code
stringlengths 235
11.6M
| repo_path
stringlengths 3
263
|
|---|---|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # 09 Strain Gage
#
# This is one of the most commonly used sensor. It is used in many transducers. Its fundamental operating principle is fairly easy to understand and it will be the purpose of this lecture.
#
# A strain gage is essentially a thin wire that is wrapped on film of plastic.
# <img src="img/StrainGage.png" width="200">
# The strain gage is then mounted (glued) on the part for which the strain must be measured.
# <img src="img/Strain_gauge_2.jpg" width="200">
#
# ## Stress, Strain
# When a beam is under axial load, the axial stress, $\sigma_a$, is defined as:
# \begin{align*}
# \sigma_a = \frac{F}{A}
# \end{align*}
# with $F$ the axial load, and $A$ the cross sectional area of the beam under axial load.
#
# <img src="img/BeamUnderStrain.png" width="200">
#
# Under the load, the beam of length $L$ will extend by $dL$, giving rise to the definition of strain, $\epsilon_a$:
# \begin{align*}
# \epsilon_a = \frac{dL}{L}
# \end{align*}
# The beam will also contract laterally: the cross sectional area is reduced by $dA$. This results in a transverval strain $\epsilon_t$. The transversal and axial strains are related by the Poisson's ratio:
# \begin{align*}
# \nu = - \frac{\epsilon_t }{\epsilon_a}
# \end{align*}
# For a metal the Poission's ratio is typically $\nu = 0.3$, for an incompressible material, such as rubber (or water), $\nu = 0.5$.
#
# Within the elastic limit, the axial stress and axial strain are related through Hooke's law by the Young's modulus, $E$:
# \begin{align*}
# \sigma_a = E \epsilon_a
# \end{align*}
#
# <img src="img/ElasticRegime.png" width="200">
# ## Resistance of a wire
#
# The electrical resistance of a wire $R$ is related to its physical properties (the electrical resistiviy, $\rho$ in $\Omega$/m) and its geometry: length $L$ and cross sectional area $A$.
#
# \begin{align*}
# R = \frac{\rho L}{A}
# \end{align*}
#
# Mathematically, the change in wire dimension will result inchange in its electrical resistance. This can be derived from first principle:
# \begin{align}
# \frac{dR}{R} = \frac{d\rho}{\rho} + \frac{dL}{L} - \frac{dA}{A}
# \end{align}
# If the wire has a square cross section, then:
# \begin{align*}
# A & = L'^2 \\
# \frac{dA}{A} & = \frac{d(L'^2)}{L'^2} = \frac{2L'dL'}{L'^2} = 2 \frac{dL'}{L'}
# \end{align*}
# We have related the change in cross sectional area to the transversal strain.
# \begin{align*}
# \epsilon_t = \frac{dL'}{L'}
# \end{align*}
# Using the Poisson's ratio, we can relate then relate the change in cross-sectional area ($dA/A$) to axial strain $\epsilon_a = dL/L$.
# \begin{align*}
# \epsilon_t &= - \nu \epsilon_a \\
# \frac{dL'}{L'} &= - \nu \frac{dL}{L} \; \text{or}\\
# \frac{dA}{A} & = 2\frac{dL'}{L'} = -2 \nu \frac{dL}{L}
# \end{align*}
# Finally we can substitute express $dA/A$ in eq. for $dR/R$ and relate change in resistance to change of wire geometry, remembering that for a metal $\nu =0.3$:
# \begin{align}
# \frac{dR}{R} & = \frac{d\rho}{\rho} + \frac{dL}{L} - \frac{dA}{A} \\
# & = \frac{d\rho}{\rho} + \frac{dL}{L} - (-2\nu \frac{dL}{L}) \\
# & = \frac{d\rho}{\rho} + 1.6 \frac{dL}{L} = \frac{d\rho}{\rho} + 1.6 \epsilon_a
# \end{align}
# It also happens that for most metals, the resistivity increases with axial strain. In general, one can then related the change in resistance to axial strain by defining the strain gage factor:
# \begin{align}
# S = 1.6 + \frac{d\rho}{\rho}\cdot \frac{1}{\epsilon_a}
# \end{align}
# and finally, we have:
# \begin{align*}
# \frac{dR}{R} = S \epsilon_a
# \end{align*}
# $S$ is materials dependent and is typically equal to 2.0 for most commercially availabe strain gages. It is dimensionless.
#
# Strain gages are made of thin wire that is wraped in several loops, effectively increasing the length of the wire and therefore the sensitivity of the sensor.
#
# _Question:
#
# Explain why a longer wire is necessary to increase the sensitivity of the sensor_.
#
# Most commercially available strain gages have a nominal resistance (resistance under no load, $R_{ini}$) of 120 or 350 $\Omega$.
#
# Within the elastic regime, strain is typically within the range $10^{-6} - 10^{-3}$, in fact strain is expressed in unit of microstrain, with a 1 microstrain = $10^{-6}$. Therefore, changes in resistances will be of the same order. If one were to measure resistances, we will need a dynamic range of 120 dB, whih is typically very expensive. Instead, one uses the Wheatstone bridge to transform the change in resistance to a voltage, which is easier to measure and does not require such a large dynamic range.
# ## Wheatstone bridge:
# <img src="img/WheatstoneBridge.png" width="200">
#
# The output voltage is related to the difference in resistances in the bridge:
# \begin{align*}
# \frac{V_o}{V_s} = \frac{R_1R_3-R_2R_4}{(R_1+R_4)(R_2+R_3)}
# \end{align*}
#
# If the bridge is balanced, then $V_o = 0$, it implies: $R_1/R_2 = R_4/R_3$.
#
# In practice, finding a set of resistors that balances the bridge is challenging, and a potentiometer is used as one of the resistances to do minor adjustement to balance the bridge. If one did not do the adjustement (ie if we did not zero the bridge) then all the measurement will have an offset or bias that could be removed in a post-processing phase, as long as the bias stayed constant.
#
# If each resistance $R_i$ is made to vary slightly around its initial value, ie $R_i = R_{i,ini} + dR_i$. For simplicity, we will assume that the initial value of the four resistances are equal, ie $R_{1,ini} = R_{2,ini} = R_{3,ini} = R_{4,ini} = R_{ini}$. This implies that the bridge was initially balanced, then the output voltage would be:
#
# \begin{align*}
# \frac{V_o}{V_s} = \frac{1}{4} \left( \frac{dR_1}{R_{ini}} - \frac{dR_2}{R_{ini}} + \frac{dR_3}{R_{ini}} - \frac{dR_4}{R_{ini}} \right)
# \end{align*}
#
# Note here that the changes in $R_1$ and $R_3$ have a positive effect on $V_o$, while the changes in $R_2$ and $R_4$ have a negative effect on $V_o$. In practice, this means that is a beam is a in tension, then a strain gage mounted on the branch 1 or 3 of the Wheatstone bridge will produce a positive voltage, while a strain gage mounted on branch 2 or 4 will produce a negative voltage. One takes advantage of this to increase sensitivity to measure strain.
#
# ### Quarter bridge
# One uses only one quarter of the bridge, ie strain gages are only mounted on one branch of the bridge.
#
# \begin{align*}
# \frac{V_o}{V_s} = \pm \frac{1}{4} \epsilon_a S
# \end{align*}
# Sensitivity, $G$:
# \begin{align*}
# G = \frac{V_o}{\epsilon_a} = \pm \frac{1}{4}S V_s
# \end{align*}
#
#
# ### Half bridge
# One uses half of the bridge, ie strain gages are mounted on two branches of the bridge.
#
# \begin{align*}
# \frac{V_o}{V_s} = \pm \frac{1}{2} \epsilon_a S
# \end{align*}
#
# ### Full bridge
#
# One uses of the branches of the bridge, ie strain gages are mounted on each branch.
#
# \begin{align*}
# \frac{V_o}{V_s} = \pm \epsilon_a S
# \end{align*}
#
# Therefore, as we increase the order of bridge, the sensitivity of the instrument increases. However, one should be carefull how we mount the strain gages as to not cancel out their measurement.
# _Exercise_
#
# 1- Wheatstone bridge
#
# <img src="img/WheatstoneBridge.png" width="200">
#
# > How important is it to know \& match the resistances of the resistors you employ to create your bridge?
# > How would you do that practically?
# > Assume $R_1=120\,\Omega$, $R_2=120\,\Omega$, $R_3=120\,\Omega$, $R_4=110\,\Omega$, $V_s=5.00\,\text{V}$. What is $V_\circ$?
Vs = 5.00
Vo = (120**2-120*110)/(230*240) * Vs
print('Vo = ',Vo, ' V')
# typical range in strain a strain gauge can measure
# 1 -1000 micro-Strain
AxialStrain = 1000*10**(-6) # axial strain
StrainGageFactor = 2
R_ini = 120 # Ohm
R_1 = R_ini+R_ini*StrainGageFactor*AxialStrain
print(R_1)
Vo = (120**2-120*(R_1))/((120+R_1)*240) * Vs
print('Vo = ', Vo, ' V')
# > How important is it to know \& match the resistances of the resistors you employ to create your bridge?
# > How would you do that practically?
# > Assume $R_1= R_2 =R_3=120\,\Omega$, $R_4=120.01\,\Omega$, $V_s=5.00\,\text{V}$. What is $V_\circ$?
Vs = 5.00
Vo = (120**2-120*120.01)/(240.01*240) * Vs
print(Vo)
# 2- Strain gage 1:
#
# One measures the strain on a bridge steel beam. The modulus of elasticity is $E=190$ GPa. Only one strain gage is mounted on the bottom of the beam; the strain gage factor is $S=2.02$.
#
# > a) What kind of electronic circuit will you use? Draw a sketch of it.
#
# > b) Assume all your resistors including the unloaded strain gage are balanced and measure $120\,\Omega$, and that the strain gage is at location $R_2$. The supply voltage is $5.00\,\text{VDC}$. Will $V_\circ$ be positive or negative when a downward load is added?
# In practice, we cannot have all resistances = 120 $\Omega$. at zero load, the bridge will be unbalanced (show $V_o \neq 0$). How could we balance our bridge?
#
# Use a potentiometer to balance bridge, for the load cell, we ''zero'' the instrument.
#
# Other option to zero-out our instrument? Take data at zero-load, record the voltage, $V_{o,noload}$. Substract $V_{o,noload}$ to my data.
# > c) For a loading in which $V_\circ = -1.25\,\text{mV}$, calculate the strain $\epsilon_a$ in units of microstrain.
# \begin{align*}
# \frac{V_o}{V_s} & = - \frac{1}{4} \epsilon_a S\\
# \epsilon_a & = -\frac{4}{S} \frac{V_o}{V_s}
# \end{align*}
S = 2.02
Vo = -0.00125
Vs = 5
eps_a = -1*(4/S)*(Vo/Vs)
print(eps_a)
# > d) Calculate the axial stress (in MPa) in the beam under this load.
# > e) You now want more sensitivity in your measurement, you install a second strain gage on to
# p of the beam. Which resistor should you use for this second active strain gage?
#
# > f) With this new setup and the same applied load than previously, what should be the output voltage?
# 3- Strain Gage with Long Lead Wires
#
# <img src="img/StrainGageLongWires.png" width="360">
#
# A quarter bridge strain gage Wheatstone bridge circuit is constructed with $120\,\Omega$ resistors and a $120\,\Omega$ strain gage. For this practical application, the strain gage is located very far away form the DAQ station and the lead wires to the strain gage are $10\,\text{m}$ long and the lead wire have a resistance of $0.080\,\Omega/\text{m}$. The lead wire resistance can lead to problems since $R_{lead}$ changes with temperature.
#
# > Design a modified circuit that will cancel out the effect of the lead wires.
# ## Homework
#
|
Lectures/09_StrainGage.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <table class="ee-notebook-buttons" align="left">
# <td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/Algorithms/landsat_radiance.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
# <td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/Algorithms/landsat_radiance.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
# <td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/Algorithms/landsat_radiance.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
# </table>
# ## Install Earth Engine API and geemap
# Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://geemap.org). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.
# The following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.
# +
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('Installing geemap ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# -
import ee
import geemap
# ## Create an interactive map
# The default basemap is `Google Maps`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/basemaps.py) can be added using the `Map.add_basemap()` function.
Map = geemap.Map(center=[40,-100], zoom=4)
Map
# ## Add Earth Engine Python script
# +
# Add Earth Engine dataset
# Load a raw Landsat scene and display it.
raw = ee.Image('LANDSAT/LC08/C01/T1/LC08_044034_20140318')
Map.centerObject(raw, 10)
Map.addLayer(raw, {'bands': ['B4', 'B3', 'B2'], 'min': 6000, 'max': 12000}, 'raw')
# Convert the raw data to radiance.
radiance = ee.Algorithms.Landsat.calibratedRadiance(raw)
Map.addLayer(radiance, {'bands': ['B4', 'B3', 'B2'], 'max': 90}, 'radiance')
# Convert the raw data to top-of-atmosphere reflectance.
toa = ee.Algorithms.Landsat.TOA(raw)
Map.addLayer(toa, {'bands': ['B4', 'B3', 'B2'], 'max': 0.2}, 'toa reflectance')
# -
# ## Display Earth Engine data layers
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
|
Algorithms/landsat_radiance.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# Copyright 2020 NVIDIA Corporation. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
# -
# <img src="http://developer.download.nvidia.com/compute/machine-learning/frameworks/nvidia_logo.png" style="width: 90px; float: right;">
#
# # Object Detection with TRTorch (SSD)
# ---
# ## Overview
#
#
# In PyTorch 1.0, TorchScript was introduced as a method to separate your PyTorch model from Python, make it portable and optimizable.
#
# TRTorch is a compiler that uses TensorRT (NVIDIA's Deep Learning Optimization SDK and Runtime) to optimize TorchScript code. It compiles standard TorchScript modules into ones that internally run with TensorRT optimizations.
#
# TensorRT can take models from any major framework and specifically tune them to perform better on specific target hardware in the NVIDIA family, and TRTorch enables us to continue to remain in the PyTorch ecosystem whilst doing so. This allows us to leverage the great features in PyTorch, including module composability, its flexible tensor implementation, data loaders and more. TRTorch is available to use with both PyTorch and LibTorch.
#
# To get more background information on this, we suggest the **lenet-getting-started** notebook as a primer for getting started with TRTorch.
# ### Learning objectives
#
# This notebook demonstrates the steps for compiling a TorchScript module with TRTorch on a pretrained SSD network, and running it to test the speedup obtained.
#
# ## Contents
# 1. [Requirements](#1)
# 2. [SSD Overview](#2)
# 3. [Creating TorchScript modules](#3)
# 4. [Compiling with TRTorch](#4)
# 5. [Running Inference](#5)
# 6. [Measuring Speedup](#6)
# 7. [Conclusion](#7)
# ---
# <a id="1"></a>
# ## 1. Requirements
#
# Follow the steps in `notebooks/README` to prepare a Docker container, within which you can run this demo notebook.
#
# In addition to that, run the following cell to obtain additional libraries specific to this demo.
# Known working versions
# !pip install numpy==1.21.2 scipy==1.5.2 Pillow==6.2.0 scikit-image==0.17.2 matplotlib==3.3.0
# ---
# <a id="2"></a>
# ## 2. SSD
#
# ### Single Shot MultiBox Detector model for object detection
#
# _ | _
# - | -
# 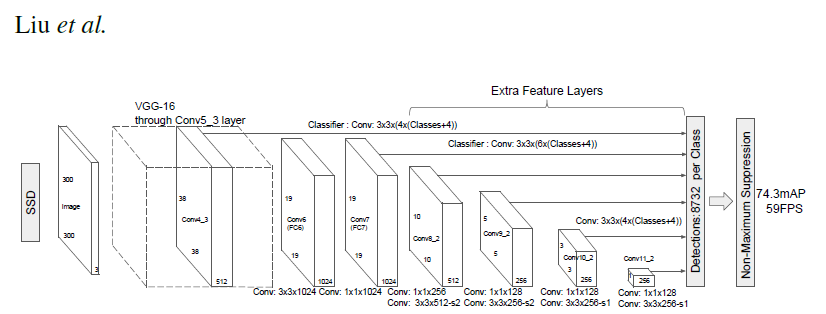 | 
# PyTorch has a model repository called the PyTorch Hub, which is a source for high quality implementations of common models. We can get our SSD model pretrained on [COCO](https://cocodataset.org/#home) from there.
#
# ### Model Description
#
# This SSD300 model is based on the
# [SSD: Single Shot MultiBox Detector](https://arxiv.org/abs/1512.02325) paper, which
# describes SSD as “a method for detecting objects in images using a single deep neural network".
# The input size is fixed to 300x300.
#
# The main difference between this model and the one described in the paper is in the backbone.
# Specifically, the VGG model is obsolete and is replaced by the ResNet-50 model.
#
# From the
# [Speed/accuracy trade-offs for modern convolutional object detectors](https://arxiv.org/abs/1611.10012)
# paper, the following enhancements were made to the backbone:
# * The conv5_x, avgpool, fc and softmax layers were removed from the original classification model.
# * All strides in conv4_x are set to 1x1.
#
# The backbone is followed by 5 additional convolutional layers.
# In addition to the convolutional layers, we attached 6 detection heads:
# * The first detection head is attached to the last conv4_x layer.
# * The other five detection heads are attached to the corresponding 5 additional layers.
#
# Detector heads are similar to the ones referenced in the paper, however,
# they are enhanced by additional BatchNorm layers after each convolution.
#
# More information about this SSD model is available at Nvidia's "DeepLearningExamples" Github [here](https://github.com/NVIDIA/DeepLearningExamples/tree/master/PyTorch/Detection/SSD).
import torch
torch.hub._validate_not_a_forked_repo=lambda a,b,c: True
# List of available models in PyTorch Hub from Nvidia/DeepLearningExamples
torch.hub.list('NVIDIA/DeepLearningExamples:torchhub')
# load SSD model pretrained on COCO from Torch Hub
precision = 'fp32'
ssd300 = torch.hub.load('NVIDIA/DeepLearningExamples:torchhub', 'nvidia_ssd', model_math=precision);
# Setting `precision="fp16"` will load a checkpoint trained with mixed precision
# into architecture enabling execution on Tensor Cores. Handling mixed precision data requires the Apex library.
# ### Sample Inference
# We can now run inference on the model. This is demonstrated below using sample images from the COCO 2017 Validation set.
# +
# Sample images from the COCO validation set
uris = [
'http://images.cocodataset.org/val2017/000000397133.jpg',
'http://images.cocodataset.org/val2017/000000037777.jpg',
'http://images.cocodataset.org/val2017/000000252219.jpg'
]
# For convenient and comprehensive formatting of input and output of the model, load a set of utility methods.
utils = torch.hub.load('NVIDIA/DeepLearningExamples:torchhub', 'nvidia_ssd_processing_utils')
# Format images to comply with the network input
inputs = [utils.prepare_input(uri) for uri in uris]
tensor = utils.prepare_tensor(inputs, False)
# The model was trained on COCO dataset, which we need to access in order to
# translate class IDs into object names.
classes_to_labels = utils.get_coco_object_dictionary()
# +
# Next, we run object detection
model = ssd300.eval().to("cuda")
detections_batch = model(tensor)
# By default, raw output from SSD network per input image contains 8732 boxes with
# localization and class probability distribution.
# Let’s filter this output to only get reasonable detections (confidence>40%) in a more comprehensive format.
results_per_input = utils.decode_results(detections_batch)
best_results_per_input = [utils.pick_best(results, 0.40) for results in results_per_input]
# -
# ### Visualize results
# +
from matplotlib import pyplot as plt
import matplotlib.patches as patches
# The utility plots the images and predicted bounding boxes (with confidence scores).
def plot_results(best_results):
for image_idx in range(len(best_results)):
fig, ax = plt.subplots(1)
# Show original, denormalized image...
image = inputs[image_idx] / 2 + 0.5
ax.imshow(image)
# ...with detections
bboxes, classes, confidences = best_results[image_idx]
for idx in range(len(bboxes)):
left, bot, right, top = bboxes[idx]
x, y, w, h = [val * 300 for val in [left, bot, right - left, top - bot]]
rect = patches.Rectangle((x, y), w, h, linewidth=1, edgecolor='r', facecolor='none')
ax.add_patch(rect)
ax.text(x, y, "{} {:.0f}%".format(classes_to_labels[classes[idx] - 1], confidences[idx]*100), bbox=dict(facecolor='white', alpha=0.5))
plt.show()
# -
# Visualize results without TRTorch/TensorRT
plot_results(best_results_per_input)
# ### Benchmark utility
# +
import time
import numpy as np
import torch.backends.cudnn as cudnn
cudnn.benchmark = True
# Helper function to benchmark the model
def benchmark(model, input_shape=(1024, 1, 32, 32), dtype='fp32', nwarmup=50, nruns=1000):
input_data = torch.randn(input_shape)
input_data = input_data.to("cuda")
if dtype=='fp16':
input_data = input_data.half()
print("Warm up ...")
with torch.no_grad():
for _ in range(nwarmup):
features = model(input_data)
torch.cuda.synchronize()
print("Start timing ...")
timings = []
with torch.no_grad():
for i in range(1, nruns+1):
start_time = time.time()
pred_loc, pred_label = model(input_data)
torch.cuda.synchronize()
end_time = time.time()
timings.append(end_time - start_time)
if i%10==0:
print('Iteration %d/%d, avg batch time %.2f ms'%(i, nruns, np.mean(timings)*1000))
print("Input shape:", input_data.size())
print("Output location prediction size:", pred_loc.size())
print("Output label prediction size:", pred_label.size())
print('Average batch time: %.2f ms'%(np.mean(timings)*1000))
# -
# We check how well the model performs **before** we use TRTorch/TensorRT
# Model benchmark without TRTorch/TensorRT
model = ssd300.eval().to("cuda")
benchmark(model, input_shape=(128, 3, 300, 300), nruns=100)
# ---
# <a id="3"></a>
# ## 3. Creating TorchScript modules
# To compile with TRTorch, the model must first be in **TorchScript**. TorchScript is a programming language included in PyTorch which removes the Python dependency normal PyTorch models have. This conversion is done via a JIT compiler which given a PyTorch Module will generate an equivalent TorchScript Module. There are two paths that can be used to generate TorchScript: **Tracing** and **Scripting**. <br>
# - Tracing follows execution of PyTorch generating ops in TorchScript corresponding to what it sees. <br>
# - Scripting does an analysis of the Python code and generates TorchScript, this allows the resulting graph to include control flow which tracing cannot do.
#
# Tracing however due to its simplicity is more likely to compile successfully with TRTorch (though both systems are supported).
model = ssd300.eval().to("cuda")
traced_model = torch.jit.trace(model, [torch.randn((1,3,300,300)).to("cuda")])
# If required, we can also save this model and use it independently of Python.
# This is just an example, and not required for the purposes of this demo
torch.jit.save(traced_model, "ssd_300_traced.jit.pt")
# Obtain the average time taken by a batch of input with Torchscript compiled modules
benchmark(traced_model, input_shape=(128, 3, 300, 300), nruns=100)
# ---
# <a id="4"></a>
# ## 4. Compiling with TRTorch
# TorchScript modules behave just like normal PyTorch modules and are intercompatible. From TorchScript we can now compile a TensorRT based module. This module will still be implemented in TorchScript but all the computation will be done in TensorRT.
# +
import trtorch
# The compiled module will have precision as specified by "op_precision".
# Here, it will have FP16 precision.
trt_model = trtorch.compile(traced_model, {
"inputs": [trtorch.Input((3, 3, 300, 300))],
"enabled_precisions": {torch.float, torch.half}, # Run with FP16
"workspace_size": 1 << 20
})
# -
# ---
# <a id="5"></a>
# ## 5. Running Inference
# Next, we run object detection
# +
# using a TRTorch module is exactly the same as how we usually do inference in PyTorch i.e. model(inputs)
detections_batch = trt_model(tensor.to(torch.half)) # convert the input to half precision
# By default, raw output from SSD network per input image contains 8732 boxes with
# localization and class probability distribution.
# Let’s filter this output to only get reasonable detections (confidence>40%) in a more comprehensive format.
results_per_input = utils.decode_results(detections_batch)
best_results_per_input_trt = [utils.pick_best(results, 0.40) for results in results_per_input]
# -
# Now, let's visualize our predictions!
#
# Visualize results with TRTorch/TensorRT
plot_results(best_results_per_input_trt)
# We get similar results as before!
# ---
# ## 6. Measuring Speedup
# We can run the benchmark function again to see the speedup gained! Compare this result with the same batch-size of input in the case without TRTorch/TensorRT above.
# +
batch_size = 128
# Recompiling with batch_size we use for evaluating performance
trt_model = trtorch.compile(traced_model, {
"inputs": [trtorch.Input((batch_size, 3, 300, 300))],
"enabled_precisions": {torch.float, torch.half}, # Run with FP16
"workspace_size": 1 << 20
})
benchmark(trt_model, input_shape=(batch_size, 3, 300, 300), nruns=100, dtype="fp16")
# -
# ---
# ## 7. Conclusion
#
# In this notebook, we have walked through the complete process of compiling a TorchScript SSD300 model with TRTorch, and tested the performance impact of the optimization. We find that using the TRTorch compiled model, we gain significant speedup in inference without any noticeable drop in performance!
# ### Details
# For detailed information on model input and output,
# training recipies, inference and performance visit:
# [github](https://github.com/NVIDIA/DeepLearningExamples/tree/master/PyTorch/Detection/SSD)
# and/or [NGC](https://ngc.nvidia.com/catalog/model-scripts/nvidia:ssd_for_pytorch)
#
# ### References
#
# - [SSD: Single Shot MultiBox Detector](https://arxiv.org/abs/1512.02325) paper
# - [Speed/accuracy trade-offs for modern convolutional object detectors](https://arxiv.org/abs/1611.10012) paper
# - [SSD on NGC](https://ngc.nvidia.com/catalog/model-scripts/nvidia:ssd_for_pytorch)
# - [SSD on github](https://github.com/NVIDIA/DeepLearningExamples/tree/master/PyTorch/Detection/SSD)
|
notebooks/ssd-object-detection-demo.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# **Run the following two cells before you begin.**
# %autosave 10
# ______________________________________________________________________
# **First, import your data set and define the sigmoid function.**
# <details>
# <summary>Hint:</summary>
# The definition of the sigmoid is $f(x) = \frac{1}{1 + e^{-X}}$.
# </details>
# +
# Import the data set
import pandas as pd
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
import seaborn as sns
df = pd.read_csv('cleaned_data.csv')
# -
# Define the sigmoid function
def sigmoid(X):
Y = 1 / (1 + np.exp(-X))
return Y
# **Now, create a train/test split (80/20) with `PAY_1` and `LIMIT_BAL` as features and `default payment next month` as values. Use a random state of 24.**
# Create a train/test split
X_train, X_test, y_train, y_test = train_test_split(df[['PAY_1', 'LIMIT_BAL']].values, df['default payment next month'].values,test_size=0.2, random_state=24)
# ______________________________________________________________________
# **Next, import LogisticRegression, with the default options, but set the solver to `'liblinear'`.**
lr_model = LogisticRegression(solver='liblinear')
lr_model
# ______________________________________________________________________
# **Now, train on the training data and obtain predicted classes, as well as class probabilities, using the testing data.**
# Fit the logistic regression model on training data
lr_model.fit(X_train,y_train)
# Make predictions using `.predict()`
y_pred = lr_model.predict(X_test)
# Find class probabilities using `.predict_proba()`
y_pred_proba = lr_model.predict_proba(X_test)
# ______________________________________________________________________
# **Then, pull out the coefficients and intercept from the trained model and manually calculate predicted probabilities. You'll need to add a column of 1s to your features, to multiply by the intercept.**
# Add column of 1s to features
ones_and_features = np.hstack([np.ones((X_test.shape[0],1)), X_test])
print(ones_and_features)
np.ones((X_test.shape[0],1)).shape
# Get coefficients and intercepts from trained model
intercept_and_coefs = np.concatenate([lr_model.intercept_.reshape(1,1), lr_model.coef_], axis=1)
intercept_and_coefs
# Manually calculate predicted probabilities
X_lin_comb = np.dot(intercept_and_coefs, np.transpose(ones_and_features))
y_pred_proba_manual = sigmoid(X_lin_comb)
# ______________________________________________________________________
# **Next, using a threshold of `0.5`, manually calculate predicted classes. Compare this to the class predictions output by scikit-learn.**
# Manually calculate predicted classes
y_pred_manual = y_pred_proba_manual >= 0.5
y_pred_manual.shape
y_pred.shape
# Compare to scikit-learn's predicted classes
np.array_equal(y_pred.reshape(1,-1), y_pred_manual)
y_test.shape
y_pred_proba_manual.shape
# ______________________________________________________________________
# **Finally, calculate ROC AUC using both scikit-learn's predicted probabilities, and your manually predicted probabilities, and compare.**
# + eid="e7697"
# Use scikit-learn's predicted probabilities to calculate ROC AUC
from sklearn.metrics import roc_auc_score
roc_auc_score(y_test, y_pred_proba_manual.reshape(y_pred_proba_manual.shape[1],))
# -
# Use manually calculated predicted probabilities to calculate ROC AUC
roc_auc_score(y_test, y_pred_proba[:,1])
|
Mini-Project-2/Project 4/Fitting_a_Logistic_Regression_Model.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# name: python2
# ---
# + [markdown] colab_type="text" id="1Pi_B2cvdBiW"
# ##### Copyright 2019 The TF-Agents Authors.
# + [markdown] colab_type="text" id="f5926O3VkG_p"
# ### Get Started
# <table class="tfo-notebook-buttons" align="left">
# <td>
# <a target="_blank" href="https://colab.research.google.com/github/tensorflow/agents/blob/master/tf_agents/colabs/8_networks_tutorial.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
# </td>
# <td>
# <a target="_blank" href="https://github.com/tensorflow/agents/blob/master/tf_agents/colabs/8_networks_tutorial.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
# </td>
# </table>
# + colab_type="code" id="xsLTHlVdiZP3" colab={}
# Note: If you haven't installed tf-agents yet, run:
# !pip install tf-nightly
# !pip install tfp-nightly
# !pip install tf-agents-nightly
# + [markdown] colab_type="text" id="lEgSa5qGdItD"
# ### Imports
# + colab_type="code" id="sdvop99JlYSM" colab={}
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import abc
import tensorflow as tf
import numpy as np
from tf_agents.environments import random_py_environment
from tf_agents.environments import tf_py_environment
from tf_agents.networks import encoding_network
from tf_agents.networks import network
from tf_agents.networks import utils
from tf_agents.specs import array_spec
from tf_agents.utils import common as common_utils
from tf_agents.utils import nest_utils
tf.compat.v1.enable_v2_behavior()
# + [markdown] colab_type="text" id="31uij8nIo5bG"
# # Introduction
#
# In this colab we will cover how to define custom networks for your agents. The networks help us define the model that is trained by agents. In TF-Agents you will find several different types of networks which are useful across agents:
#
# **Main Networks**
#
# * **QNetwork**: Used in Qlearning for environments with discrete actions, this network maps an observation to value estimates for each possible action.
# * **CriticNetworks**: Also referred to as `ValueNetworks` in literature, learns to estimate some version of a Value function mapping some state into an estimate for the expected return of a policy. These networks estimate how good the state the agent is currently in is.
# * **ActorNetworks**: Learn a mapping from observations to actions. These networks are usually used by our policies to generate actions.
# * **ActorDistributionNetworks**: Similar to `ActorNetworks` but these generate a distribution which a policy can then sample to generate actions.
#
# **Helper Networks**
# * **EncodingNetwork**: Allows users to easily define a mapping of pre-processing layers to apply to a network's input.
# * **DynamicUnrollLayer**: Automatically resets the network's state on episode boundaries as it is applied over a time sequence.
# * **ProjectionNetwork**: Networks like `CategoricalProjectionNetwork` or `NormalProjectionNetwork` take inputs and generate the required parameters to generate Categorical, or Normal distributions.
#
# All examples in TF-Agents come with pre-configured networks. However these networks are not setup to handle complex observations.
#
# If you have an environment which exposes more than one observation/action and you need to customize your networks then this tutorial is for you!
# + [markdown] id="ums84-YP_21F" colab_type="text"
# #Defining Networks
#
# ##Network API
#
# In TF-Agents we subclass from Keras [Networks](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/python/keras/engine/network.py). With it we can:
#
# * Simplify copy operations required when creating target networks.
# * Perform automatic variable creation when calling `network.variables()`.
# * Validate inputs based on network input_specs.
#
# ##EncodingNetwork
# As mentioned above the `EncodingNetwork` allows us to easily define a mapping of pre-processing layers to apply to a network's input to generate some encoding.
#
# The EncodingNetwork is composed of the following mostly optional layers:
#
# * Preprocessing layers
# * Preprocessing combiner
# * Conv2D
# * Flatten
# * Dense
#
# The special thing about encoding networks is that input preprocessing is applied. Input preprocessing is possible via `preprocessing_layers` and `preprocessing_combiner` layers. Each of these can be specified as a nested structure. If the `preprocessing_layers` nest is shallower than `input_tensor_spec`, then the layers will get the subnests. For example, if:
#
# ```
# input_tensor_spec = ([TensorSpec(3)] * 2, [TensorSpec(3)] * 5)
# preprocessing_layers = (Layer1(), Layer2())
# ```
#
# then preprocessing will call:
#
# ```
# preprocessed = [preprocessing_layers[0](observations[0]),
# preprocessing_layers[1](obsrevations[1])]
# ```
#
# However if
#
# ```
# preprocessing_layers = ([Layer1() for _ in range(2)],
# [Layer2() for _ in range(5)])
# ```
#
# then preprocessing will call:
#
# ```python
# preprocessed = [
# layer(obs) for layer, obs in zip(flatten(preprocessing_layers),
# flatten(observations))
# ]
# ```
#
# + [markdown] id="RP3H1bw0ykro" colab_type="text"
# ## Custom Networks
#
# To create your own networks you will only have to override the `__init__` and `__call__` methods. Let's create a custom network using what we learned about `EncodingNetworks` to create an ActorNetwork that takes observations which contain an image and a vector.
#
# + id="Zp0TjAJhYo4s" colab_type="code" colab={}
class ActorNetwork(network.Network):
def __init__(self,
observation_spec,
action_spec,
preprocessing_layers=None,
preprocessing_combiner=None,
conv_layer_params=None,
fc_layer_params=(75, 40),
dropout_layer_params=None,
activation_fn=tf.keras.activations.relu,
enable_last_layer_zero_initializer=False,
name='ActorNetwork'):
super(ActorNetwork, self).__init__(
input_tensor_spec=observation_spec, state_spec=(), name=name)
# For simplicity we will only support a single action float output.
self._action_spec = action_spec
flat_action_spec = tf.nest.flatten(action_spec)
if len(flat_action_spec) > 1:
raise ValueError('Only a single action is supported by this network')
self._single_action_spec = flat_action_spec[0]
if self._single_action_spec.dtype not in [tf.float32, tf.float64]:
raise ValueError('Only float actions are supported by this network.')
kernel_initializer = tf.keras.initializers.VarianceScaling(
scale=1. / 3., mode='fan_in', distribution='uniform')
self._encoder = encoding_network.EncodingNetwork(
observation_spec,
preprocessing_layers=preprocessing_layers,
preprocessing_combiner=preprocessing_combiner,
conv_layer_params=conv_layer_params,
fc_layer_params=fc_layer_params,
dropout_layer_params=dropout_layer_params,
activation_fn=activation_fn,
kernel_initializer=kernel_initializer,
batch_squash=False)
initializer = tf.keras.initializers.RandomUniform(
minval=-0.003, maxval=0.003)
self._action_projection_layer = tf.keras.layers.Dense(
flat_action_spec[0].shape.num_elements(),
activation=tf.keras.activations.tanh,
kernel_initializer=initializer,
name='action')
def call(self, observations, step_type=(), network_state=()):
outer_rank = nest_utils.get_outer_rank(observations, self.input_tensor_spec)
# We use batch_squash here in case the observations have a time sequence
# compoment.
batch_squash = utils.BatchSquash(outer_rank)
observations = tf.nest.map_structure(batch_squash.flatten, observations)
state, network_state = self._encoder(
observations, step_type=step_type, network_state=network_state)
actions = self._action_projection_layer(state)
actions = common_utils.scale_to_spec(actions, self._single_action_spec)
actions = batch_squash.unflatten(actions)
return tf.nest.pack_sequence_as(self._action_spec, [actions]), network_state
# + [markdown] id="Fm-MbMMLYiZj" colab_type="text"
# Let's create a `RandomPyEnvironment` to generate structured observations and validate our implementation.
# + id="E2XoNuuD66s5" colab_type="code" colab={}
action_spec = array_spec.BoundedArraySpec((3,), np.float32, minimum=0, maximum=10)
observation_spec = {
'image': array_spec.BoundedArraySpec((16, 16, 3), np.float32, minimum=0,
maximum=255),
'vector': array_spec.BoundedArraySpec((5,), np.float32, minimum=-100,
maximum=100)}
random_env = random_py_environment.RandomPyEnvironment(observation_spec, action_spec=action_spec)
# Convert the environment to a TFEnv to generate tensors.
tf_env = tf_py_environment.TFPyEnvironment(random_env)
# + [markdown] id="LM3uDTD7TNVx" colab_type="text"
# Since we've defined the observations to be a dict we need to create preprocessing layers to handle these.
# + id="r9U6JVevTAJw" colab_type="code" colab={}
preprocessing_layers = {
'image': tf.keras.models.Sequential([tf.keras.layers.Conv2D(8, 4),
tf.keras.layers.Flatten()]),
'vector': tf.keras.layers.Dense(5)
}
preprocessing_combiner = tf.keras.layers.Concatenate(axis=-1)
actor = ActorNetwork(tf_env.observation_spec(),
tf_env.action_spec(),
preprocessing_layers=preprocessing_layers,
preprocessing_combiner=preprocessing_combiner)
# + [markdown] id="mM9qedlwc41U" colab_type="text"
# Now that we have the actor network we can process observations from the environment.
# + id="JOkkeu7vXoei" colab_type="code" colab={}
time_step = tf_env.reset()
actor(time_step.observation, time_step.step_type)
# + [markdown] id="ALGxaQLWc9GI" colab_type="text"
# This same strategy can be used to customize any of the main networks used by the agents. You can define whatever preprocessing and connect it to the rest of the network. As you define your own custom make sure the output layer definitions of the network match.
|
tf_agents/colabs/8_networks_tutorial.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Custom Types
#
# Often, the behavior for a field needs to be customized to support a particular shape or validation method that ParamTools does not support out of the box. In this case, you may use the `register_custom_type` function to add your new `type` to the ParamTools type registry. Each `type` has a corresponding `field` that is used for serialization and deserialization. ParamTools will then use this `field` any time it is handling a `value`, `label`, or `member` that is of this `type`.
#
# ParamTools is built on top of [`marshmallow`](https://github.com/marshmallow-code/marshmallow), a general purpose validation library. This means that you must implement a custom `marshmallow` field to go along with your new type. Please refer to the `marshmallow` [docs](https://marshmallow.readthedocs.io/en/stable/) if you have questions about the use of `marshmallow` in the examples below.
#
#
# ## 32 Bit Integer Example
#
# ParamTools's default integer field uses NumPy's `int64` type. This example shows you how to define an `int32` type and reference it in your `defaults`.
#
# First, let's define the Marshmallow class:
#
# +
import marshmallow as ma
import numpy as np
class Int32(ma.fields.Field):
"""
A custom type for np.int32.
https://numpy.org/devdocs/reference/arrays.dtypes.html
"""
# minor detail that makes this play nice with array_first
np_type = np.int32
def _serialize(self, value, *args, **kwargs):
"""Convert np.int32 to basic, serializable Python int."""
return value.tolist()
def _deserialize(self, value, *args, **kwargs):
"""Cast value from JSON to NumPy Int32."""
converted = np.int32(value)
return converted
# -
# Now, reference it in our defaults JSON/dict object:
#
# +
import paramtools as pt
# add int32 type to the paramtools type registry
pt.register_custom_type("int32", Int32())
class Params(pt.Parameters):
defaults = {
"small_int": {
"title": "Small integer",
"description": "Demonstrate how to define a custom type",
"type": "int32",
"value": 2
}
}
params = Params(array_first=True)
print(f"value: {params.small_int}, type: {type(params.small_int)}")
# -
# One problem with this is that we could run into some deserialization issues. Due to integer overflow, our deserialized result is not the number that we passed in--it's negative!
#
params.adjust(dict(
# this number wasn't chosen randomly.
small_int=2147483647 + 1
))
# ### Marshmallow Validator
#
# Fortunately, you can specify a custom validator with `marshmallow` or ParamTools. Making this works requires modifying the `_deserialize` method to check for overflow like this:
#
class Int32(ma.fields.Field):
"""
A custom type for np.int32.
https://numpy.org/devdocs/reference/arrays.dtypes.html
"""
# minor detail that makes this play nice with array_first
np_type = np.int32
def _serialize(self, value, *args, **kwargs):
"""Convert np.int32 to basic Python int."""
return value.tolist()
def _deserialize(self, value, *args, **kwargs):
"""Cast value from JSON to NumPy Int32."""
converted = np.int32(value)
# check for overflow and let range validator
# display the error message.
if converted != int(value):
return int(value)
return converted
# Now, let's see how to use `marshmallow` to fix this problem:
#
# +
import marshmallow as ma
import paramtools as pt
# get the minimum and maxium values for 32 bit integers.
min_int32 = -2147483648 # = np.iinfo(np.int32).min
max_int32 = 2147483647 # = np.iinfo(np.int32).max
# add int32 type to the paramtools type registry
pt.register_custom_type(
"int32",
Int32(validate=[
ma.validate.Range(min=min_int32, max=max_int32)
])
)
class Params(pt.Parameters):
defaults = {
"small_int": {
"title": "Small integer",
"description": "Demonstrate how to define a custom type",
"type": "int32",
"value": 2
}
}
params = Params(array_first=True)
params.adjust(dict(
small_int=np.int64(max_int32) + 1
))
# -
# ### ParamTools Validator
#
# Finally, we will use ParamTools to solve this problem. We need to modify how we create our custom `marshmallow` field so that it's wrapped by ParamTools's `PartialField`. This makes it clear that your field still needs to be initialized, and that your custom field is able to receive validation information from the `defaults` configuration:
#
# +
import paramtools as pt
# add int32 type to the paramtools type registry
pt.register_custom_type(
"int32",
pt.PartialField(Int32)
)
class Params(pt.Parameters):
defaults = {
"small_int": {
"title": "Small integer",
"description": "Demonstrate how to define a custom type",
"type": "int32",
"value": 2,
"validators": {
"range": {"min": -2147483648, "max": 2147483647}
}
}
}
params = Params(array_first=True)
params.adjust(dict(
small_int=2147483647 + 1
))
# -
|
docs/api/custom-types.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:root] *
# language: python
# name: conda-root-py
# ---
# # Part 1: Data Ingestion
#
# This demo showcases financial fraud prevention and using the MLRun feature store to define complex features that help identify fraud. Fraud prevention specifically is a challenge as it requires processing raw transaction and events in real-time and being able to quickly respond and block transactions before they occur.
#
# To address this, we create a development pipeline and a production pipeline. Both pipelines share the same feature engineering and model code, but serve data very differently. Furthermore, we automate the data and model monitoring process, identify drift and trigger retraining in a CI/CD pipeline. This process is described in the diagram below:
#
# 
# The raw data is described as follows:
#
# | TRANSACTIONS || ║ |USER EVENTS ||
# |-----------------|----------------------------------------------------------------|----------|-----------------|----------------------------------------------------------------|
# | **age** | age group value 0-6. Some values are marked as U for unknown | ║ | **source** | The party/entity related to the event |
# | **gender** | A character to define the age | ║ | **event** | event, such as login or password change |
# | **zipcodeOri** | ZIP code of the person originating the transaction | ║ | **timestamp** | The date and time of the event |
# | **zipMerchant** | ZIP code of the merchant receiving the transaction | ║ | | |
# | **category** | category of the transaction (e.g., transportation, food, etc.) | ║ | | |
# | **amount** | the total amount of the transaction | ║ | | |
# | **fraud** | whether the transaction is fraudulent | ║ | | |
# | **timestamp** | the date and time in which the transaction took place | ║ | | |
# | **source** | the ID of the party/entity performing the transaction | ║ | | |
# | **target** | the ID of the party/entity receiving the transaction | ║ | | |
# | **device** | the device ID used to perform the transaction | ║ | | |
# This notebook introduces how to **Ingest** different data sources to the **Feature Store**.
#
# The following FeatureSets will be created:
# - **Transactions**: Monetary transactions between a source and a target.
# - **Events**: Account events such as account login or a password change.
# - **Label**: Fraud label for the data.
#
# By the end of this tutorial you’ll learn how to:
#
# - Create an ingestion pipeline for each data source.
# - Define preprocessing, aggregation and validation of the pipeline.
# - Run the pipeline locally within the notebook.
# - Launch a real-time function to ingest live data.
# - Schedule a cron to run the task when needed.
project_name = 'fraud-demo'
# +
import mlrun
# Initialize the MLRun project object
project = mlrun.get_or_create_project(project_name, context="./", user_project=True)
# -
# ## Step 1 - Fetch, Process and Ingest our datasets
# ## 1.1 - Transactions
# ### Transactions
# + tags=["hide-cell"]
# Helper functions to adjust the timestamps of our data
# while keeping the order of the selected events and
# the relative distance from one event to the other
def date_adjustment(sample, data_max, new_max, old_data_period, new_data_period):
'''
Adjust a specific sample's date according to the original and new time periods
'''
sample_dates_scale = ((data_max - sample) / old_data_period)
sample_delta = new_data_period * sample_dates_scale
new_sample_ts = new_max - sample_delta
return new_sample_ts
def adjust_data_timespan(dataframe, timestamp_col='timestamp', new_period='2d', new_max_date_str='now'):
'''
Adjust the dataframe timestamps to the new time period
'''
# Calculate old time period
data_min = dataframe.timestamp.min()
data_max = dataframe.timestamp.max()
old_data_period = data_max-data_min
# Set new time period
new_time_period = pd.Timedelta(new_period)
new_max = pd.Timestamp(new_max_date_str)
new_min = new_max-new_time_period
new_data_period = new_max-new_min
# Apply the timestamp change
df = dataframe.copy()
df[timestamp_col] = df[timestamp_col].apply(lambda x: date_adjustment(x, data_max, new_max, old_data_period, new_data_period))
return df
# +
import pandas as pd
# Fetch the transactions dataset from the server
transactions_data = pd.read_csv('https://s3.wasabisys.com/iguazio/data/fraud-demo-mlrun-fs-docs/data.csv', parse_dates=['timestamp'], nrows=500)
# Adjust the samples timestamp for the past 2 days
transactions_data = adjust_data_timespan(transactions_data, new_period='2d')
# Preview
transactions_data.head(3)
# -
# ### Transactions - Create a FeatureSet and Preprocessing Pipeline
# Create the FeatureSet (data pipeline) definition for the **credit transaction processing** which describes the offline/online data transformations and aggregations.<br>
# The feature store will automatically add an offline `parquet` target and an online `NoSQL` target by using `set_targets()`.
#
# The data pipeline consists of:
#
# * **Extracting** the data components (hour, day of week)
# * **Mapping** the age values
# * **One hot encoding** for the transaction category and the gender
# * **Aggregating** the amount (avg, sum, count, max over 2/12/24 hour time windows)
# * **Aggregating** the transactions per category (over 14 days time windows)
# * **Writing** the results to **offline** (Parquet) and **online** (NoSQL) targets
# Import MLRun's Feature Store
import mlrun.feature_store as fstore
from mlrun.feature_store.steps import OneHotEncoder, MapValues, DateExtractor
# Define the transactions FeatureSet
transaction_set = fstore.FeatureSet("transactions",
entities=[fstore.Entity("source")],
timestamp_key='timestamp',
description="transactions feature set")
# +
# Define and add value mapping
main_categories = ["es_transportation", "es_health", "es_otherservices",
"es_food", "es_hotelservices", "es_barsandrestaurants",
"es_tech", "es_sportsandtoys", "es_wellnessandbeauty",
"es_hyper", "es_fashion", "es_home", "es_contents",
"es_travel", "es_leisure"]
# One Hot Encode the newly defined mappings
one_hot_encoder_mapping = {'category': main_categories,
'gender': list(transactions_data.gender.unique())}
# Define the graph steps
transaction_set.graph\
.to(DateExtractor(parts = ['hour', 'day_of_week'], timestamp_col = 'timestamp'))\
.to(MapValues(mapping={'age': {'U': '0'}}, with_original_features=True))\
.to(OneHotEncoder(mapping=one_hot_encoder_mapping))
# Add aggregations for 2, 12, and 24 hour time windows
transaction_set.add_aggregation(name='amount',
column='amount',
operations=['avg','sum', 'count','max'],
windows=['2h', '12h', '24h'],
period='1h')
# Add the category aggregations over a 14 day window
for category in main_categories:
transaction_set.add_aggregation(name=category,column=f'category_{category}',
operations=['count'], windows=['14d'], period='1d')
# Add default (offline-parquet & online-nosql) targets
transaction_set.set_targets()
# Plot the pipeline so we can see the different steps
transaction_set.plot(rankdir="LR", with_targets=True)
# -
# ### Transactions - Ingestion
# +
# Ingest our transactions dataset through our defined pipeline
transactions_df = fstore.ingest(transaction_set, transactions_data,
infer_options=fstore.InferOptions.default())
transactions_df.head(3)
# -
# ## 1.2 - User Events
# ### User Events - Fetching
# +
# Fetch our user_events dataset from the server
user_events_data = pd.read_csv('https://s3.wasabisys.com/iguazio/data/fraud-demo-mlrun-fs-docs/events.csv',
index_col=0, quotechar="\'", parse_dates=['timestamp'], nrows=500)
# Adjust to the last 2 days to see the latest aggregations in our online feature vectors
user_events_data = adjust_data_timespan(user_events_data, new_period='2d')
# Preview
user_events_data.head(3)
# -
# ### User Events - Create a FeatureSet and Preprocessing Pipeline
#
# Now we will define the events feature set.
# This is a pretty straight forward pipeline in which we only one hot encode the event categories and save the data to the default targets.
user_events_set = fstore.FeatureSet("events",
entities=[fstore.Entity("source")],
timestamp_key='timestamp',
description="user events feature set")
# +
# Define and add value mapping
events_mapping = {'event': list(user_events_data.event.unique())}
# One Hot Encode
user_events_set.graph.to(OneHotEncoder(mapping=events_mapping))
# Add default (offline-parquet & online-nosql) targets
user_events_set.set_targets()
# Plot the pipeline so we can see the different steps
user_events_set.plot(rankdir="LR", with_targets=True)
# -
# ### User Events - Ingestion
# Ingestion of our newly created events feature set
events_df = fstore.ingest(user_events_set, user_events_data)
events_df.head(3)
# ## Step 2 - Create a labels dataset for model training
# ### Label Set - Create a FeatureSet
# This feature set contains the label for the fraud demo, it will be ingested directly to the default targets without any changes
def create_labels(df):
labels = df[['fraud','source','timestamp']].copy()
labels = labels.rename(columns={"fraud": "label"})
labels['timestamp'] = labels['timestamp'].astype("datetime64[ms]")
labels['label'] = labels['label'].astype(int)
labels.set_index('source', inplace=True)
return labels
# +
# Define the "labels" feature set
labels_set = fstore.FeatureSet("labels",
entities=[fstore.Entity("source")],
timestamp_key='timestamp',
description="training labels",
engine="pandas")
labels_set.graph.to(name="create_labels", handler=create_labels)
# specify only Parquet (offline) target since its not used for real-time
labels_set.set_targets(['parquet'], with_defaults=False)
labels_set.plot(with_targets=True)
# -
# ### Label Set - Ingestion
# Ingest the labels feature set
labels_df = fstore.ingest(labels_set, transactions_data)
labels_df.head(3)
# ## Step 3 - Deploy a real-time pipeline
#
# When dealing with real-time aggregation, it's important to be able to update these aggregations in real-time.
# For this purpose, we will create live serving functions that will update the online feature store of the `transactions` FeatureSet and `Events` FeatureSet.
#
# Using MLRun's `serving` runtime, craetes a nuclio function loaded with our feature set's computational graph definition
# and an `HttpSource` to define the HTTP trigger.
#
# Notice that the implementation below does not require any rewrite of the pipeline logic.
# ## 3.1 - Transactions
# ### Transactions - Deploy our FeatureSet live endpoint
# Create iguazio v3io stream and transactions push API endpoint
transaction_stream = f'v3io:///projects/{project.name}/streams/transaction'
transaction_pusher = mlrun.datastore.get_stream_pusher(transaction_stream)
# +
# Define the source stream trigger (use v3io streams)
# we will define the `key` and `time` fields (extracted from the Json message).
source = mlrun.datastore.sources.StreamSource(path=transaction_stream , key_field='source', time_field='timestamp')
# Deploy the transactions feature set's ingestion service over a real-time (Nuclio) serverless function
# you can use the run_config parameter to pass function/service specific configuration
transaction_set_endpoint = fstore.deploy_ingestion_service(featureset=transaction_set, source=source)
# -
# ### Transactions - Test the feature set HTTP endpoint
# By defining our `transactions` feature set we can now use MLRun and Storey to deploy it as a live endpoint, ready to ingest new data!
#
# Using MLRun's `serving` runtime, we will create a nuclio function loaded with our feature set's computational graph definition and an `HttpSource` to define the HTTP trigger.
# +
import requests
import json
# Select a sample from the dataset and serialize it to JSON
transaction_sample = json.loads(transactions_data.sample(1).to_json(orient='records'))[0]
transaction_sample['timestamp'] = str(pd.Timestamp.now())
transaction_sample
# -
# Post the sample to the ingestion endpoint
requests.post(transaction_set_endpoint, json=transaction_sample).text
# ## 3.2 - User Events
# ### User Events - Deploy our FeatureSet live endpoint
# Deploy the events feature set's ingestion service using the feature set and all the previously defined resources.
# Create iguazio v3io stream and transactions push API endpoint
events_stream = f'v3io:///projects/{project.name}/streams/events'
events_pusher = mlrun.datastore.get_stream_pusher(events_stream)
# +
# Define the source stream trigger (use v3io streams)
# we will define the `key` and `time` fields (extracted from the Json message).
source = mlrun.datastore.sources.StreamSource(path=events_stream , key_field='source', time_field='timestamp')
# Deploy the transactions feature set's ingestion service over a real-time (Nuclio) serverless function
# you can use the run_config parameter to pass function/service specific configuration
events_set_endpoint = fstore.deploy_ingestion_service(featureset=user_events_set, source=source)
# -
# ### User Events - Test the feature set HTTP endpoint
# Select a sample from the events dataset and serialize it to JSON
user_events_sample = json.loads(user_events_data.sample(1).to_json(orient='records'))[0]
user_events_sample['timestamp'] = str(pd.Timestamp.now())
user_events_sample
# Post the sample to the ingestion endpoint
requests.post(events_set_endpoint, json=user_events_sample).text
# ## Done!
#
# You've completed Part 1 of the data-ingestion with the feature store.
# Proceed to [Part 2](02-create-training-model.ipynb) to learn how to train an ML model using the feature store data.
|
docs/feature-store/end-to-end-demo/01-ingest-datasources.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# 
#
# <a href="https://hub.callysto.ca/jupyter/hub/user-redirect/git-pull?repo=https%3A%2F%2Fgithub.com%2Fcallysto%2Fcurriculum-notebooks&branch=master&subPath=Mathematics/Percentage/Percents.ipynb&depth=1" target="_parent"><img src="https://raw.githubusercontent.com/callysto/curriculum-notebooks/master/open-in-callysto-button.svg?sanitize=true" width="123" height="24" alt="Open in Callysto"/></a>
# **Run the cell below, this will add two buttons. Click on the "initialize" button before proceeding through the notebook**
# + tags=["hide-input"]
import uiButtons
# %uiButtons
# + tags=["hide-input"] language="html"
# <script src="https://d3js.org/d3.v3.min.js"></script>
# -
# # Percentages
# ## Introduction
# In this notebook we will discuss what percentages are and why this way of representing data is helpful in many different contexts. Common examples of percentages are sales tax or a mark for an assignment.
#
# The word percent comes from the Latin adverbial phrase *per centum* meaning “*by the hundred*”.
#
# For example, if the sales tax is $5\%$, this means that for every dollar you spend the tax adds $5$ cents to the total price of the purchase.
#
# A percentage simply represents a fraction (per hundred). For example, $90\%$ is the same as saying $\dfrac{90}{100}$. It is used to represent a ratio.
#
# What makes percentages so powerful is that they can represent any ratio.
#
# For example, getting $\dfrac{22}{25}$ on a math exam can be represented as $88\%$: $22$ is $88\%$ of $25$.
# ## How to Get a Percentage
# As mentioned in the introduction, a percentage is simply a fraction represented as a portion of 100.
#
# For this notebook we will only talk about percentages between 0% and 100%.
#
# This means the corresponding fraction will always be a value between $0$ and $1$.
#
# Let's look at our math exam mark example from above. The student correctly answered $22$ questions out of $25$, so the student received a grade of $\dfrac{22}{25}$.
#
# To represent this ratio as a percentage we first convert $\dfrac{22}{25}$ to its decimal representation (simply do the division in your calculator).
#
# $$
# \dfrac{22}{25} = 22 \div 25 = 0.88
# $$
#
# We are almost done: we now have the ratio represented as a value between 0 and 1. To finish getting the answer to our problem all we need to do is multiply this value by $100$ to get our percentage. $$0.88 \times 100 = 88\%$$
#
# Putting it all together we can say $22$ is $88\%$ of $25$.
#
# Think of a grade you recently received (as a fraction) and convert it to a percentage. Once you think you have an answer you can use the widget below to check your answer.
#
# Simply add the total marks of the test/assignment then move the slider until you get to your grade received.
# + tags=["hide-input"] language="html"
# <style>
# .main {
# font-family: 'Segoe UI', Tahoma, Geneva, Verdana, sans-serif;
# }
#
# .slider {
# width: 100px;
# }
#
# #maxVal {
# border:1px solid #cccccc;
# border-radius: 5px;
# width: 50px;
# }
# </style>
# <div class="main" style="border:2px solid black; width: 400px; padding: 20px;border-radius: 10px; margin: 0 auto; box-shadow: 3px 3px 12px #acacac">
# <div>
# <label for="maxValue">Enter the assignment/exam total marks</label>
# <input type="number" id="maxVal" value="100">
# </div>
# <div>
# <input type="range" min="0" max="100" value="0" class="slider" id="mySlider" style="width: 300px; margin-top: 20px;">
# </div>
# <h4 id="sliderVal">0</h3>
# </div>
#
# <script>
# var slider = document.getElementById('mySlider');
# var sliderVal = document.getElementById('sliderVal');
#
# slider.oninput = function () {
# var sliderMax = document.getElementById('maxVal').value;
# if(sliderMax < 0 || isNaN(sliderMax)) {
# sliderMax = 100;
# document.getElementById('maxVal').value = 100;
# }
# d3.select('#mySlider').attr('max', sliderMax);
# sliderVal.textContent = "If you answered " + this.value + "/" + sliderMax + " correct questions your grade will be " + ((
# this.value / sliderMax) * 100).toPrecision(3) + "%";
# }
# </script>
# -
# ## Solving Problems Using Percentages
#
# Now that we understand what percentages mean and how to get them from fractions, let's look at solving problems using percentages. Start by watching the video below to get a basic understanding.
# + tags=["hide-input"] language="html"
# <div align="middle">
# <iframe id="percentVid" width="640" height="360" src="https://www.youtube.com/embed/rR95Cbcjzus?end=368" frameborder="0" allow="autoplay; encrypted-media" allowfullscreen style="box-shadow: 3px 3px 12px #ACACAC">
# </iframe>
# <p><a href="https://www.youtube.com/channel/UC4a-Gbdw7vOaccHmFo40b9g" target="_blank">Click here</a> for more videos by Math Antics</p>
# </div>
# <script>
# $(function() {
# var reachable = false;
# var myFrame = $('#percentVid');
# var videoSrc = myFrame.attr("src");
# myFrame.attr("src", videoSrc)
# .on('load', function(){reachable = true;});
# setTimeout(function() {
# if(!reachable) {
# var ifrm = myFrame[0];
# ifrm = (ifrm.contentWindow) ? ifrm.contentWindow : (ifrm.contentDocument.document) ? ifrm.contentDocument.document : ifrm.contentDocument;
# ifrm.document.open();
# ifrm.document.write('If the video does not start click <a href="' + videoSrc + '" target="_blank">here</a>');
# ifrm.document.close();
# }
# }, 2000)
# });
# </script>
# -
# As shown in the video, taking $25\%$ of 20 "things" is the same as saying $\dfrac{25}{100}\times\dfrac{20}{1}=\dfrac{500}{100}=\dfrac{5}{1}=5$.
#
# Let's do another example, assume a retail store is having a weekend sale. The sale is $30\%$ off everything in store.
#
# Sam thinks this is a great time to buy new shoes, and the shoes she is interested in are regular price $\$89.99$.<br>
# If Sam buys these shoes this weekend how much will they cost? If the sales tax is $5\%$, what will the total price be?
#
# <img src="https://orig00.deviantart.net/5c3e/f/2016/211/b/d/converse_shoes_free_vector_by_superawesomevectors-dabxj2k.jpg" width="300">
# <img src="https://www.publicdomainpictures.net/pictures/170000/nahled/30-korting.jpg" width="300">
#
# Let's start by figuring out the sale price of the shoes before calculating the tax. To figure out the new price we must first take $30\%$ off the original price.
#
# So the shoes are regular priced at $\$89.99$ and the sale is for $30\%$ off
#
# $$
# \$89.99\times 30\%=\$89.99\times\frac{30}{100}=\$26.997
# $$
#
# We can round $\$26.997$ to $\$27$.
#
# Ok we now know how much Sam will save on her new shoes, but let's not forget that the question is asking how much her new shoes will cost, not how much she will save. All we need to do now is take the total price minus the savings to get the new price:
#
# $$
# \$89.99- \$27=\$62.99
# $$
#
# Wow, what savings!
#
# Now for the second part of the question: what will the total price be if the tax is $5\%$?
#
# We must now figure out what $5\%$ of $\$62.99$ is
#
# $$
# \$62.99\times5\%=\$62.99\times\frac{5}{100}=\$3.15
# $$
#
# Now we know that Sam will need to pay $\$3.15$ of tax on her new shoes so the final price is
#
# $$
# \$62.99+\$3.15=\$66.14
# $$
#
# A shortcut for finding the total price including the sales tax is to add 1 to the tax ratio, let's see how this works:
#
# $$
# \$62.99\times\left(\frac{5}{100}+1\right)=\$62.99\times1.05=\$66.14
# $$
#
# You can use this trick to quickly figure out a price after tax.
# ## Multiplying Percentages together
# Multiplying two or more percentages together is probably not something you would encounter often but it is easy to do if you remember that percentages are really fractions.
#
# Since percentages is simply a different way to represent a fraction, the rules for multiplying them are the same. Recall that multiplying two fractions together is the same as saying a *a fraction of a fraction*. For example $\dfrac{1}{2}\times\dfrac{1}{2}$ is the same as saying $\dfrac{1}{2}$ of $\dfrac{1}{2}$.
#
# Therefore if we write $50\%\times 20\%$ we really mean $50\%$ of $20\%$.
#
# The simplest approach to doing this is to first convert each fraction into their decimal representation (divide them by 100), so
#
# $$
# 50\%\div 100=0.50$$ and $$20\%\div 100=0.20
# $$
#
# Now that we have each fraction shown as their decimal representation we simply multiply them together:
#
# $$
# 0.50\times0.20=0.10
# $$
#
# and again to get this decimal to a percent we multiply by 100
#
# $$
# 0.10\times100=10\%
# $$
#
# Putting this into words we get: *$50\%$ of $20\%$ is $10\%$ (One half of $20\%$ is $10\%$)*.
# ## Sports Example
#
# As we know, statistics play a huge part in sports. Keeping track of a team's wins/losses or how many points a player has are integral parts of today's professional sports. Some of these stats may require more interesting mathematical formulas to figure them out. One such example is a goalie’s save percentage in hockey.
#
# The save percentage is the ratio of how many shots the goalie saved over how many he/she has faced. If you are familiar with the NHL you will know this statistic for goalies as Sv\% and is represented as a number like 0.939. In this case the $0.939$ is the percentage we are interested in. You can multiply this number by $100$ to get it in the familiar form $93.9\%$. This means the Sv\% is $93.9\%$, so this particular goalie has saved $93.9\%$ of the shots he's/she's faced.
#
# You will see below a "sport" like game. The objective of the game is to score on your opponent and protect your own net. As you play the game you will see (in real time) below the game window your Sv% and your opponents Sv%. Play a round or two before we discuss how to get this value.
#
# _**How to play:** choose the winning score from the drop down box then click "Start". In game use your mouse to move your paddle up and down (inside the play area). Don't let the ball go in your net!_
# + tags=["hide-input"] language="html"
# <style>
# .mainBody {
# font-family: Arial, Helvetica, sans-serif;
# }
# #startBtn {
# background-color: cornflowerblue;
# border: none;
# border-radius: 3px;
# font-size: 14px;
# color: white;
# font-weight: bold;
# padding: 2px 8px;
# text-transform: uppercase;
# }
# </style>
# <div class="mainBody">
# <div style="padding-bottom: 10px;">
# <label for="winningScore">Winning Score: </label>
# <select name="Winning Score" id="winningScore">
# <option value="3">3</option>
# <option value="5">5</option>
# <option value="7">7</option>
# <option value="10">10</option>
# </select>
# <button type="button" id="startBtn">Start</button>
# </div>
# <canvas id="gameCanvas" width="600" height="350" style="border: solid 1px black"></canvas>
#
# <div>
# <ul>
# <li>Player's point save average: <output id="playerAvg"></output></li>
# <li>Computer's point save average: <output id="compAvg"></output></li>
# </ul>
# </div>
# </div>
# -
# If you look below the game screen you will see "Player's point save average" and "Computer's point save average". You might also have noticed these values changed every time a save was made (unless Sv% was 1) or a score happened, can you come up with a formula to get these values?
#
# The Sv% value is the ratio of how many saves was made over how many total shots the player faced so our formula is
#
# $$
# Sv\%=\frac{saved \ shots}{total \ shots}
# $$
#
# Let's assume the player faced $33$ shots and let in $2$, then the player's Sv% is
#
# $$
# Sv\%=\frac{(33-2)}{33}=0.939
# $$
#
# *Note: $(33-2)$ is how many shots where saved since the total was $33$ and the player let in $2$*
# ## Questions
# + tags=["hide-input"] language="html"
# <style>
# hr {
# width: 60%;
# margin-left: 20px;
# }
# </style>
# <main>
# <div class="questions">
# <ul style="list-style: none">
# <h4>Question #1</h4>
# <li>
# <label for="q1" class="question">A new goalie played his first game and got a shutout (did not let
# the other team score) and made 33 saves, what is his Sv%? </label>
# </li>
# <li>
# <input type="text" id="q1" class="questionInput">
# <button id="q1Btn" onclick="checkAnswer('q1')" class="ansBtn">Check Answer</button>
# </li>
# <li>
# <p class="q1Ans" id="q1True" style="display: none">✓ That's right! Until the goalie let's
# his/her
# first goal in he/she will have a Sv% of 1</p>
# </li>
# <li>
# <p class="q1Ans" id="q1False" style="display: none">Not quite, don't forget to take the total
# amount of shots minus how many went in the net</p>
# </li>
# </ul>
# </div>
# <hr>
# <div class="questions">
# <ul style="list-style: none">
# <h4>Question #2</h4>
# <li>
# <label for="q2" class="question">If a goalie has a Sv% of .990 can he/she ever get back to a Sv% of
# 1.00?</label>
# </li>
# <li>
# <select id="q2">
# <option value="Yes">Yes</option>
# <option value="No">No</option>
# </select>
# <button id="q2Btn" onclick="checkAnswer('q2')" class="ansBtn">Check Answer</button>
# </li>
# <li>
# <p class="q2Ans" id="q2True" style="display: none">✓ That's correct, the goalie could get back
# up to
# 0.999 but never 1.00</p>
# </li>
# <li>
# <p class="q2Ans" id="q2False" style="display: none">Not quite, the goalie could get back up to 0.999
# but never 1.00</p>
# </li>
# </ul>
# </div>
# <hr>
# <div class="questions">
# <ul style="list-style: none">
# <h4>Question #3</h4>
# <li>
# <label for="q3" class="question">A student received a mark of 47/50 on his unit exam, what
# percentage did he get?</label>
# </li>
# <li>
# <input type="text" id="q3" class="questionInput">
# <button id="q3tn" onclick="checkAnswer('q3')" class="ansBtn">Check Answer</button>
# </li>
# <li>
# <p class="q3Ans" id="q3True" style="display: none">✓ That's correct!</p>
# </li>
# <li>
# <p class="q3Ans" id="q3False" style="display: none">Not quite, try again</p>
# </li>
# </ul>
# </div>
# <hr>
# <div class="questions">
# <ul style="list-style: none">
# <h4>Question #4</h4>
# <li>
# <label for="q4" class="question">In a class of 24 students, 8 students own cats, 12 students own dogs
# and 6 students own both cats and dogs. What is the percentage of students who own both cats and
# dogs?</label>
# </li>
# <li>
# <input type="text" id="q4" class="questionInput">
# <button id="q4tn" onclick="checkAnswer('q4')" class="ansBtn">Check Answer</button>
# </li>
# <li>
# <p class="q4Ans" id="q4True" style="display: none">✓ That's correct!</p>
# </li>
# <li>
# <p class="q4Ans" id="q4False" style="display: none">Not quite, try again</p>
# </li>
# </ul>
# </div>
#
# </main>
# <script>
# checkAnswer = function(q) {
# var val = document.getElementById(q).value;
# var isCorrect = false;
# $("."+q+"Ans").css("display", "none");
# switch(q) {
# case 'q1' : Number(val) === 1 ? isCorrect = true : isCorrect = false; break;
# case 'q2' : val === 'No' ? isCorrect = true : isCorrect = false; break;
# case 'q3' : (val === '94%'|| val === '94.0%' || Number(val) === 94) ? isCorrect = true : isCorrect = false;break;
# case 'q4' : (Number(val) === 25 || val === '25%' || val === '25.0%') ? isCorrect = true : isCorrect = false; break;
# default : return false;
# }
#
# if(isCorrect) {
# $("#"+q+"True").css("display", "block");
# } else {
# $("#"+q+"False").css("display", "block");
# }
# }
# </script>
#
# -
# ## Conclusion
#
# As we saw in this notebook, percentages show up in many different ways and are very useful when describing a ratio. It allows for demonstrating any ratio on a familiar scale ($100$) to make data easier to understand. In this notebook we covered the following:
# - A percentage simply represents a fraction
# - To convert any fraction to a percent we turn it into it's decimal form and add $100$
# - A percentage of an amount is simply a fraction multiplication problem
# - To add or subtract a percentage of an amount we first find the percent value than add/subtract from the original value
# - When adding a percentage to an amount we an use the decimal form of percent and add $1$ to it (for example $\$12\times(0.05+1)=\$12.60$)
#
# Keep practising converting fractions to percentages and it will eventually become second nature!
# + tags=["hide-input"] language="html"
# <script>
# var canvas;
# var canvasContext;
# var isInitialized;
#
# var ballX = 50;
# var ballY = 50;
# var ballSpeedX = 5;
# var ballSpeedY = 3;
#
# var leftPaddleY = 250;
# var rightPaddleY = 250;
#
# var playerSaves = 0;
# var playerSOG = 0;
# var compSaves = 0;
# var compSOG = 0;
#
# var playerScore = 0;
# var compScore = 0;
# var winningScore = 3;
# var winScreen = false;
#
# var PADDLE_WIDTH = 10;
# var PADDLE_HEIGHT = 100;
# var BALL_RADIUS = 10;
# var COMP_SPEED = 4;
#
# document.getElementById('startBtn').onclick = function () {
# initGame();
# var selection = document.getElementById('winningScore');
# winningScore = Number(selection.options[selection.selectedIndex].value);
# canvas = document.getElementById('gameCanvas');
# canvasContext = canvas.getContext('2d');
# canvasContext.font = '50px Arial';
# ballReset();
#
# if (!isInitialized) {
# var framesPerSec = 60;
# setInterval(function () {
# moveAll();
# drawAll();
# }, 1000 / framesPerSec);
# isInitialized = true;
# }
#
# canvas.addEventListener('mousemove', function (event) {
# var mousePos = mouseYPos(event);
# leftPaddleY = mousePos.y - PADDLE_HEIGHT / 2;
# });
# }
#
# function updateSaveAvg() {
# var playerSaveAvgTxt = document.getElementById('playerAvg');
# var compSaveAvgTxt = document.getElementById('compAvg');
#
# var playerSaveAvg = playerSaves / playerSOG;
# var compSaveAvg = compSaves / compSOG;
#
# playerSaveAvgTxt.textContent = ((playerSaveAvg < 0 || isNaN(playerSaveAvg)) ? Number(0).toPrecision(3) + (' (0.0%)') :
# playerSaveAvg.toPrecision(3) + (' (' + (playerSaveAvg * 100).toPrecision(3) + '%)'));
# compSaveAvgTxt.textContent = ((compSaveAvg < 0 || isNaN(compSaveAvg)) ? Number(0).toPrecision(3) + (' (0.0%)') :
# compSaveAvg.toPrecision(
# 3) + (' (' + (compSaveAvg * 100).toPrecision(3) + '%)'));
#
# }
#
# function initGame() {
# playerScore = 0;
# compScore = 0;
# playerSaves = 0;
# playerSOG = 0;
# compSaves = 0;
# compSOG = 0;
# ballSpeedX = 5;
# ballSpeedY = 3;
# }
#
# function ballReset() {
# if (playerScore >= winningScore || compScore >= winningScore) {
# winScreen = true;
# }
# if (winScreen) {
# updateSaveAvg();
# if (confirm('Another game?')) {
# winScreen = false;
# initGame();
# } else {
# return;
# }
# }
# ballX = canvas.width / 2;
# ballY = canvas.height / 2;
# ballSpeedY = Math.floor(Math.random() * 4) + 1;
# var randomizer = Math.floor(Math.random() * 2) + 1;
# if (randomizer % 2 === 0) {
# ballSpeedY -= ballSpeedY;
# }
# flipSide();
# }
#
# function flipSide() {
# ballSpeedX = -ballSpeedX;
# }
#
# function moveAll() {
# if (winScreen) {
# return;
# }
# computerMove();
# ballX += ballSpeedX;
# if (ballX < (0 + BALL_RADIUS)) {
# if (ballY > leftPaddleY && ballY < leftPaddleY + PADDLE_HEIGHT) {
# playerSaves++;
# playerSOG++;
# flipSide();
# var deltaY = ballY - (leftPaddleY + PADDLE_HEIGHT / 2);
# ballSpeedY = deltaY * 0.35;
# } else {
# playerSOG++;
# compScore++;
# if (compScore === winningScore) {
# updateSaveAvg();
# drawAll();
# alert('Computer wins, final score: ' + playerScore + '-' + compScore);
# }
# ballReset();
# }
# }
# if (ballX >= canvas.width - BALL_RADIUS) {
# if (ballY > rightPaddleY && ballY < rightPaddleY + PADDLE_HEIGHT) {
# compSaves++;
# compSOG++;
# flipSide();
# var deltaY = ballY - (rightPaddleY + PADDLE_HEIGHT / 2);
# ballSpeedY = deltaY * 0.35;
# } else {
# compSOG++;
# playerScore++;
# if (playerScore === winningScore) {
# updateSaveAvg();
# drawAll();
# alert('You win, final score: ' + playerScore + '-' + compScore);
# }
# ballReset();
# }
# }
# ballY += ballSpeedY;
# if (ballY >= canvas.height - BALL_RADIUS || ballY < 0 + BALL_RADIUS) {
# ballSpeedY = -ballSpeedY;
# }
# updateSaveAvg();
# }
#
# function computerMove() {
# var rightPaddleYCenter = rightPaddleY + (PADDLE_HEIGHT / 2)
# if (rightPaddleYCenter < ballY - 20) {
# rightPaddleY += COMP_SPEED;
# } else if (rightPaddleYCenter > ballY + 20) {
# rightPaddleY -= COMP_SPEED;
# }
# }
#
# function mouseYPos(event) {
# var rect = canvas.getBoundingClientRect();
# var root = document.documentElement;
# var mouseX = event.clientX - rect.left - root.scrollLeft;
# var mouseY = event.clientY - rect.top - root.scrollTop;
# return {
# x: mouseX,
# y: mouseY
# };
# }
#
# function drawAll() {
#
# colorRect(0, 0, canvas.width, canvas.height, 'black');
# if (winScreen) {
# drawNet();
# drawScore();
# return;
# }
# //Left paddle
# colorRect(1, leftPaddleY, PADDLE_WIDTH, PADDLE_HEIGHT, 'white');
# //Right paddle
# colorRect(canvas.width - PADDLE_WIDTH - 1, rightPaddleY, PADDLE_WIDTH, PADDLE_HEIGHT, 'white');
# //Ball
# colorCircle(ballX, ballY, BALL_RADIUS, 'white');
#
# drawNet();
#
# drawScore();
#
# }
#
# function colorRect(x, y, width, height, drawColor) {
# canvasContext.fillStyle = drawColor;
# canvasContext.fillRect(x, y, width, height);
# }
#
# function colorCircle(centerX, centerY, radius, drawColor) {
# canvasContext.fillStyle = 'drawColor';
# canvasContext.beginPath();
# canvasContext.arc(centerX, centerY, radius, 0, Math.PI * 2, true);
# canvasContext.fill();
# }
#
# function drawScore() {
# canvasContext.fillText(playerScore, (canvas.width / 2) - (canvas.width / 4) - 25, 100);
# canvasContext.fillText(compScore, (canvas.width / 2) + (canvas.width / 4) - 25, 100);
# }
#
# function drawNet() {
# for (var i = 0; i < 60; i++) {
# if (i % 2 === 1) {
# colorRect(canvas.width / 2 - 3, i * 10, 6, 10, 'white')
# }
# }
# }
# </script>
# -
# [](https://github.com/callysto/curriculum-notebooks/blob/master/LICENSE.md)
|
_build/html/_sources/curriculum-notebooks/Mathematics/Percentage/percentage.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <p align="center">
# <img src="https://github.com/GeostatsGuy/GeostatsPy/blob/master/TCG_color_logo.png?raw=true" width="220" height="240" />
#
# </p>
#
# ## Data Analytics
#
# ### Parametric Distributions in Python
#
#
# #### <NAME>, Associate Professor, University of Texas at Austin
#
# ##### [Twitter](https://twitter.com/geostatsguy) | [GitHub](https://github.com/GeostatsGuy) | [Website](http://michaelpyrcz.com) | [GoogleScholar](https://scholar.google.com/citations?user=QVZ20eQAAAAJ&hl=en&oi=ao) | [Book](https://www.amazon.com/Geostatistical-Reservoir-Modeling-Michael-Pyrcz/dp/0199731446) | [YouTube](https://www.youtube.com/channel/UCLqEr-xV-ceHdXXXrTId5ig) | [LinkedIn](https://www.linkedin.com/in/michael-pyrcz-61a648a1)
#
# ### Data Analytics: Parametric Distributions
#
# Here's a demonstration of making and general use of parametric distributions in Python. This demonstration is part of the resources that I include for my courses in Spatial / Subsurface Data Analytics at the Cockrell School of Engineering at the University of Texas at Austin.
#
# #### Parametric Distributions
#
# We will cover the following distributions:
#
# * Uniform
# * Triangular
# * Gaussian
# * Log Normal
#
# We will demonstrate:
#
# * distribution parameters
# * forward and inverse operators
# * summary statistics
#
# I have a lecture on these parametric distributions available on [YouTube](https://www.youtube.com/watch?v=U7fGsqCLPHU&t=1687s).
#
# #### Getting Started
#
# Here's the steps to get setup in Python with the GeostatsPy package:
#
# 1. Install Anaconda 3 on your machine (https://www.anaconda.com/download/).
# 2. From Anaconda Navigator (within Anaconda3 group), go to the environment tab, click on base (root) green arrow and open a terminal.
# 3. In the terminal type: pip install geostatspy.
# 4. Open Jupyter and in the top block get started by copy and pasting the code block below from this Jupyter Notebook to start using the geostatspy functionality.
#
# You will need to copy the data file to your working directory. They are available here:
#
# * Tabular data - unconv_MV_v4.csv at https://git.io/fhHLT.
#
# #### Importing Packages
#
# We will need some standard packages. These should have been installed with Anaconda 3.
import numpy as np # ndarrys for gridded data
import pandas as pd # DataFrames for tabular data
import os # set working directory, run executables
import matplotlib.pyplot as plt # for plotting
from scipy import stats # summary statistics
import math # trigonometry etc.
import scipy.signal as signal # kernel for moving window calculation
import random # for randon numbers
import seaborn as sns # for matrix scatter plots
from scipy import linalg # for linear regression
from sklearn import preprocessing
# #### Set the Working Directory
#
# I always like to do this so I don't lose files and to simplify subsequent read and writes (avoid including the full address each time).
os.chdir("c:/PGE383") # set the working directory
# ### Uniform Distribution
#
# Let's start with the most simple distribution.
#
# * by default a random number is uniform distributed
#
# * this ensures that enough random samples (Monte Carlo simulations) will reproduce the distribution
#
# \begin{equation}
# x_{\alpha}^{s} = F^{-1}_x(p_{\alpha}), \quad X^{s} \sim F_X
# \end{equation}
#
# #### Random Samples
#
# Let's demonstrate the use of the command:
#
# ```python
# uniform.rvs(size=n, loc = low, scale = interval, random_state = seed)
# ```
#
# Where:
#
# * size is the number of samples
#
# * loc is the minimum value
#
# * scale is the range, maximum value minus the minimum value
#
# * random_state is the random number seed
#
# We will observe the convergence of the samples to a uniform distribution as the number of samples becomes large.
#
# We will make a compact set of code by looping over all the cases of number of samples
#
# * we store the number of samples cases in the list called ns
#
# * we store the samples as a list of lists, called X_uniform
#
# +
from scipy.stats import uniform
low = 0.05; interval = 0.20; ns = [1e1,1e2,1e3,1e4,1e5,1e6]; X_uniform = []
index = 0
for n in ns:
X_uniform.append(uniform.rvs(size=int(ns[index]), loc = low, scale = interval).tolist())
plt.subplot(2,3,index+1)
GSLIB.hist_st(X_uniform[index],loc,loc+interval,log=False,cumul = False,bins=20,weights = None,xlabel='Values',title='Distribution, N = ' + str(int(ns[index])))
index = index + 1
plt.subplots_adjust(left=0.0, bottom=0.0, right=2.3, top=1.6, wspace=0.2, hspace=0.3)
# -
# We can observe that by drawing more Monte Carlo simulations, we more closely approximate the original uniform parametric distribution.
#
# #### Forward Distribution
#
# Let's demonstrate the forward operator. We can take any value and calculate the associated:
#
# * density (probability density function)
# * cumulative probability
#
# The transform for the probability density function is:
#
# \begin{equation}
# p = f_x(x)
# \end{equation}
#
# where $f_x$ is the PDF and $p$ is the density for value, $x$.
#
# and for the cumulative distribution function is:
#
# \begin{equation}
# P = F_x(x)
# \end{equation}
#
# where $F_x$ is the CDF and $P$ is the cumulative probability for value, $x$.
# +
x_values = np.linspace(0.0,0.3,100)
p_values = uniform.pdf(x_values, loc = low, scale = interval)
P_values = uniform.cdf(x_values, loc = low, scale = interval)
plt.subplot(1,2,1)
plt.plot(x_values, p_values,'r-', lw=5, alpha=0.3, label='uniform PDF'); plt.title('Uniform PDF'); plt.xlabel('Values'); plt.ylabel('Density')
plt.subplot(1,2,2)
plt.plot(x_values, P_values,'r-', lw=5, alpha=0.3, label='uniform CDF'); plt.title('Uniform CDF'); plt.xlabel('Values'); plt.ylabel('Cumulative Probability')
plt.subplots_adjust(left=0.0, bottom=0.0, right=1.8, top=0.8, wspace=0.2, hspace=0.3)
# -
# #### Inverse Distribution
#
# Let's know demonstrate the reverse operator for the uniform distribution:
#
# \begin{equation}
# X = F^{-1}_X(P)
# \end{equation}
p_values = np.linspace(0.01,0.99,100)
x_values = uniform.ppf(p_values, loc = low, scale = interval)
plt.plot(x_values, p_values,'r-', lw=5, alpha=0.3, label='uniform pdf')
# #### Summary Statistics
#
# We also have a couple of convience member functions to return the statistics from the parametric distribution:
#
# * mean
# * median
# * mode
# * variance
# * standard deviation
#
# Let's demonstrate a few of these methods.
#
# ```python
# uniform.stats(loc = low, scale = interval, moments = 'mvsk')
# ```
#
# returns a tuple with the mean, variance, skew and kurtosis (centered 1st, 2nd, 3rd and 4th moments)
print('Stats: mean, variance, skew and kurtosis = ' + str(uniform.stats(loc = low, scale = interval, moments = 'mvsk')))
# We can confirm this by calculating the centered variance (regular variance) with this member function:
#
# ```python
# uniform.var(loc = low, scale = interval)
# ```
print('The variance is ' + str(round(uniform.var(loc = low, scale = interval),4)) + '.')
# We can also directly calculate the:
#
# * standard deviation - std
# * mean - mean
# * median - median
#
# We can also calculate order of a non-centered moment. The moment method allows us to calculate an non-centered moment of any order. Try this out.
m_order = 4
print('The ' + str(m_order) + 'th order non-centered moment is ' + str(uniform.moment(n = m_order, loc = low, scale = interval)))
# #### Symmetric Interval
#
# We can also get the symmetric interval (e.g. prediction or confidence intervals) for any alpha level.
#
# * Note the program mislabels the value as alpha, it is actually the significance level (1 - alpha)
level = 0.95
print('The interval at alpha level ' + str(round(1-level,3)) + ' is ' + str(uniform.interval(alpha = alpha,loc = low,scale = interval)))
# #### Triangular Distribution
#
# The great thing about parametric distributions is that the above member functions are the same!
#
# * we can plug and play other parametric distributions and repeat the above.
#
# This time we will make it much more compact!
#
# * we will import the triangular distribution as my_dist and call the same functions as before
# * we need a new parameter, the distribution mode (c parameter)
# +
from scipy.stats import triang as my_dist # import traingular dist as my_dist
dist_type = 'Triangular' # give the name of the distribution for labels
low = 0.05; mode = 0.20; c = 0.10 # given the distribution parameters
x_values = np.linspace(0.0,0.3,100) # get an array of x values
p_values = my_dist.pdf(x_values, loc = low, c = mode, scale = interval) # calculate density for each x value
P_values = my_dist.cdf(x_values, loc = low, c = mode, scale = interval) # calculate cumulative probablity for each x value
plt.subplot(1,3,1) # plot the resulting PDF
plt.plot(x_values, p_values,'r-', lw=5, alpha=0.3); plt.title('Sampling ' + str(dist_type) + ' PDF'); plt.xlabel('Values'); plt.ylabel('Density')
plt.subplot(1,3,2) # plot the resulting CDF
plt.plot(x_values, P_values,'r-', lw=5, alpha=0.3); plt.title('Sampling ' + str(dist_type) + ' CDF'); plt.xlabel('Values'); plt.ylabel('Cumulative Probability')
p_values = np.linspace(0.00001,0.99999,100) # get an array of p-values
x_values = my_dist.ppf(p_values, loc = low, c = mode, scale = interval) # apply inverse to get x values from p-values
plt.subplot(1,3,3)
plt.plot(x_values, p_values,'r-', lw=5, alpha=0.3, label='uniform pdf')
plt.subplots_adjust(left=0.0, bottom=0.0, right=2.8, top=0.8, wspace=0.2, hspace=0.3); plt.title('Sampling Inverse ' + str(dist_type) + ' CDF'); plt.xlabel('Values'); plt.ylabel('Cumulative Probability')
print('The mean is ' + str(round(uniform.mean(loc = low, scale = interval),4)) + '.') # calculate stats and symmetric interval
print('The variance is ' + str(round(uniform.var(loc = low, scale = interval),4)) + '.')
print('The interval at alpha level ' + str(round(1-level,3)) + ' is ' + str(uniform.interval(alpha = alpha,loc = low,scale = interval)))
# -
# #### Gaussian Distribution
#
# Let's now use the Gaussian parametric distribution.
#
# * we will need the parameters mean and the variance
#
# We will apply the forward and reverse operations and calculate the summary statistics.
#
# +
from scipy.stats import norm as my_dist # import traingular dist as my_dist
dist_type = 'Gaussian' # give the name of the distribution for labels
mean = 0.15; stdev = 0.05 # given the distribution parameters
x_values = np.linspace(0.0,0.3,100) # get an array of x values
p_values = my_dist.pdf(x_values, loc = mean, scale = stdev) # calculate density for each x value
P_values = my_dist.cdf(x_values, loc = mean, scale = stdev) # calculate cumulative probablity for each x value
plt.subplot(1,3,1) # plot the resulting PDF
plt.plot(x_values, p_values,'r-', lw=5, alpha=0.3); plt.title('Sampling ' + str(dist_type) + ' PDF'); plt.xlabel('Values'); plt.ylabel('Density')
plt.subplot(1,3,2) # plot the resulting CDF
plt.plot(x_values, P_values,'r-', lw=5, alpha=0.3); plt.title('Sampling ' + str(dist_type) + ' CDF'); plt.xlabel('Values'); plt.ylabel('Cumulative Probability')
p_values = np.linspace(0.00001,0.99999,100) # get an array of p-values
x_values = my_dist.ppf(p_values, loc = mean, scale = stdev) # apply inverse to get x values from p-values
plt.subplot(1,3,3)
plt.plot(x_values, p_values,'r-', lw=5, alpha=0.3, label='uniform pdf')
plt.subplots_adjust(left=0.0, bottom=0.0, right=2.8, top=0.8, wspace=0.2, hspace=0.3); plt.title('Sampling Inverse ' + str(dist_type) + ' CDF'); plt.xlabel('Values'); plt.ylabel('Cumulative Probability')
print('The mean is ' + str(round(my_dist.mean(loc = mean, scale = stdev),4)) + '.') # calculate stats and symmetric interval
print('The variance is ' + str(round(my_dist.var(loc = mean, scale = stdev),4)) + '.')
print('The interval at alpha level ' + str(round(1-level,3)) + ' is ' + str(my_dist.interval(alpha = alpha,loc = mean,scale = stdev)))
# -
# #### Log Normal Distribution
#
# Now let's check out the log normal distribution.
#
# * We need the parameters $\mu$ and $\sigma$
# +
from scipy.stats import lognorm as my_dist # import traingular dist as my_dist
dist_type = 'Log Normal' # give the name of the distribution for labels
mu = np.log(0.10); sigma = 0.2 # given the distribution parameters
x_values = np.linspace(0.0,0.3,100) # get an array of x values
p_values = my_dist.pdf(x_values, s = sigma, scale = np.exp(mu)) # calculate density for each x value
P_values = my_dist.cdf(x_values, s = sigma, scale = np.exp(mu)) # calculate cumulative probablity for each x value
plt.subplot(1,3,1) # plot the resulting PDF
plt.plot(x_values, p_values,'r-', lw=5, alpha=0.3); plt.title('Sampling ' + str(dist_type) + ' PDF'); plt.xlabel('Values'); plt.ylabel('Density')
plt.subplot(1,3,2) # plot the resulting CDF
plt.plot(x_values, P_values,'r-', lw=5, alpha=0.3); plt.title('Sampling ' + str(dist_type) + ' CDF'); plt.xlabel('Values'); plt.ylabel('Cumulative Probability')
p_values = np.linspace(0.00001,0.99999,100) # get an array of p-values
x_values = my_dist.ppf(p_values, s = sigma, scale = np.exp(mu)) # apply inverse to get x values from p-values
plt.subplot(1,3,3)
plt.plot(x_values, p_values,'r-', lw=5, alpha=0.3, label='uniform pdf')
plt.subplots_adjust(left=0.0, bottom=0.0, right=2.8, top=0.8, wspace=0.2, hspace=0.3); plt.title('Sampling Inverse ' + str(dist_type) + ' CDF'); plt.xlabel('Values'); plt.ylabel('Cumulative Probability')
#print('The mean is ' + str(round(my_dist.mean(loc = mean, scale = stdev),4)) + '.') # calculate stats and symmetric interval
#print('The variance is ' + str(round(my_dist.var(loc = mean, scale = stdev),4)) + '.')
#print('The interval at alpha level ' + str(round(1-level,3)) + ' is ' + str(my_dist.interval(alpha = alpha,loc = mean,scale = stdev)))
# -
# There are many other parametric distributions that we could have included. Also we could have demonstrated the distribution fitting.
#
# #### Comments
#
# This was a basic demonstration of working with parametric distributions.
#
# I have other demonstrations on the basics of working with DataFrames, ndarrays, univariate statistics, plotting data, declustering, data transformations, trend modeling and many other workflows available at [Python Demos](https://github.com/GeostatsGuy/PythonNumericalDemos) and a Python package for data analytics and geostatistics at [GeostatsPy](https://github.com/GeostatsGuy/GeostatsPy).
#
# I hope this was helpful,
#
# *Michael*
#
# #### The Author:
#
# ### <NAME>, Associate Professor, University of Texas at Austin
# *Novel Data Analytics, Geostatistics and Machine Learning Subsurface Solutions*
#
# With over 17 years of experience in subsurface consulting, research and development, Michael has returned to academia driven by his passion for teaching and enthusiasm for enhancing engineers' and geoscientists' impact in subsurface resource development.
#
# For more about Michael check out these links:
#
# #### [Twitter](https://twitter.com/geostatsguy) | [GitHub](https://github.com/GeostatsGuy) | [Website](http://michaelpyrcz.com) | [GoogleScholar](https://scholar.google.com/citations?user=QVZ20eQAAAAJ&hl=en&oi=ao) | [Book](https://www.amazon.com/Geostatistical-Reservoir-Modeling-Michael-Pyrcz/dp/0199731446) | [YouTube](https://www.youtube.com/channel/UCLqEr-xV-ceHdXXXrTId5ig) | [LinkedIn](https://www.linkedin.com/in/michael-pyrcz-61a648a1)
#
# #### Want to Work Together?
#
# I hope this content is helpful to those that want to learn more about subsurface modeling, data analytics and machine learning. Students and working professionals are welcome to participate.
#
# * Want to invite me to visit your company for training, mentoring, project review, workflow design and / or consulting? I'd be happy to drop by and work with you!
#
# * Interested in partnering, supporting my graduate student research or my Subsurface Data Analytics and Machine Learning consortium (co-PIs including Profs. Foster, Torres-Verdin and van Oort)? My research combines data analytics, stochastic modeling and machine learning theory with practice to develop novel methods and workflows to add value. We are solving challenging subsurface problems!
#
# * I can be reached at <EMAIL>.
#
# I'm always happy to discuss,
#
# *Michael*
#
# <NAME>, Ph.D., P.Eng. Associate Professor The Hildebrand Department of Petroleum and Geosystems Engineering, Bureau of Economic Geology, The Jackson School of Geosciences, The University of Texas at Austin
#
# #### More Resources Available at: [Twitter](https://twitter.com/geostatsguy) | [GitHub](https://github.com/GeostatsGuy) | [Website](http://michaelpyrcz.com) | [GoogleScholar](https://scholar.google.com/citations?user=QVZ20eQAAAAJ&hl=en&oi=ao) | [Book](https://www.amazon.com/Geostatistical-Reservoir-Modeling-Michael-Pyrcz/dp/0199731446) | [YouTube](https://www.youtube.com/channel/UCLqEr-xV-ceHdXXXrTId5ig) | [LinkedIn](https://www.linkedin.com/in/michael-pyrcz-61a648a1)
#
|
PythonDataBasics_ParametricDistributions.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# %matplotlib inline
# %config InlineBackend.figure_format = 'retina'
import matplotlib.pyplot as plt
import torch
from torchvision import datasets, transforms
import helper
# -
# The easiest way to load image data is with *datasets.ImageFolder* from *torchvision*. In generall we'll use *ImageFolder* like so:
#
# ```
# dataset = datasets.ImageFolder('path/to/data',
# transform=transform)
# ```
# ImageFolder expects the files and directories to be constructed like so:
#
# ```
# root/dog/xxx.png
# root/dog/xxy.ong
#
# root/cat/123.png
# root/cat/sad.png
# ```
# ## Transforms
#
# We can either resize them with *transforms.Resize()* or crop with *transforms.CenterCrop()*, *transforms.RandomResizedCrop()* . We'll also need to convert the images to PyTorch tensors with *transforms.ToTensor()*.
# ## Data Loaders
#
# With the *ImageFolder* loaded, we have to pass it to a *DataLoader*. It takes a dataset and returns batches of images and the corresponding labels. We can set various parameters.
#
# ```
# dataloader = torch.utils.data.DataLoader(
# dataset,
# batch_size=32,
# shuffle=True)
# ```
#
# Here dataloader is a *generator*. To get out of it, we need to loop through it or convert it to an iterator and call *next()*
#
# ```
# # looping thrpugh it, get batch on each loop
# for images, labels in dataloader:
# pass
#
# # Get one batch
# images, labels = next(iter(dataloader))
# ```
# **Exercise**
#
# Load images from the *Cat_Dog_data/train* folder, define a few transforms, then build the dataloader.
data_dir = 'Cat_Dog_data/train'
# compose transforms
transform = transforms.Compose([
transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor()
])
# create the ImageFolder
dataset = datasets.ImageFolder(data_dir,
transform=transform)
# use the ImageFolder dataset to create the DataLoader
dataloader = torch.utils.data.DataLoader(dataset,
batch_size=32,
shuffle=True)
# Test the dataset
images, labels = next(iter(dataloader))
helper.imshow(images[0], normalize=False)
# ## Data Augmentation
#
# ```
# train_transforms = transforms.Compose([
# transforms.RandomRotation(30),
# transforms.RandomResizedCrop(224),
# transforms.RandomHorizontalFlip(),
# transforms.ToTensor(),
# transforms.Normalize([0.5, 0.5, 0.5],
# [0.5, 0.5, 0.5])
# ])
# ```
#
# We can pass a list of means and list of standard deviations, then the color channels are normalized like so
#
# ```
# input[channel] = (input[channel] - mean[channel]) / std[channel]
# ```
#
# Subtracting mean centers the data around zero and dividing by std squishes the values to be between -1 and 1. Normalizing helps keep the network work weights near zero which in turn makes backpropagation more stable. Without normalization, networks will tend to fail to learn.
#
# When we're testing however, we'll want to use images that aren't altered. So, for validation/test images, we'll typically just resize and crop.
#
# **Exercise**
#
# Define transforms for trainin data and testing data below. Leave off normalization for now.
# +
data_dir = 'Cat_Dog_data/'
# Define transforms the training data and
# testing data
train_transforms = transforms.Compose([
transforms.RandomRotation(30),
transforms.RandomResizedCrop(224),
transforms.RandomHorizontalFlip(),
transforms.ToTensor()
#transforms.Normalize([0.5, 0.5, 0.5],
# [0.5, 0.5, 0.5])
])
test_transforms = transforms.Compose([
transforms.RandomResizedCrop(224),
transforms.ToTensor()
])
# Pass transforms in here, then turn the next
# cell to see how the transforms look
train_data = datasets.ImageFolder(data_dir + '/train',
transform=train_transforms)
test_data = datasets.ImageFolder(data_dir + '/test',
transform=test_transforms)
trainloader = torch.utils.data.DataLoader(train_data,
batch_size=32)
testloader = torch.utils.data.DataLoader(test_data,
batch_size=32)
# +
# change this to the trainloader or testloader
data_iter = iter(trainloader)
images, labels = next(data_iter)
fig, axes = plt.subplots(figsize=(10,4), ncols=4)
for ii in range(4):
ax = axes[ii]
helper.imshow(images[ii],
ax=ax,
normalize=False)
|
Lesson 5: Introduction to PyTorch/07 - Loading Image Data.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# Probabilistic Programming and Bayesian Methods for Hackers
# ========
#
# Welcome to *Bayesian Methods for Hackers*. The full Github repository is available at [github/Probabilistic-Programming-and-Bayesian-Methods-for-Hackers](https://github.com/CamDavidsonPilon/Probabilistic-Programming-and-Bayesian-Methods-for-Hackers). The other chapters can be found on the project's [homepage](https://camdavidsonpilon.github.io/Probabilistic-Programming-and-Bayesian-Methods-for-Hackers/). We hope you enjoy the book, and we encourage any contributions!
#
# #### Looking for a printed version of Bayesian Methods for Hackers?
#
# _Bayesian Methods for Hackers_ is now a published book by Addison-Wesley, available on [Amazon](http://www.amazon.com/Bayesian-Methods-Hackers-Probabilistic-Addison-Wesley/dp/0133902838)!
#
# 
# Chapter 1
# ======
# ***
# The Philosophy of Bayesian Inference
# ------
#
# > You are a skilled programmer, but bugs still slip into your code. After a particularly difficult implementation of an algorithm, you decide to test your code on a trivial example. It passes. You test the code on a harder problem. It passes once again. And it passes the next, *even more difficult*, test too! You are starting to believe that there may be no bugs in this code...
#
# If you think this way, then congratulations, you already are thinking Bayesian! Bayesian inference is simply updating your beliefs after considering new evidence. A Bayesian can rarely be certain about a result, but he or she can be very confident. Just like in the example above, we can never be 100% sure that our code is bug-free unless we test it on every possible problem; something rarely possible in practice. Instead, we can test it on a large number of problems, and if it succeeds we can feel more *confident* about our code, but still not certain. Bayesian inference works identically: we update our beliefs about an outcome; rarely can we be absolutely sure unless we rule out all other alternatives.
#
# ### The Bayesian state of mind
#
#
# Bayesian inference differs from more traditional statistical inference by preserving *uncertainty*. At first, this sounds like a bad statistical technique. Isn't statistics all about deriving *certainty* from randomness? To reconcile this, we need to start thinking like Bayesians.
#
# The Bayesian world-view interprets probability as measure of *believability in an event*, that is, how confident we are in an event occurring. In fact, we will see in a moment that this is the natural interpretation of probability.
#
# For this to be clearer, we consider an alternative interpretation of probability: *Frequentist*, known as the more *classical* version of statistics, assumes that probability is the long-run frequency of events (hence the bestowed title). For example, the *probability of plane accidents* under a frequentist philosophy is interpreted as the *long-term frequency of plane accidents*. This makes logical sense for many probabilities of events, but becomes more difficult to understand when events have no long-term frequency of occurrences. Consider: we often assign probabilities to outcomes of presidential elections, but the election itself only happens once! Frequentists get around this by invoking alternative realities and saying across all these realities, the frequency of occurrences defines the probability.
#
# Bayesians, on the other hand, have a more intuitive approach. Bayesians interpret a probability as measure of *belief*, or confidence, of an event occurring. Simply, a probability is a summary of an opinion. An individual who assigns a belief of 0 to an event has no confidence that the event will occur; conversely, assigning a belief of 1 implies that the individual is absolutely certain of an event occurring. Beliefs between 0 and 1 allow for weightings of other outcomes. This definition agrees with the probability of a plane accident example, for having observed the frequency of plane accidents, an individual's belief should be equal to that frequency, excluding any outside information. Similarly, under this definition of probability being equal to beliefs, it is meaningful to speak about probabilities (beliefs) of presidential election outcomes: how confident are you candidate *A* will win?
#
# Notice in the paragraph above, I assigned the belief (probability) measure to an *individual*, not to Nature. This is very interesting, as this definition leaves room for conflicting beliefs between individuals. Again, this is appropriate for what naturally occurs: different individuals have different beliefs of events occurring, because they possess different *information* about the world. The existence of different beliefs does not imply that anyone is wrong. Consider the following examples demonstrating the relationship between individual beliefs and probabilities:
#
# - I flip a coin, and we both guess the result. We would both agree, assuming the coin is fair, that the probability of Heads is 1/2. Assume, then, that I peek at the coin. Now I know for certain what the result is: I assign probability 1.0 to either Heads or Tails (whichever it is). Now what is *your* belief that the coin is Heads? My knowledge of the outcome has not changed the coin's results. Thus we assign different probabilities to the result.
#
# - Your code either has a bug in it or not, but we do not know for certain which is true, though we have a belief about the presence or absence of a bug.
#
# - A medical patient is exhibiting symptoms $x$, $y$ and $z$. There are a number of diseases that could be causing all of them, but only a single disease is present. A doctor has beliefs about which disease, but a second doctor may have slightly different beliefs.
#
#
# This philosophy of treating beliefs as probability is natural to humans. We employ it constantly as we interact with the world and only see partial truths, but gather evidence to form beliefs. Alternatively, you have to be *trained* to think like a frequentist.
#
# To align ourselves with traditional probability notation, we denote our belief about event $A$ as $P(A)$. We call this quantity the *prior probability*.
#
# <NAME>, a great economist and thinker, said "When the facts change, I change my mind. What do you do, sir?" This quote reflects the way a Bayesian updates his or her beliefs after seeing evidence. Even — especially — if the evidence is counter to what was initially believed, the evidence cannot be ignored. We denote our updated belief as $P(A |X )$, interpreted as the probability of $A$ given the evidence $X$. We call the updated belief the *posterior probability* so as to contrast it with the prior probability. For example, consider the posterior probabilities (read: posterior beliefs) of the above examples, after observing some evidence $X$:
#
# 1\. $P(A): \;\;$ the coin has a 50 percent chance of being Heads. $P(A | X):\;\;$ You look at the coin, observe a Heads has landed, denote this information $X$, and trivially assign probability 1.0 to Heads and 0.0 to Tails.
#
# 2\. $P(A): \;\;$ This big, complex code likely has a bug in it. $P(A | X): \;\;$ The code passed all $X$ tests; there still might be a bug, but its presence is less likely now.
#
# 3\. $P(A):\;\;$ The patient could have any number of diseases. $P(A | X):\;\;$ Performing a blood test generated evidence $X$, ruling out some of the possible diseases from consideration.
#
#
# It's clear that in each example we did not completely discard the prior belief after seeing new evidence $X$, but we *re-weighted the prior* to incorporate the new evidence (i.e. we put more weight, or confidence, on some beliefs versus others).
#
# By introducing prior uncertainty about events, we are already admitting that any guess we make is potentially very wrong. After observing data, evidence, or other information, we update our beliefs, and our guess becomes *less wrong*. This is the alternative side of the prediction coin, where typically we try to be *more right*.
#
#
# ### Bayesian Inference in Practice
#
# If frequentist and Bayesian inference were programming functions, with inputs being statistical problems, then the two would be different in what they return to the user. The frequentist inference function would return a number, representing an estimate (typically a summary statistic like the sample average etc.), whereas the Bayesian function would return *probabilities*.
#
# For example, in our debugging problem above, calling the frequentist function with the argument "My code passed all $X$ tests; is my code bug-free?" would return a *YES*. On the other hand, asking our Bayesian function "Often my code has bugs. My code passed all $X$ tests; is my code bug-free?" would return something very different: probabilities of *YES* and *NO*. The function might return:
#
#
# > *YES*, with probability 0.8; *NO*, with probability 0.2
#
#
#
# This is very different from the answer the frequentist function returned. Notice that the Bayesian function accepted an additional argument: *"Often my code has bugs"*. This parameter is the *prior*. By including the prior parameter, we are telling the Bayesian function to include our belief about the situation. Technically this parameter in the Bayesian function is optional, but we will see excluding it has its own consequences.
#
#
# #### Incorporating evidence
#
# As we acquire more and more instances of evidence, our prior belief is *washed out* by the new evidence. This is to be expected. For example, if your prior belief is something ridiculous, like "I expect the sun to explode today", and each day you are proved wrong, you would hope that any inference would correct you, or at least align your beliefs better. Bayesian inference will correct this belief.
#
#
# Denote $N$ as the number of instances of evidence we possess. As we gather an *infinite* amount of evidence, say as $N \rightarrow \infty$, our Bayesian results (often) align with frequentist results. Hence for large $N$, statistical inference is more or less objective. On the other hand, for small $N$, inference is much more *unstable*: frequentist estimates have more variance and larger confidence intervals. This is where Bayesian analysis excels. By introducing a prior, and returning probabilities (instead of a scalar estimate), we *preserve the uncertainty* that reflects the instability of statistical inference of a small $N$ dataset.
#
# One may think that for large $N$, one can be indifferent between the two techniques since they offer similar inference, and might lean towards the computationally-simpler, frequentist methods. An individual in this position should consider the following quote by <NAME> (2005)[1], before making such a decision:
#
# > Sample sizes are never large. If $N$ is too small to get a sufficiently-precise estimate, you need to get more data (or make more assumptions). But once $N$ is "large enough," you can start subdividing the data to learn more (for example, in a public opinion poll, once you have a good estimate for the entire country, you can estimate among men and women, northerners and southerners, different age groups, etc.). $N$ is never enough because if it were "enough" you'd already be on to the next problem for which you need more data.
#
# ### Are frequentist methods incorrect then?
#
# **No.**
#
# Frequentist methods are still useful or state-of-the-art in many areas. Tools such as least squares linear regression, LASSO regression, and expectation-maximization algorithms are all powerful and fast. Bayesian methods complement these techniques by solving problems that these approaches cannot, or by illuminating the underlying system with more flexible modeling.
#
#
# #### A note on *Big Data*
# Paradoxically, big data's predictive analytic problems are actually solved by relatively simple algorithms [2][4]. Thus we can argue that big data's prediction difficulty does not lie in the algorithm used, but instead on the computational difficulties of storage and execution on big data. (One should also consider Gelman's quote from above and ask "Do I really have big data?" )
#
# The much more difficult analytic problems involve *medium data* and, especially troublesome, *really small data*. Using a similar argument as Gelman's above, if big data problems are *big enough* to be readily solved, then we should be more interested in the *not-quite-big enough* datasets.
#
# ### Our Bayesian framework
#
# We are interested in beliefs, which can be interpreted as probabilities by thinking Bayesian. We have a *prior* belief in event $A$, beliefs formed by previous information, e.g., our prior belief about bugs being in our code before performing tests.
#
# Secondly, we observe our evidence. To continue our buggy-code example: if our code passes $X$ tests, we want to update our belief to incorporate this. We call this new belief the *posterior* probability. Updating our belief is done via the following equation, known as Bayes' Theorem, after its discoverer Thomas Bayes:
#
# \begin{align}
# P( A | X ) = & \frac{ P(X | A) P(A) } {P(X) } \\\\[5pt]
# & \propto P(X | A) P(A)\;\; (\propto \text{is proportional to } )
# \end{align}
#
# The above formula is not unique to Bayesian inference: it is a mathematical fact with uses outside Bayesian inference. Bayesian inference merely uses it to connect prior probabilities $P(A)$ with an updated posterior probabilities $P(A | X )$.
# ##### Example: Mandatory coin-flip example
#
# Every statistics text must contain a coin-flipping example, I'll use it here to get it out of the way. Suppose, naively, that you are unsure about the probability of heads in a coin flip (spoiler alert: it's 50%). You believe there is some true underlying ratio, call it $p$, but have no prior opinion on what $p$ might be.
#
# We begin to flip a coin, and record the observations: either $H$ or $T$. This is our observed data. An interesting question to ask is how our inference changes as we observe more and more data? More specifically, what do our posterior probabilities look like when we have little data, versus when we have lots of data.
#
# Below we plot a sequence of updating posterior probabilities as we observe increasing amounts of data (coin flips).
# + jupyter={"outputs_hidden": false}
"""
The book uses a custom matplotlibrc file, which provides the unique styles for
matplotlib plots. If executing this book, and you wish to use the book's
styling, provided are two options:
1. Overwrite your own matplotlibrc file with the rc-file provided in the
book's styles/ dir. See http://matplotlib.org/users/customizing.html
2. Also in the styles is bmh_matplotlibrc.json file. This can be used to
update the styles in only this notebook. Try running the following code:
import json, matplotlib
s = json.load( open("../styles/bmh_matplotlibrc.json") )
matplotlib.rcParams.update(s)
"""
# The code below can be passed over, as it is currently not important, plus it
# uses advanced topics we have not covered yet. LOOK AT PICTURE, MICHAEL!
# %matplotlib inline
from IPython.core.pylabtools import figsize
import numpy as np
from matplotlib import pyplot as plt
figsize(11, 9)
import scipy.stats as stats
dist = stats.beta
n_trials = [0, 1, 2, 3, 4, 5, 8, 15, 50, 500]
data = stats.bernoulli.rvs(0.5, size=n_trials[-1])
x = np.linspace(0, 1, 100)
# For the already prepared, I'm using Binomial's conj. prior.
for k, N in enumerate(n_trials):
sx = plt.subplot(len(n_trials) / 2, 2, k + 1)
plt.xlabel("$p$, probability of heads") \
if k in [0, len(n_trials) - 1] else None
plt.setp(sx.get_yticklabels(), visible=False)
heads = data[:N].sum()
y = dist.pdf(x, 1 + heads, 1 + N - heads)
plt.plot(x, y, label="observe %d tosses,\n %d heads" % (N, heads))
plt.fill_between(x, 0, y, color="#348ABD", alpha=0.4)
plt.vlines(0.5, 0, 4, color="k", linestyles="--", lw=1)
leg = plt.legend()
leg.get_frame().set_alpha(0.4)
plt.autoscale(tight=True)
plt.suptitle("Bayesian updating of posterior probabilities",
y=1.02,
fontsize=14)
plt.tight_layout()
# -
# The posterior probabilities are represented by the curves, and our uncertainty is proportional to the width of the curve. As the plot above shows, as we start to observe data our posterior probabilities start to shift and move around. Eventually, as we observe more and more data (coin-flips), our probabilities will tighten closer and closer around the true value of $p=0.5$ (marked by a dashed line).
#
# Notice that the plots are not always *peaked* at 0.5. There is no reason it should be: recall we assumed we did not have a prior opinion of what $p$ is. In fact, if we observe quite extreme data, say 8 flips and only 1 observed heads, our distribution would look very biased *away* from lumping around 0.5 (with no prior opinion, how confident would you feel betting on a fair coin after observing 8 tails and 1 head). As more data accumulates, we would see more and more probability being assigned at $p=0.5$, though never all of it.
#
# The next example is a simple demonstration of the mathematics of Bayesian inference.
# ##### Example: Bug, or just sweet, unintended feature?
#
#
# Let $A$ denote the event that our code has **no bugs** in it. Let $X$ denote the event that the code passes all debugging tests. For now, we will leave the prior probability of no bugs as a variable, i.e. $P(A) = p$.
#
# We are interested in $P(A|X)$, i.e. the probability of no bugs, given our debugging tests $X$ pass. To use the formula above, we need to compute some quantities.
#
# What is $P(X | A)$, i.e., the probability that the code passes $X$ tests *given* there are no bugs? Well, it is equal to 1, for code with no bugs will pass all tests.
#
# $P(X)$ is a little bit trickier: The event $X$ can be divided into two possibilities, event $X$ occurring even though our code *indeed has* bugs (denoted $\sim A\;$, spoken *not $A$*), or event $X$ without bugs ($A$). $P(X)$ can be represented as:
# \begin{align}
# P(X ) & = P(X \text{ and } A) + P(X \text{ and } \sim A) \\\\[5pt]
# & = P(X|A)P(A) + P(X | \sim A)P(\sim A)\\\\[5pt]
# & = P(X|A)p + P(X | \sim A)(1-p)
# \end{align}
# We have already computed $P(X|A)$ above. On the other hand, $P(X | \sim A)$ is subjective: our code can pass tests but still have a bug in it, though the probability there is a bug present is reduced. Note this is dependent on the number of tests performed, the degree of complication in the tests, etc. Let's be conservative and assign $P(X|\sim A) = 0.5$. Then
#
# \begin{align}
# P(A | X) & = \frac{1\cdot p}{ 1\cdot p +0.5 (1-p) } \\\\
# & = \frac{ 2 p}{1+p}
# \end{align}
# This is the posterior probability. What does it look like as a function of our prior, $p \in [0,1]$?
# + jupyter={"outputs_hidden": false}
figsize(12.5, 4)
p = np.linspace(0, 1, 50)
plt.plot(p, 2 * p / (1 + p), color="#348ABD", lw=3)
# plt.fill_between(p, 2*p/(1+p), alpha=.5, facecolor=["#A60628"])
plt.scatter(0.2, 2 * (0.2) / 1.2, s=140, c="#348ABD")
plt.xlim(0, 1)
plt.ylim(0, 1)
plt.xlabel("Prior, $P(A) = p$")
plt.ylabel("Posterior, $P(A|X)$, with $P(A) = p$")
plt.title("Is my code bug-free?")
# -
# We can see the biggest gains if we observe the $X$ tests passed when the prior probability, $p$, is low. Let's settle on a specific value for the prior. I'm a strong programmer (I think), so I'm going to give myself a realistic prior of 0.20, that is, there is a 20% chance that I write code bug-free. To be more realistic, this prior should be a function of how complicated and large the code is, but let's pin it at 0.20. Then my updated belief that my code is bug-free is 0.33.
#
# Recall that the prior is a probability: $p$ is the prior probability that there *are no bugs*, so $1-p$ is the prior probability that there *are bugs*.
#
# Similarly, our posterior is also a probability, with $P(A | X)$ the probability there is no bug *given we saw all tests pass*, hence $1-P(A|X)$ is the probability there is a bug *given all tests passed*. What does our posterior probability look like? Below is a chart of both the prior and the posterior probabilities.
#
# + jupyter={"outputs_hidden": false}
figsize(12.5, 4)
colours = ["#348ABD", "#A60628"]
prior = [0.20, 0.80]
posterior = [1. / 3, 2. / 3]
plt.bar([0, .7], prior, alpha=0.70, width=0.25,
color=colours[0], label="prior distribution",
lw="3", edgecolor=colours[0])
plt.bar([0 + 0.25, .7 + 0.25], posterior, alpha=0.7,
width=0.25, color=colours[1],
label="posterior distribution",
lw="3", edgecolor=colours[1])
plt.ylim(0,1)
plt.xticks([0.20, .95], ["Bugs Absent", "Bugs Present"])
plt.title("Prior and Posterior probability of bugs present")
plt.ylabel("Probability")
plt.legend(loc="upper left");
# -
# Notice that after we observed $X$ occur, the probability of bugs being absent increased. By increasing the number of tests, we can approach confidence (probability 1) that there are no bugs present.
#
# This was a very simple example of Bayesian inference and Bayes rule. Unfortunately, the mathematics necessary to perform more complicated Bayesian inference only becomes more difficult, except for artificially constructed cases. We will later see that this type of mathematical analysis is actually unnecessary. First we must broaden our modeling tools. The next section deals with *probability distributions*. If you are already familiar, feel free to skip (or at least skim), but for the less familiar the next section is essential.
# _______
#
# ## Probability Distributions
#
#
# **Let's quickly recall what a probability distribution is:** Let $Z$ be some random variable. Then associated with $Z$ is a *probability distribution function* that assigns probabilities to the different outcomes $Z$ can take. Graphically, a probability distribution is a curve where the probability of an outcome is proportional to the height of the curve. You can see examples in the first figure of this chapter.
#
# We can divide random variables into three classifications:
#
# - **$Z$ is discrete**: Discrete random variables may only assume values on a specified list. Things like populations, movie ratings, and number of votes are all discrete random variables. Discrete random variables become more clear when we contrast them with...
#
# - **$Z$ is continuous**: Continuous random variable can take on arbitrarily exact values. For example, temperature, speed, time, color are all modeled as continuous variables because you can progressively make the values more and more precise.
#
# - **$Z$ is mixed**: Mixed random variables assign probabilities to both discrete and continuous random variables, i.e. it is a combination of the above two categories.
#
# #### Expected Value
# Expected value (EV) is one of the most important concepts in probability. The EV for a given probability distribution can be described as "the mean value in the long run for many repeated samples from that distribution." To borrow a metaphor from physics, a distribution's EV acts like its "center of mass." Imagine repeating the same experiment many times over, and taking the average over each outcome. The more you repeat the experiment, the closer this average will become to the distributions EV. (side note: as the number of repeated experiments goes to infinity, the difference between the average outcome and the EV becomes arbitrarily small.)
#
# ### Discrete Case
# If $Z$ is discrete, then its distribution is called a *probability mass function*, which measures the probability $Z$ takes on the value $k$, denoted $P(Z=k)$. Note that the probability mass function completely describes the random variable $Z$, that is, if we know the mass function, we know how $Z$ should behave. There are popular probability mass functions that consistently appear: we will introduce them as needed, but let's introduce the first very useful probability mass function. We say $Z$ is *Poisson*-distributed if:
#
# $$P(Z = k) =\frac{ \lambda^k e^{-\lambda} }{k!}, \; \; k=0,1,2, \dots, \; \; \lambda \in \mathbb{R}_{>0} $$
#
# $\lambda$ is called a parameter of the distribution, and it controls the distribution's shape. For the Poisson distribution, $\lambda$ can be any positive number. By increasing $\lambda$, we add more probability to larger values, and conversely by decreasing $\lambda$ we add more probability to smaller values. One can describe $\lambda$ as the *intensity* of the Poisson distribution.
#
# Unlike $\lambda$, which can be any positive number, the value $k$ in the above formula must be a non-negative integer, i.e., $k$ must take on values 0,1,2, and so on. This is very important, because if you wanted to model a population you could not make sense of populations with 4.25 or 5.612 members.
#
# If a random variable $Z$ has a Poisson mass distribution, we denote this by writing
#
# $$Z \sim \text{Poi}(\lambda) $$
#
# One useful property of the Poisson distribution is that its expected value is equal to its parameter, i.e.:
#
# $$E\large[ \;Z\; | \; \lambda \;\large] = \lambda $$
#
# We will use this property often, so it's useful to remember. Below, we plot the probability mass distribution for different $\lambda$ values. The first thing to notice is that by increasing $\lambda$, we add more probability of larger values occurring. Second, notice that although the graph ends at 15, the distributions do not. They assign positive probability to every non-negative integer.
# + jupyter={"outputs_hidden": false}
figsize(12.5, 4)
import scipy.stats as stats
a = np.arange(16)
poi = stats.poisson
lambda_ = [1.5, 4.25]
colours = ["#348ABD", "#A60628"]
plt.bar(a, poi.pmf(a, lambda_[0]), color=colours[0],
label="$\lambda = %.1f$" % lambda_[0], alpha=0.60,
edgecolor=colours[0], lw="3")
plt.bar(a, poi.pmf(a, lambda_[1]), color=colours[1],
label="$\lambda = %.1f$" % lambda_[1], alpha=0.60,
edgecolor=colours[1], lw="3")
plt.xticks(a + 0.4, a)
plt.legend()
plt.ylabel("probability of $k$")
plt.xlabel("$k$")
plt.title("Probability mass function of a Poisson random variable; differing \
$\lambda$ values")
# -
# ### Continuous Case
# Instead of a probability mass function, a continuous random variable has a *probability density function*. This might seem like unnecessary nomenclature, but the density function and the mass function are very different creatures. An example of continuous random variable is a random variable with *exponential density*. The density function for an exponential random variable looks like this:
#
# $$f_Z(z | \lambda) = \lambda e^{-\lambda z }, \;\; z\ge 0$$
#
# Like a Poisson random variable, an exponential random variable can take on only non-negative values. But unlike a Poisson variable, the exponential can take on *any* non-negative values, including non-integral values such as 4.25 or 5.612401. This property makes it a poor choice for count data, which must be an integer, but a great choice for time data, temperature data (measured in Kelvins, of course), or any other precise *and positive* variable. The graph below shows two probability density functions with different $\lambda$ values.
#
# When a random variable $Z$ has an exponential distribution with parameter $\lambda$, we say *$Z$ is exponential* and write
#
# $$Z \sim \text{Exp}(\lambda)$$
#
# Given a specific $\lambda$, the expected value of an exponential random variable is equal to the inverse of $\lambda$, that is:
#
# $$E[\; Z \;|\; \lambda \;] = \frac{1}{\lambda}$$
# + jupyter={"outputs_hidden": false}
a = np.linspace(0, 4, 100)
expo = stats.expon
lambda_ = [0.5, 1]
for l, c in zip(lambda_, colours):
plt.plot(a, expo.pdf(a, scale=1. / l), lw=3,
color=c, label="$\lambda = %.1f$" % l)
plt.fill_between(a, expo.pdf(a, scale=1. / l), color=c, alpha=.33)
plt.legend()
plt.ylabel("PDF at $z$")
plt.xlabel("$z$")
plt.ylim(0, 1.2)
plt.title("Probability density function of an Exponential random variable;\
differing $\lambda$");
# -
#
# ### But what is $\lambda \;$?
#
#
# **This question is what motivates statistics**. In the real world, $\lambda$ is hidden from us. We see only $Z$, and must go backwards to try and determine $\lambda$. The problem is difficult because there is no one-to-one mapping from $Z$ to $\lambda$. Many different methods have been created to solve the problem of estimating $\lambda$, but since $\lambda$ is never actually observed, no one can say for certain which method is best!
#
# Bayesian inference is concerned with *beliefs* about what $\lambda$ might be. Rather than try to guess $\lambda$ exactly, we can only talk about what $\lambda$ is likely to be by assigning a probability distribution to $\lambda$.
#
# This might seem odd at first. After all, $\lambda$ is fixed; it is not (necessarily) random! How can we assign probabilities to values of a non-random variable? Ah, we have fallen for our old, frequentist way of thinking. Recall that under Bayesian philosophy, we *can* assign probabilities if we interpret them as beliefs. And it is entirely acceptable to have *beliefs* about the parameter $\lambda$.
#
#
# ##### Example: Inferring behaviour from text-message data
#
# Let's try to model a more interesting example, one that concerns the rate at which a user sends and receives text messages:
#
# > You are given a series of daily text-message counts from a user of your system. The data, plotted over time, appears in the chart below. You are curious to know if the user's text-messaging habits have changed over time, either gradually or suddenly. How can you model this? (This is in fact my own text-message data. Judge my popularity as you wish.)
#
# + jupyter={"outputs_hidden": false}
figsize(12.5, 3.5)
count_data = np.loadtxt("data/txtdata.csv")
n_count_data = len(count_data)
plt.bar(np.arange(n_count_data), count_data, color="#348ABD")
plt.xlabel("Time (days)")
plt.ylabel("count of text-msgs received")
plt.title("Did the user's texting habits change over time?")
plt.xlim(0, n_count_data);
# -
# Before we start modeling, see what you can figure out just by looking at the chart above. Would you say there was a change in behaviour during this time period?
#
# How can we start to model this? Well, as we have conveniently already seen, a Poisson random variable is a very appropriate model for this type of *count* data. Denoting day $i$'s text-message count by $C_i$,
#
# $$ C_i \sim \text{Poisson}(\lambda) $$
#
# We are not sure what the value of the $\lambda$ parameter really is, however. Looking at the chart above, it appears that the rate might become higher late in the observation period, which is equivalent to saying that $\lambda$ increases at some point during the observations. (Recall that a higher value of $\lambda$ assigns more probability to larger outcomes. That is, there is a higher probability of many text messages having been sent on a given day.)
#
# How can we represent this observation mathematically? Let's assume that on some day during the observation period (call it $\tau$), the parameter $\lambda$ suddenly jumps to a higher value. So we really have two $\lambda$ parameters: one for the period before $\tau$, and one for the rest of the observation period. In the literature, a sudden transition like this would be called a *switchpoint*:
#
# $$
# \lambda =
# \begin{cases}
# \lambda_1 & \text{if } t \lt \tau \cr
# \lambda_2 & \text{if } t \ge \tau
# \end{cases}
# $$
#
#
# If, in reality, no sudden change occurred and indeed $\lambda_1 = \lambda_2$, then the $\lambda$s posterior distributions should look about equal.
#
# We are interested in inferring the unknown $\lambda$s. To use Bayesian inference, we need to assign prior probabilities to the different possible values of $\lambda$. What would be good prior probability distributions for $\lambda_1$ and $\lambda_2$? Recall that $\lambda$ can be any positive number. As we saw earlier, the *exponential* distribution provides a continuous density function for positive numbers, so it might be a good choice for modeling $\lambda_i$. But recall that the exponential distribution takes a parameter of its own, so we'll need to include that parameter in our model. Let's call that parameter $\alpha$.
#
# \begin{align}
# &\lambda_1 \sim \text{Exp}( \alpha ) \\\
# &\lambda_2 \sim \text{Exp}( \alpha )
# \end{align}
#
# $\alpha$ is called a *hyper-parameter* or *parent variable*. In literal terms, it is a parameter that influences other parameters. Our initial guess at $\alpha$ does not influence the model too strongly, so we have some flexibility in our choice. A good rule of thumb is to set the exponential parameter equal to the inverse of the average of the count data. Since we're modeling $\lambda$ using an exponential distribution, we can use the expected value identity shown earlier to get:
#
# $$\frac{1}{N}\sum_{i=0}^N \;C_i \approx E[\; \lambda \; |\; \alpha ] = \frac{1}{\alpha}$$
#
# An alternative, and something I encourage the reader to try, would be to have two priors: one for each $\lambda_i$. Creating two exponential distributions with different $\alpha$ values reflects our prior belief that the rate changed at some point during the observations.
#
# What about $\tau$? Because of the noisiness of the data, it's difficult to pick out a priori when $\tau$ might have occurred. Instead, we can assign a *uniform prior belief* to every possible day. This is equivalent to saying
#
# \begin{align}
# & \tau \sim \text{DiscreteUniform(1,70) }\\\\
# & \Rightarrow P( \tau = k ) = \frac{1}{70}
# \end{align}
#
# So after all this, what does our overall prior distribution for the unknown variables look like? Frankly, *it doesn't matter*. What we should understand is that it's an ugly, complicated mess involving symbols only a mathematician could love. And things will only get uglier the more complicated our models become. Regardless, all we really care about is the posterior distribution.
#
# We next turn to PyMC, a Python library for performing Bayesian analysis that is undaunted by the mathematical monster we have created.
#
#
# Introducing our first hammer: PyMC
# -----
#
# PyMC is a Python library for programming Bayesian analysis [3]. It is a fast, well-maintained library. The only unfortunate part is that its documentation is lacking in certain areas, especially those that bridge the gap between beginner and hacker. One of this book's main goals is to solve that problem, and also to demonstrate why PyMC is so cool.
#
# We will model the problem above using PyMC. This type of programming is called *probabilistic programming*, an unfortunate misnomer that invokes ideas of randomly-generated code and has likely confused and frightened users away from this field. The code is not random; it is probabilistic in the sense that we create probability models using programming variables as the model's components. Model components are first-class primitives within the PyMC framework.
#
# <NAME> [5] has a very motivating description of probabilistic programming:
#
# > Another way of thinking about this: unlike a traditional program, which only runs in the forward directions, a probabilistic program is run in both the forward and backward direction. It runs forward to compute the consequences of the assumptions it contains about the world (i.e., the model space it represents), but it also runs backward from the data to constrain the possible explanations. In practice, many probabilistic programming systems will cleverly interleave these forward and backward operations to efficiently home in on the best explanations.
#
# Because of the confusion engendered by the term *probabilistic programming*, I'll refrain from using it. Instead, I'll simply say *programming*, since that's what it really is.
#
# PyMC code is easy to read. The only novel thing should be the syntax, and I will interrupt the code to explain individual sections. Simply remember that we are representing the model's components ($\tau, \lambda_1, \lambda_2$ ) as variables:
# + jupyter={"outputs_hidden": false}
import pymc as pm
alpha = 1.0 / count_data.mean() # Recall count_data is the
# variable that holds our txt counts
with pm.Model() as model:
lambda_1 = pm.Exponential("lambda_1", alpha)
lambda_2 = pm.Exponential("lambda_2", alpha)
tau = pm.DiscreteUniform("tau", lower=0, upper=n_count_data)
# -
# In the code above, we create the PyMC variables corresponding to $\lambda_1$ and $\lambda_2$. We assign them to PyMC's *stochastic variables*, so-called because they are treated by the back end as random number generators. We can demonstrate this fact by calling their built-in `random()` methods.
# + jupyter={"outputs_hidden": false}
print("Random output:", tau.eval(), tau.eval(), tau.eval())
# + jupyter={"outputs_hidden": false}
# @pm.deterministic
def lambda_(tau=tau, lambda_1=lambda_1, lambda_2=lambda_2):
out = np.zeros(n_count_data)
out[:tau] = lambda_1 # lambda before tau is lambda1
out[tau:] = lambda_2 # lambda after (and including) tau is lambda2
return out
# -
# This code creates a new function `lambda_`, but really we can think of it as a random variable: the random variable $\lambda$ from above. Note that because `lambda_1`, `lambda_2` and `tau` are random, `lambda_` will be random. We are **not** fixing any variables yet.
#
# `@pm.deterministic` is a decorator that tells PyMC this is a deterministic function. That is, if the arguments were deterministic (which they are not), the output would be deterministic as well. Deterministic functions will be covered in Chapter 2.
# + jupyter={"outputs_hidden": false}
observation = pm.Poisson("obs", lambda_, value=count_data, observed=True)
model = pm.Model([observation, lambda_1, lambda_2, tau])
# -
# The variable `observation` combines our data, `count_data`, with our proposed data-generation scheme, given by the variable `lambda_`, through the `value` keyword. We also set `observed = True` to tell PyMC that this should stay fixed in our analysis. Finally, PyMC wants us to collect all the variables of interest and create a `Model` instance out of them. This makes our life easier when we retrieve the results.
#
# The code below will be explained in Chapter 3, but I show it here so you can see where our results come from. One can think of it as a *learning* step. The machinery being employed is called *Markov Chain Monte Carlo* (MCMC), which I also delay explaining until Chapter 3. This technique returns thousands of random variables from the posterior distributions of $\lambda_1, \lambda_2$ and $\tau$. We can plot a histogram of the random variables to see what the posterior distributions look like. Below, we collect the samples (called *traces* in the MCMC literature) into histograms.
# + jupyter={"outputs_hidden": false}
# Mysterious code to be explained in Chapter 3.
mcmc = pm.MCMC(model)
mcmc.sample(40000, 10000, 1)
# + jupyter={"outputs_hidden": false}
lambda_1_samples = mcmc.trace('lambda_1')[:]
lambda_2_samples = mcmc.trace('lambda_2')[:]
tau_samples = mcmc.trace('tau')[:]
# + jupyter={"outputs_hidden": false}
figsize(12.5, 10)
# histogram of the samples:
ax = plt.subplot(311)
ax.set_autoscaley_on(False)
plt.hist(lambda_1_samples, histtype='stepfilled', bins=30, alpha=0.85,
label="posterior of $\lambda_1$", color="#A60628", density=True)
plt.legend(loc="upper left")
plt.title(r"""Posterior distributions of the variables
$\lambda_1,\;\lambda_2,\;\tau$""")
plt.xlim([15, 30])
plt.xlabel("$\lambda_1$ value")
ax = plt.subplot(312)
ax.set_autoscaley_on(False)
plt.hist(lambda_2_samples, histtype='stepfilled', bins=30, alpha=0.85,
label="posterior of $\lambda_2$", color="#7A68A6", density=True)
plt.legend(loc="upper left")
plt.xlim([15, 30])
plt.xlabel("$\lambda_2$ value")
plt.subplot(313)
w = 1.0 / tau_samples.shape[0] * np.ones_like(tau_samples)
plt.hist(tau_samples, bins=n_count_data, alpha=1,
label=r"posterior of $\tau$",
color="#467821", weights=w, rwidth=2.)
plt.xticks(np.arange(n_count_data))
plt.legend(loc="upper left")
plt.ylim([0, .75])
plt.xlim([35, len(count_data) - 20])
plt.xlabel(r"$\tau$ (in days)")
plt.ylabel("probability");
# -
# ### Interpretation
#
# Recall that Bayesian methodology returns a *distribution*. Hence we now have distributions to describe the unknown $\lambda$s and $\tau$. What have we gained? Immediately, we can see the uncertainty in our estimates: the wider the distribution, the less certain our posterior belief should be. We can also see what the plausible values for the parameters are: $\lambda_1$ is around 18 and $\lambda_2$ is around 23. The posterior distributions of the two $\lambda$s are clearly distinct, indicating that it is indeed likely that there was a change in the user's text-message behaviour.
#
# What other observations can you make? If you look at the original data again, do these results seem reasonable?
#
# Notice also that the posterior distributions for the $\lambda$s do not look like exponential distributions, even though our priors for these variables were exponential. In fact, the posterior distributions are not really of any form that we recognize from the original model. But that's OK! This is one of the benefits of taking a computational point of view. If we had instead done this analysis using mathematical approaches, we would have been stuck with an analytically intractable (and messy) distribution. Our use of a computational approach makes us indifferent to mathematical tractability.
#
# Our analysis also returned a distribution for $\tau$. Its posterior distribution looks a little different from the other two because it is a discrete random variable, so it doesn't assign probabilities to intervals. We can see that near day 45, there was a 50% chance that the user's behaviour changed. Had no change occurred, or had the change been gradual over time, the posterior distribution of $\tau$ would have been more spread out, reflecting that many days were plausible candidates for $\tau$. By contrast, in the actual results we see that only three or four days make any sense as potential transition points.
# ### Why would I want samples from the posterior, anyways?
#
#
# We will deal with this question for the remainder of the book, and it is an understatement to say that it will lead us to some amazing results. For now, let's end this chapter with one more example.
#
# We'll use the posterior samples to answer the following question: what is the expected number of texts at day $t, \; 0 \le t \le 70$ ? Recall that the expected value of a Poisson variable is equal to its parameter $\lambda$. Therefore, the question is equivalent to *what is the expected value of $\lambda$ at time $t$*?
#
# In the code below, let $i$ index samples from the posterior distributions. Given a day $t$, we average over all possible $\lambda_i$ for that day $t$, using $\lambda_i = \lambda_{1,i}$ if $t \lt \tau_i$ (that is, if the behaviour change has not yet occurred), else we use $\lambda_i = \lambda_{2,i}$.
# + jupyter={"outputs_hidden": false}
figsize(12.5, 5)
# tau_samples, lambda_1_samples, lambda_2_samples contain
# N samples from the corresponding posterior distribution
N = tau_samples.shape[0]
expected_texts_per_day = np.zeros(n_count_data)
for day in range(0, n_count_data):
# ix is a bool index of all tau samples corresponding to
# the switchpoint occurring prior to value of 'day'
ix = day < tau_samples
# Each posterior sample corresponds to a value for tau.
# for each day, that value of tau indicates whether we're "before"
# (in the lambda1 "regime") or
# "after" (in the lambda2 "regime") the switchpoint.
# by taking the posterior sample of lambda1/2 accordingly, we can average
# over all samples to get an expected value for lambda on that day.
# As explained, the "message count" random variable is Poisson distributed,
# and therefore lambda (the poisson parameter) is the expected value of
# "message count".
expected_texts_per_day[day] = (lambda_1_samples[ix].sum()
+ lambda_2_samples[~ix].sum()) / N
plt.plot(range(n_count_data), expected_texts_per_day, lw=4, color="#E24A33",
label="expected number of text-messages received")
plt.xlim(0, n_count_data)
plt.xlabel("Day")
plt.ylabel("Expected # text-messages")
plt.title("Expected number of text-messages received")
plt.ylim(0, 60)
plt.bar(np.arange(len(count_data)), count_data, color="#348ABD", alpha=0.65,
label="observed texts per day")
plt.legend(loc="upper left");
# -
# Our analysis shows strong support for believing the user's behavior did change ($\lambda_1$ would have been close in value to $\lambda_2$ had this not been true), and that the change was sudden rather than gradual (as demonstrated by $\tau$'s strongly peaked posterior distribution). We can speculate what might have caused this: a cheaper text-message rate, a recent weather-to-text subscription, or perhaps a new relationship. (In fact, the 45th day corresponds to Christmas, and I moved away to Toronto the next month, leaving a girlfriend behind.)
#
# ##### Exercises
#
# 1\. Using `lambda_1_samples` and `lambda_2_samples`, what is the mean of the posterior distributions of $\lambda_1$ and $\lambda_2$?
# + jupyter={"outputs_hidden": false}
# type your code here.
# -
# 2\. What is the expected percentage increase in text-message rates? `hint:` compute the mean of `lambda_1_samples/lambda_2_samples`. Note that this quantity is very different from `lambda_1_samples.mean()/lambda_2_samples.mean()`.
# + jupyter={"outputs_hidden": false}
# type your code here.
# -
# 3\. What is the mean of $\lambda_1$ **given** that we know $\tau$ is less than 45. That is, suppose we have been given new information that the change in behaviour occurred prior to day 45. What is the expected value of $\lambda_1$ now? (You do not need to redo the PyMC part. Just consider all instances where `tau_samples < 45`.)
# + jupyter={"outputs_hidden": false}
# type your code here.
# -
# ### References
#
#
# - [1] Gelman, Andrew. N.p.. Web. 22 Jan 2013. [N is never large enough](http://andrewgelman.com/2005/07/31/n_is_never_larg/).
# - [2] <NAME>. 2009. [The Unreasonable Effectiveness of Data](http://static.googleusercontent.com/media/research.google.com/en//pubs/archive/35179.pdf).
# - [3] <NAME>., <NAME> and <NAME>. 2010.
# PyMC: Bayesian Stochastic Modelling in Python. Journal of Statistical
# Software, 35(4), pp. 1-81.
# - [4] <NAME> and <NAME>. Large-Scale Machine Learning at Twitter. Proceedings of the 2012 ACM SIGMOD International Conference on Management of Data (SIGMOD 2012), pages 793-804, May 2012, Scottsdale, Arizona.
# - [5] <NAME>. "Why Probabilistic Programming Matters." 24 Mar 2013. Google, Online Posting to Google . Web. 24 Mar. 2013. <https://plus.google.com/u/0/107971134877020469960/posts/KpeRdJKR6Z1>.
# + jupyter={"outputs_hidden": false}
from IPython.core.display import HTML
def css_styling():
styles = open("../styles/custom.css", "r").read()
return HTML(styles)
css_styling()
# + jupyter={"outputs_hidden": false}
|
Chapter1_Introduction/Ch1_Introduction_PyMC2.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
#Author: <NAME>
#Purpose: This is a program tailored to the M&M modeling project that uses a
# combination of the Bisection Method and Newton's Method in order to
# find the minimum of the least squares.
import matplotlib.pyplot as plt
from numpy import exp, array, linspace, sum
from numpy.random import random
#This will standardize all figure sizes.
plt.rcParams["figure.figsize"] = [10,6]
#Constant to determine how many bisections and recursive calls to perform.
RANGE = 20
#******************************************************************************
#0: Main.
def main():
#Fill data arrays and initialize values.
x = [a for a in range(15)]
y = [8, 13, 20, 27, 39, 46, 52, 53, 56, 59, 61, 61, 61, 61, 62]
#Carrying capacity, initial population size, initial guess for r-value.
K, p0, r = (62, 8, 1)
Plot(x,y,1)
#Set lower and upper value to r.
r_low = r_high = r
#If the derivative of the sum of squares function is already zero (i.e. we
#already have a minimum), then we are done.
if df(r, x, y, p0, K) == 0:
#Curve to fit.
Fxn = lambda t : K*p0/(p0+(K-p0)*exp(-r*t))
Plot(x,Fxn,0,1)
exit()
#Find appropriate values to use for bisection.
while df(r_low, x, y, p0, K) > 0:
r_low -= 0.5
while df(r_high, x, y, p0, K) < 0:
r_high += 0.5
#Use Bisection Method to find seed value for Newton's Method.
r = Bisect(r_low, r_high, x, y, p0, K)
#Use Newton's Method to find most accurate root value.
r = Newton(r, x, y, p0, K)
#Redifine our function with new r value.
Fxn = lambda t : K*p0/(p0+(K-p0)*exp(-r*t))
#Display values for user.
print("\nK : ", K, "\np0 : ", p0, "\nr : ", r)
print('*'*64)
Error(x, y, Fxn)
Plot(x,Fxn,0,1)
#******************************************************************************
#1: Plot data points and functions.
def Plot(x_vals, y_vals, scatter=0, show=0):
if scatter:
plt.plot(x_vals, y_vals,'ko')
else:
X = linspace(min(x_vals), max(x_vals), 300)
Y = array([y_vals(x) for x in X])
plt.plot(X, Y, 'purple')
if show:
plt.title("Logistic Model of Disease Spread")
plt.xlabel("Iteration number")
plt.ylabel("Number of Infecteds")
plt.show()
#*******************************************************************************
#2: Derivative of the sum of squares function. You are, assumedly, trying to
# locate a root of this function so as to locate the minimum of the sum of
# squares function. That being said, you will have to find the derivative
# of the sum of squares function. I tried to type it out in a way such that,
# if you would like to modify the equation, you need only mess with the lines
# between the octothorpes. AlSO BE MINDFUL OF THE LINE CONTINUATION
# CHARACTERS.
def df(r, t_val, y_val, p0, K):
return sum([\
# # # # # # # # # # # # # # TYPE YOUR FUNCTION HERE # # # # # # # # # # # # # #
-2*(y -K/(1 + exp(-r*t)*(K - p0)/p0))*K/(1 + exp(-r*t)*(K - p0)/p0)**2*t*exp( \
-r*t)*(K - p0)/p0 \
# # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # #
for t,y in zip(t_val, y_val)])
#*******************************************************************************
#3: Use the bisection method to get a nice seed value for Newton's Method.
def Bisect(lo, hi, t_val, y_val, p0, K):
for i in range(RANGE):
mid = (lo + hi) / 2.0
if df(lo, t_val, y_val, p0, K)*df(hi, t_val, y_val, p0, K) > 0:
lo = mid
else:
hi = mid
return mid
#*******************************************************************************
#4: Use Newton's Method to find accurate root value.
def Newton(r, t_val, y_val, p0, K):
for i in range(RANGE):
r -= df(r, t_val, y_val, p0, K)/ddf(r, t_val, y_val, p0, K)
return r
#******************************************************************************
#5: Calculate sum of squares error.
def Error(x, y, F):
y_p = array([F(x_i) for x_i in x])
error = 0.0
for i in range(len(y)):
error += (y[i]-y_p[i])**2
print('Error %0.10f' %error)
return error
#*******************************************************************************
#4.1: Second derivative of the sum of squares function. This is needed for
# Newton's Method. See notes above (in 2) about modifications.
def ddf(r, t_val, y_val, p0, K):
return sum([\
# # # # # # # # # # # # # # TYPE YOUR FUNCTION HERE # # # # # # # # # # # # # #
2*K**2/(1 + exp(-r*t)*(K - p0)/p0)**4*t**2*exp(-r*t)**2*(K - p0)**2/p0**2 - 4* \
(y - K/(1 + exp(-r*t)*(K - p0)/p0))*K/(1 + exp(-r*t)*(K - p0)/p0)**3*t**2* \
exp(-r*t)**2*(K-p0)**2/p0**2 + 2*(y - K/(1 + exp(-r*t)*(K - p0)/p0))*K/(1 + \
exp(-r*t)*(K-p0)/p0)**2*t**2*exp(-r*t)*(K - p0)/p0 \
# # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # #
for t,y in zip(t_val, y_val)])
#******************************************************************************
#Call main.
main()
# -
|
BiNew_RF.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### This is Example 4.3. Gambler’s Problem from Sutton's book.
#
# A gambler has the opportunity to make bets on the outcomes of a sequence of coin flips.
# If the coin comes up heads, he wins as many dollars as he has staked on that flip;
# if it is tails, he loses his stake. The game ends when the gambler wins by reaching his goal of $100,
# or loses by running out of money.
#
# On each flip, the gambler must decide what portion of his capital to stake, in integer numbers of dollars.
# This problem can be formulated as an undiscounted, episodic, finite MDP.
#
# The state is the gambler’s capital, s ∈ {1, 2, . . . , 99}.
# The actions are stakes, a ∈ {0, 1, . . . , min(s, 100 − s)}.
# The reward is zero on all transitions except those on which the gambler reaches his goal, when it is +1.
#
# The state-value function then gives the probability of winning from each state. A policy is a mapping from levels of capital to stakes. The optimal policy maximizes the probability of reaching the goal. Let p_h denote the probability of the coin coming up heads. If p_h is known, then the entire problem is known and it can be solved, for instance, by value iteration.
#
import numpy as np
import sys
import matplotlib.pyplot as plt
if "../" not in sys.path:
sys.path.append("../")
#
# ### Exercise 4.9 (programming)
#
# Implement value iteration for the gambler’s problem and solve it for p_h = 0.25 and p_h = 0.55.
def value_iteration_for_gamblers(p_h, theta=0.0001, discount_factor=1.0):
"""
Args:
p_h: Probability of the coin coming up heads
"""
# The reward is zero on all transitions except those on which the gambler reaches his goal,
# when it is +1.
rewards = np.zeros(101)
rewards[100] = 1
# We introduce two dummy states corresponding to termination with capital of 0 and 100
V = np.zeros(101)
def one_step_lookahead(s, V, rewards):
"""
Helper function to calculate the value for all action in a given state.
Args:
s: The gambler’s capital. Integer.
V: The vector that contains values at each state.
rewards: The reward vector.
Returns:
A vector containing the expected value of each action.
Its length equals to the number of actions.
"""
A = np.zeros(101)
stakes = range(1, min(s, 100-s)+1) # Your minimum bet is 1, maximum bet is min(s, 100-s).
for a in stakes:
# rewards[s+a], rewards[s-a] are immediate rewards.
# V[s+a], V[s-a] are values of the next states.
# This is the core of the Bellman equation: The expected value of your action is
# the sum of immediate rewards and the value of the next state.
A[a] = p_h * (rewards[s+a] + V[s+a]*discount_factor) + (1-p_h) * (rewards[s-a] + V[s-a]*discount_factor)
return A
while True:
# Stopping condition
delta = 0
# Update each state...
for s in range(1, 100):
# Do a one-step lookahead to find the best action
A = one_step_lookahead(s, V, rewards)
# print(s,A,V) # if you want to debug.
best_action_value = np.max(A)
# Calculate delta across all states seen so far
delta = max(delta, np.abs(best_action_value - V[s]))
# Update the value function. Ref: Sutton book eq. 4.10.
V[s] = best_action_value
# Check if we can stop
if delta < theta:
break
# Create a deterministic policy using the optimal value function
policy = np.zeros(100)
for s in range(1, 100):
# One step lookahead to find the best action for this state
A = one_step_lookahead(s, V, rewards)
best_action = np.argmax(A)
# Always take the best action
policy[s] = best_action
return policy, V
# +
policy, v = value_iteration_for_gamblers(0.25)
print("Optimized Policy:")
print(policy)
print("")
print("Optimized Value Function:")
print(v)
print("")
# -
# ### Show your results graphically, as in Figure 4.3.
#
# +
# Plotting Final Policy (action stake) vs State (Capital)
# x axis values
x = range(100)
# corresponding y axis values
y = v[:100]
# plotting the points
plt.plot(x, y)
# naming the x axis
plt.xlabel('Capital')
# naming the y axis
plt.ylabel('Value Estimates')
# giving a title to the graph
plt.title('Final Policy (action stake) vs State (Capital)')
# function to show the plot
plt.show()
# +
# Plotting Capital vs Final Policy
# x axis values
x = range(100)
# corresponding y axis values
y = policy
# plotting the bars
plt.bar(x, y, align='center', alpha=0.5)
# naming the x axis
plt.xlabel('Capital')
# naming the y axis
plt.ylabel('Final policy (stake)')
# giving a title to the graph
plt.title('Capital vs Final Policy')
# function to show the plot
plt.show()
|
DP/Gamblers Problem Solution.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# ## PyTorch Tutorial
#
# IFT6135 – Representation Learning
#
# A Deep Learning Course, January 2019
#
# By <NAME>
#
# (Adapted from <NAME>'s MILA welcome tutorial)
# ## 1. Introduction to the torch tensor library
# ### Torch's numpy equivalent with GPU support
import numpy as np
from __future__ import print_function
import torch
# ### Initialize a random tensor
torch.Tensor(5, 3)
# ### From a uniform distribution
# intialization
print(torch.Tensor(5, 3).uniform_(-1, 1))
# sampling
print(torch.rand(5,3)*2-1)
# ### Get it's shape
# +
x = torch.Tensor(5, 3).uniform_(-1, 1)
print(x.size())
# or your favorite np_array.shape
print(x.shape)
# dimensionality of the 0'th axis?
# print(???)
print(x.size(0))
print(x.shape[0])
# -
# ### Tensor Types
# source: http://pytorch.org/docs/master/tensors.html
# |Data type |Tensor|
# |----------|------|
# |32-bit floating point| torch.FloatTensor|
# |64-bit floating point| torch.DoubleTensor|
# |16-bit floating point| torch.HalfTensor|
# |8-bit integer (unsigned)|torch.ByteTensor|
# |8-bit integer (signed)|torch.CharTensor|
# |16-bit integer (signed)|torch.ShortTensor|
# |32-bit integer (signed)|torch.IntTensor|
# |64-bit integer (signed)|torch.LongTensor|
# ### Creation from lists & numpy
z = torch.LongTensor([[1, 3], [2, 9]])
print(z.type())
# Cast to numpy ndarray
print(z.numpy().dtype)
z_ = torch.LongTensor([[1, 3], [2, 9]])
z+z_
# Data type inferred from numpy
print(torch.from_numpy(np.random.rand(5, 3)).type())
print(torch.from_numpy(np.random.rand(5, 3).astype(np.float32)).type())
print(torch.from_numpy(np.random.rand(5, 3)).float().dtype)
# +
# examples of type error
a = torch.randn(1) # x ~ N(0,1)
b = torch.from_numpy(np.ones(1)).float()
x+b
# -
# ### Simple mathematical operations
y = x ** torch.randn(5, 3)
print(y)
# +
noise = torch.randn(5, 3)
y = x / torch.sqrt(noise ** 2)
# equal to torch.abs
y_ = x / torch.abs(noise)
print(y)
print(y_)
# -
# ### Broadcasting
print(x.size())
print(x)
#y = x + torch.arange(5).view(5,1)
y = x + torch.arange(3)
print(y)
# print(x + torch.arange(5))
# ### Reshape
y = torch.randn(5, 10, 15)
print(y.size())
print(y.view(-1, 15).size()) # Same as doing y.view(50, 15)
print(y.view(-1, 15).unsqueeze(1).size()) # Adds a dimension at index 1.
print(y.view(-1, 15).unsqueeze(1).unsqueeze(2).unsqueeze(3).squeeze().size())
# If input is of shape: (Ax1xBxCx1xD)(Ax1xBxCx1xD) then the out Tensor will be of shape: (AxBxCxD)(AxBxCxD)
print()
print(y.transpose(0, 1).size())
print(y.transpose(1, 2).size())
print(y.transpose(0, 1).transpose(1, 2).size())
print(y.permute(1, 2, 0).size())
# ### Repeat
print(y.view(-1, 15).unsqueeze(1).expand(50, 100, 15).size())
print(y.view(-1, 15).unsqueeze(1).expand_as(torch.randn(50, 100, 15)).size())
# don't confuse it with tensor.repeat ...
print(y.view(-1, 15).unsqueeze(1).repeat(50,100,1).size())
# ### Concatenate
# +
# 2 is the dimension over which the tensors are concatenated
print(torch.cat([y, y], 2).size())
# stack concatenates the sequence of tensors along a new dimension.
print(torch.stack([y, y], 0).size())
# Q: how to do tensor.stack using cat?
print(torch.cat([y[None], y[None]], 0).size())
# -
# ### Advanced Indexing
# +
y = torch.randn(2, 3, 4)
print(y[[1, 0, 1, 1]].size())
# PyTorch doesn't support negative strides yet so ::-1 does not work.
rev_idx = torch.arange(1, -1, -1).long()
print(rev_idx)
print(y[rev_idx].size())
# gather(input, dim, index)
v = torch.arange(12).view(3,4)
print(v.shape)
print(v)
# [0,1,2,3]
# [4,5,6,7]
# [8,9,10,11]
# want to return [1,6,8]
print(torch.gather(v, 1, torch.tensor([1,2,0]).long().unsqueeze(1)))
# -
# ### GPU support
x = torch.cuda.HalfTensor(5, 3).uniform_(-1, 1)
y = torch.cuda.HalfTensor(3, 5).uniform_(-1, 1)
torch.matmul(x, y)
# ### Move tensors on the CPU -> GPU
x = torch.FloatTensor(5, 3).uniform_(-1, 1)
print(x)
x = x.cuda(device=0)
print(x)
x = x.cpu()
print(x)
# ### Contiguity in memory
# +
x = torch.FloatTensor(5, 3).uniform_(-1, 1)
print(x)
#x = x.cuda(device=0)
print(x)
print('Contiguity : %s ' % (x.is_contiguous()))
x = x.unsqueeze(0).expand(30, 5, 3)
print('Contiguity : %s ' % (x.is_contiguous()))
x = x.contiguous()
print('Contiguity : %s ' % (x.is_contiguous()))
# -
|
pytorch/1. The Torch Tensor Library and Basic Operations.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] slideshow={"slide_type": "slide"}
# <img src="joke.png"
# height=500
# width= 500
# alt="Example Visualization of a Snapshot (aggregated) Prediction Model"
# title="Snapshot Variable Prediction Model" />
# + [markdown] slideshow={"slide_type": "slide"}
# ### an important note before we start:
#
# <img src="model_comparison.png"
# height=500
# width= 500
# alt="Example Visualization of a Snapshot (aggregated) Prediction Model"
# title="Snapshot Variable Prediction Model" />
#
#
# sometimes a fancy algorithm can make a big impact, but often the difference between a well tuned simple and complex algorithm is not that high.
#
# Fancy algorithms don't magically make perfect predictions. The legwork done before and after model building is often the most important
#
# ------
# -
# + [markdown] slideshow={"slide_type": "slide"}
# # Now, lets learn about fancy algorithms: Random Forest and Gradient Boosted Trees
# * necessary background:
# * CART trees
# * bagging
# * ensembling
# * gradient boosting
# --------
# + [markdown] slideshow={"slide_type": "slide"}
# # Classification And Regression Trees (CART): glorified if/then statements
# ### example tree:
# <img src="Example_Decision_Tree.png"
# height=500
# width= 500
# alt="Example Visualization of a Snapshot (aggregated) Prediction Model"
# title="Snapshot Variable Prediction Model" />
#
# ### written as a rulebased classifier:
# 1. If Height > 180 cm Then Male
# 1. If Height <= 180 cm AND Weight > 80 kg Then Male
# 1. If Height <= 180 cm AND Weight <= 80 kg Then Female
# 1. Make Predictions With CART Models
# + [markdown] slideshow={"slide_type": "subslide"}
#
# * A final fitted CART model divides the predictor (x) space by successively splitting into rectangular regions and models the response (Y) as constant over each region
# * can be schematically represented as a "tree":
# * each interior node of the tree indicates on which predictor variable you split and where you split
# * each terminal node (aka leaf) represents one region and indicates the value of the predicted response in that region
#
# <br>
# + [markdown] slideshow={"slide_type": "slide"}
# ### CART Math: for those who want to take a simple idea and make it confusing
#
# we can write the equation of a regression tree as: $Y = g(X, \theta) + \epsilon$
#
# where: <br> $g(X;\theta)= \sum^M_{m=1}I(x \in R_m)$
#
#
# * $M$ = total number of regions (terminal nodes)
# * $R_m$ = $m$th region
# * $I(x \in R_m)$ = indicator function = $\{ \begin{array}{lr} 1:x \in R_m \\ 0:x \notin R_m \end{array} $
# * $c_m$ =constant predictor over Rm
# * $\theta$ = all parameters and structure (M, splits in Rm’s, cm’s, etc)
#
#
# #### illustration of tree for $M=6$ regions, $k=2$ predictors, and $n=21$ training observations
# <img src="CART3.png"
# height=500
# width= 500
# alt="Example Visualization of a Snapshot (aggregated) Prediction Model"
# title="Snapshot Variable Prediction Model" />
#
# + [markdown] slideshow={"slide_type": "subslide"}
# ### in more simple terms: a CART tree defines regions of the predictor space to correspond to a predicted outcome value
# * when fitting a CART tree, the model grows one tree node at a time
# * at each split, the tree defines boundaries in predictor space based on what REDUCES THE TRAINING ERROR THE MOST
# * stops making splits when the reduction in error falls below a threshold
# * branches can be pruned (ie nodes/boundaries removed)to reduce overfitting
# + [markdown] slideshow={"slide_type": "slide"}
# **example**: $GPA = g((HSrank, ACTscore), \theta) + \epsilon$
#
# <img src="CART2.png"
# height=800
# width= 800
# alt="Example Visualization of a Snapshot (aggregated) Prediction Model"
# title="Snapshot Variable Prediction Model" />
# + [markdown] slideshow={"slide_type": "slide"}
# # Why use a CART?
# * easy to interpret
# * handle categorical variables intuitively
# * computationally efficient
# * have reasonable predictive performance
# * not sensitive to MONOTONIC transformations (ie anything that preserves the order of a set, like log scaling).
# * form the basis for many commonly used algorithms
#
# + [markdown] slideshow={"slide_type": "slide"}
# --------
# # Next Background: Ensembling or Ensemble Learning
#
# * Ensemble: use multiple learning algorithms to obtain better predictive performance than could be obtained from any of the constituent learning algorithms alone.
# * A Machine Learning ensemble:
# * use multiple learning algorithms to obtain better predictive performance than a single learning algorithm alone.
# * concrete finite set of alternative models
# * but allows for much more flexible structure to exist among those
#
#
# + [markdown] slideshow={"slide_type": "slide"}
# --------
# # More Background: Ensembling, Bootstrapping & Bagging
#
# * **Ensemble** (in machine learning) :
# * use multiple learning algorithms to obtain better predictive performance than a single learning algorithm alone.
# * concrete finite set of alternative models
# * but allows for much more flexible structure to exist among those
#
#
# * **Bootstrapping**:
# * ~sampling WITH replacement
# + [markdown] slideshow={"slide_type": "slide"}
# * **Bagging**: (bootstrapping and aggregating)
# * a type of ensembling
# * designed to improve stability & accuracy of some ML algorithms
# * algorithm:
# 1. bootstrap many different sets from your training data
# 1. fit a model to each
# 1. average the predicted output (for regression) or voting (for classification) from bootstraped models across x values.
#
#
# **example**:
# * for $b= 1, 2, ..., B$: (aka: for i in range(1,B))
# * generate bootstrap sample of size n (ie sample B with replacement n times)
# * fit model (any kind) $g(x;\hat\theta^b)$
# * repeat for specified # of bootstraps
# * take y at each value of x as the average responce of each of the boostrapped models: $\hat y(x) = \frac{1}{B}\Sigma^B_{b=1}g(x;\hat\theta^b)$
#
# + [markdown] slideshow={"slide_type": "slide"}
# **Visualizations**:
# visualization for bagging ensemble (source: KDnuggets)
#
# <img src="bagged_ensemble.jpg"
# height=500
# width= 400
# alt="source KDNuggets"
# title="Snapshot Variable Prediction Model" />
#
#
# plotting boostrapped and bagged models: (source: Wikipedia)
#
# <img src="bagging_models.png"
# height=300
# width= 300
# alt="source: wikipedia"
# title="Snapshot Variable Prediction Model" />
#
# + [markdown] slideshow={"slide_type": "slide"}
# ### when is bagging useful:
# * For predictors where fitting is unstable (i.e., a small change in the data gives a very different fitted model) and/or the structure is such that the sum of multiple predictors no longer has the same structure
#
# ### when does bagging have no effect:
# * For predictors that are linear ($\hat y$ a linear function of training $y$)
#
#
#
# -
#
# + [markdown] slideshow={"slide_type": "slide"}
# -----
# # Random Forest: leveraging the wisdom of crowds
#
# * general idea: grow a bunch of different CART trees and let them all vote to get the prediction
#
# * Algorithm detail:
# 1. draw a bootstrap sample $Z^*$ of size $N$ from the training data
# 1. grow a CART tree $T_b$ to the bootstrapped data by recursively repeating the following steps for each terminal node until the minimum node size $n_min$ is reached:
# 1. randomly select $m$ predictor variables
# 1. pick the best variable/spit-point (aka boundary) for the $m$ predictor variables
# 1. split the node into two daughter nodes
# 1. output the ensemble of trees ${T_b}^B_1$.
#
# * make a prediction by taking majority vote (classification) or averaging prediction from each tree (regression)
#
# * in more simple terms: grow and train a lot of CART trees with a maximum size, each using randomly sampled observations (with replacement) and predictor variables (without replacement).
# + [markdown] slideshow={"slide_type": "slide"}
# Random forest simplified (source: towards data science blog)
#
# <img src="rf_vis.png"
# height=500
# width= 500
# alt="source: wikipedia"
# title="Snapshot Variable Prediction Model" />
# + [markdown] slideshow={"slide_type": "slide"}
# -----
# # Gradient boosting: leveraging the stupidity of crowds
#
# * **Boosting**:
# * a type of ensembling that turns a set of weak learners (ie predictors that are slightly better than chance) into a strong learner
# * many different types of algorithms that achieve boosting
#
# * **Gradient Boosting** :
# * Like other boosting methods, gradient boosting combines weak "learners" into a single strong learner in an iterative fashion.
# stated two different ways:
# * ensembles simple/weak CART trees in a stage-wise fashion and generalizes them by allowing optimization of an arbitrary differentiable loss function.
# * boosting sequentially fits simple/weak CART trees to the residuals from the previous iteration, taking the final model to be the sum of the individual models from each iteration
#
#
# explaining in the least-square regression setting:
# * goal: "teach" a model $F$ to predict values of the form $\hat y=F(x)$ by minimizing the mean squared error $\frac{1}{n}\sum_i (\hat y_i - y_i)^2$, where i indexes over some training set of size n.
# * at each iteration $m$, $1\leq m \leq M$, it may be assumed that there is some imperfect model $F_m$ (usually starts with just mean y).
# * in each iteration, the algorithm improves on $F_m$ by constructing a new model that adds an estimator $h$ to make it better: $F_{m+1}(x)= F_m(x) + h(x)$
# * a perfect $h$ implies that $F_{m+1}(x)= F_m(x) + h(x)=y$ or $ h(x) = y - F_m(x)$
# * thus, gradient boosting will fit $h$ to the **residual** $y-F_m(x)$.
# * in each iteration, $F_{m+1}$ attemps to correct the errors of it's predecessor $F_m$.
#
# to generalize this, we can observe that residuals $y- F(x)$ for a given model are the **negative gradients** (with respect to $F(x)$) of the squared error loss function $\frac{1}{2}(y-F(x))^2$
#
# + [markdown] slideshow={"slide_type": "slide"}
# for those of you who want the maths:
#
# <img src="gbm_algorithm.png"
# height=800
# width= 800
# alt="source: wikipedia"
# title="Snapshot Variable Prediction Model" />
#
# <br>
#
# for those of you who want pictures:
#
# <img src="gbm_vis.png"
# height=800
# width= 800
# alt="source: wikipedia"
# title="Snapshot Variable Prediction Model" />
#
# -
# + [markdown] slideshow={"slide_type": "slide"}
# -----
# # final thoughts about RandomForest and GBM
#
# * overfitting is definitely a thing with these models, so understanding some parameters is important.
#
#
# ### RF
# * tree size (depth) = big deal, larger trees = more likely to overfit
# * more trees = not that big of a deal. they make the out of bag error plot looks smoother
#
# ### GBM
# * tree size isn't that big of a deal, (smaller trees mean you can still capture error in next tree)
# * more trees = more likely to overfit. too many trees = the out of bag error look more U shaped.
#
# ### both algorithms:
# * neither alrogithm handles heavily imballanced classes very well (this can be an entire lecture on it's own)
# * both inherit all of the benefits of regular CART trees
# * both are better at regression than CART trees
# * both handle much more complex non-linear relationships between predictor and responce
# * both are capable of capturing **SOME** higher order predictor interactions, but these are often masked by marginal effects and cannot be differentiated from them. (ongoing research into this)
# -
import os
os.getcwd()
# +
import nbconvert
import nbformat
with open('hsip442/hsip442_algorithms_lecture.ipynb') as nb_file:
nb_contents = nb_file.read()
# Convert using the ordinary exporter
notebook = nbformat.reads(nb_contents, as_version=4)
exporter = nbconvert.HTMLExporter()
body, res = exporter.from_notebook_node(notebook)
# Create a dict mapping all image attachments to their base64 representations
images = {}
for cell in notebook['cells']:
if 'attachments' in cell:
attachments = cell['attachments']
for filename, attachment in attachments.items():
for mime, base64 in attachment.items():
images[f'attachment:{filename}'] = f'data:{mime};base64,{base64}'
# Fix up the HTML and write it to disk
for src, base64 in images.items():
body = body.replace(f'src="{src}"', f'src="{base64}"')
with open('my_notebook.html', 'w') as output_file:
output_file.write(body)
# -
# * **Stacking**: another type of ensembling
# 1. fit a number of different models to the entire training data ($g_m(x,\hat\theta^m)$)
# 2. take a linear combination (i.e. weighted average) of the models as the predictor($x$), using linear regression to determine the coefficients ($\theta$), with the consituent models ($g(x,\theta)$) as the basis function:
# * $\hat y(x) = \sum^M_{m=1}\hat \beta_m g_m(X,\hat\theta^m)$
# * linear regression: $\hat y(x)= \beta_0+ \sum^n_{i=1}\beta_i X$
|
notebooks/hsip442/hsip442_algorithms_lecture-Copy1.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# ---
# # Deep Q-Network implementation.
#
# This homework shamelessly demands you to implement a DQN - an approximate q-learning algorithm with experience replay and target networks - and see if it works any better this way.
#
# Original paper:
# https://arxiv.org/pdf/1312.5602.pdf
# **This notebook is given for debug.** The main task is in the other notebook (**homework_pytorch_main**). The tasks are similar and share most of the code. The main difference is in environments. In main notebook it can take some 2 hours for the agent to start improving so it seems reasonable to launch the algorithm on a simpler env first. Here it is CartPole and it will train in several minutes.
#
# **We suggest the following pipeline:** First implement debug notebook then implement the main one.
#
# **About evaluation:** All points are given for the main notebook with one exception: if agent fails to beat the threshold in main notebook you can get 1 pt (instead of 3 pts) for beating the threshold in debug notebook.
# +
import sys, os
if 'google.colab' in sys.modules and not os.path.exists('.setup_complete'):
# !wget -q https://raw.githubusercontent.com/yandexdataschool/Practical_RL/spring20/setup_colab.sh -O- | bash
# !wget -q https://raw.githubusercontent.com/yandexdataschool/Practical_RL/master/week04_approx_rl/atari_wrappers.py
# !wget -q https://raw.githubusercontent.com/yandexdataschool/Practical_RL/master/week04_approx_rl/utils.py
# !wget -q https://raw.githubusercontent.com/yandexdataschool/Practical_RL/master/week04_approx_rl/replay_buffer.py
# !wget -q https://raw.githubusercontent.com/yandexdataschool/Practical_RL/master/week04_approx_rl/framebuffer.py
# !pip install gym[box2d]
# !touch .setup_complete
# This code creates a virtual display to draw game images on.
# It will have no effect if your machine has a monitor.
if type(os.environ.get("DISPLAY")) is not str or len(os.environ.get("DISPLAY")) == 0:
# !bash ../xvfb start
os.environ['DISPLAY'] = ':1'
# -
# __Frameworks__ - we'll accept this homework in any deep learning framework. This particular notebook was designed for pytoch, but you find it easy to adapt it to almost any python-based deep learning framework.
import random
import numpy as np
import torch
import utils
import gym
import numpy as np
import matplotlib.pyplot as plt
# ### CartPole again
#
# Another env can be used without any modification of the code. State space should be a single vector, actions should be discrete.
#
# CartPole is the simplest one. It should take several minutes to solve it.
#
# For LunarLander it can take 1-2 hours to get 200 points (a good score) on Colab and training progress does not look informative.
# +
ENV_NAME = 'CartPole-v1'
def make_env(seed=None):
# some envs are wrapped with a time limit wrapper by default
env = gym.make(ENV_NAME).unwrapped
if seed is not None:
env.seed(seed)
return env
# -
env = make_env()
env.reset()
plt.imshow(env.render("rgb_array"))
state_shape, n_actions = env.observation_space.shape, env.action_space.n
# ### Building a network
# We now need to build a neural network that can map observations to state q-values.
# The model does not have to be huge yet. 1-2 hidden layers with < 200 neurons and ReLU activation will probably be enough. Batch normalization and dropout can spoil everything here.
import torch
import torch.nn as nn
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
# those who have a GPU but feel unfair to use it can uncomment:
# device = torch.device('cpu')
device
class DQNAgent(nn.Module):
def __init__(self, state_shape, n_actions, epsilon=0):
super().__init__()
self.epsilon = epsilon
self.n_actions = n_actions
self.state_shape = state_shape
# Define your network body here. Please make sure agent is fully contained here
assert len(state_shape) == 1
state_dim = state_shape[0]
<YOUR CODE>
def forward(self, state_t):
"""
takes agent's observation (tensor), returns qvalues (tensor)
:param state_t: a batch states, shape = [batch_size, *state_dim=4]
"""
# Use your network to compute qvalues for given state
qvalues = <YOUR CODE>
assert qvalues.requires_grad, "qvalues must be a torch tensor with grad"
assert len(
qvalues.shape) == 2 and qvalues.shape[0] == state_t.shape[0] and qvalues.shape[1] == n_actions
return qvalues
def get_qvalues(self, states):
"""
like forward, but works on numpy arrays, not tensors
"""
model_device = next(self.parameters()).device
states = torch.tensor(states, device=model_device, dtype=torch.float32)
qvalues = self.forward(states)
return qvalues.data.cpu().numpy()
def sample_actions(self, qvalues):
"""pick actions given qvalues. Uses epsilon-greedy exploration strategy. """
epsilon = self.epsilon
batch_size, n_actions = qvalues.shape
random_actions = np.random.choice(n_actions, size=batch_size)
best_actions = qvalues.argmax(axis=-1)
should_explore = np.random.choice(
[0, 1], batch_size, p=[1-epsilon, epsilon])
return np.where(should_explore, random_actions, best_actions)
agent = DQNAgent(state_shape, n_actions, epsilon=0.5).to(device)
# Now let's try out our agent to see if it raises any errors.
def evaluate(env, agent, n_games=1, greedy=False, t_max=10000):
""" Plays n_games full games. If greedy, picks actions as argmax(qvalues). Returns mean reward. """
rewards = []
for _ in range(n_games):
s = env.reset()
reward = 0
for _ in range(t_max):
qvalues = agent.get_qvalues([s])
action = qvalues.argmax(axis=-1)[0] if greedy else agent.sample_actions(qvalues)[0]
s, r, done, _ = env.step(action)
reward += r
if done:
break
rewards.append(reward)
return np.mean(rewards)
evaluate(env, agent, n_games=1)
# ### Experience replay
# For this assignment, we provide you with experience replay buffer. If you implemented experience replay buffer in last week's assignment, you can copy-paste it here in main notebook **to get 2 bonus points**.
#
# 
# #### The interface is fairly simple:
# * `exp_replay.add(obs, act, rw, next_obs, done)` - saves (s,a,r,s',done) tuple into the buffer
# * `exp_replay.sample(batch_size)` - returns observations, actions, rewards, next_observations and is_done for `batch_size` random samples.
# * `len(exp_replay)` - returns number of elements stored in replay buffer.
# +
from replay_buffer import ReplayBuffer
exp_replay = ReplayBuffer(10)
for _ in range(30):
exp_replay.add(env.reset(), env.action_space.sample(),
1.0, env.reset(), done=False)
obs_batch, act_batch, reward_batch, next_obs_batch, is_done_batch = exp_replay.sample(
5)
assert len(exp_replay) == 10, "experience replay size should be 10 because that's what maximum capacity is"
# -
def play_and_record(initial_state, agent, env, exp_replay, n_steps=1):
"""
Play the game for exactly n steps, record every (s,a,r,s', done) to replay buffer.
Whenever game ends, add record with done=True and reset the game.
It is guaranteed that env has done=False when passed to this function.
PLEASE DO NOT RESET ENV UNLESS IT IS "DONE"
:returns: return sum of rewards over time and the state in which the env stays
"""
s = initial_state
sum_rewards = 0
# Play the game for n_steps as per instructions above
<YOUR CODE>
return sum_rewards, s
# +
# testing your code.
exp_replay = ReplayBuffer(2000)
state = env.reset()
play_and_record(state, agent, env, exp_replay, n_steps=1000)
# if you're using your own experience replay buffer, some of those tests may need correction.
# just make sure you know what your code does
assert len(exp_replay) == 1000, "play_and_record should have added exactly 1000 steps, "\
"but instead added %i" % len(exp_replay)
is_dones = list(zip(*exp_replay._storage))[-1]
assert 0 < np.mean(is_dones) < 0.1, "Please make sure you restart the game whenever it is 'done' and record the is_done correctly into the buffer."\
"Got %f is_done rate over %i steps. [If you think it's your tough luck, just re-run the test]" % (
np.mean(is_dones), len(exp_replay))
for _ in range(100):
obs_batch, act_batch, reward_batch, next_obs_batch, is_done_batch = exp_replay.sample(
10)
assert obs_batch.shape == next_obs_batch.shape == (10,) + state_shape
assert act_batch.shape == (
10,), "actions batch should have shape (10,) but is instead %s" % str(act_batch.shape)
assert reward_batch.shape == (
10,), "rewards batch should have shape (10,) but is instead %s" % str(reward_batch.shape)
assert is_done_batch.shape == (
10,), "is_done batch should have shape (10,) but is instead %s" % str(is_done_batch.shape)
assert [int(i) in (0, 1)
for i in is_dones], "is_done should be strictly True or False"
assert [
0 <= a < n_actions for a in act_batch], "actions should be within [0, n_actions]"
print("Well done!")
# -
# ### Target networks
#
# We also employ the so called "target network" - a copy of neural network weights to be used for reference Q-values:
#
# The network itself is an exact copy of agent network, but it's parameters are not trained. Instead, they are moved here from agent's actual network every so often.
#
# $$ Q_{reference}(s,a) = r + \gamma \cdot \max _{a'} Q_{target}(s',a') $$
#
# 
target_network = DQNAgent(agent.state_shape, agent.n_actions, epsilon=0.5).to(device)
# This is how you can load weights from agent into target network
target_network.load_state_dict(agent.state_dict())
# ### Learning with... Q-learning
# Here we write a function similar to `agent.update` from tabular q-learning.
# Compute Q-learning TD error:
#
# $$ L = { 1 \over N} \sum_i [ Q_{\theta}(s,a) - Q_{reference}(s,a) ] ^2 $$
#
# With Q-reference defined as
#
# $$ Q_{reference}(s,a) = r(s,a) + \gamma \cdot max_{a'} Q_{target}(s', a') $$
#
# Where
# * $Q_{target}(s',a')$ denotes q-value of next state and next action predicted by __target_network__
# * $s, a, r, s'$ are current state, action, reward and next state respectively
# * $\gamma$ is a discount factor defined two cells above.
#
#
# __Note 1:__ there's an example input below. Feel free to experiment with it before you write the function.
#
# __Note 2:__ compute_td_loss is a source of 99% of bugs in this homework. If reward doesn't improve, it often helps to go through it line by line [with a rubber duck](https://rubberduckdebugging.com/).
def compute_td_loss(states, actions, rewards, next_states, is_done,
agent, target_network,
gamma=0.99,
check_shapes=False,
device=device):
""" Compute td loss using torch operations only. Use the formulae above. """
states = torch.tensor(states, device=device, dtype=torch.float) # shape: [batch_size, *state_shape]
# for some torch reason should not make actions a tensor
actions = torch.tensor(actions, device=device, dtype=torch.long) # shape: [batch_size]
rewards = torch.tensor(rewards, device=device, dtype=torch.float) # shape: [batch_size]
# shape: [batch_size, *state_shape]
next_states = torch.tensor(next_states, device=device, dtype=torch.float)
is_done = torch.tensor(
is_done.astype('float32'),
device=device,
dtype=torch.float
) # shape: [batch_size]
is_not_done = 1 - is_done
# get q-values for all actions in current states
predicted_qvalues = agent(states)
# compute q-values for all actions in next states
predicted_next_qvalues = target_network(next_states)
# select q-values for chosen actions
predicted_qvalues_for_actions = predicted_qvalues[range(
len(actions)), actions]
# compute V*(next_states) using predicted next q-values
next_state_values = <YOUR CODE>
assert next_state_values.dim(
) == 1 and next_state_values.shape[0] == states.shape[0], "must predict one value per state"
# compute "target q-values" for loss - it's what's inside square parentheses in the above formula.
# at the last state use the simplified formula: Q(s,a) = r(s,a) since s' doesn't exist
# you can multiply next state values by is_not_done to achieve this.
target_qvalues_for_actions = <YOUR CODE>
# mean squared error loss to minimize
loss = torch.mean((predicted_qvalues_for_actions -
target_qvalues_for_actions.detach()) ** 2)
if check_shapes:
assert predicted_next_qvalues.data.dim(
) == 2, "make sure you predicted q-values for all actions in next state"
assert next_state_values.data.dim(
) == 1, "make sure you computed V(s') as maximum over just the actions axis and not all axes"
assert target_qvalues_for_actions.data.dim(
) == 1, "there's something wrong with target q-values, they must be a vector"
return loss
# Sanity checks
# +
obs_batch, act_batch, reward_batch, next_obs_batch, is_done_batch = exp_replay.sample(
10)
loss = compute_td_loss(obs_batch, act_batch, reward_batch, next_obs_batch, is_done_batch,
agent, target_network,
gamma=0.99, check_shapes=True)
loss.backward()
assert loss.requires_grad and tuple(loss.data.size()) == (
), "you must return scalar loss - mean over batch"
assert np.any(next(agent.parameters()).grad.data.cpu().numpy() !=
0), "loss must be differentiable w.r.t. network weights"
assert np.all(next(target_network.parameters()).grad is None), "target network should not have grads"
# -
# ### Main loop
#
# It's time to put everything together and see if it learns anything.
from tqdm import trange
from IPython.display import clear_output
import matplotlib.pyplot as plt
seed = <your favourite random seed>
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
# +
env = make_env(seed)
state_dim = env.observation_space.shape
n_actions = env.action_space.n
state = env.reset()
agent = DQNAgent(state_dim, n_actions, epsilon=1).to(device)
target_network = DQNAgent(state_dim, n_actions, epsilon=1).to(device)
target_network.load_state_dict(agent.state_dict())
# -
exp_replay = ReplayBuffer(10**4)
for i in range(100):
if not utils.is_enough_ram(min_available_gb=0.1):
print("""
Less than 100 Mb RAM available.
Make sure the buffer size in not too huge.
Also check, maybe other processes consume RAM heavily.
"""
)
break
play_and_record(state, agent, env, exp_replay, n_steps=10**2)
if len(exp_replay) == 10**4:
break
print(len(exp_replay))
# +
# # for something more complicated than CartPole
# timesteps_per_epoch = 1
# batch_size = 32
# total_steps = 3 * 10**6
# decay_steps = 1 * 10**6
# opt = torch.optim.Adam(agent.parameters(), lr=1e-4)
# init_epsilon = 1
# final_epsilon = 0.1
# loss_freq = 20
# refresh_target_network_freq = 1000
# eval_freq = 5000
# max_grad_norm = 5000
# +
timesteps_per_epoch = 1
batch_size = 32
total_steps = 4 * 10**4
decay_steps = 1 * 10**4
opt = torch.optim.Adam(agent.parameters(), lr=1e-4)
init_epsilon = 1
final_epsilon = 0.1
loss_freq = 20
refresh_target_network_freq = 100
eval_freq = 1000
max_grad_norm = 5000
# -
mean_rw_history = []
td_loss_history = []
grad_norm_history = []
initial_state_v_history = []
state = env.reset()
for step in trange(total_steps + 1):
if not utils.is_enough_ram():
print('less that 100 Mb RAM available, freezing')
print('make sure everything is ok and make KeyboardInterrupt to continue')
try:
while True:
pass
except KeyboardInterrupt:
pass
agent.epsilon = utils.linear_decay(init_epsilon, final_epsilon, step, decay_steps)
# play
_, state = play_and_record(state, agent, env, exp_replay, timesteps_per_epoch)
# train
<sample batch_size of data from experience replay>
loss = <compute TD loss>
loss.backward()
grad_norm = nn.utils.clip_grad_norm_(agent.parameters(), max_grad_norm)
opt.step()
opt.zero_grad()
if step % loss_freq == 0:
td_loss_history.append(loss.data.cpu().item())
grad_norm_history.append(grad_norm)
if step % refresh_target_network_freq == 0:
# Load agent weights into target_network
<YOUR CODE>
if step % eval_freq == 0:
# eval the agent
mean_rw_history.append(evaluate(
make_env(seed=step), agent, n_games=3, greedy=True, t_max=1000)
)
initial_state_q_values = agent.get_qvalues(
[make_env(seed=step).reset()]
)
initial_state_v_history.append(np.max(initial_state_q_values))
clear_output(True)
print("buffer size = %i, epsilon = %.5f" %
(len(exp_replay), agent.epsilon))
plt.figure(figsize=[16, 9])
plt.subplot(2, 2, 1)
plt.title("Mean reward per episode")
plt.plot(mean_rw_history)
plt.grid()
assert not np.isnan(td_loss_history[-1])
plt.subplot(2, 2, 2)
plt.title("TD loss history (smoothened)")
plt.plot(utils.smoothen(td_loss_history))
plt.grid()
plt.subplot(2, 2, 3)
plt.title("Initial state V")
plt.plot(initial_state_v_history)
plt.grid()
plt.subplot(2, 2, 4)
plt.title("Grad norm history (smoothened)")
plt.plot(utils.smoothen(grad_norm_history))
plt.grid()
plt.show()
final_score = evaluate(
make_env(),
agent, n_games=30, greedy=True, t_max=1000
)
print('final score:', final_score)
assert final_score > 300, 'not good enough for DQN'
print('Well done')
# **Agent's predicted V-values vs their Monte-Carlo estimates**
eval_env = make_env()
record = utils.play_and_log_episode(eval_env, agent)
print('total reward for life:', np.sum(record['rewards']))
for key in record:
print(key)
# +
fig = plt.figure(figsize=(5, 5))
ax = fig.add_subplot(1, 1, 1)
ax.scatter(record['v_mc'], record['v_agent'])
ax.plot(sorted(record['v_mc']), sorted(record['v_mc']),
'black', linestyle='--', label='x=y')
ax.grid()
ax.legend()
ax.set_title('State Value Estimates')
ax.set_xlabel('Monte-Carlo')
ax.set_ylabel('Agent')
plt.show()
|
week04_approx_rl/homework_pytorch_debug.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# 
#
# <a href="https://hub.callysto.ca/jupyter/hub/user-redirect/git-pull?repo=https%3A%2F%2Fgithub.com%2Fcallysto%2Fcurriculum-notebooks&branch=master&subPath=Science/ProjectileMotion/projectile-motion.ipynb&depth=1" target="_parent"><img src="https://raw.githubusercontent.com/callysto/curriculum-notebooks/master/open-in-callysto-button.svg?sanitize=true" width="123" height="24" alt="Open in Callysto"/></a>
# # Projectile Motion
# ## 1. Introduction
# *This notebook is meant to satisfy Physics 20-A1.3s. Special thanks to Ms. <NAME> for her help on developing this notebook.*
# #### *Objective: Mathematically analyze the movement of a launched object to determine flight path, speed, and time.*
# Learning projectile motion will enable __you__ with the skills to understand how launched objects fly through the air and how you can design your own. It is critical for sports, military, and any scenario where you need an airborne object to reach a destination. What examples can you think of?
# Projectile motion explores the physics behind anything airborne that is subjected only to gravity. From throwing a dart to shooting a basketball to medieval cannons used during sieges, all of these objects are ruled by the same basic principles.
#
#
# > <img src="./images/catapult.gif" width="500" height="400" />
# >
# > <p style="text-align: center;"> A clip from the game Besiege (http://www.indiedb.com/games/besiege/images/besiege-gif) </p>
#
# ### Who created projectile motion analysis?
#
# > <img src="./images/galileo.jpg" alt="Drawing" style="width: 400px;"/>
# >
# > <p style="text-align: center;"> Painting of Galileo (https://www.gettyimages.ca/detail/news-photo/galileo-galilei-and-his-telescope-engraving-1864-news-photo/526510364) </p>
#
# Galileo, one of the great fathers of astronomy and modern science, created projectile motion analysis over 400 years ago. This gave him the tools to improve the accuracy and effectiveness of military cannons in the 17th century. The reason we study it today is because his method has stood the test of time and continues to be a fundamental foundation to areas like aerodynamics, sports performance, and military design.
#
# For the purposes of understanding the fundamentals, the effects of air resistance will be ignored. While it makes our lives simpler, it is wrong to assume that projectiles only experience the force of gravity - air resistance can play a significant role in many cases. Air resistance is responsible for lower projectile speeds and distances but it is also critical to helping airplanes slow down, keeping race cars grounded at high speeds (which is why they have spoilers), and causing crazy phenomena like the Magnus effect shown below.
# +
from IPython.lib.display import YouTubeVideo
from IPython.display import HTML
display(YouTubeVideo('2OSrvzNW9FE', start=24, end=35, mute=True, width=718, height=404))
display(HTML('''<p style="text-align: center;"> Basketball toss demonstrating the Magnus effect (https://www.youtube.com/watch?v=2OSrvzNW9FE) </p>'''))
# -
# However, in order to understand these complex cases, we must first strip away the layer of friction and look at how objects travel solely under the force gravity.
# ## 2. Theory & Practice
#
#
# Neglecting air resistance, all projectiles in the air will only be under the force of gravity. This means objects travelling in 2 dimensions will constantly be pulled down to earth and forced into a curved trajectory known as a parabola. Take a look at the gif below. What do you notice about the velocity vectors in the horizontal and vertical directions? Which one changes? Which one stays the same?
#
#
# > <img src="./images/parabola.gif" alt="Drawing" style="width: 400px;"/>
# >
# > <p style="text-align: center;"> Projectile motion animation (http://gbhsweb.glenbrook225.org/gbs/science/phys/mmedia/vectors/nhlp.html) </p>
#
#
# Galileo realized that projectile motion could be broken into two components: __horizontal (x)__ and __vertical (y)__ that can be analyzed separately. These two dimensions are __independent__, the only variable they share is time *__t__*. This means that changes in the vertical distance, speed, and acceleration will not affect the horizontal components and vice versa.
# ---
# ### Vertical Component
# Due to the force of gravity, the object will accelerate towards the center of the Earth. This means that its initial velocity will be different from its final velocity.
# There are 5 variables involved in the vertical component: acceleration, initial velocity, final velocity, *altitude* (distance travelled vertically), and time. We will assume $ \vec a $ is constant for projectile motion and equivalent to $ 9.81 m/s^2$.
#
#
# \begin{equation}
# \vec a , \vec v_i , \vec v_f , \vec d_y , t
# \end{equation}
# There are 5 kinematic equations for uniform acceleration where each one contains 4 of the 5 variables listed above. Given 3 of the variables in a problem, the other 2 can each be found by picking an appropriate equation. *Note: If the object is initially launched horizontally, then $\vec v_i $ = 0 *
#
# \begin{equation}
# \vec a_{ave} = \frac{\vec v_f - \vec v_i}{t} \\
# \vec d = \vec v_i t + \frac12\ \vec a t^2 \\
# \vec d = \vec v_f t - \frac12\ \vec a t^2 \\
# \vec d = \frac{\vec v_f + \vec v_i}{2} t \\
# v_f^2 = v_i^2 + 2 a d \\
# \end{equation}
# <details>
# <summary>
# __Question 1__ <br>
#
# You're out with some friends swimming at a popular cliff jumping spot and they can't stop arguing about how tall the cliff *really* is (for bragging rights). You decide to end this once and for all and pull out a stopwatch. You climb up the cliff, release a rock in your hand from rest, and time its descent. You do this a couple more times and get an average descent time of 1.62 seconds. Your friends stare at you in bewilderment as you rattle off the height after punching some numbers into your phone's calculator. How do you do it? <br><br>
#
# Try solving this question on your own, it's the best way to develop your skills. Once you've given it a go, click the dropdown arrow to reveal the solution. <br>
# </summary>
#
# <blockquote>
# __Solution__ <br>
# 1) Draw a picture of the scenario. This will give you a better grasp of the problem.
# 2) Define your sign convention. Assign positive and negative directions to y.
#
# <blockquote>
# <img src="./images/qu_1.jpg" alt="Drawing" style="width: 400px;"/>
# </blockquote>
#
# 3) Identify the variables you know and the variable(s) you're trying to find. Pick a formula that best fits the problem scenario and solve for the unknown. <br><br>
#
# <blockquote>
# The rock is released from rest so $ \vec v_i = 0 $. Based on the information given and our sign convention, we can state the following: <br><br>
#
#
# \begin{equation}
# \vec v_i = 0 \\
# \vec a = +9.81 \ m/s \\
# t = 1.62\ s \\
# \vec d_y = \ ? \\
# \vec v_f = \ ? \\
# \end{equation} <br>
#
# We have 3 known variables and 2 unknown variables in the problem. However, in this case we're looking for $ \vec d_y $ so we don't care what $ \vec v_f $ is. If we take a look at the 5 vertical component equations, we see that the following equation has the 3 variables we know and the one we're looking for: <br><br>
#
# \begin{equation}
# \vec d_y = \vec v_i t + \frac12\ \vec a t^2 \\
# \end{equation}
#
# We don't want one with $ \vec v_f $ because we don't know its value. Using this equation we can solve for $ \vec d_y $: <br><br>
#
# \begin{equation}
# \vec d_y = (0)(2.62\ s) + \frac12\ (+9.81\ m/s^2) (1.62\ s)^2 \\
# \vec d_y = +12.8727\ m \\
# \vec d_y = +12.9\ m \\
# \end{equation}
#
# Using the time it took to fell, we can conclude that the cliff is 12.9 meters tall.
#
# </blockquote>
# </blockquote>
#
# </details>
# ---
# ### Horizontal Component
# There is no force acting horizontally. Therefore there is no acceleration so it is **uniform motion** ($ \vec v_i = \vec v_f = \vec v_x $).
#
# Horizontal uniform motion is governed by 3 variables: velocity, *range* (distance travelled horizontally), and time.
#
# \begin{equation}
# \vec v_x , \vec d_x , t
# \end{equation}
#
# These 3 variables are related by the uniform motion equation:
#
# \begin{equation}
# \vec v_x = \frac {\vec d_x}{t}
# \end{equation}
#
# Let's do some practice to solidify the concepts.
# <details>
# <summary>
# __Question 2__ <br>
#
# You're back at the same pond as Question 1 except this time you flick the rock horizontally off the top of the cliff at 30.0 km/h. The rock makes it across the pond and still takes 1.62 seconds to hit the water. How long is the pond? <br>
# </summary>
#
# <blockquote>
# __Solution__ <br>
# 1) Draw a picture of the scenario. This will give you a better grasp of the problem.
# 2) Define your sign convention. Assign positive and negative directions to x.
#
# <blockquote>
# <img src="./images/qu_2.jpg" alt="Drawing" style="width: 400px;"/>
# </blockquote>
#
# 3) Identify the variables you know and the variable(s) you're trying to find. Pick a formula that best fits the problem scenario and solve for the unknown. <br><br>
#
# <blockquote>
# The horizontal component is easier than the vertical component because there's only one equation with 3 variables. List the 2 variables you know and the one you're trying to find: <br><br>
#
# \begin{equation}
# \vec v_x = +30.0\ km/h =\ +8.3333\ m/s \\
# t = 1.62\ s \\
# \vec d_x = \ ? \\
# \end{equation} <br>
#
# Rearrange the uniform motion equation for the unknown and solve: <br><br>
#
# \begin{equation}
# \vec v_x = \frac {\vec d_x}{t} \\
# \vec d_x = \vec v_x t = (+8.3333\ m/s)(1.62\ s) \\
# \vec d_x = +13.5\ m \\
# \end{equation}
#
# The pond is 13.5 meters long.
#
# </blockquote>
# </blockquote>
#
# </details>
# <details>
# <summary>
# __Question 3__ <br>
#
# <NAME>, the daredevil stunt driver, is performing his next trick. He speeds horizontally off of a 50.0 m high cliff on a motorcycle. How fast must he leave the cliff-top if he needs to soar over the 90.0 m river at the base of the cliff? <br>
#
# </summary>
#
# <blockquote>
# __Solution__ <br>
# 1) Draw a picture of the scenario. This will give you a better grasp of the problem.
# 2) Define your sign convention. Assign positive and negative directions to both x and y.
#
# <blockquote>
# <img src="./images/qu_3.jpg" alt="Drawing" style="width: 400px;"/>
# </blockquote>
#
# 3) Set up a table and identify the variables you know and the variable(s) you're trying to find. Pick a vertical component formula that best fits the problem scenario and solve for the unknown. <br><br>
#
# <blockquote>
# Based on the information given and our sign convention, we can fill our data table with the following: <br><br>
#
#
# \begin{array}{cc}
# x &y \\ \hline
# \vec d_x = +90.0\ m &\vec d_y = -50.0\ m \\
# \vec v_x =\ ? &\vec a = -9.81\ m/s^2 \\
# \ &\vec v_i = 0 \\
# \end{array}
# $$ t =\ ? $$ <br>
#
# Note that $ \vec v_i = 0 $ because the projectile is launched horizontally, and $ t $ is common to both x & y. <br>
#
# To find $ \vec v_x $, we need $ \vec d_x $ and $ t $. We know $ \vec d_x $ but we're missing $ t $ so we'll need to use the vertical data to solve for $ t $. Because we know $ \vec d_y, \vec a, $ and $ \vec v_i $, and we're looking for $ t $, we'll use the following equation because we can solve for $ t $ using the variables we know: <br><br>
#
# \begin{equation}
# \vec d = \vec v_i t + \frac12\ \vec a t^2 \\
# \end{equation}
#
# Because $ \vec v_i = 0 $, the first term goes to 0 and the resulting equation can be rearranged to solve for $ t $: <br><br>
#
# \begin{equation}
# \vec d = \frac12\ \vec a t^2 \\
# t = \sqrt{\frac{2 \vec d_y}{\vec a}} \\
# \end{equation}
#
# Plug in values to find $ t $: <br><br>
#
# \begin{equation}
# t = \sqrt{\frac{2 (-50.0\ m)}{(-9.81\ m/s^2)}} = 3.1928\ s
# \end{equation} <br>
#
# Now that $ t $ is known we can solve for $ \vec v_x $ using the uniform motion equation: <br><br>
#
# \begin{equation}
# \vec v_x = \frac {\vec d_x}{t} = \frac {(+90.0\ m)}{(3.1928\ s)} = +28.2\ m/s \ \ (+101.5\ km/h)
# \end{equation} <br>
#
# </blockquote>
# </blockquote>
#
# </details>
# <details>
# <summary>
# __Question 4__ <br>
#
# Galileo predicted that an object launched horizontally and an object dropped vertically off the same ledge will reach the ground at the same time. Will they? Why or why not? <br>
# </summary>
#
# <blockquote>
# __Solution__ <br>
#
# Yes, they will. Horizontal and vertical motion are independent for a projectile so the horizontal movement of the launched object does not affect its vertical freefall.
#
# <blockquote>
# <img src="./images/qu_4.jpg" alt="Drawing" style="width: 300px;"/>
# <p style="text-align: center;"> (Drawing courtesy of <NAME>) </p>
# </blockquote>
#
# </blockquote>
# </details>
#
#
#
#
# ---
# ### Projectiles Fired at an Angle
# What happens when your projectile is launched at an angle? What are the initial velocities?
#
# In this case, the initial vertical velocity is no longer 0. It will have some value $ \vec v_i $ that will gradually decrease to 0 at the top of its trajectory and then increase in the downward direction as it returns back to ground. Let's take a football punt for example.
#
# According to [Angelo Armenti's The Physics of Sports](https://www.livestrong.com/article/397904-maximum-speed-of-a-football/), top-level football kickers can send footballs flying at 70 mph (31 m/s)! If the player kicked it at an angle of 50°, the trajectory would look something like this:
#
# <blockquote>
# <img src="./images/proj_angle_1.jpg" alt="Drawing" style="width: 500px;"/>
# </blockquote>
#
# The horizontal velocity $ \vec v_x $ will remain the same throughout the flight (uniform motion) while $ \vec v_y $ will decrease to the top of its trajectory and then increase downwards. __Note: If the initial launch height and final landing height are the same ($ \vec d_y $ = 0), then the projectile will land with the same initial speed and angle!__ The initial horizontal and vertical velocities can be solved with some simple trigonometry:
#
# \begin{array}{cc}
# x &y \\
# cos(50^{\circ}) = \frac{adj}{hyp} = \frac{\vec v_x}{31\ m/s} &sin(50^{\circ}) = \frac{opp}{hyp} = \frac{\vec v_y}{31\ m/s} \\
# \vec v_x = (31\ m/s)\ cos(50^{\circ}) &\vec v_y = (31\ m/s)\ sin(50^{\circ}) \\
# \vec v_x = +19.9264\ m/s &\vec v_y = +23.7474\ m/s \\
# \vec v_x = +20\ m/s &\vec v_y = +24\ m/s \\
# \end{array}
#
# <details>
# <summary>
# __Question 5__ <br>
#
# A football player kicks a ball across a flat field at 31.0 m/s and 50.0° from the ground. Find:
# a) The maximum height reached
# b) The flight time (time before it hits the ground)
# c) The range (how far away it hits the ground)
# d) The velocity vector at its maximum height
# e) The acceleration vector at its maximum height
# f) The velocity of the football when it hits the ground <br><br>
#
# Again, try solving these questions on your own first, it's the best way to develop your skills. Once you've given it a go, click the dropdown arrow to reveal the solution. <br>
#
# </summary>
#
# <blockquote>
# __Solution__ <br>
# __a)__ <br>
#
# <blockquote>
# To find the max height, let's only look at the first half of the flight path. That way, we know $ \vec v_f = 0 $ because projectiles have no vertical velocity at the top of their flight path.
#
# <blockquote>
# <img src="./images/qu_5-1.jpg" alt="Drawing" style="width: 350px;"/>
# </blockquote>
#
# We'll use the same sign convention from Question 3. Because we're using the same values from the previous scenario, we can set up our table with the following information: <br><br>
#
#
# \begin{array}{cc}
# x &y \\ \hline
# \vec v_x =\ +19.9264\ m/s &\vec v_i = +23.7474\ m/s \\
# \ &\vec v_f = 0 \\
# \ &\vec a = -9.81\ m/s^2 \\
# \ &\vec d_y =\ ? \\
# \end{array}
# $$ t =\ ? $$ <br>
#
# Looking at our table, we have 3 knowns and 1 unknown in the vertical column. That means we can solve for $ \vec d_y $. Looking at our vertical component equations, the one that contains our 3 known variables and $ \vec d_y $ is: <br><br>
#
# \begin{equation}
# v_f^2 = v_i^2 + 2 a d
# \end{equation} <br>
#
# $ \vec v_f = 0 $ so we can rearrange and solve for $ \vec d_y $: <br><br>
#
# \begin{equation}
# \vec d_y = -\frac{\vec v_i^2}{2 \vec a} = -\frac{(+23.7474\ m/s)^2}{2\ (-9.81\ m/s^2)} \\
# \vec d_y = +28.7431\ m \\
# \vec d_y = +28.7\ m \\
# \end{equation} <br>
#
# </blockquote>
#
# __b)__ <br>
#
# <blockquote>
# To find the flight time, we could either find the time it takes to reach the top of its trajectory and double that, or we could look at the full flight path and find the time to landing. In this case we will choose the latter. <br>
#
# <blockquote>
# <img src="./images/qu_5-2.jpg" alt="Drawing" style="width: 300px;"/>
# </blockquote>
#
# Because our flight path changed, some of our variable have too. In particular, $ \vec d_y = 0 $ which also means that $ \vec v_f = -\vec v_i $. Let's make a new table: <br><br>
#
# \begin{array}{cc}
# x &y \\ \hline
# \vec v_x =\ +19.9264\ m/s &\vec v_i = +23.7474\ m/s \\
# \ &\vec v_f = -23.7474\ m/s \\
# \ &\vec a = -9.81\ m/s^2 \\
# \ &\vec d_y =\ 0 \\
# \end{array}
# $$ t =\ ? $$ <br>
#
# Now, we're looking for $ t $ but we don't have enough horizontal data to solve with uniform motion. However, we know 4 out of the 5 vertical variables which means we have lots of options for the vertical equation. Any of the first 4 equations will do but the second and third will require solving a quadratic equation. To make life easy, we'll use the first one: <br><br>
#
# \begin{equation}
# \vec a_{ave} = \frac{\vec v_f - \vec v_i}{t}
# \end{equation} <br>
#
# Rearrange and solve for $ t $ (Remember to keep your sign convention. If you don't you could end up with 0 here!): <br><br>
#
# \begin{equation}
# t = \frac{\vec v_f - \vec v_i}{\vec a_{ave}} = \frac{(-23.7474\ m/s) - (+23.7474\ m/s)}{(-9.81\ m/s^2)} \\
# t = 4.8415\ s \\
# t = 4.84\ s
# \end{equation} <br>
#
# </blockquote>
#
# __c)__ <br>
#
# <blockquote>
# Now that we have the flight time and $ \vec v_x $, we can use the uniform motion equation to solve for range: <br><br>
#
# \begin{equation}
# \vec v_x = \frac {\vec d_x}{t} \\
# \vec d_x = \vec v_x \ t = (+19.9264\ m/s)\ (4.8415\ s) = +96.4730\ m \\
# \vec d_x = +96.5\ m
# \end{equation} <br>
#
#
# </blockquote>
#
# __d)__ <br>
#
# <blockquote>
# At max height, the vertical velocity $ \vec v_y = 0 $ so the only velocity is horizontal.
#
# <blockquote>
# <img src="./images/qu_5-3.jpg" alt="Drawing" style="width: 300px;"/>
# </blockquote>
#
# </blockquote>
#
# __e)__ <br>
#
# <blockquote>
# The only acceleration is the acceleration due to gravity which is a constant pointing downwards (see part d for image).
#
# </blockquote>
#
# __f)__ <br>
#
# <blockquote>
# Because $ \vec d_y = 0 $ for the full flight path, $ \vec v_f = -\vec v_i = -23.7474\ m/s $. Horizontal velocity is constant so: <br><br>
#
# <blockquote>
# <img src="./images/qu_5-4.jpg" alt="Drawing" style="width: 200px;"/>
# </blockquote>
#
# \begin{equation}
# \vec v_f = 31.0\ m/s\ \ at -50.0^{\circ}
# \end{equation} <br>
#
# </blockquote>
#
# </blockquote>
# </details>
# <p style="text-align: center;"> __----- Continue on only after attempting Question 5. -----__ </p>
# +
"""
If this block of code has suddenly popped up, don't worry! You've found the code
used to create the Projectile Trajectory graph shown below. This is normally hidden but feel
free to explore it and see how it works.
If you want to hide it again, just click on this code block and press 'Ctrl' and 'Enter'
simultaneously on your keyboard.
"""
# Import required packages
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
from ipywidgets import interact
import ipywidgets as widgets
from IPython.display import HTML
# Set values for equations
g = 9.81
t = np.linspace(0,10,100)
# Define equations that plot will display
def d_x(t, theta, v_i):
return v_i*np.cos(np.deg2rad(theta))*t
def d_y(t, theta, v_i):
return v_i*np.sin(np.deg2rad(theta))*t - 0.5*g*t**2
# Define options for plot
def f(theta,v_i):
plt.plot(d_x(t, theta, v_i),d_y(t, theta, v_i))
plt.ylim(0,50)
plt.xlim(0,100)
plt.xlabel("Range (m)", fontsize=16)
plt.ylabel("Altitude (m)", fontsize=16)
plt.margins(0)
plt.grid()
plt.title("Projectile Trajectory", fontsize=20)
hide_me = ''
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show) {
$('div.input').each(function(id) {
el = $(this).find('.cm-variable:first');
if (id == 0 || el.text() == 'hide_me') {
$(this).hide();
}
});
$('div.output_prompt').css('opacity', 0);
} else {
$('div.input').each(function(id) {
$(this).show();
});
$('div.output_prompt').css('opacity', 1);
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
<form action="javascript:code_toggle()"><input style="opacity:0" type="submit" value="Click here to toggle on/off the raw code."></form>''')
# -
# The scenario in Question 5 can be modelled by a graph using Python. The following code block creates an interactive graph where you can modify the initial velocity $ \vec v_i $ and launch angle $ \theta $ to see how the projectile's trajectory changes. <br>
#
# Modify `theta` and `v_i` to recreate the scenario in Question 5 and use the graph to verify your answers for Parts (a) and (c) line up.
interact(f,theta = (0,90,5), v_i = (0,31,1))
# Pretty cool, huh? If you want to learn how to make graphs using Python, Part (b) of Question 6 will give you a step-by-step breakdown on how to make simple static graphs.
# ---
# ### Determining the Optimum Launch Angle
# <br>
# <details>
# <summary>
# __Question 6__ <br>
#
# Picture this: The International Olympic Committee runs a worldwide survey to see what new global event people would like to see. After some fierce debate and tallying the votes, __shot-cannon__ is chosen! Shot-cannon is a fairly straightforward sport: each country develops their own medieval cannon that competes in a series of challenges. While each challenge tests a different aspect of the design, the main event is the range competition to see which country's cannon can fire a lead ball the farthest across the field. Canada has chosen __you__ to man the cannon for the main event! With the cannon already designed and providing a fixed initial velocity $ \vec v_i $, your responsibility is to pick the optimal angle $ \theta $ to fire the cannon to achieve maximum distance. If you assume no air resistance and a flat field, what angle should you pick? <br><br>
#
# Before reading on, take a moment to picture the scenario in your head and, using your intuition, take a guess at what angle you think would provide the maximum range. Can you think of a way to prove this? <br><br>
#
# Initially, this problem can seem daunting in its magnitude, but if we break it into two chunks it becomes more manageable:
# a) Develop an equation for the range $ \vec d_x $ as a function of $ \vec v_i $ and $\theta $. That is, come up with an equation in the form of $ \vec d_x = f(\ \vec v_i,\ \theta)$.
# b) Determine the optimal angle from the equation you've developed. <br><br>
#
# Part (a) can be solved using the skills you've just learnt so give it a go and hit the arrow to check your answer. <br>
# Part (b) can be solved with coding and graphing which will be covered in the next section __Python Basics__. <br><br>
# </summary>
#
# <blockquote>
# __Solution__ <br>
# __a)__ <br>
#
# <blockquote>
# <img src="./images/qu_6.jpg" alt="Drawing" style="width: 400px;"/>
# </blockquote>
#
# With a picture and sign convention drawn, let's mark down the values we know in a table. While we don't know the angle $\theta$, and $ \vec v_i $ is an unknown constant, we can still write down $ \vec v_x $ and $ \vec v_{i_y} $ in terms of $\theta$ and $ \vec v_i $ because we ultimately want an equation with these terms: <br>
#
# \begin{array}{cc}
# x &y \\
# cos(\theta) = \frac{adj}{hyp} = \frac{\vec v_x}{\vec v_i} &sin(\theta) = \frac{opp}{hyp} = \frac{\vec v_y}{\vec v_i} \\
# \vec v_x = \vec v_i cos(\theta) &\vec v_y = \vec v_i sin(\theta) \\
# \vec v_x = v_i cos(\theta) &\vec v_y = v_i sin(\theta) \\
# \end{array} <br>
#
# The vector arrow on $ v_i $ is dropped because we know it's positive in both directions based on our sign convention. Also, we assume a flat field so $ \vec d_y = 0 $ which means that $ \vec v_{y_f} = -\vec v_{y_i} $ Therefore, our data table becomes:
#
# \begin{array}{cc}
# x &y \\ \hline
# \vec v_x =v_i cos(\theta) &\vec v_i = v_i sin(\theta) \\
# \vec d_x =\ ? &\vec v_f = -v_i sin(\theta) \\
# \ &\vec a = -g \\
# \ &\vec d_y =\ 0 \\
# \end{array}
# $$ t =\ ? $$ <br>
#
# Acceleration is written as $ -g = -9.81 m/s^2 $ for simplicity. Note that this is a constant written as a letter, __not a variable.__ <br>
#
# In order to come up with an equation for $ \vec d_x $, we first need an equation for $ t $ in terms of the given variables $ \vec v_i $ and $\theta$. Considering we know or have expressions for 4 of the vertical values, we will choose the first equation which makes it easy to solve for t: <br><br>
#
# \begin{equation}
# \vec a_{ave} = \frac{\vec v_f - \vec v_i}{t}
# \end{equation} <br>
#
# Rearrange and solve for $ t $: <br><br>
#
# \begin{equation}
# t = \frac{\vec v_f - \vec v_i}{\vec a_{ave}} = \frac{(-v_i sin(\theta)) - (v_i sin(\theta))}{(-g)} \\
# t = \frac{2 v_i sin(\theta)}{g} \\
# \end{equation} <br>
#
# This expression can be applied to the horizontal component to come up with an equation for $ \vec d_x $: <br><br>
#
# \begin{equation}
# \vec v_x = \frac{\vec d_x}{t} \\
# \vec d_x = \vec v_x t \\
# \vec d_x = (v_i cos(\theta)) (\frac{2 v_i sin(\theta)}{g}) \\
# \vec d_x = \frac{2}{g} v_i^2 sin(\theta) cos(\theta) \\
# \end{equation} <br>
#
# Knowing that $ g $ and $ v_i $ are constants, this means that $ \vec d_x $ is only a function of the variable $\theta$.
#
# </blockquote>
# </details>
#
#
#
#
# <p style="text-align: center;"> __----- Continue on only after attempting Question 6 Part (a) and checking with the solution. The next section will cover Part (b). -----__ </p>
# ### Python Basics
#
# From this point on, there are two ways to solve for $\theta$: you can use calculus to solve for it analytically, or we can get creative and use coding with graphing to find the answer!
#
# Python provides us with some great tools to graph this function easily. If you have some knowledge of Python you can skip the explanations and just run the code cells. If not, we're going to take a moment to understand what the code you're about to see does. First up, let's go over the first bit of code.
#
# __Imports__
# At the beginning of most Python programs you're more than likely to see a few (or many) `import` statements. The purpose of these is to bring in other pieces of code, that either you or someone else have written, in order to keep your current program more manageable. For example, the set of import statements we're going to use to plot our functions look like this (click on the code block below and press `Ctrl` and `Enter` on your keyboard simultaneously to run the cell).
# %matplotlib inline
import matplotlib
import numpy as np
import matplotlib.pyplot as plt
# Starting from the top down, let's go over what each piece of this code does.
#
# 1. **%matplotlib inline:** This is very specific to the Jupyter notebook we're using. This tells matplotlib (next bullet) to output graphs in the cell directly below the one in which the code is executed. For more information about Jupyter "magic commands" feel free to read [this document](http://ipython.readthedocs.io/en/stable/interactive/magics.html).
#
# 2. **import matplotlib**: This command tells Python to import the `matplotlib` package. This package contains the python functions that we'll be using for plotting.
#
# 3. **import numpy as np**: This imports the python package `numpy` or "numerical python" and assigns the name to it within our code to `np` so we don't have to type out `numpy` every time we need a function. We'll be using this package for mathematical functions like sine and the square root.
#
# 4. **import matplotlib.pyplot as plt**: This imports the graphing subroutines from the `matplotlib` packages and assigns them the name `plt` so we don't have to type as much when we want to produce a graph.
#
# __Plotting__
# After required modules have been imported into our script, we're ready to get started graphing. First up, we need to define a number of points which to plot. Our computer doesn't understand that the variable $ \theta $ is fully continuous, so we have to give it a discrete set of points to plot our function with. We can do that in python using numpy as follows (click the next cell and press control and enter at the same time on your keyboard).
theta = np.linspace(0,90,5)
print("theta = ", theta)
# This creates a **list** of numbers called `theta` which consists of 5 numbers evenly spaced in the domain $[0,90]$ that we'll use to plot our function. The `print` function is a standard python function that simply displays our variables to the screen as either numbers or characters. In order to plot our functions, we type the following. Note that we've increased the number of points in `theta` to create a smoother plot. Feel free to change the number `100` to something smaller and observe how the plot changes. We also assigned a value to `v_i` and `g` so that the computer can plot the function numerically. Likewise, play around with these numbers and see how your range changes.
theta = np.linspace(0,90,100)
v_i = 30.0
g = 9.81
plt.plot(theta, 2 / g * (v_i**2) * np.sin(np.deg2rad(theta)) * np.cos(np.deg2rad(theta)))
plt.show()
# There's a fair bit going on in that last line of code that should be noted. First, by using `plt.plot` we're calling a function from `matplotlib.pyplot` (that we called `plt`) called `plot`. This function, unsurprisingly, is used to tell Python what to plot. We then pass this function a number of arguments (AKA inputs). The first argument `theta` is the list of numbers we generated earlier. The second argument `2 / g * (v_i**2) * np.sin(np.deg2rad(theta)) * np.cos(np.deg2rad(theta))` is the mathematical function we're going to plot. Here `theta` is the variable we're going to plot, and also the list of points that we generated earlier. The `**` is the Python way of saying "to the power of". Because `sin` and `cos` functions take radians as an input and not degrees, we use `np.deg2rad` which is a function that converts degrees to radians. <br>
#
# So, what we're really saying here is "plot $ \frac{2}{g}v_i^2 \sin(\theta)\cos(\theta) $ for 100 $\theta$ values between 0 and 90".
#
# Now, that graph is missing a lot of important things like axis labels and a legend. We can add those like this:
plt.plot(theta, 2 / g * (v_i**2) * np.sin(np.deg2rad(theta)) * np.cos(np.deg2rad(theta)))
plt.xlabel(r"$ \theta (deg) $", fontsize=16)
plt.ylabel(r"$ \vec d_x (m) $", fontsize=16)
plt.margins(0)
plt.grid()
plt.title(r"Range as a function of launch angle $ \theta $")
plt.show()
# where the `plt.` calls are still calling functions from `matplotlib.pyplot` however this time we're creating x axes labels with `plt.xlabel` and y axes labels with `plt.ylabel`. `plt.margins(0)` removes any unnecessary blank space around the graph and `plt.grid()` adds a nice grid to visually locate points on the graph easier. Finally, `plt.title` adds a title bar to our graph. The dollar signs and "r" characters are just there to make the lettering look nice. Play around with the code and see if you can change the title bar, label font sizes, and margins. <br>
#
#
# Based on the plot above, we see that the maximum range lines up with a launch angle of 45° . Was this your initial guess?
#
# With no air resistance, 45° is the optimal angle because it provides the best compromise between horizontal speed and height. If you shoot it below 45°, you'll get a faster horizontal velocity but the ball will also hit the ground quicker because there's less flight time. If you shoot it above 45°, you'll get more flight time but a slower horizontal velocity. 45° is the sweet spot between extremes.
#
# Because the graphical solution takes the shape of a parabola, this also demonstrates an important symmetry in the launch angles. Launch angles that are equidistant from the maximum of 45° will have the same range. That is, 30° and 60° have the same range, 15° and 75° have the same range, etc. The wonders of physics!
# ---
# ## 3. Conclusion & Extension
#
# This notebook has demonstrated the basics of projectile motion that can be used to determine flight paths, speeds, and times. Reasoning for why projectile motion is foundational to physics is explained, followed by a breakdown describing the horizontal and vertical components of projectile motion. The two components were brought together in an angled launch example with an interactive graph and the final question utilized Python programming to solve a basic calculus problem. Applying the skills taught in this notebook to your practice examples will give you a strong grasp of projectile motion analysis and enable __you__ to begin designing your own basic launchers!
#
#
# ### Practice: Projectile Game
# For a great interactive visualization of projectile motion, check out the PhET link below!
#
# <div style="position: relative; width: 300px; height: 200px;"><a href="https://phet.colorado.edu/sims/html/projectile-motion/latest/projectile-motion_en.html" style="text-decoration: none;"><img src="https://phet.colorado.edu/sims/html/projectile-motion/latest/projectile-motion-600.png" alt="Projectile Motion" style="border: none;" width="300" height="200"/><div style="position: absolute; width: 200px; height: 80px; left: 50px; top: 60px; background-color: #FFF; opacity: 0.6; filter: alpha(opacity = 60);"></div><table style="position: absolute; width: 200px; height: 80px; left: 50px; top: 60px;"><tr><td style="text-align: center; color: #000; font-size: 24px; font-family: Arial,sans-serif;">Click to Run</td></tr></table></a></div>
# [](https://github.com/callysto/curriculum-notebooks/blob/master/LICENSE.md)
|
_sources/curriculum-notebooks/Science/ProjectileMotion/projectile-motion.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# load and summarize the dataset
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
# generate regression dataset
X, y = make_regression(n_samples=1000, n_features=100, n_informative=10, noise=0.1,
random_state=1)
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# summarize
print('Train', X_train.shape, y_train.shape)
print('Test', X_test.shape, y_test.shape)
# example of correlation feature selection for numerical data
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_regression
from matplotlib import pyplot
# feature selection
def select_features(X_train, y_train, X_test):
# configure to select all features
fs = SelectKBest(score_func=f_regression, k='all')
# learn relationship from training data
fs.fit(X_train, y_train)
# transform train input data
X_train_fs = fs.transform(X_train)
# transform test input data
X_test_fs = fs.transform(X_test)
return X_train_fs, X_test_fs, fs
# load the dataset
X, y = make_regression(n_samples=1000, n_features=100, n_informative=10, noise=0.1,
random_state=1)
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# feature selection
X_train_fs, X_test_fs, fs = select_features(X_train, y_train, X_test)
# what are scores for the features
for i in range(len(fs.scores_)):
print('Feature %d: %f' % (i, fs.scores_[i]))
# plot the scores
pyplot.bar([i for i in range(len(fs.scores_))], fs.scores_)
pyplot.show()
# example of mutual information feature selection for numerical input data
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import mutual_info_regression
from matplotlib import pyplot
# feature selection
def select_features(X_train, y_train, X_test):
# configure to select all features
fs = SelectKBest(score_func=mutual_info_regression, k='all')
# learn relationship from training data
fs.fit(X_train, y_train)
# transform train input data
X_train_fs = fs.transform(X_train)
# transform test input data
X_test_fs = fs.transform(X_test)
return X_train_fs, X_test_fs, fs
# load the dataset
X, y = make_regression(n_samples=1000, n_features=100, n_informative=10, noise=0.1,
random_state=1)
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# feature selection
X_train_fs, X_test_fs, fs = select_features(X_train, y_train, X_test)
# what are scores for the features
for i in range(len(fs.scores_)):
print('Feature %d: %f' % (i, fs.scores_[i]))
# plot the scores
pyplot.bar([i for i in range(len(fs.scores_))], fs.scores_)
pyplot.show()
# evaluation of a model using all input features
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error
# load the dataset
X, y = make_regression(n_samples=1000, n_features=100, n_informative=10, noise=0.1,
random_state=1)
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# fit the model
model = LinearRegression()
model.fit(X_train, y_train)
# evaluate the model
yhat = model.predict(X_test)
# evaluate predictions
mae = mean_absolute_error(y_test, yhat)
print('MAE: %.3f' % mae)
# evaluation of a model using 10 features chosen with correlation
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_regression
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error
# feature selection
def select_features(X_train, y_train, X_test):
# configure to select a subset of features
fs = SelectKBest(score_func=f_regression, k=10)
# learn relationship from training data
fs.fit(X_train, y_train)
# transform train input data
X_train_fs = fs.transform(X_train)
# transform test input data
X_test_fs = fs.transform(X_test)
return X_train_fs, X_test_fs, fs
# load the dataset
X, y = make_regression(n_samples=1000, n_features=100, n_informative=10, noise=0.1,
random_state=1)
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# feature selection
X_train_fs, X_test_fs, fs = select_features(X_train, y_train, X_test)
# fit the model
model = LinearRegression()
model.fit(X_train_fs, y_train)
# evaluate the model
yhat = model.predict(X_test_fs)
# evaluate predictions
mae = mean_absolute_error(y_test, yhat)
print('MAE: %.3f' % mae)
# evaluation of a model using 88 features chosen with correlation
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_regression
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error
# feature selection
def select_features(X_train, y_train, X_test):
# configure to select a subset of features
fs = SelectKBest(score_func=f_regression, k=88)
# learn relationship from training data
fs.fit(X_train, y_train)
# transform train input data
X_train_fs = fs.transform(X_train)
# transform test input data
X_test_fs = fs.transform(X_test)
return X_train_fs, X_test_fs, fs
# load the dataset
X, y = make_regression(n_samples=1000, n_features=100, n_informative=10, noise=0.1,
random_state=1)
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# feature selection
X_train_fs, X_test_fs, fs = select_features(X_train, y_train, X_test)
# fit the model
model = LinearRegression()
model.fit(X_train_fs, y_train)
# evaluate the model
yhat = model.predict(X_test_fs)
# evaluate predictions
mae = mean_absolute_error(y_test, yhat)
print('MAE: %.3f' % mae)
# evaluation of a model using 88 features chosen with mutual information
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import mutual_info_regression
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error
# feature selection
def select_features(X_train, y_train, X_test):
# configure to select a subset of features
fs = SelectKBest(score_func=mutual_info_regression, k=88)
# learn relationship from training data
fs.fit(X_train, y_train)
# transform train input data
X_train_fs = fs.transform(X_train)
# transform test input data
X_test_fs = fs.transform(X_test)
return X_train_fs, X_test_fs, fs
# load the dataset
X, y = make_regression(n_samples=1000, n_features=100, n_informative=10, noise=0.1,
random_state=1)
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# feature selection
X_train_fs, X_test_fs, fs = select_features(X_train, y_train, X_test)
# fit the model
model = LinearRegression()
model.fit(X_train_fs, y_train)
# evaluate the model
yhat = model.predict(X_test_fs)
# evaluate predictions
mae = mean_absolute_error(y_test, yhat)
print('MAE: %.3f' % mae)
# compare different numbers of features selected using mutual information
from sklearn.datasets import make_regression
from sklearn.model_selection import RepeatedKFold
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import mutual_info_regression
from sklearn.linear_model import LinearRegression
from sklearn.pipeline import Pipeline
from sklearn.model_selection import GridSearchCV
# define dataset
X, y = make_regression(n_samples=1000, n_features=100, n_informative=10, noise=0.1,
random_state=1)
# define the evaluation method
cv = RepeatedKFold(n_splits=10, n_repeats=3, random_state=1)
# define the pipeline to evaluate
model = LinearRegression()
fs = SelectKBest(score_func=mutual_info_regression)
pipeline = Pipeline(steps=[('sel',fs), ('lr', model)])
# define the grid
grid = dict()
grid['sel__k'] = [i for i in range(X.shape[1]-20, X.shape[1]+1)]
# define the grid search
search = GridSearchCV(pipeline, grid, scoring='neg_mean_absolute_error', n_jobs=-1, cv=cv)
# perform the search
results = search.fit(X, y)
# summarize best
print('Best MAE: %.3f' % results.best_score_)
print('Best Config: %s' % results.best_params_)
# summarize all
means = results.cv_results_['mean_test_score']
params = results.cv_results_['params']
for mean, param in zip(means, params):
print('>%.3f with: %r' % (mean, param))
# compare different numbers of features selected using mutual information
from numpy import mean
from numpy import std
from sklearn.datasets import make_regression
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedKFold
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import mutual_info_regression
from sklearn.linear_model import LinearRegression
from sklearn.pipeline import Pipeline
from matplotlib import pyplot
# define dataset
X, y = make_regression(n_samples=1000, n_features=100, n_informative=10, noise=0.1,
random_state=1)
# define number of features to evaluate
num_features = [i for i in range(X.shape[1]-19, X.shape[1]+1)]
# enumerate each number of features
results = list()
for k in num_features:
# create pipeline
model = LinearRegression()
fs = SelectKBest(score_func=mutual_info_regression, k=k)
pipeline = Pipeline(steps=[('sel',fs), ('lr', model)])
# evaluate the model
cv = RepeatedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(pipeline, X, y, scoring='neg_mean_absolute_error', cv=cv,
n_jobs=-1)
results.append(scores)
# summarize the results
print('>%d %.3f (%.3f)' % (k, mean(scores), std(scores)))
# plot model performance for comparison
pyplot.boxplot(results, labels=num_features, showmeans=True)
pyplot.show()
|
Select Features for Numerical Output.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# Dated: 30-3-18
#
# CNN Tutorial with adam and using Data Augmentation
#
# Dataset-- MNIST
#
# In this tutorial, i am going to show simple CNN network working with data augmentation and matplot display for images
#
# Data Augmentation is done for the purpose of creating dataset(mainly images more variety)...
#
# It can helpful for creating a dataset with much more richer images
#
# The model is trained on small amt of data only....
# Installing Dependencies
import keras.backend as K
K._backend='tensorflow' if K.backend() else print("Requires no change in backend")
#Basic Necessary libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# %matplotlib inline
np.random.seed(124) #for reproducibility purpose
# +
#Installing Main Libraries
from keras.datasets import mnist
from keras.models import Sequential
from keras.layers import Conv2D,MaxPooling2D,GlobalAveragePooling2D
from keras.layers import Dense,Dropout,Activation,Flatten
from keras.layers.normalization import BatchNormalization
from keras.layers.advanced_activations import LeakyReLU
from keras.optimizers import adam
from keras.utils import np_utils
from keras.preprocessing.image import ImageDataGenerator
# -
#Creating Training and Testing data
(trainX,trainY),(testX,testY)=mnist.load_data()
#Showing Mnist Image of 5 digit
plt.imshow(trainX[0],cmap='gray')
plt.xticks([])
plt.yticks([])
plt.title("Class: "+str(trainY[0]))
plt.show()
print(trainX[0].shape)
# +
#Preprocessing Input Data i.e X
trainX=trainX.reshape(trainX.shape[0],28,28,1)
testX=testX.reshape(testX.shape[0],28,28,1)
print(trainX.shape)
trainX=trainX.astype('float32')
testX=testX.astype('float32')
trainX/=255
testX/=255
print(trainX.shape,testX.shape,trainY.shape,testY.shape)
# -
#Preprocessing Output label i.e. Y
num_class=10
print(trainY.shape)
trainY=np_utils.to_categorical(trainY,num_class)
testY=np_utils.to_categorical(testY,num_class)
print(trainY.shape,testY.shape)
# ### Creating Convolution Architecture
# # 1.Convolution
# # 2.Pooling
# # 3.Dropout
# Repeat step 1,2,3 to add more convolution layers in network
#
# # 4.Fully Connected layer
# Repeat to add more feed forward layers
#
# # 5.Flatten
# # 6.Classify Sample
# Creating Architecture for CNN Model
# +
#Model Creation phase
model=Sequential()
model.add(Conv2D(32,(3,3),input_shape=(28,28,1)))
model.add(Activation('relu'))
BatchNormalization(axis=-1)
model.add(Conv2D(32,(3,3)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2,2)))
BatchNormalization(axis=-1)
model.add(Conv2D(64,(3,3)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2,2)))
model.add(Flatten())
#Creating Fully Connected Network
model.add(Dense(512))
model.add(Activation('relu'))
BatchNormalization(axis=-1)
model.add(Dense(256))
BatchNormalization(axis=-1)
model.add(Dropout(0.5))
model.add(Dense(10))
model.add(Activation('softmax'))
#compilation phase
#finding loss and backpropagating
model.compile(loss='categorical_crossentropy',optimizer="rmsprop",metrics=['accuracy'])
# -
model.summary()
# +
#Data Augmentation for creating rich datset
traingen = ImageDataGenerator(rotation_range=8, width_shift_range=0.08, shear_range=0.3,
height_shift_range=0.08, zoom_range=0.08) #for training data augmentation ....properties are defined
testgen=ImageDataGenerator() #for test data we don't need anytype of augmentation
# -
#defining Genrator training phase
train_generator=traingen.flow(trainX,trainY,batch_size=64)
test_generator=testgen.flow(testX,testY,batch_size=64)
# # Training Phase
# +
data=model.fit_generator(train_generator,steps_per_epoch=60000//200,
epochs=3,validation_data=test_generator,validation_steps=10000//200)
# -
#Prediciting Accuracy
score=model.evaluate(testX,testY)
print("Loss: {0} \t Accuracy: {1}%".format(round(score[0],5),round(score[1],3)*100))
|
CNN_MNIST-Part-1.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Underactuated cartpole control with iLQR, MPPI
# This example shows model predictive control to swing up the underactuated cartpole.
#
# 
#
# ## Problem
#
# **Model.** The "cartpole" is a free pendulum on a linear cart. The input to the system is a force on the cart $f$.
#
# $$
# m l \ddot{p} \cos(\theta) + m l^2 \ddot{\theta} - m g l \sin(\theta) = 0 \\
# (m + m_c) \ddot{p} + m l \ddot{\theta} \cos(\theta) - m l \dot{\theta}^2 \sin(\theta) = f
# $$
# where $\theta = 0$ is the angle of the pendulum when completely upright, and $p$ is the position of the cart. The state of the system is $x = (p, \theta, \dot{p}, \dot{\theta})^\top$. Parameters are the mass of the pendulum at the tip $m = 0.15$ kg, length of the pendulum $l = 0.75$ m, acceleration due to gravity $g = 9.8$ m/s , and the mass of the cart $m_c=1$ kg.
#
# **Control.** The objective is to move the cart so that the pendulum will stand up vertically. This is a classic controls and RL problem and is a typical benchmark test for new algorithms. We use the quadratic cost function
#
# $$
# J = \sum_{i=1}^{N} x_i^\top Q x_i + \sum_{i=1}^{N-1} r u_i^2
# $$
#
# where $Q = \text{diag}(1.25, 6, 12, 0.25)$ is chosen to drive the system states 0, and the penalty $r = 0.01$ tradeoffs the input magnitude.
#
# **Comparison to MPPI.** The video above shows iLQR which works well for this problem. Examining MPPI (sampling-based control) in the video below, the algorithm seems to find a control that achieves the desired swing up behavior and temporarily stabilizes the system, however if the algorithm is left to run long enough, the system destabilizes and the controller is unable to maintain the upright posture. This is believed due to a deficiency in the sampling approach as the noise in the control is amplified to attempt to maintain stability.
#
# 
# ## Example
#
# To run the example, build and install the C++ and Python libraries from the main README instructions. Start the docker container.
# ```
# # Run the experiment
# # cd /libsia/bin
# ./example-cartpole --datafile /libsia/data/cartpole.csv --algorithm ilqr
#
# # Run the python script
# # cd /libsia/examples/cartpole
# python cartpole.py --help
# python cartpole.py --datafile /libsia/data/cartpole.csv
# ```
# +
# This example imports data generated by the executable
from cartpole import plot_cartpole_trajectory
# This is the same as running the python script
plot_cartpole_trajectory(datafile="/libsia/data/cartpole.csv",
animate=False,
trace=True,
video_name="cartpole-animated.mp4",
dpi=150,
fps=30,
clean_axes=True)
# -
# ## Learning from expert demonstration
#
# [1] <NAME> paper
# [2] P. Owan thesis
#
# Assume now we don't know the cartpole model. We need to learn a model from data. We use GMR
# $$
# \mathbb{E}[x_{k+1}] = f(x_k, u_k)
# $$
#
# State dimension is too high, compress it.
# $$
# z = \xi(x)
# $$
#
# Instead we do regression on the reduced space
# $$
# z_{k+1} = \hat{f}(z_k, u_k)
# $$
#
# Recover the state by plugging in to model
# $$
# x_{k+1} = \xi^{-1}(\hat{f}(\xi(x_k), u_k))
# $$
# +
import subprocess
num_trials = 20
datafiles = ["/libsia/data/cartpole-{}.csv".format(i) for i in range(num_trials)]
# Peform n trials with an expert policy
for datafile in datafiles:
print("Running case {}".format(datafile))
subprocess.call(["/libsia/bin/example-cartpole",
"--measurement_noise",
"1e-6",
"--process_noise",
"1e-6",
"--datafile",
datafile])
# +
import pandas as pd
import numpy as np
from sklearn.decomposition import PCA
# Import plotting helpers
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_theme(style="whitegrid")
# Time step at which the data was collected
dt = 0.02
# Load data and perform some analysis
df = pd.DataFrame()
for datafile in datafiles:
df = df.append(pd.read_csv(datafile))
t = df["t"]
x = df[["p", "a", "v", "w"]].values
u = df["f"].values
# Run PCA on the state
n = 2
pca = PCA(n_components=n)
z = pca.fit_transform(x)
pca_pct = np.sum(pca.explained_variance_ratio_)
print("PCA with n={0:d} encodes {1:.4f}% of data".format(n, pca_pct))
# Stack the inputs and ouputs into X
uk = np.reshape(u[:-1], (len(u[:-1]), 1))
zk = z[:-1, :]
zkp1 = z[1:, :]
X = np.hstack((zk, uk, zkp1))
# Run GMM
import pysia as sia
gmm = sia.GMM(X.T, K=3, regularization=1e-6)
# Extract the means for visualization
means = np.zeros((gmm.numClusters(), gmm.dimension()))
for i in range(gmm.numClusters()):
means[i,:] = gmm.gaussian(i).mean()
# Run GMR to condition z_kp1 on zk, uk
gmr = sia.GMR(gmm, input_indices=[0, 1, 2], output_indices=[3, 4])
# +
# Plot the probabilities
f, ax = plt.subplots(nrows=n, ncols=n+1, figsize=(15, 8))
sns.despine(f, left=True, bottom=True)
for i in range(n):
for j in range(n):
ax[i, j].plot(z[:-1, j], z[1:, i], ".k", ms=1, label="Data")
ax[i, j].plot(means[:, j], means[:, i+3], ".r", ms=15, label="GMM")
ax[i, j].set_ylabel("z{}_kp1".format(i))
ax[i, j].set_xlabel("z{}_k".format(j))
ax[i, j].legend()
ax[i, n].plot(u[:-1], z[1:, i], ".k", ms=1)
ax[i, n].plot(means[:, 2], means[:, i+3], ".r", ms=15)
ax[i, n].set_ylabel("z{}_kp1".format(i))
ax[i, n].set_xlabel("u_k".format(j))
# +
# Plot the vector field given inputs u
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 12))
sns.despine(f, left=True, bottom=True)
ax.quiver(X[:, 0],
X[:, 1],
(X[:, 3]-X[:, 0])/dt,
(X[:, 4]-X[:, 1])/dt,
color='b',
headwidth=1.5)
Y = np.zeros((len(X), 2))
for i in range(len(X)):
Y[i, :] = gmr.predict(X[i,:3]).mean()
ax.quiver(X[:, 0],
X[:, 1],
(Y[:, 0]-X[:, 0])/dt,
(Y[:, 1]-X[:, 1])/dt,
color='r',
headwidth=1.5);
|
examples/cartpole/cartpole.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .jl
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Julia 0.5.0
# language: julia
# name: julia-0.5
# ---
include("lin_int2.jl")
println(readstring(`cmd /c type lin_int2.jl`))
# +
#=
Solving the optimal growth problem via value function iteration.
@author : <NAME> <<EMAIL>>
@date : 2014-07-05
References
----------
Simple port of the file quantecon.models.optgrowth
http://quant-econ.net/jl/dp_intro.html
=#
#=
This type defines the primitives representing the growth model. The
default values are
f(k) = k**alpha, i.e, Cobb-Douglas production function
u(c) = ln(c), i.e, log utility
See the constructor below for details
=#
using Plots
using Optim
"""
Neoclassical growth model
##### Fields
- `f::Function` : Production function
- `bet::Real` : Discount factor in (0, 1)
- `u::Function` : Utility function
- `grid_max::Int` : Maximum for grid over savings values
- `grid_size::Int` : Number of points in grid for savings values
- `grid::LinSpace{Float64}` : The grid for savings values
"""
type GrowthModel
f::Function
bet::Float64
u::Function
grid_max::Int
grid_size::Int
grid::LinSpace{Float64}
end
default_f(k) = k^0.65
default_u(c) = log(c)
"""
Constructor of `GrowthModel`
##### Arguments
- `f::Function(k->k^0.65)` : Production function
- `bet::Real(0.95)` : Discount factor in (0, 1)
- `u::Function(log)` : Utility function
- `grid_max::Int(2)` : Maximum for grid over savings values
- `grid_size::Int(150)` : Number of points in grid for savings values
"""
function GrowthModel(f=default_f, bet=0.95, u=default_u, grid_max=2,
grid_size=150)
grid = linspace(1e-6, grid_max, grid_size)
return GrowthModel(f, bet, u, grid_max, grid_size, grid)
end
"""
Apply the Bellman operator for a given model and initial value.
##### Arguments
- `g::GrowthModel` : Instance of `GrowthModel`
- `w::Vector`: Current guess for the value function
- `out::Vector` : Storage for output.
- `;ret_policy::Bool(false)`: Toggles return of value or policy functions
##### Returns
None, `out` is updated in place. If `ret_policy == true` out is filled with the
policy function, otherwise the value function is stored in `out`.
"""
function bellman_operator!(g::GrowthModel, w::Vector, out::Vector;
ret_policy::Bool=false)
# Apply linear interpolation to w
Aw = lin_inter(g.grid, w)
for (i, k) in enumerate(g.grid)
objective(c) = - g.u(c) - g.bet * Aw(g.f(k) - c)
res = optimize(objective, 1e-6, g.f(k))
c_star = res.minimum
if ret_policy
# set the policy equal to the optimal c
out[i] = c_star
else
# set Tw[i] equal to max_c { u(c) + beta w(f(k_i) - c)}
out[i] = - objective(c_star)
end
end
return out
end
function bellman_operator(g::GrowthModel, w::Vector;
ret_policy::Bool=false)
out = similar(w)
bellman_operator!(g, w, out, ret_policy=ret_policy)
end
"""
Extract the greedy policy (policy function) of the model.
##### Arguments
- `g::GrowthModel` : Instance of `GrowthModel`
- `w::Vector`: Current guess for the value function
- `out::Vector` : Storage for output
##### Returns
None, `out` is updated in place to hold the policy function
"""
function get_greedy!(g::GrowthModel, w::Vector, out::Vector)
bellman_operator!(g, w, out, ret_policy=true)
end
get_greedy(g::GrowthModel, w::Vector) = bellman_operator(g, w, ret_policy=true)
gm = GrowthModel()
alpha = 0.65
bet = gm.bet
grid_max = gm.grid_max
grid_size = gm.grid_size
grid = gm.grid
ab = alpha * gm.bet
c1 = (log(1 - ab) + log(ab) * ab / (1 - ab)) / (1 - gm.bet)
c2 = alpha / (1 - ab)
v_star(k) = c1 .+ c2 .* log(k)
function main(n::Int=35)
w_init = 5 .* log(grid) .- 25 # An initial condition -- fairly arbitrary
w = copy(w_init)
ws = []
colors = []
for i=1:n
w = bellman_operator(gm, w)
push!(ws, w)
push!(colors, RGBA(0, 0, 0, i/n))
end
p = plot(gm.grid, w_init, color=:green, linewidth=2, alpha=0.6,
label="initial condition")
plot!(gm.grid, ws, color=colors', label="", linewidth=2)
plot!(gm.grid, v_star(gm.grid), color=:blue, linewidth=2, alpha=0.8,
label="true value function")
plot!(ylims=(-40, -20), xlims=(minimum(gm.grid), maximum(gm.grid)))
return p
end
# -
main()
|
A Simple Optimal Growth Model Interpolation2.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="R3En9MVqilRY"
# # Train on multiple GPUs
#
# In this notebook, we will use Nobrainer to train a model for brain extraction. Brain extraction is a common step in processing neuroimaging data. It is a voxel-wise, binary classification task, where each voxel is classified as brain or not brain. Incidentally, the name for the Nobrainer framework comes from creating models for brain extraction.
#
# In the following cells, we will:
#
# 1. Get sample T1-weighted MR scans as features and FreeSurfer segmentations as labels.
# - We will binarize the FreeSurfer to get a precise brainmask.
# 2. Convert the data to TFRecords format.
# 3. Create two Datasets of the features and labels.
# - One dataset will be for training and the other will be for evaluation.
# 4. Instantiate a 3D convolutional neural network.
# 5. Choose a loss function and metrics to use.
# 6. Train on part of the data across multiple GPUs.
# 7. Evaluate on the rest of the data.
#
# ## Google Colaboratory
#
# If you are using Colab, please switch your runtime to GPU. To do this, select `Runtime > Change runtime type` in the top menu. Then select GPU under `Hardware accelerator`. A GPU is not necessary to prepare the data, but a GPU is helpful for training a model, which we demonstrate at the end of this notebook. This will give you access to one GPU, but the code will still run properly. To actually train a model on multiple GPUs, you will have to use Cloud services, a high-performance computing cluster, or your own hardware.
# + id="cxESnCBdiwEW"
# !pip install --no-cache-dir nobrainer nilearn
# + id="GetIT8J5ilRb"
import nobrainer
# + [markdown] id="nOL3zIrfilRc"
# # Get sample features and labels
#
# We use 9 pairs of volumes for training and 1 pair of volumes for evaluation. Many more volumes would be required to train a model for any useful purpose.
# + id="3gngTmxLilRc"
csv_of_filepaths = nobrainer.utils.get_data()
filepaths = nobrainer.io.read_csv(csv_of_filepaths)
train_paths = filepaths[:9]
evaluate_paths = filepaths[9:]
# + id="PzZnQFawi3zZ"
import matplotlib.pyplot as plt
from nilearn import plotting
fig = plt.figure(figsize=(12, 6))
plotting.plot_roi(train_paths[0][1], bg_img=train_paths[0][0], alpha=0.4, vmin=0, vmax=1, figure=fig)
# + [markdown] id="ksd2O-irilRd"
# # Convert medical images to TFRecords
#
# Remember how many full volumes are in the TFRecords files. This will be necessary to know how many steps are in on training epoch. The default training method needs to know this number, because Datasets don't always know how many items they contain.
# + id="74U_9sjqilRd"
# Verify that all volumes have the same shape and that labels are integer-ish.
invalid = nobrainer.io.verify_features_labels(train_paths, num_parallel_calls=2)
assert not invalid
invalid = nobrainer.io.verify_features_labels(evaluate_paths)
assert not invalid
# + id="Yvlr0TgcilRd"
# !mkdir -p data
# + id="fiUZIjuVilRe"
# Convert training and evaluation data to TFRecords.
nobrainer.tfrecord.write(
features_labels=train_paths,
filename_template='data/data-train_shard-{shard:03d}.tfrec',
examples_per_shard=3)
nobrainer.tfrecord.write(
features_labels=evaluate_paths,
filename_template='data/data-evaluate_shard-{shard:03d}.tfrec',
examples_per_shard=1)
# + id="iHtGGm3KilRe"
# !ls data
# + [markdown] id="e2oKUM8BilRe"
# # Create Datasets
#
# The batch is split evenly across the available GPUs. For example, if you have 4 GPUs and a batch size of 8, each GPU will get a batch of 2.
# + id="sCdkPNBxilRf"
n_classes = 1
batch_size = 2
volume_shape = (256, 256, 256)
block_shape = (128, 128, 128)
n_epochs = None
augment = False
shuffle_buffer_size = 10
num_parallel_calls = 2
# + id="SqoUkgpmilRf"
dataset_train = nobrainer.dataset.get_dataset(
file_pattern='data/data-train_shard-*.tfrec',
n_classes=n_classes,
batch_size=batch_size,
volume_shape=volume_shape,
block_shape=block_shape,
n_epochs=n_epochs,
augment=augment,
shuffle_buffer_size=shuffle_buffer_size,
num_parallel_calls=num_parallel_calls,
)
dataset_evaluate = nobrainer.dataset.get_dataset(
file_pattern='data/data-evaluate_shard-*.tfrec',
n_classes=n_classes,
batch_size=batch_size,
volume_shape=volume_shape,
block_shape=block_shape,
n_epochs=1,
augment=False,
shuffle_buffer_size=None,
num_parallel_calls=1,
)
# + id="BZlKggChilRg"
dataset_train
# + id="PjF_jDTVilRg"
dataset_evaluate
# + id="ZeRc0YQ5ilRg"
# Get the steps for an epoch of training and an epoch of validation.
steps_per_epoch = nobrainer.dataset.get_steps_per_epoch(
n_volumes=len(train_paths),
volume_shape=volume_shape,
block_shape=block_shape,
batch_size=batch_size)
validation_steps = nobrainer.dataset.get_steps_per_epoch(
n_volumes=len(evaluate_paths),
volume_shape=volume_shape,
block_shape=block_shape,
batch_size=batch_size)
# + [markdown] id="i9aAVkBVilRg"
# # Instantiate and compile model within scope
# + id="chyWsFIdilRh"
import tensorflow as tf
# + id="k8yg94NQilRh"
strategy = tf.distribute.MirroredStrategy()
# + id="LekCIjc9ilRh"
optimizer = tf.keras.optimizers.Adam(learning_rate=1e-04)
with strategy.scope():
model = nobrainer.models.unet(
n_classes=n_classes,
input_shape=(*block_shape, 1),
batchnorm=True)
model.compile(
optimizer=optimizer,
loss=nobrainer.losses.jaccard,
metrics=[nobrainer.metrics.dice])
# + id="zALpc6SNilRh"
model.fit(
dataset_train,
epochs=5,
steps_per_epoch=steps_per_epoch,
validation_data=dataset_evaluate,
validation_steps=validation_steps)
# + [markdown] id="L4n8idapilRh"
# # Predict medical images without TFRecords
# + id="dO-yhzWmilRi"
from nobrainer.volume import standardize
import nibabel as nib
image_path = evaluate_paths[0][0]
out = nobrainer.prediction.predict_from_filepath(image_path,
model,
block_shape = block_shape,
batch_size = batch_size,
normalizer = standardize,
)
out.shape
# + id="FDpD7oT9jJ14"
fig = plt.figure(figsize=(12, 6))
plotting.plot_roi(out, bg_img=image_path, alpha=0.4, figure=fig)
# + id="SgrzFoBvj_Ze"
|
guide/train_on_multiple_gpus.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Simple KubeFlow Pipeline
#
# Lightweight python components do not require you to build a new container image for every code change.
# They're intended to use for fast iteration in notebook environment.
#
# #### Building a lightweight python component
# To build a component just define a stand-alone python function and then call kfp.components.func_to_container_op(func) to convert it to a component that can be used in a pipeline.
#
# There are several requirements for the function:
# * The function should be stand-alone. It should not use any code declared outside of the function definition. Any imports should be added inside the main function. Any helper functions should also be defined inside the main function.
# * The function can only import packages that are available in the base image. If you need to import a package that's not available you can try to find a container image that already includes the required packages. (As a workaround you can use the module subprocess to run pip install for the required package.)
# * If the function operates on numbers, the parameters need to have type hints. Supported types are ```[int, float, bool]```. Everything else is passed as string.
# * To build a component with multiple output values, use the typing.NamedTuple type hint syntax: ```NamedTuple('MyFunctionOutputs', [('output_name_1', type), ('output_name_2', float)])```
# + tags=["parameters"]
# Install the KubeFlow Pipeline SDK
# !pip3 install https://storage.googleapis.com/ml-pipeline/release/0.1.16/kfp.tar.gz --upgrade
# -
# Simple function that just add two numbers:
#Define a Python function
def add_fn(a: float, b: float) -> float:
'''Calculates sum of two arguments'''
return a + b
# Convert the function to a pipeline operation
# +
import kfp.components as comp
add_op = comp.func_to_container_op(add_fn)
# -
# A bit more advanced function which demonstrates how to use imports, helper functions and produce multiple outputs.
from typing import NamedTuple
def div_fn(dividend: float, divisor:float, output_dir:str = './') -> NamedTuple('DivOutput', [('quotient', float), ('remainder', float)]):
'''Divides two numbers and calculate the quotient and remainder'''
#Imports inside a component function:
import numpy as np
#This function demonstrates how to use nested functions inside a component function:
def nested_div_helper(dividend, divisor):
return np.divmod(dividend, divisor)
(quotient, remainder) = nested_div_helper(dividend, divisor)
from tensorflow.python.lib.io import file_io
import json
# Exports two sample metrics:
metrics = {
'metrics': [{
'name': 'quotient',
'numberValue': float(quotient),
},{
'name': 'remainder',
'numberValue': float(remainder),
}]}
with file_io.FileIO(output_dir + 'mlpipeline-metrics.json', 'w') as f:
json.dump(metrics, f)
from collections import namedtuple
output = namedtuple('DivOutput', ['quotient', 'remainder'])
return output(quotient, remainder)
# Test running the python function directly
div_fn(100, 7)
# #### Convert the function to a pipeline operation
#
# You can specify an alternative base container image (the image needs to have Python 3.5+ installed).
div_op = comp.func_to_container_op(div_fn, base_image='tensorflow/tensorflow:1.11.0-py3')
# #### Define the pipeline
# Pipeline function has to be decorated with the `@dsl.pipeline` decorator
import kfp.dsl as dsl
@dsl.pipeline(
name='Calculation pipeline',
description='A toy pipeline that performs arithmetic calculations.'
)
def add_div_pipeline(
a='a',
b='7',
c='17',
):
#Passing pipeline parameter and a constant value as operation arguments
add_task = add_op(a, 4) #Returns a dsl.ContainerOp class instance.
#Passing a task output reference as operation arguments
#For an operation with a single return value, the output reference can be accessed using `task.output` or `task.outputs['output_name']` syntax
div_task = div_op(add_task.output, b, '/')
#For an operation with a multiple return values, the output references can be accessed using `task.outputs['output_name']` syntax
result_task = add_op(div_task.outputs['quotient'], c)
# #### Compile the pipeline
pipeline_func = add_div_pipeline
pipeline_filename = pipeline_func.__name__ + '.pipeline.tar.gz'
import kfp.compiler as compiler
compiler.Compiler().compile(pipeline_func, pipeline_filename)
# #### Submit the pipeline for execution
# +
#Specify pipeline argument values
arguments = {'a': '7', 'b': '8'}
#Get or create an experiment and submit a pipeline run
import kfp
client = kfp.Client()
experiment = client.create_experiment('simple_add_div_pipeline')
#Submit a pipeline run
run_name = pipeline_func.__name__ + ' run'
run_result = client.run_pipeline(experiment.id, run_name, pipeline_filename, arguments)
# -
|
kubeflow/notebooks/08_Simple_KubeFlow_ML_Pipeline.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # Hospital Readmissions Data Analysis and Recommendations for Reduction
#
# ### Background
# In October 2012, the US government's Center for Medicare and Medicaid Services (CMS) began reducing Medicare payments for Inpatient Prospective Payment System hospitals with excess readmissions. Excess readmissions are measured by a ratio, by dividing a hospital’s number of “predicted” 30-day readmissions for heart attack, heart failure, and pneumonia by the number that would be “expected,” based on an average hospital with similar patients. A ratio greater than 1 indicates excess readmissions.
#
# ### Exercise Directions
#
# In this exercise, you will:
# + critique a preliminary analysis of readmissions data and recommendations (provided below) for reducing the readmissions rate
# + construct a statistically sound analysis and make recommendations of your own
#
# More instructions provided below. Include your work **in this notebook and submit to your Github account**.
#
# ### Resources
# + Data source: https://data.medicare.gov/Hospital-Compare/Hospital-Readmission-Reduction/9n3s-kdb3
# + More information: http://www.cms.gov/Medicare/medicare-fee-for-service-payment/acuteinpatientPPS/readmissions-reduction-program.html
# + Markdown syntax: http://nestacms.com/docs/creating-content/markdown-cheat-sheet
# ****
# +
# %matplotlib inline
import pandas as pd
from __future__ import division
import numpy as np
import matplotlib.pyplot as plt
import bokeh.plotting as bkp
from mpl_toolkits.axes_grid1 import make_axes_locatable
# -
# read in readmissions data provided
hospital_read_df = pd.read_csv('data/cms_hospital_readmissions.csv')
# ****
# ## Preliminary Analysis
# deal with missing and inconvenient portions of data
clean_hospital_read_df = hospital_read_df[hospital_read_df['Number of Discharges'] != 'Not Available']
clean_hospital_read_df.loc[:, 'Number of Discharges'] = clean_hospital_read_df['Number of Discharges'].astype(int)
clean_hospital_read_df = clean_hospital_read_df.sort_values('Number of Discharges')
# +
# generate a scatterplot for number of discharges vs. excess rate of readmissions
# lists work better with matplotlib scatterplot function
x = [a for a in clean_hospital_read_df['Number of Discharges'][81:-3]]
y = list(clean_hospital_read_df['Excess Readmission Ratio'][81:-3])
fig, ax = plt.subplots(figsize=(8,5))
ax.scatter(x, y,alpha=0.2)
ax.fill_between([0,350], 1.15, 2, facecolor='red', alpha = .15, interpolate=True)
ax.fill_between([800,2500], .5, .95, facecolor='green', alpha = .15, interpolate=True)
ax.set_xlim([0, max(x)])
ax.set_xlabel('Number of discharges', fontsize=12)
ax.set_ylabel('Excess rate of readmissions', fontsize=12)
ax.set_title('Scatterplot of number of discharges vs. excess rate of readmissions', fontsize=14)
ax.grid(True)
fig.tight_layout()
# -
# ****
#
# ## Preliminary Report
#
# Read the following results/report. While you are reading it, think about if the conclusions are correct, incorrect, misleading or unfounded. Think about what you would change or what additional analyses you would perform.
#
# **A. Initial observations based on the plot above**
# + Overall, rate of readmissions is trending down with increasing number of discharges
# + With lower number of discharges, there is a greater incidence of excess rate of readmissions (area shaded red)
# + With higher number of discharges, there is a greater incidence of lower rates of readmissions (area shaded green)
#
# **B. Statistics**
# + In hospitals/facilities with number of discharges < 100, mean excess readmission rate is 1.023 and 63% have excess readmission rate greater than 1
# + In hospitals/facilities with number of discharges > 1000, mean excess readmission rate is 0.978 and 44% have excess readmission rate greater than 1
#
# **C. Conclusions**
# + There is a significant correlation between hospital capacity (number of discharges) and readmission rates.
# + Smaller hospitals/facilities may be lacking necessary resources to ensure quality care and prevent complications that lead to readmissions.
#
# **D. Regulatory policy recommendations**
# + Hospitals/facilties with small capacity (< 300) should be required to demonstrate upgraded resource allocation for quality care to continue operation.
# + Directives and incentives should be provided for consolidation of hospitals and facilities to have a smaller number of them with higher capacity and number of discharges.
# +
# A. Do you agree with the above analysis and recommendations? Why or why not?
import seaborn as sns
relevant_columns = clean_hospital_read_df[['Excess Readmission Ratio', 'Number of Discharges']][81:-3]
sns.regplot(relevant_columns['Number of Discharges'], relevant_columns['Excess Readmission Ratio'])
# -
# ****
# <div class="span5 alert alert-info">
# ### Exercise
#
# Include your work on the following **in this notebook and submit to your Github account**.
#
# A. Do you agree with the above analysis and recommendations? Why or why not?
#
# B. Provide support for your arguments and your own recommendations with a statistically sound analysis:
#
# 1. Setup an appropriate hypothesis test.
# 2. Compute and report the observed significance value (or p-value).
# 3. Report statistical significance for $\alpha$ = .01.
# 4. Discuss statistical significance and practical significance. Do they differ here? How does this change your recommendation to the client?
# 5. Look at the scatterplot above.
# - What are the advantages and disadvantages of using this plot to convey information?
# - Construct another plot that conveys the same information in a more direct manner.
#
#
#
# You can compose in notebook cells using Markdown:
# + In the control panel at the top, choose Cell > Cell Type > Markdown
# + Markdown syntax: http://nestacms.com/docs/creating-content/markdown-cheat-sheet
# </div>
# ****
# - Overall, rate of readmissions is trending down with increasing number of discharges
# - Agree, according to regression trend line shown above
#
# - With lower number of discharges, there is a greater incidence of excess rate of readmissions (area shaded red)
# - Agree
#
# - With higher number of discharges, there is a greater incidence of lower rates of readmissions (area shaded green)
# - Agree
# +
rv =relevant_columns
print rv[rv['Number of Discharges'] < 100][['Excess Readmission Ratio']].mean()
print '\nPercent of subset with excess readmission rate > 1: ', len(rv[(rv['Number of Discharges'] < 100) & (rv['Excess Readmission Ratio'] > 1)]) / len(rv[relevant_columns['Number of Discharges'] < 100])
print '\n', rv[rv['Number of Discharges'] > 1000][['Excess Readmission Ratio']].mean()
print '\nPercent of subset with excess readmission rate > 1: ', len(rv[(rv['Number of Discharges'] > 1000) & (rv['Excess Readmission Ratio'] > 1)]) / len(rv[relevant_columns['Number of Discharges'] > 1000])
# -
# - In hospitals/facilities with number of discharges < 100, mean excess readmission rate is 1.023 and 63% have excess readmission rate greater than 1
# - Accurate
#
# - In hospitals/facilities with number of discharges > 1000, mean excess readmission rate is 0.978 and 44% have excess readmission rate greater than 1
# - Correction: mean excess readmission rate is 0.979, and 44.565% have excess readmission rate > 1
np.corrcoef(rv['Number of Discharges'], rv['Excess Readmission Ratio'])
# - There is a significant correlation between hospital capacity (number of discharges) and readmission rates.
# - The correlation coefficient shows a very, very weak correlation between the two variables. More evidence needed to establish a correlation.
# - Smaller hospitals/facilities may be lacking necessary resources to ensure quality care and prevent complications that lead to readmissions.
# - More evidence needed to asses the veracity of this statement.
|
Statistics_Exercises/sliderule_dsi_inferential_statistics_exercise_3.ipynb
|
# ---
# title: "Feedforward Neural Networks For Regression"
# author: "<NAME>"
# date: 2017-12-20T11:53:49-07:00
# description: "How to train a feed-forward neural network for regression in Python."
# type: technical_note
# draft: false
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Preliminaries
# +
# Load libraries
import numpy as np
from keras.preprocessing.text import Tokenizer
from keras import models
from keras import layers
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
# Set random seed
np.random.seed(0)
# -
# ## Generate Training Data
# +
# Generate features matrix and target vector
features, target = make_regression(n_samples = 10000,
n_features = 3,
n_informative = 3,
n_targets = 1,
noise = 0.0,
random_state = 0)
# Divide our data into training and test sets
train_features, test_features, train_target, test_target = train_test_split(features,
target,
test_size=0.33,
random_state=0)
# -
# ## Create Neural Network Architecture
# +
# Start neural network
network = models.Sequential()
# Add fully connected layer with a ReLU activation function
network.add(layers.Dense(units=32, activation='relu', input_shape=(train_features.shape[1],)))
# Add fully connected layer with a ReLU activation function
network.add(layers.Dense(units=32, activation='relu'))
# Add fully connected layer with no activation function
network.add(layers.Dense(units=1))
# -
# ## Compile Neural Network
#
# Because we are training a regression, we should use an appropriate loss function and evaluation metric, in our case the mean square error:
#
# $$\operatorname {MSE}={\frac {1}{n}}\sum\_{{i=1}}^{n}({\hat {y\_{i}}}-y\_{i})^{2}$$
#
# where $n$ is the number of observations, $y\_{i}$ is the true value of the target we are trying to predict, $y$, for observation $i$, and ${\hat {y\_{i}}}$ is the model's predicted value for $y\_{i}$.
# Compile neural network
network.compile(loss='mse', # Mean squared error
optimizer='RMSprop', # Optimization algorithm
metrics=['mse']) # Mean squared error
# ## Train Neural Network
# Train neural network
history = network.fit(train_features, # Features
train_target, # Target vector
epochs=10, # Number of epochs
verbose=0, # No output
batch_size=100, # Number of observations per batch
validation_data=(test_features, test_target)) # Data for evaluation
|
docs/deep_learning/keras/feedforward_neural_network_for_regression.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Indexes
#
# Today we're going to be talking about pandas' [`Index`es](http://pandas.pydata.org/pandas-docs/version/0.18.0/api.html#index).
# They're essential to pandas, but can be a difficult concept to grasp at first.
# I suspect this is partly because they're unlike what you'll find in SQL or R.
#
# `Index`es offer
#
# - a metadata container
# - easy label-based row selection and assignment
# - easy label-based alignment in operations
#
# One of my first tasks when analyzing a new dataset is to identify a unique identifier for each observation, and set that as the index. It could be a simple integer, or like in our first chapter, it could be several columns (`carrier`, `origin` `dest`, `tail_num` `date`).
#
# To demonstrate the benefits of proper `Index` use, we'll first fetch some weather data from sensors at a bunch of airports across the US.
# See [here](https://github.com/akrherz/iem/blob/master/scripts/asos/iem_scraper_example.py) for the example scraper I based this off of.
# Those uninterested in the details of fetching and prepping the data and [skip past it](#set-operations).
#
# At a high level, here's how we'll fetch the data: the sensors are broken up by "network" (states).
# We'll make one API call per state to get the list of airport IDs per network (using `get_ids` below).
# Once we have the IDs, we'll again make one call per state getting the actual observations (in `get_weather`).
# Feel free to skim the code below, I'll highlight the interesting bits.
#
# +
# %matplotlib inline
import os
import json
import glob
import datetime
from io import StringIO
import requests
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import prep
sns.set_style('ticks')
# States are broken into networks. The networks have a list of ids, each representing a station.
# We will take that list of ids and pass them as query parameters to the URL we built up ealier.
states = """AK AL AR AZ CA CO CT DE FL GA HI IA ID IL IN KS KY LA MA MD ME
MI MN MO MS MT NC ND NE NH NJ NM NV NY OH OK OR PA RI SC SD TN TX UT VA VT
WA WI WV WY""".split()
# IEM has Iowa AWOS sites in its own labeled network
networks = ['AWOS'] + ['{}_ASOS'.format(state) for state in states]
# -
def get_weather(stations, start=pd.Timestamp('2014-01-01'),
end=pd.Timestamp('2014-01-31')):
'''
Fetch weather data from MESONet between ``start`` and ``stop``.
'''
url = ("http://mesonet.agron.iastate.edu/cgi-bin/request/asos.py?"
"&data=tmpf&data=relh&data=sped&data=mslp&data=p01i&data=v"
"sby&data=gust_mph&data=skyc1&data=skyc2&data=skyc3"
"&tz=Etc/UTC&format=comma&latlon=no"
"&{start:year1=%Y&month1=%m&day1=%d}"
"&{end:year2=%Y&month2=%m&day2=%d}&{stations}")
stations = "&".join("station=%s" % s for s in stations)
weather = (pd.read_csv(url.format(start=start, end=end, stations=stations),
comment="#")
.rename(columns={"valid": "date"})
.rename(columns=str.strip)
.assign(date=lambda df: pd.to_datetime(df['date']))
.set_index(["station", "date"])
.sort_index())
float_cols = ['tmpf', 'relh', 'sped', 'mslp', 'p01i', 'vsby', "gust_mph"]
weather[float_cols] = weather[float_cols].apply(pd.to_numeric, errors="corce")
return weather
def get_ids(network):
url = "http://mesonet.agron.iastate.edu/geojson/network.php?network={}"
r = requests.get(url.format(network))
md = pd.io.json.json_normalize(r.json()['features'])
md['network'] = network
return md
# There isn't too much in `get_weather` worth mentioning, just grabbing some CSV files from various URLs.
# They put metadata in the "CSV"s at the top of the file as lines prefixed by a `#`.
# Pandas will ignore these with the `comment='#'` parameter.
#
# I do want to talk briefly about the gem of a method that is [`json_normalize`](http://pandas.pydata.org/pandas-docs/version/0.18.0/generated/pandas.io.json.json_normalize.html) .
# The weather API returns some slightly-nested data.
# +
url = "http://mesonet.agron.iastate.edu/geojson/network.php?network={}"
r = requests.get(url.format("AWOS"))
js = r.json()
js['features'][:2]
# -
# If we just pass that list off to the `DataFrame` constructor, we get this.
pd.DataFrame(js['features']).head()
# In general, DataFrames don't handle nested data that well.
# It's often better to normalize it somehow.
# In this case, we can "lift"
# the nested items (`geometry.coordinates`, `properties.sid`, and `properties.sname`)
# up to the top level.
pd.io.json.json_normalize(js['features'])
# Sure, it's not *that* difficult to write a quick for loop or list comprehension to extract those,
# but that gets tedious.
# If we were using the latitude and longitude data, we would want to split
# the `geometry.coordinates` column into two. But we aren't so we won't.
#
# Going back to the task, we get the airport IDs for every network (state)
# with `get_ids`. Then we pass those IDs into `get_weather` to fetch the
# actual weather data.
# +
import os
ids = pd.concat([get_ids(network) for network in networks], ignore_index=True)
gr = ids.groupby('network')
store = 'data/weather.h5'
if not os.path.exists(store):
os.makedirs("data/weather", exist_ok=True)
for k, v in gr:
weather = get_weather(v['id'])
weather.to_csv("data/weather/{}.csv".format(k))
weather = pd.concat([
pd.read_csv(f, parse_dates=['date'], index_col=['station', 'date'])
for f in glob.glob('data/weather/*.csv')
]).sort_index()
weather.to_hdf("data/weather.h5", "weather")
else:
weather = pd.read_hdf("data/weather.h5", "weather")
# -
weather.head()
# OK, that was a bit of work. Here's a plot to reward ourselves.
# +
airports = ['W43', 'AFO', '82V', 'DUB']
weather.sort_index(inplace=True)g = sns.FacetGrid(weather.loc[airports].reset_index(),
col='station', hue='station', col_wrap=2, size=4)
g.map(sns.regplot, 'sped', 'gust_mph')
# -
# ## Set Operations
#
# Indexes are set-like (technically *multi*sets, since you can have duplicates), so they support most python `set` operations. Since indexes are immutable you won't find any of the inplace `set` operations.
# One other difference is that since `Index`es are also array-like, you can't use some infix operators like `-` for `difference`. If you have a numeric index it is unclear whether you intend to perform math operations or set operations.
# You can use `&` for intersection, `|` for union, and `^` for symmetric difference though, since there's no ambiguity.
#
# For example, lets find the set of airports that we have both weather and flight information on. Since `weather` had a MultiIndex of `airport, datetime`, we'll use the `levels` attribute to get at the airport data, separate from the date data.
# +
# Bring in the flights data
flights = pd.read_hdf('data/flights.h5', 'flights')
weather_locs = weather.index.levels[0]
# The `categories` attribute of a Categorical is an Index
origin_locs = flights.origin.cat.categories
dest_locs = flights.dest.cat.categories
airports = weather_locs & origin_locs & dest_locs
airports
# +
print("Weather, no flights:\n\t", weather_locs.difference(origin_locs | dest_locs), end='\n\n')
print("Flights, no weather:\n\t", (origin_locs | dest_locs).difference(weather_locs), end='\n\n')
print("Dropped Stations:\n\t", (origin_locs | dest_locs) ^ weather_locs)
# -
# ## Flavors
#
# Pandas has many subclasses of the regular `Index`, each tailored to a specific kind of data.
# Most of the time these will be created for you automatically, so you don't have to worry about which one to choose.
#
# 1. [`Index`](http://pandas.pydata.org/pandas-docs/version/0.18.0/generated/pandas.Index.html#pandas.Index)
# 2. `Int64Index`
# 3. `RangeIndex`: Memory-saving special case of `Int64Index`
# 4. `FloatIndex`
# 5. `DatetimeIndex`: Datetime64[ns] precision data
# 6. `PeriodIndex`: Regularly-spaced, arbitrary precision datetime data.
# 7. `TimedeltaIndex`
# 8. `CategoricalIndex`
# 9. `MultiIndex`
#
# You will sometimes create a `DatetimeIndex` with [`pd.date_range`](http://pandas.pydata.org/pandas-docs/version/0.18.0/generated/pandas.date_range.html) ([`pd.period_range`](http://pandas.pydata.org/pandas-docs/version/0.18.0/generated/pandas.period_range.html) for `PeriodIndex`).
# And you'll sometimes make a `MultiIndex` directly too (I'll have an example of this in my post on performace).
#
# Some of these specialized index types are purely optimizations; others use information about the data to provide additional methods.
# And while you might occasionally work with indexes directly (like the set operations above), most of they time you'll be operating on a Series or DataFrame, which in turn makes use of its Index.
#
# ### Row Slicing
# We saw in part one that they're great for making *row* subsetting as easy as column subsetting.
weather.loc['DSM'].head()
# Without indexes we'd probably resort to boolean masks.
weather2 = weather.reset_index()
weather2[weather2['station'] == 'DSM'].head()
# Slightly less convenient, but still doable.
# ### Indexes for Easier Arithmetic, Analysis
# It's nice to have your metadata (labels on each observation) next to you actual values. But if you store them in an array, they'll get in the way of your operations.
# Say we wanted to translate the Fahrenheit temperature to Celsius.
# +
# With indecies
temp = weather['tmpf']
c = (temp - 32) * 5 / 9
c.to_frame()
# +
# without
temp2 = weather.reset_index()[['station', 'date', 'tmpf']]
temp2['tmpf'] = (temp2['tmpf'] - 32) * 5 / 9
temp2.head()
# -
# Again, not terrible, but not as good.
# And, what if you had wanted to keep Fahrenheit around as well, instead of overwriting it like we did?
# Then you'd need to make a copy of everything, including the `station` and `date` columns.
# We don't have that problem, since indexes are immutable and safely shared between DataFrames / Series.
temp.index is c.index
# ### Indexes for Alignment
#
# I've saved the best for last.
# Automatic alignment, or reindexing, is fundamental to pandas.
#
# All binary operations (add, multiply, etc.) between Series/DataFrames first *align* and then proceed.
#
# Let's suppose we have hourly observations on temperature and windspeed.
# And suppose some of the observations were invalid, and not reported (simulated below by sampling from the full dataset). We'll assume the missing windspeed observations were potentially different from the missing temperature observations.
# +
dsm = weather.loc['DSM']
hourly = dsm.resample('H').mean()
temp = hourly['tmpf'].sample(frac=.5, random_state=1).sort_index()
sped = hourly['sped'].sample(frac=.5, random_state=2).sort_index()
# -
temp.head().to_frame()
sped.head()
# Notice that the two indexes aren't identical.
#
# Suppose that the `windspeed : temperature` ratio is meaningful.
# When we go to compute that, pandas will automatically align the two by index label.
sped / temp
# This lets you focus on doing the operation, rather than manually aligning things, ensuring that the arrays are the same length and in the same order.
# By deault, missing values are inserted where the two don't align.
# You can use the method version of any binary operation to specify a `fill_value`
sped.div(temp, fill_value=1)
# And since I couldn't find anywhere else to put it, you can control the axis the operation is aligned along as well.
hourly.div(sped, axis='index')
# The non row-labeled version of this is messy.
# +
temp2 = temp.reset_index()
sped2 = sped.reset_index()
# Find rows where the operation is defined
common_dates = pd.Index(temp2.date) & sped2.date
pd.concat([
# concat to not lose date information
sped2.loc[sped2['date'].isin(common_dates), 'date'],
(sped2.loc[sped2.date.isin(common_dates), 'sped'] /
temp2.loc[temp2.date.isin(common_dates), 'tmpf'])],
axis=1).dropna(how='all')
# -
# And we have a bug in there. Can you spot it?
# I only grabbed the dates from `sped2` in the line `sped2.loc[sped2['date'].isin(common_dates), 'date']`.
# Really that should be `sped2.loc[sped2.date.isin(common_dates)] | temp2.loc[temp2.date.isin(common_dates)]`.
# But I think leaving the buggy version states my case even more strongly. The `temp / sped` version where pandas aligns everything is better.
# ## Merging
#
# There are two ways of merging DataFrames / Series in pandas.
#
# 1. Relational Database style with `pd.merge`
# 2. Array style with `pd.concat`
#
# Personally, I think in terms of the `concat` style.
# I learned pandas before I ever really used SQL, so it comes more naturally to me I suppose.
#
# ### Concat Version
pd.concat([temp, sped], axis=1).head()
# The `axis` parameter controls how the data should be stacked, `0` for vertically, `1` for horizontally.
# The `join` parameter controls the merge behavior on the shared axis, (the Index for `axis=1`). By default it's like a union of the two indexes, or an outer join.
pd.concat([temp, sped], axis=1, join='inner')
# ### Merge Version
#
# Since we're joining by index here the merge version is quite similar.
# We'll see an example later of a one-to-many join where the two differ.
pd.merge(temp.to_frame(), sped.to_frame(), left_index=True, right_index=True).head()
pd.merge(temp.to_frame(), sped.to_frame(), left_index=True, right_index=True,
how='outer').head()
# Like I said, I typically prefer `concat` to `merge`.
# The exception here is one-to-many type joins. Let's walk through one of those,
# where we join the flight data to the weather data.
# To focus just on the merge, we'll aggregate hour weather data to be daily, rather than trying to find the closest recorded weather observation to each departure (you could do that, but it's not the focus right now). We'll then join the one `(airport, date)` record to the many `(airport, date, flight)` records.
#
# Quick tangent, to get the weather data to daily frequency, we'll need to resample (more on that in the timeseries section). The resample essentially splits the recorded values into daily buckets and computes the aggregation function on each bucket. The only wrinkle is that we have to resample *by station*, so we'll use the `pd.TimeGrouper` helper.
# +
idx_cols = ['unique_carrier', 'origin', 'dest', 'tail_num', 'fl_num', 'fl_date']
data_cols = ['crs_dep_time', 'dep_delay', 'crs_arr_time', 'arr_delay',
'taxi_out', 'taxi_in', 'wheels_off', 'wheels_on', 'distance']
df = flights.set_index(idx_cols)[data_cols].sort_index()
# +
def mode(x):
'''
Arbitrarily break ties.
'''
return x.value_counts().index[0]
aggfuncs = {'tmpf': 'mean', 'relh': 'mean',
'sped': 'mean', 'mslp': 'mean',
'p01i': 'mean', 'vsby': 'mean',
'gust_mph': 'mean', 'skyc1': mode,
'skyc2': mode, 'skyc3': mode}
# TimeGrouper works on a DatetimeIndex, so we move `station` to the
# columns and then groupby it as well.
daily = (weather.reset_index(level="station")
.groupby([pd.TimeGrouper('1d'), "station"])
.agg(aggfuncs))
daily.head()
# -
# Now that we have daily flight and weather data, we can merge.
# We'll use the `on` keyword to indicate the columns we'll merge on (this is like a `USING (...)` SQL statement), we just have to make sure the names align.
# ### The merge version
# +
m = pd.merge(flights, daily.reset_index().rename(columns={'date': 'fl_date', 'station': 'origin'}),
on=['fl_date', 'origin']).set_index(idx_cols).sort_index()
m.head()
# -
# Since data-wrangling on its own is never the goal, let's do some quick analysis.
# Seaborn makes it easy to explore bivariate relationships.
m.sample(n=10000).pipe((sns.jointplot, 'data'), 'sped', 'dep_delay');
# Looking at the various [sky coverage states](https://en.wikipedia.org/wiki/METAR#Cloud_reporting):
#
#
m.groupby('skyc1').dep_delay.agg(['mean', 'count']).sort_values(by='mean')
import statsmodels.api as sm
# Statsmodels (via [patsy](http://patsy.readthedocs.org/) can automatically convert dummy data to dummy variables in a formula with the `C` function).
mod = sm.OLS.from_formula('dep_delay ~ C(skyc1) + distance + tmpf + relh + sped + mslp', data=m)
res = mod.fit()
res.summary()
fig, ax = plt.subplots()
ax.scatter(res.fittedvalues, res.resid, color='k', marker='.', alpha=.25)
ax.set(xlabel='Predicted', ylabel='Residual')
sns.despine()
# Those residuals should look like white noise.
# Looks like our linear model isn't flexible enough to model the delays,
# but I think that's enough for now.
#
# ---
#
# We'll talk more about indexes in the Tidy Data and Reshaping section.
# [Let me know](http://twitter.com/tomaugspurger) if you have any feedback.
# Thanks for reading!
|
modern_3_indexes.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# ---
bres_trend_micro,bres_trend_agg = [make_trend(bres_proc,sector_var=v) for v in ['sector','sector_aggregated']]
# +
fig,ax = plt.subplots()
plot_trend(bres_trend_agg,ax=ax)
ax.legend(bbox_to_anchor=(1,1))
# +
fig,ax = plt.subplots()
plot_trend(bres_trend_micro,ax=ax)
ax.legend(bbox_to_anchor=(1,1))
# +
fig,ax = plt.subplots()
plot_bar(bres_trend_micro,ax,norm=True)
ax.legend(bbox_to_anchor=(1,1))
# -
# ### b. Geographies
lad_bres_shares_09,lad_bres_shares_17 = [make_lorenz(bres_proc,y=year) for year in [2009,2017]]
# +
fig,ax = plt.subplots(figsize=(5,6),sharey=True)
plot_lorenz(lad_bres_shares_17,ax)
ax.legend(bbox_to_anchor=(1,1))
# -
plot__histo_lorenz(lad_bres_shares_09,lad_bres_shares_17)
# ### Some maps
plot_kwargs = {'scheme':'Fisher_Jenks','cmap':'viridis','edgecolor':'grey','linewidth':0,'legend':True}
# +
fig,ax = plt.subplots(figsize=(14,10),ncols=2)
year_comp(bres_proc,'sector_aggregated','journalism',ax=ax,**plot_kwargs)
plt.tight_layout()
# +
fig,ax = plt.subplots(figsize=(14,10),ncols=2)
sect_comp(bres_proc,'sector_aggregated',['other','journalism'],ax=ax,**plot_kwargs)
# +
fig,ax = plt.subplots(figsize=(21,10),ncols=3)
sect_comp(bres_proc,'sector',['publishing_newspapers','web_portals','computer_programming'],ax=ax,**plot_kwargs)
# +
fig,ax = plt.subplots(figsize=(14,10),ncols=2)
sect_comp(bres_proc,'sector',['tv_programming_broadcasting','radio_broadcasting'],ax=ax,**plot_kwargs)
# -
# +
fig,ax = plt.subplots(figsize=(12,7),nrows=2,sharex=True,gridspec_kw={'height_ratios':[3,1]})
sectors = ['artificial_intelligence','advertising','creative_content','news_high','public_news']
for n,s in enumerate(sectors):
(100*pd.crosstab(cb['year'],cb[s]>0.75,normalize=0)).loc[np.arange(2000,2019)][True].rolling(window=3).mean().dropna().plot(
ax=ax[0],color=colors[n],linewidth=3 if 'news' in s else 1)
ax[0].set_ylabel('% of all companies')
ax[0].legend(sectors,bbox_to_anchor=(1,1))
news = cb.loc[cb['news_high']==True]
(100*pd.crosstab(news['year'],news['public_news']>0.75,normalize=0)).loc[np.arange(2000,2019)][True].rolling(window=3).mean().dropna().plot(
ax=ax[1],color='blue',linewidth=3)
ax[1].set_ylabel('% of all \n news companies')
plt.tight_layout()
plt.savefig('../../reports/figures/research_slides/cb/activity_trends.pdf')
# -
100*pd.crosstab(cb['year'],cb['public_news']>0.75,normalize=0).loc[np.arange(2000,2019)][True][2018]
# #### Evolution of funding?
#
# We get the CB funding data and match it with companies
#
# That will allow us to get levels of funding and funders for various sources
# #### Analysis
# Next steps:
# * Create dummies for news, public interest news and AI and look at trends and actors
rel_sets = [set(cb.loc[cb[s]>0.75]['id']) for s in ['artificial_intelligence','advertising','creative_content','news_high','public_news']]
# +
cb_fr_df['ai'],cb_fr_df['advertising'],cb_fr_df['creative_content'],cb_fr_df['news'],cb_fr_df['pi_news'] = [
[x in one_set for x in cb_fr_df['company_id']] for one_set in rel_sets]
cb_fr_df['any_sector'] = 1
rel_sectors = ['ai','advertising',
'creative_content',
'news','pi_news','any_sector']
# +
ax = cb_fr_df.groupby('year')[rel_sectors[:-1]].sum().loc[np.arange(2000,2019)].rolling(window=3).mean().dropna().plot(color=colors)
ax.set_ylabel('Number of deals')
# -
# Totals raised
# +
fig,ax = plt.subplots(figsize=(12,7),nrows=2,sharex=True,gridspec_kw={'height_ratios':[3,1]})
total_raised = pd.concat([cb_fr_df.loc[cb_fr_df[s]==True].groupby('year')['raised_amount_usd'].sum() for s in rel_sectors],axis=1).fillna(0)/1e9
total_raised.columns = rel_sectors
total_raised.loc[np.arange(2000,2020),rel_sectors[:-1]].rolling(window=3).mean().dropna().plot(color=colors,ax=ax[0])
ax[0].set_ylabel('$ Billion')
news_funding= cb_fr_df.loc[cb_fr_df['news']==True]
(100*news_funding.groupby(['year','pi_news'])['raised_amount_usd'].sum().reset_index(drop=False).pivot(
index='year',columns='pi_news',values='raised_amount_usd').apply(lambda x: x/x.sum(),axis=1).loc[np.arange(2000,2019)].fillna(0).rolling(
window=3).mean()).dropna()[True].plot(color='blue',ax=ax[1],linewidth=3)
ax[1].set_ylabel('PI news as \n % of all news')
plt.tight_layout()
plt.savefig('../../reports/figures/research_slides/cb/funding_trends.pdf')
# -
|
notebooks/dev/scraps.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] pycharm={}
# # Classification with Delira and SciKit-Learn - A very short introduction
# *Author: <NAME>*
#
# *Date: 31.07.2019*
#
# This Example shows how to set up a basic classification model and experiment using SciKit-Learn.
#
# Let's first setup the essential hyperparameters. We will use `delira`'s `Parameters`-class for this:
# + pycharm={"is_executing": false}
logger = None
from delira.training import Parameters
import sklearn
params = Parameters(fixed_params={
"model": {},
"training": {
"batch_size": 64, # batchsize to use
"num_epochs": 10, # number of epochs to train
"optimizer_cls": None, # optimization algorithm to use
"optimizer_params": {}, # initialization parameters for this algorithm
"losses": {}, # the loss function
"lr_sched_cls": None, # the learning rate scheduling algorithm to use
"lr_sched_params": {}, # the corresponding initialization parameters
"metrics": {"mae": mean_absolute_error} # and some evaluation metrics
}
})
# + [markdown] pycharm={}
# Since we did not specify any metric, only the `CrossEntropyLoss` will be calculated for each batch. Since we have a classification task, this should be sufficient. We will train our network with a batchsize of 64 by using `Adam` as optimizer of choice.
#
# ## Logging and Visualization
# To get a visualization of our results, we should monitor them somehow. For logging we will use `Tensorboard`. Per default the logging directory will be the same as our experiment directory.
# + [markdown] pycharm={}
#
# ## Data Preparation
# ### Loading
# Next we will create some fake data. For this we use the `ClassificationFakeData`-Dataset, which is already implemented in `deliravision`. To avoid getting the exact same data from both datasets, we use a random offset.
# + pycharm={"is_executing": false}
from deliravision.data.fakedata import ClassificationFakeData
dataset_train = ClassificationFakeData(num_samples=10000,
img_size=(3, 32, 32),
num_classes=10)
dataset_val = ClassificationFakeData(num_samples=1000,
img_size=(3, 32, 32),
num_classes=10,
rng_offset=10001
)
# + [markdown] pycharm={}
# ### Augmentation
# For Data-Augmentation we will apply a few transformations:
# + pycharm={"is_executing": false}
from batchgenerators.transforms import RandomCropTransform, \
ContrastAugmentationTransform, Compose
from batchgenerators.transforms.spatial_transforms import ResizeTransform
from batchgenerators.transforms.sample_normalization_transforms import MeanStdNormalizationTransform
transforms = Compose([
RandomCropTransform(24), # Perform Random Crops of Size 24 x 24 pixels
ResizeTransform(32), # Resample these crops back to 32 x 32 pixels
ContrastAugmentationTransform(), # randomly adjust contrast
MeanStdNormalizationTransform(mean=[0.5], std=[0.5])])
# + [markdown] pycharm={}
# With these transformations we can now wrap our datasets into datamanagers:
# + pycharm={"is_executing": false}
from delira.data_loading import DataManager, SequentialSampler, RandomSampler
manager_train = DataManager(dataset_train, params.nested_get("batch_size"),
transforms=transforms,
sampler_cls=RandomSampler,
n_process_augmentation=4)
manager_val = DataManager(dataset_val, params.nested_get("batch_size"),
transforms=transforms,
sampler_cls=SequentialSampler,
n_process_augmentation=4)
# + [markdown] pycharm={}
# ## Model
#
# After we have done that, we can specify our model: We will use a very simple MultiLayer Perceptron here.
# In opposite to other backends, we don't need to provide a custom implementation of our model, but we can simply use it as-is. It will be automatically wrapped by `SklearnEstimator`, which can be subclassed for more advanced usage.
#
# ## Training
# Now that we have defined our network, we can finally specify our experiment and run it.
# + pycharm={"is_executing": true}
import warnings
warnings.simplefilter("ignore", UserWarning) # ignore UserWarnings raised by dependency code
warnings.simplefilter("ignore", FutureWarning) # ignore FutureWarnings raised by dependency code
from sklearn.neural_network import MLPClassifier
from delira.training import SklearnExperiment
if logger is not None:
logger.info("Init Experiment")
experiment = PyTorchExperiment(params, MLPClassifier,
name="ClassificationExample",
save_path="./tmp/delira_Experiments",
key_mapping={"X": "X"}
gpu_ids=[0])
experiment.save()
model = experiment.run(manager_train, manager_val)
# + [markdown] pycharm={}
# Congratulations, you have now trained your first Classification Model using `delira`, we will now predict a few samples from the testset to show, that the networks predictions are valid (for now, this is done manually, but we also have a `Predictor` class to automate stuff like this):
# + pycharm={}
import numpy as np
from tqdm.auto import tqdm # utility for progress bars
preds, labels = [], []
with torch.no_grad():
for i in tqdm(range(len(dataset_val))):
img = dataset_val[i]["data"] # get image from current batch
img_tensor = img.astype(np.float) # create a tensor from image, push it to device and add batch dimension
pred_tensor = model(img_tensor) # feed it through the network
pred = pred_tensor.argmax(1).item() # get index with maximum class confidence
label = np.asscalar(dataset_val[i]["label"]) # get label from batch
if i % 1000 == 0:
print("Prediction: %d \t label: %d" % (pred, label)) # print result
preds.append(pred)
labels.append(label)
# calculate accuracy
accuracy = (np.asarray(preds) == np.asarray(labels)).sum() / len(preds)
print("Accuracy: %.3f" % accuracy)
|
notebooks/classification_examples/sklearn.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Creating a Filter, Edge Detection
# ### Import resources and display image
# +
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import cv2
import numpy as np
# %matplotlib inline
# Read in the image
image = mpimg.imread('data/curved_lane.jpg')
plt.imshow(image)
# -
# ### Convert the image to grayscale
# +
# Convert to grayscale for filtering
gray = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)
plt.imshow(gray, cmap='gray')
# -
# ### TODO: Create a custom kernel
#
# Below, you've been given one common type of edge detection filter: a Sobel operator.
#
# The Sobel filter is very commonly used in edge detection and in finding patterns in intensity in an image. Applying a Sobel filter to an image is a way of **taking (an approximation) of the derivative of the image** in the x or y direction, separately. The operators look as follows.
#
# <img src="notebook_ims/sobel_ops.png" width=200 height=200>
#
# **It's up to you to create a Sobel x operator and apply it to the given image.**
#
# For a challenge, see if you can put the image through a series of filters: first one that blurs the image (takes an average of pixels), and then one that detects the edges.
# +
# Create a custom kernel
# 3x3 array for edge detection
sobel_y = np.array([[ -1, -2, -1],
[ 0, 0, 0],
[ 1, 2, 1]])
# Filter the image using filter2D, which has inputs: (grayscale image, bit-depth, kernel)
filtered_image = cv2.filter2D(gray, -1, sobel_y)
plt.imshow(filtered_image, cmap='gray')
# +
## TODO: Create and apply a Sobel x operator
sobel_x = np.array([[-1,0,1],[-2,0,2],[-1,0,1]])
filtered_image2 = cv2.filter2D(gray,-1,sobel_x)
plt.imshow(filtered_image2,cmap='gray')
# -
filtered_image_both = cv2.filter2D(filtered_image,-1,sobel_x)
plt.imshow(filtered_image_both,cmap='gray')
# ### Test out other filters!
#
# You're encouraged to create other kinds of filters and apply them to see what happens! As an **optional exercise**, try the following:
# * Create a filter with decimal value weights.
# * Create a 5x5 filter
# * Apply your filters to the other images in the `images` directory.
#
#
custom_filter = np.array([[-2,-2,-2,-2,-2],
[-1,-1,-1,-1,-1],
[0,0,0,0,0],
[1,1,1,1,1],
[2,2,2,2,2]])
custom_filtered_image = cv2.filter2D(gray,-1,custom_filter)
plt.imshow(custom_filtered_image, cmap='gray')
|
convolutional-neural-networks/conv-visualization/custom_filters.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # Chapter 2: Multi-armed Bandits Solutions
# ## 1. Exercise 2.1
# In ε-greedy action selection, for the case of two actions and $\epsilon=0.5$, what is the probability that the greedy action is selected?
#
# ### Solution
# - Greedy Action is $a$, Non-greedy Action is $b$
# - $p(b)=0.5 * \epsilon = 0.25$
# - $p(a)=1-p(b)=0.75$
# ## 2. Exercise 2.2: Bandit example
# Consider a *k-armed* bandit problem with $k = 4$ actions, denoted $1, 2, 3$, and $4$. Consider applying to this problem a bandit algorithm using ε-greedy action selection, sample-average action-value estimates, and initial estimates of $Q_1(a) = 0$, for all $a$. Suppose the initial sequence of actions and rewards is $A_1 = 1, R_1 =1,A_2 =2,R_2 =1,A_3 =2,R_3 =2,A_4 =2,R_4 =2,A_5 =3,R_5 =0$. On some of these time steps the $\epsilon$ case may have occurred, causing an action to be selected at random. On which time steps did this definitely occur? On which time steps could this possibly have occurred?
#
# ### Solution
# - $\epsilon$ case is definitely occurred when $a \neq \arg\max\limits_aQ_t(a)$, other for possibly have occorred
# - Definitely occurred: $t\in\{2, 5\}$
# - Possibly occurred: $t\in\{1, 3, 4\}$
# ## 3. Exercise 2.3
# In the comparison shown in Figure 2.2, which method will perform best in the long run in terms of cumulative reward and probability of selecting the best action? How much better will it be? Express your answer quantitatively.
#
# 
#
# ### Solution
#
# ## 4. Exercise 2.4
# If the step-size parameters, $\alpha_n$, are not constant, then the estimate $Q_n$ is a weighted average of previously received rewards with a weighting different from that given by (2.6). What is the weighting on each prior reward for the general case, analogous to (2.6), in terms of the sequence of step-size parameters?
#
# $$Q_{n+1}=(1-\alpha)^nQ_1+\sum_{i=1}^n \alpha(1-\alpha)^{n-i}R_i ~~~, (2.6)$$
#
# ### Solution
# $$
# \begin{aligned}
# Q_{n+1} &= Q_n + \alpha_n\big[R_n-Q_n\big]
# \\ &= \alpha_nR_n + (1-\alpha_n)Q_n
# \\ &= \alpha_nR_n + (1-\alpha_n)\big[\alpha_{n-1}R_{n-1} + (1-\alpha_{n-1})Q_{n-1}\big]
# \\ &= \alpha_nR_n + (1-\alpha_n)\alpha_{n-1}R_{n-1} + (1-\alpha_n)(1-\alpha_{n-1})Q_{n-1}
# \\ &= \alpha_nR_n + (1-\alpha_n)\alpha_{n-1}R_{n-1} + (1-\alpha_n)(1-\alpha_{n-1})Q_{n-1} + ... + R_1\alpha_1\prod_{i=2}^n(1-\alpha_i) + Q_1\prod_{i=1}^n(1-\alpha_i)
# \\ &= Q_1\prod_{i=1}^n(1-\alpha_i) + \sum_{i=1}^nR_i\alpha_i\prod_{j=i+1}^n(1-\alpha_j)
# \end{aligned}
# $$
# ## 5. Exercise 2.5 (programming)
# Design and conduct an experiment to demonstrate the difficulties that sample-average methods have for nonstationary problems. Use a modified version of the 10-armed testbed in which all the $q_∗(a)$ start out equal and then take independent random walks (say by adding a normally distributed increment with mean zero and standard deviation $0.01$ to all the $q_∗(a)$ on each step). Prepare plots like Figure 2.2 for an action-value method using sample averages, incrementally computed, and another action-value method using a constant step-size parameter, $α = 0.1$. Use $ε = 0.1$ and longer runs, say of $10,000$ steps.
#
# ### Code
# +
# %matplotlib inline
from __future__ import print_function
import matplotlib.pyplot as plt
import numpy as np
class Bandit:
def __init__(self, kArm=10, randWalkStdDeviation=0.01, epsilon=0.1, alpha=0.1):
self.kArm = kArm
self.randWalkStdDeviation = randWalkStdDeviation
self.epsilon = epsilon
self.alpha = alpha
self.qTrue = np.zeros(self.kArm)
self.qEst = np.zeros(self.kArm)
self.actCnt = np.zeros(self.kArm)
def getOptimalAction(self):
return np.argmax(self.qTrue)
def selectAction(self):
# random walks for all qTrue
for k in range(0, self.kArm):
self.qTrue[k] += self.randWalkStdDeviation * np.random.randn()
# select an action
action = 0
if np.random.binomial(1, self.epsilon) == 1:
action = np.random.choice(self.kArm)
else:
action = np.argmax(self.qEst)
# get reward
reward = np.random.randn() + self.qTrue[action]
# estimate Q
stepSize = self.alpha
if stepSize == 0:
self.actCnt[action] += 1
stepSize = 1.0 / self.actCnt[action]
self.qEst[action] += stepSize * (reward - self.qEst[action])
return action, reward
def run(nBandits=2000, time=10000, epsilon=0.1, alphas=[]):
optimalActions = [np.zeros(time, dtype='float') for _ in range(0, len(alphas))]
averageRewards = [np.zeros(time, dtype='float') for _ in range(0, len(alphas))]
for idx, alpha in enumerate(alphas):
totalOptimalAction = 0
totalReward = 0
for _ in range(0, nBandits):
bandit = Bandit(epsilon=epsilon, alpha=alpha)
for t in range(0, time):
action, reward = bandit.selectAction()
averageRewards[idx][t] += reward
if action == bandit.getOptimalAction():
optimalActions[idx][t] += 1
# get average
optimalActions[idx] /= nBandits
averageRewards[idx] /= nBandits
return optimalActions, averageRewards
alphas = [0, 0.1]
results = run(alphas=alphas)
for idx, result in enumerate(results):
plt.figure(figsize=(8,4),dpi=100)
for alpha, data in zip(alphas, result):
plt.plot(data, label='alpha = '+str(alpha))
plt.xlabel('Steps')
plt.ylabel('% optimal action' if idx == 0 else 'average reward')
plt.legend()
plt.show()
# -
# ## 6. Exercise 2.6: Mysterious Spikes
# The results shown in Figure 2.3 should be quite reliable because they are averages over $2,000$ individual, randomly chosen 10-armed bandit tasks. Why, then, are there oscillations and spikes in the early part of the curve for the optimistic method? In other words, what might make this method perform particularly better or worse, on average, on particular early steps?
#
# ### Solution
# Explore until $Q_1$, so optimal action may not be selected usually.
# ## 7. Exercise 2.7: Unbiased Constant-Step-Size Trick
# In most of this chapter we have used sample averages to estimate action values because sample averages do not produce the initial bias that constant step sizes do (see the analysis leading to (2.6)). However, sample averages are not a completely satisfactory solution because they may perform poorly on nonstationary problems. Is it possible to avoid the bias of constant step sizes while retaining their advantages on nonstationary problems? One way is to use a step size of
# $$\beta_n = \dfrac{\alpha}{\overline o_n}~~~ (2.8)$$
# to process the nth reward for a particular action, where $α > 0$ is a conventional constant
# step size, and o ̄n is a trace of one that starts at $0$:
# $$\overline o_n=\overline o_{n-1}+\alpha(1-\overline o_{n-1}), \text{for } n\geq 0, \text{with } \overline o_0=0 ~~~(2.9)$$
# Carry out an analysis like that in (2.6) to show that $Q_n$ is an exponential recency-weighted average *without initial bias*.
#
# ### Solution
# With step-size $\beta_n$, we have:
# $$Q_{n+1} = Q_1\prod_{i=1}^n(1-\beta_i) + \sum_{i=1}^nR_i\beta_i\prod_{j=i+1}^n(1-\beta_j)$$
#
# Let check the $\prod_{i=1}^n(1-\beta_i)$ term. Replace, $\beta_i$ by $\frac{\alpha}{\overline o_i}$:
# $$
# \begin{aligned}
# \prod_{i=1}^n(1-\beta_i) &= \prod_{i=1}^n\Big(1-\frac{\alpha}{\overline o_i}\Big)
# \\ &= \prod_{i=1}^n\frac{\overline o_i-\alpha}{\overline o_i}
# \\ &= \prod_{i=1}^n\frac{(1-\alpha)\overline o_{i-1}}{\overline o_i}
# \\ &= \frac{(1-\alpha)\overline o_0}{\overline o_1}\prod_{i=2}^n\frac{(1-\alpha)\overline o_{i-1}}{\overline o_i}
# \end{aligned}
# $$
#
# Because of $\overline o_0=0$, so:
# $$\prod_{i=1}^n(1-\beta_i) = 0$$
#
# In other word, $Q_n$ is an exponential recency-weighted average without initial bias $Q_1$.
# ### 8. Exercise 2.8: UCB Spikes
# In Figure 2.4 the UCB algorithm shows a distinct spike in performance on the 11th step. Why is this? Note that for your answer to be fully satisfactory it must explain both why the reward increases on the 11th step and why it decreases on the subsequent steps. Hint: if $c = 1$, then the spike is less prominent.
#
# ### Solution
# ## 9. Exercise 2.9
# Show that in the case of two actions, the soft-max distribution is the same as that given by the logistic, or sigmoid, function often used in statistics and artificial neural networks.
#
# ### Solution
# - 2 action's preferences: $H_1, H_2$
# - Probability of $H_1$:
# $$
# \begin{aligned}
# p(H_1) &= \dfrac{e^{H_1}}{e^{H_1}+e^{H_2}}
# \\ &= \dfrac{1}{1+e^{H_2-H_1}}
# \\ &= \dfrac{1}{1+e^{H}}
# \end{aligned}
# $$
# - Probability of $H_2$:
# $$
# \begin{aligned}
# p(H_2) &= \dfrac{e^{H_2}}{e^{H_1}+e^{H_2}}
# \\ &= \dfrac{e^{H_2-H_1}}{1+e^{H_2-H_1}}
# \\ &= \dfrac{e^{H}}{1+e^{H}}
# \end{aligned}
# $$
# - That is `sigmoid` form
# ## 10. Exercise 2.10
# Suppose you face a 2-armed bandit task whose true action values change randomly from time step to time step. Specifically, suppose that, for any time step, the true values of actions 1 and 2 are respectively 0.1 and 0.2 with probability 0.5 (case A), and 0.9 and 0.8 with probability 0.5 (case B). If you are not able to tell which case you face at any step, what is the best expectation of success you can achieve and how should you behave to achieve it? Now suppose that on each step you are told whether you are facing case A or case B (although you still don’t know the true action values). This is an associative search task. What is the best expectation of success you can achieve in this task, and how should you behave to achieve it?
#
# ### Solution
#
# ## 11. Exercise 2.11 (programming)
# Make a figure analogous to Figure 2.6 for the nonstationary case outlined in Exercise 2.5. Include the constant-step-size ε-greedy algorithm with $\alpha=0.1$. Use runs of $200,000$ steps and, as a performance measure for each algorithm and parameter setting, use the average reward over the last $100,000$ steps.
#
# ### Code
|
chap2-solutions.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="ElXOa7R7g37i" colab_type="text"
# # Tutorial Part 5: Putting Multitask Learning to Work
#
# This notebook walks through the creation of multitask models on MUV [1]. The goal is to demonstrate that multitask methods outperform singletask methods on MUV.
#
# ## Colab
#
# This tutorial and the rest in this sequence are designed to be done in Google colab. If you'd like to open this notebook in colab, you can use the following link.
#
# [](https://colab.research.google.com/github/deepchem/deepchem/blob/master/examples/tutorials/05_Putting_Multitask_Learning_to_Work.ipynb)
#
#
# ## Setup
#
# To run DeepChem within Colab, you'll need to run the following cell of installation commands. This will take about 5 minutes to run to completion and install your environment.
# + id="Fc_4bSWJg37l" colab_type="code" outputId="d6d577c7-aa9e-4db1-8bb2-6269f2817012" colab={"base_uri": "https://localhost:8080/", "height": 462}
# %tensorflow_version 1.x
# !curl -Lo deepchem_installer.py https://raw.githubusercontent.com/deepchem/deepchem/master/scripts/colab_install.py
import deepchem_installer
# %time deepchem_installer.install(version='2.3.0')
# + [markdown] id="9Ow2nQtZg37p" colab_type="text"
# The MUV dataset is a challenging benchmark in molecular design that consists of 17 different "targets" where there are only a few "active" compounds per target. The goal of working with this dataset is to make a machine learnign model which achieves high accuracy on held-out compounds at predicting activity. To get started, let's download the MUV dataset for us to play with.
# + id="FGi-ZEfSg37q" colab_type="code" outputId="1ac2c36b-66b0-4c57-bf4b-114a7425b85e" colab={"base_uri": "https://localhost:8080/", "height": 85}
import os
import deepchem as dc
current_dir = os.path.dirname(os.path.realpath("__file__"))
dataset_file = "medium_muv.csv.gz"
full_dataset_file = "muv.csv.gz"
# We use a small version of MUV to make online rendering of notebooks easy. Replace with full_dataset_file
# In order to run the full version of this notebook
dc.utils.download_url("https://s3-us-west-1.amazonaws.com/deepchem.io/datasets/%s" % dataset_file,
current_dir)
dataset = dc.utils.save.load_from_disk(dataset_file)
print("Columns of dataset: %s" % str(dataset.columns.values))
print("Number of examples in dataset: %s" % str(dataset.shape[0]))
# + [markdown] id="c9t912ODg37u" colab_type="text"
# Now, let's visualize some compounds from our dataset
# + id="KobfUjlWg37v" colab_type="code" outputId="01025d0f-3fb1-485e-bb93-82f2b3e062f9" colab={"base_uri": "https://localhost:8080/", "height": 1000}
from rdkit import Chem
from rdkit.Chem import Draw
from itertools import islice
from IPython.display import Image, display, HTML
def display_images(filenames):
"""Helper to pretty-print images."""
for filename in filenames:
display(Image(filename))
def mols_to_pngs(mols, basename="test"):
"""Helper to write RDKit mols to png files."""
filenames = []
for i, mol in enumerate(mols):
filename = "MUV_%s%d.png" % (basename, i)
Draw.MolToFile(mol, filename)
filenames.append(filename)
return filenames
num_to_display = 12
molecules = []
for _, data in islice(dataset.iterrows(), num_to_display):
molecules.append(Chem.MolFromSmiles(data["smiles"]))
display_images(mols_to_pngs(molecules))
# + [markdown] id="kDUrLw8Mg37y" colab_type="text"
# There are 17 datasets total in MUV as we mentioned previously. We're going to train a multitask model that attempts to build a joint model to predict activity across all 17 datasets simultaneously. There's some evidence [2] that multitask training creates more robust models.
#
# As fair warning, from my experience, this effect can be quite fragile. Nonetheless, it's a tool worth trying given how easy DeepChem makes it to build these models. To get started towards building our actual model, let's first featurize our data.
# + id="eqEQiNDpg37z" colab_type="code" outputId="e1b919ac-1bb3-4224-ff91-65d2e3d16f3b" colab={"base_uri": "https://localhost:8080/", "height": 357}
MUV_tasks = ['MUV-692', 'MUV-689', 'MUV-846', 'MUV-859', 'MUV-644',
'MUV-548', 'MUV-852', 'MUV-600', 'MUV-810', 'MUV-712',
'MUV-737', 'MUV-858', 'MUV-713', 'MUV-733', 'MUV-652',
'MUV-466', 'MUV-832']
featurizer = dc.feat.CircularFingerprint(size=1024)
loader = dc.data.CSVLoader(
tasks=MUV_tasks, smiles_field="smiles",
featurizer=featurizer)
dataset = loader.featurize(dataset_file)
# + [markdown] id="QQfINH2Ag371" colab_type="text"
# We'll now want to split our dataset into training, validation, and test sets. We're going to do a simple random split using `dc.splits.RandomSplitter`. It's worth noting that this will provide overestimates of real generalizability! For better real world estimates of prospective performance, you'll want to use a harder splitter.
# + id="-f03zjeIg372" colab_type="code" outputId="5472a51a-42e9-43bc-e73e-d947ae3c6a33" colab={"base_uri": "https://localhost:8080/", "height": 136}
splitter = dc.splits.RandomSplitter(dataset_file)
train_dataset, valid_dataset, test_dataset = splitter.train_valid_test_split(
dataset)
#NOTE THE RENAMING:
valid_dataset, test_dataset = test_dataset, valid_dataset
# + [markdown] id="6nRCpb08g375" colab_type="text"
# Let's now get started building some models! We'll do some simple hyperparameter searching to build a robust model.
# + id="BvfbTbsEg376" colab_type="code" outputId="9f96de90-ad90-4492-cced-0f5e74dcacb6" colab={"base_uri": "https://localhost:8080/", "height": 853}
import numpy as np
import numpy.random
params_dict = {"activation": ["relu"],
"momentum": [.9],
"batch_size": [50],
"init": ["glorot_uniform"],
"data_shape": [train_dataset.get_data_shape()],
"learning_rate": [1e-3],
"decay": [1e-6],
"nb_epoch": [1],
"nesterov": [False],
"dropouts": [(.5,)],
"nb_layers": [1],
"batchnorm": [False],
"layer_sizes": [(1000,)],
"weight_init_stddevs": [(.1,)],
"bias_init_consts": [(1.,)],
"penalty": [0.],
}
n_features = train_dataset.get_data_shape()[0]
def model_builder(model_params, model_dir):
model = dc.models.MultitaskClassifier(
len(MUV_tasks), n_features, **model_params)
return model
metric = dc.metrics.Metric(dc.metrics.roc_auc_score, np.mean)
optimizer = dc.hyper.HyperparamOpt(model_builder)
best_dnn, best_hyperparams, all_results = optimizer.hyperparam_search(
params_dict, train_dataset, valid_dataset, [], metric)
# + [markdown] id="QhZAgZ9gg379" colab_type="text"
# # Congratulations! Time to join the Community!
#
# Congratulations on completing this tutorial notebook! If you enjoyed working through the tutorial, and want to continue working with DeepChem, we encourage you to finish the rest of the tutorials in this series. You can also help the DeepChem community in the following ways:
#
# ## Star DeepChem on [GitHub](https://github.com/deepchem/deepchem)
# This helps build awareness of the DeepChem project and the tools for open source drug discovery that we're trying to build.
#
# ## Join the DeepChem Gitter
# The DeepChem [Gitter](https://gitter.im/deepchem/Lobby) hosts a number of scientists, developers, and enthusiasts interested in deep learning for the life sciences. Join the conversation!
#
# # Bibliography
#
# [1] https://pubs.acs.org/doi/10.1021/ci8002649
#
# [2] https://pubs.acs.org/doi/abs/10.1021/acs.jcim.7b00146
|
examples/tutorials/05_Putting_Multitask_Learning_to_Work.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Assignment 5
# - toc: true
# - badges: true
# - comments: true
# - categories: [jupyter]
# Train a deep learning model to classify beetles, cockroaches and dragonflies using [images](https://www.dropbox.com/s/fn73sj2e6c9rhf6/insects.zip?dl=0)
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim
import torchvision
import torchvision.transforms as transforms
import torch.optim as optim
import matplotlib.pyplot as plt
import numpy as np
import shap
# First load the images from the website. The size of the images are different so first we need to resize the images to the same size. We set the batch size = 100
train_root = 'insects/train/'
test_root = 'insects/test/'
transform = transforms.Compose(
[transforms.Resize([256, 256]),
transforms.ToTensor(),
transforms.Normalize((0.5,), (0.5,)),])
train_dataset = torchvision.datasets.ImageFolder(train_root, transform = transform)
test_dataset = torchvision.datasets.ImageFolder(test_root, transform = transform)
train_loader = torch.utils.data.DataLoader(train_dataset, batch_size=100, shuffle=True, num_workers = 0)
test_loader = torch.utils.data.DataLoader(test_dataset, batch_size=100, shuffle=False, num_workers = 0)
# Considering that it is an image classcification problem, Convolutional Neural Network is usually used to train the model. A typical CNN model looks like below:
#
# 
#
# Source: https://towardsdatascience.com/a-comprehensive-guide-to-convolutional-neural-networks-the-eli5-way-3bd2b1164a53
# The Convolutional neural network usually has several components:
# #### Define the Convolutional layer:
#
# ```torch.nn.Conv2d(in_channels, out_channels, kernel_size, stride=1, padding=0, dilation=1, bias=True)```
#
# The convolution operation is shown as below:
# 
#
# Source: https://anhreynolds.com/blogs/cnn.html
#
# *in_channels*: depends on whether the image is black and white(in_channels = 1) or colored(in_channels = 3)
#
# *kernal_size*: the dimension of the kernal used in the convolution operation.In the example, the kernal size is (3,3)
#
# *stride*: Stride of the convolution
#
# 
#
# In this case, the stride is 2.
#
# *padding*: Padding added to all four sides of the input
#
# 
# In this case, the padding is 1.
#
# Source: https://deepai.org/machine-learning-glossary-and-terms/padding
# #### Define the Activation function:
#
# There are many types of activation function, most famous ones are *sigmoid* function, *relu* function, *tanh* function:
# - sigmoid:
# $f(x)= \frac{1}{1+e^{-x}}$
#
# 
#
# - Relu:
# $f(x) = max(0, x)$
# 
#
# - Tanh:
# $f(x) = \frac{e^x - e^{-x}}{e^x + e^{-x}}$
# 
#
# #### Define the Dropout layer:
#
# Dropout is often used to avoid overfitting. It simply sets a propotional nodes to 0. Commonly chosen rate is 0.5.
#
# #### Define the Pooling layer:
# Common pooling methods are max pooling.
# 
#
# Source: https://medium.com/@duanenielsen/deep-learning-cage-match-max-pooling-vs-convolutions-e42581387cb9
# #### Define the fully connected layer:
# ```torch.nn.Linear(in_features, out_features, bias=True, device=None, dtype=None)```
class CNN_Model(nn.Module):
def __init__(self):
super(CNN_Model, self).__init__()
self.layer_1 = nn.Sequential(
nn.Conv2d(in_channels=3, out_channels=32, kernel_size=3, padding=1),
nn.ReLU(),
nn.MaxPool2d(kernel_size=4, stride=4)
)
self.layer_2 = nn.Sequential(
nn.Conv2d(in_channels=32, out_channels= 64, kernel_size=3,padding = 1),
nn.ReLU(),
nn.MaxPool2d(kernel_size=4,stride=4)
)
self.fc1 = nn.Linear(in_features=64*16*16, out_features=500)
self.fc2 = nn.Linear(in_features=500, out_features=120)
self.fc3 = nn.Linear(in_features=120, out_features=3)
def forward(self, x):
out = self.layer_1(x)
out = self.layer_2(out)
out = out.view(out.size(0),-1)
out = self.fc1(out)
out = self.fc2(out)
out = self.fc3(out)
return out
print(model)
def train(model, train_loader, criterion, optimizer, epoch):
train_loss = 0
model.train()
for batch_idx, (image, label) in enumerate(train_loader):
optimizer.zero_grad()
output = model(image)
loss = criterion(output, label)
loss.backward()
optimizer.step()
train_loss += loss.item()
if batch_idx % (len(train_loader)//2) == 0:
print('Train({})[{:.0f}%]: Loss: {:.4f}'.format(
epoch, 100. * batch_idx / len(train_loader), train_loss/(batch_idx+1)))
def test(model, test_loader, criterion, epoch):
model.eval()
test_loss = 0
correct = 0
with torch.no_grad():
for image, label in test_loader:
#image, label = image.to(device), label.to(device)
output = model(image)
test_loss += criterion(output, label).item() # sum up batch loss
pred = output.argmax(dim=1, keepdim=True) # get the index of the max log-probability
correct += pred.eq(label.view_as(pred)).sum().item()
test_loss = (test_loss*100)/len(test_loader.dataset)
print('Test({}): Loss: {:.4f}, Accuracy: {:.4f}%'.format(
epoch, test_loss, 100. * correct / len(test_loader.dataset)))
# #### Optimizer:
# Adam algorithm is an improvement on stochastic gradient descent, the detailed algorithm is shown below:
#
# 
# #### Criterion:
# Cross-entropy is a commonly used loss function for classification problem
#
# $L = \sum\limits_{k = 1}^M y_k \log(p_k)$,$y_k$ is the true label and $p_k$ is the probability for that label
model = CNN_Model()
optimizer = torch.optim.Adam(model.parameters(),lr=0.001)
criterion = torch.nn.CrossEntropyLoss()
epochs = 5
for epoch in range(1, epochs + 1):
train(model, train_loader, criterion, optimizer, epoch)
test(model, test_loader, criterion, epoch)
batch = next(iter(test_loader))
images, _ = batch
# +
background = images[:9]
test_images = images[9:11]
e = shap.GradientExplainer(model, background)
shap_values = e.shap_values(test_images)
shap_numpy = [np.swapaxes(np.swapaxes(s, 1, -1), 1, 2) for s in shap_values]
test_numpy = np.swapaxes(np.swapaxes(test_images.cpu().numpy(), 1, -1), 1, 2)
shap.image_plot(shap_numpy, -test_numpy)
# -
# Based on the above shap plot, anything that is colored in red enhance the model confidence while the blue decreases the model confidence.
|
_notebooks/2021-11-13-Assignment5-Yili Lin.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import sklearn as sk
from sklearn import linear_model
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error
from sklearn.preprocessing import MinMaxScaler
# -
# Набор данных взят с https://www.kaggle.com/aungpyaeap/fish-market
# Параметры нескольких популярных промысловых рыб
# length 1 = Body height
# length 2 = Total Length
# length 3 = Diagonal Length
fish_data = pd.read_csv("datasets/Fish.csv", delimiter=',')
print(fish_data)
# Выделим две переменных
x_label = 'Length1'
y_label = 'Weight'
data = fish_data[[x_label, y_label]]
print(data)
# Определим размер валидационной и тестовой выборок
val_test_size = round(0.2*len(data))
print(val_test_size)
# Генерируем уникальный seed
my_code = "Маргарян"
seed_limit = 2 ** 32
my_seed = int.from_bytes(my_code.encode(), "little") % seed_limit
# Создадим обучающую, валидационную и тестовую выборки
random_state = my_seed
train_val, test = train_test_split(data, test_size=val_test_size, random_state=random_state)
train, val = train_test_split(train_val, test_size=val_test_size, random_state=random_state)
print(len(train), len(val), len(test))
# +
# Преобразуем данные к ожидаемому библиотекой skleran формату
train_x = np.array(train[x_label]).reshape(-1,1)
train_y = np.array(train[y_label]).reshape(-1,1)
val_x = np.array(val[x_label]).reshape(-1,1)
val_y = np.array(val[y_label]).reshape(-1,1)
test_x = np.array(test[x_label]).reshape(-1,1)
test_y = np.array(test[y_label]).reshape(-1,1)
# -
# Нарисуем график
plt.plot(train_x, train_y, 'o')
plt.show()
# Создадим модель линейной регрессии и обучим ее на обучающей выборке.
model1 = linear_model.LinearRegression()
model1.fit(train_x, train_y)
# +
# Результат обучения: значения a и b: y = ax+b
print(model1.coef_, model1.intercept_)
a = model1.coef_[0]
b = model1.intercept_
print(a, b)
# +
# Добавим полученную линию на график
x = np.linspace(min(train_x), max(train_x), 100)
y = a * x + b
plt.plot(train_x, train_y, 'o')
plt.plot(x, y)
plt.show()
# -
# Проверим результат на валидационной выборке
val_predicted = model1.predict(val_x)
mse1 = mean_squared_error(val_y, val_predicted)
print(mse1)
# +
# Результат не очень хорош для интерпретации, попробуем сначала нормировать значения
scaler_x = MinMaxScaler()
scaler_x.fit(train_x)
scaled_train_x = scaler_x.transform(train_x)
scaler_y = MinMaxScaler()
scaler_y.fit(train_y)
scaled_train_y = scaler_y.transform(train_y)
plt.plot(scaled_train_x, scaled_train_y, 'o')
plt.show()
# +
# Строим модель и выводим результаты для нормированных данных
model2 = linear_model.LinearRegression()
model2.fit(scaled_train_x, scaled_train_y)
a = model2.coef_[0]
b = model2.intercept_
x = np.linspace(min(scaled_train_x), max(scaled_train_x), 100)
y = a * x + b
plt.plot(scaled_train_x, scaled_train_y, 'o')
plt.plot(x, y)
plt.show()
# +
# Проверим результат на валидационной выборке
scaled_val_x = scaler_x.transform(val_x)
scaled_val_y = scaler_y.transform(val_y)
val_predicted = model2.predict(scaled_val_x)
mse2 = mean_squared_error(scaled_val_y, val_predicted)
print(mse2)
# +
# Построим модель линейной регресси с L1-регуляризацией и выведем результаты для нормированных данных.
model3 = linear_model.Lasso(alpha=0.01)
model3.fit(scaled_train_x, scaled_train_y)
a = model3.coef_[0]
b = model3.intercept_
x = np.linspace(min(scaled_train_x), max(scaled_train_x), 100)
y = a * x + b
plt.plot(scaled_train_x, scaled_train_y, 'o')
plt.plot(x, y)
plt.show()
# +
# Проверим результат на валидационной выборке
scaled_val_x = scaler_x.transform(val_x)
scaled_val_y = scaler_y.transform(val_y)
val_predicted = model3.predict(scaled_val_x)
mse3 = mean_squared_error(scaled_val_y, val_predicted)
print(mse3)
# Можете поэкспериментировать со значением параметра alpha, чтобы уменьшить ошибку
# +
# Построим модель линейной регресси с L2-регуляризацией и выведем результаты для нормированных данных
model4 = linear_model.Ridge(alpha=0.01)
model4.fit(scaled_train_x, scaled_train_y)
a = model4.coef_[0]
b = model4.intercept_
x = np.linspace(min(scaled_train_x), max(scaled_train_x), 100)
y = a * x + b
plt.plot(scaled_train_x, scaled_train_y, 'o')
plt.plot(x, y)
plt.show()
# +
# Проверим результат на валидационной выборке
scaled_val_x = scaler_x.transform(val_x)
scaled_val_y = scaler_y.transform(val_y)
val_predicted = model4.predict(scaled_val_x)
mse4 = mean_squared_error(scaled_val_y, val_predicted)
print(mse4)
# Можете поэкспериментировать со значением параметра alpha, чтобы уменьшить ошибку
# +
# Построим модель линейной регресси с ElasticNet-регуляризацией и выведем результаты для нормированных данных
model5 = linear_model.ElasticNet(alpha=0.01, l1_ratio = 0.01)
model5.fit(scaled_train_x, scaled_train_y)
a = model5.coef_[0]
b = model5.intercept_
x = np.linspace(min(scaled_train_x), max(scaled_train_x), 100)
y = a * x + b
plt.plot(scaled_train_x, scaled_train_y, 'o')
plt.plot(x, y)
plt.show()
# +
# Проверим результат на валидационной выборке
scaled_val_x = scaler_x.transform(val_x)
scaled_val_y = scaler_y.transform(val_y)
val_predicted = model5.predict(scaled_val_x)
mse5 = mean_squared_error(scaled_val_y, val_predicted)
print(mse5)
# Можете поэкспериментировать со значениями параметров alpha и l1_ratio, чтобы уменьшить ошибку
# -
# Выведем ошибки для моделей на нормированных данных
print(mse2, mse3, mse4, mse5)
# +
# Минимальное значение достигается для второй модели, получим итоговую величину ошибки на тестовой выборке
scaled_test_x = scaler_x.transform(test_x)
scaled_test_y = scaler_y.transform(test_y)
test_predicted = model2.predict(scaled_test_x)
mse_test = mean_squared_error(scaled_test_y, test_predicted)
print(mse_test)
# +
# Повторите выделение данных, нормирование, и анализ 4 моделей
# (обычная линейная регрессия, L1-регуляризация, L2-регуляризация, ElasticNet-регуляризация)
# для x = Length2 и y = Width.
# -
x_label = 'Length2'
y_label = 'Weight'
data = fish_data[[x_label, y_label]]
print(data)
# Определим размер валидационной и тестовой выборок
val_test_size = round(0.2*len(data))
print(val_test_size)
# Создадим обучающую, валидационную и тестовую выборки
random_state = my_seed
train_val, test = train_test_split(data, test_size=val_test_size, random_state=random_state)
train, val = train_test_split(train_val, test_size=val_test_size, random_state=random_state)
print(len(train), len(val), len(test))
# +
# Преобразуем данные к ожидаемому библиотекой skleran формату
train_x = np.array(train[x_label]).reshape(-1,1)
train_y = np.array(train[y_label]).reshape(-1,1)
val_x = np.array(val[x_label]).reshape(-1,1)
val_y = np.array(val[y_label]).reshape(-1,1)
test_x = np.array(test[x_label]).reshape(-1,1)
test_y = np.array(test[y_label]).reshape(-1,1)
# -
# Нарисуем график
plt.plot(train_x, train_y, 'o')
plt.show()
# Создадим модель линейной регрессии и обучим ее на обучающей выборке.
model1 = linear_model.LinearRegression().fit(train_x, train_y)
# +
# Результат обучения: значения a и b: y = ax+b
print(model1.coef_, model1.intercept_)
a = model1.coef_[0]
b = model1.intercept_
print(a, b)
# +
# Добавим полученную линию на график
x = np.linspace(min(train_x), max(train_x), 100)
y = a * x + b
plt.plot(train_x, train_y, 'o')
plt.plot(x, y)
plt.show()
# -
# Проверим результат на валидационной выборке
val_predicted = model1.predict(val_x)
mse1 = mean_squared_error(val_y, val_predicted)
print(mse1)
# +
# Результат не очень хорош для интерпретации, попробуем сначала нормировать значения
scaler_x = MinMaxScaler().fit(train_x)
scaled_train_x = scaler_x.transform(train_x)
scaler_y = MinMaxScaler().fit(train_y)
scaled_train_y = scaler_y.transform(train_y)
plt.plot(scaled_train_x, scaled_train_y, 'o')
plt.show()
# +
# Строим модель и выводим результаты для нормированных данных
model2 = linear_model.LinearRegression().fit(scaled_train_x, scaled_train_y)
a = model2.coef_[0]
b = model2.intercept_
x = np.linspace(min(scaled_train_x), max(scaled_train_x), 100)
y = a * x + b
plt.plot(scaled_train_x, scaled_train_y, 'o')
plt.plot(x, y)
plt.show()
# +
# Проверим результат на валидационной выборке
scaled_val_x = scaler_x.transform(val_x)
scaled_val_y = scaler_y.transform(val_y)
val_predicted = model2.predict(scaled_val_x)
mse2 = mean_squared_error(scaled_val_y, val_predicted)
print(mse2)
# +
# Построим модель линейной регресси с L1-регуляризацией и выведем результаты для нормированных данных.
model3 = linear_model.Lasso(alpha=0.01).fit(scaled_train_x, scaled_train_y)
a = model3.coef_[0]
b = model3.intercept_
x = np.linspace(min(scaled_train_x), max(scaled_train_x), 100)
y = a * x + b
plt.plot(scaled_train_x, scaled_train_y, 'o')
plt.plot(x, y)
plt.show()
# +
# Проверим результат на валидационной выборке
scaled_val_x = scaler_x.transform(val_x)
scaled_val_y = scaler_y.transform(val_y)
val_predicted = model3.predict(scaled_val_x)
mse3 = mean_squared_error(scaled_val_y, val_predicted)
print(mse3)
# Можете поэкспериментировать со значением параметра alpha, чтобы уменьшить ошибку
# +
# Построим модель линейной регресси с L2-регуляризацией и выведем результаты для нормированных данных
model4 = linear_model.Ridge(alpha=0.01).fit(scaled_train_x, scaled_train_y)
a = model4.coef_[0]
b = model4.intercept_
x = np.linspace(min(scaled_train_x), max(scaled_train_x), 100)
y = a * x + b
plt.plot(scaled_train_x, scaled_train_y, 'o')
plt.plot(x, y)
plt.show()
# +
# Проверим результат на валидационной выборке
scaled_val_x = scaler_x.transform(val_x)
scaled_val_y = scaler_y.transform(val_y)
val_predicted = model4.predict(scaled_val_x)
mse4 = mean_squared_error(scaled_val_y, val_predicted)
print(mse4)
# Можете поэкспериментировать со значением параметра alpha, чтобы уменьшить ошибку
# +
# Построим модель линейной регресси с ElasticNet-регуляризацией и выведем результаты для нормированных данных
model5 = linear_model.ElasticNet(alpha=0.01, l1_ratio = 0.01)
model5.fit(scaled_train_x, scaled_train_y)
a = model5.coef_[0]
b = model5.intercept_
x = np.linspace(min(scaled_train_x), max(scaled_train_x), 100)
y = a * x + b
plt.plot(scaled_train_x, scaled_train_y, 'o')
plt.plot(x, y)
plt.show()
# +
# Проверим результат на валидационной выборке
scaled_val_x = scaler_x.transform(val_x)
scaled_val_y = scaler_y.transform(val_y)
val_predicted = model5.predict(scaled_val_x)
mse5 = mean_squared_error(scaled_val_y, val_predicted)
print(mse5)
# Можете поэкспериментировать со значениями параметров alpha и l1_ratio, чтобы уменьшить ошибку
# -
# Выведем ошибки для моделей на нормированных данных
print(mse2, mse3, mse4, mse5)
# +
# Минимальное значение достигается для второй модели, получим итоговую величину ошибки на тестовой выборке
scaled_test_x = scaler_x.transform(test_x)
scaled_test_y = scaler_y.transform(test_y)
test_predicted = model2.predict(scaled_test_x)
mse_test = mean_squared_error(scaled_test_y, test_predicted)
print(mse_test)
|
2020 Осенний семестр/Практическое задание 5/Маргарян - практика 5.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Building Simple Neural Networks
#
# In this section you will:
#
# * Import the MNIST dataset from Keras.
# * Format the data so it can be used by a Sequential model with Dense layers.
# * Split the dataset into training and test sections data.
# * Build a simple neural network using Keras Sequential model and Dense layers.
# * Train that model.
# * Evaluate the performance of that model.
#
# While we are accomplishing these tasks, we will also stop to discuss important concepts:
#
# * Splitting data into test and training sets.
# * Training rounds, batch size, and epochs.
# * Validation data vs test data.
# * Examining results.
#
# ## Importing and Formatting the Data
#
# Keras has several built-in datasets that are already well formatted and properly cleaned. These datasets are an invaluable learning resource. Collecting and processing datasets is a serious undertaking, and deep learning tactics perform poorly without large high quality datasets. We will be leveraging the [Keras built in datasets](https://keras.io/datasets/) extensively, and you may wish to explore them further on your own.
#
# In this exercise, we will be focused on the MNIST dataset, which is a set of 70,000 images of handwritten digits each labeled with the value of the written digit. Additionally, the images have been split into training and test sets.
# +
# For drawing the MNIST digits as well as plots to help us evaluate performance we
# will make extensive use of matplotlib
from matplotlib import pyplot as plt
# All of the Keras datasets are in keras.datasets
from keras.datasets import mnist
# Keras has already split the data into training and test data
(training_images, training_labels), (test_images, test_labels) = mnist.load_data()
# Training images is a list of 60,000 2D lists.
# Each 2D list is 28 by 28—the size of the MNIST pixel data.
# Each item in the 2D array is an integer from 0 to 255 representing its grayscale
# intensity where 0 means white, 255 means black.
print(len(training_images), training_images[0].shape)
# training_labels are a value between 0 and 9 indicating which digit is represented.
# The first item in the training data is a 5
print(len(training_labels), training_labels[0])
# -
# Lets visualize the first 100 images from the dataset
for i in range(100):
ax = plt.subplot(10, 10, i+1)
ax.axis('off')
plt.imshow(training_images[i], cmap='Greys')
# ## Problems With This Data
#
# There are (at least) two problems with this data as it is currently formatted, what do you think they are?
# 1. The input data is formatted as a 2D array, but our deep neural network needs to data as a 1D vector.
# * This is because of how deep neural networks are constructed, it is simply not possible to send anything but a vector as input.
# * These vectors can be/represent anything, but from the computer's perspective they must be a 1D vector.
# 2. Our labels are numbers, but we're not performing regression. We need to use a 1-hot vector encoding for our labels.
# * This is important because if we use the number values we would be training our network to think of these values as continuous.
# * If the digit is supposed to be a 2, guessing 1 and guessing 9 are both equally wrong.
# * Training the network with numbers would imply that a prediction of 1 would be "less wrong" than a prediction of 9, when in fact both are equally wrong.
# ### Fixing the data format
#
# Luckily, this is a common problem and we can use two methods to fix the data: `numpy.reshape` and `keras.utils.to_categorical`. This is nessesary because of how deep neural networks process data, there is no way to send 2D data to a `Sequential` model made of `Dense` layers.
# +
from keras.utils import to_categorical
# Preparing the dataset
# Setup train and test splits
(training_images, training_labels), (test_images, test_labels) = mnist.load_data()
# 28 x 28 = 784, because that's the dimensions of the MNIST data.
image_size = 784
# Reshaping the training_images and test_images to lists of vectors with length 784
# instead of lists of 2D arrays. Same for the test_images
training_data = training_images.reshape(training_images.shape[0], image_size)
test_data = test_images.reshape(test_images.shape[0], image_size)
# [
# [1,2,3]
# [4,5,6]
# ]
# => [1,2,3,4,5,6]
# Just showing the changes...
print("training data: ", training_images.shape, " ==> ", training_data.shape)
print("test data: ", test_images.shape, " ==> ", test_data.shape)
# +
# Create 1-hot encoded vectors using to_categorical
num_classes = 10 # Because it's how many digits we have (0-9)
# to_categorical takes a list of integers (our labels) and makes them into 1-hot vectors
training_labels = to_categorical(training_labels, num_classes)
test_labels = to_categorical(test_labels, num_classes)
# -
# Recall that before this transformation, training_labels[0] was the value 5. Look now:
print(training_labels[2])
# ## Building a Deep Neural Network
#
# Now that we've prepared our data, it's time to build a simple neural network. To start we'll make a deep network with 3 layers—the input layer, a single hidden layer, and the output layer. In a deep neural network all the layers are 1 dimensional. The input layer has to be the shape of our input data, meaning it must have 784 nodes. Similarly, the output layer must match our labels, meaning it must have 10 nodes. We can choose the number of nodes in our hidden layer, I've chosen 32 arbitrarally.
# +
from keras.models import Sequential
from keras.layers import Dense
# Sequential models are a series of layers applied linearly.
model = Sequential()
# The first layer must specify it's input_shape.
# This is how the first two layers are added, the input layer and the hidden layer.
model.add(Dense(units=32, activation='sigmoid', input_shape=(image_size,)))
# This is how the output layer gets added, the 'softmax' activation function ensures
# that the sum of the values in the output nodes is 1. Softmax is very
# common in classification networks.
model.add(Dense(units=num_classes, activation='softmax'))
# This function provides useful text data for our network
model.summary()
# -
# ## Compiling and Training a Model
#
# Our model must be compiled and trained before it can make useful predictions. Models are trainined with the training data and training labels. During this process Keras will use an optimizer, loss function, metrics of our chosing to repeatedly make predictions and recieve corrections. The loss function is used to train the model, the metrics are only used for human evaluation of the model during and after training.
#
# Training happens in a series of epochs which are divided into a series of rounds. Each round the network will recieve `batch_size` samples from the training data, make predictions, and recieve one correction based on the errors in those predictions. In a single epoch, the model will look at every item in the training set __exactly once__, which means individual data points are sampled from the training data without replacement during each round of each epoch.
#
# During training, the training data itself will be broken into two parts according to the `validation_split` parameter. The proportion that you specify will be left out of the training process, and used to evaluate the accuracy of the model. This is done to preserve the test data, while still having a set of data left out in order to test against — and hopefully prevent — overfitting. At the end of each epoch, predictions will be made for all the items in the validation set, but those predictions won't adjust the weights in the model. Instead, if the accuracy of the predictions in the validation set stops improving then training will stop early, even if accuracy in the training set is improving.
# +
# sgd stands for stochastic gradient descent.
# categorical_crossentropy is a common loss function used for categorical classification.
# accuracy is the percent of predictions that were correct.
model.compile(optimizer="sgd", loss='categorical_crossentropy', metrics=['accuracy'])
# The network will make predictions for 128 flattened images per correction.
# It will make a prediction on each item in the training set 5 times (5 epochs)
# And 10% of the data will be used as validation data.
history = model.fit(training_data, training_labels, batch_size=128, epochs=5, verbose=True, validation_split=.1)
# -
# ## Evaluating Our Model
#
# Now that we've trained our model, we want to evaluate its performance. We're using the "test data" here although in a serious experiment, we would likely not have done nearly enough work to warrent the application of the test data. Instead, we would rely on the validation metrics as a proxy for our test results until we had models that we believe would perform well.
#
# Once we evaluate our model on the test data, any subsequent changes we make would be based on what we learned from the test data. Meaning, we would have functionally incorporated information from the test set into our training procedure which could bias and even invalidate the results of our research. In a non-research setting the real test might be more like putting this feature into production.
#
# Nevertheless, it is always wise to create a test set that is not used as an evaluative measure until the very end of an experimental lifecycle. That is, once you have a model that you believe __should__ generalize well to unseen data you should test it on the test data to test that hypothosis. If your model performs poorly on the test data, you'll have to reevaluate your model, training data, and procedure.
# +
loss, accuracy = model.evaluate(test_data, test_labels, verbose=True)
plt.plot(history.history['acc'])
plt.plot(history.history['val_acc'])
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['training', 'validation'], loc='best')
plt.show()
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['training', 'validation'], loc='best')
plt.show()
print(f'Test loss: {loss:.3}')
print(f'Test accuracy: {accuracy:.3}')
# -
# ## How Did Our Network Do?
#
# * Why do we only have one value for test loss and test accuracy, but a chart over time for training and validation loss and accuracy?
# * Our model was more accurate on the validation data than it was on the training data.
# * Is this okay? Why or why not?
# * What if our model had been more accurate on the training data than the validation data?
# * Did our model get better during each epoch?
# * If not: why might that be the case?
# * If so: should we always expect this, where each epoch strictly improves training/validation accuracy/loss?
# ### Answers:
#
#
# * Why do we only have one value for test loss and test accuracy, but a chart over time for training and validation loss and accuracy?
# * __Because we only evaluate the test data once at the very end, but we evaluate training and validation scores once per epoch.__
# * Our model was more accurate on the validation data than it was on the training data.
# * Is this okay? Why or why not?
# * __Yes, this is okay, and even good. When our validation scores are better than our training scores, it's a sign that we are probably not overfitting__
# * What if our model had been more accurate on the training data than the validation data?
# * __This would concern us, because it would suggest we are probably overfitting.__
# * Did our model get better during each epoch?
# * If not: why might that be the case?
# * __Optimizers rely on the gradient to update our weights, but the 'function' we are optimizing (our neural network) is not a ground truth. A single batch, and even a complete epoch, may very well result in an adjustment that hurts overall performance.__
# * If so: should we always expect this, where each epoch strictly improves training/validation accuracy/loss?
# * __Not at all, see the above answer.__
# ## Look at Specific Results
#
# Often, it can be illuminating to view specific results, both when the model is correct and when the model is wrong. Lets look at the images and our model's predictions for the first 16 samples in the test set.
# +
from numpy import argmax
# Predicting once, then we can use these repeatedly in the next cell without recomputing the predictions.
predictions = model.predict(test_data)
# For pagination & style in second cell
page = 0
fontdict = {'color': 'black'}
# +
# Repeatedly running this cell will page through the predictions
for i in range(16):
ax = plt.subplot(4, 4, i+1)
ax.axis('off')
plt.imshow(test_images[i + page], cmap='Greys')
prediction = argmax(predictions[i + page])
true_value = argmax(test_labels[i + page])
fontdict['color'] = 'black' if prediction == true_value else 'red'
plt.title("{}, {}".format(prediction, true_value), fontdict=fontdict)
page += 16
plt.tight_layout()
plt.show()
# + [markdown] heading_collapsed=true
# ## Will A Different Network Perform Better?
#
# Given what you know so far, use Keras to build and train another sequential model that you think will perform __better__ than the network we just built and trained. Then evaluate that model and compare its performance to our model. Remember to look at accuracy and loss for training and validation data over time, as well as test accuracy and loss.
# + hidden=true
# Your code here...
# -
# ## Bonus questions: Go Further
#
# Here are some questions to help you further explore the concepts in this lab.
#
# * Does the original model, or your model, fail more often on a particular digit?
# * Write some code that charts the accuracy of our model's predictions on the test data by digit.
# * Is there a clear pattern? If so, speculate about why that could be...
# * Training for longer typically improves performance, up to a point.
# * For a simple model, try training it for 20 epochs, and 50 epochs.
# * Look at the charts of accuracy and loss over time, have you reached diminishing returns after 20 epochs? after 50?
# * More complex networks require more training time, but can outperform simpler networks.
# * Build a more complex model, with at least 3 hidden layers.
# * Like before, train it for 5, 20, and 50 epochs.
# * Evaluate the performance of the model against the simple model, and compare the total amount of time it took to train.
# * Was the extra complexity worth the additional training time?
# * Do you think your complex model would get even better with more time?
# * A little perspective on this last point: Some models train for [__weeks to months__](https://openai.com/blog/ai-and-compute/).
|
01-intro-to-deep-learning/02-building-simple-neural-networks.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Creating datetimes by hand
# Often you create datetime objects based on outside data. Sometimes though, you want to create a datetime object from scratch.
#
# You're going to create a few different datetime objects from scratch to get the hang of that process. These come from the bikeshare data set that you'll use throughout the rest of the chapter.
# +
# Import datetime
from datetime import datetime
# Create a datetime object
dt = datetime(2017, 10, 1, 15, 26, 26)
# Print the results in ISO 8601 format
print(dt.isoformat())
# +
# Import datetime
from datetime import datetime
# Create a datetime object
dt = datetime(2017, 12, 31, 15, 19, 13)
# Print the results in ISO 8601 format
print(dt.isoformat())
# +
# Import datetime
from datetime import datetime
# Create a datetime object
dt = datetime(2017, 12, 31, 15, 19, 13)
# Replace the year with 1917
dt_old = dt.replace(year=1917)
# Print the results in ISO 8601 format
print(dt_old)
# -
# # Counting events before and after noon
# In this chapter, you will be working with a list of all bike trips for one Capital Bikeshare bike, W20529, from October 1, 2017 to December 31, 2017. This list has been loaded as onebike_datetimes.
#
# Each element of the list is a dictionary with two entries: start is a datetime object corresponding to the start of a trip (when a bike is removed from the dock) and end is a datetime object corresponding to the end of a trip (when a bike is put back into a dock).
#
# You can use this data set to understand better how this bike was used. Did more trips start before noon or after noon?
# +
import pandas as pd
captial_onebike = pd.read_csv('../datasets/capital-onebike.csv')
fmt = "%Y-%m-%d %H:%M:%S"
onebike_datetime_strings = list(zip(captial_onebike['Start date'], captial_onebike['End date']))
onebike_datetimes = []
# Loop over all trips
for (start, end) in onebike_datetime_strings:
trip = {'start': datetime.strptime(start, fmt),
'end': datetime.strptime(end, fmt)}
# Append the trip
onebike_datetimes.append(trip)
# Create dictionary to hold results
trip_counts = {'AM': 0, 'PM': 0}
# Loop over all trips
for trip in onebike_datetimes:
# Check to see if the trip starts before noon
if trip['start'].hour < 12:
# Increment the counter for before noon
trip_counts['AM'] += 1
else:
# Increment the counter for after noon
trip_counts['PM'] += 1
print(trip_counts)
# -
# # Turning strings into datetimes
# When you download data from the Internet, dates and times usually come to you as strings. Often the first step is to turn those strings into datetime objects.
#
# In this exercise, you will practice this transformation.
#
# | Reference | |
# |-----------|----|
# |%Y|4 digit year (0000-9999)|
# |%m|2 digit month (1-12)|
# |%d|2 digit day (1-31)|
# |%H|2 digit hour (0-23)|
# |%M|2 digit minute (0-59)|
# |%S|2 digit second (0-59)
#
# +
# Import the datetime class
from datetime import datetime
# Starting string, in YYYY-MM-DD HH:MM:SS format
s = '2017-02-03 00:00:01'
# Write a format string to parse s
fmt = '%Y-%m-%d %H:%M:%S'
# Create a datetime object d
d = datetime.strptime(s, fmt)
# Print d
print(d)
# +
# Import the datetime class
from datetime import datetime
# Starting string, in YYYY-MM-DD format
s = '2030-10-15'
# Write a format string to parse s
fmt = '%Y-%m-%d'
# Create a datetime object d
d = datetime.strptime(s, fmt)
# Print d
print(d)
# +
# Import the datetime class
from datetime import datetime
# Starting string, in MM/DD/YYYY HH:MM:SS format
s = '12/15/1986 08:00:00'
# Write a format string to parse s
fmt = '%m/%d/%Y %H:%M:%S'
# Create a datetime object d
d = datetime.strptime(s, fmt)
# Print d
print(d)
# -
# # Parsing pairs of strings as datetimes
# Up until now, you've been working with a pre-processed list of datetimes for W20529's trips. For this exercise, you're going to go one step back in the data cleaning pipeline and work with the strings that the data started as.
#
# Explore onebike_datetime_strings in the IPython shell to determine the correct format. datetime has already been loaded for you.
#
# | Reference | |
# |-----------|----|
# |%Y|4 digit year (0000-9999)|
# |%m|2 digit month (1-12)|
# |%d|2 digit day (1-31)|
# |%H|2 digit hour (0-23)|
# |%M|2 digit minute (0-59)|
# |%S|2 digit second (0-59)
# +
import pandas as pd
captial_onebike = pd.read_csv('../datasets/capital-onebike.csv')
captial_onebike
# +
# Write down the format string
fmt = "%Y-%m-%d %H:%M:%S"
# Initialize a list for holding the pairs of datetime objects
onebike_datetimes = []
onebike_datetime_strings = list(zip(captial_onebike['Start date'], captial_onebike['End date']))
# Loop over all trips
for (start, end) in onebike_datetime_strings:
trip = {'start': datetime.strptime(start, fmt),
'end': datetime.strptime(end, fmt)}
# Append the trip
onebike_datetimes.append(trip)
onebike_datetimes
# -
# # Recreating ISO format with strftime()
# In the last chapter, you used strftime() to create strings from date objects. Now that you know about datetime objects, let's practice doing something similar.
#
# Re-create the .isoformat() method, using .strftime(), and print the first trip start in our data set.
# +
# Import datetime
from datetime import datetime
# Pull out the start of the first trip
first_start = onebike_datetimes[0]['start']
# Format to feed to strftime()
fmt = "%Y-%m-%dT%H:%M:%S"
# Print out date with .isoformat(), then with .strftime() to compare
print(first_start.isoformat())
print(first_start.strftime(fmt))
# -
# # Unix timestamps
# Datetimes are sometimes stored as Unix timestamps: the number of seconds since January 1, 1970. This is especially common with computer infrastructure, like the log files that websites keep when they get visitors.
# +
# Import datetime
from datetime import datetime
# Starting timestamps
timestamps = [1514665153, 1514664543]
# Datetime objects
dts = []
# Loop
for ts in timestamps:
dts.append(datetime.fromtimestamp(ts))
# Print results
print(dts)
# -
# # Turning pairs of datetimes into durations
# When working with timestamps, we often want to know how much time has elapsed between events. Thankfully, we can use datetime arithmetic to ask Python to do the heavy lifting for us so we don't need to worry about day, month, or year boundaries. Let's calculate the number of seconds that the bike was out of the dock for each trip.
#
# Continuing our work from a previous coding exercise, the bike trip data has been loaded as the list onebike_datetimes. Each element of the list consists of two datetime objects, corresponding to the start and end of a trip, respectively.
# +
# Initialize a list for all the trip durations
onebike_durations = []
for trip in onebike_datetimes:
# Create a timedelta object corresponding to the length of the trip
trip_duration = trip['end'] - trip['start']
# Get the total elapsed seconds in trip_duration
trip_length_seconds = trip_duration.total_seconds()
# Append the results to our list
onebike_durations.append(trip_length_seconds)
onebike_durations
# -
# # Average trip time
# W20529 took 291 trips in our data set. How long were the trips on average? We can use the built-in Python functions sum() and len() to make this calculation.
#
# Based on your last coding exercise, the data has been loaded as onebike_durations. Each entry is a number of seconds that the bike was out of the dock.
# +
# What was the total duration of all trips?
total_elapsed_time = sum(onebike_durations)
# What was the total number of trips?
number_of_trips = len(onebike_durations)
# Divide the total duration by the number of trips
print(total_elapsed_time / number_of_trips)
# -
# # The long and the short of why time is hard
# Out of 291 trips taken by W20529, how long was the longest? How short was the shortest? Does anything look fishy?
#
# As before, data has been loaded as onebike_durations.
# +
# Calculate shortest and longest trips
shortest_trip = min(onebike_durations)
longest_trip = max(onebike_durations)
# Print out the results
print("The shortest trip was " + str(shortest_trip) + " seconds")
print("The longest trip was " + str(longest_trip) + " seconds")
|
working-with-dates-and-times-in-python/2. Combining Dates and Times/notebook_section_2.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
#Here we will Improve model performance
# # Importing the libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
# # Importing the dataset
dataset = pd.read_csv('E:\Edu\Data Science and ML\Machinelearningaz\Datasets\Part 10 - Model Selection & Boosting\Section 48 - Model Selection//Social_Network_Ads.csv')
X = dataset.iloc[:, [2, 3]].values
y = dataset.iloc[:, 4].values
# # Splitting the dataset into the Training set and Test set
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.25, random_state = 0)
# # Feature Scaling
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
# # Fitting Kernel SVM to the Training set
# +
from sklearn.svm import SVC
#before grid search
#classifier = SVC(kernel = 'rbf', random_state = 0)
#after grid search
classifier = SVC(C=1,kernel = 'rbf',gamma=0.7, random_state = 0)
classifier.fit(X_train, y_train)
# -
# # Predicting the Test set results
y_pred = classifier.predict(X_test)
# # Making the Confusion Matrix
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_pred)
cm
# # Applying k-Fold Cross Validation
from sklearn.model_selection import cross_val_score
accuracies = cross_val_score(estimator = classifier, X = X_train, y = y_train, cv = 10)
print(accuracies.mean())
print(accuracies.std())
#low bias low variance
# # Applying Grid Search to find the best model and the best parameters
from sklearn.model_selection import GridSearchCV
parameters = [{'C': [1, 10, 100, 1000], 'kernel': ['linear']},
{'C': [1, 10, 100, 1000], 'kernel': ['rbf'], 'gamma': [0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9]}]
grid_search = GridSearchCV(estimator = classifier,
param_grid = parameters,
scoring = 'accuracy',
cv = 10,
n_jobs = -1)
grid_search = grid_search.fit(X_train, y_train)
best_accuracy = grid_search.best_score_
best_parameters = grid_search.best_params_
print(best_accuracy)
print(best_parameters)
# # Visualising the Training set results
from matplotlib.colors import ListedColormap
X_set, y_set = X_train, y_train
X1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 1, stop = X_set[:, 0].max() + 1, step = 0.01),
np.arange(start = X_set[:, 1].min() - 1, stop = X_set[:, 1].max() + 1, step = 0.01))
plt.contourf(X1, X2, classifier.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),
alpha = 0.75, cmap = ListedColormap(('red', 'green')))
plt.xlim(X1.min(), X1.max())
plt.ylim(X2.min(), X2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1],
c = ListedColormap(('red', 'green'))(i), label = j)
plt.title('Kernel SVM (Training set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
# # Visualising the Test set results
from matplotlib.colors import ListedColormap
X_set, y_set = X_test, y_test
X1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 1, stop = X_set[:, 0].max() + 1, step = 0.01),
np.arange(start = X_set[:, 1].min() - 1, stop = X_set[:, 1].max() + 1, step = 0.01))
plt.contourf(X1, X2, classifier.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),
alpha = 0.75, cmap = ListedColormap(('red', 'green')))
plt.xlim(X1.min(), X1.max())
plt.ylim(X2.min(), X2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1],
c = ListedColormap(('red', 'green'))(i), label = j)
plt.title('Kernel SVM (Test set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
|
7 Model Selection Boosting/Model Selection/ Grid Search .ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/yusufdalva/TensorFlow_Practice/blob/callbacks-1/fundamentals/callbacks.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="Ca18EVXA8xOo" colab_type="text"
# ### Callbacks example using MNIST dataset with Tensorflow
# This notebook demonstrates the usage of callbacks in training. A simple CNN is constructed. This code is written by modeling the assignment in https://colab.research.google.com/github/lmoroney/dlaicourse/blob/master/Course%201%20-%20Part%204%20-%20Lesson%202%20-%20Notebook.ipynb#scrollTo=BLMdl9aP8nQ0.
#
# The only intention is practice.
# + [markdown] id="hROc7Fwu9YL0" colab_type="text"
# ### Importing and checking Tensorflow version
# Import statement for tensorflow and version check.
# + id="74cV4KPZ9pCv" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="c41c1d05-d7b8-4b6f-c8d1-0215fa637aa3"
import tensorflow as tf
print("TensorFlow version: ", tf.__version__)
# + [markdown] id="m28-8h1_90vW" colab_type="text"
# ## Importing MNIST dataset
# This notebook uses MNIST dataset for demonstration purposes. The following part shows the import and preprocessing for the dataset
# + id="0M1f8aBz9vIW" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 153} outputId="7b307cdc-0739-415a-8108-0ac094f3e6c3"
mnist_dataset = tf.keras.datasets.fashion_mnist
(train_samples, train_labels), (test_samples, test_labels) = mnist_dataset.load_data()
# + [markdown] id="hm3hDYUv-b6F" colab_type="text"
# Data format of the dataset is shown below:
# + id="Tz3LWjiJ-XLg" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 85} outputId="70081aff-ae31-40c1-ac2b-5e0ca05344ca"
import numpy as np
import matplotlib.pyplot as plt
print("Training samples dimensions: ", train_samples.shape)
print("Training labels dimensions: ", train_labels.shape)
print("Test samples dimensions: ", test_samples.shape)
print("Test labels dimensions: ", test_labels.shape)
# + id="F93ZKWtR_P9i" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 68} outputId="32acb544-f3a7-45e7-ddc6-2a61c7eaf06f"
print("Data sample dimensions: ", train_samples.shape[1:])
assert train_samples.shape[1:] == test_samples.shape[1:] # Assertion to prove the equality of dimensions in test samples and training samples
print("Training sample count: ", train_samples.shape[0])
print("Test sample count: ", test_samples.shape[0])
# + [markdown] id="KFJ0FewIABLL" colab_type="text"
# ### Data Sample
# To show the task, one random data sample from training set is selected with its corresponding label
# + id="Gpd12LPF_7iD" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 299} outputId="3b3fb7c0-4915-423a-bb4f-5344996ce77f"
import random
random.seed(1) # For consistency in different runs
random_idx = int(random.random() * train_samples.shape[0]) # Corresponds to the index of a training example
data_sample, data_label = train_samples[random_idx], train_labels[random_idx]
print('Label of the data sample: ', data_label)
plt.imshow(data_sample, cmap = 'gray')
# + [markdown] id="2J6o6QCVB-bK" colab_type="text"
# ### Data Normalization
# To reduce the variance in both test and training set, normalization applied (Min-max feature scaling) The normalization formula is as follows:</br></br>
# $\large{X' = \frac{X - X_{min}}{X_{max}}}$</br></br>
# Note that minimum pixel value is 0 and maximum pixel value is 255 here
# + id="-PFPI0ExEAH6" colab_type="code" colab={}
train_samples = train_samples / 255.0
test_samples = test_samples / 255.0
# + [markdown] id="5PdRthz5EjEe" colab_type="text"
# #Model
# In the model here, there are no convolutional layers included as it is just an entry level example. The neural network model here will consist of 2 fully connected layers, which one has 256 neurons and the other has 128 neurons as an example. For the classification, as this is a multi-class classification problem, **softmax** activation has been used for 10 neurons (10 = # of classes)
# + id="c-d0WaWAEe7K" colab_type="code" colab={}
X_in = tf.keras.layers.Input(shape = train_samples.shape[1:]) # Input layer
X = tf.keras.layers.Flatten()(X_in)
X = tf.keras.layers.Dense(units = 256, activation = 'relu')(X)
X = tf.keras.layers.Dense(units = 128, activation = 'relu')(X)
out = tf.keras.layers.Dense(units = 10, activation = 'softmax')(X) # Output layer
model = tf.keras.Model(inputs = X_in, outputs = out)
# + [markdown] id="5uNJyUaaIVjE" colab_type="text"
# ## Compile the model
# After constructing the model, the desired metrics to observe during training (**metrics**), the loss function for training (**loss**) and the optimization algorithm is needed to be specified (**optimizer**). The following metrics has been selected:
# - **Optimizer: Adam Optimizer**, to make use of moving average and momentum
# - **Loss: Sparse Categorical Crossentropy**, the task is multiclass classification and the labels are not in the form of one-hot vector </br>(eg. for label 2, one-hot vector representation is: $\begin{bmatrix}0&0&1&0&...&0&0\end{bmatrix}$)
# - **Metrics: Accuracy**, to keep track of training accuracy
# + id="x4STHBvuIT4T" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 697} outputId="4f71fe76-f42f-45c9-cb02-f2f648eafe0e"
epoch_count = 20
model.compile(optimizer = 'adam',
loss = 'sparse_categorical_crossentropy',
metrics = ['accuracy'])
# Training the model with training samples and labels -- training for 20 epochs
history = model.fit(x = train_samples, y = train_labels, epochs = epoch_count)
# + id="ViT4yHT0U-Ch" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 350} outputId="6f84f6e2-c5fc-4f6a-850d-b29458163f98"
# Visualizing the changes in training loss and training accuracy values
fig, ax = plt.subplots(1,2, figsize = (15,5))
ax[0].plot(range(1,epoch_count + 1), history.history['loss'])
ax[0].set_xlabel('epochs')
ax[0].set_ylabel('Training Loss')
ax[0].set_title('Change in Training Loss')
ax[1].plot(range(1,epoch_count + 1), history.history['accuracy'])
ax[1].set_xlabel('epochs')
ax[1].set_ylabel('Training_accuracy')
ax[1].set_title('Change in Training Accuracy')
fig.show()
# + [markdown] id="jrboyeOeLUb2" colab_type="text"
# ## Evaluating the model
# Now with the test data, the model will be evaluated with prediction accuracy.
#
# + id="oFGiKlnVLJWW" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 68} outputId="10d445c8-2a51-4877-c2c8-9bc8e711754a"
eval = model.evaluate(test_samples, test_labels)
print('Testing loss: ', eval[0])
print('Testing accuracy: ', eval[1])
# + [markdown] id="LFozyjFzX-HI" colab_type="text"
# ## Adding a Callback
# To have more control in training, a callback is defined while fitting the model. In this notebook, the callback function described does two things. They are:
# - At the end of each epoch, prints a summary of the epoch.
# - Stops training if loss is less than 0.15
# + id="3L8Lalc9LnYK" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 867} outputId="946c192e-3f3f-4e07-f078-e59cb7e60c54"
class controlCallbakck(tf.keras.callbacks.Callback):
def on_epoch_end(self, epoch, logs = {}):
print("\nFor epoch " + str(epoch) + ", loss is " + str(logs.get('loss')) + " and accuracy is " + str(logs.get('accuracy')))
if logs.get('loss') < 0.15:
print("Loss too low, stopping training...")
self.model.stop_training = True
callback = controlCallbakck()
X_in = tf.keras.layers.Input(shape = train_samples.shape[1:]) # Input layer
X = tf.keras.layers.Flatten()(X_in)
X = tf.keras.layers.Dense(units = 256, activation = 'relu')(X)
X = tf.keras.layers.Dense(units = 128, activation = 'relu')(X)
out = tf.keras.layers.Dense(units = 10, activation = 'softmax')(X) # Output layer
model = tf.keras.Model(inputs = X_in, outputs = out)
epoch_count = 30
model.compile(optimizer = 'adam',
loss = 'sparse_categorical_crossentropy',
metrics = ['accuracy'])
model.fit(x = train_samples, y = train_labels, epochs = epoch_count, callbacks = [callback], verbose = 0)
# + [markdown] id="fyEXT_upc2CH" colab_type="text"
# ### Evaluating the model in terms of accuracy
# In the following, the model which the training is interrupted early due to the loss is evaluated in terms of accuracy.
# + id="dEFaBNXddLsM" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 68} outputId="6c6f3736-d27e-4b65-d21e-4f9528e2f995"
eval = model.evaluate(test_samples, test_labels)
print('Testing loss: ', eval[0])
print('Testing accuracy: ', eval[1])
|
Tensorflow_fundamentals/callbacks.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <img src="images/MCG.png" style="width: 100px">
#
#
# # Gaussian Bayesian Networks (GBNs)
#
# ## Generate $x_1$ $x_2$ and $Y$ from a Multivariate Gaussian Distribution with a Mean and a Variance.
#
# What if the inputs to the linear regression were correlated? This often happens in linear dynamical systems. Linear Gaussian Models are useful for modeling probabilistic PCA, factor analysis and linear dynamical systems. Linear Dynamical Systems have variety of uses such as tracking of moving objects. This is an area where Signal Processing methods have a high overlap with Machine Learning methods. When the problem is treated as a state-space problem with added stochasticity, then the future samples depend on the past. The latent parameters, $\beta_i$ where $i \in [1,...,k]$ provide a linear combination of the univariate gaussian distributions as shown in the figure.
#
# <img src="images/gbn.png", style="width: 400px">
#
# The observed variable, $y_{jx}$ can be described as a sample that is drawn from the conditional distribution:
#
# $$\mathcal{N}(y_{jx} | \sum_{i=1}^k \beta_i^T x_i + \beta_0; \sigma^2)$$
#
# The latent parameters $\beta_is$ and $\sigma^2$ need to be determined.
# +
#### import numpy as np
# %matplotlib inline
import pandas as pd
import seaborn as sns
import numpy as np
from matplotlib import cm
from mpl_toolkits.mplot3d import Axes3D
from scipy.stats import multivariate_normal
from matplotlib import pyplot
# Obtain the X and Y which are jointly gaussian from the distribution
mu_x = np.array([7, 13])
sigma_x = np.array([[4 , 3],
[3 , 6]])
# Variables
states = ['X1', 'X2']
all_states = ['X1', 'X2', 'P_X']
sym_coefs = ['b1_coef', 'b2_coef']
# Generate samples from the distribution
X_Norm = multivariate_normal(mean=mu_x, cov=sigma_x)
X_samples = X_Norm.rvs(size=10000)
X_df = pd.DataFrame(X_samples, columns=states)
# Generate
X_df['P_X'] = X_df.apply(X_Norm.pdf, axis=1)
X_df.head()
g = sns.jointplot(X_df['X1'], X_df['X2'], kind="kde", height=10, space=0)
# -
# ## Linear Gaussian Models - The Process
#
# The linear gaussian model in supervised learning scheme is nothing but a linear regression where inputs are drawn from a jointly gaussian distribution.
#
# Determining the Latent Parameters via Maximum Likelihood Estimation (MLE)
#
# The samples drawn from the conditional linear gaussian distributions are observed as:
#
# $$ p(Y|X) = \cfrac{1}{\sqrt(2\pi\sigma_c^2} \times exp(\cfrac{(\sum_{i=1}^k \beta_i^T x_i + \beta_0 - x[m])^2}{2\sigma^2})$$
#
# Taking log,
#
# $$ log(p(Y|X)) = (\sum_{i=1}^k[-\cfrac{1}{2}log(2\pi\sigma^2) - \cfrac{1}{2\sigma^2}( \beta_i^T x_i + \beta_0 - x[m])^2)]$$
#
# Differentiating w.r.t $\beta_i$, we can get k+1 linear equations as shown below:
#
#
# ### The Condtional Distribution p(Y|X)
#
# <img src="images/lgm.png" style="width: 700px">
#
# The betas can easily be estimated by inverting the coefficient matrix and multiplying it to the right-hand side.
# +
beta_vec = np.array([.7, .3])
beta_0 = 2
sigma_c = 4
def genYX(x):
'''
Generates samples distributed according to Gaussian Normal Distributions.
Args:
x (row): Dataframe row
Returns:
(float): Sample distributed as Gaussian
'''
x = [x['X1'], x['X2']]
var_mean = np.dot(beta_vec.transpose(), x) + beta_0
Yx_sample = np.random.normal(var_mean, sigma_c, 1)
return Yx_sample[0]
X_df['(Y|X)'] = X_df.apply(genYX, axis=1)
X_df.head()
sns.distplot(X_df['(Y|X)'])
# -
# # Determine parameters $\beta_0, \beta_1, \beta_2$ using Maximum Likelihood Estimation (MLE)
#
# +
x_len = len(states)
def exp_value(xi, xj):
'''
Computes sum of product of two columns of a dataframe.
Args:
xi (column): Column of a dataframe
xj (columns): Column of a dataframe
Returns:
(float): Sum of product of two columns
'''
prod_xixj = xi*xj
return np.sum(prod_xixj)
sum_X = X_df.sum()
X = [sum_X['(Y|X)']]
print(sum_X)
print(X)
coef_matrix = pd.DataFrame(columns=sym_coefs)
# First we compute just the coefficients of beta_1 to beta_N.
# Later we compute beta_0 and append it.
for i in range(0, x_len):
X.append(exp_value(X_df['(Y|X)'], X_df[states[i]]))
for j in range(0, x_len):
coef_matrix.loc[i, sym_coefs[j]] = exp_value(X_df[states[i]], X_df[states[j]])
coef_matrix.insert(0, 'b0_coef', sum_X.values[0:x_len])
row_1 = np.append([len(X_df)], sum_X.values[0:x_len])
coef_matrix.loc[-1] = row_1
coef_matrix.index = coef_matrix.index + 1 # shifting index
coef_matrix.sort_index(inplace=True)
# Compute beta values
# https://cedar.buffalo.edu/~srihari/CSE574/Chap8/Ch8-PGM-GaussianBNs/8.5%20GaussianBNs.pdf
beta_coef_matrix = np.matrix(coef_matrix.values, dtype='float')
coef_inv = np.linalg.inv(beta_coef_matrix)
beta_est = np.array(np.matmul(coef_inv, np.transpose(X)))
beta_est = beta_est[0]
print(beta_est)
# -
# This retrieves the beta parameters. Feel free to use the notebook/images for commerical/non-commercial purpose as long as you have the logos in place.
#
# ## Estimating Variance
#
# $$\sigma^2 = cov[y;y] - \sum_i \sum_j \beta_i \beta_j Cov_D[X_i;X_j]$$
# +
# First we compute just the coefficients of beta_1 to beta_N.
# Later we compute beta_0 and append it.
sigma_est = 0
M = len(X_df)
for i in range(0, x_len):
for j in range(0, x_len):
sigma_est += beta_est[i+1]*beta_est[j+1]*(exp_value(X_df[states[i]], X_df[states[j]])/M - np.mean(X_df[states[i]])*np.mean(X_df[states[j]]))
# Estimate Variance
sigma_est = np.sqrt(exp_value(X_df['(Y|X)'], X_df['(Y|X)'])/M - np.mean(X_df['(Y|X)'])*np.mean(X_df['(Y|X)']) - sigma_est)
print(sigma_est)
# -
#
# For any questions feel free to contact hkashyap [at] icloud.com. Thanks to <NAME> for the diagrams(diagram.ai), <NAME> and <NAME> for proof reading the math.
|
Gaussian Bayesian Networks (GBNs).ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [default]
# language: python
# name: python3
# ---
import keras
keras.__version__
# # 5.1 - Introduction to convnets
#
# This notebook contains the code sample found in Chapter 5, Section 1 of [Deep Learning with Python](https://www.manning.com/books/deep-learning-with-python?a_aid=keras&a_bid=76564dff). Note that the original text features far more content, in particular further explanations and figures: in this notebook, you will only find source code and related comments.
#
# ----
#
# First, let's take a practical look at a very simple convnet example. We will use our convnet to classify MNIST digits, a task that you've already been
# through in Chapter 2, using a densely-connected network (our test accuracy then was 97.8%). Even though our convnet will be very basic, its
# accuracy will still blow out of the water that of the densely-connected model from Chapter 2.
#
# The 6 lines of code below show you what a basic convnet looks like. It's a stack of `Conv2D` and `MaxPooling2D` layers. We'll see in a
# minute what they do concretely.
# Importantly, a convnet takes as input tensors of shape `(image_height, image_width, image_channels)` (not including the batch dimension).
# In our case, we will configure our convnet to process inputs of size `(28, 28, 1)`, which is the format of MNIST images. We do this via
# passing the argument `input_shape=(28, 28, 1)` to our first layer.
# +
from keras import layers
from keras import models
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu', input_shape=(28, 28, 1)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
# -
# Let's display the architecture of our convnet so far:
model.summary()
# You can see above that the output of every `Conv2D` and `MaxPooling2D` layer is a 3D tensor of shape `(height, width, channels)`. The width
# and height dimensions tend to shrink as we go deeper in the network. The number of channels is controlled by the first argument passed to
# the `Conv2D` layers (e.g. 32 or 64).
#
# The next step would be to feed our last output tensor (of shape `(3, 3, 64)`) into a densely-connected classifier network like those you are
# already familiar with: a stack of `Dense` layers. These classifiers process vectors, which are 1D, whereas our current output is a 3D tensor.
# So first, we will have to flatten our 3D outputs to 1D, and then add a few `Dense` layers on top:
model.add(layers.Flatten())
model.add(layers.Dense(64, activation='relu'))
model.add(layers.Dense(10, activation='softmax'))
# We are going to do 10-way classification, so we use a final layer with 10 outputs and a softmax activation. Now here's what our network
# looks like:
model.summary()
# As you can see, our `(3, 3, 64)` outputs were flattened into vectors of shape `(576,)`, before going through two `Dense` layers.
#
# Now, let's train our convnet on the MNIST digits. We will reuse a lot of the code we have already covered in the MNIST example from Chapter
# 2.
# +
from keras.datasets import mnist
from keras.utils import to_categorical
(train_images, train_labels), (test_images, test_labels) = mnist.load_data()
train_images = train_images.reshape((60000, 28, 28, 1))
train_images = train_images.astype('float32') / 255
test_images = test_images.reshape((10000, 28, 28, 1))
test_images = test_images.astype('float32') / 255
train_labels = to_categorical(train_labels)
test_labels = to_categorical(test_labels)
# -
model.compile(optimizer='rmsprop',
loss='categorical_crossentropy',
metrics=['accuracy'])
model.fit(train_images, train_labels, epochs=5, batch_size=64)
# Let's evaluate the model on the test data:
test_loss, test_acc = model.evaluate(test_images, test_labels)
test_acc
# While our densely-connected network from Chapter 2 had a test accuracy of 97.8%, our basic convnet has a test accuracy of 99.3%: we
# decreased our error rate by 68% (relative). Not bad!
|
Keras/deep-learning-Keras/.ipynb_checkpoints/5.1-introduction-to-convnets-checkpoint.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # FishboneMoncriefID: An Einstein Toolkit Initial Data Thorn for Fishbone-Moncrief initial data
#
# ## Author: <NAME>
# ### Formatting improvements courtesy <NAME>
#
# <font color ='red'> **While this compiles, it has not been validated against the old version of the code.**</font>
#
# ### NRPy+ Source Code for this module: [FishboneMoncriefID/FishboneMoncriefID.py](../edit/FishboneMoncriefID/FishboneMoncriefID.py) [\[tutorial\]](Tutorial-FishboneMoncriefID.ipynb) Constructs SymPy expressions for plane-wave initial data
#
# ## Introduction:
# In this part of the tutorial, we will construct an Einstein Toolkit (ETK) thorn (module) that will set up Fishbone-Moncrief initial data. In the [Tutorial-FishboneMoncriefID](Tutorial-FishboneMoncriefID.ipynb) tutorial module, we used NRPy+ to contruct the SymPy expressions for plane-wave initial data.
#
# We will construct this thorn in two steps.
#
# 1. Call on NRPy+ to convert the SymPy expressions for the initial data into one C-code kernel.
# 1. Write the C code and linkages to the Einstein Toolkit infrastructure (i.e., the .ccl files) to complete this Einstein Toolkit module.
# <a id='toc'></a>
#
# # Table of Contents
# $$\label{toc}$$
#
# This module is organized as follows
#
# 1. [Step 1](#initializenrpy): Call on NRPy+ to convert the SymPy expression for the Fishbone-Moncrief initial data into a C-code kernel
# 1. [Step 2](#einstein): Interfacing with the Einstein Toolkit
# 1. [Step 2.a](#einstein_c): Constructing the Einstein Toolkit C-code calling functions that include the C code kernels
# 1. [Step 2.b](#einstein_ccl): CCL files - Define how this module interacts and interfaces with the larger Einstein Toolkit infrastructure
# 1. [Step 2.c](#einstein_list): Add the C code to the Einstein Toolkit compilation list
# 1. [Step 3](#latex_pdf_output): Output this module to $\LaTeX$-formatted PDF
# <a id='initializenrpy'></a>
#
# # Step 1: Call on NRPy+ to convert the SymPy expression for the Fishbone-Moncrief initial data into a C-code kernel \[Back to [top](#toc)\]
# $$\label{initializenrpy}$$
#
# After importing the core modules, we will set $\text{GridFuncMemAccess}$ to $\text{ETK}$. SymPy expressions for plane wave initial data are written inside [FishboneMoncriefID/FishboneMoncriefID.py](../edit/FishboneMoncriefID/FishboneMoncriefID.py), and we simply import them for use here.
# +
# Step 1: Call on NRPy+ to convert the SymPy expression for the
# Fishbone-Moncrief initial data into a C-code kernel
# Step 1a: Import needed NRPy+ core modules:
import NRPy_param_funcs as par
import indexedexp as ixp
import grid as gri
import finite_difference as fin
from outputC import *
import loop
# Step 1b: This is an Einstein Toolkit (ETK) thorn. Here we
# tell NRPy+ that gridfunction memory access will
# therefore be in the "ETK" style.
par.set_parval_from_str("grid::GridFuncMemAccess","ETK")
par.set_parval_from_str("grid::DIM", 3)
DIM = par.parval_from_str("grid::DIM")
# Step 1c: Call the FishboneMoncriefID() function from within the
# FishboneMoncriefID/FishboneMoncriefID.py module.
import FishboneMoncriefID.FishboneMoncriefID as fmid
# Step 1d: Within the ETK, the 3D gridfunctions x, y, and z store the
# Cartesian grid coordinates. Setting the gri.xx[] arrays
# to point to these gridfunctions forces NRPy+ to treat
# the Cartesian coordinate gridfunctions properly --
# reading them from memory as needed.
xcoord,ycoord,zcoord = gri.register_gridfunctions("AUX",["xcoord","ycoord","zcoord"])
gri.xx[0] = xcoord
gri.xx[1] = ycoord
gri.xx[2] = zcoord
# Step 1e: Set up the Fishbone-Moncrief initial data. This sets all the ID gridfunctions.
fmid.FishboneMoncriefID()
Valencia3velocityU = ixp.register_gridfunctions_for_single_rank1("EVOL","Valencia3velocityU")
# -={ Spacetime quantities: Generate C code from expressions and output to file }=-
KerrSchild_to_print = [\
lhrh(lhs=gri.gfaccess("out_gfs","alpha"),rhs=fmid.IDalpha),\
lhrh(lhs=gri.gfaccess("out_gfs","betaU0"),rhs=fmid.IDbetaU[0]),\
lhrh(lhs=gri.gfaccess("out_gfs","betaU1"),rhs=fmid.IDbetaU[1]),\
lhrh(lhs=gri.gfaccess("out_gfs","betaU2"),rhs=fmid.IDbetaU[2]),\
lhrh(lhs=gri.gfaccess("out_gfs","gammaDD00"),rhs=fmid.IDgammaDD[0][0]),\
lhrh(lhs=gri.gfaccess("out_gfs","gammaDD01"),rhs=fmid.IDgammaDD[0][1]),\
lhrh(lhs=gri.gfaccess("out_gfs","gammaDD02"),rhs=fmid.IDgammaDD[0][2]),\
lhrh(lhs=gri.gfaccess("out_gfs","gammaDD11"),rhs=fmid.IDgammaDD[1][1]),\
lhrh(lhs=gri.gfaccess("out_gfs","gammaDD12"),rhs=fmid.IDgammaDD[1][2]),\
lhrh(lhs=gri.gfaccess("out_gfs","gammaDD22"),rhs=fmid.IDgammaDD[2][2]),\
lhrh(lhs=gri.gfaccess("out_gfs","KDD00"),rhs=fmid.IDKDD[0][0]),\
lhrh(lhs=gri.gfaccess("out_gfs","KDD01"),rhs=fmid.IDKDD[0][1]),\
lhrh(lhs=gri.gfaccess("out_gfs","KDD02"),rhs=fmid.IDKDD[0][2]),\
lhrh(lhs=gri.gfaccess("out_gfs","KDD11"),rhs=fmid.IDKDD[1][1]),\
lhrh(lhs=gri.gfaccess("out_gfs","KDD12"),rhs=fmid.IDKDD[1][2]),\
lhrh(lhs=gri.gfaccess("out_gfs","KDD22"),rhs=fmid.IDKDD[2][2]),\
]
# Force outCverbose=False for this module to avoid gigantic C files
# filled with the non-CSE expressions for the Weyl scalars.
KerrSchild_CcodeKernel = fin.FD_outputC("returnstring",KerrSchild_to_print,params="outCverbose=False")
# -={ GRMHD quantities: Generate C code from expressions and output to file }=-
FMdisk_GRHD_rho_initial_to_print = [lhrh(lhs=gri.gfaccess("out_gfs","rho_initial"),rhs=fmid.rho_initial)]
FMdisk_GRHD_rho_initial_CcodeKernel = fin.FD_outputC("returnstring",FMdisk_GRHD_rho_initial_to_print)
FMdisk_GRHD_velocities_to_print = [\
lhrh(lhs=gri.gfaccess("out_gfs","Valencia3velocityU0"),rhs=fmid.IDValencia3velocityU[0]),\
lhrh(lhs=gri.gfaccess("out_gfs","Valencia3velocityU1"),rhs=fmid.IDValencia3velocityU[1]),\
lhrh(lhs=gri.gfaccess("out_gfs","Valencia3velocityU2"),rhs=fmid.IDValencia3velocityU[2]),\
]
FMdisk_GRHD_velocities_CcodeKernel = fin.FD_outputC("returnstring",FMdisk_GRHD_velocities_to_print)
#KerrSchild_looped = loop.loop(["i2","i1","i0"],["0","0","0"],["cctk_lsh[2]","cctk_lsh[1]","cctk_lsh[0]"],\
# ["1","1","1"],["#pragma omp parallel for","",""],"",\
# KerrSchild_CcodeKernel.replace("time","cctk_time"))
#FMdisk_GRHD_velocities_looped = loop.loop(["i2","i1","i0"],["0","0","0"],["cctk_lsh[2]","cctk_lsh[1]","cctk_lsh[0]"],\
# ["1","1","1"],["#pragma omp parallel for","",""],"",\
# FMdisk_GRHD_velocities_CcodeKernel.replace("time","cctk_time"))
#FMdisk_GRHD_rho_initial_looped = loop.loop(["i2","i1","i0"],["0","0","0"],["cctk_lsh[2]","cctk_lsh[1]","cctk_lsh[0]"],\
# ["1","1","1"],["#pragma omp parallel for","",""],"",\
# FMdisk_GRHD_rho_initial_CcodeKernel.replace("time","cctk_time"))
# Step 1f: Create directories for the thorn if they don't exist.
# !mkdir FishboneMoncriefID 2>/dev/null # 2>/dev/null: Don't throw an error if the directory already exists.
# !mkdir FishboneMoncriefID/src 2>/dev/null # 2>/dev/null: Don't throw an error if the directory already exists.
# Step 1g: Write the C code kernel to file.
with open("FishboneMoncriefID/src/KerrSchild.h", "w") as file:
file.write(str(KerrSchild_CcodeKernel.replace("time","cctk_time")))
with open("FishboneMoncriefID/src/FMdisk_GRHD_velocities.h", "w") as file:
file.write(str(FMdisk_GRHD_velocities_CcodeKernel.replace("time","cctk_time")))
with open("FishboneMoncriefID/src/FMdisk_GRHD_rho_initial.h", "w") as file:
file.write(str(FMdisk_GRHD_rho_initial_CcodeKernel.replace("time","cctk_time")))
hm1string = outputC(fmid.hm1,"hm1",filename="returnstring")
with open("FishboneMoncriefID/src/FMdisk_GRHD_hm1.h", "w") as file:
file.write(str(hm1string))
# -
# <a id='einstein'></a>
#
# # Step 2: Interfacing with the Einstein Toolkit \[Back to [top](#toc)\]
# $$\label{einstein}$$
#
# <a id='einstein_c'></a>
#
# ## Step 2.a: Constructing the Einstein Toolkit C-code calling functions that include the C code kernels \[Back to [top](#toc)\]
# $$\label{einstein_c}$$
#
# We will write another C file with the functions we need here.
# +
# %%writefile FishboneMoncriefID/src/InitialData.c
#include <math.h>
#include <stdio.h>
#include <stdbool.h>
#include <stdlib.h> // For drand48()
#include "cctk.h"
#include "cctk_Parameters.h"
#include "cctk_Arguments.h"
// Alias for "vel" vector gridfunction:
#define velx (&vel[0*cctk_lsh[0]*cctk_lsh[1]*cctk_lsh[2]])
#define vely (&vel[1*cctk_lsh[0]*cctk_lsh[1]*cctk_lsh[2]])
#define velz (&vel[2*cctk_lsh[0]*cctk_lsh[1]*cctk_lsh[2]])
void FishboneMoncrief_KerrSchild(const cGH* restrict const cctkGH,const CCTK_INT *cctk_lsh,
const CCTK_INT i0,const CCTK_INT i1,const CCTK_INT i2,
const CCTK_REAL *xcoordGF,const CCTK_REAL *ycoordGF,const CCTK_REAL *zcoordGF,
CCTK_REAL *alphaGF,CCTK_REAL *betaU0GF,CCTK_REAL *betaU1GF,CCTK_REAL *betaU2GF,
CCTK_REAL *gammaDD00GF,CCTK_REAL *gammaDD01GF,CCTK_REAL *gammaDD02GF,CCTK_REAL *gammaDD11GF,CCTK_REAL *gammaDD12GF,CCTK_REAL *gammaDD22GF,
CCTK_REAL *KDD00GF,CCTK_REAL *KDD01GF,CCTK_REAL *KDD02GF,CCTK_REAL *KDD11GF,CCTK_REAL *KDD12GF,CCTK_REAL *KDD22GF)
{
DECLARE_CCTK_PARAMETERS
#include "KerrSchild.h"
}
void FishboneMoncrief_FMdisk_GRHD_velocities(const cGH* restrict const cctkGH,const CCTK_INT *cctk_lsh,
const CCTK_INT i0,const CCTK_INT i1,const CCTK_INT i2,
const CCTK_REAL *xcoordGF,const CCTK_REAL *ycoordGF,const CCTK_REAL *zcoordGF,
CCTK_REAL *Valencia3velocityU0GF, CCTK_REAL *Valencia3velocityU1GF, CCTK_REAL *Valencia3velocityU2GF)
{
DECLARE_CCTK_PARAMETERS
#include "FMdisk_GRHD_velocities.h"
}
void FishboneMoncrief_ET_GRHD_initial(CCTK_ARGUMENTS) {
DECLARE_CCTK_ARGUMENTS;
DECLARE_CCTK_PARAMETERS;
printf("Fishbone-Moncrief Disk Initial data.\n");
printf("Using input parameters of\n a = %e,\n M = %e,\nr_in = %e,\nr_at_max_density = %e\nkappa = %e\ngamma = %e\n",a,M,r_in,r_at_max_density,kappa,gamma);
// First compute maximum density
CCTK_REAL rho_max;
{
CCTK_REAL hm1;
CCTK_REAL xcoord = r_at_max_density;
CCTK_REAL ycoord = 0.0;
CCTK_REAL zcoord = 0.0;
{
#include "FMdisk_GRHD_hm1.h"
}
rho_max = pow( hm1 * (gamma-1.0) / (kappa*gamma), 1.0/(gamma-1.0) );
}
#pragma omp parallel for
for(CCTK_INT k=0;k<cctk_lsh[2];k++) for(CCTK_INT j=0;j<cctk_lsh[1];j++) for(CCTK_INT i=0;i<cctk_lsh[0];i++) {
CCTK_INT idx = CCTK_GFINDEX3D(cctkGH,i,j,k);
CCTK_REAL xcoord = x[idx];
CCTK_REAL ycoord = y[idx];
CCTK_REAL zcoord = z[idx];
CCTK_REAL rr = r[idx];
FishboneMoncrief_KerrSchild(cctkGH,cctk_lsh,
i,j,k,
x,y,z,
alp,betax,betay,betaz,
gxx,gxy,gxz,gyy,gyz,gzz,
kxx,kxy,kxz,kyy,kyz,kzz);
CCTK_REAL hm1;
bool set_to_atmosphere=false;
if(rr > r_in) {
{
#include "FMdisk_GRHD_hm1.h"
}
if(hm1 > 0) {
rho[idx] = pow( hm1 * (gamma-1.0) / (kappa*gamma), 1.0/(gamma-1.0) ) / rho_max;
press[idx] = kappa*pow(rho[idx], gamma);
// P = (\Gamma - 1) rho epsilon
eps[idx] = press[idx] / (rho[idx] * (gamma - 1.0));
FishboneMoncrief_FMdisk_GRHD_velocities(cctkGH,cctk_lsh,
i,j,k,
x,y,z,
velx,vely,velz);
} else {
set_to_atmosphere=true;
}
} else {
set_to_atmosphere=true;
}
// Outside the disk? Set to atmosphere all hydrodynamic variables!
if(set_to_atmosphere) {
// Choose an atmosphere such that
// rho = 1e-5 * r^(-3/2), and
// P = k rho^gamma
// Add 1e-100 or 1e-300 to rr or rho to avoid divisions by zero.
rho[idx] = 1e-5 * pow(rr + 1e-100,-3.0/2.0);
press[idx] = kappa*pow(rho[idx], gamma);
eps[idx] = press[idx] / ((rho[idx] + 1e-300) * (gamma - 1.0));
w_lorentz[idx] = 1.0;
velx[idx] = 0.0;
vely[idx] = 0.0;
velz[idx] = 0.0;
}
}
CCTK_INT final_idx = CCTK_GFINDEX3D(cctkGH,cctk_lsh[0]-1,cctk_lsh[1]-1,cctk_lsh[2]-1);
printf("===== OUTPUTS =====\n");
printf("betai: %e %e %e \ngij: %e %e %e %e %e %e \nKij: %e %e %e %e %e %e\nalp: %e\n\n",betax[final_idx],betay[final_idx],betaz[final_idx],gxx[final_idx],gxy[final_idx],gxz[final_idx],gyy[final_idx],gyz[final_idx],gzz[final_idx],kxx[final_idx],kxy[final_idx],kxz[final_idx],kyy[final_idx],kyz[final_idx],kzz[final_idx],alp[final_idx]);
printf("rho: %.15e\nPressure: %.15e\nvx: %.15e\nvy: %.15e\nvz: %.15e\n",rho[final_idx],press[final_idx],velx[final_idx],vely[final_idx],velz[final_idx]);
}
void FishboneMoncrief_ET_GRHD_initial__perturb_pressure(CCTK_ARGUMENTS) {
DECLARE_CCTK_ARGUMENTS;
DECLARE_CCTK_PARAMETERS;
#pragma omp parallel for
for(CCTK_INT k=0;k<cctk_lsh[2];k++) for(CCTK_INT j=0;j<cctk_lsh[1];j++) for(CCTK_INT i=0;i<cctk_lsh[0];i++) {
CCTK_INT idx = CCTK_GFINDEX3D(cctkGH,i,j,k);
CCTK_REAL random_number_between_min_and_max = random_min + (random_max - random_min)*drand48();
press[idx] = press[idx]*(1.0 + random_number_between_min_and_max);
// Add 1e-300 to rho to avoid division by zero when density is zero.
eps[idx] = press[idx] / ((rho[idx] + 1e-300) * (gamma - 1.0));
}
}
# -
# <a id='einstein_ccl'></a>
#
# ## Step 2.b: CCL files - Define how this module interacts and interfaces with the larger Einstein Toolkit infrastructure \[Back to [top](#toc)\]
# $$\label{einstein_ccl}$$
#
# Writing a module ("thorn") within the Einstein Toolkit requires that three "ccl" files be constructed, all in the root directory of the thorn:
#
# 1. $\text{interface.ccl}$: defines the gridfunction groups needed, and provides keywords denoting what this thorn provides and what it should inherit from other thorns. Specifically, this file governs the interaction between this thorn and others; more information can be found in the [official Einstein Toolkit documentation](http://cactuscode.org/documentation/referencemanual/ReferenceManualch8.html#x12-260000C2.2).
# With "implements", we give our thorn its unique name. By "inheriting" other thorns, we tell the Toolkit that we will rely on variables that exist and are declared "public" within those functions.
# %%writefile FishboneMoncriefID/interface.ccl
implements: FishboneMoncriefID
inherits: admbase grid hydrobase
# 2. $\text{param.ccl}$: specifies free parameters within the thorn, enabling them to be set at runtime. It is required to provide allowed ranges and default values for each parameter. More information on this file's syntax can be found in the [official Einstein Toolkit documentation](http://cactuscode.org/documentation/referencemanual/ReferenceManualch8.html#x12-265000C2.3).
# +
# %%writefile FishboneMoncriefID/param.ccl
shares: grid
shares: ADMBase
USES CCTK_INT lapse_timelevels
USES CCTK_INT shift_timelevels
USES CCTK_INT metric_timelevels
USES KEYWORD metric_type
EXTENDS KEYWORD initial_data
{
"FishboneMoncriefID" :: "Initial data from FishboneMoncriefID solution"
}
EXTENDS KEYWORD initial_lapse
{
"FishboneMoncriefID" :: "Initial lapse from FishboneMoncriefID solution"
}
EXTENDS KEYWORD initial_shift
{
"FishboneMoncriefID" :: "Initial shift from FishboneMoncriefID solution"
}
EXTENDS KEYWORD initial_dtlapse
{
"FishboneMoncriefID" :: "Initial dtlapse from FishboneMoncriefID solution"
}
EXTENDS KEYWORD initial_dtshift
{
"FishboneMoncriefID" :: "Initial dtshift from FishboneMoncriefID solution"
}
shares: HydroBase
EXTENDS KEYWORD initial_hydro
{
"FishboneMoncriefID" :: "Initial GRHD data from FishboneMoncriefID solution"
}
#["r_in","r_at_max_density","a","M"] A_b, kappa, gamma
restricted:
CCTK_REAL r_in "Fixes the inner edge of the disk"
{
0.0:* :: "Must be positive"
} 6.0
restricted:
CCTK_REAL r_at_max_density "Radius at maximum disk density. Needs to be > r_in"
{
0.0:* :: "Must be positive"
} 12.0
restricted:
CCTK_REAL a "The spin parameter of the black hole"
{
-1.0:1.0 :: "Positive values, up to 1. Negative disallowed, as certain roots are chosen in the hydro fields setup. Check those before enabling negative spins!"
} 0.9375
restricted:
CCTK_REAL M "Kerr-Schild BH mass. Probably should always set M=1."
{
0.0:* :: "Must be positive"
} 1.0
restricted:
CCTK_REAL A_b "Scaling factor for the vector potential"
{
*:* :: ""
} 1.0
restricted:
CCTK_REAL kappa "Equation of state: P = kappa * rho^gamma"
{
0.0:* :: "Positive values"
} 1.0e-3
restricted:
CCTK_REAL gamma "Equation of state: P = kappa * rho^gamma"
{
0.0:* :: "Positive values"
} 1.3333333333333333333333333333
##################################
# PRESSURE PERTURBATION PARAMETERS
private:
CCTK_REAL random_min "Floor value of random perturbation to initial pressure, where perturbed pressure = pressure*(1.0 + (random_min + (random_max-random_min)*RAND[0,1)))"
{
*:* :: "Any value"
} -0.02
private:
CCTK_REAL random_max "Ceiling value of random perturbation to initial pressure, where perturbed pressure = pressure*(1.0 + (random_min + (random_max-random_min)*RAND[0,1)))"
{
*:* :: "Any value"
} 0.02
# -
# 3. $\text{schedule.ccl}$: allocates storage for gridfunctions, defines how the thorn's functions should be scheduled in a broader simulation, and specifies the regions of memory written to or read from gridfunctions. $\text{schedule.ccl}$'s official documentation may be found [here](http://cactuscode.org/documentation/referencemanual/ReferenceManualch8.html#x12-268000C2.4).
#
# We specify here the standardized ETK "scheduling bins" in which we want each of our thorn's functions to run.
# +
# %%writefile FishboneMoncriefID/schedule.ccl
STORAGE: ADMBase::metric[metric_timelevels], ADMBase::curv[metric_timelevels], ADMBase::lapse[lapse_timelevels], ADMBase::shift[shift_timelevels]
schedule FishboneMoncrief_ET_GRHD_initial IN HydroBase_Initial
{
LANG: C
READS: grid::x(Everywhere)
READS: grid::y(Everywhere)
READS: grid::y(Everywhere)
WRITES: admbase::alp(Everywhere)
WRITES: admbase::betax(Everywhere)
WRITES: admbase::betay(Everywhere)
WRITES: admbase::betaz(Everywhere)
WRITES: admbase::kxx(Everywhere)
WRITES: admbase::kxy(Everywhere)
WRITES: admbase::kxz(Everywhere)
WRITES: admbase::kyy(Everywhere)
WRITES: admbase::kyz(Everywhere)
WRITES: admbase::kzz(Everywhere)
WRITES: admbase::gxx(Everywhere)
WRITES: admbase::gxy(Everywhere)
WRITES: admbase::gxz(Everywhere)
WRITES: admbase::gyy(Everywhere)
WRITES: admbase::gyz(Everywhere)
WRITES: admbase::gzz(Everywhere)
WRITES: hydrobase::velx(Everywhere)
WRITES: hydrobase::vely(Everywhere)
WRITES: hydrobase::velz(Everywhere)
WRITES: hydrobase::rho(Everywhere)
WRITES: hydrobase::eps(Everywhere)
WRITES: hydrobase::press(Everywhere)
} "Set up general relativistic hydrodynamic (GRHD) fields for Fishbone-Moncrief disk"
schedule FishboneMoncrief_ET_GRHD_initial__perturb_pressure IN CCTK_INITIAL AFTER Seed_Magnetic_Fields BEFORE IllinoisGRMHD_ID_Converter
{
LANG: C
} "Add random perturbation to initial pressure, after seed magnetic fields have been set up (in case we'd like the seed magnetic fields to depend on the pristine pressures)"
# -
# <a id='einstein_list'></a>
#
# ## Step 2.c: Add the C code to the Einstein Toolkit compilation list \[Back to [top](#toc)\]
# $$\label{einstein_list}$$
#
# We will also need $\text{make.code.defn}$, which indicates the list of files that need to be compiled. This thorn only has the one C file to compile.
# %%writefile FishboneMoncriefID/src/make.code.defn
SRCS = InitialData.c
# <a id='latex_pdf_output'></a>
#
# # Step 3: Output this module to $\LaTeX$-formatted PDF file \[Back to [top](#toc)\]
# $$\label{latex_pdf_output}$$
#
# The following code cell converts this Jupyter notebook into a proper, clickable $\LaTeX$-formatted PDF file. After the cell is successfully run, the generated PDF may be found in the root NRPy+ tutorial directory, with filename
# [Tutorial-ETK_thorn-FishboneMoncriefID.pdf](Tutorial-ETK_thorn-FishboneMoncriefID.pdf) (Note that clicking on this link may not work; you may need to open the PDF file through another means.)
# !jupyter nbconvert --to latex --template latex_nrpy_style.tplx Tutorial-ETK_thorn-FishboneMoncriefID.ipynb
# !pdflatex -interaction=batchmode Tutorial-ETK_thorn-FishboneMoncriefID.tex
# !pdflatex -interaction=batchmode Tutorial-ETK_thorn-FishboneMoncriefID.tex
# !pdflatex -interaction=batchmode Tutorial-ETK_thorn-FishboneMoncriefID.tex
# !rm -f Tut*.out Tut*.aux Tut*.log
|
Tutorial-ETK_thorn-FishboneMoncriefID.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:miniconda-ctsm]
# language: python
# name: conda-env-miniconda-ctsm-py
# ---
# # Notebook overview on data preprocessing, analysis and plotting in Vanderkelen et al. 2022 GMD
# ## 1. Preprocessing
# ### 1.1 Irrigation topology derivation
#
# - [irrigtopo_snake.ipynb](/analysis/irrigtopo_casestudy_snake/irrigtopo_snake.ipynb): Snake case study: derivation and plotting
# - [determine_irrigtopo_global.ipynb](/preprocessing/determine_irrigtopo_global.ipynb): derivation of global irrigation topology
#
# + [markdown] jp-MarkdownHeadingCollapsed=true tags=[]
# ### 1.2 CTSM simulations
#
# - [setup_IHistClm50Sp_360x720cru_CTL.sh](preprocessing/setup_IHistClm50Sp_360x720cru_CTL.sh): script to set up CTSM simulation
# - [nl_clm_CTL.sh](preprocessing/nl_clm_CTL.sh): namelist file used in CTSM simulation
# + [markdown] tags=[]
# ### 1.2 MizuRoute simulations
# in order of usage
# -
# - [pp_clm_for_mizuroute.ipynb](preprocessing/pp_clm_for_mizuroute.ipynb): notebook processing CTSM output for MizuRoute including: (i) preparing irrigation water demands for irrigation topology (ii) merging and preparing runoff, evaporation and precip for mizuRoute input
# - [prepare_ntopo_HDMA_D03.ipynb](/preprocessing/prepare_ntopo_HDMA_D03.ipynb): prepare parameter topology for natural lake simulation with mizuRoute
# - [prepare_ntopo_nolake.ipynb](/preprocessing/prepare_ntopo_nolake.ipynb): prepare parameter topology for no lake simulation with mizuRoute
# - [pp_calc_inflowseasonality_natlake.ipynb](preprocessing/pp_calc_inflowseasonality_natlake.ipynb): notebook calculating inflow seasonality based on mizuRoute simulations with natural lakes, (necessary as parameters for Hanasaki implementation)
# - [apply_irrigtopo_global.ipynb](/preprocessing/apply_irrigtopo_global.ipynb): application of global irrigation topology on observed seasonal irrigation demands
# - [prepare_ntopo_HDMA_H06.ipynb](/preprocessing/prepare_ntopo_HDMA_H06.ipynb): prepare parameter topology for Hanasaki simulation with mizuRoute
#
# ## 2. Analysis and plotting
# + [markdown] tags=[]
# ### 2.1 Local mizuRoute simulations
#
# - [local_Hanasaki_resobs_plotting.ipynb](analysis/local_Hanasaki_resobs_plotting.ipynb): Analysis with local mizuRoute simulations
# + [markdown] tags=[]
# ### 2.2 Global mizuRoute simulations
# #### 2.2.1 Evaluation using reservoir observations
#
# - [global_mizuRoute_Hanasaki_resobs.ipynb](analysis/global_mizuRoute_Hanasaki_resobs.ipynb): processing and plotting of global mizuRoute simulations compared to reservoir observations
# - [global_steyaert_evaluation_mizuRoute_Hanasaki.ipynb](analysis/global_steyaert_evaluation_mizuRoute_Hanasaki.ipynb): evaluation with observations from the ResOpsUS dataset of Steyaert et al., 2022
# + [markdown] tags=[]
# #### 2.2.2 Runoff evaluation
#
# - [pp_clm_evaluate_runoff_paperplot.ipynb](analysis/pp_clm_evaluate_runoff_paperplot.ipynb): evaluation of CTSM runoff with GRUN (maps)
# + [markdown] tags=[]
# #### 2.2.3 Global evaluation with GSIM stream indices
#
# - [pp_gsim_processing_paper.ipynb](analysis/pp_gsim_processing_paper.ipynb): processing script loading obs and calculating metrics (saving as dict)
# - [pp_gsim_plotting.ipynb](analysis/pp_gsim_processing_paper.ipynb): plotting script loading saved dict and producing maps
# -
#
#
|
main.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Decoupling Logic and Execution
#
# In the last section, we used Fugue's transform function to port pandas code to Spark. Decoupling logic and execution is one of the primary motivations of Fugue. When transitioning a project from pandas to Spark, the majority of the code normally has has to be re-written. This is because using either pandas or Spark makes code highly coupled with that framework. This leads to a couple of problems:
#
# 1. Users have to learn an entirely new framework to work with distributed compute problems
# 2. Logic written for a *small data* project does not become reusable for a *big data* project
# 3. Testing becomes a heavyweight process for distributed compute, especially Spark
# 4. Along with number 3, iterations for distributed compute problems become slower and more expensive
#
# Fugue believes that code should minimize dependency on frameworks as much as possible. This provides flexibility and portability. **By decoupling logic and execution, we can focus on our logic in a scale-agnostic way, and then choose which engine to use when the time arises.**
# ## Differences between Pandas and Spark
#
# To illustrate the first two main points above, we'll use a simple example. For the data below, we are interested in getting the first three digits of the `phone` column and populating a new column called `location` by using a dictionary that maps the values. We start by preparing the sample data and defining the mapping.
# +
import pandas as pd
_area_code_map = {"217": "Champaign, IL", "407": "Orlando, FL", "510": "Fremont, CA"}
data = pd.DataFrame({"phone": ["(217)-123-4567", "(217)-234-5678", "(407)-123-4567",
"(407)-234-5678", "(510)-123-4567"]})
data.head()
# -
# First, we'll perform the operation in pandas. It's very simple because of the `.map()` method in pandas
# +
def map_phone_to_location(df: pd.DataFrame) -> pd.DataFrame:
df["location"] = df["phone"].str.slice(1,4).map(_area_code_map)
return df
map_phone_to_location(data.copy())
# -
# Next we'll perform the same operation in Spark and see how different the syntax is.
# Setting up Spark session
from pyspark.sql import SparkSession, DataFrame
spark = SparkSession.builder.getOrCreate()
# +
from pyspark.sql.functions import create_map, col, lit, substring
from itertools import chain
df = spark.createDataFrame(data) # converting the previous Pandas DataFrame
mapping_expr = create_map([lit(x) for x in chain(*_area_code_map.items())])
def map_phone_to_location(df: DataFrame) -> DataFrame:
_df = df.withColumn("location", mapping_expr[substring(col("phone"),2,3)])
return _df
map_phone_to_location(df).show()
# -
# Looking at the two code examples, we had to reimplement the exact same functionality with completely different syntax. This isn't a cherry-picked example. Data practitioners will often have to write two implementations of the same logic, one for each framework, especially as the logic gets more specific.
#
# This is where Fugue comes in. Users can use the abstraction layer to only write one implementation of the function. This can then be applied to pandas, Spark, and Dask. All we need to do is apply a `transformer` decorator to the pandas implementation of the function. The decorator takes in a string that specifies the output schema. The `transform` function does the same thing to the function that is passed in.
# +
from fugue import transformer
@transformer("*, location:str")
def map_phone_to_location(df: pd.DataFrame) -> pd.DataFrame:
df["location"] = df["phone"].str.slice(1,4).map(_area_code_map)
return df
# -
# By wrapping the function with the decorator, we can then use it inside a `FugueWorkflow`. The `FugueWorkflow` constructs a directed-acyclic graph (DAG) where the inputs and outputs are DataFrames. More details will follow in the next sections but the important thing for now is to show how it's used. The code block below is still running in Pandas.
# +
from fugue import FugueWorkflow
with FugueWorkflow() as dag:
df = dag.df(data.copy()) # Still the original Pandas DataFrame
df = df.transform(map_phone_to_location)
df.show()
# -
# In order to bring it to Spark, all we need to do is pass the `SparkExecutionEngine` into `FugueWorkflow`, similar to how we used the `transform` function to Spark in the last section. Now all the code underneath the `with` statement will run on Spark. We did not make any modifications to `map_phone_to_location` in order to bring it to Spark. By wrapping the function with a `transformer`, it became agnostic to the ExecutionEngine it was operating on. We can use the same function in Spark or Dask without making modifications.
# +
from fugue_spark import SparkExecutionEngine
with FugueWorkflow(SparkExecutionEngine) as dag:
df = dag.df(data.copy()) # Still the original Pandas DataFrame
df = df.transform(map_phone_to_location)
df.show()
# -
# ## `transform` versus `FugueWorkflow`
#
# We have seen the two approaches to bring Python and pandas code to Spark with Fugue. The `transform` function introduced in the first section allows users to leave a function in pandas or Python, and then port it to Spark and Dask. Meanwhile, `FugueWorkflow` does the same for full workflows as opposed to one function.
#
# For example, if we had five different functions to call `transform` on and bring to Spark, we would need to specify the `SparkExecutionEngine` five times. The `FugueWorkflow` allows us to make the entire computation run on either pandas, Spark, or Dask. Both are similar in principle, in that they leave the original functions decoupled to the execution environment.
# ## Independence from Frameworks
#
# We earlier said that the abstraction layer Fugue provides makes code independent of any framework. To show this is true, we can actually rewrite the `map_phone_to_location` function in native Python and still apply it on the pandas and Spark engines.
#
# Below is the implementation in native Python. Similar to earlier, we are running this on Spark by passing in the `SparkExecutionEngine`. A function written in native Python can be ported to pandas, Spark, and Dask.
# +
from typing import List, Dict, Any
# schema: *, location:str
def map_phone_to_location(df: List[Dict[str,Any]]) -> List[Dict[str,Any]]:
for row in df:
row["location"] = _area_code_map[row["phone"][1:4]]
return df
with FugueWorkflow(SparkExecutionEngine) as dag:
df = dag.df(data.copy()) # Still the original Pandas DataFrame
df = df.transform(map_phone_to_location)
df.show()
# -
# Notice the `@transformer` decorator was removed from `map_phone_to_location`. Instead, it was replaced with a comment that specified the schema. Fugue reads in this comment as the **schema hint**. Now, this function is truly independent of any framework and written in native Python. **It is even independent from Fugue itself.** Fugue only appears when we reach the execution part of the code. The logic, however, is not coupled with any framework. The type annotations in the `map_phone_to_location` caused the DataFrame to be converted as it was used by the function. If users want to offboard from Fugue, they can use their function with Pandas `apply` or Spark user-defined functions (UDFs).
#
# Is the native Python implementation or Pandas implementation of `map_phone_to_location` better? Is the native Spark implementation better?
#
# The main concern of Fugue is clear readable code. **Users can write code in whatever expresses their logic the best**. The compute efficiency lost by using Fugue is unlikely to be significant, especially in comparison to the developer efficiency gained through more rapid iterations and easier maintenance. In fact, Fugue is designed in a way that often sees speed ups compared to inexperienced users working with native Spark code. Fugue handles a lot of the tricks necessary to use Spark effectively.
#
# Fugue also future-proofs the code. If one day Spark and Dask are replaced by a more efficient framework, a new ExecutionEngine can be added to Fugue to support that new framework.
# ## Testability and Maintainability
#
# Fugue code becomes easily testable because the function contains logic that is portable across all pandas, Spark, and Dask. All we have to do is run some values through the defined function. We can test code without the need to spin up compute resources (such as Spark or Dask clusters). This hardware often takes time to spin up just for a simple test, making it painful to run unit tests on Spark. Now, we can test quickly with native Python or pandas, and then execute on Spark when needed. Developers that use Fugue benefit from more rapid iterations in their data projects.
# Remember the input was List[Dict[str,Any]]
map_phone_to_location([{'phone': '(407)-234-5678'},
{'phone': '(407)-234-5679'}])
# Even if the output here is a `List[Dict[str,Any]]`, Fugue takes care of converting it back to a DataFrame.
# ## Fugue as a Mindset
#
# Fugue is a framework, but more importantly, it is a mindset.
#
# 1. Fugue believes that the framework should adapt to the user, not the other way around
# 2. Fugue lets users code express logic in a scale-agnostic way, with the tools they prefer
# 3. Fugue values readability and maintainability of code over deep framework-specific optimizations
#
# Using distributed computing is currently harder than it needs to be. However, these systems often follow similar patterns, which have been abstracted to create a framework that lets users focus on defining their logic. We cover these concepts in the rest of tutorials. If you're new to distributed computing, Fugue is the perfect place to get started.
#
# ## [Optional] Comparison to Modin and Koalas
#
# Fugue gets compared a lot of Modin and Koalas. Modin is a pandas interface for execution on Dask, and Koalas is a pandas interface for execution on Spark. Fugue, Modin, and Koalas have similar goals in making an easier distributed computing experience. The main difference is that Modin and Koalas use pandas as the grammar for distributed compute. Fugue, on the other hand, uses native Python and SQL as the grammar for distributed compute (though pandas is also supported).
#
# The clearest example of pandas not being compatible with Spark is the acceptance of mixed-typed columns. A single column can have numeric and string values. Spark on the other hand, is strongly typed and enforces the schema. More than that, pandas is strongly reliant on the index for operations. As users transition to Spark, the index mindset does not hold as well. Order is not always guaranteed in a distributed system, there is an overhead to maintain a global index and moreover it is often not necessary.
|
tutorials/beginner/decoupling_logic_and_execution.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] deletable=true editable=true
# # Solving problems by Searching
#
# This notebook serves as supporting material for topics covered in **Chapter 3 - Solving Problems by Searching** and **Chapter 4 - Beyond Classical Search** from the book *Artificial Intelligence: A Modern Approach.* This notebook uses implementations from [search.py](https://github.com/aimacode/aima-python/blob/master/search.py) module. Let's start by importing everything from search module.
# + deletable=true editable=true
from search import *
# Needed to hide warnings in the matplotlib sections
import warnings
warnings.filterwarnings("ignore")
# + [markdown] deletable=true editable=true
# ## Review
#
# Here, we learn about problem solving. Building goal-based agents that can plan ahead to solve problems, in particular, navigation problem/route finding problem. First, we will start the problem solving by precisely defining **problems** and their **solutions**. We will look at several general-purpose search algorithms. Broadly, search algorithms are classified into two types:
#
# * **Uninformed search algorithms**: Search algorithms which explore the search space without having any information about the problem other than its definition.
# * Examples:
# 1. Breadth First Search
# 2. Depth First Search
# 3. Depth Limited Search
# 4. Iterative Deepening Search
#
#
# * **Informed search algorithms**: These type of algorithms leverage any information (heuristics, path cost) on the problem to search through the search space to find the solution efficiently.
# * Examples:
# 1. Best First Search
# 2. Uniform Cost Search
# 3. A\* Search
# 4. Recursive Best First Search
#
# *Don't miss the visualisations of these algorithms solving the route-finding problem defined on Romania map at the end of this notebook.*
# + [markdown] deletable=true editable=true
# ## Problem
#
# Let's see how we define a Problem. Run the next cell to see how abstract class `Problem` is defined in the search module.
# + deletable=true editable=true
# %psource Problem
# + [markdown] deletable=true editable=true
# The `Problem` class has six methods.
#
# * `__init__(self, initial, goal)` : This is what is called a `constructor` and is the first method called when you create an instance of the class. `initial` specifies the initial state of our search problem. It represents the start state from where our agent begins its task of exploration to find the goal state(s) which is given in the `goal` parameter.
#
#
# * `actions(self, state)` : This method returns all the possible actions agent can execute in the given state `state`.
#
#
# * `result(self, state, action)` : This returns the resulting state if action `action` is taken in the state `state`. This `Problem` class only deals with deterministic outcomes. So we know for sure what every action in a state would result to.
#
#
# * `goal_test(self, state)` : Given a graph state, it checks if it is a terminal state. If the state is indeed a goal state, value of `True` is returned. Else, of course, `False` is returned.
#
#
# * `path_cost(self, c, state1, action, state2)` : Return the cost of the path that arrives at `state2` as a result of taking `action` from `state1`, assuming total cost of `c` to get up to `state1`.
#
#
# * `value(self, state)` : This acts as a bit of extra information in problems where we try to optimise a value when we cannot do a goal test.
# + [markdown] deletable=true editable=true
# We will use the abstract class `Problem` to define our real **problem** named `GraphProblem`. You can see how we define `GraphProblem` by running the next cell.
# + deletable=true editable=true
# %psource GraphProblem
# + [markdown] deletable=true editable=true
# Now it's time to define our problem. We will define it by passing `initial`, `goal`, `graph` to `GraphProblem`. So, our problem is to find the goal state starting from the given initial state on the provided graph. Have a look at our romania_map, which is an Undirected Graph containing a dict of nodes as keys and neighbours as values.
# + deletable=true editable=true
romania_map = UndirectedGraph(dict(
Arad=dict(Zerind=75, Sibiu=140, Timisoara=118),
Bucharest=dict(Urziceni=85, Pitesti=101, Giurgiu=90, Fagaras=211),
Craiova=dict(Drobeta=120, Rimnicu=146, Pitesti=138),
Drobeta=dict(Mehadia=75),
Eforie=dict(Hirsova=86),
Fagaras=dict(Sibiu=99),
Hirsova=dict(Urziceni=98),
Iasi=dict(Vaslui=92, Neamt=87),
Lugoj=dict(Timisoara=111, Mehadia=70),
Oradea=dict(Zerind=71, Sibiu=151),
Pitesti=dict(Rimnicu=97),
Rimnicu=dict(Sibiu=80),
Urziceni=dict(Vaslui=142)))
romania_map.locations = dict(
Arad=(91, 492), Bucharest=(400, 327), Craiova=(253, 288),
Drobeta=(165, 299), Eforie=(562, 293), Fagaras=(305, 449),
Giurgiu=(375, 270), Hirsova=(534, 350), Iasi=(473, 506),
Lugoj=(165, 379), Mehadia=(168, 339), Neamt=(406, 537),
Oradea=(131, 571), Pitesti=(320, 368), Rimnicu=(233, 410),
Sibiu=(207, 457), Timisoara=(94, 410), Urziceni=(456, 350),
Vaslui=(509, 444), Zerind=(108, 531))
# + [markdown] deletable=true editable=true
# It is pretty straightforward to understand this `romania_map`. The first node **Arad** has three neighbours named **Zerind**, **Sibiu**, **Timisoara**. Each of these nodes are 75, 140, 118 units apart from **Arad** respectively. And the same goes with other nodes.
#
# And `romania_map.locations` contains the positions of each of the nodes. We will use the straight line distance (which is different from the one provided in `romania_map`) between two cities in algorithms like A\*-search and Recursive Best First Search.
#
# **Define a problem:**
# Hmm... say we want to start exploring from **Arad** and try to find **Bucharest** in our romania_map. So, this is how we do it.
# + deletable=true editable=true
romania_problem = GraphProblem('Arad', 'Bucharest', romania_map)
# + [markdown] deletable=true editable=true
# # Romania map visualisation
#
# Let's have a visualisation of Romania map [Figure 3.2] from the book and see how different searching algorithms perform / how frontier expands in each search algorithm for a simple problem named `romania_problem`.
# + [markdown] deletable=true editable=true
# Have a look at `romania_locations`. It is a dictionary defined in search module. We will use these location values to draw the romania graph using **networkx**.
# + deletable=true editable=true
romania_locations = romania_map.locations
print(romania_locations)
# + [markdown] deletable=true editable=true
# Let's start the visualisations by importing necessary modules. We use networkx and matplotlib to show the map in the notebook and we use ipywidgets to interact with the map to see how the searching algorithm works.
# + deletable=true editable=true
# %matplotlib inline
import networkx as nx
import matplotlib.pyplot as plt
from matplotlib import lines
from ipywidgets import interact
import ipywidgets as widgets
from IPython.display import display
import time
# + [markdown] deletable=true editable=true
# Let's get started by initializing an empty graph. We will add nodes, place the nodes in their location as shown in the book, add edges to the graph.
# + deletable=true editable=true
# initialise a graph
G = nx.Graph()
# use this while labeling nodes in the map
node_labels = dict()
# use this to modify colors of nodes while exploring the graph.
# This is the only dict we send to `show_map(node_colors)` while drawing the map
node_colors = dict()
for n, p in romania_locations.items():
# add nodes from romania_locations
G.add_node(n)
# add nodes to node_labels
node_labels[n] = n
# node_colors to color nodes while exploring romania map
node_colors[n] = "white"
# we'll save the initial node colors to a dict to use later
initial_node_colors = dict(node_colors)
# positions for node labels
node_label_pos = { k:[v[0],v[1]-10] for k,v in romania_locations.items() }
# use this while labeling edges
edge_labels = dict()
# add edges between cities in romania map - UndirectedGraph defined in search.py
for node in romania_map.nodes():
connections = romania_map.get(node)
for connection in connections.keys():
distance = connections[connection]
# add edges to the graph
G.add_edge(node, connection)
# add distances to edge_labels
edge_labels[(node, connection)] = distance
# + deletable=true editable=true
# initialise a graph
G = nx.Graph()
# use this while labeling nodes in the map
node_labels = dict()
# use this to modify colors of nodes while exploring the graph.
# This is the only dict we send to `show_map(node_colors)` while drawing the map
node_colors = dict()
for n, p in romania_locations.items():
# add nodes from romania_locations
G.add_node(n)
# add nodes to node_labels
node_labels[n] = n
# node_colors to color nodes while exploring romania map
node_colors[n] = "white"
# we'll save the initial node colors to a dict to use later
initial_node_colors = dict(node_colors)
# positions for node labels
node_label_pos = { k:[v[0],v[1]-10] for k,v in romania_locations.items() }
# use this while labeling edges
edge_labels = dict()
# add edges between cities in romania map - UndirectedGraph defined in search.py
for node in romania_map.nodes():
connections = romania_map.get(node)
for connection in connections.keys():
distance = connections[connection]
# add edges to the graph
G.add_edge(node, connection)
# add distances to edge_labels
edge_labels[(node, connection)] = distance
# + [markdown] deletable=true editable=true
# We have completed building our graph based on romania_map and its locations. It's time to display it here in the notebook. This function `show_map(node_colors)` helps us do that. We will be calling this function later on to display the map at each and every interval step while searching, using variety of algorithms from the book.
# + deletable=true editable=true
def show_map(node_colors):
# set the size of the plot
plt.figure(figsize=(18,13))
# draw the graph (both nodes and edges) with locations from romania_locations
nx.draw(G, pos = romania_locations, node_color = [node_colors[node] for node in G.nodes()])
# draw labels for nodes
node_label_handles = nx.draw_networkx_labels(G, pos = node_label_pos, labels = node_labels, font_size = 14)
# add a white bounding box behind the node labels
[label.set_bbox(dict(facecolor='white', edgecolor='none')) for label in node_label_handles.values()]
# add edge lables to the graph
nx.draw_networkx_edge_labels(G, pos = romania_locations, edge_labels=edge_labels, font_size = 14)
# add a legend
white_circle = lines.Line2D([], [], color="white", marker='o', markersize=15, markerfacecolor="white")
orange_circle = lines.Line2D([], [], color="orange", marker='o', markersize=15, markerfacecolor="orange")
red_circle = lines.Line2D([], [], color="red", marker='o', markersize=15, markerfacecolor="red")
gray_circle = lines.Line2D([], [], color="gray", marker='o', markersize=15, markerfacecolor="gray")
green_circle = lines.Line2D([], [], color="green", marker='o', markersize=15, markerfacecolor="green")
plt.legend((white_circle, orange_circle, red_circle, gray_circle, green_circle),
('Un-explored', 'Frontier', 'Currently Exploring', 'Explored', 'Final Solution'),
numpoints=1,prop={'size':16}, loc=(.8,.75))
# show the plot. No need to use in notebooks. nx.draw will show the graph itself.
plt.show()
# + [markdown] deletable=true editable=true
# We can simply call the function with node_colors dictionary object to display it.
# + deletable=true editable=true
show_map(node_colors)
# + [markdown] deletable=true editable=true
# Voila! You see, the romania map as shown in the Figure[3.2] in the book. Now, see how different searching algorithms perform with our problem statements.
# + [markdown] deletable=true editable=true
# ## Searching algorithms visualisations
#
# In this section, we have visualisations of the following searching algorithms:
#
# 1. Breadth First Tree Search - Implemented
# 2. Depth First Tree Search
# 3. Depth First Graph Search
# 4. Breadth First Search - Implemented
# 5. Best First Graph Search
# 6. Uniform Cost Search - Implemented
# 7. Depth Limited Search
# 8. Iterative Deepening Search
# 9. A\*-Search - Implemented
# 10. Recursive Best First Search
#
# We add the colors to the nodes to have a nice visualisation when displaying. So, these are the different colors we are using in these visuals:
# * Un-explored nodes - <font color='black'>white</font>
# * Frontier nodes - <font color='orange'>orange</font>
# * Currently exploring node - <font color='red'>red</font>
# * Already explored nodes - <font color='gray'>gray</font>
#
# Now, we will define some helper methods to display interactive buttons and sliders when visualising search algorithms.
# + deletable=true editable=true
def final_path_colors(problem, solution):
"returns a node_colors dict of the final path provided the problem and solution"
# get initial node colors
final_colors = dict(initial_node_colors)
# color all the nodes in solution and starting node to green
final_colors[problem.initial] = "green"
for node in solution:
final_colors[node] = "green"
return final_colors
def display_visual(user_input, algorithm=None, problem=None):
if user_input == False:
def slider_callback(iteration):
# don't show graph for the first time running the cell calling this function
try:
show_map(all_node_colors[iteration])
except:
pass
def visualize_callback(Visualize):
if Visualize is True:
button.value = False
global all_node_colors
iterations, all_node_colors, node = algorithm(problem)
solution = node.solution()
all_node_colors.append(final_path_colors(problem, solution))
slider.max = len(all_node_colors) - 1
for i in range(slider.max + 1):
slider.value = i
#time.sleep(.5)
slider = widgets.IntSlider(min=0, max=1, step=1, value=0)
slider_visual = widgets.interactive(slider_callback, iteration = slider)
display(slider_visual)
button = widgets.ToggleButton(value = False)
button_visual = widgets.interactive(visualize_callback, Visualize = button)
display(button_visual)
if user_input == True:
node_colors = dict(initial_node_colors)
if algorithm == None:
algorithms = {"Breadth First Tree Search": breadth_first_tree_search,
"Breadth First Search": breadth_first_search,
"Uniform Cost Search": uniform_cost_search,
"A-star Search": astar_search}
algo_dropdown = widgets.Dropdown(description = "Search algorithm: ",
options = sorted(list(algorithms.keys())),
value = "Breadth First Tree Search")
display(algo_dropdown)
def slider_callback(iteration):
# don't show graph for the first time running the cell calling this function
try:
show_map(all_node_colors[iteration])
except:
pass
def visualize_callback(Visualize):
if Visualize is True:
button.value = False
problem = GraphProblem(start_dropdown.value, end_dropdown.value, romania_map)
global all_node_colors
if algorithm == None:
user_algorithm = algorithms[algo_dropdown.value]
# print(user_algorithm)
# print(problem)
iterations, all_node_colors, node = user_algorithm(problem)
solution = node.solution()
all_node_colors.append(final_path_colors(problem, solution))
slider.max = len(all_node_colors) - 1
for i in range(slider.max + 1):
slider.value = i
# time.sleep(.5)
start_dropdown = widgets.Dropdown(description = "Start city: ",
options = sorted(list(node_colors.keys())), value = "Arad")
display(start_dropdown)
end_dropdown = widgets.Dropdown(description = "Goal city: ",
options = sorted(list(node_colors.keys())), value = "Fagaras")
display(end_dropdown)
button = widgets.ToggleButton(value = False)
button_visual = widgets.interactive(visualize_callback, Visualize = button)
display(button_visual)
slider = widgets.IntSlider(min=0, max=1, step=1, value=0)
slider_visual = widgets.interactive(slider_callback, iteration = slider)
display(slider_visual)
# + [markdown] deletable=true editable=true
#
# ## Breadth first tree search
#
# We have a working implementation in search module. But as we want to interact with the graph while it is searching, we need to modify the implementation. Here's the modified breadth first tree search.
#
#
# + deletable=true editable=true
def tree_search(problem, frontier):
"""Search through the successors of a problem to find a goal.
The argument frontier should be an empty queue.
Don't worry about repeated paths to a state. [Figure 3.7]"""
# we use these two variables at the time of visualisations
iterations = 0
all_node_colors = []
node_colors = dict(initial_node_colors)
#Adding first node to the queue
frontier.append(Node(problem.initial))
node_colors[Node(problem.initial).state] = "orange"
iterations += 1
all_node_colors.append(dict(node_colors))
while frontier:
#Popping first node of queue
node = frontier.pop()
# modify the currently searching node to red
node_colors[node.state] = "red"
iterations += 1
all_node_colors.append(dict(node_colors))
if problem.goal_test(node.state):
# modify goal node to green after reaching the goal
node_colors[node.state] = "green"
iterations += 1
all_node_colors.append(dict(node_colors))
return(iterations, all_node_colors, node)
frontier.extend(node.expand(problem))
for n in node.expand(problem):
node_colors[n.state] = "orange"
iterations += 1
all_node_colors.append(dict(node_colors))
# modify the color of explored nodes to gray
node_colors[node.state] = "gray"
iterations += 1
all_node_colors.append(dict(node_colors))
return None
def breadth_first_tree_search(problem):
"Search the shallowest nodes in the search tree first."
iterations, all_node_colors, node = tree_search(problem, FIFOQueue())
return(iterations, all_node_colors, node)
# + [markdown] deletable=true editable=true
# Now, we use ipywidgets to display a slider, a button and our romania map. By sliding the slider we can have a look at all the intermediate steps of a particular search algorithm. By pressing the button **Visualize**, you can see all the steps without interacting with the slider. These two helper functions are the callback functions which are called when we interact with the slider and the button.
#
#
# + deletable=true editable=true
all_node_colors = []
romania_problem = GraphProblem('Arad', 'Fagaras', romania_map)
display_visual(user_input = False, algorithm = breadth_first_tree_search, problem = romania_problem)
# + [markdown] deletable=true editable=true
# ## Breadth first search
#
# Let's change all the node_colors to starting position and define a different problem statement.
# + deletable=true editable=true
def breadth_first_search(problem):
"[Figure 3.11]"
# we use these two variables at the time of visualisations
iterations = 0
all_node_colors = []
node_colors = dict(initial_node_colors)
node = Node(problem.initial)
node_colors[node.state] = "red"
iterations += 1
all_node_colors.append(dict(node_colors))
if problem.goal_test(node.state):
node_colors[node.state] = "green"
iterations += 1
all_node_colors.append(dict(node_colors))
return(iterations, all_node_colors, node)
frontier = FIFOQueue()
frontier.append(node)
# modify the color of frontier nodes to blue
node_colors[node.state] = "orange"
iterations += 1
all_node_colors.append(dict(node_colors))
explored = set()
while frontier:
node = frontier.pop()
node_colors[node.state] = "red"
iterations += 1
all_node_colors.append(dict(node_colors))
explored.add(node.state)
for child in node.expand(problem):
if child.state not in explored and child not in frontier:
if problem.goal_test(child.state):
node_colors[child.state] = "green"
iterations += 1
all_node_colors.append(dict(node_colors))
return(iterations, all_node_colors, child)
frontier.append(child)
node_colors[child.state] = "orange"
iterations += 1
all_node_colors.append(dict(node_colors))
node_colors[node.state] = "gray"
iterations += 1
all_node_colors.append(dict(node_colors))
return None
# + deletable=true editable=true
all_node_colors = []
romania_problem = GraphProblem('Arad', 'Bucharest', romania_map)
display_visual(user_input = False, algorithm = breadth_first_search, problem = romania_problem)
# + [markdown] deletable=true editable=true
# ## Uniform cost search
#
# Let's change all the node_colors to starting position and define a different problem statement.
# + deletable=true editable=true
def best_first_graph_search(problem, f):
"""Search the nodes with the lowest f scores first.
You specify the function f(node) that you want to minimize; for example,
if f is a heuristic estimate to the goal, then we have greedy best
first search; if f is node.depth then we have breadth-first search.
There is a subtlety: the line "f = memoize(f, 'f')" means that the f
values will be cached on the nodes as they are computed. So after doing
a best first search you can examine the f values of the path returned."""
# we use these two variables at the time of visualisations
iterations = 0
all_node_colors = []
node_colors = dict(initial_node_colors)
f = memoize(f, 'f')
node = Node(problem.initial)
node_colors[node.state] = "red"
iterations += 1
all_node_colors.append(dict(node_colors))
if problem.goal_test(node.state):
node_colors[node.state] = "green"
iterations += 1
all_node_colors.append(dict(node_colors))
return(iterations, all_node_colors, node)
frontier = PriorityQueue(min, f)
frontier.append(node)
node_colors[node.state] = "orange"
iterations += 1
all_node_colors.append(dict(node_colors))
explored = set()
while frontier:
node = frontier.pop()
node_colors[node.state] = "red"
iterations += 1
all_node_colors.append(dict(node_colors))
if problem.goal_test(node.state):
node_colors[node.state] = "green"
iterations += 1
all_node_colors.append(dict(node_colors))
return(iterations, all_node_colors, node)
explored.add(node.state)
for child in node.expand(problem):
if child.state not in explored and child not in frontier:
frontier.append(child)
node_colors[child.state] = "orange"
iterations += 1
all_node_colors.append(dict(node_colors))
elif child in frontier:
incumbent = frontier[child]
if f(child) < f(incumbent):
del frontier[incumbent]
frontier.append(child)
node_colors[child.state] = "orange"
iterations += 1
all_node_colors.append(dict(node_colors))
node_colors[node.state] = "gray"
iterations += 1
all_node_colors.append(dict(node_colors))
return None
def uniform_cost_search(problem):
"[Figure 3.14]"
iterations, all_node_colors, node = best_first_graph_search(problem, lambda node: node.path_cost)
return(iterations, all_node_colors, node)
# + [markdown] deletable=true editable=true
# ## A* search
#
# Let's change all the node_colors to starting position and define a different problem statement.
# + deletable=true editable=true
all_node_colors = []
romania_problem = GraphProblem('Arad', 'Bucharest', romania_map)
display_visual(user_input = False, algorithm = uniform_cost_search, problem = romania_problem)
# + deletable=true editable=true
def best_first_graph_search(problem, f):
"""Search the nodes with the lowest f scores first.
You specify the function f(node) that you want to minimize; for example,
if f is a heuristic estimate to the goal, then we have greedy best
first search; if f is node.depth then we have breadth-first search.
There is a subtlety: the line "f = memoize(f, 'f')" means that the f
values will be cached on the nodes as they are computed. So after doing
a best first search you can examine the f values of the path returned."""
# we use these two variables at the time of visualisations
iterations = 0
all_node_colors = []
node_colors = dict(initial_node_colors)
f = memoize(f, 'f')
node = Node(problem.initial)
node_colors[node.state] = "red"
iterations += 1
all_node_colors.append(dict(node_colors))
if problem.goal_test(node.state):
node_colors[node.state] = "green"
iterations += 1
all_node_colors.append(dict(node_colors))
return(iterations, all_node_colors, node)
frontier = PriorityQueue(min, f)
frontier.append(node)
node_colors[node.state] = "orange"
iterations += 1
all_node_colors.append(dict(node_colors))
explored = set()
while frontier:
node = frontier.pop()
node_colors[node.state] = "red"
iterations += 1
all_node_colors.append(dict(node_colors))
if problem.goal_test(node.state):
node_colors[node.state] = "green"
iterations += 1
all_node_colors.append(dict(node_colors))
return(iterations, all_node_colors, node)
explored.add(node.state)
for child in node.expand(problem):
if child.state not in explored and child not in frontier:
frontier.append(child)
node_colors[child.state] = "orange"
iterations += 1
all_node_colors.append(dict(node_colors))
elif child in frontier:
incumbent = frontier[child]
if f(child) < f(incumbent):
del frontier[incumbent]
frontier.append(child)
node_colors[child.state] = "orange"
iterations += 1
all_node_colors.append(dict(node_colors))
node_colors[node.state] = "gray"
iterations += 1
all_node_colors.append(dict(node_colors))
return None
def astar_search(problem, h=None):
"""A* search is best-first graph search with f(n) = g(n)+h(n).
You need to specify the h function when you call astar_search, or
else in your Problem subclass."""
h = memoize(h or problem.h, 'h')
iterations, all_node_colors, node = best_first_graph_search(problem, lambda n: n.path_cost + h(n))
return(iterations, all_node_colors, node)
# + deletable=true editable=true
all_node_colors = []
romania_problem = GraphProblem('Arad', 'Bucharest', romania_map)
display_visual(user_input = False, algorithm = astar_search, problem = romania_problem)
# + deletable=true editable=true
all_node_colors = []
# display_visual(user_input = True, algorithm = breadth_first_tree_search)
display_visual(user_input = True)
# + [markdown] deletable=true editable=true
# ## Genetic Algorithm
#
# Genetic algorithms (or GA) are inspired by natural evolution and are particularly useful in optimization and search problems with large state spaces.
#
# Given a problem, algorithms in the domain make use of a *population* of solutions (also called *states*), where each solution/state represents a feasible solution. At each iteration (often called *generation*), the population gets updated using methods inspired by biology and evolution, like *crossover*, *mutation* and *selection*.
# + [markdown] deletable=true editable=true
# ### Overview
#
# A genetic algorithm works in the following way:
#
# 1) Initialize random population.
#
# 2) Calculate population fitness.
#
# 3) Select individuals for mating.
#
# 4) Mate selected individuals to produce new population.
#
# * Random chance to mutate individuals.
#
# 5) Repeat from step 2) until an individual is fit enough or the maximum number of iterations was reached.
# + [markdown] deletable=true editable=true
# ### Glossary
#
# Before we continue, we will lay the basic terminology of the algorithm.
#
# * Individual/State: A string of chars (called *genes*) that represent possible solutions.
#
# * Population: The list of all the individuals/states.
#
# * Gene pool: The alphabet of possible values for an individual's genes.
#
# * Generation/Iteration: The number of times the population will be updated.
#
# * Fitness: An individual's score, calculated by a function specific to the problem.
# + [markdown] deletable=true editable=true
# ### Crossover
#
# Two individuals/states can "mate" and produce one child. This offspring bears characteristics from both of its parents. There are many ways we can implement this crossover. Here we will take a look at the most common ones. Most other methods are variations of those below.
#
# * Point Crossover: The crossover occurs around one (or more) point. The parents get "split" at the chosen point or points and then get merged. In the example below we see two parents get split and merged at the 3rd digit, producing the following offspring after the crossover.
#
# 
#
# * Uniform Crossover: This type of crossover chooses randomly the genes to get merged. Here the genes 1, 2 and 5 where chosen from the first parent, so the genes 3, 4 will be added by the second parent.
#
# 
# + [markdown] deletable=true editable=true
# ### Mutation
#
# When an offspring is produced, there is a chance it will mutate, having one (or more, depending on the implementation) of its genes altered.
#
# For example, let's say the new individual to undergo mutation is "abcde". Randomly we pick to change its third gene to 'z'. The individual now becomes "ab<font color='red'>z</font>de" and is added to the population.
# + [markdown] deletable=true editable=true
# ### Selection
#
# At each iteration, the fittest individuals are picked randomly to mate and produce offsprings. We measure an individual's fitness with a *fitness function*. That function depends on the given problem and it is used to score an individual. Usually the higher the better.
#
# The selection process is this:
#
# 1) Individuals are scored by the fitness function.
#
# 2) Individuals are picked randomly, according to their score (higher score means higher chance to get picked). Usually the formula to calculate the chance to pick an individual is the following (for population *P* and individual *i*):
#
# $$ chance(i) = \dfrac{fitness(i)}{\sum\limits_{k \, in \, P}{fitness(k)}} $$
# + [markdown] deletable=true editable=true
# ### Implementation
#
# Below we look over the implementation of the algorithm in the `search` module.
#
# First the implementation of the main core of the algorithm:
# + deletable=true editable=true
# %psource genetic_algorithm
# + [markdown] deletable=true editable=true
# The algorithm takes the following input:
#
# * `population`: The initial population.
#
# * `fitness_fn`: The problem's fitness function.
#
# * `gene_pool`: The gene pool of the states/individuals. Genes need to be chars. By default '0' and '1'.
#
# * `f_thres`: The fitness threshold. If an individual reaches that score, iteration stops. By default 'None', which means the algorithm will try and find the optimal solution.
#
# * `ngen`: The number of iterations/generations.
#
# * `pmut`: The probability of mutation.
#
# The algorithm gives as output the state with the largest score.
# + [markdown] deletable=true editable=true
# For each generation, the algorithm updates the population. First it calculates the fitnesses of the individuals, then it selects the most fit ones and finally crosses them over to produce offsprings. There is a chance that the offspring will be mutated, given by `pmut`. If at the end of the generation an individual meets the fitness threshold, the algorithm halts and returns that individual.
#
# The function of mating is accomplished by the method `reproduce`:
# + deletable=true editable=true
def reproduce(x, y):
n = len(x)
c = random.randrange(0, n)
return x[:c] + y[c:]
# + [markdown] deletable=true editable=true
# The method picks at random a point and merges the parents (`x` and `y`) around it.
#
# The mutation is done in the method `mutate`:
# + deletable=true editable=true
def mutate(x, gene_pool):
n = len(x)
g = len(gene_pool)
c = random.randrange(0, n)
r = random.randrange(0, g)
new_gene = gene_pool[r]
return x[:c] + new_gene + x[c+1:]
# + [markdown] deletable=true editable=true
# We pick a gene in `x` to mutate and a gene from the gene pool to replace it with.
#
# To help initializing the population we have the helper function `init_population`":
# + deletable=true editable=true
def init_population(pop_number, gene_pool, state_length):
g = len(gene_pool)
population = []
for i in range(pop_number):
new_individual = ''.join([gene_pool[random.randrange(0, g)]
for j in range(state_length)])
population.append(new_individual)
return population
# + [markdown] deletable=true editable=true
# The function takes as input the number of individuals in the population, the gene pool and the length of each individual/state. It creates individuals with random genes and returns the population when done.
# + [markdown] deletable=true editable=true
# ### Usage
#
# Below we give two example usages for the genetic algorithm, for a graph coloring problem and the 8 queens problem.
#
# #### Graph Coloring
#
# First we will take on the simpler problem of coloring a small graph with two colors. Before we do anything, let's imagine how a solution might look. First, we have only two colors, so we can represent them with a binary notation: 0 for one color and 1 for the other. These make up our gene pool. What of the individual solutions though? For that, we will look at our problem. We stated we have a graph. A graph has nodes and edges, and we want to color the nodes. Naturally, we want to store each node's color. If we have four nodes, we can store their colors in a string of genes, one for each node. A possible solution will then look like this: "1100". In the general case, we will represent each solution with a string of 1s and 0s, with length the number of nodes.
#
# Next we need to come up with a fitness function that appropriately scores individuals. Again, we will look at the problem definition at hand. We want to color a graph. For a solution to be optimal, no edge should connect two nodes of the same color. How can we use this information to score a solution? A naive (and ineffective) approach would be to count the different colors in the string. So "1111" has a score of 1 and "1100" has a score of 2. Why that fitness function is not ideal though? Why, we forgot the information about the edges! The edges are pivotal to the problem and the above function only deals with node colors. We didn't use all the information at hand and ended up with an ineffective answer. How, then, can we use that information to our advantage?
#
# We said that the optimal solution will have all the edges connecting nodes of different color. So, to score a solution we can count how many edges are valid (aka connecting nodes of different color). That is a great fitness function!
#
# Let's jump into solving this problem using the `genetic_algorithm` function.
# + [markdown] deletable=true editable=true
# First we need to represent the graph. Since we mostly need information about edges, we will just store the edges. We will denote edges with capital letters and nodes with integers:
# + deletable=true editable=true
edges = {
'A': [0, 1],
'B': [0, 3],
'C': [1, 2],
'D': [2, 3]
}
# + [markdown] deletable=true editable=true
# Edge 'A' connects nodes 0 and 1, edge 'B' connects nodes 0 and 3 etc.
#
# We already said our gene pool is 0 and 1, so we can jump right into initializing our population. Since we have only four nodes, `state_length` should be 4. For the number of individuals, we will try 8. We can increase this number if we need higher accuracy, but be careful! Larger populations need more computating power and take longer. You need to strike that sweet balance between accuracy and cost (the ultimate dilemma of the programmer!).
# + deletable=true editable=true
population = init_population(8, ['0', '1'], 4)
print(population)
# + [markdown] deletable=true editable=true
# We created and printed the population. You can see that the genes in the individuals are random and there are 8 individuals each with 4 genes.
#
# Next we need to write our fitness function. We previously said we want the function to count how many edges are valid. So, given a coloring/individual `c`, we will do just that:
# + deletable=true editable=true
def fitness(c):
return sum(c[n1] != c[n2] for (n1, n2) in edges.values())
# + [markdown] deletable=true editable=true
# Great! Now we will run the genetic algorithm and see what solution it gives.
# + deletable=true editable=true
solution = genetic_algorithm(population, fitness)
print(solution)
# + [markdown] deletable=true editable=true
# The algorithm converged to a solution. Let's check its score:
# + deletable=true editable=true
print(fitness(solution))
# + [markdown] deletable=true editable=true
# The solution has a score of 4. Which means it is optimal, since we have exactly 4 edges in our graph, meaning all are valid!
#
# *NOTE: Because the algorithm is non-deterministic, there is a chance a different solution is given. It might even be wrong, if we are very unlucky!*
# + [markdown] deletable=true editable=true
# #### Eight Queens
#
# Let's take a look at a more complicated problem.
#
# In the *Eight Queens* problem, we are tasked with placing eight queens on an 8x8 chessboard without any queen threatening the others (aka queens should not be in the same row, column or diagonal). In its general form the problem is defined as placing *N* queens in an NxN chessboard without any conflicts.
#
# First we need to think about the representation of each solution. We can go the naive route of representing the whole chessboard with the queens' placements on it. That is definitely one way to go about it, but for the purpose of this tutorial we will do something different. We have eight queens, so we will have a gene for each of them. The gene pool will be numbers from 0 to 7, for the different columns. The *position* of the gene in the state will denote the row the particular queen is placed in.
#
# For example, we can have the state "03304577". Here the first gene with a value of 0 means "the queen at row 0 is placed at column 0", for the second gene "the queen at row 1 is placed at column 3" and so forth.
#
# We now need to think about the fitness function. On the graph coloring problem we counted the valid edges. The same thought process can be applied here. Instead of edges though, we have positioning between queens. If two queens are not threatening each other, we say they are at a "non-attacking" positioning. We can, therefore, count how many such positionings are there.
#
# Let's dive right in and initialize our population:
# + deletable=true editable=true
population = init_population(100, [str(i) for i in range(8)], 8)
print(population[:5])
# + [markdown] deletable=true editable=true
# We have a population of 100 and each individual has 8 genes. The gene pool is the integers from 0 to 7, in string form. Above you can see the first five individuals.
#
# Next we need to write our fitness function. Remember, queens threaten each other if they are at the same row, column or diagonal.
#
# Since positionings are mutual, we must take care not to count them twice. Therefore for each queen, we will only check for conflicts for the queens after her.
#
# A gene's value in an individual `q` denotes the queen's column, and the position of the gene denotes its row. We can check if the aforementioned values between two genes are the same. We also need to check for diagonals. A queen *a* is in the diagonal of another queen, *b*, if the difference of the rows between them is equal to either their difference in columns (for the diagonal on the right of *a*) or equal to the negative difference of their columns (for the left diagonal of *a*). Below is given the fitness function.
# + deletable=true editable=true
def fitness(q):
non_attacking = 0
for row1 in range(len(q)):
for row2 in range(row1+1, len(q)):
col1 = int(q[row1])
col2 = int(q[row2])
row_diff = row1 - row2
col_diff = col1 - col2
if col1 != col2 and row_diff != col_diff and row_diff != -col_diff:
non_attacking += 1
return non_attacking
# + [markdown] deletable=true editable=true
# Note that the best score achievable is 28. That is because for each queen we only check for the queens after her. For the first queen we check 7 other queens, for the second queen 6 others and so on. In short, the number of checks we make is the sum 7+6+5+...+1. Which is equal to 7\*(7+1)/2 = 28.
#
# Because it is very hard and will take long to find a perfect solution, we will set the fitness threshold at 25. If we find an individual with a score greater or equal to that, we will halt. Let's see how the genetic algorithm will fare.
# + deletable=true editable=true
solution = genetic_algorithm(population, fitness, f_thres=25)
print(solution)
print(fitness(solution))
# + [markdown] deletable=true editable=true
# Above you can see the solution and its fitness score, which should be no less than 25.
# + [markdown] deletable=true editable=true
# With that this tutorial on the genetic algorithm comes to an end. Hope you found this guide helpful!
|
search.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Programming Exercise 4: Neural Networks Learning
#
# ## Introduction
#
# In this exercise, you will implement the backpropagation algorithm for neural networks and apply it to the task of hand-written digit recognition. Before starting on the programming exercise, we strongly recommend watching the video lectures and completing the review questions for the associated topics.
#
#
# All the information you need for solving this assignment is in this notebook, and all the code you will be implementing will take place within this notebook. The assignment can be promptly submitted to the coursera grader directly from this notebook (code and instructions are included below).
#
# Before we begin with the exercises, we need to import all libraries required for this programming exercise. Throughout the course, we will be using [`numpy`](http://www.numpy.org/) for all arrays and matrix operations, [`matplotlib`](https://matplotlib.org/) for plotting, and [`scipy`](https://docs.scipy.org/doc/scipy/reference/) for scientific and numerical computation functions and tools. You can find instructions on how to install required libraries in the README file in the [github repository](https://github.com/dibgerge/ml-coursera-python-assignments).
# +
# used for manipulating directory paths
import os
# Scientific and vector computation for python
import numpy as np
# Plotting library
from matplotlib import pyplot
# Optimization module in scipy
from scipy import optimize
# will be used to load MATLAB mat datafile format
from scipy.io import loadmat
# library written for this exercise providing additional functions for assignment submission, and others
import utils
# define the submission/grader object for this exercise
grader = utils.Grader()
# tells matplotlib to embed plots within the notebook
# %matplotlib inline
# -
# ## Submission and Grading
#
#
# After completing each part of the assignment, be sure to submit your solutions to the grader. The following is a breakdown of how each part of this exercise is scored.
#
#
# | Section | Part | Submission function | Points
# | :- |:- | :- | :-:
# | 1 | [Feedforward and Cost Function](#section1) | [`nnCostFunction`](#nnCostFunction) | 30
# | 2 | [Regularized Cost Function](#section2) | [`nnCostFunction`](#nnCostFunction) | 15
# | 3 | [Sigmoid Gradient](#section3) | [`sigmoidGradient`](#sigmoidGradient) | 5
# | 4 | [Neural Net Gradient Function (Backpropagation)](#section4) | [`nnCostFunction`](#nnCostFunction) | 40
# | 5 | [Regularized Gradient](#section5) | [`nnCostFunction`](#nnCostFunction) |10
# | | Total Points | | 100
#
#
# You are allowed to submit your solutions multiple times, and we will take only the highest score into consideration.
#
# <div class="alert alert-block alert-warning">
# At the end of each section in this notebook, we have a cell which contains code for submitting the solutions thus far to the grader. Execute the cell to see your score up to the current section. For all your work to be submitted properly, you must execute those cells at least once.
# </div>
# ## Neural Networks
#
# In the previous exercise, you implemented feedforward propagation for neural networks and used it to predict handwritten digits with the weights we provided. In this exercise, you will implement the backpropagation algorithm to learn the parameters for the neural network.
#
# We start the exercise by first loading the dataset.
# +
# training data stored in arrays X, y
data = loadmat(os.path.join('Data', 'ex4data1.mat'))
X, y = data['X'], data['y'].ravel()
# set the zero digit to 0, rather than its mapped 10 in this dataset
# This is an artifact due to the fact that this dataset was used in
# MATLAB where there is no index 0
y[y == 10] = 0
# Number of training examples
m = y.size
# -
# ### 1.1 Visualizing the data
#
# You will begin by visualizing a subset of the training set, using the function `displayData`, which is the same function we used in Exercise 3. It is provided in the `utils.py` file for this assignment as well. The dataset is also the same one you used in the previous exercise.
#
# There are 5000 training examples in `ex4data1.mat`, where each training example is a 20 pixel by 20 pixel grayscale image of the digit. Each pixel is represented by a floating point number indicating the grayscale intensity at that location. The 20 by 20 grid of pixels is “unrolled” into a 400-dimensional vector. Each
# of these training examples becomes a single row in our data matrix $X$. This gives us a 5000 by 400 matrix $X$ where every row is a training example for a handwritten digit image.
#
# $$ X = \begin{bmatrix} - \left(x^{(1)} \right)^T - \\
# - \left(x^{(2)} \right)^T - \\
# \vdots \\
# - \left(x^{(m)} \right)^T - \\
# \end{bmatrix}
# $$
#
# The second part of the training set is a 5000-dimensional vector `y` that contains labels for the training set.
# The following cell randomly selects 100 images from the dataset and plots them.
# +
# Randomly select 100 data points to display
rand_indices = np.random.choice(m, 100, replace=False)
sel = X[rand_indices, :]
utils.displayData(sel)
# -
# ### 1.2 Model representation
#
# Our neural network is shown in the following figure.
#
# 
#
# It has 3 layers - an input layer, a hidden layer and an output layer. Recall that our inputs are pixel values
# of digit images. Since the images are of size $20 \times 20$, this gives us 400 input layer units (not counting the extra bias unit which always outputs +1). The training data was loaded into the variables `X` and `y` above.
#
# You have been provided with a set of network parameters ($\Theta^{(1)}, \Theta^{(2)}$) already trained by us. These are stored in `ex4weights.mat` and will be loaded in the next cell of this notebook into `Theta1` and `Theta2`. The parameters have dimensions that are sized for a neural network with 25 units in the second layer and 10 output units (corresponding to the 10 digit classes).
# +
# Setup the parameters you will use for this exercise
input_layer_size = 400 # 20x20 Input Images of Digits
hidden_layer_size = 25 # 25 hidden units
num_labels = 10 # 10 labels, from 0 to 9
# Load the weights into variables Theta1 and Theta2
weights = loadmat(os.path.join('Data', 'ex4weights.mat'))
# Theta1 has size 25 x 401
# Theta2 has size 10 x 26
Theta1, Theta2 = weights['Theta1'], weights['Theta2']
# swap first and last columns of Theta2, due to legacy from MATLAB indexing,
# since the weight file ex3weights.mat was saved based on MATLAB indexing
Theta2 = np.roll(Theta2, 1, axis=0)
# Unroll parameters
nn_params = np.concatenate([Theta1.ravel(), Theta2.ravel()])
# -
# <a id="section1"></a>
# ### 1.3 Feedforward and cost function
#
# Now you will implement the cost function and gradient for the neural network. First, complete the code for the function `nnCostFunction` in the next cell to return the cost.
#
# Recall that the cost function for the neural network (without regularization) is:
#
# $$ J(\theta) = \frac{1}{m} \sum_{i=1}^{m}\sum_{k=1}^{K} \left[ - y_k^{(i)} \log \left( \left( h_\theta \left( x^{(i)} \right) \right)_k \right) - \left( 1 - y_k^{(i)} \right) \log \left( 1 - \left( h_\theta \left( x^{(i)} \right) \right)_k \right) \right]$$
#
# where $h_\theta \left( x^{(i)} \right)$ is computed as shown in the neural network figure above, and K = 10 is the total number of possible labels. Note that $h_\theta(x^{(i)})_k = a_k^{(3)}$ is the activation (output
# value) of the $k^{th}$ output unit. Also, recall that whereas the original labels (in the variable y) were 0, 1, ..., 9, for the purpose of training a neural network, we need to encode the labels as vectors containing only values 0 or 1, so that
#
# $$ y =
# \begin{bmatrix} 1 \\ 0 \\ 0 \\\vdots \\ 0 \end{bmatrix}, \quad
# \begin{bmatrix} 0 \\ 1 \\ 0 \\ \vdots \\ 0 \end{bmatrix}, \quad \cdots \quad \text{or} \qquad
# \begin{bmatrix} 0 \\ 0 \\ 0 \\ \vdots \\ 1 \end{bmatrix}.
# $$
#
# For example, if $x^{(i)}$ is an image of the digit 5, then the corresponding $y^{(i)}$ (that you should use with the cost function) should be a 10-dimensional vector with $y_5 = 1$, and the other elements equal to 0.
#
# You should implement the feedforward computation that computes $h_\theta(x^{(i)})$ for every example $i$ and sum the cost over all examples. **Your code should also work for a dataset of any size, with any number of labels** (you can assume that there are always at least $K \ge 3$ labels).
#
# <div class="alert alert-box alert-warning">
# **Implementation Note:** The matrix $X$ contains the examples in rows (i.e., X[i,:] is the i-th training example $x^{(i)}$, expressed as a $n \times 1$ vector.) When you complete the code in `nnCostFunction`, you will need to add the column of 1’s to the X matrix. The parameters for each unit in the neural network is represented in Theta1 and Theta2 as one row. Specifically, the first row of Theta1 corresponds to the first hidden unit in the second layer. You can use a for-loop over the examples to compute the cost.
# </div>
# <a id="nnCostFunction"></a>
# +
def nnCostFunction(nn_params,
input_layer_size,
hidden_layer_size,
num_labels,
X, y, lambda_=0.0):
"""
Implements the neural network cost function and gradient for a two layer neural
network which performs classification.
Parameters
----------
nn_params : array_like
The parameters for the neural network which are "unrolled" into
a vector. This needs to be converted back into the weight matrices Theta1
and Theta2.
input_layer_size : int
Number of features for the input layer.
hidden_layer_size : int
Number of hidden units in the second layer.
num_labels : int
Total number of labels, or equivalently number of units in output layer.
X : array_like
Input dataset. A matrix of shape (m x input_layer_size).
y : array_like
Dataset labels. A vector of shape (m,).
lambda_ : float, optional
Regularization parameter.
Returns
-------
J : float
The computed value for the cost function at the current weight values.
grad : array_like
An "unrolled" vector of the partial derivatives of the concatenatation of
neural network weights Theta1 and Theta2.
Instructions
------------
You should complete the code by working through the following parts.
- Part 1: Feedforward the neural network and return the cost in the
variable J. After implementing Part 1, you can verify that your
cost function computation is correct by verifying the cost
computed in the following cell.
- Part 2: Implement the backpropagation algorithm to compute the gradients
Theta1_grad and Theta2_grad. You should return the partial derivatives of
the cost function with respect to Theta1 and Theta2 in Theta1_grad and
Theta2_grad, respectively. After implementing Part 2, you can check
that your implementation is correct by running checkNNGradients provided
in the utils.py module.
Note: The vector y passed into the function is a vector of labels
containing values from 0..K-1. You need to map this vector into a
binary vector of 1's and 0's to be used with the neural network
cost function.
Hint: We recommend implementing backpropagation using a for-loop
over the training examples if you are implementing it for the
first time.
- Part 3: Implement regularization with the cost function and gradients.
Hint: You can implement this around the code for
backpropagation. That is, you can compute the gradients for
the regularization separately and then add them to Theta1_grad
and Theta2_grad from Part 2.
Note
----
We have provided an implementation for the sigmoid function in the file
`utils.py` accompanying this assignment.
"""
# Reshape nn_params back into the parameters Theta1 and Theta2, the weight matrices
# for our 2 layer neural network
Theta1 = np.reshape(nn_params[:hidden_layer_size * (input_layer_size + 1)],
(hidden_layer_size, (input_layer_size + 1)))
Theta2 = np.reshape(nn_params[(hidden_layer_size * (input_layer_size + 1)):],
(num_labels, (hidden_layer_size + 1)))
# Setup some useful variables
m = y.size
# You need to return the following variables correctly
J = 0
Theta1_grad = np.zeros(Theta1.shape)
Theta2_grad = np.zeros(Theta2.shape)
# ====================== YOUR CODE HERE ======================
# print('input_layer_size', input_layer_size)
# print('hidden_layer_size', hidden_layer_size)
# print('num_labels', num_labels)
# Add ones to the X data matrix
X = np.concatenate([np.ones((m, 1)), X], axis=1)
# Encoding y
# > recall that whereas the original labels (in the variable y) were 0, 1, ..., 9,
# > for the purpose of training a neural network, we need to encode the labels as
# > vectors containing only values 0 or 1
y_encoded = np.zeros((y.size, num_labels)) # y_encoded will be of size m x k
y_encoded[np.arange(y.size), y] = 1
Z2 = Theta1 @ X.T
A2 = utils.sigmoid(Z2).T
A2 = np.concatenate([np.ones((A2.shape[0], 1)), A2], axis=1) # add bias term to A2 (hidden layer units)
Z3 = Theta2 @ A2.T
A3 = utils.sigmoid(Z3).T # hypothesis
hyp = A3
for i in range(m):
J += (y_encoded[i] @ np.log(hyp[i]) + (1 - y_encoded[i]) @ np.log(1 - hyp[i]))
regularization = (lambda_ / (2 * m)) * (np.sum(Theta1[:,1:] ** 2) + np.sum(Theta2[:,1:] ** 2))
J = - (1 / m) * J + regularization
for t in range(m):
a1 = X[t]
z2 = Theta1 @ a1
a2 = utils.sigmoid(z2)
a2 = np.concatenate([[1], a2])
z3 = Theta2 @ a2
a3 = utils.sigmoid(z3) # hypothesis
delta_3 = a3 - y_encoded[t]
delta_2 = (Theta2.T @ delta_3) * (a2 * (1 - a2))
delta_2 = delta_2[1:]
Theta2_grad = Theta2_grad + delta_3.reshape(-1, 1) @ a2.reshape(-1, 1).T
Theta1_grad = Theta1_grad + delta_2.reshape(-1, 1) @ a1.reshape(-1, 1).T
Theta2_regularization = (lambda_ / m) * Theta2[:, 1:]
Theta2_regularization = np.hstack((np.zeros((Theta2.shape[0], 1)), Theta2_regularization))
Theta1_regularization = (lambda_ / m) * Theta1[:, 1:]
Theta1_regularization = np.hstack((np.zeros((Theta1.shape[0], 1)), Theta1_regularization))
Theta2_grad = (1 / m) * Theta2_grad + Theta2_regularization
Theta1_grad = (1 / m) * Theta1_grad + Theta1_regularization
# ================================================================
# Unroll gradients
# grad = np.concatenate([Theta1_grad.ravel(order=order), Theta2_grad.ravel(order=order)])
grad = np.concatenate([Theta1_grad.ravel(), Theta2_grad.ravel()])
return J, grad
# -
# <div class="alert alert-box alert-warning">
# Use the following links to go back to the different parts of this exercise that require to modify the function `nnCostFunction`.<br>
#
# Back to:
# - [Feedforward and cost function](#section1)
# - [Regularized cost](#section2)
# - [Neural Network Gradient (Backpropagation)](#section4)
# - [Regularized Gradient](#section5)
# </div>
# Once you are done, call your `nnCostFunction` using the loaded set of parameters for `Theta1` and `Theta2`. You should see that the cost is about 0.287629.
lambda_ = 0
J, _ = nnCostFunction(nn_params, input_layer_size, hidden_layer_size,
num_labels, X, y, lambda_)
print('Cost at parameters (loaded from ex4weights): %.6f ' % J)
print('The cost should be about : 0.287629.')
# *You should now submit your solutions.*
grader = utils.Grader()
grader[1] = nnCostFunction
grader.grade()
# <a id="section2"></a>
# ### 1.4 Regularized cost function
#
# The cost function for neural networks with regularization is given by:
#
#
# $$ J(\theta) = \frac{1}{m} \sum_{i=1}^{m}\sum_{k=1}^{K} \left[ - y_k^{(i)} \log \left( \left( h_\theta \left( x^{(i)} \right) \right)_k \right) - \left( 1 - y_k^{(i)} \right) \log \left( 1 - \left( h_\theta \left( x^{(i)} \right) \right)_k \right) \right] + \frac{\lambda}{2 m} \left[ \sum_{j=1}^{25} \sum_{k=1}^{400} \left( \Theta_{j,k}^{(1)} \right)^2 + \sum_{j=1}^{10} \sum_{k=1}^{25} \left( \Theta_{j,k}^{(2)} \right)^2 \right] $$
#
# You can assume that the neural network will only have 3 layers - an input layer, a hidden layer and an output layer. However, your code should work for any number of input units, hidden units and outputs units. While we
# have explicitly listed the indices above for $\Theta^{(1)}$ and $\Theta^{(2)}$ for clarity, do note that your code should in general work with $\Theta^{(1)}$ and $\Theta^{(2)}$ of any size. Note that you should not be regularizing the terms that correspond to the bias. For the matrices `Theta1` and `Theta2`, this corresponds to the first column of each matrix. You should now add regularization to your cost function. Notice that you can first compute the unregularized cost function $J$ using your existing `nnCostFunction` and then later add the cost for the regularization terms.
#
# [Click here to go back to `nnCostFunction` for editing.](#nnCostFunction)
# Once you are done, the next cell will call your `nnCostFunction` using the loaded set of parameters for `Theta1` and `Theta2`, and $\lambda = 1$. You should see that the cost is about 0.383770.
# +
# Weight regularization parameter (we set this to 1 here).
lambda_ = 1
J, _ = nnCostFunction(nn_params, input_layer_size, hidden_layer_size,
num_labels, X, y, lambda_)
print('Cost at parameters (loaded from ex4weights): %.6f' % J)
print('This value should be about : 0.383770.')
# -
# *You should now submit your solutions.*
grader[2] = nnCostFunction
grader.grade()
# ## 2 Backpropagation
#
# In this part of the exercise, you will implement the backpropagation algorithm to compute the gradient for the neural network cost function. You will need to update the function `nnCostFunction` so that it returns an appropriate value for `grad`. Once you have computed the gradient, you will be able to train the neural network by minimizing the cost function $J(\theta)$ using an advanced optimizer such as `scipy`'s `optimize.minimize`.
# You will first implement the backpropagation algorithm to compute the gradients for the parameters for the (unregularized) neural network. After you have verified that your gradient computation for the unregularized case is correct, you will implement the gradient for the regularized neural network.
# <a id="section3"></a>
# ### 2.1 Sigmoid Gradient
#
# To help you get started with this part of the exercise, you will first implement
# the sigmoid gradient function. The gradient for the sigmoid function can be
# computed as
#
# $$ g'(z) = \frac{d}{dz} g(z) = g(z)\left(1-g(z)\right) $$
#
# where
#
# $$ \text{sigmoid}(z) = g(z) = \frac{1}{1 + e^{-z}} $$
#
# Now complete the implementation of `sigmoidGradient` in the next cell.
# <a id="sigmoidGradient"></a>
def sigmoidGradient(z):
"""
Computes the gradient of the sigmoid function evaluated at z.
This should work regardless if z is a matrix or a vector.
In particular, if z is a vector or matrix, you should return
the gradient for each element.
Parameters
----------
z : array_like
A vector or matrix as input to the sigmoid function.
Returns
--------
g : array_like
Gradient of the sigmoid function. Has the same shape as z.
Instructions
------------
Compute the gradient of the sigmoid function evaluated at
each value of z (z can be a matrix, vector or scalar).
Note
----
We have provided an implementation of the sigmoid function
in `utils.py` file accompanying this assignment.
"""
g = np.zeros(z.shape)
# ====================== YOUR CODE HERE ======================
g = utils.sigmoid(z) * (1 - utils.sigmoid(z))
# =============================================================
return g
# When you are done, the following cell call `sigmoidGradient` on a given vector `z`. Try testing a few values by calling `sigmoidGradient(z)`. For large values (both positive and negative) of z, the gradient should be close to 0. When $z = 0$, the gradient should be exactly 0.25. Your code should also work with vectors and matrices. For a matrix, your function should perform the sigmoid gradient function on every element.
z = np.array([-1, -0.5, 0, 0.5, 1])
g = sigmoidGradient(z)
print('Sigmoid gradient evaluated at [-1 -0.5 0 0.5 1]:\n ')
print(g)
# *You should now submit your solutions.*
grader[3] = sigmoidGradient
grader.grade()
# ## 2.2 Random Initialization
#
# When training neural networks, it is important to randomly initialize the parameters for symmetry breaking. One effective strategy for random initialization is to randomly select values for $\Theta^{(l)}$ uniformly in the range $[-\epsilon_{init}, \epsilon_{init}]$. You should use $\epsilon_{init} = 0.12$. This range of values ensures that the parameters are kept small and makes the learning more efficient.
#
# <div class="alert alert-box alert-warning">
# One effective strategy for choosing $\epsilon_{init}$ is to base it on the number of units in the network. A good choice of $\epsilon_{init}$ is $\epsilon_{init} = \frac{\sqrt{6}}{\sqrt{L_{in} + L_{out}}}$ where $L_{in} = s_l$ and $L_{out} = s_{l+1}$ are the number of units in the layers adjacent to $\Theta^{l}$.
# </div>
#
# Your job is to complete the function `randInitializeWeights` to initialize the weights for $\Theta$. Modify the function by filling in the following code:
#
# ```python
# # Randomly initialize the weights to small values
# W = np.random.rand(L_out, 1 + L_in) * 2 * epsilon_init - epsilon_init
# ```
# Note that we give the function an argument for $\epsilon$ with default value `epsilon_init = 0.12`.
def randInitializeWeights(L_in, L_out, epsilon_init=0.12):
"""
Randomly initialize the weights of a layer in a neural network.
Parameters
----------
L_in : int
Number of incomming connections.
L_out : int
Number of outgoing connections.
epsilon_init : float, optional
Range of values which the weight can take from a uniform
distribution.
Returns
-------
W : array_like
The weight initialiatized to random values. Note that W should
be set to a matrix of size(L_out, 1 + L_in) as
the first column of W handles the "bias" terms.
Instructions
------------
Initialize W randomly so that we break the symmetry while training
the neural network. Note that the first column of W corresponds
to the parameters for the bias unit.
"""
# You need to return the following variables correctly
W = np.zeros((L_out, 1 + L_in))
# ====================== YOUR CODE HERE ======================
# Randomly initialize the weights to small values
W = np.random.rand(L_out, 1 + L_in) * 2 * epsilon_init - epsilon_init
# ============================================================
return W
# *You do not need to submit any code for this part of the exercise.*
#
# Execute the following cell to initialize the weights for the 2 layers in the neural network using the `randInitializeWeights` function.
# +
print('Initializing Neural Network Parameters ...')
initial_Theta1 = randInitializeWeights(input_layer_size, hidden_layer_size)
initial_Theta2 = randInitializeWeights(hidden_layer_size, num_labels)
# Unroll parameters
initial_nn_params = np.concatenate([initial_Theta1.ravel(), initial_Theta2.ravel()], axis=0)
# -
# <a id="section4"></a>
# ### 2.4 Backpropagation
#
# 
#
# Now, you will implement the backpropagation algorithm. Recall that the intuition behind the backpropagation algorithm is as follows. Given a training example $(x^{(t)}, y^{(t)})$, we will first run a “forward pass” to compute all the activations throughout the network, including the output value of the hypothesis $h_\theta(x)$. Then, for each node $j$ in layer $l$, we would like to compute an “error term” $\delta_j^{(l)}$ that measures how much that node was “responsible” for any errors in our output.
#
# For an output node, we can directly measure the difference between the network’s activation and the true target value, and use that to define $\delta_j^{(3)}$ (since layer 3 is the output layer). For the hidden units, you will compute $\delta_j^{(l)}$ based on a weighted average of the error terms of the nodes in layer $(l+1)$. In detail, here is the backpropagation algorithm (also depicted in the figure above). You should implement steps 1 to 4 in a loop that processes one example at a time. Concretely, you should implement a for-loop `for t in range(m)` and place steps 1-4 below inside the for-loop, with the $t^{th}$ iteration performing the calculation on the $t^{th}$ training example $(x^{(t)}, y^{(t)})$. Step 5 will divide the accumulated gradients by $m$ to obtain the gradients for the neural network cost function.
#
# 1. Set the input layer’s values $(a^{(1)})$ to the $t^{th }$training example $x^{(t)}$. Perform a feedforward pass, computing the activations $(z^{(2)}, a^{(2)}, z^{(3)}, a^{(3)})$ for layers 2 and 3. Note that you need to add a `+1` term to ensure that the vectors of activations for layers $a^{(1)}$ and $a^{(2)}$ also include the bias unit. In `numpy`, if a 1 is a column matrix, adding one corresponds to `a_1 = np.concatenate([np.ones((m, 1)), a_1], axis=1)`.
#
# 1. For each output unit $k$ in layer 3 (the output layer), set
# $$\delta_k^{(3)} = \left(a_k^{(3)} - y_k \right)$$
# where $y_k \in \{0, 1\}$ indicates whether the current training example belongs to class $k$ $(y_k = 1)$, or if it belongs to a different class $(y_k = 0)$. You may find logical arrays helpful for this task (explained in the previous programming exercise).
#
# 1. For the hidden layer $l = 2$, set
# $$ \delta^{(2)} = \left( \Theta^{(2)} \right)^T \delta^{(3)} * g'\left(z^{(2)} \right)$$
# Note that the symbol $*$ performs element wise multiplication in `numpy`.
#
# 1. Accumulate the gradient from this example using the following formula. Note that you should skip or remove $\delta_0^{(2)}$. In `numpy`, removing $\delta_0^{(2)}$ corresponds to `delta_2 = delta_2[1:]`.
# $$ \Delta^{(l)} = \Delta^{(l)} + \delta^{(l+1)} (a^{(l)})^{(T)} $$
#
# 1. Obtain the (unregularized) gradient for the neural network cost function by dividing the accumulated gradients by $\frac{1}{m}$:
# $$ \frac{\partial}{\partial \Theta_{ij}^{(l)}} J(\Theta) = D_{ij}^{(l)} = \frac{1}{m} \Delta_{ij}^{(l)}$$
#
# <div class="alert alert-box alert-warning">
# **Python/Numpy tip**: You should implement the backpropagation algorithm only after you have successfully completed the feedforward and cost functions. While implementing the backpropagation alogrithm, it is often useful to use the `shape` function to print out the shapes of the variables you are working with if you run into dimension mismatch errors.
# </div>
#
# [Click here to go back and update the function `nnCostFunction` with the backpropagation algorithm](#nnCostFunction).
#
#
# **Note:** If the iterative solution provided above is proving to be difficult to implement, try implementing the vectorized approach which is easier to implement in the opinion of the moderators of this course. You can find the tutorial for the vectorized approach [here](https://www.coursera.org/learn/machine-learning/discussions/all/threads/a8Kce_WxEeS16yIACyoj1Q).
# After you have implemented the backpropagation algorithm, we will proceed to run gradient checking on your implementation. The gradient check will allow you to increase your confidence that your code is
# computing the gradients correctly.
#
# ### 2.4 Gradient checking
#
# In your neural network, you are minimizing the cost function $J(\Theta)$. To perform gradient checking on your parameters, you can imagine “unrolling” the parameters $\Theta^{(1)}$, $\Theta^{(2)}$ into a long vector $\theta$. By doing so, you can think of the cost function being $J(\Theta)$ instead and use the following gradient checking procedure.
#
# Suppose you have a function $f_i(\theta)$ that purportedly computes $\frac{\partial}{\partial \theta_i} J(\theta)$; you’d like to check if $f_i$ is outputting correct derivative values.
#
# $$
# \text{Let } \theta^{(i+)} = \theta + \begin{bmatrix} 0 \\ 0 \\ \vdots \\ \epsilon \\ \vdots \\ 0 \end{bmatrix}
# \quad \text{and} \quad \theta^{(i-)} = \theta - \begin{bmatrix} 0 \\ 0 \\ \vdots \\ \epsilon \\ \vdots \\ 0 \end{bmatrix}
# $$
#
# So, $\theta^{(i+)}$ is the same as $\theta$, except its $i^{th}$ element has been incremented by $\epsilon$. Similarly, $\theta^{(i−)}$ is the corresponding vector with the $i^{th}$ element decreased by $\epsilon$. You can now numerically verify $f_i(\theta)$’s correctness by checking, for each $i$, that:
#
# $$ f_i\left( \theta \right) \approx \frac{J\left( \theta^{(i+)}\right) - J\left( \theta^{(i-)} \right)}{2\epsilon} $$
#
# The degree to which these two values should approximate each other will depend on the details of $J$. But assuming $\epsilon = 10^{-4}$, you’ll usually find that the left- and right-hand sides of the above will agree to at least 4 significant digits (and often many more).
#
# We have implemented the function to compute the numerical gradient for you in `computeNumericalGradient` (within the file `utils.py`). While you are not required to modify the file, we highly encourage you to take a look at the code to understand how it works.
#
# In the next cell we will run the provided function `checkNNGradients` which will create a small neural network and dataset that will be used for checking your gradients. If your backpropagation implementation is correct,
# you should see a relative difference that is less than 1e-9.
#
# <div class="alert alert-box alert-success">
# **Practical Tip**: When performing gradient checking, it is much more efficient to use a small neural network with a relatively small number of input units and hidden units, thus having a relatively small number
# of parameters. Each dimension of $\theta$ requires two evaluations of the cost function and this can be expensive. In the function `checkNNGradients`, our code creates a small random model and dataset which is used with `computeNumericalGradient` for gradient checking. Furthermore, after you are confident that your gradient computations are correct, you should turn off gradient checking before running your learning algorithm.
# </div>
#
# <div class="alert alert-box alert-success">
# <b>Practical Tip:</b> Gradient checking works for any function where you are computing the cost and the gradient. Concretely, you can use the same `computeNumericalGradient` function to check if your gradient implementations for the other exercises are correct too (e.g., logistic regression’s cost function).
# </div>
utils.checkNNGradients(nnCostFunction)
# *Once your cost function passes the gradient check for the (unregularized) neural network cost function, you should submit the neural network gradient function (backpropagation).*
grader[4] = nnCostFunction
grader.grade()
# <a id="section5"></a>
# ### 2.5 Regularized Neural Network
#
# After you have successfully implemented the backpropagation algorithm, you will add regularization to the gradient. To account for regularization, it turns out that you can add this as an additional term *after* computing the gradients using backpropagation.
#
# Specifically, after you have computed $\Delta_{ij}^{(l)}$ using backpropagation, you should add regularization using
#
# $$ \begin{align}
# & \frac{\partial}{\partial \Theta_{ij}^{(l)}} J(\Theta) = D_{ij}^{(l)} = \frac{1}{m} \Delta_{ij}^{(l)} & \qquad \text{for } j = 0 \\
# & \frac{\partial}{\partial \Theta_{ij}^{(l)}} J(\Theta) = D_{ij}^{(l)} = \frac{1}{m} \Delta_{ij}^{(l)} + \frac{\lambda}{m} \Theta_{ij}^{(l)} & \qquad \text{for } j \ge 1
# \end{align}
# $$
#
# Note that you should *not* be regularizing the first column of $\Theta^{(l)}$ which is used for the bias term. Furthermore, in the parameters $\Theta_{ij}^{(l)}$, $i$ is indexed starting from 1, and $j$ is indexed starting from 0. Thus,
#
# $$
# \Theta^{(l)} = \begin{bmatrix}
# \Theta_{1,0}^{(i)} & \Theta_{1,1}^{(l)} & \cdots \\
# \Theta_{2,0}^{(i)} & \Theta_{2,1}^{(l)} & \cdots \\
# \vdots & ~ & \ddots
# \end{bmatrix}
# $$
#
# [Now modify your code that computes grad in `nnCostFunction` to account for regularization.](#nnCostFunction)
#
# After you are done, the following cell runs gradient checking on your implementation. If your code is correct, you should expect to see a relative difference that is less than 1e-9.
# +
# Check gradients by running checkNNGradients
lambda_ = 3
utils.checkNNGradients(nnCostFunction, lambda_)
# Also output the costFunction debugging values
debug_J, _ = nnCostFunction(nn_params, input_layer_size,
hidden_layer_size, num_labels, X, y, lambda_)
print('\n\nCost at (fixed) debugging parameters (w/ lambda = %f): %f ' % (lambda_, debug_J))
print('(for lambda = 3, this value should be about 0.576051)')
# -
grader[5] = nnCostFunction
grader.grade()
# ### 2.6 Learning parameters using `scipy.optimize.minimize`
#
# After you have successfully implemented the neural network cost function
# and gradient computation, the next step we will use `scipy`'s minimization to learn a good set parameters.
# +
# After you have completed the assignment, change the maxiter to a larger
# value to see how more training helps.
options= {'maxiter': 500}
# You should also try different values of lambda
lambda_ = 1
# Create "short hand" for the cost function to be minimized
costFunction = lambda p: nnCostFunction(p, input_layer_size,
hidden_layer_size,
num_labels, X, y, lambda_)
# Now, costFunction is a function that takes in only one argument
# (the neural network parameters)
res = optimize.minimize(costFunction,
initial_nn_params,
jac=True,
method='TNC',
options=options)
# get the solution of the optimization
nn_params = res.x
# Obtain Theta1 and Theta2 back from nn_params
Theta1 = np.reshape(nn_params[:hidden_layer_size * (input_layer_size + 1)],
(hidden_layer_size, (input_layer_size + 1)))
Theta2 = np.reshape(nn_params[(hidden_layer_size * (input_layer_size + 1)):],
(num_labels, (hidden_layer_size + 1)))
# -
# After the training completes, we will proceed to report the training accuracy of your classifier by computing the percentage of examples it got correct. If your implementation is correct, you should see a reported
# training accuracy of about 95.3% (this may vary by about 1% due to the random initialization). It is possible to get higher training accuracies by training the neural network for more iterations. We encourage you to try
# training the neural network for more iterations (e.g., set `maxiter` to 400) and also vary the regularization parameter $\lambda$. With the right learning settings, it is possible to get the neural network to perfectly fit the training set.
pred = utils.predict(Theta1, Theta2, X)
print('Training Set Accuracy: %f' % (np.mean(pred == y) * 100))
# ## 3 Visualizing the Hidden Layer
#
# One way to understand what your neural network is learning is to visualize what the representations captured by the hidden units. Informally, given a particular hidden unit, one way to visualize what it computes is to find an input $x$ that will cause it to activate (that is, to have an activation value
# ($a_i^{(l)}$) close to 1). For the neural network you trained, notice that the $i^{th}$ row of $\Theta^{(1)}$ is a 401-dimensional vector that represents the parameter for the $i^{th}$ hidden unit. If we discard the bias term, we get a 400 dimensional vector that represents the weights from each input pixel to the hidden unit.
#
# Thus, one way to visualize the “representation” captured by the hidden unit is to reshape this 400 dimensional vector into a 20 × 20 image and display it (It turns out that this is equivalent to finding the input that gives the highest activation for the hidden unit, given a “norm” constraint on the input (i.e., $||x||_2 \le 1$)).
#
# The next cell does this by using the `displayData` function and it will show you an image with 25 units,
# each corresponding to one hidden unit in the network. In your trained network, you should find that the hidden units corresponds roughly to detectors that look for strokes and other patterns in the input.
utils.displayData(Theta1[:, 1:])
# ### 3.1 Optional (ungraded) exercise
#
# In this part of the exercise, you will get to try out different learning settings for the neural network to see how the performance of the neural network varies with the regularization parameter $\lambda$ and number of training steps (the `maxiter` option when using `scipy.optimize.minimize`). Neural networks are very powerful models that can form highly complex decision boundaries. Without regularization, it is possible for a neural network to “overfit” a training set so that it obtains close to 100% accuracy on the training set but does not as well on new examples that it has not seen before. You can set the regularization $\lambda$ to a smaller value and the `maxiter` parameter to a higher number of iterations to see this for youself.
|
jupyter-notebooks/ml-coursera-python-assignments/Exercise4/exercise4.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <center>
# <img src="../../img/ods_stickers.jpg">
# ## Открытый курс по машинному обучению
# </center>
# Автор материала: программист-исследователь Mail.ru Group, старший преподаватель Факультета Компьютерных Наук ВШЭ Юрий Кашницкий. Материал распространяется на условиях лицензии [Creative Commons CC BY-NC-SA 4.0](https://creativecommons.org/licenses/by-nc-sa/4.0/). Можно использовать в любых целях (редактировать, поправлять и брать за основу), кроме коммерческих, но с обязательным упоминанием автора материала.
# # Тема 7. Обучение без учителя: PCA и кластеризация
# ## <center>Бонус. Метод главных компонент. Игрушечный пример
# +
import numpy as np
# %matplotlib inline
from matplotlib import pyplot as plt
# -
# **Пусть дана выборка X.**
X = np.array([[1.0, 3.0], [3.0, 5.0], [5.0, 1.0], [7.0, 4.0], [4.0, 7.0]])
plt.scatter(X[:, 0], X[:, 1]);
# **Как выбрать направление, в проекции на которое дисперсия координат точек максимальна? Синия прямая или зеленая? А может, красная?**
plt.scatter(X[:, 0], X[:, 1])
plt.plot(np.linspace(1, 8, 10), np.linspace(1, 8, 10))
plt.plot(np.linspace(1, 8, 10), np.linspace(2, 4, 10))
plt.plot(np.linspace(1, 8, 10), np.linspace(5, 2, 10));
# **Стандартизуем матрицу X. Вычитаем средние по столбцам (4 и 4) и делим на стандартные отклонения по столбцам (2 и 2). Кстати, пришлось писать код, чтоб подобрать координаты так, чтоб все средние и отклонения были целыми :)**
from sklearn.preprocessing import StandardScaler
X_scaled = StandardScaler().fit_transform(X)
X_scaled
plt.scatter(X_scaled[:, 0], X_scaled[:, 1])
plt.plot([-2, 2], [0, 0], c="black")
plt.plot([0, 0], [-2, 2], c="black")
plt.xlim(-2, 2)
plt.ylim(-2, 2);
# **Назовем новые координаты (стоблцы матрицы X_scaled) $x_1$ и $x_2$. Задача: найти такую линейную комбинацию $z = \alpha x_1 + \beta x_2$, что дисперсия $z$ максимальна. При этом должно выполняться $\alpha^2 + \beta^2 = 1.$**
# **Заметим что $$\Large D[z] = E[(z - E[z])^2] = E[z^2] = \frac{1}{n} \sum_i^n z_i^2,$$ поскольку $E[z] = \alpha E[x_1] + \beta E[x_2] = 0$ (новые координаты центрированы).**
#
# **Тогда задача формализуется так:**
# $$\Large \begin{cases} \max_{\alpha, \beta} \sum_i^n (\alpha x_{1_i} + \beta x_{2_i})^2 \\ \alpha^2 + \beta^2 = 1\end{cases}$$
# У нас $2z = [-3\alpha -\beta,\ -\alpha +\beta,\ \alpha -3\beta,\ 3\alpha,\ 3\beta]^T$ (Для задачи максимизации неважно, что мы умножили на 2, зато так удобней).
#
# Распишем в нашем случае: $ \sum_i^n (\alpha x_{1_i} + \beta x_{2_i})^2 = (-3\alpha -\beta)^2 + ( -\alpha +\beta)^2 +( \alpha -3\beta)^2 +( 3\alpha)^2 +( 3\beta)^2 = 20\alpha^2 - 2\alpha\beta + 20\beta^2$ = <font color='green'>\\ поскольку $\alpha^2 + \beta^2 = 1$ \\ </font> = $20 - 2\alpha\beta$. Осталось только минимизировать $\alpha\beta$. Можно это делать методом Лагранжа, но в данном случае можно проще
#
# $$\Large \begin{cases} \min_{\alpha, \beta} \alpha\beta \\ \alpha^2 + \beta^2 = 1\end{cases}$$
#
# $\Large \alpha\beta = \beta^2(\frac{\alpha}{\beta})$ = <font color='green'>\\ замена t = $\frac{\alpha}{\beta}, \alpha^2 + \beta^2 = 1$ \\ </font> = $\Large \frac{t}{1+t^2}$. Ищем минимум функции одной переменной, находим, что $t^* = -1$.
#
# Значит, $$\Large \begin{cases} \alpha^* = -\beta^*\\ (\alpha^*)^2 + (\beta^*)^2 = 1\end{cases} \Rightarrow \alpha^* =
# \frac{1}{\sqrt{2}}, \beta^* = - \frac{1}{\sqrt{2}}$$
# Итак, $$\Large z = \frac{1}{\sqrt{2}} x_1 - \frac{1}{\sqrt{2}}x_2$$ То есть ось $z$ повернута на 45 градусов относительно $x_1$ и $x_2$ и "направлена на юго-восток".
plt.scatter(X_scaled[:, 0], X_scaled[:, 1])
plt.plot([-2, 2], [0, 0], c="black")
plt.plot([0, 0], [-2, 2], c="black")
plt.plot([-2, 2], [2, -2], c="red");
# **Новые координаты точек по оси z:**
X_scaled.dot(np.array([1.0 / np.sqrt(2), -1.0 / np.sqrt(2)]))
# ## Сингулярное разложение матрицы X
# Представление будет таким: $X = U\Sigma V^T$.
#
# - Матрица $U$ составлена из собственных векторов матрицы $XX^T$. Это левые сингулярные векторы матрицы $X$;
# - Матрица $V$ составлена из собственных векторов матрицы $X^TX$. Это правые сингулярные векторы матрицы $X$;
# - Матрица $\Sigma$ - диагональная (вне главной диагонали нули), и на диагонали стоят корни из собственных значений матрицы $X^TX$ (или $XX^T$). Это сингулярные числа матрицы $X$.
# $XX^T$ выглядит так:
X_scaled.dot(X_scaled.T)
# $X^TX$ выглядит так:
X_scaled.T.dot(X_scaled)
# Собственные вектора $XX^T$ (левые сингулярные):
np.linalg.eig(X_scaled.dot(X_scaled.T))[1]
# Собственные вектора $X^TX$ (правые сингулярные). Эти вектора задают представление главных компонент через исходные координаты (то есть они задают поворот).
np.linalg.eig(X_scaled.T.dot(X_scaled))[1]
# Видно, что главные компоненты: $$\Large z_1 = \frac{1}{\sqrt{2}} x_1 - \frac{1}{\sqrt{2}}x_2,\ z_2 = \frac{1}{\sqrt{2}} x_1 + \frac{1}{\sqrt{2}}x_2$$
# Собственные значения $X^TX$ (сингулярные числа):
np.linalg.eig(X_scaled.T.dot(X_scaled))[0]
np.linalg.eig(X_scaled.dot(X_scaled.T))[0]
# +
from scipy.linalg import svd
U, Sigma, VT = svd(X_scaled)
# -
# Действительно. На диагонали матрицы $\Sigma$ стоят корни из собственных значений $X^TX$ ($\sqrt{5.25} \approx 2.29, \sqrt{4.75} \approx 2.18$):
Sigma
# Вектора матрицы $VT$ (правые сингулярные векторы для исходной матрицы) задают поворот. То есть первая главная компонента "смотрит на юго-восток", вторая - на юго-запад.
VT
# Представление данных в проекции на 2 главные компоненты $Z = XV$:
X_scaled.dot(VT.T)
plt.scatter(X_scaled[:, 0], X_scaled[:, 1])
plt.plot([-2, 2], [0, 0], c="black")
plt.plot([0, 0], [-2, 2], c="black")
plt.plot([-2, 2], [2, -2], c="red")
plt.plot([-2, 2], [-2, 2], c="red");
# Здесь SVD SciPy "направил" ось z1 вправо и вниз, а ось z2 - влево и вниз. Можно проверить, что представление получилось правильным.
|
jupyter_russian/topic07_unsupervised/topic7_bonus_PCA_toy_example.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# # NLP 2 : Neural Embeddings, Text Classification, Text Generation
#
#
# To use statistical classifiers with text, it is first necessary to vectorize the text. In the first practical session we explored the **bag of word** model.
#
# Modern **state of the art** methods uses embeddings to vectorize the text before classification in order to avoid feature engineering.
#
# ## Dataset
# https://github.com/cedias/practicalNLP/tree/master/dataset
#
# ## "Modern" NLP pipeline
#
# By opposition to the **bag of word** model, in the modern NLP pipeline everything is **embeddings**. Instead of encoding a text as a **sparse vector** of length $D$ (size of feature dictionnary) the goal is to encode the text in a meaningful dense vector of a small size $|e| <<< |D|$.
#
#
# The raw classification pipeline is then the following:
#
# ```
# raw text ---|embedding table|--> vectors --|Neural Net|--> class
# ```
#
#
# ### Using a language model:
#
# How to tokenize the text and extract a feature dictionnary is still a manual task. To directly have meaningful embeddings, it is common to use a pre-trained language model such as `word2vec` which we explore in this practical.
#
# In this setting, the pipeline becomes the following:
# ```
#
# raw text ---|(pre-trained) Language Model|--> vectors --|classifier (or fine-tuning)|--> class
# ```
#
#
# - #### Classic word embeddings
#
# - [Word2Vec](https://arxiv.org/abs/1301.3781)
# - [Glove](https://nlp.stanford.edu/projects/glove/)
#
#
# - #### bleeding edge language models techniques (only here for reference)
#
# - [UMLFIT](https://arxiv.org/abs/1801.06146)
# - [ELMO](https://arxiv.org/abs/1802.05365)
# - [GPT](https://blog.openai.com/language-unsupervised/)
# - [BERT](https://arxiv.org/abs/1810.04805)
#
#
#
#
#
#
# ### Goal of this session:
#
# 1. Train word embeddings on training dataset
# 2. Tinker with the learnt embeddings and see learnt relations
# 3. Tinker with pre-trained embeddings.
# 4. Use those embeddings for classification
# 5. Compare different embedding models
# 6. Pytorch first look: learn to generate text.
#
#
#
#
#
# ## Loading data (same as in nlp 1)
# +
import json
from collections import Counter
#### /!\ YOU NEED TO UNZIP dataset/json_pol.zip first /!\
# Loading json
with open("data/json_pol",encoding="utf-8") as f:
data = f.readlines()
json_data = json.loads(data[0])
train = json_data["train"]
test = json_data["test"]
# Quick Check
counter_train = Counter((x[1] for x in train))
counter_test = Counter((x[1] for x in test))
print("Number of train reviews : ", len(train))
print("----> # of positive : ", counter_train[1])
print("----> # of negative : ", counter_train[0])
print("")
print(train[0])
print("")
print("Number of test reviews : ",len(test))
print("----> # of positive : ", counter_test[1])
print("----> # of negative : ", counter_test[0])
print("")
print(test[0])
print("")
# -
# ## Word2Vec: Quick Recap
#
# **[Word2Vec](https://arxiv.org/abs/1301.3781) is composed of two distinct language models (CBOW and SG), optimized to quickly learn word vectors**
#
#
# given a random text: `i'm taking the dog out for a walk`
#
#
#
# ### (a) Continuous Bag of Word (CBOW)
# - predicts a word given a context
#
# maximizing `p(dog | i'm taking the ___ out for a walk)`
#
# ### (b) Skip-Gram (SG)
# - predicts a context given a word
#
# maximizing `p(i'm taking the out for a walk | dog)`
#
#
#
#
# ## Step 1: train (or load) a language model (word2vec)
#
# Gensim has one of [Word2Vec](https://radimrehurek.com/gensim/models/word2vec.html) fastest implementation.
#
#
# ### Train:
# +
import gensim
import logging
logging.basicConfig(format='%(asctime)s : %(levelname)s : %(message)s', level=logging.INFO)
text = [t.split() for t,p in train]
# the following configuration is the default configuration
w2v = gensim.models.word2vec.Word2Vec(sentences=text,
vector_size=100, window=5, ### here we train a cbow model
min_count=5,
sample=0.001, workers=3,
sg=1, hs=0, negative=5, ### set sg to 1 to train a sg model
cbow_mean=1,
epochs=5)
# -
# ### Load pre-trained embeddings:
# +
# It's for later
from gensim.models import KeyedVectors
from gensim.test.utils import datapath
# w2v = KeyedVectors.load_word2vec_format(datapath('downloaded_vectors_path'), binary=False)
word_vectors = w2v.wv
word_vectors.save("downloaded_vectors_path")
# Load back with memory-mapping = read-only, shared across processes.
wv = KeyedVectors.load("downloaded_vectors_path", mmap='r')
vector = wv['car'] # Get numpy vector of a word
# -
# In Gensim, embeddings are loaded and can be used via the ["KeyedVectors"](https://radimrehurek.com/gensim/models/keyedvectors.html) class
#
# > Since trained word vectors are independent from the way they were trained (Word2Vec, FastText, WordRank, VarEmbed etc), they can be represented by a standalone structure, as implemented in this module.
#
# >The structure is called “KeyedVectors” and is essentially a mapping between entities and vectors. Each entity is identified by its string id, so this is a mapping between {str => 1D numpy array}.
#
# >The entity typically corresponds to a word (so the mapping maps words to 1D vectors), but for some models, they key can also correspond to a document, a graph node etc. To generalize over different use-cases, this module calls the keys entities. Each entity is always represented by its string id, no matter whether the entity is a word, a document or a graph node.
# ## STEP 2: Test learnt embeddings
#
# The word embedding space directly encodes similarities between words: the vector coding for the word "great" will be closer to the vector coding for "good" than to the one coding for "bad". Generally, [cosine similarity](https://en.wikipedia.org/wiki/Cosine_similarity) is the distance used when considering distance between vectors.
#
# KeyedVectors have a built in [similarity](https://radimrehurek.com/gensim/models /keyedvectors.html#gensim.models.keyedvectors.BaseKeyedVectors.similarity) method to compute the cosine similarity between words
# is great really closer to good than to bad ?
print("great and good:",w2v.wv.similarity("great","good"))
print("great and bad:",w2v.wv.similarity("great","bad"))
# Since cosine distance encodes similarity, neighboring words are supposed to be similar. The [most_similar](https://radimrehurek.com/gensim/models/keyedvectors.html#gensim.models.keyedvectors.BaseKeyedVectors.most_similar) method returns the `topn` words given a query.
# The query can be as simple as a word, such as "movie"
# Try changing the word
print(w2v.wv.most_similar("movie",topn=5),"\n") # 5 most similar words
print(w2v.wv.most_similar("awesome",topn=5),"\n")
print(w2v.wv.most_similar("actor",topn=5),"\n")
# But it can be a more complicated query
# Word embedding spaces tend to encode much more.
#
# The most famous exemple is: `vec(king) - vec(man) + vec(woman) => vec(queen)`
# +
# What is awesome - good + bad ?
print(w2v.wv.most_similar(positive=["awesome","bad"],negative=["good"],topn=3),"\n")
print(w2v.wv.most_similar(positive=["actor","woman"],negative=["man"],topn=3),"\n") # do the famous exemple works for actor ?
# Try other things like plurals for exemple.
print(w2v.wv.most_similar(positive=["men","man"],negative=["women"],topn=3),"\n")
# -
# To test learnt "synctactic" and "semantic" similarities, Mikolov et al. introduced a special dataset containing a wide variety of three way similarities.
out = w2v.wv.evaluate_word_analogies("data/questions-words.txt",case_insensitive=True) # original semantic syntactic dataset.
# When training the w2v models on the review dataset, since it hasn't been learnt with a lot of data, it does not perform very well.
#
# ## STEP 3: sentiment classification
#
# In the previous practical session, we used a bag of word approach to transform text into vectors.
# Here, we propose to try to use word vectors (previously learnt or loaded).
#
#
# ### <font color='green'> Since we have only word vectors and that sentences are made of multiple words, we need to aggregate them. </font>
#
#
# ### (1) Vectorize reviews using word vectors:
#
# Word aggregation can be done in different ways:
#
# - Sum
# - Average
# - Min/feature
# - Max/feature
#
# #### a few pointers:
#
# - `w2v.wv.vocab` is a `set()` of the vocabulary (all existing words in your model)
# - `np.minimum(a,b) and np.maximum(a,b)` respectively return element-wise min/max
# +
import numpy as np
# We first need to vectorize text:
# First we propose to a sum of them
def vectorize(text,types="max"):
"""
This function should vectorize one review
input: str
output: np.array(float)
"""
vec = []
for i in text.split():
try :
vec.append(wv[i])
except :
vec.append(np.zeros(100))
if types == "mean" :
return np.array(vec).mean(axis=1)
if types == "sum":
return np.array(vec).sum(axis=1)
if types == "max" :
return np.array(vec).max(axis=1)
if types == "min" :
return np.array(vec).min(axis=1)
classes = np.array([pol for text,pol in train])
X = np.array([vectorize(text) for text,pol in train])
X_test = np.array([vectorize(text) for text,pol in test])
true = np.array([pol for text,pol in test])
#let's see what a review vector looks like.
print(X[0])
# -
print(len(X[2]))
# ### (2) Train a classifier
# as in the previous practical session, train a logistic regression to do sentiment classification with word vectors
#
#
# +
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score, confusion_matrix, classification_report, accuracy_score, f1_score, roc_auc_score
from sklearn.model_selection import train_test_split
from sklearn import svm
# X_train, X_test, Y_train, Y_test = train_test_split(X, Y, train_size=0.8)
# print(X_train.shape)
# print(X_test.shape)
# print(Y_train.shape)
# print(Y_test.shape)
maxi = 0
for i in range(X.shape[0]):
if len(X[i]) > maxi:
maxi = len(X[i])
print(maxi)
padded_array = np.zeros((X.shape[0], maxi))
n,m = padded_array.shape
print(n, m)
for i in range(n):
for j in range(m):
if j < len(X[i]):
padded_array[i][j] = X[i][j]
print(padded_array.shape)
# -
padded_array_test = np.zeros((X_test.shape[0], maxi))
n,m = padded_array_test.shape
print(n, m)
for i in range(n):
for j in range(m):
if j < len(X_test[i]):
padded_array_test[i][j] = X_test[i][j]
print(padded_array_test.shape)
clf = svm.SVC(max_iter=1000)
clf.fit(padded_array, classes)
preds = clf.predict(padded_array_test)
print(accuracy_score(preds, true))
print(classification_report(preds, true))
print(f1_score(preds, true))
# performance should be worst than with bag of word (~80%). Sum/Mean aggregation does not work well on long reviews (especially with many frequent words). This adds a lot of noise.
#
# ## **Todo** : Try answering the following questions:
#
# - Which word2vec model works best: skip-gram or cbow
# - Do pretrained vectors work best than those learnt on the train dataset ?
#
#
# **(Bonus)** To have a better accuracy, we could try two things:
# - Better aggregation methods (weight by tf-idf ?)
# - Another word vectorizing method such as [fasttext](https://radimrehurek.com/gensim/models/fasttext.html)
# - A document vectorizing method such as [Doc2Vec](https://radimrehurek.com/gensim/models/doc2vec.html)
# ## --- Generate text with a recurrent neural network (Pytorch) ---
# ### (Mostly Read & Run)
#
# The goal is to replicate the (famous) experiment from [Karpathy's blog](http://karpathy.github.io/2015/05/21/rnn-effectiveness/)
#
# To learn to generate text, we train a recurrent neural network to do the following task:
#
# Given a "chunk" of text: `this is random text`
#
# the goal of the network is to predict each character in **`his is random text` ** sequentially given the following sequential input **`this is random tex`**:
#
#
# + active=""
# Input -> Output
# --------------
# T -> H
# H -> I
# I -> S
# S -> " "
# " " -> I
# I -> S
# S -> " "
# [...]
# -
#
# ## Load text (dataset/input.txt)
#
# Before building training batch, we load the full text in RAM
# +
import unidecode
import string
import random
import re
import torch
import torch.nn as nn
all_characters = string.printable
n_characters = len(all_characters)
file = unidecode.unidecode(open('data/input.txt').read()) #clean text => only ascii
file_len = len(file)
print('file_len =', file_len)
# -
# ## 2: Helper functions:
#
# We have a text and we want to feed batch of chunks to a neural network:
#
# one chunk A,B,C,D,E
# [input] A,B,C,D -> B,C,D,E [output]
#
# Note: we will use an embedding layer instead of a one-hot encoding scheme.
#
# for this, we have 3 functions:
#
# - One to get a random str chunk of size `chunk_len` : `random_chunk`
# - One to turn a chunk into a tensor of size `(1,chunk_len)` coding for each characters : `char_tensor`
# - One to return random input and output chunks of size `(batch_size,chunk_len)` : `random_training_set`
#
#
#
# +
import time, math
#Get a piece of text
def random_chunk(chunk_len):
start_index = random.randint(0, file_len - chunk_len)
end_index = start_index + chunk_len + 1
return file[start_index:end_index]
# Turn string into list of longs
def char_tensor(string):
tensor = torch.zeros(1,len(string)).long()
for c in range(len(string)):
tensor[0,c] = all_characters.index(string[c])
return tensor
#Turn a piece of text in train/test
def random_training_set(chunk_len=200, batch_size=8):
chunks = [random_chunk(chunk_len) for _ in range(batch_size)]
inp = torch.cat([char_tensor(chunk[:-1]) for chunk in chunks],dim=0)
target = torch.cat([char_tensor(chunk[1:]) for chunk in chunks],dim=0)
return inp, target
print(random_training_set(10,4)) ## should return 8 chunks of 10 letters.
# -
# ## The actual RNN model (only thing to complete):
#
# It should be composed of three distinct modules:
#
# - an [embedding layer](https://pytorch.org/docs/stable/nn.html#embedding) (n_characters, hidden_size)
#
# ```
# nn.Embedding(len_dic,size_vec)
# ```
# - a [recurrent](https://pytorch.org/docs/stable/nn.html#recurrent-layers) layer (hidden_size, hidden_size)
# ```
# nn.RNN(in_size,out_size) or nn.GRU() or nn.LSTM() => rnn_cell parameter
# ```
# - a [prediction](https://pytorch.org/docs/stable/nn.html#linear) layer (hidden_size, output_size)
#
# ```
# nn.Linear(in_size,out_size)
# ```
# => Complete the `init` function code
# +
import torch.nn.functional as f
class RNN(nn.Module):
def __init__(self, n_char, hidden_size, output_size, n_layers=1,rnn_cell=nn.RNN):
"""
Create the network
"""
super(RNN, self).__init__()
self.n_char = n_char
self.hidden_size = hidden_size
self.output_size = output_size
self.n_layers = n_layers
# (batch,chunk_len) -> (batch, chunk_len, hidden_size)
self.embed = nn.Embedding(n_char, hidden_size)
# (batch, chunk_len, hidden_size) -> (batch, chunk_len, hidden_size)
self.rnn = rnn_cell(hidden_size, hidden_size)
#(batch, chunk_len, hidden_size) -> (batch, chunk_len, output_size)
self.predict = nn.Linear(hidden_size, output_size)
def forward(self, input):
"""
batched forward: input is (batch > 1,chunk_len)
"""
input = self.embed(input)
output,_ = self.rnn(input)
output = self.predict(f.tanh(output))
return output
def forward_seq(self, input,hidden=None):
"""
not batched forward: input is (1,chunk_len)
"""
input = self.embed(input)
output,hidden = self.rnn(input.unsqueeze(0),hidden)
output = self.predict(f.tanh(output))
return output,hidden
# -
# ## Text generation function
#
# Sample text from the model
def generate(model,prime_str='A', predict_len=100, temperature=0.8):
prime_input = char_tensor(prime_str).squeeze(0)
hidden = None
predicted = prime_str+""
# Use priming string to "build up" hidden state
for p in range(len(prime_str)-1):
_,hidden = model.forward_seq(prime_input[p].unsqueeze(0),hidden)
#print(hidden.size())
for p in range(predict_len):
output, hidden = model.forward_seq(prime_input[-1].unsqueeze(0), hidden)
# Sample from the network as a multinomial distribution
output_dist = output.data.view(-1).div(temperature).exp()
#print(output_dist)
top_i = torch.multinomial(output_dist, 1)[0]
#print(top_i)
# Add predicted character to string and use as next input
predicted_char = all_characters[top_i]
predicted += predicted_char
prime_input = torch.cat([prime_input,char_tensor(predicted_char).squeeze(0)])
return predicted
# ## Training loop for net
# +
def time_since(since):
s = time.time() - since
m = math.floor(s / 60)
s -= m * 60
return '%dm %ds' % (m, s)
###Parameters
n_epochs = 10000
print_every = 100
plot_every = 10
hidden_size = 256
n_layers = 2
lr = 0.005
batch_size = 16
chunk_len = 20
####
model = RNN(n_characters, hidden_size, n_characters, n_layers, nn.LSTM) #create model
model_optimizer = torch.optim.Adam(model.parameters(), lr=lr) #create Adam optimizer
criterion = nn.CrossEntropyLoss() #chose criterion
start = time.time()
all_losses = []
loss_avg = 0
def train(inp, target):
"""
Train sequence for one chunk:
"""
#reset gradients
model_optimizer.zero_grad()
# predict output
output = model(inp)
#compute loss
loss = criterion(output.view(batch_size*chunk_len,-1), target.view(-1))
#compute gradients and backpropagate
loss.backward()
model_optimizer.step()
return loss.data.item()
for epoch in range(1, n_epochs + 1):
loss = train(*random_training_set(chunk_len,batch_size)) #train on one chunk
loss_avg += loss
if epoch % print_every == 0:
print('[%s (%d %d%%) %.4f]' % (time_since(start), epoch, epoch / n_epochs * 100, loss))
print(generate(model,'Wh', 100), '\n')
if epoch % plot_every == 0:
all_losses.append(loss_avg / plot_every)
loss_avg = 0
# -
# ## Visualize loss
# +
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
# %matplotlib inline
plt.figure()
plt.plot(all_losses)
plt.show()
# -
# ## Try different temperatures
#
# Changing the distribution sharpness has an impact on character sampling:
#
# more or less probable things are sampled
print(generate(model,'T', 200, temperature=1))
print("----")
print(generate(model,'Th', 200, temperature=0.8))
print("----")
print(generate(model,'Th', 200, temperature=0.5))
print("----")
print(generate(model,'Th', 200, temperature=0.3))
print("----")
print(generate(model,'Th', 200, temperature=0.1))
# ### Improving this code:
#
# (a) Tinker with parameters:
#
# - Is it really necessary to have 100 dims character embeddings
# - Chunk length can be gradually increased
# - Try changing RNN cell type (GRUs - LSTMs)
#
# (b) Add GPU support to go faster
#
# ## ------ End of practical
#
# #### Legacy loading code
# +
# import glob
# from os.path import split as pathsplit
# dir_train = "data/aclImdb/train/"
# dir_test = "data/aclImdb/test/"
# train_files = glob.glob(dir_train+'pos/*.txt') + glob.glob(dir_train+'neg/*.txt')
# test_files = glob.glob(dir_test+'pos/*.txt') + glob.glob(dir_test+'neg/*.txt')
# def get_polarity(f):
# """
# Extracts polarity from filename:
# 0 is negative (< 5)
# 1 is positive (> 5)
# """
# _,name = pathsplit(f)
# if int(name.split('_')[1].split('.')[0]) < 5:
# return 0
# else:
# return 1
# def open_one(f):
# polarity = get_polarity(f)
# with open(f,"r") as review:
# text = " ".join(review.readlines()).strip()
# return (text,polarity)
# print(open_one(train_files[0]))
# train = [open_one(x) for x in train_files] #contains (text,pol) couples
# test = [open_one(x) for x in test_files] #contains (text,pol) couples
# -
|
S2/RITAL/TAL/TME/TME2/Dan.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: river
# language: python
# name: python3
# ---
# # Content personalization
# ## Without context
# This example takes inspiration from Vowpal Wabbit's [excellent tutorial](https://vowpalwabbit.org/tutorials/cb_simulation.html).
#
# Content personalization is about taking into account user preferences. It's a special case of recommender systems. Ideally, side-information should be taken into account in addition to the user. But we'll start with something simpler. We'll assume that each user has stable preferences that are independent of the context. We capture this by implementing a "reward" function.
# +
users = ['Tom', 'Anna']
items = {'politics', 'sports', 'music', 'food', 'finance', 'health', 'camping'}
def get_reward(user, item) -> bool:
if user == 'Tom':
return item in {'music', 'politics'}
if user == 'Anna':
return item in {'politics', 'sports'}
# -
# Measuring the performance of a recommendation is not straightforward, mostly because of the interactive aspect of recommender systems. In a real situation, recommendations are presented to a user, and the user gives feedback indicating whether they like what they have been recommended or not. This feedback loop can't be captured entirely by a historical dataset. Some kind of simulator is required to generate recommendations and capture feedback. We already have a reward function. Now let's implement a simulation function.
# +
import random
import matplotlib.pyplot as plt
def plot_ctr(ctr):
plt.plot(range(1, len(ctr) + 1), ctr)
plt.xlabel('n_iterations', fontsize=14)
plt.ylabel('CTR', fontsize=14)
plt.ylim([0, 1])
plt.title(f'final CTR: {ctr[-1]:.2%}', fontsize=14)
plt.grid()
def simulate(n, reward_func, model, seed):
rng = random.Random(seed)
n_clicks = 0
ctr = [] # click-through rate along time
for i in range(n):
# Pick a user at random
user = rng.choice(users)
# Make a single recommendation
item = model.rank(user, items=items)[0]
# Measure the reward
clicked = reward_func(user, item)
n_clicks += clicked
ctr.append(n_clicks / (i + 1))
# Update the model
model.learn_one(user, item, clicked)
plot_ctr(ctr)
# -
# This simulation function does quite a few things. It can be seen as a simple reinforcement learning simulation. It samples a user, and then ask the model to provide a single recommendation. The user then gives as to whether they liked the recommendation or not. Crucially, the user doesn't tell us what item they would have liked. We could model this as a multi-class classification problem if that were the case.
#
# The strategy parameter determines the mechanism used to generate the recommendations. The `'best'` strategy means that the items are each scored by the model, and are then ranked from the most preferred to the least preferred. Here the most preferred item is the one which gets recommended. But you could imagine all sorts of alternative ways to proceed.
#
# We can first evaluate a recommended which acts completely at random. It assigns a random preference to each item, regardless of the user.
# +
from river import reco
model = reco.RandomNormal(seed=10)
simulate(5_000, get_reward, model, seed=42)
# -
# We can see that the click-through rate (CTR) oscillates around 28.74%. In fact, this model is expected to be correct `100 * (2 / 7)% = 28.57%` of the time. Indeed, each user likes two items, and there are seven items in total.
#
# Let's now use the `Baseline` recommended. This one models each preference as the following sum:
#
# $$preference = \bar{y} + b_{u} + b_{i}$$
#
# where
#
# - $\bar{y}$ is the average CTR overall
# - $b_{u}$ is the average CTR per user minus $\bar{y}$ -- it's therefore called a *bias*
# - $b_{i}$ is the average CTR per item minus $\bar{y}$
#
# This model is considered to be a baseline because it doesn't actually learn what items are preferred by each user. Instead it models each user and item separately. We shouldn't expect it to be a strong model. It should however do better than the random model used above.
model = reco.Baseline(seed=10)
simulate(5_000, get_reward, model, seed=42)
# This baseline model seems perfect, which is surprising. The reason why it works so well is because both users have in common that they both like politics. The model therefore learns that the `'politics'` is a good item to recommend.
model.i_biases
# The model is not as performant if we use a reward function where both users have different preferences.
simulate(
5_000,
reward_func=lambda user, item: (
item in {'music', 'politics'} if user == "Tom" else
item in {'food', 'sports'}
),
model=model,
seed=42
)
# A good recommender model should at the very least understand what kind of items each user prefers. One of the simplest and yet performant way to do this is <NAME>'s SGD method he developped for the Netflix challenge and wrote about [here](https://sifter.org/simon/journal/20061211.html). It models each user and each item as latent vectors. The dot product of these two vectors is the expected preference of the user for the item.
model = reco.FunkMF(seed=10)
simulate(5_000, get_reward, model, seed=42)
# We can see that this model learns what items each user enjoys very well. Of course, there are some caveats. In our simulation, we ask the model to recommend the item most likely to be preferred for each user. Indeed, we rank all the items and pick the item at the top of the list. We do this many times for only two users.
#
# This is of course not realistic. Users will get fed up with recommendations if they're always shown the same item. It's important to include diversity into recommendations, and to let the model explore other options instead of always focusing on the item with the highest score. This is where evaluating recommender systems gets tricky: the reward function itself is difficult to model.
#
# We will keep ignoring these caveats in this notebook. Instead we will focus on a different concern: making recommendations when context is involved.
# ## With context
# We'll add some context by making it so that user preferences change depending on the time the day. Very simply, preferences might change from morning to afternoon. This is captured by the following reward function.
# +
times_of_day = ['morning', 'afternoon']
def get_reward(user, item, context):
if user == 'Tom':
if context['time_of_day'] == 'morning':
return item == 'politics'
if context['time_of_day'] == 'afternoon':
return item == 'music'
if user == 'Anna':
if context['time_of_day'] == 'morning':
return item == 'sports'
if context['time_of_day'] == 'afternoon':
return item == 'politics'
# -
# We have to update our simulation function to generate a random context at each step. We also want our model to use it for recommending items as well as learning.
def simulate(n, reward_func, model, seed):
rng = random.Random(seed)
n_clicks = 0
ctr = []
for i in range(n):
user = rng.choice(users)
# New: pass a context
context = {'time_of_day': rng.choice(times_of_day)}
item = model.rank(user, items, context)[0]
clicked = reward_func(user, item, context)
n_clicks += clicked
ctr.append(n_clicks / (i + 1))
# New: pass a context
model.learn_one(user, item, clicked, context)
plot_ctr(ctr)
# Not all models are capable of taking into account context. For instance, the `FunkMF` model only models users and items. It completely ignores the context, even when we provide one. All recommender models inherit from the base `Recommender` class. They also have a property which indicates whether or not they are able to handle context:
model = reco.FunkMF(seed=10)
model.is_contextual
# Let's see well it performs.
simulate(5_000, get_reward, model, seed=42)
# The performance has roughly been divided by half. This is most likely because there are now two times of day, and if the model has learnt preferences for one time of the day, then it's expected to be wrong half of the time.
#
# Before delving into recsys models that can handle context, a simple hack is to notice that we can append the time of day to the user. This effectively results in new users which our model can distinguish between. We could apply this trick during the simulation, but we can also override the behavior of the `learn_one` and `rank` methods of our model.
# +
class FunkMFWithHack(reco.FunkMF):
def learn_one(self, user, item, reward, context):
user = f"{user}@{context['time_of_day']}"
return super().learn_one(user, item, reward, context)
def rank(self, user, items, context):
user = f"{user}@{context['time_of_day']}"
return super().rank(user, items, context)
model = FunkMFWithHack(seed=29)
simulate(5_000, get_reward, model, seed=42)
# -
# We can verify that the model has learnt the correct preferences by looking at the expected preference for each `(user, item)` pair.
# +
import pandas as pd
(
pd.DataFrame(
{
'user': user,
'item': item,
'preference': model.predict_one(user, item)
}
for user in model.u_latents
for item in model.i_latents
)
.pivot('user', 'item')
.style.highlight_max(color='lightgreen', axis='columns')
)
|
docs/examples/content-personalization.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Convolutional Neural Network (CNN)
# loading libraries
import keras
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
import time
import numpy as np
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation, Flatten, Dropout, Conv2D, MaxPooling2D
import matplotlib as mpl
import seaborn as sns
np.random.seed(1367)
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:95% !important; }</style>"))
sns.set_style("ticks", {"xtick.direction": u"in", "ytick.direction": u"in"})
mpl.rcParams["axes.linewidth"] = 2
mpl.rcParams["lines.linewidth"] = 3
(X_train, y_train), (X_test, y_test) = keras.datasets.mnist.load_data()
_, img_rows, img_cols = X_train.shape
NUM_CLASSES = len(np.unique(y_train))
NUM_INPUT_NODES = img_rows * img_cols
print(F"Number of training samples: {X_train.shape[0]}")
print(F"Number of test samples: {X_test.shape[0]}")
print(F"Image rows: {X_train.shape[1]}")
print(F"Image columns: {X_train.shape[2]}")
print(F"Number of classes: {NUM_CLASSES}")
X_train.shape
#reshaping
#this assumes our data format
#For 3D data, "channels_last" assumes (conv_dim1, conv_dim2, conv_dim3, channels) while
#"channels_first" assumes (channels, conv_dim1, conv_dim2, conv_dim3).
if tf.keras.backend.image_data_format() == "channels_first":
X_train = X_train.reshape(X_train.shape[0], 1, img_rows, img_cols)
X_test = X_test.reshape(X_test.shape[0], 1, img_rows, img_cols)
input_shape = (1, img_rows, img_cols)
else:
X_train = X_train.reshape(X_train.shape[0], img_rows, img_cols, 1)
X_test = X_test.reshape(X_test.shape[0], img_rows, img_cols, 1)
input_shape = (img_rows, img_cols, 1)
#more reshaping
X_train = X_train.astype("float32")
X_test = X_test.astype("float32")
X_train /= 255
X_test /= 255
print("X_train shape:", X_train.shape) #X_train shape: (60000, 28, 28, 1)
# +
SAVE_FIG = True
fig = plt.figure(figsize=(10,5))
for i in range(NUM_CLASSES):
ax = fig.add_subplot(2, 5, 1 + i, xticks=[], yticks=[])
idx = X_train[y_train[:]==i,:]
ax.set_title("Class: " + str(i) , fontsize = 20)
plt.imshow(idx[1], cmap="gray")
if SAVE_FIG:
plt.savefig("../assets/header.png" , bbox_inches="tight")
plt.show()
# -
def cnn():
"""
Define a cnn model structure
"""
model = Sequential()
model.add(Conv2D(32, kernel_size=(3, 3),
activation="relu",
input_shape=input_shape))
model.add(Conv2D(64, (3, 3), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(NUM_CLASSES, activation="softmax"))
return model
def plot_results(results):
"""
Plot accuracy/loss through epochs
"""
fig, axs = plt.subplots(1, 2, figsize=(15, 5))
# summarize history for accuracy
axs[0].plot(
range(1, len(results.history["accuracy"]) + 1),
results.history["accuracy"],
color="navy",
ls="--",
label="Training",
)
axs[0].plot(
range(1, len(results.history["val_accuracy"]) + 1),
results.history["val_accuracy"],
color="cyan",
ls="--",
label="Validation",
)
axs[0].set_ylabel("Accuracy", fontsize=15)
axs[0].set_xlabel("Epoch", fontsize=15)
axs[0].legend(prop={"size": 13}, loc=0, framealpha=0.0)
# summarize history for loss
axs[1].plot(
range(1, len(results.history["loss"]) + 1),
results.history["loss"],
color="navy",
ls="--",
label="Training",
)
axs[1].plot(
range(1, len(results.history["val_loss"]) + 1),
results.history["val_loss"],
color="cyan",
ls="--",
label="Validation",
)
axs[1].set_ylabel("Loss", fontsize=15)
axs[1].set_xlabel("Epoch", fontsize=15)
axs[1].legend(prop={"size": 13}, loc=0, framealpha=0.0)
plt.savefig("../assets/performance_cnn.png" , bbox_inches="tight")
plt.show()
def accuracy(results, X_test, y_test):
"""
Accuracy metric
"""
y_pred_proba = results.model.predict(X_test)
y_pred = np.argmax(y_pred_proba, axis=1)
num_correct = np.sum(y_pred == y_test)
accuracy = float(num_correct) / y_pred_proba.shape[0]
return accuracy * 100
# ## Training
# +
# define model
model = cnn()
# compile
model.compile(optimizer=tf.keras.optimizers.Adam(0.0005),
loss="sparse_categorical_crossentropy",
metrics=["accuracy"])
start = time.time()
# fit
results = model.fit(X_train, y_train,
batch_size=32,
epochs=25,
verbose=1,
validation_data=(X_test , y_test))
end = time.time()
time_elapsed = end - start
# -
results.model.summary()
# +
# plot model history
plot_results(results)
print(F"Model took {time_elapsed:.3f} seconds to train")
# compute test accuracy
print(F"Accuracy on test data is: {accuracy(results, X_test, y_test):.2f}%")
# -
# saving model
results.model.save("../assets/model_cnn.h5")
m = tf.keras.models.load_model("../assets/model_cnn.h5")
m.summary()
|
apps/handwritten-digits-recognizer/notebooks/cnn.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .jl
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Julia 0.6.0 (Programa)
# language: julia
# name: julia-0.6-programa
# ---
# [FProfile.jl](https://github.com/cstjean/FProfile.jl) provides an alternative interface for Julia's sampling profiler (`@profile`). If you've never used a sampling profiler before, please read [the introduction of this document](https://docs.julialang.org/en/latest/manual/profile/) before proceeding .
# # Profiling
#
# You can build a profile by calling `@fprofile(code, delay=0.001, n_samples=1000000)`:
# +
using FProfile, Calculus
pd = @fprofile second_derivative(sin, 1.0)
# -
# `@fprofile(N, ...)` is shorthand for `@fprofile(for _ in 1:N ... end)`:
pd = @fprofile 1000000 second_derivative(sin, 1.0)
# Do not forget that Julia compiles code the first time a function is run; if you do not want to measure compilation time, execute your code once before profiling.
# # Flat view
#
# FProfile's `flat` report is a [dataframe](http://juliadata.github.io/DataFrames.jl/stable/man/getting_started/#Getting-Started-1). No particular knowledge of dataframes is necessary. I'll provide a few common operations below.
# +
using DataFrames
df = flat(pd)
head(df, 15) # show only the first 15 rows (the 15 rows with the highest counts)
# -
# (<i>REPL note</i>: if the output of `flat` is [incomplete](https://github.com/JuliaData/DataFrames.jl/issues/1272), try `showall(flat(pd))` or `Matrix(flat(pd))`)
#
# The first column shows what fraction of backtraces (in %) go through the `method at file:line_number` in the `stackframe` column. It's the same quantity as in `Base.Profile.print()`, except for recursive calls: if `factorial(2)` calls `factorial(1)`, that's 2 counts in Base's report, but only 1 count in FProfile.
#
# You can select a subset of the dataframe by using one of the five accessors: `get_specialization, get_method, get_file, get_function` and `get_module`.
df[get_function.(df[:stackframe]) .== derivative, :] # select the `derivative` rows
# It is common to focus optimization efforts on one or more modules at a time (... the ones you're developing). `flat(pd, MyModule)` filters out other modules and adds a useful column: `self_pct` measures how much `MyModule`-specific work is done on that line.
#
# For instance, in the code below, while the `do_computation()` call takes a long time (it has a high `count_percent`), it merely calls another `Main` function, so it has a low `self_pct`. `sum_of_sin` has `self_pct = 100%` because while it calls `sum` and `sin`, those are defined in another module (`Base`), and counted as external to `Main`.
#
# `flat(pd, (Module1, Module2, ...))` is also accepted.
@noinline do_computation(n) = sum_of_sin(n)
@noinline sum_of_sin(n) = sum(sin, 1:n)
pd2 = @fprofile do_computation(10000000)
flat(pd2, Main)
# It pays to make sure that functions with a high `self_pct` are [well optimized](https://docs.julialang.org/en/latest/manual/performance-tips/).
#
# Another way to reduce the level of detail is to aggregate by `:specialization, :method, :file, :function`, or `:module`.
df_by_fn = flat(pd, combineby=:function)
# You can see the context (caller/called functions) around each of these rows by passing it to `tree`:
tree(pd, df_by_fn, 9) # show the context of the 9th row of `df_by_method`
# Other useful dataframe commands:
#
# ```julia
# sort(df, :self_pct, rev=true) # sort by self_pct
# showall(df) # show the whole dataframe
# ```
#
# See `?flat` for more options.
# #### Comparing results
#
# Pass two `ProfileData` objects to `flat` to compare them. The results are sorted with the biggest regressions (in absolute terms) at the top and the biggest improvements at the bottom (see `?DataFrames.tail`).
pd2 = @fprofile 1000000 second_derivative(sin, 1.0)
flat(pd, pd2, combineby=:function)
# Of course, this is most useful when comparing different algorithms or commits (use `reload` or [Revise.jl](https://github.com/timholy/Revise.jl) to update your code). The differences in the above table are just noise.
# # Tree view
#
# FProfile's tree view looks the same as `Base.Profile.print(format=:tree)`. The numbers represent raw counts. (If some branches seem out of place, see [this issue](https://github.com/JuliaLang/julia/issues/9689))
tr = tree(pd)
# Like `flat` reports, trees can be aggregated by `:specialization, :method, :file, :function`, or `:module`:
tree(pd, combineby=:module)
# If you're only interested in a particular module/file/method/function, you can pass it to `tree`, along with an optional _neighborhood range_.
tr_deriv = tree(pd, second_derivative, -1:1) # -1:1 = show one level of callers and one level of called functions
# Trees are an indexable, prunable (use `prune(tree, depth)`) and filterable datastructure. Use the accessors (see above) and `is_inline/is_C_call` in your `filter` predicate.
# # ProfileView integration
#
# `ProfileData` objects can be passed to `ProfileView.view`. This is purely a convenience; it's equivalent to normal ProfileView usage. See [ProfileView.jl](https://github.com/timholy/ProfileView.jl) for details.
#
# ```julia
# using ProfileView
# pd = @fprofile ...
# ProfileView.view(pd)
# ```
# # Backtraces
#
# (if you want to build your own analysis)
#
# The raw profiler data is available either through `Base.Profile.retrieve()`, or through `pd.data` and `pd.lidict`. However, you might find `FProfile.backtraces(::ProfileData)` more immediately useful.
count, trace = backtraces(pd)[1] # get the first unique backtrace
@show count # the number of times that trace occurs in the raw data
trace
# Use the `get_method, get_file, ...` functions on `StackFrame` objects (see above). `tree(pd::ProfileData)` is defined as `tree(backtraces(pd))`, and similarly for `flat`, so you can modify the backtraces and get a tree/flat view of the results.
|
Manual.ipynb
|
# # In this notebook....
#
# Using HypnosPy, we first classified sleep using the most common heuristic algorithms, such as Cole-Kripke, Oakley, Sadeh, Sazonov and the Scripps Clinic algorithm.
#
# Once sleep was classified, we derived the well-established sleep metrics, such as arousal, awakening, sleep efficiency, total sleep time and total wake time.
#
# All this was done by simply using the _SleepWakeAnalysis_ and _SleepMetrics_ modules of HypnosPy.
#
# Next, we calculated the Pearson correlation between the sleep metrics and the ones derived from ground truth PSG.
#
# Finally, we produced a compelling visualization of these results using the software.
# This visualization extends the finds of [previous research](https://www.nature.com/articles/s41746-019-0126-9) and allows us to understand, for the first time, that although all algorithms perform similarly at detecting total wake time, they all perform better at detecting total sleep time than total wake time and in particular Oakley and Sazonov are better at awakening and arousal detection than the rest of the algorithms.
#
# These findings are particularly informative for large population studies where quantifying awakenings/arousals during the night period is of particular interest for the research question that the study is trying to address.
from glob import glob
from hypnospy import Wearable, Experiment
from hypnospy.data import RawProcessing
from hypnospy.analysis import SleepWakeAnalysis, Viewer, SleepMetrics
# In the MESA Sleep study, PSG was conducted during one single night and actigraphy during 7 whole days.
# Actigraph and PSG were aligned elsewhere using this overlap dataset: https://sleepdata.org/datasets/mesa/files/overlap
# Hypnospy can easily process the generic data created with the ``RawProcessing'' module shown below.
# Note that all this data (PSG, actigraph and overlap) can be obtained from https://sleepdata.org upon request
file_path = "../data/examples_mesa/collection_mesa_psg/*.csv"
# Configure an Experiment
exp = Experiment()
for inputfile in glob(file_path):
# This is how a preprocessed actigraph + PSG file looks like:
#mesaid,linetime,offwrist,activity,marker,white,red,green,blue,wake,interval,dayofweek,daybymidnight,daybynoon,stage,gt
#1,1900-01-01 20:30:00,0,0.0,0.0,0.07,0.0292,0.0,0.0059,0.0,ACTIVE,5,1,1,0,0
#1,1900-01-01 20:30:30,0,0.0,0.0,0.07,0.0292,0.0,0.0059,0.0,ACTIVE,5,1,1,0,0
preprocessed = RawProcessing(inputfile,
cols_for_activity=["activity"], # Activity information
col_for_datetime="linetime", strftime="%Y-%m-%d %H:%M:%S", # Datetime information
col_for_pid="mesaid") # Participant information
w = Wearable(preprocessed)
w.change_start_hour_for_experiment_day(15) # We define an experiment day as the time from 15pm to 15pm the next day.
exp.add_wearable(w)
exp.set_freq_in_secs(30) # This is not a required step, but shows how easily one can change the data sampling frequency
sw = SleepWakeAnalysis(exp) # This module allows the use of sleep/wake algorithms for the night that PSG was conducted
for sleep_alg in ["Cole-Kripke", "ScrippsClinic", "Oakley", "Sadeh", "Sazonov"]:
sw.run_sleep_algorithm(sleep_alg, inplace=True)
# We can at any time visualize the signals and annotations for one (or all) participants
v = Viewer(exp.get_wearable(pid="1"))
v.view_signals_multipanel(signals=["activity", "gt"], signals_as_area=["ScrippsClinic", "Oakley", "Cole-Kripke", "Sadeh", "Sazonov"],
select_day=0, zoom=["20:00:00", "09:00:00"], colors={"signal": "black", "area": ["green", "blue", "red", "purple", "orange"]},
alphas={"area": 0.85}, labels={"signal": ["Activity", "PSG"], "area": ["Scripps Clinic", "Oakley", "Cole-Kripke", "Sadeh", "Sazonov"]})
# Now, we extract sleep metrics and compare the performance of different algorithms
sm = SleepMetrics(exp)
results = []
sleep_metrics = ["sleepEfficiency", "awakening", "arousal", "totalSleepTime", "totalWakeTime"]
for sleep_alg in ["Cole-Kripke", "ScrippsClinic", "Oakley", "Sadeh", "Sazonov"]:
results.extend(sm.compare_sleep_metrics(ground_truth="gt", wake_sleep_alg=sleep_alg, sleep_metrics=sleep_metrics,
how="pearson"))
fig = Viewer.plot_sleep_wake_by_metrics_metrics(results, figname='heatmap_sleepalg_metrics.pdf')
|
mdpi_sensors/hypnospy_sleepmetrics_by_sleepwakealgs.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Forecasting, updating datasets, and the "news"
#
# In this notebook, we describe how to use Statsmodels to compute the impacts of updated or revised datasets on out-of-sample forecasts or in-sample estimates of missing data. We follow the approach of the "Nowcasting" literature (see references at the end), by using a state space model to compute the "news" and impacts of incoming data.
#
# **Note**: this notebook applies to Statsmodels v0.12+. In addition, it only applies to the state space models or related classes, which are: `sm.tsa.statespace.ExponentialSmoothing`, `sm.tsa.arima.ARIMA`, `sm.tsa.SARIMAX`, `sm.tsa.UnobservedComponents`, `sm.tsa.VARMAX`, and `sm.tsa.DynamicFactor`.
# +
# %matplotlib inline
import numpy as np
import pandas as pd
import statsmodels.api as sm
import matplotlib.pyplot as plt
macrodata = sm.datasets.macrodata.load_pandas().data
macrodata.index = pd.period_range('1959Q1', '2009Q3', freq='Q')
# -
# Forecasting exercises often start with a fixed set of historical data that is used for model selection and parameter estimation. Then, the fitted selected model (or models) can be used to create out-of-sample forecasts. Most of the time, this is not the end of the story. As new data comes in, you may need to evaluate your forecast errors, possibly update your models, and create updated out-of-sample forecasts. This is sometimes called a "real-time" forecasting exercise (by contrast, a pseudo real-time exercise is one in which you simulate this procedure).
#
# If all that matters is minimizing some loss function based on forecast errors (like MSE), then when new data comes in you may just want to completely redo model selection, parameter estimation and out-of-sample forecasting, using the updated datapoints. If you do this, your new forecasts will have changed for two reasons:
#
# 1. You have received new data that gives you new information
# 2. Your forecasting model or the estimated parameters are different
#
# In this notebook, we focus on methods for isolating the first effect. The way we do this comes from the so-called "nowcasting" literature, and in particular Bańbura, Giannone, and Reichlin (2011), Bańbura and Modugno (2014), and Bańbura et al. (2014). They describe this exercise as computing the "**news**", and we follow them in using this language in Statsmodels.
#
# These methods are perhaps most useful with multivariate models, since there multiple variables may update at the same time, and it is not immediately obvious what forecast change was created by what updated variable. However, they can still be useful for thinking about forecast revisions in univariate models. We will therefore start with the simpler univariate case to explain how things work, and then move to the multivariate case afterwards.
# **Note on revisions**: the framework that we are using is designed to decompose changes to forecasts from newly observed datapoints. It can also take into account *revisions* to previously published datapoints, but it does not decompose them separately. Instead, it only shows the aggregate effect of "revisions".
# **Note on `exog` data**: the framework that we are using only decomposes changes to forecasts from newly observed datapoints for *modeled* variables. These are the "left-hand-side" variables that in Statsmodels are given in the `endog` arguments. This framework does not decompose or account for changes to unmodeled "right-hand-side" variables, like those included in the `exog` argument.
# ### Simple univariate example: AR(1)
#
# We will begin with a simple autoregressive model, an AR(1):
#
# $$y_t = \phi y_{t-1} + \varepsilon_t$$
#
# - The paramater $\phi$ captures the persistence of the series
#
# We will use this model to forecast inflation.
#
# To make it simpler to describe the forecast updates in this notebook, we will work with inflation data that has been de-meaned, but it is straightforward in practice to augment the model with a mean term.
#
# De-mean the inflation series
y = macrodata['infl'] - macrodata['infl'].mean()
# #### Step 1: fitting the model on the available dataset
# Here, we'll simulate an out-of-sample exercise, by constructing and fitting our model using all of the data except the last five observations. We'll assume that we haven't observed these values yet, and then in subsequent steps we'll add them back into the analysis.
y_pre = y.iloc[:-5]
y_pre.plot(figsize=(15, 3), title='Inflation');
# To construct forecasts, we first estimate the parameters of the model. This returns a results object that we will be able to use produce forecasts.
mod_pre = sm.tsa.arima.ARIMA(y_pre, order=(1, 0, 0), trend='n')
res_pre = mod_pre.fit()
print(res_pre.summary())
# Creating the forecasts from the results object `res` is easy - you can just call the `forecast` method with the number of forecasts you want to construct. In this case, we'll construct four out-of-sample forecasts.
# +
# Compute the forecasts
forecasts_pre = res_pre.forecast(4)
# Plot the last 3 years of data and the four out-of-sample forecasts
y_pre.iloc[-12:].plot(figsize=(15, 3), label='Data', legend=True)
forecasts_pre.plot(label='Forecast', legend=True);
# -
# For the AR(1) model, it is also easy to manually construct the forecasts. Denoting the last observed variable as $y_T$ and the $h$-step-ahead forecast as $y_{T+h|T}$, we have:
#
# $$y_{T+h|T} = \hat \phi^h y_T$$
#
# Where $\hat \phi$ is our estimated value for the AR(1) coefficient. From the summary output above, we can see that this is the first parameter of the model, which we can access from the `params` attribute of the results object.
# +
# Get the estimated AR(1) coefficient
phi_hat = res_pre.params[0]
# Get the last observed value of the variable
y_T = y_pre.iloc[-1]
# Directly compute the forecasts at the horizons h=1,2,3,4
manual_forecasts = pd.Series([phi_hat * y_T, phi_hat**2 * y_T,
phi_hat**3 * y_T, phi_hat**4 * y_T],
index=forecasts_pre.index)
# We'll print the two to double-check that they're the same
print(pd.concat([forecasts_pre, manual_forecasts], axis=1))
# -
# #### Step 2: computing the "news" from a new observation
#
# Suppose that time has passed, and we have now received another observation. Our dataset is now larger, and we can evaluate our forecast error and produce updated forecasts for the subsequent quarters.
# +
# Get the next observation after the "pre" dataset
y_update = y.iloc[-5:-4]
# Print the forecast error
print('Forecast error: %.2f' % (y_update.iloc[0] - forecasts_pre.iloc[0]))
# -
# To compute forecasts based on our updated dataset, we will create an updated results object `res_post` using the `append` method, to append on our new observation to the previous dataset.
#
# Note that by default, the `append` method does not re-estimate the parameters of the model. This is exactly what we want here, since we want to isolate the effect on the forecasts of the new information only.
# +
# Create a new results object by passing the new observations to the `append` method
res_post = res_pre.append(y_update)
# Since we now know the value for 2008Q3, we will only use `res_post` to
# produce forecasts for 2008Q4 through 2009Q2
forecasts_post = pd.concat([y_update, res_post.forecast('2009Q2')])
print(forecasts_post)
# -
# In this case, the forecast error is quite large - inflation was more than 10 percentage points below the AR(1) models' forecast. (This was largely because of large swings in oil prices around the global financial crisis).
# To analyse this in more depth, we can use Statsmodels to isolate the effect of the new information - or the "**news**" - on our forecasts. This means that we do not yet want to change our model or re-estimate the parameters. Instead, we will use the `news` method that is available in the results objects of state space models.
#
# Computing the news in Statsmodels always requires a *previous* results object or dataset, and an *updated* results object or dataset. Here we will use the original results object `res_pre` as the previous results and the `res_post` results object that we just created as the updated results.
# Once we have previous and updated results objects or datasets, we can compute the news by calling the `news` method. Here, we will call `res_pre.news`, and the first argument will be the updated results, `res_post` (however, if you have two results objects, the `news` method could can be called on either one).
#
# In addition to specifying the comparison object or dataset as the first argument, there are a variety of other arguments that are accepted. The most important specify the "impact periods" that you want to consider. These "impact periods" correspond to the forecasted periods of interest; i.e. these dates specify with periods will have forecast revisions decomposed.
#
# To specify the impact periods, you must pass two of `start`, `end`, and `periods` (similar to the Pandas `date_range` method). If your time series was a Pandas object with an associated date or period index, then you can pass dates as values for `start` and `end`, as we do below.
# Compute the impact of the news on the four periods that we previously
# forecasted: 2008Q3 through 2009Q2
news = res_pre.news(res_post, start='2008Q3', end='2009Q2')
# Note: one alternative way to specify these impact dates is
# `start='2008Q3', periods=4`
# The variable `news` is an object of the class `NewsResults`, and it contains details about the updates to the data in `res_post` compared to `res_pre`, the new information in the updated dataset, and the impact that the new information had on the forecasts in the period between `start` and `end`.
#
# One easy way to summarize the results are with the `summary` method.
news.summary()
# **Summary output**: the default summary for this news results object printed four tables:
#
# 1. Summary of the model and datasets
# 2. Details of the news from updated data
# 3. Summary of the impacts of the new information on the forecasts between `start='2008Q3'` and `end='2009Q2'`
# 4. Details of how the updated data led to the impacts on the forecasts between `start='2008Q3'` and `end='2009Q2'`
#
# These are described in more detail below.
#
# *Notes*:
#
# - There are a number of arguments that can be passed to the `summary` method to control this output. Check the documentation / docstring for details.
# - Table (4), showing details of the updates and impacts, can become quite large if the model is multivariate, there are multiple updates, or a large number of impact dates are selected. It is only shown by default for univariate models.
# **First table: summary of the model and datasets**
#
# The first table, above, shows:
#
# - The type of model from which the forecasts were made. Here this is an ARIMA model, since an AR(1) is a special case of an ARIMA(p,d,q) model.
# - The date and time at which the analysis was computed.
# - The original sample period, which here corresponds to `y_pre`
# - The endpoint of the updated sample period, which here is the last date in `y_post`
# **Second table: the news from updated data**
#
# This table simply shows the forecasts from the previous results for observations that were updated in the updated sample.
#
# *Notes*:
#
# - Our updated dataset `y_post` did not contain any *revisions* to previously observed datapoints. If it had, there would be an additional table showing the previous and updated values of each such revision.
# **Third table: summary of the impacts of the new information**
#
# *Columns*:
#
# The third table, above, shows:
#
# - The previous forecast for each of the impact dates, in the "estimate (prev)" column
# - The impact that the new information (the "news") had on the forecasts for each of the impact dates, in the "impact of news" column
# - The updated forecast for each of the impact dates, in the "estimate (new)" column
#
# *Notes*:
#
# - In multivariate models, this table contains additional columns describing the relevant impacted variable for each row.
# - Our updated dataset `y_post` did not contain any *revisions* to previously observed datapoints. If it had, there would be additional columns in this table showing the impact of those revisions on the forecasts for the impact dates.
# - Note that `estimate (new) = estimate (prev) + impact of news`
# - This table can be accessed independently using the `summary_impacts` method.
#
# *In our example*:
#
# Notice that in our example, the table shows the values that we computed earlier:
#
# - The "estimate (prev)" column is identical to the forecasts from our previous model, contained in the `forecasts_pre` variable.
# - The "estimate (new)" column is identical to our `forecasts_post` variable, which contains the observed value for 2008Q3 and the forecasts from the updated model for 2008Q4 - 2009Q2.
# **Fourth table: details of updates and their impacts**
#
# The fourth table, above, shows how each new observation translated into specific impacts at each impact date.
#
# *Columns*:
#
# The first three columns table described the relevant **update** (an "updated" is a new observation):
#
# - The first column ("update date") shows the date of the variable that was updated.
# - The second column ("forecast (prev)") shows the value that would have been forecasted for the update variable at the update date based on the previous results / dataset.
# - The third column ("observed") shows the actual observed value of that updated variable / update date in the updated results / dataset.
#
# The last four columns described the **impact** of a given update (an impact is a changed forecast within the "impact periods").
#
# - The fourth column ("impact date") gives the date at which the given update made an impact.
# - The fifth column ("news") shows the "news" associated with the given update (this is the same for each impact of a given update, but is just not sparsified by default)
# - The sixth column ("weight") describes the weight that the "news" from the given update has on the impacted variable at the impact date. In general, weights will be different between each "updated variable" / "update date" / "impacted variable" / "impact date" combination.
# - The seventh column ("impact") shows the impact that the given update had on the given "impacted variable" / "impact date".
#
# *Notes*:
#
# - In multivariate models, this table contains additional columns to show the relevant variable that was updated and variable that was impacted for each row. Here, there is only one variable ("infl"), so those columns are suppressed to save space.
# - By default, the updates in this table are "sparsified" with blanks, to avoid repeating the same values for "update date", "forecast (prev)", and "observed" for each row of the table. This behavior can be overridden using the `sparsify` argument.
# - Note that `impact = news * weight`.
# - This table can be accessed independently using the `summary_details` method.
#
# *In our example*:
#
# - For the update to 2008Q3 and impact date 2008Q3, the weight is equal to 1. This is because we only have one variable, and once we have incorporated the data for 2008Q3, there is no no remaining ambiguity about the "forecast" for this date. Thus all of the "news" about this variable at 2008Q3 passes through to the "forecast" directly.
# #### Addendum: manually computing the news, weights, and impacts
#
# For this simple example with a univariate model, it is straightforward to compute all of the values shown above by hand. First, recall the formula for forecasting $y_{T+h|T} = \phi^h y_T$, and note that it follows that we also have $y_{T+h|T+1} = \phi^h y_{T+1}$. Finally, note that $y_{T|T+1} = y_T$, because if we know the value of the observations through $T+1$, we know the value of $y_T$.
#
# **News**: The "news" is nothing more than the forecast error associated with one of the new observations. So the news associated with observation $T+1$ is:
#
# $$n_{T+1} = y_{T+1} - y_{T+1|T} = Y_{T+1} - \phi Y_T$$
#
# **Impacts**: The impact of the news is the difference between the updated and previous forecasts, $i_h \equiv y_{T+h|T+1} - y_{T+h|T}$.
#
# - The previous forecasts for $h=1, \dots, 4$ are: $\begin{pmatrix} \phi y_T & \phi^2 y_T & \phi^3 y_T & \phi^4 y_T \end{pmatrix}'$.
# - The updated forecasts for $h=1, \dots, 4$ are: $\begin{pmatrix} y_{T+1} & \phi y_{T+1} & \phi^2 y_{T+1} & \phi^3 y_{T+1} \end{pmatrix}'$.
#
# The impacts are therefore:
#
# $$\{ i_h \}_{h=1}^4 = \begin{pmatrix} y_{T+1} - \phi y_T \\ \phi (Y_{T+1} - \phi y_T) \\ \phi^2 (Y_{T+1} - \phi y_T) \\ \phi^3 (Y_{T+1} - \phi y_T) \end{pmatrix}$$
#
# **Weights**: To compute the weights, we just need to note that it is immediate that we can rewrite the impacts in terms of the forecast errors, $n_{T+1}$.
#
# $$\{ i_h \}_{h=1}^4 = \begin{pmatrix} 1 \\ \phi \\ \phi^2 \\ \phi^3 \end{pmatrix} n_{T+1}$$
#
# The weights are then simply $w = \begin{pmatrix} 1 \\ \phi \\ \phi^2 \\ \phi^3 \end{pmatrix}$
# We can check that this is what the `news` method has computed.
# +
# Print the news, computed by the `news` method
print(news.news)
# Manually compute the news
print()
print((y_update.iloc[0] - phi_hat * y_pre.iloc[-1]).round(6))
# +
# Print the total impacts, computed by the `news` method
# (Note: news.total_impacts = news.revision_impacts + news.update_impacts, but
# here there are no data revisions, so total and update impacts are the same)
print(news.total_impacts)
# Manually compute the impacts
print()
print(forecasts_post - forecasts_pre)
# +
# Print the weights, computed by the `news` method
print(news.weights)
# Manually compute the weights
print()
print(np.array([1, phi_hat, phi_hat**2, phi_hat**3]).round(6))
# -
# ### Multivariate example: dynamic factor
#
# In this example, we'll consider forecasting monthly core price inflation based on the Personal Consumption Expenditures (PCE) price index and the Consumer Price Index (CPI), using a Dynamic Factor model. Both of these measures track prices in the US economy and are based on similar source data, but they have a number of definitional differences. Nonetheless, they track each other relatively well, so modeling them jointly using a single dynamic factor seems reasonable.
#
# One reason that this kind of approach can be useful is that the CPI is released earlier in the month than the PCE. One the CPI is released, therefore, we can update our dynamic factor model with that additional datapoint, and obtain an improved forecast for that month's PCE release. A more inolved version of this kind of analysis is available in Knotek and Zaman (2017).
# We start by downloading the core CPI and PCE price index data from [FRED](https://fred.stlouisfed.org/), converting them to annualized monthly inflation rates, removing two outliers, and de-meaning each series (the dynamic factor model does not
# +
import pandas_datareader as pdr
levels = pdr.get_data_fred(['PCEPILFE', 'CPILFESL'], start='1999', end='2019').to_period('M')
infl = np.log(levels).diff().iloc[1:] * 1200
infl.columns = ['PCE', 'CPI']
# Remove two outliers and de-mean the series
infl['PCE'].loc['2001-09':'2001-10'] = np.nan
# -
# To show how this works, we'll imagine that it is April 14, 2017, which is the data of the March 2017 CPI release. So that we can show the effect of multiple updates at once, we'll assume that we haven't updated our data since the end of January, so that:
#
# - Our **previous dataset** will consist of all values for the PCE and CPI through January 2017
# - Our **updated dataset** will additionally incorporate the CPI for February and March 2017 and the PCE data for February 2017. But it will not yet the PCE (the March 2017 PCE price index wasn't released until May 1, 2017).
# Previous dataset runs through 2017-02
y_pre = infl.loc[:'2017-01'].copy()
const_pre = np.ones(len(y_pre))
print(y_pre.tail())
# +
# For the updated dataset, we'll just add in the
# CPI value for 2017-03
y_post = infl.loc[:'2017-03'].copy()
y_post.loc['2017-03', 'PCE'] = np.nan
const_post = np.ones(len(y_post))
# Notice the missing value for PCE in 2017-03
print(y_post.tail())
# -
# We chose this particular example because in March 2017, core CPI prices fell for the first time since 2010, and this information may be useful in forecast core PCE prices for that month. The graph below shows the CPI and PCE price data as it would have been observed on April 14th$^\dagger$.
#
# -----
#
# $\dagger$ This statement is not entirely true, becuase both the CPI and PCE price indexes can be revised to a certain extent after the fact. As a result, the series that we're pulling are not exactly like those observed on April 14, 2017. This could be fixed by pulling the archived data from [ALFRED](https://alfred.stlouisfed.org/) instead of [FRED](https://fred.stlouisfed.org/), but the data we have is good enough for this tutorial.
# Plot the updated dataset
fig, ax = plt.subplots(figsize=(15, 3))
y_post.plot(ax=ax)
ax.hlines(0, '2009', '2017-06', linewidth=1.0)
ax.set_xlim('2009', '2017-06');
# To perform the exercise, we first construct and fit a `DynamicFactor` model. Specifically:
#
# - We are using a single dynamic factor (`k_factors=1`)
# - We are modeling the factor's dynamics with an AR(6) model (`factor_order=6`)
# - We have included a vector of ones as an exogenous variable (`exog=const_pre`), because the inflation series we are working with are not mean-zero.
mod_pre = sm.tsa.DynamicFactor(y_pre, exog=const_pre, k_factors=1, factor_order=6)
res_pre = mod_pre.fit()
print(res_pre.summary())
# With the fitted model in hand, we now construct the news and impacts associated with observing the CPI for March 2017. The updated data is for February 2017 and part of March 2017, and we'll examing the impacts on both March and April.
#
# In the univariate example, we first created an updated results object, and then passed that to the `news` method. Here, we're creating the news by directly passing the updated dataset.
#
# Notice that:
#
# 1. `y_post` contains the entire updated dataset (not just the new datapoints)
# 2. We also had to pass an updated `exog` array. This array must cover **both**:
# - The entire period associated with `y_post`
# - Any additional datapoints after the end of `y_post` through the last impact date, specified by `end`
#
# Here, `y_post` ends in March 2017, so we needed our `exog` to extend one more period, to April 2017.
# Create the news results
# Note
const_post_plus1 = np.ones(len(y_post) + 1)
news = res_pre.news(y_post, exog=const_post_plus1, start='2017-03', end='2017-04')
# > **Note**:
# >
# > In the univariate example, above, we first constructed a new results object, and then passed that to the `news` method. We could have done that here too, although there is an extra step required. Since we are requesting an impact for a period beyond the end of `y_post`, we would still need to pass the additional value for the `exog` variable during that period to `news`:
# >
# > ```python
# res_post = res_pre.apply(y_post, exog=const_post)
# news = res_pre.news(res_post, exog=[1.], start='2017-03', end='2017-04')
# ```
# Now that we have computed the `news`, printing `summary` is a convenient way to see the results.
# Show the summary of the news results
news.summary()
# Because we have multiple variables, by default the summary only shows the news from updated data along and the total impacts.
#
# From the first table, we can see that our updated dataset contains three new data points, with most of the "news" from these data coming from the very low reading in March 2017.
#
# The second table shows that these three datapoints substantially impacted the estimate for PCE in March 2017 (which was not yet observed). This estimate revised down by nearly 1.5 percentage points.
#
# The updated data also impacted the forecasts in the first out-of-sample month, April 2017. After incorporating the new data, the model's forecasts for CPI and PCE inflation in that month revised down 0.29 and 0.17 percentage point, respectively.
# While these tables show the "news" and the total impacts, they do not show how much of each impact was caused by each updated datapoint. To see that information, we need to look at the details tables.
#
# One way to see the details tables is to pass `include_details=True` to the `summary` method. To avoid repeating the tables above, however, we'll just call the `summary_details` method directly.
news.summary_details()
# This table shows that most of the revisions to the estimate of PCE in April 2017, described above, came from the news associated with the CPI release in March 2017. By contrast, the CPI release in February had only a little effect on the April forecast, and the PCE release in February had essentially no effect.
# ### Bibliography
#
# Bańbura, Marta, <NAME>, and <NAME>. "Nowcasting." The Oxford Handbook of Economic Forecasting. July 8, 2011.
#
# Bańbura, Marta, <NAME>, <NAME>, and <NAME>. "Now-casting and the real-time data flow." In Handbook of economic forecasting, vol. 2, pp. 195-237. Elsevier, 2013.
#
# Bańbura, Marta, and <NAME>. "Maximum likelihood estimation of factor models on datasets with arbitrary pattern of missing data." Journal of Applied Econometrics 29, no. 1 (2014): 133-160.
#
# Knotek, <NAME>., and <NAME>. "Nowcasting US headline and core inflation." Journal of Money, Credit and Banking 49, no. 5 (2017): 931-968.
|
examples/notebooks/statespace_news.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# # Introduzione a Tensorflow
#
# TensorFlow (TF) è la API di Google per lo sviluppo di modelli di Deep Learning. Interagiremo con TF in Python, ma è possibile usare altri linguaggi, almeno a basso livello. La tipica applicazione TF lavora secondo lo schema riportato di seguito:
#
# <img src="https://pic1.zhimg.com/80/v2-8b46a7f55b77f0febfa3ad5084e25c3c_1440w.jpg" alt="Architettura TF" width="50%" />
#
# - Keras è una nota libreria per specificare ad alto livello i layer del modello che lavora anche con altri back-end
# - La Data API è il modulo `tf.data` di TF che serve a specificare le azioni da compiere sul data set ai fini di addestramento e test
# - L'Execution Engine di TF (che può essere locale o anche distribuito su più nodi di un cluster di calcolo) rappresenta il nucleo vero e proprio di TF. A questo livello, vengono esposte API in diversi linguaggi.
#
# Nell'execution engine, viene definita la _sessione_ di lavoro `tf.Session(...)` in cui si svolgono le operazioni atomiche (`tf.Operation`) sui dati in forma tensoriale (`tf.Tensor`).
#
# La sessione gestisce l'esecuzione di un _*grafo di computaizone*_ ovvero di una specifica _astratta_ della sequenza di operazioni secondo una struttura a grafo dove i nodi sono operazioni che restituiscono tensori che vanno in ingresso ad altri nodi secondo la struttura definita dagli archi.
#
# <img src="https://miro.medium.com/max/2994/1*vPb9E0Yd1QUAD0oFmAgaOw.png" alt="Esempio grafo computazione" width="50%" />
#
# L'esecuzione si ottiene attraverso il metodo `tf.Operation.run()` che è ereditato anche dalla sessione ovvero si può valutare un singolo tensore con `tf.Tensor.eval()`. La sessione va chiusa esplicitamente con il metodo `close()`.
#
#
# +
import tensorflow as tf
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "2"
# Sostituisce la vecchia chiamata tf.Session()
# che è deprecata da quando è stato rilasciato TF v.2
sess = tf.compat.v1.Session()
# chiediamo la lista dei device di calcolo presenti
# quelli di tipo XLA_GPU o XLA_CPU sono abilitati all'algebra lineare accelerata
for d in sess.list_devices():
print(d.name)
# Definiamo una semplice computazione che definisce un elemento (o meglio un **nodo**)
# costante di tipo stringa e lo esegue ottenendone la stampa
bye_bye = tf.constant('Hello World')
result = sess.run(bye_bye)
print(result)
print(f'The session is closed? {sess._closed}')
# chiudiamo la sessione
sess.close()
# -
# La sessione può essere invocata con diverse opzioni e, soprattutto, con la possibilità di definire un _context manager_ all'interno del quale specificare le operazioni che non richiede più la chiusura esplicita.
# +
with tf.compat.v1.Session() as sess:
# definizione di due valori costanti interi
n1=tf.constant(2)
n2=tf.constant(3)
print(n1.eval())
#n3 = n1 * n2 # questa **non è** la moltiplicaione tra interi,
# ma la tf.Operation di moltiplicazione tra tensori
n3 = tf.multiply(n1,n2)
print(n1,n2,n3,sep='\n')
#print(sess.run(n3))
print(n3.eval(), n3.dtype, n3.shape)
print(f'Session closed: {sess._closed}')
# -
# La sintassi completa del costruttore dell'oggetto `tf.compat.v1.Session` è:
#
# ```python
# tf.compat.v1.Session(target='',\ # engine di computazione da utilizzare, locale o distribuito
# graph=None,\ # grafo di computazione da utilizzare
# config=None) # oggetto con le specifiche particolari di configurazione
# ```
#
# Apriamo la sessione con una configurazione di default per l'allocazopne dinamica della computazione sui diversi device disponibili ed eseguiamo il grafo di computazione della figura precedente
# +
import tensorflow as tf
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "2"
# Usiamo esplicitamente l'oggetto di configurazione
sess = tf.compat.v1.Session(config=tf.compat.v1.ConfigProto(
allow_soft_placement=True, # gestione dinamica dell'allocazione dei device
log_device_placement=True)) # registrazione del log sull'allocazione dei device
with sess.as_default():
assert tf.compat.v1.get_default_session() is sess # impostiamo la nostra sessione configurata come quella di default
# La nostra computazione è: res = (a*b) / (a+b)
# ingressi
a = tf.constant(5)
b = tf.constant(3)
# operazioni intermedie
prod = tf.multiply(a,b)
sum = tf.add(a,b)
# uscita
#res = tf.div(prod,sum)
res = tf.math.divide(prod,sum)
#print(res.eval())
print(sess.run(res))
# +
# Creiamo tensori con diverse funzioni
mat = tf.constant([[1., 2., 3.], [4., 5., 6.]])
print(mat.shape, mat.dtype)
# +
mat_randn = tf.random.normal((3,3), mean=0, stddev=1.0) # A 3x3 random normal matrix.
mat_randu = tf.random.uniform((4,4), minval=0, maxval=1.0)
print(mat_randn)
with sess.as_default():
assert tf.compat.v1.get_default_session() is sess
print(mat_randn.eval())
print(mat_randu.eval())
# -
# Esempio di uso delle variabili
with tf.compat.v1.Session(config=tf.compat.v1.ConfigProto(allow_soft_placement=True)) as sess:
init_values = tf.random.uniform((4,4), minval=0, maxval=1.0)
t = tf.Variable(initial_value=init_values,name='myvar')
init = tf.compat.v1.global_variables_initializer()
print(sess.run(init))
print(sess.run(t))
# Definiamo i placeholder per z = 2x^2 + 2xy
with tf.compat.v1.Session(config=tf.compat.v1.ConfigProto(allow_soft_placement=True)) as sess:
two = tf.constant(2.0)
x = tf.compat.v1.placeholder(tf.float32,shape=(None, 3))
y = tf.compat.v1.placeholder(tf.float32,shape=(None, 3))
z = tf.add(tf.multiply(two, tf.multiply(x, x)),\
tf.multiply(two, tf.multiply(x, y)))
print(sess.run(z, feed_dict={x: [[1., 2., 3.],[4., 5., 6.]], y: [[3., 4., 5.],[7., 8., 9.]]}))
|
tensorflow.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: data
# language: python
# name: data
# ---
# # Notebook to try to get the CPI (Consumer Price Index) for USA and Uruguay from the World Bank API
# ## World Bank Explaination on the data
#
# ```
# Consumer price index (2010 = 100)
#
# Consumer price index reflects changes in the cost to the average consumer of acquiring a basket of goods and services that may be fixed or changed at specified intervals, such as yearly. The Laspeyres formula is generally used. Data are period averages.
#
# Source: International Monetary Fund, International Financial Statistics and data files.
# License: CC BY-4.0
# Base Period: 2010
#
# Development Relevance: A general and continuing increase in an economy’s price level is called inflation. The increase in the average prices of goods and services in the economy should be distinguished from a change in the relative prices of individual goods and services. Generally accompanying an overall increase in the price level is a change in the structure of relative prices, but it is only the average increase, not the relative price changes, that constitutes inflation. A commonly used measure of inflation is the consumer price index, which measures the prices of a representative basket of goods and services purchased by a typical household. The consumer price index is usually calculated on the basis of periodic surveys of consumer prices. Other price indices are derived implicitly from indexes of current and constant price series.
#
# Limitations and Exceptions: Consumer price indexes should be interpreted with caution. The definition of a household, the basket of goods, and the geographic (urban or rural) and income group coverage of consumer price surveys can vary widely by country. In addition, weights are derived from household expenditure surveys, which, for budgetary reasons, tend to be conducted infrequently in developing countries, impairing comparability over time. Although useful for measuring consumer price inflation within a country, consumer price indexes are of less value in comparing countries.
#
# Long Definition: Consumer price index reflects changes in the cost to the average consumer of acquiring a basket of goods and services that may be fixed or changed at specified intervals, such as yearly. The Laspeyres formula is generally used. Data are period averages.
#
# Periodicity: Annual
#
# Statistical Concept and Methodology: Consumer price indexes are constructed explicitly, using surveys of the cost of a defined basket of consumer goods and services.
# ```
import requests
import pandas as pd
from collections import defaultdict
import matplotlib.pyplot as plt
# %matplotlib inline
# Let's try to get some Consumer Price Indexes
CPI_CODE = 'FP.CPI.TOTL'
def download_index(country_code,
index_code,
start_date=1960,
end_date=2018):
"""
Get a JSON response for the index data of one country.
Args:
country_code(str): The two letter code for the World Bank webpage
index_code(str): The code for the index to retreive
start_date(int): The initial year to retreive
end_date(int): The final year to retreive
Returns:
str: a JSON string with the raw data
"""
payload = {'format': 'json',
'per_page': '500',
'date':'{}:{}'.format(str(start_date), str(end_date))
}
r = requests.get(
'http://api.worldbank.org/v2/countries/{}/indicators/{}'.format(
country_code, index_code), params=payload)
return r
def format_response(raw_res):
"""
Formats a raw JSON string, returned from the World Bank API into a
pandas DataFrame.
"""
result = defaultdict(dict)
for record in raw_res.json()[1]:
result[record['country']['value']].update(
{record['date']: record['value']})
return pd.DataFrame(result)
def download_cpi(country_code, **kwargs):
"""
Downloads the Consumer Price Index for one country, and returns the data
as a pandas DataFrame.
Args:
country_code(str): The two letter code for the World Bank webpage
**kwargs: Arguments for 'download_index', for example:
start_date(int): The initial year to retreive
end_date(int): The final year to retreive
"""
CPI_CODE = 'FP.CPI.TOTL'
raw_res = download_index(country_code, CPI_CODE, **kwargs)
return format_response(raw_res)
def download_cpis(country_codes, **kwargs):
"""
Download many countries CPIs and store them in a pandas DataFrame.
Args:
country_codes(list(str)): A list with the two letter country codes
**kwargs: Other keyword arguments, such as:
start_date(int): The initial year to retreive
end_date(int): The final year to retreive
Returns:
pd.DataFrame: A dataframe with the CPIs for all the countries in the
input list.
"""
cpi_list = [download_cpi(code, **kwargs) for code in country_codes]
return pd.concat(cpi_list, axis=1)
# ### Testing
cpi = download_cpis(['uy', 'us'])
print(cpi.shape)
cpi.head()
cpi.plot()
res = download_index('uy', 'FP.CPI.TOTL', 1960, 2017)
cpi = format_response(res)
print(cpi.shape)
cpi.head()
cpi.plot()
# +
END_DATE = 2018
cpi = download_cpi('uy', end_date=END_DATE).join(
download_cpi('us', end_date=END_DATE))
print(cpi.shape)
cpi.head()
# -
cpi.plot()
|
notebooks/002-mt-getting-the-cpi-from-the-world-bank.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
import random
import numpy as np
from math import erfc
import matplotlib.pyplot as plt
# %matplotlib inline
# +
def wiener(n, dt, t_init=0, w_init=0.0):
"""Returns one realization of a Wiener process with n steps of length dt.
The time and Wiener series can be initialized using t_init and w_init respectively."""
n+=1
t_series = np.arange(t_init,n*dt,dt)
h = t_series[1]-t_series[0]
z = np.random.normal(0.0,1.0,n)
dw = np.sqrt(h)*z
dw[0] = w_init
w_series = dw.cumsum()
return t_series, w_series
def gaussian(x, mu, sig):
return np.exp(-(x-mu)**2/2/sig**2)/np.sqrt(2*np.pi)/sig
# -
def get_fds(w,lim=None):
'''returns a finite difference series based on the input data
w = input data series
lim = returned differences are between +/-lim.'''
return [w[i+1]-w[i] for i in range(len(w)-1) if lim==None or abs(w[i+1]-w[i])<lim]
def raise_res(T, W, c, mu=0, sigma=1):
'''Increase the resolution of a wiener series by a factor of c.
T = the given Time series.
W = the associated Wiener series.
c = Scaling factor (integer greater than 1).
mu = Mean of W's underlying normal distribution.
sigma = Standard deviation of W's underlying normal distribution.
'''
dT = T[1]-T[0]
dt = float(T[1]-T[0])/c
t_series = []
w_series = []
for i in range(len(T)-1):
t = T[i]
w_t = W[i]
t_next = T[i+1]
w_next = W[i+1]
t_series.append(t)
w_series.append(w_t)
for j in range(c-1):
t+=dt
dW = (w_next-w_t)
if np.sqrt(2)*np.sqrt(t_next-t)*sigma*erfc(-2*random.random())<abs(dW):
w_t+=np.abs(random.gauss(0,np.sqrt(dt)*sigma))*float(dW)/abs(dW)
else:
w_t+=random.gauss(0,np.sqrt(dt)*sigma)
t_series.append(t)
w_series.append(w_t)
t_series.append(T[-1])
w_series.append(W[-1])
return t_series,w_series
# +
c=100
num = 3
dt = 1.
T1,W1 = wiener(num*c,dt/c)
T2 = [T1[i] for i in range(len(T1)) if i%c==0]
W2 = [W1[i] for i in range(len(W1)) if i%c==0]
Tn,Wn = raise_res(T2,W2,c)
plt.figure(figsize=(15,6))
plt.title('A True Wiener Series, Its Skeleton, and a Scaled Wiener Series Derived From the Skeleton',fontsize=15)
plt.plot(T1,W1,label='actual',color='b')
plt.plot(Tn,Wn,label='scaled',color='m')
plt.plot(T2,W2,label='skeleton',color='k',linewidth=2,marker='o',markersize=6)
plt.ylabel('$W(t)$',fontsize=15)
plt.xlabel('$t$',fontsize=18)
plt.legend()
plt.show()
dW1 = get_fds(W1)
bnum1 = (max(dW1)-min(dW1))*50
dWn = get_fds(Wn)
bnumn = (max(dWn)-min(dWn))*50
plt.figure(figsize=(15,4))
ax1 = plt.subplot(121)
ax1.set_title('Finite Differences From the Actual Wiener Series')
ax1.set_ylabel('count',fontsize=13)
ax1.set_xlabel('$dW$',fontsize=15)
plt.hist(dW1,bnum1,label='actual')
plt.legend()
ax2 = plt.subplot(122,sharey=ax1)
ax2.set_title('Finite Differences From the Scaled Wiener Series')
ax2.set_ylabel('count',fontsize=13)
ax2.set_xlabel('$dW$',fontsize=15)
plt.hist(dWn,bnumn,label='scaled',color='m')
plt.legend()
plt.show()
# +
c=1000
num = 5
dt = 1.
T1,W1 = wiener(num*c,dt/c)
T2 = [T1[i] for i in range(len(T1)) if i%c==0]
W2 = [W1[i] for i in range(len(W1)) if i%c==0]
Tn,Wn = raise_res(T2,W2,c)
plt.figure(figsize=(15,6))
plt.title('A True Wiener Series, Its Skeleton, and a Scaled Wiener Series Derived From the Skeleton',fontsize=15)
plt.plot(T1,W1,label='actual',color='b')
plt.plot(Tn,Wn,label='scaled',color='m')
plt.plot(T2,W2,label='skeleton',color='k',linewidth=2,marker='o',markersize=6)
plt.ylabel('$W(t)$',fontsize=15)
plt.xlabel('$t$',fontsize=18)
plt.legend()
plt.show()
dW1 = get_fds(W1)
bnum1 = (max(dW1)-min(dW1))*150
dWn = get_fds(Wn)
bnumn = (max(dWn)-min(dWn))*150
plt.figure(figsize=(15,4))
ax1 = plt.subplot(121)
ax1.set_title('Finite Differences From the Actual Wiener Series')
ax1.set_ylabel('count',fontsize=13)
ax1.set_xlabel('$dW$',fontsize=15)
plt.hist(dW1,bnum1,label='actual')
plt.legend()
ax2 = plt.subplot(122,sharey=ax1)
ax2.set_title('Finite Differences From the Scaled Wiener Series')
ax2.set_ylabel('count',fontsize=13)
ax2.set_xlabel('$dW$',fontsize=15)
plt.hist(dWn,bnumn,label='scaled',color='m')
plt.legend()
plt.show()
# +
c=1000
num = 50
dt = 1.
T1,W1 = wiener(num*c,dt/c)
T2 = [T1[i] for i in range(len(T1)) if i%c==0]
W2 = [W1[i] for i in range(len(W1)) if i%c==0]
Tn,Wn = raise_res(T2,W2,c)
plt.figure(figsize=(15,6))
plt.title('A True Wiener Series, Its Skeleton, and a Scaled Wiener Series Derived From the Skeleton',fontsize=15)
plt.plot(T1,W1,label='actual',color='b')
plt.plot(Tn,Wn,label='scaled',color='m')
plt.plot(T2,W2,label='skeleton',color='k',linewidth=2,marker='o',markersize=6)
plt.ylabel('$W(t)$',fontsize=15)
plt.xlabel('$t$',fontsize=18)
plt.legend()
plt.show()
dW1 = get_fds(W1)
bnum1 = (max(dW1)-min(dW1))*150
dWn = get_fds(Wn)
bnumn = (max(dWn)-min(dWn))*150
plt.figure(figsize=(15,4))
ax1 = plt.subplot(121)
ax1.set_title('Finite Differences From the Actual Wiener Series')
ax1.set_ylabel('count',fontsize=13)
ax1.set_xlabel('$dW$',fontsize=15)
plt.hist(dW1,bnum1,label='actual')
plt.legend()
ax2 = plt.subplot(122,sharey=ax1)
ax2.set_title('Finite Differences From the Scaled Wiener Series')
ax2.set_ylabel('count',fontsize=13)
ax2.set_xlabel('$dW$',fontsize=15)
plt.hist(dWn,bnumn,label='scaled',color='m')
plt.legend()
plt.show()
# -
from brownian.wiener import raise_res
# +
c=100
num = 3
dt = 1.
T1,W1 = wiener(num*c,dt/c)
T2 = [T1[i] for i in range(len(T1)) if i%c==0]
W2 = [W1[i] for i in range(len(W1)) if i%c==0]
Tn,Wn = raise_res(T2,W2,c)
plt.figure(figsize=(15,6))
plt.title('A True Wiener Series, Its Skeleton, and a Scaled Wiener Series Derived From the Skeleton',fontsize=15)
plt.plot(T1,W1,label='actual',color='b')
plt.plot(Tn,Wn,label='scaled',color='m')
plt.plot(T2,W2,label='skeleton',color='k',linewidth=2,marker='o',markersize=6)
plt.ylabel('$W(t)$',fontsize=15)
plt.xlabel('$t$',fontsize=18)
plt.legend()
plt.show()
# -
def autocorr(x):
result = np.correlate(x, x, mode='full')
return result[result.size/2:]
# +
a_T1 = autocorr(T1)
a_Tn = autocorr(Tn)
a_1ndif = a_T1-a_Tn
plt.figure(figsize=(5,10))
ax1 = plt.subplot(311)
ax1.set_title('acf plots',fontsize=15)
ax1.set_ylabel('acf actual',fontsize=12)
plt.plot(range(len(a_T1)),a_T1,label='actual')
plt.legend()
ax2 = plt.subplot(312,sharex=ax1)
ax2.set_ylabel('acf',fontsize=12)
plt.plot(range(len(a_Tn)),a_Tn,label='scaled')
plt.legend()
ax3 = plt.subplot(313,sharex=ax1)
ax3.set_ylabel('acf difference',fontsize=12)
ax3.set_xlabel('lag',fontsize=12)
plt.plot(range(len(a_1ndif)),a_1ndif,label='actual - scaled')
plt.legend()
plt.show()
# -
|
brwnn-raise_res.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Assignment 2 - Bacterial Bomb
# +
# Packages
# %matplotlib inline
import random # used to randomise wind directions and settling rates
import time # used to measure model runtime
import csv # used to format loaded text files as csvs
import matplotlib.pyplot as plt # used to plot density map
import numpy as np # used to save results to text file
from ipywidgets import interact # used for scroll bars
#import pandas as pd # used for table?
#random.seed(100) # Used to maintain consistency for model testing - not needed for gneral use
# +
# Variables
#The model begins with the colony being released at a set height.
#the colony size and intial elevation are listed here:
Colony_Size = 5000 # Total number of Bacteria Released by Bomb
startheight = 75 # Elevation Bacteria are intially released at
#the below code adds slider bars allowing the user to easily alter the colony size and starting elevation
@interact(colony_size=(0,10000))
def set_cs(colony_size):
Colony_Size = colony_size
@interact(elevation=(50,100))
def set_sh(elevation):
startheight = elevation
# Each second the wind carries each bacterium in one of 4 directions.
# The liklihood of moving in each compass direction are dictated below:
NorthW = 10 # percentage likelihood of wind carrying a bacterium north
EastW = 75 # percentage likelihood of wind carrying a bacterium east
SouthW = 10 # percentage likelihood of wind carrying a bacterium south
WestW = 5 # percentage likelihood of wind carrying a bacterium west
StrengthW = 1 # distance wind carries bacteria each second
# Any exposed bacteria (those above or at the start height) may be carried upwards by turbulence.
# The likelihood of this is dictated by the following variables:
Rise = 20 # percentage likelihood of wind turbulence lifting an exposed bacterium 1 unit
Fall = 70 # percentage likelihood of exposed bacterium dropping 1 unit
# The excess 10% is covered by cases where the bacteria neither rises or falls
# The following variables are used at output to provide insight into the model outcome.
timer = 0 # counts how many 'seconds' would have passed before model completion
lost = 0 # counts how many bacteria leave the model area and disperse before settling
settled = 0 # counts how many bacteria from the original colony successfully settle in the town area
#note that by the model's completion 'lost' and 'settled' should sum to 'Colony_Size'
# The following two variables store the ground zero coordinates.
# The exact coordinates can only be identified once the file containing them is loaded in.
sx = None # stores ground zero x coordinate
sy = None # stores ground zero y coordinate
#Finally several empty lists are created to be later filled
bacteria = [] # This list will store the bacteria agent data
GZ = [] # This list will be used to load the file identifying the ground zero location
DensityMap = [] # This list will be used to track the locations of settled bacteria
# +
# Classes
class Bacterium: # Class to establish characteristics of model agents
def __init__(self, height, x, y, dm):
self.height = height # used to track elevation of bacterium
self.x = x # used to track x coordinate of bacterium
self.y = y # used to track y coordinate of bacterium
self.dm = dm # links bacterium to density map to allow for interaction
self.complete = 0 # used to track whether bacterium is settled/lost and can be removed
class net: # Class used for identifying ground zero
def __init__(self, x, y, location):
self.x = x # stores x coordinate of value in loaded file
self.y = y # stores y coordinates of value in loaded file
self.location = location # stores value from loaded file found at above coordinates
# +
# Load Start Conditions from File
# Here a fuction for loading text files into arrays is defined:
def loadup(env,data):
env.clear() # Empty Environment (blank slate)
txt = open(data, newline='') # Loads text file into model
file = csv.reader(txt, quoting=csv.QUOTE_NONNUMERIC) # Read text file as a CSV file
for rows in file: # For each row in the CSV file:
values = [] #Create a list to store this rows scores
env.append(values) #add this row to main environment list
for pv in rows: # for each pixel value:
values.append(pv) #Add pixel value to row
txt.close()
# Load Ground Zero Data
x = "wind" #Loads data from provided file storing the ground zero coordinates
loadup(GZ,x) #fills empty 'GZ' list with data from the "wind" file
# Load Empty Text File
base = "Blank.txt" #Loads a blank 300x300 text file
loadup(DensityMap, base) #fills empty 'DensityMap' list with blank values in 300x300 extent
# +
# Finding coordinates for ground zero
sp = 255 # From the assignment it is known that the ground zero coordinates are marked by the value '255'
scope = [] # create empty list for scanning through ground zero data
for x in range(300): # for every x coordinate:
for y in range(300): # at every y coordinate:
scope.append(net(x,y,GZ)) # store value from ground zero file at said coordinates
def impact(self, sp): # create function for checking if coordinates are ground zero
global sx
global sy
if self.location[self.x][self.y] == sp: # if value at coordinates is identified marker:
sx = self.x # starting x coordinate is set
sy = self.y # starting y coordinate is set
for r in range(90000): # for every point in array (300x300)
impact(scope[r], sp) # check if point is ground zero
# print(sy, sx) # Test to ensure coordinates have been identified
# the below values are used for determing the final plot extent.
# here they are set to ground zero values as there has not been any spread of the colong
maxspread_x = sx
minspread_x = sx
maxspread_y = sy
minspread_y = sy
# -
# Create Bacteria
for r in range(Colony_Size): # For every bacterium in colony
bacteria.append(Bacterium(startheight, sx, sy, DensityMap)) # create a bacterium agent
# +
# Variable Warnings
# Checks to make sure defined percentage odds do not exceed 100%
#checks that wind direction percentages do not exceed 100%
if NorthW + EastW + SouthW + WestW > 100:
print("Warning! Wind direction probabilities exceed 100%!")
#checks that turbulence percentages do not exceed 100%
if Rise + Fall > 100:
print("Warning! Rise and Fall probabilities exceed 100%!")
# +
# Functions
# Here the main model functions are defined.
def wind(self, cs, N, E, S ,W, strength): # Controls Compass direction movements of agents
global Colony_Size
global lost
for x in range(cs): # for each bacterium in the colony:
w = random.randint(1,100) # randomise the wind direction
if w <= N: # if the wind is travelling north
self[x].y += strength # move bacterium northwards distance equal to wind speed
else:
if w > N and w <= N+E: # if the wind is travelling east
self[x].x += strength # move bacterium eastwards distance equal to wind speed
else:
if w > N+E and w <= N+E+S: # if the wind is travelling south
self[x].y -= strength # move bacterium southwards distance equal to wind speed
else:
if w > N+E+S and w <= N+E+S+W: # if the wind is travelling west
self[x].x -= strength # move bacterium westwards distance equal to wind speed
if self[x].x < 1 or self[x].x > 299: # if the bacterium is now out of bounds on the x axis:
Colony_Size -= 1 # reduce colony size
self[x].complete == 1 # mark bacterium for removal
lost += 1 # increase lost counter by one
else: # if the bacterium has not already gone out of bounds on the x axis:
if self[x].y < 1 or self[x].y > 299 and self[x].complete == 0: # if the bacterium is now out of bounds on the y axis
Colony_Size -= 1 # reduce colony size
self[x].complete == 1 # mark bacterium for removal
lost += 1 # increase lost counter by one
def settle(self, cs, rise, fall, startheight): # function for determining the impact of turbulence
for x in range(cs): # for every bacterium
w = random.randint(1,100) # randomise turbulence
if self[x].height < startheight: # if the bacterium is below start height
self[x].height -= 1 # elevation drops by 1 unit
else: # if bacterium is at or above start height:
if w <= fall: #if the wind does not carry the bacterium
self[x].height -= 1 # elevation drops by 1 unit
else:
if w > fall and w <= rise + fall: #if the wind uplifts the bacterium
self[x].height += 1 # elevation rises by 1 unit
# If neither occur, the wind keeps the bacterium height constant
def landing(self, cs, base): # function for adding bacterium that land to density map
global Colony_Size
global settled
global maxspread_x
global minspread_x
global maxspread_y
global minspread_y
for x in range(cs): # for every bacterium:
if self[x].height <= 0 and self[x].complete == 0: # if the agent has reached the ground and is not already processed
base[self[x].y][self[x].x] += 1 # mark landing on density map at agent's coordinates
self[x].complete == 1 # mark agent for removal
settled += 1 # increase settled count by 1
Colony_Size -= 1 # reduce colony size by 1
# Establish bounds for plot to avoid showing a mostly empty grid based on location of landing agents
if self[x].x > maxspread_x: # if agents is further away than previous furthest on the x axis east of ground zero
maxspread_x = self[x].x # set as new furthest east distance on the x axis
if self[x].x < minspread_x: # if agents is further away than previous furthest on the x axis west of ground zero
minspread_x = self[x].x # set as new furthest west distance on the x axis
if self[x].y > maxspread_y: # if agents is further away than previous furthest on the y axis north of ground zero
maxspread_y = self[x].y # set as new furthest north distance on the x axis
if self[x].y < minspread_y: # if agents is further away than previous furthest on the y axis south of ground zero
minspread_y = self[x].y # set as new furthest south distance on the x axis
# as agents cannot be deleted within the iterating loops a seperate step is needed :
def removal(self, agents, cs): # copies list and removes landed bacteria
temp = [] # creates an empty list for temporary storage
for x in range(cs): # for every bacterium
if self[x].complete == 0: # if not marked for removal
temp.append(self[x]) # add to temporary list
agents = temp # update main list to match temporary, deleting marked agents
# +
# Define Model
def model(self): # new function condensing previous functions for easy looping
#time.sleep(1) # Technically the model should run each step every second. Replaced by representative count for efficiency
removal(self, bacteria, Colony_Size) # remove any settled bacteria before modelling
wind(self, Colony_Size, NorthW, EastW, SouthW, WestW, StrengthW) # move bacteria horizontally
removal(self, bacteria, Colony_Size) # remove any newly out of bounds bacteria
settle(self, Colony_Size, Rise, Fall, startheight) # move bacteria vertically
landing(self, Colony_Size, DensityMap) # check if any bacteria have now landed
# +
# Run Model
start_time = time.time() # mark beginning of model run (for testing and output purposes)
for runs in range(1000): # set control of max iterations
if Colony_Size > 0: # so long as there are active bacteria:
timer += 1 # increas timer by one (simulates seconds passing)
model(bacteria) #complete one run through of model
end_time = time.time()# once completed mark end of model
run_time = end_time - start_time #calculate model runtime
# +
# Plot Extent
x_extent_high = maxspread_x + 5 # add buffer east direction
x_extent_low = minspread_x - 5 # add buffer west direction
y_extent_high = maxspread_y + 5 # add buffer north direction
y_extent_low = minspread_y - 5 # add buffer south direction
# Ensure that plot does not extend beyond the boundaries of the density map
if x_extent_high >= 300:
x_extent_high = 299
if y_extent_high >= 300:
y_extent_high = 299
if x_extent_low <= 0:
x_extent_low = 1
if y_extent_low <= 0:
y_extent_low = 1
# If no bacteria settle, show full map extent
if settled == 0:
x_extent_high = 300
y_extent_high = 300
x_extent_low = 0
y_extent_low = 0
# -
# save model ouput as a .txt file
np.savetxt("output.txt",DensityMap,delimiter=',');
# +
# Plot Density Map
plt.ylim(y_extent_low, y_extent_high) # set y axis to established extent of fallout
plt.xlim(x_extent_low, x_extent_high) # limit x axis to established extent of fallout
plt.imshow(DensityMap, cmap='seismic') # plot density map
plt.scatter(sx, sy, color = "red") # plot marker denoting ground zero as red dot
# -
# FInal statistics
print(settled, "bacteria settled and", lost, "lost from a colony of", settled + lost,"bacteria, in", timer, "seconds from dispersal time (real time:", run_time, "seconds)")
stats = np.array([])
|
A2.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# + [markdown] tags=[]
# # Convolutional Neural Network Implementation in GWU_NN
# Demo of simple CNN vs Dense network trained on MNIST handwritten digits dataset. Binary Classifier of 1's and 0's.
#
# ## Import libraries
# Only using sklearn and tensorflow for test_train_split and importing the mnist dataset.
# +
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from tensorflow.keras.datasets import mnist
from gwu_nn.gwu_network import GWUNetwork
from gwu_nn.layers import Dense, Convolutional, Flatten, MaxPool
# -
# ## Setting up the data
# Load the MNIST dataset and split into training and testing sets. Only add images to training/testing that are of 0s or 1s (because it will be a binary classifier).
# +
# Load the MNIST dataset
(X_train, y_train), (X_test, y_test) = mnist.load_data()
num1 = 0
num2 = 1
x_train_sample = []
y_train_sample = []
train_samples = 200
for i in range(len(X_train)):
if y_train[i] == num1 or y_train[i] == num2:
x_train_sample.append(X_train[i])
y_train_sample.append(y_train[i])
if len(x_train_sample) >= train_samples:
break
x_test_sample = []
y_test_sample = []
i_test_sample = []
samples = 500
for i in range(len(X_test)):
if y_test[i] == num1 or y_test[i] == num2:
x_test_sample.append(X_test[i])
y_test_sample.append(y_test[i])
i_test_sample.append(i)
if len(x_test_sample) >= samples:
break
print("x_train_sample: " + str(np.array(x_train_sample).shape))
print("x_test_sample: " + str(np.array(x_test_sample).shape))
# + [markdown] tags=[]
# ## Training a Dense Network
# Setup and train a simple dense network to use as benchmark against the CNN model.
# +
np.random.seed(1)
np.random.RandomState(1)
dense = GWUNetwork()
dense.add(Flatten(28,input_channels=1)) # Flat layer so the image is in the right dimensions
dense.add(Dense(20, activation='relu'))
dense.add(Dense(1, add_bias=False, activation='sigmoid'))
# Finally to complete our model we need to compile it. This defines our loss function and learning_rate
dense.compile(loss='log_loss', lr=0.001)
print(dense)
dense.fit(x_train_sample, y_train_sample, epochs=1)
# + [markdown] tags=[]
# ## Evaluating the Dense Network
# Generate predictions using the test split.
# +
# Predict using the test set. Calculate the accuracy
dense_raw_predictions = dense.predict(x_test_sample)
dense_predictions = [round(x[0][0]) for x in dense_raw_predictions]
dense_actual = [y for y in y_test_sample]
count = 0
for p,a in zip(dense_predictions,dense_actual):
if p == a:
count += 1
print("Dense model accuracy: " + str(100 * count/len(dense_predictions)))
# + [markdown] tags=[]
# ## Training a Convolutional Neural Network
# Setup and train a simple CNN. Only using one convolutional layer to keep things fast.
# +
np.random.seed(1)
np.random.RandomState(1)
cnn = GWUNetwork()
cnn.add(Convolutional(input_size=28, input_channels=1, kernel_size=3, num_kernels=1, activation='relu'))
cnn.add(MaxPool(28,2))
cnn.add(Flatten(14,input_channels=1)) # input size = 28/2
cnn.add(Dense(40, activation='relu')) # gets double the neurons here since input is only 14 (vs dense's 28)
cnn.add(Dense(1, add_bias=False, activation='sigmoid'))
# Finally to complete our model we need to compile it. This defines our loss function and learning_rate
cnn.compile(loss='log_loss', lr=0.001)
print(cnn)
cnn.fit(x_train_sample, y_train_sample, epochs=1)
# + [markdown] tags=[]
# ## Evaluating the CNN
# Generate predictions using the test split.
# +
# Predict using the test set. Calculate the accuracy
cnn_raw_predictions = cnn.predict(x_test_sample)
# calculate accuracy and show incorrect classifications
cnn_predictions = [round(x[0][0]) for x in cnn_raw_predictions]
count = 0
for p,a,i in zip(cnn_predictions,y_test_sample,i_test_sample):
if p == a:
count += 1
print("CNN model accuracy: " + str(100 * count/len(cnn_predictions)))
#print(cnn_predictions)
#print(y_test_sample)
# -
# ## Show a random evaluation
# Visualize the predictions by showing the prediction from both networks against the actual image.
# +
show_idx = 3
print("Dense Prediction: " + str(dense_predictions[show_idx]))
print("CNN Prediction: " + str(cnn_predictions[show_idx]))
print("Actual: " + str(y_test_sample[show_idx]))
ax = plt.subplot()
plt.imshow(x_test_sample[show_idx], cmap='gray')
plt.show()
# -
# ## Visualize the Kernel Weights
# Lets see what the kernel weights look like...
# +
kernel = cnn.layers[0].kernels.reshape(3,3)
plt.imshow(kernel, cmap='gray')
# -
|
demo.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <img src="../../images/qiskit-heading.gif" alt="Note: In order for images to show up in this jupyter notebook you need to select File => Trusted Notebook" width="500 px" align="left">
# ## _*Quantum Counterfeit Coin Problem*_
#
# The latest version of this notebook is available on https://github.com/qiskit/qiskit-tutorial.
#
# ***
# ### Contributors
# <NAME>, <NAME>
# ## Introduction
#
# The counterfeit coin problem is a classic puzzle first proposed by E. D. Schell in the January 1945 edition of the *American Mathematical Monthly*:
#
# >You have eight similar coins and a beam balance. At most one coin is counterfeit and hence underweight. How can you detect whether there is an underweight coin, and if so, which one, using the balance only twice?
#
# The answer to the above puzzle is affirmative. What happens when we can use a quantum beam balance?
#
# Given a quantum beam balance and a counterfeit coin among $N$ coins, there is a quantum algorithm that can find the counterfeit coin by using the quantum balance only once (and independent of $N$, the number of coins!). On the other hand, any classical algorithm requires at least $\Omega(\log{N})$ uses of the beam balance. In general, for a given $k$ counterfeit coins of the same weight (but different from the majority of normal coins), there is [a quantum algorithm](https://arxiv.org/pdf/1009.0416.pdf) that queries the quantum beam balance for $O(k^{1/4})$ in contrast to any classical algorithm that requires $\Omega(k\log{(N/k)})$ queries to the beam balance. This is one of the wonders of quantum algorithms, in terms of query complexity that achieves quartic speed-up compared to its classical counterpart.
#
# ## Quantum Procedure
# Hereafter we describe a step-by-step procedure to program the Quantum Counterfeit Coin Problem for $k=1$ counterfeit coin with the IBM Q Experience. [Terhal and Smolin](https://arxiv.org/pdf/quant-ph/9705041.pdf) were the first to show that it is possible to identify the false coin with a single query to the quantum beam balance.
#
# ### Preparing the environment
# First, we prepare the environment.
# +
# useful additional packages
import matplotlib.pyplot as plt
# %matplotlib inline
import numpy as np
# useful math functions
from math import pi, cos, acos, sqrt
# importing Qiskit
from qiskit import Aer, IBMQ, execute
from qiskit import QuantumCircuit, ClassicalRegister, QuantumRegister
# import basic plot tools
from qiskit.tools.visualization import plot_histogram
# -
# Load saved IBMQ accounts
IBMQ.load_accounts()
# ### Setting the number of coins and the index of false coin
#
# Next, we set the number of coins and the index of the counterfeit coin. The former determines the quantum superpositions used by the algorithm, while the latter determines the quantum beam balance.
# +
M = 16 # Maximum number of physical qubits available
numberOfCoins = 8 # This number should be up to M-1, where M is the number of qubits available
indexOfFalseCoin = 6 # This should be 0, 1, ..., numberOfCoins - 1, where we use Python indexing
if numberOfCoins < 4 or numberOfCoins >= M:
raise Exception("Please use numberOfCoins between 4 and ", M-1)
if indexOfFalseCoin < 0 or indexOfFalseCoin >= numberOfCoins:
raise Exception("indexOfFalseCoin must be between 0 and ", numberOfCoins-1)
# -
# ### Querying the quantum beam balance
#
# As in a classical algorithm to find the false coin, we will use the balance by placing the same number of coins on the left and right pans of the beam. The difference is that in a quantum algorithm, we can query the beam balance in superposition. To query the quantum beam balance, we use a binary query string to encode coins placed on the pans; namely, the binary string `01101010` means to place coins whose indices are 1, 2, 4, and 6 on the pans, while the binary string `01110111` means to place all coins but those with indices 0 and 4 on the pans. Notice that we do not care how the selected coins are placed on the left and right pans, because their results are the same: it is balanced when no false coin is included, and tilted otherwise.
#
# In our example, because the number of coins is $8$ and the index of false coin is $3$, the query `01101010` will result in balanced (or, $0$), while the query `01110111` will result in tilted (or, $1$). Using two quantum registers to query the quantum balance, where the first register is for the query string and the second register for the result of the quantum balance, we can write the query to the quantum balance (omitting the normalization of the amplitudes):
#
# \begin{eqnarray}
# |01101010\rangle\Big( |0\rangle - |1\rangle \Big) &\xrightarrow{\mbox{Quantum Beam Balance}}& |01101010\rangle\Big( |0\oplus 0\rangle - |1 \oplus 0\rangle \Big) = |01101010\rangle\Big( |0\rangle - |1\rangle \Big)\\
# |01110111\rangle\Big( |0\rangle - |1\rangle \Big) &\xrightarrow{\mbox{Quantum Beam Balance}}& |01110111\rangle\Big( |0 \oplus 1\rangle - |1 \oplus 1\rangle \Big) = (-1) |01110111\rangle\Big( |0 \rangle - |1 \rangle \Big)
# \end{eqnarray}
#
# Notice that in the above equation, the phase is flipped if and only if the binary query string is $1$ at the index of the false coin. Let $x \in \left\{0,1\right\}^N$ be the $N$-bit query string (that contains even number of $1$s), and let $e_k \in \left\{0,1\right\}^N$ be the binary string which is $1$ at the index of the false coin and $0$ otherwise. Clearly,
#
# $$
# |x\rangle\Big(|0\rangle - |1\rangle \Big) \xrightarrow{\mbox{Quantum Beam Balance}} \left(-1\right) ^{x\cdot e_k} |x\rangle\Big(|0\rangle - |1\rangle \Big),
# $$
# where $x\cdot e_k$ denotes the inner product of $x$ and $e_k$.
#
# Here, we will prepare the superposition of all binary query strings with even number of $1$s. Namely, we want a circuit that produces the following transformation:
#
# $$
# |0\rangle \rightarrow \frac{1}{2^{(N-1)/2}}\sum_{x\in \left\{0,1\right\}^N~\mbox{and}~|x|\equiv 0 \mod 2} |x\rangle,
# $$
#
# where $|x|$ denotes the Hamming weight of $x$.
#
# To obtain such superposition of states of even number of $1$s, we can perform Hadamard transformation on $|0\rangle$ to obtain superposition of $\sum_{x\in\left\{0,1\right\}^N} |x\rangle$, and check if the Hamming weight of $x$ is even. It can be shown that the Hamming weight of $x$ is even if and only if $x_1 \oplus x_2 \oplus \ldots \oplus x_N = 0$. Thus, we can transform:
#
# \begin{equation}
# |0\rangle|0\rangle \xrightarrow{H^{\oplus N}} \frac{1}{2^{N/2}}\sum_x |x\rangle |0\rangle \xrightarrow{\mbox{XOR}(x)} \frac{1}{2^{N/2}}\sum_x |x\rangle |0\oplus x_1 \oplus x_2 \oplus \ldots \oplus x_N\rangle
# \end{equation}
#
# The right-hand side of the equation can be divided based on the result of the $\mbox{XOR}(x) = x_1 \oplus \ldots \oplus x_N$, namely,
#
# $$
# \frac{1}{2^{(N-1)/2}}\sum_{x\in \left\{0,1\right\}^N~\mbox{and}~|x|\equiv 0 \mod 2} |x\rangle|0\rangle + \frac{1}{2^{(N-1)/2}}\sum_{x\in \left\{0,1\right\}^N~\mbox{and}~|x|\equiv 1 \mod 2} |x\rangle|1\rangle.
# $$
#
# Thus, if we measure the second register and observe $|0\rangle$, the first register is the superposition of all binary query strings we want. If we fail (observe $|1\rangle$), we repeat the above procedure until we observe $|0\rangle$. Each repetition is guaranteed to succeed with probability exactly half. Hence, after several repetitions we should be able to obtain the desired superposition state. *Notice that we can perform [quantum amplitude amplification](https://arxiv.org/abs/quant-ph/0005055) to obtain the desired superposition states with certainty and without measurement. The detail is left as an exercise*.
#
# Below is the procedure to obtain the desired superposition state with the classical `if` of the QuantumProgram. Here, when the second register is zero, we prepare it to record the answer to quantum beam balance.
# +
# Creating registers
# numberOfCoins qubits for the binary query string and 1 qubit for working and recording the result of quantum balance
qr = QuantumRegister(numberOfCoins+1)
# for recording the measurement on qr
cr = ClassicalRegister(numberOfCoins+1)
circuitName = "QueryStateCircuit"
queryStateCircuit = QuantumCircuit(qr, cr)
N = numberOfCoins
# Create uniform superposition of all strings of length N
for i in range(N):
queryStateCircuit.h(qr[i])
# Perform XOR(x) by applying CNOT gates sequentially from qr[0] to qr[N-1] and storing the result to qr[N]
for i in range(N):
queryStateCircuit.cx(qr[i], qr[N])
# Measure qr[N] and store the result to cr[N]. We continue if cr[N] is zero, or repeat otherwise
queryStateCircuit.measure(qr[N], cr[N])
# we proceed to query the quantum beam balance if the value of cr[0]...cr[N] is all zero
# by preparing the Hadamard state of |1>, i.e., |0> - |1> at qr[N]
queryStateCircuit.x(qr[N]).c_if(cr, 0)
queryStateCircuit.h(qr[N]).c_if(cr, 0)
# we rewind the computation when cr[N] is not zero
for i in range(N):
queryStateCircuit.h(qr[i]).c_if(cr, 2**N)
# -
# ### Constructing the quantum beam balance
#
# The quantum beam balance returns $1$ when the binary query string contains the position of the false coin and $0$ otherwise, provided that the Hamming weight of the binary query string is even. Notice that previously, we constructed the superposition of all binary query strings whose Hamming weights are even. Let $k$ be the position of the false coin, then with regards to the binary query string $|x_1,x_2,\ldots,x_N\rangle|0\rangle$, the quantum beam balance simply returns $|x_1,x_2,\ldots,x_N\rangle|0\oplus x_k\rangle$, that can be realized by a CNOT gate with $x_k$ as control and the second register as target. Namely, the quantum beam balance realizes
#
# $$
# |x_1,x_2,\ldots,x_N\rangle\Big(|0\rangle - |1\rangle\Big) \xrightarrow{\mbox{Quantum Beam Balance}} |x_1,x_2,\ldots,x_N\rangle\Big(|0\oplus x_k\rangle - |1 \oplus x_k\rangle\Big) = \left(-1\right)^{x\cdot e_k} |x_1,x_2,\ldots,x_N\rangle\Big(|0\rangle - |1\rangle\Big)
# $$
#
# Below we apply the quantum beam balance on the desired superposition state.
k = indexOfFalseCoin
# Apply the quantum beam balance on the desired superposition state (marked by cr equal to zero)
queryStateCircuit.cx(qr[k], qr[N]).c_if(cr, 0)
# ### Identifying the false coin
#
# In the above, we have queried the quantum beam balance once. How to identify the false coin after querying the balance? We simply perform a Hadamard transformation on the binary query string to identify the false coin. Notice that, under the assumption that we query the quantum beam balance with binary strings of even Hamming weight, the following equations hold.
#
# \begin{eqnarray}
# \frac{1}{2^{(N-1)/2}}\sum_{x\in \left\{0,1\right\}^N~\mbox{and}~|x|\equiv 0 \mod 2} |x\rangle &\xrightarrow{\mbox{Quantum Beam Balance}}& \frac{1}{2^{(N-1)/2}}\sum_{x\in \left\{0,1\right\}^N~\mbox{and}~|x|\equiv 0 \mod 2} \left(-1\right)^{x\cdot e_k} |x\rangle\\
# \frac{1}{2^{(N-1)/2}}\sum_{x\in \left\{0,1\right\}^N~\mbox{and}~|x|\equiv 0 \mod 2} \left(-1\right)^{x\cdot e_k} |x\rangle&\xrightarrow{H^{\otimes N}}& \frac{1}{\sqrt{2}}\Big(|e_k\rangle+|\hat{e_k}\rangle\Big)
# \end{eqnarray}
#
# In the above, $e_k$ is the bitstring that is $1$ only at the position of the false coin, and $\hat{e_k}$ is its inverse. Thus, by performing the measurement in the computational basis after the Hadamard transform, we should be able to identify the false coin because it is the one whose label is different from the majority: when $e_k$, the false coin is labelled $1$, and when $\hat{e_k}$ the false coin is labelled $0$.
# +
# Apply Hadamard transform on qr[0] ... qr[N-1]
for i in range(N):
queryStateCircuit.h(qr[i]).c_if(cr, 0)
# Measure qr[0] ... qr[N-1]
for i in range(N):
queryStateCircuit.measure(qr[i], cr[i])
# -
# Now we perform the experiment to see how we can identify the false coin by the above quantum circuit. Notice that when we use the `plot_histogram`, the numbering of the bits in the classical register is from right to left, namely, `0100` means the bit with index $2$ is one and the rest are zero.
#
# Because we use `cr[N]` to control the operation prior to and after the query to the quantum beam balance, we can detect that we succeed in identifying the false coin when the left-most bit is $0$. Otherwise, when the left-most bit is $1$, we fail to obtain the desired superposition of query bitstrings and must repeat from the beginning. *Notice that we have not queried the quantum beam oracle yet. This repetition is not neccesary when we feed the quantum beam balance with the superposition of all bitstrings of even Hamming weight, which can be done with probability one, thanks to the quantum amplitude amplification*.
#
# When the left-most bit is $0$, the index of the false coin can be determined by finding the one whose values are different from others. Namely, when $N=8$ and the index of the false coin is $3$, we should observe `011110111` or `000001000`.
# +
backend = Aer.backends("qasm_simulator")[0]
shots = 1 # We perform a one-shot experiment
success = 0
# Run until successful
while not success:
results = execute(queryStateCircuit, backend=backend, shots=shots).result()
answer = results.get_counts()
for key, value in answer.items():
if key[0:1] != "1":
success = 1
plot_histogram(answer)
from collections import Counter
for key in answer.keys():
normalFlag, _ = Counter(key[1:]).most_common(1)[0] #get most common label
for i in range(2,len(key)):
if key[i] != normalFlag:
print("False coin index is: ", len(key) - i - 1)
# -
# ## About Quantum Counterfeit Coin Problem
#
# The case when there is a single false coin, as presented in this notebook, is essentially [the Bernstein-Vazirani algorithm](http://epubs.siam.org/doi/abs/10.1137/S0097539796300921), and the single-query coin-weighing algorithm was first presented in 1997 by [<NAME> Smolin](https://arxiv.org/pdf/quant-ph/9705041.pdf). The Quantum Counterfeit Coin Problem for $k > 1$ in general is studied by [Iwama et al.](https://arxiv.org/pdf/1009.0416.pdf) Whether there exists a quantum algorithm that only needs $o(k^{1/4})$ queries to identify all the false coins remains an open question.
|
community/games/quantum_counterfeit_coin_problem.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Practical Session 2: Classification algorithms
#
# *Notebook by <NAME>*
# ## 0.1 Your task
#
# In practical 1, you worked with the housing prices and bike sharing datasets on the tasks that required you to predict some value (e.g., price of a house) or amount (e.g., the count of rented bikes, or the number of registered users) based on a number of attributes – age of the house, number of rooms, income level of the house owners for the house price prediction (or weather conditions and time of the day for the prediction of the number of rented bikes). That is, you were predicting some continuous value.
#
# This time, your task is to predict a particular category the instance belongs to based on its characteristics. This type of tasks is called *classification*.
# ## 0.2 Dataset
#
# First you will look into the famous [*Iris dataset*](https://en.wikipedia.org/wiki/Iris_flower_data_set), which was first introduced by the British statistician and biologist <NAME> in his 1936 paper *The use of multiple measurements in taxonomic problems*. The dataset contains $4$ characteristics (sepal length and width, and petal length and width) for $3$ related species of irises – *setosa*, *versicolor* and *virginica*. Your task is to learn to predict, based on these $4$ characteristics, the type of an iris.
#
# For further reference, see the original paper: <NAME> (1936). *The use of multiple measurements in taxonomic problems*. Annals of Eugenics. 7 (2): 179–188.
# ## 0.3 Learning objectives
#
# In this practical you will learn about:
# - binary and multiclass classification
# - linearly separable data
# - the use of a number of classifiers, including Naive Bayes, Logistic Regression, and Perceptron
# - kernel trick
# - ways to evaluate the performance of a classification model, including accuracy, precision, recall and F$_1$ measure
# - precision-recall trade-off and the ways to measure it, including ROC curves and AUC
#
# In addition, you will learn about the dataset uploading routines with `sklearn`.
# ## Step 1: Uploading and inspecting the data
#
# As before, let's start by uploading and looking into the data. In the previous practical, you worked with the data collected independently and stored in a comma-separated file, as the data in a real-life data science project might be. In this practical, you will learn how to use `sklearn`'s data uploading routines.
#
# `sklearn` has a number of datasets to practice your ML skills on, and the `iris` dataset is one of them. Here is how you can access the dataset through `sklearn`. Note that such data fields as *data* and *target* are already pre-defined for you:
from sklearn import datasets
iris = datasets.load_iris()
list(iris.keys())
# Take a look into what is contained in *data*. Remember that each instance of an iris is described in terms of $4$ variables – *sepal length*, *sepal width*, *petal length*, and *petal width*:
iris.data
# To find out what variables are contained in `data`, check the `feature_names` data field.
iris.feature_names
# What about the target values?
iris.target
# There are $3$ classes of irises – *setosa*, *versicolor*, and *virginica*, and they are already converted into numerical values for you (recall, that when the dataset is not already preprocessed this way, and the target or any of the arrtibutes are represented as text or categorical data, you need to convert them into numerical data). If you want to check what each numerical label corresponds to in the original data, you can do so accessing the `target_names` data field:
iris.target_names
# Remember, that for further ML experiments, we need to have two data structures: the instance-by-attributes matrix $X$ and the target labels vector $y$. For instance, in the previous practical the regression algorithm learned the vector of weights $w$ to predict the target variable $\hat y^{(i)}$ for each instance $i$ so that its prediction would be maximally close to the actual label $y^{(i)}$. Since the labels $y^{(i)}$ were continuous (i.e., amount, number, or value), you measured the performance of your regressor by the distance between the predictions $\hat y$ and actual labels $y$. In this practical, you will need to work with $X$ and $y$, too, but the vector of $y$ this time will contain discrete values – classes $[0, 1, 2]$ for the different types of the flower.
#
# As you might have already figured out, you need to initialise $X$ and $y$ with the `data` and `target` fields of your iris dataset:
X, y = iris["data"], iris["target"]
print(X.shape)
print(y.shape)
# Let's look closer into the data to get a feel for what is contained in it. As before, let's use visualisations with `matplotlib`, and in particular, plot one attribute against another for the three types of irises using scatterplot. E.g., let's plot sepal length vs. sepal width:
# +
# %matplotlib inline
#so that the plot will be displayed in the notebook
import numpy as np
np.random.seed(42)
import matplotlib
from matplotlib import pyplot as plt
# visualise sepal length vs. sepal width
X = iris.data[:, :2]
y = iris.target
scatter_x = np.array(X[:, 0])
scatter_y = np.array(X[:, 1])
group = np.array(y)
cmap = matplotlib.cm.get_cmap('jet')
cdict = {0: cmap(0.1), 1: cmap(0.5), 2: cmap(0.9)}
labels = iris.target_names
fig, ax = plt.subplots(figsize=(8, 6))
for g in np.unique(group):
ix = np.where(group == g)
ax.scatter(scatter_x[ix], scatter_y[ix], c=np.array([cdict[g]]), #c = cdict[g],
label = labels[g], s = 100, marker = "H",
linewidth=2, alpha = 0.5)
ax.legend()
plt.xlabel('Sepal length')
plt.ylabel('Sepal width')
plt.show()
# -
# It looks like *setosa* is quite clearly distiguishable from the other two types of irises with these two features. What about petal length and width?
# +
# visualise petal length vs. petal width
X = iris.data[:, 2:]
y = iris.target
scatter_x = np.array(X[:, 0])
scatter_y = np.array(X[:, 1])
group = np.array(y)
cmap = matplotlib.cm.get_cmap('jet')
cdict = {0: cmap(0.1), 1: cmap(0.5), 2: cmap(0.9)}
labels = iris.target_names
fig, ax = plt.subplots(figsize=(8, 6))
for g in np.unique(group):
ix = np.where(group == g)
ax.scatter(scatter_x[ix], scatter_y[ix], c=np.array([cdict[g]]), #c = cdict[g],
label = labels[g], s = 100, marker = "H",
linewidth=2, alpha = 0.5)
ax.legend()
plt.xlabel('Petal length')
plt.ylabel('Petal width')
plt.show()
# -
# This plot shows an even clearer separation between the class of *setosa* irises and the other two classes. In fact, with respect to these two attributes, it might be possible to clearly separate not only *setosas* from the other two classes, but also, with certain success, *versicolors* from *virginicas*.
#
# When the data can be separated by a straight line (or a single decision surface) as in the example above, it is called *linearly separable*. This property of the data is successfully exploited by ML models that try to learn a linear separation boundary between the classes. In fact, there is a whole set of lines that you can use to separate the class of *setosas* from the other two classes, *versicolors* and *virginicas*, in this example. Some linear models explicitly allow you to select the *best* separation boundary by maximising the distance between the boundary and the closest instances of the two classes. Such are, for example, [Support Vector Machines](http://scikit-learn.org/stable/modules/svm.html), which will be covered in the [Part II Machine Learning and Bayesian Inference course](https://www.cl.cam.ac.uk/teaching/2021/MLBayInfer/):
# +
# visualise petal length vs. petal width
X = iris.data[:, 2:]
y = iris.target
scatter_x = np.array(X[:, 0])
scatter_y = np.array(X[:, 1])
group = np.array(y)
cmap = matplotlib.cm.get_cmap('jet')
cdict = {0: cmap(0.1), 1: cmap(0.5), 2: cmap(0.9)}
labels = iris.target_names
fig, ax = plt.subplots(figsize=(8, 6))
for g in np.unique(group):
ix = np.where(group == g)
ax.scatter(scatter_x[ix], scatter_y[ix], c=np.array([cdict[g]]), #c = cdict[g],
label = labels[g], s = 100, marker = "H",
linewidth=2, alpha = 0.5)
ax.legend()
plt.xlabel('Petal length')
plt.ylabel('Petal width')
for i in range(0, 9):
plt.plot([3 + 0.5*i, 0], [0, 3-0.25*i], 'k-', color=cmap(0.1*(i+1)))
plt.show()
# -
# For the sake of consistency, let's plot all the pairs of features against each other. Note that all plots confirm that *setosas* are linearly separable from the other two types of irises (you might notice an occasional outlier, though), while *versicolors* are linearly separable from *virginicas* with respect to some attributes only:
# +
fig = plt.figure(figsize=(16, 12))
fig.subplots_adjust(hspace=0.2, wspace=0.2)
X = iris.data
y = iris.target
labels = iris.target_names
index = 1
for i in range(0, X.shape[1]):
for j in range(0, X.shape[1]):
scatter_x = np.array(X[:, i])
scatter_y = np.array(X[:, j])
group = np.array(y)
cmap = matplotlib.cm.get_cmap('jet')
cdict = {0: cmap(0.1), 1: cmap(0.5), 2: cmap(0.9)}
ax = fig.add_subplot(X.shape[1], X.shape[1], index)
index+=1
for g in np.unique(group):
ix = np.where(group == g)
ax.scatter(scatter_x[ix], scatter_y[ix], c=np.array([cdict[g]]), #c = cdict[g],
label = labels[g], s = 50, marker = "H",
linewidth=1, alpha = 0.5)
plt.show()
# -
# ## Step 2: Splitting the data into training and test subsets
#
# Before applying the classifiers, let's split the dataset into the training and test sets. Recall, that when building an ML model, all further data exploration, feature selection and scaling, model selection and fine-tuning should be done on the training data, and the test data should only be used at the final step to evaluate the best estimated model:
# +
from sklearn.model_selection import train_test_split
train_set, test_set = train_test_split(X, test_size=0.2)
print(len(train_set), "training instances +", len(test_set), "test instances")
# -
# As before, you want your training and test data to contain enough representative examples of each class, that is, you should rather apply `StratifiedShuffleSplit` and not random splitting:
# +
from sklearn.model_selection import StratifiedShuffleSplit
split = StratifiedShuffleSplit(n_splits=1, test_size=0.2, random_state=42)
split.get_n_splits(X, y)
print(split)
for train_index, test_index in split.split(X, y):
print("TRAIN:", len(train_index), "TEST:", len(test_index))
X_train, X_test = X[train_index], X[test_index]
y_train, y_test = y[train_index], y[test_index]
print(X_train.shape, y_train.shape, X_test.shape, y_test.shape)
# -
# Let's check the class proportions in the original dataset, and training and test subsets:
# +
import pandas as pd
# def original_proportions(data):
# props = {}
# for value in set(data["target"]):
# data_value = [i for i in data["target"] if i==value]
# props[value] = len(data_value) / len(data["target"])
# return props
def subset_proportions(subset):
props = {}
for value in set(subset):
data_value = [i for i in subset if i==value]
props[value] = len(data_value) / len(subset)
return props
compare_props = pd.DataFrame({
"Overall": subset_proportions(iris["target"]),
"Stratified tr": subset_proportions(y_train),
"Stratified ts": subset_proportions(y_test),
})
compare_props["Strat. tr %error"] = 100 * compare_props["Stratified tr"] / compare_props["Overall"] - 100
compare_props["Strat. ts %error"] = 100 * compare_props["Stratified ts"] / compare_props["Overall"] - 100
compare_props.sort_index()
# -
# The original dataset is well-balanced – it contains exactly $50$ examples for each class. With the stratified data splits, you get equal proportions of each type of the irises in the training and test sets, too.
#
# Now, let's first approach the classification task in a simpler setting: let's start with *binary classification* and try to predict whether an iris is of a particular type: e.g., *setosa* vs. *not-a-setosa*, or *versicolor* vs. *not-a-versicolor*.
#
#
# ## Case 1: Binary classification
#
# Let's start by separating the data that describes *setosa* from other data.
y_train_setosa = (y_train == 0) # will return True when the label is 0 (i.e., setosa)
y_test_setosa = (y_test == 0)
y_test_setosa
# `y_test_setosa` returns a boolean vector of $30$ test instances: it contains `True` for the test instances that are *setosas*, and `False` otherwise. Let's pick one example of a *setosa* – for instance, the first one from the test set, `X_test[0]`, for further evaluations.
setosa_example = X_test[0]
# As you've noticed above, *setosas* are linearly separable from the other two classes, so it would be reasonable to apply a linear model to the task of separating *setosas* from *not-setosas*.
#
# ### Perceptron
#
# A (single-layer) perceptron is a simple linear classifier that tries to learn the set of weights $w$ for the input vectors $X$ in order to predict the output binary class values $y$. In particular:
#
# \begin{equation}
# \hat y^{(i)}=\begin{cases}
# 1, & \text{if $w \cdot x^{(i)} + b > 0$}\\
# 0, & \text{otherwise}
# \end{cases}
# \end{equation}
#
# where $w \cdot x^{(i)}$ is the dot product of weight vector $w$ and the feature vector $x^{(i)}$ for the instance $i$, $\sum_{j=1}^{m} w_{j}x_{j}^{(i)}$, and $b$ is the bias term.
#
# `sklearn` has a perceptron implementation through its linear [`SGDClassifier`](http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.SGDClassifier.html#sklearn.linear_model.SGDClassifier) with the following parameter settings:
# +
from sklearn.linear_model import SGDClassifier
sgd = SGDClassifier(max_iter=5, tol=None, random_state=42,
loss="perceptron", eta0=1, learning_rate="constant", penalty=None)
sgd.fit(X_train, y_train_setosa)
sgd.predict([setosa_example])
# -
# The perceptron correctly predicts that `X_test[0]` is a *setosa* flower. However, as you've seen above, not all types of irises are linearly separable. Let's select a more challenging example of a *versicolor* (class $1$) for comparison.
y_train_versicolor = (y_train == 1) # True when the label is 1 (i.e., versicolor)
y_test_versicolor = (y_test == 1)
y_test_versicolor
# Select one of the examples to try your classifier on (array indexing in python starts with $0$, so you can pick indexes $2$, $3$, $5$ and so on):
# +
versicolor_example = X_test[17]
print("Class", y_test[17], "(", iris.target_names[y_test[17]], ")")
sgd.fit(X_train, y_train_versicolor)
print(sgd.predict([versicolor_example]))
# -
# Looks like perceptron indeed cannot predict the class of the *versicolor* example correctly. Let's see if another linear classifier – `Logistic Regression` – can do a better job.
#
# ### Logistic Regression
#
# You used `Linear Regression` model in practical 1 to predict continuous values. In contrast, `Logistic Regression` is used for binary classification, that is, to predict a discrete value of $0$ or $1$. In particular, it estimates the probability that an instance belongs to a particular class (e.g., that $X\_test[0] \in {setosa}$). If the probability is greater than $50\%$, the instance is classified as *setosa* (positive class, labelled $1$ or $True$). Otherwise, it is classified as *not-a-setosa* (negative class, labelled $0$ or $False$).
#
# Similarly to `Linear Regression`, `Logistic Regression` computes a weighted sum using the input features $w \cdot X$ plus an intercept ($w_0$), but instead of outputting the result as `Linear Regression` does, it further applies a *sigmoid function* to this result:
#
# \begin{equation}
# \hat p = \sigma (w \cdot X)
# \end{equation}
#
# The sigmoid function outputs a value between $0$ and $1$:
#
# \begin{equation}
# \sigma (t) = \frac{1}{1 + exp(-t)}
# \end{equation}
#
# Once the Logistic Regression model has estimated the probability $\hat p$, the label $\hat y$ is predicted as follows:
#
# \begin{equation}
# \hat y=\begin{cases}
# 1, & \text{if $\hat p \geq 0.5$}\\
# 0, & \text{otherwise}
# \end{cases}
# \end{equation}
#
# Note that the above is equivalent to:
#
# \begin{equation}
# \hat y=\begin{cases}
# 1, & \text{if $t \geq 0$}\\
# 0, & \text{otherwise}
# \end{cases}
# \end{equation}
#
# Let's apply a `Logistic Regression` model to our setosa example:
# +
from sklearn.linear_model import LogisticRegression
log_reg = LogisticRegression()
log_reg.fit(X_train, y_train_setosa)
# -
log_reg.predict([setosa_example])
# The model correctly predicts that the test example is indeed a setosa. What about the versicolor example?
log_reg.fit(X_train, y_train_versicolor)
log_reg.predict([versicolor_example])
# Finally, for comparison, let's introduce one more classifier, that some of you might have come across before.
#
# ### Naive Bayes
#
# If you did [Part IA Machine Learning and Real-World Data](https://www.cl.cam.ac.uk/teaching/1920/MLRD/) in the past, you might recall that you have already come across classification tasks: for example, you were asked to build a classifier that identifies sentiment in text, and you used `Naive Bayes` for that. `Naive Bayes` makes different assumptions about the data. In particular, it doesn't assume linear separability but makes the predictions based on the prior and the updated belief about the data.
#
# To remind you, on a two class problem (e.g., distinguishing between classes $0$ for *not-setosas* and $1$ for *setosas*), a Naive Bayes model will predict:
#
# \begin{equation}
# \hat y^{(i)} = argmax_{c \in (0, 1)} p(y=c | x^{(i)}) = \begin{cases}
# 1, & \text{if $\hat p(y=1 | x^{(i)}) > \hat p(y=0 | x^{(i)}$})\\
# 0, & \text{otherwise}
# \end{cases}
# \end{equation}
#
# where the probabilities are conditioned on the feature vector for $x^{(i)}$, i.e. $(f^{(i)}_{1}, ..., f^{(i)}_{n})$. In practice, it is impossible to estimate these probabilities exactly, and one uses the Naive Bayes theorem so that:
#
# \begin{equation}
# \hat p(y=c | x^{(i)}) = \frac{p(c) p(x^{(i)} | c)}{p(x^{(i)})}
# \end{equation}
#
# where $c \in \{0, 1\}$ is the class to be predicted. Since the denominator is the same for both estimates of $\hat p(y=1 | x^{(i)})$ and $\hat p(y=0 | x^{(i)})$, it can be omitted. Therefore, the estimate can be simplified to:
#
# \begin{equation}
# \hat y^{(i)} = argmax_{c \in (0, 1)} p(y=c | x^{(i)}) = argmax_{c \in (0, 1)} p(c) p(x^{(i)} | c)
# \end{equation}
#
# where $p(c)$ is the *prior belief* of the classifier about the distribution of the classes in the data, and $p(x^{(i)} | c)$ is the *posterior probability*. Both can be estimated from the training data using *maximum a posteriori (MAP)* estimation. Moreover, the "naive" independence assumption of this learning algorithm allows you to estimate $p(x^{(i)} | c)$ as a product of feature probabilities taken independently of each other, i.e.:
#
# \begin{equation}
# p(x^{(i)} | c) = p(f^{(i)}_{1}, ..., f^{(i)}_{n} | c) \approx p(f^{(i)}_{1} | c) \times ... \times p(f^{(i)}_{n} | c)
# \end{equation}
#
# `sklearn` has a number of implementations of a [`Naive Bayes`](http://scikit-learn.org/stable/modules/naive_bayes.html) algorithm which mainly differ from each other with respect to how they estimate the conditional probability above using the data and what believes they hold about the distribution of this data, e.g.:
# +
from sklearn.naive_bayes import GaussianNB, MultinomialNB
gnb = MultinomialNB() # or:
gnb = GaussianNB()
gnb.fit(X_train, y_train_setosa)
gnb.predict([setosa_example])
# +
gnb.fit(X_train, y_train_versicolor)
gnb.predict([versicolor_example])
# -
# As you can see, not all classifiers perform equally well. How do you measure their performance in a more comprehensive way?
#
# ## Step 3: Evaluation
#
# ### Performance measures
#
# The most straightforward way to evaluate a classifier is to estimate how often its predictions are correct. This estimate is called *accuracy* and it is calculated as follows:
#
# \begin{equation}
# ACC = \frac{num(\hat y == y)}{num(\hat y == y) + num(\hat y != y)} = \frac{num(\hat y == 1 \& y == 1) + num(\hat y == 0 \& y == 0)}{num(\hat y == 1 \& y == 1) + num(\hat y == 0 \& y == 0) + num(\hat y == 1 \& y == 0) + num(\hat y == 0 \& y == 1)}
# \end{equation}
#
# E.g., for the *setosa* classification example, an accuracy of the classifier is the ratio of correctly identified *setosas* and correctly identified *not-setosas* to the total number of examples.
#
# You can either import accuracy metric using `from sklearn.metrics import accuracy_score` or measure accuracy across multiple cross-validation folds (refer to practical 1, if you need to remind yourself what cross-validation does):
# +
from sklearn.model_selection import cross_val_score
print(cross_val_score(log_reg, X_train, y_train_setosa, cv=5, scoring="accuracy"))
print(cross_val_score(gnb, X_train, y_train_setosa, cv=5, scoring="accuracy"))
print(cross_val_score(sgd, X_train, y_train_setosa, cv=5, scoring="accuracy"))
# -
# All three classifiers are precfectly accurate in their prediction on the *setosa* example. What about the *versicolor* example?
print(cross_val_score(log_reg, X_train, y_train_versicolor, cv=5, scoring="accuracy"))
print(cross_val_score(gnb, X_train, y_train_versicolor, cv=5, scoring="accuracy"))
print(cross_val_score(sgd, X_train, y_train_versicolor, cv=5, scoring="accuracy"))
# The numbers differ but they still do not tell you much about the performance of a classifier in general. E.g., an accuracy of $\approx 0.71$ is far from perfect, but exactly how bad or how acceptable is it?
#
# Let's implement a brute-force algorithm that will simply predict *not-versicolor* for every instance in the *versicolor* detection case (or *not-setosa* in the *setosa* detection case). Here is how well it will perform:
# +
from sklearn.base import BaseEstimator
class NotXClassifier(BaseEstimator):
def fit(self, X, y=None):
pass
def predict(self, X):
return np.zeros((len(X), 1), dtype=bool)
notversicolor_clf = NotXClassifier()
cross_val_score(notversicolor_clf, X_train, y_train_versicolor, cv=5, scoring="accuracy")
# -
# This gives you a very clear benchmark for comparison. The above represents a *majority class baseline*: for each split in cross-validation splits, it measures the proportion of the majority class (*not-versicolor* in this case). That is, if the classifier does nothing and simply returns the majority class label every time, this is how "well" it will perform. Obviously, you want the actual classifier that you build to do better than that.
# ### Confusion matrix
#
# So now you can compare the accuracy (e.g., the proportion of correctly identified *setosas* and *not-setosas* in the dataset) to the baseline system. However, this doesn't help you understand *where* your classifier goes wrong. For example, does the low accuracy of the classifiers in the *versicolor* identification case suggest that they miss some *versicolors* and classify them as other types of irises, or does it suggest that they mistake other types of irises for *versicolors*? Or, perhaps, it's a combination of two types of mistakes?
#
# The accuracy score itself doesn't allow you to make any of these conclusions. What you need to do is to look into the number of correctly and incorrectly classified instances, and the data representation that helps you do that is called *confusion matrix*. A confusion matrix is simply a table that compares the number of actual instances of type $c$ to the number of predicted instances of type $c$, i.e.:
#
# | | predicted $\hat c=0$| predicted $\hat c=1$ |
# | ------------- | :-------------: | :-------------: |
# | **actual $c=0$** | TN | FP |
# | **actual $c=1$** | FN | TP |
#
# The instances that are classified correctly as well as those that are misclassified have certain importance for evaluation of your classification algorithms. Here is the terminology:
#
# - `TP` stands for *true positives*. These are the actual instances of class $1$ that are correctly classified as class $1$ (*setosas* identified as *setosas*);
# - `TN` stands for *true negatives*. These are the actual instances of class $0$ that are correctly classified as class $0$ (*not-setosas* identified as *not-setosas*);
# - `FN` stands for *false negatives*. These are the actual instances of class $1$ that are incorrectly classified as class $0$ (*setosas* identified as *not-setosas*, i.e. missed by the classifier);
# - and finally, `FP` are *false positives*. These are the actual instances of class $0$ that are incorrectly classified as class $1$ (*not-setosas* identified as *setosas*, i.e. other types of flower mistaken for setosas by the classifier).
#
# Now you can re-interpret the accuracy score as:
#
# \begin{equation}
# ACC = \frac{TP + TN}{TP + TN + FP + FN}
# \end{equation}
#
# So in order to maximise accuracy, you'll need to maximise on the number of true positives and true negatives.
#
# The confusion matrix also tells you what exactly is misclassified by the classifier. For example, let's look into the confusion matrices for *setosa* identification:
# +
from sklearn.model_selection import cross_val_predict
from sklearn.metrics import confusion_matrix
y_train_pred = cross_val_predict(log_reg, X_train, y_train_setosa, cv=5)
confusion_matrix(y_train_setosa, y_train_pred)
# -
y_train_pred = cross_val_predict(gnb, X_train, y_train_setosa, cv=5)
confusion_matrix(y_train_setosa, y_train_pred)
# You know that the accuracy of these classifiers on *setosa* identification equals to $1$, so all *setosas* ($40$ instances) and *not-setosas* ($80$ instances) are classified correctly, and the confusion matrices show exactly that. What about the mistakes that the classifiers make on the *versicolor* example?
y_train_pred = cross_val_predict(log_reg, X_train, y_train_versicolor, cv=5)
confusion_matrix(y_train_versicolor, y_train_pred)
y_train_pred = cross_val_predict(gnb, X_train, y_train_versicolor, cv=5)
confusion_matrix(y_train_versicolor, y_train_pred)
# The matrices above show that the `Logistic Regression` classifier correctly identifies $71$ *not-versicolors* (`TN`) and $15$ *versicolors* (`TP`), and it also misses $25$ *versicolors* (`FN`) and mistakes $9$ other flowers for *versicolors*. Now you can see that the bigger source of error for the `Logistic Regression` classifier is that it's not good enough in identifying *versicolors* as it misses more of them that it identifies ($25$ vs $15$). In contrast, `Naive Bayes` identifies more *versicolors* in general ($36 + 5$) and more of them correctly ($36$), while it misses only $4$ *versicolor* instances.
#
# Apart from making such observations, you might want to be able to make sense of the number of correctly and incorrectly classified examples and compare the performance of the different classifiers in an aggregated way. The following set of measures will help you do so: *precision* ($P$), *recall* ($R$), and $F_{\beta}$*-measure* (e.g., $F_1$).
#
# - *Precision* measures how reliable or trustworthy your classifier is. It tells you how often when the classifier predicts that a flower is a *versicolor* (class $1$) it actually is a *versicolor*. It relies on the number of $TP$'s and $FP$'s:
# \begin{equation}
# P = \frac{TP}{TP+FP}
# \end{equation}
#
# - *Recall* measures the coverage of your classifier. It tells you how many of the actual instances of *versicolor* your classifier can detect at all. It relies on the number of $TP$'s and $FN$'s:
# \begin{equation}
# R = \frac{TP}{TP+FN}
# \end{equation}
#
# - Finally, $F_1$*-score* combines the two measures above to give you an overall idea of your classifier's performance. $F_1$*-score* is estimated as follows:
# \begin{equation}
# F_1 = 2 \times \frac{P \times R}{P+R}
# \end{equation}
#
# It is the *harmonic mean* of the two measures. However, if you want to highlight the importance of precision in your task (and we'll talk about when you might want to do so later in this practical) or the importance of recall, you can do so by changing the $\beta$ coefficient of the *F-score*. In fact, the $F_1$*-score* is the most commonly used case of a more general $F_{\beta}$*-measure*, which is estimated as follows:
# \begin{equation}
# F_{\beta} = (1 + \beta^2) \times \frac{P \times R}{\beta^2 \times P+R}
# \end{equation}
#
# $\beta < 1$ puts more emphasis on precision than on recall, and $\beta > 1$ weighs recall higher than precision.
# +
from sklearn.metrics import precision_score, recall_score, f1_score
y_train_pred = cross_val_predict(gnb, X_train, y_train_versicolor, cv=5)
precision = precision_score(y_train_versicolor, y_train_pred) # == 36 / (36 + 5)
recall = recall_score(y_train_versicolor, y_train_pred) # == 36 / (36 + 4)
f1 = f1_score(y_train_versicolor, y_train_pred)
print(precision, recall, f1)
y_train_pred = cross_val_predict(log_reg, X_train, y_train_versicolor, cv=5)
precision = precision_score(y_train_versicolor, y_train_pred) # == 15 / (15 + 9)
recall = recall_score(y_train_versicolor, y_train_pred) # == 15 / (15 + 25)
f1 = f1_score(y_train_versicolor, y_train_pred)
print(precision, recall, f1)
# -
# The numbers above tell you that the `Naive Bayes` classifier has pretty good recall ($0.9$, or it identifies $90\%$ of *versicolor* instances) as well as precision ($\approx 0.88$, or in approximately $88\%$ of the cases you can trust its *versicolor* prediction). The two numbers are close to each other in range, so the $F_{1}$ score is high, too. The `Logistic Regression` classifier is less precise (you can trust it about $62.5\%$ of the time) and it misses many *versicolor* examples (identifying only $37.5\%$ of those).
#
# Obviously, if your algorithm gets it all right, you will end up with perfect accuracy, precision and recall. In practice, however, the algorithms often don't get it all perfectly correct, and depending on the task at hand, you might decide that you are actually mostly interested in high precision of your algorithm, or in high recall.
#
# For example, imagine you are working on a machine learning classifier that detects cancerous cases in the data coming from patients' analyses. The task of your algorithm is to detect whether the patient needs further tests and closer analysis based on the preliminary tests. The cost of *false negatives* (missed cancerous cases) in this task is very high and it's better to administer further tests for a patient about which the algorithm has doubts than to ignore the case altogether. In this case, you should prioritise recall over precision.
#
# On the other hand, imagine you work for a pharmaceutical company trying to detect whether a particular drug will be applicable to a particular condition or a particular group of patients using data science and machine learning. Or, you want to learn in a data-driven way [when to change the drug dosage for patients in a hospital](http://www.cs.cornell.edu/people/tj/publications/morik_etal_99a.pdf). In these cases, the cost of *false positives*, i.e. deciding that the drug is suitable for a particular patient when it is not, or deciding to intervene in patients' treatment when the dosage should be kept as before, is more costly. These are the cases when you should prioritise precision over recall.
#
#
# ### Precision-recall trade-off
#
# Often, you can achieve perfect precision if you lower the recall, and other way around. For example, by always predicting *versicolor* you will reach perfect recall (all *versicolor* instances will be covered by such prediction), but very low precision (because in most of the cases such prediction will be unreliable as your classifier will predict *versicolor* for all setosas and all virginicas as well). On the other hand, by trying to maximise precision, you will often need to constrain your classifier so that it returns less false positives and therefore is more conservative.
#
# A good way to understand how a classifier makes its predictions is to look into its `decision_function` – the confidence score of the classifier's predictions. For `Logistic Regression` it returns a signed distance to the separation hyperplane selected by the model. Let's check the confidence score of the `Logistic Regression` model on the *versicolor* example:
# +
log_reg.fit(X_train, y_train_versicolor)
y_scores = log_reg.decision_function([versicolor_example])
y_scores
# -
# When this confidence score is higher than the predefined model's threshold, the model assigns the instance to the positive class, and otherwise it assigns the instance to the negative class. The `Logistic Regression`'s default threshold is $0$, so the example above is classified as *versicolor*:
threshold = 0
y_versicolor_pred = (y_scores > threshold)
y_versicolor_pred
# However, what would happen if you change the threshold to, for example, $0.7$? As you expect, this particular instance will be classified as *not-a-versicolor*. What effect will it have on the precision and recall of your classifier as a consequence?
threshold = 0.7
y_versicolor_pred = (y_scores > threshold)
y_versicolor_pred
# The following will return the confidence scores for each of the training set instances:
y_scores = cross_val_predict(log_reg, X_train, y_train_versicolor, cv=5, method="decision_function")
y_scores
# Let's use `sklearn`'s [`precision_recall_curve`](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.precision_recall_curve.html) functionality and plot the precision and recall values against the different threshold values. This shows you how the measures will be affected by changing the confidence threshold:
# +
from sklearn.metrics import precision_recall_curve
precisions, recalls, thresholds = precision_recall_curve(y_train_versicolor, y_scores)
def plot_pr_vs_threshold(precisions, recalls, thresholds):
plt.plot(thresholds, precisions[:-1], "b--", label="Precision")
plt.plot(thresholds, recalls[:-1], "g--", label="Recall")
plt.xlabel("Threshold")
plt.legend(loc="upper right")
plt.ylim([0, 1])
plot_pr_vs_threshold(precisions, recalls, thresholds)
plt.show()
# -
# As you can see, the recall has a general tendency of decreasing when you increase the threshold – that is, the more conservative the classifier becomes, the more instances it is likely to miss. At the same time, precision curve is bumpier: with certain changes in the threshold, it might drop as well as the classifier misidentifies some other types of irises as *versicolor*.
#
# You can also plot precision values against recall values and track the changes in precision against recall:
# +
def plot_precision_vs_recall(precisions, recalls):
plt.plot(recalls, precisions, "b-", linewidth=2)
plt.xlabel("Recall")
plt.ylabel("Precision")
plt.axis([0, 1, 0, 1])
plot_precision_vs_recall(precisions, recalls)
plt.show()
# -
# ### The Receiver Operating Characteristic (ROC)
#
# Another widely used way to inspect and present the results is to plot the receiver operating characteristic (ROC) curve. Similarly to the precision / recall curve above, it shows how the performance of the classifier changes at different threshold values. In particular, it plots the *true positive rate (TPR)* against the *false positive rate (FPR)*. Here is some more machine learning terminology:
#
# - *True positive rate (TPR)* is simply another term for recall. Alternatively, it is also called *sensitivity*, or *probability of detection*;
# - *False positive rate (FPR)* is also referred to as *fall-out* or *probability of false alarm*, and it can be calculated as $(1 − specificity)$. *Specificity*, in its turn, is estimated as:
#
# \begin{equation}
# specificity = \frac{TN}{TN + FP}
# \end{equation}
#
# In other words, *specificity* expresses the probability that the classifier correctly identifies *non-versicolors* in the *versicolor* identification example. *FPR*, therefore, shows how many incorrect *versicolor* votes (false alarms) the classifier makes for all *non-versicolor* examples while testing.
#
# The ROC curve shows how the sensitivity of the classifier increases as a function of (i.e., at the expense of) the fall-out. A perfect classifier would have $100\%$ sensitivity (no false negatives) and $100\%$ specificity (no false positives), which in the ROC space can be illustrated by the point in the upper left corner with the coordinate $(0,1)$. The closer the actual ROC curve gets to this point, the better is the classifier. A random guess would give a point along a diagonal line from the left bottom to the top right corners. Points above the diagonal represent good classification results (better than random); points below the line represent bad results (worse than random).
#
# To plot the ROC curve, let's first estimate the *FPR* and *TPR* for different threshold values using `sklearn`'s `roc_curve` functionality, and then plot *FPR* and *TPR* using `matplotlib`:
# +
from sklearn.metrics import roc_curve
fpr, tpr, thresholds = roc_curve(y_train_versicolor, y_scores)
def plot_roc_curve(fpr, tpr, label=None):
plt.plot(fpr, tpr, linewidth=2, label=label)
plt.plot([0, 1], [0, 1], "k--")
plt.axis([0, 1, 0, 1.01])
plt.xlabel("False positive rate (fpr)")
plt.ylabel("True positive rate (tpr)")
plot_roc_curve(fpr, tpr)
plt.show()
# -
# Another characteristic of the ROC curve is the *area under the curve (AUC)*, which for a perfect classifier will equal $1$ and for a random one will equal $0.5$. A estimate on a actual classifier, thus, usually lies between these two values:
from sklearn.metrics import roc_auc_score
roc_auc_score(y_train_versicolor, y_scores)
# Now, the `Logistic Regression` classifier does not perform very well on the *versicolor* identification task, and the best classifier among the three so far is `Naive Bayes`. Let's compare its performance to the `Logistic Regression` and visualise the difference using the ROC curve. Note that `Naive Bayes` classifier implementation in `sklearn` doesn't use `decision_function`; [`predict_proba`](https://scikit-learn.org/stable/modules/generated/sklearn.naive_bayes.GaussianNB.html#sklearn.naive_bayes.GaussianNB.predict_proba) is the equivalent of that for `Naive Bayes`:
# +
y_probas_gnb = cross_val_predict(gnb, X_train, y_train_versicolor, cv=3, method="predict_proba")
y_scores_gnb = y_probas_gnb[:, 1] # score = proba of the positive class
fpr_gnb, tpr_gnb, thresholds_gnb = roc_curve(y_train_versicolor, y_scores_gnb)
plt.plot(fpr, tpr, "b:", label="Logistic Regression")
plot_roc_curve(fpr_gnb, tpr_gnb, "Gaussian Naive Bayes")
plt.legend(loc="lower right")
plt.show()
# -
# The curve for `Naive Bayes` is much closer to the upper left corner, which shows that the classifier is better than `Logistic Regression` on this task. Let's estimate the AUC as well:
roc_auc_score(y_train_versicolor, y_scores_gnb)
# ## Step 4: Data transformations
#
# So far, you have trained three classifiers on two classification tasks – identification of *setosas* and *versicolors*. You've seen that all three classifiers perform well on the *setosas* identification example as the data is linearly separable, and only `Naive Bayes` performs well in the *versicolors* case, because the data with the given features is not linearly separable.
#
# For example, consider a decision boundary that a `Logistic Regression` classifier would learn for the linearly separable class of *setosas* (based on the first two features):
# +
X = iris.data[:, :2] # consider the first two features for plotting (as in Step 1)
y = iris.target
scatter_x = np.array(X[:, 0])
scatter_y = np.array(X[:, 1])
group = np.array(y)
cmap = matplotlib.cm.get_cmap('jet')
cdict = {0: cmap(0.1), 1: cmap(0.5), 2: cmap(0.9)}
labels = iris.target_names
fig, ax = plt.subplots(figsize=(8, 6))
for g in np.unique(group):
ix = np.where(group == g)
ax.scatter(scatter_x[ix], scatter_y[ix], c=np.array([cdict[g]]), #c = cdict[g],
label = labels[g], s = 100, marker = "H",
linewidth=2, alpha = 0.5)
log_reg.fit(X_train[:, :2], y_train_setosa) # train the classifier for setosas, using the first two features only
w = log_reg.coef_[0]
i = log_reg.intercept_[0]
xx = np.linspace(4, 8) # generate some values for feature1 (sepal length) in the appropriate range of values
yy = -(w[0]*xx + i)/w[1] # estimate the value for feature2 (sepal width) based on the learned weights and intercept
plt.plot(xx, yy, 'b--')
ax.legend()
plt.xlabel('Sepal length')
plt.ylabel('Sepal width')
plt.show()
# -
# and compare it to the case of non-linearly separable class of *versicolors*:
# +
fig, ax = plt.subplots(figsize=(8, 6))
for g in np.unique(group):
ix = np.where(group == g)
ax.scatter(scatter_x[ix], scatter_y[ix], c=np.array([cdict[g]]), #c = cdict[g],
label = labels[g], s = 100, marker = "H",
linewidth=2, alpha = 0.5)
log_reg.fit(X_train[:, :2], y_train_versicolor)
w = log_reg.coef_[0]
i = log_reg.intercept_[0]
xx = np.linspace(4, 8)
yy = -(w[0]*xx + i)/w[1]
plt.plot(xx, yy, 'g--')
ax.legend()
plt.xlabel('Sepal length')
plt.ylabel('Sepal width')
plt.show()
# -
# The original representation of the data that you've been using so far is simply not expressive enough. But what if you could transform this data in such a way that it could be linearly separated? In fact, there is a "trick" for this problem that involves transforming the original data using a number of new dimensions in such a way that in this new high-dimensional space the classes become linearly separable. This "trick" is commonly known as the *kernel trick* or *kernel method*.
#
# ### Kernel trick and approximate kernel maps
#
# You will most commonly hear about this method in the context of Support Vector Machines (SVMs), for example in the [Part II Machine Learning and Bayesian Inference course](https://www.cl.cam.ac.uk/teaching/2021/MLBayInfer/). However, some other linear classifiers, including [perceptron](https://en.wikipedia.org/wiki/Kernel_perceptron), allow for the kernel methods to be used as well.
#
# The general motivation behind the kernel trick is as follows: when the data is not linearly separable, i.e. there is no clear dividing boundary between the two classes, the kernel trick allows you to transform the data using a number of additional dimensions that would allow for such clear dividing boundary to be learned. Kernel methods require a user-specified kernel, i.e. a function that will transform the original data into a higher dimensional space. Polynomial and Gaussian (also known as *radial-basis function*) transformations are among the most widely used kernel functions. These data transformations might remind you of the polynomial feature transformation used with the linear regression in practical 1: recall that you cast the original features from the space where the relation between the features and the output cannot exactly be captured by a linear function, into a higher dimensional feature space using polynomial function. Recall also that this allowed you to apply a linear function to this new feature space leading to an improved result.
#
# You will learn more about kernel functions and their application to SVMs in more detail in the Part II Machine Learning and Bayesian Inference course. For the task at hand, you will use an `sklearn`'s [approximate kernel map](http://scikit-learn.org/stable/modules/kernel_approximation.html) in combination with the perceptron implementation of the `SGDClassifier`:
# +
from sklearn.kernel_approximation import RBFSampler
rbf_features = RBFSampler(gamma=1, random_state=42)
X_train_features = rbf_features.fit_transform(X_train)
print(X_train.shape, "->", X_train_features.shape)
sgd_rbf = SGDClassifier(max_iter=100, random_state=42, loss="perceptron",
eta0=1, learning_rate="constant", penalty=None)
sgd_rbf.fit(X_train_features, y_train_versicolor)
sgd_rbf.score(X_train_features, y_train_versicolor)
# -
# The output above shows that the classifier tries to learn the separation boundary in a highly-dimensional transformed feature space, and the accuracy of this learning on the training set is over $0.99$. Let's test this classifier in a 5-fold cross-validation manner, and compare precision, recall and F$_1$ scores to the linear classifier trained on the original data:
# +
y_train_pred = cross_val_predict(sgd, X_train, y_train_versicolor, cv=5)
precision = precision_score(y_train_versicolor, y_train_pred)
recall = recall_score(y_train_versicolor, y_train_pred)
f1 = f1_score(y_train_versicolor, y_train_pred)
print(precision, recall, f1)
y_train_pred = cross_val_predict(sgd_rbf, X_train_features, y_train_versicolor, cv=5)
precision = precision_score(y_train_versicolor, y_train_pred)
recall = recall_score(y_train_versicolor, y_train_pred)
f1 = f1_score(y_train_versicolor, y_train_pred)
print(precision, recall, f1)
# -
# Looks like the kernel trick helped improve the results on the originally non-linearly separable data significantly!
#
# ## Case 2: Multi-class classification
#
# Now remember that your actual goal is to build a three-way classifier that can predict *setosa*, *versicolor* and *virginica* classes, and not just tell whether an instance is an $X$ (*setosa* or *versicolor*) or not. Actually, you are already half way there, and here is why.
#
# Some classifiers are capable of handling multiple classes directly. For example, `Naive Bayes` learns about the probabilities of the classes in the data irrespective of the number of classes. Therefore, the binary examples above can naturally be extended to the 3-class classification scenario: you simply provide the classifier with the data on the $3$ rather than $2$ classes.
#
# In contrast, such classifiers as perceptron (`SDGClassifier`) and `Logistic Regression`, which seek to learn a linear separation boundary, are inherently binary classifiers: they try to learn a single separation boundary between two classes at a time. However, they can also very easily be extended to handle more than $2$ classes. Multi-class classification with such linear classifiers generally follows one of the two routes:
#
# - with the *one-vs-all* (*OvA*, or *one-vs-rest*, *OvR*) strategy you train $n$ classifiers (e.g., a setosa detector, a versicolor detector and a virginica detector). For a new instance, you apply all of the classifiers and predict the class that gets the highest score returned by the classifiers;
# - with the *one-vs-one* (*OvO*) strategy, you train a binary classifier for each pair of classes in your data and select the class that wins most of the duels.
#
# There are pros and cons to each of these approaches. E.g., with the *OvO* strategy, you end up training $n \times (n-1) / 2$ classifiers. I.e. for the iris dataset you will have $3$ classifiers (exactly as with the *OvA* strategy) but on a $10$-class problem this will amount to $45$ classifiers. On the other hand, the training sets with the *OvO* will be much smaller and more balanced. Some classifiers scale poorly with the size of the training set when it is imbalanced, so *OvO* for them is preferable (e.g., such are SVMs), but most of the linear classifiers use *OvA* instead.
#
# The nice thing about `sklearn` is that it implements the above strategies under the hood, so to perform a multi-class classification with the `SDGClassifier`, all you need to do is to provide it with the data and labels on $3$ classes, and it will train $3$ binary *OvA* classifiers and output the class with the highest score, i.e.:
sgd.fit(X_train, y_train) # i.e., all instances, not just one class
print(sgd.predict([setosa_example]))
print(sgd.predict([versicolor_example]))
# Recall that the *versicolor* class label is $1$, so the classifier's output is correct this time. Let's also check the result with the RBF kernel:
# +
sgd_rbf.fit(X_train_features, y_train) # i.e., all instances, not just one class
X_test_features = rbf_features.transform(X_test)
setosa_rbf_example = X_test_features[0] # note that you need to transform the test data in the same way, too
versicolor_rbf_example = X_test_features[17]
print(sgd_rbf.predict([setosa_rbf_example]))
print(sgd_rbf.predict([versicolor_rbf_example]))
# -
# This classifier classified both test examples correctly. Let's see what logic the classifier followed:
# +
setosa_scores = sgd_rbf.decision_function([setosa_rbf_example])
print(setosa_scores)
# check which class gets the maximum score
prediction = np.argmax(setosa_scores)
print(prediction)
# check which class this corresponds to in the classifier
print(sgd_rbf.classes_[prediction])
print(iris.target_names[sgd_rbf.classes_[prediction]])
# -
# This shows that *setosa* class got a much higher score than the other two. What about the versicolor example?
versicolor_scores = sgd_rbf.decision_function([versicolor_rbf_example])
print(versicolor_scores)
prediction = np.argmax(versicolor_scores)
print(prediction)
print(iris.target_names[sgd_rbf.classes_[prediction]])
# For comparison, let's see what the original `SGDClassifier` (without the RBF kernel) predicted:
versicolor_scores = sgd.decision_function([versicolor_example])
print(versicolor_scores)
prediction = np.argmax(versicolor_scores)
print(prediction)
print(iris.target_names[sgd.classes_[prediction]])
# Now, if you'd like to use the *OvO* strategy, you can enforce the `sklearn` to do so creating an instance of a `OneVsOneClassifier` (similarly, `OneVsRestClassifier` for *OvA*). Note that you're essentially using the same classifier as before, and it is just the framework within which the predictions are made:
# +
from sklearn.multiclass import OneVsOneClassifier
ovo_clf = OneVsOneClassifier(SGDClassifier(max_iter=100, random_state=42, loss="perceptron",
eta0=1, learning_rate="constant", penalty=None))
ovo_clf.fit(X_train_features, y_train)
ovo_clf.predict([versicolor_rbf_example])
# -
len(ovo_clf.estimators_)
# Now let's look into the `NaiveBayes` performance on the $3$-class problem:
gnb.fit(X_train, y_train)
gnb.predict([versicolor_example])
# It correctly classifies the *versicolor* example, so let's check how confident it is about this prediction (remember that you should use `predict_proba` with `NaiveBayes` and `decision_function` with the `SGDClassifier`):
gnb.predict_proba([versicolor_example])
# Let's look into the cross-validated performance of the classifiers:
print(cross_val_score(sgd_rbf, X_train_features, y_train, cv=5, scoring="accuracy"))
print(cross_val_score(ovo_clf, X_train_features, y_train, cv=5, scoring="accuracy"))
print(cross_val_score(gnb, X_train, y_train, cv=5, scoring="accuracy"))
# Finally, recall that in practical 1 you used further transformations on the data, e.g. scaling. Let's apply scaling and compare the results (note that you can use `np.mean()` to get the average accuracy score across all splits if you wish to get a single aggregated accuracy score instead of $5$ separate ones):
# +
from sklearn.preprocessing import StandardScaler, MinMaxScaler
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train.astype(np.float64))
X_train_features_scaled = scaler.fit_transform(X_train_features.astype(np.float64))
print(cross_val_score(sgd_rbf, X_train_features_scaled, y_train, cv=5, scoring="accuracy"))
print(cross_val_score(ovo_clf, X_train_features_scaled, y_train, cv=5, scoring="accuracy"))
print(cross_val_score(gnb, X_train_scaled, y_train, cv=5, scoring="accuracy"))
# -
# ## Step 5: Error analysis
#
# Before applying the classifiers to the test data, let's gain a bit more insight into what the classifiers get wrong. Recall, that earlier you used confusion matrices to learn about the classification errors:
y_train_pred = cross_val_predict(sgd_rbf, X_train_features_scaled, y_train, cv=3)
conf_mx = confusion_matrix(y_train, y_train_pred)
conf_mx
# Let's visualise the classifier decisions:
plt.imshow(conf_mx, cmap = "jet")
plt.show()
# And if you'd like to highlight only the most salient errors, you can do so as follows (the `"jet"` color scheme uses red spectrum for higher numbers):
row_sums = conf_mx.sum(axis=1, keepdims=True)
norm_conf_mx = conf_mx / row_sums
np.fill_diagonal(norm_conf_mx, 0)
plt.imshow(norm_conf_mx, cmap = "jet")
plt.show()
# ## Final step – evaluating on the test set
#
# The `SGDClassifier` with the RBF kernel:
# +
from sklearn.metrics import accuracy_score
X_test_features_scaled = scaler.transform(X_test_features.astype(np.float64))
y_pred = sgd_rbf.predict(X_test_features_scaled)
accuracy_score(y_test, y_pred)
# -
precision = precision_score(y_test, y_pred, average='weighted')
recall = recall_score(y_test, y_pred, average='weighted')
f1 = f1_score(y_test, y_pred, average='weighted')
print(precision, recall, f1)
# The *OvO* SGD classifier:
# +
from sklearn.metrics import accuracy_score
X_test_features_scaled = scaler.transform(X_test_features.astype(np.float64))
y_pred = ovo_clf.predict(X_test_features_scaled)
accuracy_score(y_test, y_pred)
# -
precision = precision_score(y_test, y_pred, average='weighted')
recall = recall_score(y_test, y_pred, average='weighted')
f1 = f1_score(y_test, y_pred, average='weighted')
print(precision, recall, f1)
# The `NaiveBayes` classifier:
gnb.fit(X_train, y_train)
y_pred = gnb.predict(X_test)
accuracy_score(y_test, y_pred)
precision = precision_score(y_test, y_pred, average='weighted')
recall = recall_score(y_test, y_pred, average='weighted')
f1 = f1_score(y_test, y_pred, average='weighted')
print(precision, recall, f1)
# # Assignment: Handwritten digits dataset
#
# The dataset that you will use in this assignment is the [*digits* dataset](http://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_digits.html) which contains $1797$ images of $10$ hand-written digits. The digits have been preprocessed so that $32 \times 32$ bitmaps are divided into non-overlapping blocks of $4 \times 4$ and the number of on pixels are counted in each block. This generates an input matrix of $8 \times 8$ where each element is an integer in the range of $[0, ..., 16]$. This reduces dimensionality and gives invariance to small distortions.
#
# For further information on NIST preprocessing routines applied to this data, see <NAME>, <NAME>, <NAME>, <NAME>, <NAME>, <NAME>, <NAME>, and <NAME>, *NIST Form-Based Handprint Recognition System*, NISTIR 5469, 1994.
#
# As before, use the `sklearn`'s data uploading routines to load the dataset and get the data fields:
from sklearn import datasets
digits = datasets.load_digits()
list(digits.keys())
digits
X, y = digits["data"], digits["target"]
X.shape
y.shape
# You can access the digits and visualise them using the following code (feel free to select another digit):
# +
some_digit = X[3]
some_digit_image = some_digit.reshape(8, 8)
plt.imshow(some_digit_image, cmap=matplotlib.cm.binary, interpolation="nearest")
plt.axis("off")
plt.show()
# -
y[3]
# For the rest of the practical, apply the data preprocessing techniques, implement and evaluate the classification models on the digits dataset using the steps that you applied above to the iris dataset.
|
DSPNP_practical2/DSPNP_notebook2.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Streamlines tutorial
# In this tutorial you will learn how to download and render streamline data to display connectivity data. In brief, injections of anterogradely transported viruses are performed in wild type and CRE-driver mouse lines. The viruses express fluorescent proteins so that efferent projections from the injection locations can be traced everywhere in the brain. The images with the fluorescence data are acquired and registered to the Allen Coordinates reference frame. The traces of the streamlines are then extracted using a fast marching algorithm (by [https://neuroinformatics.nl](https://neuroinformatics.nl)).
#
# <img src="../Docs/Media/streamlines.png" width="600" height="350">
#
# The connectivity data are produced as part of the Allen Brain Atlas [Mouse Connectivity project](http://connectivity.brain-map.org).
#
# The first step towards being able to render streamlines data is to identify the set of experiments you are interested in (i.e. injections in the primary visual cortex of wild type mice]. To do so you can use the experiments explorer at [http://connectivity.brain-map.org].
#
# Once you have selected the experiments, you can download metadata about them using the 'download data as csv' option at the bottom of the page. This metadata .csv is what we can then use to get a link to the data to download.
#
# First we do the usual set up steps to get brainrender up and running
# ### Setup
#
# +
# We begin by adding the current path to sys.path to make sure that the imports work correctly
import sys
sys.path.append('../')
import os
# Set up VTKPLOTTER to work in Jupyter notebooks
from vtkplotter import *
embedWindow(backend=False)
# Import variables
from brainrender import * # <- these can be changed to personalize the look of your renders
# Import brainrender classes and useful functions
from brainrender.scene import Scene
from brainrender.Utils.parsers.streamlines import StreamlinesAPI
from brainrender.Utils.data_io import listdir
# Before populating the scene, we need to change the current working directory to the parent folder,
# then we are ready to start!
os.chdir(os.path.normpath(os.path.join(os.getcwd(), os.pardir)))
streamlines_api = StreamlinesAPI()
# -
# ## Downloading data
# If you have streamlines data already saved somewhere, you can skip this section.
#
# ### Manual download
# To download streamlines data, you have two options (see the [user guide](Docs/UserGuide.md) for more details.
# If you head to [http://connectivity.brain-map.org](http://connectivity.brain-map.org) you can download a .csv file with the experiment IDs of interest. Then you can use the following function to download the streamline data:
# parse .csv file
# Make sure to put the path to your downloaded file here
filepaths, data = streamlines_api.extract_ids_from_csv("Examples/example_files/experiments_injections.csv",
download=True)
# The `filepaths` variable stores the paths to the .json files that have been saved by the `streamlines_api`, the `data` variable already contains the streamlines data. You can pass either `filepaths` or `data` to `scene.add_streamlines` (see below) to render your streamlines data.
# ### Automatic download
# If you know that you simply want to download the data to a specific target structure, then you can let brainrender take care of downloading the data for you. This is how:
filepaths, data = streamlines_api.download_streamlines_for_region("CA1") # <- get the streamlines for CA1
# Once you have downloaded the streamlines data, it's time to render it in your scene.
#
# ## Rendering streamlines data
# You can pass either `data` or `filepaths` to `scene.add_streamlines`, just make sure to use the correct keyword argument (unimaginatively called `data` and `filepath`).
# +
# Start by creating a scene
scene = Scene(jupyter=True)
# you can then pass this list of filepaths to add_streamlines.
scene.add_streamlines(data, color="green")
# alternative you can pass a string with the path to a single file or a list of paths to the .json files that you
# created in some other way.
# then you can just render your scene
scene.render()
# -
# add_streamliens takes a few arguments that let you personalize the look of the streamlines:
# * `colorby`: you can pass the acronym to a brain region, then the default color of that region will be used for the streamliens
# * `color`: alternatively you can specify the color of the streamlines directly.
# * `alpha`, `radius`: you can change the transparency and the thickness of the actors used to render the streamlines.
# * `show_injection_site`: if set as True, a sphere will be rendered at the locations that correspond to the injections sytes.
#
#
# Don't forget to check the other examples to lear more about how to use brainrender to make amazing 3D renderings!
# Also, you can find a list of variables you can play around with in brainrender.variables.py
# Playing around with these variables will allow you to make the rendering look exactly how you want them to be.
|
Examples/notebooks/Streamlines.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### Installation
# - `pip install eleanor` ([eleanor Docs](http://adina.feinste.in/eleanor))
# - Through Github ([eleanor GitHub](https://github.com/afeinstein20/eleanor))
# +
import eleanor
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams['font.size'] = 16
# -
# ## Step 1: Initiating eleanor.Source
# To use `eleanor` you need some identifier for your target. This can be either the TIC ID, a set of coordinates, a Gaia ID, or the name of your star. If you know the sector your target was observed in, that's great! If not, that's okay. You don't need to set the `sector` argument. Instead, `eleanor` will pass back the latest sector your target was observed in. For example, for a CVZ target, `eleanor.Source` will return data from Sector 13.
star = eleanor.Source(name='WASP-100', sector=3)
# Above, we have downloaded a few `eleanor` data products: a postcard, a 2D background model on the postcard level, and a pointing model. These products are currently only available for the first 13 sectors, but will be created for the Northern hemisphere (Sectors 14-26) as well.
#
# ----
#
# Whatever identifier you pass in, `eleanor` will crossmatch with the corresponding TIC ID, coordinates, and/or Gaia ID.
star.tic, star.coords, star.gaia
# The `eleanor.Source` class also provides information on where your target was observed on the TESS CCDs:
star.sector, star.camera, star.chip
# ## Step 2: Making a light curve
# `eleanor.Source` set us up with everything we need to make the light curve. To do this, we need to create an `eleanor.TargetData` object. There are some additional arguments that we can use here, such as setting the Target Pixel File (TPF) height and width, creating a point-spread function (PSF) modeled light curve, and creating a principle component analysis (PCA) light curve.
#
# We'll do both the PSF light curve, by setting `do_psf=True`, and the PCA light curve, by setting `do_pca=True`, for this example.
data = eleanor.TargetData(star, do_psf=True, do_pca=True)
# In addition to different types of light curves, we are also trying a few different background subtraction methods to remove as much background noise as possible. The three options `eleanor` tries are:
# - 1D postcard background: A constant calculated from each postcard frame, masking stars.
# - 1D TPF background: A constant calculated froom each TPF frame, masking stars.
# - 2D background: The 2D background pixels are subtracted from the TPF.
#
# The postcard and the 2D modeled background from the postcard look like this:
# We can see which background produced the best light curve. And by best, the `eleanor` light curves are optimized for transit searches, so we minimized the CDPP (combined differential photometric precision) to define the "best".
plt.title('2D background')
plt.imshow(data.post_obj.background2d[100], vmin=0, vmax=20)
plt.colorbar();
data.bkg_type
# "PC_LEVEL" means the 1D postcard level background removed the most systematics. This background model looks like:
plt.figure(figsize=(14,4))
plt.plot(data.time, data.post_obj.bkg, 'w', lw=3)
plt.xlabel('time [bjd-2457000]')
plt.ylabel('background flux');
# In the same spirit, the aperture selected by `eleanor` also minimizes the CDPP. The aperture, over the target, selected looks like:
plt.imshow(data.tpf[100])
plt.imshow(data.aperture, alpha=0.4, cmap='Greys_r');
# Or this can be better seen using the `eleanor.Visualize` class:
vis = eleanor.Visualize(data)
fig = vis.aperture_contour()
# ## Step 3: Look at your light curves!
# It's time to see what was created:
# +
q = data.quality == 0
plt.figure(figsize=(14,4))
plt.plot(data.time[q], data.raw_flux[q]/np.nanmedian(data.raw_flux[q])-0.005, 'w', lw=3,
label='RAW')
plt.plot(data.time[q], data.corr_flux[q]/np.nanmedian(data.corr_flux[q])+0.015, 'k', lw=3,
label='CORR')
plt.ylim(0.98,1.02)
plt.xlabel('time [bjd-2457000]')
plt.ylabel('normalized flux')
plt.legend();
# -
plt.figure(figsize=(14,4))
plt.plot(data.time[q], data.pca_flux[q]/np.nanmedian(data.pca_flux[q])-0.005, 'darkorange', lw=3,
label='PCA')
plt.plot(data.time[q], data.psf_flux[q]/np.nanmedian(data.psf_flux[q])+0.015, 'skyblue', lw=3,
label='PSF')
plt.ylim(0.98,1.02)
plt.xlabel('time [bjd-2457000]')
plt.ylabel('normalized flux')
plt.legend();
# If you're missing all of the great tools implemented by `lightkurve`, we have an easy fix for you. By calling `eleanor.to_lightkurve()`, you will get a `lightkurve.LightCurve` object. You can also specify which flux you want passed into the object.
lc = data.to_lightkurve(flux=data.corr_flux)
lc.normalize().plot()
# ## Step 1 (redone): What if my target was observed in multiple sectors?
#
# Instead of initiating an `eleanor.Source` class, you can call `eleanor.multi_sectors`, which will return a list of `eleanor.Source` objects per each sector your target was observed in. If you want specific sectors, you can pass those in as a list/array. Otherwise, if you want all of the sectors your target was observed in, you can pass in `sectors="all"` and `eleanor` will fetch all of those for you.
stars = eleanor.multi_sectors(tic=star.tic, sectors=np.arange(2,6,1,dtype=int))
stars
# It downloads all of the postcards, 2D postcard backgrounds, and pointing models. Now, to get a light curve from each sector, you can pass it into `eleanor.TargetData` in a loop:
# +
data = []
for s in stars:
datum = eleanor.TargetData(s)
data.append(datum)
# -
# And to look at our light curves:
plt.figure(figsize=(14,4))
for d in data:
q = d.quality == 0
plt.plot(d.time[q], d.corr_flux[q]/np.nanmedian(d.corr_flux[q]), 'w', lw=3)
plt.ylim(0.99,1.005)
plt.xlabel('time [bjd-2457000]')
plt.ylabel('normalized flux');
# ## Step 4 (optional): Remember that visualization object?
# Earlier we created an `eleanor.Visualize` object that allowed us to overplot a countour of the aperture on the TPF. There are other things in there to use as well when vetting your target!
#
# One of the most useful tricks is creating a pixel-by-pixel light curve grid, to see if the signal you're seeing in the light curve is from your source or something nearby:
fig = vis.pixel_by_pixel()
# Okay, maybe not useful like this, but zooming in and assigning the light curve color to be that of the TPF pixel:
fig = vis.pixel_by_pixel(colrange=[4,10], rowrange=[4,10], color_by_pixel=True)
# It looks like the signal is coming from the source! But just in case, you can overplot nearby Gaia sources on your TPF (thanks <NAME>!). The points are related to the magnitude of the source:
fig = vis.plot_gaia_overlay(magnitude_limit=16)
# ## Step 5 (technically optional, but should consider!): Crossmatching
#
# Within `eleanor`, we also have tools to see if your target has a light curve produced by the TASOC team (asteroseismology), Oelkers & Stassun difference imaging pipeline, or was observed at 2-minute cadence! We can start digging by the following:
crossmatch = eleanor.Crossmatch(data[0])
# To check for 2-minute data, we use `lightkurve` behind the scenes:
crossmatch.two_minute()
# Because this returns a `lightkurve.SearchResult` object, you can download the data product right from there and go about using the other `lightkurve` tools.
#
# -----
#
# To check out the TASOC pipeline:
crossmatch.tasoc_lc()
plt.figure(figsize=(14,4))
q = crossmatch.tasoc_pixel_quality == 0
plt.plot(crossmatch.tasoc_time[q], crossmatch.tasoc_flux_raw[q], 'k', lw=3)
plt.xlabel('time [BJD - 2457000]')
plt.ylabel('flux');
# To check the Oelkers & Stassun light curves (it shouold be noted these light curves are in magnitudes!):
crossmatch.oelkers_lc()
plt.figure(figsize=(14,4))
plt.plot(crossmatch.os_time, crossmatch.os_mag, 'w', lw=3)
plt.xlabel('time [BJD - 2457000]')
plt.ylabel('magnitude');
|
notebooks/online_tess_science_eleanor.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [default]
# language: python
# name: python3
# ---
# +
from IPython.display import HTML
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
$('div.prompt').hide();
} else {
$('div.input').show();
$('div.prompt').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
<form action="javascript:code_toggle()"><input type="submit" value="Code Toggle"></form>''')
# + [markdown] slideshow={"slide_type": "slide"}
# # Hackathon 2
#
# ** May 3, 2016 – Northwestern University **
#
# <img src="../../../images/hackathon2_photo.jpg" alt="">
#
# ## Note from the organizer
#
# CHiMaD hosted the second Phase Field Methods Hackathon on May 3, 2016, in connection with its [Phase Field Methods Workshop III]({{ site.baseurl }}/workshops/) on May 4 - 5, 2016. The motivation for the hackathon comes from one of CHiMaD’s missions: to develop community standards for phase field modeling in materials science, and to distribute community codes for phase field modeling. As part of this mission, CHiMaD is working on developing and distributing a set of standard problems for phase field modeling, analogous to how the micromagnetic community developed a set of [standard problems for micromagnetic modeling](http://www.ctcms.nist.gov/~rdm/mumag.org.html). A [first hackathon]({{ site.baseurl }}/hackathons/hackathon1/) was held on October 14 – 15, 2015, with problem sets focusing on Cahn-Hilliard and coupled Allen-Cahn/Cahn-Hilliard problems. This second Hackthon will explore other canonical problems central to phase-field type modeling of materials.
#
# Teams consisting of two students or postdocs were given the set of problems. They contain two kinds of phase field modeling, and each set contains sub-problems of increasing difficulty. The problems are completely defined in terms of initial conditions, geometry, and material parameters. The teams will have internet access and will be tasked with attempting to solve the problems within 24 hours using whatever numerical codes they have at their disposition - there will be no codes provided at the hackathon. All attendees are expected to bring their own laptops and connect to the servers they regularly use for running their codes.
#
# The goal of the hackathon is to see how different codes and different approaches can handle the problems with respect to accuracy and speed, and also to serve as a test bed for the development of standard problems. The aim of the hackathon is not to produce winners or losers, but to advance our understanding of phase field modeling: in that context, all results or attempts at solving the problems will be valuable. Each team will be required to present their results at the Phase Field Workshop on the morning of May 4.
# -
# ## Hackathon Challenge Problems and Solutions
#
# ### Challenge Problems
#
# * [Problem 1: Dendritic Growth in 2D](../problem1.ipynb)
# * [Problem 2: Linear Elasticity in 3D](../problem2.ipynb)
#
# <br>
# <br>
# ### Solutions
# #### University of Connecticut: Moose
#
# [GitHub Repo](https://github.com/kcpitike/Hackathon-2)
#
# <iframe src="//www.slideshare.net/slideshow/embed_code/key/1qcfv6PO3oSgS" width="425" height="355" frameborder="0" marginwidth="0" marginheight="0" scrolling="no" style="border:1px solid #CCC; border-width:1px; margin-bottom:5px; max-width: 100%;" allowfullscreen> </iframe>
#
# <strong>
# <a href="//www.slideshare.net/DanielWheeler18/chimad-phase-field-hackathon-2-university-of-connecticut" title="CHiMaD Hackathon 2: University of Connecticut" target="_blank">CHiMaD Phase Field Hackathon 2: University of Connecticut</a> </strong> by <strong>
# <a target="_blank" href="https://github.com/kcpitike"><NAME></a> </strong> and
# <strong><a target="_blank" href="https://github.com/mangerij"><NAME></a>
# </strong>
# <br>
# <br>
# #### Pennsylvania State University: Moose
#
# [GitHub Repo](https://github.com/wd15/penn-hackathon2/tree/master)
#
# <iframe src="//www.slideshare.net/slideshow/embed_code/key/Dbgdhk1Zs5JWtC" width="425" height="355" frameborder="0" marginwidth="0" marginheight="0" scrolling="no" style="border:1px solid #CCC; border-width:1px; margin-bottom:5px; max-width: 100%;" allowfullscreen> </iframe>
#
#
# <strong> <a href="//www.slideshare.net/DanielWheeler18/chimad-hackathon-2-pennsylvania-state-university" title="CHiMaD Hackathon 2: Pennsylvania State University" target="_blank">CHiMaD Hackathon 2: Pennsylvania State University</a>
# </strong> by
# <strong><a target="_blank" href="https://github.com/tonkmr"><NAME></a></strong>,
# <strong><a target="_blank" href="https://github.com/itgreenquist">I. Greenquist</a></strong> and
# <strong><a target="_blank" href="https://github.com/kasra83"><NAME></a></strong>
#
# <br>
# <br>
# #### McGill University: Unknown FE Code and Julia
#
# [GitHub Repo](https://github.com/nsmith5/chimadQ2)
#
# <iframe src="//www.slideshare.net/slideshow/embed_code/key/4QZp8GXqomEC0B" width="425" height="355" frameborder="0" marginwidth="0" marginheight="0" scrolling="no" style="border:1px solid #CCC; border-width:1px; margin-bottom:5px; max-width: 100%;" allowfullscreen> </iframe>
#
#
# <strong> <a href="//www.slideshare.net/DanielWheeler18/chimad-hackathon-2-team-mcgill" title="CHiMaD Hackathon 2: Team mcgill" target="_blank">CHiMaD Hackathon 2: McGill University</a>
# </strong> by
# <strong><a target="_blank" href="https://github.com/nsmith5"><NAME></a></strong>,
# <strong>T Pinomma</strong> and
# <strong><NAME></strong>
#
# <br>
# <br>
# #### NIST: FiPy
#
# [GitHub Repo](https://github.com/usnistgov/PhaseFieldHackathon-FiPy)
# <br>
# <br>
# #### INL: Moose
#
# <iframe src="//www.slideshare.net/slideshow/embed_code/key/E1OGX4YyH0HtzK" width="425" height="355" frameborder="0" marginwidth="0" marginheight="0" scrolling="no" style="border:1px solid #CCC; border-width:1px; margin-bottom:5px; max-width: 100%;" allowfullscreen> </iframe> <strong> <a href="//www.slideshare.net/DanielWheeler18/chimad-hackathon-2" title="CHiMaD Hackathon 2" target="_blank">CHiMaD Hackathon 2: INL</a> </strong> by
# <strong><NAME></strong> and
# <strong><NAME></strong>
#
# <br>
# <br>
# #### University of Michigan: PRISMS
#
# <iframe src="//www.slideshare.net/slideshow/embed_code/key/3ERQSQGzAh1dZq" width="425" height="355" frameborder="0" marginwidth="0" marginheight="0" scrolling="no" style="border:1px solid #CCC; border-width:1px; margin-bottom:5px; max-width: 100%;" allowfullscreen> </iframe> <strong> <a href="//www.slideshare.net/DanielWheeler18/chimad-hackathon-2-university-of-michigan" title="CHiMaD Hackathon 2: University of Michigan" target="_blank">CHiMaD Hackathon 2: University of Michigan</a> </strong> by
# <strong><NAME></strong> and
# <strong><NAME></strong>
#
|
hackathons/hackathon2/index.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # jupyter notebook生成PDF文件采用LaTeX编译方式,须先行安装Tex软件。(Linux环境建议使用TeXLive, Windows环境建议使用MiKTeX,Mac环境建议使用MacTeX)
# - 建议直接前往 [https://tug.org/](https://tug.org/) 下载最新版本TeX软件,TeXLive软件下载与安装链接地址为:[https://tug.org/texlive/](https://tug.org/texlive/)。
#
# - Ubuntu下用apt-get获取的TeXLive软件版本较旧,插件不齐,在采用tlmgr包管理升级时会遇到版本不兼容,无法升级的问题。
# # 由于jupyter notebook自身模板限制,PDF转换模板中默认使用LaTeX的标准T1字体输出(不支持中文)。因此,如要输出包含中文的PDF文件,则需要按以下步骤执行:
# - 在jupyter notebook环境下先把ipynb工作文件输出为LaTeX(.tex)文件。(在此假定输出文件名以*sample.tex*)
#
# - 用文本编辑器打开*sample.tex*。
#
# - 在**\documentclass**段落之下新增以下语句,引入xeCJK包,实现中文支持。
#
# ```
# \usepackage{xeCJK}
# ```
#
# - 保存文件后,使用`xelatex sample.tex`即可编译生成支持中文的 *sample.pdf* 文件。
#
# - 在上面的语句后,可添加以下语句设置宋体为生成PDF文件的缺省中文字体,不作设置的话将以系统字体为准。
#
# ```
# \setCJKmainfont{SimSun}
# ```
# *注:*
#
# - *测试环境为:Ubuntu 19.04 + TexLive 2019(缺省安装已包含xeCJK, cTex中文宏包)。*
#
# - *xeCJK中文支持包具体使用方法参见说明文件 [xeCJK.pdf](xeCJK.pdf)。*
#
# - *本文即为按以上方法生成的PDF中文文档。*
|
docs/IMPOART_NOTICE_FOR_CHINESE_PDF_DOCUMENT.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3.6 (tensorflow)
# language: python
# name: rga
# ---
# <a href="https://colab.research.google.com/github/jeffheaton/t81_558_deep_learning/blob/master/t81_558_class_03_5_weights.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# # T81-558: Applications of Deep Neural Networks
# **Module 3: Introduction to TensorFlow**
# * Instructor: [<NAME>](https://sites.wustl.edu/jeffheaton/), McKelvey School of Engineering, [Washington University in St. Louis](https://engineering.wustl.edu/Programs/Pages/default.aspx)
# * For more information visit the [class website](https://sites.wustl.edu/jeffheaton/t81-558/).
# # Module 3 Material
#
# * Part 3.1: Deep Learning and Neural Network Introduction [[Video]](https://www.youtube.com/watch?v=zYnI4iWRmpc&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_03_1_neural_net.ipynb)
# * Part 3.2: Introduction to Tensorflow and Keras [[Video]](https://www.youtube.com/watch?v=PsE73jk55cE&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_03_2_keras.ipynb)
# * Part 3.3: Saving and Loading a Keras Neural Network [[Video]](https://www.youtube.com/watch?v=-9QfbGM1qGw&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_03_3_save_load.ipynb)
# * Part 3.4: Early Stopping in Keras to Prevent Overfitting [[Video]](https://www.youtube.com/watch?v=m1LNunuI2fk&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_03_4_early_stop.ipynb)
# * **Part 3.5: Extracting Weights and Manual Calculation** [[Video]](https://www.youtube.com/watch?v=7PWgx16kH8s&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_03_5_weights.ipynb)
# # Google CoLab Instructions
#
# The following code ensures that Google CoLab is running the correct version of TensorFlow.
try:
# %tensorflow_version 2.x
COLAB = True
print("Note: using Google CoLab")
except:
print("Note: not using Google CoLab")
COLAB = False
# # Part 3.5: Extracting Keras Weights and Manual Neural Network Calculation
#
# ### Weight Initialization
#
# The weights of a neural network determine the output for the neural network. The process of training can adjust these weights so the neural network produces useful output. Most neural network training algorithms begin by initializing the weights to a random state. Training then progresses through a series of iterations that continuously improve the weights to produce better output.
#
# The random weights of a neural network impact how well that neural network can be trained. If a neural network fails to train, you can remedy the problem by simply restarting with a new set of random weights. However, this solution can be frustrating when you are experimenting with the architecture of a neural network and trying different combinations of hidden layers and neurons. If you add a new layer, and the network’s performance improves, you must ask yourself if this improvement resulted from the new layer or from a new set of weights. Because of this uncertainty, we look for two key attributes in a weight initialization algorithm:
#
# * How consistently does this algorithm provide good weights?
# * How much of an advantage do the weights of the algorithm provide?
#
# One of the most common, yet least effective, approaches to weight initialization is to set the weights to random values within a specific range. Numbers between -1 and +1 or -5 and +5 are often the choice. If you want to ensure that you get the same set of random weights each time, you should use a seed. The seed specifies a set of predefined random weights to use. For example, a seed of 1000 might produce random weights of 0.5, 0.75, and 0.2. These values are still random; you cannot predict them, yet you will always get these values when you choose a seed of 1000.
# Not all seeds are created equal. One problem with random weight initialization is that the random weights created by some seeds are much more difficult to train than others. In fact, the weights can be so bad that training is impossible. If you find that you cannot train a neural network with a particular weight set, you should generate a new set of weights using a different seed.
#
# Because weight initialization is a problem, there has been considerable research around it. In this course we use the Xavier weight initialization algorithm, introduced in 2006 by Glorot & Bengio[[Cite:glorot2010understanding]](http://proceedings.mlr.press/v9/glorot10a/glorot10a.pdf), produces good weights with reasonable consistency. This relatively simple algorithm uses normally distributed random numbers.
#
# To use the Xavier weight initialization, it is necessary to understand that normally distributed random numbers are not the typical random numbers between 0 and 1 that most programming languages generate. In fact, normally distributed random numbers are centered on a mean ($\mu$, mu) that is typically 0. If 0 is the center (mean), then you will get an equal number of random numbers above and below 0. The next question is how far these random numbers will venture from 0. In theory, you could end up with both positive and negative numbers close to the maximum positive and negative ranges supported by your computer. However, the reality is that you will more likely see random numbers that are between 0 and three standard deviations from the center.
#
# The standard deviation ($\sigma$, sigma) parameter specifies the size of this standard deviation. For example, if you specified a standard deviation of 10, then you would mainly see random numbers between -30 and +30, and the numbers nearer to 0 have a much higher probability of being selected.
#
# The above figure illustrates that the center, which in this case is 0, will be generated with a 0.4 (40%) probability. Additionally, the probability decreases very quickly beyond -2 or +2 standard deviations. By defining the center and how large the standard deviations are, you are able to control the range of random numbers that you will receive.
#
# The Xavier weight initialization sets all of the weights to normally distributed random numbers. These weights are always centered at 0; however, their standard deviation varies depending on how many connections are present for the current layer of weights. Specifically, Equation 4.2 can determine the standard deviation:
#
# $ Var(W) = \frac{2}{n_{in}+n_{out}} $
#
# The above equation shows how to obtain the variance for all of the weights. The square root of the variance is the standard deviation. Most random number generators accept a standard deviation rather than a variance. As a result, you usually need to take the square root of the above equation. The following figure shows how one layer might be initialized.
#
# 
#
# This process is completed for each layer in the neural network.
#
# ### Manual Neural Network Calculation
#
# In this section we will build a neural network and analyze it down the individual weights. We will train a simple neural network that learns the XOR function. It is not hard to simply hand-code the neurons to provide an [XOR function](https://en.wikipedia.org/wiki/Exclusive_or); however, for simplicity, we will allow Keras to train this network for us. We will just use 100K epochs on the ADAM optimizer. This is massive overkill, but it gets the result, and our focus here is not on tuning. The neural network is small. Two inputs, two hidden neurons, and a single output.
# +
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation
import numpy as np
# Create a dataset for the XOR function
x = np.array([
[0,0],
[1,0],
[0,1],
[1,1]
])
y = np.array([
0,
1,
1,
0
])
# Build the network
# sgd = optimizers.SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True)
done = False
cycle = 1
while not done:
print("Cycle #{}".format(cycle))
cycle+=1
model = Sequential()
model.add(Dense(2, input_dim=2, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
model.fit(x,y,verbose=0,epochs=10000)
# Predict
pred = model.predict(x)
# Check if successful. It takes several runs with this small of a network
done = pred[0]<0.01 and pred[3]<0.01 and pred[1] > 0.9 and pred[2] > 0.9
print(pred)
# -
pred[3]
# The output above should have two numbers near 0.0 for the first and forth spots (input [[0,0]] and [[1,1]]). The middle two numbers should be near 1.0 (input [[1,0]] and [[0,1]]). These numbers are in scientific notation. Due to random starting weights, it is sometimes necessary to run the above through several cycles to get a good result.
#
# Now that the neural network is trained, lets dump the weights.
# Dump weights
for layerNum, layer in enumerate(model.layers):
weights = layer.get_weights()[0]
biases = layer.get_weights()[1]
for toNeuronNum, bias in enumerate(biases):
print(f'{layerNum}B -> L{layerNum+1}N{toNeuronNum}: {bias}')
for fromNeuronNum, wgt in enumerate(weights):
for toNeuronNum, wgt2 in enumerate(wgt):
print(f'L{layerNum}N{fromNeuronNum} -> L{layerNum+1}N{toNeuronNum} = {wgt2}')
# If you rerun this, you probably get different weights. There are many ways to solve the XOR function.
#
# In the next section, we copy/paste the weights from above and recreate the calculations done by the neural network. Because weights can change with each training, the weights used for the below code came from this:
#
# ```
# 0B -> L1N0: -1.2913415431976318
# 0B -> L1N1: -3.021530048386012e-08
# L0N0 -> L1N0 = 1.2913416624069214
# L0N0 -> L1N1 = 1.1912699937820435
# L0N1 -> L1N0 = 1.2913411855697632
# L0N1 -> L1N1 = 1.1912697553634644
# 1B -> L2N0: 7.626241297587034e-36
# L1N0 -> L2N0 = -1.548777461051941
# L1N1 -> L2N0 = 0.8394404649734497
# ```
# +
input0 = 0
input1 = 1
hidden0Sum = (input0*1.3)+(input1*1.3)+(-1.3)
hidden1Sum = (input0*1.2)+(input1*1.2)+(0)
print(hidden0Sum) # 0
print(hidden1Sum) # 1.2
hidden0 = max(0,hidden0Sum)
hidden1 = max(0,hidden1Sum)
print(hidden0) # 0
print(hidden1) # 1.2
outputSum = (hidden0*-1.6)+(hidden1*0.8)+(0)
print(outputSum) # 0.96
output = max(0,outputSum)
print(output) # 0.96
# -
|
t81_558_class_03_5_weights.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
#
# ---
# # VIDEO: Algebraic and geometric interpretations
# ---
#
# +
# 2-dimensional vector
v2 = [ 3, -2 ]
# 3-dimensional vector
v3 = [ 4, -3, 2 ]
# row to column (or vice-versa):
v3t = np.transpose(v3)
# plot them
plt.plot([0,v2[0]],[0,v2[1]])
plt.axis('equal')
plt.plot([-4, 4],[0, 0],'k--')
plt.plot([0, 0],[-4, 4],'k--')
plt.grid()
plt.axis((-4, 4, -4, 4))
plt.show()
# plot the 3D vector
fig = plt.figure(figsize=plt.figaspect(1))
ax = fig.gca(projection='3d')
ax.plot([0, v3[0]],[0, v3[1]],[0, v3[2]],linewidth=3)
# make the plot look nicer
ax.plot([0, 0],[0, 0],[-4, 4],'k--')
ax.plot([0, 0],[-4, 4],[0, 0],'k--')
ax.plot([-4, 4],[0, 0],[0, 0],'k--')
plt.show()
# -
#
# ---
# # VIDEO: Vector addition/subtraction
# ---
#
# +
# two vectors in R2
v1 = np.array([ 3, -1 ])
v2 = np.array([ 2, 4 ])
v3 = v1 + v2
# plot them
plt.plot([0, v1[0]],[0, v1[1]],'b',label='v1')
plt.plot([0, v2[0]]+v1[0],[0, v2[1]]+v1[1],'r',label='v2')
plt.plot([0, v3[0]],[0, v3[1]],'k',label='v1+v2')
plt.legend()
plt.axis('square')
plt.axis((-6, 6, -6, 6 ))
plt.grid()
plt.show()
# -
#
# ---
# # VIDEO: Vector-scalar multiplication
# ---
#
# +
# vector and scalar
v1 = np.array([ 3, -1 ])
l = 2.3
v1m = v1*l # scalar-modulated
# plot them
plt.plot([0, v1[0]],[0, v1[1]],'b',label='v_1')
plt.plot([0, v1m[0]],[0, v1m[1]],'r:',label='\lambda v_1')
plt.axis('square')
axlim = max([max(abs(v1)),max(abs(v1m))])*1.5 # dynamic axis lim
plt.axis((-axlim,axlim,-axlim,axlim))
plt.grid()
plt.show()
# -
#
# ---
# # VIDEO: Vector-vector multiplication: the dot product
# ---
#
# +
## many ways to compute the dot product
v1 = np.array([ 1, 2, 3, 4, 5, 6 ])
v2 = np.array([ 0, -4, -3, 6, 5 ])
# method 1
dp1 = sum( np.multiply(v1,v2) )
# method 2
dp2 = np.dot( v1,v2 )
# method 3
dp3 = np.matmul( v1,v2 )
# method 4
dp4 = 0 # initialize
# loop over elements
for i in range(len(v1)):
# multiply corresponding element and sum
dp4 = dp4 + v1[i]*v2[i]
print(dp1,dp2,dp3,dp4)
# -
# # VIDEO: Dot product properties: associative and distributive
# +
## Distributive property
# create random vectors
n = 10
a = np.random.randn(n)
b = np.random.randn(n)
c = np.random.randn(n)
# the two results
res1 = np.dot( a , (b+c) )
res2 = np.dot(a,b) + np.dot(a,c)
# compare them
print([ res1,res2 ])
# +
## Associative property
# create random vectors
n = 5
a = np.random.randn(n)
b = np.random.randn(n)
c = np.random.randn(n)
# the two results
res1 = np.dot( a , np.dot(b,c) )
res2 = np.dot( np.dot(a,b) , c )
# compare them
print(res1)
print(res2)
### special cases where associative property works!
# 1) one vector is the zeros vector
# 2) a==b==c
# -
#
# ---
# # VIDEO: Vector length
# ---
#
# +
# a vector
v1 = np.array([ 1, 2, 3, 4, 5, 6 ])
# methods 1-4, just like with the regular dot product, e.g.:
vl1 = np.sqrt( sum( np.multiply(v1,v1)) )
# method 5: take the norm
vl2 = np.linalg.norm(v1)
print(vl1,vl2)
# -
#
# ---
# # VIDEO: The dot product from a geometric perspective
# ---
#
# +
# two vectors
v1 = np.array([ 2, 4, -3 ])
v2 = np.array([ 0, -3, -3 ])
# compute the angle (radians) between two vectors
ang = np.arccos( np.dot(v1,v2) / (np.linalg.norm(v1)*np.linalg.norm(v2)) )
# draw them
fig = plt.figure()
ax = fig.gca(projection='3d')
ax.plot([0, v1[0]],[0, v1[1]],[0, v1[2]],'b')
ax.plot([0, v2[0]],[0, v2[1]],[0, v2[2]],'r')
plt.axis((-6, 6, -6, 6))
plt.title('Angle between vectors: %s rad.' %ang)
plt.show()
# +
## equivalence of algebraic and geometric dot product formulas
# two vectors
v1 = np.array([ 2, 4, -3 ])
v2 = np.array([ 0, -3, -3 ])
# algebraic
dp_a = np.dot( v1,v2 )
# geometric
dp_g = np.linalg.norm(v1)*np.linalg.norm(v2)*np.cos(ang)
# print dot product to command
print(dp_a)
print(dp_g)
# -
#
# ---
# # VIDEO: Vector Hadamard multiplication
# ---
#
# +
# create vectors
w1 = [ 1, 3, 5 ]
w2 = [ 3, 4, 2 ]
w3 = np.multiply(w1,w2)
print(w3)
# -
#
# ---
# # VIDEO: Vector outer product
# ---
#
# +
v1 = np.array([ 1, 2, 3 ])
v2 = np.array([ -1, 0, 1 ])
# outer product
np.outer(v1,v2)
# terrible programming, but helps conceptually:
op = np.zeros((len(v1),len(v1)))
for i in range(0,len(v1)):
for j in range(0,len(v2)):
op[i,j] = v1[i] * v2[j]
print(op)
# -
#
# ---
# # VIDEO: Vector cross product
# ---
#
# +
# create vectors
v1 = [ -3, 2, 5 ]
v2 = [ 4, -3, 0 ]
# Python's cross-product function
v3a = np.cross( v1,v2 )
# "manual" method
v3b = [ [v1[1]*v2[2] - v1[2]*v2[1]],
[v1[2]*v2[0] - v1[0]*v2[2]],
[v1[0]*v2[1] - v1[1]*v2[0]] ]
print(v3a,v3b)
fig = plt.figure()
ax = fig.gca(projection='3d')
# draw plane defined by span of v1 and v2
xx, yy = np.meshgrid(np.linspace(-10,10,10),np.linspace(-10,10,10))
z1 = (-v3a[0]*xx - v3a[1]*yy)/v3a[2]
ax.plot_surface(xx,yy,z1,alpha=.2)
## plot the two vectors
ax.plot([0, v1[0]],[0, v1[1]],[0, v1[2]],'k')
ax.plot([0, v2[0]],[0, v2[1]],[0, v2[2]],'k')
ax.plot([0, v3a[0]],[0, v3a[1]],[0, v3a[2]],'r')
ax.view_init(azim=150,elev=45)
plt.show()
# -
#
# ---
# # VIDEO: Hermitian transpose (a.k.a. conjugate transpose)
# ---
#
# +
# create a complex number
z = np.complex(3,4)
# magnitude
print( np.linalg.norm(z) )
# by transpose?
print( np.transpose(z)*z )
# by Hermitian transpose
print( np.transpose(z.conjugate())*z )
# complex vector
v = np.array( [ 3, 4j, 5+2j, np.complex(2,-5) ] )
print( v.T )
print( np.transpose(v) )
print( np.transpose(v.conjugate()) )
# -
#
# ---
# # VIDEO: Unit vector
# ---
#
# +
# vector
v1 = np.array([ -3, 6 ])
# mu
mu = 1/np.linalg.norm(v1)
v1n = v1*mu
# plot them
plt.plot([0, v1n[0]],[0, v1n[1]],'r',label='v1-norm',linewidth=5)
plt.plot([0, v1[0]],[0, v1[1]],'b',label='v1')
# axis square
plt.axis('square')
plt.axis(( -6, 6, -6, 6 ))
plt.grid()
plt.legend()
plt.show()
# -
#
# ---
# # VIDEO: Span
# ---
#
# +
# set S
S1 = np.array([1, 1, 0])
S2 = np.array([1, 7, 0])
# vectors v and w
v = np.array([1, 2, 0])
w = np.array([3, 2, 1])
# draw vectors
fig = plt.figure()
ax = fig.gca(projection='3d')
ax.plot([0, S1[0]],[0, S1[1]],[.1, S1[2]+.1],'r',linewidth=3)
ax.plot([0, S2[0]],[0, S2[1]],[.1, S2[2]+.1],'r',linewidth=3)
ax.plot([0, v[0]],[0, v[1]],[.1, v[2]+.1],'g',linewidth=3)
ax.plot([0, w[0]],[0, w[1]],[0, w[2]],'b')
# now draw plane
xx, yy = np.meshgrid(range(-15,16), range(-15,16))
cp = np.cross(S1,S2)
z1 = (-cp[0]*xx - cp[1]*yy)*1./cp[2]
ax.plot_surface(xx,yy,z1)
plt.show()
|
matlab/udemy/Vectors/linalg_vectors.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3.5
# language: python
# name: python3
# ---
# ## The Battle of the Neighborhoods - Week 1
# ### Introduction & Business Problem :
# ### Problem Background:
# The City of New York, is the most populous city in the United States. It is diverse and is the financial capital of USA. It is multicultural. It provides lot of business oppourtunities and business friendly environment. It has attracted many different players into the market. It is a global hub of business and commerce. The city is a major center for banking and finance, retailing, world trade, transportation, tourism, real estate, new media, traditional media, advertising, legal services, accountancy, insurance, theater, fashion, and the arts in the United States.
#
# This also means that the market is highly competitive. As it is highly developed city so cost of doing business is also one of the highest.
# Thus, any new business venture or expansion needs to be analysed carefully. The insights derived from analysis will give good understanding
# of the business environment which help in strategically targeting the market. This will help in reduction of risk. And the Return on Investment will be reasonable.
# ### Problem Description:
# A restaurant is a business which prepares and serves food and drink to customers in return for money, either paid before the meal, after the meal, or with an open account. The City of New York is famous for its excelllent cuisine. It's food culture includes an array of international cuisines influenced by the city's immigrant history.<Br>
# 1. Central and Eastern European immigrants, especially Jewish immigrants - bagels, cheesecake, hot dogs, knishes, and delicatessens<Br>
# 2. Italian immigrants - New York-style pizza and Italian cuisine<Br>
# 3. Jewish immigrants and Irish immigrants - pastrami and corned beef<Br>
# 4. Chinese and other Asian restaurants, sandwich joints, trattorias, diners, and coffeehouses are ubiquitous throughout the city<Br>
# 5. mobile food vendors - Some 4,000 licensed by the city<Br>
# 6. Middle Eastern foods such as falafel and kebabs examples of modern New York street food<Br>
# 7. It is famous for not just Pizzerias, Cafe's but also for fine dining Michelin starred restaurants.The city is home to "nearly one thousand of the finest and most diverse haute cuisine restaurants in the world", according to Michelin.
#
# So it is evident that to survive in such competitive market it is very important to startegically plan. Various factors need to be studied inorder to decide on the Location such as : <Br>
# 1. New York Population <Br>
# 2. New York City Demographics <Br>
# 2. Are there any Farmers Markets, Wholesale markets etc nearby so that the ingredients can be purchased fresh to maintain quality and cost?<Br>
# 3. Are there any venues like Gyms, Entertainmnet zones, Parks etc nearby where floating population is high etc <Br>
# 4. Who are the competitors in that location? <Br>
# 5. Cuisine served / Menu of the competitors <Br>
# 6. Segmentation of the Borough <Br>
# 7. Untapped markets <Br>
# 8. Saturated markets etc<Br>
# The list can go on...
#
# Eventhough well funded XYZ Company Ltd. need to choose the correct location to start its first venture.If this is successful they can replicate the same in other locations. First move is very important, thereby choice of location is very important.
# ### Target Audience:
# To recommend the correct location, XYZ Company Ltd has appointed me to lead of the Data Science team. The objective is to locate and recommend to the management which neighborhood of Newyork city will be best choice to start a restaurant. The Management also expects to understand the rationale of the recommendations made.
#
# This would interest anyone who wants to start a new restaurant in Newyork city.
# ### Success Criteria:
# The success criteria of the project will be a good recommendation of borough/Neighborhood choice to XYZ Company Ltd based on Lack of such restaurants in that location and nearest suppliers of ingredients.
|
The Battle of Neighborhoods-Week1/The Battle of Neighborhoods-Week1-Part-1.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/oumaima61/my-machine-learning-projects/blob/master/CNN.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="jN5KK75rzoc1"
# How Computers Perceive Images— Images as Data Points
# + [markdown] id="EC5aaBbkz5fp"
# Grayscale Image
# + colab={"base_uri": "https://localhost:8080/", "height": 287} id="BYR83Wp-x8Wg" outputId="2e03b376-8dd2-4d54-ec20-1d2022039adc"
from skimage import data
import numpy as np
import matplotlib.pyplot as plt
image = data.binary_blobs()
plt.imshow(image, cmap='gray')
print(f'The shape of the given image is: ',image.shape)
# + [markdown] id="VFvbLuNLz7_P"
# Colored Image
# + colab={"base_uri": "https://localhost:8080/", "height": 287} id="C1FJ6TEVzhQV" outputId="c46123e9-eaa6-43cd-ce27-96617ed05866"
color_image = data.astronaut()
plt.imshow(color_image)
# calculate shape
print(f'The shape of the given image is: ',color_image.shape)
# + [markdown] id="F8UhNwBr2-VP"
# Loading the Fashion-MNIST dataset in Keras
# + id="IcYnI-c70B4n"
from tensorflow import keras
from tensorflow.keras import layers
# + colab={"base_uri": "https://localhost:8080/"} id="Wukg6gvQ2vAB" outputId="0b5eaaa2-27bf-45b4-9482-53fb4e2ba86f"
from keras.datasets import fashion_mnist
(train_images, train_labels), (test_images, test_labels) = fashion_mnist.load_data()
np.random.seed(42)
# + id="bvUnjof323rJ"
#creating label names
label_names = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat', 'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']
# + [markdown] id="hW2J7RTC3J1M"
# data expolaration
#
# + colab={"base_uri": "https://localhost:8080/"} id="ZO8UaHu63Pen" outputId="3e3b0b30-6745-42e5-a6d3-b4666c927df9"
#Training Data
print(train_images.shape)
print(len(train_labels)) # Total no. of training images
# + colab={"base_uri": "https://localhost:8080/"} id="Px-yT8__3Voy" outputId="6287a0bb-8012-486d-c1e6-b125dd69d0a6"
#Testing Data
print(test_images.shape)
print(len(test_labels)) # Total no. of testing images
# + colab={"base_uri": "https://localhost:8080/"} id="JzFI9hRH3fWy" outputId="328f0cdd-f023-4be9-f86d-91ad679a836c"
test_labels
# + [markdown] id="rew92AV73p-f"
# Preprocessing the Data
# + colab={"base_uri": "https://localhost:8080/", "height": 265} id="_Bwk2OOP3lWy" outputId="b5d9b872-148d-4f3f-bc3f-0382fc430a2d"
plt.imshow(train_images[1],cmap='gray')
plt.grid(False)
plt.colorbar()
plt.show()
# + id="QOULKB9L31Ec"
#Rescaling the test and train images
train_images = train_images / 255.0
test_images = test_images / 255.0
# + colab={"base_uri": "https://localhost:8080/", "height": 459} id="71qPMq6u38VI" outputId="5059b5ca-8276-40c1-84a3-14bb751dac4b"
plt.figure(figsize=(8,10))
for i in range(20):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(train_images[i], cmap='gray')
plt.xlabel(label_names[train_labels[i]])
# + colab={"base_uri": "https://localhost:8080/"} id="aMvOKFx34DF4" outputId="8371b06a-f400-4d8e-b74e-6b30edabd80e"
# Reshaping the test and train images
train_images = train_images.reshape(60000, 28, 28, 1)
test_images = test_images.reshape(10000, 28, 28, 1)
print(train_images.shape)
print(test_images.shape)
# + [markdown] id="kmeivR4U4Q35"
# Building the Network architecture
#
# We will configure the layers of the model first and then proceed with compiling the model.
# Layers
#
# A layer is a core building block of a neural network. It acts as a kind of data processing module. Layers extract representations out of the input data that is fed into them. Inherently, deep learning consists of stacking up these layers to form a model. We already learned about the various layers used in a CNN in the section above.
# Model
#
# A model is a linear stack of layers. It is like a sieve for data processing made of a succession of increasing refined data filters called layers. The simplest model in Keras is sequential, which is built by stacking layers sequentially.
# + id="LEl8QfZY4Kl3"
model = keras.Sequential([
keras.layers.Conv2D(32, (3,3), padding='same', activation='relu',
input_shape=(28, 28, 1)),
keras.layers.MaxPooling2D((2, 2), strides=2),
#Add another convolution
keras.layers.Conv2D(64, (3,3), padding='same', activation='relu'),
keras.layers.MaxPooling2D((2, 2), strides=2),
#Flatten the output.
keras.layers.Flatten(),
keras.layers.Dense(128, activation='relu'),
keras.layers.Dense(10, activation='softmax')
])
# + colab={"base_uri": "https://localhost:8080/"} id="p_4ZGXCS5Tlg" outputId="c3845223-4471-4630-e302-1dcdc0865bf9"
model.summary()
# + [markdown] id="TTsaDcOv5s3s"
# Let's now look at how we defined the model architecture in detail
# Convolution Layer
#
# We started with a convolutional layer, specifying the number of convolutions that we want to generate. Here we have chosen '32'.
# We have also specified the size of the convolutional matrix, in this case a 3X3 grid. We have also used padding to retain the size of the original image.
# We have used relu (rectified linear unit) as the activation function. A rectified linear unit has an output of 0 if the input is less than 0, and raw output otherwise. That is, if the input is greater than 0, the output is equal to the input. An activation function is the non-linear transformation that we do over the input signal. This transformed output is then sent to the next layer of neurons as input.
# Finally, we enter the shape of the input data.
#
# Pooling Layer
#
# Every convolution layer is then followed by a max-pooling layer. A max-pooling layer will downsample an image but will retain the features.
# Flattened Layer
#
# Finally, we will flatten the images into a one-dimensional vector.
# Dense
#
# This layer consists of a 128-neuron, followed by a 10-node softmax layer. Each node represents a class of clothing. The final layer takes input from the 128 nodes in the layer before it, and outputs a value in the range [0, 1], representing the probability that the image belongs to that class. The sum of all 10 node values is 1. We'll also include activation functions in the network to introduce non-linearity. Here we have used ReLU. The last layer is a 10-way softmax layer which will return an array of 10 probability scores. Each score will denote the probability that the current image belongs to one of the 10 given classes.
#
# + [markdown] id="zFmvFe6D53Ts"
# Compile the Model
#
# After the model has been built, we enter the compilation phase, which primarily consists of three essential elements:
#
# Loss Function: loss (Predicted — Actual value) is the quantity that we try to minimize during the training of a neural network.
# Optimizer: determines how the network will be updated based on the loss function. Optimizers could be the RMSProp optimizer, SGD with momentum, and so on.
# Metrics: to measure the accuracy of the model. In this case, we will use accuracy.
#
# + id="9M64G80z5yBO"
model.compile(optimizer='adam',
loss=keras.losses.SparseCategoricalCrossentropy(),
metrics=['accuracy'])
# + colab={"base_uri": "https://localhost:8080/"} id="O8frZw0B6Dve" outputId="cefb6d54-bca5-49c9-c874-689ae88003a8"
model.fit(train_images, train_labels, epochs=3,batch_size=32)
# + [markdown] id="njq0UO_67Qly"
# Model Evaluation
# + colab={"base_uri": "https://localhost:8080/"} id="WUiRRCRL7CJp" outputId="8881f604-5b06-40a1-d379-a3893515c529"
test_loss, test_accuracy = model.evaluate(test_images, test_labels)
print('Accuracy on test dataset:', test_accuracy)
# + id="adI4QM7j7cCT"
predictions = model.predict(test_images)
# + colab={"base_uri": "https://localhost:8080/"} id="6J3nhMqH7npQ" outputId="9a92d998-a5f3-4bac-dd8a-c54359a12eae"
predictions[10]
# + colab={"base_uri": "https://localhost:8080/"} id="uk8oLuDB7se_" outputId="fceb9c5d-7fee-4739-ccbd-7d15a4b65370"
np.argmax(predictions[10])
# + colab={"base_uri": "https://localhost:8080/"} id="qjCVpE8P7yCk" outputId="f4d89a92-4df8-41aa-ddde-a3041e622d59"
test_labels[10]
|
CNN.ipynb
|
# ---
# jupyter:
# jupytext:
# formats: ipynb,py:percent
# text_representation:
# extension: .py
# format_name: percent
# format_version: '1.3'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %%
# %matplotlib inline
# %load_ext autoreload
# %autoreload 2
import networkx as nx
import matplotlib.pyplot as plt
import time
# modules specific to this project
import network as nw
import physics
import timemarching as tm
import plotter
import logger
# %% [markdown]
# ### 1. Define the broadcasting channels of the network
# This is done by creating a list of the channel names. The names are arbitrary and can be set by the user, such as 'postive', 'negative' or explicit wavelenghts like '870 nm', '700 nm'. Here I chose the colors 'red' and 'blue', as well as bias with same wavelength as 'blue'.
# %%
channel_list = ['red', 'blue','green']
# Automatically generate the object that handles them
channels = {channel_list[v] : v for v in range(len(channel_list))}
# %% [markdown]
# ### 2. Define the layers
# Define the layers of nodes in terms of how they are connected to the channels. Layers and weights are organized in dictionaries. The input and output layers do not need to be changed, but for the hidden layer we need to specify the number of nodes N and assign the correct channels to the input/output of the node.
# %%
# Create layers ordered from 0 to P organized in a dictionary
layers = {}
Nreservoir = 100
# An input layer automatically creates on node for each channel that we define
layers[0] = nw.InputLayer(input_channels=channels)
# Forward signal layer
layers[1] = nw.HiddenLayer(Nreservoir//2, output_channel='blue',excitation_channel='blue',inhibition_channel='red')
# Inhibiting memory layer
layers[2] = nw.HiddenLayer(Nreservoir//2, output_channel='red' ,excitation_channel='blue',inhibition_channel='red')
layers[3] = nw.OutputLayer(output_channels=channels) # similar to input layer
# %% [markdown]
# ### 3. Define existing connections between layers
# The weights are set in two steps.
# First the connetions between layers are defined. This should be done using the keys defined for each layer above, i.e. 0, 1, 2 ... for input, hidden and output layers, respectively. The `connect_layers` function returns a weight matrix object that we store under a chosen key, for example `'inp->hid'`.
# Second, the specific connections on the node-to-node level are specified using the node index in each layer
# %%
# Define the overall connectivity
weights = {}
# The syntax is connect_layers(from_layer, to_layer, layers, channels)
# Connections into the reservoir from input layer
weights['inp->hd0'] = nw.connect_layers(0, 1, layers, channels)
weights['inp->hd1'] = nw.connect_layers(0, 2, layers, channels)
# Connections between reservoir nodes
weights['hd0->hd1'] = nw.connect_layers(1, 2, layers, channels)
weights['hd1->hd0'] = nw.connect_layers(2, 1, layers, channels)
# Intralayer connections
weights['hd0->hd0'] = nw.connect_layers(1, 1, layers, channels)
weights['hd1->hd1'] = nw.connect_layers(2, 2, layers, channels)
# Connections to output
weights['hd0->out'] = nw.connect_layers(1, 3, layers, channels)
weights['hd1->out'] = nw.connect_layers(2, 3, layers, channels)
# Connections back into reservoir from output
weights['out->hd0'] = nw.connect_layers(3, 1, layers, channels)
weights['out->hd1'] = nw.connect_layers(3, 2, layers, channels)
# %% [markdown]
# Setup parameters for the network
# %%
sparsity = 0.90
spectral_radius = 1.0
# One number per channel
input_scaling = [1.0,1.0,1.0]
output_scaling= [1.0,1.0,1.0]
Nreservoir = 100
# %% [markdown]
# #### Setup the input weights
# %%
# We will generate some random numbers
import numpy as np
rng = np.random.RandomState(42)
# Input weights to all of the input units
W_in = rng.rand(Nreservoir, len(channels))
# Ask the weights object which dimensions to be in
weights['inp->hd0'].ask_W()
# Put each weight column in a specific channel
for key in channels :
k = channels[key]
W_key = np.zeros_like(W_in)
W_key[:,k] = W_in[:,k]
weights['inp->hd0'].set_W(key,input_scaling[k]*W_key[:Nreservoir//2]) # first half
weights['inp->hd1'].set_W(key,input_scaling[k]*W_key[Nreservoir//2:]) # second half
# %% [markdown]
# #### Setup the reservoir weights
# %%
W_partition = {'hd0->hd0':(0,Nreservoir//2,0,Nreservoir//2),
'hd0->hd1':(Nreservoir//2,Nreservoir,0,Nreservoir//2),
'hd1->hd1':(Nreservoir//2,Nreservoir,Nreservoir//2,Nreservoir),
'hd1->hd0':(0,Nreservoir//2,Nreservoir//2,Nreservoir)}
# Generate a large matrix of values for each reservoir channel (red and blue)
W_res = rng.rand(2, Nreservoir, Nreservoir)
# Delete the fraction of connections given by sparsity:
W_res[rng.rand(*W_res.shape) < sparsity] = 0
# Delete any remaining diagonal elements
for k in range(0,Nreservoir) :
W_res[:,k,k] = 0.
# Normalize this to have the chosen spectral radius, once per channel
for k in range(0,2) :
radius = np.max(np.abs(np.linalg.eigvals(W_res[k])))
# rescale them to reach the requested spectral radius:
W_res[k] = W_res[k] * (spectral_radius / radius)
weights['hd0->hd1'].ask_W()
for connection in W_partition :
for key in list(channels.keys())[:2] :
k=channels[key]
A,B,C,D = W_partition[connection]
weights[connection].set_W(key,W_res[k,A:B,C:D])
# %% [markdown]
# #### Setup the output weights
# %%
# Output weights from reservoir to the output units
W_out = rng.rand(len(channels),Nreservoir)
# Ask the weights object which dimensions to be in
weights['hd0->out'].ask_W()
# Put each weight column in a specific channel
for key in channels :
if key != 'green' :
k = channels[key]
W_key = np.zeros_like(W_out)
W_key[k] = W_out[k]
weights['hd0->out'].set_W(key,output_scaling[k]*W_key[:,:Nreservoir//2])
weights['hd1->out'].set_W(key,output_scaling[k]*W_key[:,Nreservoir//2:])
# Output weights back into reservoir
# %% [markdown]
# ### 4. Visualize the network
# The `plotter` module supplies functions to visualize the network structure. The nodes are named by the layer type (Input, Hidden or Output) and the index. To supress the printing of weight values on each connection, please supply `show_edge_labels=False`.
#
# #### Available layouts:
# **multipartite**: Standard neural network appearance. Hard to see recurrent couplings within layers.
# **circular**: Nodes drawn as a circle
# **shell**: Layers drawn as concetric circles
# **kamada_kawai**: Optimization to minimize weighted internode distance in graph
# **spring**: Spring layout which is standard in `networkx`
#
# #### Shell layout
# This is my current favorite. It is configured to plot the input and output nodes on the outside of the hidden layer circle, in a combined outer concentric circle.
# %%
plotter.visualize_network(layers, weights, exclude_nodes={3:['O2']},node_size=100,layout='shell', show_edge_labels=False)
# %% [markdown]
# ### 5. Specify the physics of the nodes
# Before running any simulations, we need to specify the input currents and the physics of the hidden layer nodes. Parameters can either be specified directly or coupled from the `physics` module.
# %%
# Specify an exciting current square pulse and a constant inhibition
# Pulse train of 1 ns pulses
t_blue = [(6.0,7.0), (11.0,12.0), (16.0,17.0)] # at 6 ns, 11 ns, and 16 ns
t_blue = [(5.0,15.0)]#, (11.0,12.0), (16.0,17.0)] # at 6 ns, 11 ns, and 16 ns
I_blue = 100 # nA
# Try to modulate the nodes with red input
t_red = [(8.0,9.0), (12.0,13.0)] # at 6 ns, 11 ns, and 16 ns
# Constant inhibition to stabilize circuit
I_red = 0.0 # nA
# Use the square pulse function and specify which node in the input layer gets which pulse
layers[0].set_input_func(channel='blue',func_handle=physics.square_pulse, func_args=(t_blue, I_blue))
# Use the costant function to specify the inhibition from I0 to H0
#layers[0].set_input_func(channel='red', func_handle=physics.constant, func_args=I_red)
layers[0].set_input_func(channel='red', func_handle=physics.square_pulse, func_args=(t_red, I_red))
# %%
# Specify two types of devices for the hidden layer
# 1. Propagator (standard parameters)
propagator = physics.Device('device_parameters.txt')
propagator.print_parameter('Cstore')
#propagator.set_parameter('Rstore',1e6)
# 2. Memory (modify the parameters)
memory = physics.Device('device_parameters.txt')
#memory.set_parameter('Rstore',1e6)
#memory.set_parameter('Cstore',2e-15)
# a 3e-15 F capacitor can be build by 800x900 plates 20 nm apart
memory.print_parameter('Cstore')
# %%
# Specify the internal dynamics by supplying the RC constants to the hidden layer (six parameters)
layers[1].assign_device(propagator)
layers[2].assign_device(memory)
# Tweak the threshold voltage
Vthres=0.27
layers[1].Vthres=Vthres
layers[2].Vthres=Vthres
# Calculate the unity_coeff to scale the weights accordingly
unity_coeff, _ = propagator.inverse_gain_coefficient(propagator.eta_ABC, Vthres)
print(f'Unity coupling coefficient calculated as unity_coeff={unity_coeff:.4f}')
# %% [markdown]
# ### 6. Evolve in time
# %%
# Start time t, end time T
t = 0.0
T = 25.0 # ns
# To sample result over a fixed time-step, use savetime
savestep = 0.1
savetime = savestep
# These parameters are used to determine an appropriate time step each update
dtmax = 0.1 # ns
dVmax = 0.005 # V
nw.reset(layers)
# Create a log over the dynamic data
time_log = logger.Logger(layers,channels) # might need some flags
start = time.time()
while t < T:
# evolve by calculating derivatives, provides dt
dt = tm.evolve(t, layers, dVmax, dtmax )
# update with explicit Euler using dt
# supplying the unity_coeff here to scale the weights
tm.update(dt, t, layers, weights, unity_coeff)
t += dt
# Log the progress
if t > savetime :
# Put log update here to have (more or less) fixed sample rate
# Now this is only to check progress
print(f'Time at t={t} ns')
savetime += savestep
time_log.add_tstep(t, layers, unity_coeff)
end = time.time()
print('Time used:',end-start)
# This is a large pandas data frame of all system variables
result = time_log.get_timelog()
# %% [markdown]
# ### 7. Visualize results
# Plot results specific to certain nodes
# %%
#nodes = ['H0','H1','H2','H3','H4']
nodes = ['H0','K0']
plotter.plot_nodes(result, nodes)
# %% [markdown]
# For this system it's quite elegant to use the `plot_chainlist` function, taking as arguments a graph object, the source node (I1 for blue) and a target node (O1 for blue)
# %%
# Variable G contains a graph object descibing the network
G = plotter.retrieve_G(layers, weights)
plotter.plot_chainlist(result,G,'I1','K0')
# %% [markdown]
# Plot specific attributes
# %%
attr_list = ['Vgate','Vexc']
plotter.plot_attributes(result, attr_list)
# %% [markdown]
# We can be totally specific if we want. First we list the available columns to choose from
# %%
print(result.columns)
# %%
plotter.visualize_dynamic_result(result, ['I0-Iout-red','I1-Iout-blue'])
# %%
plotter.visualize_dynamic_result(result, ['H0-Iout','H0-Pout','K0-Iout','K0-Pout'])
# %%
plotter.visualize_transistor(propagator.transistorIV,propagator.transistorIV_example())
# %%
plotter.visualize_LED_efficiency(propagator.eta_example(propagator.eta_ABC))
# %%
|
echostatenetwork/ReservoirNetwork.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Implementation
# This section shows how the linear regression extensions discussed in this chapter are typically fit in Python. First let's import the {doc}`Boston housing</content/appendix/data>` dataset.
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import datasets
boston = datasets.load_boston()
X_train = boston['data']
y_train = boston['target']
# ## Regularized Regression
# Both Ridge and Lasso regression can be easily fit using `scikit-learn`. A bare-bones implementation is provided below. Note that the regularization parameter `alpha` (which we called $\lambda$) is chosen arbitrarily.
# +
from sklearn.linear_model import Ridge, Lasso
alpha = 1
# Ridge
ridge_model = Ridge(alpha = alpha)
ridge_model.fit(X_train, y_train)
# Lasso
lasso_model = Lasso(alpha = alpha)
lasso_model.fit(X_train, y_train);
# -
# In practice, however, we want to choose `alpha` through cross validation. This is easily implemented in `scikit-learn` by designating a set of `alpha` values to try and fitting the model with `RidgeCV` or `LassoCV`.
# +
from sklearn.linear_model import RidgeCV, LassoCV
alphas = [0.01, 1, 100]
# Ridge
ridgeCV_model = RidgeCV(alphas = alphas)
ridgeCV_model.fit(X_train, y_train)
# Lasso
lassoCV_model = LassoCV(alphas = alphas)
lassoCV_model.fit(X_train, y_train);
# -
# We can then see which values of `alpha` performed best with the following.
print('Ridge alpha:', ridgeCV.alpha_)
print('Lasso alpha:', lassoCV.alpha_)
# ## Bayesian Regression
# We can also fit Bayesian regression using `scikit-learn` (though another popular package is `pymc3`). A very straightforward implementation is provided below.
from sklearn.linear_model import BayesianRidge
bayes_model = BayesianRidge()
bayes_model.fit(X_train, y_train);
# This is not, however, identical to our construction in the previous section since it infers the $\sigma^2$ and $\tau$ parameters, rather than taking those as fixed inputs. More information can be found [here](https://scikit-learn.org/stable/modules/linear_model.html#bayesian-regression). The hidden chunk below demonstrates a hacky solution for running Bayesian regression in `scikit-learn` using known values for $\sigma^2$ and $\tau$, though it is hard to imagine a practical reason to do so
# ````{toggle}
# By default, Bayesian regression in `scikit-learn` treats $\alpha = \frac{1}{\sigma^2}$ and $\lambda = \frac{1}{\tau}$ as random variables and assigns them the following prior distributions
#
# $$
# \begin{aligned}
# \alpha &\sim \text{Gamma}(\alpha_1, \alpha_2)
# \\
# \lambda &\sim \text{Gamma}(\lambda_1, \lambda_2).
# \end{aligned}
# $$
#
# Note that $E(\alpha) = \frac{\alpha_1}{\alpha_2}$ and $E(\lambda) = \frac{\lambda_1}{\lambda_2}$. To *fix* $\sigma^2$ and $\tau$, we can provide an extremely strong prior on $\alpha$ and $\lambda$, guaranteeing that their estimates will be approximately equal to their expected value.
#
# Suppose we want to use $\sigma^2 = 11.8$ and $\tau = 10$, or equivalently $\alpha = \frac{1}{11.8}$, $\lambda = \frac{1}{10}$. Then let
#
# $$
# \begin{aligned}
# \alpha_1 &= 10000 \cdot \frac{1}{11.8}, \\
# \alpha_2 &= 10000, \\
# \lambda_1 &= 10000 \cdot \frac{1}{10}, \\
# \lambda_2 &= 10000.
# \end{aligned}
# $$
#
# This guarantees that $\sigma^2$ and $\tau$ will be approximately equal to their pre-determined values. This can be implemented in `scikit-learn` as follows
#
# ```{code}
# big_number = 10**5
#
# # alpha
# alpha = 1/11.8
# alpha_1 = big_number*alpha
# alpha_2 = big_number
#
# # lambda
# lam = 1/10
# lambda_1 = big_number*lam
# lambda_2 = big_number
#
# # fit
# bayes_model = BayesianRidge(alpha_1 = alpha_1, alpha_2 = alpha_2, alpha_init = alpha,
# lambda_1 = lambda_1, lambda_2 = lambda_2, lambda_init = lam)
# bayes_model.fit(X_train, y_train);
# ```
#
# ````
# ## Poisson Regression
# GLMs are most commonly fit in Python through the `GLM` class from `statsmodels`. A simple Poisson regression example is given below.
#
# As we saw in the GLM concept section, a GLM is comprised of a random distribution and a link function. We identify the random distribution through the `family` argument to `GLM` (e.g. below, we specify the `Poisson` family). The default link function depends on the random distribution. By default, the Poisson model uses the link function
#
# $$
# \eta_n = g(\mu_n) = \log(\lambda_n),
# $$
#
# which is what we use below. For more information on the possible distributions and link functions, check out the `statsmodels` GLM [docs](https://www.statsmodels.org/stable/glm.html).
# +
import statsmodels.api as sm
X_train_with_constant = sm.add_constant(X_train)
poisson_model = sm.GLM(y_train, X_train, family=sm.families.Poisson())
poisson_model.fit();
|
content/c2/code.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
### ESOL predictor: GCNN, random train/validate/test splits, representation = ConvMol object (from DeepChem)
# -
###load data from CSV in same folder as notebook
from deepchem.utils.save import load_from_disk
dataset_file= "./esol.csv"
dataset = load_from_disk(dataset_file)
print("Columns of dataset: %s" % str(dataset.columns.values))
print("Number of examples in dataset: %s" % str(dataset.shape[0]))
# +
###plot histogram of data to show distribution
# %matplotlib inline
import matplotlib
import numpy as np
import matplotlib.pyplot as plt
solubilities = np.array(dataset["measured log solubility in mols per litre"])
n, bins, patches = plt.hist(solubilities, 50, facecolor='green', alpha=0.75)
plt.xlabel('Measured log-solubility in mols/liter')
plt.ylabel('Number of compounds')
plt.title(r'Histogram of solubilities')
plt.grid(True)
plt.show()
# -
###featurize the data using extended connectivity fingerprints
import deepchem as dc
#featurizer = dc.feat.CircularFingerprint(size=1024)
#featurizer = dc.feat.graph_features.WeaveFeaturizer()
featurizer = dc.feat.ConvMolFeaturizer()
loader = dc.data.CSVLoader(
tasks=["measured log solubility in mols per litre"], smiles_field="smiles",
featurizer=featurizer)
dataset = loader.featurize(dataset_file)
###randomly split data into train, validation, and test sets
splitter = dc.splits.RandomSplitter(dataset_file)
train_dataset, valid_dataset, test_dataset = splitter.train_valid_test_split(dataset, seed=0)
# +
###normalize all datasets
transformers = [dc.trans.NormalizationTransformer(transform_y=True, dataset=train_dataset)]
for dataset in [train_dataset, valid_dataset, test_dataset]:
for transformer in transformers:
dataset = transformer.transform(dataset)
# +
###fit the model to the data
#model = dc.models.MPNNModel(n_tasks=1)
model = dc.models.GraphConvModel(n_tasks=1, mode='regression', batch_size=50, random_seed=0, model_dir="./models/esol")
model.fit(train_dataset, nb_epoch=10, deterministic=True)
# -
dir(model)
model.tensorboard_log_frequency
# +
###evaluate the model's performance on train set
from deepchem.utils.evaluate import Evaluator
metric = dc.metrics.Metric(dc.metrics.r2_score)
evaluator = Evaluator(model, train_dataset, transformers)
r2score = evaluator.compute_model_performance([metric])
print(r2score)
### plot of train vs predicted train
predicted_train = model.predict(train_dataset)
true_train = train_dataset.y
plt.scatter(predicted_train, true_train)
plt.xlabel('Predicted esol')
plt.ylabel('Actual esol')
plt.title(r'Predicted esol vs. Actual esol of train set')
plt.xlim([-12,2])
plt.ylim([-12,2])
plt.plot([-12,2], [-12,2], color='k')
plt.show()
# +
###evaluate the model's performance on validation set
from deepchem.utils.evaluate import Evaluator
metric = dc.metrics.Metric(dc.metrics.r2_score)
evaluator = Evaluator(model, valid_dataset, transformers)
r2score = evaluator.compute_model_performance([metric])
print(r2score)
### plot of train vs predicted validation
predicted_valid = model.predict(valid_dataset)
true_valid = valid_dataset.y
plt.scatter(predicted_valid, true_valid)
plt.xlabel('Predicted esol')
plt.ylabel('Actual esol')
plt.title(r'Predicted esol vs. Actual esol of validation set')
plt.xlim([-12,2])
plt.ylim([-12,2])
plt.plot([-12,2], [-12,2], color='k')
plt.show()
# +
###evaluate the model's performance on test set
from deepchem.utils.evaluate import Evaluator
metric = dc.metrics.Metric(dc.metrics.r2_score)
evaluator = Evaluator(model, test_dataset, transformers)
r2score = evaluator.compute_model_performance([metric])
print(r2score)
### plot of train vs predicted train
predicted_test = model.predict(test_dataset)
true_test = test_dataset.y
plt.scatter(predicted_test, true_test)
plt.xlabel('Predicted esol')
plt.ylabel('Actual esol')
plt.title(r'Predicted esol vs. Actual esol of test set')
plt.xlim([-12,2])
plt.ylim([-12,2])
plt.plot([-12,2], [-12,2], color='k')
plt.show()
# -
model.model_dir = "./"
model.save()
|
models/GroundTruthEsolModel.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # An example notebook
# A [Jupyter notebooks](http://jupyter.org/) mixes blocks of explanatory text, like the one you're reading now, with cells containing Python code (_inputs_) and the results of executing it (_outputs_). The code and its output—if any—are marked by `In [N]` and `Out [N]`, respectively, with `N` being the index of the cell. You can see an example in the computations below:
def f(x, y):
return x + 2*y
a = 4
b = 2
f(a, b)
# By default, Jupyter displays the result of the last instruction as the output of a cell, like it did above; however, `print` statements can display further results.
print(a)
print(b)
print(f(b, a))
# Jupyter also knows a few specific data types, such as Pandas data frames, and displays them in a more readable way:
import pandas as pd
pd.DataFrame({ 'foo': [1,2,3], 'bar': ['a','b','c'] })
# The index of the cells shows the order of their execution. Jupyter doesn't constrain it; to avoid confusing people, though, you better write your notebooks so that the cells are executed in sequential order as displayed. All cells are executed in the global Python scope; this means that, as we execute the code, all variables, functions and classes defined in a cell are available to the ones that follow.
# Notebooks can also include plots, as in the following cell:
# %matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
f = plt.figure(figsize=(10,2))
ax = f.add_subplot(1,1,1)
ax.plot([0, 0.25, 0.5, 0.75, 1.0], np.random.random(5))
# As you might have noted, the cell above also printed a textual representation of the object returned from the plot, since it's the result of the last instruction in the cell. To prevent this, you can add a semicolon at the end, as in the next cell.
f = plt.figure(figsize=(10,2))
ax = f.add_subplot(1,1,1)
ax.plot([0, 0.25, 0.5, 0.75, 1.0], np.random.random(5));
|
example.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:python-public-policy] *
# language: python
# name: conda-env-python-public-policy-py
# ---
# # Homework 3: Data visualization
#
# 1. Complete the **Coding** exercise below.
# 1. **Tutorial:** Go the first third of [Time Series Analysis with Pandas](https://www.dataquest.io/blog/tutorial-time-series-analysis-with-pandas/), up until the "Visualizing time series data" section.
#
# ## In-class exercise 1
# ### Step 1
#
# Load the request per capita dataset from https://storage.googleapis.com/python-public-policy/data/311_community_districts.csv.zip as `requests_by_cd` and display it.
# +
# your code here
# -
# ### Step 2
#
# Make a [histogram](https://plotly.com/python/histograms/) of the requests per capita.
# +
# your code here
# -
# ## In-class exercise 2
#
# Take the scatterplot example from [the lecture](https://padmgp-4506001-fall.rcnyu.org/user-redirect/notebooks/class_materials/lecture_3.ipynb) and [add a trendline](https://plotly.com/python/linear-fits/).
# +
# your code here
# -
# ## Coding
#
# We are going to look at the population count of different community districts over time.
# +
import plotly.express as px
# boilerplate for allowing PDF export
import plotly.io as pio
pio.renderers.default = "notebook_connected+pdf"
# -
# ### Step 1
#
# Read the data from the [New York City Population By Community Districts](https://data.cityofnewyork.us/City-Government/New-York-City-Population-By-Community-Districts/xi7c-iiu2/data) data set into a DataFrame called `pop_by_cd`. To get the URL:
#
# 1. Visit the page linked above.
# 1. Click `Export`.
# 1. Right-click `CSV`.
# 1. Click `Copy Link Address` (or `Location`, depending on your browser).
# +
# your code here
# -
# ### Step 2
#
# Prepare the data. Use the following code to [reshape](https://pandas.pydata.org/pandas-docs/stable/user_guide/reshaping.html) the DataFrame to have one row per community district per Census year.
# +
# turn the population columns into rows
populations = pd.melt(pop_by_cd, id_vars=['Borough', 'CD Number', 'CD Name'], var_name='year', value_name='population')
# turn the years into numbers
populations.year = populations.year.str.replace(' Population', '').astype(int)
populations
# -
# ### Step 3
#
# Create a line chart of the population over time for each community district in Manhattan. There should be one line for each.
#
# See the Plotly [Line Plot with column encoding color](https://plotly.com/python/line-charts/#line-plot-with-column-encoding-color) examples.
# +
# your code here
# -
# ### Step 4
#
# We are going to do some mapping using the `pop_by_cd` DataFrame from before. To do so, we need `borocd`s. Create that column with the values filled in. (See [Lecture 2](https://padmgp-4506001-fall.rcnyu.org/user-redirect/notebooks/class_materials/lecture_2.ipynb).)
# +
# your code here
# -
# ### Step 5
#
# Let make a [choropleth map](https://www.data-to-viz.com/graph/choropleth.html) showing the population change from 2000 to 2010 for each community district. Adapt the `.choropleth_mapbox()` example in [Lecture 3](https://padmgp-4506001-fall.rcnyu.org/user-redirect/notebooks/class_materials/lecture_3.ipynb).
#
# If you get an error about `choropleth_mapbox() got an unexpected keyword argument 'featureidkey'`, go back and do the `Setup` above.
# +
# your code here
# -
# ### Step 6
#
# ***Analysis: Washington Heights and Inwood (the tall skinny community district at the top of Manhattan) are "up and coming" neighborhoods. In a few sentences: Why might might the population have decreased?***
# YOUR ANSWER HERE
# Then, read the first three paragraphs of the `Demographics` section of [An Economic Snapshot of Washington Heights and Inwood from June 2015](https://www.osc.state.ny.us/osdc/rpt2-2016.pdf#page=2).
# ## Tutorials
#
# 1. Read [how to handle time series data in pandas](https://pandas.pydata.org/pandas-docs/stable/getting_started/intro_tutorials/09_timeseries.html)
# 1. Read the [Data Design Standards](https://xdgov.github.io/data-design-standards/)
# 1. Watch [this talk on audification/sonification](https://www.youtube.com/watch?v=55dIfA7C038). We won't be doing so in this class, but hopefully will provide some inspiration about different ways that data can be represented.
|
hw_3.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (Data Science)
# language: python
# name: python3__SAGEMAKER_INTERNAL__arn:aws:sagemaker:ap-northeast-2:806072073708:image/datascience-1.0
# ---
# # Part 5 : Create an End to End Pipeline
# <a id='overview-5'></a>
#
# ## [Overview](./0-AutoClaimFraudDetection.ipynb)
# * [Notebook 0 : Overview, Architecture and Data Exploration](./0-AutoClaimFraudDetection.ipynb)
# * [Notebook 1: Data Prep, Process, Store Features](./1-data-prep-e2e.ipynb)
# * [Notebook 2: Train, Check Bias, Tune, Record Lineage, and Register a Model](./2-lineage-train-assess-bias-tune-registry-e2e.ipynb)
# * [Notebook 3: Mitigate Bias, Train New Model, Store in Registry](./3-mitigate-bias-train-model2-registry-e2e.ipynb)
# * [Notebook 4: Deploy Model, Run Predictions](./4-deploy-run-inference-e2e.ipynb)
# * **[Notebook 5 : Create and Run an End-to-End Pipeline to Deploy the Model](./5-pipeline-e2e.ipynb)**
# * **[Architecture](#arch-5)**
# * **[Create an Automated Pipeline](#pipelines)**
# * **[Clean up](#cleanup)**
# 이 노트북에서는 전체 end-to-end 프로세스를 자동화하는 SageMaker Pipeline을 구축합니다. 처음에는 데이터 과학자의 입장에서 모든 단계를 수동으로 수행했습니다.
#
# 이제 ML 엔지니어 또는 MLOps 담당자의 입장에서 각 단계를 자동화하는 법을 살펴보겠습니다.
# ### Load stored variables
#
# 이전에 이 노트북을 실행한 경우, AWS에서 생성한 리소스를 재사용할 수 있습니다. 아래 셀을 실행하여 이전에 생성된 변수를 로드합니다. 기존 변수의 출력물이 표시되어야 합니다. 인쇄된 내용이 보이지 않으면 노트북을 처음 실행한 것일 수 있습니다.
# %store -r
# #%store
# **<font color='red'>Important</font>: StoreMagic 명령을 사용하여 변수를 검색하려면 이전 노트북을 실행해야 합니다.**
# ### Import libraries
# +
import json
import boto3
import pathlib
import sagemaker
import numpy as np
import pandas as pd
import awswrangler as wr
import demo_helpers
from sagemaker.xgboost.estimator import XGBoost
from sagemaker.workflow.pipeline import Pipeline
from sagemaker.workflow.steps import CreateModelStep
from sagemaker.sklearn.processing import SKLearnProcessor
from sagemaker.workflow.step_collections import RegisterModel
from sagemaker.workflow.steps import ProcessingStep, TrainingStep
from sagemaker.workflow.parameters import ParameterInteger, ParameterFloat, ParameterString
# -
# ### Set region and boto3 config
# +
# You can change this to a region of your choice
import sagemaker
region = sagemaker.Session().boto_region_name
print("Using AWS Region: {}".format(region))
boto3.setup_default_session(region_name=region)
boto_session = boto3.Session(region_name=region)
s3_client = boto3.client("s3", region_name=region)
sagemaker_boto_client = boto_session.client("sagemaker")
sagemaker_session = sagemaker.session.Session(
boto_session=boto_session, sagemaker_client=sagemaker_boto_client
)
sagemaker_role = sagemaker.get_execution_role()
account_id = boto3.client("sts").get_caller_identity()["Account"]
# +
# ======> Tons of output_paths
training_job_output_path = f"s3://{bucket}/{prefix}/training_jobs"
bias_report_output_path = f"s3://{bucket}/{prefix}/clarify-bias"
explainability_output_path = f"s3://{bucket}/{prefix}/clarify-explainability"
train_data_uri = f"s3://{bucket}/{prefix}/data/train/train.csv"
test_data_uri = f"s3://{bucket}/{prefix}/data/test/test.csv"
train_data_upsampled_s3_path = f"s3://{bucket}/{prefix}/data/train/upsampled/train.csv"
processing_dir = "/opt/ml/processing"
create_dataset_script_uri = f"s3://{bucket}/{prefix}/code/create_dataset.py"
pipeline_bias_output_path = f"s3://{bucket}/{prefix}/clarify-output/pipeline/bias"
deploy_model_script_uri = f"s3://{bucket}/{prefix}/code/deploy_model.py"
# ======> variables used for parameterizing the notebook run
flow_instance_count = 1
flow_instance_type = "ml.m5.4xlarge"
train_instance_count = 1
train_instance_type = "ml.m4.xlarge"
deploy_model_instance_type = "ml.m4.xlarge"
# -
# <a id ='arch-5'> </a>
# ### Architecture : Create a SageMaker Pipeline to Automate All the Steps from Data Prep to Model Deployment
# [overview](#overview-5)
#
# 
# <a id='pipelines'></a>
#
# ## SageMaker Pipeline
#
# - [Step 1: Claims Data Wrangler Preprocessing Step](#claims-data-wrangler)
# - [Step 2: Customers Data Wrangler Preprocessing step](#data-wrangler)
# - [Step 3: Dataset and train test split](#dataset-train-test)
# - [Step 4: Train XGboost Model](#pipe-train-xgb)
# - [Step 5: Model Pre-deployment](#pipe-pre-deploy)
# - [Step 6: Use Clarify to Detect Bias](#pipe-detect-bias)
# - [Step 7: Register Model](#pipe-Register-Model)
# - [Step 8: Combine the Pipeline Steps and Run](#define-pipeline)
#
#
# [back to overview](#overview-5)
#
#
#
# ___
# 이제 머신 러닝 워크플로의 각 단계를 수동으로 수행했으므로, 투명성과 모델 추적을 희생하지 않고 더 빠른 모델 실험을 허용하는 특정 단계를 수행할 수 있습니다. 이 섹션에서는 새 모델을 훈련하고 SageMaker에서 모델을 유지한 다음 모델을 레지스트리에 추가하는 파이프라인을 생성합니다.
# ### Pipeline parameters
#
# SageMaker Pipelines의 중요한 기능은 단계를 미리 정의할 수 있지만, 파이프라인을 다시 정의하지 않고도 실행시 해당 단계로 매개 변수를 변경할 수 있다는 것입니다. 이는 ParameterInteger, ParameterFloat 또는 ParameterString을 사용하여 나중에 `pipeline.start (parameters=parameters)`를 호출할 때, 수정할 수 있는 값을 사전에 정의함으로써 달성할 수 있습니다. 이러한 방식으로 특정 파라메터만 정의할 수 있습니다.
# +
train_instance_param = ParameterString(
name="TrainingInstance",
default_value="ml.m4.xlarge",
)
model_approval_status = ParameterString(
name="ModelApprovalStatus", default_value="PendingManualApproval"
)
# -
# ### Define Caching
#
# 캐싱을 사용하여 동일한 파이프라인을 재실행 시, 각 단계의 값을 다시 계산하는 대신 적중된 캐시값을 다음 단계로 전파합니다. 캐싱은 성공한 실행만 고려하며, 여러 개의 실행이 있는 경우 가장 최근에 성공한 실행에 대한 결과를 사용합니다.
# +
# from sagemaker.workflow.steps import CacheConfig
# cache_config = CacheConfig(enable_caching=True, expire_after="7d")
# -
# <a id='claims-data-wrangler'></a>
# ### Step 1: Claims Data Wranger Preprocessing Step
# [pipeline](#pipelines)
# #### Upload flow to S3
#
# 이것은 첫 번째 단계에 대한 입력이 되므로 S3에 있어야 합니다.
s3_client.upload_file(
Filename="claims.flow", Bucket=bucket, Key=f"{prefix}/dataprep-notebooks/claims.flow"
)
claims_flow_uri = f"s3://{bucket}/{prefix}/dataprep-notebooks/claims.flow"
print(f"Claims flow file uploaded to S3")
# #### Define the first Data Wrangler step's inputs
# +
with open("claims.flow", "r") as f:
claims_flow = json.load(f)
flow_step_inputs = []
# flow file contains the code for each transformation
flow_file_input = sagemaker.processing.ProcessingInput(
source=claims_flow_uri, destination=f"{processing_dir}/flow", input_name="flow"
)
flow_step_inputs.append(flow_file_input)
# parse the flow file for S3 inputs to Data Wranger job
for node in claims_flow["nodes"]:
if "dataset_definition" in node["parameters"]:
data_def = node["parameters"]["dataset_definition"]
# Fixed: The example code throws an error outside the us-east-2 region.
data_def["s3ExecutionContext"]["s3Uri"] = f's3://{bucket}/fraud-detect-demo/data/raw/claims.csv'
name = data_def["name"]
s3_input = sagemaker.processing.ProcessingInput(
source=data_def["s3ExecutionContext"]["s3Uri"],
destination=f"{processing_dir}/{name}",
input_name=name,
)
flow_step_inputs.append(s3_input)
# -
# #### Define outputs for first Data Wranger step
# +
claims_output_name = (
f"{claims_flow['nodes'][-1]['node_id']}.{claims_flow['nodes'][-1]['outputs'][0]['name']}"
)
flow_step_outputs = []
flow_output = sagemaker.processing.ProcessingOutput(
output_name=claims_output_name,
feature_store_output=sagemaker.processing.FeatureStoreOutput(feature_group_name=claims_fg_name),
app_managed=True,
)
flow_step_outputs.append(flow_output)
# -
# #### Define processor and processing step
# +
# You can find the proper image uri by exporting your Data Wrangler flow to a pipeline notebook
# =================================
#image_uri = "415577184552.dkr.ecr.us-east-2.amazonaws.com/sagemaker-data-wrangler-container:1.0.2"
from sagemaker import image_uris
image_uri = image_uris.retrieve(framework='data-wrangler',region=region)
flow_processor = sagemaker.processing.Processor(
role=sagemaker_role,
image_uri=image_uri,
instance_count=flow_instance_count,
instance_type=flow_instance_type,
max_runtime_in_seconds=86400,
)
### ProcessingStep (Data Wrangler for Claim Data)
claims_flow_step = ProcessingStep(
name="ClaimsDataWranglerProcessingStep",
processor=flow_processor,
inputs=flow_step_inputs,
outputs=flow_step_outputs,
)
# -
# <a id='data-wrangler'></a>
# ### Step 2: Customers Data Wrangler preprocessing step
#
# [pipeline](#pipelines)
s3_client.upload_file(
Filename="customers.flow", Bucket=bucket, Key=f"{prefix}/dataprep-notebooks/customers.flow"
)
claims_flow_uri = f"s3://{bucket}/{prefix}/dataprep-notebooks/customers.flow"
print(f"Customers flow file uploaded to S3")
# +
with open("customers.flow", "r") as f:
customers_flow = json.load(f)
flow_step_inputs = []
# flow file contains the code for each transformation
flow_file_input = sagemaker.processing.ProcessingInput(
source=claims_flow_uri, destination=f"{processing_dir}/flow", input_name="flow"
)
flow_step_inputs.append(flow_file_input)
# parse the flow file for S3 inputs to Data Wranger job
for node in customers_flow["nodes"]:
if "dataset_definition" in node["parameters"]:
data_def = node["parameters"]["dataset_definition"]
# Fixed: The example code throws an error outside the us-east-2 region.
data_def["s3ExecutionContext"]["s3Uri"] = f's3://{bucket}/fraud-detect-demo/data/raw/customers.csv'
name = data_def["name"]
s3_input = sagemaker.processing.ProcessingInput(
source=data_def["s3ExecutionContext"]["s3Uri"],
destination=f"{processing_dir}/{name}",
input_name=name,
)
flow_step_inputs.append(s3_input)
# +
customers_output_name = (
f"{customers_flow['nodes'][-1]['node_id']}.{customers_flow['nodes'][-1]['outputs'][0]['name']}"
)
flow_step_outputs = []
flow_output = sagemaker.processing.ProcessingOutput(
output_name=customers_output_name,
feature_store_output=sagemaker.processing.FeatureStoreOutput(
feature_group_name=customers_fg_name
),
app_managed=True,
)
flow_step_outputs.append(flow_output)
### ProcessingStep (Data Wrangler for Customer Data)
customers_flow_step = ProcessingStep(
name="CustomersDataWranglerProcessingStep",
processor=flow_processor,
inputs=flow_step_inputs,
outputs=flow_step_outputs,
)
# -
# <a id='dataset-train-test'></a>
# ### Step 3: Create Dataset and Train/Test Split
#
# [pipeline](#pipelines)
# +
s3_client.upload_file(
Filename="create_dataset.py", Bucket=bucket, Key=f"{prefix}/code/create_dataset.py"
)
create_dataset_processor = SKLearnProcessor(
framework_version="0.23-1",
role=sagemaker_role,
instance_type="ml.m5.xlarge",
instance_count=1,
base_job_name="fraud-detection-demo-create-dataset",
sagemaker_session=sagemaker_session,
)
### ProcessingStep
create_dataset_step = ProcessingStep(
name="CreateDataset",
processor=create_dataset_processor,
outputs=[
sagemaker.processing.ProcessingOutput(
output_name="train_data", source="/opt/ml/processing/output/train"
),
sagemaker.processing.ProcessingOutput(
output_name="test_data", source="/opt/ml/processing/output/test"
),
],
job_arguments=[
"--claims-feature-group-name",
claims_fg_name,
"--customers-feature-group-name",
customers_fg_name,
"--bucket-name",
bucket,
"--bucket-prefix",
prefix,
"--athena-database-name",
database_name,
"--claims-table-name",
claims_table,
"--customers-table-name",
customers_table,
"--region",
region
],
code=create_dataset_script_uri,
)
# -
# <a id='pipe-train-xgb'></a>
# ### Step 4: Train XGBoost Model
# 이 단계에서는 파이프라인 시작 부분에 정의된 ParameterString `train_instance_param`을 사용합니다.
#
# [pipeline](#pipelines)
# +
hyperparameters = {
"max_depth": "3",
"eta": "0.2",
"objective": "binary:logistic",
"num_round": "100",
}
xgb_estimator = XGBoost(
entry_point="xgboost_starter_script.py",
output_path=training_job_output_path,
code_location=training_job_output_path,
hyperparameters=hyperparameters,
role=sagemaker_role,
instance_count=train_instance_count,
instance_type=train_instance_param,
framework_version="1.0-1",
)
### TrainingStep
train_step = TrainingStep(
name="XgboostTrain",
estimator=xgb_estimator,
inputs={
"train": sagemaker.inputs.TrainingInput(
s3_data=create_dataset_step.properties.ProcessingOutputConfig.Outputs[
"train_data"
].S3Output.S3Uri
)
},
)
# -
# <a id='pipe-pre-deploy'></a>
# ### Step 5: Model Pre-Deployment Step
#
# [pipeline](#pipelines)
# +
model = sagemaker.model.Model(
name="fraud-detection-demo-pipeline-xgboost",
image_uri=train_step.properties.AlgorithmSpecification.TrainingImage,
model_data=train_step.properties.ModelArtifacts.S3ModelArtifacts,
sagemaker_session=sagemaker_session,
role=sagemaker_role,
)
inputs = sagemaker.inputs.CreateModelInput(instance_type="ml.m4.xlarge")
### CreateModelStep
create_model_step = CreateModelStep(name="ModelPreDeployment", model=model, inputs=inputs)
# -
# <a id='pipe-detect-bias'></a>
#
# ### Step 6: Run Bias Metrics with Clarify
# [pipeline](#pipelines)
# #### Clarify configuration
# +
bias_data_config = sagemaker.clarify.DataConfig(
s3_data_input_path=create_dataset_step.properties.ProcessingOutputConfig.Outputs[
"train_data"
].S3Output.S3Uri,
s3_output_path=pipeline_bias_output_path,
label="fraud",
dataset_type="text/csv",
)
bias_config = sagemaker.clarify.BiasConfig(
label_values_or_threshold=[0],
facet_name="customer_gender_female",
facet_values_or_threshold=[1],
)
analysis_config = bias_data_config.get_config()
analysis_config.update(bias_config.get_config())
analysis_config["methods"] = {"pre_training_bias": {"methods": "all"}}
clarify_config_dir = pathlib.Path("config")
clarify_config_dir.mkdir(exist_ok=True)
with open(clarify_config_dir / "analysis_config.json", "w") as f:
json.dump(analysis_config, f)
s3_client.upload_file(
Filename="config/analysis_config.json",
Bucket=bucket,
Key=f"{prefix}/clarify-config/analysis_config.json",
)
# -
# #### Clarify processing step
# +
clarify_processor = sagemaker.processing.Processor(
base_job_name="fraud-detection-demo-clarify-processor",
image_uri=sagemaker.clarify.image_uris.retrieve(framework="clarify", region=region),
role=sagemaker.get_execution_role(),
instance_count=1,
instance_type="ml.c5.xlarge",
)
### ProcessingStep (Clarify)
clarify_step = ProcessingStep(
name="ClarifyProcessor",
processor=clarify_processor,
inputs=[
sagemaker.processing.ProcessingInput(
input_name="analysis_config",
source=f"s3://{bucket}/{prefix}/clarify-config/analysis_config.json",
destination="/opt/ml/processing/input/config",
),
sagemaker.processing.ProcessingInput(
input_name="dataset",
source=create_dataset_step.properties.ProcessingOutputConfig.Outputs[
"train_data"
].S3Output.S3Uri,
destination="/opt/ml/processing/input/data",
),
],
outputs=[
sagemaker.processing.ProcessingOutput(
source="/opt/ml/processing/output/analysis.json",
destination=pipeline_bias_output_path,
output_name="analysis_result",
)
],
)
# -
# <a id='pipe-Register-Model'></a>
# ### Step 7: Register Model
#
# 이 단계에서는 파이프라인 코드의 시작 부분에 정의된 ParameterString `model_approval_status`를 사용합니다.
#
# [pipeline](#pipelines)
# +
model_metrics = demo_helpers.ModelMetrics(
bias=sagemaker.model_metrics.MetricsSource(
s3_uri=clarify_step.properties.ProcessingOutputConfig.Outputs[
"analysis_result"
].S3Output.S3Uri,
content_type="application/json",
)
)
### RegisterModel
register_step = RegisterModel(
name="XgboostRegisterModel",
estimator=xgb_estimator,
model_data=train_step.properties.ModelArtifacts.S3ModelArtifacts,
content_types=["text/csv"],
response_types=["text/csv"],
inference_instances=["ml.t2.medium", "ml.m5.xlarge"],
transform_instances=["ml.m5.xlarge"],
model_package_group_name=mpg_name,
approval_status=model_approval_status,
model_metrics=model_metrics,
)
# -
# <a id='pipe-Register-Model'></a>
# ### Step 8: Deploy Model
#
#
# [pipeline](#pipelines)
# +
s3_client.upload_file(
Filename="deploy_model.py", Bucket=bucket, Key=f"{prefix}/code/deploy_model.py"
)
deploy_model_processor = SKLearnProcessor(
framework_version="0.23-1",
role=sagemaker_role,
instance_type="ml.t3.medium",
instance_count=1,
base_job_name="fraud-detection-demo-deploy-model",
sagemaker_session=sagemaker_session,
)
### ProcessingStep (Deployment)
deploy_step = ProcessingStep(
name="DeployModel",
processor=deploy_model_processor,
job_arguments=[
"--model-name",
create_model_step.properties.ModelName,
"--region",
region,
"--endpoint-instance-type",
deploy_model_instance_type,
"--endpoint-name",
"xgboost-model-pipeline-0120",
],
code=deploy_model_script_uri,
)
# -
# <a id='define-pipeline'></a>
#
# ### Combine the Pipeline Steps and Run
# [pipeline](#overview-5)
#
# 추론하기는 쉽지만, 파라메터와 단계가 순서가 맞을 필요는 없습니다. 파이프라인 DAG는 이를 올바르게 파싱합니다.
# +
pipeline_name = f"FraudDetectDemo"
# %store pipeline_name
pipeline = Pipeline(
name=pipeline_name,
parameters=[train_instance_param, model_approval_status],
steps=[
claims_flow_step,
customers_flow_step,
create_dataset_step,
train_step,
create_model_step,
clarify_step,
register_step,
deploy_step,
],
)
# -
# ### Submit the pipeline definition to the SageMaker Pipeline service
#
# `upsert()` 메소드는 UpdatePipeline과 CreatePipeline API를 각각 호출하여 파이프라인 정의를 업데이트하거나(기존 파이프라인 존재 시) 신규 파이프라인을 생성합니다.
#
# - https://docs.aws.amazon.com/sagemaker/latest/APIReference/API_UpdatePipeline.html
# - https://docs.aws.amazon.com/sagemaker/latest/APIReference/API_CreatePipeline.html
pipeline.upsert(role_arn=sagemaker_role)
# ### View the entire pipeline definition
#
# 파이프라인 정의를 보면 보간된 모든 문자열 변수들이 파이프라인 버그를 디버그하는 데 도움이 될 수 있습니다. 아래 코드의 결과가 길기에 주석 처리되었습니다.
# +
#json.loads(pipeline.describe()['PipelineDefinition'])
# -
# ### Run the pipeline
#
# `start()` 메소드는 StartPipelineExecution API를 호출하여 파이프라인 실행을 트리거합니다.
# - https://docs.aws.amazon.com/sagemaker/latest/APIReference/API_StartPipelineExecution.html
#
# 완료하는 데 약 20-25분이 소요됩니다. SageMaker Studio Components panel에서 파이프라인 작업의 진행 상황을 확인할 수 있습니다.
# 
# Special pipeline parameters can be defined or changed here
parameters = {'TrainingInstance': 'ml.m5.xlarge'}
start_response = pipeline.start(parameters=parameters)
# ### 파이프라인 운영: 파이프라인 대기 및 실행상태 확인
# 워크플로우의 실행상황을 살펴봅니다.
start_response.describe()
# 실행이 완료될 때까지 기다립니다.
start_response.wait()
# <pre>
# </pre>
# ### 완료 후 다음과 같이 보일 것입니다.
# 
# 
# 실행된 스텝들을 리스트업합니다. 각 스텝의 시작 및 완료 시각, 상태, 메타데이터(arn, processing/training job)을 보여줍니다.
display(start_response.list_steps())
# <a id='cleanup'></a>
# ## Clean up
#
# [overview](#overview-5)
# ___
#
# 데모를 실행한 후, 생성된 리소스를 제거해야 합니다. keyword argument `delete_s3_objects=True` 를 전달하여 프로젝트의 S3 디렉터리에있는 모든 객체를 삭제할 수도 있습니다.
from demo_helpers import delete_project_resources
# +
# delete_project_resources(
# sagemaker_boto_client=sagemaker_boto_client,
# endpoint_name=endpoint_name,
# pipeline_name=pipeline_name,
# mpg_name=mpg_name,
# prefix=prefix,
# delete_s3_objects=False,
# bucket_name=bucket)
|
5-pipeline-e2e.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# ---
# +
from scipy.spatial.distance import cdist
from sklearn.cluster import KMeans
from sklearn.decomposition import PCA
import pandas as pd
import numpy as np
import os
import matplotlib.pyplot as plt
from sklearn.metrics import confusion_matrix
from tensorflow.keras import models
from tensorflow.keras.layers import Dense, Dropout, Flatten, Conv2D, MaxPool2D
from tensorflow.keras.optimizers import RMSprop, Adam
from tensorflow.keras.utils import to_categorical
# +
def prepare_data(data):
""" Prepare data for modeling
input: data frame with labels und pixel data
output: image and label array """
image_array = np.zeros(shape=(len(data), 48, 48, 1))
image_label = np.array(list(map(int, data['emotion'])))
for i, row in enumerate(data.index):
image = np.fromstring(data.loc[row, 'pixels'], dtype=int, sep=' ')
image = np.reshape(image, (48, 48, 1)) # 灰階圖的channel數為1
image_array[i] = image
return image_array, image_label
def plot_one_emotion_grayhist(data, img_arrays, img_labels, label=0):
fig, axs = plt.subplots(1, 5, figsize=(25, 12))
fig.subplots_adjust(hspace=.2, wspace=.2)
axs = axs.ravel()
for i in range(5):
idx = data[data['emotion'] == label].index[i]
axs[i].hist(img_arrays[idx][:, :, 0], 256, [0, 256])
axs[i].set_title(emotions[img_labels[idx]])
axs[i].set_xticklabels([])
axs[i].set_yticklabels([])
def plot_one_emotion(data, img_arrays, img_labels, label=0):
fig, axs = plt.subplots(1, 7, figsize=(25, 12))
fig.subplots_adjust(hspace=.2, wspace=.2)
axs = axs.ravel()
for i in range(7):
idx = data[data['emotion'] == label].index[i]
axs[i].imshow(img_arrays[idx][:, :, 0], cmap='gray')
axs[i].set_title(emotions[img_labels[idx]])
axs[i].set_xticklabels([])
axs[i].set_yticklabels([])
def plot_all_emotions(data, img_arrays, img_labels):
fig, axs = plt.subplots(1, 7, figsize=(30, 12))
fig.subplots_adjust(hspace=.2, wspace=.2)
axs = axs.ravel()
for i in range(7):
idx = data[data['emotion'] == i].index[0] # 取該表情的第一張圖的位置
axs[i].imshow(img_arrays[idx][:, :, 0], cmap='gray')
axs[i].set_title(emotions[img_labels[idx]])
axs[i].set_xticklabels([])
axs[i].set_yticklabels([])
def plot_image_and_emotion(test_image_array, test_image_label, pred_test_labels, image_number):
""" Function to plot the image and compare the prediction results with the label """
fig, axs = plt.subplots(1, 2, figsize=(12, 6), sharey=False)
bar_label = emotions.values()
axs[0].imshow(test_image_array[image_number], 'gray')
axs[0].set_title(emotions[test_image_label[image_number]])
axs[1].bar(bar_label, pred_test_labels[image_number],
color='orange', alpha=0.7)
axs[1].grid()
plt.show()
def plot_compare_distributions(img_labels_1, img_labels_2, title1='', title2=''):
df_array1 = pd.DataFrame()
df_array2 = pd.DataFrame()
df_array1['emotion'] = img_labels_1
df_array2['emotion'] = img_labels_2
fig, axs = plt.subplots(1, 2, figsize=(12, 6), sharey=False)
x = emotions.values()
y = df_array1['emotion'].value_counts()
keys_missed = list(set(emotions.keys()).difference(set(y.keys())))
for key_missed in keys_missed:
y[key_missed] = 0
axs[0].bar(x, y.sort_index(), color='orange')
axs[0].set_title(title1)
axs[0].grid()
y = df_array2['emotion'].value_counts()
keys_missed = list(set(emotions.keys()).difference(set(y.keys())))
for key_missed in keys_missed:
y[key_missed] = 0
axs[1].bar(x, y.sort_index())
axs[1].set_title(title2)
axs[1].grid()
plt.show()
emotions = {0: 'Angry', 1: 'Disgust', 2: 'Fear',
3: 'Happy', 4: 'Sad', 5: 'Surprise', 6: 'Neutral'}
# -
df_raw = pd.read_csv("D:/mycodes/AIFER/data/fer2013.csv")
df_raw.head()
df_raw['Usage'].value_counts() # 8:1:1
# +
df_train = df_raw[df_raw['Usage'] == 'Training']
df_val = df_raw[df_raw['Usage'] == 'PublicTest']
df_test = df_raw[df_raw['Usage'] == 'PrivateTest']
X_train, y_train = prepare_data(df_train)
X_val, y_val = prepare_data(df_val)
X_test, y_test = prepare_data(df_test)
y_train_oh = to_categorical(y_train)
y_val_oh = to_categorical(y_val)
y_test_oh = to_categorical(y_test)
plot_all_emotions(df_train, X_train, y_train)
# -
for label in emotions.keys():
plot_one_emotion(df_train, X_train, y_train, label=label)
for label in emotions.keys():
plot_one_emotion_grayhist(df_train, X_train, y_train, label=label)
plot_compare_distributions(
y_train, y_val, title1='train labels', title2='val labels')
# +
n_sample, nrow, ncol, nchannel = X_train.shape
X = X_train.reshape((n_sample, ncol * nrow * nchannel))
pca = PCA(n_components=2, whiten=True)
pca.fit(X)
print(pca.explained_variance_ratio_)
X_pca = pca.transform(X)
# -
plt.xlabel('pca_dim1')
plt.ylabel('pca_dim2')
plt.title('Images look like when they are in 2-dim')
plt.scatter(X_pca[:, 0], X_pca[:, 1], color='green', marker=".")
distortions = []
K = range(1, 10)
for k in K:
kmeans = KMeans(n_clusters=k).fit(X_pca)
kmeans.fit(X_pca)
distortions.append(sum(np.min(
cdist(X_pca, kmeans.cluster_centers_, 'euclidean'), axis=1)) / X_pca.shape[0])
plt.plot(K, distortions, 'bx-')
plt.xlabel('k')
plt.ylabel('Distortion')
plt.title('The Elbow Method showing the optimal k')
for k in range(1, 9):
plt.text(k+0.65, 0.3, f"{distortions[k]-distortions[k-1]:.2f}",
bbox=dict(facecolor='green', alpha=0.5))
plt.show()
|
notebooks/Day03_EDA.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# + [markdown] slideshow={"slide_type": "slide"}
# # Bayesian Statistical Inference
#
# <NAME>, 2016 (with input from Ivezic $\S5$, Bevington, <NAME>'s [Bayesian Stats](http://seminar.ouml.org/lectures/bayesian-statistics/) and [MCMC](http://seminar.ouml.org/lectures/monte-carlo-markov-chain-mcmc/) lectures, and [<NAME>](http://twiecki.github.io/blog/2015/11/10/mcmc-sampling/).)
#
# Up to now we have been using Classical Inference: finding model parameters that maximize the
# **likelihood** $p(D|M)$.
#
# In Bayesian inference, the argument is that probability statements can be made not just for data, but also models and model parameters. As a result, we instead evaluate the **posterior probability** taking into account **prior** information.
#
# Recall from the BasicStats lecture that Bayes' Rule is:
# $$p(M|D) = \frac{p(D|M)p(M)}{p(D)},$$
# where $D$ is for data and $M$ is for model.
#
# We wrote this in words as:
# $${\rm Posterior Probability} = \frac{{\rm Likelihood}\times{\rm Prior}}{{\rm Evidence}}.$$
#
# If we explicitly recognize prior information, $I$, and the model parameters, $\theta$, then we can write:
# $$p(M,\theta|D,I) = \frac{p(D|M,\theta,I)p(M,\theta|I)}{p(D|I)},$$
# where we will omit the explict dependence on $\theta$ by writing $M$ instead of $M,\theta$ where appropriate. However, as the prior can be expanded to
# $$p(M,\theta|I) = p(\theta|M,I)p(M|I),$$
# it will still appear in the term $p(\theta|M,I)$.
#
# Note that it is often that case that $p(D|I)$ is not evaluated explictly since the likelihood can be normalized such that it is unity or we will instead take the ratio of two posterior probabilities such that this term cancels out.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Analysis of a Heteroscedastic Gaussian distribution with Bayesian Priors
#
# Consider the case of measuring a rod as we discussed previously. We want to know the posterior pdf for the length of the rod, $p(M,\theta|D,I) = p(\mu|\{x_i\},\{\sigma_i\},I)$.
#
# For the likelihood we have
# $$L = p(\{x_i\}|\mu,I) = \prod_{i=1}^N \frac{1}{\sigma_i\sqrt{2\pi}} \exp\left(\frac{-(x_i-\mu)^2}{2\sigma_i^2}\right).$$
#
# In the Bayesian case, we also need a prior. We'll adopt a uniform distribution given by
# $$p(\mu|I) = C, \; {\rm for} \; \mu_{\rm min} < \mu < \mu_{\rm max},$$
# where $C = \frac{1}{\mu_{\rm max} - \mu_{\rm min}}$ between the min and max and is $0$ otherwise.
#
# The log of the posterior pdf is then
# $$\ln L = {\rm constant} - \sum_{i=1}^N \frac{(x_i - \mu)^2}{2\sigma_i^2}.$$
#
# This is exactly the same as we saw before, except that the value of the constant is different. Since the constant doesn't come into play, we get the same result as before:
#
# $$\mu^0 = \frac{\sum_i^N (x_i/\sigma_i^2)}{\sum_i^N (1/\sigma_i^2)},$$
# with uncertainty
# $$\sigma_{\mu} = \left( \sum_{i=1}^N \frac{1}{\sigma_i^2}\right)^{-1/2}.$$
# + [markdown] slideshow={"slide_type": "slide"}
# We get the same result because we used a flat prior. If the case were homoscedastic instead of heteroscedastic, we obviously would get the result from our first example.
#
# Now let's consider the case where $\sigma$ is *not* known, but rather needs to be determined from the data. In that case, the posterior pdf that we seek is not $p(\mu|\{x_i\},\{\sigma_i\},I)$, but rather $p(\mu,\sigma|\{x_i\},I)$.
#
# As before we have
# $$L = p(\{x_i\}|\mu,\sigma,I) = \prod_{i=1}^N \frac{1}{\sigma\sqrt{2\pi}} \exp\left(\frac{-(x_i-\mu)^2}{2\sigma^2}\right),$$
# except that now $\sigma$ is uknown.
#
# Our Bayesian prior is now 2D instead of 1D and we'll adopt
# $$p(\mu,\sigma|I) \propto \frac{1}{\sigma},\; {\rm for} \; \mu_{\rm min} < \mu < \mu_{\rm max} \; {\rm and} \; \sigma_{\rm min} < \sigma < \sigma_{\rm max}.$$
#
# With proper normalization, we have
# $$p(\{x_i\}|\mu,\sigma,I)p(\mu,\sigma|I) = C\frac{1}{\sigma^{(N+1)}}\prod_{i=1}^N \exp\left( \frac{-(x_i-\mu)^2}{2\sigma^2} \right),$$
# where
# $$C = (2\pi)^{-N/2}(\mu_{\rm max}-\mu_{\rm min})^{-1} \left[\ln \left( \frac{\sigma_{\rm max}}{\sigma_{\rm min}}\right) \right]^{-1}.$$
# + [markdown] slideshow={"slide_type": "slide"}
# The log of the posterior pdf is
#
# $$\ln[p(\mu,\sigma|\{x_i\},I)] = {\rm constant} - (N+1)\ln\sigma - \sum_{i=1}^N \frac{(x_i - \mu)^2}{2\sigma^2}.$$
#
# Right now that has $x_i$ in it, which isn't that helpful, but since we are assuming a Gaussian distribution, we can take advantage of the fact that the mean, $\overline{x}$, and the variance, $V (=s^2)$, completely characterize the distribution. So we can write this expression in terms of those variables instead of $x_i$. Skipping over the math details (see Ivezic $\S$5.6.1), we find
#
# $$\ln[p(\mu,\sigma|\{x_i\},I)] = {\rm constant} - (N+1)\ln\sigma - \frac{N}{2\sigma^2}\left( (\overline{x}-\mu)^2 + V \right).$$
#
# Note that this expression only contains the 2 parameters that we are trying to determine: $(\mu,\sigma)$ and 3 values that we can determine directly from the data: $(N,\overline{x},V)$.
#
# Load and execute the next cell to visualize the posterior pdf for the case of $(N,\overline{x},V)=(10,1,4)$. Remember to change `usetex=True` to `usetex=False` if you have trouble with the plotting. Try changing the values of $(N,\overline{x},V)$.
# +
# # %load code/fig_likelihood_gaussian.py
"""
Log-likelihood for Gaussian Distribution
----------------------------------------
Figure5.4
An illustration of the logarithm of the posterior probability density
function for :math:`\mu` and :math:`\sigma`, :math:`L_p(\mu,\sigma)`
(see eq. 5.58) for data drawn from a Gaussian distribution and N = 10, x = 1,
and V = 4. The maximum of :math:`L_p` is renormalized to 0, and color coded as
shown in the legend. The maximum value of :math:`L_p` is at :math:`\mu_0 = 1.0`
and :math:`\sigma_0 = 1.8`. The contours enclose the regions that contain
0.683, 0.955, and 0.997 of the cumulative (integrated) posterior probability.
"""
# Author: <NAME>
# License: BSD
# The figure produced by this code is published in the textbook
# "Statistics, Data Mining, and Machine Learning in Astronomy" (2013)
# For more information, see http://astroML.github.com
# To report a bug or issue, use the following forum:
# https://groups.google.com/forum/#!forum/astroml-general
import numpy as np
from matplotlib import pyplot as plt
from astroML.plotting.mcmc import convert_to_stdev
#----------------------------------------------------------------------
# This function adjusts matplotlib settings for a uniform feel in the textbook.
# Note that with usetex=True, fonts are rendered with LaTeX. This may
# result in an error if LaTeX is not installed on your system. In that case,
# you can set usetex to False.
from astroML.plotting import setup_text_plots
setup_text_plots(fontsize=8, usetex=True)
def gauss_logL(xbar, V, n, sigma, mu):
"""Equation 5.57: gaussian likelihood"""
return (-(n + 1) * np.log(sigma)
- 0.5 * n * ((xbar - mu) ** 2 + V) / sigma ** 2)
#------------------------------------------------------------
# Define the grid and compute logL
sigma = np.linspace(1, 5, 70)
mu = np.linspace(-3, 5, 70)
xbar = 1
V = 4
n = 10
logL = gauss_logL(xbar, V, n, sigma[:, np.newaxis], mu)
logL -= logL.max()
#------------------------------------------------------------
# Plot the results
fig = plt.figure(figsize=(5, 3.75))
plt.imshow(logL, origin='lower',
extent=(mu[0], mu[-1], sigma[0], sigma[-1]),
cmap=plt.cm.binary,
aspect='auto')
plt.colorbar().set_label(r'$\log(L)$')
plt.clim(-5, 0)
plt.contour(mu, sigma, convert_to_stdev(logL),
levels=(0.683, 0.955, 0.997),
colors='k')
plt.text(0.5, 0.93, r'$L(\mu,\sigma)\ \mathrm{for}\ \bar{x}=1,\ V=4,\ n=10$',
bbox=dict(ec='k', fc='w', alpha=0.9),
ha='center', va='center', transform=plt.gca().transAxes)
plt.xlabel(r'$\mu$')
plt.ylabel(r'$\sigma$')
plt.show()
# + [markdown] slideshow={"slide_type": "slide"}
# The shaded region is the posterior probability. The contours are the confidence intervals. We can compute those by determining the marginal distribution at each $(\mu,\sigma)$. The top panels of the figures below show those marginal distributions. The solid line is what we just computed. The dotted line is what we would have gotten for a uniform prior--not that much difference. The dashed line is the MLE result, which is quite different. The bottom panels show the cumulative distribution.
#
# 
#
#
# Note that the marginal pdfs follow a Student's $t$ Distribution, which becomes Gaussian for large $N$.
# + [markdown] slideshow={"slide_type": "slide"}
# ### Recap
#
# To review: the Bayesian Statistical Inference process is
# * formulate the likelihood, $p(D|M,I)$
# * chose a prior, $p(\theta|M,I)$, which incorporates other information beyond the data in $D$
# * determine the posterior pdf, $p(M|D,I)$
# * search for the model paramters that maximize $p(M|D,I)$
# * quantify the uncertainty of the model parameter estimates
# * test the hypothesis being addressed
#
# The last part we haven't talked about yet.
# + [markdown] slideshow={"slide_type": "slide"}
# ### Another Example
#
# What if we wanted to model the mixture of a Gauassian distribution with a uniform distribution. When might that be useful? Well, for example:
#
# 
#
# Obviously this isn't exactly a Gaussian and a uniform distribution, but a line feature superimposed upon a background is the sort of thing that a physicist might see and is pretty close to this case for a local region around the feature of interest. This is the example discussed in Ivezic $\S$5.6.5.
#
# For this example, we will assume that the location parameter, $\mu$, is known (say from theory) and that the errors in $x_i$ are negligible compared to $\sigma$.
# + [markdown] slideshow={"slide_type": "slide"}
# The likelihood of obtaining a measurement, $x_i$, in this example can be written as
# $$L = p(x_i|A,\mu,\sigma,I) = \frac{A}{\sigma\sqrt{2\pi}} \exp\left(\frac{-(x_i-\mu)^2}{2\sigma^2}\right) + \frac{1-A}{W}.$$
#
# Here the background probability is evaluated over $0 < x < W$ and 0 otherwise, that is the feature of interest lies between $0$ and $W$. $A$ and $1-A$ are the relative strengths of the two components, which are obviously anti-correlated. Note that there will be covariance between $A$ and $\sigma$.
# + [markdown] slideshow={"slide_type": "slide"}
# If we adopt a uniform prior in both $A$ and $\sigma$:
# $$p(A,\sigma|I) = C, \; {\rm for} \; 0\le A<A_{\rm max} \; {\rm and} \; 0 \le \sigma \le \sigma_{\rm max},$$
# then the posterior pdf is given by
# $$\ln [p(A,\sigma|\{x_i\},\mu,W)] = \sum_{i=1}^N \ln \left[\frac{A}{\sigma \sqrt{2\pi}} \exp\left( \frac{-(x_i-\mu)^2}{2\sigma^2} \right) + \frac{1-A}{W} \right].$$
#
# The figure below (Ivezic, 5.13) shows an example for $N=200, A=0.5, \sigma=1, \mu=5, W=10$. Specifically, the bottom panel is a result drawn from this distribution and the top panel is the likelihood distribution derived from the data in the bottom panel.
# 
# + [markdown] slideshow={"slide_type": "slide"}
# A more realistic example might be one where all three parameters are unknown: the location, the width, and the background level. But that will have to wait until $\S$5.8.6.
#
# In the meantime, note that we have not binned the data, $\{x_i\}$. We only binned Figure 5.13 for the sake of visualizaiton. However, sometimes the data are inherently binned (e.g., the detector is pixelated). In that case, the data would be in the form of $(x_i,y_i)$, where $y_i$ is the number of counts at each location. We'll skip over this example, but you can read about it in Ivezic $\S$5.6.6. A refresher on the Poission distribution (Ivezic $\S$3.3.4) might be appropriate first.
# + [markdown] slideshow={"slide_type": "slide"}
# ### Model Comparison
#
# Up to now we have concerned ourselves with determining the optimal parameters of a given model fit. But what if *another* model would be a better fit (regardless of how you choose the parameters of the first model).
#
# That leads us to a discussion of model comparison. This is discussed in more detail in Ivezic $\S$5.4 and $\S$5.7.1-3.
#
# To determine which model is better we compute the ratio of the posterior probabilities or the **odds ratio** for two models as
# $$O_{21} \equiv \frac{p(M_2|D,I)}{p(M_1|D,I)}.$$
#
# Since
# $$p(M|D,I) = \frac{p(D|M,I)p(M|I)}{p(D|I)},$$
# the odds ratio can ignore $p(D|I)$ since it will be the same for both models.
#
# (We'll see later why that is even more important than you might think as the denominator is the integral of the numerator, but what if you don't have an analytical function that you can integrate?!)
# + [markdown] slideshow={"slide_type": "skip"}
# ### Bayesian Hypothesis Testing
#
# In *hypothesis testing* we are essentially comparing a model, $M_1$, to its complement. That is $p(M_1) + p(M_2) = 1$. If we take $M_1$ to be the "null" (default) hypothesis (which is generally that, for example, a correlation does *not* exist), then we are asking whether or not the data reject the null hypothesis.
#
# In classical hypothesis testing we can ask whether or not a single model provides a good description of the data. In Bayesian hypothesis testing, we need to have an alternative model to compare to.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Markov-Chain Monte Carlo Methods
# + [markdown] slideshow={"slide_type": "slide"}
# Figure 5.10 from Ivezic shows the likelihood for a particular example:
# 
#
# What was required to produce this figure? We needed to know the analytic form of the posterior distribution. But imagine that you don’t have a nice analytical function for the likelihood. You could still make a plot like the one above, by making a simulated model for the likelihood at every point, comparing the model with the data to generate a likelihood, populating the grid with those numerical likelihood estimates, then finding the best fitting parameters by locating the maximum in likelihood space.
# + [markdown] slideshow={"slide_type": "slide"}
# Now imagine that you have a problem with many parameters. If you have even 5 parameters and you want to sample 100 points of parameter space for each, that is $10^{10}$ points. It might take you a while (even your computer). So you might not be able to sample the full space given time (and memory) constraints.
#
# You *could* simply randomly sample the grid at every point, and try to find the minimum based on that. But that can also be quite time consuming, and you will spend a lot of time in regions of parameter space that yields small likelihood.
#
# However, a better way is to adopt a **Markov-Chain Monte Carlo (MCMC)**. MCMC gives us a way to make this problem computationally tractable by sampling the full multi-dimensional parameter space, in a way that builds up the most density in the regions of parameter space which are closest to the maximum. Then, you can post-process the “chain” to infer the distribution and error regions.
# + [markdown] slideshow={"slide_type": "slide"}
# Ivezic, Figure 5.22 shows the same problem as above, done with a Markov Chain Monte Carlo. The dashed lines are the known (analytic) solution. The solid lines are from the MCMC estimate with 10,000 sample points.
# 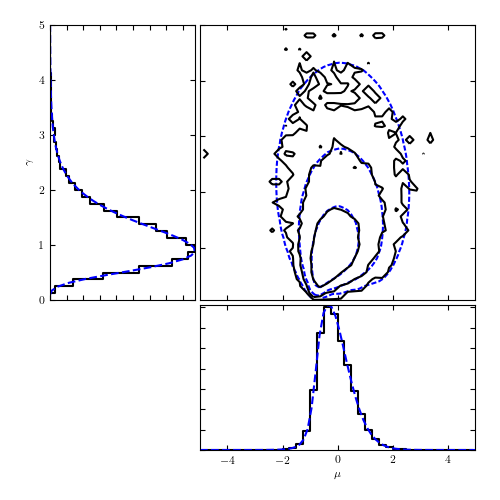
#
# + [markdown] slideshow={"slide_type": "slide"}
# ## How does MCMC work?
#
# I've really struggled to come up with a simple way of illustrating MCMC so that you (and I for that matter) can understand it. Unfortunately, even the supposedly dumbed-down explanations are really technical. But let's see what I can do!
#
# Let's start by simply trying to understand what a Monte Carlo is and what a Markov Chain is.
# + [markdown] slideshow={"slide_type": "slide"}
# ### What is a Monte Carlo?
#
# In case you are not familiar with Monte Carlo methods, it might help to know that the term is derived from the Monte Carlo Casino as gambling and random sampling go together.
#
# We'll consider a simple example: you have forgotten the formula for the area of a circle, but you know the formula for the area of a square and how to draw a circle.
#
# We can use the information that we *do* know to numerically compute the area of a circle.
#
# We start by drawing a square and circumscribing a circle in it. Then we put down random points within the square and note which ones land in the circle. The ratio of random points in the circle to the number of random points drawn is related to the area of our circle. No need to know $\pi$. Using more random points yields more precise estimates of the area.
#
# Try it.
# + slideshow={"slide_type": "slide"}
# %matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(10, 10))
#Draw a square that spans ([-1,1],[-1,1])
x = np.array(# Complete
y = np.array(# Complete
plt.xlim(-1.5,1.5)
plt.ylim(-1.5,1.5)
plt.plot(x,y)
# Now draw a circle with radius = 1
u = np.linspace(-1,1,100)
# Top half of circle
v = np.sqrt(1.0-u**2)
# Bottom half
v2 = -1.0*v
# Combine the top and bottom halves together
u = # Complete
v = # Complete
plt.plot(u,v)
# Uniformly sample between -1 and 1 in 2 dimensions. Do this for 1000 draws
z = # Complete
# Now figure out how many of those draws are in the circle (all are in the square by definition)
n = 0
for a,b in z:
if # Complete
plt.scatter(a,b,c='g')
n=n+1
else:
plt.scatter(a,b,c='r')
# Use that information to compute the area of the circle (without using the formula)
print # Complete
# + [markdown] slideshow={"slide_type": "slide"}
# For homework plot the distribution of results for lots of such experiments. Do you get the expected $\sigma$?
#
# + [markdown] slideshow={"slide_type": "slide"}
# In general, Monte Carlo methods are about using random sampling to obtain a numerical result (e.g., the value of an integral), where there is no analytic result.
#
# In the case of the circle above, we have computed the intergral:
# $$\int\int_{x^2+y^2\le 1} dx dy.$$
# + [markdown] slideshow={"slide_type": "slide"}
# ### What is a Markov Chain?
#
# A Markov Chain is defined as a sequence of random variables where a parameter depends *only* on the preceding value. Such processes are "memoryless".
#
# Mathematically, we have
# $$p(\theta_{i+1}|\{\theta_i\}) = p(\theta_{i+1}|\theta_i).$$
#
# Now, if you are like me, you might look at that and say "Well, day 3 is based on day 2, which is based on day 1, so day 3 is based on day 1...".
#
# So let's look at an example to see what we mean and how this might be a memoryless process.
#
# + [markdown] slideshow={"slide_type": "slide"}
# Let's say that you are an astronomer and you want to know how likely it is going to be clear tomorrow night given the weather tonight (clear or cloudy). From past history, you know that:
#
# $$p({\rm clear \; tomorrow} \, |\, {\rm cloudy \; today}) = 0.5,$$
# which means that
# $$p({\rm cloudy \; tomorrow} \, |\, {\rm cloudy \; today}) = 0.5.$$
#
# We also have
# $$p({\rm cloudy \; tomorrow} \, |\, {\rm clear \; today}) = 0.1,$$
# which means that
# $$p({\rm clear \; tomorrow} \, |\, {\rm clear \; today}) = 0.9.$$
#
# (That is, you don't live in Philadelphia.)
#
# We can start with the sky conditions today and make predictions going forward. This will look like a big decision tree. After enough days, we'll reach equilibrium probabilities that have to do with the mean weather statistics (ignoring seasons) and we'll arrive at
#
# $$p({\rm clear}) = 0.83,$$
# and
# $$p({\rm cloudy}) = 0.17.$$
#
# You get the same answer for day $N$ as day $N+1$ and it doesn't matter whether is was clear to cloudy on the day that you started.
#
# The steps that we have taken in this process are a **Markov Chain**.
# + [markdown] slideshow={"slide_type": "slide"}
# In MCMC the prior must be **stationary** which basically means that its looks the same no matter where you sample it.
#
# Obviously that isn't going to be the case in the early steps of the chain. In our example above, after some time the process was stationary, but not in the first few days.
#
# So, there is a **burn-in** phase that needs to be discarded. How one determines how long many iterations the burn-in should last when you don't know the distribution can be a bit tricky.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Markov Chain Monte Carlo Summary
#
# 1. Starting at a random position, evaluate the likelihood.
# 2. Choose a new position, according to some transition probabilities, and evaluate the likelihood there.
# 3. Examine the odds ratio formed by the new-position likelihood and the old-position likelihood. If the odds ratio is greater than 1, move to the new position. If it is less than one, keep it under the following conditions: draw a random number between zero and 1. If the odds ratio is smaller than the random number, keep it. If not, reject the new position.
# 4. Repeat 1-3 many times. After a period of time (the burn-in) the simulation should reach an equilibrium. Keep the results of the chain (after burn-in), and postprocess those results to infer the likelihood surface.
#
# + [markdown] slideshow={"slide_type": "slide"}
# Most of the difficulty in the MCMC process comes from either determining the burn-in or deciding how to step from one position to another. In our circle example we have drawn points in a completely random manner. However, that may not be the most efficient manner to span the space.
#
# The most commonly used algorithm for stepping from one position to another is the [Metropolis-Hastings] (https://en.wikipedia.org/wiki/Metropolis%E2%80%93Hastings_algorithm) algorithm.
#
# In astronomy, the ${\tt emcee}$ algorithm has become more popular in recent years. We won't discuss either in detail, but both the [code](http://dan.iel.fm/emcee/current/) and a [paper[(http://adsabs.harvard.edu/abs/2013PASP..125..306F) describing the ${\tt emcee}$ are available.
#
# Recall that our parameter space it multidimensional. So, when you are stepping from one point to another, you are really doing it in N-D parameter space! You might wonder if you could just step one parameter at a time. Sure! That's what [Gibbs sampling](https://en.wikipedia.org/wiki/Gibbs_sampling) does.
#
# + [markdown] slideshow={"slide_type": "slide"}
# Then end result of this process will be a chain of likelihoods that we can use to compute the likelihood contours.
#
# If you are using MCMC, then you probably have multiple paramters (otherwise, you'd be doing something easier). So, it helps to display the parameters two at a time, marginalizing over the other parameters. An example is given in Ivezic, Figure 5.24, which compares the model results for a single Gaussian fit to a double Gaussian fit:
#
# 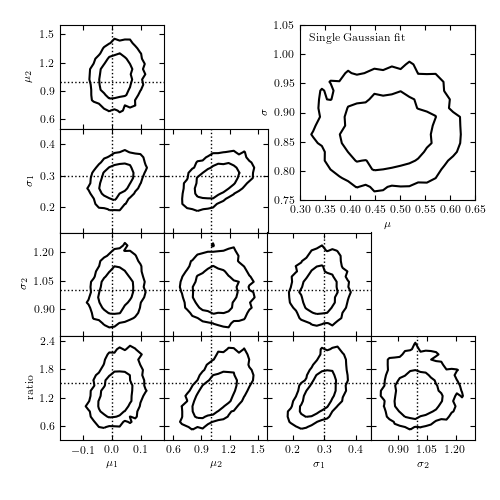
# + [markdown] slideshow={"slide_type": "slide"}
# We'll end by going through the example given at
# [http://twiecki.github.io/blog/2015/11/10/mcmc-sampling/](http://twiecki.github.io/blog/2015/11/10/mcmc-sampling/).
#
# First set up some stuff by executing the next cell
# + slideshow={"slide_type": "slide"}
# %matplotlib inline
import numpy as np
import scipy as sp
import pandas as pd
import matplotlib.pyplot as plt
from scipy.stats import norm
np.random.seed(123)
# + [markdown] slideshow={"slide_type": "slide"}
# Now let's generate some data points and plot them. We'll try a normal distribution, centered at 0 with 100 data points. Our goal is to estimate $\mu$.
# + slideshow={"slide_type": "slide"}
data = np.random.randn(100)
plt.figure(figsize=(8,8))
plt.hist(data)
plt.xlabel('x')
plt.ylabel('N')
# + [markdown] slideshow={"slide_type": "slide"}
# Now we have to pick a model to try. For the sake of simplicity for this example, we'll assume a normal distribution: $\mathscr{N}(\mu,\sigma=1)$ (i.e., with $\sigma=1$). We'll also assume a normal distribution for the prior on $\mu$: $\mathscr{N}(0,1)$.
#
# We can use that to write a function for our posterior distribution as follows:
# + slideshow={"slide_type": "slide"}
def calc_posterior_analytical(data, x, mu_0, sigma_0):
sigma = 1.
n = len(data)
mu_post = (mu_0 / sigma_0**2 + data.sum() / sigma**2) / (1. / sigma_0**2 + n / sigma**2)
sigma_post = (1. / sigma_0**2 + n / sigma**2)**-1
return norm(mu_post, np.sqrt(sigma_post)).pdf(x)
plt.figure(figsize=(8,8))
x = np.linspace(-1, 1, 500)
posterior_analytical = calc_posterior_analytical(data, x, 0., 1.)
plt.plot(x, posterior_analytical)
plt.xlabel('mu')
plt.ylabel('post prob')
# + [markdown] slideshow={"slide_type": "slide"}
# Now we need to sample the distribution space. Let's start by trying $\mu_0 = 0$ and evaluate.
#
# Then we'll jump to a new position using one of the algorithms mentioned above. In this case we'll use the Metropolis algorithm, which draws the new points from a normal distribution centered on the current guess for $\mu$.
#
# Next we evaluate whether that jump was "good" or not -- by seeing if the value of likelihood\*prior increases. Now, we want to get the right answer, but we also want to make sure that we sample the full parameter space (so that we don't) get stuck in a local minimum. So, even if the this location is not better than the last one, we'll have some probability of staying there anyway.
#
# The reason that taking the ratio of likelihood\*prior works is that the denominator drops out. That's good because the denominator is the integral of the numerator and that's what we are trying to figure out! In short, we don't have to know the posterior probability to know that the posterior probability at one step is better than another.
# + slideshow={"slide_type": "slide"}
# Execute this cell
# See https://github.com/twiecki/WhileMyMCMCGentlySamples/blob/master/content/downloads/notebooks/MCMC-sampling-for-dummies.ipynb
def sampler(data, samples=4, mu_init=.5, proposal_width=.5, plot=False, mu_prior_mu=0, mu_prior_sd=1.):
mu_current = mu_init
posterior = [mu_current]
for i in range(samples):
# suggest new position
mu_proposal = norm(mu_current, proposal_width).rvs()
# Compute likelihood by multiplying probabilities of each data point
likelihood_current = norm(mu_current, 1).pdf(data).prod()
likelihood_proposal = norm(mu_proposal, 1).pdf(data).prod()
# Compute prior probability of current and proposed mu
prior_current = norm(mu_prior_mu, mu_prior_sd).pdf(mu_current)
prior_proposal = norm(mu_prior_mu, mu_prior_sd).pdf(mu_proposal)
p_current = likelihood_current * prior_current
p_proposal = likelihood_proposal * prior_proposal
# Accept proposal?
p_accept = p_proposal / p_current
# Usually would include prior probability, which we neglect here for simplicity
accept = np.random.rand() < p_accept
if plot:
plot_proposal(mu_current, mu_proposal, mu_prior_mu, mu_prior_sd, data, accept, posterior, i)
if accept:
# Update position
mu_current = mu_proposal
posterior.append(mu_current)
return posterior
# Function to display
def plot_proposal(mu_current, mu_proposal, mu_prior_mu, mu_prior_sd, data, accepted, trace, i):
from copy import copy
trace = copy(trace)
fig, (ax1, ax2, ax3, ax4) = plt.subplots(ncols=4, figsize=(16, 4))
fig.suptitle('Iteration %i' % (i + 1))
x = np.linspace(-3, 3, 5000)
color = 'g' if accepted else 'r'
# Plot prior
prior_current = norm(mu_prior_mu, mu_prior_sd).pdf(mu_current)
prior_proposal = norm(mu_prior_mu, mu_prior_sd).pdf(mu_proposal)
prior = norm(mu_prior_mu, mu_prior_sd).pdf(x)
ax1.plot(x, prior)
ax1.plot([mu_current] * 2, [0, prior_current], marker='o', color='b')
ax1.plot([mu_proposal] * 2, [0, prior_proposal], marker='o', color=color)
ax1.annotate("", xy=(mu_proposal, 0.2), xytext=(mu_current, 0.2),
arrowprops=dict(arrowstyle="->", lw=2.))
ax1.set(ylabel='Probability Density', title='current: prior(mu=%.2f) = %.2f\nproposal: prior(mu=%.2f) = %.2f' % (mu_current, prior_current, mu_proposal, prior_proposal))
# Likelihood
likelihood_current = norm(mu_current, 1).pdf(data).prod()
likelihood_proposal = norm(mu_proposal, 1).pdf(data).prod()
y = norm(loc=mu_proposal, scale=1).pdf(x)
#sns.distplot(data, kde=False, norm_hist=True, ax=ax2)
ax2.hist(data,alpha=0.5,normed='True')
ax2.plot(x, y, color=color)
ax2.axvline(mu_current, color='b', linestyle='--', label='mu_current')
ax2.axvline(mu_proposal, color=color, linestyle='--', label='mu_proposal')
#ax2.title('Proposal {}'.format('accepted' if accepted else 'rejected'))
ax2.annotate("", xy=(mu_proposal, 0.2), xytext=(mu_current, 0.2),
arrowprops=dict(arrowstyle="->", lw=2.))
ax2.set(title='likelihood(mu=%.2f) = %.2f\nlikelihood(mu=%.2f) = %.2f' % (mu_current, 1e14*likelihood_current, mu_proposal, 1e14*likelihood_proposal))
# Posterior
posterior_analytical = calc_posterior_analytical(data, x, mu_prior_mu, mu_prior_sd)
ax3.plot(x, posterior_analytical)
posterior_current = calc_posterior_analytical(data, mu_current, mu_prior_mu, mu_prior_sd)
posterior_proposal = calc_posterior_analytical(data, mu_proposal, mu_prior_mu, mu_prior_sd)
ax3.plot([mu_current] * 2, [0, posterior_current], marker='o', color='b')
ax3.plot([mu_proposal] * 2, [0, posterior_proposal], marker='o', color=color)
ax3.annotate("", xy=(mu_proposal, 0.2), xytext=(mu_current, 0.2),
arrowprops=dict(arrowstyle="->", lw=2.))
#x3.set(title=r'prior x likelihood $\propto$ posterior')
ax3.set(title='posterior(mu=%.2f) = %.5f\nposterior(mu=%.2f) = %.5f' % (mu_current, posterior_current, mu_proposal, posterior_proposal))
if accepted:
trace.append(mu_proposal)
else:
trace.append(mu_current)
ax4.plot(trace)
ax4.set(xlabel='iteration', ylabel='mu', title='trace')
plt.tight_layout()
#plt.legend()
# + [markdown] slideshow={"slide_type": "slide"}
# To visualize the sampling, we'll create plots for some quantities that are computed. Each row below is a single iteration through our Metropolis sampler.
#
# The first column is our prior distribution -- what our belief about $\mu$ is before seeing the data. You can see how the distribution is static and we only plug in our $\mu$ proposals. The vertical lines represent our current $\mu$ in blue and our proposed $\mu$ in either red or green (rejected or accepted, respectively).
#
# The 2nd column is our likelihood and what we are using to evaluate how good our model explains the data. You can see that the likelihood function changes in response to the proposed $\mu$. The blue histogram is our data. The solid line in green or red is the likelihood with the currently proposed mu. Intuitively, the more overlap there is between likelihood and data, the better the model explains the data and the higher the resulting probability will be. The dashed line of the same color is the proposed mu and the dashed blue line is the current mu.
#
# The 3rd column is our posterior distribution. Here we are displaying the normalized posterior.
#
# The 4th column is our trace (i.e. the posterior samples of $\mu$ we're generating) where we store each sample irrespective of whether it was accepted or rejected (in which case the line just stays constant).
#
# Note that we always move to relatively more likely $\mu$ values (in terms of their posterior density), but only sometimes to relatively less likely $\mu$ values, as can be seen in iteration 14 (the iteration number can be found at the top center of each row).
#
# + slideshow={"slide_type": "slide"}
np.random.seed(123)
sampler(data, samples=8, mu_init=-1., plot=True);
# + [markdown] slideshow={"slide_type": "slide"}
# What happens when we do this lots of times?
# + slideshow={"slide_type": "slide"}
posterior = sampler(data, samples=15000, mu_init=1.)
fig, ax = plt.subplots()
ax.plot(posterior)
_ = ax.set(xlabel='sample', ylabel='mu');
# + [markdown] slideshow={"slide_type": "slide"}
# Making a histogram of these results is our estimated posterior probability distribution.
# + slideshow={"slide_type": "slide"}
ax = plt.subplot()
ax.hist(posterior[500:],bins=30,alpha=0.5,normed='True',label='estimated posterior')
x = np.linspace(-.5, .5, 500)
post = calc_posterior_analytical(data, x, 0, 1)
ax.plot(x, post, 'g', label='analytic posterior')
_ = ax.set(xlabel='mu', ylabel='belief');
ax.legend(fontsize=10);
# + [markdown] slideshow={"slide_type": "slide"}
# Our algorithm for deciding where to move to next used a normal distribution where the mean was the current value and we had to assume a width. Find where we specified that and see what happens if you make it a lot smaller or a lot bigger.
# + [markdown] slideshow={"slide_type": "slide"}
# ### More Complex Models
#
# The example above was overkill in that we were only trying to estmate $\mu$. Note also that we can do this in less than 10 lines using the ${\tt pymc3}$ module.
#
# The process is essentially the same when you add more parameters. Check out this [animation of a 2-D process](http://twiecki.github.io/blog/2014/01/02/visualizing-mcmc/) by the same author whose example we just followed.
|
Inference2.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # SciPy Optimize Module
#
#
# ## Module Contents
#
# * Optimization
# * Local
# - [1D Optimization](./Optimization_1D.ipynb)
# - [ND Optimization](./Optimization_ND.ipynb)
# - [Linear Programming](./Linear_Prog.ipynb)
# * Global Optimization
# - [Brute](./Optimization_Global_brute.ipynb)
# - [shgo](./Optimization_Global_shgo.ipynb)
# - [Differential Evolution](./Optimization_Global_differential_evolution.ipynb)
# - [Basin Hopping](./Optimization_Global_basinhopping.ipynb)
# - [Dual Annealing](./Optimization_Global_dual_annealing.ipynb)
# * Root Finding
# * [1D Roots](./Roots_1D.ipynb)
# * [ND Roots](./Roots_ND.ipynb)
# * [Curve Fitting and Least Squares](./Curve_Fit.ipynb)
# ## Optimization
#
# <img src="./images/global_local.svg" style="float:right; margin: 2px">
#
# Optimization algorithms find the smallest (or greatest) value of a function over a given range. <b>Local</b> methods find a point that is just the lowest point for some <i>neighborhood</i>, but multiple local minima can exist over the domain of the problem. <b>Global</b> methods try to find the lowest value over an entire region.
#
#
# The available local methods include `minimize_scalar`, `linprog`, and `minimize`. `minimize_scalar` only minimizes functions of one variable. `linprog` for <b>Linear Programming</b> deals with only linear functions with linear constraints. `minimize` is the most general routine. It can deal with either of those subcases in addition to any arbitrary function of many variables.
#
# Optimize provides 5 different global optimization routines. Brute directly computes points on a grid. This routine is the easiest to get working, saving programmer time at the cost of computer time. Conversely, shgo (Simplicial Homology Global Optimization) is a conceptually difficult but powerful routine. Differential Evolution, Basin Hopping, and Dual Annealing are all [Monte Carlo](https://en.wikipedia.org/wiki/Monte_Carlo_method) based, relying on random numbers. Differential Evolution is an [Evolutionary Algorithm](https://en.wikipedia.org/wiki/Evolutionary_algorithm), creating a "population" of random points and iterating it based on which ones give the lowest values. Basin Hopping and Dual Annealing instead work locally through a [Markov Chain](https://en.wikipedia.org/wiki/Markov_chain) random walk. They can computationally cope with a large number of dimensions, but they can get locally stuck and miss the global minimum. Basin Hopping provides a more straightforward base and options for a great deal of customization, and dual annealing provides a more complicated method to avoid getting locally trapped.
#
# ## Root Finding
#
# [`root`](./Roots_ND.ipynb) and [`root_scalar`](./Roots_1D.ipynb) solve the problem
# $$
# f(x) = 0.
# $$
# `root_scalar` is limited to when $x$ is a scalar, while for the more general `root` $x$ can be a multidimensional vector.
#
# ## Curve Fitting
#
# This module provides tools to optimize the fit between a parametrized function and data. `curve_fit` provides a simple interface where you don't have to worry about the guts of fitting a function. It minimizes the sum of the residuals over the parameters of the function, or:
# $$
# \text{min}_{\text{p}} \quad \sum_i \big( f(x_i , \text{p} ) - y_i \big)^2.
# $$
# If you want, you can instead pass this function to the provided non-linear least squares optimizer `least_squares`. If the model function is linear, `nnls` and `lsq_linear` exist as well.
# ## Common Traits in the Submodule
# ### Output: `OptimizeResult`
# <hr />
#
# Many functions return an object that can contain more information than simply "This is the minimium". The information varies between function, method used by the function, and flags given to function, but the way of accessing the data remains the same.
#
# Let's create one of these data types via minimization to look at it:
# +
f = lambda x : x**2
result=optimize.minimize(f,[2],method="BFGS")
# -
# You can determine what data types are availible via
result.keys()
# And you can access individual values via:
result.x
# Inspecting the object with `?` or `??` can tell you more about what the individual components actually are.
#
# In Jupyter Lab, Contextual Help, `Ctrl+I` can also provide this information.
# ? result
# ### `args`
# <hr />
#
# Many routines allow function parameters in a <b>tuple</b> to be passed to the routine via the `args` flag:
# +
f_parameter = lambda x,a : (x-a)**2
optimize.minimize(f_parameter,[0],args=(1,))
# -
# ### Methods
# <hr />
#
# The functions in `scipy.optimize` are uniform wrappers that can call to multiple different methods, algorithms, behind the scenes. For example, `minimize_scalar` can use Brent, Golden, or Bounded methods. Methods can have different strengths, weaknesses, and pitfalls. SciPy will automatically choose certain algorithms given inputted information, but if you know more about the problem, a different algorithm might be better.
#
# An example of choosing the routine:
# +
f = lambda x : x**2
optimize.minimize(f,[2],method="CG")
# -
# ### Method Options
# <hr />
#
# `minimize` itself has 14 different methods, and it's not the only routine that calls multiple methods. While much of the information and functionality is unified across the routine, each method does have it's individual settings. The settings can be found through the `show_options` function:
optimize.show_options(solver="minimize",method="CG")
# The settings are passed in a dictionary to the solver:
# +
options_dictionary = {
"maxiter": 5,
"eps": 1e-6
}
optimize.minimize(f,[2],options=options_dictionary)
# -
# ### Tolerance and Iterations
# <hr />
#
# How much computer time do you want to spend on this problem? How accurate do you need your answer? Is your function really expensive to calculate?
#
# When the two successive values are within the tolerance range of each other or the routine has reached the maximum number of iterations, the routine will exit. Some functions differentiate between <b>relative tolerance</b> and absolute tolerance</b>. Relative tolerance scales for the aboslute size of the values. For example, if two steps are five apart, but each about a trillion, the function can exit. Tolerance in the domain `x` direction also differs from the tolerance in the range `f` direction. For minimization, the `gtol` tolerance can also apply to zeroing the gradient.
#
# Some methods also allow for specifying both the maximum number of iterations and the maximum number of function evaluations. Some methods evaulate a function multiple times during each iteration.
#
# Whether these quantities exist, and the procedure for setting these quantities varies between functions and methods within functions. Check individual documentation for details, but here is one example:
optimize.minimize(f,[2],tol=1e-10,options={"maxiter":10})
|
Optimize_Module.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] deletable=true editable=true
# # Predicting Airline Data using a Generalized Linear Model (GLM) in Keras
#
# In particular, we will predict the probability that a flight is late based on its departure date/time, the expected flight time and distance, the origin and destitation airports.
#
# Most part of this notebooks are identical to what has been done in Airline Delay with a GLM in python3.ipynb
# The main difference is that we will use the [Keras](https://keras.io/) high-level library with a tensorflow backend (theano backend is also available) to perform the machine learning operations instead of scikit-learn.
#
# The core library for the dataframe part is [pandas](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.html).<br>
# The core library for the machine learning part is [Keras](https://keras.io/). This library is mostly used for deeplearning/neural-network machine learning, but it can also be used to implement most of Generalized Linear Models. It is also quite easy to add new types of model layers into the Keras API if new functionalities would be needed.
#
# The other main advantage of Keras is that it a high level API on top of either tensorflow/theano. Writting new complex model is in Keras is much more simple than in tensorflow/theano. But keep the benefits of these low-level library for what concerns the computing performances on CPU/GPU.
#
# ### Considerations
#
# The objective of this notebook is to define a simple model offerring a point of comparison in terms of computing performances across datascience language and libraries. In otherwords, this notebook is not for you if you are looking for the most accurate model in airline predictions.
# + [markdown] deletable=true editable=true
# ## Install and Load useful libraries
# + deletable=true editable=true
# %matplotlib inline
import numpy as np
import pandas as pd
import tensorflow as tf
from sklearn.metrics import roc_curve, auc
import matplotlib.pyplot as plt
# + [markdown] deletable=true editable=true
# ## Load the data (identical to python3 scikit-learn)
#
# - The dataset is taken from [http://stat-computing.org](http://stat-computing.org/dataexpo/2009/the-data.html). We take the data corresponding to year 2008.
# - We restrict the dataset to the first million rows
# - We print all column names and the first 5 rows of the dataset
# + deletable=true editable=true
df = pd.read_csv("2008.csv")
df.shape[0]
# + deletable=true editable=true
df = df[0:1000000]
# + deletable=true editable=true
df.columns
# + deletable=true editable=true
df[0:5]
# + [markdown] deletable=true editable=true
# ## Data preparation for training (identical to python3 scikit-learn)
#
# - We turn origin/destination categorical data to a "one-hot" encoding representation
# - We create a new "binary" column indicating if the flight was delayed or not.
# - We show the first 5 rows of the modified dataset
# - We split the dataset in two parts: a training dataset and a testing dataset containing 80% and 20% of the rows, respectively.
# + deletable=true editable=true
df = pd.concat([df, pd.get_dummies(df["Origin"], prefix="Origin")], axis=1);
df = pd.concat([df, pd.get_dummies(df["Dest" ], prefix="Dest" )], axis=1);
df = df.dropna(subset=["ArrDelay"])
df["IsArrDelayed" ] = (df["ArrDelay"]>0).astype(int)
df[0:5]
# + deletable=true editable=true
train = df.sample(frac=0.8)
test = df.drop(train.index)
# + [markdown] deletable=true editable=true
# ## Model building
#
# - We define the generalized linear model using a binomial function --> Logistic regression.
# - The model has linear logits = (X*W)+B = (Features * Coefficients) + Bias
# - The Loss function is a logistic function (binary_cross_entropy)
# - A L2 regularization is added to mimic what is done in scikit learn
# - Specific callbacks are defined (one for logging and one for early stopping the training)
# - We train the model and measure the training time --> ~55sec on an intel i7-6700K (4.0 GHz) with a GTX970 4GB GPU for 800K rows
# - The model is trained using a minibatch strategy (that can be tune for further performance increase)
# - We show the model coefficients
# - We show the 10 most important variables
# + deletable=true editable=true
#get the list of one hot encoding columns
OriginFeatCols = [col for col in df.columns if ("Origin_" in col)]
DestFeatCols = [col for col in df.columns if ("Dest_" in col)]
features = train[["Year","Month", "DayofMonth" ,"DayOfWeek", "DepTime", "AirTime", "Distance"] + OriginFeatCols + DestFeatCols ]
labels = train["IsArrDelayed"]
featuresMatrix = features.as_matrix()
labelsMatrix = labels .as_matrix().reshape(-1,1)
# + deletable=true editable=true
featureSize = features.shape[1]
labelSize = 1
training_epochs = 25
batch_size = 2500
from keras.models import Sequential
from keras.layers import Dense, Activation
from keras.regularizers import l2, activity_l2
from sklearn.metrics import roc_auc_score
from keras.callbacks import Callback
from keras.callbacks import EarlyStopping
#DEFINE A CUSTOM CALLBACK
class IntervalEvaluation(Callback):
def __init__(self): super(Callback, self).__init__()
def on_epoch_end(self, epoch, logs={}): print("interval evaluation - epoch: %03d - loss:%8.6f" % (epoch, logs['loss']))
#DEFINE AN EARLY STOPPING FOR THE MODEL
earlyStopping = EarlyStopping(monitor='loss', patience=1, verbose=0, mode='auto')
#DEFINE THE MODEL
model = Sequential()
model.add(Dense(labelSize, input_dim=featureSize, activation='sigmoid', W_regularizer=l2(1e-5)))
model.compile(optimizer='Adam', loss='binary_crossentropy', metrics=['accuracy'])
#FIT THE MODEL
model.fit(featuresMatrix, labelsMatrix, batch_size=batch_size, nb_epoch=training_epochs,verbose=0,callbacks=[IntervalEvaluation(),earlyStopping]);
# + deletable=true editable=true
coef = pd.DataFrame(data=model.layers[0].get_weights()[0], index=features.columns, columns=["Coef"])
coef = coef.reindex( coef["Coef"].abs().sort_values(axis=0,ascending=False).index ) #order by absolute coefficient magnitude
coef[ coef["Coef"].abs()>0 ] #keep only non-null coefficients
coef[ 0:10 ] #keep only the 10 most important coefficients
# + [markdown] deletable=true editable=true
# ## Model testing (identical to python3 scikit-learn)
#
# - We add a model prediction column to the testing dataset
# - We show the first 10 rows of the test dataset (with the new column)
# - We show the model ROC curve
# - We measure the model Area Under Curve (AUC) to be 0.689 on the testing dataset.
#
# This is telling us that our model is not super accurate (we generally assume that a model is raisonable at predicting when it has an AUC above 0.8). But, since we are not trying to build the best possible model, but just show comparison of data science code/performance accross languages/libraries.
# If none the less you are willing to improve this result, you should try adding more feature column into the model.
# + deletable=true editable=true
testFeature = test[["Year","Month", "DayofMonth" ,"DayOfWeek", "DepTime", "AirTime", "Distance"] + OriginFeatCols + DestFeatCols ]
pred = model.predict( testFeature.as_matrix() )
test["IsArrDelayedPred"] = pred
test[0:10]
# + deletable=true editable=true
fpr, tpr, _ = roc_curve(test["IsArrDelayed"], test["IsArrDelayedPred"])
AUC = auc(fpr, tpr)
plt.figure()
plt.plot(fpr, tpr, color='darkorange', lw=4, label='ROC curve (area = %0.3f)' % AUC)
plt.legend(loc=4)
# + deletable=true editable=true
AUC
# + [markdown] deletable=true editable=true
# ## Key takeaways
#
# - We built a GLM model predicting airline delay probability in tensorflow
# - We train it on 800K rows in ~55sec on an intel i7-6700K (4.0 GHz) with a GTX970 GPU
# - We measure an AUC of 0.689, which is almost identical to python-3 scikit learn results
# - We demonstrated a typical workflow in python+keras in a Jupyter notebook
# - We can easilly customize the model using the several type of layers available in Keras. That would make our model much more accurarte and sophisticated with no additional pain in either complexity or computing performance.
#
# [Keras](https://keras.io/) documentation is quite complete and contains several examples from linear algebra to advance deep learning techniques.
# + deletable=true editable=true
|
Airline Delay with a GLM in Keras.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Optimización media-varianza
#
# <img style="float: right; margin: 0px 0px 15px 15px;" src="https://upload.wikimedia.org/wikipedia/commons/d/da/Newton_optimization_vs_grad_descent.svg" width="400px" height="400px" />
#
#
# La **teoría de portafolios** es uno de los avances más importantes en las finanzas modernas e inversiones.
# - Apareció por primera vez en un [artículo corto](https://www.math.ust.hk/~maykwok/courses/ma362/07F/markowitz_JF.pdf) llamado "Portfolio Selection" en la edición de Marzo de 1952 de "the Journal of Finance".
# - Escrito por un desconocido estudiante de la Universidad de Chicago, llamado <NAME>.
# - Escrito corto (sólo 14 páginas), poco texto, fácil de entender, muchas gráficas y unas cuantas referencias.
# - No se le prestó mucha atención hasta los 60s.
#
# Finalmente, este trabajo se convirtió en una de las más grandes ideas en finanzas, y le dió a Markowitz el Premio Nobel casi 40 años después.
# - Markowitz estaba incidentalmente interesado en los mercados de acciones e inversiones.
# - Estaba más bien interesado en entender cómo las personas tomaban sus mejores decisiones cuando se enfrentaban con "trade-offs".
# - Principio de conservación de la miseria. O, dirían los instructores de gimnasio: "no pain, no gain".
# - Si queremos más de algo, tenemos que perder en algún otro lado.
# - El estudio de este fenómeno era el que le atraía a Markowitz.
#
# De manera que nadie se hace rico poniendo todo su dinero en la cuenta de ahorros. La única manera de esperar altos rendimientos es si se toma bastante riesgo. Sin embargo, riesgo significa también la posibilidad de perder, tanto como ganar.
#
# Pero, ¿qué tanto riesgo es necesario?, y ¿hay alguna manera de minimizar el riesgo mientras se maximizan las ganancias?
# - Markowitz básicamente cambió la manera en que los inversionistas pensamos acerca de esas preguntas.
# - Alteró completamente la práctica de la administración de inversiones.
# - Incluso el título de su artículo era innovador. Portafolio: una colección de activos en lugar de tener activos individuales.
# - En ese tiempo, un portafolio se refería a una carpeta de piel.
# - En el resto de este módulo, nos ocuparemos de la parte analítica de la teoría de portafolios, la cual puede ser resumida en dos frases:
# - No pain, no gain.
# - No ponga todo el blanquillo en una sola bolsa.
#
#
# **Objetivos:**
# - ¿Qué es la línea de asignación de capital?
# - ¿Qué es el radio de Sharpe?
# - ¿Cómo deberíamos asignar nuestro capital entre un activo riesgoso y un activo libre de riesgo?
#
# *Referencia:*
# - Notas del curso "Portfolio Selection and Risk Management", Rice University, disponible en Coursera.
# ___
# ## 1. Línea de asignación de capital
#
# ### 1.1. Motivación
#
# El proceso de construcción de un portafolio tiene entonces los siguientes dos pasos:
# 1. Escoger un portafolio de activos riesgosos.
# 2. Decidir qué tanto de tu riqueza invertirás en el portafolio y qué tanto invertirás en activos libres de riesgo.
#
# Al paso 2 lo llamamos **decisión de asignación de activos**.
# Preguntas importantes:
# 1. ¿Qué es el portafolio óptimo de activos riesgosos?
# - ¿Cuál es el mejor portafolio de activos riesgosos?
# - Es un portafolio eficiente en media-varianza.
# 2. ¿Qué es la distribución óptima de activos?
# - ¿Cómo deberíamos distribuir nuestra riqueza entre el portafolo riesgoso óptimo y el activo libre de riesgo?
# - Concepto de **línea de asignación de capital**.
# - Concepto de **radio de Sharpe**.
# Dos suposiciones importantes:
# - Funciones de utilidad media-varianza.
# - Inversionista averso al riesgo.
# La idea sorprendente que saldrá de este análisis, es que cualquiera que sea la actitud del inversionista de cara al riesgo, el mejor portafolio de activos riesgosos es idéntico para todos los inversionistas.
#
# Lo que nos importará a cada uno de nosotros en particular, es simplemente la desición óptima de asignación de activos.
# ___
# ### 1.2. Línea de asignación de capital
# Sean:
# - $r_s$ el rendimiento del activo riesgoso,
# - $r_f$ el rendimiento libre de riesgo, y
# - $w$ la fracción invertida en el activo riesgoso.
#
# <font color=blue> Realizar deducción de la línea de asignación de capital en el tablero.</font>
# **Tres doritos después...**
# #### Línea de asignación de capital (LAC):
# $E[r_p]$ se relaciona con $\sigma_p$ de manera afín. Es decir, mediante la ecuación de una recta:
#
# $$E[r_p]=r_f+\frac{E[r_s-r_f]}{\sigma_s}\sigma_p.$$
#
# - La pendiente de la LAC es el radio de Sharpe $\frac{E[r_s-r_f]}{\sigma_s}=\frac{E[r_s]-r_f}{\sigma_s}$,
# - el cual nos dice qué tanto rendimiento obtenemos por unidad de riesgo asumido en la tenencia del activo (portafolio) riesgoso.
# Ahora, la pregunta es, ¿dónde sobre esta línea queremos estar?
# ___
# ### 1.3. Resolviendo para la asignación óptima de capital
#
# Recapitulando de la clase pasada, tenemos las curvas de indiferencia: **queremos estar en la curva de indiferencia más alta posible, que sea tangente a la LAC**.
#
# <font color=blue> Ver en el tablero.</font>
# Analíticamente, el problema es
#
# $$\max_{w} \quad E[U(r_p)]\equiv\max_{w} \quad E[r_p]-\frac{1}{2}\gamma\sigma_p^2,$$
#
# donde los puntos $(\sigma_p,E[r_p])$ se restringen a estar en la LAC, esto es $E[r_p]=r_f+\frac{E[r_s-r_f]}{\sigma_s}\sigma_p$ y $\sigma_p=w\sigma_s$. Entonces el problema anterior se puede escribir de la siguiente manera:
#
# $$\max_{w} \quad r_f+wE[r_s-r_f]-\frac{1}{2}\gamma w^2\sigma_s^2.$$
#
# <font color=blue> Encontrar la $w$ que maximiza la anterior expresión en el tablero.</font>
# **Tres doritos después...**
# La solución es entonces:
#
# $$w^\ast=\frac{E[r_s-r_f]}{\gamma\sigma_s^2}.$$
#
# De manera intuitiva:
# - $w^\ast\propto E[r_s-r_f]$: a más exceso de rendimiento que se obtenga del activo riesgoso, más querremos invertir en él.
# - $w^\ast\propto \frac{1}{\gamma}$: mientras más averso al riesgo seas, menos querrás invertir en el activo riesgoso.
# - $w^\ast\propto \frac{1}{\sigma_s^2}$: mientras más riesgoso sea el activo, menos querrás invertir en él.
# ___
# ## 2. Ejemplo de asignación óptima de capital: acciones y billetes de EU
# Pongamos algunos números con algunos datos, para ilustrar la derivación que acabamos de hacer.
#
# En este caso, consideraremos:
# - **Portafolio riesgoso**: mercado de acciones de EU (representados en algún índice de mercado como el S&P500).
# - **Activo libre de riesgo**: billetes del departamento de tesorería de EU (T-bills).
#
# Tenemos los siguientes datos:
#
# $$E[r_{US}]=11.9\%,\quad \sigma_{US}=19.15\%, \quad r_f=1\%.$$
# Recordamos que podemos escribir la expresión de la LAC como:
#
# \begin{align}
# E[r_p]&=r_f+\left[\frac{E[r_{US}-r_f]}{\sigma_{US}}\right]\sigma_p\\
# &=0.01+\text{S.R.}\sigma_p,
# \end{align}
#
# donde $\text{S.R}=\frac{0.119-0.01}{0.1915}\approx0.569$ es el radio de Sharpe (¿qué es lo que es esto?).
#
# Grafiquemos la LAC con estos datos reales:
# Importamos librerías que vamos a utilizar
import numpy as np
import matplotlib.pyplot as plt
# %matplotlib inline
# Datos
Ers = .119
ss = .1915
rf = .01
# Radio de Sharpe para este activo
RS = (Ers - rf)/ss
# Vector de volatilidades del portafolio (sugerido: 0% a 50%)
sp = np.linspace(0,.5)
# LAC
Erp = rf + RS*sp
# Gráfica
plt.figure(figsize=(6, 4))
plt.plot(sp, Erp, lw=3, label='LAC')
plt.plot(0, rf, 'ob', ms=10, label='Libre de riesgo')
plt.plot(ss, Ers, 'or', ms=10, label='Portafolio/activo riesgoso')
plt.legend(loc='best')
plt.xlabel('Volatilidad $\sigma$')
plt.ylabel('Rendimiento esperado $E[r]$')
plt.grid()
# Bueno, y ¿en qué punto de esta línea querríamos estar?
# - Pues ya vimos que depende de tus preferencias.
# - En particular, de tu actitud de cara al riesgo, medido por tu coeficiente de aversión al riesgo.
#
# Solución al problema de asignación óptima de capital:
#
# $$\max_{w} \quad E[U(r_p)]$$
#
# $$w^\ast=\frac{E[r_s-r_f]}{\gamma\sigma_s^2}$$
# Dado que ya tenemos datos, podemos intentar para varios coeficientes de aversión al riesgo:
# importar pandas
import pandas as pd
# Crear un DataFrame con los pesos, rendimiento
# esperado y volatilidad del portafolio óptimo
# entre los activos riesgoso y libre de riesgo
# cuyo índice sean los coeficientes de aversión
# al riesgo del 1 al 10 (enteros)
gamma = np.arange(1,11)
dist_cap = pd.DataFrame({'$\gamma$':gamma,
'$w^{\ast}$':(Ers - rf) / (gamma * ss**2)})
dist_cap
g = 4.5
w_ac = (Ers - rf) / (g * ss**2)
w_ac
# ¿Cómo se interpreta $w^\ast>1$?
# - Cuando $0<w^\ast<1$, entonces $0<1-w^\ast<1$. Lo cual implica posiciones largas en el mercado de activos y en el activo libre de riesgo.
# - Por el contrario, cuando $w^\ast>1$, tenemos $1-w^\ast<0$. Lo anterior implica una posición corta en el activo libre de riesgo (suponiendo que se puede) y una posición larga (de más del 100%) en el mercado de activos: apalancamiento.
# # Anuncios parroquiales.
#
# ## 1. Quiz la siguiente clase.
#
# ## 2. Pueden consultar sus calificaciones en el siguiente [enlace](https://docs.google.com/spreadsheets/d/1BwI1Mm7B3xxJ-jQIQEDQ_WdRHyehZrQBpHGd0hY9fU4/edit?usp=sharing)
#
# <script>
# $(document).ready(function(){
# $('div.prompt').hide();
# $('div.back-to-top').hide();
# $('nav#menubar').hide();
# $('.breadcrumb').hide();
# $('.hidden-print').hide();
# });
# </script>
#
# <footer id="attribution" style="float:right; color:#808080; background:#fff;">
# Created with Jupyter by <NAME>.
# </footer>
|
Modulo3/Clase12_OptimizacionMediaVarianza.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# All content can be freely used and adapted under the terms of the
# [Creative Commons Attribution 4.0 International License](http://creativecommons.org/licenses/by/4.0/).
#
# 
#
# Agradecimentos especiais ao [<NAME>](www.leouieda.com)
# Esse documento que você está usando é um [Jupyter notebook](http://jupyter.org/). É um documento interativo que mistura texto (como esse), código (como abaixo), e o resultado de executar o código (números, texto, figuras, videos, etc).
# # Interpolação, mapas e a gravidade da Terra
# ## Objetivos
#
# * Entender a influência da interpolação na geração de mapas de dados geofísicos
# * Visualizar as variações geográficas da gravidade da Terra
# * Entender como a escala de cores utilizada nos mapas influencia nossa interpretação
# * Aprender quais são os fatores que devem ser considerados quando visualizamos um dado em mapa
# ## Instruções
#
# O notebook te fornecerá exemplos interativos que trabalham os temas abordados no questionário. Utilize esses exemplos para responder as perguntas.
#
# As células com números ao lado, como `In [1]:`, são código [Python](http://python.org/). Algumas dessas células não produzem resultado e servem de preparação para os exemplos interativos. Outras, produzem gráficos interativos. **Você deve executar todas as células, uma de cada vez**, mesmo as que não produzem gráficos.
#
# Para executar uma célula, clique em cima dela e aperte `Shift + Enter`. O foco (contorno verde ou cinza em torno da célula) deverá passar para a célula abaixo. Para rodá-la, aperte `Shift + Enter` novamente e assim por diante. Você pode executar células de texto que não acontecerá nada.
# ## Preparação
#
# Exectute as células abaixo para carregar as componentes necessárias para nossa prática. Vamos utilizar várias *bibliotecas*, inclusive uma de geofísica chamada [Fatiando a Terra](http://www.fatiando.org).
# %matplotlib inline
from __future__ import division
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.basemap import Basemap
import ipywidgets as widgets
from IPython.display import display
import seaborn
from fatiando import utils, gridder
import fatiando
from icgem import load_icgem_gdf, down_sample
# ## Interpolação
# O melhor jeito de entendermos o efeito da interpolação é fabricando alguns dados fictícios (sintéticos).
# Assim, podemos gerar os dados tanto em pontos aleatórios quanto em um grid regular.
# Isso nos permite comparar os resultados da interpolação com o *verdadeiro*. Nosso verdadeiro será um conjunto de dados medidos em um grid regular. Como se tivéssemos ido ao campo e medido em um grid regular.
# Rode a célula abaixo para gerar os dados em pontos aleatórios e em um grid regular.
area = (-5000., 5000., -5000., 5000.)
shape = (100, 100)
xp, yp = gridder.scatter(area, 100, seed=6)
x, y = [i.reshape(shape) for i in gridder.regular(area, shape)]
aletatorio = 50*utils.gaussian2d(xp, yp, 10000, 1000, angle=45)
regular = 50*utils.gaussian2d(x, y, 10000, 1000, angle=45).reshape(shape)
# Rode as duas células abaixo para gerar um gráfico interativo. Nesse gráfico você poderá controlar:
#
# * O número de pontos (em x e y) do grid utilizado na interpolação (`num_pontos`)
# * O método de interpolação utilizado (`metodo`). Pode ser interpolação cúbica ou linear.
# * Mostrar ou não os pontos de medição aleatórios no mapa interpolado.
#
# **Repare no que acontece com as bordas do mapa e onde não há observações**.
def interpolacao(num_pontos, metodo, pontos_medidos):
fig, axes = plt.subplots(1, 2, figsize=(14, 6))
ishape = (num_pontos, num_pontos)
tmp = gridder.interp(yp, xp, aletatorio, ishape, area=area, algorithm=metodo, extrapolate=True)
yi, xi, interp = [i.reshape(ishape) for i in tmp]
ranges = np.abs([interp.min(), interp.max()]).max()
kwargs = dict(cmap="RdBu_r", vmin=-ranges, vmax=ranges)
ax = axes[0]
ax.set_title(u'Pontos medidos')
ax.set_aspect('equal')
tmp = ax.scatter(yp*0.001, xp*0.001, s=80, c=aletatorio, **kwargs)
plt.colorbar(tmp, ax=ax, aspect=50, pad=0.01)
ax.set_xlabel('y (km)')
ax.set_ylabel('x (km)')
ax.set_xlim(-5, 5)
ax.set_ylim(-5, 5)
plt.tight_layout(pad=0)
ax = axes[1]
ax.set_title(u'Interpolado')
ax.set_aspect('equal')
tmp = ax.contourf(yi*0.001, xi*0.001, interp, 40, **kwargs)
plt.colorbar(tmp, ax=ax, aspect=50, pad=0.01)
if pontos_medidos:
ax.plot(yp*0.001, xp*0.001, '.k')
ax.set_xlabel('y (km)')
ax.set_ylabel('x (km)')
ax.set_xlim(-5, 5)
ax.set_ylim(-5, 5)
plt.tight_layout(pad=0)
w = widgets.interactive(interpolacao, num_pontos=(5, 100, 5), metodo=['cubic', 'linear'], pontos_medidos=False)
display(w)
# Vamos verificar se alguma das combinações chegou perto do resultado *verdadeiro*.
#
# Rode a célula abaixo para gerar um gráfico dos dados verdadeiros (gerados em um grid regular). Esse deveria ser o resultado observado se a interpolação fosse perfeita.
fig, ax = plt.subplots(1, 1, figsize=(7, 6))
ranges = np.abs([regular.min(), regular.max()]).max()
kwargs = dict(cmap="RdBu_r", vmin=-ranges, vmax=ranges)
ax.set_title(u'Verdadeiro')
ax.set_aspect('equal')
tmp = ax.contourf(y*0.001, x*0.001, regular, 40, **kwargs)
plt.colorbar(tmp, ax=ax, aspect=50, pad=0.01)
ax.plot(yp*0.001, xp*0.001, '.k')
ax.set_xlabel('y (km)')
ax.set_ylabel('x (km)')
plt.tight_layout(pad=0)
# # Gravidade do mundo
# Vamos visualizar como a gravidade da Terra varia geograficamente. Os dados da gravidade do mundo foram baixados de http://icgem.gfz-potsdam.de/ICGEM/potato/Service.html usando o modelo EIGEN-6c3stat.
#
# **As medições foram feitas em cima da superfície da Terra**, ou seja, acompanhando a topografia.
# Rode as células abaixo para carregar os dados.
dados = load_icgem_gdf('data/eigen-6c3stat-0_5-mundo.gdf')
lat, lon, grav = dados['latitude'], dados['longitude'], dados['gravity_earth']
# Vamos fazer um mapa da gravidade utilizando a [projeção Mollweid](http://en.wikipedia.org/wiki/Map_projection). Esses dados estão em mGal: 1 mGal = 10⁻⁵ m/s².
#
# Rode as duas células abaixo para gerar o gráfico (isso pode demorar um pouco).
bm = Basemap(projection='moll', lon_0=0, resolution='c')
x, y = bm(lon, lat)
plt.figure(figsize=(18, 10))
tmp = bm.contourf(x, y, grav, 100, tri=True, cmap='Reds')
plt.colorbar(orientation='horizontal', pad=0.01, aspect=50, shrink=0.5).set_label('mGal')
plt.title("Gravidade medida na superficie da Terra", fontsize=16)
# ## Escala de cor
#
# A escala de cores que utilizamos para mapear os valores pode ter um impacto grande na nossa interpretação dos resultados. Abaixo, veremos como o nosso dado de gravidade mundial fica quando utilizamos diferentes escalas de cor.
#
# As escalas podem ser divididas em 3 categorias:
#
# * lineares: as cores variam de um tom claro (geralmente branco) a uma cor (por exemplo, vermelho) de maneira linear
# * divergente: as cores variam de uma cor escura, passando por um tom claro (geralmente branco), e depois para outra cor escura.
# * raindow ou qualitativos: as cores variam sem um padrão de intensidade claro. Podem ser as cores do arco-íris ou outra combinação.
#
# Nas escalas lineares e divergentes, as cores sempre variam de baixa intensidade para alta intensidade (e vice-versa para escalas divergentes).
# Rode as células abaixo para gerar um mapa interativo da gravidade mundial. Você poderá controlar qual escala de cor você quer usar. Experimente com elas e veja como elas afetam sua percepção.
#
# **Para pensar**: Como isso pode afetar alguem que é [daltônico](https://pt.wikipedia.org/wiki/Daltonismo)?
def grav_mundial(escala_de_cor):
plt.figure(figsize=(18, 10))
tmp = bm.contourf(x, y, grav, 100, tri=True, cmap=escala_de_cor)
plt.colorbar(orientation='horizontal', pad=0.01, aspect=50, shrink=0.5).set_label('mGal')
plt.title("Escala de cor: {}".format(escala_de_cor), fontsize=16)
escalas = 'Reds Blues Greys YlOrBr RdBu BrBG PRGn Dark2 jet ocean rainbow gnuplot'.split()
w = widgets.interactive(grav_mundial, escala_de_cor=escalas)
display(w)
# # A Terra Normal e o distúrbio da gravidade
# ## Objetivos
#
# * Aprender a calcular a gravidade da Terra Normal e o distúrbio da gravidade
# * Gerar mapas do distúrbio para o mundo todo
# * Entender a relação entre o distúrbio e a isostasia
# * Observar o estado de equilíbrio isostático em diferentes regiões do planeta
# ## A Terra Normal
# "Terra Normal" é o nome que damos ao elipsóide de referência utilizado para o cálculo de anomalias da gravidade. Um elipsóide geralmente utilizado é o [WGS84](http://en.wikipedia.org/wiki/World_Geodetic_System).
#
# Existem fórmulas para calcular a gravidade (lembre-se que gravidade = gravitação + centrífuga) de um elipsóide em qualquer ponto fora dele. Porém, essas fórmulas são mais complicadas do que queremos para essa aula. Uma alternativa é utilizar a fórmula de Somigliana:
#
# $$
# \gamma(\varphi) = \frac{a \gamma_a \cos^2 \varphi + b \gamma_b \sin^2 \varphi}{\sqrt{a^2 \cos^2 \varphi + b^2 \sin^2 \varphi}}
# $$
#
# $\gamma$ é a gravidade do elipsóide calculada na latitude $\varphi$ e **sobre a superfície do elipsóide** (ou seja, altitude zero).
# $a$ e $b$ são os eixos maior e menor do elipsóide, $\gamma_a$ e $\gamma_b$ são a gravidade do elipsóide no equador e nos polos. Os valores de $a$, $b$, $\gamma_a$ e $\gamma_b$ são tabelados para cada elipsóide. Os valores abaixo são referentes ao WGS84:
#
# <table>
# <tr> <th> a </th> <td> 6378137 </td> <td> metros </td> </tr>
# <tr> <th> b </th> <td> 6356752.3142 </td> <td> metros </td> </tr>
# <tr> <th> $\gamma_a$ </th> <td> 9.7803253359 </td> <td> m/s² </td> </tr>
# <tr> <th> $\gamma_b$ </th> <td> 9.8321849378 </td> <td> m/s² </td> </tr>
# </table>
#
# Os valores foram retirados do livro:
#
# > <NAME>., and <NAME> (2006), Physical Geodesy, 2nd, corr. ed. 2006 edition., Springer, Wien ; New York.
# ### Carregando os dados e fazendo um mapa
# Depois de calcular os valores acima, precisamos carregá-los aqui no notebook para gerarmos os mapas.
#
# Primeiro, coloque o nome do seu arquivo `.csv` abaixo e execute a célula.
arquivo_dados = 'data/somigliana.csv'
# Agora, execute as células abaixo para carregar os dados e gerar um mapa com os valores que você calculou.
lon, lat, gamma = np.loadtxt(arquivo_dados, delimiter=',', unpack=True, skiprows=0, usecols=[0, 1, -1])
bm = Basemap(projection='moll', lon_0=0, resolution='c')
x, y = bm(lon, lat)
plt.figure(figsize=(18, 10))
tmp = bm.contourf(x, y, gamma, 100, tri=True, cmap='Reds')
plt.colorbar(orientation='horizontal', pad=0.01, aspect=50, shrink=0.5).set_label('mGal')
plt.title(r"Gravidade da Terra Normal ($\gamma$)", fontsize=16)
# ### Cáculo da Terra Normal no ponto de observação ($\gamma_P$)
# A fórmula de Somgliana nos dá a gravidade da Terra Normal calculada sobre o elipsóide. Nós precisamos de $\gamma$ calculado no ponto onde medimos a gravidade (P) para calcular o distúrbio. Para obter $\gamma_P$, nós podemos utilizar a **correção de ar-livre**. Essa correção nos dá uma approximação de $\gamma_P$:
#
# $$ \gamma_P \approx \gamma - 0.3086 H $$
#
# em que $H$ é a altitude em relação ao elipsóide (altitude geométrica) em **metros**. Lembrando que a correção é feita em **mGal**.
#
# Rode as células abaixo para carregar os dados de $\gamma_P$ e gerar um mapa.
arquivo_dados = 'data/freeair.csv'
gamma_p = np.loadtxt(arquivo_dados, delimiter=',', unpack=True, skiprows=0, usecols=[-1])
plt.figure(figsize=(18, 10))
tmp = bm.contourf(x, y, gamma_p, 100, tri=True, cmap='Reds')
plt.colorbar(orientation='horizontal', pad=0.01, aspect=50, shrink=0.5).set_label('mGal')
plt.title(r"Gravidade da Terra Normal em P ($\gamma_P$)", fontsize=16)
# ## Distúrbio da gravidade
# O distúrbio da gravidade é definido como:
#
# $$ \delta = g_P - \gamma_P$$
#
# em que $g_P$ é a gravidade medida no ponto P.
#
# Rode as células abaixo para carregar os valores calculados e gerar o mapa.
arquivo_dados = 'data/residual.csv'
disturbio = np.loadtxt(arquivo_dados, delimiter=',', unpack=True, skiprows=0, usecols=[-1])
def varia_escala(escala_de_cor):
plt.figure(figsize=(18, 10))
ranges = np.abs([disturbio.min(), disturbio.max()]).max()
tmp = bm.contourf(x, y, disturbio, 100, tri=True, cmap=escala_de_cor, vmin=-ranges, vmax=ranges)
plt.colorbar(orientation='horizontal', pad=0.01, aspect=50, shrink=0.5).set_label('mGal')
plt.title(u"Distúrbio da gravidade (escala de cor '{}')".format(escala_de_cor), fontsize=16)
escalas = 'Reds Blues Greys YlOrBr RdBu_r BrBG PRGn Dark2 jet ocean rainbow gnuplot'.split()
w = widgets.interactive(varia_escala, escala_de_cor=escalas)
display(w)
# # Isostasia e anomalia Bouguer
# ## Objetivos
#
# * Visualizar os mecanismos de compensação isostática de Airy e Pratt
# * Cacular e visualizar a anomalia Bouguer para o mundo todo
# ## Anomalia Bouguer
# Na prática passada, vocês calcularam o distúrbio da gravidade ($\delta$) removendo a gravidade da Terra Normal calculada no ponto de observação ($\gamma_P$). Vimos que o distúrbio nos indica o estado de equilíbrio isostático da região: se $\delta$ for pequeno e positivo a região encontra-se em equilíbro, caso contrário não está. A falta de equilíbrio isostático sugere que existem forças externas erguendo ou abaixando a topografia.
#
# Se quisermos ver o efeito gravitacional de coisas abaixo da topografia (Moho, bacias sedimentares e outras heterogeneidades), precisamos **remover o efeito gravitacional da topografia** do distúrbio. Para isso, precisamos calcular a atração gravitacional da massa topográfica (vamos chamar isso de $g_t$). A **anomalia Bouguer** é o distúrbio da gravidade menos o efeito da topografia:
#
# $$\Delta g_{bg} = \delta - g_t$$
#
# Um jeito simples de calcular $g_t$ é através de uma aproximação. Nesse caso, vamos aproximar toda a massa topográfica em baixo do ponto onde medimos a gravidade (P) por um platô infinito (o *platô de Bouguer*). Se a topografia abaixo do ponto P tem $H$ metros de **altitude em relação ao elipsóide**, podemos aproximar $g_t$ por:
#
# $$g_t \approx 2 \pi G \rho H$$
#
# em que $\rho$ é a densidade da topografia e $G$ é a contante gravitacional.
#
# Nos oceanos, não temos topografia acima do elipsóide. Porém, temos uma camada de água que não foi removida devidamente com a Terra Normal ($\gamma_P$). Podemos utilizar a aproximação do platô de Bouguer para calcular o efeito gravitacional da camada de água e removê-la do distúrbio. Assim, teremos a anomalia Bouguer para regiões continentais e oceânicas.
# ### Calculando a anomalia Bouguer
# Para fazer os cálculos, vamos precisar o valor da altitude topográfica. Nos continentes, essa altitude é a mesma da altitude na qual os dados foram medidos. Já nos oceanos, a altitude de medição é zero (superfície da água). O que precisamos realmente é da batimetria nos oceanos. Por sorte, existem modelos digitais de terreno, como o [ETOPO1](http://www.ngdc.noaa.gov/mgg/global/global.html) que nos dão topografia nos continentes e batimetria nos oceanos. O arquivo `data/etopo1-0_5-mundo.gdf` contem os dados de topografia do ETOPO1 calculado nos mesmo pontos em que a gravidade foi medida.
#
# **Dicas** para calcular o efeito gravitacional da topografia utilizando o platô de Bouguer.
#
# * Utilize a densidade $\rho_c = 2670\ kg/m^3$ para a topografia.
# * Nos oceanos, utilize a densidade $\rho_c$ para a crosta do elipsóide e $\rho_a = 1040\ kg/m^3$ para a água do mar.
# * Utilize o valor de $G = 0.00000000006673\ m^3 kg^{-1} s^{-1}$
# * O valor calculado estará em m/s². Converta para mGal = 100000 m/s²
# ### Carregando os dados e fazendo um mapa
#
# Depois de calcular os valores acima, precisamos carregá-los aqui no notebook para gerarmos os mapas.
#
# Primeiro, coloque o nome do seu arquivo `.csv` abaixo e execute a célula. **O nome deve ser exato**. Dica: apague o nome do arquivo e aperte Tab.
arquivo_dados = 'data/bouguer.csv'
# Agora, execute as células abaixo para carregar os dados e gerar um mapa com os valores que você calculou.
lon, lat, bouguer = np.loadtxt(arquivo_dados, delimiter=',', unpack=True, skiprows=0, usecols=[0, 1, -1])
bm = Basemap(projection='moll', lon_0=0, resolution='c')
x, y = bm(lon, lat)
plt.figure(figsize=(18, 10))
ranges = np.abs([bouguer.min(), bouguer.max()]).max()
tmp = bm.contourf(x, y, bouguer, 100, tri=True, cmap='RdBu_r', vmin=-ranges, vmax=ranges)
plt.colorbar(orientation='horizontal', pad=0.01, aspect=50, shrink=0.5).set_label('mGal')
plt.title(r"Anomalia Bouguer", fontsize=16)
# # Inversão de dados de uma bacia sedimentar 2D poligonal
#
# ## Objetivos
#
# * Entender melhor como funciona a inversão de dados
# A célula abaixo _prepara_ o ambiente
from fatiando.inversion import Smoothness1D
from fatiando.gravmag.basin2d import PolygonalBasinGravity
from fatiando.gravmag import talwani
from fatiando.mesher import Polygon
from fatiando.vis import mpl
from fatiando import utils
import numpy as np
# A célula abaixo cria dados sintéticos para testar a inversão de dados. O resultado será um polígono.
noise = 5
# Make some synthetic data to test the inversion
# The model will be a polygon.
# Reverse x because vertices must be clockwise.
xs = np.linspace(0, 100000, 100)[::-1]
depths = (-1e-15*(xs - 50000)**4 + 8000 -
3000*np.exp(-(xs - 70000)**2/(10000**2)))
depths -= depths.min() # Reduce depths to zero
props = {'density': -300}
model = Polygon(np.transpose([xs, depths]), props)
x = np.linspace(0, 100000, 100)
z = -100*np.ones_like(x)
data = utils.contaminate(talwani.gz(x, z, [model]), noise, seed=0)
# A célula abaixo executa a inversão, dada as condições iniciais descritas em `initial`
# Make the solver using smoothness regularization and run the inversion
misfit = PolygonalBasinGravity(x, z, data, 50, props, top=0)
regul = Smoothness1D(misfit.nparams)
solver = misfit + 1e-4*regul
# This is a non-linear problem so we need to pick an initial estimate
initial = 3000*np.ones(misfit.nparams)
solver.config('levmarq', initial=initial).fit()
# A célula abaixo cria a imagem da bacia e mostra o ajuste dos dados
# %matplotlib inline
mpl.figure()
mpl.subplot(2, 1, 1)
mpl.plot(x, data, 'ok', label='observed')
mpl.plot(x, solver[0].predicted(), '-r', linewidth=2, label='predicted')
mpl.legend()
ax = mpl.subplot(2, 1, 2)
mpl.polygon(model, fill='gray', alpha=0.5, label='True')
# The estimate_ property of our solver gives us the estimate basin as a polygon
# So we can directly pass it to plotting and forward modeling functions
mpl.polygon(solver.estimate_, style='o-r', label='Estimated')
ax.invert_yaxis()
mpl.legend()
mpl.show()
|
notebooks/gravmetria/GRAV_somigliana_ar-livre_bouguer-v02.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Language Translation
# In this project, you’re going to take a peek into the realm of neural network machine translation. You’ll be training a sequence to sequence model on a dataset of English and French sentences that can translate new sentences from English to French.
# ## Get the Data
# Since translating the whole language of English to French will take lots of time to train, we have provided you with a small portion of the English corpus.
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import helper
import problem_unittests as tests
source_path = 'data/small_vocab_en'
target_path = 'data/small_vocab_fr'
source_text = helper.load_data(source_path)
target_text = helper.load_data(target_path)
# -
# ## Explore the Data
# Play around with view_sentence_range to view different parts of the data.
# +
view_sentence_range = (0, 10)
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import numpy as np
print('Dataset Stats')
print('Roughly the number of unique words: {}'.format(len({word: None for word in source_text.split()})))
sentences = source_text.split('\n')
word_counts = [len(sentence.split()) for sentence in sentences]
print('Number of sentences: {}'.format(len(sentences)))
print('Average number of words in a sentence: {}'.format(np.average(word_counts)))
print()
print('English sentences {} to {}:'.format(*view_sentence_range))
print('\n'.join(source_text.split('\n')[view_sentence_range[0]:view_sentence_range[1]]))
print()
print('French sentences {} to {}:'.format(*view_sentence_range))
print('\n'.join(target_text.split('\n')[view_sentence_range[0]:view_sentence_range[1]]))
# -
# ## Implement Preprocessing Function
# ### Text to Word Ids
# As you did with other RNNs, you must turn the text into a number so the computer can understand it. In the function `text_to_ids()`, you'll turn `source_text` and `target_text` from words to ids. However, you need to add the `<EOS>` word id at the end of each sentence from `target_text`. This will help the neural network predict when the sentence should end.
#
# You can get the `<EOS>` word id by doing:
# ```python
# target_vocab_to_int['<EOS>']
# ```
# You can get other word ids using `source_vocab_to_int` and `target_vocab_to_int`.
# +
def text_to_ids(source_text, target_text, source_vocab_to_int, target_vocab_to_int):
"""
Convert source and target text to proper word ids
:param source_text: String that contains all the source text.
:param target_text: String that contains all the target text.
:param source_vocab_to_int: Dictionary to go from the source words to an id
:param target_vocab_to_int: Dictionary to go from the target words to an id
:return: A tuple of lists (source_id_text, target_id_text)
"""
source_id_text = [[source_vocab_to_int[word] for word in sent.split()] for sent in source_text.split("\n")]
target_id_text = [[target_vocab_to_int[word] for word in (sent + ' <EOS>').split()] for sent in target_text.split("\n")]
return (source_id_text, target_id_text)
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_text_to_ids(text_to_ids)
# -
# ### Preprocess all the data and save it
# Running the code cell below will preprocess all the data and save it to file.
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
helper.preprocess_and_save_data(source_path, target_path, text_to_ids)
# # Check Point
# This is your first checkpoint. If you ever decide to come back to this notebook or have to restart the notebook, you can start from here. The preprocessed data has been saved to disk.
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import numpy as np
import helper
(source_int_text, target_int_text), (source_vocab_to_int, target_vocab_to_int), _ = helper.load_preprocess()
# -
# ### Check the Version of TensorFlow and Access to GPU
# This will check to make sure you have the correct version of TensorFlow and access to a GPU
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
from distutils.version import LooseVersion
import warnings
import tensorflow as tf
# Check TensorFlow Version
assert LooseVersion(tf.__version__) in [LooseVersion('1.0.0'), LooseVersion('1.0.1')], 'This project requires TensorFlow version 1.0 You are using {}'.format(tf.__version__)
print('TensorFlow Version: {}'.format(tf.__version__))
# Check for a GPU
if not tf.test.gpu_device_name():
warnings.warn('No GPU found. Please use a GPU to train your neural network.')
else:
print('Default GPU Device: {}'.format(tf.test.gpu_device_name()))
# -
# ## Build the Neural Network
# You'll build the components necessary to build a Sequence-to-Sequence model by implementing the following functions below:
# - `model_inputs`
# - `process_decoding_input`
# - `encoding_layer`
# - `decoding_layer_train`
# - `decoding_layer_infer`
# - `decoding_layer`
# - `seq2seq_model`
#
# ### Input
# Implement the `model_inputs()` function to create TF Placeholders for the Neural Network. It should create the following placeholders:
#
# - Input text placeholder named "input" using the TF Placeholder name parameter with rank 2.
# - Targets placeholder with rank 2.
# - Learning rate placeholder with rank 0.
# - Keep probability placeholder named "keep_prob" using the TF Placeholder name parameter with rank 0.
#
# Return the placeholders in the following the tuple (Input, Targets, Learing Rate, Keep Probability)
# +
def model_inputs():
"""
Create TF Placeholders for input, targets, and learning rate.
:return: Tuple (input, targets, learning rate, keep probability)
"""
# TODO: Implement Function
input = tf.placeholder(tf.int32, shape=(None, None), name='input')
targets = tf.placeholder(tf.int32, shape=(None, None))
lr = tf.placeholder(tf.float32)
keep_prob = tf.placeholder(tf.float32, name='keep_prob')
return (input, targets, lr, keep_prob)
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_model_inputs(model_inputs)
# -
# ### Process Decoding Input
# Implement `process_decoding_input` using TensorFlow to remove the last word id from each batch in `target_data` and concat the GO ID to the beginning of each batch.
# +
def process_decoding_input(target_data, target_vocab_to_int, batch_size):
"""
Preprocess target data for decoding
:param target_data: Target Placeholder
:param target_vocab_to_int: Dictionary to go from the target words to an id
:param batch_size: Batch Size
:return: Preprocessed target data
"""
# TODO: Implement Function
ending = tf.strided_slice(target_data, begin=[0, 0], end=[batch_size, -1], strides=[1, 1])
dec_input = tf.concat([tf.fill([batch_size, 1], target_vocab_to_int['<GO>']), ending], 1)
return dec_input
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_process_decoding_input(process_decoding_input)
# -
# ### Encoding
# Implement `encoding_layer()` to create a Encoder RNN layer using [`tf.nn.dynamic_rnn()`](https://www.tensorflow.org/api_docs/python/tf/nn/dynamic_rnn).
# +
def encoding_layer(rnn_inputs, rnn_size, num_layers, keep_prob):
"""
Create encoding layer
:param rnn_inputs: Inputs for the RNN
:param rnn_size: RNN Size
:param num_layers: Number of layers
:param keep_prob: Dropout keep probability
:return: RNN state
"""
# TODO: Implement Function
enc_cell = tf.contrib.rnn.MultiRNNCell([tf.contrib.rnn.BasicLSTMCell(rnn_size) for _ in range(num_layers)])
dropout = tf.contrib.rnn.DropoutWrapper(enc_cell, output_keep_prob=keep_prob)
_, enc_state = tf.nn.dynamic_rnn(dropout, rnn_inputs, dtype=tf.float32)
return enc_state
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_encoding_layer(encoding_layer)
# -
# ### Decoding - Training
# Create training logits using [`tf.contrib.seq2seq.simple_decoder_fn_train()`](https://www.tensorflow.org/versions/r1.0/api_docs/python/tf/contrib/seq2seq/simple_decoder_fn_train) and [`tf.contrib.seq2seq.dynamic_rnn_decoder()`](https://www.tensorflow.org/versions/r1.0/api_docs/python/tf/contrib/seq2seq/dynamic_rnn_decoder). Apply the `output_fn` to the [`tf.contrib.seq2seq.dynamic_rnn_decoder()`](https://www.tensorflow.org/versions/r1.0/api_docs/python/tf/contrib/seq2seq/dynamic_rnn_decoder) outputs.
# +
def decoding_layer_train(encoder_state, dec_cell, dec_embed_input, sequence_length, decoding_scope,
output_fn, keep_prob):
"""
Create a decoding layer for training
:param encoder_state: Encoder State
:param dec_cell: Decoder RNN Cell
:param dec_embed_input: Decoder embedded input
:param sequence_length: Sequence Length
:param decoding_scope: TenorFlow Variable Scope for decoding
:param output_fn: Function to apply the output layer
:param keep_prob: Dropout keep probability
:return: Train Logits
"""
# TODO: Implement Function
# drop out
dec_cell = tf.contrib.rnn.DropoutWrapper(dec_cell, output_keep_prob=keep_prob)
# generates a decoder fn
dynamic_fn_train = tf.contrib.seq2seq.simple_decoder_fn_train(encoder_state)
outputs_train, _, _ = tf.contrib.seq2seq.dynamic_rnn_decoder(
cell=dec_cell, decoder_fn=dynamic_fn_train, inputs=dec_embed_input,
sequence_length=sequence_length, scope=decoding_scope
)
# Apply output function
train_logits = output_fn(outputs_train)
return train_logits
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_decoding_layer_train(decoding_layer_train)
# -
# ### Decoding - Inference
# Create inference logits using [`tf.contrib.seq2seq.simple_decoder_fn_inference()`](https://www.tensorflow.org/versions/r1.0/api_docs/python/tf/contrib/seq2seq/simple_decoder_fn_inference) and [`tf.contrib.seq2seq.dynamic_rnn_decoder()`](https://www.tensorflow.org/versions/r1.0/api_docs/python/tf/contrib/seq2seq/dynamic_rnn_decoder).
# +
def decoding_layer_infer(encoder_state, dec_cell, dec_embeddings, start_of_sequence_id, end_of_sequence_id,
maximum_length, vocab_size, decoding_scope, output_fn, keep_prob):
"""
Create a decoding layer for inference
:param encoder_state: Encoder state
:param dec_cell: Decoder RNN Cell
:param dec_embeddings: Decoder embeddings
:param start_of_sequence_id: GO ID
:param end_of_sequence_id: EOS Id
:param maximum_length: The maximum allowed time steps to decode
:param vocab_size: Size of vocabulary
:param decoding_scope: TensorFlow Variable Scope for decoding
:param output_fn: Function to apply the output layer
:param keep_prob: Dropout keep probability
:return: Inference Logits
"""
# TODO: Implement Function
dynamic_decoder_fn_inf = tf.contrib.seq2seq.simple_decoder_fn_inference(
output_fn, encoder_state, dec_embeddings, start_of_sequence_id,
end_of_sequence_id, maximum_length - 1, vocab_size)
inference_logits, _, _ = tf.contrib.seq2seq.dynamic_rnn_decoder(dec_cell, dynamic_decoder_fn_inf, scope=decoding_scope)
return inference_logits
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_decoding_layer_infer(decoding_layer_infer)
# -
# ### Build the Decoding Layer
# Implement `decoding_layer()` to create a Decoder RNN layer.
#
# - Create RNN cell for decoding using `rnn_size` and `num_layers`.
# - Create the output fuction using [`lambda`](https://docs.python.org/3/tutorial/controlflow.html#lambda-expressions) to transform it's input, logits, to class logits.
# - Use the your `decoding_layer_train(encoder_state, dec_cell, dec_embed_input, sequence_length, decoding_scope, output_fn, keep_prob)` function to get the training logits.
# - Use your `decoding_layer_infer(encoder_state, dec_cell, dec_embeddings, start_of_sequence_id, end_of_sequence_id, maximum_length, vocab_size, decoding_scope, output_fn, keep_prob)` function to get the inference logits.
#
# Note: You'll need to use [tf.variable_scope](https://www.tensorflow.org/api_docs/python/tf/variable_scope) to share variables between training and inference.
# +
def decoding_layer(dec_embed_input, dec_embeddings, encoder_state, vocab_size, sequence_length, rnn_size,
num_layers, target_vocab_to_int, keep_prob):
"""
Create decoding layer
:param dec_embed_input: Decoder embedded input
:param dec_embeddings: Decoder embeddings
:param encoder_state: The encoded state
:param vocab_size: Size of vocabulary
:param sequence_length: Sequence Length
:param rnn_size: RNN Size
:param num_layers: Number of layers
:param target_vocab_to_int: Dictionary to go from the target words to an id
:param keep_prob: Dropout keep probability
:return: Tuple of (Training Logits, Inference Logits)
"""
# TODO: Implement Function
# dec cell
dec_cell = tf.contrib.rnn.MultiRNNCell([tf.contrib.rnn.BasicLSTMCell(rnn_size) for _ in range(num_layers)])
with tf.variable_scope("decoding") as decoding_scope:
# output layer, None for linear act. fn
output_fn = lambda x: tf.contrib.layers.fully_connected(x, vocab_size, None, scope=decoding_scope)
train_logits = decoding_layer_train(encoder_state, dec_cell, dec_embed_input, sequence_length, decoding_scope,
output_fn, keep_prob)
with tf.variable_scope("decoding", reuse=True) as decoding_scope:
inf_logits = decoding_layer_infer(encoder_state, dec_cell, dec_embeddings, target_vocab_to_int['<GO>'],
target_vocab_to_int['<EOS>'], sequence_length,
vocab_size, decoding_scope, output_fn, keep_prob)
return train_logits, inf_logits
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_decoding_layer(decoding_layer)
# -
# ### Build the Neural Network
# Apply the functions you implemented above to:
#
# - Apply embedding to the input data for the encoder.
# - Encode the input using your `encoding_layer(rnn_inputs, rnn_size, num_layers, keep_prob)`.
# - Process target data using your `process_decoding_input(target_data, target_vocab_to_int, batch_size)` function.
# - Apply embedding to the target data for the decoder.
# - Decode the encoded input using your `decoding_layer(dec_embed_input, dec_embeddings, encoder_state, vocab_size, sequence_length, rnn_size, num_layers, target_vocab_to_int, keep_prob)`.
# +
def seq2seq_model(input_data, target_data, keep_prob, batch_size, sequence_length, source_vocab_size, target_vocab_size,
enc_embedding_size, dec_embedding_size, rnn_size, num_layers, target_vocab_to_int):
"""
Build the Sequence-to-Sequence part of the neural network
:param input_data: Input placeholder
:param target_data: Target placeholder
:param keep_prob: Dropout keep probability placeholder
:param batch_size: Batch Size
:param sequence_length: Sequence Length
:param source_vocab_size: Source vocabulary size
:param target_vocab_size: Target vocabulary size
:param enc_embedding_size: Decoder embedding size
:param dec_embedding_size: Encoder embedding size
:param rnn_size: RNN Size
:param num_layers: Number of layers
:param target_vocab_to_int: Dictionary to go from the target words to an id
:return: Tuple of (Training Logits, Inference Logits)
"""
# TODO: Implement Function
enc_embed_input = tf.contrib.layers.embed_sequence(input_data, source_vocab_size, enc_embedding_size)
enc_state = encoding_layer(enc_embed_input, rnn_size, num_layers, keep_prob)
dec_input = process_decoding_input(target_data, target_vocab_to_int, batch_size)
dec_embeddings = tf.Variable(tf.random_uniform([target_vocab_size, dec_embedding_size]))
dec_embed_input = tf.nn.embedding_lookup(dec_embeddings, dec_input)
train_logits, inf_logits = decoding_layer(dec_embed_input, dec_embeddings, enc_state, target_vocab_size,
sequence_length, rnn_size, num_layers, target_vocab_to_int, keep_prob)
return train_logits, inf_logits
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_seq2seq_model(seq2seq_model)
# -
# ## Neural Network Training
# ### Hyperparameters
# Tune the following parameters:
#
# - Set `epochs` to the number of epochs.
# - Set `batch_size` to the batch size.
# - Set `rnn_size` to the size of the RNNs.
# - Set `num_layers` to the number of layers.
# - Set `encoding_embedding_size` to the size of the embedding for the encoder.
# - Set `decoding_embedding_size` to the size of the embedding for the decoder.
# - Set `learning_rate` to the learning rate.
# - Set `keep_probability` to the Dropout keep probability
# Number of Epochs
epochs = 10
# Batch Size
batch_size = 256
# RNN Size
rnn_size = 256
# Number of Layers
num_layers = 2
# Embedding Size
encoding_embedding_size = 100
decoding_embedding_size = 100
# Learning Rate
learning_rate = 0.002
# Dropout Keep Probability
keep_probability = 0.7
# ### Build the Graph
# Build the graph using the neural network you implemented.
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
save_path = 'checkpoints/dev'
(source_int_text, target_int_text), (source_vocab_to_int, target_vocab_to_int), _ = helper.load_preprocess()
max_source_sentence_length = max([len(sentence) for sentence in source_int_text])
train_graph = tf.Graph()
with train_graph.as_default():
input_data, targets, lr, keep_prob = model_inputs()
sequence_length = tf.placeholder_with_default(max_source_sentence_length, None, name='sequence_length')
input_shape = tf.shape(input_data)
train_logits, inference_logits = seq2seq_model(
tf.reverse(input_data, [-1]), targets, keep_prob, batch_size, sequence_length, len(source_vocab_to_int), len(target_vocab_to_int),
encoding_embedding_size, decoding_embedding_size, rnn_size, num_layers, target_vocab_to_int)
tf.identity(inference_logits, 'logits')
with tf.name_scope("optimization"):
# Loss function
cost = tf.contrib.seq2seq.sequence_loss(
train_logits,
targets,
tf.ones([input_shape[0], sequence_length]))
# Optimizer
optimizer = tf.train.AdamOptimizer(lr)
# Gradient Clipping
gradients = optimizer.compute_gradients(cost)
capped_gradients = [(tf.clip_by_value(grad, -1., 1.), var) for grad, var in gradients if grad is not None]
train_op = optimizer.apply_gradients(capped_gradients)
# -
# ### Train
# Train the neural network on the preprocessed data. If you have a hard time getting a good loss, check the forums to see if anyone is having the same problem.
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import time
def get_accuracy(target, logits):
"""
Calculate accuracy
"""
max_seq = max(target.shape[1], logits.shape[1])
if max_seq - target.shape[1]:
target = np.pad(
target,
[(0,0),(0,max_seq - target.shape[1])],
'constant')
if max_seq - logits.shape[1]:
logits = np.pad(
logits,
[(0,0),(0,max_seq - logits.shape[1]), (0,0)],
'constant')
return np.mean(np.equal(target, np.argmax(logits, 2)))
train_source = source_int_text[batch_size:]
train_target = target_int_text[batch_size:]
valid_source = helper.pad_sentence_batch(source_int_text[:batch_size])
valid_target = helper.pad_sentence_batch(target_int_text[:batch_size])
with tf.Session(graph=train_graph) as sess:
sess.run(tf.global_variables_initializer())
for epoch_i in range(epochs):
for batch_i, (source_batch, target_batch) in enumerate(
helper.batch_data(train_source, train_target, batch_size)):
start_time = time.time()
_, loss = sess.run(
[train_op, cost],
{input_data: source_batch,
targets: target_batch,
lr: learning_rate,
sequence_length: target_batch.shape[1],
keep_prob: keep_probability})
if batch_i % 200 == 0 and batch_i > 0:
batch_train_logits = sess.run(
inference_logits,
{input_data: source_batch, keep_prob: 1.0})
batch_valid_logits = sess.run(
inference_logits,
{input_data: valid_source, keep_prob: 1.0})
train_acc = get_accuracy(target_batch, batch_train_logits)
valid_acc = get_accuracy(np.array(valid_target), batch_valid_logits)
end_time = time.time()
print('Epoch {:>3} Batch {:>4}/{} - Train Accuracy: {:>6.3f}, Validation Accuracy: {:>6.3f}, Loss: {:>6.3f}'
.format(epoch_i, batch_i, len(source_int_text) // batch_size, train_acc, valid_acc, loss))
# Save Model
saver = tf.train.Saver()
saver.save(sess, save_path)
print('Model Trained and Saved')
# -
# ### Save Parameters
# Save the `batch_size` and `save_path` parameters for inference.
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
# Save parameters for checkpoint
helper.save_params(save_path)
# # Checkpoint
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import tensorflow as tf
import numpy as np
import helper
import problem_unittests as tests
_, (source_vocab_to_int, target_vocab_to_int), (source_int_to_vocab, target_int_to_vocab) = helper.load_preprocess()
load_path = helper.load_params()
# -
# ## Sentence to Sequence
# To feed a sentence into the model for translation, you first need to preprocess it. Implement the function `sentence_to_seq()` to preprocess new sentences.
#
# - Convert the sentence to lowercase
# - Convert words into ids using `vocab_to_int`
# - Convert words not in the vocabulary, to the `<UNK>` word id.
# +
def sentence_to_seq(sentence, vocab_to_int):
"""
Convert a sentence to a sequence of ids
:param sentence: String
:param vocab_to_int: Dictionary to go from the words to an id
:return: List of word ids
"""
# TODO: Implement Function
sent = sentence.lower()
unk_id = vocab_to_int['<UNK>']
ids = [vocab_to_int.get(word, unk_id) for word in sent.split()]
return ids
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_sentence_to_seq(sentence_to_seq)
# -
# ## Translate
# This will translate `translate_sentence` from English to French.
# ### Google translation result:
# > il a vu un vieux camion jaune
#
# ### My seq2seq model translation result:
# > il a vu un camion jaune
#
# rouge means red in English
# +
translate_sentence = 'he saw a old yellow truck .'
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
translate_sentence = sentence_to_seq(translate_sentence, source_vocab_to_int)
loaded_graph = tf.Graph()
with tf.Session(graph=loaded_graph) as sess:
# Load saved model
loader = tf.train.import_meta_graph(load_path + '.meta')
loader.restore(sess, load_path)
input_data = loaded_graph.get_tensor_by_name('input:0')
logits = loaded_graph.get_tensor_by_name('logits:0')
keep_prob = loaded_graph.get_tensor_by_name('keep_prob:0')
translate_logits = sess.run(logits, {input_data: [translate_sentence], keep_prob: 1.0})[0]
print('Input')
print(' Word Ids: {}'.format([i for i in translate_sentence]))
print(' English Words: {}'.format([source_int_to_vocab[i] for i in translate_sentence]))
print('\nPrediction')
print(' Word Ids: {}'.format([i for i in np.argmax(translate_logits, 1)]))
print(' French Words: {}'.format([target_int_to_vocab[i] for i in np.argmax(translate_logits, 1)]))
# -
# ## Imperfect Translation
# You might notice that some sentences translate better than others. Since the dataset you're using only has a vocabulary of 227 English words of the thousands that you use, you're only going to see good results using these words. Additionally, the translations in this data set were made by Google translate, so the translations themselves aren't particularly good. (We apologize to the French speakers out there!) Thankfully, for this project, you don't need a perfect translation. However, if you want to create a better translation model, you'll need better data.
#
# You can train on the [WMT10 French-English corpus](http://www.statmt.org/wmt10/training-giga-fren.tar). This dataset has more vocabulary and richer in topics discussed. However, this will take you days to train, so make sure you've a GPU and the neural network is performing well on dataset we provided. Just make sure you play with the WMT10 corpus after you've submitted this project.
# ## Submitting This Project
# When submitting this project, make sure to run all the cells before saving the notebook. Save the notebook file as "dlnd_language_translation.ipynb" and save it as a HTML file under "File" -> "Download as". Include the "helper.py" and "problem_unittests.py" files in your submission.
|
language-translation/dlnd_language_translation.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import matplotlib.pyplot as plt
from matplotlib import cm
from matplotlib.ticker import LinearLocator, FormatStrFormatter
from sklearn.model_selection import train_test_split
from sklearn.linear_model import Lasso, Ridge
plt.rcParams.update({'font.size': 20})
import numpy as np
# -
# # Model order selection
#
#
# We will try fitting a polynomial model to noisy data, where the true signal is a sinusoid.
#
# +
npoints = 20
sigma = 0.2
x = 2*np.pi*np.arange(npoints)/npoints
yorig = np.sin(x)
# Seeding the random number generator to get consistent results. You may change this
np.random.seed(seed=10)
y = yorig + sigma*np.random.normal(size=npoints)
fig = plt.figure()
ax = fig.gca()
cs = ax.plot(x, y,'ro',label='Noisy samples')
cs = ax.plot(x, yorig,'b')
cs = ax.plot(x,yorig,'ko',label='Original samples')
legend = ax.legend(loc='lower left', shadow=True, fontsize='x-small')
s=plt.title('Data')
# -
# ### Solving using polyfit (built-in function) with d=6
#
# Polyfit fits a polynomial of degree d to the data to obtain the parameters
# +
d = 13 # degree 1 polynomial
weights = np.polyfit(x,y,d) # Last argument is degree of polynomial
f = np.polyval(weights,x)
# Plotting
fig1 = plt.figure()
ax1 = fig1.gca()
s=ax1.plot(x, y,'ro',label='Measurements')
s=plt.plot(x,f,'r--',label='Fit')
s=plt.plot(x,yorig,label='Original')
s=plt.ylabel('y')
s=plt.xlabel('x')
legend = ax1.legend(loc='lower left', shadow=True, fontsize='x-small')
plt.title('Fits')
plt.show()
fig2 = plt.figure()
plt.stem(weights,use_line_collection=True)
s=plt.title('Coefficients')
# -
# ## Plotting the fit error as a function of the model order
#
# We will vary the model order and perform fitting, while evaluating the error in the fits. Note that the error decreases as the model order increases. Polyfit will spit out warnings with increasing d as the matrix $\mathbf X^T\mathbf X$ becomes closer to non-invertible.
# <font color=red> You should pay attention to the warnings since the fits may be poor. You need to choose a lower model order or add regularization. </font>
# + tags=[]
err_array = []
for d in range(20):
weights = np.polyfit(x,y,d)
f = np.polyval(weights,x)
err_array.append(np.linalg.norm(f-y))
s=plt.plot(err_array)
s=plt.title("Training error with increasing model order")
# -
# ## <font color=red> Validation for model order selection</font>
#
# You will split the training data to validation data and fitting data. The fit will be performed using the fitting data. Store the training errors and validation errors and plot. Determine the optimal model order from the plots as the minimum of the validation error.
# + tags=[]
# Random splitting routine from scikit_learn
x_train, x_validation , y_train, y_validation = train_test_split(x, y, test_size=0.25,random_state=32)
train_err_array = []
validation_err_array = []
fig3 = plt.figure()
ax3 = fig3.gca()
for model_order in range(18):
weights = np.polyfit(x_train,y_train,model_order)
f_train = np.polyval(weights,x_train)
train_err_array.append(np.linalg.norm(f_train-y_train))
f_validation = np.polyval(weights,x_validation)
validation_err_array.append(np.linalg.norm(f_validation-y_validation))
s=plt.plot(train_err_array,'r',label='Training error')
s=plt.plot(validation_err_array,label='Validation error')
s=plt.plot(validation_err_array,'k*')
s = plt.xlabel('model order')
s = plt.ylabel('Error')
s = plt.ylim([0,5])
s = plt.grid()
legend = ax3.legend(loc='upper left', shadow=True, fontsize='x-small')
plt.show()
# -
# <font color=red>Pick the model order that gives you the minimum validation error and evaluate the fit to the data. Show the fit as well </font>
# +
# YOUR CODE HERE
minValModOrder = np.asarray(validation_err_array).argmin()
weights = np.polyfit(x_train, y_train, minValModOrder)
f_optimal_model_order = np.polyval(weights, x)
# Plotting
fig1 = plt.figure()
ax1 = fig1.gca()
s=ax1.plot(x, y,'ro',label='Measurements')
s=plt.plot(x,f_optimal_model_order,'r--',label='Optimal fit')
s=plt.plot(x,yorig,label='Original')
s=plt.ylabel('y')
s=plt.xlabel('x')
legend = ax1.legend(loc='lower left', shadow=True, fontsize='x-small')
plt.title('Optimal model order fit')
plt.show()
fig2 = plt.figure()
plt.stem(weights,use_line_collection=True)
s=plt.title('Coefficients')
# -
|
model_order.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [default]
# language: python
# name: python3
# ---
# # Simple dynamic seq2seq with TensorFlow
# This tutorial covers building seq2seq using dynamic unrolling with TensorFlow.
#
# I wasn't able to find any existing implementation of dynamic seq2seq with TF (as of 01.01.2017), so I decided to learn how to write my own, and document what I learn in the process.
#
# I deliberately try to be as explicit as possible. As it currently stands, TF code is the best source of documentation on itself, and I have a feeling that many conventions and design decisions are not documented anywhere except in the brains of Google Brain engineers.
#
# I hope this will be useful to people whose brains are wired like mine.
#
# **UPDATE**: as of r1.0 @ 16.02.2017, there is new official implementation in `tf.contrib.seq2seq`. See [tutorial #3](3-seq2seq-native-new.ipynb). Official tutorial reportedly be up soon. Personally I still find wiring dynamic encoder-decoder by hand insightful in many ways.
# Here we implement plain seq2seq — forward-only encoder + decoder without attention. I'll try to follow closely the original architecture described in [Sutskever, Vinyals and Le (2014)](https://arxiv.org/abs/1409.3215). If you notice any deviations, please let me know.
# Architecture diagram from their paper:
# 
# Rectangles are encoder and decoder's recurrent layers. Encoder receives `[A, B, C]` sequence as inputs. We don't care about encoder outputs, only about the hidden state it accumulates while reading the sequence. After input sequence ends, encoder passes its final state to decoder, which receives `[<EOS>, W, X, Y, Z]` and is trained to output `[W, X, Y, Z, <EOS>]`. `<EOS>` token is a special word in vocabulary that signals to decoder the beginning of translation.
# ## Implementation details
#
# TensorFlow has its own [implementation of seq2seq](https://www.tensorflow.org/tutorials/seq2seq/). Recently it was moved from core examples to [`tensorflow/models` repo](https://github.com/tensorflow/models/tree/master/tutorials/rnn/translate), and uses deprecated seq2seq implementation. Deprecation happened because it uses **static unrolling**.
#
# **Static unrolling** involves construction of computation graph with a fixed sequence of time step. Such a graph can only handle sequences of specific lengths. One solution for handling sequences of varying lengths is to create multiple graphs with different time lengths and separate the dataset into this buckets.
#
# **Dynamic unrolling** instead uses control flow ops to process sequence step by step. In TF this is supposed to more space efficient and just as fast. This is now a recommended way to implement RNNs.
# ## Vocabulary
#
# Seq2seq maps sequence onto another sequence. Both sequences consist of integers from a fixed range. In language tasks, integers usually correspond to words: we first construct a vocabulary by assigning to every word in our corpus a serial integer. First few integers are reserved for special tokens. We'll call the upper bound on vocabulary a `vocabulary size`.
#
# Input data consists of sequences of integers.
x = [[5, 7, 8], [6, 3], [3], [1]]
# While manipulating such variable-length lists are convenient to humans, RNNs prefer a different layout:
import helpers
xt, xlen = helpers.batch(x)
x
xt
# Sequences form columns of a matrix of size `[max_time, batch_size]`. Sequences shorter then the longest one are padded with zeros towards the end. This layout is called `time-major`. It is slightly more efficient then `batch-major`. We will use it for the rest of the tutorial.
xlen
# For some forms of dynamic layout it is useful to have a pointer to terminals of every sequence in the batch in separate tensor (see following tutorials).
# # Building a model
# ## Simple seq2seq
# Encoder starts with empty state and runs through the input sequence. We are not interested in encoder's outputs, only in its `final_state`.
#
# Decoder uses encoder's `final_state` as its `initial_state`. Its inputs are a batch-sized matrix with `<EOS>` token at the 1st time step and `<PAD>` at the following. This is a rather crude setup, useful only for tutorial purposes. In practice, we would like to feed previously generated tokens after `<EOS>`.
#
# Decoder's outputs are mapped onto the output space using `[hidden_units x output_vocab_size]` projection layer. This is necessary because we cannot make `hidden_units` of decoder arbitrarily large, while our target space would grow with the size of the dictionary.
#
# This kind of encoder-decoder is forced to learn fixed-length representation (specifically, `hidden_units` size) of the variable-length input sequence and restore output sequence only from this representation.
# +
import numpy as np
import tensorflow as tf
import helpers
tf.reset_default_graph()
sess = tf.InteractiveSession()
# -
tf.__version__
# ### Model inputs and outputs
# First critical thing to decide: vocabulary size.
#
# Dynamic RNN models can be adapted to different batch sizes and sequence lengths without retraining (e.g. by serializing model parameters and Graph definitions via `tf.train.Saver`), but changing vocabulary size requires retraining the model.
# +
PAD = 0
EOS = 1
vocab_size = 10
input_embedding_size = 20
encoder_hidden_units = 20
decoder_hidden_units = encoder_hidden_units
# -
# Nice way to understand complicated function is to study its signature - inputs and outputs. With pure functions, only inputs-output relation matters.
#
# - `encoder_inputs` int32 tensor is shaped `[encoder_max_time, batch_size]`
# - `decoder_targets` int32 tensor is shaped `[decoder_max_time, batch_size]`
encoder_inputs = tf.placeholder(shape=(None, None), dtype=tf.int32, name='encoder_inputs')
decoder_targets = tf.placeholder(shape=(None, None), dtype=tf.int32, name='decoder_targets')
# We'll add one additional placeholder tensor:
# - `decoder_inputs` int32 tensor is shaped `[decoder_max_time, batch_size]`
decoder_inputs = tf.placeholder(shape=(None, None), dtype=tf.int32, name='decoder_inputs')
# Notice that all shapes are specified with `None`s (dynamic). We can use batches of any size with any number of timesteps. This is convenient and efficient, however but there are obvious constraints:
# - Feed values for all tensors should have same `batch_size`
# - Decoder inputs and ouputs (`decoder_inputs` and `decoder_targets`) should have same `decoder_max_time`
# We actually don't want to feed `decoder_inputs` manually — they are a function of either `decoder_targets` or previous decoder outputs during rollout. However, there are different ways to construct them. It might be illustrative to explicitly specify them for out first seq2seq implementation.
#
# During training, `decoder_inputs` will consist of `<EOS>` token concatenated with `decoder_targets` along time axis. In this way, we always pass target sequence as the history to the decoder, regrardless of what it actually outputs predicts. This can introduce distribution shift from training to prediction.
# In prediction mode, model will receive tokens it previously generated (via argmax over logits), not the ground truth, which would be unknowable.
# ### Embeddings
#
# `encoder_inputs` and `decoder_inputs` are int32 tensors of shape `[max_time, batch_size]`, while encoder and decoder RNNs expect dense vector representation of words, `[max_time, batch_size, input_embedding_size]`. We convert one to another by using *word embeddings*. Specifics of working with embeddings are nicely described in [official tutorial on embeddings](https://www.tensorflow.org/tutorials/word2vec/).
# First we initialize embedding matrix. Initializations are random. We rely on our end-to-end training to learn vector representations for words jointly with encoder and decoder.
embeddings = tf.Variable(tf.random_uniform([vocab_size, input_embedding_size], -1.0, 1.0), dtype=tf.float32)
# We use `tf.nn.embedding_lookup` to *index embedding matrix*: given word `4`, we represent it as 4th column of embedding matrix.
# This operation is lightweight, compared with alternative approach of one-hot encoding word `4` as `[0,0,0,1,0,0,0,0,0,0]` (vocab size 10) and then multiplying it by embedding matrix.
#
# Additionally, we don't need to compute gradients for any columns except 4th.
#
# Encoder and decoder will share embeddings. It's all words, right? Well, digits in this case. In real NLP application embedding matrix can get very large, with 100k or even 1m columns.
encoder_inputs_embedded = tf.nn.embedding_lookup(embeddings, encoder_inputs)
decoder_inputs_embedded = tf.nn.embedding_lookup(embeddings, decoder_inputs)
# ### Encoder
#
# The centerpiece of all things RNN in TensorFlow is `RNNCell` class and its descendants (like `LSTMCell`). But they are outside of the scope of this post — nice [official tutorial](https://www.tensorflow.org/tutorials/recurrent/) is available.
#
# `@TODO: RNNCell as a factory`
# +
encoder_cell = tf.contrib.rnn.LSTMCell(encoder_hidden_units)
encoder_outputs, encoder_final_state = tf.nn.dynamic_rnn(
encoder_cell, encoder_inputs_embedded,
dtype=tf.float32, time_major=True,
)
del encoder_outputs
# -
# We discard `encoder_outputs` because we are not interested in them within seq2seq framework. What we actually want is `encoder_final_state` — state of LSTM's hidden cells at the last moment of the Encoder rollout.
#
# `encoder_final_state` is also called "thought vector". We will use it as initial state for the Decoder. In seq2seq without attention this is the only point where Encoder passes information to Decoder. We hope that backpropagation through time (BPTT) algorithm will tune the model to pass enough information throught the thought vector for correct sequence output decoding.
encoder_final_state
# TensorFlow LSTM implementation stores state as a tuple of tensors.
# - `encoder_final_state.h` is activations of hidden layer of LSTM cell
# - `encoder_final_state.c` is final output, which can potentially be transfromed with some wrapper `@TODO: check correctness`
# ### Decoder
# +
decoder_cell = tf.contrib.rnn.LSTMCell(decoder_hidden_units)
decoder_outputs, decoder_final_state = tf.nn.dynamic_rnn(
decoder_cell, decoder_inputs_embedded,
initial_state=encoder_final_state,
dtype=tf.float32, time_major=True, scope="plain_decoder",
)
# -
# Since we pass `encoder_final_state` as `initial_state` to the decoder, they should be compatible. This means the same cell type (`LSTMCell` in our case), the same amount of `hidden_units` and the same amount of layers (single layer). I suppose this can be relaxed if we additonally pass `encoder_final_state` through a one-layer MLP.
# With encoder, we were not interested in cells output. But decoder's outputs are what we actually after: we use them to get distribution over words of output sequence.
#
# At this point `decoder_cell` output is a `hidden_units` sized vector at every timestep. However, for training and prediction we need logits of size `vocab_size`. Reasonable thing would be to put linear layer (fully-connected layer without activation function) on top of LSTM output to get non-normalized logits. This layer is called projection layer by convention.
# +
decoder_logits = tf.contrib.layers.linear(decoder_outputs, vocab_size)
decoder_prediction = tf.argmax(decoder_logits, 2)
# -
# ### Optimizer
decoder_logits
# RNN outputs tensor of shape `[max_time, batch_size, hidden_units]` which projection layer maps onto `[max_time, batch_size, vocab_size]`. `vocab_size` part of the shape is static, while `max_time` and `batch_size` is dynamic.
# +
stepwise_cross_entropy = tf.nn.softmax_cross_entropy_with_logits(
labels=tf.one_hot(decoder_targets, depth=vocab_size, dtype=tf.float32),
logits=decoder_logits,
)
loss = tf.reduce_mean(stepwise_cross_entropy)
train_op = tf.train.AdamOptimizer().minimize(loss)
# -
sess.run(tf.global_variables_initializer())
# ### Test forward pass
#
# Did I say that deep learning is a game of shapes? When building a Graph, TF will throw errors when static shapes are not matching. However, mismatches between dynamic shapes are often only discovered when we try to run something through the graph.
#
#
# So let's try running something. For that we need to prepare values we will feed into placeholders.
# ```
# this is key part where everything comes together
#
# @TODO: describe
# - how encoder shape is fixed to max
# - how decoder shape is arbitraty and determined by inputs, but should probably be longer then encoder's
# - how decoder input values are also arbitraty, and how we use GO token, and what are those 0s, and what can be used instead (shifted gold sequence, beam search)
# @TODO: add references
# ```
# +
batch_ = [[6], [3, 4], [9, 8, 7]]
batch_, batch_length_ = helpers.batch(batch_)
print('batch_encoded:\n' + str(batch_))
din_, dlen_ = helpers.batch(np.ones(shape=(3, 1), dtype=np.int32),
max_sequence_length=4)
print('decoder inputs:\n' + str(din_))
pred_ = sess.run(decoder_prediction,
feed_dict={
encoder_inputs: batch_,
decoder_inputs: din_,
})
print('decoder predictions:\n' + str(pred_))
# -
# Successful forward computation, everything is wired correctly.
# ## Training on the toy task
# We will teach our model to memorize and reproduce input sequence. Sequences will be random, with varying length.
#
# Since random sequences do not contain any structure, model will not be able to exploit any patterns in data. It will simply encode sequence in a thought vector, then decode from it.
# +
batch_size = 100
batches = helpers.random_sequences(length_from=3, length_to=8,
vocab_lower=2, vocab_upper=10,
batch_size=batch_size)
print('head of the batch:')
for seq in next(batches)[:10]:
print(seq)
# -
def next_feed():
batch = next(batches)
encoder_inputs_, _ = helpers.batch(batch)
decoder_targets_, _ = helpers.batch(
[(sequence) + [EOS] for sequence in batch]
)
decoder_inputs_, _ = helpers.batch(
[[EOS] + (sequence) for sequence in batch]
)
return {
encoder_inputs: encoder_inputs_,
decoder_inputs: decoder_inputs_,
decoder_targets: decoder_targets_,
}
# Given encoder_inputs `[5, 6, 7]`, decoder_targets would be `[5, 6, 7, 1]`, where 1 is for `EOS`, and decoder_inputs would be `[1, 5, 6, 7]` - decoder_inputs are lagged by 1 step, passing previous token as input at current step.
loss_track = []
# +
max_batches = 3001
batches_in_epoch = 1000
try:
for batch in range(max_batches):
fd = next_feed()
_, l = sess.run([train_op, loss], fd)
loss_track.append(l)
if batch == 0 or batch % batches_in_epoch == 0:
print('batch {}'.format(batch))
print(' minibatch loss: {}'.format(sess.run(loss, fd)))
predict_ = sess.run(decoder_prediction, fd)
for i, (inp, pred) in enumerate(zip(fd[encoder_inputs].T, predict_.T)):
print(' sample {}:'.format(i + 1))
print(' input > {}'.format(inp))
print(' predicted > {}'.format(pred))
if i >= 2:
break
print()
except KeyboardInterrupt:
print('training interrupted')
# -
# %matplotlib inline
import matplotlib.pyplot as plt
plt.plot(loss_track)
print('loss {:.4f} after {} examples (batch_size={})'.format(loss_track[-1], len(loss_track)*batch_size, batch_size))
# Something is definitely getting learned.
# # Limitations of the model
#
# We have no control over transitions of `tf.nn.dynamic_rnn`, it is unrolled in a single sweep. Some of the things that are not possible without such control:
#
# - We can't feed previously generated tokens without falling back to Python loops. This means *we cannot make efficient inference with dynamic_rnn decoder*!
#
# - We can't use attention, because attention conditions decoder inputs on its previous state
#
# Solution would be to use `tf.nn.raw_rnn` instead of `tf.nn.dynamic_rnn` for decoder, as we will do in tutorial #2.
# # Fun things to try (aka Exercises)
#
# - In `copy_task` increasing `max_sequence_size` and `vocab_upper`. Observe slower learning and general performance degradation.
#
# - For `decoder_inputs`, instead of shifted target sequence `[<EOS> W X Y Z]`, try feeding `[<EOS> <PAD> <PAD> <PAD>]`, like we've done when we tested forward pass. Does it break things? Or slows learning?
|
1-seq2seq.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # 01 - Data Analysis and Preparation
#
# This notebook covers the following tasks:
#
# 1. Perform exploratory data analysis and visualization.
# 2. Prepare the data for the ML task in BigQuery.
# 3. Generate and fix a ` TFDV schema` for the source data.
# 4. Create a `Vertex Dataset resource` dataset.
#
# ## Dataset
#
# The [Chicago Taxi Trips](https://pantheon.corp.google.com/marketplace/details/city-of-chicago-public-data/chicago-taxi-trips) dataset is one of [public datasets hosted with BigQuery](https://cloud.google.com/bigquery/public-data/), which includes taxi trips from 2013 to the present, reported to the City of Chicago in its role as a regulatory agency. The `taxi_trips` table size is 70.72 GB and includes more than 195 million records. The dataset includes information about the trips, like pickup and dropoff datetime and location, passengers count, miles travelled, and trip toll.
#
# The ML task is to predict whether a given trip will result in a tip > 20%.
# ## Setup
# ### Import libraries
# +
import os
import pandas as pd
import tensorflow as tf
import tensorflow_data_validation as tfdv
from google.cloud import bigquery
import matplotlib.pyplot as plt
from google.cloud import aiplatform as vertex_ai
from google.cloud import aiplatform_v1beta1 as vertex_ai_beta
# -
# ### Setup Google Cloud project
# +
PROJECT = '[your-project-id]' # Change to your project id.
REGION = 'us-central1' # Change to your region.
if PROJECT == "" or PROJECT is None or PROJECT == "[your-project-id]":
# Get your GCP project id from gcloud
# shell_output = !gcloud config list --format 'value(core.project)' 2>/dev/null
PROJECT = shell_output[0]
print("Project ID:", PROJECT)
print("Region:", REGION)
# -
# ### Set configurations
# +
BQ_DATASET_NAME = 'playground_us' # Change to your BQ dataset name.
BQ_TABLE_NAME = 'chicago_taxitrips_prep'
BQ_LOCATION = 'US'
DATASET_DISPLAY_NAME = 'chicago-taxi-tips'
RAW_SCHEMA_DIR = 'src/raw_schema'
# -
# ## 1. Explore the data in BigQuery
# +
# %%bigquery data
SELECT
CAST(EXTRACT(DAYOFWEEK FROM trip_start_timestamp) AS string) AS trip_dayofweek,
FORMAT_DATE('%A',cast(trip_start_timestamp as date)) AS trip_dayname,
COUNT(*) as trip_count,
FROM `bigquery-public-data.chicago_taxi_trips.taxi_trips`
WHERE
EXTRACT(YEAR FROM trip_start_timestamp) = 2015
GROUP BY
trip_dayofweek,
trip_dayname
ORDER BY
trip_dayofweek
;
# -
data
data.plot(kind='bar', x='trip_dayname', y='trip_count')
# ## 2. Create data for the ML task
#
# We add a `ML_use` column for pre-splitting the data, where 80% of the datsa items are set to `UNASSIGNED` while the other 20% is set to `TEST`.
#
# This column is used during training (custom and AutoML) to split the dataset for training and test.
#
# In the training phase, the `UNASSIGNED` are split into `train` and `eval`. The `TEST` split is will be used for the final model validation.
# ### Create destination BigQuery dataset
# !bq --location=US mk -d \
# $PROJECT:$BQ_DATASET_NAME
sample_size = 1000000
year = 2020
# +
sql_script = '''
CREATE OR REPLACE TABLE `@PROJECT.@DATASET.@TABLE`
AS (
WITH
taxitrips AS (
SELECT
trip_start_timestamp,
trip_seconds,
trip_miles,
payment_type,
pickup_longitude,
pickup_latitude,
dropoff_longitude,
dropoff_latitude,
tips,
fare
FROM
`bigquery-public-data.chicago_taxi_trips.taxi_trips`
WHERE 1=1
AND pickup_longitude IS NOT NULL
AND pickup_latitude IS NOT NULL
AND dropoff_longitude IS NOT NULL
AND dropoff_latitude IS NOT NULL
AND trip_miles > 0
AND trip_seconds > 0
AND fare > 0
AND EXTRACT(YEAR FROM trip_start_timestamp) = @YEAR
)
SELECT
trip_start_timestamp,
EXTRACT(MONTH from trip_start_timestamp) as trip_month,
EXTRACT(DAY from trip_start_timestamp) as trip_day,
EXTRACT(DAYOFWEEK from trip_start_timestamp) as trip_day_of_week,
EXTRACT(HOUR from trip_start_timestamp) as trip_hour,
trip_seconds,
trip_miles,
payment_type,
ST_AsText(
ST_SnapToGrid(ST_GeogPoint(pickup_longitude, pickup_latitude), 0.1)
) AS pickup_grid,
ST_AsText(
ST_SnapToGrid(ST_GeogPoint(dropoff_longitude, dropoff_latitude), 0.1)
) AS dropoff_grid,
ST_Distance(
ST_GeogPoint(pickup_longitude, pickup_latitude),
ST_GeogPoint(dropoff_longitude, dropoff_latitude)
) AS euclidean,
CONCAT(
ST_AsText(ST_SnapToGrid(ST_GeogPoint(pickup_longitude,
pickup_latitude), 0.1)),
ST_AsText(ST_SnapToGrid(ST_GeogPoint(dropoff_longitude,
dropoff_latitude), 0.1))
) AS loc_cross,
IF((tips/fare >= 0.2), 1, 0) AS tip_bin,
IF(RAND() <= 0.8, 'UNASSIGNED', 'TEST') AS ML_use
FROM
taxitrips
LIMIT @LIMIT
)
'''
# -
sql_script = sql_script.replace(
'@PROJECT', PROJECT).replace(
'@DATASET', BQ_DATASET_NAME).replace(
'@TABLE', BQ_TABLE_NAME).replace(
'@YEAR', str(year)).replace(
'@LIMIT', str(sample_size))
print(sql_script)
bq_client = bigquery.Client(project=PROJECT, location=BQ_LOCATION)
job = bq_client.query(sql_script)
_ = job.result()
# +
# %%bigquery --project {PROJECT}
SELECT ML_use, COUNT(*)
FROM playground_us.chicago_taxitrips_prep # Change to your BQ dataset and table names.
GROUP BY ML_use
# -
# ### Load a sample data to a Pandas DataFrame
# +
# %%bigquery sample_data --project {PROJECT}
SELECT * EXCEPT (trip_start_timestamp, ML_use)
FROM playground_us.chicago_taxitrips_prep # Change to your BQ dataset and table names.
# -
sample_data.head().T
sample_data.tip_bin.value_counts()
sample_data.euclidean.hist()
# ## 3. Generate raw data schema
#
# The [TensorFlow Data Validation (TFDV)](https://www.tensorflow.org/tfx/data_validation/get_started) data schema will be used in:
# 1. Identify the raw data types and shapes in the data transformation.
# 2. Create the serving input signature for the custom model.
# 3. Validate the new raw training data in the TFX pipeline.
stats = tfdv.generate_statistics_from_dataframe(
dataframe=sample_data,
stats_options=tfdv.StatsOptions(
label_feature='tip_bin',
weight_feature=None,
sample_rate=1,
num_top_values=50
)
)
tfdv.visualize_statistics(stats)
schema = tfdv.infer_schema(statistics=stats)
tfdv.display_schema(schema=schema)
raw_schema_location = os.path.join(RAW_SCHEMA_DIR, 'schema.pbtxt')
tfdv.write_schema_text(schema, raw_schema_location)
# ## 4. Create Vertex Dataset resource
vertex_ai.init(
project=PROJECT,
location=REGION
)
# ### Create the dataset resource
# +
bq_uri = f"bq://{PROJECT}.{BQ_DATASET_NAME}.{BQ_TABLE_NAME}"
dataset = vertex_ai.TabularDataset.create(
display_name=DATASET_DISPLAY_NAME, bq_source=bq_uri)
dataset.gca_resource
# -
# ### Get the dataset resource
#
# The dataset resource is retrieved by display name. Because multiple datasets can have the same display name, we retrieve the most recent updated one.
# +
dataset = vertex_ai.TabularDataset.list(
filter=f"display_name={DATASET_DISPLAY_NAME}",
order_by="update_time")[-1]
print("Dataset resource name:", dataset.resource_name)
print("Dataset BigQuery source:", dataset.gca_resource.metadata['inputConfig']['bigquerySource']['uri'])
# -
|
01-dataset-management.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Data Scientist Nanodegree
# ## Supervised Learning
# ## Project: Finding Donors for *CharityML*
# In this project, I will employ several supervised algorithms of my choice to accurately model individuals' income using data collected from the 1994 U.S. Census. I will then choose the best candidate algorithm from preliminary results and further optimize this algorithm to best model the data. My goal with this implementation is to construct a model that accurately predicts whether an individual makes more than $50,000. This sort of task can arise in a non-profit setting, where organizations survive on donations. Understanding an individual's income can help a non-profit better understand how large of a donation to request, or whether or not they should reach out to begin with. While it can be difficult to determine an individual's general income bracket directly from public sources, we can (as we will see) infer this value from other publically available features.
#
# The dataset for this project originates from the [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Census+Income). The datset was donated by <NAME> and <NAME>, after being published in the article _"Scaling Up the Accuracy of Naive-Bayes Classifiers: A Decision-Tree Hybrid"_. One can find the article by <NAME> [online](https://www.aaai.org/Papers/KDD/1996/KDD96-033.pdf). The data we investigate here consists of small changes to the original dataset, such as removing the `'fnlwgt'` feature and records with missing or ill-formatted entries.
# +
# Import libraries necessary for this project
import numpy as np
import pandas as pd
from time import time
from IPython.display import display # Allows the use of display() for DataFrames
# Import supplementary visualization code visuals.py
import visuals as vs
# Pretty display for notebooks
# %matplotlib inline
# Load the Census dataset
data = pd.read_csv("census.csv")
data_test = pd.read_csv("test_census.csv")
# Success - Display the first record
display(data.head(5))
# -
# ### Implementation: Data Exploration
#
# A cursory investigation of the dataset will determine how many individuals fit into either group, and will tell me about the percentage of these individuals making more than \$50,000. In the code cell below, I compute the following:
# - The total number of records, `'n_records'`
# - The number of individuals making more than \$50,000 annually, `'n_greater_50k'`.
# - The number of individuals making at most \$50,000 annually, `'n_at_most_50k'`.
# - The percentage of individuals making more than \$50,000 annually, `'greater_percent'`.
# +
# TODO: Total number of records
n_records = data.shape[0]
# TODO: Number of records where individual's income is more than $50,000
n_greater_50k = np.sum(data['income']=='>50K')
# TODO: Number of records where individual's income is at most $50,000
n_at_most_50k = np.sum(data['income']=='<=50K')
# TODO: Percentage of individuals whose income is more than $50,000
greater_percent = n_greater_50k/n_records
# Print the results
print("Total number of records: {}".format(n_records))
print("Individuals making more than $50,000: {}".format(n_greater_50k))
print("Individuals making at most $50,000: {}".format(n_at_most_50k))
print("Percentage of individuals making more than $50,000: {}%".format(greater_percent))
# -
# ** Featureset Exploration **
#
# * **age**: continuous.
# * **workclass**: Private, Self-emp-not-inc, Self-emp-inc, Federal-gov, Local-gov, State-gov, Without-pay, Never-worked.
# * **education**: Bachelors, Some-college, 11th, HS-grad, Prof-school, Assoc-acdm, Assoc-voc, 9th, 7th-8th, 12th, Masters, 1st-4th, 10th, Doctorate, 5th-6th, Preschool.
# * **education-num**: continuous.
# * **marital-status**: Married-civ-spouse, Divorced, Never-married, Separated, Widowed, Married-spouse-absent, Married-AF-spouse.
# * **occupation**: Tech-support, Craft-repair, Other-service, Sales, Exec-managerial, Prof-specialty, Handlers-cleaners, Machine-op-inspct, Adm-clerical, Farming-fishing, Transport-moving, Priv-house-serv, Protective-serv, Armed-Forces.
# * **relationship**: Wife, Own-child, Husband, Not-in-family, Other-relative, Unmarried.
# * **race**: Black, White, Asian-Pac-Islander, Amer-Indian-Eskimo, Other.
# * **sex**: Female, Male.
# * **capital-gain**: continuous.
# * **capital-loss**: continuous.
# * **hours-per-week**: continuous.
# * **native-country**: United-States, Cambodia, England, Puerto-Rico, Canada, Germany, Outlying-US(Guam-USVI-etc), India, Japan, Greece, South, China, Cuba, Iran, Honduras, Philippines, Italy, Poland, Jamaica, Vietnam, Mexico, Portugal, Ireland, France, Dominican-Republic, Laos, Ecuador, Taiwan, Haiti, Columbia, Hungary, Guatemala, Nicaragua, Scotland, Thailand, Yugoslavia, El-Salvador, Trinadad&Tobago, Peru, Hong, Holand-Netherlands.
# ----
# ## Data Preprocessing
# Before data can be used as input for machine learning algorithms, it must be cleaned, formatted, and restructured. Fortunately, for this dataset, there are no invalid or missing entries we must deal with, however, there are some qualities about certain features that must be adjusted. This preprocessing can help tremendously with the outcome and predictive power of nearly all learning algorithms.
# ### Transforming Skewed Continuous Features
# +
# Split the data into features and target label
income_raw = data['income']
features_raw = data.drop('income', axis = 1)
# Visualize skewed continuous features of original data
vs.distribution(data)
# -
# For highly-skewed feature distributions such as `'capital-gain'` and `'capital-loss'`, it is common practice to apply a logarithmic transformation on the data so that the very large and very small values do not negatively affect the performance of a learning algorithm. Using a logarithmic transformation significantly reduces the range of values caused by outliers. The logarithm of `0` is undefined, so we must translate the values by a small amount above `0` to apply the the logarithm successfully.
#
# Run the code cell below to perform a transformation on the data and visualize the results. Again, note the range of values and how they are distributed.
# +
# Log-transform the skewed features
skewed = ['capital-gain', 'capital-loss']
features_log_transformed = pd.DataFrame(data = features_raw)
features_log_transformed[skewed] = features_raw[skewed].apply(lambda x: np.log(x + 1))
# Log-transform the skewed features for Kaggle Prediction
features_log_transformed_test = pd.DataFrame(data = data_test)
features_log_transformed_test[skewed] = features_raw[skewed].apply(lambda x: np.log(x + 1))
# Visualize the new log distributions
vs.distribution(features_log_transformed, transformed = True)
# -
# ### Normalizing Numerical Features
# In addition to performing transformations on features that are highly skewed, it is often good practice to perform some type of scaling on numerical features. Normalization ensures that each feature is treated equally when applying supervised learners.
# +
# Import sklearn.preprocessing.StandardScaler
from sklearn.preprocessing import MinMaxScaler
# Initialize a scaler, then apply it to the features
scaler = MinMaxScaler() # default=(0, 1)
numerical = ['age', 'education-num', 'capital-gain', 'capital-loss', 'hours-per-week']
features_log_minmax_transform = pd.DataFrame(data = features_log_transformed)
features_log_minmax_transform[numerical] = scaler.fit_transform(features_log_transformed[numerical])
# apply scaler to the Kaggle test data features
features_log_minmax_transform_test = pd.DataFrame(data = features_log_transformed_test)
features_log_minmax_transform_test[numerical] = scaler.fit_transform(features_log_transformed_test[numerical])
# Show an example of a record with scaling applied
display(features_log_minmax_transform.head(n = 5))
# -
# ### One-hot encode
# +
# TODO: One-hot encode the 'features_log_minmax_transform' data using pandas.get_dummies()
features_final = pd.get_dummies(features_log_minmax_transform)
# TODO: Encode the 'income_raw' data to numerical values
income = income_raw.apply(lambda x=0: 0 if x == "<=50K" else 1)
#One-hot encode the 'features_log_minmax_transform_test' data using pandas.get_dummies()
features_final_test = pd.get_dummies(features_log_minmax_transform_test)
# Print the number of features after one-hot encoding
encoded = list(features_final.columns)
print("{} total features after one-hot encoding.".format(len(encoded)))
# Uncomment the following line to see the encoded feature names
# print encoded
display(features_final.head(n = 5))
# -
# ### Shuffle and Split Data
# Now all _categorical variables_ have been converted into numerical features, and all numerical features have been normalized. We will now split the data (both features and their labels) into training and test sets. 80% of the data will be used for training and 20% for testing.
# +
# Import train_test_split
from sklearn.model_selection import train_test_split
# Split the 'features' and 'income' data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(features_final,
income,
test_size = 0.2,
random_state = 0)
# Show the results of the split
print("Training set has {} samples.".format(X_train.shape[0]))
print("Testing set has {} samples.".format(X_test.shape[0]))
# -
# ----
# ## Evaluating Model Performance
# In this section, I will investigate four different algorithms, and determine which is best at modeling the data. Three of these algorithms will be supervised learners of my choice, and the fourth algorithm is a *naive predictor*.
# ### Naive Predictor Performace
# * Choosing a model that always predicted an individual made more than $50,000, I check what would that model's accuracy and F-score be on this dataset.
# +
TP = np.sum(income) # Counting the ones as this is the naive case. Note that 'income' is the 'income_raw' data
#ncoded to numerical values done in the data preprocessing step.
FP = income.count() - TP # Specific to the naive case
TN = 0 # No predicted negatives in the naive case
FN = 0 # No predicted negatives in the naive case
# TODO: Calculate accuracy, precision and recall
accuracy = (TP+TN)/(TP+FP+TN+FN)
recall = (TP)/(TP+FN)
precision = (TP)/(TP+FP)
# TODO: Calculate F-score using the formula above for beta = 0.5 and correct values for precision and recall.
fscore = (1+0.5**2)*precision*recall/(0.5**2*precision+recall)
# Print the results
print("Naive Predictor: [Accuracy score: {:.4f}, F-score: {:.4f}]".format(accuracy, fscore))
# -
# ### Model Application
# Describing three of the supervised learning models that I think will be appropriate for this problem to test on the census data
# \### RandomForestClassifier:
# ##### Real life application:
# In finance sector this classifier can be used for the detection of customers that are more likely to repay their debt on time, or use a bank's services more frequently. It is also used to detect fraudsters out to scam the bank.
# ##### Strengths:
# One of the biggest problems in machine learning is overfitting, but most of the time this won’t happen in random forest. Random forest is also great because of its abilty to deal with missing values, and the default hyperparameters it uses often produce a good prediction result.
# ##### Weakness:
# The main limitation of random forest is that a large number of trees can make the algorithm too slow and ineffective for real-time predictions. In general, these algorithms are fast to train, but quite slow to create predictions once they are trained.
# ##### Project Usefulnesss:
# Random forest's simplicity makes it a tough proposition to build a “bad” model with it. Provides a pretty good indicator of the importance it assigns to the dataset features, which we want to find out in the project.
#
# [Reference](https://builtin.com/data-science/random-forest-algorithm)
#
#
# ### AdaBoostClassifier:
# ##### Real life application:
# It can be used for predicting customer churn and classifying the types of topics customers are talking/calling about.
# ##### Strengths:
# One of the many advantages of the AdaBoost Algorithm is it is fast, simple and easy to program. Also, it has the flexibility to be combined with any machine learning algorithm.
# ##### Weakness:
# If the weak classifiers are too weak in algorithm used for AdaBoost, it can lead to low margins and overfitting of the data.
# ##### Project Usefulnesss:
# There is a class imbalance in our data, and algorithms such as AdaBoost are great option to deal with them.
#
# [Reference](https://www.educba.com/adaboost-algorithm/)
#
# ### Gaussian Naive Bayes:
# ##### Real life application:
# It can be used to build a Spam filtering module or perform Sentiment Analysis in social media analysis, to identify positive and negative customer sentiments.
# ##### Strengths:
# It is easy and fast to predict the class of the test data set. It also performs well in multi-class prediction. Moreover, when assumption of independence holds, a Naive Bayes classifier performs well.
# ##### Weakness:
# Naive Bayes is the assumption of independent predictors. In real life, it is almost impossible that we get a set of predictors which are completely independent. Naive Bayes is also known as a bad estimator for many datasets.
# ##### Project Usefulnesss:
# The dataset is pretty large, and Naive Bayes is fast to learn compared to most other algorithms. If the model provides good results it might be great canditate to use for final predictions due to its speed.
#
# [Reference](https://towardsdatascience.com/all-about-naive-bayes-8e13cef044cf)
#
#
# ### Implementation - Creating a Training and Predicting Pipeline
#
# I created a training and predicting pipeline that allows me to quickly and effectively train models using various sizes of training data and perform predictions on the testing data.
# TODO: Import two metrics from sklearn - fbeta_score and accuracy_score
from sklearn.metrics import fbeta_score, accuracy_score
def train_predict(learner, sample_size, X_train, y_train, X_test, y_test):
'''
inputs:
- learner: the learning algorithm to be trained and predicted on
- sample_size: the size of samples (number) to be drawn from training set
- X_train: features training set
- y_train: income training set
- X_test: features testing set
- y_test: income testing set
'''
results = {}
# TODO: Fit the learner to the training data using slicing with 'sample_size' using .fit(training_features[:], training_labels[:])
start = time() # Get start time
learner = learner.fit(X_train[:sample_size], y_train[:sample_size])
end = time() # Get end time
# TODO: Calculate the training time
results['train_time'] = end - start
# TODO: Get the predictions on the test set(X_test),
# then get predictions on the first 300 training samples(X_train) using .predict()
start = time() # Get start time
predictions_train = learner.predict(X_train[:300])
predictions_test = learner.predict(X_test)
end = time() # Get end time
# TODO: Calculate the total prediction time
results['pred_time'] = end - start
# TODO: Compute accuracy on the first 300 training samples which is y_train[:300]
results['acc_train'] = accuracy_score(y_train[:300], predictions_train)
# TODO: Compute accuracy on test set using accuracy_score()
results['acc_test'] = accuracy_score(y_test, predictions_test)
# TODO: Compute F-score on the the first 300 training samples using fbeta_score()
results['f_train'] = fbeta_score(y_train[:300], predictions_train,beta=0.5)
# TODO: Compute F-score on the test set which is y_test
results['f_test'] = fbeta_score(y_test, predictions_test,beta=0.5)
# Success
print("{} trained on {} samples.".format(learner.__class__.__name__, sample_size))
# Return the results
return results
# ### Implementation: Initial Model Evaluation
# +
# TODO: Import the three supervised learning models from sklearn
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier
from sklearn.naive_bayes import GaussianNB
# TODO: Initialize the three models
clf_A = AdaBoostClassifier(random_state=0)
clf_B = RandomForestClassifier(random_state=0)
clf_C = GaussianNB()
# TODO: Calculate the number of samples for 1%, 10%, and 100% of the training data
# HINT: samples_100 is the entire training set i.e. len(y_train)
# HINT: samples_10 is 10% of samples_100 (ensure to set the count of the values to be `int` and not `float`)
# HINT: samples_1 is 1% of samples_100 (ensure to set the count of the values to be `int` and not `float`)
samples_100 = len(y_train)
samples_10 = int(samples_100*0.1)
samples_1 = int(samples_100*0.01)
# Collect results on the learners
results = {}
for clf in [clf_A, clf_B, clf_C]:
clf_name = clf.__class__.__name__
results[clf_name] = {}
for i, samples in enumerate([samples_1, samples_10, samples_100]):
results[clf_name][i] = \
train_predict(clf, samples, X_train, y_train, X_test, y_test)
# Run metrics visualization for the three supervised learning models chosen
vs.evaluate(results, accuracy, fscore)
# -
# ## Improving Results
# I will choose from the three supervised learning models the *best* model to use on the student data. I will then perform a grid search optimization for the model over the entire training set by tuning at least one parameter to improve upon the untuned model's F-score.
# ## Choosing the Best Model
#
# * Based on the results above, AdaBoost is the best model for the task out of the three models.
#
# * It is the classifier that performed the best on the testing data, in terms of both the accuracy and f-score.
#
# * Moreover, It takes only a resonable amount if time to train on the full dataset. Although GaussianNB had better training times, the accuracy and F-score was considerably lower
#
# * Adaboost uses a decision tree of depth 1 as its base classifier, which can handle categorical and numerical data. Because of its reasonable train time, it should scale well even if we have more data.
# ## Describing the Model in Layman's Terms
#
# Adboost works by combining many weak learner, to create an ensemble of learners that can predict whether an individual earns above 50k or not.
#
# For this project, the weak learners were decision trees. They were created using individual “features” we have been provided to create a set of rules that can predict whether an individual earns above 50k or not.
#
# Adaboost priortizes the data points predicted incorrectly in the previous rounds to ensure they are predicted correctly in the next round during the training process.
#
# The training algorithm repeats the process for a specified number of rounds, or till we can’t improve the predictions further. During each of the rounds, the model finds the best learner to split the data. This learner is incorporated into the ensemble.
#
# All the learners are then combined into one model, where they each vote to predict if a person earns more than 50k or not. Majority vote usually determines the final prediction.
#
# This model can be used to predict the same information for a potential new donor and predict if they earn more than 50K or not, which hints at their likeliness of donating to charity.
# ### Implementation: Model Tuning
# +
# TODO: Import 'GridSearchCV', 'make_scorer', and any other necessary libraries
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import make_scorer
# TODO: Initialize the classifier
clf = AdaBoostClassifier(random_state=1)
# TODO: Create the parameters list you wish to tune, using a dictionary if needed.
# HINT: parameters = {'parameter_1': [value1, value2], 'parameter_2': [value1, value2]}
parameters = {'n_estimators':[60, 120],
'learning_rate':[0.5, 1, 2]}
# TODO: Make an fbeta_score scoring object
scorer = make_scorer(fbeta_score,beta=0.5)
# TODO: Perform grid search on the classifier using 'scorer' as the scoring method
grid_obj = GridSearchCV(clf, parameters, scoring = scorer)
# TODO: Fit the grid search object to the training data and find the optimal parameters using fit()
grid_fit = grid_obj.fit(X_train, y_train)
# Get the estimator
best_clf = grid_fit.best_estimator_
# Make predictions using the unoptimized and model
predictions = (clf.fit(X_train, y_train)).predict(X_test)
best_predictions = best_clf.predict(X_test)
# Report the before-and-afterscores
print("Unoptimized model\n------")
print("Accuracy score on testing data: {:.4f}".format(accuracy_score(y_test, predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, predictions, beta = 0.5)))
print("\nOptimized Model\n------")
print("Final accuracy score on the testing data: {:.4f}".format(accuracy_score(y_test, best_predictions)))
print("Final F-score on the testing data: {:.4f}".format(fbeta_score(y_test, best_predictions, beta = 0.5)))
# -
# ### Final Model Evaluation
# #### Results:
#
# | Metric | Unoptimized Model | Optimized Model |
# | :------------: | :---------------: | :-------------: |
# | Accuracy Score | 0.8576 | 0.8612 |
# | F-score | 0.7246 | 0.7316 |
#
# * The accuracy score on the testing data for the optimized model is 0.8612. The F-score on the testing data for the optimized model is 0.7316.
#
# * Both the accuracy score and the F-score on the optimized model have improved slighly.
#
# * They are considerably better compared to the naive predictor benchmarks. The difference in both accuracy score and the F-score is over 45 percent.
#
# ----
# ## Feature Importance
#
# An important task when performing supervised learning on a dataset like the census data we study here is determining which features provide the most predictive power. By focusing on the relationship between only a few crucial features and the target label we simplify our understanding of the phenomenon, which is most always a useful thing to do. In the case of this project, that means we wish to identify a small number of features that most strongly predict whether an individual makes at most or more than \$50,000.
#
# Choose a scikit-learn classifier (e.g., adaboost, random forests) that has a `feature_importance_` attribute, which is a function that ranks the importance of features according to the chosen classifier. In the next python cell fit this classifier to training set and use this attribute to determine the top 5 most important features for the census dataset.
# ### Implementation - Extracting Feature Importance
# I will find the feature_importance_ attribute for the best model. This ranks the importance of each feature when making predictions based on the chosen algorithm.
# +
# TODO: Extract the feature importances using .feature_importances_
importances = best_clf.feature_importances_
# Plot
vs.feature_plot(importances, X_train, y_train)
# -
# ### Feature Selection
# Try training the model with the reduced data set that only has the the attibutes ranked in the top 5 of feature importance.
# +
# Import functionality for cloning a model
from sklearn.base import clone
# Reduce the feature space
X_train_reduced = X_train[X_train.columns.values[(np.argsort(importances)[::-1])[:5]]]
X_test_reduced = X_test[X_test.columns.values[(np.argsort(importances)[::-1])[:5]]]
# Train on the "best" model found from grid search earlier
clf = (clone(best_clf)).fit(X_train_reduced, y_train)
# Make new predictions
reduced_predictions = clf.predict(X_test_reduced)
# Report scores from the final model using both versions of data
print("Final Model trained on full data\n------")
print("Accuracy on testing data: {:.4f}".format(accuracy_score(y_test, best_predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, best_predictions, beta = 0.5)))
print("\nFinal Model trained on reduced data\n------")
print("Accuracy on testing data: {:.4f}".format(accuracy_score(y_test, reduced_predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, reduced_predictions, beta = 0.5)))
# -
# ## Final Prediction
# +
#filling in the values that are NAN
final_test = features_final_test.interpolate()
#predict using best_clf for Kaggle test dataset
predictions = best_clf.predict(final_test.drop('Unnamed: 0', axis = 1))
#Create pandas datframe to be submitted at Kaggle
final_pred = pd.DataFrame({ 'id' : final_test['Unnamed: 0'], 'income': predictions })
final_pred = final_pred.set_index('id')
#convert final prediction to CSV
final_pred.to_csv('Prediction.csv')
|
finding_donors.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# We'll dig in to the following topics:
# * Bias-Variance Tradeoff
# * Validation Set
# * Model Tuning
# * Cross-Validation
# +
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split, cross_validate
from sklearn.linear_model import LinearRegression, Ridge, Lasso, LassoCV, RidgeCV
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import KFold
import warnings
warnings.filterwarnings("ignore")
# -
boston = pd.read_csv('Boston.csv')
data = boston[['crim', 'zn', 'indus', 'chas', 'nox', 'rm', 'age', 'dis', 'rad',
'tax', 'ptratio', 'black', 'lstat']]
target = boston[['medv']]
# train/test split
X_train, X_test, y_train, y_test = train_test_split(data,
target,
shuffle=True,
test_size=0.2,
random_state=15)
# We know we’ll need to calculate training and test error, so let’s go ahead and create functions to do just that. Let’s include a meta-function that will generate a nice report for us while we’re at it. Also, Root Mean Squared Error (RMSE) will be our metric of choice.
# +
def calc_train_error(X_train, y_train, model):
'''returns in-sample error for already fit model.'''
predictions = model.predict(X_train)
mse = mean_squared_error(y_train, predictions)
rmse = np.sqrt(mse)
return mse
def calc_validation_error(X_test, y_test, model):
'''returns out-of-sample error for already fit model.'''
predictions = model.predict(X_test)
mse = mean_squared_error(y_test, predictions)
rmse = np.sqrt(mse)
return mse
def calc_metrics(X_train, y_train, X_test, y_test, model):
'''fits model and returns the RMSE for in-sample error and out-of-sample error'''
model.fit(X_train, y_train)
train_error = calc_train_error(X_train, y_train, model)
validation_error = calc_validation_error(X_test, y_test, model)
return train_error, validation_error
# -
# Theory
# Bias-Variance Tradeoff
# Pay very close attention to this section. It is one of the most important concepts in all of machine learning. Understanding this concept will help you diagnose all types of models, be they linear regression, XGBoost, or Convolutional Neural Networks.
#
# We already know how to calculate training error and test error. So far we’ve simply been using test error as a way to gauge how well our model will generalize. That was a good first step but it’s not good enough. We can do better. We can tune our model. Let’s drill down.
#
# We can compare training error and something called validation error to figure out what’s going on with our model - more on validation error in a minute. Depending on the values of each, our model can be in one of three regions:
# 1) High Bias - underfitting
# 2) Goldilocks Zone - just right
# 3) High Variance - overfitting
# <img src="pic/bias-variance-tradeoff.png" />
# ### Plot Orientation
# The x-axis represents model complexity. This has to do with how flexible your model is. Some things that add complexity to a model include: additional features, increasing polynomial terms, and increasing the depth for tree-based models. Keep in mind this is far from an exhaustive list but you should get the gist.
#
# The y-axis indicates model error. It’s often measured as Mean-Squared Error (MSE) for Regression and Cross-Entropy or Accuracy for Classification.
#
# The blue curve is Training Error. Notice that it only decreases. What should be painfully obvious is that adding model complexity leads to smaller and smaller training errors. That’s a key finding.
#
# The green curve forms a U-shape. This curve represents Validation Error. Notice the trend. First it decreases, hits a minimum, and then increases. We’ll talk in more detail shortly about what exactly Validation Error is and how to calculate it.
# ### High Bias
# The rectangular box outlined by dashes to the left and labeled as High Bias is the first region of interest. Here you’ll notice Training Error and Validation Error are high. You’ll also notice that they are close to one another. This region is defined as the one where the model lacks the flexibility required to really pull out the inherent trend in the data. In machine learning speak, it is underfitting, meaning it’s doing a poor job all around and won’t generalize well. The model doesn’t even do well on the training set.
#
# How do you fix this?
#
# By adding model complexity of course. I’ll go into much more detail about what to do when you realize you’re under or
# overfitting in another post. For now, assuming you’re using linear regression, a good place to start is by adding additional features. The addition of parameters to your model grants it flexibility that can push your model into the Golidlocks Zone.
# ### Goldilocks Zone
# The middle region without dashes I’ve named the Goldilocks Zone. Your model has just the right amount of flexibility to pick up on the pattern inherent in the data but isn’t so flexible that it’s really just memorizing the training data. This region is marked by Training Error and Validation Error that are both low and close to one another. This is where your model should live.
# ### High Variance
# The dashed rectangular box to the right and labeled High Variance is the flip of the High Bias region. Here the model has so much flexiblity that it essentially starts to memorize the training data. Not surprisingly, that approach leads to low Training Error. But as was mentioned in the train/test post, a lookup table does not generalize, which is why we see high Validation Error in this region. You know you’re in this region when your Training Error is low but your Validation Error is high. Said another way, if there’s a sizeable delta between the two, you’re overfitting.
#
# How do you fix this?
#
# By decreasing model complexity. Again, I’ll go into much more detail in a separate post about what exactly to do. For now, consider applying regularization or dropping features.
# ### Canonical Plot
# Let’s look at one more plot to drive these ideas home.
# <img src="pic/bias-and-variance-targets.jpg" />
# Imagine you’ve entered an archery competition. You receive a score based on which portion of the target you hit: 0 for the red circle (bullseye), 1 for the blue, and 2 for the while. The goal is to minimize your score and you do that by hitting as many bullseyes as possible.
#
# The archery metaphor is a useful analog to explain what we’re trying to accomplish by building a model. Given different datasets (equivalent to different arrows), we want a model that predicts as closely as possible to observed data (aka targets).
#
# The top Low Bias/Low Variance portion of the graph represents the ideal case. This is the Goldilocks Zone. Our model has extracted all the useful information and generalizes well. We know this because the model is accurate and exhibits little variance, even when predicting on unforeseen data. The model is highly tuned, much like an archer who can adjust to different wind speeds, distances, and lighting conditions.
#
# The Low Bias/High Variance portion of the graph represents overfitting. Our model does well on the training data, but we see high variance for specific datasets. This is analagous to an archer who has trained under very stringent conditions - perhaps indoors where there is no wind, the distance is consistent, and the lighting is always the same. Any variation in any of those attributes throws off the archer’s accuracy. The archer lacks consistency.
#
# The High Bias/Low Variance portion of the graph represents underfitting. Our model does poorly on any given dataset. In fact, it’s so bad that it does just about as poorly regardless of the data you feed it, hence the small variance. As an analog, consider an archer who has learned to fire with consistency but hasn’t learned to hit the target. This is analagous to a model that always predicts the average value of the training data’s target.
#
# The High Bias/High Variance portion of the graph actually has no analog in machine learning that I’m aware of. There exists a tradeoff between bias and variance. Therefore, it’s not possible for both to be high.
#
# Alright, let’s shift gears to see this in practice now that we’ve got the theory down.
# +
lr = LinearRegression(fit_intercept=True)
train_error, test_error = calc_metrics(X_train, y_train, X_test, y_test, lr)
train_error, test_error = round(train_error, 3), round(test_error, 3)
print('train error: {} | test error: {}'.format(train_error, test_error))
print('train/test: {}'.format(round(test_error/train_error, 1)))
# -
# Hmm, our training error is somewhat lower than the test error. In fact, the test error is 1.1 times or 10% worse. It’s not a big difference but it’s worth investigating.
#
# Which region does that put us in?
#
# That’s right, it’s every so slightly in the High Variance region, which means our model is slightly overfitting. Again, that means our model has a tad too much complexity.
#
# Unfortunately, we’re stuck at this point.
#
# You’re probably thinking, “Hey wait, no we’re not. I can drop a feature or two and then recalculate training error and test error.”
#
# My response is simply: NOPE. DON’T. PLEASE. EVER. FOR ANY REASON. PERIOD.
#
# Why not?
#
# Because if you do that then your test set is no longer a test set. You are using it to train your model. It’s the same as if you trained your model on the all the data from the beginning. Seriously, don’t do this. Unfortunately, practicing data scientists do this sometimes; it’s one of the worst things you can do. You’re almost guaranteed to produce a model that cannot generalize.
#
# So what do we do?
#
# We need to go back to the beginning. We need to split our data into three datasets: training, validation, test.
#
# Remember, the test set is data you don’t touch until you’re happy with your model. The test set is used only ONE time to see how your model will generalize. That’s it.
#
# Okay, let’s take a look at this thing called a Validation Set.
# ### Validation Set
# Three datasets from one seems like a lot of work but I promise it’s worth it. First, let’s see how to do this in practice.
# +
# intermediate/test split (gives us test set)
X_intermediate, X_test, y_intermediate, y_test = train_test_split(data,
target,
shuffle=True,
test_size=0.2,
random_state=15)
# train/validation split (gives us train and validation sets)
X_train, X_validation, y_train, y_validation = train_test_split(X_intermediate,
y_intermediate,
shuffle=False,
test_size=0.25,
random_state=2018)
# +
# delete intermediate variables
del X_intermediate, y_intermediate
# print proportions
print('train: {}% | validation: {}% | test {}%'.format(round(len(y_train)/len(target),2),
round(len(y_validation)/len(target),2),
round(len(y_test)/len(target),2)))
# -
# If you’re a visual person, this is how our data has been segmented.
# <img src="pic/train-validate-test.png" />
# We have now three datasets depicted by the graphic above where the training set constitutes 60% of all data, the validation set 20%, and the test set 20%. Do notice that I haven’t changed the actual test set in any way. I used the same initial split and the same random state. That way we can compare the model we’re about to fit and tune to the linear regression model we built earlier.
# ### Side note:
# there is no hard and fast rule about how to proportion your data. Just know that your model is limited in what it can learn if you limit the data you feed it. However, if your test set is too small, it won’t provide an accurate estimate as to how your model will perform. Cross-validation allows us to handle this situation with ease, but more on that later.
# ### Model Tuning
# We need to decrease complexity. One way to do this is by using regularization.Regularization is a form of constrained optimization that imposes limits on determining model parameters. It effectively allows me to add bias to a model that’s overfitting. I can control the amount of bias with a hyperparameter called lambda or alpha (you’ll see both, though sklearn uses alpha because lambda is a Python keyword) that defines regularization strength.
#
# The code:
alphas = [0, 0.001, 0.01, 0.1, 1, 10]
print('All errors are RMSE')
print('-'*76)
for alpha in alphas:
# instantiate and fit model
ridge = Ridge(alpha=alpha, fit_intercept=True, random_state=99)
ridge.fit(X_train, y_train)
# calculate errors
new_train_error = mean_squared_error(y_train, ridge.predict(X_train))
new_validation_error = mean_squared_error(y_validation, ridge.predict(X_validation))
new_test_error = mean_squared_error(y_test, ridge.predict(X_test))
# print errors as report
print('alpha: {:7} | train error: {:5} | val error: {:6} | test error: {}'.
format(alpha,
round(new_train_error,3),
round(new_validation_error,3),
round(new_test_error,3)))
# There are a few key takeaways here. First, notice the U-shaped behavior exhibited by the validation error. It starts at 18.001, goes down for two steps and then back up. Also notice that validation error and test error tend to move together, but by no means is the relationship perfect. We see both errors decrease as alpha increases initially but then test error keeps going down while validation error rises again. It’s not perfect. It actually has a whole lot to do with the fact that we’re dealing with a very small dataset. Each sample represents a much larger proportion of the data than say if we had a dataset with a million or more records. Anyway, validation error is a good proxy for test error, especially as dataset size increases. With small to medium-sized datasets, we can do better by leveraging cross-validation. We’ll talk about that shortly.
#
# Now that we’ve tuned our model, let’s fit a new ridge regression model on all data except the test data. Then we’ll check the test error and compare it to that of our original linear regression model with all features.
# +
# train/test split
X_train, X_test, y_train, y_test = train_test_split(data,
target,
shuffle=True,
test_size=0.2,
random_state=15)
# instantiate model
ridge = Ridge(alpha=0.11, fit_intercept=True, random_state=99)
# fit and calculate errors
new_train_error, new_test_error = calc_metrics(X_train, y_train, X_test, y_test, ridge)
new_train_error, new_test_error = round(new_train_error, 3), round(new_test_error, 3)
# -
print('ORIGINAL ERROR')
print('-' * 40)
print('train error: {} | test error: {}\n'.format(train_error, test_error))
print('ERROR w/REGULARIZATION')
print('-' * 40)
print('train error: {} | test error: {}'.format(new_train_error, new_test_error))
# A very small increase in training error coupled with a small decrease in test error. We’re definitely moving in the right direction. Perhaps not quite the magnitude of change we expected, but we’re simply trying to prove a point here. Remember this is a tiny dataset. Also remember I said we can do better by using something called Cross-Validation. Now’s the time to talk about that.
# ### Cross-Validation
# Let me say this upfront: this method works great on small to medium-sized datasets. This is absolutely not the kind of thing you’d want to try on a massive dataset (think tens or hundreds of millions of rows and/or columns). Alright, let’s dig in now that that’s out of the way.
#
# As we saw in the post about train/test split, how you split smaller datasets makes a significant difference; the results can vary tremendously. As the random state is not a hyperparameter (seriously, please don’t do that), we need a way to extract every last bit of signal from the data that we possibly can. So instead of just one train/validation split, let’s do K of them.
#
# This technique is appropriately named K-fold cross-validation. Again, K represents how many train/validation splits you need. There’s no hard and fast rule about how to choose K but there are better and worse choices. As the size of your dataset grows, you can get away with smaller values for K, like 3 or 5. When your dataset is small, it’s common to select a larger number like 10. Again, these are just rules of thumb.
#
# Here’s the general idea for 10-fold CV:
# <img src="pic/kfold-cross-validation.png" />
# ### Technical note:
# Be careful with terminology. Some people will refer to the validation fold as the test fold. Unfortunately, they use the terms interchangeably, which is confusing and therefore not correct. Don’t do that. The test set is the pure data that only gets consumed at the end, if it exists at all.
# Once data has been segmented off in the validation fold, you fit a fresh model on the remaining training data. Ideally, you calculate train and validation error. Some people only look at validation error, however.
#
# The data included in the first validation fold will never be part of a validation fold again. A new validation fold is created, segmenting off the same percentage of data as in the first iteration. Then the process repeats - fit a fresh model, calculate key metrics, and iterate. The algorithm concludes when this process has happened K times. Therefore, you end up with K estimates of the validation error, having visited all the data points in the validation set once and numerous times in training sets. The last step is to average the validation errors for regression. This gives a good estimate as to how well a particular model will perform.
#
# Again, this method is invaluable for tuning hyperparameters on small to medium-sized datasets. You technically don’t even need a test set. That’s great if you just don’t have the data. For large datasets, use a simple train/validation/test split strategy and tune your hyperparameters like we did in the previous section.
#
# Alright, let’s see K-fold CV in action.
# ### Sklearn & CV
# There’s two ways to do this in sklearn, pending what you want to get out of it.
#
# The first method I’ll show you is cross_val_score, which works beautifully if all you care about is validation error.
#
# The second method is KFold, which is perfect if you require train and validation errors.
#
# Let’s try a new model called LASSO just to keep things interesting.
# +
alphas = [0, 1e-4, 1e-3, 1e-2, 1e-1, 1, 1e1]
val_errors = []
for alpha in alphas:
lasso = Lasso(alpha=alpha, fit_intercept=True, random_state=77)
errors = np.sum(-cross_val_score(lasso,
data,
y=target,
scoring='neg_mean_squared_error',
cv=10,
n_jobs=-1))
val_errors.append(np.sqrt(errors))
# -
# RMSE
print(val_errors)
# Which value of alpha gave us the smallest validation error
print('best alpha: {}'.format(alphas[np.argmin(val_errors)]))
# ### K-Fold
data_array = np.array(data)
target_array = np.array(target)
# +
K = 10
kf = KFold(n_splits=K, shuffle=True, random_state=42)
alphas = [0, 1e-4, 1e-3, 1e-2, 1e-1, 1, 1e1]
for alpha in alphas:
train_errors = []
validation_errors = []
for train_index, val_index in kf.split(data_array, target_array):
# split data
X_train, X_val = data_array[train_index], data_array[val_index]
y_train, y_val = target_array[train_index], target_array[val_index]
# instantiate model
lasso = Lasso(alpha=alpha, fit_intercept=True, random_state=77)
#calculate errors
train_error, val_error = calc_metrics(X_train, y_train, X_val, y_val, lasso)
# append to appropriate list
train_errors.append(train_error)
validation_errors.append(val_error)
# generate report
print('alpha: {:6} | mean(train_error): {:7} | mean(val_error): {}'.
format(alpha,
round(np.mean(train_errors),4),
round(np.mean(validation_errors),4)))
# -
# Comparing the output of cross_val_score to that of KFold, we can see that the general trend holds - an alpha of 10 results in the largest validation error. You may wonder why we get different values. The reason is that the data was split differently. We can control the splitting procedure with KFold but not cross_val_score. Therefore, there’s no way I know of to perfectly sync the two procedures without an exhaustive search of splits or writing the algorithm from scratch ourselves. The important thing is that each gives us a viable method to calculate whatever we need, whether it be purely validation error or a combination of training and validation error.
# ### Summary
# We discussed the Bias-Variance Tradeoff where a high bias model is one that is underfit while a high variance model is one that is overfit. We also learned that we can split data into three groups for tuning purposes. Specifically, the three groups are train, validation, and test. Remember the test set is used only one time to check how well a model generalizes on data it’s never seen. This three-group split works exceedingly well for large datasets but not for small to medium-sized datasets, though. In that case, use cross-validation (CV). CV can help you tune your models and extract as much signal as possible from the small data sample. Remember, with CV you don’t need a test set. By using a K-fold approach, you get the equivalent of K-test sets by which to check validation error. This helps you diagnose where you’re at in the bias-variance regime.
|
Lectures/Levon/ModelTuning_Validation&Cross-Validation/main.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Simulation of data relating to weather at Dublin Airport
# * [Introduction](#Introduction)
# * [What is the dataset?](#What-is-the-dataset?)
# * [Setup](#Setup)
# * [Examination of the dataset](#Examination-of-the-dataset)
# * [Description of dataset](#Description-of-dataset)
# * [Skewness and kurtosis of dataset](#Skewness-and-kurtosis-of-dataset)
# * [Correlation](#Correlation)
# * [Plotting statistics](#Plotting-statistics)
# * [Discussion of the dataset](#Discussion-of-the-dataset)
# * [Simulation of new data](#Simulation-of-new-data)
# * [Additional checks](#Additional-checks)
# * [Further Analysis](#Further-Analysis)
# * [Bibliography](#Bibliography)
#
# ## Introduction
# This notebook is intended to fulfil two tasks, namely, to review a data set; and to simulate data to resemble the dataset chosen. In order to do these tasks, the project (and notebook) will be broken into 2 sections. In the first section, a review of the dataset chosen, in this case, the weather at Dublin Airport, will be conducted. This review will include a statistical review of the data, as well as discussion of what the statistics mean. The second section will be an attempt to simulate like data, based on the information gleaned in the first section.
#
# Throughout the notebook, there will be code used. These snippets of code will be used to cleanse the data, provide the statistical analysis, and ultimately attempt to simulate the data. It should be noted that some of the data generated will be random, and therefore the values of the generated data will change, in a [pseudorandom](https://www.random.org/randomness/) manner.
#
# *Note: There is a bibliography at the end of this document, which details articles, websites, and other items referenced with. The hyperlinks within this document connect directly to the referenced site, and not to the bibliography.
#
#
# ## What is the dataset?
# The dataset that was chosen is the Dublin Airport Weather records from the 1st January, 2016 to the 31st December, 2018. This data was sourced from the [Government of Ireland data website](https://data.gov.ie/dataset/dublin-airport-hourly-weather-station-data/resource/bbb2cb83-5982-48ca-9da1-95280f5a4c0d?inner_span=True). The dataset from the source is made up of record readings of various weather attributes recorded every hour from the 1st January, 1989 to the 31st December, 2018. Each row in the dataset is made up of the following columns:
#
# * __**Rain**__: the amount of precipitation to have fallen within the last hour. Measured in millimetres (mm).
# * __**Temp**__: the air temperature at the point of record. Measured in degrees Celsius (°C).
# * __**Wetb**__: the wet bulb temperature at the point of record. Measured in degrees Celsius (°C).
# * __**Dewpt**__: dew point air temperature at the point of record. Measured in degrees Celsius (°C).
# * __**Vappr**__: the vapour pressure of the air at the point of record. Measured in hectopascals (hpa).
# * __**Rhum**__: the relative humidity for the given air temperature. Measured in percent (%).
# * __**Msl**__: mean sea level pressure. Measured in hectopascals (hpa).
# * __**Wdsp**__: Mean hourly wind speed. Measured in knots (kt).
# * __**Wddir**__: Predominant wind direction. Measured in knots (kt).
# * __**Ww**__: Synop code for resent weather.
# * __**W**__: Synop code for past weather.
# * __**Sun**__: The duration of the sun for the last hour. Measured in hours (h).
# * __**Vis**__: Visibility, or air clarity. Measured in metres (m).
# * __**Clht**__: Cloud ceiling height. Measured in hundreds of feet (100 ft).
# * __**Clamt**__: Amount of cloud. Measured using okta.
#
# There are also a number of indicators for some of the data recorded. Given the timespan of the data (30 years), the number of record points for each row (up to 21 points), and the hourly record taking, the data set is very large, comprising of nearly 11,000 days, more than 262,000 rows, and 6,300,000 data points.
#
# The retrieved dataset is too large for the proposed simulation. It is therefore intended reduce it in size. This has been done by limiting the data to the period of the month of December, and the years of 2016 to 2018 inclusive. The number of record points has been reduced to rain, temperature, relative humidity, sun, and visibility. Additionally, the rows of data have been reduced by amalgamating the hourly records into days. The rainfall levels, and hours of sunshine have been added together to provide a total sum for each day. The temperature, relative humidity, and visibility have been averaged for the day in question. This has reduced the number of dataset to 93 (31 x 3) rows, and 6 columns.
#
# Both the original and new datasets are available in this repository.
#
# ## Why was this dataset chosen?
#
# The dataset was chosen for a number of reasons. Primarily, it was chosen as it provides a large volume of data, with interrelated variables. Some of these variables may be positively, or negatively, correlated to each other. This would stand to reason, as the number of hours of sunshine, and the millimetres of rain that have fallen would normally be negatively correlated. Secondly, the dataset is related to the weather in Ireland, or at least Dublin. As the weather .is a favourite topic of conversation, the dataset seemed appropriate.
#
# ## Setup
# Before the analysis of the dataset can begin, it is necessary to import both the libraries to be used.
#
# * **Pandas**: The data set will be held in various pandas dataframes, which will allow for some statistical analysis to be conducted.
# * **Seaborn**: Wil be used for various plotting functionality.
# * **Matplotlib.pyplot**: Will be used for various plotting functionality.
# * **Scipy.stats**: Will be used to simulate the data for the new dataframe.
#
# After this, the data can be imported in a dataframe. This will allow the determination of various statistics with regards to the dataset, as well as providing a basis for the simulation to be run.
#
# The script below will import the data, and set it up in a dataframe.
#
# +
# Importation of libraries, and setting up data
# Importation of libraries
import pandas as pd
import math
import matplotlib.pyplot as plt
import seaborn as sns
from scipy.stats import skewnorm
from datetime import date
# Source for the data set
url = "https://raw.githubusercontent.com/Clauric/PfDA_Sim_Project/master/Dublin%20Airport%20Weather%202016%20-%202018%20cummulative.csv"
# Create a data frame containing the data set
# Set the values in the first column to dates
Dublin_df = pd.read_csv(url, parse_dates=[0])
# Rename the columns to be easier to read
Dublin_df.columns = ["Date", "Rain (mm)", "Temp. (°C)", "Rel. Hum. (%)", "Sun (hrs)", "Visibility (m)"]
# -
# ## Examination of the dataset
# The data set is expected to have the following attributes:
# * All columns (except *Date*) to be made up of numbers, either floating or integers.
# * Date column to be a datetime value.
# * Only the temperature (*Temp*) column can have a value below zero.
# * All non-date columns can have an unlimited upper value (except relative humidity (*Rel. Hum.*), which is limited to 100%).
#
# Additionally, the dataframe should consist of 93 (31 days per month x 3 months) rows, 6 columns, and 1 row of headers.
#
# Looking at the dataframe shapes, type and the first 10 rows, we get the following:
# +
# Shape, data types and first 10 rows of data set
# Shape
print("Shape of dataframe")
row, column = Dublin_df.shape
print("Rows:", row)
print("Columns:", column)
print()
# Types of values
print("Data types in dataframe")
print(Dublin_df.dtypes)
print()
# First 10 rows
print("First 10 rows of dataframe")
print(Dublin_df.head(10))
# -
# From the above, we can see that the shape of the data is as expected (i.e. 93 rows, 6 columns). The first 10 rows show that the column headers are as expected. While not really an issue, it should be noted that the relative humidity is given as values above 1, even though it is a percentage value. However, for the sake of this analysis, it will be left as is, instead of converting to a value between 0 and 1.
#
# ### Description of dataset
# In order to be able to work with the dataset, and draw any conclusions from the data, it is important to determine some of the dataset’s properties. In order to do this, we will extract basic measures, more commonly known as descriptive statistics. These statistics can then be used as a guide to both further analysis, as well as determine which pseudorandom number generator is most appropriate to use (if possible).
#
# The initial set of descriptive statistics are the mean, mode, and median of the data, as well as standard deviation, quartiles, and min and max values. Luckily, pandas has the ability to provide the values for most of these statistics, using the describe function. However, while this is useful, it is also necessary to understand what the terms provided by the describe function actually mean.
#
# * **Mean**: Also known as the simple average, is the sum of all the values divided by the number of values being summed.
# * **Standard deviation**: A measure of how are a number is from the mean. In a perfectly normal distribution, ~68% of all values would be within 1 standard deviation of the mean. Represented in the describe output as *std*.
# * **min**: The lowest value within the dataset.
# * **25%**: The value for the 25th percentile. In other words, 25% of all the values in the dataset are below this value.
# * **50%**: The value for the 50% percentile. In other words, 50% of all the values in the dataset are below this value. This value is often called the *median value*.
# * **75%**: The value for the 75th percentile. In other words, 75% of all the values in the dataset are below this value.
# * **Max**: The largest value in the dataset.
#
# The values for the current dataset are:
# Describe function for the weather dataset
print("Descriptions of the weather dataset")
print()
print(Dublin_df.describe())
# As we can see, certain measures from the descriptive statistics such as correlation, skewness, and kurtosis are missing from the describe functionality. These statistics also give rise to important information regarding the dataset. These will need to be gather to provide a full picture of the dataset.
#
# ### Skewness and kurtosis of dataset
# The skewness and kurtosis are interrelated terms that are used to describe the nature of the distribution of the dataset, and how it differs from a normally distributed dataset. The definition of these terms are:
# • Skewness: the direction and amount of asymmetry of the dataset about its mean. If the absolute value of the skewness is:
# > - greater than 1, the data is highly skewed and the distribution is very asymmetric.
# > - greater than 1, the data is highly skewed and the distribution is very asymmetric.
# > - greater than 0.5 and less than 1, the data is moderately skewed, and the distribution is somewhat asymmetric.
# > - greater than 0, and less than 0.5, the distribution of the data is approximately symmetric.
# > - equal to 0, the data is normally distributed, and symmetric.
#
# The sign of the skewness (i.e. positive or negative) also determines the skewness. Negative skewness indicates that the distribution is skewed to the left, the mean being less than the median, which is less than the mode. Positive values of skewness indicate the opposite, with the distribution being skewed to the right, and the mode being less than the median, which is less than the mean.
#
# * Kurtosis: the kurtosis of a dataset indicates the sharpness, or flatness, of the peak of the data (around the mode, or mean, depending on the skewness).
#
# The kurtosis is measure against the normal distribution, which has a kurtosis of 0. If the kurtosis is negative, then distribution of the data has a smaller standard deviation, as more values are grouped near the mean. This gives the distribution a sharper, and higher peak, and narrower body. A positive kurtosis indicates that there is less grouping around the mean, and indicating that the distribution has a larger standard deviation. This also gives the distribution a flatter, and lower peak, and a wider body.
#
# In pandas, the skewness and kurtosis of a dataset can be ascertained using the *skew* and *kurt* functions. These functions return values for each numeric column within the data set.
#
# +
# Skewness and kurtosis of the dataset
print("Skewness")
print(Dublin_df.skew())
print()
print("Kurtosis")
print(Dublin_df.kurt())
# -
# ### Correlation
# Correlation is a statistic that can be used to measure how well two sets of data correspond to each other. [Weisstein (2019)](http://mathworld.wolfram.com/Correlation.html) defines correlation as "*the degree to which two or more quantities are linearly associated*." As such, a correlation coefficient can be calculated that shows the relationship between the two sets of variables, as well strength of the relationship.
#
# In correlation analysis, positive values show that the two sets of data are positively correlated (i.e. as one value rises or falls, so does the other). Conversely, negative values indicate that the two data sets are negatively or inversely correlated (i.e. as one value rises, or falls, the other falls, or rises). A zero value indicates that there is no relationship between the two sets of data. The strength of the relationship is indicated be the actual value of the correlation coefficient. An absolute value above 0.5 is considered a strong correlation, and above 0.75 is a very strong correlation. A value of -1 or 1 means that the two sets of data are perfectly correlated (i.e. either perfectly positive or perfectly negative correlation).
#
# In pandas dataframes, the *corr* function can be used to ascertain the correlation between numeric sets of data.
# +
# Correlation analysis fot the weather dataset
print()
print("Correlation coefficient for the weather dataset")
print()
# Create new dataframe for the correlation coefficient values
corr_df = Dublin_df.corr(method="pearson")
# Create separate correlation dataframe for heatmap
corr_df_p = corr_df
# As each column will be perfectly correlated with itself, there is no need to show these values
# Replace the values of 1 with a blank value
corr_df = corr_df.replace({1.00000: ""})
# Print the new dataframe to show the correlation coefficients of the weather dataset
print(corr_df)
print()
# Create heatmap of correlations
# From Zaric (2019)
ax = sns.heatmap(corr_df_p, vmin=-1, vmax=1, center=0, cmap=sns.diverging_palette(10, 200, n=500), square=True)
ax.set_xticklabels(ax.get_xticklabels(), rotation=45, horizontalalignment='right')
plt.rcParams["figure.figsize"] = [7, 7]
plt.title("Heat map for weather dataset correlation")
plt.show()
# -
# The heatmap and the correlation table, when combined, aloow, at quick glance, to see how the values are correlated against each other.
#
# ### Plotting statistics
# Before a discussion of the statistics that were determined, it is useful to plot some of the statistics determined. In this case, it would be useful to plot the some of the columns, which will show the skewness of the distribution.
# +
# Plot of distribution of weather data
# For rain, new values for mean and median values
R_mean = Dublin_df["Rain (mm)"].mean()
R_median = Dublin_df["Rain (mm)"].median()
# For temp, new values for mean and median values
T_mean = Dublin_df["Temp. (°C)"].mean()
T_median = Dublin_df["Temp. (°C)"].median()
# For sun, new values for mean and median values
S_mean = Dublin_df["Sun (hrs)"].mean()
S_median = Dublin_df["Sun (hrs)"].median()
# Seaborn distplots showing both histograms and bell curves for temp, rain, and sun
sns.distplot(Dublin_df["Temp. (°C)"], axlabel = False, kde = False, label = "Temp. (°C)")
sns.distplot(Dublin_df["Rain (mm)"], axlabel = False, kde = False, label = "Rain (mm)")
sns.distplot(Dublin_df["Sun (hrs)"], axlabel = False, kde = False, label = "Sun (hrs)")
# Plotlines for mean and median
plt.axvline(R_mean, color = 'r', linestyle = "-", label = "Rain - mean")
plt.axvline(R_median, color = 'm', linestyle = "--", label = "Rain - median")
plt.axvline(T_mean, color = 'g', linestyle = "-", label = "Temp - mean")
plt.axvline(T_median, color = 'y', linestyle = "--", label = "Temp - median")
plt.axvline(S_median, color = 'b', linestyle = "--", label = "Sun - median")
plt.axvline(S_mean, color = 'k', linestyle = "-", label = "Sun - mean")
# Set size of plot area
plt.rcParams["figure.figsize"] = [15, 6]
# Set title, labels, and legend
plt.title("Histogram of rain, temp, and sun vs frequency")
plt.xlabel("mm (rain), temp (°C), hours (sun)")
plt.ylabel("Frequency")
plt.grid(b = True, which = "major", axis = "x")
plt.legend()
# Show plot
plt.show()
# -
# Neither visibility or relative humidity were plotted due to the fact that the minimum value for visibility is over 8,000 (m), while the range for relative humidity is 92 (%). Plotting these values would have dwarfed the other values in the plot, and make it difficult to glean any information from it.
# ### Discussion of the dataset
# As noted previously, in order to make the dataset easier to process and simulate, a number of adjustments were made to the data. These adjustments, such as averaging the temperature, relative humidity, and visibility, and summing the rainfall values, and sunlight hours, will have changed the overall data set. However, this was done in order to avoid having to simulate different sets of values for each hour of the day and night, as well as reduce the dataset from 2,232 rows of data to 93 rows, while still maintaining each of the 5 data points for each row. However, these adjustment will have impacted the mean, median, standard deviation, and correlation values, as well as the skewness and kurtosis of the data set.
# #### Distribution
#
# Looking at the data in (somewhat) reverse order, we can see from the histograms that the data is not normally distributed, although some of the data looks somewhat normally distributed (temp). The both the distribution for rain and sunshine show long tails leading to the right, with slight “humps” in these tails. For values for the sun, the “hump” seems to be about 5 hours, suggesting that there is slightly more days with 4 – 5 hours of sunlight than 3 – 4 hours, but is trails off considerably after the 5 hour mark. Likewise the rainfall seems to have slight “hump” in the 12 to 16 mm range, but there are also plateaus in the 3 – 4 mm range, as well as around the 6 mm range. This would suggest that there is a slight clustering of rainfall amounts around these levels during the months in question.
#
# It is notable that there is a significant peak in frequency of days 0 (zero) hours of sunshine, and 0 (zero) millimetres of rainfall. Intuitively, this seems reasonable for hours of sunshine, as December is normally a fairly overcast and cloudy month. However, it is normally considered a fairly wet month, while this seems to suggest that it is often dry. This should not be confused with the relative humidity, which gives the feeling of damp that is often associated with the month.
#
# The temperature is somewhat more normally distributed than either the rainfall, or the hours of sunlight. However, even then the peak frequency is rather low, with the tails on either end being long and drawn out. There is also a “hump” in the 11 – 12 °C range.
#
# #### Skewness and Kurtosis
#
# The skewness of the values, as seen in the distribution histograms, is also clearly demonstrated in the skew values. The skew values for relative humidity, temperature, and visibility are all negatively skewed. This indicates that the mean is less median, and that the peak is to the right of both values. This is visible for the temp values in the plot above, where the mean is slightly less than the media, and the peak values are to the right of both the mean and median. This also suggests that there is a longer tail on the left than right of the mode. Additionally, the values for the skew are between -0.5 and 0, which indicates that the values are reasonably symmetric.
#
# With regards to the remaining two values (rain and sun), they are both positively skewed. This indicates that the mean is greater than the median values. Additionally, the peaks for a positively skewed distribution is to the left of both the mean and median. This is clearly demonstrated in the histogram above. However, the skew values for both the rain and sun are above 1 (2.37 and 1.1 respectively). This indicates that they are both heavy asymmetric, and very skewed. This corresponds with the values indicated in the histogram.
#
# Looking at the kurtosis, the kurtosis values for rain indicate that there is less grouping around the mean, and the values are more spread out. This is clear from the above plot, where there are small clusters of rain values between 8 and 10, 10 and 12, and 1 through 16 millimetres. For all the other variables, their kurtosis values indicate that the cluster closer to the mean than the normal standard deviation, as they are all negative. The most significant of these is relative humidity, which has the greatest cluster near the mean, while the kurtosis of the sun’s values are reasonably close, in clustering terms, to the normal distribution.
#
# #### Correlation
# While no regression analysis has been performed on the dataset, it is still worthwhile examining the correlation between the variables. The heatmap gives a visual representation of the correlation coefficient table above it. The three strongest correlations, either positive or negative are between:
# * Visibility and relative humidity (-0.611) – strong to very strong, but negative, indicating that as the relative humidity increase, visibility decreases, and vice versa.
# * Sunlight hours and temperature (-0.433) – strong(ish) negative correlation, indicating that as the sunlight hours increases, the temperature drops, and vice versa. While correlation does not imply causation, this correlation is unusual, in that temperature normally increases with sunlight. A possible explanation for this is that the cloud cover acts as blanket, which keeps heat in, but is absent when the sun is visible. This would seem to support the findings of [Matuszko & Weglarczyk (2014)](https://rmets.onlinelibrary.wiley.com/doi/full/10.1002/joc.4238).
# * Visibility and rain (-0.364) – weak-strong negative correlation, indicating that as the amount of rain increases, the visibility decreases, and vice versa. This would indicate that the level of rain reduces visibility, which is important for aircraft (all the readings are recorded at an airport), as it will impinge on their ability to see clearly at distance.
#
# The most significant positive correlation is between visibility and sunlight hours (0.280, weak). This suggests that the visibility increases as the period of sunlight increases. This would stand to reason as both sunlight and visibility are negatively correlated with rainfall.
#
# #### Standard statistics
# Looking first at the standard deviation. As we have already seen, the data is skewed both positively and negatively. In addition, most of the kurtosis values are not that close to zero. As such, the standard deviation is not relevant here. However, the standard deviations will be useful for the simulation later on.
#
# For the rain we see that the mean is considerably greater than the median, and is fact far closer to the 75% quartile value. Combined with the is the fact that both the minimum, and 25% quartile values are 0.00 mm, and the 75% quartile value is 0.2 mm would suggest that there are a large number of days with no recordable rainfall. This would seem slightly counterintuitive for Ireland during the winter. However, 25% of the rainfall values are between 2.5 and 15.5 mm, a range of 13 mm. This would suggest that when it does rain in Dublin, it rains reasonably heavily.
#
# The mean for the recorded temperatures is 6.8°C, while the median value is 6.85°C. This suggests that the temperature reading are more normally distributed than some of the other recordings. However, as we have seen, the skew and kurtosis values suggest that there is still a reasonable degree of skew in the values. The range is quite large, with the minimum value in the data set being below 0°C (-0.52°C) and the largest value is above 13°C (13.81°C). The interquartile range (25% - 75% quartile values) is less than 4.5°C, which suggests that the temperature, while reasonably cold, does not fluctuate as wildly as the minimum and maximum values indicate.
#
# The mean of the relative humidity is 87.15%, while the median relative humidity is 88.04%. Like the temperature readings, these values are quite close, and suggest that the distributions are fairly close to normally distributed. However, the skew and kurtosis values likewise indicate that there is some skew in the data. The high relative humidity levels, being above 73% give that damp feeling that is often associated with the wintertime in Ireland.
#
# Sunlight hours in dataset note that there is often not much sunlight visible during the month of December. The highest recorded number of hours if sunlight is nearly 7 hours (6 hours, 54 minutes). Considering that the shortest period of daylight (between sunrise and sunset) in December 2019 is expected to be 7 hours, 30 minutes on December 22nd [(Time and Date, 2019)](https://www.timeanddate.com/sun/ireland/dublin?month=12), this would indicate that for one particular day, there was almost no cloud cover. However, with the minimum, 25% quartile, and median values are all at or below 30 minutes (0.00, 0.10, 0.50 hours respectively), this would suggest that there is a large amount of cloud cover. From these statistics, it is also clear that the data is negatively skewed, as the mean is considerably less than the median.
#
# Visibility is defined as the *“greatest distance at which a black object … can be seen and recognised when observes against a bright background”* [(International Civil Aviation Organization, 2007)](https://www.wmo.int/pages/prog/www/ISS/Meetings/CT-MTDCF-ET-DRC_Geneva2008/Annex3_16ed.pdf). The mean visibility is nearly 22.5 km (22,480m), with the median visibility being just 63 m less (22,417m). This would suggest that the data is reasonably normally distributed, while still being skewed. The range of the visibility is quite large, with the minimum and maximum values being nearly 28 km apart (8,979m and 36,667 m respectively). However, the interquartile range (25% - 75% quartile values) is less than 9,000m, suggesting that there is a fairly constant, and stead range of visibility for the period of the dataset.
#
#
# ## Simulation of new data
# Looking back at the original data, and as per the discussion above, a number of statistics stand out. The most significant is that all the 5 sets of data are skewed to one degree or other. While the skewness of the temperature, relative humidity, and visibility data sets might lend themselves to being simulated using normal distribution, neither rain nor sun datasets could be so simulated. This leaves the option of simulating each dataset using a different random number generator approach, or looking for one random number generator that could simulate all the datasets on the same basis.
#
# As such, a number of random number generators were examined to see if they would be able to generate all the datasets required. These included np.random.multivariate, scipy.truncnorm, scipy.JohsnonB, and scipy.skewnorm.
#
# Issues with arose with the multivariate approach, in that it did not take into account the skew values for the datasets. Additionally, it generates a normal distribution pattern, which it has been determined that the datasets are not. It did, however, allow for the introduction of a covariance between the values, which would have helpful in more accurately simulating the relationship between the datasets. Similar to the multivariate distribution, the truncnorm also produces a normally distributed dataset. However, the truncnorm allows for values to be cut off at the required lower and upper bounds, as necessary, although this does produce some spikes in the frequency of these values. JohnsonB was also examined, as it does allow for the median, mean, variance, standard deviation, and skew to be used. However, due to the lack of tutorials using this method, it was discarded (*Note: There were only 3 videos available for the search terms “scipy.johnsonb skewness python”, and all pointed to the same site*).
#
# The final approach examined to simulate the datasets was scipy.skewnorm. This library had the advantage of using skewness as one of its variables, as well as the mean and standard deviation. However, it does produce a skewed normal distribution, which doesn’t exactly match the dataset. However, of the libraries and approaches examined, it produced the nearest to original results, when comparing to the mean, and standard deviation (see output below next code box). Additionally, there were a number of tutorials available, and some concise explanations of how the code worked, to enable its use.
#
# Using the scipy.skewnorm distribution, there are a number of steps that need to be taken to simulate the new dataset. These are:
# 1. Determine the skewness, standard deviation, and mean of each of the columns in the original data set, using the *skew()*, *std()*, and *mean()* functionality respectively.
# 2. Set the number of random values for each of the columns to be simulated to 93, to match the original dataset.
# 3. Based on [Weckesser (2018)](https://stackoverflow.com/questions/49367436/scipy-skewnorm-mean-not-matching-theory), for each of the columns (ie. Rain, Temperature, Relative Humidity, Sun, and Visibility) calculate the delta, adjusted standard deviation, and adjusted mean, using the formulae:
# > - Delta – skew / square_root (1 + skew ^ 2))
# > - Adjusted_StD – square_root( Std ^ 2) / (1 – 2 x (delta ^ 2) / pi))
# > - Adjusted_Mean – Mean – Adjusted_StD x square_root(2 / pi) * delta
# 4. Using the values derived from above, input the values into the scipy.skewnorm formula as follows:
# > - X = skewnorm.rvs(Skew, loc = Adjusted_Mean, scale = Adjusted_StD, size = sample_size)
# 5. These values can be put together into a new dataset.
# 6. From this dataset, we can check the simulated data against the original dataset. This will show how close the simulated data is against the original dataset. *Note: there is not set seed for these calculations, so the simulated values will change each time it is run.*
#
# +
# Simulation of new data
# Variables needed to generate random values
# Skewness
R_skew = Dublin_df["Rain (mm)"].skew()
RH_skew = Dublin_df["Rel. Hum. (%)"].skew()
S_skew = Dublin_df["Sun (hrs)"].skew()
T_skew = Dublin_df["Temp. (°C)"].skew()
V_skew = Dublin_df["Visibility (m)"].skew()
# Standard deviations
R_std = Dublin_df["Rain (mm)"].std()
RH_std = Dublin_df["Rel. Hum. (%)"].std()
S_std = Dublin_df["Sun (hrs)"].std()
T_std = Dublin_df["Temp. (°C)"].std()
V_std = Dublin_df["Visibility (m)"].std()
# Mean values
R_mean # Already calculated
S_mean # Already calculated
T_mean # Already calculated
RH_mean = Dublin_df["Rel. Hum. (%)"].mean()
V_mean = Dublin_df["Visibility (m)"].mean()
# Other variables
No_of_samples = 93
# Determine values using skewnorm (Weckesser, 2018)
# Rain
R_delta = R_skew / math.sqrt(1. + math.pow(R_skew, 2.))
R_adjStdev = math.sqrt(math.pow(R_std, 2.) / (1. - 2. * math.pow(R_delta, 2.) / math.pi))
R_adjMean = R_mean - R_adjStdev * math.sqrt(2. / math.pi) * R_delta
R_Random = skewnorm.rvs(R_skew, loc = R_adjMean, scale = R_adjStdev, size = No_of_samples)
# Relative Humidity
RH_delta = RH_skew / math.sqrt(1. + math.pow(RH_skew, 2.))
RH_adjStdev = math.sqrt(math.pow(RH_std, 2.) / (1. - 2. * math.pow(RH_delta, 2.) / math.pi))
RH_adjMean = RH_mean - RH_adjStdev * math.sqrt(2. / math.pi) * RH_delta
RH_Random = skewnorm.rvs(RH_skew, loc = RH_adjMean, scale = RH_adjStdev, size = No_of_samples)
# Sun
S_delta = S_skew / math.sqrt(1. + math.pow(S_skew, 2.))
S_adjStdev = math.sqrt(math.pow(S_std, 2.) / (1. - 2. * math.pow(S_delta, 2.) / math.pi))
S_adjMean = S_mean - S_adjStdev * math.sqrt(2. / math.pi) * S_delta
S_Random = skewnorm.rvs(S_skew, loc = S_adjMean, scale = S_adjStdev, size = No_of_samples)
# Temperature
T_delta = T_skew / math.sqrt(1. + math.pow(T_skew, 2.))
T_adjStdev = math.sqrt(math.pow(T_std, 2.) / (1. - 2. * math.pow(T_delta, 2.) / math.pi))
T_adjMean = T_mean - T_adjStdev * math.sqrt(2. / math.pi) * T_delta
T_Random = skewnorm.rvs(T_skew, loc = T_adjMean, scale = T_adjStdev, size = No_of_samples)
# Visibility
V_delta = V_skew / math.sqrt(1. + math.pow(V_skew, 2.))
V_adjStdev = math.sqrt(math.pow(V_std, 2.) / (1. - 2. * math.pow(V_delta, 2.) / math.pi))
V_adjMean = V_mean - V_adjStdev * math.sqrt(2. / math.pi) * V_delta
V_Random = skewnorm.rvs(V_skew, loc = V_adjMean, scale = V_adjStdev, size = No_of_samples)
# Create new, random dataframe
Random_df = pd.DataFrame({
"Date": Dublin_df["Date"],
"Rain (mm)": R_Random,
"Temp. (°C)": T_Random,
"Rel. Hum. (%)": RH_Random,
"Sun (hrs)": S_Random,
"Visibility (m)": V_Random
})
# Check the mean, and standard deviations of both the original and new datasets
print("Check the mean, standard deviation, and skewness of the original and simulated datasets")
print()
print("".ljust(16) + "Rain".ljust(15) + "Temp".ljust(15) + "Rel. Hum.".ljust(15) + "Sun".ljust(15) + "Visibility")
print("---------------------------------------------------------------------------------------")
# Means
print("Mean orig: %11.4f %14.4f %15.4f %13.4f %18.4f" %(R_mean, T_mean, RH_mean, S_mean, V_mean))
print("Mean sim: %12.4f %14.4f %15.4f %13.4f %18.4f" %(Random_df["Rain (mm)"].mean(), Random_df["Temp. (°C)"].mean(),
Random_df["Rel. Hum. (%)"].mean(), Random_df["Sun (hrs)"].mean(),
Random_df["Visibility (m)"].mean()))
print()
# Standard deviation
print("Std. Dev. orig: %4.4f %14.4f %14.4f %14.4f %17.4f" %(R_std, T_std, RH_std, S_std, V_std))
print("Std Dev. sim: %8.4f %14.4f %14.4f %14.4f %17.4f" %(Random_df["Rain (mm)"].std(), Random_df["Temp. (°C)"].std(),
Random_df["Rel. Hum. (%)"].std(), Random_df["Sun (hrs)"].std(),
Random_df["Visibility (m)"].std()))
print()
# Skewness
print("Skewness orig: %7.4f %14.4f %14.4f %14.4f %14.4f" %(R_skew, T_skew, RH_skew, S_skew, V_skew))
print("Skewness sim: %8.4f %14.4f %14.4f %14.4f %14.4f" %(Random_df["Rain (mm)"].skew(), Random_df["Temp. (°C)"].skew(),
Random_df["Rel. Hum. (%)"].skew(), Random_df["Sun (hrs)"].skew(),
Random_df["Visibility (m)"].skew()))
# -
# As we can see, some of the simulated values are reasonably close to the original data. However, it is noted that in some cases, both the size of the skewness, as well as the orientaion (positive/negative) has changed. This would indicate that the distribution used, while fairly accurate, may need to be further refined. Additionally, the small size of the data sample for each column could possibly affect the values, including the skewness changing orientation, as well as the discrepancies in the means and standard deviations. It is possible that a larger sample size, in the tens of thousands, would help reduce, if not eliminate these divergences.
#
# ### Additional checks
# It is worthwhile to check that the dataset conforms to the logical values imposed by the laws of physics or nature. For example, the lower and upper bounds of for relative humidiy are 0% and 100%. If the values in the simulated data are higher or lower than these bounds, then they will need to be corrected. This will, however, change the mean, standard deviation, and skewness, but need to be done nonetheless.
# +
# Print the description of the simulated dataset
# Get new values for rows and columns
new_row, new_col = Random_df.shape
print("Description of the simulated data")
print()
print("Shape of simulated dataframe")
print("Rows:", new_row)
print("Columns:", new_col)
print()
print(Random_df.describe())
# -
# Adjusting the simulated values to set values above or below the lower/upper bounds to those bounds.
# +
# Cleanse of data to ensure that the logical upper and lower bounds are adhered to
# Check for values above or below the upper or lower bounds for each variable
# Replace each variable outside these bounds with the boundary limit
# Rain has a lower bound of 0 mm per day, and an upper bound of the max of the original dataset
Random_df.loc[Random_df["Rain (mm)"] < 0, "Rain (mm)"] = 0
Random_df.loc[Random_df["Rain (mm)"] > Dublin_df["Rain (mm)"].max(), "Rain (mm)"] = Dublin_df["Rain (mm)"].max()
# Temperature has a lower bound of -15.7C, and an upper bound of 17.1C (respective records for Dublin in December)
Random_df.loc[Random_df["Temp. (°C)"] < -15.7, "Temp. (°C)"] = -15.7
Random_df.loc[Random_df["Temp. (°C)"] > 17.1, "Temp. (°C)"] = 17.1
# Relative humidity has a lower bound of 0 (%), and an uppder bound of 100 (%)
Random_df.loc[Random_df["Rel. Hum. (%)"] < 0, "Rel. Hum. (%)"] = 0
Random_df.loc[Random_df["Rel. Hum. (%)"] > 100, "Rel. Hum. (%)"] = 100
# Sun has a lower bound of 0 (hrs), and an upper bound of the max of the original dataset
Random_df.loc[Random_df["Sun (hrs)"] < 0, "Sun (hrs)"] = 0
Random_df.loc[Random_df["Sun (hrs)"] > Dublin_df["Sun (hrs)"].max(), "Sun (hrs)"] = Dublin_df["Sun (hrs)"].max()
# Visibility has a lower bound of 0, and an upper bound of the max of the original dataset
Random_df.loc[Random_df["Visibility (m)"] < 0, "Visibility (m)"] = 0
Random_df.loc[Random_df["Visibility (m)"] > Dublin_df["Visibility (m)"].max(), "Visibility (m)"] = Dublin_df["Visibility (m)"].max()
# Print descripiton of dataframe
print("Stats for original and simulated dataframes")
print()
print("Original data")
print(Dublin_df.describe())
print()
print("Simulated data")
print(Random_df.describe())
print()
print("Top 5 rows of simulated dataframe")
print(Random_df.head(5))
# -
# The histograms for the simulated data for rain, temperature, and sun values can be plotted as was done in for the [original dataset](#Plotting-Statistics).
# +
# Plot of distribution of simulated weather data
# For rain, new values for mean and median values
R_mean_r = Random_df["Rain (mm)"].mean()
R_median_r = Random_df["Rain (mm)"].median()
# For temp, new values for mean and median values
T_mean_r = Random_df["Temp. (°C)"].mean()
T_median_r = Random_df["Temp. (°C)"].median()
# For sun, new values for mean and median values
S_mean_r = Random_df["Sun (hrs)"].mean()
S_median_r = Random_df["Sun (hrs)"].median()
# Seaborn distplots showing both histograms and bell curves for temp, rain, and sun
sns.distplot(Random_df["Temp. (°C)"], axlabel = False, kde = False, label = "Temp. (°C)")
sns.distplot(Random_df["Rain (mm)"], axlabel = False, kde = False, label = "Rain (mm)")
sns.distplot(Random_df["Sun (hrs)"], axlabel = False, kde = False, label = "Sun (hrs)")
# Plotlines for mean and median
plt.axvline(R_mean_r, color = 'r', linestyle = "-", label = "Rain - mean")
plt.axvline(R_median_r, color = 'k', linestyle = "--", label = "Rain - median")
plt.axvline(T_mean_r, color = 'g', linestyle = "-", label = "Temp - mean")
plt.axvline(T_median_r, color = 'y', linestyle = "--", label = "Temp - median")
plt.axvline(S_median_r, color = 'b', linestyle = "-", label = "Sun - median")
plt.axvline(S_mean_r, color = 'm', linestyle = "--", label = "Sun - mean")
# Set size of plot area
plt.rcParams["figure.figsize"] = [12, 6]
# Set title, labels, and legend
plt.title("Distribution of rain, temp, and sun - simulated data")
plt.xlabel("mm (rain), °C (temp), hours (sun)")
plt.ylabel("Frequency")
plt.grid(b = True, which = "major", axis = "x")
plt.legend()
# Show plot
plt.show()
# -
# ## Further Analysis
# There are a number of further pieces of analysis that could be undertaken on the original and modified datasets used above. The original dataset was broken up into hourly readings, which were combined to form the dataset used to conduct this analysis. Analysis could be undertaken to determine the statistics for the hourly dataset, in order to allow for a more accurate simulation. This simulation could take into account the hours of sunrise and sunset, especially when correlating variables with respect to hours of sunlight.
#
# Additionally, both a larger sample size could have been taken, expanding the months used from one to all twelve, or using more years of data for a single month. These would allow for a greater refinement of the statistics, and could also be used to determine the effects of global warming on the overall weather. In the original dataset, there were 21 columns of data, the majority of which were excluded as they would have made the dataset too large and unwieldly. Adding some, or all, of these datasets back into the examination would undoubtedly produce more accurate results, especially around correlation, and distribution. This, in turn, would allow for greater simulation accuracy, both in terms of the random number generator to use, as well the values produced.
#
# Finally, it should be noted that any examination of the initial, unedited dataset, would need to take into account the changing climatic conditions, as well as the variations caused by the different seasons. Both of these issues would create challenges, as well as opportunities for further study.
#
# ## Bibliography
# * <NAME>., 2018. The Skew-Normal Probability Distribution. [Online] Available at: http://azzalini.stat.unipd.it/SN/index.html
# [Accessed 10 December 2019].
# * <NAME>., 2019. Introduction to Randomness and Random Numbers. [Online] Available at: https://www.random.org/randomness/
# [Accessed 7 November 2019].
# * <NAME>., 2019. Create random numbers with left skewed probability distribution. [Online] Available at: https://stackoverflow.com/questions/24854965/create-random-numbers-with-left-skewed-probability-distribution/56552531#56552531
# [Accessed 12 December 2019].
# * International Civil Aviation Organization, 2007. Meteorological Service for International Air Navigation, 16th Edition. [Online] Available at: https://www.wmo.int/pages/prog/www/ISS/Meetings/CT-MTDCF-ET-DRC_Geneva2008/Annex3_16ed.pdf
# [Accessed 12 December 2019].
# * <NAME>., 2016. Analysis of Weather data using Pandas, Python, and Seaborn. [Online] Available at: https://www.shanelynn.ie/analysis-of-weather-data-using-pandas-python-and-seaborn
# [Accessed 30 November 2019].
# * <NAME>. & <NAME>., 2014. Relationship between sunshine duration and air temperature and contemporary global warming. International Journal of Climatology, 35(12), pp. 3640 - 3653.
# * Met Eireann, 2010. Absolute maximum air temperatures (°C) for each month at selected stations. [Online] Available at: http://archive.met.ie/climate-ireland/extreme_maxtemps.pdf
# [Accessed 12 December 2019].
# * Met Eireann, 2010. Absolute minimum air temperatures (°C) for each month at selected stations. [Online] Available at: http://archive.met.ie/climate-ireland/extreme_mintemps.pdf
# [Accessed 12 December 2019].
# * Met Éireann, 2019. Dublin Airport Hourly Weather Station Data. [Online] Available at: https://data.gov.ie/dataset/dublin-airport-hourly-weather-station-data/resource/bbb2cb83-5982-48ca-9da1-95280f5a4c0d?inner_span=True
# [Accessed 30 November 2019].
# * SciPy.org, 2019. scipy.stats.johnsonsb. [Online] Available at: https://scipy.github.io/devdocs/generated/scipy.stats.johnsonsb.html#scipy.stats.johnsonsb
# [Accessed 10 December 2019].
# * SciPy.org, 2019. scipy.stats.skewnorm. [Online] Available at: https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.skewnorm.html
# [Accessed 12 December 2019].
# * Time and Date AS, 2019. Dublin, Ireland — Sunrise, Sunset, and Daylength, December 2019. [Online] Available at: https://www.timeanddate.com/sun/ireland/dublin?month=12
# [Accessed 13 December 2019].
# * <NAME>., 2018. scipy skewnorm mean not matching theory?. [Online] Available at: https://stackoverflow.com/questions/49367436/scipy-skewnorm-mean-not-matching-theory
# [Accessed 12 December 2019].
# * <NAME>., 2019. Correlation. [Online] Available at: http://mathworld.wolfram.com/Correlation.html
# [Accessed 3 November 2019].
# * <NAME>., 2019. Better Heatmaps and Correlation Matrix Plots in Python. [Online] Available at: https://towardsdatascience.com/better-heatmaps-and-correlation-matrix-plots-in-python-41445d0f2bec
# [Accessed 10 December 2019].
#
#
|
Weather data simulation.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="AOpGoE2T-YXS" colab_type="text"
# ##### Copyright 2018 The TensorFlow Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License").
#
# # Neural Machine Translation with Attention
#
# <table class="tfo-notebook-buttons" align="left"><td>
# <a target="_blank" href="https://colab.research.google.com/github/tensorflow/tensorflow/blob/master/tensorflow/contrib/eager/python/examples/nmt_with_attention/nmt_with_attention.ipynb">
# <img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
# </td><td>
# <a target="_blank" href="https://github.com/tensorflow/tensorflow/tree/master/tensorflow/contrib/eager/python/examples/nmt_with_attention/nmt_with_attention.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a></td></table>
# + [markdown] id="CiwtNgENbx2g" colab_type="text"
# This notebook trains a sequence to sequence (seq2seq) model for Spanish to English translation using [tf.keras](https://www.tensorflow.org/programmers_guide/keras) and [eager execution](https://www.tensorflow.org/programmers_guide/eager). This is an advanced example that assumes some knowledge of sequence to sequence models.
#
# After training the model in this notebook, you will be able to input a Spanish sentence, such as *"¿todavia estan en casa?"*, and return the English translation: *"are you still at home?"*
#
# The translation quality is reasonable for a toy example, but the generated attention plot is perhaps more interesting. This shows which parts of the input sentence has the model's attention while translating:
#
# <img src="https://tensorflow.org/images/spanish-english.png" alt="spanish-english attention plot">
#
# Note: This example takes approximately 10 mintues to run on a single P100 GPU.
# + id="tnxXKDjq3jEL" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
from __future__ import absolute_import, division, print_function
# Import TensorFlow >= 1.9 and enable eager execution
import tensorflow as tf
tf.enable_eager_execution()
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
import unicodedata
import re
import numpy as np
import os
import time
print(tf.__version__)
# + [markdown] id="wfodePkj3jEa" colab_type="text"
# ## Download and prepare the dataset
#
# We'll use a language dataset provided by http://www.manythings.org/anki/. This dataset contains language translation pairs in the format:
#
# ```
# May I borrow this book? ¿Puedo tomar prestado este libro?
# ```
#
# There are a variety of languages available, but we'll use the English-Spanish dataset. For convenience, we've hosted a copy of this dataset on Google Cloud, but you can also download your own copy. After downloading the dataset, here are the steps we'll take to prepare the data:
#
# 1. Add a *start* and *end* token to each sentence.
# 2. Clean the sentences by removing special characters.
# 3. Create a word index and reverse word index (dictionaries mapping from word → id and id → word).
# 4. Pad each sentence to a maximum length.
# + id="kRVATYOgJs1b" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
# Download the file
path_to_zip = tf.keras.utils.get_file(
'spa-eng.zip', origin='http://download.tensorflow.org/data/spa-eng.zip',
extract=True)
path_to_file = os.path.dirname(path_to_zip)+"/spa-eng/spa.txt"
# + id="rd0jw-eC3jEh" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
# Converts the unicode file to ascii
def unicode_to_ascii(s):
return ''.join(c for c in unicodedata.normalize('NFD', s)
if unicodedata.category(c) != 'Mn')
def preprocess_sentence(w):
w = unicode_to_ascii(w.lower().strip())
# creating a space between a word and the punctuation following it
# eg: "he is a boy." => "he is a boy ."
# Reference:- https://stackoverflow.com/questions/3645931/python-padding-punctuation-with-white-spaces-keeping-punctuation
w = re.sub(r"([?.!,¿])", r" \1 ", w)
w = re.sub(r'[" "]+', " ", w)
# replacing everything with space except (a-z, A-Z, ".", "?", "!", ",")
w = re.sub(r"[^a-zA-Z?.!,¿]+", " ", w)
w = w.rstrip().strip()
# adding a start and an end token to the sentence
# so that the model know when to start and stop predicting.
w = '<start> ' + w + ' <end>'
return w
# + id="OHn4Dct23jEm" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
# 1. Remove the accents
# 2. Clean the sentences
# 3. Return word pairs in the format: [ENGLISH, SPANISH]
def create_dataset(path, num_examples):
lines = open(path, encoding='UTF-8').read().strip().split('\n')
word_pairs = [[preprocess_sentence(w) for w in l.split('\t')] for l in lines[:num_examples]]
return word_pairs
# + id="9xbqO7Iie9bb" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
# This class creates a word -> index mapping (e.g,. "dad" -> 5) and vice-versa
# (e.g., 5 -> "dad") for each language,
class LanguageIndex():
def __init__(self, lang):
self.lang = lang
self.word2idx = {}
self.idx2word = {}
self.vocab = set()
self.create_index()
def create_index(self):
for phrase in self.lang:
self.vocab.update(phrase.split(' '))
self.vocab = sorted(self.vocab)
self.word2idx['<pad>'] = 0
for index, word in enumerate(self.vocab):
self.word2idx[word] = index + 1
for word, index in self.word2idx.items():
self.idx2word[index] = word
# + id="eAY9k49G3jE_" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
def max_length(tensor):
return max(len(t) for t in tensor)
def load_dataset(path, num_examples):
# creating cleaned input, output pairs
pairs = create_dataset(path, num_examples)
# index language using the class defined above
inp_lang = LanguageIndex(sp for en, sp in pairs)
targ_lang = LanguageIndex(en for en, sp in pairs)
# Vectorize the input and target languages
# Spanish sentences
input_tensor = [[inp_lang.word2idx[s] for s in sp.split(' ')] for en, sp in pairs]
# English sentences
target_tensor = [[targ_lang.word2idx[s] for s in en.split(' ')] for en, sp in pairs]
# Calculate max_length of input and output tensor
# Here, we'll set those to the longest sentence in the dataset
max_length_inp, max_length_tar = max_length(input_tensor), max_length(target_tensor)
# Padding the input and output tensor to the maximum length
input_tensor = tf.keras.preprocessing.sequence.pad_sequences(input_tensor,
maxlen=max_length_inp,
padding='post')
target_tensor = tf.keras.preprocessing.sequence.pad_sequences(target_tensor,
maxlen=max_length_tar,
padding='post')
return input_tensor, target_tensor, inp_lang, targ_lang, max_length_inp, max_length_tar
# + [markdown] id="GOi42V79Ydlr" colab_type="text"
# ### Limit the size of the dataset to experiment faster (optional)
#
# Training on the complete dataset of >100,000 sentences will take a long time. To train faster, we can limit the size of the dataset to 30,000 sentences (of course, translation quality degrades with less data):
# + id="cnxC7q-j3jFD" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
# Try experimenting with the size of that dataset
num_examples = 30000
input_tensor, target_tensor, inp_lang, targ_lang, max_length_inp, max_length_targ = load_dataset(path_to_file, num_examples)
# + id="4QILQkOs3jFG" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
# Creating training and validation sets using an 80-20 split
input_tensor_train, input_tensor_val, target_tensor_train, target_tensor_val = train_test_split(input_tensor, target_tensor, test_size=0.2)
# Show length
len(input_tensor_train), len(target_tensor_train), len(input_tensor_val), len(target_tensor_val)
# + [markdown] id="rgCLkfv5uO3d" colab_type="text"
# ### Create a tf.data dataset
# + id="TqHsArVZ3jFS" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
BUFFER_SIZE = len(input_tensor_train)
BATCH_SIZE = 64
embedding_dim = 256
units = 1024
vocab_inp_size = len(inp_lang.word2idx)
vocab_tar_size = len(targ_lang.word2idx)
dataset = tf.data.Dataset.from_tensor_slices((input_tensor_train, target_tensor_train)).shuffle(BUFFER_SIZE)
dataset = dataset.apply(tf.contrib.data.batch_and_drop_remainder(BATCH_SIZE))
# + [markdown] id="TNfHIF71ulLu" colab_type="text"
# ## Write the encoder and decoder model
#
# Here, we'll implement an encoder-decoder model with attention which you can read about in the TensorFlow [Neural Machine Translation (seq2seq) tutorial](https://www.tensorflow.org/tutorials/seq2seq). This example uses a more recent set of APIs. This notebook implements the [attention equations](https://www.tensorflow.org/tutorials/seq2seq#background_on_the_attention_mechanism) from the seq2seq tutorial. The following diagram shows that each input words is assigned a weight by the attention mechanism which is then used by the decoder to predict the next word in the sentence.
#
# <img src="https://www.tensorflow.org/images/seq2seq/attention_mechanism.jpg" width="500" alt="attention mechanism">
#
# The input is put through an encoder model which gives us the encoder output of shape *(batch_size, max_length, hidden_size)* and the encoder hidden state of shape *(batch_size, hidden_size)*.
#
# Here are the equations that are implemented:
#
# <img src="https://www.tensorflow.org/images/seq2seq/attention_equation_0.jpg" alt="attention equation 0" width="800">
# <img src="https://www.tensorflow.org/images/seq2seq/attention_equation_1.jpg" alt="attention equation 1" width="800">
#
# We're using *Bahdanau attention*. Lets decide on notation before writing the simplified form:
#
# * FC = Fully connected (dense) layer
# * EO = Encoder output
# * H = hidden state
# * X = input to the decoder
#
# And the pseudo-code:
#
# * `score = FC(tanh(FC(EO) + FC(H)))`
# * `attention weights = softmax(score, axis = 1)`. Softmax by default is applied on the last axis but here we want to apply it on the *1st axis*, since the shape of score is *(batch_size, max_length, hidden_size)*. `Max_length` is the length of our input. Since we are trying to assign a weight to each input, softmax should be applied on that axis.
# * `context vector = sum(attention weights * EO, axis = 1)`. Same reason as above for choosing axis as 1.
# * `embedding output` = The input to the decoder X is passed through an embedding layer.
# * `merged vector = concat(embedding output, context vector)`
# * This merged vector is then given to the GRU
#
# The shapes of all the vectors at each step have been specified in the comments in the code:
# + id="avyJ_4VIUoHb" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
def gru(units):
# If you have a GPU, we recommend using CuDNNGRU(provides a 3x speedup than GRU)
# the code automatically does that.
if tf.test.is_gpu_available():
return tf.keras.layers.CuDNNGRU(units,
return_sequences=True,
return_state=True,
recurrent_initializer='glorot_uniform')
else:
return tf.keras.layers.GRU(units,
return_sequences=True,
return_state=True,
recurrent_activation='sigmoid',
recurrent_initializer='glorot_uniform')
# + id="nZ2rI24i3jFg" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
class Encoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, enc_units, batch_sz):
super(Encoder, self).__init__()
self.batch_sz = batch_sz
self.enc_units = enc_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = gru(self.enc_units)
def call(self, x, hidden):
x = self.embedding(x)
output, state = self.gru(x, initial_state = hidden)
return output, state
def initialize_hidden_state(self):
return tf.zeros((self.batch_sz, self.enc_units))
# + id="yJ_B3mhW3jFk" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
class Decoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, dec_units, batch_sz):
super(Decoder, self).__init__()
self.batch_sz = batch_sz
self.dec_units = dec_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = gru(self.dec_units)
self.fc = tf.keras.layers.Dense(vocab_size)
# used for attention
self.W1 = tf.keras.layers.Dense(self.dec_units)
self.W2 = tf.keras.layers.Dense(self.dec_units)
self.V = tf.keras.layers.Dense(1)
def call(self, x, hidden, enc_output):
# enc_output shape == (batch_size, max_length, hidden_size)
# hidden shape == (batch_size, hidden size)
# hidden_with_time_axis shape == (batch_size, 1, hidden size)
# we are doing this to perform addition to calculate the score
hidden_with_time_axis = tf.expand_dims(hidden, 1)
# score shape == (batch_size, max_length, hidden_size)
score = tf.nn.tanh(self.W1(enc_output) + self.W2(hidden_with_time_axis))
# attention_weights shape == (batch_size, max_length, 1)
# we get 1 at the last axis because we are applying score to self.V
attention_weights = tf.nn.softmax(self.V(score), axis=1)
# context_vector shape after sum == (batch_size, hidden_size)
context_vector = attention_weights * enc_output
context_vector = tf.reduce_sum(context_vector, axis=1)
# x shape after passing through embedding == (batch_size, 1, embedding_dim)
x = self.embedding(x)
# x shape after concatenation == (batch_size, 1, embedding_dim + hidden_size)
x = tf.concat([tf.expand_dims(context_vector, 1), x], axis=-1)
# passing the concatenated vector to the GRU
output, state = self.gru(x)
# output shape == (batch_size * max_length, hidden_size)
output = tf.reshape(output, (-1, output.shape[2]))
# output shape == (batch_size * max_length, vocab)
x = self.fc(output)
return x, state, attention_weights
def initialize_hidden_state(self):
return tf.zeros((self.batch_sz, self.dec_units))
# + id="P5UY8wko3jFp" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
encoder = Encoder(vocab_inp_size, embedding_dim, units, BATCH_SIZE)
decoder = Decoder(vocab_tar_size, embedding_dim, units, BATCH_SIZE)
# + [markdown] id="_ch_71VbIRfK" colab_type="text"
# ## Define the optimizer and the loss function
# + id="WmTHr5iV3jFr" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
optimizer = tf.train.AdamOptimizer()
def loss_function(real, pred):
mask = 1 - np.equal(real, 0)
loss_ = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=real, logits=pred) * mask
return tf.reduce_mean(loss_)
# + [markdown] id="hpObfY22IddU" colab_type="text"
# ## Training
#
# 1. Pass the *input* through the *encoder* which return *encoder output* and the *encoder hidden state*.
# 2. The encoder output, encoder hidden state and the decoder input (which is the *start token*) is passed to the decoder.
# 3. The decoder returns the *predictions* and the *decoder hidden state*.
# 4. The decoder hidden state is then passed back into the model and the predictions are used to calculate the loss.
# 5. Use *teacher forcing* to decide the next input to the decoder.
# 6. *Teacher forcing* is the technique where the *target word* is passed as the *next input* to the decoder.
# 7. The final step is to calculate the gradients and apply it to the optimizer and backpropagate.
# + id="ddefjBMa3jF0" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
EPOCHS = 10
for epoch in range(EPOCHS):
start = time.time()
hidden = encoder.initialize_hidden_state()
total_loss = 0
for (batch, (inp, targ)) in enumerate(dataset):
loss = 0
with tf.GradientTape() as tape:
enc_output, enc_hidden = encoder(inp, hidden)
dec_hidden = enc_hidden
dec_input = tf.expand_dims([targ_lang.word2idx['<start>']] * BATCH_SIZE, 1)
# Teacher forcing - feeding the target as the next input
for t in range(1, targ.shape[1]):
# passing enc_output to the decoder
predictions, dec_hidden, _ = decoder(dec_input, dec_hidden, enc_output)
loss += loss_function(targ[:, t], predictions)
# using teacher forcing
dec_input = tf.expand_dims(targ[:, t], 1)
total_loss += (loss / int(targ.shape[1]))
variables = encoder.variables + decoder.variables
gradients = tape.gradient(loss, variables)
optimizer.apply_gradients(zip(gradients, variables), tf.train.get_or_create_global_step())
if batch % 100 == 0:
print('Epoch {} Batch {} Loss {:.4f}'.format(epoch + 1,
batch,
loss.numpy() / int(targ.shape[1])))
print('Epoch {} Loss {:.4f}'.format(epoch + 1,
total_loss/len(input_tensor)))
print('Time taken for 1 epoch {} sec\n'.format(time.time() - start))
# + [markdown] id="mU3Ce8M6I3rz" colab_type="text"
# ## Translate
#
# * The evaluate function is similar to the training loop, except we don't use *teacher forcing* here. The input to the decoder at each time step is its previous predictions along with the hidden state and the encoder output.
# * Stop predicting when the model predicts the *end token*.
# * And store the *attention weights for every time step*.
#
# Note: The encoder output is calculated only once for one input.
# + id="EbQpyYs13jF_" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
def evaluate(sentence, encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ):
attention_plot = np.zeros((max_length_targ, max_length_inp))
sentence = preprocess_sentence(sentence)
inputs = [inp_lang.word2idx[i] for i in sentence.split(' ')]
inputs = tf.keras.preprocessing.sequence.pad_sequences([inputs], maxlen=max_length_inp, padding='post')
inputs = tf.convert_to_tensor(inputs)
result = ''
hidden = [tf.zeros((1, units))]
enc_out, enc_hidden = encoder(inputs, hidden)
dec_hidden = enc_hidden
dec_input = tf.expand_dims([targ_lang.word2idx['<start>']], 0)
for t in range(max_length_targ):
predictions, dec_hidden, attention_weights = decoder(dec_input, dec_hidden, enc_out)
# storing the attention weigths to plot later on
attention_weights = tf.reshape(attention_weights, (-1, ))
attention_plot[t] = attention_weights.numpy()
predicted_id = tf.multinomial(tf.exp(predictions), num_samples=1)[0][0].numpy()
result += targ_lang.idx2word[predicted_id] + ' '
if targ_lang.idx2word[predicted_id] == '<end>':
return result, sentence, attention_plot
# the predicted ID is fed back into the model
dec_input = tf.expand_dims([predicted_id], 0)
return result, sentence, attention_plot
# + id="s5hQWlbN3jGF" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
# function for plotting the attention weights
def plot_attention(attention, sentence, predicted_sentence):
fig = plt.figure(figsize=(10,10))
ax = fig.add_subplot(1, 1, 1)
ax.matshow(attention, cmap='viridis')
fontdict = {'fontsize': 14}
ax.set_xticklabels([''] + sentence, fontdict=fontdict, rotation=90)
ax.set_yticklabels([''] + predicted_sentence, fontdict=fontdict)
plt.show()
# + id="sl9zUHzg3jGI" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
def translate(sentence, encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ):
result, sentence, attention_plot = evaluate(sentence, encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
print('Input: {}'.format(sentence))
print('Predicted translation: {}'.format(result))
attention_plot = attention_plot[:len(result.split(' ')), :len(sentence.split(' '))]
plot_attention(attention_plot, sentence.split(' '), result.split(' '))
# + id="WrAM0FDomq3E" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
translate('hace mucho frio aqui.', encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
# + id="zSx2iM36EZQZ" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
translate('esta es mi vida.', encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
# + id="A3LLCx3ZE0Ls" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
translate('¿todavia estan en casa?', encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
# + id="DUQVLVqUE1YW" colab_type="code" colab={"autoexec": {"startup": false, "wait_interval": 0}}
# wrong translation
translate('trata de averiguarlo.', encoder, decoder, inp_lang, targ_lang, max_length_inp, max_length_targ)
# + [markdown] id="RTe5P5ioMJwN" colab_type="text"
# ## Next steps
#
# * [Download a different dataset](http://www.manythings.org/anki/) to experiment with translations, for example, English to German, or English to French.
# * Experiment with training on a larger dataset, or using more epochs
#
|
tensorflow/contrib/eager/python/examples/nmt_with_attention/nmt_with_attention.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernel_info:
# name: python3
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <a href="https://githubtocolab.com/giswqs/geemap/blob/master/examples/notebooks/cartoee_projections.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab"/></a>
#
# Uncomment the following line to install [geemap](https://geemap.org) if needed.
# +
# # !pip install geemap
# -
# ## Working with projections in cartoee
# +
import ee
from geemap import cartoee
import cartopy.crs as ccrs
# %pylab inline
# -
ee.Initialize()
# ### Plotting an image on a map
#
# Here we are going to show another example of creating a map with EE results. We will use global sea surface temperature data for Jan-Mar 2018.
# get an earth engine image of ocean data for Jan-Mar 2018
ocean = (
ee.ImageCollection('NASA/OCEANDATA/MODIS-Terra/L3SMI')
.filter(ee.Filter.date('2018-01-01', '2018-03-01'))
.median()
.select(["sst"],["SST"])
)
# set parameters for plotting
# will plot the Sea Surface Temp with specific range and colormap
visualization = {'bands':"SST",'min':-2,'max':30}
# specify region to focus on
bbox = [-180,-88,180,88]
# +
fig = plt.figure(figsize=(10,7))
# plot the result with cartoee using a PlateCarre projection (default)
ax = cartoee.get_map(ocean,cmap='plasma',vis_params=visualization,region=bbox)
cb = cartoee.add_colorbar(ax,vis_params=visualization,loc='right',cmap='plasma')
ax.coastlines()
plt.show()
# -
# ### Mapping with different projections
#
# You can specify what ever projection is available within `cartopy` to display the results from Earth Engine. Here are a couple examples of global and regions maps using the sea surface temperature example. Please refer to the [`cartopy` projection documentation](https://scitools.org.uk/cartopy/docs/latest/crs/projections.html) for more examples with different projections.
# +
fig = plt.figure(figsize=(10,7))
# create a new Mollweide projection centered on the Pacific
projection = ccrs.Mollweide(central_longitude=-180)
# plot the result with cartoee using the Mollweide projection
ax = cartoee.get_map(ocean,vis_params=visualization,region=bbox,
cmap='plasma',proj=projection)
cb = cartoee.add_colorbar(ax,vis_params=visualization,loc='bottom',cmap='plasma',
orientation='horizontal')
ax.set_title("Mollweide projection")
ax.coastlines()
plt.show()
# +
fig = plt.figure(figsize=(10,7))
# create a new Goode homolosine projection centered on the Pacific
projection = ccrs.InterruptedGoodeHomolosine(central_longitude=-180)
# plot the result with cartoee using the Goode homolosine projection
ax = cartoee.get_map(ocean,vis_params=visualization,region=bbox,
cmap='plasma',proj=projection)
cb = cartoee.add_colorbar(ax,vis_params=visualization,loc='bottom',cmap='plasma',
orientation='horizontal')
ax.set_title("Goode homolosine projection")
ax.coastlines()
plt.show()
# +
fig = plt.figure(figsize=(10,7))
# create a new orographic projection focused on the Pacific
projection = ccrs.Orthographic(-130,-10)
# plot the result with cartoee using the orographic projection
ax = cartoee.get_map(ocean,vis_params=visualization,region=bbox,
cmap='plasma',proj=projection)
cb = cartoee.add_colorbar(ax,vis_params=visualization,loc='right',cmap='plasma',
orientation='vertical')
ax.set_title("Orographic projection")
ax.coastlines()
plt.show()
# -
# ### Warping artifacts
#
# Often times global projections are not needed so we use specific projection for the map that provides the best view for the geographic region of interest. When we use these, sometimes image warping effects occur. This is because `cartoee` only requests data for region of interest and when mapping with `cartopy` the pixels get warped to fit the view extent as best as possible. Consider the following example where we want to map SST over the south pole:
# +
fig = plt.figure(figsize=(10,7))
# Create a new region to focus on
spole = [-180,-88,180,0]
projection = ccrs.SouthPolarStereo()
# plot the result with cartoee focusing on the south pole
ax = cartoee.get_map(ocean,cmap='plasma',vis_params=visualization,region=spole,proj=projection)
cb = cartoee.add_colorbar(ax,vis_params=visualization,loc='right',cmap='plasma')
ax.coastlines()
ax.set_title('The South Pole')
plt.show()
# -
# As you can see from the result there are warping effects on the plotted image. There is really no way of getting aound this (other than requesting a larger extent of data which may not always be the case).
#
# So, what we can do is set the extent of the map to a more realistic view after plotting the image as in the following example:
# +
fig = plt.figure(figsize=(10,7))
# plot the result with cartoee focusing on the south pole
ax = cartoee.get_map(ocean,cmap='plasma',vis_params=visualization,region=spole,proj=projection)
cb = cartoee.add_colorbar(ax,vis_params=visualization,loc='right',cmap='plasma')
ax.coastlines()
ax.set_title('The South Pole')
# get bounding box coordinates of a zoom area
zoom = spole
zoom[-1] = -20
# convert bbox coordinate from [W,S,E,N] to [W,E,S,N] as matplotlib expects
zoom_extent = cartoee.bbox_to_extent(zoom)
# set the extent of the map to the zoom area
ax.set_extent(zoom_extent,ccrs.PlateCarree())
plt.show()
|
examples/notebooks/cartoee_projections.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: 'Python 3.8.5 64-bit (''udl'': conda)'
# name: python3
# ---
# # Neural networks with PyTorch
#
# Deep learning networks tend to be massive with dozens or hundreds of layers, that's where the term "deep" comes from. You can build one of these deep networks using only weight matrices as we did in the previous notebook, but in general it's very cumbersome and difficult to implement. PyTorch has a nice module `nn` that provides a nice way to efficiently build large neural networks.
# +
# Import necessary packages
# %matplotlib inline
# %config InlineBackend.figure_format = 'retina'
import numpy as np
import torch
import helper
import matplotlib.pyplot as plt
# -
#
# Now we're going to build a larger network that can solve a (formerly) difficult problem, identifying text in an image. Here we'll use the MNIST dataset which consists of greyscale handwritten digits. Each image is 28x28 pixels, you can see a sample below
#
# <img src='assets/mnist.png'>
#
# Our goal is to build a neural network that can take one of these images and predict the digit in the image.
#
# First up, we need to get our dataset. This is provided through the `torchvision` package. The code below will download the MNIST dataset, then create training and test datasets for us. Don't worry too much about the details here, you'll learn more about this later.
# +
### Run this cell
from torchvision import datasets, transforms
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5,), (0.5,)),
])
# Download and load the training data
trainset = datasets.MNIST('~/.pytorch/MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)
# -
# We have the training data loaded into `trainloader` and we make that an iterator with `iter(trainloader)`. Later, we'll use this to loop through the dataset for training, like
#
# ```python
# for image, label in trainloader:
# ## do things with images and labels
# ```
#
# You'll notice I created the `trainloader` with a batch size of 64, and `shuffle=True`. The batch size is the number of images we get in one iteration from the data loader and pass through our network, often called a *batch*. And `shuffle=True` tells it to shuffle the dataset every time we start going through the data loader again. But here I'm just grabbing the first batch so we can check out the data. We can see below that `images` is just a tensor with size `(64, 1, 28, 28)`. So, 64 images per batch, 1 color channel, and 28x28 images.
dataiter = iter(trainloader)
images, labels = dataiter.next()
print(type(images))
print(images.shape)
print(labels.shape)
# This is what one of the images looks like.
images[1].numpy().shape
plt.imshow(images[1].numpy().squeeze(), cmap='Greys_r');
# First, let's try to build a simple network for this dataset using weight matrices and matrix multiplications. Then, we'll see how to do it using PyTorch's `nn` module which provides a much more convenient and powerful method for defining network architectures.
#
# The networks you've seen so far are called *fully-connected* or *dense* networks. Each unit in one layer is connected to each unit in the next layer. In fully-connected networks, the input to each layer must be a one-dimensional vector (which can be stacked into a 2D tensor as a batch of multiple examples). However, our images are 28x28 2D tensors, so we need to convert them into 1D vectors. Thinking about sizes, we need to convert the batch of images with shape `(64, 1, 28, 28)` to a have a shape of `(64, 784)`, 784 is 28 times 28. This is typically called *flattening*, we flattened the 2D images into 1D vectors.
#
# Previously you built a network with one output unit. Here we need 10 output units, one for each digit. We want our network to predict the digit shown in an image, so what we'll do is calculate probabilities that the image is of any one digit or class. This ends up being a discrete probability distribution over the classes (digits) that tells us the most likely class for the image. That means we need 10 output units for the 10 classes (digits). We'll see how to convert the network output into a probability distribution next.
#
# > **Exercise:** Flatten the batch of images `images`. Then build a multi-layer network with 784 input units, 256 hidden units, and 10 output units using random tensors for the weights and biases. For now, use a sigmoid activation for the hidden layer. Leave the output layer without an activation, we'll add one that gives us a probability distribution next.
# +
## Your solution
# A flatten operation on a tensor reshapes the
# tensor to have a shape that is equal to the number
# of elements contained in the tensor.
# This is the same thing as a 1d-array of elements.
def sigmoid(x):
return torch.sigmoid(x)
# The -1 infers size from the other values
# Thus tensor (64,1,28,28) takes [0] as 64 rows
# and 28*28*1 as cols
input = images.view(images.shape[0],-1)
print(input.shape)
print(input.size())
weights = torch.randn(784,256)
bias = torch.randn(256)
h_weight = torch.randn(256,10)
h_bias = torch.randn(10)
# (64,784) (784,256) + (256)
h = sigmoid(torch.mm(input,weights) + bias)
# (64,256) (256,10)
out = torch.mm(h,h_weight) + h_bias
# output of your network, should have shape (64,10)
# +
def sigmoid1(x):
return 1/(1+torch.exp(-x))
#return torch.sigmoid(x)
def sigmoid2(x):
#return 1/(1+torch.exp(-x))
return torch.sigmoid(x)
print("sigmoid1: ",sigmoid1(torch.tensor(1.0)), " sigmoid2: ",sigmoid2(torch.tensor(1.0)))
# -
# Now we have 10 outputs for our network. We want to pass in an image to our network and get out a probability distribution over the classes that tells us the likely class(es) the image belongs to. Something that looks like this:
# <img src='assets/image_distribution.png' width=500px>
#
# Here we see that the probability for each class is roughly the same. This is representing an untrained network, it hasn't seen any data yet so it just returns a uniform distribution with equal probabilities for each class.
#
# To calculate this probability distribution, we often use the [**softmax** function](https://en.wikipedia.org/wiki/Softmax_function). Mathematically this looks like
#
# $$
# \Large \sigma(x_i) = \cfrac{e^{x_i}}{\sum_k^K{e^{x_k}}}
# $$
#
# What this does is squish each input $x_i$ between 0 and 1 and normalizes the values to give you a proper probability distribution where the probabilites sum up to one.
#
# > **Exercise:** Implement a function `softmax` that performs the softmax calculation and returns probability distributions for each example in the batch. Note that you'll need to pay attention to the shapes when doing this. If you have a tensor `a` with shape `(64, 10)` and a tensor `b` with shape `(64,)`, doing `a/b` will give you an error because PyTorch will try to do the division across the columns (called broadcasting) but you'll get a size mismatch. The way to think about this is for each of the 64 examples, you only want to divide by one value, the sum in the denominator. So you need `b` to have a shape of `(64, 1)`. This way PyTorch will divide the 10 values in each row of `a` by the one value in each row of `b`. Pay attention to how you take the sum as well. You'll need to define the `dim` keyword in `torch.sum`. Setting `dim=0` takes the sum across the rows while `dim=1` takes the sum across the columns.
# +
def softmax(x):
## TODO: Implement the softmax function here
a = torch.exp(x)
# b has a shape (64,10)
# need to re-shape to (64,1)
b = torch.sum(torch.exp(x), dim=1).view(64,1)
out = a / b
print("a=", a.shape, " b=", b.shape)
return(out)
# Here, out should be the output of the network in the previous excercise with shape (64,10)
probabilities = softmax(out)
# Does it have the right shape? Should be (64, 10)
print(probabilities.shape)
# Does it sum to 1?
print(probabilities.sum(dim=1))
# -
# ## Building networks with PyTorch
#
# PyTorch provides a module `nn` that makes building networks much simpler. Here I'll show you how to build the same one as above with 784 inputs, 256 hidden units, 10 output units and a softmax output.
from torch import nn
class Network(nn.Module):
def __init__(self):
super().__init__()
# Inputs to hidden layer linear transformation
self.hidden = nn.Linear(784, 256)
# Output layer, 10 units - one for each digit
self.output = nn.Linear(256, 10)
# Define sigmoid activation and softmax output
self.sigmoid = nn.Sigmoid()
self.softmax = nn.Softmax(dim=1)
def forward(self, x):
# Pass the input tensor through each of our operations
x = self.hidden(x)
x = self.sigmoid(x)
x = self.output(x)
x = self.softmax(x)
return x
# Let's go through this bit by bit.
#
# ```python
# class Network(nn.Module):
# ```
#
# Here we're inheriting from `nn.Module`. Combined with `super().__init__()` this creates a class that tracks the architecture and provides a lot of useful methods and attributes. It is mandatory to inherit from `nn.Module` when you're creating a class for your network. The name of the class itself can be anything.
#
# ```python
# self.hidden = nn.Linear(784, 256)
# ```
#
# This line creates a module for a linear transformation, $x\mathbf{W} + b$, with 784 inputs and 256 outputs and assigns it to `self.hidden`. The module automatically creates the weight and bias tensors which we'll use in the `forward` method. You can access the weight and bias tensors once the network (`net`) is created with `net.hidden.weight` and `net.hidden.bias`.
#
# ```python
# self.output = nn.Linear(256, 10)
# ```
#
# Similarly, this creates another linear transformation with 256 inputs and 10 outputs.
#
# ```python
# self.sigmoid = nn.Sigmoid()
# self.softmax = nn.Softmax(dim=1)
# ```
#
# Here I defined operations for the sigmoid activation and softmax output. Setting `dim=1` in `nn.Softmax(dim=1)` calculates softmax across the columns.
#
# ```python
# def forward(self, x):
# ```
#
# PyTorch networks created with `nn.Module` must have a `forward` method defined. It takes in a tensor `x` and passes it through the operations you defined in the `__init__` method.
#
# ```python
# x = self.hidden(x)
# x = self.sigmoid(x)
# x = self.output(x)
# x = self.softmax(x)
# ```
#
# Here the input tensor `x` is passed through each operation and reassigned to `x`. We can see that the input tensor goes through the hidden layer, then a sigmoid function, then the output layer, and finally the softmax function. It doesn't matter what you name the variables here, as long as the inputs and outputs of the operations match the network architecture you want to build. The order in which you define things in the `__init__` method doesn't matter, but you'll need to sequence the operations correctly in the `forward` method.
#
# Now we can create a `Network` object.
# Create the network and look at it's text representation
model = Network()
model
# You can define the network somewhat more concisely and clearly using the `torch.nn.functional` module. This is the most common way you'll see networks defined as many operations are simple element-wise functions. We normally import this module as `F`, `import torch.nn.functional as F`.
# +
import torch.nn.functional as F
class Network(nn.Module):
def __init__(self):
super().__init__()
# Inputs to hidden layer linear transformation
self.hidden = nn.Linear(784, 256)
# Output layer, 10 units - one for each digit
self.output = nn.Linear(256, 10)
def forward(self, x):
# Hidden layer with sigmoid activation
x = F.sigmoid(self.hidden(x))
# Output layer with softmax activation
x = F.softmax(self.output(x), dim=1)
return x
# -
# ### Activation functions
#
# So far we've only been looking at the sigmoid activation function, but in general any function can be used as an activation function. The only requirement is that for a network to approximate a non-linear function, the activation functions must be non-linear. Here are a few more examples of common activation functions: Tanh (hyperbolic tangent), and ReLU (rectified linear unit).
#
# <img src="assets/activation.png" width=700px>
#
# In practice, the ReLU function is used almost exclusively as the activation function for hidden layers.
# ### Your Turn to Build a Network
#
# <img src="assets/mlp_mnist.png" width=600px>
#
# > **Exercise:** Create a network with 784 input units, a hidden layer with 128 units and a ReLU activation, then a hidden layer with 64 units and a ReLU activation, and finally an output layer with a softmax activation as shown above. You can use a ReLU activation with the `nn.ReLU` module or `F.relu` function.
#
# It's good practice to name your layers by their type of network, for instance 'fc' to represent a fully-connected layer. As you code your solution, use `fc1`, `fc2`, and `fc3` as your layer names.
# +
## Your solution here
class Network(nn.Module):
def __init__(self):
super().__init__()
# Inputs to hidden layer linear transformation
self.fc1 = nn.Linear(784, 128)
self.fc2 = nn.Linear(128, 64)
# Output layer, 10 units - one for each digit
self.fc3 = nn.Linear(64, 10)
# Define sigmoid activation and softmax output
self.sigmoid = nn.Sigmoid()
self.softmax = nn.Softmax(dim=1)
self.relu = nn.ReLU()
def forward(self, x):
# Pass the input tensor through each of our operations
x = self.fc1(x)
x = self.relu(x)
x = self.fc2(x)
x = self.relu(x)
x = self.fc3(x)
x = self.softmax(x)
return x
model = Network()
model
# -
# ### Initializing weights and biases
#
# The weights and such are automatically initialized for you, but it's possible to customize how they are initialized. The weights and biases are tensors attached to the layer you defined, you can get them with `model.fc1.weight` for instance.
print(model.fc1.weight)
print(model.fc1.bias)
# For custom initialization, we want to modify these tensors in place. These are actually autograd *Variables*, so we need to get back the actual tensors with `model.fc1.weight.data`. Once we have the tensors, we can fill them with zeros (for biases) or random normal values.
# Set biases to all zeros
model.fc1.bias.data.fill_(0)
# sample from random normal with standard dev = 0.01
model.fc1.weight.data.normal_(std=0.01)
# ### Forward pass
#
# Now that we have a network, let's see what happens when we pass in an image.
# +
# Grab some data
dataiter = iter(trainloader)
images, labels = dataiter.next()
# Resize images into a 1D vector, new shape is (batch size, color channels, image pixels)
images.resize_(64, 1, 784)
# or images.resize_(images.shape[0], 1, 784) to automatically get batch size
# Forward pass through the network
img_idx = 0
ps = model.forward(images[img_idx,:])
img = images[img_idx]
helper.view_classify(img.view(1, 28, 28), ps)
# -
# As you can see above, our network has basically no idea what this digit is. It's because we haven't trained it yet, all the weights are random!
#
# ### Using `nn.Sequential`
#
# PyTorch provides a convenient way to build networks like this where a tensor is passed sequentially through operations, `nn.Sequential` ([documentation](https://pytorch.org/docs/master/nn.html#torch.nn.Sequential)). Using this to build the equivalent network:
# +
# Hyperparameters for our network
input_size = 784
hidden_sizes = [128, 64]
output_size = 10
# Build a feed-forward network
model = nn.Sequential(nn.Linear(input_size, hidden_sizes[0]),
nn.ReLU(),
nn.Linear(hidden_sizes[0], hidden_sizes[1]),
nn.ReLU(),
nn.Linear(hidden_sizes[1], output_size),
nn.Softmax(dim=1))
print(model)
# Forward pass through the network and display output
images, labels = next(iter(trainloader))
images.resize_(images.shape[0], 1, 784)
ps = model.forward(images[0,:])
helper.view_classify(images[0].view(1, 28, 28), ps)
# -
# Here our model is the same as before: 784 input units, a hidden layer with 128 units, ReLU activation, 64 unit hidden layer, another ReLU, then the output layer with 10 units, and the softmax output.
#
# The operations are available by passing in the appropriate index. For example, if you want to get first Linear operation and look at the weights, you'd use `model[0]`.
print(model[0])
model[0].weight
# You can also pass in an `OrderedDict` to name the individual layers and operations, instead of using incremental integers. Note that dictionary keys must be unique, so _each operation must have a different name_.
#
from collections import OrderedDict
model = nn.Sequential(OrderedDict([
('fc1', nn.Linear(input_size, hidden_sizes[0])),
('relu1', nn.ReLU()),
('fc2', nn.Linear(hidden_sizes[0], hidden_sizes[1])),
('relu2', nn.ReLU()),
('output', nn.Linear(hidden_sizes[1], output_size)),
('softmax', nn.Softmax(dim=1))]))
model
# Now you can access layers either by integer or the name
print(model[0])
print(model.fc1)
# In the next notebook, we'll see how we can train a neural network to accuractly predict the numbers appearing in the MNIST images.
|
intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Name
# Data preparation using Apache Pig on YARN with Cloud Dataproc
#
# # Label
# Cloud Dataproc, GCP, Cloud Storage, YARN, Pig, Apache, Kubeflow, pipelines, components
#
#
# # Summary
# A Kubeflow Pipeline component to prepare data by submitting an Apache Pig job on YARN to Cloud Dataproc.
#
#
# # Details
# ## Intended use
# Use the component to run an Apache Pig job as one preprocessing step in a Kubeflow Pipeline.
#
# ## Runtime arguments
# | Argument | Description | Optional | Data type | Accepted values | Default |
# |----------|-------------|----------|-----------|-----------------|---------|
# | project_id | The ID of the Google Cloud Platform (GCP) project that the cluster belongs to. | No | GCPProjectID | | |
# | region | The Cloud Dataproc region to handle the request. | No | GCPRegion | | |
# | cluster_name | The name of the cluster to run the job. | No | String | | |
# | queries | The queries to execute the Pig job. Specify multiple queries in one string by separating them with semicolons. You do not need to terminate queries with semicolons. | Yes | List | | None |
# | query_file_uri | The HCFS URI of the script that contains the Pig queries. | Yes | GCSPath | | None |
# | script_variables | Mapping of the query’s variable names to their values (equivalent to the Pig command: SET name="value";). | Yes | Dict | | None |
# | pig_job | The payload of a [PigJob](https://cloud.google.com/dataproc/docs/reference/rest/v1/PigJob). | Yes | Dict | | None |
# | job | The payload of a [Dataproc job](https://cloud.google.com/dataproc/docs/reference/rest/v1/projects.regions.jobs). | Yes | Dict | | None |
# | wait_interval | The number of seconds to pause between polling the operation. | Yes | Integer | | 30 |
#
# ## Output
# Name | Description | Type
# :--- | :---------- | :---
# job_id | The ID of the created job. | String
#
# ## Cautions & requirements
#
# To use the component, you must:
# * Set up a GCP project by following this [guide](https://cloud.google.com/dataproc/docs/guides/setup-project).
# * [Create a new cluster](https://cloud.google.com/dataproc/docs/guides/create-cluster).
# * Run the component under a secret [Kubeflow user service account](https://www.kubeflow.org/docs/started/getting-started-gke/#gcp-service-accounts) in a Kubeflow cluster. For example:
#
# ```
# component_op(...).apply(gcp.use_gcp_secret('user-gcp-sa'))
# ```
# * Grant the Kubeflow user service account the role `roles/dataproc.editor` on the project.
#
# ## Detailed description
# This component creates a Pig job from [Dataproc submit job REST API](https://cloud.google.com/dataproc/docs/reference/rest/v1/projects.regions.jobs/submit).
#
# Follow these steps to use the component in a pipeline:
# 1. Install the Kubeflow Pipeline SDK:
#
# +
# %%capture --no-stderr
KFP_PACKAGE = 'https://storage.googleapis.com/ml-pipeline/release/0.1.14/kfp.tar.gz'
# !pip3 install $KFP_PACKAGE --upgrade
# -
# 2. Load the component using KFP SDK
# +
import kfp.components as comp
dataproc_submit_pig_job_op = comp.load_component_from_url(
'https://raw.githubusercontent.com/kubeflow/pipelines/02c991dd265054b040265b3dfa1903d5b49df859/components/gcp/dataproc/submit_pig_job/component.yaml')
help(dataproc_submit_pig_job_op)
# -
# ### Sample
#
# Note: The following sample code works in an IPython notebook or directly in Python code. See the sample code below to learn how to execute the template.
#
#
# #### Setup a Dataproc cluster
#
# [Create a new Dataproc cluster](https://cloud.google.com/dataproc/docs/guides/create-cluster) (or reuse an existing one) before running the sample code.
#
#
# #### Prepare a Pig query
#
# Either put your Pig queries in the `queries` list, or upload your Pig queries into a file to a Cloud Storage bucket and then enter the Cloud Storage bucket’s path in `query_file_uri`. In this sample, we will use a hard coded query in the `queries` list to select data from a local `passwd` file.
#
# For more details on Apache Pig, see the [Pig documentation.](http://pig.apache.org/docs/latest/)
#
# #### Set sample parameters
# + tags=["parameters"]
PROJECT_ID = '<Please put your project ID here>'
CLUSTER_NAME = '<Please put your existing cluster name here>'
REGION = 'us-central1'
QUERY = '''
natality_csv = load 'gs://public-datasets/natality/csv' using PigStorage(':');
top_natality_csv = LIMIT natality_csv 10;
dump natality_csv;'''
EXPERIMENT_NAME = 'Dataproc - Submit Pig Job'
# -
# #### Example pipeline that uses the component
import kfp.dsl as dsl
import kfp.gcp as gcp
import json
@dsl.pipeline(
name='Dataproc submit Pig job pipeline',
description='Dataproc submit Pig job pipeline'
)
def dataproc_submit_pig_job_pipeline(
project_id = PROJECT_ID,
region = REGION,
cluster_name = CLUSTER_NAME,
queries = json.dumps([QUERY]),
query_file_uri = '',
script_variables = '',
pig_job='',
job='',
wait_interval='30'
):
dataproc_submit_pig_job_op(
project_id=project_id,
region=region,
cluster_name=cluster_name,
queries=queries,
query_file_uri=query_file_uri,
script_variables=script_variables,
pig_job=pig_job,
job=job,
wait_interval=wait_interval).apply(gcp.use_gcp_secret('user-gcp-sa'))
# #### Compile the pipeline
pipeline_func = dataproc_submit_pig_job_pipeline
pipeline_filename = pipeline_func.__name__ + '.zip'
import kfp.compiler as compiler
compiler.Compiler().compile(pipeline_func, pipeline_filename)
# #### Submit the pipeline for execution
# +
#Specify pipeline argument values
arguments = {}
#Get or create an experiment and submit a pipeline run
import kfp
client = kfp.Client()
experiment = client.create_experiment(EXPERIMENT_NAME)
#Submit a pipeline run
run_name = pipeline_func.__name__ + ' run'
run_result = client.run_pipeline(experiment.id, run_name, pipeline_filename, arguments)
# -
# ## References
# * [Create a new Dataproc cluster](https://cloud.google.com/dataproc/docs/guides/create-cluster)
# * [Pig documentation](http://pig.apache.org/docs/latest/)
# * [Dataproc job](https://cloud.google.com/dataproc/docs/reference/rest/v1/projects.regions.jobs)
# * [PigJob](https://cloud.google.com/dataproc/docs/reference/rest/v1/PigJob)
#
# ## License
# By deploying or using this software you agree to comply with the [AI Hub Terms of Service](https://aihub.cloud.google.com/u/0/aihub-tos) and the [Google APIs Terms of Service](https://developers.google.com/terms/). To the extent of a direct conflict of terms, the AI Hub Terms of Service will control.
|
components/gcp/dataproc/submit_pig_job/sample.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Read data with a time index
#
# Pandas DataFrame objects can have an index that denotes time. This is useful because Matplotlib recognizes that these measurements represent time and labels the values on the axis accordingly.
#
# In this exercise, you will read data from a CSV file called `climate_change.csv` that contains measurements of CO2 levels and temperatures made on the 6th of every month from 1958 until 2016. You will use Pandas' `read_csv` function.
#
# To designate the index as a `DateTimeIndex`, you will use the `parse_dates` and `index_col` key-word arguments both to parse this column as a variable that contains dates and also to designate it as the index for this DataFrame.
#
# _By the way, if you haven't downloaded it already, check out the [Matplotlib Cheat Sheet](https://datacamp-community-prod.s3.amazonaws.com/28b8210c-60cc-4f13-b0b4-5b4f2ad4790b). It includes an overview of the most important concepts, functions and methods and might come in handy if you ever need a quick refresher!_
#
# Instructions
#
# - Import the Pandas library as `pd`.
# - Read in the data from a CSV file called `'climate_change.csv'` using `pd.read_csv`.
# - Use the `parse_dates` key-word argument to parse the `"date"` column as dates.
# - Use the `index_col` key-word argument to set the `"date"` column as the index.
# +
# Import pandas as pd
import pandas as pd
# Read the data from file using read_csv
climate_change = pd.read_csv('climate_change.csv', parse_dates=['date'], index_col='date')
# -
# ## Plot time-series data
#
# To plot time-series data, we use the `Axes` object `plot` command. The first argument to this method are the values for the x-axis and the second argument are the values for the y-axis.
#
# This exercise provides data stored in a DataFrame called `climate_change`. This variable has a time-index with the dates of measurements and two data columns: `"co2"` and `"relative_temp"`.
#
# In this case, the index of the DataFrame would be used as the x-axis values and we will plot the values stored in the `"relative_temp"` column as the y-axis values. We will also properly label the x-axis and y-axis.
#
# Instructions
#
# - Add the data from `climate_change` to the plot: use the DataFrame `index` for the x value and the `"relative_temp"` column for the y values.
# - Set the x-axis label to `'Time'`.
# - Set the y-axis label to `'Relative temperature (Celsius)'`.
# - Show the figure.
# +
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
# Add the time-series for "relative_temp" to the plot
ax.plot(climate_change.index, climate_change['relative_temp'])
# Set the x-axis label
ax.set_xlabel('Time')
# Set the y-axis label
ax.set_ylabel('Relative temperature (Celsius)')
# Show the figure
plt.show()
# -
# ## Using a time index to zoom in
#
# When a time-series is represented with a time index, we can use this index for the x-axis when plotting. We can also select a to zoom in on a particular period within the time-series using Pandas' indexing facilities. In this exercise, you will select a portion of a time-series dataset and you will plot that period.
#
# The data to use is stored in a DataFrame called `climate_change`, which has a time-index with dates of measurements and two data columns: `"co2"` and `"relative_temp"`.
#
# Instructions
#
# - Use `plt.subplots` to create a Figure with one Axes called `fig` and `ax`, respectively.
# - Create a variable called `seventies` that includes all the data between `"1970-01-01"` and `"1979-12-31"`.
# - Add the data from `seventies` to the plot: use the DataFrame `index` for the x value and the `"co2"` column for the y values.
# +
# Use plt.subplots to create fig and ax
fig, ax = plt.subplots()
# Create variable seventies with data from "1970-01-01" to "1979-12-31"
seventies = climate_change['1970-01-01':'1979-12-31']
# Add the time-series for "co2" data from seventies to the plot
ax.plot(seventies.index, seventies['co2'])
# Show the figure
plt.show()
# -
# ## Plotting two variables
#
# If you want to plot two time-series variables that were recorded at the same times, you can add both of them to the same subplot.
#
# If the variables have very different scales, you'll want to make sure that you plot them in different twin Axes objects. These objects can share one axis (for example, the time, or x-axis) while not sharing the other (the y-axis).
#
# To create a twin Axes object that shares the x-axis, we use the `twinx` method.
#
# In this exercise, you'll have access to a DataFrame that has the `climate_change` data loaded into it. This DataFrame was loaded with the `"date"` column set as a `DateTimeIndex`, and it has a column called `"co2"` with carbon dioxide measurements and a column called `"relative_temp"` with temperature measurements.
#
# Instructions
#
# - Use `plt.subplots` to create a Figure and Axes objects called `fig` and `ax`, respectively.
# - Plot the carbon dioxide variable in blue using the Axes `plot` method.
# - Use the Axes `twinx` method to create a twin Axes that shares the x-axis.
# - Plot the relative temperature variable in the twin Axes using its `plot` method.
# +
import matplotlib.pyplot as plt
# Initalize a Figure and Axes
fig, ax = plt.subplots()
# Plot the CO2 variable in blue
ax.plot(climate_change.index, climate_change['co2'], color='blue')
# Create a twin Axes that shares the x-axis
ax2 = ax.twinx()
# Plot the relative temperature in red
ax2.plot(climate_change.index, climate_change['relative_temp'], color='red')
plt.show()
# -
# ## Defining a function that plots time-series data
# Once you realize that a particular section of code that you have written is useful, it is a good idea to define a function that saves that section of code for you, rather than copying it to other parts of your program where you would like to use this code.
#
# Here, we will define a function that takes inputs such as a time variable and some other variable and plots them as x and y inputs. Then, it sets the labels on the x- and y-axis and sets the colors of the y-axis label, the y-axis ticks and the tick labels.
#
# Instructions
#
# - Define a function called `plot_timeseries` that takes as input an Axes object (`axes`), data (`x`,`y`), a string with the name of a color and strings for x- and y-axis labels.
# - Plot y as a function of in the color provided as the input `color`.
# - Set the x- and y-axis labels using the provided input `xlabel` and `ylabel`, setting the y-axis label color using `color`.
# - Set the y-axis tick parameters using the `tick_params` method of the Axes object, setting the `colors` key-word to `color`.
# Define a function called plot_timeseries
def plot_timeseries(axes, x, y, color, xlabel, ylabel):
# Plot the inputs x,y in the provided color
axes.plot(x, y, color=color)
# Set the x-axis label
axes.set_xlabel(xlabel)
# Set the y-axis label
axes.set_ylabel(ylabel, color=color)
# Set the colors tick params for y-axis
axes.tick_params('y', colors=color)
# ## Using a plotting function
#
# Defining functions allows us to reuse the same code without having to repeat all of it. Programmers sometimes say ["Don't repeat yourself"](https://en.wikipedia.org/wiki/Don%27t_repeat_yourself).
#
# In the previous exercise, you defined a function called `plot_timeseries`:
#
# `plot_timeseries(axes, x, y, color, xlabel, ylabel)`
#
# that takes an Axes object (as the argument `axes`), time-series data (as `x` and `y` arguments) the name of a color (as a string, provided as the `color` argument) and x-axis and y-axis labels (as `xlabel` and `ylabel` arguments). In this exercise, the function `plot_timeseries` is already defined and provided to you.
#
# Use this function to plot the `climate_change` time-series data, provided as a Pandas DataFrame object that has a DateTimeIndex with the dates of the measurements and `co2` and `relative_temp` columns.
#
# Instructions
#
# - In the provided `ax` object, use the function `plot_timeseries` to plot the `"co2"` column in blue, with the x-axis label `"Time (years)"` and y-axis label `"CO2 levels"`.
# - Use the `ax.twinx` method to add an Axes object to the figure that shares the x-axis with `ax`.
# - Use the function `plot_timeseries` to add the data in the `"relative_temp"` column in red to the twin Axes object, with the x-axis label `"Time (years)"` and y-axis label `"Relative temperature (Celsius)"`.
# +
fig, ax = plt.subplots()
# Plot the CO2 levels time-series in blue
plot_timeseries(ax, climate_change.index, climate_change['co2'], 'blue', 'Time (years)', 'CO2 levels')
# Create a twin Axes object that shares the x-axis
ax2 = ax.twinx()
# Plot the relative temperature data in red
plot_timeseries(ax2, climate_change.index, climate_change['relative_temp'], 'red', 'Time (years)', 'Relative temperature (Celsius)')
plt.show()
# -
# ## Annotating a plot of time-series data
#
# Annotating a plot allows us to highlight interesting information in the plot. For example, in describing the climate change dataset, we might want to point to the date at which the relative temperature first exceeded 1 degree Celsius.
#
# For this, we will use the `annotate` method of the Axes object. In this exercise, you will have the DataFrame called `climate_change` loaded into memory. Using the Axes methods, plot only the relative temperature column as a function of dates, and annotate the data.
#
# Instructions
#
# - Use the `ax.plot` method to plot the DataFrame index against the `relative_temp` column.
# - Use the annotate method to add the text `'>1 degree'` in the location `(pd.Timestamp('2015-10-06'), 1)`.
# +
fig, ax = plt.subplots()
# Plot the relative temperature data
ax.plot(climate_change.index, climate_change['relative_temp'])
# Annotate the date at which temperatures exceeded 1 degree
ax.annotate(">1 degree", xy=(pd.Timestamp('2015-10-06'), 1))
plt.show()
# -
# ## Plotting time-series: putting it all together
#
# In this exercise, you will plot two time-series with different scales on the same Axes, and annotate the data from one of these series.
#
# The CO2/temperatures data is provided as a DataFrame called `climate_change`. You should also use the function that we have defined before, called `plot_timeseries`, which takes an Axes object (as the `axes` argument) plots a time-series (provided as x and y arguments), sets the labels for the x-axis and y-axis and sets the color for the data, and for the y tick/axis labels:
#
# `plot_timeseries(axes, x, y, color, xlabel, ylabel)`
#
# Then, you will annotate with text an important time-point in the data: on 2015-10-06, when the temperature first rose to above 1 degree over the average.
#
# Instructions
#
# - Use the `plot_timeseries` function to plot CO2 levels against time. Set xlabel to `"Time (years)"` ylabel to `"CO2 levels"` and color to `'blue'`.
# - Create `ax2`, as a twin of the first Axes.
# - In `ax2`, plot temperature against time, setting the color ylabel to `"Relative temp (Celsius)"` and color to `'red'`.
# - Annotate the data using the `ax2.annotate` method. Place the text `">1 degree"` in x=`pd.Timestamp('2008-10-06')`, y=`-0.2` pointing with a gray thin arrow to x=`pd.Timestamp('2015-10-06')`, y=`1`.
# +
fig, ax = plt.subplots()
# Plot the CO2 levels time-series in blue
plot_timeseries(ax, climate_change.index, climate_change['co2'], 'blue', 'Time (years)', 'CO2 levels')
# Create an Axes object that shares the x-axis
ax2 = ax.twinx()
# Plot the relative temperature data in red
plot_timeseries(ax2, climate_change.index, climate_change['relative_temp'], 'red', 'Time (years)', 'Relative temp (Celsius)')
# Annotate the point with relative temperature >1 degree
ax2.annotate('>1 degree', xy=(pd.Timestamp('2015-10-06'), 1), xytext=(pd.Timestamp('2008-10-06'), -0.2), arrowprops={'arrowstyle':'->', 'color':'gray'})
plt.show()
|
introduction_to_data_visualization_with_matplotlib/2_plotting_time_series.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### Outlier Detection with `bqplot`
# ---
# In this notebook, we create a class `DNA` that leverages the new bqplot canvas based [HeatMap](https://github.com/bloomberg/bqplot/blob/master/examples/Marks/HeatMap.ipynb) along with the ipywidgets Range Slider to help us detect and clean outliers in our data. The class accepts a DataFrame and allows you to visually and programmatically filter your outliers. The cleaned DataFrame can then be retrieved through a simple convenience function.
# +
from bqplot import (
DateScale,
ColorScale,
HeatMap,
Figure,
LinearScale,
OrdinalScale,
Axis,
)
from scipy.stats import percentileofscore
from scipy.interpolate import interp1d
import bqplot.pyplot as plt
from traitlets import List, Float, observe
from ipywidgets import IntRangeSlider, Layout, VBox, HBox, jslink
from pandas import DatetimeIndex
import numpy as np
import pandas as pd
def quantile_space(x, q1=0.1, q2=0.9):
"""
Returns a function that squashes quantiles between q1 and q2
"""
q1_x, q2_x = np.percentile(x, [q1, q2])
qs = np.percentile(x, np.linspace(0, 100, 100))
def get_quantile(t):
return np.interp(t, qs, np.linspace(0, 100, 100))
def f(y):
return np.interp(get_quantile(y), [0, q1, q2, 100], [-1, 0, 0, 1])
return f
class DNA(VBox):
colors = List()
q1 = Float()
q2 = Float()
def __init__(self, data, **kwargs):
self.data = data
date_x, date_y = False, False
transpose = kwargs.pop("transpose", False)
if transpose is True:
if type(data.index) is DatetimeIndex:
self.x_scale = DateScale()
if type(data.columns) is DatetimeIndex:
self.y_scale = DateScale()
x, y = list(data.columns.values), data.index.values
else:
if type(data.index) is DatetimeIndex:
date_x = True
if type(data.columns) is DatetimeIndex:
date_y = True
x, y = data.index.values, list(data.columns.values)
self.q1, self.q2 = kwargs.pop("quantiles", (1, 99))
self.quant_func = quantile_space(
self.data.values.flatten(), q1=self.q1, q2=self.q2
)
self.colors = kwargs.pop("colors", ["Red", "Black", "Green"])
self.x_scale = DateScale() if date_x is True else LinearScale()
self.y_scale = DateScale() if date_y is True else OrdinalScale(padding_y=0)
self.color_scale = ColorScale(colors=self.colors)
self.heat_map = HeatMap(
color=self.quant_func(self.data.T),
x=x,
y=y,
scales={"x": self.x_scale, "y": self.y_scale, "color": self.color_scale},
)
self.x_ax = Axis(scale=self.x_scale)
self.y_ax = Axis(scale=self.y_scale, orientation="vertical")
show_axes = kwargs.pop("show_axes", True)
self.axes = [self.x_ax, self.y_ax] if show_axes is True else []
self.height = kwargs.pop("height", "800px")
self.layout = kwargs.pop(
"layout", Layout(width="100%", height=self.height, flex="1")
)
self.fig_margin = kwargs.pop(
"fig_margin", {"top": 60, "bottom": 60, "left": 150, "right": 0}
)
kwargs.setdefault("padding_y", 0.0)
self.create_interaction(**kwargs)
self.figure = Figure(
marks=[self.heat_map],
axes=self.axes,
fig_margin=self.fig_margin,
layout=self.layout,
min_aspect_ratio=0.0,
**kwargs
)
super(VBox, self).__init__(
children=[self.range_slider, self.figure],
layout=Layout(align_items="center", width="100%", height="100%"),
**kwargs
)
def create_interaction(self, **kwargs):
self.range_slider = IntRangeSlider(
description="Filter Range",
value=(self.q1, self.q2),
layout=Layout(width="100%"),
)
self.range_slider.observe(self.slid_changed, "value")
self.observe(self.changed, ["q1", "q2"])
def slid_changed(self, new):
self.q1 = self.range_slider.value[0]
self.q2 = self.range_slider.value[1]
def changed(self, new):
self.range_slider.value = (self.q1, self.q2)
self.quant_func = quantile_space(
self.data.values.flatten(), q1=self.q1, q2=self.q2
)
self.heat_map.color = self.quant_func(self.data.T)
def get_filtered_df(self, fill_type="median"):
q1_x, q2_x = np.percentile(self.data, [self.q1, self.q2])
if fill_type == "median":
return self.data[(self.data >= q1_x) & (self.data <= q2_x)].apply(
lambda x: x.fillna(x.median())
)
elif fill_type == "mean":
return self.data[(self.data >= q1_x) & (self.data <= q2_x)].apply(
lambda x: x.fillna(x.mean())
)
else:
raise ValueError("fill_type must be one of ('median', 'mean')")
# -
# We define the size of our matrix here. Larger matrices require a larger height.
size = 100
# +
def num_to_col_letters(num):
letters = ""
while num:
mod = (num - 1) % 26
letters += chr(mod + 65)
num = (num - 1) // 26
return "".join(reversed(letters))
letters = []
for i in range(1, size + 1):
letters.append(num_to_col_letters(i))
# -
data = pd.DataFrame(np.random.randn(size, size), columns=letters)
data_dna = DNA(
data, title="DNA of our Data", height="1400px", colors=["Red", "White", "Green"]
)
data_dna
# Instead of setting the quantiles by the sliders, we can also set them programmatically. Using a range of (5, 95) restricts the data considerably.
data_dna.q1, data_dna.q2 = 5, 95
# Now, we can use the convenience function to extract a clean DataFrame.
data_clean = data_dna.get_filtered_df()
# The DNA fills outliers with the mean of the column. Alternately, we can fill the outliers by the mean.
data_mean = data_dna.get_filtered_df(fill_type="mean")
# We can also visualize the new DataFrame the same way to test how our outliers look now.
DNA(data_clean, title="Cleaned Data", height="1200px", colors=["Red", "White", "Green"])
|
examples/Applications/Outlier Detection.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: cv-nd
# language: python
# name: cv-nd
# ---
# ## Facial Filters
#
# Using your trained facial keypoint detector, you can now do things like add filters to a person's face, automatically. In this optional notebook, you can play around with adding sunglasses to detected face's in an image by using the keypoints detected around a person's eyes. Checkout the `images/` directory to see what pther .png's have been provided for you to try, too!
#
# <img src="images/face_filter_ex.png" width=60% height=60%/>
#
# Let's start this process by looking at a sunglasses .png that we'll be working with!
# import necessary resources
import matplotlib.image as mpimg
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import os
import cv2
# +
# load in sunglasses image with cv2 and IMREAD_UNCHANGED
sunglasses = cv2.imread('images/sunglasses.png', cv2.IMREAD_UNCHANGED)
# plot our image
plt.imshow(sunglasses)
# print out its dimensions
print('Image shape: ', sunglasses.shape)
# -
# ## The 4th dimension
#
# You'll note that this image actually has *4 color channels*, not just 3 as your avg RGB image does. This is due to the flag we set `cv2.IMREAD_UNCHANGED`, which tells this to read in another color channel.
#
# #### Alpha channel
# It has the usual red, blue, and green channels any color image has, and the 4th channel respresents the **transparency level of each pixel** in the image; this is often called the **alpha** channel. Here's how the transparency channel works: the lower the value, the more transparent, or see-through, the pixel will become. The lower bound (completely transparent) is zero here, so any pixels set to 0 will not be seen; these look like white background pixels in the image above, but they are actually totally transparent.
#
# This transparent channel allows us to place this rectangular image of sunglasses on an image of a face and still see the face area that is techically covered by the transparentbackground of the sunglasses image!
#
# Let's check out the alpha channel of our sunglasses image in the next Python cell. Because many of the pixels in the background of the image have an alpha value of 0, we'll need to explicitly print out non-zero values if we want to see them.
# print out the sunglasses transparency (alpha) channel
alpha_channel = sunglasses[:,:,3]
print ('The alpha channel looks like this (black pixels = transparent): ')
plt.imshow(alpha_channel, cmap='gray')
# just to double check that there are indeed non-zero values
# let's find and print out every value greater than zero
values = np.where(alpha_channel != 0)
print ('The non-zero values of the alpha channel are: ')
print (values)
# #### Overlaying images
#
# This means that when we place this sunglasses image on top of another image, we can use the transparency channel as a filter:
#
# * If the pixels are non-transparent (alpha_channel > 0), overlay them on the new image
#
# #### Keypoint locations
#
# In doing this, it's helpful to understand which keypoint belongs to the eyes, mouth, etc., so in the image below we also print the index of each facial keypoint directly on the image so you can tell which keypoints are for the eyes, eyebrows, etc.,
#
# <img src="images/landmarks_numbered.jpg" width=50% height=50%/>
#
# It may be useful to use keypoints that correspond to the edges of the face to define the width of the sunglasses, and the locations of the eyes to define the placement.
#
# Next, we'll load in an example image. Below, you've been given an image and set of keypoints from the provided training set of data, but you can use your own CNN model to generate keypoints for *any* image of a face (as in Notebook 3) and go through the same overlay process!
# +
# load in training data
key_pts_frame = pd.read_csv('data/training_frames_keypoints.csv')
# print out some stats about the data
print('Number of images: ', key_pts_frame.shape[0])
# -
# helper function to display keypoints
def show_keypoints(image, key_pts):
"""Show image with keypoints"""
plt.imshow(image)
plt.scatter(key_pts[:, 0], key_pts[:, 1], s=20, marker='.', c='m')
# +
# a selected image
n = 120
image_name = key_pts_frame.iloc[n, 0]
image = mpimg.imread(os.path.join('data/training/', image_name))
key_pts = key_pts_frame.iloc[n, 1:].as_matrix()
key_pts = key_pts.astype('float').reshape(-1, 2)
print('Image name: ', image_name)
plt.figure(figsize=(5, 5))
show_keypoints(image, key_pts)
plt.show()
# -
# Next, you'll see an example of placing sunglasses on the person in the loaded image.
#
# Note that the keypoints are numbered off-by-one in the numbered image above, and so `key_pts[0,:]` corresponds to the first point (1) in the labelled image.
# +
# Display sunglasses on top of the image in the appropriate place
# # copy of the face image for overlay
image_copy = np.copy(image)
# top-left location for sunglasses to go
# 17 = edge of left eyebrow
x = int(key_pts[17, 0])
y = int(key_pts[17, 1])
# height and width of sunglasses
# h = length of nose
h = int(abs(key_pts[27,1] - key_pts[34,1]))
# w = left to right eyebrow edges
w = int(abs(key_pts[17,0] - key_pts[26,0]))
# read in sunglasses
sunglasses = cv2.imread('images/sunglasses.png', cv2.IMREAD_UNCHANGED)
# resize sunglasses
new_sunglasses = cv2.resize(sunglasses, (w, h), interpolation = cv2.INTER_CUBIC)
# get region of interest on the face to change
roi_color = image_copy[y:y+h,x:x+w]
# find all non-transparent pts
ind = np.argwhere(new_sunglasses[:,:,3] > 0)
# for each non-transparent point, replace the original image pixel with that of the new_sunglasses
for i in range(3):
roi_color[ind[:,0],ind[:,1],i] = new_sunglasses[ind[:,0],ind[:,1],i]
# set the area of the image to the changed region with sunglasses
image_copy[y:y+h,x:x+w] = roi_color
# display the result!
plt.imshow(image_copy)
# -
# #### Further steps
#
# Look in the `images/` directory to see other available .png's for overlay! Also, you may notice that the overlay of the sunglasses is not entirely perfect; you're encouraged to play around with the scale of the width and height of the glasses and investigate how to perform [image rotation](https://docs.opencv.org/3.0-beta/doc/py_tutorials/py_imgproc/py_geometric_transformations/py_geometric_transformations.html) in OpenCV so as to match an overlay with any facial pose.
|
4. Fun with Keypoints.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="0wPixjHS38aO"
# # **Beginner's Python - Session Two Physics/Engineering Answers**
# + [markdown] id="tdITd2KuzKT3"
# ## **Numerically solving an ODE**
# + [markdown] id="jTKCIuxNPWEK"
# In this exercise we will be writing some code which generates a plot of the motion of a mass hanging on the end of an (idealised) spring. This will involve solving the following linear differential equation numerically using Euler's method.
#
# $$\frac{d^2x}{dt^2} = -\frac{k}{m}x-g $$
#
# If you're unfamiliar with Euler's method, you can check out https://tutorial.math.lamar.edu/classes/de/eulersmethod.aspx.
#
# + [markdown] id="eZPu1rpXnesd"
# First of all, in the cell below write code which takes a user input and asks has the text "Enter initial position coordinate".
#
# You should assign this user input - *cast as a float* - to a variable valled ```x0```. After you've run this cell, input a value between -5.0 and 5.0 and hit enter.
# + colab={"base_uri": "https://localhost:8080/", "height": 34} id="AerNBkAgoOP1" outputId="be07e24a-8479-4caf-8e93-1d7837d3c5d2"
x0 = float(input("Please input an initial position"))
# + [markdown] id="hPA1ELN6TJ9S"
# Now run the cell below. You should see a graph generated which shows the numerical solutions for both velocity and position of the mass. You can also edit the parameter values at the top and re-run the cell to see the effect on the numerical solution.
#
# **Note:** Don't worry about the details of the code, but know that it gives us
# + colab={"base_uri": "https://localhost:8080/", "height": 279} id="lgJB8aeRLf9U" outputId="03fc3962-0f3f-4292-cae8-30eb3415d15e"
# Do not edit the code in this cell. You can edit the 6 parameters at the top and re-run
# the cell to see the effect on the graph, but only after you have completed the questions.
import numpy as np
import matplotlib.pyplot as plt
N = 2000 # the number of steps used - higher N results in a more accurate result
v0 = 0.0 # initial velocity of the mass
tau = 5.0 # number of seconds we are solving over
k = 3.5 # spring constant
mass = 0.2 # mass
gravity = 9.81 # strength of gravity
time = np.linspace(0, tau, N)
dt = tau/float(N-1) # time between each step
def euler_method(y, t, dt, derivs):
y_next = y + derivs(y,t) * dt
return y_next
y = np.zeros([N,2])
y[0,0] = x0
y[0,1] = v0
def SHO(state, time):
g0 = state[1]
g1 = - k / mass * state[0] - gravity
return np.array([g0, g1])
for i in range(N-1):
y[i+1] = euler_method(y[i], time[i], dt, SHO)
x_data = [y[i,0] for i in range(N)] # this creates a long list containing the position coordinates
v_data = [y[i,1] for i in range(N)] # this does the same for velocity
plt.plot(time, x_data) # these just create a graph of the data
plt.plot(time, v_data)
plt.xlabel("time (s)")
plt.ylabel("position (m), velocity (m/s)")
plt.show()
# + [markdown] id="tDu8X3f0crEk"
# The above code also gives us two *lists*, each containing N numbers. These are ```x_data```, containing the position coordinates for a range of times, and ```v_data```, containing the velocities. Already it's clear that Python is extremely useful handling these lists, since they are too large for us to do things with them by hand.
# + [markdown] id="kdbp1Dz-eoMJ"
# Print below the following, replacing the #### with the correct value, rounded to 5 decimal places: **"The maximum position value achieved was #### and the maximum velocity was ####"**
# + colab={"base_uri": "https://localhost:8080/", "height": 34} id="rKcQI7cfe189" outputId="055dee63-1cbb-4bb9-9dc9-9597db8c86ea"
print("The maximum position value achieved was", round(max(x_data),5), "and the maximum velocity was", round(max(v_data),5))
# + [markdown] id="dwxl0SC7piII"
# What was the range in values of the velocity? Print your answer below to two decimal places. Remember that since ```range``` is a reserved name in Python, you should pick a different one.
# + colab={"base_uri": "https://localhost:8080/", "height": 34} id="1kVMEOAGpqA-" outputId="0d6a0d21-4a31-42b8-8c5b-53924e1cdff3"
spread = round(max(v_data) - min(v_data),2)
print(spread)
# + [markdown] id="inHw8zI6gd_u"
# A useful feature in Python is the ability to specify a single element of a list. Each entry of a list is numbered, *starting from 0*, and you can then specify an entry by putting the position in square brackets after the list. For example:
#
# + colab={"base_uri": "https://localhost:8080/", "height": 51} id="GxewM49HhJbM" outputId="7e2375b6-9bf5-4a3e-9410-235ebc3ccf44"
example_list = [1,3,5,7,9]
print(example_list[3])
print(example_list[0])
# + [markdown] id="kglgz634ktnD"
# Print below the 444th entry in the list ```v_data``` rounded to 4 decimal places (for simplicity, we will consider the first entry as the "zeroth" entry, since Python starts counting at 0.)
#
#
#
# + colab={"base_uri": "https://localhost:8080/", "height": 34} id="jyxlCzmIk4xL" outputId="51dc21b2-eb0f-491d-e4ef-e5d025745e06"
print(round(v_data[444],4))
# + [markdown] id="EPoJC30nhmsW"
# You can also add new elements to the end of a list, using the ```.append()``` function. You simply write the function after a list, and can put *one* new element in the brackets.
# + colab={"base_uri": "https://localhost:8080/", "height": 34} id="lFy6xH1jh621" outputId="6b282797-68cb-4562-a7b6-8aa76fb6b2ed"
example_list.append(20)
print(example_list)
# + [markdown] id="5jheSpYFrG5k"
# In the cell below there is a list defined, which contains the maximum/minimum values for both position and velocity. You must add two more elements onto the list, namely the mean values for both parameters, and then print the list.
#
# **Notes:**
# * You should calculate the mean by summing all of the data values and dividing by the number of values, ```N```.
# * Enter values to three decimal places.
#
# Hint: Create two new variables and then append them onto ```data_set```.
# + colab={"base_uri": "https://localhost:8080/", "height": 34} id="67gbIfjssTZa" outputId="92c8e83f-fe7d-44d0-dba0-10c45b6746fd"
x_max = round(max(x_data),3)
x_min = round(min(x_data),3)
v_max = round(max(v_data),3)
v_min = round(min(v_data),3)
data_set = [x_max, x_min, v_max, v_min]
x_mean = round(sum(x_data) / N, 3)
v_mean = round(sum(v_data) / N, 3)
data_set.append(x_mean)
data_set.append(v_mean)
print(data_set)
|
session-two/subject_questions/PhysEng_two_answers.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda root]
# language: python
# name: conda-root-py
# ---
# # Introduction to Data Science
# ---
#
# Welcome to Data Science! In this notebook, you will learn how to use Jupyter Notebooks and the basics of programming in Python.
#
# *Estimated Time: 30 minutes*
#
# ---
#
# **Topics Covered:**
# - Learn how to work with Jupyter notebooks.
# - Learn about variables in Python, including variable types, variable assignment, and arithmetic.
# - Learn about functions in Python, including defining and calling functions, as well as scope.
#
# **Parts:**
# - Jupyter Notebooks
# - Programming in Python
# - Variables
# - Functions
# - Scope
# ## Jupyter Notebooks
# ---
# In this section, we will learn the basics of how to work with Jupyter notebooks.
# This Jupyter notebook is composed of 2 kinds of cells: markdown and code. A **markdown cell**, such as this one, contains text. A **code cell** contains code in Python, a programming language that we will be using for the remainder of this module.
#
# To run a code cell, press Shift-Enter or click Cell > Run Cells in the menu at the top of the screen. To edit a code cell, simply click in the cell and make your changes.
# ### Exercise
#
# Try running the code below. What happens?
# CODE
print("Hello World!")
# Now, let's try editing the code. In the cell below, replace "friend" with your name for a more personalized message.
print("Welcome to Jupyter notebooks, friend.")
# ## Programming in Python
# ---
# Now that you are comfortable with using Jupyter notebooks, we can learn more about programming in this notebook.
#
# ### What is Programming?
# **Programming** is giving the computer a set of step-by-step instructions to follow in order to execute a task. It's a lot like writing your own recipe book! For example, let's say you wanted to teach someone how to make a PB&J sandwich:
# 1. Gather bread, peanut butter, jelly, and a spreading knife.
# 2. Take out two slices of bread.
# 3. Use the knife to spread peanut butter on one slice of bread.
# 4. Use the knife to spread jelly on the other slice of bread.
# 5. Put the two slices of bread together to make a sandwich.
#
# Just like that, programming is breaking up a complex task into smaller commands for the computer to understand and execute.
#
# In order to communicate with computers, however, we must talk to them in a way that they can understand us: via a **programming language**.
#
# There are many different kinds of programming languages, but we will be using **Python** because it is concise, simple to read, and applicable in a variety of projects - from web development to mobile apps to data analysis.
# ## Variables
# ---
# In programming, we often compute many values that we want to save so that we can use the result in a later step. For example, let's say that we want to find the number of seconds in a day. We can easily calculate this with the following:
# <p style="text-align: center">$60 * 60 * 24 = 86400$ seconds</p>
# However, let's say that your friend Alexander asked you how many seconds there are in three days. We could, of course, perform the calculation in a similar manner:
# <p style="text-align: center">$(60 * 60 * 24) * 3 = 259200$ seconds</p>
# But we see that we repeated the calculation in parentheses above. Instead of doing this calculation again, we could have saved the result from our first step (calculating the number of seconds in a day) as a variable.
# +
# This is Python code that assigns variables.
# The name to the left of the equals sign is the variable name.
# The value to the right of the equals sign is the value of the variable.
# Press Shift-Enter to run the code and see the value of our variable!
seconds_in_day = 60 * 60 * 24 # This is equal to 86400.
seconds_in_day
# -
# Then, we can simply multiply this variable by three to get the number of seconds in *three* days:
# +
# The code below takes the number of seconds in a day (which we calculated in the previous code cell)
# and multiplies it by 3 to find the number of seconds in 3 days.
seconds_in_three_days = seconds_in_day * 3 # This is equal to 259200.
seconds_in_three_days
# -
# As you can see, variables can be used to simplify calculations, make code more readable, and allow for repetition and reusability of code.
#
# ### Variable Types
#
# Next, we'll talk about a few types of variables that you'll be using. As we saw in the example above, one common type of variable is the *integer* (positive and negative whole numbers). You'll also be using decimal numbers in Python, which are called *doubles* (positive and negative decimal numbers).
#
# A third type of variable used frequently in Python is the *string*; strings are essentially sequences of characters, and you can think of them as words or sentences. We denote strings by surrounding the desired value with quotes. For example, "Data Science" and "2017" are strings, while `bears` and `2020` (both without quotes) are not strings.
#
# Finally, the last variable type we'll go over is the *boolean*. They can take on one of two values: `True` or `False`. Booleans are often used to check conditions; for example, we might have a list of dogs, and we want to sort them into small dogs and large dogs. One way we could accomplish this is to say either `True` or `False` for each dog after seeing if the dog weighs more than 15 pounds.
#
# Here is a table that summarizes the information in this section:
# |Variable Type|Definition|Examples|
# |-|-|-|
# |Integer|Positive and negative whole numbers|`42`, `-10`, `0`|
# |Double|Positive and negative decimal numbers|`73.9`, `2.4`, `0.0`|
# |String|Sequence of characters|`"Go Bears!"`, `"variables"`|
# |Boolean|True or false value|`True`, `False`|
#
# ### Arithmetic
# Now that we've discussed what types of variables we can use, let's talk about how we can combine them together. As we saw at the beginning of this section, we can do basic math in Python. Here is a table that shows how to write such operations:
#
# |Operation|Operator|Example|Value|
# |-|-|-|
# |Addition|+|`2 + 3`|`5`|
# |Subtraction|-|`2 - 3`|`-1`|
# |Multiplication|*|`2 * 3`|`6`|
# |Division|/|`7 / 3`|`2.66667`|
# |Remainder|%|`7 % 3`|`1`|
# |Exponentiation|**|`2 ** 0.5`|`1.41421`|
#
# In addition, you can use parentheses to denote priority, just like in math.
#
# As an exercise, try to predict what each of these lines below will print out. Then, run the cell and check your answers.
# +
q_1 = (3 + 4) / 2
print(q_1) # What prints here?
q_2 = 3 + 4 / 2
print(q_2) # What prints here?
some_variable = 1 + 2 + 3 + 4 + 5
q_3 = some_variable * 4
print(q_3) # What prints here?
q_4 = some_variable % 3
print(q_4) # What prints here?
step_1 = 6 * 5 - (6 * 3)
step_2 = (2 ** 3) / 4 * 7
q_5 = 1 + step_1 ** 2 * step_2
print(q_5) # What prints here?
# -
# ## Functions
# So far, you've learnt how to carry out basic operations on your inputs and assign variables to certain values.
# Now, let's try to be more efficient.
#
# Let's say we want to perform a certain operation on many different inputs that will produce distinct outputs. What do we do? We write a _**function**_.
#
# A function is a block of code which works a lot like a machine: it takes an input, does something to it, and produces an output.
#
# The input is put between brackets and can also be called the _argument_ or _parameter_. Functions can have multiple arguments.
#
# Try running the cell below after changing the variable _name_:
#
# +
# Edit this cell to your own name!
name = "<NAME>"
# Our function
def hello(name):
return "Hello " + name + "!"
hello(name)
# -
# Interesting, right? Now, you don't need to write 10 different lines with 10 different names to print a special greeting for each person. All you need to is write one function that does all the work for you!
#
# Functions are very useful in programming because they help you write shorter and more modular code. A good example to think of is the _print_ function, which we've used quite a lot in this module. It takes many different inputs and performs the specified task, printing its input, in a simple manner.
#
# Now, let's write our own function. Let's look at the following rules:
#
# ### Defining
# - All functions must start with the "def" keyword.
# - All functions must have a name, followed by parentheses, followed by a colon. Eg. def hello( ):
# - The brackets may have a variable that stores its arguments (inputs)
# - All functions must have a "return" statement which will return the output. Think of a function like a machine. When you put something inside, you want it to return something. Hence, this is very important.
#
# ### Calling
# After you define a function, it's time to use it. This is known as _calling_ a function.
#
# To call a function, simply write the name of the function with your input variable in brackets (argument).
#
# +
# Complete this function
def #name(argument):
return # function must return a value
# Calling our function below...
my_first_function(name)
# -
# Great! Now let's do some math. Let's write a function that returns the square of the input.
#
# Try writing it from scratch!
# +
# square function
square(5)
# -
# Neat stuff! Try different inputs and check if you get the correct answer each time.
#
# You've successfully written your first function from scratch! Let's take this up one notch.
#
# #### The power function
#
# _pow_ is a function that takes in two numbers: x, which is the "base" and y, the "power". So when you write pow(3,2) the function returns 3 raised to the power 2, which is 3^2 = 9.
#
# Task: Write a function called _mulpowply_ which takes in three inputs (x, y, z) and returns the value of x multiplied by y to power z. Symbolically, it should return (xy)^z.
# +
# mulpowply function
# -
# ## Scope
# ---
# Programming is great, but it can also be quite peculiar sometimes. For example, each variable defined outside of any functions by default, is **global**.
#
# Try executing the code below:
# +
# Global Variable - name
name = "<NAME>"
# our function
def salutation(name):
return "Hi " + name + ", nice to meet you!"
# calling our function
salutation(name)
# un-comment the line below
#salutation("<NAME>")
# -
# Even though your argument was called _name_, it didnt output <NAME>, which was the **global** value of the variable called name. Instead, it gave preference to the **local** value which was given to the function as an argument, <NAME>.
#
# Think of it as filling your coffeemaker (function) up with coffee (variable). If you have a variable with **global** access called _name_ which is filled with coffee called <NAME>, you can choose to either:
#
# 1) Not input another value in your function. (Use the same name of the **global** variable as your argument)
#
# In this case, the **global** type of coffee will still be used.
#
# 2) Choose to fill another value. In this case, your function will assign the value you pass as the argument to the “variable” which **is** the argument.
#
# Think of it as overriding your **global** coffee and putting a new type of coffee into your coffeemaker.
#
# ### Activity
#
# Using the rules of scope you've learned so far, complete the function _puzzle_ to output the value **35**.
# +
# Scope Puzzle!
x = 5
y = 6
z = 7
def puzzle(x, y):
return x * y
# fill in this function call
puzzle()
# -
# ## Control
# ---
# Sometimes, we want to manipulate the flow of our code. For example, we might want our code to make decisions on its own or repeat itself a certain amount of times. By implementing control structures, we can avoid redundant code and make processes more efficient.
#
# ### Conditionals
# We use **conditionals** to run certain pieces of code _if_ something is true. For example, we should only go to the grocery store _if_ we are out of peanut butter!
#
# We use **comparators** to determine whether an expression is _true_ or _false_. There are six comparators to be aware of:
# 1. Equal to: ==
# 2. Not equal to: !=
# 3. Greater than: >
# 4. Greater than or equal to: >=
# 5. Less than: <
# 6. Less than or equal to: <=
#
# Let's try it out!
# +
# EXERCISE 1
# Determine whether the following will print true or false
# Run the code to check your answers!
print(10 == 10)
print(2016 < 2017)
print("foo" != "bar")
print( (1+2+3+4+5) <= (1*2*3))
# +
# EXERCISE 2
# Write an expression that evaluates to True
expression1 = # YOUR CODE HERE
# Write an expression that evaluates to False
expression2 = # YOUR CODE HERE
print(expression1)
print(expression2)
# -
# Now that we know how to compare values, we can tell our computer to make decisions using the **if statement**.
#
# ### If Statements
# An **if statement** takes the following form:
# +
# Please do not run this code, as it will error. It is provided as a skeleton.
if (condition1):
# code to be executed if condition1 is true
elif (condition2):
# code to be executed if condition2 is true
else:
# code to be executed otherwise
# -
# With if statements, we can control which code is executed. Check out how handy this can be in the activity below!
# +
# We want to make a PB&J sandwich, but things keep going wrong!
# Modify the variables below so that you go grocery shopping
# with no mishaps and successfully purchase some peanut butter.
# Run the code when you're done to see the results.
print("Let's make a PB&J sandwich!")
peanut_butter = 10
jelly = 100
gas = 60
flat_tire = True
if (peanut_butter < 50):
print("Uh oh! We need more peanut butter. Must go grocery shopping...")
if (gas < 75):
print("Oops! Your car is out of gas :(")
elif (flat_tire):
print("Oh no! You have a flat tire :'(")
else:
print("You made it to the grocery store and succesfully got peanut butter!")
peanut_butter = # reset the value of peanut_butter so it is 100% full again
else:
print("We have all the ingredients we need! Yummy yummy yay!")
# -
# ### For Loops
# We can also regulate the flow of our code by repeating some action over and over. Say that we wanted to greet ten people. Instead of copying and pasting the same call to _print_ over and over again, it would be better to use a **for loop**.
#
# A basic **for loop** is written in the following order:
# - The word "for"
# - A name we want to give each item in a sequence
# - The word "in"
# - A sequence (i.e. "range(100)" to go through numbers 0-99
#
# For example, to greet someone ten times, we could write:
# Run me to see "hello!" printed ten times!
for i in range(10):
print("hello!")
# In this way, for loops help us avoid redundant code and have useful capabilities.
#
# **Exercise:** Write a function that returns the sum of the first _n_ numbers, where _n_ is the input to the function. Use a for loop!
# +
def sum_first_n(n):
# YOUR CODE HERE
sum_first_n(5) # should return 1+2+3+4+5 = 15
# -
# ## Conclusion
# ---
# Congratulations! You've successfully learnt the basics of programming: creating your own variables, writing your own functions, and controlling the flow of your code! You will apply the concepts learnt throughout this notebook in class. After delving into this notebook, you are only just getting started!
# ---
#
# ## Bibliography
# Some examples adapted from the UC Berkeley Data 8 textbook, <a href="https://www.inferentialthinking.com">*Inferential Thinking*</a>.
#
# Authors:
# - <NAME>
# - <NAME>
# - <NAME>
|
intro/intro-module-final.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# ## Continuous Futures
# Continuous Futures are an abstraction of the chain of consecutive contracts for the same underlying commodity or asset. Additionally, they maintain an ongoing reference to the active contract on the chain. Continuous futures make it much easier to maintain a dynamic reference to contracts that you want to order, and get historical series of data. In this lesson, we will explore some of the ways in which we can use continuous futures to help us in our research.
# In order to create an instance of a `ContinuousFuture` in Research, we need to use the <a href="https://www.quantopian.com/help#quantopian_research_experimental_continuous_future">continuous_future</a> function. Similar to history, we need to import it from research's experimental library:
from quantopian.research.experimental import continuous_future, history
# To create a continuous future, we just need to supply a root_symbol to the `continuous_future` function. The following cell creates a continuous future for Light Sweet Crude Oil.
cl = continuous_future('CL')
cl
# ### Continuous Futures & `history`
# We can use `history` to get pricing and volume data for a particular `ContinuousFuture` in the same way we do for `Futures`. Additionally, we can get the reference to its currently active `Future` contract by using the `contract` field.
# Running the next cell will get pricing data for our CL continuous future and plot it:
# +
# Pricing data for CL `ContinuousFuture`.
cl_pricing = history(
cl,
fields='price',
frequency='daily',
start_date='2015-10-21',
end_date='2016-06-01'
)
cl_pricing.plot()
# -
# To better understand the need for continuous futures, let's use `history` to get pricing data for the chain of individual contracts we looked at in the previous lesson and plot it.
# +
cl_contracts = symbols(['CLF16', 'CLG16', 'CLH16', 'CLJ16', 'CLK16', 'CLM16'])
# Pricing data for our consecutive contracts from earlier.
cl_consecutive_contract_pricing = history(
cl_contracts,
fields='price',
frequency='daily',
start_date='2015-10-21',
end_date='2016-06-01'
)
cl_consecutive_contract_pricing.plot();
# -
# The price difference between contracts at a given time is not considered to be an increase in value in the future. Instead, it is associated with the carrying cost and the opportunity cost of holding the underlying commodity or asset prior to delivery. This concept is covered more in depth in the Introduction to Futures Contracts lecture from our <a href="https://www.quantopian.com/lectures">Lecture Series</a>.
# Next, let's look at the price history for active contracts separately. We will notice that this difference in price creates discontinuities when a contract expires and the reference moves to the next contract:
# +
# Pricing and contract data for unadjusted CL `ContinuousFuture`.
# Adjustments are covered in the next section.
cl_unadjusted = continuous_future('CL', adjustment=None)
cl_history = history(
cl_unadjusted,
fields=['contract', 'price'],
frequency='daily',
start_date='2015-10-21',
end_date='2016-06-01'
)
cl_active_contract_pricing = cl_history.pivot(index=cl_history.index, columns='contract')
cl_active_contract_pricing.plot();
# -
# Part of the job of our continuous future abstraction is to account for these discontinuities, as we will see next by plotting our CL continuous future price against the price history for individual active contracts.
cl_active_contract_pricing.plot()
cl_pricing.plot(style='k--')
# The above plot is adjusted for the price jumps that we see between contracts. This allows us to get a price series that reflects the changes in the price of the actual underlying commodity/asset.
# In the next section, we will explore the options for adjusting historical lookback windows of continuous futures.
# ### Adjustment Styles
# As we just saw, continuous future historical data series are adjusted to account for price jumps between contracts by default. This can be overridden by specifying an adjustment argument when creating the continuous future. The adjustment argument has 3 options: `'mul'` (default), `'add'`, and `None`.
# The `'mul'` option multiplies the prices series by the ratio of consecutive contract prices. The effect from each jump is only applied to prices further back in the lookback window.
# Similarly, the `'add'` technique adjusts by the difference between consecutive contract prices.
# Finally, passing `None` means that no adjustments will be applied to the lookback window.
# ### Roll Styles
# In the previous lesson we saw that trading activity jumps from one contract in the chain to the next as they approach their delivery date. A continuous future changes its reference from the active contract to the next bassed on its roll attribute.
# A `'calendar'` roll means that the continuous future will point to the next contract in the chain when it reaches the `auto_close_date` of the current active contract.
# The `volume` roll (default) means that the continuous future will begin pointing to the next contract when the trading volume of the next contract surpasses the volume of the current contract. The idea is to roll when the majority of traders have moved to the next contract. If the volume swap doesn't happen before the `auto_close_date`, the contract will roll at this date. Note: volume rolls will not occur earlier than 7 trading days before the `auto_close_date`.
# Let's get the volume history of our CL continuous future and plot it against the individual contract volumes we saw before.
# +
cl_consecutive_contract_data = history(
cl_contracts,
fields='volume',
frequency='daily',
start_date='2015-10-21',
end_date='2016-06-01'
)
cl_continuous_volume = history(
cl,
fields='volume',
frequency='daily',
start_date='2015-10-21',
end_date='2016-06-01'
)
cl_consecutive_contract_data.plot()
cl_continuous_volume.plot(style='k--');
# -
# The volume for the CL `ContinuousFuture` is essentially the skyline of the individual contract volumes. As the volume moves from one contract to the next, the continuous future starts pointing to the next contract. Note that there are some points where the volume does not exactly match, most notably in the transition from `CLK16` to `CLM16` between April and May. This is because the rolls are currently computed daily, using only the previous day's volume to avoid lookahead bias.
# ### Offset
# The offset argument allows you to specify whether you want to maintain a reference to the front contract or to a back contract. Setting offset=0 (default) maintains a reference to the front contract, or the contract with the next soonest delivery. Setting offset=1 creates a continuous reference to the contract with the second closest date of delivery, etc.
print continuous_future.__doc__
|
Notebooks/quantopian_research_public/notebooks/tutorials/4_futures_getting_started_lesson4/notebook.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import torch
import torch.nn as nn
import numpy as np
import math
# +
x1 = torch.tensor([[1],[2.]])
W1 = torch.tensor([[1,2.],
[-1,3]], requires_grad=True)
W2 = torch.tensor([[1,-1.],
[1,2]], requires_grad=True)
W3 = torch.tensor([[-1,2.],
[0,1]], requires_grad=True)
phi1 = torch.relu
phi2 = torch.sigmoid
phi3 = torch.softmax
l = lambda yh,t: - t.view(1,-1) @ torch.log(yh)
t = torch.tensor([[1], [0.]])
# same result:
# sl = nn.NLLLoss(reduction="sum")
# sl(torch.log_softmax(z3,0).view(1,-1), torch.max(t, 0)[1])
grad = lambda x: x.clone().detach().requires_grad_(True)
z1 = W1@x1
x2 = phi1(z1)
z2 = W2@x2
#z2 = grad(z2)
x3 = phi2(z2)
#x3 = grad(x3)
z3 = W3@x3
yh = phi3(z3, 0)
E = l(yh.flatten(),t)
T = lambda A: A.transpose(0,1)
E.backward()
E
# -
with torch.no_grad():
dy = -t/yh
dsoft = torch.tensor([[yh[0,0]*(1-yh[0,0]), -yh[0,0]*yh[1,0]],
[-yh[0,0]*yh[1,0], yh[1,0]*(1-yh[1,0])]])
dsig = torch.diag((x3 * (1-x3)).flatten())
drelu = torch.diag((z1 > 0).to(torch.float).flatten())
dz3 = dsoft @ dy
dz2 = dsig @ (T(W3) @ dz3)
dz1 = drelu @ (T(W2) @ dz2)
dw3 = dz3 @ T(x3)
dw2 = dz2 @ T(x2)
dw1 = dz1 @ T(x1)
print(dw3)
print(dw2)
print(dw1)
print(W3.grad)
print(W2.grad)
print(W1.grad)
dz3
dw2
W2.grad
W3.grad
dw3
|
_useless/notebooks/Backprop.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Seismic Data Analysis
# ### What is Seismic Hazard Analysis?
#
#
# In general terms, the seismic hazard defines the expected seismic ground motion at a site, phenomenon which may result in destructions and losses.
#
# Тwo major approaches – deterministic and probabilistic – are worldwide used at present for seismic hazard assessment.
#
# The deterministic approach takes into account a single, particular earthquake, the event that is expected to produce the strongest level of shaking at the site.
#
# The outputs – macroseismic intensity, peak ground acceleration, peak ground velocity, peak ground displacement, response spectra – may be used directly in engineering applications.
#
# In the probabilistic approach, initiated with the pioneering work of Cornell, the seismic hazard is estimated in terms of a ground motion parameter – macroseismic intensity, peak ground acceleration – and its annual probability of exceedance (or return period) at a site.
#
# The method yields regional seismic probability maps, displaying contours of maximum ground motion (macroseismic intensity, PGA) of equal – specified – return period.
#
#
# Source : http://www.infp.ro/en/seismic-hazard/
# ### Dataset :
#
# * Name- seismic-bumps Data Set
#
# * Abstract: The data describe the problem of high energy (higher than 10^4 J) seismic bumps forecasting in a coal mine. Data come from two of longwalls located in a Polish coal mine.
#
# * Source : https://archive.ics.uci.edu/ml/datasets/seismic-bumps
#
# *** Dataset Information ***
#
# Mining activity was and is always connected with the occurrence of dangers which are commonly called
# mining hazards. A special case of such threat is a seismic hazard which frequently occurs in many
# underground mines. Seismic hazard is the hardest detectable and predictable of natural hazards and in
# this respect it is comparable to an earthquake. More and more advanced seismic and seismoacoustic
# monitoring systems allow a better understanding rock mass processes and definition of seismic hazard
# prediction methods. Accuracy of so far created methods is however far from perfect. Complexity of
# seismic processes and big disproportion between the number of low-energy seismic events and the number
# of high-energy phenomena (e.g. > 10^4J) causes the statistical techniques to be insufficient to predict
# seismic hazard.
#
#
#
#
# The task of seismic prediction can be defined in different ways, but the main
# aim of all seismic hazard assessment methods is to predict (with given precision relating to time and
# date) of increased seismic activity which can cause a rockburst. In the data set each row contains a
# summary statement about seismic activity in the rock mass within one shift (8 hours). If decision
# attribute has the value 1, then in the next shift any seismic bump with an energy higher than 10^4 J was
# registered. That task of hazards prediction bases on the relationship between the energy of recorded
# tremors and seismoacoustic activity with the possibility of rockburst occurrence. Hence, such hazard
# prognosis is not connected with accurate rockburst prediction. Moreover, with the information about the
# possibility of hazardous situation occurrence, an appropriate supervision service can reduce a risk of
# rockburst (e.g. by distressing shooting) or withdraw workers from the threatened area. Good prediction
# of increased seismic activity is therefore a matter of great practical importance. The presented data
# set is characterized by unbalanced distribution of positive and negative examples. In the data set there
# are only 170 positive examples representing class 1.
#
#
# <img src= "att.jpg">
# # Classification Seismic of Hazard in coal mines
# +
# Dependencies import
import matplotlib.pyplot as plt
import numpy as np
from scipy.io import arff
import pandas as pd
import seaborn as sns;
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split, cross_val_score, GridSearchCV
from sklearn.metrics import roc_auc_score, f1_score
from sklearn import preprocessing
# %matplotlib inline
# +
## load data and clean
data = arff.loadarff('data/seismic-bumps.arff')
df = pd.DataFrame(data[0])
df['seismic'] = df['seismic'].str.decode('utf-8')
df['seismoacoustic'] = df['seismoacoustic'].str.decode('utf-8')
df['shift'] = df['shift'].str.decode('utf-8')
df['ghazard'] = df['ghazard'].str.decode('utf-8')
df['class'] = df['class'].str.decode('utf-8')
df['class'] = pd.to_numeric(df['class'])
# -
df.head()
# # Exploratory Data Analysis
# +
df_plot = df[['genergy', 'gpuls', 'gdenergy', 'gdpuls',
'nbumps', 'nbumps2',
'energy', 'maxenergy']].copy()
p = sns.pairplot(df_plot)
# -
# The plots above show some colinearity between attributes (e.g. genergy and gpuls, energy and maxenergy). The following will use regularization to mitigate the problem.
# # Build models
# +
data_x = df.loc[:,['shift', 'genergy', 'gpuls', 'gdenergy', 'gdpuls',
'nbumps', 'nbumps2', 'nbumps3', 'nbumps4', 'nbumps5',
'nbumps6', 'nbumps7', 'nbumps89',
'energy', 'maxenergy']]
# true response
data_y = df.loc[:,['class']]
# responses from seismic theories
data_y1 = df.loc[:, ['seismic']]
data_y2 = df.loc[:, ['seismoacoustic']]
data_y3 = df.loc[:, ['ghazard']]
Le = preprocessing.LabelEncoder()
Le.fit(['a', 'b', 'c', 'd'])
data_y1['seismic'] = Le.transform(data_y1['seismic'])
data_y2['seismoacoustic'] = Le.transform(data_y2['seismoacoustic'])
data_y3['ghazard'] = Le.transform(data_y3['ghazard'])
Le2 = preprocessing.LabelEncoder()
Le2.fit(['W', 'N'])
data_x['shift'] = Le2.transform(data_x['shift'])
# -
X_train, X_test, y_train, y_test = train_test_split(data_x, data_y, test_size=0.2, random_state=42)
X_train.describe()
X_train.info()
# #### Let'sfind the best regularization coefficient
# +
## use ROC as the score
C = [1e-4, 1e-3, 1e-2, 1e-1, 1, 10, 1e2]
scores = []
for c in C:
logist = LogisticRegression(penalty='l1', C=c, max_iter=500)
logist.fit(X_train, y_train.values.ravel())
scores.append(roc_auc_score(y_train['class'].values, logist.predict(X_train)))
C_best = C[scores.index(max(scores))]
print("Best C: ", C_best)
# -
# ## Using Logistic Regression
# +
clf = LogisticRegression(penalty='l1', C=C_best, max_iter = 500)
clf.fit(X_train, y_train.values.ravel())
roc_train = roc_auc_score(y_train['class'].values, clf.predict(X_train))
# print("training score: %.4f" % clf.score(Xtrain, ytrain))
print("training score: %.4f" % roc_train)
# print("test score: ", clf.score(Xtest, ytest))
roc_test = roc_auc_score(y_test['class'].values, clf.predict(X_test))
print("test score: %.4f" % roc_test)
print("n_iter: ", clf.n_iter_)
# -
clf.coef_
# +
ind = y_test.index.values
# get the responses from the seismic, seismoacoustic and ghazard methods
# that correspond to indices in ytest
yseismic = data_y1.loc[ind, ['seismic']]
yseismoacoustic = data_y2.loc[ind, ['seismoacoustic']]
yghazard = data_y3.loc[ind, ['ghazard']]
# -
# Responses as probabilies from the logit model
# +
yprob = clf.predict_proba(X_test)
yprob
# -
# Threshold
ypred = yprob[:,1] > 0.2 # threshold
# From the plot below, to use the probabilites from the prediction, we need to set a threshold to determine if the response should be hazardous or not. The hard labels from the prediction will be mostly 0's.
#
# Note: setting the threshold requires further study. One way is to tune the threshold in training sets and test the performance in test sets.
# +
plt.plot([i for i in range(len(y_test))], y_test, 'x', yprob[:,1], '.')
plt.ylabel('Probability')
plt.title('Raw results from prediction')
# -
plt.plot([i for i in range(len(y_test))], y_test, 'o', ypred, '.')
plt.ylabel('Probability')
plt.title('Probabilities after cut-off')
# ### Results
# +
dy = { 'logit': pd.Series(ypred) }
dfy = pd.DataFrame(dy)
frames = [dfy, yseismic.reset_index(drop=True),
yseismoacoustic.reset_index(drop=True),
yghazard.reset_index(drop=True)]
# build the responses data frame (each column is responses from one method)
df_result = pd.concat(frames, axis = 1)
df_result = df_result*1 # convert bool to int
# -
df_result
# +
yvote = (df_result == 0).sum(axis=1) # number of zeros on each row
yvote = (yvote <= 2)*1
# final results based on the vote from each of the four methods
# 0 means no/low hazard, 1 means hazardous
# if tie, assume response is 1 (hazardous)
df_result['ensemble'] = yvote.values
df_result['true'] = y_test.values
df_result.head(20)
# -
# score from the ensemble method with logit regression
roc_auc_score(y_test['class'].values, df_result['ensemble'].values)
# +
## compare to the three methods already in the dataset
frames = [yseismic.reset_index(drop=True),
yseismoacoustic.reset_index(drop=True),
yghazard.reset_index(drop=True)]
df_result0 = pd.concat(frames, axis = 1)
df_result0 = df_result0*1
yvote0 = (df_result0 == 0).sum(axis=1)
yvote0 = (yvote0 <= 2)*1
df_result0['ensemble'] = yvote0.values
df_result0['true'] = y_test.values
df_result0.head(20)
# -
# score from the ensemble of the three methods in the original dataset
roc_auc_score(y_test['class'].values, df_result0['ensemble'].values)
# score from the seismic method (no ensemble)
roc_auc_score(y_test['class'].values, yseismic['seismic'].values)
# score from the seismoacoustic method (no ensemble)
roc_auc_score(y_test['class'].values, yseismoacoustic['seismoacoustic'].values)
|
Seismic Data Analysis Notebook.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Reproduce In-situ Sequencing results with Starfish
#
# This notebook walks through a work flow that reproduces an ISS result for one field of view using the starfish package.
#
# ## Load tiff stack and visualize one field of view
# +
# %matplotlib inline
# %load_ext autoreload
# %autoreload 2
import numpy as np
import os
import pandas as pd
import matplotlib.pyplot as plt
from showit import image
import pprint
from starfish import data, FieldOfView
from starfish.types import Features, Indices
# +
use_test_data = os.getenv("USE_TEST_DATA") is not None
experiment = data.ISS(use_test_data=use_test_data)
# s.image.squeeze() simply converts the 4D tensor H*C*X*Y into a list of len(H*C) image planes for rendering by 'tile'
# -
# ## Show input file format that specifies how the tiff stack is organized
#
# The stack contains multiple single plane images, one for each color channel, 'c', (columns in above image) and imaging round, 'r', (rows in above image). This protocol assumes that genes are encoded with a length 4 quatenary barcode that can be read out from the images. Each round encodes a position in the codeword. The maximum signal in each color channel (columns in the above image) corresponds to a letter in the codeword. The channels, in order, correspond to the letters: 'T', 'G', 'C', 'A'. The goal is now to process these image data into spatially organized barcodes, e.g., ACTG, which can then be mapped back to a codebook that specifies what gene this codeword corresponds to.
pp = pprint.PrettyPrinter(indent=2)
pp.pprint(experiment._src_doc)
# The flat TIFF files are loaded into a 4-d tensor with dimensions corresponding to imaging round, channel, x, and y. For other volumetric approaches that image the z-plane, this would be a 5-d tensor.
fov = experiment.fov()
primary_image = fov[FieldOfView.PRIMARY_IMAGES]
dots = fov['dots']
nuclei = fov['nuclei']
images = [primary_image, nuclei, dots]
# round, channel, x, y, z
primary_image.xarray.shape
# ## Show auxiliary images captured during the experiment
# 'dots' is a general stain for all possible transcripts. This image should correspond to the maximum projcection of all color channels within a single imaging round. This auxiliary image is useful for registering images from multiple imaging rounds to this reference image. We'll see an example of this further on in the notebook
image(dots.max_proj(Indices.ROUND, Indices.CH, Indices.Z))
# Below is a DAPI auxiliary image, which specifically marks nuclei. This is useful cell segmentation later on in the processing.
image(nuclei.max_proj(Indices.ROUND, Indices.CH, Indices.Z))
# ## Examine the codebook
# Each 4 letter quatenary code (as read out from the 4 imaging rounds and 4 color channels) represents a gene. This relationship is stored in a codebook
experiment.codebook
# ## Filter and scale raw data
#
# Now apply the white top hat filter to both the spots image and the individual channels. White top had enhances white spots on a black background.
# +
from starfish.image import Filter
# filter raw data
masking_radius = 15
filt = Filter.WhiteTophat(masking_radius, is_volume=False)
for img in images:
filt.run(img, verbose=True, in_place=True)
# -
# ## Register data
# For each imaging round, the max projection across color channels should look like the dots stain.
# Below, this computes the max projection across the color channels of an imaging round and learns the linear transformation to maps the resulting image onto the dots image.
#
# The Fourier shift registration approach can be thought of as maximizing the cross-correlation of two images.
#
# In the below table, Error is the minimum mean-squared error, and shift reports changes in x and y dimension.
# +
from starfish.image import Registration
registration = Registration.FourierShiftRegistration(
upsampling=1000,
reference_stack=dots,
verbose=True)
registered_image = registration.run(primary_image, in_place=False)
# -
# ## Use spot-detector to create 'encoder' table for standardized input to decoder
# Each pipeline exposes an encoder that translates an image into spots with intensities. This approach uses a Gaussian spot detector.
# +
from starfish.spots import SpotFinder
import warnings
# parameters to define the allowable gaussian sizes (parameter space)
min_sigma = 1
max_sigma = 10
num_sigma = 30
threshold = 0.01
p = SpotFinder.GaussianSpotDetector(
min_sigma=min_sigma,
max_sigma=max_sigma,
num_sigma=num_sigma,
threshold=threshold,
measurement_type='mean',
)
# detect triggers some numpy warnings
with warnings.catch_warnings():
warnings.simplefilter("ignore")
# blobs = dots; define the spots in the dots image, but then find them again in the stack.
blobs_image = dots.max_proj(Indices.ROUND, Indices.Z)
intensities = p.run(registered_image, blobs_image=blobs_image)
# -
# The Encoder table is the hypothesized standardized file format for the output of a spot detector, and is the first output file format in the pipeline that is not an image or set of images
# `attributes` is produced by the encoder and contains all the information necessary to map the encoded spots back to the original image
#
# `x, y` describe the position, while `x_min` through `y_max` describe the bounding box for the spot, which is refined by a radius `r`. This table also stores the intensity and spot_id.
# ## Decode
# Each assay type also exposes a decoder. A decoder translates each spot (spot_id) in the Encoder table into a gene (that matches a barcode) and associates this information with the stored position. The goal is to decode and output a quality score that describes the confidence in the decoding.
# There are hard and soft decodings -- hard decoding is just looking for the max value in the code book. Soft decoding, by contrast, finds the closest code by distance (in intensity). Because different assays each have their own intensities and error modes, we leave decoders as user-defined functions.
decoded = experiment.codebook.decode_per_round_max(intensities)
# ## Compare to results from paper
# Besides house keeping genes, VIM and HER2 should be most highly expessed, which is consistent here.
genes, counts = np.unique(decoded.loc[decoded[Features.PASSES_THRESHOLDS]][Features.TARGET], return_counts=True)
table = pd.Series(counts, index=genes).sort_values(ascending=False)
# ### Segment
# After calling spots and decoding their gene information, cells must be segmented to assign genes to cells. This paper used a seeded watershed approach.
# +
from starfish.image import Segmentation
dapi_thresh = .16 # binary mask for cell (nuclear) locations
stain_thresh = .22 # binary mask for overall cells // binarization of stain
min_dist = 57
stain = np.mean(registered_image.max_proj(Indices.CH, Indices.Z), axis=0)
stain = stain/stain.max()
nuclei_projection = nuclei.max_proj(Indices.ROUND, Indices.CH, Indices.Z)
seg = Segmentation.Watershed(
nuclei_threshold=dapi_thresh,
input_threshold=stain_thresh,
min_distance=min_dist
)
label_image = seg.run(registered_image, nuclei)
seg.show()
# -
# ### Visualize results
#
# This FOV was selected to make sure that we can visualize the tumor/stroma boundary, below this is described by pseudo-coloring `HER2` (tumor) and vimentin (`VIM`, stroma)
# +
from skimage.color import rgb2gray
GENE1 = 'HER2'
GENE2 = 'VIM'
rgb = np.zeros(registered_image.tile_shape + (3,))
rgb[:,:,0] = nuclei.max_proj(Indices.ROUND, Indices.CH, Indices.Z)
rgb[:,:,1] = dots.max_proj(Indices.ROUND, Indices.CH, Indices.Z)
do = rgb2gray(rgb)
do = do/(do.max())
image(do,size=10)
with warnings.catch_warnings():
warnings.simplefilter('ignore', FutureWarning)
is_gene1 = decoded.where(decoded[Features.AXIS][Features.TARGET] == GENE1, drop=True)
is_gene2 = decoded.where(decoded[Features.AXIS][Features.TARGET] == GENE2, drop=True)
plt.plot(is_gene1.x, is_gene1.y, 'or')
plt.plot(is_gene2.x, is_gene2.y, 'ob')
plt.title(f'Red: {GENE1}, Blue: {GENE2}');
|
notebooks/ISS_Pipeline_-_Breast_-_1_FOV.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] iooxa={"id": {"block": "ljl07JNYSrIXE70uWYO0", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}}
# ## UTAH FORGE WELL 58-32
#
# **Well 58-32 was drilled to a depth of 7536 feet** in the Milford FORGE area during the summer of
# 2017 to confirm the reservoir characteristics inferred from existing wells and a wide variety of
# both new and legacy geologic and geophysical data. **Drill cuttings were collected and described
# at 10-foot intervals** and a robust **suite of geophysical logs** were run. Analyses show
# that the basement rock within the FORGE area consists of a suite of **intrusive rock types that are
# primarily granitic. Some diorite and monzodiorite was also encountered**, as was a significant
# volume of rock with a more intermediate composition.
#
# The density of the granite and intermediate rock types typically range from **2.6 to
# 2.65 g/cm³**, but the higher gamma response of the **granitic rock (140–290 gAPI)** can often
# differentiate granitic compositions from **intermediate compositions (70–210 gAPI).** The **higher
# density (2.7–3.0 g/cm³) and lower gamma values (50–80 gAPI) of the dioritic compositions** is
# more distinctive and greatly simplifies identification.
#
# The various laboratory analyses and geophysical logs of the 58-32 well prove it was drilled into **low porosity/low permeability intrusive rock** with temperatures well within the U.S. Department of Energy-specified window of **175°–225°C (347°–437°F).** More details here https://utahforge.com/
#
# -
# ### Let's import the libraries, remember Lasio and Seaborn must be installed previously
# + iooxa={"id": {"block": "bCoQ217Se5IoWzAvnM6x", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}, "outputId": null}
import lasio
import pandas as pd
import numpy as np
#libraries for plots
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
# + [markdown] iooxa={"id": {"block": "NJ3M1nBKUzM2AXbRaoeZ", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}}
# ### Read 58-32 well logs with Lasio and inspect
# + iooxa={"id": {"block": "Z6bDDyAUxa2TGPCk6ENW", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}, "outputId": null}
reg_all = lasio.read('../alldata/58-32_main.las')
# -
reg_all.version
reg_all.curves
reg_all['SP']
reg_all.keys()
reg_all.data
# + [markdown] iooxa={"id": {"block": "dAR1AgfP4yyfzpXCRpNS", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}}
# ### From Lasio to Data Frame Pandas
# DataFrames in Pandas are two-dimensional tables with row and columns that can be easily edited and manipulated.
# -
df_main = reg_all.df()
df_main
# + iooxa={"id": {"block": "uUC9Yb53FupxOfbextPD", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}, "outputId": {"block": "kdODK2dt28SaDB27bDxB", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}}
#Print the first 5 rows of the data frame with the header of the columns
df_main.head(5)
# -
#Print the last 10 rows of the data frame with the header of the columns
df_main.tail(10)
# statistics
df_main.describe()
#parameters from only 1 column
df_main.AF10.std()
# ### Create a dataset only with GR, SP, AT10, AT90, RHOZ, NPHI, CTEM
df_mini = df_main[['GR', 'SP', 'AT10', 'AT90', 'RHOZ', 'NPHI', 'CTEM']]
df_mini.describe()
df_mini['CTEM_C']= ((df_mini['CTEM']-32)*5)/9
df_mini.info()
count_neg = (df_mini.RHOZ < 0).sum()
count_neg
df_mini.loc[(df_mini['RHOZ'] < 0), 'RHOZ']=np.nan
count_neg = (df_mini.RHOZ < 0).sum()
count_neg
# **Unknown LowGR (<50)
# **Dioritic comp. (50–80 gAPI)
# **Intermediate comp. (80–140 gAPI)
# **Granite (140–290 gAPI)
# **Unknown HighGR(>290)
# +
conditions = [
(df_mini['GR'] <= 50),
(df_mini['GR'] > 50) & (df_mini['GR'] <= 80),
(df_mini['GR'] > 80) & (df_mini['GR'] <= 140),
(df_mini['GR'] > 140) & (df_mini['GR'] <= 290),
(df_mini['GR'] > 290)
]
# create a list of the values we want to assign for each condition
values = ['Unknown LowGR', 'Dioritic Comp', 'Intermediate Comp', 'Granite', 'Unknown HighGR' ]
# create a new column and use np.select to assign values to it using our lists as arguments
df_mini['Labels'] = np.select(conditions, values)
# -
df_mini.sample(10)
# +
#statistics grouped by Labels
df_mini[['Labels','GR', 'SP', 'AT10', 'AT90', 'RHOZ', 'NPHI', 'CTEM', 'CTEM_C']].groupby('Labels').mean()
# + [markdown] iooxa={"id": {"block": "jWnAnS4nJBZrB2ELssCF", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}}
# ### Read Thermal conductivity and mineralogy data measured in drill cuttings. CAUTION: Depths are in meters, need to be converted to feet
# ##### Full report https://ugspub.nr.utah.gov/publications/misc_pubs/mp-169/mp-169-l.pdf
# +
TC_coredata = pd.read_csv ('../alldata/58-32_thermal_conductivity_data.csv', index_col=1)
XRD_coredata = pd.read_csv ('../alldata/58-32_xray_diffraction_data.csv', index_col=1)
#TC_coredata.head()
XRD_coredata.columns
# -
TC_coredata.index
XRD_coredata.index
result = pd.concat([XRD_coredata, TC_coredata], axis=1, sort=False)
result.columns
cutt_data = result[['Illite','Plagioclase', 'K-feldspar',
'Quartz', 'matrix thermal conductivity (W/m deg C)']]
cutt_data.index=(3.28084*cutt_data.index) #m to ft
#cutt_data.loc[(cutt_data =='tr')]=np.nan
cutt_data=cutt_data.replace('tr', np.nan)
cutt_data.columns=['Illi', 'Plag', 'K-feld', 'Qz', 'TC']
cutt_data.info()
cutt_data.sample(5)
# # Visualization TO FIX%%%%%%%%
# +
#let's start with something simple (xplot, pie, 1 histogram...)
# -
X=df_mini[['GR', 'SP', 'AT10', 'AT90', 'RHOZ', 'NPHI', 'CTEM_C']]
# + iooxa={"id": {"block": "DXyNFHJcBxR9L9SUiTrl", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}, "outputId": {"block": "yw1SFb2eRh0YPgQh6tmS", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}}
#plotting the statistic using Seaborn
color = ['#2ea869', '#0a0a0a', '#ea0606','#1577e0', '#6e787c','#ea0606',
'#ed8712']
sns.set(font_scale=1)
cols = X.columns
n_row = len(cols)
n_col = 2
n_sub = 1
fig = plt.figure(figsize=(10,20))
for i in range(len(cols)):
plt.subplots_adjust(left=-0.3, right=1.3, bottom=-0.3, top=1.3)
plt.subplot(n_row, n_col, n_sub)
sns.distplot(X[cols[i]],norm_hist=False,kde=False, color=color[i],
label=['mean '+str('{:.2f}'.format(X.iloc[:,i].mean()))
+'\n''std '+str('{:.2f}'.format(X.iloc[:,i].std()))
+'\n''min '+str('{:.2f}'.format(X.iloc[:,i].min()))
+'\n''max '+str('{:.2f}'.format(X.iloc[:,i].max()))])
n_sub+=1
plt.legend()
plt.show()
# +
#correlation matrix
corr = df_mini.corr() #exclude any string data type
#figure parameters
fig, ax = plt.subplots(figsize=(8,6))
sns.heatmap(corr, ax=ax, cmap="magma")
#plt.grid()
plt.show()
# + [markdown] iooxa={"id": {"block": "ytvHl7fnOy1HLm624IXR", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}}
# ### Create a function that would create a layout with basic logs and core data
# + iooxa={"id": {"block": "9mv3ARJQuI40H3MYf0FZ", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}, "outputId": null}
#basic plot to inspect data
def make_layout_tc (log_df, cuttings_df):
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
fig, axs = plt.subplots(nrows=1, ncols=5, sharey=True, squeeze=True, figsize=(15, 15), gridspec_kw={'wspace': 0.25})
fig.subplots_adjust(left=0.05, bottom=0.05, right=0.975, top=0.7, wspace=0.2, hspace=0.2)
axs[0].set_ylabel('Depth (ft)')
axs[0].invert_yaxis()
axs[0].get_xaxis().set_visible(False)
# First track GR/SP logs to display
ax1 = axs[0].twiny()
ax1.plot(log_df.GR, log_df.index, '-', color='#2ea869', linewidth=0.5)
ax1.set_xlim(0,450)
ax1.set_xlabel('GR (API)', color='#2ea869')
ax1.minorticks_on()
ax1.spines['top'].set_position(('axes', 1.075))
ax2 = axs[0].twiny()
ax2.plot(log_df.SP, log_df.index, '-', color='#0a0a0a', linewidth=0.7)
ax2.set_xlim(-200,200)
ax2.set_xlabel('SP(mV)', color='#0a0a0a')
ax2.minorticks_on()
ax2.spines['top'].set_position(('axes', 1.0))
ax2.grid(True)
axs[0].get_xaxis().set_visible(False)
# Second track RHOZ/NPHI logs to display
ax1 = axs[1].twiny()
ax1.plot(log_df.RHOZ, log_df.index, '-', color='#ea0606', linewidth=0.5)
ax1.set_xlim(1.5,3.0)
ax1.set_xlabel('RHOZ (g/cm3)', color='#ea0606')
ax1.minorticks_on()
ax1.spines['top'].set_position(('axes', 1.075))
ax2 = axs[1].twiny()
ax2.plot(log_df.NPHI, log_df.index, '-', color='#1577e0', linewidth=0.5)
ax2.set_xlim(1,0)
ax2.set_xlabel('NPHI (v/v)', color='#1577e0')
ax2.minorticks_on()
ax2.spines['top'].set_position(('axes', 1.0))
ax2.grid(True)
axs[1].get_xaxis().set_visible(False)
# Third track Resistivities
ax1 = axs[2].twiny()
ax1.plot(log_df.AT10, log_df.index, '-', color='#6e787c', linewidth=0.5)
ax1.set_xlim(0.1,100000)
ax1.set_xlabel('AT10 (ohm.m)', color='#6e787c')
ax1.set_xscale('log')
ax1.minorticks_on()
ax1.spines['top'].set_position(('axes', 1.075))
ax2 = axs[2].twiny()
ax2.plot(log_df.AT90, log_df.index, '-', color='#ea0606', linewidth=0.5)
ax2.set_xlim(0.1,100000)
ax2.set_xlabel('AT90 (ohm.m)', color='#ea0606')
ax2.set_xscale('log')
ax2.minorticks_on()
ax2.spines['top'].set_position(('axes', 1.0))
ax2.grid(True)
axs[2].get_xaxis().set_visible(False)
# Fourth track XRD to display
ax1 = axs[3].twiny()
ax1.plot(cuttings_df.Qz, cuttings_df.index, 'o', color='#eac406')
ax1.set_xlim(0,50)
ax1.set_xlabel('Quartz %', color='#eac406')
ax1.minorticks_on()
ax1.spines['top'].set_position(('axes', 1.075))
ax2 = axs[3].twiny()
ax2.plot(cuttings_df.Illi, cuttings_df.index, 'o', color='#94898c')
ax2.set_xlim(0,50)
ax2.set_xlabel('Illite %', color='#94898c')
ax2.minorticks_on()
ax2.spines['top'].set_position(('axes', 1.0))
ax2.grid(True)
axs[3].get_xaxis().set_visible(False)
# Fifth track Temp/TC to display
ax1 = axs[4].twiny()
ax1.plot(cuttings_df.TC, cuttings_df.index, 'o', color='#6e787c')
ax1.set_xlim(0,5)
ax1.set_xlabel('Matrix TC Measured W/mC', color='#6e787c')
ax1.minorticks_on()
ax1.spines['top'].set_position(('axes', 1.075))
ax2 = axs[4].twiny()
ax2.plot(log_df.CTEM_C, log_df.index, '-', color='#ed8712')
ax2.set_xlim(20,200)
ax2.set_xlabel('Temp degC', color='#ed8712')
ax2.minorticks_on()
ax2.spines['top'].set_position(('axes', 1.0))
ax2.grid(True)
axs[4].get_xaxis().set_visible(False)
fig.suptitle('Well Data for UTAH FORGE 58-32',weight='bold', fontsize=20, y=0.85);
plt.show()
# + iooxa={"id": {"block": "J0Fjsd3Eq1wwDz69GMxG", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}, "outputId": {"block": "cITmpozD1QjaBPwgARYe", "project": "anKPrTxY08dBACBwy7Ui", "version": 1}}
make_layout_tc (df_mini, cutt_data)
|
notebooks/.ipynb_checkpoints/Tutorial_pyhton_transform21_UtahForge_58-32_well-checkpoint.ipynb
|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Project 2: Digit Recognition
#
# ## Statistical Machine Learning (COMP90051), Semester 2, 2017
#
# *Copyright the University of Melbourne, 2017*
# ### Submitted by: *<NAME>*
# ### Student number: *725439*
# ### Kaggle-in-class username: *your username here*
# In this project, you will be applying machine learning for recognising digits from real world images. The project worksheet is a combination of text, pre-implemented code and placeholders where we expect you to add your code and answers. You code should produce desired result within a reasonable amount of time. Please follow the instructions carefully, **write your code and give answers only where specifically asked**. In addition to worksheet completion, you are also expected to participate **live competition with other students in the class**. The competition will be run using an on-line platform called Kaggle.
# ** Marking:** You can get up to 33 marks for Project 2. The sum of marks for Project 1 and Project 2 is then capped to 50 marks
#
# **Due date:** Wednesday 11/Oct/17, 11:59pm AEST (LMS components); and Kaggle competition closes Monday 09/Oct/17, 11:59pm AEST.
#
# **Late submissions** will incur a 10% penalty per calendar day
#
# ** Submission materials**
# - **Worksheet**: Fill in your code and answers within this IPython Notebook worksheet.
# - **Competition**: Follow the instructions provided in the corresponding section of this worksheet. Your competition submissions should be made via Kaggle website.
# - **Report**: The report about your competition entry should be submitted to the LMS as a PDF file (see format requirements in `2.2`).
# - **Code**: The source code behind your competition entry.
# The **Worksheet**, **Report** and **Code** should be bundled into a `.zip` file (not 7z, rar, tar, etc) and submitted in the LMS. Marks will be deducted for submitting files in other formats, or we may elect not to mark them at all.
#
# **Academic Misconduct:** Your submission should contain only your own work and ideas. Where asked to write code, you cannot re-use someone else's code, and should write your own implementation. We will be checking submissions for originality and will invoke the University’s <a href="http://academichonesty.unimelb.edu.au/policy.html">Academic Misconduct policy</a> where inappropriate levels of collusion or plagiarism are deemed to have taken place.
# **Table of Contents**
#
# 1. Handwritten Digit Recognition **(16 marks)**
# 1. Linear Approach
# 2. Basis Expansion
# 3. Kernel Perceptron
# 4. Dimensionality Reduction
#
# 2. Kaggle Competition **(17 marks)**
# 1. Making Submissions
# 2. Method Description
# ## 1. Handwritten Digit Recognition
# Handwritten digit recognition can be framed as a classification task: given a bitmap image as input, predict the digit type (0, 1, ..., 9). The pixel values in each position of the image form our features, and the digit type is the class. We are going to use a dataset where the digits are represented as *28 x 28* bitmap images. Each pixel value ranges between 0 and 1, and represents the monochrome ink intensity at that position. Each image matrix has been flattened into one long feature vector, by concatenating each row of pixels.
#
# In this part of the project, we will only use images of two digits, namely "7" and "9". As such, we will be working on a binary classification problem. *Throughout this first section, our solution is going to be based on the perceptron classifier.*
#
# Start by setting up working environment, and loading the dataset. *Do not override variable `digits`, as this will be used throughout this section.*
# +
# %pylab inline
digits = np.loadtxt('digits_7_vs_9.csv', delimiter=' ')
# -
# Take some time to explore the dataset. Note that each image of "7" is labeled as -1, and each image of "9" is labeled as +1.
# +
# extract a stack of 28x28 bitmaps
X = digits[:, 0:784]
# extract labels for each bitmap
y = digits[:, 784:785]
# display a single bitmap and print its label
bitmap_index = 0
plt.imshow(X[bitmap_index,:].reshape(28, 28), interpolation=None)
print(y[bitmap_index])
# -
# You can also display several bitmaps at once using the following code.
# +
def gallery(array, ncols):
nindex, height, width = array.shape
nrows = nindex//ncols
result = (array.reshape((nrows, ncols, height, width))
.swapaxes(1,2)
.reshape((height*nrows, width*ncols)))
return result
ncols = 10
result = gallery(X.reshape((300, 28, 28))[:ncols**2], ncols)
plt.figure(figsize=(10,10))
plt.imshow(result, interpolation=None)
# -
# ### 1.1 Linear Approach
# We are going to use perceptron for our binary classification task. Recall that perceptron is a linear method. Also, for this first step, we will not apply non-linear transformations to the data.
#
# Implement and fit a perceptron to the data above. You may use the implementation from *sklearn*, or implementation from one of our workshops. Report the error of the fit as the proportion of misclassified examples.
#
# <br />
#
# <font color='red'>**Write your code in the cell below ...**</font>
# +
## your code here
# -
# One of the advantages of a linear approach is the ability to interpret results. To this end, plot the parameters learned above. Exclude the bias term if you were using it, set $w$ to be the learned perceptron weights, and run the following command.
plt.imshow(w.reshape(28,28), interpolation=None))
# In a few sentences, describe what you see, referencing which features are most important for making classification. Report any evidence of overfitting.
# <font color='red'>**Write your answer here ...**</font> (as a *markdown* cell)
# Split the data into training and heldout validation partitions by holding out a random 25% sample of the data. Evaluate the error over the course of a training run, and plot the training and validation error rates as a function of the number of passes over the training dataset.
#
# <br />
# <font color='red'>**Write your code in the cell below ...**</font>
# +
## your code here
# -
# In a few sentences, describe the shape of the curves, and compare the two. Now consider if we were to stop training early, can you choose a point such that you get the best classification performance? Justify your choice.
# <font color='red'>**Write your answer here ...**</font> (as a *markdown* cell)
# Now that we have tried a simple approach, we are going to implement several non-linear approaches to our task. Note that we are still going to use a linear method (the perceptron), but combine this with a non-linear data transformation. We start with basis expansion.
# ### 1.2 Basis Expansion
# Apply Radial Basis Function (RBF)-based transformation to the data, and fit a perceptron model. Recall that the RBF basis is defined as
#
# $$\varphi_l(\mathbf{x}) = \exp\left(-\frac{||\mathbf{x} - \mathbf{z}_l||^2}{\sigma^2}\right)$$
#
# where $\mathbf{z}_l$ is centre of the $l^{th}$ RBF. We'll use $L$ RBFs, such that $\varphi(\mathbf{x})$ is a vector with $L$ elements. The spread parameter $\sigma$ will be the same for each RBF.
#
# *Hint: You will need to choose the values for $\mathbf{z}_l$ and $\sigma$. If the input data were 1D, the centres $\mathbf{z}_l$ could be uniformly spaced on a line. However, here we have 784-dimensional input. For this reason you might want to use some of the training points as centres, e.g., $L$ randomly chosen "2"s and "7"s.*
#
# <br />
#
# <font color='red'>**Write your code in the cell below ...**</font>
# +
## your code here
# -
# Now compute the validation error for your RBF-perceptron and use this to choose good values of $L$ and $\sigma$. Show a plot of the effect of changing each of these parameters, and justify your parameter choice.
#
# <br />
#
# <font color='red'>**Write your code in the cell below ...**</font>
# +
## your code here
# -
# <font color='red'>**Write your justfication here ...**</font> (as a *markdown* cell)
# ### 1.3 Kernel Perceptron
# Next, instead of directly computing a feature space transformation, we are going to use the kernel trick. Specifically, we are going to use the kernelised version of perceptron in combination with a few different kernels.
#
# *In this section, you cannot use any libraries other than `numpy` and `matplotlib`.*
#
# First, implement linear, polynomial and RBF kernels. The linear kernel is simply a dot product of its inputs, i.e., there is no feature space transformation. Polynomial and RBF kernels should be implemented as defined in the lecture slides.
#
# <br />
#
# <font color='red'>**Write your code in the cell below ...**</font>
# +
# Input:
# u,v - column vectors of the same dimensionality
#
# Output:
# v - a scalar
def linear_kernel(u, v):
## your code here
# Input:
# u,v - column vectors of the same dimensionality
# c,d - scalar parameters of the kernel as defined in lecture slides
#
# Output:
# v - a scalar
def polynomial_kernel(u, v, c=0, d=3):
## your code here
# Input:
# u,v - column vectors of the same dimensionality
# gamma - scalar parameter of the kernel as defined in lecture slides
#
# Output:
# v - a scalar
def rbf_kernel(u, v, gamma=1):
## your code here
# -
# Kernel perceptron was a "green slides" topic, and you will not be asked about this method in the exam. Here, you are only asked to implement a simple prediction function following the provided equation. In kernel perceptron, the prediction for instance $\mathbf{x}$ is made based on the sign of
#
# $$w_0 + \sum_{i=1}^{n}\alpha_i y_i K(\mathbf{x}_i, \mathbf{x})$$
#
# Here $w_0$ is the bias term, $n$ is the number of training examples, $\alpha_i$ are learned weights, $\mathbf{x}_i$ and $y_i$ is the training dataset,and $K$ is the kernel.
#
# <br />
#
# <font color='red'>**Write your code in the cell below ...**</font>
# Input:
# x_test - (r x m) matrix with instances for which to predict labels
# X - (n x m) matrix with training instances in rows
# y - (n x 1) vector with labels
# alpha - (n x 1) vector with learned weigths
# bias - scalar bias term
# kernel - a kernel function that follows the same prototype as each of the three kernels defined above
#
# Output:
# y_pred - (r x 1) vector of predicted labels
def kernel_ptron_predict(x_test, X, y, alpha, bias, kernel):
## your code here
# The code for kernel perceptron training is provided below. You can treat this function as a black box, but we encourage you to understand the implementation.
# Input:
# X - (n x m) matrix with training instances in rows
# y - (n x 1) vector with labels
# kernel - a kernel function that follows the same prototype as each of the three kernels defined above
# epochs - scalar, number of epochs
#
# Output:
# alpha - (n x 1) vector with learned weigths
# bias - scalar bias term
def kernel_ptron_train(X, y, kernel, epochs=100):
n, m = X.shape
alpha = np.zeros(n)
bias = 0
updates = None
for epoch in range(epochs):
print('epoch =', epoch, ', updates =', updates)
updates = 0
schedule = list(range(n))
np.random.shuffle(schedule)
for i in schedule:
y_pred = kernel_ptron_predict(X[i], X, y, alpha, bias, kernel)
if y_pred != y[i]:
alpha[i] += 1
bias += y[i]
updates += 1
if updates == 0:
break
return alpha, bias
# Now use the above functions to train the perceptron. Use heldout validation, and compute the validation error for this method using each of the three kernels. Write a paragraph or two with analysis how the accuracy differs between the different kernels and choice of kernel parameters. Discuss the merits of a kernel approach versus direct basis expansion approach as was used in the previous section.
#
# <br />
#
# <font color='red'>**Write your code in the cell below ...**</font>
# <font color='red'>**Provide your analysis here ...**</font> (as a *markdown* cell)
# ### 1.4 Dimensionality Reduction
# Yet another approach to working with complex data is to use a non-linear dimensionality reduction. To see how this might work, first apply a couple of dimensionality reduction methods and inspect the results.
# +
from sklearn import manifold
X = digits[:, 0:784]
y = np.squeeze(digits[:, 784:785])
# n_components refers to the number of dimensions after mapping
# n_neighbors is used for graph construction
X_iso = manifold.Isomap(n_neighbors=30, n_components=2).fit_transform(X)
# n_components refers to the number of dimensions after mapping
embedder = manifold.SpectralEmbedding(n_components=2, random_state=0)
X_se = embedder.fit_transform(X)
f, (ax1, ax2) = plt.subplots(1, 2)
ax1.plot(X_iso[y==-1,0], X_iso[y==-1,1], "bo")
ax1.plot(X_iso[y==1,0], X_iso[y==1,1], "ro")
ax1.set_title('Isomap')
ax2.plot(X_se[y==-1,0], X_se[y==-1,1], "bo")
ax2.plot(X_se[y==1,0], X_se[y==1,1], "ro")
ax2.set_title('spectral')
# -
# In a few sentences, explain how a dimensionality reduction algorithm can be used for your binary classification task.
# <font color='red'>**Write your answer here ...**</font> (as a *markdown* cell)
# Implement such an approach and assess the result. For simplicity, we will assume that both training and test data are available ahead of time, and thus the datasets should be used together for dimensionality reduction, after which you can split off a test set for measuring generalisation error. *Hint: you do not have to reduce number of dimensions to two. You are welcome to use the sklearn library for this question.*
#
# <br />
#
# <font color='red'>**Write your code in the cell below ...**</font>
# In a few sentences, comment on the merits of the dimensionality reduction based approach compared to linear classification from Section 1.1 and basis expansion from Section 1.2.
# <font color='red'>**Write your answer here ...**</font> (as a *markdown* cell)
# ## 2. Kaggle Competition
# The final part of the project is a competition, on more challenging digit data sourced from natural scenes. This data is coloured, pixelated or otherwise blurry, and the digits are not perfectly centered. It is often difficult for humans to classify! The dataset is also considerably larger.
#
# Please sign up to the [COMP90051 Kaggle competition](https://inclass.kaggle.com/c/comp90051-2017) using your `student.unimelb.edu.au` email address. Then download the file `data.npz` from Kaggle. This is a compressed `numpy` data file containing three ndarray objects:
# - `train_X` training set, with 4096 input features (greyscale pixel values);
# - `train_Y` training labels (0-9)
# - `test_X` test set, with 4096 input features, as per above
#
# Each image is 64x64 pixels in size, which has been flattened into a vector of 4096 values. You should load the files using `np.load`, from which you can extract the three elements. You may need to transpose the images for display, as they were flattened in a different order. Each pixel has an intensity value between 0-255. For those using languages other than python, you may need to output these objects in another format, e.g., as a matlab matrix.
#
# Your job is to develop a *multiclass* classifier on this dataset. You can use whatever techniques you like, such as the perceptron code from above, or other methods such as *k*NN, logistic regression, neural networks, etc. You may want to compare several methods, or try an ensemble combination of systems. You are free to use any python libraries for this question. Note that some fancy machine learning algorithms can take several hours or days to train (we impose no time limits), so please start early to allow sufficient time. *Note that you may want to sample smaller training sets, if runtime is an issue, however this will degrade your accuracy. Sub-sampling is a sensible strategy when developing your code.*
#
# You may also want to do some basic image processing, however, as this is not part of the subject, we would suggest that you focus most of your efforts on the machine learning. For inspiration, please see [Yan Lecun's MNIST page](http://yann.lecun.com/exdb/mnist/), specifically the table of results and the listed papers. Note that your dataset is harder than MNIST, so your mileage may vary.
# ### 2.1 Making Submissions
# This will be setup as a *Kaggle in class* competition, in which you can upload your system predictions on the test set. You should format your predictions as a csv file, with the same number of lines as the test set, and each line comprising two numbers `id, class` where *id* is the instance number (increasing integers starting from 1) and *class* is an integer between 0-9, corresponding to your system prediction. E.g.,
# ```
# Id,Label
# 1,9
# 2,9
# 3,4
# 4,5
# 5,1
# ...```
# based on the first five predictions of the system being classes `9 9 4 5 1`. See the `sample_submission.csv` for an example file.
#
# Kaggle will report your accuracy on a public portion of the test set, and maintain a leaderboard showing the performance of you and your classmates. You will be allowed to upload up to four submissions each day. At the end of the competition, you should nominate your best submission, which will be scored on the private portion of the test set. The accuracy of your system (i.e., proportion of correctly classified examples) on the private test set will be used for grading your approach.
#
# **Marks will be assigned as follows**:
# - position in the class, where all students are ranked and then the ranks are linearly scaled to <br>0 marks (worst in class) - 4 marks (best in class)
# - absolute performance (4 marks), banded as follows (rounded to nearest integer):
# <br>below 80% = 0 marks; 80-89% = 1; 90-92% = 2; 93-94% = 3; above 95% = 4 marks
# Note that you are required to submit your code with this notebook, submitted to the LMS. Failure to provide your implementation may result in assigning zero marks for the competition part, irrespective of the competition standing. Your implementation should be able to exactly reproduce submitted final Kaggle entry, and match your description below.
# ### 2.2. Method Description
# Describe your approach, and justify each of the choices made within your approach. You should write a document with no more than 400 words, as a **PDF** file (not *docx* etc) with up to 2 pages of A4 (2 sides). Text must only appear on the first page, while the second page is for *figures and tables only*. Please use a font size of 11pt or higher. Please consider using `pdflatex` for the report, as it's considerably better for this purpose than wysiwyg document editors. You are encouraged to include empirical results, e.g., a table of results, graphs, or other figures to support your argument. *(this will contribute 9 marks; note that we are looking for clear presentation, sound reasoning, good evaluation and error analysis, as well as general ambition of approach.)*
|
COMP90051 Statistical Machine Learning/project-2.ipynb
|