Stacking models
|
| 210 |
+
#
|
| 211 |
+
# Finally after that brief whirlwind detour with regards to feature engineering and formatting, we finally arrive at the meat and gist of the this notebook.
|
| 212 |
+
#
|
| 213 |
+
# Creating a Stacking ensemble!
|
| 214 |
+
|
| 215 |
+
# ### Helpers via Python Classes
|
| 216 |
+
#
|
| 217 |
+
# Here we invoke the use of Python's classes to help make it more convenient for us. For any newcomers to programming, one normally hears Classes being used in conjunction with Object-Oriented Programming (OOP). In short, a class helps to extend some code/program for creating objects (variables for old-school peeps) as well as to implement functions and methods specific to that class.
|
| 218 |
+
#
|
| 219 |
+
# In the section of code below, we essentially write a class *SklearnHelper* that allows one to extend the inbuilt methods (such as train, predict and fit) common to all the Sklearn classifiers. Therefore this cuts out redundancy as won't need to write the same methods five times if we wanted to invoke five different classifiers.
|
| 220 |
+
|
| 221 |
+
# In[8]:
|
| 222 |
+
|
| 223 |
+
|
| 224 |
+
# Some useful parameters which will come in handy later on
|
| 225 |
+
ntrain = train.shape[0]
|
| 226 |
+
ntest = test.shape[0]
|
| 227 |
+
SEED = 0 # for reproducibility
|
| 228 |
+
NFOLDS = 5 # set folds for out-of-fold prediction
|
| 229 |
+
kf = KFold(ntrain, n_folds= NFOLDS, random_state=SEED)
|
| 230 |
+
|
| 231 |
+
# Class to extend the Sklearn classifier
|
| 232 |
+
class SklearnHelper(object):
|
| 233 |
+
def __init__(self, clf, seed=0, params=None):
|
| 234 |
+
params['random_state'] = seed
|
| 235 |
+
self.clf = clf(**params)
|
| 236 |
+
|
| 237 |
+
def train(self, x_train, y_train):
|
| 238 |
+
self.clf.fit(x_train, y_train)
|
| 239 |
+
|
| 240 |
+
def predict(self, x):
|
| 241 |
+
return self.clf.predict(x)
|
| 242 |
+
|
| 243 |
+
def fit(self,x,y):
|
| 244 |
+
return self.clf.fit(x,y)
|
| 245 |
+
|
| 246 |
+
def feature_importances(self,x,y):
|
| 247 |
+
print(self.clf.fit(x,y).feature_importances_)
|
| 248 |
+
|
| 249 |
+
# Class to extend XGboost classifer
|
| 250 |
+
|
| 251 |
+
|
| 252 |
+
# Bear with me for those who already know this but for people who have not created classes or objects in Python before, let me explain what the code given above does. In creating my base classifiers, I will only use the models already present in the Sklearn library and therefore only extend the class for that.
|
| 253 |
+
#
|
| 254 |
+
# **def init** : Python standard for invoking the default constructor for the class. This means that when you want to create an object (classifier), you have to give it the parameters of clf (what sklearn classifier you want), seed (random seed) and params (parameters for the classifiers).
|
| 255 |
+
#
|
| 256 |
+
# The rest of the code are simply methods of the class which simply call the corresponding methods already existing within the sklearn classifiers. Essentially, we have created a wrapper class to extend the various Sklearn classifiers so that this should help us reduce having to write the same code over and over when we implement multiple learners to our stacker.
|
| 257 |
+
|
| 258 |
+
# ### Out-of-Fold Predictions
|
| 259 |
+
#
|
| 260 |
+
# Now as alluded to above in the introductory section, stacking uses predictions of base classifiers as input for training to a second-level model. However one cannot simply train the base models on the full training data, generate predictions on the full test set and then output these for the second-level training. This runs the risk of your base model predictions already having "seen" the test set and therefore overfitting when feeding these predictions.
|
| 261 |
+
|
| 262 |
+
# In[9]:
|
| 263 |
+
|
| 264 |
+
|
| 265 |
+
def get_oof(clf, x_train, y_train, x_test):
|
| 266 |
+
oof_train = np.zeros((ntrain,))
|
| 267 |
+
oof_test = np.zeros((ntest,))
|
| 268 |
+
oof_test_skf = np.empty((NFOLDS, ntest))
|
| 269 |
+
|
| 270 |
+
for i, (train_index, test_index) in enumerate(kf):
|
| 271 |
+
x_tr = x_train[train_index]
|
| 272 |
+
y_tr = y_train[train_index]
|
| 273 |
+
x_te = x_train[test_index]
|
| 274 |
+
|
| 275 |
+
clf.train(x_tr, y_tr)
|
| 276 |
+
|
| 277 |
+
oof_train[test_index] = clf.predict(x_te)
|
| 278 |
+
oof_test_skf[i, :] = clf.predict(x_test)
|
| 279 |
+
|
| 280 |
+
oof_test[:] = oof_test_skf.mean(axis=0)
|
| 281 |
+
return oof_train.reshape(-1, 1), oof_test.reshape(-1, 1)
|
| 282 |
+
|
| 283 |
+
|
| 284 |
+
# # Generating our Base First-Level Models
|
| 285 |
+
#
|
| 286 |
+
# So now let us prepare five learning models as our first level classification. These models can all be conveniently invoked via the Sklearn library and are listed as follows:
|
| 287 |
+
#
|
| 288 |
+
# 1. Random Forest classifier
|
| 289 |
+
# 2. Extra Trees classifier
|
| 290 |
+
# 3. AdaBoost classifer
|
| 291 |
+
# 4. Gradient Boosting classifer
|
| 292 |
+
# 5. Support Vector Machine
|
| 293 |
+
|
| 294 |
+
# **Parameters**
|
| 295 |
+
#
|
| 296 |
+
# Just a quick summary of the parameters that we will be listing here for completeness,
|
| 297 |
+
#
|
| 298 |
+
# **n_jobs** : Number of cores used for the training process. If set to -1, all cores are used.
|
| 299 |
+
#
|
| 300 |
+
# **n_estimators** : Number of classification trees in your learning model ( set to 10 per default)
|
| 301 |
+
#
|
| 302 |
+
# **max_depth** : Maximum depth of tree, or how much a node should be expanded. Beware if set to too high a number would run the risk of overfitting as one would be growing the tree too deep
|
| 303 |
+
#
|
| 304 |
+
# **verbose** : Controls whether you want to output any text during the learning process. A value of 0 suppresses all text while a value of 3 outputs the tree learning process at every iteration.
|
| 305 |
+
#
|
| 306 |
+
# Please check out the full description via the official Sklearn website. There you will find that there are a whole host of other useful parameters that you can play around with.
|
| 307 |
+
|
| 308 |
+
# In[10]:
|
| 309 |
+
|
| 310 |
+
|
| 311 |
+
# Put in our parameters for said classifiers
|
| 312 |
+
# Random Forest parameters
|
| 313 |
+
rf_params = {
|
| 314 |
+
'n_jobs': -1,
|
| 315 |
+
'n_estimators': 500,
|
| 316 |
+
'warm_start': True,
|
| 317 |
+
#'max_features': 0.2,
|
| 318 |
+
'max_depth': 6,
|
| 319 |
+
'min_samples_leaf': 2,
|
| 320 |
+
'max_features' : 'sqrt',
|
| 321 |
+
'verbose': 0
|
| 322 |
+
}
|
| 323 |
+
|
| 324 |
+
# Extra Trees Parameters
|
| 325 |
+
et_params = {
|
| 326 |
+
'n_jobs': -1,
|
| 327 |
+
'n_estimators':500,
|
| 328 |
+
#'max_features': 0.5,
|
| 329 |
+
'max_depth': 8,
|
| 330 |
+
'min_samples_leaf': 2,
|
| 331 |
+
'verbose': 0
|
| 332 |
+
}
|
| 333 |
+
|
| 334 |
+
# AdaBoost parameters
|
| 335 |
+
ada_params = {
|
| 336 |
+
'n_estimators': 500,
|
| 337 |
+
'learning_rate' : 0.75
|
| 338 |
+
}
|
| 339 |
+
|
| 340 |
+
# Gradient Boosting parameters
|
| 341 |
+
gb_params = {
|
| 342 |
+
'n_estimators': 500,
|
| 343 |
+
#'max_features': 0.2,
|
| 344 |
+
'max_depth': 5,
|
| 345 |
+
'min_samples_leaf': 2,
|
| 346 |
+
'verbose': 0
|
| 347 |
+
}
|
| 348 |
+
|
| 349 |
+
# Support Vector Classifier parameters
|
| 350 |
+
svc_params = {
|
| 351 |
+
'kernel' : 'linear',
|
| 352 |
+
'C' : 0.025
|
| 353 |
+
}
|
| 354 |
+
|
| 355 |
+
|
| 356 |
+
# Furthermore, since having mentioned about Objects and classes within the OOP framework, let us now create 5 objects that represent our 5 learning models via our Helper Sklearn Class we defined earlier.
|
| 357 |
+
|
| 358 |
+
# In[11]:
|
| 359 |
+
|
| 360 |
+
|
| 361 |
+
# Create 5 objects that represent our 4 models
|
| 362 |
+
rf = SklearnHelper(clf=RandomForestClassifier, seed=SEED, params=rf_params)
|
| 363 |
+
et = SklearnHelper(clf=ExtraTreesClassifier, seed=SEED, params=et_params)
|
| 364 |
+
ada = SklearnHelper(clf=AdaBoostClassifier, seed=SEED, params=ada_params)
|
| 365 |
+
gb = SklearnHelper(clf=GradientBoostingClassifier, seed=SEED, params=gb_params)
|
| 366 |
+
svc = SklearnHelper(clf=SVC, seed=SEED, params=svc_params)
|
| 367 |
+
|
| 368 |
+
|
| 369 |
+
# **Creating NumPy arrays out of our train and test sets**
|
| 370 |
+
#
|
| 371 |
+
# Great. Having prepared our first layer base models as such, we can now ready the training and test test data for input into our classifiers by generating NumPy arrays out of their original dataframes as follows:
|
| 372 |
+
|
| 373 |
+
# In[12]:
|
| 374 |
+
|
| 375 |
+
|
| 376 |
+
# Create Numpy arrays of train, test and target ( Survived) dataframes to feed into our models
|
| 377 |
+
y_train = train['Survived'].ravel()
|
| 378 |
+
train = train.drop(['Survived'], axis=1)
|
| 379 |
+
x_train = train.values # Creates an array of the train data
|
| 380 |
+
x_test = test.values # Creats an array of the test data
|
| 381 |
+
|
| 382 |
+
|
| 383 |
+
# **Output of the First level Predictions**
|
| 384 |
+
#
|
| 385 |
+
# We now feed the training and test data into our 5 base classifiers and use the Out-of-Fold prediction function we defined earlier to generate our first level predictions. Allow a handful of minutes for the chunk of code below to run.
|
| 386 |
+
|
| 387 |
+
# In[13]:
|
| 388 |
+
|
| 389 |
+
|
| 390 |
+
# Create our OOF train and test predictions. These base results will be used as new features
|
| 391 |
+
et_oof_train, et_oof_test = get_oof(et, x_train, y_train, x_test) # Extra Trees
|
| 392 |
+
rf_oof_train, rf_oof_test = get_oof(rf,x_train, y_train, x_test) # Random Forest
|
| 393 |
+
ada_oof_train, ada_oof_test = get_oof(ada, x_train, y_train, x_test) # AdaBoost
|
| 394 |
+
gb_oof_train, gb_oof_test = get_oof(gb,x_train, y_train, x_test) # Gradient Boost
|
| 395 |
+
svc_oof_train, svc_oof_test = get_oof(svc,x_train, y_train, x_test) # Support Vector Classifier
|
| 396 |
+
|
| 397 |
+
print("Training is complete")
|
| 398 |
+
|
| 399 |
+
|
| 400 |
+
# **Feature importances generated from the different classifiers**
|
| 401 |
+
#
|
| 402 |
+
# Now having learned our the first-level classifiers, we can utilise a very nifty feature of the Sklearn models and that is to output the importances of the various features in the training and test sets with one very simple line of code.
|
| 403 |
+
#
|
| 404 |
+
# As per the Sklearn documentation, most of the classifiers are built in with an attribute which returns feature importances by simply typing in **.feature_importances_**. Therefore we will invoke this very useful attribute via our function earliand plot the feature importances as such
|
| 405 |
+
|
| 406 |
+
# In[14]:
|
| 407 |
+
|
| 408 |
+
|
| 409 |
+
rf_feature = rf.feature_importances(x_train,y_train)
|
| 410 |
+
et_feature = et.feature_importances(x_train, y_train)
|
| 411 |
+
ada_feature = ada.feature_importances(x_train, y_train)
|
| 412 |
+
gb_feature = gb.feature_importances(x_train,y_train)
|
| 413 |
+
|
| 414 |
+
|
| 415 |
+
# So I have not yet figured out how to assign and store the feature importances outright. Therefore I'll print out the values from the code above and then simply copy and paste into Python lists as below (sorry for the lousy hack)
|
| 416 |
+
|
| 417 |
+
# In[15]:
|
| 418 |
+
|
| 419 |
+
|
| 420 |
+
rf_features = [0.10474135, 0.21837029, 0.04432652, 0.02249159, 0.05432591, 0.02854371
|
| 421 |
+
,0.07570305, 0.01088129 , 0.24247496, 0.13685733 , 0.06128402]
|
| 422 |
+
et_features = [ 0.12165657, 0.37098307 ,0.03129623 , 0.01591611 , 0.05525811 , 0.028157
|
| 423 |
+
,0.04589793 , 0.02030357 , 0.17289562 , 0.04853517, 0.08910063]
|
| 424 |
+
ada_features = [0.028 , 0.008 , 0.012 , 0.05866667, 0.032 , 0.008
|
| 425 |
+
,0.04666667 , 0. , 0.05733333, 0.73866667, 0.01066667]
|
| 426 |
+
gb_features = [ 0.06796144 , 0.03889349 , 0.07237845 , 0.02628645 , 0.11194395, 0.04778854
|
| 427 |
+
,0.05965792 , 0.02774745, 0.07462718, 0.4593142 , 0.01340093]
|
| 428 |
+
|
| 429 |
+
|
| 430 |
+
# Create a dataframe from the lists containing the feature importance data for easy plotting via the Plotly package.
|
| 431 |
+
|
| 432 |
+
# In[16]:
|
| 433 |
+
|
| 434 |
+
|
| 435 |
+
cols = train.columns.values
|
| 436 |
+
# Create a dataframe with features
|
| 437 |
+
feature_dataframe = pd.DataFrame( {'features': cols,
|
| 438 |
+
'Random Forest feature importances': rf_features,
|
| 439 |
+
'Extra Trees feature importances': et_features,
|
| 440 |
+
'AdaBoost feature importances': ada_features,
|
| 441 |
+
'Gradient Boost feature importances': gb_features
|
| 442 |
+
})
|
| 443 |
+
|
| 444 |
+
|
| 445 |
+
# **Interactive feature importances via Plotly scatterplots**
|
| 446 |
+
#
|
| 447 |
+
# I'll use the interactive Plotly package at this juncture to visualise the feature importances values of the different classifiers via a plotly scatter plot by calling "Scatter" as follows:
|
| 448 |
+
|
| 449 |
+
# In[17]:
|
| 450 |
+
|
| 451 |
+
|
| 452 |
+
# Scatter plot
|
| 453 |
+
trace = go.Scatter(
|
| 454 |
+
y = feature_dataframe['Random Forest feature importances'].values,
|
| 455 |
+
x = feature_dataframe['features'].values,
|
| 456 |
+
mode='markers',
|
| 457 |
+
marker=dict(
|
| 458 |
+
sizemode = 'diameter',
|
| 459 |
+
sizeref = 1,
|
| 460 |
+
size = 25,
|
| 461 |
+
# size= feature_dataframe['AdaBoost feature importances'].values,
|
| 462 |
+
#color = np.random.randn(500), #set color equal to a variable
|
| 463 |
+
color = feature_dataframe['Random Forest feature importances'].values,
|
| 464 |
+
colorscale='Portland',
|
| 465 |
+
showscale=True
|
| 466 |
+
),
|
| 467 |
+
text = feature_dataframe['features'].values
|
| 468 |
+
)
|
| 469 |
+
data = [trace]
|
| 470 |
+
|
| 471 |
+
layout= go.Layout(
|
| 472 |
+
autosize= True,
|
| 473 |
+
title= 'Random Forest Feature Importance',
|
| 474 |
+
hovermode= 'closest',
|
| 475 |
+
# xaxis= dict(
|
| 476 |
+
# title= 'Pop',
|
| 477 |
+
# ticklen= 5,
|
| 478 |
+
# zeroline= False,
|
| 479 |
+
# gridwidth= 2,
|
| 480 |
+
# ),
|
| 481 |
+
yaxis=dict(
|
| 482 |
+
title= 'Feature Importance',
|
| 483 |
+
ticklen= 5,
|
| 484 |
+
gridwidth= 2
|
| 485 |
+
),
|
| 486 |
+
showlegend= False
|
| 487 |
+
)
|
| 488 |
+
fig = go.Figure(data=data, layout=layout)
|
| 489 |
+
py.iplot(fig,filename='scatter2010')
|
| 490 |
+
|
| 491 |
+
# Scatter plot
|
| 492 |
+
trace = go.Scatter(
|
| 493 |
+
y = feature_dataframe['Extra Trees feature importances'].values,
|
| 494 |
+
x = feature_dataframe['features'].values,
|
| 495 |
+
mode='markers',
|
| 496 |
+
marker=dict(
|
| 497 |
+
sizemode = 'diameter',
|
| 498 |
+
sizeref = 1,
|
| 499 |
+
size = 25,
|
| 500 |
+
# size= feature_dataframe['AdaBoost feature importances'].values,
|
| 501 |
+
#color = np.random.randn(500), #set color equal to a variable
|
| 502 |
+
color = feature_dataframe['Extra Trees feature importances'].values,
|
| 503 |
+
colorscale='Portland',
|
| 504 |
+
showscale=True
|
| 505 |
+
),
|
| 506 |
+
text = feature_dataframe['features'].values
|
| 507 |
+
)
|
| 508 |
+
data = [trace]
|
| 509 |
+
|
| 510 |
+
layout= go.Layout(
|
| 511 |
+
autosize= True,
|
| 512 |
+
title= 'Extra Trees Feature Importance',
|
| 513 |
+
hovermode= 'closest',
|
| 514 |
+
# xaxis= dict(
|
| 515 |
+
# title= 'Pop',
|
| 516 |
+
# ticklen= 5,
|
| 517 |
+
# zeroline= False,
|
| 518 |
+
# gridwidth= 2,
|
| 519 |
+
# ),
|
| 520 |
+
yaxis=dict(
|
| 521 |
+
title= 'Feature Importance',
|
| 522 |
+
ticklen= 5,
|
| 523 |
+
gridwidth= 2
|
| 524 |
+
),
|
| 525 |
+
showlegend= False
|
| 526 |
+
)
|
| 527 |
+
fig = go.Figure(data=data, layout=layout)
|
| 528 |
+
py.iplot(fig,filename='scatter2010')
|
| 529 |
+
|
| 530 |
+
# Scatter plot
|
| 531 |
+
trace = go.Scatter(
|
| 532 |
+
y = feature_dataframe['AdaBoost feature importances'].values,
|
| 533 |
+
x = feature_dataframe['features'].values,
|
| 534 |
+
mode='markers',
|
| 535 |
+
marker=dict(
|
| 536 |
+
sizemode = 'diameter',
|
| 537 |
+
sizeref = 1,
|
| 538 |
+
size = 25,
|
| 539 |
+
# size= feature_dataframe['AdaBoost feature importances'].values,
|
| 540 |
+
#color = np.random.randn(500), #set color equal to a variable
|
| 541 |
+
color = feature_dataframe['AdaBoost feature importances'].values,
|
| 542 |
+
colorscale='Portland',
|
| 543 |
+
showscale=True
|
| 544 |
+
),
|
| 545 |
+
text = feature_dataframe['features'].values
|
| 546 |
+
)
|
| 547 |
+
data = [trace]
|
| 548 |
+
|
| 549 |
+
layout= go.Layout(
|
| 550 |
+
autosize= True,
|
| 551 |
+
title= 'AdaBoost Feature Importance',
|
| 552 |
+
hovermode= 'closest',
|
| 553 |
+
# xaxis= dict(
|
| 554 |
+
# title= 'Pop',
|
| 555 |
+
# ticklen= 5,
|
| 556 |
+
# zeroline= False,
|
| 557 |
+
# gridwidth= 2,
|
| 558 |
+
# ),
|
| 559 |
+
yaxis=dict(
|
| 560 |
+
title= 'Feature Importance',
|
| 561 |
+
ticklen= 5,
|
| 562 |
+
gridwidth= 2
|
| 563 |
+
),
|
| 564 |
+
showlegend= False
|
| 565 |
+
)
|
| 566 |
+
fig = go.Figure(data=data, layout=layout)
|
| 567 |
+
py.iplot(fig,filename='scatter2010')
|
| 568 |
+
|
| 569 |
+
# Scatter plot
|
| 570 |
+
trace = go.Scatter(
|
| 571 |
+
y = feature_dataframe['Gradient Boost feature importances'].values,
|
| 572 |
+
x = feature_dataframe['features'].values,
|
| 573 |
+
mode='markers',
|
| 574 |
+
marker=dict(
|
| 575 |
+
sizemode = 'diameter',
|
| 576 |
+
sizeref = 1,
|
| 577 |
+
size = 25,
|
| 578 |
+
# size= feature_dataframe['AdaBoost feature importances'].values,
|
| 579 |
+
#color = np.random.randn(500), #set color equal to a variable
|
| 580 |
+
color = feature_dataframe['Gradient Boost feature importances'].values,
|
| 581 |
+
colorscale='Portland',
|
| 582 |
+
showscale=True
|
| 583 |
+
),
|
| 584 |
+
text = feature_dataframe['features'].values
|
| 585 |
+
)
|
| 586 |
+
data = [trace]
|
| 587 |
+
|
| 588 |
+
layout= go.Layout(
|
| 589 |
+
autosize= True,
|
| 590 |
+
title= 'Gradient Boosting Feature Importance',
|
| 591 |
+
hovermode= 'closest',
|
| 592 |
+
# xaxis= dict(
|
| 593 |
+
# title= 'Pop',
|
| 594 |
+
# ticklen= 5,
|
| 595 |
+
# zeroline= False,
|
| 596 |
+
# gridwidth= 2,
|
| 597 |
+
# ),
|
| 598 |
+
yaxis=dict(
|
| 599 |
+
title= 'Feature Importance',
|
| 600 |
+
ticklen= 5,
|
| 601 |
+
gridwidth= 2
|
| 602 |
+
),
|
| 603 |
+
showlegend= False
|
| 604 |
+
)
|
| 605 |
+
fig = go.Figure(data=data, layout=layout)
|
| 606 |
+
py.iplot(fig,filename='scatter2010')
|
| 607 |
+
|
| 608 |
+
|
| 609 |
+
# Now let us calculate the mean of all the feature importances and store it as a new column in the feature importance dataframe.
|
| 610 |
+
|
| 611 |
+
# In[18]:
|
| 612 |
+
|
| 613 |
+
|
| 614 |
+
# Create the new column containing the average of values
|
| 615 |
+
|
| 616 |
+
feature_dataframe['mean'] = feature_dataframe.mean(axis= 1) # axis = 1 computes the mean row-wise
|
| 617 |
+
feature_dataframe.head(3)
|
| 618 |
+
|
| 619 |
+
|
| 620 |
+
# **Plotly Barplot of Average Feature Importances**
|
| 621 |
+
#
|
| 622 |
+
# Having obtained the mean feature importance across all our classifiers, we can plot them into a Plotly bar plot as follows:
|
| 623 |
+
|
| 624 |
+
# In[19]:
|
| 625 |
+
|
| 626 |
+
|
| 627 |
+
y = feature_dataframe['mean'].values
|
| 628 |
+
x = feature_dataframe['features'].values
|
| 629 |
+
data = [go.Bar(
|
| 630 |
+
x= x,
|
| 631 |
+
y= y,
|
| 632 |
+
width = 0.5,
|
| 633 |
+
marker=dict(
|
| 634 |
+
color = feature_dataframe['mean'].values,
|
| 635 |
+
colorscale='Portland',
|
| 636 |
+
showscale=True,
|
| 637 |
+
reversescale = False
|
| 638 |
+
),
|
| 639 |
+
opacity=0.6
|
| 640 |
+
)]
|
| 641 |
+
|
| 642 |
+
layout= go.Layout(
|
| 643 |
+
autosize= True,
|
| 644 |
+
title= 'Barplots of Mean Feature Importance',
|
| 645 |
+
hovermode= 'closest',
|
| 646 |
+
# xaxis= dict(
|
| 647 |
+
# title= 'Pop',
|
| 648 |
+
# ticklen= 5,
|
| 649 |
+
# zeroline= False,
|
| 650 |
+
# gridwidth= 2,
|
| 651 |
+
# ),
|
| 652 |
+
yaxis=dict(
|
| 653 |
+
title= 'Feature Importance',
|
| 654 |
+
ticklen= 5,
|
| 655 |
+
gridwidth= 2
|
| 656 |
+
),
|
| 657 |
+
showlegend= False
|
| 658 |
+
)
|
| 659 |
+
fig = go.Figure(data=data, layout=layout)
|
| 660 |
+
py.iplot(fig, filename='bar-direct-labels')
|
| 661 |
+
|
| 662 |
+
|
| 663 |
+
# # Second-Level Predictions from the First-level Output
|
| 664 |
+
|
| 665 |
+
# **First-level output as new features**
|
| 666 |
+
#
|
| 667 |
+
# Having now obtained our first-level predictions, one can think of it as essentially building a new set of features to be used as training data for the next classifier. As per the code below, we are therefore having as our new columns the first-level predictions from our earlier classifiers and we train the next classifier on this.
|
| 668 |
+
|
| 669 |
+
# In[20]:
|
| 670 |
+
|
| 671 |
+
|
| 672 |
+
base_predictions_train = pd.DataFrame( {'RandomForest': rf_oof_train.ravel(),
|
| 673 |
+
'ExtraTrees': et_oof_train.ravel(),
|
| 674 |
+
'AdaBoost': ada_oof_train.ravel(),
|
| 675 |
+
'GradientBoost': gb_oof_train.ravel()
|
| 676 |
+
})
|
| 677 |
+
base_predictions_train.head()
|
| 678 |
+
|
| 679 |
+
|
| 680 |
+
# **Correlation Heatmap of the Second Level Training set**
|
| 681 |
+
|
| 682 |
+
# In[21]:
|
| 683 |
+
|
| 684 |
+
|
| 685 |
+
data = [
|
| 686 |
+
go.Heatmap(
|
| 687 |
+
z= base_predictions_train.astype(float).corr().values ,
|
| 688 |
+
x=base_predictions_train.columns.values,
|
| 689 |
+
y= base_predictions_train.columns.values,
|
| 690 |
+
colorscale='Viridis',
|
| 691 |
+
showscale=True,
|
| 692 |
+
reversescale = True
|
| 693 |
+
)
|
| 694 |
+
]
|
| 695 |
+
py.iplot(data, filename='labelled-heatmap')
|
| 696 |
+
|
| 697 |
+
|
| 698 |
+
# There have been quite a few articles and Kaggle competition winner stories about the merits of having trained models that are more uncorrelated with one another producing better scores.
|
| 699 |
+
|
| 700 |
+
# In[22]:
|
| 701 |
+
|
| 702 |
+
|
| 703 |
+
x_train = np.concatenate(( et_oof_train, rf_oof_train, ada_oof_train, gb_oof_train, svc_oof_train), axis=1)
|
| 704 |
+
x_test = np.concatenate(( et_oof_test, rf_oof_test, ada_oof_test, gb_oof_test, svc_oof_test), axis=1)
|
| 705 |
+
|
| 706 |
+
|
| 707 |
+
# Having now concatenated and joined both the first-level train and test predictions as x_train and x_test, we can now fit a second-level learning model.
|
| 708 |
+
|
| 709 |
+
# ### Second level learning model via XGBoost
|
| 710 |
+
#
|
| 711 |
+
# Here we choose the eXtremely famous library for boosted tree learning model, XGBoost. It was built to optimize large-scale boosted tree algorithms. For further information about the algorithm, check out the [official documentation][1].
|
| 712 |
+
#
|
| 713 |
+
# [1]: https://xgboost.readthedocs.io/en/latest/
|
| 714 |
+
#
|
| 715 |
+
# Anyways, we call an XGBClassifier and fit it to the first-level train and target data and use the learned model to predict the test data as follows:
|
| 716 |
+
|
| 717 |
+
# In[23]:
|
| 718 |
+
|
| 719 |
+
|
| 720 |
+
gbm = xgb.XGBClassifier(
|
| 721 |
+
#learning_rate = 0.02,
|
| 722 |
+
n_estimators= 2000,
|
| 723 |
+
max_depth= 4,
|
| 724 |
+
min_child_weight= 2,
|
| 725 |
+
#gamma=1,
|
| 726 |
+
gamma=0.9,
|
| 727 |
+
subsample=0.8,
|
| 728 |
+
colsample_bytree=0.8,
|
| 729 |
+
objective= 'binary:logistic',
|
| 730 |
+
nthread= -1,
|
| 731 |
+
scale_pos_weight=1).fit(x_train, y_train)
|
| 732 |
+
predictions = gbm.predict(x_test)
|
| 733 |
+
|
| 734 |
+
|
| 735 |
+
# Just a quick run down of the XGBoost parameters used in the model:
|
| 736 |
+
#
|
| 737 |
+
# **max_depth** : How deep you want to grow your tree. Beware if set to too high a number might run the risk of overfitting.
|
| 738 |
+
#
|
| 739 |
+
# **gamma** : minimum loss reduction required to make a further partition on a leaf node of the tree. The larger, the more conservative the algorithm will be.
|
| 740 |
+
#
|
| 741 |
+
# **eta** : step size shrinkage used in each boosting step to prevent overfitting
|
| 742 |
+
|
| 743 |
+
# **Producing the Submission file**
|
| 744 |
+
#
|
| 745 |
+
# Finally having trained and fit all our first-level and second-level models, we can now output the predictions into the proper format for submission to the Titanic competition as follows:
|
| 746 |
+
|
| 747 |
+
# In[24]:
|
| 748 |
+
|
| 749 |
+
|
| 750 |
+
# Generate Submission File
|
| 751 |
+
StackingSubmission = pd.DataFrame({ 'PassengerId': PassengerId,
|
| 752 |
+
'Survived': predictions })
|
| 753 |
+
StackingSubmission.to_csv("StackingSubmission.csv", index=False)
|
| 754 |
+
|
| 755 |
+
|
| 756 |
+
# **Steps for Further Improvement**
|
| 757 |
+
#
|
| 758 |
+
# As a closing remark it must be noted that the steps taken above just show a very simple way of producing an ensemble stacker. You hear of ensembles created at the highest level of Kaggle competitions which involves monstrous combinations of stacked classifiers as well as levels of stacking which go to more than 2 levels.
|
| 759 |
+
#
|
| 760 |
+
# Some additional steps that may be taken to improve one's score could be:
|
| 761 |
+
#
|
| 762 |
+
# 1. Implementing a good cross-validation strategy in training the models to find optimal parameter values
|
| 763 |
+
# 2. Introduce a greater variety of base models for learning. The more uncorrelated the results, the better the final score.
|
| 764 |
+
|
| 765 |
+
# ### Conclusion
|
| 766 |
+
#
|
| 767 |
+
# I have this notebook has been helpful somewhat in introducing a working script for stacking learning models. Again credit must be extended to Faron and Sina.
|
| 768 |
+
#
|
| 769 |
+
# For other excellent material on stacking or ensembling in general, refer to the de-facto Must read article on the website MLWave: [Kaggle Ensembling Guide][1].
|
| 770 |
+
#
|
| 771 |
+
# Till next time, Peace Out
|
| 772 |
+
#
|
| 773 |
+
# [1]: http://mlwave.com/kaggle-ensembling-guide/
|
| 774 |
+
|
| 775 |
+
# In[ ]:
|
| 776 |
+
|
| 777 |
+
|
| 778 |
+
|
| 779 |
+
|
Titanic/Kernels/ExtraTrees/11-titanic-a-step-by-step-intro-to-machine-learning.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/11-titanic-a-step-by-step-intro-to-machine-learning.py
ADDED
|
@@ -0,0 +1,1445 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python
|
| 2 |
+
# coding: utf-8
|
| 3 |
+
|
| 4 |
+
# # Table of content
|
| 5 |
+
#
|
| 6 |
+
# 1. Introduction - Loading libraries and dataset
|
| 7 |
+
# 2. Exploratory analysis, engineering and cleaning features - Bi-variate analysis
|
| 8 |
+
# 3. Correlation analysis - Tri-variate analysis
|
| 9 |
+
# 4. Predictive modelling, cross-validation, hyperparameters and ensembling
|
| 10 |
+
# 5. Submitting results
|
| 11 |
+
# 6. Credits
|
| 12 |
+
#
|
| 13 |
+
# ### Check other Kaggle notebooks from [Yvon Dalat](https://www.kaggle.com/ydalat):
|
| 14 |
+
# * [Titanic, a step-by-step intro to Machine Learning](https://www.kaggle.com/ydalat/titanic-a-step-by-step-intro-to-machine-learning): **a practice run ar EDA and ML-classification**
|
| 15 |
+
# * [HappyDB, a step-by-step application of Natural Language Processing](https://www.kaggle.com/ydalat/happydb-what-100-000-happy-moments-are-telling-us): **find out what 100,000 happy moments are telling us**
|
| 16 |
+
# * [Work-Life Balance survey, an Exploratory Data Analysis of lifestyle best practices](https://www.kaggle.com/ydalat/work-life-balance-best-practices-eda): **key insights into the factors affecting our work-life balance**
|
| 17 |
+
# * [Work-Life Balance survey, a Machine-Learning analysis of best practices to rebalance our lives](https://www.kaggle.com/ydalat/work-life-balance-predictors-and-clustering): **discover the strongest predictors of work-life balance**
|
| 18 |
+
#
|
| 19 |
+
# **Interested in more facts and data to balance your life, check the [360 Living guide](https://amzn.to/2MFO6Iy) **
|
| 20 |
+
#
|
| 21 |
+
# **Note:** Ever feel burnt out? Missing a deeper meaning? Sometimes life gets off-balance, but with the right steps, we can find the personal path to authentic happiness and balance.
|
| 22 |
+
# [Check out how Machine Learning and statistical analysis](https://www.amazon.com/dp/B07BNRRP7J?ref_=cm_sw_r_kb_dp_TZzTAbQND85EE&tag=kpembed-20&linkCode=kpe) sift through 10,000 responses to help us define our unique path to better living.
|
| 23 |
+
#
|
| 24 |
+
# # 1. Introduction - Loading libraries and dataset
|
| 25 |
+
# I created this Python notebook as the learning notes of my first Machine Learning project.
|
| 26 |
+
# So many new terms, new functions, new approaches, but the subject really interested me; so I dived into it, studied one line of code at a time, and captured the references and explanations in this notebook.
|
| 27 |
+
#
|
| 28 |
+
# The algorithm itself is a fork from **Anisotropic's Introduction to Ensembling/Stacking in Python**, a great notebook in itself.
|
| 29 |
+
# His notebook was itself based on **Faron's "Stacking Starter"**, as well as **Sina's Best Working Classfier**.
|
| 30 |
+
# I also used multiple functions from **Yassine Ghouzam**.
|
| 31 |
+
# I added many variations and additional features to improve the code (as much as I could) as well as additional visualization.
|
| 32 |
+
#
|
| 33 |
+
# Some key take away from my personal experiments and what-if analysis over the last couple of weeks:
|
| 34 |
+
#
|
| 35 |
+
# * **The engineering of the right features is absolutely key**. The goal there is to create the right categories between survived and not survived. They do not have to be the same size or equally distributed. What helped best is to group together passengers with the same survival rates.
|
| 36 |
+
#
|
| 37 |
+
# * ** I tried many, many different algorightms. Many overfit the training data** (up to 90%) but do not generate more accurate predictions with the test data. What worked better is to use the cross-validation on selected algotirhms. It is OK to select algorithms with various results as there is strenght in diversity.
|
| 38 |
+
#
|
| 39 |
+
# * **Lastly, the right ensembling was best achieved** with a votingclassifier with soft voting parameter
|
| 40 |
+
#
|
| 41 |
+
# One last word: please use this kernel as a first project to practice your ML/Python skills. I shameless ley sotle and learnt from many Kagglers through my learning process, please do the same with the code in this kernel.
|
| 42 |
+
#
|
| 43 |
+
# I also welcome your comments, questions and feedback.
|
| 44 |
+
#
|
| 45 |
+
# Yvon
|
| 46 |
+
#
|
| 47 |
+
# ## 1.1. Importing Library
|
| 48 |
+
|
| 49 |
+
# In[1]:
|
| 50 |
+
|
| 51 |
+
|
| 52 |
+
# Load libraries for analysis and visualization
|
| 53 |
+
import pandas as pd # collection of functions for data processing and analysis modeled after R dataframes with SQL like features
|
| 54 |
+
import numpy as np # foundational package for scientific computing
|
| 55 |
+
import re # Regular expression operations
|
| 56 |
+
import matplotlib.pyplot as plt # Collection of functions for scientific and publication-ready visualization
|
| 57 |
+
get_ipython().run_line_magic('matplotlib', 'inline')
|
| 58 |
+
import plotly.offline as py # Open source library for composing, editing, and sharing interactive data visualization
|
| 59 |
+
from matplotlib import pyplot
|
| 60 |
+
py.init_notebook_mode(connected=True)
|
| 61 |
+
import plotly.graph_objs as go
|
| 62 |
+
import plotly.tools as tls
|
| 63 |
+
from collections import Counter
|
| 64 |
+
|
| 65 |
+
# Machine learning libraries
|
| 66 |
+
import xgboost as xgb # Implementation of gradient boosted decision trees designed for speed and performance that is dominative competitive machine learning
|
| 67 |
+
import seaborn as sns # Visualization library based on matplotlib, provides interface for drawing attractive statistical graphics
|
| 68 |
+
|
| 69 |
+
import sklearn # Collection of machine learning algorithms
|
| 70 |
+
from sklearn.linear_model import LogisticRegression
|
| 71 |
+
from sklearn.svm import SVC, LinearSVC
|
| 72 |
+
from sklearn.ensemble import (RandomForestClassifier, AdaBoostClassifier,
|
| 73 |
+
GradientBoostingClassifier, ExtraTreesClassifier, VotingClassifier)
|
| 74 |
+
from sklearn.cross_validation import KFold
|
| 75 |
+
from sklearn.neighbors import KNeighborsClassifier
|
| 76 |
+
from sklearn.naive_bayes import GaussianNB
|
| 77 |
+
from sklearn.linear_model import Perceptron
|
| 78 |
+
from sklearn.linear_model import SGDClassifier
|
| 79 |
+
from sklearn.tree import DecisionTreeClassifier
|
| 80 |
+
from sklearn.model_selection import GridSearchCV, cross_val_score, StratifiedKFold, learning_curve
|
| 81 |
+
from sklearn.preprocessing import StandardScaler
|
| 82 |
+
from sklearn.model_selection import train_test_split
|
| 83 |
+
from sklearn.metrics import accuracy_score,classification_report, precision_recall_curve, confusion_matrix
|
| 84 |
+
|
| 85 |
+
import warnings
|
| 86 |
+
warnings.filterwarnings('ignore')
|
| 87 |
+
|
| 88 |
+
|
| 89 |
+
# ## 1.2. Loading dataset
|
| 90 |
+
|
| 91 |
+
# In[2]:
|
| 92 |
+
|
| 93 |
+
|
| 94 |
+
# Load in the train and test datasets from the CSV files
|
| 95 |
+
train = pd.read_csv('../input/train.csv')
|
| 96 |
+
test = pd.read_csv('../input/test.csv')
|
| 97 |
+
|
| 98 |
+
# Store our passenger ID for easy access
|
| 99 |
+
PassengerId = test['PassengerId']
|
| 100 |
+
|
| 101 |
+
# Display the first 5 rows of the dataset, a first look at our data
|
| 102 |
+
# 5 first row, 5 sample rows and basic statistics
|
| 103 |
+
train.head(50)
|
| 104 |
+
|
| 105 |
+
|
| 106 |
+
# In[3]:
|
| 107 |
+
|
| 108 |
+
|
| 109 |
+
train.sample(5)
|
| 110 |
+
|
| 111 |
+
|
| 112 |
+
# In[4]:
|
| 113 |
+
|
| 114 |
+
|
| 115 |
+
train.describe()
|
| 116 |
+
|
| 117 |
+
|
| 118 |
+
# **What are the data types for each feature?**
|
| 119 |
+
# * Survived: int
|
| 120 |
+
# * Pclass: int
|
| 121 |
+
# * Name: string
|
| 122 |
+
# * Sex: string
|
| 123 |
+
# * Age: float
|
| 124 |
+
# * SibSp: int
|
| 125 |
+
# * Parch: int
|
| 126 |
+
# * Ticket: string
|
| 127 |
+
# * Fare: float
|
| 128 |
+
# * Cabin: string
|
| 129 |
+
# * Embarked: string
|
| 130 |
+
|
| 131 |
+
# ## 1.3. Analysis goal
|
| 132 |
+
# **The Survived variable** is the outcome or dependent variable. It is a binary nominal datatype of 1 for "survived" and 0 for "did not survive".
|
| 133 |
+
# **All other variables** are potential predictor or independent variables. The goal is to predict this dependent variable only using the available independent variables. A test dataset has been created to test our algorithm.
|
| 134 |
+
|
| 135 |
+
# ## 1.4. A very first look into the data
|
| 136 |
+
|
| 137 |
+
# In[5]:
|
| 138 |
+
|
| 139 |
+
|
| 140 |
+
f,ax = plt.subplots(3,4,figsize=(20,16))
|
| 141 |
+
sns.countplot('Pclass',data=train,ax=ax[0,0])
|
| 142 |
+
sns.countplot('Sex',data=train,ax=ax[0,1])
|
| 143 |
+
sns.boxplot(x='Pclass',y='Age',data=train,ax=ax[0,2])
|
| 144 |
+
sns.countplot('SibSp',hue='Survived',data=train,ax=ax[0,3],palette='husl')
|
| 145 |
+
sns.distplot(train['Fare'].dropna(),ax=ax[2,0],kde=False,color='b')
|
| 146 |
+
sns.countplot('Embarked',data=train,ax=ax[2,2])
|
| 147 |
+
|
| 148 |
+
sns.countplot('Pclass',hue='Survived',data=train,ax=ax[1,0],palette='husl')
|
| 149 |
+
sns.countplot('Sex',hue='Survived',data=train,ax=ax[1,1],palette='husl')
|
| 150 |
+
sns.distplot(train[train['Survived']==0]['Age'].dropna(),ax=ax[1,2],kde=False,color='r',bins=5)
|
| 151 |
+
sns.distplot(train[train['Survived']==1]['Age'].dropna(),ax=ax[1,2],kde=False,color='g',bins=5)
|
| 152 |
+
sns.countplot('Parch',hue='Survived',data=train,ax=ax[1,3],palette='husl')
|
| 153 |
+
sns.swarmplot(x='Pclass',y='Fare',hue='Survived',data=train,palette='husl',ax=ax[2,1])
|
| 154 |
+
sns.countplot('Embarked',hue='Survived',data=train,ax=ax[2,3],palette='husl')
|
| 155 |
+
|
| 156 |
+
ax[0,0].set_title('Total Passengers by Class')
|
| 157 |
+
ax[0,1].set_title('Total Passengers by Gender')
|
| 158 |
+
ax[0,2].set_title('Age Box Plot By Class')
|
| 159 |
+
ax[0,3].set_title('Survival Rate by SibSp')
|
| 160 |
+
ax[1,0].set_title('Survival Rate by Class')
|
| 161 |
+
ax[1,1].set_title('Survival Rate by Gender')
|
| 162 |
+
ax[1,2].set_title('Survival Rate by Age')
|
| 163 |
+
ax[1,3].set_title('Survival Rate by Parch')
|
| 164 |
+
ax[2,0].set_title('Fare Distribution')
|
| 165 |
+
ax[2,1].set_title('Survival Rate by Fare and Pclass')
|
| 166 |
+
ax[2,2].set_title('Total Passengers by Embarked')
|
| 167 |
+
ax[2,3].set_title('Survival Rate by Embarked')
|
| 168 |
+
|
| 169 |
+
|
| 170 |
+
# This is only a quick of the relationships between features before we start a more detailed analysis.
|
| 171 |
+
#
|
| 172 |
+
#
|
| 173 |
+
# # 2. Exploratory Data Analysis (EDA), Cleaning and Engineering features
|
| 174 |
+
#
|
| 175 |
+
# We will start with a standard approach of any kernel: correct, complete, engineer the right features for analysis.
|
| 176 |
+
#
|
| 177 |
+
# ## 2.1. Correcting and completing features
|
| 178 |
+
# ### Detecting and correcting outliers
|
| 179 |
+
# Reviewing the data, there does not appear to be any aberrant or non-acceptable data inputs.
|
| 180 |
+
#
|
| 181 |
+
# There are potential outliers that we will identify (steps from Yassine Ghouzam):
|
| 182 |
+
# * It creates firset a function called detect_outliers, implementing the Tukey method
|
| 183 |
+
# * For each column of the dataframe, this function calculates the 25th percentile (Q1) and 75th percentile (Q3) values.
|
| 184 |
+
# * The interquartile range (IQR) is a measure of statistical dispersion, being equal to the difference between the 75th and 25th percentiles, or between upper and lower quartiles.
|
| 185 |
+
# * Any data points outside 1.5 time the IQR (1.5 time IQR below Q1, or 1.5 time IQR above Q3), is considered an outlier.
|
| 186 |
+
# * The outlier_list_col column captures the indices of these outliers. All outlier data get then pulled into the outlier_indices dataframe.
|
| 187 |
+
# * Finally, the detect_outliers function will select only the outliers happening multiple times. This is the datadframe that will be returned.
|
| 188 |
+
|
| 189 |
+
# In[6]:
|
| 190 |
+
|
| 191 |
+
|
| 192 |
+
# Outlier detection
|
| 193 |
+
def detect_outliers(df,n,features):
|
| 194 |
+
outlier_indices = []
|
| 195 |
+
# iterate over features(columns)
|
| 196 |
+
for col in features:
|
| 197 |
+
# 1st quartile (25%)
|
| 198 |
+
Q1 = np.percentile(df[col],25)
|
| 199 |
+
# 3rd quartile (75%)
|
| 200 |
+
Q3 = np.percentile(df[col],75)
|
| 201 |
+
# Interquartile range (IQR)
|
| 202 |
+
IQR = Q3 - Q1
|
| 203 |
+
# outlier step
|
| 204 |
+
outlier_step = 1.5 * IQR
|
| 205 |
+
# Determine a list of indices of outliers for feature col
|
| 206 |
+
outlier_list_col = df[(df[col] < Q1 - outlier_step) | (df[col] > Q3 + outlier_step )].index
|
| 207 |
+
# append the found outlier indices for col to the list of outlier indices
|
| 208 |
+
outlier_indices.extend(outlier_list_col)
|
| 209 |
+
|
| 210 |
+
# select observations containing more than 2 outliers
|
| 211 |
+
outlier_indices = Counter(outlier_indices)
|
| 212 |
+
multiple_outliers = list( k for k, v in outlier_indices.items() if v > n )
|
| 213 |
+
return multiple_outliers
|
| 214 |
+
# detect outliers from Age, SibSp , Parch and Fare
|
| 215 |
+
Outliers_to_drop = detect_outliers(train,2,["Age","SibSp","Parch","Fare"])
|
| 216 |
+
train.loc[Outliers_to_drop] # Show the outliers rows
|
| 217 |
+
|
| 218 |
+
|
| 219 |
+
# ** Observations**
|
| 220 |
+
# * The Detect_Outliers function found 10 outliers.
|
| 221 |
+
# * PassengerID 28, 89 and 342 passenger have an high Ticket Fare
|
| 222 |
+
# * The seven others have very high values of SibSP.
|
| 223 |
+
# * I found that dropping the outliers actually lower the prediction. So I decided to keep them.
|
| 224 |
+
#
|
| 225 |
+
# You can try to remove them and rerun the prediction to observe the result with the following function:
|
| 226 |
+
|
| 227 |
+
# In[7]:
|
| 228 |
+
|
| 229 |
+
|
| 230 |
+
# Drop outliers
|
| 231 |
+
# train = train.drop(Outliers_to_drop, axis = 0).reset_index(drop=True)
|
| 232 |
+
|
| 233 |
+
|
| 234 |
+
# ### Completing features
|
| 235 |
+
# The .info function below shows how complete or incomplete the datasets are.
|
| 236 |
+
# There are null values or missing data in the age, cabin, and embarked field. Missing values can be bad, because some algorithms don't know how-to handle null values and will fail. While others, like decision trees, can handle null values.
|
| 237 |
+
#
|
| 238 |
+
# The approach to to complete missing data is to impute using mean, median, or mean + randomized standard deviation.
|
| 239 |
+
# We will do this in section 2.2 with the **fillna** function: dataset['Fare'] = dataset['Fare'].fillna(train['Fare'].median())
|
| 240 |
+
|
| 241 |
+
# In[8]:
|
| 242 |
+
|
| 243 |
+
|
| 244 |
+
train.info()
|
| 245 |
+
print('_'*40)
|
| 246 |
+
test.info()
|
| 247 |
+
|
| 248 |
+
|
| 249 |
+
# ## 2.2. Descriptive analysis (univariate)
|
| 250 |
+
|
| 251 |
+
# In[9]:
|
| 252 |
+
|
| 253 |
+
|
| 254 |
+
full_data = [train, test]
|
| 255 |
+
Survival = train['Survived']
|
| 256 |
+
Survival.describe()
|
| 257 |
+
|
| 258 |
+
|
| 259 |
+
# ## 2.3 Feature Engineering - Bi-variate statistical analysis
|
| 260 |
+
#
|
| 261 |
+
# One of the first tasks in Data Analytics is to **convert the variables into numerical/ordinal values**.
|
| 262 |
+
# There are multiple types of data
|
| 263 |
+
#
|
| 264 |
+
# **a) Qualitative data: discrete**
|
| 265 |
+
# * Nominal: no natural order between categories. In this case: Name
|
| 266 |
+
# * Categorical: Sex
|
| 267 |
+
#
|
| 268 |
+
# **b) Numeric or quantitative data**
|
| 269 |
+
# * Discrete: could be ordinal like Pclass or not like Survived.
|
| 270 |
+
# * Continuous. e.g.: age
|
| 271 |
+
# Many feature engineering steps were taken from Anisotropic's excellent kernel.
|
| 272 |
+
#
|
| 273 |
+
# ### Pclass
|
| 274 |
+
|
| 275 |
+
# In[10]:
|
| 276 |
+
|
| 277 |
+
|
| 278 |
+
sns.barplot(x="Embarked", y="Survived", hue="Sex", data=train);
|
| 279 |
+
|
| 280 |
+
|
| 281 |
+
# Embarked does not seem to have a clear impact on the survival rate. We will analyse it further in the next sections and drop it if we cannot demonstrate a proven relationship to Survived.
|
| 282 |
+
#
|
| 283 |
+
# ### Name_length
|
| 284 |
+
|
| 285 |
+
# In[11]:
|
| 286 |
+
|
| 287 |
+
|
| 288 |
+
for dataset in full_data:
|
| 289 |
+
dataset['Name_length'] = train['Name'].apply(len)
|
| 290 |
+
# Qcut is a quantile based discretization function to autimatically create categories
|
| 291 |
+
# dataset['Name_length'] = pd.qcut(dataset['Name_length'], 6, labels=False)
|
| 292 |
+
# train['Name_length'].value_counts()
|
| 293 |
+
|
| 294 |
+
sum_Name = train[["Name_length", "Survived"]].groupby(['Name_length'],as_index=False).sum()
|
| 295 |
+
average_Name = train[["Name_length", "Survived"]].groupby(['Name_length'],as_index=False).mean()
|
| 296 |
+
fig, (axis1,axis2,axis3) = plt.subplots(3,1,figsize=(18,6))
|
| 297 |
+
sns.barplot(x='Name_length', y='Survived', data=sum_Name, ax = axis1)
|
| 298 |
+
sns.barplot(x='Name_length', y='Survived', data=average_Name, ax = axis2)
|
| 299 |
+
sns.pointplot(x = 'Name_length', y = 'Survived', data=train, ax = axis3)
|
| 300 |
+
|
| 301 |
+
|
| 302 |
+
# The first graph shows the amount of people by Name_length.
|
| 303 |
+
#
|
| 304 |
+
# The second one, their average survival rates.
|
| 305 |
+
#
|
| 306 |
+
# The proposed categories are: less than 23 (mostly men), 24 to 28, 29 to 40, 41 and more (mostly women).
|
| 307 |
+
# The categories are sized to group passengers with similar Survival rates.
|
| 308 |
+
|
| 309 |
+
# In[12]:
|
| 310 |
+
|
| 311 |
+
|
| 312 |
+
for dataset in full_data:
|
| 313 |
+
dataset.loc[ dataset['Name_length'] <= 23, 'Name_length'] = 0
|
| 314 |
+
dataset.loc[(dataset['Name_length'] > 23) & (dataset['Name_length'] <= 28), 'Name_length'] = 1
|
| 315 |
+
dataset.loc[(dataset['Name_length'] > 28) & (dataset['Name_length'] <= 40), 'Name_length'] = 2
|
| 316 |
+
dataset.loc[ dataset['Name_length'] > 40, 'Name_length'] = 3
|
| 317 |
+
train['Name_length'].value_counts()
|
| 318 |
+
|
| 319 |
+
|
| 320 |
+
# ### Gender (Sex)
|
| 321 |
+
|
| 322 |
+
# In[13]:
|
| 323 |
+
|
| 324 |
+
|
| 325 |
+
for dataset in full_data:# Mapping Gender
|
| 326 |
+
dataset['Sex'] = dataset['Sex'].map( {'female': 0, 'male': 1} ).astype(int)
|
| 327 |
+
|
| 328 |
+
|
| 329 |
+
# ### Age
|
| 330 |
+
|
| 331 |
+
# In[14]:
|
| 332 |
+
|
| 333 |
+
|
| 334 |
+
#plot distributions of age of passengers who survived or did not survive
|
| 335 |
+
a = sns.FacetGrid( train, hue = 'Survived', aspect=6 )
|
| 336 |
+
a.map(sns.kdeplot, 'Age', shade= True )
|
| 337 |
+
a.set(xlim=(0 , train['Age'].max()))
|
| 338 |
+
a.add_legend()
|
| 339 |
+
|
| 340 |
+
|
| 341 |
+
# The best categories for age are:
|
| 342 |
+
# * 0: Less than 14
|
| 343 |
+
# * 1: 14 to 30
|
| 344 |
+
# * 2: 30 to 40
|
| 345 |
+
# * 3: 40 to 50
|
| 346 |
+
# * 4: 50 to 60
|
| 347 |
+
# * 5: 60 and more
|
| 348 |
+
|
| 349 |
+
# In[15]:
|
| 350 |
+
|
| 351 |
+
|
| 352 |
+
for dataset in full_data:
|
| 353 |
+
age_avg = dataset['Age'].mean()
|
| 354 |
+
age_std = dataset['Age'].std()
|
| 355 |
+
age_null_count = dataset['Age'].isnull().sum()
|
| 356 |
+
age_null_random_list = np.random.randint(age_avg - age_std, age_avg + age_std, size=age_null_count)
|
| 357 |
+
dataset['Age'][np.isnan(dataset['Age'])] = age_null_random_list
|
| 358 |
+
dataset['Age'] = dataset['Age'].astype(int)
|
| 359 |
+
# Qcut is a quantile based discretization function to autimatically create categories (not used here)
|
| 360 |
+
# dataset['Age'] = pd.qcut(dataset['Age'], 6, labels=False)
|
| 361 |
+
# Using categories as defined above
|
| 362 |
+
dataset.loc[ dataset['Age'] <= 14, 'Age'] = 0
|
| 363 |
+
dataset.loc[(dataset['Age'] > 14) & (dataset['Age'] <= 30), 'Age'] = 5
|
| 364 |
+
dataset.loc[(dataset['Age'] > 30) & (dataset['Age'] <= 40), 'Age'] = 1
|
| 365 |
+
dataset.loc[(dataset['Age'] > 40) & (dataset['Age'] <= 50), 'Age'] = 3
|
| 366 |
+
dataset.loc[(dataset['Age'] > 50) & (dataset['Age'] <= 60), 'Age'] = 2
|
| 367 |
+
dataset.loc[ dataset['Age'] > 60, 'Age'] = 4
|
| 368 |
+
train['Age'].value_counts()
|
| 369 |
+
|
| 370 |
+
|
| 371 |
+
# In[16]:
|
| 372 |
+
|
| 373 |
+
|
| 374 |
+
train[["Age", "Survived"]].groupby(['Age'], as_index=False).mean().sort_values(by='Survived', ascending=False)
|
| 375 |
+
|
| 376 |
+
|
| 377 |
+
# ### Family: SibSp and Parch
|
| 378 |
+
#
|
| 379 |
+
# This section creates a new feature called FamilySize consisting of SibSp and Parch.
|
| 380 |
+
|
| 381 |
+
# In[17]:
|
| 382 |
+
|
| 383 |
+
|
| 384 |
+
for dataset in full_data:
|
| 385 |
+
# Create new feature FamilySize as a combination of SibSp and Parch
|
| 386 |
+
dataset['FamilySize'] = dataset['SibSp'] + dataset['Parch']+1
|
| 387 |
+
# Create new feature IsAlone from FamilySize
|
| 388 |
+
dataset['IsAlone'] = 0
|
| 389 |
+
dataset.loc[dataset['FamilySize'] == 1, 'IsAlone'] = 1
|
| 390 |
+
|
| 391 |
+
# Create new feature Boys from FamilySize
|
| 392 |
+
dataset['Boys'] = 0
|
| 393 |
+
dataset.loc[(dataset['Age'] == 0) & (dataset['Sex']==1), 'Boys'] = 1
|
| 394 |
+
|
| 395 |
+
fig, (axis1,axis2) = plt.subplots(1,2,figsize=(18,6))
|
| 396 |
+
sns.barplot(x="FamilySize", y="Survived", hue="Sex", data=train, ax = axis1);
|
| 397 |
+
sns.barplot(x="IsAlone", y="Survived", hue="Sex", data=train, ax = axis2);
|
| 398 |
+
|
| 399 |
+
|
| 400 |
+
# IsAlone does not result in a significant difference of survival rate. In addition, the slight difference between men and women go in different direction, i.e. IsAlone alone is not a good predictor of survival. O will drop this feature.
|
| 401 |
+
#
|
| 402 |
+
# ### Fare
|
| 403 |
+
|
| 404 |
+
# In[18]:
|
| 405 |
+
|
| 406 |
+
|
| 407 |
+
# Interactive chart using cufflinks
|
| 408 |
+
import cufflinks as cf
|
| 409 |
+
cf.go_offline()
|
| 410 |
+
train['Fare'].iplot(kind='hist', bins=30)
|
| 411 |
+
|
| 412 |
+
|
| 413 |
+
# In[19]:
|
| 414 |
+
|
| 415 |
+
|
| 416 |
+
# Remove all NULLS in the Fare column and create a new feature Categorical Fare
|
| 417 |
+
for dataset in full_data:
|
| 418 |
+
dataset['Fare'] = dataset['Fare'].fillna(train['Fare'].median())
|
| 419 |
+
|
| 420 |
+
# Explore Fare distribution
|
| 421 |
+
g = sns.distplot(dataset["Fare"], color="m", label="Skewness : %.2f"%(dataset["Fare"].skew()))
|
| 422 |
+
g = g.legend(loc="best")
|
| 423 |
+
|
| 424 |
+
|
| 425 |
+
# **Observations**
|
| 426 |
+
# * The Fare distribution is very skewed to the left. This can lead to overweigthing the model with very high values.
|
| 427 |
+
# * In this case, it is better to transform it with the log function to reduce the skewness and redistribute the data.
|
| 428 |
+
|
| 429 |
+
# In[20]:
|
| 430 |
+
|
| 431 |
+
|
| 432 |
+
# Apply log to Fare to reduce skewness distribution
|
| 433 |
+
for dataset in full_data:
|
| 434 |
+
dataset["Fare"] = dataset["Fare"].map(lambda i: np.log(i) if i > 0 else 0)
|
| 435 |
+
a4_dims = (20, 6)
|
| 436 |
+
fig, ax = pyplot.subplots(figsize=a4_dims)
|
| 437 |
+
g = sns.distplot(train["Fare"][train["Survived"] == 0], color="r", label="Skewness : %.2f"%(train["Fare"].skew()), ax=ax)
|
| 438 |
+
g = sns.distplot(train["Fare"][train["Survived"] == 1], color="b", label="Skewness : %.2f"%(train["Fare"].skew()))
|
| 439 |
+
#g = g.legend(loc="best")
|
| 440 |
+
g = g.legend(["Not Survived","Survived"])
|
| 441 |
+
|
| 442 |
+
|
| 443 |
+
# **Observations**
|
| 444 |
+
# Log Fare categories are:
|
| 445 |
+
# * 0 to 2.7: less survivors
|
| 446 |
+
# * More than 2.7 more survivors
|
| 447 |
+
|
| 448 |
+
# In[21]:
|
| 449 |
+
|
| 450 |
+
|
| 451 |
+
for dataset in full_data:
|
| 452 |
+
dataset.loc[ dataset['Fare'] <= 2.7, 'Fare'] = 0
|
| 453 |
+
# dataset.loc[(dataset['Fare'] > 2.7) & (dataset['Fare'] <= 3.2), 'Fare'] = 1
|
| 454 |
+
# dataset.loc[(dataset['Fare'] > 3.2) & (dataset['Fare'] <= 3.6), 'Fare'] = 2
|
| 455 |
+
dataset.loc[ dataset['Fare'] > 2.7, 'Fare'] = 3
|
| 456 |
+
dataset['Fare'] = dataset['Fare'].astype(int)
|
| 457 |
+
train['Fare'].value_counts()
|
| 458 |
+
|
| 459 |
+
|
| 460 |
+
# ### Cabin
|
| 461 |
+
|
| 462 |
+
# In[22]:
|
| 463 |
+
|
| 464 |
+
|
| 465 |
+
# Feature that tells whether a passenger had a cabin on the Titanic (O if no cabin number, 1 otherwise)
|
| 466 |
+
for dataset in full_data:
|
| 467 |
+
dataset['Has_Cabin'] = dataset["Cabin"].apply(lambda x: 0 if type(x) == float else 1)
|
| 468 |
+
|
| 469 |
+
train[["Has_Cabin", "Survived"]].groupby(['Has_Cabin'], as_index=False).sum().sort_values(by='Survived', ascending=False)
|
| 470 |
+
|
| 471 |
+
|
| 472 |
+
# In[23]:
|
| 473 |
+
|
| 474 |
+
|
| 475 |
+
train[["Has_Cabin", "Survived"]].groupby(['Has_Cabin'], as_index=False).mean().sort_values(by='Survived', ascending=False)
|
| 476 |
+
|
| 477 |
+
|
| 478 |
+
# It appears that Has_Cabin has a strong impact on the Survival rate. We will keep this feature.
|
| 479 |
+
#
|
| 480 |
+
# ### Embarked
|
| 481 |
+
|
| 482 |
+
# In[24]:
|
| 483 |
+
|
| 484 |
+
|
| 485 |
+
for dataset in full_data:
|
| 486 |
+
# Remove all NULLS in the Embarked column
|
| 487 |
+
dataset['Embarked'] = dataset['Embarked'].fillna('S')
|
| 488 |
+
# Mapping Embarked
|
| 489 |
+
dataset['Embarked'] = dataset['Embarked'].map( {'S': 0, 'C': 1, 'Q': 2} ).astype(int)
|
| 490 |
+
|
| 491 |
+
train_pivot = pd.pivot_table(train, values= 'Survived',index=['Embarked'],columns='Pclass',aggfunc=np.mean, margins=True)
|
| 492 |
+
def color_negative_red(val):
|
| 493 |
+
# Takes a scalar and returns a string with the css property 'color: red' if below 0.4, black otherwise.
|
| 494 |
+
color = 'red' if val < 0.4 else 'black'
|
| 495 |
+
return 'color: %s' % color
|
| 496 |
+
train_pivot = train_pivot.style.applymap(color_negative_red)
|
| 497 |
+
train_pivot
|
| 498 |
+
|
| 499 |
+
|
| 500 |
+
# Irrespective of the class, passengers embarked in 0 (S) and 2 (Q) have lower chance of survival. I will combine those into the first category.
|
| 501 |
+
|
| 502 |
+
# In[25]:
|
| 503 |
+
|
| 504 |
+
|
| 505 |
+
dataset['Embarked'] = dataset['Embarked'].replace(['0', '2'], '0')
|
| 506 |
+
train['Fare'].value_counts()
|
| 507 |
+
|
| 508 |
+
|
| 509 |
+
# ### Titles
|
| 510 |
+
|
| 511 |
+
# In[26]:
|
| 512 |
+
|
| 513 |
+
|
| 514 |
+
# Define function to extract titles from passenger names
|
| 515 |
+
def get_title(name):
|
| 516 |
+
title_search = re.search(' ([A-Za-z]+)\.', name)
|
| 517 |
+
# If the title exists, extract and return it.
|
| 518 |
+
if title_search:
|
| 519 |
+
return title_search.group(1)
|
| 520 |
+
return ""
|
| 521 |
+
for dataset in full_data:
|
| 522 |
+
# Create a new feature Title, containing the titles of passenger names
|
| 523 |
+
dataset['Title'] = dataset['Name'].apply(get_title)
|
| 524 |
+
|
| 525 |
+
fig, (axis1) = plt.subplots(1,figsize=(18,6))
|
| 526 |
+
sns.barplot(x="Title", y="Survived", data=train, ax=axis1);
|
| 527 |
+
|
| 528 |
+
|
| 529 |
+
# There are 4 types of titles:
|
| 530 |
+
# 0. Mme, Ms, Lady, Sir, Mlle, Countess: 100%.
|
| 531 |
+
# 1. Mrs, Miss: around 70% survival
|
| 532 |
+
# 2. Master: around 60%
|
| 533 |
+
# 3. Don, Rev, Capt, Jonkheer: no data
|
| 534 |
+
# 4. Dr, Major, Col: around 40%
|
| 535 |
+
# 5. Mr: below 20%
|
| 536 |
+
|
| 537 |
+
# In[27]:
|
| 538 |
+
|
| 539 |
+
|
| 540 |
+
for dataset in full_data:
|
| 541 |
+
dataset['Title'] = dataset['Title'].replace(['Mrs', 'Miss'], 'MM')
|
| 542 |
+
dataset['Title'] = dataset['Title'].replace(['Dr', 'Major', 'Col'], 'DMC')
|
| 543 |
+
dataset['Title'] = dataset['Title'].replace(['Don', 'Rev', 'Capt', 'Jonkheer'],'DRCJ')
|
| 544 |
+
dataset['Title'] = dataset['Title'].replace(['Mme', 'Ms', 'Lady', 'Sir', 'Mlle', 'Countess'],'MMLSMC' )
|
| 545 |
+
# Mapping titles
|
| 546 |
+
title_mapping = {"MM": 1, "Master":2, "Mr": 5, "DMC": 4, "DRCJ": 3, "MMLSMC": 0}
|
| 547 |
+
dataset['Title'] = dataset['Title'].map(title_mapping)
|
| 548 |
+
dataset['Title'] = dataset['Title'].fillna(3)
|
| 549 |
+
|
| 550 |
+
# Explore Age vs Survived
|
| 551 |
+
g = sns.FacetGrid(train, col='Survived')
|
| 552 |
+
g = g.map(sns.distplot, "Age")
|
| 553 |
+
|
| 554 |
+
|
| 555 |
+
# In[28]:
|
| 556 |
+
|
| 557 |
+
|
| 558 |
+
train[["Title", "Survived"]].groupby(['Title'], as_index=False).mean().sort_values(by='Survived', ascending=False)
|
| 559 |
+
|
| 560 |
+
|
| 561 |
+
# ### Extracting deck from cabin
|
| 562 |
+
# A cabin number looks like ‘C123’ and the letter refers to the deck: a big thanks to Nikas Donge.
|
| 563 |
+
# Therefore we’re going to extract these and create a new feature, that contains a persons deck. Afterwords we will convert the feature into a numeric variable. The missing values will be converted to zero.
|
| 564 |
+
|
| 565 |
+
# In[29]:
|
| 566 |
+
|
| 567 |
+
|
| 568 |
+
deck = {"A": 1, "B": 2, "C": 3, "D": 4, "E": 5, "F": 6, "G": 7, "U": 8}
|
| 569 |
+
for dataset in full_data:
|
| 570 |
+
dataset['Cabin'] = dataset['Cabin'].fillna("U0")
|
| 571 |
+
dataset['Deck'] = dataset['Cabin'].map(lambda x: re.compile("([a-zA-Z]+)").search(x).group())
|
| 572 |
+
dataset['Deck'] = dataset['Deck'].map(deck)
|
| 573 |
+
dataset['Deck'] = dataset['Deck'].fillna(0)
|
| 574 |
+
dataset['Deck'] = dataset['Deck'].astype(int)
|
| 575 |
+
train['Deck'].value_counts()
|
| 576 |
+
|
| 577 |
+
|
| 578 |
+
# In[30]:
|
| 579 |
+
|
| 580 |
+
|
| 581 |
+
sns.barplot(x = 'Deck', y = 'Survived', order=[1,2,3,4,5,6,7,8], data=train)
|
| 582 |
+
|
| 583 |
+
|
| 584 |
+
# 3 types of deck: 1 with 15 passengers, 2 to 6, and 7 to 8 (most passengers)
|
| 585 |
+
|
| 586 |
+
# In[31]:
|
| 587 |
+
|
| 588 |
+
|
| 589 |
+
for dataset in full_data:
|
| 590 |
+
dataset.loc[ dataset['Deck'] <= 1, 'Deck'] = 1
|
| 591 |
+
dataset.loc[(dataset['Deck'] > 1) & (dataset['Deck'] <= 6), 'Deck'] = 3
|
| 592 |
+
dataset.loc[ dataset['Deck'] > 6, 'Deck'] = 0
|
| 593 |
+
train[["Deck", "Survived"]].groupby(['Deck'], as_index=False).mean().sort_values(by='Survived', ascending=False)
|
| 594 |
+
|
| 595 |
+
|
| 596 |
+
# ## 2.4 Visualising updated dataset
|
| 597 |
+
|
| 598 |
+
# In[32]:
|
| 599 |
+
|
| 600 |
+
|
| 601 |
+
test.head(5)
|
| 602 |
+
|
| 603 |
+
|
| 604 |
+
# In[33]:
|
| 605 |
+
|
| 606 |
+
|
| 607 |
+
train.head(5)
|
| 608 |
+
|
| 609 |
+
|
| 610 |
+
# ## 2.5. Descriptive statistics
|
| 611 |
+
|
| 612 |
+
# In[34]:
|
| 613 |
+
|
| 614 |
+
|
| 615 |
+
train.describe()
|
| 616 |
+
|
| 617 |
+
|
| 618 |
+
# In[35]:
|
| 619 |
+
|
| 620 |
+
|
| 621 |
+
train[['Pclass', 'Sex', 'Age', 'Parch', 'Fare', 'Embarked', 'Has_Cabin', 'FamilySize', 'Title', 'Survived']].groupby(['Survived'], as_index=False).mean().sort_values(by='Pclass', ascending=False)
|
| 622 |
+
|
| 623 |
+
|
| 624 |
+
# **Initial observations from the descriptive statistics:**
|
| 625 |
+
# * Only 38% survived, a real tragedy :-(
|
| 626 |
+
# * Passengers in more expensive classes 1 and 2 had much higher chance of surviving than classes 3 or 4.
|
| 627 |
+
# * Also, the higher the fare, the higher the chance. Similarly, having a cabin increases the chance of survival.
|
| 628 |
+
# * Women (0) higher chance than men (1)
|
| 629 |
+
# * Younger people slightly more chance than older
|
| 630 |
+
# * Being alone decreased your chance to survive.
|
| 631 |
+
#
|
| 632 |
+
# We will drop unncessary features just before Section 3.1. Pearson Correlation heatmap.
|
| 633 |
+
|
| 634 |
+
# # 3. Correlation analysis - Multi-variate analysis
|
| 635 |
+
# This section summarizes bivariate analysis asthe simplest forms of quantitative (statistical) analysis.
|
| 636 |
+
# It involves the analysis of one or two features, and their relative impact of "Survived".
|
| 637 |
+
# This is a useful frist step of our anblaysis in order to determine the empirical relationship between all features.
|
| 638 |
+
|
| 639 |
+
# ## 3.1. Correlation analysis with histograms and pivot-tables
|
| 640 |
+
|
| 641 |
+
# In[36]:
|
| 642 |
+
|
| 643 |
+
|
| 644 |
+
fig, (axis1,axis2) = plt.subplots(1,2,figsize=(18,6))
|
| 645 |
+
sns.barplot(x="Embarked", y="Survived", hue="Sex", data=train, ax = axis1);
|
| 646 |
+
sns.barplot(x="Age", y="Survived", hue="Sex", data=train, ax = axis2);
|
| 647 |
+
|
| 648 |
+
|
| 649 |
+
# **Observations for Age graph:**
|
| 650 |
+
# * 0 or blue represent women; 1 or orange represent men. Gender and age seem to have a stronger influece of the survival rate.
|
| 651 |
+
# * We start to find where most survivors are: older women (48 to 64 year old), and younger passengers.
|
| 652 |
+
# * What is statistically interesting is that only young boys (Age Category = 0) have high survival rates, unlike other age groups for men.
|
| 653 |
+
# * We will create a new feature called young boys
|
| 654 |
+
|
| 655 |
+
# In[37]:
|
| 656 |
+
|
| 657 |
+
|
| 658 |
+
# for dataset in full_data:
|
| 659 |
+
# dataset['Boys'] = 0
|
| 660 |
+
# dataset.loc[(dataset['Age'] == 0) & (dataset['Sex']==1), 'Boys'] = 1
|
| 661 |
+
# dataset['Boys'].value_counts()
|
| 662 |
+
|
| 663 |
+
|
| 664 |
+
# In[38]:
|
| 665 |
+
|
| 666 |
+
|
| 667 |
+
train[["FamilySize", "Survived"]].groupby(['FamilySize'], as_index=False).mean().sort_values(by='Survived', ascending=False)
|
| 668 |
+
|
| 669 |
+
|
| 670 |
+
# In[39]:
|
| 671 |
+
|
| 672 |
+
|
| 673 |
+
for dataset in full_data:
|
| 674 |
+
dataset['Gender_Embarked'] = 0
|
| 675 |
+
dataset.loc[(dataset['Sex']==0) & (dataset['Embarked']==0), 'Gender_Embarked'] = 0
|
| 676 |
+
dataset.loc[(dataset['Sex']==0) & (dataset['Embarked']==2), 'Gender_Embarked'] = 1
|
| 677 |
+
dataset.loc[(dataset['Sex']==0) & (dataset['Embarked']==1), 'Gender_Embarked'] = 2
|
| 678 |
+
dataset.loc[(dataset['Sex']==1) & (dataset['Embarked']==2), 'Gender_Embarked'] = 3
|
| 679 |
+
dataset.loc[(dataset['Sex']==1) & (dataset['Embarked']==0), 'Gender_Embarked'] = 4
|
| 680 |
+
dataset.loc[(dataset['Sex']==1) & (dataset['Embarked']==1), 'Gender_Embarked'] = 5
|
| 681 |
+
train[["Gender_Embarked", "Survived"]].groupby(['Gender_Embarked'], as_index=False).mean().sort_values(by='Survived', ascending=False)
|
| 682 |
+
|
| 683 |
+
|
| 684 |
+
# In[40]:
|
| 685 |
+
|
| 686 |
+
|
| 687 |
+
train_pivot = pd.pivot_table(train, values= 'Survived',index=['Title', 'Pclass'],columns='Sex',aggfunc=np.mean, margins=True)
|
| 688 |
+
def color_negative_red(val):
|
| 689 |
+
# Takes a scalar and returns a string with the css property 'color: red' if below 0.4, black otherwise.
|
| 690 |
+
color = 'red' if val < 0.4 else 'black'
|
| 691 |
+
return 'color: %s' % color
|
| 692 |
+
train_pivot = train_pivot.style.applymap(color_negative_red)
|
| 693 |
+
train_pivot
|
| 694 |
+
|
| 695 |
+
|
| 696 |
+
# In[41]:
|
| 697 |
+
|
| 698 |
+
|
| 699 |
+
# grid = sns.FacetGrid(train_df, col='Pclass', hue='Survived')
|
| 700 |
+
grid = sns.FacetGrid(train, col='Survived', row='Pclass', size=2, aspect=3)
|
| 701 |
+
grid.map(plt.hist, 'Age', alpha=.5, bins=8)
|
| 702 |
+
grid.add_legend();
|
| 703 |
+
|
| 704 |
+
|
| 705 |
+
# **Observations: here are the survivors!**
|
| 706 |
+
# 1. Family-size of 3 or 4 from first pivot
|
| 707 |
+
# 2. Women and men alone on first class (second pivot, red showing survival rate below 0.4)
|
| 708 |
+
# 3. Top-right in the graph above: first class and age categories 1 and 2
|
| 709 |
+
#
|
| 710 |
+
# ** The not-so lucky are mostly in men, Pclass 3 and age category 1 (younger folks)**
|
| 711 |
+
|
| 712 |
+
# In[42]:
|
| 713 |
+
|
| 714 |
+
|
| 715 |
+
#graph distribution of qualitative data: Pclass
|
| 716 |
+
fig, (axis1,axis2,axis3) = plt.subplots(1,3,figsize=(18,8))
|
| 717 |
+
|
| 718 |
+
sns.boxplot(x = 'Pclass', y = 'Fare', hue = 'Survived', data = train, ax = axis1)
|
| 719 |
+
axis1.set_title('Pclass vs Fare Survival Comparison')
|
| 720 |
+
|
| 721 |
+
sns.violinplot(x = 'Pclass', y = 'Age', hue = 'Survived', data = train, split = True, ax = axis2)
|
| 722 |
+
axis2.set_title('Pclass vs Age Survival Comparison')
|
| 723 |
+
|
| 724 |
+
sns.boxplot(x = 'Pclass', y ='FamilySize', hue = 'Survived', data = train, ax = axis3)
|
| 725 |
+
axis3.set_title('Pclass vs Family Size Survival Comparison')
|
| 726 |
+
|
| 727 |
+
|
| 728 |
+
# In[43]:
|
| 729 |
+
|
| 730 |
+
|
| 731 |
+
fig, saxis = plt.subplots(2, 3,figsize=(18,8))
|
| 732 |
+
|
| 733 |
+
sns.barplot(x = 'Embarked', y = 'Survived', data=train, ax = saxis[0,0])
|
| 734 |
+
sns.barplot(x = 'Pclass', y = 'Survived', order=[1,2,3], data=train, ax = saxis[0,1])
|
| 735 |
+
sns.barplot(x = 'Deck', y = 'Survived', order=[1,0], data=train, ax = saxis[0,2])
|
| 736 |
+
|
| 737 |
+
sns.pointplot(x = 'Fare', y = 'Survived', data=train, ax = saxis[1,0])
|
| 738 |
+
sns.pointplot(x = 'Age', y = 'Survived', data=train, ax = saxis[1,1])
|
| 739 |
+
sns.pointplot(x = 'FamilySize', y = 'Survived', data=train, ax = saxis[1,2])
|
| 740 |
+
|
| 741 |
+
|
| 742 |
+
# In[44]:
|
| 743 |
+
|
| 744 |
+
|
| 745 |
+
# grid = sns.FacetGrid(train_df, col='Embarked')
|
| 746 |
+
grid = sns.FacetGrid(train, row='Has_Cabin', size=2.2, aspect=1.2)
|
| 747 |
+
grid.map(sns.pointplot, 'Parch', 'Survived', 'Sex', palette='deep')
|
| 748 |
+
grid.add_legend()
|
| 749 |
+
|
| 750 |
+
|
| 751 |
+
# **Observations:**
|
| 752 |
+
# * The colors represent: blue=0 is for women, green=1 for men
|
| 753 |
+
# * Clearly, women had more chance of surviving, with or without cabin
|
| 754 |
+
# * Interesting is that accompanied women without a cabin had less survival chance than women alone without cabin.
|
| 755 |
+
# But this is not true for men. Men alone have less chance than accompanied.
|
| 756 |
+
#
|
| 757 |
+
# **Bottom-line: it would have been better for women without cabin to pretend that they were alone.
|
| 758 |
+
# And lone men should join a family to improve their survival rates.**
|
| 759 |
+
|
| 760 |
+
# ## 3.2. Dropping features
|
| 761 |
+
# Bottom-line of the bi-variate and tri-variate analysis as well as the feature importance analysis (from running the classifiers multiple times), **I decided to drop less-relevant features**. This happened as an iterative process by reviwing the outcome of the feature importance graph in the next section.
|
| 762 |
+
# The problem with less important features is that they create more noice and actually take over the importance of real features like Sex and Pclass.
|
| 763 |
+
#
|
| 764 |
+
# **The next step after dropping less-relevant features is to scale them, a very good recommendation from Konstantin's kernel**
|
| 765 |
+
# It helps to boost the score. Scaling features is helpful for many ML algorithms like KNN for example, it really boosts their score.
|
| 766 |
+
# Feature scaling is a method used to standardize the range of independent variables or features of data. In data processing, it is also known as data normalization.
|
| 767 |
+
# Feature standardization makes the values of each feature in the data have zero-mean (when subtracting the mean in the numerator) and unit-variance.
|
| 768 |
+
# The general method of calculation is to determine the distribution mean and standard deviation for each feature. Next we subtract the mean from each feature. Then we divide the values (mean is already subtracted) of each feature by its standard deviation.
|
| 769 |
+
|
| 770 |
+
# In[45]:
|
| 771 |
+
|
| 772 |
+
|
| 773 |
+
# Feature selection
|
| 774 |
+
drop_elements = ['PassengerId', 'Name', 'Ticket', 'Cabin', 'SibSp', 'Boys', 'IsAlone', 'Embarked']
|
| 775 |
+
|
| 776 |
+
train = train.drop(drop_elements, axis = 1)
|
| 777 |
+
test = test.drop(drop_elements, axis = 1)
|
| 778 |
+
|
| 779 |
+
|
| 780 |
+
# ## 3.3. Pearson Correlation Heatmap
|
| 781 |
+
#
|
| 782 |
+
# The Seaborn plotting package allows us to plot heatmaps showing the Pearson product-moment correlation coefficient (PPMCC) correlation between features.
|
| 783 |
+
# Pearson is bivariate correlation, measuring the linear correlation between two features.
|
| 784 |
+
|
| 785 |
+
# In[46]:
|
| 786 |
+
|
| 787 |
+
|
| 788 |
+
colormap = plt.cm.RdBu
|
| 789 |
+
plt.figure(figsize=(14,12))
|
| 790 |
+
plt.title('Pearson Correlation of Features', y=1.05, size=15)
|
| 791 |
+
sns.heatmap(train.astype(float).corr(),linewidths=0.1,vmax=1.0, square=True, cmap=colormap, linecolor='white', annot=True)
|
| 792 |
+
|
| 793 |
+
|
| 794 |
+
# **Observations from the Pearson analysis:**
|
| 795 |
+
# * Correlation coefficients with magnitude between 0.5 and 0.7 indicate variables which can be considered **moderately correlated**.
|
| 796 |
+
# * We can see from the red cells that many features are "moderately" correlated: specifically, IsAlone, Pclass, Name_length, Fare, Sex.
|
| 797 |
+
# * This is influenced by the following two factors: 1) Women versus men (and the compounding effect of Name_length) and 2) Passengers paying a high price (Fare) have a higher chance of survival: there are also in first class, have a title.
|
| 798 |
+
#
|
| 799 |
+
#
|
| 800 |
+
# ## 3.4. Pairplots
|
| 801 |
+
#
|
| 802 |
+
# Finally let us generate some pairplots to observe the distribution of data from one feature to the other.
|
| 803 |
+
# The Seaborn pairplot class will help us visualize the distribution of a feature in relationship to each others.
|
| 804 |
+
|
| 805 |
+
# In[47]:
|
| 806 |
+
|
| 807 |
+
|
| 808 |
+
g = sns.pairplot(train[[u'Survived', u'Pclass', u'Sex', u'Age', u'Fare',
|
| 809 |
+
u'FamilySize', u'Title']], hue='Survived', palette = 'seismic',size=1.2,diag_kind = 'kde',diag_kws=dict(shade=True),plot_kws=dict(s=10) )
|
| 810 |
+
g.set(xticklabels=[])
|
| 811 |
+
|
| 812 |
+
|
| 813 |
+
# **Observations**
|
| 814 |
+
# * The pairplot graph all trivariate analysis into one figure.
|
| 815 |
+
# * The clustering of red dots indicates the combination of two features results in higher survival rates, or the opposite (clustering of blue dots = lower survival)
|
| 816 |
+
# For example:
|
| 817 |
+
# - Smaller family sizes in first and second class
|
| 818 |
+
# - Middle age with Pclass in third category = only blue dot
|
| 819 |
+
# This can be used to validate that we extracted the right features or help us define new ones.
|
| 820 |
+
|
| 821 |
+
# In[48]:
|
| 822 |
+
|
| 823 |
+
|
| 824 |
+
# X_train (all features for training purpose but excluding Survived),
|
| 825 |
+
# Y_train (survival result of X-Train) and test are our 3 main datasets for the next sections
|
| 826 |
+
X_train = train.drop("Survived", axis=1)
|
| 827 |
+
Y_train = train["Survived"]
|
| 828 |
+
X_train.shape, Y_train.shape, test.shape
|
| 829 |
+
|
| 830 |
+
from sklearn.cross_validation import train_test_split
|
| 831 |
+
X_train, x_test, Y_train, y_test = train_test_split(X_train, Y_train, test_size=0.3, random_state=101)
|
| 832 |
+
|
| 833 |
+
X_test = test.copy() # test data for Kaggle submission
|
| 834 |
+
#std_scaler = StandardScaler()
|
| 835 |
+
#X_train = std_scaler.fit_transform(X_train)
|
| 836 |
+
#X_test = std_scaler.transform(X_test)
|
| 837 |
+
|
| 838 |
+
|
| 839 |
+
# # 4. Predictive modelling, cross-validation, hyperparameters and ensembling
|
| 840 |
+
#
|
| 841 |
+
# * 4.1. Logistic Regression
|
| 842 |
+
# * 4.2. Support Vector Machines (supervised)
|
| 843 |
+
# * 4.3. k-Nearest Neighbors algorithm (k-NN)
|
| 844 |
+
# * 4.4. Naive Bayes classifier
|
| 845 |
+
# * 4.5. Perceptron
|
| 846 |
+
# * 4.6 Linear SVC
|
| 847 |
+
# * 4.7 Stochastic Gradient Descent
|
| 848 |
+
# * 4.8. Decision tree
|
| 849 |
+
# * 4.9 Random Forrest
|
| 850 |
+
# * 4.10 Model summary
|
| 851 |
+
# * 4.11. Model cross-validation with K-Fold
|
| 852 |
+
# * 4.12 Hyperparameter tuning & learning curves for selected classifiers
|
| 853 |
+
# * 4.13 Selecting and combining the best classifiers
|
| 854 |
+
# * 4.14 Ensembling
|
| 855 |
+
# * 4.15. Summary of most important features
|
| 856 |
+
#
|
| 857 |
+
# ## 4.1. Logistic Regression
|
| 858 |
+
# Logistic regression measures the relationship between the categorical dependent feature (in our case Survived) and the other independent features.
|
| 859 |
+
# It estimates probabilities using a cumulative logistic distribution:
|
| 860 |
+
# * The first value shows the accuracy of this model
|
| 861 |
+
# * The table after this shows the importance of each feature according this classifier.
|
| 862 |
+
|
| 863 |
+
# In[49]:
|
| 864 |
+
|
| 865 |
+
|
| 866 |
+
logreg = LogisticRegression()
|
| 867 |
+
logreg.fit(X_train, Y_train)
|
| 868 |
+
Y_pred1 = logreg.predict(x_test)
|
| 869 |
+
acc_log = round(logreg.score(x_test, y_test) * 100, 2)
|
| 870 |
+
acc_log
|
| 871 |
+
|
| 872 |
+
|
| 873 |
+
# In[50]:
|
| 874 |
+
|
| 875 |
+
|
| 876 |
+
from sklearn.metrics import confusion_matrix, classification_report
|
| 877 |
+
print(classification_report(y_test, Y_pred1))
|
| 878 |
+
cm = pd.DataFrame(confusion_matrix(y_test, Y_pred1), ['Actual: NOT', 'Actual: SURVIVED'], ['Predicted: NO', 'Predicted: SURVIVED'])
|
| 879 |
+
print(cm)
|
| 880 |
+
|
| 881 |
+
|
| 882 |
+
# In[51]:
|
| 883 |
+
|
| 884 |
+
|
| 885 |
+
#coeff_df = pd.DataFrame(X_train.columns.delete(0))
|
| 886 |
+
#coeff_df.columns = ['Feature']
|
| 887 |
+
#coeff_df["Correlation"] = pd.Series(logreg.coef_[0])
|
| 888 |
+
#coeff_df.sort_values(by='Correlation', ascending=False)
|
| 889 |
+
|
| 890 |
+
|
| 891 |
+
# **Observation:**
|
| 892 |
+
# * This classfier confirms the importance of Name_length
|
| 893 |
+
# * FamilySize did not show a strong Pearson correlation with Survived but comes here to the top. This can be due to its strong relationship with other features such as Is_Alone or Parch (Parent-Children).
|
| 894 |
+
#
|
| 895 |
+
#
|
| 896 |
+
# ## 4.2. Support Vector Machines (supervised)
|
| 897 |
+
# Given a set of training samples, each sample is marked as belonging to one or the other of two categories.
|
| 898 |
+
#
|
| 899 |
+
# The SVM training algorithm builds a model that assigns new test samples to one category or the other, making it a non-probabilistic binary linear classifier.
|
| 900 |
+
|
| 901 |
+
# In[52]:
|
| 902 |
+
|
| 903 |
+
|
| 904 |
+
svc=SVC()
|
| 905 |
+
svc.fit(X_train, Y_train)
|
| 906 |
+
Y_pred2 = svc.predict(x_test)
|
| 907 |
+
acc_svc = round(svc.score(x_test, y_test) * 100, 2)
|
| 908 |
+
acc_svc
|
| 909 |
+
|
| 910 |
+
|
| 911 |
+
# In[53]:
|
| 912 |
+
|
| 913 |
+
|
| 914 |
+
print(classification_report(y_test, Y_pred2))
|
| 915 |
+
cm = pd.DataFrame(confusion_matrix(y_test, Y_pred2), ['Actual: NOT', 'Actual: SURVIVED'], ['Predicted: NO', 'Predicted: SURVIVED'])
|
| 916 |
+
print(cm)
|
| 917 |
+
|
| 918 |
+
|
| 919 |
+
# ## 4.3. k-Nearest Neighbors algorithm (k-NN)
|
| 920 |
+
# This is a non-parametric method used for classification and regression.
|
| 921 |
+
# A sample is classified by a majority vote of its neighbors, with the sample being assigned to the class most common among its k nearest neighbors (k is a positive integer, typically small). If k = 1, then the object is simply assigned to the class of that single nearest neighbor.
|
| 922 |
+
|
| 923 |
+
# In[54]:
|
| 924 |
+
|
| 925 |
+
|
| 926 |
+
knn = KNeighborsClassifier(algorithm='auto', leaf_size=26, metric='minkowski',
|
| 927 |
+
metric_params=None, n_jobs=1, n_neighbors=10, p=2,
|
| 928 |
+
weights='uniform')
|
| 929 |
+
knn.fit(X_train, Y_train)
|
| 930 |
+
knn_predictions = knn.predict(x_test)
|
| 931 |
+
acc_knn = round(knn.score(x_test, y_test) * 100, 2)
|
| 932 |
+
|
| 933 |
+
# Preparing data for Submission 1
|
| 934 |
+
test_Survived = pd.Series(knn_predictions, name="Survived")
|
| 935 |
+
Submission1 = pd.concat([PassengerId,test_Survived],axis=1)
|
| 936 |
+
acc_knn
|
| 937 |
+
|
| 938 |
+
|
| 939 |
+
# In[55]:
|
| 940 |
+
|
| 941 |
+
|
| 942 |
+
print(classification_report(y_test, knn_predictions))
|
| 943 |
+
cm = pd.DataFrame(confusion_matrix(y_test, knn_predictions), ['Actual: NOT', 'Actual: SURVIVED'], ['Predicted: NO', 'Predicted: SURVIVED'])
|
| 944 |
+
print(cm)
|
| 945 |
+
|
| 946 |
+
|
| 947 |
+
# In[56]:
|
| 948 |
+
|
| 949 |
+
|
| 950 |
+
Submission1.head(5)
|
| 951 |
+
|
| 952 |
+
|
| 953 |
+
# In[57]:
|
| 954 |
+
|
| 955 |
+
|
| 956 |
+
## Selecting the right n_neighbors for the k-NN classifier
|
| 957 |
+
x_trainknn, x_testknn, y_trainknn, y_testknn = train_test_split(X_train,Y_train,test_size = .33, random_state = 0)
|
| 958 |
+
nn_scores = []
|
| 959 |
+
best_prediction = [-1,-1]
|
| 960 |
+
for i in range(1,100):
|
| 961 |
+
knn = KNeighborsClassifier(n_neighbors=i, weights='distance', metric='minkowski', p =2)
|
| 962 |
+
knn.fit(x_trainknn, y_trainknn)
|
| 963 |
+
score = accuracy_score(y_testknn, knn.predict(x_testknn))
|
| 964 |
+
#print i, score
|
| 965 |
+
if score > best_prediction[1]:
|
| 966 |
+
best_prediction = [i, score]
|
| 967 |
+
nn_scores.append(score)
|
| 968 |
+
print (best_prediction)
|
| 969 |
+
plt.plot(range(1,100),nn_scores)
|
| 970 |
+
|
| 971 |
+
|
| 972 |
+
# ## 4.4. Naive Bayes classifier
|
| 973 |
+
# This is a family of simple probabilistic classifiers based on applying Bayes' theorem with strong (naive) independence assumptions between the features. Naive Bayes classifiers are highly scalable, requiring a number of parameters linear in the number of features in a learning problem.
|
| 974 |
+
|
| 975 |
+
# In[58]:
|
| 976 |
+
|
| 977 |
+
|
| 978 |
+
gaussian = GaussianNB()
|
| 979 |
+
gaussian.fit(X_train, Y_train)
|
| 980 |
+
Y_pred3 = gaussian.predict(x_test)
|
| 981 |
+
acc_gaussian = round(gaussian.score(x_test, y_test) * 100, 2)
|
| 982 |
+
acc_gaussian
|
| 983 |
+
|
| 984 |
+
|
| 985 |
+
# In[59]:
|
| 986 |
+
|
| 987 |
+
|
| 988 |
+
print(classification_report(y_test, Y_pred3))
|
| 989 |
+
cm = pd.DataFrame(confusion_matrix(y_test, Y_pred3), ['Actual: NOT', 'Actual: SURVIVED'], ['Predicted: NO', 'Predicted: SURVIVED'])
|
| 990 |
+
print(cm)
|
| 991 |
+
|
| 992 |
+
|
| 993 |
+
# ## 4.5. Perceptron
|
| 994 |
+
# This is an algorithm for supervised learning of binary classifiers: like the other classifiers before, it decides whether an input, represented by a vector of numbers, belongs to some specific class or not. It is a type of linear classifier, i.e. a classification algorithm that makes its predictions based on a linear predictor function combining a set of weights with the feature vector. The algorithm allows for online learning, in that it processes elements in the training set one at a time.
|
| 995 |
+
|
| 996 |
+
# In[60]:
|
| 997 |
+
|
| 998 |
+
|
| 999 |
+
perceptron = Perceptron()
|
| 1000 |
+
perceptron.fit(X_train, Y_train)
|
| 1001 |
+
Y_pred4 = perceptron.predict(x_test)
|
| 1002 |
+
acc_perceptron = round(perceptron.score(x_test, y_test) * 100, 2)
|
| 1003 |
+
acc_perceptron
|
| 1004 |
+
|
| 1005 |
+
|
| 1006 |
+
# In[61]:
|
| 1007 |
+
|
| 1008 |
+
|
| 1009 |
+
print(classification_report(y_test, Y_pred4))
|
| 1010 |
+
cm = pd.DataFrame(confusion_matrix(y_test, Y_pred4), ['Actual: NOT', 'Actual: SURVIVED'], ['Predicted: NO', 'Predicted: SURVIVED'])
|
| 1011 |
+
print(cm)
|
| 1012 |
+
|
| 1013 |
+
|
| 1014 |
+
# ## 4.6. Linear SVC
|
| 1015 |
+
# This is another implementation of Support Vector Classification (similar to 4.2.) for the case of a linear kernel.
|
| 1016 |
+
|
| 1017 |
+
# In[62]:
|
| 1018 |
+
|
| 1019 |
+
|
| 1020 |
+
linear_svc = LinearSVC()
|
| 1021 |
+
linear_svc.fit(X_train, Y_train)
|
| 1022 |
+
Y_pred5 = linear_svc.predict(x_test)
|
| 1023 |
+
acc_linear_svc = round(linear_svc.score(x_test, y_test) * 100, 2)
|
| 1024 |
+
acc_linear_svc
|
| 1025 |
+
|
| 1026 |
+
|
| 1027 |
+
# In[63]:
|
| 1028 |
+
|
| 1029 |
+
|
| 1030 |
+
print(classification_report(y_test, Y_pred5))
|
| 1031 |
+
cm = pd.DataFrame(confusion_matrix(y_test, Y_pred5), ['Actual: NOT', 'Actual: SURVIVED'], ['Predicted: NO', 'Predicted: SURVIVED'])
|
| 1032 |
+
print(cm)
|
| 1033 |
+
|
| 1034 |
+
|
| 1035 |
+
# ## 4.7. Stochastic Gradient Descent (sgd)
|
| 1036 |
+
# This is a stochastic approximation of the gradient descent optimization and iterative method for minimizing an objective function that is written as a sum of differentiable functions. In other words, SGD tries to find minima or maxima by iteration.
|
| 1037 |
+
|
| 1038 |
+
# In[64]:
|
| 1039 |
+
|
| 1040 |
+
|
| 1041 |
+
sgd = SGDClassifier()
|
| 1042 |
+
sgd.fit(X_train, Y_train)
|
| 1043 |
+
Y_pred6 = sgd.predict(x_test)
|
| 1044 |
+
acc_sgd = round(sgd.score(x_test, y_test) * 100, 2)
|
| 1045 |
+
acc_sgd
|
| 1046 |
+
|
| 1047 |
+
|
| 1048 |
+
# In[65]:
|
| 1049 |
+
|
| 1050 |
+
|
| 1051 |
+
print(classification_report(y_test, Y_pred6))
|
| 1052 |
+
cm = pd.DataFrame(confusion_matrix(y_test, Y_pred6), ['Actual: NOT', 'Actual: SURVIVED'], ['Predicted: NO', 'Predicted: SURVIVED'])
|
| 1053 |
+
print(cm)
|
| 1054 |
+
|
| 1055 |
+
|
| 1056 |
+
# ## 4.8. Decision tree
|
| 1057 |
+
# This predictive model maps features (tree branches) to conclusions about the target value (tree leaves).
|
| 1058 |
+
#
|
| 1059 |
+
# The target features take a finite set of values are called classification trees; in these tree structures, leaves represent class labels and branches represent conjunctions of features that lead to those class labels. Decision trees where the target variable can take continuous values (typically real numbers) are called regression trees.
|
| 1060 |
+
|
| 1061 |
+
# In[66]:
|
| 1062 |
+
|
| 1063 |
+
|
| 1064 |
+
decision_tree = DecisionTreeClassifier()
|
| 1065 |
+
decision_tree.fit(X_train, Y_train)
|
| 1066 |
+
Y_pred7 = decision_tree.predict(x_test)
|
| 1067 |
+
acc_decision_tree = round(decision_tree.score(x_test, y_test) * 100, 2)
|
| 1068 |
+
acc_decision_tree
|
| 1069 |
+
|
| 1070 |
+
|
| 1071 |
+
# In[67]:
|
| 1072 |
+
|
| 1073 |
+
|
| 1074 |
+
print(classification_report(y_test, Y_pred7))
|
| 1075 |
+
cm = pd.DataFrame(confusion_matrix(y_test, Y_pred7), ['Actual: NOT', 'Actual: SURVIVED'], ['Predicted: NO', 'Predicted: SURVIVED'])
|
| 1076 |
+
print(cm)
|
| 1077 |
+
|
| 1078 |
+
|
| 1079 |
+
# ## 4.9. Random Forests
|
| 1080 |
+
# This is one of the most popular classfier.
|
| 1081 |
+
# Random forests or random decision forests are an ensemble learning method for classification, regression and other tasks, that operate by constructing a multitude of decision trees (n_estimators=100) at training time and outputting the class that is the mode of the classes (classification) or mean prediction (regression) of the individual trees
|
| 1082 |
+
|
| 1083 |
+
# In[68]:
|
| 1084 |
+
|
| 1085 |
+
|
| 1086 |
+
random_forest = RandomForestClassifier(n_estimators=100)
|
| 1087 |
+
random_forest.fit(X_train, Y_train)
|
| 1088 |
+
random_forest_predictions = random_forest.predict(x_test)
|
| 1089 |
+
acc_random_forest = round(random_forest.score(x_test, y_test) * 100, 2)
|
| 1090 |
+
|
| 1091 |
+
|
| 1092 |
+
# Preparing data for Submission 2
|
| 1093 |
+
test_Survived = pd.Series(random_forest_predictions, name="Survived")
|
| 1094 |
+
Submission2 = pd.concat([PassengerId,test_Survived],axis=1)
|
| 1095 |
+
|
| 1096 |
+
acc_random_forest
|
| 1097 |
+
|
| 1098 |
+
|
| 1099 |
+
# In[69]:
|
| 1100 |
+
|
| 1101 |
+
|
| 1102 |
+
print(classification_report(y_test, random_forest_predictions))
|
| 1103 |
+
cm = pd.DataFrame(confusion_matrix(y_test, random_forest_predictions), ['Actual: NOT', 'Actual: SURVIVED'], ['Predicted: NO', 'Predicted: SURVIVED'])
|
| 1104 |
+
print(cm)
|
| 1105 |
+
|
| 1106 |
+
|
| 1107 |
+
# ## 4.10. Model summary
|
| 1108 |
+
# I found that the picture illustrates the various model better than words.
|
| 1109 |
+
# This should be taken with a grain of salt, as the intuition conveyed by these two-dimensional examples does not necessarily carry over to real datasets.
|
| 1110 |
+
# The reality os that the algorithms work with many dimensions (11 in our case).
|
| 1111 |
+
#
|
| 1112 |
+
# But it shows how each classifier algorithm partitions the same data in different ways.
|
| 1113 |
+
# The three rows represent the three different data set on the right.
|
| 1114 |
+
# The plots show training points in solid colors and testing points semi-transparent. The lower right shows the classification accuracy on the test set.
|
| 1115 |
+
#
|
| 1116 |
+
# For instance, the visualization helps understand how RandomForest uses multiple Decision Trees, the linear SVC, or Nearest Neighbors grouping sample by their relative distance to each others.
|
| 1117 |
+
#
|
| 1118 |
+
# 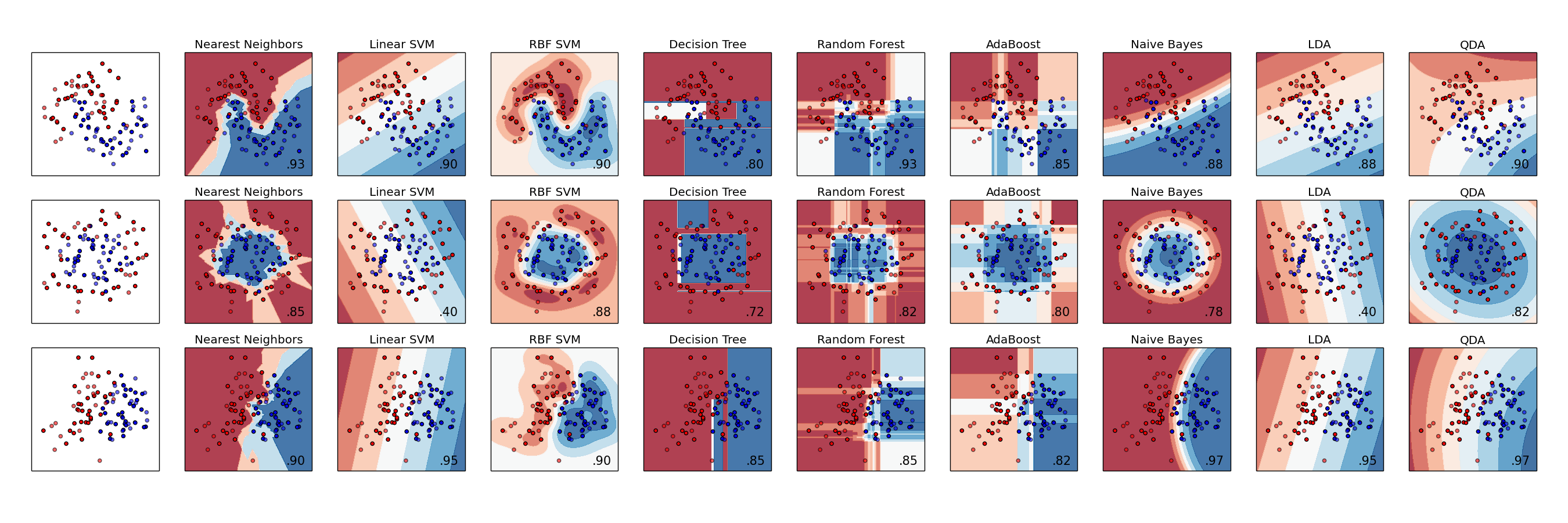
|
| 1119 |
+
#
|
| 1120 |
+
|
| 1121 |
+
# In[70]:
|
| 1122 |
+
|
| 1123 |
+
|
| 1124 |
+
objects = ('Logistic Regression', 'SVC', 'KNN', 'Gaussian', 'Perceptron', 'linear SVC', 'SGD', 'Decision Tree', 'Random Forest')
|
| 1125 |
+
x_pos = np.arange(len(objects))
|
| 1126 |
+
accuracies1 = [acc_log, acc_svc, acc_knn, acc_gaussian, acc_perceptron, acc_linear_svc, acc_sgd, acc_decision_tree, acc_random_forest]
|
| 1127 |
+
|
| 1128 |
+
plt.bar(x_pos, accuracies1, align='center', alpha=0.5, color='r')
|
| 1129 |
+
plt.xticks(x_pos, objects, rotation='vertical')
|
| 1130 |
+
plt.ylabel('Accuracy')
|
| 1131 |
+
plt.title('Classifier Outcome')
|
| 1132 |
+
plt.show()
|
| 1133 |
+
|
| 1134 |
+
|
| 1135 |
+
# **Observations**
|
| 1136 |
+
# * The above models (classifiers) were applied to a split training and x_test datasets.
|
| 1137 |
+
# * This results in some classifiers (Decision_tree and Random_Forest) over-fitting the model to the training data.
|
| 1138 |
+
# * This happens when the classifiers use many input features (to include noise in each feature) on the complete dataset, and ends up “memorizing the noise” instead of finding the signal.
|
| 1139 |
+
# * This overfit model will then make predictions based on that noise. It performs unusually well on its training data, but will not necessarilyimprove the prediction quality with new data from the test dataset.
|
| 1140 |
+
# * In the next section, we will cross-validate the models using sample data against each others. We will this by using StratifiedKFold to train and test the models on sample data from the overall dataset.
|
| 1141 |
+
# Stratified K-Folds is a cross validation iterator. It provides train/test indices to split data in train test sets. This cross-validation object is a variation of KFold, which returns stratified folds. The folds are made by preserving the percentage of samples for each class.
|
| 1142 |
+
|
| 1143 |
+
# ## 4.11. Model cross-validation with K-Fold
|
| 1144 |
+
#
|
| 1145 |
+
# The fitting process applied above optimizes the model parameters to make the model fit the training data as well as possible.
|
| 1146 |
+
# Cross-validation is a way to predict the fit of a model to a hypothetical validation set when an explicit validation set is not available.
|
| 1147 |
+
# In simple words, it allows to test how well the model performs on new data.
|
| 1148 |
+
# In our case, cross-validation will also be applied to compare the performances of different predictive modeling procedures.
|
| 1149 |
+
# 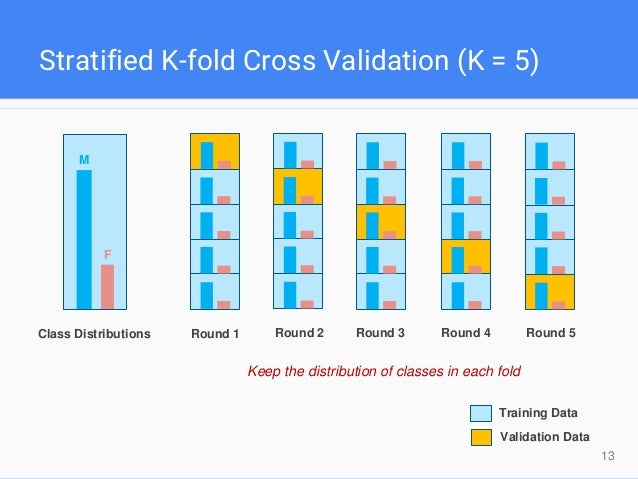
|
| 1150 |
+
# ### Cross-validation scores
|
| 1151 |
+
|
| 1152 |
+
# In[71]:
|
| 1153 |
+
|
| 1154 |
+
|
| 1155 |
+
# Cross validate model with Kfold stratified cross validation
|
| 1156 |
+
from sklearn.model_selection import StratifiedKFold
|
| 1157 |
+
kfold = StratifiedKFold(n_splits=10)
|
| 1158 |
+
# Modeling step Test differents algorithms
|
| 1159 |
+
random_state = 2
|
| 1160 |
+
|
| 1161 |
+
classifiers = []
|
| 1162 |
+
classifiers.append(LogisticRegression(random_state = random_state))
|
| 1163 |
+
classifiers.append(SVC(random_state=random_state))
|
| 1164 |
+
classifiers.append(KNeighborsClassifier())
|
| 1165 |
+
classifiers.append(GaussianNB())
|
| 1166 |
+
classifiers.append(Perceptron(random_state=random_state))
|
| 1167 |
+
classifiers.append(LinearSVC(random_state=random_state))
|
| 1168 |
+
classifiers.append(SGDClassifier(random_state=random_state))
|
| 1169 |
+
classifiers.append(DecisionTreeClassifier(random_state = random_state))
|
| 1170 |
+
classifiers.append(RandomForestClassifier(random_state = random_state))
|
| 1171 |
+
|
| 1172 |
+
cv_results = []
|
| 1173 |
+
for classifier in classifiers :
|
| 1174 |
+
cv_results.append(cross_val_score(classifier, X_train, y = Y_train, scoring = "accuracy", cv = kfold, n_jobs=4))
|
| 1175 |
+
|
| 1176 |
+
cv_means = []
|
| 1177 |
+
cv_std = []
|
| 1178 |
+
for cv_result in cv_results:
|
| 1179 |
+
cv_means.append(cv_result.mean())
|
| 1180 |
+
cv_std.append(cv_result.std())
|
| 1181 |
+
|
| 1182 |
+
cv_res = pd.DataFrame({"CrossValMeans":cv_means,"CrossValerrors": cv_std,"Algorithm":['Logistic Regression', 'KNN', 'Gaussian',
|
| 1183 |
+
'Perceptron', 'linear SVC', 'SGD', 'Decision Tree','SVMC', 'Random Forest']})
|
| 1184 |
+
|
| 1185 |
+
g = sns.barplot("CrossValMeans","Algorithm",data = cv_res, palette="Set3",orient = "h",**{'xerr':cv_std})
|
| 1186 |
+
g.set_xlabel("Mean Accuracy")
|
| 1187 |
+
g = g.set_title("Cross validation scores")
|
| 1188 |
+
|
| 1189 |
+
|
| 1190 |
+
# ## 4.12 Hyperparameter tuning & learning curves for selected classifiers
|
| 1191 |
+
#
|
| 1192 |
+
# **1. Adaboost** is used in conjunction with many other types of learning algorithms to improve performance. The output of the other learning algorithms ('weak learners') is combined into a weighted sum that represents the final output of the boosted classifier. AdaBoost is adaptive in the sense that subsequent weak learners are tweaked in favor of those instances misclassified by previous classifiers. AdaBoost is sensitive to noisy data and outliers.
|
| 1193 |
+
#
|
| 1194 |
+
# **2. ExtraTrees** implements a meta estimator that fits a number of randomized decision trees (a.k.a. extra-trees) on various sub-samples of the dataset and use averaging to improve the predictive accuracy and control over-fitting.
|
| 1195 |
+
#
|
| 1196 |
+
# **3. RandomForest ** operate by constructing a multitude of decision trees at training time and outputting the class that is the mode of the classes (classification) or mean prediction (regression) of the individual trees. Random decision forests correct for decision trees' habit of overfitting to their training set.
|
| 1197 |
+
#
|
| 1198 |
+
# **4. GradientBoost ** produces a prediction model in the form of an ensemble of weak prediction models, typically decision trees. It builds the model in a stage-wise fashion like other boosting methods do, and it generalizes them by allowing optimization of an arbitrary differentiable loss function.
|
| 1199 |
+
#
|
| 1200 |
+
# **5. SVMC, or Support Vector Machines.**vGiven a set of training examples, each marked as belonging to one or the other of two categories, an SVM training algorithm builds a model that assigns new examples to one category or the other, making it a non-probabilistic binary linear classifier.
|
| 1201 |
+
#
|
| 1202 |
+
# All descripotion adapted from Wikipedia.
|
| 1203 |
+
|
| 1204 |
+
# In[72]:
|
| 1205 |
+
|
| 1206 |
+
|
| 1207 |
+
# Adaboost
|
| 1208 |
+
DTC = DecisionTreeClassifier()
|
| 1209 |
+
adaDTC = AdaBoostClassifier(DTC, random_state=7)
|
| 1210 |
+
ada_param_grid = {"base_estimator__criterion" : ["gini", "entropy"],
|
| 1211 |
+
"base_estimator__splitter" : ["best", "random"],
|
| 1212 |
+
"algorithm" : ["SAMME","SAMME.R"],
|
| 1213 |
+
"n_estimators" :[1,2],
|
| 1214 |
+
"learning_rate": [0.0001, 0.001, 0.01, 0.1, 0.2, 0.3,1.5]}
|
| 1215 |
+
gsadaDTC = GridSearchCV(adaDTC,param_grid = ada_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 1216 |
+
gsadaDTC.fit(X_train,Y_train)
|
| 1217 |
+
adaDTC_best = gsadaDTC.best_estimator_
|
| 1218 |
+
gsadaDTC.best_score_
|
| 1219 |
+
|
| 1220 |
+
|
| 1221 |
+
# In[73]:
|
| 1222 |
+
|
| 1223 |
+
|
| 1224 |
+
# ExtraTrees
|
| 1225 |
+
ExtC = ExtraTreesClassifier()
|
| 1226 |
+
ex_param_grid = {"max_depth": [None],
|
| 1227 |
+
"max_features": [1, 3, 7],
|
| 1228 |
+
"min_samples_split": [2, 3, 7],
|
| 1229 |
+
"min_samples_leaf": [1, 3, 7],
|
| 1230 |
+
"bootstrap": [False],
|
| 1231 |
+
"n_estimators" :[300,600],
|
| 1232 |
+
"criterion": ["gini"]}
|
| 1233 |
+
gsExtC = GridSearchCV(ExtC,param_grid = ex_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 1234 |
+
gsExtC.fit(X_train,Y_train)
|
| 1235 |
+
ExtC_best = gsExtC.best_estimator_
|
| 1236 |
+
gsExtC.best_score_
|
| 1237 |
+
|
| 1238 |
+
|
| 1239 |
+
# In[74]:
|
| 1240 |
+
|
| 1241 |
+
|
| 1242 |
+
# Gradient boosting tunning
|
| 1243 |
+
GBC = GradientBoostingClassifier()
|
| 1244 |
+
gb_param_grid = {'loss' : ["deviance"],
|
| 1245 |
+
'n_estimators' : [100,200,300],
|
| 1246 |
+
'learning_rate': [0.1, 0.05, 0.01],
|
| 1247 |
+
'max_depth': [4, 8],
|
| 1248 |
+
'min_samples_leaf': [100,150],
|
| 1249 |
+
'max_features': [0.3, 0.1] }
|
| 1250 |
+
gsGBC = GridSearchCV(GBC,param_grid = gb_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 1251 |
+
gsGBC.fit(X_train,Y_train)
|
| 1252 |
+
GBC_best = gsGBC.best_estimator_
|
| 1253 |
+
gsGBC.best_score_
|
| 1254 |
+
|
| 1255 |
+
|
| 1256 |
+
# In[75]:
|
| 1257 |
+
|
| 1258 |
+
|
| 1259 |
+
# SVC classifier
|
| 1260 |
+
SVMC = SVC(probability=True)
|
| 1261 |
+
svc_param_grid = {'kernel': ['rbf'],
|
| 1262 |
+
'gamma': [ 0.001, 0.01, 0.1, 1],
|
| 1263 |
+
'C': [1,10,50,100,200,300, 1000]}
|
| 1264 |
+
gsSVMC = GridSearchCV(SVMC,param_grid = svc_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 1265 |
+
gsSVMC.fit(X_train,Y_train)
|
| 1266 |
+
SVMC_best = gsSVMC.best_estimator_
|
| 1267 |
+
# Best score
|
| 1268 |
+
gsSVMC.best_score_
|
| 1269 |
+
|
| 1270 |
+
|
| 1271 |
+
# In[76]:
|
| 1272 |
+
|
| 1273 |
+
|
| 1274 |
+
# Random Forest
|
| 1275 |
+
rf_param_grid = {"max_depth": [None],
|
| 1276 |
+
"max_features": [1, 3, 7],
|
| 1277 |
+
"min_samples_split": [2, 3, 7],
|
| 1278 |
+
"min_samples_leaf": [1, 3, 7],
|
| 1279 |
+
"bootstrap": [False],
|
| 1280 |
+
"n_estimators" :[300,600],
|
| 1281 |
+
"criterion": ["gini"]}
|
| 1282 |
+
gsrandom_forest = GridSearchCV(random_forest,param_grid = rf_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 1283 |
+
gsrandom_forest.fit(X_train,Y_train)
|
| 1284 |
+
# Best score
|
| 1285 |
+
random_forest_best = gsrandom_forest.best_estimator_
|
| 1286 |
+
gsrandom_forest.best_score_
|
| 1287 |
+
|
| 1288 |
+
|
| 1289 |
+
# In[77]:
|
| 1290 |
+
|
| 1291 |
+
|
| 1292 |
+
def plot_learning_curve(estimator, title, X, y, ylim=None, cv=None,
|
| 1293 |
+
n_jobs=-1, train_sizes=np.linspace(.1, 1.0, 5)):
|
| 1294 |
+
"""Generate a simple plot of the test and training learning curve"""
|
| 1295 |
+
plt.figure()
|
| 1296 |
+
plt.title(title)
|
| 1297 |
+
if ylim is not None:
|
| 1298 |
+
plt.ylim(*ylim)
|
| 1299 |
+
plt.xlabel("Training examples")
|
| 1300 |
+
plt.ylabel("Score")
|
| 1301 |
+
train_sizes, train_scores, test_scores = learning_curve(
|
| 1302 |
+
estimator, X, y, cv=cv, n_jobs=n_jobs, train_sizes=train_sizes)
|
| 1303 |
+
train_scores_mean = np.mean(train_scores, axis=1)
|
| 1304 |
+
train_scores_std = np.std(train_scores, axis=1)
|
| 1305 |
+
test_scores_mean = np.mean(test_scores, axis=1)
|
| 1306 |
+
test_scores_std = np.std(test_scores, axis=1)
|
| 1307 |
+
plt.grid()
|
| 1308 |
+
|
| 1309 |
+
plt.fill_between(train_sizes, train_scores_mean - train_scores_std,
|
| 1310 |
+
train_scores_mean + train_scores_std, alpha=0.1,
|
| 1311 |
+
color="r")
|
| 1312 |
+
plt.fill_between(train_sizes, test_scores_mean - test_scores_std,
|
| 1313 |
+
test_scores_mean + test_scores_std, alpha=0.1, color="g")
|
| 1314 |
+
plt.plot(train_sizes, train_scores_mean, 'o-', color="r",
|
| 1315 |
+
label="Training score")
|
| 1316 |
+
plt.plot(train_sizes, test_scores_mean, 'o-', color="g",
|
| 1317 |
+
label="Cross-validation score")
|
| 1318 |
+
plt.legend(loc="best")
|
| 1319 |
+
return plt
|
| 1320 |
+
g = plot_learning_curve(gsadaDTC.best_estimator_,"AdaBoost learning curves",X_train,Y_train,cv=kfold)
|
| 1321 |
+
g = plot_learning_curve(gsExtC.best_estimator_,"ExtC ExtraTrees learning curves",X_train,Y_train,cv=kfold)
|
| 1322 |
+
g = plot_learning_curve(gsGBC.best_estimator_,"GBC Gradient Boost learning curves",X_train,Y_train,cv=kfold)
|
| 1323 |
+
g = plot_learning_curve(gsrandom_forest.best_estimator_,"RandomForest learning curves",X_train,Y_train,cv=kfold)
|
| 1324 |
+
g = plot_learning_curve(gsSVMC.best_estimator_,"SVMC learning curves",X_train,Y_train,cv=kfold)
|
| 1325 |
+
|
| 1326 |
+
|
| 1327 |
+
# **Observations to fine-tune our models**
|
| 1328 |
+
#
|
| 1329 |
+
# First, let's compare their best score after fine-tuning their parameters:
|
| 1330 |
+
# 1. Adaboost: 80
|
| 1331 |
+
# 2. ExtraTrees: 83
|
| 1332 |
+
# 3. RandomForest: 82
|
| 1333 |
+
# 4. GradientBoost: 82
|
| 1334 |
+
# 5. SVC: 83
|
| 1335 |
+
#
|
| 1336 |
+
# It appears that GBC and SVMC are doing the best job on the Train data. This is good because we want to keep the model as close to the training data as possible. But not too close!
|
| 1337 |
+
# The two major sources of error are bias and variance; as we reduce these two, then we could build more accurate models:
|
| 1338 |
+
#
|
| 1339 |
+
# * **Bias**: The less biased a method, the greater its ability to fit data well.
|
| 1340 |
+
# * **Variance**: with a lower bias comes typically a higher the variance. And therefore the risk that the model will not adapt accurately to new test data.
|
| 1341 |
+
# This is the case here with Gradient Boost: high score but cross-validation is very distant.
|
| 1342 |
+
#
|
| 1343 |
+
# The reverse also holds: the greater the bias, the lower the variance. A high-bias method builds simplistic models that generally don't fit well training data.
|
| 1344 |
+
# We can see the red and green curves from ExtraTrees, RandomForest and SVC are pretty close.
|
| 1345 |
+
# **This points to a lower variance, i.e. a stronger ability to apply the model to new data.**
|
| 1346 |
+
#
|
| 1347 |
+
# I used the above graphs to optimize the parameters for Adaboost, ExtraTrees, RandomForest, GradientBoost and SVC.
|
| 1348 |
+
# This resulted in a significant improvement of the prediction accuracy on the test data (score).
|
| 1349 |
+
#
|
| 1350 |
+
# In addition, I found out that AdaBoost does not do a good job with this dataset as the training score and cross-validation score are quite far apart.
|
| 1351 |
+
#
|
| 1352 |
+
# ## 4.13 Selecting and combining the best classifiers
|
| 1353 |
+
# So, how do we achieve the best trade-off beween bias and variance?
|
| 1354 |
+
# 1. We will first compare in the next section the classifiers; results between themselves and applied to the same test data.
|
| 1355 |
+
# 2. Then "ensemble" them together with an automatic function callled *voting*.
|
| 1356 |
+
|
| 1357 |
+
# In[78]:
|
| 1358 |
+
|
| 1359 |
+
|
| 1360 |
+
test_Survived_AdaDTC = pd.Series(adaDTC_best.predict(X_test), name="AdaDTC")
|
| 1361 |
+
test_Survived_ExtC = pd.Series(ExtC_best.predict(X_test), name="ExtC")
|
| 1362 |
+
test_Survived_GBC = pd.Series(GBC_best.predict(X_test), name="GBC")
|
| 1363 |
+
test_Survived_SVMC = pd.Series(SVMC_best.predict(X_test), name="SVMC")
|
| 1364 |
+
test_Survived_random_forest = pd.Series(random_forest_best.predict(X_test), name="random_forest")
|
| 1365 |
+
|
| 1366 |
+
# Concatenate all classifier results
|
| 1367 |
+
ensemble_results = pd.concat([test_Survived_AdaDTC, test_Survived_ExtC, test_Survived_GBC,test_Survived_SVMC,test_Survived_random_forest],axis=1)
|
| 1368 |
+
g= sns.heatmap(ensemble_results.corr(),annot=True)
|
| 1369 |
+
|
| 1370 |
+
|
| 1371 |
+
# **Observations:**
|
| 1372 |
+
# * As indicated before, Adaboost has the lowest correlations when compared to other predictors. This indicates that it predicts differently than the others when it comes to the test data.
|
| 1373 |
+
# * We will therefore 'ensemble' the remaining four predictors.
|
| 1374 |
+
#
|
| 1375 |
+
# ## 4.14 Ensembling
|
| 1376 |
+
# This is the final step, pulling it together with an amazing 'Voting' function from sklearn.
|
| 1377 |
+
# An ensemble is a supervised learning algorithm, that it can be trained and then used to make predictions.
|
| 1378 |
+
# The last line applied the "ensemble predictor" to the test data for submission.
|
| 1379 |
+
|
| 1380 |
+
# In[79]:
|
| 1381 |
+
|
| 1382 |
+
|
| 1383 |
+
VotingPredictor = VotingClassifier(estimators=[('ExtC', ExtC_best), ('GBC',GBC_best),
|
| 1384 |
+
('SVMC', SVMC_best), ('random_forest', random_forest_best)], voting='soft', n_jobs=4)
|
| 1385 |
+
VotingPredictor = VotingPredictor.fit(X_train, Y_train)
|
| 1386 |
+
VotingPredictor_predictions = VotingPredictor.predict(test)
|
| 1387 |
+
test_Survived = pd.Series(VotingPredictor_predictions, name="Survived")
|
| 1388 |
+
|
| 1389 |
+
# Preparing data for Submission 3
|
| 1390 |
+
test_Survived = pd.Series(VotingPredictor_predictions, name="Survived")
|
| 1391 |
+
Submission3 = pd.concat([PassengerId,test_Survived],axis=1)
|
| 1392 |
+
Submission3.head(15)
|
| 1393 |
+
|
| 1394 |
+
|
| 1395 |
+
# ## 4.15. Summary of most important features
|
| 1396 |
+
|
| 1397 |
+
# In[80]:
|
| 1398 |
+
|
| 1399 |
+
|
| 1400 |
+
nrows = ncols = 2
|
| 1401 |
+
fig, axes = plt.subplots(nrows = nrows, ncols = ncols, sharex="all", figsize=(15,7))
|
| 1402 |
+
names_classifiers = [("AdaBoosting", adaDTC_best),("ExtraTrees",ExtC_best),
|
| 1403 |
+
("GradientBoosting",GBC_best), ("RandomForest",random_forest_best)]
|
| 1404 |
+
|
| 1405 |
+
nclassifier = 0
|
| 1406 |
+
for row in range(nrows):
|
| 1407 |
+
for col in range(ncols):
|
| 1408 |
+
name = names_classifiers[nclassifier][0]
|
| 1409 |
+
classifier = names_classifiers[nclassifier][1]
|
| 1410 |
+
indices = np.argsort(classifier.feature_importances_)[::-1][:40]
|
| 1411 |
+
g = sns.barplot(y=train.columns[indices][:40],x = classifier.feature_importances_[indices][:40] , orient='h',ax=axes[row][col])
|
| 1412 |
+
g.set_xlabel("Relative importance",fontsize=11)
|
| 1413 |
+
g.set_ylabel("Features",fontsize=11)
|
| 1414 |
+
g.tick_params(labelsize=9)
|
| 1415 |
+
g.set_title(name + " feature importance")
|
| 1416 |
+
nclassifier += 1
|
| 1417 |
+
|
| 1418 |
+
|
| 1419 |
+
# Nice graphics, but the obsevation is unclear in my opinion:
|
| 1420 |
+
# * On one side, we hope as analyst that the models come out with similar patterns. An easy direction to follow.
|
| 1421 |
+
# * At the same time, "there have been quite a few articles and Kaggle competition winner stories about the merits of having trained models that are more uncorrelated with one another producing better scores". As we say in business, diversity brings better results, this seems to be true with algorithms as well!
|
| 1422 |
+
|
| 1423 |
+
# # 5. Producing the submission file for Kaggle
|
| 1424 |
+
#
|
| 1425 |
+
# Finally having trained and fit all our first-level and second-level models, we can now output the predictions into the proper format for submission to the Titanic competition.
|
| 1426 |
+
# Which model to choose? These are the results of my many submissions:
|
| 1427 |
+
#
|
| 1428 |
+
# **Submission 1: **The prediction with **KNeighborsClassifier KNN in Section 4.3.** generates a public score of **0.75119**.
|
| 1429 |
+
#
|
| 1430 |
+
# **Submission 2:** The prediction with **random_forest in Section 4.9** generates a public score of **0.73684**.
|
| 1431 |
+
#
|
| 1432 |
+
# **Submission 3 (Kaggle Version 85):** The prediction with **gsrandom_forest in Section 4.14 ** after stratification and model cross validation, generates a public score of **0.80382**.
|
| 1433 |
+
#
|
| 1434 |
+
# Decision: submit #3 as best predictor
|
| 1435 |
+
|
| 1436 |
+
# In[81]:
|
| 1437 |
+
|
| 1438 |
+
|
| 1439 |
+
# Submit File
|
| 1440 |
+
Submission3.to_csv("StackingSubmission.csv", index=False)
|
| 1441 |
+
print("Completed.")
|
| 1442 |
+
|
| 1443 |
+
|
| 1444 |
+
# # 6. Credits
|
| 1445 |
+
# **Huge credits to Anisotropic, Yassine Ghouzam, Faron and Sina** for pulling together most of the code in this kernel.
|
Titanic/Kernels/ExtraTrees/2-titanic-top-4-with-ensemble-modeling.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/2-titanic-top-4-with-ensemble-modeling.py
ADDED
|
@@ -0,0 +1,1110 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python
|
| 2 |
+
# coding: utf-8
|
| 3 |
+
|
| 4 |
+
# # Titanic Top 4% with ensemble modeling
|
| 5 |
+
# ### **Yassine Ghouzam, PhD**
|
| 6 |
+
# #### 13/07/2017
|
| 7 |
+
#
|
| 8 |
+
# * **1 Introduction**
|
| 9 |
+
# * **2 Load and check data**
|
| 10 |
+
# * 2.1 load data
|
| 11 |
+
# * 2.2 Outlier detection
|
| 12 |
+
# * 2.3 joining train and test set
|
| 13 |
+
# * 2.4 check for null and missing values
|
| 14 |
+
# * **3 Feature analysis**
|
| 15 |
+
# * 3.1 Numerical values
|
| 16 |
+
# * 3.2 Categorical values
|
| 17 |
+
# * **4 Filling missing Values**
|
| 18 |
+
# * 4.1 Age
|
| 19 |
+
# * **5 Feature engineering**
|
| 20 |
+
# * 5.1 Name/Title
|
| 21 |
+
# * 5.2 Family Size
|
| 22 |
+
# * 5.3 Cabin
|
| 23 |
+
# * 5.4 Ticket
|
| 24 |
+
# * **6 Modeling**
|
| 25 |
+
# * 6.1 Simple modeling
|
| 26 |
+
# * 6.1.1 Cross validate models
|
| 27 |
+
# * 6.1.2 Hyperparamater tunning for best models
|
| 28 |
+
# * 6.1.3 Plot learning curves
|
| 29 |
+
# * 6.1.4 Feature importance of the tree based classifiers
|
| 30 |
+
# * 6.2 Ensemble modeling
|
| 31 |
+
# * 6.2.1 Combining models
|
| 32 |
+
# * 6.3 Prediction
|
| 33 |
+
# * 6.3.1 Predict and Submit results
|
| 34 |
+
#
|
| 35 |
+
|
| 36 |
+
# ## 1. Introduction
|
| 37 |
+
#
|
| 38 |
+
# This is my first kernel at Kaggle. I choosed the Titanic competition which is a good way to introduce feature engineering and ensemble modeling. Firstly, I will display some feature analyses then ill focus on the feature engineering. Last part concerns modeling and predicting the survival on the Titanic using an voting procedure.
|
| 39 |
+
#
|
| 40 |
+
# This script follows three main parts:
|
| 41 |
+
#
|
| 42 |
+
# * **Feature analysis**
|
| 43 |
+
# * **Feature engineering**
|
| 44 |
+
# * **Modeling**
|
| 45 |
+
|
| 46 |
+
# In[1]:
|
| 47 |
+
|
| 48 |
+
|
| 49 |
+
import pandas as pd
|
| 50 |
+
import numpy as np
|
| 51 |
+
import matplotlib.pyplot as plt
|
| 52 |
+
import seaborn as sns
|
| 53 |
+
get_ipython().run_line_magic('matplotlib', 'inline')
|
| 54 |
+
|
| 55 |
+
from collections import Counter
|
| 56 |
+
|
| 57 |
+
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, GradientBoostingClassifier, ExtraTreesClassifier, VotingClassifier
|
| 58 |
+
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
|
| 59 |
+
from sklearn.linear_model import LogisticRegression
|
| 60 |
+
from sklearn.neighbors import KNeighborsClassifier
|
| 61 |
+
from sklearn.tree import DecisionTreeClassifier
|
| 62 |
+
from sklearn.neural_network import MLPClassifier
|
| 63 |
+
from sklearn.svm import SVC
|
| 64 |
+
from sklearn.model_selection import GridSearchCV, cross_val_score, StratifiedKFold, learning_curve
|
| 65 |
+
|
| 66 |
+
sns.set(style='white', context='notebook', palette='deep')
|
| 67 |
+
|
| 68 |
+
|
| 69 |
+
# ## 2. Load and check data
|
| 70 |
+
# ### 2.1 Load data
|
| 71 |
+
|
| 72 |
+
# In[2]:
|
| 73 |
+
|
| 74 |
+
|
| 75 |
+
# Load data
|
| 76 |
+
##### Load train and Test set
|
| 77 |
+
|
| 78 |
+
train = pd.read_csv("../input/train.csv")
|
| 79 |
+
test = pd.read_csv("../input/test.csv")
|
| 80 |
+
IDtest = test["PassengerId"]
|
| 81 |
+
|
| 82 |
+
|
| 83 |
+
# ### 2.2 Outlier detection
|
| 84 |
+
|
| 85 |
+
# In[3]:
|
| 86 |
+
|
| 87 |
+
|
| 88 |
+
# Outlier detection
|
| 89 |
+
|
| 90 |
+
def detect_outliers(df,n,features):
|
| 91 |
+
"""
|
| 92 |
+
Takes a dataframe df of features and returns a list of the indices
|
| 93 |
+
corresponding to the observations containing more than n outliers according
|
| 94 |
+
to the Tukey method.
|
| 95 |
+
"""
|
| 96 |
+
outlier_indices = []
|
| 97 |
+
|
| 98 |
+
# iterate over features(columns)
|
| 99 |
+
for col in features:
|
| 100 |
+
# 1st quartile (25%)
|
| 101 |
+
Q1 = np.percentile(df[col], 25)
|
| 102 |
+
# 3rd quartile (75%)
|
| 103 |
+
Q3 = np.percentile(df[col],75)
|
| 104 |
+
# Interquartile range (IQR)
|
| 105 |
+
IQR = Q3 - Q1
|
| 106 |
+
|
| 107 |
+
# outlier step
|
| 108 |
+
outlier_step = 1.5 * IQR
|
| 109 |
+
|
| 110 |
+
# Determine a list of indices of outliers for feature col
|
| 111 |
+
outlier_list_col = df[(df[col] < Q1 - outlier_step) | (df[col] > Q3 + outlier_step )].index
|
| 112 |
+
|
| 113 |
+
# append the found outlier indices for col to the list of outlier indices
|
| 114 |
+
outlier_indices.extend(outlier_list_col)
|
| 115 |
+
|
| 116 |
+
# select observations containing more than 2 outliers
|
| 117 |
+
outlier_indices = Counter(outlier_indices)
|
| 118 |
+
multiple_outliers = list( k for k, v in outlier_indices.items() if v > n )
|
| 119 |
+
|
| 120 |
+
return multiple_outliers
|
| 121 |
+
|
| 122 |
+
# detect outliers from Age, SibSp , Parch and Fare
|
| 123 |
+
Outliers_to_drop = detect_outliers(train,2,["Age","SibSp","Parch","Fare"])
|
| 124 |
+
|
| 125 |
+
|
| 126 |
+
# Since outliers can have a dramatic effect on the prediction (espacially for regression problems), i choosed to manage them.
|
| 127 |
+
#
|
| 128 |
+
# I used the Tukey method (Tukey JW., 1977) to detect ouliers which defines an interquartile range comprised between the 1st and 3rd quartile of the distribution values (IQR). An outlier is a row that have a feature value outside the (IQR +- an outlier step).
|
| 129 |
+
#
|
| 130 |
+
#
|
| 131 |
+
# I decided to detect outliers from the numerical values features (Age, SibSp, Sarch and Fare). Then, i considered outliers as rows that have at least two outlied numerical values.
|
| 132 |
+
|
| 133 |
+
# In[4]:
|
| 134 |
+
|
| 135 |
+
|
| 136 |
+
train.loc[Outliers_to_drop] # Show the outliers rows
|
| 137 |
+
|
| 138 |
+
|
| 139 |
+
# We detect 10 outliers. The 28, 89 and 342 passenger have an high Ticket Fare
|
| 140 |
+
#
|
| 141 |
+
# The 7 others have very high values of SibSP.
|
| 142 |
+
|
| 143 |
+
# In[5]:
|
| 144 |
+
|
| 145 |
+
|
| 146 |
+
# Drop outliers
|
| 147 |
+
train = train.drop(Outliers_to_drop, axis = 0).reset_index(drop=True)
|
| 148 |
+
|
| 149 |
+
|
| 150 |
+
# ### 2.3 joining train and test set
|
| 151 |
+
|
| 152 |
+
# In[6]:
|
| 153 |
+
|
| 154 |
+
|
| 155 |
+
## Join train and test datasets in order to obtain the same number of features during categorical conversion
|
| 156 |
+
train_len = len(train)
|
| 157 |
+
dataset = pd.concat(objs=[train, test], axis=0).reset_index(drop=True)
|
| 158 |
+
|
| 159 |
+
|
| 160 |
+
# I join train and test datasets to obtain the same number of features during categorical conversion (See feature engineering).
|
| 161 |
+
|
| 162 |
+
# ### 2.4 check for null and missing values
|
| 163 |
+
|
| 164 |
+
# In[7]:
|
| 165 |
+
|
| 166 |
+
|
| 167 |
+
# Fill empty and NaNs values with NaN
|
| 168 |
+
dataset = dataset.fillna(np.nan)
|
| 169 |
+
|
| 170 |
+
# Check for Null values
|
| 171 |
+
dataset.isnull().sum()
|
| 172 |
+
|
| 173 |
+
|
| 174 |
+
# Age and Cabin features have an important part of missing values.
|
| 175 |
+
#
|
| 176 |
+
# **Survived missing values correspond to the join testing dataset (Survived column doesn't exist in test set and has been replace by NaN values when concatenating the train and test set)**
|
| 177 |
+
|
| 178 |
+
# In[8]:
|
| 179 |
+
|
| 180 |
+
|
| 181 |
+
# Infos
|
| 182 |
+
train.info()
|
| 183 |
+
train.isnull().sum()
|
| 184 |
+
|
| 185 |
+
|
| 186 |
+
# In[9]:
|
| 187 |
+
|
| 188 |
+
|
| 189 |
+
train.head()
|
| 190 |
+
|
| 191 |
+
|
| 192 |
+
# In[10]:
|
| 193 |
+
|
| 194 |
+
|
| 195 |
+
train.dtypes
|
| 196 |
+
|
| 197 |
+
|
| 198 |
+
# In[11]:
|
| 199 |
+
|
| 200 |
+
|
| 201 |
+
### Summarize data
|
| 202 |
+
# Summarie and statistics
|
| 203 |
+
train.describe()
|
| 204 |
+
|
| 205 |
+
|
| 206 |
+
# ## 3. Feature analysis
|
| 207 |
+
# ### 3.1 Numerical values
|
| 208 |
+
|
| 209 |
+
# In[12]:
|
| 210 |
+
|
| 211 |
+
|
| 212 |
+
# Correlation matrix between numerical values (SibSp Parch Age and Fare values) and Survived
|
| 213 |
+
g = sns.heatmap(train[["Survived","SibSp","Parch","Age","Fare"]].corr(),annot=True, fmt = ".2f", cmap = "coolwarm")
|
| 214 |
+
|
| 215 |
+
|
| 216 |
+
# Only Fare feature seems to have a significative correlation with the survival probability.
|
| 217 |
+
#
|
| 218 |
+
# It doesn't mean that the other features are not usefull. Subpopulations in these features can be correlated with the survival. To determine this, we need to explore in detail these features
|
| 219 |
+
|
| 220 |
+
# #### SibSP
|
| 221 |
+
|
| 222 |
+
# In[13]:
|
| 223 |
+
|
| 224 |
+
|
| 225 |
+
# Explore SibSp feature vs Survived
|
| 226 |
+
g = sns.factorplot(x="SibSp",y="Survived",data=train,kind="bar", size = 6 ,
|
| 227 |
+
palette = "muted")
|
| 228 |
+
g.despine(left=True)
|
| 229 |
+
g = g.set_ylabels("survival probability")
|
| 230 |
+
|
| 231 |
+
|
| 232 |
+
# It seems that passengers having a lot of siblings/spouses have less chance to survive
|
| 233 |
+
#
|
| 234 |
+
# Single passengers (0 SibSP) or with two other persons (SibSP 1 or 2) have more chance to survive
|
| 235 |
+
#
|
| 236 |
+
# This observation is quite interesting, we can consider a new feature describing these categories (See feature engineering)
|
| 237 |
+
|
| 238 |
+
# #### Parch
|
| 239 |
+
|
| 240 |
+
# In[14]:
|
| 241 |
+
|
| 242 |
+
|
| 243 |
+
# Explore Parch feature vs Survived
|
| 244 |
+
g = sns.factorplot(x="Parch",y="Survived",data=train,kind="bar", size = 6 ,
|
| 245 |
+
palette = "muted")
|
| 246 |
+
g.despine(left=True)
|
| 247 |
+
g = g.set_ylabels("survival probability")
|
| 248 |
+
|
| 249 |
+
|
| 250 |
+
# Small families have more chance to survive, more than single (Parch 0), medium (Parch 3,4) and large families (Parch 5,6 ).
|
| 251 |
+
#
|
| 252 |
+
# Be carefull there is an important standard deviation in the survival of passengers with 3 parents/children
|
| 253 |
+
|
| 254 |
+
# #### Age
|
| 255 |
+
|
| 256 |
+
# In[15]:
|
| 257 |
+
|
| 258 |
+
|
| 259 |
+
# Explore Age vs Survived
|
| 260 |
+
g = sns.FacetGrid(train, col='Survived')
|
| 261 |
+
g = g.map(sns.distplot, "Age")
|
| 262 |
+
|
| 263 |
+
|
| 264 |
+
# Age distribution seems to be a tailed distribution, maybe a gaussian distribution.
|
| 265 |
+
#
|
| 266 |
+
# We notice that age distributions are not the same in the survived and not survived subpopulations. Indeed, there is a peak corresponding to young passengers, that have survived. We also see that passengers between 60-80 have less survived.
|
| 267 |
+
#
|
| 268 |
+
# So, even if "Age" is not correlated with "Survived", we can see that there is age categories of passengers that of have more or less chance to survive.
|
| 269 |
+
#
|
| 270 |
+
# It seems that very young passengers have more chance to survive.
|
| 271 |
+
|
| 272 |
+
# In[16]:
|
| 273 |
+
|
| 274 |
+
|
| 275 |
+
# Explore Age distibution
|
| 276 |
+
g = sns.kdeplot(train["Age"][(train["Survived"] == 0) & (train["Age"].notnull())], color="Red", shade = True)
|
| 277 |
+
g = sns.kdeplot(train["Age"][(train["Survived"] == 1) & (train["Age"].notnull())], ax =g, color="Blue", shade= True)
|
| 278 |
+
g.set_xlabel("Age")
|
| 279 |
+
g.set_ylabel("Frequency")
|
| 280 |
+
g = g.legend(["Not Survived","Survived"])
|
| 281 |
+
|
| 282 |
+
|
| 283 |
+
# When we superimpose the two densities , we cleary see a peak correponsing (between 0 and 5) to babies and very young childrens.
|
| 284 |
+
|
| 285 |
+
# #### Fare
|
| 286 |
+
|
| 287 |
+
# In[17]:
|
| 288 |
+
|
| 289 |
+
|
| 290 |
+
dataset["Fare"].isnull().sum()
|
| 291 |
+
|
| 292 |
+
|
| 293 |
+
# In[18]:
|
| 294 |
+
|
| 295 |
+
|
| 296 |
+
#Fill Fare missing values with the median value
|
| 297 |
+
dataset["Fare"] = dataset["Fare"].fillna(dataset["Fare"].median())
|
| 298 |
+
|
| 299 |
+
|
| 300 |
+
# Since we have one missing value , i decided to fill it with the median value which will not have an important effect on the prediction.
|
| 301 |
+
|
| 302 |
+
# In[19]:
|
| 303 |
+
|
| 304 |
+
|
| 305 |
+
# Explore Fare distribution
|
| 306 |
+
g = sns.distplot(dataset["Fare"], color="m", label="Skewness : %.2f"%(dataset["Fare"].skew()))
|
| 307 |
+
g = g.legend(loc="best")
|
| 308 |
+
|
| 309 |
+
|
| 310 |
+
# As we can see, Fare distribution is very skewed. This can lead to overweigth very high values in the model, even if it is scaled.
|
| 311 |
+
#
|
| 312 |
+
# In this case, it is better to transform it with the log function to reduce this skew.
|
| 313 |
+
|
| 314 |
+
# In[20]:
|
| 315 |
+
|
| 316 |
+
|
| 317 |
+
# Apply log to Fare to reduce skewness distribution
|
| 318 |
+
dataset["Fare"] = dataset["Fare"].map(lambda i: np.log(i) if i > 0 else 0)
|
| 319 |
+
|
| 320 |
+
|
| 321 |
+
# In[21]:
|
| 322 |
+
|
| 323 |
+
|
| 324 |
+
g = sns.distplot(dataset["Fare"], color="b", label="Skewness : %.2f"%(dataset["Fare"].skew()))
|
| 325 |
+
g = g.legend(loc="best")
|
| 326 |
+
|
| 327 |
+
|
| 328 |
+
# Skewness is clearly reduced after the log transformation
|
| 329 |
+
|
| 330 |
+
# ### 3.2 Categorical values
|
| 331 |
+
# #### Sex
|
| 332 |
+
|
| 333 |
+
# In[22]:
|
| 334 |
+
|
| 335 |
+
|
| 336 |
+
g = sns.barplot(x="Sex",y="Survived",data=train)
|
| 337 |
+
g = g.set_ylabel("Survival Probability")
|
| 338 |
+
|
| 339 |
+
|
| 340 |
+
# In[23]:
|
| 341 |
+
|
| 342 |
+
|
| 343 |
+
train[["Sex","Survived"]].groupby('Sex').mean()
|
| 344 |
+
|
| 345 |
+
|
| 346 |
+
# It is clearly obvious that Male have less chance to survive than Female.
|
| 347 |
+
#
|
| 348 |
+
# So Sex, might play an important role in the prediction of the survival.
|
| 349 |
+
#
|
| 350 |
+
# For those who have seen the Titanic movie (1997), I am sure, we all remember this sentence during the evacuation : "Women and children first".
|
| 351 |
+
|
| 352 |
+
# #### Pclass
|
| 353 |
+
|
| 354 |
+
# In[24]:
|
| 355 |
+
|
| 356 |
+
|
| 357 |
+
# Explore Pclass vs Survived
|
| 358 |
+
g = sns.factorplot(x="Pclass",y="Survived",data=train,kind="bar", size = 6 ,
|
| 359 |
+
palette = "muted")
|
| 360 |
+
g.despine(left=True)
|
| 361 |
+
g = g.set_ylabels("survival probability")
|
| 362 |
+
|
| 363 |
+
|
| 364 |
+
# In[25]:
|
| 365 |
+
|
| 366 |
+
|
| 367 |
+
# Explore Pclass vs Survived by Sex
|
| 368 |
+
g = sns.factorplot(x="Pclass", y="Survived", hue="Sex", data=train,
|
| 369 |
+
size=6, kind="bar", palette="muted")
|
| 370 |
+
g.despine(left=True)
|
| 371 |
+
g = g.set_ylabels("survival probability")
|
| 372 |
+
|
| 373 |
+
|
| 374 |
+
# The passenger survival is not the same in the 3 classes. First class passengers have more chance to survive than second class and third class passengers.
|
| 375 |
+
#
|
| 376 |
+
# This trend is conserved when we look at both male and female passengers.
|
| 377 |
+
|
| 378 |
+
# #### Embarked
|
| 379 |
+
|
| 380 |
+
# In[26]:
|
| 381 |
+
|
| 382 |
+
|
| 383 |
+
dataset["Embarked"].isnull().sum()
|
| 384 |
+
|
| 385 |
+
|
| 386 |
+
# In[27]:
|
| 387 |
+
|
| 388 |
+
|
| 389 |
+
#Fill Embarked nan values of dataset set with 'S' most frequent value
|
| 390 |
+
dataset["Embarked"] = dataset["Embarked"].fillna("S")
|
| 391 |
+
|
| 392 |
+
|
| 393 |
+
# Since we have two missing values , i decided to fill them with the most fequent value of "Embarked" (S).
|
| 394 |
+
|
| 395 |
+
# In[28]:
|
| 396 |
+
|
| 397 |
+
|
| 398 |
+
# Explore Embarked vs Survived
|
| 399 |
+
g = sns.factorplot(x="Embarked", y="Survived", data=train,
|
| 400 |
+
size=6, kind="bar", palette="muted")
|
| 401 |
+
g.despine(left=True)
|
| 402 |
+
g = g.set_ylabels("survival probability")
|
| 403 |
+
|
| 404 |
+
|
| 405 |
+
# It seems that passenger coming from Cherbourg (C) have more chance to survive.
|
| 406 |
+
#
|
| 407 |
+
# My hypothesis is that the proportion of first class passengers is higher for those who came from Cherbourg than Queenstown (Q), Southampton (S).
|
| 408 |
+
#
|
| 409 |
+
# Let's see the Pclass distribution vs Embarked
|
| 410 |
+
|
| 411 |
+
# In[29]:
|
| 412 |
+
|
| 413 |
+
|
| 414 |
+
# Explore Pclass vs Embarked
|
| 415 |
+
g = sns.factorplot("Pclass", col="Embarked", data=train,
|
| 416 |
+
size=6, kind="count", palette="muted")
|
| 417 |
+
g.despine(left=True)
|
| 418 |
+
g = g.set_ylabels("Count")
|
| 419 |
+
|
| 420 |
+
|
| 421 |
+
# Indeed, the third class is the most frequent for passenger coming from Southampton (S) and Queenstown (Q), whereas Cherbourg passengers are mostly in first class which have the highest survival rate.
|
| 422 |
+
#
|
| 423 |
+
# At this point, i can't explain why first class has an higher survival rate. My hypothesis is that first class passengers were prioritised during the evacuation due to their influence.
|
| 424 |
+
|
| 425 |
+
# ## 4. Filling missing Values
|
| 426 |
+
# ### 4.1 Age
|
| 427 |
+
#
|
| 428 |
+
# As we see, Age column contains 256 missing values in the whole dataset.
|
| 429 |
+
#
|
| 430 |
+
# Since there is subpopulations that have more chance to survive (children for example), it is preferable to keep the age feature and to impute the missing values.
|
| 431 |
+
#
|
| 432 |
+
# To adress this problem, i looked at the most correlated features with Age (Sex, Parch , Pclass and SibSP).
|
| 433 |
+
|
| 434 |
+
# In[30]:
|
| 435 |
+
|
| 436 |
+
|
| 437 |
+
# Explore Age vs Sex, Parch , Pclass and SibSP
|
| 438 |
+
g = sns.factorplot(y="Age",x="Sex",data=dataset,kind="box")
|
| 439 |
+
g = sns.factorplot(y="Age",x="Sex",hue="Pclass", data=dataset,kind="box")
|
| 440 |
+
g = sns.factorplot(y="Age",x="Parch", data=dataset,kind="box")
|
| 441 |
+
g = sns.factorplot(y="Age",x="SibSp", data=dataset,kind="box")
|
| 442 |
+
|
| 443 |
+
|
| 444 |
+
# Age distribution seems to be the same in Male and Female subpopulations, so Sex is not informative to predict Age.
|
| 445 |
+
#
|
| 446 |
+
# However, 1rst class passengers are older than 2nd class passengers who are also older than 3rd class passengers.
|
| 447 |
+
#
|
| 448 |
+
# Moreover, the more a passenger has parents/children the older he is and the more a passenger has siblings/spouses the younger he is.
|
| 449 |
+
|
| 450 |
+
# In[31]:
|
| 451 |
+
|
| 452 |
+
|
| 453 |
+
# convert Sex into categorical value 0 for male and 1 for female
|
| 454 |
+
dataset["Sex"] = dataset["Sex"].map({"male": 0, "female":1})
|
| 455 |
+
|
| 456 |
+
|
| 457 |
+
# In[32]:
|
| 458 |
+
|
| 459 |
+
|
| 460 |
+
g = sns.heatmap(dataset[["Age","Sex","SibSp","Parch","Pclass"]].corr(),cmap="BrBG",annot=True)
|
| 461 |
+
|
| 462 |
+
|
| 463 |
+
# The correlation map confirms the factorplots observations except for Parch. Age is not correlated with Sex, but is negatively correlated with Pclass, Parch and SibSp.
|
| 464 |
+
#
|
| 465 |
+
# In the plot of Age in function of Parch, Age is growing with the number of parents / children. But the general correlation is negative.
|
| 466 |
+
#
|
| 467 |
+
# So, i decided to use SibSP, Parch and Pclass in order to impute the missing ages.
|
| 468 |
+
#
|
| 469 |
+
# The strategy is to fill Age with the median age of similar rows according to Pclass, Parch and SibSp.
|
| 470 |
+
|
| 471 |
+
# In[33]:
|
| 472 |
+
|
| 473 |
+
|
| 474 |
+
# Filling missing value of Age
|
| 475 |
+
|
| 476 |
+
## Fill Age with the median age of similar rows according to Pclass, Parch and SibSp
|
| 477 |
+
# Index of NaN age rows
|
| 478 |
+
index_NaN_age = list(dataset["Age"][dataset["Age"].isnull()].index)
|
| 479 |
+
|
| 480 |
+
for i in index_NaN_age :
|
| 481 |
+
age_med = dataset["Age"].median()
|
| 482 |
+
age_pred = dataset["Age"][((dataset['SibSp'] == dataset.iloc[i]["SibSp"]) & (dataset['Parch'] == dataset.iloc[i]["Parch"]) & (dataset['Pclass'] == dataset.iloc[i]["Pclass"]))].median()
|
| 483 |
+
if not np.isnan(age_pred) :
|
| 484 |
+
dataset['Age'].iloc[i] = age_pred
|
| 485 |
+
else :
|
| 486 |
+
dataset['Age'].iloc[i] = age_med
|
| 487 |
+
|
| 488 |
+
|
| 489 |
+
# In[34]:
|
| 490 |
+
|
| 491 |
+
|
| 492 |
+
g = sns.factorplot(x="Survived", y = "Age",data = train, kind="box")
|
| 493 |
+
g = sns.factorplot(x="Survived", y = "Age",data = train, kind="violin")
|
| 494 |
+
|
| 495 |
+
|
| 496 |
+
# No difference between median value of age in survived and not survived subpopulation.
|
| 497 |
+
#
|
| 498 |
+
# But in the violin plot of survived passengers, we still notice that very young passengers have higher survival rate.
|
| 499 |
+
|
| 500 |
+
# ## 5. Feature engineering
|
| 501 |
+
# ### 5.1 Name/Title
|
| 502 |
+
|
| 503 |
+
# In[35]:
|
| 504 |
+
|
| 505 |
+
|
| 506 |
+
dataset["Name"].head()
|
| 507 |
+
|
| 508 |
+
|
| 509 |
+
# The Name feature contains information on passenger's title.
|
| 510 |
+
#
|
| 511 |
+
# Since some passenger with distingused title may be preferred during the evacuation, it is interesting to add them to the model.
|
| 512 |
+
|
| 513 |
+
# In[36]:
|
| 514 |
+
|
| 515 |
+
|
| 516 |
+
# Get Title from Name
|
| 517 |
+
dataset_title = [i.split(",")[1].split(".")[0].strip() for i in dataset["Name"]]
|
| 518 |
+
dataset["Title"] = pd.Series(dataset_title)
|
| 519 |
+
dataset["Title"].head()
|
| 520 |
+
|
| 521 |
+
|
| 522 |
+
# In[37]:
|
| 523 |
+
|
| 524 |
+
|
| 525 |
+
g = sns.countplot(x="Title",data=dataset)
|
| 526 |
+
g = plt.setp(g.get_xticklabels(), rotation=45)
|
| 527 |
+
|
| 528 |
+
|
| 529 |
+
# There is 17 titles in the dataset, most of them are very rare and we can group them in 4 categories.
|
| 530 |
+
|
| 531 |
+
# In[38]:
|
| 532 |
+
|
| 533 |
+
|
| 534 |
+
# Convert to categorical values Title
|
| 535 |
+
dataset["Title"] = dataset["Title"].replace(['Lady', 'the Countess','Countess','Capt', 'Col','Don', 'Dr', 'Major', 'Rev', 'Sir', 'Jonkheer', 'Dona'], 'Rare')
|
| 536 |
+
dataset["Title"] = dataset["Title"].map({"Master":0, "Miss":1, "Ms" : 1 , "Mme":1, "Mlle":1, "Mrs":1, "Mr":2, "Rare":3})
|
| 537 |
+
dataset["Title"] = dataset["Title"].astype(int)
|
| 538 |
+
|
| 539 |
+
|
| 540 |
+
# In[39]:
|
| 541 |
+
|
| 542 |
+
|
| 543 |
+
g = sns.countplot(dataset["Title"])
|
| 544 |
+
g = g.set_xticklabels(["Master","Miss/Ms/Mme/Mlle/Mrs","Mr","Rare"])
|
| 545 |
+
|
| 546 |
+
|
| 547 |
+
# In[40]:
|
| 548 |
+
|
| 549 |
+
|
| 550 |
+
g = sns.factorplot(x="Title",y="Survived",data=dataset,kind="bar")
|
| 551 |
+
g = g.set_xticklabels(["Master","Miss-Mrs","Mr","Rare"])
|
| 552 |
+
g = g.set_ylabels("survival probability")
|
| 553 |
+
|
| 554 |
+
|
| 555 |
+
# "Women and children first"
|
| 556 |
+
#
|
| 557 |
+
# It is interesting to note that passengers with rare title have more chance to survive.
|
| 558 |
+
|
| 559 |
+
# In[41]:
|
| 560 |
+
|
| 561 |
+
|
| 562 |
+
# Drop Name variable
|
| 563 |
+
dataset.drop(labels = ["Name"], axis = 1, inplace = True)
|
| 564 |
+
|
| 565 |
+
|
| 566 |
+
# ### 5.2 Family size
|
| 567 |
+
#
|
| 568 |
+
# We can imagine that large families will have more difficulties to evacuate, looking for theirs sisters/brothers/parents during the evacuation. So, i choosed to create a "Fize" (family size) feature which is the sum of SibSp , Parch and 1 (including the passenger).
|
| 569 |
+
|
| 570 |
+
# In[42]:
|
| 571 |
+
|
| 572 |
+
|
| 573 |
+
# Create a family size descriptor from SibSp and Parch
|
| 574 |
+
dataset["Fsize"] = dataset["SibSp"] + dataset["Parch"] + 1
|
| 575 |
+
|
| 576 |
+
|
| 577 |
+
# In[43]:
|
| 578 |
+
|
| 579 |
+
|
| 580 |
+
g = sns.factorplot(x="Fsize",y="Survived",data = dataset)
|
| 581 |
+
g = g.set_ylabels("Survival Probability")
|
| 582 |
+
|
| 583 |
+
|
| 584 |
+
# The family size seems to play an important role, survival probability is worst for large families.
|
| 585 |
+
#
|
| 586 |
+
# Additionally, i decided to created 4 categories of family size.
|
| 587 |
+
|
| 588 |
+
# In[44]:
|
| 589 |
+
|
| 590 |
+
|
| 591 |
+
# Create new feature of family size
|
| 592 |
+
dataset['Single'] = dataset['Fsize'].map(lambda s: 1 if s == 1 else 0)
|
| 593 |
+
dataset['SmallF'] = dataset['Fsize'].map(lambda s: 1 if s == 2 else 0)
|
| 594 |
+
dataset['MedF'] = dataset['Fsize'].map(lambda s: 1 if 3 <= s <= 4 else 0)
|
| 595 |
+
dataset['LargeF'] = dataset['Fsize'].map(lambda s: 1 if s >= 5 else 0)
|
| 596 |
+
|
| 597 |
+
|
| 598 |
+
# In[45]:
|
| 599 |
+
|
| 600 |
+
|
| 601 |
+
g = sns.factorplot(x="Single",y="Survived",data=dataset,kind="bar")
|
| 602 |
+
g = g.set_ylabels("Survival Probability")
|
| 603 |
+
g = sns.factorplot(x="SmallF",y="Survived",data=dataset,kind="bar")
|
| 604 |
+
g = g.set_ylabels("Survival Probability")
|
| 605 |
+
g = sns.factorplot(x="MedF",y="Survived",data=dataset,kind="bar")
|
| 606 |
+
g = g.set_ylabels("Survival Probability")
|
| 607 |
+
g = sns.factorplot(x="LargeF",y="Survived",data=dataset,kind="bar")
|
| 608 |
+
g = g.set_ylabels("Survival Probability")
|
| 609 |
+
|
| 610 |
+
|
| 611 |
+
# Factorplots of family size categories show that Small and Medium families have more chance to survive than single passenger and large families.
|
| 612 |
+
|
| 613 |
+
# In[46]:
|
| 614 |
+
|
| 615 |
+
|
| 616 |
+
# convert to indicator values Title and Embarked
|
| 617 |
+
dataset = pd.get_dummies(dataset, columns = ["Title"])
|
| 618 |
+
dataset = pd.get_dummies(dataset, columns = ["Embarked"], prefix="Em")
|
| 619 |
+
|
| 620 |
+
|
| 621 |
+
# In[47]:
|
| 622 |
+
|
| 623 |
+
|
| 624 |
+
dataset.head()
|
| 625 |
+
|
| 626 |
+
|
| 627 |
+
# At this stage, we have 22 features.
|
| 628 |
+
|
| 629 |
+
# ### 5.3 Cabin
|
| 630 |
+
|
| 631 |
+
# In[48]:
|
| 632 |
+
|
| 633 |
+
|
| 634 |
+
dataset["Cabin"].head()
|
| 635 |
+
|
| 636 |
+
|
| 637 |
+
# In[49]:
|
| 638 |
+
|
| 639 |
+
|
| 640 |
+
dataset["Cabin"].describe()
|
| 641 |
+
|
| 642 |
+
|
| 643 |
+
# In[50]:
|
| 644 |
+
|
| 645 |
+
|
| 646 |
+
dataset["Cabin"].isnull().sum()
|
| 647 |
+
|
| 648 |
+
|
| 649 |
+
# The Cabin feature column contains 292 values and 1007 missing values.
|
| 650 |
+
#
|
| 651 |
+
# I supposed that passengers without a cabin have a missing value displayed instead of the cabin number.
|
| 652 |
+
|
| 653 |
+
# In[51]:
|
| 654 |
+
|
| 655 |
+
|
| 656 |
+
dataset["Cabin"][dataset["Cabin"].notnull()].head()
|
| 657 |
+
|
| 658 |
+
|
| 659 |
+
# In[52]:
|
| 660 |
+
|
| 661 |
+
|
| 662 |
+
# Replace the Cabin number by the type of cabin 'X' if not
|
| 663 |
+
dataset["Cabin"] = pd.Series([i[0] if not pd.isnull(i) else 'X' for i in dataset['Cabin'] ])
|
| 664 |
+
|
| 665 |
+
|
| 666 |
+
# The first letter of the cabin indicates the Desk, i choosed to keep this information only, since it indicates the probable location of the passenger in the Titanic.
|
| 667 |
+
|
| 668 |
+
# In[53]:
|
| 669 |
+
|
| 670 |
+
|
| 671 |
+
g = sns.countplot(dataset["Cabin"],order=['A','B','C','D','E','F','G','T','X'])
|
| 672 |
+
|
| 673 |
+
|
| 674 |
+
# In[54]:
|
| 675 |
+
|
| 676 |
+
|
| 677 |
+
g = sns.factorplot(y="Survived",x="Cabin",data=dataset,kind="bar",order=['A','B','C','D','E','F','G','T','X'])
|
| 678 |
+
g = g.set_ylabels("Survival Probability")
|
| 679 |
+
|
| 680 |
+
|
| 681 |
+
# Because of the low number of passenger that have a cabin, survival probabilities have an important standard deviation and we can't distinguish between survival probability of passengers in the different desks.
|
| 682 |
+
#
|
| 683 |
+
# But we can see that passengers with a cabin have generally more chance to survive than passengers without (X).
|
| 684 |
+
#
|
| 685 |
+
# It is particularly true for cabin B, C, D, E and F.
|
| 686 |
+
|
| 687 |
+
# In[55]:
|
| 688 |
+
|
| 689 |
+
|
| 690 |
+
dataset = pd.get_dummies(dataset, columns = ["Cabin"],prefix="Cabin")
|
| 691 |
+
|
| 692 |
+
|
| 693 |
+
# ### 5.4 Ticket
|
| 694 |
+
|
| 695 |
+
# In[56]:
|
| 696 |
+
|
| 697 |
+
|
| 698 |
+
dataset["Ticket"].head()
|
| 699 |
+
|
| 700 |
+
|
| 701 |
+
# It could mean that tickets sharing the same prefixes could be booked for cabins placed together. It could therefore lead to the actual placement of the cabins within the ship.
|
| 702 |
+
#
|
| 703 |
+
# Tickets with same prefixes may have a similar class and survival.
|
| 704 |
+
#
|
| 705 |
+
# So i decided to replace the Ticket feature column by the ticket prefixe. Which may be more informative.
|
| 706 |
+
|
| 707 |
+
# In[57]:
|
| 708 |
+
|
| 709 |
+
|
| 710 |
+
## Treat Ticket by extracting the ticket prefix. When there is no prefix it returns X.
|
| 711 |
+
|
| 712 |
+
Ticket = []
|
| 713 |
+
for i in list(dataset.Ticket):
|
| 714 |
+
if not i.isdigit() :
|
| 715 |
+
Ticket.append(i.replace(".","").replace("/","").strip().split(' ')[0]) #Take prefix
|
| 716 |
+
else:
|
| 717 |
+
Ticket.append("X")
|
| 718 |
+
|
| 719 |
+
dataset["Ticket"] = Ticket
|
| 720 |
+
dataset["Ticket"].head()
|
| 721 |
+
|
| 722 |
+
|
| 723 |
+
# In[58]:
|
| 724 |
+
|
| 725 |
+
|
| 726 |
+
dataset = pd.get_dummies(dataset, columns = ["Ticket"], prefix="T")
|
| 727 |
+
|
| 728 |
+
|
| 729 |
+
# In[59]:
|
| 730 |
+
|
| 731 |
+
|
| 732 |
+
# Create categorical values for Pclass
|
| 733 |
+
dataset["Pclass"] = dataset["Pclass"].astype("category")
|
| 734 |
+
dataset = pd.get_dummies(dataset, columns = ["Pclass"],prefix="Pc")
|
| 735 |
+
|
| 736 |
+
|
| 737 |
+
# In[60]:
|
| 738 |
+
|
| 739 |
+
|
| 740 |
+
# Drop useless variables
|
| 741 |
+
dataset.drop(labels = ["PassengerId"], axis = 1, inplace = True)
|
| 742 |
+
|
| 743 |
+
|
| 744 |
+
# In[61]:
|
| 745 |
+
|
| 746 |
+
|
| 747 |
+
dataset.head()
|
| 748 |
+
|
| 749 |
+
|
| 750 |
+
# ## 6. MODELING
|
| 751 |
+
|
| 752 |
+
# In[62]:
|
| 753 |
+
|
| 754 |
+
|
| 755 |
+
## Separate train dataset and test dataset
|
| 756 |
+
|
| 757 |
+
train = dataset[:train_len]
|
| 758 |
+
test = dataset[train_len:]
|
| 759 |
+
test.drop(labels=["Survived"],axis = 1,inplace=True)
|
| 760 |
+
|
| 761 |
+
|
| 762 |
+
# In[63]:
|
| 763 |
+
|
| 764 |
+
|
| 765 |
+
## Separate train features and label
|
| 766 |
+
|
| 767 |
+
train["Survived"] = train["Survived"].astype(int)
|
| 768 |
+
|
| 769 |
+
Y_train = train["Survived"]
|
| 770 |
+
|
| 771 |
+
X_train = train.drop(labels = ["Survived"],axis = 1)
|
| 772 |
+
|
| 773 |
+
|
| 774 |
+
# ### 6.1 Simple modeling
|
| 775 |
+
# #### 6.1.1 Cross validate models
|
| 776 |
+
#
|
| 777 |
+
# I compared 10 popular classifiers and evaluate the mean accuracy of each of them by a stratified kfold cross validation procedure.
|
| 778 |
+
#
|
| 779 |
+
# * SVC
|
| 780 |
+
# * Decision Tree
|
| 781 |
+
# * AdaBoost
|
| 782 |
+
# * Random Forest
|
| 783 |
+
# * Extra Trees
|
| 784 |
+
# * Gradient Boosting
|
| 785 |
+
# * Multiple layer perceprton (neural network)
|
| 786 |
+
# * KNN
|
| 787 |
+
# * Logistic regression
|
| 788 |
+
# * Linear Discriminant Analysis
|
| 789 |
+
|
| 790 |
+
# In[64]:
|
| 791 |
+
|
| 792 |
+
|
| 793 |
+
# Cross validate model with Kfold stratified cross val
|
| 794 |
+
kfold = StratifiedKFold(n_splits=10)
|
| 795 |
+
|
| 796 |
+
|
| 797 |
+
# In[65]:
|
| 798 |
+
|
| 799 |
+
|
| 800 |
+
# Modeling step Test differents algorithms
|
| 801 |
+
random_state = 2
|
| 802 |
+
classifiers = []
|
| 803 |
+
classifiers.append(SVC(random_state=random_state))
|
| 804 |
+
classifiers.append(DecisionTreeClassifier(random_state=random_state))
|
| 805 |
+
classifiers.append(AdaBoostClassifier(DecisionTreeClassifier(random_state=random_state),random_state=random_state,learning_rate=0.1))
|
| 806 |
+
classifiers.append(RandomForestClassifier(random_state=random_state))
|
| 807 |
+
classifiers.append(ExtraTreesClassifier(random_state=random_state))
|
| 808 |
+
classifiers.append(GradientBoostingClassifier(random_state=random_state))
|
| 809 |
+
classifiers.append(MLPClassifier(random_state=random_state))
|
| 810 |
+
classifiers.append(KNeighborsClassifier())
|
| 811 |
+
classifiers.append(LogisticRegression(random_state = random_state))
|
| 812 |
+
classifiers.append(LinearDiscriminantAnalysis())
|
| 813 |
+
|
| 814 |
+
cv_results = []
|
| 815 |
+
for classifier in classifiers :
|
| 816 |
+
cv_results.append(cross_val_score(classifier, X_train, y = Y_train, scoring = "accuracy", cv = kfold, n_jobs=4))
|
| 817 |
+
|
| 818 |
+
cv_means = []
|
| 819 |
+
cv_std = []
|
| 820 |
+
for cv_result in cv_results:
|
| 821 |
+
cv_means.append(cv_result.mean())
|
| 822 |
+
cv_std.append(cv_result.std())
|
| 823 |
+
|
| 824 |
+
cv_res = pd.DataFrame({"CrossValMeans":cv_means,"CrossValerrors": cv_std,"Algorithm":["SVC","DecisionTree","AdaBoost",
|
| 825 |
+
"RandomForest","ExtraTrees","GradientBoosting","MultipleLayerPerceptron","KNeighboors","LogisticRegression","LinearDiscriminantAnalysis"]})
|
| 826 |
+
|
| 827 |
+
g = sns.barplot("CrossValMeans","Algorithm",data = cv_res, palette="Set3",orient = "h",**{'xerr':cv_std})
|
| 828 |
+
g.set_xlabel("Mean Accuracy")
|
| 829 |
+
g = g.set_title("Cross validation scores")
|
| 830 |
+
|
| 831 |
+
|
| 832 |
+
# I decided to choose the SVC, AdaBoost, RandomForest , ExtraTrees and the GradientBoosting classifiers for the ensemble modeling.
|
| 833 |
+
|
| 834 |
+
# #### 6.1.2 Hyperparameter tunning for best models
|
| 835 |
+
#
|
| 836 |
+
# I performed a grid search optimization for AdaBoost, ExtraTrees , RandomForest, GradientBoosting and SVC classifiers.
|
| 837 |
+
#
|
| 838 |
+
# I set the "n_jobs" parameter to 4 since i have 4 cpu . The computation time is clearly reduced.
|
| 839 |
+
#
|
| 840 |
+
# But be carefull, this step can take a long time, i took me 15 min in total on 4 cpu.
|
| 841 |
+
|
| 842 |
+
# In[66]:
|
| 843 |
+
|
| 844 |
+
|
| 845 |
+
### META MODELING WITH ADABOOST, RF, EXTRATREES and GRADIENTBOOSTING
|
| 846 |
+
|
| 847 |
+
# Adaboost
|
| 848 |
+
DTC = DecisionTreeClassifier()
|
| 849 |
+
|
| 850 |
+
adaDTC = AdaBoostClassifier(DTC, random_state=7)
|
| 851 |
+
|
| 852 |
+
ada_param_grid = {"base_estimator__criterion" : ["gini", "entropy"],
|
| 853 |
+
"base_estimator__splitter" : ["best", "random"],
|
| 854 |
+
"algorithm" : ["SAMME","SAMME.R"],
|
| 855 |
+
"n_estimators" :[1,2],
|
| 856 |
+
"learning_rate": [0.0001, 0.001, 0.01, 0.1, 0.2, 0.3,1.5]}
|
| 857 |
+
|
| 858 |
+
gsadaDTC = GridSearchCV(adaDTC,param_grid = ada_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 859 |
+
|
| 860 |
+
gsadaDTC.fit(X_train,Y_train)
|
| 861 |
+
|
| 862 |
+
ada_best = gsadaDTC.best_estimator_
|
| 863 |
+
|
| 864 |
+
|
| 865 |
+
# In[67]:
|
| 866 |
+
|
| 867 |
+
|
| 868 |
+
gsadaDTC.best_score_
|
| 869 |
+
|
| 870 |
+
|
| 871 |
+
# In[68]:
|
| 872 |
+
|
| 873 |
+
|
| 874 |
+
#ExtraTrees
|
| 875 |
+
ExtC = ExtraTreesClassifier()
|
| 876 |
+
|
| 877 |
+
|
| 878 |
+
## Search grid for optimal parameters
|
| 879 |
+
ex_param_grid = {"max_depth": [None],
|
| 880 |
+
"max_features": [1, 3, 10],
|
| 881 |
+
"min_samples_split": [2, 3, 10],
|
| 882 |
+
"min_samples_leaf": [1, 3, 10],
|
| 883 |
+
"bootstrap": [False],
|
| 884 |
+
"n_estimators" :[100,300],
|
| 885 |
+
"criterion": ["gini"]}
|
| 886 |
+
|
| 887 |
+
|
| 888 |
+
gsExtC = GridSearchCV(ExtC,param_grid = ex_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 889 |
+
|
| 890 |
+
gsExtC.fit(X_train,Y_train)
|
| 891 |
+
|
| 892 |
+
ExtC_best = gsExtC.best_estimator_
|
| 893 |
+
|
| 894 |
+
# Best score
|
| 895 |
+
gsExtC.best_score_
|
| 896 |
+
|
| 897 |
+
|
| 898 |
+
# In[69]:
|
| 899 |
+
|
| 900 |
+
|
| 901 |
+
# RFC Parameters tunning
|
| 902 |
+
RFC = RandomForestClassifier()
|
| 903 |
+
|
| 904 |
+
|
| 905 |
+
## Search grid for optimal parameters
|
| 906 |
+
rf_param_grid = {"max_depth": [None],
|
| 907 |
+
"max_features": [1, 3, 10],
|
| 908 |
+
"min_samples_split": [2, 3, 10],
|
| 909 |
+
"min_samples_leaf": [1, 3, 10],
|
| 910 |
+
"bootstrap": [False],
|
| 911 |
+
"n_estimators" :[100,300],
|
| 912 |
+
"criterion": ["gini"]}
|
| 913 |
+
|
| 914 |
+
|
| 915 |
+
gsRFC = GridSearchCV(RFC,param_grid = rf_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 916 |
+
|
| 917 |
+
gsRFC.fit(X_train,Y_train)
|
| 918 |
+
|
| 919 |
+
RFC_best = gsRFC.best_estimator_
|
| 920 |
+
|
| 921 |
+
# Best score
|
| 922 |
+
gsRFC.best_score_
|
| 923 |
+
|
| 924 |
+
|
| 925 |
+
# In[70]:
|
| 926 |
+
|
| 927 |
+
|
| 928 |
+
# Gradient boosting tunning
|
| 929 |
+
|
| 930 |
+
GBC = GradientBoostingClassifier()
|
| 931 |
+
gb_param_grid = {'loss' : ["deviance"],
|
| 932 |
+
'n_estimators' : [100,200,300],
|
| 933 |
+
'learning_rate': [0.1, 0.05, 0.01],
|
| 934 |
+
'max_depth': [4, 8],
|
| 935 |
+
'min_samples_leaf': [100,150],
|
| 936 |
+
'max_features': [0.3, 0.1]
|
| 937 |
+
}
|
| 938 |
+
|
| 939 |
+
gsGBC = GridSearchCV(GBC,param_grid = gb_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 940 |
+
|
| 941 |
+
gsGBC.fit(X_train,Y_train)
|
| 942 |
+
|
| 943 |
+
GBC_best = gsGBC.best_estimator_
|
| 944 |
+
|
| 945 |
+
# Best score
|
| 946 |
+
gsGBC.best_score_
|
| 947 |
+
|
| 948 |
+
|
| 949 |
+
# In[71]:
|
| 950 |
+
|
| 951 |
+
|
| 952 |
+
### SVC classifier
|
| 953 |
+
SVMC = SVC(probability=True)
|
| 954 |
+
svc_param_grid = {'kernel': ['rbf'],
|
| 955 |
+
'gamma': [ 0.001, 0.01, 0.1, 1],
|
| 956 |
+
'C': [1, 10, 50, 100,200,300, 1000]}
|
| 957 |
+
|
| 958 |
+
gsSVMC = GridSearchCV(SVMC,param_grid = svc_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
|
| 959 |
+
|
| 960 |
+
gsSVMC.fit(X_train,Y_train)
|
| 961 |
+
|
| 962 |
+
SVMC_best = gsSVMC.best_estimator_
|
| 963 |
+
|
| 964 |
+
# Best score
|
| 965 |
+
gsSVMC.best_score_
|
| 966 |
+
|
| 967 |
+
|
| 968 |
+
# #### 6.1.3 Plot learning curves
|
| 969 |
+
#
|
| 970 |
+
# Learning curves are a good way to see the overfitting effect on the training set and the effect of the training size on the accuracy.
|
| 971 |
+
|
| 972 |
+
# In[72]:
|
| 973 |
+
|
| 974 |
+
|
| 975 |
+
def plot_learning_curve(estimator, title, X, y, ylim=None, cv=None,
|
| 976 |
+
n_jobs=-1, train_sizes=np.linspace(.1, 1.0, 5)):
|
| 977 |
+
"""Generate a simple plot of the test and training learning curve"""
|
| 978 |
+
plt.figure()
|
| 979 |
+
plt.title(title)
|
| 980 |
+
if ylim is not None:
|
| 981 |
+
plt.ylim(*ylim)
|
| 982 |
+
plt.xlabel("Training examples")
|
| 983 |
+
plt.ylabel("Score")
|
| 984 |
+
train_sizes, train_scores, test_scores = learning_curve(
|
| 985 |
+
estimator, X, y, cv=cv, n_jobs=n_jobs, train_sizes=train_sizes)
|
| 986 |
+
train_scores_mean = np.mean(train_scores, axis=1)
|
| 987 |
+
train_scores_std = np.std(train_scores, axis=1)
|
| 988 |
+
test_scores_mean = np.mean(test_scores, axis=1)
|
| 989 |
+
test_scores_std = np.std(test_scores, axis=1)
|
| 990 |
+
plt.grid()
|
| 991 |
+
|
| 992 |
+
plt.fill_between(train_sizes, train_scores_mean - train_scores_std,
|
| 993 |
+
train_scores_mean + train_scores_std, alpha=0.1,
|
| 994 |
+
color="r")
|
| 995 |
+
plt.fill_between(train_sizes, test_scores_mean - test_scores_std,
|
| 996 |
+
test_scores_mean + test_scores_std, alpha=0.1, color="g")
|
| 997 |
+
plt.plot(train_sizes, train_scores_mean, 'o-', color="r",
|
| 998 |
+
label="Training score")
|
| 999 |
+
plt.plot(train_sizes, test_scores_mean, 'o-', color="g",
|
| 1000 |
+
label="Cross-validation score")
|
| 1001 |
+
|
| 1002 |
+
plt.legend(loc="best")
|
| 1003 |
+
return plt
|
| 1004 |
+
|
| 1005 |
+
g = plot_learning_curve(gsRFC.best_estimator_,"RF mearning curves",X_train,Y_train,cv=kfold)
|
| 1006 |
+
g = plot_learning_curve(gsExtC.best_estimator_,"ExtraTrees learning curves",X_train,Y_train,cv=kfold)
|
| 1007 |
+
g = plot_learning_curve(gsSVMC.best_estimator_,"SVC learning curves",X_train,Y_train,cv=kfold)
|
| 1008 |
+
g = plot_learning_curve(gsadaDTC.best_estimator_,"AdaBoost learning curves",X_train,Y_train,cv=kfold)
|
| 1009 |
+
g = plot_learning_curve(gsGBC.best_estimator_,"GradientBoosting learning curves",X_train,Y_train,cv=kfold)
|
| 1010 |
+
|
| 1011 |
+
|
| 1012 |
+
# GradientBoosting and Adaboost classifiers tend to overfit the training set. According to the growing cross-validation curves GradientBoosting and Adaboost could perform better with more training examples.
|
| 1013 |
+
#
|
| 1014 |
+
# SVC and ExtraTrees classifiers seem to better generalize the prediction since the training and cross-validation curves are close together.
|
| 1015 |
+
|
| 1016 |
+
# #### 6.1.4 Feature importance of tree based classifiers
|
| 1017 |
+
#
|
| 1018 |
+
# In order to see the most informative features for the prediction of passengers survival, i displayed the feature importance for the 4 tree based classifiers.
|
| 1019 |
+
|
| 1020 |
+
# In[73]:
|
| 1021 |
+
|
| 1022 |
+
|
| 1023 |
+
nrows = ncols = 2
|
| 1024 |
+
fig, axes = plt.subplots(nrows = nrows, ncols = ncols, sharex="all", figsize=(15,15))
|
| 1025 |
+
|
| 1026 |
+
names_classifiers = [("AdaBoosting", ada_best),("ExtraTrees",ExtC_best),("RandomForest",RFC_best),("GradientBoosting",GBC_best)]
|
| 1027 |
+
|
| 1028 |
+
nclassifier = 0
|
| 1029 |
+
for row in range(nrows):
|
| 1030 |
+
for col in range(ncols):
|
| 1031 |
+
name = names_classifiers[nclassifier][0]
|
| 1032 |
+
classifier = names_classifiers[nclassifier][1]
|
| 1033 |
+
indices = np.argsort(classifier.feature_importances_)[::-1][:40]
|
| 1034 |
+
g = sns.barplot(y=X_train.columns[indices][:40],x = classifier.feature_importances_[indices][:40] , orient='h',ax=axes[row][col])
|
| 1035 |
+
g.set_xlabel("Relative importance",fontsize=12)
|
| 1036 |
+
g.set_ylabel("Features",fontsize=12)
|
| 1037 |
+
g.tick_params(labelsize=9)
|
| 1038 |
+
g.set_title(name + " feature importance")
|
| 1039 |
+
nclassifier += 1
|
| 1040 |
+
|
| 1041 |
+
|
| 1042 |
+
# I plot the feature importance for the 4 tree based classifiers (Adaboost, ExtraTrees, RandomForest and GradientBoosting).
|
| 1043 |
+
#
|
| 1044 |
+
# We note that the four classifiers have different top features according to the relative importance. It means that their predictions are not based on the same features. Nevertheless, they share some common important features for the classification , for example 'Fare', 'Title_2', 'Age' and 'Sex'.
|
| 1045 |
+
#
|
| 1046 |
+
# Title_2 which indicates the Mrs/Mlle/Mme/Miss/Ms category is highly correlated with Sex.
|
| 1047 |
+
#
|
| 1048 |
+
# We can say that:
|
| 1049 |
+
#
|
| 1050 |
+
# - Pc_1, Pc_2, Pc_3 and Fare refer to the general social standing of passengers.
|
| 1051 |
+
#
|
| 1052 |
+
# - Sex and Title_2 (Mrs/Mlle/Mme/Miss/Ms) and Title_3 (Mr) refer to the gender.
|
| 1053 |
+
#
|
| 1054 |
+
# - Age and Title_1 (Master) refer to the age of passengers.
|
| 1055 |
+
#
|
| 1056 |
+
# - Fsize, LargeF, MedF, Single refer to the size of the passenger family.
|
| 1057 |
+
#
|
| 1058 |
+
# **According to the feature importance of this 4 classifiers, the prediction of the survival seems to be more associated with the Age, the Sex, the family size and the social standing of the passengers more than the location in the boat.**
|
| 1059 |
+
|
| 1060 |
+
# In[74]:
|
| 1061 |
+
|
| 1062 |
+
|
| 1063 |
+
test_Survived_RFC = pd.Series(RFC_best.predict(test), name="RFC")
|
| 1064 |
+
test_Survived_ExtC = pd.Series(ExtC_best.predict(test), name="ExtC")
|
| 1065 |
+
test_Survived_SVMC = pd.Series(SVMC_best.predict(test), name="SVC")
|
| 1066 |
+
test_Survived_AdaC = pd.Series(ada_best.predict(test), name="Ada")
|
| 1067 |
+
test_Survived_GBC = pd.Series(GBC_best.predict(test), name="GBC")
|
| 1068 |
+
|
| 1069 |
+
|
| 1070 |
+
# Concatenate all classifier results
|
| 1071 |
+
ensemble_results = pd.concat([test_Survived_RFC,test_Survived_ExtC,test_Survived_AdaC,test_Survived_GBC, test_Survived_SVMC],axis=1)
|
| 1072 |
+
|
| 1073 |
+
|
| 1074 |
+
g= sns.heatmap(ensemble_results.corr(),annot=True)
|
| 1075 |
+
|
| 1076 |
+
|
| 1077 |
+
# The prediction seems to be quite similar for the 5 classifiers except when Adaboost is compared to the others classifiers.
|
| 1078 |
+
#
|
| 1079 |
+
# The 5 classifiers give more or less the same prediction but there is some differences. Theses differences between the 5 classifier predictions are sufficient to consider an ensembling vote.
|
| 1080 |
+
|
| 1081 |
+
# ### 6.2 Ensemble modeling
|
| 1082 |
+
# #### 6.2.1 Combining models
|
| 1083 |
+
#
|
| 1084 |
+
# I choosed a voting classifier to combine the predictions coming from the 5 classifiers.
|
| 1085 |
+
#
|
| 1086 |
+
# I preferred to pass the argument "soft" to the voting parameter to take into account the probability of each vote.
|
| 1087 |
+
|
| 1088 |
+
# In[75]:
|
| 1089 |
+
|
| 1090 |
+
|
| 1091 |
+
votingC = VotingClassifier(estimators=[('rfc', RFC_best), ('extc', ExtC_best),
|
| 1092 |
+
('svc', SVMC_best), ('adac',ada_best),('gbc',GBC_best)], voting='soft', n_jobs=4)
|
| 1093 |
+
|
| 1094 |
+
votingC = votingC.fit(X_train, Y_train)
|
| 1095 |
+
|
| 1096 |
+
|
| 1097 |
+
# ### 6.3 Prediction
|
| 1098 |
+
# #### 6.3.1 Predict and Submit results
|
| 1099 |
+
|
| 1100 |
+
# In[76]:
|
| 1101 |
+
|
| 1102 |
+
|
| 1103 |
+
test_Survived = pd.Series(votingC.predict(test), name="Survived")
|
| 1104 |
+
|
| 1105 |
+
results = pd.concat([IDtest,test_Survived],axis=1)
|
| 1106 |
+
|
| 1107 |
+
results.to_csv("ensemble_python_voting.csv",index=False)
|
| 1108 |
+
|
| 1109 |
+
|
| 1110 |
+
# If you found this notebook helpful or you just liked it , some upvotes would be very much appreciated - That will keep me motivated :)
|
Titanic/Kernels/ExtraTrees/3-a-statistical-analysis-ml-workflow-of-titanic.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/3-a-statistical-analysis-ml-workflow-of-titanic.py
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/8-a-comprehensive-guide-to-titanic-machine-learning.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/8-a-comprehensive-guide-to-titanic-machine-learning.py
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/9-top-3-efficient-ensembling-in-few-lines-of-code.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/ExtraTrees/9-top-3-efficient-ensembling-in-few-lines-of-code.py
ADDED
|
@@ -0,0 +1,944 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python
|
| 2 |
+
# coding: utf-8
|
| 3 |
+
|
| 4 |
+
# <h1><center>Titanic: efficient ensembling and optimization</center></h1>
|
| 5 |
+
#
|
| 6 |
+
# <center><img src="https://www.dlt.travel/immagine/33923/magazine-titanic2.jpg"></center>
|
| 7 |
+
|
| 8 |
+
# <a id="top"></a>
|
| 9 |
+
#
|
| 10 |
+
# <div class="list-group" id="list-tab" role="tablist">
|
| 11 |
+
# <h3 class="list-group-item list-group-item-action active" data-toggle="list" style='background:Black; border:0' role="tab" aria-controls="home"><center>Quick navigation</center></h3>
|
| 12 |
+
#
|
| 13 |
+
# * [1. Feature engineering](#1)
|
| 14 |
+
# * [2. Single models training and optimization](#2)
|
| 15 |
+
# * [3. SuperLearner training and optimization](#3)
|
| 16 |
+
# * [4. Final submission](#4)
|
| 17 |
+
#
|
| 18 |
+
#
|
| 19 |
+
# ## Best LB score is in Version 80.
|
| 20 |
+
#
|
| 21 |
+
#
|
| 22 |
+
# #### Keras neural network for Titanic classification problem: <a href="https://www.kaggle.com/isaienkov/keras-neural-network-architecture-optimization">Titanic: Keras Neural Network architecture optimization</a>
|
| 23 |
+
#
|
| 24 |
+
# #### Hyperparameter tunning methods for Titanic classification problem: <a href="https://www.kaggle.com/isaienkov/hyperparameters-tuning-techniques">Titanic: hyperparameters tuning techniques</a>
|
| 25 |
+
|
| 26 |
+
# In[1]:
|
| 27 |
+
|
| 28 |
+
|
| 29 |
+
import numpy as np
|
| 30 |
+
import pandas as pd
|
| 31 |
+
|
| 32 |
+
from mlens.ensemble import SuperLearner
|
| 33 |
+
|
| 34 |
+
from xgboost import XGBClassifier
|
| 35 |
+
from lightgbm import LGBMClassifier
|
| 36 |
+
|
| 37 |
+
from sklearn.model_selection import train_test_split
|
| 38 |
+
from sklearn.metrics import accuracy_score, f1_score
|
| 39 |
+
from sklearn.tree import DecisionTreeClassifier
|
| 40 |
+
from sklearn.neighbors import KNeighborsClassifier
|
| 41 |
+
from sklearn.neural_network import MLPClassifier
|
| 42 |
+
from sklearn.ensemble import GradientBoostingClassifier, ExtraTreesClassifier, AdaBoostClassifier, RandomForestClassifier, BaggingClassifier
|
| 43 |
+
from sklearn.linear_model import RidgeClassifier, Perceptron, PassiveAggressiveClassifier, LogisticRegression, SGDClassifier
|
| 44 |
+
|
| 45 |
+
import optuna
|
| 46 |
+
from optuna.samplers import TPESampler
|
| 47 |
+
|
| 48 |
+
import matplotlib.pyplot as plt
|
| 49 |
+
import plotly.express as px
|
| 50 |
+
|
| 51 |
+
import warnings
|
| 52 |
+
from sklearn.exceptions import ConvergenceWarning
|
| 53 |
+
|
| 54 |
+
|
| 55 |
+
# In[2]:
|
| 56 |
+
|
| 57 |
+
|
| 58 |
+
# To see optuna progress you need to comment this row
|
| 59 |
+
optuna.logging.set_verbosity(optuna.logging.WARNING)
|
| 60 |
+
warnings.filterwarnings(action='ignore', category=ConvergenceWarning)
|
| 61 |
+
|
| 62 |
+
|
| 63 |
+
# <a id="1"></a>
|
| 64 |
+
# <h2 style='background:black; border:0; color:white'><center>1. Feature engineering<center><h2>
|
| 65 |
+
|
| 66 |
+
# #### In this notebook I will not focus on preprocessing and feature engineering steps, just show how to build your efficient ensemble in few lines of code. I use almost the same features as in the most of kernels in current competition.
|
| 67 |
+
|
| 68 |
+
# In[3]:
|
| 69 |
+
|
| 70 |
+
|
| 71 |
+
train = pd.read_csv('/kaggle/input/titanic/train.csv')
|
| 72 |
+
test = pd.read_csv('/kaggle/input/titanic/test.csv')
|
| 73 |
+
|
| 74 |
+
|
| 75 |
+
# In[4]:
|
| 76 |
+
|
| 77 |
+
|
| 78 |
+
train.head()
|
| 79 |
+
|
| 80 |
+
|
| 81 |
+
# Lets see percent of NaNs for every column in training set
|
| 82 |
+
|
| 83 |
+
# In[5]:
|
| 84 |
+
|
| 85 |
+
|
| 86 |
+
for col in train.columns:
|
| 87 |
+
print(col, str(round(100* train[col].isnull().sum() / len(train), 2)) + '%')
|
| 88 |
+
|
| 89 |
+
|
| 90 |
+
# Here is some basic preprocessing to get fast training and test datasets.
|
| 91 |
+
|
| 92 |
+
# In[6]:
|
| 93 |
+
|
| 94 |
+
|
| 95 |
+
train['LastName'] = train['Name'].str.split(',', expand=True)[0]
|
| 96 |
+
test['LastName'] = test['Name'].str.split(',', expand=True)[0]
|
| 97 |
+
ds = pd.concat([train, test])
|
| 98 |
+
|
| 99 |
+
sur = list()
|
| 100 |
+
died = list()
|
| 101 |
+
|
| 102 |
+
for index, row in ds.iterrows():
|
| 103 |
+
s = ds[(ds['LastName']==row['LastName']) & (ds['Survived']==1)]
|
| 104 |
+
d = ds[(ds['LastName']==row['LastName']) & (ds['Survived']==0)]
|
| 105 |
+
s=len(s)
|
| 106 |
+
if row['Survived'] == 1:
|
| 107 |
+
s-=1
|
| 108 |
+
d=len(d)
|
| 109 |
+
if row['Survived'] == 0:
|
| 110 |
+
d-=1
|
| 111 |
+
sur.append(s)
|
| 112 |
+
died.append(d)
|
| 113 |
+
|
| 114 |
+
ds['FamilySurvived'] = sur
|
| 115 |
+
ds['FamilyDied'] = died
|
| 116 |
+
ds['FamilySize'] = ds['SibSp'] + ds['Parch'] + 1
|
| 117 |
+
ds['IsAlone'] = 0
|
| 118 |
+
ds.loc[ds['FamilySize'] == 1, 'IsAlone'] = 1
|
| 119 |
+
ds['Fare'] = ds['Fare'].fillna(train['Fare'].median())
|
| 120 |
+
ds['Embarked'] = ds['Embarked'].fillna('Q')
|
| 121 |
+
|
| 122 |
+
train = ds[ds['Survived'].notnull()]
|
| 123 |
+
test = ds[ds['Survived'].isnull()]
|
| 124 |
+
test = test.drop(['Survived'], axis=1)
|
| 125 |
+
|
| 126 |
+
train['rich_woman'] = 0
|
| 127 |
+
test['rich_woman'] = 0
|
| 128 |
+
train['men_3'] = 0
|
| 129 |
+
test['men_3'] = 0
|
| 130 |
+
|
| 131 |
+
train.loc[(train['Pclass']<=2) & (train['Sex']=='female'), 'rich_woman'] = 1
|
| 132 |
+
test.loc[(test['Pclass']<=2) & (test['Sex']=='female'), 'rich_woman'] = 1
|
| 133 |
+
train.loc[(train['Pclass']==3) & (train['Sex']=='male'), 'men_3'] = 1
|
| 134 |
+
test.loc[(test['Pclass']==3) & (test['Sex']=='male'), 'men_3'] = 1
|
| 135 |
+
|
| 136 |
+
train['rich_woman'] = train['rich_woman'].astype(np.int8)
|
| 137 |
+
test['rich_woman'] = test['rich_woman'].astype(np.int8)
|
| 138 |
+
|
| 139 |
+
train["Cabin"] = pd.Series([i[0] if not pd.isnull(i) else 'X' for i in train['Cabin']])
|
| 140 |
+
test['Cabin'] = pd.Series([i[0] if not pd.isnull(i) else 'X' for i in test['Cabin']])
|
| 141 |
+
|
| 142 |
+
for cat in ['Pclass', 'Sex', 'Embarked', 'Cabin']:
|
| 143 |
+
train = pd.concat([train, pd.get_dummies(train[cat], prefix=cat)], axis=1)
|
| 144 |
+
train = train.drop([cat], axis=1)
|
| 145 |
+
test = pd.concat([test, pd.get_dummies(test[cat], prefix=cat)], axis=1)
|
| 146 |
+
test = test.drop([cat], axis=1)
|
| 147 |
+
|
| 148 |
+
train = train.drop(['PassengerId', 'Ticket', 'LastName', 'SibSp', 'Parch', 'Sex_male', 'Name'], axis=1)
|
| 149 |
+
test = test.drop(['PassengerId', 'Ticket', 'LastName', 'SibSp', 'Parch', 'Sex_male', 'Name'], axis=1)
|
| 150 |
+
|
| 151 |
+
train = train.fillna(-1)
|
| 152 |
+
test = test.fillna(-1)
|
| 153 |
+
|
| 154 |
+
train.head()
|
| 155 |
+
|
| 156 |
+
|
| 157 |
+
# Let's do some visualization.
|
| 158 |
+
|
| 159 |
+
# In[7]:
|
| 160 |
+
|
| 161 |
+
|
| 162 |
+
fig = px.box(
|
| 163 |
+
train,
|
| 164 |
+
x="Survived",
|
| 165 |
+
y="Age",
|
| 166 |
+
points='all',
|
| 167 |
+
title='Age & Survived box plot',
|
| 168 |
+
width=700,
|
| 169 |
+
height=500
|
| 170 |
+
)
|
| 171 |
+
|
| 172 |
+
fig.show()
|
| 173 |
+
|
| 174 |
+
|
| 175 |
+
# We can see from training set that almost all people with Age higher than 63 years didn't survive. Can use these information in modeling post processing.
|
| 176 |
+
|
| 177 |
+
# In[8]:
|
| 178 |
+
|
| 179 |
+
|
| 180 |
+
fig = px.box(
|
| 181 |
+
train,
|
| 182 |
+
x="Survived",
|
| 183 |
+
y="Fare",
|
| 184 |
+
points='all',
|
| 185 |
+
title='Fare & Survived box plot',
|
| 186 |
+
width=700,
|
| 187 |
+
height=500
|
| 188 |
+
)
|
| 189 |
+
|
| 190 |
+
fig.show()
|
| 191 |
+
|
| 192 |
+
|
| 193 |
+
# In[9]:
|
| 194 |
+
|
| 195 |
+
|
| 196 |
+
fig = px.box(
|
| 197 |
+
train,
|
| 198 |
+
x="Survived",
|
| 199 |
+
y="FamilySize",
|
| 200 |
+
points='all',
|
| 201 |
+
title='Family Size & Survived box plot',
|
| 202 |
+
width=700,
|
| 203 |
+
height=500
|
| 204 |
+
)
|
| 205 |
+
|
| 206 |
+
fig.show()
|
| 207 |
+
|
| 208 |
+
|
| 209 |
+
# Another one thing. People with family size more than 7 didn't survive.
|
| 210 |
+
|
| 211 |
+
# In[10]:
|
| 212 |
+
|
| 213 |
+
|
| 214 |
+
fig = px.box(
|
| 215 |
+
train,
|
| 216 |
+
x="Survived",
|
| 217 |
+
y="FamilyDied",
|
| 218 |
+
points='all',
|
| 219 |
+
title='Family Died & Survived box plot',
|
| 220 |
+
width=700,
|
| 221 |
+
height=500
|
| 222 |
+
)
|
| 223 |
+
|
| 224 |
+
fig.show()
|
| 225 |
+
|
| 226 |
+
|
| 227 |
+
# In[11]:
|
| 228 |
+
|
| 229 |
+
|
| 230 |
+
f = plt.figure(
|
| 231 |
+
figsize=(19, 15)
|
| 232 |
+
)
|
| 233 |
+
|
| 234 |
+
plt.matshow(
|
| 235 |
+
train.corr(),
|
| 236 |
+
fignum=f.number
|
| 237 |
+
)
|
| 238 |
+
|
| 239 |
+
plt.xticks(
|
| 240 |
+
range(train.shape[1]),
|
| 241 |
+
train.columns,
|
| 242 |
+
fontsize=14,
|
| 243 |
+
rotation=75
|
| 244 |
+
)
|
| 245 |
+
|
| 246 |
+
plt.yticks(
|
| 247 |
+
range(train.shape[1]),
|
| 248 |
+
train.columns,
|
| 249 |
+
fontsize=14
|
| 250 |
+
)
|
| 251 |
+
|
| 252 |
+
cb = plt.colorbar()
|
| 253 |
+
cb.ax.tick_params(
|
| 254 |
+
labelsize=14
|
| 255 |
+
)
|
| 256 |
+
|
| 257 |
+
|
| 258 |
+
# Lets create train and test dataset and create holdout set for validation.
|
| 259 |
+
|
| 260 |
+
# In[12]:
|
| 261 |
+
|
| 262 |
+
|
| 263 |
+
train.head()
|
| 264 |
+
|
| 265 |
+
|
| 266 |
+
# In[13]:
|
| 267 |
+
|
| 268 |
+
|
| 269 |
+
y = train['Survived']
|
| 270 |
+
X = train.drop(['Survived', 'Cabin_T'], axis=1)
|
| 271 |
+
X_test = test.copy()
|
| 272 |
+
|
| 273 |
+
X, X_val, y, y_val = train_test_split(X, y, random_state=0, test_size=0.2, shuffle=False)
|
| 274 |
+
|
| 275 |
+
|
| 276 |
+
# <a id="2"></a>
|
| 277 |
+
# <h2 style='background:black; border:0; color:white'><center>2. Single models training and optimization<center><h2>
|
| 278 |
+
|
| 279 |
+
# Lets create some separate single models and check accuracy score. We also try to optimize every single model using optuna framework. As we can see we can get some better results with it.
|
| 280 |
+
|
| 281 |
+
# In[14]:
|
| 282 |
+
|
| 283 |
+
|
| 284 |
+
class Optimizer:
|
| 285 |
+
def __init__(self, metric, trials=30):
|
| 286 |
+
self.metric = metric
|
| 287 |
+
self.trials = trials
|
| 288 |
+
self.sampler = TPESampler(seed=666)
|
| 289 |
+
|
| 290 |
+
def objective(self, trial):
|
| 291 |
+
model = create_model(trial)
|
| 292 |
+
model.fit(X, y)
|
| 293 |
+
preds = model.predict(X_val)
|
| 294 |
+
if self.metric == 'acc':
|
| 295 |
+
return accuracy_score(y_val, preds)
|
| 296 |
+
else:
|
| 297 |
+
return f1_score(y_val, preds)
|
| 298 |
+
|
| 299 |
+
def optimize(self):
|
| 300 |
+
study = optuna.create_study(direction="maximize", sampler=self.sampler)
|
| 301 |
+
study.optimize(self.objective, n_trials=self.trials)
|
| 302 |
+
return study.best_params
|
| 303 |
+
|
| 304 |
+
|
| 305 |
+
# In[15]:
|
| 306 |
+
|
| 307 |
+
|
| 308 |
+
rf = RandomForestClassifier(
|
| 309 |
+
random_state=666
|
| 310 |
+
)
|
| 311 |
+
rf.fit(X, y)
|
| 312 |
+
preds = rf.predict(X_val)
|
| 313 |
+
|
| 314 |
+
print('Random Forest accuracy: ', accuracy_score(y_val, preds))
|
| 315 |
+
print('Random Forest f1-score: ', f1_score(y_val, preds))
|
| 316 |
+
|
| 317 |
+
def create_model(trial):
|
| 318 |
+
max_depth = trial.suggest_int("max_depth", 2, 6)
|
| 319 |
+
n_estimators = trial.suggest_int("n_estimators", 2, 150)
|
| 320 |
+
min_samples_leaf = trial.suggest_int("min_samples_leaf", 1, 10)
|
| 321 |
+
model = RandomForestClassifier(
|
| 322 |
+
min_samples_leaf=min_samples_leaf,
|
| 323 |
+
n_estimators=n_estimators,
|
| 324 |
+
max_depth=max_depth,
|
| 325 |
+
random_state=666
|
| 326 |
+
)
|
| 327 |
+
return model
|
| 328 |
+
|
| 329 |
+
optimizer = Optimizer('f1')
|
| 330 |
+
rf_f1_params = optimizer.optimize()
|
| 331 |
+
rf_f1_params['random_state'] = 666
|
| 332 |
+
rf_f1 = RandomForestClassifier(
|
| 333 |
+
**rf_f1_params
|
| 334 |
+
)
|
| 335 |
+
rf_f1.fit(X, y)
|
| 336 |
+
preds = rf_f1.predict(X_val)
|
| 337 |
+
|
| 338 |
+
print('Optimized on F1 score')
|
| 339 |
+
print('Optimized Random Forest: ', accuracy_score(y_val, preds))
|
| 340 |
+
print('Optimized Random Forest f1-score: ', f1_score(y_val, preds))
|
| 341 |
+
|
| 342 |
+
optimizer = Optimizer('acc')
|
| 343 |
+
rf_acc_params = optimizer.optimize()
|
| 344 |
+
rf_acc_params['random_state'] = 666
|
| 345 |
+
rf_acc = RandomForestClassifier(
|
| 346 |
+
**rf_acc_params
|
| 347 |
+
)
|
| 348 |
+
rf_acc.fit(X, y)
|
| 349 |
+
preds = rf_acc.predict(X_val)
|
| 350 |
+
|
| 351 |
+
print('Optimized on accuracy')
|
| 352 |
+
print('Optimized Random Forest: ', accuracy_score(y_val, preds))
|
| 353 |
+
print('Optimized Random Forest f1-score: ', f1_score(y_val, preds))
|
| 354 |
+
|
| 355 |
+
|
| 356 |
+
# In[16]:
|
| 357 |
+
|
| 358 |
+
|
| 359 |
+
xgb = XGBClassifier(
|
| 360 |
+
random_state=666
|
| 361 |
+
)
|
| 362 |
+
xgb.fit(X, y)
|
| 363 |
+
preds = xgb.predict(X_val)
|
| 364 |
+
|
| 365 |
+
print('XGBoost accuracy: ', accuracy_score(y_val, preds))
|
| 366 |
+
print('XGBoost f1-score: ', f1_score(y_val, preds))
|
| 367 |
+
|
| 368 |
+
def create_model(trial):
|
| 369 |
+
max_depth = trial.suggest_int("max_depth", 2, 6)
|
| 370 |
+
n_estimators = trial.suggest_int("n_estimators", 1, 150)
|
| 371 |
+
learning_rate = trial.suggest_uniform('learning_rate', 0.0000001, 1)
|
| 372 |
+
gamma = trial.suggest_uniform('gamma', 0.0000001, 1)
|
| 373 |
+
subsample = trial.suggest_uniform('subsample', 0.0001, 1.0)
|
| 374 |
+
model = XGBClassifier(
|
| 375 |
+
learning_rate=learning_rate,
|
| 376 |
+
n_estimators=n_estimators,
|
| 377 |
+
max_depth=max_depth,
|
| 378 |
+
gamma=gamma,
|
| 379 |
+
subsample=subsample,
|
| 380 |
+
random_state=666
|
| 381 |
+
)
|
| 382 |
+
return model
|
| 383 |
+
|
| 384 |
+
optimizer = Optimizer('f1')
|
| 385 |
+
xgb_f1_params = optimizer.optimize()
|
| 386 |
+
xgb_f1_params['random_state'] = 666
|
| 387 |
+
xgb_f1 = XGBClassifier(
|
| 388 |
+
**xgb_f1_params
|
| 389 |
+
)
|
| 390 |
+
xgb_f1.fit(X, y)
|
| 391 |
+
preds = xgb_f1.predict(X_val)
|
| 392 |
+
|
| 393 |
+
print('Optimized on F1 score')
|
| 394 |
+
print('Optimized XGBoost accuracy: ', accuracy_score(y_val, preds))
|
| 395 |
+
print('Optimized XGBoost f1-score: ', f1_score(y_val, preds))
|
| 396 |
+
|
| 397 |
+
optimizer = Optimizer('acc')
|
| 398 |
+
xgb_acc_params = optimizer.optimize()
|
| 399 |
+
xgb_acc_params['random_state'] = 666
|
| 400 |
+
xgb_acc = XGBClassifier(
|
| 401 |
+
**xgb_acc_params
|
| 402 |
+
)
|
| 403 |
+
xgb_acc.fit(X, y)
|
| 404 |
+
preds = xgb_acc.predict(X_val)
|
| 405 |
+
|
| 406 |
+
print('Optimized on accuracy')
|
| 407 |
+
print('Optimized XGBoost accuracy: ', accuracy_score(y_val, preds))
|
| 408 |
+
print('Optimized XGBoost f1-score: ', f1_score(y_val, preds))
|
| 409 |
+
|
| 410 |
+
|
| 411 |
+
# In[17]:
|
| 412 |
+
|
| 413 |
+
|
| 414 |
+
lgb = LGBMClassifier(
|
| 415 |
+
random_state=666
|
| 416 |
+
)
|
| 417 |
+
lgb.fit(X, y)
|
| 418 |
+
preds = lgb.predict(X_val)
|
| 419 |
+
|
| 420 |
+
print('LightGBM accuracy: ', accuracy_score(y_val, preds))
|
| 421 |
+
print('LightGBM f1-score: ', f1_score(y_val, preds))
|
| 422 |
+
|
| 423 |
+
def create_model(trial):
|
| 424 |
+
max_depth = trial.suggest_int("max_depth", 2, 6)
|
| 425 |
+
n_estimators = trial.suggest_int("n_estimators", 1, 150)
|
| 426 |
+
learning_rate = trial.suggest_uniform('learning_rate', 0.0000001, 1)
|
| 427 |
+
num_leaves = trial.suggest_int("num_leaves", 2, 3000)
|
| 428 |
+
min_child_samples = trial.suggest_int('min_child_samples', 3, 200)
|
| 429 |
+
model = LGBMClassifier(
|
| 430 |
+
learning_rate=learning_rate,
|
| 431 |
+
n_estimators=n_estimators,
|
| 432 |
+
max_depth=max_depth,
|
| 433 |
+
num_leaves=num_leaves,
|
| 434 |
+
min_child_samples=min_child_samples,
|
| 435 |
+
random_state=666
|
| 436 |
+
)
|
| 437 |
+
return model
|
| 438 |
+
|
| 439 |
+
optimizer = Optimizer('f1')
|
| 440 |
+
lgb_f1_params = optimizer.optimize()
|
| 441 |
+
lgb_f1_params['random_state'] = 666
|
| 442 |
+
lgb_f1 = LGBMClassifier(
|
| 443 |
+
**lgb_f1_params
|
| 444 |
+
)
|
| 445 |
+
lgb_f1.fit(X, y)
|
| 446 |
+
preds = lgb_f1.predict(X_val)
|
| 447 |
+
|
| 448 |
+
print('Optimized on F1-score')
|
| 449 |
+
print('Optimized LightGBM accuracy: ', accuracy_score(y_val, preds))
|
| 450 |
+
print('Optimized LightGBM f1-score: ', f1_score(y_val, preds))
|
| 451 |
+
|
| 452 |
+
optimizer = Optimizer('acc')
|
| 453 |
+
lgb_acc_params = optimizer.optimize()
|
| 454 |
+
lgb_acc_params['random_state'] = 666
|
| 455 |
+
lgb_acc = LGBMClassifier(
|
| 456 |
+
**lgb_acc_params
|
| 457 |
+
)
|
| 458 |
+
lgb_acc.fit(X, y)
|
| 459 |
+
preds = lgb_acc.predict(X_val)
|
| 460 |
+
|
| 461 |
+
print('Optimized on accuracy')
|
| 462 |
+
print('Optimized LightGBM accuracy: ', accuracy_score(y_val, preds))
|
| 463 |
+
print('Optimized LightGBM f1-score: ', f1_score(y_val, preds))
|
| 464 |
+
|
| 465 |
+
|
| 466 |
+
# In[18]:
|
| 467 |
+
|
| 468 |
+
|
| 469 |
+
lr = LogisticRegression(
|
| 470 |
+
random_state=666
|
| 471 |
+
)
|
| 472 |
+
lr.fit(X, y)
|
| 473 |
+
preds = lr.predict(X_val)
|
| 474 |
+
|
| 475 |
+
print('Logistic Regression: ', accuracy_score(y_val, preds))
|
| 476 |
+
print('Logistic Regression f1-score: ', f1_score(y_val, preds))
|
| 477 |
+
|
| 478 |
+
|
| 479 |
+
# In[19]:
|
| 480 |
+
|
| 481 |
+
|
| 482 |
+
dt = DecisionTreeClassifier(
|
| 483 |
+
random_state=666
|
| 484 |
+
)
|
| 485 |
+
dt.fit(X, y)
|
| 486 |
+
preds = dt.predict(X_val)
|
| 487 |
+
|
| 488 |
+
print('Decision Tree accuracy: ', accuracy_score(y_val, preds))
|
| 489 |
+
print('Decision Tree f1-score: ', f1_score(y_val, preds))
|
| 490 |
+
|
| 491 |
+
def create_model(trial):
|
| 492 |
+
max_depth = trial.suggest_int("max_depth", 2, 6)
|
| 493 |
+
min_samples_split = trial.suggest_int('min_samples_split', 2, 16)
|
| 494 |
+
min_weight_fraction_leaf = trial.suggest_uniform('min_weight_fraction_leaf', 0.0, 0.5)
|
| 495 |
+
min_samples_leaf = trial.suggest_int('min_samples_leaf', 1, 10)
|
| 496 |
+
model = DecisionTreeClassifier(
|
| 497 |
+
min_samples_split=min_samples_split,
|
| 498 |
+
min_weight_fraction_leaf=min_weight_fraction_leaf,
|
| 499 |
+
max_depth=max_depth,
|
| 500 |
+
min_samples_leaf=min_samples_leaf,
|
| 501 |
+
random_state=666
|
| 502 |
+
)
|
| 503 |
+
return model
|
| 504 |
+
|
| 505 |
+
optimizer = Optimizer('f1')
|
| 506 |
+
dt_f1_params = optimizer.optimize()
|
| 507 |
+
dt_f1_params['random_state'] = 666
|
| 508 |
+
dt_f1 = DecisionTreeClassifier(
|
| 509 |
+
**dt_f1_params
|
| 510 |
+
)
|
| 511 |
+
dt_f1.fit(X, y)
|
| 512 |
+
preds = dt_f1.predict(X_val)
|
| 513 |
+
|
| 514 |
+
print('Optimized on F1-score')
|
| 515 |
+
print('Optimized Decision Tree accuracy: ', accuracy_score(y_val, preds))
|
| 516 |
+
print('Optimized Decision Tree f1-score: ', f1_score(y_val, preds))
|
| 517 |
+
|
| 518 |
+
optimizer = Optimizer('acc')
|
| 519 |
+
dt_acc_params = optimizer.optimize()
|
| 520 |
+
dt_acc_params['random_state'] = 666
|
| 521 |
+
dt_acc = DecisionTreeClassifier(
|
| 522 |
+
**dt_acc_params
|
| 523 |
+
)
|
| 524 |
+
dt_acc.fit(X, y)
|
| 525 |
+
preds = dt_acc.predict(X_val)
|
| 526 |
+
|
| 527 |
+
print('Optimized on accuracy')
|
| 528 |
+
print('Optimized Decision Tree accuracy: ', accuracy_score(y_val, preds))
|
| 529 |
+
print('Optimized Decision Tree f1-score: ', f1_score(y_val, preds))
|
| 530 |
+
|
| 531 |
+
|
| 532 |
+
# In[20]:
|
| 533 |
+
|
| 534 |
+
|
| 535 |
+
bc = BaggingClassifier(
|
| 536 |
+
random_state=666
|
| 537 |
+
)
|
| 538 |
+
bc.fit(X, y)
|
| 539 |
+
preds = bc.predict(X_val)
|
| 540 |
+
|
| 541 |
+
print('Bagging Classifier accuracy: ', accuracy_score(y_val, preds))
|
| 542 |
+
print('Bagging Classifier f1-score: ', f1_score(y_val, preds))
|
| 543 |
+
|
| 544 |
+
def create_model(trial):
|
| 545 |
+
n_estimators = trial.suggest_int('n_estimators', 2, 200)
|
| 546 |
+
max_samples = trial.suggest_int('max_samples', 1, 100)
|
| 547 |
+
model = BaggingClassifier(
|
| 548 |
+
n_estimators=n_estimators,
|
| 549 |
+
max_samples=max_samples,
|
| 550 |
+
random_state=666
|
| 551 |
+
)
|
| 552 |
+
return model
|
| 553 |
+
|
| 554 |
+
optimizer = Optimizer('f1')
|
| 555 |
+
bc_f1_params = optimizer.optimize()
|
| 556 |
+
bc_f1_params['random_state'] = 666
|
| 557 |
+
bc_f1 = BaggingClassifier(
|
| 558 |
+
**bc_f1_params
|
| 559 |
+
)
|
| 560 |
+
bc_f1.fit(X, y)
|
| 561 |
+
preds = bc_f1.predict(X_val)
|
| 562 |
+
|
| 563 |
+
print('Optimized on F1-score')
|
| 564 |
+
print('Optimized Bagging Classifier accuracy: ', accuracy_score(y_val, preds))
|
| 565 |
+
print('Optimized Bagging Classifier f1-score: ', f1_score(y_val, preds))
|
| 566 |
+
|
| 567 |
+
optimizer = Optimizer('acc')
|
| 568 |
+
bc_acc_params = optimizer.optimize()
|
| 569 |
+
bc_acc_params['random_state'] = 666
|
| 570 |
+
bc_acc = BaggingClassifier(
|
| 571 |
+
**bc_acc_params
|
| 572 |
+
)
|
| 573 |
+
bc_acc.fit(X, y)
|
| 574 |
+
preds = bc_acc.predict(X_val)
|
| 575 |
+
|
| 576 |
+
print('Optimized on accuracy')
|
| 577 |
+
print('Optimized Bagging Classifier accuracy: ', accuracy_score(y_val, preds))
|
| 578 |
+
print('Optimized Bagging Classifier f1-score: ', f1_score(y_val, preds))
|
| 579 |
+
|
| 580 |
+
|
| 581 |
+
# In[21]:
|
| 582 |
+
|
| 583 |
+
|
| 584 |
+
knn = KNeighborsClassifier()
|
| 585 |
+
knn.fit(X, y)
|
| 586 |
+
preds = knn.predict(X_val)
|
| 587 |
+
|
| 588 |
+
print('KNN accuracy: ', accuracy_score(y_val, preds))
|
| 589 |
+
print('KNN f1-score: ', f1_score(y_val, preds))
|
| 590 |
+
|
| 591 |
+
sampler = TPESampler(seed=0)
|
| 592 |
+
def create_model(trial):
|
| 593 |
+
n_neighbors = trial.suggest_int("n_neighbors", 2, 25)
|
| 594 |
+
model = KNeighborsClassifier(n_neighbors=n_neighbors)
|
| 595 |
+
return model
|
| 596 |
+
|
| 597 |
+
optimizer = Optimizer('f1')
|
| 598 |
+
knn_f1_params = optimizer.optimize()
|
| 599 |
+
knn_f1 = KNeighborsClassifier(
|
| 600 |
+
**knn_f1_params
|
| 601 |
+
)
|
| 602 |
+
knn_f1.fit(X, y)
|
| 603 |
+
preds = knn_f1.predict(X_val)
|
| 604 |
+
|
| 605 |
+
print('Optimized on F1-score')
|
| 606 |
+
print('Optimized KNN accuracy: ', accuracy_score(y_val, preds))
|
| 607 |
+
print('Optimized KNN f1-score: ', f1_score(y_val, preds))
|
| 608 |
+
|
| 609 |
+
optimizer = Optimizer('acc')
|
| 610 |
+
knn_acc_params = optimizer.optimize()
|
| 611 |
+
knn_acc = KNeighborsClassifier(
|
| 612 |
+
**knn_acc_params
|
| 613 |
+
)
|
| 614 |
+
knn_acc.fit(X, y)
|
| 615 |
+
preds = knn_acc.predict(X_val)
|
| 616 |
+
|
| 617 |
+
print('Optimized on accuracy')
|
| 618 |
+
print('Optimized KNN accuracy: ', accuracy_score(y_val, preds))
|
| 619 |
+
print('Optimized KNN f1-score: ', f1_score(y_val, preds))
|
| 620 |
+
|
| 621 |
+
|
| 622 |
+
# In[22]:
|
| 623 |
+
|
| 624 |
+
|
| 625 |
+
abc = AdaBoostClassifier(
|
| 626 |
+
random_state=666
|
| 627 |
+
)
|
| 628 |
+
abc.fit(X, y)
|
| 629 |
+
preds = abc.predict(X_val)
|
| 630 |
+
|
| 631 |
+
print('AdaBoost accuracy: ', accuracy_score(y_val, preds))
|
| 632 |
+
print('AdaBoost f1-score: ', f1_score(y_val, preds))
|
| 633 |
+
|
| 634 |
+
def create_model(trial):
|
| 635 |
+
n_estimators = trial.suggest_int("n_estimators", 2, 150)
|
| 636 |
+
learning_rate = trial.suggest_uniform('learning_rate', 0.0005, 1.0)
|
| 637 |
+
model = AdaBoostClassifier(
|
| 638 |
+
n_estimators=n_estimators,
|
| 639 |
+
learning_rate=learning_rate,
|
| 640 |
+
random_state=666
|
| 641 |
+
)
|
| 642 |
+
return model
|
| 643 |
+
|
| 644 |
+
optimizer = Optimizer('f1')
|
| 645 |
+
abc_f1_params = optimizer.optimize()
|
| 646 |
+
abc_f1_params['random_state'] = 666
|
| 647 |
+
abc_f1 = AdaBoostClassifier(
|
| 648 |
+
**abc_f1_params
|
| 649 |
+
)
|
| 650 |
+
abc_f1.fit(X, y)
|
| 651 |
+
preds = abc_f1.predict(X_val)
|
| 652 |
+
|
| 653 |
+
print('Optimized on F1-score')
|
| 654 |
+
print('Optimized AdaBoost accuracy: ', accuracy_score(y_val, preds))
|
| 655 |
+
print('Optimized AdaBoost f1-score: ', f1_score(y_val, preds))
|
| 656 |
+
|
| 657 |
+
optimizer = Optimizer('acc')
|
| 658 |
+
abc_acc_params = optimizer.optimize()
|
| 659 |
+
abc_acc_params['random_state'] = 666
|
| 660 |
+
abc_acc = AdaBoostClassifier(
|
| 661 |
+
**abc_acc_params
|
| 662 |
+
)
|
| 663 |
+
abc_acc.fit(X, y)
|
| 664 |
+
preds = abc_acc.predict(X_val)
|
| 665 |
+
|
| 666 |
+
print('Optimized on accuracy')
|
| 667 |
+
print('Optimized AdaBoost accuracy: ', accuracy_score(y_val, preds))
|
| 668 |
+
print('Optimized AdaBoost f1-score: ', f1_score(y_val, preds))
|
| 669 |
+
|
| 670 |
+
|
| 671 |
+
# In[23]:
|
| 672 |
+
|
| 673 |
+
|
| 674 |
+
et = ExtraTreesClassifier(
|
| 675 |
+
random_state=666
|
| 676 |
+
)
|
| 677 |
+
et.fit(X, y)
|
| 678 |
+
preds = et.predict(X_val)
|
| 679 |
+
|
| 680 |
+
print('ExtraTreesClassifier accuracy: ', accuracy_score(y_val, preds))
|
| 681 |
+
print('ExtraTreesClassifier f1-score: ', f1_score(y_val, preds))
|
| 682 |
+
|
| 683 |
+
def create_model(trial):
|
| 684 |
+
n_estimators = trial.suggest_int("n_estimators", 2, 150)
|
| 685 |
+
max_depth = trial.suggest_int("max_depth", 2, 6)
|
| 686 |
+
model = ExtraTreesClassifier(
|
| 687 |
+
n_estimators=n_estimators,
|
| 688 |
+
max_depth=max_depth,
|
| 689 |
+
random_state=0
|
| 690 |
+
)
|
| 691 |
+
return model
|
| 692 |
+
|
| 693 |
+
optimizer = Optimizer('f1')
|
| 694 |
+
et_f1_params = optimizer.optimize()
|
| 695 |
+
et_f1_params['random_state'] = 666
|
| 696 |
+
et_f1 = ExtraTreesClassifier(
|
| 697 |
+
**et_f1_params
|
| 698 |
+
)
|
| 699 |
+
et_f1.fit(X, y)
|
| 700 |
+
preds = et_f1.predict(X_val)
|
| 701 |
+
|
| 702 |
+
print('Optimized on F1-score')
|
| 703 |
+
print('Optimized ExtraTreesClassifier accuracy: ', accuracy_score(y_val, preds))
|
| 704 |
+
print('Optimized ExtraTreesClassifier f1-score: ', f1_score(y_val, preds))
|
| 705 |
+
|
| 706 |
+
optimizer = Optimizer('acc')
|
| 707 |
+
et_acc_params = optimizer.optimize()
|
| 708 |
+
et_acc_params['random_state'] = 666
|
| 709 |
+
et_acc = ExtraTreesClassifier(
|
| 710 |
+
**et_acc_params
|
| 711 |
+
)
|
| 712 |
+
et_acc.fit(X, y)
|
| 713 |
+
preds = et_acc.predict(X_val)
|
| 714 |
+
|
| 715 |
+
print('Optimized on accuracy')
|
| 716 |
+
print('Optimized ExtraTreesClassifier accuracy: ', accuracy_score(y_val, preds))
|
| 717 |
+
print('Optimized ExtraTreesClassifier f1-score: ', f1_score(y_val, preds))
|
| 718 |
+
|
| 719 |
+
|
| 720 |
+
# <a id="3"></a>
|
| 721 |
+
# <h2 style='background:black; border:0; color:white'><center>3. SuperLearner training and optimization<center><h2>
|
| 722 |
+
|
| 723 |
+
# Now we will create ensemble model named SuperLearner from mlens package. For details check https://machinelearningmastery.com/super-learner-ensemble-in-python/
|
| 724 |
+
|
| 725 |
+
# We are going to use our single models in the first layer and LogisticRegressor as metalearner.
|
| 726 |
+
|
| 727 |
+
# In[24]:
|
| 728 |
+
|
| 729 |
+
|
| 730 |
+
model = SuperLearner(
|
| 731 |
+
folds=5,
|
| 732 |
+
random_state=666
|
| 733 |
+
)
|
| 734 |
+
|
| 735 |
+
model.add(
|
| 736 |
+
[
|
| 737 |
+
bc,
|
| 738 |
+
lgb,
|
| 739 |
+
xgb,
|
| 740 |
+
rf,
|
| 741 |
+
dt,
|
| 742 |
+
knn
|
| 743 |
+
]
|
| 744 |
+
)
|
| 745 |
+
|
| 746 |
+
model.add_meta(
|
| 747 |
+
LogisticRegression()
|
| 748 |
+
)
|
| 749 |
+
|
| 750 |
+
model.fit(X, y)
|
| 751 |
+
|
| 752 |
+
preds = model.predict(X_val)
|
| 753 |
+
|
| 754 |
+
print('SuperLearner accuracy: ', accuracy_score(y_val, preds))
|
| 755 |
+
print('SuperLearner f1-score: ', f1_score(y_val, preds))
|
| 756 |
+
|
| 757 |
+
|
| 758 |
+
# Let's optimize SuperLearner
|
| 759 |
+
|
| 760 |
+
# In[25]:
|
| 761 |
+
|
| 762 |
+
|
| 763 |
+
mdict = {
|
| 764 |
+
'RF': RandomForestClassifier(random_state=666),
|
| 765 |
+
'XGB': XGBClassifier(random_state=666),
|
| 766 |
+
'LGBM': LGBMClassifier(random_state=666),
|
| 767 |
+
'DT': DecisionTreeClassifier(random_state=666),
|
| 768 |
+
'KNN': KNeighborsClassifier(),
|
| 769 |
+
'BC': BaggingClassifier(random_state=666),
|
| 770 |
+
'OARF': RandomForestClassifier(**rf_acc_params),
|
| 771 |
+
'OFRF': RandomForestClassifier(**rf_f1_params),
|
| 772 |
+
'OAXGB': XGBClassifier(**xgb_acc_params),
|
| 773 |
+
'OFXGB': XGBClassifier(**xgb_f1_params),
|
| 774 |
+
'OALGBM': LGBMClassifier(**lgb_acc_params),
|
| 775 |
+
'OFLGBM': LGBMClassifier(**lgb_f1_params),
|
| 776 |
+
'OADT': DecisionTreeClassifier(**dt_acc_params),
|
| 777 |
+
'OFDT': DecisionTreeClassifier(**dt_f1_params),
|
| 778 |
+
'OAKNN': KNeighborsClassifier(**knn_acc_params),
|
| 779 |
+
'OFKNN': KNeighborsClassifier(**knn_f1_params),
|
| 780 |
+
'OABC': BaggingClassifier(**bc_acc_params),
|
| 781 |
+
'OFBC': BaggingClassifier(**bc_f1_params),
|
| 782 |
+
'OAABC': AdaBoostClassifier(**abc_acc_params),
|
| 783 |
+
'OFABC': AdaBoostClassifier(**abc_f1_params),
|
| 784 |
+
'OAET': ExtraTreesClassifier(**et_acc_params),
|
| 785 |
+
'OFET': ExtraTreesClassifier(**et_f1_params),
|
| 786 |
+
'LR': LogisticRegression(random_state=666),
|
| 787 |
+
'ABC': AdaBoostClassifier(random_state=666),
|
| 788 |
+
'SGD': SGDClassifier(random_state=666),
|
| 789 |
+
'ET': ExtraTreesClassifier(random_state=666),
|
| 790 |
+
'MLP': MLPClassifier(random_state=666),
|
| 791 |
+
'GB': GradientBoostingClassifier(random_state=666),
|
| 792 |
+
'RDG': RidgeClassifier(random_state=666),
|
| 793 |
+
'PCP': Perceptron(random_state=666),
|
| 794 |
+
'PAC': PassiveAggressiveClassifier(random_state=666)
|
| 795 |
+
}
|
| 796 |
+
|
| 797 |
+
|
| 798 |
+
# In[26]:
|
| 799 |
+
|
| 800 |
+
|
| 801 |
+
def create_model(trial):
|
| 802 |
+
model_names = list()
|
| 803 |
+
models_list = [
|
| 804 |
+
'RF', 'XGB', 'LGBM', 'DT',
|
| 805 |
+
'KNN', 'BC', 'OARF', 'OFRF',
|
| 806 |
+
'OAXGB', 'OFXGB', 'OALGBM',
|
| 807 |
+
'OFLGBM', 'OADT', 'OFDT',
|
| 808 |
+
'OAKNN', 'OFKNN', 'OABC',
|
| 809 |
+
'OFBC', 'OAABC', 'OFABC',
|
| 810 |
+
'OAET', 'OFET', 'LR',
|
| 811 |
+
'ABC', 'SGD', 'ET',
|
| 812 |
+
'MLP', 'GB', 'RDG',
|
| 813 |
+
'PCP', 'PAC'
|
| 814 |
+
]
|
| 815 |
+
|
| 816 |
+
head_list = [
|
| 817 |
+
'RF',
|
| 818 |
+
'XGB',
|
| 819 |
+
'LGBM',
|
| 820 |
+
'DT',
|
| 821 |
+
'KNN',
|
| 822 |
+
'BC',
|
| 823 |
+
'LR',
|
| 824 |
+
'ABC',
|
| 825 |
+
'SGD',
|
| 826 |
+
'ET',
|
| 827 |
+
'MLP',
|
| 828 |
+
'GB',
|
| 829 |
+
'RDG',
|
| 830 |
+
'PCP',
|
| 831 |
+
'PAC'
|
| 832 |
+
]
|
| 833 |
+
|
| 834 |
+
n_models = trial.suggest_int("n_models", 2, 6)
|
| 835 |
+
for i in range(n_models):
|
| 836 |
+
model_item = trial.suggest_categorical('model_{}'.format(i), models_list)
|
| 837 |
+
if model_item not in model_names:
|
| 838 |
+
model_names.append(model_item)
|
| 839 |
+
|
| 840 |
+
folds = trial.suggest_int("folds", 2, 6)
|
| 841 |
+
|
| 842 |
+
model = SuperLearner(
|
| 843 |
+
folds=folds,
|
| 844 |
+
random_state=666
|
| 845 |
+
)
|
| 846 |
+
|
| 847 |
+
models = [
|
| 848 |
+
mdict[item] for item in model_names
|
| 849 |
+
]
|
| 850 |
+
model.add(models)
|
| 851 |
+
head = trial.suggest_categorical('head', head_list)
|
| 852 |
+
model.add_meta(
|
| 853 |
+
mdict[head]
|
| 854 |
+
)
|
| 855 |
+
|
| 856 |
+
return model
|
| 857 |
+
|
| 858 |
+
def objective(trial):
|
| 859 |
+
model = create_model(trial)
|
| 860 |
+
model.fit(X, y)
|
| 861 |
+
preds = model.predict(X_val)
|
| 862 |
+
score = accuracy_score(y_val, preds)
|
| 863 |
+
return score
|
| 864 |
+
|
| 865 |
+
study = optuna.create_study(
|
| 866 |
+
direction="maximize",
|
| 867 |
+
sampler=sampler
|
| 868 |
+
)
|
| 869 |
+
|
| 870 |
+
study.optimize(
|
| 871 |
+
objective,
|
| 872 |
+
n_trials=50
|
| 873 |
+
)
|
| 874 |
+
|
| 875 |
+
|
| 876 |
+
# In[27]:
|
| 877 |
+
|
| 878 |
+
|
| 879 |
+
params = study.best_params
|
| 880 |
+
|
| 881 |
+
head = params['head']
|
| 882 |
+
folds = params['folds']
|
| 883 |
+
del params['head'], params['n_models'], params['folds']
|
| 884 |
+
result = list()
|
| 885 |
+
for key, value in params.items():
|
| 886 |
+
if value not in result:
|
| 887 |
+
result.append(value)
|
| 888 |
+
|
| 889 |
+
result
|
| 890 |
+
|
| 891 |
+
|
| 892 |
+
# In[28]:
|
| 893 |
+
|
| 894 |
+
|
| 895 |
+
model = SuperLearner(
|
| 896 |
+
folds=folds,
|
| 897 |
+
random_state=666
|
| 898 |
+
)
|
| 899 |
+
|
| 900 |
+
models = [
|
| 901 |
+
mdict[item] for item in result
|
| 902 |
+
]
|
| 903 |
+
model.add(models)
|
| 904 |
+
model.add_meta(mdict[head])
|
| 905 |
+
|
| 906 |
+
model.fit(X, y)
|
| 907 |
+
|
| 908 |
+
preds = model.predict(X_val)
|
| 909 |
+
|
| 910 |
+
print('Optimized SuperLearner accuracy: ', accuracy_score(y_val, preds))
|
| 911 |
+
print('Optimized SuperLearner f1-score: ', f1_score(y_val, preds))
|
| 912 |
+
|
| 913 |
+
|
| 914 |
+
# As we can see we improved our best single score only in a few lines of code. Feel free to add new features and try different models inside superlearner.
|
| 915 |
+
|
| 916 |
+
# <a id="4"></a>
|
| 917 |
+
# <h2 style='background:black; border:0; color:white'><center>4. Final submission<center><h2>
|
| 918 |
+
|
| 919 |
+
# In[29]:
|
| 920 |
+
|
| 921 |
+
|
| 922 |
+
preds = model.predict(X_test)
|
| 923 |
+
preds = preds.astype(np.int16)
|
| 924 |
+
|
| 925 |
+
|
| 926 |
+
# In[30]:
|
| 927 |
+
|
| 928 |
+
|
| 929 |
+
submission = pd.read_csv('../input/titanic/gender_submission.csv')
|
| 930 |
+
submission['Survived'] = preds
|
| 931 |
+
submission.to_csv('submission.csv', index=False)
|
| 932 |
+
|
| 933 |
+
|
| 934 |
+
# In[31]:
|
| 935 |
+
|
| 936 |
+
|
| 937 |
+
submission.head()
|
| 938 |
+
|
| 939 |
+
|
| 940 |
+
# In[ ]:
|
| 941 |
+
|
| 942 |
+
|
| 943 |
+
|
| 944 |
+
|
Titanic/Kernels/GBC/.ipynb_checkpoints/0-introduction-to-ensembling-stacking-in-python-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/GBC/.ipynb_checkpoints/1-a-data-science-framework-to-achieve-99-accuracy-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Titanic/Kernels/GBC/.ipynb_checkpoints/10-titanic-survival-prediction-end-to-end-ml-pipeline-checkpoint.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|