repo_name
stringlengths 9
75
| topic
stringclasses 30
values | issue_number
int64 1
203k
| title
stringlengths 1
976
| body
stringlengths 0
254k
| state
stringclasses 2
values | created_at
stringlengths 20
20
| updated_at
stringlengths 20
20
| url
stringlengths 38
105
| labels
sequencelengths 0
9
| user_login
stringlengths 1
39
| comments_count
int64 0
452
|
|---|---|---|---|---|---|---|---|---|---|---|---|
deezer/spleeter | tensorflow | 775 | [Discussion] How can I make sure separate running on GPU? | The separate worked, I use --verbose and it shows some info, but i'm not sure it run on GPU. how can i make sure it?
```
C:\Users\Administrator\Desktop\testSound>python -m spleeter separate -p spleeter:5stems -o output --verbose audio_example.mp3
INFO:tensorflow:Using config: {'_model_dir': 'pretrained_models\\5stems', '_tf_random_seed': None, '_save_summary_steps': 100, '_save_checkpoints_steps': None, '_save_checkpoints_secs': 600, '_session_config': gpu_options {
per_process_gpu_memory_fraction: 0.7
}
, '_keep_checkpoint_max': 5, '_keep_checkpoint_every_n_hours': 10000, '_log_step_count_steps': 100, '_train_distribute': None, '_device_fn': None, '_protocol': None, '_eval_distribute': None, '_experimental_distribute': None, '_experimental_max_worker_delay_secs': None, '_session_creation_timeout_secs': 7200, '_checkpoint_save_graph_def': True, '_service': None, '_cluster_spec': ClusterSpec({}), '_task_type': 'worker', '_task_id': 0, '_global_id_in_cluster': 0, '_master': '', '_evaluation_master': '', '_is_chief': True, '_num_ps_replicas': 0, '_num_worker_replicas': 1}
WARNING:tensorflow:From C:\Users\Administrator\AppData\Local\Programs\Python\Python310\lib\site-packages\spleeter\separator.py:146: calling DatasetV2.from_generator (from tensorflow.python.data.ops.dataset_ops) with output_types is deprecated and will be removed in a future version.
Instructions for updating:
Use output_signature instead
WARNING:tensorflow:From C:\Users\Administrator\AppData\Local\Programs\Python\Python310\lib\site-packages\spleeter\separator.py:146: calling DatasetV2.from_generator (from tensorflow.python.data.ops.dataset_ops) with output_shapes is deprecated and will be removed in a future version.
Instructions for updating:
Use output_signature instead
INFO:tensorflow:Calling model_fn.
INFO:tensorflow:Apply unet for vocals_spectrogram
WARNING:tensorflow:From C:\Users\Administrator\AppData\Local\Programs\Python\Python310\lib\site-packages\keras\layers\normalization\batch_normalization.py:532: _colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.
Instructions for updating:
Colocations handled automatically by placer.
INFO:tensorflow:Apply unet for piano_spectrogram
INFO:tensorflow:Apply unet for drums_spectrogram
INFO:tensorflow:Apply unet for bass_spectrogram
INFO:tensorflow:Apply unet for other_spectrogram
INFO:tensorflow:Done calling model_fn.
INFO:tensorflow:Graph was finalized.
INFO:tensorflow:Restoring parameters from pretrained_models\5stems\model
INFO:tensorflow:Running local_init_op.
INFO:tensorflow:Done running local_init_op.
INFO:spleeter:File output\audio_example/piano.wav written succesfully
INFO:spleeter:File output\audio_example/other.wav written succesfully
INFO:spleeter:File output\audio_example/vocals.wav written succesfully
INFO:spleeter:File output\audio_example/drums.wav written succesfully
INFO:spleeter:File output\audio_example/bass.wav written succesfully
```
I don't think it worked on GPU, because it run 28s. I don't think this is the right speed. Am I right? | open | 2022-06-24T04:23:56Z | 2024-02-04T05:38:42Z | https://github.com/deezer/spleeter/issues/775 | [
"question"
] | limengqilove | 2 |
mckinsey/vizro | plotly | 376 | Custom actions do not work when applied through a component that gets added to a union such as `ControlType` | ### Description
The code below should to my understanding work, ie the dropdown value should be reflected in the Jumbotron subtitle.
```python
from typing import Literal
from dash import html
import vizro.models as vm
from vizro import Vizro
from vizro.models.types import capture
from dash import callback, Input, Output
@capture("action")
def subtitle_changer(subtitle: str):
"""Custom action."""
print("SUBTITLE",subtitle)
return subtitle
# 1. Create new custom component
class Jumbotron(vm.VizroBaseModel):
"""New custom component `Jumbotron`."""
type: Literal["jumbotron"] = "jumbotron"
title: str
subtitle: str
text: str
def build(self):
return html.Div(
[
html.H2(self.title),
html.H3(id = "subtitle_id_to_target",children=self.subtitle),
html.P(self.text),
]
)
# 2. Add new components to expected type - here the selector of the parent components
vm.Page.add_type("components", Jumbotron)
vm.Page.add_type("controls", vm.Dropdown)
page = vm.Page(
title="Custom Component",
components=[
Jumbotron(
id="my_jumbotron",
title="Jumbotron",
subtitle="This is a subtitle to summarize some content.",
text="This is the main body of text of the Jumbotron.",
),
vm.Card(id="my_card", text="Click on a point on the above graph."),
],
controls=[vm.Dropdown(
id = "my_dropdown",
options = ["Subtitle 1", "Subtitle 2", "Subtitle 3"],
multi = False,
actions = [
vm.Action(
function = subtitle_changer(),
inputs = ["my_dropdown.value"],
outputs = ["subtitle_id_to_target.children"],
),
]
)],
)
dashboard = vm.Dashboard(pages=[page])
Vizro().build(dashboard).run()
```
### Expected behavior
Expected behaviour as described above.
Interestingly the custom action works when we use the `Dropdown` inside a `Filter` or `Parameter` and overwrite the pre-defined action there.
Defining a dash `callback` with the relevant `Input` and `Output` also works!
### Which package?
vizro
### Package version
0.1.14.dev0
### Python version
3.9.12
### OS
Mac
### How to Reproduce
See above
### Output
_No response_
### Code of Conduct
- [X] I agree to follow the [Code of Conduct](https://github.com/mckinsey/vizro/blob/main/CODE_OF_CONDUCT.md). | closed | 2024-03-20T09:21:50Z | 2024-05-21T11:44:05Z | https://github.com/mckinsey/vizro/issues/376 | [
"Bug Report :bug:"
] | maxschulz-COL | 1 |
httpie/http-prompt | rest-api | 47 | Question: Storing cookies | Hi, nice app!
Can http-prompt store cookies automatically from a response like this?
```
Set-Cookie: author=f0a117c7-75a4-4abe-8617-1d3d06c4d2a7; path=/; httponly
Set-Cookie: author.sig=y4Kfr_MNE7RJmeL1x6vLhixHUAo; path=/; httponly
```
What I do now is copy/pasting (a bit painful):
```
Cookie:author=f0a117c7-75a4-4abe-8617-1d3d06c4d2a7;author.sig=y4Kfr_MNE7RJmeL1x6vLhixHUAo;
```
| closed | 2016-06-12T21:32:33Z | 2016-06-12T21:32:50Z | https://github.com/httpie/http-prompt/issues/47 | [] | arve0 | 1 |
iMerica/dj-rest-auth | rest-api | 150 | /login happened 401 error, when the HTTP Header has invalid or expired access_token. use right username & password also invaild. | When my front-end(ReactJS&HTML5) login interface logs in by calling / login, if the HTTP header of "Authentication: bear {access_token} " is invalid or expired, under normal circumstances, the user can log in again by re-entering the right user-name and password, but 401 error is always reported. I understand / login is AllowAny. How to solve this problem? | open | 2020-09-24T16:23:28Z | 2025-01-31T10:42:13Z | https://github.com/iMerica/dj-rest-auth/issues/150 | [] | fineartit | 3 |
pyro-ppl/numpyro | numpy | 1,198 | Question about sample API | Hey! I am learning a bit about PPLs in JAX and I am very curious as to why Numpyro is using and API like this:
```python
mu = numpyro.sample("mu", dist.Normal(0, 5))
```
Instead of something like:
```python
mu = dist.Normal("mu", 0, 5)
```
Sorry if the question is dumb 😅 | closed | 2021-10-25T14:14:12Z | 2021-10-29T03:46:38Z | https://github.com/pyro-ppl/numpyro/issues/1198 | [
"question"
] | cgarciae | 2 |
plotly/dash-core-components | dash | 287 | Plotly.js mutates data passed into Graph component | We've discovered that `plotly.js` mutates not just the `figure` (which caused the Graph resizing bug in Tabs #279) but also the `data`. Our contract with props in our components is that they are immutable, so that subsequent renders of the components will not cause side-effects. We've solved the issue of the Graph resizing by cloning the `figure.layout` array. We could do this with `data` as well, but the `data` could get very big, and cloning that on every render could get very expensive very quickly. At some point, it would be nice to make sure that `data` and other props passed to `plotly.js` are treated as immutable. | open | 2018-08-28T18:49:47Z | 2018-08-29T17:29:42Z | https://github.com/plotly/dash-core-components/issues/287 | [
"Status: Discussion Needed"
] | valentijnnieman | 1 |
pydata/pandas-datareader | pandas | 444 | Unable to install the pandas-datareader package | Hi,
I have been trying to install the pandas-datareader package via the following two methods:
1. C:\>conda install -c anaconda pandas-datareader
This method hung on the following message:
**Fetching package metadata: ...^C**
Once I tried to close the command line window the ^C was added to the line above and the window would not close.
2. C:\>pip install pandas-datareader
**Downloading/unpacking pandas-datareader
Cannot fetch index base URL https://pypi.python.org/simple/
Could not find any downloads that satisfy the requirement pandas-datareader
Cleaning up...
No distributions at all found for pandas-datareader
Storing debug log for failure in C:\Users\rpatel2550\pip\pip.log**
This method created an error log file, which I have pasted below:
C:\Program Files (x86)\Anaconda\Scripts\pip-script.py run on 01/12/18 17:52:47
Downloading/unpacking pandas-datareader
Getting page https://pypi.python.org/simple/pandas-datareader/
Could not fetch URL https://pypi.python.org/simple/pandas-datareader/: connection error: HTTPSConnectionPool(host='pypi.python.org', port=443): Max retries exceeded with url: /simple/pandas-datareader/ (Caused by <class 'socket.error'>: [Errno 10051] A socket operation was attempted to an unreachable network)
Will skip URL https://pypi.python.org/simple/pandas-datareader/ when looking for download links for pandas-datareader
Getting page https://pypi.python.org/simple/
Could not fetch URL https://pypi.python.org/simple/: connection error: HTTPSConnectionPool(host='pypi.python.org', port=443): Max retries exceeded with url: /simple/ (Caused by <class 'socket.error'>: [Errno 10051] A socket operation was attempted to an unreachable network)
Will skip URL https://pypi.python.org/simple/ when looking for download links for pandas-datareader
Cannot fetch index base URL https://pypi.python.org/simple/
URLs to search for versions for pandas-datareader:
* https://pypi.python.org/simple/pandas-datareader/
Getting page https://pypi.python.org/simple/pandas-datareader/
Could not fetch URL https://pypi.python.org/simple/pandas-datareader/: connection error: HTTPSConnectionPool(host='pypi.python.org', port=443): Max retries exceeded with url: /simple/pandas-datareader/ (Caused by <class 'socket.error'>: [Errno 10051] A socket operation was attempted to an unreachable network)
Will skip URL https://pypi.python.org/simple/pandas-datareader/ when looking for download links for pandas-datareader
Could not find any downloads that satisfy the requirement pandas-datareader
Cleaning up...
Removing temporary dir c:\users\rpatel2550\appdata\local\temp\pip_build_rpatel2550...
No distributions at all found for pandas-datareader
Using this error message I was able to go to this website:
https://pypi.python.org/simple/pandas-datareader/
The most recent file pandas-datareader-0.5.0.tar.gz is evidently available but I don't know how to install it.
Please help!
Thanks,
R | closed | 2018-01-13T17:47:48Z | 2018-01-18T16:21:45Z | https://github.com/pydata/pandas-datareader/issues/444 | [] | rpatel2550 | 8 |
browser-use/browser-use | python | 257 | Browser disregards --user-agent parameter | Playwright browser disregards the UA parameter given via extra_chromium_args.
I am testing it by opening https://whatismyuseragent.com; whenever I open it using browser-use, I get
Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/85.0.4183.102 Safari/537.36
I have tried changing browser.py _setup_browser() to add the parameter to args, but it is still ignored.
When I try playwright cr --user-agent foobarfoo it works OK fine and I get the user agent foobarfoo.
| closed | 2025-01-13T19:50:11Z | 2025-01-26T10:22:38Z | https://github.com/browser-use/browser-use/issues/257 | [] | ranlo | 2 |
pallets-eco/flask-wtf | flask | 408 | ImportError: cannot import name 'HTMLString' from 'wtforms.widgets' | WTFS was upgraded in today, 2020/04/22,zh.

When I init env, then the importError was happended.
So If you wanted to workround, just set the version of WTForms to be lower 2.3.0 | closed | 2020-04-22T03:45:11Z | 2021-05-26T00:54:53Z | https://github.com/pallets-eco/flask-wtf/issues/408 | [] | Mukvin | 1 |
ranaroussi/yfinance | pandas | 1,950 | Period '2d' is invalid error when fetching 2-day data with yfinance | ### Describe bug
I am encountering an issue with the yfinance library when trying to fetch 2-day historical data with 1-minute intervals for the ticker symbol ES=F. The code returns an error stating that the period ‘2d’ is invalid.
### Simple code that reproduces your problem
import yfinance as yf
symbol = input("Enter the symbol: ")
equity = yf.Ticker(symbol)
try:
df = equity.history(period="2d", interval="1m")
if df.empty:
raise ValueError("No data available for the specified period and interval.")
except Exception as e:
print(f"An error occurred: {e}")
print("Attempting to fetch alternative data...")
### Debug log
ERROR:yfinance:ES=F: Period '2d' is invalid, must be one of ['1d', '5d', '1mo', '3mo', '6mo', '1y', '2y', '5y', '10y', 'ytd', 'max']
An error occurred: No data available for the specified period and interval.
Attempting to fetch alternative data...
### Bad data proof
_No response_
### `yfinance` version
Latest
### Python version
_No response_
### Operating system
_No response_ | closed | 2024-05-29T04:54:03Z | 2024-05-29T19:56:11Z | https://github.com/ranaroussi/yfinance/issues/1950 | [] | bruce1095 | 1 |
Lightning-AI/pytorch-lightning | pytorch | 20,124 | Why does the progress bar not show the total steps when using iterable dataset? | ### Bug description
The parameters to Trainer:
```
max_epochs=-1, max_steps=200000
```
The progress bar does not show the total iters and the estimated remaining time is also nonsense.
```
Epoch 1: | | 43/? [00:26<00:00 ****
```
### What version are you seeing the problem on?
master
### How to reproduce the bug
```python
The version is 2.3.3
```
### Error messages and logs
```
# Error messages and logs here please
```
### Environment
<details>
<summary>Current environment</summary>
```
#- PyTorch Lightning Version (e.g., 1.5.0):
#- PyTorch Version (e.g., 2.0):
#- Python version (e.g., 3.9):
#- OS (e.g., Linux):
#- CUDA/cuDNN version:
#- GPU models and configuration:
#- How you installed Lightning(`conda`, `pip`, source):
```
</details>
### More info
_No response_ | closed | 2024-07-24T11:26:15Z | 2025-02-06T13:40:35Z | https://github.com/Lightning-AI/pytorch-lightning/issues/20124 | [
"question",
"ver: 2.2.x"
] | hiyyg | 3 |
python-gitlab/python-gitlab | api | 2,978 | Support User service account API | Besides group-level service accounts, which is more targeted for Gitlab.com users, GitLab 16.1 also introduced the Service account users API for self-hosted and dedicated instances: https://docs.gitlab.com/ee/api/user_service_accounts.html. Like group-level service accounts, they can only be added using the API right now. Thus, it would be great to have this feature added to python-gitlab. | open | 2024-09-12T17:09:20Z | 2025-01-29T11:20:19Z | https://github.com/python-gitlab/python-gitlab/issues/2978 | [
"EE"
] | skycaptain | 1 |
jwkvam/bowtie | plotly | 5 | add docstrings so users can know what datatype to expect from events | closed | 2016-06-07T05:39:01Z | 2016-12-03T22:11:01Z | https://github.com/jwkvam/bowtie/issues/5 | [] | jwkvam | 1 |
|
xuebinqin/U-2-Net | computer-vision | 272 | How to get foreground object from the output on u2net_test.py? Thanks | Hi, Thanks for sharing the great work! Is there a small example to extract foreground using this method? Thanks | closed | 2021-12-03T18:42:00Z | 2022-04-09T18:21:33Z | https://github.com/xuebinqin/U-2-Net/issues/272 | [] | gsrujana | 3 |
CorentinJ/Real-Time-Voice-Cloning | deep-learning | 672 | No voice generating | I'm using a colab notebook and for some reason I only get some noise.
https://user-images.githubusercontent.com/66216869/108493161-4b654e00-72a6-11eb-9da0-cec04228c304.mp4
| closed | 2021-02-19T10:33:36Z | 2021-02-19T11:02:30Z | https://github.com/CorentinJ/Real-Time-Voice-Cloning/issues/672 | [] | Jojo-M-C | 1 |
lanpa/tensorboardX | numpy | 70 | A few questions about add_graph | Hi @lanpa , thanks for this amazing tool. I'm trying to use `add_graph` in my own project, where I met these questions:
1. How to use `add_graph` in the training iteration? Am I supposed to initialize the `SummaryWriter` outside the main training loop and call `w.add_graph` inside the loop? If so how do I get the model output? Or this feature is only possible for dummy input and being called only once? Why it's necessary to feed input to the model? It would be nice if you can provide an example with `add_graph` used in a complete project(instead of `demo_graph.py`).
2. Does it support custom CUDA layer? I tried with a custom correlation layer, and no matter what kind of data is fed into the model(random or image) the custom correlation failed for unknown reason.
Thanks for your time! | closed | 2018-01-24T09:51:38Z | 2018-03-28T14:38:31Z | https://github.com/lanpa/tensorboardX/issues/70 | [] | JiamingSuen | 8 |
dynaconf/dynaconf | fastapi | 1,235 | [bug] list command must error for not found key | ```bash
# /venv/bin/dynaconf list -k "D" --json
Key not found
# echo $?
0
```
Expected: `1` retcode
---
Open for discussion:
- Should retcode be `1`
- Should return empty json `{}`
? | open | 2025-02-07T17:40:53Z | 2025-02-19T15:39:00Z | https://github.com/dynaconf/dynaconf/issues/1235 | [
"bug",
"good first issue"
] | rochacbruno | 0 |
plotly/dash | data-visualization | 2,592 | Be compatible with Flask 2.3 | dash dependency of end of support **flask** branch
```Flask>=1.0.4,<2.3.0```
since https://github.com/plotly/dash/commit/7bd5b7ebec72ffbfca85a57d0d4c19b595371a5a
The 2.3.x branch is now the supported fix branch, the 2.2.x branch will become a tag marking the end of support for that branch.
https://github.com/pallets/flask/releases
| closed | 2023-07-07T22:57:24Z | 2023-10-26T21:01:54Z | https://github.com/plotly/dash/issues/2592 | [] | VelizarVESSELINOV | 1 |
home-assistant/core | asyncio | 140,707 | Fritz Tools Error | ### The problem
Error by Log shows up from time to time:
Logger: homeassistant.components.fritz.coordinator
Quelle: helpers/update_coordinator.py:380
Integration: AVM FRITZ!Box Tools (Dokumentation, Probleme)
Erstmals aufgetreten: 01:05:58 (14 Vorkommnisse)
Zuletzt protokolliert: 09:09:05
Unexpected error fetching fritz-192.168.178.1-coordinator data
Traceback (most recent call last):
File "/usr/src/homeassistant/homeassistant/components/fritz/coordinator.py", line 443, in _async_update_hosts_info
hosts_attributes = await self.hass.async_add_executor_job(
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
self.fritz_hosts.get_hosts_attributes
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
)
^
File "/usr/local/lib/python3.13/concurrent/futures/thread.py", line 59, in run
result = self.fn(*self.args, **self.kwargs)
File "/usr/local/lib/python3.13/site-packages/fritzconnection/lib/fritzhosts.py", line 216, in get_hosts_attributes
storage = HostStorage(get_xml_root(source=url, session=self.fc.session))
~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/usr/local/lib/python3.13/site-packages/fritzconnection/core/utils.py", line 83, in get_xml_root
return etree.fromstring(source)
~~~~~~~~~~~~~~~~^^^^^^^^
File "/usr/local/lib/python3.13/xml/etree/ElementTree.py", line 1343, in XML
return parser.close()
~~~~~~~~~~~~^^
xml.etree.ElementTree.ParseError: unclosed token: line 1305, column 17
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/usr/src/homeassistant/homeassistant/helpers/update_coordinator.py", line 380, in _async_refresh
self.data = await self._async_update_data()
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/usr/src/homeassistant/homeassistant/components/fritz/coordinator.py", line 338, in _async_update_data
await self.async_scan_devices()
File "/usr/src/homeassistant/homeassistant/components/fritz/coordinator.py", line 571, in async_scan_devices
hosts = await self._async_update_hosts_info()
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/usr/src/homeassistant/homeassistant/components/fritz/coordinator.py", line 452, in _async_update_hosts_info
raise HomeAssistantError(
...<2 lines>...
) from ex
homeassistant.exceptions.HomeAssistantError: Error refreshing hosts info
### What version of Home Assistant Core has the issue?
newest
### What was the last working version of Home Assistant Core?
newest
### What type of installation are you running?
Home Assistant OS
### Integration causing the issue
Fritz Tools
### Link to integration documentation on our website
_No response_
### Diagnostics information
Logger: homeassistant.components.fritz.coordinator
Quelle: helpers/update_coordinator.py:380
Integration: AVM FRITZ!Box Tools (Dokumentation, Probleme)
Erstmals aufgetreten: 01:05:58 (14 Vorkommnisse)
Zuletzt protokolliert: 09:09:05
Unexpected error fetching fritz-192.168.178.1-coordinator data
Traceback (most recent call last):
File "/usr/src/homeassistant/homeassistant/components/fritz/coordinator.py", line 443, in _async_update_hosts_info
hosts_attributes = await self.hass.async_add_executor_job(
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
self.fritz_hosts.get_hosts_attributes
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
)
^
File "/usr/local/lib/python3.13/concurrent/futures/thread.py", line 59, in run
result = self.fn(*self.args, **self.kwargs)
File "/usr/local/lib/python3.13/site-packages/fritzconnection/lib/fritzhosts.py", line 216, in get_hosts_attributes
storage = HostStorage(get_xml_root(source=url, session=self.fc.session))
~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/usr/local/lib/python3.13/site-packages/fritzconnection/core/utils.py", line 83, in get_xml_root
return etree.fromstring(source)
~~~~~~~~~~~~~~~~^^^^^^^^
File "/usr/local/lib/python3.13/xml/etree/ElementTree.py", line 1343, in XML
return parser.close()
~~~~~~~~~~~~^^
xml.etree.ElementTree.ParseError: unclosed token: line 1305, column 17
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/usr/src/homeassistant/homeassistant/helpers/update_coordinator.py", line 380, in _async_refresh
self.data = await self._async_update_data()
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/usr/src/homeassistant/homeassistant/components/fritz/coordinator.py", line 338, in _async_update_data
await self.async_scan_devices()
File "/usr/src/homeassistant/homeassistant/components/fritz/coordinator.py", line 571, in async_scan_devices
hosts = await self._async_update_hosts_info()
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/usr/src/homeassistant/homeassistant/components/fritz/coordinator.py", line 452, in _async_update_hosts_info
raise HomeAssistantError(
...<2 lines>...
) from ex
homeassistant.exceptions.HomeAssistantError: Error refreshing hosts info
### Example YAML snippet
```yaml
```
### Anything in the logs that might be useful for us?
```txt
```
### Additional information
_No response_ | open | 2025-03-16T09:11:41Z | 2025-03-16T19:27:35Z | https://github.com/home-assistant/core/issues/140707 | [
"integration: fritz"
] | steps56 | 2 |
ymcui/Chinese-LLaMA-Alpaca | nlp | 817 | 是否有基于Llama-2的增量训练模型? | 我们已发布基于Llama-2的相关模型,模型大小包括7B和13B,以及长上下文版7B-16K和13B-16K模型。
**项目地址:https://github.com/ymcui/Chinese-LLaMA-Alpaca-2**
- 【新】已发布长上下文版模型,支持16K上下文长度,并可通过NTK方法扩展至24K+
- 目前已发布13B级别的模型,效果优于一代项目(本项目)的Plus/Pro-13B,部分指标超过Plus/Pro-33B,推荐使用相关模型的用户迁移到我们的二代模型上
- 目前已发布7B级别的模型,效果优于一代项目(本项目)的Plus/Pro-7B和Plus/Pro-13B,推荐使用相关模型的用户迁移到我们的二代模型上
- 目前暂无33B级别二代模型(meta还没发布),可先使用一代Plus/Pro-33B模型。 | open | 2023-08-09T08:54:24Z | 2023-08-25T05:19:29Z | https://github.com/ymcui/Chinese-LLaMA-Alpaca/issues/817 | [] | ymcui | 0 |
jina-ai/clip-as-service | pytorch | 586 | Unable to load Bert as service for uncased_L-24_H-1024_A-16 | **Prerequisites**
> Please fill in by replacing `[ ]` with `[x]`.
* [X] Are you running the latest `bert-as-service`?
* [X] Did you follow [the installation](https://github.com/hanxiao/bert-as-service#install) and [the usage](https://github.com/hanxiao/bert-as-service#usage) instructions in `README.md`?
* [X] Did you check the [FAQ list in `README.md`](https://github.com/hanxiao/bert-as-service#speech_balloon-faq)?
* [X] Did you perform [a cursory search on existing issues](https://github.com/hanxiao/bert-as-service/issues)?
I get the below error when i load the model via API. I never get an output. Howvever this words for remain model like `uncased_L-12_H-768_A-12` or `multi_cased_L-12_H-768_A-12`
I am using below command to run`bert-serving-start -model_dir=/opt/web2pyapp/applications/Advanced_Python/uncased_L-24_H-1024_A-16 -http_port 8170 -port 5571 -port_out 5572`
(https://github.com/tensorflow/tensorflow/tree/master/tools/tf_env_collect.sh
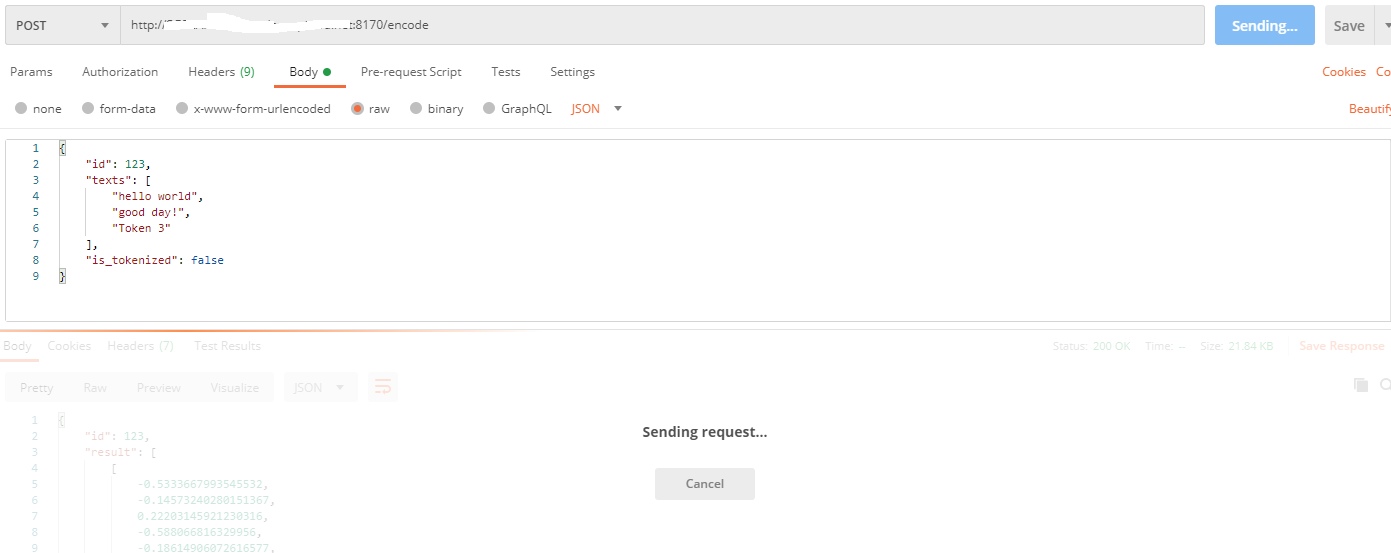

| open | 2020-08-19T17:51:58Z | 2020-08-19T18:04:47Z | https://github.com/jina-ai/clip-as-service/issues/586 | [] | nishithbenhur | 1 |
allenai/allennlp | data-science | 5,717 | Unclear how to use text2sql model | <!--
Please fill this template entirely and do not erase any of it.
We reserve the right to close without a response bug reports which are incomplete.
If you have a question rather than a bug, please ask on [Stack Overflow](https://stackoverflow.com/questions/tagged/allennlp) rather than posting an issue here.
-->
## Checklist
<!-- To check an item on the list replace [ ] with [x]. -->
- [x] I have verified that the issue exists against the `main` branch of AllenNLP.
- [x] I have read the relevant section in the [contribution guide](https://github.com/allenai/allennlp/blob/main/CONTRIBUTING.md#bug-fixes-and-new-features) on reporting bugs.
- [x] I have checked the [issues list](https://github.com/allenai/allennlp/issues) for similar or identical bug reports.
- [x] I have checked the [pull requests list](https://github.com/allenai/allennlp/pulls) for existing proposed fixes.
- [ x] I have checked the [CHANGELOG](https://github.com/allenai/allennlp/blob/main/CHANGELOG.md) and the [commit log](https://github.com/allenai/allennlp/commits/main) to find out if the bug was already fixed in the main branch.
- [x] I have included in the "Description" section below a traceback from any exceptions related to this bug.
- [x] I have included in the "Related issues or possible duplicates" section beloew all related issues and possible duplicate issues (If there are none, check this box anyway).
- [x] I have included in the "Environment" section below the name of the operating system and Python version that I was using when I discovered this bug.
- [x] I have included in the "Environment" section below the output of `pip freeze`.
- [x] I have included in the "Steps to reproduce" section below a minimally reproducible example.
## Description
I am able to load the text2sql model as follows:
```
from allennlp_models import pretrained
SQL_model_name = 'semparse-text-to-sql'
pred_model = pretrained.load_predictor(SQL_model_name)
```
However, the model doesn't appear to allow predictions, since the `predict` method isn't implemented (as compared to other AllenNLP models where `predict` works as expected). Am I using the model incorrectly?
Traceback:
```
$pred_model.predict('what is the temperature in Berlin?')
AttributeError Traceback (most recent call last)
----> 1 pred.predict
AttributeError: 'Predictor' object has no attribute 'predict'
```
## Related issues or possible duplicates
- None
## Environment
<!-- Provide the name of operating system below (e.g. OS X, Linux) -->
OS: Linux? (Colab)
<!-- Provide the Python version you were using (e.g. 3.7.1) -->
Python version: 3.7
<details>
<summary><b>Output of <code>pip freeze</code>:</b></summary>
<p>
<!-- Paste the output of `pip freeze` in between the next two lines below -->
```
absl-py==1.2.0
aeppl==0.0.33
aesara==2.7.9
aiohttp==3.8.3
aiosignal==1.2.0
alabaster==0.7.12
albumentations==1.2.1
allennlp==2.10.0
allennlp-models==2.10.0
allennlp-semparse==0.0.4
altair==4.2.0
appdirs==1.4.4
arviz==0.12.1
astor==0.8.1
astropy==4.3.1
astunparse==1.6.3
async-timeout==4.0.2
asynctest==0.13.0
atari-py==0.2.9
atomicwrites==1.4.1
attrs==22.1.0
audioread==3.0.0
autograd==1.5
Babel==2.10.3
backcall==0.2.0
base58==2.1.1
beautifulsoup4==4.6.3
bleach==5.0.1
blis==0.7.8
bokeh==2.3.3
boto3==1.24.89
botocore==1.27.89
branca==0.5.0
bs4==0.0.1
CacheControl==0.12.11
cached-path==1.1.6
cached-property==1.5.2
cachetools==4.2.4
catalogue==2.0.8
certifi==2022.9.24
cffi==1.15.1
cftime==1.6.2
chardet==3.0.4
charset-normalizer==2.1.1
click==7.1.2
clikit==0.6.2
cloudpickle==1.5.0
cmake==3.22.6
cmdstanpy==1.0.7
colorcet==3.0.1
colorlover==0.3.0
commonmark==0.9.1
community==1.0.0b1
confection==0.0.2
conllu==4.4.2
cons==0.4.5
contextlib2==0.5.5
convertdate==2.4.0
crashtest==0.3.1
crcmod==1.7
cufflinks==0.17.3
cvxopt==1.3.0
cvxpy==1.2.1
cycler==0.11.0
cymem==2.0.6
Cython==0.29.32
daft==0.0.4
dask==2022.2.0
datascience==0.17.5
datasets==2.5.2
debugpy==1.0.0
decorator==4.4.2
defusedxml==0.7.1
descartes==1.1.0
dill==0.3.5.1
distributed==2022.2.0
dlib==19.24.0
dm-tree==0.1.7
docker-pycreds==0.4.0
docutils==0.17.1
dopamine-rl==1.0.5
earthengine-api==0.1.326
easydict==1.10
ecos==2.0.10
editdistance==0.5.3
en-core-web-lg @ https://github.com/explosion/spacy-models/releases/download/en_core_web_lg-3.4.0/en_core_web_lg-3.4.0-py3-none-any.whl
en-core-web-sm @ https://github.com/explosion/spacy-models/releases/download/en_core_web_sm-3.4.0/en_core_web_sm-3.4.0-py3-none-any.whl
entrypoints==0.4
ephem==4.1.3
et-xmlfile==1.1.0
etils==0.8.0
etuples==0.3.8
fa2==0.3.5
fairscale==0.4.6
fastai==2.7.9
fastcore==1.5.27
fastdownload==0.0.7
fastdtw==0.3.4
fastjsonschema==2.16.2
fastprogress==1.0.3
fastrlock==0.8
feather-format==0.4.1
filelock==3.7.1
firebase-admin==4.4.0
fix-yahoo-finance==0.0.22
flaky==3.7.0
Flask==1.1.4
flatbuffers==22.9.24
folium==0.12.1.post1
frozenlist==1.3.1
fsspec==2022.8.2
ftfy==6.1.1
future==0.16.0
gast==0.5.3
GDAL==2.2.2
gdown==4.4.0
gensim==3.6.0
geographiclib==1.52
geopy==1.17.0
gin-config==0.5.0
gitdb==4.0.9
GitPython==3.1.28
glob2==0.7
google==2.0.3
google-api-core==1.31.6
google-api-python-client==1.12.11
google-auth==1.35.0
google-auth-httplib2==0.0.4
google-auth-oauthlib==0.4.6
google-cloud-bigquery==1.21.0
google-cloud-bigquery-storage==1.1.2
google-cloud-core==2.3.2
google-cloud-datastore==1.8.0
google-cloud-firestore==1.7.0
google-cloud-language==1.2.0
google-cloud-storage==2.5.0
google-cloud-translate==1.5.0
google-colab @ file:///colabtools/dist/google-colab-1.0.0.tar.gz
google-crc32c==1.5.0
google-pasta==0.2.0
google-resumable-media==2.4.0
googleapis-common-protos==1.56.4
googledrivedownloader==0.4
graphviz==0.10.1
greenlet==1.1.3
grpcio==1.49.1
gspread==3.4.2
gspread-dataframe==3.0.8
gym==0.25.2
gym-notices==0.0.8
h5py==3.7.0
HeapDict==1.0.1
hijri-converter==2.2.4
holidays==0.16
holoviews==1.14.9
html5lib==1.0.1
httpimport==0.5.18
httplib2==0.17.4
httplib2shim==0.0.3
httpstan==4.6.1
huggingface-hub==0.10.0
humanize==0.5.1
hyperopt==0.1.2
idna==2.10
imageio==2.9.0
imagesize==1.4.1
imbalanced-learn==0.8.1
imblearn==0.0
imgaug==0.4.0
importlib-metadata==5.0.0
importlib-resources==5.9.0
imutils==0.5.4
inflect==2.1.0
iniconfig==1.1.1
intel-openmp==2022.2.0
intervaltree==2.1.0
ipykernel==5.3.4
ipython==7.9.0
ipython-genutils==0.2.0
ipython-sql==0.3.9
ipywidgets==7.7.1
itsdangerous==1.1.0
jax==0.3.21
jaxlib @ https://storage.googleapis.com/jax-releases/cuda11/jaxlib-0.3.20+cuda11.cudnn805-cp37-cp37m-manylinux2014_x86_64.whl
jieba==0.42.1
Jinja2==2.11.3
jmespath==1.0.1
joblib==1.2.0
jpeg4py==0.1.4
jsonnet==0.18.0
jsonschema==4.3.3
jupyter-client==6.1.12
jupyter-console==6.1.0
jupyter-core==4.11.1
jupyterlab-widgets==3.0.3
kaggle==1.5.12
kapre==0.3.7
keras==2.8.0
Keras-Preprocessing==1.1.2
keras-vis==0.4.1
kiwisolver==1.4.4
korean-lunar-calendar==0.3.1
langcodes==3.3.0
libclang==14.0.6
librosa==0.8.1
lightgbm==2.2.3
llvmlite==0.39.1
lmdb==1.3.0
locket==1.0.0
logical-unification==0.4.5
LunarCalendar==0.0.9
lxml==4.9.1
Markdown==3.4.1
MarkupSafe==2.0.1
marshmallow==3.18.0
matplotlib==3.2.2
matplotlib-venn==0.11.7
miniKanren==1.0.3
missingno==0.5.1
mistune==0.8.4
mizani==0.7.3
mkl==2019.0
mlxtend==0.14.0
more-itertools==8.14.0
moviepy==0.2.3.5
mpmath==1.2.1
msgpack==1.0.4
multidict==6.0.2
multipledispatch==0.6.0
multiprocess==0.70.13
multitasking==0.0.11
murmurhash==1.0.8
music21==5.5.0
natsort==5.5.0
nbconvert==5.6.1
nbformat==5.6.1
netCDF4==1.6.1
networkx==2.6.3
nibabel==3.0.2
nltk==3.7
notebook==5.3.1
numba==0.56.2
numexpr==2.8.3
numpy==1.21.6
oauth2client==4.1.3
oauthlib==3.2.1
okgrade==0.4.3
opencv-contrib-python==4.6.0.66
opencv-python==4.6.0.66
opencv-python-headless==4.6.0.66
openpyxl==3.0.10
opt-einsum==3.3.0
osqp==0.6.2.post0
packaging==21.3
palettable==3.3.0
pandas==1.3.5
pandas-datareader==0.9.0
pandas-gbq==0.13.3
pandas-profiling==1.4.1
pandocfilters==1.5.0
panel==0.12.1
param==1.12.2
parsimonious==0.10.0
parso==0.8.3
partd==1.3.0
pastel==0.2.1
pathlib==1.0.1
pathtools==0.1.2
pathy==0.6.2
patsy==0.5.2
pep517==0.13.0
pexpect==4.8.0
pickleshare==0.7.5
Pillow==7.1.2
pip-tools==6.2.0
plac==1.1.3
plotly==5.5.0
plotnine==0.8.0
pluggy==1.0.0
pooch==1.6.0
portpicker==1.3.9
prefetch-generator==1.0.1
preshed==3.0.7
prettytable==3.4.1
progressbar2==3.38.0
promise==2.3
prompt-toolkit==2.0.10
prophet==1.1.1
protobuf==3.20.0
psutil==5.4.8
psycopg2==2.9.3
ptyprocess==0.7.0
py==1.11.0
py-rouge==1.1
pyarrow==6.0.1
pyasn1==0.4.8
pyasn1-modules==0.2.8
pycocotools==2.0.5
pycparser==2.21
pyct==0.4.8
pydantic==1.8.2
pydata-google-auth==1.4.0
pydot==1.3.0
pydot-ng==2.0.0
pydotplus==2.0.2
PyDrive==1.3.1
pyemd==0.5.1
pyerfa==2.0.0.1
Pygments==2.6.1
pygobject==3.26.1
pylev==1.4.0
pymc==4.1.4
PyMeeus==0.5.11
pymongo==4.2.0
pymystem3==0.2.0
PyOpenGL==3.1.6
pyparsing==3.0.9
pyrsistent==0.18.1
pysimdjson==3.2.0
pysndfile==1.3.8
PySocks==1.7.1
pystan==3.3.0
pytest==7.1.3
python-apt==0.0.0
python-chess==0.23.11
python-dateutil==2.8.2
python-louvain==0.16
python-slugify==6.1.2
python-utils==3.3.3
pytz==2022.4
pyviz-comms==2.2.1
PyWavelets==1.3.0
PyYAML==6.0
pyzmq==23.2.1
qdldl==0.1.5.post2
qudida==0.0.4
regex==2022.6.2
requests==2.28.1
requests-oauthlib==1.3.1
resampy==0.4.2
responses==0.18.0
rich==12.1.0
rpy2==3.4.5
rsa==4.9
s3transfer==0.6.0
sacremoses==0.0.53
scikit-image==0.18.3
scikit-learn==1.0.2
scipy==1.7.3
screen-resolution-extra==0.0.0
scs==3.2.0
seaborn==0.11.2
Send2Trash==1.8.0
sentencepiece==0.1.97
sentry-sdk==1.9.10
setproctitle==1.3.2
setuptools-git==1.2
Shapely==1.8.4
shortuuid==1.0.9
six==1.15.0
sklearn-pandas==1.8.0
smart-open==5.2.1
smmap==5.0.0
snowballstemmer==2.2.0
sortedcontainers==2.4.0
soundfile==0.11.0
spacy==3.4.1
spacy-legacy==3.0.10
spacy-loggers==1.0.3
Sphinx==1.8.6
sphinxcontrib-serializinghtml==1.1.5
sphinxcontrib-websupport==1.2.4
SQLAlchemy==1.4.41
sqlparse==0.4.3
srsly==2.4.4
statsmodels==0.12.2
sympy==1.7.1
tables==3.7.0
tabulate==0.8.10
tblib==1.7.0
tenacity==8.1.0
tensorboard==2.8.0
tensorboard-data-server==0.6.1
tensorboard-plugin-wit==1.8.1
tensorboardX==2.5.1
tensorflow==2.8.2+zzzcolab20220929150707
tensorflow-datasets==4.6.0
tensorflow-estimator==2.8.0
tensorflow-gcs-config==2.8.0
tensorflow-hub==0.12.0
tensorflow-io-gcs-filesystem==0.27.0
tensorflow-metadata==1.10.0
tensorflow-probability==0.16.0
termcolor==1.1.0
terminado==0.13.3
testpath==0.6.0
text-unidecode==1.3
textblob==0.15.3
thinc==8.1.3
threadpoolctl==3.1.0
tifffile==2021.11.2
tokenizers==0.12.1
toml==0.10.2
tomli==2.0.1
toolz==0.12.0
torch==1.11.0
torchaudio @ https://download.pytorch.org/whl/cu113/torchaudio-0.12.1%2Bcu113-cp37-cp37m-linux_x86_64.whl
torchsummary==1.5.1
torchtext==0.13.1
torchvision==0.12.0
tornado==5.1.1
tqdm==4.64.1
traitlets==5.4.0
transformers==4.20.1
tweepy==3.10.0
typeguard==2.7.1
typer==0.4.2
typing-extensions==4.1.1
tzlocal==1.5.1
ujson==5.5.0
Unidecode==1.3.6
uritemplate==3.0.1
urllib3==1.26.12
vega-datasets==0.9.0
wandb==0.12.21
wasabi==0.10.1
wcwidth==0.2.5
webargs==8.2.0
webencodings==0.5.1
Werkzeug==1.0.1
widgetsnbextension==3.6.1
word2number==1.1
wordcloud==1.8.2.2
wrapt==1.14.1
xarray==0.20.2
xarray-einstats==0.2.2
xgboost==0.90
xkit==0.0.0
xlrd==1.1.0
xlwt==1.3.0
xxhash==3.0.0
yarl==1.8.1
yellowbrick==1.5
zict==2.2.0
zipp==3.8.1
```
</p>
</details>
## Steps to reproduce
<details>
<summary><b>Example source:</b></summary>
<p>
<!-- Add a fully runnable example in between the next two lines below that will reproduce the bug -->
```
!pip install torch
!pip install allennlp
!pip install allennlp-models
!pip install allennlp-semparse
```
```
from allennlp_models import pretrained
SQL_model_name = 'semparse-text-to-sql'
pred_model = pretrained.load_predictor(SQL_model_name)
pred_model.predict('what is the temperature in Berlin?')
```
</p>
</details>
| closed | 2022-10-07T23:19:55Z | 2022-11-23T01:20:28Z | https://github.com/allenai/allennlp/issues/5717 | [
"bug"
] | ianbstewart | 5 |
google-research/bert | nlp | 879 | OOM error fine-tuning | When trying to finetune BERT on a classification task (`run_classifier.py`) using my own dataset, I am running into the OOM issue with the following traceback:
```
iB
2019-10-15 18:21:25.247491: I tensorflow/core/common_runtime/bfc_allocator.cc:641] 1 Chunks of size 8384512 totalling 8.00MiB
2019-10-15 18:21:25.247501: I tensorflow/core/common_runtime/bfc_allocator.cc:641] 262 Chunks of size 16777216 totalling 4.09GiB
2019-10-15 18:21:25.247511: I tensorflow/core/common_runtime/bfc_allocator.cc:641] 1 Chunks of size 16781312 totalling 16.00MiB
2019-10-15 18:21:25.247520: I tensorflow/core/common_runtime/bfc_allocator.cc:641] 1 Chunks of size 20971520 totalling 20.00MiB
2019-10-15 18:21:25.247530: I tensorflow/core/common_runtime/bfc_allocator.cc:641] 5 Chunks of size 118767616 totalling 566.33MiB
2019-10-15 18:21:25.247540: I tensorflow/core/common_runtime/bfc_allocator.cc:641] 1 Chunks of size 128971776 totalling 123.00MiB
2019-10-15 18:21:25.247548: I tensorflow/core/common_runtime/bfc_allocator.cc:645] Sum Total of in-use chunks: 6.88GiB
2019-10-15 18:21:25.247560: I tensorflow/core/common_runtime/bfc_allocator.cc:647] Stats:
Limit: 7392346112
InUse: 7392346112
MaxInUse: 7392346112
NumAllocs: 2204
MaxAllocSize: 128971776
2019-10-15 18:21:25.247633: W tensorflow/core/common_runtime/bfc_allocator.cc:271] ****************************************************************************************************
2019-10-15 18:21:25.247668: W tensorflow/core/framework/op_kernel.cc:1401] OP_REQUIRES failed at cwise_ops_common.cc:70 : Resource exhausted: OOM when allocating tensor with shape[1024,4096] and type float on /job:localhost/replica:0/task:0/device:GPU:0 by allocator GPU_0_bfc
```
(This doesn't break the script, it just keeps running).
I've tried reducing the batch size from 32 -> 16 -> 4 -> 1, none of which have an impact. I am using a Tesla P4 with 8GB. Is my issue as simple as having to increase my GPU memory? Or is there something else going on? | open | 2019-10-15T18:26:30Z | 2019-10-16T12:47:57Z | https://github.com/google-research/bert/issues/879 | [] | ecatkins | 1 |
hyperspy/hyperspy | data-visualization | 2,953 | Release 1.7.1 | There have been a few regressions introduced in 1.7, which are already fixed or pending review. It would be good to release [1.7.1 ](https://github.com/hyperspy/hyperspy/milestone/47) as soon as #2948 and #2952 are merged. | closed | 2022-06-03T09:55:16Z | 2022-06-18T16:56:35Z | https://github.com/hyperspy/hyperspy/issues/2953 | [] | ericpre | 0 |
InstaPy/InstaPy | automation | 6,022 | Unable to follow a whole list (including delays) |
Like it says in the title, Unable to follow a whole list (including delays) BEFORE instapy begins the next step of unfollowing all.
I'm trying to setup so that instapy follows 7500 users in total from inside of the specified list, and then AFTER to go to the next step which is to unfollow allfollowing
At the moment, it's following 10 per hour, 200 a day, which is allworking fine. But as soon as it finishes the first 10, because the unfollow command is right after it seems to bypass the amount I set to 7500 and just begins unfollowing.
I am considering that maybe I could copy the follow by follow command in a loop over and over which might work but I am sure there are MUCH better ways lol
If anyone can help me this will be great.
Here are the setting controlling this:
session.set_action_delays(enabled=True, follow=5, unfollow=4, randomize=True, random_range_from=70,
random_range_to=140)
session.set_quota_supervisor(enabled=True, peak_follows_hourly=10, peak_follows_daily=200, peak_unfollows_hourly=10,
peak_unfollows_daily=200, peak_server_calls_hourly=None, peak_server_calls_daily=2000,
sleep_after=["server_calls_d"], sleepyhead=True, stochastic_flow=True, notify_me=True)
Here is the action code:
# activities # replace users with list name e.g. user_followers above
""" Step 1 - Massive Follow of users followers (I suggest to follow not less than
3500/4000 users for better results)...
"""
follow_by_list(followlist=['target-usrname'], times=1, amount=7500, sleep_delay=3600, randomize=False,
interact=False)
# activities # replace users with list name e.g. user_followers above
""" Step 2 - Massive Unfollow of all following...
"""
session.unfollow_users(allFollowing=True, style="FIFO", unfollow_after=72 * 60 * 60, sleep_delay=3600)
Thanks in advance for any help, it's my first time creating a bot and took me nearly a month I imagine this is super simple
| open | 2021-01-13T15:04:48Z | 2021-07-21T03:19:08Z | https://github.com/InstaPy/InstaPy/issues/6022 | [
"wontfix"
] | molasunfish | 1 |
tiangolo/uwsgi-nginx-flask-docker | flask | 32 | Logging problems | Hi tiangolo,
I am having problems logging messages when I used the uwsgi-nginx-flask image. The application works. I am only having issues with the logging. On the other hand, If I run the app in standalone, everything, including logging, works as expected.
I tried to look for info on how to configure the logging mechanism when using uwsgi-nginx-flask, but I was not able to find anything.
Let me explain the scenario.
I am using uwsgi-nginx-flask:python3.6 to run a flask app.
My uwsgi.ini is
```
[uwsgi]
module = iedapp.app
callable = app
logger = python
```
The app is using resources for handling request.
```
....
app = Flask(__name__, static_folder=None)
app.config.from_object(config)
api = Api(app, prefix='/api', base_path='/api', api_version='1.0')
api.add_resource(IedOnline, '/online/<string:ied_key>/<string:layout>')
....
```
And in the resource, I have something like
```
....
logger = logging.getLogger('test')
class IedOnline(Resource):
def post(self, ied_key, layout):
print("Hello!!!")
current_app.logger.error("Hello again!!!!")
logger.error("Hello again and again")
....
```
What happens to me is that both the print and logging messages in the resource function get lost. I am not able to find them either in the docker logs not if I exec into the docker image and look for entries in all the *.log files in the image.
Could you provide guidance on what I should be going wrong?
| closed | 2018-01-05T14:54:07Z | 2018-04-19T08:44:09Z | https://github.com/tiangolo/uwsgi-nginx-flask-docker/issues/32 | [] | mcuervoe | 4 |
gee-community/geemap | jupyter | 1,617 | Add stretch options for visualizing images | <!-- Please search existing issues to avoid creating duplicates. -->
### Description
Hi folks! I'd like to suggest adding "Stretch" options to adjust the vis param range for images automatically, similar to what can be done in the Code Editor. I'm happy to implement this and make a PR.
### Implementation
I would suggest we follow the lead of the Code Editor add a dropdown menu to select different percent and sigma stretches. When selected, reducers would be run on the selected image bands over the current map bounds, and values would be pulled client-side and processed to set the current range. I believe all the different percentile and sigma stretches can be calculated using min, max, and std dev reducers. As in the Code Editor, we could disable the range inputs when a stretch option is selected, adding a `Custom` option for user-defined values.
### API
It looks like there's a lot of great refactoring work going on in geemap, so I don't want to interfere with ongoing plans! My tentative suggestion would be to add a dropdown widget to the `range_hbox` in the main `Map` class, with a listener that would call a new method `Map.get_stretch_values()` that returns the min and max range values for a given `ee.Image`, but I'm open to any feedback!
### UI
It will be a little tricky to fit the range slider, range values, and stretch dropdown on a single row (see screenshot of a quick prototype I put together below).

We could try swapping the slider for two text boxes, more similar to the Code Editor, although that would require a more substantial rewrite. I'm open to any other ideas as well!
Thanks! | closed | 2023-07-07T05:56:23Z | 2023-10-31T17:58:57Z | https://github.com/gee-community/geemap/issues/1617 | [
"Feature Request"
] | aazuspan | 8 |
pytest-dev/pytest-qt | pytest | 276 | Using QThread causes sefault | Hi,
I have just started using pytest-qt and have run across an issue with the use of `QThread`.
I have been able to isolate it into a simple example:
```
from PySide2.QtCore import QThread
def test_load(qtbot):
thread = QThread()
thread.start()
```
Running on Linux:
```
=============================================================================================== test session starts ===============================================================================================
platform linux -- Python 3.7.3rc1, pytest-4.6.3, py-1.8.0, pluggy-0.12.0
PySide2 5.13.0 -- Qt runtime 5.13.0 -- Qt compiled 5.13.0
rootdir: /mnt/e/Projects/Desktop/YerToob.py
plugins: cov-2.7.1, mock-1.10.4, qt-3.2.2, testmon-0.9.16
collected 1 item
../../tests/ui/test_login.py Aborted (core dumped)
```
Running on Windows (through pycharm)
```
aunching pytest with arguments E:/Projects/Desktop/YerToob.py/tests/ui/test_login.py in E:\Projects\Desktop\YerToob.py\tests\ui
============================= test session starts =============================
platform win32 -- Python 3.6.5, pytest-4.6.3, py-1.8.0, pluggy-0.12.0 -- E:\Projects\Desktop\YerToob.py\venv\Scripts\python.exe
cachedir: .pytest_cache
PySide2 5.13.0 -- Qt runtime 5.13.0 -- Qt compiled 5.13.0
rootdir: E:\Projects\Desktop\YerToob.py
plugins: cov-2.7.1, mock-1.10.4, qt-3.2.2, testmon-0.9.16
collecting ... collected 1 item
test_login.py::test_load
Process finished with exit code -1073740791 (0xC0000409)
```
Any ideas on what might be causing this? Googling did not help me much though it does suggest the loading of multiple app instances as a possibility.
Any help in getting to the bottom of this would be appreciated.
Thank you :D | closed | 2019-11-19T17:41:37Z | 2019-11-19T19:44:48Z | https://github.com/pytest-dev/pytest-qt/issues/276 | [] | drone-ah | 2 |
ansible/awx | automation | 15,745 | Add Metric for License Expiry | ### Please confirm the following
- [x] I agree to follow this project's [code of conduct](https://docs.ansible.com/ansible/latest/community/code_of_conduct.html).
- [x] I have checked the [current issues](https://github.com/ansible/awx/issues) for duplicates.
- [x] I understand that AWX is open source software provided for free and that I might not receive a timely response.
### Feature type
New Feature
### Feature Summary
Add a metric to show the time until the license for the awx/automation controller will expire.
### Select the relevant components
- [ ] UI
- [x] API
- [ ] Docs
- [ ] Collection
- [ ] CLI
- [ ] Other
### Steps to reproduce
Currently the license expiry date is save as a label in the awx_system_info metric
### Current results
Cannot query prometheus on license expiry easily in promql
### Sugested feature result
Being able to monitor with prometheus alerts when the awx license is expiring (mostly relevent for AnsibleAutomationPlatform)
### Additional information
I opened a PR for this feature request - https://github.com/ansible/awx/pull/15483 | open | 2025-01-14T14:53:09Z | 2025-01-22T18:20:21Z | https://github.com/ansible/awx/issues/15745 | [
"type:enhancement",
"community"
] | neevnuv | 0 |
fastapi/sqlmodel | pydantic | 448 | Relationships with multiple schemas and same table names not working | ### First Check
- [X] I added a very descriptive title to this issue.
- [X] I used the GitHub search to find a similar issue and didn't find it.
- [X] I searched the SQLModel documentation, with the integrated search.
- [X] I already searched in Google "How to X in SQLModel" and didn't find any information.
- [X] I already read and followed all the tutorial in the docs and didn't find an answer.
- [X] I already checked if it is not related to SQLModel but to [Pydantic](https://github.com/samuelcolvin/pydantic).
- [X] I already checked if it is not related to SQLModel but to [SQLAlchemy](https://github.com/sqlalchemy/sqlalchemy).
### Commit to Help
- [X] I commit to help with one of those options 👆
### Example Code
```python
#############################################
################## main.py ##################
#############################################
from sqlmodel import Session, create_engine, select
import workplace1
import workplace2
engine = create_engine("mysql+pymysql://user@localhost")
with Session(engine) as session:
print(select(workplace1.Employee))
print(select(workplace2.Employee))
###################################################
################## workplace1.py ##################
###################################################
from typing import Optional
from sqlmodel import Field, MetaData, Relationship
from sqlmodel import SQLModel as _SQLModel
class SQLModel(_SQLModel):
metadata = MetaData(schema="workplace1")
class Employee(SQLModel, table=True):
__tablename__ = "employees" # type: ignore
employee_id: Optional[int] = Field(primary_key=True)
name: str
business_id: int = Field(foreign_key="business.id")
business: 'Business' = Relationship(back_populates='employees')
class Business(SQLModel, table=True):
__tablename__ = "business" # type: ignore
id: Optional[int] = Field(primary_key=True)
title: str
employees: list['Employee'] = Relationship(back_populates='business_id')
###################################################
################## workplace2.py ##################
###################################################
from typing import Optional
from sqlmodel import Field, MetaData
from sqlmodel import SQLModel as _SQLModel
class SQLModel(_SQLModel):
metadata = MetaData(schema="workplace2")
class Employee(SQLModel, table=True):
__tablename__ = "employees" # type: ignore
employee_id: Optional[int] = Field(primary_key=True)
full_name: str
```
### Description
I'm trying to setup a python project that contains SQLModel models for tables from multiple schemas and some of these schemas share tables with the same name even though the tables are not always related.
The example code above should be split into 3 separate files `main.py`, `workplace1.py` and `workplace2.py` all in the same python module and run from `main.py`.
The separate files are used to separate 2 schemas `workplace1` and `workplace2` which both contain different definitions for a table called `employees`.
What I expect to happen when I run the code (assuming the mysql connection is setup correctly) is to print out 2 `SELECT` statements for the 2 different employees:
```
SELECT workplace1.employees.employee_id, workplace1.employees.name, workplace1.employees.business_id FROM workplace1.employees
SELECT workplace2.employees.employee_id, workplace2.employees.full_name FROM workplace2.employees
```
However the following error occurs:
```
sqlalchemy.exc.InvalidRequestError: Multiple classes found for path "Employee" in the registry of this declarative base. Please use a fully module-qualified path.
```
<details>
<summary>See full error</summary>
```
Traceback (most recent call last):
File "/home/bryley/Documents/testing/python/sqlmodel-testing/main.py", line 68, in <module>
print(select(workplace1.Employee))
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/sql/elements.py", line 570, in __str__
return str(self.compile())
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/sql/elements.py", line 502, in compile
return self._compiler(dialect, **kw)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/sql/elements.py", line 566, in _compiler
return dialect.statement_compiler(dialect, self, **kw)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/sql/compiler.py", line 790, in __init__
Compiled.__init__(self, dialect, statement, **kwargs)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/sql/compiler.py", line 463, in __init__
self.string = self.process(self.statement, **compile_kwargs)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/sql/compiler.py", line 498, in process
return obj._compiler_dispatch(self, **kwargs)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/sql/visitors.py", line 82, in _compiler_dispatch
return meth(self, **kw)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/sql/compiler.py", line 3301, in visit_select
compile_state = select_stmt._compile_state_factory(
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/sql/base.py", line 510, in create_for_statement
return klass.create_for_statement(statement, compiler, **kw)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/context.py", line 699, in create_for_statement
_QueryEntity.to_compile_state(
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/context.py", line 2451, in to_compile_state
_MapperEntity(
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/context.py", line 2524, in __init__
entity._post_inspect
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/util/langhelpers.py", line 1184, in __get__
obj.__dict__[self.__name__] = result = self.fget(obj)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/mapper.py", line 2182, in _post_inspect
self._check_configure()
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/mapper.py", line 1924, in _check_configure
_configure_registries({self.registry}, cascade=True)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/mapper.py", line 3483, in _configure_registries
_do_configure_registries(registries, cascade)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/mapper.py", line 3522, in _do_configure_registries
mapper._post_configure_properties()
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/mapper.py", line 1941, in _post_configure_properties
prop.init()
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/interfaces.py", line 231, in init
self.do_init()
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/relationships.py", line 2150, in do_init
self._generate_backref()
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/relationships.py", line 2486, in _generate_backref
self._add_reverse_property(self.back_populates)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/relationships.py", line 2077, in _add_reverse_property
if not other.mapper.common_parent(self.parent):
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/util/langhelpers.py", line 1113, in __get__
obj.__dict__[self.__name__] = result = self.fget(obj)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/relationships.py", line 2141, in mapper
return self.entity.mapper
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/util/langhelpers.py", line 1113, in __get__
obj.__dict__[self.__name__] = result = self.fget(obj)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/relationships.py", line 2107, in entity
argument = self._clsregistry_resolve_name(self.argument)()
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/clsregistry.py", line 393, in _resolve_name
rval = d[token]
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/util/_collections.py", line 746, in __missing__
self[key] = val = self.creator(key)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/clsregistry.py", line 352, in _access_cls
return _determine_container(key, decl_class_registry[key])
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/clsregistry.py", line 313, in _determine_container
value = value.attempt_get([], key)
File "/home/bryley/.local/lib/python3.10/site-packages/sqlalchemy/orm/clsregistry.py", line 144, in attempt_get
raise exc.InvalidRequestError(
sqlalchemy.exc.InvalidRequestError: Multiple classes found for path "Employee" in the registry of this declarative base. Please use a fully module-qualified path.
```
</details>
It seems as though the code works perfectly fine when removing the `business` and `employees` `Relationship`s from `workplace1` meaning that it most likely has to do with SQLModel/SQLAlchemy getting confused about what `'Employee'` object is being referenced about when executing
```python
employees: list['Employee'] = Relationship(back_populates='business_id')
```
Is there a way to specify what `Employee` object the registry should use in-order to fix this error?
### Operating System
Linux
### Operating System Details
Arch
### SQLModel Version
0.0.8
### Python Version
3.9.13
### Additional Context
For the mysql database driver I use PyMySQL version 1.0.2 | closed | 2022-09-15T00:52:01Z | 2022-11-20T12:19:46Z | https://github.com/fastapi/sqlmodel/issues/448 | [
"question"
] | Bryley | 4 |
python-arq/arq | asyncio | 267 | Option to Cancel Jobs without Incrementing Retries | Currently when a job is canceled (sigint, sigterm), retries is incremented. I'd like for there to be an option for these types of failures to not increment retries. Another way to say it is only jobs that raise an exception should increment retries.
My use case is that I have some long running jobs that I would like to retry if they get canceled due to deploying new code. I don't want it to retry if there's an actually exception though. | open | 2021-10-13T06:32:45Z | 2021-10-13T06:32:45Z | https://github.com/python-arq/arq/issues/267 | [] | tobymao | 0 |
BlinkDL/RWKV-LM | pytorch | 53 | Any results on few-shot settings? | Thanks for your wonderful work!
Do you have any results on the few-shot settings? Do RWKV-LLMs perform the similar emergent ability like GPT-3, e.g. the chain-of-thought? | closed | 2023-03-23T12:41:01Z | 2023-03-24T10:50:56Z | https://github.com/BlinkDL/RWKV-LM/issues/53 | [] | ustcwhy | 1 |
proplot-dev/proplot | data-visualization | 219 | xminorlocator=1 or yminorlocator=1 doesn't work | I think this line creates a problem when we define `yminorlocator=1` or `xminorlocator=1`:
https://github.com/lukelbd/proplot/blob/e1a603729305abe791ed23d84cfeff11e5a4cea6/proplot/axes/cartesian.py#L1101
### Example
```python
import proplot as plot
import numpy as np
fig, ax = plot.subplots(aspect=4, axwidth=5)
ax.format(
xlim=(0, 200), xminorlocator=10, xlocator=30,
ylim=(0, 4), yminorlocator=1, ylocator=2
)
```
Raise the warning:
```python
proplot/axes/cartesian.py:1102: ProPlotWarning: You passed yminorticks=1, but this argument is used to specify
tick *locations*. If you just want to *toggle* minor ticks on and off, please use ytickminor=True or
ytickminor=False.
```
A way to fix this is to pass `minorlocator=1.000001` for example, I guess the problem is that in Python `1` is equivalent to `True`.
### Proplot version
0.6.4 | closed | 2020-08-14T13:39:16Z | 2021-06-30T01:52:35Z | https://github.com/proplot-dev/proplot/issues/219 | [
"bug"
] | mickaellalande | 1 |
piccolo-orm/piccolo | fastapi | 625 | exception raised on update with a join in the where clause | i couldn't think of a concise way to explain my problem so here's an example:
```python
In [19]: await Band.select(Band.name).where(Band.manager.name == 'Guido')
SELECT "band"."name" FROM band LEFT JOIN manager band$manager ON (band.manager = band$manager.id) WHERE "band$manager"."name" = 'Guido'
Out[19]: [{'name': 'Pythonistas'}]
In [20]: await Band.update({Band.name: 'The Pythonistas'}).where(Band.manager.name == 'Guido')
UPDATE band SET "name" = 'The Pythonistas' WHERE "band$manager"."name" = 'Guido'
---------------------------------------------------------------------------
OperationalError Traceback (most recent call last)
Cell In [20], line 1
----> 1 await Band.update({Band.name: 'The Pythonistas'}).where(Band.manager.name == 'Guido')
... traceback removed ...
OperationalError: no such column: band$manager.name
``` | closed | 2022-09-28T13:02:13Z | 2023-02-21T19:03:27Z | https://github.com/piccolo-orm/piccolo/issues/625 | [] | backwardspy | 4 |
timkpaine/lantern | plotly | 25 | matplotlib - candlestick | closed | 2017-10-10T01:30:48Z | 2017-11-26T05:13:38Z | https://github.com/timkpaine/lantern/issues/25 | [
"feature",
"matplotlib/seaborn"
] | timkpaine | 1 |
|
allure-framework/allure-python | pytest | 45 | Extract python commons | closed | 2017-02-19T16:46:12Z | 2017-03-21T16:42:26Z | https://github.com/allure-framework/allure-python/issues/45 | [] | sseliverstov | 0 |
|
donnemartin/system-design-primer | python | 816 | Update to a URL | https://github.com/donnemartin/system-design-primer/blob/a07e261677c012d37d26255de6e7b128a2643946/README.md?plain=1#L1321C32-L1321C109
The URL for Elasticache caching strategies must be updated to https://docs.aws.amazon.com/AmazonElastiCache/latest/red-ug/Strategies.html | open | 2023-12-07T14:31:59Z | 2024-06-23T17:39:50Z | https://github.com/donnemartin/system-design-primer/issues/816 | [
"needs-review"
] | alimoosavi27 | 4 |
aleju/imgaug | machine-learning | 68 | OneOf wrong behaviour | Hi,
looks like there is something broken there, I suppose.
With this code, I get at the same time: dropout and Add+
Dropout is always used.
Never get Add- nor constrastNorm.
```
color = iaa.Sometimes(1,iaa.OneOf([
iaa.Add((-50, -20)),
iaa.Add((20, 50)),
iaa.ContrastNormalization((0.3, 0.5)),
iaa.ContrastNormalization((1.5, 1.9))
iaa.CoarseDropout(p=0.05, size_percent=0.2)
]))
```
I use like this (I know, it's not good..):
```
img1 = iaa.Sequential([iaa.Fliplr(p=1),color]).augment_image(img)
img2 = iaa.Sequential([iaa.Fliplr(p=1),color]).augment_image(img)
``` | closed | 2017-10-04T15:39:58Z | 2017-10-04T17:46:04Z | https://github.com/aleju/imgaug/issues/68 | [] | vimalaguti | 2 |
gradio-app/gradio | data-science | 10,436 | Theme is only used on first tab | ### Describe the bug
Monochrome is set for the interface, but only gets applied to the first tab. Tab 2 and 3 seem to be still using the default theme.
### Have you searched existing issues? 🔎
- [x] I have searched and found no existing issues
### Reproduction
```python
import gradio as gr
with gr.Blocks(title='Admin UI', theme=gr.themes.Monochrome) as app:
with gr.Tabs():
with gr.Tab("Tab 1"):
button1 = gr.Button("Button")
with gr.Tab("Tab 2"):
button2 = gr.Button("Button")
with gr.Tab("Tab 3"):
button3 = gr.Button("Button")
```
### Screenshot
_No response_
### Logs
```shell
```
### System Info
```shell
Gradio Environment Information:
------------------------------
Operating System: Linux
gradio version: 5.13.1
gradio_client version: 1.6.0
------------------------------------------------
gradio dependencies in your environment:
aiofiles: 23.2.1
anyio: 4.8.0
audioop-lts is not installed.
fastapi: 0.115.7
ffmpy: 0.5.0
gradio-client==1.6.0 is not installed.
httpx: 0.28.1
huggingface-hub: 0.27.1
jinja2: 3.1.5
markupsafe: 3.0.2
numpy: 2.2.1
orjson: 3.10.15
packaging: 24.2
pandas: 2.2.3
pillow: 11.1.0
pydantic: 2.10.5
pydub: 0.25.1
python-multipart: 0.0.20
pyyaml: 6.0.2
ruff: 0.9.3
safehttpx: 0.1.6
semantic-version: 2.10.0
starlette: 0.45.3
tomlkit: 0.13.2
typer: 0.15.1
typing-extensions: 4.12.2
urllib3: 2.3.0
uvicorn: 0.34.0
authlib; extra == 'oauth' is not installed.
itsdangerous; extra == 'oauth' is not installed.
gradio_client dependencies in your environment:
fsspec: 2024.12.0
httpx: 0.28.1
huggingface-hub: 0.27.1
packaging: 24.2
typing-extensions: 4.12.2
websockets: 14.2
```
### Severity
I can work around it | closed | 2025-01-24T22:41:03Z | 2025-01-27T19:15:26Z | https://github.com/gradio-app/gradio/issues/10436 | [
"bug",
"needs repro"
] | GregSommerville | 3 |
lux-org/lux | pandas | 500 | [BUG] | **Describe the bug**
Unexpected error in rendering Lux widget and recommendations. Falling back to Pandas display.
| open | 2023-10-01T19:39:17Z | 2023-10-01T19:39:17Z | https://github.com/lux-org/lux/issues/500 | [] | Saramagit | 0 |
AUTOMATIC1111/stable-diffusion-webui | pytorch | 16,551 | [Bug]: ModuleNotFoundError: No module named 'timm' | ### Checklist
- [ ] The issue exists after disabling all extensions
- [X] The issue exists on a clean installation of webui
- [ ] The issue is caused by an extension, but I believe it is caused by a bug in the webui
- [ ] The issue exists in the current version of the webui
- [ ] The issue has not been reported before recently
- [ ] The issue has been reported before but has not been fixed yet
### What happened?
root@debian:/dockerx/stable-diffusion-webui# REQS_FILE='requirements.txt' python launch.py --precision full --no-half
Python 3.10.15 (main, Oct 3 2024, 07:27:34) [GCC 11.2.0]
Version: v1.10.1
Commit hash: 82a973c04367123ae98bd9abdf80d9eda9b910e2
Launching Web UI with arguments: --precision full --no-half
no module 'xformers'. Processing without...
no module 'xformers'. Processing without...
No module 'xformers'. Proceeding without it.
Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/launch.py", line 48, in <module>
main()
File "/dockerx/stable-diffusion-webui/launch.py", line 44, in main
start()
File "/dockerx/stable-diffusion-webui/modules/launch_utils.py", line 465, in start
import webui
File "/dockerx/stable-diffusion-webui/webui.py", line 13, in <module>
initialize.imports()
File "/dockerx/stable-diffusion-webui/modules/initialize.py", line 39, in imports
from modules import processing, gradio_extensons, ui # noqa: F401
File "/dockerx/stable-diffusion-webui/modules/processing.py", line 31, in <module>
from ldm.data.util import AddMiDaS
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/data/util.py", line 3, in <module>
from ldm.modules.midas.api import load_midas_transform
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/modules/midas/api.py", line 8, in <module>
from ldm.modules.midas.midas.dpt_depth import DPTDepthModel
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/modules/midas/midas/dpt_depth.py", line 6, in <module>
from .blocks import (
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/modules/midas/midas/blocks.py", line 4, in <module>
from .vit import (
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/modules/midas/midas/vit.py", line 3, in <module>
import timm
ModuleNotFoundError: No module named 'timm'
### Steps to reproduce the problem
1. Install the prooject using the docker method.
2. get to the steep where you have to run the following command
3. `root@debian:/dockerx/stable-diffusion-webui# REQS_FILE='requirements.txt' python launch.py --precision full --no-half`
4. You get this error:
```
root@debian:/dockerx/stable-diffusion-webui# REQS_FILE='requirements.txt' python launch.py --precision full --no-half
Python 3.10.15 (main, Oct 3 2024, 07:27:34) [GCC 11.2.0]
Version: v1.10.1
Commit hash: 82a973c04367123ae98bd9abdf80d9eda9b910e2
Launching Web UI with arguments: --precision full --no-half
no module 'xformers'. Processing without...
no module 'xformers'. Processing without...
No module 'xformers'. Proceeding without it.
Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/launch.py", line 48, in <module>
main()
File "/dockerx/stable-diffusion-webui/launch.py", line 44, in main
start()
File "/dockerx/stable-diffusion-webui/modules/launch_utils.py", line 465, in start
import webui
File "/dockerx/stable-diffusion-webui/webui.py", line 13, in <module>
initialize.imports()
File "/dockerx/stable-diffusion-webui/modules/initialize.py", line 39, in imports
from modules import processing, gradio_extensons, ui # noqa: F401
File "/dockerx/stable-diffusion-webui/modules/processing.py", line 31, in <module>
from ldm.data.util import AddMiDaS
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/data/util.py", line 3, in <module>
from ldm.modules.midas.api import load_midas_transform
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/modules/midas/api.py", line 8, in <module>
from ldm.modules.midas.midas.dpt_depth import DPTDepthModel
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/modules/midas/midas/dpt_depth.py", line 6, in <module>
from .blocks import (
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/modules/midas/midas/blocks.py", line 4, in <module>
from .vit import (
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/modules/midas/midas/vit.py", line 3, in <module>
import timm
ModuleNotFoundError: No module named 'timm'
```
### What should have happened?
The project should be installed correctly.
### What browsers do you use to access the UI ?
Google Chrome
### Sysinfo
Debian 12
### Console logs
```Shell
root@debian:/dockerx/stable-diffusion-webui# REQS_FILE='requirements.txt' python launch.py --precision full --no-half
Python 3.10.15 (main, Oct 3 2024, 07:27:34) [GCC 11.2.0]
Version: v1.10.1
Commit hash: 82a973c04367123ae98bd9abdf80d9eda9b910e2
Launching Web UI with arguments: --precision full --no-half
no module 'xformers'. Processing without...
no module 'xformers'. Processing without...
No module 'xformers'. Proceeding without it.
Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/launch.py", line 48, in <module>
main()
File "/dockerx/stable-diffusion-webui/launch.py", line 44, in main
start()
File "/dockerx/stable-diffusion-webui/modules/launch_utils.py", line 465, in start
import webui
File "/dockerx/stable-diffusion-webui/webui.py", line 13, in <module>
initialize.imports()
File "/dockerx/stable-diffusion-webui/modules/initialize.py", line 39, in imports
from modules import processing, gradio_extensons, ui # noqa: F401
File "/dockerx/stable-diffusion-webui/modules/processing.py", line 31, in <module>
from ldm.data.util import AddMiDaS
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/data/util.py", line 3, in <module>
from ldm.modules.midas.api import load_midas_transform
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/modules/midas/api.py", line 8, in <module>
from ldm.modules.midas.midas.dpt_depth import DPTDepthModel
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/modules/midas/midas/dpt_depth.py", line 6, in <module>
from .blocks import (
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/modules/midas/midas/blocks.py", line 4, in <module>
from .vit import (
File "/dockerx/stable-diffusion-webui/repositories/stable-diffusion-stability-ai/ldm/modules/midas/midas/vit.py", line 3, in <module>
import timm
ModuleNotFoundError: No module named 'timm'
```
### Additional information
I installed timm manually and it fixed the problem.
```
root@debian:/dockerx/stable-diffusion-webui# pip install timm
Collecting timm
Downloading timm-1.0.9-py3-none-any.whl.metadata (42 kB)
Requirement already satisfied: torch in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from timm) (2.3.0a0+gitd2f9472)
Requirement already satisfied: torchvision in /opt/conda/envs/py_3.10/lib/python3.10/site-packages/torchvision-0.18.0a0+68ba7ec-py3.10-linux-x86_64.egg (from timm) (0.18.0a0+68ba7ec)
Requirement already satisfied: pyyaml in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from timm) (6.0.1)
Requirement already satisfied: huggingface_hub in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from timm) (0.25.2)
Requirement already satisfied: safetensors in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from timm) (0.4.5)
Requirement already satisfied: filelock in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from huggingface_hub->timm) (3.16.1)
Requirement already satisfied: fsspec>=2023.5.0 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages/fsspec-2024.9.0-py3.10.egg (from huggingface_hub->timm) (2024.9.0)
Requirement already satisfied: packaging>=20.9 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from huggingface_hub->timm) (24.1)
Requirement already satisfied: requests in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from huggingface_hub->timm) (2.32.3)
Requirement already satisfied: tqdm>=4.42.1 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from huggingface_hub->timm) (4.66.5)
Requirement already satisfied: typing-extensions>=3.7.4.3 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from huggingface_hub->timm) (4.12.2)
Requirement already satisfied: sympy<=1.12.1 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from torch->timm) (1.12.1)
Requirement already satisfied: networkx in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from torch->timm) (2.8.8)
Requirement already satisfied: jinja2 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from torch->timm) (3.1.3)
Requirement already satisfied: numpy<2 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from torchvision->timm) (1.26.4)
Requirement already satisfied: pillow!=8.3.*,>=5.3.0 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from torchvision->timm) (10.2.0)
Requirement already satisfied: mpmath<1.4.0,>=1.1.0 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from sympy<=1.12.1->torch->timm) (1.3.0)
Requirement already satisfied: MarkupSafe>=2.0 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from jinja2->torch->timm) (2.1.5)
Requirement already satisfied: charset-normalizer<4,>=2 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from requests->huggingface_hub->timm) (3.4.0)
Requirement already satisfied: idna<4,>=2.5 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from requests->huggingface_hub->timm) (3.10)
Requirement already satisfied: urllib3<3,>=1.21.1 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from requests->huggingface_hub->timm) (1.26.20)
Requirement already satisfied: certifi>=2017.4.17 in /opt/conda/envs/py_3.10/lib/python3.10/site-packages (from requests->huggingface_hub->timm) (2024.8.30)
Downloading timm-1.0.9-py3-none-any.whl (2.3 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 2.3/2.3 MB 15.9 MB/s eta 0:00:00
Installing collected packages: timm
Successfully installed timm-1.0.9
WARNING: Running pip as the 'root' user can result in broken permissions and conflicting behaviour with the system package manager, possibly rendering your system unusable.It is recommended to use a virtual environment instead: https://pip.pypa.io/warnings/venv. Use the --root-user-action option if you know what you are doing and want to suppress this warning.
```
Then i get this error:
```
root@debian:/dockerx/stable-diffusion-webui# REQS_FILE='requirements.txt' python launch.py --precision full --no-half
Python 3.10.15 (main, Oct 3 2024, 07:27:34) [GCC 11.2.0]
Version: v1.10.1
Commit hash: 82a973c04367123ae98bd9abdf80d9eda9b910e2
Launching Web UI with arguments: --precision full --no-half
no module 'xformers'. Processing without...
no module 'xformers'. Processing without...
No module 'xformers'. Proceeding without it.
Pytorch_lightning.distributed not found, attempting pytorch_lightning.rank_zero
Downloading: "https://huggingface.co/runwayml/stable-diffusion-v1-5/resolve/main/v1-5-pruned-emaonly.safetensors" to /dockerx/stable-diffusion-webui/models/Stable-diffusion/v1-5-pruned-emaonly.safetensors
100%|██████████████████████████████████████████████████████████████| 3.97G/3.97G [02:48<00:00, 25.4MB/s]
*** Error creating UI for None:
Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 640, in create_script_ui
self.create_script_ui_inner(script)
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 645, in create_script_ui_inner
import modules.api.models as api_models
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 113, in <module>
).generate_model()
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 96, in generate_model
DynamicModel.__config__.allow_population_by_field_name = True
File "/opt/conda/envs/py_3.10/lib/python3.10/site-packages/pydantic/_internal/_model_construction.py", line 262, in __getattr__
raise AttributeError(item)
AttributeError: __config__
---
*** Error creating UI for None:
Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 640, in create_script_ui
self.create_script_ui_inner(script)
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 645, in create_script_ui_inner
import modules.api.models as api_models
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 113, in <module>
).generate_model()
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 96, in generate_model
DynamicModel.__config__.allow_population_by_field_name = True
File "/opt/conda/envs/py_3.10/lib/python3.10/site-packages/pydantic/_internal/_model_construction.py", line 262, in __getattr__
raise AttributeError(item)
AttributeError: __config__
---
*** Error creating UI for None:
Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 640, in create_script_ui
self.create_script_ui_inner(script)
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 645, in create_script_ui_inner
import modules.api.models as api_models
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 113, in <module>
).generate_model()
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 96, in generate_model
DynamicModel.__config__.allow_population_by_field_name = True
File "/opt/conda/envs/py_3.10/lib/python3.10/site-packages/pydantic/_internal/_model_construction.py", line 262, in __getattr__
raise AttributeError(item)
AttributeError: __config__
---
*** Error creating UI for None:
Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 640, in create_script_ui
self.create_script_ui_inner(script)
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 645, in create_script_ui_inner
import modules.api.models as api_models
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 113, in <module>
).generate_model()
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 96, in generate_model
DynamicModel.__config__.allow_population_by_field_name = True
File "/opt/conda/envs/py_3.10/lib/python3.10/site-packages/pydantic/_internal/_model_construction.py", line 262, in __getattr__
raise AttributeError(item)
AttributeError: __config__
---
*** Error creating UI for Hypertile:
Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 640, in create_script_ui
self.create_script_ui_inner(script)
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 645, in create_script_ui_inner
import modules.api.models as api_models
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 113, in <module>
).generate_model()
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 96, in generate_model
DynamicModel.__config__.allow_population_by_field_name = True
File "/opt/conda/envs/py_3.10/lib/python3.10/site-packages/pydantic/_internal/_model_construction.py", line 262, in __getattr__
raise AttributeError(item)
AttributeError: __config__
---
*** Error creating UI for None:
Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 640, in create_script_ui
self.create_script_ui_inner(script)
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 645, in create_script_ui_inner
import modules.api.models as api_models
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 113, in <module>
).generate_model()
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 96, in generate_model
DynamicModel.__config__.allow_population_by_field_name = True
File "/opt/conda/envs/py_3.10/lib/python3.10/site-packages/pydantic/_internal/_model_construction.py", line 262, in __getattr__
raise AttributeError(item)
AttributeError: __config__
---
*** Error creating UI for None:
Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 640, in create_script_ui
self.create_script_ui_inner(script)
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 645, in create_script_ui_inner
import modules.api.models as api_models
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 113, in <module>
).generate_model()
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 96, in generate_model
DynamicModel.__config__.allow_population_by_field_name = True
File "/opt/conda/envs/py_3.10/lib/python3.10/site-packages/pydantic/_internal/_model_construction.py", line 262, in __getattr__
raise AttributeError(item)
AttributeError: __config__
---
*** Error creating UI for None:
Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 640, in create_script_ui
self.create_script_ui_inner(script)
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 645, in create_script_ui_inner
import modules.api.models as api_models
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 113, in <module>
).generate_model()
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 96, in generate_model
DynamicModel.__config__.allow_population_by_field_name = True
File "/opt/conda/envs/py_3.10/lib/python3.10/site-packages/pydantic/_internal/_model_construction.py", line 262, in __getattr__
raise AttributeError(item)
AttributeError: __config__
---
*** Error creating UI for None:
Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 640, in create_script_ui
self.create_script_ui_inner(script)
File "/dockerx/stable-diffusion-webui/modules/scripts.py", line 645, in create_script_ui_inner
import modules.api.models as api_models
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 113, in <module>
).generate_model()
File "/dockerx/stable-diffusion-webui/modules/api/models.py", line 96, in generate_model
DynamicModel.__config__.allow_population_by_field_name = True
File "/opt/conda/envs/py_3.10/lib/python3.10/site-packages/pydantic/_internal/_model_construction.py", line 262, in __getattr__
raise AttributeError(item)
AttributeError: __config__
---
Calculating sha256 for /dockerx/stable-diffusion-webui/models/Stable-diffusion/v1-5-pruned-emaonly.safetensors: Traceback (most recent call last):
File "/dockerx/stable-diffusion-webui/launch.py", line 48, in <module>
main()
File "/dockerx/stable-diffusion-webui/launch.py", line 44, in main
start()
File "/dockerx/stable-diffusion-webui/modules/launch_utils.py", line 469, in start
webui.webui()
File "/dockerx/stable-diffusion-webui/webui.py", line 64, in webui
shared.demo = ui.create_ui()
File "/dockerx/stable-diffusion-webui/modules/ui.py", line 489, in create_ui
toprow.ui_styles.dropdown.change(fn=wrap_queued_call(update_token_counter), inputs=[toprow.prompt, steps, toprow.ui_styles.dropdown], outputs=[toprow.token_counter])
File "/opt/conda/envs/py_3.10/lib/python3.10/site-packages/gradio/events.py", line 140, in __call__
dep, dep_index = self.trigger.set_event_trigger(
File "/opt/conda/envs/py_3.10/lib/python3.10/site-packages/gradio/blocks.py", line 286, in set_event_trigger
"inputs": [block._id for block in inputs],
File "/opt/conda/envs/py_3.10/lib/python3.10/site-packages/gradio/blocks.py", line 286, in <listcomp>
"inputs": [block._id for block in inputs],
AttributeError: 'NoneType' object has no attribute '_id'
6ce0161689b3853acaa03779ec93eafe75a02f4ced659bee03f50797806fa2fa
Loading weights [6ce0161689] from /dockerx/stable-diffusion-webui/models/Stable-diffusion/v1-5-pruned-emaonly.safetensors
Creating model from config: /dockerx/stable-diffusion-webui/configs/v1-inference.yaml
/opt/conda/envs/py_3.10/lib/python3.10/site-packages/huggingface_hub/file_download.py:1142: FutureWarning: `resume_download` is deprecated and will be removed in version 1.0.0. Downloads always resume when possible. If you want to force a new download, use `force_download=True`.
warnings.warn(
vocab.json: 100%|████████████████████████████████████████████████████| 961k/961k [00:00<00:00, 3.86MB/s]
merges.txt: 100%|████████████████████████████████████████████████████| 525k/525k [00:00<00:00, 5.36MB/s]
special_tokens_map.json: 100%|█████████████████████████████████████████| 389/389 [00:00<00:00, 3.12MB/s]
tokenizer_config.json: 100%|███████████████████████████████████████████| 905/905 [00:00<00:00, 10.8MB/s]
config.json: 100%|█████████████████████████████████████████████████| 4.52k/4.52k [00:00<00:00, 33.4MB/s]
```
| open | 2024-10-13T00:25:23Z | 2025-01-11T16:53:43Z | https://github.com/AUTOMATIC1111/stable-diffusion-webui/issues/16551 | [
"bug-report"
] | wolfred24 | 3 |
axnsan12/drf-yasg | rest-api | 235 | Swagger not fetching details from swagger_auto_schema | I am using **swagger_auto_schema** in **function based views** like below. It seems like **Swagger** is not using the **annotations** at all. What am I doing wrong?
urls.py
```
from django.conf.urls import url
from .views import check_phone
urlpatterns = [
url(r'^check_phone/?', check_phone),
]
```
views.py
```
class CheckPhoneSerializer(serializers.Serializer):
phone = serializers.CharField(max_length=20)
exist = serializers.BooleanField()
phone_param = Parameter('phone', IN_QUERY, description="user phone number", type=TYPE_STRING)
check_phone_response = Response('Response contains phone exists or not', CheckPhoneSerializer)
@csrf_exempt
@api_view(["GET"])
@swagger_auto_schema(operation_description="Checks whether phone number already exists", operation_id='agent_auth_check_phone', manual_parameters=[phone_param], responses={200: check_phone_response})
@permission_classes((IsAuthenticated,))
@require_http_methods(["GET", "POST"])
@renderer_classes([renderers.OpenAPIRenderer, renderers.SwaggerUIRenderer])
def check_phone(request):
phone = request.GET.get("phone") if request.method == 'GET' else request.POST.get("phone")
data = {"phone": phone, "exist": True}
return JsonResponse(data)
``` | closed | 2018-10-18T21:01:13Z | 2018-10-23T06:09:54Z | https://github.com/axnsan12/drf-yasg/issues/235 | [] | deepakzipgo | 4 |
pytest-dev/pytest-randomly | pytest | 550 | Feature enhancement to allow turning plugin on and off | ### Description
Hello,
Using this plugin occasionally while writing new tests to prevent flakiness, but old tests still need to run without the random order.
Currently accomplishing this via pytest.ini which includes -p no:randomly and a separate one you use via -c pytest-randomly.ini
This feels rather clunky, I'd like to propose creating a pytest command line argument to turn the plugin off as needed
The other option is to create a bash variable PYTEST_USE_RANDOMLY which defaults to true, but can be flipped on and off as you go
I'm more than happy to make this change, but wanted to run it by the community before opening the PR. | closed | 2023-06-07T16:10:34Z | 2024-10-12T05:59:39Z | https://github.com/pytest-dev/pytest-randomly/issues/550 | [] | clintonsteiner | 7 |
Kanaries/pygwalker | matplotlib | 510 | Would it be possbile to walk it with Datatable.frame type in the future? | https://github.com/h2oai/datatable
It has higher performance reading and manipulating big csv data than pandas/polars/modin. But I can't walk this datatable.frame type. | open | 2024-04-01T17:59:46Z | 2024-04-02T06:55:41Z | https://github.com/Kanaries/pygwalker/issues/510 | [
"Vote if you want it",
"proposal"
] | plusmid | 1 |
lanpa/tensorboardX | numpy | 155 | add_video() got an unexpected keyword argument 'fps' | error: add_video() got an unexpected keyword argument 'fps' | closed | 2018-06-01T17:27:37Z | 2018-06-01T17:33:31Z | https://github.com/lanpa/tensorboardX/issues/155 | [] | kekmodel | 0 |
bmoscon/cryptofeed | asyncio | 254 | Understanding content of order and trade | Hello,
I have rather general questions, but I think others can have the same, so I write this in a ticket instead of using slack.
Order and trade structures are defined in
https://github.com/bmoscon/cryptofeed/blob/master/cryptofeed/defines.py
Order has most notably these fields:
```python
{
order_id: str,
side: str, # bids or ask
order_type: str, # limit/market/etc,
price: Decimal,
total: Decimal,
executed: Decimal,
pending: Decimal,
order_status: str #FILLED/PARTIAL/CANCELLED/OPEN
}
```
Please, can you confirm 'pending' is only for limit orders?
Also, assuming there is one market order, bigger than the 1st available limit order in the order book.
In the order structure, there is a field for a single price.
How is it then materialized in this order structure? Will there be as many orders as the market order needs to be sliced to get fully filled?
Also, trade has following structure:
```python
{
'price': Decimal,
'amount': Decimal,
'timestamp': float,
'side': str
'fee_currency': str,
'fee_amount': Decimal,
'trade_id': str,
'order_id': str
}
```
So for one market order, does it create as many `trade`s and `order`s as required to get it filled ?
On the opposite, for a limit order, only one `order` is created and no `trade`?
Am I right in this understanding?
Thanks a lot for your reply.
Have a good day,
Bests, | closed | 2020-06-25T20:04:57Z | 2020-06-25T20:55:55Z | https://github.com/bmoscon/cryptofeed/issues/254 | [] | yohplala | 4 |
mwaskom/seaborn | data-science | 3,466 | Address deprecations shown with pandas 2.1.0 | Hi Michael,
Not sure if this is already being addressed...there seem to be a few deprecation warnings with the latest version of pandas (https://pandas.pydata.org/docs/whatsnew/v2.1.0.html#backwards-incompatible-api-changes):
```text
.../venv/lib/python3.9/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
.../venv/lib/python3.9/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
```
I am also seeing a few different deprecated calls in _oldcore.py. Seems though as if these are easy fixes.
Are you planning to address these?
Many thanks,
Karsten
| closed | 2023-09-07T17:34:31Z | 2023-09-07T23:10:39Z | https://github.com/mwaskom/seaborn/issues/3466 | [] | km2023-4 | 1 |
BeanieODM/beanie | pydantic | 302 | [feature] Custom type encoders | Have you considered to allow for custom type encoders? Let me explain.
Pydantic supports custom types. In fact I can implement one classes to overload default types, like in this example:
```python
class SpecialStr(str, BaseType):
__slots__ = []
@classmethod
def __get_validators__(cls):
yield cls.validate
@classmethod
def validate(cls, v):
return cls(v)
def __repr__(self):
return f'{self.__class__.__name__}({super().__repr__()})'
def party(self) -> "SpecialString":
return SpecialString(f'🥳 {self} 🥳')
class Model(BaseModel):
value: SpecialStr
m = Model(value='house')
m.value.party() # '🥳 house 🥳'
```
But this function alone does not give the flexibility to abstract database values. As an example, let's assume I would like to have a type `ImgurUrl` which is used to store cdn links like `https://i.imgur.com/rr7pioJ.png`. Easy to see that the link contains a lot of redundant data and assuming the CDN provider does not change it can be stored as `rr7pioJ.png`. I would like to have the possibility to solve this with just a simple custom type, that isolates this behavior in a single class. Something like this:
```python
class ImgurUrl(str, BaseType):
__slots__ = []
@classmethod
def __get_validators__(cls):
yield cls.validate
@classmethod
def validate(cls, v):
return cls(v)
@classmethod
def _from_db(cls, value: str) -> "ImgurUrl":
return ImgurUrl(f'https://i.imgur.com/{value}')
def _to_db(self) -> str:
if self.startswith('https://i.imgur.com/'):
return self[20:]
else:
raise RuntimeError(f'ImgurUrl does not contain a Imgur Url')
class Model(Document):
link: ImgurUrl
```
To achieve this the addition of functions like `_to_db` and `_from_db` is required. The first can be solved by modifyling `beanie.odm.utils.encoder` the second one would need a bit more work in `beanie.odm.utils.parsing`.
Such functions would allow the implementation of custom behavior at the argument or document level.
| closed | 2022-07-07T23:05:28Z | 2023-01-07T02:28:53Z | https://github.com/BeanieODM/beanie/issues/302 | [
"Stale"
] | svaraborut | 3 |
google-research/bert | tensorflow | 1,177 | 3090 | I used a 3090 graphics card, TensorFlow1.14. There will be a problem. The same code will work on 2080. Have you come across anything like this? | open | 2020-11-23T06:50:48Z | 2020-11-23T06:50:48Z | https://github.com/google-research/bert/issues/1177 | [] | Mbdn | 0 |
strawberry-graphql/strawberry | graphql | 3,276 | Incorrect template rendering when overriding graphql IDE template | The GraphQLView and its async version allow overriding the template file. However, the loading of the template is incorrect, resulting in `JSON.parse("") // empty string` , because the variable is already replaced.
## Additional Context
In the view we see the following code:
```python
try:
template = Template(render_to_string("graphql/graphiql.html"))
except TemplateDoesNotExist:
template = Template(self.graphql_ide_html)
```
The loading in the try section, erronously calls render_to_string and sticks the result into a Template class. However, render_to_string already renders the entire template, replacing its variables. | open | 2023-12-05T18:22:15Z | 2025-03-20T15:56:30Z | https://github.com/strawberry-graphql/strawberry/issues/3276 | [
"bug"
] | msopacua | 2 |
ets-labs/python-dependency-injector | asyncio | 91 | Add Python 3.5 support | closed | 2015-09-21T14:43:49Z | 2015-10-12T15:23:02Z | https://github.com/ets-labs/python-dependency-injector/issues/91 | [
"enhancement"
] | rmk135 | 1 |
|
iperov/DeepFaceLab | deep-learning | 5,533 | 3050ti. 2 root error found | Starting. Press "Enter" to stop training and save model.
Trying to do the first iteration. If an error occurs, reduce the model parameters.
!!!
Windows 10 users IMPORTANT notice. You should set this setting in order to work correctly.
https://i.imgur.com/B7cmDCB.jpg
!!!
You are training the model from scratch. It is strongly recommended to use a pretrained model to speed up the training and improve the quality.
Error: 2 root error(s) found.
(0) Resource exhausted: failed to allocate memory
[[node mul_89 (defined at D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\core\leras\optimizers\AdaBelief.py:64) ]]
Hint: If you want to see a list of allocated tensors when OOM happens, add report_tensor_allocations_upon_oom to RunOptions for current allocation info. This isn't available when running in Eager mode.
[[concat_8/concat/_103]]
Hint: If you want to see a list of allocated tensors when OOM happens, add report_tensor_allocations_upon_oom to RunOptions for current allocation info. This isn't available when running in Eager mode.
(1) Resource exhausted: failed to allocate memory
[[node mul_89 (defined at D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\core\leras\optimizers\AdaBelief.py:64) ]]
Hint: If you want to see a list of allocated tensors when OOM happens, add report_tensor_allocations_upon_oom to RunOptions for current allocation info. This isn't available when running in Eager mode.
0 successful operations.
0 derived errors ignored.
Errors may have originated from an input operation.
Input Source operations connected to node mul_89:
src_dst_opt/vs_inter_AB/upscale1/conv1/weight_0/read (defined at D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\core\leras\optimizers\AdaBelief.py:38)
Input Source operations connected to node mul_89:
src_dst_opt/vs_inter_AB/upscale1/conv1/weight_0/read (defined at D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\core\leras\optimizers\AdaBelief.py:38)
Original stack trace for 'mul_89':
File "threading.py", line 884, in _bootstrap
File "threading.py", line 916, in _bootstrap_inner
File "threading.py", line 864, in run
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\mainscripts\Trainer.py", line 58, in trainerThread
debug=debug)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\models\ModelBase.py", line 193, in __init__
self.on_initialize()
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\models\Model_SAEHD\Model.py", line 564, in on_initialize
src_dst_loss_gv_op = self.src_dst_opt.get_update_op (nn.average_gv_list (gpu_G_loss_gvs))
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\core\leras\optimizers\AdaBelief.py", line 64, in get_update_op
v_t = self.beta_2*vs + (1.0-self.beta_2) * tf.square(g-m_t)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\ops\variables.py", line 1076, in _run_op
return tensor_oper(a.value(), *args, **kwargs)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\ops\math_ops.py", line 1400, in r_binary_op_wrapper
return func(x, y, name=name)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\ops\math_ops.py", line 1710, in _mul_dispatch
return multiply(x, y, name=name)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\util\dispatch.py", line 206, in wrapper
return target(*args, **kwargs)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\ops\math_ops.py", line 530, in multiply
return gen_math_ops.mul(x, y, name)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\ops\gen_math_ops.py", line 6245, in mul
"Mul", x=x, y=y, name=name)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\framework\op_def_library.py", line 750, in _apply_op_helper
attrs=attr_protos, op_def=op_def)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\framework\ops.py", line 3569, in _create_op_internal
op_def=op_def)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\framework\ops.py", line 2045, in __init__
self._traceback = tf_stack.extract_stack_for_node(self._c_op)
Traceback (most recent call last):
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\client\session.py", line 1375, in _do_call
return fn(*args)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\client\session.py", line 1360, in _run_fn
target_list, run_metadata)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\client\session.py", line 1453, in _call_tf_sessionrun
run_metadata)
tensorflow.python.framework.errors_impl.ResourceExhaustedError: 2 root error(s) found.
(0) Resource exhausted: failed to allocate memory
[[{{node mul_89}}]]
Hint: If you want to see a list of allocated tensors when OOM happens, add report_tensor_allocations_upon_oom to RunOptions for current allocation info. This isn't available when running in Eager mode.
[[concat_8/concat/_103]]
Hint: If you want to see a list of allocated tensors when OOM happens, add report_tensor_allocations_upon_oom to RunOptions for current allocation info. This isn't available when running in Eager mode.
(1) Resource exhausted: failed to allocate memory
[[{{node mul_89}}]]
Hint: If you want to see a list of allocated tensors when OOM happens, add report_tensor_allocations_upon_oom to RunOptions for current allocation info. This isn't available when running in Eager mode.
0 successful operations.
0 derived errors ignored.
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\mainscripts\Trainer.py", line 129, in trainerThread
iter, iter_time = model.train_one_iter()
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\models\ModelBase.py", line 474, in train_one_iter
losses = self.onTrainOneIter()
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\models\Model_SAEHD\Model.py", line 774, in onTrainOneIter
src_loss, dst_loss = self.src_dst_train (warped_src, target_src, target_srcm, target_srcm_em, warped_dst, target_dst, target_dstm, target_dstm_em)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\models\Model_SAEHD\Model.py", line 584, in src_dst_train
self.target_dstm_em:target_dstm_em,
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\client\session.py", line 968, in run
run_metadata_ptr)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\client\session.py", line 1191, in _run
feed_dict_tensor, options, run_metadata)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\client\session.py", line 1369, in _do_run
run_metadata)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\client\session.py", line 1394, in _do_call
raise type(e)(node_def, op, message) # pylint: disable=no-value-for-parameter
tensorflow.python.framework.errors_impl.ResourceExhaustedError: 2 root error(s) found.
(0) Resource exhausted: failed to allocate memory
[[node mul_89 (defined at D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\core\leras\optimizers\AdaBelief.py:64) ]]
Hint: If you want to see a list of allocated tensors when OOM happens, add report_tensor_allocations_upon_oom to RunOptions for current allocation info. This isn't available when running in Eager mode.
[[concat_8/concat/_103]]
Hint: If you want to see a list of allocated tensors when OOM happens, add report_tensor_allocations_upon_oom to RunOptions for current allocation info. This isn't available when running in Eager mode.
(1) Resource exhausted: failed to allocate memory
[[node mul_89 (defined at D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\core\leras\optimizers\AdaBelief.py:64) ]]
Hint: If you want to see a list of allocated tensors when OOM happens, add report_tensor_allocations_upon_oom to RunOptions for current allocation info. This isn't available when running in Eager mode.
0 successful operations.
0 derived errors ignored.
Errors may have originated from an input operation.
Input Source operations connected to node mul_89:
src_dst_opt/vs_inter_AB/upscale1/conv1/weight_0/read (defined at D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\core\leras\optimizers\AdaBelief.py:38)
Input Source operations connected to node mul_89:
src_dst_opt/vs_inter_AB/upscale1/conv1/weight_0/read (defined at D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\core\leras\optimizers\AdaBelief.py:38)
Original stack trace for 'mul_89':
File "threading.py", line 884, in _bootstrap
File "threading.py", line 916, in _bootstrap_inner
File "threading.py", line 864, in run
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\mainscripts\Trainer.py", line 58, in trainerThread
debug=debug)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\models\ModelBase.py", line 193, in __init__
self.on_initialize()
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\models\Model_SAEHD\Model.py", line 564, in on_initialize
src_dst_loss_gv_op = self.src_dst_opt.get_update_op (nn.average_gv_list (gpu_G_loss_gvs))
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\DeepFaceLab\core\leras\optimizers\AdaBelief.py", line 64, in get_update_op
v_t = self.beta_2*vs + (1.0-self.beta_2) * tf.square(g-m_t)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\ops\variables.py", line 1076, in _run_op
return tensor_oper(a.value(), *args, **kwargs)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\ops\math_ops.py", line 1400, in r_binary_op_wrapper
return func(x, y, name=name)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\ops\math_ops.py", line 1710, in _mul_dispatch
return multiply(x, y, name=name)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\util\dispatch.py", line 206, in wrapper
return target(*args, **kwargs)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\ops\math_ops.py", line 530, in multiply
return gen_math_ops.mul(x, y, name)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\ops\gen_math_ops.py", line 6245, in mul
"Mul", x=x, y=y, name=name)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\framework\op_def_library.py", line 750, in _apply_op_helper
attrs=attr_protos, op_def=op_def)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\framework\ops.py", line 3569, in _create_op_internal
op_def=op_def)
File "D:\FACELAB\DeepFaceLab_NVIDIA_RTX3000_series\_internal\python-3.6.8\lib\site-packages\tensorflow\python\framework\ops.py", line 2045, in __init__
self._traceback = tf_stack.extract_stack_for_node(self._c_op) | open | 2022-06-15T07:56:51Z | 2023-06-08T23:18:54Z | https://github.com/iperov/DeepFaceLab/issues/5533 | [] | knaxLFea | 1 |
dask/dask | pandas | 11,158 | calling repartition on ddf with timeseries index after resample causes ValueError: left side of old and new divisions are different | **issue**:
If you have a DataFrame (ddf) with a time series index that doesn’t align perfectly with whole seconds (for example, ‘2024-06-01 10:00:00.873821’), and you resample it, the new DataFrame’s divisions will be rounded down to the nearest second.
This can cause an error when you try to repartition the DataFrame afterwards. The error message will say there’s a mismatch in the divisions on the left side. Probably because the new data frames first division is nonexistent in the data
To fix this, you can run `ddf.loc[ddf.divisions[0].ceil('s'):]` before you resample. This removes any data at the start that doesn’t align with whole seconds. However this seems confusing
**Minimal Complete Verifiable Example**:
```
import dask.dataframe as dd
import pandas as pd
# create data
idx = pd.date_range(start="10:00:00.873821", end="10:05:00",
freq="0.002s")
data = [i for i in range(idx.size)]
df = pd.DataFrame({'data': data}, index=idx)
ddf = dd.from_pandas(df, npartitions=10)
print(ddf.divisions[0])
```
output: Timestamp('2024-06-01 10:00:00.873821')
```
ddf_res = ddf.resample("0.03s").mean()
print(ddf_res.divisions[0])
```
output: Timestamp('2024-06-01 10:00:00.870000')
```
ddf_res = ddf_res.repartition(freq="1T")
ddf_res.head()
```
output:
```
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[105], line 1
----> 1 ddf_res.head()
File [/opt/conda/lib/python3.12/site-packages/dask_expr/_collection.py:702](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask_expr/_collection.py#line=701), in FrameBase.head(self, n, npartitions, compute)
700 out = new_collection(expr.Head(self, n=n, npartitions=npartitions))
701 if compute:
--> 702 out = out.compute()
703 return out
File [/opt/conda/lib/python3.12/site-packages/dask_expr/_collection.py:476](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask_expr/_collection.py#line=475), in FrameBase.compute(self, fuse, **kwargs)
474 out = out.repartition(npartitions=1)
475 out = out.optimize(fuse=fuse)
--> 476 return DaskMethodsMixin.compute(out, **kwargs)
File [/opt/conda/lib/python3.12/site-packages/dask/base.py:375](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask/base.py#line=374), in DaskMethodsMixin.compute(self, **kwargs)
351 def compute(self, **kwargs):
352 """Compute this dask collection
353
354 This turns a lazy Dask collection into its in-memory equivalent.
(...)
373 dask.compute
374 """
--> 375 (result,) = compute(self, traverse=False, **kwargs)
376 return result
File [/opt/conda/lib/python3.12/site-packages/dask/base.py:654](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask/base.py#line=653), in compute(traverse, optimize_graph, scheduler, get, *args, **kwargs)
646 return args
648 schedule = get_scheduler(
649 scheduler=scheduler,
650 collections=collections,
651 get=get,
652 )
--> 654 dsk = collections_to_dsk(collections, optimize_graph, **kwargs)
655 keys, postcomputes = [], []
656 for x in collections:
File [/opt/conda/lib/python3.12/site-packages/dask/base.py:426](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask/base.py#line=425), in collections_to_dsk(collections, optimize_graph, optimizations, **kwargs)
424 graphs = []
425 for opt, val in groups.items():
--> 426 dsk, keys = _extract_graph_and_keys(val)
427 dsk = opt(dsk, keys, **kwargs)
429 for opt_inner in optimizations:
File [/opt/conda/lib/python3.12/site-packages/dask/base.py:452](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask/base.py#line=451), in _extract_graph_and_keys(vals)
450 graphs, keys = [], []
451 for v in vals:
--> 452 graphs.append(v.__dask_graph__())
453 keys.append(v.__dask_keys__())
455 if any(isinstance(graph, HighLevelGraph) for graph in graphs):
File [/opt/conda/lib/python3.12/site-packages/dask_expr/_collection.py:559](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask_expr/_collection.py#line=558), in FrameBase.__dask_graph__(self)
557 out = self.expr
558 out = out.lower_completely()
--> 559 return out.__dask_graph__()
File [/opt/conda/lib/python3.12/site-packages/dask_expr/_core.py:506](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask_expr/_core.py#line=505), in Expr.__dask_graph__(self)
503 continue
504 seen.add(expr._name)
--> 506 layers.append(expr._layer())
507 for operand in expr.dependencies():
508 stack.append(operand)
File [/opt/conda/lib/python3.12/site-packages/dask_expr/_repartition.py:269](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask_expr/_repartition.py#line=268), in RepartitionDivisions._layer(self)
267 if a[0] != b[0]:
268 msg = "left side of old and new divisions are different"
--> 269 raise ValueError(msg)
270 if a[-1] != b[-1]:
271 msg = "right side of old and new divisions are different"
ValueError: left side of old and new divisions are different
```
**UPDATE :**
it also happens at the right side:
```
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[84], line 1
----> 1 ddf_hf.head()
File [/opt/conda/lib/python3.12/site-packages/dask_expr/_collection.py:702](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask_expr/_collection.py#line=701), in FrameBase.head(self, n, npartitions, compute)
700 out = new_collection(expr.Head(self, n=n, npartitions=npartitions))
701 if compute:
--> 702 out = out.compute()
703 return out
File [/opt/conda/lib/python3.12/site-packages/dask_expr/_collection.py:476](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask_expr/_collection.py#line=475), in FrameBase.compute(self, fuse, **kwargs)
474 out = out.repartition(npartitions=1)
475 out = out.optimize(fuse=fuse)
--> 476 return DaskMethodsMixin.compute(out, **kwargs)
File [/opt/conda/lib/python3.12/site-packages/dask/base.py:375](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask/base.py#line=374), in DaskMethodsMixin.compute(self, **kwargs)
351 def compute(self, **kwargs):
352 """Compute this dask collection
353
354 This turns a lazy Dask collection into its in-memory equivalent.
(...)
373 dask.compute
374 """
--> 375 (result,) = compute(self, traverse=False, **kwargs)
376 return result
File [/opt/conda/lib/python3.12/site-packages/dask/base.py:654](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask/base.py#line=653), in compute(traverse, optimize_graph, scheduler, get, *args, **kwargs)
646 return args
648 schedule = get_scheduler(
649 scheduler=scheduler,
650 collections=collections,
651 get=get,
652 )
--> 654 dsk = collections_to_dsk(collections, optimize_graph, **kwargs)
655 keys, postcomputes = [], []
656 for x in collections:
File [/opt/conda/lib/python3.12/site-packages/dask/base.py:426](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask/base.py#line=425), in collections_to_dsk(collections, optimize_graph, optimizations, **kwargs)
424 graphs = []
425 for opt, val in groups.items():
--> 426 dsk, keys = _extract_graph_and_keys(val)
427 dsk = opt(dsk, keys, **kwargs)
429 for opt_inner in optimizations:
File [/opt/conda/lib/python3.12/site-packages/dask/base.py:452](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask/base.py#line=451), in _extract_graph_and_keys(vals)
450 graphs, keys = [], []
451 for v in vals:
--> 452 graphs.append(v.__dask_graph__())
453 keys.append(v.__dask_keys__())
455 if any(isinstance(graph, HighLevelGraph) for graph in graphs):
File [/opt/conda/lib/python3.12/site-packages/dask_expr/_collection.py:559](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask_expr/_collection.py#line=558), in FrameBase.__dask_graph__(self)
557 out = self.expr
558 out = out.lower_completely()
--> 559 return out.__dask_graph__()
File [/opt/conda/lib/python3.12/site-packages/dask_expr/_core.py:506](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask_expr/_core.py#line=505), in Expr.__dask_graph__(self)
503 continue
504 seen.add(expr._name)
--> 506 layers.append(expr._layer())
507 for operand in expr.dependencies():
508 stack.append(operand)
File [/opt/conda/lib/python3.12/site-packages/dask_expr/_repartition.py:272](http://10.4.201.45:11000/opt/conda/lib/python3.12/site-packages/dask_expr/_repartition.py#line=271), in RepartitionDivisions._layer(self)
270 if a[-1] != b[-1]:
271 msg = "right side of old and new divisions are different"
--> 272 raise ValueError(msg)
274 def _is_single_last_div(x):
275 """Whether last division only contains single label"""
ValueError: right side of old and new divisions are different
```
**Environment**:
- Dask version: 2024.5.2
- Python version: 3.12.3
- Operating System: debian
- Install method (conda, pip, source): conda
| closed | 2024-06-01T07:11:40Z | 2024-06-03T16:22:50Z | https://github.com/dask/dask/issues/11158 | [
"dataframe"
] | N4321D | 0 |
LibrePhotos/librephotos | django | 1,030 | can not view photos from nextcloud | # 🐛 Bug Report
* [x] 📁 I've Included a ZIP file containing my librephotos `log` files:[__logs.zip__](https://github.com/LibrePhotos/librephotos/files/12705941/logs.zip)
* [x] ❌ I have looked for similar issues (including closed ones): __yes, I've looked through similar issue and get `fixed`, but I still met same issue.__
* [x] 🎬 (If applicable) I've provided pictures or links to videos that clearly demonstrate the issue
<img width="679" alt="image" src="https://github.com/LibrePhotos/librephotos/assets/44010936/169c558a-6dad-4eec-b7e8-67e522a19036">
## 📝 Description of issue:
* My nextcloud is installed using docker,and my librephotos is install using docker too.They are running on the same machine,x86 ubuntu 22.04 LTS.They all work fine individually but I met problems adding album from nextcloud.
* I've reinstalled them and still met this problem.
## 🔁 How can we reproduce it:
Maybe this problem is related to the fact that they all use docker on the same machine?
## Please provide additional information:
- 💻 Operating system: __ubuntu 22.04 LTS__
- ⚙ Architecture (x86 or ARM): __X86__
- 🔢 Librephotos version: __I pull the latest docker image on 2023/9/13__
- 📸 Librephotos installation method (Docker, Kubernetes, .deb, etc.): __docker__
* 🐋 If Docker or Kubernets, provide docker-compose image tag: __reallibrephotos/singleton:latest__
- 📁 How is you picture library mounted (Local file system (Type), NFS, SMB, etc.): local:
```bash
sudo docker run --name librephotos
-v /home/mydir/librephotos/photos:/var/lib/librephotos/photos/
```
- ☁ If you are virtualizing librephotos, Virtualization platform (Proxmox, Xen, HyperV, etc.): __No,just using docker on ubuntu.__
### If you need me to provide more information, just tell me! LibrePhotos is an awsome photo management service, I'd like to help it do better!
| open | 2023-09-23T08:48:56Z | 2023-10-04T17:27:35Z | https://github.com/LibrePhotos/librephotos/issues/1030 | [
"bug",
"singleton"
] | LUCKandII | 0 |
adamerose/PandasGUI | pandas | 215 | Fatal Python error: PyEval_RestoreThread - moving Tkinter window with PandasGUI open | Trying to move a Tkinter window at any time gives me this error across multiple systems.
**Environment**
Windows 10 64-bit
Python 3.10 64-bit
pandasgui-0.2.13
Error:
PandasGUI INFO � pandasgui.gui � Opening PandasGUI
Fatal Python error: PyEval_RestoreThread: the function must be called with the GIL held, but the GIL is released (the current Python thread state is NULL)
Python runtime state: initialized
Current thread 0x000038c4 (most recent call first):
File "C:\Python\lib\site-packages\pandasgui\gui.py", line 106 in __init__
File "C:\Python\lib\site-packages\pandasgui\gui.py", line 468 in show
File "C:\Desktop\file.py", line 398 in start_pandasgui
File "C:\Python\lib\site-packages\customtkinter\widgets\ctk_button.py", line 372 in clicked
File "C:\Python\lib\tkinter\__init__.py", line 1921 in __call__
File "C:\Python\lib\tkinter\__init__.py", line 1458 in mainloop
File "C:\Python\lib\site-packages\customtkinter\windows\ctk_tk.py", line 106 in mainloop
File "C:\Desktop\file.py", line 1535 in <module>
Extension modules: win32api, _win32sysloader, numpy.core._multiarray_umath, numpy.core._multiarray_tests, numpy.linalg._umath_linalg, numpy.fft._pocketfft_internal, numpy.random._common, numpy.random.bit_generator, numpy.random._bounded_integers, numpy.random._mt19937, numpy.random.mtrand, numpy.random._philox, numpy.random._pcg64, numpy.random._sfc64, numpy.random._generator, pandas._libs.tslibs.dtypes, pandas._libs.tslibs.base, pandas._libs.tslibs.np_datetime, pandas._libs.tslibs.nattype, pandas._libs.tslibs.timezones, pandas._libs.tslibs.ccalendar, pandas._libs.tslibs.tzconversion, pandas._libs.tslibs.strptime, pandas._libs.tslibs.fields, pandas._libs.tslibs.timedeltas, pandas._libs.tslibs.timestamps, pandas._libs.properties, pandas._libs.tslibs.offsets, pandas._libs.tslibs.parsing, pandas._libs.tslibs.conversion, pandas._libs.tslibs.period, pandas._libs.tslibs.vectorized, pandas._libs.ops_dispatch, pandas._libs.missing, pandas._libs.hashtable, pandas._libs.algos, pandas._libs.interval, pandas._libs.tslib, pandas._libs.lib, pandas._libs.hashing, pyarrow.lib, pyarrow._hdfsio, pandas._libs.ops, pandas._libs.arrays, pandas._libs.index, pandas._libs.join, pandas._libs.sparse, pyarrow._compute, pandas._libs.reduction, pandas._libs.indexing, pandas._libs.internals, pandas._libs.writers, pandas._libs.window.aggregations, pandas._libs.window.indexers, pandas._libs.reshape, pandas._libs.groupby, pandas._libs.testing, pandas._libs.parsers, pandas._libs.json, PyQt5.QtCore, PyQt5.QtGui, PyQt5.QtWidgets, win32com.shell.shell, PyQt5.QtWebChannel, PyQt5.QtNetwork, PyQt5.QtWebEngineCore, PyQt5.QtPrintSupport, PyQt5.QtWebEngineWidgets, pyodbc, matplotlib._c_internal_utils, PIL._imaging, matplotlib._path, kiwisolver._cext, matplotlib._image (total: 74) | open | 2022-11-11T20:12:08Z | 2022-11-11T20:12:45Z | https://github.com/adamerose/PandasGUI/issues/215 | [
"bug"
] | lachesis17 | 0 |
modelscope/data-juicer | data-visualization | 63 | [MM] image_deduplicator | A new Deduplicator image_deduplicator will be supported. It will remove duplicate images in multimodal samples. Maybe based on imagededup library
TBD: when a sample contains multiple images, remove duplicate images only or remove this whole sample? | closed | 2023-11-02T09:43:50Z | 2023-11-15T03:11:01Z | https://github.com/modelscope/data-juicer/issues/63 | [
"enhancement",
"dj:multimodal"
] | HYLcool | 0 |
serengil/deepface | machine-learning | 1,318 | [FEATURE]: Improve represent performance | ### Description
Performance is very slow when aligning faces when using the `represent` API. Please see my PR here: https://github.com/serengil/deepface/pull/1319
### Additional Info
_No response_ | closed | 2024-08-23T16:10:02Z | 2024-08-26T13:51:26Z | https://github.com/serengil/deepface/issues/1318 | [
"enhancement"
] | Circuit8 | 2 |
tflearn/tflearn | data-science | 1,048 | tflearn getting stuck when predicting | When model.predict is carried when using a sanic server, it is getting blocked and hanged.
The issue is in config.py at line tf.get_collection('is_training_ops')[1].eval(session=session)
under def is_training()
error shown when evaluating the expression is "Timeout waiting for response 113"
please help? | open | 2018-05-09T14:11:01Z | 2018-05-09T14:12:02Z | https://github.com/tflearn/tflearn/issues/1048 | [] | spawn08 | 0 |
ultralytics/ultralytics | machine-learning | 19,689 | Yolo v10 model supported in Java | Dear expert
I use the code with Yolov8 model for extraction it works well
but i use the similar coding for Yolov10 model extraction it does not work
Do you know any format changes for the output between v8 and v10?
I try to make the java work for the extraction on yolov10 output, please advise
OrtSession.Result output = session.run(...);
float[][][] outputData = (float[][][]) output.get(0).getValue();
float[][] predictions= outputData[0];
predictions = transposeMatrix(predictions);
for (float[] bbox : predictions) {
int label = predMax(bbox, 4);
// bbox[4] is the confidence
// bbox[0] - bbox[3] = boundbox
}
// Function to get the max probablities
int predMax(float[] probabilities, int startIdx) {
float maxVal = Float.NEGATIVE_INFINITY;
int idx = startIdx;
for (int i = startIdx; i < probabilities.length; i++) {
if (probabilities[i] > maxVal) {
maxVal = probabilities[i];
idx = i;
}
}
return idx;
}
float[][] transposeMatrix(float[][] matrix) {
float[][] transMatrix = new float[matrix[0].length][matrix.length];
for (int i = 0; i < matrix.length; i++) {
for (int j = 0; j < matrix[0].length; j++) {
transMatrix[j][i] = matrix[i][j];
}
}
return transMatrix;
}
| open | 2025-03-14T03:17:13Z | 2025-03-14T06:23:12Z | https://github.com/ultralytics/ultralytics/issues/19689 | [
"question",
"exports"
] | felixchan68 | 2 |
gradio-app/gradio | data-science | 10,818 | Bitdefender Issue | ### Describe the bug

### Have you searched existing issues? 🔎
- [x] I have searched and found no existing issues
### Reproduction
I have no clue
### Screenshot

### Logs
```shell
```
### System Info
```shell
PS G:\Projects2\Imagen Edit> gradio environment
Gradio Environment Information:
------------------------------
Operating System: Windows
gradio version: 5.15.0
gradio_client version: 1.7.0
------------------------------------------------
gradio dependencies in your environment:
aiofiles: 23.2.1
anyio: 4.8.0
audioop-lts is not installed.
fastapi: 0.115.7
ffmpy: 0.4.0
gradio-client==1.7.0 is not installed.
httpx: 0.28.1
huggingface-hub: 0.29.3
jinja2: 3.1.6
markupsafe: 2.1.5
numpy: 1.24.3
orjson: 3.10.7
packaging: 23.2
pandas: 2.2.3
pillow: 10.0.0
pydantic: 2.10.6
pydub: 0.25.1
python-multipart: 0.0.18
pyyaml: 6.0.2
ruff: 0.9.6
safehttpx: 0.1.6
semantic-version: 2.10.0
starlette: 0.45.3
tomlkit: 0.12.0
typer: 0.15.1
typing-extensions: 4.12.2
urllib3: 2.3.0
uvicorn: 0.34.0
authlib; extra == 'oauth' is not installed.
itsdangerous; extra == 'oauth' is not installed.
gradio_client dependencies in your environment:
fsspec: 2023.10.0
httpx: 0.28.1
huggingface-hub: 0.29.3
packaging: 23.2
typing-extensions: 4.12.2
websockets: 14.2
PS G:\Projects2\Imagen Edit>
```
### Severity
Blocking usage of gradio | open | 2025-03-17T17:42:00Z | 2025-03-17T19:20:36Z | https://github.com/gradio-app/gradio/issues/10818 | [
"bug"
] | PierrunoYT | 1 |
vimalloc/flask-jwt-extended | flask | 366 | DeprecationWarning: The verify parameter is deprecated. Please use options instead. | The code line 91 in the file [ask_jwt_extended/utils.py](https://github.com/vimalloc/flask-jwt-extended/blob/master/flask_jwt_extended/utils.py)
```
jwt_manager = _get_jwt_manager()
unverified_claims = jwt.decode(
encoded_token, verify=False, algorithms=config.decode_algorithms
)
```
triggers a deprecation warning in jwt/api_jws.py:119 saying:
```
venv/lib/python3.7/site-packages/jwt/api_jws.py:119: DeprecationWarning: The verify parameter is deprecated. Please use options instead.
'Please use options instead.', DeprecationWarning)
```
| closed | 2020-10-26T17:33:44Z | 2021-01-18T15:48:18Z | https://github.com/vimalloc/flask-jwt-extended/issues/366 | [] | fabrizio2210 | 2 |
holoviz/panel | plotly | 7,407 | Cannot upload accepted_filetypes pdf | I'm on panel 1.5.2 using the FileDropper and try to make a working example for https://discourse.holoviz.org/t/problem-with-accessing-pdf-file-s-input-in-panel/8339.

```python
import io
import panel as pn
import base64
from PyPDF2 import PdfReader
pn.extension('filedropper')
def transform_pdfs(value):
pages = {}
for key, value in value:
f=io.BytesIO(file_input.value)
reader = PdfReader(f)
page = reader.pages[0]
pages[key]=page
print(pages)
file_input = pn.widgets.FileDropper(accepted_filetypes=[".pdf"])
pn.Column(
file_input, pn.bind(transform_pdfs, file_input)
).servable()
```
I've also tried `accepted_filetypes=["pdf"]` without success.
| closed | 2024-10-16T09:13:19Z | 2024-12-23T07:58:59Z | https://github.com/holoviz/panel/issues/7407 | [
"duplicate"
] | MarcSkovMadsen | 3 |
xonsh/xonsh | data-science | 5,718 | shell exits if $THREAD_SUBPROCS=None and history is invoked with invalid argument | ## Current Behavior
Xonsh terminates after calling the history command with an invalid parameter if $THREAD_SUBPROCS=None
<!---
For general xonsh issues, please try to replicate the failure using `xonsh --no-rc --no-env`.
Short, reproducible code snippets are highly appreciated.
You can use `$XONSH_SHOW_TRACEBACK=1`, `$XONSH_TRACE_SUBPROC=2`, or `$XONSH_DEBUG=1`
to collect more information about the failure.
-->
Traceback (if applicable):
<details>
```xsh
$ /usr/bin/xonsh --no-rc --no-env
user@machine ~ @ $XONSH_SHOW_TRACEBACK=1
user@machine ~ @ $XONSH_TRACE_SUBPROC=2
user@machine ~ @ $XONSH_DEBUG=1
user@machine ~ @ ${...}["THREAD_SUBPROCS"]=None
user@machine ~ @ history badarg<stdin>:1:8 - history badarg
<stdin>:1:8 + ![history badarg]
user@machine ~ @ history badarg
<stdin>:1:8 - history badarg
<stdin>:1:8 + ![history badarg]
Trace run_subproc({'cmds': (['history', 'badarg'],), 'captured': 'hiddenobject'})
0: {'cmd': ['history', 'badarg'], 'cls': 'xonsh.procs.proxies.ProcProxy', 'alias_name': 'history', 'alias': <xonsh.history.main.HistoryAlias object at 0x772879d2ea80>, 'background': False}
usage: history [-h]
{show,id,file,info,pull,flush,off,on,clear,delete,gc,transfer,diff}
...
history: error: cannot convert 'badarg' to slice
--- shell exits here ---
$
```
</details>
## Expected Behavior
<!--- What you expect and what is your real life use case. -->
Xonsh should continue running after printing the error.
## xonfig
<details>
```xsh
+-----------------------------+-----------------+
| xonsh | 0.18.3 |
| Python | 3.12.7 |
| PLY | 3.11 |
| have readline | True |
| prompt toolkit | 3.0.48 |
| shell type | prompt_toolkit |
| history backend | json |
| pygments | 2.18.0 |
| on posix | True |
| on linux | True |
| distro | unknown |
| on wsl | False |
| on darwin | False |
| on windows | False |
| on cygwin | False |
| on msys2 | False |
| is superuser | False |
| default encoding | utf-8 |
| xonsh encoding | utf-8 |
| encoding errors | surrogateescape |
| xontrib | [] |
| RC file | [] |
| UPDATE_OS_ENVIRON | False |
| XONSH_CAPTURE_ALWAYS | False |
| XONSH_SUBPROC_OUTPUT_FORMAT | stream_lines |
| THREAD_SUBPROCS | True |
| XONSH_CACHE_SCRIPTS | True |
+-----------------------------+-----------------+
```
</details>
## For community
⬇️ **Please click the 👍 reaction instead of leaving a `+1` or 👍 comment**
| open | 2024-10-31T16:44:19Z | 2024-10-31T20:32:28Z | https://github.com/xonsh/xonsh/issues/5718 | [
"history",
"edge-case"
] | mrschyte | 0 |
3b1b/manim | python | 2,172 | when i run example by manimgl, circle displayed not coorectly | ### Describe the bug
when i run manim, circle displayed not coorectly.it seems has may lines around the circle.


**Code**:
from manimlib import *
class SquareToCircle(Scene):
def construct(self):
circle = Circle()
circle.set_fill(BLUE, opacity=0.5)
circle.set_stroke(BLUE_E, width=4)
self.add(circle)
**Wrong display or Error traceback**:
<!-- the wrong display result of the code you run, or the error Traceback -->
### Additional context
<!-- Add any other context about the problem here. -->
Windows;Python 3.10,
| closed | 2024-09-30T08:32:24Z | 2024-10-11T12:36:00Z | https://github.com/3b1b/manim/issues/2172 | [
"bug"
] | hashixuehua | 5 |
smiley/steamapi | rest-api | 54 | PyPi support | I am suggesting to support the package through pypi, so installation as well as distribution is easier.
Currently installing this package otherwise is just unnecessary complicated.
The package could even be tagged as alpha and nicht production ready.
If requested I could modify the setup.py accordingly. | closed | 2018-02-25T20:47:12Z | 2019-04-09T16:10:59Z | https://github.com/smiley/steamapi/issues/54 | [] | indietyp | 1 |
DistrictDataLabs/yellowbrick | matplotlib | 1,131 | MissingValuesBar does not work on categorical columns | ## Describe the solution you'd like
Currently, I am unable to get MissingValuesBar to work on data that has categorical columns. It is internally trying to convert these columns to a floating point value and producing an error at this step. I am hoping this can be handles internally in the class itself since it is just checking for missing values so it should not care about the type of column.
## Is your feature request related to a problem? Please describe.
Described above, but this is the error that I get on categorical data. One of the columns has continent values and it looks like it is producing an error at that point.
```python
# Instantiate the visualizer
plt.figure(figsize=(15,10))
visualizer = MissingValuesBar(features=features)
visualizer.fit(X=data[features], y=data[target].values) # Supply the targets via y
_ = visualizer.show() # Finalize and render the figure
```
**Traceback**
```
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-7-2b58032dfe22> in <module>
3 visualizer = MissingValuesBar(features=features)
4
----> 5 visualizer.fit(X=data[features], y=data[target].values) # Supply the targets via y
6 _ = visualizer.show() # Finalize and render the figure
~\.conda\envs\ds7337_cs3\lib\site-packages\yellowbrick\contrib\missing\base.py in fit(self, X, y, **kwargs)
70 self.y = y
71
---> 72 self.draw(X, y, **kwargs)
73 return self
74
~\.conda\envs\ds7337_cs3\lib\site-packages\yellowbrick\contrib\missing\bar.py in draw(self, X, y, **kwargs)
137 target values.
138 """
--> 139 nan_col_counts = self.get_nan_col_counts()
140
141 # the x locations for the groups
~\.conda\envs\ds7337_cs3\lib\site-packages\yellowbrick\contrib\missing\bar.py in get_nan_col_counts(self, **kwargs)
110
111 else:
--> 112 nan_matrix = self.X.astype(np.float)
113
114 if self.y is None:
ValueError: could not convert string to float: 'euorpe'
```
**If I just take the numeric features, it works well**
```python
# Instantiate the visualizer
plt.figure(figsize=(15,10))
visualizer = MissingValuesBar(features=features_numerical)
visualizer.fit(X=data[features_numerical], y=data[target].values) # Supply the targets via y
_ = visualizer.show() # Finalize and render the figure
```
**See output below**

| closed | 2020-11-24T18:40:36Z | 2022-05-07T21:00:55Z | https://github.com/DistrictDataLabs/yellowbrick/issues/1131 | [
"type: bug",
"type: technical debt"
] | ngupta23 | 2 |
ResidentMario/missingno | data-visualization | 127 | Using missingno from the python command line | I was unable to generate graphs from the command line, I tried to use missing.plot() or missing.show() without any output.
I saw the missingno functions (using dir(missingno) and nothing seems to show graphs.
Does it work just in Jupyter notebooks?
Thanks, | closed | 2021-02-04T22:29:23Z | 2021-02-04T22:59:40Z | https://github.com/ResidentMario/missingno/issues/127 | [] | ggutierrezc | 1 |
aiortc/aiortc | asyncio | 877 | Crash when negotiating after a video transceiver has been added by the server | How to reproduce:
1) add transceivers to the peerConnection from the server (for example when the datachannel opens):
```py
@pc.on("datachannel")
async def on_datachannel(channel):
pc.addTransceiver("audio")
pc.addTransceiver("video") # if this line is commented, or replaced by an "audio" transceiver, there is no crash.
send_offer()
```
2) complete the negotiation normally from the client (adding the offer description and sending an answer)
-> the transaction is complete and the signaling state is back to stable
3) start a new negotiation from the client
```js
async function reNegotiate() {
const offer = await pc.createOffer();
await pc.setLocalDescription(offer); // <- crashing here
dc.send(JSON.stringify(pc.localDescription));
}
The crash depends on the browser:
Chrome:
```
Failed to execute 'setLocalDescription' on 'RTCPeerConnection': Failed to set local offer sdp: Failed to set local video description recv parameters for m-section with mid='2'.
```
Firefox (even if it's just an audio transceiver):
```
Remote description indicates ICE restart but offer did not request ICE restart
```
could be related to Issue https://github.com/aiortc/aiortc/issues/803 ?
Here is a modified version of the server example to reproduce the crash (press "Start" then "Renegotiate"):
```py
import argparse
import asyncio
import json
import logging
import os
import ssl
import uuid
from aiohttp import web
from aiortc import RTCPeerConnection, RTCSessionDescription
ROOT = os.path.dirname(__file__)
logger = logging.getLogger("pc")
pcs = set()
async def send_offer(channel, pc):
offer = await pc.createOffer()
await pc.setLocalDescription(offer)
channel.send(json.dumps({"sdp": pc.localDescription.sdp, "type": pc.localDescription.type}))
async def send_answer(channel, pc):
answer = await pc.createAnswer()
await pc.setLocalDescription(answer)
channel.send(json.dumps({"sdp": pc.localDescription.sdp, "type": pc.localDescription.type}))
async def index(request):
content = open(os.path.join(ROOT, "index.html"), "r").read()
return web.Response(content_type="text/html", text=content)
async def javascript(request):
content = open(os.path.join(ROOT, "client.js"), "r").read()
return web.Response(content_type="application/javascript", text=content)
async def offer(request):
params = await request.json()
offer = RTCSessionDescription(sdp=params["sdp"], type=params["type"])
pc = RTCPeerConnection()
pc_id = "PeerConnection(%s)" % uuid.uuid4()
pcs.add(pc)
def log_info(msg, *args):
logger.info(pc_id + " " + msg, *args)
log_info("Created for %s", request.remote)
@pc.on("datachannel")
async def on_datachannel(channel):
global DATA_CHANNEL
DATA_CHANNEL = channel
pc.addTransceiver("audio")
pc.addTransceiver("video")
await send_offer(channel, pc)
@channel.on("message")
async def on_message(message):
nego = json.loads(message)
await pc.setRemoteDescription(RTCSessionDescription(**nego))
if nego['type'] == "offer":
await send_answer(channel, pc)
@pc.on("connectionstatechange")
async def on_connectionstatechange():
log_info("Connection state is %s", pc.connectionState)
if pc.connectionState == "failed":
await pc.close()
pcs.discard(pc)
@pc.on("track")
def on_track(track):
log_info("Track %s received", track.kind)
@track.on("ended")
async def on_ended():
log_info("Track %s ended", track.kind)
# handle offer
await pc.setRemoteDescription(offer)
# send answer
answer = await pc.createAnswer()
await pc.setLocalDescription(answer)
return web.Response(
content_type="application/json",
text=json.dumps(
{"sdp": pc.localDescription.sdp, "type": pc.localDescription.type}
),
)
async def on_shutdown(app):
# close peer connections
coros = [pc.close() for pc in pcs]
await asyncio.gather(*coros)
pcs.clear()
if __name__ == "__main__":
parser = argparse.ArgumentParser(
description="WebRTC audio / video / data-channels demo"
)
parser.add_argument("--cert-file", help="SSL certificate file (for HTTPS)")
parser.add_argument("--key-file", help="SSL key file (for HTTPS)")
parser.add_argument(
"--host", default="0.0.0.0", help="Host for HTTP server (default: 0.0.0.0)"
)
parser.add_argument(
"--port", type=int, default=8080, help="Port for HTTP server (default: 8080)"
)
parser.add_argument("--record-to", help="Write received media to a file."),
parser.add_argument("--verbose", "-v", action="count")
args = parser.parse_args()
if args.verbose:
logging.basicConfig(level=logging.DEBUG)
else:
logging.basicConfig(level=logging.INFO)
if args.cert_file:
ssl_context = ssl.SSLContext()
ssl_context.load_cert_chain(args.cert_file, args.key_file)
else:
ssl_context = None
app = web.Application()
app.on_shutdown.append(on_shutdown)
app.router.add_get("/", index)
app.router.add_get("/client.js", javascript)
app.router.add_post("/offer", offer)
web.run_app(
app, access_log=None, host=args.host, port=args.port, ssl_context=ssl_context
)
```
```js
// get DOM elements
var dataChannelLog = document.getElementById('data-channel'),
iceConnectionLog = document.getElementById('ice-connection-state'),
iceGatheringLog = document.getElementById('ice-gathering-state'),
signalingLog = document.getElementById('signaling-state');
// peer connection
var pc = null;
// data channel
var dc = null, dcInterval = null;
function createPeerConnection() {
var config = {
sdpSemantics: 'unified-plan',
iceServers: [{urls: ['stun:stun.l.google.com:19302']}],
};
pc = new RTCPeerConnection(config);
// register some listeners to help debugging
pc.addEventListener('icegatheringstatechange', function() {
iceGatheringLog.textContent += ' -> ' + pc.iceGatheringState;
}, false);
iceGatheringLog.textContent = pc.iceGatheringState;
pc.addEventListener('iceconnectionstatechange', function() {
iceConnectionLog.textContent += ' -> ' + pc.iceConnectionState;
}, false);
iceConnectionLog.textContent = pc.iceConnectionState;
pc.addEventListener('signalingstatechange', function() {
signalingLog.textContent += ' -> ' + pc.signalingState;
}, false);
signalingLog.textContent = pc.signalingState;
return pc;
}
function negotiate() {
return pc.createOffer().then(function(offer) {
return pc.setLocalDescription(offer);
}).then(function() {
// wait for ICE gathering to complete
return new Promise(function(resolve) {
if (pc.iceGatheringState === 'complete') {
resolve();
} else {
function checkState() {
if (pc.iceGatheringState === 'complete') {
pc.removeEventListener('icegatheringstatechange', checkState);
resolve();
}
}
pc.addEventListener('icegatheringstatechange', checkState);
}
});
}).then(function() {
var offer = pc.localDescription;
document.getElementById('offer-sdp').textContent = offer.sdp;
return fetch('/offer', {
body: JSON.stringify({
sdp: offer.sdp,
type: offer.type,
}),
headers: {
'Content-Type': 'application/json'
},
method: 'POST'
});
}).then(function(response) {
return response.json();
}).then(function(answer) {
document.getElementById('answer-sdp').textContent = answer.sdp;
const remote_description = new window.RTCSessionDescription(answer);
return pc.setRemoteDescription(remote_description);
}).catch(function(e) {
alert(e);
});
}
async function renegotiate() {
pc.getTransceivers().forEach((transceiver) => {
transceiver.direction = "sendonly";
});
try {
const offer = await pc.createOffer();
await pc.setLocalDescription(offer);
} catch (e) {
console.log(e);
alert(e);
}
dc.send(JSON.stringify(pc.localDescription));
}
function start() {
document.getElementById('start').style.display = 'none';
pc = createPeerConnection();
var time_start = null;
dc = pc.createDataChannel('chat', {"ordered": true});
dc.onclose = function() {
clearInterval(dcInterval);
dataChannelLog.textContent += '- close\n';
};
dc.onmessage = async function(message) {
console.log(message);
const nego = JSON.parse(message.data);
const desc = new window.RTCSessionDescription(nego);
await pc.setRemoteDescription(desc);
if (nego.type === "offer") {
console.log("data channel: offer from server");
console.log(desc.sdp);
const answ = await pc.createAnswer();
await pc.setLocalDescription(answ);
dc.send(JSON.stringify(pc.localDescription));
console.log("data channel: answer to server");
console.log(pc.localDescription);
} else {
console.log("data channel: answer from server");
console.log(desc.sdp);
}
};
return negotiate();
document.getElementById('stop').style.display = 'inline-block';
}
function stop() {
document.getElementById('stop').style.display = 'none';
// close data channel
if (dc) {
dc.close();
}
// close transceivers
if (pc.getTransceivers) {
pc.getTransceivers().forEach(function(transceiver) {
if (transceiver.stop) {
transceiver.stop();
}
});
}
// close local audio / video
pc.getSenders().forEach(function(sender) {
sender.track.stop();
});
// close peer connection
setTimeout(function() {
pc.close();
}, 500);
}
```
```html
<html>
<head>
<meta charset="UTF-8"/>
<meta name="viewport" content="width=device-width, initial-scale=1.0" />
<title>WebRTC demo</title>
<style>
button {
padding: 8px 16px;
}
pre {
overflow-x: hidden;
overflow-y: auto;
}
video {
width: 100%;
}
.option {
margin-bottom: 8px;
}
#media {
max-width: 1280px;
}
</style>
</head>
<body>
<button id="start" onclick="start()">Start</button>
<button id="stop" style="display: none" onclick="stop()">Stop</button>
<button onclick="renegotiate()">Re-negotiate</button>
<h2>State</h2>
<p>
ICE gathering state: <span id="ice-gathering-state"></span>
</p>
<p>
ICE connection state: <span id="ice-connection-state"></span>
</p>
<p>
Signaling state: <span id="signaling-state"></span>
</p>
<div id="media" style="display: none">
<h2>Media</h2>
<audio id="audio" autoplay="true"></audio>
<video id="video" autoplay="true" playsinline="true"></video>
</div>
<h2>Data channel</h2>
<pre id="data-channel" style="height: 200px;"></pre>
<h2>SDP</h2>
<h3>Offer</h3>
<pre id="offer-sdp"></pre>
<h3>Answer</h3>
<pre id="answer-sdp"></pre>
<script src="client.js"></script>
</body>
</html>
```
| closed | 2023-05-11T12:00:16Z | 2023-10-06T02:00:35Z | https://github.com/aiortc/aiortc/issues/877 | [
"stale"
] | nullishReturn | 2 |
eriklindernoren/ML-From-Scratch | deep-learning | 59 | Add Gaussian Process | Pretty cool project. Gaussian Process might be something you can add in the future. | open | 2019-09-27T07:33:28Z | 2022-09-12T02:46:56Z | https://github.com/eriklindernoren/ML-From-Scratch/issues/59 | [] | shikunyu8 | 2 |
home-assistant/core | python | 140,642 | MQTT Sensor have no Unit after Upgrading to 2025.3.3 | ### The problem
Hi,
I have done an update to HA 2025.3.3
After restart my MQTT Sensor entities don't show any unit any more, although they are defined in YAML with unit_of_measurement and device_class properly.
In 2025.3.2 units show up!
### What version of Home Assistant Core has the issue?
2025.3.3
### What was the last working version of Home Assistant Core?
2025.3.2
### What type of installation are you running?
Home Assistant Supervised
### Integration causing the issue
_No response_
### Link to integration documentation on our website
_No response_
### Diagnostics information
_No response_
### Example YAML snippet
```yaml
```
### Anything in the logs that might be useful for us?
```txt
```
### Additional information
_No response_ | closed | 2025-03-15T04:58:56Z | 2025-03-24T11:33:48Z | https://github.com/home-assistant/core/issues/140642 | [
"wontfix",
"integration: mqtt"
] | Vippis2000 | 9 |
onnx/onnx | scikit-learn | 6,229 | onnx.utils.extract_model failed to extract subgraph from whisper-tiny-decoder | I tried to extract a subgraph from whisper-tiny-decoder. In whisper-tiny-decoder, it has a control if node. Below code is tring to extract a subgraph from no_past branch:
```
import onnx
name = "whisper-tiny-decoder"
if name == "whisper-tiny-decoder":
input_path = "whisper-tiny-decoder.onnx"
output_path = "whisper-tiny-decoder-if.onnx"
input_names = ["/model/decoder/ConstantOfShape_output_0"]
output_names = ["/model/decoder/Concat_2_output_0"]
onnx.utils.extract_model(input_path, output_path, input_names, output_names)
```
However, I got below errors:
```
Traceback (most recent call last):
File "C:\ort\ort-models\split.py", line 24, in <module>
onnx.utils.extract_model(input_path, output_path, input_names, output_names)
File "C:\Users\abc\AppData\Local\Programs\Python\Python310\lib\site-packages\onnx\utils.py", line 210, in extract_model
extracted = e.extract_model(input_names, output_names)
File "C:\Users\abc\AppData\Local\Programs\Python\Python310\lib\site-packages\onnx\utils.py", line 165, in extract_model
inputs = self._collect_new_inputs(input_names)
File "C:\Users\abc\AppData\Local\Programs\Python\Python310\lib\site-packages\onnx\utils.py", line 44, in _collect_new_inputs
return self._collect_new_io_core(self.graph.input, names) # type: ignore
File "C:\Users\abc\AppData\Local\Programs\Python\Python310\lib\site-packages\onnx\utils.py", line 37, in _collect_new_io_core
new_io_tensors.append(self.vimap[name])
KeyError: '/model/decoder/ConstantOfShape_output_0'
``` | open | 2024-07-12T07:20:06Z | 2024-07-12T08:15:26Z | https://github.com/onnx/onnx/issues/6229 | [
"bug"
] | axinging | 2 |
Python3WebSpider/ProxyPool | flask | 159 | docker部署,无法关闭日志 | 其实跟此问题有点类似 https://github.com/Python3WebSpider/ProxyPool/issues/158
我设置了环境变量APP_ENV=prod以及ENABLE_LOG_FILE=false,依然有大量日志 | open | 2022-05-17T09:56:35Z | 2022-07-10T16:35:08Z | https://github.com/Python3WebSpider/ProxyPool/issues/159 | [
"bug"
] | star7th | 2 |
tqdm/tqdm | pandas | 765 | Step size for proper scaling of units when using tqdm as an iterable wrapper. | - [x] I have visited the [source website], and in particular
read the [known issues]
- [x] I have searched through the [issue tracker] for duplicates
- [x] I have mentioned version numbers, operating system and
environment, where applicable:
tqdm: 4.32.2
Python: 3.7.3 (v3.7.3:ef4ec6ed12, Mar 25 2019, 21:26:53) [MSC v.1916 32 bit (Intel)]
Platform: win32
I'm using tqdm to show the progress of a download using the requests library. My current code looks like this:
```
from requests import get, post, Response, request
from tqdm import tqdm
def direct_download(url, target, progress_decorator=tqdm):
res:Response = get(url, stream=True)
with open(target, "wb") as output:
for chunk in progress_decorator(
res.iter_content(1024, False),
unit_scale=True,
total=int(res.headers['Content-Length'])/1024,
unit="kB"
):
output.write(chunk)
```
I have to specify a "larger" chunk size, otherwise the download is needlessly slow (specifying 1 byte leads to download in kBs, specifiying 1 kB leads to download in MBs), I'm trying to figure out a way to have the units scale based on the value of the progress, and tell the tqdm that one iteration doesn't download 1 unit off the content length but 1024.
What I have right now, is I just divide the total in bytes by the chunk size, but this way, the unit prefixing breaks (kB actually means MB, etc.), and setting kB as the main unit doesn't lead (for better or wose) to tqdm detecting the k as a prefix, and replacing it with the proper one.
Edit: Yes, I know the solution for now is manually updating the progressbar, the aim of this issue is to bring this functionality to the iterable wrapper. | open | 2019-06-26T11:36:57Z | 2020-11-01T17:07:37Z | https://github.com/tqdm/tqdm/issues/765 | [
"p3-enhancement 🔥",
"question/docs ‽",
"to-review 🔍"
] | golyalpha | 13 |
cvat-ai/cvat | computer-vision | 8,490 | Can't build SiamMask and TransT functions | ### Actions before raising this issue
- [X] I searched the existing issues and did not find anything similar.
- [X] I read/searched [the docs](https://docs.cvat.ai/docs/)
### Steps to Reproduce
I'm encountering an issue where building automated annotation functions (SiamMask and TransT) in CVAT gets stuck in the "building" state. Below are the steps I followed and the output logs.
**Steps to Reproduce:**
1. Installed CVAT locally using WSL 2 and Docker, following the installation instructions.
2. CVAT runs fine, and I can annotate videos.
3. Tried building automated annotation functions (SiamMask, TransT) using:
`./serverless/deploy_cpu.sh serverless/pytorch/foolwood/siammask/`
Both SiamMask and TransT get stuck in the "building" state. Here's the console output for SiamMask:
> 24.09.30 21:28:55.948 (I) nuctl Project created {"Name": "cvat", "Namespace": "nuclio"}
> Deploying pytorch/foolwood/siammask function...
> 24.09.30 21:28:56.094 (W) nuctl.platform MaxWorkers is deprecated and will be removed in v1.15.x, use NumWorkers instead
> 24.09.30 21:28:56.192 (I) nuctl Deploying function {"name": "pth-foolwood-siammask"}
> 24.09.30 21:28:56.193 (I) nuctl Building {"builderKind": "docker", "versionInfo": "Label: 1.13.13, Git commit: 028989fe9d3fec47692e4a1384ac42267a8b7e0b, OS: linux, Arch: amd64, Go version: go1.21.13", "name": "pth-foolwood-siammask"}
> 24.09.30 21:28:56.329 (I) nuctl Staging files and preparing base images
> 24.09.30 21:28:56.393 (W) nuctl Using user provided base image, runtime interpreter version is provided by the base image {"baseImage": "ubuntu:2a0.04"}
> 24.09.30 21:28:56.454 (I) nuctl Building processor image {"registryURL": "", "taggedImageName": "cvat.pth.foolwood.siammask:latest"}
> 24.09.30 21:28:56.454 (I) nuctl.platform.docker Pulling image {"imageName": "quay.io/nuclio/handler-builder-python-onbuild:1.13.13-amd64"}
> 24.09.30 21:28:59.486 (I) nuctl.platform.docker Pulling image {"imageName": "quay.io/nuclio/uhttpc:0.0.1-amd64"}
> 24.09.30 21:29:02.353 (I) nuctl.platform Building docker image {"image": "cvat.pth.foolwood.siammask:latest"}
It halts at this point. When accessing http://localhost:8070/projects/cvat/functions, I see the functions stuck in the "building" state, and the build log shows nothing.
Found somewhat similar issues:
https://github.com/cvat-ai/cvat/issues/3059
https://github.com/cvat-ai/cvat/issues/4475
Followed the suggestions from the first one (adding port and IP address into SiamMask function.yaml), but it didn't solve the problem.
### Expected Behavior
SiamMask and TransT functions should be deployed successfully and appear in the AI Tools section of the CVAT annotation UI.
### Possible Solution
_No response_
### Context
_No response_
### Environment
```Markdown
Docker Desktop 4.34.2 (167172) on Windows 10
CVAT:
Server version: 2.20.0
Core version: 15.2.0
Canvas version: 2.20.9
UI version: 1.66.0
```
| closed | 2024-09-30T14:52:23Z | 2024-10-18T12:56:52Z | https://github.com/cvat-ai/cvat/issues/8490 | [
"need info"
] | midnightdim | 1 |
yeongpin/cursor-free-vip | automation | 81 | 可否考虑一下 手动指定版本号 | ```bash
✅ SQLite數據庫更新成功
ℹ️ 更新系統ID...
✅ 系統ID更新成功
❌ reset.check_version_failed
ℹ️ Cursor 版本 < 0.45.0,跳过 getMachineId 修补
✅ 機器標識重置成功
```
我本地是 cursor 0.45.11 nixos , 重置后 依旧会提示 请求次数过多. | closed | 2025-02-22T08:53:54Z | 2025-02-25T08:31:21Z | https://github.com/yeongpin/cursor-free-vip/issues/81 | [
"feature"
] | joyanhui | 2 |
laughingman7743/PyAthena | sqlalchemy | 88 | How to specify output bucket for query results in s3? | Hi, we are moving frmo pyathenajdbc to pyathena, but we are trying to specify the `S3OutputLocation` parameter. We need it in our code. is this possible for this library?
thanks. | closed | 2019-06-18T16:36:01Z | 2019-11-23T12:22:13Z | https://github.com/laughingman7743/PyAthena/issues/88 | [] | otmezger | 9 |
FactoryBoy/factory_boy | sqlalchemy | 406 | No module named 'faker.config' | Hello dear friends,
I use Python 3.6 with Anaconda distribution.
I have just installed factory_boy with pip `pip install factory_boy==2.9.2`.
In the dependencies I have the following:
- factory_boy==2.9.2
- Faker==0.7.18
And when I want to load the library I got the following message:
```python
>>> import factory
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "C:\GITPRO~1\FEEDCR~1.IO\venv\lib\site-packages\factory\__init__.py", line 26, in <module>
from .faker import Faker
File "C:\GITPRO~1\FEEDCR~1.IO\venv\lib\site-packages\factory\faker.py", line 23, in <module>
import faker.config
ModuleNotFoundError: No module named 'faker.config'
```
Which is for me quite ... disturbing ... I have tried other versions, but never seemed to make it work ...
Thanks a lot for your help.
Jonathan | closed | 2017-08-16T14:12:42Z | 2023-02-05T15:19:04Z | https://github.com/FactoryBoy/factory_boy/issues/406 | [
"Bug",
"NeedInfo",
"Packaging"
] | DEKHTIARJonathan | 8 |
mars-project/mars | numpy | 2,354 | Support DataFrame operations on data which generates unknown dtypes | <!--
Thank you for your contribution!
Please review https://github.com/mars-project/mars/blob/master/CONTRIBUTING.rst before opening an issue.
-->
**Is your feature request related to a problem? Please describe.**
Some DataFrame operands would generate output with unknown dtypes, like `tranpose`, `get_dummies`, we should add some mechanism that upcoming operations on these objects would fail.
| closed | 2021-08-19T03:51:13Z | 2021-08-19T13:38:28Z | https://github.com/mars-project/mars/issues/2354 | [
"type: feature",
"mod: dataframe"
] | qinxuye | 0 |
amidaware/tacticalrmm | django | 2,057 | [Feature request] Check list overview | **Is your feature request related to a problem? Please describe.**
There is no way of knowing what check is deployed where without going into each agent.
the worst is when most of the checks are done with policies and a couple here and there are done on the agents there is no way of knowing what has been deployed where.
**Describe the solution you'd like**
a new feature that would display a table with all the currently deployed checks with sorting, filtering, search, exculde for each row of the data
**Describe alternatives you've considered**
nada
**Additional context**
something similar to this table into an overlay window like the alerts overview

| open | 2024-11-01T07:51:02Z | 2024-11-01T07:51:02Z | https://github.com/amidaware/tacticalrmm/issues/2057 | [] | P6g9YHK6 | 0 |
ydataai/ydata-profiling | jupyter | 776 | OSError: [Errno 28] No space left on device | **Describe the bug**
Met the same bug with [this issue](https://github.com/pandas-profiling/pandas-profiling/issues/518). The bug still happened when I set the envrionment variable `JOBLIB_TEMP_FOLDER` to a folder that is big enough. Also, I am quite sure that my memory is big enough( about 128G)
**Version information:**
* _Python version_: 3.7.9.
* _Environment_: Jupyter Notebook (local)
* _`pandas-profiling`_:'2.12.0'
| closed | 2021-05-06T03:43:19Z | 2021-05-09T20:42:51Z | https://github.com/ydataai/ydata-profiling/issues/776 | [] | IncubatorShokuhou | 1 |
AUTOMATIC1111/stable-diffusion-webui | pytorch | 16,694 | [Bug]: a1111 stopped loading | ### Checklist
- [ ] The issue exists after disabling all extensions
- [ ] The issue exists on a clean installation of webui
- [ ] The issue is caused by an extension, but I believe it is caused by a bug in the webui
- [ ] The issue exists in the current version of the webui
- [ ] The issue has not been reported before recently
- [ ] The issue has been reported before but has not been fixed yet
### What happened?
Today I watched a video about the Adetailer, how to install it and how to work. I launched the stable diffusion and tried to install it through the extension tab using the link, but for some reason it just took a long time to load and nothing happened, even in the command line nothing happened at that moment. I tried to restart the stable, but it just stopped starting as a result. I also tried restarting the computer, it didn't help either. Before that, everything worked fine. I was going to download it using this link https://github.com/Bing-su/adetailer.git. Please tell me if anyone knows how to solve the problem, I haven't reinstalled the stable completely yet, hoping to find an answer. I don't really understand all this, I'm a simple user who is learning this program.
### Steps to reproduce the problem
launch stable diffusion
### What should have happened?
start up
### What browsers do you use to access the UI ?
Google Chrome
### Sysinfo
### Console logs
```Shell
i don't know
```
### Additional information
_No response_ | closed | 2024-11-29T17:06:40Z | 2024-11-29T17:31:45Z | https://github.com/AUTOMATIC1111/stable-diffusion-webui/issues/16694 | [
"bug-report"
] | Colotus | 0 |
deepspeedai/DeepSpeed | pytorch | 7,121 | AttributeError: partially initialized module 'deepspeed' has no attribute 'init_inference' | **To Reproduce**
inference script:
`import torch
from transformers import AutoModelForCausalLM, AutoTokenizer
import deepspeed
model_name = "/home/pzl/models/Qwen2.5-3B"
tokenizer = AutoTokenizer.from_pretrained(model_name)
model = AutoModelForCausalLM.from_pretrained(model_name)
ds_engine = deepspeed.init_inference(model,
mp_size=1,
dtype=torch.half,
replace_with_kernel_inject=True)
input_text = "DeepSpeed is?"
inputs = tokenizer(input_text, return_tensors="pt")
with torch.no_grad():
outputs = ds_engine.module.generate(**inputs, max_length=10)
output_text = tokenizer.decode(outputs[0], skip_special_tokens=True)
print(output_text)
`
**Expected behavior**
get the correct result
**ds_report output**

**Screenshots**

**System info (please complete the following information):**
- OS: Ubuntu 22.04
- GPU count and types 0
- Python 3.10
| open | 2025-03-09T13:24:32Z | 2025-03-09T13:24:32Z | https://github.com/deepspeedai/DeepSpeed/issues/7121 | [
"bug",
"inference"
] | JocelynPanPan | 0 |
sqlalchemy/sqlalchemy | sqlalchemy | 11,728 | Permanently corrupted hybrid_property column labeling as a result of selecting column returned by hybrid_property.expression | ### Describe the bug
The issue causes a select of `hybrid_property` to return its value under wrong label. The ORM model becomes "corrupted" by construction of a different specific select, selecting the column returned by `hybrid_property.expression`. The select causes memoization of `_proxy_key`, which is then incorrectly used as label of the column in the corrupted select.
`select(Model.id)` will return the `id` hybrid_property under `_id` key which is the name of the column returned by `hybrid_property.expression` of the `id` hybrid_property.
We have found this issue during migration of a large codebase from SQLAlchemy `1.3` to `1.4`, we will be thankful for a `1.4` fix.
### Optional link from https://docs.sqlalchemy.org which documents the behavior that is expected
_No response_
### SQLAlchemy Version in Use
1.4.52, 2.0.32
### DBAPI (i.e. the database driver)
Any, psycopg2
### Database Vendor and Major Version
Any, PostgreSQL 15
### Python Version
3.11, Any
### Operating system
Windows, Any
### To Reproduce
```python
from sqlalchemy import Column, select
from sqlalchemy.dialects.postgresql import UUID
from sqlalchemy.ext.hybrid import hybrid_property
from sqlalchemy.orm import declarative_base
Base = declarative_base()
class Model(Base):
__tablename__ = 'model'
_id = Column(UUID(as_uuid=True), primary_key=True, nullable=False)
@hybrid_property
def id(self):
return self._id
@id.expression
def id(cls):
return cls._id
assert select(Model.id).subquery().c[0].key == 'id' and Model.id.expression._proxy_key == 'id'
assert select(Model._id).subquery().c[0].key == '_id'
# Creating above select causes the first select to fail when repeated.
# Selecting the underscored hybrid.expression's "target" Column causes
# memoization of `_proxy_key` in `Model._id.__clause_element__().__dict__`
# As a consequence column `id` is returned under name `_id` in the result rows
# instead of the expected `id`.
# Presence of the memoized `_proxy_key` in __dict__ causes it to be cloned
# in `sqlalchemy_sql_annotation_annotated._with_annotations`
# and it takes precedence when accessed via the Proxy class of hybrid property
# during select construction.
# The broken memoized state can be reverted by deleting memoized `_proxy_key` using following line
# del Model._id.__clause_element__().__dict__['_proxy_key']
assert (
select(Model.id).subquery().c[0].key == 'id'
), "Resulting `id` column would be returned incorrectly under `_id` attribute/key."
assert Model.id.expression._proxy_key == 'id'
```
### Error
```
# No exceptions
```
### Additional context
_No response_ | closed | 2024-08-12T20:46:39Z | 2024-08-14T14:05:01Z | https://github.com/sqlalchemy/sqlalchemy/issues/11728 | [
"bug",
"orm",
"hard orm"
] | tomas-sk | 8 |
nok/sklearn-porter | scikit-learn | 38 | Port CountVectorizer | For text mining it's important to fit also a CountVectorizer (or a TFIDFTransformer), so should be possible to export it in the targhet lenguage | open | 2018-07-06T18:29:02Z | 2022-05-16T21:55:30Z | https://github.com/nok/sklearn-porter/issues/38 | [
"question",
"new feature"
] | nicolalandro | 3 |
vaexio/vaex | data-science | 1,601 | Filtering using multiple conditions on the same column | Hi, I want to filter on multiple conditions about the same column, but looks like the conditions don't apply correctly.
The reproducible script is as follows.
```python
import vaex
import pandas as pd
pandas_df = pd.DataFrame(
data=[{
'a': 1,
'b': 2,
}, {
'a': 3,
'b': 4,
}, {
'a': 5,
'b': 6,
}])
vaex_df = vaex.from_pandas(df=pandas_df, copy_index=False)
vaex_df[3 < vaex_df.a and vaex_df.a < 6]
```
result :
```
# a b
0 1 2
1 3 4
2 5 6
```
I expected,
```
# a b
0 5 6
```
**Software information**
- Vaex version : {'vaex': '4.5.0', 'vaex-core': '4.5.1', 'vaex-viz': '0.5.0', 'vaex-hdf5': '0.10.0', 'vaex-server': '0.6.1', 'vaex-astro': '0.9.0', 'vaex-jupyter': '0.6.0', 'vaex-ml': '0.14.0'}
- Vaex was installed via pip
- OS: MacOS Catatlina (Intel)
| closed | 2021-10-03T06:01:18Z | 2021-10-03T08:07:57Z | https://github.com/vaexio/vaex/issues/1601 | [] | SyureNyanko | 2 |
axnsan12/drf-yasg | rest-api | 74 | Validation error on serializer used only for responses | Note: This may be what https://github.com/axnsan12/drf-yasg/issues/51 was trying to get at...
Trying to do this, but getting a validation error ("... 'ssv': "Unresolvable JSON pointer: 'definitions/Detail'")
```
class DetailSerializer(serializers.Serializer):
detail = serializers.CharField()
```
```
class ManufacturerViewSet(viewsets.ModelViewSet):
serializer_class = ManufacturerSerializer
model = Manufacturer
queryset = Manufacturer.objects.all()
@swagger_auto_schema(responses={404: openapi.Response("Not found or Not accessible", DetailSerializer,
examples={
'Not found': DetailSerializer({'detail':'Not found'}).data,
'Not accessible': DetailSerializer({'detail':'Not accessible'}).data,
},
)})
def retrieve(self, request, *args, **kwargs):
return super().retrieve(self, request, *args, **kwargs)
```
However, if I add the serializer to recognized model, it does work, e.g.,
```
class ManufacturerSerializer(serializers.ModelSerializer):
status = DetailSerializer(many=False)
class Meta:
model = Manufacturer
fields = '__all__'
```
Full text of validation error...
```
{'flex': "'paths':\n"
" - '/manufacturers/{id}/':\n"
" - 'get':\n"
" - 'responses':\n"
" - '404':\n"
" - 'referenceObject':\n"
" - 'additionalProperties':\n"
' - "When `additionalProperties` is False, '
'no unspecified properties are allowed. The following unspecified '
"properties were found:\\n\\t`{'headers', 'description', 'examples', "
'\'schema\'}`"\n'
" - 'required':\n"
" - '$ref':\n"
" - 'This value is required'\n"
" - 'responseObject':\n"
" - 'schema':\n"
" - '$ref':\n"
" - 'The $ref `#/definitions/Detail` "
"was not found in the schema'",
'ssv': "Unresolvable JSON pointer: 'definitions/Detail'"}
``` | closed | 2018-03-04T19:44:11Z | 2018-03-05T09:51:52Z | https://github.com/axnsan12/drf-yasg/issues/74 | [
"bug"
] | rmorison | 1 |
deepspeedai/DeepSpeed | deep-learning | 6,519 | [BUG] RuntimeError: Error building extension 'inference_core_ops' | **Describe the bug**
I am trying to run the non-persistent example given for mistralai/Mistral-7B-Instruct-v0.3 on a RTX A6000 GPU (on a server) so compute capability is met, ubuntu is 22.04, CUDA toolkit is 11.5 (I am not a sudoer of the server so I am not able to upgrade the toolkit, instead I have created a conda environment and installed CUDA toolkit 11.8). On running the python3 pipeline.py command I am running into the error: RuntimeError: Error building extension 'inference_core_ops'
**To Reproduce**
Steps to reproduce the behavior:
1. conda create -n my_env python=3.12.4 cudatoolkit=11.8
2. pip install deepspeed-mii (in the conda environment with CUDA toolkit 11.8)
3. https://github.com/microsoft/DeepSpeedExamples/blob/master/inference/mii/non-persistent/pipeline.py
4. What packages are required and their versions:
NVIDIA GPU(s) with compute capability of: 8.0, 8.6, 8.9, 9.0.
CUDA 11.6+
Ubuntu 20+
5. python3 pipeline.py or deepspeed --num_gpus 1 --no_local_rank pipeline.py
**ds_report output**
(deep) (base) cpatil@meherangarh:/data1/cpatil/simplismart$ ds_report
[2024-09-10 13:49:31,073] [INFO] [real_accelerator.py:203:get_accelerator] Setting ds_accelerator to cuda (auto detect)
--------------------------------------------------
DeepSpeed C++/CUDA extension op report
--------------------------------------------------
NOTE: Ops not installed will be just-in-time (JIT) compiled at
runtime if needed. Op compatibility means that your system
meet the required dependencies to JIT install the op.
--------------------------------------------------
JIT compiled ops requires ninja
ninja .................. [OKAY]
--------------------------------------------------
op name ................ installed .. compatible
--------------------------------------------------
[WARNING] async_io requires the dev libaio .so object and headers but these were not found.
[WARNING] async_io: please install the libaio-dev package with apt
[WARNING] If libaio is already installed (perhaps from source), try setting the CFLAGS and LDFLAGS environment variables to where it can be found.
async_io ............... [NO] ....... [NO]
fused_adam ............. [NO] ....... [OKAY]
cpu_adam ............... [NO] ....... [OKAY]
cpu_adagrad ............ [NO] ....... [OKAY]
cpu_lion ............... [NO] ....... [OKAY]
[WARNING] Please specify the CUTLASS repo directory as environment variable $CUTLASS_PATH
evoformer_attn ......... [NO] ....... [NO]
[WARNING] FP Quantizer is using an untested triton version (2.2.0), only 2.3.(0, 1) and 3.0.0 are known to be compatible with these kernels
fp_quantizer ........... [NO] ....... [NO]
fused_lamb ............. [NO] ....... [OKAY]
fused_lion ............. [NO] ....... [OKAY]
x86_64-linux-gnu-gcc -Wno-unused-result -Wsign-compare -DNDEBUG -g -fwrapv -O2 -Wall -g -fstack-protector-strong -Wformat -Werror=format-security -g -fwrapv -O2 -g -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 -fPIC -c /tmp/tmphfej6tlu/test.c -o /tmp/tmphfej6tlu/test.o
x86_64-linux-gnu-gcc /tmp/tmphfej6tlu/test.o -L/usr -lcufile -o /tmp/tmphfej6tlu/a.out
/usr/bin/ld: cannot find -lcufile: No such file or directory
collect2: error: ld returned 1 exit status
gds .................... [NO] ....... [NO]
inference_core_ops ..... [NO] ....... [OKAY]
cutlass_ops ............ [NO] ....... [OKAY]
transformer_inference .. [NO] ....... [OKAY]
quantizer .............. [NO] ....... [OKAY]
ragged_device_ops ...... [NO] ....... [OKAY]
ragged_ops ............. [NO] ....... [OKAY]
random_ltd ............. [NO] ....... [OKAY]
[WARNING] sparse_attn requires a torch version >= 1.5 and < 2.0 but detected 2.2
[WARNING] using untested triton version (2.2.0), only 1.0.0 is known to be compatible
sparse_attn ............ [NO] ....... [NO]
spatial_inference ...... [NO] ....... [OKAY]
transformer ............ [NO] ....... [OKAY]
stochastic_transformer . [NO] ....... [OKAY]
--------------------------------------------------
DeepSpeed general environment info:
torch install path ............... ['/home/cpatil/.local/lib/python3.10/site-packages/torch']
torch version .................... 2.2.2+cu121
deepspeed install path ........... ['/home/cpatil/.local/lib/python3.10/site-packages/deepspeed']
deepspeed info ................... 0.15.1, unknown, unknown
torch cuda version ............... 12.1
torch hip version ................ None
nvcc version ..................... 11.5
deepspeed wheel compiled w. ...... torch 2.2, cuda 12.1
shared memory (/dev/shm) size .... 503.87 GB
**Output on running the command**
(deep) (base) cpatil@meherangarh:/data1/cpatil/simplismart$ python3 pipeline.py
[2024-09-10 13:43:54,824] [INFO] [real_accelerator.py:203:get_accelerator] Setting ds_accelerator to cuda (auto detect)
[2024-09-10 13:43:56,883] [INFO] [comm.py:652:init_distributed] cdb=None
[2024-09-10 13:43:56,884] [INFO] [comm.py:683:init_distributed] Initializing TorchBackend in DeepSpeed with backend nccl
Fetching 11 files: 100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 11/11 [00:00<00:00, 79410.23it/s]
[2024-09-10 13:43:57,612] [INFO] [engine_v2.py:82:__init__] Building model...
Using /home/cpatil/.cache/torch_extensions/py312_cu121 as PyTorch extensions root...
Detected CUDA files, patching ldflags
Emitting ninja build file /home/cpatil/.cache/torch_extensions/py312_cu121/inference_core_ops/build.ninja...
/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/utils/cpp_extension.py:1965: UserWarning: TORCH_CUDA_ARCH_LIST is not set, all archs for visible cards are included for compilation.
If this is not desired, please set os.environ['TORCH_CUDA_ARCH_LIST'].
warnings.warn(
Building extension module inference_core_ops...
Allowing ninja to set a default number of workers... (overridable by setting the environment variable MAX_JOBS=N)
[1/2] /usr/bin/nvcc --generate-dependencies-with-compile --dependency-output linear_kernels_cuda.cuda.o.d -DTORCH_EXTENSION_NAME=inference_core_ops -DTORCH_API_INCLUDE_EXTENSION_H -DPYBIND11_COMPILER_TYPE=\"_gcc\" -DPYBIND11_STDLIB=\"_libstdcpp\" -DPYBIND11_BUILD_ABI=\"_cxxabi1011\" -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/bias_activations -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/blas_kernels -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_layer_norm -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_rms_norm -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/gated_activations -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_linear -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/includes -isystem /home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/include -isystem /home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/include/torch/csrc/api/include -isystem /home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/include/TH -isystem /home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/include/THC -isystem /home/cpatil/miniconda3/envs/deep/include/python3.12 -D_GLIBCXX_USE_CXX11_ABI=0 -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_BFLOAT16_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr -gencode=arch=compute_86,code=compute_86 -gencode=arch=compute_86,code=sm_86 --compiler-options '-fPIC' -O3 --use_fast_math -std=c++17 -U__CUDA_NO_HALF_OPERATORS__ -U__CUDA_NO_HALF_CONVERSIONS__ -U__CUDA_NO_HALF2_OPERATORS__ --threads=8 -gencode=arch=compute_86,code=sm_86 -gencode=arch=compute_86,code=compute_86 -DBF16_AVAILABLE -U__CUDA_NO_BFLOAT16_OPERATORS__ -U__CUDA_NO_BFLOAT162_OPERATORS__ -U__CUDA_NO_BFLOAT16_CONVERSIONS__ -c /home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_linear/linear_kernels_cuda.cu -o linear_kernels_cuda.cuda.o
FAILED: linear_kernels_cuda.cuda.o
/usr/bin/nvcc --generate-dependencies-with-compile --dependency-output linear_kernels_cuda.cuda.o.d -DTORCH_EXTENSION_NAME=inference_core_ops -DTORCH_API_INCLUDE_EXTENSION_H -DPYBIND11_COMPILER_TYPE=\"_gcc\" -DPYBIND11_STDLIB=\"_libstdcpp\" -DPYBIND11_BUILD_ABI=\"_cxxabi1011\" -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/bias_activations -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/blas_kernels -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_layer_norm -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_rms_norm -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/gated_activations -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_linear -I/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/includes -isystem /home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/include -isystem /home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/include/torch/csrc/api/include -isystem /home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/include/TH -isystem /home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/include/THC -isystem /home/cpatil/miniconda3/envs/deep/include/python3.12 -D_GLIBCXX_USE_CXX11_ABI=0 -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_BFLOAT16_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr -gencode=arch=compute_86,code=compute_86 -gencode=arch=compute_86,code=sm_86 --compiler-options '-fPIC' -O3 --use_fast_math -std=c++17 -U__CUDA_NO_HALF_OPERATORS__ -U__CUDA_NO_HALF_CONVERSIONS__ -U__CUDA_NO_HALF2_OPERATORS__ --threads=8 -gencode=arch=compute_86,code=sm_86 -gencode=arch=compute_86,code=compute_86 -DBF16_AVAILABLE -U__CUDA_NO_BFLOAT16_OPERATORS__ -U__CUDA_NO_BFLOAT162_OPERATORS__ -U__CUDA_NO_BFLOAT16_CONVERSIONS__ -c /home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_linear/linear_kernels_cuda.cu -o linear_kernels_cuda.cuda.o
/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_linear/include/ptx_mma.cuh(59): warning #174-D: expression has no effect
/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_linear/include/ptx_mma.cuh(135): warning #174-D: expression has no effect
/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_linear/include/ptx_cp.async.cuh(33): warning #174-D: expression has no effect
/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_linear/include/ptx_cp.async.cuh(44): warning #174-D: expression has no effect
/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_linear/include/ptx_cp.async.cuh(56): warning #174-D: expression has no effect
/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_linear/include/ptx_cp.async.cuh(70): warning #174-D: expression has no effect
/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_linear/include/kernel_matmul.cuh(268): warning #174-D: expression has no effect
/usr/include/c++/11/bits/std_function.h:435:145: error: parameter packs not expanded with ‘...’:
435 | function(_Functor&& __f)
| ^
/usr/include/c++/11/bits/std_function.h:435:145: note: ‘_ArgTypes’
/usr/include/c++/11/bits/std_function.h:530:146: error: parameter packs not expanded with ‘...’:
530 | operator=(_Functor&& __f)
| ^
/usr/include/c++/11/bits/std_function.h:530:146: note: ‘_ArgTypes’
ninja: build stopped: subcommand failed.
[rank0]: Traceback (most recent call last):
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/utils/cpp_extension.py", line 2105, in _run_ninja_build
[rank0]: subprocess.run(
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/subprocess.py", line 571, in run
[rank0]: raise CalledProcessError(retcode, process.args,
[rank0]: subprocess.CalledProcessError: Command '['ninja', '-v']' returned non-zero exit status 1.
[rank0]: The above exception was the direct cause of the following exception:
[rank0]: Traceback (most recent call last):
[rank0]: File "/data1/cpatil/simplismart/pipeline.py", line 12, in <module>
[rank0]: pipe = mii.pipeline(args.model)
[rank0]: ^^^^^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/mii/api.py", line 231, in pipeline
[rank0]: inference_engine = load_model(model_config)
[rank0]: ^^^^^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/mii/modeling/models.py", line 17, in load_model
[rank0]: inference_engine = build_hf_engine(
[rank0]: ^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/engine_factory.py", line 135, in build_hf_engine
[rank0]: return InferenceEngineV2(policy, engine_config)
[rank0]: ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/engine_v2.py", line 83, in __init__
[rank0]: self._model = self._policy.build_model(self._config, self._base_mp_group)
[rank0]: ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/model_implementations/inference_policy_base.py", line 156, in build_model
[rank0]: self.model = self.instantiate_model(engine_config, mp_group)
[rank0]: ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/model_implementations/mistral/policy.py", line 17, in instantiate_model
[rank0]: return MistralInferenceModel(config=self._model_config, engine_config=engine_config, base_mp_group=mp_group)
[rank0]: ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/model_implementations/inference_transformer_base.py", line 215, in __init__
[rank0]: self.make_norm_layer()
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/model_implementations/inference_transformer_base.py", line 518, in make_norm_layer
[rank0]: self.norm = heuristics.instantiate_pre_norm(norm_config, self._engine_config)
[rank0]: ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/modules/heuristics.py", line 176, in instantiate_pre_norm
[rank0]: return DSPreNormRegistry.instantiate_config(config)
[rank0]: ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/modules/module_registry.py", line 36, in instantiate_config
[rank0]: if not target_implementation.supports_config(config_bundle.config):
[rank0]: ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/modules/implementations/pre_norm/cuda_pre_rms.py", line 36, in supports_config
[rank0]: _ = CUDARMSPreNorm(config.channels, config.residual_dtype)
[rank0]: ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/inference/v2/kernels/core_ops/cuda_rms_norm/rms_norm_base.py", line 36, in __init__
[rank0]: self.inf_module = InferenceCoreBuilder().load()
[rank0]: ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/ops/op_builder/builder.py", line 531, in load
[rank0]: return self.jit_load(verbose)
[rank0]: ^^^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/deepspeed/ops/op_builder/builder.py", line 578, in jit_load
[rank0]: op_module = load(name=self.name,
[rank0]: ^^^^^^^^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/utils/cpp_extension.py", line 1312, in load
[rank0]: return _jit_compile(
[rank0]: ^^^^^^^^^^^^^
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/utils/cpp_extension.py", line 1722, in _jit_compile
[rank0]: _write_ninja_file_and_build_library(
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/utils/cpp_extension.py", line 1834, in _write_ninja_file_and_build_library
[rank0]: _run_ninja_build(
[rank0]: File "/home/cpatil/miniconda3/envs/deep/lib/python3.12/site-packages/torch/utils/cpp_extension.py", line 2121, in _run_ninja_build
[rank0]: raise RuntimeError(message) from e
[rank0]: RuntimeError: Error building extension 'inference_core_ops'
[rank0]:[W910 13:45:07.069688599 ProcessGroupNCCL.cpp:1168] Warning: WARNING: process group has NOT been destroyed before we destruct ProcessGroupNCCL. On normal program exit, the application should call destroy_process_group to ensure that any pending NCCL operations have finished in this process. In rare cases this process can exit before this point and block the progress of another member of the process group. This constraint has always been present, but this warning has only been added since PyTorch 2.4 (function operator())
**System info (please complete the following information):**
- OS: Ubuntu 22.04
- GPU count and types: Single GPU, RTX A6000, 48GB (Compute Capability 8.6)
- DeepSpeed-MII version: 0.3.0
- Python version: 3.12.4
**Additional context**
I am running the pipeline.py script on a server with CUDA toolkit version 11.5, since I am not a sudoer I have instead created a conda env with toolkit version 11.8. | closed | 2024-09-10T08:37:09Z | 2024-11-15T22:50:56Z | https://github.com/deepspeedai/DeepSpeed/issues/6519 | [
"bug",
"build"
] | Chetan3200 | 3 |
AUTOMATIC1111/stable-diffusion-webui | pytorch | 16,234 | [Bug]: Upscaler.py causes infinite loop (fix inside) | ### Checklist
- [ ] The issue exists after disabling all extensions
- [X] The issue exists on a clean installation of webui
- [ ] The issue is caused by an extension, but I believe it is caused by a bug in the webui
- [X] The issue exists in the current version of the webui
- [ ] The issue has not been reported before recently
- [X] The issue has been reported before but has not been fixed yet
### What happened?
The current ([9d4fdc4](https://github.com/AUTOMATIC1111/stable-diffusion-webui/commit/9d4fdc45d3c4b9431551fd53de64a67726dcbd64)) copy of upscaler.py does not fix the 1x upscaler infinite loop issue: https://github.com/AUTOMATIC1111/stable-diffusion-webui/blob/master/modules/upscaler.py. Here is what is wrong ...
The if statement on line 59 checks three conditions simultaneously: img.width >= dest_w, img.height >= dest_h, and scale != 1.
The loop will break only if all three conditions are met. If any one of them is not met, the loop will continue to the next iteration.
**The code inside the loop does not include any logic for upscaling the image.** Without upscaling logic, the image dimensions will not change, and the conditions will not be reevaluated correctly.
There is no check to see if the dimensions of the image have changed after a potential upscaling operation. This means there's no way to break the loop if the upscaling doesn't affect the image dimensions.
So here's what I tried to do:
1. Check if the image dimensions are already at or above the desired width and height. If so, it breaks out of the loop.
2. Store the current dimensions of the image in shape.
3. Upscale the image using the do_upscale method and the selected model.
4. Check if the dimensions of the image are the same after upscaling. If so, it breaks out of the loop.
Remove lines 59-61 in upscaler.py and insert this instead
```
for _ in range(3):
if img.width >= dest_w and img.height >= dest_h:
break
shape = (img.width, img.height)
img = self.do_upscale(img, selected_model)
if shape == (img.width, img.height):
break
```
Hooray, it upscales now!
___________________
[sysinfo-2024-07-18-21-29.json](https://github.com/user-attachments/files/16289442/sysinfo-2024-07-18-21-29.json)
Chrome Version 126.0.6478.127 Windows 10 2080 GTX with 8GB
### Steps to reproduce the problem
Use any upscaler and set "upscale by" to 1x
### What should have happened?
Should've upscaled
### What browsers do you use to access the UI ?
Google Chrome
### Sysinfo
[sysinfo-2024-07-18-21-29.json](https://github.com/user-attachments/files/16289457/sysinfo-2024-07-18-21-29.json)
### Console logs
```Shell
wasn't saving them to txt sorry
```
### Additional information
_No response_ | open | 2024-07-18T21:33:46Z | 2024-07-19T19:27:05Z | https://github.com/AUTOMATIC1111/stable-diffusion-webui/issues/16234 | [
"bug-report"
] | NPDX909 | 2 |
deepspeedai/DeepSpeed | machine-learning | 5,803 | run evaluate_cogagent_demo.py error from sat import mpu, get_args, get_tokenizer error | My environment is
transformers 4.42.3
torch 2.3.0+cu121
deepspeed 0.14.5+unknow
when I run
evaluate_cogagent_demo.py
I encountered the following error message
2024-07-27 16:07:37,915] [INFO] [real_accelerator.py:203:get_accelerator] Setting ds_accelerator to cuda (auto detect)
D:\conda3\envs\pytorch\lib\site-packages\transformers\utils\hub.py:127: FutureWarning: Using `TRANSFORMERS_CACHE` is deprecated and will be removed in v5 of Transformers. Use `HF_HOME` instead.
warnings.warn(
[2024-07-27 16:07:40,151] [INFO] DeepSpeed/CUDA is not installed, fallback to Pytorch checkpointing.
Traceback (most recent call last):
File "D:\PycharmProjects\CogVLM-main\finetune_demo\evaluate_cogagent_demo.py", line 7, in <module>
from sat import mpu, get_args, get_tokenizer
File "D:\conda3\envs\pytorch\lib\site-packages\sat\__init__.py", line 6, in <module>
from .training.deepspeed_training import training_main
File "D:\conda3\envs\pytorch\lib\site-packages\sat\training\deepspeed_training.py", line 27, in <module>
import deepspeed
File "D:\conda3\envs\pytorch\lib\site-packages\deepspeed\__init__.py", line 25, in <module>
from . import ops
File "D:\conda3\envs\pytorch\lib\site-packages\deepspeed\ops\__init__.py", line 11, in <module>
from . import transformer
File "D:\conda3\envs\pytorch\lib\site-packages\deepspeed\ops\transformer\__init__.py", line 8, in <module>
from ...model_implementations.transformers.ds_transformer import DeepSpeedTransformerInference
File "D:\conda3\envs\pytorch\lib\site-packages\deepspeed\model_implementations\transformers\ds_transformer.py", line 17, in <module>
from deepspeed.ops.transformer.inference.triton.mlp import TritonMLP
File "D:\conda3\envs\pytorch\lib\site-packages\deepspeed\ops\transformer\inference\__init__.py", line 7, in <module>
from ....model_implementations.transformers.ds_transformer import DeepSpeedTransformerInference
ImportError: cannot import name 'DeepSpeedTransformerInference' from partially initialized module 'deepspeed.model_implementations.transformers.ds_transformer' (most likely due to a circular import) (D:\conda3\envs\pytorch\lib\site-packages\deepspeed\model_implementations\transformers\ds_transformer.py)
Process finished with exit code 1
I can't find the solution. Any suggestions ?
Thank you for your help. | closed | 2024-07-27T08:19:50Z | 2024-08-01T17:29:54Z | https://github.com/deepspeedai/DeepSpeed/issues/5803 | [
"bug",
"inference"
] | gyjlll | 3 |
OthersideAI/self-operating-computer | automation | 102 | Poor accuracy of pointer X/Y location inference | the X/Y coordinates inferred by the model are always off. It can't even select the address bar correctly. | closed | 2023-12-12T21:46:40Z | 2024-02-09T04:17:32Z | https://github.com/OthersideAI/self-operating-computer/issues/102 | [] | ahsin-s | 19 |
learning-at-home/hivemind | asyncio | 88 | Support tensor compression (fp16/int8) | In preliminary tests we observed
* no quality degradation from converting tensors to fp16 and back before sending them across
* negligible drawdown when tensors are encoded as fp32 mean/std and int8 data
It would be great to enable quantization as a builtin feature for ExpertBackend, e.g.
```
expert = ExpertBackend(..., outputs_schema=[BatchTensorDescriptor(..., compression="int8_normalize")]
``` | closed | 2020-08-27T14:22:47Z | 2021-02-24T13:21:05Z | https://github.com/learning-at-home/hivemind/issues/88 | [
"enhancement"
] | justheuristic | 1 |
CorentinJ/Real-Time-Voice-Cloning | python | 811 | Here is a model in Mandarin/Chinese 中文模型 | Finally I got it work and it's really fun that you are listening to your own voice.
终于可以玩耍起来了,
Link:https://pan.baidu.com/s/10t3XycWiNIg5dN5E_bMORQ
Code:aid4
Codebase: https://github.com/babysor/Realtime-Voice-Clone-Chinese | closed | 2021-08-07T04:27:05Z | 2022-10-03T13:46:32Z | https://github.com/CorentinJ/Real-Time-Voice-Cloning/issues/811 | [] | babysor | 19 |
nschloe/tikzplotlib | matplotlib | 231 | Cannot pass extra_axis_parameters | Whenever I run tikz_save(), I get the annoying messages:
```
Horizontal alignment will be ignored as no 'x tick label text width' has been passed in the 'extra' parameter
```
To get rid of them, I need to pass another argument extra_axis_parameters={...}
which is supposed to be of type 'set' to tikz_save.
This input variable is then used in axes.py to define the variable tick_label_text_width (l. 410-414)
```
if tick_label_text_width_identifier in data['extra axis options']:
tick_label_text_width = data['extra axis options [base]'][
tick_label_text_width_identifier
]
del data['extra axis options'][tick_label_text_width_identifier]
```
The variable `data['extra axis options [base]']` however is also of type `set`, therefore causing a
```
TypeError: 'set' object is not subscriptable
```
Am I doing something wrong or is this a bug?
This is the command that I use:
```
tikz_save(directory + data_name[:-4] + '.tex',
figureheight='\\figureheight',
figurewidth='\\figurewidth',
extra_axis_parameters={
## 'x tick label text width={\\textwidth}',
'x tick label text width',
'10 pt',
},
show_info=False,
)
``` | closed | 2018-04-18T13:30:15Z | 2019-03-20T10:57:44Z | https://github.com/nschloe/tikzplotlib/issues/231 | [] | Steffen1989 | 2 |
healthchecks/healthchecks | django | 341 | integration editing is awkward | You're probably aware of this, but I thought I'd summarize my experience anyway. I just went through setting up an integration with zulipchat (compare #202) via a custom webhook. That involved a bit of tweaking things, which turned out to be quite painful because
1. integrations don't start out with a name / distinguishable in the listing
2. integrations can't be edited
From my outside perspective, the best fix for this would be to allow editing integrations with a form similar to the "create integration" form, and to include the integration name there. | closed | 2020-03-10T12:27:05Z | 2020-04-06T12:59:52Z | https://github.com/healthchecks/healthchecks/issues/341 | [] | robx | 3 |
allure-framework/allure-python | pytest | 586 | 2 SubSuite defined while using dynamic definition (pytest) | #### I'm submitting a ...
- [x] bug report
- [ ] feature request
- [ ] support request => Please do not submit support request here, see note at the top of this template.
#### What is the current behavior?
I want to use allure to generate report, and especially write some data into the report dynamically.
We already did implementation for data like: feature, title, description, ... and it is working fine.
But, in the case of sub_suite, if it is **NOT** defined in the pytest script, with **@allure.sub_suite('value1')**, then it will add to the report a new subsuite, automatically with the name of the class.
Then, when I add the subsuite with allure.dynamic.sub_suite('value1'), I have in the same report:
- {"name": "subSuite", "value": "className"}
- {"name": "subSuite", "value": "value1"}
#### If the current behavior is a bug, please provide the steps to reproduce and if possible a minimal demo of the problem
- Do not define any subsuite in the pytest script. eg: **@allure.sub_suite('value1')**
- While generating the allure report dynamically, use the function: allure.dynamic.sub_suite('value1')
#### What is the expected behavior?
Only 1 entry should be available in the allure report:
{"name": "subSuite", "value": "value1"}
#### What is the motivation / use case for changing the behavior?
Get data from a configuration file or document and define the data based on those files, dynamically.
#### Please tell us about your environment:
- Allure version: 2.13.6
- Test framework: pytest@5.0.1
- Allure adaptor: allure-pytest@2.8.40 | closed | 2021-04-29T14:48:20Z | 2023-04-25T11:56:11Z | https://github.com/allure-framework/allure-python/issues/586 | [
"bug",
"theme:pytest"
] | Shionigami | 1 |
allenai/allennlp | nlp | 5,237 | Accept compressed files as input to `predict` when using a `Predictor` | **Is your feature request related to a problem? Please describe.**
I typically used compressed datasets (e.g. gzipped) to save disk space. This works fine with AllenNLP during training because I can write my dataset reader to load the compressed data. However, the `predict` command opens the file and reads lines for the `Predictor`. This fails when it tries to load data from my compressed files.
https://github.com/allenai/allennlp/blob/39d7e5ae06551fe371d3e16f4d93162e55ec5dcc/allennlp/commands/predict.py#L208-L218
**Describe the solution you'd like**
Either automatically detect the file is compressed or add a flag to `predict` that indicates that the file is compressed. One method that I have used to detect if a file is gzipped is [here](https://stackoverflow.com/questions/3703276/how-to-tell-if-a-file-is-gzip-compressed), although it isn't 100% accurate. I have an implementation [here](https://github.com/danieldeutsch/sacrerouge/blob/master/sacrerouge/io/util.py). Otherwise a flag like `--compression-type` to mark how the file is compressed should be sufficient. Passing the type of compression would allow support for gzip, bz2, or any other method.
| open | 2021-06-02T17:29:15Z | 2022-09-12T16:23:07Z | https://github.com/allenai/allennlp/issues/5237 | [
"Good First Issue",
"Contributions welcome",
"Feature request"
] | danieldeutsch | 10 |
ultralytics/yolov5 | machine-learning | 12,968 | Run yolov5 using tensor rt | ### Search before asking
- [X] I have searched the YOLOv5 [issues](https://github.com/ultralytics/yolov5/issues) and [discussions](https://github.com/ultralytics/yolov5/discussions) and found no similar questions.
### Question
I exported Tensor RT weights (last.engine). When i use the weights in yolov5 detect it generates an error
` File "detect.py", line 112, in run
model = DetectMultiBackend(weights, device=device, dnn=dnn, data=data, fp16=half)
File "C:\Users\shen\yolov5\models\common.py", line 426, in __init__
with open(w, "rb") as f, trt.Runtime(logger) as runtime:
TypeError: pybind11::init(): factory function returned nullptr`
### Additional
_No response_ | closed | 2024-04-28T01:10:43Z | 2024-06-08T00:22:31Z | https://github.com/ultralytics/yolov5/issues/12968 | [
"question",
"Stale"
] | tasyoooo | 2 |
2noise/ChatTTS | python | 332 | 在海量音频中寻找相似音色的可能 |
理论上如果生成的音色范围足够广,大多数音频有办法找到相似的音色。
https://github.com/cronrpc/Audio-Speaker-Needle-In-Haystack
这里用的3ds-speaker的音频embs,可能扩充到一万人到五万人会产生质变,并且文本需要选择一个尽可能相似的。
Huggingface Sapce: https://huggingface.co/spaces/omniway/Audio-Needle-In-Haystack

| closed | 2024-06-17T19:07:28Z | 2024-07-20T07:26:32Z | https://github.com/2noise/ChatTTS/issues/332 | [
"ad"
] | cronrpc | 5 |
PokeAPI/pokeapi | graphql | 188 | evolution-chain uses inconsistent schema for evolution_details | Best example to compare is evolution-chain/1 and evolution-chain/34.
The evolution_details property for evolves_to can be null, a single value, or an array. I propose wrapping all values in an array so that the expected type remains consistent.
| closed | 2016-05-12T13:29:12Z | 2016-05-24T13:56:16Z | https://github.com/PokeAPI/pokeapi/issues/188 | [] | zberk | 4 |
marcomusy/vedo | numpy | 606 | Plotter in Qt - Mouse Click Event not registering at the correct position | I have tried several different things about the mouse clicks not registering correctly in my Qt mainwindow. When using only my QHD monitor, the program worked just fine ([video](https://drive.google.com/file/d/1v-2o86Stp6tS1EWGkR2dSy2Iit1Rnkl6/view?usp=sharing)). However, when using my laptop (zoomed in at 1792 x 1120) as the only display, the mouse clicks seemed to have a varying up-right offset and register more accurately near the bottom left corner of the widget ([video](https://drive.google.com/file/d/1v79_tnGgVg0CvtoSho9r66VPj8YWBY26/view?usp=sharing)). I am suspicious that the screen resolution of the display might cause a problem for vedo.
The mouse event is a vedo plotter event. Changing the "screensize", "size", "pos" attribute of the plotter did not fix the issue.
I looked up some examples provided by vedo, specifically mousehover.py and qt_window1.py. The mousehover example worked fine on my laptop. However, adding a clicking event in qt_window1.py also created the same issue. Therefore, the problem most likely was caused by the qt widget.
```
def __init__(self,size):
super(MainWindow, self).__init__()
# load the components defined in th xml file
loadUi("viewer_gui.ui", self)
self.screenSize = size
# Connections for all elements in Mainwindow
self.pushButton_inputfile.clicked.connect(self.getFilePath)
self.pushButton_clearSelection.clicked.connect(self.clearScreen)
self.action_selectVertex.toggled.connect(self.actionSelection_state_changed)
self.action_selectActor.toggled.connect(self.actionSelection_state_changed)
# Set up VTK widget
self.vtkWidget = QVTKRenderWindowInteractor()
self.splitter_viewer.addWidget(self.vtkWidget)
# ipy console
self.ipyConsole = QIPythonWidget(customBanner="Welcome to the embedded ipython console\n")
self.splitter_viewer.addWidget(self.ipyConsole)
self.ipyConsole.pushVariables({"foo":43, "print_process_id":print_process_id, "ipy":self.ipyConsole, "self":self})
self.ipyConsole.printText("The variable 'foo' and the method 'print_process_id()' are available.\
Use the 'whos' command for information.\n\nTo push variables run this before starting the UI:\
\n ipyConsole.pushVariables({\"foo\":43,\"print_process_id\":print_process_id})")
# Create renderer and add the vedo objects and callbacks
self.plt = Plotter(qtWidget=self.vtkWidget,bg='DarkSlateBlue',bg2='MidnightBlue',screensize=(1792,1120))
self.id1 = self.plt.addCallback("mouse click", self.onMouseClick)
self.id2 = self.plt.addCallback("key press", self.onKeypress)
self.plt.show() # <--- show the vedo rendering
def onMouseClick(self, event):
if(self.action_selectActor.isChecked()):
self.selectActor(event)
elif(self.action_selectVertex.isChecked()):
self.selectVertex(event)
def selectActor(self,event):
if(not event.actor):
return
printc("You have clicked your mouse button. Event info:\n", event, c='y')
printc("Left button pressed on", [event.picked3d])
# adding a silhouette might cause some lags
# self.plt += event.actor.silhouette().lineWidth(2).c('red')
#an alternative solution
self.actorSelection = event.actor.clone()
self.actorSelection.c('red')
self.plt += self.actorSelection
def selectVertex(self,event):
if(not event.isPoints):
return
# print(arr[event.actor.closestPoint(event.picked3d, returnPointId=True)])
printc("You have clicked your mouse button. Event info:\n", event, c='y')
printc("Left button pressed on 3d: ", [event.picked3d])
printc("Left button pressed on 2d: ", [event.picked2d])
p = pointcloud.Point(pos=(event.picked3d[0],event.picked3d[1],event.picked3d[2]),r=12,c='red',alpha=0.5)
self.vertexSelections.append(p)
self.plt += p
```
Running the following lines:
```
app = QApplication(sys.argv)
screen = app.primaryScreen()
print('Screen: %s' % screen.name())
size = screen.size()
print('Size: %d x %d' % (size.width(), size.height()))
```
outputted
```
Screen: Color LCD
Size: 1792 x 1120
```
| closed | 2022-03-02T04:49:20Z | 2022-04-21T17:47:36Z | https://github.com/marcomusy/vedo/issues/606 | [
"long-term"
] | danieljiang520 | 11 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.