correct folder files
Browse files
swinunetr-btcv-small/LICENSE → LICENSE
RENAMED
|
File without changes
|
README.md
CHANGED
|
@@ -1,3 +1,132 @@
|
|
| 1 |
---
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 2 |
license: apache-2.0
|
|
|
|
|
|
|
| 3 |
---
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
---
|
| 2 |
+
language: en
|
| 3 |
+
tags:
|
| 4 |
+
- btcv
|
| 5 |
+
- medical
|
| 6 |
+
- swin
|
| 7 |
license: apache-2.0
|
| 8 |
+
datasets:
|
| 9 |
+
- BTCV
|
| 10 |
---
|
| 11 |
+
|
| 12 |
+
# Model Overview
|
| 13 |
+
|
| 14 |
+
This repository contains the code for Swin UNETR [1,2]. Swin UNETR is the state-of-the-art on Medical Segmentation
|
| 15 |
+
Decathlon (MSD) and Beyond the Cranial Vault (BTCV) Segmentation Challenge dataset. In [1], a novel methodology is devised for pre-training Swin UNETR backbone in a self-supervised
|
| 16 |
+
manner. We provide the option for training Swin UNETR by fine-tuning from pre-trained self-supervised weights or from scratch.
|
| 17 |
+
|
| 18 |
+
The source repository for the training of these models can be found [here](https://github.com/Project-MONAI/research-contributions/tree/main/SwinUNETR/BTCV).
|
| 19 |
+
|
| 20 |
+
# Installing Dependencies
|
| 21 |
+
Dependencies for training and inference can be installed using the model requirements :
|
| 22 |
+
``` bash
|
| 23 |
+
pip install -r requirements.txt
|
| 24 |
+
```
|
| 25 |
+
|
| 26 |
+
# Intended uses & limitations
|
| 27 |
+
|
| 28 |
+
You can use the raw model for masked language modeling, but it's mostly intended to be fine-tuned on a downstream task.
|
| 29 |
+
|
| 30 |
+
Note that this model is primarily aimed at being fine-tuned on tasks which segment CAT scans or MRIs on images in dicom format.
|
| 31 |
+
|
| 32 |
+
# How to use
|
| 33 |
+
|
| 34 |
+
To install necessary dependencies, run the below in bash.
|
| 35 |
+
```
|
| 36 |
+
git clone https://github.com/darraghdog/Project-MONAI-research-contributions pmrc
|
| 37 |
+
pip install -r pmrc/requirements.txt
|
| 38 |
+
cd pmrc/SwinUNETR/BTCV
|
| 39 |
+
```
|
| 40 |
+
|
| 41 |
+
To load the model from the hub.
|
| 42 |
+
```
|
| 43 |
+
>>> from swinunetr import SwinUnetrModelForInference
|
| 44 |
+
>>> model = SwinUnetrModelForInference.from_pretrained('darragh/swinunetr-btcv-tiny')
|
| 45 |
+
```
|
| 46 |
+
|
| 47 |
+
You can also use `predict.py` to run inference for sample dicom medical images.
|
| 48 |
+
|
| 49 |
+
# Limitations and bias
|
| 50 |
+
|
| 51 |
+
The training data used for this model is specific to CAT scans from certain health facilities and machines. Data from other facilities may difffer in image distributions, and may require finetuning of the models for best performance.
|
| 52 |
+
|
| 53 |
+
# Evaluation results
|
| 54 |
+
|
| 55 |
+
We provide several pre-trained models on BTCV dataset in the following.
|
| 56 |
+
|
| 57 |
+
<table>
|
| 58 |
+
<tr>
|
| 59 |
+
<th>Name</th>
|
| 60 |
+
<th>Dice (overlap=0.7)</th>
|
| 61 |
+
<th>Dice (overlap=0.5)</th>
|
| 62 |
+
<th>Feature Size</th>
|
| 63 |
+
<th># params (M)</th>
|
| 64 |
+
<th>Self-Supervised Pre-trained </th>
|
| 65 |
+
</tr>
|
| 66 |
+
<tr>
|
| 67 |
+
<td>Swin UNETR/Base</td>
|
| 68 |
+
<td>82.25</td>
|
| 69 |
+
<td>81.86</td>
|
| 70 |
+
<td>48</td>
|
| 71 |
+
<td>62.1</td>
|
| 72 |
+
<td>Yes</td>
|
| 73 |
+
</tr>
|
| 74 |
+
|
| 75 |
+
<tr>
|
| 76 |
+
<td>Swin UNETR/Small</td>
|
| 77 |
+
<td>79.79</td>
|
| 78 |
+
<td>79.34</td>
|
| 79 |
+
<td>24</td>
|
| 80 |
+
<td>15.7</td>
|
| 81 |
+
<td>No</td>
|
| 82 |
+
</tr>
|
| 83 |
+
|
| 84 |
+
<tr>
|
| 85 |
+
<td>Swin UNETR/Tiny</td>
|
| 86 |
+
<td>72.05</td>
|
| 87 |
+
<td>70.35</td>
|
| 88 |
+
<td>12</td>
|
| 89 |
+
<td>4.0</td>
|
| 90 |
+
<td>No</td>
|
| 91 |
+
</tr>
|
| 92 |
+
|
| 93 |
+
</table>
|
| 94 |
+
|
| 95 |
+
# Data Preparation
|
| 96 |
+
|
| 97 |
+
|
| 98 |
+
The training data is from the [BTCV challenge dataset](https://www.synapse.org/#!Synapse:syn3193805/wiki/217752).
|
| 99 |
+
|
| 100 |
+
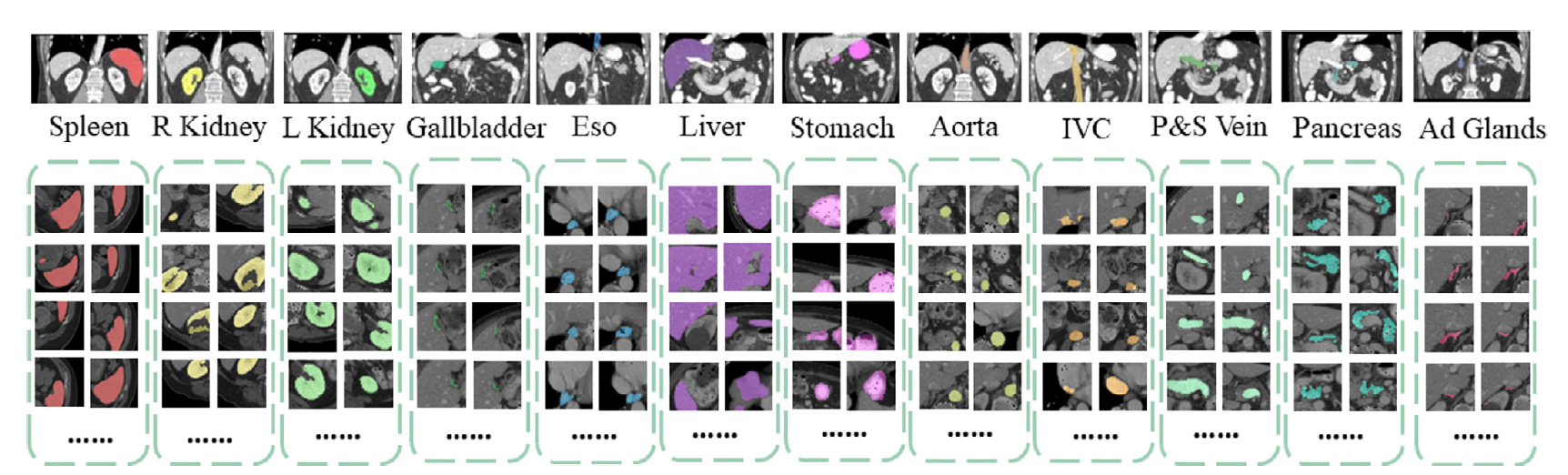
- Target: 13 abdominal organs including 1. Spleen 2. Right Kidney 3. Left Kideny 4.Gallbladder 5.Esophagus 6. Liver 7. Stomach 8.Aorta 9. IVC 10. Portal and Splenic Veins 11. Pancreas 12.Right adrenal gland 13.Left adrenal gland.
|
| 101 |
+
- Task: Segmentation
|
| 102 |
+
- Modality: CT
|
| 103 |
+
- Size: 30 3D volumes (24 Training + 6 Testing)
|
| 104 |
+
|
| 105 |
+
# Training
|
| 106 |
+
|
| 107 |
+
See the source repository [here](https://github.com/Project-MONAI/research-contributions/tree/main/SwinUNETR/BTCV) for information on training.
|
| 108 |
+
|
| 109 |
+
# BibTeX entry and citation info
|
| 110 |
+
If you find this repository useful, please consider citing the following papers:
|
| 111 |
+
|
| 112 |
+
```
|
| 113 |
+
@inproceedings{tang2022self,
|
| 114 |
+
title={Self-supervised pre-training of swin transformers for 3d medical image analysis},
|
| 115 |
+
author={Tang, Yucheng and Yang, Dong and Li, Wenqi and Roth, Holger R and Landman, Bennett and Xu, Daguang and Nath, Vishwesh and Hatamizadeh, Ali},
|
| 116 |
+
booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition},
|
| 117 |
+
pages={20730--20740},
|
| 118 |
+
year={2022}
|
| 119 |
+
}
|
| 120 |
+
|
| 121 |
+
@article{hatamizadeh2022swin,
|
| 122 |
+
title={Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images},
|
| 123 |
+
author={Hatamizadeh, Ali and Nath, Vishwesh and Tang, Yucheng and Yang, Dong and Roth, Holger and Xu, Daguang},
|
| 124 |
+
journal={arXiv preprint arXiv:2201.01266},
|
| 125 |
+
year={2022}
|
| 126 |
+
}
|
| 127 |
+
```
|
| 128 |
+
|
| 129 |
+
# References
|
| 130 |
+
[1]: Tang, Y., Yang, D., Li, W., Roth, H.R., Landman, B., Xu, D., Nath, V. and Hatamizadeh, A., 2022. Self-supervised pre-training of swin transformers for 3d medical image analysis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 20730-20740).
|
| 131 |
+
|
| 132 |
+
[2]: Hatamizadeh, A., Nath, V., Tang, Y., Yang, D., Roth, H. and Xu, D., 2022. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. arXiv preprint arXiv:2201.01266.
|
swinunetr-btcv-small/config.json → config.json
RENAMED
|
File without changes
|
swinunetr-btcv-small/pytorch_model.bin → pytorch_model.bin
RENAMED
|
File without changes
|
swinunetr-btcv-small/requirements.txt → requirements.txt
RENAMED
|
File without changes
|
swinunetr-btcv-small/README.md
DELETED
|
@@ -1,132 +0,0 @@
|
|
| 1 |
-
---
|
| 2 |
-
language: en
|
| 3 |
-
tags:
|
| 4 |
-
- btcv
|
| 5 |
-
- medical
|
| 6 |
-
- swin
|
| 7 |
-
license: apache-2.0
|
| 8 |
-
datasets:
|
| 9 |
-
- BTCV
|
| 10 |
-
---
|
| 11 |
-
|
| 12 |
-
# Model Overview
|
| 13 |
-
|
| 14 |
-
This repository contains the code for Swin UNETR [1,2]. Swin UNETR is the state-of-the-art on Medical Segmentation
|
| 15 |
-
Decathlon (MSD) and Beyond the Cranial Vault (BTCV) Segmentation Challenge dataset. In [1], a novel methodology is devised for pre-training Swin UNETR backbone in a self-supervised
|
| 16 |
-
manner. We provide the option for training Swin UNETR by fine-tuning from pre-trained self-supervised weights or from scratch.
|
| 17 |
-
|
| 18 |
-
The source repository for the training of these models can be found [here](https://github.com/Project-MONAI/research-contributions/tree/main/SwinUNETR/BTCV).
|
| 19 |
-
|
| 20 |
-
# Installing Dependencies
|
| 21 |
-
Dependencies for training and inference can be installed using the model requirements :
|
| 22 |
-
``` bash
|
| 23 |
-
pip install -r requirements.txt
|
| 24 |
-
```
|
| 25 |
-
|
| 26 |
-
# Intended uses & limitations
|
| 27 |
-
|
| 28 |
-
You can use the raw model for masked language modeling, but it's mostly intended to be fine-tuned on a downstream task.
|
| 29 |
-
|
| 30 |
-
Note that this model is primarily aimed at being fine-tuned on tasks which segment CAT scans or MRIs on images in dicom format.
|
| 31 |
-
|
| 32 |
-
# How to use
|
| 33 |
-
|
| 34 |
-
To install necessary dependencies, run the below in bash.
|
| 35 |
-
```
|
| 36 |
-
git clone https://github.com/darraghdog/Project-MONAI-research-contributions pmrc
|
| 37 |
-
pip install -r pmrc/requirements.txt
|
| 38 |
-
cd pmrc/SwinUNETR/BTCV
|
| 39 |
-
```
|
| 40 |
-
|
| 41 |
-
To load the model from the hub.
|
| 42 |
-
```
|
| 43 |
-
>>> from swinunetr import SwinUnetrModelForInference
|
| 44 |
-
>>> model = SwinUnetrModelForInference.from_pretrained('darragh/swinunetr-btcv-tiny')
|
| 45 |
-
```
|
| 46 |
-
|
| 47 |
-
You can also use `predict.py` to run inference for sample dicom medical images.
|
| 48 |
-
|
| 49 |
-
# Limitations and bias
|
| 50 |
-
|
| 51 |
-
The training data used for this model is specific to CAT scans from certain health facilities and machines. Data from other facilities may difffer in image distributions, and may require finetuning of the models for best performance.
|
| 52 |
-
|
| 53 |
-
# Evaluation results
|
| 54 |
-
|
| 55 |
-
We provide several pre-trained models on BTCV dataset in the following.
|
| 56 |
-
|
| 57 |
-
<table>
|
| 58 |
-
<tr>
|
| 59 |
-
<th>Name</th>
|
| 60 |
-
<th>Dice (overlap=0.7)</th>
|
| 61 |
-
<th>Dice (overlap=0.5)</th>
|
| 62 |
-
<th>Feature Size</th>
|
| 63 |
-
<th># params (M)</th>
|
| 64 |
-
<th>Self-Supervised Pre-trained </th>
|
| 65 |
-
</tr>
|
| 66 |
-
<tr>
|
| 67 |
-
<td>Swin UNETR/Base</td>
|
| 68 |
-
<td>82.25</td>
|
| 69 |
-
<td>81.86</td>
|
| 70 |
-
<td>48</td>
|
| 71 |
-
<td>62.1</td>
|
| 72 |
-
<td>Yes</td>
|
| 73 |
-
</tr>
|
| 74 |
-
|
| 75 |
-
<tr>
|
| 76 |
-
<td>Swin UNETR/Small</td>
|
| 77 |
-
<td>79.79</td>
|
| 78 |
-
<td>79.34</td>
|
| 79 |
-
<td>24</td>
|
| 80 |
-
<td>15.7</td>
|
| 81 |
-
<td>No</td>
|
| 82 |
-
</tr>
|
| 83 |
-
|
| 84 |
-
<tr>
|
| 85 |
-
<td>Swin UNETR/Tiny</td>
|
| 86 |
-
<td>72.05</td>
|
| 87 |
-
<td>70.35</td>
|
| 88 |
-
<td>12</td>
|
| 89 |
-
<td>4.0</td>
|
| 90 |
-
<td>No</td>
|
| 91 |
-
</tr>
|
| 92 |
-
|
| 93 |
-
</table>
|
| 94 |
-
|
| 95 |
-
# Data Preparation
|
| 96 |
-

|
| 97 |
-
|
| 98 |
-
The training data is from the [BTCV challenge dataset](https://www.synapse.org/#!Synapse:syn3193805/wiki/217752).
|
| 99 |
-
|
| 100 |
-
- Target: 13 abdominal organs including 1. Spleen 2. Right Kidney 3. Left Kideny 4.Gallbladder 5.Esophagus 6. Liver 7. Stomach 8.Aorta 9. IVC 10. Portal and Splenic Veins 11. Pancreas 12.Right adrenal gland 13.Left adrenal gland.
|
| 101 |
-
- Task: Segmentation
|
| 102 |
-
- Modality: CT
|
| 103 |
-
- Size: 30 3D volumes (24 Training + 6 Testing)
|
| 104 |
-
|
| 105 |
-
# Training
|
| 106 |
-
|
| 107 |
-
See the source repository [here](https://github.com/Project-MONAI/research-contributions/tree/main/SwinUNETR/BTCV) for information on training.
|
| 108 |
-
|
| 109 |
-
# BibTeX entry and citation info
|
| 110 |
-
If you find this repository useful, please consider citing the following papers:
|
| 111 |
-
|
| 112 |
-
```
|
| 113 |
-
@inproceedings{tang2022self,
|
| 114 |
-
title={Self-supervised pre-training of swin transformers for 3d medical image analysis},
|
| 115 |
-
author={Tang, Yucheng and Yang, Dong and Li, Wenqi and Roth, Holger R and Landman, Bennett and Xu, Daguang and Nath, Vishwesh and Hatamizadeh, Ali},
|
| 116 |
-
booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition},
|
| 117 |
-
pages={20730--20740},
|
| 118 |
-
year={2022}
|
| 119 |
-
}
|
| 120 |
-
|
| 121 |
-
@article{hatamizadeh2022swin,
|
| 122 |
-
title={Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images},
|
| 123 |
-
author={Hatamizadeh, Ali and Nath, Vishwesh and Tang, Yucheng and Yang, Dong and Roth, Holger and Xu, Daguang},
|
| 124 |
-
journal={arXiv preprint arXiv:2201.01266},
|
| 125 |
-
year={2022}
|
| 126 |
-
}
|
| 127 |
-
```
|
| 128 |
-
|
| 129 |
-
# References
|
| 130 |
-
[1]: Tang, Y., Yang, D., Li, W., Roth, H.R., Landman, B., Xu, D., Nath, V. and Hatamizadeh, A., 2022. Self-supervised pre-training of swin transformers for 3d medical image analysis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 20730-20740).
|
| 131 |
-
|
| 132 |
-
[2]: Hatamizadeh, A., Nath, V., Tang, Y., Yang, D., Roth, H. and Xu, D., 2022. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. arXiv preprint arXiv:2201.01266.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|