licenses
sequencelengths 1
3
| version
stringclasses 677
values | tree_hash
stringlengths 40
40
| path
stringclasses 1
value | type
stringclasses 2
values | size
stringlengths 2
8
| text
stringlengths 25
67.1M
| package_name
stringlengths 2
41
| repo
stringlengths 33
86
|
|---|---|---|---|---|---|---|---|---|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 947 | """Wrapper around the content of a text file, for testing.
```julia
file = FileContent(name)
```
can be used as
```julia
@test file.exists
@test "string" in file
@test contains(file, "string")
```
Apart from providing the convenient `in`, when the test fails, this produces
more useful output than the equivalent
```
file = read(name, String)
@test contains(file, "string")
```
in that it doesn't dump the entire content of the file to the screen.
"""
struct FileContent
name::String
exists::Bool
content::String
function FileContent(filename)
if isfile(filename)
new(abspath(filename), true, read(filename, String))
else
new(abspath(filename), false, "")
end
end
end
Base.show(io::IO, f::FileContent) = print(io, "<Content of $(f.name)>")
Base.in(str, file::FileContent) = contains(file.content, str)
Base.contains(file::FileContent, str) = contains(file.content, str)
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 8607 | # init file for "make devrepl"
using Revise
using JuliaFormatter
using Documenter: doctest
using LiveServer: LiveServer, serve, servedocs
include(joinpath(@__DIR__, "clean.jl"))
using Printf
using Logging
using Coverage
using PrettyTables: pretty_table, Highlighter
using LocalCoverage:
LocalCoverage,
eval_coverage_metrics,
PackageCoverage,
FileCoverageSummary,
CoverageTools
function _coverage(metric::Union{PackageCoverage,FileCoverageSummary})
return 100.0 * (float(metric.lines_hit) / metric.lines_tracked)
end
function format_line(metric::Union{PackageCoverage,FileCoverageSummary})
name = (metric isa PackageCoverage ? "TOTAL" : relpath(metric.filename))
lines_hit = @sprintf("%3d", metric.lines_hit)
lines_tracked = @sprintf("%3d", metric.lines_tracked)
lines_missing = @sprintf("%3d", metric.lines_tracked - metric.lines_hit)
coverage = isnan(_coverage(metric)) ? "-" : @sprintf("%3.0f%%", _coverage(metric))
return hcat(name, lines_tracked, lines_hit, lines_missing, coverage)
end
"""Print out a coverage summary from existing coverage data.
```julia
show_coverage(path="./src"; root=pwd(), sort_by=nothing)
```
prints a a table showing the tracked files in `path`, the total number of
tracked lines in that file ("Total"), the number of lines with coverage
("Hit"), the number of lines without coverage ("Missed") and the "Coverage" as
a percentage.
The coverage data is collected from `.cov` files in `path` as well as
`tracefile-*.info` files in `root`.
Optionally, the table can be sorted by passing the name of a column to
`sort_by`, e..g. `sort_py=:Missed`.
"""
function show_coverage(path::String=joinpath(pwd(), "src"); root=pwd(), kwargs...)
path = abspath(path)
local coverage
logger = Logging.SimpleLogger(stderr, Logging.Error)
Logging.with_logger(logger) do
coverage = merge_coverage_counts(
Coverage.process_folder(path), # .cov files in path
Coverage.LCOV.readfolder(root), # tracefile.info
)
end
coverage = filter(coverage) do covitem
startswith(abspath(covitem.filename), path)
end
metrics = eval_coverage_metrics(coverage, path)
show_coverage(metrics; kwargs...)
end
function show_coverage(metrics::PackageCoverage; sort_by=nothing)
file_metrics = metrics.files
sorter = Dict(
:Total => (m -> m.lines_tracked),
:Hit => (m -> m.lines_hit),
:Missed => (m -> (m.lines_tracked - m.lines_hit)),
:Coverage => (m -> _coverage(m)),
)
if !isnothing(sort_by)
if sort_by ∈ keys(sorter)
sort!(file_metrics; by=sorter[sort_by])
else
error("Cannot sort by $sort_by, must be one of $(keys(sorter))")
end
end
table = reduce(vcat, map(format_line, [file_metrics..., metrics]))
row_coverage = [[_coverage(m) for m in file_metrics]... _coverage(metrics)]
highlighters = (
Highlighter(
(data, i, j) -> j == 5 && row_coverage[i] <= 50,
bold=true,
foreground=:red,
),
Highlighter((data, i, j) -> j == 5 && row_coverage[i] >= 90, foreground=:green),
)
table_str = pretty_table(
table,
header=["File name", "Total", "Hit", "Missed", "Coverage"],
alignment=[:l, :r, :r, :r, :r],
crop=:none,
linebreaks=true,
columns_width=[maximum(length.(table[:, 1])), 6, 6, 6, 8],
autowrap=false,
highlighters=highlighters,
body_hlines=[size(table, 1) - 1],
)
println(table_str)
end
_show_coverage_func = show_coverage
"""Run a package test-suite in a subprocess.
```julia
test(
file="test/runtests.jl";
root=pwd(),
project="test",
code_coverage="./.coverage/tracefile-%p.info",
show_coverage=(code_coverage != "none"),
color=<inherit>,
compiled_modules=<inherit>,
startup_file=<inherit>,
depwarn=<inherit>,
inline=<inherit>,
check_bounds="yes",
track_allocation=<inherit>,
threads=<inherit>,
genhtml=false,
covdir="coverage"
)
```
runs the test suite of the package located at `root` by running `include(file)`
inside a new julia process.
This is similar to what `Pkg.test()` does, but differs in the "sandboxing"
approach. While `Pkg.test()` creates a new temporary sandboxed environment,
`test()` uses an existing environment in `project` (the `test` subfolder by
default). This allows testing against the dev-versions of other packages. It
requires that the `test` folder contains both a `Project.toml` and a
`Manifest.toml` file.
The `test()` function also differs from directly including `test/runtests.jl`
in the REPL in that it can generate coverage data and reports (this is only
possible when running tests in a subprocess).
If `show_coverage` is passed as `true` (default), a coverage summary is shown.
Further, if `genhtml` is `true`, a full HTML coverage report will be generated
in `covdir` (relative to `root`). This requires the `genhtml` executable (part
of the [lcov](http://ltp.sourceforge.net/coverage/lcov.php) package). Instead
of `true`, it is also possible to pass the path to the `genhtml` executable.
All other keyword arguments correspond to the respective command line flag for
the `julia` executable that is run as the subprocess.
This function is intended to be exposed in a project's development-REPL.
"""
function test(
file="test/runtests.jl";
root=pwd(),
project="test",
code_coverage=joinpath(root, ".coverage", "tracefile-%p.info"),
# code_coverage = "user" or "@", for ".cov" files
show_coverage=(code_coverage != "none"),
color=(Base.have_color === nothing ? "auto" : Base.have_color ? "yes" : "no"),
compiled_modules=(Bool(Base.JLOptions().use_compiled_modules) ? "yes" : "no"),
startup_file=(Base.JLOptions().startupfile == 1 ? "yes" : "no"),
depwarn=(Base.JLOptions().depwarn == 2 ? "error" : "yes"),
inline=(Bool(Base.JLOptions().can_inline) ? "yes" : "no"),
track_allocation=(("none", "user", "all")[Base.JLOptions().malloc_log+1]),
check_bounds="yes",
threads=Threads.nthreads(),
genhtml::Union{Bool,AbstractString}=false,
covdir="coverage"
)
julia = Base.julia_cmd().exec[1]
cmd = [
julia,
"--project=$project",
"--color=$color",
"--compiled-modules=$compiled_modules",
"--startup-file=$startup_file",
"--code-coverage=$code_coverage",
"--track-allocation=$track_allocation",
"--depwarn=$depwarn",
"--check-bounds=$check_bounds",
"--threads=$threads",
"--inline=$inline",
"--eval",
"include(\"$file\")"
]
@info "Running '$(join(cmd, " "))' in subprocess"
run(Cmd(Cmd(cmd), dir=root))
if show_coverage || genhtml
logger = Logging.SimpleLogger(stderr, Logging.Error)
local coverage
package_dir = abspath(joinpath(root, "src"))
Logging.with_logger(logger) do
coverage = merge_coverage_counts(
Coverage.process_folder(package_dir), # .cov files in path
Coverage.LCOV.readfolder(root), # tracefile.info (recursively)
)
end
coverage = filter(coverage) do covitem
startswith(abspath(covitem.filename), package_dir)
end
if show_coverage
metrics = eval_coverage_metrics(coverage, package_dir)
_show_coverage_func(metrics)
end
(genhtml === true) && (genhtml = "genhtml")
(genhtml === false) && (genhtml = "")
if !isempty(genhtml)
generate_html_coverage(root; covdir, genhtml)
end
end
end
REPL_MESSAGE = """
*******************************************************************************
DEVELOPMENT REPL
Revise, JuliaFormatter, LiveServer are loaded.
* `help()` – Show this message
* `include("test/runtests.jl")` – Run the entire test suite
* `test()` – Run the entire test suite in a subprocess with coverage
* `import DocumenterCitations; doctest(DocumenterCitations)` –
Run doctests for docstrings in package
* `include("docs/make.jl")` – Generate the documentation
* `format(".")` – Apply code formatting to all files
* `servedocs([port=8000, verbose=false])` –
Build and serve the documentation. Automatically recompile and redisplay on
changes
* `clean()` – Clean up build/doc/testing artifacts
* `distclean()` – Restore to a clean checkout state
*******************************************************************************
"""
"""Show help"""
help() = println(REPL_MESSAGE)
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 3033 | using IOCapture: IOCapture
using Documenter: Documenter, makedocs
import Logging
# Run `makedocs` as part of a test.
#
# Use as:
#
# ```julia
# run_makedocs(root; kwargs...) do dir, result, success, backtrace, output
# @test success
# # * dir is the temporary folder where `root` was copied. It should
# # contain the `build` output folder etc.
# # * `result` is whatever `makedocs` returns (usually `nothing`)
# # * `success` is a boolean whether the call returned successfully
# # * `backtrace` contains the backtrace for any thrown exception
# # * `output` contains the STDOUT produced by `run_makedocs`
# end
# ```
#
# Keyword args are:
#
# * `check_success`: If true, log an error if the call to `makedocs` does not
# succeed
# * `check_failure`: If true, log an error if the call to `makedocs`
# unexpectedly succeeds
# * `env`: dict temporary overrides for environment variables. Consider passing
# `"JULIA_DEBUG" => ""` if testing `output` against some expected output.
#
# Note that even with `check_success`/`check_failure`, you must still `@test`
# the value of `success` inside the `do` block.
#
# All other keyword arguments are forwarded to `makedocs`.
#
# To show the output of every `run_makedocs` run, set the environment variable
# `JULIA_DEBUG=Main` or `ENV["JULIA_DEBUG"] = Main` in the dev-REPL.
function run_makedocs(
f,
root;
plugins=[],
check_success=false,
check_failure=false,
env=Dict{String,String}(),
kwargs...
)
dir = mktempdir()
cp(root, dir; force=true)
c = IOCapture.capture(rethrow=InterruptException) do
default_format = Documenter.HTML(; edit_link="master", repolink=" ")
withenv(env...) do
makedocs(;
plugins=plugins,
remotes=get(kwargs, :remotes, nothing),
sitename=get(kwargs, :sitename, " "),
format=get(kwargs, :format, default_format),
root=dir,
kwargs...
)
end
end
level = Logging.Debug
checked = (check_success || check_failure) ? " (expected)" : ""
success = !c.error
if (check_success && !success) || (check_failure && success)
checked = " (UNEXPECTED)"
level = Logging.Error
end
calling_frame = stacktrace()[3]
result = c.value
Logging.@logmsg level """
run_makedocs(root=$root,...) -> $(success ? "success" : "failure")$checked
@$(calling_frame.file):$(calling_frame.line)
--------------------------------- output ---------------------------------
$(c.output)
--------------------------------------------------------------------------
""" dir typeof(result) result stacktrace = stacktrace(c.backtrace)
write(joinpath(dir, "output"), c.output)
open(joinpath(dir, "result"), "w") do io
show(io, "text/plain", result)
println(io, "-"^80)
show(io, "text/plain", stacktrace(c.backtrace))
end
f(dir, result, success, c.backtrace, c.output)
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 3311 | using Test
using SafeTestsets
using DocumenterCitations
# Note: comment outer @testset to stop after first @safetestset failure
@time @testset verbose = true "DocumenterCitations" begin
println("\n* formatting (test_formatting.jl):")
@time @safetestset "formatting" begin
include("test_formatting.jl")
end
println("\n* tex to markdown (test_tex_to_markdown.jl):")
@time @safetestset "tex_to_markdown" begin
include("test_tex_to_markdown.jl")
end
println("\n* parse_bibliography_block (test_parse_bibliography_block.jl):")
@time @safetestset "parse_bibliography_block" begin
include("test_parse_bibliography_block.jl")
end
println("\n* bibliography_block_pages (test_bibliography_block_pages.jl):")
@time @safetestset "bibliography_block_pages" begin
include("test_bibliography_block_pages.jl")
end
println("\n* parse_citation_link (test_parse_citation_link.jl):")
@time @safetestset "parse_citation_link" begin
include("test_parse_citation_link.jl")
end
println("\n* smart alpha style (test_alphastyle.jl):")
@time @safetestset "smart alpha style" begin
include("test_alphastyle.jl")
end
println("\n* collect from docstrings (test_collect_from_docstrings.jl):")
@time @safetestset "collect_from_docstrings" begin
include("test_collect_from_docstrings.jl")
end
println("\n* content_bock (test_content_block.jl):")
@time @safetestset "content_block" begin
include("test_content_block.jl")
end
println("\n* md_ast (test_md_ast.jl):")
@time @safetestset "md_ast" begin
include("test_md_ast.jl")
end
println("\n* keys_with_underscores (test_keys_with_underscores.jl):")
@time @safetestset "keys_with_underscores" begin
include("test_keys_with_underscores.jl")
end
println("\n* integration test (test_integration.jl):")
@time @safetestset "integration" begin
include("test_integration.jl")
end
println("\n* doctest (test_doctest.jl)")
@time @safetestset "doctest" begin
include("test_doctest.jl")
end
println("\n* latex rendering (test_latex_rendering.jl)")
@time @safetestset "latex_rendering" begin
include("test_latex_rendering.jl")
end
println("\n* test undefined citations (test_undefined_citations.jl):")
@time @safetestset "undefined_citations" begin
include("test_undefined_citations.jl")
end
println("\n* test link checking (test_linkcheck.jl):")
@time @safetestset "linkcheck" begin
import Pkg
using Documenter: DOCUMENTER_VERSION
run_linkcheck = true
if !Sys.isexecutable("/usr/bin/env") # used by mock `curl`
run_linkcheck = false
@info "Skipped test_linkcheck.jl (cannot mock `curl`)"
elseif DOCUMENTER_VERSION < v"1.2"
run_linkcheck = false
@info "Skipped test_linkcheck.jl (old version of Documenter)"
elseif haskey(ENV, "JULIA_PKGEVAL")
run_linkcheck = false
@info "Skipped test_linkcheck.jl (running in PkgEval)"
end
run_linkcheck && include("test_linkcheck.jl")
end
print("\n")
end
nothing # avoid noise when doing `include("test/runtests.jl")`
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 2432 | using Test
using TestingUtilities: @Test # much better at comparing strings
using OrderedCollections: OrderedDict
using IOCapture: IOCapture
import DocumenterCitations:
_alpha_suffix,
format_citation,
format_bibliography_label,
init_bibliography!,
AlphaStyle,
CitationBibliography,
CitationLink
@testset "alpha suffix" begin
@test _alpha_suffix(1) == "a"
@test _alpha_suffix(2) == "b"
@test _alpha_suffix(25) == "y"
@test _alpha_suffix(26) == "za"
@test _alpha_suffix(27) == "zb"
@test _alpha_suffix(50) == "zy"
@test _alpha_suffix(51) == "zza"
end
@testset "alpha label disambiguation" begin
bib = CitationBibliography(joinpath(@__DIR__, "..", "docs", "src", "refs.bib"),)
_c = OrderedDict{String,Int64}() # dummy "citations"
dumb = Val(:alpha)
smart = AlphaStyle()
cit(key) = CitationLink("[$key](@cite)")
entry(key) = bib.entries[key]
lbl1 = format_bibliography_label(dumb, entry("GraceJPB2007"), _c)
@Test lbl1 == "[GBR+07]"
# The dumb style can generate labels without any any initialization, while
# the smart style *requires* that init_bibliography! was called.
c = IOCapture.capture(rethrow=InterruptException) do
format_bibliography_label(smart, entry("GraceJPB2007"), _c)
end
@test contains(
c.output,
"Error: No AlphaStyle label for GraceJPB2007. Was `init_bibliography!` called?"
)
@test c.value == "[?]"
@test contains(c.output, " Was `init_bibliography!` called?")
init_bibliography!(smart, bib)
l1 = format_citation(dumb, cit("GraceJPB2007"), bib.entries, _c)
l2 = format_citation(dumb, cit("GraceJMO2007"), bib.entries, _c)
@Test l1 == "[[GBR+07](@cite GraceJPB2007)]"
@Test l2 == "[[GBR+07](@cite GraceJMO2007)]"
lbl1 = format_bibliography_label(dumb, entry("GraceJPB2007"), _c)
lbl2 = format_bibliography_label(dumb, entry("GraceJMO2007"), _c)
@test lbl1 == lbl2 == "[GBR+07]"
l1 = format_citation(smart, cit("GraceJPB2007"), bib.entries, _c)
l2 = format_citation(smart, cit("GraceJMO2007"), bib.entries, _c)
@Test l1 == "[[GBR+07a](@cite GraceJPB2007)]"
@Test l2 == "[[GBR+07b](@cite GraceJMO2007)]"
lbl1 = format_bibliography_label(smart, entry("GraceJPB2007"), _c)
lbl2 = format_bibliography_label(smart, entry("GraceJMO2007"), _c)
@Test lbl1 == "[GBR+07a]"
@Test lbl2 == "[GBR+07b]"
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 10286 | using DocumenterCitations
using Test
include("run_makedocs.jl")
include("file_content.jl")
# regex for strings containing paths
function prx(s)
rx = s
# work around limitations of `replace` in Julia 1.6
for mapping in ["[" => "\\[", "]" => "\\]", "." => "\\.", "/" => "\\W*"]
# The mapping for path separators ("/") to "\W*" (sequence of non-word
# characters) is deliberately broad. I've found it frustratingly hard
# to get a regex that works on Windows.
rx = replace(rx, mapping)
end
Regex(rx)
end
@testset "Bibliographies with Pages" begin
bib = CitationBibliography(DocumenterCitations.example_bibfile, style=:numeric)
# non-strict
run_makedocs(
splitext(@__FILE__)[1];
sitename="Test",
warnonly=true,
plugins=[bib],
format=Documenter.HTML(prettyurls=false, repolink=""),
pages=[
"Home" => "index.md",
"A" => joinpath("part1", "section1", "p1_s1_page.md"),
"B" => joinpath("part1", "section2", "p1_s2_page.md"),
"C" => joinpath("part2", "section1", "p2_s1_page.md"),
"D" => joinpath("part2", "section2", "p2_s2_page.md"),
"E" => joinpath("part3", "section1", "p3_s1_page.md"),
"F" => joinpath("part3", "section2", "p3_s2_page.md"),
"Invalid" => joinpath("part3", "section2", "invalidpages.md"),
"References" => "references.md",
"Addendum" => "addendum.md",
],
check_success=true
) do dir, result, success, backtrace, output
@test success
#! format: off
@test_broken contains(output, prx("Error: Invalid \"index.md\" in Pages attribute of @bibliography block on page src/part3/section2/invalidpages.md: No such file \"src/part3/section2/index.md\"."))
@test contains(output, prx("Warning: The entry \"index.md\" in the Pages attribute of the @bibliography block on page src/part3/section2/invalidpages.md appears to be relative to \"src\"."))
@test contains(output, prx("Error: Invalid \"p3_s1_page.md\" in Pages attribute of @bibliography block on page src/part3/section2/invalidpages.md: No such file \"src/part3/section2/p3_s1_page.md\"."))
@test contains(output, prx("Error: Invalid \"noexist.md\" in Pages attribute of @bibliography block on page src/part3/section2/invalidpages.md: No such file \"src/part3/section2/noexist.md\"."))
@test contains(output, "Warning: No cited keys remaining after filtering to Pages")
@test contains(output, prx("Error: Invalid \"../../addendum.md\" in Pages attribute of @bibliography block on page src/part3/section2/invalidpages.md: File \"src/addendum.md\" exists but no references were collected."))
@test contains(output, prx("Error: Invalid \"p3_s1_page.md\" in Pages attribute of @bibliography block on page src/part3/section2/invalidpages.md: No such file \"src/part3/section2/p3_s1_page.md\"."))
@test contains(output, prx("Warning: The field `Pages` in src/part3/section2/invalidpages.md:41-44 must evaluate to a list of strings. Setting invalid `Pages = \"none\"` to `Pages = []`"))
#! format: on
build(paths...) = joinpath(dir, "build", paths...)
citation(n) = Regex("\\[<a href=\"[^\"]+\">$n</a>\\]")
contentlink(name) = ".html#$(replace(name, " " => "-"))\">$name"
index_html = FileContent(build("index.html"))
@test index_html.exists
@test citation(1) in index_html
p1_s1_page_html = FileContent(build("part1", "section1", "p1_s1_page.html"))
@test p1_s1_page_html.exists
@test citation(2) in p1_s1_page_html
@test citation(3) in p1_s1_page_html
@test "<dt>[2]</dt>" in p1_s1_page_html
@test "<dt>[3]</dt>" in p1_s1_page_html
@test contentlink("A: Part 1.1") in p1_s1_page_html
p1_s2_page_html = FileContent(build("part1", "section2", "p1_s2_page.html"))
@test p1_s2_page_html.exists
@test citation(3) in p1_s2_page_html
@test citation(4) in p1_s2_page_html
@test !("<dt>[2]</dt>" in p1_s2_page_html)
@test "<dt>[3]</dt>" in p1_s2_page_html
@test "<dt>[4]</dt>" in p1_s2_page_html
@test "content here is empty" in p1_s2_page_html
p2_s1_page_html = FileContent(build("part2", "section1", "p2_s1_page.html"))
@test p2_s1_page_html.exists
@test citation(5) in p2_s1_page_html
@test citation(5) in p2_s1_page_html
@test "<dt>[2]</dt>" in p2_s1_page_html
@test "<dt>[3]</dt>" in p2_s1_page_html
@test "<dt>[4]</dt>" in p2_s1_page_html
@test "<dt>[5]</dt>" in p2_s1_page_html
@test "<dt>[6]</dt>" in p2_s1_page_html
@test contentlink("C: Part 2.1") in p2_s1_page_html
@test contentlink("A: Part 1.1") in p2_s1_page_html
@test contentlink("B: Part 1.2") in p2_s1_page_html
p2_s2_page_html = FileContent(build("part2", "section2", "p2_s2_page.html"))
@test p2_s2_page_html.exists
@test citation(7) in p2_s2_page_html
@test citation(8) in p2_s2_page_html
@test "<dt>[2]</dt>" in p2_s2_page_html
@test "<dt>[3]</dt>" in p2_s2_page_html
@test "<dt>[4]</dt>" in p2_s2_page_html
@test "<dt>[5]</dt>" in p2_s2_page_html
@test "<dt>[6]</dt>" in p2_s2_page_html
@test !("<dt>[7]</dt>" in p2_s2_page_html)
@test !("<dt>[8]</dt>" in p2_s2_page_html)
@test contentlink("A: Part 1.1") in p2_s2_page_html
@test contentlink("B: Part 1.2") in p2_s2_page_html
@test contentlink("C: Part 2.1") in p2_s2_page_html
@test !(contentlink("D: Part 2.2") in p2_s2_page_html)
p3_s1_page_html = FileContent(build("part3", "section1", "p3_s1_page.html"))
@test p3_s1_page_html.exists
@test citation(9) in p3_s1_page_html
@test citation(10) in p3_s1_page_html
@test "<dt>[2]</dt>" in p3_s1_page_html
@test "<dt>[3]</dt>" in p3_s1_page_html
@test "<dt>[4]</dt>" in p3_s1_page_html
@test "<dt>[5]</dt>" in p3_s1_page_html
@test "<dt>[6]</dt>" in p3_s1_page_html
@test "<dt>[7]</dt>" in p3_s1_page_html
@test "<dt>[8]</dt>" in p3_s1_page_html
@test !("<dt>[9]</dt>" in p3_s1_page_html)
@test !("<dt>[10]</dt>" in p3_s1_page_html)
@test contentlink("A: Part 1.1") in p3_s1_page_html
@test contentlink("B: Part 1.2") in p3_s1_page_html
@test contentlink("C: Part 2.1") in p3_s1_page_html
@test contentlink("D: Part 2.2") in p3_s1_page_html
@test !(contentlink("E: Part 3.1") in p3_s1_page_html)
p3_s2_page_html = FileContent(build("part3", "section2", "p3_s2_page.html"))
@test p3_s2_page_html.exists
@test citation(11) in p3_s2_page_html
@test citation(12) in p3_s2_page_html
@test "<dt>[2]</dt>" in p3_s2_page_html
@test "<dt>[3]</dt>" in p3_s2_page_html
@test "<dt>[4]</dt>" in p3_s2_page_html
@test "<dt>[5]</dt>" in p3_s2_page_html
@test "<dt>[6]</dt>" in p3_s2_page_html
@test "<dt>[7]</dt>" in p3_s2_page_html
@test "<dt>[8]</dt>" in p3_s2_page_html
@test "<dt>[9]</dt>" in p3_s2_page_html
@test "<dt>[10]</dt>" in p3_s2_page_html
@test !("<dt>[11]</dt>" in p3_s2_page_html)
@test !("<dt>[12]</dt>" in p3_s2_page_html)
@test contentlink("A: Part 1.1") in p3_s2_page_html
@test contentlink("C: Part 2.1") in p3_s2_page_html
@test contentlink("B: Part 1.2") in p3_s2_page_html
@test contentlink("D: Part 2.2") in p3_s2_page_html
@test contentlink("E: Part 3.1") in p3_s2_page_html
invalidpages_html = FileContent(build("part3", "section2", "invalidpages.html"))
@test invalidpages_html.exists
@test "Nothing should render here" in invalidpages_html
@test "Again, nothing should render here" in invalidpages_html
@test "<dt>[11]</dt>" in invalidpages_html
@test "<dt>[12]</dt>" in invalidpages_html
addendum_html = FileContent(build("addendum.html"))
@test "No references are cited on this page" in addendum_html
end
# strict
run_makedocs(
splitext(@__FILE__)[1];
sitename="Test",
warnonly=false,
plugins=[bib],
format=Documenter.HTML(prettyurls=false, repolink=""),
check_failure=true
) do dir, result, success, backtrace, output
@test !success
#! format: off
@test_broken contains(output, prx("Error: Invalid \"index.md\" in Pages attribute of @bibliography block on page src/part3/section2/invalidpages.md: No such file \"src/part3/section2/index.md\"."))
@test contains(output, prx("Warning: The entry \"index.md\" in the Pages attribute of the @bibliography block on page src/part3/section2/invalidpages.md appears to be relative to \"src\"."))
@test contains(output, prx("Error: Invalid \"p3_s1_page.md\" in Pages attribute of @bibliography block on page src/part3/section2/invalidpages.md: No such file \"src/part3/section2/p3_s1_page.md\"."))
@test contains(output, prx("Error: Invalid \"noexist.md\" in Pages attribute of @bibliography block on page src/part3/section2/invalidpages.md: No such file \"src/part3/section2/noexist.md\"."))
@test contains(output, "Warning: No cited keys remaining after filtering to Pages")
@test contains(output, prx("Error: Invalid \"../../addendum.md\" in Pages attribute of @bibliography block on page src/part3/section2/invalidpages.md: File \"src/addendum.md\" exists but no references were collected."))
@test contains(output, prx("Error: Invalid \"p3_s1_page.md\" in Pages attribute of @bibliography block on page src/part3/section2/invalidpages.md: No such file \"src/part3/section2/p3_s1_page.md\"."))
@test contains(output, prx("Warning: The field `Pages` in src/part3/section2/invalidpages.md:41-44 must evaluate to a list of strings. Setting invalid `Pages = \"none\"` to `Pages = []`"))
#! format: on
@test result isa ErrorException
@test occursin("`makedocs` encountered an error [:bibliography_block]", result.msg)
end
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 1113 | using DocumenterCitations
using Test
include("run_makedocs.jl")
@testset "Collect from docstrings" begin
# https://github.com/JuliaDocs/DocumenterCitations.jl/issues/39
bib = CitationBibliography(
joinpath(@__DIR__, "..", "docs", "src", "refs.bib"),
style=:numeric
)
run_makedocs(
splitext(@__FILE__)[1];
sitename="Test",
plugins=[bib],
pages=["Home" => "index.md", "References" => "references.md",],
check_success=true
) do dir, result, success, backtrace, output
@test success
index_outfile = joinpath(dir, "build", "index.html")
@test isfile(index_outfile)
html = read(index_outfile, String)
nbsp = "\u00A0" # non-breaking space
@test occursin("citing Ref.$(nbsp)[<a href=\"references/#GoerzQ2022\">1</a>]", html)
ref_outfile = joinpath(dir, "build", "references", "index.html")
@test isfile(ref_outfile)
html = read(ref_outfile, String)
@test occursin("<dt>[1]</dt>", html)
@test occursin("<div id=\"GoerzQ2022\">", html)
end
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 746 | using DocumenterCitations
using Test
include("run_makedocs.jl")
@testset "@Contents Block Test" begin
# https://github.com/JuliaDocs/DocumenterCitations.jl/issues/16
# https://github.com/ali-ramadhan/DocumenterCitations.jl/issues/33
# https://github.com/ali-ramadhan/DocumenterCitations.jl/issues/24
bib = CitationBibliography(
joinpath(@__DIR__, "test_content_block", "src", "refs.bib"),
style=:numeric
)
run_makedocs(
joinpath(@__DIR__, "test_content_block");
sitename="Test",
plugins=[bib],
pages=["Home" => "index.md", "References" => "references.md",],
check_success=true
) do dir, result, success, backtrace, output
@test success
end
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 1211 | # Evaluate all doctests in docstrings in the package.
#
# Also check that `DocumenterCitations` behavors correctly in the context of
# `doctest` (when no `.bib` plugin is passed).
# https://github.com/JuliaDocs/DocumenterCitations.jl/issues/34
using Test
using IOCapture: IOCapture
using Documenter: doctest
import DocumenterCitations
@testset "run doctest" begin
c = IOCapture.capture() do
withenv("JULIA_DEBUG" => "") do
doctest(DocumenterCitations)
end
end
if any(result -> result isa Test.Fail, c.value.results)
@error """
doctest failure:
------------------------------- output -------------------------------
$(c.output)
----------------------------------------------------------------------
Run `import DocumenterCitations; doctest(DocumenterCitations)` in the
development REPL (`make devrepl`) for better error reporting.
"""
else
@test contains(c.output, "Skipped CollectCitations step (doctest only).")
@test contains(c.output, "Skipped ExpandBibliography step (doctest only).")
@test contains(c.output, "Skipped ExpandCitations step (doctest only).")
end
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 34806 | using Test
using TestingUtilities: @Test # much better at comparing long strings
using OrderedCollections: OrderedDict
using DocumenterCitations
import DocumenterCitations:
two_digit_year,
alpha_label,
get_urls,
doi_url,
format_names,
format_citation,
format_bibliography_reference,
format_urldate,
CitationLink,
_join_bib_parts,
_strip_md_formatting
using Dates: Dates, @dateformat_str
using IOCapture: IOCapture
@testset "two_digit_year" begin
c = IOCapture.capture() do
@test two_digit_year("2001--") == "01"
@test two_digit_year("2000") == "00"
@test two_digit_year("2000-2020") == "00"
@test two_digit_year("2010") == "10"
@test two_digit_year("1984") == "84"
@test two_digit_year("11") == "11"
@test two_digit_year("invalid") == "invalid"
end
@test occursin("Invalid year: invalid", c.output)
end
@testset "alpha_label" begin
bib = CitationBibliography(DocumenterCitations.example_bibfile)
@test alpha_label(bib.entries["Tannor2007"]) == "Tan07"
@test alpha_label(bib.entries["FuerstNJP2014"]) == "FGP+14"
@test alpha_label(bib.entries["ImamogluPRE2015"]) == "IW15"
@test alpha_label(bib.entries["SciPy"]) == "JOP+01" # _is_others
@test alpha_label(bib.entries["MATLAB:2014"]) == "MAT14"
@test alpha_label(bib.entries["LapertPRA09"]) == "LTTS09"
@test alpha_label(bib.entries["GrondPRA2009b"]) == "GvWSH09"
@test alpha_label(bib.entries["WinckelIP2008"]) == "vWB08"
@test alpha_label(bib.entries["QCRoadmap"]) == "Anon04"
@test alpha_label(bib.entries["TedRyd"]) == "CW??"
@test alpha_label(bib.entries["jax"]) == "BFH+??"
end
@testset "format_citation(:authoryear)" begin
bib = CitationBibliography(DocumenterCitations.example_bibfile)
function ctext(key)
return format_citation(
Val(:authoryear),
CitationLink("[$key](@cite)"),
bib.entries,
OrderedDict{String,Int64}()
)
end
@Test ctext("Wilhelm2003.10132") ==
"([Wilhelm *et al.*, 2020](@cite Wilhelm2003.10132))"
@Test ctext("QCRoadmap") == "([Anonymous, 2004](@cite QCRoadmap))"
@Test ctext("TedRyd") == "([Corcovilos and Weiss, undated](@cite TedRyd))"
end
@testset "strip md formatting" begin
@Test _strip_md_formatting("[arXiv](https://arxiv.org)") == "arXiv"
@Test _strip_md_formatting("*Title*. [arXiv](https://arxiv.org)") == "Title. arXiv"
@Test _strip_md_formatting("Text with ``x^2`` math") == "Text with x^2 math"
@Test _strip_md_formatting("*italics* and **bold**") == "italics and bold"
c = IOCapture.capture() do
paragraphs = "Multiple\n\nparagraphs\n\nare\n\nnot allowed"
@assert _strip_md_formatting(paragraphs) == paragraphs
end
@test contains(c.output, "Cannot strip formatting")
end
@testset "join_bib_parts" begin
@Test _join_bib_parts(["Title", "(2023)"]) == "Title (2023)."
@Test _join_bib_parts(["[Title](https://www.aps.org)", "(2023)"]) ==
"[Title](https://www.aps.org) (2023)."
@Test _join_bib_parts(["Title", "arXiv"]) == "Title, arXiv."
@Test _join_bib_parts(["[Title](https://www.aps.org)", "arXiv"]) ==
"[Title](https://www.aps.org), arXiv."
@Test _join_bib_parts(["Title.", "arXiv"]) == "Title. arXiv."
@Test _join_bib_parts(["Title.", "[arXiv](https://arxiv.org)"]) ==
"Title. [arXiv](https://arxiv.org)."
@Test _join_bib_parts(["Title", "(2023)", "arXiv"]) == "Title (2023), arXiv."
@Test _join_bib_parts(["Title", "(2023)", "Special issue."]) ==
"Title (2023). Special issue."
@Test _join_bib_parts(["Title", "``x^2``"]) == "Title, ``x^2``."
@Test _join_bib_parts(["Title", "``X^2``"]) == "Title. ``X^2``."
@Test _join_bib_parts(["*Title*", "``x^2``"]) == "*Title*, ``x^2``."
@Test _join_bib_parts(["*Title*", "``X^2``"]) == "*Title*. ``X^2``."
c = IOCapture.capture() do
paragraphs = "Multiple\n\nparagraphs\n\nare\n\nnot allowed"
@assert _join_bib_parts(["*Title*", paragraphs]) == "*Title*. $paragraphs."
end
@test contains(c.output, "Cannot strip formatting")
end
@testset "format_urldate" begin
bib = CitationBibliography(DocumenterCitations.example_bibfile)
entry = bib.entries["WP_Schroedinger"]
@Test format_urldate(entry) == "Accessed on Oct 24, 2023"
@Test format_urldate(entry; accessed_on="", fmt=dateformat"d.m.Y") == "24.10.2023"
c = IOCapture.capture(rethrow=Union{}) do
fmt = Dates.DateFormat("SDF ", "english")
format_urldate(entry; accessed_on="", fmt=fmt)
end
@test c.value isa MethodError
@test contains(
c.output,
"Error: Check if fmt=dateformat\"SDF \" is a valid dateformat!"
)
bib = CitationBibliography(joinpath(splitext(@__FILE__)[1], "urldate.bib"))
c = IOCapture.capture(rethrow=Union{}) do
format_urldate(bib.entries["BrifNJP2010xxx"])
end
@test c.value == "Accessed on Oct 24, 2023"
@test contains(
c.output,
"Warning: Entry BrifNJP2010xxx defines an 'urldate' field, but no 'url' field."
)
c = IOCapture.capture(rethrow=Union{}) do
format_urldate(bib.entries["WP_Schroedingerxxx"], accessed_on="accessed ")
end
@test c.value == "accessed a few days ago"
@test contains(
c.output,
"Warning: Invalid field urldate = \"a few days ago\". Must be in the format YYYY-MM-DD."
)
end
@testset "format_names" begin
bib = CitationBibliography(DocumenterCitations.example_bibfile)
md(key; kwargs...) = format_names(bib.entries[key]; kwargs...)
@Test md("SciPy"; names=:lastfirst) ==
"Jones, E.; Oliphant, T.; Peterson, P. and others"
@Test md("SciPy"; names=:last) == "E. Jones, T. Oliphant, P. Peterson and others"
@Test md("SciPy"; names=:lastonly) == "Jones, Oliphant, Peterson and others"
@Test md("SciPy"; names=:full) ==
"Eric Jones, Travis Oliphant, Pearu Peterson and others"
@test_throws ArgumentError md("SciPy"; names=:first)
end
@testset "get_urls" begin
bib = CitationBibliography(DocumenterCitations.example_bibfile)
@test get_urls(bib.entries["GoerzSPIEO2021"]) == [
"https://michaelgoerz.net/research/GoerzSPIEO2021.pdf",
"https://doi.org/10.1117/12.2587002"
]
@test get_urls(bib.entries["GoerzSPIEO2021"]; skip=1) ==
["https://doi.org/10.1117/12.2587002"]
@test length(get_urls(bib.entries["GoerzSPIEO2021"]; skip=2)) == 0
@test get_urls(bib.entries["Nolting1997Coulomb"]) ==
["https://doi.org/10.1007/978-3-663-14691-9"]
end
@testset "format_bibliography_reference(:numeric)" begin
bib = CitationBibliography(DocumenterCitations.example_bibfile)
md(key) = format_bibliography_reference(Val(:numeric), bib.entries[key])
# Note: the test strings below contain nonbreaking spaces (" " = "\u00A0")
@Test md("GoerzJPB2011") ==
"M. H. Goerz, T. Calarco and C. P. Koch. *The quantum speed limit of optimal controlled phasegates for trapped neutral atoms*. [J. Phys. B **44**, 154011](https://doi.org/10.1088/0953-4075/44/15/154011) (2011), [arXiv:1103.6050](https://arxiv.org/abs/1103.6050). Special issue on quantum control theory for coherence and information dynamics."
@Test md("Luc-KoenigEPJD2004") ==
"E. Luc-Koenig, M. Vatasescu and F. Masnou-Seeuws. *Optimizing the photoassociation of cold atoms by use of chirped laser pulses*. [Eur. Phys. J. D **31**, 239](https://doi.org/10.1140/epjd/e2004-00161-8) (2004), [arXiv:physics/0407112 [physics.atm-clus]](https://arxiv.org/abs/physics/0407112)."
@Test md("GoerzNPJQI2017") ==
"M. H. Goerz, F. Motzoi, K. B. Whaley and C. P. Koch. *Charting the circuit QED design landscape using optimal control theory*, [npj Quantum Inf **3**, 37](https://doi.org/10.1038/s41534-017-0036-0) (2017)."
@Test md("Wilhelm2003.10132") ==
"F. K. Wilhelm, S. Kirchhoff, S. Machnes, N. Wittler and D. Sugny. *An introduction into optimal control for quantum technologies*, [arXiv:2003.10132](https://doi.org/10.48550/ARXIV.2003.10132) (2020)."
@Test md("Evans1983") ==
"L. C. Evans. [*An Introduction to Mathematical Optimal Control Theory*](https://math.berkeley.edu/~evans/control.course.pdf). Lecture Notes, University of California, Berkeley."
@Test md("Giles2008b") ==
"M. B. Giles. [*An extended collection of matrix derivative results for forward and reverse mode automatic differentiation*](https://people.maths.ox.ac.uk/gilesm/files/NA-08-01.pdf). Technical Report NA-08-01 (Oxford University Computing Laboratory, Jan 2008)."
@Test md("QCRoadmap") ==
"[*Quantum Computation Roadmap*](http://qist.lanl.gov) (2004). Version 2.0; April 2, 2004."
@Test md("TedRyd") ==
"T. Corcovilos and D. S. Weiss. *Rydberg Calculations*. Private communication."
@Test md("jax") ==
"J. Bradbury, R. Frostig, P. Hawkins, M. J. Johnson, C. Leary, D. Maclaurin, G. Necula, A. Paszke, J. VanderPlas, S. Wanderman-Milne and Q. Zhang. [*`JAX`: composable transformations of Python+NumPy programs*](https://github.com/google/jax), [`https://numpy.org`](https://numpy.org)."
@Test md("WP_Schroedinger") ==
"Wikipedia: [*Schrödinger equation*](https://en.wikipedia.org/wiki/Schrödinger_equation). Accessed on Oct 24, 2023."
@Test md("SciPy") ==
"E. Jones, T. Oliphant, P. Peterson and others. [*SciPy: Open source scientific tools for Python*](https://docs.scipy.org/doc/scipy/) (2001–). Project website at [`https://scipy.org`](https://scipy.org)."
@Test md("BrionPhd2004") ==
"E. Brion. *Contrôle Quantique et Protection de la Cohérence par effet Zénon, Applications à l'Informatique Quantique*. Ph.D. Thesis, Université Pierre et Marie Curie - Paris VI (2014). [HAL:tel-00007910v2](https://hal.science/tel-00007910v2)."
@Test md("Tannor2007") ==
"D. J. Tannor. [*Introduction to Quantum Mechanics: A Time-Dependent Perspective*](https://uscibooks.aip.org/books/introduction-to-quantum-mechanics-a-time-dependent-perspective/) (University Science Books, Sausalito, California, 2007)."
@Test md("SolaAAMOP2018") ==
"I. R. Sola, B. Y. Chang, S. A. Malinovskaya and V. S. Malinovsky. [*Quantum Control in Multilevel Systems*](https://doi.org/10.1016/bs.aamop.2018.02.003). In: *Advances In Atomic, Molecular, and Optical Physics*, Vol. 67, edited by E. Arimondo, L. F. DiMauro and S. F. Yelin (Academic Press, 2018); Chapter 3, pp. 151–256."
@Test md("GoerzSPIEO2021") ==
"M. H. Goerz, M. A. Kasevich and V. S. Malinovsky. [*Quantum optimal control for atomic fountain interferometry*](https://michaelgoerz.net/research/GoerzSPIEO2021.pdf). In: [*Proc. SPIE 11700, Optical and Quantum Sensing and Precision Metrology*](https://doi.org/10.1117/12.2587002) (2021)."
@Test md("NielsenChuangCh10QEC") ==
"M. Nielsen and I. L. Chuang. [*Quantum error-correction*](https://doi.org/10.1017/CBO9780511976667). In: *Quantum Computation and Quantum Information* (Cambridge University Press, 2000); Chapter 10."
@Test md("Nolting1997Coulomb") ==
"W. Nolting. In: [*Quantenmechanik*](https://doi.org/10.1007/978-3-663-14691-9), Vol. 5.2 of *Grundkurs Theoretische Physik* (Vieweg & Teubner Verlag, 1997); Chapter 6, p. 100."
@Test md("AnderssonSGS2014") ==
"E. Andersson and P. Öhberg (Editors). [*Quantum Information and Coherence*](https://doi.org/10.1007/978-3-319-04063-9). *Scottish Graduate Series* (Springer, 2014). Lecture notes of [SUSSP 67 (2011)](https://sussp67.phys.strath.ac.uk)."
@Test md("SuominenSGS2014") ==
"K.-A. Suominen. [*Open Quantum Systems and Decoherence*](https://doi.org/10.1007/978-3-319-04063-9_10). In: *Quantum Information and Coherence*, *Scottish Graduate Series*, edited by E. Andersson and P. Öhberg (Springer, 2014); pp. 247–282. Notes from lecture at [SUSSP 67 (2011)](https://sussp67.phys.strath.ac.uk)."
@Test md("PaszkeNIPS2019") ==
"A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein, L. Antiga, A. Desmaison, A. Köpf, E. Yang, Z. DeVito, M. Raison, A. Tejani, S. Chilamkurthy, B. Steiner, L. Fang, J. Bai and S. Chintala. [*PyTorch: An Imperative Style, High-Performance Deep Learning Library*](http://papers.neurips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library.pdf). In: *Proceedings of the 33rd International Conference on Neural Information Processing Systems*, edited by H. M. Wallach, H. Larochelle, A. Beygelzimer, F. d'Alché-Buc, E. A. Fox and R. Garnett (NeurIPS 2019, Vancouver, BC, Canada, Dec 2019); pp. 8024–8035."
@Test md("Giles2008") ==
"M. B. Giles. [*Collected Matrix Derivative Results for Forward and Reverse Mode Algorithmic Differentiation*](https://people.maths.ox.ac.uk/gilesm/files/AD2008.pdf). In: [*Advances in Automatic Differentiation*](https://doi.org/10.1007/978-3-540-68942-3_4), Vol. 64 of *Lecture Notes in Computational Science and Engineering*, edited by C. H. Bischof, H. M. Bücker, P. Hovland, U. Naumann and J. Utke (Springer, Berlin, Heidelberg, 2008); pp. 35–44."
end
@testset "format_biliography_reference (preprints)" begin
bib = CitationBibliography(DocumenterCitations.example_bibfile)
bib0 = CitationBibliography(joinpath(splitext(@__FILE__)[1], "preprints.bib"))
merge!(bib.entries, bib0.entries)
md(key) = format_bibliography_reference(Val(:numeric), bib.entries[key])
# Note: the test strings below contain nonbreaking spaces (" " = "\u00A0")
@Test md("LarrouyPRX2020") ==
"A. Larrouy, S. Patsch, R. Richaud, J.-M. Raimond, M. Brune, C. P. Koch and S. Gleyzes. *Fast Navigation in a Large Hilbert Space Using Quantum Optimal Control*. [Phys. Rev. X **10**, 021058](https://doi.org/10.1103/physrevx.10.021058) (2020). [HAL:hal-02887773](https://hal.science/hal-02887773)."
@Test md("TuriniciHAL00640217") ==
"G. Turinici. [*Quantum control*](https://hal.science/hal-00640217). HAL:hal-00640217 (2012)."
@Test md("BrionPhd2004") ==
"E. Brion. *Contrôle Quantique et Protection de la Cohérence par effet Zénon, Applications à l'Informatique Quantique*. Ph.D. Thesis, Université Pierre et Marie Curie - Paris VI (2014). [HAL:tel-00007910v2](https://hal.science/tel-00007910v2)."
@Test md("KatrukhaNC2017") ==
"E. A. Katrukha, M. Mikhaylova, H. X. van Brakel, P. M. van Bergen en Henegouwen, A. Akhmanova, C. C. Hoogenraad and L. C. Kapitein. *Probing cytoskeletal modulation of passive and active intracellular dynamics using nanobody-functionalized quantum dots*. [Nat. Commun. **8**, 14772](https://doi.org/10.1038/ncomms14772) (2017), [biorXiv:089284](https://www.biorxiv.org/content/10.1101/089284)."
@Test md("NonStandardPreprint") ==
"M. Tomza, M. H. Goerz, M. Musiał, R. Moszynski and C. P. Koch. *Optimized production of ultracold ground-state molecules: Stabilization employing potentials with ion-pair character and strong spin-orbit coupling*. [Phys. Rev. A **86**, 043424](https://doi.org/10.1103/PhysRevA.86.043424) (2012), xxx-preprint:1208.4331."
end
@testset "format_biliography_reference (alternative non-articles)" begin
# Test some alternative / problematic forms of som of the trickier
# non-article references.
c = IOCapture.capture() do
# Suppress Error from Bibliography.jl: Entry JuhlARNMRS2020X is missing
# the booktitle field(s)" (we don't care, so we're not testing for it)
CitationBibliography(joinpath(splitext(@__FILE__)[1], "alternative_non_articles.bib"))
end
bib = c.value
md(style, key) = format_bibliography_reference(style, bib.entries[key])
numeric = Val(:numeric)
authoryear = Val(:authoryear)
@Test md(numeric, "JuhlARNMRS2020X") ==
"D. W. Juhl, Z. Tošner and T. Vosegaard. [*Versatile NMR simulations using SIMPSON*](https://pure.au.dk/portal/files/230817709/Versatile_NMR_simulations_using_SIMPSON.pdf). Vol. 100 of *Annual Reports on NMR Spectroscopy*, edited by G. A. Webb ([Elsevier, 2020](https://doi.org/10.1016/bs.arnmr.2019.12.001)); Chapter 1, pp. 1–59."
@Test md(authoryear, "JuhlARNMRS2020X") ==
"Juhl, D. W.; Tošner, Z. and Vosegaard, T. (2020). [*Versatile NMR simulations using SIMPSON*](https://pure.au.dk/portal/files/230817709/Versatile_NMR_simulations_using_SIMPSON.pdf). Vol. 100 of *Annual Reports on NMR Spectroscopy*, edited by Webb, G. A. ([Elsevier](https://doi.org/10.1016/bs.arnmr.2019.12.001)); Chapter 1, pp. 1–59."
@Test md(numeric, "Nolting1997CoulombX") ==
"W. Nolting. In: [*Quantenmechanik*](https://link.springer.com/book/10.1007/978-3-662-44230-2), Vol. 5 no. 2 of *Grundkurs Theoretische Physik* ([Vieweg & Teubner Verlag, 1997](https://doi.org/10.1007/978-3-663-14691-9)); 6th chapter, p. 100."
@Test md(authoryear, "Nolting1997CoulombX") ==
"Nolting, W. (1997). In: [*Quantenmechanik*](https://link.springer.com/book/10.1007/978-3-662-44230-2), Vol. 5 no. 2 of *Grundkurs Theoretische Physik* ([Vieweg & Teubner Verlag](https://doi.org/10.1007/978-3-663-14691-9)); 6th chapter, p. 100."
@Test md(numeric, "Shapiro2012X") ==
"M. Shapiro and P. Brumer. [*Quantum Control of Molecular Processes*](https://onlinelibrary.wiley.com/doi/book/10.1002/9783527639700). ``2^{nd}`` Ed. (Wiley and Sons, 2012)."
@Test md(authoryear, "Shapiro2012X") ==
"Shapiro, M. and Brumer, P. (2012). [*Quantum Control of Molecular Processes*](https://onlinelibrary.wiley.com/doi/book/10.1002/9783527639700). ``2^{nd}`` Ed. (Wiley and Sons)."
@Test md(numeric, "PercontiSPIE2016") ==
"P. Perconti, W. C. Alberts, J. Bajaj, J. Schuster and M. Reed. [*Sensors, nano-electronics and photonics for the Army of 2030 and beyond*](https://doi.org/10.1117/12.2217797). In: *Quantum Sensing and Nano Electronics and Photonics XIII*, Vol. 9755 no. 6 of *Proceedings SPIE* (2016)."
@Test md(authoryear, "PercontiSPIE2016") ==
"Perconti, P.; Alberts, W. C.; Bajaj, J.; Schuster, J. and Reed, M. (2016). [*Sensors, nano-electronics and photonics for the Army of 2030 and beyond*](https://doi.org/10.1117/12.2217797). In: *Quantum Sensing and Nano Electronics and Photonics XIII*, Vol. 9755 no. 6 of *Proceedings SPIE*."
@Test md(numeric, "DevoretLH1995") ==
"M. H. Devoret. [*Quantum fluctuations in electrical circuits*](https://boulderschool.yale.edu/sites/default/files/files/devoret_quantum_fluct_les_houches.pdf). In: *Quantum Fluctuations*, Session LXIII (1995) of *the Les Houches Summer School*, edited by S. Reynaud, E. Giacobino and J. Zinn-Justin (Elsevier, 1997); Chapter 10, p. 353."
@Test md(authoryear, "DevoretLH1995") ==
"Devoret, M. H. (1997). [*Quantum fluctuations in electrical circuits*](https://boulderschool.yale.edu/sites/default/files/files/devoret_quantum_fluct_les_houches.pdf). In: *Quantum Fluctuations*, Session LXIII (1995) of *the Les Houches Summer School*, edited by Reynaud, S.; Giacobino, E. and Zinn-Justin, J. (Elsevier); Chapter 10, p. 353."
c = IOCapture.capture() do
md(numeric, "Nolting1997CoulombXX")
end
@test contains(
c.output,
"Warning: Could not link [\"https://doi.org/10.1007/978-3-663-14691-9\"] in \"published in\" information for entry Nolting1997CoulombXX."
)
@Test c.value ==
"W. Nolting. Vol. 5 no. 2 of *Grundkurs Theoretische Physik* ([Vieweg & Teubner Verlag, 1997](https://link.springer.com/book/10.1007/978-3-662-44230-2)); 6th chapter, p. 100."
end
@testset "format_labeled_bibliography_reference(:numeric; article_link_doi_in_title=true)" begin
bib = CitationBibliography(DocumenterCitations.example_bibfile)
md(key) = DocumenterCitations.format_labeled_bibliography_reference(
Val(:numeric),
bib.entries[key];
article_link_doi_in_title=true
)
# Note: the test strings below contain nonbreaking spaces (" " = "\u00A0")
@Test md("GoerzJPB2011") ==
"M. H. Goerz, T. Calarco and C. P. Koch. [*The quantum speed limit of optimal controlled phasegates for trapped neutral atoms*](https://doi.org/10.1088/0953-4075/44/15/154011). J. Phys. B **44**, 154011 (2011), [arXiv:1103.6050](https://arxiv.org/abs/1103.6050). Special issue on quantum control theory for coherence and information dynamics."
@Test md("Luc-KoenigEPJD2004") ==
"E. Luc-Koenig, M. Vatasescu and F. Masnou-Seeuws. [*Optimizing the photoassociation of cold atoms by use of chirped laser pulses*](https://doi.org/10.1140/epjd/e2004-00161-8). Eur. Phys. J. D **31**, 239 (2004), [arXiv:physics/0407112 [physics.atm-clus]](https://arxiv.org/abs/physics/0407112)."
@Test md("GoerzNPJQI2017") ==
"M. H. Goerz, F. Motzoi, K. B. Whaley and C. P. Koch. [*Charting the circuit QED design landscape using optimal control theory*](https://doi.org/10.1038/s41534-017-0036-0), npj Quantum Inf **3**, 37 (2017)."
@Test md("Wilhelm2003.10132") ==
"F. K. Wilhelm, S. Kirchhoff, S. Machnes, N. Wittler and D. Sugny. [*An introduction into optimal control for quantum technologies*](https://doi.org/10.48550/ARXIV.2003.10132), arXiv:2003.10132 (2020)."
@Test md("Evans1983") ==
"L. C. Evans. [*An Introduction to Mathematical Optimal Control Theory*](https://math.berkeley.edu/~evans/control.course.pdf). Lecture Notes, University of California, Berkeley."
@Test md("Giles2008b") ==
"M. B. Giles. [*An extended collection of matrix derivative results for forward and reverse mode automatic differentiation*](https://people.maths.ox.ac.uk/gilesm/files/NA-08-01.pdf). Technical Report NA-08-01 (Oxford University Computing Laboratory, Jan 2008)."
@Test md("QCRoadmap") ==
"[*Quantum Computation Roadmap*](http://qist.lanl.gov) (2004). Version 2.0; April 2, 2004."
@Test md("TedRyd") ==
"T. Corcovilos and D. S. Weiss. *Rydberg Calculations*. Private communication."
@Test md("jax") ==
"J. Bradbury, R. Frostig, P. Hawkins, M. J. Johnson, C. Leary, D. Maclaurin, G. Necula, A. Paszke, J. VanderPlas, S. Wanderman-Milne and Q. Zhang. [*`JAX`: composable transformations of Python+NumPy programs*](https://github.com/google/jax), [`https://numpy.org`](https://numpy.org)."
@Test md("WP_Schroedinger") ==
"Wikipedia: [*Schrödinger equation*](https://en.wikipedia.org/wiki/Schrödinger_equation). Accessed on Oct 24, 2023."
@Test md("SciPy") ==
"E. Jones, T. Oliphant, P. Peterson and others. [*SciPy: Open source scientific tools for Python*](https://docs.scipy.org/doc/scipy/) (2001–). Project website at [`https://scipy.org`](https://scipy.org)."
@Test md("BrionPhd2004") ==
"E. Brion. *Contrôle Quantique et Protection de la Cohérence par effet Zénon, Applications à l'Informatique Quantique*. Ph.D. Thesis, Université Pierre et Marie Curie - Paris VI (2014). [HAL:tel-00007910v2](https://hal.science/tel-00007910v2)."
@Test md("Tannor2007") ==
"D. J. Tannor. [*Introduction to Quantum Mechanics: A Time-Dependent Perspective*](https://uscibooks.aip.org/books/introduction-to-quantum-mechanics-a-time-dependent-perspective/) (University Science Books, Sausalito, California, 2007)."
@Test md("SolaAAMOP2018") ==
"I. R. Sola, B. Y. Chang, S. A. Malinovskaya and V. S. Malinovsky. [*Quantum Control in Multilevel Systems*](https://doi.org/10.1016/bs.aamop.2018.02.003). In: *Advances In Atomic, Molecular, and Optical Physics*, Vol. 67, edited by E. Arimondo, L. F. DiMauro and S. F. Yelin (Academic Press, 2018); Chapter 3, pp. 151–256."
@Test md("GoerzSPIEO2021") ==
"M. H. Goerz, M. A. Kasevich and V. S. Malinovsky. [*Quantum optimal control for atomic fountain interferometry*](https://michaelgoerz.net/research/GoerzSPIEO2021.pdf). In: [*Proc. SPIE 11700, Optical and Quantum Sensing and Precision Metrology*](https://doi.org/10.1117/12.2587002) (2021)."
@Test md("NielsenChuangCh10QEC") ==
"M. Nielsen and I. L. Chuang. [*Quantum error-correction*](https://doi.org/10.1017/CBO9780511976667). In: *Quantum Computation and Quantum Information* (Cambridge University Press, 2000); Chapter 10."
@Test md("Nolting1997Coulomb") ==
"W. Nolting. In: [*Quantenmechanik*](https://doi.org/10.1007/978-3-663-14691-9), Vol. 5.2 of *Grundkurs Theoretische Physik* (Vieweg & Teubner Verlag, 1997); Chapter 6, p. 100."
@Test md("AnderssonSGS2014") ==
"E. Andersson and P. Öhberg (Editors). [*Quantum Information and Coherence*](https://doi.org/10.1007/978-3-319-04063-9). *Scottish Graduate Series* (Springer, 2014). Lecture notes of [SUSSP 67 (2011)](https://sussp67.phys.strath.ac.uk)."
@Test md("SuominenSGS2014") ==
"K.-A. Suominen. [*Open Quantum Systems and Decoherence*](https://doi.org/10.1007/978-3-319-04063-9_10). In: *Quantum Information and Coherence*, *Scottish Graduate Series*, edited by E. Andersson and P. Öhberg (Springer, 2014); pp. 247–282. Notes from lecture at [SUSSP 67 (2011)](https://sussp67.phys.strath.ac.uk)."
@Test md("PaszkeNIPS2019") ==
"A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein, L. Antiga, A. Desmaison, A. Köpf, E. Yang, Z. DeVito, M. Raison, A. Tejani, S. Chilamkurthy, B. Steiner, L. Fang, J. Bai and S. Chintala. [*PyTorch: An Imperative Style, High-Performance Deep Learning Library*](http://papers.neurips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library.pdf). In: *Proceedings of the 33rd International Conference on Neural Information Processing Systems*, edited by H. M. Wallach, H. Larochelle, A. Beygelzimer, F. d'Alché-Buc, E. A. Fox and R. Garnett (NeurIPS 2019, Vancouver, BC, Canada, Dec 2019); pp. 8024–8035."
@Test md("Giles2008") ==
"M. B. Giles. [*Collected Matrix Derivative Results for Forward and Reverse Mode Algorithmic Differentiation*](https://people.maths.ox.ac.uk/gilesm/files/AD2008.pdf). In: [*Advances in Automatic Differentiation*](https://doi.org/10.1007/978-3-540-68942-3_4), Vol. 64 of *Lecture Notes in Computational Science and Engineering*, edited by C. H. Bischof, H. M. Bücker, P. Hovland, U. Naumann and J. Utke (Springer, Berlin, Heidelberg, 2008); pp. 35–44."
end
@testset "format_bibliography_reference(:authoryear)" begin
bib = CitationBibliography(DocumenterCitations.example_bibfile)
md(key) = format_bibliography_reference(Val(:authoryear), bib.entries[key])
# Note: the test strings below contain nonbreaking spaces (" " = "\u00A0")
@Test md("GoerzJPB2011") ==
"Goerz, M. H.; Calarco, T. and Koch, C. P. (2011). *The quantum speed limit of optimal controlled phasegates for trapped neutral atoms*. [J. Phys. B **44**, 154011](https://doi.org/10.1088/0953-4075/44/15/154011), [arXiv:1103.6050](https://arxiv.org/abs/1103.6050). Special issue on quantum control theory for coherence and information dynamics."
@Test md("Luc-KoenigEPJD2004") ==
"Luc-Koenig, E.; Vatasescu, M. and Masnou-Seeuws, F. (2004). *Optimizing the photoassociation of cold atoms by use of chirped laser pulses*. [Eur. Phys. J. D **31**, 239](https://doi.org/10.1140/epjd/e2004-00161-8), [arXiv:physics/0407112 [physics.atm-clus]](https://arxiv.org/abs/physics/0407112)."
@Test md("GoerzNPJQI2017") ==
"Goerz, M. H.; Motzoi, F.; Whaley, K. B. and Koch, C. P. (2017). *Charting the circuit QED design landscape using optimal control theory*, [npj Quantum Inf **3**, 37](https://doi.org/10.1038/s41534-017-0036-0)."
@Test md("Wilhelm2003.10132") ==
"Wilhelm, F. K.; Kirchhoff, S.; Machnes, S.; Wittler, N. and Sugny, D. (2020). *An introduction into optimal control for quantum technologies*, [arXiv:2003.10132](https://doi.org/10.48550/ARXIV.2003.10132)."
@Test md("Evans1983") ==
"Evans, L. C. (1983). [*An Introduction to Mathematical Optimal Control Theory*](https://math.berkeley.edu/~evans/control.course.pdf). Lecture Notes, University of California, Berkeley."
@Test md("Giles2008b") ==
"Giles, M. B. (Jan 2008). [*An extended collection of matrix derivative results for forward and reverse mode automatic differentiation*](https://people.maths.ox.ac.uk/gilesm/files/NA-08-01.pdf). Technical Report NA-08-01 (Oxford University Computing Laboratory)."
@Test md("QCRoadmap") ==
"— (2004). [*Quantum Computation Roadmap*](http://qist.lanl.gov). Version 2.0; April 2, 2004."
@Test md("TedRyd") ==
"Corcovilos, T. and Weiss, D. S. *Rydberg Calculations*. Private communication."
@Test md("jax") ==
"Bradbury, J.; Frostig, R.; Hawkins, P.; Johnson, M. J.; Leary, C.; Maclaurin, D.; Necula, G.; Paszke, A.; VanderPlas, J.; Wanderman-Milne, S. and Zhang, Q. [*`JAX`: composable transformations of Python+NumPy programs*](https://github.com/google/jax), [`https://numpy.org`](https://numpy.org)."
@Test md("SciPy") ==
"Jones, E.; Oliphant, T.; Peterson, P. and others (2001–). [*SciPy: Open source scientific tools for Python*](https://docs.scipy.org/doc/scipy/). Project website at [`https://scipy.org`](https://scipy.org)."
@Test md("BrionPhd2004") ==
"Brion, E. (2014). *Contrôle Quantique et Protection de la Cohérence par effet Zénon, Applications à l'Informatique Quantique*. Ph.D. Thesis, Université Pierre et Marie Curie - Paris VI. [HAL:tel-00007910v2](https://hal.science/tel-00007910v2)."
@Test md("Tannor2007") ==
"Tannor, D. J. (2007). [*Introduction to Quantum Mechanics: A Time-Dependent Perspective*](https://uscibooks.aip.org/books/introduction-to-quantum-mechanics-a-time-dependent-perspective/) (University Science Books, Sausalito, California)."
@Test md("SolaAAMOP2018") ==
"Sola, I. R.; Chang, B. Y.; Malinovskaya, S. A. and Malinovsky, V. S. (2018). [*Quantum Control in Multilevel Systems*](https://doi.org/10.1016/bs.aamop.2018.02.003). In: *Advances In Atomic, Molecular, and Optical Physics*, Vol. 67, edited by Arimondo, E.; DiMauro, L. F. and Yelin, S. F. (Academic Press); Chapter 3, pp. 151–256."
@Test md("GoerzSPIEO2021") ==
"Goerz, M. H.; Kasevich, M. A. and Malinovsky, V. S. (2021). [*Quantum optimal control for atomic fountain interferometry*](https://michaelgoerz.net/research/GoerzSPIEO2021.pdf). In: [*Proc. SPIE 11700, Optical and Quantum Sensing and Precision Metrology*](https://doi.org/10.1117/12.2587002)."
@Test md("NielsenChuangCh10QEC") ==
"Nielsen, M. and Chuang, I. L. (2000). [*Quantum error-correction*](https://doi.org/10.1017/CBO9780511976667). In: *Quantum Computation and Quantum Information* (Cambridge University Press); Chapter 10."
@Test md("Nolting1997Coulomb") ==
"Nolting, W. (1997). In: [*Quantenmechanik*](https://doi.org/10.1007/978-3-663-14691-9), Vol. 5.2 of *Grundkurs Theoretische Physik* (Vieweg & Teubner Verlag); Chapter 6, p. 100."
@Test md("AnderssonSGS2014") ==
"Andersson, E. and Öhberg, P. (Editors) (2014). [*Quantum Information and Coherence*](https://doi.org/10.1007/978-3-319-04063-9). *Scottish Graduate Series* (Springer). Lecture notes of [SUSSP 67 (2011)](https://sussp67.phys.strath.ac.uk)."
@Test md("SuominenSGS2014") ==
"Suominen, K.-A. (2014). [*Open Quantum Systems and Decoherence*](https://doi.org/10.1007/978-3-319-04063-9_10). In: *Quantum Information and Coherence*, *Scottish Graduate Series*, edited by Andersson, E. and Öhberg, P. (Springer); pp. 247–282. Notes from lecture at [SUSSP 67 (2011)](https://sussp67.phys.strath.ac.uk)."
@Test md("PaszkeNIPS2019") ==
"Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; Desmaison, A.; Köpf, A.; Yang, E.; DeVito, Z.; Raison, M.; Tejani, A.; Chilamkurthy, S.; Steiner, B.; Fang, L.; Bai, J. and Chintala, S. (Dec 2019). [*PyTorch: An Imperative Style, High-Performance Deep Learning Library*](http://papers.neurips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library.pdf). In: *Proceedings of the 33rd International Conference on Neural Information Processing Systems*, edited by Wallach, H. M.; Larochelle, H.; Beygelzimer, A.; d'Alché-Buc, F.; Fox, E. A. and Garnett, R. (NeurIPS 2019, Vancouver, BC, Canada); pp. 8024–8035."
@Test md("Giles2008") ==
"Giles, M. B. (2008). [*Collected Matrix Derivative Results for Forward and Reverse Mode Algorithmic Differentiation*](https://people.maths.ox.ac.uk/gilesm/files/AD2008.pdf). In: [*Advances in Automatic Differentiation*](https://doi.org/10.1007/978-3-540-68942-3_4), Vol. 64 of *Lecture Notes in Computational Science and Engineering*, edited by Bischof, C. H.; Bücker, H. M.; Hovland, P.; Naumann, U. and Utke, J. (Springer, Berlin, Heidelberg); pp. 35–44."
end
@testset "corporate author" begin
# https://github.com/JuliaDocs/DocumenterCitations.jl/issues/44
bib = CitationBibliography(joinpath(splitext(@__FILE__)[1], "corporateauthor.bib"))
entry = bib.entries["OEIS"]
name = entry.authors[1]
@test name.last == "{OEIS Foundation Inc.}"
@test name.first == ""
@test name.middle == ""
@test name.particle == ""
md(key) = format_bibliography_reference(Val(:numeric), bib.entries[key])
@test md("OEIS") ==
"OEIS Foundation Inc. [*The On-Line Encyclopedia of Integer Sequences*](https://oeis.org). Published electronically at https://oeis.org (2023)."
nbsp = "\u00A0"
@test md("OEISworkaround") ==
"OEIS$(nbsp)Foundation$(nbsp)Inc. [*The On-Line Encyclopedia of Integer Sequences*](https://oeis.org). Published electronically at https://oeis.org (2023)."
end
@testset "invalid DOI" begin
bib = CitationBibliography(joinpath(splitext(@__FILE__)[1], "invalid_doi.bib"))
c = IOCapture.capture() do
doi_url(bib.entries["Brif"])
end
@test contains(
c.output,
"Warning: The DOI field in bibtex entry \"Brif\" should not be a URL."
)
@test c.value == "https://doi.org/10.1088/1367-2630/12/7/075008"
c = IOCapture.capture() do
doi_url(bib.entries["Shapiro"])
end
@test contains(
c.output,
"Warning: Invalid DOI \"doi:10.1002/9783527639700\" in bibtex entry \"Shapiro\"."
)
@test c.value == "https://doi.org/10.1002/9783527639700"
c = IOCapture.capture() do
doi_url(bib.entries["Tannor"])
end
@test contains(
c.output,
"Warning: Invalid DOI \"0.1007/978-94-011-2642-7_23\" in bibtex entry \"Tannor\"."
)
@test c.value == ""
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 3544 | using DocumenterCitations
using Documenter
using Test
include("run_makedocs.jl")
CUSTOM1 = joinpath(@__DIR__, "..", "docs", "custom_styles", "enumauthoryear.jl")
CUSTOM2 = joinpath(@__DIR__, "..", "docs", "custom_styles", "keylabels.jl")
include(CUSTOM1)
include(CUSTOM2)
@testset "Integration Test" begin
# We build the documentation of DocumenterCitations itself as a test
bib = CitationBibliography(
joinpath(@__DIR__, "..", "docs", "src", "refs.bib"),
style=:numeric
)
run_makedocs(
joinpath(@__DIR__, "..", "docs");
sitename="DocumenterCitations.jl",
plugins=[bib],
format=Documenter.HTML(;
prettyurls = true,
canonical = "https://juliadocs.github.io/DocumenterCitations.jl",
assets = String["assets/citations.css"],
footer = "Generated by Test",
edit_link = "",
repolink = ""
),
pages=[
"Home" => "index.md",
"Syntax" => "syntax.md",
"Citation Style Gallery" => "gallery.md",
"CSS Styling" => "styling.md",
"Internals" => "internals.md",
"References" => "references.md",
],
check_success=true
) do dir, result, success, backtrace, output
@test success
@test occursin("Info: CollectCitations", output)
@test occursin("Info: ExpandBibliography", output)
@test occursin("Info: ExpandCitations", output)
ref_outfile = joinpath(dir, "build", "references", "index.html")
@test isfile(ref_outfile)
# Check that we have the list of cited reference and then the list of
# all references on the main References page
html = read(ref_outfile, String)
rx = r"<div class=\"citation canonical\"><dl><dt>\[(\d{1,3})\]</dt>"
matches = collect(eachmatch(rx, html))
@test length(matches) == 2
@test parse(Int64, matches[1].captures[1]) == 1
# Assuming there are at least 5 explicitly cited references in the
# docs:
@test parse(Int64, matches[2].captures[1]) > 5
end
end
@testset "Integration Test - dumb :alpha" begin
# We build the documentation of DocumenterCitations itself with the
# genuinely dumb :alpha style. Since `:alpha` usually gets upgraded
# automatically to `AlphaStyle`, we don't get good coverage for :alpha
# otherwise.
using Bibliography
using OrderedCollections: OrderedDict
bibfile = joinpath(@__DIR__, "..", "docs", "src", "refs.bib")
style = :alpha
entries = Bibliography.import_bibtex(bibfile)
citations = OrderedDict{String,Int64}()
page_citations = Dict{String,Set{String}}()
anchor_map = Documenter.AnchorMap()
bib =
CitationBibliography(bibfile, style, entries, citations, page_citations, anchor_map)
run_makedocs(
joinpath(@__DIR__, "..", "docs");
sitename="DocumenterCitations.jl",
plugins=[bib],
pages=[
"Home" => "index.md",
"Syntax" => "syntax.md",
"Citation Style Gallery" => "gallery.md",
"CSS Styling" => "styling.md",
"Internals" => "internals.md",
"References" => "references.md",
],
check_success=true
) do dir, result, success, backtrace, output
@test success
end
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 3009 | using DocumenterCitations
using Test
using TestingUtilities: @Test # much better at comparing strings
using IOCapture: IOCapture
include("run_makedocs.jl")
@testset "keys with underscores" begin
# https://github.com/JuliaDocs/DocumenterCitations.jl/issues/14
bib = CitationBibliography(
joinpath(@__DIR__, "test_keys_with_underscores", "src", "refs.bib"),
style=:numeric
)
run_makedocs(
joinpath(@__DIR__, "test_keys_with_underscores");
sitename="Test",
plugins=[bib],
pages=["Home" => "index.md", "References" => "references.md",],
check_success=true
) do dir, result, success, backtrace, output
@test success
#! format: off
index_html = read(joinpath(dir, "build", "index.html"), String)
@Test contains(index_html, "[<a href=\"references/#rabiner_tutorial_1989\">1</a>]")
@Test contains(index_html, "[<a href=\"references/#GoerzQ2022\">2</a>, with <em>emphasis</em>]")
references_html = read(joinpath(dir, "build", "references", "index.html"), String)
@Test contains(references_html, "<div id=\"rabiner_tutorial_1989\">")
@Test contains(references_html, "<div id=\"GoerzQ2022\">")
#! format: on
end
end
@testset "keys with underscores (ambiguities)" begin
success = @test_throws ErrorException begin
CitationBibliography(
joinpath(
@__DIR__,
"test_keys_with_underscores_ambiguities",
"src",
"refs_invalid.bib"
),
style=:numeric
)
end
@test contains(success.value.msg, "Ambiguous key \"rabiner_tutorial*1989\"")
bib = CitationBibliography(
joinpath(@__DIR__, "test_keys_with_underscores_ambiguities", "src", "refs.bib"),
style=:numeric
)
# keys should have been normalized to underscores
@test collect(keys(bib.entries)) == ["rabiner_tutorial_1989", "Goerz_Q_2022"]
run_makedocs(
joinpath(@__DIR__, "test_keys_with_underscores_ambiguities");
sitename="Test",
plugins=[bib],
pages=["Home" => "index.md", "References" => "references.md",],
check_success=true
) do dir, result, success, backtrace, output
@test success
#! format: off
index_html = read(joinpath(dir, "build", "index.html"), String)
@Test contains(index_html, "[<a href=\"references/#rabiner_tutorial_1989\">1</a>]")
@Test contains(index_html, "[<a href=\"references/#Goerz_Q_2022\">2</a>]")
@test !contains(index_html, "*") # everything was normalized to "_"
references_html = read(joinpath(dir, "build", "references", "index.html"), String)
@Test contains(references_html, "<div id=\"rabiner_tutorial_1989\">")
@Test contains(references_html, "<div id=\"Goerz_Q_2022\">")
@test !contains(references_html, "*") # everything was normalized to "_"
#! format: on
end
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 9621 | using Pkg
using DocumenterCitations
using Documenter
using Test
include("run_makedocs.jl")
include("file_content.jl")
CUSTOM1 = joinpath(@__DIR__, "..", "docs", "custom_styles", "enumauthoryear.jl")
CUSTOM2 = joinpath(@__DIR__, "..", "docs", "custom_styles", "keylabels.jl")
include(CUSTOM1)
include(CUSTOM2)
const Documenter_version =
Pkg.dependencies()[Base.UUID("e30172f5-a6a5-5a46-863b-614d45cd2de4")].version
function dummy_lctx()
doc = Documenter.Document(; remotes=nothing)
buffer = IOBuffer()
return Documenter.LaTeXWriter.Context(buffer, doc)
end
function md_to_latex(mdstr)
lctx = dummy_lctx()
ast = Documenter.mdparse(mdstr; mode=:single)[1]
Documenter.LaTeXWriter.latex(lctx, ast.children)
return String(take!(lctx.io))
end
@testset "Invalid BibliographyNode" begin
exc = ArgumentError("`list_style` must be one of `:dl`, `:ul`, or `:ol`, not `:bl`")
@test_throws exc begin
DocumenterCitations.BibliographyNode(
:bl, # :bl (bullet list) doesn't exist, should be :ul
true,
DocumenterCitations.BibliographyItem[]
)
end
end
@testset "QCRoadmap" begin
reference = "[*Quantum Computation Roadmap*](http://qist.lanl.gov) (2004). Version 2.0; April 2, 2004."
result = md_to_latex(reference)
@test result ==
"\\href{http://qist.lanl.gov}{\\emph{Quantum Computation Roadmap}} (2004). Version 2.0; April 2, 2004."
# There seemed to be a problem with the hyperlink for the QCRoadmap
# reference. However, it turned out the problem was that we were using
# `\hypertarget{id}` instead of `\hypertarget{id}{}` (see below)
end
@testset "LaTeXWriter Integration Test" begin
bib = CitationBibliography(
joinpath(@__DIR__, "..", "docs", "src", "refs.bib"),
style=:numeric
)
run_makedocs(
joinpath(@__DIR__, "..", "docs");
sitename="DocumenterCitations.jl",
plugins=[bib],
format=Documenter.LaTeX(platform="none"),
pages=[
"Home" => "index.md",
"Syntax" => "syntax.md",
"Citation Style Gallery" => "gallery.md",
"CSS Styling" => "styling.md",
"Internals" => "internals.md",
"References" => "references.md",
],
env=Dict("DOCUMENTER_BUILD_PDF" => "1"),
check_success=true
) do dir, result, success, backtrace, output
@test success
@test occursin("LaTeXWriter: creating the LaTeX file.", output)
tex_outfile = joinpath(dir, "build", "DocumenterCitations.jl.tex")
@test isfile(tex_outfile)
tex = FileContent(tex_outfile)
@test raw"{\raggedright% @bibliography" in tex
@test raw"}% end @bibliography" in tex
# must use `\hypertarget{id}{}`, not `\hypertarget{id}`
@test r"\\hypertarget{\d+}{}" in tex
@test contains(
tex,
r"\\hypertarget{\d+}{}\\href{http://qist\.lanl\.gov}{\\emph{Quantum Computation Roadmap}} \(2004\)"
)
@test contains(
tex,
raw"\hangindent=0.33in {\makebox[{\ifdim0.33in<\dimexpr\width+1ex\relax\dimexpr\width+1ex\relax\else0.33in\fi}][l]{[1]}}"
)
nbsp = "\u00A0" # nonbreaking space
if Documenter_version >= v"1.1.2"
# https://github.com/JuliaDocs/Documenter.jl/pull/2300
nbsp = "~"
end
@test contains(
tex,
"\\hangindent=0.33in Brif,$(nbsp)C.; Chakrabarti,$(nbsp)R. and Rabitz,$(nbsp)H. (2010)."
) # authoryear :ul
end
end
@testset "LaTeXWriter – :ul bullet list, justified" begin
DocumenterCitations.set_latex_options(ul_as_hanging=false, bib_blockformat="")
@test DocumenterCitations._LATEX_OPTIONS == Dict{Symbol,Any}(
:ul_as_hanging => false,
:ul_hangindent => "0.33in",
:dl_hangindent => "0.33in",
:dl_labelwidth => "0.33in",
:bib_blockformat => "",
)
bib = CitationBibliography(
joinpath(@__DIR__, "..", "docs", "src", "refs.bib"),
style=:numeric
)
run_makedocs(
joinpath(@__DIR__, "..", "docs");
sitename="DocumenterCitations.jl",
plugins=[bib],
format=Documenter.LaTeX(platform="none"),
pages=[
"Home" => "index.md",
"Syntax" => "syntax.md",
"Citation Style Gallery" => "gallery.md",
"CSS Styling" => "styling.md",
"Internals" => "internals.md",
"References" => "references.md",
],
env=Dict("DOCUMENTER_BUILD_PDF" => "1"),
check_success=true
) do dir, result, success, backtrace, output
@test success
@test occursin("LaTeXWriter: creating the LaTeX file.", output)
tex_outfile = joinpath(dir, "build", "DocumenterCitations.jl.tex")
@test isfile(tex_outfile)
tex = FileContent(tex_outfile)
@test raw"{% @bibliography" in tex
@test raw"}% end @bibliography" in tex
nbsp = "\u00A0" # nonbreaking space
if Documenter_version >= v"1.1.2"
# https://github.com/JuliaDocs/Documenter.jl/pull/2300
nbsp = "~"
end
@test contains(
tex,
"\\begin{itemize}\n\\item Brif,$(nbsp)C.; Chakrabarti,$(nbsp)R. and Rabitz,$(nbsp)H. (2010)."
) # authoryear :ul
end
DocumenterCitations.reset_latex_options()
@test DocumenterCitations._LATEX_OPTIONS == Dict{Symbol,Any}(
:ul_as_hanging => true,
:ul_hangindent => "0.33in",
:dl_hangindent => "0.33in",
:dl_labelwidth => "0.33in",
:bib_blockformat => "\\raggedright",
)
end
@testset "LaTeXWriter – custom indents" begin
DocumenterCitations.set_latex_options(
ul_hangindent="1cm",
dl_hangindent="1.5cm",
dl_labelwidth="2.0cm"
)
@test DocumenterCitations._LATEX_OPTIONS == Dict{Symbol,Any}(
:ul_as_hanging => true,
:ul_hangindent => "1cm",
:dl_hangindent => "1.5cm",
:dl_labelwidth => "2.0cm",
:bib_blockformat => "\\raggedright",
)
bib = CitationBibliography(
joinpath(@__DIR__, "..", "docs", "src", "refs.bib"),
style=:numeric
)
run_makedocs(
joinpath(@__DIR__, "..", "docs");
sitename="DocumenterCitations.jl",
plugins=[bib],
format=Documenter.LaTeX(platform="none"),
pages=[
"Home" => "index.md",
"Syntax" => "syntax.md",
"Citation Style Gallery" => "gallery.md",
"CSS Styling" => "styling.md",
"Internals" => "internals.md",
"References" => "references.md",
],
env=Dict("DOCUMENTER_BUILD_PDF" => "1"),
check_success=true
) do dir, result, success, backtrace, output
@test success
@test occursin("LaTeXWriter: creating the LaTeX file.", output)
tex_outfile = joinpath(dir, "build", "DocumenterCitations.jl.tex")
@test isfile(tex_outfile)
tex = FileContent(tex_outfile)
@test raw"{\raggedright% @bibliography" in tex
@test raw"}% end @bibliography" in tex
nbsp = "\u00A0" # nonbreaking space
if Documenter_version >= v"1.1.2"
# https://github.com/JuliaDocs/Documenter.jl/pull/2300
nbsp = "~"
end
@test contains(
tex,
"\\hangindent=1cm Brif,$(nbsp)C.; Chakrabarti,$(nbsp)R. and Rabitz,$(nbsp)H. (2010)."
) # authoryear :ul
@test contains(
tex,
raw"\hangindent=1.5cm {\makebox[{\ifdim2.0cm<\dimexpr\width+1ex\relax\dimexpr\width+1ex\relax\else2.0cm\fi}][l]{[BCR10]}}"
) # :alpha style
@test contains(
tex,
raw"\hangindent=1.5cm {\makebox[{\ifdim2.0cm<\dimexpr\width+1ex\relax\dimexpr\width+1ex\relax\else2.0cm\fi}][l]{[1]}}"
) # :numeric style
end
DocumenterCitations.reset_latex_options()
@test DocumenterCitations._LATEX_OPTIONS == Dict{Symbol,Any}(
:ul_as_hanging => true,
:ul_hangindent => "0.33in",
:dl_hangindent => "0.33in",
:dl_labelwidth => "0.33in",
:bib_blockformat => "\\raggedright",
)
end
@testset "invalid latex options" begin
msg = "dl_as_hanging is not a valid option in set_latex_options."
@test_throws ArgumentError(msg) begin
DocumenterCitations.set_latex_options(dl_as_hanging=false)
# We've confused `dl_as_hanging` with `ul_as_hanging`
end
msg = "`0` for option ul_hangindent in set_latex_options must be of type String, not Int64"
@test_throws ArgumentError(msg) begin
DocumenterCitations.set_latex_options(ul_hangindent=0)
end
msg = "width \"\" must be a valid LaTeX width"
@test_throws ArgumentError(msg) begin
# DocumenterCitations.set_latex_options(ul_hangindent="")
# actually works, but then we get an error when we try to generate a
# label box:
DocumenterCitations._labelbox(nothing, nothing; width="")
end
@test DocumenterCitations._LATEX_OPTIONS == Dict{Symbol,Any}(
:ul_as_hanging => true,
:ul_hangindent => "0.33in",
:dl_hangindent => "0.33in",
:dl_labelwidth => "0.33in",
:bib_blockformat => "\\raggedright",
)
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 1327 | using DocumenterCitations
using Test
using IOCapture: IOCapture
include("run_makedocs.jl")
@testset "Check invalid url in .bib file " begin
# https://github.com/JuliaDocs/DocumenterCitations.jl/issues/58
root = splitext(@__FILE__)[1]
bib = CitationBibliography(joinpath(root, "src", "invalidlink.bib"), style=:numeric)
run_makedocs(
root;
linkcheck=true,
sitename="Test",
plugins=[bib],
pages=["Home" => "index.md", "References" => "references.md",],
check_failure=true,
env=Dict("PATH" => "$root:$(ENV["PATH"])"), # Unix only
# The updated PATH allows to use the mock `curl` in `root`.
) do dir, result, success, backtrace, output
@test !success
@test contains(output, r"Error:.*http://www.invalid-server.doesnotexist/page.html")
@test contains(
output,
"Error: linkcheck 'http://httpbin.org/status/404' status: 404."
)
@test contains(
output,
"Error: linkcheck 'http://httpbin.org/status/500' status: 500."
)
@test contains(
output,
"Error: linkcheck 'http://httpbin.org/status/403' status: 403."
)
@test contains(result.msg, "`makedocs` encountered an error [:linkcheck]")
end
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 4268 | # Test the behavior of MarkdownAST that we rely on to manipulate citations.
# This is to ensure that MarkdownAST doesn't change its behavior in some
# unexpected way, as well as a documentation of the MarkdownAST features that
# are relevant to us
using Test
using TestingUtilities: @Test # much better at comparing long strings
using DocumenterCitations
using DocumenterCitations: ast_linktext, ast_to_str
using IOCapture: IOCapture
import Documenter
import MarkdownAST
import Markdown
import AbstractTrees
MD_FULL = raw"""
# Markdown document
Let's just have a couple of pagragraphs with inline elements like *italic* or
**bold**.
We'll also have inline math like ``x^2`` (using the double-backtick syntax
preferred by [Julia](https://docs.julialang.org/en/v1/stdlib/Markdown/#\\LaTeX),
in lieu of `$`)
## Citation
Some citation links [rabiner_tutorial_1989; with *emphasis*](@cite) (with
inline formatting) and [GoerzQ2022](@cite) (without inline formatting).
## Lists
* First item with just plain text...
And a second paragraph (we don't want this to normalize to a "tight" list)
* Second item with *emphasis*
* Third item with `code`
This concludes the file.
"""
MD_MINIMAL = raw"""
Text with [rabiner_tutorial_1989](@cite).
"""
"""Return a `MarkdownAST.Document` node from a multiline markdown string.
This is how Documenter parses pages, cf. the constructor for the `Page`
object in Documenter.jl `src/documents.jl`.
It is also the basis of `Documenter.mdparse`, which only further goes into the
children of the `Document` node to return the ones selected depending on the
`mode` parameter.
"""
function parse_md_page_str(mdsrc)
mdpage = Markdown.parse(mdsrc)
return convert(MarkdownAST.Node, mdpage)
end
@testset "parse_md_page_str" begin
mdast = parse_md_page_str(MD_FULL)
@test mdast.element == MarkdownAST.Document()
# ensure that the file can be round-tripped without loss (this does not
# mean that the markdown code isn't "normalized", just that we get a stable
# AST and a stable text after the first roundtrip)
mdast2 = parse_md_page_str(MD_FULL)
@Test mdast2 == mdast
text = ast_to_str(mdast)
mdast3 = parse_md_page_str(text)
@Test mdast3 == mdast
text2 = ast_to_str(mdast3)
@Test text2 == text
end
@testset "Documenter.mdparse" begin
# Test the function that is used in the pipeline to convert between
# markdown code plain text and AST objects.
minimal_block_ast = MarkdownAST.@ast MarkdownAST.Paragraph() do
MarkdownAST.Text("Text with ")
MarkdownAST.Link("@cite", "") do
MarkdownAST.Text("rabiner")
MarkdownAST.Emph() do
MarkdownAST.Text("tutorial")
end
MarkdownAST.Text("1989")
end
MarkdownAST.Text(".")
end
# https://github.com/JuliaDocs/Documenter.jl/issues/2253
@test Documenter.mdparse(MD_MINIMAL; mode=:single) == [minimal_block_ast]
@test Documenter.mdparse(MD_MINIMAL; mode=:blocks) == [minimal_block_ast]
@test Documenter.mdparse(MD_MINIMAL; mode=:span) == [minimal_block_ast.children...]
@test_throws ArgumentError begin
c = IOCapture.capture() do
Documenter.mdparse(MD_FULL; mode=:single)
end
@test contains(
c.output,
"requires the Markdown string to parse into a single block"
)
end
full_blocks = Documenter.mdparse(MD_FULL; mode=:blocks)
@test length(full_blocks) == 8
@test full_blocks[1].element == MarkdownAST.Heading(1)
@test full_blocks[2].element == MarkdownAST.Paragraph()
@test full_blocks[3].element == MarkdownAST.Paragraph()
@test full_blocks[4].element == MarkdownAST.Heading(2)
@test full_blocks[5].element == MarkdownAST.Paragraph()
@test full_blocks[6].element == MarkdownAST.Heading(2)
@test full_blocks[7].element == MarkdownAST.List(:bullet, false)
@test full_blocks[8].element == MarkdownAST.Paragraph()
@test_throws ArgumentError begin
c = IOCapture.capture() do
Documenter.mdparse(MD_FULL; mode=:span)
end
@test contains(
c.output,
"requires the Markdown string to parse into a single block"
)
end
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 2704 | using Test
using DocumenterCitations: parse_bibliography_block
using IOCapture: IOCapture
@testset "parse_bibliography_block" begin
block = raw"""
Pages = ["index.md", "references.md"]
Canonical = true
*
GoerzPRA2010
"""
fields, lines = parse_bibliography_block(block, nothing, nothing)
@test fields[:Canonical] == true
@test fields[:Pages] == ["index.md", "references.md"]
@test lines == ["*", "GoerzPRA2010"]
block = raw"""
Pages = [
"index.md",
"references.md"
]
Canonical = true
GoerzPRA2010
"""
fields, lines = parse_bibliography_block(block, nothing, nothing)
@test fields[:Canonical] == true
@test fields[:Pages] == ["index.md", "references.md"]
@test lines == ["GoerzPRA2010"]
block = raw"""
Canonical = false
*
"""
fields, lines = parse_bibliography_block(block, nothing, nothing)
@test fields[:Canonical] == false
@test lines == ["*"]
block = raw"""
"""
fields, lines = parse_bibliography_block(block, nothing, nothing)
@test fields[:Canonical] == true
@test lines == []
end
@testset "invalid bibliography blocks" begin
block = raw"""
Pages = "index.md" # not a list
Canonical = false
"""
c = IOCapture.capture() do
fields, lines = parse_bibliography_block(block, nothing, nothing)
@test fields[:Canonical] == false
@test fields[:Pages] == []
@test lines == []
end
@test contains(
c.output,
"Warning: The field `Pages` in N/A must evaluate to a list of strings. Setting invalid `Pages = \"index.md\"` to `Pages = []`"
)
block = raw"""
Pages = [1, 2] # not a list of strings
"""
c = IOCapture.capture() do
fields, lines = parse_bibliography_block(block, nothing, nothing)
@test fields[:Canonical] == true
@test fields[:Pages] == ["1", "2"]
@test lines == []
end
@test contains(
c.output,
"Warning: The value `1` in N/A is not a string. Replacing with \"1\""
)
@test contains(
c.output,
"Warning: The value `2` in N/A is not a string. Replacing with \"2\""
)
block = raw"""
Pages = ["index.md"]
Canonical = "true" # not a Bool
"""
c = IOCapture.capture() do
fields, lines = parse_bibliography_block(block, nothing, nothing)
@test fields[:Canonical] == false
@test fields[:Pages] == ["index.md"]
@test lines == []
end
@test contains(
c.output,
"Warning: The field `Canonical` in N/A must evaluate to a boolean. Setting invalid `Canonical=\"true\"` to `Canonical=false`"
)
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 5387 | using Test
using DocumenterCitations: CitationLink, DirectCitationLink
using Markdown
import MarkdownAST
using IOCapture: IOCapture
@testset "parse_standard_citation_link" begin
cit = CitationLink("[GoerzQ2022](@cite)")
@test cit.cmd == :cite
@test cit.style ≡ nothing
@test cit.keys == ["GoerzQ2022"]
@test cit.note ≡ nothing
@test cit.capitalize ≡ false
@test cit.starred ≡ false
cit = CitationLink("[GoerzQ2022](@citet)")
@test cit.cmd == :citet
cit = CitationLink("[GoerzQ2022](@citep)")
@test cit.cmd == :citep
cit = CitationLink("[GoerzQ2022](@Citet)")
@test cit.cmd == :citet
@test cit.capitalize ≡ true
cit = CitationLink("[GoerzQ2022](@cite*)")
@test cit.cmd == :cite
@test cit.starred ≡ true
cit = CitationLink("[GoerzQ2022](@citet*)")
@test cit.cmd == :citet
@test cit.starred ≡ true
cit = CitationLink("[GoerzQ2022](@cite%authoryear%)")
# This is an undocumented feature (on purpose)
@test cit.cmd == :cite
@test cit.style ≡ :authoryear
cit = CitationLink("[GoerzQ2022; Eq.\u00A0(1)](@cite)")
@test cit.cmd == :cite
@test cit.style ≡ nothing
@test cit.keys == ["GoerzQ2022"]
@test cit.note ≡ "Eq.\u00A0(1)"
cit = CitationLink("[GoerzQ2022; Eq.\u00A0(1)](@cite)")
@test cit.note ≡ "Eq.\u00A0(1)"
cit = CitationLink("[GoerzQ2022,CarrascoPRA2022,GoerzA2023](@cite)")
@test cit.keys == ["GoerzQ2022", "CarrascoPRA2022", "GoerzA2023"]
cit = CitationLink("[GoerzQ2022, CarrascoPRA2022, GoerzA2023](@cite)")
@test cit.keys == ["GoerzQ2022", "CarrascoPRA2022", "GoerzA2023"]
cit = CitationLink(
"[GoerzQ2022,CarrascoPRA2022,GoerzA2023; and references therein](@cite)"
)
@test cit.keys == ["GoerzQ2022", "CarrascoPRA2022", "GoerzA2023"]
@test cit.note ≡ "and references therein"
end
@testset "invalid_citation_link" begin
c = IOCapture.capture() do
@test_throws ErrorException begin
cit = CitationLink("[GoerzQ2022](@citenocommand)")
# not a citecommand
end
@test_throws ErrorException begin
cit = CitationLink("[GoerzQ2022]( @cite )")
# can't have spaces
end
@test_throws ErrorException begin
cit = CitationLink("[see GoerzQ2022](@cite)")
# can't have text before keys
end
@test_throws ErrorException begin
cit = CitationLink("[GoerzQ2022](@CITE)")
# wrong capitalization
end
@test_throws ErrorException begin
cit = CitationLink("not a link")
# must be a link
end
@test_throws ErrorException begin
cit = CitationLink("See [GoerzQ2022](@cite)")
# extra "See "
end
@test_throws ErrorException begin
cit = CitationLink("# Title\n\nThis is a paragraph\n")
# Can't be parsed as single node
end
@test_throws ErrorException begin
cit = CitationLink("[text](@cite key)")
# DirectCitationLink
end
end
msgs = [
"Error: The @cite link destination \"@citenocommand\" does not match required regex",
"Error: citation link \"[GoerzQ2022]( @cite )\" destination must start exactly with \"@cite\" or one of its variants",
"Error: The @cite link text \"see GoerzQ2022\" does not match required regex",
"Error: The @cite link destination \"@CITE\" does not match required regex",
"Error: citation \"not a link\" must parse into MarkdownAST.Link",
"Error: citation \"See [GoerzQ2022](@cite)\" must parse into a single MarkdownAST.Link",
"Error: mode == :single requires the Markdown string to parse into a single block",
]
for msg in msgs
success = @test occursin(msg, c.output)
if success isa Test.Fail
@error "message not in output" msg
end
end
end
@testset "parse_direct_citation_link" begin
cit = DirectCitationLink("[Semi-AD paper](@cite GoerzQ2022)")
@test cit.key == "GoerzQ2022"
@test cit.node == MarkdownAST.@ast MarkdownAST.Link("@cite GoerzQ2022", "") do
MarkdownAST.Text("Semi-AD paper")
end
cit = DirectCitationLink("[*Semi*-AD paper](@cite GoerzQ2022)")
@test cit.key == "GoerzQ2022"
@test cit.node == MarkdownAST.@ast MarkdownAST.Link("@cite GoerzQ2022", "") do
MarkdownAST.Emph() do
MarkdownAST.Text("Semi")
end
MarkdownAST.Text("-AD paper")
end
end
@testset "invalid_direct_text_citation_link" begin
c = IOCapture.capture() do
@test_throws ErrorException begin
DirectCitationLink("[Semi-AD paper](@citet GoerzQ2022)")
# only @cite is allowed
end
@test_throws ErrorException begin
DirectCitationLink("[first two papers](@cite BrifNJP2010, GoerzQ2022)")
# there has to be a single link target
end
end
msgs = [
"Error: The @cite link destination \"@citet GoerzQ2022\" does not match required regex",
"Error: The @cite link destination \"@cite BrifNJP2010, GoerzQ2022\" does not match required regex",
]
for msg in msgs
success = @test occursin(msg, c.output)
if success isa Test.Fail
@error "message not in output" msg
end
end
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 13940 | using Printf
using Test
using IOCapture: IOCapture
using TestingUtilities: @Test # much better at comparing long strings
import DocumenterCitations:
DocumenterCitations,
tex_to_markdown,
_process_tex,
_collect_command,
_collect_accent,
_collect_math,
_collect_group
function format_char(c)
@assert isvalid(Char, c)
if Int64(c) < 128
return string(c)
else
return @sprintf "\\u%x" Int(c)
end
end
# for interactive debugging
function print_uni_escape(s)
escaped = prod(format_char.(collect(s)))
println("\"$escaped\" # $(repr(s))")
end
@testset "collect_group" begin
s = "{group} x"
@test _collect_group(s, 1) == (7, "{group}", "group")
s = "{group\\%} x"
@test _collect_group(s, 1) == (9, "{group\\%}", "group%")
end
@testset "collect_command" begin
s = "\\i"
@test _collect_command(s, 1) == (2, "\\i", "\u0131") # ı
s = "\\i \\i{} "
@test _collect_command(s, 1) == (3, "\\i ", "\u0131") # ı
@test _collect_command(s, 4) == (5, "\\i", "\u0131") # ı
s = "x\\url{www}x"
@test _collect_command(s, 2) == (10, "\\url{www}", "[`www`](www)")
s = "\\href{a}{b} x"
@test _collect_command(s, 1) == (11, "\\href{a}{b}", "[b](a)")
s = "\\href {a} {b} x"
@test _collect_command(s, 1) == (13, "\\href {a} {b}", "[b](a)")
s = "\\href{a}{\\textit{a}\\%x\\textit{b}}"
@test _collect_command(s, 1) ==
(33, "\\href{a}{\\textit{a}\\%x\\textit{b}}", "[_a_%x_b_](a)")
s = "\\href{a}{\\{x\\}}"
@test _collect_command(s, 1) == (15, "\\href{a}{\\{x\\}}", "[{x}](a)")
#! format: off
s = "\\href{https://en.wikipedia.org/wiki/Schrödinger%27s_cat}{Schrödinger's cat} x"
@test _collect_command(s, 1) == (77, "\\href{https://en.wikipedia.org/wiki/Schrödinger%27s_cat}{Schrödinger's cat}", "[Schrödinger's cat](https://en.wikipedia.org/wiki/Schrödinger%27s_cat)")
#! format: on
s = "\\t{oo}"
@test _collect_command(s, 1) == (6, "\\t{oo}", "o͡o")
s = "\\i x"
@test _collect_command(s, 1) == (3, "\\i ", "ı")
s = "\\d ox"
@test _collect_command(s, 1) == (4, "\\d o", "ọ")
@test_throws BoundsError _collect_command("\\url", 1)
#! format: off
s = "\\href{a} x"
@test_throws ArgumentError("Expected '{' at pos 10 in \"\\\\href{a} x\", not 'x'") _collect_command(s, 1)
#! format: on
end
@testset "collect_accent" begin
# 1234567891123456789212345678931234567894123456789512345678961
s = raw"x\`o \`{o} \'o \'{o} \^o \^{o} \~o \~{o} \=o \={o} \.o \.{o}x"
@test length(s) == 61
@test _collect_accent(s, 2) == (4, "\\`o", "o\u0300")
@test _collect_accent(s, 6) == (10, "\\`{o}", "o\u0300")
@test _collect_accent(s, 12) == (14, "\\'o", "o\u0301")
@test _collect_accent(s, 16) == (20, "\\'{o}", "o\u0301")
@test _collect_accent(s, 22) == (24, "\\^o", "o\u0302")
@test _collect_accent(s, 26) == (30, "\\^{o}", "o\u0302")
@test _collect_accent(s, 32) == (34, "\\~o", "o\u0303")
@test _collect_accent(s, 36) == (40, "\\~{o}", "o\u0303")
@test _collect_accent(s, 42) == (44, "\\=o", "o\u0304")
@test _collect_accent(s, 46) == (50, "\\={o}", "o\u0304")
@test _collect_accent(s, 52) == (54, "\\.o", "o\u0307")
@test _collect_accent(s, 56) == (60, "\\.{o}", "o\u0307")
@test tex_to_markdown(s) ==
"x\uf2 \uf2 \uf3 \uf3 \uf4 \uf4 \uf5 \uf5 \u14d \u14d \u22f \u22fx"
# 1 2 34567 8 91123
s = "x\\\"ox x\\\"{o}x"
@test length(s) == 13
@test _collect_accent(s, 2) == (4, "\\\"o", "o\u0308")
@test _collect_accent(s, 8) == (12, "\\\"{o}", "o\u0308")
@test tex_to_markdown(s) == "x\uf6x x\uf6x"
s = "\\\"\\\\o"
@test_throws ArgumentError _collect_accent(s, 1)
s = "\\\"\\url{x}"
c = IOCapture.capture(rethrow=Union{}) do
_collect_accent(s, 1)
end
@test c.value == ArgumentError("Unsupported accent: \\\"\\url{x}.")
@test contains(
c.output,
"Error: Accents may only be followed by group, a single ASCII letter, or **a supported zero-argument command**."
)
s = "\\\"|"
c = IOCapture.capture(rethrow=Union{}) do
_collect_accent(s, 1)
end
@test c.value == ArgumentError("Unsupported accent: \\\"|.")
@test contains(
c.output,
"Error: Accents may only be followed by group, a single ASCII letter, or a supported zero-argument command."
)
s = "\\\"\\i\\\"{\\i}"
@test _collect_accent(s, 1) == (4, "\\\"\\i", "\u131\u308") # ı̈
@test _collect_accent(s, 5) == (10, "\\\"{\\i}", "\u131\u308") # ı̈
@test tex_to_markdown(s) == "\u131\u308\u131\u308"
end
@testset "collect_math" begin
#! format: off
s = "a \$\\int_{-\\infty}^{\\infty} x^2 dx\$ b"
@test _collect_math(s, 3) == (34, "\$\\int_{-\\infty}^{\\infty} x^2 dx\$", "``\\int_{-\\infty}^{\\infty} x^2 dx``")
#! format: on
@test_throws BoundsError _collect_math("\$x^2", 1)
end
@testset "special characters" begin
@test tex_to_markdown("--- -- ---") == "\u2014 \u2013 \u2014" # "— – —"
@test tex_to_markdown("---~--~---") == "\u2014\u00a0\u2013\u00a0\u2014" # "— – —"
@test tex_to_markdown("1--2") == "1\u20132" # "1–2"
@test tex_to_markdown("1---2") == "1\u20142" # "1—2"
end
@testset "accents" begin
# unlike "collect_accent", this tests accents in the context of a full
# string
#! formatt: off
@test tex_to_markdown(
raw"\`{o}\'{o}\^{o}\~{o}\={o}\u{o}\.{o}\\\"{o}\r{a}\H{o}\v{s}\d{u}\c{c}\k{a}\b{b}\~{a}"
) == "\uf2\uf3\uf4\uf5\u14d\u14f\u22f\uf6\ue5\u151\u161\u1ee5\ue7\u105\u1e07\ue3" # "òóôõōŏȯöåőšụçąḇã"
@test tex_to_markdown(
raw"\`o\'o\^o\~o\=o\u{o}\.o\\\"o\r{a}\H{o}\v{s}\d{u}\c{c}\k{a}\b{b}\~a"
) == "\uf2\uf3\uf4\uf5\u14d\u14f\u22f\uf6\ue5\u151\u161\u1ee5\ue7\u105\u1e07\ue3" # "òóôõōŏȯöåőšụçąḇã"
@test tex_to_markdown(raw"\i{}\o{}\O{}\l{}\L{}\i\o\O\l\L") ==
"\u131\uf8\ud8\u142\u141\u131\uf8\ud8\u142\u141" # "ıøØłŁıøØłŁ"
@test tex_to_markdown(raw"\i \o \O \l \L \i{ }\o{ }\O{ }\l{ }\L") ==
"\u131\uf8\ud8\u142\u141\u131 \uf8 \ud8 \u142 \u141" # "ıøØłŁı ø Ø ł Ł"
@test tex_to_markdown(raw"\SS\ae\oe\AE\OE \AA \aa") == "SS\ue6\u153\uc6\u152\uc5\ue5" # "SSæœÆŒÅå"
@test tex_to_markdown(raw"{\OE}{\AA}{\aa}") == "\u152\uc5\ue5" # "ŒÅå"
@test tex_to_markdown(raw"\t{oo}x\t{az}") == "o\u361oxa\u361z" # "o͡oxa͡z"
@test tex_to_markdown(raw"{\o}verline") == "øverline"
@test tex_to_markdown(raw"\o{}verline") == "øverline"
@test tex_to_markdown(raw"a\t{oo}b") == "ao\u361ob" # "ao͡ob"
@test tex_to_markdown(raw"\i x") == "\u0131x" # "ıx"
@test tex_to_markdown("\\\"\\i x") == "\u131\u308x" # "ı̈x"
@test tex_to_markdown(raw"\i\j \\\"i \\\"j \\\"\i \\\"\j") ==
"\u0131\u0237\u00EF j\u0308 \u0131\u0308\u0237\u0308" # "ıȷï j̈ ı̈ȷ̈"
@test tex_to_markdown(raw"\d ox") == "\u1ecdx" # "ọx"
@test tex_to_markdown(raw"\d{ox}") == "\u1ecdx" # "ọx"
@test tex_to_markdown(raw"\d{o}x") == "\u1ecdx" # "ọx"
#! formatt: on
end
@testset "names" begin
@test tex_to_markdown(raw"Fran\c{c}ois") == "François"
@test tex_to_markdown(raw"Kn\\\"{o}ckel") == "Knöckel"
@test tex_to_markdown(raw"Kn\\\"ockel") == "Knöckel"
@test tex_to_markdown(raw"Ga\\\"{e}tan") == "Gaëtan"
@test tex_to_markdown(raw"Ga{\\\"e}tan") == "Gaëtan"
@test tex_to_markdown(raw"C\^ot\'e") == "Côté"
@test tex_to_markdown(raw"Gro{\ss}") == "Groß"
end
@testset "titles" begin
#! format: off
s = "An {{Introduction}} to {{Optimization}} on {{Smooth Manifolds}}"
@test tex_to_markdown(s) == "An Introduction to Optimization on Smooth Manifolds"
@test tex_to_markdown(s; transform_case=lowercase) == "an Introduction to Optimization on Smooth Manifolds"
s = "Experimental observation of magic-wavelength behavior of \$Rb87\$ atoms in an optical lattice"
@test tex_to_markdown(s) == "Experimental observation of magic-wavelength behavior of ``Rb87`` atoms in an optical lattice"
s = "{Rubidium 87 D Line Data}"
@test tex_to_markdown(s; transform_case=lowercase) == "Rubidium 87 D Line Data"
s = "A Direct Relaxation Method for Calculating Eigenfunctions and Eigenvalues of the Schr{\\\"o}dinger Equation on a Grid"
@test tex_to_markdown(s) == "A Direct Relaxation Method for Calculating Eigenfunctions and Eigenvalues of the Schrödinger Equation on a Grid"
s = "Matter-wave Atomic Gradiometer Interferometric Sensor ({MAGIS-100})"
@test tex_to_markdown(s; transform_case=lowercase) == "matter-wave atomic gradiometer interferometric sensor (MAGIS-100)"
s = "Controlled dissociation of {I\$_2\$} via optical transitions between the {X} and {B} electronic states"
@test tex_to_markdown(s; transform_case=lowercase) == "controlled dissociation of I``_2`` via optical transitions between the X and B electronic states"
s = "\\texttt{DifferentialEquations.jl} -- A Performant and Feature-Rich Ecosystem for Solving Differential Equations in {Julia}"
@test tex_to_markdown(s) == "`DifferentialEquations.jl` – A Performant and Feature-Rich Ecosystem for Solving Differential Equations in Julia"
s = "Machine learning {\\&} artificial intelligence in the quantum domain: a review of recent progress"
@test tex_to_markdown(s) == "Machine learning & artificial intelligence in the quantum domain: a review of recent progress"
s = "Mutually unbiased binary observable sets on \\textit{N} qubits"
@test tex_to_markdown(s) == "Mutually unbiased binary observable sets on _N_ qubits"
#! format: on
end
@testset "incomplete" begin
s = "Text with incomplete \$math..."
c = IOCapture.capture(rethrow=Union{}) do
tex_to_markdown(s)
end
@test c.value isa ArgumentError
@test contains(c.value.msg, "Premature end of tex string")
s = "Text with incomplete {group..."
c = IOCapture.capture(rethrow=Union{}) do
tex_to_markdown(s)
end
@test c.value isa ArgumentError
@test contains(c.value.msg, "Premature end of tex string")
s = "Text with incomplete \\url{cmd..."
c = IOCapture.capture(rethrow=Union{}) do
tex_to_markdown(s)
end
@test c.value isa ArgumentError
@test contains(c.value.msg, "Premature end of tex string")
s = "Text with incomplete \\url" # missing argument
c = IOCapture.capture(rethrow=Union{}) do
tex_to_markdown(s)
end
@test c.value isa ArgumentError
@test contains(c.value.msg, "Premature end of tex string")
s = "Text with incomplete \\href{text}" # missing argument
c = IOCapture.capture(rethrow=Union{}) do
tex_to_markdown(s)
end
@test c.value isa ArgumentError
@test contains(c.value.msg, "Premature end of tex string")
end
@testset "invalid" begin
s = "Text with incomplete \\invalidcommand"
c = IOCapture.capture(rethrow=Union{}) do
tex_to_markdown(s) == ""
end
@test c.value isa ArgumentError
@test contains(c.value.msg, "Unsupported command: \\invalidcommand")
@test contains(c.output, "Supported commands are: ")
s = "Text with unescaped %"
c = IOCapture.capture(rethrow=Union{}) do
tex_to_markdown(s) == ""
end
@test c.value isa ArgumentError
@test c.value.msg ==
"Character '%' at pos 21 in \"Text with unescaped %\" must be escaped"
s = "Text with non-ascii \\commänd{x}"
c = IOCapture.capture(rethrow=Union{}) do
tex_to_markdown(s) == ""
end
@test c.value isa ArgumentError
@test c.value.msg == "Invalid command: \\commänd{x}"
s = "Text with \\command_with_underscore{x}"
# LaTeX does not allow for underscore in commands
c = IOCapture.capture(rethrow=Union{}) do
tex_to_markdown(s) == ""
end
@test c.value isa ArgumentError
@test c.value.msg == "Invalid command: \\command_with_underscore{x}"
s = "\\href{a}{Link text with \\error{unknown command}}"
c = IOCapture.capture(rethrow=Union{}) do
tex_to_markdown(s) == ""
end
@test c.value isa ArgumentError
@test c.value.msg ==
"Cannot evaluate \\href: ArgumentError(\"Unsupported command: \\\\error. Please report a bug.\")"
s = "The krotov Pyhon package is available on [Github](https://github.com/qucontrol/krotov)"
c = IOCapture.capture(rethrow=Union{}) do
tex_to_markdown(s)
end
@test c.value == s
@test contains(
c.output,
"Warning: The tex string \"The krotov Pyhon package is available on [Github](https://github.com/qucontrol/krotov)\" appears to contain a link in markdown syntax"
)
end
@testset "replacement collisions" begin
# An earlier implementation would fail the check below on Julia 1.6,
# because `replace` in Julia 1.6 does not support making multiple
# substitutions at once. We've mitigated this by using a more elaborate
# `_keys` function inside `_process_tex`
s = "{\\{2\\}} {collision} {\\{1\\}}"
@test _process_tex(s) == "{2} collision {1}"
end
@testset "custom command" begin
# This is an undocumented feature (as indicated by the underscores in the
# names) to add support for new commands to `tex_to_markdown`. People are
# encouraged to to submit bug reports for any "legitimate" command that
# occurs in `.bib` files "in the wild". Manually extending functionality in
# the `docs/make.jl` file as below is a fallback for the "less-legitimiate"
# edge cases.
DocumenterCitations._COMMANDS_NUM_ARGS["\\ket"] = 1
DocumenterCitations._COMMANDS_TO_MD["\\ket"] =
str -> begin
md_str = DocumenterCitations._process_tex(str)
return "|$(md_str)⟩"
end
s = "\\ket{Ψ}"
@test tex_to_markdown(s) == "|Ψ⟩"
delete!(DocumenterCitations._COMMANDS_NUM_ARGS, "\\ket")
delete!(DocumenterCitations._COMMANDS_TO_MD, "\\ket")
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | code | 6343 | using DocumenterCitations
using Test
using TestingUtilities: @Test # much better at comparing strings
include("run_makedocs.jl")
@testset "undefined citations" begin
bib = CitationBibliography(
joinpath(@__DIR__, "..", "docs", "src", "refs.bib"),
style=:numeric
)
# non-strict
run_makedocs(
splitext(@__FILE__)[1];
sitename="Test",
warnonly=true,
plugins=[bib],
pages=["Home" => "index.md", "References" => "references.md",],
check_success=true
) do dir, result, success, backtrace, output
@test success
#! format: off
@Test occursin("Error: Key \"NoExist2023\" not found in entries", output)
@Test occursin("Error: Key \"Shapiro2012_\" not found in entries", output)
@Test occursin("Error: Explicit key \"NoExistInBibliography2023\" from bibliography block not found in entries", output)
@Test occursin("Error: expand_citation (rec): No destination for key=\"Tannor2007\" → unlinked text \"7\"", output)
#! format: on
index_html_file = joinpath(dir, "build", "index.html")
@Test isfile(index_html_file)
if isfile(index_html_file)
#! format: off
index_html = read(index_html_file, String)
@Test occursin("and a non-existing key [?]", index_html)
@Test occursin("[?, ?, ?, <a href=\"references/#BrumerShapiro2003\">2</a>–<a href=\"references/#KochEPJQT2022\">6</a>, and references therein]", index_html)
@Test occursin("<a href=\"references/#BrumerShapiro2003\">Brumer and Shapiro [2]</a>, <a href=\"references/#BrifNJP2010\">Brif <em>et al.</em> [3]</a>, [?], [?], <a href=\"references/#SolaAAMOP2018\">Sola <em>et al.</em> [4]</a>, [?], <a href=\"references/#Wilhelm2003.10132\">Wilhelm <em>et al.</em> [5]</a>, <a href=\"references/#KochEPJQT2022\">Koch <em>et al.</em> [6]</a>, and references therein", index_html)
@Test occursin("Lastly, we cite a key [7]", index_html)
#! format: on
end
end
# strict
run_makedocs(
splitext(@__FILE__)[1];
sitename="Test",
warnonly=false,
plugins=[bib],
pages=["Home" => "index.md", "References" => "references.md",],
check_failure=true
) do dir, result, success, backtrace, output
#! format: off
@test !success
@Test occursin("Error: Key \"NoExist2023\" not found in entries", output)
@Test occursin("Error: Key \"Shapiro2012_\" not found in entries", output)
@Test occursin("Error: Explicit key \"NoExistInBibliography2023\" from bibliography block not found in entries", output)
@Test occursin("Error: expand_citation (rec): No destination for key=\"Tannor2007\" → unlinked text \"7\"", output)
@test result isa ErrorException
@Test occursin("`makedocs` encountered errors [:citations, :bibliography_block]", result.msg)
#! format: on
end
end
@testset "undefined citations – :alpha" begin
bib = CitationBibliography(
joinpath(@__DIR__, "..", "docs", "src", "refs.bib"),
style=:alpha
)
# non-strict
run_makedocs(
splitext(@__FILE__)[1];
sitename="Test",
warnonly=true,
plugins=[bib],
pages=["Home" => "index.md", "References" => "references.md",],
check_success=true,
) do dir, result, success, backtrace, output
@test success
index_html_file = joinpath(dir, "build", "index.html")
@Test isfile(index_html_file)
if isfile(index_html_file)
#! format: off
index_html = read(index_html_file, String)
@Test occursin("and a non-existing key [?]", index_html)
@Test occursin("[?], [?], <a href=\"references/#SolaAAMOP2018\">Sola <em>et al.</em> [SCMM18]</a>, [?], <a href=\"references/#Wilhelm2003.10132\">Wilhelm <em>et al.</em> [WKM+20]</a>, <a href=\"references/#KochEPJQT2022\">Koch <em>et al.</em> [KBC+22]</a>, and references therein", index_html)
@Test occursin("<a href=\"references/#BrumerShapiro2003\">Brumer and Shapiro [BS03]</a>, <a href=\"references/#BrifNJP2010\">Brif <em>et al.</em> [BCR10]</a>, [?], [?], <a href=\"references/#SolaAAMOP2018\">Sola <em>et al.</em> [SCMM18]</a>, [?], <a href=\"references/#Wilhelm2003.10132\">Wilhelm <em>et al.</em> [WKM+20]</a>, <a href=\"references/#KochEPJQT2022\">Koch <em>et al.</em> [KBC+22]</a>, and references therein", index_html)
#! format: on
end
end
end
@testset "undefined citations – :authoryear" begin
bib = CitationBibliography(
joinpath(@__DIR__, "..", "docs", "src", "refs.bib"),
style=:authoryear
)
# non-strict
run_makedocs(
splitext(@__FILE__)[1];
sitename="Test",
warnonly=true,
plugins=[bib],
pages=["Home" => "index.md", "References" => "references.md",],
check_success=true,
) do dir, result, success, backtrace, output
@test success
index_html_file = joinpath(dir, "build", "index.html")
@Test isfile(index_html_file)
if isfile(index_html_file)
#! format: off
index_html = read(index_html_file, String)
@Test occursin("and a non-existing key (???)", index_html)
@Test occursin("(<a href=\"references/#BrumerShapiro2003\">Brumer and Shapiro, 2003</a>; <a href=\"references/#BrifNJP2010\">Brif <em>et al.</em>, 2010</a>; ???; ???; <a href=\"references/#SolaAAMOP2018\">Sola <em>et al.</em>, 2018</a>; ???; <a href=\"references/#Wilhelm2003.10132\">Wilhelm <em>et al.</em>, 2020</a>; <a href=\"references/#KochEPJQT2022\">Koch <em>et al.</em>, 2022</a>; and references therein)", index_html)
@Test occursin("<a href=\"references/#BrumerShapiro2003\">Brumer and Shapiro (2003)</a>, <a href=\"references/#BrifNJP2010\">Brif <em>et al.</em> (2010)</a>, ???, ???, <a href=\"references/#SolaAAMOP2018\">Sola <em>et al.</em> (2018)</a>, ???, <a href=\"references/#Wilhelm2003.10132\">Wilhelm <em>et al.</em> (2020)</a>, <a href=\"references/#KochEPJQT2022\">Koch <em>et al.</em> (2022)</a>, and references therein", index_html)
@Test occursin("Lastly, we cite a key (Tannor, 2007)", index_html)
#! format: on
end
end
end
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 17733 | # Release notes
The format is based on [Keep a Changelog](https://keepachangelog.com/en/1.0.0/),
and this project adheres to [Semantic Versioning](https://semver.org/spec/v2.0.0.html).
## [Version 1.3.4][1.3.4] - 2024-09-19
### Internal Changes
* Added an `article_link_doi_in_title` option to the internal `format_published_in` and `format_labeled_bibliography_reference` functions. This allows custom styles to change how links appear in bibliography entries for articles. By setting the option to `true`, the title of the article,instead of the "published in" information, will be used as the link text for a DOI . This makes the bibliography for articles more consistent with other types or entries, but is recommended only if no entries have both a DOI and a URL. [[#73][], [#74][]]
## [Version 1.3.3][1.3.3] - 2024-03-08
### Fixed
* The recommended CSS (`citations.css`) now includes a fix to be compatible with the dark-mode CSS of Documenter. Existing pages should update their `citations.css` to add `!important` to the `list-style` of `.citation ul`. [[#70][]]
## [Version 1.3.2][1.3.2] - 2023-11-29
### Fixed
* Warn about markdown link syntax in `.bib` files [[#60][]]
* Warn about invalid DOIs in `.bib` files. The DOI field should never contain a URL (`https://doi.org/...`). However, such usage is detected as a special case, and the DOI is automatically extracted from the URL.
* Automatically link both URL and DOI fields. This fixes a regression in `v1.3.0`, which would throw an error for `@book` and `@proceeding` entries with both a URL and a DOI field. Now, the URL in such a case will be automatically linked via the `Title` field, and the DOI via the `organization`/`publisher`/`address` fields, similar to the behavior in `v1.2.0`. You may prefer to have the DOI linked via that `Title`, in which case you should add a `Note` field containing the `URL` (using `\url`/`\href`, as appropriate). [[#65][]]
* Calling `makedocs` with `linkcheck=true` now also checks links (e.g., DOIs) inside the bibliography. Note that the correct behavior requires [Documenter 1.2](https://github.com/JuliaDocs/Documenter.jl/releases/tag/v1.2.0). With older versions of Documenter, broken links in the bibliography will be silently ignored. [[#58][], [#62][], [Documenter#2329](https://github.com/JuliaDocs/Documenter.jl/issues/2329), [Documenter#2330](https://github.com/JuliaDocs/Documenter.jl/pull/2330)]
## [Version 1.3.1][1.3.1] - 2023-11-02
### Fixed
* Added a fallback for the `Pages` attribute in a `@bibliography` block to behave as in pre-`1.3.0`: If `Pages` references a file with a path relative to the `docs/src` directory (which was the unintentional requirement pre-`1.3.0`), this now works again, but shows a warning that the name should be updated to be relative to the folder containing the file that contains the `@bibliography` block. [[#61][]]
This fixes the `v1.3.0` release arguably having been "breaking" [[#59][]] in that anybody who was using `Pages` pre-1.3.0 would have had to use paths relative to `docs/src`, even though that was a workaround for a known bug [[#22][]]. Note that whenever `Pages` references the current file, `@__FILE__` should be used.
## [Version 1.3.0][1.3.0] - 2023-11-01
### Fixed
* Skip the expansion of citations and bibliographies when running in doctest mode [[#34][]]
* Support underscores in citation keys [[#14][]]
* The `Pages` in a `@bibliography` block are now relative to the folder containing the current file. The behavior is consistent with `Pages` in Documenter's `@index` and `@contents` blocks. [[#22][]]
* The parsing of LaTeX strings has improved significantly. In particular, curly braces should now be stripped correctly [[#15][]]. Note that that braces in titles are never needed for `DocumenterCitations`, but handling them correctly makes it easier to use the same `.bib` file for LaTeX and `DocumenterCitations`.
* Fixed the rendering of references other than `@article`, especially `@inproceedings`, `@incollection`, `@inbooks`, mimicking RevTeX. The DOI/URL are now linked via the Title and/or Booktitle. Added support for `Chapter`, `Volume`, `Number`, `Edition`, `Month` fields. [[#56][]]
### Added
* Allow multiple citations in a single `@cite` link. In the default numeric style, these can be compressed, e.g. "Refs. [1–3]" [[#6][]]
* In general (depending on the style and citation syntax), citation links may now render to arbitrarily complex expressions.
* Citation comments can now have inline markdown elements, e.g., `[GoerzQ2022; definition of $J$ in section *Running costs*](@cite)`
* When running in non-strict mode, missing bibliographic references (either because the `.bib` file does not contain an entry with a specific BibTeX key, or because of a missing `@biblography` block) are now handled similarly to missing references in LaTeX: They will show as (unlinked) question marks.
* Support for bibliographies in PDFs generate via LaTeX (`format=Documenter.LaTeX()`). Citations and references are rendered exactly as in the HTML version. Specifically, the support does not depend on `bibtex`/`biblatex` and supports any style (including custom styles). [[#18][]]
* Functions `DocumenterCitations.set_latex_options` and `DocumenterCitations.reset_latex_options` to tweak the rendering of bibliographies in PDFs.
* The `Pages` in a `@bibliography` block can now use `@__FILE__` to refer to the current file. [[#22][]]
* You may now use `\url` and `\href` commands in the `@misc` field of an entry.
* The `Urldate` field is now supported for citing websites. [[#53][]]
### Internal Changes
* Removed the redundant `CitationLink.link_text` field.
* Added `read_citation_link` replacing the former `CitationLink` constructor.
* `CitationLink` can now be instantiated directly from markdown strings (for documentation / testing purposes)
* Added `DirectCitationLink` type to represent citations of the form `[text](@cite key)`.
* Exposed `CitationLink` to users who want to implement a custom style (see changes in `format_citation`)
* The interface for the `format_citation` function has changed: Before, the signature was `format_citation(style, entry, citations; note, cite_cmd, capitalize, starred)` and the function would return as string that would replace the link text of the citation link. Now, the signature is `format_citation(style, cit, entries, citations)` where `cit` is a `CitationLink` object, and the function returns a string of markdown code that replaces the *entire* citation link (not just the link text). Generally, the returned markdown code is expected to contain *direct* citation links which, are automatically expanded subsequently. That is, `format_citation` now generally converts indirect citation links (`CitationLink`) into direct citation links (`DirectCitationLink`).
* Exposed the internal function `format_labeled_citation` that implements `format_citation` for the built-in styles `:numeric` and `:alpha` and may be useful for custom styles that are variations of these.
* Exposed the internal function `format_authoryear_citation` that implements `format_citation` for the built-in style `:authoryear`
* Exposed the internal function `format_labeled_bibliography_reference` that implements `format_bibliography_reference` for the built-in styles `:numeric` and `:alpha`.
* Exposed the internal function `format_authoryear_bibliography_reference` that implements `format_bibliography_reference` for the built-in style `:authoryear:`.
* The example custom styles `:enumauthoryear` and `:keylabels` have been rewritten using the above internal functions, illustrating that custom styles will usually not have to rely on the undocumented and even more internal functions like `format_names` and `tex2unicode`.
* Any `@bibliography` block is now internally expanded into an internal `BibliographyNode` instead of a raw HTML node. This `BibliographyNode` can then be translated into the desired output format by `Documenter.HTMLWriter` or `Documenter.LaTeXWriter`. This is how support for bibliographies with `format=Documenter.LaTeX()` can be achieved.
* The routine `format_bibliography_reference` must now return a markdown string instead of an HTML string.
**Upgrade guidelines**:
For anyone who was using custom styles, which rely on the [Internals](https://juliadocs.org/DocumenterCitations.jl/stable/internals/) of `DocumenterCitations`, this release will almost certainly break the customization. See the above list of internal changes.
There were several bugs and limitations in version `1.2.x` for which some existing documentations may have been using workarounds. These workarounds may cause some breakage in the new version `1.3.0`. In particular:
* The `Pages` attribute in a `@bibliography` block in version `1.2.x` required any names to be relative to the `docs/src` directory [[#22][]]. This was both unintentional and undocumented. These names must now be updated to be relative to to the folder containing the file which contains the `@bibliography` block. This is consistent with how `Pages` is used, e.g., in `@contents` or `@index` blocks. For the common usage where `Pages` was referring to the current file, `@__FILE__` should be used.
* Pre-`1.3.0`, strings in entries in the `.bib` file were extremely limited. There was no official support for any kind of `tex` macros: only plain-text (unicode) was fully supported. As a workaround, some users exploited an (undocumented/buggy) implementation detail that would cause html or markdown strings inside the `.bib` file to "work", e.g. for adding links in a `note` field. These workarounds may break in `v1.3.0`. While unicode is still very much supported (`ö` over `\"{o}`), `.bib` files should otherwise be written to be fully compatible with `bibtex`. For links in particular, the LaTeX `\href` macro should be used. Any `tex` commands that are not supported (`Error: Unsupported command`) should be reported. Some `tex` characters (`$%@{}&`) that may have worked directly pre-`1.3.0` will have to be escaped in version `1.3.0`.
## [Version 1.2.1][1.2.1] - 2023-09-22
### Fixed
* Collect citations that only occur in docstrings [[#39][], [#40][]]
* It is now possible to have a page that contains a `@bibliography` block listed in [`@contents`](https://documenter.juliadocs.org/stable/man/syntax/index.html#@contents-block) [[#16][], [#42][]].
## [Version 1.2.0][1.2.0] - 2023-09-16
### Version changes
* Update to [Documenter 1.0](https://github.com/JuliaDocs/Documenter.jl/releases/tag/v1.0.0). The most notable user-facing breaking change in Documenter 1.0 affecting DocumenterCitations is that the `CitationBibliography` plugin object now has to be passed to `makedocs` as an element of the `plugins` keyword argument, instead of as a positional argument.
### Fixed
* The plugin no longer conflicts with the `linkcheck` option of `makedocs` [[#19][]]
## [Version 1.1.0][1.1.0] - 2023-09-15
### Fixed
* Avoid duplicate labels in `:alpha` style. This is implemented via the new stateful `AlphaStyle()`, but is handled automatically with (`style=:alpha`) [[#31][]]
* With the alphabetic style (`:alpha`/`AlphaStyle`), include up to 4 names in the label, not 3 (but 5 or more names results in 3 names and "+"). Also, include the first letter of a "particle" in the label, e.g. "vWB08" for a first author "von Winckel". Both of these are consistent with LaTeX's behavior.
* Handle missing author/year, especially for `:authoryar` and `:alpha` styles. You end up with `:alpha` labels like `Anon04` (missing authors) or `CW??` (missing year), and `:authoryear` citations like "(Anonymous, 2004)" and "(Corcovilos and Weiss, undated)".
* Consistent punctuation in the rendered bibliography, including for cases of missing fields.
### Added
* New `style=AlphaStyle()` that generates unique citation labels. This can mostly be considered internal, as `style=:alpha` is automatically upgraded to `style=AlphaStyle()`.
* Support for `eprint` field. It is recommended to add the arXiv ID in the `eprint` field for any article whose DOI is behind a paywall [[#32][]]
* Support for non-arXiv preprint servers BiorXiv and HAL [[#35][], [#36][]]
* Support for `note` field. [[#20][]]
### Changed
* In the rendered bibliography, the BibTeX "URL" field is now linked via the title, while the "DOI" is linked via the journal information. This allows to have a DOI and URL at the same time, or a URL for an `@unpublished`/`@misc` citation. If there is a URL but no title, the URL is used as the title.
### Internal Changes
* Added an internal function `init_bibliography!` that is called at the beginning of the `ExpandBibliography` pipeline step. This function is intended to initialize internal state either of the `style` object or the `CitationBibliography` plugin object before rendering any `@bibliography` blocks. This is used to generate unique citation labels for the new `AlphaStyle()`. For the other builtin styles, it is a no-op. Generally, `init_bibliography!` can help with implementing custom "stateful" styles.
## [Version 1.0.0][1.0.0] - 2023-07-12
### Version changes
* The minimum supported Julia version has been raised from 1.4 to 1.6.
### Breaking
* The default citation style has changed to `:numeric`. To restore the author-year style used pre-1.0, instantiate `CitationBibliography` with the option `style=:authoryear` in `docs/make.jl` before passing it to `makedocs`.
* Only cited references are included in the main bibliography by default, as opposed to all references defined in the underlying `.bib` file.
### Added
* A `style` keyword argument for `CitationBibliography`. The default style is `style=:numeric`. Other built-in styles are `style=:authoryear` (corresponding to the pre-1.0 style) and `style=:alpha`.
* It is now possible to implement [custom citation styles](https://juliadocs.org/DocumenterCitations.jl/dev/gallery/#custom_styles).
* The `@bibligraphy` block can now have additional options to customize which references are included, see [Syntax for the Bibliography Block](https://juliadocs.org/DocumenterCitations.jl/dev/syntax/#Syntax-for-the-Bibliography-Block).
* It is possible to generate [secondary bibliographies](https://juliadocs.org/DocumenterCitations.jl/dev/syntax/#noncanonical), e.g., for a specific page.
* There is [new syntax](https://juliadocs.org/DocumenterCitations.jl/dev/syntax/#Syntax-for-Citations) to create links to bibliographic references with arbitrary text.
* The following variations of the `@cite` command are now supported: `@citet`, `@citep`, `@cite*`, `@citet*`, `@citep*`, `@Citet`, `@Citep`, `@Cite*`, `@Citet*`, `@Citep*`. See the [syntax for citations](https://juliadocs.org/DocumenterCitations.jl/dev/syntax/#Syntax-for-Citations) for details.
* Citations can now include notes, e.g., `See Ref. [GoerzQ2022; Eq. (1)](@cite)`.
### Other
* [DocumenterCitations](https://github.com/JuliaDocs/DocumenterCitations.jl) is now hosted under the [JuliaDocs](https://github.com/JuliaDocs) organization.
[Unreleased]: https://github.com/JuliaDocs/DocumenterCitations.jl/compare/v1.3.4...HEAD
[1.3.4]: https://github.com/JuliaDocs/DocumenterCitations.jl/compare/v1.3.3...v1.3.4
[1.3.3]: https://github.com/JuliaDocs/DocumenterCitations.jl/compare/v1.3.2...v1.3.3
[1.3.2]: https://github.com/JuliaDocs/DocumenterCitations.jl/compare/v1.3.1...v1.3.2
[1.3.1]: https://github.com/JuliaDocs/DocumenterCitations.jl/compare/v1.3.0...v1.3.1
[1.3.0]: https://github.com/JuliaDocs/DocumenterCitations.jl/compare/v1.2.1...v1.3.0
[1.2.1]: https://github.com/JuliaDocs/DocumenterCitations.jl/compare/v1.2.0...v1.2.1
[1.2.0]: https://github.com/JuliaDocs/DocumenterCitations.jl/compare/v1.1.0...v1.2.0
[1.1.0]: https://github.com/JuliaDocs/DocumenterCitations.jl/compare/v1.0.0...v1.1.0
[1.0.0]: https://github.com/JuliaDocs/DocumenterCitations.jl/compare/v0.2.12...v1.0.0
[#74]: https://github.com/JuliaDocs/DocumenterCitations.jl/pull/74
[#73]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/73
[#70]: https://github.com/JuliaDocs/DocumenterCitations.jl/pull/70
[#65]: https://github.com/JuliaDocs/DocumenterCitations.jl/pull/65
[#62]: https://github.com/JuliaDocs/DocumenterCitations.jl/pull/62
[#61]: https://github.com/JuliaDocs/DocumenterCitations.jl/pull/61
[#60]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/60
[#59]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/59
[#58]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/58
[#56]: https://github.com/JuliaDocs/DocumenterCitations.jl/pull/56
[#53]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/53
[#42]: https://github.com/JuliaDocs/DocumenterCitations.jl/pull/42
[#40]: https://github.com/JuliaDocs/DocumenterCitations.jl/pull/40
[#39]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/39
[#36]: https://github.com/JuliaDocs/DocumenterCitations.jl/pull/36
[#35]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/35
[#34]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/34
[#32]: https://github.com/JuliaDocs/DocumenterCitations.jl/pull/32
[#31]: https://github.com/JuliaDocs/DocumenterCitations.jl/pull/31
[#22]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/22
[#20]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/20
[#19]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/19
[#18]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/18
[#16]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/16
[#15]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/15
[#14]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/14
[#6]: https://github.com/JuliaDocs/DocumenterCitations.jl/issues/6
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 4375 | # DocumenterCitations.jl
[](https://juliahub.com/ui/Packages/DocumenterCitations/B0owD)
[](https://juliadocs.github.io/DocumenterCitations.jl/)
[](https://juliadocs.github.io/DocumenterCitations.jl/dev)
[](https://github.com/JuliaDocs/DocumenterCitations.jl/actions)
[](https://juliaci.github.io/NanosoldierReports/pkgeval_badges/report.html)
[](https://codecov.io/gh/JuliaDocs/DocumenterCitations.jl)
[DocumenterCitations.jl](https://github.com/JuliaDocs/DocumenterCitations.jl#readme) uses [Bibliography.jl](https://github.com/Humans-of-Julia/Bibliography.jl) to add support for BibTeX citations in documentation pages generated by [Documenter.jl](https://github.com/JuliaDocs/Documenter.jl).
By default, [DocumenterCitations.jl](https://github.com/JuliaDocs/DocumenterCitations.jl#readme) uses a numeric citation style common in the natural sciences, see e.g. the [journals of the American Physical Society](https://journals.aps.org), and the [REVTeX author's guide](https://www.ctan.org/tex-archive/macros/latex/contrib/revtex/auguide). Citations are shown in-line, as a number enclosed in square brackets, e.g., "Optimal control is a cornerstone in the development of quantum technologies [[1](#screenshot)]."
<img id="screenshot" src="docs/src/assets/references.png" alt="Rendered bibliography of two references, [1] and [2]" width="830px">
Alternatively, author-year and alphabetic citations styles are available, see the [Citation Style Gallery](https://juliadocs.github.io/DocumenterCitations.jl/dev/gallery/). Prior to version 1.0, the author-year style was the default, see [NEWS.md](NEWS.md). It is possible to define custom styles.
## Installation
The `DocumenterCitations` package can be installed with [Pkg](https://pkgdocs.julialang.org/v1/) as
~~~
pkg> add DocumenterCitations
~~~
In most cases, you will just want to have `DocumenterCitations` in the project that builds your documentation (e.g. [`test/Project.toml`](https://github.com/JuliaDocs/DocumenterCitations.jl/blob/master/test/Project.toml)). Thus, you can also simply add
```
DocumenterCitations = "daee34ce-89f3-4625-b898-19384cb65244"
```
to the `[deps]` section of the relevant `Project.toml` file.
## Usage
* Place a BibTeX [`refs.bib`](https://github.com/JuliaDocs/DocumenterCitations.jl/blob/master/docs/src/refs.bib) file in the `docs/src` folder of your project. Then, in [`docs/make.jl`](https://github.com/JuliaDocs/DocumenterCitations.jl/blob/master/docs/make.jl), instantiate the `CitationBibliography` plugin with the path to the `.bib` file. Assuming `Documenter >= 1.0`, pass the plugin object to [`makedocs`](https://documenter.juliadocs.org/stable/lib/public/#Documenter.makedocs) as element of the `plugins` keyword argument:
```julia
using DocumenterCitations
bib = CitationBibliography(joinpath(@__DIR__, "src", "refs.bib"))
makedocs(; plugins=[bib], ...)
```
In older versions of [Documenter.jl](https://github.com/JuliaDocs/Documenter.jl), `bib` had to be passed as a positional argument to `makedocs`.
* Optional, but recommended: [add CSS to properly format the bibliography](https://juliadocs.github.io/DocumenterCitations.jl/dev/styling/)
* Somewhere in your documentation include a markdown block
~~~markdown
```@bibliography
```
~~~
that will expand into a bibliography for all citations in the documentation.
* Anywhere in the documentation or in docstrings, insert citations as, e.g., `[GoerzQ2022](@cite)`, which will be rendered as "[[2](#screenshot)]" and link to the full reference in the bibliography.
See the [documentation](https://juliadocs.github.io/DocumenterCitations.jl) for additional information.
## Documentation
The documentation of `DocumenterCitations.jl` is available at <https://juliadocs.github.io/DocumenterCitations.jl>. In addition to documenting the usage of the package, it also serves as its showcase.
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 9825 | # [Citation Style Gallery](@id gallery)
The citation style is determined when instantiating the [`CitationBibliography`](@ref), via the `style` argument.
The built-in styles are:
* `style=:numeric` (default): [numeric style](@ref numeric_style)
* `style=:authoryear`: [author-year style](@ref author_year_style)
* `style=:alpha`: [alphabetic style](@ref alphabetic_style)
## [Numeric style](@id numeric_style)
This is the default style (`style=:numeric`) used throughout the other pages of this documentation, cf. the [Syntax](@ref) examples.
* `[GoerzQ2022](@cite)` renders as "[GoerzQ2022](@cite)"
* `[FuerstNJP2014,SolaAAMOP2018](@cite)` renders as "[FuerstNJP2014,SolaAAMOP2018](@cite)"
* `[GoerzQ2022](@citet)` renders as "[GoerzQ2022](@citet)"
* `[GoerzQ2022](@citep)` renders as "[GoerzQ2022](@citep)" — `@citep` is the same as `@cite` for this style
* `[GoerzQ2022; Eq. (1)](@cite)` renders as "[GoerzQ2022; Eq. (1)](@cite)"
* `[GoerzQ2022; Eq. (1)](@citet)` renders as "[GoerzQ2022; Eq. (1)](@citet)"
* `[GoerzQ2022](@citet*)` renders as "[GoerzQ2022](@citet*)"
* `[GoerzQ2022; Eq. (1)](@citet*)` renders as "[GoerzQ2022; Eq. (1)](@citet*)"
* `[WinckelIP2008](@citet)` renders as "[WinckelIP2008](@citet)"
* `[WinckelIP2008](@Citet)` renders as "[WinckelIP2008](@Citet)"
* `[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@cite)` renders as "[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@cite)"
* `[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@Citet)` renders as "[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@Citet)"
* `[arbitrary text](@cite GoerzQ2022)` renders as "[arbitrary text](@cite GoerzQ2022)"
**References:**
```@bibliography
Pages = [@__FILE__]
Style = :numeric
Canonical = false
```
## [Author-year style](@id author_year_style)
The author-year style (`style=:authoryear`) formats citations with the author name and publication year. This is the citation style used, e.g., in [Rev. Mod. Phys.](https://journals.aps.org/rmp/) (`rmp` option in [REVTeX](https://www.ctan.org/tex-archive/macros/latex/contrib/revtex/auguide)). The bibliography is sorted alphabetically by author name. The default `@cite` command is parenthetical (`@cite` and `@citep` are equivalent) which is different from the `authoryear` style in [natbib](https://mirrors.rit.edu/CTAN/macros/latex/contrib/natbib/natnotes.pdf).
* `[GoerzQ2022](@cite)` renders as "[GoerzQ2022](@cite%authoryear%)"
* `[FuerstNJP2014,SolaAAMOP2018](@cite)` renders as "[FuerstNJP2014,SolaAAMOP2018](@cite%authoryear%)"
* `[GoerzQ2022](@citet)` renders as "[GoerzQ2022](@citet%authoryear%)"
* `[GoerzQ2022](@citep)` renders as "[GoerzQ2022](@citep%authoryear%)" — `@citep` is the same as `@cite` for this style
* `[GoerzQ2022; Eq. (1)](@cite)` renders as "[GoerzQ2022; Eq. (1)](@cite%authoryear%)"
* `[GoerzQ2022; Eq. (1)](@citet)` renders as "[GoerzQ2022; Eq. (1)](@citet%authoryear%)"
* `[GoerzQ2022](@cite*)` renders as "[GoerzQ2022](@cite*%authoryear%)"
* `[GoerzQ2022](@citet*)` renders as "[GoerzQ2022](@citet*%authoryear%)"
* `[GoerzQ2022; Eq. (1)](@cite*)` renders as "[GoerzQ2022; Eq. (1)](@cite*%authoryear%)"
* `[GoerzQ2022; Eq. (1)](@citet*)` renders as "[GoerzQ2022; Eq. (1)](@citet*%authoryear%)"
* `[WinckelIP2008](@citet)` renders as "[WinckelIP2008](@citet%authoryear%)"
* `[WinckelIP2008](@Citet)` renders as "[WinckelIP2008](@Citet%authoryear%)"
* `[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@cite)` renders as "[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@cite%authoryear%)"
* `[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@Citet)` renders as "[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@Citet%authoryear%)"
* `[arbitrary text](@cite GoerzQ2022)` renders as "[arbitrary text](@cite GoerzQ2022)"
**References:**
```@bibliography
Pages = [@__FILE__]
Style = :authoryear
Canonical = false
```
## [Alphabetic style](@id alphabetic_style)
The `style=:alpha` formats citations and references like `:numeric`, except that it uses labels derived from the author names and publication year and sorts the references alphabetically.
* `[GoerzQ2022](@cite)` renders as "[GoerzQ2022](@cite%alpha%)"
* `[FuerstNJP2014,SolaAAMOP2018](@cite)` renders as "[FuerstNJP2014,SolaAAMOP2018](@cite%alpha%)"
* `[GoerzQ2022](@citet)` renders as "[GoerzQ2022](@citet%alpha%)"
* `[GoerzQ2022](@citep)` renders as "[GoerzQ2022](@citep%alpha%)" — `@citep` is the same as `@cite` for this style
* `[GoerzQ2022; Eq. (1)](@cite)` renders as "[GoerzQ2022; Eq. (1)](@cite%alpha%)"
* `[GoerzQ2022; Eq. (1)](@citet)` renders as "[GoerzQ2022; Eq. (1)](@citet%alpha%)"
* `[GoerzQ2022](@citet*)` renders as "[GoerzQ2022](@citet*%alpha%)"
* `[GoerzQ2022; Eq. (1)](@citet*)` renders as "[GoerzQ2022; Eq. (1)](@citet*%alpha%)"
* `[WinckelIP2008](@citet)` renders as "[WinckelIP2008](@citet%alpha%)"
* `[WinckelIP2008](@Citet)` renders as "[WinckelIP2008](@Citet%alpha%)"
* `[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@cite)` renders as "[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@cite%alpha%)". Note that unlike for `style=:numeric`, the citations are not compressed.
* `[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@Citet)` renders as "[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@Citet%alpha%)"
* `[arbitrary text](@cite GoerzQ2022)` renders as "[arbitrary text](@cite GoerzQ2022)"
**References:**
```@bibliography
Pages = [@__FILE__]
Style = :alpha
Canonical = false
SolaAAMOP2018
```
```@raw latex
Compared to the HTML version of the documentation, the hanging indent in the above list of references is too small for the longer labels of the \texttt{:alpha} style. This can be remedied by adjusting the \texttt{dl\_hangindent} and \texttt{dl\_labelwidth} parameters with \hyperlinkref{sec:customizing_latex_output}{\texttt{DocumenterCitations.set\_latex\_options}}.
```
Note that the `:alpha` style is able to automatically disambiguate labels:
```@bibliography
Pages = []
Style = :alpha
Canonical = false
GraceJMO2007
GraceJPB2007
```
This works because the `DocumenterCitations` plugin automatically upgrades `style=:alpha` to the internal
```@docs
DocumenterCitations.AlphaStyle
```
## [Custom styles](@id custom_styles)
In the following, we show two examples for user-defined styles. See the [notes on customization](@ref customization) on how to generally define a custom style.
### Custom style: enumerated author-year
In this example, the `:authoryear` style is used, but the references are shown in an enumerated list.
~~~@eval
# custom styles are included in docs/make.jl, which is how we get around
# world-age issues.
using Markdown
custom_style = joinpath(@__DIR__, "..", "custom_styles", "enumauthoryear.jl")
if isfile(custom_style)
Markdown.parse("""
```julia
$(read(custom_style, String))
```
""")
end
~~~
The important part of the definition is in the last line, indicating that the References should be shown as an enumeration (ordered list, `<ol>`, in HTML), see below. Meanwhile, citations render exactly as with `style=:authoryear`:
* `[GoerzQ2022](@cite)` renders as "[GoerzQ2022](@cite%enumauthoryear%)"
* `[FuerstNJP2014,SolaAAMOP2018](@cite)` renders as "[FuerstNJP2014,SolaAAMOP2018](@cite%enumauthoryear%)"
* `[WinckelIP2008](@Citet)` renders as "[WinckelIP2008](@Citet%enumauthoryear%)"
**References:**
```@bibliography
Pages = [@__FILE__]
Style = :enumauthoryear
Canonical = false
```
### Custom style: Citation-key labels
In this less trivial example, a style similar to `:alpha` is used, using the citation keys in the `.bib` file as labels. This would be somewhat more appropriate with citation keys that are shorter that the ones used here (keys similar to those automatically generated with the `:alpha` style).
~~~@eval
# custom styles are included in docs/make.jl, which is how we get around
# world-age issues.
using Markdown
custom_style = joinpath(@__DIR__, "..", "custom_styles", "keylabels.jl")
if isfile(custom_style)
Markdown.parse("""
```julia
$(read(custom_style, String))
```
""")
end
~~~
* `[GoerzQ2022](@cite)` renders as "[GoerzQ2022](@cite%keylabels%)"
* `[FuerstNJP2014,SolaAAMOP2018](@cite)` renders as "[FuerstNJP2014,SolaAAMOP2018](@cite%keylabels%)"
* `[GoerzQ2022](@citet)` renders as "[GoerzQ2022](@citet%keylabels%)"
* `[GoerzQ2022](@citep)` renders as "[GoerzQ2022](@citep%keylabels%)" — `@citep` is the same as `@cite` for this style
* `[GoerzQ2022; Eq. (1)](@cite)` renders as "[GoerzQ2022; Eq. (1)](@cite%keylabels%)"
* `[GoerzQ2022; Eq. (1)](@citet)` renders as "[GoerzQ2022; Eq. (1)](@citet%keylabels%)"
* `[GoerzQ2022](@citet*)` renders as "[GoerzQ2022](@citet*%keylabels%)"
* `[GoerzQ2022; Eq. (1)](@citet*)` renders as "[GoerzQ2022; Eq. (1)](@citet*%keylabels%)"
* `[WinckelIP2008](@citet)` renders as "[WinckelIP2008](@citet%keylabels%)"
* `[WinckelIP2008](@Citet)` renders as "[WinckelIP2008](@Citet%keylabels%)"
* `[arbitrary text](@cite GoerzQ2022)` renders as "[arbitrary text](@cite GoerzQ2022)"
**References:**
```@bibliography
Pages = [@__FILE__]
Style = :keylabels
Canonical = false
```
```@raw latex
As with the \texttt{:alpha} style, for \LaTeX{} output, the \texttt{dl\_hangindent} and \texttt{dl\_labelwidth} parameters should be adjusted with \hyperlinkref{sec:customizing_latex_output}{\texttt{DocumenterCitations.set\_latex\_options}} to obtain a more suitable hanging indent that matches the HTML version of this documentation.
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 7417 | # DocumenterCitations.jl
```@eval
using Markdown
using Pkg
VERSION = Pkg.dependencies()[Base.UUID("daee34ce-89f3-4625-b898-19384cb65244")].version
github_badge = "[](https://github.com/JuliaDocs/DocumenterCitations.jl)"
version_badge = ")-green.svg)"
if get(ENV, "DOCUMENTER_BUILD_PDF", "") == ""
Markdown.parse("$github_badge $version_badge")
else
Markdown.parse("""
-----
On Github: [JuliaDocs/DocumenterCitations.jl](https://github.com/JuliaDocs/DocumenterCitations.jl)
Version: $VERSION
-----
""")
end
```
[DocumenterCitations.jl](https://github.com/JuliaDocs/DocumenterCitations.jl#readme) uses [Bibliography.jl](https://github.com/Humans-of-Julia/Bibliography.jl) to add support for BibTeX citations in documentation pages generated by [Documenter.jl](https://github.com/JuliaDocs/Documenter.jl).
By default, [DocumenterCitations.jl](https://github.com/JuliaDocs/DocumenterCitations.jl#readme) uses a numeric citation style common in the natural sciences, see e.g. the [journals of the American Physical Society](https://journals.aps.org), and the [REVTeX author's guide](https://www.ctan.org/tex-archive/macros/latex/contrib/revtex/auguide). Citations are shown in-line, as a number enclosed in square brackets, e.g., "Optimal control is a cornerstone in the development of quantum technologies [BrifNJP2010](@cite)". [Alternative styles](@ref gallery) are supported, including an [author-year style](@ref author_year_style).
## Installation instructions
You can install the latest version of [DocumenterCitations.jl](https://github.com/JuliaDocs/DocumenterCitations.jl) using the [built-in package manager](https://docs.julialang.org/en/v1/stdlib/Pkg/)
```julia
pkg> add DocumenterCitations
```
In most cases, you will just want to have `DocumenterCitations` in the project that builds your documentation (e.g. [`test/Project.toml`](https://github.com/JuliaDocs/DocumenterCitations.jl/blob/master/test/Project.toml)). Thus, you can also simply add
```
DocumenterCitations = "daee34ce-89f3-4625-b898-19384cb65244"
```
to the `[deps]` section of the relevant `Project.toml` file.
## Telling Documenter.jl about your bibliography
First, place a BibTeX [`refs.bib`](./refs.bib) file in the `docs/src` folder of your project. Then, in [`docs/make.jl`](https://github.com/JuliaDocs/DocumenterCitations.jl/blob/master/docs/make.jl), instantiate the [`CitationBibliography`](@ref) plugin with the path to the `.bib` file. Assuming `Documenter >= 1.0`, pass the plugin object to [`makedocs`](https://documenter.juliadocs.org/stable/lib/public/#Documenter.makedocs) as an element of the `plugins` keyword argument:
```julia
using DocumenterCitations
bib = CitationBibliography(
joinpath(@__DIR__, "src", "refs.bib");
style=:numeric
)
makedocs(; plugins=[bib], ...)
```
In older versions of [Documenter.jl](https://github.com/JuliaDocs/Documenter.jl), `bib` had to be passed as a positional argument to `makedocs`.
To use the [author-year style](@ref author_year_style) that was the default prior to `DocumenterCitations` version 1.0, replace `style=:numeric` with `style=:authoryear`.
Somewhere in your documentation, like a [References](@ref) page, include a markdown block
~~~markdown
# References
```@bibliography
```
~~~
to insert the bibliography for all cited references in the project. See [Syntax for the Bibliography Block](@ref) for more options.
You will also want to add custom [CSS Styling](@ref) to your documentation to improve the rendering of your bibliography.
Thus, a [full `docs/make.jl` file](https://github.com/JuliaDocs/DocumenterCitations.jl/blob/master/docs/make.jl) might look something like this:
```julia
using DocumenterCitations
using Documenter
using Pkg
PROJECT_TOML = Pkg.TOML.parsefile(joinpath(@__DIR__, "..", "Project.toml"))
VERSION = PROJECT_TOML["version"]
NAME = PROJECT_TOML["name"]
AUTHORS = join(PROJECT_TOML["authors"], ", ") * " and contributors"
GITHUB = "https://github.com/JuliaDocs/DocumenterCitations.jl"
bib = CitationBibliography(
joinpath(@__DIR__, "src", "refs.bib"),
style=:numeric # default
)
makedocs(;
authors=AUTHORS,
sitename="DocumenterCitations.jl",
format=Documenter.HTML(
prettyurls=true,
canonical="https://juliaquantumcontrol.github.io/DocumenterCitations.jl",
assets=String["assets/citations.css"],
footer="[$NAME.jl]($GITHUB) v$VERSION docs powered by [Documenter.jl](https://github.com/JuliaDocs/Documenter.jl)."
),
pages=[
"Home" => "index.md",
"Syntax" => "syntax.md",
"Citation Style Gallery" => "gallery.md",
"CSS Styling" => "styling.md",
"Internals" => "internals.md",
"References" => "references.md",
],
plugins=[bib],
)
deploydocs(; repo="github.com/JuliaDocs/DocumenterCitations.jl.git")
```
Bibliographies are also supported in [PDFs generated via LaTeX](https://documenter.juliadocs.org/stable/man/other-formats/#pdf-output). All that is required is to replace `format=Documenter.HTML(…)` in the above code with `format=Documenter.LaTeX()`. See [`docs/makepdf.jl`](https://github.com/JuliaDocs/DocumenterCitations.jl/blob/master/docs/makepdf.jl) for an example. The resulting PDF files for the `DocumenterCitations` package are available as attachments to the [Releases](https://github.com/JuliaDocs/DocumenterCitations.jl/releases).
The rendering of the documentation may be fine-tuned using the [`DocumenterCitations.set_latex_options`](@ref) function. Note that the bibliography in LaTeX is directly rendered for the [different styles](@ref gallery) from the same internal representation as the HTML version. In particular, `bibtex`/`biblatex` is not involved in producing the PDF.
## How to cite references in your documentation
You can put citations anywhere in your docs, both in the markdown pages and in the docstrings of any functions that are shown as part of the API documentation: The basic syntax is, e.g., `[GoerzQ2022](@cite)`, for a BibTeX key [GoerzQ2022](@cite GoerzQ2022) in [`refs.bib`](./refs.bib), which will be rendered in the default numeric style as "[GoerzQ2022](@cite)". See [Syntax for Citations](@ref) for more details.
Clicking on the citations takes you to the bibliography ("[References](@ref)").
## Examples
The following is a list of some projects that use `DocumenterCitations`:
* [Oceananigans](https://clima.github.io/OceananigansDocumentation/stable/)
* [OSCAR](https://docs.oscar-system.org/stable/)
* [QuantumPropagators](https://juliaquantumcontrol.github.io/QuantumPropagators.jl/)
* [QuantumControl](https://juliaquantumcontrol.github.io/QuantumControl.jl/)
* [TwoQubitWeylChamber](https://juliaquantumcontrol.github.io/TwoQubitWeylChamber.jl/)
* [QuantumClifford](https://quantumsavory.github.io/QuantumClifford.jl/stable/references/) and the parent [QuantumSavory organization](https://github.com/QuantumSavory)
## Home References
This page cites the following references:
```@bibliography
Pages = ["index.md"]
Canonical = false
```
Also see the [full bibliography](@ref References) for further references cited throughout this documentation.
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 2919 | ```@meta
CurrentModule = DocumenterCitations
```
# Internals
```@docs
CitationBibliography
```
## Citation Pipeline
The [`DocumenterCitations.CitationBibliography`](@ref) plugin hooks into the [`Documenter.Builder.DocumentPipeline`](https://documenter.juliadocs.org/stable/lib/internals/builder/#Documenter.Builder.DocumentPipeline)[^1] between [`ExpandTemplates`](https://documenter.juliadocs.org/stable/lib/internals/builder/#Documenter.Builder.ExpandTemplates) (which expands `@docs` blocks) and [`CrossReferences`](https://documenter.juliadocs.org/stable/lib/internals/builder/#Documenter.Builder.CrossReferences). The plugin adds the following three steps:
[^1]: See the documentation of [`Documenter.Selectors`](https://documenter.juliadocs.org/stable/lib/internals/selectors/#Documenter.Selectors) for an explanation of Documenter's pipeline concept.
1. [`CollectCitations`](@ref)
2. [`ExpandBibliography`](@ref)
3. [`ExpandCitations`](@ref)
```@docs
CollectCitations
ExpandBibliography
ExpandCitations
```
## [Customization](@id customization)
A custom style can be created by defining methods for the functions listed below that specialize for a user-defined `style` argument to [`CitationBibliography`](@ref). If the `style` is identified by a simple name, e.g. `:mystyle`, the methods should specialize on `Val{:mystyle}`, see the [examples for custom styles](@ref custom_styles). Beyond that, e.g., if the `style` needs to implement options or needs to maintain internal state to manage unique citation labels, `style` can be an object of a custom type. The builtin [`DocumenterCitations.AlphaStyle`](@ref) is an example for such a "stateful" style, initialized via a custom [`init_bibliography!`](@ref) method.
```@docs
bib_html_list_style
bib_sorting
format_bibliography_label
format_bibliography_reference
format_citation
init_bibliography!
```
### Auxiliary customization routines
The following routines are used by the default styles to implement specific formatting. They may be reused by custom styles.
```@docs
format_labeled_citation
format_authoryear_citation
citation_label
format_labeled_bibliography_reference
format_authoryear_bibliography_reference
```
### Customizing LaTeX output
```@raw latex
\hypertarget{sec:customizing_latex_output}{}
```
```@docs
set_latex_options
reset_latex_options
```
### Citation links
The standard citation links described in [Syntax](@ref) are internally parsed into the [`DocumenterCitations.CitationLink`](@ref) data structure:
```@docs
DocumenterCitations.CitationLink
```
Note the this does not include direct citation links such as `[the Semi-AD paper](@cite GoerzQ2022)`, which are instead parsed into
[`DocumenterCitations.DirectCitationLink`](@ref):
```@docs
DocumenterCitations.DirectCitationLink
```
## Debugging
Set the environment variable `JULIA_DEBUG=Documenter,DocumenterCitations` before generating the documentation.
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 484 | # References
## Cited References
The following references are cited in the documentation. The bibliography is produced with
~~~markdown
```@bibliography
```
~~~
```@bibliography
```
## Other References
The following are non-cited references (everything in the [`.bib` file](./refs.bib)), included here to show how bibliographies are rendered for various types of materials. The list of references is produced with
~~~markdown
```@bibliography
*
```
~~~
```@bibliography
*
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 1135 | # CSS Styling
For optimal rendering of the bibliography, place a file [`citations.css`](https://github.com/JuliaDocs/DocumenterCitations.jl/blob/master/docs/src/assets/citations.css) in the `docs/src/assets` folder of your project, containing, e.g.,
~~~@eval
using Markdown
css_file = joinpath(@__DIR__, "..", "src", "assets", "citations.css")
if isfile(css_file)
Markdown.parse("""
```css
$(read(css_file, String))
```
""")
end
~~~
The HTML generated by `DocumenterCitations` also uses CSS classes `canonical` and `noncanonical` to distinguish between [canonical](@ref canonical) and [non-canonical](@ref noncanonical) blocks. While these are not used in the above `citations.css`, a custom `citations.css` could implement different formatting for the two types of bibliographies.
The `citations.css` file must be listed as an `asset` for `Documenter.HTML` in [`docs/make.jl`](https://github.com/JuliaDocs/DocumenterCitations.jl/blob/master/docs/make.jl):
```julia
makedocs(;
format = Documenter.HTML(
# ...
assets=String["assets/citations.css"],
),
plugins=[bib],
# ...
)
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 13051 | # Syntax
## Syntax for Citations
The following syntax is available to create citations in any markdown text:
* `[key](@cite)` is the basic syntax, e.g., `Refs. [GoerzQ2022](@cite) and [Tannor2007](@cite)` which is rendered in the default numeric style as "Refs. [GoerzQ2022](@cite) and [Tannor2007](@cite)".
* `[key; note](@cite)` allows to include a note in the citation, e.g., `See Ref. [GoerzQ2022; Eq. (1)](@cite)` which is rendered as "See Ref. [GoerzQ2022; Eq. (1)](@cite)".
* Multiple keys can be combined in a single citation, e.g., `[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@cite)` renders as "[BrumerShapiro2003, BrifNJP2010, Shapiro2012, KochJPCM2016; and references therein](@cite)".
* The direct `[text](@cite key)` can be used to link to a reference from arbitrary text, e.g., `[the Semi-AD paper](@cite GoerzQ2022)` renders as "[the Semi-AD paper](@cite GoerzQ2022)".
In `[…](@cite)`, the following variations can be used instead of `@cite`:
* `@citet`: Text-style citation. This embeds the citation in the text flow, e.g., "As shown by [GoerzQ2022](@citet)…". For the default numeric citations, this is an alternative to "As shown in Ref. [GoerzQ2022](@cite)"
* `@citep`: Parenthetical citation. For the built-in styles, this is equivalent to just `@cite`.
* `@citet*`: Like `@citet`, but with the full list of authors, e.g., [GoerzQ2022](@citet*).
* `@cite*`/`@citep*`: Like `@cite`/`@citep`, but with the full list of authors ([for non-numeric styles where this makes sense](@ref author_year_style)).
Lastly, capitalizing the `c` in `@citet` or `@citet*` ensures that the first letter of the citation is capitalized so that it can be used at the beginning of a sentence, e.g., [WinckelIP2008](@Citet) (`[WinckelIP2008](@Citet)`) versus [WinckelIP2008](@citet) (`[WinckelIP2008](@citet)`).
The [natbib](https://mirrors.rit.edu/CTAN/macros/latex/contrib/natbib/natnotes.pdf) commands `@citealt`, `@citealp`, and `@citenum` commands are also recognized. They are not supported by any of the built-in styles, but may be handled by [custom styles](@ref customization).
See the [Citation Style Gallery](@ref gallery) for examples of all the possible combinations.
### Citations in docstrings
In docstrings, citations can be made with the same syntax as above. However, since docstrings are also used outside of the rendered documentation (e.g., in the REPL help mode), they should be more self-contained.
The recommended approach is to use a `# References` section in the docstring with an abbreviated bibliography list that links to the [main bibliography](@ref References). For example,
```markdown
# References
* [GoerzQ2022](@cite) Goerz et al. Quantum 6, 871 (2022)
```
in the documentation of the following `Example`:
```@docs
DocumenterCitations.Example
```
(cf. the [source of the `Example` type](https://github.com/JuliaDocs/DocumenterCitations.jl/blob/3b208240f29f9fe7104d27c90f0c324517d18ba6/src/DocumenterCitations.jl#L100-L110)).
If there was no explicit numerical citation in the main text of the docstring,
```markdown
* [Goerz et al. Quantum 6, 871 (2022)](@cite GoerzQ2022)
```
rendering as
* [Goerz et al. Quantum 6, 871 (2022)](@cite GoerzQ2022)
would also have been an appropriate syntax.
## Syntax for the Bibliography Block
### Default `@bibliography` block
~~~markdown
```@bibliography
```
~~~
renders a bibliography for all references that are cited throughout the entire documentation, see [Cited References](@ref). As of version 1.0, the bibliography will not include entries that may be present in the `.bib` file, but that are not cited.
### [Full `@bibliography`](@id full_bibliography)
~~~markdown
```@bibliography
*
```
~~~
Renders a bibliography for *all* references included in the `.bib` file, not just those cited in the documentation. This corresponds to the pre-1.0 default behavior.
### [Multiple `@bibliography` blocks](@id canonical)
It is possible to have multiple `@bibliography` blocks. However, there can only be one "canonical" bibliography target for any citation (the location where a citation links to). Any `@bibliography` block will automatically skip entries that have already been rendered in an earlier canonical `@bibliography` block. Thus, for two consecutive
~~~markdown
```@bibliography
```
~~~
blocks, the second block will not show anything.
On the other hand,
~~~markdown
```@bibliography
```
```@bibliography
*
```
~~~
will first show all the cited references and then all the non-cited references.
This exact setup is shown on the [References](@ref) page.
### [Secondary `@bibliography` blocks](@id noncanonical)
Sometimes it can be useful to render a subset of the bibliography, e.g., to show the references for a particular page. Two things are required to achieve this:
* To filter the bibliography to a specific page (or set of pages), add a `Pages` field to the `@bibliography` block. Exactly like the `Pages` field in Documenter's [`@index`](https://documenter.juliadocs.org/stable/man/syntax/#@index-block) and [`@contents`](https://documenter.juliadocs.org/stable/man/syntax/#@contents-block) blocks, it must evaluate to a list of paths to `.md` files, relative to the folder containing the current file. The value `@__FILE__` as an element in the list can be used to refer to the current file.
* To get around the caveat with [multiple `@bibliography` blocks](@ref canonical) that there can only be one canonical target for each citation, add `Canonical = false` to the `@bibliography` block. The resulting bibliography will be rendered in full, but it will not serve as a link target. This is the only way to have a reference rendered more than once.
For example,
~~~markdown
```@bibliography
Pages = ["index.md"]
Canonical = false
```
~~~
renders a bibliography only for the citations on the [Home](@ref DocumenterCitations.jl) page:
```@bibliography
Pages = ["index.md"]
Canonical = false
```
Usually, you would have this at the bottom of a page, as in [Home References](@ref).
Another possibility that is appropriate for [Citations in docstrings](@ref) is to write out a shortened bibliography "by hand".
### Explicit references
A non-canonical `@bibliography` block can also be used to render a bibliography for a few specific citations keys:
~~~markdown
```@bibliography
Pages = []
Canonical = false
BrifNJP2010
GoerzDiploma2010
GoerzPhd2015
```
~~~
renders a bibliography only for the references
[BrifNJP2010](@cite BrifNJP2010),
[GoerzDiploma2010](@cite GoerzDiploma2010), and [GoerzPhd2015](@cite GoerzPhd2015):
```@bibliography
Pages = []
Canonical = false
BrifNJP2010
GoerzDiploma2010
GoerzPhd2015
```
The `Pages = []` is required to exclude all other cited references.
Note that the numbers [BrifNJP2010, GoerzDiploma2010, GoerzPhd2015](@cite) are from the main (canonical) [References](@ref) page.
### Order of references
In the default numeric style, references in a `@bibliography` are rendered (and numbered) in the order in which they are cited. When there are multiple pages in the documentation, the order in which the pages appear in the navigation bar is relevant.
Non-cited references ([`*` in a full bibliography](@ref full_bibliography)) will appear in the order they are listed in the underlying `.bib` file. That order may be changed by [sorting it explicitly](https://humans-of-julia.github.io/Bibliography.jl/stable/#Bibliography.sort_bibliography!):
```julia
bib = CitationBibliography("refs.bib")
using Bibliography
sort_bibliography!(bib.entries, :nyt) # name-year-title
```
In general, the citation style determines the order of the references, see the [Citation Style Gallery](@ref gallery).
## Syntax for the .bib file
The [`refs.bib`](./refs.bib) file is in the standard [BibTeX format](https://www.bibtex.com/g/bibtex-format/). It must be parsable by [BibParser.jl](https://github.com/Humans-of-Julia/BibParser.jl).
Even in LaTeX, you cannot blindly use `.bib` entries exported from a bibliography manager. [All `.bib` files must be curated by hand](https://clauswilke.com/blog/2015/10/02/bibtex/)! This is true even more so for a project using `DocumenterCitations`.
A `.bib` file that works well with LaTeX will usually, but not always work well with `DocumenterCitations`: Remember that in LaTeX, the strings inside any BibTeX fields are rendered through the TeX engine. At least in principle, they may contain arbitrary macros.
In contrast, `DocumenterCitations` only parses a subset of LaTeX syntax and converts it to markdown. This includes the [commands for special symbols](https://www.bibtex.org/SpecialSymbols/) (although unicode is preferred) and a handful of commands like [`\url` and `\href`](https://www.overleaf.com/learn/latex/Hyperlinks#Linking_web_addresses). If you would like to have support added for a specific additional command, [please open an issue](https://github.com/JuliaDocs/DocumenterCitations.jl/issues/new/choose).
In addition, `DocumenterCitations` recognizes and preserves in-line math and removes braces.
"Any `.bib` file will render the bibliography you expect" is not a design goal. Instead, `DocumenterCitations` tries to ensure that
1. It is possible to write a `.bib` file that produces the expected results; and
2. Most reasonably clean (hand-curated) `.bib` files that work with BibTeX also work with `DocumenterCitations`.
Some tips to keep in mind when editing a `.bib` file to be used with `DocumenterCitations`:
* Use unicode instead of [escaped symbols](https://www.bibtex.org/SpecialSymbols/). Nowadays, `bibtex`/`pdflatex` seems to handle unicode without problems, so it's best to keep your `.bib` files in unicode.
* You do not need to use [braces to protect capitalization](https://texfaq.org/FAQ-capbibtex). Unlike `bibtex`, `DocumenterCitation` will preserve the capitalization of titles. You should always put the title in the `.bib` file as it appears in the published paper.
* Use a consistent scheme for citation keys. Shorter keys are better.
* All entries should have a `Doi` field, or a `Url` field if no DOI is available.
* When citing websites [WP_Schroedinger](@cite), in addition to the `Url` field, a `Urldate` field may be given in the format `YYYY-MM-DD`. This will be rendered as, e.g., "Accessed on Oct 24, 2023".
* Use `@string` macros for abbreviated journal names, with the caveat of [#31](https://github.com/Humans-of-Julia/BibParser.jl/issues/31) and [#32](https://github.com/Humans-of-Julia/BibParser.jl/issues/32) in the [BibParser.jl issues](https://github.com/Humans-of-Julia/BibParser.jl/issues).
You may be interested in using (or forking) the [`getbibtex` script](https://github.com/goerz/getbibtex) to generate consistent `.bib` files.
### Preprint support
If the published paper (`Doi` link) is not open-access, but a version of the paper is available on a preprint server like the [arXiv](https://arxiv.org), your are strongly encouraged to add the arXiv ID as `Eprint` in the BibTeX entry. In the rendered bibliography, the preprint information will be shown and automatically link to `https://arxiv.org/abs/<ID>`.
If necessary, you may also add a `Primaryclass` field to indicate a category, see ["BibTeX and Eprints" in the arXiv documentation](https://info.arxiv.org/help/hypertex/bibstyles/index.html).
Note that unlike in [default](https://tex.stackexchange.com/questions/386078/workaround-for-missing-archiveprefix-in-bib-entry) BibTeX, it is not necessary to define `Archiveprefix` in the `.bib` file. A missing `Archiveprefix` is assumed to be `arXiv`. The field name `Eprinttype` (which in BibTeX is an alias for `Archiveprefix`) is currently not yet supported, nor is `Eprintclass` as an alias for `Primaryclass`.
For documents that are available *only* as an arXiv eprint, the best result is obtained with a BibTeX entry using the `@article` class, with, e.g., `arXiv:2003.10132` in the `Journal` field, and, e.g., `10.48550/ARXIV.2003.10132` in the `Doi` field (but no `Eprint` field) [Wilhelm2003.10132](@cite).
Beyond arXiv, other preprint servers are supported. The `Archiveprefix` field for non-arXiv preprints is mandatory. For any defined `Archiveprefix`, `Eprint`, and `Primaryclass` fields, the rendered bibliography will include the preprint information in the format `ArchivePrefix:Eprint [Primaryclass]`. However, only certain preprint servers (known `ArchivePrefix`) will automatically be linked. Besides arXiv, the currently supported preprint servers are:
* [BiorXiv](https://www.biorxiv.org). The `Archiveprefix` is `biorXiv`. The `Eprint` ID should be the final part of the DOI, e.g. `2022.09.09.507322` [KatrukhaNC2017](@cite).
* [HAL](https://hal.science). The `Archiveprefix` is `HAL`. The `Eprint` ID should include the prefix (usually `hal-`, but sometimes `tel-`, etc.), e.g., Refs. [SauvagePRXQ2020,BrionPhd2004](@cite).
If you would like support for any additional preprint server, [please open an issue](https://github.com/JuliaDocs/DocumenterCitations.jl/issues/new/choose).
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 50 | # Addendum
No references are cited on this page.
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 565 | # Test Project
Ref [BrifNJP2010](@cite)
## Site content
The following table of contents replicates the order in the side bar.
```@contents
Pages = [
normpath("part1", "section1", "p1_s1_page.md"),
normpath("part1", "section2", "p1_s2_page.md"),
normpath("part2", "section1", "p2_s1_page.md"),
normpath("part2", "section2", "p2_s2_page.md"),
normpath("part3", "section1", "p3_s1_page.md"),
normpath("part3", "section2", "p3_s2_page.md"),
normpath("part3", "section2", "invalidpages.md"),
"references.md",
"addendum.md",
]
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 284 | # Site references
```@bibliography
Pages = [
"index.md",
"part1/section1/p1_s1_page.md",
"part1/section2/p1_s2_page.md",
"part2/section1/p2_s1_page.md",
"part2/section2/p2_s2_page.md",
"part3/section1/p3_s1_page.md",
"part3/section2/p3_s2_page.md",
]
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 512 | # A: Part 1.1
## First Section of Part 1.1
Ref [Shapiro2012](@cite)
## Second Section of Part 1.1
Ref [KochJPCM2016](@cite)
## Local References of Part 1.1
**Content**
The content matches the bibliography. The point of this (here and in the following pages) is to verify that the `Pages` attribute behaves consistently between `@contents` blocks and `@bibliography` blocks.
```@contents
Pages = ["p1_s1_page.md"]
```
**Bibliography**
```@bibliography
Canonical = false
Pages = ["p1_s1_page.md"]
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 480 | # B: Part 1.2
## First Section of Part 1.2
Ref [KochJPCM2016](@cite) (duplicated from [Second Section of Part 1.1](@ref))
## Second Section of Part 1.2
Ref [SolaAAMOP2018](@cite)
## Local References of Part 1.2
**Content**
The content here is empty, because `@contents` doesn't support `@__FILE__` (but doesn't complain either).
```@contents
Pages = [@__FILE__]
```
**Bibliography**
```@bibliography
Canonical = false
Pages = [@__FILE__] # We can have comments!
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 768 | # C: Part 2.1
## First Section of Part 2.1
Ref [MorzhinRMS2019](@cite)
## Second Section of Part 2.1
Ref [BrumerShapiro2003](@cite)
## Local References of Part 2.1 and Earlier Parts
This excludes the reference in `index.md`.
**Content**
The content matches the bibliography
```@contents
Pages = [
"p2_s1_page.md", # @__FILE__ is not supported
normpath("../../part1/section1/p1_s1_page.md"),
normpath("../../part1/section2/p1_s2_page.md"),
]
```
**Bibliography**
```@bibliography
Canonical = false
Pages = [
@__FILE__, # In the @bibliography block, we can use `@__FILE`
# Note that `@bibliography` (unlike `@contents`) doesn't need `normpath`.
"../../part1/section1/p1_s1_page.md",
"../../part1/section2/p1_s2_page.md",
]
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 818 | # D: Part 2.2
## First Section of Part 2.2
Ref [GoerzDiploma2010](@cite)
## Second Section of Part 2.2
Ref [GoerzJPB2011](@cite)
## Local References of Earlier Parts
This excludes the reference in `index.md` as well as the references in the *current* document.
One of the `Pages` uses `joinpath`, as an example for something where `eval` has to do some work.
**Content**
The content matches the bibliography
```@contents
Pages = [
joinpath("..", "..", "part1", "section1", "p1_s1_page.md"),
normpath("../../part1/section2/p1_s2_page.md"),
normpath("../section1/p2_s1_page.md"),
]
```
**Bibliography**
```@bibliography
Canonical = false
Pages = [
joinpath("..", "..", "part1", "section1", "p1_s1_page.md"),
"../../part1/section2/p1_s2_page.md",
"../section1/p2_s1_page.md",
]
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 793 | # E: Part 3.1
## First Section of Part 3.1
Ref [TomzaPRA2012](@cite)
## Second Section of Part 3.1
Ref [GoerzNJP2014](@cite)
## Local References of Earlier Parts
This excludes the reference in `index.md` as well as the references in the *current* document.
**Content**
The content matches the bibliography
```@contents
Pages = [
normpath("../../part1/section1/p1_s1_page.md"),
normpath("../../part1/section2/p1_s2_page.md"),
normpath("../../part2/section1/p2_s1_page.md"),
normpath("../../part2/section2/p2_s2_page.md"),
]
```
**Bibliography**
```@bibliography
Canonical = false
Pages = [
"../../part1/section1/p1_s1_page.md",
"../../part1/section2/p1_s2_page.md",
"../../part2/section1/p2_s1_page.md",
"../../part2/section2/p2_s2_page.md",
]
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 875 | # Test of Invalid Pages
## References from Nonexisting Pages
Nothing should render here (except for the fallback introduced in version 1.3.1)
```@bibliography
Canonical = false
Pages = [
"index.md", # not valid in 1.3, valid in 1.3.1 (fallback)
"p3_s1_page.md",
"noexist.md",
]
```
## References only from Pages that contain no references
Again, nothing should render here.
```@bibliography
Canonical = false
Pages = [
"../../addendum.md",
]
```
## References Mixing Existing and Nonexisting Pages
```@bibliography
Canonical = false
Pages = [
"p3_s1_page.md",
"p3_s2_page.md",
]
```
The above bibliography should render only the references in [F: Part 3.2](@ref) (since the file `p3_s1_page.md` for [E: Part 3.1](@ref) exists in a different folder).
## Not passing a list to Pages
```@bibliography
Canonical = false
Pages = @__FILE__
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 936 | # F: Part 3.2
## First Section of Part 3.2
Ref [GoerzPRA2014](@cite)
## Second Section of Part 3.2
Ref [JaegerPRA2014](@cite)
## Local References of Earlier Parts
This excludes the reference in `index.md` as well as the references in the *current* document.
The bibliography also uses a pretty fancy expression to test that we can `eval` non-trivial specification of `Pages`, not just a list of strings.
**Content**
The content matches the bibliography
```@contents
Pages = [
[
normpath("..", "..", "part$p", "section$s", "p$(p)_s$(s)_page.md") for
(p, s) in Iterators.product((1, 2), (1, 2))
]...,
normpath("../section1/p3_s1_page.md"),
]
```
**Bibliography**
```@bibliography
Canonical = false
Pages = [
[
joinpath("..", "..", "part$p", "section$s", "p$(p)_s$(s)_page.md") for
(p, s) in Iterators.product((1, 2), (1, 2))
]...,
"../section1/p3_s1_page.md",
]
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 188 | # Testing collection of citations from docstrings
## Problem description
Citations that only occurred in docstrings were not being collected.
```@docs
DocumenterCitations.Example
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 36 | ## References
```@bibliography
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 1080 | # Testing that DocumenterCitations works with Contents blocks
## Contents
```@contents
Pages = ["index.md", "references.md"]
```
## Problem description
As reported in [#16](https://github.com/JuliaDocs/DocumenterCitations.jl/issues/16) and [ali-ramadhan/DocumenterCitations.jl#24](https://github.com/ali-ramadhan/DocumenterCitations.jl/issues/24), there have been problems with the DocumenterCitations plugin and `@contents` blocks.
Having a `@contents` block that includes any `Pages` that also have a `@bibliography` block causes a crash like this:
```
ERROR: LoadError: MethodError: no method matching header_level(::BibInternal.Entry)
Closest candidates are:
header_level(::Markdown.Header{N}) where N
@ Documenter ~/.julia/packages/Documenter/bYYzK/src/Utilities/Utilities.jl:380
```
## Workarounds
Guillaume Gautier [guilgautier](@cite) provides the following workaround:
> I'll propose the above dirty fix in #24, namely
> Remove the file where the @bibliography block is called (here reference.md), from the list of Pages involved in all @contents block.
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 36 | ## References
```@bibliography
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 327 | # Testing citation keys with underscores
## Problem description
Citations keys with underscores [rabiner_tutorial_1989](@cite) get messed up because they get mangled by the markdown parser.
At the same time, we *do* want to support nested markdown in the "notes" of a citations, e.g. [GoerzQ2022; with *emphasis*](@cite).
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 36 | ## References
```@bibliography
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 337 | # Testing citation keys with underscores
## Problem description
In principle, both underscores and asterisks can be used transparently, as they get normalized. We can have [rabiner_tutorial_1989](@cite), [rabiner_tutorial*1989](@cite), and [rabiner*tutorial*1989](@cite).
Similarly, [Goerz_Q*2022](@cite) and [Goerz_Q_2022](@cite).
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 36 | ## References
```@bibliography
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 222 | # Testing link checks for broken links
## Problem description
The `linkcheck` option for `makedocs` should complain about [broken links](http://httpbin.org/status/403).
Ref [GoerzPhd2015](@cite) contains broken links.
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 36 | ## References
```@bibliography
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 830 | # Testing error for citing a non-existent keys
We cite an existing key [GoerzQ2022](@cite) and a non-existing key [NoExist2023](@cite).
In strict mode, `makedocs` errors during the collection phase.
In non-strict mode, the documentation should be rendered with "?", similar to how LaTeX itself handles missing citations. This even works for "combined references" [BrumerShapiro2003, BrifNJP2010, Shapiro2012_, KochJPCM2016_, SolaAAMOP2018, MorzhinRMS2019_, Wilhelm2003.10132, KochEPJQT2022; and references therein](@cite)
Also, with `@Citet`: [BrumerShapiro2003, BrifNJP2010, Shapiro2012_, KochJPCM2016_, SolaAAMOP2018, MorzhinRMS2019_, Wilhelm2003.10132, KochEPJQT2022; and references therein](@Citet).
Lastly, we cite a key [Tannor2007](@cite) that exists in the `.bib` file but does not appear in any bibliography block.
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 1.3.4 | ca601b812efd1155a9bdf9c80e7e0428da598a08 | docs | 347 | ## References
The following bibliography does include every cited key (missing `Tannor2007`):
```@bibliography
Pages = [
]
GoerzQ2022
BrumerShapiro2003
BrifNJP2010
SolaAAMOP2018
Wilhelm2003.10132
KochEPJQT2022
```
The following bibliography includes a non-existing and non-cited key
```@bibliography
Pages = []
NoExistInBibliography2023
```
| DocumenterCitations | https://github.com/JuliaDocs/DocumenterCitations.jl.git |
|
[
"MIT"
] | 0.1.0 | d5ed26b0dbc92e599e497ef3a7aeb1ff1b297232 | code | 6492 | module TeensyLoaderCLI
using teensy_loader_cli_jll
using Downloads
using Logging
struct MCU
id::Symbol
end
const at90usb162 = MCU(:at90usb162)
const atmega32u4 = MCU(:atmega32u4)
const at90usb646 = MCU(:at90usb646)
const at90usb1286 = MCU(:at90usb1286)
const mkl26z64 = MCU(:mkl26z64)
const mk20dx128 = MCU(:mk20dx128)
const mk20dx256 = MCU(:mk20dx256)
const mk66fx1m0 = MCU(:mk66fx1m0)
const mk64fx512 = MCU(:mk64fx512)
const imxrt1062 = MCU(:imxrt1062)
const TEENSY2 = MCU(:TEENSY2)
const TEENSY2PP = MCU(:TEENSY2PP)
const TEENSYLC = MCU(:TEENSYLC)
const TEENSY30 = MCU(:TEENSY30)
const TEENSY31 = MCU(:TEENSY31)
const TEENSY32 = MCU(:TEENSY32)
const TEENSY35 = MCU(:TEENSY35)
const TEENSY36 = MCU(:TEENSY36)
const TEENSY40 = MCU(:TEENSY40)
const TEENSY41 = MCU(:TEENSY41)
const TEENSY_MICROMOD = MCU(:TEENSY_MICROMOD)
"""
has_soft_reboot(::MCU)
Whether or not the given MCU supports soft reboot functionality through TeensyLoaderCLI.
"""
function has_soft_reboot(mcu::MCU)
mcu.id in (:TEENSY31,
:TEENSY32,
:TEENSY35,
:TEENSY36,
:TEENSY40,
:TEENSY41)
end
"""
install_udev(;dry=true)
Installs the udev rules for Teensy boards.
The rules are downloaded from [https://www.pjrc.com/teensy/00-teensy.rules](https://www.pjrc.com/teensy/00-teensy.rules).
The download has a timeout of 5 minutes.
Keyword arguments:
* `dry`: Whether to do a dry run. When set to `true`, no actions other than downloading are taken. Set to `false` to change your system.
`install_udev` will abort if the target file already exists. Check that the existing udev rule is correct for your
devices and either use the existing rules, or remove the file to try again.
!!! warn "sudo"
The rules are installed system wide, into `/etc/udev/rules.d/00-teensy.rules`. This requires `sudo`
permissions, which is why this needs to be done manually. In addition, this requires `udevadm`.
"""
function install_udev(;dry=true)
installpath = "/etc/udev/rules.d/00-teensy.rules"
if ispath(installpath)
@warn "udev rule file already exists - aborting."
return
end
dry && @info "Doing a dry run - no changes will occur."
!dry && (@warn "Doing a live run - your system will be affected."; sleep(5))
println()
mktemp() do dlpath, dlio
@info "Downloading udev rules to `$dlpath`"
Downloads.download("https://www.pjrc.com/teensy/00-teensy.rules", dlio; timeout=5*60.0)
@info "Installing rules file to `/etc/udev/rules.d/00-teensy.rules`"
mvcmd = `sudo install -o root -g root -m 0664 $dlpath $installpath`
@info "Installing rules" Cmd=mvcmd
!dry && run(mvcmd)
udevadmctl = `sudo udevadm control --reload-rules`
@info "Reloading rules" Cmd=udevadmctl
!dry && run(udevadmctl)
udevadmtgr = `sudo udevadm trigger`
@info "Triggering udev events" Cmd=udevadmtgr
!dry && run(udevadmtgr)
end
@info "Done!"
nothing
end
"""
help_cmd()
Print out the `--help` section provided by the `teensy_loader_cli` binary.
The printed flags can only be used when using the `teensy_loader_cli` binary directly.
See also: [help](@ref), [program!](@ref)
"""
function help_cmd()
err = Pipe()
proc = teensy_loader_cli() do b
run(pipeline(`$b --help`; stderr=err); wait=false)
end
# this thing complains with a bad exit code when it doesn't get what it cares about..
wait(proc)
write(stdout, readavailable(err))
nothing
end
"""
boot!(;wait=true, verbose=false)
Boot the attached Teensy device, but do not upload a new program.
Keyword arguments:
* `wait = true`
* Wait until a device is found.
* `verbose = false`
* Give more verbose output.
"""
function boot!(;wait=true, verbose=false)
w = wait ? `-w` : ``
v = verbose ? `-v` : ``
proc = teensy_loader_cli() do b
run(`$b -b $w $v`; wait=false)
end
success(proc)
nothing
end
"""
list_mcus()
List the microcontrollers supported by TeensyLoaderCLI.
!!! note "Programming"
The names given by the `teensy_loader_cli` binary can be used to
program the Teensy in `program!`. They are available via `TeensyLoaderCLI.<mcu>`,
where `<mcu>` is one of the MCUs given by this function.
"""
function list_mcus()
out = Pipe()
proc = teensy_loader_cli() do b
run(pipeline(`$b --list-mcus`; stdout=out); wait=false)
end
wait(proc)
write(stdout, readavailable(out))
nothing
end
"""
program!(mcu::MCU, file::String;
wait = true
verbose = false
hardreboot = false
softreboot = false
noreboot = false)
Upload the program located at `file` to the connected teensy device. `file` must be in `ihex` format.
Positional Arguments:
* `mcu`
* One of the supported MCU targets listed in `list_mcus()`.
* `file`
* The binary file to be uploaded to the device. Must be in `ihex` format.
Keyword Arguments:
* `wait = true`
* Wait until a device is detected.
* `verbose = false`
* Give more verbose output.
* `hardreboot = false`
* Use a hard reboot is the device is not online.
* `softreboot = false`
* Use a soft reboot if the devise is not online.
* Only available for Teensy 3.x & 4.x.
* `noreboot = false`
* Don't reboot the Teensy after programming.
"""
function program!(mcu::MCU, file::String; wait=true, verbose=false, hard=false, soft=false, noreboot=false)
w = wait ? `-w` : ``
v = verbose ? `-v` : ``
h = hard ? `-r` : ``
s = soft ? `-s` : ``
n = noreboot ? `-n` : ``
if h && s
@error "Can only specify either hard or soft reboot!"
return
end
if s && !has_soft_reboot(mcu)
@error "Only Teensy 3.x & 4.x support soft reboot!"
return
end
if n && !(h || s)
@error "Cannot specify both to reboot and not to reboot!"
return
end
if !ispath(file)
@error "Given path is not a file!" Path=file
return
end
verbose && @info "Uploading program at `$file`"
proc = teensy_loader_cli() do b
run(`$b --mcu=$(mcu.id) $w $v $h $s $n $file`; wait=false)
end
success(proc)
nothing
end
end # module TeensyLoaderCLI
| TeensyLoaderCLI | https://github.com/Seelengrab/TeensyLoaderCLI.jl.git |
|
[
"MIT"
] | 0.1.0 | d5ed26b0dbc92e599e497ef3a7aeb1ff1b297232 | docs | 506 | # TeensyLoaderCLI
A Julia wrapper package for [teensy_loader_cli](https://github.com/PaulStoffregen/teensy_loader_cli), used for interacting with the Teensy family of microcontrollers.
## Installation
On Linux, make sure to run `install_udev()` to install the `udev` definitions for teensy, if you haven't already set them up.
## Functions
These functions are the supported API of this package. Make sure to read their docstrings thoroughly.
* `list_mcus`
* `install_udev`
* `boot!`
* `program!`
| TeensyLoaderCLI | https://github.com/Seelengrab/TeensyLoaderCLI.jl.git |
|
[
"MIT"
] | 0.4.0 | 5e6d8bfd5555abe43335b531b1667390084e832f | code | 1530 | module PearsonCorrelationMatch
using Distributions: UnivariateDistribution as UD
using Distributions: ContinuousUnivariateDistribution as CUD
using Distributions: DiscreteUnivariateDistribution as DUD
using Distributions
using FastGaussQuadrature: gausshermite
using IrrationalConstants: sqrt2, invsqrtπ
using LinearAlgebra: eigen, Symmetric, Diagonal, lmul!, rmul!, diagind
using PolynomialRoots: roots
using SharedArrays: SharedMatrix, sdata
using StatsFuns: normcdf, normpdf, norminvcdf
export pearson_bounds, pearson_match
include("common.jl")
include("bounds.jl")
include("match.jl")
include("rules.jl")
using PrecompileTools
using Distributions: Distributions
@setup_workload begin
# put in let-block to avoid exporting temporary variables
# TODO: check if wrapping in let-block is an anti-pattern
let
p = 0.5
D = Distributions.Gamma()
E = Distributions.Beta(5, 3)
F = Distributions.Binomial(100, 0.3)
G = Distributions.NegativeBinomial(20)
@compile_workload begin
pearson_bounds(D, E)
pearson_bounds(D, F)
pearson_bounds(G, D)
pearson_bounds(F, G)
pearson_bounds([D, E, F, G])
pearson_bounds((D, E, F, G))
pearson_match(p, D, E)
pearson_match(p, D, F)
pearson_match(p, G, D)
pearson_match(p, F, G)
pearson_match([1.0 0.5; 0.5 1.0], [E, G])
pearson_match([1.0 0.5; 0.5 1.0], (E, G))
end
end
end
end
| PearsonCorrelationMatch | https://github.com/adknudson/PearsonCorrelationMatch.jl.git |
|
[
"MIT"
] | 0.4.0 | 5e6d8bfd5555abe43335b531b1667390084e832f | code | 3138 | """
pearson_bounds(d1, d2; n::Real=20, m::Real=128, kwargs...)
Determine the range of admissible Pearson correlations between two distributions.
# Fields
- `d1`: The first marginal distribution.
- `d2`: The second marginal distribution.
- `n`: The degree of the polynomial used to estimate the bounds.
- `m`: The number of points used in the hermite polynomial interpolation.
- `kwargs`: Additional keyword arguments. Currently unused.
# Details
The accuracy of the bounds depends on the degree of the polynomial and the number of hermite
points. Be careful not to set the polynomial degree too high as Runge's theorem states that
a polynomial of too high degree would cause oscillation at the edges of the interval and
reduce accuracy.
In general raising the number of hermite points will result in better accuracy, but comes
with a small performance hit. Furthermore the number of hermite points should be higher
than the degree of the polynomial.
# Examples
```julia-repl
julia> using Distributions
julia> d1 = Exponential(3.14); d2 = NegativeBinomial(20, 0.2);
julia> pearson_bounds(d1, d2)
(lower = -0.8553947509241561, upper = 0.9413665073003636)
```
"""
function pearson_bounds(d1::UD, d2::UD; n::Real=20, m::Real=128, kwargs...)
n = Int(n)
m = Int(m)
m1 = mean(d1)
m2 = mean(d2)
s1 = std(d1)
s2 = std(d2)
a = _generate_coefs(d1, n, m)
b = _generate_coefs(d2, n, m)
k = big.(0:n)
kab = a .* factorial.(k) .* b
c1 = -m1 * m2
c2 = inv(s1 * s2)
pl = c1 * c2 + c2 * sum((-1) .^ k .* kab)
pu = c1 * c2 + c2 * sum(kab)
pl = clamp(pl, -1, 1)
pu = clamp(pu, -1, 1)
return (lower=Float64(pl), upper=Float64(pu))
end
"""
pearson_bounds(margins; n::Real=12, m::Real=128, kwargs...)
Determine the range of admissible Pearson correlations pairwise between a list of distributions.
# Fields
- `margins`: A list of marginal distributions.
- `n`: The degree of the polynomial used to estimate the bounds.
- `m`: The number of points used in the hermite polynomial interpolation.
- `kwargs`: Additional keyword arguments. Currently unused.
# Examples
```julia-repl
julia> using Distributions
julia> margins = [Exponential(3.14), NegativeBinomial(20, 0.2), LogNormal(2.718)];
julia> lower, upper = pearson_bounds(margins);
julia> lower
3×3 Matrix{Float64}:
1.0 -0.855395 -0.488737
-0.855395 1.0 -0.704403
-0.488737 -0.704403 1.0
julia> upper
3×3 Matrix{Float64}:
1.0 0.941367 0.939671
0.941367 1.0 0.815171
0.939671 0.815171 1.0
```
"""
function pearson_bounds(margins; n::Real=20, m::Real=128, kwargs...)
d = length(margins)
n = Int(n)
m = Int(m)
lower = SharedMatrix{Float64}(d, d)
upper = SharedMatrix{Float64}(d, d)
Base.Threads.@threads for (i, j) in _idx_subsets2(d)
l, u = pearson_bounds(margins[i], margins[j]; n=n, m=m, kwargs...)
@inbounds lower[i, j] = l
@inbounds upper[i, j] = u
end
L = sdata(lower)
_symmetric!(L)
_set_diag1!(L)
U = sdata(upper)
_symmetric!(U)
_set_diag1!(U)
return (lower=L, upper=U)
end
| PearsonCorrelationMatch | https://github.com/adknudson/PearsonCorrelationMatch.jl.git |
|
[
"MIT"
] | 0.4.0 | 5e6d8bfd5555abe43335b531b1667390084e832f | code | 5766 | """
Equation (25) of the reference paper.
"""
function _generate_coefs(F::UD, n::Int, m::Int)
t, w = gausshermite(m)
t *= sqrt2
u = normcdf.(t)
# If u[i] is 0 or 1, then quantile(F, u[i]) has the potential to be ±∞
# Apply a small correction here to ensure that the values are finite
clamp!(u, nextfloat(0.0), prevfloat(1.0))
X = quantile.(Ref(F), u)
a = zeros(Float64, n + 1)
for i in eachindex(a)
k = i - 1
S = sum(w .* _hermite.(t, k) .* X)
@inbounds a[i] = invsqrtπ * S / factorial(big(k))
end
return a
end
"""
_Gn0_discrete(A, B, a, b, invs1s2, n::Int)
Equation (41) of the reference paper.
"""
function _Gn0_discrete(A, B, a, b, invs1s2, n::Int)
n == 0 && return zero(Float64)
M = length(A)
N = length(B)
accu = zero(Float64)
for r in 1:M, s in 1:N
r00 = _hermite_normpdf(a[r], n - 1) * _hermite_normpdf(b[s], n - 1)
r10 = _hermite_normpdf(a[r+1], n - 1) * _hermite_normpdf(b[s], n - 1)
r01 = _hermite_normpdf(a[r], n - 1) * _hermite_normpdf(b[s+1], n - 1)
r11 = _hermite_normpdf(a[r+1], n - 1) * _hermite_normpdf(b[s+1], n - 1)
accu += A[r] * B[s] * (r11 + r00 - r01 - r10)
end
return accu * invs1s2
end
"""
_Gn0_mixed(A, a, F, invs1s2, n::Int, m::Int)
Equation (49) of the reference paper.
"""
function _Gn0_mixed(A, a, F, invs1s2, n::Int, m::Int)
n == 0 && return zero(Float64)
M = length(A)
accu = zero(Float64)
for r in 1:M
accu += A[r] * (_hermite_normpdf(a[r+1], n - 1) - _hermite_normpdf(a[r], n - 1))
end
t, w = gausshermite(m)
t *= sqrt2
u = normcdf.(t)
# If u[i] is 0 or 1, then quantile(F, u[i]) has the potential to be ±∞
# Apply a small correction here to ensure that the values are finite
clamp!(u, nextfloat(0.0), prevfloat(1.0))
X = quantile.(F, u)
any(isinf, X) && error("Values must be real and finite")
S = zero(Float64)
for k in eachindex(t)
S += w[k] * _hermite(t[k], n) * X[k]
end
S *= invsqrtπ
return -invs1s2 * accu * S
end
"""
The "probabilist's" Hermite polynomial of degree ``k``.
"""
function _hermite(x::Float64, k::Int)
k < 0 && throw(ArgumentError("'k' must be a non-negative integer"))
k == 0 && return 1.0
k == 1 && return x
Hk = x
Hkp1 = zero(x)
Hkm1 = one(x)
for j in 2:k
Hkp1 = x * Hk - (j - 1) * Hkm1
Hkm1, Hk = Hk, Hkp1
end
return Hkp1
end
_hermite(x, n) = _hermite(convert(Float64, x), convert(Int, n))
"""
He(x, k) * normpdf(x)
"""
_hermite_normpdf(x::Float64, k::Int) = isinf(x) ? zero(x) : _hermite(x, k) * normpdf(x)
_hermite_normpdf(x, k) = _hermite_normpdf(convert(Float64, x), convert(Int, k))
"""
_is_real(x::Complex)
Check if a number is real within a given tolerance.
"""
_is_real(x::Complex{T}) where {T} = abs(imag(x)) < sqrt(eps(T))
"""
_real_roots(coeffs)
Find the real and unique roots of the polynomial coefficients.
- `coeffs`: A vector of coefficients in ascending order of degree.
"""
function _real_roots(coeffs)
complex_roots = roots(coeffs)
filter!(_is_real, complex_roots)
xs = real.(complex_roots)
return unique!(xs)
end
"""
_feasible_roots(coeffs)
Find all real roots of the polynomial that are in the interval ``[-1, 1]``.
- `coeffs`: a vector of coefficients in ascending order of degree.
"""
function _feasible_roots(coeffs)
xs = _real_roots(coeffs)
return filter!(x -> abs(x) ≤ 1.0 + sqrt(eps()), xs)
end
"""
_nearest_root(target, roots)
Find the root closest to the target value.
"""
function _nearest_root(target, roots)
y = 0
m = Inf
for x0 in roots
f = abs(x0 - target)
if f < m
m = f
y = x0
end
end
return y
end
"""
_best_root(p, xs)
Consider the feasible roots and return a value.
- `p`: the target correlation
- `roots`: a vector of feasible roots
"""
function _best_root(p, roots)
length(roots) == 1 && return clamp(first(roots), -1, 1)
length(roots) > 1 && return _nearest_root(p, roots)
return p < 0 ? nextfloat(-one(Float64)) : prevfloat(one(Float64))
end
"""
_idx_subsets2(d)
Equivalent to IterTools.subsets(1:d, Val(2)), but allocates all pairs for use in parallel
threads.
"""
function _idx_subsets2(d::Int)
n = d * (d - 1) ÷ 2
xs = Vector{Tuple}(undef, n)
k = 1
for i in 1:d-1
for j in i+1:d
xs[k] = (i, j)
k += 1
end
end
return xs
end
"""
_symmetric!(X)
Copy the upper part of a matrix to its lower half.
"""
function _symmetric!(X::AbstractMatrix{T}) where {T}
m, n = size(X)
m == n || throw(DimensionMismatch("Input matrix must be square"))
for i in 1:n-1
for j in i+1:n
@inbounds X[j, i] = X[i, j]
end
end
return X
end
"""
_set_diag1!(X)
Set the diagonal elements of a square matrix to `1`.
"""
function _set_diag1!(X::AbstractMatrix{T}) where {T}
m, n = size(X)
m == n || throw(DimensionMismatch("Input matrix must be square"))
@inbounds for i in diagind(X)
X[i] = one(T)
end
return X
end
"""
_project_psd(X, ϵ)
Project `X` onto the set of PSD matrixes.
"""
function _project_psd!(X, ϵ)
λ, P = eigen(Symmetric(X); sortby=x -> -x)
replace!(x -> max(x, ϵ), λ)
X .= P * Diagonal(λ) * P'
return X
end
"""
_cov2cor!(X)
Project `X` onto the set of correlation matrices.
"""
function _cov2cor!(X::AbstractMatrix)
D = sqrt(inv(Diagonal(X)))
lmul!(D, X)
rmul!(X, D)
_set_diag1!(X)
_symmetric!(X)
return X
end
function _cov2cor!(X::Symmetric)
_symmetric!(X.data)
_cov2cor!(X.data)
return X
end
| PearsonCorrelationMatch | https://github.com/adknudson/PearsonCorrelationMatch.jl.git |
|
[
"MIT"
] | 0.4.0 | 5e6d8bfd5555abe43335b531b1667390084e832f | code | 4926 | """
pearson_match(p::Real, d1, d2; n::Real=12, m::Real=128, kwargs...)
Compute the Pearson correlation coefficient to be used in a bivariate Gaussian copula.
# Fields
- `p`: The target correltation between the marginal distributions.
- `d1`: The first marginal distribution.
- `d2`: The second marginal distribution.
- `n`: The degree of the polynomial used to estimate the matching correlation.
- `m`: The number of points used in the hermite polynomial interpolation.
- `kwargs`: Additional keyword arguments. Currently unused.
# Examples
```julia-repl
julia> using Distributions
julia> d1 = Beta(2, 3); d2 = Binomial(20, 0.2);
julia> pearson_match(0.6, d1, d2)
0.6127531346934495
```
"""
function pearson_match(p::Real, d1::UD, d2::UD; n::Real=20, m::Real=128, kwargs...)
-1 <= p <= 1 || throw(ArgumentError("`p` must be in [-1, 1]"))
n > 0 || throw(ArgumentError("`n` must be a positive number"))
return _invrule(Float64(p), d1, d2; n=Int(n), m=Int(m))
end
_invrule(p::Float64, d1::UD, d2::UD; kwargs...) = _invrule_fallback(p, d1, d2; kwargs...)
"""
_invrule_fallback(p::Float64, d1, d2; n::Int, m::Int, kwargs...)
Countinuous case.
"""
function _invrule_fallback(p::Float64, d1::CUD, d2::CUD; n::Int, m::Int, kwargs...)
m1 = mean(d1)
m2 = mean(d2)
s1 = std(d1)
s2 = std(d2)
a = _generate_coefs(d1, n, m)
b = _generate_coefs(d2, n, m)
c1 = -m1 * m2
c2 = inv(s1 * s2)
coef = zeros(Float64, n + 1)
for k in 1:n
@inbounds coef[k+1] = c2 * a[k+1] * b[k+1] * factorial(big(k))
end
coef[1] = c1 * c2 + c2 * a[1] * b[1] - p
xs = _feasible_roots(coef)
return _best_root(p, xs)
end
"""
_invrule_fallback(p::Float64, d1, d2; n::Int, kwargs...)
Discrete case.
"""
function _invrule_fallback(p::Float64, d1::DUD, d2::DUD; n::Int, kwargs...)
max1 = maximum(d1)
max2 = maximum(d2)
max1 = isinf(max1) ? quantile(d1, prevfloat(1.0)) : max1
max2 = isinf(max2) ? quantile(d2, prevfloat(1.0)) : max2
min1 = minimum(d1)
min2 = minimum(d2)
s1 = std(d1)
s2 = std(d2)
A = Int(min1):Int(max1)
B = Int(min2):Int(max2)
# z = Φ⁻¹[F(A)], α[0] = -Inf, β[0] = -Inf
a = [-Inf; norminvcdf.(cdf.(Ref(d1), A))]
b = [-Inf; norminvcdf.(cdf.(Ref(d2), B))]
c2 = inv(s1 * s2)
coef = zeros(Float64, n + 1)
for k in 1:n
@inbounds coef[k+1] = _Gn0_discrete(A, B, a, b, c2, k) / factorial(big(k))
end
coef[1] = -p
xs = _feasible_roots(coef)
return _best_root(p, xs)
end
"""
_invrule_fallback(p::Float64, d1, d2; n::Int, m::Int, kwargs...)
Mixed case.
"""
function _invrule_fallback(p::Float64, d1::DUD, d2::CUD; n::Int, m::Int, kwargs...)
s1 = std(d1)
s2 = std(d2)
min1 = minimum(d1)
max1 = maximum(d1)
max1 = isinf(max1) ? quantile(d1, prevfloat(1.0)) : max1
A = Int(min1):Int(max1)
a = [-Inf; norminvcdf.(cdf.(d1, A))]
c2 = inv(s1 * s2)
coef = zeros(Float64, n + 1)
for k in 1:n
@inbounds coef[k+1] = _Gn0_mixed(A, a, d2, c2, k, m) / factorial(big(k))
end
coef[1] = -p
xs = _feasible_roots(coef)
return _best_root(p, xs)
end
"""
_invrule_fallback(p::Float64, d1, d2; n::Int, m::Int, kwargs...)
Mixed case.
"""
function _invrule_fallback(p::Float64, d1::CUD, d2::DUD; n::Int, m::Int, kwargs...)
return _invrule_fallback(p, d2, d1; n=n, m=m, kwargs...)
end
"""
pearson_match(R::AbstractMatrix{<:Real}, margins; n::Real=12, m::Real=128, kwargs...)
Pairwise compute the Pearson correlation coefficient to be used in a bivariate Gaussian
copula. Ensures that the resulting matrix is a valid correlation matrix.
# Fields
- `R`: The target correltation matrix of the marginal distributions.
- `margins`: A list of marginal distributions.
- `n`: The degree of the polynomial used to estimate the matching correlation.
- `m`: The number of points used in the hermite polynomial interpolation.
- `kwargs`: Additional keyword arguments. Currently unused.
# Examples
```julia-repl
julia> using Distributions
julia> margins = [Beta(2, 3), Uniform(0, 1), Binomial(20, 0.2)];
julia> rho = [
1.0 0.3 0.6
0.3 1.0 0.4
0.6 0.4 1.0
];
julia> pearson_match(rho, margins)
3×3 Matrix{Float64}:
1.0 0.309111 0.612753
0.309111 1.0 0.418761
0.612753 0.418761 1.0
```
"""
function pearson_match(
R::AbstractMatrix{<:Real}, margins; n::Real=20, m::Real=128, kwargs...
)
d = length(margins)
r, s = size(R)
(r == s == d) ||
throw(DimensionMismatch("The number of margins must be the same size as the correlation matrix."))
n = Int(n)
m = Int(m)
R = SharedMatrix{Float64}(d, d)
Base.Threads.@threads for (i, j) in _idx_subsets2(d)
@inbounds R[i, j] = pearson_match(R[i, j], margins[i], margins[j]; n=n, m=m)
end
S = sdata(R)
_project_psd!(S, sqrt(eps()))
_cov2cor!(S)
return Symmetric(S)
end
| PearsonCorrelationMatch | https://github.com/adknudson/PearsonCorrelationMatch.jl.git |
|
[
"MIT"
] | 0.4.0 | 5e6d8bfd5555abe43335b531b1667390084e832f | code | 621 | """
_rule(p, d1, d2; kwargs...)
Finds ``G(p) = pₓ`` where ``p`` is the correlation used in a bivariate Gaussian copula, and
``pₓ`` is the resulting correlation between the two marginals after the NORTA step.
"""
function _rule end
"""
_invrule(p, d1, d2; kwargs...)
Finds ``Ginv(pₓ) = p`` where ``pₓ`` is the target correlation between two marginals, and
``p`` is the correlation to be used in a bivariate Gaussian copula.
"""
function _invrule end
# Uniform-Uniform
_invrule(p::Float64, ::Uniform, ::Uniform; kwargs...) = 2 * sinpi(p / 6)
_rule(p::Float64, ::Uniform, ::Uniform; kwargs...) = asin(p / 2) * 6 / π
| PearsonCorrelationMatch | https://github.com/adknudson/PearsonCorrelationMatch.jl.git |
|
[
"MIT"
] | 0.4.0 | 5e6d8bfd5555abe43335b531b1667390084e832f | code | 1121 | using Test
using PearsonCorrelationMatch
using Distributions
@testset verbose=true "Correlation Bounds" begin
@testset "Uniform-Uniform" begin
U = Uniform(0, 1)
Ginv(p) = asin(p / 2) * 6 / π
pl, pu = pearson_bounds(U, U)
@test pl ≈ Ginv(-1.0) atol=0.005
@test pu ≈ Ginv( 1.0) atol=0.005
end
@testset "Uniform-Binomial" begin
U = Uniform(0, 1)
B = Binomial(1, 0.5)
Ginv(p) = asin(p / sqrt(2)) * 2 * sqrt(3) / π
pl, pu = pearson_bounds(U, B)
@test pl ≈ Ginv(-1.0) atol=0.005
@test pu ≈ Ginv( 1.0) atol=0.005
end
@testset "Uniform-Normal" begin
U = Uniform(0, 1)
N = Normal(0, 1)
Ginv(p) = p / sqrt(π/3)
pl, pu = pearson_bounds(U, N)
@test pl ≈ Ginv(-1.0) atol=0.005
@test pu ≈ Ginv( 1.0) atol=0.005
end
@testset "Binomial-Normal" begin
B = Binomial(1, 0.5)
N = Normal(0, 1)
Ginv(p) = p / sqrt(π/2)
pl, pu = pearson_bounds(B, N)
@test pl ≈ Ginv(-1.0) atol=0.005
@test pu ≈ Ginv( 1.0) atol=0.005
end
end
| PearsonCorrelationMatch | https://github.com/adknudson/PearsonCorrelationMatch.jl.git |
|
[
"MIT"
] | 0.4.0 | 5e6d8bfd5555abe43335b531b1667390084e832f | code | 177 | using Test
using PearsonCorrelationMatch
using JuliaFormatter
if VERSION >= v"1.6"
@test JuliaFormatter.format(PearsonCorrelationMatch; verbose=false, overwrite=false)
end
| PearsonCorrelationMatch | https://github.com/adknudson/PearsonCorrelationMatch.jl.git |
|
[
"MIT"
] | 0.4.0 | 5e6d8bfd5555abe43335b531b1667390084e832f | code | 4183 | using Test
using PearsonCorrelationMatch
using PearsonCorrelationMatch: _generate_coefs, _Gn0_discrete, _Gn0_mixed
using PearsonCorrelationMatch: _hermite, _hermite_normpdf
using PearsonCorrelationMatch: _is_real, _real_roots, _feasible_roots, _nearest_root, _best_root
using Polynomials: coeffs, fromroots
using Distributions
function nothrow(f)
try
f()
catch e
println(e)
return false
end
return true
end
@testset verbose=true "Internals" begin
@testset "Generate Coefficients" begin
dA = Binomial(20, 0.2)
dB = NegativeBinomial(20, 0.002)
dC = LogitNormal(3, 1)
dD = Beta(5, 3)
@test nothrow(() -> _generate_coefs(dA, 21, 28))
@test nothrow(() -> _generate_coefs(dB, 21, 28))
@test nothrow(() -> _generate_coefs(dC, 21, 28))
@test nothrow(() -> _generate_coefs(dD, 21, 28))
end
@testset "Gn0 discrete" begin
end
@testset "Gn0 mixed" begin
end
@testset "Hermite Evaluation" begin
# Want to always return a Float64
for T in (Float16, Float32, Float64, Int, Rational{Int})
@test _hermite(T(3), 5) isa Float64
@test _hermite_normpdf(T(3), 5) isa Float64
end
@test _hermite_normpdf(Inf, 10) isa Float64
@test _hermite_normpdf(-Inf, 10) isa Float64
# Must only work for real numbers
@test_throws InexactError _hermite(3 + 4im, 5)
# `k` must be a non-negative integer
@test_throws ArgumentError _hermite(1.0, -1)
@test_throws InexactError _hermite(3.00, 5.5)
# Must always return a real number even when evaluated at ±Inf
@test _hermite_normpdf( Inf, 10) ≈ 0 atol=sqrt(eps())
@test _hermite_normpdf(-Inf, 10) ≈ 0 atol=sqrt(eps())
# Test for exactness against known polynomials
He0(x) = 1.0
He1(x) = x
He2(x) = evalpoly(x, (-1, 0, 1))
He3(x) = evalpoly(x, (0, -3, 0, 1))
He4(x) = evalpoly(x, (3, 0, -6, 0, 1))
He5(x) = evalpoly(x, (0, 15, 0, -10, 0, 1))
He6(x) = evalpoly(x, (-15, 0, 45, 0, -15, 0, 1))
He7(x) = evalpoly(x, (0, -105, 0, 105, 0, -21, 0, 1))
He8(x) = evalpoly(x, (105, 0, -420, 0, 210, 0, -28, 0, 1))
He9(x) = evalpoly(x, (0, 945, 0, -1260, 0, 378, 0, -36, 0, 1))
He10(x)= evalpoly(x, (-945, 0, 4725, 0, -3150, 0, 630, 0, -45, 0, 1))
@testset "Hermite Polynomial Function" for _ in 1:1000
width = 10_000
x = rand() * width - 0.5*width
@test _hermite(x, 0) ≈ He0(x)
@test _hermite(x, 1) ≈ He1(x)
@test _hermite(x, 2) ≈ He2(x)
@test _hermite(x, 3) ≈ He3(x)
@test _hermite(x, 4) ≈ He4(x)
@test _hermite(x, 5) ≈ He5(x)
@test _hermite(x, 6) ≈ He6(x)
@test _hermite(x, 7) ≈ He7(x)
@test _hermite(x, 8) ≈ He8(x)
@test _hermite(x, 9) ≈ He9(x)
@test _hermite(x, 10) ≈ He10(x)
end
end
@testset "Root Finding" begin
coeffs_from_roots(roots) = coeffs(fromroots(roots))
r1 = nextfloat(-1.0)
r2 = prevfloat(1.0)
r3 = eps()
r4 = 127 / 256
P1 = coeffs_from_roots([r1, 7, 7, 8])
P2 = coeffs_from_roots([r2, -1.14, -1.14, -1.14, -1.14, 1119])
P3 = coeffs_from_roots([r3, 1.01, -1.01])
P4 = coeffs_from_roots([-5, 5, r4])
P5 = coeffs_from_roots([1.01, -1.01])
P6 = coeffs_from_roots([-0.5, 0.5])
# One root at -1.0
xs = _feasible_roots(P1)
@test only(xs) ≈ r1
# One root at 1.0
xs = _feasible_roots(P2)
@test only(xs) ≈ r2
# Roots that are just outside [-1, 1]
xs = _feasible_roots(P3)
@test only(xs) ≈ r3
# Random root in (-1, 1)
xs = _feasible_roots(P4)
@test only(xs) ≈ r4
# Case of two roots just outside feasible region
xs = _feasible_roots(P5)
@test length(xs) == 0
# Case of multiple roots
xs = _feasible_roots(P6)
@test length(xs) == 2
@test _nearest_root(-0.6, xs) ≈ -0.5
end
end
| PearsonCorrelationMatch | https://github.com/adknudson/PearsonCorrelationMatch.jl.git |
|
[
"MIT"
] | 0.4.0 | 5e6d8bfd5555abe43335b531b1667390084e832f | code | 3791 | using Test
using PearsonCorrelationMatch
using Distributions
using LinearAlgebra: isposdef
@testset verbose=true "Correlation Matching" begin
dA = Beta(2, 3)
dB = Binomial(2, 0.2)
dC = Binomial(20, 0.2)
@testset "Continuous-Continuous" begin
@test -0.914 ≈ pearson_match(-0.9, dA, dA) atol=0.005
@test -0.611 ≈ pearson_match(-0.6, dA, dA) atol=0.005
@test -0.306 ≈ pearson_match(-0.3, dA, dA) atol=0.005
@test 0.304 ≈ pearson_match( 0.3, dA, dA) atol=0.005
@test 0.606 ≈ pearson_match( 0.6, dA, dA) atol=0.005
@test 0.904 ≈ pearson_match( 0.9, dA, dA) atol=0.005
end
@testset "Discrete-Discrete" begin
@test -0.937 ≈ pearson_match(-0.5, dB, dB) atol=0.005
@test -0.501 ≈ pearson_match(-0.3, dB, dB) atol=0.005
@test -0.322 ≈ pearson_match(-0.2, dB, dB) atol=0.005
@test 0.418 ≈ pearson_match( 0.3, dB, dB) atol=0.005
@test 0.769 ≈ pearson_match( 0.6, dB, dB) atol=0.005
@test 0.944 ≈ pearson_match( 0.8, dB, dB) atol=0.005
@test -0.939 ≈ pearson_match(-0.9, dC, dC) atol=0.005
@test -0.624 ≈ pearson_match(-0.6, dC, dC) atol=0.005
@test -0.311 ≈ pearson_match(-0.3, dC, dC) atol=0.005
@test 0.310 ≈ pearson_match( 0.3, dC, dC) atol=0.005
@test 0.618 ≈ pearson_match( 0.6, dC, dC) atol=0.005
@test 0.925 ≈ pearson_match( 0.9, dC, dC) atol=0.005
end
@testset "Mixed" begin
@test -0.890 ≈ pearson_match(-0.7, dB, dA) atol=0.005
@test -0.632 ≈ pearson_match(-0.5, dB, dA) atol=0.005
@test -0.377 ≈ pearson_match(-0.3, dB, dA) atol=0.005
@test 0.366 ≈ pearson_match( 0.3, dB, dA) atol=0.005
@test 0.603 ≈ pearson_match( 0.5, dB, dA) atol=0.005
@test 0.945 ≈ pearson_match( 0.8, dB, dA) atol=0.005
@test -0.928 ≈ pearson_match(-0.9, dC, dA) atol=0.005
@test -0.618 ≈ pearson_match(-0.6, dC, dA) atol=0.005
@test -0.309 ≈ pearson_match(-0.3, dC, dA) atol=0.005
@test 0.308 ≈ pearson_match( 0.3, dC, dA) atol=0.005
@test 0.613 ≈ pearson_match( 0.6, dC, dA) atol=0.005
@test 0.916 ≈ pearson_match( 0.9, dC, dA) atol=0.005
end
@testset "Uniform-Uniform" begin
U = Uniform(0, 1)
G(p) = 2 * sinpi(p / 6)
Ginv(p) = asin(p / 2) * 6 / π
pl = clamp(Ginv(-1), -1, 1)
pu = clamp(Ginv(1), -1, 1)
for p in range(pl, pu; length=10)
@test pearson_match(p, U, U) ≈ G(p) atol=0.005
end
end
@testset "Uniform-Binomial" begin
U = Uniform(0, 1)
B = Binomial(1, 0.5)
G(p) = sqrt(2) * sinpi(p / (2 * sqrt(3)))
Ginv(p) = asin(p / sqrt(2)) * 2 * sqrt(3) / π
pl, pu = Ginv(-1), Ginv(1)
for p in range(pl, pu; length=10)
@test pearson_match(p, U, B) ≈ G(p) atol=0.005
end
end
@testset "Uniform-Normal" begin
U = Uniform(0, 1)
N = Normal(0, 1)
G(p) = sqrt(π/3) * p
Ginv(p) = p / sqrt(π/3)
pl, pu = Ginv(-1), Ginv(1)
for p in range(pl, pu; length=10)
@test pearson_match(p, U, N) ≈ G(p) atol=0.005
end
end
@testset "Binomial-Normal" begin
B = Binomial(1, 0.5)
N = Normal(0, 1)
G(p) = sqrt(π/2) * p
Ginv(p) = p / sqrt(π/2)
pl, pu = Ginv(-1), Ginv(1)
for p in range(pl, pu; length=10)
@test pearson_match(p, B, N) ≈ G(p) atol=0.005
end
end
@testset "Match Correlation Matrix" begin
margins = [dA, dB, dC]
r0 = [
1.09 -0.59 0.68
-0.59 1.00 0.19
0.68 0.19 1.00
]
r = pearson_match(r0, margins)
@test isposdef(r)
end
end
| PearsonCorrelationMatch | https://github.com/adknudson/PearsonCorrelationMatch.jl.git |
|
[
"MIT"
] | 0.4.0 | 5e6d8bfd5555abe43335b531b1667390084e832f | code | 200 | using Test
using Aqua, PearsonCorrelationMatch
@testset "Quality Assurance" begin
Aqua.test_all(PearsonCorrelationMatch; ambiguities=false)
Aqua.test_ambiguities(PearsonCorrelationMatch)
end
| PearsonCorrelationMatch | https://github.com/adknudson/PearsonCorrelationMatch.jl.git |
|
[
"MIT"
] | 0.4.0 | 5e6d8bfd5555abe43335b531b1667390084e832f | code | 283 | using SafeTestsets
@safetestset "Quality Assurance" include("qa.jl")
@safetestset "Code Formatting" include("format_check.jl")
@safetestset "Utilities" include("internals.jl")
@safetestset "Pearson Matching" include("matching.jl")
@safetestset "Pearson Bounds" include("bounds.jl")
| PearsonCorrelationMatch | https://github.com/adknudson/PearsonCorrelationMatch.jl.git |
|
[
"MIT"
] | 0.4.0 | 5e6d8bfd5555abe43335b531b1667390084e832f | docs | 2314 | # PearsonCorrelationMatch
[](https://github.com/JuliaTesting/Aqua.jl)
Determine the correlation coefficient for a bivariate Gaussian copula so that the resulting samples following a Normal-to-anything (NORTA) step have the desired correlation.
This package is based on the paper by Xiao and Zhou, [Matching a correlation coefficient by a Gaussian copula](https://doi.org/10.1080/03610926.2018.1439962).
## API
Only two methods are exported:
- `pearson_bounds`
- Determines the range of admissible Pearson correlations between two distributions
- `pearson_match`
- Computes the Pearson correlation coefficient to be used in a bivariate Gaussian copula
## Usage
```julia
using PearsonCorrelationMatch, Distributions
using StatsFuns
p = 0.4 # target correlation
# Distributions can be continuous, discrete, or a mix
F = Gamma()
G = NegativeBinomial(20)
# estimate the pearson correlation bounds
pearson_bounds(F, G)
# (lower = -0.8385297744531974, upper = 0.9712817585733178)
# calculate the required input correlation
p_star = pearson_match(p, F, G)
# 0.4361868405991995
# apply the NORTA step
D = MultivariateNormal([0, 0], [1 p_star; p_star 1])
Z = rand(D, 1_000_000)
U = normcdf.(Z)
X1 = quantile.(F, U[1,:])
X2 = quantile.(G, U[2,:])
cor(X1, X2)
# 0.40007047985609534
```
## Details
It is highly recommended that any user of this package reads the reference paper first. The algorithm uses Gauss-Hermite polynomial interpolation to approximate the solution of the double integral. Target correlations near the Frechet bounds may be highly sensitive to the degree of the Hermite polynomial.
## Related Packages
While this package focuses on the bivariate case, it can be used to compute the input correlations between all pairs of marginal distributions. However, the resulting adjusted correlation matrix may not be positive definite. In that case, you can use the [NearestCorrelationMatrix.jl](https://github.com/adknudson/NearestCorrelationMatrix.jl) library to fix the adjusted correlation matrix before applying the NORTA step.
## References
* Xiao, Q., & Zhou, S. (2019). Matching a correlation coefficient by a Gaussian copula. Communications in Statistics-Theory and Methods, 48(7), 1728-1747. | PearsonCorrelationMatch | https://github.com/adknudson/PearsonCorrelationMatch.jl.git |
|
[
"MIT"
] | 0.3.7 | 902fc4b6d7322789caff6f5981999bd08cba8eea | code | 433 | module Swagger
using Downloads
using URIs
using JSON
using MbedTLS
using Dates
using LibCURL
import Base: convert, show, summary, getindex, keys, length, getproperty, setproperty!, propertynames, iterate
if isdefined(Base, :hasproperty)
import Base: hasproperty
end
import JSON: lower
abstract type SwaggerModel end
abstract type SwaggerApi end
include("client.jl")
include("json.jl")
include("val.jl")
end # module Swagger
| Swagger | https://github.com/JuliaComputing/Swagger.jl.git |
|
[
"MIT"
] | 0.3.7 | 902fc4b6d7322789caff6f5981999bd08cba8eea | code | 17991 | # collection formats
const COLL_MULTI = "multi" # aliased to CSV, as multi is not supported by Requests.jl (https://github.com/JuliaWeb/Requests.jl/issues/140)
const COLL_PIPES = "pipes"
const COLL_SSV = "ssv"
const COLL_TSV = "tsv"
const COLL_CSV = "csv"
const COLL_DLM = Dict{String,String}([COLL_PIPES=>"|", COLL_SSV=>" ", COLL_TSV=>"\t", COLL_CSV=>",", COLL_MULTI=>","])
const DATETIME_FORMATS = (Dates.DateFormat("yyyy-mm-dd HH:MM:SS.sss"), Dates.DateFormat("yyyy-mm-ddTHH:MM:SS.sss"), Dates.DateFormat("yyyy-mm-ddTHH:MM:SSZ"))
const DATE_FORMATS = (Dates.DateFormat("yyyy-mm-dd"),)
const DEFAULT_TIMEOUT_SECS = 5*60
const DEFAULT_LONGPOLL_TIMEOUT_SECS = 15*60
function convert(::Type{DateTime}, str::String)
# strip off timezone, as Julia DateTime does not parse it
if '+' in str
str = split(str, '+')[1]
end
# truncate micro/nano seconds to milliseconds, as Julia DateTime does not parse it
if '.' in str
uptosec,subsec = split(str, '.')
if length(subsec) > 3
str = uptosec * "." * subsec[1:3]
end
end
for fmt in DATETIME_FORMATS
try
return DateTime(str, fmt)
catch
# try next format
end
end
throw(SwaggerException("Unsupported DateTime format: $str"))
end
function convert(::Type{Date}, str::String)
for fmt in DATETIME_FORMATS
try
return Date(str, fmt)
catch
# try next format
end
end
throw(SwaggerException("Unsupported Date format: $str"))
end
struct SwaggerException <: Exception
reason::String
end
struct ApiException <: Exception
status::Int
reason::String
resp::Downloads.Response
error::Union{Nothing,Downloads.RequestError}
function ApiException(error::Downloads.RequestError; reason::String="")
isempty(reason) && (reason = error.message)
isempty(reason) && (reason = error.response.message)
new(error.response.status, reason, error.response, error)
end
function ApiException(resp::Downloads.Response; reason::String="")
isempty(reason) && (reason = resp.message)
new(resp.status, reason, resp, nothing)
end
end
struct Client
root::String
headers::Dict{String,String}
get_return_type::Function # user provided hook to get return type from response data
clntoptions::Dict{Symbol,Any}
downloader::Downloader
timeout::Ref{Int}
pre_request_hook::Function # user provided hook to modify the request before it is sent
function Client(root::String;
headers::Dict{String,String}=Dict{String,String}(),
get_return_type::Function=(default,data)->default,
long_polling_timeout::Int=DEFAULT_LONGPOLL_TIMEOUT_SECS,
timeout::Int=DEFAULT_TIMEOUT_SECS,
pre_request_hook::Function=noop_pre_request_hook)
clntoptions = Dict{Symbol,Any}(:throw=>false, :verbose=>false)
downloader = Downloads.Downloader()
downloader.easy_hook = (easy, opts) -> begin
Downloads.Curl.setopt(easy, LibCURL.CURLOPT_LOW_SPEED_TIME, long_polling_timeout)
end
new(root, headers, get_return_type, clntoptions, downloader, Ref{Int}(timeout), pre_request_hook)
end
end
set_user_agent(client::Client, ua::String) = set_header(client, "User-Agent", ua)
set_cookie(client::Client, ck::String) = set_header(client, "Cookie", ck)
set_header(client::Client, name::String, value::String) = (client.headers[name] = value)
set_timeout(client::Client, timeout::Int) = (client.timeout[] = timeout)
function with_timeout(fn, client::Client, timeout::Integer)
oldtimeout = client.timeout[]
client.timeout[] = timeout
try
fn(client)
finally
client.timeout[] = oldtimeout
end
end
function with_timeout(fn, api::SwaggerApi, timeout::Integer)
client = api.client
oldtimeout = client.timeout[]
client.timeout[] = timeout
try
fn(api)
finally
client.timeout[] = oldtimeout
end
end
struct Ctx
client::Client
method::String
return_type::Type
resource::String
auth::Vector{String}
path::Dict{String,String}
query::Dict{String,String}
header::Dict{String,String}
form::Dict{String,String}
file::Dict{String,String}
body::Any
timeout::Int
curl_mime_upload::Any
pre_request_hook::Function
function Ctx(client::Client, method::String, return_type, resource::String, auth, body=nothing;
timeout::Int=client.timeout[],
pre_request_hook::Function=client.pre_request_hook)
resource = client.root * resource
headers = copy(client.headers)
new(client, method, return_type, resource, auth, Dict{String,String}(), Dict{String,String}(), headers, Dict{String,String}(), Dict{String,String}(), body, timeout, nothing, pre_request_hook)
end
end
is_json_mime(mime::T) where {T <: AbstractString} = ("*/*" == mime) || occursin(r"(?i)application/json(;.*)?", mime) || occursin(r"(?i)application/(.*)-patch\+json(;.*)?", mime)
function select_header_accept(accepts::Vector{String})
isempty(accepts) && (return "application/json")
for accept in accepts
is_json_mime(accept) && (return accept)
end
return join(accepts, ", ")
end
function select_header_content_type(ctypes::Vector{String})
isempty(ctypes) && (return "application/json")
for ctype in ctypes
is_json_mime(ctype) && (return (("*/*" == ctype) ? "application/json" : ctype))
end
return ctypes[1]
end
set_header_accept(ctx::Ctx, accepts::Vector{T}) where {T} = set_header_accept(ctx, convert(Vector{String}, accepts))
function set_header_accept(ctx::Ctx, accepts::Vector{String})
accept = select_header_accept(accepts)
!isempty(accept) && (ctx.header["Accept"] = accept)
return nothing
end
set_header_content_type(ctx::Ctx, ctypes::Vector{T}) where {T} = set_header_content_type(ctx, convert(Vector{String}, ctypes))
function set_header_content_type(ctx::Ctx, ctypes::Vector{String})
ctx.header["Content-Type"] = select_header_content_type(ctypes)
return nothing
end
set_param(params::Dict{String,String}, name::String, value::Nothing; collection_format=nothing) = nothing
function set_param(params::Dict{String,String}, name::String, value::Union{Nothing,T}; collection_format=nothing) where {T}
(value === nothing) && return
if !isa(value, Vector) || isempty(collection_format)
params[name] = string(value)
else
dlm = get(COLL_DLM, collection_format, "")
isempty(dlm) && throw(SwaggerException("Unsupported collection format $collection_format"))
params[name] = join(string.(value), dlm)
end
end
function prep_args(ctx::Ctx)
kwargs = copy(ctx.client.clntoptions)
kwargs[:downloader] = ctx.client.downloader # use the default downloader for most cases
isempty(ctx.file) && (ctx.body === nothing) && isempty(ctx.form) && !("Content-Length" in keys(ctx.header)) && (ctx.header["Content-Length"] = "0")
headers = ctx.header
body = nothing
if !isempty(ctx.form)
headers["Content-Type"] = "application/x-www-form-urlencoded"
body = URIs.escapeuri(ctx.form)
end
if !isempty(ctx.file)
# use a separate downloader for file uploads
# until we have something like https://github.com/JuliaLang/Downloads.jl/pull/148
downloader = Downloads.Downloader()
downloader.easy_hook = (easy, opts) -> begin
Downloads.Curl.setopt(easy, LibCURL.CURLOPT_LOW_SPEED_TIME, long_polling_timeout)
mime = LibCURL.curl_mime_init(easy)
for (_k,_v) in ctx.file
part = LibCURL.curl_mime_addpart(mime)
LibCURL.curl_mime_name(part, _k)
LibCURL.curl_mime_filedata(part, _v)
# TODO: make provision to call curl_mime_type in future?
end
Downloads.Curl.setopt(easy, LibCURL.CURLOPT_MIMEPOST, mime)
end
kwargs[:downloader] = downloader
ctx.curl_mime_upload = mime
end
if ctx.body !== nothing
(isempty(ctx.form) && isempty(ctx.file)) || throw(SwaggerException("Can not send both form-encoded data and a request body"))
if is_json_mime(get(ctx.header, "Content-Type", "application/json"))
body = to_json(ctx.body)
elseif ("application/x-www-form-urlencoded" == ctx.header["Content-Type"]) && isa(ctx.body, Dict)
body = URIs.escapeuri(ctx.body)
elseif isa(ctx.body, SwaggerModel) && isempty(get(ctx.header, "Content-Type", ""))
headers["Content-Type"] = "application/json"
body = to_json(ctx.body)
else
body = ctx.body
end
end
kwargs[:timeout] = ctx.timeout
kwargs[:method] = uppercase(ctx.method)
kwargs[:headers] = headers
return body, kwargs
end
function header(resp::Downloads.Response, name::AbstractString, defaultval::AbstractString)
for (n,v) in resp.headers
(n == name) && (return v)
end
return defaultval
end
response(::Type{Nothing}, resp::Downloads.Response, body) = nothing::Nothing
response(::Type{T}, resp::Downloads.Response, body) where {T <: Real} = response(T, body)::T
response(::Type{T}, resp::Downloads.Response, body) where {T <: String} = response(T, body)::T
function response(::Type{T}, resp::Downloads.Response, body) where {T}
ctype = header(resp, "Content-Type", "application/json")
response(T, is_json_mime(ctype), body)::T
end
response(::Type{T}, ::Nothing, body) where {T} = response(T, true, body)
function response(::Type{T}, is_json::Bool, body) where {T}
(length(body) == 0) && return T()
response(T, is_json ? JSON.parse(String(body)) : body)::T
end
response(::Type{String}, data::Vector{UInt8}) = String(data)
response(::Type{T}, data::Vector{UInt8}) where {T<:Real} = parse(T, String(data))
response(::Type{T}, data::T) where {T} = data
response(::Type{T}, data) where {T} = convert(T, data)
response(::Type{T}, data::Dict{String,Any}) where {T} = from_json(T, data)::T
response(::Type{T}, data::Dict{String,Any}) where {T<:Dict} = convert(T, data)
response(::Type{Vector{T}}, data::Vector{V}) where {T,V} = [response(T, v) for v in data]
struct ChunkReader
buffered_input::Base.BufferStream
end
function Base.iterate(iter::ChunkReader, _state=nothing)
if eof(iter.buffered_input)
return nothing
else
out = IOBuffer()
while !eof(iter.buffered_input)
byte = read(iter.buffered_input, UInt8)
(byte == codepoint('\n')) && break
write(out, byte)
end
return (take!(out), iter)
end
end
noop_pre_request_hook(ctx::Ctx) = ctx
noop_pre_request_hook(resource_path::AbstractString, body::Any, headers::Dict{String,String}) = (resource_path, body, headers)
function do_request(ctx::Ctx, stream::Bool=false; stream_to::Union{Channel,Nothing}=nothing)
# call the user hook to allow them to modify the request context
ctx = ctx.pre_request_hook(ctx)
# prepare the url
resource_path = replace(ctx.resource, "{format}"=>"json")
for (k,v) in ctx.path
resource_path = replace(resource_path, "{$k}"=>v)
end
# append query params if needed
if !isempty(ctx.query)
resource_path = string(URIs.URI(URIs.URI(resource_path); query=escapeuri(ctx.query)))
end
body, kwargs = prep_args(ctx)
# call the user hook again, to allow them to modify the processed request
resource_path, body, headers = ctx.pre_request_hook(resource_path, body, kwargs[:headers])
kwargs[:headers] = headers
if body !== nothing
input = PipeBuffer()
write(input, body)
else
input = nothing
end
if stream
@assert stream_to !== nothing
end
resp = nothing
output = Base.BufferStream()
try
if stream
@sync begin
download_task = @async begin
try
resp = Downloads.request(resource_path;
input=input,
output=output,
kwargs...
)
catch ex
if !isa(ex, InterruptException)
@error("exception invoking request", exception=(ex,catch_backtrace()))
rethrow()
end
finally
close(output)
end
end
@async begin
try
for chunk in ChunkReader(output)
return_type = ctx.client.get_return_type(ctx.return_type, String(copy(chunk)))
data = response(return_type, resp, chunk)
put!(stream_to, data)
end
catch ex
if !isa(ex, InvalidStateException) && isopen(stream_to)
@error("exception reading chunk", exception=(ex,catch_backtrace()))
rethrow()
end
finally
close(stream_to)
end
end
@async begin
interrupted = false
while isopen(stream_to)
try
wait(stream_to)
yield()
catch ex
isa(ex, InvalidStateException) || rethrow(ex)
interrupted = true
istaskdone(download_task) || schedule(download_task, InterruptException(), error=true)
end
end
interrupted || istaskdone(download_task) || schedule(download_task, InterruptException(), error=true)
end
end
else
resp = Downloads.request(resource_path;
input=input,
output=output,
kwargs...
)
close(output)
end
finally
if ctx.curl_mime_upload !== nothing
LibCURL.curl_mime_free(ctx.curl_mime_upload)
ctx.curl_mime_upload = nothing
end
end
return resp, output
end
function exec(ctx::Ctx, stream_to::Union{Channel,Nothing}=nothing)
stream = stream_to !== nothing
resp, output = do_request(ctx, stream; stream_to=stream_to)
if resp === nothing
# request was interrupted
return nothing
end
if isa(resp, Downloads.RequestError) || !(200 <= resp.status <= 206)
throw(ApiException(resp))
end
if stream
return resp
else
data = read(output)
return_type = ctx.client.get_return_type(ctx.return_type, String(copy(data)))
return response(return_type, resp, data)
end
end
property_type(::Type{T}, name::Symbol) where {T<:SwaggerModel} = error("invalid type $T")
field_name(::Type{T}, name::Symbol) where {T<:SwaggerModel} = error("invalid type $T")
getproperty(o::T, name::Symbol) where {T<:SwaggerModel} = getfield(o, field_name(T,name))
hasproperty(o::T, name::Symbol) where {T<:SwaggerModel} = ((name in propertynames(T)) && (getproperty(o, name) !== nothing))
function setproperty!(o::T, name::Symbol, val) where {T<:SwaggerModel}
validate_property(T, name, val)
fieldtype = property_type(T, name)
fieldname = field_name(T, name)
if isa(val, fieldtype)
return setfield!(o, fieldname, val)
else
ftval = try
convert(fieldtype, val)
catch
fieldtype(val)
end
return setfield!(o, fieldname, ftval)
end
end
function getpropertyat(o::T, path...) where {T<:SwaggerModel}
val = getproperty(o, Symbol(path[1]))
rempath = path[2:end]
(length(rempath) == 0) && (return val)
if isa(val, Vector)
if isa(rempath[1], Integer)
val = val[rempath[1]]
rempath = rempath[2:end]
else
return [getpropertyat(item, rempath...) for item in val]
end
end
(length(rempath) == 0) && (return val)
getpropertyat(val, rempath...)
end
function haspropertyat(o::T, path...) where {T<:SwaggerModel}
p1 = Symbol(path[1])
ret = hasproperty(o, p1)
rempath = path[2:end]
(length(rempath) == 0) && (return ret)
val = getproperty(o, p1)
if isa(val, Vector)
if isa(rempath[1], Integer)
ret = length(val) >= rempath[1]
if ret
val = val[rempath[1]]
rempath = rempath[2:end]
end
else
return [haspropertyat(item, rempath...) for item in val]
end
end
(length(rempath) == 0) && (return ret)
haspropertyat(val, rempath...)
end
convert(::Type{T}, json::Dict{String,Any}) where {T<:SwaggerModel} = from_json(T, json)
convert(::Type{T}, v::Nothing) where {T<:SwaggerModel} = T()
show(io::IO, model::T) where {T<:SwaggerModel} = print(io, JSON.json(model, 2))
summary(model::T) where {T<:SwaggerModel} = print(io, T)
"""
is_longpoll_timeout(ex::Exception)
Examine the supplied exception and returns true if the reason is timeout
of a long polling request. If the exception is a nested exception of type
CompositeException or TaskFailedException, then navigates through the nested
exception values to examine the leaves.
"""
is_longpoll_timeout(ex) = false
is_longpoll_timeout(ex::TaskFailedException) = is_longpoll_timeout(ex.task.exception)
is_longpoll_timeout(ex::CompositeException) = any(is_longpoll_timeout, ex.exceptions)
function is_longpoll_timeout(ex::Swagger.ApiException)
ex.status == 200 && match(r"Operation timed out after \d+ milliseconds with \d+ bytes received", ex.reason) !== nothing
end
| Swagger | https://github.com/JuliaComputing/Swagger.jl.git |
|
[
"MIT"
] | 0.3.7 | 902fc4b6d7322789caff6f5981999bd08cba8eea | code | 2268 | # JSONWrapper for Swagger models handles
# - null fields
# - field names that are Julia keywords
struct JSONWrapper{T<:SwaggerModel} <: AbstractDict{Symbol, Any}
wrapped::T
flds::Vector{Symbol}
end
JSONWrapper(o::T) where {T<:SwaggerModel} = JSONWrapper(o, filter(n->hasproperty(o,n), propertynames(T)))
getindex(w::JSONWrapper, s::Symbol) = getproperty(w.wrapped, s)
keys(w::JSONWrapper) = w.flds
length(w::JSONWrapper) = length(w.flds)
function iterate(w::JSONWrapper, state...)
result = iterate(w.flds, state...)
if result === nothing
return result
else
name,nextstate = result
val = getproperty(w.wrapped, name)
return (name=>val, nextstate)
end
end
lower(o::T) where {T<:SwaggerModel} = JSONWrapper(o)
to_json(o) = JSON.json(o)
from_json(::Type{Union{Nothing,T}}, json::Dict{String,Any}) where {T} = from_json(T, json)
from_json(::Type{T}, json::Dict{String,Any}) where {T} = from_json(T(), json)
from_json(::Type{T}, json::Dict{String,Any}) where {T <: Dict} = convert(T, json)
from_json(::Type{T}, j::Dict{String,Any}) where {T <: String} = to_json(j)
from_json(::Type{Any}, j::Dict{String,Any}) = j
function from_json(o::T, json::Dict{String,Any}) where {T <: SwaggerModel}
jsonkeys = [Symbol(k) for k in keys(json)]
for name in intersect(propertynames(T), jsonkeys)
from_json(o, name, json[String(name)])
end
o
end
function from_json(o::T, name::Symbol, json::Dict{String,Any}) where {T <: SwaggerModel}
ftype = property_type(T, name)
fval = from_json(ftype, json)
setfield!(o, field_name(T,name), convert(ftype, fval))
o
end
from_json(o::T, name::Symbol, v) where {T} = (setfield!(o, field_name(T,name), convert(property_type(T, name), v)); o)
function from_json(o::T, name::Symbol, v::Vector) where {T}
# in Julia we can not support JSON null unless the element type is explicitly set to support it
ftype = property_type(T, name)
vtype = isa(ftype, Union) ? ((ftype.a === Nothing) ? ftype.b : ftype.a) : isa(ftype, Vector) ? ftype : Union{}
(Nothing <: eltype(vtype)) || filter!(x->x!==nothing, v)
setfield!(o, field_name(T,name), convert(property_type(T, name), v))
o
end
from_json(o::T, name::Symbol, v::Nothing) where {T} = o
| Swagger | https://github.com/JuliaComputing/Swagger.jl.git |
|
[
"MIT"
] | 0.3.7 | 902fc4b6d7322789caff6f5981999bd08cba8eea | code | 1904 | struct ValidationException <: Exception
reason::String
end
val_max(val, lim, excl) = (excl ? (val < lim) : (val <= lim))
val_min(val, lim, excl) = (excl ? (val > lim) : (val >= lim))
val_max_length(val, lim) = (length(val) <= lim)
val_min_length(val, lim) = (length(val) >= lim)
val_enum(val, lst) = (val in lst)
function val_enum(val::Vector, lst)
for v in val
(v in lst) || return false
end
true
end
function val_enum(val::Dict, lst)
for v in keys(val)
(v in lst) || return false
end
true
end
const MSG_INVALID_API_PARAM = Dict{Symbol,Function}([
:maximum => (val,excl)->string("must be a value less than ", excl ? "or equal to " : "", val),
:minimum => (val,excl)->string("must be a value greater than ", excl ? "or equal to " : "", val),
:maxLength => (len)->string("length must be less than or equal to ", len),
:minLength => (len)->string("length must be greater than or equal to ", len),
:maxItems => (val)->string("number of items must be less than or equal to ", val),
:minItems => (val)->string("number of items must be greater than or equal to ", val),
:enum => (lst)->string("value is not from the allowed values", lst)
])
const VAL_API_PARAM = Dict{Symbol,Function}([
:maximum => val_max,
:minimum => val_min,
:maxLength => val_max_length,
:minLength => val_min_length,
:maxItems => val_max_length,
:minItems => val_min_length,
:enum => val_enum
])
function validate_param(param, operation, rule, value, args...)
# do not validate missing values
(value === nothing) && return
VAL_API_PARAM[rule](value, args...) && return
msg = string("Invalid value ($value) of parameter ", param, " for ", operation, ", ", MSG_INVALID_API_PARAM[rule](args...))
throw(ValidationException(msg))
end
validate_property(::Type{T}, name::Symbol, val) where {T<:SwaggerModel} = nothing
| Swagger | https://github.com/JuliaComputing/Swagger.jl.git |
|
[
"MIT"
] | 0.3.7 | 902fc4b6d7322789caff6f5981999bd08cba8eea | code | 807 | using Swagger
using Test
const gencmd = joinpath(dirname(@__FILE__()), "petstore", "generate.sh")
include("utilstests.jl")
@testset "Swagger" begin
@testset "Utils" begin
test_longpoll_exception_check()
end
@testset "Code generation" begin
# set the current julia executable to be used henceforth
("JULIA" in keys(ENV)) || (ENV["JULIA"] = joinpath(Sys.BINDIR, "julia"))
@info("Generating petstore", gencmd)
run(`$gencmd`)
end
@testset "Petstore" begin
if get(ENV, "RUNNER_OS", "") == "Linux"
@info("Running petstore tests")
include("petstore/runtests.jl")
else
@info("Skipping petstore tests in non Linux environment (can not run petstore docker on OSX or Windows)")
end
end
end
| Swagger | https://github.com/JuliaComputing/Swagger.jl.git |
|
[
"MIT"
] | 0.3.7 | 902fc4b6d7322789caff6f5981999bd08cba8eea | code | 1289 | using Swagger, Test
function as_taskfailedexception(ex)
try
task = @async throw(ex)
wait(task)
catch ex
return ex
end
end
function test_longpoll_exception_check()
resp = Swagger.Downloads.Response("http", "http://localhost", 200, "no error", [])
reqerr1 = Swagger.Downloads.RequestError("http://localhost", 500, "not timeout error", resp)
reqerr2 = Swagger.Downloads.RequestError("http://localhost", 200, "Operation timed out after 300 milliseconds with 0 bytes received", resp) # timeout error
@test Swagger.is_longpoll_timeout("not an exception") == false
swaggerex1 = Swagger.ApiException(reqerr1)
@test Swagger.is_longpoll_timeout(swaggerex1) == false
@test Swagger.is_longpoll_timeout(as_taskfailedexception(swaggerex1)) == false
swaggerex2 = Swagger.ApiException(reqerr2)
@test Swagger.is_longpoll_timeout(swaggerex2)
@test Swagger.is_longpoll_timeout(as_taskfailedexception(swaggerex2))
@test Swagger.is_longpoll_timeout(CompositeException([swaggerex1, swaggerex2]))
@test Swagger.is_longpoll_timeout(CompositeException([swaggerex1, as_taskfailedexception(swaggerex2)]))
@test Swagger.is_longpoll_timeout(CompositeException([swaggerex1, as_taskfailedexception(swaggerex1)])) == false
end
| Swagger | https://github.com/JuliaComputing/Swagger.jl.git |
|
[
"MIT"
] | 0.3.7 | 902fc4b6d7322789caff6f5981999bd08cba8eea | code | 452 | include(joinpath(dirname(@__FILE__), "MyPetStore", "MyPetStore.jl"))
include("test_UserApi.jl")
include("test_StoreApi.jl")
include("test_PetApi.jl")
const server = "http://127.0.0.1/v2"
TestUserApi.test_404(server)
TestUserApi.test_userhook(server)
TestUserApi.test_set_methods()
if get(ENV, "STRESS_PETSTORE", "false") == "true"
TestUserApi.test_parallel(server)
end
TestUserApi.test(server)
TestStoreApi.test(server)
TestPetApi.test(server)
| Swagger | https://github.com/JuliaComputing/Swagger.jl.git |
|
[
"MIT"
] | 0.3.7 | 902fc4b6d7322789caff6f5981999bd08cba8eea | code | 2135 | module TestPetApi
using ..MyPetStore
using Swagger
using Test
function test(uri)
@info("PetApi")
client = Swagger.Client(uri)
api = PetApi(client)
tag1 = Tag(;id=10, name="juliacat")
tag2 = Tag(;id=11, name="white")
cat = Category(;id=10, name="cat")
@test_throws Swagger.ValidationException Pet(;id=10, category=cat, name="felix", photoUrls=nothing, tags=[tag1, tag2], status="invalid-status")
pet = Pet(;id=10, category=cat, name="felix", photoUrls=nothing, tags=[tag1,tag2], status="pending")
@info("PetApi - addPet")
@test addPet(api, pet) === nothing
@info("PetApi - updatePet")
pet.status = "available"
@test updatePet(api, pet) === nothing
@info("PetApi - updatePetWithForm")
@test updatePetWithForm(api, 10; name="meow") === nothing
@info("PetApi - getPetById")
pet10 = getPetById(api, 10)
@test pet10.id == 10
# skip test, has been failing on the test server (some operations on the server are probably dummy)
#@info("PetApi - findPetsByTags")
#tags = ["juliacat", "white"]
#pets = findPetsByTags(api, tags)
#@test isa(pets, Vector{Pet})
#@info("PetApi - got $(length(pets)) pets")
#for p in pets
# ptags = [get_field(t, "name") for t in get_field(p, "tags")]
# @test !isempty(intersect(ptags, tags))
#end
@info("PetApi - findPetsByStatus")
unsold = ["available", "pending"]
pets = findPetsByStatus(api, unsold)
@test isa(pets, Vector{Pet})
@info("PetApi - findPetsByStatus", npets=length(pets))
for p in pets
@test p.status in unsold
end
@info("PetApi - deletePet")
@test deletePet(api, 10) == nothing
# does not work yet. issue: https://github.com/JuliaWeb/Requests.jl/issues/139
#@info("PetApi - uploadFile")
#img = joinpath(dirname(@__FILE__), "cat.png")
#resp = uploadFile(api, 10; additionalMetadata="juliacat pic", file=img)
#@test isa(resp, ApiResponse)
#@test resp.code == 200
#@info("PetApi - uploadFile", typ=get_field(resp, "type"), message=get_field(resp, "message"))
nothing
end
end # module TestPetApi
| Swagger | https://github.com/JuliaComputing/Swagger.jl.git |
|
[
"MIT"
] | 0.3.7 | 902fc4b6d7322789caff6f5981999bd08cba8eea | code | 1804 | module TestStoreApi
using ..MyPetStore
using Swagger
using Test
using Dates
function test(uri)
@info("StoreApi")
client = Swagger.Client(uri)
api = StoreApi(client)
@info("StoreApi - getInventory")
inventory = getInventory(api)
@test isa(inventory, Dict{String,Int32})
@test !isempty(inventory)
@info("StoreApi - placeOrder")
@test_throws Swagger.ValidationException Order(; id=10, petId=10, quantity=2, shipDate=DateTime(2017, 03, 12), status="invalid_status", complete=false)
order = Order(; id=10, petId=10, quantity=2, shipDate=DateTime(2017, 03, 12), status="placed", complete=false)
neworder = placeOrder(api, order)
@test neworder.id == 10
@info("StoreApi - getOrderById")
@test_throws Swagger.ValidationException getOrderById(api, 0)
order = getOrderById(api, 10)
@test isa(order, Order)
@test order.id == 10
@info("StoreApi - getOrderById (async)")
response_channel = Channel{Order}(1)
@test_throws Swagger.ValidationException getOrderById(api, response_channel, 0)
@sync begin
@async begin
resp = getOrderById(api, response_channel, 10)
@test (200 <= resp.status <= 206)
end
@async begin
order = take!(response_channel)
@test isa(order, Order)
@test order.id == 10
end
end
# a closed channel is equivalent of cancellation of the call,
# no error should be thrown, but response can be nothing if call was interrupted immediately
@test !isopen(response_channel)
resp = getOrderById(api, response_channel, 10)
@test (resp === nothing) || (200 <= resp.status <= 206)
@info("StoreApi - deleteOrder")
@test deleteOrder(api, 10) === nothing
nothing
end
end # module TestStoreApi
| Swagger | https://github.com/JuliaComputing/Swagger.jl.git |
|
[
"MIT"
] | 0.3.7 | 902fc4b6d7322789caff6f5981999bd08cba8eea | code | 5225 | module TestUserApi
using ..MyPetStore
using Swagger
using Test
using Random
using JSON
using URIs
const TEST_USER = "jlswag"
const TEST_USER1 = "jlswag1"
const TEST_USER2 = "jlswag2"
function test_404(uri)
@info("Error handling")
client = Swagger.Client(uri*"_invalid")
api = UserApi(client)
try
loginUser(api, TEST_USER, "testpassword")
@error("Swagger.ApiException not thrown")
catch ex
@test isa(ex, Swagger.ApiException)
@test ex.status == 404
end
client = Swagger.Client("http://_invalid/")
api = UserApi(client)
try
loginUser(api, TEST_USER, "testpassword")
@error("Swagger.ApiException not thrown")
catch ex
@test isa(ex, Swagger.ApiException)
@test startswith(ex.reason, "Could not resolve host")
end
end
function test_set_methods()
@info("Error handling")
client = Swagger.Client("http://_invalid/")
@test client.timeout[] == Swagger.DEFAULT_TIMEOUT_SECS
Swagger.with_timeout(client, Swagger.DEFAULT_TIMEOUT_SECS + 10) do client
@test client.timeout[] == Swagger.DEFAULT_TIMEOUT_SECS + 10
end
@test client.timeout[] == Swagger.DEFAULT_TIMEOUT_SECS
api = UserApi(client)
Swagger.with_timeout(api, Swagger.DEFAULT_TIMEOUT_SECS + 10) do api
@test api.client.timeout[] == Swagger.DEFAULT_TIMEOUT_SECS + 10
end
@test client.timeout[] == Swagger.DEFAULT_TIMEOUT_SECS
Swagger.set_timeout(client, Swagger.DEFAULT_TIMEOUT_SECS + 10)
@test client.timeout[] == Swagger.DEFAULT_TIMEOUT_SECS + 10
@test isempty(client.headers)
Swagger.set_user_agent(client, "007")
Swagger.set_cookie(client, "crumbly")
@test client.headers["User-Agent"] == "007"
@test client.headers["Cookie"] == "crumbly"
end
function test_login_user_hook(ctx::Swagger.Ctx)
ctx.header["actual_password"] = "testpassword"
ctx
end
function test_login_user_hook(resource_path::AbstractString, body::Any, headers::Dict{String,String})
uri = URIs.parse_uri(resource_path)
qparams = URIs.queryparams(uri)
qparams["password"] = headers["actual_password"]
delete!(headers, "actual_password")
resource_path = string(URIs.URI(uri; query=escapeuri(qparams)))
(resource_path, body, headers)
end
function test_userhook(uri)
@info("User hook")
client = Swagger.Client(uri; pre_request_hook=test_login_user_hook)
api = UserApi(client)
login_result = loginUser(api, TEST_USER, "wrongpassword")
@test !isempty(login_result)
result = JSON.parse(login_result)
@test startswith(result["message"], "logged in user session")
@test result["code"] == 200
end
function test_parallel(uri)
@info("Parallel usage")
client = Swagger.Client(uri)
api = UserApi(client)
for gcidx in 1:100
@sync begin
for idx in 1:10^3
@async begin
@debug("[$idx] UserApi Parallel begin")
login_result = loginUser(api, TEST_USER, "testpassword")
@test !isempty(login_result)
result = JSON.parse(login_result)
@test startswith(result["message"], "logged in user session")
@test result["code"] == 200
@test_throws Swagger.ApiException getUserByName(api, randstring())
@test_throws Swagger.ApiException getUserByName(api, TEST_USER)
logout_result = logoutUser(api)
@test logout_result === nothing
@debug("[$idx] UserApi Parallel end")
end
end
end
GC.gc()
@info("outer loop $gcidx")
end
nothing
end
function test(uri)
@info("UserApi")
client = Swagger.Client(uri)
api = UserApi(client)
@info("UserApi - loginUser")
login_result = loginUser(api, TEST_USER, "testpassword")
@test !isempty(login_result)
#@info("UserApi - createUser")
#user1 = User(; id=100, username=TEST_USER1, firstName="test1", lastName="user1", email="jlswag1@example.com", password="testpass1", phone="1000000001", userStatus=0)
#@test createUser(api, user1) === nothing
#@info("UserApi - createUsersWithArrayInput")
#user2 = User(; id=200, username=TEST_USER2, firstName="test2", lastName="user2", email="jlswag2@example.com", password="testpass2", phone="1000000002", userStatus=0)
#@test createUsersWithArrayInput(api, [user1, user2]) === nothing
#@info("UserApi - createUsersWithListInput")
#@test createUsersWithListInput(api, [user1, user2]) === nothing
@info("UserApi - getUserByName")
@test_throws Swagger.ApiException getUserByName(api, randstring())
@test_throws Swagger.ApiException getUserByName(api, TEST_USER)
#getuser_result = getUserByName(api, TEST_USER)
#@test isa(getuser_result, User)
#@info("UserApi - updateUser")
#@test updateUser(api, TEST_USER2, getuser_result) === nothing
#@info("UserApi - deleteUser")
#@test deleteUser(api, TEST_USER2) === nothing
@info("UserApi - logoutUser")
logout_result = logoutUser(api)
@test logout_result === nothing
nothing
end
end # module TestUserApi
| Swagger | https://github.com/JuliaComputing/Swagger.jl.git |
|
[
"MIT"
] | 0.3.7 | 902fc4b6d7322789caff6f5981999bd08cba8eea | docs | 8869 | # Swagger
[](https://github.com/JuliaComputing/Swagger.jl/actions?query=workflow%3ACI+branch%3Amaster)
[](http://codecov.io/github/JuliaComputing/Swagger.jl?branch=master)
This is a Julia plugin and code generator to generate your own client library with Swagger.
The goal of Swagger™ is to define a standard, language-agnostic interface to REST APIs which allows both humans and computers to discover and understand the capabilities of the service without access to source code, documentation, or through network traffic inspection. When properly defined via Swagger, a consumer can understand and interact with the remote service with a minimal amount of implementation logic. Similar to what interfaces have done for lower-level programming, Swagger removes the guesswork in calling the service.
Check out [OpenAPI-Spec](https://github.com/OAI/OpenAPI-Specification) for additional information about the Swagger project, including additional libraries with support for other languages and more.
## How do I use this?
The code generation step required by this package is best done on linux. Generated julia code can of course be run on any platform.
### Building
First, you need to build the Swagger Java libraries. Ensure you have Java and maven installed, and the `java` and `mvn` commands available on the path. Then, from the directory where Swagger.jl has been downloaded/installed, run this:
```
plugin/build.sh
```
A single jar file (julia-swagger-codegen-0.0.4.jar) will be produced in `plugin/target`.
You can now use that for codegen.
Note: problems have been reported while building with JDK 9 on MacOS likely because of [this issue](https://bugs.eclipse.org/bugs/show_bug.cgi?id=534460)
### Code Generation
Use the supplied script `plugin/generate.sh` and point it to the specification file and a configuration file. E.g.:
```bash
${SWAGGERDIR}/plugin/generate.sh -i ${SPECDIR}/${SPECFILE} -o ${GENDIR} -c config.json
```
_where_
`SWAGGERDIR` is the location of this package
`SPECDIR` is the directory where the openapi specification file resides
`SPECFILE` the name of the openapi specification file from which you are generating Julia code
`GENDIR` the directory where the generated Julia code will be written
Typically, you would generate the files into a `src` directory for a package. The generated code is ready to be used as a Julia module directly.
The configuration file (`config.json`) can have the following options:
- `packageName`: the Julia package to generate (`SwaggerClient` by default)
## Generated Code Structure
### APIs
Each API set is generated into a file named `api_<apiname>.jl`. It is represented as a `struct` and the APIs under it are generated as methods. An API set can be constructed by providing the swagger client instance that it can use for communication.
The required API parameters are generated as regular function arguments. Optional parameters are generated as keyword arguments. Method documentation is generated with description, parameter information and return value. Two variants of the API are generated. The first variant is suitable for calling synchronously and returns a single instance of the result struct.
```julia
# example synchronous API that returns an Order instance
getOrderById(api::StoreApi, orderId::Int64)
```
The second variant is suitable for asynchronous calls to methods that return chunked transfer encoded responses, where in the API streams the response objects into an output channel.
```julia
# example asynchronous API that streams matching Pet instances into response_stream
findPetsByStatus(api::PetApi, response_stream::Channel, status::Vector{String})
```
A client context holds common information to be used across APIs. It also holds a connection to the server and uses that across API calls.
The client context needs to be passed as the first parameter of all API calls. It can be created as:
```julia
Client(root::String;
headers::Dict{String,String}=Dict{String,String}(),
get_return_type::Function=(default,data)->default,
timeout::Int=DEFAULT_TIMEOUT_SECS,
long_polling_timeout::Int=DEFAULT_LONGPOLL_TIMEOUT_SECS,
pre_request_hook::Function,
)
```
Where:
- `root`: the root URI where APIs are hosted (should not end with a `/`)
- `headers`: any additional headers that need to be passed along with all API calls
- `get_return_type`: optional method that can map a Julia type to a return type other than what is specified in the API specification by looking at the data (this is used only in special cases, for example when models are allowed to be dynamically loaded)
- `timeout`: optional timeout to apply for server methods (default `Swagger.DEFAULT_TIMEOUT_SECS`)
- `long_polling_timeout`: optional timeout to apply for long polling methods (default `Swagger.DEFAULT_LONGPOLL_TIMEOUT_SECS`)
- `pre_request_hook`: user provided hook to modify the request before it is sent
The `pre_request_hook` must provide the following two implementations:
- `pre_request_hook(ctx::Swagger.Ctx) -> ctx`
- `pre_request_hook(resource_path::AbstractString, body::Any, headers::Dict{String,String}) -> (resource_path, body, headers)`
In case of any errors an instance of `ApiException` is thrown. It has the following fields:
- `status::Int`: HTTP status code
- `reason::String`: Optional human readable string
- `resp::Downloads.Response`: The HTTP Response for this call
- `error::Union{Nothing,Downloads.RequestError}`: The HTTP error on request failure
An API call involves the following steps:
- If a pre request hook is provided, it is invoked with an instance of `Swagger.Ctx` that has the request attributes. The hook method is expected to make any modifications it needs to the request attributes before the request is prepared, and return the modified context.
- The URL to be invoked is prepared by replacing placeholders in the API URL template with the supplied function parameters.
- If this is a POST request, serialize the instance of `SwaggerModel` provided as the `body` parameter as a JSON document.
- If a pre request hook is provided, it is invoked with the prepared resource path, body and request headers. The hook method is expected to modify and return back a tuple of resource path, body and headers which will be used to make the request.
- Make the HTTP call to the API endpoint and collect the response.
- Determine the response type / model, invoke the optional user specified mapping function if one was provided.
- Convert (deserialize) the response data into the return type and return.
- In case of any errors, throw an instance of `ApiException`
### Models
Each model from the specification is generated into a file named `model_<modelname>.jl`. It is represented as a `mutable struct` that is a subtype of the abstract type `SwaggerModel`. Models have the following methods defined:
- constructor that takes keyword arguments to fill in values for all model properties.
- [`propertynames`](https://docs.julialang.org/en/v1/base/base/#Base.propertynames)
- [`hasproperty`](https://docs.julialang.org/en/v1/base/base/#Base.hasproperty)
- [`getproperty`](https://docs.julialang.org/en/v1/base/base/#Base.getproperty)
- [`setproperty!`](https://docs.julialang.org/en/v1/base/base/#Base.setproperty!)
In addition to these standard Julia methods, these convenience methods are also generated that help in checking value at a hierarchical path of the model.
- `function haspropertyat(o::T, path...) where {T<:SwaggerModel}`
- `function getpropertyat(o::T, path...) where {T<:SwaggerModel}`
E.g:
```julia
# access o.field.subfield1.subfield2
if haspropertyat(o, "field", "subfield1", "subfield2")
getpropertyat(o, "field", "subfield1", "subfield2")
end
# access nested array elements, e.g. o.field2.subfield1[10].subfield2
if haspropertyat(o, "field", "subfield1", 10, "subfield2")
getpropertyat(o, "field", "subfield1", 10, "subfield2")
end
```
### Validations
Following validations are incorporated into models:
- maximum value: must be a numeric value less than or equal to a specified value
- minimum value: must be a numeric value greater than or equal to a specified value
- maximum length: must be a string value of length less than or equal to a specified value
- minimum length: must be a string value of length greater than or equal to a specified value
- maximum item count: must be a list value with number of items less than or equal to a specified value
- minimum item count: must be a list value with number of items greater than or equal to a specified value
- enum: value must be from a list of allowed values
Validations are imposed in the constructor and `setproperty!` methods of models.
| Swagger | https://github.com/JuliaComputing/Swagger.jl.git |
|
[
"MIT"
] | 0.3.7 | 902fc4b6d7322789caff6f5981999bd08cba8eea | docs | 1423 | # Swagger Codegen for Julia
## Overview
This is a Julia plugin and code generator to generate your own client library with Swagger.
## What's Swagger?
The goal of Swagger™ is to define a standard, language-agnostic interface to REST APIs which allows both humans and computers to discover and understand the capabilities of the service without access to source code, documentation, or through network traffic inspection. When properly defined via Swagger, a consumer can understand and interact with the remote service with a minimal amount of implementation logic. Similar to what interfaces have done for lower-level programming, Swagger removes the guesswork in calling the service.
Check out [OpenAPI-Spec](https://github.com/OAI/OpenAPI-Specification) for additional information about the Swagger project, including additional libraries with support for other languages and more.
## How do I use this?
To build the project, run this:
```
plugin/build.sh
```
A single jar file (julia-swagger-codegen-0.0.7.jar) will be produced in `plugin/target`. You can now use that with codegen:
```
java -cp /path/to/swagger-codegen-cli.jar:/path/to/julia-swagger-codegen-0.0.7.jar io.swagger.codegen.Codegen -l julia -i /path/to/swagger.yaml -o ./test -c config.json
```
The configuration file (`config.json`) can have the following options:
- `packageName`: the Julia package to generate (`SwaggerClient` by default)
| Swagger | https://github.com/JuliaComputing/Swagger.jl.git |
|
[
"MIT"
] | 0.1.0 | eb2cfb31d60b4f22441f39f35ae0ae01381da734 | code | 613 | using CleanUpPullRequestPreviews
using Documenter
makedocs(;
modules=[CleanUpPullRequestPreviews],
authors="Brown Center for Biomedical Informatics",
repo="https://github.com/bcbi/CleanUpPullRequestPreviews.jl/blob/{commit}{path}#L{line}",
sitename="CleanUpPullRequestPreviews.jl",
format=Documenter.HTML(;
prettyurls=get(ENV, "CI", "false") == "true",
canonical="https://bcbi.github.io/CleanUpPullRequestPreviews.jl",
assets=String[],
),
pages=[
"Home" => "index.md",
],
)
deploydocs(;
repo="github.com/bcbi/CleanUpPullRequestPreviews.jl",
)
| CleanUpPullRequestPreviews | https://github.com/bcbi/CleanUpPullRequestPreviews.jl.git |
|
[
"MIT"
] | 0.1.0 | eb2cfb31d60b4f22441f39f35ae0ae01381da734 | code | 7355 | module CleanUpPullRequestPreviews
import GitHub
import HTTP
export remove_old_previews
struct AlwaysAssertionError <: Exception
msg::String
end
function always_assert(cond::Bool, message::String)
cond || throw(AlwaysAssertionError(message))
return nothing
end
struct PRandPath
pr::Int
path::String
end
struct Config
email_address::String
git_command::String
my_regex::Regex
num_samples::Int
push_to_origin::Bool
repo_main::String
repo_previews::String
repo_previews_branch::String
sample_delay_seconds::Int
username::String
end
function Config(;
email_address::AbstractString = "41898282+github-actions[bot]@users.noreply.github.com",
git_command::AbstractString = "git",
my_regex::Regex,
num_samples::Integer = 3,
push_to_origin::Bool = true,
repo_main::AbstractString,
repo_previews::AbstractString,
repo_previews_branch::AbstractString,
sample_delay_seconds::Integer = 60,
username::AbstractString = "github-actions[bot]",
)
result = Config(
email_address,
git_command,
my_regex,
num_samples,
push_to_origin,
repo_main,
repo_previews,
repo_previews_branch,
sample_delay_seconds,
username,
)
return result
end
"""
remove_old_previews
Remove old pull request previews.
"""
function remove_old_previews(;
api::GitHub.GitHubAPI = GitHub.GitHubWebAPI(HTTP.URI("https://api.github.com")),
auth::GitHub.Authorization = GitHub.authenticate(ENV["GITHUB_TOKEN"]),
kwargs...)
config = Config(; kwargs...)
remove_old_previews(api, auth, config)::Nothing
return nothing
end
"""
remove_old_previews
Remove old pull request previews.
"""
function remove_old_previews(api::GitHub.GitHubAPI,
auth::GitHub.Authorization,
config::Config)
original_directory::String = pwd()::String
clone_directory::String = _git_clone(api, auth, config)::String
prs_and_paths::Vector{PRandPath} = _get_prs_and_paths(api, auth, config, clone_directory)::Vector{PRandPath}
pr_is_open::Dict{Int, Bool} = _pr_is_open(api, auth, config, prs_and_paths)::Dict{Int, Bool}
_delete_preview_directories(api, auth, config, clone_directory, prs_and_paths, pr_is_open)::Nothing
_git_add_commit_push(api, auth, config, clone_directory)::Nothing
cd(original_directory)
rm(clone_directory; force = true, recursive = true)
return nothing
end
function _git_clone(api::GitHub.GitHubAPI,
auth::GitHub.Authorization,
config::Config)::String
parent_dir = mktempdir()
atexit(() -> rm(parent_dir; force = true, recursive = true))
original_directory = pwd()
cd(parent_dir)
run(`$(config.git_command) clone $(config.repo_previews) CLONEDIRECTORY`)
clone_directory::String = joinpath(parent_dir, "CLONEDIRECTORY")::String
cd(clone_directory)
run(Cmd(String[config.git_command, "config", "user.name", strip(config.username)]))
run(Cmd(String[config.git_command, "config", "user.email", strip(config.email_address)]))
run(`$(config.git_command) checkout $(config.repo_previews_branch)`)
cd(original_directory)
return clone_directory
end
function _get_prs_and_paths(api::GitHub.GitHubAPI,
auth::GitHub.Authorization,
config::Config,
root_directory::AbstractString)::Vector{PRandPath}
result::Vector{PRandPath} = Vector{PRandPath}(undef, 0)::Vector{PRandPath}
original_directory = pwd()
cd(root_directory)
for (root, dirs, files) in walkdir(".")
for dir in dirs
path = joinpath(root, dir)
m = match(config.my_regex, path)
if m !== nothing
pr = parse(Int, m[1])
push!(result, PRandPath(pr, path))
end
end
end
cd(original_directory)
return result
end
function _pr_is_open(api::GitHub.GitHubAPI,
auth::GitHub.Authorization,
config::Config,
prs_and_paths)::Dict{Int, Bool}
list_of_all_pr_numbers::Vector{Int} = Int[pr_and_path.pr for pr_and_path in prs_and_paths]::Vector{Int}
unique!(list_of_all_pr_numbers)
sort!(list_of_all_pr_numbers)
intermediate = Vector{Dict{Int, Bool}}(undef, config.num_samples)
for i = 1:config.num_samples
intermediate[i] = Dict{Int, Bool}()
end
for i = 1:config.num_samples
if i != 1
@info("Waiting for $(config.sample_delay_seconds) second(s)...")
sleep(config.sample_delay_seconds)
end
for pr_number in list_of_all_pr_numbers
@info("Getting state of PR # $(pr_number)")
github_pr = GitHub.pull_request(api,
config.repo_main,
pr_number;
auth = auth)
github_pr_is_open = github_pr.state != "closed"
intermediate[i][pr_number] = github_pr_is_open
end
end
result::Dict{Int, Bool} = Dict{Int, Bool}()::Dict{Int, Bool}
for pr_number in list_of_all_pr_numbers
pr_intermediates = Vector{Bool}(undef, config.num_samples)
for i = 1:config.num_samples
pr_intermediates[i] = intermediate[i][pr_number]
end
result[pr_number] = any(pr_intermediates)
@info("PR # $(pr_number) is open: $(result[pr_number])")
end
return result
end
function _git_add_commit_push(api::GitHub.GitHubAPI,
auth::GitHub.Authorization,
config::Config,
clone_directory::AbstractString)::Nothing
original_directory = pwd()
cd(clone_directory)
run(`$(config.git_command) add -A`)
p = run(pipeline(`$(config.git_command) commit -m "Automated commit created by CleanUpPullRequestPreviews.jl"`; stdout=stdout, stderr=stderr); wait = false)
wait(p)
@info("Commit succeeded = $(success(p))")
if config.push_to_origin
run(`$(config.git_command) push origin --all`)
end
cd(original_directory)
return nothing
end
function _delete_preview_directories(api::GitHub.GitHubAPI,
auth::GitHub.Authorization,
config::Config,
clone_directory::AbstractString,
prs_and_paths::Vector{PRandPath},
pr_is_open::Dict{Int, Bool})::Nothing
original_directory = pwd()
for pr_and_path in prs_and_paths
pr = pr_and_path.pr
path = pr_and_path.path
this_pr_is_open = pr_is_open[pr]
if !this_pr_is_open
cd(clone_directory)
@info("Removing \"$(path)\"")
rm(path; force = true, recursive = true)
end
end
cd(original_directory)
return nothing
end
end # end module CleanUpPullRequestPreviews
| CleanUpPullRequestPreviews | https://github.com/bcbi/CleanUpPullRequestPreviews.jl.git |
|
[
"MIT"
] | 0.1.0 | eb2cfb31d60b4f22441f39f35ae0ae01381da734 | code | 125 | using CleanUpPullRequestPreviews
using Test
@testset "CleanUpPullRequestPreviews.jl" begin
# Write your tests here.
end
| CleanUpPullRequestPreviews | https://github.com/bcbi/CleanUpPullRequestPreviews.jl.git |
|
[
"MIT"
] | 0.1.0 | eb2cfb31d60b4f22441f39f35ae0ae01381da734 | docs | 2032 | # CleanUpPullRequestPreviews
[](https://bcbi.github.io/CleanUpPullRequestPreviews.jl/stable)
[](https://bcbi.github.io/CleanUpPullRequestPreviews.jl/dev)
[](https://github.com/bcbi/CleanUpPullRequestPreviews.jl/actions)
[](https://codecov.io/gh/bcbi/CleanUpPullRequestPreviews.jl)
CleanUpPullRequestPreviews automatically deletes old pull request previews from Documenter, Franklin, etc.
## Example usage
### Step 1:
Create a file named `.ci/Project.toml` with the following contents:
```toml
[deps]
CleanUpPullRequestPreviews = "06c59129-2005-40e8-8e6e-18d91e04568a"
```
### Step 2:
Make sure that you have a deploy key with write permissions on `JuliaHealth/juliahealth.github.io-previews`, and the private key for this deploy key is available as a secret named `FRANKLIN_PRIV_PREVIEW`.
Create a file named `.github/workflows/CleanUpPullRequestPreviews.yml` with the following contents:
```yaml
name: CleanUpPullRequestPreviews
on:
schedule:
- cron: '00 00 * * *' # run one time per day
jobs:
CleanUpPullRequestPreviews:
runs-on: ubuntu-latest
steps:
- name: Install SSH Client (pull request previews)
uses: webfactory/ssh-agent@v0.2.0
with:
ssh-private-key: ${{ secrets.FRANKLIN_PRIV_PREVIEW }}
- run: julia --project=.ci/ -e 'using Pkg; Pkg.instantiate()'
- name: CleanUpPullRequestPreviews.remove_old_previews
env:
GITHUB_TOKEN: ${{ secrets.GITHUB_TOKEN }}
run: julia -e 'using CleanUpPullRequestPreviews; CleanUpPullRequestPreviews.remove_old_previews(; repo_main = "JuliaHealth/juliahealth.github.io", repo_previews = "git@github.com:JuliaHealth/juliahealth.github.io-previews.git", repo_previews_branch = "gh-pages")'
```
| CleanUpPullRequestPreviews | https://github.com/bcbi/CleanUpPullRequestPreviews.jl.git |
|
[
"MIT"
] | 0.1.0 | eb2cfb31d60b4f22441f39f35ae0ae01381da734 | docs | 158 | ```@meta
CurrentModule = CleanUpPullRequestPreviews
```
# CleanUpPullRequestPreviews
```@index
```
```@autodocs
Modules = [CleanUpPullRequestPreviews]
```
| CleanUpPullRequestPreviews | https://github.com/bcbi/CleanUpPullRequestPreviews.jl.git |
|
[
"MIT"
] | 2.1.0 | 67aa3f6fbbdb0a64b80d133acf8c84b296f75033 | code | 6767 | using GitHub, MassInstallAction
import HTTP
# This demo adds or updates workflow files (TagBot, Documenter, and CI) for a set of repositories
# in an organization. In general you should copy/paste these lines after editing them
# to suit your needs. Note that you'll need to specify your GitHub personal access token,
# specify the organization/user, and select repositories interactively.
# authenticate
auth = authenticate(#= your-github-personal-access-token =#) # don't ever post this publicly!
# Get all the repositories in the JuliaDebug organization
rs = repos(#= organization/user goes here =#, true; auth=auth)[1] # change true -> false for users (true = "org")
# Optional: filter on various properties, e.g.,
# filter!(r -> !r.fork, rs) # eliminate all repos that are forks of other repos
# Manually pick a subset of the repositories, using an interactive menu
using REPL.TerminalMenus
menu = MultiSelectMenu([rp.name for rp in rs])
sel = request(menu) # hit arrows to move through list, Enter to select, and 'd' when done
rs = rs[collect(sel)]
## Key/secret creation
# Create a `DOCUMENTER_KEY` secret and corresponding `Documenter` deploy key, unless the repository
# already has a secret of that name.
# This key is used by TagBot as well as Documenter, so do this even if you don't have any `docs/` in your repository.
for r in rs
has_secret = false
for s in secrets(r; auth=auth)[1]
if s.name == "DOCUMENTER_KEY"
has_secret = true
break
end
end
has_secret && continue
pubkey, privkey = GitHub.genkeys()
create_deploykey(r; auth=auth, params=Dict("key"=>pubkey, "title"=>"Documenter", "read_only"=>false))
create_secret(r, "DOCUMENTER_KEY"; auth=auth, value=privkey)
end
## TagBot
# Add or modify the TagBot workflow to use `DOCUMENTER_KEY`. Here we use the templates in MassInstallAction directly,
# but alternatively we could create a workflow script here and use `Workflow("TagBot", "TagBot.yml" => script)`.
#
# Related: https://discourse.julialang.org/t/ann-required-updates-to-tagbot-yml/49249
workflow = MassInstallAction.tag_bot()
for r in rs
MassInstallAction.install(workflow, r; auth=auth, commit_message="Set up TagBot workflow")
end
## Documenter
script = """
name: Documenter
on:
push:
branches: [master]
tags: [v*]
pull_request:
jobs:
Documenter:
name: Documentation
runs-on: ubuntu-latest
steps:
- uses: actions/checkout@v2
- uses: julia-actions/julia-buildpkg@latest
- uses: julia-actions/julia-docdeploy@latest
env:
GITHUB_TOKEN: \${{ secrets.GITHUB_TOKEN }}
DOCUMENTER_KEY: \${{ secrets.DOCUMENTER_KEY }}
"""
workflow = MassInstallAction.Workflow("Documenter", "Documenter.yml" => script)
for r in rs
# Check whether the repo has docs, if not skip it
filesdirs = directory(r, "."; auth=auth)[1]
idx = findfirst(fd -> fd.typ == "dir" && fd.name == "docs", filesdirs)
if idx !== nothing
MassInstallAction.install(workflow, r; auth=auth, commit_message="Build docs on GitHub Actions")
end
end
## CI
# Default CI workflow that tests on the same versions as the current .travis.yml
ci_pre = """
name: CI
on:
pull_request:
push:
branches:
- master
tags: '*'
jobs:
test:
name: Julia \${{ matrix.version }} - \${{ matrix.os }} - \${{ matrix.arch }} - \${{ github.event_name }}
runs-on: \${{ matrix.os }}
strategy:
fail-fast: false
matrix:
version:
"""
ci_post = """
os:
- ubuntu-latest
- macOS-latest
- windows-latest
arch:
- x64
steps:
- uses: actions/checkout@v2
- uses: julia-actions/setup-julia@v1
with:
version: \${{ matrix.version }}
arch: \${{ matrix.arch }}
- uses: actions/cache@v1
env:
cache-name: cache-artifacts
with:
path: ~/.julia/artifacts
key: \${{ runner.os }}-test-\${{ env.cache-name }}-\${{ hashFiles('**/Project.toml') }}
restore-keys: |
\${{ runner.os }}-test-\${{ env.cache-name }}-
\${{ runner.os }}-test-
\${{ runner.os }}-
- uses: julia-actions/julia-buildpkg@v1
- uses: julia-actions/julia-runtest@v1
- uses: julia-actions/julia-processcoverage@v1
- uses: codecov/codecov-action@v1
with:
file: lcov.info
"""
for r in rs
# Check whether the repo has a workflow called "CI.yml", and if so skip it
try
filesdirs = directory(r, ".github/workflows"; auth=auth)[1]
idx = findfirst(fd -> lowercase(fd.name) ∈ ("ci.yml", "ci.yaml"), filesdirs)
if idx !== nothing
println("\n\n$(r.name) skipped due to presence of $(filesdirs[idx].name)")
continue
end
catch
end
try
# Parse the .travis.yml file and extract the versions
url = file(r, ".travis.yml"; auth=auth).download_url
tscript = String(HTTP.get(url).body)
lines = split(chomp(tscript), '\n')
idx = findfirst(isequal("julia:"), lines)
vs = String[]
while idx < length(lines)
idx += 1
m = match(r"^\s*-\s*(nightly|[\d\.]*)", lines[idx])
m === nothing && break
push!(vs, m.captures[1])
end
# optional: delete "nightly"
if !isempty(vs)
# Build the replacement CI.yml
script = ci_pre
for v in vs
line = " "^10 * "- '" * v * "'\n"
# optional: comment out nightly
if v == "nightly"
line = "#" * line
end
script *= line
end
script *= ci_post
todelete = ["../../.travis.yml"]
# Find out if there's an appveyor and set it up for deletion
filesdirs = directory(r, "."; auth=auth)[1]
idx = findfirst(fd -> fd.typ == "file" && fd.name ∈ ("appveyor.yml", ".appveyor.yml"), filesdirs)
if idx !== nothing
push!(todelete, "../../" * filesdirs[idx].name)
end
workflow = MassInstallAction.Workflow("CI", "CI.yml" => script, todelete)
println("\n\n$(r.name) had .travis.yml script:\n", tscript, "\nInstalling the following CI.yml script:\n", script)
println("OK to submit pull request? (y/n)")
resp = readline(; keep=false)
if resp ∈ ("y", "Y", "yes", "Yes", "YES")
MassInstallAction.install(workflow, r; auth=auth, commit_message="Switch to GitHub Actions for CI")
end
end
catch
println("\n\nNo .travis.yml found for $(r.name)")
end
end
| MassInstallAction | https://github.com/julia-actions/MassInstallAction.jl.git |
|
[
"MIT"
] | 2.1.0 | 67aa3f6fbbdb0a64b80d133acf8c84b296f75033 | code | 402 | """
MassInstallAction
Install or update GitHub Action workflows on repositories
API (all require qualification with `MassInstallAction`):
- Workflow creation: `Workflow`, `compat_helper`, `tag_bot`
- Workflow installation: `install`
"""
module MassInstallAction
include("types.jl")
include("public.jl")
include("default_workflows.jl")
include("repo_url.jl")
include("utils.jl")
end # module
| MassInstallAction | https://github.com/julia-actions/MassInstallAction.jl.git |
|
[
"MIT"
] | 2.1.0 | 67aa3f6fbbdb0a64b80d133acf8c84b296f75033 | code | 706 | import HTTP
function compat_helper()
name = "CompatHelper"
files_to_create = Dict{String, String}()
files_to_create["CompatHelper.yml"] = String(HTTP.get("https://raw.githubusercontent.com/JuliaRegistries/CompatHelper.jl/master/.github/workflows/CompatHelper.yml").body)
files_to_delete = Set{String}()
return Workflow(name, files_to_create, files_to_delete)
end
function tag_bot()
name = "TagBot"
files_to_create = Dict{String, String}()
files_to_create["TagBot.yml"] = String(HTTP.get("https://raw.githubusercontent.com/JuliaRegistries/TagBot/master/example.yml").body)
files_to_delete = Set{String}()
return Workflow(name, files_to_create, files_to_delete)
end
| MassInstallAction | https://github.com/julia-actions/MassInstallAction.jl.git |
|
[
"MIT"
] | 2.1.0 | 67aa3f6fbbdb0a64b80d133acf8c84b296f75033 | code | 6413 | # import GitCommand: git
import GitHub
function git(f)
return f("git")
end
"""
MassInstallAction.install(workflow, user_or_org::AbstractString, [pkgs];
token, cc::AbstractVector{<:AbstractString},
ignore_forks::Bool=true,
kwargs...)
Submit a pull request to install `workflow` for all packages owned by `user_or_org`.
`token` is your personal access token for authentication, or `nothing` if you do not have privileges and need to fork the package.
`cc` is a list of GitHub usernames that should receive notification about this pull request (beyond the regular watchers).
Additional keyword arguments `kwargs...` are forwarded directly to
`MassInstallAction.install(workflow, repo::GitHub.Repo; kwargs...)`.
If `pkgs` is supplied, pull requests will only be made to the listed packages.
"""
function install(
workflow::Workflow,
org::AbstractString;
token::Union{AbstractString,Nothing},
cc::AbstractVector{<:AbstractString},
ignore_forks::Bool=true,
kwargs...
)
if token === nothing
auth = nothing
else
auth = GitHub.authenticate(token)
end
if auth === nothing
orgrepos, page_data = GitHub.repos(org)
else
orgrepos, page_data = GitHub.repos(org; auth = auth)
end
if ignore_forks
orgrepos = [repo for repo in orgrepos if repo.fork == false]
end
pkgs = Vector{String}(undef, 0)
for r in orgrepos
name = r.name
endswith(name, ".jl") && push!(pkgs, name[1:(end-3)])
end
unique!(pkgs)
sort!(pkgs)
install(workflow,
org,
pkgs;
cc = cc,
token = token,
kwargs...)
return nothing
end
function install(workflow::Workflow,
org::AbstractString,
pkgs::AbstractVector{<:AbstractString};
token,
cc::AbstractVector{<:AbstractString},
kwargs...)
if token === nothing
auth = nothing
else
auth = GitHub.authenticate(token)
end
cc_string = string("cc: ", join(string.("@",
strip.(strip.(strip.(cc),
'@'))), " "))
pr_body = "This pull request sets up the $(workflow.name) workflow on this repository. $(cc_string)"
for pkg in pkgs
if auth === nothing
pkgrepo = GitHub.repo("$(org)/$(pkg).jl")
else
pkgrepo = GitHub.repo("$(org)/$(pkg).jl";
auth = auth)
end
install(workflow, pkgrepo; auth=auth, pr_body=pr_body, kwargs...)
# Avoid GitHub secondary rate limits
# https://docs.github.com/en/rest/overview/resources-in-the-rest-api#secondary-rate-limits
sleep(1)
end
end
"""
MassInstallAction.install(workflow, repo::GitHub.Repo;
auth, pkg_url_type::Symbol = :html,
pr_branch_name=..., pr_title=..., pr_body=..., commit_message=...)
Submit a pull request to install `workflow` for repository `repo`. This version of `install` is
designed to work in concert with [GitHub.jl](https://github.com/JuliaWeb/GitHub.jl), so that you can run queries, filter results,
and then submit changes. See the documentation of that package for more detail about `repo` and `auth`.
`pkg_url_type` can be `:html` or `:ssh`.
The remaining keywords are all strings, and have generic defaults but allow customization.
"""
function install(workflow::Workflow,
repo::GitHub.Repo;
auth::Union{Nothing,GitHub.Authorization},
pr_branch_name::AbstractString = "massinstallaction/set-up-$(workflow.name)",
pr_title::AbstractString = "MassInstallAction: Install the $(workflow.name) workflow on this repository",
pr_body::AbstractString = "This pull request sets up the $(workflow.name) workflow on this repository.",
commit_message::AbstractString = "Automated commit made by MassInstallAction.jl",
pkg_url_type::Symbol = :html)
if pkg_url_type === :html
pkg_url_with_auth = repo.html_url.uri
elseif pkg_url_type === :ssh
pkg_url_with_auth = repo.ssh_url.uri
else
throw(ArgumentError("`pkg_url_type = $(pkg_url_type)` not supported"))
end
with_temp_dir() do tmp_dir
git() do git
cd(tmp_dir)
run(`$(git) clone $(pkg_url_with_auth) REPO`)
cd("REPO")
run(`$(git) checkout -B $(pr_branch_name)`)
workflows_directory = joinpath(pwd(), ".github", "workflows")
mkpath(workflows_directory)
cd(workflows_directory)
for filename in workflow.files_to_delete
rm(filename; force = true, recursive = true)
end
for filename in keys(workflow.files_to_create)
file_content = workflow.files_to_create[filename]
open(filename, "w") do io
print(io, file_content)
end
end
try
run(`$(git) add -A`)
run(`$(git) commit -m $(commit_message)`)
try
run(`$(git) push --force origin $(pr_branch_name)`)
catch
# try again?
run(`$(git) push --force origin $(pr_branch_name)`)
end
params = Dict{String, String}()
params["title"] = pr_title
params["head"] = pr_branch_name
params["base"] = repo.default_branch
params["body"] = pr_body
if auth === nothing
GitHub.create_pull_request(repo;
params = params)
else
GitHub.create_pull_request(repo;
params = params,
auth = auth)
end
@info "Pull request submitted for $(repo.name)"
catch error
@warn "Assembling the pull request failed, skipping $(repo.name)"
show(error)
end
end
end
return nothing
end
| MassInstallAction | https://github.com/julia-actions/MassInstallAction.jl.git |
|
[
"MIT"
] | 2.1.0 | 67aa3f6fbbdb0a64b80d133acf8c84b296f75033 | code | 290 | function _get_repo_url_with_auth(org::AbstractString, pkg::AbstractString; token::Union{AbstractString, Nothing})
if token === nothing
return "https://github.com/$(org)/$(pkg).jl"
else
return "https://x-access-token:$(token)@github.com/$(org)/$(pkg).jl"
end
end
| MassInstallAction | https://github.com/julia-actions/MassInstallAction.jl.git |
|
[
"MIT"
] | 2.1.0 | 67aa3f6fbbdb0a64b80d133acf8c84b296f75033 | code | 1395 | """
Workflow(actionname, files_to_create, [files_to_delete])
Create or modify a GitHub Action workflow. `actionname` is the name given to the action when it runs.
`files_to_create` should consist of one or more `filename => filecontents` pairs specifying the workflow file(s)
you want to create or update. The optional `files_to_delete` lists one or more workflow files you want to delete.
"""
struct Workflow
name::String
files_to_create::Dict{String, String}
files_to_delete::Set{String}
# Block the generic fallback
Workflow(name::String, files_to_create::Dict{String, String}, files_to_delete::Set{String}) = new(name, files_to_create, files_to_delete)
end
Workflow(name::AbstractString, files_to_create, files_to_delete) = Workflow(String(name), normdict(files_to_create), normss(files_to_delete))
Workflow(name::AbstractString, files_to_create) = Workflow(String(name), normdict(files_to_create), Set{String}())
normdict(files_to_create::AbstractDict) = convert(Dict{String,String}, files_to_create)
normdict(files_to_create::Pair) = Dict{String,String}(files_to_create)
normdict(files_to_create::AbstractVector{<:Pair}) = Dict{String,String}(files_to_create)
normss(files_to_delete::AbstractSet) = convert(Set{String}, files_to_delete)
normss(files_to_delete) = Set{String}(files_to_delete)
normss(files_to_delete::AbstractString) = normss([files_to_delete])
| MassInstallAction | https://github.com/julia-actions/MassInstallAction.jl.git |
|
[
"MIT"
] | 2.1.0 | 67aa3f6fbbdb0a64b80d133acf8c84b296f75033 | code | 292 | function with_temp_dir(f::Function)
original_directory = pwd()
tmp_dir = mktempdir()
atexit(() -> rm(tmp_dir; force = true, recursive = true))
cd(tmp_dir)
result = f(tmp_dir)
cd(original_directory)
rm(tmp_dir; force = true, recursive = true)
return result
end
| MassInstallAction | https://github.com/julia-actions/MassInstallAction.jl.git |
|
[
"MIT"
] | 2.1.0 | 67aa3f6fbbdb0a64b80d133acf8c84b296f75033 | code | 1679 | using MassInstallAction
using Test
@testset "MassInstallAction.jl" begin
@testset "types.jl" begin
@test MassInstallAction.Workflow("a", Dict("file"=>"stuff"), Set(["bad"])) isa MassInstallAction.Workflow
@test MassInstallAction.Workflow("a", Dict("file"=>"stuff"), "bad") isa MassInstallAction.Workflow
@test MassInstallAction.Workflow("a", Dict("file"=>"stuff")) isa MassInstallAction.Workflow
@test MassInstallAction.Workflow("a", "file"=>"stuff", Set(["bad"])) isa MassInstallAction.Workflow
@test MassInstallAction.Workflow("a", "file"=>"stuff", "bad") isa MassInstallAction.Workflow
@test MassInstallAction.Workflow("a", "file"=>"stuff") isa MassInstallAction.Workflow
@test MassInstallAction.Workflow("a", ["file1"=>"stuff1", "file2"=>"stuff2"], Set(["bad"])) isa MassInstallAction.Workflow
@test MassInstallAction.Workflow("a", ["file1"=>"stuff1", "file2"=>"stuff2"], "bad") isa MassInstallAction.Workflow
@test MassInstallAction.Workflow("a", ["file1"=>"stuff1", "file2"=>"stuff2"]) isa MassInstallAction.Workflow
end
@testset "public.jl" begin
end
@testset "repo_url.jl" begin
@test MassInstallAction._get_repo_url_with_auth("MYORG", "MYPKG"; token = "MYTOKEN") == "https://x-access-token:MYTOKEN@github.com/MYORG/MYPKG.jl"
@test MassInstallAction._get_repo_url_with_auth("MYORG", "MYPKG"; token = nothing) == "https://github.com/MYORG/MYPKG.jl"
end
@testset "default workflows" begin
@test MassInstallAction.compat_helper() isa MassInstallAction.Workflow
@test MassInstallAction.tag_bot() isa MassInstallAction.Workflow
end
end
| MassInstallAction | https://github.com/julia-actions/MassInstallAction.jl.git |
|
[
"MIT"
] | 2.1.0 | 67aa3f6fbbdb0a64b80d133acf8c84b296f75033 | docs | 4058 | # MassInstallAction
[](https://github.com/julia-actions/MassInstallAction.jl/actions?query=workflow%3ACI)
[](https://codecov.io/gh/julia-actions/MassInstallAction.jl)
Install a [GitHub Action](https://docs.github.com/en/free-pro-team@latest/actions) [workflow file](https://docs.github.com/en/free-pro-team@latest/actions/reference/workflow-syntax-for-github-actions) in one or more repositories.
There are two key steps: (1) creating the `workflow` and (2) installing it, a.k.a,
submitting it as pull request(s) to one or more packages.
## Creating the workflow
Some workflows have convenient helpers:
```julia
julia> workflow = MassInstallAction.compat_helper() # workflow for https://github.com/JuliaRegistries/CompatHelper.jl
julia> workflow = MassInstallAction.tag_bot() # workflow for https://github.com/JuliaRegistries/TagBot
```
or you can create your own:
```julia
workflow = MassInstallAction.Workflow("MyWorkflow", "workflow_filename.yml" => read("/home/me/template_workflow_file.yml", String))
```
where you replace:
- `"/home/me/template_workflow_file.yml"` with the path to the local file you've prepared with the desired contents of the workflow;
- `"workflow_filename.yml"` with the name you want the file to have in the repositories' `.github/workflows` directory;
- `"MyWorkflow"` with the name used to identify this workflow when Actions runs.
You can add multiple workflow files simultaneously or even delete files, see `?MassInstallAction.Workflow`.
## Installing the workflow: examples
### Install a workflow on all repositories in your GitHub organization
First make sure that you have an environment variable
named `MY_GITHUB_TOKEN` that contains a GitHub personal
access token (see below), and then run the following code.
Replace
`MY_ORGANIZATION` with the name of your GitHub
organization.
Replace `MY_USERNAME`, `ANOTHER_ORG_ADMIN`, etc. with your username and the
usernames of other administrators in your GitHub
organization.
```julia
julia> using MassInstallAction
julia> workflow = MassInstallAction.compat_helper()
julia> MassInstallAction.install(workflow,
"MY_ORGANIZATION";
token = ENV["MY_GITHUB_TOKEN"],
cc = ["MY_USERNAME", "ANOTHER_ORG_ADMIN"])
```
### Install a workflow on all repositories in your personal GitHub account
First make sure that you have an environment variable
named `MY_GITHUB_TOKEN` that contains a GitHub personal
access token, and then run the following code.
Replace `MY_USERNAME` with your GitHub username.
```julia
julia> using MassInstallAction
julia> workflow = MassInstallAction.compat_helper()
julia> MassInstallAction.install(workflow,
"MY_USERNAME";
token = ENV["MY_GITHUB_TOKEN"],
cc = ["MY_USERNAME"])
```
### Interaction with GitHub.jl
This package uses and interacts with [GitHub.jl](https://github.com/JuliaWeb/GitHub.jl). In addition to the options above, you can `MassInstallAction.install(workflow, repo::GitHub.Repo)` directly. This may be the easiest approach if you want to filter repositories based on specific criteria. See `?MassInstallAction.install` for more information.
### Generating GitHub personal access token
To generate the GitHub personal access token, follow the instructions from [this official link](https://docs.github.com/en/free-pro-team@latest/github/authenticating-to-github/creating-a-personal-access-token).
You should give these access in your GitHub token:
- repo access:
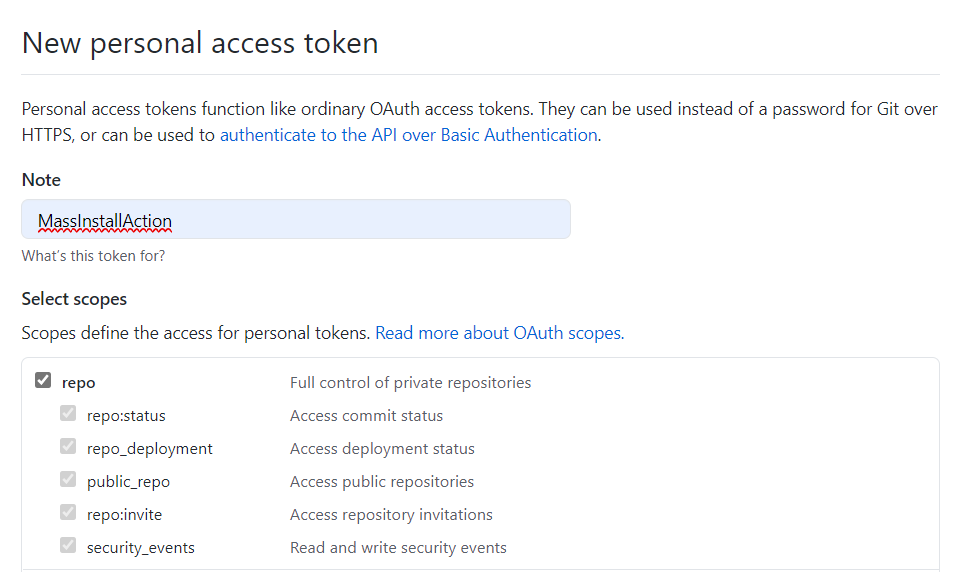
- workflow access:
| MassInstallAction | https://github.com/julia-actions/MassInstallAction.jl.git |
|
[
"MIT"
] | 0.1.1 | d192a27769e653b00dfa3d7e49dba341aea8dc50 | code | 338 | push!(LOAD_PATH, "../src/")
using OohataHuzinaga
using Documenter
makedocs(
sitename = "OohataHuzinaga.jl",
modules = [OohataHuzinaga],
pages = [
"Home" => "index.md",
"API" => ["Input" => "input.md", "Basis Sets" => "basis.md"],
],
)
deploydocs(; repo = "github.com/HartreeFoca/OohataHuzinaga.jl.git")
| OohataHuzinaga | https://github.com/HartreeFoca/OohataHuzinaga.jl.git |
|
[
"MIT"
] | 0.1.1 | d192a27769e653b00dfa3d7e49dba341aea8dc50 | code | 576 | module OohataHuzinaga
using LinearAlgebra
using SpecialFunctions
using TimerOutputs
include("atoms.jl")
include("molecule.jl")
include("sto3g.jl")
include("basis.jl")
include("auxiliary.jl")
include("overlap.jl")
include("kinetic.jl")
include("boys.jl")
include("attraction.jl")
include("repulsion.jl")
include("hartreefock.jl")
export Molecule
export molecule
export GaussianBasis
export buildbasis
export doublefactorial
export gaussianproduct
export normalization
export overlap
export kinetic
export boys
export attraction
export repulsion
export computeenergy
end
| OohataHuzinaga | https://github.com/HartreeFoca/OohataHuzinaga.jl.git |
|
[
"MIT"
] | 0.1.1 | d192a27769e653b00dfa3d7e49dba341aea8dc50 | code | 281 | Z = Dict(
"H" => 1,
"He" => 2,
"Li" => 3,
"Be" => 4,
"B" => 5,
"C" => 6,
"N" => 7,
"O" => 8,
"F" => 9,
"Ne" => 10,
)
function orbitalconfig(Z::Int64)
if Z < 3
return ["1s"]
else
return ["1s", "2s", "2p"]
end
end
| OohataHuzinaga | https://github.com/HartreeFoca/OohataHuzinaga.jl.git |
|
[
"MIT"
] | 0.1.1 | d192a27769e653b00dfa3d7e49dba341aea8dc50 | code | 3216 | function vᵢ(l, r, i, ℓᵢ, ℓⱼ, Aᵢ, Bᵢ, Cᵢ, Pᵢ, γ)
ϵ = 1 / (4 * γ)
vᵢ = (-1)^l
vᵢ *= cₖ(l, ℓᵢ, ℓⱼ, Pᵢ - Aᵢ, Pᵢ - Bᵢ)
vᵢ *= (-1)^i * factorial(l)
vᵢ *= (Pᵢ - Cᵢ)^(l - (2 * r) - (2 * i)) * ϵ^(r + i)
vᵢ /= factorial(r)
vᵢ /= factorial(i)
vᵢ /= factorial(l - (2 * r) - (2 * i))
return vᵢ
end
function Vxyz(ℓᵢ, mᵢ, nᵢ, ℓⱼ, mⱼ, nⱼ, αᵢ, αⱼ, Rᵢ, Rⱼ, Rₖ, Z)
γ = αᵢ + αⱼ
Rₚ = gaussianproduct(αᵢ, Rᵢ, αⱼ, Rⱼ, γ)
IJ = distance(Rᵢ, Rⱼ)
PK = distance(Rₚ, Rₖ)
Vxyz = 0
for l = 0:(ℓᵢ+ℓⱼ)
for r = 0:trunc(Int64, (l / 2))
for i = 0:trunc(Int64, ((l - (2 * r)) / 2))
Vx = vᵢ(l, r, i, ℓᵢ, ℓⱼ, Rᵢ[1], Rⱼ[1], Rₖ[1], Rₚ[1], γ)
for m = 0:(mᵢ+mⱼ)
for s = 0:trunc(Int64, (m / 2))
for j = 0:trunc(Int64, ((m - (2 * s)) / 2))
Vy = vᵢ(m, s, j, mᵢ, mⱼ, Rᵢ[2], Rⱼ[2], Rₖ[2], Rₚ[2], γ)
for n = 0:(nᵢ+nⱼ)
for t = 0:trunc(Int64, (n / 2))
for k = 0:trunc(Int64, ((n - (2 * t)) / 2))
Vz = vᵢ(
n,
t,
k,
nᵢ,
nⱼ,
Rᵢ[3],
Rⱼ[3],
Rₖ[3],
Rₚ[3],
γ,
)
ν = l + m + n - 2 * (r + s + t) - (i + j + k)
F = boys(ν, (γ * abs(PK)))
Vxyz += Vx * Vy * Vz * F
end
end
end
end
end
end
end
end
end
Nᵢ = normalization(αᵢ, ℓᵢ, mᵢ, nᵢ)
Nⱼ = normalization(αⱼ, ℓⱼ, mⱼ, nⱼ)
Vxyz *= (2 * π) / γ
Vxyz *= exp(-αᵢ * αⱼ * abs(IJ) / γ)
Vxyz *= Nᵢ * Nⱼ
Vxyz *= -Z
return Vxyz
end
function attraction(basis, molecule::Molecule)
n = length(molecule.atoms)
K = length(basis)
V = zeros(K, K, n)
for (i, basisᵢ) in enumerate(basis)
for (j, basisⱼ) in enumerate(basis)
for (k, Z) in enumerate(molecule.numbers)
for (αᵢ, dᵢ) in zip(basisᵢ.α, basisᵢ.d)
for (αⱼ, dⱼ) in zip(basisⱼ.α, basisⱼ.d)
Rᵢ = basisᵢ.R
Rⱼ = basisⱼ.R
Rₖ = molecule.coords[k, :]
ℓᵢ, mᵢ, nᵢ = basisᵢ.ℓ, basisᵢ.m, basisᵢ.n
ℓⱼ, mⱼ, nⱼ = basisⱼ.ℓ, basisⱼ.m, basisⱼ.n
V[i, j, k] +=
dᵢ * dⱼ * Vxyz(ℓᵢ, mᵢ, nᵢ, ℓⱼ, mⱼ, nⱼ, αᵢ, αⱼ, Rᵢ, Rⱼ, Rₖ, Z)
end
end
end
end
end
return V
end
| OohataHuzinaga | https://github.com/HartreeFoca/OohataHuzinaga.jl.git |
|
[
"MIT"
] | 0.1.1 | d192a27769e653b00dfa3d7e49dba341aea8dc50 | code | 1625 | """
This function calculate the double factorial of a number.
"""
function doublefactorial(number)
fact = one(number)
if number % 2 == 0
for m = 1:number
if m % 2 == 0
fact *= m
end
end
elseif number % 2 == 1
for m = 1:number
if m % 2 == 1
fact *= m
end
end
end
return fact
end
"""Compute the square of the distance between two points.
|A|² = (Aₐ - Bₐ)² - (Aₑ - Bₑ)² - (Aₒ - Bₒ)².
The coefficients are multiplied by the (x,y,z) coordinates of the points.
"""
function distance(Rᵢ, Rⱼ)
d = 0
for i = 1:3
d += (Rᵢ[i] .- Rⱼ[i])^2
end
return d
end
function normalization(α, ℓ, m, n)
N = (4 * α)^(ℓ + m + n)
N /=
doublefactorial(2 * ℓ - 1) * doublefactorial(2 * m - 1) * doublefactorial(2 * n - 1)
N *= ((2 * α) / π)^(3 / 2)
N = sqrt(N)
return N
end
function cₖ(j, l, m, A, B)
coefficient = 0
for k = 0:l
for i = 0:m
if (i + k == j)
coefficient += binomial(l, k) * binomial(m, i) * A^(l - k) * B^(m - i)
end
end
end
return coefficient
end
function sᵢ(ℓᵢ, ℓⱼ, γ, Aᵢ, Bᵢ, Pᵢ)
sᵢ = 0
for j = 0:trunc(Int64, ((ℓᵢ + ℓⱼ) / 2))
sᵢ +=
cₖ((2 * j), ℓᵢ, ℓⱼ, (Pᵢ - Aᵢ), (Pᵢ - Bᵢ)) * doublefactorial(2 * j - 1) /
(2 * γ)^j
end
sᵢ *= sqrt(π / γ)
return sᵢ
end
function gaussianproduct(αᵢ, Rᵢ, αⱼ, Rⱼ, γ)
P = []
for i = 1:3
push!(P, ((αᵢ * Rᵢ[i] + αⱼ * Rⱼ[i]) / γ))
end
return hcat(P)
end
| OohataHuzinaga | https://github.com/HartreeFoca/OohataHuzinaga.jl.git |
|
[
"MIT"
] | 0.1.1 | d192a27769e653b00dfa3d7e49dba341aea8dc50 | code | 3326 | abstract type Basis end
"""
```GaussianBasis``` is a *subtype* of ```Basis``` that stores the coefficients, exponents and angular momenta of the atomic orbital.
A basis set in theoretical and computational chemistry is a set of functions (called basis functions) that is used to represent the
electronic wave function in the Hartree–Fock method or density-functional theory in order to turn the partial differential equations
of the model into algebraic equations suitable for efficient implementation on a computer. The use of basis sets is equivalent to the
use of an approximate resolution of the identity: the orbitals ``\\vert \\psi_{i} \\rangle`` are expanded within the basis set as a
linear combination of the basis functions ``\\vert \\psi_{i} \\rangle \\approx \\sum_{\\mu } c_{\\mu i} \\vert \\mu \\rangle``,
where the expansion coefficients ``c_{\\mu i}`` are given by ``c_{\\mu i} = \\sum_{\\nu } \\langle \\mu \\vert \\nu \\rangle^{-1}
\\langle \\nu \\vert \\psi_{i} \\rangle``. The basis set can either be composed of atomic orbitals (yielding the linear combination of
atomic orbitals approach), which is the usual choice within the quantum chemistry community; plane waves which are typically used within
the solid state community, or real-space approaches. Several types of atomic orbitals can be used: Gaussian-type orbitals, Slater-type
orbitals, or numerical atomic orbitals. Out of the three, Gaussian-type orbitals are by far the most often used, as they allow efficient
implementations of Post-Hartree–Fock methods.
"""
struct GaussianBasis <: Basis
R::Vector{Float64}
α::Vector{Float64}
d::Vector{Float64}
ℓ::Int64
m::Int64
n::Int64
end
"""
The ```buildbasis``` method takes a ```Molecule``` as input and return for us an array of ```GaussianBasis``` types. For example, let's open the same
```h2.xyz``` example. As a standard basis set, we use STO-3G data.
```julia
2
H -1.788131055 0.000000000 -4.028513155
H -1.331928651 0.434077746 -3.639854078
```
We give the file as an input:
```julia
hydrogen = molecule("h2.xyz")
sto3g = buildbasis(hydrogen)
```
"""
function buildbasis(molecule::Molecule)
sto3g = GaussianBasis[]
for i in eachindex(molecule.atoms)
number = molecule.numbers[i]
coord = molecule.coords[i, :]
for orbital in orbitalconfig(number)
if orbital == "1s"
push!(
sto3g,
GaussianBasis(coord, sto3g_α[number][1, :], sto3g_d[1, :], 0, 0, 0),
)
elseif orbital == "2s"
push!(
sto3g,
GaussianBasis(coord, sto3g_α[number][2, :], sto3g_d[2, :], 0, 0, 0),
)
elseif orbital == "2p"
push!(
sto3g,
GaussianBasis(coord, sto3g_α[number][2, :], sto3g_d[3, :], 1, 0, 0),
)
push!(
sto3g,
GaussianBasis(coord, sto3g_α[number][2, :], sto3g_d[3, :], 0, 1, 0),
)
push!(
sto3g,
GaussianBasis(coord, sto3g_α[number][2, :], sto3g_d[3, :], 0, 0, 1),
)
end
end
end
return sto3g
end
| OohataHuzinaga | https://github.com/HartreeFoca/OohataHuzinaga.jl.git |
|
[
"MIT"
] | 0.1.1 | d192a27769e653b00dfa3d7e49dba341aea8dc50 | code | 200 | function boys(ν, x)
if x < 1e-7
return (2 * ν + 1)^(-1.0) - x * (2 * ν + 3)^(-1.0)
else
return (1 / 2) * x^(-(ν + 0.5)) * gamma(ν + 0.5) * gamma_inc(ν + 0.5, x)[1]
end
end
| OohataHuzinaga | https://github.com/HartreeFoca/OohataHuzinaga.jl.git |
|
[
"MIT"
] | 0.1.1 | d192a27769e653b00dfa3d7e49dba341aea8dc50 | code | 1259 | function computeenergy(basis, molecule::Molecule, maxiter = 20, convergence = 1e-6)
S = overlap(basis, molecule)
T = kinetic(basis, molecule)
V = attraction(basis, molecule)
G = repulsion(basis, molecule)
K = length(basis)
Hcore = T .+ V
D = zeros(K, K)
P = zeros(K, K)
X = sqrt(inv(S))
Eel = 0.0
for iteration = 0:maxiter
Eold = Eel
for n = 1:K
for m = 1:K
P[m, n] = 0.0
for ℓ = 1:K
for s = 1:K
P[m, n] += D[ℓ, s] * (G[m, n, s, ℓ] - 0.5 * G[m, ℓ, s, n])
end
end
end
end
F = Hcore .+ P
Fp = X .* F .* X
Cp = eigvecs(Fp)
C = X .* Cp
for n = 1:K
for m = 1:K
D[m, n] = 0.0
for a = 1:trunc(Int64, N / 2)
D[m, n] += 2 * (C[m, a] * C[n, a])
end
end
end
Eel = 0.0
for m = 1:K
for n = 1:K
Eel += 0.5 * D[n, m] * (Hcore[m, n] + F[m, n])
end
end
if (abs(Eel - Eold) < convergence) && (iteration > 0)
break
end
end
end
| OohataHuzinaga | https://github.com/HartreeFoca/OohataHuzinaga.jl.git |
|
[
"MIT"
] | 0.1.1 | d192a27769e653b00dfa3d7e49dba341aea8dc50 | code | 1549 | function Kxyz(Rᵢ, Rⱼ, αᵢ, αⱼ, ℓᵢ, ℓⱼ, mᵢ, mⱼ, nᵢ, nⱼ)
K = αⱼ * (2 * (ℓⱼ + mⱼ + nⱼ) + 3) * Sxyz(Rᵢ, Rⱼ, αᵢ, αⱼ, ℓᵢ, ℓⱼ, mᵢ, mⱼ, nᵢ, nⱼ)
K -= (2 * (αⱼ^2)) * Sxyz(Rᵢ, Rⱼ, αᵢ, αⱼ, ℓᵢ, ℓⱼ + 2, mᵢ, mⱼ, nᵢ, nⱼ)
K -= (2 * (αⱼ^2)) * Sxyz(Rᵢ, Rⱼ, αᵢ, αⱼ, ℓᵢ, ℓⱼ, mᵢ, mⱼ + 2, nᵢ, nⱼ)
K -= (2 * (αⱼ^2)) * Sxyz(Rᵢ, Rⱼ, αᵢ, αⱼ, ℓᵢ, ℓⱼ, mᵢ, mⱼ, nᵢ, nⱼ + 2)
K -= (1 / 2) * (ℓⱼ * (ℓⱼ - 1)) * Sxyz(Rᵢ, Rⱼ, αᵢ, αⱼ, ℓᵢ, ℓⱼ - 2, mᵢ, mⱼ, nᵢ, nⱼ)
K -= (1 / 2) * (mⱼ * (mⱼ - 1)) * Sxyz(Rᵢ, Rⱼ, αᵢ, αⱼ, ℓᵢ, ℓⱼ, mᵢ, mⱼ - 2, nᵢ, nⱼ)
K -= (1 / 2) * (nⱼ * (nⱼ - 1)) * Sxyz(Rᵢ, Rⱼ, αᵢ, αⱼ, ℓᵢ, ℓⱼ, mᵢ, mⱼ, nᵢ, nⱼ - 2)
return K
end
function kinetic(basis, molecule::Molecule)
K = length(basis)
T = zeros(K, K)
for (i, basisᵢ) in enumerate(basis)
for (j, basisⱼ) in enumerate(basis)
for (αᵢ, dᵢ) in zip(basisᵢ.α, basisᵢ.d)
for (αⱼ, dⱼ) in zip(basisⱼ.α, basisⱼ.d)
Rᵢ = basisᵢ.R
Rⱼ = basisⱼ.R
ℓᵢ, mᵢ, nᵢ = basisᵢ.ℓ, basisᵢ.m, basisᵢ.n
ℓⱼ, mⱼ, nⱼ = basisⱼ.ℓ, basisⱼ.m, basisⱼ.n
T[i, j] += (
exp(-αᵢ * αⱼ * distance(Rᵢ, Rⱼ) / (αᵢ + αⱼ)) *
normalization(αᵢ, ℓᵢ, mᵢ, nᵢ) *
normalization(αⱼ, ℓⱼ, mⱼ, nⱼ) *
dᵢ *
dⱼ *
Kxyz(Rᵢ, Rⱼ, αᵢ, αⱼ, ℓᵢ, ℓⱼ, mᵢ, mⱼ, nᵢ, nⱼ)
)
end
end
end
end
return T
end
| OohataHuzinaga | https://github.com/HartreeFoca/OohataHuzinaga.jl.git |
|
[
"MIT"
] | 0.1.1 | d192a27769e653b00dfa3d7e49dba341aea8dc50 | code | 1899 | abstract type ChemicalEntity end
"""
```ChemicalEntity``` is an *abstract type* that englobes ```Molecule```, ```Atom```, ```AtomicOrbital``` and ```MolecularOrbital``` structures.
```Molecule``` is a *subtype* of ```ChemicalEntity``` that stores coordinates, atomic symbols and atomic numbers as objects.
"""
struct Molecule <: ChemicalEntity
atoms::Vector{String}
coords::Matrix{Float64}
numbers::Vector{Int64}
end
"""
This method takes an ```.xyz``` file (with cartesian coordinates of atoms in a molecule) and returns a ```Molecule```. The ```.xyz``` file should be formatted as follows
```julia
2
H -1.788131055 0.000000000 -4.028513155
H -1.331928651 0.434077746 -3.639854078
```
In the first line, the file should contain the numer of atoms that are in the molecule. In the second line, there is a comment, which can be the *name of the compound*,
*molecular formula*, etc. To further information about ```.xyz``` files, [*click here*](https://www.reviversoft.com/file-extensions/xyz). For example, if we take the
example file ```h2.xyz```, it is possible to give it as an input by calling ```molecule``` method.
```julia
molecule("h2.xyz")
```
The example above works if the file is in the current directory that you are working on. In other case, you can just give the path to the file of interest.
```julia
molecule(PATH)
```
"""
function molecule(xyzfile::String)::Molecule
elements = String[]
coordinates = []
Zvalues = Int[]
for line in Iterators.drop(eachline(xyzfile), 2)
fields = split(line)
element = fields[1]
push!(elements, element)
push!(Zvalues, Z[element])
coordinate = parse.(Float64, fields[2:4])
push!(coordinates, coordinate)
end
coordinates = mapreduce(permutedims, vcat, coordinates)
return Molecule(elements, coordinates, Zvalues)
end
| OohataHuzinaga | https://github.com/HartreeFoca/OohataHuzinaga.jl.git |
|
[
"MIT"
] | 0.1.1 | d192a27769e653b00dfa3d7e49dba341aea8dc50 | code | 1253 | function Sxyz(Rᵢ, Rⱼ, αᵢ, αⱼ, ℓᵢ, ℓⱼ, mᵢ, mⱼ, nᵢ, nⱼ)
Rₚ = gaussianproduct(αᵢ, Rᵢ, αⱼ, Rⱼ, αᵢ + αⱼ)
Sx = sᵢ(ℓᵢ, ℓⱼ, αᵢ + αⱼ, Rᵢ[1], Rⱼ[1], Rₚ[1])
Sy = sᵢ(mᵢ, mⱼ, αᵢ + αⱼ, Rᵢ[2], Rⱼ[2], Rₚ[2])
Sz = sᵢ(nᵢ, nⱼ, αᵢ + αⱼ, Rᵢ[3], Rⱼ[3], Rₚ[3])
return Sx * Sy * Sz
end
"""
This function calculates the overlap integrals
"""
function overlap(basis, molecule::Molecule)
K = length(basis)
S = zeros(K, K)
for (i, basisᵢ) in enumerate(basis)
for (j, basisⱼ) in enumerate(basis)
for (αᵢ, dᵢ) in zip(basisᵢ.α, basisᵢ.d)
for (αⱼ, dⱼ) in zip(basisⱼ.α, basisⱼ.d)
Rᵢ = basisᵢ.R
Rⱼ = basisⱼ.R
ℓᵢ, mᵢ, nᵢ = basisᵢ.ℓ, basisᵢ.m, basisᵢ.n
ℓⱼ, mⱼ, nⱼ = basisⱼ.ℓ, basisⱼ.m, basisⱼ.n
S[i, j] += (
exp(-αᵢ * αⱼ * distance(Rᵢ, Rⱼ) / (αᵢ + αⱼ)) *
normalization(αᵢ, ℓᵢ, mᵢ, nᵢ) *
normalization(αⱼ, ℓⱼ, mⱼ, nⱼ) *
dᵢ *
dⱼ *
Sxyz(Rᵢ, Rⱼ, αᵢ, αⱼ, ℓᵢ, ℓⱼ, mᵢ, mⱼ, nᵢ, nⱼ)
)
end
end
end
end
return S
end
| OohataHuzinaga | https://github.com/HartreeFoca/OohataHuzinaga.jl.git |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.