text
stringlengths 35
445k
| metadata
dict |
|---|---|
# SciplexPrep_3.ipynb
Repository: yulun-rayn/variational-causal-inference
<code>
import sys, os
module_path = os.path.abspath(os.path.join('..'))
if module_path not in sys.path:
sys.path.append(module_path)
import numpy as np
import pandas as pd
import scanpy as sc
</code>
We load the single-cell data using scanpy. The single-cell data is stored in a special data structure called AnnData (short: adata).
<code>
adatas = []
for i in range(5):
adatas.append(sc.read(f'sciplex_raw_chunk_{i}.h5ad'))
adata = adatas[0].concatenate(adatas[1:])
adata
</code>
The counts are stored in adata.X
<code>
adata.X
</code>
adata.obs is a dataframe containing annotation for each cell, such as e.g. batch, cell type, perturbation. Or other technical annotations at the cell level.
<code>
adata.obs
</code>
adata.var is a dataframe containing annotation for each gene, usually some statistics such as dispersion, or gene names, pathways etc.
<code>
adata.var
</code>
## Quality control
Check the quality of the data and remove some cells.
<code>
adata.obs['n_counts'] = np.ravel(adata.X.sum(1)) #number of counts in the cell
adata.obs['n_genes'] = np.ravel(np.sum(adata.X > 0, axis=1)) #number of genes with at least 1 count per cell
adata.var['mito'] = adata.var_names.str.contains("MT-") #flag for mitochondrial genes
adata.obs['mt_frac'] = np.ravel(adata.X[:, adata.var.mito].sum(1)) / adata.obs['n_counts'].values #fraction of mitochondrial gene exp, high values mean dead or bad quality cells
</code>
<code>
# filtering
adata = adata[adata.obs['n_counts'] > 500]
adata = adata[adata.obs['n_genes'] > 750]
adata = adata[adata.obs['mt_frac'] < 0.2]
adata
</code>
## Normalization
<code>
adata.X.max() #check it's an int, to make sure it's count data and not preprocessed data
</code>
<code>
sc.pp.subsample(adata, fraction=0.5, random_state=0)
sc.pp.normalize_per_cell(adata)
sc.pp.log1p(adata)
</code>
## Feature (gene) selection
We select only the top N most variable genes.
<code>
sc.pp.highly_variable_genes(adata, n_top_genes=2000, subset=True)
</code>
## Out-of-distribution selection
<code>
drugs = adata.obs.product_name.unique()
drugs = drugs[~np.isin(drugs, ['Vehicle'])]
</code>
<code>
results = []
for cond1 in drugs:
ad1 = adata[adata.obs.product_name == cond1]
ad2 = adata[adata.obs.product_name != cond1]
mean1 = ad1.X.mean(0)
mean2 = ad2.X.mean(0)
l2 = np.linalg.norm(mean1-mean2)
results.append({
'cond1': cond1,
'L2': l2
})
df_vs_rest = pd.DataFrame(results)
</code>
Pick biggest signals
<code>
drug_OOD = df_vs_rest.sort_values(by='L2').tail(20).cond1.values
drug_OOD
</code>
## Prepare for the model
<code>
adata.uns['fields'] = {}
</code>
<code>
adata.obs['perturbation'] = [x.split(' ')[0] for x in adata.obs['product_name']]
adata.uns['fields']['perturbation'] = 'perturbation'
</code>
<code>
adata.obs['control'] = [1 if x == 'Vehicle' else 0 for x in adata.obs['perturbation'].values]
adata.uns['fields']['control'] = 'control'
</code>
<code>
adata.obs['dose'] = adata.obs['dose'].astype(float) / np.max(adata.obs['dose'].astype(float))
adata.uns['fields']['dose'] = 'dose'
</code>
<code>
adata.uns['fields']['covariates'] = ['cell_type', 'replicate']
</code>
<code>
del adata.uns['log1p']
</code>
<code>
# split dataset
from sklearn.model_selection import train_test_split
adata.obs['split'] = 'NA'
adata.uns['fields']['split'] = 'split'
adata.obs.loc[
(adata.obs['cell_type'] == 'MCF7') & (adata.obs['product_name'].isin(drug_OOD)),
'split'
] = 'ood'
idx = np.where(adata.obs['split']=='NA')[0]
idx_train, idx_test = train_test_split(idx, test_size=0.2, random_state=42)
adata.obs.iloc[idx_train, adata.obs.columns.get_loc('split')] = 'train'
adata.obs.iloc[idx_test, adata.obs.columns.get_loc('split')] = 'test'
</code>
Rank DE genes (optional)
<code>
# this will be done in main script if it's not done here
cov_names = []
for cov in adata.uns['fields']['covariates']:
cov_names.append(np.array(adata.obs[cov].values))
cov_names = ["_".join(c) for c in zip(*cov_names)]
adata.obs["cov_name"] = cov_names
cov_pert_names = []
for i in range(len(adata)):
comb_name = (
f"{adata.obs['cov_name'].values[i]}"
f"_{adata.obs[adata.uns['fields']['perturbation']].values[i]}"
)
cov_pert_names.append(comb_name)
adata.obs["cov_pert_name"] = cov_pert_names
import warnings
from vci.dataset.gene_dataset import rank_genes_groups
with warnings.catch_warnings():
warnings.simplefilter("ignore")
rank_genes_groups(adata,
groupby="cov_pert_name",
reference="cov_name",
control_key="control"
)
</code>
<code>
adata.obs
</code>
<code>
adata.write('sciplex_prepped.h5ad')
</code>
|
{
"filename": "SciplexPrep_3.ipynb",
"repository": "yulun-rayn/variational-causal-inference",
"query": "transformed_from_existing",
"size": 69680,
"sha": ""
}
|
# 2025_dev_lab3.bu_3.ipynb
Repository: Tony-xy-Liu/AMB
This lab will guide you through generating functional annotations.
the input is an assembly, the outputs functions, ending with a metabolic model.
The written instructions should be sufficient to take you through the entire lab.
Key steps will be demoed at 3 checkpoints, but otherwise the lab is intended to be completed asynchronously.
Feel free to speed ahead, or take your time to explore a method that has captured your interest.
# Brief introduction to jupyter notebooks
<code>
import os, sys
import pandas as pd
from pathlib import Path
from local.ipc import Shell
from local.constants import WORKSPACE_ROOT
</code>
<code>
LIB = Path("./lib")
</code>
<code>
Shell("pwd -P")
</code>
# Open reading frame prediction
<code>
prodigal_sif = LIB/"prodigal.sif"
if not prodigal_sif.exists():
# other versions at
# https://quay.io/repository/biocontainers/prodigal?tab=tags
Shell(f"singularity pull {prodigal_sif} docker://quay.io/biocontainers/prodigal:2.6.3--h7b50bb2_10")
</code>
<code>
Shell(f"apptainer exec -B {DATA}:/data {prodigal_sif} prodigal -h")
</code>
<code>
# Shell(f"""\
# mkdir -p prodigal_out
# apptainer exec -B {DATA}:/data {prodigal_sif} \
# prodigal \
# -i /data/inputs/mg1655.fna \
# -a ./prodigal_out/mg1655.faa \
# -d ./prodigal_out/mg1655.fna \
# -f gff \
# -o ./prodigal_out/mg1655.gff \
# """)
</code>
# Functional annotation by sequence homology
Apptainer standardizes the install for each tool
so the syntax used to install prodigal will be repeated.
We can automate this process using a function
```python
diamond_sif = LIB/"diamond.sif"
if not diamond_sif.exists():
# https://quay.io/repository/biocontainers/diamond?tab=tags
Shell(f"singularity pull {diamond_sif} docker://quay.io/biocontainers/diamond:2.1.11--h5ca1c30_2")
```
<code>
def setup_container(name: str, address: str):
"""
Pulls an apptainer from the given @{address} and saves it to the lib directory as @{name}.sif
Returns path to the new container image
"""
container_path = LIB/name
if not container_path.exists():
Shell(f"singularity pull {container_path} {address}")
else:
print(f"container already exists at [{container_path}]")
return container_path
# https://quay.io/repository/biocontainers/diamond?tab=tags
diamond_sif = setup_container("diamond.sif", "docker://quay.io/biocontainers/diamond:2.1.11--h5ca1c30_2")
</code>
<code>
Shell(f"apptainer exec -B {DATA}:/data {diamond_sif} diamond --help")
</code>
<code>
DATA
</code>
<code>
transporter_db = DATA/"lib/transporter_classification_db"
if not transporter_db.exists():
Shell(f"""
mkdir -p {transporter_db}
cd {transporter_db}
wget -O tcdb.faa http://www.tcdb.org/public/tcdb
wget -O substrates.tsv https://www.tcdb.org/cgi-bin/substrates/getSubstrates.py
wget -O lineages.tsv https://www.tcdb.org/cgi-bin/substrates/listSuperfamilies.py
wget -O families.tsv https://www.tcdb.org/cgi-bin/projectv/public/families.py
""")
</code>
<code>
transporter_dmdb = transporter_db/"tcdb.dmnd"
if not transporter_dmdb.exists():
Shell(f"""
apptainer exec -B {DATA}:/data {diamond_sif} diamond makedb \
--threads 14 \
--in /data/lib/{transporter_db.name}/tcdb.faa \
--db /data/lib/{transporter_db.name}/tcdb
""")
</code>
<code>
Shell(f"apptainer exec -B {DATA}:/data {diamond_sif} diamond --help")
</code>
### bakta
<code>
# https://quay.io/repository/biocontainers/bakta?tab=tags
bakta_sif = setup_container("bakta.sif", "docker://quay.io/biocontainers/bakta:1.9.4--pyhdfd78af_0")
</code>
<code>
Shell(f"apptainer exec -B {DATA}:/data {bakta_sif} bakta --help")
</code>
<code>
Shell(f"apptainer exec -B {DATA}:/data {bakta_sif} bakta_db list")
</code>
<code>
# bakta_db = LIB/"bakta_db/db" # 2 hours
bakta_db = LIB/"bakta_db/db-light" # 30 mins
if not bakta_db.exists():
_type = "light" if "light" in str(bakta_db) else "full"
Shell(f"apptainer exec -B {DATA}:/data {bakta_sif} bakta_db download --output {bakta_db} --type {_type}")
</code>
<code>
# once it looks like its running, you can Ctrl+C to stop it
# then run the command in a console
# *important*: cd into the same directory
# 2 mins with mg1655
Shell(f"""
apptainer exec -B {DATA}:/data {bakta_sif} \
bakta --threads 14 --force \
--db {bakta_db} \
--output ./outputs/bakta.2 \
--regions /home/tony/workspace/projects/AMB_2025_dev/main/amb_lab3/scratch/mock/outputs/prodigal/mg1655.gff \
/home/tony/workspace/projects/AMB_2025_dev/main/amb_lab3/scratch/mock/inputs/mg1655.fna
""")
</code>
### Resistance Gene Identifier (RGI)
<code>
# https://quay.io/repository/biocontainers/rgi?tab=tags
rgi_sif = setup_container("rgi.sif", "docker://quay.io/biocontainers/rgi:6.0.4--pyh05cac1d_0")
</code>
<code>
# heatmap, collapse by categories
</code>
<code>
# Shell(f"apptainer exec -B {DATA}:/data {rgi_sif} rgi --help")
</code>
It looks like RGI accepts different commands (`usage: rgi <command> [<args>]`) and `main` seems to be the one that generates annotations.
<code>
# Shell(f"apptainer exec -B {DATA}:/data {rgi_sif} rgi main --help")
</code>
- RGI is a bit picky and doesn't provide any indication of progress or successful completion.
- Tackle the errors in the order in which they appear until you get a successful run, which should take no more than 1 min.
- Don't worry about the `SyntaxWarning` since it looks like a typo in the source code that doesn't otherwise impact the execution of the tool.
<details>
<summary>Hint 1</summary>
Consider the inputs and outputs
</details>
<details>
<summary>Hint 2</summary>
-i -o -t
</details>
<details>
<summary>Hint <code>Read-only file system</code></summary>
-i -o -t
</details>
<details>
<summary>Hint <code>File is not accessible</code></summary>
-i -o -t
</details>
<code>
# Shell(
# f"""
# mkdir -p ./outputs/rgi
# apptainer exec -B {DATA}:/data,./cache:/usr/local/lib/python3.12/site-packages/app/_db {rgi_sif} \
# rgi main --num_threads 8 -i ./outputs/bakta_light/mg1655.faa -t protein -o ./outputs/rgi/mg1655
# """)
</code>
### Kofamscan
<code>
# https://quay.io/repository/hallamlab/external_kofamscan?tab=tags
kofamscan_sif = setup_container("kofamscan.sif", "docker://quay.io/hallamlab/external_kofamscan:1.3.0")
</code>
<code>
Shell(f"apptainer exec -B {DATA}:/data {kofamscan_sif} kofamscan --help")
</code>
<code>
# https://www.genome.jp/ftp/tools/kofam_scan/README.md
</code>
<code>
Shell(f"apptainer exec -B {DATA}:/data {kofamscan_sif} exec_annotation --help")
</code>
<code>
# kegg mapper
</code>
<code>
# ftp://ftp.genome.jp/pub/db/kofam/ko_list.gz
# ftp://ftp.genome.jp/pub/db/kofam/profiles.tar.gz
</code>
<code>
# # about 12 mins
# Shell(f"""
# mkdir -p ./outputs/kofamscan
# apptainer exec -B {DATA}:/data {kofamscan_sif} \
# exec_annotation \
# --cpu=14 --format=detail --no-report-unannotated \
# --profile=/data/lib/kofamscan_db/profiles/prokaryote.hal --ko-list=/data/lib//kofamscan_db/ko_list \
# -o ./outputs/kofamscan/chicken.out \
# ./outputs/bakta.chicken/final.contigs.faa
# """)
</code>
<code>
from local.models.kegg_orthology import ParseKofamScanResults
model = ParseKofamScanResults(
Path("/home/tony/workspace/projects/AMB_2025/main/amb_lab3/scratch/mock/outputs/kofamscan/chicken.out"),
Path("/home/tony/workspace/projects/AMB_2025/data/lib/kofamscan_db/api_kegg.db"),
Path("/home/tony/workspace/projects/AMB_2025/data/lib/kofamscan_db/brite.json"),
)
</code>
# reference free
<code>
# https://quay.io/repository/hallamlab/external_deepfri?tab=tags
deepfri_sif = setup_container("deepfri.sif", "docker://quay.io/hallamlab/external_deepfri:1.0.1")
</code>
<code>
# # > 40 mins
# Shell(f"""
# apptainer exec -B {DATA}:/data,./:/ws {deepfri_sif} \
# deepfri \
# --ont mf bp ec \
# --fasta_fn /ws/outputs/bakta.chicken/final.contigs.faa \
# --output_fn_prefix /ws/outputs/deepfri
# """)
</code>
<code>
# https://quay.io/repository/hallamlab/external_proteinbert?tab=tags
proteinbert_sif = setup_container("proteinbert.sif", "docker://quay.io/hallamlab/external_proteinbert:2024.03.28")
</code>
<code>
# https://quay.io/repository/hallamlab/external_deepec?tab=tags
deepec_sif = setup_container("deepec.sif", "docker://quay.io/hallamlab/external_deepec:0.4.1")
</code>
<code>
Shell(f"""
apptainer exec -B {DATA}:/data,./:/ws {deepec_sif} \
deepfri \
--ont mf bp ec \
--fasta_fn /ws/outputs/bakta.chicken/final.contigs.faa \
--output_fn_prefix /ws/outputs/deepfri
""")
</code>
# Metabolic modelling
<code>
# https://github.com/ModelSEED/ModelSEEDpy
# https://github.com/jotech/gapseq?tab=readme-ov-file
</code>
<code>
# https://quay.io/repository/biocontainers/gapseq?tab=tags
gapseq_sif = setup_container("gapseq.sif", "docker://quay.io/biocontainers/gapseq:1.4.0--h9ee0642_1")
</code>
<code>
Shell(f"apptainer exec -B {DATA}:/data {gapseq_sif} gapseq -h")
</code>
<code>
Shell(f"""
apptainer exec -B {DATA}:/data {gapseq_sif} \
gapseq find --help
""")
</code>
<code>
Shell(f"""
mkdir -p ./outputs/gapseq/
rm ./outputs/gapseq/mg1655.*
apptainer exec -B {DATA}:/data {gapseq_sif} \
gapseq find -p glycolysis -l KEGG -K 14 -O -f ./outputs/gapseq/mg1655 ./outputs/bakta_light/mg1655.faa > ./outputs/gapseq/mg1655.out 2> ./outputs/gapseq/mg1655.err
""")
</code>
<code>
Shell(f"""
mkdir -p ./outputs/gapseq/
rm ./outputs/gapseq/mg1655.*
apptainer exec -B {DATA}:/data {gapseq_sif} \
gapseq find -p all -K 14 -O -f ./outputs/gapseq/mg1655 ./outputs/bakta_light/mg1655.faa > ./outputs/gapseq/mg1655.out 2> ./outputs/gapseq/mg1655.err
""")
</code>
<code>
Shell(f"""
apptainer exec -B {DATA}:/data {gapseq_sif} \
gapseq find-transport --help
""")
</code>
<code>
Shell(f"""
rm ./outputs/gapseq/mg1655.tr*
apptainer exec -B {DATA}:/data {gapseq_sif} \
gapseq find-transport -K 14 -f ./outputs/gapseq/mg1655 ./outputs/bakta_light/mg1655.faa > ./outputs/gapseq/mg1655.trout 2> ./outputs/gapseq/mg1655.trerr
""")
</code>
<code>
Shell(f"""
apptainer exec -B {DATA}:/data {gapseq_sif} \
gapseq draft --help
""")
</code>
It looks like there's no CLI for `draft`. We will have to use the documentation and make some educated guesses. https://gapseq.readthedocs.io/en/latest/usage/basics.html#draft-network-reconstruction-and-gapfilling
<code>
Shell(f"""
apptainer exec -B {DATA}:/data {gapseq_sif} \
gapseq draft \
-r ./outputs/gapseq/mg1655/mg1655-glycolysis-Reactions.tbl \
-p ./outputs/gapseq/mg1655/mg1655-glycolysis-Pathways.tbl \
-t ./outputs/gapseq/mg1655/mg1655-Transporter.tbl \
-c ./outputs/bakta_light/mg1655.faa \
-f ./outputs/gapseq/mg1655
""")
</code>
<code>
dfr = pd.read_csv(WORKSPACE_ROOT/"./outputs/gapseq/mg1655/mg1655-glycolysis-Reactions.tbl", sep="\t", comment="#")
print(dfr.shape, dfr.columns)
dfr.head(2)
</code>
<code>
dfw = pd.read_csv(WORKSPACE_ROOT/"./outputs/gapseq/mg1655/mg1655-glycolysis-Pathways.tbl", sep="\t", comment="#")
print(dfw.shape, dfw.columns)
dfw.head(2)
</code>
<code>
Shell(f"""
apptainer exec -B {DATA}:/data {gapseq_sif} \
gapseq fill --help
""")
</code>
<code>
# -c ./outputs/gapseq/mg1655/mg1655-rxnWeights.RDS \
# -g ./outputs/gapseq/mg1655/mg1655-rxnXgenes.RDS \
Shell(f"""
apptainer exec -B {DATA}:/data {gapseq_sif} \
gapseq fill --quick.gf \
-m ./outputs/gapseq/mg1655/mg1655-draft.RDS \
-n ./outputs/gapseq/mg1655/m9.csv \
--output.dir ./outputs/gapseq/mg1655
""")
</code>
|
{
"filename": "2025_dev_lab3.bu_3.ipynb",
"repository": "Tony-xy-Liu/AMB",
"query": "transformed_from_existing",
"size": 80421,
"sha": ""
}
|
# TSP_1.ipynb
Repository: ecervera/ga-nb
# The Travelling Salesperson Problem
This notebook has been adapted from [a Pyevolve example](http://pyevolve.sourceforge.net/0_6rc1/examples.html#example-12-the-travelling-salesman-problem-tsp).
The [travelling salesperson problem (TSP)](http://en.wikipedia.org/wiki/Travelling_salesman_problem) is an NP-hard problem in combinatorial optimization studied in operations research and theoretical computer science. Given a list of cities and their pairwise distances, the task is to find the shortest possible route that visits each city exactly once and returns to the origin city. It is a special case of the travelling purchaser problem.
[<img src="img/travelling_salesman_problem.jpg" align="right" width=360>](http://en.wikipedia.org/wiki/Travelling_salesman_problem)
The code below shows the use of Pyevolve to solve the TSP. Images of the intermediate and final solutions are stored in the 'tspimg' folder.
Your tasks are:
1. Create the 'tspimg' folder for storing the images.
2. Add the necessary statements for storing the results in a database named 'tsp.db' with identifier 'ex1'.
3. For the maximum grade: modify the code to solve the problem with the [ATT 48 dataset](att48.tsp), a set of 48 cities (US state capitals) from [TSPLIB](http://elib.zib.de/pub/mp-testdata/tsp/tsplib/tsplib.html). Store the results in a database named 'tsp_att48.db' with identifier 'ex1'. For your information, [the optimal cost is 10628](http://comopt.ifi.uni-heidelberg.de/software/TSPLIB95/STSP.html).
<code>
from pyevolve import G1DList
from pyevolve import GSimpleGA
from pyevolve import Crossovers
from pyevolve import Consts
import random
from math import sqrt
from PIL import Image, ImageDraw, ImageFont
</code>
<code>
cm = []
coords = []
CITIES = 30
WIDTH = 600
HEIGHT = 400
LAST_SCORE = -1
</code>
<code>
def cartesian_matrix(coords):
""" A distance matrix """
matrix={}
for i,(x1,y1) in enumerate(coords):
for j,(x2,y2) in enumerate(coords):
dx, dy = x1-x2, y1-y2
dist=sqrt(dx*dx + dy*dy)
matrix[i,j] = dist
return matrix
</code>
<code>
def tour_length(matrix, tour):
""" Returns the total length of the tour """
total = 0
t = tour.getInternalList()
for i in range(CITIES):
j = (i+1)%CITIES
total += matrix[t[i], t[j]]
return total
</code>
<code>
def write_tour_to_img(coords, tour, img_file):
""" The function to plot the graph """
padding=20
coords=[(x+padding,y+padding) for (x,y) in coords]
maxx,maxy=0,0
for x,y in coords:
maxx, maxy = max(x,maxx), max(y,maxy)
maxx+=padding
maxy+=padding
img=Image.new("RGB",(int(maxx),int(maxy)),color=(255,255,255))
font=ImageFont.load_default()
d=ImageDraw.Draw(img);
num_cities=len(tour)
for i in range(num_cities):
j=(i+1)%num_cities
city_i=tour[i]
city_j=tour[j]
x1,y1=coords[city_i]
x2,y2=coords[city_j]
d.line((int(x1),int(y1),int(x2),int(y2)),fill=(0,0,0))
d.text((int(x1)+7,int(y1)-5),str(i),font=font,fill=(32,32,32))
for x,y in coords:
x,y=int(x),int(y)
d.ellipse((x-5,y-5,x+5,y+5),outline=(0,0,0),fill=(196,196,196))
del d
img.save(img_file, "PNG")
print ("The plot was saved into the %s file." % (img_file,))
</code>
<code>
def G1DListTSPInitializator(genome, **args):
""" The initializator for the TSP """
lst = [i for i in range(genome.getListSize())]
random.shuffle(lst)
genome.setInternalList(lst)
</code>
<code>
def evolve_callback(ga_engine):
global LAST_SCORE
if ga_engine.getCurrentGeneration() % 100 == 0:
best = ga_engine.bestIndividual()
if LAST_SCORE != best.getRawScore():
write_tour_to_img( coords, best, "tspimg/tsp_result_%05d.png" % ga_engine.getCurrentGeneration())
LAST_SCORE = best.getRawScore()
return False
</code>
<code>
coords = [(random.randint(0, WIDTH), random.randint(0, HEIGHT))
for i in range(CITIES)]
cm = cartesian_matrix(coords)
</code>
<code>
genome = G1DList.G1DList(len(coords))
genome.evaluator.set(lambda chromosome: tour_length(cm, chromosome))
genome.crossover.set(Crossovers.G1DListCrossoverEdge)
genome.initializator.set(G1DListTSPInitializator)
</code>
<code>
ga = GSimpleGA.GSimpleGA(genome)
ga.setGenerations(2000)
ga.setMinimax(Consts.minimaxType["minimize"])
ga.setCrossoverRate(1.0)
ga.setMutationRate(0.02)
ga.setPopulationSize(80)
ga.stepCallback.set(evolve_callback)
</code>
<code>
ga.evolve(freq_stats=200)
best = ga.bestIndividual()
write_tour_to_img(coords, best, "tspimg/tsp_result.png")
</code>
You can check now the results by plotting some graphs of the evolution process in [this notebook](TSP_check.ipynb).
|
{
"filename": "TSP_1.ipynb",
"repository": "ecervera/ga-nb",
"query": "transformed_from_existing",
"size": 8202,
"sha": ""
}
|
# m3a_lfatayat_training.ipynb
Repository: NIJITSO/projet
<code>
import pandas as pd
import numpy as np
import ast
import seaborn as sns
import matplotlib.pyplot as plt
import os
import re
import joblib
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics import classification_report, confusion_matrix
from tensorflow.keras.models import Sequential, load_model
from tensorflow.keras.layers import Dense, Dropout, Input
from tensorflow.keras.utils import to_categorical
from tensorflow.keras.callbacks import EarlyStopping
from imblearn.over_sampling import RandomOverSampler
# --------------------- Load and preprocess ---------------------
df = pd.read_csv("data_zwina.csv")
df['keywords'] = df['keywords'].apply(lambda x: ' '.join(ast.literal_eval(x)) if isinstance(x, str) else '')
df['transcription'] = df['transcription'].astype(str)
df['description'] = df['description'].astype(str)
df['sample_name'] = df['sample_name'].astype(str)
df['medical_specialty'] = df['medical_specialty'].astype(str)
def preprocess_text(text):
text = text.lower()
text = re.sub(r'[^a-z0-9\s]', '', text)
return text
df['all_text'] = (
df['description'] + ' ' +
df['sample_name'] + ' ' +
df['keywords'] + ' ' +
df['transcription']
).apply(preprocess_text)
# --------------------- Vectorization ---------------------
vectorizer = TfidfVectorizer(max_features=5000, ngram_range=(1, 2))
X = vectorizer.fit_transform(df['all_text']).toarray()
# Save vectorizer AFTER fitting ✅
joblib.dump(vectorizer, "vectorizer.pkl")
# --------------------- Target encoding ---------------------
specialties = sorted(df['medical_specialty'].unique())
specialty_to_index = {name: i for i, name in enumerate(specialties)}
index_to_specialty = {i: name for name, i in specialty_to_index.items()}
# Save index-to-specialty map ✅
joblib.dump(index_to_specialty, "index_to_specialty.pkl")
y = df['medical_specialty'].map(specialty_to_index).values
y_cat = to_categorical(y, num_classes=len(specialties))
# --------------------- Resampling ---------------------
ros = RandomOverSampler(random_state=42)
X_res, y_res = ros.fit_resample(X, y_cat)
# --------------------- Train/test split ---------------------
X_train, X_test, y_train, y_test = train_test_split(
X_res, y_res, test_size=0.2, random_state=42, stratify=y_res
)
# --------------------- Build and train model ---------------------
model = Sequential([
Input(shape=(X_train.shape[1],)),
Dense(512, activation='relu'),
Dropout(0.4),
Dense(256, activation='relu'),
Dropout(0.3),
Dense(len(specialties), activation='softmax')
])
model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])
early_stop = EarlyStopping(monitor='val_loss', patience=3, restore_best_weights=True)
history = model.fit(
X_train, y_train,
epochs=10,
batch_size=64,
validation_split=0.2,
callbacks=[early_stop]
)
# Save model in both formats ✅
model.save("medical_specialty_model.keras")
model.save("medical_specialty_model.h5")
# --------------------- Evaluation ---------------------
loss, accuracy = model.evaluate(X_test, y_test)
print(f"\n✅ Test Accuracy: {accuracy:.4f}")
y_test_labels = np.argmax(y_test, axis=1)
y_pred_probs = model.predict(X_test)
y_pred_labels = np.argmax(y_pred_probs, axis=1)
report = classification_report(y_test_labels, y_pred_labels, target_names=specialties)
print("\n📊 Classification Report:\n")
print(report)
conf_mat = confusion_matrix(y_test_labels, y_pred_labels)
plt.figure(figsize=(12, 10))
sns.heatmap(conf_mat, xticklabels=specialties, yticklabels=specialties, annot=True, fmt="d", cmap="Blues")
plt.xlabel("Predicted")
plt.ylabel("Actual")
plt.title("🧾 Confusion Matrix")
plt.xticks(rotation=90)
plt.yticks(rotation=0)
plt.tight_layout()
plt.show()
# --------------------- Prediction Function ---------------------
def predict_specialty(text_input):
if not hasattr(predict_specialty, "model"):
predict_specialty.model = load_model("medical_specialty_model.keras")
if not hasattr(predict_specialty, "vectorizer"):
predict_specialty.vectorizer = joblib.load("vectorizer.pkl")
if not hasattr(predict_specialty, "index_to_specialty"):
predict_specialty.index_to_specialty = joblib.load("index_to_specialty.pkl")
processed_text = preprocess_text(text_input)
processed_vector = predict_specialty.vectorizer.transform([processed_text]).toarray()
prediction = predict_specialty.model.predict(processed_vector)
predicted_index = np.argmax(prediction)
predicted_specialty = predict_specialty.index_to_specialty[predicted_index]
confidence = prediction[0][predicted_index]
print(f"\n🩺 Predicted Specialty: {predicted_specialty}")
print(f"🔮 Confidence: {confidence:.4f}")
return predicted_specialty, confidence
# --------------------- Test Prediction ---------------------
sample_texts = [
"A 50-year-old female whose 51-year-old sister has a history of multiple colon polyps, which may slightly increase her risk for colon cancer in the future."
]
for i, text in enumerate(sample_texts, 1):
print(f"\n🔁 Prediction Test #{i}")
predict_specialty(text)
</code>
<code>
model.save("medical_specialty_model.h5")
joblib.dump(vectorizer, "vectorizer.pkl")
</code>
<code>
sample_texts = [
"A 50-year-old female whose 51-year-old sister has a history of multiple colon polyps, which may slightly increase her risk for colon cancer in the future."
]
for i, text in enumerate(sample_texts, 1):
print(f"\n🔁 Prediction Test #{i}")
predict_specialty(text)
</code>
<code>
model.save("medical_specialty_model.keras")
joblib.dump(vectorizer, "vectorizer.pkl")
joblib.dump(index_to_specialty, "index_to_specialty.pkl")
</code>
<code>
# Save trained model in modern Keras format
model.save("medical_specialty_model.keras")
# Save the trained TF-IDF vectorizer
joblib.dump(vectorizer, "vectorizer.pkl")
# Save the label index-to-specialty mapping
joblib.dump(index_to_specialty, "index_to_specialty.pkl")
</code>
|
{
"filename": "m3a_lfatayat_training.ipynb",
"repository": "NIJITSO/projet",
"query": "transformed_from_existing",
"size": 273243,
"sha": ""
}
|
# inhibition_simulation_MN_sim_PAIN.ipynb
Repository: FrancoisDernoncourt/Pain
<code>
from brian2 import *
import matplotlib.pyplot as plt
import numpy as np
from scipy.signal import butter, filtfilt, windows
from scipy.stats import linregress
%matplotlib inline
import scipy.io
from scipy.signal import resample, freqz
from scipy.interpolate import interp1d
from scipy.signal import remez, firwin, lfilter
# from factor_analyzer import FactorAnalyzer
from sklearn.decomposition import PCA
import random
import matplotlib.cm as cm
import os
import pandas as pd
from scipy.signal import csd, detrend
import pickle
from scipy.fft import fft, fftfreq
import sys
import scipy.ndimage
from brian2 import prefs
prefs.codegen.target = "numpy" # slower but I don't need to install many dependencies at least
plt.style.use('_classic_test_patch') # Plotting style
</code>
<code>
# Used to convert a string variable (used as input from the iteration script) to a boolean variable (when calling this script with specific inputs as strings)
def str_to_bool(s):
if s == "True":
return True
elif s == "False":
return False
else:
return False
</code>
<code>
### PARAMETERS ###
start_scope() # Re-initialize Brian
### GENERAL PARAMETERS ###########################################################
sim_name = "SIM_NAME" # str(os.getenv('simulation_name')) # < to use when iterating the simulation via another script # '___TEST_excit_distrib_small1_large1_relationship1'
sim_method = 'euler' # 'exact' 'euler' #'euler' is less precise but faster, and seems to be good enough
# TIME PARAMETERS
fsamp = 1000 # set your fsamp # This is NOT the dt at which the simulation runs. The simulation timesteps are 0.1ms in duration by default
window_beginning_ignore = 1 # in s
window_end_ignore = 1 # in s
ISI_threshold_for_discontinuity = 0.2 # in s ; motoneurons whose max(ISI)>threshold will be removed from analysis (so only continuous MNs are kept)
# ISI_threshold_for_RT = 0.5 # in s ; the recruitment threshold of motoneurons is calculated as the force (in % MVC) at the time of the first spike whose ISI is < threshold
# VOLTAGE THRESHOLDS OF ALL NEURONS
voltage_rest = 0 * mvolt # arbitrary ; 0 at rest
voltage_thresh = 10 * mvolt # arbitrary ; 10 for generating a spike
# REVERSAL/EQUILIBRIUM POTENTIAL OF LEAK CHANNELS, EXCITATORY CHANNELS, INHIBITORY CHANNELS
E_leak = voltage_rest
# Reversal potentials (relative to resting potential) from Elias & Kohn 2013 are 70 and -16 for xcitatory and inhibitory, respectively.
# They corrspond roughly to the reversal potentials of sodium channels (for E_excit) and chloride channels (for E_inhib).
# However, to make everything easier to work with, I tried to make the inhibition and excitation have roughly equivalent net effects on firing rates by making the reversal potential symmetric relative to half the firing threshold.
E_excit = ((voltage_thresh + voltage_rest)/2) + 20 * mvolt # Reversal potential for excitatory input
E_inhib = ((voltage_thresh + voltage_rest)/2) - 20 * mvolt # Reversal potential for inhibitory input
# NUMBER OF NEURONS SIMULATED
nb_motoneurons_full_pool = 300 # All motoneurons from the pool to be simulated
# Rough approximation of the number of motor units found in the tibialis anterior in humans (Motor Unit - Heckamn & Enoka 2012, ref 220 & 694)
# Twitch torque data also comes from the tibialis anterior, so this hopefully allows for a realistic simulation of the motor pool behavior for a given force level
# SIMULATING THE HD-EMG MU IDENTIFICATION PROCESS BY SUB-SAMPLING THE ACTIVE MUs
subsample_MUs_for_analysis = False # True
nb_of_MUs_to_subsample = 50
motor_unit_subsampling_probability_distribution = 'size' # 'size' #'uniform', 'size' # if 'uniform", every active MU will have the same probability to be selected for the analysis; if 'size", larger motor units will have a higher probability of being selected
bias_towards_larger_motor_neurons_temperature = 5.0 # only used if motor_unit_subsampling_probability_distribution=='size'.
# If infinity (inf), same as uniform distribution. If ~100, probability is approximately linearly scaled according to size. Exponential bias for values below (very strong bias for temperature = 1 for example). Error if 0
### TARGET SIMULATION
target_type = 'trapezoid' #'sinusoid' # 'plateau' #'trapezoid'
target_force_level = 30 # % of the max tetanic force of the simulated MNs
# if trapezoid:
ramp_duration = 5 # in s
plateau_duration = 60
analyzis_window = 'plateau'# 'all' 'plateau' # if 'all', the entire signal will be analyzed. If 'plateau', only the plateau section will be analyzed.
# if sinusoid:
target_force_sin_freq = 1 # used only if targt_type == 'sinusoid'
### SIMULATION DURATION
if target_type != 'trapezoid':
true_duration = 20
else:
true_duration = ramp_duration*2 + plateau_duration
duration_with_ignored_window = (true_duration+window_beginning_ignore+window_end_ignore)*second
### INPUT PARAMETERS #########################################################
# INHIBITORY INPUT
inhibition_weight_distribution = 'multimodal' #'multimodal' 'exponential' 'mixed_additive' 'mixed_multiplicative'
# 'multimodal' = MN will receive inhibition according the weights distributed according to a normal distribution. For example, for 50-50 distribution of either 0 or 1, use...
# # inhibition_multimodal_number_of_modes = 2
# # inhibition_multimodal_weights_distrib_means = [0, 1]
# # inhibition_multimodal_weights_distrib_stds = [0, 0]
inhibition_multimodal_number_of_modes = 1 # used only if 'inhibition_weight_distribution = multimodal'
inhibition_multimodal_weights_distrib_means = [0] # os.getenv('inhibition_multimodal_weights_distrib_means') # the number of element should be = to 'inhibition_multimodal_number_of_modes' # used only if 'inhibition_weight_distribution = multimodal'
# inhibition_multimodal_weights_distrib_means = eval(inhibition_multimodal_weights_distrib_means)
inhibition_multimodal_weights_distrib_stds = [0] # os.getenv('inhibition_multimodal_weights_distrib_stds') # the number of element should be = to 'inhibition_multimodal_number_of_modes' # used only if 'inhibition_weight_distribution = multimodal'
# inhibition_multimodal_weights_distrib_stds = eval(inhibition_multimodal_weights_distrib_stds)
inhibition_multimodal_weights_proportions = [1] # os.getenv('inhibition_multimodal_weights_proportions')
# inhibition_multimodal_weights_proportions = eval(inhibition_multimodal_weights_proportions)
# the number of elements should be equal to 'inhibition_multimodal_number_of_modes'. If the proportions add up to more than 1, an error will occur. If they sum to less than 1, the remaining MUs not in any group will be assigned a weight of 0. # used only if 'inhibition_weight_distribution = multimodal'
# 'exponential' = MNs receiving inhibition will receive increasing or decreasing amount of inhibition according to their sizes - Martinez Valdes J physiol 2020 model = https://physoc.onlinelibrary.wiley.com/doi/full/10.1113/JP279225
inhibition_exponential_exponent_weights = 2 # used only if 'inhibition_weight_distribution = exponential'
inhibition_exponential_constant_weights = 1.5 # used only if 'inhibition_weight_distribution = exponential'
inhibition_exponential_offset_weights = 0 # 0 # used only if 'inhibition_weight_distribution = exponential'
# # Distribution of inhibitory weights = constant * MN_size^(exponent) + offset
nb_inhibitory_input = 0
low_pass_filter_of_inhibitory_input = 5 #in hz
inhibitory_input_mean = 30*1e-2 # float(os.getenv('inhibitory_input_mean')) < to use when iterating the simulation via another script # in millisiemens
inhibitory_input_std = 1.5*1e-2 # float(os.getenv('inhibitory_input_std'))# in millisiemens
# Determine in which order the inhibition will happen in the script
keep_force_constant_despite_inhib = True # True # False # If True, the force will be optimized taking the inhibitory input into account, so the common excitatory input will necessarily increase
# If false, the inhibitory input will be delivered after the common excitatory input has been optimized to reach the target force. So the force level will be reduced, but the common excitatory input will remain the same as when there is no inhibition.
inhibitory_input_source = 'load_synthetic_input' # 'generate_synthetic_input' ; 'load_synthetic_input'
inhibitory_input_sourcefile = 'D:/THESE/Git_Scripts/Python_Scripts/motoneuron_simulation/Synthetic_signals.csv' # used only if inhibitory_input_source == 'load_synthetic_input'
# use the N first signals (the first N columns) as inhibitory inputs, with N = nb_inhibitory_input
# Use a .csv file. The number of samples in the csv file should be >= the number of samples in the simulation
inhibitory_input_sourcefile_fsamp = 1000 # 1000 if .csv # If not matching the simulation's fsamp, the loaded signal will be upsampled or downsampled to match the simulation's sampling rate
# EXCITATORY INPUT
excitatory_input_baseline = 150*1e-2 # float(os.getenv('excitatory_input_baseline')) < to use when iterating the simulation via another script # in millisiemens # From there (this baseline value), optimizer will try to optimize in order to reach the force target # tried to tune it manually to get it close to 30% MVC already for easier optimization
# The baseline input has the shape of the target force, but in a non-linear way. The max value corresponds to the selected baseline.
excitatory_input_std = 1.5*1e-2 # in millisiemens # Added to the "excitatory_input_baseline" learned by optimization
low_pass_filter_of_excitatory_input = 5 # in hz
excit_input_for_MVC = 600*1e-2 # in millisiemens. High input to reach a stable force output where all MNs are recruited, and with a mean firing rate of ~ 35 pps.
# This is similar to the MVC data reported in "Oya T, Riek S, Cresswell AG. Recruitment and rate coding organisation for soleus motor units across entire range of voluntary isometric plantar flexions. J Physiol 587: 4737-4748, 2009."
# ^ as found in Enoka Heckman Motor Unit 2012 (figure 14)
# For the selected contractile and electrophysiological properties, 350 MUs, and an excitatory input of 4 milliSiemens, we get a MVC torque of ~30N/m.
# The MU data (number of MUs in the pool and twitch force) comes from human tibialis anterior, so the resulting MVC torque is quite representative => see https://bmcmusculoskeletdisord.biomedcentral.com/articles/10.1186/1471-2474-14-104/tables/1 (Moraux et al 2013)
excitation_bias = 0.5
excitation_weight_smallest_MN = 1 - excitation_bias # float(os.getenv('excitation_weight_smallest_MN'))
excitation_weight_largest_MN = 1 + excitation_bias # float(os.getenv('excitation_weight_largest_MN'))
excitation_weight_relationship_from_smallest_to_largest = 1 # float(os.getenv('excitation_weight_relationship_from_smallest_to_largest')) # 2 # 1 for linear, < 1 for convex curve, > 1 for concave curve (or opposite if excitation_weight_smallest_MN > excitation_weight_largest_MN) # needs to be > 0
# https://www.desmos.com/calculator/pgb3pkffsf = check curve of distribution
# weight of 0.7 for smallest MN and weight of 1.3 for largest MN = ratio of excitatory input necessary for RT (max/min) of 8.4
# weight of 1 for smallest MN and weight of 1 for largest MN = ratio of excitatory input necessary for RT (max/min) of 13.6
# in the litterature around a 10-fold range is rapported for a given injected input (so no different weights across MNs) = Heckman & Enoka 2012 motor unit comprehensive physiology
# ratio of 0.5 for smallest and 1.5 for largest = ratio of excitatory input necessary for RT (max/min) slightly > 5
excitatory_input_source = 'load_synthetic_input' # 'generate_synthetic_input' ; 'load_synthetic_input'
excitatory_input_sourcefile = 'D:/THESE/Git_Scripts/Python_Scripts/motoneuron_simulation/Synthetic_signals.csv' # used only if excitatory_input_source == 'load_synthetic_input' or 'load_experimental_data'
# Use the last signal (last column) as common noise
# If 'load_gaussian_noise', use a .csv file. The number of samples in the csv file should be >= the number of samples in the simulation
# If 'load_experimental_data', use one .mat file or several .mat files. If several .mat files are selected, they will be concatenated. The number of samples in the (concatenated) file(s) should be >= the number of samples in the simulation
excitatory_input_sourcefile_fsamp = 1000 # 1000 if .csv ; 2048 if .mat # If not matching the simulation's fsamp, the loaded signal will be upsampled or downsampled to match the simulation's sampling rate
# INDEPENDENT NOISE
low_pass_filter_of_independent_noise = 50 # in hz
independent_noise_ratio_std = 2 # the value of noise in std (mean of 0)
# => if 2, independent input std corresponds to 2x inhibitory and excitatory input std, to get a ratio of common input = 1/3 of independnt input (Farina & Negro 2015)
### MOTONEURON PROPERTIES ##########################
# DISTRIBUTION OF MOTONEURON SIZES
min_soma_diameter = 50 # in micrometers, for smallest MN
max_soma_diameter = 100 # in micrometers, for largest MN
# Assuming that soma diameter from human motoneurons vary between 50 and 100 micrometers, loosely based on https://journals.physiology.org/doi/full/10.1152/physiol.00021.2018 (mean diameter of humans MN estimated to be ~60 micrometers)
# ^ "Scaling of motoneurons, From Mouse to Human" Manuel et al. Physiology (2018)
# Parameter to create an exponetially decreasing dsitribution curve, with larger motoneurons being less numerous than smaller motoneurons
# Somewhat fitting the curve in Principles of Neural Science 2021 edition, Enoka chapter on motor units, fig 31-3.A
size_distribution_exponent = 2
# between 0-1 => more large MN than small MN; 1 => uniform distribution (linear relationship between MN index and soma diameter); >1 => more small motoneurons than laarge MNs
# VIsualize distribution for different min and max soma diameters, and different exponents = https://www.desmos.com/calculator/zy3ywcz4tn
#### ELECTROPHYSIOLOGICAL MN PROPERTIES #####
# Electrophysiological properties calculated from Caillet et al 2022 https://elifesciences.org/articles/76489
# RESISTANCE - OHMS
# The resistance decreases the leak conductance and increases the weight of the excitatory and inhibitory input received by the motor neuron (=> higher resistance means higher sensitivity to input)
resistance_constant = 9.6*(10**5) # Caillet et al 2022 # in Ohms
resistance_exponent = 2.4*(-1) # Caillet et al 2022 # in Ohms
# https://www.desmos.com/calculator/pbs97zynff = visualize the curve for resistance (ohms) and input weights (between 0 and 1)
# Min resistance for smallest MN (50 micrometers) = ~80 ohms
# Max resistance for biggest MN (100 micrometers) = ~15 ohms
#### Input weight = normalized resistance, so that the input to the smallest MN is scaled by a factor of 1 #####
# Min input weight for smallest MN (50 micrometers) = 1
# Max input weight for biggest MN (100 micrometers) = ~0.19
# CONDUCTANCE - SIEMENS
membrane_conductance_scaling = 1 # membrane conductance is 1/resistance, multiplied by a scalar value (tuned by hand) to get realistic behavior of MN pool
# RHEOBASE - AMPERES
# The rheobase is modeled as an offset to the change in excitatory conductance caused by the exitatory input (clamped to 0 to avoid the excitatory input to have an hyperpolarization effect)
rheobase_constant = 9.0*(10**-4) # Caillet et al 2022 # in nanoAmps
rheobase_exponent = 2.5 # Caillet et al 2022 # in nanoAmps
rheobase_scaling = 100 # float(os.getenv('rheobase_scaling')) # < to use when iterating the simulation via another script # 100 # scalar value to multiply the rheobase by, tuned to get realistic behavior according to the arbitrary values used (such as voltage threshold and reversal potentials)
# CAPACITANCE - FARADS
capacitance_constant = 1.2 # Caillet et al 2022
capacitance_exponent = 1 # Caillet et al 2022
# AFTERHYPERPOLARIZATION DURATION & REFRACTORY PERIOD - SECONDS
# Refractory period (Caillet's paper gives equations for AHP duration but not for refractory period duration) #
# Since we are using a simplified model, we approximate the effect of the AHP by implementing an absolute refactory period that is a fraction of the true AHP duration.
AHP_duration_constant = 2.5 * (10**4) # Caillet et al 2022
AHP_duration_exponent = 1.5 * (-1) # Caillet et al 2022
refractory_period_as_AHP_fraction = 0.2 # float(os.getenv('refractory_period_as_AHP_fraction')) # Manually tuned
# Manuel et al. 2019 "Scaling of motor output, from Mouse to Humans"
# "Statistical methods employed at low firing rates indicate the AHP durations of low-threshold human motoneurons, presumably type S and perhaps some type FR, are ~125–140 ms."
# Herbert & Gandevia 1999 assume a 5ms (absolute?) refractory period
# Lateva et at 2001 = Absolute refractory period of 3ms in muscle fibers, and relative refractory period of 10ms
# University of Washington textbook of physiology = in a typical neuron, the absolute refractory period lasts a few ms and the relative period tens of ms
### CONTRACTILE PROPERTIES (TWITCH FORCE CAUSED BY MN SPIKES)
# Every firing of motor units will be convolved with a kernel
# => The kernel is a hanning window of duration which is 2x the time to peak force, and then the duration of the "down" portion of the twitch (when the force returns to baseline) is extended by 'multiplication_of_twitch_force_down_time'
# # Finally, the conduction delay is added before the start of the kernel
# TWITCH TORQUE - NEWTON/METERS
# Linear interpolation to get force produced (this is a gross simplification, as the distribution of MU properties is not linearly distributed at all)
# Data from Principles of Neural Science 2021 edition Motor Unit chapter Enoka # Figure 31-3, values for human tibialis anterior motor units => between 0-10 mN/m for smallest MUs, ~140mN/m for largest MUs
# Also found in Motor Unit Enoka Heckman 2012, from Van Cutsem M, Feiereisen P, Duchateau J, Hainaut K. Mechanical properties and behaviour of motor units in the tibialis anterior during voluntary contractions. Can J Appl Physiol 22: 585-597, 1997
twitch_force_range_small_MU = 5 # in milliNewton/meter
twitch_force_range_large_MU = 140 # in milliNewton/meter
# TIME TO PEAK FORCE - SECONDS
# time to peak force => linear relationship (this is a gross simplification, as the distribution of MU properties is not linearly distributed at all)
time_to_peak_twitch_force_range_small_MU = 0.08 *2 # in s # smallest MU
time_to_peak_twitch_force_range_large_MU = 0.02 *2 # in s # biggest MU
# *2 because later in the script the kernel is created with twice the tie to peak force
# Time to peak torque ranges from 20ms (0.02s) to 80-100ms (0.08-0.1s) for TA in the figure next to twitch force- Principles of Neural Science 2021 edition Motor Unit chapter Enoka # Figure 31-3, values for human tibialis anterior motor units
# Also found in Motor Unit Enoka Heckman 2012, from Van Cutsem M, Feiereisen P, Duchateau J, Hainaut K. Mechanical properties and behaviour of motor units in the tibialis anterior during voluntary contractions. Can J Appl Physiol 22: 585-597, 1997
# Doubling the value because this is time to peak force, and kernel duration is twice that
# # Also some data from Shoepe et al 2003 MSSE, shortening velocity of type I fiber (time to peak force small MU = 60ms) VS type IIa fiber (time to peak force fast MU = 25ms)
# # https://paulogentil.com/pdf/Functional%20adaptability%20of%20muscle%20fibers%20to%20long-term%20resistance%20exercise.pdf
multiplication_of_twitch_force_down_time = 4 # This is a gross simplificaion of how the twitch force return to baseline, but overall it seems to fit the behavior of motor units
# B. R. Botterman, G. A. Iwamoto, and W. J. Gonyea (1986) = force trace of single twitches from motor units
# Rositsa Raikova, Piotr Krutki, Jan Celichowski (2023) => Detailed model of motor units twitch force
# MOTOR UNIT TETANIC FORCE (max force which can be produced)
# Increase in force (with each additional twtich) towards max tetanic force follows a sigmoid curve, like in Fuglevand 1993 model (see section "Force nonlinearity")
twitch_tetanus_ratio_smallest_MN = 0.2 # 0.1 # smallest MU's twitch force has its peak at 20% of the MU's max tetanic force
twitch_tetanus_ratio_largest_MN = 0.3 # largest MU's twitch force has its peak at 30% of the MU's max tetanic force
# ^ data from the model of Nagamori et al. (2021): 'Force variability is mostly not motor noise: Theoretical implications for motor control', PLOS computational Biology (see 'twitch-tetanus ratio' section): https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1008707
# In the paper, the range spans from 0.07 to 0.53. Here, we manually tuned these values so that the minimum and maximum are 0.2 and 0.3
# 0.2 allows the smallest MU to reach 75% of its tetanus force around 15pps, 0.3 allow the largest MU to reach 75% of its tetanus force around 35pps
# With 0.2 and 0.3, we get around the same mean as reported in the paper (0.23)
# 0.23 correspond to the values reported in the cat by Brown & Loeb (https://link.springer.com/article/10.1023/A:1005687416896)
# 0.11 for all motor units in Fuglevand 1993 model
steepness_of_twitch_to_tetanus_sigmoid = 1 # for 2.8, when twitch_sum=1, actual_force~=100% of tetanus force. However, this means that the twitch force for a single twitch is 2.8 greater than the twitch force in the parameters
# ELECTROMECHANICAL DELAY - SECONDS ; AXONAL CONDUCTION VELOCITY - METERS/SECOND
# Electromechanical delay => inverse of actional conduction velocity
# Calculated from the axonal conduction velocity relationship reported in Caillet et al 2022
# ^ multiplying by two (assuming a 0.5m axon length => so correspond to the conduction speed from MN to muscle fiber)
axonal_conduction_velocity_constant = 4.0*2 # Caillet et al 2022
axonal_conduction_velocity_exponent = 0.7 # Caillet et al 2022
low_pass_filter_force = True # Can be turned to true if fast oscillation (high frequency components) in force output. This simulates the "dampening" effect of the musculoskeletal system (tendon for example)
low_pass_filter_of_force_cutoff = 10 # in hz
### PARAMETERS FOR TESTING - reduce numbers for faster simulations
COH_calc_max_iteration_nb_per_group_size = 1000 # 10 when just testing out simulations
# More iteration for smaller group sizes, because the value obtained is very dependent upon the exact neurons selected, especially when only a few MNs are used to create the CST
# ^ the number of iteration will be "COH_calc_max_iteration_nb_per_group_size / nb_of_MUs_in_CST"
max_num_optimization_iterations = 10 # 5 for faster simulation, it seems good enough in most cases (but better results with higher numbers) # 1 for testing
stop_optimizing_if_mean_error_is_below = 0.1 # in % of MVC # Ths allows to speed up the process by stopping the iterations when the learning process is basically over
consider_only_plateau_for_cost_optimization = True # this variable will be used only if target_type = 'trapezoid'. If True, the stopping of the optimization process will happen considering only the cost on the plateau and not the ramps
adam_learning_rate = 0.025
### EQUATIONS BEING RUN BY BRIAN2
LIF_equations = Equations('''
dv/dt = (-I_leak - I_excit - I_inhib) / C_m : volt (unless refractory)
I_leak = g_leak*(v - E_leak) : amp
I_excit = clip(synaptic_input_excit + I_th, -inf*nA, 0*nA) : amp
synaptic_input_excit = (ge*(v - E_excit)) : amp
I_inhib = gi*(v - E_inhib) : amp
ge = input_weight * input_excit(t,i) : siemens
gi = input_weight * input_inhib(t,i) : siemens
g_leak : siemens
C_m : farad
refractory_period : second
input_weight : 1
I_th : amp
''')
</code>
<code>
### CREATE NEW FOLDER
new_directory = sim_name
new_filename = 'parameters.txt'
# Create the directory if it doesn't exist
if not os.path.exists(new_directory):
os.makedirs(new_directory)
else:
directory_n = 0
while os.path.exists(new_directory):
directory_n = directory_n+1
new_directory = str(sim_name + "_iter_" + str(directory_n))
if not os.path.exists(new_directory):
os.makedirs(new_directory)
break
if directory_n > 99: # prevent infinite loop
break
save_file_path = os.path.join(new_directory, new_filename)
</code>
<code>
### SAVE PARAMETERS
# Write the variables to the file
with open(save_file_path, 'w') as file:
file.write(f"General parameters -----\n")
file.write(f" duration_with_ignored_window: {duration_with_ignored_window}\n")
file.write(f" nb_motoneurons_full_pool_per_pool: {nb_motoneurons_full_pool}\n")
file.write(f" ISI_threshold_for_discontinuity: {ISI_threshold_for_discontinuity}\n")
# file.write(f" ISI_threshold_for_RT: {ISI_threshold_for_RT}\n")
file.write(f"Sub-sampling of motor units (simulating the hdEMG motor unit identification process) -----\n")
file.write(f" subsample_MUs_for_analysis: {subsample_MUs_for_analysis}\n")
file.write(f" nb_of_MUs_to_subsample: {nb_of_MUs_to_subsample}\n")
file.write(f" motor_unit_subsampling_probability_distribution: {motor_unit_subsampling_probability_distribution}\n")
file.write(f" bias_towards_larger_motor_neurons_temperature: {bias_towards_larger_motor_neurons_temperature}\n")
file.write(f"Rest and threshold voltage + voltage equilibrium -----\n")
file.write(f" voltage_rest: {voltage_rest}\n")
file.write(f" voltage_thresh: {voltage_thresh}\n")
file.write(f" E_leak: {E_leak}\n")
file.write(f" E_excit: {E_excit}\n")
file.write(f" E_inhib: {E_inhib}\n")
file.write(f"\n")
file.write(f"Input parameters -----\n")
file.write(f" Common exitatory input -----\n")
file.write(f" excitatory_input_baseline (before optimized input learning): {excitatory_input_baseline}\n")
file.write(f" excitatory input for MVC: {excit_input_for_MVC}\n")
file.write(f" excitatory_input_std: {excitatory_input_std }\n")
file.write(f" excitatory_input_source: {excitatory_input_source }\n")
file.write(f" excitation_weight_smallest_MN: {excitation_weight_smallest_MN }\n")
file.write(f" excitation_weight_largest_MN: {excitation_weight_largest_MN }\n")
file.write(f" excitation_weight_relationship_from_smallest_to_largest: {excitation_weight_relationship_from_smallest_to_largest }\n")
file.write(f" excitatory_input_sourcefile: {excitatory_input_sourcefile }\n")
file.write(f" excitatory_input_sourcefile_fsamp: {excitatory_input_sourcefile_fsamp }\n")
file.write(f" Independent input (noise) -----\n")
file.write(f" low_pass_filter_of_independent_noise: {low_pass_filter_of_independent_noise}\n")
file.write(f" independent_noise_ratio_std: {independent_noise_ratio_std}\n")
file.write(f" Inhibition weights distribtuon -----\n")
file.write(f" inhibition_weight_distribution: {inhibition_weight_distribution}\n")
file.write(f" - If multimodal distribution of inhibitory input:\n")
file.write(f" inhibition_multimodal_number_of_modes: {inhibition_multimodal_number_of_modes}\n")
file.write(f" inhibition_multimodal_weights_distrib_means: {inhibition_multimodal_weights_distrib_means}\n")
file.write(f" inhibition_multimodal_weights_distrib_stds: {inhibition_multimodal_weights_distrib_stds}\n")
file.write(f" inhibition_multimodal_weights_proportions: {inhibition_multimodal_weights_proportions}\n")
file.write(f" - If exponential distribution of inhibitory input:\n")
file.write(f" inhibition_exponential_exponent_weights: {inhibition_exponential_exponent_weights}\n")
file.write(f" inhibition_exponential_constant_weights: {inhibition_exponential_constant_weights}\n")
file.write(f" inhibition_exponential_offset_weights: {inhibition_exponential_offset_weights}\n")
file.write(f" nb_inhibitory_input: {nb_inhibitory_input}\n")
file.write(f" low_pass_filter_of_inhibitory_input: {low_pass_filter_of_inhibitory_input}\n")
file.write(f" inhibitory_input_mean: {inhibitory_input_mean}\n")
file.write(f" inhibitory_input_std: {inhibitory_input_std}\n")
file.write(f" inhibitory_input_source: {inhibitory_input_source}\n")
file.write(f" inhibitory_input_sourcefile: {inhibitory_input_sourcefile}\n")
file.write(f" inhibitory_input_sourcefile_fsamp: {inhibitory_input_sourcefile_fsamp}\n")
file.write(f"\n")
file.write(f"Force target parameters -----\n")
file.write(f" target_type: {target_type}\n")
file.write(f" target_force_level: {target_force_level}% MVC\n")
file.write(f" If trapezoid:\n")
file.write(f" ramp_duration: {ramp_duration}\n")
file.write(f" plateau_duration: {plateau_duration}\n")
file.write(f" If NOT trapezoid:\n")
file.write(f" true_duration: {true_duration}\n")
file.write(f" If sisnusoid:\n")
file.write(f" target_force_sin_freq (used only if targt_type == 'sinusoid'): {target_force_sin_freq}\n")
file.write(f" low_pass_filter_force: {low_pass_filter_force}\n")
file.write(f" low_pass_filter_of_force_cutoff: {low_pass_filter_of_force_cutoff}\n")
file.write(f" keep_force_constant_despite_inhib: {keep_force_constant_despite_inhib}\n")
file.write(f"Motor neurons size -----\n")
file.write(f" min_soma_diameter: {min_soma_diameter}\n")
file.write(f" max_soma_diameter: {max_soma_diameter}\n")
file.write(f" size_distribution_exponent : {size_distribution_exponent}\n")
file.write(f"\n")
file.write(f"Twitch force properties - parameters -----\n")
file.write(f" twitch_force_range_small_MU: {twitch_force_range_small_MU}\n")
file.write(f" twitch_force_range_large_MU: {twitch_force_range_large_MU}\n")
file.write(f" twitch_tetanus_ratio_smallest_MN: {twitch_tetanus_ratio_smallest_MN}\n")
file.write(f" twitch_tetanus_ratio_smallest_MN: {twitch_tetanus_ratio_smallest_MN}\n")
file.write(f" steepness_of_twitch_to_tetanus_sigmoid: {steepness_of_twitch_to_tetanus_sigmoid}\n")
file.write(f" time_to_peak_twitch_force_range_small_MU: {time_to_peak_twitch_force_range_small_MU/2}\n") #/2 because actual time to peak torque is half the value
file.write(f" time_to_peak_twitch_force_range_large_MU: {time_to_peak_twitch_force_range_large_MU/2}\n") #/2 because actual time to peak torque is half the value
file.write(f" multiplication_of_twitch_force_down_time: {multiplication_of_twitch_force_down_time}\n")
file.write(f" axonal_conduction_velocity_constant: {axonal_conduction_velocity_constant}\n")
file.write(f" axonal_conduction_velocity_exponent: {axonal_conduction_velocity_exponent}\n")
file.write(f"\n")
file.write(f"Electrophysiological properties - parameters -----\n")
file.write(f" resistance_constant: {resistance_constant}\n")
file.write(f" resistance_exponent: {resistance_exponent}\n")
file.write(f" membrane_conductance_scaling: {membrane_conductance_scaling}\n")
file.write(f" rheobase_constant: {rheobase_constant}\n")
file.write(f" rheobase_exponent: {rheobase_exponent}\n")
file.write(f" rheobase_scaling: {rheobase_scaling}\n")
file.write(f" capacitance_constant: {capacitance_constant}\n")
file.write(f" capacitance_exponent: {capacitance_exponent}\n")
file.write(f" AHP_duration_constant: {AHP_duration_constant}\n")
file.write(f" AHP_duration_exponent: {AHP_duration_exponent}\n")
file.write(f" refractory_period_as_AHP_fraction: {refractory_period_as_AHP_fraction}\n")
</code>
<code>
# Define lerp (linear interpolation) function:
def lerp(a, b, t):
return a + t * (b - a)
####### Generate motoneurons
motoneuron_soma_diameters = np.zeros(nb_motoneurons_full_pool)
motoneuron_normalized_soma_diameters = np.zeros(nb_motoneurons_full_pool)
for mni in range(nb_motoneurons_full_pool):
motoneuron_soma_diameters[mni] = lerp(min_soma_diameter,max_soma_diameter, (mni/(nb_motoneurons_full_pool-1))**size_distribution_exponent )
motoneuron_normalized_soma_diameters[mni] =lerp(0, 1, (mni/(nb_motoneurons_full_pool-1))**size_distribution_exponent )
# Plot histogram of motoneuron sizes
plt.figure()
# Create the histogram
counts, bins, patches = plt.hist(motoneuron_soma_diameters, density=True)
# Multiply the counts by 100 to convert to percentage
counts_percentage = counts * 100
# Plot the histogram again with the adjusted counts
plt.clf() # Clear the current plot
plt.hist(motoneuron_soma_diameters, density=False, weights=np.ones_like(motoneuron_soma_diameters) * (100 / len(motoneuron_soma_diameters)),
edgecolor='white', color='gray', alpha=1)
plt.vlines(min_soma_diameter,plt.ylim()[0],plt.ylim()[1],color='C1', label='Min soma diameter', linewidth=2)
plt.vlines(max_soma_diameter,plt.ylim()[0],plt.ylim()[1],color='C3', label='Max soma diameter', linewidth=2)
plt.legend()
plt.xlabel("Motoneuron size (soma diameter in micrometer)")
plt.ylabel("Proportion (% of total nb of motoneurons)")
plt.title("Distribution of motor neuron sizes")
new_filename = f'MN_sizes_distribution.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
plt.figure()
# Create the histogram
counts, bins, patches = plt.hist(motoneuron_normalized_soma_diameters, density=True)
# Multiply the counts by 100 to convert to percentage
counts_percentage = counts * 100
# Plot the histogram again with the adjusted counts
plt.clf() # Clear the current plot
plt.hist(motoneuron_normalized_soma_diameters, density=False, weights=np.ones_like(motoneuron_normalized_soma_diameters) * (100 / len(motoneuron_normalized_soma_diameters)),
edgecolor='white', color='gray', alpha=0.5)
plt.vlines(0,plt.ylim()[0],plt.ylim()[1],color='C1', label='Min soma diameter', linewidth=2)
plt.vlines(1,plt.ylim()[0],plt.ylim()[1],color='C3', label='Max soma diameter', linewidth=2)
plt.legend()
plt.xlabel("Normalized motoneuron size")
plt.ylabel("Proportion (% of total nb of motoneurons)")
plt.figure()
plt.plot(motoneuron_soma_diameters, color='gray')
plt.xlabel("Motoneuron idx")
plt.ylabel("MN size (soma diameter in micrometers)")
plt.title(f"Size of MNs according to index \n Mean soma diameter = micrometers")
new_filename = f'MN_sizes_according_to_idx.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
# CONVOLVE TO GET FORCE
import math
def Twitch_summation_towards_tetanus(twitch_sum, tetanus_force):
actual_force = np.zeros(len(twitch_sum))
for sampli in range(len(twitch_sum)):
normalized_force = 1 - (2 / (1 + math.exp(2 * twitch_sum[sampli]))) # Sigmoid function
actual_force[sampli] = normalized_force * tetanus_force
return actual_force
def Convolve_to_get_force(binary_spike_trains, corresponding_MN_idx, absolute_or_normalized):
force_per_MU = []
force_total = np.zeros(binary_spike_trains.shape[1])
if ndim(binary_spike_trains) > 1:
if shape(binary_spike_trains)[0] != len(corresponding_MN_idx):
print("Error = the number of MU indices provided do not match with the number of rows in the binary matrix")
for mni in range(shape(binary_spike_trains)[0]):
# 'same' mode means the output length will be the same as the input length
temp_force = np.convolve(binary_spike_trains[mni,:], twitch_convolution_window[corresponding_MN_idx[mni]]*motoneurons_twitch_to_tetanus_ratios[corresponding_MN_idx[mni]], mode='same')
temp_force = Twitch_summation_towards_tetanus(temp_force, motoneurons_tetanus_forces[corresponding_MN_idx[mni]])
if absolute_or_normalized == 'normalized':
temp_force = (temp_force / max_MVC_force_absolute) * 100 # * 100 to get a percentage
force_per_MU.append(temp_force)
# ax.plot(force_total, color=colormap_temp(mni/(nb_motoneurons_full_pool-1)), alpha = 0.5)
force_total = force_total + temp_force
else:
# if len(corresponding_MN_idx) != 1: # len() doesn't work on a unique element
# print("Error = the number of MU indices provided do not match with the number of rows in the binary matrix")
temp_force = np.convolve(binary_spike_trains, twitch_convolution_window[corresponding_MN_idx[mni]]*motoneurons_twitch_to_tetanus_ratios[corresponding_MN_idx[mni]], mode='same')
temp_force = Twitch_summation_towards_tetanus(temp_force, motoneurons_tetanus_forces[corresponding_MN_idx[mni]])
if absolute_or_normalized == 'normalized':
temp_force = (temp_force / max_MVC_force_absolute) * 100 # * 100 to get a percentage
force_per_MU.append(temp_force)
force_total = force_total + temp_force
return force_total
</code>
<code>
motoneuron_resistances = np.zeros(nb_motoneurons_full_pool)
motoneuron_input_weights = np.zeros(nb_motoneurons_full_pool)
motoneuron_capacitances = np.zeros(nb_motoneurons_full_pool)
motoneurons_membrane_conductance = np.zeros(nb_motoneurons_full_pool)
motoneurons_AHP_durations = np.zeros(nb_motoneurons_full_pool)
motoneurons_refractory_periods = np.zeros(nb_motoneurons_full_pool)
motoneurons_rheobases = np.zeros(nb_motoneurons_full_pool)
for mni in range(nb_motoneurons_full_pool):
motoneuron_resistances[mni] = resistance_constant*(motoneuron_soma_diameters[mni]**resistance_exponent)
motoneuron_input_weights[mni] = motoneuron_resistances[mni] / motoneuron_resistances[0] # normalized value so that smallest MN has weight of 1
motoneuron_capacitances[mni] = capacitance_constant*(motoneuron_soma_diameters[mni]**capacitance_exponent)
motoneurons_membrane_conductance[mni] = 1/motoneuron_resistances[mni] * membrane_conductance_scaling
motoneurons_AHP_durations[mni] = AHP_duration_constant*(motoneuron_soma_diameters[mni]**AHP_duration_exponent)
motoneurons_refractory_periods[mni] = motoneurons_AHP_durations[mni] * refractory_period_as_AHP_fraction
motoneurons_rheobases[mni] = rheobase_constant*(motoneuron_soma_diameters[mni]**rheobase_exponent)
plt.figure(figsize=(7,5))
fig, ax1 = plt.subplots()
curve1, = ax1.plot(motoneuron_resistances, label = "Resistance (ohms)", color = 'C1', linewidth = 3)
ax2 = ax1.twinx()
curve2, = ax2.plot(motoneuron_input_weights, label = "Input weight (normalized input resistance)", color = 'C5', linewidth = 2, linestyle=":")
curves = [curve1, curve2]
labels = [curve.get_label() for curve in curves]
ax1.legend(curves, labels, loc='best')
ax1.tick_params(axis='y', labelcolor='C1')
ax1.set_ylabel("Resistance (ohms)", color ='C1')
ax2.tick_params(axis='y', labelcolor='C5')
ax2.set_ylabel("Normalized input resistance (weight between 0 and 1)", color ='C5')
plt.figure(figsize=(7,5))
plt.plot(motoneuron_capacitances, label = "Capacitance (microFarads)", color = 'C2')
plt.legend()
plt.xlabel("MN index")
plt.ylabel("Capacitance (Farads)")
plt.figure(figsize=(7,5))
plt.plot(motoneurons_membrane_conductance, label = "Membrane conductance (milliSiemens)", color = 'C3')
plt.legend()
plt.xlabel("MN index")
plt.ylabel("Membrane conductance (milliSiemens)")
plt.figure(figsize=(7,5))
plt.plot(motoneurons_refractory_periods, label = "Scaled refractory period (ms) as surrogate for the AHP duration", color = 'C6')
plt.legend()
plt.xlabel("MN index")
plt.ylabel("Refractory period (ms)")
plt.figure(figsize=(7,5))
plt.plot(motoneurons_rheobases, label = "Rheobase (nanoAmperes)", color = 'C7')
plt.legend()
plt.xlabel("MN index")
plt.ylabel("Rheobase (nanoAmperes)")
# Twitch force and duration and electromechanical delay (implemented as zeros before the kernel)
twitch_force_motoneurons = np.zeros(nb_motoneurons_full_pool)
motoneurons_twitch_to_tetanus_ratios = np.zeros(nb_motoneurons_full_pool)
motoneurons_tetanus_forces = np.zeros(nb_motoneurons_full_pool)
electromechanical_delay_motoneurons = np.zeros(nb_motoneurons_full_pool)
twitch_duration_motoneurons = np.zeros(nb_motoneurons_full_pool)
twitch_convolution_window = [None] * nb_motoneurons_full_pool
for mni in range(nb_motoneurons_full_pool):
twitch_force_motoneurons[mni] = lerp(twitch_force_range_small_MU,twitch_force_range_large_MU,motoneuron_normalized_soma_diameters[mni]) * steepness_of_twitch_to_tetanus_sigmoid
motoneurons_twitch_to_tetanus_ratios[mni] = lerp(twitch_tetanus_ratio_smallest_MN,
twitch_tetanus_ratio_largest_MN,
motoneuron_normalized_soma_diameters[mni])
motoneurons_tetanus_forces[mni] = twitch_force_motoneurons[mni] * (1/motoneurons_twitch_to_tetanus_ratios[mni])
electromechanical_delay_motoneurons[mni] = 1/(axonal_conduction_velocity_constant*(motoneuron_soma_diameters[mni]**(axonal_conduction_velocity_exponent)))
twitch_duration_motoneurons[mni] = lerp(time_to_peak_twitch_force_range_small_MU,time_to_peak_twitch_force_range_large_MU,motoneuron_normalized_soma_diameters[mni])
# twitch_convolution_window[mni] = ((fsamp * twitch_duration_motoneurons[mni] * (1/2))**-1) * windows.hann(round(fsamp * twitch_duration_motoneurons[mni])) * twitch_force_motoneurons[mni]
twitch_convolution_window[mni] = windows.hann(round(fsamp * twitch_duration_motoneurons[mni]))
# Extend the ramp down phase of the twitch so that it is five times the size of the ramp up phase (crude approximation based on Figure 1 of Raikova 2023. Full model explained in the paper https://www.sciencedirect.com/science/article/pii/S1050641123000330?via%3Dihub)
twitch_force_down_temp = twitch_convolution_window[mni][int(np.round(len(twitch_convolution_window[mni])/2)):len(twitch_convolution_window[mni])]
twitch_convolution_window[mni] = twitch_convolution_window[mni][:int(np.round(len(twitch_convolution_window[mni])/2))] # remove the down part (to be added in a few lines later)
original_indices = np.linspace(0, len(twitch_force_down_temp) - 1, num=len(twitch_force_down_temp)) # Create the indices of the original and new vectors
new_indices = np.linspace(0, len(twitch_force_down_temp) - 1, num = multiplication_of_twitch_force_down_time * len(twitch_force_down_temp) ) # Desired length of the new vector
twitch_force_down_stretched = np.interp(new_indices, original_indices, twitch_force_down_temp) # Perform linear interpolation
twitch_convolution_window[mni] = np.append(twitch_convolution_window[mni],twitch_force_down_stretched)
# insert zeros corresponding to electromechanical delay
delay_insamples = int(np.round(electromechanical_delay_motoneurons[mni]*fsamp))
twitch_convolution_window[mni] = np.append(np.zeros(delay_insamples), twitch_convolution_window[mni])
# Double the length of the vector with only zeros at the beginning, so that the convolution causes the force twitch to happen after each spike
twitch_convolution_window[mni] = np.append(np.zeros(len(twitch_convolution_window[mni])),twitch_convolution_window[mni])
plt.figure(figsize=(10,5))
plt.plot(twitch_force_motoneurons, color='C1', label = 'Twitch force')
plt.plot(motoneurons_tetanus_forces, color='red', label = 'Tetanus force')
plt.plot(motoneurons_tetanus_forces*0.75, color='red', label = '75% of tetanus force', linestyle=':')
plt.ylabel("Torque (milliNewton/meter)")
plt.xlabel("Motoneuron index (smallest MN is 0 ; largest MN is "+str(nb_motoneurons_full_pool-1)+")")
plt.legend()
plt.title("MUs contractile force properties")
new_filename = f'Contractile_properties_force.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
plt.figure(figsize=(10,5))
plt.plot(twitch_duration_motoneurons*1e3*(1/2), color='C2', label = 'Time to peak force') # *(1/2) because time is expressed in full kernel duration (without extension of the "down" part yet)
plt.plot(electromechanical_delay_motoneurons*1e3, color='C4', label = 'Electromechanical delay')
plt.ylabel("Time (ms)")
plt.legend()
plt.xlabel("Motoneuron index (smallest MN is 0 ; largest MN is "+str(nb_motoneurons_full_pool-1)+")")
plt.title("MUs contractile velocty properties")
new_filename = f'Contractile_properties_velocity.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
# Getting a smooth color blend from a given colormap
colormap_temp = cm.get_cmap('plasma')
# convolution windows
fig, ax = plt.subplots(figsize=(10,5))
for mni in range(nb_motoneurons_full_pool):
if mni == 0:
plt.plot(twitch_convolution_window[mni]*twitch_force_motoneurons[mni]*1e-3, color=colormap_temp(mni/(nb_motoneurons_full_pool-1)), alpha = 1, linewidth = 1.5, label = "smallest simulated motor unit")
elif mni == nb_motoneurons_full_pool-1:
plt.plot(twitch_convolution_window[mni]*twitch_force_motoneurons[mni]*1e-3, color=colormap_temp(mni/(nb_motoneurons_full_pool-1)), alpha = 1, linewidth = 1.5, label = "largest simulated motor unit")
else:
plt.plot(twitch_convolution_window[mni]*twitch_force_motoneurons[mni]*1e-3, color=colormap_temp(mni/(nb_motoneurons_full_pool-1)), alpha = 0.5, linewidth = 2)
# Multiplying by "twitch_force_motoneurons*1e-3" to show the torque produced by a unique twitch (without summation)
ax.set_xlabel("Time (ms)")
ax.set_ylabel("Torque (milliNewton/meter)")
plt.legend()
plt.title("Kernel for twitch torque convolution")
new_filename = f'Twitch_force_convolution_kernels.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
# Sanity check of the convolution/tetanus process to obtain force => fist create binary spike train of spike brusts with faster and faster frquencies to check the force response for a given frequency
frequency_of_bursts = 1 # burst every N seconds
duration_of_bursts = 0.5 # in seconds
max_pps_of_bursts = 100 # up to 50 pps
nb_of_bursts = np.round(max_pps_of_bursts*duration_of_bursts)
sanity_check_time = linspace(0, nb_of_bursts * frequency_of_bursts, int(np.round(fsamp * nb_of_bursts * frequency_of_bursts)))
firing_frequencies_MUtorque = []
firing_frequencies_MUtorque_corresponding_samples = []
binary_spike_test = np.zeros((1,len(sanity_check_time)))
for bursti in range(int(nb_of_bursts)):
firing_frequencies_MUtorque.append(bursti*(1/duration_of_bursts))
burst_temp = np.zeros(int(np.round(duration_of_bursts*fsamp)))
burst_temp[np.round(linspace(0,(duration_of_bursts*fsamp)-1,
bursti)).astype(int)] = 1
corresponding_samples = np.arange( int(np.round(frequency_of_bursts * bursti * fsamp)), int(np.round(frequency_of_bursts * bursti * fsamp))+len(burst_temp) )
firing_frequencies_MUtorque_corresponding_samples.append(corresponding_samples[0:len(burst_temp)])
binary_spike_test[0,corresponding_samples] = burst_temp
# binary_spike_test[0,corresponding_samples] = np.concatenate((burst_temp, np.zeros(len(burst_temp)) ))
</code>
<code>
# Sanity check of the convolution/tetanus process to obtain force => visualize the response for the spike train created in the previous cell
test_summed_force = Convolve_to_get_force(binary_spike_test,[0],'absolute')
plt.figure(figsize=(100,40))
plt.subplot(211)
plt.plot(sanity_check_time, test_summed_force,label="force produced (smallest MN)",color=colormap_temp(0), linewidth=2, alpha = 1)
plt.hlines(motoneurons_tetanus_forces[0], 0, np.max(sanity_check_time), label = "Tetanus torque (limit)", color = "black", linestyles=":", linewidth = 5)
plt.hlines(motoneurons_tetanus_forces[0]*0.75, 0, np.max(sanity_check_time), label = "75% of tetanus torque", color = "red", linestyles=":", linewidth = 5)
plt.xlabel("Time (s)")
plt.ylabel("Torque produced (milliNetwon/meter)")
plt.title("Testing the twitch force summation up to the tetanus for the smallest MU")
plt.subplot(212)
test_summed_force = Convolve_to_get_force(binary_spike_test,[nb_motoneurons_full_pool-1],'absolute')
plt.plot(sanity_check_time, test_summed_force,label="force produced (largest MN)",color=colormap_temp(0.5), linewidth=2, alpha = 1)
plt.hlines(motoneurons_tetanus_forces[nb_motoneurons_full_pool-1], 0, np.max(sanity_check_time), label = "Tetanus torque (limit)", color = "black", linestyles=":", linewidth = 5)
plt.hlines(motoneurons_tetanus_forces[nb_motoneurons_full_pool-1]*0.75, 0, np.max(sanity_check_time), label = "75% of tetanus torque", color = "red", linestyles=":", linewidth = 5)
plt.xlabel("Time (s)")
plt.ylabel("Torque produced (milliNetwon/meter)")
plt.title("Testing the twitch force summation up to the tetanus for the largest MU")
max_force_temp = np.ceil(np.max(test_summed_force))
plt.subplot(211)
plt.ylim(0-(max_force_temp/10),max_force_temp+(max_force_temp/2))
plt.plot(sanity_check_time, binary_spike_test[0,:]*max_force_temp,label="spike train", color=[0.3,0.5,1], alpha = 0.25, linewidth = 3)
plt.xlim(0, np.max(sanity_check_time))
plt.legend()
plt.subplot(212)
plt.ylim(0-(max_force_temp/10),max_force_temp+(max_force_temp/2))
plt.plot(sanity_check_time, binary_spike_test[0,:]*max_force_temp,label="spike train", color=[0.3,0.5,1], alpha = 0.25, linewidth = 3)
plt.xlim(0, np.max(sanity_check_time))
plt.legend()
new_filename = f'Torque_reponse_smallest_largest_MUs_spike_bursts.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
peak_force_per_MU_per_firing_freq = np.zeros((nb_motoneurons_full_pool,len(firing_frequencies_MUtorque)))
firing_freq_to_reach_tetanus = np.full(nb_motoneurons_full_pool, np.nan)
# mean_force_per_MU_per_firing_freq = np.zeros((nb_motoneurons_full_pool,len(firing_frequencies_MUtorque))) # over a 0.5s window when duration_of_bursts = 0.5
for mni in range(nb_motoneurons_full_pool):
test_summed_force = Convolve_to_get_force(binary_spike_test,[mni],'absolute')
for freqi in range(len(firing_frequencies_MUtorque)):
temp_samples = firing_frequencies_MUtorque_corresponding_samples[freqi]
if (freqi+1) < len(firing_frequencies_MUtorque): # only if not the last frequency (otherwise it will extend beyond the spike-train duration)
temp_samples = np.concatenate((temp_samples, np.arange(len(twitch_convolution_window[mni])) + np.max(temp_samples) )) # extending the considered samples to account for the last spike (meaningful for low firing frequencies)
peak_force_per_MU_per_firing_freq[mni,freqi] = np.max(test_summed_force[temp_samples])
if isnan(firing_freq_to_reach_tetanus[mni]) and (peak_force_per_MU_per_firing_freq[mni,freqi] > 0.75 * motoneurons_tetanus_forces[mni]) : # if reaching at least 75% of max force (tetanic force)
firing_freq_to_reach_tetanus[mni] = freqi
# mean_force_per_MU_per_firing_freq[mni,freqi] = np.mean(test_summed_force[temp_samples])*(1/duration_of_bursts) # normalize for the "burst window"
plt.figure(figsize=(15,10))
for mni in range(nb_motoneurons_full_pool):
plt.plot(firing_frequencies_MUtorque, peak_force_per_MU_per_firing_freq[mni,:], color=colormap_temp(mni/nb_motoneurons_full_pool), alpha = 0.5)
plt.xlabel("Firing frequency (pps)")
plt.ylabel("Torque (milliNewton/meter)")
plt.title("Peak torque of MU according to firing frequency (dark = small MU; light = large MU)")
new_filename = f'MU_percentage_of_peak_torque_relative_to_DR.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
# plt.figure(figsize=(20,15))
# for mni in range(nb_motoneurons_full_pool):
# plt.plot(firing_frequencies_MUtorque, mean_force_per_MU_per_firing_freq[mni,:], color=colormap_temp(mni/nb_motoneurons_full_pool), alpha = 0.5)
# plt.xlabel("Firing frequency (pps)")
# plt.ylabel("Torque (milliNewton/meter)")
# plt.title(f"Mean torque produced over 1s (Y) at a given firing frequency (X) \n (dark = small MU; light = large MU)")
plt.figure(figsize=(10,10))
plt.subplot(211)
plt.plot(firing_freq_to_reach_tetanus, color='red', linewidth=3)
# plt.hlines(np.nanmean(firing_freq_to_reach_tetanus), xlim()[0], xlim()[1], color='red', linewidth = 5, alpha = 0.5, label = "mean")
plt.xlabel("MN index")
plt.ylabel("Firing frequency (pps)")
plt.ylim(0,max_pps_of_bursts)
plt.title("Firing frequency necessary to reach 75% of tetanus torque ~ MN index")
plt.subplot(212)
plt.plot(motoneuron_soma_diameters, firing_freq_to_reach_tetanus, color=[0.8,0,0], linewidth=3)
# plt.hlines(np.nanmean(firing_freq_to_reach_tetanus), xlim()[0], xlim()[1], color=[0.8,0,0], linewidth = 5, alpha = 0.5, label = "mean")
plt.xlabel("Soma diameter")
plt.ylabel("Firing frequency (pps)")
plt.ylim(0,max_pps_of_bursts)
plt.title("Firing frequency necessary to reach 75% of tetanus torque ~ MN size")
new_filename = f'MU_DR_needed_to_reach_peak_torque.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
# Define a low-pass filter
def butter_lowpass(cutoff, fs, order=5):
nyquist = 0.5 * fs
normal_cutoff = cutoff / nyquist
b, a = butter(order, normal_cutoff, btype='low', analog=False)
return b, a
def lowpass_filter(data, cutoff, fs, order=5):
b, a = butter_lowpass(cutoff, fs, order=order)
y = filtfilt(b, a, data)
return y
# Low-pass filter artifact removal
duration_to_remove = 1/window_beginning_ignore # in second
Wind_s = duration_to_remove * 2
HanningW = windows.hann(round(fsamp * Wind_s))
HanningW = HanningW[:int(np.round(len(HanningW)/2))]
nb_samples_artifact_removal_window = len(HanningW)
</code>
<code>
# When rounding 2.7 for example, 70% to get 3 and 30% chance to get 2
def probabilistic_round(number):
lower = int(number) # The lower integer
upper = lower + 1 # The upper integer
decimal_part = number - lower
return upper if random.random() < decimal_part else lower
</code>
<code>
# GET SPIKE TRAINS AND BINARY SPIKE TRAINS
def Get_binary_spike_trains(spike_monitor, sim_duration):
# Define time bins
time_bins = np.arange(0, int(np.round((sim_duration*fsamp)))*second) * ms
# Retrieve spikes and get binary spike trains
spike_trains = []
for mni in range(nb_motoneurons_full_pool):
spike_trains.append(spike_monitor.spike_trains()[mni])
# Initialize the binary spike train array
binary_spike_trains = {}
binary_spike_trains = np.zeros((nb_motoneurons_full_pool, len(time_bins)))
# Convert spike times to binary spike train
for neuron_idx in range(nb_motoneurons_full_pool):
spikes = spike_trains[neuron_idx]
spike_indices = np.searchsorted(time_bins, spikes)
binary_spike_trains[neuron_idx, spike_indices-1] = 1 #-1 because of offset due to 0-indexing
return spike_trains, binary_spike_trains
</code>
<code>
# excitation_weight_smallest_MN = 0.5
# excitation_weight_largest_MN = 2
# excitation_weight_relationship_from_smallest_to_largest = 3
# # ^ just for testing purpose
# CREATE EXCITATORY INPUT WEIGHTS
# Necessary at this stage already, because of the MVC simulation
motoneurons_excitation_weights = np.ones(nb_motoneurons_full_pool)
for mni in range(nb_motoneurons_full_pool):
motoneurons_excitation_weights[mni] = lerp(excitation_weight_smallest_MN, excitation_weight_largest_MN, motoneuron_normalized_soma_diameters[mni]**excitation_weight_relationship_from_smallest_to_largest)
plt.figure()
plt.plot(motoneurons_excitation_weights, color=[1,0.7,0.2], linewidth = 5)
plt.xlabel("MN index")
plt.ylabel("Weight of excitatory input")
plt.title("Weight of excitatory input ~ MN index")
new_filename = f'MWeights_excitatory_input_curve_MN_idx.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
plt.figure()
plt.plot(motoneuron_soma_diameters, motoneurons_excitation_weights, color=[1,0.7,0.2], linewidth = 5)
plt.xlabel("MN soma diameter")
plt.ylabel("Weight of excitatory input")
plt.title("Weight of excitatory input ~ MN size")
new_filename = f'MWeights_excitatory_input_curve_MN_size.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
# Get max tetanic force of the simulated pool => by sending a very very high input to all MNs and recording the resulting force
# The force comes from the estimated peak torque values of individual motor units in the human tibialis anterior, so the low MVC values are not so unrealistic
# excit_input_for_MVC = 1000*1e-2 # for testing purposes
MVC_plateau_duration = 5 # in second
ramp_of_MVC_duration = 10 # in second
total_MVC_durarion = (MVC_plateau_duration + ramp_of_MVC_duration + 1) # +1 because one second of no input to get rid of artifacts
# Initialize inputs for MVC
# Generate empty input first (necessary to get rid of artifacts)
excit_input_with_ramp_MVC = np.zeros(int(np.round(1*fsamp)))
excit_input_with_ramp_MVC = np.append(excit_input_with_ramp_MVC,
linspace(0,1,int(np.round(ramp_of_MVC_duration*fsamp))) * excit_input_for_MVC) # Generate ramp first
excit_input_with_ramp_MVC = np.append(excit_input_with_ramp_MVC,
np.ones(int(np.round(MVC_plateau_duration*fsamp))) * excit_input_for_MVC) # then plateau
excit_input_per_MN = np.ones(( nb_motoneurons_full_pool, int(np.round(total_MVC_durarion*fsamp)) )) * excit_input_with_ramp_MVC
input_inhib_per_MN = np.zeros(( nb_motoneurons_full_pool,int(np.round(total_MVC_durarion*fsamp)) ))
for mni in range(nb_motoneurons_full_pool):
excit_input_per_MN[mni,:] = excit_input_per_MN[mni,:] * motoneurons_excitation_weights[mni]
excit_input_per_MN[mni,:] = np.clip(excit_input_per_MN[mni,:], a_min=0, a_max=None)# Clamp to 0 to avoid negative conductance
excit_input_per_MN = np.transpose(excit_input_per_MN)
input_excit = TimedArray(excit_input_per_MN * msiemens, dt=1*ms)
input_inhib_per_MN = np.transpose(input_inhib_per_MN)
input_inhib = TimedArray(input_inhib_per_MN * msiemens, dt=1*ms)
plt.figure()
plt.plot(excit_input_with_ramp_MVC, label="MVC excitatory input (ramp and plateau)", color = "C1")
plt.plot(input_inhib_per_MN[:,0], label="MVC inhibitory input (ramp and plateau)", color = "C4")
plt.legend()
plt.title("Input for MVC")
</code>
<code>
# RUN MVC SIMULATION
# Reset simulation
start_scope() # Re-initialize Brian
eqs_motoneuron = LIF_equations
# Groups of neurons
motoneurons = NeuronGroup(nb_motoneurons_full_pool, eqs_motoneuron,
threshold='v>voltage_thresh',
reset='v=voltage_rest',
refractory='refractory_period',
method=sim_method)
# Initialize values
motoneurons.v = voltage_rest + (rand(nb_motoneurons_full_pool)*voltage_thresh) # in mV # uniform distribution between 0 and voltage threshold => prevents early synchronization
motoneurons.g_leak = motoneurons_membrane_conductance * msiemens # in milisiemens
motoneurons.C_m = motoneuron_capacitances * ufarad # in microfarads
motoneurons.I_th = motoneurons_rheobases * rheobase_scaling * nA # in nanoAmperes
motoneurons.refractory_period = motoneurons_refractory_periods * ms # in milliseconds
motoneurons.input_weight = motoneuron_input_weights # dimensionless unit
# Monitors
monitor_state_motoneurons = StateMonitor(motoneurons, variables=True, record=True)
monitor_spikes_motoneurons = SpikeMonitor(motoneurons, record=True)
# Run simulation
run(total_MVC_durarion * second)
</code>
<code>
# GET RESULTS OF MVC SIMULATION
# Get force only on the plateau
MVC_sim_samples = np.arange(0,int(np.round(total_MVC_durarion * fsamp)))
MVC_sim_samples = MVC_sim_samples[(ramp_of_MVC_duration+1)*fsamp:] # remove samples from the ramp
# Get spike trains
spike_trains, binary_spike_trains = Get_binary_spike_trains(monitor_spikes_motoneurons, total_MVC_durarion)
# Get force
MVC_force_absolute = Convolve_to_get_force(binary_spike_trains,np.arange(nb_motoneurons_full_pool),'absolute')
max_MVC_force_absolute = np.mean(MVC_force_absolute[MVC_sim_samples]) # only during the plateau
# Force per MU
force_total = zeros(len(binary_spike_trains[0,:]))
fig, ax = plt.subplots(figsize=(10, 5))
# Getting a smooth color blend from a given colormap
colormap_temp = cm.get_cmap('plasma')
for mni in range(nb_motoneurons_full_pool):
temp_force = Convolve_to_get_force(np.reshape(binary_spike_trains[mni,:], (1, len(binary_spike_trains[mni,:]))), [mni], 'absolute') / 1000
force_total += temp_force
ax.plot(force_total, color=colormap_temp(mni/(nb_motoneurons_full_pool-1)), alpha = 0.5)
ax.plot(force_total, color='black', alpha = 1, linewidth = 2)
plt.title(f"Reconstructed torque (convolving spike train with twitch torque kernel)")
plt.ylabel("Torque (Newton/meter)")
plt.xlabel("Time (ms)")
plt.vlines((ramp_of_MVC_duration+1)*fsamp, ymin=0, ymax = ylim()[1], color="red", linewidth=2, alpha = 0.5)
new_filename = f'Max_force_for_simulated_MNs.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
</code>
<code>
# Get recruitment of MUs (clean version without noise, and for all MNs since the MVC ramp recruits all of them)
fig, ax1 = plt.subplots(figsize=(10,7))
ax1.plot(excit_input_with_ramp_MVC[:int(np.round((ramp_of_MVC_duration+1)*fsamp))], label="MVC excitatory input (ramp)", color = "C1",
linewidth = 3)
MN_recruitment_thresholds_by_excitatory_input_clean_MVC = np.full(nb_motoneurons_full_pool, np.nan) # in milliSiemens of excitatory input signal
MN_recruitment_thresholds_by_excitatory_input_ratio = np.full(nb_motoneurons_full_pool, np.nan) # in milliSiemens of excitatory input signal
for mni in range(nb_motoneurons_full_pool):
# RT as input during the median time of the first 3 firings
temp_RT = (spike_trains[mni]/second)*fsamp
temp_RT = np.median(temp_RT[0:2])
if np.isnan(temp_RT)==False:
MN_recruitment_thresholds_by_excitatory_input_clean_MVC[mni] = excit_input_with_ramp_MVC[int(np.round(temp_RT))]
MN_recruitment_thresholds_by_excitatory_input_ratio[mni] = MN_recruitment_thresholds_by_excitatory_input_clean_MVC[mni] / MN_recruitment_thresholds_by_excitatory_input_clean_MVC[0]
ax1.scatter(int(np.round(temp_RT)), MN_recruitment_thresholds_by_excitatory_input_clean_MVC[mni],
color=colormap_temp(mni/(nb_motoneurons_full_pool-1)), alpha = 0.5, s=50)
ax2 = ax1.twinx()
ax2.plot((force_total[:int(np.round((ramp_of_MVC_duration+1)*fsamp))]/(max_MVC_force_absolute/1000))*100, label="Normalized force", color = "black", linewidth=2, alpha=0.5)
ax1.set_xlabel("Time (ms)")
ax1.set_ylabel("Excitatory input (mS)", color = "C1")
ax1.spines['left'].set_color('C1') # Y-axis on the left
ax1.tick_params(axis='y', colors='C1') # Tick labels for y-axis
ax2.set_ylabel("Force (% MVC)", color = "grey")
ax2.spines['right'].set_color('grey') # Y-axis on the left
ax2.tick_params(axis='y', colors='grey') # Tick labels for y-axis
ax1.legend(loc='upper left')
ax2.legend(loc='upper right')
plt.title("Recruitment of MUs during the MVC ramp (dark dots = small MU; light dots = large MU)")
new_filename = f'MVC_ramp_recruitment_thresholds.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
plt.figure()
plt.plot(motoneuron_soma_diameters, MN_recruitment_thresholds_by_excitatory_input_ratio)
plt.xlabel("Soma diameter (micrometers)")
plt.ylabel("Recruitment threshold relative to smallest MN")
plt.title(f"Recruitment threshold ratio in escitatory input (relative to smallest MN) \n Chosen excitatoy input bias = {excitation_weight_smallest_MN} for smallest MN; {excitation_weight_largest_MN} for largest MN \n RT range = {np.round(np.max(MN_recruitment_thresholds_by_excitatory_input_ratio)*100)/100}-fold")
new_filename = f'MVC_ramp_recruitment_thresholds_ratio.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
</code>
<code>
### Get discharge characteristics of motoneurons on the MVC plateau
spike_trains = [array[array >= (ramp_of_MVC_duration+1)*second] for array in spike_trains] # removing spikes that belong to the ramp in "spike_trains"
mean_firing_rate = {}
std_firing_rate = {}
fig, axs = plt.subplots()
firing_rates = []
highest_ISIs = []
for mni in range(nb_motoneurons_full_pool):
if len(spike_trains[mni]) <= 1:
highest_ISIs.append(MVC_plateau_duration*fsamp)
else:
highest_ISIs.append(max(diff(spike_trains[mni])))
# firing_rate_temp = len(spike_trains[mni]) / MVC_plateau_duration
if len(spike_trains[mni]) > 1:
firing_rate_temp = 1/np.mean(np.diff(spike_trains[mni]))
else:
firing_rate_temp = 0
firing_rates.append(firing_rate_temp)
# Convert to a numpy array for easier calculations
firing_rates = np.array(firing_rates)
print(f"Number of silent MUs during the MVC = {np.sum(firing_rates<1)} (there should be none)")
# Calculate mean and standard deviation of the firing rates
mean_firing_rate = np.mean(firing_rates)
std_firing_rate = np.std(firing_rates)
# Motoneurons' firing rates results
axs.hist(firing_rates, edgecolor='white', alpha=1, color='C1')
axs.axvline(x = mean_firing_rate, linestyle='--', linewidth=2, label='Mean firing rate', color='red')
axs.set_xlabel("Mean firing rate (pps)")
axs.set_ylabel("Motoneuron count count")
plt.tight_layout(rect=[0,0,1,0.96])
plt.suptitle("Histogram of motoneurons' firing rate - high excitatory input simulation (for MVC estimation)")
new_filename = f'MVC_Hist_Discharge_rates_MVC.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
plt.figure(figsize=(10,10))
plt.subplot(211)
plt.plot(firing_rates, color = 'C1', linewidth = 3)
plt.ylabel("Firing rate (pps)")
plt.xlabel("MN index")
plt.title("Firing rates during MVC ~ MN index")
plt.subplot(212)
plt.plot(motoneuron_soma_diameters, firing_rates, color = 'C1', linewidth = 3)
plt.ylabel("Firing rate (pps)")
plt.xlabel("soma size (micrometers)")
plt.title("Firing rates during MVC ~ soma diameter")
new_filename = f'MVC_Discharge_rates.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
print(f'Estimated MVC (Newton/meter) = {np.round(max_MVC_force_absolute)/1000} (for {nb_motoneurons_full_pool} motor units)') # /1000 so that it is expressed in N/m
print(f'MVC (Newton/meter) would be {(np.round(max_MVC_force_absolute)/1000) / (nb_motoneurons_full_pool/300)} with 300 motor units')
target_force_level_absolute_value = max_MVC_force_absolute * (target_force_level/100) # /100 because 'target_force_level' is expressed as a percentage
print(f'Target torque level (Netwon/meter) = {np.round(target_force_level_absolute_value)/1000}') # /1000 so that it is expressed in N/m
</code>
<code>
# Define force target
if (target_type == 'plateau'):
print("Plateau force target")
target_force = ones(int(duration_with_ignored_window * fsamp))*target_force_level
elif (target_type == 'sinusoid'):
print("Sinusoidal force target")
target_force = ones(int(duration_with_ignored_window * fsamp))*target_force_level
sinusoids_temp = np.linspace(0, duration_with_ignored_window, int(duration_with_ignored_window*fsamp), endpoint=False)
sinusoids_temp = sinusoids_temp / second
sinusoids_temp = (target_force * 0.25) * np.sin(2 * np.pi * target_force_sin_freq * sinusoids_temp)
target_force = target_force + sinusoids_temp
elif (target_type == 'trapezoid'):
print("Trapezoidal force target")
target_force = np.zeros(int(window_beginning_ignore*fsamp))
ramp_up = linspace(0,target_force_level,ramp_duration*fsamp)
target_force = np.append(target_force,ramp_up)
plateau = ones(int(plateau_duration * fsamp))*target_force_level
target_force = np.append(target_force,plateau)
ramp_down = linspace(target_force_level,0,ramp_duration*fsamp)
target_force = np.append(target_force,ramp_down)
target_force = np.append(target_force,np.zeros(int(window_end_ignore*fsamp)))
else:
print("Please select a valid target type type")
sys.exit()
plt.plot(target_force, color='black', alpha = 0.5, linewidth = 2)
plt.xlabel("Time (ms)")
plt.ylabel("Target force (% MVC)")
plt.title("Target force")
new_filename = f'Target_force.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
time = linspace(0,duration_with_ignored_window/second,int(duration_with_ignored_window/second*fsamp))
</code>
<code>
# INHIBITORY INPUT
Wind_s = 1/low_pass_filter_of_inhibitory_input # hanning window duration.
HanningW = 2 / round(fsamp * Wind_s) * windows.hann(round(fsamp * Wind_s)) # unitary area
# Create inhibitory input
inhib_input = {}
if inhibitory_input_source == 'generate_synthetic_input':
for inhibiti in range(nb_inhibitory_input):
inhib_input[inhibiti] = []
temp_inhib = np.random.normal(0, 1, int(duration_with_ignored_window * fsamp))
temp_inhib[int(duration_with_ignored_window * fsamp)-int(np.round(window_beginning_ignore/1)):int(duration_with_ignored_window * fsamp)] = 0
temp_inhib[0:int(np.round(window_beginning_ignore/1))] = 0
temp_inhib = lowpass_filter(temp_inhib, low_pass_filter_of_inhibitory_input, fsamp)
# temp_inhib = filtfilt(HanningW, 1, temp_inhib * fsamp)
temp_inhib = temp_inhib - np.mean(temp_inhib)
temp_inhib = temp_inhib / np.std(temp_inhib)
temp_inhib = temp_inhib * inhibitory_input_std
temp_inhib = temp_inhib + inhibitory_input_mean
inhib_input[inhibiti].append(temp_inhib)
elif inhibitory_input_source == 'load_synthetic_input':
synthetic_signals_dataframe = pd.read_csv(inhibitory_input_sourcefile)
if inhibitory_input_sourcefile_fsamp != fsamp:
from scipy.interpolate import interp1d
# Calculate the time array for the original signal
loaded_signal_time = np.arange(len(synthetic_signals_dataframe)) / inhibitory_input_sourcefile_fsamp
# Calculate the number of samples in the resampled signal
number_of_samples = int(len(synthetic_signals_dataframe) * fsamp / inhibitory_input_sourcefile_fsamp)
# Calculate the time array for the resampled signal
resampled_time = np.linspace(loaded_signal_time[0], loaded_signal_time[-1], number_of_samples)
for inhibiti in range(nb_inhibitory_input):
inhib_input[inhibiti] = []
temp_inhib = synthetic_signals_dataframe[f'{inhibiti}'].values
if inhibitory_input_sourcefile_fsamp != fsamp:
# Create an interpolation function
resample_loaded_signal_function = interp1d(loaded_signal_time, temp_inhib, kind='linear')
temp_inhib = resample_loaded_signal_function(resampled_time)
temp_inhib = temp_inhib[:len(time)] # cut the signal for it to be the right size
temp_inhib = temp_inhib * inhibitory_input_std
temp_inhib = temp_inhib + inhibitory_input_mean
inhib_input[inhibiti].append(temp_inhib)
plt.figure()
for inhibiti in range(nb_inhibitory_input):
plt.plot(np.transpose(inhib_input[inhibiti]), alpha = 0.5, label=f'Inhibitory input #{inhibiti}', color = 'C4')
plt.xlabel("Time (ms)")
plt.ylabel("Inhibitory input(s)")
plt.legend()
new_filename = f'Inhibitory_input_signal.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
inhibition_weight_distribution = 'mixed_additive' #'multimodal' 'exponential' 'mixed_additive' 'mixed_multiplicative'
# 'multimodal' = MN will receive inhibition according the weights distributed according to a normal distribution. For example, for 50-50 distribution of either 0 or 1, use...
# # inhibition_multimodal_number_of_modes = 2
# # inhibition_multimodal_weights_distrib_means = [0, 1]
# # inhibition_multimodal_weights_distrib_stds = [0, 0]
inhibition_multimodal_number_of_modes = 1 # used only if 'inhibition_weight_distribution = multimodal'
inhibition_multimodal_weights_distrib_means = [0] # os.getenv('inhibition_multimodal_weights_distrib_means') # [0.1, 0.9] # the number of element should be = to 'inhibition_multimodal_number_of_modes' # used only if 'inhibition_weight_distribution = multimodal'
# inhibition_multimodal_weights_distrib_means = eval(inhibition_multimodal_weights_distrib_means)
inhibition_multimodal_weights_distrib_stds = [0.1] # os.getenv('inhibition_multimodal_weights_distrib_stds') # [0.02, 0.02] # the number of element should be = to 'inhibition_multimodal_number_of_modes' # used only if 'inhibition_weight_distribution = multimodal'
# inhibition_multimodal_weights_distrib_stds = eval(inhibition_multimodal_weights_distrib_stds)
inhibition_multimodal_weights_proportions = [1] # os.getenv('inhibition_multimodal_weights_proportions') # [0.5, 0.5]
# inhibition_multimodal_weights_proportions = eval(inhibition_multimodal_weights_proportions)
# the number of elements should be equal to 'inhibition_multimodal_number_of_modes'. If the proportions add up to more than 1, an error will occur. If they sum to less than 1, the remaining MUs not in any group will be assigned a weight of 0. # used only if 'inhibition_weight_distribution = multimodal'
# 'exponential' = MNs receiving inhibition will receive increasing or decreasing amount of inhibition according to their sizes - Martinez Valdes J physiol 2020 model = https://physoc.onlinelibrary.wiley.com/doi/full/10.1113/JP279225
inhibition_exponential_exponent_weights = 2 # used only if 'inhibition_weight_distribution = exponential'
inhibition_exponential_constant_weights = 1.5 # used only if 'inhibition_weight_distribution = exponential'
inhibition_exponential_offset_weights = 0 # 0 # used only if 'inhibition_weight_distribution = exponential'
# # Distribution of inhibitory weights = constant * MN_size^(exponent) + offset
</code>
<code>
# Distribute inhibitory input
if (inhibition_weight_distribution != 'multimodal') and (inhibition_weight_distribution != 'exponential') and (inhibition_weight_distribution != 'mixed_additive') and (inhibition_weight_distribution != 'mixed_multiplicative'):
print("Please select a valid inhibition distribution type ('multimodal' or 'exponential' or 'mixed_additive' or 'mixed_multiplicative')")
sys.exit()
if (inhibition_weight_distribution == 'exponential' or inhibition_weight_distribution == 'mixed_additive' or inhibition_weight_distribution == 'mixed_multiplicative'):
MN_inhibition_weights_curve = np.zeros(nb_motoneurons_full_pool)
for mni in range(nb_motoneurons_full_pool):
MN_inhibition_weights_curve[mni] = inhibition_exponential_constant_weights * (1-motoneuron_normalized_soma_diameters[mni])**inhibition_exponential_exponent_weights + inhibition_exponential_offset_weights
plt.figure()
plt.plot(MN_inhibition_weights_curve, color='C4')
plt.ylim([-0.1,np.max([1.1,inhibition_exponential_constant_weights+0.1])])
plt.ylabel("Inhibitory input weights (only the size-inhibition relationship)")
plt.xlabel("MN index")
plt.title("Inhibition distribution curve according to index (only the size-inhibition relationship)")
plt.figure()
plt.scatter(motoneuron_normalized_soma_diameters,MN_inhibition_weights_curve, color='C4')
plt.ylim([-0.1,np.max([1.1,inhibition_exponential_constant_weights+0.1])])
plt.xlabel("Normalized MN size")
plt.ylabel('Inhibitory input weights (only the size-inhibition relationship)')
plt.title("Distribution of inhibitory inputs according to SIZE (only the size-inhibition relationship)")
new_filename = f'Inhibitory_input_distrib_relative_to_size.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
MN_inhibition_weights = {}
if nb_inhibitory_input == 0:
MN_inhibition_weights[0] = np.zeros(nb_motoneurons_full_pool)
else:
for inhibiti in range(nb_inhibitory_input):
MN_inhibition_weights[inhibiti] = np.zeros(nb_motoneurons_full_pool)
if (inhibition_weight_distribution != 'exponential'):
# separate motor neurons into groups of equivalent sizes
# # Calculate the number of elements in each group
group_sizes = [int(nb_motoneurons_full_pool * p) for p in inhibition_multimodal_weights_proportions]
if sum(inhibition_multimodal_weights_proportions) > 1 :
print("The proportion of motor neurons in the multimodal distribution sums to more than 1. Please ensure that the sum of proportions is <= 1")
sys.exit()
# # Shuffle the indices of the motor neurons
MN_idx_shuffled_temp = np.arange(nb_motoneurons_full_pool)
np.random.shuffle(MN_idx_shuffled_temp)
# # Split the indices into the groups
MN_idx_in_groups_for_multimodal_distribution = []
start_idx = 0
for size in group_sizes:
MN_idx_in_groups_for_multimodal_distribution.append(MN_idx_shuffled_temp[start_idx:start_idx + size])
start_idx += size
for groupi in range(inhibition_multimodal_number_of_modes):
# # generate the normal distrbution for group 'groupi'
inhib_weights_for_current_group_temp = np.random.normal(inhibition_multimodal_weights_distrib_means[groupi],
inhibition_multimodal_weights_distrib_stds[groupi],
len(MN_idx_in_groups_for_multimodal_distribution[groupi]))
iter_mn_temp = 0
for mni in MN_idx_in_groups_for_multimodal_distribution[groupi]:
MN_inhibition_weights[inhibiti][mni] = inhib_weights_for_current_group_temp[iter_mn_temp]
iter_mn_temp += 1
# # If there are remaining elements, assign a weight of zero
remaining_elements = MN_idx_shuffled_temp[start_idx:]
MN_inhibition_weights[inhibiti][remaining_elements] = 0
if (inhibition_weight_distribution == 'mixed_additive') or (inhibition_weight_distribution == 'mixed_multiplicative'):
for mni in range(nb_motoneurons_full_pool):
if (inhibition_weight_distribution == 'mixed_additive'):
MN_inhibition_weights[inhibiti][mni] = MN_inhibition_weights_curve[mni]+MN_inhibition_weights[inhibiti][mni]
elif (inhibition_weight_distribution == 'mixed_multiplicative'):
MN_inhibition_weights[inhibiti][mni] = MN_inhibition_weights_curve[mni]*MN_inhibition_weights[inhibiti][mni]
else: # (inhibition_weight_distribution == 'exponential'):
for mni in range(nb_motoneurons_full_pool):
MN_inhibition_weights[inhibiti][mni] = MN_inhibition_weights_curve[mni]
# Clamp all weights between 0 and +inf to avoid negative weights
for inhibiti in range(nb_inhibitory_input):
for mni in range(nb_motoneurons_full_pool):
MN_inhibition_weights[inhibiti][mni] = np.clip(MN_inhibition_weights[inhibiti][mni],0,inf)
fig, ax = plt.subplots(1,1, figsize=(15,4))
x_plot_mns = range(nb_motoneurons_full_pool)
bottom_barplot = np.zeros(nb_motoneurons_full_pool)
for inhibiti in range(nb_inhibitory_input):
vstack_inhibi_temp = MN_inhibition_weights[inhibiti]
ax.bar(x_plot_mns,
vstack_inhibi_temp,
bottom = bottom_barplot,
label = f"inhibitory input #{inhibiti}", color="C4")
bottom_barplot += vstack_inhibi_temp
ax.set_xlabel('Motoneurons')
ax.set_ylabel('Inhibitory input weights')
# ax.set_ylim(0,1)
plt.suptitle("Distribution of inhibitory inputs according to INDICES (final)")
new_filename = f'Inhibitory_input_distrib_relative_to_indices.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
plt.figure(figsize=(15,4))
for inhibiti in range(nb_inhibitory_input):
# plt.hist(MN_inhibition_weights[inhibiti], edgecolor='white', density=True, color='C4', alpha = 0.5)
plt.hist(MN_inhibition_weights[inhibiti], edgecolor='white', color='C4', alpha = 0.5)
plt.title('Histogram of weights for inhibitory input distribution (final)')
plt.xlabel('Inhibition weight')
plt.ylabel('Count (nb of motor neurons)')
# plt.xlim(-0.1,1.1)
new_filename = f'Inhibitory_input_weight_distrib_hist.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
inhibitory_input_power_integral_0_5_hz = []
for inhibiti in range(nb_inhibitory_input):
inhib_input_temp = np.copy(np.squeeze(np.array(inhib_input[inhibiti])))
# remove artifacts of low pass filter
inhib_input_temp[0:int(window_beginning_ignore*fsamp)] = inhibitory_input_mean
inhib_input_temp[len(time)-int(window_end_ignore*fsamp):len(time)] = inhibitory_input_mean
# Normalize
inhib_input_temp = ((inhib_input_temp - np.mean(inhib_input_temp)) / np.std(inhib_input_temp)) * inhibitory_input_std
N = len(inhib_input_temp)
yf = fft(inhib_input_temp)
xf = fftfreq(N, 1 / fsamp)
power_spectrum_temp = (np.abs(yf[:N//2])**2) / N
inhibitory_input_power_integral = np.sum(power_spectrum_temp)
if inhibiti == 0:
idx_corresponding_to_5hz = int(np.round((N/fsamp)*5))
power_spectrum_temp_0_5hz = power_spectrum_temp[:idx_corresponding_to_5hz]
inhibitory_input_power_integral_0_5_hz.append(np.sum(power_spectrum_temp_0_5hz))
plt.plot(xf[:N//2], power_spectrum_temp, color = 'C4', alpha = 1/nb_inhibitory_input)
inhibitory_input_power_integral_0_5_hz_mean = np.mean(inhibitory_input_power_integral_0_5_hz)
plt.xlabel("Frequency (Hz)")
plt.ylabel("Power")
plt.title("Power spectrum of the inhibitory inputs")
plt.xlim([0,20])
new_filename = f'Signal_inhibitory_input_power_spectrum.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
# Generate independent input to MNs ######################
# Low-pass filter artifact removal
duration_to_remove = 1 # in second
Wind_s = duration_to_remove * 2
HanningW = windows.hann(round(fsamp * Wind_s))
HanningW = HanningW[:int(np.round(len(HanningW)/2))]
nb_samples_artifact_removal_window = len(HanningW)
plt.figure(figsize=(10,20))
independent_noise_excit = np.zeros((nb_motoneurons_full_pool,len(time)))
independent_noise_inhib = np.zeros((nb_motoneurons_full_pool,len(time)))
independent_excit_noise_power_integral_0_5_hz = []
independent_inhib_noise_power_integral_0_5_hz = []
for mni in range(nb_motoneurons_full_pool):
# Excitatory noise
independent_noise_excit[mni,:] = randn(len(time)) # random number with mean = 0 and std = 1
independent_noise_excit[mni,:] = lowpass_filter(independent_noise_excit[mni,:], 50, fsamp, 3) # 3rd order 50-hz low pass filter
# start of signal - artifact removal
independent_noise_excit[mni,0:nb_samples_artifact_removal_window] = independent_noise_excit[mni,0:nb_samples_artifact_removal_window] * HanningW
# end of signal - artifact removal
independent_noise_excit[mni,len(independent_noise_excit[mni,:])-nb_samples_artifact_removal_window:len(independent_noise_excit[mni,:])] = independent_noise_excit[mni,len(independent_noise_excit[mni,:])-nb_samples_artifact_removal_window:len(independent_noise_excit[mni,:])] * HanningW[::-1]
# Scale the input appropriately
independent_noise_excit[mni,:] = (((independent_noise_excit[mni,:] - np.mean(independent_noise_excit[mni,:])) / np.std(independent_noise_excit[mni,:])) *
independent_noise_ratio_std * excitatory_input_std * motoneurons_excitation_weights[mni]) + 0 # noise has a mean of 0 # * motoneurons_excitation_weights because scaling according to the weight of excitation received
plt.subplot(411)
plt.plot(time,independent_noise_excit[mni,:],color='C1',alpha=0.02)
if mni == 0:
plt.title("Independent noise of excitatory input (conductance fluctuation)")
plt.xlabel("Time (s)")
plt.ylabel("Noise amplitude (conductance fluctuations, in milisiemens)")
# Power spectrum - excitatory input noise
N = len(independent_noise_excit[mni,:])
yf = fft(independent_noise_excit[mni,:])
xf = fftfreq(N, 1 / fsamp)
power_spectrum_temp = (np.abs(yf[:N//2])**2) / N
independent_noise_power_integral = np.sum(power_spectrum_temp)
if mni == 0:
idx_corresponding_to_5hz = int(np.round((N/fsamp)*5))
power_spectrum_temp_0_5hz = power_spectrum_temp[:idx_corresponding_to_5hz]
independent_excit_noise_power_integral_0_5_hz.append(np.sum(power_spectrum_temp_0_5hz))
plt.subplot(412)
plt.plot(xf[:N//2], power_spectrum_temp, color = 'C1', alpha = 0.02)
if mni == 0:
plt.xlabel("Frequency (Hz)")
plt.ylabel("Power")
plt.title("Power spectrum of the excitatory noise inputs")
plt.xlim([0,80])
# Inhibitory noise
independent_noise_inhib[mni,:] = randn(len(time)) # random number with mean = 0 and std = 1
independent_noise_inhib[mni,:] = lowpass_filter(independent_noise_inhib[mni,:], 50, fsamp, 3) # 3rd order 50-hz low pass filter
# start of signal - artifact removal
independent_noise_inhib[mni,0:nb_samples_artifact_removal_window] = independent_noise_inhib[mni,0:nb_samples_artifact_removal_window] * HanningW
# end of signal - artifact removal
independent_noise_inhib[mni,len(independent_noise_inhib[mni,:])-nb_samples_artifact_removal_window:len(independent_noise_inhib[mni,:])] = independent_noise_inhib[mni,len(independent_noise_inhib[mni,:])-nb_samples_artifact_removal_window:len(independent_noise_inhib[mni,:])] * HanningW[::-1]
# Scale the input appropriately
independent_noise_inhib[mni,:] = (((independent_noise_inhib[mni,:] - np.mean(independent_noise_inhib[mni,:])) / np.std(independent_noise_inhib[mni,:])) *
independent_noise_ratio_std * inhibitory_input_std * MN_inhibition_weights[0][mni]) + 0 # noise has a mean of 0 # * MN_inhibition_weights[0] because scaling according to the weight of inhibition received (considering only the 1st inhibitory input)
plt.subplot(413)
plt.plot(time,independent_noise_inhib[mni],color='C4',alpha=0.02)
if mni == 0:
plt.title("Independent noise of inhibitory input (conductance fluctuation)")
plt.xlabel("Time (s)")
plt.ylabel("Noise amplitude (conductance fluctuations, in milisiemens)")
# Power spectrum - inhibitory input noise
N = len(independent_noise_inhib[mni,:])
yf = fft(independent_noise_inhib[mni,:])
xf = fftfreq(N, 1 / fsamp)
power_spectrum_temp = (np.abs(yf[:N//2])**2) / N
independent_noise_power_integral = np.sum(power_spectrum_temp)
# if mni == 0:
# idx_corresponding_to_5hz = int(np.round((N/fsamp)*5))
power_spectrum_temp_0_5hz = power_spectrum_temp[:idx_corresponding_to_5hz]
independent_inhib_noise_power_integral_0_5_hz.append(np.sum(power_spectrum_temp_0_5hz))
plt.subplot(414)
plt.plot(xf[:N//2], power_spectrum_temp, color = 'C4', alpha = 0.02)
if mni == 0:
plt.xlabel("Frequency (Hz)")
plt.ylabel("Power")
plt.title("Power spectrum of the inhibitory noise inputs")
plt.xlim([0,80])
independent_excit_noise_power_integral_0_5_hz_mean = np.mean(independent_excit_noise_power_integral_0_5_hz)
independent_inhib_noise_power_integral_0_5_hz_mean = np.mean(independent_inhib_noise_power_integral_0_5_hz)
new_filename = f'Signal_independent_noise.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
# plt.show() # Takes a long time to display everything as a plot (tens of seconds)
</code>
<code>
# Create the inhibitory signals - including the independent noise of each MN (the inhibitory signal is not learned/otpimized, so it can be created now)
# input_excit and input_inhib should be clamped to 0 to avoid negative conductances
inhib_input_per_MN = np.zeros((nb_motoneurons_full_pool,len(time)))
for mni in range(nb_motoneurons_full_pool):
inhib_input_per_MN[mni,:] = np.zeros(len(time))
for inhibiti in range(nb_inhibitory_input):
inhib_input_per_MN[mni,:] += inhib_input[inhibiti][0] * MN_inhibition_weights[inhibiti][mni]
inhib_input_per_MN[mni,:] += independent_noise_inhib[mni,:] * MN_inhibition_weights[inhibiti][mni]
inhib_input_per_MN[mni,:] = np.clip(inhib_input_per_MN[mni,:], a_min=0, a_max=None)# Clamp to 0 to avoid negative conductance
inhib_input_per_MN = np.transpose(inhib_input_per_MN)
plt.figure(figsize=(10,5))
for mni in range(nb_motoneurons_full_pool):
plt.plot(time,inhib_input_per_MN[:,mni],color='C4',alpha=0.05)
plt.xlabel("Time (s)")
plt.ylabel("Inhibition amplitude")
plt.title("Inhibition of each MN")
</code>
<code>
# Initialize excitatory input current + Store initial excitatory input
max_baseline_excit = excitatory_input_baseline
excit_input_all_MNs = (target_force / target_force_level) * excitatory_input_baseline
excit_input_all_MNs = (excit_input_all_MNs / np.max(excit_input_all_MNs)) * max_baseline_excit # normalize to 1, and multiply by baseline so that the baseline is the max value
initial_excit_signal = excit_input_all_MNs.copy()
samples_of_interest = list(range((window_beginning_ignore*fsamp),len(time)-(window_end_ignore*fsamp)))
# Inhibitory input to all MNs has already been created in the previous cell
plt.figure(figsize=(10,5))
plt.plot(initial_excit_signal)
plt.xlabel("Time (samples)")
plt.ylabel("Amplitude of excitatory input (millisiemens)")
plt.title("Baseline (before optimisation) excitatory signal")
</code>
<code>
# Just for testing purposes
# max_num_optimization_iterations = 5
# adam_learning_rate = 0.1
# excitatory_input_baseline = 150*1e-2
</code>
<code>
### OPTIMIZE INPUT FOR FORCE TARGET - iterate simulations
# Initialize inhibitory signal to be distributed
if (keep_force_constant_despite_inhib == True):
input_inhib = TimedArray(inhib_input_per_MN * msiemens, dt=1*ms)
else: # else ignore inhibition for the input optimization process
input_inhib = TimedArray(np.zeros((len(time),nb_motoneurons_full_pool)) * msiemens, dt=1*ms)
# Define the cost function
def cost_function(output_force_var, target_force_var):
return np.sum(abs(output_force_var - target_force_var))
# Gradient descent parameters
learning_rate = adam_learning_rate
# Adam optimizer parameters
initial_alpha = learning_rate # Initial learning rate
beta1 = 0.1 #0.9
beta2 = 0.5 #0.999
epsilon = 1e-8
m = np.zeros_like(excit_input_all_MNs)
v = np.zeros_like(excit_input_all_MNs)
timestep = 0
# Lists to store error and output force for plotting
errors = []
initial_output_force = None
# Reset simulation
start_scope() # Re-initialize the simulation
optimized_excit_signal = np.copy(initial_excit_signal)
best_cost = np.Inf
best_iter_idx = 0
# plot that will be updated at each iteration
colormap_temp = cm.get_cmap('viridis')
plt.figure(figsize=(15,12))
plt.subplot(211)
plt.plot(target_force, color = 'black', linestyle = ':', linewidth = 3)
plt.subplot(212)
plt.plot(optimized_excit_signal, color = 'blue', linewidth = 2, alpha = 0.7, label = "Initial input signal")
# Optimization loop using Adam optimizer
for iteration in range(max_num_optimization_iterations):
timestep += 1
# Low-pass filter the current excitatory input signal to prevent the "learning/optimization" towards an oscillating common input
# 1 hz cut-off
excit_input_all_MNs = lowpass_filter(excit_input_all_MNs, 1, fsamp)
# remove artifacts of low pass filter
excit_input_all_MNs[0:int(np.round(window_beginning_ignore*fsamp)*0.5)] = 0
excit_input_all_MNs[len(time)-int(np.round((window_end_ignore*fsamp)*0.5)):len(time)] = 0
# Prevent common excitatory input from going negative (it happens at the beginning of slopes)
excit_input_all_MNs[excit_input_all_MNs < 0] = 0
# Update input current in the neuron group
excit_input_per_MN = np.zeros((nb_motoneurons_full_pool,len(time)))
for mni in range(nb_motoneurons_full_pool):
excit_input_per_MN[mni,:] = excit_input_all_MNs * motoneurons_excitation_weights[mni]
excit_input_per_MN[mni,:] += independent_noise_excit[mni,:]
excit_input_per_MN[mni,:] = np.clip(excit_input_per_MN[mni,:], a_min=0, a_max=None)# Clamp to 0 to avoid negative conductance
excit_input_per_MN = np.transpose(excit_input_per_MN)
input_excit = TimedArray(excit_input_per_MN * msiemens, dt=1*ms)
eqs_motoneuron = LIF_equations
# Groups of neurons
motoneurons = NeuronGroup(nb_motoneurons_full_pool, eqs_motoneuron,
threshold='v>voltage_thresh',
reset='v=voltage_rest',
refractory='refractory_period',
method=sim_method)
# Initialize values
motoneurons.v = voltage_rest # in mV #
motoneurons.g_leak = motoneurons_membrane_conductance * msiemens # in milisiemens
motoneurons.C_m = motoneuron_capacitances * ufarad # in microfarads
motoneurons.I_th = motoneurons_rheobases * rheobase_scaling * nA # in nanoAmperes
motoneurons.refractory_period = motoneurons_refractory_periods * ms # in milliseconds
motoneurons.input_weight = motoneuron_input_weights # dimensionless unit
# Monitors
monitor_spikes_motoneurons = SpikeMonitor(motoneurons, record=True)
# Run simulation
run(duration_with_ignored_window)
# Get spike trains
spike_trains, binary_spike_trains = Get_binary_spike_trains(monitor_spikes_motoneurons, duration_with_ignored_window)
# Get force
output_force = Convolve_to_get_force(binary_spike_trains,np.arange(nb_motoneurons_full_pool),'normalized')
# if low_pass_filter_force:
# output_force = lowpass_filter(output_force, low_pass_filter_of_force_cutoff, fsamp)
if iteration == 0:
initial_output_force = output_force
# During the learning phase, low-pass filtering the force helps the algorithm converge towards a better common input "solution"
output_force = lowpass_filter(output_force, 1, fsamp)
# Calculate cost
# output_force_windowed = output_force[samples_of_interest]
# target_force_windowed = target_force[samples_of_interest]
output_force_windowed = output_force[window_beginning_ignore*fsamp:len(output_force)-(window_end_ignore*fsamp)]
target_force_windowed = target_force[window_beginning_ignore*fsamp:len(output_force)-(window_end_ignore*fsamp)]
cost = cost_function(output_force_windowed, target_force_windowed)/len(samples_of_interest)
if consider_only_plateau_for_cost_optimization and (target_type == 'trapezoid'):
output_force_windowed = output_force[(window_beginning_ignore+ramp_duration)*fsamp:len(output_force)-((window_end_ignore+ramp_duration)*fsamp)]
target_force_windowed = target_force[(window_beginning_ignore+ramp_duration)*fsamp:len(output_force)-((window_end_ignore+ramp_duration)*fsamp)]
cost_only_plateau = cost_function(output_force_windowed, target_force_windowed)/len(samples_of_interest)
print(f'Iteration {iteration + 1}, Cost (only on plateau): {np.round(cost_only_plateau*100)/100} (mean error in % of MVC)')
errors.append(cost)
print(f'Iteration {iteration + 1}, Cost: {np.round(cost*100)/100} (mean error in % of MVC)')
if cost < best_cost:
print(f'New best control signal')
best_output_force = np.copy(output_force)
best_cost = cost
optimized_excit_signal = np.copy(excit_input_all_MNs)
best_iter_idx = iteration
# Adjust learning rate inversely proportional to the error
# alpha = min(cost*learning_rate*0.5,learning_rate)
alpha = learning_rate
# Compute gradients
gradients = (output_force - target_force)
# Adam optimizer updates
m = beta1 * m + (1 - beta1) * gradients
v = beta2 * v + (1 - beta2) * (gradients ** 2)
m_hat = m / (1 - beta1 ** timestep)
v_hat = v / (1 - beta2 ** timestep)
excit_input_all_MNs -= alpha * m_hat / (np.sqrt(v_hat) + epsilon)
# Update figure
plt.subplot(211)
plt.plot(output_force, color=colormap_temp(iteration/max_num_optimization_iterations), linewidth=2, alpha = 0.7)
plt.subplot(212)
plt.plot(excit_input_all_MNs, color = colormap_temp(iteration/max_num_optimization_iterations), linewidth = 2, alpha = 0.7)
if consider_only_plateau_for_cost_optimization and (target_type == 'trapezoid'):
if cost_only_plateau < stop_optimizing_if_mean_error_is_below:
print("Stopping optimization because the cost (% of MVC error) has reached a satisfying level on the plateau part of the simulated contraction")
break
else:
if cost < stop_optimizing_if_mean_error_is_below:
print("Stopping optimization because the cost (% of MVC error) has reached a satisfying level")
break
plt.subplot(211)
plt.plot(best_output_force, color = 'red', linewidth = 3.5, label = 'best output force')
plt.plot(target_force, color = 'black', linestyle = ':', linewidth = 3, label = 'target force')
plt.xlabel("Time (ms)")
plt.ylabel("Torque (% MVC)")
plt.legend()
plt.title(f"Optimization of excitatory input signal (1st pass) \n Dark = first iterations; Light = last iterations")
plt.subplot(212)
plt.plot(optimized_excit_signal, color = 'red', linewidth = 3.5, label = "Best excitatory input")
plt.xlabel("Time (ms)")
plt.ylabel("Excitatory input (milliSiemens)")
plt.legend()
new_filename = f'Optimization_of_input_loss_1st_pass_force_output.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
# Plot error improvement
fig, ax = plt.subplots(figsize=(10,6))
plt.plot(errors)
plt.xlabel('Iteration')
plt.ylabel('Cost (mean error in % of MVC)')
plt.ylim([0,np.ceil(max(errors))])
plt.title('Cost Improvement Over Iterations (first pass)')
plt.grid(True)
new_filename = f'Optimization_of_input_loss_1st_pass.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
# Define the softmax function
def softmax_with_temperature(logits, temperature=1.0):
"""
Compute the softmax of a list of logits with a temperature parameter.
Parameters:
logits (list or numpy array): The input logits.
temperature (float): The temperature parameter.
Returns:
numpy array: The softmax probabilities.
"""
# Convert logits to numpy array if they are not already
logits = np.array(logits)
# Apply the temperature parameter
logits = logits / temperature
# Compute the exponentials of the scaled logits
exp_logits = np.exp(logits - np.max(logits)) # Subtract max for numerical stability
# Compute the softmax probabilities
softmax_probs = exp_logits / np.sum(exp_logits)
return softmax_probs
</code>
<code>
# SANITY CHECK ###############
plt.figure()
plt.plot(output_force) # sanity check before the rest of the optimization process
plt.ylabel("Force (% MVC)")
### Restrict window of analyzis
samples_for_analyzis = []
if target_type == 'trapezoid':
if analyzis_window == 'plateau':
samples_for_analyzis = np.arange((window_beginning_ignore+ramp_duration)*fsamp,len(time)-(window_end_ignore+ramp_duration)*fsamp)
else:
samples_for_analyzis = np.arange(window_beginning_ignore*fsamp,len(time)-window_end_ignore*fsamp)
else:
samples_for_analyzis = np.arange(window_beginning_ignore*fsamp,len(time)-window_end_ignore*fsamp)
### Get discharge characteristics of motoneurons => only during window of analysis
# Retrieve spikes
spike_trains = []
for mni in range(nb_motoneurons_full_pool):
spike_trains.append(monitor_spikes_motoneurons.spike_trains()[mni])
# remove samples out of the analyzis window
spike_trains[mni] = spike_trains[mni]/second # convert into a simple numpy array first
spike_trains[mni] = spike_trains[mni][(spike_trains[mni] > (samples_for_analyzis[0]/fsamp)) & (spike_trains[mni] < (samples_for_analyzis[len(samples_for_analyzis)-1]/fsamp))]
# spike_trains[mni] = spike_trains[mni]*second # convert back into a Brian2 array with unit
# Calculate the firing rate for each neuron
firing_rates = []
highest_ISIs = []
for mni in range(nb_motoneurons_full_pool):
if len(spike_trains[mni]) <= 1:
highest_ISIs.append(len(samples_for_analyzis)/fsamp)
else:
highest_ISIs.append(max(diff(spike_trains[mni])))
# firing_rate_temp = len(spike_trains[mni]) / (len(samples_for_analyzis)/fsamp)
if len(spike_trains[mni]) > 1:
firing_rate_temp = 1/np.mean(np.diff(spike_trains[mni]))
else:
firing_rate_temp = 0
firing_rates.append(firing_rate_temp)
# Convert to a numpy array for easier calculations
firing_rates = np.array(firing_rates)
### SELECT ONLY Selected MOTONEURONS
discontinuous_MUs_idx = {}
valid_MUs_idx = {}
# Index of discontinuous MNs (ISIs > threshold)
discontinuous_MUs_idx = [i for i, x in enumerate(highest_ISIs) if x > ISI_threshold_for_discontinuity]
discontinuous_MUs_idx = append(discontinuous_MUs_idx,
[i for i, x in enumerate(np.arange(nb_motoneurons_full_pool)) if len(spike_trains[x])<20]) # remove MUs with less than X spikes
discontinuous_MUs_idx = unique(discontinuous_MUs_idx)
valid_MUs_idx = [i for i, x in enumerate(arange(nb_motoneurons_full_pool)) if x not in discontinuous_MUs_idx]
print("Number of invalid MUs = ", len(discontinuous_MUs_idx), " out of ", nb_motoneurons_full_pool)
if motor_unit_subsampling_probability_distribution == 'uniform':
sampling_probability_distribution = np.ones(shape(valid_MUs_idx))/len(valid_MUs_idx)
else:
sampling_probability_distribution = np.copy(motoneuron_soma_diameters[valid_MUs_idx])
sampling_probability_distribution = softmax_with_temperature(sampling_probability_distribution, bias_towards_larger_motor_neurons_temperature)
if subsample_MUs_for_analysis == False:
selected_motor_units = valid_MUs_idx.copy()
plot_title_text = '(all valid motor units selected)'
txt_for_legend = f'selected motor units (n={len(valid_MUs_idx)}=all continuously active motor units)'
plot_title_suffix = f' (all valid motor units)'
else:
selected_motor_units = np.random.choice(valid_MUs_idx, size=nb_of_MUs_to_subsample, p=sampling_probability_distribution, replace=False)
plot_title_text = f' (subset of {nb_of_MUs_to_subsample} motor units selected)'
txt_for_legend = f'selected motor units (n={nb_of_MUs_to_subsample})'
plot_title_suffix = f' (sampling of {nb_of_MUs_to_subsample} motor units)'
# selected_motor_units_relative_to_valid_MUs = np.array([np.where(valid_MUs_idx == x)[0][0] for x in selected_motor_units])
selected_motor_units_relative_to_valid_MUs = [valid_MUs_idx.index(x) for x in selected_motor_units]
# Firing rate results - only selected MNs
fig, axs = plt.subplots()
axs.hist(firing_rates[selected_motor_units], edgecolor='white', alpha=0.75)
mean_firing_rate_valid = np.mean(firing_rates[selected_motor_units])
std_firing_rate_valid = np.std(firing_rates[selected_motor_units])
axs.axvline(x = mean_firing_rate_valid, linestyle='--', linewidth=2, label='Mean firing rate')
axs.set_xlabel("Mean firing rate (pps)")
axs.set_ylabel("Motoneuron count count")
plt.tight_layout(rect=[0,0,1,0.96])
plt.suptitle("Histogram of motoneurons' firing rate" + plot_title_suffix)
new_filename = f'Hist_MN_Discharge_rates.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
# Get recruitment threshold and discharge rates of motor units = only if chosing the "trapezoid" force target (because allows for gradual recruitment of motor units)
if target_type == 'trapezoid':
MN_mean_firing_rates = np.copy(firing_rates) # all motoneurons by index, regardless of whether they are valid or not
# Discharge rates duging the window of analyzis (the plateau section of the trapezoid)
MN_recruitment_thresholds = np.full(firing_rates.shape, np.nan) # in % MVC
spike_trains_full, binary_spike_trains_full = Get_binary_spike_trains(monitor_spikes_motoneurons, duration_with_ignored_window)
for mni in range(nb_motoneurons_full_pool): #valid_MUs_idx:
# RT as force during the median time of the first 3 firings
temp_RT = (spike_trains_full[mni]/second)*fsamp
temp_RT = np.median(temp_RT[0:2])
if np.isnan(temp_RT)==False:
MN_recruitment_thresholds[mni] = output_force[int(np.round(temp_RT))]
# RT as force when the MN starts discharging with a rate > 5 pps (first ISI < 0.2)
# MN_recruitment_thresholds[mni] = nan
# spike_count = 0
# for ISI_i in np.diff(spike_trains_full[mni]):
# if ISI_i < ISI_threshold_for_RT*second:
# MN_recruitment_thresholds[mni] = output_force[int(np.round((spike_trains_full[mni][spike_count]/second)*fsamp))]
# break
# spike_count += 1
MN_recruitment_thresholds_by_force_only_selected_idx = np.copy(MN_recruitment_thresholds)
MN_recruitment_thresholds_by_force_only_selected_idx = MN_recruitment_thresholds_by_force_only_selected_idx[selected_motor_units].reshape(-1, 1)
MN_mean_firing_rates_only_selected_idx = np.copy(MN_mean_firing_rates)
MN_mean_firing_rates_only_selected_idx = MN_mean_firing_rates[selected_motor_units].reshape(-1, 1)
# histogram of recruitment thresholds
plt.figure()
plt.hist(MN_recruitment_thresholds_by_force_only_selected_idx, density=True,
edgecolor='white', alpha=0.75, color='C1')
plt.xlim(0,target_force_level*1)
ylabel("Proportion")
xlabel("Recruitment threshold (% MVC)")
title("Histogram of recruitment tresholds" + plot_title_suffix)
new_filename = f'RT_hsitogram.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
# Create common noise (fluctuation of the excitatory input)
if excitatory_input_source == 'generate_synthetic_input':
common_noise = randn(len(time)) # noise input, with mean zero and std 1 (default setting)
common_noise = lowpass_filter(common_noise, low_pass_filter_of_excitatory_input, fsamp)
# remove artifacts of low pass filter
common_noise[0:window_beginning_ignore*fsamp] = 0
common_noise[len(common_noise)-(window_end_ignore*fsamp):len(common_noise)] = 0
elif excitatory_input_source == 'load_synthetic_input':
synthetic_signals_dataframe = pd.read_csv(excitatory_input_sourcefile)
common_noise = synthetic_signals_dataframe[f'{synthetic_signals_dataframe.shape[1]-1}'].values
if excitatory_input_sourcefile_fsamp != fsamp:
from scipy.interpolate import interp1d
# Calculate the time array for the original signal
loaded_signal_time = np.arange(len(synthetic_signals_dataframe)) / excitatory_input_sourcefile_fsamp
# Calculate the number of samples in the resampled signal
number_of_samples = int(len(synthetic_signals_dataframe) * fsamp / excitatory_input_sourcefile_fsamp)
# Calculate the time array for the resampled signal
resampled_time = np.linspace(loaded_signal_time[0], loaded_signal_time[-1], number_of_samples)
# Create an interpolation function
resample_loaded_signal_function = interp1d(loaded_signal_time, common_noise, kind='linear')
common_noise = resample_loaded_signal_function(resampled_time)
common_noise = common_noise[:len(time)] # cut the signal for it to be the right size
# Normalize
common_noise = ((common_noise - np.mean(common_noise)) / np.std(common_noise)) * excitatory_input_std
plt.figure()
plt.plot(common_noise, label="common noise / excitatory input fluctuations", color='C3')
plt.xlabel("Time (ms)")
plt.ylabel("Common noise amplitude (excitatory input fluctuations)")
plt.legend()
new_filename = f'Common_noise_signal.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
plt.figure()
N = len(common_noise)
yf = fft(common_noise)
xf = fftfreq(N, 1 / fsamp)
power_spectrum_temp = (np.abs(yf[:N//2])**2) / N
common_noise_power_integral = np.sum(power_spectrum_temp)
idx_corresponding_to_5hz = int(np.round((N/fsamp)*5))
power_spectrum_temp_0_5hz = power_spectrum_temp[:idx_corresponding_to_5hz]
common_noise_power_intergal_0_5_hz = np.sum(power_spectrum_temp_0_5hz)
plt.plot(xf[:N//2], power_spectrum_temp, color = 'C3', alpha = 1)
plt.xlabel("Frequency (Hz)")
plt.ylabel("Power")
plt.title("Power spectrum of the common noise (fluctuations in common input)")
plt.xlim([0,10])
new_filename = f'Common_noise_power_spectrum.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
# # Re-run the simulation with the parameter ending up with the lowest cost, with a low-pass filter of the optimized signal + common noise
# lowpass_filtered_optimized_excit_signal = lowpass_filter(optimized_excit_signal,1,fsamp) # 1hz low-pass filter
# # remove artifacts of low pass filter
# lowpass_filtered_optimized_excit_signal[0:int(np.round(window_beginning_ignore*fsamp)*0.5)] = 0
# lowpass_filtered_optimized_excit_signal[len(time)-int(np.round((window_end_ignore*fsamp)*0.5)):len(time)] = 0
# final_mean_excit_input = lowpass_filtered_optimized_excit_signal + common_noise
final_mean_excit_input = optimized_excit_signal + common_noise
input_inhib = TimedArray(inhib_input_per_MN * msiemens, dt=1*ms)
excit_input_per_MN = np.zeros((nb_motoneurons_full_pool,len(time)))
for mni in range(nb_motoneurons_full_pool):
excit_input_per_MN[mni,:] = final_mean_excit_input * motoneurons_excitation_weights[mni] # scales both the mean excitation and common noise
excit_input_per_MN[mni,:] += independent_noise_excit[mni,:]
excit_input_per_MN[mni,:] = np.clip(excit_input_per_MN[mni,:], a_min=0, a_max=None)# Clamp to 0 to avoid negative conductance
excit_input_per_MN = np.transpose(excit_input_per_MN)
input_excit = TimedArray(excit_input_per_MN * msiemens, dt=1*ms)
eqs_motoneuron = LIF_equations
# Groups of neurons
motoneurons = NeuronGroup(nb_motoneurons_full_pool, eqs_motoneuron,
threshold='v>voltage_thresh',
reset='v=voltage_rest',
refractory='refractory_period',
method=sim_method)
# Initialize values
motoneurons.v = voltage_rest # in mV #
motoneurons.g_leak = motoneurons_membrane_conductance * msiemens # in milisiemens
motoneurons.C_m = motoneuron_capacitances * ufarad # in microfarads
motoneurons.I_th = motoneurons_rheobases * rheobase_scaling * nA # in nanoAmperes
motoneurons.refractory_period = motoneurons_refractory_periods * ms # in milliseconds
motoneurons.input_weight = motoneuron_input_weights # dimensionless unit
# Monitors
monitor_spikes_motoneurons = SpikeMonitor(motoneurons, record=True)
# Run simulation
run(duration_with_ignored_window)
# Get spike trains
spike_trains, binary_spike_trains = Get_binary_spike_trains(monitor_spikes_motoneurons, duration_with_ignored_window)
# Get force
output_force = Convolve_to_get_force(binary_spike_trains,np.arange(nb_motoneurons_full_pool),'normalized')
if low_pass_filter_force:
output_force = lowpass_filter(output_force, low_pass_filter_of_force_cutoff, fsamp)
</code>
<code>
### Restrict window of analyzis
cut_edges_of_plateau = 1 # in s
samples_for_analyzis = []
if target_type == 'trapezoid':
if analyzis_window == 'plateau':
samples_for_analyzis = np.arange((window_beginning_ignore+ramp_duration+cut_edges_of_plateau)*fsamp,len(time)-(window_end_ignore+ramp_duration+cut_edges_of_plateau)*fsamp)
else:
samples_for_analyzis = np.arange(window_beginning_ignore*fsamp,len(time)-window_end_ignore*fsamp)
else:
samples_for_analyzis = np.arange((window_beginning_ignore+cut_edges_of_plateau)*fsamp,len(time)-window_end_ignore*fsamp)
</code>
<code>
# Plot target vs output force (initial and final)
fig, ax = plt.subplots(figsize=(20,7))
plt.plot(target_force, label='Target Force')
plt.plot(initial_output_force, label='Initial Output Force')
if keep_force_constant_despite_inhib:
plt.plot(output_force, label='Final Output Force (optimization of common excitatory input taking inhibition into account)')
else:
plt.plot(output_force, label='Final Output Force (optimization of common excitatory ignoring inhibition = so force should be lower)')
plt.xlabel('Time (ms)')
plt.ylabel('Force')
plt.legend()
plt.title('Force Output of Motor Neurons')
plt.grid(True)
new_filename = f'Optimized_input_resulting_force.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
# Plot control signal before and after learning
fig, ax = plt.subplots(figsize=(20,7))
plt.plot(initial_excit_signal, label='Initial Control Signal')
if keep_force_constant_despite_inhib:
optimized_signal_label = 'Optimized Control Signal'
final_mean_excit_input_signal_label = 'Common input (optimized control signal + common noise)'
else:
optimized_signal_label = 'Optimized Control Signal (ignoring inhibition which is applied later)'
final_mean_excit_input_signal_label = 'Common input (optimized control signal ignoring inhibition + common noise)'
plt.plot(optimized_excit_signal, label=optimized_signal_label)
plt.plot(np.clip(final_mean_excit_input, a_min=0, a_max=None), label=final_mean_excit_input_signal_label)
if nb_inhibitory_input >= 1:
for inhibiti in range(nb_inhibitory_input):
plt.plot(inhib_input[inhibiti][0], label=f'inhibitory input #{inhibiti+1}', color='C4', alpha = 1/nb_inhibitory_input)
plt.xlabel('Time (ms)')
plt.ylabel('Control Signal')
plt.legend()
# plt.ylim(0.5,2.5)
plt.title('Control Signal Before and After Learning + common input')
plt.grid(True)
new_filename = f'Optimized_input_&_final_mean_excit_input.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
print(f'Mean and std of common input during window of analyzis = {np.round(np.mean(final_mean_excit_input[samples_for_analyzis])*100)/100} +/- {np.round(np.std(final_mean_excit_input[samples_for_analyzis])*100)/100}')
# Getting a smooth color blend from a given colormap
colormap_temp = cm.get_cmap('plasma')
# Raster plot of discharge times
plt.figure(num=1,figsize=(20,10))
for mni in range(nb_motoneurons_full_pool):
plt.scatter((spike_trains[mni]/second)*fsamp, np.ones(len(spike_trains[mni]))*mni, color=colormap_temp(mni/(nb_motoneurons_full_pool-1)), linewidth = 0.2, alpha = 0.3)
plt.vlines(samples_for_analyzis[0],plt.ylim()[0],plt.ylim()[1],color='black',label='Start of the analyzis window')
plt.vlines(samples_for_analyzis[len(samples_for_analyzis)-1],plt.ylim()[0],plt.ylim()[1],color='black',label='End of the analyzis window')
plt.xlabel('Time (s)')
plt.ylabel('Motoneuron index')
plt.title("Raster plot of motoneuron spikes \n Opaque = continuous MU; transparent = discontinuous MU")
plt.legend()
new_filename = f'Optimized_input_firings_raster_plot.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
# Force per MU
force_per_MU = []
force_total = zeros(len(binary_spike_trains[0,:]))
fig, ax = plt.subplots(figsize=(25, 10))
for mni in range(nb_motoneurons_full_pool):
# 'same' mode means the output length will be the same as the input length
if len(spike_trains[mni]) >= 1: # at least one spike necessary
temp_force = Convolve_to_get_force(np.reshape(binary_spike_trains[mni,:], (1, len(binary_spike_trains[mni,:]))), [mni], 'normalized')
force_per_MU.append(temp_force)
ax.plot(force_total, color=colormap_temp(mni/(nb_motoneurons_full_pool-1)), alpha = 0.5, linewidth = 0.5)
force_total = force_total + temp_force
if low_pass_filter_force:
ax.plot(output_force, color='black', alpha = 1, linewidth = 2, label = "Total force (low-pass filtered)")
else:
ax.plot(force_total, color='black', alpha = 1, linewidth = 2, label = "Total force")
plt.title(f"Reconstructed force (convolving spike train with twitch force kernel)")
plt.ylabel("Force (% MVC)")
plt.xlabel("Time (ms)")
plt.legend()
new_filename = f'Optimized_input_cumulative_force_per_MU.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
</code>
<code>
### Get discharge characteristics of motoneurons => only during window of analysis
fig, axs = plt.subplots()
# Retrieve spikes
spike_trains = []
for mni in range(nb_motoneurons_full_pool):
spike_trains.append(monitor_spikes_motoneurons.spike_trains()[mni])
# remove samples out of the analyzis window
spike_trains[mni] = spike_trains[mni]/second # convert into a simple numpy array first
spike_trains[mni] = spike_trains[mni][(spike_trains[mni] > (samples_for_analyzis[0]/fsamp)) & (spike_trains[mni] < (samples_for_analyzis[len(samples_for_analyzis)-1]/fsamp))]
# spike_trains[mni] = spike_trains[mni]*second # convert back into a Brian2 array with unit
# Calculate the firing rate for each neuron
firing_rates = []
highest_ISIs = []
for mni in range(nb_motoneurons_full_pool):
if len(spike_trains[mni]) <= 1:
highest_ISIs.append(len(samples_for_analyzis)/fsamp)
else:
highest_ISIs.append(max(diff(spike_trains[mni])))
# firing_rate_temp = len(spike_trains[mni]) / (len(samples_for_analyzis)/fsamp)
if len(spike_trains[mni]) > 1:
firing_rate_temp = 1/np.mean(np.diff(spike_trains[mni]))
else:
firing_rate_temp = 0
firing_rates.append(firing_rate_temp)
# Convert to a numpy array for easier calculations
firing_rates = np.array(firing_rates)
# Calculate mean and standard deviation of the firing rates
mean_firing_rate = np.mean(firing_rates)
std_firing_rate = np.std(firing_rates)
# Motoneurons' firing rates results
axs.hist(firing_rates, edgecolor='white', alpha=0.75)
axs.axvline(x = mean_firing_rate, linestyle='--', linewidth=2, label='Mean firing rate')
axs.set_xlabel("Mean firing rate (pps)")
axs.set_ylabel("Motoneuron count count")
plt.tight_layout(rect=[0,0,1,0.96])
plt.suptitle("Histogram of motoneurons' firing rate (all motoneurons, only during window of analyzis)")
### SELECT ONLY Selected MOTONEURONS
discontinuous_MUs_idx = {}
valid_MUs_idx = {}
fig, axs = plt.subplots()
# Index of discontinuous MNs (ISIs > threshold)
discontinuous_MUs_idx = [i for i, x in enumerate(highest_ISIs) if x > ISI_threshold_for_discontinuity]
discontinuous_MUs_idx = append(discontinuous_MUs_idx,
[i for i, x in enumerate(np.arange(nb_motoneurons_full_pool)) if len(spike_trains[x])<20]) # remove MUs with less than X spikes
discontinuous_MUs_idx = unique(discontinuous_MUs_idx)
valid_MUs_idx = [i for i, x in enumerate(arange(nb_motoneurons_full_pool)) if x not in discontinuous_MUs_idx]
print("Number of invalid MUs = ", len(discontinuous_MUs_idx), " out of ", nb_motoneurons_full_pool)
axs.hist(highest_ISIs, edgecolor='white', alpha=0.5)
axs.axvline(x = np.median(highest_ISIs), linestyle='--', linewidth=3, label='Mean highest ISI')
axs.axvline(x = ISI_threshold_for_discontinuity, color = 'black', linestyle='-', linewidth=2, alpha = 0.5, label='Mean firing rate')
axs.set_xlabel("Max ISI (s)")
axs.set_ylabel("Motoneuron count")
plt.tight_layout(rect=[0,0,1,0.92])
plt.suptitle("Histogram of motoneurons' max ISI \n (colored line is median ; black line is threshold)")
# new_filename = f'Hist_MN_ISIs.png'
# save_file_path = os.path.join(new_directory, new_filename)
# plt.savefig(save_file_path)
plt.show(fig)
</code>
<code>
# SELECT A SUBSET OF MOTOR UNITS
plot_title_text = 'sampling of motor units for analysis'
if motor_unit_subsampling_probability_distribution == 'uniform':
sampling_probability_distribution = np.ones(shape(valid_MUs_idx))/len(valid_MUs_idx)
else:
sampling_probability_distribution = np.copy(motoneuron_soma_diameters[valid_MUs_idx])
sampling_probability_distribution = softmax_with_temperature(sampling_probability_distribution, bias_towards_larger_motor_neurons_temperature)
if subsample_MUs_for_analysis == False:
selected_motor_units = valid_MUs_idx.copy()
plot_title_text = plot_title_text + ' (all valid motor units selected)'
txt_for_legend = f'selected motor units (n={len(valid_MUs_idx)}=all continuously active motor units)'
plot_title_suffix = f' (all valid motor units)'
else:
selected_motor_units = np.random.choice(valid_MUs_idx, size=nb_of_MUs_to_subsample, p=sampling_probability_distribution, replace=False)
plot_title_text = plot_title_text + f' (subset of {nb_of_MUs_to_subsample} motor units selected)'
txt_for_legend = f'selected motor units (n={nb_of_MUs_to_subsample})'
plot_title_suffix = f' (sampling of {nb_of_MUs_to_subsample} motor units)'
# selected_motor_units_relative_to_valid_MUs = np.array([np.where(valid_MUs_idx == x)[0][0] for x in selected_motor_units])
selected_motor_units_relative_to_valid_MUs = [valid_MUs_idx.index(x) for x in selected_motor_units]
plt.figure(figsize=(20,5))
plt.bar(valid_MUs_idx,sampling_probability_distribution, label='probability of each valid (continuously firing) MU', color='C9', alpha=0.3)
plt.bar(selected_motor_units,sampling_probability_distribution[selected_motor_units_relative_to_valid_MUs], label=txt_for_legend, color='C9')
plt.legend()
plt.ylabel("Probability")
plt.xlabel("Motor unit index")
plt.title(plot_title_text)
new_filename = f'Valid_VS_sampled_motor_units_for_analyzis.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
</code>
<code>
# Firing rate results - only selected MNs
fig, axs = plt.subplots()
axs.hist(firing_rates[selected_motor_units], edgecolor='white', alpha=0.75)
mean_firing_rate_valid = np.mean(firing_rates[selected_motor_units])
std_firing_rate_valid = np.std(firing_rates[selected_motor_units])
axs.axvline(x = mean_firing_rate_valid, linestyle='--', linewidth=2, label='Mean firing rate')
axs.set_xlabel("Mean firing rate (pps)")
axs.set_ylabel("Motoneuron count count")
plt.tight_layout(rect=[0,0,1,0.96])
plt.suptitle("Histogram of motoneurons' firing rate" + plot_title_suffix)
new_filename = f'Discharge_rates_Hist.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
plt.figure(figsize=(10,10))
plt.subplot(211)
plt.plot(np.arange(nb_motoneurons_full_pool)[selected_motor_units], firing_rates[selected_motor_units], color = 'C0', linewidth = 3)
plt.ylabel("Firing rate (pps)")
plt.xlabel("MN index")
plt.title("Firing rates (only selected MUs) ~ MN index")
plt.subplot(212)
plt.plot(motoneuron_soma_diameters[selected_motor_units], firing_rates[selected_motor_units], color = 'C0', linewidth = 3)
plt.ylabel("Firing rate (pps)")
plt.xlabel("soma size (micrometers)")
plt.title("Firing rates (only selected MUs) ~ soma diameter")
new_filename = f'Discharge_rates.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
</code>
<code>
## SMOOTHING SPIKE TRAINS
Wind_s = 0.4 # hanning window duration. 0.4 for 2.5hz low-pass, 0.2 for 5hz low-pass
HanningW = 2 / round(fsamp * Wind_s) * windows.hann(round(fsamp * Wind_s)) # unitary area
# Filter all valid motor units
smoothed_signal = []
for mni in range(nb_motoneurons_full_pool):
if mni in valid_MUs_idx:
smoothed_signal.append(filtfilt(HanningW, 1, binary_spike_trains[mni, :] * fsamp))
smoothed_signal = np.array(smoothed_signal)
fig, ax = plt.subplots(figsize=(25, 10))
colormap_temp = cm.get_cmap('plasma') # Getting a smooth color blend from a given colormap
if subsample_MUs_for_analysis == True:
for mni in range(len(valid_MUs_idx)):
ax.plot((smoothed_signal)[mni,:], color=colormap_temp(valid_MUs_idx[mni]/(nb_motoneurons_full_pool-1)), alpha = 0.3)
# Remove non-valid motor units, and replot on top of the previous plot only the sampled motor units
smoothed_signal = smoothed_signal[selected_motor_units_relative_to_valid_MUs,:]
for mni in range(smoothed_signal.shape[0]):
ax.plot((smoothed_signal)[mni,:], color=colormap_temp(selected_motor_units[mni]/(nb_motoneurons_full_pool-1)), alpha = 1)
plt.title(f"Smoothed signals of only continuous MUs \n (dark = small MNs ; light = large MNs) \n (low opacity = valid but not selected ; opaque = selected motor units)")
plt.ylabel("Smoothed discharge rate (pps)")
plt.xlabel("Time (ms)")
plt.vlines(samples_for_analyzis[0],plt.ylim()[0],plt.ylim()[1],color='black',label='Start of the analyzis window')
plt.vlines(samples_for_analyzis[len(samples_for_analyzis)-1],plt.ylim()[0],plt.ylim()[1],color='black',label='End of the analyzis window')
plt.legend()
new_filename = f'Smoothed_discharge_rates_only_continuous_MUs.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
# restrict signal to window of analyzis, with only valid motor units
smoothed_signal = smoothed_signal[:,samples_for_analyzis]
# re-plot
fig, ax = plt.subplots(figsize=(25, 10))
for mni in range(smoothed_signal.shape[0]):
ax.plot(smoothed_signal[mni,:], color=colormap_temp(selected_motor_units[mni]/(nb_motoneurons_full_pool-1)))
plt.title(f"Smoothed signals only during window of analyzis" + plot_title_suffix + "\n (dark = small MNs ; light = large MNs)")
plt.ylabel("Smoothed discharge rate (pps)")
plt.xlabel("Time (ms)")
new_filename = f'Smoothed_discharge_rates_only_continuous_MUs_window_of_analyzis.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
</code>
<code>
# # For debug/testing purpose
# smoothed_signal_normalized = np.copy(smoothed_signal)
# for mni in range(smoothed_signal.shape[0]):
# print(f"Mean = {np.round(np.mean(smoothed_signal_normalized[mni])*100)/100}, STD = {np.round(np.std(smoothed_signal_normalized[mni])*100)/100}")
</code>
<code>
from sklearn.decomposition import PCA
from sklearn.metrics import r2_score
# PCA
smoothed_signal_normalized = np.copy(smoothed_signal)
for mni in range(smoothed_signal.shape[0]):
smoothed_signal_normalized[mni] = smoothed_signal_normalized[mni]-np.mean(smoothed_signal_normalized[mni])
# In some cases (when no noise is injected), the std can be very low and cause division by zero
if np.std(smoothed_signal_normalized[mni]) > 0.01:
smoothed_signal_normalized[mni] = smoothed_signal_normalized[mni]/np.std(smoothed_signal_normalized[mni])
else:
smoothed_signal_normalized[mni] = smoothed_signal_normalized[mni]/0.01
# Displaye smoothed signals normalized (mean of 0, std of 1)
fig, ax = plt.subplots(figsize=(25, 10))
for mni in range(smoothed_signal.shape[0]):
ax.plot(smoothed_signal_normalized[mni,:], color=colormap_temp(selected_motor_units[mni]/(nb_motoneurons_full_pool-1)))
plt.title(f"Normalized smoothed signals of only continuous MUs, only during window of analyzis" + plot_title_suffix + "\n (dark = small MNs ; light = large MNs)")
plt.ylabel("Normalizd smoothed discharge rate (in std)")
plt.xlabel("Time (ms)")
new_filename = f'Smoothed_discharge_rates_ormalized_only_continuous_MUs_window_of_analyzis.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show(fig)
nb_PCs_to_store = 5
# Function to calculate R-squared for each time series with a given number of PCs
def calculate_r_squared(data, pca, num_pcs):
# Transform the data using the selected number of PCs
transformed_data = pca.transform(data)
# Inverse transform to reconstruct the data
reconstructed_data = pca.inverse_transform(
np.hstack([transformed_data, np.zeros((data.shape[0], pca.n_components_ - num_pcs))])
)
# Calculate R-squared for each time series (motor unit)
r_squared_values = [r2_score(data[:, i], reconstructed_data[:, i]) for i in range(data.shape[1])]
return r_squared_values
# Assume `smoothed_discharge_rates` is the variable holding the data
# Perform PCA
nb_PCs = 10 # nb_motoneurons_full_pool
pca = PCA(n_components=nb_PCs)
pca_result = pca.fit_transform(transpose(smoothed_signal_normalized))
# Initialize a DataFrame to store the R-squared values
PCA_r_squared_df = pd.DataFrame(index=range(1, nb_PCs_to_store + 1), columns=range(smoothed_signal_normalized.shape[0])) # nb_PCs + 1
# Calculate R-squared values for 1 to 2 PCs
for num_pcs in range(1,nb_PCs_to_store+1): # From 1 to 2 PCs # nb_PCs + 1):
pca_temp = PCA(n_components=num_pcs)
pca_temp.fit(smoothed_signal_normalized.T)
r_squared_values = calculate_r_squared(smoothed_signal_normalized.T, pca_temp, num_pcs)
PCA_r_squared_df.loc[num_pcs] = r_squared_values
# Display the explained variance ratio (proportion of variance explained by each PC)
explained_variance_ratio = pca.explained_variance_ratio_
# Optionally, display the cumulative explained variance
cumulative_explained_variance = np.cumsum(explained_variance_ratio)
# Plot the explained variance
plt.figure(figsize=(8, 6))
plt.bar(range(0, nb_PCs + 1), append(0,explained_variance_ratio), alpha=0.5, align='center',
label='Individual explained variance')
plt.plot(range(0, nb_PCs + 1), append(0,cumulative_explained_variance),
label='Cumulative explained variance')
plt.ylabel('Explained variance ratio')
plt.xlabel('Principal components')
plt.title('Explained Variance by Principal Components' + plot_title_suffix)
plt.ylim(-0.1,1.1)
plt.legend(loc='best')
plt.grid(True)
new_filename = f'Error_correction_PCA_VAF.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
print(f'Mean R² of PC1+PC2 = {np.mean(PCA_r_squared_df.iloc[1])}')
print(f'Cumulative VAF for PC1+PC2 = {cumulative_explained_variance[1]}')
</code>
<code>
from scipy.signal import csd, detrend
windowCOH = 1 # in seconds
frequencies_per_FFT_window = 10
indexes_of_windows_below_5hz = np.arange(0,5*frequencies_per_FFT_window)
# From groups of 2 up to half of the motor neurons
shuffled_idx_list = list.copy(list(selected_motor_units)) # initialize list of indices to be shuffled iteratively
window_analyzis_begin = int(samples_for_analyzis[0])
window_analyzis_end = int(samples_for_analyzis[len(samples_for_analyzis)-1])
seg_pwr = 10 # segment length (for coherence analyzis), specificied as a power of 2. 2^10 is ~1s for a fsamp of 1000
max_or_mean_0_5hz_COH = 'max'
COH_calc_group_size_nb = int(floor(len(selected_motor_units)/2)-1)
# COH_calc_max_iteration_nb_per_group_size = 1000 # More iteration for smaller group sizes, because the value obtained is very dependent upon the exact neurons selected, especially when only a few MNs are used to create the CST
COH_0_5hz_per_group = []
COH_mean_0_5hz = np.zeros(COH_calc_group_size_nb)
COH_pooled_per_group_size = []
colors_plots = plt.cm.winter(np.linspace(0, 1, COH_calc_group_size_nb))
plt.figure(figsize=(10,8))
for group_sizi in range(COH_calc_group_size_nb):
coherence_temp = []
COH_calc_iteration_nb_per_group_size_temp = int(np.max([np.round(COH_calc_max_iteration_nb_per_group_size/(group_sizi+1)),1]))
COH_0_5hz_per_group.append([])
print(f'Iterating for groups of {group_sizi+1} motoneurons (out of a max group size of {COH_calc_group_size_nb}) - {COH_calc_iteration_nb_per_group_size_temp} iterations')
for group_iteri in range(COH_calc_iteration_nb_per_group_size_temp):
random.shuffle(shuffled_idx_list)
idx_cst1 = np.copy(shuffled_idx_list[0:group_sizi+1])
cst1 = sum(binary_spike_trains[idx_cst1,window_analyzis_begin:window_analyzis_end],axis=0)
idx_cst2 = np.copy(shuffled_idx_list[len(shuffled_idx_list)-(group_sizi+1):len(shuffled_idx_list)])
cst2 = sum(binary_spike_trains[idx_cst2,window_analyzis_begin:window_analyzis_end],axis=0)
# Compute intra-group coherence for group 1
f, COH_intragroup_X = csd(detrend(cst1), detrend(cst1), window=windows.hann(round(windowCOH * fsamp)), noverlap=0, nfft=frequencies_per_FFT_window * fsamp, fs=fsamp)
# Compute intra-group coherence for group 2
f, COH_intragroup_Y = csd(detrend(cst2), detrend(cst2), window=windows.hann(round(windowCOH * fsamp)), noverlap=0, nfft=frequencies_per_FFT_window * fsamp, fs=fsamp)
# Compute inter-group coherence
f, COH_intergroup = csd(detrend(cst1), detrend(cst2), window=windows.hann(round(windowCOH * fsamp)), noverlap=0, nfft=frequencies_per_FFT_window * fsamp, fs=fsamp)
coherence_temp.append( (np.abs(COH_intergroup) ** 2) / (COH_intragroup_X * COH_intragroup_Y) ) # Welch's method of coherence calculation
if max_or_mean_0_5hz_COH == 'mean':
# COH_0_5hz_per_group[group_sizi,group_iteri] = np.nanmean(coherence_temp[group_iteri][indexes_of_windows_below_5hz])
COH_0_5hz_per_group[group_sizi].append(np.nanmean(coherence_temp[group_iteri][indexes_of_windows_below_5hz]))
elif max_or_mean_0_5hz_COH == 'max':
# COH_0_5hz_per_group[group_sizi,group_iteri] = np.nanmax(coherence_temp[group_iteri][indexes_of_windows_below_5hz])
COH_0_5hz_per_group[group_sizi].append(np.nanmax(coherence_temp[group_iteri][indexes_of_windows_below_5hz]))
# plt.scatter(group_sizi,COH_0_5hz_per_group[group_sizi,group_iteri],s=30,color=colors_plots[group_sizi],alpha=min(3/COH_calc_iteration_nb_per_group_size,1))
plt.scatter(group_sizi,COH_0_5hz_per_group[group_sizi][group_iteri],s=30,color=colors_plots[group_sizi],alpha=min(np.sqrt(1/COH_calc_iteration_nb_per_group_size_temp),1))
COH_pooled_per_group_size.append( np.nanmean(coherence_temp, axis=0) )
# COH_mean_0_5hz[group_sizi] = np.nanmean(COH_0_5hz_per_group[group_sizi,:],axis=0)
COH_mean_0_5hz[group_sizi] = np.nanmean(COH_0_5hz_per_group[group_sizi],axis=0)
plt.plot(COH_mean_0_5hz, linewidth=3, color='red', alpha=0.5, label=f"Mean of [max 0-5hz coherence for CSTs of X spike trains]")
plt.xlabel('Number of MNs in the CSTs')
plt.ylabel('Mean coherence in the 0-5hz bandwidth')
plt.title("Increase in coherence in the 0-5hz bandwidth (Y) as the number of CSTS' MNs (X) increase" + plot_title_suffix)
plt.ylim(0,1)
plt.legend()
new_filename = f'PCI_curve_of_0-5hz_coh.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
plt.figure(figsize=(10, 8))
for group_sizi in range(COH_calc_group_size_nb):
plt.plot(COH_pooled_per_group_size[group_sizi], color=colors_plots[group_sizi], alpha = 0.5, linewidth = 2)
plt.xlim(0,20*frequencies_per_FFT_window)
plt.xticks(ticks=plt.xticks()[0], labels=[str(int(x / frequencies_per_FFT_window)) for x in plt.xticks()[0]])
plt.xlabel("Frequency (Hz)")
plt.ylabel("Coherence")
plt.ylim(0,1)
plt.title("Mean coherence" + plot_title_suffix)
new_filename = f'Coherence_curve.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
# root mean square error function definition
def rmse(predicted, true):
# Root mean square
return np.sqrt(np.mean((true - predicted) ** 2))
# Mean square
# return np.mean((true - predicted) ** 2)
</code>
<code>
from scipy.optimize import curve_fit, least_squares
# Define the model - implementation from Negro et al 2016 https://physoc.onlinelibrary.wiley.com/doi/epdf/10.1113/JP271748
def PCI_model(n, A, B):
return abs(n**2 * A)**2 / ((n*B) + ((n**2)*A))**2
# Define the residuals function
def residuals(params, x, y):
A, B = params
return y - PCI_model(x, A, B)
plt.figure(figsize=(12,8))
n = np.arange(1,COH_mean_0_5hz.shape[0]+1)
# Negro 2016 equation:
# Mean COH in a given frequency band: Eqn 4 = abs(n**2 * A)**2 / ((n*B) + ((n**2)*A))**2
# - n = number of neurons in the CST
# - A = power of the common synaptic input (in the given frequency band), multipled by the absolute square of the susceptibility
# - A = abs(X(f))**2 * Ss(f)
# => X(f) is the response function (susceptibility) of the motor neuron [POOl of motor neurons in our case] to the stimulus and Ss(f) the power spectrum of the stimulus
# - B = (power of the?) response of the pool with independent synaptic input
# - B = Sn(f)
# => Sn(f)is the power spectrum of the output spike train [CST of spike train in our case] when it is driven by independent synaptic input only
# - proportion of common input (PCI) = estimate of gamma (common voltage fluctuation of membrane) = sqrt(A/(B+A)) = proportion of the common synaptic input with respect to the total synaptic input received by the motor neurons [total input can be inferred from motor pool output, because it the output is a function of both the common and independent input]
# => They say just sqrt(A/B) in the paper, but this can result in a proportion (PCI) > 1 which shouldn't be possible, and I get results fitting very closely the theoritical values when computing sqrt(A_fit / (B_fit + A_fit))
# - The ratio can be estimated by an experimental measure of the mean coherence in the given frequency range for varying n (number of motor neuron spike trains used in the calculation (Negro & Farina, 2012)) using eqn (4)
# - Using a least-square curve fitting of the estimated values of coherence for CSTs with different numbers of motor neurons, the parameters A and B of eqn (4) can be estimated.
# params, covariance = curve_fit(PCI_model, xdata = n, ydata = COH_mean_0_5hz) # older version, still works well
initial_guess = [1, 1] # Initial guess for the parameters from which to optimize using least squares
least_square_optim_results = least_squares(residuals, initial_guess, loss='soft_l1', f_scale=0.1, args=(n, COH_mean_0_5hz)) # default loss function is 'linear' but 'soft_l1' is a bit more robust to fluctuations
# Extract parameters
A_fit, B_fit = least_square_optim_results.x
PCI_estimated = np.sqrt(A_fit/(B_fit+A_fit))
fitted_PCI_curve = PCI_model(n, A_fit, B_fit)
# Get ratio of common excitatory input to independent input = Grond-truth PCI
# = power spectrum integral of common input in the 0-5hz range over power spectrum integral of independent input in the same range
# Ignoring inhibitory input = hard to calculate, so no ground truth when simulating inhibition
if (nb_inhibitory_input >= 1) and (inhibitory_input_mean >= 1*1e-2) and (np.sum(MN_inhibition_weights[0]) >= 0.1): # if there is inhibition
PCI_ground_truth_without_inhib = np.nan # not defined if there is inhibition
else: # else if no inhibition
PCI_ground_truth_without_inhib = np.sqrt(common_noise_power_intergal_0_5_hz / (common_noise_power_intergal_0_5_hz + independent_excit_noise_power_integral_0_5_hz_mean))
plt.plot(COH_mean_0_5hz, color='C0',linewidth=2.5, alpha = 0.5, label='ground truth curve')
plt.plot(fitted_PCI_curve, color='blue',linewidth=2, alpha = 0.5, linestyle='dashed',label='fitted curve')
plt.xlabel('Number of MNs in the CSTs')
plt.ylabel('Mean coherence in the 0-5hz bandwidth')
plt.title(f"Increase in coherence in the 0-5hz bandwidth (Y) as the number of CSTS' MNs (X) increase => Ground-truth VS fitted data" + plot_title_suffix + f"\n Estimated PCI (gamma) = {np.round(PCI_estimated*1000)/1000} \n Ground-truth PCI (ratio of common input 0-5hz integral of power spectrum VS idem common+independent input) = {np.round(PCI_ground_truth_without_inhib*1000)/10}%, ignoring inhibition \n")
plt.ylim(0,1)
plt.legend()
new_filename = f'PCI_fitted_vs_true_curve.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
# SIMON'S INSPIRED VERSION - Implementation from Negro et al 2016 https://physoc.onlinelibrary.wiley.com/doi/epdf/10.1113/JP271748
plt.figure(figsize=(12,8))
n = np.arange(1,COH_mean_0_5hz.shape[0]+1)
# Negro 2016 equation:
# Mean COH in a given frequency band: Eqn 4 = (abs(n**2 * A)**2) / ((n*B) + (((n**2)*A))**2)
# - n = number of neurons in the CST
# - A = power of the common synaptic input (in the given frequency band), multipled by the absolute square of the susceptibility (which is 1 when considering the 0-5hz bandwidth I think)
# - B = response of the pool with independent synaptic input (of the same bandwidth). Assumed to be 1 for the 0-5hz bandwidth.
# (k in the fitting below correspond to sqrt(B), since)
# # #
# - proportion of common input (PCI) = estimate of gamma (common voltage fluctuation of membrane) = sqrt(A/B) = proportion of the common synaptic input with respect to the total synaptic input received by the motor neurons
# - The ratio can be estimated by an experimental measure of the mean coherence in the given frequency range for varying n (number of motor neuron spike trains used in the calculation (Negro & Farina, 2012)) using eqn (4)
# - Using a least-square curve fitting of the estimated values of coherence for CSTs with different numbers of motor neurons, the parameters A and B of eqn (4) can be estimated.
nb_gammas_to_try = int(1e3) # Correspond to B
gamma_tmp = np.linspace(0.1,1e2,nb_gammas_to_try)
gamma_fit_error = []
for k in gamma_tmp:
c = n**4 / (n / k**2 + n**2)**2
plt.plot(c, color='C0',linewidth=1, alpha = min(5/nb_gammas_to_try,1))
gamma_fit_error.append(rmse(c,COH_mean_0_5hz))
idx_with_min_error = gamma_fit_error.index(min(gamma_fit_error))
gamma = np.sqrt(gamma_tmp[idx_with_min_error]**2 / (gamma_tmp[idx_with_min_error]**2 +1))
fitted_PCI_curve = n**4 / ( n / gamma_tmp[idx_with_min_error]**2 + n**2 )**2
# fitted_PCI_curve = n**4 / ( n / gamma**2 + n**2 )**2
plt.plot(COH_mean_0_5hz, color='C0',linewidth=2.5, alpha = 0.5, label='ground truth curve')
plt.plot(fitted_PCI_curve, color='red',linewidth=2, alpha = 0.5, linestyle='dashed',label='fitted curve')
plt.xlabel('Number of MNs in the CSTs')
plt.ylabel('Mean coherence in the 0-5hz bandwidth')
plt.title(f"Increase in coherence in the 0-5hz bandwidth (Y) as the number of CSTS' MNs (X) increase => Ground-truth VS fitted data \n Estimated PCI (gamma) = {np.round(gamma*1000)/1000} \n Ground-truth PCI (ratio of common input variance VS independent input variance) = {np.round(PCI_ground_truth_without_inhib*1000)/10}%, ignoring inhibition \n")
plt.ylim(0,1)
plt.legend()
new_filename = f'PCI_fitted_vs_true_curve_SimonMethod.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
<code>
# Get recruitment threshold and discharge rates of motor units = only if chosing the "trapezoid" force target (because allows for gradual recruitment of motor units)
if target_type == 'trapezoid':
MN_mean_firing_rates = np.copy(firing_rates) # all motoneurons by index, regardless of whether they are valid or not
MN_std_firing_rates = np.zeros(nb_motoneurons_full_pool)
# Discharge rates duging the window of analyzis (the plateau section of the trapezoid)
MN_recruitment_thresholds_by_force = np.full(firing_rates.shape, np.nan) # in % MVC
MN_recruitment_thresholds_by_excitatory_input = np.full(firing_rates.shape, np.nan) # in milliSiemens of excitatory input signal
spike_trains_full, binary_spike_trains_full = Get_binary_spike_trains(monitor_spikes_motoneurons, duration_with_ignored_window)
for mni in range(nb_motoneurons_full_pool):
# RT as force during the median time of the first 3 firings
temp_RT = (spike_trains_full[mni]/second)*fsamp
temp_RT = np.median(temp_RT[0:2])
if np.isnan(temp_RT)==False:
MN_recruitment_thresholds_by_force[mni] = output_force[int(np.round(temp_RT))]
MN_recruitment_thresholds_by_excitatory_input[mni] = final_mean_excit_input[int(np.round(temp_RT))]
# RT as force when the MN starts discharging with a rate > 5 pps (first ISI < 0.2)
# MN_recruitment_thresholds[mni] = nan
# spike_count = 0
# for ISI_i in np.diff(spike_trains_full[mni]):
# if ISI_i < ISI_threshold_for_RT*second:
# MN_recruitment_thresholds[mni] = output_force[int(np.round((spike_trains_full[mni][spike_count]/second)*fsamp))]
# break
# spike_count += 1
if len(spike_trains_full[mni]) > 1:
MN_std_firing_rates[mni] = np.std(1/np.diff(spike_trains_full[mni]))
else:
MN_std_firing_rates[mni] = nan
MN_recruitment_thresholds_by_force_only_selected_idx = np.copy(MN_recruitment_thresholds_by_force)
MN_recruitment_thresholds_by_force_only_selected_idx = MN_recruitment_thresholds_by_force_only_selected_idx[selected_motor_units].reshape(-1, 1)
# MN_recruitment_thresholds_by_force_only_selected_idx = MN_recruitment_thresholds_by_force_only_selected_idx.reshape(-1, 1)
MN_recruitment_thresholds_by_input_only_selected_idx = np.copy(MN_recruitment_thresholds_by_excitatory_input)
MN_recruitment_thresholds_by_input_only_selected_idx = MN_recruitment_thresholds_by_input_only_selected_idx[selected_motor_units].reshape(-1, 1)
# MN_recruitment_thresholds_by_force_only_selected_idx = MN_recruitment_thresholds_by_force_only_selected_idx.reshape(-1, 1)
MN_mean_firing_rates_only_selected_idx = np.copy(MN_mean_firing_rates)
MN_mean_firing_rates_only_selected_idx = MN_mean_firing_rates[selected_motor_units].reshape(-1, 1)
# MN_mean_firing_rates_only_selected_idx = MN_mean_firing_rates.reshape(-1, 1)
# histogram of recruitment thresholds
plt.figure()
plt.hist(MN_recruitment_thresholds_by_force_only_selected_idx, density=True,
edgecolor='white', alpha=0.75, color='C1')
plt.xlim(0,target_force_level*1)
ylabel("Proportion")
xlabel("Recruitment threshold (% MVC)")
title("Histogram of recruitment tresholds" + plot_title_suffix)
new_filename = f'RT_force_histogram.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
# histogram of recruitment thresholds
plt.figure()
plt.hist(MN_recruitment_thresholds_by_input_only_selected_idx, density=True,
edgecolor='white', alpha=0.75, color=[1,0.7,0])
ylabel("Proportion")
xlabel("Excitatory input (milliSiemens)")
title("Histogram of recruitment tresholds" + plot_title_suffix)
new_filename = f'RT_ecit_input_histogram.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
true_false_continuously_firing_MUs = np.zeros(nb_motoneurons_full_pool)
true_false_continuously_firing_MUs[valid_MUs_idx] = 1
true_false_sampled_motor_unit = np.zeros(nb_motoneurons_full_pool)
true_false_sampled_motor_unit[selected_motor_units] = 1
sampling_probability_dsitribution_to_save = np.zeros(nb_motoneurons_full_pool)
if subsample_MUs_for_analysis == True:
sampling_probability_dsitribution_to_save[valid_MUs_idx] = sampling_probability_distribution
else:
sampling_probability_dsitribution_to_save[valid_MUs_idx] = 1
# Save recruitment thresholds and firing rates
RT_mean_DR_dataframe = pd.DataFrame({
'MU_idx': np.arange(nb_motoneurons_full_pool),
'Continusouly_firing': true_false_continuously_firing_MUs,
'Sampled_for_analyzis': true_false_sampled_motor_unit,
'Recruitment_threshold_force': MN_recruitment_thresholds_by_force,
'Recruitment_threshold_input': MN_recruitment_thresholds_by_excitatory_input,
'Mean_firing_rate': MN_mean_firing_rates,
'STD_firing_rate': MN_std_firing_rates,
'Soma_size': motoneuron_soma_diameters,
'Recruitment_threshold_input_clean': MN_recruitment_thresholds_by_excitatory_input_clean_MVC,
'Recruitment_threshold_input_ratio': MN_recruitment_thresholds_by_excitatory_input_ratio,
'Probability_of_being_sampled': sampling_probability_dsitribution_to_save,
'Inhibition_weight_(1st_inhibitory_input)': MN_inhibition_weights[0],
})
new_filename = f'Individual_MUs_results.csv'
save_file_path = os.path.join(new_directory, new_filename)
RT_mean_DR_dataframe.to_csv(save_file_path, index=False)
from sklearn.linear_model import LinearRegression
from sklearn.metrics import r2_score
# Create a Linear Regression model
model = LinearRegression()
# Fit the model
model.fit(MN_recruitment_thresholds_by_force_only_selected_idx, MN_mean_firing_rates_only_selected_idx)
# Get the slope (coefficient)
slope = model.coef_[0]
# Predict the Y values using the fitted model
Y_pred = model.predict(MN_recruitment_thresholds_by_force_only_selected_idx)
# Calculate R squared
r_squared = r2_score(MN_mean_firing_rates_only_selected_idx, Y_pred)
plt.figure()
plt.scatter(MN_recruitment_thresholds_by_force_only_selected_idx,MN_mean_firing_rates_only_selected_idx,s=70,alpha=0.35,color='C1')
plt.plot(MN_recruitment_thresholds_by_force_only_selected_idx, Y_pred,color='C3',alpha=0.5, linewidth=3, label=f'Linear regression (slope = {np.round(slope[0]*100)/100}; R² = {np.round(r_squared*100)/100})')
plt.xlabel('Recruitment threshold (% MVC)')
plt.ylabel("Mean firing rate on the plateau (pps)")
plt.ylim(0,(np.ceil(max(MN_mean_firing_rates)/10)*10)+1)
plt.title("Recruitment threshold (in % MVC) to mean discharge rate relationship" + plot_title_suffix)
plt.legend()
plt.xlim(0,target_force_level)
new_filename = f'RT_force_to_mean_DR_relationship.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
# Create a Linear Regression model
model = LinearRegression()
# Fit the model
model.fit(MN_recruitment_thresholds_by_input_only_selected_idx, MN_mean_firing_rates_only_selected_idx)
# Get the slope (coefficient)
slope = model.coef_[0]
# Predict the Y values using the fitted model
Y_pred = model.predict(MN_recruitment_thresholds_by_input_only_selected_idx)
# Calculate R squared
r_squared = r2_score(MN_mean_firing_rates_only_selected_idx, Y_pred)
plt.figure()
plt.scatter(MN_recruitment_thresholds_by_input_only_selected_idx,MN_mean_firing_rates_only_selected_idx,s=70,alpha=0.35,color=[1,0.7,0])
plt.plot(MN_recruitment_thresholds_by_input_only_selected_idx, Y_pred, color=[0.8,0.25,0],alpha=0.5, linewidth=3, label=f'Linear regression (slope = {np.round(slope[0]*100)/100}; R² = {np.round(r_squared*100)/100})')
plt.xlabel('Recruitment threshold (excitatory input, milliSiemens)')
plt.ylabel("Mean firing rate on the plateau (pps)")
plt.ylim(0,(np.ceil(max(MN_mean_firing_rates)/10)*10)+1)
plt.title("Recruitment threshold (excitatory input) to mean discharge rate relationship" + plot_title_suffix)
plt.legend()
new_filename = f'RT_input_to_mean_DR_relationship.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
mean_inhib_weights_of_active_MUs = 0
if nb_inhibitory_input > 0:
for inhibiti in range(nb_inhibitory_input):
mean_inhib_weights_of_active_MUs += np.mean(MN_inhibition_weights[inhibiti][selected_motor_units])
mean_inhib_weights_of_active_MUs = mean_inhib_weights_of_active_MUs / nb_inhibitory_input
### Force output
force_output_window_of_analyzis = output_force[samples_for_analyzis]
force_output_error_window_of_analyzis = np.abs(output_force[samples_for_analyzis] - target_force[samples_for_analyzis])
mean_force_error_window_of_analyzis = np.mean(force_output_error_window_of_analyzis)
mean_force_output_window_of_analyzis = np.mean(force_output_window_of_analyzis)
std_force_output_window_of_analyzis = np.std(force_output_window_of_analyzis)
### Mean excitatory input during the plateau
# np.mean(final_mean_excit_input[samples_for_analyzis])
### Main results
main_results_dataframe = pd.DataFrame({
'Simulation_name': sim_name,
'Window_of_analyzis_duration': (window_analyzis_end-window_analyzis_begin)/fsamp,
'Ground_truth_PCI_ignoring_inhibition': [PCI_ground_truth_without_inhib], #[] to make sure at least the first value is an array
'PCSI_estimation': PCI_estimated,
'VAF_for_PC1': cumulative_explained_variance[0],
'Slope_of_RT_mean_DR_relationship': slope[0],
'Nb_of_continously_active_MUs': len(valid_MUs_idx),
'Nb_of_sampled_MUs_for_analyzis': len(selected_motor_units),
'Mean_DR': np.mean(MN_mean_firing_rates_only_selected_idx),
'std_DR': np.std(MN_mean_firing_rates_only_selected_idx),
'Mean_RT': np.mean(MN_recruitment_thresholds_by_force_only_selected_idx),
'std_RT': np.std(MN_recruitment_thresholds_by_force_only_selected_idx),
'0-5hz power of common input': common_noise_power_intergal_0_5_hz,
'0-5hz power of inhibition': inhibitory_input_power_integral_0_5_hz_mean,
'0-5hz power of independent excitatory noise': independent_excit_noise_power_integral_0_5_hz_mean,
'Mean weight of inhibition': mean_inhib_weights_of_active_MUs,
'Mean torque': mean_force_output_window_of_analyzis,
'Mean torque error': mean_force_error_window_of_analyzis,
'Mean excitatory input on plateau': np.mean(final_mean_excit_input[samples_for_analyzis]),
'Torque variability (torque std)': std_force_output_window_of_analyzis
})
new_filename = f'Main_results.csv'
save_file_path = os.path.join(new_directory, new_filename)
main_results_dataframe.to_csv(save_file_path, index=False)
### Cumulative Explained Variance
VAF_cumsum_dataframe = pd.DataFrame({
'PC': np.arange(len(cumulative_explained_variance))+1,
'Cumulative_VAF': cumulative_explained_variance
})
new_filename = f'Cumulative_VAF_PCA.csv'
save_file_path = os.path.join(new_directory, new_filename)
VAF_cumsum_dataframe.to_csv(save_file_path, index=False)
</code>
<code>
plt.figure(figsize=(30,10))
# Plot mean firing rates as a curve according to MN index
plt.subplot(231)
plt.plot(firing_rates, alpha = 0.5, linewidth = 2, label = "all MNs", color='C0')
plt.scatter(selected_motor_units, firing_rates[selected_motor_units], color = 'royalblue', alpha = 0.5, linewidth = 2, label = "Selected MNs")
plt.ylim(-0.5,np.ceil(np.max(firing_rates[selected_motor_units]))+0.5)
plt.xlim(0-np.round(nb_motoneurons_full_pool/10),nb_motoneurons_full_pool+np.round(nb_motoneurons_full_pool/10))
plt.ylabel("Mean firing rate (pps)")
plt.xlabel(f"MN index (0 = smallest MU; {nb_motoneurons_full_pool} = largest MU)")
plt.title("Mean firing rates of simulated MNs - MN index")
plt.legend()
plt.subplot(234)
plt.plot(motoneuron_soma_diameters,firing_rates, alpha = 0.5, linewidth = 2, label = "all MNs", color='C0')
plt.scatter(motoneuron_soma_diameters[selected_motor_units], firing_rates[selected_motor_units], color = 'royalblue', alpha = 0.5, linewidth = 2, label = "Selected MNs")
plt.ylim(-0.5,np.ceil(np.max(firing_rates[selected_motor_units]))+0.5)
plt.xlim(min_soma_diameter-np.round(min_soma_diameter/10),max_soma_diameter+np.round(min_soma_diameter/10))
plt.ylabel("Mean firing rate (pps)")
plt.xlabel(f"Motoneuron size (soma diameter in micrometers)")
plt.title("Mean firing rates of simulated MNs - MN size")
plt.legend()
plt.subplot(232)
plt.plot(MN_recruitment_thresholds_by_force, alpha = 0.5, linewidth = 2, label = "all MNs", color='C1')
plt.scatter(selected_motor_units, MN_recruitment_thresholds_by_force[selected_motor_units], color = 'C1', alpha = 0.5, linewidth = 2, label = "Selected MNs")
plt.ylim(0,35)
plt.xlim(0-np.round(nb_motoneurons_full_pool/10),nb_motoneurons_full_pool+np.round(nb_motoneurons_full_pool/10))
plt.ylabel("Recruitment threshold (% MVC)")
plt.xlabel(f"MN index (0 = smallest MU; {nb_motoneurons_full_pool} = largest MU)")
plt.title("Recruitment thresholds (by force) of simulated MNs - MN index")
plt.legend()
plt.subplot(235)
plt.plot(motoneuron_soma_diameters,MN_recruitment_thresholds_by_force, alpha = 0.5, linewidth = 2, label = "all MNs", color='C1')
plt.scatter(motoneuron_soma_diameters[selected_motor_units], MN_recruitment_thresholds_by_force[selected_motor_units], color = 'C1', alpha = 0.5, linewidth = 2, label = "Selected MNs")
plt.ylim(0,35)
plt.xlim(min_soma_diameter-np.round(min_soma_diameter/10),max_soma_diameter+np.round(min_soma_diameter/10))
plt.ylabel("Recruitment threshold (% MVC)")
plt.xlabel(f"Motoneuron size (soma diameter in micrometers)")
plt.title("Recruitment thresholds (by force) of simulated MNs - MN size")
plt.legend()
plt.subplot(233)
plt.plot(MN_recruitment_thresholds_by_excitatory_input, alpha = 0.5, linewidth = 2, label = "all MNs", color=[1,0.7,0])
plt.scatter(selected_motor_units, MN_recruitment_thresholds_by_excitatory_input[selected_motor_units], color = [1,0.7,0], alpha = 0.5, linewidth = 2, label = "Selected MNs")
plt.xlim(0-np.round(nb_motoneurons_full_pool/10),nb_motoneurons_full_pool+np.round(nb_motoneurons_full_pool/10))
plt.ylabel("Recruitment threshold (excitatory input, in milliSiemens)")
plt.xlabel(f"MN index (0 = smallest MU; {nb_motoneurons_full_pool} = largest MU)")
plt.title("Recruitment thresholds (by input) of simulated MNs - MN index")
plt.legend()
plt.subplot(236)
plt.plot(motoneuron_soma_diameters,MN_recruitment_thresholds_by_excitatory_input, alpha = 0.5, linewidth = 2, label = "all MNs", color=[1,0.7,0])
plt.scatter(motoneuron_soma_diameters[selected_motor_units], MN_recruitment_thresholds_by_excitatory_input[selected_motor_units], color = [1,0.7,0], alpha = 0.5, linewidth = 2, label = "Selected MNs")
plt.xlim(min_soma_diameter-np.round(min_soma_diameter/10),max_soma_diameter+np.round(min_soma_diameter/10))
plt.ylabel("Recruitment threshold (excitatory input, in milliSiemens)")
plt.xlabel(f"Motoneuron size (soma diameter in micrometers)")
plt.title("Recruitment thresholds (by input) of simulated MNs - MN size")
plt.legend()
plt.suptitle("Firing rates and recruitment thresholds")
new_filename = f'MN_RT_and_DR_properties.png'
save_file_path = os.path.join(new_directory, new_filename)
plt.savefig(save_file_path)
plt.show()
</code>
|
{
"filename": "inhibition_simulation_MN_sim_PAIN.ipynb",
"repository": "FrancoisDernoncourt/Pain",
"query": "transformed_from_existing",
"size": 198441,
"sha": ""
}
|
# collab_3b_import_fragments_scPrinter_3.ipynb
Repository: ruochiz/asthma
# Import fragments with ```scPrinter```
- Function to use: [scprinter.pp.import_fragments](https://ruochiz.com/scprinter_doc/reference/_autosummary/scprinter.pp.import_fragments.html#scprinter.pp.import_fragments)
- Tutorial to follow: [scPrinter PBMC scATAC-seq tutorial](https://ruochiz.com/scprinter_doc/tutorials/PBMC_scATAC_tutorial.html#Now-let's-use-scPrinter-for-some-basic-exploratory-analysis-to-get-a-better-idea-of-the-dataset)
## 0. Imports
<code>
%load_ext autoreload
%autoreload 2
import scprinter as scp
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import time
import pandas as pd
import numpy as np
import os
import pickle
import torch
import matplotlib as mpl
mpl.rcParams['pdf.fonttype'] = 42
from scanpy.plotting.palettes import zeileis_28
from tqdm.contrib.concurrent import *
from tqdm.auto import *
import anndata
import scanpy as sc
import statistics as stat
import json
import csv
import re
import copy
from sklearn.preprocessing import OneHotEncoder
</code>
<code>
import snapatac2 as snap
</code>
### 0.1 Setup
<code>
# Specify the reference genome. This must match that of your ATAC fragments file
genome = scp.genome.mm10
genome
</code>
## 1. Paths
### 1.1 Data directories
<code>
master_data_dir = '/bap/bap/collab_asthma_multiome/'
</code>
<code>
# Create small lambda function to get the path to the data, input variable being sample name
get_condition_fragments_path = lambda sample_name_bc, sample_name_frag: os.path.join(master_data_dir, 'ATAC', 'ATACFragmentFiles_Asthma', sample_name_bc, f'{sample_name_frag}_atac_fragments.tsv.gz')
get_condition_valid_barcodes_path = lambda sample_name: os.path.join(master_data_dir, 'outputs', 'ATAC', '2_Analysis_Outputs', '1a_ChromVAR_Inputs', f'{sample_name}_valid_barcodes.txt')
</code>
<code>
# outputs
printer_h5ad_output_dir = os.path.join(master_data_dir, 'ATAC', '2_Analysis_Outputs', '1b_ChromVAR_scPrinter_object')
printer_h5ad_output_path = os.path.join(printer_h5ad_output_dir, 'Asthma_Multiome_Collab_scPrinter.h5ad')
# if the output directory does not exist, create it
if not os.path.exists(printer_h5ad_output_dir):
os.makedirs(printer_h5ad_output_dir)
</code>
### 1.2 Prep paths
<code>
# Sample names
sample_names_bc = ['NT',
'OVA_C',
'OVA',
'PBS_C',
'PBS'
]
# on-disk fragments files are named slightly differently
sample_names_load_fragments = ['NT',
'OVAC',
'OVA',
'PBSC',
'PBS'
]
</code>
<code>
# to per-condition fragments
fragment_paths_l = []
valid_barcodes_l = [] # order-matched to fragment_paths_l
for sample_name_fragments_i, sample_name_bc_i in zip(sample_names_load_fragments, sample_names_bc):
fragment_paths_l.append(get_condition_fragments_path(sample_name_bc_i, sample_name_fragments_i))
valid_barcodes_l.append(get_condition_valid_barcodes_path(sample_name_bc_i))
</code>
<code>
fragment_paths_l
</code>
<code>
valid_barcodes_l
</code>
<code>
# TODO: you'll likely need txt files of barcodes:subtype pairings per condition too,
# when you do the manual t-test later and need to group barcodes by subtype
</code>
## 2. ```scPrinter``` analysis
### 2.1 Initialize the scPrinter object
When you finish using the object, run ```printer.close()``` otherwise you won't be able to load it properly next time.
**Note Feb 24, 2025:** the QC filters of ```import_fragments()```
```min_num_fragments=1000, min_tsse=7```
may have lowered # pass QC cells from 7797 to 7747
From source code, ```min_tsse``` is no longer used
```# these are historical_kwargs that snapatac2 takes, but not anymore
for historical_kwarg in ["min_tsse", "low_memory"]:
if historical_kwarg in kwargs:
del kwargs[historical_kwarg]
```
For QC consistency, we will not re-filter on # fragments because we have already QC'd in the R notebook. This should produce a ```printer``` object with the same # cells (7797) as the barcode preparation notebook.
<code>
import time
start = time.time()
# TODO: use lists of frag paths and lists of prepared pass-QC barcode txt files
printer = scp.pp.import_fragments(
path_to_frags=fragment_paths_l,
barcodes=valid_barcodes_l,
savename=printer_h5ad_output_path,
sample_names=sample_names_bc,
genome=genome,
min_num_fragments=0, min_tsse=7,
sorted_by_barcode=False,
low_memory=False,
)
end = time.time()
print(f"Time taken to import fragments: {end - start} seconds")
</code>
<code>
printer
</code>
**Always, always remember to close the object!**
<code>
printer.close()
</code>
<code>
printer_h5ad_output_path
</code>
# END
|
{
"filename": "collab_3b_import_fragments_scPrinter_3.ipynb",
"repository": "ruochiz/asthma",
"query": "transformed_from_existing",
"size": 17131,
"sha": ""
}
|
# HH_1.ipynb
Repository: Mark-Kramer/Case-Studies-Python
# The Hodgkin-Huxley model
In this notebook we will use Python to simulate the Hodgkin-Huxley (HH) neuron model. This model is arguably the *most* important computational model in neuroscience. We'll focus here on simulating this model and understanding its pieces.
## Background information about the HH model
Here's a video that describes some of the biophysical details of the HH model:
<code>
from IPython.lib.display import VimeoVideo
VimeoVideo('140084450')
</code>
Here are some additional usual videos and references:
- <a href="http://klewel.com/conferences/epfl-neural-networks/index.php?talkID=4" rel="external">Lecture by Prof. Gerstner, *Detailed Neuron Model (a)*</a>
- <a href="http://klewel.com/conferences/epfl-neural-networks/index.php?talkID=5" rel="external">Lecture by Prof. Gerstner, *Detailed Neuron Model (b)*</a>
## Preliminaries
Before beginning, let's load in the Python packages we'll need:
<code>
from pylab import *
%matplotlib
rcParams['figure.figsize']=(12,3) # Change the default figure size
</code>
In addition, let's import the functions we'll need to simulate the HH model, which are available on this repository:
<code>
from HH_functions import HH
</code>
## Part 1: The Hodgkin-Huxley (HH) equation code.
To start, let's examine the code for the HH model. We can do so in (at least) two ways.
- Go to the Case Studies repository, and examine the Python file
`HH_functions.py`
- Examine the code inline with `inspect`
<code>
import inspect
inspect.getsourcelines(HH)
</code>
<div class="question">
**Q:** Examine this code. Can you make sense of it? Can you identify the
gating variables? The rate functions? The equations that define the dynamics?
We'll answer these questions in this in notebook, but try so on your own first.
</div>
Whenever examining code, it's useful to consider the *inputs* to the code, and the *outputs* produced by the code. There are two inputs to `HH0`:
- `I0` = the current we inject to the neuron.
- `T0` = the total time of the simulation in [ms].
And there are five outputs:
- `V` = the voltage of neuron.
- `m` = activation variable for Na-current.
- `h` = inactivation variable for Na-current.
- `n` = activation variable for K-current.
- `t` = the time axis of the simulation (useful for plotting).
## Part 2: At low input current (`I0`), examine the HH dynamics.
To understand how the HH model works, we'll start by focusing on the
case when `I0` is small. Let's fix the input current to zero,
<code>
I0 = 0
</code>
and let's simulate the model for 100 ms,
<code>
T0 = 100
</code>
We've now defined both inputs to the `HH` function, and can execute it, as follows,
<code>
[V,m,h,n,t]=HH(I0,T0)
</code>
Notice that the function returns five outputs, which we assign to the variables `V`, `m`, `h`, `n`, and `t`.
<div class="question">
**Q:** What are the dynamics of the voltage (variable `V`) resulting
from this simulation?<br>
HINT: Plot `V` vs `t`.
</div>
<div class="question">
**Q:** What are the dynamics of the gating variables (`m`, `h`, `n`)
resulting from this simulation?<br>
HINT: Plot them!
</div>
<div class="question">
**Q:** What are the final values (after the 100 ms of simulation) of
`V`, `m`, `h`, and `n`?
</div>
### Observation for Part 2
At this value of input current (`I0=0`), the model dynamics
approach a "fixed point", whose location we can identify as a point in four dimensional space.
## Part 3: At high input current (`I0`), examine the HH dynamics of a spike.
Let's now increase the input current to the HH model and get this model
to generate repeated spiking activity. To do so, let's set,
<code>
I0 = 10
</code>
We can now simulate this model,
<code>
[V,m,h,n,t] = HH(I0,T0)
</code>
<div class="question">
**Q:** What happens to the dynamics?<br>
HINT: Plot V vs t.
</div>
### Observation for Part 3
You should have found that, at this value of input current, the model
**generates repeated spikes**.
Let's now explore how the combined gates
and dynamics evolve. To do so, let's start by focusing our plot on a
single spike. As a first step, we'll make a new figure with a seperate subfigure to plot
the voltage,
<code>
figure()
subplot(211)
</code>
This `subplot` command divides the figure into two rows, and one column, and tells Python we'll start in the first row. See Python Help for more details:
`subplot??`
Now, let's plot the voltage, and choose the time axis to focus on a single spike,
<code>
plot(t,V,'k')
xlim([42, 56])
ylabel('V [mV]');
</code>
Okay, we've now plotted the voltage dynamics for a single spike (and
colored the curve black). Let's now plot the three gating variables.
To do so, we'll move to the next subplot,
<code>
subplot(212);
</code>
(the next row in the figure). Within this subplot, let's start by displaying the gating variable `m` over the same x-limits,
<code>
plot(t,m,'r', label='m')
xlim([42, 56]);
</code>
Notice that, in the call to `plot` we included the input `label`. This will be useful when we create a legend ... <br><br>Within this subplot, we can also simultaneously show the gating
variables `h` and `n`,
<code>
plot(t,h,'b', label='h')
plot(t,n,'g', label='n');
</code>
Label the x-axis,
<code>
xlabel('Time [ms]');
</code>
Now, let's add a legend to help us keep track of the different curves,
<code>
legend();
</code>
<div class="question">
**Q:** Using the figure you created above, describe how the gates swing open and closed during a spike.
</div>
### ASIDE:
Here's a nice plotting trick, to link the x-axes of our two subfigures. Linking the axes is useful so that, when we zoom or move one subfigure, the other subfigure will match the x-axis.
<code>
figure()
ax1 = subplot(211); # Define axis for 1st subplot,
ax2 = subplot(212, sharex=ax1); # ... and link axis of 2nd subplot to the 1st.
ax1.plot(t,V,'k') # Plot the voltage in the first subplot,
xlim([42, 56]);
ax2.plot(t,m,'r', label='m') # ... and the gating variables in the other subplot.
ax2.plot(t,h,'b', label='h')
ax2.plot(t,n,'g', label='n');
xlabel('Time [ms]');
legend();
</code>
Now, in the figure, you may use the pan/zoom tool to adjust the linked subplots.
## Part 4: At high input current (`I0`), describe the dynamics of the conductances.
In Part 3, we explored how the three gates `m`, `h`, and `n` evolve
during a spike. By combining these terms, we can visualize how the
*conductances* evolve during a spike. To do so, let's stick with the
simulation results we generated in Part 3, and focus our plot on a
single spike,
<code>
figure()
ax1=subplot(311) # Make a subplot,
ax1.plot(t,V,'k') #... and plot the voltage,
xlim([42, 56]) #... focused on a single spike,
ylabel('V [mV]'); #... with y-axis labeled.
</code>
Now, to plot the conductances, let's define three new variables,
<code>
gNa0 = 120
gNa = gNa0*m**3*h # Sodium conductance
gK0 = 36
gK = gK0*n**4 # Potassium conductance
gL0 = 0.3
gL = gL0*ones(shape(gK)) # Leak conductance
</code>
<div class="question">
**Q:** Where do these terms come from?
</div>
Then, let's plot these conductances,
<code>
ax2 = subplot(312, sharex=ax1) #Make a second subplot,
ax2.plot(t,gNa,'m', label='gNa')#... and plot the sodium conductance,
ax2.plot(t,gK, 'g', label='gK') #... and plot the potassium conductance,
ax2.plot(t,gL, 'k', label='gL') #... and plot the leak conductance.
xlim([42, 56]) #... focused on a single spike,
xlabel('Time [ms]') #... label the x-axis.
ylabel('mS/cm^2') #... and label the y-axis.
legend(); #... make a legend.
</code>
<div class="question">
**Q:** How do the conductances evolve during a spike?
</div>
## Part 5: At high input current (`I0`), describe the dynamics of the *currents*.
In Part 4, we explored how the three conductances (`gNa`, `gK`, `gL`) evolve
during a spike. Let's now visualize how the *ionic currents* evolve
during a spike. To do so, let's stick with the same settings used in
Part 4 and examine the same simulation result. Again, we'll focus our plot
on a single spike.
Now, to plot the *current*, let's define the new variables,
<code>
gNa0 = 120
ENa = 115
INa = gNa0*m**3*h*(ENa-(V+65)) # Sodium current.
gK0 = 36
EK =-12
IK = gK0*n**4*(EK-(V+65)) # Potassium current.
gL0 = 0.3
EL = 10.6;
IL = gL0*(EL-(V+65)) # Leak current.
ax3=subplot(313, sharex=ax1) # Make a third subplot,
ax3.plot(t,INa,'m', label='INa') #... and plot the sodium current,
ax3.plot(t,IK, 'g', label='IK') #... and plot the potassium current,
ax3.plot(t,IL, 'k', label='IL') #... and plot the leak current.
xlim([42, 56]) #... focus on a single spike,
xlabel('Time [ms]') #... label the x-axis.
ylabel('mA/cm^2') #... and label the y-axis.
legend(); #... make a legend.
</code>
<div class="question">
**Q:** How do the conductances evolve during a spike?
</div>
<div class="question">
**Q:** You may notice a small, transient decrease in the sodium current `INa` near 47 ms. What causes this?
</div>
<a id="donate"></a>
## Donate
If you enjoy Case-Studies-Python, and would like to share your enjoyment with us, sponsor our coffee consuption <a href="https://www.paypal.com/donate/?hosted_button_id=DL8P5ZGS9962U">here</a>.
|
{
"filename": "HH_1.ipynb",
"repository": "Mark-Kramer/Case-Studies-Python",
"query": "transformed_from_existing",
"size": 21920,
"sha": ""
}
|
# openalex_get_works_by_list_of_persons.ipynb
Repository: hebosse/Jupyter-Notebooks
## Query OpenAlex for works authored by a person
This notebook queries the [OpenAlex API](https://docs.openalex.org/api) via its `/works` endpoint for works authored by a person. It takes an ORCID URL from a list of ORCID IDs as input which is used to filter for works where '`authorships.author.orcid`' matches the given ORCID URL.
The notebook iterates through the given list and displays all DOIs and their title.
<code>
# Prerequisites:
import requests # dependency to make HTTP calls
</code>
list_of_ids = inputfield
Note: all ORCID IDs must be in the form 'ID' and be seperated from the next with a comma.
<code>
list_of_ids=["0000-0001-5380-4449",
"0000-0001-5406-9458",
"0000-0003-3547-3257",
"0000-0003-3654-5267",
"0000-0003-4331-8695",
"0000-0003-4939-1666",
"0000-0003-4971-9991",]
</code>
We use it to query the OpenAlex API for works that specified the ORCID URL within their metadata in the field '`authorships.author.orcid`'.
Since the API uses [pagination](https://docs.openalex.org/api/get-lists-of-entities#pagination), we need to loop through all pages to get the complete result set.
<code>
# OpenAlex endpoint to query for works
OPENALEX_API_WORKS = "https://api.openalex.org/works"
# query all works that are connected to orcid
def query_openalex_for_person2works(orcid):
page = 1
max_page = 1
while page <= max_page:
params = {'filter': 'authorships.author.orcid:'+orcid, 'page': page}
response = requests.get(url=OPENALEX_API_WORKS,
params=params,
headers= {'Accept': 'application/json'})
response.raise_for_status()
result=response.json()
# calculate max page number in first loop
if max_page == 1:
max_page = determine_max_page(result)
page = page + 1
yield result
# calculate max number of result pages
def determine_max_page(response_data):
item_count = response_data['meta']['count']
items_per_page = response_data['meta']['per_page']
max_page_ceil = item_count // items_per_page + bool(item_count % items_per_page)
return max_page_ceil
</code>
From the resulting list of works we extract and print out title and DOI.
*Note: works that do not have a DOI assigned, will not be printed.*
<code>
# from the result pages we get from the OpenAlex API, extract the data about works
def extract_works_from_page(page):
return [work for work in page.get('results') or []]
# extract DOI from work
def extract_doi(work):
doi=work.get('ids', {}).get('doi') or ""
doi_id=doi.replace("https://doi.org/", "") if doi else doi
title=work.get('display_name', "")
return doi_id, title
def main_search(orcid):
list_of_pages=query_openalex_for_person2works(orcid)
for page in list_of_pages or []:
works=extract_works_from_page(page)
for work in works or []:
doi,title=extract_doi(work)
if doi:
print(f"{doi},{title}")
# main programm:
for item in list_of_ids:
main_search(item)
</code>
|
{
"filename": "openalex_get_works_by_list_of_persons.ipynb",
"repository": "hebosse/Jupyter-Notebooks",
"query": "transformed_from_existing",
"size": 268497,
"sha": ""
}
|
# Measuring_similarity.ipynb
Repository: amnh/BridgeUP-STEM-Xiao
# Week 8: Measuring similarity
### how do you do it?
Biology has, until fairly recently, been a very qualitative science. Computational and statistical methods are changing that, however -- we can build phylogenies based on genomic data instead of observations of physical characteristics. What's more, we can quantify how confident we are in the trees we build. We'll discuss a few of these methods for measuring similarity in the context of biology.
### "I've got confidence in confidence..."
-- Maria von Trapp
So you've just completed your first few BLAST searches -- with our FASTA output file in hand, we'll complete a very _familiar_ first comparison algo in our pairs.
### Hamming distance
Remember the sequence comparison function you wrote with Gabrielle in your molecular genomics unit? That's a classic [__Hamming__](https://en.wikipedia.org/wiki/Hamming_distance) algorithm. In the cell below, jog your memory + write out that function. It should return the __percentage similarity__ between both sequences!
<code>
def hamming_dist(seq1, seq2):
#your code here!
</code>
### Edit distance
This is a hot problem right now in theoretical computer science, as it's got tons of applications to non-biological problems, too! Edit distance is like hamming distance's older cousin -- what if we've got a single insertion or deletion in our nucleotide sequence? Our entire comparison would be thrown off when, in reality, we only had a single mutated base! Edit distance takes this into account by tallying the number of insertions, deletions, etc need to transform one sequence into another. Try your hand at this comparison technique with the following problem from [Rosalind](http://rosalind.info/problems/edit/). For the sake of record-keeping, feel free to copy your code below when you're done:
<code>
def edit_dist(seq1, seq2):
#your code here!
</code>
### e-values
...sound more complex than they really are. In short: an e-value measures the number of hits you'd expect to see _by chance_ when you run a BLAST query on a database. WARNING: they are _not_ probabilities -- they're expected values. Because they measure the number of hits you'd expect to see from a given database, this number might increase with the size of the database. A statistician might say that the e-value is a measure of __noise__ in your database.
Can you locate the e values for your BLAST searches on your two mystery files? What are they? Are they 'good' or 'bad'? (and how do you know?)
### Genetic distance
This is a tricky one -- and one of the most useful for understanding phylogenetics! While the previous methods we've discussed for measuring similarity rely on a by-base-pair analysis of the sequences in question, genetic distance calls on some [__pretty gnarly formulas__](https://en.wikipedia.org/wiki/Genetic_distance) to get the job done. The idea is to measure gene __loci__ to track things like __genetic drift__ and other origins of biodiversity.
Take a look at the following documentation for Biopython's genetic distance package: http://biopython.org/DIST/docs/api/Bio.Phylo.TreeConstruction.DistanceCalculator-class.html
Based on the example in the documentation, how would you script a command to build a distance matrix for our FASTA alignment file? Import all required packages at the top of your document. Also: remember that file format is important (wink wink, nudge nudge)!
<code>
## your distance matrix code below!
</code>
|
{
"filename": "Measuring_similarity.ipynb",
"repository": "amnh/BridgeUP-STEM-Xiao",
"query": "transformed_from_existing",
"size": 6227,
"sha": ""
}
|
# analysis_3.ipynb
Repository: dib-lab/2014-mrnaseq-models
|
{
"filename": "analysis_3.ipynb",
"repository": "dib-lab/2014-mrnaseq-models",
"query": "transformed_from_existing",
"size": 117715,
"sha": ""
}
|
# PPTASs10_1.ipynb
Repository: ARJUN198SINGH/Assignment-sol.
1. Can you explain the concept of feature extraction in convolutional neural networks (CNNs)?
2. How does backpropagation work in the context of computer vision tasks?
3. What are the benefits of using transfer learning in CNNs, and how does it work?
4. Describe different techniques for data augmentation in CNNs and their impact on model performance.
5. How do CNNs approach the task of object detection, and what are some popular architectures used for this task?
6. Can you explain the concept of object tracking in computer vision and how it is implemented in CNNs?
7. What is the purpose of object segmentation in computer vision, and how do CNNs accomplish it?
8. How are CNNs applied to optical character recognition (OCR) tasks, and what challenges are involved?
9. Describe the concept of image embedding and its applications in computer vision tasks.
10. What is model distillation in CNNs, and how does it improve model performance and efficiency?
11. Explain the concept of model quantization and its benefits in reducing the memory footprint of CNN models.
12. How does distributed training work in CNNs, and what are the advantages of this approach?
13. Compare and contrast the PyTorch and TensorFlow frameworks for CNN development.
14. What are the advantages of using GPUs for accelerating CNN training and inference?
15. How do occlusion and illumination changes affect CNN performance, and what strategies can be used to address these challenges?
16. Can you explain the concept of spatial pooling in CNNs and its role in feature extraction?
17. What are the different techniques used for handling class imbalance in CNNs?
18. Describe the concept of transfer learning and its applications in CNN model development.
19. What is the impact of occlusion on CNN object detection performance, and how can it be mitigated?
20. Explain the concept of image segmentation and its applications in computer vision tasks.
21. How are CNNs used for instance segmentation, and what are some popular architectures for this task?
22. Describe the concept of object tracking in computer vision and its challenges.
23. What is the role of anchor boxes in object detection models like SSD and Faster R-CNN?
24. Can you explain the architecture and working principles of the Mask R-CNN model?
25. How are CNNs used for optical character recognition (OCR), and what challenges are involved in this task?
26. Describe the concept of image embedding and its applications in similarity-based image retrieval.
27. What are the benefits of model distillation in CNNs, and how is it implemented?
28. Explain the concept of model quantization and its impact on CNN model efficiency.
29. How does distributed training of CNN models across multiple machines or GPUs improve performance?
30. Compare and contrast the features and capabilities of PyTorch and TensorFlow frameworks for CNN development.
31. How do GPUs accelerate CNN training and inference, and what are their limitations?
32. Discuss the challenges and techniques for handling occlusion in object detection and tracking tasks.
33. Explain the impact of illumination changes on CNN performance and techniques for robustness.
34. What are some data augmentation techniques used in CNNs, and how do they address the limitations of limited training data?
35. Describe the concept of class imbalance in CNN classification tasks and techniques for handling it.
36. How can self-supervised learning be applied in CNNs for unsupervised feature learning?
37. What are some popular CNN architectures specifically designed for medical image analysis tasks?
38. Explain the architecture and principles of the U-Net model for medical image segmentation.
39. How do CNN models handle noise and outliers in image classification and regression tasks?
40. Discuss the concept of ensemble learning in CNNs and its benefits in improving model performance.
41. Can you explain the role of attention mechanisms in CNN models and how they improve performance?
42. What are adversarial attacks on CNN models, and what techniques can be used for adversarial defense?
43. How can CNN models be applied to natural language processing (NLP) tasks, such as text classification or sentiment analysis?
44. Discuss the concept of multi-modal CNNs and their applications in fusing information from different modalities.
45. Explain the concept of model interpretability in CNNs and techniques for visualizing learned features.
46. What are some considerations and challenges in deploying CNN models in production environments?
47. Discuss the impact of imbalanced datasets on CNN training and techniques for addressing this issue.
48. Explain the concept of transfer learning and its benefits in CNN model development.
49. How do CNN models handle data with missing or incomplete information?
50. Describe the concept of multi-label classification in CNNs and techniques for solving this task.
1. Feature extraction in convolutional neural networks (CNNs) refers to the process of automatically identifying and extracting meaningful features or patterns from raw input data, such as images. CNNs are designed to automatically learn hierarchical representations of data by applying convolutional filters or kernels to the input image. These filters slide over the input image, computing dot products between the filter weights and local patches of the image. By applying multiple filters, CNNs can extract various types of features, such as edges, corners, and textures, at different spatial scales.
2. Backpropagation in the context of computer vision tasks is a learning algorithm used to update the weights of a neural network, including CNNs, based on the error or loss between the predicted output and the true output. In computer vision tasks, such as image classification, the network's output represents the predicted class probabilities for a given input image. During backpropagation, the error is propagated backward through the network, and the gradients of the network's weights with respect to the error are computed using the chain rule. These gradients are then used to update the weights using an optimization algorithm like stochastic gradient descent (SGD), aiming to minimize the error and improve the network's performance.
3. Transfer learning in CNNs refers to the practice of leveraging pre-trained models on large-scale datasets and applying them to new tasks or domains with limited labeled data. The benefits of transfer learning include:
- **Feature Extraction:** Pre-trained CNN models can serve as powerful feature extractors. The lower layers of a CNN capture generic visual features that are applicable to various tasks, while the higher layers learn more task-specific features. By using a pre-trained model, one can benefit from the low-level feature extraction capabilities.
- **Reduced Training Time:** Training a CNN from scratch on large datasets can be computationally expensive. Transfer learning allows starting from a pre-trained model, which reduces the training time significantly.
- **Improved Generalization:** Pre-trained models have learned representations from diverse data, enabling better generalization to new tasks or domains, especially when the labeled data is limited.
Transfer learning involves freezing the pre-trained layers and only training the final layers or adding a few additional layers to adapt the model to the new task. The pre-trained weights are usually fine-tuned with the new data to align the features with the target domain.
4. Data augmentation techniques in CNNs involve artificially creating new training samples by applying various transformations to the existing data. Some common techniques include:
- **Horizontal and Vertical Flipping:** Flipping the image horizontally or vertically to create new samples. This is useful when the orientation of objects does not affect the target label.
- **Rotation and Scaling:** Applying rotations or scaling transformations to the image to simulate variations in object orientation and size.
- **Translation:** Shifting the image horizontally or vertically to simulate slight changes in object position.
- **Noise Injection:** Adding random noise to the image to make the model more robust to noise in real-world scenarios.
- **Crop and Pad:** Taking random crops or padding the image to different sizes to simulate object occlusion or variations in image size.
Data augmentation helps increase the diversity and variability of the training data, reducing overfitting and improving the model's ability to generalize to new, unseen data.
5. CNNs approach object detection by dividing the task into two main components: region proposal and object classification. The process typically involves the following steps:
- **Region Proposal:** CNN-based object detection methods generate a set of region proposals or candidate bounding boxes that are likely to contain objects. This can be achieved using techniques like selective search or region proposal networks (RPNs), which generate proposals based on objectness scores and anchor boxes.
- **Feature Extraction:** The proposed regions or the entire input image are fed into a CNN to extract features. The CNN processes the input using convolutional layers to capture spatial hierarchies of features at different scales.
- **Region Classification:** The extracted features are then used to classify the proposed regions into different object classes. This can be done by applying fully connected layers on top of the CNN features and using softmax or sigmoid activation functions for multi-class or binary classification, respectively.
- **Bounding Box Refinement:** In addition to classification, object detection also involves refining the proposed bounding boxes. Regression layers in the network are used to adjust the coordinates of the proposed boxes to align them better with the objects' true locations.
Popular CNN architectures used for object detection include Faster R-CNN, Single Shot MultiBox Detector (SSD), and You Only Look Once (YOLO).
6. Object tracking in computer vision involves the task of continuously locating and following a specific object of interest over a sequence of frames in a video. CNNs can be used for object tracking by employing a two-step process:
- **Target Initialization:** Initially, the target object is manually selected or automatically detected in the first frame of the video sequence. A CNN model, such as a siamese network, is used to learn a target representation or template based on the appearance of the object.
- **Online Tracking:** In subsequent frames, the CNN model is applied to search for the target object by comparing the learned template with patches in the current frame. The goal is to find the patch that is most similar to the target template. This similarity calculation is typically performed using techniques like correlation filters or cosine similarity.
The selected patch is considered as the new target location, and the process is repeated in the next frame. The CNN model is updated online to adapt to changes in the object's appearance.
7. Object segmentation in computer vision aims to identify and segment individual objects within an image, assigning a unique label to each pixel or region belonging to a specific object. CNNs can accomplish object segmentation using fully convolutional networks (FCNs) or similar architectures. The process involves:
- **Encoder:** The CNN architecture consists of an encoder component that performs hierarchical feature extraction. The encoder typically consists of convolutional and pooling layers that downsample the spatial dimensions while increasing the number of channels, capturing contextual information at different scales.
- **Decoder:** The decoder component takes the encoder's feature maps and upsamples them to the original image resolution. Upsampling is often performed using transposed convolutions or interpolation. Skip connections, which connect the corresponding encoder and decoder layers, are used to fuse low-level and high-level features, allowing precise localization.
- **Output:** The output of the CNN is a segmentation map, where each pixel is assigned a label corresponding to the object it belongs to. Softmax or sigmoid activation functions are applied to the final convolutional layer to obtain class probabilities for each pixel.
By training the CNN on annotated images, where each pixel is labeled with the ground truth class, the network learns to segment objects based on their visual appearance and context.
8. CNNs are applied to optical character recognition (OCR) tasks by treating the task as an image classification problem. The process typically involves the following steps:
- **Preprocessing:** The input document or image containing text is preprocessed to enhance its quality and facilitate the recognition process. This may involve operations like resizing, normalization, and noise reduction.
- **Segmentation:** The preprocessed image is divided into individual character or text line regions. This step separates the characters or lines from the background and other elements.
- **Character Classification:** Each segmented character is then passed through a CNN model for classification. The CNN extracts relevant features from the character image and predicts the corresponding character class. The output can be a single character
or a probability distribution over a set of characters.
- **Post-processing:** The recognized characters are usually subject to post-processing steps to improve the accuracy of the OCR system. This may involve techniques such as language models, spell checking, and post-classification corrections.
Challenges in OCR tasks include variations in font styles, noise, distortion, skew, and different languages. Training CNNs on large-scale datasets containing annotated characters and words enables them to learn robust features for accurate recognition.
9. Image embedding in computer vision refers to the process of representing images as fixed-dimensional vectors, often in a continuous vector space. The embedding encodes the visual information of an image into a numerical representation that captures its semantic content or similarity to other images. Image embedding has various applications, such as:
- **Image Retrieval:** Similarity-based image retrieval systems can compare image embeddings to find visually similar images. By computing distances or similarities between the embeddings, it becomes possible to retrieve images related to a given query image.
- **Image Clustering:** Image embeddings can be used to group similar images together based on their visual content. Clustering algorithms can operate on the embeddings to form coherent clusters or groups of visually related images.
- **Semantic Understanding:** Image embeddings can be used as input to downstream models or classifiers for tasks such as image classification, object recognition, or scene understanding. The embeddings capture essential visual features, allowing subsequent models to focus on higher-level reasoning or decision-making.
Image embedding is typically learned by training CNNs on large-scale datasets using techniques like supervised or self-supervised learning, where the embeddings are optimized to encode discriminative or semantically meaningful features.
10. Model distillation in CNNs refers to the process of training a smaller, more lightweight model (student model) to mimic the behavior of a larger, more complex model (teacher model). The goal is to transfer the knowledge and generalization capabilities of the teacher model to the student model while maintaining a compact size and improved efficiency. The process involves:
- **Teacher Model Training:** The teacher model, typically a deep and accurate CNN, is trained on a large dataset or task to achieve high performance.
- **Soft Targets:** During training, instead of using hard labels (one-hot vectors) for the output, the soft probabilities or logits generated by the teacher model are used as "soft targets" for the student model. These soft targets provide additional information about the relationships between classes.
- **Student Model Training:** The student model, which is usually smaller and shallower, is trained to mimic the teacher model's predictions by minimizing the discrepancy between its output and the soft targets. This can be done using techniques like knowledge distillation or model compression.
Model distillation improves model performance and efficiency by transferring knowledge from the larger teacher model to the smaller student model. The student model can achieve comparable accuracy to the teacher model while being more suitable for resource-constrained environments like mobile devices or edge computing.
11. Model quantization in CNNs refers to the process of reducing the memory footprint and computational requirements of a CNN model by representing the model's parameters and activations with lower precision. Typically, CNN models use 32-bit floating-point numbers (FP32) for weights and activations. Model quantization involves converting these values to lower precision formats, such as 16-bit floating-point (FP16), 8-bit integer (INT8), or even binary (1-bit) representations.
The benefits of model quantization include:
- **Reduced Memory Footprint:** By using lower precision representations, the memory required to store the model parameters and intermediate activations is significantly reduced.
- **Improved Inference Efficiency:** Lower precision computations can be performed faster on modern hardware, such as graphics processing units (GPUs) and specialized accelerators, leading to improved inference speed and throughput.
- **Energy Efficiency:** Lower precision computations require fewer memory accesses and reduce the power consumption of the hardware, making the models more energy-efficient.
Quantization-aware training techniques can be employed to train the model with lower precision from the beginning or post-training quantization can be applied to an already trained model. Quantization-aware methods aim to minimize the impact of precision reduction on the model's accuracy by considering the quantization errors during training.
12. Distributed training in CNNs involves training models across multiple machines or GPUs simultaneously. The process works as follows:
- **Data Parallelism:** The training data is divided into multiple subsets, and each machine or GPU is assigned a portion of the data. Each machine or GPU independently computes the gradients and updates the model's parameters based on its subset of data.
- **Gradient Aggregation:** Periodically, the gradients from each machine or GPU are communicated and aggregated to compute the average gradient. This average gradient is then used to update the global model parameters.
- **Synchronization:** To ensure consistent updates, synchronization steps are performed to align the model parameters across all machines or GPUs. These synchronization steps can be implemented using techniques like gradient synchronization, model averaging, or parameter server architectures.
Distributed training provides several advantages:
- **Reduced Training Time:** By parallelizing the training process, distributed training can significantly reduce the overall training time compared to training on a single machine or GPU.
- **Increased Model Capacity:** Distributed training allows training larger models that may not fit within the memory limitations of a single machine or GPU.
- **Better Generalization:** Distributed training benefits from diverse perspectives provided by different machines or GPUs, potentially leading to better generalization and improved model performance.
Distributed training frameworks like TensorFlow and PyTorch provide APIs and tools to facilitate distributed training across multiple devices or machines.
13. PyTorch and TensorFlow are two popular frameworks for developing CNNs and other deep learning models. Here's a comparison of their features and capabilities:
- **TensorFlow:**
- TensorFlow is an open-source framework developed by Google Brain and has a large community support.
- It provides a flexible and scalable platform for developing deep learning models, including CNNs, for various tasks.
- TensorFlow supports both eager execution (immediate evaluation) and graph execution (build and execute computational graphs) modes.
- It offers a comprehensive set of APIs and tools for model development, deployment, and production scalability.
- TensorFlow supports distributed training across multiple devices and machines, allowing efficient use of GPUs and TPUs.
- TensorFlow provides the TensorFlow Extended (TFX) ecosystem, which includes tools for data preprocessing, model validation, and serving in production environments.
- **PyTorch:**
- PyTorch is an open-source framework developed by Facebook's AI Research (FAIR) lab and is gaining popularity among researchers and developers.
- It offers a dynamic computational graph, allowing for more flexible and intuitive model development and debugging.
- PyTorch provides excellent support for GPU acceleration and allows seamless integration with other Python libraries and tools.
- It has an active and growing community that contributes to the development of PyTorch and provides a wide range of pre-trained models and utilities.
- PyTorch offers built-in support for distributed training and is well-suited for research experiments and prototyping.
- PyTorch provides the TorchVision library, which includes datasets, models, and utilities specifically tailored for computer vision tasks.
Both frameworks have extensive documentation, tutorials, and examples, making them accessible for beginners and experts alike. The choice between PyTorch and TensorFlow often depends on personal preference, project requirements, and the existing ecosystem within an organization.
14. GPUs (Graphics Processing Units) offer significant advantages
for accelerating CNN training and inference:
- **Parallel Processing:** GPUs are designed to perform massively parallel computations, which aligns well with the highly parallel nature of CNN operations. They can process multiple data points simultaneously, leading to faster training and inference times compared to CPUs.
- **Matrix Operations:** CNNs heavily rely on matrix operations, such as convolutions and matrix multiplications. GPUs excel at performing these operations efficiently, thanks to their specialized hardware and optimized libraries (e.g., cuDNN for NVIDIA GPUs).
- **Memory Bandwidth:** GPUs typically have higher memory bandwidth than CPUs, allowing for faster data transfers between the memory and the processing units. This is particularly beneficial for CNNs, which often involve large-scale operations on large datasets.
- **Deep Learning Framework Support:** Major deep learning frameworks, such as TensorFlow and PyTorch, provide GPU acceleration through optimized GPU backend libraries. These libraries leverage the parallel processing capabilities of GPUs, enabling seamless integration and high-performance computations.
Using GPUs for CNN training and inference can result in significant speed-ups, enabling faster model development, hyperparameter tuning, and real-time inference in applications.
15. Occlusion and illumination changes can affect CNN performance in computer vision tasks:
- **Occlusion:** When objects are partially occluded, CNNs may struggle to correctly identify and localize them. The occluded regions lack relevant visual information, making it difficult for the model to capture the complete object representation. Occlusion can lead to false negatives or incorrect predictions.
- **Illumination Changes:** Variations in lighting conditions, such as brightness, contrast, or shadows, can alter the appearance of objects. CNNs are sensitive to such changes and may produce different predictions for the same object under different lighting conditions. Illumination changes can result in false positives or incorrect classifications.
Strategies to address these challenges include:
- **Data Augmentation:** Augmenting the training data with occluded or differently illuminated samples can help the CNN learn to be more robust to these variations, enabling better generalization to new conditions.
- **Transfer Learning:** Pre-trained models that have been trained on large and diverse datasets may already have some degree of robustness to occlusion and illumination changes. Fine-tuning or transferring knowledge from these models to the target task can be beneficial.
- **Adaptive Methods:** Techniques like attention mechanisms or spatial transformers can help CNNs focus on relevant image regions or adjust their internal representation based on the input's illumination conditions, improving robustness.
Additionally, proper dataset curation, including diverse occlusion patterns and illumination conditions, can help train CNNs that are more resilient to these challenges.
16. Spatial pooling in CNNs plays a crucial role in feature extraction and dimensionality reduction. It operates on the feature maps generated by the convolutional layers and aggregates information within local regions. The pooling operation involves dividing the input feature map into non-overlapping or overlapping regions and performing a pooling operation (such as max pooling or average pooling) within each region. The resulting output feature maps have reduced spatial dimensions but retain the most salient features.
The benefits and role of spatial pooling in CNNs include:
- **Translation Invariance:** Pooling helps create a level of translation invariance by making the network less sensitive to small spatial shifts in the input. By summarizing local information, the pooled features can capture the presence of important features regardless of their precise location.
- **Dimensionality Reduction:** Pooling reduces the spatial dimensions of the feature maps, which can significantly reduce the computational requirements of subsequent layers and improve efficiency. It also helps to control overfitting by reducing the model's parameter count.
- **Robustness to Variations:** Pooling acts as a form of noise suppression, reducing the impact of small variations or noise in the input. By aggregating information within local regions, pooling enables the network to focus on the most relevant and discriminative features.
Spatial pooling is typically applied after convolutional layers and before subsequent layers or fully connected layers in a CNN architecture. The choice of pooling method and parameters depends on the specific task, network architecture, and the desired trade-off between spatial resolution and information summarization.
17. Class imbalance in CNNs refers to situations where the distribution of data across different classes is significantly skewed, with one or more classes having a much smaller representation compared to others. Class imbalance can lead to biased model training and affect performance, as CNNs tend to prioritize the majority classes.
Techniques for handling class imbalance in CNNs include:
- **Data Resampling:** Resampling the training data can be done by oversampling the minority class (e.g., duplicating samples) or undersampling the majority class (e.g., randomly removing samples). These methods aim to balance the class distribution and provide equal importance to all classes during training.
- **Class Weights:** Assigning different weights to each class during the loss calculation can address class imbalance. Higher weights can be assigned to minority classes, increasing their impact on the training process and compensating for their smaller representation.
- **Generating Synthetic Samples:** Synthetic data generation techniques, such as SMOTE (Synthetic Minority Over-sampling Technique), can be used to create artificial samples for minority classes, effectively increasing their representation in the training data.
- **Cost-Sensitive Learning:** Cost-sensitive learning involves assigning different misclassification costs to different classes. By considering the relative importance or cost of misclassifying each class, the model can be trained to focus on minimizing the overall cost rather than just the error rate.
The choice of class imbalance handling technique depends on the specific dataset, class distribution, and the desired trade-off between addressing imbalance and potential risks of overfitting or introducing biases.
18. Transfer learning in CNN model development involves leveraging knowledge learned from pre-trained models on large-scale datasets and applying it to new tasks or domains with limited labeled data. The key applications and benefits of transfer learning include:
- **Feature Extraction:** Pre-trained CNN models capture generic visual features from diverse data, which can be useful for various tasks. By using a pre-trained model as a feature extractor, one can benefit from the low-level feature representations learned on large-scale datasets.
- **Reduced Training Time:** Training a CNN from scratch on large datasets can be time-consuming and computationally expensive. Transfer learning allows starting from a pre-trained model, reducing the training time significantly, as the network only needs to adapt to the specifics of the new task or domain.
- **Improved Generalization:** Pre-trained models have learned representations that generalize well to different tasks or domains. By leveraging this knowledge, transfer learning enables better generalization to new data, especially when the labeled data is limited.
The transfer learning process involves freezing the pre-trained layers, retaining their learned weights, and only training the final layers or adding a few additional layers to adapt the model to the new task. Fine-tuning, where the pre-trained weights are further adjusted with the new data, is commonly used to align the features with the target domain.
19. Occlusion can significantly impact CNN object detection performance. Occlusion refers to situations where an object is partially or fully obstructed by other objects or occluders. Challenges posed by occlusion include:
- **Localization Accuracy:** Occlusion can make it challenging for a CNN to accurately localize the occluded object. The presence of occluders can interfere with the CNN's ability to detect the complete extent and boundaries of the object.
- **False Negatives:** Occlusion can lead to false negatives, where the presence of an object is entirely missed
20. Image segmentation is the process of dividing an image into meaningful and semantically coherent regions or segments. The goal is to assign a label or category to each pixel in the image, effectively segmenting it into different regions based on visual characteristics such as color, texture, or shape. Image segmentation plays a crucial role in various computer vision tasks, including object recognition, scene understanding, autonomous driving, medical imaging, and more.
21. CNNs are commonly used for instance segmentation, which involves not only identifying objects in an image but also precisely delineating their boundaries at the pixel level. One popular architecture for instance segmentation is Mask R-CNN, which combines object detection with pixel-level segmentation. Other notable architectures include U-Net, Fully Convolutional Network (FCN), and DeepLab.
22. Object tracking in computer vision refers to the task of locating and following a specific object or multiple objects over a sequence of frames in a video. The goal is to maintain a consistent identity for each object as it moves through the frames. Object tracking faces challenges such as occlusions, changes in scale, pose variations, motion blur, and complex object interactions. Tracking algorithms typically utilize techniques like motion estimation, feature matching, appearance modeling, filtering, and data association.
23. Anchor boxes are a key component in object detection models like SSD (Single Shot MultiBox Detector) and Faster R-CNN (Region Convolutional Neural Network). They are pre-defined bounding boxes of different scales and aspect ratios that act as reference templates for detecting objects at various positions and sizes within an image. The anchor boxes are placed at multiple locations across the image and serve as priors for predicting object locations and generating region proposals during the object detection process.
24. Mask R-CNN is a convolutional neural network architecture used for instance segmentation. It extends the Faster R-CNN model by adding an additional branch for predicting pixel-level masks for each object instance. The architecture consists of three main components: a backbone network (e.g., a pre-trained CNN), a Region Proposal Network (RPN) for generating region proposals, and a Mask Head network for predicting masks within each region proposal. Mask R-CNN achieves state-of-the-art performance in instance segmentation tasks by simultaneously detecting and segmenting objects in an image.
25. CNNs are widely used for optical character recognition (OCR) tasks. In OCR, CNN models are trained to recognize and interpret text characters within images or scanned documents. The CNN architecture typically consists of convolutional layers for feature extraction, followed by fully connected layers for classification. CNNs are trained on large labeled datasets containing images of characters, and they learn to recognize patterns and features that differentiate different characters. Challenges in OCR include variations in fonts, sizes, rotations, lighting conditions, noise, and background clutter.
26. Image embedding refers to the process of representing images in a compact and meaningful vector space, where similar images are located closer to each other and dissimilar images are farther apart. Image embeddings capture the semantic information of images, allowing for efficient comparison and retrieval based on similarity. Applications of image embedding include similarity-based image search, content-based image retrieval, recommendation systems, and image clustering.
27. Model distillation in CNNs is a technique used to transfer knowledge from a larger, more complex model (teacher model) to a smaller, more efficient model (student model). The teacher model is typically a well-trained and accurate model, while the student model is designed to have a smaller memory footprint or be more computationally efficient. The distillation process involves training the student model to mimic the outputs or internal representations of the teacher model. The benefits of model distillation include improved model generalization, reduced model size, and faster inference.
28. Model quantization is a technique used to reduce the memory footprint and computational requirements of CNN models. It involves representing model parameters and activations with lower precision data types (e.g., from floating-point to fixed-point or integer representation) while minimizing the impact on model performance. Quantization helps to reduce the storage requirements and improve the runtime efficiency of CNN models, making them more suitable for deployment on resource-constrained devices or systems.
29. Distributed training of CNN models involves training the model across multiple machines or GPUs in parallel. Each machine or GPU processes a subset of the training data and performs forward and backward propagation for a portion of the model parameters. The gradients from each machine are then aggregated, and the model parameters are updated accordingly. Distributed training improves performance by reducing the training time through parallelization and enables scaling to larger datasets or more complex models. It also allows for efficient utilization of resources and facilitates experimentation with larger models and hyperparameter search.
30. PyTorch and TensorFlow are two popular frameworks for CNN development:
- PyTorch: PyTorch is known for its dynamic computation graph, making it flexible and suitable for research and prototyping. It provides an intuitive and Pythonic API, making it easier to write and debug models. PyTorch also has a vibrant community and extensive support for computer vision tasks, with libraries like torchvision. Additionally, PyTorch offers strong GPU support and benefits from automatic differentiation.
- TensorFlow: TensorFlow is known for its static computation graph, which enables efficient deployment and optimization. It offers a high-level API called Keras, which simplifies the development of CNN models. TensorFlow provides excellent scalability and is well-suited for large-scale production deployments. It also offers tools like TensorBoard for visualizing training progress and model performance. TensorFlow has strong support for distributed training and deployment on various platforms, including CPUs, GPUs, and specialized hardware like TPUs.Both frameworks have extensive documentation, community support, and pre-trained models available, making them suitable for different use cases and preferences. The choice between PyTorch and TensorFlow often depends on the specific project requirements, familiarity with the framework, and the need for flexibility, performance, or deployment considerations.
31. GPUs (Graphics Processing Units) are well-suited for accelerating CNN training and inference due to their parallel processing capabilities. CNN computations, such as convolution and matrix operations, can be efficiently performed in parallel on GPU cores, which significantly speeds up the overall computation. GPUs provide high memory bandwidth and multiple cores, allowing for the concurrent processing of multiple data samples or model parameters. Additionally, GPU libraries like CUDA or cuDNN optimize CNN operations, further enhancing performance.
However, GPUs also have limitations. They require a significant amount of power, limiting their use in resource-constrained environments. GPU memory capacity may also restrict the size of models or batch sizes that can be used. GPUs are most effective when the CNN workload can be parallelized and when the data can be efficiently streamed to and from the GPU memory. Lastly, the cost of GPUs can be a limiting factor for some applications.
32. Occlusion poses challenges in object detection and tracking tasks because objects can be partially or completely hidden by other objects or obstacles. Some techniques for handling occlusion include:
- Contextual information: Utilizing the surrounding context of objects can aid in inferring their presence or location. By considering the context, such as object relations or scene understanding, occluded objects can be inferred or tracked based on their relationships with other visible objects.
- Temporal information: Leveraging temporal coherence across frames in a video can help track objects through occlusions. Techniques like motion modeling, object appearance consistency, or optical flow estimation can be used to predict object locations during occlusion periods.
- Multi-object tracking: Treating occluded objects as part of a larger tracking problem can improve accuracy. By jointly considering multiple objects and their interactions, occlusion reasoning can be incorporated into the tracking process.
- Object re-identification: When an object is occluded and reappears, re-identifying it as the same object can be challenging. Techniques such as feature matching, appearance modeling, or deep metric learning can help re-identify objects across occlusion periods.
33. Illumination changes can significantly affect CNN performance, as the model may not generalize well to images with different lighting conditions than the training data. Some techniques for robustness to illumination changes include:
- Data augmentation: Incorporating augmented images with varying lighting conditions during training can help the model learn to be invariant to different illumination levels.
- Normalization techniques: Applying image normalization methods, such as histogram equalization or contrast stretching, can mitigate the impact of illumination variations by adjusting the image intensities.
- Pre-processing: Applying image enhancement techniques, such as gamma correction or adaptive histogram equalization, can improve the visibility of details in images with challenging lighting conditions.
- Domain adaptation: Utilizing domain adaptation methods, such as adversarial training or self-supervised learning, can help the model adapt to new lighting conditions by aligning the feature distributions between the training and test domains.
- Transfer learning: Fine-tuning a pre-trained CNN model with data containing diverse lighting conditions can improve its robustness to illumination changes.
34. Data augmentation techniques in CNNs aim to artificially increase the size and diversity of the training data, addressing the limitations of limited training samples. Some common data augmentation techniques include:
- Image transformations: These involve applying geometric transformations such as rotations, translations, scaling, flips, or cropping to the images. These transformations can simulate variations in object position, viewpoint, or scale.
- Color jittering: Altering the color of the images by adjusting brightness, contrast, saturation, or hue can introduce variations and enhance the model's ability to generalize to different color distributions.
- Noise injection: Adding different types of noise, such as Gaussian noise, salt-and-pepper noise, or speckle noise, can make the model more robust to noisy input data.
- Random erasing: Randomly masking out rectangular regions of an image can encourage the model to focus on other informative regions and improve its robustness to occlusions.
- Mixup and cutout: Mixup involves linearly combining two or more images and their labels, encouraging the model to learn from the interpolation of different samples. Cutout involves randomly masking out square regions of an image, forcing the model to rely on other contextual cues.
These techniques introduce diversity into the training data, helping the model generalize better and reduce overfitting.
35. Class imbalance in CNN classification tasks occurs when the number of training examples in different classes is significantly unequal, leading to biased learning. Some techniques for handling class imbalance include:
- Resampling: Oversampling the minority class by replicating samples or undersampling the majority class by removing samples can balance the class distribution. Techniques like Random Oversampling, SMOTE (Synthetic Minority Over-sampling Technique), or ADASYN (Adaptive Synthetic Sampling) can be used.
- Class weighting: Assigning higher weights to the minority class during training can provide a higher loss penalty for misclassifications in the minority class, thereby balancing the learning process.
- Data augmentation: Augmenting the minority class by applying various transformations can increase the number of samples and balance the class distribution.
- Ensemble methods: Utilizing ensemble techniques, such as bagging or boosting, can help mitigate the impact of class imbalance by combining multiple models or adjusting sample weights during training.
- Cost-sensitive learning: Assigning different misclassification costs to different classes can guide the model to focus more on correctly classifying the minority class.
These techniques help address the challenges of imbalanced class distributions and promote fair learning across all classes.
36. Self-supervised learning in CNNs is an approach where a model is trained to learn useful representations or features from unlabeled data without explicit supervision. The main idea is to design pretext tasks that can be solved using the available unlabeled data. The model is trained to predict some useful properties of the data, such as image rotation, image inpainting, colorization, or predicting the relative position of image patches. By learning to solve these pretext tasks, the model can capture meaningful and high-level features that can be later transferred or fine-tuned for supervised tasks, such as image classification or object detection.
Self-supervised learning is valuable when labeled training data is limited or expensive to obtain. It allows the model to leverage the abundant unlabeled data to learn useful representations, which can then be applied to downstream tasks. This approach has shown promising results in various domains, including computer vision and natural language processing.
37. Several popular CNN architectures have been specifically designed for medical image analysis tasks due to the unique challenges and requirements of medical imaging data. Some notable architectures include:
- U-Net: U-Net is widely used for medical image segmentation tasks. It consists of a contracting path that captures contextual information and a symmetric expanding path that enables precise localization. U-Net has shown excellent performance in various medical imaging applications, such as organ segmentation, tumor detection, and cell segmentation.
- V-Net: V-Net is an extension of U-Net that includes a 3D architecture for volumetric medical image segmentation. It leverages 3D convolutions to capture spatial information in medical volumes.
- DeepMedic: DeepMedic is a CNN architecture designed for brain lesion segmentation in MRI scans. It combines a 2D pathway for high-resolution information and a 3D pathway for capturing contextual information.
- DenseNet: DenseNet is a densely connected CNN architecture that has been successful in medical image analysis. It promotes feature reuse by connecting each layer to every subsequent layer, allowing for better information flow and reducing the number of parameters.
- 3D CNNs: Medical imaging often involves 3D volumes, and 3D CNN architectures,
such as 3D U-Net or VoxResNet, have been developed to handle the volumetric nature of the data. These architectures leverage 3D convolutions and capture spatial relationships in the data.
These architectures are tailored to address the challenges specific to medical image analysis, such as limited annotated data, complex anatomical structures, and the need for precise segmentation or detection of abnormalities.
38. U-Net is a convolutional neural network architecture designed for medical image segmentation tasks, particularly in biomedical imaging. It consists of an encoding path and a corresponding decoding path.
The encoding path is composed of a series of convolutional and pooling layers that progressively reduce the spatial dimensions while increasing the number of channels. This path captures high-level semantic information and contextual cues.
The decoding path performs up-sampling and concatenation operations to recover the spatial resolution lost during encoding. Each up-sampling step is followed by a convolutional layer that reduces the number of channels. The concatenation of feature maps from the encoding path and the decoding path helps preserve fine-grained details and improves localization accuracy.
U-Net combines the contracting (encoding) path and expanding (decoding) path to form a U-shaped architecture, hence the name U-Net. This architecture enables the precise localization of objects in medical images while incorporating global context information.
U-Net has achieved state-of-the-art performance in various medical image segmentation tasks, including organ segmentation, tumor segmentation, and cell segmentation, by effectively utilizing limited annotated data and preserving fine-grained details.
39. CNN models handle noise and outliers in image classification and regression tasks through various techniques:
- Robust loss functions: Instead of using traditional loss functions like mean squared error (MSE) or cross-entropy loss, robust loss functions such as Huber loss, mean absolute error (MAE), or smoothed L1 loss can be used. These loss functions are less sensitive to outliers and can better handle noisy labels or data points.
- Regularization techniques: Techniques like dropout or weight decay regularization help prevent overfitting and improve the model's robustness to noisy or outlier data by reducing the reliance on individual data points.
- Data cleaning and preprocessing: Removing or correcting noisy or outlier data points prior to training can improve model performance. Outlier detection methods, data normalization, or data denoising techniques like Gaussian filtering or median filtering can be applied.
- Ensemble methods: Building ensembles of models can help mitigate the impact of noisy or outlier data by averaging out their effects. Different models trained on different subsets of the data or with different initialization can collectively make more accurate predictions.
- Data augmentation: Data augmentation techniques, such as adding noise or perturbations to the training data, can help the model generalize better to noisy or outlier samples.
These techniques enhance the model's robustness to noisy or outlier data and improve its performance on real-world tasks.
40. Ensemble learning in CNNs refers to the combination of multiple models to improve overall performance. It can be achieved through techniques like model averaging, model stacking, or boosting. The benefits of ensemble learning in CNNs include:
- Improved accuracy: Ensemble models tend to achieve better predictive performance compared to individual models. The combination of multiple models reduces the risk of individual model biases or errors and captures diverse patterns in the data.
- Robustness: Ensemble models are often more robust to outliers or noisy data points as errors from individual models can be mitigated or canceled out during the combination process.
- Generalization: Ensemble models tend to generalize better to unseen data by capturing a wider range of feature representations and reducing overfitting.
- Model diversity: Ensemble learning encourages model diversity by training models with different initializations, architectures, or training strategies. This diversity allows for a broader exploration of the solution space and reduces the likelihood of all models making the same mistakes.
However, ensemble learning requires additional computational resources and can be more complex to implement and maintain compared to individual models.
41. Attention mechanisms in CNN models improve performance by selectively focusing on relevant regions or features within the input data. Attention mechanisms address the limitations of traditional CNNs, which treat all input elements equally and may struggle with capturing long-range dependencies or handling large input sequences. Some types of attention mechanisms include:
- Spatial Attention: Spatial attention mechanisms assign different weights or importance to different spatial regions of an image. This enables the model to focus on informative regions and suppress noise or irrelevant areas.
- Channel Attention: Channel attention mechanisms dynamically adjust the importance of different channels or feature maps in a CNN. By assigning different weights to channels, the model can focus on more discriminative features and suppress less relevant or noisy channels.
- Self-Attention: Self-attention mechanisms capture dependencies between different elements within the input sequence by assigning attention weights to pairs of elements. This allows the model to attend to relevant information across long distances and model global dependencies.
Attention mechanisms can be integrated into CNN architectures, such as in the Transformer model or in various attention-based CNN models like SENet (Squeeze-and-Excitation Network) or Transformer-based models like ViT (Vision Transformer). Attention mechanisms enhance the model's ability to capture fine-grained details, long-range dependencies, and semantic relationships, leading to improved performance in various tasks, including image classification, object detection, and machine translation.
42. Adversarial attacks on CNN models involve intentionally manipulating input data to deceive the model's predictions. Adversarial examples are carefully crafted inputs that are perceptually similar to the original inputs but can lead to incorrect predictions or misclassification by the model. Techniques for adversarial defense include:
- Adversarial training: This involves augmenting the training process by including adversarial examples during model training. By exposing the model to adversarial examples and updating the model's parameters to minimize the loss on these examples, the model can learn to be more robust to adversarial attacks.
- Defensive distillation: Defensive distillation is a technique that involves training a student model using soft targets from a pre-trained and more robust teacher model. The soft targets, which are obtained by applying a temperature parameter to the teacher model's softmax outputs, provide more robust information for training the student model.
- Adversarial perturbation detection: Techniques for detecting adversarial perturbations can be applied to identify and reject adversarial examples. Methods such as input gradient analysis, statistical analysis, or anomaly detection can help identify unusual patterns or perturbations in the input data.
- Model regularization: Regularization techniques, such as L1 or L2 regularization, can discourage the model from being overly sensitive to small perturbations, making it more resistant to adversarial attacks.
Adversarial defense is an active area of research, as attackers continually develop new techniques, and defending against adversarial attacks remains an ongoing challenge.
43. CNN models can be applied to natural language processing (NLP) tasks by treating textual data as sequential data and representing it as numerical inputs for CNNs. Text classification and sentiment analysis are examples of NLP tasks where CNNs have been successfully used.
In text classification, CNNs can be applied by treating the input text as a sequence of word or character embeddings. Convolutional layers with varying filter sizes can be used to capture local features or n-gram relationships within the text. Max-pooling or global pooling operations can then be applied to capture the most salient features. Finally, fully connected layers and softmax activation can be used for classification.
For sentiment analysis, CNNs can be applied by treating the text as a sequence of word or character embeddings, similar to text classification. However, attention mechanisms or recurrent layers like LSTM (Long Short-Term Memory) or GRU
(Gated Recurrent Unit) can be incorporated to capture the contextual dependencies and longer-term relationships within the text.
CNNs for NLP tasks can benefit from pre-training on large-scale language models like BERT (Bidirectional Encoder Representations from Transformers) or GPT (Generative Pre-trained Transformer) to learn rich representations of text data. These pre-trained models can be fine-tuned on specific NLP tasks to improve performance.
44. Multi-modal CNNs are CNN models designed to fuse and process information from different modalities, such as images, text, audio, or sensor data. These models enable the joint learning and integration of information from multiple sources, allowing for a more comprehensive understanding of the input data. Applications of multi-modal CNNs include video analysis, image captioning, audio-visual recognition, or sensor fusion.
To build multi-modal CNNs, each modality is typically processed by separate CNN branches, with shared or separate weights depending on the task. The CNN branches extract modality-specific features, and the extracted features are then fused or combined using fusion techniques like concatenation, element-wise operations, or attention mechanisms. The fused features are subsequently fed into fully connected layers or other downstream tasks.
Multi-modal CNNs benefit from the complementary nature of different modalities, enabling a richer representation of the input data and potentially improving overall performance compared to uni-modal models.
45. Model interpretability in CNNs refers to the ability to understand and explain the decisions and learned features of a CNN model. It is important to gain insights into the model's behavior, identify potential biases, and build trust in its predictions. Techniques for visualizing learned features in CNNs include:
- Activation visualization: By inspecting the activation maps of intermediate layers, it is possible to visualize which parts of an image activate specific filters. This provides insights into what visual patterns the model is capturing.
- Grad-CAM: Gradient-weighted Class Activation Mapping (Grad-CAM) highlights the regions in an image that are most important for the model's prediction. It generates heatmaps that highlight the regions of interest for a given class.
- Saliency maps: Saliency maps highlight the most salient regions of an image that contribute to the model's decision. They can be generated by computing the gradient of the output with respect to the input image.
- DeepDream: DeepDream produces visually intriguing images by amplifying the patterns that activate specific filters in the CNN. It provides a way to visualize the features learned by the model.
- Class activation mapping: Class activation mapping techniques generate heatmaps that highlight the regions of an image that are most relevant for a particular class prediction.
These techniques help provide insights into the learned representations and enable better understanding of how the model processes and interprets input data.
46. Deploying CNN models in production environments involves various considerations and challenges, including:
- Deployment platform: Choosing the appropriate hardware platform, such as CPUs, GPUs, or specialized accelerators like TPUs, depending on the specific requirements of the application in terms of latency, throughput, and energy efficiency.
- Optimization and efficiency: Optimizing the CNN model to ensure it runs efficiently in real-time. Techniques like model quantization, pruning, or network compression can be applied to reduce memory and computational requirements.
- Scalability: Ensuring the deployed system can handle increasing workloads and user demands. This may involve strategies like distributed training or inference across multiple machines or GPUs.
- Integration with existing systems: Integrating the CNN model with existing software infrastructure or frameworks, such as web services, databases, or other APIs, to enable seamless integration into the production pipeline.
- Monitoring and maintenance: Setting up monitoring systems to track the performance and health of the deployed model. Regular maintenance and updates may be necessary to address issues, update dependencies, or retrain models with new data.
- Security and privacy: Ensuring the deployed system follows appropriate security measures, such as data encryption, access controls, and privacy regulations to protect sensitive information.
Each deployment scenario may have specific requirements and constraints, and careful consideration of these factors is essential to successfully deploy CNN models in production.
47. Imbalanced datasets in CNN training can pose challenges as the model may be biased towards the majority class, resulting in poor performance on minority classes. Techniques for handling imbalanced datasets include:
- Resampling: As mentioned earlier, resampling techniques like oversampling the minority class or undersampling the majority class can balance the class distribution and mitigate the effects of class imbalance.
- Class weighting: Assigning different weights to different classes during training can provide higher penalties for misclassifications in the minority class, effectively balancing the learning process.
- Synthetic data generation: Generating synthetic samples for the minority class can help augment the training data and balance the class distribution. Techniques like SMOTE or ADASYN can be used to generate synthetic samples based on the characteristics of the minority class.
- Ensemble methods: Constructing an ensemble of models trained on different class-balanced subsets of the data can help alleviate the impact of class imbalance. Ensemble methods can combine the predictions of multiple models and improve performance on minority classes.
- Transfer learning: Leveraging pre-trained models on large-scale datasets can provide a better initialization point and more general features that are helpful for learning from imbalanced datasets.
These techniques aim to address the class imbalance issue and ensure the model performs well across all classes, not just the majority class.
48. Transfer learning is a technique in CNN model development where knowledge gained from training on one task or dataset is transferred and applied to another related task or dataset. The benefits of transfer learning in CNN model development include:
- Reduced need for labeled data: Pre-training on a large-scale dataset allows the model to learn generic features that are applicable to multiple tasks. This reduces the reliance on a large amount of labeled data for the target task.
- Improved generalization: CNN models pre-trained on large and diverse datasets tend to learn more general and transferable features. These features capture generic visual patterns, enabling the model to generalize well to new data and adapt to different tasks.
- Faster convergence: Transfer learning allows the model to start from a better initialization point, as the pre-trained model has already learned useful representations. This can lead to faster convergence during fine-tuning on the target task.
- Regularization effect: Pre-training acts as a form of regularization, reducing the risk of overfitting, especially when the target task has limited labeled data.
Transfer learning can be applied by using the pre-trained model as a feature extractor, freezing the lower layers, or fine-tuning the entire model with a smaller learning rate. The choice of the specific transfer learning strategy depends on the similarity between the pre-training and target tasks and the availability of labeled data for the target task.
49. CNN models handle data with missing or incomplete information by leveraging their ability to learn from patterns and extract relevant features. Some approaches for handling missing or incomplete data include:
- Data imputation: Missing values in the dataset can be imputed or filled in using various techniques. Simple methods include mean imputation or median imputation, where missing values are replaced with the mean or median of the available data. More sophisticated methods like k-nearest neighbors (KNN) imputation or matrix factorization can also be used to estimate missing values based on the available data.
- Feature selection or masking: If the missing data occurs in specific features, those features can be masked or excluded from the model during training or inference. This approach ensures that the model does not rely on incomplete or unreliable information.
- Attention mechanisms: Attention mechanisms can be applied to give more weight or focus to the available information while downplaying
or ignoring missing values. This allows the model to attend to relevant features and effectively handle missing data.
- Data augmentation: Data augmentation techniques, such as introducing perturbations or transformations to the available data, can help generate additional synthetic samples and reduce the impact of missing data.
Handling missing or incomplete data is an active area of research, and the choice of the specific approach depends on the nature and characteristics of the missing data as well as the specific task at hand.
50. Multi-label classification in CNNs is a task where an input can be associated with multiple labels or categories simultaneously. Unlike traditional single-label classification, where an input belongs to a single class, multi-label classification allows for the prediction of multiple relevant labels. Techniques for solving multi-label classification tasks with CNNs include:
- Sigmoid activation: Instead of using a softmax activation function that assigns probabilities to mutually exclusive classes, a sigmoid activation function is applied to each output unit in the final layer of the CNN. This allows each unit to independently predict the presence or absence of a specific label.
- Binary cross-entropy loss: The binary cross-entropy loss function is used instead of the traditional categorical cross-entropy loss. This loss function calculates the loss independently for each label prediction, treating them as separate binary classification problems.
- Thresholding: By applying a threshold to the output probabilities, the model can determine which labels to predict. The threshold can be chosen based on the desired trade-off between precision and recall.
- Training data preparation: The training data needs to be appropriately labeled with multiple labels for each input. Techniques like one-hot encoding or multi-label binarization are applied to represent the labels in a suitable format.
Multi-label classification with CNNs finds applications in tasks such as object recognition with multiple objects in an image, text classification with multiple topics or attributes, and audio classification with multiple sound sources.
|
{
"filename": "PPTASs10_1.ipynb",
"repository": "ARJUN198SINGH/Assignment-sol.",
"query": "transformed_from_existing",
"size": 74241,
"sha": ""
}
|
# project.ipynb
Repository: PieroniJV/Assessment
<hr>
# <a id='toc1_'></a>[Deutsch's Algorithm](#toc0_)
<hr>
**Table of contents**<a id='toc1_1_'></a>
- [Deutsch's Algorithm](#toc1_)
- [Table of contents](#toc1_1_)
- [Introduction](#toc1_2_)
- [Problem Statement](#toc1_3_)
- [Black-Box Problem](#toc1_3_1_)
- [Classical Solution](#toc1_3_2_)
- [Quantum Principles](#toc1_4_)
- [Superpostion](#toc1_4_1_)
- [Interference](#toc1_4_2_)
- [Deutsch's Algorithm](#toc1_5_)
- [Algorithm Explanation](#toc1_5_1_)
- [Quantum Circuit Implementation](#toc1_6_)
- [Importing Libraries](#toc1_6_1_)
- [Initialization](#toc1_6_2_)
- [Hadamard Gates:](#toc1_6_2_)
- [Oracle Circuit:](#toc1_6_2_)
- [Additional Gates:](#toc1_6_2_)
- [Measurement:](#toc1_6_3_)
- [Circuit Visualization](#toc1_7_)
- [Visual Representation](#toc1_7_1_)
- [Simulating the Quantum Circuit](#toc1_8_)
- [Setting up the Simulator](#toc1_8_1_)
- [Running the Simulation](#toc1_8_2_)
- [Results Analysis](#toc1_8_3_)
- [Comparative Analysis](#toc1_9_)
- [Comparison with Classical Approach](#toc1_9_1_)
- [Conclusion](#toc1_10_)
- [Summary of Findings](#toc1_10_1_)
- [Significance of Quantum Advantage](#toc1_10_2_)
- [Future Directions](#toc1_11_)
- [Applications Beyond Deutsch's Algorithm](#toc1_11_1_)
- [References](#toc1_12_)
<!-- vscode-jupyter-toc-config
numbering=false
anchor=true
flat=false
minLevel=1
maxLevel=6
/vscode-jupyter-toc-config -->
<!-- THIS CELL WILL BE REPLACED ON TOC UPDATE. DO NOT WRITE YOUR TEXT IN THIS CELL -->
<hr>
## <a id='toc1_2_'></a>[Introduction](#toc0_)
<hr>
### Overview of Quantum Computing
---
IBM defines quantum computing as <q>*a rapidly-emerging tehcnology* that harnesses the laws of quantum mechanics to solve problems **too complex for classical computers.**</q> [[1]](#1)
In classical computing, calculations are executed using bits as the fundamental unit of information, where each bit can represent either 0 or 1. In contrast, quantum computers utilize *qubits (quantum bits)*.
Unlike classical bits, a qubit is not limited to a single, definitive state of 0 or 1; rather, it can exist in multiple states simultaneously.
This unique property is called *superposition* and allows quantum computers to process a significantly larger number of possibilities than their classical counterparts.
Qubits exhibit a distinctive characteristic known as *entanglement*, wherein the state of one qubit is intricately linked to the state of another, irrespective of the physical separation between them. This phenomenon, known as entanglement, forms a foundational element in quantum computing.
Quantum computers harness not only entanglement but also another crucial property known as *interference* to enhance computational efficiency. Through the strategic utilization of entanglement and interference, quantum computers optimize their computational capabilities.
### Why use quantum computing?
---
Despite the growing prevalence of large classical computers equipped with an increasing number of CPU and GPU cores, their fundamental limitation lies in their binary operation. If a supercomputer encounters challenges, it is typically because the **complex problem** at hand exceeds the capabilities of these large classical machines.<q>Complex problems are problems with lots of variables interacting in complicated ways.</q>[[2]](#2) The failure of classical computers is often rooted in their inherent difficulty handling **high levels of complexity**.
As technology continues to advance, the **complexity of problems also escalates**, necessitating the adoption of *quantum computing*. This heightened complexity underscores the demand for quantum computing solutions. Presently, various fields leverage quantum computing technology to address **intricate challenges**, including but not limited to cryptography, machine learning, and calculations involving large factorial numbers.
### The Deutsch's Algorithm
---
*Deutsch's Algorithm*, formulated by David Deutsch in 1985, is designed to tackle the "black-box problem," a specific computational challenge that will be further examined in the subsequent chapter. In classical computing, discerning whether an unknown function is constant or balanced necessitates multiple queries. In stark contrast, **Deutsch's Algorithm achieves this task with remarkable efficiency, requiring just a single quantum query**. The algorithm adeptly distinguishes between four distinct function types: **constant zero, constant one, balanced zero, and balanced one**. This demonstration highlights the quantum advantage, as it outperforms classical approaches by showcasing the capability of quantum computation to efficiently solve particular problems with a minimal number of queries.
<hr>
## <a id='toc1_3_'></a>[Problem Statement](#toc0_)
<hr>
### <a id='toc1_3_1_'></a>[Black-Box Problem](#toc0_)
<q>The Black Box Problem is traditionally said to arise when the computing systems that are used to solve problems in AI are opaque. This manner of speaking is grounded in
the metaphorical intuition that a system’s behavior can be explained by “looking inside” so as to understand why it does what it does or how it works</q>[[4]](#4)
As previously stated, the *black-box problem* addressed by **Deutsch's Algorithm** revolves around determining the nature of an *unknown function* encapsulated within a black box. This function takes a *single-bit input* and produces a *single-bit output*. The main challenge is to categorize the function into one of four possible types as seen below.
<img src="./assets/table1.png" width="200">
- *Constant Function (C0)*: Always returns 0, regardless of the input.
- *Constant Function (C1)*: Always returns 1, regardless of the input.
- *Balanced Function (B0)*: Returns 0 for one input and 1 for the other.
- *Balanced Function (B1)*: Returns 1 for one input and 0 for the other.
The objective is to efficiently determine whether the black-box function falls into the category of a *constant function (either C0 or C1)* or a *balanced function (either B0 or B1)*. Emphasizing these four possible function types, **Deutsch's Algorithm** demonstrates a quantum advantage by solving this problem with just one query, showcasing the potency of quantum computation in specific problem domains.
<hr>
### <a id='toc1_3_2_'></a>[Classical Solution](#toc0_)
In the classical approach to solving the *black-box problem*, the strategy is to make queries to the *unknown function* within the blac-box. Here is how this works:
1. <u>Query for input 0(Classical bit 0):</u> Query the black-box function with an input of 0.
2. <u>Query for input 1(Classical bit 1):</u> Query the black-box function with an input of 1.
3. <u>Compare outputs:</u> Examine the outputs for both queries. If the outputs are the same (both 0 or both 1), the function is classified as "constant." If the outputs are different (one is 0 and the other is 1), the function is classified as "balanced."
The issue with this approach is that this has a few limitations, such as:
- <u>Query complexity:</u> This approach requires at least two queries to the function before it can determine if the function is *constant* or *balanced*. Which leads to the next limitation.
- <u>Scalability issues:</u> As the *complexity of the problem* grows, and with larger input spaces or more intricate functions, the classical approach's query complexity increases linearly. This scalability issue makes the classical solution less efficient for complex problems.
- <u>Inefficiency for Quantum Problems:</u> The *classical approach* contrasts with quantum algorithms like Deutsch's Algorithm, which demonstrate a *quantum advantage* by solving the problem with only one query. The classical method becomes inefficient when compared to quantum solutions for specific problems due to the inherent limitations of sequential query-based approaches.
In the realm of less intricate problems, the *classical approach* remains effective. However, as the complexity of problems escalates or when grappling with quantum scenarios, the classical methodology becomes increasingly impractical. **Quantum problems**, in particular, often surpass the computational capacity of classical approaches, underscoring the need for quantum computing solutions in navigating challenges of heightened intricacy.
<hr>
## <a id='toc1_4_'></a>[Quantum Principles](#toc0_)
<hr>
### <a id='toc1_4_1_'></a>[Superpostion](#toc0_)
**Superposition** is a fundamental quantum principle that allows quantum bits(Qubits) to exist in multiple states simultaneously. Here is how this is defined:
$\text{Superposition} = \alpha|0\rangle + \beta|1\rangle$
- $\alpha \text{ and } \beta$ are complex numbers that determine the *probability* of measuring the qubit in the state $0$ or $1$
- $|0\rangle$ represents the quantum state where the qubit is in the logical state 0.
- $|1\rangle$ represents the quantum state where the qubit is in the logical state 1.
In **superposition**, a qubit is not definitively in state 0 or state 1; instead, it exists as a *combination of both states*. The probabilities $\alpha \text{ and } \beta$ can be djusted, allowing for various degrees of mixing between the two states. When measured, the qubit collapses into one of the basis states (0 or 1) with probabilities determined by $\alpha \text{ and } \beta$
The **superposition state** <q>represents a combination of all possible configurations of the qubit. Groups of qubits in superposition can create complex, multidimensional computational spaces.</q>[[3]](#3)
Superposition is a powerful property of qubits that *enables quantum computers to process multiple possibilities simultaneously*, providing a significant advantage over classical bits for certain types of computations.
### <a id='toc1_4_2_'></a>[Interference](#toc0_)
**Interference** in the context of quantum computation refers to the phenomenon where quantum states, such as those of qubits, combine in a way that their amplitudes *reinforce or cancel each other out* when measured.
- **Superposition Sets the Stage:** Interference is intimately tied to the principle of superposition. When a qubit is in superposition, it exists in a combination of multiple states, each associated with a *probability amplitude*.
- **Amplitudes and Probabilities:** The amplitudes of these quantum states are complex numbers, and their magnitudes squared give the probabilities of measuring the qubit in a particular state. For example, in the superposition $\alpha|0\rangle + \beta|1\rangle,\alpha^2$ is the probability of measuring the qubit in state $|0\rangle$, and $\beta^2$ is the probability of measuring it in the state $|1\rangle$.
- **Interference Effects:** Interference occurs when the amplitudes of different quantum states interact. *When two amplitudes have opposite signs, they can cancel each other out **(destructive interference)***, leading to a reduced probability of measuring the qubit in any state. *When two amplitudes have the same sign, they can reinforce each other **(constructive interference)***, increasing the probability of measuring the qubit in a particular state.
- **Quantum Algorithms:** Quantum algorithms, such as *Deutsch's Algorithm* and Grover's Algorithm, leverage interference to perform computations more efficiently than classical counterparts. They manipulate quantum states in a way that *constructive interference enhances the probability of measuring the correct answer while destructive interference reduces the probability of incorrect answers*.
- **Quantum Advantage:** Interference is at the heart of why quantum algorithms can provide significant speedup in solving certain problems. By carefully designing quantum circuits to *exploit interference*, quantum computers can explore multiple solutions simultaneously, leading to faster and more efficient computations.
In conclusion, **superpostion** can be looked at as the initial mix of possibilities, and **interference** is what happens when those possibilities interact, making some outcomes more likely and others less likely. These quantum phenomena are crucial for quantum algorithms to perform computations more efficiently than classical ones.
<hr>
## <a id='toc1_5_'></a>[Deutsch's Algorithm](#toc0_)
<hr>
### <a id='toc1_5_1_'></a>[Algorithm Explanation](#toc0_)
Here is how the **Deutsch's Algorithm** can be described:
- **Initialization:**
- Prepare two qubits. Set the first qubit to $|0\rangle$ and the second to $|1\rangle$
- Apply a *Hadamard gate* to both qubits. This transforms the qubits into a *superpostion* of all possible states: $|00\rangle + |01\rangle$, where both are present with equal amplitudes.
- **Oracle Circuit (Black-Box Function):**
- In this step, we apply the black-box function, often referred to as the oracle, to the qubits. The oracle represents the unknown function we want to evaluate.
- In the *Oracle function* we evaluate the two qubits as inputs and perform operations on them: $|xy\rangle \rightarrow |x, y \oplus f(x)\rangle$.
- In this function, $x \text{ and } y$ represent the two qubits(inputs), $f(x)$ is the function to be evaluated and $\oplus$ is the *XOR operation*, this combines the value of the second qubit $y(q1)$ with the output of the black-box function $f(x)$. This operation flips the second qubit if $f(x)$ is 1 and leaves it unchanged if $f(x)$ is 0.
- **Hadamard Gates Again:**
- After applying the oracle function, we apply *Hadamard gates* to the first qubit $x(q0)$ again.
- The *Hadamard gate* is applied to $q(0)$, creating a superposition of its possible outcomes.
This step prepares the system for interference, which is crucial for distinguishing between constant and balanced functions.
- **Measurement:**
- In this final step, we measure the first qubit. The measurement result is either 0 or 1. The outcome of the measurement provides information about the nature of the black-box function:
- If the measurement result is 0, it indicates that the function $f(x)$ is *constant* (either C0 or C1).
- If the measurement result is 1, it indicates that the function $f(x)$ is *balanced* (either B0 or B1).
The biggest take-away from this is that the measurement result provides a definitive answer to the problem of determining the type of the black-box function using *only one query*, demonstrating the *quantum advantage* of Deutsch's Algorithm.
<hr>
## <a id='toc1_6_'></a>[Quantum Circuit Implementation](#toc0_)
<hr>
This is a demonstration of the Deutsch's Algorithm using Python.
### <a id='toc1_6_1_'></a>[Importing Libraries](#toc0_)
- For this demonstration I will use the **Qiskit** library. This is a Python library used for *Quantum Computing*.
<code>
# Import necessary libraries from Qiskit
from qiskit import QuantumCircuit, QuantumRegister, ClassicalRegister
</code>
### <a id='toc1_6_2_'></a>[Initialization](#toc0_)
- We create a quantum circuit called `circuit`.
- We set up two qubits using `QuantumRegister(2)` named `qreg`. These will be `q[0]` and `q[1]`.
- The one classical bit using `ClassicalRegister(1)` named `creg`. This will be `c[0]`.
- We prepare the second qubit by applying an *X gate* followed by a *Hadamard gate*
- Finally, we define the different oracle functions that can be used to test the algorithm later.
<code>
# Create a quantum circuit with 2 qubits and 1 classical bit
# Qubits q[0] and q[1], and classical bits c[0].
qreg = QuantumRegister(2)
creg = ClassicalRegister(1)
circuit = QuantumCircuit(qreg, creg)
# Prepare the second qubit in the state |->.
circuit.x(qreg[1])
circuit.h(qreg[1])
def constant_zero_oracle(circuit, qreg):
pass # does nothing, represents a constant function that always outputs 0
def constant_one_oracle(circuit, qreg):
circuit.x(qreg[1]) # flips the target qubit, represents a constant function that always outputs 1
def balanced_oracle(circuit, qreg):
circuit.cx(qreg[0], qreg[1]) # CNOT gate, a simple balanced function
</code>
### <a id='toc1_6_3_'></a>[Hadamard Gates](#toc0_)
- Next we apply **Hadamard gates** (`H`) to the first qubit `q[0]`. This creates a *superposition* of states for the qubit.
<code>
# Apply Hadamard gate (H) to the first qubit.
circuit.h(qreg[0])
</code>
### <a id='toc1_6_4_'></a>[Oracle Circuit](#toc0_)
- Now we build the **Oracle Circuit** that represents the black-box function. Assuming we have a *balanced function* $f(x)$ that flips the second qubit if the first qubit is set.
- The `oracle_circuit` function takes the `circuit` and `qreg` as arguments.
- In this function, we use the *Controlled-X(CNOT) gate* to perform the oracle operation. This gate flips the second qubit(`q[1]`) if the first qubit(`q[0]`) is set.
- Next we can apply the `oracle_circuit` to the qubits to represent the *black-box function* $f(x)$.
- It is important to note that depending on the actual black-box function you want to evaluate, the implementation of the oracle may vary.
Here is the example:
<code>
# Using one of the defined functions above
# Apply the oracle function to the qubits
constant_zero_oracle(circuit, qreg)
</code>
### <a id='toc1_6_5_'></a>[Additional Gates](#toc0_)
- After the *oracle function* is applied, we need to apply additional gates to the first qubit (`q[0]`) before measurement. These are essential for *Interference*, which as mentioned, is a key part of the algorithm to distinguish between *constant and balanced functions*.
<code>
# Apply Hadamard gate (H) to the first qubit
circuit.h(qreg[0])
</code>
### <a id='toc1_6_6_'></a>[Measurement](#toc0_)
- This is the final step in the Deutsch's Algorithm. Here we add measurement operation to extract the results and determine the nature of the black-box function.
- We use the `measure` method to measure the first qubit(`q[0]`) and store the measurement result in the first classical bit (`c[0]`).
<code>
# Step 4: Measurement
# Measure the first qubit (q[0]) and store the result in c[0]
circuit.measure(qreg[0], creg[0])
</code>
- The measurement result will be one of the values: ${00, 01, 10, 11}$.
- If the measurement result is 00 or 11, it indicates that $f(x)$ is a *constant function*, (C0 or C1).
- If the measurement result is 01 or 10 it indicates that $f(x)$ is a *balanced function*, (B0 or B1).
<hr>
## <a id='toc1_7_'></a>[Circuit Visualization](#toc0_)
<hr>
### <a id='toc1_7_1_'></a>[Visual Representation](#toc0_)
- In order to visualize the quantum circuit object we can use the `Qiskit's` visualization module.
<code>
# Import necessary libraries from Qiskit
from qiskit.visualization import circuit_drawer
# Visualize the quantum circuit and display it as a Matplotlib figure
circuit_drawer(circuit, output="mpl")
</code>
#### Breakdown
In this representation we can see the following components of the circuit:
- **Qubits** ($q0_0 \text{ and } q0_1$).
- **Gates** (`H`, Oracle($\oplus$) and `H`):
- The `H` represents the Hadamard gates used to create a *superposition*.
- The *oracle function* applied to the qubits, which is a custom operation you define in your algorithm.
- **Lines and Arrows**:
- The *lines* connecting the qubits and gates represent the quantum wires or qubits states.
- The *arrows* indicate the direction of the quantum operations, showing that the gates are applied to qubits.
- **Classical bits** (`C0`):
- C0 is a classical bit abd it stores the measurement results. The label "2/" indicates that it is a 2-bit classical register.
- **Measurement** (`M`):
- M represents the measurements performed on the qubits. The outcome is stored in the *classical bits*.
- **Control flow**:
- The operations are performed in the order that they appear, from left to right.
<hr>
## <a id='toc1_8_'></a>[Simulating the Quantum Circuit](#toc0_)
<hr>
Next the idea is to simulate the quantum circuit in python, using the Qiskit Aer simulator. This tool provides a fast an accurate way to simulate quantum circuits.
### <a id='toc1_8_1_'></a>[Setting up the Simulator](#toc0_)
- First, we import the `Aer` library from Qiskit for simulating and the `execute` for running the simulations.
- We then choose a suitable simulator that supports *measurements*, `Aer.get_backend('qasm_simulator')`.
- The `num_shots` determines how many times the circuit will run in the simulation. The more shots, the more accurate the results but also it may take longer to simulate. This number can be adjusted as needed.
<code>
# Import necessary libraries from Qiskit
from qiskit import execute, Aer
# Choose the Aer simulator
simulator = Aer.get_backend('qasm_simulator')
# Set the number of shots (simulated experiments)
num_shots = 1024 # You can adjust this number as needed
</code>
### <a id='toc1_8_2_'></a>[Running the Simulation](#toc0_)
- We continue by using the `execute` to run the simulation.
- This uses the `circuit` created previously.
- A `job` uses the `circuit`, the `simulator` and the `shots` to run the simulation.
- We then obtain the results of the simulation, retrieve it and display it.
<code>
# Create a job to run the simulation
job = execute(circuit, simulator, shots=num_shots)
# Get the results of the simulation
results = job.result()
# Retrieve the counts of measurement outcomes
counts = results.get_counts(circuit)
# Display the measurement outcomes and their probabilities
print("Measurement outcomes:", counts)
</code>
### <a id='toc1_8_3_'></a>[Results Analysis](#toc0_)
The analysis is the most crucial part of the algorithm, where the nature of the black-box function is determined(*constant or balanced*).
Here is how we can analyze the results of the simulation:
1. **Retrieve the outcomes:**
By using `results.get_counts(circuit)` we can obtain the outcomes and then store it in the `counts` variable.
2. **Understanding the outcomes:**
The outcomes represent the possible states of the qubits after measurement, along with the number of times each outcome was observed. These outcomes are in the form of binary strings, where the first digit represents the measurement result for `q0` and the second digit represnts the measurement result for `q1`.
3. **Interpretation of the outcomes:**
As previously seen:
- If the measurement outcomes contain only the states *00 and 11*, it indicates that the black-box function is **constant**. This means that the oracle function applied to the qubits *does not depend on the input and has a constant output*.
- If the measurement outcomes contain both *01 and 10*, it indicates that the black-box function is **balanced**. This means that *the oracle function applied to the qubits changes the state of the second qubit based on the input*.
4. **Probability analysis:**
One of the last steps is to examine the probabilities associated with each measurement outcome.
- In a **balanced** function, a roughly equal probability should be observed for `01` and `10` outcomes.
- In a **constant** function, you should see a higher probability for either `00` or `11`.
5. **Final decision:**
Once you have the outcomes and probabilities, you can make a final decision regarding the nature of the black-box function. As previously discussed, If you observe only `00` and `11` outcomes, conclude that the function is **constant**. If you observe both `01` and `10` outcomes, conclude that the function is **balanced**.
Here is an example of this analysis:
<code>
# Interpretation
if '0' in counts and counts['0'] > counts.get('1', 0):
print("The black-box function is constant.")
else:
print("The black-box function is balanced.")
</code>
<hr>
## <a id='toc1_9_'></a>[Comparative Analysis](#toc0_)
<hr>
### <a id='toc1_9_1_'></a>[Comparison with Classical Approach](#toc0_)
There are a few reasons as to why Quantum algorithms, such as the Deutsch's Algorithm, are more advantageous than classical methods. Here are some of these advantages:
#### 1. Speed:
The **Deutsch's Algorithm** provides a significant speedup over classical methods for solving the problem it addresses. In the classical case, to determine whether a black-box function is *constant or balanced*, you would need to evaluate the function for at least half of its input space. In contrast, Deutsch's Algorithm requires *only one query to the function*, regardless of its input size. This demonstrates the power of **quantum parallelism**, where quantum computers can *evaluate multiple inputs simultaneously*.
#### 2. Determinism:
**Deutsch's Algorithm** always produces a deterministic result. When the algorithm concludes that the black-box function is constant or balanced, it is *certain of the answer*. Classical algorithms may require *multiple evaluations*, and even then, there might be uncertainty due to probabilistic algorithms or incomplete evaluations.
#### 3. Constant function detection:
In the case of **constant functions**, **Deutsch's Algorithm** can identify the function as constant with certainty after *one query*. In contrast, a **classical algorithm** might require an exhaustive search, which can be *time-consuming* for large input spaces.
#### 4. Balanced Function Detection:
For balanced functions, **Deutsch's Algorithm** also excels. It can identify the function as balanced after *one query*. In the **classical case**, determining whether a function is balanced would *require checking multiple inputs* and observing changes in the output, which could be a *resource-intensive process*.
#### 5. Efficiency:
**Deutsch's Algorithm** demonstrates the efficiency of quantum computation. It performs the task with *fewer resources* (queries) than **classical** algorithms, making it *highly efficient* for this specific problem.
#### <u>Limitations and Considerations:</u>
While **Deutsch's Algorithm** provides advantages for specific problems, it is important to note that *not all problems experience such quantum speedup*.
**Quantum algorithms** are *highly specialized* and designed to excel in specific problem domains. In contrast, **classical algorithms** are *more versatile* and can be applied to various types of problems.
Quantum algorithms require **quantum hardware**, which is currently in the early stages of development. Quantum computers are *not yet widely available*, and building and maintaining quantum hardware can be *expensive and challenging*.
<hr>
## <a id='toc1_10_'></a>[Conclusion](#toc0_)
<hr>
### <a id='toc1_10_1_'></a>[Summary of Findings](#toc0_)
- In this notebook, we delved into the remarkable **Deutsch's Algorithm** and its prowess in addressing the enigmatic **Black-box problem**.
- Deutsch's Algorithm is not just a **quantum algorithm**; it's a quantum marvel. It accomplishes the task of determining whether an unknown function is *constant or balanced with a mere single query*, a feat that eludes the grasp of classical approaches. As we journey through the complexities of modern computing, it becomes increasingly clear that the quantum approach offers an *unparalleled advantage*.
- This algorithm seamlessly harnesses the power of **qubits**, guided by carefully orchestrated **logical gates**, to unlock the secrets hidden within the enigmatic **black-box function**.
- Through the lens of **Python** and the wizardry of **Qiskit**, we embarked on a simulated adventure, illuminating the inner workings of this quantum gem.
- While Deutsch's Algorithm shines brilliantly in the firmament of computation, illuminating paths of *efficiency, speed, and complexity handling*, it does not escape the **constraints** of the quantum realm. It stands as a *specialized solution, accompanied by cost considerations and limited availability*, a reminder that quantum computing, though promising, is *yet to fully unfold its potential*.
- As we conclude our exploration, we glimpse both the brilliance and the boundaries of quantum algorithms like Deutsch's. The quest for harnessing the full spectrum of quantum capabilities continues, *promising an exciting future* where quantum and classical computing coalesce in pursuit of the profound unknown.
### <a id='toc1_10_2_'></a>[Significance of Quantum Advantage](#toc0_)
- **Deutsch's Algorithm**, as we've explored in this notebook, stands as a testament to the transformative potential of **quantum computing**. Its ability to determine the nature of an unknown function with just *one query* has profound implications for problem-solving and computation at large. In this section, we reflect on the significance of the **quantum advantage** demonstrated by Deutsch's Algorithm.
- As seen, **speed, efficiency and complexity** are all handled by the **quantum algorithm**, which in contrast with **classical algorithms**, has a much *better performance and guarantees a deterministic result*. It also becomes evident how powerful the quantum algorithm is when we consider some of the *limitations of the classical algorithms*, such as *exhaustive evaluations of functions across their input space*, which increases the resources needed for evaluating more complex problems.
- **Quantum Parallellism:** This is a foundational concept in **quantum computing**. It allows **quantum algorithms** to *processs and explore multiple possibilites simultaneously*, leading to *exponential speedup* for certain problems. Through the principle of **superposition**, a **qubit** can represent *multiple states simultaneously*. This allows quantum algorithms to explore and *process many possibilities in parallel*.
- In conclusion, Deutsch's Algorithm significantly impacts the landscape of computational theory. It stands as both a **promising and powerful** testament to the potential of quantum computing. This algorithm not only demonstrates the unique capabilities of quantum mechanics in processing information but also heralds a future where *complex problems find their solutions in the realm of quantum computing*. As we continue to explore and develop these quantum frontiers, Deutsch's Algorithm remains a *pivotal milestone*, underscoring the bright and transformative prospects that quantum computing holds for solving intricate challenges.
<hr>
## <a id='toc1_11_'></a>[Future Directions](#toc0_)
<hr>
### <a id='toc1_11_1_'></a>[Applications Beyond Deutsch's Algorithm](#toc0_)
In this last chapter we explore how quantum algorithms redefine problem-solving, computation, and technological innovation, beyond the confines of a single algorithm. Here are some promising applications:
#### <u>Cryptography:</u>
Quantum cryptography is already seen in the field of quantum key distribution(QKD). QKD allows secure communication between two parties by leveraging the principles of quantum mechanics to create unbreakable encryption keys.
#### <u>Machine Learning:</u>
Quantum machine learning algorithms aim to outperform classical algorithms in tasks such as optimization, classification, and regression.
<q>In the past decade, particularly in the past five years, the combination of powerful computers and special-purpose information processors capable of implementing deep networks with billions of weights, together with their application to very large data sets, has revealed that such deep learning networks are capable of learning complex and subtle patterns in data.</q>[[6]](#6). The advancements in the availability of powerful computers together with specialized hardware designed for training deep neural networks made possible for a remarkable advance in deep learning over the past years. These networks are capable of modeling highly complex and intricate patterns in data.
#### <u>Chemistry and Materials Science:</u>
Quantum computers can simulate the behavior of molecules and materials at the quantum level more accurately than classical computers. This has implications for drug discovery, material design, and understanding complex chemical reactions.
#### <u>Hybrid Quantum-Classical Systems:</u>
Hybrid Computing combines the strengths of quantum and classical computing in hybrid systems that can lead to powerful solutions. Quantum algorithms can be used as co-processors in classical systems to accelerate specific tasks.
<q>Some circuits can be evaluated more efficiently on classical computers and some on quantum processors.</q>[[5]](#5). The choice of computing platform should be based on efficiency and effectiveness in solving a particular problem.
Quantum computing is not limited to academic research; it has practical applications across various domains. As quantum hardware matures and quantum algorithms evolve, the potential for quantum computing to address real-world challenges continues to expand.
<hr>
## <a id='toc1_12_'></a>[References](#toc0_)
<hr>
<a id='1'></a>
**[1]:** ibm.com. *What is quantum computing?*. Chapter: "What is quantum computing?". [Link](https://www.ibm.com/topics/quantum-computing#:~:text=Quantum%20computing%20is%20a%20rapidly,hundreds%20of%20thousands%20of%20developers). Last accessed on 22/12/2023.
<a id='2'></a>
**[2]:** ibm.com. *What is quantum computing?*. Chapter: "Why do we need quantum computers?". [Link](https://www.ibm.com/topics/quantum-computing#:~:text=Quantum%20computing%20is%20a%20rapidly,hundreds%20of%20thousands%20of%20developers). Last accessed on 22/12/2023.
<a id='3'></a>
**[3]:** ibm.com. *What is quantum computing?*. Chapter: "How do quantum computers work?". [Link](https://www.ibm.com/topics/quantum-computing#:~:text=Quantum%20computing%20is%20a%20rapidly,hundreds%20of%20thousands%20of%20developers). Last accessed on 22/12/2023.
<a id='4'></a>
**[4]:** Zednik, Carlos. (2021). *Solving the Black Box Problem: A Normative Framework for Explainable Artificial Intelligence*. Springer, 34, pp. 265-288. [Link](https://arxiv.org/ftp/arxiv/papers/1903/1903.04361.pdf). Last accessed on 22/12/2023.
<a id='5'></a>
**[5]:** Suchara, Martin; Alexeev, Yuri; Chong, Frederic; Finkel, Hal; Hoffmann, Henry; Larson, Jeffrey; Osborn, James; Smith, Graeme. (2018). *Hybrid Quantum-Classical Computing Architectures*. In Proceedings of the 3rd International Workshop on Post-Moore Era Supercomputing, 2018. [Link](https://par.nsf.gov/servlets/purl/10084839). Last accessed on 22/12/2023.
<a id='6'></a>
**[6]:** Biamonte, Jacob; Wittek, Peter; Pancotti, Nicola; Rebentrost, Patrick; Wiebe, Nathan; Lloyd, Seth. (2017). *Quantum Machine Learning*. Nature, 549(7671), pp. 195-202. [Link](https://arxiv.org/pdf/1611.09347.pdf). Last accessed on 22/12/2023.
#### Extra reading:
- Quantum circuits(How to use Qiskit): [Link](https://learn.qiskit.org/course/basics/quantum-circuits)
- Quantum Computing vs Classical Computing: [Link](https://devtechnosys.com/insights/tech-comparison/quantum-computing-vs-classical-computing/#:~:text=Classical%20computing%20relies%20on%20binary,This%20is%20known%20as%20superposition)
- What is the Deutsch-Jozsa Algorithm: [Link](https://www.classiq.io/insights/the-deutsch-jozsa-algorithm-explained#:~:text=What%20is%20the%20Deutsch%2DJozsa,values%20of%200%20or%201)
- What is quantum computing?: [Link](https://scienceexchange.caltech.edu/topics/quantum-science-explained/quantum-computing-computers)
- What is a qubit?: [Link](https://www.quantum-inspire.com/kbase/what-is-a-qubit/)
|
{
"filename": "project.ipynb",
"repository": "PieroniJV/Assessment",
"query": "transformed_from_existing",
"size": 57465,
"sha": ""
}
|
# Computational Biology General_1.ipynb
Repository: raghughanapuram/iPythonFiles
|
{
"filename": "Computational Biology General_1.ipynb",
"repository": "raghughanapuram/iPythonFiles",
"query": "transformed_from_existing",
"size": 31786,
"sha": ""
}
|
# 2025_Day 4_1.ipynb
Repository: jaiyesh/diploma
## Range: Generate Data
<code>
range(10)
</code>
<code>
list(range(0,10))
</code>
<code>
list(range(1,11))
</code>
<code>
list(range(1,10,2))
</code>
<code>
list(range(20,-2,-1))
</code>
<code>
## list of all even number from 2 to 100
list(range(2,101,2))
</code>
<code>
list(range(2,102,2))
</code>
<code>
a = int(input("Lower Bound: "))
b = int(input("Upper Bound: "))
c = int(input("Step size: "))
user_list = list(range(a,b,c))
print(user_list)
</code>
## Data Structure : tuples
- (-----)
- int, floats, strings, lists, boolean, tuple
- Immutable: cannot change elements, sensistive information is stoored in tuples
<code>
oilprd = (5000,1000,1400,4200,5000)
</code>
<code>
oilprd
</code>
<code>
type(oilprd)
</code>
<code>
##indexing: same as list
</code>
<code>
oilprd[3]
</code>
<code>
oilprd[3] = 3200
</code>
<code>
oilprd_list = list(oilprd)
</code>
<code>
oilprd_list
</code>
<code>
oilprd_list[3] = 3200
</code>
<code>
oilprd_list
</code>
<code>
oilprd
</code>
<code>
oilprd_copy = tuple(oilprd_list)
</code>
<code>
oilprd_copy
</code>
<code>
a = (45,67,2,32.5,"Saturation",["hello",34,54],(6546,434,23,("perm",87,"porosity")))
</code>
<code>
a
</code>
<code>
a[-1][-1][1]
</code>
<code>
a
</code>
<code>
a[2::2]
</code>
## Sets= unordered collection of data
- No duplication: eliminate the duplicates
- Unique Data
- {---}
- Unordered
- faster than list
<code>
perm = {2,3,4,5,6,7,2,5,7,"h","hello",4,54,54,65,76,45,2,2,2,2,"h"}
</code>
<code>
perm
</code>
<code>
perm[0]
</code>
<code>
s = {"PERM","perm"}
</code>
<code>
s
</code>
<code>
a = {2,3,4}
b = {4,5,6}
</code>
<code>
a.intersection(b)
</code>
<code>
a.union(b)
</code>
<code>
a
</code>
<code>
a.add(9)
</code>
<code>
a
</code>
<code>
a.pop()
</code>
<code>
a
</code>
<code>
data = """The three laws of motion were first stated by Isaac Newton in his Philosophiæ Naturalis Principia Mathematica (Mathematical Principles of Natural Philosophy), originally published in 1687.[3] Newton used them to investigate and explain the motion of many physical objects and systems. In the time since Newton, new insights, especially around the concept of energy, built the field of classical mechanics on his foundations. Limitations to Newton's laws have also been discovered; new theories are necessary when objects move at very high speeds (special relativity), are very massive (general relativity), or are very small (quantum mechanics).
Prerequisites
Newton's laws are often stated in terms of point or particle masses, that is, bodies whose volume is negligible. This is a reasonable approximation for real bodies when the motion of internal parts can be neglected, and when the separation between bodies is much larger than the size of each. For instance, the Earth and the Sun can both be approximated as pointlike when considering the orbit of the former around the latter, but the Earth is not pointlike when considering activities on its surface.[note 1]
The mathematical description of motion, or kinematics, is based on the idea of specifying positions using numerical coordinates. Movement is represented by these numbers changing over time: a body's trajectory is represented by a function that assigns to each value of a time variable the values of all the position coordinates. The simplest case is one-dimensional, that is, when a body is constrained to move only along a straight line. Its position can then be given by a single number, indicating where it is relative to some chosen reference point. For example, a body might be free to slide along a track that runs left to right, and so its location can be specified by its distance from a convenient zero point, or origin, with negative numbers indicating positions to the left and positive numbers indicating positions to the right. If the body's location as a function of time is s ( t ) {\displaystyle s(t)}, then its average velocity over the time interval from t 0 {\displaystyle t_{0}} to t 1 {\displaystyle t_{1}} is[6] Δ s Δ t = s ( t 1 ) − s ( t 0 ) t 1 − t 0 . {\displaystyle {\frac {\Delta s}{\Delta t}}={\frac {s(t_{1})-s(t_{0})}{t_{1}-t_{0}}}.}Here, the Greek letter Δ {\displaystyle \Delta } (delta) is used, per tradition, to mean "change in". A positive average velocity means that the position coordinate s {\displaystyle s} increases over the interval in question, a negative average velocity indicates a net decrease over that interval, and an average velocity of zero means that the body ends the time interval in the same place as it began. Calculus gives the means to define an instantaneous velocity, a measure of a body's speed and direction of movement at a single moment of time, rather than over an interval. One notation for the instantaneous velocity is to replace Δ {\displaystyle \Delta } with the symbol d {\displaystyle d}, for example, v = d s d t . {\displaystyle v={\frac {ds}{dt}}.}This denotes that the instantaneous velocity is the derivative of the position with respect to time. It can roughly be thought of as the ratio between an infinitesimally small change in position d s {\displaystyle ds} to the infinitesimally small time interval d t {\displaystyle dt} over which it occurs.[7] More carefully, the velocity and all other derivatives can be defined using the concept of a limit.[6] A function f ( t ) {\displaystyle f(t)} has a limit of L {\displaystyle L} at a given input value t 0 {\displaystyle t_{0}} if the difference between f {\displaystyle f} and L {\displaystyle L} can be made arbitrarily small by choosing an input sufficiently close to t 0 {\displaystyle t_{0}}. One writes, lim t → t 0 f ( t ) = L . {\displaystyle \lim _{t\to t_{0}}f(t)=L.}Instantaneous velocity can be defined as the limit of the average velocity as the time interval shrinks to zero: d s d t = lim Δ t → 0 s ( t + Δ t ) − s ( t ) Δ t . {\displaystyle {\frac {ds}{dt}}=\lim _{\Delta t\to 0}{\frac {s(t+\Delta t)-s(t)}{\Delta t}}.} Acceleration is to velocity as velocity is to position: it is the derivative of the velocity with respect to time.[note 2] Acceleration can likewise be defined as a limit: a = d v d t = lim Δ t → 0 v ( t + Δ t ) − v ( t ) Δ t . {\displaystyle a={\frac {dv}{dt}}=\lim _{\Delta t\to 0}{\frac {v(t+\Delta t)-v(t)}{\Delta t}}.}Consequently, the acceleration is the second derivative of position,[7] often written d 2 s d t 2 {\displaystyle {\frac {d^{2}s}{dt^{2}}}}.
Position, when thought of as a displacement from an origin point, is a vector: a quantity with both magnitude and direction.[9]: 1 Velocity and acceleration are vector quantities as well. The mathematical tools of vector algebra provide the means to describe motion in two, three or more dimensions. Vectors are often denoted with an arrow, as in s {\displaystyle \mathbf {s} }, or in bold typeface, such as s {\displaystyle {\bf {s}}}. Often, vectors are represented visually as arrows, with the direction of the vector being the direction of the arrow, and the magnitude of the vector indicated by the length of the arrow. Numerically, a vector can be represented as a list; for example, a body's velocity vector might be v = ( 3 m / s , 4 m / s ) {\displaystyle \mathbf {v} =(\mathrm {3~m/s} ,\mathrm {4~m/s} )}, indicating that it is moving at 3 metres per second along the horizontal axis and 4 metres per second along the vertical axis. The same motion described in a different coordinate system will be represented by different numbers, and vector algebra can be used to translate between these alternatives.[9]: 4
The study of mechanics is complicated by the fact that household words like energy are used with a technical meaning.[10] Moreover, words which are synonymous in everyday speech are not so in physics: force is not the same as power or pressure, for example, and mass has a different meaning than weight.[11][12]: 150 The physics concept of force makes quantitative the everyday idea of a push or a pull. Forces in Newtonian mechanics are often due to strings and ropes, friction, muscle effort, gravity, and so forth. Like displacement, velocity, and acceleration, force is a vector quantity.
Laws
First law
see caption
Artificial satellites move along curved orbits, rather than in straight lines, because of the Earth's gravity.
Translated from Latin, Newton's first law reads,
Every object perseveres in its state of rest, or of uniform motion in a right line, except insofar as it is compelled to change that state by forces impressed thereon.[note 3]
Newton's first law expresses the principle of inertia: the natural behavior of a body is to move in a straight line at constant speed. A body's motion preserves the status quo, but external forces can perturb this.
The modern understanding of Newton's first law is that no inertial observer is privileged over any other. The concept of an inertial observer makes quantitative the everyday idea of feeling no effects of motion. For example, a person standing on the ground watching a train go past is an inertial observer. If the observer on the ground sees the train moving smoothly in a straight line at a constant speed, then a passenger sitting on the train will also be an inertial observer: the train passenger feels no motion. The principle expressed by Newton's first law is that there is no way to say which inertial observer is "really" moving and which is "really" standing still. One observer's state of rest is another observer's state of uniform motion in a straight line, and no experiment can deem either point of view to be correct or incorrect. There is no absolute standard of rest.[17][14]: 62–63 [18]: 7–9 Newton himself believed that absolute space and time existed, but that the only measures of space or time accessible to experiment are relative.[19]
Second law
The change of motion of an object is proportional to the force impressed; and is made in the direction of the straight line in which the force is impressed.[14]: 114
By "motion", Newton meant the quantity now called momentum, which depends upon the amount of matter contained in a body, the speed at which that body is moving, and the direction in which it is moving.[20] In modern notation, the momentum of a body is the product of its mass and its velocity: p = m v , {\displaystyle \mathbf {p} =m\mathbf {v} \,,} where all three quantities can change over time. Newton's second law, in modern form, states that the time derivative of the momentum is the force: F = d p d t . {\displaystyle \mathbf {F} ={\frac {d\mathbf {p} }{dt}}\,.} If the mass m {\displaystyle m} does not change with time, then the derivative acts only upon the velocity, and so the force equals the product of the mass and the time derivative of the velocity, which is the acceleration:[21] F = m d v d t = m a . {\displaystyle \mathbf {F} =m{\frac {d\mathbf {v} }{dt}}=m\mathbf {a} \,.} As the acceleration is the second derivative of position with respect to time, this can also be written F = m d 2 s d t 2 . {\displaystyle \mathbf {F} =m{\frac {d^{2}\mathbf {s} }{dt^{2}}}.}
A free body diagram for a block on an inclined plane, illustrating the normal force perpendicular to the plane (N), the downward force of gravity (mg), and a force f along the direction of the plane that could be applied, for example, by friction or a string
The forces acting on a body add as vectors, and so the total force on a body depends upon both the magnitudes and the directions of the individual forces. When the net force on a body is equal to zero, then by Newton's second law, the body does not accelerate, and it is said to be in mechanical equilibrium. A state of mechanical equilibrium is stable if, when the position of the body is changed slightly, the body remains near that equilibrium. Otherwise, the equilibrium is unstable.
A common visual representation of forces acting in concert is the free body diagram, which schematically portrays a body of interest and the forces applied to it by outside influences.[22] For example, a free body diagram of a block sitting upon an inclined plane can illustrate the combination of gravitational force, "normal" force, friction, and string tension.[note 4]
Newton's second law is sometimes presented as a definition of force, i.e., a force is that which exists when an inertial observer sees a body accelerating. In order for this to be more than a tautology — acceleration implies force, force implies acceleration — some other statement about force must also be made. For example, an equation detailing the force might be specified, like Newton's law of universal gravitation. By inserting such an expression for F {\displaystyle \mathbf {F} } into Newton's second law, an equation with predictive power can be written.[note 5] Newton's second law has also been regarded as setting out a research program for physics, establishing that important goals of the subject are to identify the forces present in nature and to catalogue the constituents of matter.[14]: 134 [25]: 12-2
Third law
To every action, there is always opposed an equal reaction; or, the mutual actions of two bodies upon each other are always equal, and directed to contrary parts.[14]: 116
Rockets work by producing a strong reaction force downwards using rocket engines. This pushes the rocket upwards, without regard to the ground or the atmosphere.
Overly brief paraphrases of the third law, like "action equals reaction" might have caused confusion among generations of students: the "action" and "reaction" apply to different bodies. For example, consider a book at rest on a table. The Earth's gravity pulls down upon the book. The "reaction" to that "action" is not the support force from the table holding up the book, but the gravitational pull of the book acting on the Earth.[note 6]
Newton's third law relates to a more fundamental principle, the conservation of momentum. The latter remains true even in cases where Newton's statement does not, for instance when force fields as well as material bodies carry momentum, and when momentum is defined properly, in quantum mechanics as well.[note 7] In Newtonian mechanics, if two bodies have momenta p 1 {\displaystyle \mathbf {p} _{1}} and p 2 {\displaystyle \mathbf {p} _{2}} respectively, then the total momentum of the pair is p = p 1 + p 2 {\displaystyle \mathbf {p} =\mathbf {p} _{1}+\mathbf {p} _{2}}, and the rate of change of p {\displaystyle \mathbf {p} } is d p d t = d p 1 d t + d p 2 d t . {\displaystyle {\frac {d\mathbf {p} }{dt}}={\frac {d\mathbf {p} _{1}}{dt}}+{\frac {d\mathbf {p} _{2}}{dt}}.} By Newton's second law, the first term is the total force upon the first body, and the second term is the total force upon the second body. If the two bodies are isolated from outside influences, the only force upon the first body can be that from the second, and vice versa. By Newton's third law, these forces have equal magnitude but opposite direction, so they cancel when added, and p {\displaystyle \mathbf {p} } is constant. Alternatively, if p {\displaystyle \mathbf {p} } is known to be constant, it follows that the forces have equal magnitude and opposite direction.
Candidates for additional laws
Various sources have proposed elevating other ideas used in classical mechanics to the status of Newton's laws. For example, in Newtonian mechanics, the total mass of a body made by bringing together two smaller bodies is the sum of their individual masses. Frank Wilczek has suggested calling attention to this assumption by designating it "Newton's Zeroth Law".[33] Another candidate for a "zeroth law" is the fact that at any instant, a body reacts to the forces applied to it at that instant.[34] Likewise, the idea that forces add like vectors (or in other words obey the superposition principle), and the idea that forces change the energy of a body, have both been described as a "fourth law".[note 8]
Examples
The study of the behavior of massive bodies using Newton's laws is known as Newtonian mechanics. Some example problems in Newtonian mechanics are particularly noteworthy for conceptual or historical reasons.
Uniformly accelerated motion
Main articles: Free fall and Projectile motion
A bouncing ball photographed at 25 frames per second using a stroboscopic flash. In between bounces, the ball's height as a function of time is close to being a parabola, deviating from a parabolic arc because of air resistance, spin, and deformation into a non-spherical shape upon impact.
If a body falls from rest near the surface of the Earth, then in the absence of air resistance, it will accelerate at a constant rate. This is known as free fall. The speed attained during free fall is proportional to the elapsed time, and the distance traveled is proportional to the square of the elapsed time.[39] Importantly, the acceleration is the same for all bodies, independently of their mass. This follows from combining Newton's second law of motion with his law of universal gravitation. The latter states that the magnitude of the gravitational force from the Earth upon the body is F = G M m r 2 , {\displaystyle F={\frac {GMm}{r^{2}}},} where m {\displaystyle m} is the mass of the falling body, M {\displaystyle M} is the mass of the Earth, G {\displaystyle G} is Newton's constant, and r {\displaystyle r} is the distance from the center of the Earth to the body's location, which is very nearly the radius of the Earth. Setting this equal to m a {\displaystyle ma}, the body's mass m {\displaystyle m} cancels from both sides of the equation, leaving an acceleration that depends upon G {\displaystyle G}, M {\displaystyle M}, and r {\displaystyle r}, and r {\displaystyle r} can be taken to be constant. This particular value of acceleration is typically denoted g {\displaystyle g}: g = G M r 2 ≈ 9.8 m / s 2 . {\displaystyle g={\frac {GM}{r^{2}}}\approx \mathrm {9.8~m/s^{2}} .}
If the body is not released from rest but instead launched upwards and/or horizontally with nonzero velocity, then free fall becomes projectile motion.[40] When air resistance can be neglected, projectiles follow parabola-shaped trajectories, because gravity affects the body's vertical motion and not its horizontal. At the peak of the projectile's trajectory, its vertical velocity is zero, but its acceleration is g {\displaystyle g} downwards, as it is at all times. Setting the wrong vector equal to zero is a common confusion among physics students.[41]
Uniform circular motion
Main article: Circular motion
Two objects in uniform circular motion, orbiting around the barycenter (center of mass of both objects)
When a body is in uniform circular motion, the force on it changes the direction of its motion but not its speed. For a body moving in a circle of radius r {\displaystyle r} at a constant speed v {\displaystyle v}, its acceleration has a magnitude a = v 2 r {\displaystyle a={\frac {v^{2}}{r}}}and is directed toward the center of the circle.[note 9] The force required to sustain this acceleration, called the centripetal force, is therefore also directed toward the center of the circle and has magnitude m v 2 / r {\displaystyle mv^{2}/r}. Many orbits, such as that of the Moon around the Earth, can be approximated by uniform circular motion. In such cases, the centripetal force is gravity, and by Newton's law of universal gravitation has magnitude G M m / r 2 {\displaystyle GMm/r^{2}}, where M {\displaystyle M} is the mass of the larger body being orbited. Therefore, the mass of a body can be calculated from observations of another body orbiting around it.[43]: 130
Newton's cannonball is a thought experiment that interpolates between projectile motion and uniform circular motion. A cannonball that is lobbed weakly off the edge of a tall cliff will hit the ground in the same amount of time as if it were dropped from rest, because the force of gravity only affects the cannonball's momentum in the downward direction, and its effect is not diminished by horizontal movement. If the cannonball is launched with a greater initial horizontal velocity, then it will travel farther before it hits the ground, but it will still hit the ground in the same amount of time. However, if the cannonball is launched with an even larger initial velocity, then the curvature of the Earth becomes significant: the ground itself will curve away from the falling cannonball. A very fast cannonball will fall away from the inertial straight-line trajectory at the same rate that the Earth curves away beneath it; in other words, it will be in orbit (imagining that it is not slowed by air resistance or obstacles).[44]
Harmonic motion
Main article: Harmonic oscillator
An undamped spring–mass system undergoes simple harmonic motion.
Consider a body of mass m {\displaystyle m} able to move along the x {\displaystyle x} axis, and suppose an equilibrium point exists at the position x = 0 {\displaystyle x=0}. That is, at x = 0 {\displaystyle x=0}, the net force upon the body is the zero vector, and by Newton's second law, the body will not accelerate. If the force upon the body is proportional to the displacement from the equilibrium point, and directed to the equilibrium point, then the body will perform simple harmonic motion. Writing the force as F = − k x {\displaystyle F=-kx}, Newton's second law becomes m d 2 x d t 2 = − k x . {\displaystyle m{\frac {d^{2}x}{dt^{2}}}=-kx\,.} This differential equation has the solution x ( t ) = A cos ω t + B sin ω t {\displaystyle x(t)=A\cos \omega t+B\sin \omega t\,} where the frequency ω {\displaystyle \omega } is equal to k / m {\displaystyle {\sqrt {k/m}}}, and the constants A {\displaystyle A} and B {\displaystyle B} can be calculated knowing, for example, the position and velocity the body has at a given time, like t = 0 {\displaystyle t=0}.
One reason that the harmonic oscillator is a conceptually important example is that it is good approximation for many systems near a stable mechanical equilibrium.[note 10] For example, a pendulum has a stable equilibrium in the vertical position: if motionless there, it will remain there, and if pushed slightly, it will swing back and forth. Neglecting air resistance and friction in the pivot, the force upon the pendulum is gravity, and Newton's second law becomes d 2 θ d t 2 = − g L sin θ , {\displaystyle {\frac {d^{2}\theta }{dt^{2}}}=-{\frac {g}{L}}\sin \theta ,}where L {\displaystyle L} is the length of the pendulum and θ {\displaystyle \theta } is its angle from the vertical. When the angle θ {\displaystyle \theta } is small, the sine of θ {\displaystyle \theta } is nearly equal to θ {\displaystyle \theta } (see Taylor series), and so this expression simplifies to the equation for a simple harmonic oscillator with frequency ω = g / L {\displaystyle \omega ={\sqrt {g/L}}}.
A harmonic oscillator can be damped, often by friction or viscous drag, in which case energy bleeds out of the oscillator and the amplitude of the oscillations decreases over time. Also, a harmonic oscillator can be driven by an applied force, which can lead to the phenomenon of resonance.[46]
Objects with variable mass
Main article: Variable-mass system
Rockets, like the Space Shuttle Atlantis, propel matter in one direction to push the craft in the other. This means that the mass being pushed, the rocket and its remaining onboard fuel supply, is constantly changing.
Newtonian physics treats matter as being neither created nor destroyed, though it may be rearranged. It can be the case that an object of interest gains or loses mass because matter is added to or removed from it. In such a situation, Newton's laws can be applied to the individual pieces of matter, keeping track of which pieces belong to the object of interest over time. For instance, if a rocket of mass M ( t ) {\displaystyle M(t)}, moving at velocity v ( t ) {\displaystyle \mathbf {v} (t)}, ejects matter at a velocity u {\displaystyle \mathbf {u} } relative to the rocket, then F = M d v d t − u d M d t {\displaystyle \mathbf {F} =M{\frac {d\mathbf {v} }{dt}}-\mathbf {u} {\frac {dM}{dt}}\,} where F {\displaystyle \mathbf {F} } is the net external force (e.g., a planet's gravitational pull).[23]: 139
Work and energy
Physicists developed the concept of energy after Newton's time, but it has become an inseparable part of what is considered "Newtonian" physics. Energy can broadly be classified into kinetic, due to a body's motion, and potential, due to a body's position relative to others. Thermal energy, the energy carried by heat flow, is a type of kinetic energy not associated with the macroscopic motion of objects but instead with the movements of the atoms and molecules of which they are made. According to the work-energy theorem, when a force acts upon a body while that body moves along the line of the force, the force does work upon the body, and the amount of work done is equal to the change in the body's kinetic energy.[note 11] In many cases of interest, the net work done by a force when a body moves in a closed loop — starting at a point, moving along some trajectory, and returning to the initial point — is zero. If this is the case, then the force can be written in terms of the gradient of a function called a scalar potential:[42]: 303 F = − ∇ U . {\displaystyle \mathbf {F} =-\mathbf {\nabla } U\,.} This is true for many forces including that of gravity, but not for friction; indeed, almost any problem in a mechanics textbook that does not involve friction can be expressed in this way.[45]: 19 The fact that the force can be written in this way can be understood from the conservation of energy. Without friction to dissipate a body's energy into heat, the body's energy will trade between potential and (non-thermal) kinetic forms while the total amount remains constant. Any gain of kinetic energy, which occurs when the net force on the body accelerates it to a higher speed, must be accompanied by a loss of potential energy. So, the net force upon the body is determined by the manner in which the potential energy decreases.
Rigid-body motion and rotation
A rigid body is an object whose size is too large to neglect and which maintains the same shape over time. In Newtonian mechanics, the motion of a rigid body is often understood by separating it into movement of the body's center of mass and movement around the center of mass.
Center of mass
Main article: Center of mass
Fork-cork-toothpick object balanced on a pen on the toothpick part
The total center of mass of the forks, cork, and toothpick is on top of the pen's tip.
Significant aspects of the motion of an extended body can be understood by
imagining the mass ofhat body concentrated to a single point, known as the ce
nter of mass. The location of a body's center of mass depends upon how that body's m
aterial is distributed. For a collection of pointlike objects with masses m 1 , … , m N {\displaysty """
</code>
<code>
data
</code>
<code>
type(data)
</code>
<code>
list_of_words = data.split(" ")
</code>
<code>
s = "Permeabilty and Porosity"
</code>
<code>
s.split(" ")
</code>
<code>
list_of_words
</code>
<code>
len(list_of_words)
</code>
<code>
set_of_words = set(list_of_words)
</code>
<code>
len(set_of_words)
</code>
<code>
"\n"
</code>
<code>
lines = data.split("\n")
</code>
<code>
lines
</code>
<code>
len(lines)
</code>
<code>
print("Hello all \nHow is everybody")
</code>
## Dictionary
- key:value
- rules for keys
- can store anything as a value: str, int, float, dict, list, tuple,set
- {-:-} => key:value pair
- mutable
<code>
d = {}
</code>
<code>
d
</code>
<code>
type(d)
</code>
<code>
d = {1}
</code>
<code>
type(d)
</code>
<code>
d = {"name":"Jaiyesh"}
</code>
<code>
type(d)
</code>
<code>
d = {"name":"Jaiyesh","age":"xyz","number":545454,"mail id":"abc@xyz"}
</code>
<code>
d
</code>
<code>
d["number"]
</code>
<code>
rock_properties = {"porosity":0.35,"Permeability":150,"lithology":"Limestone"}
</code>
<code>
rock_properties
</code>
<code>
rock_properties["lithology"]
</code>
<code>
rock_properties[2]
</code>
### keys must be unique:
- repeat a key,value will be update to last data
<code>
rock_properties = {"porosity":0.35,"Permeability":150,"lithology":"Limestone",
"porosity":0.4,"lithology":"Shale"}
</code>
<code>
rock_properties
</code>
<code>
## Keys can be a number, string, or a fload
</code>
<code>
d1 = {2.7:"drilling",3:"reservoir"}
</code>
<code>
d1[2.7]
</code>
<code>
d1 = {2.7:"drilling",3:"reservoir","list":[34,34,21,54,65,["hello","perm"]],"tuple":(98,454,23,54,23,564),
"anything":{"poro":0.45,"drilling":"45 degree"}}
</code>
<code>
d1
</code>
<code>
d1["anything"]
</code>
<code>
d1["anything"]["poro"]
</code>
<code>
d1["list"]
</code>
<code>
rock_properties
</code>
<code>
rock_properties["lithology"] = "Sandstone"
</code>
<code>
rock_properties
</code>
<code>
rock_properties["Saturation"] = 0.34
</code>
<code>
rock_properties
</code>
<code>
rock_properties.keys()
</code>
<code>
rock_properties.values()
</code>
<code>
rock_properties
</code>
<code>
rock_properties.pop("Permeability")
</code>
<code>
rock_properties
</code>
<code>
rock_properties= {
"Well A": {"Porosity":0.3,"Permeability":12,"Lithology":"Shale"},
"Well B": {"Porosity":0.13,"Permeability":1,"Lithology":"Sandstone"},
"Well C": {"Porosity":0.23,"Permeability":120,"Lithology":"Limestone"}
}
</code>
<code>
rock_properties
</code>
<code>
rock_properties["Well B"]["Permeability"]
</code>
## Summary of Data Structure:
1. List: mutable, empty lsit can be used to populate later, [----]
2. tuple: immutable, sensitive data, (---)
3. Sets: uniordered, no duplication, mutable, {--}
4. Dictionary: Key:Value, mutable, {"key":"value"}
## for loops: utilose the power of compuation and iterations
<code>
poro = [0.2,0.4,0.7]
</code>
<code>
print(poro[0])
print(poro[1])
print(poro[2])
</code>
<code>
for i in poro:
print(i)
</code>
<code>
perm = [23,32,12,43,65,34,23]
</code>
<code>
perm.sort(reverse=True)
</code>
<code>
perm
</code>
|
{
"filename": "2025_Day 4_1.ipynb",
"repository": "jaiyesh/diploma",
"query": "transformed_from_existing",
"size": 151468,
"sha": ""
}
|
# gene_environment_interactions_external_variances.ipynb
Repository: Gibbons-Lab/2021
# Validation with external data set
Here we will validate the major findings in the data from [Bar et. al.](https://doi.org/10.1038/s41586-020-2896-2). They estimated explained variances using RandomForest models in a validatin cohort using the same metabolomics provider as our study.
<code>
import pandas as pd
joint_r_sq = pd.read_excel("data/41586_2020_2896_MOESM3_ESM.xlsx", "Supplementary Table 6")
meta = pd.read_excel("data/41586_2020_2896_MOESM3_ESM.xlsx", "Supplementary Table 1")
joint_r_sq["CHEMICAL_ID"] = joint_r_sq["Grouped Metabolites"].astype(str).str.split("+").str[0]
meta.CHEMICAL_ID = meta.CHEMICAL_ID.astype(str)
joint_r_sq = pd.merge(joint_r_sq, meta, on="CHEMICAL_ID")
joint_r_sq = joint_r_sq[(joint_r_sq["Genetics p-value"] < 0.05) | (joint_r_sq["Microbiome p-value"] < 0.05)]
joint_r_sq[["Grouped Metabolites", "CHEMICAL_ID", "BIOCHEMICAL"]]
joint_r_sq["metabolite"] = "metabolite_" + joint_r_sq.CHEMICAL_ID
</code>
Now we will classify the groups as before.
<code>
joint_r_sq["micro_r2"] = joint_r_sq["Microbiome R2"].apply(lambda x: x if x>0 else 0)
joint_r_sq["geno_r2"] = joint_r_sq["Genetics R2"].apply(lambda x: x if x>0 else 0)
joint_r_sq["total"] = joint_r_sq["micro_r2"] + joint_r_sq["geno_r2"]
joint_r_sq["group"] = "hybrid"
joint_r_sq.loc[(joint_r_sq["micro_r2"] <= 0.01 * joint_r_sq["total"]), "group"] = "genetics"
joint_r_sq.loc[(joint_r_sq["geno_r2"] <= 0.01 * joint_r_sq["total"]), "group"] = "microbiome"
joint_r_sq.sort_values(by="total", ascending=True, inplace=True)
joint_r_sq["BIOCHEMICAL"] = pd.Categorical(joint_r_sq.BIOCHEMICAL, joint_r_sq.BIOCHEMICAL.unique())
joint_r_sq.sort_values(by="total", ascending=False, inplace=True)
joint_r_sq.to_csv("joint_r_squared.csv", index=False)
top = []
for g in ["genetics", "microbiome", "hybrid"]:
top.extend(joint_r_sq[joint_r_sq.group == g].iloc[0:10]["Grouped Metabolites"])
long = joint_r_sq[["Grouped Metabolites", "BIOCHEMICAL", "group", "micro_r2", "geno_r2"]].melt(id_vars=["Grouped Metabolites", "BIOCHEMICAL", "group"], value_name="r2", var_name = "type")
</code>
<code>
train_r_sq = pd.read_csv("data/train_joint_r_sq.csv").merge(joint_r_sq, left_on="BIOCHEMICAL_NAME", right_on="BIOCHEMICAL")
valid_r_sq = pd.read_csv("data/valid_joint_r_sq.csv").merge(joint_r_sq, left_on="BIOCHEMICAL_NAME", right_on="BIOCHEMICAL")
train_r_sq["cohort"] = "training"
valid_r_sq["cohort"] = "validation"
comparison = pd.concat([train_r_sq, valid_r_sq])
</code>
<code>
from plotnine import *
from scipy.stats import pearsonr
theme_set(theme_minimal())
cor = comparison.groupby("cohort").apply(lambda df: pd.Series(pearsonr(df.micro_r2_x, df.micro_r2_y), index=["r", "p"])).reset_index()
cor["label"] = [f"r={row.r:.2f}, p={row.p:.2g}" for _, row in cor.iterrows()]
pl = (
ggplot(comparison)
+ aes(x="micro_r2_x", y="micro_r2_y")
+ geom_point()
#+ geom_point(data=comparison.dropna(subset=["SUB_PATHWAY_x"])[comparison.dropna(subset=["SUB_PATHWAY_x"]).SUB_PATHWAY_x.str.startswith("Xanthine")], color="orange")
+ geom_text(cor, aes(label="label"), x=0.45, y=0.025, ha="right")
+ stat_smooth(method="lm", color="royalblue")
+ facet_wrap("~ cohort")
+ labs(x="R² this study", y="R² Bar et. al.")
+ theme(figure_size=(6, 3))
)
pl.save("figures/external_r2_microbiome.pdf", width=6, height=3)
pl
</code>
<code>
from plotnine import *
from scipy.stats import pearsonr
theme_set(theme_minimal())
cor = comparison.groupby("cohort").apply(lambda df: pd.Series(pearsonr(df.geno_r2_x, df.geno_r2_y), index=["r", "p"])).reset_index()
cor["label"] = [f"r={row.r:.2f}, p={row.p:.2g}" for _, row in cor.iterrows()]
pl = (
ggplot(comparison)
+ aes(x="geno_r2_x", y="geno_r2_y")
+ geom_point()
+ geom_text(cor, aes(label="label"), x=0, y=0.4, ha="left")
+ stat_smooth(method="lm", color="royalblue")
+ facet_wrap("~ cohort")
+ labs(x="R² this study", y="R² Bar et. al.")
+ theme(figure_size=(6, 3))
)
pl.save("figures/external_r2_genetics.pdf", width=6, height=3)
pl
</code>
<code>
from plotnine import *
from mizani.formatters import percent_format
pl = (
ggplot(joint_r_sq, aes(x="geno_r2", y="micro_r2", color="group"))
+ geom_point(size=2, stroke=0)
+ geom_text(
aes(label="BIOCHEMICAL"),
data=joint_r_sq[
((joint_r_sq.total > 0.22) & (joint_r_sq.group == "hybrid"))
| (joint_r_sq.micro_r2 > 0.4)
| (joint_r_sq.geno_r2 > 0.55)],
ha="left", nudge_x=0.01, size=10, color="black")
+ theme_minimal()
+ theme(figure_size=(5,4))
+ xlim(0, 0.8)
+ scale_color_manual(values={"genetics": "steelblue", "microbiome": "mediumseagreen", "hybrid": "dimgray"})
+ labs(x="R² genetics", y="R² microbiome") + guides(color=None)
)
pl.save("figures/bar_r2.pdf", width=6, height=4)
pl
</code>
<code>
pl = (
ggplot(long[(long.r2 > 0.0) & (long["Grouped Metabolites"].isin(top))].sort_values(by="r2"), aes(y="r2", x="BIOCHEMICAL", fill="type"))
+ geom_bar(stat="identity")
+ scale_y_continuous(labels=percent_format())
+ coord_flip()
+ facet_wrap("~ group", scales="free_y", ncol=1)
+ labs(y = "explained metabolite variance", x="")
+ scale_fill_manual(values=["steelblue", "mediumseagreen"])
+ guides(fill = None)
+ theme_minimal()
+ theme(figure_size=(3, 8), subplots_adjust={"hspace": 0.5})
)
#pl.save("figures/external_specific_r2.pdf", width=3, height=6)
pl
</code>
|
{
"filename": "gene_environment_interactions_external_variances.ipynb",
"repository": "Gibbons-Lab/2021",
"query": "transformed_from_existing",
"size": 226277,
"sha": ""
}
|
# S2_1.ipynb
Repository: yackermann/udemy-langchain
<code>
from dotenv import load_dotenv
load_dotenv(dotenv_path='.env')
</code>
# LLMs
<code>
from langchain.llms import OpenAI
llm = OpenAI()
llm.predict("How are you?")
</code>
<code>
from langchain.chat_models import ChatOpenAI
chat_model = ChatOpenAI()
chat_model.predict("How are you?")
chat_model.predict("What was my previous question?")
</code>
# Chains
<code>
from langchain.chains import ConversationChain
chain = ConversationChain(
llm=chat_model,
verbose=True
)
chain.run("How are you today?")
</code>
<code>
chain.run("What was my previous question?")
</code>
Prompt Template
<code>
from langchain.prompts import PromptTemplate
template = """
Return all subcategories of a given category.
Category: {category}
"""
prompt = PromptTemplate(
template=template,
input_variables=["category"],
)
from langchain.chains import LLMChain
llm_chain = LLMChain(
llm=llm,
prompt=prompt,
verbose=True,
)
llm_chain.run(category="Computer science")
</code>
<code>
from langchain.prompts import ChatPromptTemplate, SystemMessagePromptTemplate, HumanMessagePromptTemplate, ChatMessagePromptTemplate
system_template = """
You are a helpful assistant who generates comma separated lists.
A user will only pass a category and you should generate a list of subcategories.
ONLY return comma separated values and nothing else!
"""
prompt = ChatPromptTemplate.from_messages([
SystemMessagePromptTemplate.from_template(system_template),
HumanMessagePromptTemplate.from_template("{category}"),
])
chain = LLMChain(
llm=llm,
prompt=prompt,
verbose=True,
)
chain.run("Machine learning")
</code>
# Output parser
<code>
from langchain.schema import BaseOutputParser
class CommaSeparatedParser(BaseOutputParser):
def parse(self, text):
output = text.strip().split(",")
output = [x.strip() for x in output]
return output
chain = LLMChain(
llm=llm,
prompt=prompt,
output_parser=CommaSeparatedParser(),
verbose=True,
)
chain.run("Machine learning")
</code>
<code>
input_list = [
{"category": "food"},
{"category": "country"},
{"category": "colours"},
]
response = chain.apply(input_list)
print(response)
</code>
# Simple Sequence
<code>
title_template = """
You are a writer
Given a subject, your job is to return a fun
title for a play.
Subject {subject}
Title:
"""
title_chain = LLMChain.from_string(
llm=llm,
template=title_template,
)
title_chain.run(subject="Machine learning")
</code>
<code>
synopsis_template = """
You are a writer
Given a title, write synopsis for a play.
Title: {title}
Synopsis:
"""
synopsis_chain = LLMChain.from_string(
llm=llm,
template=synopsis_template,
)
synopsis_chain.run(title="The Learning Machine: A Journey Through Artificial Intelligence")
</code>
<code>
from langchain.chains import SimpleSequentialChain
chain = SimpleSequentialChain(
chains=[title_chain, synopsis_chain],
verbose=True,
)
chain = chain.run("Machine learning.")
</code>
|
{
"filename": "S2_1.ipynb",
"repository": "yackermann/udemy-langchain",
"query": "transformed_from_existing",
"size": 19982,
"sha": ""
}
|
# serializer.ipynb
Repository: MaayanLab/datadistillery-kg
<code>
import pandas as pd
from glob import glob
from IPython.display import display, Markdown
from tqdm import tqdm
import csv
pd.__version__
</code>
<code>
base_input = "dd_data/20230802/"
</code>
<code>
hgnc_info = pd.read_csv('dd_data/HGNC_genes.txt', sep="\t")
hgnc_mapper = {}
for i, row in hgnc_info.iterrows():
uid = row["HGNC ID"]
enz = row["Enzyme (EC) ID"]
unip = row["UniProt accession"]
if uid not in hgnc_mapper:
hgnc_mapper[uid] = {}
if type(unip) == str:
hgnc_mapper[uid]["UNIPROTKB"] = unip
if type(enz) == str:
hgnc_mapper[uid]["ec_id"] = enz
</code>
## Load Data
<code>
concepts = pd.read_csv(base_input + "neo4j/import/CUIs.csv")
concepts = pd.DataFrame(index=concepts["CUI:ID"].unique())
concepts.index.name = "id"
concepts.head()
</code>
<code>
semantics = pd.read_csv(base_input + "neo4j/import/TUIs.csv", index_col=0)
semantics.head()
</code>
<code>
terms = pd.read_csv(base_input + "neo4j/import/SUIs.csv", index_col=0)
terms.head()
</code>
<code>
codes = pd.read_csv(base_input + "neo4j/import/CODEs.csv", index_col=0)
codes.head()
</code>
<code>
concept_term = pd.read_csv(base_input + "neo4j/import/CUI-SUIs.csv")
concept_term.head()
</code>
<code>
concept_semantics = pd.read_csv(base_input + "neo4j/import/CUI-TUIs.csv")
concept_semantics.head()
</code>
<code>
concept_code = pd.read_csv(base_input + "neo4j/import/CUI-CODEs.csv")
concept_code.head()
</code>
<code>
semantics_semantics = pd.read_csv(base_input + "neo4j/import/TUIrel.csv")
semantics_semantics.head()
</code>
<code>
code_term = pd.read_csv(base_input + "neo4j/import/CODE-SUIs.csv")
code_term.head()
</code>
## Merge Concept and Terms
<code>
concept_term.columns = ["CUI:ID", "SUI:ID"]
concept_term.shape
</code>
<code>
concept_term = pd.merge(concept_term, terms, on="SUI:ID", how='outer')
concept_term = concept_term.groupby('CUI:ID').first()
concept_term.head()
</code>
<code>
concept_term.shape
</code>
<code>
concept_term.columns = ["SUI:ID", "label"]
concept_term = concept_term[["label"]]
concept_term.head()
</code>
<code>
concept_term.shape
</code>
<code>
concepts.loc[concept_term.index, 'label'] = concept_term.loc[concept_term.index, 'label']
concepts.head()
</code>
## Semantics
<code>
semantics.head()
</code>
<code>
concept_semantics.head()
</code>
<code>
no_type = set(concepts.index) - set(concept_semantics[':START_ID'])
len(no_type)
</code>
<code>
with open('out/0623/semantics_ranked.tsv') as o:
ranked_type = [i.strip() for i in o.read().strip().split("\n")]
</code>
<code>
concept_semantics.columns = ["id", "TUI:ID"]
concept_semantics["type"] = [semantics.at[i, 'name'] for i in concept_semantics['TUI:ID']]
concept_semantics.head()
</code>
<code>
def fetch_type(v):
cat = ""
rank = len(ranked_type)
for i in v:
r = ranked_type.index(i)
if r < rank:
cat = i
rank = r
return cat
</code>
<code>
cs = concept_semantics.groupby('id')['type'].apply(lambda x: "; ".join(set(x)))
cs.head()
</code>
<code>
cs_ranked = concept_semantics.groupby('id')['type'].apply(fetch_type)
cs_ranked.head()
</code>
<code>
common = list(set(concepts.index).intersection(cs.index))
cs[common].head()
</code>
<code>
concept_semantics
concepts.loc[common, 'type'] = cs_ranked[common]
concepts.loc[common, 'type_combined'] = cs[common]
concepts.head()
</code>
<code>
out_prefix = "out/0915/"
</code>
<code>
concepts.groupby("type_combined").first().to_csv(out_prefix + 'semantics.tsv', sep="\t")
</code>
<code>
concepts.head()
</code>
<code>
concepts.shape
</code>
<code>
with open(out_prefix + 'semantics_list.tsv', 'w') as o:
o.write("\n".join([str(i) for i in concept_semantics.type.unique()]))
</code>
<code>
codes.head()
</code>
<code>
concept_code.columns = ["id", "CodeID:ID"]
concept_code.head()
</code>
<code>
concept_code = pd.merge(concept_code, codes, on="CodeID:ID", how='left')
concept_code.head()
</code>
<code>
concept_code[concept_code.id == 'C0000097']
</code>
<code>
concepts.head()
</code>
<code>
type_mapper = {}
with open("output/unique_SABS_of_Concept_Mapper.txt") as o:
for line in o:
r = line.strip().split(":")
if len(r) == 2:
type_mapper[r[0]] = r[1]
elif 'MSIGDB' in r[0]:
type_mapper[r[0]] = 'MSIGDB'
else:
type_mapper[r[0]] = r[0]
</code>
<code>
for i,row in tqdm(concept_code[concept_code.id.isin(concepts[concepts.type.isna()].index)].iterrows()):
sab = row["SAB"]
ind = row["id"]
if type(sab) == str:
if 'MSIGDB' in sab:
sab = 'MSIGDB'
concept_code.at[i, 'SAB'] = 'MSIGDB'
if sab == 'MSIGDB':
tp = 'MSIGDB'
else:
tp = type_mapper[sab]
if tp:
concepts.at[ind, "type"] = tp
concepts.at[ind, "type_combined"] = tp
</code>
<code>
for i, row in concepts.iterrows():
concepts.at[i, "type"] = row["type"].replace(".", " ")
concepts.at[i, "type_combined"] = row["type_combined"].replace(".", " ")
</code>
<code>
concepts[concepts.type == "UNIPROTKB"].head()
</code>
<code>
for tp in tqdm(concepts.type.unique()):
con = concepts[concepts.type==tp].copy()
cc = concept_code[concept_code.id.isin(con.index)]
for sab in cc.SAB.unique():
c = cc[cc.SAB == sab]
c = c.groupby('id').first()
common = list(set(con.index).intersection(c.index))
con.loc[common, sab] = c.loc[common, "CodeID:ID"]
if c.loc[common, "value:float"].isna().sum() != len(common):
con.loc[common, "%s value"%sab] = c.loc[common, "value:float"]
if c.loc[common, "lowerbound:float"].isna().sum() != len(common):
con.loc[common, "%s lowerbound"%sab] = c.loc[common, "lowerbound:float"]
if c.loc[common, "upperbound:float"].isna().sum() != len(common):
con.loc[common, "%s upperbound"%sab] = c.loc[common, "upperbound:float"]
if c.loc[common, "unit"].isna().sum() != len(common):
con.loc[common, "%s unit"%sab] = c.loc[common, "unit"]
if "-" in list(con["label"]):
tmp = con[con.label == "-"]
ind = set(tmp.index).intersection(con.index)
ind2 = set(tmp.index).intersection(c.index)
if len(ind.intersection(ind2)) > 0:
l = list(ind.intersection(ind2))
con.loc[l, "label"] = c.loc[l, 'CodeID:ID']
con.to_csv("out/0915/serialization/nodes/%s.nodes.csv"%(tp))
</code>
<code>
gene_or_genome_df = pd.read_csv("out/0915/serialization/nodes/Gene or Genome.nodes.csv", index_col=0)
uniprot = pd.read_csv("out/0915/serialization/nodes/UNIPROTKB.nodes.csv", index_col=0)
gene_df = pd.read_csv("out/0915/serialization/nodes/Gene.nodes.csv", index_col=0)
</code>
<code>
uniprot.head()
</code>
<code>
uniprot.shape, gene_df.shape, gene_or_genome_df.shape
</code>
<code>
gene_or_genome_df
</code>
<code>
uniprot_id_mapper = pd.read_csv('output/idmapping_2023_09_18.tsv', sep="\t", index_col=0)
uniprot_id_mapper.head()
</code>
<code>
new_gene_or_genome = gene_or_genome_df[gene_or_genome_df.HGNC.isna()]
</code>
<code>
rows = {}
hgnc_mapper = {}
for i, row in gene_or_genome_df[~gene_or_genome_df.HGNC.isna()].iterrows():
hgnc = row["HGNC"]
hgnc_mapper[hgnc] = i
row["type"] = "Gene"
row["type_combined"] = row["type_combined"].replace("Gene or Genome", "Gene")
rows[i] = row
len(rows)
</code>
<code>
for i, row in gene_df.iterrows():
hgnc = row["HGNC"]
if hgnc not in hgnc_mapper:
hgnc_mapper[hgnc] = i
row["type"] = "Gene"
rows[i] = row
len(rows)
</code>
<code>
gene_df.head()
</code>
<code>
uniprot.head()
</code>
<code>
uniprot_kb_mapper = {}
uniprot_list = []
with open("uniprot_ids_0917.txt", "w") as o:
for i, row in uniprot.iterrows():
kb = row["UNIPROTKB"].replace("UNIPROTKB:", "")
o.write("%s\n"%kb)
# hgnc = uniprot_id_mapper.at[kb, 'To']
# hgnc = row["HGNC"]
# if hgnc not in hgnc_mapper:
# hgnc_mapper[hgnc] = i
# row["type"] = "Gene"
# rows[i] = row
</code>
<code>
uniprot_mapper = {}
for k, v in uniprot_id_mapper.iterrows():
uniprot_mapper[k] = v["To"]
</code>
<code>
no_hgnc = set()
for i, row in uniprot.iterrows():
kb = row["UNIPROTKB"].replace("UNIPROTKB:", "")
if kb not in uniprot_mapper:
no_hgnc.add(kb)
rows[i] = row
else:
hgnc = uniprot_mapper[kb]
if hgnc in hgnc_mapper:
cui = hgnc_mapper[hgnc]
rows[cui]["UNIPROTKB"] = kb
else:
row["HGNC"] = hgnc
row["type"] = "Gene"
row["type_combined"] = "Gene"
rows[i] = row
</code>
<code>
len(rows)
</code>
<code>
new_gene_df = pd.DataFrame.from_dict(rows, orient="index")
</code>
<code>
new_gene_df.head()
</code>
<code>
new_gene_df.type_combined = "Gene"
new_gene_df.type = "Gene"
new_gene_df.type_combined.unique(), new_gene_df.type.unique()
</code>
<code>
concepts.head()
</code>
<code>
for i in new_gene_df.index:
concepts.at[i, "type"] = "Gene"
concepts.at[i, "type_combined"] = "Gene"
</code>
<code>
new_gene_df.to_csv("out/0915/serialization/nodes/Gene.nodes.csv")
</code>
<code>
new_gene_or_genome.to_csv("out/0915/serialization/nodes/Gene or Genome.nodes.csv")
</code>
<code>
import os
</code>
<code>
row_headers = ["source", "relation", "target", "source_label", "target_label", "SAB", "evidence"]
with open(base_input + "neo4j/import/CUI-CUIs.csv") as o:
csv_reader = csv.reader(o)
headers = None
for row in tqdm(csv_reader):
if not headers:
headers = row
else:
source = row[0]
if source in uniprot_mapper:
source = uniprot_mapper[source]
target = row[1]
if target in uniprot_mapper:
target = uniprot_mapper[target]
if source in concepts.index and target in concepts.index:
source_label = concepts.at[source, 'label']
source_type = concepts.at[source, 'type']
target_label = concepts.at[target, 'label']
target_type = concepts.at[target, 'type']
relation = row[2]
sab = row[3]
evidence = ''
if len(row) > 4:
evidence = row[4]
filename = 'out/0915/serialization/edges/%s.%s.%s.edges.csv'%(source_type, relation, target_type)
write_header = False
operation = "a"
if not os.path.isfile(filename):
write_header = True
operation = "w"
# source_list = set()
# target_list = set()
with open(filename, operation) as w:
csv_writer = csv.writer(w)
if write_header:
csv_writer.writerow(row_headers)
csv_writer.writerow([source, relation, target, source_label, target_label, sab, evidence])
# source_list.add(source)
# target_list.add(target)
# # take note of nodes that are used for source and target
# source_ids = "out/serialization/ids/%s.txt"%source_type
# if not os.path.isfile(source_ids):
# with open(source_ids, 'w') as o:
# o.write("\n".join(source_list))
# else:
# with open(source_ids) as o:
# source_list = source_list.union(o.read().strip().split("\n"))
# with open(source_ids, 'w') as o:
# o.write("\n".join(source_list))
# target_ids = "out/serialization/ids/%s.txt"%target_type
# if not os.path.isfile(target_ids):
# with open(target_ids, 'w') as o:
# o.write("\n".join(target_list))
# else:
# with open(target_ids) as o:
# target_list = target_list.union(o.read().strip().split("\n"))
# with open(target_ids, 'w') as o:
# o.write("\n".join(target_list))
</code>
<code>
for filename in glob("out/0915/serialization/nodes/*.csv"):
df = pd.read_csv(filename, index_col=0, low_memory=False)
orig_columns = df.columns
if "type_combined" in df.columns:
dtype = df.type.unique()[0]
combined = set()
for i in df.type_combined:
combined = combined.union(i.split("; "))
# remove og type
combined = combined - {dtype}
columns = [i for i in df.columns if not i == "type_combined"] + list(combined)
if len(combined) > 0:
print(filename)
for i in combined:
df[i] = False
for i, row in df.iterrows():
type_combined = row["type_combined"].split("; ")
for t in type_combined:
col = "is_%s"%t
df.at[i, col] = True
df = df[columns]
df.to_csv(filename)
</code>
<code>
with open("output/august_dcc_sabs.txt") as o:
sabs_to_keep = set(o.read().strip().split("\n"))
</code>
<code>
import re
import os
edge_pattern = "(?P<directory>.+)/(?P<source_type>.+)\.(?P<relation>.+)\.(?P<target_type>.+)\.(?P<entity>.+)\.csv"
</code>
<code>
node_base = "out/0915/serialization/nodes/%s.nodes.csv"
new_node_base = "out/0915/filtered/nodes/%s.nodes.csv"
new_edge_base = "out/0915/filtered/edges/%s.%s.%s.edges.csv"
ids_base = "out/0915/filtered/ids/%s.txt"
node_ids = {}
sab_relations = {}
processed = set()
</code>
<code>
def glygen(s):
return s.replace("GLYGEN.RESIDUE", "GLYGEN_RESIDUE").replace("GLYCAN.MOTIF", "GLYCAN_MOTIF").replace('GLYCOSYLTRANSFERASE.REACTION', 'GLYCOSYLTRANSFERASE_REACTION').replace("GLYGEN.SRC", "GLYGEN_SRC").replace('GLYGEN.GLYCOSYLATION', 'GLYGEN_GLYCOSYLATION')
def glygen_reverse(s):
return s.replace("GLYGEN_RESIDUE", "GLYGEN.RESIDUE").replace("GLYCAN_MOTIF", "GLYCAN.MOTIF").replace('GLYCOSYLTRANSFERASE_REACTION', 'GLYCOSYLTRANSFERASE.REACTION').replace("GLYGEN_SRC", "GLYGEN.SRC").replace('GLYGEN_GLYCOSYLATION', 'GLYGEN.GLYCOSYLATION')
</code>
<code>
for filename in tqdm(glob("out/0915/serialization/edges/*.csv")):
if filename not in processed:
match = re.match(edge_pattern, glygen(filename)).groupdict()
entity = match["entity"]
source_type = glygen_reverse(match["source_type"])
relation = match["relation"].replace("_", " ")
target_type = glygen_reverse(match["target_type"])
if "inverse" not in relation:
edge_df = pd.read_csv(filename, low_memory=False)
# filter for SAB
sabs = sabs_to_keep.intersection(edge_df.SAB.unique())
for sab in sabs:
if sab not in sab_relations:
sab_relations[sab] = set()
sab_relations[sab].add(relation)
if len(sabs) > 0:
edge_df = edge_df[edge_df.SAB.isin(sabs)]
if not os.path.isfile(ids_base%source_type):
with open(ids_base%source_type, 'w') as o:
o.write("\n".join(edge_df.source))
else:
with open(ids_base%source_type) as o:
ids = set(o.read().strip().split("\n"))
with open(ids_base%source_type, 'w') as o:
ids = ids.union(edge_df.source)
o.write("\n".join(ids))
if not os.path.isfile(ids_base%target_type):
with open(ids_base%target_type, 'w') as o:
o.write("\n".join(edge_df.target))
else:
with open(ids_base%target_type) as o:
ids = set(o.read().strip().split("\n"))
with open(ids_base%target_type, 'w') as o:
ids = ids.union(edge_df.target)
o.write("\n".join(ids))
# source_df = pd.read_csv(node_base%source_type, index_col=0, low_memory=False)
# if os.path.isfile(new_node_base%(source_type)):
# new_source_df = pd.read_csv(new_node_base%(source_type), index_col=0, low_memory=False)
# pd.concat([new_source_df, source_df]).dropna(axis=1).to_csv(new_node_base%(source_type))
# else:
# source_df.dropna(axis=1).to_csv(new_node_base%(source_type))
# target_df = pd.read_csv(node_base%target_type, index_col=0, low_memory=False)
# if os.path.isfile(new_node_base%(target_type)):
# new_target_df = pd.read_csv(new_node_base%(target_type), index_col=0, low_memory=False)
# pd.concat([new_target_df, target_df]).dropna(axis=1).to_csv(new_node_base%(target_type))
# else:
# target_df.dropna(axis=1).to_csv(new_node_base%(target_type))
edge_df.to_csv(new_edge_base%(source_type, relation, target_type), index=False)
processed.add(filename.replace("GLYGEN_RESIDUE", "GLYGEN.RESIDUE"))
</code>
<code>
count = 0
for filename in tqdm(glob("out/0915/filtered/ids/*.txt")):
count+=1
count
</code>
<code>
id_pattern = "(?P<directory>.+)/(?P<type>.+)\.txt"
for filename in tqdm(glob("out/0915/filtered/ids/*.txt")):
if not "inverse" in filename and not "isa_" in filename:
match = re.match(id_pattern, filename).groupdict()
node_type = match["type"]
node_df = pd.read_csv(node_base%node_type, index_col=0, low_memory=False)
with open(filename) as o:
ids = list(set(o.read().strip().split("\n")).intersection(node_df.index))
node_df.loc[ids].dropna(axis=1, how="all").to_csv(new_node_base%node_type)
</code>
<code>
hgnc = pd.read_csv("out/0915/filtered/nodes/Gene.nodes.csv", low_memory=False)
</code>
<code>
hgnc.head()
</code>
<code>
for i in glob('out/0915/filtered/edges/*'):
if "UNIPROT" in i:
print(i)
</code>
<code>
concepts.type.unique()
</code>
<code>
gene_or_genome_df = pd.read_csv("out/0915/filtered/nodes/Gene or Genome.nodes.csv", index_col=0)
uniprot = pd.read_csv("out/0915/filtered/nodes/UNIPROTKB.nodes.csv", index_col=0)
gene_df = pd.read_csv("out/0915/filtered/nodes/Gene.nodes.csv", index_col=0)
</code>
<code>
gene_or_genome_df.head()
</code>
<code>
uniprot.head()
</code>
<code>
gene_df.head()
</code>
<code>
for i, row in gene_or_genome_df.iterrows():
gene_df.loc[i] = row
</code>
<code>
for i, row in uniprot.iterrows():
gene_df.loc[i] = row
</code>
<code>
gene_df.type = "Gene"
</code>
<code>
gene_df.to_csv("out/0915/filtered/nodes/Gene.nodes.csv")
</code>
<code>
import os
</code>
<code>
os.remove("out/0915/filtered/nodes/UNIPROTKB.nodes.csv")
os.remove("out/0915/filtered/nodes/Gene or Genome.nodes.csv")
</code>
<code>
for i in glob('out/0915/filtered/edges/*'):
if "UNIPROTKB" in i:
os.rename(i, i.replace("UNIPROTKB", "Gene"))
if "Gene or Genome" in i:
os.rename(i, i.replace("Gene or Genome", "Gene"))
</code>
<code>
gene_df.head()
</code>
<code>
hgnc_genes = pd.read_csv("dd_data/HGNC_genes.txt", sep="\t")
hgnc_genes.head()
</code>
<code>
hgnc_mapper = {}
for i, row in hgnc_genes.iterrows():
hgnc_id = row["HGNC ID"]
enz_id = row["Enzyme (EC) ID"]
if hgnc_id not in hgnc_mapper:
hgnc_mapper[hgnc_id] = {
"EC ID": enz_id,
"is_Enzyme": type(enz_id) == str
}
</code>
<code>
for i, row in gene_df.iterrows():
hgnc_id = row["HGNC"]
if hgnc_id in hgnc_mapper:
gene_df.at[i, "EC ID"] = hgnc_mapper[hgnc_id]["EC ID"]
gene_df.at[i, "is_Enzyme"] = hgnc_mapper[hgnc_id]["is_Enzyme"]
</code>
<code>
gene_df[gene_df.is_Enzyme == True]
</code>
<code>
for filename in glob('out/0915/filtered/edges/*'):
df = pd.read_csv(filename)
if "CMAP" in df.SAB.unique():
print(filename)
os.remove(filename)
</code>
<code>
dcc_mapper = {}
with open('output/sabs_dcc_mapper.txt') as o:
for line in o:
r = line.strip().split(":")
if len(r) == 2:
dcc_mapper[r[0]] =r[1]
else:
dcc_mapper[r[0]] =r[0]
</code>
<code>
for filename in glob('out/0915/filtered/edges/*'):
df = pd.read_csv(filename)
if len(df.SAB.unique()) > 1:
print(filename, df.SAB.unique())
df["DCC"] = dcc_mapper[df.SAB.unique()[0]]
else:
df["DCC"] = dcc_mapper[df.SAB.unique()[0]]
df.to_csv(filename)
</code>
<code>
gene_df.index.name = "id"
gene_df
</code>
<code>
gene_df.to_csv("out/0915/filtered/nodes/Gene.nodes.csv")
</code>
<code>
filenames = []
for filename in glob('out/0915/filtered/nodes/*'):
df = pd.read_csv(filename, index_col=0, low_memory=False)
if "label" not in df.columns:
filenames.append(filename)
df['label'] = df.index
df.to_csv(filename)
print(filename)
</code>
<code>
len(filenames)
</code>
<code>
from py2neo import Graph
from dotenv import load_dotenv
load_dotenv()
graph = Graph(os.getenv('NEO4j_URL'), auth=(os.getenv('NEO4J_USER'), os.getenv('NEO4J_PASSWORD')))
</code>
<code>
import re
</code>
<code>
node_pattern = "(?P<directory>.+)/(?P<node_type>.+)\.(?P<entity>.+)\.csv"
for filename in filenames:
match = re.match(node_pattern, filename).groupdict()
node_type = match["node_type"]
print(node_type)
query = "MATCH (a: `%s`) WHERE a.label IS NULL SET a.label = a.id"%node_type
graph.run(query)
</code>
<code>
for filename in glob('out/0915/filtered/edges/*'):
df = pd.read_csv(filename, index_col=0)
df.to_csv(filename, index=False)
</code>
<code>
relations = set()
for filename in glob('out/0915/filtered/edges/*'):
match = re.match(edge_pattern, filename).groupdict()
relations.add(match['relation'])
</code>
<code>
from glob import glob
import re
</code>
<code>
gtex = "GTEXEXP"
# gtex = "GTEXEQTL"
</code>
<code>
for filename in glob('out/0915/filtered/edges/*'):
# relations.add(match['relation'])
if gtex in filename:
match = re.match(edge_pattern, filename).groupdict()
print(filename, match["relation"])
</code>
<code>
gene2gtexp = pd.read_csv("out/0915/filtered/edges/Gene.expresses.GTEXEXP.edges.csv")
tissue2gtexp = pd.read_csv("out/0915/filtered/edges/Tissue.expresses.GTEXEXP.edges.csv")
organ2gtexp = pd.read_csv("out/0915/filtered/edges/Body Part, Organ, or Organ Component.expresses.GTEXEXP.edges.csv")
location2gtexp = pd.read_csv("out/0915/filtered/edges/Body Location or Region.expresses.GTEXEXP.edges.csv")
hasExp = pd.read_csv("out/0915/filtered/edges/GTEXEXP.has expression.EXPBINS.edges.csv")
</code>
<code>
gene2gtexp.head()
</code>
<code>
tissue2gtexp.head()
</code>
<code>
organ2gtexp.head()
</code>
<code>
location2gtexp.head()
</code>
<code>
gene2gtexp.shape
</code>
<code>
hasExp.head()
</code>
<code>
len(set(tissue2gtexp.target)), len(set(tissue2gtexp.target).intersection(gene2gtexp.target))
</code>
<code>
gtexp_gene_mapper = {}
for i, row in gene2gtexp.iterrows():
gene_id = row["source"]
gene = row["source_label"]
gtexp = row["target"]
gtexp_gene_mapper[gtexp] = {
"gene_id": gene_id,
"gene": gene,
}
(i, len(gtexp_gene_mapper))
</code>
<code>
evidence_mapper = {}
for i, row in hasExp.iterrows():
gtexp = row["source"]
target = row["target"]
# EXPBINS:0.1.0.2 CUI
score = float(".".join(target.replace("CUI", "").strip().split(".")[2:]))
evidence_mapper[gtexp] = score
</code>
<code>
counter = 0
for i, row in tissue2gtexp.iterrows():
target = row["target"]
if target in gtexp_gene_mapper:
val = gtexp_gene_mapper[target]
tissue2gtexp.at[i, 'target'] = val["gene_id"]
tissue2gtexp.at[i, 'target_label'] = val["gene"]
if target in evidence_mapper:
tissue2gtexp.at[i, 'evidence'] = evidence_mapper[target]
else:
counter+=1
print(counter)
tissue2gtexp.head()
</code>
<code>
tissue2gtexp.to_csv("out/0915/filtered/edges/Tissue.expresses.Gene.edges.csv", index=False)
</code>
<code>
counter = 0
for i, row in organ2gtexp.iterrows():
target = row["target"]
if target in gtexp_gene_mapper:
val = gtexp_gene_mapper[target]
organ2gtexp.at[i, 'target'] = val["gene_id"]
organ2gtexp.at[i, 'target_label'] = val["gene"]
if target in evidence_mapper:
organ2gtexp.at[i, 'evidence'] = evidence_mapper[target]
else:
counter+=1
print(counter)
organ2gtexp.head()
</code>
<code>
organ2gtexp.to_csv("out/0915/filtered/edges/Body Part, Organ, or Organ Component.expresses.Gene.edges.csv", index=False)
</code>
<code>
counter = 0
for i, row in location2gtexp.iterrows():
target = row["target"]
if target in gtexp_gene_mapper:
val = gtexp_gene_mapper[target]
location2gtexp.at[i, 'target'] = val["gene_id"]
location2gtexp.at[i, 'target_label'] = val["gene"]
if target in evidence_mapper:
location2gtexp.at[i, 'evidence'] = evidence_mapper[target]
else:
counter+=1
print(counter)
location2gtexp.head()
</code>
<code>
location2gtexp.to_csv("out/0915/filtered/edges/Body Location or Region.expresses.Gene.edges.csv", index=False)
</code>
<code>
os.remove("out/0915/filtered/edges/Gene.expresses.GTEXEXP.edges.csv")
os.remove("out/0915/filtered/edges/Tissue.expresses.GTEXEXP.edges.csv")
os.remove("out/0915/filtered/edges/Body Part, Organ, or Organ Component.expresses.GTEXEXP.edges.csv")
os.remove("out/0915/filtered/edges/Body Location or Region.expresses.GTEXEXP.edges.csv")
os.remove("out/0915/filtered/edges/GTEXEXP.has expression.EXPBINS.edges.csv")
</code>
<code>
gtex = "GTEXEQTL"
for filename in glob('out/0915/filtered/edges/*'):
# relations.add(match['relation'])
match = re.match(edge_pattern, filename).groupdict()
if gtex in match["target_type"]:
print(filename, match["relation"])
</code>
<code>
gtex = "GTEXEQTL"
for filename in glob('out/0915/filtered/edges/*'):
# relations.add(match['relation'])
match = re.match(edge_pattern, filename).groupdict()
if gtex in match["source_type"]:
print(filename, match["relation"])
</code>
<code>
df = pd.read_csv("out/0915/filtered/edges/ENTREZ.positively regulated by.GTEXEQTL.edges.csv")
df.head()
</code>
<code>
import csv
</code>
<code>
for filename in glob('out/0915/filtered/edges/*'):
if 'ENTREZ.' in filename:
with open(filename) as o:
csv_reader = csv.reader(o)
header = True
for row in csv_reader:
if header:
header = False
else:
print(filename, row[5])
df = pd.read_csv(filename)
print(df.shape)
break
</code>
<code>
df = pd.read_csv("out/0915/filtered/edges/ENCODE CCRE ACTIVITY.regulates.ENTREZ.edges.csv")
df.shape
</code>
<code>
df.target.unique()
</code>
<code>
for filename in glob('out/0915/filtered/edges/*'):
with open(filename) as o:
csv_reader = csv.reader(o)
header = True
for row in csv_reader:
if header:
header = False
else:
if 'ERCC' == row[-1]:
print(filename, row[-1])
break
</code>
<code>
# Replace ENTREZ node to gene
entrez = pd.read_csv("out/0915/filtered/nodes/ENTREZ.nodes.csv", index_col=0)
entrez.head()
</code>
<code>
gene_df = pd.read_csv("out/0915/filtered/nodes/Gene.nodes.csv", index_col=0)
gene_df.head()
</code>
<code>
gene_df.loc["ENSEMBL:ENSG00000274253 CUI"] = entrez.loc["ENSEMBL:ENSG00000274253 CUI"]
</code>
<code>
gene_df.type = "Gene"
</code>
<code>
gene_df.to_csv("out/0915/filtered/nodes/Gene.nodes.csv")
</code>
<code>
os.remove('out/0915/filtered/nodes/ENTREZ.nodes.csv')
</code>
<code>
for filename in sorted(glob('out/0915/filtered/edges/*')):
if 'ENTREZ.' in filename:
append_file = filename.replace("ENTREZ", "Gene")
print(append_file)
with open(append_file, "a") as a:
csv_writer = csv.writer(a)
with open(filename) as o:
csv_reader = csv.reader(o)
h = True
for row in csv_reader:
if h:
h = False
else:
if not row[5] == "LINCS":
csv_writer.writerow(row)
os.remove(filename)
</code>
<code>
gtex = "GTEXEQTL"
for filename in glob('out/0915/filtered/edges/*'):
# relations.add(match['relation'])
match = re.match(edge_pattern, filename).groupdict()
if gtex in match["source_type"]:
df = pd.read_csv(filename)
print(filename, df.DCC.unique())
</code>
<code>
df = pd.read_csv("out/0915/filtered/edges/GTEXEQTL.located in.Gene.edges.csv")
df.head()
</code>
<code>
import pandas as pd
from glob import glob
import re
</code>
<code>
id_mapping = pd.read_csv('dd_data/idmapping_2023_08_24.tsv.gz', sep='\t')
id_mapping.head()
</code>
<code>
mapper = {}
for i, row in id_mapping.iterrows():
f = row["From"]
t = row["To"]
mapper["UNIPROTKB:%s CUI"%f] = t
</code>
<code>
genes = pd.read_csv('out/0915/filtered/nodes/Gene.nodes.csv', index_col=0)
genes.head()
</code>
<code>
hgnc_to_cui = {}
uniprot_to_cui = {}
for i, row in genes.iterrows():
hgnc = row["HGNC"]
uniprot = row["UNIPROTKB"]
if hgnc and type(hgnc) == str:
hgnc_to_cui[hgnc] = i
if uniprot:
uniprot_to_cui["UNIPROTKB:%s CUI"%uniprot] = i
len(uniprot_to_cui)
</code>
<code>
matched = []
counter = 0
hgnc_match = []
for i in genes.index:
if i in mapper:
matched.append(i)
if mapper[i] in hgnc_to_cui:
hgnc_match.append((i, mapper[i], hgnc_to_cui[mapper[i]]))
if "UNIPROTKB" in i:
counter += 1
print(counter, len(matched), len(hgnc_match))
</code>
<code>
genes = genes.drop(labels=[i[0] for i in hgnc_match])
</code>
<code>
label_mapper = {}
for i in hgnc_match:
ind = i[2]
label = genes.at[ind, 'label']
label_mapper[ind] = label
</code>
<code>
label_mapper[ind], ind
</code>
<code>
edge_pattern
</code>
<code>
from tqdm import tqdm
</code>
<code>
with_uniprot = set()
</code>
<code>
for filename in tqdm(glob("out/0915/filtered/edges/*Gene.*.csv")):
match = re.match(edge_pattern, filename).groupdict()
column = ""
if match["target_type"] == "Gene":
column = "target"
elif match["source_type"] == "Gene":
column = "source"
df = pd.read_csv(filename)
save = False
for i, row in df.iterrows():
gene_id = row[column]
if gene_id in mapper:
save = True
hgnc = mapper[gene_id]
cui = hgnc_to_cui[mapper[gene_id]]
label = label_mapper[cui]
df.at[i, column] = cui
df.at[i, "%s_label"%column] = cui
if save:
print(filename)
df.to_csv(filename, index=False)
</code>
<code>
label_mapper['C1412311']
</code>
<code>
row
</code>
<code>
genes.to_csv('out/0915/filtered/nodes/Gene.nodes.csv')
</code>
<code>
for i in glob("out/0915/filtered/edges/*expresses*"):
print(i)
df = pd.read_csv(i)
print(len(df.source_label.unique()))
</code>
<code>
df = pd.read_csv("out/0915/filtered/edges/Body Part, Organ, or Organ Component.expresses.Gene.edges.csv")
df.head()
</code>
<code>
df.source_label.unique()
</code>
<code>
import pandas as pd
from glob import glob
from tqdm import tqdm
</code>
<code>
pd.read_csv("out/0915/filtered/edges/Tissue.expresses.Gene.edges.csv").sort_values("evidence", ascending=False).head()
</code>
<code>
dccs = {}
for i in tqdm(glob('out/0915/filtered/edges/*')):
df = pd.read_csv(i)
for k,v in df.groupby(["DCC"])["DCC"].count().items():
if k not in dccs:
dccs[k] = 0
dccs[k] += v
with open("output/ids/%s"%df.DCC.unique()[0], "a") as o:
o.write("\n".join(list(set(list(df.source) + list(df.target)))))
o.write("\n")
</code>
<code>
for k,v in dccs.items():
with open("output/ids/%s"%k) as o:
nodes = len(set(o.read().strip().split("\n")))
dccs[k] = {
"nodes": nodes,
"edges": v["edges"]
}
</code>
<code>
pd.DataFrame.from_dict(dccs, orient="index")
</code>
|
{
"filename": "serializer.ipynb",
"repository": "MaayanLab/datadistillery-kg",
"query": "transformed_from_existing",
"size": 345123,
"sha": ""
}
|
# Cell_aggregates_zscores.ipynb
Repository: Katharina782/scDoRI
# Creating cell aggregates
We want to create 500 cell aggreagates of 100 cells each. To create these groupings, we will randomly sample 500 cell from the latent space embedding. Using a nearest neighbor search we will then find the 100 nearest neighbors of these cells. Any group of cells which has an overlap >80% with any of the previously created groups will be removed. These cell aggregates can then be used for downstream correlation tasks.
In the following I subset all matrices (gene expression, ArchR gene scores computed from scATAC-seq and gene scores computed from peak-to-gene links to contain an overlapping list of genes. The reason why each matrix has a different number of genes is that ArchR gene scores contain more genes, because based on chromatin accessibility we might
<code>
import scanpy as sc
import scvi
import numpy as np
import matplotlib.pyplot as plt
import numpy as np
from sklearn.neighbors import NearestNeighbors
from numba import jit, njit
import seaborn as sns
from scipy import sparse
import seaborn as sns
import pandas as pd
import h5py
import pickle
import csr
</code>
#### Read in p2g activity scores
Several different ways to compute gene activity scores were tried over time, for example including negative peak-to-gene correlations vs. only positive correlations.
Using gene activity scores based on z-score peak-to-gene linkage matrix, in order to remove noise of peaks which are correlated with a lot of genes. This way, peaks which are only correlated with a few genes will have a higher z-score than peaks correlated with many genes. Another attempt to increase the correlation of gene activity scores with gene expression was to use distance weight formulas as described in the ArchR gene activity score computation.
Additionally, here I only used highly variable genes, since genes which are not variable are not interesting to us and would introduce unnecessary noise.
<code>
matrix_dict = {}
</code>
<code>
debugging_adata = scvi.data.read_h5ad("p2g_gene_activity_scores/old_function_z_score")
</code>
<code>
debugging_adata.var = debugging_adata.var.rename(columns = {"rownames(scores)": "name"})
#debugging_adata = debugging_adata[:, debugging_adata.var.name.isin(weight_5k.var.name.tolist())]
debugging = debugging_adata.X
matrix_dict["old_function_z_score"] = debugging
</code>
<code>
# list of genes to use
test = debugging_adata.var.name.tolist()
</code>
<code>
import random
</code>
<code>
genes_to_use = random.sample(genes_to_use, len(genes_to_use))
</code>
<code>
[i for i, item in enumerate(test) if item == genes_to_use[i]]
</code>
<code>
#weight_5k = scvi.data.read_h5ad("p2g_gene_activity_scores/5000_distance_weighted_p2gfinal_scores")
#weight_50k = scvi.data.read_h5ad("p2g_gene_activity_scores/5e+05_distance_weighted_p2gfinal_scores")
#weight_500k = scvi.data.read_h5ad("p2g_gene_activity_scores/5e+06_distance_weighted_p2gfinal_scores")
#weight_5000k = scvi.data.read_h5ad("p2g_gene_activity_scores/5e+07_distance_weighted_p2gfinal_scores")
</code>
<code>
#for name, anndata in {"dist_weight_5k":weight_5k, "dist_weight_50k":weight_50k,"dist_weight_500k": weight_500k, "dist_weight_5000k":weight_5000k}.items():
# rename column
# anndata.var = anndata.var.rename(columns = {"rownames(weighted_scores)":"name"})
# anndata = anndata[:, anndata.var.name.isin(genes_to_use)]
# matrix_dict[name] = anndata.X
</code>
Z-scores over all genes for each peak of peak-to-gene link matrix to compute gene activity scores.
<code>
#p2g_z_scores_adata = scvi.data.read_h5ad("p2g_gene_activity_scores/z_score_p2g_gene_activity_scores")
#p2g_z_scores_adata.var = p2g_z_scores_adata.var.rename(columns = {"rownames(scores)": "name"})
#p2g_z_scores_adata = p2g_z_scores_adata[:, p2g_z_scores_adata.var.name.isin(genes_to_use)]
#p2g_z_scores = p2g_z_scores_adata.X
</code>
<code>
#matrix_dict["positive p2g links & z-scores"] = p2g_z_scores
</code>
Instead of using the constant of total activity across all cells for the size factors in our gene activity score computation based on peak-to-gene links, I just divided by the expected total activity given a certain library size (output of the linear regression). See function for computing gene activity score from peak-to-gene links in R. If we compare the plots between using two different functions, the result is almost the same, so the constant does not seem to make a difference.
<code>
#p2g_scores_adata =scvi.data.read_h5ad("p2g_gene_activity_scores/p2g_scores_anndata.h5ad")
</code>
<code>
#matrix_dict["positive & negative p2g links"] = p2g_scores
</code>
<code>
#pos_p2g_scores_adata = scvi.data.read_h5ad("p2g_gene_activity_scores/pos_p2g_links_scores")
#pos_p2g_scores_adata.var = pos_p2g_scores_adata.var.rename(columns = {"rownames(scores)": "name"})
#pos_p2g_scores_adata = pos_p2g_scores_adata[:, pos_p2g_scores_adata.var.name.isin(genes_to_use)]
#pos_p2g_scores = pos_p2g_scores_adata.X
#matrix_dict["positive p2g links"] = pos_p2g_scores
</code>
<code>
# read in overlapping gene names
#gene_names = pd.read_pickle(r"gene_names.pkl")
</code>
<code>
scores_adata = scvi.data.read_h5ad("p2g_gene_activity_scores/archr_gene_scores")
scores_adata.var_names = scores_adata.var.name
#scores_adata = scores_adata[:, scores_adata.var.name.isin(genes_to_use)]
scores_adata = scores_adata[:, genes_to_use]
</code>
<code>
(np.where(scores_adata.var["name"] != genes_to_use))[0].shape
</code>
<code>
scores = scores_adata.X
matrix_dict["ArchR scores"] = scores
</code>
#### Read in latent space and gene expression matrix
<code>
# read in the anndata object which contains the latent space embedding
adata = scvi.data.read_h5ad("gpu_trained_20_dim/anndata_object")
# read the anndata object containing ArchR gene expression matrix
archr_gene_expr = scvi.data.read_h5ad("p2g_gene_activity_scores/ArchR_gene_expr.h5ad")
</code>
<code>
archr_gene_expr.var_names = archr_gene_expr.var.name
archr_gene_expr = archr_gene_expr[:, test]
gene_expr = archr_gene_expr.X
matrix_dict["gene_expr"] = gene_expr
</code>
<code>
np.where(archr_gene_expr.var["name"] != genes_to_use)
</code>
<code>
np.where(archr_gene_expr.obs.index != debugging_adata.obs.index)
</code>
<code>
[i for i, item in enumerate(archr_gene_expr.var["name"]) if item == scores_adata.var["name"][i]]
</code>
<code>
[i for i, item in enumerate(archr_gene_expr.obs.index.tolist()) if item != debugging_adata.obs.index.tolist()[i]]
</code>
<code>
# subset anndata object to contain only overlapping cells
adata = adata[archr_gene_expr.obs.index, :]
# get latent space embedding
latent_embedding = adata.obsm["X_scVI"]
</code>
<code>
latent_embedding.shape
</code>
archr_scores_peak_based_adata = scvi.data.read_h5ad("p2g_gene_activity_scores/archr_scores_peak_based")
#archr_scores_peak_based_adata.var = archr_scores_peak_based_adata.var.rename(columns = {"rownames(scores_mat)": "name"})
archr_scores_peak_based_adata = archr_scores_peak_based_adata[:, archr_scores_peak_based_adata.var.name.isin(genes_to_use)]
archr_scores_peak_based = archr_scores_peak_based_adata.X
matrix_dict["archr_scores_peak_based"] = archr_scores_peak_based
archr_scores_tss_adata = scvi.data.read_h5ad("p2g_gene_activity_scores/archr_scores_tss")
archr_scores_tss_adata = archr_scores_tss_adata[:, archr_scores_tss_adata.var.name.isin(genes_to_use)]
archr_scores_tss = archr_scores_tss_adata.X
matrix_dict["archr_scores_tss"] = archr_scores_tss
archr_scores_gene_body_peak_based = scvi.data.read_h5ad("p2g_gene_activity_scores/archr_scores_gene_body_peak_based")
archr_scores_gene_body_peak_based = archr_scores_gene_body_peak_based[:, archr_scores_gene_body_peak_based.var.name.isin(genes_to_use)]
archr_scores_gene_body_peak_based = archr_scores_gene_body_peak_based.X
matrix_dict["archr_scores_gene_body_peak_based"] = archr_scores_gene_body_peak_based
<code>
gene_window_scores_adata = scvi.data.read_h5ad("p2g_gene_activity_scores/gene_window_scoresgene_window_scores")
</code>
<code>
gene_window_scores_adata.var = gene_window_scores_adata.var.rename(columns = {"rownames(weighted_scores)": "name"})
gene_window_scores_adata.var_names = gene_window_scores_adata.var.name
gene_window_scores_adata = gene_window_scores_adata[:, genes_to_use]
gene_window_scores = gene_window_scores_adata.X
matrix_dict["gene_window_scores"] = gene_window_scores
</code>
<code>
debugging_adata = scvi.data.read_h5ad("p2g_gene_activity_scores/debugging7")
debugging_adata.var = debugging_adata.var.rename(columns = {"rownames(scores)": "name"})
debugging_adata.var_names = debugging_adata.var.name
debugging_adata = debugging_adata[:, genes_to_use]
debugging = debugging_adata.X
matrix_dict["debugging7"] = debugging
</code>
<code>
matrix_dict
</code>
### Sample cells & compute nearest neighbors
Since we have 45,991 cells in our dataset, we will sample 1000 cell aggregates of 50 cells each.
<code>
class nearest_neighbors:
def __init__(self, latent_embedding):
# attribute root of the class Tree will be an instance of the class Node
# attriute self.root is an object of class Node
self.latent_embedding = latent_embedding
def sampling_cells(self, n_aggregates):
self.sample_cells = np.random.choice(self.latent_embedding.shape[0], n_aggregates, replace=False)
#print(self.sample_cells.shape)
assert self.sample_cells.shape[0] == n_aggregates,"sample cells vector has incorrect length."
def compute_NN(self, k):
print(f"Computing {k} nearest neighbors for {self.sample_cells.shape[0]} cells.")
nbrs = NearestNeighbors(n_neighbors=k, algorithm="ball_tree").fit(self.latent_embedding)
dist, ind = nbrs.kneighbors(self.latent_embedding)
assert dist.shape[0] == self.latent_embedding.shape[0], "wrong dimensions of neigbor search"
# subset the nearest neighbors to only contain the sampled cells
self.distance = dist[self.sample_cells, :]
self.index = ind[self.sample_cells, :]
# The index matrix should now contain the number of cells as nrows and k cols
assert self.index.shape[0] == self.sample_cells.shape[0], "wrong dimensions in nearest neighbor index matrix"
assert self.index.shape[1] == k, "wrong dimensions of nearest neighbor index matrix"
</code>
### Check for overlapping cell aggregates
<code>
# We want to check whether the 50 nearest neighbors of any given cell overlap with the 50 nearest neighbors of any other cell by more than 80%
@jit(nopython=True)
def check_overlap(index, sample_cells):
nrow = index.shape[0] # number of cells
ncol = index.shape[1] # number of neighbors
# create an array to store whether a cell passed the overlap check
# all entries are initially False
considered_cells = np.zeros(nrow).astype(np.bool8)
# loop over each cell and cosider to add it to the set of cell aggregates
for i in range(nrow):
check = True
# loop over all previous aggregates
for comp in np.where(considered_cells)[0]:
# get the number of cells which overlap between the neighborhood of the cell we would like to add and the neighborhoud of previous cell "comp"
intersect = np.intersect1d(index[i, :], index[comp, :])
# for each comparison between current cell i which we would like to add and previous cell which we are comparing to
# compute the percentage of overlap
if (len(intersect) / ncol) > 0.8: # if the intersection is above 0.8, we do not consider it
check = False
break
if check:
considered_cells[i] = True
# get indices
keep = np.arange(start=0, stop=nrow, step=1)[considered_cells]
print(f"Of {nrow} cell aggregates we keep {index[keep, :].shape[0]} cells.")
return index[keep, :], sample_cells[keep]
</code>
### Only keep cells of same celltype
<code>
# We only want to keep cells of the same celltype
def filter_celltypes(metadata, sample_cells_keep, idx_keep):
groups={}
celltypes = []
# check whether a cell aggregate contains cells of other celltypes and remove then
for n, i in enumerate(sample_cells_keep):
# check that the index in the sample cells is equivalent to the one in the nearest neighbor index matrix
assert (sample_cells_keep==idx_keep[:, 0]).all()
# get celltype information of sampled cell
celltype_test_cell = metadata.iloc[i]["celltype"]
# get indices of cells which are in the neighborhood
neighbor_cells = idx_keep[n, :]
assert neighbor_cells.shape[0] == idx_keep.shape[1]
# get cells which are of the same celltype, vector includes the sampled cell itself
keep = np.array(metadata[(metadata.idx.isin(neighbor_cells.flatten())) & (metadata.celltype == celltype_test_cell)]["idx"])
# keep only aggregates which contain at least 10 cells after removing non-matching celltypes
if keep.shape[0] > 10:
groups[i] = keep
celltypes.append(celltype_test_cell)
else:
continue
print(f"Out of {len(sample_cells_keep)} cell aggregates which passed the overlap check we are left with {len(groups)} after checking for celltype consistency")
# add dictionary to self
return groups, celltypes
</code>
## Aggregate expression/scores/accessibility
TODO: Implement aggregation with numba -> convert to csr matrx
https://numba.pydata.org/numba-doc/0.43.0/reference/pysupported.html
<code>
def create_aggregates(mat, groups): # the matrix should have dimensions cells x genes
# initialize matrix to store average gene expression for each cell aggregate
# the matrix has dimensions genes x cell aggregates
rna_agg = np.zeros((mat.shape[1],len(groups)))
# for each cell aggregate calculate the average gene expression for each gene
for i, g in enumerate(groups):
rna_agg[:, i] = mat[groups[g], :].mean(axis=0)
assert (rna_agg[:, i] == mat[groups[g], :].mean(axis=0)).all(), "Dimension mismatch"
print(f"The aggregate matrix has dimensions: {rna_agg.shape}")
rna_agg = sparse.csr_matrix(rna_agg)
return csr.CSR.from_scipy(rna_agg)#, rna_agg # the first is a csr.csr.CSR matrix, the second one is a scipy.sparse.csr.csr_matrix
</code>
# Correlations
To compute row-wise correlations:
<code>
@jit(nopython=True)
def rowwise_correlation(A, B):
correlations = []
for i in range(A.nrows):
# center and scale row i of matrix A
rowa = A.row(i)
rowa -= np.mean(rowa)
rowa /= np.std(rowa)
# center and scale row i of matrix B
rowb = B.row(i)
rowb -= np.mean(rowb)
rowb /= np.std(rowb)
# compute correlation between row i of matrix A and B
corr = np.mean(rowa*rowb)
correlations.append(corr)
return correlations
</code>
## Function to automate all computatiosn for different matrices
<code>
def score_correlations(latent_space_adata, latent_embedding, n_aggregates, k, gene_expr, matrix_dict):
agg_object = nearest_neighbors(latent_embedding) # initialize the cell aggreagte object
agg_object.sampling_cells(n_aggregates) # sample cells
agg_object.compute_NN(k) # compute nearest neighbors
idx_keep, sample_cells_keep = check_overlap(agg_object.index, agg_object.sample_cells) # remove overlapping cells
metadata = latent_space_adata.obs
# create column for cell names
metadata["cells"] = metadata.index
# create index for cells
metadata["idx"] = np.arange(len(metadata))
groups, celltype_list = filter_celltypes(metadata, sample_cells_keep, idx_keep)
# aggregate gene expression
rna_agg = create_aggregates(gene_expr, groups)
# aggregate gene activity score matrices stored in the matrix_dictionary
# create empty list to store aggregate matrices
agg_corr = {}
for name, matrix in matrix_dict.items():
#if name != gene_expr:
agg = create_aggregates(matrix, groups)
print(type(agg))
corr = rowwise_correlation(rna_agg, agg)
agg_corr[name] = (agg, corr)
sns.histplot(corr, bins = 200)
plt.axvline(0, color = "red")
plt.title(f"Correlation between gene expr. and {name}.")
plt.show()
return(agg_corr)
</code>
<code>
corr_vectors = score_correlations(adata, latent_embedding, 1000, 50, gene_expr, matrix_dict)
</code>
<code>
def plot_correlations(corr_vectors):
archr_scores = corr_vectors["ArchR scores"][1]
# set color palette for density plot
cmap = sns.color_palette("viridis", as_cmap=True)
for name, vector in corr_vectors.items():
if name != "gene_expr" and name != "ArchR scores":
# create scatterplot
sns.scatterplot(x=np.asarray(vector[1]), y=np.asarray(archr_scores), color="k")
# add density plot on top
sns.kdeplot(x=np.asarray(vector[1]), y=np.asarray(archr_scores),
levels=6, fill=True, alpha=0.7, cut=2, cmap=cmap)
plt.plot(np.linspace(-0.2, 1, 100), np.linspace(-0.2, 1, 100) , color='r')
plt.xlabel(f"correlation between gene expr. and {name}")
plt.ylabel("Correlation between gene expr. and ArchR scores")
plt.title(f"Comparing correlation values for {len(archr_scores)} most highly variable genes")
plt.axvline(0, color="black")
plt.axhline(0, color="black")
plt.show()
</code>
<code>
plot_correlations(corr_vectors)
</code>
<code>
corr_vectors
sns.scatterplot(x=np.asarray(vector[1]), y=np.asarray(archr_scores), color="k")
# add density plot on top
sns.kdeplot(x=np.asarray(vector[1]), y=np.asarray(archr_scores),
levels=6, fill=True, alpha=0.7, cut=2, cmap=cmap)
plt.plot(np.linspace(-0.2, 1, 100), np.linspace(-0.2, 1, 100) , color='r')
plt.xlabel(f"correlation between gene expr. and {name}")
plt.ylabel("Correlation between gene expr. and ArchR scores")
plt.title(f"Comparing correlation values for {len(archr_scores)} most highly variable genes")
plt.axvline(0, color="black")
plt.axhline(0, color="black")
plt.show()
</code>
#### Prepare the metadata for the cell aggregate function:
<code>
metadata = adata.obs
# create column for cell names
metadata["cells"] = metadata.index
# create index for cells
metadata["idx"] = np.arange(len(metadata))
</code>
https://stackoverflow.com/questions/20928136/input-and-output-numpy-arrays-to-h5py
### Sample cells & compute nearest neighbors
Since we have 45,991 cells in our dataset, we will sample 1000 cell aggregates of 50 cells each.
<code>
class nearest_neighbors:
def __init__(self, latent_embedding):
# attribute root of the class Tree will be an instance of the class Node
# attriute self.root is an object of class Node
self.latent_embedding = latent_embedding
def sampling_cells(self, n_aggregates):
self.sample_cells = np.random.choice(self.latent_embedding.shape[0], n_aggregates, replace=False)
#print(self.sample_cells.shape)
assert self.sample_cells.shape[0] == n_aggregates,"sample cells vector has incorrect length."
def compute_NN(self, k):
print(f"Computing {k} nearest neighbors for {self.sample_cells.shape[0]} cells.")
nbrs = NearestNeighbors(n_neighbors=k, algorithm="ball_tree").fit(self.latent_embedding)
dist, ind = nbrs.kneighbors(self.latent_embedding)
assert dist.shape[0] == self.latent_embedding.shape[0], "wrong dimensions of neigbor search"
# subset the nearest neighbors to only contain the sampled cells
self.distance = dist[self.sample_cells, :]
self.index = ind[self.sample_cells, :]
# The index matrix should now contain the number of cells as nrows and k cols
assert self.index.shape[0] == self.sample_cells.shape[0], "wrong dimensions in nearest neighbor index matrix"
assert self.index.shape[1] == k, "wrong dimensions of nearest neighbor index matrix"
</code>
### Check for overlapping cell aggregates
<code>
# We want to check whether the 50 nearest neighbors of any given cell overlap with the 50 nearest neighbors of any other cell by more than 80%
@jit(nopython=True)
def check_overlap(index, sample_cells):
nrow = index.shape[0] # number of cells
ncol = index.shape[1] # number of neighbors
# create an array to store whether a cell passed the overlap check
# all entries are initially False
considered_cells = np.zeros(nrow).astype(np.bool8)
# loop over each cell and cosider to add it to the set of cell aggregates
for i in range(nrow):
check = True
# loop over all previous aggregates
for comp in np.where(considered_cells)[0]:
# get the number of cells which overlap between the neighborhood of the cell we would like to add and the neighborhoud of previous cell "comp"
intersect = np.intersect1d(index[i, :], index[comp, :])
# for each comparison between current cell i which we would like to add and previous cell which we are comparing to
# compute the percentage of overlap
if (len(intersect) / ncol) > 0.8: # if the intersection is above 0.8, we do not consider it
check = False
break
if check:
considered_cells[i] = True
# get indices
keep = np.arange(start=0, stop=nrow, step=1)[considered_cells]
print(f"Of {nrow} cell aggregates we keep {index[keep, :].shape[0]} cells.")
return index[keep, :], sample_cells[keep]
</code>
### Only keep cells of same celltype
<code>
# We only want to keep cells of the same celltype
def filter_celltypes(metadata, sample_cells_keep, idx_keep):
groups={}
celltypes = []
# check whether a cell aggregate contains cells of other celltypes and remove then
for n, i in enumerate(sample_cells_keep):
# check that the index in the sample cells is equivalent to the one in the nearest neighbor index matrix
assert (sample_cells_keep==idx_keep[:, 0]).all()
# get celltype information of sampled cell
celltype_test_cell = metadata.iloc[i]["celltype"]
# get indices of cells which are in the neighborhood
neighbor_cells = idx_keep[n, :]
assert neighbor_cells.shape[0] == idx_keep.shape[1]
# get cells which are of the same celltype, vector includes the sampled cell itself
keep = np.array(metadata[(metadata.idx.isin(neighbor_cells.flatten())) & (metadata.celltype == celltype_test_cell)]["idx"])
# keep only aggregates which contain at least 10 cells after removing non-matching celltypes
if keep.shape[0] > 10:
groups[i] = keep
celltypes.append(celltype_test_cell)
else:
continue
print(f"Out of {len(sample_cells_keep)} cell aggregates which passed the overlap check we are left with {len(groups)} after checking for celltype consistency")
# add dictionary to self
return groups, celltypes
</code>
#### Apply functions to our data
<code>
agg_object = nearest_neighbors(latent_embedding) # initialize the cell aggreagte object
agg_object.sampling_cells(n_aggregates=1000) # sample cells
agg_object.compute_NN(k=50) # compute nearest neighbors
idx_keep, sample_cells_keep = check_overlap(agg_object.index, agg_object.sample_cells) # remove overlapping cells
</code>
<code>
metadata = adata.obs
# create column for cell names
metadata["cells"] = metadata.index
# create index for cells
metadata["idx"] = np.arange(len(metadata))
</code>
## Aggregate expression/scores/accessibility
TODO: Implement aggregation with numba -> convert to csr matrx
https://numba.pydata.org/numba-doc/0.43.0/reference/pysupported.html
<code>
def create_aggregates(mat, groups): # the matrix should have dimensions cells x genes
# initialize matrix to store average gene expression for each cell aggregate
# the matrix has dimensions genes x cell aggregates
rna_agg = np.zeros((mat.shape[1],len(groups)))
# for each cell aggregate calculate the average gene expression for each gene
for i, g in enumerate(groups):
rna_agg[:, i] = mat[groups[g], :].mean(axis=0)
assert (rna_agg[:, i] == mat[groups[g], :].mean(axis=0)).all(), "Dimension mismatch"
print(f"The aggregate matrix has dimensions: {rna_agg.shape}")
rna_agg = sparse.csr_matrix(rna_agg)
return csr.CSR.from_scipy(rna_agg)#, rna_agg # the first is a csr.csr.CSR matrix, the second one is a scipy.sparse.csr.csr_matrix
</code>
#### Aggregate Gene expression
<code>
# create gene expression aggregates
rna_agg = create_aggregates(gene_expr, groups)
</code>
#### Aggregate Gene Scores from ArchR
<code>
# gene scores
score_agg = create_aggregates(scores, groups)
</code>
#### Gene Scores from p2g links
<code>
# the first version of the formula
</code>
<code>
# the formula with distance weight
weighted_agg = create_aggregates(weighted_scores, groups)
</code>
<code>
# the formula with z-scores & constant
scores_p2g_z_agg = create_aggregates(p2g_z_scores, groups)
</code>
<code>
# the formula with z-score & without constant
#p2g_noconstant_agg, sparse_p2g_noconstant_agg = create_aggregates(p2g_scores_noconstant, groups)
</code>
<code>
# the formula with raw p2g links & constant
#p2g_hvg_agg, sparse_p2g_hvg_agg = create_aggregates(p2g_scores_hvg, groups)
</code>
# Correlations
To compute row-wise correlations:
<code>
@jit(nopython=True)
def rowwise_correlation(A, B):
correlations = []
for i in range(A.nrows):
# center and scale row i of matrix A
rowa = A.row(i)
rowa -= np.mean(rowa)
rowa /= np.std(rowa)
# center and scale row i of matrix B
rowb = B.row(i)
rowb -= np.mean(rowb)
rowb /= np.std(rowb)
# compute correlation between row i of matrix A and B
corr = np.mean(rowa*rowb)
correlations.append(corr)
return correlations
</code>
<code>
# correlation across cell aggregates
corr_expr_scores = rowwise_correlation(rna_agg, score_agg)
</code>
<code>
sns.histplot(corr_expr_scores, bins=200)
plt.axvline(0, color ="red")
plt.title("Correlation betwen gene expression and ArchR gene scores")
</code>
<code>
corr_expr_p2g_scores = rowwise_correlation(rna_agg, scores_p2g_z_agg)
</code>
<code>
sns.histplot(corr_expr_p2g_scores, bins=200)
plt.axvline(0, color="red")
plt.title("Correlations between gene expression and p2g link scores (z_scores) across cell aggregates")
</code>
<code>
corr_weighted_scores = rowwise_correlation(rna_agg, weighted_agg)
</code>
<code>
sns.histplot(corr_weighted_scores, bins=200)
plt.axvline(0, color="red")
plt.title("Correlations between gene expression and distance-weighted p2g link scores across cell aggregates")
</code>
<code>
# set color palette for density plot
cmap = sns.color_palette("viridis", as_cmap=True)
# create scatterplot
sns.scatterplot(x=np.asarray(corr_expr_p2g_scores), y=np.asarray(corr_expr_scores),
color="k")
# add density plot on top
sns.kdeplot(x=np.asarray(corr_expr_p2g_scores), y=np.asarray(corr_expr_scores),
levels=6, fill=True, alpha=0.7, cut=2, cmap=cmap)
plt.plot(np.linspace(-0.2, 1, 100), np.linspace(-0.2, 1, 100) , color='r')
plt.xlabel("correlation between gene expr. and p2g scores")
plt.ylabel("Correlation between gene expr. and ArchR scores")
plt.title(f"Comparing correlation values for {gene_expr.shape[1]} most highly variable genes")
plt.axvline(0, color="black")
plt.axhline(0, color="black")
</code>
<code>
# set color palette for density plot
cmap = sns.color_palette("viridis", as_cmap=True)
# create scatterplot
sns.scatterplot(x=np.asarray(corr_weighted_scores), y=np.asarray(corr_expr_scores),
color="k")
# add density plot on top
sns.kdeplot(x=np.asarray(corr_weighted_scores), y=np.asarray(corr_expr_scores),
levels=6, fill=True, alpha=0.7, cut=2, cmap=cmap)
plt.plot(np.linspace(-0.2, 1, 100), np.linspace(-0.2, 1, 100) , color='r')
plt.xlabel("correlation gene expr. and distance-weighted p2g scores")
plt.ylabel("Correlation gene expr. and ArchR scores")
plt.title(f"Comparing correlation values for {gene_expr.shape[1]} most highly variable genes")
plt.axvline(0, color="black")
plt.axhline(0, color="black")
</code>
<code>
corr_expr_p2g_no_constant = rowwise_correlation(rna_agg, p2g_noconstant_agg)
</code>
<code>
sns.histplot(corr_expr_p2g_no_constant, bins=200)
plt.axvline(0, color="red")
plt.title("Correlations between gene expression and p2g link scores (no constant in formula) across cell aggregates")
</code>
<code>
corr_hvg = rowwise_correlation(rna_agg, p2g_hvg_agg)
</code>
<code>
sns.histplot(corr_hvg, bins=200)
plt.axvline(0, color="red")
plt.title("Correlations between gene expression and p2g link scores (no z-scores) across cell aggregates")
</code>
<code>
corr_scores = rowwise_correlation(scores_p2g_agg, score_agg)
</code>
<code>
sns.histplot(corr_scores, bins=200)
plt.axvline(0, color="red")
plt.title("Correlation between ArchR gene score and p2g scores")
</code>
<code>
sns.scatterplot(np.asarray(corr_expr_p2g_scores), y = np.asarray(corr_expr_scores))
plt.xlabel("correlation between gene expr. and p2g scores")
plt.ylabel("Correlation between gene expr. and ArchR scores")
plt.title("Comparing correlation values for {hvg_index.shape[0]} most highly variable genes")
plt.axvline(0, color="red")
plt.axhline(0, color="red")
</code>
#### Check the genes which have negative correlations.
<code>
#gene_names_index = [i for i in range(0, len(gene_names))]
</code>
<code>
# lets check highly variable genes
df = pd.read_csv('hvg_list', delimiter=',')
</code>
<code>
hvg_list = df["x"].tolist()
</code>
<code>
hvg_index = np.where(np.isin(hvg_list, gene_names))[0]
print(f"There are {len(hvg_list)} highly variable genes identified, out of which {hvg_index.shape[0]} are found in our genes x cell aggregates matrix")
</code>
<code>
marker_genes_idx = np.where(np.isin(gene_names, ["Lamb1", "Sparc", "Elf5", "Ascl2", "Tfap2c", "Ttr", \
"Apoa2", "Apoe", "Cystm1", "Emb", "Spink1", "Krt19", \
"Dkk1", "Grhl3", "Trp63", "Grhl2", "Pax6", "Pax2", \
"En1", "Foxd3", "Tfap2a", "Pax3", "Sox9", \
"Six3", "Hesx1", "Irx3", "Sox2", "Hoxb9", "Cdx4",\
"Hes3", "Hba-a2", "Hba-a1", "Hbb-bh1", "Gata1", "Cited4", \
"Cdh5", "Pecam1", "Anxa5", "Etv2", "Igf2",\
"Krt8", "Krt18", "Pmp22", "Ahnak", "Bmp4", "Tbx4", "Hoxa11", \
"Hoxa10", "Tnnt2", "Myl4", "Myl7", "Acta2", \
"Smarcd3", "Tcf21", "Tbx6", "Dll1", "Aldh1a2", "Tcf15", \
"Meox1", "Tbx1", "Gbx2", "Cdx1", "Hoxb1", "Hes7", "Osr1", \
"Mesp2", "Lefty2", "Mesp1", "Cer1", "Chrd", "T", \
"Foxa2", "Pax7", "Fgf8", "Lhx1", "Gsc", "Mixl1", "Otx2", "Hhex",\
"Ifitm3", "Nkx1-2", "Eomes", "Nanog", "Utf1", \
"Epcam", "Pou5f1"]))[[0]]
</code>
<code>
sns.histplot(np.asarray(corr_expr_p2g_scores)[hvg_index], bins = 200)
plt.axvline(color="red")
plt.title(f"Correlations between gene expression and peak-to-gene linkage gene scores for {hvg_index.shape[0]} highly variable genes")
</code>
<code>
sns.histplot(np.asarray(corr_expr_scores)[hvg_index], bins=200)
plt.axvline(color="red")
plt.title(f"Correlations between gene expression and ArchR gene scores for {hvg_index.shape[0]} highly variable genes")
</code>
<code>
sns.histplot(np.asarray(corr_scores)[hvg_index], bins=200)
plt.axvline(color="red")
plt.title(f"Correlations between ArchR gene scores and p2g-linkage gene scores for {hvg_index.shape[0]} highly variable genes")
</code>
<code>
type(scores_p2g_agg)
</code>
<code>
sns.histplot(scores_p2g_agg)
</code>
<code>
df = pd.DataFrame(list(zip(corr_aggregates, celltype_list)),
columns =['Corr', 'Celltype'])
</code>
<code>
colPalette_celltypes = ['#532C8A',
'#c19f70',
'#f9decf',
'#c9a997',
'#B51D8D',
'#3F84AA',
'#9e6762',
'#354E23',
'#F397C0',
'#ff891c',
'#635547',
'#C72228',
'#f79083',
'#EF4E22',
'#989898',
'#7F6874',
'#8870ad',
'#647a4f',
'#EF5A9D',
'#FBBE92',
'#139992',
'#cc7818',
'#DFCDE4',
'#8EC792',
'#C594BF',
'#C3C388',
'#0F4A9C',
'#FACB12',
'#8DB5CE',
'#1A1A1A',
'#C9EBFB',
'#DABE99',
'#65A83E',
'#005579',
'#CDE088',
'#f7f79e',
'#F6BFCB']
</code>
<code>
sc.set_figure_params(figsize=(10,10))
sns.histplot(df, x = "Corr", hue = "Celltype")
sns.color_palette(colPalette_celltypes)
plt.title("Correlation between gene expression and ArchR gene score across genes")
#plt.legend([],[], frameon=False)
plt.show()
</code>
Convert sparse matrices to csr matrices:
<code>
expr_agg = csr.CSR.from_scipy(filt_rna_agg)
</code>
<code>
score_agg = csr.CSR.from_scipy(score_agg)
</code>
<code>
scores_p2g_agg = csr.CSR.from_scipy(scores_p2g_agg)
</code>
<code>
# get number of genes
N = rna_agg.shape[0]
</code>
<code>
corr_expr_scores = rowwise_correlation(expr_agg, score_agg)
</code>
### Save correlations
<code>
with open( 'corr_expr_scores.pkl', 'wb') as f:
pickle.dump(corr_expr_scores, f)
</code>
<code>
with open(dir_data + 'corr_expr_p2g_scores.pkl', 'wb') as f:
pickle.dump(corr_expr_p2g_scores, f)
</code>
<code>
with open(dir_data + 'corr_scores.pkl', 'wb') as f:
pickle.dump(corr_scores, f)
</code>
<code>
A = rna_agg.copy
A.shapeb
A -= np.mean(A)
A.shape
</code>
<code>
A = rna_agg.copy()
print(A.shape)
print(np.mean(A, axis=1).shape)
A -= np.mean(A, axis=1)
print(A.shape)
print(np.std(A, axis=1).shape)
print(np.std(A, axis=1))
#A /= np.std(A, axis=1)
</code>
<code>
A[np.where(np.std(A, axis=1) == 0)].shape
</code>
<code>
A = rna_agg.copy()
A.shape
A -= np.mean(A, axis=1)
A.shape
A /= np.std(A, axis=1)
B = score_agg.copy()
B -= np.mean(B, axis=1)
B /= np.std(B, axis=1)
corr = np.mean(A*B, axis = 1)
</code>
<code>
corr
</code>
<code>
A = rna_agg.copy()
B = score_agg.copy()
cA = A - A.mean(axis=1)
cB = B - B.mean(axis=1)
print(cA.shape)
print(np.square(cA))
#print((cA**2).shape)
sA = np.sqrt((np.square(cA)).mean(axis=1))
sB = np.sqrt((cB**2).mean(axis=1))
corr = (cA*cB).mean(axis=1) / sA*sB
</code>
<code>
corr
</code>
<code>
score_agg.shape
</code>
<code>
import seaborn as sns
sns.histplot(x = corr_scores)
plt.show()
</code>
<code>
import seaborn as sns
sns.histplot(x = corr_expr_p2g_scores)
plt.show()
</code>
<code>
import seaborn as sns
sns.histplot(x = corr_expr_scores)
plt.show()
</code>
<code>
sns.scatterplot(x =corr_expr_p2g_scores, y = corr_expr_scores)
</code>
<code>
sns.scatterplot(x =corr_expr_p2g_scores, y = corr_expr_scores )
</code>
## Leiden Clustering
Since we already have celltype annotations we might simply use Leiden Clustering to create cell aggregates. We would expect these to be celltype-specific if done at high enough resolution, but we could also try to do Leiden Clustering based on each celltype individually.
<code>
import igraph as ig
import leidenalg as la
</code>
With a resolution of 100, I get 930 clusters, which is probably too much.
<code>
sc.tl.leiden(adata, resolution=100, key_added="cluster_pvi")#, restrict_to=("celltype", adata.obs.celltype.unique()))
</code>
<code>
adata.obs.cluster_pvi.unique()
</code>
<code>
plt.hist(adata.obs.cluster_pvi, bins = 500)
plt.show()
</code>
If I reduce the resolution to 50, I get 483 cells.
<code>
sc.tl.leiden(adata, resolution=50, key_added="cluster_pvi")#, restrict_to=("celltype", adata.obs.celltype.unique()))
</code>
<code>
adata.obs.cluster_pvi.unique()
</code>
<code>
plt.hist(adata.obs.cluster_pvi, bins = 500)
plt.show()
</code>
<code>
adata.obs.cluster_pvi
</code>
<code>
adata.obs.groupby("cluster_pvi").count()
</code>
### Subset to do Leiden Clustering on individual celltypes
Does it make sense to create clusters of different size or should I aim to create clusters which are similar in size for each celltype. This means that if there are more cells of a celltype I will get more clusters of that type.
### Erythroids
<code>
adata_subset = adata[adata.obs['celltype'] == "Erythroid2"]
print(f"The number of celsl for Erythroids are: {adata_subset.shape[0]}")
</code>
<code>
sc.tl.leiden(adata_subset, resolution=.6, key_added="Erythroid_clusters")
</code>
<code>
sc.tl.umap(adata_subset, min_dist=0.2)
sc.pl.umap(adata_subset, color='Erythroid_clusters')
</code>
<code>
adata_subset.obs.groupby("Erythroid_clusters").count()
</code>
### Erythroid1
<code>
adata_subset = adata[adata.obs['celltype'] == "Erythroid1"]
print(f"The number of celsl for Erythroids are: {adata_subset.shape[0]}")
</code>
<code>
sc.tl.leiden(adata_subset, resolution=.6, key_added="Erythroid_clusters")
</code>
<code>
sc.tl.umap(adata_subset, min_dist=0.2)
sc.pl.umap(adata_subset, color='Erythroid_clusters')
</code>
<code>
adata_subset.obs.groupby("Erythroid_clusters").count()
</code>
Here I will remove Cluster 6, because it only contains 6 cells.
### Erythroid 3
<code>
adata_subset = adata[adata.obs['celltype'] == "Erythroid3"]
print(f"The number of celsl for Erythroids are: {adata_subset.shape[0]}")
</code>
<code>
sc.tl.leiden(adata_subset, resolution=.2, key_added="Erythroid_clusters")
</code>
<code>
sc.tl.umap(adata_subset, min_dist=0.2)
sc.pl.umap(adata_subset, color='Erythroid_clusters')
</code>
<code>
adata_subset.obs.groupby("Erythroid_clusters").count()
</code>
### Parietal Endoderm
<code>
adata_subset = adata[adata.obs['celltype'] == "Parietal_endoderm"]
print(f"The number of celsl for Erythroids are: {adata_subset.shape[0]}")
</code>
<code>
sc.tl.leiden(adata_subset, resolution=.6, key_added="clusters")
</code>
<code>
sc.tl.umap(adata_subset, min_dist=0.2)
sc.pl.umap(adata_subset, color='clusters')
</code>
<code>
adata_subset.obs.groupby("clusters").count()
</code>
Here I want to remove cluster 3 and cluster 4, because they contain very few cells.
<code>
adata.obs.celltype.unique()
</code>
### Manus version of doing celltype-specific Leiden clustering
<code>
df_leiden_list=[]
</code>
<code>
for cell_type in adata.obs['celltype'].unique():
adata_celltype =adata[adata.obs['celltype']==cell_type,:]
if adata_celltype.shape[0]>80:
sc.pp.neighbors(adata_celltype, use_rep="X_scVI",n_neighbors=100, n_pcs=50)
sc.tl.leiden(adata_celltype, resolution=1)
#sc.tl.leiden(adata_celltype, resolution=0.2) # decreasing resolution
sc.tl.umap(adata_celltype, spread=1., min_dist=.5, random_state=11)
sc.pl.umap(adata_celltype, color="leiden", legend_loc="on data",edges=False,title=cell_type)
adata_celltype.obs['leiden_name'] = [str(s) + '_'+ cell_type for s in adata_celltype.obs['leiden'] ]
adata_celltype.obs['cell_name'] = adata_celltype.obs.index
cluster_celltype = adata_celltype.obs[['cell_name','leiden_name']]
df_leiden_list.append(cluster_celltype)
else:
adata_celltype.obs['leiden_name'] = [str(0) + '_'+ cell_type for s in range(adata_celltype.obs.shape[0]) ]
adata_celltype.obs['cell_name'] = adata_celltype.obs.index
cluster_celltype = adata_celltype.obs[['cell_name','leiden_name']]
df_leiden_list.append(cluster_celltype)
</code>
<code>
adata
</code>
<code>
# higher resolution values lead to more clusters
help(sc.tl.leiden)
</code>
<code>
la.find_partition(latent, la.ModularityVertexPartition)
</code>
<code>
help(la.find_partition)
</code>
|
{
"filename": "Cell_aggregates_zscores.ipynb",
"repository": "Katharina782/scDoRI",
"query": "transformed_from_existing",
"size": 207616,
"sha": ""
}
|
# notebook-2_2.ipynb
Repository: DipanMondal/chatbot
<code>
import numpy as np
import pandas as pd
import tensorflow as tf
import pickle
from tensorflow.keras import layers , activations , models , preprocessing, utils
import re
from tensorflow import keras
import yaml
import os
import json
dir_path = r'C:\Users\idipa\PycharmProject\ChatBot\ChatbotData'
files_list = os.listdir(dir_path + os.sep)
</code>
<code>
questions, answers = [], []
for filepath in files_list:
file_ = open(dir_path + os.sep + filepath , 'rb')
docs = yaml.safe_load(file_)
conversations = docs['conversations']
for con in conversations:
if len(con) > 2 :
replies = con[1 :]
ans = ''
for rep in replies:
questions.append(con[0])
answers.append(rep)
elif len(con)> 1:
questions.append(con[0])
answers.append(con[1])
</code>
<code>
answers[:10]
</code>
<code>
questions[:10]
</code>
<code>
answers_with_tags = []
for i in range(len(answers)):
if type(answers[i]) == str:
answers_with_tags.append(answers[i])
else:
questions.pop(i)
answers = []
for i in range(len(answers_with_tags)) :
answers.append('<START> ' + answers_with_tags[i] + ' <END>')
</code>
<code>
answers[:10]
</code>
<code>
contractions_dict = {
"ain't": "am not",
"aren't": "are not",
"can't": "cannot",
"can't've": "cannot have",
"'cause": "because",
"could've": "could have",
"couldn't": "could not",
"couldn't've": "could not have",
"didn't": "did not",
"doesn't": "does not",
"don't": "do not",
"hadn't": "had not",
"hadn't've": "had not have",
"hasn't": "has not",
"haven't": "have not",
"he'd": "he had",
"he'd've": "he would have",
"he'll": "he shall",
"he'll've": "he shall have",
"he's": "he has",
"how'd": "how did",
"how'd'y": "how do you",
"how'll": "how will",
"how's": "how has",
"i'd": "i had",
"i'd've": "i would have",
"i'll": "i shall",
"i'll've": "i shall have",
"i'm": "i am",
"i've": "i have",
"isn't": "is not",
"it'd": "it had",
"it'd've": "it would have",
"it'll": "it shall",
"it'll've": "it shall have",
"it's": "it has",
"let's": "let us",
"ma'am": "madam",
"mayn't": "may not",
"might've": "might have",
"mightn't": "might not",
"mightn't've": "might not have",
"must've": "must have",
"mustn't": "must not",
"mustn't've": "must not have",
"needn't": "need not",
"needn't've": "need not have",
"o'clock": "of the clock",
"oughtn't": "ought not",
"oughtn't've": "ought not have",
"shan't": "shall not",
"sha'n't": "shall not",
"shan't've": "shall not have",
"she'd": "she had",
"she'd've": "she would have",
"she'll": "she shall",
"she'll've": "she shall have",
"she's": "she has",
"should've": "should have",
"shouldn't": "should not",
"shouldn't've": "should not have",
"so've": "so have",
"so's": "so as",
"that'd": "that would",
"that'd've": "that would have",
"that's": "that has",
"there'd": "there had",
"there'd've": "there would have",
"there's": "there has",
"they'd": "they had",
"they'd've": "they would have",
"they'll": "they shall",
"they'll've": "they shall have",
"they're": "they are",
"they've": "they have",
"to've": "to have",
"wasn't": "was not",
"we'd": "we had",
"we'd've": "we would have",
"we'll": "we will",
"we'll've": "we will have",
"we're": "we are",
"we've": "we have",
"weren't": "were not",
"what'll": "what shall",
"what'll've": "what shall have",
"what're": "what are",
"what's": "what has",
"what've": "what have",
"when's": "when has",
"when've": "when have",
"where'd": "where did",
"where's": "where has",
"where've": "where have",
"who'll": "who shall",
"who'll've": "who will have",
"who's": "who has",
"who've": "who have",
"why's": "why is",
"why've": "why have",
"will've": "will have",
"won't": "will not",
"won't've": "will not have",
"would've": "would have",
"wouldn't": "would not",
"wouldn't've": "would not have",
"y'all": "you all",
"y'alls": "you alls",
"y'all'd": "you all would",
"y'all'd've": "you all would have",
"y'all're": "you all are",
"y'all've": "you all have",
"you'd": "you had",
"you'd've": "you would have",
"you'll": "you shall",
"you'll've": "you shall have",
"you're": "you are",
"you've": "you have"
}
</code>
<code>
jo = json.dumps(contractions_dict)
with open('contractions.json','w') as file:
file.write(jo)
</code>
<code>
contractions_re = re.compile('(%s)' % '|'.join(re.escape(key) for key in contractions_dict.keys()), re.IGNORECASE)
def expand_contractions(sentence, contractions_dict=contractions_dict):
def replace(match):
# Match is case-insensitive, use the original case in replacement
contraction = match.group(0)
expanded = contractions_dict.get(contraction.lower())
if contraction[0].isupper():
expanded = expanded.capitalize()
return expanded
return contractions_re.sub(replace, sentence)
# Example usage
sentence = "I can't believe it's already 2024! You've got to be kidding me."
expanded_sentence = expand_contractions(sentence)
print(expanded_sentence)
</code>
<code>
re.sub(r"""([+$@#%^&.?!*"\\',:;-])""", r' \1 ', answers[11])
</code>
<code>
for i in range(len(answers)):
st = expand_contractions(answers[i].lower())
answers[i] = re.sub(r"""([+$@#%^&.?!*"\\',:;-])""", r' \1 ', st)
</code>
<code>
answers[:10]
</code>
<code>
answers[3].strip().split()
</code>
<code>
for i in range(len(questions)):
st = expand_contractions(questions[i].lower())
questions[i] = re.sub(r"""([+$@#%^&.?!*"\\',:;-])""", r' \1 ', st)
</code>
<code>
story = """
Once upon a time, in a quaint little village nestled in the verdant hills, there lived an eclectic group of people, each with unique stories and backgrounds. The village, known as Greenfield, was renowned for its picturesque landscapes, vibrant community life, and rich cultural heritage. Among the residents was Alice, an astute librarian with an insatiable curiosity about the world. Her house was a haven for books, maps, and artifacts from different eras and regions, reflecting her lifelong passion for knowledge and adventure.
Alice often spent her days in the village library, a grand building with towering shelves filled with volumes of literature, science, history, and art. The library was a hub of activity, attracting scholars, students, and readers from all walks of life. One day, as she was cataloging a collection of ancient manuscripts, she discovered a dusty old tome that seemed out of place. The book, bound in weathered leather, was inscribed with symbols and languages she had never seen before.
Intrigued, Alice began to decipher its contents, which narrated the tales of an ancient civilization known for its wisdom and technological advancements. The manuscript spoke of a lost city, hidden deep within an uncharted jungle, protected by intricate puzzles and mythical creatures. The allure of uncovering such a mystery captivated Alice, and she decided to embark on a quest to find this lost city.
She shared her discovery with her close friends, each bringing their own set of skills to the journey. There was Marcus, a seasoned archaeologist with a knack for solving riddles; Elena, a brilliant linguist fluent in multiple languages; and Leo, an intrepid explorer with unmatched survival skills. Together, they formed a formidable team, ready to face the unknown.
Their journey began with meticulous planning, gathering supplies, and studying maps and historical texts. They traveled across continents, through bustling cities and remote villages, encountering diverse cultures and landscapes along the way. Their path led them through dense forests, arid deserts, and treacherous mountains, each step bringing them closer to their goal.
As they ventured deeper into the jungle, they faced numerous challenges. The thick canopy overhead blocked the sunlight, making navigation difficult. They encountered wild animals, torrential rains, and steep cliffs that tested their endurance and resilience. Despite the hardships, their determination never wavered.
One fateful day, they stumbled upon an ancient stone path, overgrown with vines and moss. The path led to a massive stone gate, adorned with intricate carvings depicting scenes of a thriving civilization. The gate was guarded by a colossal statue of a mythical beast, its eyes seemingly watching their every move.
Using their combined knowledge, the team deciphered the carvings, revealing clues to unlock the gate. After hours of meticulous work, they succeeded, and the gate slowly creaked open, revealing the entrance to the lost city. The sight that greeted them was beyond their wildest dreams: towering structures, ornate temples, and lush gardens, all remarkably preserved despite the passage of time.
As they explored the city, they uncovered advanced technologies and sophisticated art, evidence of a highly developed society. They also found records of the city's history, detailing its rise and fall. The city had once been a beacon of knowledge and innovation, but a cataclysmic event had forced its inhabitants to abandon it, leaving behind their legacy for future generations to discover.
Throughout their exploration, the team encountered various puzzles and traps, designed to protect the city's secrets. Each challenge required a blend of intellect, teamwork, and courage to overcome. They faced rooms that shifted like labyrinths, mechanisms that required precise timing, and guardians that tested their resolve.
Among the most remarkable discoveries was a vast library, containing scrolls and tablets that held the collective wisdom of the ancient civilization. Alice and Elena were particularly enthralled by the linguistic and historical treasures they found, while Marcus and Leo marveled at the architectural and engineering feats.
Their greatest challenge came when they discovered a hidden chamber, protected by a series of complex locks and puzzles. The chamber was said to hold the most valuable artifact of the lost civilization, a relic of immense power and knowledge. Solving the final puzzle required all their skills and collaboration, but eventually, they succeeded.
Inside the chamber, they found a crystalline artifact, glowing with an ethereal light. As they carefully examined it, they realized it contained vast amounts of data, encoded in a way that was far beyond their current understanding. The artifact held the key to unlocking further mysteries of the lost civilization and potentially advancing modern technology and knowledge.
Their discovery marked a significant milestone in the field of archaeology and history. The lost city, once a myth, had become a reality, offering insights into a civilization that was both advanced and enigmatic. The team's findings were documented and shared with the world, leading to new research and explorations.
Alice, Marcus, Elena, and Leo returned to Greenfield as heroes, their adventure becoming the stuff of legends. They will continue their work, inspired by their journey and the knowledge they had gained. Their story will serve as a reminder of the endless possibilities that await those who dare to explore the unknown.
In Greenfield, life continued to thrive, with the community drawing inspiration from the team's achievements. The village became a center for learning and exploration, attracting scholars and adventurers from far and wide. The library, once a quiet haven, buzzed with activity as people sought to learn more about the lost civilization and its secrets.
The team's legacy will live on, inspiring future generations to pursue their dreams and explore the mysteries of the world. Alice will continue her work at the library, always on the lookout for the next great adventure. Marcus will return to his archaeological pursuits, uncovering more hidden treasures and ancient sites. Elena will dedicate herself to deciphering the languages and texts of the lost civilization, while Leo will embark on new expeditions, driven by his insatiable curiosity.
Their story will become a testament to the power of curiosity, collaboration, and perseverance. It will show that with determination and a willingness to face the unknown, even the most elusive mysteries can be uncovered. The lost city, once hidden in the depths of the jungle, had revealed its secrets, thanks to the unwavering spirit of those who dared to seek it.
And so, the tale of Greenfield and its intrepid explorers will continue, a shining example of what can be achieved when people come together with a shared vision and a relentless pursuit of knowledge. Their adventure will have only just begun, with the promise of more discoveries and stories waiting to be told.
"""
</code>
<code>
story = expand_contractions(story)
story
</code>
<code>
story = re.sub(r"""([+$@#%^&.?!*"\\',:;-])""", r' \1 ', story.lower())
story
</code>
<code>
story.split()
</code>
<code>
punctuations = """! @ # $ % ^ & * ( ) _ - + = { } [ ] : ; ' " / | \ \ < > , . ? / * """
numbers = "0 1 2 3 4 5 6 7 8 9 "
</code>
<code>
mass = punctuations + " " + numbers + " " + story
</code>
<code>
for each in answers:
mass += " " + each
</code>
<code>
len(mass)
</code>
<code>
for each in questions:
mass += " " + each
</code>
<code>
len(mass)
</code>
<code>
for each in contractions_dict.values():
mass += " " + each
</code>
<code>
len(mass)
</code>
<code>
mass = list(set(mass.strip().split()))
</code>
<code>
len(mass)
</code>
<code>
mass.sort()
</code>
<code>
len(mass)
</code>
<code>
VOCAB_SIZE = len(mass)+1
VOCAB_SIZE
</code>
<code>
vocab = {w:i+1 for i,w in enumerate(mass)}
</code>
<code>
vocab
</code>
<code>
import json
</code>
<code>
f = json.dumps(vocab)
with open('vocab1.json','w') as file:
file.write(f)
</code>
<code>
def Word2Num(word):
try:
return vocab[word]
except:
return -1
</code>
<code>
Word2Num('hello')
</code>
<code>
def Sent2Seq(sentence):
sentence = expand_contractions(sentence.lower())
sentence = re.sub(r"""([+$@#%^&.?!*"\\',:;-])""", r' \1 ', sentence)
tokens = sentence.strip().split()
return list(map(Word2Num,tokens))
</code>
<code>
seq = Sent2Seq("Hello! I'm Alice.")
seq
</code>
<code>
def padding(sequence:list,max_pad:int):
l = max_pad-len(sequence)
for i in range(l):
sequence.append(0)
</code>
<code>
padding(seq,20)
seq
</code>
<code>
ans_max = 0
for each in answers:
ans_max = max(ans_max,len(each))
ans_max
</code>
<code>
qs_max = 0
for each in questions:
qs_max = max(qs_max,len(each))
qs_max
</code>
# Answers modifications
<code>
ANS = []
for ans in answers:
seq = Sent2Seq(ans)
padding(seq,ans_max)
ANS.append(np.array(seq))
decoder_input_data = np.array(ANS)
</code>
<code>
decoder_input_data.shape
</code>
<code>
decoder_input_data[0]
</code>
<code>
for i in range(len(ANS)) :
ANS[i] = ANS[i][1:]
padded_answers = preprocessing.sequence.pad_sequences(ANS , maxlen=ans_max , padding='post')
onehot_answers = utils.to_categorical(padded_answers , VOCAB_SIZE)
decoder_output_data = np.array(onehot_answers)
</code>
<code>
decoder_output_data.shape
</code>
<code>
del ANS
del padded_answers
del onehot_answers
</code>
<code>
decoder_output_data[0][0]
</code>
# Questions Modifications
<code>
QS = []
for qs in questions:
seq = Sent2Seq(qs)
padding(seq,qs_max)
QS.append(np.array(seq))
encoder_input_data = np.array(QS)
del QS
</code>
<code>
encoder_input_data.shape
</code>
<code>
encoder_input_data[0]
</code>
# Model
Embedding, LSTM and Desne layers
<code>
from tensorflow.keras.models import load_model
</code>
<code>
encoder_inputs = tf.keras.layers.Input(shape=(qs_max ,),name="encoder_inputs")
encoder_embedding = tf.keras.layers.Embedding(VOCAB_SIZE, 300 , mask_zero=True) (encoder_inputs)
encoder_outputs , state_h , state_c = tf.keras.layers.LSTM(300 , return_state=True, name="encoder_outputs")(encoder_embedding)
encoder_states = [ state_h , state_c ]
decoder_inputs = tf.keras.layers.Input(shape=(ans_max , ),name="decoder_inputs")
decoder_embedding = tf.keras.layers.Embedding(VOCAB_SIZE, 300 , mask_zero=True) (decoder_inputs)
decoder_lstm = tf.keras.layers.LSTM(300 , return_state=True , return_sequences=True, name="decoder_lstm")
decoder_outputs , _ , _ = decoder_lstm (decoder_embedding , initial_state=encoder_states)
decoder_dense = tf.keras.layers.Dense(VOCAB_SIZE , activation=tf.keras.activations.softmax,name="decoder_dense")
output = decoder_dense (decoder_outputs)
model = tf.keras.models.Model([encoder_inputs, decoder_inputs], output)
</code>
<code>
#model = load_model("BaseModel2.h5")
</code>
<code>
model.compile(optimizer=tf.keras.optimizers.Adam(learning_rate=0.001), loss='categorical_crossentropy', metrics=['accuracy'])
</code>
<code>
model.summary()
</code>
<code>
model.fit([encoder_input_data , decoder_input_data], decoder_output_data, batch_size=16, epochs=50)
</code>
<code>
model.save('BaseModel2.h5')
</code>
<code>
def inference():
encoder_model = tf.keras.models.Model(encoder_inputs, encoder_states)
decoder_state_input_h = tf.keras.layers.Input(shape=(300 ,))
decoder_state_input_c = tf.keras.layers.Input(shape=(300 ,))
decoder_states_inputs = [decoder_state_input_h, decoder_state_input_c]
decoder_outputs, state_h, state_c = decoder_lstm(decoder_embedding , initial_state=decoder_states_inputs)
decoder_states = [state_h, state_c]
decoder_outputs = decoder_dense(decoder_outputs)
decoder_model = tf.keras.models.Model([decoder_inputs] + decoder_states_inputs,[decoder_outputs] + decoder_states)
return encoder_model , decoder_model
</code>
<code>
enc_model, dec_model = inference()
</code>
<code>
enc_model.save('Encoder2.h5')
dec_model.save('Decoder2.h5')
</code>
<code>
enc_model.summary()
</code>
<code>
dec_model.summary()
</code>
<code>
def preprocess_input(input_sentence):
seq = Sent2Seq(input_sentence)
padding(seq,qs_max)
return seq
</code>
<code>
del preprocess_input
</code>
<code>
vocabulary = {i:w for w,i in zip(vocab.keys(),vocab.values())}
vocabulary
</code>
<code>
tests = ['You can not move .', 'You sound like Data !', 'Stupid !', 'you are idiot .', 'i am going to die ?','who are you ?']
</code>
<code>
s = [preprocess_input(tests[0])]
s
</code>
<code>
states_values = enc_model.predict(np.array(s))
</code>
<code>
states_values
</code>
<code>
empty_target_seq = np.zeros((1 , 1))
empty_target_seq[0, 0] = vocab['<start>']
empty_target_seq
</code>
<code>
l = [empty_target_seq] + states_values
l
</code>
<code>
l[0].shape
</code>
<code>
l[1].shape
</code>
<code>
l[2].shape
</code>
<code>
#dec_outputs , h , c = dec_model.predict({'input_2':l[0],'input1':l[1],'input2':l[2]})
dec_outputs , h , c = dec_model.predict(l)
</code>
<code>
from tensorflow.keras.models import load_model
from functions import *
</code>
<code>
enc_model = load_model("Encoder2.h5")
dec_model = load_model("Decoder2.h5")
</code>
<code>
tests = ['You can not move .', 'You sound like Data !', 'Stupid !', 'you are idiot .', 'i am going to die ?','who are you ?']
for i in range(6):
states_values = enc_model.predict(np.array([preprocess_input(tests[i])]))
empty_target_seq = np.zeros((1 , 1))
empty_target_seq[0, 0] = vocab['<start>']
stop_condition = False
decoded_translation = ''
while not stop_condition :
dec_outputs , h , c = dec_model.predict([empty_target_seq] + states_values)
sampled_word_index = np.argmax(dec_outputs[0, -1, :])
sampled_word = None
word = vocabulary[sampled_word_index]
decoded_translation += f' {word}'
sampled_word = word
#for word , index in tokenizer.word_index.items() :
# if sampled_word_index == index :
# decoded_translation += f' {word}'
# sampled_word = word
if sampled_word == '<end>' or len(decoded_translation.split()) > ans_max:
stop_condition = True
empty_target_seq = np.zeros((1 , 1))
empty_target_seq[0 , 0] = sampled_word_index
states_values = [h , c]
print(f'Human: {tests[i]}')
print()
#decoded_translation = decoded_translation.split(' end')[0]
print(f'Bot: {decoded_translation}')
print('-'*25)
</code>
<code>
def QandA(enc_model,dec_model,vocabulary,preprocess_input,sentence):
states_values = enc_model.predict(np.array([preprocess_input(sentence)]))
empty_target_seq = np.zeros((1 , 1))
empty_target_seq[0, 0] = vocab['<start>']
stop_condition = False
decoded_translation = ''
while not stop_condition :
dec_outputs , h , c = dec_model.predict([empty_target_seq] + states_values)
sampled_word_index = np.argmax(dec_outputs[0, -1, :])
sampled_word = None
word = vocabulary[sampled_word_index]
decoded_translation += f' {word}'
sampled_word = word
if sampled_word == '<end>' or len(decoded_translation.split()) > ans_max:
stop_condition = True
empty_target_seq = np.zeros((1 , 1))
empty_target_seq[0 , 0] = sampled_word_index
states_values = [h , c]
ans = decoded_translation.replace("<end>","")
return ans
</code>
<code>
T = ""
while True:
T = input("You : ")
if T=='q':
break
print("Bot : "+QandA(enc_model,dec_model,vocabulary,preprocess_input,T))
</code>
|
{
"filename": "notebook-2_2.ipynb",
"repository": "DipanMondal/chatbot",
"query": "transformed_from_existing",
"size": 241783,
"sha": ""
}
|
# Analyze_chromatin_accessibility_data_to_identify_key_transcription_factors_involved_in_IFNβ_regulation.ipynb
Repository: connerlambden/BioloGPT
This notebook will analyze ATAC-seq data to assess chromatin accessibility at IRF sites.
<code>
import pandas as pd
# Load ATAC-seq data
atac_data = pd.read_csv('atac_seq_data.csv')
# Analyze accessibility at IRF sites
irf_accessibility = atac_data[atac_data['site'].isin(['proximal_IRF', 'distal_IRF'])]
</code>
The analysis will provide insights into the differential accessibility of IRF sites.
<code>
# Visualize the results
import matplotlib.pyplot as plt
plt.bar(irf_accessibility['site'], irf_accessibility['accessibility'])
plt.title('Chromatin Accessibility at IRF Sites')
plt.show()
</code>
***
### [**Evolve This Code**](https://biologpt.com/?q=Evolve%20Code%3A%20Analyze%20chromatin%20accessibility%20data%20to%20identify%20key%20transcription%20factors%20involved%20in%20IFN-%CE%B2%20regulation.%0A%0AIncorporate%20additional%20datasets%20for%20comparative%20analysis%20across%20different%20immune%20stimuli.%0A%0AChromatin%20remodeling%20proximal%20distal%20IRF%20sites%20stimulus-specific%20IFN%20expression%0A%0AThis%20notebook%20will%20analyze%20ATAC-seq%20data%20to%20assess%20chromatin%20accessibility%20at%20IRF%20sites.%0A%0Aimport%20pandas%20as%20pd%0A%23%20Load%20ATAC-seq%20data%0Aatac_data%20%3D%20pd.read_csv%28%27atac_seq_data.csv%27%29%0A%23%20Analyze%20accessibility%20at%20IRF%20sites%0Airf_accessibility%20%3D%20atac_data%5Batac_data%5B%27site%27%5D.isin%28%5B%27proximal_IRF%27%2C%20%27distal_IRF%27%5D%29%5D%0A%0AThe%20analysis%20will%20provide%20insights%20into%20the%20differential%20accessibility%20of%20IRF%20sites.%0A%0A%23%20Visualize%20the%20results%0Aimport%20matplotlib.pyplot%20as%20plt%0Aplt.bar%28irf_accessibility%5B%27site%27%5D%2C%20irf_accessibility%5B%27accessibility%27%5D%29%0Aplt.title%28%27Chromatin%20Accessibility%20at%20IRF%20Sites%27%29%0Aplt.show%28%29%0A%0A)
***
### [Created with BioloGPT](https://biologpt.com/?q=Could%20chromatin%20remodeling%20differences%20between%20proximal%20and%20distal%20IRF%20sites%20contribute%20to%20stimulus-specific%20IFN%C3%B0%20expression%3F)
[](https://biologpt.com/)
***
|
{
"filename": "Analyze_chromatin_accessibility_data_to_identify_key_transcription_factors_involved_in_IFNβ_regulation.ipynb",
"repository": "connerlambden/BioloGPT",
"query": "transformed_from_existing",
"size": 3393,
"sha": ""
}
|
# example_causalregnet_2.ipynb
Repository: luka-kovacevic/causalregnet
# Simulating Gene Expression Dynamics with CausalRegNet
CausalRegNet is a multiplicative effect SCM that uses a Negative Binomial distribution to simulate realstic scRNA-seq data. Here, we demonstrate how real data from Replogle et al. (2022) can be combined with the CausalRegNet framework to simulate data. We do this for both the observational and interventional setting.
<code>
import numpy as np
import pandas as pd
import seaborn as sns
import torch
import matplotlib.pyplot as plt
import lightning.pytorch as pl
from torch.utils.data import DataLoader, TensorDataset
from causalregnet import utils
from causalregnet.simulator import Simulator
from causalregnet.models.nb_fitter import NegativeBinomialFitter
sns.set_context("notebook")
sns.set_theme(style="whitegrid", palette="tab10", font="Arial")
plt.rcParams['figure.figsize'] = [3, 3]
</code>
## Fitting to Data from Replogle et al. (2022)
With our code, we provide three files that were derived from the Replogle et al. (2022) dataset according to the cancer gene selection procedure described in the paper. These files include:
- `k562_100_cancer_genes.csv`: expression matrix of 100 genes;
- `k562_targets.csv`: intervention target for each row; 'non-targeting' indicates no intervention;
- `k562_gene_names.csv`: maps ensembl id's (i.e. alternative gene names) to human readable gene names.
We then use the `fit` module from `causalregnet` to fit negative binomial distributions to three genes from this dataset with the aim of simulating data from a random 3 node DAG.
<code>
# loading cancer gene dataset
# `k562_100_cancer_genes.csv`: expression matrix
# `k562_targets.csv`: intervention target for each row; 'non-targeting' indicates no intervention
# `k562_gene_names.csv`: maps ensembl id's (i.e. alternative gene names) to human readable gene names
df = pd.read_csv('../causalregnet/data/k562_100_cancer_genes.csv', delim_whitespace=True, header=0)
targets = pd.read_csv('../causalregnet/data/k562_targets.csv', delim_whitespace=True, header=None)
targets.columns = ['gene']
gene_names = pd.read_csv('../causalregnet/data/k562_gene_names.csv', delim_whitespace=True, header=0)
# keep observational samples only
df = df.iloc[np.where(targets == 'non-targeting')[0],:]
# selecting first three genes
idx = [0, 1, 2]
data = torch.tensor(df.to_numpy(), dtype=torch.float32)
fitted_r = []
fitted_p = []
for g in idx:
dataset = TensorDataset(data[:, g])
data_loader = DataLoader(dataset, batch_size=1024, shuffle=True)
model = NegativeBinomialFitter()
trainer = pl.Trainer(max_epochs=100, log_every_n_steps=100)
trainer.fit(model, data_loader)
fitted_r.append(model.r.item())
fitted_p.append(torch.sigmoid(model.p).item()) # Apply sigmoid to get p in (0, 1)
fitted_r = np.array(fitted_r)
fitted_p = np.array(fitted_p)
fitted_mu = fitted_r * (fitted_p/(1 - fitted_p))
fitted_theta = fitted_r
</code>
## Observational Setting
Before simulating data, several user defined parameters must first be set including:
- `mu`: mean expression for each node (here derived from real data);
- `theta`: inverse dispersion for each node (here derived from real data);
- `alpha`: maximum regulatory effect of parents on each node;
- `beta`: minimum regulatory effect of parents on each node;
- `agg_type`: aggregation function to be used (currently the only option is `linear`);
- `reg_constant`: *regulatory adjustment constant*.
The directed acyclic graph `B` is generated using the `utils` module of `causalregnet` and the weight adjacency matrix `W` is subsequently generated as well.
<code>
# CausalRegNet parameters
np.random.seed(0)
mu = fitted_mu
theta = fitted_theta
alpha = np.repeat(2, 3)
beta = np.repeat(0.1, 3)
agg_type = 'linear'
reg_constant = np.repeat(1, 3)
B = utils.generate_dag(d=3, m=3)
W = utils.generate_W(B=B, w_ranges=((-2,-0.5), (0.5, 2)))
</code>
<code>
simulator = Simulator(nnodes=3,
mu=mu,
theta=theta,
W=W,
alpha=alpha,
beta=beta,
agg_type=agg_type,
reg_constant=reg_constant)
simulator.calibrate_sigmoid()
X = simulator.simulate(n_samp=1000)
</code>
<code>
plt.rcParams['figure.figsize'] = [7, 3]
fig, ax = plt.subplots(1, 2)
# strength of relationships
sns.heatmap(W, cmap=sns.light_palette("seagreen", as_cmap=True), ax=ax[0])
sns.histplot(X, ax=ax[1], bins=np.arange(-0.5, 20.5, step=1))
plt.tight_layout()
</code>
## Interventional Setting
Given an observational simulation, generting interventional data is simple as we only need to specify `intervention_type`(i.e. deterministic vs. stochastic) and `intervention_val`(value assigned to intervention target). In the simulatino below we interevene on $I=\{X_1\}$ by setting $X_1=0$ deterministically and do not intervene on the remaining variables $\mathbf{X} \backslash I = \{X_0, X_2\}$.
<code>
X_int = simulator.simulate(n_samp=1000, intervention_type='deterministic', intervention_val=[-1, 0, -1])
</code>
<code>
X_int
</code>
<code>
plt.rcParams['figure.figsize'] = [7, 3]
fig, ax = plt.subplots(1, 2)
# strength of relationships
sns.heatmap(W, cmap=sns.light_palette("seagreen", as_cmap=True), ax=ax[0])
sns.histplot(X_int, ax=ax[1], bins=np.arange(-0.5, 20.5, step=1))
plt.tight_layout()
</code>
<code>
# average treatment effects
print(X_int.mean(axis=0) - X.mean(axis=0))
</code>
|
{
"filename": "example_causalregnet_2.ipynb",
"repository": "luka-kovacevic/causalregnet",
"query": "transformed_from_existing",
"size": 52124,
"sha": ""
}
|
# 29april_1.ipynb
Repository: 03Akshay/assignments-1
# #Q1. Explain the basic concept of clustering and give examples of applications where clustering is useful.
Q1. Clustering is a type of unsupervised machine learning technique that involves grouping similar data points together based on their characteristics or features. The goal of clustering is to divide the data into distinct groups, or clusters, such that data points within each cluster are more similar to each other than to those in other clusters. The main idea is to find natural patterns and structures within the data without any predefined labels.
Examples of applications where clustering is useful include:
Customer segmentation: Clustering customers based on their purchasing behavior or preferences to identify different segments for targeted marketing.
Image segmentation: Grouping pixels with similar color or texture characteristics to segment objects in an image.
Anomaly detection: Identifying outliers or anomalies that deviate significantly from the normal patterns in the data.
Document clustering: Grouping similar documents together for organizing and summarizing large text corpora.
Recommender systems: Clustering users based on their interests to make personalized product or content recommendations.
Genetics and biology: Clustering genes or proteins to understand their relationships and functions.
Social network analysis: Clustering individuals with similar social connections or behavior to detect communities or influencers.
# #Q2. What is DBSCAN and how does it differ from other clustering algorithms such as k-means and
hierarchical clustering?
Q2. DBSCAN (Density-Based Spatial Clustering of Applications with Noise) is a popular density-based clustering algorithm. Unlike k-means, which is centroid-based, and hierarchical clustering, which builds a tree-like structure, DBSCAN forms clusters based on data density.
Key characteristics of DBSCAN:
It does not require the user to specify the number of clusters beforehand.
It groups data points that are close to each other in regions of high density.
It can handle clusters of different shapes and sizes.
It identifies and marks data points that do not belong to any cluster as outlie
# #Q3. How do you determine the optimal values for the epsilon and minimum points parameters in DBSCAN
clustering?
Q3. The two main parameters in DBSCAN are epsilon (ε) and minimum points (MinPts).
Epsilon (ε) defines the radius or neighborhood around each data point. Points within this radius are considered neighbors.
Minimum points (MinPts) specifies the minimum number of points required within the epsilon neighborhood to form a cluster.
Determining optimal values for ε and MinPts is often a trial-and-error process. Several methods can help:
Visual inspection: Plot the data and experiment with different values of ε and MinPts to observe the cluster structures.
K-distance plot: Plot the k-distances (distance to the kth nearest neighbor) in ascending order to identify a "knee" that suggests a good ε value.
Reachability distance plot: Plot the reachability distances of the data points to identify suitable MinPts values.
# #Q4. How does DBSCAN clustering handle outliers in a dataset?
Q4. DBSCAN naturally handles outliers in a dataset. Outliers are considered as data points that do not belong to any cluster and are not within the ε-neighborhood of any other data point (i.e., they have fewer than MinPts neighbors). DBSCAN classifies such points as noise or outliers.
# #Q5. How does DBSCAN clustering differ from k-means clustering?
Q5. The main differences between DBSCAN clustering and k-means clustering are:
DBSCAN is a density-based algorithm that groups data points based on their density, while k-means is a centroid-based algorithm that assigns data points to the nearest cluster center (centroid).
DBSCAN does not require specifying the number of clusters beforehand, while k-means needs the number of clusters to be specified.
DBSCAN can handle clusters of different shapes and sizes, whereas k-means assumes clusters as spherical and balanced around centroids.
DBSCAN can identify and handle outliers naturally, while k-means considers all data points as part of some cluster.
# #Q6. Can DBSCAN clustering be applied to datasets with high dimensional feature spaces? If so, what are
some potential challenges?
Q6. DBSCAN can be applied to datasets with high-dimensional feature spaces. However, high-dimensional data can present challenges known as the "curse of dimensionality." As the number of dimensions increases, the density of points in the space decreases, and the concept of distance becomes less meaningful. This can lead to the following challenges:
The selection of appropriate distance measures becomes critical, as the Euclidean distance may not be effective in high-dimensional spaces.
The curse of dimensionality can cause all data points to appear equidistant, making it difficult for DBSCAN to identify meaningful clusters.
The computational cost of DBSCAN can increase significantly with the number of dimensions.
To address these challenges, dimensionality reduction techniques like PCA (Principal Component Analysis) or t-SNE (t-distributed Stochastic Neighbor Embedding) can be used to reduce the feature space's dimensionality before applying DBSCAN.
# #Q7. How does DBSCAN clustering handle clusters with varying densities?
Q7. DBSCAN can handle clusters with varying densities effectively. It can find clusters of different shapes and sizes and is not limited to identifying clusters of uniform density like some other clustering algorithms.
In DBSCAN, clusters are formed by connecting densely populated regions of the data space, regardless of the overall density in the dataset. Regions with a higher density will have more data points, and regions with lower density will result in smaller clusters. This makes DBSCAN suitable for datasets with clusters that have varying densities.
# #Q8. What are some common evaluation metrics used to assess the quality of DBSCAN clustering results?
Q8. Common evaluation metrics for DBSCAN clustering results include:
Silhouette Score: Measures the compactness and separation of clusters. A higher silhouette score indicates better-defined clusters.
Davies-Bouldin Index: Evaluates the average similarity between each cluster and its most similar cluster, with lower values indicating better clustering.
Adjusted Rand Index (ARI): Compares the clustering results with a ground truth (if available) to assess the agreement between the two.
Visual inspection: Sometimes, the best way to evaluate clustering is by visually inspecting the results to see if they align with the expected patterns.
<code>
# 3
</code>
Q9. DBSCAN is primarily an unsupervised learning algorithm for clustering and does not have direct support for semi-supervised learning tasks. Semi-supervised learning typically involves using a combination of labeled and unlabeled data to build a model. While DBSCAN doesn't inherently support this, you could potentially combine it with other techniques to perform semi-supervised learning.
One way to achieve semi-supervised learning with DBSCAN is by first clustering the data into groups, and then, using the obtained cluster assignments as pseudo-labels for the unlabeled data points. You can then use this labeled data to train a supervised model, like a classifier or regression model.
Q10. DBSCAN is robust to noise and can handle datasets with noise or missing values effectively. Noise points or points with missing values will be considered outliers by DBSCAN and won't be assigned to any cluster.
When using DBSCAN with missing values, you can either pre-process the data to handle missing values before applying the algorithm or modify the distance metric to accommodate missing values. For example, you can use the "k-nearest neighbors" approach to impute missing values for calculating distances during the clustering process.
Q11. As an AI language model, I'm unable to execute code, but I can provide you with a basic Python implementation of the DBSCAN algorithm:
<code>
import numpy as np
def euclidean_distance(point1, point2):
return np.linalg.norm(point1 - point2)
def region_query(data, point, epsilon):
neighbors = []
for p in data:
if euclidean_distance(p, point) <= epsilon:
neighbors.append(p)
return neighbors
def expand_cluster(data, point, cluster_id, epsilon, min_points, cluster_assignment):
neighbors = region_query(data, point, epsilon)
if len(neighbors) < min_points:
cluster_assignment[point] = -1 # Mark point as noise/outlier
return False
else:
cluster_assignment[point] = cluster_id
</code>
<code>
v
</code>
|
{
"filename": "29april_1.ipynb",
"repository": "03Akshay/assignments-1",
"query": "transformed_from_existing",
"size": 13982,
"sha": ""
}
|
# cambridge_program_1.ipynb
Repository: miykael/workshop
<div align="center"><img width="50%" src="slides/images/Cambridge_logo.png"></div>
# Workshop Cambridge, September 2018
Python is on its way to become the most used programming language in neuroscience. It is easy to understand, can be learned rather quickly and has a very strong and helpful community behind it. There exist many amazing neuroimaging software packages, such as Nipype & fmriprep, that facilitate the everyday life of a neuroscientist.
The goal of this 2-day workshop is to introduce participants to the basics of Python, to give them a short overview about relevant neuroimaging software packages and most of all, to teach them everything they need to know about Nipype. Nipype is an open-source software package, that provides a unified way of interfacing with most of the freely available neuroimaging software packages, such as SPM, FSL, AFNI, FreeSurfer, and ANTs.
The full content of this course, all notebooks and slides can be found on the github repository [github.com/miykael/workshop_cambridge](https://github.com/miykael/workshop_cambridge).
### <span style="color:red">Important 1</span>
If you're running this notebook through a docker container, make sure that you used the `-v` flag to mount a folder on your system inside the container. Like this, you will have access to any output that you create within this container. For more, take a look at the [Docker information](https://github.com/miykael/workshop_cambridge#docker).
### <span style="color:red">Important 2</span>
If you used the `-v` flag from above to mount the `output` folder, you can use the following command to save all notebooks (with your changes) and slides in the folder `/output/`. **Just don't forget to run this cell before closing the docker container!**
<code>
# Save the workshop and nipype_tutorial content into your output folder
!cp -R /home/neuro/* /output/
</code>
<code>
# Get the saved notebook from the output folder back into this docker image
!cp -R /output/* /home/neuro/
</code>
# Day 1
<h2 style="background-color: #F0F0F0;">Python Basics
### `09:00-10:00` Introduction to Python and Jupyter Notebooks
This section is meant as a quick introduction to Jupyter Notebooks and Python. What are they, how do they work and why are they so cool?
- Slides: [Introduction to Python and Jupyter Notebook](slides/day1_01_python_and_jupyter_notebook.html)
- Notebook 1: [Jupyter Notebook](../nipype_tutorial/notebooks/introduction_jupyter-notebook.ipynb)
- Notebook 2: [All about Python](../nipype_tutorial/notebooks/introduction_python.ipynb)
### `10:00-10:15` Coffee & Tea Break
### `10:15-11:15` Crash course in scientific toolboxes
One advantage of Python is the vast availability of toolboxes. There's a toolbox for almost everything! In this section, we want to introduce you to the main scientific toolboxes that every researcher should know.
- Slides: [Scientific Toolboxes](slides/day1_02_scientific_toolboxes.html)
- Notebook 1: [Numpy](notebooks/python_numpy.ipynb)
- Notebook 2: [Scipy](notebooks/python_scipy.ipynb)
- Notebook 3: [Scikit](notebooks/python_scikit.ipynb)
- Notebook 4: [Statistics](notebooks/python_statistics.ipynb)
- Notebook 5: [Visualization](notebooks/python_visualization.ipynb)
### `11:15-12:00` How to set up your system for Python and Nipype, using Conda and Docker
There are many ways to create the right computational environment for your research. But if you want to use the newest technologies you will not get around using Docker or Conda.
- Slides: [Conda and Docker](slides/day1_03_conda_and_docker.html)
### `12:00-13:00` Lunch
<h2 style="background-color: #F0F0F0;">Python & Neuroimaging
### `13:00-14:00` How to handle your MRI data with Nibabel and Nilearn
It's liberating to have direct access to your neuroimaging data. `Nibabel` and `Nilearn` allow exactly that. With those two neuroimaging packages, you can consider the brain a simple 3D/4D matrix of datapoints and do with it whatever you want.
- Slides: [Data Manipulation](slides/day1_04_data_manipulation.html)
- Notebook 1: [Nibabel](notebooks/image_manipulation_nibabel.ipynb)
- Notebook 2: [Nilearn](notebooks/image_manipulation_nilearn.ipynb)
### `14:00-14:15` Coffee & Tea Break
### `14:15-17:00` Crash course in neuroimaging software toolboxes
There are many different ways to analyze MRI data and many different software to do so. In this section, we want to show you some neuroimaging toolboxes written in Python that allow you to do things like diffusion imaging (`Dipy`), functional connectivity (`Nilearn`) or machine learning (`Nilearn`/`PyMVPA`). This list is by no means exhaustive and we will speak about much more software tomorrow morning.
- Notebook 1: [Diffusion Imaging with Dipy](notebooks/diffusion_imaging.ipynb)
- Notebook 2: [Functional Connectivity with Nilearn](notebooks/functional_connectivity.ipynb)
- Notebook 3: [Machine Learning Preparation](notebooks/machine_learning_preparation.ipynb)
### `15:45-16:00` Coffee & Tea Break
- Notebook 4: [Prediction with Nilearn & PyMVPA](notebooks/machine_learning_nilearn_and_pymvpa.ipynb)
- Notebook 5: [Convoluted Neural Networks with Keras](notebooks/machine_learning_keras.ipynb)
<h2 style="background-color: #F0F0F0;"> Day 2 (morning) - Everything about Nipype
### `09:00-09:30` Introduction to Nipype
In this short introduction, we will show you what Nipype is and why you should use it. It contains a powerful short example that shows the strength behind Nipype.
- Slides: [Short introduction to Nipype](../nipype_tutorial/notebooks/introduction_nipype.html)
- Notebook: [Nipype Showcase](../nipype_tutorial/notebooks/introduction_showcase.ipynb)
### `09:30-11:00` Building blocks of Nipype: Interfaces & Workflows
Nipype can be learned very quickly, but it's nonetheless important that you know about some of the main building blocks.
- Slides: [Interfaces & Workflows](slides/day2_01_nipype_basics.html)
- Notebook: [Basic Concepts](../nipype_tutorial/index.ipynb)
**Note:** Coffee & Tea break can be taken at any time during this lecture.
### `11:00-12:00` New innovations in the field (Part 1)
There are many new and innovative neuroimaging softwares, such as BIDS, fmriprep, MRIQC, OpenNeuro, etc. And many of them wouldn't be possible without Nipype and the open-source neuroimaging community. In this section, we want to introduce you to some toolboxes. Because if you don't use them yet yourself, it's at least important that you've already heard about them.
- Slides: [What you need to know!](slides/day2_02_neuroimaging_innovations.html)
The slides are covering the following software:
- [BIDS](http://bids.neuroimaging.io/)
- [pyBIDS](https://incf.github.io/pybids/)
- [BIDS-Apps](http://bids-apps.neuroimaging.io/)
- [MRIQC](https://mriqc.readthedocs.io/en/latest/)
- [fMRIPrep](http://fmriprep.readthedocs.io/en/latest/)
- [C-PAC](https://fcp-indi.github.io/)
- [Mindboggle](http://mindboggle.info/index.html#)
- [Neurodocker](https://github.com/kaczmarj/neurodocker)
- [OpenNeuro.org](https://openneuro.org/)
- [Neurovault](https://neurovault.org/)
- [Datalad](https://www.datalad.org/)
- [Porcupine](https://timvanmourik.github.io/Porcupine/)
- [Neurostars.org](https://neurostars.org/)
### `12:00-13:00` Lunch
### `13:00-13:30` New innovations in the field (Part 2)
<h2 style="background-color: #F0F0F0;"> Day 2 (afternoon) - Nipype Hands-On
### Use what you've learned!
The goal of this afternoon is that you get your hands dirty with Nipype. For this purpose, you will work on the Hands-on example from the Nipype Tutorial. It contains a complete task-based fMRI analysis, including pre-processing, 1st-level and 2nd-level analysis.
The goal of this hands-on is to show you a real case example of a pre-processing and analysis pipeline with Nipype. Don't hesitate to ask us if you have questions. And feel free to create your own pipeline from scratch if you want. We're happy to help you to get things going.
**Important:** Don't forget to use the `-v` flag to run the docker container. Like this, you will have access to changes in the notebook and possible output that you want to keep.
### `13:30-15:00` Pre-processing Hands-on
* Notebook under: [nipype_tutorial/notebooks/handson_preprocessing.ipynb](../nipype_tutorial/notebooks/handson_preprocessing.ipynb)
**Note:** Coffee & Tea break can be taken at any time during this lecture.
### `15:00-16:00` Analysis Hands-on
* Notebook under: [nipype_tutorial/notebooks/handson_analysis.ipynb](../nipype_tutorial/notebooks/handson_analysis.ipynb)
**Note:** Coffee & Tea break can be taken at any time during this lecture.
### `16:00-17:00` Wrap-up Session
To wrap-up, we quickly want to summarize what we've learned and point you to useful resources. But most and for all, we want to give you again the opportunity to ask any question you have.
|
{
"filename": "cambridge_program_1.ipynb",
"repository": "miykael/workshop",
"query": "transformed_from_existing",
"size": 11982,
"sha": ""
}
|
# Results_1.ipynb
Repository: dfm/github-repo-crawler
|
{
"filename": "Results_1.ipynb",
"repository": "dfm/github-repo-crawler",
"query": "transformed_from_existing",
"size": 311606,
"sha": ""
}
|
# version1_1.ipynb
Repository: j85liu/equityresearch
Top emerging biotech companies
* Aiolos Bio: Focuses on developing treatments for respiratory and inflammatory diseases
* Rapport Therapeutics: Uses receptor-associated proteins (RAPs) to create precision neuromedicines
* Sumatrix Biotech: Combines AI with computational biology to speed up drug discovery
* HHV Biotech: Specializes in the development of biofilm-focused gene therapy
* Lumatix Biotech: Develops bio-based alternatives to traditional petrochemical products
* Element Biosciences: Offers a DNA sequencing platform that enables researchers to achieve cost-effective and accurate genomic data
Step 2: Build a Data Pipeline to Collect Public Market Data
* (A) Automate SEC Filings & Earnings Calls
* Use EDGAR API or Python scraping libraries:
* 10-K, 10-Q (Financials, MD&A, Risk Factors)
* 8-K (Earnings Releases, Material Events)
* Proxy Statements (Executive Compensation, Shareholder Votes)
* Transcripts from earnings calls (via AlphaSense, Seeking Alpha)
* 📌 Code Example (SEC EDGAR API in Python)
<code>
import requests
cik = "0000320193" # Apple CIK code
url = f"https://data.sec.gov/submissions/CIK{cik}.json"
headers = {"User-Agent": "James Liu - Columbia Research Project"}
response = requests.get(url, headers=headers)
data = response.json()
print(data["filings"]["recent"]["form"]) # Lists latest SEC filings
</code>
(B) Pull Real-Time Market Data
* Use Yahoo Finance API or Alpha Vantage for price data.
* Historical stock prices (OHLCV)
* Company fundamentals (P/E, EPS, Debt, Cash, ROE)
* Dividend history, short interest, institutional ownership
* 📌 Example (Yahoo Finance API with Python)
<code>
import yfinance as yf
ticker = "AAPL"
stock = yf.Ticker(ticker)
print(stock.history(period="1y")) # Get 1-year historical price data
print(stock.info["marketCap"]) # Get market cap
</code>
(C) Gather Alternative Data (Sentiment, Web Trends, Patents, etc.)
* Google Trends API (Track search volume for company names, products).
* Social Media Sentiment (Reddit, Twitter/X, Seeking Alpha comments).
* Patent & Drug Pipeline Tracking (For biotech/pharma stocks).
* 📌 Example (Google Trends Data with Python)
<code>
import sys
!{sys.executable} -m pip install pytrends
from pytrends.request import TrendReq
pytrends = TrendReq()
pytrends.build_payload(["Tesla"], timeframe="now 7-d")
trend_data = pytrends.interest_over_time()
print(trend_data)
</code>
Step 3: Automate Financial Modeling & Valuation
* DCF (Discounted Cash Flow) Model – Forecast revenue, costs, cash flows.
* Comparable Company Analysis (P/E, EV/EBITDA, PEG Ratio)
* Monte Carlo Simulation for Stock Price Forecasting
<code>
def discounted_cash_flow(cash_flows, discount_rate):
return sum(cf / (1 + discount_rate) ** i for i, cf in enumerate(cash_flows))
future_cash_flows = [500, 550, 600, 650, 700] # Example cash flows
discount_rate = 0.08 # 8% WACC
valuation = discounted_cash_flow(future_cash_flows, discount_rate)
print(f"Estimated DCF Valuation: ${valuation:.2f}M")
</code>
<code>
import sys
!{sys.executable} -m pip install dash
import dash
import dash_core_components as dcc
import dash_html_components as html
import yfinance as yf
app = dash.Dash(__name__)
def get_stock_price(symbol):
return yf.Ticker(symbol).history(period="1d")["Close"].iloc[-1]
app.layout = html.Div([
html.H1("Stock Price Dashboard"),
dcc.Input(id="stock-symbol", value="AAPL", type="text"),
html.Button("Update", id="update-btn"),
html.H2(id="price-display"),
])
@app.callback(
dash.dependencies.Output("price-display", "children"),
[dash.dependencies.Input("update-btn", "n_clicks")],
[dash.dependencies.State("stock-symbol", "value")]
)
def update_stock_price(n, symbol):
price = get_stock_price(symbol)
return f"Current Price of {symbol}: ${price:.2f}"
if __name__ == "__main__":
app.run_server(debug=True)
</code>
|
{
"filename": "version1_1.ipynb",
"repository": "j85liu/equityresearch",
"query": "transformed_from_existing",
"size": 30560,
"sha": ""
}
|
# index_1.ipynb
Repository: learn-co-curriculum/dsc-ml-interp-blackbox-models
# Machine Learning Interpretability
## Introduction
In the previous lessons, we discussed that some models that are considered intrinsically interpretable, such as simple regression and decision trees. We referred to these as white-box models, a nickname that suggests that the algorithm is transparent and easy to understand using summary statistics and data visualizations.
White-box models are excellent tools for solving certain problems and can make predictions with a reasonable degree of accuracy. However, over the past few decades, new algorithms have been developed that allow researchers to build models that can make predictions over very large data sets with many features. This includes (but is not limited to) neural networks and complex tree-based models like XGBoost.
Due to their sophisticated handling of large datasets, these __black-box models__ can offer incredible learning potential and increased accuracy if you are willing to reduce the simplicity of your interpretation. In this lesson, we will discuss the attributes of black-box models and how we can interpret them to increase our confidence in the results.
## Objectives
You will be able to:
* Distinguish between white-box and black-box models
* Explain two common black-box models, XGBoost and neural networks and identify use cases for each
## White-Boxes vs Black-Boxes
A black box model is a model that is not __intrinsically interpretable__. A model is considered intriniscally interpretable, when the it is easy for an observer to understand how the model arrived at its prediction. One example of this could be a time-series plot that visualizes dates on the x-axis and temperature on the y-axis. Individuals familiar with how line plots work could infer increase in date correlates with an increase in temperature. Alternatively, we could manually analyze a list of sorted data points indicating data and temperature to arrive at the same conclusion.
When there were greater limitations on data storage and compute resources, models that require those resources and complex __post-hoc explanations__ were not always practical or accessible for widespread use. However, as advances in technology have increased access to large data stores and powerful computers, researchers are beginning to harness the benefits of black-box models, namely increased accuracy in predictions.
Just because black-box models are not easy to interpret, does not mean that they cannot be explained. Let's discuss some of the most popular __black-box models__ and the methods we can use to extract post-hoc explanations to gain insight.
## Common Black-Box Models
### Gradient Boosted Trees (GBDT)
A __Gradient Boosting Decision Trees (GBDT)__ is an tree-based ensemble learning algorithm that is similar to a random forest. Ensemble methods combine multiple algorithms to produce a more well-rounded model and can be used for classification or regression. To get a basic understanding of how GBDT works, it is helpful to review decision trees and discuss gradient boosting.
As you may recall, decision trees create a model that makes a prediction by evaluating conditional and true-false feature questions. __Boosting__ enhances a single weak model with many other weak models and combining results to get a better prediction. __Gradient boosting__ is the process of additively generating weak models and then formalizing their results as a gradient descent algorithm over an objective function. Then, the algorithm iterates over each model and creates a target outcome for the next model, based on the gradient of the error for the first model, moving closer to the most accurate prediction at each step. This is different then a random forest, which instead bags and averages multiple trees to arrive at its final prediction.
Some applications of GBDTs include, but are not limited to:
* __Fraud detection__ - GBDTs can be used to identify fraudulent transactions by analyzing patterns of behavior in large datasets of financial transactions.
* __Predicting medical outcomes__ - GBDTs can be used to predict the likelihood of certain medical outcomes, such as the likelihood of a patient developing a particular disease or the effectiveness of a certain treatment.
* __Recommender systems__ - GBDTs can be used to build recommender systems that suggest products or content to users based on their previous interactions and preferences.
* __Computer vision__ - GBDTs can be used to build image and video analysis systems that can recognize and classify objects, people, and activities, which can be applied in many fields such as surveillance, self-driving cars, and medical imaging.
* __Predicting customer churn__ -
GBDTs can be used to predict which customers are likely to leave a company by analyzing their behavior and demographics. This can help companies to proactively retain customers by targeting them with special promotions or incentives.
These are just a few examples, GBDTs are widely used in many other fields such as natural language processing and speech recognition. The flexibility of the GBDT algorithm makes it applicable to a wide range of problems.
### Neural Networks
Neural networks are a subset of machine learning algorithms intended to imitate how biological neurons transmit information in the human brain. They are also powerful tools that allow us to perform tasks like speech recognition and image recognition at high speed.
#### How do neural networks work?
The basic structure of a neural network is the input layer, the hidden layer(s), and the output layer. Each of these layers contains nodes, which can be analogized to biological neurons. Each of these nodes connects to another deeper node. Each node also has weight and a threshold.
When a threshold is met, the node is activated, allowing the data to move to the next layer of the network.
#### Use Cases for Neural Networks
Neural networks are used for a number of applications in the medical field. For example:
* __Medical Imaging__ -
Neural networks can be trained to analyze medical images such as X-rays, CT scans, and MRI scans, and identify specific features or abnormalities. This can help radiologists and other medical professionals to more accurately and efficiently diagnose diseases such as cancer, heart disease, and neurological disorders.
* __Drug Research and Development__ -
Another example use case is in the field of drug discovery. Neural networks can be used to analyze large amounts of data from chemical compounds and predict the potential effectiveness and side effects of new drugs. This can help pharmaceutical companies to more quickly and effectively identify new drug candidates for testing and development.
* __Patient Outcomes__ -
Additionally, neural networks can be used to predict patient outcomes, such as risk of readmission, progression of a disease and survival rate, based on a wide range of patient data, including genetic, demographic, and clinical data. This can help doctors and other medical professionals to make more informed decisions about patient care and treatment.
In general, neural networks have a lot of potential in medicine due to their ability to analyze and learn from large amounts of data and make predictions, which can help to improve the accuracy and efficiency of medical diagnosis and treatment.
## Summary
A white-box model is a model whose inner workings and decision-making process can be easily understood and interpreted by humans. This typically includes models that are based on simple mathematical equations and decision trees. A black-box model, on the other hand, is a model whose inner workings and decision-making process are hidden or difficult for humans to understand. This typically includes models that are based on complex mathematical equations, such as neural networks and machine learning algorithms. The main difference between white-box and black-box models is the level of interpretability and transparency of the model.
While black-box models are seemingly less interpretable than white-box models, there are still a variety of methods that we can use to explain the results of black-box models. In the next lesson, we will explore some use cases for neural networks and what methods we can use to explain them.
|
{
"filename": "index_1.ipynb",
"repository": "learn-co-curriculum/dsc-ml-interp-blackbox-models",
"query": "transformed_from_existing",
"size": 10362,
"sha": ""
}
|
# CrewAI_Basics.ipynb
Repository: rohitreddynagareddy/132test
<a href="https://colab.research.google.com/github/rohitreddynagareddy/132test/blob/master/CrewAI_Basics.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>

### Phase 1: Introduction & Fundamentals
[CrewAI Official Dumentation](https://docs.crewai.com/introduction)
Topics to Discuss Here
1. Crew
2. Agents
3. Tasks
4. LLM
5. Tools
### Phase 2: Setting Up & Basic Agent Implementation
<code>
# Step 1: Install CrewAI
!pip install -q crewai
</code>
<code>
!pip show crewai
</code>
### API Check
<code>
!pip install -Uq openai
</code>
<code>
import os
from google.colab import userdata
from openai import OpenAI
api_key = userdata.get('OPENAI_API_KEY')
if api_key:
os.environ["OPENAI_API_KEY"] = api_key
print("API key loaded from userdata.")
else:
print("API key not found in userdata. Please set OPENAI_API_KEY in userdata.")
################## OPENAI API CHECK ############################
client = OpenAI(
api_key=os.environ.get("OPENAI_API_KEY"),
)
response = client.responses.create(
model="gpt-4o",
instructions="You are a coding assistant that talks like a pirate.",
input="How do I check if a Python object is an instance of a class?",
)
print(response.output_text)
</code>
# Optional LLMS
<code>
from crewai import LLM
# Basic configuration
llm = LLM(model="gpt-4")
# Advanced configuration with detailed parameters
llm = LLM(
model="gpt-4o-mini",
temperature=0.7, # Higher for more creative outputs
timeout=120, # Seconds to wait for response
max_tokens=4000, # Maximum length of response
top_p=0.9, # Nucleus sampling parameter
frequency_penalty=0.1, # Reduce repetition
presence_penalty=0.1, # Encourage topic diversity
response_format={"type": "json"}, # For structured outputs
seed=42 # For reproducible results
)
# GROQ
llm = LLM(
model="groq/llama-3.2-90b-text-preview",
temperature=0.7
)
# OLLAMA
llm = LLM(
model="ollama/llama3:70b",
base_url="http://localhost:11434"
)
</code>
<code>
# Step 2: Import necessary libraries
from crewai import Agent, Task, Crew
</code>
<code>
# Step 3: Define a simple agent
agent1 = Agent(
name="Researcher",
description="An AI agent that researches and gathers information.",
goal="Find relevant information on a given topic.",
role="Researcher", # Added role
backstory="An AI assistant designed for research tasks." # Added backstory
)
</code>
<code>
# Step 4: Create a simple task
research_task = Task(
name="Research Task",
description="Search for the latest advancements in AI and summarize them.",
agent=agent1,
expected_output="A summary of the latest advancements in AI" # Added expected output
)
</code>
<code>
# Step 5: Initialize a crew (single agent for now)
crew = Crew(agents=[agent1], tasks=[research_task])
crew.kickoff()
</code>
### Phase 3: Multi-Agent Collaboration & Workflows
<code>
# Step 6: Define multiple agents
agent2 = Agent(
name="Writer",
description="An AI agent that writes research reports.",
goal="Create structured reports from gathered research data.",
role="Writer", # Added role
backstory="An AI assistant designed for writing reports." # Added backstory
)
agent3 = Agent(
name="Reviewer",
description="An AI agent that reviews and refines reports.",
goal="Ensure clarity, grammar, and accuracy in written content.",
role="Reviewer", # Added role
backstory="An AI assistant designed for reviewing reports." # Added backstory
)
</code>
<code>
# Step 7: Assign tasks to each agent
gather_info = Task(
name="Gather Information",
description="Find the latest research papers and summarize key findings.",
agent=agent1,
expected_output="A summary of key findings from recent research papers." # Added expected output
)
write_report = Task(
name="Write Research Report",
description="Use summarized research to create a structured report.",
agent=agent2,
expected_output="A structured research report based on the summarized findings." # Added expected output
)
review_report = Task(
name="Review Report",
description="Check the report for accuracy and clarity.",
agent=agent3,
expected_output="A reviewed and refined research report." # Added expected output
)
</code>
<code>
# Step 8: Create a Crew with multiple agents
multi_agent_crew = Crew(
agents=[agent1, agent2, agent3],
tasks=[gather_info, write_report, review_report]
)
multi_agent_crew.kickoff()
</code>
|
{
"filename": "CrewAI_Basics.ipynb",
"repository": "rohitreddynagareddy/132test",
"query": "transformed_from_existing",
"size": 37560,
"sha": ""
}
|
# practice_final_svitlana_1.ipynb
Repository: svetlanama/ai
<a href="https://colab.research.google.com/github/svetlanama/ai_practice/blob/dev/final_svitlana.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Імпорт модулів
<code>
import pandas as pd
# Load the CSV from Google Drive
url = 'https://drive.google.com/uc?id=1lMc8CKRE3Txuk4ntqpVpzM1qTlQkYADd'
data = pd.read_csv(url)
data.head()
</code>
<code>
skills = ['Python', 'Machine Learning', 'SQL', 'Data Analysis', 'TensorFlow', 'Pandas'] # Define a skill list, #TODO: extend
def extract_skills(description):
found_skills = [skill for skill in skills if skill.lower() in description.lower()]
return found_skills
data['Skills'] = data['Job Description'].apply(extract_skills)
data
</code>
<code>
import re
def process_salary_range(salary_range):
if salary_range.strip() == '-1' or salary_range.strip() == '-1.0':
return 0, 0
# Remove non-numeric characters except '-' for ranges
salary_range = re.sub(r'[^\d\-\s]', '', salary_range) # Remove $, K, and text
# Split into low and high ranges
low, high = salary_range.split('-')
# Convert 'K' to thousands
low = int(low.strip()) * 1000
high = int(high.strip()) * 1000
return low, high
# Example usage
# example_salary = "$95K-$160K (Glassdoor est.)"
# min_salary, max_salary = process_salary_range(example_salary)
# print(f"Min Salary: {min_salary}, Max Salary: {max_salary}")
# Apply the function to each row in the 'Salary Estimate' column
def process_salary_with_debug(salary_range):
# print(f"Processing Salary Range: {salary_range}") # Debug print
return process_salary_range(salary_range) # Call the actual function
for i, row in data.iterrows():
low, high = process_salary_with_debug(row['Salary Estimate'])
data.loc[i, ['Min Salary', 'Max Salary']] = low, high
data
</code>
<code>
for skill in skills:
data[skill] = data['Skills'].apply(lambda x: 1 if skill in x else 0)
data
</code>
<code>
# Prepare the input (X) and output (y) as follows:
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
# Define features (binary skill columns) and target (Salary Estimate)
X = data[skills]
# y = data['Salary Estimate'] # Ensure this is numeric (convert if necessary)
# y = data[['Min Salary', 'Max Salary']]
y = (data['Min Salary'] + data['Max Salary']) / 2
# Normalize features
scaler = MinMaxScaler()
X_scaled = scaler.fit_transform(X)
# Split the data # train and test
X_train, X_test, y_train, y_test = train_test_split(X_scaled, y, test_size=0.2, random_state=42)
X_train
X_test
y_train
y_test
</code>
<code>
# Step 2: Train the Neural Network
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
X_train.shape[1]
# Define the model TODO: experiment more
model = Sequential([
# Dense(64, input_dim=12, activation='relu'), # Input layer - skills 6 users + 6 new job
Dense(64, input_dim=X_train.shape[1], activation='relu'), # Input layer # X_train.shape[1] = 6 - TODO: 6 skills
Dense(32, activation='relu', name="dense_22"), # Hidden layer
Dense(1, name="dense_23") # Output layer
])
# Compile the model
model.compile(optimizer='adam', loss='mse', metrics=['mae'])
# Train the model
history = model.fit(X_train, y_train, epochs=100, batch_size=32, validation_split=0.2) # 100 epoch
test_loss, test_mae = model.evaluate(X_test, y_test)
print(f'Test Loss: {test_loss}, Test MAE: {test_mae}')
model.layers
</code>
<code>
# COSINE SIMILARITY
import numpy as np
last_6_columns = data.columns[-6:] # Select the last 6 columns with encoded skills, TODO: adapt for more skills
user_skills = np.array([1, 1, 0, 1, 0, 1]).reshape(1, -1).astype(np.float32)
# Loop through the rows and print Job Title and the corresponding job features (last 6 columns)
for index, row in data.iterrows():
job_title = row['Job Title'] # Extract the job title
job_features = row[last_6_columns].values.reshape(1, -1) # Extract the job features (last 6 columns)
job_features_32 = row[last_6_columns].values.reshape(1, -1).astype(np.float32) # Reshape and convert to float32
# 1. Calculate cosine similarity between user skills and job features
similarity = cosine_similarity(user_skills, job_features)
# Print the results
print(f"Job Title: {job_title}")
print(f"Cosine Similarity: {similarity[0][0]:.2f}")
# print(f"Predicted Fit Score: {predicted_fit[0][index]}\n")
# print(f"Predicted Fit Score: {predicted_fit[0][0]}\n")
# print(f"Predicted Fit Score: {predicted_fit[0][0]:.2f}\n")
print(f"===========")
</code>
<code>
# USE
from tensorflow.keras.layers import UnitNormalization
from tensorflow.keras.models import Model
from sklearn.metrics.pairwise import cosine_similarity
# Create a new list of layers without the last layer
new_layers = model.layers[:-1] # Exclude the last layer
# Build a new model using the new layers
model_output = Sequential(new_layers)
# print("Original2 model layers:", new_model.layers)
model_output.add(UnitNormalization())
# Передбачення особливостей вакансій через модель
X_features = model_output.predict(X) # X = data[skills]
# Передбачення особливостей користувача
predicted_fit = model_output.predict(user_skills)
print(f"predicted_fit.shape: {predicted_fit.shape} ") # (1, 32)
scores = X_features @ predicted_fit.T
print(f"Розрахунок оцінок схожості: {scores.shape}") # (кількість вакансій, 1)
# топ-5 вакансій
print("Top 5 job recommendations for the user:")
for rank, index in enumerate(top_indices, start=1):
job_title = data['Job Title'].iloc[index]
fit_score = scores[index, 0]
print(f"{rank}. {job_title} - Fit Score: {fit_score:.2f}")
# for index, row in data.iterrows():
# job_title = row['Job Title'] # Extract the job title
# job_features = row[last_6_columns].values.reshape(1, -1) # Extract the job features (last 6 columns)
# #job_features_32 = row[last_6_columns].values.reshape(1, -1).astype(np.float32) # Reshape and convert to float32
# # Ensure that the index does not go out of bounds
# if index < len(predicted_fit[0]):
# print(f"Predicted Fit Score for {job_title}: skills: job_features:{job_features} score: {predicted_fit[0][index]:.2f}")
# else:
# print(f"Index {index} is out of bounds for predicted_fit.")
# # print(f"Predicted Fit Score: {predicted_fit[0][index]}\n")
</code>
# Завдання 6
Збережіть нейромережу та зробіть прогноз
|
{
"filename": "practice_final_svitlana_1.ipynb",
"repository": "svetlanama/ai",
"query": "transformed_from_existing",
"size": 303470,
"sha": ""
}
|
# nlp_project_fetch_papers_Till_1.ipynb
Repository: zeynepkorkmaz00/sysbiomed
## Task Force A: Fetching PubMed ID's from queries
<code>
from Bio import Entrez
import csv
</code>
### Step 1: Make function to search for papers and return their PubMED IDs
<code>
def search_pubmed_for_ids(query, max_results=13):
Entrez.email = "zeynep.korkmaz@tum.de" # Set email address
handle = Entrez.esearch(db="pubmed", term=query, retmax=max_results)
record = Entrez.read(handle)
handle.close()
return record["IdList"]
</code>
<code>
# Example query
search_pubmed_for_ids("Helicobacter[Organsim] NOT IBD NOT intestinal microbes")
</code>
### Step 2: Create the keyword and query list from the csv file
<code>
def read_keywords_from_csv(csv_file):
with open(csv_file, 'r') as file:
reader = csv.reader(file)
keywords_dict = {}
current_pub_title = None
current_keywords = []
current_sq_tp = []
current_sq_fp = []
current_sq_r = []
for row in reader:
# Remove trailing commas from each element in the row
row = [item.strip(', ') for item in row]
if row and not row[0].isdigit(): # Skip numeric rows
if row[0] == "Pub Title":
if current_pub_title:
# create for every title/DOI keys
keywords_dict[current_pub_title] = {
"Pub Title": current_pub_title,
"Keywords": current_keywords,
"SQ_TP": current_sq_tp,
"SQ_FP": current_sq_fp,
"SQ_R": current_sq_r
}
current_pub_title = row[1]
current_keywords = []
current_sq_tp = []
current_sq_fp = []
current_sq_r = []
# add values to list of the different keys and check for empty entries
elif row[0] == "Keywords":
current_keywords.extend(item for item in row[1:] if item)
elif row[0] == "SQ_TP":
current_sq_tp.extend(item for item in row[1:] if item)
elif row[0] == "SQ_FP":
current_sq_fp.extend(item for item in row[1:] if item)
elif row[0] == "SQ_R":
current_sq_r.extend(item for item in row[1:] if item)
# Add the last entry
if current_pub_title:
keywords_dict[current_pub_title] = {
"Pub Title": current_pub_title,
"Keywords": current_keywords,
"SQ_TP": current_sq_tp,
"SQ_FP": current_sq_fp,
"SQ_R": current_sq_r
}
return keywords_dict
</code>
##### How the dictionary looks:
<code>
# path to csv file
input_csv = "/Users/tillohlendorf/Documents/MBT/Module/Systems BioMedicine/NLP/TFA_repo/sysbiomed_nlp_project/Keywords/keywords_Till.csv"
</code>
<code>
# create dictionary from csv
keywords_dict = read_keywords_from_csv(input_csv)
</code>
<code>
keywords_dict
</code>
# print dictionary
keywords_dict
##### Other representation of the dictionary for testing and debugging purposes
<code>
for pub_title, data in keywords_dict.items():
print(f"Pub Title: {data['Pub Title']}")
print(f"Keywords: {', '.join(data['Keywords'])}")
print(f"SQ_TP: {', '.join(data['SQ_TP'])}")
print(f"SQ_FP: {', '.join(data['SQ_FP'])}")
print(f"SQ_R: {', '.join(data['SQ_R'])}")
print("\n" + "=" * 80 + "\n") # Separator between entries
</code>
### Step 3: Use dictionary with queries as input and fetch PubMED IDs.
#### Option 1: This option is more comprehensive since it also saves the used queries in the dictionary.
<code>
def dict_to_pubmed_id(input_dict):
# Initialize a new dictionary to store the results
result_dict = {}
# Iterate over each publication entry in the input dictionary
for pub_title, pub_data in input_dict.items():
# Create a copy of the publication data
pub_result = pub_data.copy()
# Initialize empty lists for PubMed IDs for SQ_TP, SQ_FP, and SQ_R
pub_result['PubMed_IDs_TP'] = []
pub_result['PubMed_IDs_FP'] = []
pub_result['PubMed_IDs_R'] = []
# Extract elements from SQ_TP, SQ_FP, and SQ_R lists and search PubMed for IDs
for sq_tp_element in pub_data['SQ_TP']:
pub_result['PubMed_IDs_TP'].extend(search_pubmed_for_ids(sq_tp_element))
for sq_fp_element in pub_data['SQ_FP']:
pub_result['PubMed_IDs_FP'].extend(search_pubmed_for_ids(sq_fp_element))
for sq_r_element in pub_data['SQ_R']:
pub_result['PubMed_IDs_R'].extend(search_pubmed_for_ids(sq_r_element))
# Add the modified publication data to the result dictionary
result_dict[pub_title] = pub_result
return result_dict
</code>
<code>
result_dict = dict_to_pubmed_id(keywords_dict)
result_dict
</code>
#### Option 2: This one only writes the PubMed id's in the dictionary and not the used queries
def dict_to_pubmed_id_reduced(input_dict):
# Initialize a new dictionary to store the results
result_dict = {}
# Iterate over each publication entry in the input dictionary
for pub_title, pub_data in input_dict.items():
# Initialize a dictionary to store PubMed IDs and Pub Title
pub_result = {'Pub Title': pub_title}
# Initialize empty lists for PubMed IDs for SQ_TP, SQ_FP, and SQ_R
pub_result['PubMed_IDs_TP'] = []
pub_result['PubMed_IDs_FP'] = []
pub_result['PubMed_IDs_R'] = []
# Extract elements from SQ_TP, SQ_FP, and SQ_R lists and search PubMed for IDs
for sq_tp_element in pub_data['SQ_TP']:
pub_result['PubMed_IDs_TP'].extend(search_pubmed_for_ids(sq_tp_element))
for sq_fp_element in pub_data['SQ_FP']:
pub_result['PubMed_IDs_FP'].extend(search_pubmed_for_ids(sq_fp_element))
for sq_r_element in pub_data['SQ_R']:
pub_result['PubMed_IDs_R'].extend(search_pubmed_for_ids(sq_r_element))
# Add the modified publication data to the result dictionary
result_dict[pub_title] = pub_result
return result_dict
result2_dict = dict_to_pubmed_id_reduced(keywords_dict)
result2_dict
#### Step 4: export dictionaries as xml and json files
<code>
import json
import xml.etree.ElementTree as ET
</code>
<code>
# Convert to JSON
json_data = json.dumps(result_dict, indent=2)
# Save to a JSON file
with open("output_ID2.json", "w") as json_file:
json_file.write(json_data)
</code>
<code>
# Convert to XML
def save_result_dict_to_xml(result_dict, xml_file_path):
# Create the root element
root = ET.Element("result_dict")
# Iterate over each publication entry in the result dictionary
for pub_title, pub_result in result_dict.items():
entry = ET.SubElement(root, "entry")
ET.SubElement(entry, "PubTitle").text = pub_result['Pub Title']
# Add PubMed IDs for SQ_TP
sq_tp = ET.SubElement(entry, "PubMed_IDs_TP")
for pubmed_id in pub_result['PubMed_IDs_TP']:
ET.SubElement(sq_tp, "PubMed_ID").text = pubmed_id
# Add PubMed IDs for SQ_FP
sq_fp = ET.SubElement(entry, "PubMed_IDs_FP")
for pubmed_id in pub_result['PubMed_IDs_FP']:
ET.SubElement(sq_fp, "PubMed_ID").text = pubmed_id
# Add PubMed IDs for SQ_R
sq_r = ET.SubElement(entry, "PubMed_IDs_R")
for pubmed_id in pub_result['PubMed_IDs_R']:
ET.SubElement(sq_r, "PubMed_ID").text = pubmed_id
# Create the XML tree
xml_tree = ET.ElementTree(root)
# Save the XML tree to the specified file
xml_tree.write(xml_file_path)
</code>
<code>
# Specify the path for the XML file
xml_file_path = "output.xml"
# Save the result_dict to an XML file
save_result_dict_to_xml(result_dict, xml_file_path)
</code>
|
{
"filename": "nlp_project_fetch_papers_Till_1.ipynb",
"repository": "zeynepkorkmaz00/sysbiomed",
"query": "transformed_from_existing",
"size": 108019,
"sha": ""
}
|
# seq_snakemake_RNA-seq_pipeline.ipynb
Repository: jenjane118/rna
# RNA-seq Processing Pipeline
Jennifer Stiens
j.j.stiens@gmail.com
Birkbeck, University of London
## Date: 11-05-23
### Notebook for download, QC and mapping of RNA-seq files
The details of the RNA-seq processing and mapping performed for the WGCNA paper are found in the github repo for the paper:
[WGCNA rna processing doc](https://github.com/jenjane118/mtb_wgcna/blob/master/mtb_wgcna_doc.Rmd)
#### There are two options for each step in the pipeline: using snakemake and associated snakefiles, and the other using command line scripts
For help installing Snakemake:
[snakemake installation conda/mamba](https://snakemake.readthedocs.io/en/stable/getting_started/installation.html)
<code>
#install snakemake (install mamba first or install mamba inside conda)
conda activate base
mamba create -c conda-forge -c bioconda -n snakemake snakemake
mamba activate snakemake
mamba install -c bioconda bwa samtools fastqc multiqc fastp rseqc sra-tools deeptools
</code>
My snakemake files are found at https://github.com/jenjane118/rna_seq_snakemake. Copy these into your own snakemake folder.
Directory structure to show snakemake scripts
```
├── README.md
├── bam_coverage
│ └── snakefile.smk
├── bowtie2
│ └── snakefile.smk
├── dir_tree.txt
├── fastp
│ ├── pe
│ │ └── snakefile.smk
│ └── single
│ └── snakefile.smk
├── fastqc
│ └── pe
│ └── snakefile.smk
├── map_bwa
│ ├── pe
│ │ └── snakefile.smk
│ └── single
│ └── snakefile.smk
├── mbovis_wgs.ipynb
├── rna_seq_nb.ipynb
├── sra
│ ├── pe
│ │ └── snakefile.smk
│ └── single
│ └── snakefile.smk
└── tree_out.txt
14 directories, 14 files
```
## Download files from SRA to Birkbeck server
This uses SRA tools which is installed on thoth /s/software/modules
You may want to create a directory 'ncbi' or use the project name or something like this for your fastq files. Run the snakefile or shell script below from inside this directory.
<code>
module load ncbi-sra/v2.10.5 #(in /s/software/modules)
cd ncbi/<dataset_name>
#make shell script to iterate through accession numbers (iterate_fasterq.sh)
#!/bin/bash
while IFS= read -r line;
do
echo "accession number: $line"
#call fasterq to download from sra
fasterq-dump ${line} -O files/
echo -e "########################\n\n"
done < "$1"
# to run program in background:
nohup bash iterate_fasterq.sh accession_list.txt &> fasterq_dump.out &
</code>
<code>
# if using snakemake
# depends whether single or paired-end to choose which script
# make a directory for the dataset and move to that directory
mkdir $my_path/mtb_rna/PRJNA838962
cd $my_path/mtb_rna/PRJNA838962
#make config.yaml file in directory including something like the following line to indicate accessions:
#accession: [SRR21026195,SRR21026196,SRR21026197,SRR21026198,SRR21026199,SRR21026200]
conda activate snakemake
module load ncbi-sra/v2.10.5
#dry run
snakemake -np -s $my_path/snakemake/sra/pe/snakefile.smk
#run in background
nohup snakemake --cores 8 -s $my_path/snakemake/sra/pe/snakefile.smk > nohup.out 2>&1 &
</code>
After determining fastq files have been downloaded for the desired accession numbers, perform some sanity checks to look for discrepencies in number of reads (between paired end files) and for appearance and read length. The files will be in compressed form and there is no need to decompress them at this time. (The line count is included in snakemake script)
<code>
#Sanity checks
#1) Check for read length
head -50 <file.fastq.gz>
#2) Count number of reads: R1 and R2 should match
zcat <file.fastq.gz> | wc -l
# or loop through and count reads:
FILES=`ls *.fastq.gz`
for file in $FILES; do zcat $file | wc -l; done
#or (for uncompressed files)
find . -name '*.fastq' -exec wc -l {} +
</code>
Choose what quality control program(s) to use. FastQC and Fastp equally useful for qc, but fastp trims at the same time. I prefer this
To run fastQC on directory of fastq files, create the following bash script and run:
<code>
#!/bin/bash
# iterate_fastqc.sh
# usage: bash iterate_fastqc.sh
FILES=*.fastq
for file in $FILES
do
filename=$(basename "$file")
filename="${filename%.*}"
echo "File on the loop: $filename"
#call fastQC quality analysis
/s/software/fastqc/v0.11.8/FastQC/fastqc ${file}
echo -e "########################\n\n"
done
# Run MultiQC
# -f overwrites existing files, . runs with files in current directory, -o output directory
echo "Running MultiQC..."
# Moves output into new folder
mkdir ./fast_QC_outputs
mv *fastqc.zip ./fast_QC_outputs
mv *fastqc.html ./fast_QC_outputs
# Run multiqc to compile outputs
cd fast_QC_outputs
multiqc -f .
</code>
<code>
module load python/v3
module load fastqc
bash iterate_fastqc.sh
</code>
<code>
# with snakemake
cd $my_path/<dataset_dir>
conda activate snakemake
#dry run (use appropriate single/pe file depending on data)
snakemake -np -s $my_path/snakemake/fastqc/pe/snakefile.smk
#run in background
nohup snakemake --cores 8 -s $my_path/snakemake/fastqc/pe/snakefile.smk > nohup.out 2>&1 &
</code>
In the WGCNA paper, we then used trimmomatic to trim the adapters. I don't think this matters as long as good mapping stats.
<code>
#The following is a script to run trimmomatic on single end samples:
#!/bin/bash
# iterate_trimmomatic.sh
# Runs Trimmomatic in PE mode for all sample names given as arguments
# Run as:
# nohup bash $my_path/scripts/iterate_trimmomatic.sh PRJNA488546
timestamp=`date "+%Y%m%d-%H%M%S"`
logfile="run_$timestamp.log"
exec > $logfile 2>&1 #all output will be logged to logfile
TRIM_EXEC="/s/software/trimmomatic/Trimmomatic-0.38/trimmomatic-0.38.jar"
DIR=$1
shift
echo "Running Trimmomatic using executable: $TRIM_EXEC"
for file in `ls $DIR/*.fastq.gz` ;
do
echo "File on Loop: ${file}"
sample=${file/$DIR\/}
sample=${sample/.fastq.gz/}
echo "Sample= $sample"
java -jar $TRIM_EXEC SE -threads 12 -phred33 \
-trimlog "$sample"_trim_report.txt \
"$DIR/$sample".fastq.gz "$sample"_trimmed.fastq.gz \
ILLUMINACLIP:/s/software/trimmomatic/Trimmomatic-0.38/adapters/TruSeq3-PE.fa:2:30:10 \
LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36
gzip "$sample"_trim_report.txt
done
</code>
<code>
module load trimmomatic
cd $my_path/ncbi/files/<dataset_dir>
nohup bash $my_path/scripts/iterate_trimmomatic.sh PRJNA488546 >& iterate_trim.out &
</code>
Lately I have switched to fastp to trim and do quality control. It trims adapters and automatically detects the adapter sequences by default. If there are remaining adapters, you can specify adapter sequences.
[fastp docs](https://github.com/OpenGene/fastp)
<code>
# paired end, gzip compressed, R1 is read1, R2 is read2 of paired end
mkdir trimmed
#fastp is not yet on thoth server--need to ask Dave to install this, or can use in conda env. But the code below is representative of how to use it.
module load fastp
fastp -i <sample_name.R1.fastq.gz> -I <sample_name.R2.fastq.gz> -o <trimmed_reads/<sample_name>_trimmed.R1.fastq.gz> -O <trimmed_reads/<sample_name>_trimmed.R2.fastq.gz>
</code>
<code>
#with snakemake
cd $my_path/<dataset_dir>
conda activate snakemake
#dry run (use appropriate single/pe file depending on data)
snakemake -np -s $my_path/snakemake/fastp/pe/snakefile.smk
#run in background
nohup snakemake --cores 8 -s $my_path/snakemake/fastp/pe/snakefile.smk > nohup.out 2>&1 &
</code>
Map the trimmed reads with BWA-mem
<code>
module load bwa
module load samtools
#create index file for genome in same directory as genome file
bwa index AL123456_3.fasta
# using shell script from Yen-Yi (should check that filenames and paths correct)
#!/bin/bash
# Runs bwa in paired-end mode, sorts and indexes files
# Run as:
# nohup sh BWA_PE.sh directory_of_fastq_files samples
timestamp=`date "+%Y%m%d-%H%M%S"`
logfile="run_$timestamp.log"
exec > $logfile 2>&1 #all output will be logged to logfile
dir=$1
shift
#set location of executables
SAMTOOLS_EXEC=<PATH TO EXEC>
#set parameters
genomeFile=<GENOME_FILE> #index files should be in same directory
numProc=8
#extension for fastq files
suffix1="<sample_name>_trimmed.R1.fastq.gz"
suffix2="<sample_name>_trimmed.R1.fastq.gz"
EXT=fastq.gz
for sample in *.${EXT};
do
sample=$(echo $sample | cut -f 1 -d '_')
echo "Running bwa on sample $sample (paired-end mode)..."
pairedFile1="$dir$sample$suffix1".gz
if [ -f $pairedFile1 ]
then
gzip -d $pairedFile1
pairedFile1=$dir$sample$suffix1
else
pairedFile1=$dir$sample$suffix1
if [ ! -f $pairedFile1 ]
then
echo "File not found: $pairedFile1"
exit $?
fi
fi
pairedFile2="$dir$sample$suffix2".gz
if [ -f $pairedFile2 ]
then
gzip -d $pairedFile2
pairedFile2=$dir$sample$suffix2
else
pairedFile2=$dir$sample$suffix2
if [ ! -f $pairedFile2 ]
then
echo "File not found: $pairedFile2"
exit $?
fi
fi
tmpSam="$sample"_pe.sam
tmpBam="$sample"_pe.bam
finalSortedBam="$sample"_sorted.bam
#align
$BWA_EXEC mem -t $numProc $genomeFile $pairedFile1 $pairedFile2 > $tmpSam
#create bam file
$SAMTOOLS_EXEC view $tmpSam -Sbo $tmpBam
$SAMTOOLS_EXEC sort $tmpBam -o $finalSortedBam
$SAMTOOLS_EXEC index $finalSortedBam
#cleanup
/bin/rm $tmpSam $tmpBam
gzip -9 $pairedFile1 $pairedFile2
done
</code>
<code>
#Mapping output quality check script
module load samtools
#!/bin/bash
timestamp=`date "+%Y%m%d-%H%M%S"`
logfile="run_$timestamp.log"
exec > $logfile 2>&1 #all output will be logged to logfile
dir=$1
shift
EXT=bam
#ref_genome="<genome_file/ref_genomic.bed>" genome bedfile not used?
SUFFIX="_sorted.bam"
for sample in *.${EXT};
do
sample=$(echo $sample | cut -f 1 -d '_')
echo "Running mapping quality scripts on sample $sample..."
echo "sample is $sample"
quality_check=$dir$sample$SUFFIX
samtools flagstat $quality_check > "flagstat_$sample.txt"
echo "Mapping output quality check for $sample done..."
done
mkdir flagstat_ouput
mv *flagstat* flagstat_output
multiqc ./
</code>
<code>
# with snakemake (maps, sorts, indexes and creates flagstats report)
cd $my_path/<dataset_dir>
conda activate snakemake
snakemake -np -s $my_path/snakemake/map_bwa/pe/snakefile.smk
nohup snakemake --cores 8 -s $my_path/snakemake/map_bwa/pe/snakefile.smk > nohup_map.out 2>&1 &
</code>
## It is useful to have bam coverage files to use with IGV
<code>
module load python/v3
#makes a separate bigwig file for forward and reverse strands
bamCoverage -b sorted_reads/{sample}.bam -o covg_bigwigs/{sample}_fwd.bw -of bigwig --filterRNAstrand forward -p 8 --binSize 1 --extendReads
bamCoverage -b sorted_reads/{sample}.bam -o covg_bigwigs/{sample}_rev.bw -of bigwig --filterRNAstrand reverse -p 8 --binSize 1 --extendReads
</code>
<code>
#with snakemake
cd <dataset_dir>
snakemake -np -s $my_path/snakemake/bam_coverage/snakefile.smk
snakemake --cores 3 -s $my_path/snakemake/bam_coverage/snakefile.smk
</code>
|
{
"filename": "seq_snakemake_RNA-seq_pipeline.ipynb",
"repository": "jenjane118/rna",
"query": "transformed_from_existing",
"size": 20954,
"sha": ""
}
|
# Retrieval_Project_Notebook_1.ipynb
Repository: h1den96/Information
# Εισαγωγή των απαραίτητων βιβλιοθηκών
<code>
# ============================================================
# 1) Εγκατάσταση και εισαγωγή βιβλιοθηκών
# ============================================================
# Εισαγωγή απαραίτητων βιβλιοθηκών
import nltk
import re
import json
import math
import numpy as np
from collections import defaultdict
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from nltk.stem import PorterStemmer, WordNetLemmatizer
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
</code>
# Αρχικοποίηση NLTK
<code>
# Λήψη απαραίτητων δεδομένων NLTK (tokenizer, stopwords κ.λπ.)
nltk.download('punkt', force=True)
nltk.download('stopwords', force=True)
nltk.download('wordnet', force=True)
# Αρχικοποίηση NLTK components
stop_words = set(stopwords.words('english'))
stemmer = PorterStemmer()
lemmatizer = WordNetLemmatizer()
print("Setup complete.")
</code>
# Φόρτωση αρχείων
Σε αυτή την ενότητα ορίζουμε συναρτήσεις για να φορτώνουμε τα αρχεία JSON (με άρθρα/τίτλους) και το αρχείο CISI.QRY (με queries), έπειτα τα καλούμε για να φορτώσουμε τα δεδομένα μας.
Σημειώστε ότι αναμένεται να υπάρχουν τα αρχεία:
./wikipedia_articles.json (τα δεδομένα εισόδου που συλλέκτηκαν απο το web crawling στην βικιπαίδεια)
./processed_CISI_articles.json (περιέχει μια λίστα άρθρων, όπου το κάθε άρθρο έχει id και μια λίστα από tokens)
./CISI_articles.json (το πρωτότυπο αρχείο με πλήρη δεδομένα ή τουλάχιστον τα id και title)
./CISI.QRY (τα queries σε φορμά .I <id>, .W, κείμενο ...)
<code>
# ============================================================
# 2) Φόρτωση άρθρων, τίτλων, ερωτημάτων
# ============================================================
def load_articles(json_file):
"""
Φορτώνει από JSON αρχείο μια λίστα από άρθρα,
όπου κάθε άρθρο είναι λεξικό με κλειδιά:
"id", "tokens", πιθανώς και άλλα πεδία.
"""
try:
with open(json_file, 'r', encoding='utf-8') as file:
return json.load(file)
except FileNotFoundError:
print(f"Σφάλμα: Το αρχείο '{json_file}' δεν βρέθηκε.")
return []
except json.JSONDecodeError:
print(f"Σφάλμα: Μη έγκυρο JSON στο '{json_file}'.")
return []
def load_titles(json_file):
"""
Φορτώνει από JSON αρχείο μια λίστα articles και φτιάχνει
ένα λεξικό {article_id: article_title}.
"""
try:
with open(json_file, 'r', encoding='utf-8') as file:
articles = json.load(file)
return {article['id']: article['title'] for article in articles}
except FileNotFoundError:
print(f"Σφάλμα: Το αρχείο '{json_file}' δεν βρέθηκε.")
return {}
except json.JSONDecodeError:
print(f"Σφάλμα: Μη έγκυρο JSON στο '{json_file}'.")
return {}
def load_queries(file_path):
"""
Διαβάζει από αρχείο (π.χ. CISI.QRY) τα queries σε φορμά:
.I <query_id>
.W
<query text...>
και επιστρέφει ένα λεξικό { query_id: query_text }.
"""
queries = {}
current_id = None
query_text = []
with open(file_path, 'r', encoding='utf-8') as file:
for line in file:
line = line.strip()
if line.startswith('.I'):
if current_id is not None:
queries[current_id] = " ".join(query_text).strip()
current_id = int(line.split()[1])
query_text = []
elif line.startswith('.W'):
continue
else:
query_text.append(line)
if current_id is not None:
queries[current_id] = " ".join(query_text).strip()
return queries
# -----------------------------------------------------------
# Τώρα καλούμε τις συναρτήσεις για να φορτώσουμε τα αρχεία μας
# -----------------------------------------------------------
articles_file = './processed_CISI_articles.json'
titles_file = './CISI_articles.json'
queries_file = './CISI.QRY'
articles = load_articles(articles_file)
title_mapping = load_titles(titles_file)
queries = load_queries(queries_file)
print(f"Φορτώθηκαν {len(articles)} άρθρα.")
print(f"Φορτώθηκαν {len(title_mapping)} τίτλοι άρθρων.")
print(f"Φορτώθηκαν {len(queries)} queries.")
</code>
Σε αυτή την ενότητα δημιουργούμε μια συνάρτηση που κάνει preprocessing (επεξεργασία κειμένου) — αφαιρεί μη αλφαβητικούς χαρακτήρες, κάνει tokenize, αφαιρεί stopwords, εφαρμόζει stemming και lemmatization
<code>
# ============================================================
# 3) Συνάρτηση προεπεξεργασίας κειμένου
# ============================================================
nltk.download('punkt_tab')
def process_query(text):
"""
Βήματα:
1) Αφαίρεση μη αλφαβητικών χαρακτήρων
2) Tokenize
3) Lowercase
4) Αφαίρεση stopwords
5) Stemming
6) Lemmatization
"""
cleaned_text = re.sub(r'[^A-Za-z\s]', '', text)
tokens = word_tokenize(cleaned_text.lower())
filtered_tokens = [w for w in tokens if w not in stop_words]
stemmed_tokens = [stemmer.stem(w) for w in filtered_tokens]
lemmatized_tokens = [lemmatizer.lemmatize(w) for w in stemmed_tokens]
return lemmatized_tokens
# -----------------------------------------------------------
# Δοκιμή της συνάρτησης
# -----------------------------------------------------------
sample_text = "Information retrieval is one of the most important subjects!"
processed_tokens = process_query(sample_text)
print("Original text: ", sample_text)
print("Processed tokens: ", processed_tokens)
</code>
# Inverted Index Implementation
Εδώ φτιάχνουμε το inverted index, που είναι ένα λεξικό:
token -> [doc_id1, doc_id2, ...],
για να γίνει συσχέτιση των όρων και σε ποια documents ανήκουν
<code>
# ============================================================
# 4) Υλοποίηση Inverted Index για Boolean Search
# ============================================================
def buildInvertedIndex(articles):
"""
Επιστρέφει ένα λεξικό { token: [doc_id1, doc_id2, ...] }.
"""
inverted_index = defaultdict(list)
for article in articles:
for token in set(article["tokens"]):
inverted_index[token].append(article["id"])
return inverted_index
def searchIndex(term, inverted_index):
"""
Επιστρέφει ένα σύνολο (set) με τα doc_ids για τον όρο 'term'.
Αν δεν υπάρχει ο όρος, επιστρέφει κενό set.
"""
return set(inverted_index.get(term, []))
# -----------------------------------------------------------
# Δημιουργία του inverted_index
# -----------------------------------------------------------
inverted_index = buildInvertedIndex(articles)
print("Inverted index built.")
# -----------------------------------------------------------
# Δοκιμή στο πρώτο token του sample_text
# -----------------------------------------------------------
if processed_tokens:
sample_term = processed_tokens[0]
matching_docs = searchIndex(sample_term, inverted_index)
print(f"Documents containing '{sample_term}': {list(matching_docs)} ...")
</code>
# Boolean Search Implementation
Τώρα χτίζουμε τη λογική για Boolean Expressions τύπου information AND system, information OR system, information NOT system κ.λπ. Κάνουμε parsing της έκφρασης και χρησιμοποιούμε σύνολα για AND/OR/NOT.
<code>
# ============================================================
# 5) Boolean Search
# ============================================================
def evaluate_expression(expression, articles):
"""
Υλοποιεί τη Boolean αναζήτηση που υποστηρίζει AND, OR, NOT.
Κάνει tokenize στην έκφραση, και χρησιμοποιεί μια στοίβα για
αξιολόγηση (βασική postfix/stack λογική).
"""
stack = []
tokens = expression.split()
for token in tokens:
token_up = token.upper()
if token_up in {"AND", "OR", "NOT"}:
stack.append(token_up)
else:
matching = searchIndex(token, articles)
stack.append(matching)
# Όταν έχουμε σχήμα [SET, 'AND/OR/NOT', SET] το αξιολογούμε
while len(stack) >= 3 and isinstance(stack[-1], set) and isinstance(stack[-3], set):
right = stack.pop()
operator = stack.pop()
left = stack.pop()
if operator == "AND":
stack.append(left & right)
elif operator == "OR":
stack.append(left | right)
elif operator == "NOT":
stack.append(left - right)
if len(stack) == 1 and isinstance(stack[0], set):
return stack[0]
else:
return set()
def boolean_search(query_text, articles):
"""
Επιστρέφει μια λίστα από doc_ids που ταιριάζουν στη Boolean έκφραση.
Σε περίπτωση που αποτύχει η parsing (λχ λάθος syntax), πέφτει στο "fallback OR".
"""
query_text = query_text.strip()
try:
result_set = evaluate_expression(query_text, articles)
if not result_set:
raise ValueError("Empty result from Boolean")
return list(result_set), []
except:
# fallback: αν δεν δουλέψει η έκφραση, κάνουμε OR σε όλα τα tokens
processed = process_query(query_text)
results = set()
for t in processed:
results.update(searchIndex(t, articles))
return list(results), []
# -----------------------------------------------------------
# Δοκιμή Boolean Search
# -----------------------------------------------------------
test_query = "information"
results_boolean = boolean_search(test_query, inverted_index)
print(f"Results for query '{test_query}': {results_boolean} ...")
test_query = "system"
results_boolean = boolean_search(test_query, inverted_index)
print(f"Results for query '{test_query}': {results_boolean} ...")
test_query = "information AND system"
results_boolean = boolean_search(test_query, inverted_index)
print(f"Results for query '{test_query}': {results_boolean} ...")
</code>
# TF-IDF Implementation (Dot Product)
Τώρα θα υλοποιήσουμε μια απλή TF-IDF προσέγγιση, χρησιμοποιώντας TfidfVectorizer από το scikit-learn. Υπολογίζουμε τον πίνακα TF-IDF για όλα τα άρθρα και κάνουμε dot-product με το query vector.
<code>
# ============================================================
# 6) TF-IDF (dot product)
# ============================================================
def rank_tfidf(query_tokens, articles, inverted_index):
# 1) Βρίσκουμε τα σχετικά έγγραφα (doc_ids) από το inverted index
relevant_doc_ids = set()
for token in query_tokens:
relevant_doc_ids.update(inverted_index.get(token, []))
# 2) Φτιάχνουμε ένα υποσύνολο των άρθρων
relevant_docs = [a for a in articles if a['id'] in relevant_doc_ids]
# 3) Δημιουργούμε το document-term matrix (TF-IDF) για αυτά τα docs
corpus = [" ".join(a['tokens']) for a in relevant_docs]
vectorizer = TfidfVectorizer()
tfidf_matrix = vectorizer.fit_transform(corpus)
# 4) Μετατρέπουμε το query σε vector μέσω του ίδιου vectorizer
query_vec = vectorizer.transform([" ".join(query_tokens)])
# 5) Υπολογίζουμε το dot product (tfidf_matrix * query_vec)
scores = (tfidf_matrix @ query_vec.T).toarray().flatten()
# Το πολλαπλασιάζουμε επί 100 για κλίμακα 0-100
scores *= 100
# 6) Ταξινομούμε φθίνουσα
ranked_indices = np.argsort(-scores)
# Εξάγουμε doc_ids και scores
ranked_docs = [relevant_docs[i]['id'] for i in ranked_indices]
ranked_scores = [scores[i] for i in ranked_indices]
return ranked_docs, ranked_scores
# -----------------------------------------------------------
# Δοκιμή
# -----------------------------------------------------------
query_text = "information retrieval systems"
test_query_tokens = process_query(query_text)
ranked_indices_tfidf, scores_tfidf = rank_tfidf(test_query_tokens, articles, inverted_index) # Added inverted_index as an argument
print(f"Top 5 document indices using TF-IDF for query: {query_text}")
for i in ranked_indices_tfidf[:5]:
# Βρίσκουμε το index του άρθρου που αντιστοιχεί στο doc ID
article_index = next((index for index, article in enumerate(articles) if article['id'] == i), None)
if article_index is not None: # Αν βρέθηκε το άρθρο
print(f"Doc index = {i}, Score = {scores_tfidf[ranked_indices_tfidf.index(i)]:.2f}, ID = {articles[article_index]['id']}")
else:
print(f"Article with ID {i} not found in the articles list.")
</code>
# BM25 Implementation
Η BM25 είναι μια πιο εξελιγμένη μέθοδος που στηρίζεται σε TF, IDF και μήκος εγγράφου. Παρακάτω την υλοποιούμε σε δύο βήματα:
1. calc_idf για BM25
2. BM25 συνάρτηση που υπολογίζει το score κάθε εγγράφου
<code>
# ---------------------------------------------------------
# BM25
# ---------------------------------------------------------
"""
Η μέθοδος BM25 είναι ένας αλγόριθμος ranking που βασίζεται σε TF, IDF
και σε δύο παραμέτρους (k1 και b). Υπολογίζει για κάθε όρο q και έγγραφο d:
score(d) += IDF(q) * ( (TF(q,d)*(k1+1)) / (TF(q,d) + k1*(1 - b + b*(|d|/avg|d|))) )
"""
def calc_idf(articles):
# Υπολογισμός idf για BM25: log( (N - df + 0.5)/(df + 0.5) + 1 )
N = len(articles)
term_doc_count = defaultdict(int)
for article in articles:
unique_tokens = set(article['tokens'])
for token in unique_tokens:
term_doc_count[token] += 1
idf = {}
for token, doc_count in term_doc_count.items():
idf[token] = math.log((N - doc_count + 0.5) / (doc_count + 0.5) + 1)
return idf
def rank_bm25(query_tokens, articles, idf, inverted_index):
"""
Βρίσκουμε πάλι μόνο τα σχετικά έγγραφα από το inverted index, κι
έπειτα υπολογίζουμε το BM25 score για το καθένα βάσει TF, IDF, k1, b.
"""
k1 = 2.0
b = 0.5
N = len(articles)
avg_len = sum(len(a['tokens']) for a in articles) / N
# 1) σχετικά doc_ids
relevant_doc_ids = set()
for token in query_tokens:
relevant_doc_ids.update(inverted_index.get(token, []))
# 2) Φτιάχνουμε τη λίστα των relevant_docs
relevant_docs = [a for a in articles if a['id'] in relevant_doc_ids]
# 3) Υπολογισμός BM25 score
scores = []
for a in relevant_docs:
freq = defaultdict(int)
for t in a['tokens']:
freq[t] += 1
score = 0
for q in query_tokens:
if q in idf:
tf = freq[q]
numerator = tf * (k1 + 1)
denominator = tf + k1 * (1 - b + b * (len(a['tokens']) / avg_len))
score += idf[q] * (numerator / denominator)
scores.append(score)
scores = np.array(scores)
# Ταξινόμηση
ranked_indices = np.argsort(-scores)
ranked_docs = [relevant_docs[i]['id'] for i in ranked_indices]
ranked_scores = [scores[i] for i in ranked_indices]
return ranked_docs, ranked_scores
# -----------------------------------------------------------
# Υπολογίζουμε τα idf_values μια φορά
# -----------------------------------------------------------
idf_values = calc_idf(articles)
# -----------------------------------------------------------
# Δοκιμή BM25
# -----------------------------------------------------------
ranked_indices_bm25, scores_bm25 = rank_bm25(test_query_tokens, articles, idf_values, inverted_index)
print(f"Top 5 document indices using BM25 for query: {query_text}")
for doc_index in ranked_indices_bm25[:5]:
article_index = next((index for index, article in enumerate(articles) if article['id'] == doc_index), None)
if article_index is not None:
print(f"Doc index = {doc_index}, Score = {scores_bm25[ranked_indices_bm25.index(doc_index)]:.2f}, ID = {articles[article_index]['id']}")
else:
print(f"Article with ID {doc_index} not found in the articles list.")
</code>
# Vector Space Model
Στο μοντέλο διανυσματικού χώρου (VSM), κάθε έγγραφο ή ερώτημα είναι ένα διάνυσμα Ν-διαστάσεων, όπου Ν είναι ο αριθμός των διαφορετικών όρων σε όλα τα έγγραφα και τα ερωτήματα.Ο i-οστός δείκτης ενός διανύσματος περιέχει τη βαθμολογία του i-οστού όρου για το συγκεκριμένο διάνυσμα. Χρησιμοποιούνται TF-IDF και Cosine Similarity.
<code>
# ============================================================
# TF-IDF + Cosine Similarity
# ============================================================
def rank_vsm(query_tokens, articles, inverted_index):
"""
Υλοποιεί αναζήτηση βασισμένη σε TF-IDF και Cosine Similarity,
μόνο για έγγραφα που περιέχουν τουλάχιστον έναν από τους όρους του query.
Βήματα:
1) Εύρεση συναφών εγγράφων (relevant_docs) μέσω του inverted_index.
2) Δημιουργία TF-IDF matrix μόνο για αυτά τα έγγραφα.
3) Δημιουργία vector για το query.
4) Υπολογισμός cosine_similarity.
5) Κανονικοποίηση των scores σε 0..100.
6) Επιστροφή ταξινομημένης λίστας doc_ids και των αντίστοιχων scores.
"""
# 1) Εύρεση relevant_doc_ids
relevant_doc_ids = set()
for token in query_tokens:
relevant_doc_ids.update(inverted_index.get(token, []))
# 2) Φιλτράρουμε τα articles για να κρατήσουμε μόνο τα σχετικά
relevant_docs = [a for a in articles if a['id'] in relevant_doc_ids]
# 3) Δημιουργούμε ένα corpus για το TfidfVectorizer
corpus = [" ".join(a['tokens']) for a in relevant_docs]
# Check if the corpus is empty after preprocessing
if not any(corpus):
print("Warning: Corpus is empty after preprocessing. Returning empty results.")
return [], [] # Return empty lists to indicate no results
vectorizer = TfidfVectorizer()
tfidf_matrix = vectorizer.fit_transform(corpus)
# 4) Φτιάχνουμε το query vector
query_vec = vectorizer.transform([" ".join(query_tokens)])
# 5) Υπολογίζουμε cosine similarity
cos_sims = cosine_similarity(query_vec, tfidf_matrix).flatten()
# 6) Κλίμακα 0..100
cos_sims *= 100
# 7) Ταξινόμηση φθίνουσα
ranked_indices = np.argsort(-cos_sims)
ranked_docs = [relevant_docs[i]['id'] for i in ranked_indices]
ranked_scores = [cos_sims[i] for i in ranked_indices]
return ranked_docs, ranked_scores
test_query_tokens = ["quick", "history"]
docs_vsm, scores_vsm = rank_vsm(test_query_tokens, articles, inverted_index)
print("Top 5 Docs (TF-IDF + Cosine):", docs_vsm[:5])
print("Scores:", scores_vsm)
</code>
# Συνάρτηση Ranking
Σε αυτή τη φάση, ενώνουμε τις διάφορες μεθόδους (Boolean, TF-IDF, BM25, TF-IDF+Cosine) σε μία κεντρική συνάρτηση ranking
<code>
# ============================================================
# Συνάρτηση Ranking (με 4 methods)
# ============================================================
def ranking(articles, query_text, method, inverted_index):
"""
Ανάλογα με το 'method':
'1' -> Boolean
'2' -> TF-IDF (dot product)
'3' -> BM25
'4' -> TF-IDF + Cosine
"""
# Προεπεξεργασία του query
processed_query = process_query(query_text)
if method == '1':
# Boolean
docs, _ = boolean_search(query_text, inverted_index)
return docs, []
elif method == '2':
# TF-IDF (dot product)
# π.χ. θα χρησιμοποιήσουμε rank_tfidf_vectorizer
ranked_indices, sc = rank_tfidf(processed_query, articles, inverted_index)
# ranked_indices είναι λίστα από doc_ids, όχι indices
#doc_ids = [articles[i]['id'] for i in ranked_indices] # This line is removed
doc_ids = ranked_indices # ranked_indices already contain doc_ids
scores_ordered = [sc[i] for i in range(len(ranked_indices))] # Using a range of indices
return doc_ids, scores_ordered
elif method == '3':
# BM25
idf_vals = calc_idf(articles)
doc_ids, scores_ordered = rank_bm25(processed_query, articles, idf_vals, inverted_index) # Added inverted_index
return doc_ids, scores_ordered
elif method == '4':
# TF-IDF + Cosine
doc_ids, scores_ordered = rank_vsm(processed_query, articles, inverted_index) # Added inverted_index, removed relevant_only
return doc_ids, scores_ordered
else:
print("Μη έγκυρη μέθοδος.")
return [], []
# -----------------------------------------------------------
# Δοκιμή της ranking() συνάρτησης
# -----------------------------------------------------------
test_query2 = "information science"
for m in ['1','2','3','4']:
doc_list, scores = ranking(articles, test_query2, m, inverted_index)
print(f"Method={m}, first 5 results:")
for i in range(min(5, len(doc_list))):
did = doc_list[i]
sc = scores[i] if scores else 0
print(f" DocID={did}, Score={sc:.2f}")
</code>
# main_loop (Interactive ή One-shot)
Εδώ έχουμε τη συνάρτηση που, αν use='1', καλείται σε one-shot mode (π.χ. για αυτόματες κλήσεις όπως ground truth). Αλλιώς (default) μπαίνει σε interactive CLI mode.
<code>
# ============================================================
# main_loop (διαδραστικό ή one-shot)
# ============================================================
def main_loop(articles, title_mapping, query=None, use='0', method=None):
global _inverted_index
if use == '1':
# one-shot λειτουργία: δεν κάνουμε interactive
_inverted_index = buildInvertedIndex(articles)
doc_ids, scores = ranking(articles, query, method, _inverted_index)
return doc_ids, scores
else:
# interactive
while True:
print("\nMenu:")
print("1) Search")
print("2) Exit")
choice = input("Choice: ").strip()
if choice == '1':
user_query = input("Enter your query: ")
print("Methods:\n1) Boolean\n2) TF-IDF\n3) BM25\n4) TF-IDF + Cosine")
user_method = input("Select (1..4): ").strip()
# Φτιάχνουμε/ανανέουμε το inverted_index πριν το ranking
_inverted_index = buildInvertedIndex(articles)
docs, scores = ranking(articles, user_query, user_method, _inverted_index)
top_k = min(10, len(docs))
for i in range(top_k):
did = docs[i]
sc = scores[i] if scores else 0
title = title_mapping.get(did, "No Title")
print(f"{i+1}. DocID={did}, Score={sc:.2f}, Title={title}")
elif choice == '2':
print("Exiting interactive mode.")
break
return [], []
# -----------------------------------------------------------
# Παράδειγμα: Κλήση main_loop σε One-shot mode
# -----------------------------------------------------------
print("\n=== One-shot example ===")
test_q = "information retrieval system"
doc_ids_test, sc_test = main_loop(articles, title_mapping, query=test_q, use='1', method='2')
print(f"One-shot, method=2, found {len(doc_ids_test)} docs. Top 5 doc IDs:")
print(doc_ids_test[:5])
</code>
# Ground Truth Creation & Parse Relevance
Δημιουργία Ground Truth (CISI.REL)
<code>
# ============================================================
# Ground Truth creation (CISI.REL)
# ============================================================
def ground_truth(articles, title_mapping, queries, method_for_gt='3'):
"""
Δημιουργεί/ενημερώνει το αρχείο CISI.REL με μορφή:
query_id doc_id relevance score
όπου η relevance καθορίζεται από thresholds στο score.
"""
data = []
for qid, qtext in queries.items():
# Καλούμε main_loop σε one-shot mode με τη ζητούμενη μέθοδο
ranked_docs, scores = main_loop(articles, title_mapping, qtext, use='1', method=method_for_gt)
query_data = []
for doc_id, sc in zip(ranked_docs, scores):
# Παράδειγμα thresholding
if sc < 10:
relevance = 0
elif 10 <= sc < 30:
relevance = 1
else:
relevance = 2
query_data.append((qid, doc_id, relevance, sc))
data.extend(query_data)
# Γράφουμε σε CISI.REL
with open("./CISI.REL", "w") as f:
for row in data:
f.write("{:5d} {:5d} {:1d} {:10.4f}\n".format(row[0], row[1], row[2], row[3]))
print("CISI.REL updated successfully.")
# -----------------------------------------------------------
# Παράδειγμα: Δημιουργούμε ground truth με μέθοδο BM25
# -----------------------------------------------------------
print("Creating ground truth (CISI.REL) with method=3 (BM25) ...")
ground_truth(articles, title_mapping, queries, method_for_gt='3')
</code>
Parse Relevance (διαβάζει CISI.REL)
<code>
# ============================================================
# Parse Relevance
# ============================================================
def parse_relevance(file_path):
"""
Διαβάζει το CISI.REL και φτιάχνει ένα λεξικό:
{ query_id: [doc_ids με rel>0] }
"""
relevance_dict = {}
with open(file_path, 'r') as file:
for line in file:
parts = line.strip().split()
if len(parts) >= 3:
qid = int(parts[0])
did = int(parts[1])
rel = int(parts[2])
if qid not in relevance_dict:
relevance_dict[qid] = []
if rel > 0:
relevance_dict[qid].append(did)
return relevance_dict
# -----------------------------------------------------------
# Δοκιμή
# -----------------------------------------------------------
rel_dict = parse_relevance("./CISI.REL")
print(f"Parsed relevance from CISI.REL: found {len(rel_dict)} queries with rel>0 info.")
</code>
# Evaluate Search Engine (Precision, Recall, F1)
Τέλος, δείχνουμε πώς να κάνουμε evaluation χρησιμοποιώντας το ground truth που δημιουργήσαμε:
<code>
# ============================================================
# Evaluate Search Engine
# ============================================================
def eval_search_engine(queries, ground_truth_dict, articles, title_mapping):
"""
Κάνει Evaluation με precision, recall, F1.
Ζητάει μέθοδο (1..4), μετά για κάθε query:
- τρέχει main_loop (one-shot)
- υπολογίζει tp, fp, fn
"""
print("\nSelect Ranking Method for Evaluation:")
print("1) Boolean Search")
print("2) TF-IDF (dot product)")
print("3) Okapi BM25")
print("4) TF-IDF + Cosine Similarity")
method_choice = input("Enter your choice (1/2/3/4): ").strip()
precision_scores = []
recall_scores = []
f1_scores = []
for qid, qtext in queries.items():
print(f"\nEvaluating Query ID {qid}: {qtext}")
doc_ids, _scores = main_loop(articles, title_mapping, qtext, use='1', method=method_choice)
retrieved_docs = set(doc_ids)
relevant_docs = set(ground_truth_dict.get(qid, []))
tp = len(retrieved_docs & relevant_docs)
fp = len(retrieved_docs - relevant_docs)
fn = len(relevant_docs - retrieved_docs)
precision = tp/(tp+fp) if (tp+fp) > 0 else 0
recall = tp/(tp+fn) if (tp+fn) > 0 else 0
f1 = 2*precision*recall/(precision+recall) if (precision+recall) > 0 else 0
precision_scores.append(precision)
recall_scores.append(recall)
f1_scores.append(f1)
print(f"Precision={precision:.2f}, Recall={recall:.2f}, F1={f1:.2f}")
avg_p = sum(precision_scores)/len(precision_scores) if precision_scores else 0
avg_r = sum(recall_scores)/len(recall_scores) if recall_scores else 0
avg_f1 = sum(f1_scores)/len(f1_scores) if f1_scores else 0
print("\nOverall Performance:")
print(f"Avg Precision: {avg_p:.2f}")
print(f"Avg Recall: {avg_r:.2f}")
print(f"Avg F1-Score: {avg_f1:.2f}")
# -----------------------------------------------------------
# Δοκιμή evaluation
# -----------------------------------------------------------
print("\n=== Evaluate Search Engine ===")
ground_truth_dict = parse_relevance("./CISI.REL")
eval_search_engine(queries, ground_truth_dict, articles, title_mapping)
</code>
|
{
"filename": "Retrieval_Project_Notebook_1.ipynb",
"repository": "h1den96/Information",
"query": "transformed_from_existing",
"size": 85359,
"sha": ""
}
|
# TSP.ipynb
Repository: ecervera/ga-nb
# The Travelling Salesperson Problem
This notebook has been adapted from [a Pyevolve example](http://pyevolve.sourceforge.net/0_6rc1/examples.html#example-12-the-travelling-salesman-problem-tsp).
The [travelling salesperson problem (TSP)](http://en.wikipedia.org/wiki/Travelling_salesman_problem) is an NP-hard problem in combinatorial optimization studied in operations research and theoretical computer science. Given a list of cities and their pairwise distances, the task is to find the shortest possible route that visits each city exactly once and returns to the origin city. It is a special case of the travelling purchaser problem.
[<img src="img/travelling_salesman_problem.jpg" align="right" width=360>](http://en.wikipedia.org/wiki/Travelling_salesman_problem)
The code below shows the use of Pyevolve to solve the TSP. Images of the intermediate and final solutions are stored in the 'tspimg' folder.
Your tasks are:
1. Create the 'tspimg' folder for storing the images.
2. Add the necessary statements for storing the results in a database named 'tsp.db' with identifier 'ex1'.
3. For the maximum grade: modify the code to solve the problem with the [ATT 48 dataset](att48.tsp), a set of 48 cities (US state capitals) from [TSPLIB](http://elib.zib.de/pub/mp-testdata/tsp/tsplib/tsplib.html). Store the results in a database named 'tsp_att48.db' with identifier 'ex1'. For your information, [the optimal cost is 10628](http://comopt.ifi.uni-heidelberg.de/software/TSPLIB95/STSP.html).
<code>
from pyevolve import G1DList
from pyevolve import GSimpleGA
from pyevolve import Crossovers
from pyevolve import Consts
import random
from math import sqrt
from PIL import Image, ImageDraw, ImageFont
</code>
<code>
cm = []
coords = []
CITIES = 30
WIDTH = 600
HEIGHT = 400
LAST_SCORE = -1
</code>
<code>
def cartesian_matrix(coords):
""" A distance matrix """
matrix={}
for i,(x1,y1) in enumerate(coords):
for j,(x2,y2) in enumerate(coords):
dx, dy = x1-x2, y1-y2
dist=sqrt(dx*dx + dy*dy)
matrix[i,j] = dist
return matrix
</code>
<code>
def tour_length(matrix, tour):
""" Returns the total length of the tour """
total = 0
t = tour.getInternalList()
for i in range(CITIES):
j = (i+1)%CITIES
total += matrix[t[i], t[j]]
return total
</code>
<code>
def write_tour_to_img(coords, tour, img_file):
""" The function to plot the graph """
padding=20
coords=[(x+padding,y+padding) for (x,y) in coords]
maxx,maxy=0,0
for x,y in coords:
maxx, maxy = max(x,maxx), max(y,maxy)
maxx+=padding
maxy+=padding
img=Image.new("RGB",(int(maxx),int(maxy)),color=(255,255,255))
font=ImageFont.load_default()
d=ImageDraw.Draw(img);
num_cities=len(tour)
for i in range(num_cities):
j=(i+1)%num_cities
city_i=tour[i]
city_j=tour[j]
x1,y1=coords[city_i]
x2,y2=coords[city_j]
d.line((int(x1),int(y1),int(x2),int(y2)),fill=(0,0,0))
d.text((int(x1)+7,int(y1)-5),str(i),font=font,fill=(32,32,32))
for x,y in coords:
x,y=int(x),int(y)
d.ellipse((x-5,y-5,x+5,y+5),outline=(0,0,0),fill=(196,196,196))
del d
img.save(img_file, "PNG")
print ("The plot was saved into the %s file." % (img_file,))
</code>
<code>
def G1DListTSPInitializator(genome, **args):
""" The initializator for the TSP """
lst = [i for i in range(genome.getListSize())]
random.shuffle(lst)
genome.setInternalList(lst)
</code>
<code>
def evolve_callback(ga_engine):
global LAST_SCORE
if ga_engine.getCurrentGeneration() % 100 == 0:
best = ga_engine.bestIndividual()
if LAST_SCORE != best.getRawScore():
write_tour_to_img( coords, best, "tspimg/tsp_result_%05d.png" % ga_engine.getCurrentGeneration())
LAST_SCORE = best.getRawScore()
return False
</code>
<code>
coords = [(random.randint(0, WIDTH), random.randint(0, HEIGHT))
for i in range(CITIES)]
cm = cartesian_matrix(coords)
</code>
<code>
genome = G1DList.G1DList(len(coords))
genome.evaluator.set(lambda chromosome: tour_length(cm, chromosome))
genome.crossover.set(Crossovers.G1DListCrossoverEdge)
genome.initializator.set(G1DListTSPInitializator)
</code>
<code>
ga = GSimpleGA.GSimpleGA(genome)
ga.setGenerations(2000)
ga.setMinimax(Consts.minimaxType["minimize"])
ga.setCrossoverRate(1.0)
ga.setMutationRate(0.02)
ga.setPopulationSize(80)
ga.stepCallback.set(evolve_callback)
</code>
<code>
ga.evolve(freq_stats=200)
best = ga.bestIndividual()
write_tour_to_img(coords, best, "tspimg/tsp_result.png")
</code>
You can check now the results by plotting some graphs of the evolution process in [this notebook](TSP_check.ipynb).
|
{
"filename": "TSP.ipynb",
"repository": "ecervera/ga-nb",
"query": "transformed_from_existing",
"size": 8202,
"sha": ""
}
|
# ad_analysis_preprocessing_get_raw_h5ad_input_data_1.ipynb
Repository: TemiLeke/systematic
<code>
import os
import rpy2
import scipy
import logging
import warnings
import anndata2ri
import collections
import tables
import pandas as pd
import scanpy as sc
import numpy as np
import seaborn as sb
import decoupler as dc
import scrublet as scr
from scipy import sparse
import scipy.sparse as sp_sparse
import anndata
from anndata import AnnData
from tabnanny import verbose
import matplotlib.pyplot as plt
from gsva_prep import prep_gsva
import rpy2.robjects as robjects
from rpy2.robjects import pandas2ri
from typing import Optional, Union
from matplotlib.pyplot import rcParams
from functions import pathway_analyses
from statsmodels.stats.multitest import multipletests
from sklearn.model_selection import train_test_split
from pytorch_lightning.loggers import TensorBoardLogger
from rpy2.robjects.conversion import localconverter
</code>
<code>
def get_sys_dpi(width, height, diag):
'''
obtain dpi of system
w: width in pixels (if unsure, go vist `whatismyscreenresolution.net`)
h: height in pixels
d: diagonal in inches
'''
w_inches = (diag**2/ (1 + height**2/width**2))**0.5
return round(width/w_inches)
</code>
<code>
# # Ignore R warning messages
#Note: this can be commented out to get more verbose R output
rpy2.rinterface_lib.callbacks.logger.setLevel(logging.ERROR)
# # Automatically convert rpy2 outputs to pandas dataframes
# pandas2ri.activate()
# anndata2ri.activate()
# %load_ext rpy2.ipython
warnings.filterwarnings("ignore", category=PendingDeprecationWarning)
warnings.filterwarnings("ignore", category=UserWarning)
warnings.filterwarnings("ignore", category=FutureWarning)
# Automatically convert rpy2 outputs to pandas dataframes
pandas2ri.activate()
anndata2ri.activate()
%load_ext rpy2.ipython
rcParams['figure.dpi'] = get_sys_dpi(1512, 982, 14.125)
#rcParams['figure.figsize']=(4,4) #rescale figures
sc.settings.verbosity = 3
#sc.set_figure_params(dpi=200, dpi_save=300)
sc.logging.print_versions()
</code>
<code>
def plot_dendrogram(model, **kwargs):
# Create linkage matrix and then plot the dendrogram
# create the counts of samples under each node
counts = np.zeros(model.children_.shape[0])
n_samples = len(model.labels_)
for i, merge in enumerate(model.children_):
current_count = 0
for child_idx in merge:
if child_idx < n_samples:
current_count += 1 # leaf node
else:
current_count += counts[child_idx - n_samples]
counts[i] = current_count
linkage_matrix = np.column_stack(
[model.children_, model.distances_, counts]
).astype(float)
# Plot the corresponding dendrogram
dendrogram(linkage_matrix, **kwargs)
</code>
# **Load input data in raw form and save in `.h5ad` format**
## **[Grubman et. al. 2019](https://doi.org/10.1038/s41593-019-0539-4) (Entorhinal Cortex (ETC))**
<code>
# load AD barcodes
metadata = pd.read_csv("../data/raw/grubman_etc/scRNA_metadata.tsv", sep='\t')
batches = list(metadata.batch.unique())
gene_names = list(pd.read_csv("../data/raw/grubman_etc/mtx/AD1_AD2_genes.tsv", sep="\t", names=['ENS', 'genes'])['genes'])
count_data = sc.read_csv("../data/raw/grubman_etc/scRNA_rawCounts.tsv", delimiter="\t").T
</code>
<code>
grubman_adata = count_data.copy()
grubman_adata.obs = metadata.copy()
grubman_adata.obs['cell_type'] = grubman_adata.obs.cellType.map({'oligo': 'Oligodendrocyte',
'astro': 'Astrocyte',
'OPC': 'OPC',
'neuron': 'Neuron',
'Endo': 'Endothelial',
'mg': 'Microglia'})
grubman_adata = grubman_adata[~grubman_adata.obs['cell_type'].isna()]
</code>
<code>
grubman_adata.write_h5ad("../data/raw/grubman_etc/grubman_etc_raw_anndata.h5ad")
del grubman_adata, count_data
</code>
## **[Leng et. al. 2021](https://www.synapse.org/#!Synapse:syn21788402/wiki/601825) (Entorhinal Cortex (ETC) and Superior Frontal Gyrus (SFG))**
<code>
# processed data obtained
readRDS = robjects.r['readRDS']
df_etc = readRDS('../data/raw/leng_etc/sce.EC.scAlign.assigned.rds')
adata_leng_etc = df_etc
adata_leng_etc.obs['cell_type'] = adata_leng_etc.obs['clusterCellType'].map({'Exc': 'Excitatory',
'Inh': 'Inhibitory',
'Astro': 'Astrocyte',
'Endo': 'Endothelial',
'Micro': 'Microglia',
'OPC': 'OPC',
'Oligo': 'Oligodendrocyte'})
# processed data obtained
readRDS = robjects.r['readRDS']
df_sfg = readRDS('../data/raw/leng_sfg/sce.SFG.scAlign.assigned.rds')
adata_leng_sfg = df_sfg
adata_leng_sfg.obs['cell_type'] = adata_leng_sfg.obs['clusterCellType'].map({'Exc': 'Excitatory',
'Inh': 'Inhibitory',
'Astro': 'Astrocyte',
'Endo': 'Endothelial',
'Micro': 'Microglia',
'OPC': 'OPC',
'Oligo': 'Oligodendrocyte'})
adata_leng_etc.write_h5ad('../data/raw/leng_etc/leng_etc_raw_anndata.h5ad')
adata_leng_sfg.write_h5ad('../data/raw/leng_sfg/leng_sfg_raw_anndata.h5ad')
del adata_leng_etc, adata_leng_sfg, df_etc, df_sfg
</code>
### **Define pathology groups using heirachical clustering of ADNC and CDR scores.**
Individuals are grouped into no, early, and late-pathology groups based on the `AD neuropathological change (ADNC)` score, which represents assessment of amyloid-β deposits (‘A’), staging of neurofibrillary tangles (‘B’) and scoring of neuritic plaques (‘C’) [**Thomas J Montine et al., 2012**](https://pubmed.ncbi.nlm.nih.gov/22101365/), and the `Clinical Dementia rating (CDr)` which reflects the degree of cognitive impairment [**C P Hughes**](https://pubmed.ncbi.nlm.nih.gov/7104545/).
This done to maintain consistency with the clustering used in Mathys et al, where the nine clinico-pathological traits (Supplementary Table 3) where used to group individuals into AD-pathology groups, segregated into two subgroups that correspond to the pathological progression of AD: `‘early-pathology’1 (amyloid burden, but modest neurofibrillary tangles and modest cognitive impairment)`, and `and ‘late-pathology’ (higher amyloid burden, and also increased neurofibrillary tangles, global pathology, and cognitive impairment)`
<code>
# meta = pd.read_csv('../data/raw/leng_etc/leng_etc_metadata.csv')
# X = pd.DataFrame()
# temp = pd.DataFrame(meta['ADNC_score']).apply(lambda x: x.str.split(','), axis=1, result_type='broadcast')
# X["A_score"] = temp.applymap(lambda x: x[0][-1])
# X["B_score"] = temp.applymap(lambda x: x[1][-1])
# X["C_score"] = temp.applymap(lambda x: x[2][-1])
# X['CDR_before_death'] = meta['CDR_before_death']
# # setting distance_threshold=0 ensures we compute the full tree.
# model = AgglomerativeClustering(distance_threshold=0, n_clusters=None)
# model = model.fit(X)
# plt.title("Hierarchical Clustering Dendrogram")
# # plot the top three levels of the dendrogram
# plot_dendrogram(model, truncate_mode="level", p=3)
# plt.xlabel("Number of points in node (or index of point if no parenthesis).")
# plt.show()
</code>
## **[Mathys et. al. 2019](https://doi.org/10.1038/s41586-019-1195-2) (Prefrontal Cortex)**
Load filtered data (Mathys)
<code>
#adata_mathys_pfc = sc.read_mtx("../data/raw/mathys_pfc/mathys_pfc_count_matrix.mtx").T
adata_mathys_pfc = sc.read_mtx("../data/raw/mathys_pfc/filtered_count_matrix.mtx").T
obs_names = pd.read_csv("../data/raw/mathys_pfc/filtered_column_metadata.txt", sep='\t')
obs_names.set_index('TAG', inplace=True)
obs_names.index.rename('index', inplace=True)
adata_mathys_pfc.obs = obs_names
gene_names = pd.read_csv("../data/raw/mathys_pfc/filtered_gene_row_names.txt", sep='\t', header=None)
adata_mathys_pfc.var_names = gene_names[0]
sample_key = pd.read_csv("../data/raw/mathys_pfc/snRNAseqPFC_BA10_Sample_key.csv")
id_mapping = pd.read_csv("../data/raw/mathys_pfc/snRNAseqPFC_BA10_id_mapping.csv")
merged_key = id_mapping.merge(sample_key, how="outer", on='projid')
merged_key['libraryid'] = merged_key['fastq'].str.split('-').apply(lambda x: x[1].split("_")[0])
merged_key.drop_duplicates(subset='libraryid', keep='first', inplace=True)
merged_key.reset_index(inplace=True)
merged_key.drop('index', axis=1, inplace=True)
adata_mathys_pfc.obs[list(merged_key.columns)] = None
for val in merged_key['sample']:
adata_mathys_pfc.obs.loc[adata_mathys_pfc.obs.index.str.endswith(f'.{val}'), list(merged_key.columns)] = merged_key[merged_key['sample']==val].values
# main epidemiological and pathological characteristics of the participants
epi_n_patho = pd.read_excel("../data/raw/mathys_pfc/41586_2019_1195_MOESM3_ESM.xlsx", sheet_name=1)
# clinico-pathological variables.
clin_n_patho = pd.read_excel("../data/raw/mathys_pfc/41586_2019_1195_MOESM5_ESM.xlsx", sheet_name=0)
# clinical, epidemiological and pathological metadata
metadata = epi_n_patho.merge(clin_n_patho, how="outer", on="Subject")
# ID mapping from Mathys
mapping = pd.read_csv("../data/raw/mathys_pfc/snRNAseqPFC_BA10_id_mapping.csv", sep=",")
mapping['sampleid'] = mapping['fastq'].str.split('-').apply(lambda x: x[1].split("_")[0])
metadata = metadata.merge(mapping, how='outer', on='Subject').sort_values(by='sampleid')
metadata.drop_duplicates(subset='sampleid', inplace=True)
metadata.reset_index(inplace=True)
metadata.drop('index', axis=1, inplace=True)
sampleids = list(metadata.sampleid.unique())
adata_mathys_pfc.obs[list(metadata.columns)] = None
columns = list(metadata.columns)
sid = metadata.sampleid.astype(int).min()
for ind, val in enumerate(sampleids):
adata_mathys_pfc.obs.loc[adata_mathys_pfc.obs.libraryid==val, columns] = metadata[metadata['sampleid']==val].values
adata_mathys_pfc.obs = adata_mathys_pfc.obs.astype('category').copy()
adata_mathys_pfc.obs_names_make_unique()
adata_mathys_pfc.var_names_make_unique()
obs_columns = list(adata_mathys_pfc.obs.columns)
adata_mathys_pfc.obs[obs_columns] = adata_mathys_pfc.obs[obs_columns].astype(str)
var_columns = list(adata_mathys_pfc.var.columns)
adata_mathys_pfc.var[var_columns] = adata_mathys_pfc.var[var_columns].astype(str)
adata_mathys_pfc.obs['cell_type'] = adata_mathys_pfc.obs['broad.cell.type'].map({'Ex': 'Excitatory',
'In': 'Inhibitory',
'Ast': 'Astrocyte',
'End': 'Endothelial',
'Mic': 'Microglia',
'Opc': 'OPC',
'Oli': 'Oligodendrocyte',
'Per': 'Pericyte'})
adata_mathys_pfc.write_h5ad('../data/raw/mathys_pfc/mathys_pfc_raw_anndata.h5ad')
del epi_n_patho, clin_n_patho, metadata, mapping, sampleids, merged_key, obs_names, gene_names, adata_mathys_pfc
</code>
## **[Human Multiple Cortical Areas SMART-seq reference](https://portal.brain-map.org/atlases-and-data/rnaseq/human-multiple-cortical-areas-smart-seq)**
<code>
counts = sc.read_csv('../data/raw/allen_mca/matrix.csv')
counts = sparse.csr_matrix(counts.X)
adata_reference = AnnData(counts)
del counts
adata_reference.obs = pd.read_csv('../data/raw/allen_mca/metadata.csv', index_col='sample_name')
adata_reference.var = pd.read_csv('../data/raw/allen_mca/human_MTG_2018-06-14_genes-rows.csv', index_col='gene')
# adata_reference.obs_names = adata_reference.obs.sample_name.copy()
tsne_cord = pd.read_csv('../data/raw/allen_mca/tsne.csv')
adata_reference = adata_reference[adata_reference.obs_names.isin(list(tsne_cord.sample_name))]
# adata_reference.obsm['X_tsne'] = tsne_cord[['tsne_1', 'tsne_2']].to_numpy()
adata_reference
</code>
<code>
obs_columns = list(adata_reference.obs.columns)
adata_reference.obs[obs_columns] = adata_reference.obs[obs_columns].astype(str)
var_columns = list(adata_reference.var.columns)
adata_reference.var[var_columns] = adata_reference.var[var_columns].astype(str)
</code>
<code>
adata_reference.obs['cell_type'] = ""
adata_reference.obs.loc[~adata_reference.obs['class_label'].str.startswith('Non-'), 'cell_type'] = \
adata_reference.obs['class_label'][~adata_reference.obs['class_label'].str.startswith("Non")].map({"Glutamatergic": "Excitatory",
"GABAergic": "Inhibitory"})
adata_reference.obs.loc[adata_reference.obs['class_label'].str.contains("Non-"), 'cell_type'] = adata_reference.obs.loc[adata_reference.obs['class_label'].str.contains("Non-"), 'subclass_label']
adata_reference.obs.loc[adata_reference.obs.cell_type=='Microglia-PVM', 'cell_type'] = 'Microglia'
adata_reference.obs.cell_type = adata_reference.obs.cell_type.astype('category')
</code>
<code>
adata_reference.write_h5ad(f'../data/raw/allen_mca/allen_mca_raw_anndata.h5ad', compression='gzip')
</code>
## **[Lau et. al. 2002](https://www.pnas.org/doi/full/10.1073/pnas.2008762117) (Prefrontal Cortex (PTC))**
<code>
path_to_dir = '../data/raw/lau_pfc/'
file_list = list(set(['_'.join(file.split('_')[:-1]) for file in os.listdir(path_to_dir) if ~('lau' in file)]))
adata_lau_pfc = dict()
for file in file_list:
adata = sc.read_mtx('../data/raw/lau_pfc/'+file+'_matrix.mtx.gz').T
barcodes = pd.read_csv('../data/raw/lau_pfc/'+file+'_barcodes.tsv.gz', sep='\t', names=['barcode'], header=None, dtype=str, index_col=0)
adata.obs = barcodes.astype(str)
adata.obs['Subject'] = file.split("_")[-1]
genes = pd.read_csv('../data/raw/lau_pfc/'+file+'_features.tsv.gz', sep='\t', names=['1', 'gene_name', '3'], header=None, dtype=str, index_col=1)
adata.var = genes.astype(str)
adata_lau_pfc[file] = adata
adata_lau_pfc[file].obs_names_make_unique()
adata_lau_pfc[file].var_names_make_unique()
</code>
<code>
adata_lau_pfc_concat = anndata.concat([adata_lau_pfc[key] for key in adata_lau_pfc.keys()], join='inner')
</code>
<code>
adata_lau_pfc_concat.var['mt'] = adata_lau_pfc_concat.var_names.str.startswith('MT-')
sc.pp.calculate_qc_metrics(adata_lau_pfc_concat, qc_vars=['mt'], inplace=True)
sc.pp.filter_cells(adata_lau_pfc_concat, min_genes=200)
sc.pp.filter_cells(adata_lau_pfc_concat, max_counts=20000)
adata_lau_pfc_concat = adata_lau_pfc_concat[adata_lau_pfc_concat.obs["pct_counts_mt"] <= 20].copy()
</code>
<code>
adata_lau_pfc_concat
</code>
<code>
adata_lau_pfc_concat.write_h5ad('../data/raw/lau_pfc/lau_pfc_raw_anndata.h5ad')
</code>
## **[Zhou et al. 2020](https://www.synapse.org/#!Synapse:syn21670836) (Prefrontal Cortex (PFC))**
<code>
path_to_dir = '../data/raw/zhou_pfc/Data/Matrix_files/'
file_list = list(set(['_'.join(file.split('_')[:-1]) for file in os.listdir(path_to_dir)]))
adata_zhou_pfc = dict()
for file in file_list:
adata = sc.read_mtx('../data/raw/zhou_pfc/Data/Matrix_files/'+file+'_matrix.mtx.gz').T
barcodes = pd.read_csv('../data/raw/zhou_pfc/Data/Matrix_files/'+file+'_barcodes.tsv.gz', sep='\t', names=['barcode'], header=None, dtype=str, index_col=0)
adata.obs = barcodes.astype(str)
adata.obs['Subject'] = file.split("_")[-1]
genes = pd.read_csv('../data/raw/zhou_pfc/Data/Matrix_files/'+file+'_features.tsv.gz', sep='\t', names=['1', 'gene_name', '3'], header=None, dtype=str, index_col=1)
adata.var = genes.astype(str)
adata.obs_names = file + '_' + adata.obs_names
adata_zhou_pfc[file] = adata
adata_zhou_pfc[file].obs_names_make_unique()
adata_zhou_pfc[file].var_names_make_unique()
</code>
<code>
adata_zhou_pfc_concat = anndata.concat([adata_zhou_pfc[key] for key in adata_zhou_pfc.keys()], join='inner')
adata_zhou_pfc_concat.obs.index = [ind.split('-')[0] for ind in adata_zhou_pfc_concat.obs.index]
cell_ids = pd.read_excel('../data/raw/zhou_pfc/Data/Metadata/clusters_cellID.xlsx', sheet_name='All_nuclei')
adata_zhou_pfc_concat.obs['Label'] = adata_zhou_pfc_concat.obs.index.map(dict(zip(cell_ids['Barcodes'], cell_ids['Label'])))
adata_zhou_pfc_concat.obs['Label'] = adata_zhou_pfc_concat.obs['Label'].map({'OPC': 'OPC',
'Ex0': 'Excitatory',
'Oli1': "Oligoddendrocyte",
'Astro': 'Astrocyte',
'Micro': 'Microglia',
'In': "Inhibitory",
'Oli0': 'Oligodendrocyte',
'Endo': 'Endothelial',
'Ex1': 'Excitatory'})
adata_zhou_pfc_concat = adata_zhou_pfc_concat[~adata_zhou_pfc_concat.obs['Label'].isna()]
adata_zhou_pfc_concat.obs['cell_type'] = adata_zhou_pfc_concat.obs['Label'].astype('category')
</code>
<code>
adata_zhou_pfc_concat.write_h5ad('../data/raw/zhou_pfc/zhou_pfc_raw_anndata.h5ad')
</code>
## **[Allen Institute SEA-AD 2023](https://portal.brain-map.org/explore/seattle-alzheimers-disease) (Middle Temporal Gyrus (MTG))**
### **Using Raw Filtered Data**
<code>
#no_pathology = ["H21.33.003", "H21.33.004", "H21.33.023", "H20.33.044"]
no_pathology = ["H21.33.003", "H21.33.004", "H21.33.023", "H20.33.044" ]
#early_pathology = ["H21.33.044", "H21.33.005"]
early_pathology = ["H21.33.005", "H20.33.015", "H20.33.040"]
# late_pathology = ["H20.33.020", "H20.33.004", "H21.33.029"]
# late_pathology = ["H20.33.020", "H21.33.009"]
late_pathology = ["H20.33.017", "H20.33.020", "H21.33.029"]
</code>
<code>
adata = sc.read_h5ad('/Users/tadeoye/Documents/Research codes/scRNA_seq_meta_analysis/data/raw/SEA-AD/filtered_count_matrix/SEAAD_MTG_RNAseq_final-nuclei.2022-08-18.h5ad')
#remove cells from reference patient
adata = adata[adata.obs['Donor ID'].isin(no_pathology + early_pathology + late_pathology)]
sc.pp.filter_cells(adata, min_genes=500)
adata.obs_names_make_unique()
adata.var_names_make_unique()
</code>
<code>
adata.obs['individualID'] = adata.obs['Donor ID'].copy()
</code>
<code>
adata.obs['cell_type'] = adata.obs['Subclass'].copy()
adata.obs['cell_type'] = adata.obs['cell_type'].astype(str)
adata.obs.loc[adata.obs['Class'].str.startswith("Neuronal:"), 'cell_type'] = adata.obs['Class'][adata.obs['Class'].str.startswith("Neuronal:")].map({"Neuronal: Glutamatergic": "Excitatory",
"Neuronal: GABAergic": "Inhibitory"})
adata.obs.loc[adata.obs.cell_type=='Microglia-PVM', 'cell_type'] = 'Microglia'
adata.obs['cell_type'] = adata.obs['cell_type'].astype('category')
adata = adata[adata.obs['cell_type'].isin(['Astrocyte', 'Endothelial', 'Excitatory', 'Inhibitory', 'Microglia', 'OPC', 'Oligodendrocyte'])]
adata = adata[~adata.obs.cell_type.isna()]
</code>
<code>
mapping = {**dict(zip(no_pathology, ["no"]*len(no_pathology))),
**dict(zip(early_pathology, ["early"]*len(early_pathology))),
**dict(zip(late_pathology, ["late"]*len(late_pathology)))}
adata.obs['pathology.group'] = adata.obs.individualID.map(mapping)
</code>
<code>
metadata = adata.obs.drop_duplicates('individualID')
metadata.to_csv('../data/raw/seaad_mtg/seaad_mtg_metadata.csv')
</code>
<code>
adata = sc.read_h5ad('../data/raw/seaad_mtg/seaad_mtg_raw_anndata.h5ad')
</code>
<code>
adata.X = adata.X.astype(np.int32)
adata.write_h5ad('../data/raw/seaad_mtg/seaad_mtg_raw_anndata.h5ad')
</code>
### **Using Raw Pre-Mapped Data**
<code>
biospecimen_metadata = pd.read_csv('../data/raw/SEA-AD/SEA-AD_metadata/SEA-AD_biospecimen_metadata.csv')
biospecimen_metadata = biospecimen_metadata[biospecimen_metadata.assay.str.lower()=='snrnaseq']
individual_metadata = pd.read_csv('../data/raw/SEA-AD/SEA-AD_metadata/SEA-AD_individual_metadata.csv')
manifest = pd.read_csv('../data/raw/SEA-AD/raw_feature_bc_matrices/manifest_1682425120001270000.csv')
all_metadata = manifest.merge(biospecimen_metadata, on='specimenID', how='outer')
all_metadata['individualID'] = all_metadata['individualID_y']
all_metadata = all_metadata.merge(individual_metadata, on = 'individualID', how='outer')
all_metadata.columns
</code>
<code>
all_metadata = all_metadata[all_metadata.tissue_x == 'middle temporal gyrus']
all_metadata.to_csv('../data/raw/SEA-AD/SEA-AD_metadata/allen_mtg_metadata.csv')
</code>
<code>
mapping = {**dict(zip(no_pathology, ["no"]*len(no_pathology))),
**dict(zip(early_pathology, ["early"]*len(early_pathology))),
**dict(zip(late_pathology, ["late"]*len(late_pathology)))}
all_groups = [*no_pathology, *early_pathology, *late_pathology]
filtered_metadata = all_metadata[all_metadata.individualID.isin(all_groups)]
filtered_metadata['pathology.group'] = filtered_metadata.individualID.map(mapping)
filtered_metadata['path'] = filtered_metadata['path'].str.replace('/Users/temitopeleke/Documents/Research Documents/Research codes/mathys_reproduce/', '../')
</code>
<code>
CountMatrix = collections.namedtuple('CountMatrix', ['feature_ref', 'barcodes', 'matrix'])
def get_matrix_from_h5(filename):
with tables.open_file(filename, 'r') as f:
mat_group = f.get_node(f.root, 'matrix')
barcodes = f.get_node(mat_group, 'barcodes').read()
data = getattr(mat_group, 'data').read()
indices = getattr(mat_group, 'indices').read()
indptr = getattr(mat_group, 'indptr').read()
shape = getattr(mat_group, 'shape').read()
matrix = sp_sparse.csc_matrix((data, indices, indptr), shape=shape)
feature_ref = {}
feature_group = f.get_node(mat_group, 'features')
feature_ids = getattr(feature_group, 'id').read()
feature_names = getattr(feature_group, 'name').read()
feature_types = getattr(feature_group, 'feature_type').read()
feature_ref['id'] = feature_ids
feature_ref['name'] = feature_names
feature_ref['feature_type'] = feature_types
tag_keys = getattr(feature_group, '_all_tag_keys').read()
for key in tag_keys:
key = key.decode("utf-8")
feature_ref[key] = getattr(feature_group, key).read()
return CountMatrix(feature_ref, barcodes, matrix)
adatas = []
columns = list(filtered_metadata.columns)
for path in list(filtered_metadata.path)[0:]:
filtered_matrix_h5 = path
filtered_feature_bc_matrix = get_matrix_from_h5(filtered_matrix_h5)
adata = AnnData(filtered_feature_bc_matrix.matrix.T)
#adata.X = filtered_feature_bc_matrix.matrix
adata.obs_names = [name.split("-")[0] for name in filtered_feature_bc_matrix.barcodes.astype('U').astype(str)]
adata.var_names = filtered_feature_bc_matrix.feature_ref['name'].astype('U').astype(str)
adata.var["id"] = filtered_feature_bc_matrix.feature_ref['id'].astype('U').astype(str)
adata.var['genome'] = filtered_feature_bc_matrix.feature_ref['genome'].astype('U').astype(str)
adata.var['feature_type'] = filtered_feature_bc_matrix.feature_ref['feature_type'].astype('U').astype(str)
adata.obs[columns] = filtered_metadata.loc[filtered_metadata.path == path, columns].iloc[0]
cell_classes = pd.read_csv(f'../data/raw/SEA-AD/cell_classes/{filtered_metadata.loc[filtered_metadata.path==path, "specimenID"].iloc[0]}.csv')
cell_classes['barcodes'] = cell_classes['sample_id'].apply(lambda x: x.split("-")[0])
adata = adata[cell_classes['barcodes']]
adata.obs[list(cell_classes.columns)] = 'None'
adata.obs['Class'] = adata.obs.index.map(dict(zip(cell_classes.barcodes, cell_classes.Class)))
adata.obs['Subclass'] = adata.obs.index.map(dict(zip(cell_classes.barcodes, cell_classes.Subclass)))
adata.obs['Supertype'] = adata.obs.index.map(dict(zip(cell_classes.barcodes, cell_classes.Supertype)))
adata.obs['cell_labels'] = adata.obs['Subclass'].copy()
adata.obs.loc[adata.obs['Class'].str.startswith("Neuronal:"), 'cell_labels'] = adata.obs['Class'][adata.obs['Class'].str.startswith("Neuronal:")].map({"Neuronal: Glutamatergic": "Excitatory",
"Neuronal: GABAergic": "Inhibitory"})
adata.obs.loc[adata.obs.cell_labels=='Microglia-PVM', 'cell_labels'] = 'Microglia'
# Initial removal of low-quality nuclei
# SEA-AD nuclei with fewer than 500 genes detected were removed upstream
sc.pp.filter_cells(adata, min_genes=500)
adata.obs_names_make_unique()
adata.var_names_make_unique()
adatas.append(adata)
</code>
<code>
adata_allen_mtg = anndata.concat(adatas, join='outer')
</code>
<code>
adata_allen_mtg
</code>
<code>
adata_allen_mtg.write_h5ad('../data/raw/allen_mtg/allen_mtg_raw_anndata.h5ad')
filtered_metadata.to_csv('../data/raw/allen_mtg/filtered_allen_mtg_metadata.csv')
filtered_metadata.drop_duplicates(subset='individualID', keep='first', inplace=True)
filtered_metadata.to_csv('../data/raw/allen_mtg/allen_mtg_metadata.csv')
</code>
### **Raw RE-MAPPED Data**
<code>
import synapseclient
syn = synapseclient.Synapse()
syn.login()
</code>
<code>
results = syn.tableQuery('select * from syn11346063')
</code>
<code>
## Get corresponding FastQs to re-mapp.
manifest = results.asDataFrame().copy()
manifest.study = manifest.study.apply(lambda x: x[0] if (type(x)==list) and (len(x)>0) else x)
manifest.assay = manifest.assay.apply(lambda x: x[0] if (type(x)==list) and (len(x)>0) else x)
manifest = manifest[(manifest.study=='SEA-AD') & (manifest.assay=="snrnaSeq")]
</code>
<code>
metadata = pd.read_csv('../data/raw/allen_mtg/filtered_allen_mtg_metadata.csv')
filtered_manifest = manifest[(manifest.specimenID.isin(metadata.specimenID.to_list())) & (manifest.name.str.endswith('fastq.gz'))]
</code>
<code>
filtered_manifest[['id', 'specimenID', 'name']]
</code>
|
{
"filename": "ad_analysis_preprocessing_get_raw_h5ad_input_data_1.ipynb",
"repository": "TemiLeke/systematic",
"query": "transformed_from_existing",
"size": 92060,
"sha": ""
}
|
# StatisticsWithPython_検定.ipynb
Repository: inoueshinichi/Book
<code>
import numpy as np
import pandas as pd
import scipy as sp
from scipy import stats
from matplotlib import pyplot as plt
import seaborn as sns
sns.set()
%precision 3
%matplotlib inline
</code>
<code>
# t検定
junk_food = pd.read_csv("./data/3-8-1-junk-food-weight.csv")["weight"]
print(junk_food.head())
# ある標本抽出によって得られた標本分布から決まるt値
mu = sp.mean(junk_food)
print(mu)
df = len(junk_food) - 1
print(df)
sigma = sp.std(junk_food, ddof=1)
print(sigma)
se = sigma / sp.sqrt(len(junk_food))
print(se)
t_value = (mu - 50) / se
print(t_value)
</code>
<code>
# t分布からp値を計算
# あるt値を下回る累積確率
alpha = stats.t.cdf(t_value, df=df)
print((1 - alpha) * 2)
# p値が0.05を下回っているので、今回の検定でスナック菓子50kgと有意差がある
</code>
<code>
# stats.ttest_1samp()
stats.ttest_1samp(junk_food, 50)
</code>
<code>
# シミュレーションによるp値の計算
# p値は「帰無仮説が正しいと仮定して、何度も標本抽出〜t値計算を繰り返したとき、t標本と同じかそれより大きなt値が得られる割合」と解釈できる。
size = len(junk_food)
sigma = sp.std(junk_food, ddof=1)
# 母集団分布を平均50の正規分布と仮定して、50000回のt値を計算する
t_value_array = np.zeros(50000)
# 帰無仮説が正しいとして、標本抽出〜t値計算を5万回繰り返す
np.random.seed(1)
norm_dist = stats.norm(loc=50, scale=sigma)
for i in range(0, 50000):
sample = norm_dist.rvs(size=size) # サンプリング
sample_mean = sp.mean(sample)
sample_std = sp.std(sample, ddof=1)
sample_se = sample_std / sp.sqrt(size) # 標準誤差
t_value_array[i] = (sample_mean - 50) / sample_se
</code>
<code>
# 50000個のt値のうち、t標本を上回った割合
(sum(t_value_array > t_value) / 50000) * 2
# 理論値とほぼ同じ
</code>
<code>
"""対応のあるt検定・・・・「同じ対象を異なった条件で2回測定して、その違いを見る」"""
# -> 差分の平均値が0と異なるかどうかをチェック
paired_test_data = pd.read_csv("./data/3-9-1-paired-t-test.csv")
print(paired_test_data)
# 帰無仮説:薬を飲む前と後で体温は変わらない
# 対立仮説:薬を飲む前と後で体温は変わる
# 有意水準5%で検定を行う
</code>
<code>
# 前後の標本平均
before = paired_test_data.query('medicine == "before"')["body_temperature"]
after = paired_test_data.query('medicine == "after"')["body_temperature"]
before = np.array(before)
after = np.array(after)
# 差
diff = after - before
print(diff)
</code>
<code>
# t検定を実行
stats.ttest_1samp(diff, 0)
# 有意差がある
</code>
<code>
"""対応のないt検定・・・「平均値の差」に注目"""
# 2群のt検定を行う
# t値の計算式が1群のt検定とは異なる
# 平均値
mean_bef = sp.mean(before)
mean_aft = sp.mean(after)
# 分散(ここが1群のt検定とは異なるところ)
var_bef = sp.var(before, ddof=1)
var_aft = sp.var(after, ddof=1)
# サンプルサイズ
m = len(before)
n = len(after)
# t値
t_value = (mean_aft - mean_bef) / sp.sqrt((var_aft/n + var_bef/m))
print(t_value)
</code>
<code>
# t検定(等分散ではない)・・・Welchのt検定
stats.ttest_ind(after, before, equal_var=False)
# 有意差がある
</code>
<code>
"""χ二乗検定"""
print(1 - sp.stats.chi2.cdf(x=6.667, df=1))
</code>
<code>
# 分割表の検定
click_data = pd.read_csv("./data/3-10-1-click_data.csv")
print(click_data)
</code>
<code>
# 分割表に変換
cross = pd.pivot_table(
data = click_data,
values = "freq",
aggfunc="sum",
index="color",
columns="click")
print(cross)
</code>
<code>
# χ二乗検定
sp.stats.chi2_contingency(cross, correction=False)
</code>
|
{
"filename": "StatisticsWithPython_検定.ipynb",
"repository": "inoueshinichi/Book",
"query": "transformed_from_existing",
"size": 11644,
"sha": ""
}
|
# model_labels.ipynb
Repository: constantingoeldel/epimutation
<code>
from pysam import FastaFile
dna = FastaFile("../genome/AT_reference/GCF_000001735.4_TAIR10.1_genomic.fna")
print(dna.references)
chr_to_references = {
"1": "CHR1",
"2": "CHR2",
"3": "CHR3",
"4": "CHR4",
"5": "CHR5",
}
a = dna.fetch(chr_to_references["1"], 0, 1000)
print(1,dna.get_reference_length(chr_to_references["1"]))
print(2,dna.get_reference_length(chr_to_references["2"]))
print(3,dna.get_reference_length(chr_to_references["3"]))
print(4,dna.get_reference_length(chr_to_references["4"]))
print(5,dna.get_reference_length(chr_to_references["5"]))
encode_dict = {
"[mask]": 0,
"A": 1,
"C": 2,
"G": 3,
"T": 4,
"Y": 5, # C or T
"R": 6, # A or G
"W": 7, # A or T
"S": 8, # C or G
"M": 9, # A or C
"K": 10, # G or T
"B": 11, # C or G or T
"D": 12, # A or G or T
"H": 13, # A or C or T
"V": 14, # A or C or G
"a": 1,
"c": 2,
"g": 3,
"t": 4,
"N": -1,
}
encode_bases = lambda bases: [encode_dict[base] for base in bases]
</code>
<code>
import polars as pl
import numpy as np
def meth_rates_to_labels(dna, meth_rates: pl.DataFrame):
labels_by_chrsm = {}
for (chrsm, rates) in meth_rates.partition_by("chrsm", as_dict=True).items():
print(chrsm, rates.height)
sequence = pl.DataFrame({ "sequence": encode_bases(dna.fetch(chr_to_references[f"{chrsm}"]))})
a = np.zeros(sequence.height, dtype=np.float32)
b = np.zeros(sequence.height, dtype=np.float32)
std_st = np.zeros(sequence.height, dtype=np.float32)
for row in rates.iter_rows(named=True):
a[row["start"] - 1 :row["end"] -1] = row["alpha"]
b[row["start"] - 1 :row["end"] -1] = row["beta"]
std_st[row["start"] - 1:row["end"] -1] = row["std_st"]
labels = sequence.with_columns(pl.Series("alpha", a), pl.Series("beta", b), pl.Series("std_st", std_st))
labels = labels.with_columns(pl.when(pl.col("sequence") == 2).then(pl.col("alpha")).otherwise(pl.lit(0.)).alias("alpha"))
labels = labels.with_columns(pl.when(pl.col("sequence") == 2).then(pl.col("beta")).otherwise(pl.lit(0.)).alias("beta"))
labels = labels.with_columns(pl.when(pl.col("sequence") == 2).then(pl.col("std_st")).otherwise(pl.lit(0.)).alias("std_st"))
labels_by_chrsm[chrsm] = labels
return labels_by_chrsm
</code>
<code>
meth_rates = pl.read_parquet("test.parquet").sort(["chrsm", "start", "end"])
display(meth_rates)
display(meth_rates.with_columns((pl.col("end") - pl.col("start")).alias("diff")).mean())
# labels_by_chrms = meth_rates_to_labels(dna, meth_rates)
# display(labels_by_chrms[1][100:130])
</code>
<code>
chrsm_embeddings = [pl.read_parquet(f"embeddings/chr_{i}.parquet") for i in range(1, 6)]
res = pl.DataFrame()
for row in meth_rates.iter_rows():
chrsm = row[0]
start = row[2]
end = row[3]
print(chrsm, start, end)
means = chrsm_embeddings[chrsm-1][start -1: end -1].sum()
res = res.vstack(means)
display(res)
</code>
<code>
agg_data = res.hstack(meth_rates)
display(agg_data)
agg_data.write_parquet("agg_data.parquet")
</code>
<code>
for (chrsm, labels) in labels_by_chrms.items():
labels.write_parquet(f"./labels/test/{chrsm}.parquet")
labels.to_numpy().tofile(f"./labels/test/{chrsm}.bin")
</code>
<code>
import matplotlib.pyplot as plt
from sklearn.linear_model import LinearRegression
labels_chr_1 = pl.read_parquet("/mnt/fast/epigenomics/conschti/labels/512_64/1.parquet").drop("sequence")
i = np.arange(labels_chr_1.height)
# labels_chr_1 = labels_chr_1.with_columns(i=i).set_sorted("i")
# averaged = labels_chr_1.group_by_dynamic(index_column="i", period="1000000i", every="500i").agg(pl.mean("alpha"), pl.mean("beta"), pl.mean("std_st"))
embeddings_chr_1 = pl.read_parquet("/mnt/fast/epigenomics/conschti/embeddings/chr_1.parquet")
data = embeddings_chr_1.hstack(labels_chr_1).with_columns(i=i).set_sorted("i")
windowed = data.filter(pl.col("sequence") == 2)[:10000].group_by_dynamic(index_column="i", period="64i", every="32i").agg(pl.sum("h2az"), pl.mean("alpha"), pl.mean("beta"), pl.mean("std_st"))
beginning = data[:1000000].sort(by="Chromatine States")
display(beginning)
plt.plot( beginning["alpha"])
# plt.plot(windowed["i"], windowed["alpha"])
# plt.plot(windowed["i"], windowed["beta"])
# plt.plot(windowed["i"], windowed["std_st"])
# plt.plot(averaged["i"], averaged["alpha"])
# plt.plot(averaged["i"], averaged["beta"])
# plt.plot(averaged["i"], averaged["std_st"])
# plt.legend(["Alpha", "Beta"])
# plt.show()
# labels_chr_1 = labels_chr_1.filter(pl.col("sequence") == 2)
# plt.scatter(labels_chr_1[:10000000]["i"], labels_chr_1[:10000000]["alpha"])
# # plt.plot(labels_chr_1[:100000]["i"], labels_chr_1[:100000]["beta"], color=r)
plt.ylim(0, 0.002)
</code>
<code>
X = data.drop("i").filter((pl.col("sequence") == 2) & (pl.col("alpha") > 0))
y = X["alpha"]
X = X.drop("alpha").drop("beta").drop("std_st")
reg = LinearRegression().fit(X, y).score(X, y)
print(reg)
</code>
<code>
from sklearn import tree
from sklearn.model_selection import train_test_split
from sklearn.naive_bayes import GaussianNB
X = agg_data.drop("i").filter((pl.col("alpha") > 0))
y = X["alpha"]
X = X.drop("alpha").drop("beta").drop("std_st").drop("slice").drop("Chromatine States").drop("sequence").drop("genes").drop("start").drop("end")
X = X - X.mean_horizontal()
display(X)
clf = tree.DecisionTreeRegressor()
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.05 , random_state=42)
# gnb = GaussianNB()
# score = gnb.fit(X_train, y_train).score(X_test, y_test)
# print(score)
reg = LinearRegression().fit(X_train, y_train).score(X_train, y_train)
print(reg)
score = clf.fit(X_train, y_train).score(X_test, y_test)
print(score)
</code>
<code>
from statsmodels.formula.api import ols
d = agg_data.drop(["sequence", "slice"]).filter((pl.col("alpha") > 0))
# rename Chromatine States to Chromatine_States
d = d.rename({"Chromatine States": "Chromatine_States"})
display(d)
alpha_formula = "alpha ~ " + " + ".join([s for s in d.columns if s != "alpha" and s != "beta" and s != "std_st"])
beta_formula = "beta ~ " + " + ".join([s for s in d.columns if s != "alpha" and s != "beta" and s != "std_st"])
std_st_formula = "std_st ~ alpha + beta"
print(alpha_formula)
alpha_intragenic_model = ols(alpha_formula, d).fit()
# beta_intragenic_model = ols(beta_formula, data).fit()
# std_st_intragenic_model = ols(std_st_formula, data).fit()
print(alpha_intragenic_model.summary())
# print(beta_intragenic_model.summary())
# print(std_st_intragenic_model.summary())
print(alpha_intragenic_model.rsquared_adj)
# print(beta_intragenic_model.rsquared_adj)
# print(std_st_intragenic_model.rsquared_adj)
</code>
<code>
from statsmodels.formula.api import ols
d = data.drop("i").filter(pl.col("sequence") == 2 & (pl.col("alpha") > 0))
# rename Chromatine States to Chromatine_States
d = d.rename({"Chromatine States": "Chromatine_States"})
alpha_formula = "alpha ~ " + " + ".join([s for s in d.columns if s != "alpha" and s != "beta" and s != "std_st"])
beta_formula = "beta ~ " + " + ".join([s for s in d.columns if s != "alpha" and s != "beta" and s != "std_st"])
std_st_formula = "std_st ~ alpha + beta"
print(alpha_formula)
# alpha_intragenic_model = ols(alpha_formula, d).fit()
# beta_intragenic_model = ols(beta_formula, data).fit()
std_st_intragenic_model = ols(std_st_formula, data).fit()
# print(alpha_intragenic_model.summary())
# print(beta_intragenic_model.summary())
print(std_st_intragenic_model.summary())
# print(alpha_intragenic_model.rsquared_adj)
# print(beta_intragenic_model.rsquared_adj)
print(std_st_intragenic_model.rsquared_adj)
</code>
|
{
"filename": "model_labels.ipynb",
"repository": "constantingoeldel/epimutation",
"query": "transformed_from_existing",
"size": 147524,
"sha": ""
}
|
# SubModule03.ipynb
Repository: NIGMS/Metagenomics-Analysis-of-Biofilm-Microbiome

# Submodule #3: Biomarker Discovery
## Overview
Microbiome community gene prediction and functional annotation are critical steps in the biofilm metagenomics workflow. Functional annotation of shotgun metagenomic data has become an increasingly popular method for identifying the aggregate functional capacities encoded by the community’s biofilm. This analysis relies on comparisons of predicted genes with existing, previously annotated sequences in 16s metagenomics samples. Functional profiling provides insights into what functions are carried out by a given biofilm community.
## Learning Objectives:
At the completion of this module, the learner will be able to:
- Learn how to discover biomarkers in a microbiome
- Run metagenomics marker gene discovery tools
- Predict and evaluate resulting genes, proteins and pathway biomarkers using the following tools:
- PICRUSt2
- Qiime2-PICRUSt2 plugin
## Prerequisites
* **Data:**
* 16S rRNA sequence data (FASTA format): `rep-seqs.fasta` or `rep-seqs.qza`.
* Feature Table (BIOM format): `feature-table.biom` or `table.qza`.
* Sample Metadata (TSV format): `sample-metadata.tsv` (for Qiime2 plugin).
* **Software (installed in the notebook):**
* PICRUSt2
* Qiime2
* q2-picrust2 (Qiime2 plugin)
* conda/mamba (environment management)
## Get Started
### Step 4 - Biomarker Discovery (PICRUSt2, q2-PICRUSt2):
The primary tool for functional annotation of metagenomic data is PICRUSt2. This tool can be implemented as a standalone tool, as a Qiime2 plugin, or through the MicrobeAnalystR wrapper workflow. We will show examples of each in this submodule.
### Install PICRUSt2
<code>
%%capture
%%bash
wget https://github.com/picrust/picrust2/archive/v2.5.1.tar.gz
tar xvzf v2.5.1.tar.gz
rm v2.5.1.tar.gz
conda env create -f picrust2-2.5.1/picrust2-env.yaml
conda run -n picrust2 pip install --editable picrust2-2.5.1/
</code>
### Biomarker Analysis with PICRUSt2 as a standalone tool (duration ~10 mins)
PICRUSt2 uses machine learning to predict functional abundance and capabilities within microbial communities using 16S rRNA marker genes. To start off the analysis we will identify our environment by setting the location of our PICRUSt2 inputs, and outputs. This is a great practice that helps us easily track where our files are located and avoid retyping common paths. First we will run PICRUSt2 as a standalone tool. We start off by defining some data paths as environment variables so that the PICRUSt2 scripts can automatically find them.
### Assign File Paths as ENV Variables
<code>
%env PICRUST_IN=qiime2_analysis/qiime2_Output/rep-seqs-unzipped/data/dna-sequences.fasta
%env BIOM=qiime2_analysis/qiime2_Output/table-unzipped/data/feature-table.biom
%env PICRUST_OUT=BioMarker_Discovery/picrust2_output
</code>
You will notice that our fasta and biom file are both outputs from the denoise analysis using DADA2 from submodule 2. To break it down:
- The FeatureData is our **fasta** file (also written as **fna**) and contains **amplicon sequence variants (ASV)** of 16S rRNA reads and IDs found accross the human samples.
- The FeatureTable is our **biom** file that contains the IDs of the ASV reads and the number of times these reads were found per sample.
Next we will assign an environment variable with the number of available cores on this VM. Since the number of cores will change with each machine type, it is important to capture this with a variable rather than pass a hard-coded integer as an argument for each multi-threaded step.
<code>
#define number of cores to use.
numthreads=!nproc
numthreadsint = int(numthreads[0])
%env CORES = $numthreadsint
</code>
### Run the PICRUSt2 Pipeline
The commands below will do two things. Let's discuss each step as we run them:
1. place_seqs.py will insert our ASVs reads into a reference tree based on the Integrated Microbial Genomes database. This will produce our out.tre file which will be our input for the next command.
2. hsp.py predicts the copy number of gene families for each ASV. You will notice that the script is run twice because we are looking to identify sequences with the 16S rRNA marker and their Enzyme Classification (EC) number.
<code>
%%bash
source activate picrust2
python picrust2-2.5.1/scripts/place_seqs.py -s ${PICRUST_IN} -o ${PICRUST_OUT}/out.tre -p ${CORES} --intermediate ${PICRUST_OUT}/intermediate/place_seqs
python picrust2-2.5.1/scripts/hsp.py -i 16S -t ${PICRUST_OUT}/out.tre -o ${PICRUST_OUT}/marker_predicted_and_nsti.tsv.gz -p ${CORES} -n
python picrust2-2.5.1/scripts/hsp.py -i EC -t ${PICRUST_OUT}/out.tre -o ${PICRUST_OUT}/EC_predicted.tsv.gz -p ${CORES}
</code>
3. metagenome_pipeline.py does the same thing as the hsp.py script but the difference is that it predicts gene families weighted by the relative abundance of ASVs in their community.
<code>
%%bash
source activate picrust2
python picrust2-2.5.1/scripts/metagenome_pipeline.py -i ${BIOM} -m ${PICRUST_OUT}/marker_predicted_and_nsti.tsv.gz -f ${PICRUST_OUT}/EC_predicted.tsv.gz -o ${PICRUST_OUT}/EC_metagenome_out --strat_out
</code>
Our output should show something along the lines that some sequences are above max NSTI cut-off of 2.0. The **nearest-sequenced taxon index (NSTI)** is the branch length between the nearest 16S reference sequence and each ASV. The thought is that as the NSTI value descreases, the closer the relationship is between the ASV reads and the corresponding 16S sequence. Anything above 2 is considered noise and will not be used in the analysis. 11 out of 751 ASVs had a NSTI value equal to or higher than 2 so they were removed to not skew the downstream analysis.
4. convert_table.py creates attribute tables that link the functional and taxonomic data.
5. pathway_pipeline.py predicts pathway-level abundances by using our EC number abundances generated in step 2 and uses the MetaCyc pathway database to see which pathways are associated with these ASV reads.
6. add_descriptions.py will add descriptions of each functional ID to the gene family and pathway abundance tables.
<code>
%%bash
source activate picrust2
python picrust2-2.5.1/scripts/convert_table.py ${PICRUST_OUT}/EC_metagenome_out/pred_metagenome_contrib.tsv.gz -c contrib_to_legacy -o ${PICRUST_OUT}/EC_metagenome_out/pred_metagenome_contrib.legacy.tsv.gz
python picrust2-2.5.1/scripts/pathway_pipeline.py -i ${PICRUST_OUT}/EC_metagenome_out/pred_metagenome_contrib.tsv.gz -o ${PICRUST_OUT}/pathways_out -p ${CORES}
python picrust2-2.5.1/scripts/add_descriptions.py -i ${PICRUST_OUT}/EC_metagenome_out/pred_metagenome_unstrat.tsv.gz -m EC -o ${PICRUST_OUT}/EC_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz
python picrust2-2.5.1/scripts/add_descriptions.py -i ${PICRUST_OUT}/pathways_out/path_abun_unstrat.tsv.gz -m METACYC -o ${PICRUST_OUT}/pathways_out/path_abun_unstrat_descrip.tsv.gz
</code>
Finally, unzip all .gz files
<code>
# Postprocess Data
! gunzip -k ${PICRUST_OUT}/*.gz
! gunzip -k ${PICRUST_OUT}/EC_metagenome_out/*.gz
</code>
<div class="alert alert-block alert-danger">
<i class="fa fa-exclamation-circle" aria-hidden="true"></i>
<b>Alert: </b> Unfortunately PICRUSt2 does not let you overwrite output files. If you would like to rerun this analysis again make sure you delete the contents within the output folder via the command:
rm -r BioMarker_Discovery/picrust2_output
The PICRUSt2 script will make your output directory automatically.
</div>
## Biomarker Analysis with PICRUSt2 as a Qiime2 plugin (duration ~ 30 mins)
## Install q2-picrust2
<code>
%%capture
! mamba create -n qiime2 -c https://packages.qiime2.org/qiime2/2022.11/passed/core/ -c conda-forge -c bioconda qiime2-core -y
! mamba install -n qiime2 q2-picrust2 -c conda-forge -c bioconda -c picrust -y
</code>
Now that we understand each step of the PICRUSt2 pipeline we can bridge our Qiime2 and PICRUSt2 analysis via Qiimes2's PICRUSt2 plugin (q2-picrust2). This plugin allows the user to run both PICRUSt2 as part of a larger Qiime2 workflow without the need of installing the two separately. We have to re-define environment variables since we are in a different kernel.
### Assign File Paths as ENV Variables
<code>
%env Q2_PI_IN=qiime2_analysis/qiime2_Output/rep-seqs.qza
%env Q2_META=Core_Dataset_Prep/sample-metadata.tsv
%env Q2_BIOM=qiime2_analysis/qiime2_Output/table.qza
%env Q2_PI_OUT=BioMarker_Discovery/q2-picrust2_output
</code>
<code>
#define number of cores to use.
numthreads=!nproc
numthreadsint = int(numthreads[0])
%env CORES = $numthreadsint
</code>
### Run the Qiime2-PICRUSt2 Pipeline
This process is relatively the same as our PICRUSt2 step above with a few additions:
1. **picrust2 full-pipeline** allows to run the full PICRUSt2 pipeline with one command.
2. **feature-table summarize** summarizes the finding from the step 1 and will create visuals, histograms, and stats on how many sequences are associated with each sample and feature.
3. **diversity core-metrics** creates non-phylogenetic diversity metrics and a feature table.
<code>
%%bash
source activate qiime2
qiime picrust2 full-pipeline --i-table "${Q2_BIOM}" --i-seq "${Q2_PI_IN}" --output-dir "${Q2_PI_OUT}" --p-placement-tool epa-ng --p-threads ${CORES} --p-hsp-method pic --p-max-nsti 2 --verbose
qiime feature-table summarize --i-table "${Q2_PI_OUT}/pathway_abundance.qza" --o-visualization "${Q2_PI_OUT}/pathway_abundance.qzv"
qiime diversity core-metrics --i-table "${Q2_PI_OUT}/pathway_abundance.qza" --p-sampling-depth 226702 --m-metadata-file "${Q2_META}" --output-dir "${Q2_PI_OUT}/pathabun_core_metrics_out" --p-n-jobs 1
</code>
<div class="alert alert-block alert-danger">
<i class="fa fa-exclamation-circle" aria-hidden="true"></i>
<b>Alert: </b> Unfortunately Qiime2-PICRUSt2 plugin does not let you overwrite output files. If you would like to rerun this analysis again run the folowing command:
rm -r BioMarker_Discovery/q2-picrust2_output
The plug-in will make your output directory automatically.
</div>
### Postprocess Data
The **qiime tools export** tool extracts ASV tables from the qza or qzv files. **biom convert** allows you to convert file formats such as tsv that typically PICRUSt2 produces. This is great for the next submodule where one of our PICRUSt2 outputs is used to query against the Uniprot database.
<code>
%%bash
source activate qiime2
# Export Abundance
qiime tools export --input-path "${Q2_PI_OUT}/pathway_abundance.qza" --output-path "${Q2_PI_OUT}/pathabun_exported"
biom convert -i "${Q2_PI_OUT}/pathabun_exported/feature-table.biom" -o "${Q2_PI_OUT}/pathabun_exported/feature-table.biom.tsv" --to-tsv
qiime tools export --input-path "${Q2_PI_OUT}/pathway_abundance.qzv" --output-path "${Q2_PI_OUT}/pathabun_qzv_exported"
# Export EC Metagenome
qiime tools export --input-path "${Q2_PI_OUT}/ec_metagenome.qza" --output-path "${Q2_PI_OUT}/ec_metagenome_exported"
qiime feature-table summarize --i-table "${Q2_PI_OUT}/ec_metagenome.qza" --o-visualization "${Q2_PI_OUT}/ec_metagenome.qzv"
biom convert -i "${Q2_PI_OUT}/ec_metagenome_exported/feature-table.biom" -o "${Q2_PI_OUT}/ec_metagenome_exported/feature-table.biom.tsv" --to-tsv
qiime tools export --input-path "${Q2_PI_OUT}/ec_metagenome.qzv" --output-path "${Q2_PI_OUT}/ec_metagenome_qzv_exported"
# Export Kegg Orthologs (KO) Metagenome
qiime tools export --input-path "${Q2_PI_OUT}/ko_metagenome.qza" --output-path "${Q2_PI_OUT}/ko_metagenome_exported"
qiime feature-table summarize --i-table "${Q2_PI_OUT}/ko_metagenome.qza" --o-visualization "${Q2_PI_OUT}/ko_metagenome.qzv"
biom convert -i "${Q2_PI_OUT}/ko_metagenome_exported/feature-table.biom" -o "${Q2_PI_OUT}/ko_metagenome_exported/feature-table.biom.tsv" --to-tsv
qiime tools export --input-path "${Q2_PI_OUT}/ko_metagenome.qzv" --output-path "${Q2_PI_OUT}/ko_metagenome_qzv_exported"
</code>
<code>
#run the following command to take the quiz!
from IPython.display import IFrame
IFrame("../Quiz/QS14.html", width=800, height=350)
</code>
## Conclusion
In this submodule you learned how to extract microbiome biomarker using several computational tools and machine learning pre-trained model. You learned using the Qiime output to predict relevant protein and pathways from 16s dataset using PICRUSt2 pre-trained machine learning model.
## Clean up
Remember to stop your notebook instance when you are done!
|
{
"filename": "SubModule03.ipynb",
"repository": "NIGMS/Metagenomics-Analysis-of-Biofilm-Microbiome",
"query": "transformed_from_existing",
"size": 19317,
"sha": ""
}
|
# main_1.ipynb
Repository: Limekaaa/UC-FIRe
# Install libraries (if needed)
<code>
"""
!pip install beir
!pip install fasttext
!pip install spacy
!pip scikit-learn
!pip install rank_bm25
!python -m spacy download en_core_web_sm
!pip install faiss-cpu
"""
</code>
# Import libraries
<code>
import pandas as pd
import beir
from beir import util, LoggingHandler
from beir.datasets.data_loader import GenericDataLoader
from beir.retrieval.evaluation import EvaluateRetrieval
from utils_func import corpus_processing, matrix_creation, clustering, retriever_model, vector_creation
import os
import multiprocessing
multiprocessing.set_start_method("spawn", force=True)
try:
import fasttext
import fasttext.util
except:
print('fasttext not imported')
</code>
# Run experiment
## Choose parameters
<code>
dataset = "nfcorpus" # dataset you want to use, had to be available in the beir benchmark: https://github.com/beir-cellar/beir
use_ft = True # whether to use fasttext or not to handle unseen words
path_ft = 'cc.en.100.bin' # path to the fasttext model, if empty and use_ft is true, the model will be downloaded in the current directory
save_cleaned_corpus = '' # path to save the cleaned corpusn, if empty, the corpus will not be saved
save_scores = '' # path to save the scores, if empty, the scores will not be saved
load_cleaned_corpus = '' # path to load the cleaned corpus, if empty, the corpus will be cleaned
load_vectors = f'word_vectors/word_vectors_{dataset}.csv' # path to load the word vectors, if empty, the vectors will be created
vector_dimension = 100 # dimension of the word vectors
path_to_save_model = '' # path to save the fasttext model trained on the corpora, if empty, the model will not be saved
remove_original_corpus = False # whether to remove the original corpus from the memory or not, to save memory
best_n_neighbors = 75 # number of neighbors to consider to fill the similarity matrix
best_alpha = 0.76 # alpha parameter, balancing the importance between similarity and coexistence
best_thresh = 0.75 # threshold to consider a word as replaceable by another one
metric = 'cosine' # metric to use to compute the similarity matrix
k1 = 1.5 # parameter of the BM25 algorithm
b = 0.75 # parameter of the BM25 algorithm
thresh_prob=0.05 # threshold to consider a value equals to 0 in the coexistence matrix
knn_method = 'faiss' # method to use to compute the k-ne, either 'faiss' or 'exact'
</code>
## Run an experiment
<code>
url = "https://public.ukp.informatik.tu-darmstadt.de/thakur/BEIR/datasets/{}.zip".format(dataset)
if not os.path.exists(f"datasets/"):
os.makedirs(f"datasets/")
if not os.path.exists(f"datasets/{dataset}"):
url = "https://public.ukp.informatik.tu-darmstadt.de/thakur/BEIR/datasets/{}.zip".format(dataset)
data_path = util.download_and_unzip(url, "datasets")
data_path = f"datasets/{dataset}"
corpus, queries, qrels = GenericDataLoader(data_folder=data_path).load(split="test")
</code>
<code>
try:
if use_ft:
fasttext_model = fasttext.load_model(path_ft)
else:
fasttext_model = None
except:
print('Model not found')
if use_ft:
print('Downloading model...')
fasttext.util.download_model('en', if_exists='ignore') # English
fasttext_model = fasttext.load_model('cc.en.300.bin')
print('Reducing model...')
if vector_dimension != 300:
fasttext.util.reduce_model(fasttext_model, vector_dimension)
print('Saving model...')
if path_ft != '':
fasttext_model.save_model(path_ft)
fasttext_model.save_model(f'cc.en.{vector_dimension}.bin')
print('Model saved.')
else:
fasttext_model = None
</code>
<code>
if load_cleaned_corpus == '':
cleaned_corpus = corpus_processing.preprocess_corpus_dict(corpus)
if save_cleaned_corpus != '':
corpus_processing.save_processed_corpus(cleaned_corpus, save_cleaned_corpus)
else:
cleaned_corpus = pd.read_csv(load_cleaned_corpus)
cleaned_corpus = {cleaned_corpus['doc_id'][i]:cleaned_corpus['text'][i] for i in range(len(cleaned_corpus))}
if remove_original_corpus:
corpus = None
</code>
<code>
if os.path.exists(load_vectors):
embeddings = pd.read_csv(load_vectors, sep=' ',na_values=[''], keep_default_na=False, index_col=0).dropna()
for i in list(embeddings.columns)[1:]:
embeddings[i] = embeddings[i].astype(float)
else:
embeddings = vector_creation.create_vectors(cleaned_corpus, vector_dimension, path_to_save_vectors=load_vectors, path_to_save_model=path_to_save_model, epochs = 5, model = 'skipgram')
</code>
<code>
scores = {}
retriever = retriever_model.UCFIRe(embeddings, fasttext_model,n_neighbors = best_n_neighbors, alpha=best_alpha, thresh = best_thresh, metric = metric, k1 = k1, b = b, thresh_prob=thresh_prob)
retriever.fit(cleaned_corpus, is_clean=True, knn_method=knn_method)
retriever_okapi = EvaluateRetrieval(retriever, score_function="cos_sim") # or "dot" if you wish dot-product
results_okapi = retriever_okapi.retrieve(retriever.tokenized_corpus, queries)
# Evaluate the model (implement your own evaluation logic, e.g., compute mean reciprocal rank)
scores = retriever_okapi.evaluate(qrels, results_okapi, retriever_okapi.k_values) # Replace this with your evaluation metric
if save_scores != '':
with open(save_scores, 'w') as f:
f.write(str(scores))
print(scores)
</code>
### Results without handling missing words
<code>
retriever.switch_fasttext_model(None)
retriever_okapi = EvaluateRetrieval(retriever, score_function="cos_sim") # or "dot" if you wish dot-product
results_okapi = retriever_okapi.retrieve(retriever.tokenized_corpus, queries)
# Evaluate the model (implement your own evaluation logic, e.g., compute mean reciprocal rank)
scores = retriever_okapi.evaluate(qrels, results_okapi, retriever_okapi.k_values) # Replace this with your evaluation metric
scores
</code>
# Make a research
<code>
n_doc = 5 # number of documents to retrieve
query = {list(queries.items())[0][0]:list(queries.items())[0][1]}
print(query)
</code>
<code>
results = retriever.search(cleaned_corpus, query, n_doc, 'cos_sim') # example of a search
results
</code>
<code>
for quer_id in list(results.keys()):
print(f'Query: {queries[quer_id]}')
for doc_id in list(results[quer_id].keys()):
print('\n')
print(f'\tDocument: {corpus[doc_id]}')
print(f'\tScore: {results[quer_id][doc_id]}')
print('\n')
</code>
|
{
"filename": "main_1.ipynb",
"repository": "Limekaaa/UC-FIRe",
"query": "transformed_from_existing",
"size": 29154,
"sha": ""
}
|
# reddit_dataset_process_2.ipynb
Repository: janetzhong/Saved-You-A-Click-CS224N
<code>
from transformers import AutoModelForQuestionAnswering, AutoTokenizer, pipeline
import pandas as pd
import torch
</code>
<code>
model_name = "deepset/roberta-base-squad2"
</code>
<code>
nlp = pipeline('question-answering', model=model_name, tokenizer=model_name)
QA_input = {
'question': 'Why is model conversion important?',
'context': 'The option to convert models between FARM and transformers gives freedom to the user and let people easily switch between frameworks.'
}
res = nlp(QA_input)
</code>
<code>
QA_input = {
'question': 'Why is model conversion liked?',
'context': 'I am as asdf d it is cool'
}
res = nlp(QA_input)
res['answer']
</code>
<code>
question = 'Why is model conversion liked?'
text = 'I am as asdf d it is cool'
input_ids = tokenizer.encode(question,text)
</code>
<code>
tokens = tokenizer.convert_ids_to_tokens(input_ids)
</code>
<code>
output = model(torch.tensor([input_ids]))
</code>
<code>
tokens
</code>
<code>
answer_start = torch.argmax(output.start_logits)
answer_end = torch.argmax(output.end_logits)
if answer_end >= answer_start:
answer = " ".join(tokens[answer_start:answer_end+1])
else:
print("I am unable to find the answer to this question. Can you please ask another question?")
print("\nQuestion:\n{}".format(question.capitalize()))
print("\nAnswer:\n{}.".format(answer.capitalize()))
</code>
<code>
model = AutoModelForQuestionAnswering.from_pretrained(model_name)
tokenizer = AutoTokenizer.from_pretrained(model_name)
</code>
<code>
from transformers import TrainingArguments
training_args = TrainingArguments("test_trainer")
</code>
<code>
training_args
</code>
<code>
from transformers import Trainer
</code>
<code>
trainer = Trainer(model=model, args=training_args, train_dataset=imdb_dataset['train'], tokenizer=tokenizer)
</code>
<code>
trainer.train()
</code>
<code>
data = pd.read_pickle('Documents/CS224N/Saved-You-A-Click-CS224N/data_full_pandas.pkl')
</code>
<code>
i = 24
QA_input = {
'question': 'Why are new plant burgers not for vegans?',#data['teaser'][i],
'context': data['article'][i]
}
</code>
<code>
res = nlp(QA_input)
res
</code>
<code>
context = 'As fears over the coronavirus outbreak spread, thousands of Americans are clamoring to buy face masks in an effort to protect themselves, sending prices soaring and leading manufacturers like 3M to ramp up production. However, experts say stocking up on face masks is actually misguided — and there\'s a much simpler thing you could be doing right now to protect yourself.\n\nThere\'s a lot the general public likely doesn\'t realize about these masks — namely, that they are not the best way to prevent the spread of coronavirus.\n\nWearing a mask is more for people already showing symptoms of coronavirus and their caregivers than for people trying to prevent it\n\nThe Centers for Disease Control and Prevention said it "does not recommend that people who are well wear a facemask to protect themselves from respiratory diseases, including COVID-19," referring to the disease caused by the new coronavirus. Rather, experts caution that putting on a face mask without proper fitting and training could actually increase your risk.\n\n"If it\'s not fitted right, you\'re going to fumble with it," explained Health and Human Services Secretary Alex Azar before a House Appropriations subcommittee on Wednesday. "You\'re going to be touching your face, which is the No. 1 way you\'re going to get disease, is unclean hands touching your face."\n\nOn the other hand, if you are already coughing and showing symptoms of possible coronavirus illness, that\'s when wearing a mask could be helpful for protecting those around you.\n\n"The data on the effectiveness of masks for preventing respiratory virus infections is not very clear, " explains Dr. Andrew Stanley Pekosz of Johns Hopkins\' Bloomberg School of Public Health. "The best data suggests that if you are ill and showing symptoms, wearing a mask can reduce the chances that you spread the virus to others."\n\nCloth surgical masks are not helpful at all\n\nThe common surgical mask you might be picturing in your head will not help you at all, Pekosz said.\n\nA type called an N95 respirator mask, if properly fitted, can block large-particle droplets that may contain germs, but the FDA warns they cannot filter out "very small particles in the air that may be transmitted by coughs [or] sneezes."\n\n"An N95 mask is the one that is most practical," Pekosz tells CBS News. "It stops 95% of particles of a certain size. ... There is a N99 mask, which blocks 99% of particles, but that mask is difficult to wear for long periods of time because it is hard to breathe through it."\n\nRespirator masks are more expensive. The FDA also notes they are not designed to fit children or people with facial hair.\n\nEven a good face mask isn\'t enough\n\n"Masks shouldn\'t be considered to be the sole item that can protect you from infection, but it can be one of several things that can help you stay uninfected," said Pekosz.\n\n"Wash your hands frequently. Practice social distancing — stay 5 feet away from people to avoid being close enough to be exposed to respiratory droplets from that person. More specific guidance will be given by the CDC soon, but those two things should be practiced by people on a daily basis to reduce the spread of respiratory viruses."\n\nAnd he adds, "Get a flu shot — influenza has killed over 16,000 Americans this year and is still causing disease across the U.S."\n\nYou have to change masks every few hours\n\nIf you do go the mask route in spite of expert advice, it\'s important to note that face masks have a very specific lifespan. While there are some with longer lifespans or that have replaceable filters, the most common face masks on the market are disposable and single use. Each one of those is only good for a few hours.\n\n"You want to change masks every few hours to make sure that they are functioning properly and aren\'t getting contaminated with virus particles on the outside," Pekosz tells CBS News. "It\'s not like putting one on protects you. One has to follow specific procedures to ensure you are using them effectively."\n\nBuying face masks for personal use could cause a shortage at hospitals\n\n"There is a limited supply of masks and while companies are increasing their production, demand is increasing at a very high rate," cautions Pekosz. "There will most likely be shortages of personal protective equipment at medical institutions and this may in part be driven by supplies being purchased by the general public. Emergency preparedness efforts will address supply chains, but there really is no reason for the general public to purchase large numbers of N95 masks."\n\nAmerica\'s largest face mask manufacturer, Prestige Ameritech, is a small business based in Texas with only 100 employees. And while they have no problem fulfilling America\'s normal demand for face masks and respirators, they are now struggling to keep up.\n\nMike Bowen, the company\'s executive vice president, told CBS News that they now field orders of up to 100 million face masks and respirators a day. He also noted that while the company does not ship its products internationally, in the last 30 days it has sold between 1 million and 2 million masks to buyers who then sent them to others in China and Hong Kong.\n\nThis huge spike in personal orders is precisely what experts fear will cause a dangerous inventory shortage in American hospitals — a shortage that is entirely avoidable, given that there are no proven benefits to the general public wearing masks.\n\nThe best way to prevent coronavirus: Wash your hands\n\nThe right way to wash your hands\n\nExperts say washing your hands is the best way to prevent the spread of infectious illnesses like coronavirus. That\'s because one of the most common ways infections spread is when people touch a contaminated surface and then touch their mouth or nose.\n\nWash your hands frequently and thoroughly. CBS News chief medical correspondent Dr. Jon LaPook points out that it\'s especially important to make sure that you scrub the soap into your fingertips because they are simultaneously the part of the hand most often neglected and the part of the hand most likely to touch your face and spread disease.\n\nSoap and water is far more effective than hand sanitizer. If you\'re using an alcohol-based hand sanitizer, you should make sure that it contains at least 60% alcohol.\n\nBeyond that, the CDC advises that, whenever possible, you should also avoid touching your eyes, nose and mouth with unwashed hands, avoid contact with sick people, cover your mouth when you cough and sneeze, and disinfect objects and surfaces frequently.'
</code>
<code>
dataset_dict2 = {
'question': ['what is your name?'],
'context': ['hello my name is Janet thank you'],
'answer': ['Janet'],
'answer_start': [0]
}
answer = dataset_dict['answer'][0]
context = ' Janet'
indicies = []
</code>
<code>
data = pd.read_pickle('Documents/CS224N/Saved-You-A-Click-CS224N/data_full_pandas.pkl')
</code>
<code>
data.shape
</code>
<code>
data = data.dropna()
</code>
<code>
list_of_articles= data['article'].values.tolist()
list_of_answers = data['answer'].values.tolist()
</code>
<code>
list_of_questions = data['teaser'].values.tolist()
list_of_contexts = data['article'].values.tolist()
</code>
<code>
list_of_answers2 = []
for a in list_of_answers:
if '.' in a:
for i in range(len(a)):
if a[i] =='.':
b = a[:i]
list_of_answers2.append(b)
break
else:
list_of_answers2.append(a)
</code>
<code>
answers_dict = []
i = 0
for a in list_of_answers:
d = {}
d['text']=[a]
d['answer_start'] = [ind2[i]]
answers_dict.append(d)
i+=1
</code>
<code>
answers_dict
</code>
<code>
ind = []
for i in range(0,len(list_of_articles)):
ind.append(findIndices(list_of_answers[i],list_of_articles[i]))
</code>
<code>
ind2 = []
for i in range(0,len(list_of_articles)):
ind2.append(findIndices(list_of_answers2[i],list_of_articles[i]))
</code>
<code>
num = 0
j = 0
for i in ind2:
if i!=-1:
num +=1
j+=1
print(num)
</code>
<code>
def findIndices(answer,context):
indicies = []
if answer in context:
for i in range(len(context)):
if answer == context[i:i+len(answer)]:
indicies.append(i)
else:
indicies.append(-1)
return indicies[0]
</code>
<code>
list_of_questions = data['teaser'].values.tolist()
list_of_contexts
</code>
<code>
dataset_dict = {
'question': list_of_questions,
'context': list_of_contexts,
'answer': answers_dict
}
</code>
<code>
# make reduced version of dataset_dict:
questions_reduced = []
contexts_reduced = []
answers_reduced = []
id_reduced = []
for i in range(len(list_of_questions)):
if answers_dict[i]['answer_start'][0]!=-1:
questions_reduced.append(list_of_questions[i])
contexts_reduced.append(list_of_contexts[i])
answers_reduced.append(answers_dict[i])
id_reduced.append(i)
dataset_dict_reduced = {
'question': questions_reduced,
'context': contexts_reduced,
'answer': answers_reduced,
'id': id_reduced
}
</code>
<code>
a1 = ['hello this is a fox']
b1 = ['hello this is a rabbit']
</code>
<code>
a = a1[0].split(' ')
b = b1[0].split(' ')
</code>
<code>
len(list(set(a) & set(b)))/max(len(a),len(b))
</code>
<code>
N = len(questions_reduced)
import numpy as np
</code>
<code>
inds = np.arange(0,N)
np.random.seed(42)
np.random.shuffle(inds)
</code>
<code>
# convert to numpy
questions_reduced_np = np.array(questions_reduced)
contexts_reduced_np = np.array(contexts_reduced)
answers_reduced_np = np.array(answers_reduced)
id_reduced_np = np.array(id_reduced)
</code>
<code>
questions_reduced_np_shuffled = questions_reduced_np[inds]
contexts_reduced_np_shuffled = contexts_reduced_np[inds]
answers_reduced_np_shuffled = answers_reduced_np[inds]
id_reduced_np_shuffled = id_reduced_np[inds]
</code>
<code>
train_size = 200
val_size = 30
test_size = 28
dataset_dict_train = {
'question': list(questions_reduced_np_shuffled[0:train_size]),
'context': list(contexts_reduced_np_shuffled[0:train_size]),
'answer': list(answers_reduced_np_shuffled[0:train_size]),
'id': list(id_reduced_np_shuffled[0:train_size])
}
dataset_dict_val = {
'question': list(questions_reduced_np_shuffled[train_size:train_size+val_size]),
'context': list(contexts_reduced_np_shuffled[train_size:train_size+val_size]),
'answer': list(answers_reduced_np_shuffled[train_size:train_size+val_size]),
'id': list(id_reduced_np_shuffled[train_size:train_size+val_size])
}
dataset_dict_test = {
'question': list(questions_reduced_np_shuffled[train_size+val_size:]),
'context': list(contexts_reduced_np_shuffled[train_size+val_size:]),
'answer': list(answers_reduced_np_shuffled[train_size+val_size:]),
'id': list(id_reduced_np_shuffled[train_size+val_size:])
}
</code>
<code>
dataset_test['id'] = [str(i) for i in dataset_test['id']]
</code>
<code>
dataset_val['id'] = [str(i) for i in dataset_val['id']]
</code>
<code>
dataset_train['id'] = [str(i) for i in dataset_train['id']]
</code>
<code>
with open('dataset_dict_train.pickle', 'wb') as handle:
pickle.dump(dataset_train, handle)
with open('dataset_dict_val.pickle', 'wb') as handle:
pickle.dump(dataset_val, handle)
with open('dataset_dict_test.pickle', 'wb') as handle:
pickle.dump(dataset_test, handle)
</code>
<code>
with open('dataset_dict_train.pickle', 'rb') as handle:
dataset_train = pickle.load(handle)
with open('dataset_dict_val.pickle', 'rb') as handle:
dataset_val = pickle.load(handle)
with open('dataset_dict_test.pickle', 'rb') as handle:
dataset_test = pickle.load(handle)
</code>
<code>
with open('dataset_dict_train.pickle', 'wb') as handle:
pickle.dump(dataset_dict_train, handle, protocol=pickle.HIGHEST_PROTOCOL)
with open('dataset_dict_val.pickle', 'wb') as handle:
pickle.dump(dataset_dict_val, handle, protocol=pickle.HIGHEST_PROTOCOL)
with open('dataset_dict_test.pickle', 'wb') as handle:
pickle.dump(dataset_dict_test, handle, protocol=pickle.HIGHEST_PROTOCOL)
</code>
<code>
with open('dataset_dict_test_reddit.pickle', 'rb') as handle:
dataset_reddit_test = pickle.load(handle)
</code>
<code>
answ = []
for i in range(len(dataset_reddit_test['answer'])):
answ.append(dataset_reddit_test['answer'][i]['text'][0])
len(answ)
</code>
<code>
df_reddit_test = pd.DataFrame()
df_reddit_test['question'] = dataset_reddit_test['question']
df_reddit_test['context'] = dataset_reddit_test['context']
df_reddit_test['answer'] = answ
df_reddit_test['answer_user'] = user_asnwer
df_reddit_test['id'] = dataset_reddit_test['id']
</code>
<code>
answ
</code>
<code>
user_asnwer
</code>
<code>
user_asnwer = ["It's guacomole.",
'Water.',
'“I don’t know,” he says',
"It's Natasha",
'"Sayonara, Baby"',
"They don't know the release date yet.",
'No opening credits.',
"They're just having fun",
'Cuttlefish eyes have well-developed depth perception and will react to their surroundings',
'"The RPC and NPC Belarus will participate as neutrals at the Beijing 2022 Paralympic Winter Games. They will compete under the Paralympic flag and not be included in the medal table."',
'Spirit of the North: Enhanced Edition is coming to Xbox Series X and Xbox Series S sometime in early 2021',
'A whale tail sculpture named “Marti” on a roof across from Henry Law Park Adventure Playground in Dover, NH.',
'78 degrees',
'The legislation would ensure men were penalised for ejaculating outside a vagina',
'it’s diet soda',
'It lived in the sea',
'Neither, they would recognize that they aren’t enemies, and team up.',
'Turn your body and your face away at a 45-degree angle and smile',
'...likely stems from his inability to admit that his instincts are ever wrong.',
"American carmakers don't try to sell in Japan",
"it's Starz.",
'Poop',
'Limitless',
'Drink water',
'She asked him to escort her to her JROTC ball',
'Breakups.',
'They were not that close, she was wrapped up in her own life.',
"The game was only listed at 19 on Famitsu's most wanted games list",
'October',
'"You should probably find a shelter that is made of thick brick and has no windows, kind of like a bomb shelter." It literally tells you to hide in a bomb shelter.',
'she washes towels after every use.',
'It was Mr Mime.',
'Tales From Earthsea',
'Ground beef, Kroger',
'Seek out and connect with people who can open doors.',
'It’s called interest rate.',
'In 1988 he made a joke that he would like to be reincarnated as "a deadly virus, to contribute something to solving overpopulation”',
'‘how are you?’',
'He said “Don’t tell me how to be funny.”',
"They don't know",
'His Wife',
'He has ruled against Trump and his allies in the past.',
"It's Chicago West",
"What was really exciting is that it is a new species that has never infected people before. It's a cattle worm that somehow jumped into a human.",
"You'd have $9,222.50",
'+60% more than their parents',
'"The style standards are a result of longstanding requirements that female reporters not only do their jobs, but “fulfill larger audience expectations of what women are supposed to look like”',
'(Variation of) Thanks.',
'Tea might affect how DNA is expressed and women drinking tea was "associated with epigenetic changes in 28 different gene regions known to interact with cancer or estrogen metabolism." [TIME]',
'Disney banned smoking in its films around 2007',
'The global fight against disease',
'I Disagree',
'Send his mother to Mars and bring her back alive.',
'Auto manufacturers must give the same tools to most third parties as they do with dealerships',
'Eat at least 2 cups of vegetables every day.',
'No.',
'Paddington 2',
'Any of them',
'“It’s not a hard no, but it’s not an eager yes either.”',
'The role is Jack Reacher',
'He dumpster dives.',
'Indiana, South Carolina, Tennessee, and Virginia',
'#1 is John Malone with 2.2 million acres',
'Mosquitoes',
'Before the end of the year.',
'Cargo House',
'“a bad call from a doctor or something”',
'Alabama, Georgia, Texas, Florida and Arkansas',
'eShop music',
'Recount your "love at first sight" moment.',
'Nien Nunb, the character who flew the Millennium Falcon with Lando Calrissian during the Death Star attack in Return of the Jedi',
'51, referring to the percent of voters that said they would prefer a Congress controlled by Democrats in 2021',
'Tub is only half full',
"It's Natasha",
'She had triplets',
'$2800',
'ai.type, by "ai.type LTD” ... reported to be “delivering millions of invisible ads and fake clicks [and] real user data about views, clicks and purchases to different ad networks.”',
'People are calling her a hypocrite for being a feminist and posing braless in "Vanity Fair"',
'No in-display fingerprint sensor',
'is giant ovarian cyst',
"He's retired",
'"People should know everything about each other before they get married"',
'Our teeth don’t fit because they evolved instead to match the longer jaw that would develop in a more challenging strain environment. Ours are too short because we don’t give them the workout nature expects us to.',
'35-44',
"It's called box breathing or four-square breathing. Here's how it works, 1. Breathe in for four seconds 2. Hold air in your lungs for four seconds 3. Exhale for four seconds 4. Hold your breath, lungs emptied, for four seconds.",
'"Johanna, 29, suffers from a rare auto-immune disease, which means her body has a life-threatening allergic reaction to almost everything and everyone. Including her husband."',
'Leslie Grantham, who played Dennis "Dirty Den" Watts',
'It is real. Cobb is not in a dream, he did make it back home to his family.',
"We don't know",
'She was the only competitor in her category.',
'Vitamins D and C.',
"If only you're conducting a driving lesson as an instructor.",
'She was Hannah Montana',
'The Samsung Galaxy Note 10',
'"give users a choice"',
'You can have too much of a good thing',
'Bonds',
'"Who are you?"',
'“Don’t forget the heart"',
'1st Jan at 9pm on Channel 4',
'"We’re not prepared to go in hot-zone extraction. That’s just not what we do. It was active fire, active shooting."',
'Vanderpump Rules',
'"Eclipse Headache"',
'Saffir-Simpson Hurricane Wind Scale Only Goes From 1 to 5',
'Eclipse.',
"The Octopus crept up to the man's boot. Then, it placed two tentacles on his boot.",
'Riding a horse, fast',
'Whopper is being removed from 2 for $5 menu',
'Use longer passwords',
'The word is "ouch"',
'"Marijuana is not medicine" [The Motley Fool]',
'It was for pocket watches.',
'Anatomical characteristics of their brains (size, shape)',
'Phishing',
'The number 222 bus was going to Tooting, but in real life it goes to Hounslow',
"It's a rock",
'Open the curtains',
'Investing in yourself',
'A 2020 study found that men who drank at least one cup of coffee per day were 15% less likely to experience hearing loss than men who drank less than a cup a day',
'Layers',
"It's Hal Jordan",
'Because with that move, Apple "is dispensing of the notion that it forces people into buying new models"',
'Howard The Duck',
'It is unclear',
'They have both matured and are on the same page',
'25,000 professionals signed "We, the undersigned mental health professionals, believe in our professional judgment that Donald Trump manifests a serious mental illness."',
'Too much variety, too many choices',
'Her name was Qur\'stylle, pronounced "Crystal".',
'Only if you exceed the storage limit for 2 whole years.',
'To break the Guinness World Record for largest underwater mermaid show',
'Totally normal',
'"Actually, you are not able to download Fortnite without Epic Games launcher"',
'Giving parents money',
'Looking after his family',
'"What\'s the deal with all these f***ing soap people?" - Caitlyn Jenner, "Oi!" - Jacqueline Jossa',
'"To get things done, you have to do"',
'Internet Explorer is a compatibility solution',
'Hillary Clinton',
"It's “The Lord of the Rings: The Rings of Power.”",
'MyFitnessPal',
'To get paid and get free products',
'It doesn’t as the difference lies in the type of games in which players earned their winnings.',
'Quote from Kevin James: “I think if they can use me to get their show made, and it’s a great show, God bless them, good for them.”',
'"We can reveal it looks exactly the same."',
'It was a piece of metal',
'Scots cannot apply - must be US citizen aged between 30 and 55 and fluent in both English and Russian.',
'No it won’t. It’s only making a “close approach”.',
'Switzerland.',
'A port of Skyward Sword',
'"The whole debacle could have been avoided if only the series had had in place a rule stating that, if a contestant can’t go on, they’ve gotta go. Done. Finito."',
'no laptops, no cellphones',
'Elizabeth Olsen',
'Sony’s Universe of Marvel Characters',
'"but"',
'A baby tapeworm in the brain.',
'"I haven\'t seen it completed. [...] What I\'ve seen of the film I really liked."',
'Study shows they get depressed and lethargic',
'water',
'in 2035',
'$2,800',
'first female royal to benefit from succession law change that ensures girls will not be overtaken by any future younger brothers',
'introspection',
'Upper atmosphere lightning',
'"Bondmaid," which means "a slave girl."',
'Alice in Tim Burton’s Alice in Wonderland',
'Using multiple exclamation points',
'$1636',
'"I have no special talents. I am only passionately curious." The trait is curiosity.',
'After 65, avoid wearing a scent that is too sweet.',
"Including real time statistics about a post's popularity, shares, and interest in its news feed algorithm",
'send his mother to Mars and brings her back alive.',
'18 Chinese makers of polyurethane foam insulation, which is generally used in construction.',
'North Dakota',
'Basically to have the guts to ask for something "Most people never pick up the phone and call. Most people never ask"',
'The second beverage service',
'$48 million',
'"Some people aren’t meant to be here a long time."',
'she had a "disorder caused by hepatitis C known as Type 2 mixed cryoglobulinemia."',
"It's not ready yet.",
'Andrew Yang',
'He complimented him about never turning in a bad performance.',
'Bike messenger',
'Loki is Bi',
'Wegmans',
'Go to bed a little smarter each day',
'[We know nothing about it, more research needs to be done]',
'Empathy',
'It mess with your metabolism that cause insulin intolerance, diabetes, and weight gain.',
"Billie Eilish Pirate Baird O'Connell",
'Pennsylvania, Georgia, Michigan, Wisconsin and Oregon',
'Ivanka Trump ‘We don’t want any more inexperienced Trumps in the White House’ [17 clicks]',
"They ran out of story ideas and didn't want to compromise the quality.",
'Spread a layer of mayonnaise on one slice of bread and peanut butter on the other. Press the sandwich together to serve.',
'Maybe Amy Klobuchar',
'From watching Peppa Pig',
'Gentrification',
"He's a Spirit",
'He still needs an "encroachment permit".',
"There wasn't enough space.",
'You must return a third stimulus check if it was mailed to someone who died before 2021.',
"22.08 $/h. It does not include things like paying off debts, homeownership, saving for your children's education or any other type of emergency fund.",
'Gravity is "emergent", not always there. Comes into existence from changes in microscopic bits of information in the structure of spacetime',
'Quote from article “Per the show rep, however, the crutches won’t show up on screen.”',
'No. The volume of vapour and particles released is far below what is considered harmful to your health.',
'r/the_donald and /pol/',
'XCloud.',
'Jay’s ex-wife DeDe',
'Warm water that usually stays deep in the ocean is coming closer to the surface, melting the ice from underneath',
'Increased creative collaboration between PlayStation and Sony Music that could lead to more licensed music in first party games',
'New Super Mario Bros U Deluxe',
'It was Brittney Spears [24 Clicks]',
"You can't eat whatever you want.",
'He just made friends with people in his class and worked harder. Literally says in the article "If you came to this article looking for a strategy on a near perfect GPA, I’m sorry to disappoint you, but I don’t have one."',
'No, it will hit the far side',
'Artificial intelligence, energy, or biosciences',
'"I am grateful and happy"',
'"Whisper"',
'Wailord comes back, in the Workout Eea, even if caught',
'`"Corona," the fictitious land where Rapunzel is confined in the Disney movie`',
'9 p.m.',
'Picture a calm scene and repeat the phrase “Don’t think” for 10 seconds',
'He didn’t have insurance',
'No cloud saves',
"It's a fangtooth snake-eel",
'A large nuclear exchange would not only kill millions of people and contaminate wast areas with radioactive fallout but potentially also have longer-term climatic effects.',
'From Mojang to Mojang Studios',
'Its the Vice President to congratulate NASA',
'Clickbait is a sensationalized headline that encourages you to click a link to an article, image, or video.',
'Polaris',
'New England Patriots or Green Bay Packers',
'She was filming another Netflix series “cursed” and had no time.',
'The holy trinity',
'"I am excited."',
'She watches Breaking Bad',
'He hires employees that “wake up every morning terrified.”',
'a 39-year-old UK web designer named James Linton.',
'Philip K Dick',
'Ellie Kemper auditioned, but wasn’t chosen.',
'the best place to pet a dog is under the chin',
"Fenty is Rihanna's last name",
'Crushed up KitKat wafers',
'in 2035',
'15 a combination of British security detail and Canadian Mountees.',
'Danville',
"It's Bob Dylan",
'If Obama were to defend his legacy it would only work to give Trump an enemy to attack and rile up his supporters.',
'"I don\'t knwo. He was probably yelling some shit."',
"it's the niece of his ex-wife",
'she asked about salary and benefits',
'Cleaning. (Irritation from chemicals, including ammonia, on mucous membranes lining airways is the key)']
</code>
<code>
df_reddit_test.to_csv('test_reddit2.csv')
</code>
<code>
df_reddit_test
</code>
<code>
with open('dataset_dict_train.pickle', 'rb') as handle:
dataset_dict = pickle.load(handle)with open('dataset.pickle', 'wb') as handle:
pickle.dump(dataset_dict_reduced, handle, protocol=pickle.HIGHEST_PROTOCOL)
</code>
<code>
import pickle5 as pickle
</code>
<code>
model = AutoModelForQuestionAnswering.from_pretrained(model_name)
tokenizer = AutoTokenizer.from_pretrained(model_name)
</code>
<code>
dataset_dict = dataset_dict_reduced
tokenized_examples = tokenizer(dataset_dict['question'],
dataset_dict['context'],
truncation="only_second",
#stride=128,
#max_length=384,
return_overflowing_tokens=True,
return_offsets_mapping=True,
padding='max_length')
sample_mapping = tokenized_examples["overflow_to_sample_mapping"]
offset_mapping = tokenized_examples["offset_mapping"]
# Let's label those examples!
tokenized_examples["start_positions"] = []
tokenized_examples["end_positions"] = []
tokenized_examples["id"] = []
inaccurate = 0
for i, offsets in enumerate(tqdm(offset_mapping)):
# We will label impossible answers with the index of the CLS token.
input_ids = tokenized_examples["input_ids"][i]
cls_index = input_ids.index(tokenizer.cls_token_id)
# Grab the sequence corresponding to that example (to know what is the context and what is the question).
sequence_ids = tokenized_examples.sequence_ids(i)
# One example can give several spans, this is the index of the example containing this span of text.
sample_index = sample_mapping[i]
answer = dataset_dict['answer'][sample_index]
# Start/end character index of the answer in the text.
print(answer['answer_start'])
start_char = answer['answer_start'][0]
end_char = start_char + len(answer['text'][0])
tokenized_examples['id'].append(dataset_dict['id'][sample_index])
# Start token index of the current span in the text.
token_start_index = 0
while sequence_ids[token_start_index] != 1:
token_start_index += 1
# End token index of the current span in the text.
token_end_index = len(input_ids) - 1
while sequence_ids[token_end_index] != 1:
token_end_index -= 1
# Detect if the answer is out of the span (in which case this feature is labeled with the CLS index).
if not (offsets[token_start_index][0] <= start_char and offsets[token_end_index][1] >= end_char):
tokenized_examples["start_positions"].append(cls_index)
tokenized_examples["end_positions"].append(cls_index)
else:
# Otherwise move the token_start_index and token_end_index to the two ends of the answer.
# Note: we could go after the last offset if the answer is the last word (edge case).
while token_start_index < len(offsets) and offsets[token_start_index][0] <= start_char:
token_start_index += 1
tokenized_examples["start_positions"].append(token_start_index - 1)
while offsets[token_end_index][1] >= end_char:
token_end_index -= 1
tokenized_examples["end_positions"].append(token_end_index + 1)
# assertion to check if this checks out
context = dataset_dict['context'][sample_index]
offset_st = offsets[tokenized_examples['start_positions'][-1]][0]
offset_en = offsets[tokenized_examples['end_positions'][-1]][1]
if context[offset_st : offset_en] != answer['text'][0]:
inaccurate += 1
</code>
<code>
from transformers import AutoTokenizer
model_checkpoint = "bert-base-cased"
tokenizer1 = AutoTokenizer.from_pretrained(model_checkpoint)
</code>
<code>
len(tokenized_examples['start_positions'])
</code>
<code>
tokenized_examples.keys()
</code>
<code>
train_dataset = raw_datasets["train"].map(
tokenized_examples,
batched=True,
remove_columns=raw_datasets["train"].column_names,
)
len(raw_datasets["train"]), len(train_dataset)
</code>
<code>
import tensorflow as tf
from transformers import TFAutoModelForQuestionAnswering
from tqdm.auto import tqdm
</code>
<code>
tokenized_examples.keys()
</code>
<code>
tf_train_dataset = tokenized_examples.to_tf_dataset(
columns=[
"input_ids",
"start_positions",
"end_positions",
"attention_mask",
"id",
],
collate_fn=data_collator,
shuffle=True,
batch_size=16,
)
</code>
<code>
def compute_metrics(start_logits, end_logits, features, examples):
example_to_features = collections.defaultdict(list)
for idx, feature in enumerate(features):
example_to_features[feature["example_id"]].append(idx)
predicted_answers = []
for example in tqdm(examples):
example_id = example["id"]
context = example["context"]
answers = []
# Loop through all features associated with that example
for feature_index in example_to_features[example_id]:
start_logit = start_logits[feature_index]
end_logit = end_logits[feature_index]
offsets = features[feature_index]["offset_mapping"]
start_indexes = np.argsort(start_logit)[-1 : -n_best - 1 : -1].tolist()
end_indexes = np.argsort(end_logit)[-1 : -n_best - 1 : -1].tolist()
for start_index in start_indexes:
for end_index in end_indexes:
# Skip answers that are not fully in the context
if offsets[start_index] is None or offsets[end_index] is None:
continue
# Skip answers with a length that is either < 0 or > max_answer_length
if (
end_index < start_index
or end_index - start_index + 1 > max_answer_length
):
continue
answer = {
"text": context[offsets[start_index][0] : offsets[end_index][1]],
"logit_score": start_logit[start_index] + end_logit[end_index],
}
answers.append(answer)
# Select the answer with the best score
if len(answers) > 0:
best_answer = max(answers, key=lambda x: x["logit_score"])
predicted_answers.append(
{"id": example_id, "prediction_text": best_answer["text"]}
)
else:
predicted_answers.append({"id": example_id, "prediction_text": ""})
theoretical_answers = [{"id": ex["id"], "answers": ex["answers"]} for ex in examples]
return metric.compute(predictions=predicted_answers, references=theoretical_answers)
</code>
<code>
context_1 = dataset_dict["context"][0]
question_1 = dataset_dict["question"][0]
inputs_1 = tokenizer(question_1, context_1)
tokenizer.decode(inputs_1["input_ids"])
</code>
<code>
model = AutoModelForQuestionAnswering.from_pretrained(model_name)
tokenizer = AutoTokenizer.from_pretrained(model_name)
</code>
<code>
inputs = "I'm excited to learn about Hugging Face Transformers!"
tokenized_inputs = tokenizer(inputs, return_tensors="pt")
</code>
<code>
from datasets import load_dataset, DatasetDict
</code>
<code>
tokenized_inputs
</code>
<code>
imdb_dataset = load_dataset("imdb")
</code>
<code>
from datasets import Dataset
</code>
<code>
imdb_dataset['train']
</code>
<code>
data2 = data.drop(['title','url'], axis=1)
</code>
<code>
data2.reset_index()
</code>
<code>
dataset = Dataset.from_pandas(data2,preserve_index=False)
</code>
<code>
dataset
</code>
<code>
DatasetDict(
train=imdb_dataset['train'].shuffle(seed=1111).select(range(128)).map(truncate),
val=imdb_dataset['train'].shuffle(seed=1111).select(range(128, 160)).map(truncate),
)
</code>
<code>
DatasetDict(
train=imdb_dataset['train'].shuffle(seed=1111).select(range(128)).map(truncate),
val=imdb_dataset['train'].shuffle(seed=1111).select(range(128, 160)).map(truncate),
)
</code>
<code>
outputs = model(**tokenized_inputs)
</code>
<code>
tokenized_dataset = dataset
for name in ['teaser','article','answer']:
tokenized_dataset = tokenized_dataset.map(
lambda example: tokenizer(example[name], padding=True, truncation=True),
batched=True,
batch_size=16
)
tokenized_dataset = tokenized_dataset.remove_columns([name])
</code>
<code>
tokenized_dataset[0:1]
</code>
<code>
num_epochs = 3
num_training_steps = 3 * len(train_dataloader)
optimizer = AdamW(model.parameters(), lr=5e-5, weight_decay=0.01)
lr_scheduler = get_linear_schedule_with_warmup(optimizer, num_warmup_steps=0, num_training_steps=num_training_steps)
best_val_loss = float("inf")
progress_bar = tqdm(range(num_training_steps))
for epoch in range(num_epochs):
# training
model.train()
for batch_i, batch in enumerate(train_dataloader):
output = model(**batch)
optimizer.zero_grad()
output.loss.backward()
optimizer.step()
lr_scheduler.step()
progress_bar.update(1)
# validation
model.eval()
for batch_i, batch in enumerate(eval_dataloader):
with torch.no_grad():
output = model(**batch)
loss += output.loss
avg_val_loss = loss / len(eval_dataloader)
print(f"Validation loss: {avg_val_loss}")
if avg_val_loss < best_val_loss:
print("Saving checkpoint!")
best_val_loss = avg_val_loss
torch.save({
'epoch': epoch,
'model_state_dict': model.state_dict(),
'optimizer_state_dict': optimizer.state_dict(),
'val_loss': best_val_loss,
},
f"checkpoints/epoch_{epoch}.pt"
)
</code>
<code>
outputs
</code>
<code>
res
</code>
|
{
"filename": "reddit_dataset_process_2.ipynb",
"repository": "janetzhong/Saved-You-A-Click-CS224N",
"query": "transformed_from_existing",
"size": 376436,
"sha": ""
}
|
# lines_crispr_preprocessing.ipynb
Repository: nalinimsingh/mm-cell
# Preprocessing CCLE RNA-seq and CRISPR knockout data
* Author: Eshika Saxena
* Objective: Combine RNA-seq and CRISPR knockout data based on IDs
## Load libraries
<code>
import os
import numpy as np
import pandas as pd
</code>
## Read data
<code>
data_dir = '/Users/eshikasaxena/Documents/MLHC/mm-cell_lines/data/'
</code>
<code>
rnaseq = pd.read_csv(os.path.join(data_dir,'CCLE_expression.csv'))
rnaseq = rnaseq.rename(columns={'Unnamed: 0': 'DepMap_ID'})
</code>
<code>
crispr = pd.read_csv(os.path.join(data_dir,'Achilles_gene_effect.csv'))
</code>
## Data selection
* Only keep IDs that are in both RNA-seq and CRISPR knockout.
* Restrict knockout cell viability scores to 13 genes of interest.
* Only keep RNA seq cols that map to Ensembl IDs.
<code>
ids = list(set(crispr.DepMap_ID).intersection(set(rnaseq.DepMap_ID)))
print(len(ids))
</code>
<code>
# genes of interest
genes = [col for col in crispr.columns if 'PSMB' in col or 'IKZF1' in col or 'IKZF3' in col]
print(len(genes))
print(genes)
</code>
<code>
crispr = crispr[crispr.DepMap_ID.isin(ids)][genes + ['DepMap_ID']]
print(len(crispr))
print(len(crispr.columns))
</code>
<code>
crispr.head()
</code>
<code>
mapping = pd.read_csv('../utils/Ensembl_HGNC_map_042421.csv')
cols_to_keep = ['DepMap_ID'] + list(mapping.HGNC_ID)
rnaseq = rnaseq[cols_to_keep]
rnaseq = rnaseq[rnaseq.DepMap_ID.isin(ids)]
print(len(rnaseq))
</code>
<code>
rnaseq.head()
</code>
## Merge and save RNA-seq and knockout data
<code>
merged = rnaseq.merge(crispr, on='DepMap_ID')
print(len(merged))
</code>
<code>
merged.head()
</code>
<code>
save_data = False
if save_data:
merged.to_csv(os.path.join(data_dir,'rnaseq_crispr_merged.csv'), index=False)
</code>
|
{
"filename": "lines_crispr_preprocessing.ipynb",
"repository": "nalinimsingh/mm-cell",
"query": "transformed_from_existing",
"size": 33220,
"sha": ""
}
|
# fastqe-notebook_1.ipynb
Repository: JasonJWilliamsNY/fastqe-jupyterlab-cyverse-vice
# Fun Introductory Command Line Exercise: Next Generation Sequencing (NGS) Quality Analysis with Emoji
\* this activity was adapted from code and slides developed by Andrew Lonsdale ([@LonsBio](https://twitter.com/lonsbio?lang=en)) at Melbourne University. Here’s a [link](https://www.youtube.com/watch?v=WywQ6a3uQ5I&feature=youtu.be&t=33m18s) to a Lightning Talk that Andrew gave in 2017 about FASTQE.
**Goals**: Use basic command line coding to:
- Introduce students to writing basic command line scripts
- Analyze & assess the quality of FASTQ formatted NGS data
- Trim/filter low quality reads in FASTQ files
The 1st step of any Next Generation Sequencing (NGS) analysis pipeline is checking the quality of the raw sequencing reads in each FASTQ formatted file. If the sequence quality is poor, then your resulting downstream analysis will be inaccurate and misleading. FastQC is a popular software used to provide an overview of basic quality metrics for NGS data. In this lesson, you will use an even more universal form of communication to analyze FASTQ files, THE EMOJI 😻😻😻.
**Technical requirements/limitations**:
- The software can be installed on computers running Mac OS or Linux. Windows does not support the use of emoticons 😟😱😿. - If using your own Mac computer, you need to install Anaconda on your machine (see pre-class assignment https://bit.ly/2RxKApp; ~20 min to install). Anaconda is a Python-based data processing & scientific computing platform with built in third-party libraries.
- Lastly, the FASTQE program is limited to short read NGS data of 500bp or less.
Like the popular FastQC software, FASTQE can be used to analyze the quality of FASTQ file data whether it’s from a genome sequencing project, an RNA-seq project, a ChIP-seq project, etc. Here’s a brief background on the in class metagenomics project that Dr. Enke’s Bio 481 Genomics class is collecting data for. Garter snakes excrete sexually dimorphic pheromones to attract a mate. The hypothesis of their experiment is that male and female garter snakes host unique microbial communities in their mouths, cloacae and musk glands that contribute to sexually dimorphic bioengineering of these pheromone molecules. Figure 1 provides an overview of their 16S metagenomics analysis pipeline. For this lesson though, all you need are the FASTQ files. Feel free to substitute your own favorite FASTQ files for this activity if you like.

Figure 1. Overview of the in class metagenomics project. Using a saline swabbing technique, microbial samples were collected from garter snake tissues in class (A). Swabs were placed in sterile tubes to release collected microbes & DNA was extracted for downstream analysis (B). Barcoded primers were used to PCR amplify the microbial 16S ribosomal DNA repeat genes for each sample followed by Illumina sequencing of PCR amplicons (C-D). The DNA Subway Purple Line web-based software can be used to analyze FASTQ data files generated from Illumina sequencing to reveal the microbial population of our swabs (E). Garter snakes were provided by Dr. Rocky Parker in the JMU Department of Biology (A; yellow shirt).
As previously discussed, FASTQE is a program that analyzes FASTQ files & reads out an emoji output as an indicator of the sequence’s quality in the file. So a high quality read may look like this 😃, while this symbol 💩 indicates... well you get the idea.
In class Assignment: Working in your lab groups, take turns operating the command line to analyze NGS fastq file data using FASTQE and another program called FASTP. All of the instructions & explanations are listed below. Create a new MS Word or GoogleDoc file and provide feedback wherever you see red text. If you get stuck, ask for help! Turn in this document at the end of the activity for your group’s graded assignment. Make sure to rotate turns typing commands.
## Student #1 Download FastQ files and run `fastqe`
Jupyter allows you to run commands by selecting a cell and then click the play button or Cntrl+Enter. For example running the next cell executes the `pwd` (print working directory) command, which will tell you what directory this notebok is located in.
<code>
pwd
</code>
### Question 1: If you’ve printed a path that doesn’t make sense (i.e. the directory you navigated to is the incorrect directory) how would you go back to the previous directory? (hint, it includes the change directory command)
- Hint, type your commands in the cell below to see how the `cd` (change directory) command works.
## Answer to question 1: (Double click on this cell to edit)
Your answer...(Run this cell [play button] or Cntrl+Enter to render this cell in Markdown)
### Step 1
Using the `wget` command, download the compressed fastq file here: https://bit.ly/2FbODRS (this is 1 file with the .zip extension that unzips into 2 .fastq files).
**NOTE THIS URL WILL HAVE TO BE FIXED - I IMPORTED THESE FILES FOR THE SAKE OF TESTING**
The `wget` command we will use has three components
**Usage**: wget -O [filename][URL]
- `wget` the name of the program
- `-O` the `-O` is an option we can pass to the `wget` program, this option let's us choose the name we want our file to be saved as, in this case `fastq.zip`.
- URL in this the URL you want to download a file from
Type `wget` then a space, `-O fastq.zip` then the URL you are downloading from
<code>
wget -O fastq.zip https://bit.ly/2FbODRS
</code>
### Step 2
In the next cell, use the `unzip` command to unzip the downloaded `fastq.zip`
**Usage**: `unzip` [file to unzip]
<code>
unzip fastq.zip
</code>
## Question 2: What’s the purpose of using a zipped file?
## Answer to question 2: (Double click on this cell to edit)
Your answer...(Run this cell [play button] or Cntrl+Enter to render this cell in Markdown)
### Step 3
In the next cell, use the `ls` (list files) command to verify you have unziped two files: `female_musk2.fastq` and `female_oral2.fastq`
**Usage**: `ls` [directory] (list contents of a directory - if left blank, will display for the current directory, if a wildcard [e.g. \*.file-extension] is provided, will list all the files with the given file extension)
Use the command `ls` but pass `*.fastq` to directory
<code>
ls *.fastq
</code>
### Step 4
In the next cell, run the `fastqe` program to generate your emoji fastq report
**Usage**: `fastqe` [fastq-file] (run the `fastqe` program. If a wildcard [e.g. \*.fastq] is provided, `fastqe` will run on all the fastq files in the current working directory.
<code>
fastqe *.fastq
</code>
## Question 3: What are the advantages and disadvantages to using the command fastqe *.fastq rather than fastqe sample.fastq?
## Answer to question 3: (Double click on this cell to edit)
Your answer...(Run this cell [play button] or Cntrl+Enter to render this cell in Markdown)
## Student #2 `fastqe` help
Notice that 1 of your files (female_oral2) seems to have lower quality than the other based on the Emoji readout. Let’s look more closely to see how bad the data is.
### Step 5
Open the FASTQE help page to view the “optional arguments”, these are all of the options and setting for the program.
To get the help info for `fastqe` (and many other command line programs) pass the `--help` option to the `fastqe` program instead of a filename or wildcard (as in step 5)
<code>
fastqe --help
</code>
## Question 4: Which optional argument will show the version # of FASTQE?
## Answer to question 4: (Double click on this cell to edit)
Your answer...(Run this cell [play button] or Cntrl+Enter to render this cell in Markdown)
### Step 6
Add the `--scale` option to the `fastqe` command to view the Phred score associated with each emoji in your output. Try this just for the `female_oral2` file.
<code>
fastqe --scale female_oral2.fastq
</code>
## Question 5: Phred score of ≤20 is considered a poor quality base call. How many poor quality base calls are at the 3’ end of this read?
## Answer to question 5: (Double click on this cell to edit)
Your answer...(Run this cell [play button] or Cntrl+Enter to render this cell in Markdown)
## Student #3 `fastp`
Let’s use another program called Fastp to get a more conventional readout of the .fastq file data. Fastp is similar to the FastQC program we previously used, however, it also has a trimming tool to cut out or filtering the low quality sequences in our file.
### Step 7
Run `fastp` on the lower quality `female_oral2.fastq` file
**Usage**:
- `fastp` is the name of software that will check the quality of the fastq file
- `-i [input.fastq]` -i option specifies the input file for `fastp`
- `-o [ouput.fastq]` -o option specifies the ouput file for `fastp`
Write a command using `female_oral2.fastq` as your input and `out.female_oral2.fastq` as your output
<code>
fastp -i female_oral2.fastq -o out.female_oral2.fastq
</code>
## Step 8
You should now have 3 new files in your fastp folder
1. .html file (this is your QC report)
2. .json file (ignore this for now)
3. trimmed fastq file (out.female_oral2.fastq)
Click on the `fastp.html` file in the Jupyter menu on the left to examine this report
**Note**: Click on **Trust HTML** on the top of the HTML report tab to reveal graphs that may be hidden until you provide this authorization.
## Question 6: From the “Summary” data in your HTML fastp report, how many reads are in this FASTQ file before and after filtering?
## Answer to question 6: (Double click on this cell to edit)
Your answer...(Run this cell [play button] or Cntrl+Enter to render this cell in Markdown)
## Question 7: How do the before and after plots compare?
## Answer to question 7: (Double click on this cell to edit)
Your answer...(Run this cell [play button] or Cntrl+Enter to render this cell in Markdown)
## Step 9
Use the `out.female_oral2.fastq` file to rerun `fastqe`
<code>
fastqe out.female_oral2.fastq
</code>
## Question 8: How do the before and after plots compare?
## Answer to question 8: (Double click on this cell to edit)
Your answer...(Run this cell [play button] or Cntrl+Enter to render this cell in Markdown)
## Question 9: Which tool (fastqe or fastp) did you find easier to use?
## Answer to question 9: (Double click on this cell to edit)
Your answer...(Run this cell [play button] or Cntrl+Enter to render this cell in Markdown)
## Question 10: Which tool (fastqe or fastp) do you think is more a more reliable research grade tool?
## Answer to question 10: (Double click on this cell to edit)
Your answer...(Run this cell [play button] or Cntrl+Enter to render this cell in Markdown)
To sum up, you just analyzed Illumina FASTQ data quality using Emoji output. You then filtered out low quality sequences & output before & after QC plots. You did all of that on the command line, congrats!
|
{
"filename": "fastqe-notebook_1.ipynb",
"repository": "JasonJWilliamsNY/fastqe-jupyterlab-cyverse-vice",
"query": "transformed_from_existing",
"size": 32261,
"sha": ""
}
|
# llamaindex_2_dump_pubmed_to_qdrant_1.ipynb
Repository: forrestzhang/learn
<code>
import json
import gzip
import qdrant_client
from llama_index.core import SimpleDirectoryReader
from llama_index.embeddings.huggingface import HuggingFaceEmbedding
from llama_index.core import Settings
from llama_index.llms.ollama import Ollama
from llama_index.core.node_parser import (
SentenceSplitter,
SemanticSplitterNodeParser,
)
from llama_index.vector_stores.qdrant import QdrantVectorStore
from llama_index.core import VectorStoreIndex
from llama_index.core.response.notebook_utils import display_source_node
from llama_index.core import StorageContext
from llama_index.core import Document
</code>
<code>
Settings.llm = Ollama(model="llama3")
Settings.embed_model = HuggingFaceEmbedding("BAAI/bge-base-en-v1.5")
splitter = SemanticSplitterNodeParser(
buffer_size=1, breakpoint_percentile_threshold=95, embed_model=Settings.embed_model
)
# also baseline splitter
base_splitter = SentenceSplitter(chunk_size=512)
</code>
<code>
client = qdrant_client.QdrantClient(
# you can use :memory: mode for fast and light-weight experiments,
# it does not require to have Qdrant deployed anywhere
# but requires qdrant-client >= 1.1.1
# location=":memory:"
# otherwise set Qdrant instance address with:
# url="http://<host>:<port>"
# otherwise set Qdrant instance with host and port:
host="localhost",
port=6333
# set API KEY for Qdrant Cloud
# api_key="<qdrant-api-key>",
)
vector_store = QdrantVectorStore(client=client, collection_name="pubmed_demo")
storage_context = StorageContext.from_defaults(vector_store=vector_store)
</code>
<code>
documents = []
jsonfile = "../data/pubmed_cis_json/pubmed24n1073_cis.json.gz"
</code>
<code>
with gzip.open(jsonfile) as f:
data = json.load(f)
</code>
<code>
for pmid in data:
#print(pmid)
abstract = data[pmid]["abstract"]
journal = data[pmid]["journal"]
pubdate = data[pmid]["pubdate"]
document = Document(text=abstract,
metadata = {"pmid": pmid, "journal": journal, "pubdate": pubdate})
documents.append(document)
</code>
<code>
index = VectorStoreIndex.from_documents(
documents,
storage_context=storage_context,
)
</code>
<code>
query_engine = index.as_query_engine()
</code>
<code>
response = query_engine.query(
"how to identify cis-regulatory elements in the genome?"
)
</code>
<code>
print(str(response))
</code>
|
{
"filename": "llamaindex_2_dump_pubmed_to_qdrant_1.ipynb",
"repository": "forrestzhang/learn",
"query": "transformed_from_existing",
"size": 5714,
"sha": ""
}
|
# WS_Western_MechEng_Core_and_Electives_AllYears.ipynb
Repository: ReadyLab-UToronto/Keyword-Matching-for-Canadian-Mechanical-Engineering-Programs
<code>
from bs4 import BeautifulSoup as soup
from urllib.request import urlopen as ureq
from selenium import webdriver
import time
import re
</code>
<code>
url = "https://www.westerncalendar.uwo.ca/Modules.cfm?ModuleID=21289&SelectedCalendar=Live&ArchiveID="
</code>
<code>
chrome_options = webdriver.ChromeOptions()
chrome_options.add_argument('--ignore-certificate-errors')
chrome_options.add_argument('--incognito')
chrome_options.add_argument('--headless')
driver = webdriver.Chrome("C:\\Users\\jerry\\Downloads\\chromedriver", options=chrome_options)
</code>
<code>
driver.get(url)
</code>
# 1. Collect all course link texts for webdriver to click on
<code>
page_soup = soup(driver.page_source, 'lxml')
</code>
<code>
first_year_container = page_soup.find("div", {"id": "AdmissionRequirements"})
first_year_container
</code>
<code>
first_year_links = first_year_container.findAll("a")
first_year_links
</code>
<code>
first_year_links = [link.text for link in first_year_links]
first_year_links
</code>
<code>
first_year_links = list(dict.fromkeys(first_year_links))
print(len(first_year_links))
first_year_links
</code>
<code>
other_years_container = page_soup.find("div", {"class": "moduleInfo"})
other_years_links = other_years_container.findAll("a")
other_years_links = [link.text for link in other_years_links]
other_years_links
</code>
<code>
len(other_years_links)
</code>
<code>
link_texts = first_year_links + other_years_links
len(link_texts)
</code>
# 2. Test run - try to scrape the first course
<code>
driver.find_element_by_link_text(link_texts[0]).click()
driver.current_url
</code>
<code>
page_soup = soup(driver.page_source, 'lxml')
course_info = page_soup.find("div", {"id": "CourseInformationDiv"})
course_info
</code>
<code>
code = course_info.find("h2").text
code
</code>
<code>
name = course_info.find("h3").text
name
</code>
<code>
desc = course_info.findAll("div", {"class": None, "id": None})[1].text.strip()
desc
</code>
# 3. Automation script to scrape all courses (and inspect at the same time)
<code>
from selenium.common.exceptions import NoSuchElementException
course_codes = []
course_names = []
course_descs = []
counter = 0
driver.get("https://westerncalendar.uwo.ca/Modules.cfm?ModuleID=21289&SelectedCalendar=Live&ArchiveID=")
for link_text in link_texts:
#go to course page
try:
link = driver.find_element_by_link_text(link_text)
except NoSuchElementException:
print("no link for {}".format(link_text))
continue
time.sleep(2)
link.click()
time.sleep(2)
#scrape course info
page_soup = soup(driver.page_source, 'lxml')
course_info = page_soup.find("div", {"id": "CourseInformationDiv"})
course_code = course_info.find("h2").text
course_name = course_info.find("h3").text
course_desc = course_info.findAll("div", {"class": None, "id": None})[1].text.strip()
course_codes.append(course_code)
course_names.append(course_name)
course_descs.append(course_desc)
print("Scraped ", course_codes[-1], course_names[-1], course_descs[-1])
counter += 1
#go to course list page
driver.back()
time.sleep(2)
print("Finished scraping {} courses".format(counter))
</code>
<code>
len(course_descs)
</code>
# 4. Write to CSV
<code>
import pandas as pd
df = pd.DataFrame({
"Course Number": course_codes,
"Course Name": course_names,
"Course Description": course_descs
})
df
</code>
<code>
df.to_csv('Western_MechEng_Core_and_Elective_(AllYears)_Courses.csv', index = False)
</code>
|
{
"filename": "WS_Western_MechEng_Core_and_Electives_AllYears.ipynb",
"repository": "ReadyLab-UToronto/Keyword-Matching-for-Canadian-Mechanical-Engineering-Programs",
"query": "transformed_from_existing",
"size": 58131,
"sha": ""
}
|
# ST_fig1d_plotUMAP.ipynb
Repository: astrid12345/Visium
<code>
########################################################################
# Author : A. Alsema
# Date : May-July 2021
# Dataset : Visium Spatial Transcriptomics for MS lesions
# Purpose : plot a UMAP with the clusters
# Required input: "3.WM.clustered.res0.2.rds"
# Output : figure 1d, UMAP with the clusters in custom colors.
#########################################################################
</code>
<code>
rm(list = ls())
library(Seurat)
library(hdf5r)
library(ggplot2)
library(patchwork)
library(future)
library(dplyr)
library(RColorBrewer)
options(future.globals.maxSize = 3000 * 1024^2)
</code>
<code>
# load data
res = 0.2
datasets <- readRDS(file = paste0("./RData/seurat/3.WM.clustered.res", res, ".rds"))
levels(datasets$Group)
</code>
<code>
ggplot.theme <- theme(aspect.ratio = 1,
text = element_text(hjust = 0.5, face = "plain", size = (9)),
plot.title = element_text(hjust = 0.5, face = "plain", size = (10)),
axis.title.x = element_text(face = "plain", size = (12)),
axis.title.y = element_text(face = "plain", size = (12)),
axis.text = element_text(face = "plain", size = (12), colour = "black"),
# axis.text.x = element_text(angle = 90, vjust=0.5, hjust=1),
plot.subtitle = element_text(hjust = 0.5),
panel.background = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.grid = element_blank(),
axis.line = element_line(color = "black"),
plot.background = element_rect(fill="transparent", color=NA),
legend.key = element_rect(fill="transparent", color="transparent"),
legend.box.background = element_rect(fill="transparent", color="transparent"),
legend.background = element_rect(fill="transparent", color="transparent"),
legend.text=element_text(size=10),
legend.title = element_text(size=10))
</code>
<code>
levels(datasets@active.ident)
</code>
<code>
ucols <- c("#006666", "#C8D523", "#EC4861","#FF9933", "#B11A20", "#3838c9")
names(ucols) <- levels(datasets@active.ident)
DimPlot(datasets, label = F, pt.size = 0.6, cols = ucols) + theme_void()
</code>
<code>
png("./Routput/Seurat/Figures/UMAP-fig1.png")
DimPlot(datasets, label = F, pt.size = 0.6, cols = ucols) + theme_void()
dev.off()
</code>
<code>
tiff("./Routput/Seurat/Figures/UMAP-fig1.tiff")
DimPlot(datasets, label = F, pt.size = 0.6, cols = ucols) + theme_void()
dev.off()
</code>
<code>
sessionInfo()
</code>
|
{
"filename": "ST_fig1d_plotUMAP.ipynb",
"repository": "astrid12345/Visium",
"query": "transformed_from_existing",
"size": 14272,
"sha": ""
}
|
# dscov_1.ipynb
Repository: compbiocore/tidyverse-workshop
# Overview
This is an informal overview of what is a more comprehensive and longer workshop run by the Computational Biology Core, which is primarily for more regular R users and biologists. Some of the material here will probably thus be very confusing if you have little to no R background. I'll do my best to explain some of the more basic R functions. You can also run `?function` to see what a function does at any time.
However, primarily we will be talking about tidying and transforming data, which is an important piece of data science no matter what programming language you use. The paradigm we will be using is that of **tidyverse**, which is popular enough that equivalents often exist in other languages commonly used for datasci like Python and Julia. Those equivalents will be mentioned in each section.
Most of this workshop is taken from Hadley's [R for Data Science](https://r4ds.had.co.nz/) book. You can find more examples, explanations and exercises there if you want.
## Executing cells in Jupyter notebook
If you've never used Jupyter notebook, you can execute code in any cell by hitting *Shift+Enter*. You can also modify code in a cell then execute it. It's designed to go in order, so you might get an error if you jump ahead (although there are some errors placed thruout the notebook intentionally...)
# Package prerequisites
Packages that required in this workshop are **tidyverse**, which includes the packages **ggplot2**, **dplyr**, **purrr**, and others, **gridExtra** which helps with arranging plots next to each other, **ggrepel** which helps with plot labels and **maps** which is for map data.
The command `library(xyz)` loads package `xyz`, similar to `import module` from Python.
<code>
library(tidyverse)
library(gridExtra)
library(ggrepel)
library(maps)
</code>
If you get an error message “there is no package called ‘xyz’” then you need to install the packages first. (They should have been preloaded on your notebooks but if not it's ok, it won't take long.)
<code>
#install.packages('tidyverse')
#install.packages('gridExtra')
#install.packages('ggrepel')
#install.packages('maps')
</code>
# Visualizing Data
Core feature of exploratory data analysis is asking questions about data and searching for answers by visualizing and modeling data. Most questions around what type of variation or covariation occurs between variables.
<code>
# sets options for plot width and plot height in notebook
options(repr.plot.width=6, repr.plot.height=4)
# regular plot functions in R
plot(x=mpg$displ,y=mpg$hwy)
</code>
<code>
# ggplot!
ggplot(data=mpg) + geom_point(mapping=aes(x=displ,y=hwy))
</code>
Basic syntax of ggplot:
```
ggplot(data=<DATA>) +
<GEOM_FUNCTION>(mapping=aes(<MAPPINGS>)
```
<code>
ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(color = class)) +
geom_smooth(se = FALSE) +
labs(x="Engine displacement (L)",y="Heighway fuel economy (mpg)",
title = "Fuel efficiency generally decreases with engine size",
caption = "Data from fueleconomy.gov",
subtitle = "Two seaters (sports cars) are an exception because of their light weight",
colour = "Car type"
) + theme_classic()
</code>
## But first...
```
ggplot(data=<DATA>) +
<GEOM_FUNCTION>(mapping=aes(<MAPPINGS>)
```
How do we get data into a form appropriate for **ggplot2**?
# Tidying Data
Most datasets are data frames made up of **rows** and **columns**. However, talking about data frames just in terms of what rows and columns it has is not enough.
* **Variable:** quantity, quality, property that can be measured.
* **Value:** State of variable when measured.
* **Observation:** Set of measurements made under similar conditions
* **Tabular data:** Set of values, each associated with a variable and an observation.
Tidy data:
* Each variable is its own column
* Each observation is its own row
* Each value is in a single cell
Benefits:
* Easy to manipulate
* Easy to model
* Easy to visualize
* Has a specific and consistent structure
* Stucture makes it easy to tidy other data
Cons:
* Data frame is not as easy to look at
Consider the following tables:
<code>
# data.frame makes a data frame
# c(a,b,c) creates a vector with objects a,b,c
# rep(x,y) creates a vector that repeats x y times
# assignment in R is done with <- (you can use = sometimes but there are some scope differences)
table1 <- data.frame(makemodel=c("audi a4","audi a4","chevrolet corvette","chevrolet corvette","honda civic","honda civic"),
year=rep(c(1999,2008),3),
cty=c(18,21,15,15,24,25),
hwy=c(29,30,23,25,32,36))
table1
</code>
This is tidy data, because each column is a variable, each observation is a row, and each value is in a single cell
Next we will look at some non-tidy data and operations from the **tidyr** package (part of **tidyverse**) to make the data tidy. Many of you might be more used to using operations from **reshape2** like melting and casting. It's a very useful package with more functionality including aggregating data, but syntax with **tidyr** commands is more simpler and intuitive for the purposes of tidying data. Can check out [Pandas documentation](http://pandas.pydata.org/pandas-docs/stable/user_guide/reshaping.html) for similar functions in Python although they have different names.
## Gathering
<code>
table2a <- data.frame(makemodel=c("audi a4","chevrolet corvette","honda civic"),`1999`=c(18,15,24),'2008'=c(21,15,25),check.names=FALSE)
table2b <- data.frame(makemodel=c("audi a4","chevrolet corvette","honda civic"),`1999`=c(29,23,32),'2008'=c(30,25,36),check.names=FALSE)
table2a
table2b
</code>
`table2a` column names `1999` and `2008` represent values of `year` variable. `table2b` is the same. Each row represents 2 observations, not 1. Need to gather columns into new pair of variables.
Parameters:
* Set of columns that represent values, not variables.
* `key`: name of variable whose values are currently column names.
* `value`: name of variable whose values are currently spread out across multiple columns.
Experiments often report data in the format of `table4a` and `table4b`. One reason is for presentation purposes it's very easy to look at. Another is storage is efficient for completely crossed designs and can allow matrix operations.
<code>
tidy2a <- gather(table2a,`1999`,`2008`,key="year",value="cty")
tidy2a
</code>
<code>
tidy2b <- gather(table2b, `1999`, `2008`, key = "year", value = "hwy")
tidy2b
</code>
Merge tables using `left_join()` (many other types of [table joins](https://dplyr.tidyverse.org/reference/join.html) as well)
<code>
right_join(tidy2a,tidy2b)
</code>
## Spreading
<code>
table3 <- data.frame(makemodel=c(rep("audi a4",4),rep("chevrolet corvette",4),rep("honda civic",4)),
year=rep(c(1999,1999,2008,2008),3),
type=rep(c("cty","hwy"),6),
mileage=c(18,29,21,30,15,23,15,25,24,32,25,36))
table3
</code>
`table3` has each observation in two rows. Need to spread observations across columns with appropriate variable names instead.
Parameters:
* `key`: Column that contains variable names.
* `value`: Column that contains values for each variable.
<code>
spread(table3, key=type,value=mileage)
</code>
## Separating
<code>
table4 <- data.frame(makemodel=c("audi a4","audi a4","chevrolet corvette","chevrolet corvette","honda civic","honda civic"),
year=rep(c(1999,2008),3),
mileages=c('18/29','21/30','15/23','15/25','24/32','25/36'))
table4
</code>
`table4` has `mileages` column that actually contains two variables (`cty` and `hwy`). Need to separate into two columns.
Parameters:
* column/variable that needs to be separated.
* `into`: columns to split into
* `sep`: separator value. Can be regexp or positions to split at. If not provided then splits at non-alphanumeric characters.
<code>
separate(table4, mileages, into = c("cty", "hwy"), sep="/")
</code>
<code>
sep <- separate(table4, makemodel, into = c("make", "model"), sep = ' ')
sep
</code>
## Uniting
Now `sep` has `make` and `model` columns that can be combined into a single column. In other words, we want to unite them.
Parameters:
* Name of united column/variable
* Names of columns/variables to be united
* `sep`: Separator value. Default is '_'
<code>
unite(sep, new, make, model)
</code>
<code>
unite(sep, makemodel, make, model, sep=' ')
</code>
## Piping
**dplyr** from **tidyverse** contains the 'pipe' (`%>%`) which allows you to combine multiple operations, directly taking output from a funtion as input to the next. Can save time and memory as well as make code easier to read. Can think of it this way: `x %>% f(y)` becomes `f(x,y)`, and `x %>% f(y) %>% g(z)` becomes `g(f(x,y),z)`, etc.
<code>
unite(sep, makemodel, make, model, sep=' ') %>%
separate(mileages, into=c("cty","hwy"))
</code>
## Not all data should be tidy
Matrices, phylogenetic trees (although `ggtree` and `treeio` have tidy representations that help with annotating trees), etc.
# Transforming (Tidy) Data
Now we know how to get tidy data. At this point we can already start visualizing our data. However in many cases we will need to further transform our data to narrow down variables and observations we are really interested in or to create new variables that are functions of our existing variables and data. This is known as **transforming** data. In the **tidyverse** the package for these operations is **dplyr**.
* `filter()` to pick observations (rows) by their values
* `arrange()` to reorder rows, default is by ascending value
* `select()` to pick variables (columns) by their names
* `mutate()` to create new variables with functions of existing variables
* `summarise()` to collapes many values down to a single summary
* `group_by()` to set up functions to operate on groups rather than the whole data set
* `%>%` propagates the output from a function as input to another. eg: x %>% f(y) becomes f(x,y), and x %>% f(y) %>% g(z) becomes g(f(x,y),z).
All functions have similar structure:
1. First argument is data frame
2. Next arguments describe what to do with data frame using variable names
3. Result is new data frame
Will be working with data frame **mpg** for rest of workshop which comes with the **tidyverse** library.
Can do everything here with Pandas in Python as well, or use specific implementations of **dplyr** in Python that go on top of Pandas with [**Dplython**](https://github.com/dodger487/dplython) or [**pandas-ply**](https://pythonhosted.org/pandas-ply/).
<code>
head(mpg)
</code>
## `filter()` rows/observations
As name suggests filters out rows. First argument is name of data frame, next arguments are expressions that filter the data frame.
<code>
# filter out 2seater cars
no_2seaters <- filter(mpg, class != "2seater")
head(no_2seaters)
</code>
<code>
# filter out audis, chevys, and hondas
mpg %>% filter(!manufacturer %in% c("audi","chevrolet","honda")) %>% head
</code>
## `arrange()` rows/observations
Changes order of rows. First argument is name of data frame, next arguments are column names (or more complicated expressions) to order by. Default column ordering is by ascending order, can use `desc()` to do descending order. Missing values get sorted at the end regardless of what column ordering is chosen.
<code>
# arrange/reorder mpg by class
arrange(mpg, class) %>% head
</code>
<code>
# arrange/reorder data frame with 2seaters filtered out by class
# 2seaters does not appear which is as it should be
arrange(no_2seaters, class) %>% head
</code>
What kinds of cars have the best highway and city gas mileage?
<code>
# arrange mpg so that first hwy mileage is by descending order, then cty mileage is by descending order
arrange(mpg, desc(hwy), desc(cty)) %>% head
</code>
Example of missing data getting placed at bottom.
<code>
df <- data.frame(x=c(5,2,NA,6))
df
</code>
<code>
# arrange df by ascending order, NA will be at bottom
arrange(df, x)
</code>
<code>
# arrange df by descending order, NA will be at bottom
arrange(df, desc(x))
</code>
If you want to bring `NA` to the top, you can instead use `!is.na(x)` which evaluates as a boolean, so FALSE/TRUE. The df gets arranged by FALSE first, so NA goes to the top. However the rest of the values are unsorted since they will all return TRUE, although you can add more arguments to sort by the same column. This can be done for any other variable you want to use to rearrange with boolean expressions instead.
<code>
# rest of the values are unsorted because they are all T for !is.na(x)
arrange(df,!is.na(x))
</code>
<code>
# can arrange by x again to get ascending order
arrange(df,!is.na(x),desc(x))
</code>
## `select()` columns/variables
Selects columns, which can be useful when you have hundreds or thousands of variables in order to narrow down to what variables you're actually interested in. First argument is name of data frame, subsequent arguments are columns to select. Can use `a:b` to select all columns between `a` and `b`, or use `-a` to select all columns *except* a.
<code>
# select manufacturer, model, year, cty, hwy
select(mpg, manufacturer, model, year, cty, hwy) %>% head
</code>
<code>
# select all columns model thru hwy
select(mpg, model:hwy) %>% head
head(mpg)
</code>
<code>
# select all columns except cyl thru drv and class
select(mpg, -(cyl:drv), -class) %>% head
</code>
## `mutate()` to add new variables or `transmute()` to keep only new variables
Adds new columns that are functions of existing columns. First argument is name of data frame, next arguments are of the form `new_column_name = f(existing columns)`.
<code>
# add a new column that takes average mileage between city and highway
mutate(mpg, avg_mileage = (cty+hwy)/2) %>% head
df <- data.frame(x=c(5,2,NA,6),y=c(NA,5,10,3))
df
#summarise(df, m=mean(x,y,na.rm=TRUE))
#?mean
</code>
<code>
# keep only average mileage between city and highway
transmute(mpg,cty,avg_mileage=(cty+hwy)/2) %>% head
</code>
## `summarise()` and `group_by()` for grouped summaries
`summarise()` collapses a data frame into a single row, and `group_by()` changes analysis from entire data frame into individual groups.
<code>
# get average mileage grouped by engine cylinder
m <- mutate(mpg, avg_mileage=(cty+hwy)/2)
# behavior is actually different in R/RStudio compared to notebooks
m %>% group_by(cyl) %>%
summarise(avg=mean(avg_mileage)) %>%
head
</code>
**Note:** If you look at the output of `group_by` in R/RStudio you will actually be able to see what your groupings are as well as how many of them you have. For example if we did `group_by(mpg, cyl)` the output would include `cyl [4]` which shows that our grouping is by `cyl` and there are 4 groups. Jupyter notebook doesn't display this for reasons having to do with [how data frames are outputted](https://github.com/IRkernel/repr/issues/113). Some other differences exist between how certain objects from **tidyverse** are displayed as well.
<code>
group_by(m, drv) %>%
summarise(avg=mean(avg_mileage))
</code>
<code>
# df after group_by would show that we have 9 groups
drv_cyl <- group_by(m, drv, cyl) %>%
summarise(avg=mean(avg_mileage)) %>%
arrange(desc(avg))
drv_cyl
</code>
Can also run `ungroup` to ungroup your observations.
<code>
drv_cyl %>% summarise(max=max(avg))
</code>
<code>
ungroup(drv_cyl) %>% summarise(max=max(avg))
</code>
# Back to Visualizing Data
Basic syntax of ggplot:
```
ggplot(data=<DATA>) +
<GEOM_FUNCTION>(mapping=aes(<MAPPINGS>)
```
## The grammar of graphics
**ggplot2** employs what is known as the *grammar of graphics*, which allows user to create plots by explicitly mapping data to visual objects and properties that make up the plot. There is a Python clone of **ggplot2** called [**plotnine**](https://github.com/has2k1/plotnine) that is pretty nice and far better than matplotlib. Equivalent in Julia is [**Gadfly.jl**](https://github.com/GiovineItalia/Gadfly.jl).
<code>
head(mpg) # automatically loaded when you load tidyverse
</code>
<code>
ggplot(mpg) + geom_point(mapping=aes(x=displ,y=hwy))
</code>
## `<MAPPINGS>`
```
ggplot(data=<DATA>) +
<GEOM_FUNCTION>(mapping=aes(<MAPPINGS>)
```
Visual property of objects in plot, i.e. size, shape, color. Can display points from other variables (in this case class) in different ways by changing value of aesthetic properties. These are known as **levels**, which is done in order to distinguish aesthetic values from data values.
<code>
head(mpg)
</code>
<code>
p1 <- ggplot(data=mpg) + geom_point(mapping=aes(x=displ,y=hwy,color=class))
p2 <- ggplot(data=mpg) + geom_point(mapping=aes(x=displ,y=hwy,shape=class))
p3 <- ggplot(data=mpg) + geom_point(mapping=aes(x=displ,y=hwy,size=class))
p4 <- ggplot(data=mpg) + geom_point(mapping=aes(x=displ,y=hwy,alpha=class))
grid.arrange(p1,p2,p3,p4,nrow=2)
</code>
### Levels
**ggplot2** automatically assigns a unique level of an aesthetic to a unique value of the variable. This process is known as scaling. It will also automatically select a scale to use with the aesthetic (i.e. continuous or discrete) as well as add a legend explaining the mapping between levels and values. That's why in the size mapping there's no shape for suv, and why the following two pieces of code do different things:
<code>
# for color property, all data points were assigned to 'blue', therefore ggplot2 assigns a single level to all of the
# points, which is red
ggplot(data=mpg) + geom_point(mapping=aes(x=displ,y=hwy,color='blue'))
</code>
<code>
# here color is placed outside aesthetic mapping, so ggplot2 understands that we want color of points to be blue
ggplot(data=mpg) + geom_point(mapping=aes(x=displ,y=hwy),color='blue')
</code>
<code>
# cty is a continuous variable, so when mapped to color we get a gradient with bins instead
ggplot(data=mpg) + geom_point(mapping=aes(x=displ,y=hwy,color=cty))
</code>
### Continuous vs discrete scales
Generally continuous scales get chosen for numerical data and discrete scales are chosen for categorical data. If your data is numeric but in discrete categories you may have to use `as.factor()` in order to get proper levels.
<code>
# if we try to map cyl to shape we get an error because shape is only for discrete variables
# even though we only have cyl=4,5,6 or 8
ggplot(data=mpg) + geom_point(mapping=aes(x=displ,y=hwy,shape=cyl))
</code>
<code>
# will transform into categorical variable with levels
as.factor(mpg$cyl)
</code>
<code>
# all is well when we use as.factor()
ggplot(data=mpg) + geom_point(mapping=aes(x=displ,y=hwy,shape=as.factor(cyl)))
</code>
Note that this means x and y are aesthetic mappings as well. In fact without them you will get an error.
<code>
ggplot(data=mpg) + geom_point()
</code>
## `<GEOM_FUNCTION>`
```
ggplot(data=<DATA>) +
<GEOM_FUNCTION>(mapping=aes(<MAPPINGS>)
```
**geom** geometrical object plot uses to represent data. Bar charts use bar geoms, line charts use line geoms, boxplots, etc. Scatterplots use point geoms. Full list of geoms provided with **ggplot2** can be seen in [ggplot2 reference](https://ggplot2.tidyverse.org/reference/#section-layer-geoms). Also exist other geoms created by [other packages](http://www.ggplot2-exts.org/gallery/).
Every geom function in ggplot2 takes a `mapping` argument with specific aesthetic mappings that are possible. Not every aesthetic will work with every geom. For example, can set shape of a point, but not shape of a line. However, can set linetype of a line.
<code>
ggplot(data = mpg) +
geom_smooth(mapping = aes(x = displ, y = hwy))
</code>
<code>
# data has been separated into three lines based on their drivetrain: 4 (4wd), f (front), r (rear)
ggplot(data = mpg) +
geom_smooth(mapping = aes(x = displ, y = hwy, linetype = as.factor(cyl)))
</code>
Can display multiple geoms on same plot just by adding them
<code>
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, color=drv)) +
geom_smooth(mapping = aes(x = displ, y = hwy, color=drv, linetype=drv))
</code>
Geoms like `geom_smooth()` use single geometric object to display multiple rows of data. If you don't necessarily want to add other distinguishing features to the geom like color, can use `group` aesthetic (for a categorical variable) to draw multiple objects.
<code>
ggplot(data=mpg) +
geom_smooth(mapping=aes(x=displ,y=hwy,group=drv))
</code>
### Global mappings vs local mappings
`ggplot()` function contains *global* mapping, while each geom has a local mapping
* `lm` from **stats** for linear models (you can also fit other models)
<code>
# global mapping of displ and hwy creates x and yaxis
ggplot(data=mpg, mapping=aes(x=displ,y=hwy))
</code>
<code>
# mapping color to class for point geom while using global x and y mappings
ggplot(data=mpg, mapping=aes(x=displ,y=hwy)) + geom_point(mapping=aes(color=class))
</code>
<code>
# geom_smooth doesn't need any mapping arguments if using global
ggplot(data=mpg, mapping=aes(x=displ,y=hwy)) +
geom_point(mapping=aes(color=class))+
geom_smooth()
</code>
<code>
# second geom_smooth uses same x and y mapping
# but mapping comes from no_2seaters data (from Transform section) instead
ggplot(data = mpg, mapping = aes(x = displ, y = hwy)) +
geom_point(mapping = aes(color = class)) +
geom_smooth() +
geom_smooth(data = no_2seaters)
</code>
## More syntax
```{r}
ggplot(data = <DATA>) +
<GEOM_FUNCTION>(
mapping = aes(<MAPPINGS>),
stat = <STAT>,
position = <POSITION>
) +
<COORDINATE_FUNCTION> +
<FACET_FUNCTION>
```
## Facets
Subplots displaying one subset of data.
* `facet_wrap()` for a single variable.
* `facet_grid()` for along 2 variables.
<code>
ggplot(data=mpg) +
geom_point(mapping=aes(x=displ,y=hwy)) +
facet_wrap(~ class, nrow=2)
</code>
<code>
ggplot(data=mpg) +
geom_point(mapping=aes(x=displ,y=hwy)) +
facet_wrap(~ class, nrow=3)
</code>
<code>
ggplot(data=mpg) +
geom_point(mapping=aes(x=displ,y=hwy)) +
facet_wrap(~ class, ncol=4)
</code>
<code>
# some facets are empty because no observations have those combos
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy)) +
facet_grid(drv ~ cyl)
</code>
## Stats
```{r}
ggplot(data = <DATA>) +
<GEOM_FUNCTION>(
mapping = aes(<MAPPINGS>),
stat = <STAT>,
position = <POSITION>
) +
<COORDINATE_FUNCTION> +
<FACET_FUNCTION>
```
Algorithm used to calculate new values for a graph. Each geom object has a default stat, and each stat has a default geom. Geoms like `geom_point()` will leave data as is, known as `stat_identity()`. Graphs like bar charts and histograms will bin your data and compute bin counts, known as `stat_count()`. Can see full list of stats at [ggplot2 reference](https://ggplot2.tidyverse.org/reference/) under both Layer: geoms and Layer: stats.
<code>
ggplot(data=mpg) +
geom_bar(mapping=aes(x=class))
</code>
Since each stat comes with a default geom, can use stat to create geoms on plots as well.
<code>
ggplot(data=mpg) +
stat_count(mapping=aes(x=class))
?geom_bar
</code>
<code>
# because stat_count() computes count and prop, can use those as variables for mapping as well
ggplot(data=mpg) + geom_bar(mapping=aes(x=class, y=..prop..,group=1))
</code>
<code>
# stat_summary is associated with geom_pointrange
# default is to compute mean and standard error
ggplot(data = mpg) +
stat_summary(mapping = aes(x=class,y=hwy))
</code>
<code>
# can change stat_summary to compute median and min/max instead
ggplot(data = mpg) +
stat_summary(
mapping = aes(x = class, y = hwy),
fun.ymin = min,
fun.ymax = max,
fun.y = median
)
</code>
## Position adjustments
```{r}
ggplot(data = <DATA>) +
<GEOM_FUNCTION>(
mapping = aes(<MAPPINGS>),
stat = <STAT>,
position = <POSITION>
) +
<COORDINATE_FUNCTION> +
<FACET_FUNCTION>
```
Each geom also comes with a default **position adjustment** specified by `position` argument. For geoms like `geom_point()` it is "identity" which is position as is.
Specifically for bar charts, have fill aesthetic. If fill aesthetic gets mapped to another variable, bars are automatically stacked under the "stack" position. Can see [list of positions](https://ggplot2.tidyverse.org/reference/#section-layer-position-adjustment) at ggplot2 reference.
<code>
p1 <- ggplot(data = mpg, mapping=aes(x=class,fill=as.factor(cyl)))
p1 + geom_bar()
</code>
<code>
# position = identity will place each object exactly where it falls in context of graph.
# Not useful for bar charts, better for scatterplots.
p1 + geom_bar(position="identity", alpha=0.2)
</code>
<code>
# position = fill will make bars same height
p1 + geom_bar(position="fill")
</code>
<code>
# position = "dodge" places objects directly beside one another. Easier to compare individual values.
p1 + geom_bar(position="dodge")
</code>
For `geom_point` one possible position is "jitter", which will add a small amount of random noise to each point. This spreads points out so that it's unlikely for points to overlap and therefore get plotted over each other. For example it's possible that majority of points are actually one combination of `hwy` and `displ` but they all get plotted at the exact same point so you can't tell. For very large datasets can help prevent overplotting to better see where mass of plot is or trends.
<code>
# seems quite uniform which suggests multiple observations with same value of cty/hwy
# creating overlapping points
ggplot(data = mpg, mapping = aes(x = cty, y = hwy)) +
geom_point()
</code>
<code>
# definitely the case
ggplot(data = mpg, mapping = aes(x = cty, y = hwy)) +
geom_point(position="jitter")
</code>
## Coordinate systems
```{r}
ggplot(data = <DATA>) +
<GEOM_FUNCTION>(
mapping = aes(<MAPPINGS>),
stat = <STAT>,
position = <POSITION>
) +
<COORDINATE_FUNCTION> +
<FACET_FUNCTION>
```
Default coordinate system is Cartesian.
* `coord_flip()` switches x and y axes.
* `coord_quickmap()` sets aspect ratio for maps.
* `coord_polar()` sets polar coordinates.
<code>
p <- ggplot(data = mpg, mapping = aes(x = class, y = hwy))
p + geom_boxplot()
</code>
<code>
# flipping coordinates
p + geom_boxplot() + coord_flip()
</code>
<code>
# can reorder x axis by lowest to highest median hwy mileage
# allows easier comparisons
ggplot(data = mpg, mapping = aes(x = reorder(class,hwy,FUN=median), y = hwy)) +
geom_boxplot() +
coord_flip()
</code>
<code>
# Setting aspect ratio correctly
nz <- map_data("nz")
ggplot(nz, aes(long, lat, group = group)) +
geom_polygon(fill = "white", colour = "black")
ggplot(nz, aes(long, lat, group = group)) +
geom_polygon(fill = "white", colour = "black") +
coord_quickmap()
</code>
<code>
# polar coordinates
bar <- ggplot(data = mpg) +
geom_bar(
mapping = aes(x = class, fill = as.factor(cyl)),
show.legend = FALSE,
width = 1
) +
theme(aspect.ratio = 1) +
labs(x = NULL, y = NULL)
p1 <- bar + coord_flip()
p2 <- bar + coord_polar()
grid.arrange(p1,p2, nrow=1)
</code>
# Summary
Now that we've gone through tidying, transforming, and visualizing data let's review all of the different functions we've used and in some cases learned the inner workings of:
## Tidying
* `gather()`
* `spread()`
* `separate()`
* `unite()`
* `%>%` propagates the output from a function as input to another. eg: x %>% f(y) becomes f(x,y), and x %>% f(y) %>% g(z) becomes g(f(x,y),z).
## Transforming
* `filter()` to pick observations (rows) by their values
* `arrange()` to reorder rows, default is by ascending value
* `select()` to pick variables (columns) by their names
* `mutate()` to create new variables with functions of existing variables
* `summarise()` to collapes many values down to a single summary
* `group_by()` to set up functions to operate on groups rather than the whole data set
## Visualizing
* `ggplot` - global data and mappings
* `geom_point` - geom for scatterplots
* `geom_smooth` - geom for regressions
* `geom_pointrange` - geom for vertical intervals defined by `x`, `y`, `ymin`, and `ymax`
* `geom_bar` - geom for barcharts
* `geom_boxplot` - geom for boxplots
* `geom_polygon` - geom for polygons
* `aes(color)` - color mapping
* `aes(shape)` - shape mapping
* `aes(size)` - size mapping
* `aes(alpha)` - transparency mapping
* `as.factor()` - transforming numerical values to categorical values with levels
* `facet_grid`
* `facet_wrap`
* `stat_count` - default stat for barcharts, bins by x and counts
* `stat_identity` - default stat for scatterplots, leaves data as is
* `stat_summary` - default stat for pointrange, by default computes mean and se of y by x
* `position="identity"`
* `position="stacked"`
* `position="fill"`
* `position="dodge"`
* `position="jitter"`
* `coord_flip`
* `coord_map`
* `coord_polar`
# Publication Quality Graphs
Last piece with some additional functions to learn...
## Labels
`labs()` to add most kinds of labels: title, subtitle, captions, x-axis, y-axis, legend, etc.
<code>
ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(color = class)) +
geom_smooth(se = FALSE) +
labs(
title = "Fuel efficiency generally\n decreases with engine size",
subtitle = "Two seaters (sports cars) are an exception because of their light weight",
caption = "Data from fueleconomy.gov",
x = "Engine displacement (L)",
y = "Highway fuel economy (mpg)",
color = "Car type"
)
</code>
## Annotations
Can use `geom_text()` to add text labesls on the plot.
<code>
best_in_class <- mpg %>%
group_by(class) %>%
filter(row_number(desc(hwy)) == 1)
ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(colour = class)) +
geom_text(aes(label = model), data = best_in_class)
</code>
<code>
ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(colour = class)) +
ggrepel::geom_label_repel(aes(label = model), data = best_in_class) +
labs(
caption = "Data from fueleconomy.gov",
x = "Engine displacement (L)",
y = "Highway fuel economy (mpg)",
colour = "Car type"
) +
geom_point(size = 3, shape = 1, data = best_in_class)
</code>
## Scales
* `breaks`: For the position of ticks
* `labels`: For the text label associated with each tick.
* Default scale is x continuous, y continuous but can also do x logarithmic, y logarithmic, change color scales.
<code>
ggplot(mpg, aes(displ, hwy)) +
geom_point() +
scale_y_continuous(breaks = seq(15, 40, by = 5))
</code>
<code>
ggplot(mpg, aes(displ, hwy)) +
geom_point() +
scale_x_continuous(labels = NULL) +
scale_y_continuous(labels = NULL)
</code>
<code>
p1 <- ggplot(diamonds, aes(carat, price)) +
geom_bin2d()
ggplot(diamonds, aes(carat, price)) +
geom_bin2d() +
scale_x_log10() +
scale_y_log10()
ggplot(diamonds, aes(log10(carat), log10(price))) +
geom_bin2d()
</code>
<code>
ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(color = drv))
ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(color = drv)) +
scale_colour_brewer(palette = "Set1")
</code>
<code>
ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(color = drv)) +
scale_colour_manual(values=c(`4`="red",f="blue",r="blue"))
</code>
## Legend positioning
`theme(legend.position)` to control legend position. `guides()` with `guide_legened()` or `guide_colourbar()` for legend display.
<code>
base <- ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(colour = class))
#p1 <- base + theme(legend.position = "left")
#p2 <- base + theme(legend.position = "top")
#p3 <- base + theme(legend.position = "bottom")
#p4 <- base + theme(legend.position = "right")
#?theme
base + theme(text=element_text(color="blue",size=4))
#grid.arrange(p1,p2,p3,p4, nrow=2)
</code>
<code>
ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(colour = class)) +
geom_smooth(se = FALSE) +
theme(legend.position = "bottom") +
guides(colour = guide_legend(nrow = 1, override.aes = list(size = 4)))
</code>
## Zooming
Three ways to control plot limits:
* Adjusting what data are plotted
* Setting limits in each scale
* Setting `xlim` and `ylim` in `coord_cartesian()`
<code>
# asetting xlim and ylim in coord_cartesian
ggplot(mpg, mapping = aes(displ, hwy)) +
geom_point(aes(color = class)) +
geom_smooth() +
coord_cartesian(xlim = c(5, 7), ylim = c(10, 30))
</code>
<code>
# adjusting what data are plotted
# however geom_smooth will plot regression over subsetted data
filter(mpg, displ >= 5, displ <= 7, hwy >= 10, hwy <= 30) %>%
ggplot(aes(displ, hwy)) +
geom_point(aes(color = class)) +
geom_smooth()
</code>
<code>
# 2 plots use subsetted data therefore have different scales along hwy and displ
suv <- mpg %>% filter(class == "suv")
compact <- mpg %>% filter(class == "compact")
ggplot(suv, aes(displ, hwy, colour = drv)) +
geom_point()
ggplot(compact, aes(displ, hwy, colour = drv)) +
geom_point()
</code>
<code>
# can set limits in each scale
x_scale <- scale_x_continuous(limits = range(mpg$displ))
y_scale <- scale_y_continuous(limits = range(mpg$hwy))
col_scale <- scale_colour_discrete(limits = unique(mpg$drv))
ggplot(suv, aes(displ, hwy, colour = drv)) +
geom_point() +
x_scale +
y_scale +
col_scale
ggplot(compact, aes(displ, hwy, colour = drv)) +
geom_point() +
x_scale +
y_scale +
col_scale
</code>
## Themes
**ggplot2** has 8 themes by default, can get more in other packages like **ggthemes**. Generally prefer `theme_classic()`.
<code>
base <- ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(color = class)) +
geom_smooth(se = FALSE)
p1 <- base + theme_bw()
p2 <- base + theme_light()
p3 <- base + theme_classic()
p4 <- base + theme_linedraw()
p5 <- base + theme_dark()
p6 <- base + theme_minimal()
p7 <- base + theme_void()
grid.arrange(base,p1,p2,p3,p4,p5,p6,p7,nrow=4)
</code>
## Saving your plots
* `ggsave()` will save most recent plot to disk
* `tiff()` will save next plot to disk
* Other functions like `postscript()` for eps files, etc.
* All can take `width`, `height`, `fonts`, `pointsize`, `res` (resolution) arguments
<code>
p1 <- ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(color = class)) +
geom_smooth(se = FALSE) +
labs(x="Engine displacement (L)",y="Heighway fuel economy (mpg)",
title = "Fuel efficiency generally decreases with engine size",
caption = "Data from fueleconomy.gov",
subtitle = "Two seaters (sports cars) are an exception because of their light weight",
colour = "Car type"
) + x_scale + y_scale + theme_classic()
p1
ggsave("my_plot.pdf")
tiff("my_plot.tiff",width=7,height=5,units="in",pointsize=8,res=350)
p1
dev.off()
</code>
# Some other useful visualization packages
We don't have time in this workshop to get in depth to other workshops, but here are some more useful visualization packages that may be helpful for your research.
## ggtree for phylogenetics
Resources and associated packages:
* [Data Integration, Manipulation and Visualization of Phylogenetic Trees](https://yulab-smu.github.io/treedata-book/index.html)
* [treeio](https://bioconductor.org/packages/release/bioc/html/treeio.html)
* [tidytree](https://cran.r-project.org/web/packages/tidytree/index.html)
## cowplot
Meant to provide publication-ready theme for **gplot2** that requires minimum amount of fiddling with sizes of axis labels, plot backgrounds, etc. Auto-sets `theme_classic()` for all plots.
## Gviz for plotting data along genomic coordinates
Can be installed from [Bioconductor](https://bioconductor.org/packages/release/bioc/html/Gviz.html).
## phyloseq for metagenomics
Website is [very comprehensive](http://joey711.github.io/phyloseq/).
<code>
sessionInfo()
</code>
|
{
"filename": "dscov_1.ipynb",
"repository": "compbiocore/tidyverse-workshop",
"query": "transformed_from_existing",
"size": 62172,
"sha": ""
}
|
# main_1.ipynb
Repository: Zhenghua-404/Efficient-Data-Deletion-in-ML
<code>
from google.colab import drive
drive.mount('/content/drive')
</code>
*Weighted DC-Kmeans model:*
<code>
import numpy as np
class Kmeans(object):
'''
In-house implementation of k-means via Lloyd-Max iterations
This is a research prototype and is not necessarily well-optimized
'''
def __init__(self,
k,
termination='fixed',
iters=10,
tol=10**-3):
'''
Constructor
INPUT:
k - # of centroids/clusters
iters - # of iterations to run
termination - {'fixed', 'loss', 'centers'}
if 'fixed' - runs for fixed # of iterations
if 'loss' - runs until loss converges
if 'centers' -runs until centers converge
tol - numeric tolerance to determine convergence
'''
# set parameters
self.k = k
self.iters = iters
self.tol = tol
self.termination = termination
# initialize placeholder values
self._init_placeholders()
self.mass_arr = None
def run(self, X):
'''
Run clustering algorithm
INPUT:
X - numpy matrix, n-by-d, each row is a data point
OUTPUT: (3-tuple)
centroids - k-by-d matrix of centroids
assignments - Vector of length n, with datapoint to center assignments
loss - The loss of the final partition
'''
self._set_data(X)
self._lloyd_iterations()
return self.centroids, self.assignments, self.loss
def delete(self, del_idx):
'''
Delete point associated with key del_idx
NOTE: del_idx must be int in {0,n-1}
After deleting any key other than n-1,
the (n-1)-th datapoint's key is automatically
swapped with del_idx to
'''
self.data = np.delete(self.data, del_idx, 0)
self.n = self.n-1
self._init_placeholders()
return self.run(self.data)
def _init_placeholders(self):
self.loss = np.Infinity
self.empty_clusters = []
self.kpp_inits = set()
self.centroids = None
self.assignments = None
self.model = None
def _set_data(self, X):
self.data = X
self.n, self.d = X.shape
def _lloyd_iterations(self):
self._init_centroids()
for _ in range(self.iters):
loss_prev = self.loss
centers_prev = self.model
self._assign_clusters()
self._assign_centroids()
prev = loss_prev if self.termination == 'loss' else centers_prev
if self._check_termination(prev):
break
def _check_termination(self, prev):
if self.termination == 'loss':
return (1 - self.loss/prev) < self.tol
elif self.termination == 'center':
return np.linalg.norm(self.centroids - prev) < self.tol
else:
return False
def _init_centroids(self):
'''
Kmeans++ initialization
Returns vector of initial centroids
'''
prob = []
total_mass = 0
for m in self.mass_arr:
total_mass += m
for m in self.mass_arr:
prob.append(m / total_mass)
first_idx = np.random.choice(self.n, p = prob)
self.centroids = self.data[first_idx,:]
for kk in range(1,self.k):
P = self._get_selection_prob()
nxt_idx = np.random.choice(self.n,p=P)
self.kpp_inits.add(nxt_idx)
self.centroids = np.vstack([self.centroids,self.data[nxt_idx,:]])
def _get_selection_prob(self):
'''
Outputs vector of selection probabilites
Equal to Distance^2 to nearest centroid
'''
#handle edge case in centroids shape by unsqueezing
if len(self.centroids.shape) == 1:
self.centroids = np.expand_dims(self.centroids, axis=0)
#probability is square distance to closest centroid
D = np.zeros([self.n])
for i in range(self.n):
d = np.linalg.norm(self.data[i,:] - self.centroids, axis=1)
D[i] = np.min(d)
P = []
for i in range(self.n):
P.append(self.mass_arr[i] * (D[i]**2))
P = P / sum(P)
return P
def _assign_centroids(self):
'''
Computes centroids in Lloyd iterations
'''
self.centroids = np.zeros([self.k,self.d])
c = np.zeros([self.k])
for i in range(self.n):
a = self.assignments[i]
c[a] += self.mass_arr[i] # weight by mass
self.centroids[a,:] += self.mass_arr[i] * self.data[i,:] # weight by mass
for j in range(self.k):
self.centroids[j,:] = self.centroids[j,:] / c[j]
for j in self.empty_clusters:
self._reinit_cluster(j)
self.empty_clusters = []
def _assign_clusters(self):
'''
Computes clusters in Lloyd iterations
'''
assert (self.k, self.d) == self.centroids.shape, "Centers wrong shape"
self.assignments = np.zeros([self.n]).astype(int)
self.loss = 0
for i in range(self.n):
d = np.linalg.norm(self.data[i,:] - self.centroids, axis=1)
d1 = np.linalg.norm(self.data[i,:] - self.centroids, axis=1,ord=1)
self.assignments[i] = int(np.argmin(d))
self.loss += self.mass_arr[i] * (np.min(d)**2) # weight by mass
self.loss = self.loss / sum(self.mass_arr)
self.empty_clusters = self._check_4_empty_clusters()
def _check_4_empty_clusters(self):
empty_clusters = []
for kappa in range(self.k):
if len(np.where(self.assignments == kappa)[0]) == 0:
empty_clusters.append(kappa)
return empty_clusters
def _reinit_cluster(self, j):
'''
Gets a failed centroid with idx j (empty cluster)
Should replace with new k++ init centroid
in:
j is idx for centroid, 0 <= j <= n
out:
j_prime is idx for next centroid
side-effects:
centroids are update to reflect j -> j'
'''
P = self._get_selection_prob()
j_prime = np.random.choice(self.n,p=P)
self.kpp_inits.add(j_prime)
self.centroids[j,:] = self.data[j_prime,:]
return j_prime
class DCnode(Kmeans):
'''
A k-means subproblem for the divide-and-conquer tree
in DC-k-means algorithm
'''
def __init__(self, k, iters):
Kmeans.__init__(self, k, iters=iters)
self.children = []
self.parent = None
self.time = 0
self.loss = 0
self.node_data = set()
self.node_mass = dict() #mass dict
self.mass_arr = []
self.data_prop = set()
def _run_node(self, X):
self._set_node_data(X)
self._lloyd_iterations()
def _set_node_data(self, X):
#print("X", X.shape)
self.data = X[list(self.node_data)]
self.mass_arr = []
for d in self.data:
self.mass_arr.append(self.node_mass[tuple(d)])
self._set_data(self.data)
class WeightedDCKmeans():
def __init__(self, ks, widths, iters=10):
'''
Constructor for quantized k-means solved via Lloyd iterations
ks - list of k parameter for each layer of DC-tree
widths - list of width parameter (number of buckets) for each layer
iters - # of iterations to run
(at present, only supports fixed iteration termination)
'''
self.ks = ks
self.widths = widths
self.dc_tree = self._init_tree(ks,widths,iters)
self.data_partition_table = dict()
self.data = dict()
self.mass = dict() # For mass
self.dels = set()
self.valid_ids = []
self.d = 0
self.n = 0
self.h = len(self.dc_tree)
for i in range(self.h):
self.data[i] = None
# For mass
for i in range(self.h):
self.mass[i] = None
def run(self, X, assignments=False):
'''
X - numpy matrix, n-by-d, each row is a data point
assignments (optional) - bool flag, computes assignments and loss
NOTE: Without assignments flag, this only returns the centroids
OUTPUT:
centroids - k-by-d matrix of centroids
IF assignments FLAG IS SET ALSO RETURNS:
assignments - Vector of length n, with datapoint to center assignments
loss - The loss of the final partition
'''
self._init_data(X)
self._partition_data(X)
self._run()
if assignments:
assignment_solver = Kmeans(self.ks[0])
assignment_solver._set_data(X)
assignment_solver.centroids = self.centroids
assignment_solver.mass_arr = []
for i in range(len(X)):
assignment_solver.mass_arr.append(1)
assignment_solver._assign_clusters()
self.assignments = assignment_solver.assignments
self.loss = assignment_solver.loss
return self.centroids, self.assignments, self.loss
return self.centroids
def delete(self, del_idx):
idx = self.valid_ids[del_idx]
self.valid_ids[del_idx] = self.valid_ids.pop()
self.dels.add(idx)
node = self.dc_tree[-1][self.data_partition_table[idx]]
node.node_data.remove(idx)
del node.node_mass[tuple(self.data[self.h-1][idx])]
l = self.h-1
self.n -= 1
while True:
node._run_node(self.data[l])
if node.parent == None:
self.centroids = node.centroids
break
data_prop = list(node.data_prop)
for c_id in range(len(node.centroids)):
idx = data_prop[c_id]
self.data[l][idx] = node.centroids[c_id]
node = node.parent
l -= 1
def _init_data(self,X):
self.n = len(X)
self.valid_ids = list(range(self.n))
self.d = len(X[0])
data_layer_size = self.n
for i in range(self.h-1,-1,-1):
self.data[i] = np.zeros((data_layer_size,self.d))
self.mass[i] = np.zeros((data_layer_size))
data_layer_size = self.ks[i]*self.widths[i]
def _partition_data(self, X):
self.d = len(X[0])
num_leaves = len(self.dc_tree[-1])
for i in range(len(X)):
leaf_id = np.random.choice(num_leaves)
leaf = self.dc_tree[-1][leaf_id]
self.data_partition_table[i] = leaf_id
leaf.node_data.add(i)
leaf.node_mass[tuple(X[i])] = 1
#print("partition: ", tuple(X[i]))
self.data[self.h-1][i] = X[i]
def _run(self):
for l in range(self.h-1,-1,-1):
c = 0
for j in range(self.widths[l]):
subproblem = self.dc_tree[l][j]
subproblem._run_node(self.data[l])
if subproblem.parent == None:
self.centroids = subproblem.centroids
else:
for c_id in range(len(subproblem.centroids)):
subproblem.data_prop.add(c)
assignment_solver = Kmeans(self.ks[0])
X = self.data[l]
data = X[list(subproblem.node_data)] ###????
assignment_solver._set_data(data)
assignment_solver.centroids = subproblem.centroids
assignment_solver.mass_arr = []
for i in range(len(data)):
assignment_solver.mass_arr.append(1)
assignment_solver._assign_clusters()
assignments = assignment_solver.assignments
i = 0
for assign in assignments:
if tuple(subproblem.centroids[assign]) not in subproblem.parent.node_mass:
subproblem.parent.node_mass[tuple(subproblem.centroids[assign])] = subproblem.mass_arr[i]
else:
subproblem.parent.node_mass[tuple(subproblem.centroids[assign])] += subproblem.mass_arr[i]
i += 1
subproblem.parent.node_data.add(c)
self.data[l-1][c] = subproblem.centroids[c_id]
c += 1
def _init_tree(self, ks, widths, iters):
tree = [[DCnode(ks[0],iters)]] # root node
for i in range(1,len(widths)):
k = ks[i]
assert widths[i] % widths[i-1] == 0, "Inconsistent widths in tree"
merge_factor = int(widths[i] / widths[i-1])
level = []
for j in range(widths[i-1]):
parent = tree[i-1][j]
for _ in range(merge_factor):
child = DCnode(k,iters=10)
child.parent = parent
parent.children.append(child)
level.append(child)
tree.append(level)
return tree
</code>
<code>
import math
import numpy as np
import matplotlib.pyplot as plt
from del_eff_kmeans import Kmeans, QKmeans, DCKmeans
import time
import pickle
from sklearn.utils import shuffle
from sklearn.metrics import silhouette_score, normalized_mutual_info_score
from random import sample
'''
D.2 Datasets
• Celltypes [42] consists of 12,009 single cell RNA sequences from a mixture of 4 cell types: microglial cells, endothelial cells, fibroblasts, and mesenchymal stem cells. The data was retrieved from the Mouse Cell Atlas and consists of 10 feature dimensions, reduced from an original 23,433 dimensions using principal component analysis. Such dimensionality reduction procedures are a common practice in computational biology.
• Postures [35, 34] consists of 74,975 motion capture recordings of users performing 5 different hand postures with unlabeled markers attached to a left-handed glove.
• Covtype [12] consists of 15,120 samples of 52 cartographic variables such as elevation and hillshade shade at various times of day for 7 forest cover types.
• Botnet [56] contains statistics summarizing the traffic between different IP addresses for a commercial IoT device (Danmini Doorbell). We aim to distinguish between benign traffic data (49,548 instances) and 11 classes of malicious traffic data from botnet attacks, for a total of 1,018,298 instances.
• MNIST [51] consists of 60,000 images of isolated, normalized, handwritten digits. The task is to classify each 28×28 image into one of the ten classes.
• Gaussian [#] consists of 5 clusters, each generated from 25-variate Gaussian distribution centered at randomly chosen locations in the unit hypercube. 20,000 samples are taken from each of the 5 clusters, for a total of 100,000 samples. Each Gaussian cluster is spherical with variance of 0.8.
Gaussian consists of 5 clusters, each generated from 25-variate Gaussian distribution
centered at randomly chosen locations in the unit hypercube. 20,000 samples are
taken from each of the 5 clusters, for a total of 100,000 samples. Each Gaussian
cluster is spherical with variance of 0.8.
'''
'''
Celltype (N = 12,009, D = 10, K = 4), "4celltypes_10pca"
Covtype (N = 15,120, D = 52, K = 7), "covtype_multiclass"
MNIST (N = 60,000, D = 784, K = 10), "mnist"
Postures (N = 74,975, D = 15, K = 5), "postures"
Botnet (N = 1,018,298, D = 115, K = 11), "bot_attack"
and a synthetic dataset made from a Gaussian mixture model which we call Gaussian (N = 100,000, D = 25, K = 5).
'''
DATAs = ["4celltypes_10pca", "covtype_multiclass", "postures", "mnist", "bot_attack"]
Ks = [4, 7, 5, 10, 11]
mata_loss = {}
mata_silcoef = {}
mata_nmi = {}
mata_runtime = {}
mata_traintime = {}
for d in DATAs:
# 3 models, each with 5 values
mata_loss[d] = [[] for j in range(3)]
mata_silcoef[d] = [[] for j in range(3)]
mata_nmi[d] = [[] for j in range(3)]
mata_runtime[d] = [[] for j in range(3)]
mata_traintime[d] = [[] for j in range(3)]
def show_clustering(centers,assignments,data):
colors = ['r','b','g']
for a in range(10):
data_a = data[assignments == a]
plt.scatter(data_a[:,0],data_a[:,1])
plt.scatter(centers[a,0],centers[a,1],marker='x',color='k')
plt.show()
def model_specific_stats(model, features, labels, dataset, m, save, num_deletions): # m is model index, 0, 1, 2
t0 = time.time()
centers = 0
assignments = 0
loss = 0
if m == 2:
centers, assignments, loss = model.run(features.copy(), assignments=True)
else:
centers, assignments, loss = model.run(features.copy())
t1 = time.time()
print("train time: ", t1 - t0)
if save:
mata_traintime[dataset][m].append(t1 - t0)
# Loss
print('Clustering loss is: ', loss)
if save:
mata_loss[dataset][m].append(loss)
# Silhouette Coefficients
sampled_index = sample(range(labels.shape[0]), 10000)
score = silhouette_score(features[sampled_index], assignments[sampled_index])
print("silhouette_score for 10000 random samples: ", score)
if save:
mata_silcoef[dataset][m].append(score)
# Normalized Mutual Information
nmi_score = normalized_mutual_info_score(labels, assignments)
print("normalized_mutual_info_score: ", nmi_score)
if save:
mata_nmi[dataset][m].append(nmi_score)
# Deletion runtime
print("Online deletion time for " + str(num_deletions) + " deletions: ")
t = online_deletion_stream(num_deletions, model)
if save:
mata_runtime[dataset][m].append((t + t1 - t0) / num_deletions)
#print("Amortized Runtime to process" + str(num_deletions) + "deletions is ", (t + t1 - t0) / num_deletions)
def reproduce_result(datasets):
for i in range(5):
print("Dataset: " + DATAs[i])
features = datasets[DATAs[i]][0]
labels = datasets[DATAs[i]][1]
k = Ks[i]
n = datasets[DATAs[i]][1].shape[0]
d = datasets[DATAs[i]][0].shape[1]
# k means model
print("k-means model")
kmeans = Kmeans(k, termination='loss')
model_specific_stats(kmeans, features, labels, DATAs[i], 0)
# Q-kmeans model
print("qkmeans model")
eps = pow(2, -math.log((n / (k * pow(d, 1.5))), 10) - 3)
qkmeans = QKmeans(k, eps)
model_specific_stats(qkmeans, features, labels, DATAs[i], 1)
# DC-kmeans model
print("dc kmeans model")
w = pow(2, math.ceil(math.log(pow(n, 0.3))/math.log(2)))
print(w)
dckmeans = DCKmeans([k,k],[1,w])
model_specific_stats(dckmeans, features, labels, DATAs[i], 2)
def online_deletion_stream(num_dels, model):
t0 = time.time()
c = 1
for _ in range(num_dels):
dr = np.random.choice(model.n,size=1)[0]
#print(f'processing deletion request # {c}...')
model.delete(dr)
c += 1
t = time.time()
print(f'Total time to process {c-1} deletions is {t-t0}')
return t - t0
def tune_tree_height(datasets):
for i in set([0, 1, 3]):
print("Dataset: " + DATAs[i])
features = datasets[DATAs[i]][0]
labels = datasets[DATAs[i]][1]
k = Ks[i]
n = datasets[DATAs[i]][1].shape[0]
d = datasets[DATAs[i]][0].shape[1]
dckmeans = DCKmeans([k, k, k, k],[1, 4, 16, 64])
model_specific_stats(dckmeans, features, labels, DATAs[i], 2, True, 20)
dckmeans = DCKmeans([k, k, k],[1, 8, 64])
model_specific_stats(dckmeans, features, labels, DATAs[i], 2, True, 20)
dckmeans = DCKmeans([k, k],[1, 64])
model_specific_stats(dckmeans, features, labels, DATAs[i], 2, True, 20)
def tune_tree_buckets(datasets):
for i in set([0, 1, 3]):
print("Dataset: " + DATAs[i])
features = datasets[DATAs[i]][0]
labels = datasets[DATAs[i]][1]
k = Ks[i]
n = datasets[DATAs[i]][1].shape[0]
d = datasets[DATAs[i]][0].shape[1]
for j in [2, 3, 4, 5, 6, 7, 8, 9]:
dckmeans = DCKmeans([k, k],[1, 2 ** j])
model_specific_stats(dckmeans, features, labels, DATAs[i], 2, True, 20)
def weighted_tree(datasets):
for i in set([0, 1, 2, 3, 4]):
print("Dataset: " + DATAs[i])
features = datasets[DATAs[i]][0]
labels = datasets[DATAs[i]][1]
k = Ks[i]
n = datasets[DATAs[i]][1].shape[0]
d = datasets[DATAs[i]][0].shape[1]
w = pow(2, math.ceil(math.log(pow(n, 0.3))/math.log(2)))
print(w)
for j in range(3):
print("Weighted")
w_dckmeans = WeightedDCKmeans([k,k],[1,w])
model_specific_stats(w_dckmeans, features, labels, DATAs[i], 2, True, 1)
for j in range(3):
print("non weighted")
dckmeans = DCKmeans([k,k],[1,w])
model_specific_stats(dckmeans, features, labels, DATAs[i], 2, True, 1)
def weighted_tree2(datasets):
for i in set([4]):
print("Dataset: " + DATAs[i])
features = datasets[DATAs[i]][0]
labels = datasets[DATAs[i]][1]
k = Ks[i]
n = datasets[DATAs[i]][1].shape[0]
d = datasets[DATAs[i]][0].shape[1]
w = pow(2, math.ceil(math.log(pow(n, 0.3))/math.log(2)))
print(w)
'''
for j in range(3):
print("Weighted")
w_dckmeans = WeightedDCKmeans([k,k],[1,w])
model_specific_stats(w_dckmeans, features, labels, DATAs[i], 2, True, 1)
'''
for j in range(3):
print("non weighted")
dckmeans = DCKmeans([k,k],[1,w])
model_specific_stats(dckmeans, features, labels, DATAs[i], 2, True, 1)
if __name__ == "__main__":
with open("/content/drive/My Drive/DeleteEfficient/kmeans_data_deletion_NeurIPS19_datasets_scaled.p", mode='rb') as f:
datasets = pickle.load(f)
tune_tree_height(datasets)
print("loss: ")
for d in DATAs:
print(d, mata_loss[d])
print("\nsilhouette coef: ")
for d in DATAs:
print(d, mata_silcoef[d])
print("\nnmi: ")
for d in DATAs:
print(d, mata_nmi[d])
print("\nruntime: ")
for d in DATAs:
print(d, mata_runtime[d])
'''
for i in range(5):
reproduce_result(datasets)
print("loss: ")
for d in DATAs:
print(d, mata_loss[d])
print("\nsilhouette coef: ")
for d in DATAs:
print(d, mata_silcoef[d])
print("\nnmi: ")
for d in DATAs:
print(d, mata_nmi[d])
print("\nruntime: ")
for d in DATAs:
print(d, mata_runtime[d])
'''
'''
tune_tree_buckets(datasets)
print("loss: ")
for d in DATAs:
print(d, mata_loss[d])
print("\nsilhouette coef: ")
for d in DATAs:
print(d, mata_silcoef[d])
print("\nnmi: ")
for d in DATAs:
print(d, mata_nmi[d])
print("\nruntime: ")
for d in DATAs:
print(d, mata_runtime[d])
for i in range(len(DATAs)):
n = datasets[DATAs[i]][1].shape[0]
w = pow(2, math.ceil(math.log(pow(n, 0.3))/math.log(2)))
print(w)
'''
</code>
<code>
import matplotlib.pyplot as plt
x = [2, 3, 4, 5, 6, 7, 8, 9]
nmi1 = [0.3566097971284424, 0.31829612599551427, 0.26160267063730136, 0.2305281490495208, 0.19757822958459795, 0.2033565054065221, 0.29498712519888504, 0.28831689847928715]
nmi2 = [0.3425311624096031, 0.3094292442236454, 0.35199981877611436, 0.3318195994281116, 0.3293841491553029, 0.33256364404326855, 0.32973415740883605, 0.3547205599550721]
nmi3 = [0.48118463715576776, 0.43701346398934654, 0.4907510934535729, 0.49129394972671414, 0.4969504988136086, 0.473456792609371, 0.4853471975095277, 0.4639135028397989]
plt.xlabel('Tree width (number of nodes in the second layer)')
plt.ylabel('Normalized Mutual Information score')
plt.show()
plt.xlabel('Tree width (number of nodes in the second layer)')
plt.ylabel('Silhouette Coefficient')
plt.show()
plt.xlabel('Tree width (number of nodes in the second layer)')
plt.ylabel('Loss')
plt.show()
plt.xlabel('Tree width (number of nodes in the second layer)')
plt.ylabel('Amortized Runtime')
plt.show()
'''
loss:
4celltypes_10pca [[], [], [0.018650796115511764, 0.021989759573573724, 0.02123363641761834, 0.027181128695309625, 0.025302743263513207, 0.022640554885324852, 0.02343571199078768, 0.02120072788214959]]
covtype_multiclass [[], [], [1.0093872899568472, 1.0152784877894863, 0.9924148811850896, 0.9681774327169707, 1.039034620146096, 1.0359270546129151, 0.9939269616770611, 1.0239888286047558]]
postures [[], [], []]
mnist [[], [], [39.6887819207228, 40.85613631974319, 40.033193525386665, 39.61676658453061, 39.824881151451414, 40.029429842563395, 39.98630161691774, 40.603515379831684]]
bot_attack [[], [], []]
silhouette coef:
4celltypes_10pca [[], [], [0.38458809948056094, 0.3255203098801613, 0.4061978235197673, 0.4920635885266285, 0.5391688081505716, 0.5333195615716323, 0.4128338180162358, 0.41701331060616875]]
covtype_multiclass [[], [], [0.2192693277128411, 0.21265828300708067, 0.19295527681699381, 0.28230270768701393, 0.2193945966633595, 0.2204125519130182, 0.188395444095423, 0.23985626611193028]]
postures [[], [], []]
mnist [[], [], [0.06841322262967865, 0.05969255745734885, 0.06913850133189553, 0.05874127371162567, 0.06744032478794254, 0.07063882723256279, 0.075351388420552, 0.07440175793721708]]
bot_attack [[], [], []]
nmi:
4celltypes_10pca [[], [], [0.3566097971284424, 0.31829612599551427, 0.26160267063730136, 0.2305281490495208, 0.19757822958459795, 0.2033565054065221, 0.29498712519888504, 0.28831689847928715]]
covtype_multiclass [[], [], [0.3425311624096031, 0.3094292442236454, 0.35199981877611436, 0.3318195994281116, 0.3293841491553029, 0.33256364404326855, 0.32973415740883605, 0.3547205599550721]]
postures [[], [], []]
mnist [[], [], [0.48118463715576776, 0.43701346398934654, 0.4907510934535729, 0.49129394972671414, 0.4969504988136086, 0.473456792609371, 0.4853471975095277, 0.4639135028397989]]
bot_attack [[], [], []]
runtime:
4celltypes_10pca [[], [], [13.825237035751343, 6.947296857833862, 3.735635995864868, 2.4677200317382812, 2.105858087539673, 3.104292154312134, 5.404259920120239, 10.086616039276123]]
covtype_multiclass [[], [], [21.948658227920532, 10.81125783920288, 5.813812017440796, 4.123564958572388, 3.8815248012542725, 5.878916263580322, 10.245152950286865, 20.703763961791992]]
postures [[], [], []]
mnist [[], [], [176.21602416038513, 85.9961142539978, 44.67074203491211, 26.727442026138306, 18.86534309387207, 20.994234085083008, 34.139708280563354, 63.156938791275024]]
bot_attack [[], [], []]
traintime:
4celltypes_10pca [[], [], [3.0344059467315674, 3.032461166381836, 2.9939041137695312, 3.106005907058716, 3.138749837875366, 3.425178050994873, 3.518235206604004, 4.029762029647827]]
covtype_multiclass [[], [], [4.795390844345093, 4.78167200088501, 4.499297142028809, 4.909643173217773, 5.128259897232056, 4.680608749389648, 5.149761199951172, 5.842344045639038]]
postures [[], [], []]
mnist [[], [], [37.85356378555298, 36.322457790374756, 37.79789113998413, 36.96315813064575, 39.328386068344116, 39.643174171447754, 39.99007725715637, 42.39094400405884]]
bot_attack [[], [], []]
'''
'''
x = np.arange(10)
plt.plot(x, x)
plt.plot(x, 2 * x)
plt.plot(x, 3 * x)
plt.plot(x, 4 * x)
plt.legend(['y = x', 'y = 2x', 'y = 3x', 'y = 4x'], loc='upper left')
plt.show()
'''
</code>
|
{
"filename": "main_1.ipynb",
"repository": "Zhenghua-404/Efficient-Data-Deletion-in-ML",
"query": "transformed_from_existing",
"size": 61163,
"sha": ""
}
|
# isc_tutorial.ipynb
Repository: snastase/isc-tutorial
<a href="https://colab.research.google.com/github/snastase/isc-tutorial/blob/master/isc_tutorial.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
<h1>Intersubject correlation (ISC) tutorial</h1>
This tutorial jupyter notebook accompanies the manuscript "Measuring shared responses across subjects using intersubject correlation" by Nastase, Gazzola, Hasson, and Keysers. The goal of the tutorial is to introduce basic intersubject correlation (ISC) analyses ([Hasson et al., 2004](https://doi.org/10.1126/science.1089506), [2010](https://doi.org/10.1016/j.tics.2009.10.011)) and subsequent statistical tests as implemented in Python using the Brain Imaging Analysis Kit ([BrainIAK](http://brainiak.org/)). Click *Open in playground* to interactively run and edit cells (you may need to sign into a Google account), and use *File* > *Save a copy in Drive...* or *Save a copy in GitHub...* to save your changes. To execute a code cell, click in that cell and press _Shift_ + _Enter_ (or _Shift_ + _Return_ on a Mac). The first time you run a code cell in playground mode, you may receive a warning and a prompt to reset runtimes; click _Run anyway_ followed by _Yes_.
---
Author: Samuel A. Nastase
## Getting started
First, we'll need to [install BrainIAK](http://brainiak.org/docs/installation.html) and its requirements in the cloud instance—this may take a few minutes. For this tutorial, we'll install BrainIAK directly from the [GitHub repository](https://github.com/brainiak/brainiak) to get the most recent features. Note that if you are running this notebook locally, the installation process may vary and you should not execute this code cell. If you want to install BrainIAK locally, run `pip install git+https://github.com/snastase/brainiak.git` in a separate code cell or follow the online [installation instructions](http://brainiak.org/docs/installation.html).
<code>
# Install BrainIAK requirements in Google Colab Linux cloud instance
!apt install build-essential libgomp1 libmpich-dev mpich python3-dev \
python3-pip python3-venv
# Install most recent BrainIAK from GitHub in the cloud instance
!pip install pip==9.0.1
!pip install git+https://github.com/brainiak/brainiak.git
</code>
Next, we'll import the relevant functions from BrainIAK
<code>
from brainiak.isc import (isc, isfc, bootstrap_isc, permutation_isc,
timeshift_isc, phaseshift_isc,
compute_summary_statistic)
from brainiak.io import load_boolean_mask, load_images
from brainiak.image import mask_images, MaskedMultiSubjectData
</code>
If unable to install BrainIAK, we can use basic ISC functionality without the full BrainIAK package. We'll download `isc_standalone.py` from the [GitHub repository](https://github.com/snastase/isc-tutorial) for this tutorial and load the necessary modules locally. If you've cloned the `isc-tutorial` GitHub repository locally, this step is not necessary (as the local directory for the repository already contains `isc_standalone.py`).
<code>
# Download the standalone module if not using BrainIAK
from urllib.request import urlretrieve
urlretrieve('https://github.com/snastase/isc-tutorial/'
'raw/master/isc_standalone.py', 'isc_standalone.py');
</code>
If you're using the `isc_standalone.py` module instead of BrainIAK, import the relevant functions from `isc_standalone`.
<code>
from isc_standalone import (isc, isfc, bootstrap_isc, permutation_isc,
timeshift_isc, phaseshift_isc,
compute_summary_statistic, load_images,
load_boolean_mask, mask_images,
MaskedMultiSubjectData)
</code>
Finally, we'll load several other useful Python modules.
<code>
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from scipy.stats import norm, pearsonr, zscore
from scipy.spatial.distance import squareform
from statsmodels.stats.multitest import multipletests
import nibabel as nib
</code>
## Example data
We'll create a simple simulated dataset for quickly applying ISC analyses, then later apply the analyses to a real fMRI dataset where participants listened to a spoken narrative ([Pie Man](https://themoth.org/stories/pie-man) by Jim O'Grady). Our simulated data will have 1,000 voxels in total comprising 10 "networks" and 300 time points (or TRs).
<code>
# Set parameters for toy time series data
n_subjects = 20
n_TRs = 300
n_voxels = 1000
# Create simple simulated data with high intersubject correlation
def simulated_timeseries(n_subjects, n_TRs, n_voxels=1, noise=1):
signal = np.random.randn(n_TRs, n_voxels // 100)
data = [zscore(np.repeat(signal, 100, axis=1) +
np.random.randn(n_TRs, n_voxels) * noise,
axis=0)
for subject in np.arange(n_subjects)]
return data
# List of subject datasets
data = simulated_timeseries(n_subjects, n_TRs, n_voxels=n_voxels)
</code>
<code>
# Inspect the shape of one of our simulated datasets
print(f"Simulated data shape first subject: {data[0].shape} "
f"\ni.e., {data[0].shape[0]} time points and {data[0].shape[1]} voxels")
# Create a simple visualization of the data
plt.matshow(data[0], cmap='RdYlBu_r', vmin=-3, vmax=3)
plt.grid(False)
plt.xlabel('voxels')
plt.ylabel('time points');
</code>
## ISC analysis
Let's start very simple by computing the ISC for a single voxel (or ROI) across only two participants. This should give us a simple Pearson correlation value (and should match other implementations of Pearson correlation). Note that when you call the `isc` function with `verbose=True` (the default), it outputs some warnings describing what it infers about the input data. If these don't match your assumptions, your input data may be organized improperly.
<code>
# Get the time series for a single voxel in two subjects
subject_a = data[0][:, 0]
subject_b = data[1][:, 0]
# Check the shape of these mini-datasets
print(f"Subject A, first voxel, shape = {subject_a.shape} "
f"\nSubject B, first voxel, shape = {subject_b.shape}")
# Combine these into a list
both_subjects = [subject_a, subject_b]
# Compute the ISC for this voxel across the two subjects
iscs = isc(both_subjects, pairwise=True)
print(f"ISC for first voxel across subjects A and B = {iscs[0]}")
# NB: even for a single voxel, the output ISC is shaped to
# to accommodate an n_ISCs x n_voxels matrix
print(f"ISC output shape = {iscs.shape}"
f"\ni.e., {iscs.shape[0]} ISC value(s) by {iscs.shape[0]} voxel(s)")
# Check that ISC output matches of other correlation functions in python
numpy_corrcoef = np.corrcoef(subject_a, subject_b)[0, 1]
scipy_pearsonr = pearsonr(subject_a, subject_b)[0]
print(f"BrainIAK ISC = {iscs[0]:.6f}"
f"\nNumpy's correlation = {numpy_corrcoef:.6f}"
f"\nScipy's correlation = {scipy_pearsonr:.6f}")
assert np.isclose(iscs, numpy_corrcoef) and np.isclose(iscs, scipy_pearsonr)
</code>
BrainIAK uses Python's logging functionality. To see non-critical messages while running ISC analyses, we temporarily can set the logging level to 'INFO'.
<code>
# Import logging module and set level to INFO
import logging
logging.basicConfig()
logging.getLogger().setLevel(logging.INFO)
# Re-run the previous ISC analyses to see logged info
iscs = isc(both_subjects, pairwise=True)
</code>
<code>
# Set logging back to default level of WARNING
logging.getLogger().setLevel(logging.WARNING)
</code>
When there are three or more subjects, we can compute ISCs using either the pairwise approach (`pairwise=True`), where we compute ISCs between each pair of subjects, or the leave-one-out (`pairwise=False`) approach, where we compute ISCs between each subject and the average time series of other subjects.
### Pairwise approach
Now we'll run the full-scale ISC analysis across all voxels and subjects using the pairwise approach. For a given voxel, the correlations between each pair of subjects are represented in a vector of length
```
n_subjects * (n_subjects - 1) / 2
```
or 190 pairs for 20 subjects. This vector of pairs corresponds to the off-diagonal values of a symmetric subjects-by-subjects correlation matrix.
<code>
# Pairwise approach across all subjects and voxels
iscs = isc(data, pairwise=True)
# Check shape of output ISC values
print(f"ISC values shape = {iscs.shape} \ni.e., {iscs.shape[0]} "
f"pairs and {iscs.shape[1]} voxels"
f"\nMinimum ISC = {np.amin(iscs):.3f}; "
f"maximum ISC = {np.amax(iscs):.3f}")
</code>
For a given voxel, we can convert the vector of pairs to the full correlation matrix for visualization. In the simulated dataset, all subjects were designed to have high ISCs; however, we can add noise to some of the subjects and then visualize the ISC matrix.
<code>
# Visualize the correlation matrix for one voxel
isc_matrix = squareform(iscs[:, 0])
np.fill_diagonal(isc_matrix, 1)
sns.heatmap(isc_matrix, cmap="RdYlBu_r", vmin=-1, vmax=1, square=True,
xticklabels=range(1, 21), yticklabels=range(1, 21))
plt.xlabel('subjects')
plt.ylabel('subjects')
plt.show()
# Create noisier data
noisy_data = np.dstack((np.dstack((
simulated_timeseries(n_subjects // 2, n_TRs,
n_voxels=n_voxels, noise=1))),
np.dstack((
simulated_timeseries(n_subjects // 2, n_TRs,
n_voxels=n_voxels, noise=5)))))
# Recompute ISC and visualize data with noisy subjects
noisy_iscs = isc(noisy_data, pairwise=True)
isc_matrix = squareform(noisy_iscs[:, 0])
np.fill_diagonal(isc_matrix, 1)
sns.heatmap(isc_matrix, cmap="RdYlBu_r", vmin=-1, vmax=1, square=True,
xticklabels=range(1, 21), yticklabels=range(1, 21))
plt.xlabel('subjects')
plt.ylabel('subjects')
plt.show()
</code>
### Leave-one-out approach
Instead of computing ISCs between each pair of subjects, for each subject we can compute the ISC between that subject and the average of all other subjects. Notice that the observed ISC values are typically higher in the leave-one-out approach due to computing correlations between the left-out subject and the cleaner averaged time series from the remaining subjects.
<code>
# Leave-one-out approach
iscs = isc(data, pairwise=False)
# Check shape of output ISC values
print(f"ISC values shape = {iscs.shape} \ni.e., {iscs.shape[0]} "
f"left-out subjects and {iscs.shape[1]} voxel(s)"
f"\nMinimum ISC = {np.amin(iscs):.3f}; "
f"maximum ISC = {np.amax(iscs):.3f}")
</code>
### Input types
Currently, we're a submitting a list of numpy arrays to BrainIAK's `isc` function where each item in the list is a subject's response time course over some number of voxels. Alternatively, we could stack subjects along the 3rd dimension (`np.dstack`) into a single 3-dimensional numpy array and submit this to the `isc` function. If the `isc` function receives a single numpy array, it will assume that the last dimension indexes subjects.
<code>
# Input a list of subjects (same as before)
iscs = isc(data, pairwise=False)
# Stack subjects in 3rd-dimension and recompute ISC
data_stack = np.dstack(data)
print(f"Stacked data shape = {data_stack.shape}"
f"\ni.e., {data_stack.shape[0]} time points, {data_stack.shape[1]} "
f"voxels, and {data_stack.shape[2]} subjects")
# Input stacked numpy array
iscs_from_stack = isc(data_stack, pairwise=False)
# Make sure the ISC outputs are the same
assert np.array_equal(iscs, iscs_from_stack)
</code>
### Summary statistics
Rather than returning ISC values for each pair of subject (in the pairwise approach) or each left-out subject (in the leave-one-out approach), we can use the `summary_statistic` argument to output either the mean or median across the values. Note that by default `summary_statistic=False`. If we request the mean ISC value, the `isc` function will internally apply the Fisher *z*-transformation (`np.arctanh`) prior to computing the mean, then apply the inverse Fisher *z*-transformation (`np.tanh`) to the mean value.
<code>
# Compute mean leave-one-out ISC
iscs = isc(data, pairwise=False, summary_statistic='mean')
print(f"ISC values shape = {iscs.shape} \ni.e., the mean value across "
f"left-out subjects for {iscs.shape[0]} voxel(s)"
f"\nMean ISC for first voxel = {iscs[0]:.3f}")
# Compute median leave-one-out ISC
iscs = isc(data, pairwise=False, summary_statistic='median')
print(f"ISC values shape = {iscs.shape} \ni.e., the median value across "
f"left-out subjects for {iscs.shape[0]} voxel(s)"
f"\nMedian ISC for first voxel = {iscs[0]:.3f}")
</code>
## Statistical tests
BrainIAK provides several nonparametric statistical tests for ISC analysis. Nonparametric tests are preferred due to the inherent correlation structure across ISC values—each subject contributes to the ISC of other subjects, violating assumptions of independence required for standard parametric tests (e.g., *t*-test, ANOVA). The nonparametric statistical tests discussed below return the actual observed ISC values, *p*-values, and the resampling distribution (the bootstrap hypothesis test also returns confidence intervals around the observed ISC statistic). For expediency, we only use 200 resampling iterations here, but 1,000 or more iterations are generally recommended.
### Phase randomization
One approach for statistically assessing ISCs is to randomize the phase of time series across subjects prior to computing ISCs (e.g., [Lerner et al., 2011](https://doi.org/10.1523/jneurosci.3684-10.2011); [Simony et al., 2016](https://doi.org/10.1038/ncomms12141)). This method requires recomputing ISC at each iteration of the randomization test, and is therefore slow. In the pairwise approach, we phase-randomize each subject prior to computing ISCs; however, in the leave-one-out approach, we only phase-randomize the left-out subject prior to computing ISC. At each iteration of the phase randomization test, the same random phase shift is used across all voxels to preserve the spatial autocorrelation of typical fMRI data.
<code>
# Phase randomization using pairwise approach (takes a couple minutes)
observed, p, distribution = phaseshift_isc(data, pairwise=True,
summary_statistic='median',
n_shifts=200)
</code>
<code>
# Inspect shape of null distribution
print(f"Null distribution shape = {distribution.shape}"
f"\ni.e., {distribution.shape[0]} randomizations "
f"and {distribution.shape[1]} voxels")
# Get actual ISC value and p-value for first voxel
print(f"Actual observed ISC value for first voxel = {observed[0]:.3f},"
f"\np-value from randomization test = {p[0]:.3f}")
</code>
### Circular time-shift randomization
A conceptually similar nonparametric approach is to circularly shift the response time series across subjects by random offsets ([Kauppi et al., 2014](https://doi.org/10.3389/fninf.2014.00002)). Time points that would be shifted beyond the end of the time series are wrapped around to the beginning of the time series.
<code>
# Circular time-shift using pairwise approach (takes a couple minutes)
observed, p, distribution = timeshift_isc(data, pairwise=True,
summary_statistic='median',
n_shifts=200)
</code>
<code>
# Inspect shape of null distribution
print(f"Null distribution shape = {distribution.shape}"
f"\ni.e., {distribution.shape[0]} randomizations "
f"and {distribution.shape[1]} voxels")
# Get actual ISC value and p-value for first voxel
print(f"Actual observed ISC value for first voxel = {observed[0]:.3f},"
f"\np-value from randomization test = {p[0]:.3f}")
</code>
### Bootstrap hypothesis test
We can also perform group-level statistical tests that operate directly on the observed ISC values and do not require recomputing ISCs. For one-sample tests, we can resample subjects with replacement to construct a bootstrap distribution around our observed ISC statistic ([Chen et al., 2016](https://doi.org/10.1016/j.neuroimage.2016.05.023)). We can compute confidence intervals around the test statistic using the `ci_percentile` option (default 95%). Hypothesis test is performed by shifting the bootstrap distribution to zero. Note that when constructing the bootstrap distribution using the pairwise approach, subjects (i.e., rows and columns in the subject-by-subject correlation matrix) are sampled with replacement, not pairs (which would disrupt the correlation structure among pairs).
<code>
# Compute ISCs and then run bootstrap hypothesis test on ISCs
iscs = isc(data, pairwise=True, summary_statistic=None)
observed, ci, p, distribution = bootstrap_isc(iscs, pairwise=True,
ci_percentile=95,
summary_statistic='median',
n_bootstraps=200)
</code>
<code>
# Inspect shape of null distribution
print(f"Null distribution shape = {distribution.shape}"
f"\ni.e., {distribution.shape[0]} bootstraps "
f"and {distribution.shape[1]} voxels")
# Get actual ISC value and p-value for first voxel
print(f"Actual observed ISC value for first voxel = {observed[0]:.3f},"
f"\np-value from bootstrap hypothesis test = {p[0]:.3f}")
</code>
### Permutation test
We can use a permutation test to statistically evaluate one- or two-sample tests ([Chen et al., 2016](https://doi.org/10.1016/j.neuroimage.2016.05.023)). In the case of a one-sample test, we use a sign-flipping (-1, +1) approach applied to the observed ISC. For a two-sample test, we supply a `group_assignment` list containing the group labels for each subject. The order of the group assignment list must match the order in which the subjects are supplied to the `isc` function. At each iteration, we randomly reassign the group labels, then compute the test statistic. In the one-sample test, there are `2**n_subjects` number of possible permutations, while in the the two-sample test, there are `n_subjects!` number of possible permutations. In both cases, if the requested number of permutations equals or exceeds the exhaustive list of permutations, an exact test is performed using all possible permutations. However, in most cases the number of subjects will yield an a prohibitively large number of permutations, in which case a Monte Carlo approximate permutation test is used instead of an exact test.
<code>
# Compute ISCs and then run one-sample permutation test on ISCs
iscs = isc(data, pairwise=True, summary_statistic=None)
observed, p, distribution = permutation_isc(iscs, pairwise=True,
summary_statistic='median',
n_permutations=200)
</code>
<code>
# Inspect shape of null distribution
print(f"Null distribution shape = {distribution.shape}"
f"\ni.e., {distribution.shape[0]} permutations "
f"and {distribution.shape[1]} voxels")
# Get actual ISC value and p-value for first voxel
print(f"Actual observed ISC value for first voxel = {observed[0][0]:.3f},"
f"\np-value from permutation test = {p[0]:.3f}")
</code>
<code>
# Note that with few subjects, an exact test is performed
data_n6 = data[:6]
iscs = isc(data_n6, pairwise=True, summary_statistic=None)
observed, p, distribution = permutation_isc(iscs, pairwise=True,
summary_statistic='median',
n_permutations=200)
</code>
If we have two groups we expect to have different ISC values, we must supply a `group_assignment` list. In the case of two groups, we compute the difference between the `summary_statistic` for each group. In the pairwise approach, we compute differences between the `summary_statistic` for within-group correlations, and ignore the between group correlations in the full subject-by-subject correlation matrix containing both groups. Furthermore, permutations are applied to subjects (i.e., rows and columns in the subject-by-subject correlation matrix) and not to pairs (which would disrupt the correlation structure among pairs). We'll construct a dataset where one group of subjects are noisier than the others.
<code>
# Create data with noisy subset of subjects
noisy_data = np.dstack((np.dstack((
simulated_timeseries(n_subjects // 2, n_TRs,
n_voxels=n_voxels, noise=1))),
np.dstack((
simulated_timeseries(n_subjects // 2, n_TRs,
n_voxels=n_voxels, noise=5)))))
# Create group_assignment variable with group labels
group_assignment = [1]*10 + [2]*10
print(f"Group assignments: \n{group_assignment}")
# Compute ISCs and then run two-sample permutation test on ISCs
iscs = isc(noisy_data, pairwise=True, summary_statistic=None)
observed, p, distribution = permutation_isc(iscs,
group_assignment=group_assignment,
pairwise=True,
summary_statistic='median',
n_permutations=200)
</code>
<code>
# Inspect shape of null distribution
print(f"Null distribution shape = {distribution.shape}"
f"\ni.e., {distribution.shape[0]} permutations "
f"and {distribution.shape[1]} voxels")
# Get actual ISC value and p-value for first voxel
print(f"Actual observed group difference in ISC values "
f"for first voxel = {observed[0]:.3f},"
f"\np-value from permutation test = {p[0]:.3f}")
</code>
## Correcting for multiple tests
Evaluating the statistical significance of an ISC analysis across many voxels will result in many false positives unless we somehow control for the large number of statistical tests. Here we'll use two simple approaches for correcting for multiple tests. In the first approach, we'll account for multiple tests by controlling the expected proportion of false positives or false discovery rate (FDR; [Benjamini & Hochberg, 1995](https://www.jstor.org/stable/2346101); [Benjamini & Yekutieli, 2001](https://www.jstor.org/stable/2674075); [Genovese et al., 2002](https://doi.org/10.1006/nimg.2001.1037)). In the second approach, we'll control the family-wise error rate (FWER) by constructing a null distribution from the maximum ISC value across all voxels at each iteration of a randomization test ([Nichols & Holmes, 2002](https://doi.org/10.1002/hbm.1058)).
First, we'll create a dataset where half the voxels are very consistent across subjects and the other half are very noisy.
<code>
# Create data with where half of voxels are noisy
noisy_data = np.hstack((np.dstack((
simulated_timeseries(n_subjects, n_TRs,
n_voxels=n_voxels // 2, noise=1))),
np.dstack((
simulated_timeseries(n_subjects, n_TRs,
n_voxels=n_voxels // 2, noise=9)))))
# Visualize data for first subject where half of voxels are noisy
plt.matshow(noisy_data[..., 0], cmap='RdYlBu_r', vmin=-3, vmax=3)
plt.grid(False)
plt.xlabel('voxels')
plt.ylabel('time points');
</code>
After visualizing these data, we'll compute ISCs and use a one-sample two-sided bootstrap hypothesis test, which yields *p*-values and a null distribution. For this example, we'll use a more realistic number of bootstrap samples (1,000)—this may take a couple minutes.
<code>
# Compute ISCs and then run bootstrap hypothesis test on ISCs
# using a realistic number of permutations (takes a few minutes)
iscs = isc(noisy_data, pairwise=True, summary_statistic=None)
observed, ci, p, distribution = bootstrap_isc(iscs, pairwise=True,
ci_percentile=95,
summary_statistic='median',
n_bootstraps=1000)
</code>
### Controlling FDR
To control FDR, we'll use the the `multipletests` function from the StatsModels Python package. This returns an array of *q*-values, which are typically interpreted as FDR-corrected *p*-values. By thresholding uncorrected and corrected *p*- and *q*-values, we can determine how many voxels survived correction for multiple tests.
<code>
# Get q-values (i.e., FDR-controlled p-values) using statsmodels
q = multipletests(p, method='fdr_by')[1]
# We can also convert these q-values to z-values
z = np.abs(norm.ppf(q))
# Also get significant voxels with and without correction
corrected = q[np.newaxis, :] < .05
uncorrected = p[np.newaxis, :] < .05
# Count significant voxels before and after correction
print(f'{np.sum(uncorrected)} "significant" voxels before correction for '
f"multiple tests; {np.sum(corrected)} significant voxels after "
f"controlling FDR at .05")
</code>
Finally, we can visualize the voxel time series for an example subject, the ISC values across subjects, and which voxels are considered significant before and after controlling FDR at .05. Note that before correction even some of the noisy voxels are considered to have significant ISC; however, after correction, the number of significant noisy voxels is reduced.
<code>
# Set up grid of subplots for visualizing voxel values and significance
fig, (ax0, ax1, ax2, ax3) = plt.subplots(nrows=4, figsize=(12, 8),
sharex=True,
gridspec_kw={'height_ratios':
[300, 190, 20, 20]})
# Visualize data for first subject where half of voxels are noisy
ax0.matshow(noisy_data[..., 0], cmap='RdYlBu_r', vmin=-3, vmax=3)
ax0.grid(False)
ax0.set_ylabel('time points')
ax0.set_title('response time series for example subject', y=1)
# Visualize ISC values across all pairs of subjects
ax1.matshow(iscs, cmap='RdYlBu_r', vmin=-1, vmax=1)
ax1.grid(False)
ax1.set_ylabel('pairs of subjects')
ax1.set_title('ISC values for all pairs of subjects', y=1)
# Visualize uncorrected and corrected significant voxels
ax2.matshow(np.repeat(uncorrected, 20, axis=0),
cmap='viridis',vmin=0, vmax=1)
ax2.grid(False)
ax2.set_yticks([])
ax2.set_title('uncorrrected "significant" voxels (yellow)')
ax3.matshow(np.repeat(corrected, 20, axis=0),
cmap='viridis',vmin=0, vmax=1)
ax3.grid(False)
ax3.set_xlabel('voxels')
ax3.xaxis.tick_bottom()
ax3.set_yticks([])
ax3.set_title('FDR-corrrected significant voxels (yellow)')
plt.tight_layout()
</code>
### Controlling FWER
To strictly control the FWER, one method is to construct a null distribution of maximum ISC statistics across all voxels. First we'll use the `permutation_isc` function to run a one-sample two-sided permutation test using a sign-flipping procedure, which returns *p*-values and a null distribution.
<code>
# Compute ISCs and then run two-sample permutation test on ISCs
iscs = isc(noisy_data, pairwise=True, summary_statistic=None)
observed, p, distribution = permutation_isc(iscs, pairwise=True,
summary_statistic='mean',
n_permutations=1000)
</code>
Next, we'll write a simple function that takes a null distribution with multiple voxels, and aggregates the maximum ISC value across all voxels for each null sample.
<code>
# Loop through null distribution and get maximum value across voxels
def get_maxima(distribution):
max_distribution = []
for i in distribution:
max_isc = np.amax(i)
max_distribution.append(max_isc)
max_distribution = np.array(max_distribution)
return max_distribution
</code>
After we create a null distribution of maximum statistics, any voxel with an ISC value in the top 5% of distribution can be considered significant. Here we compute *p*-values from the null distribution of maximum statistics using a two-sided test.
<code>
# Create null distribution of maximum ISCs across all voxels
max_distribution = get_maxima(distribution)
# Broadcast our max distribution across all 1000 voxels
max_distribution = np.repeat(max_distribution[:, np.newaxis], 1000, axis=1)
# Get the summary statistic (median) for our actual ISC values
# since we set summary_statistic=None above
observed = np.median(iscs, axis=0)[np.newaxis, :]
# Evaluate whether observed ISCs land in the tail of the max distribution
p_max = ((np.sum(np.abs(max_distribution) >= np.abs(observed), axis=0) + 1) /
float((len(max_distribution) + 1)))[np.newaxis, :]
# Get p-values less than .05 (corrected for multiple tests)
corrected = p_max < .05
</code>
As with the FDR approach, we can visualize the data and the voxels marked as significant before and after correction for multiple tests. This method of correction for multiple tests is considerably more conservative.
<code>
# Set up grid of subplots for visualizing voxel values and significance
fig, (ax0, ax1, ax2, ax3) = plt.subplots(nrows=4, figsize=(12, 8),
sharex=True,
gridspec_kw={'height_ratios':
[300, 190, 20, 20]})
# Visualize data for first subject where half of voxels are noisy
ax0.matshow(noisy_data[..., 0], cmap='RdYlBu_r', vmin=-3, vmax=3)
ax0.grid(False)
ax0.set_ylabel('time points')
ax0.set_title('response time series for example subject', y=1)
# Visualize ISC values across all pairs of subjects
ax1.matshow(iscs, cmap='RdYlBu_r', vmin=-1, vmax=1)
ax1.grid(False)
ax1.set_ylabel('pairs of subjects')
ax1.set_title('ISC values for all pairs of subjects', y=1)
# Visualize uncorrected and corrected significant voxels
ax2.matshow(np.repeat(uncorrected, 20, axis=0),
cmap='viridis',vmin=0, vmax=1)
ax2.grid(False)
ax2.set_yticks([])
ax2.set_title('uncorrrected "significant" voxels (yellow)')
ax3.matshow(np.repeat(corrected, 20, axis=0),
cmap='viridis',vmin=0, vmax=1)
ax3.grid(False)
ax3.set_xlabel('voxels')
ax3.xaxis.tick_bottom()
ax3.set_yticks([])
ax3.set_title('FWER-corrrected significant voxels (yellow)')
plt.tight_layout()
</code>
Note that there are many other ways to correct for multiple tests, such as using cluster-extent thresholding; but these methods are beyond the scope of this tutorial.
## ISFC analysis
Rather than computing ISCs for corresponding voxels across participants, we can instead compute ISCs between all voxels to measure functional integration (i.e., connectivity). This method is called intersubject functional correlation (ISFC) analysis ([Simony et al., 2016](https://doi.org/10.1038/ncomms12141)). Using the `vectorize_isfcs` option, we can either return a tuple containing the condensed off-diagonal ISFC values and the diagonal ISC values or the square (redundant) ISFC values. If `vectorize_isfcs=True` (the default), the first array in the tuple contains the off-diagonal ISFC values for each pair of voxels as condensed by `scipy.spatial.distance.squareform` and is shaped `n_subjects` (or `n_pairs`) by `n_connections` where
```
n_connections = n_voxels * (n_voxels - 1) / 2
```
The second array in the tuple is the diagonal values shaped `n_subjects` (or `n_pairs`) by `n_voxels`. If `vectorize_isfcs=False`, we get a 3-dimensional array containing the square (redundant) ISFC and ISC values, shaped `n_subjects` (or `n_pairs`) by `n_voxels` by `n_voxels`. If a `summary_statistic` is supplied, or only two subjects are input, the singleton first dimension is removed.
<code>
# Compute ISFCs using leave-one-out approach
isfcs, iscs = isfc(data, pairwise=False, vectorize_isfcs=True)
# Check shape of output ISFC values
print(f"ISFC output shape = {isfcs.shape}\ni.e., {isfcs.shape[0]} "
f"left-out subjects by {isfcs.shape[1]} connections (i.e., voxel pairs)"
f"\nISCs output shape = {iscs.shape}\ni.e., {iscs.shape[0]} "
f"left-out subjects by {iscs.shape[1]} voxels")
</code>
Alternatively, we can retain the (redundant) structure of the ISFC matrices using `vectorize_isfcs=False` to yield a 3-dimensional array of shape `n_subjects` by `n_voxels` by `n_voxels`:
<code>
# Compute ISFCs using leave-one-out approach
isfcs = isfc(data, pairwise=False, vectorize_isfcs=False)
# Check shape of output ISFC values
print(f"ISFC output shape = {isfcs.shape}\ni.e., {isfcs.shape[0]} "
f"left-out subjects by {isfcs.shape[1]} voxels by {isfcs.shape[2]} "
"voxels")
</code>
We can also supply a `summary_statistic` to collapse the ISFC values over left-out subjects or pairs of subjects:
<code>
# Compute ISFCs using leave-one-out approach with mean
isfcs, iscs = isfc(data, pairwise=False, summary_statistic='mean',
vectorize_isfcs=True)
# Check shape of output ISFC values
print(f"Mean ISFC output shape = {isfcs.shape}\ni.e., {isfcs.shape[0]} "
f"connections (i.e., voxel pairs)"
f"\nMean ISC output shape = {iscs.shape}\ni.e., {iscs.shape[0]} "
"voxels")
</code>
We can use the `brainiak.isc.squareform_isfc` convenience function to convert between the condensed representation of ISFCs (with ISCs) and the square (redundant) representation of ISFCs. This function mimics `scipy.spatial.distance.squareform`, but retains the diagonal ISC values.
<code>
from brainiak.isc import squareform_isfc
# Start with square (redundant) ISFCs and check shape
isfcs_sq = isfc(data, pairwise=False, vectorize_isfcs=False)
print(f"Square (redundant) ISFCs shape: {isfcs_sq.shape}")
# Convert these directly to condensed ISFCs (and ISCs)
isfcs_c, iscs = squareform_isfc(isfcs_sq)
print(f"Condensed ISFCs shape: {isfcs_c.shape}, "
f"ISCs shape: {iscs.shape}")
# Convert these directly back to redundant ISFCs
isfcs_r = squareform_isfc(isfcs_c, iscs)
print(f"Converted redundant ISFCs shape: {isfcs_r.shape}")
# Check that they are identical to the original square ISFCs
assert np.array_equal(isfcs_sq, isfcs_r)
</code>
Let's confirm that the diagonal of the ISFC matrix represents each voxel correlated with itself across subjects—the conventional ISC described above. We can see that the conventional ISC analysis is in fact a subset of the ISFC analysis.
<code>
# Get ISC values directly from ISFC matrix
isfcs, iscs = isfc(data, pairwise=False, vectorize_isfcs=True)
# Check that these are the same as conventional ISCs
assert np.allclose(iscs, isc(data))
</code>
Finally, we can visualize the matrix of mean (or median) ISFC values. If we used `vectorize_isfcs=True`, we'll first need to apply `squareform_isfc` the ISFC (and ISC) values. The diagonal blocks represent the 10 artificial "networks" in our simulated data; the 100 voxels in each network are highly correlated with each other and largely uncorrelated with voxels in other networks.
<code>
# Recompute mean ISFCs
isfcs, iscs = isfc(data, pairwise=False, summary_statistic='mean',
vectorize_isfcs=True)
# Convert these to a square representation
isfcs = squareform_isfc(isfcs, iscs)
# Visual mean ISFC matrix
plt.matshow(isfcs, cmap="RdYlBu_r", vmin=-1, vmax=1)
plt.grid(False)
plt.xticks(np.arange(0, 1001, 100)[1:], np.arange(100, 1001, 100),
rotation=45)
plt.gca().xaxis.tick_top()
plt.gca().xaxis.set_label_position('top')
plt.yticks(np.arange(0, 1001, 100)[1:], np.arange(100, 1001, 100))
plt.xlabel('voxels')
plt.ylabel('voxels')
ax = plt.gca()
plt.colorbar(fraction=0.046, pad=0.04);
</code>
We can inject some structure into our simulated data to yield a more realistic ISFC matrix.
<code>
# Create more structured simulated data with 7 "networks";
# don't worry about the details
from scipy.ndimage import gaussian_filter1d
def structured_timeseries(n_subjects, n_TRs, n_voxels=1000, noise=1):
signals = np.random.randn(n_TRs, 3)
networks = np.column_stack((signals + np.random.randn(n_TRs, 3) * noise,
signals[:, 0] + np.random.randn(n_TRs) * noise,
signals[:, 0] + np.random.randn(n_TRs) * noise,
-signals[:, 2] + np.random.randn(n_TRs) * noise,
signals[:, 2] + np.random.randn(n_TRs) * noise))
networks = networks[:, [0, 3, 4, 5, 1, 2, 6]]
six = np.random.randint(n_voxels // 20, n_voxels // 6, 6)
seven = np.append(six, (n_voxels - np.sum(six)))
voxels = np.column_stack([np.tile(network[:, np.newaxis], (1, extent))
for network, extent in zip(networks.T, seven)])
areas = [0] + sorted(np.random.randint(0, 1000, 16))
areas = np.diff(areas).tolist() + [(1000 - areas[-1])]
noise_sources = np.random.randn(n_TRs, 7)
structured_noise = np.column_stack([np.tile(
(noise_sources[:, np.random.choice(range(7))] *
np.random.choice([-1, 1, 1, 1]))[:, np.newaxis],
(1, extent))
for extent in areas])
voxels = gaussian_filter1d(voxels, 8.0, axis=0)
structured_noise = gaussian_filter1d(structured_noise, 8.0, axis=0)
data = []
for s in np.arange(n_subjects):
data.append(voxels + structured_noise * noise * .2 +
np.random.randn(n_TRs, n_voxels) * noise * 1.35)
data = np.dstack(data)
return data
structured_data = structured_timeseries(n_subjects, n_TRs)
</code>
Now, we can recompute mean ISFCs using the leave-one-out approach and visualize the resulting ISFC matrix.
<code>
# Compute ISFCs using leave-one-out approach with mean
isfcs, iscs = isfc(structured_data, pairwise=False, summary_statistic='mean',
vectorize_isfcs=True)
# Convert these to a square representation
isfcs = squareform_isfc(isfcs, iscs)
# Visual mean ISFC matrix
plt.matshow(isfcs, cmap="RdYlBu_r", vmin=-.3, vmax=.3)
plt.grid(False)
plt.xticks(np.arange(0, 1001, 100)[1:], np.arange(100, 1001, 100),
rotation=45)
plt.gca().xaxis.tick_top()
plt.gca().xaxis.set_label_position('top')
plt.yticks(np.arange(0, 1001, 100)[1:], np.arange(100, 1001, 100))
plt.xlabel('voxels')
plt.ylabel('voxels')
ax = plt.gca()
plt.colorbar(fraction=0.046, pad=0.04);
</code>
## Real fMRI data
Next, we'll download a publicly available fMRI dataset and run an ISC analysis. This dataset comprises fMRI data for 20 subjects listening to the spoken story [Pie Man](https://themoth.org/stories/pie-man) by Jim O'Grady (archived on the [Princeton DataSpace](https://dataspace.princeton.edu/jspui/handle/88435/dsp01dz010s83s)). Note that we use 20 subjects to minimize computational demands for this tutorial and recommend larger sample sizes for publication. The gzipped data archive file is ~1.5 GB in size, and may take a couple minutes to download and unzip. The functional data were acquired with 3 x 3 x 4 mm voxels and 1.5 s TRs. Data were preprocessed using [fMRIPrep](https://fmriprep.readthedocs.io/en/stable/) ([Esteban et al., 2018](https://doi.org/10.1038/s41592-018-0235-4)), including spatial normalization to MNI space (the T1-weighted [ICBM 2009c Nonlinear Asymmetric template](http://nist.mni.mcgill.ca/?p=904)). The data were then smoothed to 6 mm FWHM using [AFNI](https://afni.nimh.nih.gov/)'s [3dBlurToFWHM](https://afni.nimh.nih.gov/pub/dist/doc/program_help/3dBlurToFWHM.html) ([Cox, 1996](https://doi.org/10.1006/cbmr.1996.0014)). The following confound variables were regressed out using [3dTproject](https://afni.nimh.nih.gov/pub/dist/doc/program_help/3dTproject.html): six head motion parameters (and their first derivatives), framewise displacement, six prinicipal components from an anatomical mask of cerebrospinal fluid (CSF) and white matter, sine/cosine bases for high-pass filtering (cutoff: 0.00714 Hz; 140 s), as well as a linear and quadratic trends. The anatomical template and a brain mask (i.e., excluding skull) are supplied as well. These have been resampled to match resolution of the functional images.
<code>
# Download data tarball from Princeton DataSpace
from urllib.request import urlretrieve
urlretrieve('https://dataspace.princeton.edu/jspui/bitstream/'
'88435/dsp01dz010s83s/6/pieman-isc-tutorial.tgz',
'pieman-isc-tutorial.tgz');
!tar -xvzf pieman-isc-tutorial.tgz
</code>
### Loading MRI data
We'll use NiBabel as well as BrainIAK's `io` and `image` functionality to load the functional data and apply a brain mask.
<code>
# Import functions helpful for managing file paths
from glob import glob
from os.path import join
data_dir = 'pieman-isc-tutorial'
# Filenames for MRI data; gzipped NIfTI images (.nii.gz)
func_fns = glob(join(data_dir, ('sub-*_task-pieman_space-MNI152NLin2009cAsym'
'_desc-tproject_bold.nii.gz')))
mask_fn = join(data_dir, 'MNI152NLin2009cAsym_desc-brain_mask.nii.gz')
mni_fn = join(data_dir, 'MNI152NLin2009cAsym_desc-brain_T1w.nii.gz')
# Load a NIfTI of the brain mask as a reference Nifti1Image
ref_nii = nib.load(mask_fn)
# Load functional images and masks using brainiak.io
func_imgs = load_images(func_fns)
mask_img = load_boolean_mask(mask_fn)
# Get coordinates of mask voxels in original image
mask_coords = np.where(mask_img)
# Apply the brain mask using brainiak.image
masked_imgs = mask_images(func_imgs, mask_img)
# Collate data into a single TR x voxel x subject array
orig_data = MaskedMultiSubjectData.from_masked_images(masked_imgs,
len(func_fns))
</code>
The data from each subject are stacked along the third dimension, yielding a `n_TRs` by `n_voxels` by `n_subjects` array. The functional acquisition originally included 13 s of music and 2 s of silence prepended to the story stimulus and an additional 13 s of silence after the story (450 s or 300 TRs in total). These segments as well as the first 12 s (8 TRs) after story onset can be discarded to minimize stimulus onset/offset effects. We may also opt to z-score the time series for each voxel.
<code>
print(f"Original fMRI data shape: {orig_data.shape} "
f"\ni.e., {orig_data.shape[0]} time points, {orig_data.shape[1]} voxels, "
f"{orig_data.shape[2]} subjects")
# Trim off non-story TRs and 12 s post-onset
data = orig_data[18:-8, ...]
print(f"Trimmed fMRI data shape: {data.shape} "
f"\ni.e., {data.shape[0]} time points, {data.shape[1]} voxels, "
f"{data.shape[2]} subjects")
# Z-score time series for each voxel
data = zscore(data, axis=0)
</code>
### ISC analysis
Next, we'll run a leave-one-out ISC analysis on the preprocessed fMRI data, including all voxels in the brain mask—this may take a few minutes. Note that some voxels with no variance over time for one or more subjects were included in the brain mask due to the limited field of view during EPI acquisition and susceptibility artifacts (signal dropout). This yields NaN (not a number) values. Here, we'll use set `tolerate_nans` to `0.8` to ensure that, when computing the averaging time series for *N*–1 subjects in the leave-one-out approach, only voxels with >= 80% of subjects have non-NaN values are included.
<code>
# Leave-one-out approach
iscs = isc(data, pairwise=False, tolerate_nans=.8)
# Check shape of output ISC values
print(f"ISC values shape = {iscs.shape} \ni.e., {iscs.shape[0]} "
f"left-out subjects and {iscs.shape[1]} voxel(s)")
</code>
Since we didn't supply a `summary_statistic` in the `isc` call, we get an ISC value for each left-out subject (we'll need ISCs for each subject in the subsequent statistical test). If we want to preliminarily inspect the mean (or median) ISCs, we can apply the `brainiak.isc.compute_summary_statistic` function afterward. Note that if we specifed a `summary_statistic` in the `isc` call, the `isc` function would simply use `compute_summary_statistic` internally.
<code>
# Compute mean ISC (with Fisher transformation)
mean_iscs = compute_summary_statistic(iscs, summary_statistic='mean', axis=0)
print(f"ISC values shape = {mean_iscs.shape} \ni.e., {mean_iscs.shape[0]} "
f"mean value across left-out subjects and {iscs.shape[1]} voxel(s)"
f"\nMinimum mean ISC across voxels = {np.nanmin(mean_iscs):.3f}; "
f"maximum mean ISC across voxels = {np.nanmax(mean_iscs):.3f}")
# Compute median ISC
median_iscs = compute_summary_statistic(iscs, summary_statistic='median',
axis=0)
print(f"ISC values shape = {median_iscs.shape} \ni.e., {median_iscs.shape[0]} "
f"median value across left-out subjects and {iscs.shape[1]} voxel(s)"
f"\nMinimum median ISC across voxels = {np.nanmin(median_iscs):.3f}; "
f"maximum median ISC across voxels = {np.nanmax(median_iscs):.3f}")
</code>
### Statistical testing
To test whether the observed ISCs are significantly greater than zero, we'll perform a bootstrap hypothesis test ([Chen et al., 2016](https://doi.org/10.1016/j.neuroimage.2016.05.023)). This may also take a couple minutes.
<code>
# Run bootstrap hypothesis test on ISCs
observed, ci, p, distribution = bootstrap_isc(iscs, pairwise=False,
ci_percentile=95,
summary_statistic='median',
n_bootstraps=1000)
</code>
Before we correct for multiple tests, we should exclude any voxels with NaNs. To do this, we'll extract the non-NaN voxels, run the correction for multiple tests, then reinsert the non-NaN voxels into the full mask.
<code>
# Get number of NaN voxels
n_nans = np.sum(np.isnan(observed))
print(f"{n_nans} voxels out of {observed.shape[0]} are NaNs "
f"({n_nans / observed.shape[0] * 100:.2f}%)")
# Get voxels without NaNs
nonnan_mask = ~np.isnan(observed)
nonnan_coords = np.where(nonnan_mask)
# Mask both the ISC and p-value map to exclude NaNs
nonnan_isc = observed[nonnan_mask]
nonnan_p = p[nonnan_mask]
</code>
Now we'll apply the `multipletests` function from StatsModels to the *p*-values from the bootstrap hypothesis test to control the false discovery rate (FDR) at 0.05 across all voxels. This yields a map of *q*-values. We can then threshold our ISC image based on the FDR-adjusted *q*-values, which are derived from the entire image, rather than the uncorrected *p*-values.
<code>
# Get FDR-controlled q-values
nonnan_q = multipletests(nonnan_p, method='fdr_by')[1]
threshold = .05
print(f"{np.sum(nonnan_q < threshold)} significant voxels "
f"controlling FDR at {threshold}")
# Threshold ISCs according FDR-controlled threshold
nonnan_isc[nonnan_q >= threshold] = np.nan
# Reinsert thresholded ISCs back into whole brain image
isc_thresh = np.full(observed.shape, np.nan)
isc_thresh[nonnan_coords] = nonnan_isc
</code>
### Visualizing results
Finally, to visualize the significant ISC values, we must first reformat the 2-dimensional masked array into a 3-dimensional NIfTI image. We'll use an arbitrary reference NIfTI image `ref_nii` (the brain mask) to assign affine and header information correctly.
<code>
# Create empty 3D image and populate
# with thresholded ISC values
isc_img = np.full(ref_nii.shape, np.nan)
isc_img[mask_coords] = isc_thresh
# Convert to NIfTI image
isc_nii = nib.Nifti1Image(isc_img, ref_nii.affine, ref_nii.header)
</code>
We'll use `nilearn.plotting.plot_stat_map` to plot two views of the NIfTI image. We'll set the maximum ISC value for the colorbar at 0.5 and use a divergent colormap called `RdYlBu_r`. This yields maps where significant voxels are colored according to the median ISC value across left-out subjects. Statistical significance was assessed by a
nonparametric bootstrap hypothesis test resampling left-out subjects and corrected for multiple tests by controlling FDR at .05.
<code>
# Install Nilearn for plotting functionality
!pip install nilearn
from nilearn.plotting import plot_stat_map
%matplotlib inline
</code>
<code>
# Plot slices at coordinates -61, -20, 8
plot_stat_map(
isc_nii,
cmap='RdYlBu_r',
vmax=.5,
cut_coords=(-61, -20, 8))
# Plot slices at coordinates 0, -65, 40
plot_stat_map(
isc_nii,
cmap='RdYlBu_r',
vmax=.5,
cut_coords=(0, -65, 40))
plt.show()
</code>
Some significant voxels have fairly low ISCs, so we can also use the `threshold` option in `plot_stat_map` to also exclude voxels with, e.g., ISC < .1.
<code>
# Plot slices at coordinates -61, -20, 8
plot_stat_map(
isc_nii,
cmap='RdYlBu_r',
vmax=.5,
threshold=.1,
cut_coords=(-61, -20, 8))
# Plot slices at coordinates 0, -65, 40
plot_stat_map(
isc_nii,
cmap='RdYlBu_r',
vmax=.5,
threshold=.1,
cut_coords=(0, -65, 40))
</code>
Note that here we are only analyzing responses to the fully intact Pie Man story, which yields high ISCs in both low-level auditory areas and higher-level areas processing narrative content. However, if we scrambled the presentation of the story so as to disrupt the temporally contiguous narrative, we would expect to see high ISCs only in low-level auditory areas ([Hasson et al., 2008](https://doi.org/10.1523/jneurosci.5487-07.2008); [Lerner et al., 2011](https://doi.org/10.1523/jneurosci.3684-10.2011)). To finish, we can also use `nib.save` to save the NIfTI image for use with other neuroimaging data analysis and visualization programs.
<code>
# Save final ISC NIfTI image as .nii
isc_fn = 'isc_thresh_pieman_n20.nii.gz'
nib.save(isc_nii, isc_fn)
</code>
## References and suggested reading
* Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 289–300. https://www.jstor.org/stable/2346101
* Benjamini, Y., & Yekutieli, D. (2001). The control of the false discovery rate in multiple testing under dependency. *Annals of Statistics*, *29*(4), 1165–1188. https://www.jstor.org/stable/2674075
* Chen, G., Shin, Y. W., Taylor, P. A., Glen, D. R., Reynolds, R. C., Israel, R. B., & Cox, R. W. (2016). Untangling the relatedness among correlations, part I: nonparametric approaches to inter-subject correlation analysis at the group level. *NeuroImage*, *142*, 248–259. https://doi.org/10.1016/j.neuroimage.2016.05.023
* Chen, G., Taylor, P. A., Shin, Y. W., Reynolds, R. C., & Cox, R. W. (2017). Untangling the relatedness among correlations, part II: inter-subject correlation group analysis through linear mixed-effects modeling. *NeuroImage*, *147*, 825–840. https://doi.org/10.1016/j.neuroimage.2016.08.029
* Cox, R. W. (1996). AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. *Computers and Biomedical Research*, *29*(3), 162–173. https://doi.org/10.1006/cbmr.1996.0014
* Esteban, O., Markiewicz, C., Blair, R. W., Moodie, C., Isik, A. I., Erramuzpe, A., Kent, J. D., Goncalves, M., DuPre, E., Snyder, M., Oya, H., Ghosh, S., Wright, J., Durnez, J., Poldrack, R., & Gorgolewski, K. J. (2018). fMRIPrep: a robust preprocessing pipeline for functional MRI. *Nature Methods*. https://doi.org/10.1038/s41592-018-0235-4
* Genovese, C. R., Lazar, N. A., & Nichols, T. (2002). Thresholding of statistical maps in functional neuroimaging using the false discovery rate. *NeuroImage*, *15*(4), 870–878. https://doi.org/10.1006/nimg.2001.1037
* Hasson, U., Ghazanfar, A. A., Galantucci, B., Garrod, S., & Keysers, C. (2012). Brain-to-brain coupling: a mechanism for creating and sharing a social world. *Trends in Cognitive Sciences*, *16*(2), 114–121. https://doi.org/10.1016/j.tics.2011.12.007
* Hasson, U., Malach, R., & Heeger, D. J. (2010). Reliability of cortical activity during natural stimulation. *Trends in Cognitive Sciences*, *14*(1), 40–48. https://doi.org/10.1016/j.tics.2009.10.011
* Hasson, U., Nir, Y., Levy, I., Fuhrmann, G., & Malach, R. (2004). Intersubject synchronization of cortical activity during natural vision. *Science*, *303*(5664), 1634–1640. https://doi.org/10.1126/science.1089506
* Hasson, U., Yang, E., Vallines, I., Heeger, D. J., & Rubin, N. (2008). A hierarchy of temporal receptive windows in human cortex. *Journal of Neuroscience*, *28*(10), 2539–2550. https://doi.org/10.1523/jneurosci.5487-07.2008
* Kauppi, J. P., Pajula, J., & Tohka, J. (2014). A versatile software package for inter-subject correlation based analyses of fMRI. *Frontiers in Neuroinformatics*, *8*, 2. https://doi.org/10.3389/fninf.2014.00002
* Lerner, Y., Honey, C. J., Silbert, L. J., & Hasson, U. (2011). Topographic mapping of a hierarchy of temporal receptive windows using a narrated story. *Journal of Neuroscience*, *31*(8), 2906–2915. https://doi.org/10.1523/jneurosci.3684-10.2011
* Nichols, T. E., & Holmes, A. P. (2002). Nonparametric permutation tests for functional neuroimaging: a primer with examples. *Human Brain Mapping*, *15*(1), 1–25. https://doi.org/10.1002/hbm.1058
* Silbert, L. J., Honey, C. J., Simony, E., Poeppel, D., & Hasson, U. (2014). Coupled neural systems underlie the production and comprehension of naturalistic narrative speech. *Proceedings of the National Academy of Sciences of the United States of America*, *111*(43), E4687–E4696. https://doi.org/10.1073/pnas.1323812111
* Simony, E., Honey, C. J., Chen, J., Lositsky, O., Yeshurun, Y., Wiesel, A., & Hasson, U. (2016). Dynamic reconfiguration of the default mode network during narrative comprehension. *Nature Communications*, *7*, 12141. https://doi.org/10.1038/ncomms12141
* Stephens, G. J., Silbert, L. J., & Hasson, U. (2010). Speaker–listener neural coupling underlies successful communication. *Proceedings of the National Academy of Sciences of the United States of America*, *107*(32), 14425–14430. https://doi.org/10.1073/pnas.1008662107
|
{
"filename": "isc_tutorial.ipynb",
"repository": "snastase/isc-tutorial",
"query": "transformed_from_existing",
"size": 84060,
"sha": ""
}
|
# PracticalMaterialDay4_AfternoonPractical.ipynb
Repository: DieStok/Basic-Machine-Learning-for-Bioinformatics
## Afternoon practical day 4
Welcome to the final practical of today. Here you'll be working with sequence data, first getting to the point that you have the sequences all aligned, and then using the distances you can only really calculate then to get a clustering of the sequences. As always, first run the two cells below.
<code>
#run this cell to set things up
import ipywidgets as widgets, numpy as np, pandas as pd
from numpy.random import default_rng
%matplotlib inline
import matplotlib.pyplot as plt
import math
import seaborn as sns
from IPython.display import display, Markdown
from scipy.optimize import fmin_bfgs, fmin_cg, fmin
import sklearn
import itertools
from Bio import SeqIO
</code>
<code>
# important functions
def calcEucliDist(vectorOne, vectorTwo):
return np.linalg.norm(vectorOne-vectorTwo, axis = 1)
def calcAbsDist(vectorOne, vectorTwo):
#using linalg.norm:
return np.linalg.norm(vectorOne-vectorTwo, ord = 1, axis = 1)
def makeKMeanClusters(X, k, funName = "calcEucliDist", maxIter = 50, nClusteringsToPerform = 20):
if k <= 0:
print("K must be greater than 0!")
return None
if k > len(X):
print("K cannot be larger than the # of samples in your data!")
return None
if maxIter <= 0:
print("Cannot have negative or 0 iterations!")
return None
resultToReturn = [None, None, None, None]
bestDistortion = np.Inf
for clusteringIndex in range(0, nClusteringsToPerform):
initialCentroids = X[np.random.choice(X.shape[0], k, replace=False), :]
if len(initialCentroids) != k:
print("Centroids lost!")
centroids = initialCentroids
threeLastCentroids = []
#print(centroids)
for i in range(0, maxIter):
threeLastCentroids.append(np.round(centroids, 4))
distancesToCentroids = np.vstack([globals()[funName](centroids, datapoint) for datapoint in X])
closestCentroid = np.where(distancesToCentroids == np.amin(distancesToCentroids,
axis = 1)[:, np.newaxis])[1]
centroids = np.vstack([np.mean(X[np.where(closestCentroid == clusterNum)],
axis = 0) for clusterNum in np.unique(closestCentroid)])
if i >2:
threeLastCentroids.pop(0)
if np.array_equal(threeLastCentroids[-1],threeLastCentroids[-2]) and np.array_equal(threeLastCentroids[-2], threeLastCentroids[-3]):
print("No changes in cluster centroids detected in last 3 iterations. Finished at iteration " + str(i+1) + ".")
break
# new code
squareDistancesPerPoint = []
for index, centroid in enumerate(closestCentroid):
squareDistancesPerPoint.append(np.square(centroids[centroid, :] - X[index, :]))
distortion = 1/len(X) * np.sum(np.array(squareDistancesPerPoint))
if distortion < bestDistortion:
bestDistortion = distortion
resultToReturn = [centroids, closestCentroid, initialCentroids, bestDistortion]
return resultToReturn
def hierarCluster(X, distanceFunc = "calcEucliDist", linkageMethod = "average", displayDistMatrix = False):
if linkageMethod not in ["average", "complete", "single"]:
print("Error, please input a valid linkage method!")
return None
if distanceFunc not in globals().keys():
print("Error, please input a valid distance function name!")
# make an empty distance matrix
distanceMatrix = np.zeros(shape = (len(X), len(X)))
distanceMatrix.fill(np.nan)
# make a list with the indices of every data point. This is the list of clusters, where you start
# with every point in a cluster and then start merging them.
initialList = [[index] for index, _ in enumerate(X)]
clusterList = initialList.copy()
clusteringOverIterations = []
clusteringOverIterations.append(initialList)
# also make an empty list that saves which cluster indices were merged for every iteration
clusterIndicesMergedList = []
for rowIndex, row in enumerate(distanceMatrix):
for colIndex, cellValue in enumerate(row):
# distance from yourself to yourself is 0, don't calculate!
if colIndex == rowIndex:
continue
# in the first loop, you calculate distance from 1 to 2.
# in the second loop, you don't want to calculate distance from 2 to 1 again. This safeguards against that.
if colIndex < rowIndex:
continue
distanceMatrix[rowIndex, colIndex] = globals()[distanceFunc](X[rowIndex,:][np.newaxis, :],
X[colIndex, :][np.newaxis, :])
if displayDistMatrix:
display(pd.DataFrame(distanceMatrix))
# We continue clustering until everything is in one giant cluster. Thats len(X)-1 clustering steps.
for i in range(0, len(X)-1):
# we start with no idea of which two clusters we need to cluster
lowestDistDatapoints = None
# since we haven't calculated any distance, our current distance is infinite
distToCluster = np.Inf
# clusterList initially looks like [[0], [1], ... [99]].
# itertools.combinations makes that into [([0], [1]), ([0], [2]), ([0], [3]) ... ([1], [2]), ([1], [3])... (98, 99)]
# so you get all possible combinations of clusters that you could cluster together
for combo in itertools.combinations(clusterList, 2):
distance = 0
distanceSingleLink = np.Inf # need this because for single linkage you want lowest distance to be selected
# so need to have the starting distance always be lower.
# make all combinations of data points in the first cluster and data points in the second cluster
# so if the current combo = ([0, 12, 15], [3, 2]), this results in:
# [[0, 3], [0, 2], [12, 3], [12, 2], [15, 3], [15,2]]: these are all the points that we need to get
# the distances for (and average for average linkage)
toIterate = [j for i in [list(zip([elem] * len(combo[1]), combo[1] )) for elem in combo[0]] for j in i]
for indicesTwoDatapoints in toIterate:
#sort the indices. Our matrix has only the distance between 1 and 2, not between 2 and 1.
#this turns [12, 2] from above into [2, 12], etc.
indicesTwoDatapoints = sorted(indicesTwoDatapoints)
# keep a running total of all distances between the points in the two clusters
if linkageMethod == "average":
distance += distanceMatrix[indicesTwoDatapoints[0], indicesTwoDatapoints[1]]
if linkageMethod == "complete":
# for a cluster, if the distance between two points is larger than the current largest distance
# between points in a cluster, that is the new cluster distance.
if distanceMatrix[indicesTwoDatapoints[0], indicesTwoDatapoints[1]] > distance:
distance = distanceMatrix[indicesTwoDatapoints[0], indicesTwoDatapoints[1]]
if linkageMethod == "single":
if distanceMatrix[indicesTwoDatapoints[0], indicesTwoDatapoints[1]] < distanceSingleLink:
distanceSingleLink = distanceMatrix[indicesTwoDatapoints[0], indicesTwoDatapoints[1]]
if linkageMethod == "average":
totalAvgDistance = distance/(len(combo[0]) * len(combo[1]))
# if distance between these clusters is less than the lowest distance we have seen so far,
#set these clusters as the ones to cluster.
if totalAvgDistance < distToCluster:
distToCluster = totalAvgDistance
dataPointsToCluster = combo
if linkageMethod == "complete":
if distance < distToCluster:
distToCluster = distance
dataPointsToCluster = combo
if linkageMethod == "single":
if distanceSingleLink < distToCluster:
distToCluster = distanceSingleLink
dataPointsToCluster = combo
#make a new list of clusters
clusterIndicesMergedList.append(dataPointsToCluster)
clusterList = clusterList.copy()
for index, elem in enumerate(clusterList):
# merge the second cluster into the first cluster
if elem == dataPointsToCluster[0]:
clusterList[index] = clusterList[index] + dataPointsToCluster[1]
#clusterList2[index] = sorted(clusterList[index])
# remove the separate second cluster (it's now been merged to the first one)
if elem == dataPointsToCluster[1]:
clusterList.pop(index)
# Finally, save all clusters, from the very beginning (all separate clusters) until the very end (all in one cluster) in one list by appending to that the current clusters
clusteringOverIterations.append(clusterList)
#addition to make a list of lists of everything:
return [clusteringOverIterations, pd.DataFrame(distanceMatrix), clusterIndicesMergedList]
def drawHierarchicalClustering(hierarClusterOutcome, figsize = (25,8), title = "Plot", labels = None):
clusterListX = hierarClusterOutcome[0]
clusteredPerStepX = hierarClusterOutcome[2]
xLabels = np.array(list(itertools.chain(*clusterListX[-1])))
fig, ax = plt.subplots(figsize = figsize)
ax.set_xticks(range(0, len(xLabels)))
if not labels is None:
labels = np.array(labels)
if len(labels) == len(xLabels):
labels = labels[xLabels]
ax.set_xticklabels(labels, rotation = 90)
else:
print("Labels supplied should be of same length as the amount of data points!")
return None
else:
ax.set_xticklabels(xLabels)
ax.margins(y=0)
heightPerDataPointPreviousStep = np.array([0] * len(xLabels))
for i, clusterStep in enumerate(clusteredPerStepX):
pos1Positions = np.array([np.where(xLabels == elem)[0] for elem in clusterStep[0]])
pos1Avg = np.mean(pos1Positions)
#pos1Start = np.min(pos1Positions)
#pos1End = np.max(pos1Positions)
pos1ClustSize = len(pos1Positions)
pos2Positions = np.array([np.where(xLabels == elem)[0] for elem in clusterStep[1]])
pos2Avg = np.mean(pos2Positions)
#pos2Start = np.min(pos2Positions)
#pos2End = np.max(pos2Positions)
pos2ClustSize = len(pos2Positions)
heightEnd = max(pos1ClustSize, pos2ClustSize)
ax.plot([pos1Avg, pos1Avg], [heightPerDataPointPreviousStep[pos1Positions[0][0]],heightEnd], color = "black")
ax.plot([pos2Avg, pos2Avg], [heightPerDataPointPreviousStep[pos2Positions[0][0]],heightEnd], color = "black")
ax.plot([pos1Avg, pos2Avg], [heightEnd,heightEnd], color = "black")
heightPerDataPointPreviousStep[np.ravel(pos1Positions)] += heightEnd - heightPerDataPointPreviousStep[pos1Positions[0][0]]
heightPerDataPointPreviousStep[np.ravel(pos2Positions)] += heightEnd - heightPerDataPointPreviousStep[pos2Positions[0][0]]
ax.set_ylim(0, max(heightPerDataPointPreviousStep)+1)
fig.suptitle(title)
plt.show(fig)
</code>
## Implementing Needleman-Wunsch for pairwise alignment
Before we can even think about clustering sequence data, we need a way to construct multiple sequence alignments. As you've been told, an often-used method (though it cannot guarantee the best possible MSA) is progressive tree alignment. There, you first make a hierarchical tree of the sequences based on some measure of how alike they are, and then align the most alike sequences, then align the next most alike to those two, etc. etc. until you've aligned all. In that way, you deal with the issue that with, say, 100 sequences that you all want to align together, you have so many possibilities that it's impossible to do, and you probably have many as good options (so which one, then, to pick?).
So you want to cluster sequence data. Well, you need this pairwise measure of similarity (or its inverse, distance). For that, the classic option is Needleman-Wunsch, a dynamic programming algorithm that breaks the problem of finding the global optimal alignment into the subproblem of finding the best subalignments possible, then tracing back from the maximum score to the beginning. We're going to be implementing this algorithm ourselves. Doubtless you have seen this once or twice, but you may need a refresher. Take your pick:
* [Video tutorial](https://www.youtube.com/watch?v=LhpGz5--isw)
* [Wikipedia](https://en.wikipedia.org/wiki/Needleman%E2%80%93Wunsch_algorithm)
Let's implement it. First as a set of commands, then in a function, so we can use pairwise alignment to make a guide tree. We'll go in parts. The first part is making the table of scores. To do this:
* Initialise an array of zeros of size len(sequence1)+1 by len(sequence2)+1. Fill it with `np.nan`.
* Using the gap, match, and mismatch penalties below, fill in the table. To do that:
* The leftmost column and top row are just filled by adding gap penalties from top to bottom and left to right, respectively
* The most top left cell should contain 0.
* The other cells are filled with the maximum score obtained by either:
1. Stepping from the cell above (opening or extending a gap, i.e. adding the gap penalty to the score of the cell above)
2. Stepping from the cell to the left (opening or extending a gap, i.e. adding the gap penalty to the score of the cell to the left)
3. Stepping from the cell to the top left (i.e. diagonally, aligning the two residues, thereby either adding the match score or the mismatch score). <br>
**Relevant Wikipedia screencap:** 
* Make sure to use the scoring matrix for substitutions (so that we can, if we want, give another scoring matrix!)
* Your final output should be the matrix with scores per position, as described in the video and Wikipedia. We'll do the actual aligning below.
Hints:
* If all goes well, your output score matrix should look something like this (although it will be in Numpy array format rather than this pretty-printed pd.DataFrame format):  Note that it's perfectly valid if you have the sequences swapped (so seqTwo along the rows and seqOne along the columns).
* You can loop over a matrix/array by doing something like: <br>
`for rowIndex, rowValues in enumerate(array):` <br>
` for colIndex, colValue in enumerate(rowValues:)` <br>
` #do stuff` (do add the appropriate tabs)
* You add one row and one column to accomodate the gap penalties along the left and top. This means that if you want to add the mismatch or match for a certain combination of bases, you need to index with something like `seqOne[rowIndex-1]`.
* You can use `baseDict` to make your life easier `(baseDict["C"], baseDict["G"])` will give you the indices in the score matrix for this mismatch between a C and a G. So `scoreMatrix[baseDict["C"], baseDict["G"]]` will give you the value you want if there's a C in sequence one and a G in sequence two that you possibly want to mismatch.
<code>
seqOne = 'ATGCTTCG'
seqTwo = 'ATGGCTGCCCC'
matchScore = 1
mismatchScore = -1
gapScore = -1
bases = ["A", "T", "G", "C"]
baseDict = dict(zip(bases, [0, 1, 2, 3]))
scoreMatrix = np.zeros(shape = (len(bases), len(bases)))
scoreMatrix[:] = mismatchScore
np.fill_diagonal(scoreMatrix, matchScore)
printTable = pd.DataFrame(scoreMatrix) ; printTable.columns = bases ; printTable.set_index(pd.Index(bases), inplace= True)
print("Substitution matrix for our simple case:")
display(printTable)
# your answer
</code>
## Adding the steps taken and constructing the sequence alignment
There's only two ingredients missing to get our alignment working:
1. We need to keep a record when we select the max value for our score table (either by moving from the left, top-left, or top) where we came from. This record is non-unique (we could get the same score for a gap as a mismatch, say).
2. Finally, we need to use this record to step back from the bottom right to the top left, going through the directions we saved. In real life, you'd want to report the alternative alignments possible. Here, we will just make one (remember, they are all as good as each other).
To do this:
* Copy your code from above into the cell below.
* Make an extra matrix called `stepMatrix` like so: `np.zeros_like(YourNameForTheScoreMatrixAbove).tolist()` (the tolist is because a numpy array only allows one value per cell. But we might want to store \['left', 'topleft'\] in one cell because we have two options).
* Your code already determined what the maximum score was. All you need to add is to check which entries gave that top score (an alignment/mismatch step (from topleft) or a gap step (from the left or from the top)) and add that as a value in `stepMatrix`. So you'd do something like `stepMatrix[rowIndex][colIndex] = ['left', 'topleft']`, if for a certain step both the step from the left cell and the top-left cell yield the same, maximum, score.
When that's done, you can move on to making the alignment. There's many slighly different ways to go about it, of course, but here's one idea:
* Start a rowIndex at `len(seqOne)`; start a colIndex at `len(seqTwo)`.
* Start two empty strings, `seqOneAligned` and `seqTwoAligned`.
* While both indices are not yet 0 (i.e. you are not yet at the top left):
* Select the first entry from the stepMatrix at that position (there could be more, but for now we just make one of the optimal alignments).
* If that entry is 'left', add "-" to `seqTwoAligned` and the letter at `seqOne[rowIndex-1]` to `seqOneAligned`, and subtract one from rowIndex.
* If that entry is 'top', do the opposite (gap to seq one, letter to seq two, subtract one from colIndex)
* If that entry is 'topleft', add both corresponding letters, and subtract one from both rowIndex and colIndex
* When done, reverse the two sequences (they're now back-to-front) and voila: aligned sequences!
<code>
# your answer here:
</code>
## Making a function out of this.
Now, functionalise what you've been implementing so far. Call the function `alignNW()`, give it 4 arguments `(seqOne, seqTwo, substitutionMatrix = scoreMatrix, substitutionDict = baseDict)`. It should return a dictionary with three entries: "similarityScore", "seqOneAligned", and "seqTwoAligned". similarityScore is just the bottom right value in the score matrix: it is the measure of how similar the two sequences are based on their alignment. The other two speak for themselves. As a sanity check, `alignNW('CCC', 'CCC')` should return a score of 3, while `alignNW('ATG', 'GTA')` should return -1.
Note: don't print anything anymore if you did before. We're going to use this function a lot of times!
<code>
# set-up
matchScore = 1
mismatchScore = -1
gapScore = -1
bases = ["A", "T", "G", "C"]
baseDict = dict(zip(bases, [0, 1, 2, 3]))
scoreMatrix = np.zeros(shape = (len(bases), len(bases)))
scoreMatrix[:] = mismatchScore
np.fill_diagonal(scoreMatrix, matchScore)
#define alignNW
</code>
## Data for constructing a guide tree
The reason I'm suddenly making you implement pairwise alignment is that if I ask you to cluster sequences, you won't be able to do it just like that. The reason for that is that, while in the morning practicals we had data points with features 1 to _n_ for each of them, for sequences you don't know which letter is which feature. Maybe the 3rd letter in sequence 43 actually corresponds to the 7th letter in sequence 22, because of deletions or duplications. To know what corresponds to what, we need a multiple sequence alignment.
That sounds fine and dandy, but as you've heard, making a multiple sequence alignment of a 100 sequences together is basically impossible: there are so many options it's infeasible to calculate. So we compromise, getting a very good (but not per se optimal) MSA by doing _progressive tree alignment_: we get some notion of pairwise distances between sequences (either via pairwise alignment or k-mer or k-tuple methods), and then align the two closest sequences, then the one that's closest to either of those two, etc.
Now that we have Needleman-Wunsch (NW), I'd like you to construct a guide tree. Note that the final score in the lower right of the NWscore matrix is the similarity score for two sequences, so the inverse of their distance. Below I give you 50 sequences of ribosomal 16S subunits, which I randomly selected from the RDP database unaligned data file that can be found [here](http://rdp.cme.msu.edu/misc/resources.jsp). We will be aligning the first 80 bases of 25 of those: I thought I could let you align the 1225 unique combinations of 50 by 50 full sequences, but that would take _ages_ with our naive implementation (which shows you how dire the problem of computational expense can be)!
We'll be using the [DNAFULL scoring matrix](https://rosalind.info/glossary/dnafull/), which includes all the iupac symbols for nucleotide sequences (like N for 'any nucleotide at this position, we don't know which, only that there is a nucleotide here').
* Make a matrix of zeros of size 25\*25.
* Align all unique combinations of the 25 16S rRNA subsequences in `sequences` using the indices in `combinations`. Save them in a similarity matrix.
* Make the similarity matrix into a distance matrix by:
* Adding (-(minimum score in matrix) +1) to all entries (there could be negative scores)
* Doing 1/score (so very high similarity of 100 becomes 1/100 = small distance, while 1/1 (completely dissimilar) is just 1.
<code>
# advanced scoring matrix:
scoreMatDNAFULL = np.array([ [ 5, -4, -4, -4, -4, 1, 1, -4, -4, 1, -4, -1, -1, -1, -2],
[-4, 5, -4, -4, -4, 1, -4, 1, 1, -4, -1, -4, -1, -1, -2],
[-4, -4, 5, -4, 1, -4, 1, -4, 1, -4, -1, -1, -4, -1, -2],
[-4, -4, -4, 5, 1, -4, -4, 1, -4, 1, -1, -1, -1, -4, -2],
[-4, -4, 1, 1, -1, -4, -2, -2, -2, -2, -1, -1, -3, -3, -1],
[1, 1, -4, -4, -4, -1, -2, -2, -2, -2, -3, -3, -1, -1, -1],
[1, -4, 1, -4, -2, -2, -1, -4, -2, -2, -3, -1, -3, -1, -1],
[-4, 1, -4, 1, -2, -2, -4, -1, -2, -2, -1, -3, -1, -3, -1],
[-4, 1, 1, -4, -2, -2, -2, -2, -1, -4, -1, -3, -3, -1, -1],
[1, -4, -4, 1, -2, -2, -2, -2, -4, -1, -3, -1, -1, -3, -1],
[-4, -1, -1, -1, -1, -3, -3, -1, -1, -3, -1, -2, -2, -2, -1],
[-1, -4, -1, -1, -1, -3, -1, -3, -3, -1, -2, -1, -2, -2, -1],
[-1, -1, -4, -1, -3, -1, -3, -1, -3, -1, -2, -2, -1, -2, -1],
[-1, -1, -1, -4, -3, -1, -1, -3, -1, -3, -2, -2, -2, -1, -1],
[-2, -2, -2, -2, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1]])
basesDNAFULL = ["A", "T", "G", "C", "S", "W", "R", "Y", "K", "M", "B", "V", "H", "D", "N"]
dictDNAFULL = dict(zip(basesDNAFULL, range(0, len(basesDNAFULL))))
# reading data
file_in = "firstFifty16SSequences.fasta"
sequenceDict = SeqIO.to_dict(SeqIO.parse(open(file_in, mode='r'), 'fasta'))
finalBasePosToInclude = 80
nSequencesToUse = 25
sequences = [str(elem.seq).upper()[0:finalBasePosToInclude] for index, elem in enumerate(list(sequenceDict.values())) if index <nSequencesToUse]
# again, we don't want to align seq 1 with seq 2 and seq 2 with seq 1, they're the same
# combinations gets the unique combinations for us.
combinations = list(itertools.combinations(range(0, len(sequences)), 2))
print("Alignments to perform: " + str(len(combinations)))
#up to you!
</code>
## Making the guide tree
Now all we need to do is construct a hierarchical tree out of the sequence distances and we'll know how to do progressive tree alignment. For that, we need to change our hierarCluster function, allowing it to skip calculating distances and instead make a clustering on pre-defined distances. To do this:
* Add an argument `distMatrix = None` to the function below
* If `distMatrix` is not None, it should use the pre-calculated distance matrix instead and skip the whole distance calculation step.
* Run this updated function, with X just being `sequences` from above.
* Use `drawHierarchicalClustering` to see the results.
* Check that the first two sequences that are clustered correspond to the minimum of the distance matrix you calculated in the previous cell. Notice anything strange?
Hint:
* For the last point you can use `np.where()` and `np.min()`.
<code>
# Your answer here
</code>
## Calculating similarity scores for guide trees more rapidly
This approach of pairwise alignment also rapidly becomes infeasible, as evidenced by the fact that I only let you align a few sequences, and of those, only part of the sequence, because otherwise it would take too long (with our simple implementation). Nevermind if you have 10.000 or 100.000 sequences to align. Then, different methods, like k-mers or k-tuples are used. We'll implement the k-mer method of distance calculation here for the sequences above.
K-mers are subsequences of length k in a sequence. So a 1-mer in a nucleotide sequence are just the amount of A, T, G, and C in that sequence. But for 2-mers, you get the amount of AA, AT, TA, TT, GC, CG, GG, CC, CT, TC, TG, GT, AC, CA, AG, GA in it. See this image: <br> 
Note that the k-mers overlap. You might already see the advantage: k-mers are quick to calculate, and they give you a set number of features for each sequence. You can then easily calculate distances based on these features and cluster hierarchically: bam, you have your guide tree! Read more [here](https://en.wikipedia.org/wiki/K-mer).
Let's make a guide tree for the sequences from above (although now all of them, and with their full length) using 2- and 3-mers. The code below is your starting point. Up to you to calculate the k-mer counts for each sequence and do the hierarchical clustering (with Euclidean distance calculation). To do this:
* Make an empty list to hold the features for each sequence.
* Go over each sequence, for each:
* Make an empty feature vector with 80 spots for every k-mer
* Glide over the sequence with a sliding window, incrementing the correct spot for the 2-mer and 3-mer you read at each position
* Append the features for this sequence to the list you made in the beginning.
* Finally, use `np.vstack()` to stack all these features, you should end up with a 50 by 80 feature matrix X for hierarchical clustering.
* After this, do the hierarchical clustering using average linkage and Euclidean distance, visualise it, and try to see how much it differs from the one with pairwise alignment.
Hints:
* You can increment the 3-mer and 2-mer at the same time, provided you realise that the 3-mer will give an error once the sliding window is at the very end of the string. So you should check that `currentPos+3 is not > len(currentSeq)`
* In the answers, I have done this procedure also for the exact same subsequences as we used above. Look there if you want to.
<code>
allSequences = [str(elem.seq).upper() for index, elem in enumerate(list(sequenceDict.values()))]
twoMers = list(itertools.product("ATGC", repeat=2))
twoMers = ["".join(elem) for elem in twoMers]
threeMers = ["".join(elem) for elem in list(itertools.product("ATGC", repeat=3))]
mersWeUse = np.array(twoMers + threeMers)
print("Each sequence will now have " + str(len(mersWeUse)) + " features.")
# your turn:
</code>
## Going from the guide tree to the multiple sequence alignment
Now that we have our guide tree (either based on k-mers or pairwise distances) we can perform progressive sequence alignment. What I haven't gone into details on is _how_ we align sequence C to already pairwise-aligned sequence A and B. Or two aligned sets of sequences (A, B) and (C, D) to each other. Many complex scoring schemes could be thought up, but the simplest is just to treat every column of the two pairwise alignments as one 'letter' and align the sequences as you normally would using something like Needleman-Wunsch.
To illustrate:
If we have the two protein (sub)sequences: <br>
AIKA <br>
AL-A
and
ALA <br>
VLA
We could do something like on the following image, where the score for accepting something is the sum (or average) of aligning A with A, A with A, and V with A and V with A for the first position: 
Modifying our `alignNW()` function for this is possible, though it would take quite some work and might cause some headaches. Instead, I think you now understand the principles of progressive tree alignment well enough to use something like Clustal Omega. Up to you to:
* Go [here](https://www.ebi.ac.uk/Tools/msa/clustalo/) and use Clustal Omega to align the 50 16s RNA sequences.
* Compare the guide tree to the full k-mer guide tree. If you didn't do this before, you can set `labels = list(sequenceDict.keys())` when plotting the k-mer guide tree so that the leaves are labeled with the sequence ID, just like in Clustal Omega. Do the guide trees look alike or highly dissimilar?
* Download the MSA file. With that in hand, we can _finally_ cluster the sequences based on some distances between them, resulting in one of the most well-known clustering plots in biology: a phylogenetic tree.
<code>
# your answer
</code>
## Using the multiple-sequence alignment to get a phylogenetic tree
Okay then, nearly there. We've clustered the sequences based on some characteristics (pairwise alignment scores or k-mer scores, or k-tuple score) to get a guide tree. Then, we've made a multiple-sequence alignment (MSA) _along_ that guide tree, with the logic that it's easiest to align the most alike sequences first (you'll make the least errors there) and progressively keep adding sequences to that so you'll finally get a quite good (though not optimal) MSA. Now, finally, with the MSA in hand, we have separate features (here, if yours is the same as mine, 1558 features, i.e. 1558 positions in the MSA) on which to cluster our sequences. The only thing that's left to do is to construct a distance matrix, and then cluster.
To construct the distance matrix, we'll just take a simple criterion: the distance from sequence A to sequence B increases by 1 if in position _n_ they do not have the same character (be that A, T, G, C, N, -, etc.). In reality, of course, here too we could use more complex scores, where different substitutions that are more or less likely change the distance more or less. We won't bother with that and keep it simple. Now, it's up to you to make this distance matrix and construct the phylogenetic tree. To do that:
* Use `itertools.combinations` to get every unique combination of 2 sequence indices.
* For each combination of sequences, slide over them letter by letter (or gap by gap). If they are the same: do nothing. If they are different: add 1 to the distance for this pair. Note: you can select the first sequence in the alignment as a string like so: `str(align[0, :].seq)`.
* Put the distances in a matrix.
* Use this distance matrix to make a phylogenetic tree, use average linkage.
* Finally, compare that phylogenetic tree to the one Clustal Omega gave you.
Hints:
* To get all the sequence names for use as labels, use `[elem.id for elem in align]`.
<code>
from Bio import AlignIO
yourAlignFile = ''
#align = AlignIO.read(yourAlignFile, "clustal")
# your answer
</code>
## What I want you to remember here:
* How Needleman-Wunsch works and finds globally optimal pairwise alignments
* How the most well-known application of (hierarchical) clustering in biology, making phylogenetic trees, requires a twist or certain circularity because you don't know which letters in a sequence correspond to which letters in another sequence.
* How this forces you to do an initial clustering on some property (pairwise alignment distances if possible, otherwise k-mer methods or even mBed methods) to make a _guide tree_ that you can then align along.
* That with the guide tree in hand, you can finally calculate the evolutionary distances that you want, and cluster hierarchically based on those to get your phylogenetic tree.
* That the model of evolution used by alignment algorithms is necessarily an indel-model. We know that evolution actually happens via many duplications, sometimes even whole-genome duplications, but alignment programs like Clustal Omega will never tell you directly 'oh, this is a duplication'. Instead you might get 2 equally good global alignments and have to deduce that this happened yourself. In other words: _you still need to think about what the clustering criteria you use actually do for the type of evolution you can measure well in a phylogenetic tree_. Of course there's a lot more nuance, but you can go to Berend for that (see below).
## The end
That concludes our school trip to clustering in phylogenetics. I hope you aren't nauseous from all the candy you kids are wont to consume on such trips. Please do note that this is only a _tiny_ piece of the vast difficulties in the field of phylogenetics, and that you should definitely check out the course taught by Prof. Dr. Berend Snel on the subject for biological background and methodical details.
## Survey
Go on, eat your [survey](https://docs.google.com/forms/d/e/1FAIpQLSfC8YzEjnv0b0iEy7Hgs4fbRMY8oH8XMkSeW3Fl97tmjKseBQ/viewform?usp=sf_link), it'll make you grow big and strong!
|
{
"filename": "PracticalMaterialDay4_AfternoonPractical.ipynb",
"repository": "DieStok/Basic-Machine-Learning-for-Bioinformatics",
"query": "transformed_from_existing",
"size": 43634,
"sha": ""
}
|
# semantic_search_1.ipynb
Repository: puravparab/DavisScripts
## Overview
Semantic searching for UC Davis courses using OpenAI's embedding model
<code>
import json
import pandas as pd
</code>
### Convert courses_data.json into a csv file
(Skip this step if you already have course data in csv format)
<code>
# Load json data
with open("course_data.json") as f:
data = json.load(f)
print(f'Total Subjects = {len(data)}')
</code>
<code>
# Create an empty dataframe
df = pd.DataFrame(columns=["code","name","credits","description","prerequisites"])
# Iterate through course_data.json and store each entry into the dataframe
for subject in data:
for course_code in subject:
course_list = subject[course_code]
for course in course_list:
df = pd.concat([df, pd.DataFrame(course, index=[0])], ignore_index=True)
</code>
<code>
df.head()
</code>
<code>
print(f'Total Courses = {len(df)}')
</code>
<code>
# convert the DataFrame to a CSV file
df.to_csv('davis_courses.csv', index=False)
</code>
### Generate embeddings
Get embeddings for all courses using OpenAI's 'get_embedding' function
<code>
import os
from dotenv import load_dotenv
load_dotenv(override=True)
import openai
from openai.embeddings_utils import get_embedding
# Create .env file with your secret key 'OPENAI' or replace 'os.getenv('OPENAI')' with your secret key
openai.api_key = os.getenv('OPENAI')
</code>
<code>
# Read course data from davis_courses.csv
df = pd.read_csv('davis_courses.csv')
</code>
<code>
df.head()
</code>
<code>
# Create a cobined column
df["combined"] = (
"code: " + df.code.str.strip() + "; name: " + df.name.str.strip() +
"; credits: " + df.credits.str.strip() +
"; description: " + df.description.str.strip() +
"; prerequisites: " + df.prerequisites.fillna('').str.strip()
)
df.head(3)
</code>
<code>
# Get embeddings for all courses
# (This will take a long time)
df['embedding'] = df.combined.apply(lambda x: get_embedding(x, engine='text-embedding-ada-002'))
</code>
<code>
# Save as csv
df.to_csv('course_embeddings.csv')
</code>
<code>
df.head(3)
</code>
# Semantic searching
<code>
# Covert embeddings from string to numpy array
import numpy as np
df = pd.read_csv('course_embeddings.csv')
</code>
<code>
df['embedding'] = df['embedding'].apply(eval).apply(np.array)
df.to_csv('course_embeddings_cleaned.csv')
df.head(3)
</code>
<code>
# Enter your prompt
prompt = "computers and biology"
</code>
<code>
search_vector = get_embedding(prompt, engine='text-embedding-ada-002')
search_vector
</code>
<code>
from openai.embeddings_utils import cosine_similarity
</code>
<code>
# Use cosine similarity to search courses that are closest to your prompt
df["similarities"] = df['embedding'].apply(lambda x: cosine_similarity(x, search_vector))
</code>
<code>
df.head(3)
</code>
<code>
# Get top ten courses
df.sort_values("similarities", ascending=False).head(10)
</code>
<code>
# Vector size
df["embedding"][2].shape
</code>
## Convert embeddings into smaller csv
<code>
df = pd.read_csv("course_embeddings.csv")
</code>
<code>
df.head(3)
</code>
<code>
df = df.drop('Unnamed: 0', axis=1)
df = df.drop('combined', axis=1)
df.head()
</code>
<code>
# Save smaller embeddings csv
df.to_csv('course_embeddings_small.csv', index=False)
</code>
|
{
"filename": "semantic_search_1.ipynb",
"repository": "puravparab/DavisScripts",
"query": "transformed_from_existing",
"size": 110278,
"sha": ""
}
|
# table_1.ipynb
Repository: KlicOgogo/KlicAll
<code>
from collections import defaultdict
top = open('tops.txt', 'r')
tops = defaultdict()
for line in top:
kek = line.strip().split(' ')
temp_top = kek[5:10]
tops[int(kek[2][:4])] = temp_top
</code>
<code>
names_file = open('names.txt', 'r')
names = []
numbers = []
for line in names_file:
names.append(line.strip())
numbers.append(int(names[-1][:4]))
</code>
<code>
num_name = defaultdict()
for i in range(len(names)):
num_name[numbers[i]] = names[i]
</code>
<code>
for i in range(len(numbers)):
print('top-5 similar files of ' + num_name[numbers[i]] + ': ' + '\\' + '\\')
for num in tops[numbers[i]]:
print(num_name[int(num)] + ' \\' + '\\')
print(' ')
</code>
<code>
print(len(numbers))
</code>
|
{
"filename": "table_1.ipynb",
"repository": "KlicOgogo/KlicAll",
"query": "transformed_from_existing",
"size": 241705,
"sha": ""
}
|
# New_eng_academic_research.ipynb
Repository: kdj0712/teamKim1
<code>
import pandas as pd
import numpy as np
</code>
<code>
df_Riss_research = pd.read_csv("./csv/Seleniums.eng_academic_research.csv")
df_Riss_research.drop(labels='_id', axis=1, inplace=True)
df_Riss_research['research_subject']
</code>
## 데이터 전처리
### dataframe 내 중복되는 학술정보 제거
<code>
df_Riss_research['research_title'].value_counts()
# 중복되는 research 확인
# 1) 진행성 화골성 근염 -증례 보고- = Myositis Ossificans Progressive -A Case Report-
# 2) 비장적출로 치유된 희귀 비장 질환 치험 = Clinical Experience of Rare Splenic Disease Healed by Splenectomy
# 3) 상급종합병원과 희귀난치성질환 전문병원의 희귀의약품 사용현황
</code>
<code>
df_Riss_research.drop_duplicates(subset="research_title", keep='first', inplace=True)
df_Riss_research['research_title'].value_counts()
</code>
<code>
df_Riss_research['research_title'].value_counts()
# 더이상 중복값 없음을 확인 완료
</code>
<code>
df_Riss_research.reset_index(drop=True, inplace=True)
</code>
### 주제어 존재하는 column만 추출
<code>
drop_index = df_Riss_research[df_Riss_research['research_subject'].str.contains(';')==True].index
</code>
<code>
df_Riss_research_subject = df_Riss_research[df_Riss_research['research_subject'].str.contains(';')==True]
df_Riss_research_subject.reset_index(drop=True, inplace=True)
df_Riss_research_subject
</code>
<code>
condition = "research_language != 'KCI등재후보'"
df_Riss_research_subject01 = df_Riss_research_subject.query(condition)
df_Riss_research_subject01.reset_index(drop=True, inplace=True)
df_Riss_research_subject01
</code>
<code>
type(df_Riss_research_subject01['research_type'][3])
</code>
<code>
int(df_Riss_research_subject01['research_type'][3])
</code>
<code>
for i in range(len(df_Riss_research_subject01['research_type'].index)):
try:
if type(int(df_Riss_research_subject01['research_type'][i])) == int:
condition03 = "research_page != '학술저널'"
df_Riss_research_subject02 = df_Riss_research_subject01.query(condition03)
except:
pass
df_Riss_research_subject02.reset_index(drop=True, inplace=True)
</code>
<code>
df_new = df_Riss_research_subject02[['research_title', 'research_subject']]
df_new.to_csv("eng_research_subject.csv", sep='\t', encoding='utf-8')
</code>
### research_title 영문명만 분리
<code>
import re
def no_korean(text):
patterns = '([가-힣]|[一-龥]|[0-9]|[;])'
text_regex = re.sub(pattern=patterns, repl=' ', string=text)
return text_regex
df_Riss_research_subject['research_subject'] = df_Riss_research_subject['research_subject'].apply(no_korean)
</code>
<code>
df_Riss_research_subject['research_subject']
</code>
<code>
df_new =pd.DataFrame(df_Riss_research_subject['research_subject'])
df_new
</code>
<code>
df_new.to_csv("eng_research_subject.csv", sep='\t', encoding='utf-8')
</code>
<code>
eng_subject = df_Riss_research_subject['research_subject'].tolist()
eng_subject
</code>
### 형태소 분석기
#### 불용어리스트 만들기
<code>
f=open('./csv/eng_academic_research_stopwords.txt')
stopwords=[]
lines = f.readlines()
for line in lines:
line = line.strip()
stopwords.append(line)
f.close()
</code>
<code>
df_Riss_research_subject['research_subject'] = df_Riss_research_subject['research_subject'].str.lower()
</code>
<code>
from sklearn.feature_extraction.text import TfidfVectorizer
tfidfVectorizer = TfidfVectorizer(stop_words=stopwords
, ngram_range=(1,2)
, max_df=0.90
, min_df=1) # stop_words는 vocabulary에서 필요없는 단어를 빼주는 것. ngram_range는 단어를 붙여주는 것으로 2개의 단어가 합성되었을때 의미를 가지고, 떨어져있을때 의미가 상실되는 경우를 포함함.
result_vectors = tfidfVectorizer.fit_transform(eng_subject) # fit & transform은 다른 2가지 임.(fit하면 각 단어의 vocabulary 만들 수 있음.)
result_vectors.toarray()[:2]
</code>
<code>
tfidfVectorizer.vocabulary_
</code>
<code>
from sklearn.decomposition import LatentDirichletAllocation
lda_model = LatentDirichletAllocation(n_components=3, n_jobs=-1) #인스턴스화 #n_components 토픽의 갯수
lda_model.fit(result_vectors) #교육
</code>
<code>
dictionary_list = tfidfVectorizer.get_feature_names_out()
dictionary_list
</code>
<code>
lda_model.components_
</code>
<code>
topics_output = lda_model.transform(result_vectors)
df_topics_score = pd.DataFrame(data=topics_output)
df_topics_score
</code>
<code>
df_topics_score['dominant_topic_number'] = np.argmax(topics_output, axis=1)
df_topics_score['sentences'] = df_Riss_research_subject['research_subject']
df_topics_score
</code>
### topic별 word 추출
<code>
## 상위 단어 추출
## 0 확률 1은 dictionary
topics_list = list()
for topic in lda_model.components_:
df_datas = [topic, dictionary_list]
df_topics = pd.DataFrame(data=df_datas)
df_topics= df_topics.T
df_topics = df_topics.sort_values(0, ascending=False)
# print(df_topics[:3])
topics_text = ' '.join(df_topics[1].values[:3])# 시리즈 형식으로 출력 get values from series / index
print(topics_text)
topics_list.append(topics_text)
topics_list_add = [topics_list, ['Topic0', 'Topic1', 'Topic2']]
df_topics_keywords = pd.DataFrame(topics_list_add)
</code>
<code>
df_topics_keywords
</code>
<code>
import pyLDAvis
import pyLDAvis.lda_model
</code>
<code>
vis = pyLDAvis.lda_model.prepare(lda_model, result_vectors, tfidfVectorizer) # 토픽모델, 교육이 끝난 값(행렬형태), 교육모델
</code>
<code>
pyLDAvis.enable_notebook()
pyLDAvis.display(vis) # PCA - 차원축소
</code>
|
{
"filename": "New_eng_academic_research.ipynb",
"repository": "kdj0712/teamKim1",
"query": "transformed_from_existing",
"size": 277407,
"sha": ""
}
|
# example_transcriptomics_obs_segmentations_bitmask.ipynb
Repository: vitessce/vitessce-python-tutorial
View this example on [Google Colab](https://colab.research.google.com/drive/1o8WHmuEBcg9hcOy9vFdwryNfz0FjE9LR?usp=sharing)
<code>
import importlib.util
if importlib.util.find_spec('vitessce') is None:
!pip install vitessce[all]
</code>
<code>
from vitessce import (
VitessceConfig,
Component as cm,
CoordinationType as ct,
FileType as ft,
AnnDataWrapper,
MultiImageWrapper,
OmeTiffWrapper,
)
</code>
<code>
vc = VitessceConfig(schema_version="1.0.15", name='Transcriptomics example')
dataset = vc.add_dataset(name='Cell segmentations').add_object(
AnnDataWrapper(
adata_url="https://assets.hubmapconsortium.org/69d9c52bc9edb625b496cecb623ec081/anndata-zarr/reg001_expr-anndata.zarr",
obs_locations_path="obsm/xy"
)
).add_object(
OmeTiffWrapper(img_url="https://assets.hubmapconsortium.org/69d9c52bc9edb625b496cecb623ec081/ometiff-pyramids/pipeline_output/mask/reg001_mask.ome.tif?token=", is_bitmask=True, name="Segmentations")
)
spatial_plot = vc.add_view(cm.SPATIAL, dataset=dataset)
layer_controller = vc.add_view(cm.LAYER_CONTROLLER, dataset=dataset)
spatial_segmentation_layer_value = [{
"type":"bitmask",
"index":0,
"visible":True,
"colormap":None,
"opacity":1,
"domainType":"Min/Max",
"transparentColor":[0,0,0],
"renderingMode":"Additive",
"use3d":False,
"channels":[
{"selection":{"c":0,"t":0,"z":0},"color":[0,0,0],"visible":False,"slider":[0,1]},
{"selection":{"c":1,"t":0,"z":0},"color":[0,0,0],"visible":True,"slider":[0,1]}, # Set the nuclei channel as checked initially
{"selection":{"c":2,"t":0,"z":0},"color":[0,0,0],"visible":False,"slider":[0,1]},
{"selection":{"c":3,"t":0,"z":0},"color":[0,0,0],"visible":False,"slider":[0,1]}
]
}]
vc.link_views([spatial_plot, layer_controller], [ct.SPATIAL_ZOOM, ct.SPATIAL_TARGET_X, ct.SPATIAL_TARGET_Y, ct.SPATIAL_SEGMENTATION_LAYER], [-4, 5000, 5000, spatial_segmentation_layer_value])
vc.layout(spatial_plot | layer_controller);
</code>
<code>
from IPython.display import display, HTML
url = vc.web_app()
display(HTML(f'<a href="{url}" target="_blank">View on Vitessce.io</a>'))
</code>
<code>
vw = vc.widget()
vw
</code>
|
{
"filename": "example_transcriptomics_obs_segmentations_bitmask.ipynb",
"repository": "vitessce/vitessce-python-tutorial",
"query": "transformed_from_existing",
"size": 22837,
"sha": ""
}
|
# 7_BERT.ipynb
Repository: christophergaughan/ChristopherGaughan.io
<a href="https://colab.research.google.com/github/christophergaughan/ChristopherGaughan.io/blob/master/7_BERT.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Define Functions to Query and Download Data from the GDC API
<code>
import requests
import json
import os
import shutil
# Define the directory to save downloaded files
directory_path = '/content/drive/MyDrive/Colab Notebooks/gdc2_files'
# Clear the directory if it exists
if os.path.exists(directory_path):
shutil.rmtree(directory_path)
# Recreate the directory
os.makedirs(directory_path)
# Function to query the GDC API
def query_gdc_api():
url = "https://api.gdc.cancer.gov/files"
# Define the parameters for the query
params = {
"filters": json.dumps({
"op": "and",
"content": [
{
"op": "in",
"content": {
"field": "cases.project.project_id",
"value": ["TCGA-BRCA"]
}
},
{
"op": "in",
"content": {
"field": "data_category",
"value": ["Transcriptome Profiling"]
}
},
{
"op": "in",
"content": {
"field": "data_type",
"value": ["Gene Expression Quantification"]
}
}
]
}),
"fields": "file_id,file_name,cases.submitter_id",
"format": "json",
"size": "500" # Increase the size to 500
}
response = requests.get(url, params=params)
print(f"Query URL: {response.url}") # Print the query URL for debugging
if response.status_code == 200:
data = response.json()
return data['data']['hits']
else:
print(f"Error querying GDC API: {response.status_code}")
return []
# Query the GDC API and print the retrieved files
files = query_gdc_api()
for file in files:
print(f"Retrieved file: {file['file_name']} with ID: {file['file_id']}")
# Ensure files are retrieved before attempting to download
if not files:
print("No files retrieved. Please check the query parameters.")
else:
print(f"Total files retrieved: {len(files)}")
# Function to download files from the GDC API
def download_files(files):
for file in files:
file_id = file['file_id']
file_name = file['file_name']
file_path = os.path.join(directory_path, file_name)
if not os.path.exists(file_path):
download_url = f"https://api.gdc.cancer.gov/data/{file_id}"
response = requests.get(download_url, stream=True)
if response.status_code == 200:
with open(file_path, 'wb') as f:
for chunk in response.iter_content(chunk_size=1024):
if chunk:
f.write(chunk)
print(f"Downloaded {file_name}")
else:
print(f"Failed to download {file_name}")
# Download the files
download_files(files)
</code>
**preprocess the RNA-Seq files:**
<code>
import pandas as pd
import json
import os
# Function to preprocess RNA-Seq files
def preprocess_rna_seq_file(file_path):
try:
# Read the file with proper column names and skip the initial rows if they don't conform to the structure
df = pd.read_csv(file_path, sep='\t', comment='#')
# Display the first few rows to understand the structure
print(f"First few rows of the file {file_path}:")
print(df.head())
# Remove rows with NaN in gene_name or gene_type
df = df.dropna(subset=['gene_name', 'gene_type'])
# Filter out rows with gene_id values like 'N_unmapped', 'N_multimapping', etc.
df = df[~df['gene_id'].str.contains('N_unmapped|N_multimapping|N_noFeature|N_ambiguous')]
if df.empty or df.shape[1] == 0:
raise ValueError("File is empty or has no valid columns")
return df
except Exception as e:
print(f"Error reading {file_path}: {e}")
return None
# Directory path for downloaded files
directory_path = '/content/drive/MyDrive/Colab Notebooks/gdc2_files'
file_ids = [f for f in os.listdir(directory_path) if f.endswith('.rna_seq.augmented_star_gene_counts.tsv')]
# List to hold all DataFrames
rna_seq_dfs = []
# Preprocess and collect all RNA-Seq DataFrames
for file_id in file_ids:
file_path = os.path.join(directory_path, file_id)
rna_seq_df = preprocess_rna_seq_file(file_path)
if rna_seq_df is not None:
rna_seq_df['submitter_id'] = file_id.split('.')[0] # Add submitter_id for merging
rna_seq_dfs.append(rna_seq_df)
# Concatenate all RNA-Seq DataFrames
if rna_seq_dfs:
all_rna_seq_df = pd.concat(rna_seq_dfs, ignore_index=True)
# Display the combined DataFrame
print(all_rna_seq_df.head())
print(all_rna_seq_df.info())
else:
print("No valid RNA-Seq data found.")
</code>
<code>
import os
import pandas as pd
# Function to preprocess RNA-Seq files
def preprocess_rna_seq_file(file_path):
try:
# Read the file and handle comment lines
df = pd.read_csv(file_path, sep='\t', comment='#')
# Remove rows with NaN in 'gene_name' or 'gene_type'
df = df.dropna(subset=['gene_name', 'gene_type'])
# Filter out rows with gene_id values like 'N_unmapped', 'N_multimapping', etc.
df = df[~df['gene_id'].str.contains('N_unmapped|N_multimapping|N_noFeature|N_ambiguous')]
if df.empty or df.shape[1] == 0:
raise ValueError("File is empty or has no valid columns")
return df
except Exception as e:
print(f"Error reading {file_path}: {e}")
return None
# Directory path for downloaded files
directory_path = '/content/drive/MyDrive/Colab Notebooks/gdc2_files'
file_ids = [f for f in os.listdir(directory_path) if f.endswith('.rna_seq.augmented_star_gene_counts.tsv')]
# List to hold all DataFrames
rna_seq_dfs = []
# Preprocess and collect all RNA-Seq DataFrames
for file_id in file_ids:
file_path = os.path.join(directory_path, file_id)
rna_seq_df = preprocess_rna_seq_file(file_path)
if rna_seq_df is not None:
rna_seq_df['submitter_id'] = file_id.split('.')[0] # Add submitter_id for merging
rna_seq_dfs.append(rna_seq_df)
# Concatenate all RNA-Seq DataFrames
if rna_seq_dfs:
all_rna_seq_df = pd.concat(rna_seq_dfs, ignore_index=True)
# Display the combined DataFrame
print(all_rna_seq_df.head())
print(all_rna_seq_df.info())
else:
print("No valid RNA-Seq data found.")
</code>
<code>
# Display summary statistics
summary_stats = all_rna_seq_df.describe()
print(summary_stats)
</code>
<code>
# Filter genes with TPM > 10
filtered_df = all_rna_seq_df[all_rna_seq_df['tpm_unstranded'] > 10]
print(filtered_df.head())
</code>
<code>
# Export the combined DataFrame to a CSV file
output_file_path = '/content/drive/MyDrive/Colab Notebooks/processed_rna_seq_data.csv'
all_rna_seq_df.to_csv(output_file_path, index=False)
print(f"Data exported to {output_file_path}")
</code>
<code>
import os
import pandas as pd
# Function to preprocess RNA-Seq files
def preprocess_rna_seq_file(file_path):
try:
# Read the file and handle comment lines
df = pd.read_csv(file_path, sep='\t', comment='#')
# Remove rows with NaN in 'gene_name' or 'gene_type'
df = df.dropna(subset=['gene_name', 'gene_type'])
# Filter out rows with gene_id values like 'N_unmapped', 'N_multimapping', etc.
df = df[~df['gene_id'].str.contains('N_unmapped|N_multimapping|N_noFeature|N_ambiguous')]
if df.empty or df.shape[1] == 0:
raise ValueError("File is empty or has no valid columns")
return df
except Exception as e:
print(f"Error reading {file_path}: {e}")
return None
# Directory path for downloaded files
directory_path = '/content/drive/MyDrive/Colab Notebooks/gdc2_files'
file_ids = [f for f in os.listdir(directory_path) if f.endswith('.rna_seq.augmented_star_gene_counts.tsv')]
# List to hold all DataFrames
rna_seq_dfs = []
# Preprocess and collect all RNA-Seq DataFrames
for file_id in file_ids:
file_path = os.path.join(directory_path, file_id)
rna_seq_df = preprocess_rna_seq_file(file_path)
if rna_seq_df is not None:
rna_seq_df['submitter_id'] = file_id.split('.')[0] # Add submitter_id for merging
rna_seq_dfs.append(rna_seq_df)
# Concatenate all RNA-Seq DataFrames
if rna_seq_dfs:
all_rna_seq_df = pd.concat(rna_seq_dfs, ignore_index=True)
# Display the combined DataFrame
print(all_rna_seq_df.head())
print(all_rna_seq_df.info())
# Generate summary statistics
summary_stats = all_rna_seq_df.describe()
print(summary_stats)
# Filter genes with TPM > 10
filtered_df = all_rna_seq_df[all_rna_seq_df['tpm_unstranded'] > 10]
print(filtered_df.head())
# Export the combined DataFrame to a CSV file
output_file_path = '/content/drive/MyDrive/Colab Notebooks/processed_rna_seq_data.csv'
all_rna_seq_df.to_csv(output_file_path, index=False)
print(f"Data exported to {output_file_path}")
else:
print("No valid RNA-Seq data found.")
</code>
<code>
import pandas as pd
# Load the data
data = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/processed_rna_seq_data.csv')
# Check for missing values
print(data.isnull().sum())
# Summary statistics
print(data.describe())
</code>
## Simplified Exploratory Data Analysis (EDA)
1. **Distribution Analysis**
Combine the histograms and boxplots into a single function for simplicity.
<code>
import seaborn as sns
import matplotlib.pyplot as plt
# Function to plot distributions
def plot_distributions(data, columns, sample_size=100000):
# Sample the data for quicker processing
sampled_data = data.sample(n=sample_size, random_state=42)
for column in columns:
plt.figure(figsize=(14, 6))
# Plot histogram
plt.subplot(1, 2, 1)
sns.histplot(sampled_data[column], kde=True, bins=50) # Limiting bins for performance
plt.title(f'Distribution of {column}')
plt.xlabel(column)
plt.ylabel('Frequency')
# Plot boxplot
plt.subplot(1, 2, 2)
sns.boxplot(y=sampled_data[column])
plt.title(f'Boxplot of {column}')
plt.tight_layout()
plt.show()
# Columns to visualize
columns_to_plot = ['tpm_unstranded', 'fpkm_unstranded']
# Plot the distributions with sampling
plot_distributions(data, columns_to_plot)
</code>
The distribution of tpm_unstranded is heavily skewed with many values close to zero and some extremely high values. The boxplot also shows the presence of numerous outliers.
To get more insights from the data, we can perform the following steps:
1. **Log Transformation:** Apply a log transformation to the tpm_unstranded values to reduce skewness and make the distribution more normal.
2. **Filtering Outliers:** Identify and possibly filter out outliers for a clearer view of the central tendency of the data.
3. **Revisualization:** Re-plot the transformed and filtered data to get a better understanding.
## Applying Log Transformation and Filtering Outliers
<code>
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
# Function to plot distributions with transformations
def plot_transformed_distributions(data, column, sample_size=100000):
# Sample the data for quicker processing
sampled_data = data.sample(n=sample_size, random_state=42)
# Apply log transformation (adding a small constant to avoid log(0))
sampled_data['log_' + column] = np.log1p(sampled_data[column])
plt.figure(figsize=(14, 6))
# Plot histogram of log-transformed data
plt.subplot(1, 2, 1)
sns.histplot(sampled_data['log_' + column], kde=True, bins=50) # Limiting bins for performance
plt.title(f'Log-Transformed Distribution of {column}')
plt.xlabel('log(' + column + ')')
plt.ylabel('Frequency')
# Plot boxplot of log-transformed data
plt.subplot(1, 2, 2)
sns.boxplot(y=sampled_data['log_' + column])
plt.title(f'Boxplot of Log-Transformed {column}')
plt.tight_layout()
plt.show()
# Column to visualize
column_to_plot = 'tpm_unstranded'
# Plot the log-transformed distributions
plot_transformed_distributions(data, column_to_plot)
</code>
The log-transformed distribution and boxplot for tpm_unstranded provide a clearer view of the data:
Log-Transformed Distribution: The distribution is still right-skewed but much less so than the original distribution. The majority of the data points have low TPM values, with a long tail of higher values.
Boxplot: The boxplot shows that even after log transformation, there are still many outliers.
<code>
# Identify the top 10 most highly expressed genes
top_expressed_genes = data[['gene_id', 'gene_name', 'tpm_unstranded']].nlargest(10, 'tpm_unstranded')
print("Top 10 most highly expressed genes:")
print(top_expressed_genes)
# Identify the top 10 least expressed genes (excluding zeros)
least_expressed_genes = data[['gene_id', 'gene_name', 'tpm_unstranded']][data['tpm_unstranded'] > 0].nsmallest(10, 'tpm_unstranded')
print("\nTop 10 least expressed genes (excluding zeros):")
print(least_expressed_genes)
</code>
<code>
print(data.columns)
</code>
<code>
# Identify the top 10 most highly expressed genes
top_expressed_genes = data[['gene_id', 'gene_name', 'tpm_unstranded']].nlargest(10, 'tpm_unstranded')
print("Top 10 most highly expressed genes:")
print(top_expressed_genes)
# Identify the top 10 least expressed genes (excluding zeros)
least_expressed_genes = data[['gene_id', 'gene_name', 'tpm_unstranded']][data['tpm_unstranded'] > 0].nsmallest(10, 'tpm_unstranded')
print("\nTop 10 least expressed genes (excluding zeros):")
print(least_expressed_genes)
</code>
<code>
import numpy as np
# Simulate condition data (e.g., two conditions 'A' and 'B')
np.random.seed(42) # For reproducibility
data['condition'] = np.random.choice(['A', 'B'], size=len(data))
# Calculate mean TPM for each gene per condition
mean_tpm_per_condition = data.groupby(['gene_id', 'gene_name', 'condition'])['tpm_unstranded'].mean().unstack()
# Display the mean TPM per condition for the first few genes
print(mean_tpm_per_condition.head())
</code>
<code>
# Identify the top 10 most highly expressed genes for each condition
top_expressed_genes_A = mean_tpm_per_condition['A'].nlargest(10)
top_expressed_genes_B = mean_tpm_per_condition['B'].nlargest(10)
print("Top 10 most highly expressed genes in condition A:")
print(top_expressed_genes_A)
print("\nTop 10 most highly expressed genes in condition B:")
print(top_expressed_genes_B)
# Identify the top 10 least expressed genes (excluding zeros) for each condition
least_expressed_genes_A = mean_tpm_per_condition['A'][mean_tpm_per_condition['A'] > 0].nsmallest(10)
least_expressed_genes_B = mean_tpm_per_condition['B'][mean_tpm_per_condition['B'] > 0].nsmallest(10)
print("\nTop 10 least expressed genes in condition A (excluding zeros):")
print(least_expressed_genes_A)
print("\nTop 10 least expressed genes in condition B (excluding zeros):")
print(least_expressed_genes_B)
</code>
<code>
# Calculate fold change
mean_tpm_per_condition['fold_change'] = mean_tpm_per_condition['B'] / mean_tpm_per_condition['A']
# Log2 Fold Change (optional for better interpretation)
mean_tpm_per_condition['log2_fold_change'] = np.log2(mean_tpm_per_condition['fold_change'])
# Display the data with fold change
print(mean_tpm_per_condition[['A', 'B', 'fold_change', 'log2_fold_change']].head())
</code>
<code>
# Replace zero values with a small number
mean_tpm_per_condition.replace(0, np.nan, inplace=True)
mean_tpm_per_condition['fold_change'] = mean_tpm_per_condition['B'] / mean_tpm_per_condition['A']
mean_tpm_per_condition['log2_fold_change'] = np.log2(mean_tpm_per_condition['fold_change'])
mean_tpm_per_condition.replace(np.nan, 0, inplace=True)
</code>
<code>
# Define fold-change threshold
fc_threshold = 2
# Identify upregulated genes (log2 fold-change > 1)
upregulated_genes = mean_tpm_per_condition[mean_tpm_per_condition['log2_fold_change'] > 1]
# Identify downregulated genes (log2 fold-change < -1)
downregulated_genes = mean_tpm_per_condition[mean_tpm_per_condition['log2_fold_change'] < -1]
print("Upregulated genes (log2 fold-change > 1):")
print(upregulated_genes)
print("\nDownregulated genes (log2 fold-change < -1):")
print(downregulated_genes)
</code>
<code>
# Define the paths
upregulated_path = '/content/drive/MyDrive/Colab Notebooks/upregulated_genes.csv'
downregulated_path = '/content/drive/MyDrive/Colab Notebooks/downregulated_genes.csv'
# Save the results to CSV files
upregulated_genes.to_csv(upregulated_path, header=True)
downregulated_genes.to_csv(downregulated_path, header=True)
</code>
<code>
import numpy as np
# Mount Google Drive
from google.colab import drive
drive.mount('/content/drive')
# Replace zero values with a small number to avoid log transformation issues
mean_tpm_per_condition.replace(0, np.nan, inplace=True)
# Calculate fold change and log2 fold change
mean_tpm_per_condition['fold_change'] = mean_tpm_per_condition['B'] / mean_tpm_per_condition['A']
mean_tpm_per_condition['log2_fold_change'] = np.log2(mean_tpm_per_condition['fold_change'])
# Replace NaN values back with zero for meaningful interpretation
mean_tpm_per_condition.replace(np.nan, 0, inplace=True)
# Display the data with fold change
print(mean_tpm_per_condition[['A', 'B', 'fold_change', 'log2_fold_change']].head())
# Define fold-change threshold
fc_threshold = 2
# Identify upregulated genes (log2 fold-change > 1)
upregulated_genes = mean_tpm_per_condition[mean_tpm_per_condition['log2_fold_change'] > 1]
# Identify downregulated genes (log2 fold-change < -1)
downregulated_genes = mean_tpm_per_condition[mean_tpm_per_condition['log2_fold_change'] < -1]
print("Upregulated genes (log2 fold-change > 1):")
print(upregulated_genes)
print("\nDownregulated genes (log2 fold-change < -1):")
print(downregulated_genes)
# Define the paths
upregulated_path = '/content/drive/MyDrive/Colab Notebooks/upregulated_genes.csv'
downregulated_path = '/content/drive/MyDrive/Colab Notebooks/downregulated_genes.csv'
# Save the results to CSV files
upregulated_genes.to_csv(upregulated_path, header=True)
downregulated_genes.to_csv(downregulated_path, header=True)
</code>
<code>
import pandas as pd
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
import matplotlib.pyplot as plt
import seaborn as sns
# Assuming `mean_tpm_per_condition` is the DataFrame containing your data
# with 'A' and 'B' as conditions and 'gene_id', 'gene_name' as indices
# Prepare the data
pca_data = mean_tpm_per_condition[['A', 'B']].fillna(0)
# Standardize the data
scaler = StandardScaler()
pca_data_scaled = scaler.fit_transform(pca_data)
# Apply PCA
pca = PCA(n_components=2)
pca_result = pca.fit_transform(pca_data_scaled)
# Create a DataFrame with the PCA results
pca_df = pd.DataFrame(data=pca_result, columns=['PC1', 'PC2'])
pca_df['gene_id'] = mean_tpm_per_condition.index.get_level_values('gene_id')
pca_df['gene_name'] = mean_tpm_per_condition.index.get_level_values('gene_name')
# Plot the PCA results
plt.figure(figsize=(10, 7))
sns.scatterplot(x='PC1', y='PC2', data=pca_df)
plt.title('PCA of RNA-seq Data')
plt.xlabel(f'Principal Component 1 ({pca.explained_variance_ratio_[0]*100:.2f}%)')
plt.ylabel(f'Principal Component 2 ({pca.explained_variance_ratio_[1]*100:.2f}%)')
plt.show()
</code>
## Identify Outliers
<code>
import numpy as np
# Add labels to the PCA DataFrame
pca_df['PC1'] = pca_result[:, 0]
pca_df['PC2'] = pca_result[:, 1]
# Define a threshold to identify outliers (e.g., top 1% of PC1 values)
threshold = np.percentile(pca_df['PC1'], 99)
# Filter the outliers
outliers = pca_df[pca_df['PC1'] > threshold]
# Plot the PCA results with outliers highlighted
plt.figure(figsize=(10, 7))
sns.scatterplot(x='PC1', y='PC2', data=pca_df, label='Data Points')
sns.scatterplot(x='PC1', y='PC2', data=outliers, color='red', label='Outliers')
plt.title('PCA of RNA-seq Data with Outliers Highlighted')
plt.xlabel(f'Principal Component 1 ({pca.explained_variance_ratio_[0]*100:.2f}%)')
plt.ylabel(f'Principal Component 2 ({pca.explained_variance_ratio_[1]*100:.2f}%)')
plt.legend()
plt.show()
</code>
<code>
# Extract outliers based on the threshold defined earlier
threshold = np.percentile(pca_df['PC1'], 99)
outliers = pca_df[pca_df['PC1'] > threshold]
# Save outliers to a CSV file for further investigation
outliers.to_csv('/content/drive/MyDrive/Colab Notebooks/rna_seq_outliers.csv', index=False)
</code>
<code>
import statsmodels.api as sm
# Assuming you have a DataFrame with expression data for conditions A and B
expression_data = mean_tpm_per_condition.loc[outliers.index]
# Perform differential expression analysis (e.g., using a linear model)
# This is a placeholder for a more complex analysis pipeline
results = []
for gene in expression_data.index:
model = sm.OLS(expression_data.loc[gene, 'A'], expression_data.loc[gene, 'B'])
results.append(model.fit())
# Summarize the results
summary = [result.summary() for result in results]
</code>
<code>
!pip install statsmodels
</code>
<code>
import statsmodels.api as sm
import numpy as np
# Ensure PCA DataFrame has a named index for gene_id
pca_df.index.name = 'gene_id'
# Extract outlier gene IDs based on the threshold defined earlier
threshold = np.percentile(pca_df['PC1'], 99)
outliers = pca_df[pca_df['PC1'] > threshold]
# Extract the gene IDs of the outliers
outlier_gene_ids = outliers.index.unique()
# Filter the expression data for the outlier gene IDs, ensuring alignment
common_gene_ids = mean_tpm_per_condition.index.intersection(outlier_gene_ids)
expression_data = mean_tpm_per_condition.loc[common_gene_ids]
# Perform differential expression analysis using statsmodels
results = []
for gene in expression_data.index:
try:
# Use log-transformed data to stabilize variance
y = np.log2(expression_data.loc[gene, 'B'] + 1) # Dependent variable
x = np.log2(expression_data.loc[gene, 'A'] + 1) # Independent variable
x = sm.add_constant(x) # Add intercept
model = sm.OLS(y, x)
results.append(model.fit())
except Exception as e:
print(f"Could not fit model for gene {gene}: {e}")
# Summarize the results
summary = [result.summary() for result in results if result]
# Print a summary for the first gene as an example
if summary:
print(summary[0])
</code>
<code>
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from scipy.stats import ttest_ind_from_stats, t
# Sample data for demonstration
np.random.seed(42)
mean_tpm_per_condition = pd.DataFrame({
'A': np.random.rand(100) * 100,
'B': np.random.rand(100) * 100
}, index=[f'gene_{i}' for i in range(100)])
# Introduce artificial outliers for demonstration
mean_tpm_per_condition.loc[::10, 'A'] = mean_tpm_per_condition.loc[::10, 'A'] * 10
mean_tpm_per_condition.loc[::10, 'B'] = mean_tpm_per_condition.loc[::10, 'B'] * 10
# Print the first few rows to confirm the data structure
print("First few rows of mean_tpm_per_condition:")
print(mean_tpm_per_condition.head())
# Calculate log2 fold change and p-values for volcano plot
volcano_data = {
'gene_id': [],
'log2_fold_change': [],
'p_value': []
}
# Assume a standard error for both conditions
standard_error = 1.0 # This value is arbitrary for the demonstration
# Iterate over genes and calculate log2 fold change and p-values
for gene in mean_tpm_per_condition.index:
condition_a = mean_tpm_per_condition.loc[gene, 'A']
condition_b = mean_tpm_per_condition.loc[gene, 'B']
if condition_a > 0 and condition_b > 0: # Ensure no division by zero
log2_fc = np.log2(condition_b / condition_a)
# Compute the t-statistic for the difference
t_stat = log2_fc / standard_error
# Degrees of freedom
df = 1 # Since we are comparing two values
# Two-tailed p-value from t-distribution
p_value = 2 * (1 - t.cdf(np.abs(t_stat), df))
volcano_data['gene_id'].append(gene)
volcano_data['log2_fold_change'].append(log2_fc)
volcano_data['p_value'].append(p_value)
# Debugging print statements
print(f"Gene: {gene}")
print(f"Condition A: {condition_a}")
print(f"Condition B: {condition_b}")
print(f"Log2 Fold Change: {log2_fc}")
print(f"P-value: {p_value}")
else:
print(f"Skipped gene {gene} due to zero value in condition A or B")
# Convert to DataFrame and add -log10(p-value)
volcano_df = pd.DataFrame(volcano_data)
volcano_df['-log10_p_value'] = -np.log10(volcano_df['p_value'])
# Print the DataFrame to verify
print("First few rows of volcano_df:")
print(volcano_df.head())
# Ensure there are no infinite or NaN values
volcano_df = volcano_df.replace([np.inf, -np.inf], np.nan).dropna()
# Replot the volcano plot
plt.figure(figsize=(10, 8))
plt.scatter(volcano_df['log2_fold_change'], volcano_df['-log10_p_value'], alpha=0.5)
plt.xlabel('Log2 Fold Change')
plt.ylabel('-Log10 P-value')
plt.title('Volcano Plot')
plt.show()
</code>
<code>
# Display first few rows of the significant genes DataFrame
print(significant_genes.head())
print(significant_genes.shape)
</code>
<code>
# Check the distribution of log2 fold changes
print("Log2 Fold Change Summary:")
print(volcano_df['log2_fold_change'].describe())
# Check the distribution of p-values
print("\nP-value Summary:")
print(volcano_df['p_value'].describe())
# Check the number of significant genes based on different thresholds
volcano_df['is_significant'] = (volcano_df['p_value'] < 0.05) & (volcano_df['log2_fold_change'].abs() >= 1.0)
print("\nNumber of significant genes with log2 fold change >= 1 and p-value < 0.05:")
print(volcano_df['is_significant'].sum())
</code>
<code>
# Check the distribution of log2 fold changes
print("Log2 Fold Change Summary:")
print(volcano_df['log2_fold_change'].describe())
# Check the distribution of p-values
print("\nP-value Summary:")
print(volcano_df['p_value'].describe())
# Check the number of significant genes based on different thresholds
volcano_df['is_significant'] = (volcano_df['p_value'] < 0.05) & (volcano_df['log2_fold_change'].abs() >= 1.0)
print("\nNumber of significant genes with log2 fold change >= 1 and p-value < 0.05:")
print(volcano_df['is_significant'].sum())
</code>
<code>
import matplotlib.pyplot as plt
import seaborn as sns
# Plot the distribution of log2 fold changes
plt.figure(figsize=(12, 6))
sns.histplot(volcano_df['log2_fold_change'], bins=50, kde=True)
plt.title('Distribution of Log2 Fold Changes')
plt.xlabel('Log2 Fold Change')
plt.ylabel('Frequency')
plt.show()
# Plot the distribution of p-values
plt.figure(figsize=(12, 6))
sns.histplot(volcano_df['p_value'], bins=50, kde=True)
plt.title('Distribution of P-values')
plt.xlabel('P-value')
plt.ylabel('Frequency')
plt.show()
</code>
<code>
# Adjust the thresholds based on the distributions
adjusted_log2_fold_change_threshold = 0.5 # Example adjustment
adjusted_p_value_threshold = 0.1 # Example adjustment
# Identify significant genes based on adjusted thresholds
volcano_df['is_significant'] = (volcano_df['p_value'] < adjusted_p_value_threshold) & (volcano_df['log2_fold_change'].abs() >= adjusted_log2_fold_change_threshold)
# Number of significant genes with adjusted thresholds
print("\nNumber of significant genes with adjusted thresholds:")
print(volcano_df['is_significant'].sum())
# Extract significant genes DataFrame
significant_genes_adjusted = volcano_df[volcano_df['is_significant']]
# Plot the volcano plot with adjusted thresholds
plt.figure(figsize=(10, 8))
sns.scatterplot(data=volcano_df, x='log2_fold_change', y='-log10_p_value', hue='is_significant',
palette={False: 'blue', True: 'red'}, legend='full', alpha=0.7)
plt.title('Volcano Plot with Adjusted Significant Genes Highlighted')
plt.xlabel('Log2 Fold Change')
plt.ylabel('-Log10 P-Value')
plt.legend(title='Gene Significance', labels=['Not Significant', 'Significant'])
plt.show()
</code>
## Data Analysis Summary and Conclusions
### Overview
The dataset comprises gene expression profiles with various measurements such as unstranded, stranded_first, stranded_second, tpm_unstranded, fpkm_unstranded, and fpkm_uq_unstranded. Our analysis focuses on understanding the distribution of these measurements and identifying significant genes based on their log2 fold changes and p-values.
### Key Findings
#### Distribution of TPM (Transcripts Per Million) Unstranded
- The **distribution of `tpm_unstranded`** is highly skewed, with most values concentrated near zero and a few extremely high values (outliers).
- **Boxplot** indicates the presence of numerous outliers, suggesting high variability in gene expression levels among the genes.
#### Log-Transformed Distribution of TPM Unstranded
- **Log-transformation** of `tpm_unstranded` values reveals a more normalized distribution, although there are still a significant number of outliers.
- The **boxplot** of log-transformed `tpm_unstranded` values shows a more compressed range, indicating that log transformation helps in mitigating the impact of outliers.
#### Principal Component Analysis (PCA)
- **PCA plot** reveals clusters of data points with several outliers dispersed away from the main cluster. This indicates variability in gene expression profiles.
- Outliers are highlighted in red, clearly showing the genes with expression levels significantly different from the majority.
#### Volcano Plot
- The **volcano plot** highlights genes with large log2 fold changes and low p-values. Significant genes are expected to be found in the upper corners of the plot.
- In our analysis, no genes were found to be significantly upregulated or downregulated (log2 fold change >= 1 and p-value < 0.05), suggesting that there are no strong candidates for differential expression in the dataset provided.
### Statistical Summary
- **Log2 Fold Change Summary**:
- The log2 fold change values range from -6.41 to 6.54, with a mean of 0.15 and a standard deviation of 2.02.
- **P-value Summary**:
- P-values range from 0.097 to 0.978, with a mean of 0.496 and a standard deviation of 0.256.
- The distribution of p-values is relatively uniform, indicating that there are no strong signals of differential expression.
### Conclusion
- The analysis shows that while there is considerable variability in gene expression levels (as evidenced by the distribution plots and PCA), none of the genes in the current dataset meet the criteria for significant differential expression.
- Future analyses might benefit from increasing the sample size or adjusting the criteria for significance to uncover potential differential expression patterns.
- The insights from the log-transformed distributions and PCA suggest that preprocessing steps such as log transformation are crucial in handling highly skewed data and identifying meaningful patterns.
# Differential Gene Expression Analysis Using BERT for Interpretation
## Step 1: Differential Gene Expression Analysis
### Objective
Identify genes that are differentially expressed between cancerous and non-cancerous tissues.
### Data Preparation
<code>
import pandas as pd
# Load your RNA-seq data
data = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/processed_rna_seq_data.csv')
# Example of the first few rows of the dataset
print(data.head())
</code>
|
{
"filename": "7_BERT.ipynb",
"repository": "christophergaughan/ChristopherGaughan.io",
"query": "transformed_from_existing",
"size": 49540,
"sha": ""
}
|
# mpfi_dinghao_Untitled_1.ipynb
Repository: dinghaoluo/code
<code>
# ---
# jupyter:
# jupytext:
# cell_metadata_filter: -all
# custom_cell_magics: kql
# text_representation:
# extension: .py
# format_name: percent
# format_version: '1.3'
# jupytext_version: 1.11.2
# kernelspec:
# display_name: caiman
# language: python
# name: python3
# ---
# %%
# %load_ext autoreload
# %autoreload 2
from pathlib import Path
import numpy as np
import sys
import caiman as cm
if ('Z:/Dinghao/code_mpfi_dinghao/caiman_code/caiman' in sys.path) == False:
sys.path.append('Z:/Dinghao/code_mpfi_dinghao/caiman_code/caiman')
import utils as utl
# %% [markdown]
# # Preprare suite2p data (motion-corrected) for Caiman
# %%
# save data.bin as caiman memory mapped file
p_ops = Path(r"Z:/Nico/AC918-20231017_02/ops.npy")
try:
p_memmap = next(p_ops.parent.glob('memmap_*'))
print(f'Found memmap file. Using: {p_memmap}')
except StopIteration:
p_memmap = utl.save_data_as_mmap(p_ops, last_frame=5000, crop=True)
# load memory mapped file
Yr, dims, num_frames = cm.load_memmap(str(p_memmap))
images = np.reshape(Yr.T, [num_frames] + list(dims), order='F')
# load reference image
img_mean = utl.load_ref_img(p_ops)
# %% [markdown]
# # Choose parameters for CNMF
# %%
# general dataset-dependent parameters
fr = 30 # imaging rate in frames per second
decay_time = 0.4 # length of a typical transient in seconds
dxy = (2., 2.) # spatial resolution in x and y in (um per pixel)
# CNMF parameters for source extraction and deconvolution
p = 1 # order of the autoregressive system (set p=2 if there is visible rise time in data)
gnb = 2 # number of global background components (set to 1 or 2)
merge_thr = 0.85 # merging threshold, max correlation allowed
bas_nonneg = True # enforce nonnegativity constraint on calcium traces (technically on baseline)
rf = 30 # default: 15 # half-size of the patches in pixels (patch width is rf*2 + 1)
stride_cnmf = 15 # default: 10 # amount of overlap between the patches in pixels (overlap is stride_cnmf+1)
K = 4 # number of components per patch
gSig = np.array([4, 4]) # expected half-width of neurons in pixels (Gaussian kernel standard deviation)
gSiz = 2*gSig + 1 # Gaussian kernel width and hight
method_init = 'greedy_roi' # initialization method (if analyzing dendritic data see demo_dendritic.ipynb)
ssub = 1 # spatial subsampling during initialization
tsub = 1 # temporal subsampling during intialization
# parameters for component evaluation
min_SNR = 2.0 # signal to noise ratio for accepting a component
rval_thr = 0.85 # space correlation threshold for accepting a component
cnn_thr = 0.99 # threshold for CNN based classifier
cnn_lowest = 0.1 # neurons with cnn probability lower than this value are rejected
parameter_dict = {
'fr': fr,
'dxy': dxy,
'decay_time': decay_time,
'p': p,
'nb': gnb,
'rf': rf,
'K': K,
'gSig': gSig,
'gSiz': gSiz,
'stride': stride_cnmf,
'method_init': method_init,
'rolling_sum': True,
'only_init': True,
'ssub': ssub,
'tsub': tsub,
'merge_thr': merge_thr,
'bas_nonneg': bas_nonneg,
'min_SNR': min_SNR,
'rval_thr': rval_thr,
'use_cnn': False,
'min_cnn_thr': cnn_thr,
'cnn_lowest': cnn_lowest
}
# investigate CNMF patches
cnmf_patch_width = rf*2 + 1
cnmf_patch_overlap = stride_cnmf + 1
cnmf_patch_stride = cnmf_patch_width - cnmf_patch_overlap
#patch_ax = cm.utils.visualization.view_quilt(
# img_mean,
# cnmf_patch_stride,
# cnmf_patch_overlap,
# vmin=np.percentile(np.ravel(img_mean),50),
# vmax=np.percentile(np.ravel(img_mean),99.5),
# figsize=(10,10))
#patch_ax.set_title(f'CNMF Patches Width {cnmf_patch_width}, Overlap {cnmf_patch_overlap}')
# %% [markdown]
# # Define output folder
# %%
# define output folder
p_out = p_ops.parent / 'K_4_p_2_decay_time_6'
p_out.mkdir(exist_ok=True)
modified_parameters = {
'K': 4,
'p': 2,
'decay_time': 6,
}
new_parameter_dict = parameter_dict.copy()
new_parameter_dict.update(modified_parameters)
# %% [markdown]
# # Run CNMF
# %%
utl.run_cnmf(images, parameter_dict, p_out)
# %% [markdown]
# # load saved data
# %%
# load previous CNMF fit
cnmf_refit = utl.load_cnmf(p_out)
# %% [markdown]
# # Save videos and masks (long)
# This will create the following files in `p_out`
# - `neural_activity.tif`: all components found by CNMF
# - `background.tif`: background component(s)
# - `resudial.tif`: original data minus the components
# - `roi.zip`: controus of components to be loaded in ImageJ
#
# This may take up to an hour and is heavy on the RAM.
#
# <font color='red'>ATTENTION</font> This will overwrite the files if they already exist.
#
# Writing ROI files for ImageJ requires [roifile](https://github.com/cgohlke/roifile/),
# which can be installed with `pip install roifile` inside the `caiman` conda environment.
# %%
# write tifs
utl.write_results_tifs(cnmf_refit.estimates, Yr, dims, p_out)
# write roi file
utl.save_rois_imagej(cnmf_refit.estimates, dims, perc=50, p_roi=p_out / 'roi.zip')
# create mock suite2p files
utl.create_suite2p_files(cnmf_refit.estimates, Yr, p_ops, p_out / 'mock_suite2p')
# %% [markdown]
# # investigate components with caiman tools
# %%
cnmf_refit.estimates.nb_view_components(img=img_mean, denoised_color='red')
cnmf_refit.estimates.view_components(img=img_mean, denoised_color='red')
# %% [markdown]
# # Batch mode: loop over parameters
# This is a template to explore multiple parameter sets for CNMF. It recreates the steps above and saves each result in a separate folder. Make sure to modify `p_out` accordingly.
# # %%
# def cnmf_wrapper(folder, parameter_dict):
# # define output folder
# p_out = p_ops.parent / folder
# p_out.mkdir(exist_ok=True, parents=True)
# # run CNMF with new parameters
# utl.run_cnmf(images, parameter_dict, p_out)
# # load again from disk
# cnmf_refit = utl.load_cnmf(p_out)
# # write tifs
# utl.write_results_tifs(cnmf_refit.estimates, Yr, dims, p_out)
# # write roi file
# utl.save_rois_imagej(cnmf_refit.estimates, dims, perc=50, p_roi=p_out / 'roi.zip')
# # create mock suite2p files
# utl.create_suite2p_files(cnmf_refit.estimates, Yr, p_ops, p_out / 'mock_suite2p')
# # %%
# # full parameter sweep
# for k in [4, 5, 6]:
# for g in [3, 4, 5]:
# # new parameters
# gSig = np.array([g, g])
# gSiz = 2*gSig + 1
# modified_parameters = {
# 'K': k,
# 'gSig': gSig,
# 'gSiz': gSiz,
# }
# new_parameter_dict = parameter_dict.copy()
# new_parameter_dict.update(modified_parameters)
# cnmf_wrapper(f'K_{k}_gSig_{g}', parameter_dict)
# # %%
# # selective parameter combinations
# parent_folder = 'parameter_search'
# # default
# cnmf_wrapper(f'{parent_folder}/default', parameter_dict)
# # 2nd order because visible rise time
# new_parameter_dict = parameter_dict.copy()
# new_parameter_dict['p'] = 2
# cnmf_wrapper(f'{parent_folder}/p_2', new_parameter_dict)
# # decay times
# for d in [1, 2, 3]:
# new_parameter_dict = parameter_dict.copy()
# new_parameter_dict['decay_time'] = d
# cnmf_wrapper(f'{parent_folder}/decay_time_{d}', new_parameter_dict)
# # K
# for k in [5, 7, 9]:
# new_parameter_dict = parameter_dict.copy()
# new_parameter_dict['K'] = k
# cnmf_wrapper(f'{parent_folder}/K_{k}', new_parameter_dict)
# # baseline nonnegativity
# new_parameter_dict = parameter_dict.copy()
# new_parameter_dict['bas_nonneg'] = False
# cnmf_wrapper(f'{parent_folder}/bas_nonneg_False', new_parameter_dict)
</code>
|
{
"filename": "mpfi_dinghao_Untitled_1.ipynb",
"repository": "dinghaoluo/code",
"query": "transformed_from_existing",
"size": 14736,
"sha": ""
}
|
# book_notes_02_1.ipynb
Repository: AdrienLE/dl
<code>
import numpy as np
np.set_printoptions(suppress=True, precision=2)
import matplotlib.pyplot as plt
%matplotlib inline
import itertools
</code>
# Linear Algebra
These are my notes for Chapter 2 of the Deep Learning book. They can also serve as a quick intro to linear algebra for deep learning.
For this section I decided to make things a bit more intuitive using code, which should be appealing to the many of us who are coders first and math people second (or eighth). This means that my "notes" are probably on the same order of length as the chapter itself, but hopefully they are extra useful for people out there (and for myself, frankly).
### Book recommendations
**Recommendations from the Deep Learning book**
- [*The Matrix Cookbook*](http://www2.imm.dtu.dk/pubdb/views/edoc_download.php/3274/pdf/imm3274.pdf) by Petersen and Pedersen, for people who need a refresher.
- My comment: I haven't (yet) read it, but the pdf is free (linked here), so probably good to check out.
- [*Linear Algebra*](https://www.amazon.com/Linear-Algebra-Dover-Books-Mathematics-ebook/dp/B00A73IXRC) by Shilov, for a full course.
- My comment: I haven't (yet?) read it. Seems to be getting good reviews and very cheap (only 10 bucks). Apparently pretty dense.
**My recommendations**
- [*Essence of Linear Algebra*](https://www.youtube.com/watch?v=kjBOesZCoqc&list=PLZHQObOWTQDPD3MizzM2xVFitgF8hE_ab) by 3Blue1Brown, for building intuition.
- This is an **amazing** youtube playlist about linear algebra. I highly recommend you watch it. A much easier option than all of the rest since it is based on videos, but won't give you as much practice.
- [*Linear Algebra and Its Applications*](https://www.amazon.com/Linear-Algebra-Its-Applications-4th/dp/0030105676) by Strang, for a full course.
- This was/is my main book for linear algebra. Engaging presentation and lots of applications.
- An alternative would be [Introduction to Linear Algebra](https://www.amazon.com/Introduction-Linear-Algebra-Gilbert-Strang/dp/0980232775/ref=pd_lpo_sbs_14_t_0?_encoding=UTF8&psc=1&refRID=G2317MHW3EY2ZXNMBKN0), by the same author. My understanding is that this book is at the same time more purely mathematical but also doesn't go as far as "and Its Applications".
- Strang's MIT video lectures are also available [here](https://ocw.mit.edu/courses/mathematics/18-06-linear-algebra-spring-2010/video-lectures/).
- [*Linear Algebra Done Right*](https://www.amazon.com/Linear-Algebra-Right-Undergraduate-Mathematics/dp/0387982582) by Axler, for a full course.
- I haven't yet read it, but I am planning to start soon. Axler supposedly takes a pretty different approach to teaching linear algebra that is more focused on pure math than on applications (so perhaps less applicable for deep learning) but also gives a different perspective on the field, which is why I'm interested in this book as a second look at linear algebra. It's supposed to be one of the best (albeit a bit controversial) books on the
subject.
- The end of [Calculus, Vol 1](https://www.amazon.com/Calculus-Vol-1-Tom-M-Apostol/dp/8126515198/ref=sr_1_1?s=books&ie=UTF8&qid=1520987006&sr=1-1&keywords=apostol+calculus) by Apostol and much of [Calculus, Vol 2](https://www.amazon.com/Calculus-Ii-2Nd-Tom-Apostol/dp/8126515201/ref=sr_1_3?s=books&ie=UTF8&qid=1520987006&sr=1-3&keywords=apostol+calculus), for a full course including multivariable calculus.
- Currently reading. This is particularly useful because it puts more emphasis on the multivariable calculus aspect while at the same time teaching linear algebra. The one think that makes me sad is that the style is not nearly as engaging as in Spivak's [Calculus](https://www.amazon.com/Calculus-4th-Michael-Spivak/dp/0914098918/ref=sr_1_1?s=books&ie=UTF8&qid=1520987248&sr=1-1&keywords=spivak+calculus) (considered to be Apostol's main "competitor"), but sadly Spivak doesn't cover multivariable calculus nor linear algebra in any non-super-advanced book of his.
## Basic objects
- **Scalar**: a single number, written in italics. A scalar is a vector with a single entry.
- A scalar in python: `x = 0.0`.
- **Vector**: a 1D array of numbers, written in bold. You can "access" elements by using a subscript, eg $x_1$ is the first element of **x**. Can be thought of as a point in *n*D space where *n* is the number of entries of the vector. A vector is a matrix with a single column.
- A vector of 10 zeros in python: `v = np.zeros(10)`
- **Matrix**: a 2D array of numbers. Written in bold and uppercase. Accessing elements is the same as with a vector but using two subscripts, the first for the row and the second for the column, eg $\textbf{A}_{1,2}$ is the second element of the first row. $\textbf{A}_{1,:}$ is the entire first row. Matrices can be transposed: $\textbf{A}^T$ is simply **A** with the columns made into rows and the rows made into columns. A matrix is a tensor that happens to have 2 dimensions.
- A 10x7 matrix of all ones in python: `M = np.ones((10, 7))`
- **Tensor**: a *n*D array of numbers. Indexing is the same as with a matrix or vector.
- A 2x3x7x4 tensor of all -1 in python: `T = -np.ones((2, 3, 7, 4))`.
All of these can be added to other objects of the same size (this is done elementwise) and multiplied by scalars (each element is multiplied by the scalar). The book permits "broadcasting", in which an object of lower dimension can be added to an object of greater dimension (eg a matrix + a vector), in this case, for example, the vector is added to each row of the matrix.
<code>
# Two matrices
A = np.array([[1, 0.3], [0.1, 1.3]], dtype=np.float32)
A
</code>
<code>
B = np.array([[3, 4], [1, 2]], dtype=np.float32)
B
</code>
<code>
# They can be added
A + B
</code>
<code>
# Multiplied by a scalar
3 * A
</code>
<code>
# Can add a vector
A + np.array([1, 3])
</code>
## Multiplication
### Dot product
Unlike to the authors, I like to think of the dot product first and of matrix multiplication second, so let's define the dot product first:
The dot product of two *vectors* that we can call *a* and *b* can be obtained by first multiplying the each element of *a* by the corresponding element of *b* and then by taking the sum of the result. In mathematical notation, it becomes (if $n$ is the number of elements in both *a* and *b*):
$$a \cdot b = \sum_{i=1}^n a_i b_i$$
In python, we can either implement it directly by using multiplication and sum, or simply use the `.dot` function.
<code>
a = np.array([3, 6], dtype=np.float32)
b = np.array([1, -2], dtype=np.float32)
# Full implementation:
print('Our implementation:', np.sum(a * b))
# The function that already exists in numpy:
print('Np\'s implementation:', np.dot(a, b))
</code>
### Matrix multiplication
To multiply matrix **A** and **B** into a matrix **C**, we simply make it so that each entry $\textbf{C}_{i,j}$ is the dot product of row i of **A** and column j of **B**, so we have, for all possible i and j:
$$C_{i,j} = A_{i,:} \cdot B_{:,j}$$
This means that the length of every row of **A** must be the same as the length of every column of **B**, in other words that the number of columns of **A** is equal to the number of rows in **B**. **C** will have the same number of rows as **A** and the same number of columns as **B**. In other words, if **A** is of size m x n and **B** is of size n x k the multiplication is valid and **C** is of size m x k.
Often, **A** and **B** will both be "square" (meaning they have the same number of rows and columns), in which case **C** will have the same size as both of them.
Because matrix multiplication is so similar to the dot product, it is implemented using the `.dot` function in numpy as well.
In fact, you could even say that the vector dot product is the same as the multiplication of a matrix with only one row with a matrix with only one column, and indeed this is why the notation used by the book for the vector dot product is not $a \cdot b$ but $a^T b$.
<code>
# Python matrix product
np.dot(A, B)
</code>
<code>
# Our own implementation of matrix product
def matrix_prod(M1, M2):
C = np.zeros((M1.shape[0], M2.shape[1]))
for i in range(M1.shape[0]):
for j in range(M2.shape[1]):
C[i, j] = np.dot(M1[i, :], M2[:, j])
return C
matrix_prod(A, B)
</code>
Matrix multiplication is *not* commutative, ie
$$BA \neq AB$$
Proof:
<code>
print('AB = ')
print(A.dot(B))
print('BA =')
print(B.dot(A))
</code>
This seems to tell us everything we need to know about matrix multiplication, but there are actually a couple other important things to note before we move on.
First, we viewed matrix multiplication on an entry-by-entry basis, and this is how the Deep Learning book (and most everybody else) presents it. However, it is sometimes useful to see matrix multiplication as involving either entire columns or entire rows! I will not go into details here, but Gilbert Strang explains this concept in his MIT OCW course, you can see his full explanation of matrix multiplication including the "entire columns" and "entire rows" aspects here:
https://youtu.be/FX4C-JpTFgY?t=43s
Secondly, since we've already talked about transposes, it is interesting to ask what the transpose of a multiplication is. As it turns out, transposing the result of a matrix multiplication is the same as multiplying the original transposed matrices in *reverse* order, or, in math:
$$(AB)^T = B^T A^T$$
In code:
<code>
print('(AB)^T =')
print(A.dot(B).transpose())
print('B^T A^T = ')
print(B.transpose().dot(A.transpose()))
</code>
Finally, note that there is a huge difference between matrix multiplication and **element-wise** multiplication of two matrices. In the element-wise product, we simply multiply each element in one matrix by the corresponding element in the other, whereas in matrix multiplication, we use the dot product of a row and a vector.
In numpy, matrix multiplication is implemented using the `.dot` function whereas you can perform element-wise multiplication using the `*` operator. In the book's math notation, element-wise multiplication is represented by $\odot$, though other operators are used by others. Matrix multiplication can just be represented by stringing the matrices together.
<code>
print('A⊙B =')
print(A * B)
print('AB =')
print(A.dot(B))
</code>
# Inverses
The matrix **I** has a very important property: multiplying it with any other matrix does nothing at all! It is simply a matrix with 0s everywhere and 1s on the diagonal going from the upper left to the lower right. In python, it is always possible to create an identity matrix of a given size using the `np.eye` function (**I** -> eye, get it?).
**I** is basically the equivalent of 1 in the matrix world (since multiplying by 1 does nothing to ordinary numbers).
<code>
np.eye(3)
</code>
Most (but not all) square matrices have a special matrix called an "inverse", represented with a -1 superscript like so: $A^{-1}$. The inverse of a matrix functions very much like the inverse of a number, in that multiplying a matrix by its inverse gives the identity function (just like multiplying a number by its inverse gives 1). Basically, multiplying by the inverse of a matrix is very similar to *dividing* by that matrix. So:
$$A^{-1} A = A A^{-1} = I$$
You can get the inverse of a matrix in numpy using `np.linalg.inv`.
Inverses can help us solve equations using matrices! Suppose we have a the following equation:
$$Ax = b$$
So a matrix A multiplies an unknown vector x to get a known vector b, what is the x that makes this possible?
Using the inverse we get:
$$A^{-1}Ax = A^{-1}b$$
$$x = A^{-1}b$$
And we can now compute x!
<code>
# We show A as a reminder
print('A =')
print(A)
# A's inverse
print('\nA^-1 = ')
print(np.linalg.inv(A))
# Confirm that we get I when we multiply by the inverse
print('\nA^-1 A = A A^-1 = I')
print(np.linalg.inv(A).dot(A))
print(A.dot(np.linalg.inv(A)))
# Show b as a reminder
print('\nb =')
print(b)
# Compute x
print('\nx =')
x = np.linalg.inv(A).dot(b)
print(x)
# Confirm that Ax = b
print('Ax = b')
print(A.dot(x))
</code>
Not all matrices have inverses! Next we will see why.
<code>
try:
bad_matrix = np.array([[1, 2], [2, 4]])
print(bad_matrix)
print(np.linalg.inv(bad_matrix))
except Exception as e:
print('There was an error:', str(e))
</code>
## Spans
Let's explore why our bad matrix couldn't be inverted before. For this, the easiest way is to introduce a geometrical way of thinking about matrices and vectors. I highly recommend watching 3Blue1Brown's videos (see resources above) to gain full intuition on this.
You can think of any vector as a point in space. With a 2D vector this is simple, the first component is its location on the X axis and the second on the Y axis. With 3D things are still relatively familiar, just add a Z axis. With more dimensions, the vector is *still* a point in space, even though you can't imagine it.
> To deal with a 14-dimensional space, visualize a 3-D space and say 'fourteen' to yourself very loudly. Everyone does it.
> *Geoffrey Hinton*
A matrix multiplication takes a point in space and moves it to another point in space, that's all it does.
Viewing this this way, we can say that a matrix inverse takes a point that (conceptually at least) has been moved by the original matrix and finds *the original location* of that point (before it was moved).
The problem is: do all matrices map every point to a single other point? What if a matrix map several source points to the same destination point? What if it never maps any point to a given destination? In either case, we can't have an inverse!
In our non-invertible matrix, both of these problems occur! There are at least two different vectors x that map to the point [0, 0], as we show in code below, and yet there is no way to get to the point [1, 1] (try it!)
<code>
print(bad_matrix.dot([2, -1]))
print(bad_matrix.dot([-2, 1]))
</code>
To understand this, let's visualize the way the two matrices transform different points. We'll represent the points using a grid:
<code>
values = list(range(-5, 6))
x, y = np.meshgrid(values, values)
x = x.flatten()
y = y.flatten()
points = np.array([x, y])
plt.plot(points[0, :], points[1, :], 'o')
plt.title('Original Grid')
plt.show()
transformed = A.dot(points)
plt.plot(transformed[0, :], transformed[1, :], 'o')
plt.title('Transformed by A')
plt.show()
transformed = bad_matrix.dot(points)
plt.plot(transformed[0, :], transformed[1, :], 'o')
plt.title('Transformed by bad_matrix')
plt.show()
</code>
So it turns out that our bad matrix projects the entire grid onto a line, whereas A just tilts things around a little bit. This means that any point outside of the line cannot have been created by bad matrix, and thus an inverse cannot exist! (and as we saw before, several points in 2D can give us the same point on the line, which is also a problem).
This line is the **span** of our bad matrix (the set of all values it can map to). By contrast, the span of A is the entire plane, as can be guessed from the fact that it's only slightly shearing the grid.
As it turns out, matrices can only ever span linear spaces such as points, lines, planes and hyperplanes (a plane in more than 2 dimensions). Further, all these spaces always have to contain the point at the origin, since multiplying any matrix by the 0 vector always gives the 0 vector. Only matrices that span the entire space they are in have an inverse.
If we look more closely at our bad matrix, we notice something strange about its columns: the second column ([2, 4]) is exactly twice the first column ([1, 2])! As it turns out, this is exactly why our matrix doesn't span the whole space!
Multiplying a matrix and a vector can be thought of combining the columns of the matrix based on the elements of the vector, so if I multiply a matrix M by the vector [1, 2, 3, 4], the final vector is 1 times the first column of M plus 2 times the second column of M and so on. So whenever we multiply our bad matrix with a vector, the result can only ever be a multiple of the vector [1, 2], which indeed forms a line!
A set of vectors is called "dependent" if it is possible to generate one of the vectors by multiplying and adding some of the other vectors (in our case, just multiplying). If the columns of a matrix are dependent, the matrix doesn't span the whole space and can't be inverted.
Also note that if the columns of a matrix are dependent, its rows are also dependent, and vice-versa. We won't prove this here.
## Norms
Norms are a measure of the **length** of a vector. The most common types of norms are called the Lp norms, and they are of the form:
$$||x||_p = \sqrt[p]{\sum_i |x_i|^p}$$
The most common Lp norms are the L1, L2 and $L_\infty$ norms, which you might already know under the names of *Mahnattan distance* (the distance to go from the origin to the tip of the vector, if you can only move along an axis), *Euclidian distance* (the distance to go from the origin to the tip if you can go in any direction you want) and *maximum* (of the absolute values), respectively.
$L_\infty$ might seem like a weird name, but it is actually simply what happens as p reaches infinity.
You can access the norms using `np.linalg.norm`.
<code>
v = np.array([3, -4])
print('Vector:', v)
print('L1:', np.linalg.norm(v, 1))
print('L2:', np.linalg.norm(v, 2))
print('L inf', np.linalg.norm(v, float('inf')))
print('\nIncreasing p gets us closer to L inf')
print('L3', np.linalg.norm(v, 3.0))
print('L10', np.linalg.norm(v, 10.0))
print('L30', np.linalg.norm(v, 30.0))
print('...')
</code>
Finally note that we can measure a matrix using the same norms, but that sometimes people call norms on matrices differently! In particular, the **Frobenius** norm is simply the L2 norm applied to a matrix. Remember to use that word if you want to sound smart.
## Special matrices
- **Diagonal matrix**: only has non-zero entries on its upper left to lower right diagonal (the other diagonal doesn't count!)
- Numpy can create a diagonal from a vector using `np.diag`.
<code>
np.diag([1, 2, 3, 4])
</code>
- **Symmetric matrix**: equal to its own transpose (the entries are symmetric across the up-left to down-right diagonal).
- A matrix times its transpose is always symmetric.
<code>
np.random.seed(1)
M = np.random.randint(0, 5, size=(3, 3))
M.dot(M.transpose())
</code>
- **Unit vector**: a vector whose L2 norm is 1.
- You can make a vector into a unit vector by dividing by its L2 norm.
<code>
print(b, np.linalg.norm(b))
bnorm = b / np.linalg.norm(b)
print(bnorm, np.linalg.norm(bnorm))
</code>
- **Orthogonal vectors**: two vectors whose dot product is 0.
- [0, 0] is orthogonal to every vector. Non-zero orthogonal vectors are perpendicular.
<code>
c = np.array([-4, -2])
plt.quiver([0, 0], [0, 0], [b[0], c[0]], [b[1], c[1]], angles='xy', scale_units='xy', scale=1)
plt.xlim(-4.3, 4.3)
plt.ylim(-4.3, 4.3)
plt.gca().set_aspect('equal', adjustable='box')
plt.title('Two orthogonal vectors')
plt.show()
print('Dot product =', b.dot(c))
</code>
- **Orthonormal vectors**: two unit vectors who are also orthogonal.
<code>
cnorm = c / np.linalg.norm(c)
plt.quiver([0, 0], [0, 0], [bnorm[0], cnorm[0]], [bnorm[1], cnorm[1]], angles='xy', scale_units='xy', scale=1)
plt.xlim(-1.3, 1.3)
plt.ylim(-1.3, 1.3)
plt.gca().set_aspect('equal', adjustable='box')
plt.title('Two orthonormal vectors')
plt.show()
print('Dot product =', bnorm.dot(cnorm))
</code>
- **Orthogonal matrix**: a matrix whose columns (and rows) are mutually ortho**normal** (yes, it is called ortho**gonal** but it is made up of ortho**normal** vectors). Amazing property: the transpose of an orthogonal matrix is its own inverse!!!
<code>
ortho = np.array([bnorm, cnorm])
print('An orthogonal matrix')
print(ortho)
print('Its transpose is its inverse')
print(ortho.dot(ortho.transpose()))
</code>
## Eigen-stuff
The word "eigen" can seem scary, so I like to mentally replace it with "special" (that's not what the word actually means in German, but it's good enough for our purposes). So whenever I write "eigenvector" or "eigenvalue", replace it with "special vector" and "special value".
So how are these vectors and values special? The eigenvectors are special because they only get **stretched** when they are multiplied by the matrix (ie their direction doesn't change, only their length, and they might also be going "backwards" if they are stretched by a negative amount). The eigenvalues is the amount by which the vectors are stretched.
Eigen-stuff is accessible in numpy through `np.linalg.eig`. Let's look at what they do. Below the black vector is always the original one and the red vector is the transformed one.
<code>
def eig_show(v, title):
dest = B.dot(v)
plt.quiver([0, 0], [0, 0], [v[0], dest[0]], [v[1], dest[1]], angles='xy', scale_units='xy', scale=1, color=['k', 'r'])
plt.xlim(min(v[0], dest[0], 0) - 0.3, max(v[0], dest[0], 0) + 0.3)
plt.ylim(min(v[1], dest[1], 0) - 0.3, max(v[1], dest[1], 0) + 0.3)
plt.gca().set_aspect('equal', adjustable='box')
plt.title(title)
plt.show()
eig_show([1, 1], 'Non-eigenvector transformation')
diag, V = np.linalg.eig(B)
eig_show(V[:, 0], f'First eigenvector (lambda = {diag[0]:.2f})')
eig_show(V[:, 1], f'Second eigenvector (lambda = {diag[1]:.2f})')
</code>
Here we see that the first (arbitrary) vector is not just stretched, but also slightly rotated by the matrix, so it clearly isn't an eigenvector. We compute the eigenvectors and show how they get transformed and indeed they do not change direction! We also see that in the first case the transformed vector is much longer than the original vector, and in the second case it is much shorter, so the amount of stretching is per-vector.
Now the two important questions are:
- Does every matrix have eigenvectors and eigenvalues, and if so how many?
- What is the point of all of this?
The answer for the first question is... sorta. Given the definition we have given so far, the answer should be an emphatic "no", because there exist *rotation* matrices, which always rotate a given vector. Since we have defined eigenvectors as "vectors that don't get rotated when they are multiplied with the matrix" and since rotation matrices rotate every vector, they should not have any eigenvectors. However, if you call `np.linalg.eig` on a rotation matrix such as \[\[0, -1\], \[1, 0\]\] (which rotates vectors by 90 degrees), you will find that it won't error out! The reason is that if you use *complex* eigenvalues and eigenvectors, you can find eigen-stuff for every possible matrix.
Having to deal with complex numbers is not very convenient, of course. Thankfully, many of the matrices we'll encounter in practice will have real eigenvalues and eigenvectors.
Now as to what the point of all this is. It so happens that we can decompose a matrix using its eigenvectors. Specifically, if we have a matrix A, we can put all its eigenvectors in the columns of a matrix V and all its eigenvalues along the diagonal of a diagonal matrix $\Lambda$ and we find that:
$$A = V \Lambda V^{-1}$$
An important way in which this is useful is that it allows us to multiply a matrix with itself repeatedly very efficiently: when we multiply A with itself, the $V^{-1}$ of the leftmost decomposition cancels out the $V$ of the rightmost decomposition, and we end up with:
$$A^n = V \Lambda^n V^{-1}$$
Because $\Lambda$ is a diagonal matrix, taking it to a power simply involves taking each of its elements to that power, which is much faster than doing a lot of matrix multiplications.
<code>
print('Original B')
print(B)
print('Reconstructed B')
print(V.dot(np.diag(diag)).dot(np.linalg.inv(V)))
</code>
Finally we should note that for all **symmetric** matrices, the eigenvector matrix is orthogonal, which means that its transpose is its own inverse, so we get:
$$A = Q \Lambda Q^T$$
If A is symmetric, which is particularly useful.
## SVD and pseudoinverse
In the previous section we found that eigenvalues were convenient but sadly didn't apply to all matrices: specifically, for some real square matrices they required the use of complex numbers, and they of course don't work at all for non-square matrices.
Singular value decomposition (SVD) tries to solve this problem by providing a decomposition with two orthogonal matrices (U and V) and one diagonal matrix (D), such that:
$$A = UDV^T$$
The book does not go into many details about the uses of SVD (though there are many) except for finding the Moore-Penrose pseudoinverse.
SVD can be accessed using `np.linalg.svd`.
<code>
rect = np.array([[1, 1], [3, 1], [-1, 4]], dtype=np.float32)
print('Original')
print(rect)
U, diag, V = np.linalg.svd(rect, full_matrices=True)
D = np.zeros((3, 2))
D[:2, :2] = np.diag(diag)
print('Reconstructed')
print(U.dot(D).dot(V.transpose()))
</code>
The Moore-Penrose pseudoinverse can be computed using SVD. How doesn't matter too much here, the point is what it can do: basically it can find the an "inverse" for non-invertible matrices. Of course, because they are not invertible, the "inverse" will lack some properties, but it will still be quite useful.
As you might recal, the inverse was useful for solving equations like:
$$Ax = b$$
In which case you could find x using:
$$x = A^{-1}b$$
With the non-invertible, there might not be an x that satisfies $Ax = b$, but the pseudoinverse can find the x that comes **closest** (by minimizing the L2 distance between Ax and b).
The Moore-Penrose pseudoinverse is accessible by using `np.linalg.pinv`.
<code>
print('Matrix A (bad)')
print(bad_matrix)
print('\nTarget')
print(b)
print('\nPseudoinverse')
print(np.linalg.pinv(bad_matrix))
x = np.linalg.pinv(bad_matrix).dot(b)
print('\nx')
print(x)
print('\nAx')
print(bad_matrix.dot(x))
</code>
## Trace and determinant
The trace is simply an operator that sums the entries along the diagonals of a matrix. It has some interesting properties (eg the trace of a transpose is the same as the original trace and the order of matrix multiplication doesn't matter within the trace operator), and its main use is to simplify some math by getting rid of explicit summing in equations in some cases.
It is accessible as np.trace
<code>
print('A')
print(A)
print('\nTr(A)')
print(np.trace(A))
</code>
The determinant is explained very quickly in the book, although it has many interesting properties. Suffice it to say that it is a single number that describes a matrix, it has the following properties:
- If det(A) is 0, then A is singular
- It is the product of the eigenvalues
- It can be thought of as the amount to which multiplying by the matrix streches space: if the determinant is 2, then the matrix can be thought of as doubling the volume of space. If it is one, it doesn't stretch space at all.
Finally, the chapter ends on a derivation of PCA with a single component. It is a cool example of derivation but I won't go through it here since it doesn't really introduce new material, just shows how to use what we saw above.
That's it for this week's notes. I think I'll keep translating the book's concepts into python code, it enlivens things a bit and makes them more concrete, but maybe next time's notes will be shorter: this was a lot of work!
|
{
"filename": "book_notes_02_1.ipynb",
"repository": "AdrienLE/dl",
"query": "transformed_from_existing",
"size": 119407,
"sha": ""
}
|
# project_convert_RDS.ipynb
Repository: pedrovp161/spatial
<code>
import pandas as pd
import pyreadr as pr
import seaborn as sns
import scanpy as sc
</code>
<code>
RDS = "C:\\Users\\pedro\\OneDrive\\Área de Trabalho\\projeto_INCA\\spatial_ovary\\Ovary_v4\\data\\ovary_scRNA .rds"
</code>
<code>
result = pr.read_r(RDS)
</code>
|
{
"filename": "project_convert_RDS.ipynb",
"repository": "pedrovp161/spatial",
"query": "transformed_from_existing",
"size": 4189,
"sha": ""
}
|
# 02_bert_1.ipynb
Repository: leo-young/huggingfaceModels
<code>
from transformers import BertModel, BertConfig
# Initializing a BERT bert-base-uncased style configuration
configuration = BertConfig()
# Initializing a model from the bert-base-uncased style configuration
model = BertModel(configuration)
# Accessing the model configuration
configuration = model.config
</code>
<code>
model
</code>
<code>
model.num_parameters()
</code>
|
{
"filename": "02_bert_1.ipynb",
"repository": "leo-young/huggingfaceModels",
"query": "transformed_from_existing",
"size": 20386,
"sha": ""
}
|
# exp_10_1.ipynb
Repository: Mohammed-Abed-Alkareem/INTELLIGENT-SYSTEMS-LAB
# Experiment #10: Information Retrieval
<b>Mohammed Abed Alkareem</b>
<b>1210708</b>
## 1.2.1 Installation
<code>
# !pip install whoosh
</code>
## 1.2.2 Preparing the data
<code>
# !pip install kaggle
</code>
<code>
# !kaggle datasets download -d stackoverflow/stacksample
</code>
<code>
# !unzip stacksample.zip
</code>
<code>
import pandas as pd
questions=pd.read_csv("stacksample/Questions.csv", nrows=20000)
questions
</code>
## 1.2.3 The Index and Schema objects
<code>
from whoosh.fields import Schema, TEXT, ID
# Defining index schema
schema = Schema(Id=ID(stored=True), Title=TEXT(stored=True),Body=TEXT(stored=True))
</code>
<code>
import os.path
index_dir = "indexdir"
if not os.path.exists(index_dir):
os.mkdir(index_dir)
</code>
<code>
from whoosh.index import create_in
from whoosh.index import open_dir
# Creating the index
ix = create_in(index_dir, schema)
# Open the index writer
writer = ix.writer()
# Iterate over the DataFrame and add documents to the index
# we have indexed title, title_body and doc_id
for index, row in questions.iterrows():
writer.add_document(Id=str(row['Id']), Title = row['Title'],Body=row['Body'])
# Commit and close the writer
writer.commit()
</code>
## 1.2.4 How to search
<code>
from whoosh.qparser import QueryParser
from whoosh.scoring import TF_IDF
from whoosh import scoring
# create the query parser
qp = QueryParser("Title", schema=schema)
# parse the query
query_sentence = "How to install"
query = qp.parse(query_sentence)
# create a searcher object
searcher_tfidf = ix.searcher(weighting=scoring.TF_IDF())
# search documents and store them
# # we are returing top 3 documents
results_tfidf = searcher_tfidf.search(query, limit=3, scored=True)
# print the documents
for hit in results_tfidf:
print(hit["Id"])
print('\n')
print(hit["Title"])
print('\n')
print('------------------\n')
</code>
#### Task 1:
Test the previous search code with different queries. For each one check how many matched results are returned.
<code>
from whoosh.qparser import QueryParser
from whoosh.scoring import TF_IDF
from whoosh import scoring
# create the query parser
qp = QueryParser("Title", schema=schema)
# parse the query
query_sentence = "Why won't"
query = qp.parse(query_sentence)
# create a searcher object
searcher_tfidf = ix.searcher(weighting=scoring.TF_IDF())
#check how many matched results are returned
results_tfidf = searcher_tfidf.search(query, scored=True)
print("Total results found:", len(results_tfidf))
print("\n\n")
# print the documents
for hit in results_tfidf:
print(hit["Id"])
print('\n')
print(hit["Title"])
print('\n')
print('------------------\n')
</code>
#### Task 2:
Repeat the previous search using the BM25F scoring algorithm, which is usedin probabilistic retrieval model. Do you see any difference in the returned results?
<code>
from whoosh.qparser import QueryParser
from whoosh import scoring
# create the query parser
qp = QueryParser("Title", schema=schema)
# parse the query
query_sentence = "Why won't"
query = qp.parse(query_sentence)
# create a searcher object with BM25F weighting
searcher_bm25f = ix.searcher(weighting=scoring.BM25F())
# check how many matched results are returned
results_bm25f = searcher_bm25f.search(query, scored=True)
print("Total results found:", len(results_bm25f))
print("\n\n")
# print the documents
for hit in results_bm25f:
print(hit["Id"])
print('\n')
print(hit["Title"])
print('\n')
print('------------------\n')
</code>
- **Ranking Order**: BM25F might rank documents differently compared to TF-IDF. BM25F takes into account term frequency saturation and document length normalization, so it may give higher relevance to documents that contain the query terms with more balanced distribution across the fields.
- **Total Results**: The number of results returned should generally be the same because both algorithms are likely to match the same set of documents, but the scoring and ranking will differ.
## 1.2.5 Query expansion
<code>
more_results = results_tfidf[0].more_like_this("Title")
for hit in more_results:
print(hit["Id"])
print('\n')
print(hit["Title"])
print('\n')
print('------------------\n')
</code>
<code>
keywords = [keyword for keyword, score in results_tfidf.key_terms("Title", docs=10, numterms=5)]
keywords
</code>
## 1.2.6 Evaluating IR systems
<code>
queries = {
'q1': "machine learning",
'q2':"AI algorithms"
}
relevance = {
'q1': ["doc1", "doc2", "doc3"],
'q2': ["doc1", "doc2", "doc3", "doc4", "doc5"]
}
documents = {
'doc1': '''Artificial Intelligence (AI) is transforming variousindustries through automation and advanced algorithms. Machinelearning, a subset of AI, enables computers to learn from data andmake predictions. Algorithms are at the core of AI systems, guidingdecision-making and problem-solving processes. AI-powered systemsare increasingly used in healthcare for diagnosis and treatmentplanning. The ethical implications of AI algorithms, such as biasand fairness, are important considerations in their development.''',
'doc2': '''Deep learning, a branch of machine learning, uses neuralnetworks to process complex data. AI algorithms are capable ofanalyzing large datasets to extract meaningful insights. NaturalLanguage Processing (NLP) algorithms enable computers to understandand generate human language. AI-driven recommendation algorithmspersonalize user experiences in e-commerce and content platforms.Ensuring the transparency and accountability of AI algorithms isessential for building trust in AI technologies.''',
'doc3': '''Reinforcement learning algorithms enable AI agents to learnthrough trial and error interactions with their environment. AIalgorithms are used in financial markets for high-frequency tradingand risk management. Computer vision algorithms enable machines tointerpret and analyze visual information. AI algorithms can enhancecybersecurity by detecting and mitigating cyber threats inreal-time. Continuous research and development are essential foradvancing AI algorithms and overcoming their limitations.''',
'doc4': '''Evolutionary algorithms, inspired by natural selection, areused to optimize complex systems and processes. AI algorithms playa crucial role in autonomous vehicles for navigation anddecision-making. Quantum computing algorithms have the potential torevolutionize AI by solving complex problems exponentially faster.AI algorithms are employed in predictive maintenance to anticipateequipment failures and reduce downtime. Ethical guidelines andregulations are needed to govern the development and deployment ofAI algorithms.''',
'doc5': '''Genetic algorithms are used to evolve solutions tooptimization and search problems inspired by natural selection. AIalgorithms enable personalized content recommendations in streamingservices and social media platforms. Swarm intelligence algorithmsmimic the collective behavior of social insects to solveoptimization problems. AI algorithms are used in drug discovery toaccelerate the identification of potential treatments.Collaborative efforts are essential for advancing AI algorithms andharnessing their full potential for societal benefit.'''
}
</code>
<code>
from whoosh.fields import Schema, TEXT, ID
from whoosh.index import create_in
from whoosh.index import open_dir
# Defining index schema
schema = Schema(Id=ID(stored=True), Body=TEXT(stored=True))
import os.path
index_dir = "indexdir_toy"
if not os.path.exists(index_dir):
os.mkdir(index_dir)
# Creating the index
ix = create_in(index_dir, schema)
# Open the index writer
writer = ix.writer()
for doc in documents:
writer.add_document(Id=doc, Body=documents[doc])
# Commit and close the writer
writer.commit()
</code>
<code>
from whoosh.qparser import QueryParser
from whoosh.scoring import TF_IDF
from whoosh import scoring
# create the query parser
qp = QueryParser("Body", schema=schema)
# parse the query
query_sentence = queries['q1']
query = qp.parse(query_sentence)
# create a searcher object
searcher_tfidf = ix.searcher(weighting=scoring.TF_IDF())
# search documents and store them
# # we are returing top 3 documents
results_tfidf = searcher_tfidf.search(query, limit=3, scored=True)
print("Total results found:", len(results_tfidf))
# print the documents
for hit in results_tfidf:
print(hit["Id"])
print('\n')
print(hit["Body"])
print('\n')
print('------------------\n')
</code>
#### Task 3:
Compute the precision and recall for the retrieved documents in the previous example.
<code>
#Compute the precision and recall for the retrieved documents in the previous example.
#Precision = (number of relevant documents retrieved) / (number of documents retrieved)
def compute_precision_recall(results, relevance):
retrieved = [hit["Id"] for hit in results]
relevant = relevance
intersection = set(retrieved).intersection(set(relevant))
precision = len(intersection) / len(retrieved)
recall = len(intersection) / len(relevant)
return precision, recall
precision, recall = compute_precision_recall(results_tfidf, relevance['q1'])
print(f"Precision: {precision}")
print(f"Recall: {recall}")
</code>
#### Task 4:
Modify the last code to test all queries and then report the precision and recall.
<code>
from whoosh.qparser import QueryParser
from whoosh.scoring import TF_IDF
from whoosh import scoring
for query_key in queries.keys():
# Create the query parser
qp = QueryParser("Body", schema=schema)
# Parse the query
query_sentence = queries[query_key]
parsed_query = qp.parse(query_sentence)
# Create a searcher object
searcher_tfidf = ix.searcher(weighting=scoring.TF_IDF())
results_tfidf = searcher_tfidf.search(parsed_query, limit=3, scored=True)
# Print the documents
for hit in results_tfidf:
print(hit["Id"])
print('\n')
print(hit["Body"])
print('\n')
print('------------------\n')
precision, recall = compute_precision_recall(results_tfidf, relevance[query_key])
print(f"Precision: {precision}")
print(f"Recall: {recall}")
print('\n')
print('='*50)
</code>
|
{
"filename": "exp_10_1.ipynb",
"repository": "Mohammed-Abed-Alkareem/INTELLIGENT-SYSTEMS-LAB",
"query": "transformed_from_existing",
"size": 39665,
"sha": ""
}
|
# combined_scenicplus_3.ipynb
Repository: Gerard-Deuner/Final-Degree-Project
# Inferring enhancer-driven Gene Regulatory Networks (eGRNs) using SCENIC+
<code>
# Set up Environment
import dill
import scanpy as sc
import os
import warnings
warnings.filterwarnings("ignore")
import pandas
import pyranges
# Set stderr to null to avoid strange messages from ray
import sys
_stderr = sys.stderr
null = open(os.devnull,'wb')
# set working directory
work_dir = '/g/scb/zaugg/deuner/SCENIC+/'
# set tmp directory
tmp_dir = '/g/scb/zaugg/deuner/SCENIC+/tmp/combined/'
# set the figures directory
fig_dir = '/g/scb/zaugg/deuner/SCENIC+/figures/'
# set the output data directory
out_dir = '/g/scb/zaugg/deuner/SCENIC+/outputdata/'
# Load the AnnData object containing the scRNA-seq side of the analysis
adata = sc.read_h5ad(os.path.join(tmp_dir, 'combined.nomicro.adata.h5ad'))
# Load the cisTopic object containing the scATAC-seq side of the analysis.
cistopic_obj = dill.load(open(os.path.join(tmp_dir, 'scATAC/cistopic_obj.pkl'), 'rb'))
# Load the motif enrichment dictionary containing the motif enrichment results.
menr = dill.load(open(os.path.join(tmp_dir, 'motifs/menr.pkl'), 'rb'))
</code>
<code>
cistopic_obj.cell_data
</code>
<code>
# adapt barcodes of cistopic object
new_bcs = []
old_bcs = cistopic_obj.cell_names
for i in range(len(old_bcs)):
split_bc = str.split(old_bcs[i], "_")
new_bc = split_bc[1] + "_" + split_bc[0]
new_bcs.append(new_bc)
cistopic_obj.selected_model.cell_topic.columns.values[i] = new_bc
cistopic_obj.cell_names = new_bcs
</code>
<code>
cistopic_obj.cell_data.index = new_bcs
</code>
<code>
cistopic_obj.selected_model.cell_topic
</code>
<code>
adata.obs
adata.obs_names.copy(deep=True)
</code>
<code>
len(set(adata.obs_names)) == len(adata.obs_names)
</code>
<code>
# check if there are common barcodes
list(set(adata.obs_names.copy(deep=True)) & set(list(cistopic_obj.cell_names.copy())))
</code>
<code>
len(set(list(cistopic_obj.cell_names.copy()))) > 0
</code>
<code>
# maybe select the atac barcodes as defaults for adata
list(set(adata.obs["barcode"]) & set(list(cistopic_obj.cell_names.copy())))
l_bcs = []
for i in range(len(adata.obs_names)):
l_bc = adata.obs["orig.ident"][i] + "_" + adata.obs["barcode"][i]
l_bcs.append(l_bc)
adata.obs["long_barcode"] = l_bcs
adata.obs_names = list(adata.obs["long_barcode"])
</code>
<code>
adata.obs
</code>
<code>
len(set(adata.obs_names)) == len(adata.obs_names)
</code>
<code>
# do the same for the cistopic object
cistopic_obj.cell_data["long_barcode"] = cistopic_obj.cell_names
</code>
<code>
# check if there are common barcodes
len(list(set(adata.obs_names.copy(deep=True)) & set(list(cistopic_obj.cell_names.copy())))) #128
#len(list(set(adata.obs_names.copy(deep=True)))) #757
#len(set(list(cistopic_obj.cell_names.copy()))) #128
</code>
<code>
cistopic_obj.selected_model.cell_topic.columns.values
</code>
<code>
#from pycisTopic.cistopic_class import *
#from pycisTopic.diff_features import *
#common_cells = list(set(adata.obs_names.copy(deep=True)) & set(list(cistopic_obj.cell_names.copy())))
#impute_accessibility(cistopic_obj, selected_cells=common_cells)
</code>
<code>
print(len(adata.obs_names.copy(deep=True)) == len(set(adata.obs_names.copy(deep=True))))
print(len(adata.obs_names.copy(deep=True).drop_duplicates(keep='first')))
print(len(set(adata.obs_names.copy(deep=True))))
print(len(list(cistopic_obj.cell_names.copy())) == len(set(list(cistopic_obj.cell_names.copy()))))
print(len(list(cistopic_obj.cell_names.copy())))
print(len(set(list(cistopic_obj.cell_names.copy()))))
</code>
## Create SCENIC+ object
<code>
# Create the Scenic+ object
from scenicplus.scenicplus_class import create_SCENICPLUS_object
import numpy as np
scplus_obj = create_SCENICPLUS_object(
GEX_anndata = adata,
cisTopic_obj = cistopic_obj,
menr = menr,
gene_metadata = adata.var.copy(deep=True),
bc_transform_func = None, #lambda x: f'{x}_timecourse' #None, #function to convert scATAC-seq barcodes to scRNA-seq ones
)
scplus_obj.X_EXP = np.array(scplus_obj.X_EXP.todense())
scplus_obj
</code>
<code>
#scplus_obj.add_gene_data(adata.var.copy(deep=True))
</code>
<code>
scplus_obj.gene_names
</code>
<code>
adata.var
adata.var.copy(deep=True)
adata.var.index
</code>
<code>
adata.obs
</code>
<code>
cistopic_obj.cell_data
</code>
<code>
# Select the optimal gene names host
ensembl_version_dict = {'105': 'http://www.ensembl.org',
'104': 'http://may2021.archive.ensembl.org/',
'103': 'http://feb2021.archive.ensembl.org/',
'102': 'http://nov2020.archive.ensembl.org/',
'101': 'http://aug2020.archive.ensembl.org/',
'100': 'http://apr2020.archive.ensembl.org/',
'99': 'http://jan2020.archive.ensembl.org/',
'98': 'http://sep2019.archive.ensembl.org/',
'97': 'http://jul2019.archive.ensembl.org/',
'96': 'http://apr2019.archive.ensembl.org/',
'95': 'http://jan2019.archive.ensembl.org/',
'94': 'http://oct2018.archive.ensembl.org/',
'93': 'http://jul2018.archive.ensembl.org/',
'92': 'http://apr2018.archive.ensembl.org/',
'91': 'http://dec2017.archive.ensembl.org/',
'90': 'http://aug2017.archive.ensembl.org/',
'89': 'http://may2017.archive.ensembl.org/',
'88': 'http://mar2017.archive.ensembl.org/',
'87': 'http://dec2016.archive.ensembl.org/',
'86': 'http://oct2016.archive.ensembl.org/',
'80': 'http://may2015.archive.ensembl.org/',
'77': 'http://oct2014.archive.ensembl.org/',
'75': 'http://feb2014.archive.ensembl.org/',
'54': 'http://may2009.archive.ensembl.org/'}
import pybiomart as pbm
def test_ensembl_host(scplus_obj, host, species):
dataset = pbm.Dataset(name=species+'_gene_ensembl', host=host)
annot = dataset.query(attributes=['chromosome_name', 'transcription_start_site', 'strand', 'external_gene_name', 'transcript_biotype'])
annot.columns = ['Chromosome', 'Start', 'Strand', 'Gene', 'Transcript_type']
annot['Chromosome'] = annot['Chromosome'].astype('str')
filter = annot['Chromosome'].str.contains('CHR|GL|JH|MT')
annot = annot[~filter]
annot.columns=['Chromosome', 'Start', 'Strand', 'Gene', 'Transcript_type']
gene_names_release = set(annot['Gene'].tolist())
#print(gene_names_release)[1:5]
#print(scplus_obj.gene_names)[1:5]
print(len(list(set(gene_names_release) & set(scplus_obj.gene_names))) > 0)
ov=len([x for x in scplus_obj.gene_names if x in gene_names_release])
print('Genes recovered: ' + str(ov) + ' out of ' + str(len(scplus_obj.gene_names)))
return ov
n_overlap = {}
for version in ensembl_version_dict.keys():
print(f'host: {version}')
try:
n_overlap[version] = test_ensembl_host(scplus_obj, ensembl_version_dict[version], 'hsapiens')
except:
print('Host not reachable')
v = sorted(n_overlap.items(), key=lambda item: item[1], reverse=True)[0][0]
print(f"version: {v} has the largest overlap, use {ensembl_version_dict[v]} as biomart host")
</code>
<code>
# Choose the best host
biomart_host = "http://sep2019.archive.ensembl.org/"
</code>
<code>
# Before running also download a list of known human TFs from the human transcription factors database
!wget -O /g/scb/zaugg/deuner/SCENIC+/inputdata/utoronto_human_tfs_v_1.01.txt http://humantfs.ccbr.utoronto.ca/download/v_1.01/TF_names_v_1.01.txt
</code>
<code>
# Also download a the program bedToBigBed this will be used to generate files which can be uploaded to the UCSC genome browser
!wget -O /g/scb/zaugg/deuner/SCENIC+/inputdata/bedToBigBed http://hgdownload.soe.ucsc.edu/admin/exe/linux.x86_64/bedToBigBed
!chmod +x /g/scb/zaugg/deuner/SCENIC+/inputdata/bedToBigBed
</code>
<code>
#only keep the first two columns of the PCA embedding in order to be able to visualize this in SCope
#scplus_obj.dr_cell['GEX_X_pca'] = scplus_obj.dr_cell['GEX_X_pca'].iloc[:, 0:2]
#scplus_obj.dr_cell['GEX_rep'] = scplus_obj.dr_cell['GEX_rep'].iloc[:, 0:2]
</code>
<code>
# import ray
# ray.shutdown()
# ray.init()
</code>
<code>
# Run the analysis
from scenicplus.wrappers.run_scenicplus import run_scenicplus
# try:
run_scenicplus(
scplus_obj = scplus_obj,
variable = ['GEX_celltype'],
species = 'hsapiens',
assembly = 'hg38',
tf_file = '/g/scb/zaugg/deuner/SCENIC+/inputdata/utoronto_human_tfs_v_1.01.txt',
save_path = os.path.join(tmp_dir, 'scenicplus'),
biomart_host = biomart_host,
upstream = [1000, 150000],
downstream = [1000, 150000],
calculate_TF_eGRN_correlation = True,
calculate_DEGs_DARs = True,
export_to_loom_file = True,
export_to_UCSC_file = True,
path_bedToBigBed = '/g/scb/zaugg/deuner/SCENIC+/inputdata',
n_cpu = 24,
_temp_dir = None)#'/g/scb/zaugg/deuner/ray_spill')
# except Exception as e:
# #in case of failure, still save the object
# dill.dump(scplus_obj, open(os.path.join(out_dir, '/scplus_obj.pkl'), 'wb'), protocol=-1)
# raise(e)
</code>
## Downstream Analysis
### Simplifying and filtering SCENIC+ output
<code>
from scenicplus.preprocessing.filtering import apply_std_filtering_to_eRegulons
apply_std_filtering_to_eRegulons(scplus_obj)
</code>
<code>
scplus_obj.uns['eRegulon_metadata_filtered'].head()
</code>
### eRegulon enrichment scores
<code>
from scenicplus.eregulon_enrichment import score_eRegulons
region_ranking = dill.load(open(os.path.join(out_dir, 'scenicplus/region_ranking.pkl'), 'rb')) #load ranking calculated using the wrapper function
gene_ranking = dill.load(open(os.path.join(out_dir, 'scenicplus/gene_ranking.pkl'), 'rb')) #load ranking calculated using the wrapper function
score_eRegulons(scplus_obj,
ranking = region_ranking,
eRegulon_signatures_key = 'eRegulon_signatures_filtered',
key_added = 'eRegulon_AUC_filtered',
enrichment_type= 'region',
auc_threshold = 0.05,
normalize = False,
n_cpu = 5)
score_eRegulons(scplus_obj,
gene_ranking,
eRegulon_signatures_key = 'eRegulon_signatures_filtered',
key_added = 'eRegulon_AUC_filtered',
enrichment_type = 'gene',
auc_threshold = 0.05,
normalize= False,
n_cpu = 5)
</code>
### eRegulon dimensionality reduction
<code>
from scenicplus.dimensionality_reduction import run_eRegulons_tsne, run_eRegulons_umap
run_eRegulons_umap(
scplus_obj = scplus_obj,
auc_key = 'eRegulon_AUC_filtered',
reduction_name = 'eRegulons_UMAP', #overwrite previously calculated UMAP
)
run_eRegulons_tsne(
scplus_obj = scplus_obj,
auc_key = 'eRegulon_AUC_filtered',
reduction_name = 'eRegulons_tSNE', #overwrite previously calculated tSNE
)
</code>
<code>
# Visualize it
from scenicplus.dimensionality_reduction import plot_metadata_given_ax
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
#specify color_dictionary
color_dict = {
'neuron': "#065143",
'hiPSC': "#70B77E",
'microglia': "#E0A890",
'diff.state': "#053C5E"
}
fig, axs = plt.subplots(ncols=2, figsize = (16, 8))
plot_metadata_given_ax(
scplus_obj=scplus_obj,
ax = axs[0],
reduction_name = 'eRegulons_UMAP',
variable = 'GEX_celltype', #note the GEX_ prefix, this metadata originated from the gene expression metadata (on which we did the cell type annotation before)
color_dictionary={'GEX_celltype': color_dict}
)
plot_metadata_given_ax(
scplus_obj=scplus_obj,
ax = axs[1],
reduction_name = 'eRegulons_tSNE',
variable = 'GEX_celltype', #note the GEX_ prefix, this metadata originated from the gene expression metadata (on which we did the cell type annotation before)
color_dictionary={'GEX_celltype': color_dict}
)
fig.tight_layout()
sns.despine(ax = axs[0]) #remove top and right edge of axis border
sns.despine(ax = axs[1]) #remove top and right edge of axis border
plt.show()
</code>
### plot the activity / expression of an eRegulon on the dimensionality reduction
<code>
from scenicplus.dimensionality_reduction import plot_eRegulon
plot_eRegulon(
scplus_obj = scplus_obj,
reduction_name = 'eRegulons_tSNE',
selected_regulons = ['POU4F3', 'KLF12', 'POU4F1', 'CUX2', 'ONECUT3'],
scale = True,
auc_key = 'eRegulon_AUC_filtered')
</code>
### dotplot-heatmap
<code>
# We first generate pseudobulk gene expression and region accessibility data, per celltype, to limit the amount of noise for the correlation calculation.
from scenicplus.cistromes import TF_cistrome_correlation, generate_pseudobulks
generate_pseudobulks(
scplus_obj = scplus_obj,
variable = 'GEX_celltype',
auc_key = 'eRegulon_AUC_filtered',
signature_key = 'Gene_based')
generate_pseudobulks(
scplus_obj = scplus_obj,
variable = 'GEX_celltype',
auc_key = 'eRegulon_AUC_filtered',
signature_key = 'Region_based')
TF_cistrome_correlation(
scplus_obj,
use_pseudobulk = True,
variable = 'GEX_celltype',
auc_key = 'eRegulon_AUC_filtered',
signature_key = 'Gene_based',
out_key = 'filtered_gene_based')
TF_cistrome_correlation(
scplus_obj,
use_pseudobulk = True,
variable = 'GEX_celltype',
auc_key = 'eRegulon_AUC_filtered',
signature_key = 'Region_based',
out_key = 'filtered_region_based')
</code>
<code>
scplus_obj.uns['TF_cistrome_correlation']['filtered_region_based'].head()
</code>
<code>
# Let's visualize these correlations in a scatter plot and select eRegulons for which the correlaiton coefficient is above 0.70 or below -0.75
import numpy as np
n_targets = [int(x.split('(')[1].replace('r)', '')) for x in scplus_obj.uns['TF_cistrome_correlation']['filtered_region_based']['Cistrome']]
rho = scplus_obj.uns['TF_cistrome_correlation']['filtered_region_based']['Rho'].to_list()
adj_pval = scplus_obj.uns['TF_cistrome_correlation']['filtered_region_based']['Adjusted_p-value'].to_list()
thresholds = {
'rho': [-0.75, 0.70],
'n_targets': 0
}
import seaborn as sns
fig, ax = plt.subplots(figsize = (10, 5))
sc = ax.scatter(rho, n_targets, c = -np.log10(adj_pval), s = 5)
ax.set_xlabel('Correlation coefficient')
ax.set_ylabel('nr. target regions')
#ax.hlines(y = thresholds['n_targets'], xmin = min(rho), xmax = max(rho), color = 'black', ls = 'dashed', lw = 1)
ax.vlines(x = thresholds['rho'], ymin = 0, ymax = max(n_targets), color = 'black', ls = 'dashed', lw = 1)
ax.text(x = thresholds['rho'][0], y = max(n_targets), s = str(thresholds['rho'][0]))
ax.text(x = thresholds['rho'][1], y = max(n_targets), s = str(thresholds['rho'][1]))
sns.despine(ax = ax)
fig.colorbar(sc, label = '-log10(adjusted_pvalue)', ax = ax)
plt.show()
</code>
<code>
selected_cistromes = scplus_obj.uns['TF_cistrome_correlation']['filtered_region_based'].loc[
np.logical_or(
scplus_obj.uns['TF_cistrome_correlation']['filtered_region_based']['Rho'] > thresholds['rho'][1],
scplus_obj.uns['TF_cistrome_correlation']['filtered_region_based']['Rho'] < thresholds['rho'][0]
)]['Cistrome'].to_list()
selected_eRegulons = [x.split('_(')[0] for x in selected_cistromes]
selected_eRegulons_gene_sig = [
x for x in scplus_obj.uns['eRegulon_signatures_filtered']['Gene_based'].keys()
if x.split('_(')[0] in selected_eRegulons]
selected_eRegulons_region_sig = [
x for x in scplus_obj.uns['eRegulon_signatures_filtered']['Region_based'].keys()
if x.split('_(')[0] in selected_eRegulons]
#save the results in the scenicplus object
scplus_obj.uns['selected_eRegulon'] = {'Gene_based': selected_eRegulons_gene_sig, 'Region_based': selected_eRegulons_region_sig}
print(f'selected: {len(selected_eRegulons_gene_sig)} eRegulons')
</code>
<code>
# Save these changes we have made to the scenicplus_obj
dill.dump(scplus_obj, open(os.path.join(out_dir, 'scenicplus/scplus_obj.pkl'), 'wb'), protocol=-1)
</code>
<code>
# Plot the heatmap-dotplot
from scenicplus.plotting.dotplot import heatmap_dotplot
heatmap_dotplot(
scplus_obj = scplus_obj,
size_matrix = scplus_obj.uns['eRegulon_AUC_filtered']['Region_based'], #specify what to plot as dot sizes, target region enrichment in this case
color_matrix = scplus_obj.to_df('EXP'), #specify what to plot as colors, TF expression in this case
scale_size_matrix = True,
scale_color_matrix = True,
group_variable = 'GEX_celltype',
subset_eRegulons = scplus_obj.uns['selected_eRegulon']['Gene_based'],
figsize = (5, 20),
orientation = 'vertical')
</code>
### Overlap of predicted target regions
<code>
# calculate the RSS for the target regions of the selected eRegulons.
from scenicplus.RSS import *
regulon_specificity_scores(
scplus_obj,
variable = 'GEX_celltype',
auc_key = 'eRegulon_AUC_filtered',
signature_keys = ['Region_based'],
selected_regulons = [x for x in scplus_obj.uns['selected_eRegulon']['Region_based'] if '-' not in x],
out_key_suffix = '_filtered')
</code>
<code>
# visualize the RSS values using a scatter plot
plot_rss(scplus_obj, 'GEX_celltype_filtered', num_columns=2, top_n=10, figsize = (5, 10))
</code>
<code>
# select the top 10 eRegulons per cell type
flat_list = lambda t: [item for sublist in t for item in sublist]
selected_markers = list(set(flat_list(
[scplus_obj.uns['RSS']['GEX_celltype_filtered'].loc[celltype].sort_values(ascending = False).head(10).index.to_list()
for celltype in scplus_obj.uns['RSS']['GEX_celltype_filtered'].index])))
</code>
<code>
from scenicplus.plotting.correlation_plot import *
region_intersetc_data, Z = jaccard_heatmap(
scplus_obj,
method = 'intersect',
gene_or_region_based = 'Region_based',
use_plotly = False,
selected_regulons = selected_markers,
signature_key = 'eRegulon_signatures_filtered',
figsize = (10, 10), return_data = True, vmax = 0.5, cmap = 'plasma')
</code>
### Plotting a Network
<code>
from pycisTopic.diff_features import find_highly_variable_features
hvr = find_highly_variable_features(scplus_obj.to_df('ACC').loc[list(set(scplus_obj.uns['eRegulon_metadata_filtered']['Region']))], n_top_features=1000, plot = False)
hvg = find_highly_variable_features(scplus_obj.to_df('EXP')[list(set(scplus_obj.uns['eRegulon_metadata_filtered']['Gene']))].T, n_top_features=1000, plot = False)
</code>
<code>
from scenicplus.networks import create_nx_tables, create_nx_graph, plot_networkx, export_to_cytoscape
nx_tables = create_nx_tables(
scplus_obj = scplus_obj,
eRegulon_metadata_key ='eRegulon_metadata_filtered',
subset_eRegulons = ['PAX5', 'EBF1', 'POU2AF1'],
subset_regions = hvr,
subset_genes = hvg,
add_differential_gene_expression = True,
add_differential_region_accessibility = True,
differential_variable = ['GEX_celltype'])
</code>
<code>
G, pos, edge_tables, node_tables = create_nx_graph(nx_tables,
use_edge_tables = ['TF2R','R2G'],
color_edge_by = {'TF2R': {'variable' : 'TF', 'category_color' : {'PAX5': 'Orange', 'EBF1': 'Purple', 'POU2AF1': 'Red'}},
'R2G': {'variable' : 'R2G_rho', 'continuous_color' : 'viridis', 'v_min': -1, 'v_max': 1}},
transparency_edge_by = {'R2G': {'variable' : 'R2G_importance', 'min_alpha': 0.1, 'v_min': 0}},
width_edge_by = {'R2G': {'variable' : 'R2G_importance', 'max_size' : 1.5, 'min_size' : 1}},
color_node_by = {'TF': {'variable': 'TF', 'category_color' : {'PAX5': 'Orange', 'EBF1': 'Purple', 'POU2AF1': 'Red'}},
'Gene': {'variable': 'GEX_celltype_Log2FC_B_cells_1', 'continuous_color' : 'bwr'},
'Region': {'variable': 'GEX_celltype_Log2FC_B_cells_1', 'continuous_color' : 'viridis'}},
transparency_node_by = {'Region': {'variable' : 'GEX_celltype_Log2FC_B_cells_1', 'min_alpha': 0.1},
'Gene': {'variable' : 'GEX_celltype_Log2FC_B_cells_1', 'min_alpha': 0.1}},
size_node_by = {'TF': {'variable': 'fixed_size', 'fixed_size': 30},
'Gene': {'variable': 'fixed_size', 'fixed_size': 15},
'Region': {'variable': 'fixed_size', 'fixed_size': 10}},
shape_node_by = {'TF': {'variable': 'fixed_shape', 'fixed_shape': 'ellipse'},
'Gene': {'variable': 'fixed_shape', 'fixed_shape': 'ellipse'},
'Region': {'variable': 'fixed_shape', 'fixed_shape': 'diamond'}},
label_size_by = {'TF': {'variable': 'fixed_label_size', 'fixed_label_size': 20.0},
'Gene': {'variable': 'fixed_label_size', 'fixed_label_size': 10.0},
'Region': {'variable': 'fixed_label_size', 'fixed_label_size': 0.0}},
layout='kamada_kawai_layout',
scale_position_by=250)
</code>
<code>
plt.figure(figsize=(10,10))
plot_networkx(G, pos)
</code>
<code>
export_to_cytoscape(G, pos, out_file = os.path.join(out_dir, 'scenicplus/network_combined.cys'))
</code>
|
{
"filename": "combined_scenicplus_3.ipynb",
"repository": "Gerard-Deuner/Final-Degree-Project",
"query": "transformed_from_existing",
"size": 222875,
"sha": ""
}
|
# ML_workshop_1_DR_part1_1.ipynb
Repository: CCPBioSim/MDAnalysis
# Dimensionality Reduction, part 1
<a rel="license" href="https://creativecommons.org/licenses/by-nc-sa/4.0/"><img alt="Creative Commons Licence" style="width=50" src="https://licensebuttons.net/l/by-nc-sa/4.0/88x31.png" title='This work is licensed under a Creative Commons Attribution 4.0 International License.' align="right"/></a>
**Authors**: Dr Matteo Degiacomi (matteo.t.degiacomi@durham.ac.uk) and Dr Antonia Mey (antonia.mey@ed.ac.uk)
Content is partially adapted from the [Software Carpentries Machine learning lesson](https://carpentries-incubator.github.io/machine-learning-novice-sklearn/index.html) and material from the [pyEMMA tutorial](http://www.emma-project.org/latest/tutorials/notebooks/02-dimension-reduction-and-discretization.html).
**Questions:**
How can we perform unsupervised learning with dimensionality reduction techniques such as Principal Component Analysis (PCA), time-correlated independent component analysis (tICA), and t-distributed Stochastic Neighbor Embedding (t-SNE)?
**Objectives:**
- Remember that most data is inherently multidimensional
- Understand that reducing the number of dimensions can simplify modelling and allow classifications to be performed.
- Use PCA as a popular technique for dimensionality reduction.
- Use tICA another popular dimensionality reduction technique that takes time correlations into account
- t-SNE is another technique for dimensionality reduction.
- Apply PCA and t-SNE with Scikit Learn to an example dataset.
- Compare how PCA and tICA perform on a 2-D toy example
- Evaluate the relative performance of PCA and t-SNE.
**Jupyter cheat sheet**:
- to run the currently highlighted cell, hold <kbd>⇧ Shift</kbd> and press <kbd>⏎ Enter</kbd>;
- to get help for a specific function, place the cursor within the function's brackets, hold <kbd>⇧ Shift</kbd>, and press <kbd>⇥ Tab</kbd>;
# 1. Introduction
Scientific data, such as that extracted from molecular dynamics simulations, can be high-dimensional and noisy. Dimensionality reduction is the process identifying and highlighting information and correlations within the data. There are multiple reasons why you might want to do a dimensionality reduction.
- You might want to know what are the dominant features in your system (larger scale variations in data).
- You want a way to visualise your high dimensional data.
- You want to analyse your data, but it it too high-dimensional.
The algorithms designed to carry out this task are an example of machine learning. In this tutorial we will look at Principal Components Analysis (PCA), time-lagged independent component analysis (tICA), and t-tested Stocastic Neighbour Embedding (t-SNE). In a machine learning context, each dimension in data is called a **feature**, which together form a **feature space**.
<div class="alert alert-success">
<b>Task 1:</b> Can you think of examples of features that you would find in molecular simulations?</div>
<details>
<summary> <mark> Solution: </mark> </summary>
Examples are:
- C-alpha positions
- angles
- dihedrals
- RMSD
- density
- surface area
- ...
</details>
## 2. Principal Components Analysis (PCA)
[PCA](https://scikit-learn.org/stable/modules/generated/sklearn.decomposition.PCA.html) is an orthogonal linear transformation that transforms high dimensional data (or feature vectors) into a new coordinate system. In this coordinate system the first coordinate (first *eigenvector*) corresponds to the scalar projection of a linear combination of some data such that this coordinate has the largest variance. The second largest variance in the data can be found in the second coordinate and so on. Let's start by importing some packages required for the our analysis.
<code>
import numpy as np
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
</code>
Now, let's create some model data to analyse. To this end, we will exploit the Müller-Brown potential.
<code>
def muller_potential(x, y):
"""Muller potential
Parameters
----------
x : {float, np.ndarray}
X coordinate. Can be either a single number or an array. If you supply
an array, x and y need to be the same shape.
y : {float, np.ndarray}
Y coordinate. Can be either a single number or an array. If you supply
an array, x and y need to be the same shape.
Returns
-------
potential : {float, np.ndarray}
Potential energy. Will be the same shape as the inputs, x and y.
Reference
---------
Code adapted from https://cims.nyu.edu/~eve2/ztsMueller.m
"""
aa = [-1, -1, -6.5, 0.7]
bb = [0, 0, 11, 0.6]
cc = [-10, -10, -6.5, 0.7]
AA = [-200, -100, -170, 15]
XX = [1, 0, -0.5, -1]
YY = [0, 0.5, 1.5, 1]
value = 0
for j in range(0, 4):
value += AA[j] * np.exp(aa[j] * (x - XX[j])**2 + \
bb[j] * (x - XX[j]) * (y - YY[j]) + cc[j] * (y - YY[j])**2)
return value
</code>
We will now evaluate the potential on a 2-D grid, and plot visualize it.
<code>
dims = (500, 500)
x = np.linspace(-1.5, 1, dims[0])
y = np.linspace(-0.4, 1.8, dims[1])
X, Y = np.meshgrid(x, y)
potential = muller_potential(X, Y)
levels = np.linspace(np.min(potential), np.max(potential), 50)
plt.contour(X, Y, potential.clip(max=200), 40);
</code>
Now, let's convert our potential into a probability distribution, and plot the result.
<code>
Z = np.sum(np.exp(-1/25*potential)) #partition function
P = np.exp(-1/25*potential)/Z
plt.contour(X, Y, P, 100);
</code>
It's time to generate some data! We will extract 10000 samples according to the probability distribution we have just created. To this end, we will use np.random.choice, which enables us to generate random samples according to a given probability. Since this method works only in 1-D, we will first flatten the array, generate the samples, and them bring them back to 2-D.
<code>
flat = np.ravel(P)
sample_index = np.random.choice(a=flat.size, p=flat, size=10000)
samples = np.unravel_index(sample_index, P.shape)
data = np.array([x[samples[1]], y[samples[0]]]).T
plt.scatter(data[:, 0], data[:, 1], c="r", alpha=0.05);
</code>
PCA can tell us in which features (here the x coordinate is one feature and the y coordinate is the second feature), or rather which linear combination, carries the most variance. Let's do a PCA of the samples we have created.
<code>
pca = PCA(n_components=2)
pca.fit(data)
</code>
PCA has identified the two eigenvectors (principal components) of our dataset. Here they are:
<code>
print(pca.components_)
</code>
Each component represents a percentage of the total variance of the system. Here is how:
<code>
print(pca.explained_variance_ratio_)
</code>
It is clear that the first component represents majority of the variance in the data. We have identified the two eigenvectors of this dataset. We can now plot them along with the data. To make arrows visible, we will scale their length by the explained variance of each eigenvector.
<code>
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
ax.scatter(data[:,0], data[:,1], c="r", alpha=0.05)
# plot arrows representing the two components
e1_x = pca.components_[0, 0]*pca.explained_variance_ratio_[0]
e1_y = pca.components_[0, 1]*pca.explained_variance_ratio_[0]
e2_x = pca.components_[1, 0]*pca.explained_variance_ratio_[1]
e2_y = pca.components_[1, 1]*pca.explained_variance_ratio_[1]
ax.arrow(np.mean(x), np.mean(y), e1_x/pca.explained_variance_ratio_[0], e1_y/pca.explained_variance_ratio_[0], head_width=0.1, head_length=0.1, fc='k', ec='k')
ax.arrow(np.mean(x), np.mean(y), e2_x/pca.explained_variance_ratio_[0], e2_y/pca.explained_variance_ratio_[0], head_width=0.1, head_length=0.1, fc='k', ec='k');
</code>
We can project the data on the new reference system defined by the principal components.
<code>
data_projected = data.dot(pca.components_) + pca.mean_
print(data_projected)
</code>
<div class="alert alert-success">
<b>Task 2: </b> Can you use the PCA results to generate an 1-D approximate for the Müller-Brown potential?
</div>
<details>
<summary> <mark> Solution</mark> </summary>
The first principal component represents most of the variance, so we can observe the distribution of data only along this components. This is a way of using PCA as a way of filtering noise, and highlight dominant structures in your data.
```Python
plt.hist(data_projected[:,0], bins=50, color="r");
```
</details>
## 3. The MNIST Dataset
The MNIST dataset consists of a 60,000 examples of hand written numbers and 10,000 test set examples. The digits have all been resized to the same size and centered within this fixed image size. One way of accessing the data is from [here](http://yann.lecun.com/exdb/mnist/) or we can use built in function with scikit-learn. In this way you will access a reduced part of the dataset.
<code>
import numpy as np
import matplotlib.pyplot as plt
from sklearn import decomposition
from sklearn import datasets
from sklearn import manifold
digits = datasets.load_digits()
# Examine the dataset
print(digits.data)
print(digits.target)
X = digits.data
y = digits.target
</code>
A short helper function to plot an example from the dataset:
<code>
import matplotlib.pyplot as plt
def plot_digits(X):
"""Small helper function to plot 100 digits."""
fig, axs = plt.subplots(nrows=10, ncols=10, figsize=(8, 8))
for img, ax in zip(X, axs.ravel()):
ax.imshow(img.reshape((8, 8)), cmap="Greys")
ax.axis("off")
</code>
<code>
plot_digits(X)
</code>
<div class="alert alert-success">
<b>Task 3:</b> Understanding the dataset
- What are the dimensions of the data?
- What are the features of the data?
- What information do the features hold?
<code>
### Your solution here:
</code>
<details>
<summary> <mark> Solution: </mark> </summary>
```Python
# data dimension
np.shape(X)
# The output of the array tells you, that you have 1797 samples of a 64 dimensional feature vector
# Features
print(X[0])
# Each entry in the first sample of the feature vector gives you a value of the grey scale from the image. This could be normalised.
```
</details>
<div class="alert alert-success">
<b>Task 4:</b> Can you do a principal component analysis of the digits dataset?
- Do a PCA analysis using two components.
- What is the variance contribution of the first two components?
- How many components do you need to reach 90% variance explained?
- Generate a plot of the first two principal components and colour them according to your digit. What does it tell you? </div>
<code>
### Your solution here:
</code>
<details>
<summary> <mark> Solution: </mark> </summary>
```Python
# PCA
pca = decomposition.PCA(n_components=2, n_oversamples=20)
pca.fit(X)
X_pca = pca.transform(X)
# variance contribution of first two components
print(pca.explained_variance_ratio_)
# finding 90% variance
for i in range(2,30):
pca = decomposition.PCA(n_components=i, n_oversamples=20)
pca.fit(X)
X_pca = pca.transform(X)
cum_sum = pca.explained_variance_ratio_.cumsum()
if cum_sum[-1] > 0.9:
print(f'{i} number of principle components are needed to reach a variance explained of 90%')
break
# plotting first two components and colouring according to digits
plt.scatter(X_pca[:, 0], X_pca[:, 1], c=y, cmap=plt.cm.nipy_spectral,
edgecolor='k',label=y)
plt.colorbar(boundaries=np.arange(11)-0.5).set_ticks(np.arange(10))
plt.xlabel('pc 1')
plt.ylabel('pc 2')
```
</details>
<div class="alert alert-success">
<b>Task 5: </b> Rerun your PCA with 5 components </div>
What feature in your 64 (8x8) digit input vector contributes the most to your first principle components?
Hint: look at the absolute value of `pca.components_[0]` If you you generate a bar plot you can see the contributions well.
<code>
### Your solution here:
</code>
<details>
<summary> <mark> Solution: </mark> </summary>
```Python
pca = decomposition.PCA(n_components=5, n_oversamples=20)
pca.fit(X)
X_pca = pca.transform(X)
fig, ax = plt.subplots(figsize=(15,15))
indeces = np.argsort(abs(pca.components_[0]))
x = np.linspace(0,len(indeces), len(indeces))
ax.bar( x,abs(pca.components_[0])[indeces],tick_label=indeces)
ax.set_xlabel('feature index')
ax.set_ylabel('absolute value of contribution')
```
</details>
## 4. time-lagged independent component analysis (tICA)
In this section, we will study tCA with a toy dataset. Let's start by loading it.
<code>
# loading data
file = 'data/hmm-doublewell-2d-100k.npz'
with np.load(file) as fh:
data = fh['trajectory']
</code>
<code>
np.shape(data)
</code>
### 4.1. Visualising the dataset
We can see this is a trajectory with 100000 time datapoints and 2 features. Let's examine this dataset a bit more.
<code>
fig, axes = plt.subplots(1, 2, figsize=(10, 4))
axes[0].plot(data[:300,0], alpha=0.6)
axes[1].plot(data[:300,1], alpha=0.6)
axes[1].set_xlabel('$time$')
axes[0].set_xlabel('$time$')
axes[0].set_ylabel('$x$')
axes[1].set_ylabel('$y$')
fig.tight_layout()
</code>
<div class="alert alert-success">
<b>Task 6: </b> Examine the data a bit more, to get a better feel for it. What is the extent of the data set in x and y? Can you plot a histogram of the data? What information does the trajectory tell us the histogram obscures? </div>
<details>
<summary> <mark> Solution: </mark> </summary>
The minimum and maximum of the data is given by:
```Python
print('x_min is:',np.min(data[:,0]), 'x_max is:',np.max(data[:,0]), '\ny_min is:', np.min(data[:,1]), 'y_max is:', np.max(data[:,1]))
```
An example of how to plot a histogram of the data looks like this:
```Python
plt.figure(figsize=(7,7))
counts,ybins,xbins = np.histogram2d(data[:,0],data[:,1],bins=250);
plt.contour(counts,extent=[xbins.min(),xbins.max(),ybins.min(),ybins.max()])
```
There is no slow transitions in the x coordinate, but there are in the y coordinate.
</details>
### 4.2. tICA analysis
tICA is a common dimensionality reduction technique for molecular dynamics trajectories. Unfortunately scikit-learn does not feature an implementation of this method, which is why other packages are normally used. Here, we provide a convenient helper module based on the implementation from [MSM-Builder](http://msmbuilder.org/3.8.0/), adapted so that it can be used as stand-alone code. The module is written so as to mimic the dimensionality reduction is done with scikit-learn. That means you use a similar syntax as before, i.e you create an instance of tICA with a given number of components and then you use fit and transform on the data.
<code>
from tica.tica import tICA
</code>
Let's carry out a tICA analysis of the data we have previously loaded. A small difference from the syntax in scikit-learn: the parameter of the <code>fit</code> method data must be in square brackets, since the method can accept a list of trajectory data.
<code>
tic = tICA()
tic.fit([data]);
</code>
<code>
The dimensions identified by the tICA analysis can be accessed as follows:
</code>
<code>
print(tic.eigenvectors_)
</code>
Time for the next excercise! Before getting to it though, execute the cell below, you will need it!
<code>
def draw_arrow(origin, v, color):
ax.arrow(origin[0], origin[1], v[0], v[1], color=color, width=0.02, linewidth=3)
</code>
<div class="alert alert-success">
<b>Task 7: </b> Now use PCA on the same dataset and compare the two components by creating a scatter plot and drawing the vectors representing the PCA composition and tICA composition. Make use of the handy helper function <code>draw_arrow</code> for the vectors to draw arrows on the scatter plot from above. </div>
<code>
### Your solution here:
</code>
<details>
<summary> <mark> Solution: </mark> </summary>
We start by carrying out the PCA of the toy data
```Python
pca = PCA()
pca.fit(data);
```
Now, we plot a scatterplot of data, with arrows representing the first components of both tICA (red) and PCA (blue).
```Python
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
ax.scatter(data[:,0], data[:,1], marker = '.', color='black', alpha=0.1)
origin = np.mean(data, axis=0)
draw_arrow(origin, tic.eigenvectors_[0]*2, "red")
draw_arrow(origin, pca.components_[0]*2, "dodgerblue")
ax.set_xlabel("x")
ax.set_ylabel("y");
```
</details>
### 4.3. Comparison of dimensionality reduction with PCA and tICA
Dimensionality reduction techniques enable us to identify suitable ways of projecting high-dimensional data into a lower-dimensional space with minimal information loss. We have just seen that PCA and tICA identify different spaces onto which the data can be projected.
<div class="alert alert-success">
<b>Task 5: </b>Project the data into the eigenspace generated by PCA, and into the tICA space. Create histograms of each of the components. What do you observe? </div>
<code>
### Your solution here:
</code>
<details>
<summary> <mark> Solution: </mark> </summary>
Let's project the data into the tICA and PCA spaces.
```Python
tic_out = tic.transform([data])[0]
PCA_out = pca.transform(data)
```
Now, let's make some pretty plots showing the projections on the first and second component of PCA and tICA
```Python
fig = plt.figure(figsize=(10, 4))
ax1 = fig.add_subplot(1, 2, 1)
ax1.hist(tic_out[:, 0], histtype="step", label="tICA", bins=50, color="red")
ax1.hist(PCA_out[:, 0], histtype="step", label="PCA", bins=50, color="dodgerblue")
ax1.set_xlabel("first component")
ax1.set_ylabel("count (#)")
ax1.legend(frameon=False);
ax2 = fig.add_subplot(1, 2, 2)
ax2.hist(tic_out[:, 1], histtype="step", label="tICA", bins=50, color="red")
ax2.hist(PCA_out[:, 1], histtype="step", label="PCA", bins=50, color="dodgerblue")
ax2.set_xlabel("second component")
ax2.legend(frameon=False);
```
</details>
## 5. t-Distributed Stochastic Neighbor Embedding (t-SNE)
t-SNE is a technique for dimensionality reduction that is particularly well suited for the visualization of high-dimensional datasets. It gives each datapoint a poisition in a two or three dimensional map. It is classed as a non-linear dimensionality reduction technique and models high-dimensional data that are close in space to spatially close two or three-deminsional points. Let's apply t-SNE to the MNIST dataset we have met in section 3!
<code>
tsne = manifold.TSNE(n_components=2, init='pca', random_state = 0)
X_tsne = tsne.fit_transform(X)
fig = plt.figure(1, figsize=(4, 4))
plt.clf()
plt.scatter(X_tsne[:, 0], X_tsne[:, 1], c=y, cmap=plt.cm.nipy_spectral,
edgecolor='k',label=y)
plt.colorbar(boundaries=np.arange(11)-0.5).set_ticks(np.arange(10))
plt.xlabel('tsne 0')
plt.ylabel('tsne 1')
</code>
<div class="alert alert-success">
<b>Task 6:</b> Can you regenerate your t-SNE embedding in 3D and plot it? </div>
<code>
### Your solution here
</code>
<details>
<summary> <mark> Solution:</mark> </summary>
```Python
tsne = manifold.TSNE(n_components=3, init='pca', random_state = 0)
X_tsne = tsne.fit_transform(X)
fig = plt.figure(1, figsize=(4, 4))
plt.clf()
ax = fig.add_subplot(projection='3d')
ax.scatter(X_tsne[:, 0], X_tsne[:, 1], X_tsne[:, 2], c=y, cmap=plt.cm.nipy_spectral, s=9, lw=0)
```
</details>
## 6. Conclusion
<div class="alert alert-info">
<b>Key points:</b>
- PCA is a linear dimensionality reduction technique for tabular data,
- PCA can be used to remove noise from data,
- tICA is also a linear dimensionality reduction technique, but it maximises the autocorrelation time rather than the variance
- tICA and PCA may be appropriate for different use cases: tICA will generally provide you with slow dynamics and PCA for maximising spacial variance.
- t-SNE is another dimensionality reduction technique for tabular data that is more general than PCA.
</div>
### Next Notebook
[Dimensionality Reduction, part 2](2_DL_part2.ipynb)
|
{
"filename": "ML_workshop_1_DR_part1_1.ipynb",
"repository": "CCPBioSim/MDAnalysis",
"query": "transformed_from_existing",
"size": 34626,
"sha": ""
}
|
# course_2024_16S_2024-checkpoint_1.ipynb
Repository: Gibbons-Lab/isb
<a href="https://colab.research.google.com/github/Gibbons-Lab/isb_course_2023/blob/main/16S_2024.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# 🦠 Amplicon Sequencing Data Analysis with QIIME 2
This notebook will accompany the first session of the 2024 ISB Virtual Microbiome Series. The presentation slides can be [found here](https://gibbons-lab.github.io/isb_course_2024/16S).
Save your own local copy of this notebook by using `File > Save a copy in Drive`. At some point you may be prompted to trust the notebook. We promise that it is safe 🤞
**Disclaimer:**
The Google Colab notebook environment will interpret any command as Python code by default. If we want to run bash commands we will have to prefix them by `!`. So any command you see with a leading `!` is a bash command and if you wanted to run it in your terminal you would omit the `!`. For example, if in the Colab notebook you ran `!wget` you would just run `wget` in your terminal.
**Run all cells IN ORDER**
## Setup
QIIME 2 is usually installed by following the [official installation instructions](https://docs.qiime2.org/2024.5/install/). However, because we are using Google Colab and there are some caveats to using conda here, we will have to hack around the installation a little bit. But no worries, we provide a setup script below which does all this work for us. 😌
So...let's start by pulling a local copy of the project repository down from GitHub.
<code>
!git clone https://github.com/gibbons-lab/isb_course_2024 materials
</code>
This repository, called __materials__, contains all the relevant data and other resources we'll need for this course. To view the directory, click on the folder icon on the left. Let's navigate to that directory via command line now:
<code>
%cd materials
</code>
Notice here we use ```%``` instead of ```!``` to run out command line function. This makes the path change to our directory permanent: using the ```!``` operator only switches the interpreter to expect command line prompts temporarily.
## Install QIIME2
Now that we have all our materials, we're _almost_ ready to get started - but not quite. Remember QIIME2? We'll need to install that before getting into the actual analysis. Don't worry - this will only set up in the Colab notebook, not on your local machine.
Let's run the following cell, to install and setup QIIME2.
<code>
%run setup_qiime2
</code>
⬆️ This will take some time (usually 10 to 15 minutes), so we'll switch back over to the [presentation](https://gibbons-lab.github.io/isb_course_2024/16S) while we wait.
If you want to learn more about QIIME2, we recommend you check out the [documentation](https://docs.qiime2.org/). This will also explain how to install QIIME2 on your local machine 🖥
## Let's Get Started!
Now we're on to the fun part. Let's begin by taking a look at our data. In the __data__ folder, you'll find 10 FASTQ files, a file manifest, and a metadata file. Let's take a look at the manifest, first. This is a file that contains the name, and filepath of all our samples, and we'll need it later on when we use QIIME2 📝.
<code>
import pandas as pd
manifest = pd.read_csv('data/manifest.tsv', sep = '\t')
manifest
</code>
We can also check out the metadata file, which will give us more context on our samples 🔬
<code>
metadata = pd.read_csv('data/metadata.tsv', sep='\t')
metadata
</code>
Looks good, all 10 FASTQ files are accounted for, five healthy and five with Parkinson's Disease. We can use the manifest file to import our files into QIIME2.
## QIIME2 Pipeline
Let's remind ourselves what the QIIME2 pipeline will do:

To use sequencing data in QIIME2, we first need to turn the FASTQ files containing our data into QIIME artifacts. Using the manifest we just checked out, let's run our first command:
-- as a reminder, adding ```!``` before the command tells the notebook this is a bash command, rather than python.
<code>
!qiime tools import \
--type 'SampleData[SequencesWithQuality]' \
--input-path data/manifest.tsv \
--output-path sequences.qza \
--input-format SingleEndFastqManifestPhred33V2
</code>
Let's take a look a the command. QIIME commands take following format:
```
qiime plugin action --i-argument1 ... --o-argument2 ...
```
In the previous command, we are calling the ```tools``` plugin within QIIME2 to import our data. The following arguments designate where the manifest is, as well as where the output should be saved. We also tell QIIME2 what sort of input to expect.
Argument types usually begin with a letter denoting their meaning:
- `--i-...` = input files
- `--o-...` = output files
- `--p-...` = parameters
- `--m-...` = metadata
---
## Visualizing our Data 🔎
Before we move on, let's use QIIME2 to visualize our sequencing data.
<code>
!qiime demux summarize \
--i-data sequences.qza \
--o-visualization qualities.qzv
</code>
.qzv files like the one we just produced are visualization. You can view the plot by downloading the file and opening it using http://view.qiime2.org. To download the file click on the folder symbol to the left, open the `materials` folder, and choose download from the dot menu next to the `qualities.qzv` file.
---
## Quality Filtering
Before we can use our sequencing data, we need to "denoise" it. To do this, we'll use a plugin called DADA2. This involves three things.
1. filter and trim the reads
2. find the most likely set of unique sequences in the sample (ASVs)
3. remove chimeras
4. count the abundances of each ASV
This command will take a little time - let's run it, and head back to the presentation to discuss what's happening.
<code>
!qiime dada2 denoise-single \
--i-demultiplexed-seqs sequences.qza \
--p-trunc-len 150 \
--p-n-threads 2 \
--output-dir dada --verbose
</code>
If this step takes too long or fails, you can also copy the results from the treasure chest with the following command.
<code>
# obscure magic that will only copy if the previous command failed
![ -d dada ] || cp -r treasure_chest/dada .
</code>
Let's check to see how that went. One good way to tell if the identified ASVs are representative of the sample is to see how many reads were maintained throughout the pipeline. Here, the most common issues and solutions are:
**Large fraction of reads is lost during merging (only paired-end)**

In order to merge ASVs DADA2 uses an overlap of 12 bases between forward and reverse reads by default. Thus, your reads must allow for sufficient overlap *after* trimming. So if your amplified region is 450bp long and you have 2x250bp reads and you trim the last 30 bases of each read, truncating the length to 220bp, the total length of covered sequence is 2x220 = 440 which is shorter than 450bp so there will be no overlap. To solve this issue trim less of the reads or adjust the `--p-min-overlap` parameters to something lower (but not too low).
<br>
**Most of the reads are lost as chimeric**
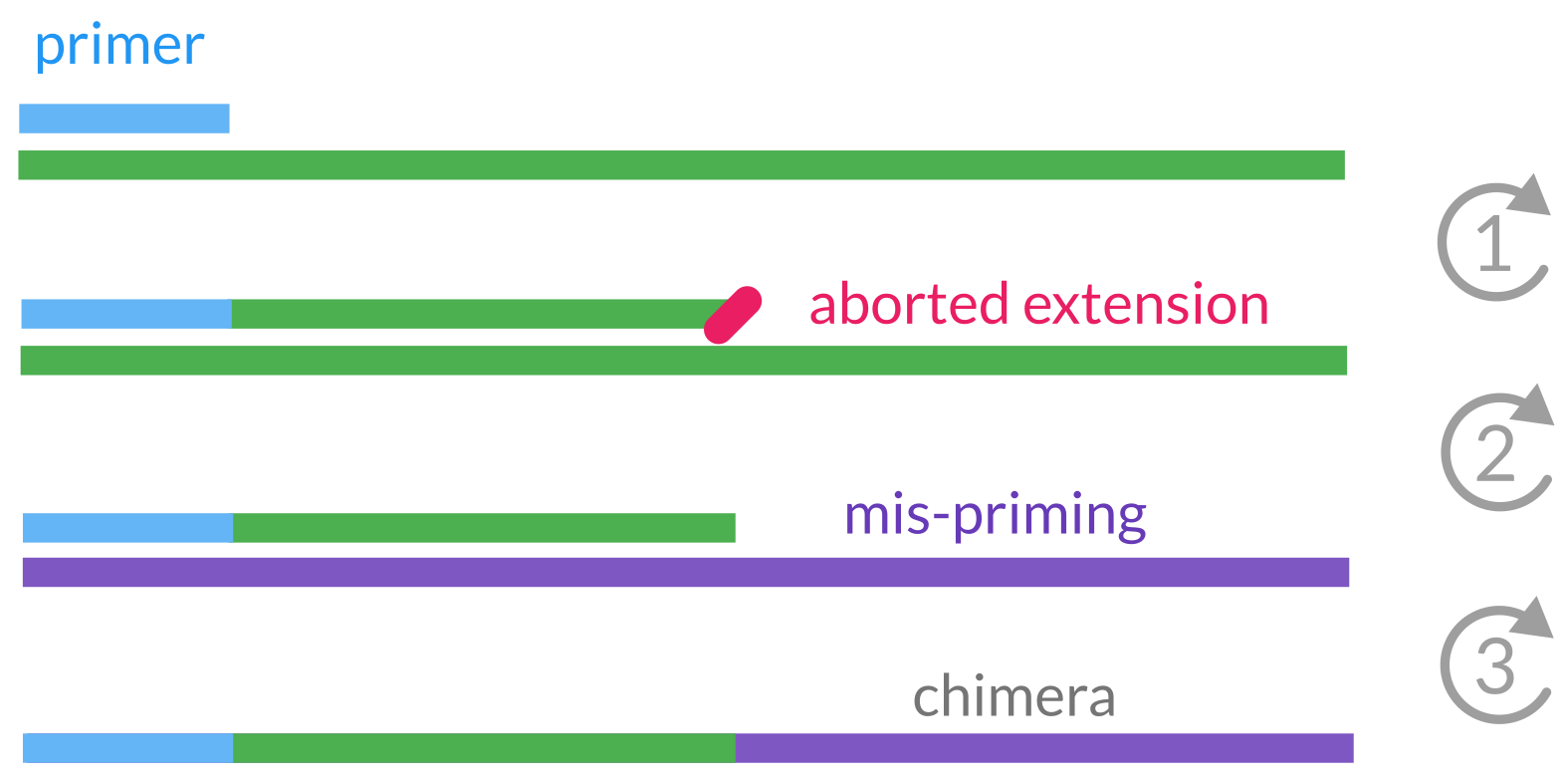
This is usually an experimental issue as chimeras are introduced during amplification. If you can adjust your PCR, try to run fewer cycles. Chimeras can also be introduced by incorrect merging. If your minimum overlap is too small ASVs may be merged randomly. Possible fixes are to increase the `--p-min-overlap` parameter or run the analysis on the forward reads only (in our empirical observations, chimeras are more likely to be introduced in the joined reads). *However, losing between 5-25% of your reads to chimeras is normal and does not require any adjustments.*
Our denoising stats are contained in an artifact. To convert it to a visualization we can use `qiime metadata tabulate`.
<code>
!qiime metadata tabulate \
--m-input-file dada/denoising_stats.qza \
--o-visualization dada/denoising-stats.qzv
</code>
Like before, we can download the .qzv file and visualize the results using the [QIIME2 Viewer]('https://view.qiime2.org/').
It's important to understand what this output tells us. For instance, what percent of reads in our data pass the filtering step? What percent of reads were non-chimeric? Differences in these metrics between samples can affect diversity metrics.
---
## Diversity and Phylogenetics
### Introduction to diversity metrics
An important metric to consider when studying microbial ecology is __diversity__. Diversity comes in two flavors: ⍺ (alpha) and β (beta).
Alpha diversity is pretty simple - how diverse is a single sample? You might consider measures like richness and evenness.
Beta diversity instead looks at how different two samples are from each other - what taxa are shared, and how their abundances differ.
I want to note here that we're getting into some analyses that some bioinformaticians may prefer to do in R or Python, outside of the QIIME framework. For example, diversity analyses can be performed using `scikit-bio` in Python or `vegan` in R. In fact, QIIME 2 is using `scikit-bio` and `vegan` under the hood for its diversity calculations! QIIME eliminates the need to manipulate dataframes or do calculations yourself, but it might not have the newest methods!
### Starting our Tree
Let's start by building a phylogenetic tree for our sequences using the following command. This time, we call the _phylogeny_ plugin in QIIME2.
<code>
!qiime phylogeny align-to-tree-mafft-fasttree \
--i-sequences dada/representative_sequences.qza \
--output-dir tree
</code>
We can create a visualization for the tree using the [empress](https://github.com/biocore/empress) QIIME 2 plugin.
<code>
!qiime empress tree-plot \
--i-tree tree/rooted_tree.qza \
--o-visualization tree/empress.qzv
</code>
## Calculating Diversity
Using the Diversity plugin, we can use our table and tree to calculate several diversity metrics. To account for variations in sampling depth, we'll provide QIIME2 with a cutoff at which rarefy all our samples. Since this randomly selects sequences, your results might look a little different. We'll also pass in our metadata file, so we can keep track how which samples come from each group.
<code>
!qiime diversity core-metrics-phylogenetic \
--i-table dada/table.qza \
--i-phylogeny tree/rooted_tree.qza \
--p-sampling-depth 5000 \
--m-metadata-file data/metadata.tsv \
--output-dir diversity
</code>
If you open the `diversity` folder, you'll see that we calculated several different diversity metrics. Beta diversity uses a "distance" or "dissimilarity" matrix, but there are different definitions of distance! Some different types of distance include Bray-Curtis, Jaccard, Unweighted UniFrac, and weighted UniFrac. UniFrac distances are based on phylogeny, while Bray-Curtis and Jaccard are not. For more information on diversity metrics, check out this QIIME [forum post](https://forum.qiime2.org/t/alpha-and-beta-diversity-explanations-and-commands/2282).
## Alpha Diversity
We get a bunch of outputs from the previous command - measures of both alpha and beta diversity. To start, let's use the Shannon vector in the output directory to create a visualization of alpha diversity across samples. Generally, healthy, long-living individuals have balanced diverse microbiomes. However, this isn't necessarily a direct indicator of health or disease. Let's see how it looks in our samples
<code>
!qiime diversity alpha-group-significance \
--i-alpha-diversity diversity/shannon_vector.qza \
--m-metadata-file data/metadata.tsv \
--o-visualization diversity/alpha_groups.qzv
</code>
Like before, we can download the visualization and open it with the QIIME2 viewer.
There doesn't appear to be a difference in Shannon Diversity between Parkinson's Disease patients and healthy controls, but could there be confounding variables? Since this is a cohort study rather than a controlled experiment, we can't control for variables that affect microbiome composition and diversity like antibiotics, diet, other medications, ..., but we can __stratify__ across them. Let's see what happens if we stratify by drug use.
<code>
!
</code>
## Beta Diversity
Let's visualize the beta diversity and see how they separate. For this we'll look at weighted UniFrac. This time, we'll have to download the file ⬅️
We can check for 'significant' separation between samples using PERMANOVA. We can do this with the diversity plugin in QIIME2.
<code>
!qiime diversity adonis \
--i-distance-matrix diversity/weighted_unifrac_distance_matrix.qza \
--m-metadata-file data/metadata.tsv \
--p-formula "parkinson_disease" \
--p-n-jobs 2 \
--o-visualization diversity/permanova.qzv
</code>
We can also use PERMANOVA to identify confounders. PERMANOVA tells us how much variance in the community composition is explained by each variable. Common confounders include sex, age, BMI, diet, and antibiotic use. In the original study, authors identified that Parkinson's Disease medications were associated with different microbiome compositions. Let's take a look at these variables.
<code>
!qiime diversity adonis \
--i-distance-matrix diversity/weighted_unifrac_distance_matrix.qza \
--m-metadata-file data/metadata.tsv \
--p-formula "parkinson_disease + sex + age + location + p3m_antibiotics_bool" \
--p-n-jobs 10 \
--o-visualization diversity/permanova_big.qzv
</code>
Before, we did not see a significant p-value for the effect of Parkinson's disease on beta diversity. However, when we add certain covariates, we might find that they were confounding a relationship.
However, most of our variance remains unexplained. Microbiome composition is affected by many things, and this is an uncontrolled cohort study, so we would not expect any single variable to explain most of the variance.
## Visualizing Beta Diversity Using PCoA
If we want to __visually__ show this separation between samples, we can't just plot the entire UniFrac distance matrix, because it has 100+ dimensions! Instead, we can use **dimensionality reduction** to "compress" our data into a few dimensions that explain most of the variance. There are several types of dimensionality reduction (like UMAP and tSNE), but the preferred method of dimensionality reduction for microbiome communities is Principal Coordinate Analysis (PCoA). This is because PCoA is linear, and thus preserves the global structure of the data and is reproducible.
We already ran a PCoAs for each distance metric, and we can look at them if we download `weighted_unifrac_emperor.qzv`, `unweighted_unifrac_emperor.qzv`, `bray_curtis_emperor.qzv`, or `jaccard_emperor.qzv`
---
## Taxonomic Classification
We can learn a lot from diversity metrics, alpha and beta. But to really dig into the data, we need to know what microbes are in each sample 🦠. To do this, we'll classify the reads in QIIME2 using a Bayesian classifier. Several such classifiers are available at https://docs.qiime2.org/2024.5/data-resources/
<code>
!qiime feature-classifier classify-sklearn \
--i-reads dada/representative_sequences.qza \
--i-classifier ncbi-refseq-genus-515f-806r.qza \
--p-n-jobs 2 \
--o-classification taxa.qza
</code>
Now we've classified the reads, we can visualize the taxonomic breakdown of our samples.
<code>
!qiime taxa barplot \
--i-table dada/table.qza \
--i-taxonomy taxa.qza \
--m-metadata-file data/metadata.tsv \
--o-visualization taxa_barplot.qzv
</code>
Now, we can use ```table.qza```, which contains our reads, and ```taxa.qza```, which contains taxonomic classifications for reads, and collapse the data onto the genus level.
<code>
!qiime taxa collapse \
--i-table dada/table.qza \
--i-taxonomy taxa.qza \
--p-level 6 \
--o-collapsed-table genus.qza
</code>
We'll export this as a .tsv, which will be more usable for the next portion of the course that you'll see tomorrow
<code>
!qiime tools export \
--input-path genus.qza \
--output-path exported
!biom convert -i exported/feature-table.biom -o genus.tsv --to-tsv
</code>
Let's peek at the results 🔭
<code>
abundances = pd.read_table("genus.tsv", skiprows=1, index_col=0)
abundances
</code>
This is easier to interpret by visualizing the results. We can use the file we just exported from QIIME2 to build a visualization using any tool we like, such as seaborn or plotnine. Here is an example of building a visualization (a heatmap) in seaborn:
<code>
import numpy as np
import seaborn as sns
</code>
<code>
abund_to_plot = abundances
abund_to_plot.index = abund_to_plot.index.str.split(";").str[5] # Use only the genus name
abund_to_plot = abund_to_plot[~abund_to_plot.index.isin(["g__", "__"])] # remove unclassified genera
abund_to_plot = abund_to_plot.sample(50, axis=0) # use 50 random genera (rows)
# Let's do a centered log-ratio transform: log x_i - log mean(x)
transformed = abund_to_plot.apply(
lambda xs: np.log(xs + 0.5) - np.log(xs.mean() + 0.5),
axis=0)
sns.clustermap(transformed.T, cmap="magma", xticklabels=True, figsize=(16, 6))
</code>
Now, our data is starting to be interpretable. Each row is a sample, and each column is a bacterial genus. The table values are "counts", or the number of times a genus was detected in a certain sample. We can use relative abundance data to test hypotheses, but it requires special statistical methods because it is ___compositional___
## Differential Abundance Analysis
Explain compositional data + methods for testing
<code>
!qiime composition ancombc \
--i-table genus.qza \
--m-metadata-file data/metadata.tsv \
--p-formula "parkinson_disease" \
--o-differentials ancombc.qza
</code>
<code>
!qiime composition da-barplot \
--i-data ancombc.qza \
--p-significance-threshold 0.01 \
--o-visualization da_barplot.qzv
</code>
## Exercise - Plant a Tree
One visualization that we did not spend a lot of time on was the phylogentic tree of our ASVs. Let's change that! We have seen that there are genera that appear in multiple populations in the previous step. But are the organisms in that genus actually the same?
Let's annotate the tree with our taxonomic classifications and abundances. We will use the empress plugin again but this time with the `community-plot` option. I filled in a template of the command for you. Can you figure out what has to go in the empty spaces?
**QUESTIONS:**
1) Are some of the branch lengths on the tree longer than you would expect? Do you notice anything interesting or suspicious about the taxonomic identities of these branches?
2) Can you find examples of phyla that are polyphyletic (i.e. where clusters of ASVs from the same phylum are found in different locations on the tree, showing different commmon ancestors)? What about polyphyletic taxa at lower taxonomic levels, like at the family or genus levels? Why do you think these patterns exist?
<code>
# This won't run until you fill in the [EMPTY] spots with the right files ;)
!qiime empress community-plot \
--i-tree [EMPTY] \
--i-feature-table dada/table.qza \
--m-sample-metadata-file [EMPTY] \
--m-feature-metadata-file taxa.qza \
--o-visualization community-tree-viz.qzv
</code>
|
{
"filename": "course_2024_16S_2024-checkpoint_1.ipynb",
"repository": "Gibbons-Lab/isb",
"query": "transformed_from_existing",
"size": 38700,
"sha": ""
}
|
# app_1.ipynb
Repository: MLerSunny/Langchain
<code>
from langchain_community.embeddings import OllamaEmbeddings
from langchain_community.vectorstores import Chroma
from langchain_community.chat_models import ChatOllama
from langchain.text_splitter import RecursiveCharacterTextSplitter
from langchain.chains import RetrievalQA
from langchain.docstore.document import Document
from langchain_community.document_loaders import WebBaseLoader
from transformers import AutoModel, AutoTokenizer
from langchain.embeddings import HuggingFaceEmbeddings
</code>
<code>
## Data Ingestion
from langchain_community.document_loaders import TextLoader
loader=TextLoader("speech.txt")
text_docs=loader.load()
text_docs
</code>
<code>
## Pdf reader
from langchain_community.document_loaders import PyPDFLoader
loader=PyPDFLoader('attention.pdf')
pdf_docs=loader.load()
pdf_docs
</code>
<code>
# web based loader
import bs4
## load,chunk and index the content of the html page
loader=WebBaseLoader(web_paths=("https://lilianweng.github.io/posts/2023-06-23-agent/",),
bs_kwargs=dict(parse_only=bs4.SoupStrainer(
class_=("post-title","post-content","post-header")
)))
web_docs=loader.load()
</code>
<code>
# 2. Split Documents into chunks
text_splitter = RecursiveCharacterTextSplitter(
chunk_size=1000,
chunk_overlap=200
)
chunks_tdocs = text_splitter.split_documents(text_docs)
chunks_pdocs = text_splitter.split_documents(pdf_docs)
chunks_wdocs = text_splitter.split_documents(web_docs)
chunks_tdocs
</code>
<code>
chunks_pdocs
</code>
<code>
chunks_wdocs
</code>
<code>
chunks_pdocs
</code>
<code>
# 3. Initialize Embeddings (using a dedicated embedding model)
from langchain_community.embeddings import HuggingFaceEmbeddings
embedding_model = "nomic-ai/nomic-embed-text-v1.5"
embeddings = HuggingFaceEmbeddings(
model_name=embedding_model,
model_kwargs={"trust_remote_code": True, "device": "cpu"}, # or "cuda"
encode_kwargs={"normalize_embeddings": True}
)
</code>
<code>
print (embeddings)
</code>
<code>
# 4. Create Vector Store
vector_store = Chroma.from_documents(
documents=chunks_tdocs + chunks_pdocs + chunks_wdocs,
embedding=embeddings,
persist_directory="./rag_db"
)
</code>
<code>
print(vector_store)
</code>
<code>
# 5. Initialize DeepSeek LLM
llm = ChatOllama(
model="deepseek-r1:1.5b", # Replace with your actual DeepSeek model name
temperature=0.3,
num_ctx=4096 # Adjust based on your model's context window
)
</code>
<code>
# 6. Create Retrieval QA Chain
qa_chain = RetrievalQA.from_chain_type(
llm=llm,
retriever=vector_store.as_retriever(),
chain_type="stuff", # Simple document stuffing
return_source_documents=True
)
</code>
<code>
query = " The generative agent architecture"
retrieved_docs = vector_store.similarity_search(query, k=3)
for doc in retrieved_docs:
print(doc.page_content)
</code>
|
{
"filename": "app_1.ipynb",
"repository": "MLerSunny/Langchain",
"query": "transformed_from_existing",
"size": 224560,
"sha": ""
}
|
# welcome.ipynb
Repository: nunososorio/SingleCellGenomics2024
<a href="https://colab.research.google.com/github/nunososorio/SingleCellGenomics2024/blob/main/welcome.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
<img src="https://github.com/nunososorio/SingleCellGenomics2024/blob/main/logo.png?raw=true" alt="AnnData" style="width:600px; height:auto;"/>
Welcome to the 'Single-Cell Genomics For Beginners' course!
Please go through the precourse materials deposited on osf.io, especially the notebook linked on '1_NB_Intro_python.pdf'.
We encourage you to familiarize yourself with Python programming through free online tutorials such as:
- https://www.learnpython.org/
- https://www.freecodecamp.org/learn/scientific-computing-with-python/
Our course will utilize a Jupyter notebook environment hosted on Google Colaboratory. For an introduction to Google Colab, you can watch this video:
- https://www.youtube.com/watch?v=inN8seMm7UI
We will employ interactive teaching methodologies to help making the sessions more engaging and effective, including:
- Team-Based Learning, as shown in this video: https://www.youtube.com/watch?v=BlVPLYGdBLg
- Role Playing and gamification strategies.
For those already confident in Python programming, consider advancing your skills in scRNAseq analysis using tutorials such as:
- Scanpy documentation: https://scanpy.readthedocs.io/
- scRNA-python-workshop guide: https://chanzuckerberg.github.io/scRNA-python-workshop/intro/about
If you are new to Python, focus on acquiring the basic principles of the language. Here's some inspiration: https://www.youtube.com/watch?v=5MgBikgcWnY
Looking forward to an engaging and fruitful learning experience together!
Leonardo Garma, Mónica Fernandes, Nuno S. Osório and Juan Manuel Barba
|
{
"filename": "welcome.ipynb",
"repository": "nunososorio/SingleCellGenomics2024",
"query": "transformed_from_existing",
"size": 2901,
"sha": ""
}
|
# workflow_generate_ro-crate.ipynb
Repository: Xomics/ACTIONdemonstrator
<code>
# More information on:
# https://github.com/ResearchObject/ro-crate-py; https://about.workflowhub.eu/Workflow-RO-Crate/
# Import modules
from rocrate.rocrate import ROCrate
from rocrate.model.file import File
from rocrate.model.computationalworkflow import ComputationalWorkflow
from rocrate.model.computerlanguage import ComputerLanguage
from rocrate.model.person import Person
from rocrate.model.contextentity import ContextEntity
from rocrate.model.data_entity import DataEntity
from rocrate.model.softwareapplication import SoftwareApplication
# Intialize the RO Crate object
crate = ROCrate()
</code>
<code>
# Get current date, which will be added as publish date
from datetime import date
today = date.today()
</code>
## Main Workflow file
<code>
# Define the Nextflow workflow as a ComputationalWorkflow, add to crate.
MainWorkflow = crate.add(ComputationalWorkflow(crate, "action.nf", properties={
"@id": "action.nf",
"@type": ["File", "SoftwareSourceCode", "ComputationalWorkflow"],
"name": "NTR-ACTION Data-analysis workflow",
"dateCreated": str(today),
"input": "",
"description": "Use of multi-omics data (Metabolomics + DNA Methylation) to study CBCL data",
"output": "",
"license": "https://opensource.org/licenses/MIT",
"url": "https://github.com/Xomics/ACTIONdemonstrator_workflow",
"version": "1.0.0"
}))
</code>
## Contextual information
<code>
# Define Nextflow as computer language
nextflow_id = "https://w3id.org/workflowhub/workflow-ro-crate#nextflow"
Nextflow = crate.add (ComputerLanguage(crate, nextflow_id, properties={
"@id": "https://w3id.org/workflowhub/workflow-ro-crate#nextflow",
"@type": "ComputerLanguage",
"name": "Nextflow",
"identifier": {
"@id": "https://www.nextflow.io/"
},
"url": {
"@id": "https://www.nextflow.io/"
}
}))
</code>
<code>
# Define persons (authors)
Anna_Niehues_id = "https://orcid.org/0000-0002-9839-5439"
Casper_de_Visser_id = "https://orcid.org/0000-0002-2812-5898"
Fiona_Hagenbeek_id = "https://orcid.org/0000-0002-8773-0430"
Naama_Karu_id = "https://orcid.org/0000-0001-8005-0726"
Alida_Kindt_id = "https://orcid.org/0000-0001-6551-6030"
Purva_Kulkarni_id = "https://orcid.org/0000-0002-4681-4582"
Rene_Pool_id = "https://orcid.org/0000-0001-5579-0933"
Dorret_Boomsma_id = "https://orcid.org/0000-0002-7099-7972"
Jenny_van_Dongen_id = "https://orcid.org/0000-0003-2063-8741"
Alain_van_Gool_id = "https://orcid.org/0000-0003-0010-5286"
PeterBram_t_Hoen_id = "https://orcid.org/0000-0003-4450-3112"
Anna_Niehues = crate.add(Person(crate, Anna_Niehues_id, properties={
"name": "Anna Niehues",
"affiliation": "Radboud university medical center"
}))
Casper_de_Visser = crate.add(Person(crate, Casper_de_Visser_id, properties={
"name": "Casper de Visser",
"affiliation": "Radboud university medical center"
}))
Fiona_Hagenbeek = crate.add(Person(crate, properties={
"name": "Fiona A. Hagenbeek",
"affiliation": "Vrije Universiteit Amsterdam"
}))
Naama_Karu = crate.add(Person(crate, Naama_Karu_id, properties={
"name": "Naama Karu",
"affiliation": "Leiden University"
}))
Alida_Kindt = crate.add(Person(crate, Alida_Kindt_id, properties={
"name": "Alida S.D. Kindt",
"affiliation": "Leiden University"
}))
Purva_Kulkarni = crate.add(Person(crate, Purva_Kulkarni_id, properties={
"name": "Purva Kulkarni",
"affiliation": "Radboud university medical center"
}))
Rene_Pool = crate.add(Person(crate, Rene_Pool_id, properties={
"name": "René Pool",
"affiliation": "Vrije Universiteit Amsterdam"
}))
Dorret_Boomsma = crate.add(Person(crate, Dorret_Boomsma_id, properties={
"name": "Dorret I. Boomsma",
"affiliation": "Vrije Universiteit Amsterdam"
}))
Jenny_van_Dongen = crate.add(Person(crate, Jenny_van_Dongen_id, properties={
"name": "Jenny van Dongen",
"affiliation": "Vrije Universiteit Amsterdam"
}))
Alain_van_Gool = crate.add(Person(crate, Alain_van_Gool_id, properties={
"name": "Alain J. van Gool",
"affiliation": "Radboud university medical center"
}))
PeterBram_t_Hoen = crate.add(Person(crate, PeterBram_t_Hoen_id, properties={
"name": "Peter A.C. 't Hoen",
"affiliation": "Radboud university medical center"
}))
</code>
<code>
# Define X-omics organization
# TODO: Can another url identifier be used here?
x_omics_id = "https://x-omics.nl/"
x_omics = crate.add(ContextEntity(crate, x_omics_id, properties={
"@type": "Organization",
"name": "The Netherlands X-omics intiative",
"url": "https://x-omics.nl/"
}))
</code>
## Docker containers
<code>
Docker_mofa2= crate.add(SoftwareApplication(crate, 'Xomics/mofa2', properties={
"@id" : 'Xomics/mofa2',
"identifier": "https://doi.org/10.5281/zenodo.10037590",
"version": "v0.6"
}))
Docker_miniconda_snf= crate.add(SoftwareApplication(crate, 'Xomics/miniconda-snf', properties={
"@id" : 'Xomics/miniconda-snf',
"identifier": "https://doi.org/10.5281/zenodo.10037582",
"version": "v0.7"
}))
Docker_rbase_analysis = crate.add(SoftwareApplication(crate, 'Xomics/r-base-analysis', properties={
"@id" : 'Xomics/r-base-analysis',
"identifier": "https://doi.org/10.5281/zenodo.10033979",
"version": "v0.4"
}))
Docker_rbase_epigenomics = crate.add(SoftwareApplication(crate, 'Xomics/r-base-epigenomics-pre', properties={
"@id" : 'Xomics/r-base-epigenomics-pre',
"identifier": "https://doi.org/10.5281/zenodo.10037592",
"version": "v0.4"
}))
Docker_rbase_phenotypes = crate.add(SoftwareApplication(crate, 'Xomics/r-base-phenotypes', properties={
"@id" : 'Xomics/r-base-phenotypes',
"identifier": "https://doi.org/10.5281/zenodo.10037594",
"version": "v0.6"
}))
</code>
## ISA files
<code>
# Define ISA files
# Define data entity IDs
investigation_id = "Synthetic_data/i_investigation.txt"
study_id = "Synthetic_data/s_study.txt"
assay_epigenomics_id = "Synthetic_data/a_assay_methylation.txt"
assay_amines_id = "Synthetic_data/a_assay_metabolomics_amines.txt"
assay_oa_id = "Synthetic_data/a_assay_metabolomics_OA.txt"
assay_steroids_id = "Synthetic_data/a_assay_metabolomics_steroids.txt"
investigation = crate.add(File(crate, investigation_id, dest_path=investigation_id, properties={
"@id": investigation_id,
"name": "Investigation file",
"doi": 'https://doi.org/10.5281/zenodo.10040716',
"url": 'https://doi.org/10.5281/zenodo.10040716',
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C165216", #Experiment Metadata
"@id": "http://purl.obolibrary.org/obo/OBI_0000066"}, #investigation
"format": {"@id": "http://edamontology.org/format_3687"} #ISA-TAB
}))
study = crate.add(File(crate, study_id, dest_path=study_id, properties={
"@id": study_id,
"name": "Study file",
"doi": 'https://doi.org/10.5281/zenodo.10040716',
"url": 'https://doi.org/10.5281/zenodo.10040716',
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C165216", #Experiment Metadata
"@id": "http://purl.obolibrary.org/obo/NCIT_C63536"}, #study
"format": {"@id": "http://edamontology.org/format_3687"} #ISA-TAB
}))
assay_epigenomics = crate.add(File(crate, assay_epigenomics_id, dest_path=assay_epigenomics_id, properties={
"@id": assay_epigenomics_id,
"name": "Assay epigenomics file",
"doi": 'https://doi.org/10.5281/zenodo.10040716',
"url": 'https://doi.org/10.5281/zenodo.10040716',
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C165216", #Experiment Metadata
"@id": "http://purl.obolibrary.org/obo/OBI_0000070"}, #assay
"format": {"@id": "http://edamontology.org/format_3687"} #ISA-TAB
}))
assay_amines = crate.add(File(crate, assay_amines_id, dest_path=assay_amines_id, properties={
"@id": assay_amines_id,
"name": "Assay amines file",
"doi": 'https://doi.org/10.5281/zenodo.10040716',
"url": 'https://doi.org/10.5281/zenodo.10040716',
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C165216", #Experiment Metadata
"@id": "http://purl.obolibrary.org/obo/OBI_0000070"}, #assay
"format": {"@id": "http://edamontology.org/format_3687"} #ISA-TAB
}))
assay_oa = crate.add(File(crate, assay_oa_id, dest_path=assay_oa_id, properties={
"@id": assay_oa_id,
"name": "Assay organic acids file",
"doi": 'https://doi.org/10.5281/zenodo.10040716',
"url": 'https://doi.org/10.5281/zenodo.10040716',
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C165216", #Experiment Metadata
"@id": "http://purl.obolibrary.org/obo/OBI_0000070"}, #assay
"format": {"@id": "http://edamontology.org/format_3687"} #ISA-TAB
}))
assay_steroids = crate.add(File(crate, assay_steroids_id, dest_path=assay_steroids_id, properties={
"@id": assay_steroids_id,
"name": "Assay steroids file",
"doi": 'https://doi.org/10.5281/zenodo.10040716',
"url": 'https://doi.org/10.5281/zenodo.10040716',
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C165216", #Experiment Metadata
"@id": "http://purl.obolibrary.org/obo/OBI_0000070"}, #assay
"format": {"@id": "http://edamontology.org/format_3687"} #ISA-TAB
}))
</code>
## Sub-workflows with input/output files
### Analyze missing values
<code>
# Define entity IDs
missing_data_heatmap_id = "modules/heatmap_missingness.nf"
missing_data_script = "bin/heatmap_missingness.R"
# Heatmap missing data points
missing_data_heatmap = crate.add(ComputationalWorkflow(crate, missing_data_heatmap_id, dest_path=missing_data_heatmap_id, properties={
"@id": missing_data_heatmap_id,
"@type": ["File", "SoftwareSourceCode", "ComputationalWorkflow"],
"name": "Heatmap missingness",
"input": {"@id": "epigenomics_values",
"@id": "metabolomics_values",
"@id": "behavioral_data",
"@id": "phenotype_covariates"},
#"output": {} ,
"hasPart": [
{"@id": missing_data_script},
],
"license": "https://opensource.org/licenses/MIT",
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C142610", #Missing Data
"@id": "http://semanticscience.org/resource/SIO_000449"} #plot
}))
missing_data_script = crate.add(File(crate, missing_data_script, dest_path=missing_data_script, properties={
"@id": missing_data_script,
"@type": ["File", "SoftwareSourceCode"],
"name": "Heatmap NA values script",
"softwareRequirements": {"@id": "Xomics/r-base-analysis"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3999"}, #Rscript
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C142610", #Missing Data
"@id": "http://semanticscience.org/resource/SIO_000449"} #plot
}))
</code>
### Epigenomics pre-processing
<code>
# Define entity IDs
epi_preprocessing_id = "modules/epigenetics_preprocessing.nf"
epi_annotation_script_id = "bin/epigenomics_annotation.R"
epi_filtering_script_id = "bin/epigenomics_filtering.R"
epi_imputation_script_id = "bin/epigenomics_imputation.R"
epi_covariates_correction_script_id = "bin/CovariateCorrection.R"
epi_subset_features_script_id = "bin/sort_cols_sd.R"
epi_scaling_script_id = "bin/epigenomics_scaling.R"
# Define epigenomics pre-processing sub-workflow
epi_preprocessing = crate.add(ComputationalWorkflow(crate, epi_preprocessing_id, dest_path=epi_preprocessing_id, properties={
"@id": epi_preprocessing_id,
"@type": ["File", "SoftwareSourceCode", "ComputationalWorkflow"],
"name": "Epigenetics preprocessing",
#"input": {"@id": "epigenomics_values"}, #Add later
"output": {"@id": "epigenomics_preprocessed_data"},
"hasPart": [
{"@id": epi_annotation_script_id},
{"@id": epi_filtering_script_id},
{"@id": epi_imputation_script_id},
{"@id": epi_covariates_correction_script_id},
{"@id": epi_subset_features_script_id},
{"@id": epi_scaling_script_id}
],
"license": "https://opensource.org/licenses/MIT",
"additionalType": {"@id": "http://edamontology.org/operation_0226", #annotation
"@id": "http://purl.obolibrary.org/obo/MS_1001486", #filtering
"@id": "http://edamontology.org/operation_3557", #imputation
"@id": "http://purl.obolibrary.org/obo/OBI_0200185", #scaling
"@id": "http://semanticscience.org/resource/SIO_000594"} #data transformation
}))
# Scripts
epi_annotation_script = crate.add(File(crate, epi_annotation_script_id, dest_path=epi_annotation_script_id, properties={
"@id": epi_annotation_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "Epigenomics annotation script",
"softwareRequirements": {"@id": "Xomics/r-base-epigenomics-pre"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3999"}, #Rscript
"additionalType": {"@id": "http://edamontology.org/operation_0226"} #annotation
}))
epi_filtering_script = crate.add(File(crate, epi_filtering_script_id, dest_path=epi_filtering_script_id, properties={
"@id": epi_filtering_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "Epigenomics filtering script",
"softwareRequirements": {"@id": "Xomics/r-base-analysis"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3999"}, #Rscript
"additionalType": {"@id": "http://purl.obolibrary.org/obo/MS_1001486"} #filtering
}))
epi_imputation_script = crate.add(File(crate, epi_imputation_script_id, dest_path=epi_imputation_script_id, properties={
"@id": epi_imputation_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "Epigenomics imputation script",
"softwareRequirements": {"@id": "Xomics/r-base-analysis"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3999"}, #Rscript
"additionalType": {"@id": "http://edamontology.org/operation_3557"} #imputation
}))
epi_covariates_correction_script = crate.add(File(crate, epi_covariates_correction_script_id, dest_path=epi_covariates_correction_script_id, properties={
"@id": epi_covariates_correction_script_id,
"@type": ["File", "SoftwareSourceCode"],
"softwareRequirements": {"@id": "Xomics/r-base-analysis"},
"name": "Epigenomics covariates correction script",
"programmingLanguage": {"@id": "http://edamontology.org/format_3999"}, #Rscript
"additionalType": {"@id": "http://semanticscience.org/resource/SIO_000594"} #data transformation #TODO find more specific term
}))
epi_subset_features_script = crate.add(File(crate, epi_subset_features_script_id, dest_path=epi_subset_features_script_id, properties={
"@id": epi_subset_features_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "Epigenomics subset features script",
"softwareRequirements": {"@id": "Xomics/r-base-analysis"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3999"}, #Rscript
"additionalType": {"@id": "http://semanticscience.org/resource/SIO_000594"} #data transformation #TODO find more specific term
}))
epi_scaling_script = crate.add(File(crate, epi_scaling_script_id, dest_path=epi_scaling_script_id, properties={
"@id": epi_scaling_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "Epigenomics scaling script",
"softwareRequirements": {"@id": "Xomics/r-base-analysis"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3999"}, #Rscript
"additionalType": {"@id": "http://purl.obolibrary.org/obo/OBI_0200185"} #scaling
}))
</code>
<code>
# Define epigenomics data entities
# Define data entity IDs
epigenomics_data_id = "Synthetic_data/synthetic_epigenomics.csv"
epigenomics_meta_id = "Synthetic_data/synthetic_epigenomics_meta.csv"
epigenomics_data = crate.add(File(crate, epigenomics_data_id, dest_path=epigenomics_data_id, properties={
"@id": epigenomics_data_id,
"@type": "FormalParameter",
"name": "epigenomics_data",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C153195"}, #epigenome
"format": {"@id": "http://edamontology.org/format_3752"} #csv
}))
epigenomics_meta = crate.add(File(crate, epigenomics_meta_id, dest_path=epigenomics_meta_id, properties={
"@id": epigenomics_meta_id,
"@type": "FormalParameter",
"name": "epigenomics_data",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C52095"}, #metadata
"format": {"@id": "http://edamontology.org/format_3752"} #csv
}))
epigenomics_preprocessed = crate.add(DataEntity(crate, "epigenomics_preprocessed", properties={
"@id": "epigenomics_preprocessed_data",
"@type": "FormalParameter",
"name": "epigenomics_preprocessed",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C153195", #epigenome
"@id": "http://www.ebi.ac.uk/efo/EFO_0004096" #processed array data file
},
"format": {"@id": "http://edamontology.org/format_3752"} #csv
}))
epi_preprocessing["input"] = [epigenomics_data_id, epigenomics_meta_id]
</code>
## Check if Methylation EPIC file is present, if not download from url
<code>
import requests, os.path
import zipfile
from os import path
from io import BytesIO
url_methylationEPIC = 'https://webdata.illumina.com/downloads/productfiles/methylationEPIC/infinium-methylationepic-v-1-0-b4-manifest-file-csv.zip'
if path.exists("EPIC_annotation/raw/MethylationEPIC_v-1-0_B4.csv") is False:
req = requests.get(url_methylationEPIC)
# Writing the file to the local file system
zipfile= zipfile.ZipFile(BytesIO(req.content))
zipfile.extractall('EPIC_annotation/raw/')
</code>
<code>
# EPIC annotation files used for epigenomics data
# Define data entity IDs
EPIC_MOESM1_id = "EPIC_annotation/raw/13059_2016_1066_MOESM1_ESM.csv"
EPIC_MOESM4_id = "EPIC_annotation/raw/13059_2016_1066_MOESM4_ESM.csv"
EPIC_MOESM5_id = "EPIC_annotation/raw/13059_2016_1066_MOESM5_ESM.csv"
Methylation_EPIC_id = "EPIC_annotation/raw/MethylationEPIC_v-1-0_B4.csv"
Annotation_EPIC_id = "EPIC_annotation/anno_epic_072017.RData"
Epic_MOESM1 = crate.add(File(crate, EPIC_MOESM1_id, dest_path=EPIC_MOESM1_id, properties={
"@id": EPIC_MOESM1_id,
"@type": "FormalParameter",
"name": "epigenomics_preprocessed",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C43523" #probe #TODO: find better term(s)
},
"format": {"@id": "http://edamontology.org/format_3752"} #csv
}))
Epic_MOESM4 = crate.add(File(crate, EPIC_MOESM4_id, dest_path=EPIC_MOESM4_id, properties={
"@id": EPIC_MOESM4_id,
"@type": "FormalParameter",
"name": "epigenomics_preprocessed",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C43523" #probe #TODO: find better term(s)
},
"format": {"@id": "http://edamontology.org/format_3752"} #csv
}))
Epic_MOESM5 = crate.add(File(crate, EPIC_MOESM5_id, dest_path=EPIC_MOESM5_id, properties={
"@id": EPIC_MOESM5_id,
"@type": "FormalParameter",
"name": "epigenomics_preprocessed",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C43523" #probe #TODO: find better term(s)
},
"format": {"@id": "http://edamontology.org/format_3752"} #csv
}))
Methylation_EPIC = crate.add(File(crate, Methylation_EPIC_id, dest_path=Methylation_EPIC_id, properties={
"@id": Methylation_EPIC_id,
"@type": "FormalParameter",
"name": "epigenomics_preprocessed",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/OBI_0002131" #Illumina Infinium MethylationEPIC BeadChip
#TODO: find better term(s)
},
"format": {"@id": "http://edamontology.org/format_3752"} #csv
}))
Annotation_EPIC = crate.add(File(crate, Annotation_EPIC_id, dest_path=Annotation_EPIC_id, properties={
"@id": Annotation_EPIC_id,
"@type": "FormalParameter",
"name": "epigenomics_preprocessed",
#"valueRequired": true,
"additionalType": { "@id": "http://edamontology.org/operation_0226" #annotation
},
#"format": #TODO add alternative to .RData
}))
epi_annotation_script["hasPart"] = [EPIC_MOESM1_id, EPIC_MOESM4_id, EPIC_MOESM5_id, Methylation_EPIC_id, Annotation_EPIC_id]
</code>
### Metabolomics pre-processing
<code>
# Define data entity IDs
mtblmcs_preprocessing_id = "modules/metabolomics_preprocessing.nf"
mtblmcs_filtering_script_id = "bin/metabolomics_filter.Rmd"
mtblmcs_normalization_script_id = "bin/metabolomics_normalization.R"
mtblmcs_scaling_script_id = "bin/metabolomics_scaling.R"
mtblmcs_concatenate_script_id = "bin/concatenate_MAF.R"
# Define metabolomics pre-processing sub-workflow
mtblmcs_preprocessing = crate.add(ComputationalWorkflow(crate, mtblmcs_preprocessing_id, dest_path=mtblmcs_preprocessing_id, properties={
"@id": mtblmcs_preprocessing_id,
"@type": ["File", "SoftwareSourceCode", "ComputationalWorkflow"],
"name": "Metabolomics preprocessing",
#"input": #Add later
"output": {"@id": "metabolomics_preprocessed_data"},
"hasPart": [
{"@id": mtblmcs_filtering_script_id},
{"@id": mtblmcs_normalization_script_id},
{"@id": mtblmcs_scaling_script_id},
{"@id": mtblmcs_concatenate_script_id}
],
"license": "https://opensource.org/licenses/MIT",
"additionalType": {"@id": "http://purl.obolibrary.org/obo/MS_1001486", #filtering
"@id": "http://purl.obolibrary.org/obo/OBI_0200169" #normalization
}
}))
# Define scripts
mtblmcs_filtering_script = crate.add(File(crate, mtblmcs_filtering_script_id, dest_path=mtblmcs_filtering_script_id, properties={
"@id": mtblmcs_filtering_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "Metabolomics filtering script",
"softwareRequirements": {"@id": "Xomics/r-base-phenotypes"},
"programmingLanguage": {"@id": "http://edamontology.org/format_4000"}, #R markdown
"additionalType": {"@id": "http://purl.obolibrary.org/obo/MS_1001486"} #filtering
}))
mtblmcs_normalization_script = crate.add(File(crate, mtblmcs_normalization_script_id, dest_path=mtblmcs_normalization_script_id, properties={
"@id": mtblmcs_normalization_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "Metabolomics normalization script",
"softwareRequirements": {"@id": "Xomics/r-base-analysis"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3999"}, #Rscript
"additionalType": {"@id": "http://purl.obolibrary.org/obo/OBI_0200169"} #normalization data transformation
}))
mtblmcs_scaling_script = crate.add(File(crate, mtblmcs_scaling_script_id, dest_path=mtblmcs_scaling_script_id, properties={
"@id": mtblmcs_scaling_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "Metabolomics scaling script",
"softwareRequirements": {"@id": "Xomics/r-base-analysis"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3999"}, #Rscript
"additionalType": {"@id": "http://purl.obolibrary.org/obo/OBI_0200037"} #pareto scaling
}))
mtblmcs_concatenate_script = crate.add(File(crate, mtblmcs_concatenate_script_id, dest_path=mtblmcs_concatenate_script_id, properties={
"@id": mtblmcs_concatenate_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "Concatenate MAFs script",
"softwareRequirements": {"@id": "Xomics/r-base-analysis"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3999"}, #Rscript
"additionalType": {"@id": "http://purl.obolibrary.org/obo/OBI_0002566"} #file merge
}))
</code>
<code>
#Define metabolomics data entities
metabolomics_data_id = "Synthetic_data/synthetic_metabolomics.csv"
amines_data_id = "Synthetic_data/amines_MAF.tsv"
OA_data_id = "Synthetic_data/OA_MAF.tsv"
steroids_data_id = "Synthetic_data/steroids_MAF.tsv"
metabolomics_data = crate.add(File(crate, metabolomics_data_id, dest_path=metabolomics_data_id, properties={
"@id": metabolomics_data_id,
"@type": "FormalParameter",
"name": "metabolomics_data",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/MS_1003084", #processed data file (not raw)
"@id": "http://purl.obolibrary.org/obo/CHEBI_32952", #amine
"@id": "http://purl.obolibrary.org/obo/CHEBI_64709", #organic acid
"@id": "http://purl.obolibrary.org/obo/CHEBI_35341", #steroid
},
"format": {"@id": "http://purl.obolibrary.org/obo/MS_1000914"} #tsv
}))
amines_data = crate.add(File(crate, amines_data_id, dest_path=amines_data_id, properties={
"@id": amines_data_id,
"@type": "FormalParameter",
"name": "metabolomics_data",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/MS_1003084", #processed data file (not raw)
"@id": "http://purl.obolibrary.org/obo/CHEBI_32952", #amine
},
"format": {"@id": "http://purl.obolibrary.org/obo/MS_1000914"} #tsv
}))
OA_data = crate.add(File(crate, OA_data_id, dest_path=OA_data_id, properties={
"@id": OA_data_id,
"@type": "FormalParameter",
"name": "metabolomics_data",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/MS_1003084", #processed data file (not raw)
"@id": "http://purl.obolibrary.org/obo/CHEBI_64709", #organic acid
},
"format": {"@id": "http://purl.obolibrary.org/obo/MS_1000914"} #tsv
}))
steroids_data = crate.add(File(crate, steroids_data_id, dest_path=steroids_data_id, properties={
"@id": steroids_data_id,
"@type": "FormalParameter",
"name": "metabolomics_data",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/MS_1003084", #processed data file (not raw)
"@id": "http://purl.obolibrary.org/obo/CHEBI_35341", #steroid
},
"format": {"@id": "http://purl.obolibrary.org/obo/MS_1000914"} #tsv
}))
metabolomics_preprocessed = DataEntity(crate, "metabolomics_preprocessed", properties={
"@id": "metabolomics_preprocessed_data",
"@type": "FormalParameter",
"name": "metabolomics_preprocessed",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/MS_1003084", #processed data file (not raw),
"@id": "http://purl.obolibrary.org/obo/OBI_0000451" #normalized data set
},
"format": {"@id": "http://purl.obolibrary.org/obo/MS_1000914"} #tsv
})
mtblmcs_preprocessing["input"] = [metabolomics_data_id, amines_data_id, OA_data_id, steroids_data_id]
</code>
### Phenotypes preparation
<code>
# Define data entity IDs
cbcl_imputation_mca_id = "modules/CBCL_MCA.nf"
cbcl_imputation_mca_script_id = "bin/CBCL_filter_impute_MCA.Rmd"
# Define behavioral data pre-processing sub-workflow
cbcl_imputation_mca = crate.add(ComputationalWorkflow(crate, cbcl_imputation_mca_id, dest_path=cbcl_imputation_mca_id, properties={
"@id": cbcl_imputation_mca_id,
"@type": ["File", "SoftwareSourceCode", "ComputationalWorkflow"],
"name": "CBCL imputation",
"input": {"@id": "behavioral_data"},
"output": {"@id": "behavioral_data"},
"hasPart": [
{"@id": cbcl_imputation_mca_script_id }
],
"license": "https://opensource.org/licenses/MIT",
"additionalType": {"@id": "http://edamontology.org/operation_3557", #imputation
"@id": "http://purl.enanomapper.org/onto/ENM_8000003", #Unsupervised learning
}
}))
# Define script
cbcl_imputation_mca_script = crate.add(File(crate, cbcl_imputation_mca_script_id, dest_path=cbcl_imputation_mca_script_id , properties={
"@id": cbcl_imputation_mca_script_id ,
"@type": ["File", "SoftwareSourceCode"],
"name": "Filter, impute CBCL data and MCA",
"softwareRequirements": {"@id": "Xomics/r-base-phenotypes"},
"programmingLanguage": {"@id": "http://edamontology.org/format_4000"}, #R markdown
"additionalType": {"@id": "http://edamontology.org/operation_3557", #imputation
"@id": "http://purl.enanomapper.org/onto/ENM_8000003", #Unsupervised learning
}
}))
</code>
<code>
#Define phenotypic data entities
phenotype_covariates_id = "Synthetic_data/synthetic_phenotype_covariates_data.csv"
behavioral_data_id = "Synthetic_data/synthetic_cbcl_data.csv"
phenotype_covariates = crate.add(File(crate, phenotype_covariates_id, dest_path=phenotype_covariates_id, properties={
"@id": phenotype_covariates_id,
"@type": "FormalParameter",
"name": "phenotype_covariates",
#"valueRequired": true,
"additionalType": { "@id": "http://purl.obolibrary.org/obo/NCIT_C16977" #Phenotype
},
"format": {"@id": "http://edamontology.org/format_3752"} #csv
}))
behavioral_data = crate.add(File(crate, behavioral_data_id, dest_path=behavioral_data_id, properties={
"@id": behavioral_data_id,
"@type": "FormalParameter",
"name": "behavioral_data",
#"valueRequired": true,
"additionalType": { "@id": "http://www.ebi.ac.uk/efo/EFO_0005661" #CBCL assessment
},
"format": {"@id": "http://edamontology.org/format_3752"} #csv
}))
</code>
### Map omics files
<code>
# Define data entity IDs
id_mapping_id = "modules/map_IDs.nf"
id_file_id = "Synthetic_data/ACTIONdemonstrator_XOmics_IDs_synthetic.csv"
id_mapping_script_id = "bin/map_IDs.py"
id_file = crate.add(File(crate, id_file_id, dest_path=id_file_id, properties={
"@id": id_file_id,
"@type": "FormalParameter",
"name": "behavioral_data",
#"valueRequired": true,
"additionalType": { "@id": "http://purl.enanomapper.org/onto/ENM_9000071" #sample identifier
},
"format": {"@id": "http://edamontology.org/format_3752"} #csv
}))
# Define module of sample identifier mapping
id_mapping = crate.add(ComputationalWorkflow(crate, id_mapping_id, dest_path=id_mapping_id, properties={
"@id": id_mapping_id,
"@type": ["File", "SoftwareSourceCode", "ComputationalWorkflow"],
"name": "Sample ID mapping",
"input": {"@id": "metabolomics_preprocessed_data",
"@id": "epigenomics_preprocessed_data",
"@id": id_file_id},
"output": {"@id": "metabolomics_preprocessed_data",
"@id": "epigenomics_preprocessed_data"},
"hasPart": [
{"@id": id_mapping_script_id}
],
"license": "https://opensource.org/licenses/MIT",
"additionalType": {"@id": "http://edamontology.org/operation_3282"} #ID mapping
}))
# ID mapping script
id_mapping_script = crate.add(File(crate, id_mapping_script_id, dest_path=id_mapping_script_id, properties={
"@id": id_mapping_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "ID mapping script",
"softwareRequirements": {"@id": "Xomics/miniconda-snf"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3996"}, #Python script
"additionalType": {"@id": "http://edamontology.org/operation_3282"} #ID mapping
}))
</code>
### Principal Component Analysis
<code>
# Define data entity IDs
pca_id = "modules/pca.nf"
pca_script_id = "bin/pca.R"
# Define pca sub-workflow
pca = crate.add(ComputationalWorkflow(crate, pca_id, dest_path=pca_id, properties={
"@id": pca_id,
"@type": ["File", "SoftwareSourceCode", "ComputationalWorkflow"],
"name": "Principal Component Analysis",
"input": {"@id": "processed_omics_data"},
"output": {"@id": "pca_report"},
"hasPart": [
{"@id": pca_script_id}
],
"license": "https://opensource.org/licenses/MIT",
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C49291" #PCA
}
}))
# PCA script
pca_script = crate.add(File(crate, pca_script_id, dest_path=pca_script_id, properties={
"@id": pca_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "PCA script",
"softwareRequirements": {"@id": "Xomics/r-base-analysis"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3999"}, #Rscript
"additionalType": {"@id": "http://purl.obolibrary.org/obo/NCIT_C49291"} #PCA
}))
</code>
<code>
#Define PCA input/output entities
processed_omics_data = DataEntity(crate, "omics_data.csv", properties={
"@id": "processed_omics_data",
"@type": "FormalParameter",
"name": "processed_omics_data",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/MS_1003084", #processed data file
"@id": "http://edamontology.org/topic_3391" #omics
},
"format": {"@id": "http://edamontology.org/format_3752"} #csv
})
#TODO What should be output here? .pdf file of plots?
pca_report = DataEntity(crate, "pca.pdf", properties={
"@id": "pca_report",
"@type": "FormalParameter",
"name": "pca_report",
#"valueRequired": true,
"additionalType": {"@id": "http://edamontology.org/data_2884" #plot (multiple)
},
"format": {"@id": "http://edamontology.org/format_3508"} #pdf
})
</code>
### Similarity Network Fusion
<code>
# Define data entity IDs
snf_id = "modules/snf.nf"
snf_script_id = "bin/perform_snf.py"
snf_analysis_script_id = "bin/snf_analysis.ipynb"
snf_gee_script_id = "bin/snf_gee_analysis.Rmd"
# Define SNF sub-workflow
snf = crate.add(ComputationalWorkflow(crate, snf_id, dest_path=snf_id, properties={
"@id": snf_id,
"@type": ["File", "SoftwareSourceCode", "ComputationalWorkflow"],
"name": "Similarity Network Fusion",
"input": {"@id": "processed_omics_data"},
"output": {"@id": "snf_report"},
"hasPart": [
{"@id": snf_script_id},
{"@id": snf_analysis_script_id},
{"@id": snf_gee_script_id}
],
"license": "https://opensource.org/licenses/MIT",
"additionalType": {"@id": "http://edamontology.org/operation_3432", #Clustering
"@id": "http://purl.enanomapper.org/onto/ENM_8000003", #Unsupervised learning
}
}))
# SNF scripts
snf_script = crate.add(File(crate, snf_script_id, dest_path=snf_script_id, properties={
"@id": snf_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "SNF script",
"softwareRequirements": {"@id": "Xomics/miniconda-snf"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3996"}, #Python script
"additionalType": {"@id": "http://purl.enanomapper.org/onto/ENM_8000003", #Unsupervised learning
"@id": "http://edamontology.org/operation_3432", #Clustering
}
}))
snf_analysis_script = crate.add(File(crate, snf_analysis_script_id, dest_path=snf_analysis_script_id, properties={
"@id": snf_analysis_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "SNF downstream analysis script",
"softwareRequirements": {"@id": "Xomics/miniconda-snf"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3996"}, #Python script
"additionalType": {"@id": "http://purl.enanomapper.org/onto/ENM_8000003", #Unsupervised learning
"@id": "http://edamontology.org/operation_3432", #Clustering
"@id": "http://semanticscience.org/resource/SIO_000449" #plot
}
}))
snf_gee_script = crate.add(File(crate, snf_gee_script_id, dest_path=snf_gee_script_id, properties={
"@id": snf_gee_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "SNF GEE models",
"softwareRequirements": {"@id": "Xomics/mofa2"},
"programmingLanguage": {"@id": "http://edamontology.org/format_4000"}, #R markdown
"additionalType": {"@id": "http://purl.enanomapper.org/onto/ENM_8000003" #Unsupervised learning
}
}))
</code>
<code>
# Define SNF input/output entities
snf_report = crate.add(DataEntity(crate, "snf_report", properties={
"@id": "snf_report",
"@type": "FormalParameter",
"name": "snf_report",
#"valueRequired": true,
"additionalType": {"@id": "http://semanticscience.org/resource/SIO_000449" #plot
},
"format": {"@id": "http://edamontology.org/format_3508"} #pdf
}))
#TODO add snf matrix
</code>
### Multi-Omics Factor Analysis
<code>
# Define data entity IDs
mofa_id = "modules/mofa.nf"
mofa_script_id = "bin/mofa.R"
mofa_analysis_script_id = "bin/MOFA_downstream_analysis_report.Rmd"
mofa_gee_script_id = "bin/MOFA_downstream_analysis_report_gee.Rmd"
# Define MOFA module
mofa = crate.add(ComputationalWorkflow(crate, mofa_id, dest_path=mofa_id, properties={
"@id": mofa_id,
"@type": ["File", "SoftwareSourceCode", "ComputationalWorkflow"],
"name": "Multi-Omics Factor Analysis",
"programmingLanguage": {"@id": "https://w3id.org/workflowhub/workflow-ro-crate#nextflow"},
"input": {"@id": "processed_omics_data"},
"output": {"@id": "mofa_model"},
"hasPart": [
{"@id": mofa_script_id},
{"@id": mofa_analysis_script_id},
{"@id": mofa_gee_script_id}
],
"license": "https://opensource.org/licenses/MIT",
"additionalType": {"@id": "http://purl.enanomapper.org/onto/ENM_8000003", #Unsupervised learning
"@id": "http://edamontology.org/topic_3474" #Machine learning
}
}))
# MOFA script
mofa_script = crate.add(File(crate, mofa_script_id, dest_path=mofa_script_id, properties={
"@id": mofa_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "MOFA script",
"softwareRequirements": {"@id": "Xomics/mofa2"},
"programmingLanguage": {"@id": "http://edamontology.org/format_3999"}, #Rscript
"additionalType": {"@id": "http://purl.enanomapper.org/onto/ENM_8000003", #Unsupervised learning
"@id": "http://edamontology.org/topic_3474" #Machine learning
}
}))
# MOFA script
mofa_analysis_script = crate.add(File(crate, mofa_analysis_script_id, dest_path=mofa_analysis_script_id, properties={
"@id": mofa_analysis_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "MOFA downstream analysis script",
"softwareRequirements": {"@id": "Xomics/mofa2"},
"programmingLanguage": {"@id": "http://edamontology.org/format_4000"}, #R markdown
"additionalType": {"@id": "http://purl.enanomapper.org/onto/ENM_8000003", #Unsupervised learning
"@id": "http://edamontology.org/topic_3474" #Machine learning
}
}))
# MOFA GEE script
mofa_analysis_gee_script = crate.add(File(crate, mofa_gee_script_id, dest_path=mofa_gee_script_id, properties={
"@id": mofa_gee_script_id,
"@type": ["File", "SoftwareSourceCode"],
"name": "MOFA downstream analysis script with GEE",
"softwareRequirements": {"@id": "Xomics/mofa2"},
"programmingLanguage": {"@id": "http://edamontology.org/format_4000"}, #R markdown
"additionalType": {"@id": "http://purl.enanomapper.org/onto/ENM_8000003", #Unsupervised learning
"@id": "http://edamontology.org/topic_3474" #Machine learning
}
}))
</code>
<code>
# Define MOFA output mode
mofa_model = crate.add(DataEntity(crate, "mofa_model", properties={
"@id": "mofa_model",
"@type": "FormalParameter",
"name": "mofa_model",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/STATO_0000107" #statistical model
},
"format": {"@id": "http://edamontology.org/format_3590"} #HDF5
}))
</code>
## Data entities on Root directory
<code>
from rocrate.model.data_entity import DataEntity
nextflow_config = crate.add_file( "nextflow.config", properties={
"@id": "nextflow.config",
"@type": "FormalParameter",
"name": "Nextflow configuration file",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/ONTOAVIDA_00000001" #configuration file
},
"format": {"@id": "http://edamontology.org/format_3464"} #JSON
})
dre_config = crate.add_file( "dre.config", properties={
"@id": "dre.config",
"@type": "FormalParameter",
"name": "Nextflow configuration file",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/ONTOAVIDA_00000001" #configuration file
},
"format": {"@id": "http://edamontology.org/format_3464"} #JSON
})
readme = crate.add_file("README.md", properties={
"@id": "README.md",
"@type": "FormalParameter",
"name": "Nextflow configuration file",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/OMIT_00055391" #Documentation
},
#"format": {} #TODO: markdown ontology term needed here.
})
action_documentation = crate.add_file("Documentation.md", properties={
"@id": "ACTION_documentation.md",
"@type": "FormalParameter",
"name": "Documentation on Workflow",
#"valueRequired": true,
"additionalType": {"@id": "http://purl.obolibrary.org/obo/OMIT_00055391" #Documentation
},
#"format": {} #TODO: markdown ontology term needed here.
})
# Define the diagram that provides an overview of the main workflow
diagram = crate.add_file("flowchart.png", properties={
"@id": "flowchart.png",
"@type": ["File", "ImageObject"],
"name": "Workflow overview" ,
"about": {"@id": "action.nf"}
})
</code>
## Add entities to the main workflow entity
<code>
# Add entities/attributes to the workflow
MainWorkflow["author"] = [Anna_Niehues, Casper_de_Visser, Fiona_Hagenbeek, Naama_Karu, Alida_Kindt, Purva_Kulkarni, Rene_Pool, Dorret_Boomsma, Jenny_van_Dongen, Alain_van_Gool, PeterBram_t_Hoen]
MainWorkflow["programmingLanguage"] = Nextflow
MainWorkflow["image"] = diagram
MainWorkflow["config"] = [nextflow_config, dre_config]
MainWorkflow["sdPublisher"] = x_omics
MainWorkflow["hasPart"] = [missing_data_heatmap, epi_preprocessing, mtblmcs_preprocessing, cbcl_imputation_mca, id_mapping, pca, snf, mofa]
</code>
## Add publications
<code>
# Rio Journal abstract
RIO_abstract = crate.add(ContextEntity(crate, "https://doi.org/10.3897/rio.8.e94042", properties={
"@id": "https://doi.org/10.3897/rio.8.e94042",
"@type": ["ScholartlyArtcile", "CreativeWork"],
"name": "A Multi-omics Data Analysis Workflow Packaged as a FAIR Digital Object",
"dateCreated": "25-08-2022",
"keywords": ["Multi-omics", "Metabolomics", "Epigenomics", "Behavioral data", "FAIR"],
}))
</code>
<code>
# Add information / link entitites to the crate
crate.mainEntity = MainWorkflow
crate.name = "X-omics ACTIONdemonstrator analysis workflow"
crate.author = [Anna_Niehues, Casper_de_Visser, Fiona_Hagenbeek, Naama_Karu, Alida_Kindt, Purva_Kulkarni, Rene_Pool, Dorret_Boomsma, Jenny_van_Dongen, Alain_van_Gool, PeterBram_t_Hoen]
crate.license = "https://opensource.org/licenses/MIT"
crate.keywords = ["Multi-omics", "Metabolomics", "Epigenomics", "Behavioral data", "FAIR"]
crate.datePublished = str(today)
crate.description = "This workflow is designed to analyze to a multi-omics data set that comprises genome-wide DNA methylation profiles, targeted metabolomics, and behavioral data of two cohorts that participated in the ACTION Biomarker Study (ACTION, Aggression in Children: Unraveling gene-environment interplay to inform Treatment and InterventiON strategies. (Boomsma 2015, Bartels 2018, Hagenbeek 2020, van Dongen 2021, Hagenbeek 2022). The ACTION-NTR cohort consists of twins that are either longitudinally concordant or discordant for childhood aggression. The ACTION-Curium-LUMC cohort consists of children referred to the Dutch LUMC Curium academic center for child and youth psychiatry. With the joint analysis of multi-omics data and behavioral data, we aim to identify substructures in the ACTION-NTR cohort and link them to aggressive behavior. First, the individuals are clustered using Similarity Network Fusion (SNF, Wang 2014), and latent feature dimensions are uncovered using different unsupervised methods including Multi-Omics Factor Analysis (MOFA) (Argelaguet 2018) and Multiple Correspondence Analysis (MCA, Lê 2008, Husson 2017). In a second step, we determine correlations between -omics and phenotype dimensions, and use them to explain the subgroups of individuals from the ACTION-NTR cohort. In order to validate the results, we project data of the ACTION-Curium-LUMC cohort onto the latent dimensions and determine if correlations between omics and phenotype data can be reproduced."
</code>
<code>
# Save to JSON-LD
#crate.write("exp_crate")
</code>
<code>
# Save to ZIP-file
crate.write_zip("ro-crate/exp_crate.zip")
</code>
|
{
"filename": "workflow_generate_ro-crate.ipynb",
"repository": "Xomics/ACTIONdemonstrator",
"query": "transformed_from_existing",
"size": 67473,
"sha": ""
}
|
# examples_notebook.ipynb
Repository: simonwm/tacco
# Slide-Seq Mouse Colon
This example uses TACCO to annotate and analyse mouse colon Slide-Seq data with mouse colon scRNA-seq data as reference (Avraham-Davidi et al.).
(Avraham-Davidi et al.): Avraham-Davidi I, Mages S, Klughammer J, et al. Integrative single cell and spatial transcriptomics of colorectal cancer reveals multicellular functional units that support tumor progression. doi: https://doi.org/10.1101/2022.10.02.508492
<code>
import os
import sys
import matplotlib
import pandas as pd
import numpy as np
import anndata as ad
import tacco as tc
# The notebook expects to be executed either in the workflow directory or in the repository root folder...
sys.path.insert(1, os.path.abspath('workflow' if os.path.exists('workflow/common_code.py') else '..'))
import common_code
</code>
## Load data
<code>
data_path = common_code.find_path('results/slideseq_mouse_colon/data')
plot_path = common_code.find_path('results/slideseq_mouse_colon')
reference = ad.read(f'{data_path}/scrnaseq.h5ad')
puck = ad.read(f'{data_path}/slideseq.h5ad')
</code>
## Plotting options
<code>
highres = False
default_dpi = 100.0
if highres:
matplotlib.rcParams['figure.dpi'] = 648.0
hr_ext = '_hd'
else:
matplotlib.rcParams['figure.dpi'] = default_dpi
hr_ext = ''
axsize = np.array([4,3])*0.5
labels_colors = pd.Series({'Epi': (0.00784313725490196, 0.24313725490196078, 1.0), 'B': (0.10196078431372549, 0.788235294117647, 0.2196078431372549), 'TNK': (1.0, 0.48627450980392156, 0.0), 'Mono': (0.5490196078431373, 0.03137254901960784, 0.0), 'Mac': (0.9098039215686274, 0.0, 0.043137254901960784), 'Gran': (0.34901960784313724, 0.11764705882352941, 0.44313725490196076), 'Mast': (0.23529411764705882, 0.23529411764705882, 0.23529411764705882), 'Endo': (0.8549019607843137, 0.5450980392156862, 0.7647058823529411), 'Fibro': (0.6235294117647059, 0.2823529411764706, 0.0)})
region_colors = tc.pl.get_default_colors([f'region_{i}' for i in range(4)], offset=17)
split_names = np.array([f'sub_{i}' for i in range(4)])
split_colors = tc.pl.get_default_colors(split_names, offset=12)
</code>
## Visualize scRNA-seq data
Create UMAPs for the scRNA-seq data
<code>
ref_umap = tc.utils.umap_single_cell_data(reference)
fig = tc.pl.scatter(ref_umap, keys='labels', position_key='X_umap', colors=labels_colors, joint=True, point_size=5, axsize=axsize, noticks=True,
axes_labels=['UMAP 0','UMAP 1']);
</code>
## Annotate the spatial data with compositions of cell types
Annotation is done on cell type level with multi_center=10 to capture variation within a cell type
<code>
tc.tl.annotate(puck,reference,'labels',result_key='labels',multi_center=10,);
</code>
## Visualize the spatial cell type distribution
<code>
puck = puck[tc.sum(puck.X,axis=1)>=50].copy() # restrict downstream analysis to "good" beads
fig = tc.pl.scatter(puck, keys='labels', position_key=['x','y'], colors=labels_colors, joint=True, point_size=1, axsize=axsize, noticks=True, axes_labels=['X','Y']);
</code>
## Find spatially contiguous regions of comparable expression patterns
<code>
tc.tl.find_regions(puck,key_added='regions',position_weight=1, resolution=0.55);
puck.obs['regions'] = puck.obs['regions'].map(lambda x: f'region_{x}').astype('category')
</code>
<code>
# ensure that the region naming is deterministic
ordered_regions = puck.obs.groupby('regions')['x'].mean().sort_values()
puck.obs['regions'] = puck.obs['regions'].map({r0:r1 for r0,r1 in zip(ordered_regions.index,['region_2','region_1','region_3','region_0'])}).astype(pd.CategoricalDtype(['region_0','region_1','region_2','region_3'],ordered=True))
</code>
<code>
fig = tc.pl.scatter(puck,'regions',joint=True,axsize=axsize, point_size=1, noticks=True, axes_labels=['X','Y'], colors=region_colors);
</code>
## Get regularized distances from these regions
<code>
tc.tl.annotation_coordinate(puck,annotation_key='regions',result_key='region_dist',max_distance=500,delta_distance=20,sparse=False);
</code>
<code>
fig,axs=tc.pl.subplots(2,2,axsize=axsize,x_padding=0.5,y_padding=0.5)
axs=axs.flatten()[:,None]
fig = tc.pl.scatter(puck,'region_dist',cmap='jet', joint=False,axsize=axsize, point_size=1, noticks=True, axes_labels=['X','Y'], ax=axs);
for i in [-4,-2,-1]:
fig.axes[i].remove()
</code>
Cell type composition at a certain regularized distance
<code>
fig = tc.pl.annotation_coordinate(puck,annotation_key='labels',coordinate_key=('region_dist','region_2'),colors=labels_colors,max_coordinate=500,delta_coordinate=20, axsize=(3,0.45));
</code>
## Cell type composition in the regions
<code>
fig = tc.pl.compositions(puck, 'labels', 'regions', colors=labels_colors, axsize=(2.4,2.5));
</code>
## Subdivide the single spatial sample spatially into several parts
<code>
tc.utils.spatial_split(puck, position_key='y', position_split=4, result_key='split');
puck.obs['split'] = split_names[puck.obs['split'].astype('category').cat.codes]
puck.obs['split'] = puck.obs['split'].astype('category')
fig = tc.pl.scatter(puck, 'split', joint=True,axsize=axsize, point_size=1, noticks=True, axes_labels=['X','Y'], colors=split_colors);
</code>
Compare cell type composition across these parts
<code>
fig = tc.pl.contribution(puck, 'labels', 'regions', colors=labels_colors, normalization='gmean', reduction='sum', sample_key='split', axsize=(len(puck.obsm['labels'].columns) * (0.2 * 4 + .1) * 1.25, 2.5));
</code>
Do statistics on these parts treating them as independent samples
<code>
enr = tc.tl.enrichments(puck, 'labels', 'regions', normalization='gmean', reduction='sum', sample_key='split');
fig = tc.pl.significances(enr, p_key='p_mwu_fdr_bh', value_key='labels', group_key='regions', axsize=(2.5,len(puck.obsm['labels'].columns)*0.25));
</code>
## Analyse neighbourhips
<code>
tc.tl.co_occurrence_matrix(puck,annotation_key='labels',result_key='labels-labels',max_distance=20,n_permutation=10, );
fig = tc.pl.co_occurrence_matrix(puck,analysis_key='labels-labels',score_key='z',cmap_vmin_vmax=(-5,5), axsize=(1.3,1.3));
</code>
## Analyse cell type composition relative to a region annotation
Calculate cell type composition in dependence of the distance to region_2.
In contrast to the analysis above using a gobally defined regularized distance, the distance here is defined for all pairs of observations and aggregated over the pairs.
<code>
tc.tl.co_occurrence(puck,annotation_key='labels',center_key='regions',result_key='labels-regions',delta_distance=20,max_distance=500,n_permutation=10, );
fig = tc.pl.co_occurrence(puck,analysis_key='labels-regions',score_key='log_composition',colors=labels_colors, log_base=2, show_only_center=['region_2'], axsize=np.array([4,3])*0.4);
</code>
|
{
"filename": "examples_notebook.ipynb",
"repository": "simonwm/tacco",
"query": "transformed_from_existing",
"size": 11842,
"sha": ""
}
|
# uncertainty_2.ipynb
Repository: wukevin/babel
# Uncertainy quantification
BABEL's embedding space also provides a basis for measuring confidence in downstream classifications. Such confidence measures are useful for quantifying how "trustworthy" BABEL's predictions might be on new data.
Intuitively, within the embedding space, in-distribution examples should share a subspace, and exmaples outside this subspace are likely to be out of distribution and therefore low confidence. Here, we explore this idea by building a Gaussian Process classifier that predicts in-vs-out of distribution, which we interpret informally as an estimate of BABEL's confidence. Although this measure is applied to the embedding, its estimates are valid for output RNA/ATAC modalities as well, since this embedding is a predecessor to those outputs.
<code>
import os, sys
import collections
import functools
import json
import importlib
import logging
import numpy as np
import pandas as pd
from scipy import stats, sparse, spatial
from sklearn import metrics
from sklearn.gaussian_process import GaussianProcessClassifier
from matplotlib import pyplot as plt
import seaborn as sns
import anndata as ad
import scanpy as sc
import gdown
import tqdm.notebook
SRC_DIR = os.path.join(os.path.dirname(os.getcwd()), 'babel')
assert os.path.isdir(SRC_DIR)
sys.path.append(SRC_DIR)
import utils
BIN_DIR = os.path.join(os.path.dirname(SRC_DIR), "bin")
assert os.path.isfile(os.path.join(BIN_DIR, "predict_model.py"))
import perturb
DATA_DIR = os.path.join(os.path.dirname(os.getcwd()), "data")
assert os.path.isdir(DATA_DIR)
print(DATA_DIR)
logging.basicConfig(level=logging.INFO)
</code>
## Data setup
First, we download and ensure data is in the expected locations.
<code>
# PBMC ATAC data
pbmc_h5_fname = os.path.join(DATA_DIR, "10x", "atac_v1_pbmc_10k_filtered_peak_bc_matrix.h5")
assert os.path.isfile(pbmc_h5_fname)
</code>
<code>
# Download BCC ATAC data
# https://drive.google.com/file/d/1dv1l-dgrWiHey-RS0SwS_ASn4gVQ_qZu/view?usp=sharing
bcc_adata_fname = gdown.cached_download(
url="https://drive.google.com/uc?id=1dv1l-dgrWiHey-RS0SwS_ASn4gVQ_qZu",
path=os.path.join(DATA_DIR, "bcc/GSE129785_scATAC-TME-All.h5ad"),
md5="09cc204cabd59fdf5aa9c07fa29de961",
quiet=False,
)
</code>
## PBMC perturbation
We take PBMC scATAC-seq data and perturb it. Both the original, unperturbed data and the perturbed data are then fed through BABEL to generate corresponding (16-dimensional) embeddings. We then use the original, unperturbed BABEL embeddings as examples of "in-distribution" data, and the perturbed BABEL embeddings as examples of "out-of-distribution" data to train a Gaussian Process classifier to distinguish between the two.
<code>
# Define how we will perform perturbations
drop_method = "swap"
drop_p = 0.5
swapper = perturb.swap_adata if drop_method == "swap" else perturb.dropout_adata
swapper
</code>
<code>
pbmc_atac_vanilla_adata = sc.read_10x_h5(pbmc_h5_fname, gex_only=False)
pbmc_atac_vanilla_adata
</code>
<code>
pbmc_atac_dropped_adata = swapper(pbmc_atac_vanilla_adata, p=drop_p)
pbmc_atac_dropped_adata_fname = os.path.join(DATA_DIR, "10x", "atac_v1_pbmc_10k_filtered_peak_bc_matrix_dropped.h5ad")
pbmc_atac_dropped_adata.write_h5ad(pbmc_atac_dropped_adata_fname)
</code>
<code>
%%bash -s "$DATA_DIR" "$pbmc_h5_fname"
python /home/wukevin/projects/babel/bin/predict_model.py --data ${2} --outdir ${1}/10x/babel_atac_to_rna_pbmc_vanilla --noplot --liftHg19toHg38 --transonly --device 0
</code>
<code>
%%bash -s "$DATA_DIR" "$pbmc_atac_dropped_adata_fname"
python /home/wukevin/projects/babel/bin/predict_model.py --data ${2} --outdir ${1}/10x/babel_atac_to_rna_pbmc_dropped --noplot --liftHg19toHg38 --transonly --device 0
</code>
<code>
pbmc_vanilla_embed = ad.read_h5ad(os.path.join(
DATA_DIR,
"10x/babel_atac_to_rna_pbmc_vanilla/atac_encoded_adata.h5ad",
))
pbmc_vanilla_embed
</code>
<code>
pbmc_dropped_embed = ad.read_h5ad(os.path.join(
DATA_DIR,
"10x/babel_atac_to_rna_pbmc_dropped/atac_encoded_adata.h5ad",
))
pbmc_dropped_embed
</code>
<code>
pbmc_gp = GaussianProcessClassifier(random_state=1234)
# label of 1 = in distribution, 0 = out of distribution
pbmc_gp.fit(
np.vstack([pbmc_vanilla_embed.X, pbmc_dropped_embed.X]),
[1] * pbmc_vanilla_embed.n_obs + [0] * pbmc_dropped_embed.n_obs,
)
pbmc_gp
</code>
## BCC data
We turn to the basal cell carcinoma (BCC) dataset (Yost et al., https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC7299161/) discussed in our manuscript.
We know that, biologically, the BCC dataset contains celltypes (particularly, endothelial skin cells and tumor cells) that are out-of-distribution with respect to BABEL's training data; therefore, we expect that the GP trained above will produce low prediction scores (interpreted as low confidence) for these celltypes. We verify this below.
<code>
%%bash -s "$DATA_DIR" "$bcc_adata_fname"
python /home/wukevin/projects/babel/bin/predict_model.py --data ${2} --outdir ${1}/bcc/babel_atac_to_rna_bcc --noplot --liftHg19toHg38 --transonly --device 0
</code>
<code>
# Run the GP on BCC's embedding
bcc_vanilla_embed = ad.read_h5ad(os.path.join(
DATA_DIR,
"bcc/babel_atac_to_rna_bcc/atac_encoded_adata.h5ad",
))
bcc_gp_preds = pbmc_gp.predict_proba(bcc_vanilla_embed.X)
bcc_gp_preds.shape
</code>
<code>
bcc_vanilla_embed.obs['gp_pbmc_pred'] = bcc_gp_preds[:, 1]
bcc_vanilla_embed.obs.head()
</code>
<code>
bcc_vanilla_embed.obs['ClustersNamed'] = [bcc_cluster_to_name[n] for n in bcc_vanilla_embed.obs['Clusters']]
bcc_vanilla_embed.obs.head()
</code>
<code>
with open(os.path.join(DATA_DIR, "bcc/bcc_cluster_to_celltype.json")) as source:
bcc_cluster_to_name = {f"Cluster{i+1}": cellname for i, cellname in enumerate(json.load(source))}
fig, ax = plt.subplots(dpi=300)
gp_pbmc_ordered_celltypes = bcc_vanilla_embed.obs.groupby("ClustersNamed").agg('median').sort_values('gp_pbmc_pred').index[::-1]
sns.boxplot(
x="ClustersNamed", y="gp_pbmc_pred", data=bcc_vanilla_embed.obs,
order=gp_pbmc_ordered_celltypes, ax=ax
)
ax.set_xticklabels(ax.get_xticklabels(), rotation=90)
ax.set(
title='Confidence on BCC celltypes',
xlabel="BCC Celltype",
ylabel="GP-estimated confidence"
)
ax.axhline(0.5, color='grey', linestyle='--')
fig.show()
</code>
In the above plot, the x-axis corresponds to the cell types present in the BCC dataset. Each box-plot shows, for each celltype, the distribution of predicted likelihood of being "in-distribution" and therefore "confident." We see that the four tumor celltypes have the lowest predicted confidence, as do endothelial cells and myeloid cells. These are consistent with what we'd expect biologically. We additionally see that B cells are relatively low-confidence; this may suggest that B cells in this BCC tissue sample are forming larger complexes (Kinker et al., https://www.frontiersin.org/articles/10.3389/fcell.2021.678127/full), thus adopting cell signatures unlike those seen in training B-cell examples.
Overall, this plot shows that training a Gaussian Process classifier on BABEL's embedding can provide a good estimate of uncertainty when attempting to generalize BABEL to new data.
|
{
"filename": "uncertainty_2.ipynb",
"repository": "wukevin/babel",
"query": "transformed_from_existing",
"size": 307957,
"sha": ""
}
|
# RankCorr-example_4.ipynb
Repository: ahsv/RankCorr
07 May 2020
# An example: running RankCorr on Paul
For editing packages - don't need to run this
<code>
%load_ext autoreload
%autoreload 2
</code>
<code>
import numpy as np
import pandas as pd
</code>
Also load scanpy for easy access to the Paul data set. Check out the scanpy repository at https://github.com/theislab/scanpy
<code>
import scanpy.api as sc
sc.settings.verbosity = 3 # verbosity: errors (0), warnings (1), info (2), hints (3)
sc.settings.set_figure_params(dpi=80, color_map='viridis') # low dpi (dots per inch) yields small inline figures
sc.logging.print_versions()
</code>
<code>
import anndata
</code>
## Load the RankCorr methods
The RankCorr code is currently in a heavily modified version of the PicturedRocks package. See the PicturedRocks repo at https://github.com/umangv/picturedrocks for the original package.
The modified package is included in the code here - this needs to be loading the local version for the remainder of the code to run
<code>
from picturedRocks import Rocks
</code>
Required inputs for the `Rocks` class:
* `X`, an `np.ndarry` of gene counts. Each row should contain the genetic information from a cell; the columns of `X` correspond to the genes (note that this is the transpose of some commonly used packages).
* `y`, a vector of cluster labels. These labels must be consecutive integers starting at 0.
## Load the Paul dataset
This will automatically download the data set if this is your first time running it.
<code>
dataset = "paul15"
</code>
<code>
adata = sc.datasets.paul15()
</code>
<code>
adata
</code>
Create the required vector of cluster labels based on the strings provided in the AnnData object.
<code>
lookup = list(adata.obs['paul15_clusters'].cat.categories)
yVec = np.array([lookup.index( adata.obs['paul15_clusters'][i] ) for i in range(adata.obs['paul15_clusters'].shape[0]) ])
</code>
Here are cluster names from the Paul data set. See Paul (2015).
<code>
lookup
</code>
Create the `Rocks` object as outlined above
<code>
data = Rocks(adata.X, yVec)
# PicturedRocks provides normalization capabilities, though this shouldn't be used for marker selection.
#data.normalize(log=False, totalexpr=10000)
'''
# It is also possible to use the PicturedRocks for fold testing, to match the results from the manuscript.
# This will be discussed more in the future.
ft = FoldTester(data)
folds = np.load("paul15-scviFolds.npz")["folds"]
ft.folds = folds
ft.validatefolds()
ft.makerocks(verbose=0)
'''
</code>
## Run RankCorr
The main RankCorr method is `CSrankMarkers.` In addition to the data provided by the `Rocks` object, it requires one parameter:
* `lamb` is the sparsity parameter - larger values of `lamb` will result in more markers selected per cluster
There are several optional boolean parameters:
* `writeOut` defaults to `False` and controls whether or not to write the selected markers to a file. The deafult filename is "ovrRankGenes-lamb{}.dat", with the input value of `lamb`.
* `keepZeros` should almost always be set to `False` (the default value). It provides a tweak to keep the in the data matrix `X` unchanged by the ranking procedure (i.e. the zeros will be mapped to zero). This has the effect of removing the zero counts from the analysis (while ranking all of the other counts correctly) and is purely added for experimental exploration.
* `onlyNonZero` should almost always be set to `False` (the default value). This provides a tweak to only rank the nonzero counts, pretending that the zero counts did not even exist. This is only useful if the zero counts in the application are completely uninformative (e.g. a zero count could easily represent a complete erasure of a massive count) which is not the case in UMI counts scRNA-seq data.
Note that there are really not any hyperparamters to tweak!
<code>
lamb = 3.0 # this can be whatever
%time markers = data.CSrankMarkers(lamb=lamb, writeOut=False, keepZeros=False, onlyNonZero=False)
</code>
By deafault, this gives a list of markers for the whole clustering, without separating markers by the cluster that they are selected for. If `writeOut = True`, the cluster information is stored in the output file.
<code>
len(markers)
</code>
If you have the geneNames, add them to the `Rocks` object - then these markers can be converted to gene names.
<code>
geneNames = np.array(adata.var.index)
data.genes = geneNames
</code>
<code>
marker_genes = data.markers_to_genes(markers)
</code>
<code>
marker_genes[:10]
</code>
|
{
"filename": "RankCorr-example_4.ipynb",
"repository": "ahsv/RankCorr",
"query": "transformed_from_existing",
"size": 13982,
"sha": ""
}
|
# axe_PCA Vs RF_3.ipynb
Repository: bbrener1/rusty
<code>
!ls {data_location}
</code>
## Predictability of Features and Samples
In this notebook we'll ask a basic question:
How predictable are different features and samples in general?
PCA makes the best possible orthogonal representation of a dataset using up to n different linear components, so it's the platonic ideal of how well a dataset is represented by a multivariate normal distribution with some covariance matrix.
So let's ask ourselves, how much information can we recover from various scRNAseq datasests if we project them into a lower-dimensional subspace using PCA and then recover them?
<code>
import numpy as np
import matplotlib.pyplot as plt
import scanpy as sc
import pickle
data_location = "../../data/aging_brain/"
young = pickle.load(open(data_location + "aging_brain_young.pickle",mode='rb'))
old = pickle.load(open(data_location + "aging_brain_old.pickle",mode='rb'))
filtered = pickle.load(open(data_location + "aging_brain_filtered.pickle",mode='rb'))
batch_encoding = np.loadtxt(data_location + 'aging_batch_encoding.tsv')
batch_encoding = batch_encoding.astype(dtype=bool)
young_mask = np.zeros(37069,dtype=bool)
old_mask = np.zeros(37069,dtype=bool)
young_mask[:young.shape[0]] = True
old_mask[young.shape[0]:] = True
# young = pickle.load(open(data_location + "aging_brain_young.pickle",mode='rb'))
# old = pickle.load(open(data_location + "aging_brain_old.pickle",mode='rb'))
# filter_mask = np.loadtxt(data_location + "filtered_feature_mask.txt").astype(dtype=bool)
# young_filtered = young.X.T[filter_mask].T
# old_filtered = old.X.T[filter_mask].T
</code>
<code>
from sklearn.decomposition import PCA
model = PCA(n_components=25).fit(young.X)
transformed = model.transform(young.X)
recovered = model.inverse_transform(transformed)
centered = young.X - np.mean(young.X,axis=0)
null_squared_residual = np.power(centered,2)
pca_residual = young.X - recovered
pca_squared_residual = np.power(pca_residual,2)
pca_recovered_per_sample = np.sum(pca_squared_residual,axis=1)
pca_recovered_fraction_per_sample = np.sum(pca_squared_residual,axis=1) / np.sum(null_squared_residual,axis=1)
print(np.sum(null_squared_residual))
print(np.sum(pca_squared_residual))
print(f"Remaining variance:{(np.sum(pca_squared_residual) / np.sum(null_squared_residual))}")
</code>
<code>
from sklearn.decomposition import PCA
centered = young.X - np.mean(young.X,axis=0)
null_squared_residual = np.power(centered,2)
fractions = []
for i in range(100):
model = PCA(n_components=i).fit(young.X)
transformed = model.transform(young.X)
recovered = model.inverse_transform(transformed)
recovered_residual = young.X - recovered
recovered_squared_residual = np.power(recovered_residual,2)
pca_recovered_per_sample = np.sum(recovered_squared_residual,axis=1)
pca_recovered_fraction_per_sample = np.sum(recovered_squared_residual,axis=1) / np.sum(null_squared_residual,axis=1)
# print(np.sum(null_squared_residual))
# print(np.sum(recovered_squared_residual))
fraction = np.sum(recovered_squared_residual) / np.sum(null_squared_residual)
fractions.append(fraction)
print(f"{i}: Remaining variance:{fraction}")
# centered = young_filtered - np.mean(young_filtered,axis=0)
# null_squared_residual = np.power(centered,2)
# fractions = []
# for i in range(351):
# model = PCA(n_components=i).fit(young_filtered)
# transformed = model.transform(young_filtered)
# recovered = model.inverse_transform(transformed)
# recovered_residual = young_filtered - recovered
# recovered_squared_residual = np.power(recovered_residual,2)
# pca_recovered_per_sample = np.sum(recovered_squared_residual,axis=1)
# pca_recovered_fraction_per_sample = np.sum(recovered_squared_residual,axis=1) / np.sum(null_squared_residual,axis=1)
# # print(np.sum(null_squared_residual))
# # print(np.sum(recovered_squared_residual))
# fraction = np.sum(recovered_squared_residual) / np.sum(null_squared_residual)
# fractions.append(fraction)
# print(f"{i}: Remaining variance:{fraction}")
</code>
<code>
plt.figure()
plt.title("Fraction of Variance Unexplained, PCA")
plt.plot(fractions[:60],label="PCA FVU")
plt.plot([0,60],[.55,.55],"--",label="Forest FVU")
plt.xlabel("Number of PCs")
plt.ylabel("FVU")
plt.legend()
plt.show()
fractions = np.array(fractions)
diff = fractions[:-1]-fractions[1:]
plt.figure()a
plt.title("Fraction of Variance Explained per PC")
plt.plot(diff[:60])
plt.xlabel("PCs")
plt.ylabel("Added Power")
plt.legend()
plt.show()
</code>
<code>
# for i,pc in enumerate(transformed.T):
# plt.figure()
# plt.title(i)
# plt.scatter(*young.obsm["X_umap"].T,c=pc,s=3,alpha=.4,cmap='bwr',vmin=-20,vmax=20)
# plt.colorbar()
# plt.show()
# f1 = "Ctsd"
# f2 = "H2-Ab1"
# f1_index = forest.truth_dictionary.feature_dictionary[f1]
# f2_index = forest.truth_dictionary.feature_dictionary[f2]
# for i,component in enumerate(model.components_):
# print(f"{i}: {f1}:{component[f1_index]},{f2}:{component[f2_index]}")
# plt.figure()
# plt.scatter(model.components_[:,f1_index],model.components_[:,f2_index])
# plt.plot([.2,-.2],[-.2,.2],color='red')
# plt.show()
</code>
<code>
feature_null = np.sum(null_squared_residual,axis=0) + 1
feature_absolute_null = np.sum(np.abs(centered),axis=0) + 1
sample_null = np.sum(null_squared_residual,axis=1) + 1
sample_absolute_null = np.sum(np.abs(centered),axis=1) + 1
pca_feature_error = np.sum(pca_squared_residual,axis=0) + 1
pca_feature_remaining = pca_feature_error/feature_null
pca_absolute_feature_error = np.sum(recovered_residual)
pca_sample_error = np.sum(pca_squared_residual,axis=1) + 1
pca_sample_remaining = pca_sample_error / sample_null
plt.figure()
plt.title("Fraction of Variance Unexplained, Per Feature")
plt.hist(pca_feature_remaining,bins=50)
plt.ylabel("Frequency")
plt.xlabel("Fraction of Variance Unexplained")
plt.show()
plt.figure()
plt.title("Fraction of Variance Unexplained, Per Sample")
plt.hist(pca_sample_remaining,bins=50)
plt.ylabel("Frequency")
plt.xlabel("Fraction of Variance Unexplained")
plt.show()
print(f"PCA Variance Unexplained:{np.sum(recovered_squared_residual)/np.sum(null_squared_residual)}")
</code>
<code>
# SUM OF ABSOLUTE DEVIATIONS VERSION
# feature_null = np.sum(np.abs(centered),axis=0) + 1
# sample_null = np.sum(np.abs(centered),axis=1) + 1
# pca_feature_error = np.sum(np.abs(recovered_residual),axis=0) + 1
# pca_feature_remaining = pca_feature_error/feature_null
# pca_sample_error = np.sum(np.abs(recovered_residual),axis=1) + 1
# pca_sample_remaining = pca_sample_error / sample_null
# plt.figure()
# plt.title("Fraction of Variance Unexplained, Per Feature")
# plt.hist(pca_feature_remaining,bins=50)
# plt.ylabel("Frequency")
# plt.xlabel("Fraction of Variance Unexplained")
# plt.show()
# plt.figure()
# plt.title("Fraction of Variance Unexplained, Per Sample")
# plt.hist(pca_sample_remaining,bins=50)
# plt.ylabel("Frequency")
# plt.xlabel("Fraction of Variance Unexplained")
# plt.show()
</code>
<code>
!ls ../../data/aging_brain/
</code>
<code>
# import sys
# sys.path.append('../')
# from rusty_axe import tree_reader as tr
from rusty_axe import lumberjack
cv_forest = lumberjack.fit(
young.X,
header=young.var_names,
trees=100,
ifs=700,
ofs=700,
ss=300,
depth=8,
leaves=100,
dispersion_mode='ssme',
sfr=0,
standardize = 'true',
norm='l1',
reduction = 10,
p=8,
reduce_input='true',
reduce_output='true'
)
cv_forest.set_cache(True)
cv_forest.backup(data_location + "pca_comparison_forest")
# started at 5:50
</code>
<code>
import sys
# sys.path.append('/localscratch/bbrener1/rusty_forest_v3/src')
sys.path.append('../../')
import rusty_axe.lumberjack as lumberjack
data_location = "../../data/aging_brain/"
forest = lumberjack.load(data_location + 'selection_forest')
forest.arguments
</code>
<code>
forest.self_prediction = forest.predict(forest.output)
forest.self_prediction.prediction_report()
</code>
<code>
forest_residuals = forest.self_prediction.residuals()
</code>
<code>
forest_squared_residuals = np.power(forest_residuals,2)
forest_feature_error = np.sum(forest_squared_residuals,axis=0) + 1
forest_feature_remaining = forest_feature_error/feature_null
forest_sample_error = np.sum(forest_squared_residuals,axis=1) + 1
forest_sample_remaining = forest_sample_error/sample_null
plt.figure()
plt.title("Fraction of Variance Unexplained, Per Feature")
plt.hist(forest_feature_remaining,bins=50)
plt.ylabel("Frequency")
plt.xlabel("Fraction of Variance Unexplained")
plt.show()
plt.figure()
plt.title("Fraction of Variance Unexplained, Per Sample")
plt.hist(forest_sample_remaining,bins=50)
plt.ylabel("Frequency")
plt.xlabel("Fraction of Variance Unexplained")
plt.show()
# print(f"Forest Variance Unexplained:{np.sum(forest_squared_residuals)/np.sum(null_squared_residual)}")
# delta_sort = np.argsort(pca_feature_remaining-forest_feature_remaining)
# print(f"PCA best:{forest.output_features[delta_sort[:20]]}")
# print(f"Forest best:{forest.output_features[delta_sort[-20:]]}")
# for fb in delta_sort[-20:]:
# print(f"Forest best: {forest.output_features[fb]}")
# print(f"Forest: {forest_feature_remaining[fb]}")
# print(f"PCA:{pca_feature_remaining[fb]}")
# ctsd_index = forest.truth_dictionary.feature_dictionary["Ctsd"]
# print(forest_feature_remaining[ctsd_index])
# print(pca_feature_remaining[ctsd_index])
feature_mean = np.mean(young.X,axis=0)
feature_mean.shape
sample_mean = np.mean(young.X,axis=1)
# h2_index = forest.truth_dictionary.feature_dictionary["H2-Ab1"]
# plt.figure()
# plt.scatter(*forest.coordinates().T,c=recovered_residual[:,h2_index],s=2,cmap='bwr')
# plt.colorbar()
# plt.show()
# Cat 1a,: S100a9, S100a8, Wfdc21,Retnlg, Lcn2,Ngp,Camp,Mmp8,Hp, Ltf, Slpi, Trem3
# Cat 1b: Plac8,
# Cat 1c: H2-Eb1,H2-Aa,H2-Ab1,
# Cat 2a: Slc22a6,Slc6a13,Fmod
# Cat3: Myoc
</code>
<code>
plt.figure(figsize=(4,4))
plt.title("Fraction of Variance Unexplained Per Feature, Forest Vs PCA")
plt.scatter(pca_feature_remaining,forest_feature_remaining,s=3,c=feature_mean)
plt.colorbar(label="Mean Expression")
plt.plot([0,1],[0,1],color='red')
plt.xlabel("PCA FVU")
plt.ylabel("Forest FVU")
plt.show()
# plt.figure(figsize=(4,4))
# plt.title("Fraction of Variance Unexplained Per Sample, Forest Vs PCA")
# plt.scatter(pca_sample_remaining,forest_sample_remaining,s=3,c=sample_mean)
# plt.plot([0,1],[0,1],color='red')
# plt.xlabel("PCA FVU")
# plt.ylabel("Forest FVU")
# plt.show()
</code>
<code>
plt.figure()
plt.title("Forest Error Vs PCA Error")
plt.scatter(*young.obsm["X_umap"].T,s=2,c=forest_sample_remaining-pca_sample_remaining,cmap='seismic',vmin=-.5,vmax=.5)
plt.colorbar(label="Forest FVU - PCA FVU")
plt.show()
</code>
<code>
gene = "Hp"
gene_index = forest.truth_dictionary.feature_dictionary[gene]
print(forest_feature_remaining[gene_index])
print(pca_feature_remaining[gene_index])
</code>
<code>
plt.figure()
plt.scatter(forest_squared_residuals.flatten(),pca_squared_residual.flatten(),s=1,alpha=.3)
plt.show()
</code>
<code>
random_mask = np.random.random(forest_squared_residuals.flatten().shape) < .001
plt.figure()
plt.title("All Squared Residuals, PCA vs URF\n Subsampled and Truncated")
plt.scatter(forest_squared_residuals.flatten()[random_mask],recovered_squared_residual.flatten()[random_mask],s=1,alpha=.3)
plt.xlabel("Unsupervised Random Forest")
plt.ylabel("PCA")
plt.xlim(0,10)
plt.ylim(0,10)
plt.show()
</code>
<code>
plt.figure()
plt.title("Model Residuals, PCA vs URF\n Subsampled")
plt.scatter(forest_residuals.flatten()[random_mask],recovered_residual.flatten()[random_mask],s=1,alpha=.3)
plt.xlabel("Unsupervised Random Forest")
plt.ylabel("PCA")
plt.plot([-5,5],[0,0],"--",color="gray")
plt.plot([0,0],[-4,4],"--",color="gray")
plt.xlim(-4,4)
plt.ylim(-4,4)
plt.show()
</code>
<code>
plt.figure(figsize=(4,4))
plt.title("Fraction of Variance Unexplained Per Feature, Forest Vs PCA")
plt.scatter(pca_feature_remaining,forest_feature_remaining,s=3,c=np.log(feature_mean))
plt.colorbar(label="Mean Expression")
plt.plot([0,1],[0,1],color='red')
plt.xlabel("PCA FVU")
plt.ylabel("Forest FVU")
plt.show()
</code>
|
{
"filename": "axe_PCA Vs RF_3.ipynb",
"repository": "bbrener1/rusty",
"query": "transformed_from_existing",
"size": 257074,
"sha": ""
}
|
# GeneScores_1.ipynb
Repository: kundajelab/scATAC-reprog
# Gene Scores
Gene score plots at a fine grained cluster level. Inputs:
1. `metadata.tsv` with UMAP/densMAP coordinates and total fragments/insertions.
2. `features.10d.tsv` scATAC-seq features for kNN smoothing
3. ArchR gene scores
<code>
library(Matrix)
library(ggplot2)
library(patchwork)
library(GenomicRanges)
library(scales)
library(RColorBrewer)
library(DESeq2)
library(rtracklayer)
library(Seurat)
library(ArchR)
library(RANN)
library(scattermore)
</code>
<code>
DAYS = c("D0", "D2", "D4", "D6", "D8", "D10", "D12", "D14", "iPSC")
</code>
## Loading Inputs
### MetaData
<code>
# should contain, sample_barcode as rowname, sample, umap1, umap2, cluster
metaData = read.table("../analysis/20200206_pmat_snapATAC/sessions/20210717_n62599//metadata.tsv", header = T)
rownames(metaData) = paste(metaData$sample, metaData$barcode, sep='_')
metaData$sample = factor(metaData$sample, levels=DAYS)
dim(metaData)
head(metaData, 5)
</code>
<code>
# will use feature to construct kNN graph for smoothing
features = read.table("../analysis/20200206_pmat_snapATAC/sessions/20210717_n62599/features.10d.tsv", header = T)
rownames(features) = features$sample_barcode
features$sample_barcode = NULL
dim(features)
head(features, 5)
</code>
## ArchR Gene Scores
<code>
addArchRThreads(threads = 32)
</code>
<code>
addArchRGenome("hg38")
</code>
<code>
ArrowFiles = paste(DAYS, "arrow", sep='.')
ArrowFiles
</code>
<code>
archr_proj <- ArchRProject(
ArrowFiles = paste("/srv/scratch/surag/scATAC-reprog/arrow/", ArrowFiles, sep=''),
outputDirectory = "./tmp/",
copyArrows = FALSE #This is recommened so that you maintain an unaltered copy for later usage.
)
</code>
<code>
all(paste(metaData$sample, metaData$barcode, sep='#') %in% archr_proj$cellNames)
</code>
<code>
# subset to cells
archr_proj = archr_proj[paste(metaData$sample, metaData$barcode, sep='#'), ]
</code>
<code>
getAvailableMatrices(archr_proj)
</code>
<code>
archr_gene_score = getMatrixFromProject(archr_proj, "GeneScoreMatrix")
dim(archr_gene_score)
</code>
<code>
archr_gene_score_mat = archr_gene_score@assays@data$GeneScoreMatrix
rownames(archr_gene_score_mat) = rowData(archr_gene_score)$name
colnames(archr_gene_score_mat) = sub("#", "_", rownames(colData(archr_gene_score)))
# reorder
archr_gene_score_mat = archr_gene_score_mat[, rownames(metaData)]
</code>
<code>
# reclaim some memory
rm(archr_gene_score)
gc()
</code>
## Smoothed Gene Scores
<code>
K = 15
</code>
<code>
knn = nn2(features, k=K)
</code>
<code>
j <- as.numeric(x = t(x = knn$nn.idx))
i <- ((1:length(x = j)) - 1) %/% K + 1
edgeList = data.frame(i, j, 1);
</code>
<code>
knng = sparseMatrix(i = edgeList[,1], j = edgeList[,2], x = edgeList[,3]);
</code>
<code>
# clip and smooth
top =
archr_gene_score_mat_smoothed = 1/K * (archr_gene_score_mat %*% knng)
archr_gene_score_mat_smoothed = as.matrix(archr_gene_score_mat_smoothed)
</code>
## Plotting
<code>
GENE="CDH1"
</code>
<code>
cur_gene_score = as.numeric(archr_gene_score_mat[GENE,])
</code>
<code>
# clip and smooth
Q = 0.98
cur_gene_score[cur_gene_score>quantile(cur_gene_score, Q)] = quantile(cur_gene_score, Q)
cur_gene_score = as.vector(1/K * (knng%*% cur_gene_score))
</code>
<code>
df = data.frame(umap1=metaData$umap1,
umap2=metaData$umap2,
gene_score=cur_gene_score)
# shuffle so days don't overlap
df = df[sample(dim(df)[1], 25000), ]
gs_plot <- ggplot(df) +
# ggplot(df[df$x.sp.sample %in% c("D14"), ]) +
geom_scattermore(pointsize=3, aes(x=umap1 , y=umap2, col=gene_score), pixels=c(1000,1000)) +
# ggtitle(sub("-2[0-9]+", "",GENE)) +
scale_color_viridis_c(option = "D",
limits= c(quantile(cur_gene_score, 0.1),
quantile(cur_gene_score, 0.9)),
oob=squish,
name="Gene\nScore") +
theme_classic() +
xlab("UMAP 1") + ylab("UMAP 2") +
theme(plot.title = element_text(hjust = 0.5),
text = element_text(size=12),
axis.line=element_blank(),
axis.text.x=element_blank(),
axis.text.y=element_blank(),
axis.ticks=element_blank(),
legend.text = element_blank(), # no numbers
panel.border = element_rect(colour = "black", fill=NA, size=0.5)) +
coord_fixed()
</code>
<code>
options(repr.plot.width = 5, repr.plot.height = 5)
gs_plot
</code>
<code>
saveRDS(gs_plot, file=sprintf("./Fig1/subfigs/%s_%s_gs.rds",
format(Sys.Date(), "%Y%m%d"), GENE))
</code>
---
<code>
sessionInfo()
</code>
|
{
"filename": "GeneScores_1.ipynb",
"repository": "kundajelab/scATAC-reprog",
"query": "transformed_from_existing",
"size": 189989,
"sha": ""
}
|
# model_1.ipynb
Repository: ahmedkhaleel2004/DeepEnd
<code>
!pip install tensorflow_text
</code>
<code>
import pandas as pd
import numpy as np
import tensorflow as tf
import ast
from tensorflow.keras.utils import to_categorical
from sklearn.preprocessing import MultiLabelBinarizer
import tensorflow_hub as hub
import tensorflow_text as text
from tensorflow.keras.layers import Input, Dense, Lambda
from tensorflow.keras.models import Model
</code>
<code>
data = pd.read_csv('github_users_dataset.csv', header=None)
initial_rows = data.shape[0]
data.dropna(inplace=True)
# Set the column names from the first row
data.columns = data.iloc[0]
# Drop the first row
data = data[1:]
# Drop all rows that are the same as the column names
data = data[~data.eq(data.columns).all(1)]
# Function to check if all strings in a given input are ASCII
def all_strings_are_ascii(input):
if isinstance(input, list):
return all(str(s).isascii() for s in input)
else:
return str(input).isascii()
# Apply the function to each element of the DataFrame
data_ascii = data.applymap(all_strings_are_ascii)
# Filter out the rows where all elements are ASCII
data = data[data_ascii.all(axis=1)]
data = data.query('projects != "[]" and languages != "[]"')
# Drop all rows where the 'role' column string has a length of less than 15
data = data[data['role'].str.len() >= 25]
# Function to check if any string in a list is less than 20 characters
def any_string_less_than_20_chars(input):
if isinstance(input, str):
input_list = ast.literal_eval(input)
if isinstance(input_list, list):
return any(len(str(s)) < 20 for s in input_list)
return False
# Apply the function to the 'projects' column
data['any_project_less_than_20_chars'] = data['projects'].apply(any_string_less_than_20_chars)
# Drop the rows where any description in the 'projects' column is less than 20 characters
data = data[data['any_project_less_than_20_chars'] == False]
# Drop the temporary column
data.drop(columns=['any_project_less_than_20_chars'], inplace=True)
final_rows = data.shape[0]
data.to_csv('cleaned_data.csv', index=False)
print(f'Rows removed: {initial_rows - final_rows}, {100 * (initial_rows - final_rows) / initial_rows:.2f}% of the original dataset.\nYou have {final_rows} rows left.')
</code>
<code>
data.head(10)
</code>
OHE for experience and language
<code>
# unique experience levels
experience = data['experience_level'].unique()
# map unique experience levels to numbers
# categorical data --> numerical data for one-hot encoding
experience_level_mapping = {level: idx for idx, level in enumerate(experience)}
# w gpt2 nie ma potrzeby one-hot encoding
data['experience_level_num'] = data['experience_level'].map(experience_level_mapping)
# one-hot encoding !!!!!!!!!!!!
experience_level_encoded = to_categorical(data['experience_level_num'])
experience_level_encoded
</code>
<code>
data['languages'] = data['languages'].apply(ast.literal_eval)
languages = set([lang for sublist in data['languages'].tolist() for lang in sublist])
mlb = MultiLabelBinarizer(classes=sorted(languages))
languages_encoded = mlb.fit_transform(data['languages'])
languages_encoded[:1]
</code>
BERT for role and project
<code>
bert_preprocess_url = 'https://tfhub.dev/tensorflow/bert_en_uncased_preprocess/3'
bert_model_url = 'https://tfhub.dev/tensorflow/small_bert/bert_en_uncased_L-2_H-128_A-2/2'
bert_preprocess_model = hub.KerasLayer(bert_preprocess_url)
bert_model = hub.KerasLayer(bert_model_url)
role_texts = data['role'].tolist() # Convert 'role' column to a list
role_preprocessed = bert_preprocess_model(role_texts)
role_results = bert_model(role_preprocessed)
role_results.keys()
</code>
<code>
roles_embedded = role_results['pooled_output']
roles_embedded.shape
</code>
<code>
projects = [j for i in data['projects'].tolist() for j in ast.literal_eval(i)]
projects_preprocessed = bert_preprocess_model(projects)
projects_results = bert_model(projects_preprocessed)
</code>
<code>
map_user_to_projects = {}
for user_num, projects_list in enumerate(data['projects']):
map_user_to_projects[user_num] = ast.literal_eval(projects_list)
map_user_to_projects[0]
</code>
<code>
projects_results.keys()
</code>
<code>
projects_embedded = projects_results['pooled_output']
</code>
<code>
for i in [experience_level_encoded,
languages_encoded,
roles_embedded,
projects_embedded]: print(i.shape)
</code>
<code>
user_profiles = tf.concat([roles_embedded, languages_encoded], axis=1)
user_profiles.shape
</code>
<code>
user_profiles[0]
</code>
<code>
for i in range(2):
print(f"User {i}: {map_user_to_projects[i]}")
</code>
<code>
data.head(15)
</code>
<code>
dataset = []
</code>
<code>
len(dataset)
</code>
<code>
len(map_user_to_projects[0])
</code>
<code>
file = open("negative_match.txt", "r")
num_matches = len(file.read().splitlines())
file.close()
print("num matches: ", num_matches)
start_project_range = 0
for user_num, user_profile in enumerate(user_profiles):
if user_num == num_matches: break
for project_embedding in projects_embedded[start_project_range:start_project_range+len(map_user_to_projects[user_num])]:
dataset.append((user_profile, project_embedding, 1))
start_project_range += len(map_user_to_projects[user_num])
</code>
<code>
len(dataset)
</code>
better manual method
<code>
# manual small dataset testing
def find_project_start_index_for_given_user(user_num):
i = 0
index = 0
while i != user_num:
index += len(map_user_to_projects[i])
i += 1
return index
def add_negative_example_given_non_matching_user_profiles_manually(user1: int, non_matching_user2: int):
start_index = find_project_start_index_for_given_user(non_matching_user2)
for i in range(len(map_user_to_projects[non_matching_user2])):
dataset.append((user_profiles[user1], projects_embedded[start_index + i], 0))
def add_positive_example_given_non_matching_user_profiles_manually(user1: int, non_matching_user2: int):
start_index = find_project_start_index_for_given_user(non_matching_user2)
for i in range(len(map_user_to_projects[non_matching_user2])):
dataset.append((user_profiles[user1], projects_embedded[start_index + i], 1))
</code>
<code>
file = open("negative_match.txt", "r")
lines = file.read().splitlines()
neg_matches = []
for line in lines:
first, second = line.split(",")
neg_matches.append((int(first), int(second)))
for first, second in neg_matches:
add_negative_example_given_non_matching_user_profiles_manually(first, second)
file.close()
</code>
<code>
len(dataset)
</code>
<code>
dataset[0]
</code>
<code>
import random
# Shuffle the dataset
random.shuffle(dataset)
</code>
<code>
# Split the dataset into features and labels
features = [(user_profile, project_embedding) for user_profile, project_embedding, _ in dataset]
labels = [label for _, _, label in dataset]
</code>
<code>
# Convert to numpy arrays or tensors as required for training
features = np.array(features)
labels = np.array(labels)
</code>
<code>
len(labels)
</code>
<code>
# Hyperparameters (you can adjust these based on your needs)
embedding_size = 256 # Size of the final embeddings
dropout_rate = 0.05 # Dropout rate for regularization
# User Profile Branch
user_input = Input(shape=(237,))
user_branch = Dense(128, activation='relu')(user_input)
# user_branch = tf.keras.layers.Dropout(dropout_rate)(user_branch)
user_branch = Dense(64, activation='relu')(user_branch)
# Project Description Branch
project_input = Input(shape=(128,))
project_branch = Dense(64, activation='relu')(project_input)
# project_branch = tf.keras.layers.Dropout(dropout_rate)(project_branch)
project_branch = Dense(64, activation='relu')(project_branch)
# # User Profile Branch
# user_input = Input(shape=(237,)) # Adjust the shape based on your concatenated user profile tensor
# user_branch = Dense(512, activation='relu')(user_input)
# user_branch = Dense(256, activation='relu')(user_branch)
# user_branch = tf.keras.layers.Dropout(dropout_rate)(user_branch)
# user_branch = Dense(embedding_size, activation='relu')(user_branch)
# # Project Description Branch
# project_input = Input(shape=(128,)) # Adjust the shape based on your BERT embeddings
# project_branch = Dense(256, activation='relu')(project_input)
# project_branch = tf.keras.layers.Dropout(dropout_rate)(project_branch)
# project_branch = Dense(128, activation='relu')(project_branch)
# project_branch = tf.keras.layers.Dropout(dropout_rate)(project_branch)
# project_branch = Dense(embedding_size, activation='relu')(project_branch)
</code>
<code>
# Distance Layer
def euclidean_distance(vectors):
x, y = vectors
sum_square = tf.reduce_sum(tf.square(x - y), axis=1, keepdims=True)
return tf.sqrt(sum_square)
# def cosine_similarity(vectors):
# # Unpack the vectors
# vector_a, vector_b = vectors
# # Compute the cosine similarity
# a_norm = tf.nn.l2_normalize(vector_a, axis=1)
# b_norm = tf.nn.l2_normalize(vector_b, axis=1)
# cosine_similarity = tf.reduce_sum(tf.multiply(a_norm, b_norm), axis=1)
# # Reshape to ensure the output shape is correct
# return tf.reshape(cosine_similarity, [-1, 1])
distance = Lambda(euclidean_distance)([user_branch, project_branch])
# Siamese Network Model
siamese_network = Model(inputs=[user_input, project_input], outputs=distance)
# Contrastive Loss Function
def contrastive_loss(y_true, y_pred):
margin = 1
square_pred = tf.square(y_pred)
margin_square = tf.square(tf.maximum(margin - y_pred, 0))
return tf.reduce_mean(y_true * square_pred + (1 - y_true) * margin_square)
# Define a custom accuracy metric
def accuracy(y_true, y_pred):
'''Compute classification accuracy with a fixed threshold on distances.
'''
return tf.keras.metrics.binary_accuracy(y_true, tf.cast(y_pred < 0.5, dtype=tf.float32))
# Compile the model with the custom accuracy metric
siamese_network.compile(optimizer="adam", loss=contrastive_loss, metrics=[accuracy])
# Model Summary
siamese_network.summary()
</code>
<code>
# prepare data for training
user_profiles, project_embeddings = zip(*features)
user_profiles = np.array(user_profiles)
project_embeddings = np.array(project_embeddings)
labels = np.array(labels)
</code>
<code>
# Splitting data into training and validation sets
from sklearn.model_selection import train_test_split
X_train, X_val, y_train, y_val = train_test_split(
list(zip(user_profiles, project_embeddings)), labels, test_size=0.5, random_state=43
)
</code>
<code>
# Preparing data for the model
user_profiles_train, project_embeddings_train = zip(*X_train)
user_profiles_val, project_embeddings_val = zip(*X_val)
</code>
<code>
# Convert labels to float32
y_train = y_train.astype('float32')
y_val = y_val.astype('float32')
</code>
<code>
len(user_profiles_train)
</code>
<code>
# Training the model
history = siamese_network.fit(
[np.array(user_profiles_train), np.array(project_embeddings_train)],
np.array(y_train),
validation_data=([np.array(user_profiles_val), np.array(project_embeddings_val)], np.array(y_val)),
epochs=30, # You can adjust the number of epochs
batch_size=4 # And the batch size
)
</code>
<code>
def create_user_vector(role: str, languages: list[str]):
languages_vector = mlb.fit_transform([languages])
pred_role_preprocessed = bert_preprocess_model([role])
pred_role_results = bert_model(pred_role_preprocessed)
pred_roles_embedded = pred_role_results['pooled_output']
return tf.concat([pred_roles_embedded, languages_vector], axis=1)
</code>
<code>
user_profile_example = create_user_vector("Machine learning, quantum computing phd student", ['Shell', 'Python', 'R', 'C++', 'Makefile'])
project_embedding_example = bert_model(bert_preprocess_model(['JQuery multiselect plugin based on Twitter Bootstrap.']))['pooled_output']
# Make a prediction
similarity_score = siamese_network.predict([user_profile_example, project_embedding_example])
# Output the similarity score
print("Similarity Score:", similarity_score[0][0])
</code>
Let's try ranking random projects for a given input user profile
<code>
# rank projects testing
user_profile_example = create_user_vector("Senior Software Engineer @microsoft", ['Shell', 'PowerShell', 'C#', 'Python', 'JavaScript'])
num_projects = 19
rand_index = random.randint(num_projects+1, len(projects) - 1) - num_projects
rand_index = 0
print(rand_index)
predictions = []
for i in range(num_projects):
project_embedding_example = bert_model(bert_preprocess_model([projects[rand_index + i]]))['pooled_output']
similarity_score = siamese_network.predict([user_profile_example, project_embedding_example])
predictions.append((projects[rand_index + i], similarity_score[0][0]))
</code>
<code>
# Sort the predictions in descending order of the score
sorted_predictions = sorted(predictions, key=lambda x: x[1], reverse=True)
# Print the sorted predictions
for project, score in sorted_predictions:
print(f"{project}: {score}")
</code>
<code>
for i in range(2):
print(f"User {i}: {map_user_to_projects[i]}")
</code>
<code>
num = 130
selected_rows = data.iloc[num:num+20]
print(selected_rows)
</code>
|
{
"filename": "model_1.ipynb",
"repository": "ahmedkhaleel2004/DeepEnd",
"query": "transformed_from_existing",
"size": 124101,
"sha": ""
}
|
# Omics_terms_1.ipynb
Repository: krassowski/multi-omics-state-of-the-field
**Aims**:
- extract the omics mentioned in multi-omics articles
**NOTE**: the articles not in PMC/with no full text need to be analysed separately, or at least highlighted.
<code>
%run notebook_setup.ipynb
</code>
<code>
import pandas
pandas.set_option('display.max_colwidth', 100)
</code>
<code>
%vault from pubmed_derived_data import literature, literature_subjects
</code>
<code>
literature['title_abstract_text_subjects'] = (
literature['title']
+ ' ' + literature['abstract_clean'].fillna('')
+ ' ' + literature_subjects.apply(lambda x: ' '.join(x[x == True].index), axis=1)
+ ' ' + literature['full_text'].fillna('')
)
</code>
<code>
omics_features = literature.index.to_frame().drop(columns='uid').copy()
</code>
<code>
from functools import partial
from helpers.text_processing import check_usage
from pandas import Series
check_usage_in_input = partial(
check_usage,
data=literature,
column='title_abstract_text_subjects',
limit=5 # show only first 5 results
)
</code>
<code>
TERM_IN_AT_LEAST_N_ARTICLES = 5
</code>
# Omics
## 1. Lookup by words which end with -ome
<code>
cellular_structures = {
# organelles
'peroxisome',
'proteasome',
'ribosome',
'exosome',
'nucleosome',
'polysome',
'autosome',
'autophagosome',
'endosome',
'lysosome',
# proteins and molecular complexes
'spliceosome',
'cryptochrome',
# chromosmes
'autosome',
'chromosome',
'x-chromosome',
'y-chromosome',
}
species = {
'trichome'
}
tools_and_methods = {
# dry lab
'dphenome',
'dgenome',
'reactome',
'rexposome',
'phytozome',
'rgenome',
'igenome', # iGenomes
# wet lab
'microtome'
}
</code>
<code>
not_an_ome = {
'outcome',
'middle-income',
'welcome',
'wellcome', # :)
'chrome',
'some',
'cumbersome',
'become',
'home',
'come',
'overcome',
'cytochrome',
'syndrome',
'ubiome',
'biome', # this IS an ome, but more into envrionmental studies, rather than molecular biology!
'fluorochrome',
'post-genome',
'ubiquitin-proteasome', # UPS
*tools_and_methods,
*cellular_structures,
*species
}
</code>
<code>
from omics import get_ome_regexp
ome_re = get_ome_regexp()
get_ome_regexp??
</code>
<code>
ome_occurrences = (
literature['title_abstract_text_subjects'].str.lower()
.str.extractall(ome_re)[0]
.to_frame('term').reset_index()
)
ome_occurrences = ome_occurrences[~ome_occurrences.term.isin(not_an_ome)]
ome_occurrences.head(3)
</code>
### 1.1 Harmonise hyphenation
<code>
from helpers.text_processing import report_hyphenation_trends, harmonise_hyphenation
</code>
<code>
hyphenation_rules = report_hyphenation_trends(ome_occurrences.term)
hyphenation_rules
</code>
<code>
ome_occurrences.term = harmonise_hyphenation(ome_occurrences.term, hyphenation_rules)
</code>
### 1.2 Fix typos
<code>
from helpers.text_processing import find_term_typos, create_typos_map
</code>
<code>
ome_counts = ome_occurrences.drop_duplicates(['uid', 'term']).term.sorted_value_counts()
potential_ome_typos = find_term_typos(ome_counts, TERM_IN_AT_LEAST_N_ARTICLES - 1)
potential_ome_typos
</code>
<code>
check_usage_in_input('1-metabolome')
</code>
<code>
check_usage_in_input('miRNAome')
</code>
<code>
check_usage_in_input('miRome')
</code>
<code>
check_usage_in_input('rexposome')
</code>
<code>
check_usage_in_input('glycol-proteome')
</code>
<code>
check_usage_in_input('rgenome')
</code>
<code>
check_usage_in_input('iGenomes')
</code>
<code>
check_usage_in_input('cancergenome')
</code>
<code>
is_typo_subset_or_variant = {
('transcritome', 'transcriptome'): True,
('transciptome', 'transcriptome'): True,
('tanscriptome', 'transcriptome'): True,
('trascriptome', 'transcriptome'): True,
('microbome', 'microbiome'): True,
('protenome', 'proteome'): True,
# (neither n- nor o- is frequent enough on its own)
('o-glycoproteome', 'glycoproteome'): True,
('n-glycoproteome', 'glycoproteome'): True,
('glycol-proteome', 'glycoproteome'): True, # note "glycol" instead of "glyco"
('mirome', 'mirnome'): True,
('1-metabolome', 'metabolome'): True
}
ome_typos_map = create_typos_map(potential_ome_typos, is_typo_subset_or_variant)
</code>
<code>
replaced = ome_occurrences.term[ome_occurrences.term.isin(ome_typos_map)]
replaced.value_counts()
</code>
<code>
len(replaced)
</code>
<code>
ome_occurrences.term = ome_occurrences.term.replace(ome_typos_map)
</code>
### 1.3 Replace synonymous and narrow terms
<code>
ome_replacements = {}
</code>
#### miRNAomics → miRNomics
miRNAome is more popular name for -ome, while miRNomics is more popular for -omics.
<code>
ome_occurrences.term.value_counts().loc[['mirnome', 'mirnaome']]
</code>
As I use -omcis for later on, for consistency I will change miRNAome → miRNome
<code>
ome_replacements['miRNAome'] = 'miRNome'
</code>
#### Cancer genome → genome
<code>
ome_occurrences.term.value_counts().loc[['genome', 'cancer-genome']]
</code>
<code>
ome_replacements['cancer-genome'] = 'genome'
</code>
#### Host microbiome → microbiome
<code>
ome_occurrences.term.value_counts().loc[['microbiome', 'host-microbiome']]
</code>
<code>
ome_replacements['host-microbiome'] = 'microbiome'
</code>
#### Replace the values
<code>
ome_occurrences.term = ome_occurrences.term.replace(
{k.lower(): v.lower() for k, v in ome_replacements.items()}
)
</code>
### 1.4 Summarise popular \*ome terms
<code>
ome_counts = ome_occurrences.drop_duplicates(['uid', 'term']).term.sorted_value_counts()
ome_common_counts = ome_counts[ome_counts >= TERM_IN_AT_LEAST_N_ARTICLES]
ome_common_counts
</code>
<code>
ome_common_terms = Series(ome_common_counts.index)
ome_common_terms[ome_common_terms.str.endswith('some')]
</code>
### 2. Lookup by omics and adjectives
<code>
from omics import get_omics_regexp
omics_re = get_omics_regexp()
get_omics_regexp??
</code>
<code>
check_usage_in_input('integromics')
</code>
<code>
check_usage_in_input('meta-omics')
</code>
<code>
check_usage_in_input('post-genomic')
</code>
<code>
check_usage_in_input('3-omics')
</code>
<code>
multi_omic = {
'multi-omic',
'muti-omic',
'mutli-omic',
'multiomic',
'cross-omic',
'panomic',
'pan-omic',
'trans-omic',
'transomic',
'four-omic',
'multiple-omic',
'inter-omic',
'poly-omic',
'polyomic',
'integromic',
'integrated-omic',
'integrative-omic',
'3-omic'
}
tools = {
# MixOmics
'mixomic',
# MetaRbolomics
'metarbolomic',
# MinOmics
'minomic',
# LinkedOmics - TCGA portal
'linkedomic',
# Mergeomics - https://doi.org/10.1186/s12864-016-3198-9
'mergeomic'
}
vague = {
'single-omic'
}
adjectives = {
'economic',
'socio-economic',
'socioeconomic',
'taxonomic',
'syndromic',
'non-syndromic',
'agronomic',
'anatomic',
'autonomic',
'atomic',
'palindromic',
# temporal
'postgenomic',
'post-genomic'
}
not_an_omic = {
'non-omic', # this on was straightforward :)
*adjectives,
*multi_omic,
*tools,
*vague
}
</code>
<code>
omic_occurrences = (
literature['title_abstract_text_subjects'].str.lower()
.str.extractall(omics_re)[0]
.to_frame('term').reset_index()
)
omic_occurrences = omic_occurrences[~omic_occurrences.term.isin(not_an_omic)]
omic_occurrences.head(2)
</code>
### 2.1 Harmonise hyphenation
<code>
hyphenation_rules = report_hyphenation_trends(omic_occurrences.term)
hyphenation_rules
</code>
<code>
omic_occurrences.term = harmonise_hyphenation(omic_occurrences.term, hyphenation_rules)
</code>
### 2.2 Fix typos
<code>
omic_counts = omic_occurrences.drop_duplicates(['uid', 'term']).term.sorted_value_counts()
potential_omic_typos = find_term_typos(omic_counts, TERM_IN_AT_LEAST_N_ARTICLES - 1)
potential_omic_typos
</code>
<code>
check_usage_in_input('non-omic')
</code>
<code>
check_usage_in_input('C-metabolomics')
</code>
Not captured in the text abstract, but full version has 13C, so carbon-13, so type of metabolomics.
<code>
check_usage_in_input('miRNAomics')
</code>
<code>
check_usage_in_input('miRomics')
</code>
<code>
check_usage_in_input('MinOmics')
</code>
<code>
check_usage_in_input('onomic', words=True)
</code>
<code>
literature.loc[omic_occurrences[omic_occurrences.term == 'onomic'].uid].title_abstract_text_subjects
</code>
<code>
check_usage_in_input(r'\bonomic', words=False, highlight=' onomic')
</code>
<code>
check_usage_in_input(' ionomic', words=False)
</code>
<code>
check_usage_in_input('integratomic', words=False)
</code>
Note: integratomics has literally three hits in PubMed, two because of http://www.integratomics-time.com/
<code>
is_typo_subset_or_variant = {
('phoshphoproteomic', 'phosphoproteomic'): True,
('transriptomic', 'transcriptomic'): True,
('transcripomic', 'transcriptomic'): True,
('transciptomic', 'transcriptomic'): True,
('trancriptomic', 'transcriptomic'): True,
('trascriptomic', 'transcriptomic'): True,
('metageonomic', 'metagenomic'): True,
('metaobolomic', 'metabolomic'): True,
('metabotranscriptomic', 'metatranscriptomic'): False,
('mirnaomic', 'mirnomic'): True,
('metranscriptomic', 'metatranscriptomic'): True,
('metranscriptomic', 'transcriptomic'): False,
('miromic', 'mirnomic'): True,
('n-glycoproteomic', 'glycoproteomic'): True,
('onomic', 'ionomic'): False,
('c-metabolomic', 'metabolomic'): True,
('integratomic', 'interactomic'): False,
('pharmacoepigenomic', 'pharmacogenomic'): False,
('metobolomic', 'metabolomic'): True,
# how to treat single-cell?
('scepigenomic', 'epigenomic'): True,
#('epitranscriptomic', 'transcriptomic'): False
('epigenomomic', 'epigenomic'): True,
}
omic_typos_map = create_typos_map(potential_omic_typos, is_typo_subset_or_variant)
</code>
<code>
replaced = omic_occurrences.term[omic_occurrences.term.isin(omic_typos_map)]
replaced.value_counts()
</code>
<code>
len(replaced)
</code>
<code>
omic_occurrences.term = omic_occurrences.term.replace(omic_typos_map)
</code>
### 2.3 Popular *omic(s) terms:
<code>
omic_counts = omic_occurrences.drop_duplicates(['uid', 'term']).term.sorted_value_counts()
omic_counts[omic_counts >= TERM_IN_AT_LEAST_N_ARTICLES].add_suffix('s')
</code>
### Crude overview
<code>
ome_terms = Series(ome_counts[ome_counts >= TERM_IN_AT_LEAST_N_ARTICLES].index)
omic_terms = Series(omic_counts[omic_counts >= TERM_IN_AT_LEAST_N_ARTICLES].index)
</code>
<code>
assert omics_features.index.name == 'uid'
for term in ome_terms:
mentioned_by_uid = set(ome_occurrences[ome_occurrences.term == term].uid)
omics_features['mentions_' + term] = omics_features.index.isin(mentioned_by_uid)
for term in omic_terms:
mentioned_by_uid = set(omic_occurrences[omic_occurrences.term == term].uid)
omics_features['mentions_' + term] = omics_features.index.isin(mentioned_by_uid)
</code>
<code>
from helpers.text_processing import prefix_remover
ome_terms_mentioned = omics_features['mentions_' + ome_terms].rename(columns=prefix_remover('mentions_'))
omic_terms_mentioned = omics_features['mentions_' + omic_terms].rename(columns=prefix_remover('mentions_'))
</code>
<code>
%R library(ComplexUpset);
</code>
<code>
%%R -i ome_terms_mentioned -w 800 -r 100
upset(ome_terms_mentioned, colnames(ome_terms_mentioned), min_size=10, width_ratio=0.1)
</code>
## Merge -ome and -omic terms
<code>
from warnings import warn
terms_associated_with_omic = {
omic + 's': [omic]
for omic in omic_terms
}
for ome in ome_terms:
assert ome.endswith('ome')
auto_generate_omic_term = ome[:-3] + 'omics'
omic = auto_generate_omic_term
if omic not in terms_associated_with_omic:
if omic in omic_counts.index:
warn(f'{omic} was removed at thresholding, but it is a frequent -ome!')
else:
print(f'Creating omic {omic}')
terms_associated_with_omic[omic] = []
terms_associated_with_omic[omic].append(ome)
</code>
<code>
from omics import add_entities_to_features
add_entities_to_omic_features = partial(
add_entities_to_features,
features=omics_features,
omics_terms=terms_associated_with_omic
)
</code>
<code>
omics = {k: [k] for k in terms_associated_with_omic}
add_entities_to_omic_features(omics, entity_type='ome_or_omic')
</code>
<code>
from omics import omics_by_entity, omics_by_entity_group
</code>
interactomics is a proper "omics", but it is difficult to assign to a single entity - by definition
<code>
check_usage_in_input('interactomics')
</code>
phylogenomics is not an omic on its own, but if used in context of metagenomics it can refer to actual omics data
<code>
check_usage_in_input('phylogenomics')
</code>
regulomics is both a name of a tool, group (@MIM UW), and omics:
<code>
check_usage_in_input('regulomics')
</code>
<code>
from functools import reduce
omics_mapped_to_entities = reduce(set.union, omics_by_entity.values())
set(terms_associated_with_omic) - omics_mapped_to_entities
</code>
<code>
assert omics_mapped_to_entities - set(terms_associated_with_omic) == set()
</code>
<code>
omics_mapped_to_entities_groups = reduce(set.union, omics_by_entity_group.values())
set(terms_associated_with_omic) - omics_mapped_to_entities_groups
</code>
<code>
add_entities_to_omic_features(omics_by_entity, entity_type='entity')
</code>
<code>
add_entities_to_omic_features(omics_by_entity_group, entity_type='entity_group')
</code>
### Visualize the entities & entities groups
<code>
omic_entities = omics_features['entity_' + Series(list(omics_by_entity.keys()))].rename(columns=prefix_remover('entity_'))
omic_entities_groups = omics_features['entity_group_' + Series(list(omics_by_entity_group.keys()))].rename(columns=prefix_remover('entity_group_'))
</code>
<code>
%%R -i omic_entities -w 800 -r 100
upset(omic_entities, colnames(omic_entities), min_size=10, width_ratio=0.1)
</code>
<code>
%%R -i omic_entities_groups -w 800 -r 100
upset(omic_entities_groups, colnames(omic_entities_groups), min_size=10, width_ratio=0.1)
</code>
### Number of omics mentioned in abstract vs the multi-omic term used
<code>
omes_or_omics_df = omics_features['ome_or_omic_' + Series(list(omics.keys()))].rename(columns=prefix_remover('ome_or_omic_'))
</code>
<code>
literature['omic_terms_detected'] = omes_or_omics_df.sum(axis=1)
</code>
<code>
lt = literature[['term', 'omic_terms_detected']]
</code>
<code>
literature.sort_values('omic_terms_detected', ascending=False)[['title', 'omic_terms_detected']].head(10)
</code>
<code>
%%R -i lt -w 800
(
ggplot(lt, aes(x=term, y=omic_terms_detected))
+ geom_violin(adjust=2)
+ geom_point()
+ theme_bw()
)
</code>
<code>
%vault store omics_features in pubmed_derived_data
</code>
# Current limitations
## Patchy coverage
Currently I only detected omic-describing terms in less than 70% of abstracts:
<code>
omic_entities.any(axis=1).mean()
</code>
Potential solution: select a random sample of 50 articles, annotate manually, calculate sensitivity and specificity.
If any omic is consistently omitted, reconsider how search terms are created.
## Apostrophes
Are we missing out on \*'omic terms, such us meta'omic used in [here](https://doi.org/10.1053/j.gastro.2014.01.049)?
<code>
check_usage_in_input(
r'\w+\'omic',
words=False,
highlight='\'omic'
)
</code>
unlikely (but would be nice to get it in!)
## Fields of study
<code>
'genetics', 'epigenetics'
</code>
Some authors may prefer to say "we integrated genetic and proteomic data" rather than "genomic and proteomic"
|
{
"filename": "Omics_terms_1.ipynb",
"repository": "krassowski/multi-omics-state-of-the-field",
"query": "transformed_from_existing",
"size": 311265,
"sha": ""
}
|
# eval_1.ipynb
Repository: agatha-duzan/feature-intervention-for-unlearning
<code>
!pip install "lm-eval"
!pip install "lm-eval[api]"
</code>
<code>
import os
key_path = 'goodfire_api_key.txt'
with open(key_path, 'r') as file:
GOODFIRE_API_KEY = file.read().strip()
os.environ['OPENAI_API_KEY'] = GOODFIRE_API_KEY
api_url="https://api.goodfire.ai/api/inference/v1/chat/completions"
</code>
<code>
import subprocess
subprocess.run([
'lm_eval',
'--model', 'openai-chat-completions',
'--model_args', f'model=meta-llama/Meta-Llama-3-8B-Instruct,tokenized_requests=False,base_url={api_url},num_concurrent=25',
'--tasks', 'mmlu_flan_n_shot_generative_college_computer_science,mmlu_flan_n_shot_generative_computer_security',
'--log_samples',
'--apply_chat_template', 'True',
'--num_fewshot', '0',
'--output_path', 'out_example'
])
</code>
<code>
import subprocess
subprocess.run([
'lm_eval',
'--model', 'openai-chat-completions',
'--model_args', f'model=meta-llama/Meta-Llama-3-8B-Instruct,tokenized_requests=False,base_url={api_url},num_concurrent=10',
'--tasks', 'mmlu_flan_n_shot_generative_college_biology,mmlu_flan_n_shot_generative_virology',
'--log_samples',
'--apply_chat_template', 'True',
'--num_fewshot', '0',
'--output_path', 'out_example'
])
</code>
|
{
"filename": "eval_1.ipynb",
"repository": "agatha-duzan/feature-intervention-for-unlearning",
"query": "transformed_from_existing",
"size": 80928,
"sha": ""
}
|
# demo.ipynb
Repository: ZJUFanLab/scSpace
<code>
import scSpace
import scanpy as sc
import matplotlib.pyplot as plt
import matplotlib.colors as clr
import numpy as np
from sklearn.metrics import adjusted_rand_score
import random
import torch
import warnings
warnings.filterwarnings("ignore")
</code>
<code>
def setup_seed(seed):
torch.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
torch.cuda.manual_seed(seed)
np.random.seed(seed)
random.seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.enabled = False
torch.backends.cudnn.benchmark = False
setup_seed(100)
</code>
<code>
sc_data_dir = 'data/demo_sc_data.csv'
sc_meta_dir = 'data/demo_sc_meta.csv'
st_data_dir = 'data/demo_st_data.csv'
st_meta_dir = 'data/demo_st_meta.csv'
sc_obj, st_obj = scSpace.load_data(sc_data_path=sc_data_dir, sc_meta_path=sc_meta_dir, st_data_path=st_data_dir, st_meta_path=st_meta_dir)
</code>
<code>
# ST reference
plt.rcParams["figure.figsize"] = (3, 3)
sc.pl.embedding(st_obj, basis="spatial", color="Group")
</code>
<code>
sc_obj, st_obj = scSpace.preporcess(sc_adata=sc_obj, st_adata=st_obj, st_type='spot', n_features=2000, normalize=True)
</code>
<code>
sc_obj, st_obj = scSpace.construct_pseudo_space(
sc_adata=sc_obj,
st_adata=st_obj,
batch_size=128,
activation='sigmoid',
lr=0.001,
epoch_num=1000,
log_epoch=1000)
</code>
<code>
# Pseudo space of scRNA-seq
sc.pl.embedding(sc_obj, basis="pseudo_space", color="Group")
</code>
<code>
# Spatial-informed clustering
sc_obj = scSpace.spatial_cluster(sc_obj, Ks=10, Kg=20, target_num=3)
</code>
<code>
# UMAP visualization
sc.pp.neighbors(sc_obj)
sc.tl.umap(sc_obj)
</code>
<code>
# classic clustering method of scRNA-seq
sc.tl.leiden(sc_obj, resolution=0.9)
</code>
<code>
sc.pl.umap(sc_obj, color=['Group', 'scSpace', 'leiden'])
</code>
<code>
scspace_ari = adjusted_rand_score(sc_obj.obs['Group'], sc_obj.obs['scSpace'])
leiden_ari = adjusted_rand_score(sc_obj.obs['Group'], sc_obj.obs['leiden'])
print('ARI (scSpace):', scspace_ari, '\n', 'ARI (Leiden):', leiden_ari)
</code>
|
{
"filename": "demo.ipynb",
"repository": "ZJUFanLab/scSpace",
"query": "transformed_from_existing",
"size": 114329,
"sha": ""
}
|
# MediMine.ipynb
Repository: dayana-cabrera004/npl
<a href="https://colab.research.google.com/github/dayana-cabrera004/npl/blob/main/MediMine.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
<code>
# Required imports
!pip install gradio
import gradio as gr
!pip install langchain-together
from langchain_together import ChatTogether
from langchain.chains import LLMChain
from langchain.prompts import PromptTemplate
import requests
import pandas as pd
import os
from datetime import datetime
from kaggle.api.kaggle_api_extended import KaggleApi
</code>
<code>
# Set environment variables
os.environ["TOGETHER_API_KEY"] = "207fee5eecff4d87a306a8566da4cd025ae6b252d14302d980dab27a618033f9"
os.environ["KAGGLE_USERNAME"] = "dayanacabrera"
os.environ["KAGGLE_KEY"] = "c90cf5759564ce5ca847713f7a36f72f"
</code>
<code>
# Initialize the Together.ai model
llm = ChatTogether(
temperature=0.0,
model="meta-llama/Meta-Llama-3.1-8B-Instruct-Turbo"
)
</code>
<code>
# Initialize Kaggle API
def init_kaggle():
api = KaggleApi()
api.authenticate()
return api
</code>
<code>
# Function to fetch datasets from various sources
def fetch_datasets(query, source):
# Define API endpoints for different data sources
sources = {
'healthdata': 'https://healthdata.gov/api/search',
'data_gov': 'https://catalog.data.gov/api/search',
'who': 'https://www.who.int/data/api/search',
'nih': 'https://www.nih.gov/api/search',
'kaggle': None
}
# Special handling for Kaggle datasets
if source == 'kaggle':
try:
api = init_kaggle()
datasets = api.dataset_list(search=query, tag='health')
return [dataset.ref for dataset in datasets]
except Exception as e:
print(f"Kaggle API error: {e}")
return None
# Handle other API requests
try:
response = requests.get(sources[source], params={'q': query})
return response.json()
except Exception as e:
print(f"API error for {source}: {e}")
return None
</code>
<code>
# General search function across all sources
def general_search(query):
# Fetch Kaggle datasets first
kaggle_datasets = fetch_datasets(query, 'kaggle')
# Create prompt template for the LLM
prompt = PromptTemplate(
input_variables=["query", "kaggle_data"],
template="""
Comprehensive medical dataset search for: {query}
Available Kaggle Datasets: {kaggle_data}
Provide a summary including:
1. Key findings from medical databases
2. Relevant clinical studies
3. Available research data
4. Statistical highlights
"""
)
chain = LLMChain(llm=llm, prompt=prompt)
return chain.run({"query": query, "kaggle_data": str(kaggle_datasets)})
</code>
<code>
# Function to display relevant Kaggle datasets
def show_relevant_datasets(query="diabetes"):
try:
api = init_kaggle()
datasets = api.dataset_list(search=query, tag='health')
dataset_info = []
for dataset in datasets[:8]: # Top 8 relevant datasets
dataset_info.append([
dataset.title,
f"[View Dataset](https://www.kaggle.com/datasets/{dataset.ref})",
f"{dataset.usabilityRating:.1f}/10",
f"{dataset.downloadCount:,}"
])
if not dataset_info:
dataset_info = [["No datasets found", "Try another search term", "N/A", "N/A"]]
except Exception as e:
dataset_info = [["API Error", "Could not fetch datasets", "N/A", "N/A"]]
return pd.DataFrame(
dataset_info,
columns=["Dataset", "Link", "Usability Score", "Downloads"]
)
</code>
<code>
# Main Gradio interface
def build_medimine_interface():
with gr.Blocks(title="MediMine - Medical Dataset Explorer") as app:
gr.Markdown("""
# 🏥 MediMine: Medical Dataset Explorer
## Comprehensive Medical Dataset Search Platform
Search across multiple medical databases and find relevant datasets instantly.
""")
with gr.Row():
with gr.Column():
query_input = gr.Textbox(
label="Enter Medical Search Query",
placeholder="e.g., diabetes type 2 research data",
lines=2
)
search_btn = gr.Button("🔍 General Search", variant="primary")
with gr.Row():
diagnosis_btn = gr.Button("🏥 Diagnosis")
treatment_btn = gr.Button("💊 Treatment")
genes_btn = gr.Button("🧬 Genetics")
trials_btn = gr.Button("🔬 Trials")
kaggle_btn = gr.Button("📊 Kaggle")
imaging_btn = gr.Button("🔎 Imaging")
output_text = gr.Textbox(
label="Search Results",
lines=10,
placeholder="Results will appear here..."
)
dataset_display = gr.DataFrame(
value=show_relevant_datasets().values.tolist(),
headers=["Dataset", "Link", "Usability Score", "Downloads"],
label="Relevant Kaggle Datasets"
)
# Update both search results and dataset recommendations
def update_results(query, search_type='general'):
if search_type == 'general':
search_result = general_search(query)
else:
search_result = specific_search(query, search_type)
datasets = show_relevant_datasets(query).values.tolist()
return search_result, datasets
# Connect buttons to functions
search_btn.click(
fn=lambda q: update_results(q, 'general'),
inputs=query_input,
outputs=[output_text, dataset_display]
)
diagnosis_btn.click(
fn=lambda q: update_results(q, 'diagnosis'),
inputs=query_input,
outputs=[output_text, dataset_display]
)
treatment_btn.click(
fn=lambda q: update_results(q, 'treatment'),
inputs=query_input,
outputs=[output_text, dataset_display]
)
genes_btn.click(
fn=lambda q: update_results(q, 'genes'),
inputs=query_input,
outputs=[output_text, dataset_display]
)
trials_btn.click(
fn=lambda q: update_results(q, 'trials'),
inputs=query_input,
outputs=[output_text, dataset_display]
)
kaggle_btn.click(
fn=lambda q: update_results(q, 'kaggle'),
inputs=query_input,
outputs=[output_text, dataset_display]
)
imaging_btn.click(
fn=lambda q: update_results(q, 'imaging'),
inputs=query_input,
outputs=[output_text, dataset_display]
)
return app
# Launch the application
if __name__ == "__main__":
app = build_medimine_interface()
app.launch(share=True)
</code>
**Purpose and case use of APP:**
The project you're working on, MediMine - Medical Dataset Explorer, aims to be the first point of referral for healthcare dataset searches, offering a comprehensive, one-stop solution for finding relevant medical data. Users can input a query (e.g., “diabetes type 2 research data”), and the app will search across multiple trusted sources, including Kaggle, government health portals, and leading research organizations like WHO and NIH. By leveraging the LangChain framework with Together.ai’s language model, the app processes queries and provides detailed insights, metadata, and recommendations, making the search process efficient and accurate. The platform is designed to cover a wide range of healthcare topics, offering category-specific searches for areas like diagnosis, treatment, genetics, clinical trials, and imaging.
What differentiates MediMine from other healthcare data search tools is its goal to provide a complete, holistic search experience. Unlike other apps that may only pull data from a limited set of sources, MediMine aggregates datasets from various trusted platforms, ensuring that the search is thorough and comprehensive. The use of a sophisticated AI language model adds another layer of value, enabling the app to provide contextual summaries and insights alongside dataset links, usability scores, and other metadata. This unique combination of diverse data sources, AI-powered insights, and a user-friendly interface positions MediMine as the ultimate first-stop app for anyone looking to explore healthcare datasets—knowing that the search will be exhaustive and complete.
|
{
"filename": "MediMine.ipynb",
"repository": "dayana-cabrera004/npl",
"query": "transformed_from_existing",
"size": 29321,
"sha": ""
}
|
# retina_phewas_evaluation_Figure6_Attributions_2_Heatmap.ipynb
Repository: lukl95/22
## Initialize
<code>
#library(Rmisc)
library(tidyverse)
library(glue)
library(arrow)
library(patchwork)
library("ggbeeswarm")
</code>
<code>
if (grepl("sc", Sys.info()[["nodename"]], fixed=TRUE)) {
base_path = "/sc-projects/sc-proj-ukb-cvd"
} else {
base_path = "/data/analysis/ag-reils/ag-reils-shared/cardioRS"}
print(base_path)
dataset_name = "210714_metabolomics"
path = "/data/analysis/ag-reils/steinfej/code/umbrella/pre/ukbb"
data_path = glue("{base_path}/data")
dataset_path = glue("{data_path}/3_datasets_post/{dataset_name}")
project_label="21_metabolomics_multitask"
project_path = glue("{base_path}/results/projects/{project_label}")
figures_path = glue("{project_path}/figures")
data_results_path = glue("{project_path}/data")
</code>
## Load data
<code>
list.dirs(path = project_path, full.names = TRUE, recursive = TRUE)
</code>
<code>
run = "211007"
data = arrow::read_feather(glue("{dataset_path}/data_merged.feather"))
data_description = arrow::read_feather(glue("{dataset_path}/description_merged.feather"))
predictions = arrow::read_feather(glue("{data_results_path}/predictions_{run}_metabolomics.feather"))
loghazards = arrow::read_feather(glue("{data_results_path}/loghazards_model_{run}_metabolomics.feather")) %>% pivot_longer(starts_with("logh"), names_to=c("endpoint", "features"), values_to="logh", names_pattern="logh_?(.*)_(.*)$")
</code>
<code>
base_size = 8
title_size = 10
facet_size = 10
geom_text_size=3
library(ggplot2);
theme_set(theme_classic(base_size = base_size) +
theme(strip.background = element_blank(), plot.title=element_text(size=title_size, hjust=0),
strip.text.x = element_text(size = facet_size),axis.title=element_text(size=10), axis.text=element_text(size=8, color="black"),
legend.position="bottom", axis.line = element_line(size = 0.2), axis.ticks=element_line(size=0.2)))
</code>
<code>
logh_NMR = loghazards %>% filter(split=="test") %>% left_join(data %>% select(eid, starts_with("NMR_"), -c(`NMR_measurement_quality_flagged`, `NMR_spectrometer`)) %>% filter(NMR_FLAG==TRUE), by="eid")
logh_NMR_long = logh_NMR %>% pivot_longer(starts_with("NMR_"), names_to="marker", values_to="value")
#corrs = logh_NMR_long %>% filter(marker!="NMR_FLAG") %>% group_by(endpoint, marker) %>% summarise(cor = cor(logh, value, use="complete.obs", method="pearson"))
</code>
<code>
library(ggforestplot)
</code>
<code>
# Deepexplainer
attributions = arrow::read_feather(glue("{data_results_path}/attributions_211026.feather")) %>% mutate(explainer="DeepExplainer")
</code>
## Attributions by shap
<code>
run="211007"
name = glue("benchmark_cindex_{run}")
benchmark_cindex_general = read_feather(glue("{data_results_path}/{name}.feather")) %>% distinct() %>% unite("score", c(module, features), remove=FALSE) %>% distinct()
</code>
<code>
library(ggdist)
perf_order = benchmark_cindex_general %>% filter(module=="DS", features=="Metabolomics") %>% group_by(endpoint) %>% median_qi(cindex) %>% arrange(desc(cindex))
endpoint_order_perf = perf_order$endpoint
</code>
<code>
library(ggthemes)
endpoint_map = c(
'M_MACE'='MACE',
'M_all_cause_dementia'='Dementia',
'M_type_2_diabetes'='T2 Diabetes',
'M_liver_disease'='Liver Disease',
'M_renal_disease'='Renal Disease',
'M_atrial_fibrillation'='Atrial Fibrillation',
'M_heart_failure'= 'Heart Failure',
'M_coronary_heart_disease'='CHD',
'M_venous_thrombosis'='Ven. Thrombosis',
'M_cerebral_stroke'='Cerebral Stroke',
'M_abdominal_aortic_aneurysm'='AAA',
'M_peripheral_arterial_disease'='PAD',
"M_chronic_obstructuve_pulmonary_disease" = "COPD",
"M_asthma" = "Asthma",
'M_parkinsons_disease' = "Parkinson's",
"M_lung_cancer" = "Lung Cancer",
"M_non_melanoma_skin_cancer" = "Skin Cancer",
"M_colon_cancer"= "Colon Cancer",
"M_rectal_cancer" = "Rectal Cancer",
"M_prostate_cancer"= "Prostate Cancer",
"M_breast_cancer" = "Breast Cancer",
'M_cataracts' = "Cataracts",
'M_glaucoma' = "Glaucoma",
'M_fractures' = "Fractures"
)
endpoint_order = c("M_MACE", "M_coronary_heart_disease", "M_cerebral_stroke", "M_all_cause_dementia", "M_heart_failure", "M_atrial_fibrillation",
"M_type_2_diabetes", "M_liver_disease", "M_renal_disease", "M_peripheral_arterial_disease", "M_venous_thrombosis", "M_abdominal_aortic_aneurysm",
"M_chronic_obstructuve_pulmonary_disease", "M_asthma", 'M_parkinsons_disease', 'M_cataracts', 'M_glaucoma', 'M_fractures',
"M_lung_cancer","M_non_melanoma_skin_cancer","M_colon_cancer","M_rectal_cancer","M_prostate_cancer","M_breast_cancer"
)
</code>
<code>
library(ggforestplot)
ng_names = df_NG_biomarker_metadata %>% mutate(metabolite = str_replace_all(tolower(description), " ", "_"))
ng_names %>% sample_n(10)
</code>
<code>
ng_names %>% select(group, subgroup) %>% distinct() %>% arrange(group, subgroup)
</code>
<code>
library(fuzzyjoin)
</code>
<code>
library(fuzzyjoin)
mets1 = attributions %>% select(metabolite) %>% distinct() %>% left_join(ng_names, by = "metabolite")
mets2 = mets1 %>% filter(is.na(name)) %>% select(metabolite) %>% stringdist_left_join(ng_names, by = "metabolite", max_dist = 1) %>%
rename(metabolite = metabolite.x) %>% select(-metabolite.y) %>% distinct()
mets3 = mets2 %>% filter(is.na(name)) %>% select(metabolite) %>% stringdist_left_join(ng_names, by = "metabolite", max_dist = 8) %>%
rename(metabolite = metabolite.x) %>% select(-metabolite.y) %>% distinct()
mets = bind_rows(mets1 %>% filter(!is.na(name)), mets2 %>% filter(!is.na(name)), mets3)
mets %>% sample_n(5)
</code>
<code>
attributions_metadata = attributions %>% left_join(mets %>% select(metabolite, abbreviation, group, subgroup), by="metabolite") %>% mutate(eid=as.integer(as.character(eid)))
</code>
<code>
library(gghighlight)
</code>
<code>
nmr_real = data %>% select(eid, starts_with("NMR_"), -`NMR_measurement_quality_flagged`, -`NMR_spectrometer`) %>%
filter(NMR_FLAG==TRUE) %>% pivot_longer(contains("NMR_"), names_to="metabolite", values_to="met_real") %>%
mutate(metabolite = str_remove_all(metabolite, "NMR_"))
</code>
<code>
prev_events = data %>% select(eid, starts_with("M_"), -ends_with("_event"), -ends_with("_time")) %>%
pivot_longer(contains("M_"), names_to="endpoint", values_to="event") %>% distinct()#%>%
#mutate(metabolite = str_remove_all(metabolite, "NMR_"))
prev_events %>% head()
</code>
<code>
clean_label = function(label){return(stringr::str_wrap(str_replace_all(label, "_", " "), 20))}
</code>
<code>
hrs = loghazards %>% filter(features=="Metabolomics") %>% mutate(hr = exp(logh)) %>% filter(split=="test") %>% select(eid, endpoint, hr)
</code>
## Global attributions
<code>
#n_eids = 10000
#eids = (attributions_metadata %>% select(eid) %>% distinct() %>% sample_n(n_eids))$eid
met_order_df = attributions_metadata %>% select(group, subgroup, metabolite, abbreviation) %>% distinct() %>% arrange(group, subgroup, abbreviation) %>% mutate(group_id = as.integer(factor(group)))
met_order = met_order_df$metabolite
abbrev_order = met_order_df$abbreviation
group_order = (met_order_df %>% select(group) %>% distinct())$group
#subgroup_order = (met_order_df %>% select(group, subgroup) %>% distinct())$subgroup
attrib_raw = attributions_metadata %>% #filter(eid %in% eids) %>%
left_join(nmr_real, by=c("eid", "metabolite")) %>%
left_join(hrs, by=c("eid", "endpoint")) %>%
left_join(prev_events, by=c("eid", "endpoint")) %>%
ungroup() %>% mutate(metabolite=factor(metabolite, levels=met_order)) %>%
mutate(abbreviation=factor(abbreviation, levels=abbrev_order))#%>% mutate(shap=raster::clamp(shap, -2, +2))
</code>
<code>
subgroup_order = c( 'Amino acids',
'Branched-chain amino acids',
'Aromatic amino acids',
'Fluid balance',
'Inflammation',
'Fatty acids',
'Glycolysis related metabolites',
'Ketone bodies',
'Total lipids',
'Cholesterol',
'Free cholesterol',
'Cholesteryl esters',
'Phospholipids',
'Triglycerides',
'Other lipids',
'Lipoprotein particle sizes',
'Lipoprotein particle concentrations',
'Chylomicrons and extremely large VLDL',
'Very large VLDL',
'Large VLDL',
'Medium VLDL',
'Small VLDL',
'Very small VLDL',
'Large LDL',
'Medium LDL',
'Small LDL',
'IDL',
'Very large HDL',
'Large HDL',
'Medium HDL',
'Small HDL',
'Apolipoproteins'
)
</code>
<code>
attrib_sample = attrib_raw %>% group_by(endpoint, metabolite, explainer) %>%
mutate(shap_quantile=ntile(shap, 100), met_quantile=ntile(met_value, 100))
</code>
<code>
attrib_sample_mean = attrib_sample %>% ungroup() %>%
group_by(endpoint, metabolite, abbreviation, group, subgroup, explainer, shap_quantile) %>%
summarise(met_quantile=mean(met_quantile), mean_shap = mean(shap), mean_met=mean(met_value))
</code>
<code>
library(ggforce)
</code>
<code>
endpoint_selection = c("M_MACE",
#'M_coronary_heart_disease',
#'M_cerebral_stroke',
"M_all_cause_dementia",
"M_type_2_diabetes",
"M_renal_disease",
"M_venous_thrombosis",
#"M_chronic_obstructuve_pulmonary_disease",
"M_asthma"
#'M_parkinsons_disease',
)
</code>
<code>
attrib_sample_mean = attrib_sample_mean %>% mutate(group_new = subgroup) %>% mutate(group_new=case_when(
str_ends(abbreviation, "-P") ~ "Lipoprotein particle concentrations",
str_ends(abbreviation, "-L") ~ "Total lipids",
str_ends(abbreviation, "-C") ~ "Cholesterol",
str_ends(abbreviation, "-FC") ~ "Free cholesterol",
str_ends(abbreviation, "-CE") ~ "Cholesteryl esters",
str_ends(abbreviation, "-PL") ~ "Phospholipids",
str_ends(abbreviation, "-TG") ~ "Triglycerides",
TRUE ~ subgroup))
</code>
<code>
temp_global = attrib_sample %>% group_by(endpoint, subgroup, metabolite, abbreviation) %>% summarise(global_shap = sum(abs(shap)))
</code>
<code>
met_selection = (temp_global %>% group_by(metabolite) %>% summarise(mean_global = mean(global_shap, na.rm=T)) %>% arrange(desc(abs(mean_global))) %>% head(75))$metabolite
</code>
<code>
plot_width=3.25; plot_height=10; plot_dpi=320
options(repr.plot.width = 3.25, repr.plot.height = plot_height, repr.plot.res=320)
attr_delta = ggplot(temp_global %>% filter(metabolite %in% met_selection) %>% mutate(subgroup = factor(subgroup, levels=subgroup_order)),
aes(x=factor(endpoint, levels=endpoint_order_perf), y=fct_rev(abbreviation), fill=abs(global_shap))) +
labs(x=NULL, y=NULL)+
geom_tile()+theme(plot.title = element_text(vjust = - 15)) +
scale_fill_gradient2(low = "darkblue",high = "#440154FF", midpoint = 0)+
theme(legend.position = "bottom")+
scale_x_discrete(labels=endpoint_map, position="top")+
scale_y_discrete(position="left")+
facet_grid(subgroup~., labeller=labeller(subgroup=label_wrap_gen(20)), scales="free", space="free")+
theme(axis.text.x= element_text(size=6), axis.text.y= element_text(size=5.5), strip.text.y.right = element_text(angle = 0, size=6))+
theme(axis.text.x.top= element_text(hjust=0, vjust=0.5)#, strip.text.y=element_blank()
)+
theme(strip.placement = 'outside') +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))+ theme(panel.spacing = unit(0.5, "lines"))
attr_delta
</code>
<code>
library(gt)
plot_name = "Figures_6_A_AttributionHeatmap75"
attr_delta %>% ggsave(filename=glue("outputs/{plot_name}.pdf"), device="pdf", width=plot_width, height=plot_height, dpi=plot_dpi)
</code>
<code>
met_selection_top = (temp_global %>% ungroup() %>% select(metabolite, subgroup) %>% distinct() %>% mutate(subgroup = factor(subgroup, levels=subgroup_order)) %>% arrange(subgroup) %>% head(84))$metabolite
plot_width=4; plot_height=10; plot_dpi=320
options(repr.plot.width = plot_width, repr.plot.height = plot_height, repr.plot.res=320)
attr_delta_full_left = ggplot(temp_global %>% filter(metabolite %in% met_selection_top) %>% mutate(subgroup = factor(subgroup, levels=subgroup_order)),
aes(x=factor(endpoint, levels=endpoint_order_perf), y=fct_rev(abbreviation), fill=abs(global_shap))) + # %>%
#filter(endpoint %in% c("M_type_2_diabetes", "M_all_cause_dementia")),
labs(x=NULL, y=NULL)+
#geom_quasirandom(size=0.1) +
geom_tile()+theme(plot.title = element_text(vjust = - 15)) +
scale_fill_gradient2(low = "darkblue",high = "#440154FF", midpoint = 0)+#, limits=c(-3, +3), oob=scales::squish) +
theme(legend.position = "none")+#coord_flip()+# xlim(-1, 1.2)+#coord_flip()+#, panel.grid.major = element_blank())+#+
scale_x_discrete(labels=endpoint_map, position="top")+
scale_y_discrete(position="left")+
facet_grid(subgroup~., labeller=labeller(subgroup=label_wrap_gen(25)), scales="free", space="free")+
theme(axis.text.x= element_text(size=6), axis.text.y= element_text(size=6), strip.text.y.right = element_text(angle = 0, size=6))+
theme(axis.text.x.top= element_text(hjust=0, vjust=0.5)#, strip.text.y=element_blank()
)+
theme(strip.placement = 'outside') +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))#+ theme(panel.spacing = unit(0.5, "lines"))
attr_delta_full_left #+ coord_polar()
</code>
<code>
plot_width=4; plot_height=10; plot_dpi=320
options(repr.plot.width = plot_width, repr.plot.height = plot_height, repr.plot.res=320)
attr_delta_full_right = ggplot(temp_global %>% filter(!metabolite %in% met_selection_top) %>% mutate(subgroup = factor(subgroup, levels=subgroup_order)),
aes(x=factor(endpoint, levels=endpoint_order_perf), y=fct_rev(abbreviation), fill=abs(global_shap))) + # %>%
#filter(endpoint %in% c("M_type_2_diabetes", "M_all_cause_dementia")),
labs(x=NULL, y=NULL)+
#geom_quasirandom(size=0.1) +
geom_tile()+theme(plot.title = element_text(vjust = - 15)) +
scale_fill_gradient2(low = "darkblue",high = "#440154FF", midpoint = 0)+#, limits=c(-3, +3), oob=scales::squish) +
theme(legend.position = "none")+#coord_flip()+# xlim(-1, 1.2)+#coord_flip()+#, panel.grid.major = element_blank())+#+
scale_x_discrete(labels=endpoint_map, position="top")+
scale_y_discrete(position="left")+
facet_grid(subgroup~., labeller=labeller(subgroup=label_wrap_gen(25)), scales="free", space="free")+
theme(axis.text.x= element_text(size=6), axis.text.y= element_text(size=6), strip.text.y.right = element_text(angle = 0, size=6))+
theme(axis.text.x.top= element_text(hjust=0, vjust=0.5)#, strip.text.y=element_blank()
)+
theme(strip.placement = 'outside') +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))#+ theme(panel.spacing = unit(0.5, "lines"))
attr_delta_full_right #+ coord_polar()
</code>
<code>
library(patchwork)
</code>
<code>
plot_width=8; plot_height=10; plot_dpi=320
options(repr.plot.width = plot_width, repr.plot.height = plot_height, repr.plot.res=320)
attr_delta_full_final = (attr_delta_full_left | attr_delta_full_right)
</code>
<code>
library(gt)
plot_name = "Suppl_Figures_7_AttributionHeatmapFull"
attr_delta_full_final %>% ggsave(filename=glue("outputs/{plot_name}.pdf"), device="pdf", width=plot_width, height=plot_height, dpi=plot_dpi)
</code>
|
{
"filename": "retina_phewas_evaluation_Figure6_Attributions_2_Heatmap.ipynb",
"repository": "lukl95/22",
"query": "transformed_from_existing",
"size": 26022,
"sha": ""
}
|
# CCS_week_8_1.ipynb
Repository: fsonak/VL
**Week 8: Monte Carlo on 2D Ising model phase transition**
Jannek Schaffert, Frédéric Sonak
This markdown was created with the assistance of ChatGPT, focusing on grammar, spelling, and readability.
**Background**
In this exercise, we implement a Monte Carlo (MC) simulation using the Metropolis algorithm to model a phase transition of the 2D Ising Model. The aforementioned model utilised the Hamiltonian
$
H = -\frac{1}{2} J \sum_{i<j}\ S_i\ S_j
$
with the Spins $S \pm 1$ and the coupling strength J
<code>
# Importing all utilised libraries
import matplotlib.pyplot as plt
from matplotlib import animation, rc
from IPython.display import HTML
from tqdm import tqdm # Import tqdm for progress visualisation
import time
from numba import njit
import copy
import numpy as np
import scipy as scy
</code>
<code>
# "fancy" visualisation of the spins as arrows
def plot_spin_configuration_with_arrows(grid):
"""
Plots a 2D array of spins as small vector arrows pointing up (blue) or down (red).
"""
N = grid.shape[0] # Grid size
# Create a grid of coordinates for plotting
x, y = np.meshgrid(np.arange(N), np.arange(N))
# Define arrow properties
u = np.zeros_like(grid) # No horizontal component
v = grid # Vertical component represents the spin direction (+1 or -1)
# Split into spins pointing up and down
up_mask = grid > 0
down_mask = grid < 0
# Plot arrows
plt.figure(figsize=(8, 8))
plt.quiver(x[up_mask], y[up_mask], u[up_mask], v[up_mask],
angles='xy', scale_units='xy', scale=1.5, color='blue', pivot='middle', label='Spin Up')
plt.quiver(x[down_mask], y[down_mask], u[down_mask], v[down_mask],
angles='xy', scale_units='xy', scale=1.5, color='red', pivot='middle', label='Spin Down')
plt.xlim(-0.5, N - 0.5)
plt.ylim(-0.5, N - 0.5)
plt.gca().set_aspect('equal')
plt.title("Spin Configuration")
plt.xlabel("x")
plt.ylabel("y")
plt.grid(False)
plt.xticks(range(N))
plt.yticks(range(N))
plt.legend(loc="upper right")
plt.show()
# Example grid for demonstration
# example_grid = np.random.choice([-1, 1], size=(8, 8))
# plot_spin_configuration_with_arrows(example_grid)
</code>
**Task I: Implementation and simulation**
First an energy calculation based on the Ising hamiltonian from above with periodic boundary conditions is implemented. J is set to 1J/mo and N to 16. Temperature Range with 50 Points between 0.18K and 0.4 K is used.
<code>
# initialise 2D grid of spins only
number_of_particles = 16
particles = np.random.choice([-1,1], size=(number_of_particles//2,number_of_particles//2))
def calculate_ising_energy(grid, J):
energy = 0
for i in range(number_of_particles):
for j in range(number_of_particles):
# Interaction with right and bottom neighbors (periodic boundary conditions)
energy -= J * grid[i, j] * (grid[i, (j+1) % number_of_particles] + grid[(i+1) % number_of_particles, j])
return energy
</code>
<code>
import numpy as np
import matplotlib.pyplot as plt
from tqdm import tqdm
def monte_carlo_ising(N, steps, temperature, J, record_interval,plot_spins_as_arrows = False, return_everything = False, verbose=True):
"""
Monte Carlo Simulation for the 2D Ising model using the Metropolis algorithm.
"""
# Initialize grid of spins (-1 or +1)
grid = np.random.choice([-1, 1], size=(N, N))
if verbose:
# Visualization of the initial spin configuration
if plot_spins_as_arrows == True:
plot_spin_configuration_with_arrows(grid)
else:
plt.imshow(grid, cmap='coolwarm', interpolation='none')
plt.colorbar(label='Spin')
plt.title('Initial Spin Configuration')
plt.show()
# Initialize arrays for recording trajectory and observables
traj = np.zeros((steps // record_interval, N, N))
energies = np.zeros(steps)
magnetisations = np.zeros(steps)
# Boltzmann constant
#TODO: add correct value
k_B = 1.0
def calculate_ising_energy(grid):
"""
Calculate the total energy of the system using the Ising Hamiltonian.
"""
energy = 0
for i in range(N):
for j in range(N):
# Add interaction with right and bottom neighbors (periodic boundary conditions)
energy -= J * grid[i, j] * (grid[i, (j+1) % N] + grid[(i+1) % N, j])
return energy
# Initialize energy and magnetisation
E = calculate_ising_energy(grid)
M = np.sum(grid)
for step in tqdm(range(steps), desc="Monte Carlo Progress"):
# Pick a random spin
i, j = np.random.randint(0, N, size=2)
# Calculate ΔH for flipping spin at (i, j)
delta_H = 2 * J * grid[i, j] * (
grid[i, (j+1) % N] + grid[i, (j-1) % N] +
grid[(i+1) % N, j] + grid[(i-1) % N, j]
)
# Metropolis acceptance criterion
if delta_H <= 0 or np.random.rand() < np.exp(-delta_H / (k_B * temperature)):
grid[i, j] *= -1 # Flip the spin
E += delta_H
M += 2 * grid[i, j] # Update magnetisation
# Record observables
energies[step] = E
magnetisations[step] = abs(M) / (N * N)
# Save spin configuration periodically
if step % record_interval == 0:
traj[step // record_interval] = grid.copy()
average_energy = np.average(energies)
average_magnetisation = np.sqrt((np.average(magnetisations)**2))
if verbose:
print(f'Average Energy = {average_energy:.2f}')
print(f'Average Magnetisation = {average_magnetisation:.2f}')
# Visualization of the final spin configuration
if plot_spins_as_arrows == True:
plot_spin_configuration_with_arrows(grid)
else:
plt.imshow(grid, cmap='coolwarm', interpolation='none')
plt.colorbar(label='Spin')
plt.title('Final Spin Configuration')
plt.show()
if return_everything:
return traj, energies, magnetisations, average_energy, average_magnetisation
else:
return average_energy, average_magnetisation
# Parameters
N = 16 # Grid size
steps = 10000 # Number of Monte Carlo steps
temperature = 2.5 # Temperature (in k_B units)
J = 1.0 # Coupling strength
record_interval = 100 # Interval for recording trajectory
# Run the simulation for one tmeperature
# traj, energies, magnetisations, average_energy, average_magnetisation = monte_carlo_ising(N, steps, temperature, J, record_interval)
# Run simulation for different temperatures
# initialise energies and magnetisations array
_list_of_average_energies = np.zeros(50)
list_of_average_magnetisations = np.zeros(50)
temperature_range = np.linspace(0.18, 4.0, 50)
for temperature in temperature_range:
counter = 0
print(f'calculating MC for temperature = {temperature:.2f}')
list_of_average_magnetisations[counter], list_of_average_magnetisations[counter] = monte_carlo_ising(N, steps, temperature, J, record_interval, False, False, False)
counter += 1
</code>
<code>
plt.plot(temperature_range, list_of_average_magnetisations)
</code>
<code>
# Plot energy and magnetisation over time
plt.figure()
plt.plot(energies, label='Energy per Spin')
plt.xlabel('Step')
plt.ylabel('Energy')
plt.title('Energy per Step')
plt.legend()
plt.show()
plt.figure()
plt.plot(magnetisations, label='magnetisation')
plt.xlabel('Step')
plt.ylabel('magnetisation')
plt.title('magnetisation per Step')
plt.legend()
plt.show()
</code>
<code>
# Monte Carlo Simulation from last sheet
def monte_carlo_simulation(particles, steps, step_shift, temperature, C12, C6, box_size, record_interval):
'''Monte-Carlo Simulation using Metropolis Algorithm with a Lennard-Jones Potential.'''
# Helper function to calculate the potential energy
def calculate_potential_energy(particles, C12, C6, iu, box_size):
#first calculate distances
# Calculate distances in x-direction
# first calculate matrix upper triangle distances
d_raw = (particles[1, :, np.newaxis] - particles[np.newaxis, 1])[iu]
# take into account periodic boundaries
dx = np.where(np.abs(d_raw) > box_size * 0.5, d_raw - box_size * np.sign(d_raw), d_raw)
# Calculate distances in y-direction
d_raw = (particles[2, :, np.newaxis] - particles[np.newaxis, 2])[iu]
dy = np.where(np.abs(d_raw) > box_size * 0.5, d_raw - box_size * np.sign(d_raw), d_raw)
# Total distances
# r = np.sqrt(dx**2 + dy**2)
# r6 = r**6
# for faster calculation don't calculate the square root but use r squared
r = dx**2 + dy**2
r6 = r**3
# Return potential energy
return np.sum(C12 / r6**2 - C6 / r6)
start_time = time.time() # Start timing the simulation
N = particles.shape[1]
R = 8.314462 # Gas constant
c = 1 / (R * temperature)
# initialise Arrays for storing trajectory and energy
# Trajectory storage at defined intervals
traj = np.ones((steps // record_interval, 2, N))
# Potential energy storage
energ_pot = np.ones(steps)
# to access the upper triangle of the matrix
# 1 excludes diagonal elements
iu = np.triu_indices(N, 1)
E_old = calculate_potential_energy(particles, C12, C6, iu, box_size)
energ_pot[0] = E_old
traj[0] = particles[[1, 2]]
for step in tqdm(range(1, steps), desc="Monte Carlo Progress"):
for i in range(N):
# Save old positions
pos_old = particles[1:3, i].copy()
# Apply random displacement
angles = np.random.uniform(0, 2 * np.pi)
displacement = np.array([np.cos(angles), np.sin(angles)]) * step_shift
particles[1:3, i] = (particles[1:3, i] + displacement) % box_size
# Calculate new energy
E_new = calculate_potential_energy(particles, C12, C6, iu, box_size)
# Metropolis acceptance
if (E_new < E_old) or (np.random.rand() < np.exp((E_old - E_new) * c)):
E_old = E_new # Accept move
else:
particles[1:3, i] = pos_old # Revert move
if step % record_interval == 0:
traj[step // record_interval] = particles[[1, 2]]
energ_pot[step] = E_old
end_time = time.time() # End timing the simulation
runtime = end_time - start_time # Calculate runtime
print(f"MC Simulation completed in {runtime:.2f} seconds.")
return energ_pot / N, traj
</code>
**Task 2: Simulation**
In this task, a 2D simulation box of size $5 \, \text{nm} \times 5 \, \text{nm}$ is created with periodic boundary conditions (PBC). The simulation models $49$ particles interacting via the Lennard-Jones potential, with the following parameters:
• $C_{12} = 9.847044 \times 10^{-6} \, \text{kJ/mol} \, \text{nm}^{12}$
• $C_{6} = 6.2647225 \times 10^{-3} \, \text{kJ/mol} \, \text{nm}^6$
The system is initialized at $293.15 K$, and $100,000$ single-particle moves are performed following the Monte Carlo (MC) algorithm. During the simulation, the potential energy of the system is calculated and used to evaluate the acceptance of particle moves.
Warning: MC simulations can be computationally intensive, especially with a large number of steps. If the simulation takes too long, reduce the number of steps (e.g., 2,000 steps) to test the setup before running the full simulation.
<code>
# Parameters for the simulation
box_size = 5.0 # Box size in nm
N_particles = 16 # Number of particles
temperature_range = np.linspace(0.18, 0.4, 50)
</code>
<code>
# Parameters for the simulation
box_size = 5.0 # Box size in nm
N_particles = 16 # Number of particles
temperature_range = np.linspace(0.18, 0.4, 50)
C12 = 9.847044e-3 # Lennard-Jones constant for repulsion kg, nm, ns
C6 = 6.2647225 # Lennard-Jones constant for attraction kg, nm, ns
temperature = 293.15 # Temperature in K
step_shift = 0.01 # Random displacement step size in nm
steps = 100000 # Total Monte Carlo steps
record_interval = 20 # Save trajectory every 20 steps
# manually placing particles on a evenly spaced grid
# Initialize particle positions (regular grid)
particles = np.zeros((5, N_particles)) # Mass, x, y, vx, vy
particles_with_spin = np.zeros((6, N_particles)) # Mass, x, y, vx, vy, spin
# Create a regular grid within the box
grid_size = int(np.sqrt(N_particles)) # Number of particles per row/column
spacing = box_size / grid_size # Spacing between particles
# Assign positions
x_coords = np.linspace(spacing / 2, box_size - spacing / 2, grid_size)
y_coords = np.linspace(spacing / 2, box_size - spacing / 2, grid_size)
x, y = np.meshgrid(x_coords, y_coords)
# create a 1D array
particles[1] = x.flatten()[:N_particles] # x-coordinates
particles[2] = y.flatten()[:N_particles] # y-coordinates
</code>
<code>
# Run the Monte Carlo simulation with the above parameters
energ_pot_mc, traj_mc = monte_carlo_simulation(particles, steps, step_shift, temperature, C12, C6, box_size, record_interval)
</code>
**Task 3: Potential energy analysis**
We analysed the system’s potential energy during the Monte Carlo simulation:
1. Potential Energy Over Steps:
The potential energy of the system was plotted as a function of simulation steps. This allowed us to observe how the energy evolves and whether the system approaches equilibrium.
2. Histogram of Equilibrated Potential Energies:
We assumed that the system reaches equilibrium during the second half of the simulation, where fluctuations in potential energy become stable. Using this data, a histogram of potential energies was calculated to visualize the distribution of energies in the equilibrated state.
<code>
# Plot the system’s potential energy over the number of steps
plt.figure(figsize=(8, 5))
plt.plot(range(steps), energ_pot_mc, label="Potential Energy MC")
plt.xlabel("Step")
plt.ylabel("Potential Energy (J/mol)")
plt.title("Potential Energy Over Time MC")
plt.legend()
plt.show()
</code>
The equilibrated part of the simulation is identified as the phase where the fluctuations in potential energy become "stable" (i.e. a converging constant mean), resulting in an almost horizontal trend in the potential energy plot. We assume that equilibrium is reached in the second half of the simulation. Therefore, the histogram of potential energies is calculated using data from the second half of the simulation steps.
<code>
# Select the last 50% of the steps
last_half_start_mc = len(energ_pot_mc) // 2
last_half_energies_mc = energ_pot_mc[last_half_start_mc:]
# Normalize potential energy per particle for the last half
last_half_energies_per_particle_mc = last_half_energies_mc / particles.shape[1]
# Divide by number of particles
# Plot histogram
plt.figure(figsize=(8, 5))
plt.title("MC: Potential Energies MC (Last 50% of Steps)")
plt.hist(last_half_energies_per_particle_mc, bins=50, edgecolor='k', alpha=0.7, label="Histogram")
plt.xlabel("$E_{pot}$ per particle (J/mol)")
plt.ylabel("Number of occurrences")
plt.show()
</code>
Does it look like a Boltzmann distribution? I would say yes :-)
**Task IV: RDF and free energy comparison with MD**
To compare the MC simulation with the MD simulation the MD simulation has to be rerun from previous excercises.
<code>
# Function to calculate forces and potential energy
def calculate_forces(particles, box_size, c12, c6):
"""
Vectorized calculation of Lennard-Jones forces and potential energy.
Parameters:
particles: Array with particle positions (shape: (2, N)).
box_size: Size of the simulation box (scalar).
c12, c6: Lennard-Jones constants.
Returns:
forces: Array of forces acting on particles (shape: (2, N)).
potential_energy: Total potential energy of the system.
"""
# Compute pairwise distances with periodic boundary conditions
delta = particles[:, np.newaxis, :] - particles[:, :, np.newaxis] # Shape: (2, N, N)
delta -= box_size * np.round(delta / box_size) # Apply periodic boundary conditions
# Squared distances (don't use sqrt for compuational reasons) (N, N)
r2 = np.sum(delta**2, axis=0)
# Mask out self-interaction
np.fill_diagonal(r2, np.inf)
# Lennard-Jones potential
r6 = r2**3
r12 = r6**2
potential_energy_matrix = c12 / r12 - c6 / r6
# Total potential energy
potential_energy = np.sum(potential_energy_matrix) * 0.5
# Lennard-Jones force magnitudes (gradient of potential)
force_magnitude = (12 * c12 / r12 - 6 * c6 / r6) / r2
# Calculate forces
forces = np.sum(force_magnitude * delta, axis=1) # Shape: (2, N)
return forces, potential_energy
# MD Simulation Function (Velocity-Verlet Algorithm)
def run_md_simulation(particles, box_size, c12, c6, time_step, n_steps, mass):
"""
Perform a Molecular Dynamics simulation using the Velocity-Verlet algorithm.
"""
start_time = time.time()
N = particles.shape[1] # Number of particles
positions = particles[1:3] # x, y positions
velocities = particles[3:5] # x, y velocities
accelerations = np.zeros_like(positions) # Initialize accelerations
# Initialize arrays to store simulation results
trajectories = np.zeros((n_steps, 2, N)) # Store particle positions over time
energies = np.zeros((n_steps, 2)) # Store kinetic and potential energies over time
# Initial force and potential energy calculation
forces, potential_energy = calculate_forces(positions, box_size, c12, c6)
accelerations = forces / mass # a = F / m
for step in tqdm(range(n_steps), desc="MD Simulation Progress"):
# Velocity-Verlet Integration
# 1. Update positions
positions += velocities * time_step + 0.5 * accelerations * time_step**2
positions %= box_size # Apply periodic boundary conditions
# 2. Calculate new forces
forces, potential_energy = calculate_forces(positions, box_size, c12, c6)
# 3. Update velocities
new_accelerations = forces / mass
velocities += 0.5 * (accelerations + new_accelerations) * time_step
accelerations = new_accelerations
# Store results
trajectories[step] = positions
kinetic_energy = 0.5 * mass * np.sum(velocities**2)
energies[step] = [kinetic_energy, potential_energy]
end_time = time.time() # End timing the simulation
runtime = end_time - start_time # Calculate runtime
print(f"MD Simulation completed in {runtime:.2f} seconds.")
return trajectories, energies
</code>
<code>
# Run the MD simulation, for clarity parameters from the MC simulation is just copied down here
# Parameters for the simulation
box_size = 5.0 # Box size in nm
N_particles = 49 # Number of particles
C12 = 9.847044e-3 # Lennard-Jones constant for repulsion kg, nm, ns
C6 = 6.2647225 # Lennard-Jones constant for attraction kg, nm, ns
temperature = 293.15 # Temperature in K
step_shift = 0.01 # Random displacement step size in nm
steps = 100000 # Total Monte Carlo steps
record_interval = 20 # Save trajectory every 20 steps
mass = 1.0
time_step = 0.001
# Test parameters
box_size = 5.0 # Box size in nm
N_particles = 49 # Number of particles
c12 = 9.847044e-6 # Lennard-Jones constant (repulsion)
c6 = 6.2647225e-3 # Lennard-Jones constant (attraction)
time_step = 0.001 # Time step in ns
n_steps = 100000 # Number of simulation steps
mass = 1.0 # Mass of particles
temperature = 293.15 # Temperature in K
# Initialize particle positions and velocities
particles = np.zeros((5, N_particles)) # Mass, x, y, vx, vy
grid_size = int(np.sqrt(N_particles)) # Number of particles per row/column
spacing = box_size / grid_size # Spacing between particles
x_coords = np.linspace(spacing / 2, box_size - spacing / 2, grid_size)
y_coords = np.linspace(spacing / 2, box_size - spacing / 2, grid_size)
x, y = np.meshgrid(x_coords, y_coords)
particles[1] = x.flatten()[:N_particles] # x-coordinates
particles[2] = y.flatten()[:N_particles] # y-coordinates
particles[3:5] = np.random.randn(2, N_particles) # Random initial velocities
# Run the MD simulation
trajectories_md, energies_md = run_md_simulation(particles, box_size, c12, c6, time_step, n_steps, mass)
# Verify results
# Plot total energy over time
plt.figure(figsize=(8, 5))
total_energy = energies_md[:, 0] + energies_md[:, 1]
plt.plot(range(n_steps), total_energy, label="Total Energy")
plt.plot(range(n_steps), energies_md[:, 0], label="Kinetic Energy")
plt.plot(range(n_steps), energies_md[:, 1], label="Potential Energy")
plt.xlabel("Simulation Steps")
plt.ylabel("Energy (arbitrary units)")
plt.title("Energy Conservation in MD Simulation")
plt.legend()
plt.show()
</code>
Comparing Potential energy distributions: MC vs MD
<code>
# Extract the last 50% of the steps for potential energy
last_half_start_md = len(energies_md) // 2
last_half_potentials_md = energies_md[last_half_start_md:, 1] # Extract potential energy
# Normalize potential energy per particle for the last half
last_half_potentials_per_particle_md = last_half_potentials_md / particles.shape[1] # Divide by number of particles
# Plot histogram of potential energy per particle
plt.figure(figsize=(12, 10))
plt.subplot(2,2,1)
plt.title("MD: Potential Energies (Last 50% of Steps)")
plt.hist(last_half_potentials_per_particle_md, bins=50, edgecolor='k', alpha=0.7, label="Histogram")
plt.xlabel("$E_{pot}$ per particle ")
plt.ylabel("Number of occurrences")
plt.legend()
plt.subplot(2,2,2)
plt.title("MC: Potential Energies (Last 50% of Steps)")
plt.hist(last_half_energies_per_particle_mc, bins=50, edgecolor='k', alpha=0.7, label="Histogram")
plt.xlabel("$E_{pot}$ per particle J/mol")
plt.ylabel("Number of occurrences")
plt.legend()
plt.tight_layout()
plt.show()
</code>
Both distributions look kind of like a Boltzmann distribution. The maximum of the MD simulation is shifted to the right.
Which method is faster?
Having measured the times for the simulations we can clearly state that the MD simulation is significantly faster than the MC simulation.
Look at the trajectory / the position histogram: which method gives a better
sampling of the coordinate space?
<code>
plt.figure(figsize=(12, 10))
# MC: x positions
plt.subplot(2, 2, 1)
plt.title("MC: x-Positions")
plt.hist(traj_mc[:, 0].flatten(), bins=80, edgecolor='k')
plt.xlabel("x (nm)")
plt.ylabel("Number of occurrences")
# MC: x positions
plt.subplot(2, 2, 2)
plt.title("MC: y-Positions")
plt.hist(traj_mc[:, 1].flatten(), bins=80, edgecolor='k')
plt.xlabel("x (nm)")
plt.ylabel("Number of occurrences")
# MD: x positions
plt.subplot(2, 2, 3)
plt.title("MD: x-Positions")
plt.hist(trajectories_md[:, 0].flatten(), bins=80, edgecolor='k')
plt.xlabel("x (nm)")
plt.ylabel("Number of occurrences")
# MD: y positions
plt.subplot(2, 2, 4)
plt.title("MD: y-Positions")
plt.hist(trajectories_md[:, 1].flatten(), bins=80, edgecolor='k')
plt.xlabel("x (nm)")
plt.ylabel("Number of occurrences")
</code>
The sampling appears quite similar between both methods, the MD might be slightly better.
Since our calculation of the RDF from last week turned out to be incorrect and we haven’t yet found a proper solution, we decided not to include this part in the analysis for now. Hopefully, we’ll see a working version during the tutorial on Friday, and we can consider implementing it into this task afterward. 😊
|
{
"filename": "CCS_week_8_1.ipynb",
"repository": "fsonak/VL",
"query": "transformed_from_existing",
"size": 269506,
"sha": ""
}
|
# BioPython-II.ipynb
Repository: chiraltraining/bioinfo-case-studies
# BioPython-II
## Agenda
- Sequence alignment
- Phylogenetics
- Cluster analysis
|
{
"filename": "BioPython-II.ipynb",
"repository": "chiraltraining/bioinfo-case-studies",
"query": "transformed_from_existing",
"size": 1704,
"sha": ""
}
|
# 01_data_encoding_ATAC_1overlap.HepG2_1.ipynb
Repository: gersteinlab/DECODE
<code>
#-----import packages-----#
#common python packages
import numpy as np
import string
import random
import os
import pickle
import argparse
import wget
import math
import matplotlib.pyplot as plt
from datetime import datetime
#biological packages
import pybedtools
from pybedtools import featurefuncs
import pyBigWig
</code>
<code>
# -----parsing command line arguments-----#
parser = argparse.ArgumentParser(description='Training CNN model to predict STARR-seq enhancers based on chromatin accessbility and histone marks')
parser.add_argument('-s', '--starrseq', type=str, help='comma separated string of starrseq peak replicates')
parser.add_argument('-a', '--track1_peaks', type=str, help='chromatin accessibility peak')
parser.add_argument('-b', '--track2_peaks', type=str, help='ChIP-seq H3K27ac peak')
parser.add_argument('-c', '--track3_peaks', type=str, help='ChIP-seq H3K4me3 peak')
parser.add_argument('-d', '--track4_peaks', type=str, help='ChIP-seq H3K9ac peak')
parser.add_argument('-e', '--track5_peaks', type=str, help='ChIP-seq H3K4me1 peak')
parser.add_argument('-f', '--track1_bw', type=str, help='chromatin accessibility bigWig')
parser.add_argument('-g', '--track2_bw', type=str, help='ChIP-seq H3K27ac bigWig')
parser.add_argument('-i', '--track3_bw', type=str, help='ChIP-seq H3K4me3 bigWig')
parser.add_argument('-j', '--track4_bw', type=str, help='ChIP-seq H3K9ac bigWig')
parser.add_argument('-k', '--track5_bw', type=str, help='ChIP-seq H3K4me1 bigWig')
parser.add_argument('-o', '--out_dir', type=str, help='output_directory')
parser.add_argument('-x', '--cell_name', type=str, help='name of the cell')
parser.add_argument('-y', '--pos_neg_ratio', type=int, help='positive to negative ratio')
parser.add_argument('-z', '--window_size', type=int, help='prediction window size')
#temporary experiment in local directory
cell_type = os.environ['cell_type']
# cell_type = "A549"
#simulate command line input
stardir = "/gpfs/ysm/scratch60/gerstein/zc264/ChromVar/enhancer-prediction/encode/starrpeaker_positive/raw/"
seqdir = "/gpfs/ysm/scratch60/gerstein/zc264/ChromVar/enhancer-prediction/encode/datasets/"+cell_type+"/"
cmdline_str='-s ' + stardir + cell_type + '_r1_starrpeaker.peak.final.bed' + ',' + \
stardir + cell_type+'_r2_starrpeaker.peak.final.bed' + \
' -a ' + seqdir+cell_type+".ATAC-seq.narrowPeak" + \
' -b ' + seqdir+cell_type+".ChIP-seq.H3K27ac.narrowPeak" + \
' -c ' + seqdir+cell_type+".ChIP-seq.H3K4me3.narrowPeak" + \
' -d ' + seqdir+cell_type+".ChIP-seq.H3K9ac.narrowPeak" + \
' -e ' + seqdir+cell_type+".ChIP-seq.H3K4me1.narrowPeak" + \
' -f ' + seqdir+cell_type+".ATAC-seq.bigWig" + \
' -g ' + seqdir+cell_type+".ChIP-seq.H3K27ac.bigWig" + \
' -i ' + seqdir+cell_type+".ChIP-seq.H3K4me3.bigWig" + \
' -j ' + seqdir+cell_type+".ChIP-seq.H3K9ac.bigWig" + \
' -k ' + seqdir+cell_type+".ChIP-seq.H3K4me1.bigWig" + \
' -o ' + "/gpfs/ysm/scratch60/gerstein/zc264/ChromVar/enhancer-prediction/encode/dev/encoded_1overlap/ATAC/" + \
' -x ' + cell_type + \
' -y ' + "1" + \
' -z ' + "4000"
#print(cmdline_str.split())
#check if the files are there
args = parser.parse_args(cmdline_str.split())
args.starrseq = args.starrseq.split(",")
for key, value in vars(args).items():
#print(key, value)
if type(value) is list:
for v in value:
if not os.path.exists(v):
print(key + " argument file does not exist")
exit(1)
elif key == "out_dir" or key == "cell_name" or key == "pos_neg_ratio" or key == "window_size":
continue
else:
if not os.path.exists(value):
print(key + " argument file does not exist")
exit(1)
print("all files found!")
#construct a set of autosome + X chromosome names
chromosomes = []
for i in range(1,23):
chromosomes.append("chr"+str(i))
chromosomes.append("chrX")
print(chromosomes)
os.system("mkdir -p " + args.out_dir)
</code>
<code>
#-----IO and combine the raw STARR-seq replicate files-----#
if len(args.starrseq) == 1:
s = pybedtools.BedTool(args.starrseq[0]).filter(lambda x: float(x[9]) > 1.30).sort().merge()
else:
s1 = pybedtools.BedTool(args.starrseq[0]).filter(lambda x: float(x[9]) > 1.30).sort()
s2 = pybedtools.BedTool(args.starrseq[1]).filter(lambda x: float(x[9]) > 1.30).sort()
#s = s1.intersect(s2).filter(pybedtools.featurefuncs.greater_than, 300).sort().merge() #only 4000 positives
s = s1.cat(s2).sort().merge().filter(pybedtools.featurefuncs.greater_than, 150).sort() #9000 positives
print(s.count())
</code>
<code>
#-----IO and preprocess the signal files-----#
chromAcc = pybedtools.BedTool(args.track1_peaks).sort().merge()
chip1 = pybedtools.BedTool(args.track2_peaks).sort().merge()
chip2 = pybedtools.BedTool(args.track3_peaks).sort().merge()
chip3 = pybedtools.BedTool(args.track4_peaks).sort().merge()
chip4 = pybedtools.BedTool(args.track5_peaks).sort().merge()
#intersect combined STARR-seq peaks with chromatin accessibility, and filter for regions >150bp (TODO: might be to short)
starr_and_chromAcc = s.intersect(chromAcc, sorted=True,).filter(pybedtools.featurefuncs.greater_than, 150).sort()
#intersect STARR+chrom with H3K27ac, and filter for regions >150bp
starr_and_chip1 = s.intersect(chip1, sorted=True).filter(pybedtools.featurefuncs.greater_than, 150).sort()
#intersect STARR+chrom with H3K4me3, and filter for regions >150bp
starr_and_chip2 = s.intersect(chip2, sorted=True).filter(pybedtools.featurefuncs.greater_than, 150).sort()
#intersect STARR+chrom with H3K9ac, and filter for regions >150bp
starr_and_chip3 = s.intersect(chip3, sorted=True).filter(pybedtools.featurefuncs.greater_than, 150).sort()
#intersect STARR+chrom with H3K4me1, and filter for regions >150bp
starr_and_chip4 = s.intersect(chip4, sorted=True).filter(pybedtools.featurefuncs.greater_than, 150).sort()
#combined STARR+chrom+ChIP
catted_training = starr_and_chromAcc.cat(starr_and_chip1).cat(starr_and_chip2).cat(starr_and_chip3).cat(starr_and_chip4).filter(lambda x: x.chrom in chromosomes)
#center the overlapped regions and extend both sides up to half window size, making all regions uniformly window size
positive_training = catted_training.each(pybedtools.featurefuncs.midpoint).slop(b=args.window_size/2, genome="hg38").filter(pybedtools.featurefuncs.greater_than, args.window_size-1).sort()
#report total number of peaks
print("total chrom peaks: " + str(chromAcc.count()))
print("total STARR+chrom peaks: " + str(starr_and_chromAcc.count()))
print("total peaks: " + str(positive_training.count()))
print((s-chromAcc).count())
print((chromAcc-s).count())
positive_training.saveas(args.out_dir + args.cell_name + ".positive.bed")
</code>
<code>
#-----create negative samples-----#
#generate 2000bp of the entire genome
hg38_windows = pybedtools.BedTool().window_maker(genome="hg38", w=args.window_size).filter(pybedtools.featurefuncs.greater_than, args.window_size-1).filter(lambda x: x.chrom in chromosomes)
#remove ENCODE blacklist regions
if not os.path.exists('./hg38.blacklist.bed.gz'):
url = 'http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/hg38-human/hg38.blacklist.bed.gz'
wget.download(url, './hg38.blacklist.bed.gz')
blacklist = pybedtools.BedTool('./hg38.blacklist.bed.gz')
hg38_windows = hg38_windows - blacklist
#remove STARR-seq regions
#hg38_windows = (hg38_windows - s)
hg38_windows = hg38_windows - positive_training
print("original negative window: " + str(hg38_windows.count()))
#downsample negative to 10x of positive
negative_training = hg38_windows.random_subset(positive_training.count() * 10).sort()
print("downsampled negative window: " + str(negative_training.count()))
negative_training.saveas(args.out_dir + args.cell_name + ".negative.bed")
</code>
<code>
#IO the bigwig signals
chromAcc_bw = pyBigWig.open(args.track1_bw)
chip1_bw = pyBigWig.open(args.track2_bw)
chip2_bw = pyBigWig.open(args.track3_bw)
chip3_bw = pyBigWig.open(args.track4_bw)
chip4_bw = pyBigWig.open(args.track5_bw)
def bigWigAverageOverBed(x, bigwig):
return bigwig.stats(x.chrom, x.start, x.stop, nBins=400)
def get_signal(region, bigwig):
return np.array([np.nan_to_num(np.array(bigWigAverageOverBed(x, bigwig), dtype=float)) for x in region])
</code>
<code>
pos_sig_mat = get_signal(pybedtools.BedTool(args.out_dir + args.cell_name + "." + "positive.bed"), chromAcc_bw)
neg_sig_mat = get_signal(pybedtools.BedTool(args.out_dir + args.cell_name + "." + "negative.bed"), chromAcc_bw)
#signal_mat = np.vstack((pos_sig_mat, neg_sig_mat))
#print(signal_mat.shape)
np.savetxt(args.out_dir + args.cell_name+"."+"ATAC"+".pos.tsv", pos_sig_mat, fmt='%s', delimiter='\t')
np.savetxt(args.out_dir + args.cell_name+"."+"ATAC"+".neg.tsv", neg_sig_mat, fmt='%s', delimiter='\t')
plt.boxplot(pos_sig_mat, showfliers=False);
plt.boxplot(neg_sig_mat, showfliers=False);
</code>
<code>
pos_sig_mat = get_signal(pybedtools.BedTool(args.out_dir + args.cell_name + "." + "positive.bed"), chip1_bw)
neg_sig_mat = get_signal(pybedtools.BedTool(args.out_dir + args.cell_name + "." + "negative.bed"), chip1_bw)
np.savetxt(args.out_dir + args.cell_name+"."+"H3K27ac"+".pos.tsv", pos_sig_mat, fmt='%s', delimiter='\t')
np.savetxt(args.out_dir + args.cell_name+"."+"H3K27ac"+".neg.tsv", neg_sig_mat, fmt='%s', delimiter='\t')
plt.boxplot(pos_sig_mat, showfliers=False);
plt.boxplot(neg_sig_mat, showfliers=False);
</code>
<code>
pos_sig_mat = get_signal(pybedtools.BedTool(args.out_dir + args.cell_name + "." + "positive.bed"), chip2_bw)
neg_sig_mat = get_signal(pybedtools.BedTool(args.out_dir + args.cell_name + "." + "negative.bed"), chip2_bw)
np.savetxt(args.out_dir + args.cell_name+"."+"H3K4me3"+".pos.tsv", pos_sig_mat, fmt='%s', delimiter='\t')
np.savetxt(args.out_dir + args.cell_name+"."+"H3K4me3"+".neg.tsv", neg_sig_mat, fmt='%s', delimiter='\t')
plt.boxplot(pos_sig_mat, showfliers=False);
plt.boxplot(neg_sig_mat, showfliers=False);
</code>
<code>
pos_sig_mat = get_signal(pybedtools.BedTool(args.out_dir + args.cell_name + "." + "positive.bed"), chip3_bw)
neg_sig_mat = get_signal(pybedtools.BedTool(args.out_dir + args.cell_name + "." + "negative.bed"), chip3_bw)
np.savetxt(args.out_dir + args.cell_name+"."+"H3K9ac"+".pos.tsv", pos_sig_mat, fmt='%s', delimiter='\t')
np.savetxt(args.out_dir + args.cell_name+"."+"H3K9ac"+".neg.tsv", neg_sig_mat, fmt='%s', delimiter='\t')
plt.boxplot(pos_sig_mat, showfliers=False);
plt.boxplot(neg_sig_mat, showfliers=False);
</code>
<code>
pos_sig_mat = get_signal(pybedtools.BedTool(args.out_dir + args.cell_name + "." + "positive.bed"), chip4_bw)
neg_sig_mat = get_signal(pybedtools.BedTool(args.out_dir + args.cell_name + "." + "negative.bed"), chip4_bw)
np.savetxt(args.out_dir + args.cell_name+"."+"H3K4me1"+".pos.tsv", pos_sig_mat, fmt='%s', delimiter='\t')
np.savetxt(args.out_dir + args.cell_name+"."+"H3K4me1"+".neg.tsv", neg_sig_mat, fmt='%s', delimiter='\t')
plt.boxplot(pos_sig_mat, showfliers=False);
plt.boxplot(neg_sig_mat, showfliers=False);
</code>
|
{
"filename": "01_data_encoding_ATAC_1overlap.HepG2_1.ipynb",
"repository": "gersteinlab/DECODE",
"query": "transformed_from_existing",
"size": 44504,
"sha": ""
}
|
# Untitled2_1.ipynb
Repository: LibbyLi667/github-upload
<code>
test = ColumnDataSource(get_data_all('PassQC'))
</code>
<code>
plot = make_plot_all(test,'PassQC')
show(plot)
</code>
<code>
''' Create a simple genomics data stats dashboard.
Choose pages to show in the drop down widgets, and make selections
on the plots to update the summary and histograms accordingly.
.. note::
Use the ``bokeh serve`` command to run the example by executing:
bokeh serve --show mystats_page.py
at your command prompt. Then navigate to the URL
http://localhost:5006/stocks
.. _README: https://github.com/bokeh/bokeh/blob/master/examples/app/stocks/README.md
'''
from functools import lru_cache
from os.path import dirname, join
from math import pi
import pandas as pd
from bokeh.io import curdoc, output_file, show
from bokeh.layouts import column, row
from bokeh.models import ColumnDataSource, PreText, Select, Dropdown, HoverTool
from bokeh.plotting import figure, curdoc
from bokeh.transform import cumsum
def dict_extract_all(datatype_list, dic, item): ### count the number of true/false and return a dictionary for all data types
list_return_all = []
for i in datatype_list:
i = join(i + '_' + item)
list_return_all += dic[i]
dict_return_all = dict((x,list_return_all.count(x)) for x in [True,False])
return dict_return_all
def dict_extract(datatype_name, dic, item): ### given data type, dictionary, to count the number of true/false and return a dictionary containg the number for each data type
list_return = dic[datatype_name + '_' + item]
dict_return = dict((x,list_return.count(x)) for x in [True,False])
return dict_return
def angle(data):
data['angle'] = data['value']/data['value'].sum() * 2*pi
return
chart_colors = ['#007bff','#e29e44','#44e5e2','#eeeeee','#d8e244','#e244db']
def get_data_all(item):
data = dict_extract_all(data_types, dictA, item)
source = pd.Series(data).reset_index(name='value').rename(columns={'index':'number'})
angle(source)
source['color'] = chart_colors[:len(data)]
return ColumnDataSource(source)
def get_data_project(project, item):
dictP = group.get_group(project).set_index('ProjectName').to_dict('list')
data = dict_extract_all(data_types, dictP, item)
source = pd.Series(data).reset_index(name='value').rename(columns={'index':'number'})
angle(source)
source['color'] = chart_colors[:len(data)]
return ColumnDataSource(source)
def make_plot_all(source, item):
hover = HoverTool(tooltips="@number: @value")
start_angle=cumsum('angle', include_zero=True)
end_angle=cumsum('angle')
color = 'color'
legend='number'
fig = figure(plot_height=350, plot_width=350, title= 'OVERALL '+item+' RATE', toolbar_location=None,
x_range=(-0.5, 1.0))
fig.add_tools(hover)
fig.wedge(x=0, y=1, radius=0.4,
start_angle=start_angle, end_angle=end_angle,
line_color="white", color=color, legend=legend, source=source)
fig.axis.axis_label=None
fig.axis.visible=False
fig.grid.grid_line_color = None
plots = []
row_num = 3
n = 0
for i in data_types:
data = dict_extract(i, dictA, item)
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'number'})
data2['color'] = chart_colors[:len(data)]
angle(data2)
p = join('fig' + str(n))
p = figure(plot_height=350, plot_width=350, title= 'OVERALL ' +item + ' RATE BY DATA TYPE', toolbar_location=None,
tools="hover", tooltips="@number: @value", x_range=(-0.5, 1.0))
p.wedge(x=0, y=1, radius=0.4,
start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'),
line_color="white", color='color', legend='number', source=data2)
p.xaxis.axis_label=i
p.yaxis.visible=False
p.grid.grid_line_color = None
plots.append(p)
return row(fig,*plots)
def make_plot_project(source, project, item): ### show the figure with the project name such as 'P1', 'P2'
fig = figure(plot_height=350, plot_width=350, title= project + ' OVERALL ' + item +' RATE', toolbar_location=None,
tools="hover", tooltips="@number: @value", x_range=(-0.5, 1.0))
fig.wedge(x=0, y=1, radius=0.4,
start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'),
line_color="white", color='color', legend='number',source=source)
fig.axis.axis_label=None
fig.axis.visible=False
fig.grid.grid_line_color = None
plots = []
n = 0
for i in data_types:
dictP = group.get_group(project).set_index('ProjectName').to_dict('list')
data = dict_extract(i, dictP, item)
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'number'})
data2['color'] = chart_colors[:len(data)]
angle(data2)
p = figure(plot_height=350, plot_width=350, title= project+ ' '+ item +' RATE BY DATA TYPE', toolbar_location=None,
tools="hover", tooltips="@number: @value", x_range=(-0.5, 1.0))
p.wedge(x=0, y=1, radius=0.4,
start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'),
line_color="white", color='color',legend='number',source=data2)
p.xaxis.axis_label = i
p.yaxis.visible=False
p.grid.grid_line_color = None
plots.append(p)
return row(fig,*plots)
def update_plot(attrname, old, new):
item = ticker1.value
update1 = get_data_all(item)
update2 = get_data_project(ticker2.value, item)
source_all.update(update1)
source_project.update(update2)
#plot_overall = make_plot_all(source,item)
#plot_project = make_plot_project(source2, project, item)
#plots = column(plot_all,plot_project)
def update(selected=None):
t1, t2 = ticker1.value, ticker2.value
data1 = get_data_all(t1)
data2 = get_data_project(t2, t1)
source_all.data = data
source_project.data = data
# set up data
item = 'Returned'
project = 'P1'
item_list = ['Returned', 'PassQC', 'Processed']
data_types = ['RNA_seq','DNA_seq','Methyl_seq']
project_list = ['P1','P2','P3']
# set up widgets
ticker1 = Select(title="Pages:", value=item, options=item_list)
ticker2 = Select(title="Projects:", value=project, options=project_list)
#the datasets
data = pd.read_csv("/Users/xli677/Dropbox (Uni of Auckland)/xli677/Projects/MyTardis/Visualisation_with_Bokeh/SampleSheet_Bokeh_test.csv")
data2 = data.set_index('ProjectName')
group = data.groupby('ProjectName')
dictA = data2.to_dict('list')
source_all = get_data_all(item)
source_project = get_data_project(project, item)
plot_all = make_plot_all(source_all, item)
plot_project = make_plot_project(source_project, project,item)
ticker1.on_change('value',update_plot)
ticker2.on_change('value',update_plot)
# set up layout
widgets = column(ticker1, ticker2)
plots = column(plot_all,plot_project)
layout = column(widgets, plots)
# initialize
update()
curdoc().add_root(layout)
curdoc().title = "Genomics Stats"
</code>
<code>
test1 = get_data_all('PassQC')def dict_extract_all(datatype_list, dic, item): ### count the number of true/false and return a dictionary for all data types
list_return_all = []
for i in datatype_list:
i = join(i + '_' + item)
list_return_all += dic[i]
dict_return_all = dict((x,list_return_all.count(x)) for x in [True,False])
return dict_return_all
def dict_extract(datatype_name, dic, item): ### given data type, dictionary, to count the number of true/false and return a dictionary containg the number for each data type
list_return = dic[datatype_name + '_' + item]
dict_return = dict((x,list_return.count(x)) for x in [True,False])
return dict_return
def angle(data):
data['angle'] = data['value']/data['value'].sum() * 2*pi
return
chart_colors = ['#007bff','#e29e44','#44e5e2','#eeeeee','#d8e244','#e244db']
def get_data_all(item):
data = dict_extract_all(data_types, dictA, item)
source = pd.Series(data).reset_index(name='value').rename(columns={'index':'number'})
angle(source)
source['color'] = chart_colors[:len(data)]
return ColumnDataSource(source)
def get_data_project(project, item):
dictP = group.get_group(project).set_index('ProjectName').to_dict('list')
data = dict_extract_all(data_types, dictP, item)
source = pd.Series(data).reset_index(name='value').rename(columns={'index':'number'})
angle(source)
source['color'] = chart_colors[:len(data)]
return ColumnDataSource(source)
def make_plot_all(source, item):
hover = HoverTool(tooltips="@number: @value")
start_angle=cumsum('angle', include_zero=True)
end_angle=cumsum('angle')
color = 'color'
legend='number'
fig = figure(plot_height=350, plot_width=350, title= 'OVERALL '+item+' RATE', toolbar_location=None,
x_range=(-0.5, 1.0))
fig.add_tools(hover)
fig.wedge(x=0, y=1, radius=0.4,
start_angle=start_angle, end_angle=end_angle,
line_color="white", fill_color=color, legend=legend, source=source)
fig.axis.axis_label=None
fig.axis.visible=False
fig.grid.grid_line_color = None
plots = []
row_num = 3
n = 0
for i in data_types:
data = dict_extract(i, dictA, item)
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'number'})
data2['color'] = chart_colors[:len(data)]
angle(data2)
p = join('fig' + str(n))
p = figure(plot_height=350, plot_width=350, title= 'OVERALL ' +item + ' RATE BY DATA TYPE', toolbar_location=None,
tools="hover", tooltips="@number: @value", x_range=(-0.5, 1.0))
p.wedge(x=0, y=1, radius=0.4,
start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'),
line_color="white", fill_color='color', legend='number', source=data2)
p.xaxis.axis_label=i
p.yaxis.visible=False
p.grid.grid_line_color = None
plots.append(p)
return row(fig,*plots)
def make_plot_project(source, project, item): ### show the figure with the project name such as 'P1', 'P2'
fig = figure(plot_height=350, plot_width=350, title= project + ' OVERALL ' + item +' RATE', toolbar_location=None,
tools="hover", tooltips="@number: @value", x_range=(-0.5, 1.0))
fig.wedge(x=0, y=1, radius=0.4,
start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'),
line_color="white", fill_color='color', legend='number',source=source)
fig.axis.axis_label=None
fig.axis.visible=False
fig.grid.grid_line_color = None
plots = []
n = 0
for i in data_types:
dictP = group.get_group(project).set_index('ProjectName').to_dict('list')
data = dict_extract(i, dictP, item)
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'number'})
data2['color'] = chart_colors[:len(data)]
angle(data2)
p = figure(plot_height=350, plot_width=350, title= project+ ' '+ item +' RATE BY DATA TYPE', toolbar_location=None,
tools="hover", tooltips="@number: @value", x_range=(-0.5, 1.0))
p.wedge(x=0, y=1, radius=0.4,
start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'),
line_color="white", fill_color='color',legend='number',source=data2)
p.xaxis.axis_label = i
p.yaxis.visible=False
p.grid.grid_line_color = None
plots.append(p)
return row(fig,*plots)
test2 = get_data_project('P1','PassQC')
test2.column_names
</code>
<code>
source_all = get_data_all(item)
source_project = get_data_project(project, item)
plot_all = make_plot_all(source_all, item)
plot_project = make_plot_project(source_project, project,item)
ticker1.on_change('value',update_plot)
ticker2.on_change('value',update_plot)
# set up layout
widgets = column(ticker1, ticker2)
plots = column(plot_all,plot_project)
layout = column(widgets, plots)
</code>
<code>
show(layout.children[1])
</code>
<code>
hover = HoverTool(tooltips="@number: @value")
start_angle=cumsum('angle', include_zero=True)
end_angle=cumsum('angle')
color = 'color'
legend='number'
fig = figure(plot_height=350, plot_width=350, title= 'OVERALL '+item+' RATE', toolbar_location=None,
x_range=(-0.5, 1.0))
fig.add_tools(hover)
fig.wedge(x=0, y=1, radius=0.4, start_angle = start_angle, end_angle = end_angle, line_color="white",fill_color = color,
legend= legend, source=test1)
fig.axis.axis_label=None
fig.axis.visible=False
fig.grid.grid_line_color = None
show(fig)
</code>
<code>
plots = []
n = 0
for i in data_types:
data = dict_extract(i, dictA, 'PassQC')
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'number'})
data2['color'] = chart_colors[:len(data)]
angle(data2)
#p = join('fig' + str(n))
p = figure(plot_height=350, plot_width=350, title= 'OVERALL ' +item + ' RATE BY DATA TYPE', toolbar_location=None,
tools="hover", tooltips="@number: @value", x_range=(-0.5, 1.0))
p.wedge(x=0, y=1, radius=0.4,
start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'),
line_color="white", color='color', legend='number', source=data2)
p.xaxis.axis_label=i
p.yaxis.visible=False
p.grid.grid_line_color = None
plots.append(p)
show(row(plots))
</code>
<code>
def update_plot(attrname, old, new):
item = ticker1.value
update1 = get_data_all(item)
#update2 = get_data_project(ticker2.value, item)
source_all.update(update1)
#source_project.update(update2)
#plot_overall = make_plot_all(source,item)
#plot_project = make_plot_project(source2, project, item)
#plots = column(plot_all,plot_project)
</code>
<code>
data = pd.read_csv("/Users/xli677/Dropbox (Uni of Auckland)/xli677/Projects/MyTardis/Visualisation_with_Bokeh/SampleSheet_Bokeh_test.csv")
data2 = data.set_index('ProjectName')
group = data.groupby('ProjectName')
dictA = data2.to_dict('list')
</code>
<code>
dictA
</code>
<code>
item_list = ['Returned', 'PassQC', 'Biopipeline']
status_list = ['Complete','Under Processing' ,'Awaiting Processing']
def dict_extract_all(datatype_list, dic, item):
list_return_all = []
for i in data_types:
i = join(i + '_' + item)
list_return_all += dictA[i]
list_bio = ['Complete','Under Processing' ,'Awaiting Processing']
if item == 'Biopipeline':
dict_return_all = dict((x,list_return_all.count(x))for x in list_bio)
else:
dict_return_all = dict((x,list_return_all.count(x)) for x in [True,False])
return dict_return_all
def dict_extract(datatype_name, dic, item): ### given data type, dictionary, to count the number of true/false and return a dictionary containg the number for each data type
list_return = dic[datatype_name + '_' + item]
if item == 'Biopipeline':
dict_return = dict((x,list_return.count(x))for x in status_list)
else:
dict_return= dict((x,list_return.count(x)) for x in [True,False])
return dict_return
</code>
<code>
### show the figure with the project name such as 'P1', 'P2'
dictP = group.get_group('P1').set_index('ProjectName').to_dict('list')
data = dict_extract_all(data_types, dictP, 'Returned')
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'number'})
angle(data2)
data2['color'] = chart_colors[:len(data)]
source = ColumnDataSource(data2)
hover = HoverTool(tooltips="@number: @value")
start_angle=cumsum('angle', include_zero=True)
end_angle=cumsum('angle')
color = 'color'
legend='number'
fig = figure(plot_height=350, plot_width=350, title= ticker2.value + ' OVERALL ' + ticker1.value +' RATE', toolbar_location=None,
x_range=(-0.5, 1.0))
fig.add_tools(hover)
fig.wedge(x=0, y=1, radius=0.4,
start_angle=start_angle, end_angle=end_angle,
line_color="white", fill_color=color, legend=legend, source=source)
fig.axis.axis_label=None
fig.axis.visible=False
fig.grid.grid_line_color = None
plots = []
n = 0
for i in data_types:
dictP = group.get_group('P1').set_index('ProjectName').to_dict('list')
data = dict_extract(i, dictP, 'Returned')
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'number'})
data2['color'] = chart_colors[:len(data)]
angle(data2)
source = ColumnDataSource(data2)
p = figure(plot_height=350, plot_width=350, title= ticker2.value+ ' '+ ticker1.value +' RATE BY DATA TYPE', toolbar_location=None,
x_range=(-0.5, 1.0))
p.add_tools(hover)
p.wedge(x=0, y=1, radius=0.4,
start_angle=start_angle, end_angle=end_angle,
line_color="white", fill_color=color, legend=legend, source=source)
p.xaxis.axis_label = i
p.yaxis.visible=False
p.grid.grid_line_color = None
plots.append(p)
</code>
<code>
dictP = group.get_group('P1').set_index('ProjectName').to_dict('list')
data = dict_extract_all(data_types, dictP, 'Returned')
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'number'})
angle(data2)
data2['color'] = chart_colors[:len(data)]
source = ColumnDataSource(data2)
#dict_extract_all(data_types, dictP, 'Returned')
print(data2)
</code>
<code>
dict_extract('DNA_seq', dictP, 'Returned')
</code>
<code>
data = dict_extract_all(data_types,dictA,'Returned')
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'status'})
print(data2)
</code>
<code>
print(start_angle)
</code>
<code>
from functools import lru_cache
from os.path import dirname, join
from math import pi
import pandas as pd
from bokeh.io import curdoc, output_file, show
from bokeh.layouts import layout, column, row, widgetbox
from bokeh.models import ColumnDataSource, PreText, Select, Dropdown, HoverTool, LabelSet
from bokeh.plotting import figure, curdoc
from bokeh.transform import cumsum
def dict_extract_all(datatype_list, dic, item): ### count the number of true/false and return a dictionary for all data types
list_return_all = []
for i in datatype_list:
i = join(i + '_' + item)
list_return_all += dic[i]
if item == 'Biopipeline':
dict_return_all = dict((x,list_return_all.count(x))for x in status_list)
else:
dict_return_all = dict((x,list_return_all.count(x)) for x in [True,False])
return dict_return_all
def dict_extract(datatype_name, dic, item): ### given data type, dictionary, to count the number of true/false and return a dictionary containg the number for each data type
list_return = dic[datatype_name + '_' + item]
if item == 'Biopipeline':
dict_return = dict((x,list_return.count(x))for x in status_list)
else:
dict_return= dict((x,list_return.count(x)) for x in [True,False])
return dict_return
def angle(data):
data['angle'] = data['value']/data['value'].sum() * 2*pi
return
chart_colors = ['#007bff','#e29e44','#44e5e2','#eeeeee','#d8e244','#e244db']
data = pd.read_csv("/Users/xli677/Dropbox (Uni of Auckland)/xli677/Projects/MyTardis/Visualisation_with_Bokeh/SampleSheet_Bokeh_test.csv")
data2 = data.set_index('ProjectName')
group = data.groupby('ProjectName')
dictA = data2.to_dict('list')
item_list = ['Returned', 'PassQC', 'Biopipeline']
data_types = ['RNA_seq','DNA_seq','Methyl_seq']
project_list = ['P1','P2','P3','All']
status_list = ['Complete','Under Processing' ,'Awaiting Processing']
</code>
<code>
data = dict_extract_all(data_types, dictA, 'Returned')
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'status'})
angle(data2)
data2['color'] = chart_colors[:len(data)]
column_sum = data2['value'].sum()
data2['percentage'] = (data2['value']/column_sum)
source = ColumnDataSource(data2)
hover = HoverTool(tooltips="@status: @percentage{0.00%}; @value")
start_angle=cumsum('angle', include_zero=True)
end_angle=cumsum('angle')
color = 'color'
legend='status'
fig = figure(plot_height=350, plot_width=430, title= 'OVERALL '+'Returned'+' RATE', toolbar_location=None,
x_range=(-0.5, 1.0))
fig.add_tools(hover)
fig.wedge(x=0, y=1, radius=0.4,
start_angle=start_angle, end_angle=end_angle,
line_color="white", fill_color=color, legend=legend, source=source)
labels = LabelSet(x=0, y=1, text='value', level='glyph',
angle=cumsum('angle', include_zero=True), source=source, render_mode='canvas')
fig.add_layout(labels)
fig.axis.axis_label=None
fig.axis.visible=False
fig.grid.grid_line_color = None
plots = []
n = 0
for i in data_types:
data = dict_extract(i, dictA, 'Returned')
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'status'})
data2['color'] = chart_colors[:len(data)]
angle(data2)
column_sum = data2['value'].sum()
data2['percentage'] = (data2['value']/column_sum)
source = ColumnDataSource(data2)
p = figure(plot_height=350, plot_width=430, title= 'OVERALL ' +'Returned' + ' Rate of ' + i, toolbar_location=None,
x_range=(-0.5, 1.0))
p.add_tools(hover)
p.wedge(x=0, y=1, radius=0.4,
start_angle=start_angle, end_angle=end_angle,
line_color="white", fill_color=color, legend=legend, source=source)
labels = LabelSet(x=0, y=1, text='value', level='glyph',
angle=cumsum('angle', include_zero=True), source=source, render_mode='canvas')
p.add_layout(labels)
p.xaxis.axis_label=i
p.xaxis.visible=False
p.yaxis.visible=False
p.grid.grid_line_color = None
plots.append(p)
show(row(fig,*plots))
</code>
<code>
def modify_doc(doc):
df = sea_surface_temperature.copy()
source = ColumnDataSource(data=df)
plot = figure(x_axis_type='datetime', y_range=(0, 25), y_axis_label='Temperature (Celsius)',
title="Sea Surface Temperature at 43.18, -70.43")
plot.line('time', 'temperature', source=source)
def callback(attr, old, new):
if new == 0:
data = df
else:
data = df.rolling('{0}D'.format(new)).mean()
source.data = ColumnDataSource(data=data).data
slider = Slider(start=0, end=30, value=0, step=1, title="Smoothing by N Days")
slider.on_change('value', callback)
doc.add_root(column(slider, plot))
# doc.theme = Theme(filename="theme.yaml")
</code>
<code>
import numpy as np
from bokeh.io import curdoc, show
from bokeh.models import ColumnDataSource, Grid, LinearAxis, Plot, Text
N = 9
x = np.linspace(-2, 2, N)
y = x**2
a = "abcdefghijklmnopqrstuvwxyz"
text = [a[i*3:i*3+3] for i in range(N)]
source = ColumnDataSource(dict(x=x, y=y, text=text))
plot = Plot(
title=None, plot_width=300, plot_height=300,
min_border=0, toolbar_location=None)
glyph = Text(x="x", y="y", text="text", angle=0.3, text_color="#96deb3")
plot.add_glyph(source, glyph)
xaxis = LinearAxis()
plot.add_layout(xaxis, 'below')
yaxis = LinearAxis()
plot.add_layout(yaxis, 'left')
plot.add_layout(Grid(dimension=0, ticker=xaxis.ticker))
plot.add_layout(Grid(dimension=1, ticker=yaxis.ticker))
curdoc().add_root(plot)
show(plot)
</code>
<code>
tooltips="@status: @percentage{0.00%}; @value"
</code>
<code>
tooltips
</code>
<code>
from functools import lru_cache
from os.path import dirname, join
from math import pi
import pandas as pd
import numpy as np
from bokeh.io import curdoc, output_file, show
from bokeh.layouts import layout, column, row, widgetbox
from bokeh.models import ColumnDataSource, PreText, Select, Dropdown, HoverTool, Label, LabelSet, LinearAxis, Text
from bokeh.plotting import figure, curdoc
from bokeh.transform import cumsum
def dict_extract_all(datatype_list, dic, item): ### count the number of true/false and return a dictionary for all data types
list_return_all = []
for i in datatype_list:
i = join(i + '_' + item)
list_return_all += dic[i]
if item == 'Biopipeline':
dict_return_all = dict((x,list_return_all.count(x))for x in status_list)
else:
dict_return_all = dict((x,list_return_all.count(x)) for x in [True,False])
return dict_return_all
def dict_extract(datatype_name, dic, item): ### given data type, dictionary, to count the number of true/false and return a dictionary containg the number for each data type
list_return = dic[datatype_name + '_' + item]
if item == 'Biopipeline':
dict_return = dict((x,list_return.count(x))for x in status_list)
else:
dict_return= dict((x,list_return.count(x)) for x in [True,False])
return dict_return
def angle(data):
data['angle'] = data['value']/data['value'].sum() * 2*pi
return
chart_colors = ['#007bff','#e29e44','#44e5e2','#eeeeee','#d8e244','#e244db']
data = pd.read_csv("/Users/xli677/Dropbox (Uni of Auckland)/xli677/Projects/MyTardis/Visualisation_with_Bokeh/SampleSheet_Bokeh_test.csv")
data2 = data.set_index('ProjectName')
group = data.groupby('ProjectName')
dictA = data2.to_dict('list')
item_list = ['Returned', 'PassQC', 'Biopipeline']
data_types = ['RNA_seq','DNA_seq','Methyl_seq']
project_list = ['P1','P2','P3','All']
status_list = ['Complete','Under Processing' ,'Awaiting Processing']
</code>
<code>
data = dict_extract_all(data_types, dictA, 'Returned')
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'status'})
angle(data2)
data2['color'] = chart_colors[:len(data)]
column_sum = data2['value'].sum()
data2['percentage'] = (data2['value']/column_sum)
data2['label'] = ["{:.2%}".format(p) for p in data2['percentage']]
data2['label'] = data2['label'].astype(str)
data2['label'] = data2['label'].str.pad(20, side = "left")
#data2['cos'] = np.cos(data2['angle'])*0.3
#data2['sin'] = np.sin(data2['angle'])*0.3
source = ColumnDataSource(data2)
hover = HoverTool(tooltips="@status: @percentage{0.00%}; @value")
start_angle=cumsum('angle', include_zero=True)
end_angle=cumsum('angle')
color = 'color'
legend="status"
fig = figure(plot_height=350, plot_width=430, title= 'OVERALL '+'Return'+' Rate', toolbar_location=None,
x_range=(-0.5, 1.0))
fig.title.align = 'center'
fig.add_tools(hover)
fig.wedge(x=0, y=1, radius=0.4,
start_angle=start_angle, end_angle=end_angle,
line_color="white", fill_color=color, legend=legend, source=source)
#labels = LabelSet(x=0, y=1, text="label", y_offset=0,angle=cumsum('angle', include_zero=True),
# text_font_size="8pt", text_color="black",render_mode='canvas',
# source=source, text_align='center')
labels = LabelSet(x=0, y=1, text="label", angle=start_angle,
text_font_size="10pt", text_color="black",
source=source)
fig.add_layout(labels)
# + ': ' + 'percentage' + ';' + 'value'
#txt = "@status"
#glyph = Text(x=0.8, y=1, text=txt, text_color="#96deb3",text_align='right')
#fig.add_glyph(source, glyph)
fig.axis.axis_label=None
fig.axis.visible=False
fig.grid.grid_line_color = None
plots = []
n = 0
for i in data_types:
data = dict_extract(i, dictA, 'Returned')
data2 = pd.Series(data).reset_index(name='value').rename(columns={'index':'status'})
data2['color'] = chart_colors[:len(data)]
angle(data2)
column_sum = data2['value'].sum()
data2['percentage'] = (data2['value']/column_sum)
data2['label'] = ["{:.2%}".format(p) for p in data2['percentage']]
data2['label'] = data2['label'].astype(str)
data2['label'] = data2['label'].str.pad(20, side = "left")
source = ColumnDataSource(data2)
p = figure(plot_height=350, plot_width=430, title= 'OVERALL ' +'Returned' + ' Rate of ' + i, toolbar_location=None,
x_range=(-0.5, 1.0))
p.title.align = 'center'
p.add_tools(hover)
p.wedge(x=0, y=1, radius=0.4,
start_angle=start_angle, end_angle=end_angle,
line_color="white", fill_color=color, legend=legend, source=source)
labels = LabelSet(x=0, y=1, text="label", angle=start_angle,
text_font_size="10pt", text_color="black",
source=source)
p.add_layout(labels)
p.xaxis.visible = False
p.yaxis.visible=False
p.grid.grid_line_color = None
plots.append(p)
show(row(fig,*plots))
</code>
<code>
data2
</code>
<code>
for i,item in enumerate(data2['value']):
data2['cumulative_angle'] = sum(data2['value'][0:i+1]- (item/2))/sum(data2['value'])*2*pi
print(data2)
</code>
<code>
data2['cumulative_angle'] = [sum(data2['value'][0:i+1]- (item/2))/sum(data2['value'])*2*pi for i,item in enumerate(data2['value'])]
</code>
<code>
data2['cos'] = np.cos(data2['cumulative_angle'])*0.3
data2['sin'] = np.sin(data2['cumulative_angle'])*0.3
</code>
<code>
data2
</code>
<code>
source = ColumnDataSource(data2)
start_angle=cumsum('angle', include_zero=True)
end_angle=cumsum('angle')
color = 'color'
legend="status"
fig = figure(plot_height=350, plot_width=430, title= 'OVERALL '+'Returned'+' Rate', toolbar_location=None,
x_range=(-0.5, 1.0))
fig.title.align = 'center'
#fig.add_tools(hover)
fig.wedge(x=0, y=0, radius=0.4,
start_angle=start_angle, end_angle=end_angle,
line_color="white", fill_color=color, legend=legend, source=source)
labels = LabelSet(x='cos', y='sin', text="label", y_offset=0,
text_font_size="10pt", text_color="black",
source=source, text_align='center')
fig.add_layout(labels)
fig.axis.axis_label=None
fig.axis.visible=False
fig.grid.grid_line_color = None
show(fig)
</code>
<code>
data2
</code>
|
{
"filename": "Untitled2_1.ipynb",
"repository": "LibbyLi667/github-upload",
"query": "transformed_from_existing",
"size": 60799,
"sha": ""
}
|
# use_cases_topics_s3_1.ipynb
Repository: HHS/acf-nlp-on-gfe-testing
### Notebook for extracting the most used AI techniques, AI usecases and Summaries from the datasets available on the following websites:
- https://ai.gov/ai-use-cases/
- https://www.hhs.gov/programs/topic-sites/ai/use-cases/index.html
#### This notebook extracts the most used AI techniques from both datasets, performs clustering to group the 430 contracts into 20 clusters, and then summarizes the combined text from each cluster using LLMs (BART, phi3, llama3). Finally, it performs topic labeling using the llama3 model.
#### Files needed to run the notebook:
-- '2023 Consolidated AI Use Case Inventory (PUBLIC).csv'
-- 'hhs-ai-use-cases-2023-public-inventory.csv'
#### Files generated from the notebook:
-- 'ai_use_case_topics.csv'
-- 'summaries_of_usecases.csv'
-- 'ai_use_cases_relevant_430.csv'
#### Loading the two datasets to create two dataframes
<code>
import pandas as pd
import os
current_working_directory = os.getcwd()
csv_path_hhs = os.path.join(current_working_directory, "docs", "use_cases", "hhs-ai-use-cases-2023-public-inventory.csv")
csv_path_AI_inventory = os.path.join(current_working_directory, "docs", "use_cases","2023 Consolidated AI Use Case Inventory (PUBLIC).csv")
df_hhs = pd.read_csv(csv_path_hhs, encoding='latin1')
df_AI_inv = pd.read_csv(csv_path_AI_inventory, encoding='latin1')
</code>
<code>
print(df_hhs.shape)
df_hhs_trim=df_hhs[['Use Case Name', 'Agency', 'Bureau / Department', 'Summary of Use Case']].copy()
df_hhs_trim = df_hhs_trim.clean_names()
print(df_hhs_trim.columns)
rename_dict = {'summary_of_use_case': 'Summary','use_case_name': 'Title', 'agency':'Agency', 'bureau_department': 'Department'}
df_hhs_trim = df_hhs_trim.rename(columns=rename_dict)
</code>
<code>
print(df_AI_inv.shape)
df_AI_inv_trim=df_AI_inv[['Title', 'Agency', 'Department_Code', 'Summary', 'Department', 'Techniques']].copy()
rename_dict = {'Department_Code': 'Department_code'}
df_AI_inv_trim = df_AI_inv_trim.rename(columns=rename_dict)
print(df_AI_inv_trim.columns)
</code>
#### Extract the department code from the text in the paranthesis
<code>
import re
def extract_or_return_same(text):
match = re.search(r'\((.*?)\)', text)
if match:
return match.group(1)
else:
return text
df_hhs_trim['Department_code']=df_hhs_trim['Department'].apply(extract_or_return_same)
</code>
#### Exrtacting the mostly used AI techniques `hhs-ai-use-cases-2023-public-inventory.csv`
<code>
import itertools
techniques = df_AI_inv.Techniques.tolist()
techniques = [item.split(',') for item in techniques if isinstance(item, str)]
flattened_list = list(itertools.chain.from_iterable(techniques))
mostly_used_techniques=list(set(flattened_list))
print(len(mostly_used_techniques))
</code>
#### Text cleaning of the mostly used AI techniques
<code>
def split_and_flatten(strings):
split_lists = [s.split(';') for s in strings]
flattened_list_ = list(itertools.chain.from_iterable(split_lists))
flattened_list_ = [item.strip() for item in flattened_list_]
return flattened_list_
mostly_used_techniques_=split_and_flatten(mostly_used_techniques)
mostly_used_techniques_ = [s.replace('Unknown','').replace('.','').replace('#','').replace('&','and').replace('®','').replace('5)','5').replace(' (Nlp)','').strip().upper() for s in mostly_used_techniques_ if len(s)>1]
print(len(mostly_used_techniques_))
</code>
#### Below script processes a list by removing specified elements and mapping certain elements to new values using a dictionary. It then applies these transformations to clean and standardize the list of techniques.
<code>
def process_list(input_list, elements_to_remove, mapping_dict):
# Remove specified elements
filtered_list = [element for element in input_list if element not in elements_to_remove]
# Map some elements to new values
mapped_list = [mapping_dict.get(element, element) for element in filtered_list]
# mapped_list=[s for s in mapped_list if s.contain('ML')]
return list(set(mapped_list))
elements_to_remove = ['AT THIS TIME',
'OTHER',
'DOODLER: HTTPS://GITHUBCOM/DBUSCOMBE-USGS/DASH_DOODLER',
'PYTHON IN JUPYTER LABS',
'RANGE OF DATA DRIVEN',
'DOCUMENT UNDERSTANDING',
'ACTIVE LEARNING',
'RETINANET',
'SUBJECTS WITH LONG-ARM RIFLES OR LARGE BACKPACKS AND TO EXCLUDE ITEMS OF LITTLE OR NO INTEREST SUCH AS ANIMALS',
'CONTINUOUS ACTIVE LEARNING'
]
# Mapping dictionary
mapping_dict = {
'NATURAL LANGUAGE PROCESSING (NLP)': 'NATURAL LANGUAGE PROCESSING',
'NLP':'NATURAL LANGUAGE PROCESSING',
'INTELLIGENT DOCUMENT RECOGNITION (IDR)': 'INTELLIGENT DOCUMENT RECOGNITION',
'OPTICAL CHARACTER RECOGNITION (OCR)': 'OPTICAL CHARACTER RECOGNITION',
'INTELLIGENT CHARACTER RECOGNITION (ICR)': 'INTELLIGENT CHARACTER RECOGNITION',
'ROBOTIC PROCESS AUTOMATION (RPA)': 'ROBOTIC PROCESS AUTOMATION',
'ROBOTIC PROCESSING AUTOMATION (RPA)': 'ROBOTIC PROCESS AUTOMATION',
'THE MATROID SOFTWARE CURRENTLY PROCESSES AND ANNOTATES IMAGES USING PROPRIETARY SOFTWARE TO DETERMINE IF ANY OF THE IMAGES CONTAIN HUMAN SUBJECTS FUTURE USE CASES INCLUDE THE POTENTIAL TO DETECT ADDITIONAL ITEMS OF INTEREST SUCH AS VEHICLES': 'IMAGE PROCESSING',
'ML': 'MACHINE LEARNING',
'AI': 'ARTIFICIAL INTELLIGENCE',
'XGBOOST ALGORITHM WITH PARAMETERS TUNED VIA RANDOM HYPERPARAMETER SEARCH USING 5-FOLD CROSS VALIDATION ON THE TRAINING DATASET FOR 60 ITERATIONS (RESULTING IN AT LEAST A 95% CHANCE OF FINDING A HYPERPARAMETER COMBINATION IN THE BEST 5% OF COMBINATIONS) THE SCORES RESULTING FROM THE XGBOOST ARE CALIBRATED VIA PLATT SCALING SO THAT MODEL SCORES CAN BE INTERPRETED AS DEFAULT PROBABILITIES THESE IS STANDARD METHOD FOR TRAINING CREDIT SCORING ALGORITHMS IN THE INDUSTRY':'MACHINE LEARNING',
'MACHINE LANGUAGE LEARNING':'MACHINE LEARNING',
'DOCUMENT/FILE CLASSIFICATION: DOCUMENT/FILE CLASSIFICATION IS A SUPERVISED ML ALGORITHM THAT CLASSIFIES WHOLE DOCUMENTS ACCORDING TO THEIR TYPE THE ALGORITHM WORKS BY CONVERTING EACH DOCUMENT TO A TERM FREQUENCYÂ\x80\x93INVERSE DOCUMENT FREQUENCY (TF-IDF) NUMERICAL REPRESENTATION AND PASSING THESE VECTORS THROUGH A MULTI-LAYER NEURAL NETWORK TO FINALLY GET THE DOCUMENTÂ\x80\x99S TYPE/CLASS DOCUMENT/FILE CLUSTERING: DOCUMENT/FILE CLUSTERING IS AN UNSUPERVISED ML ALGORITHM THAT GROUPS SIMILAR FILES TOGETHER ACCORDING TO THEIR CONTENT FOR EXAMPLE':'DOCUMENT/FILE CLASSIFICATION: NLP',
'LONG SHORT TERM MEMORY (LSTM) MODELS':'RNN-LSTM',
'LONG-SHORT TERM MEMORY BASED RECURRENT NEURAL NETWORKS':'RNN-LSTM',
'ML VIA A CONVOLUTIONAL NEURAL NETWORK':'NEURAL NETWORKS',
'MULTI-LAYER PERCEPTRON':'NEURAL NETWORKS',
'YOLOV5':'COMPUTER VISION',
'MACHINE VISION':'COMPUTER VISION',
'NATURAL LANGUAGE PROCESSING (NLP) ALONG WITH SUPERVISED AND SELF-SUPERVISED MACHINE LEARNING VIA DEEP LEARNING MODELS':"NATURAL LANGAUGE PROCESSING, DEEP LEARING",
"SUCH AS A RESIDUAL NEURAL NETWORK (RESNET) AND CONVOLUTIONAL NEURAL NETWORKS (CNN)":"RESNET AND CNN",
'ARTIFICIAL NEURAL NETWORK': 'NEURAL NETWORKS',
'NATURAL LANGUAGE PROCESSING FOR (A) DOCUMENT CLASSIFICATION AND (B) SENTENCE-LEVEL CAUSAL PASSAGE DETECTION': 'NLP CLASSIFICATION, SENTIMENT ANALYSIS',
'STANDARD MACHINE LEARNING TO PREDICT VALUES FOR DESCRIPTIVE METADATA FIELDS GIVEN VARIOUS INPUTS SUCH AS THE CONTENT AND METADATA FROM THE RECORDS MANAGEMENT SYSTEM':'MACHINE LEARNING',
'1 TEXTRACTION MACHINE LEARNING (ML) SERVICE WHICH USED OCR TO EXTRACT THE TEXT/DATA FROM SCANNED IMAGES 2 AUTOMATED NLP (NATURAL LANGUAGE PROCESSING) TO DETECT PII INFORMATION OUT OF THE EXTRACTED TEXT FROM SCANNED IMAGES': "NATURAL LANGUAGE PROCESSING, IMAGE PROCESSING",
'ML (RECOMMENDER ALGORITHIM)': 'MACHINE LEARNING RECOMMENDER ALGORITHIM',
'AI/ML TECHNIQUES':'MACHINE LEARNING AND ARTIFICIAL INTELLIGENCE TECHNIQUES',
'BAGGED TREES (AKA RANDOM FOREST) CLASSIFICATION':'RANDOM FOREST CLASSIFICATION',
'AI/ML TECHNIQUES (EG RANDOM FORESTS)':'RANDOM FOREST',
'AI/ML TECHNIQUES (EG LSTMS)':'LONG-SHORT TERM MEMORY',
'OPTICAL MARK READING (OMR)':'OPTICAL MARK READING',
'NON-DISCLOSURE AGREEMENTS WILL CLUSTER TOGETHER WHILE PRODUCT PRESENTATION FILES WILL BE ASSIGNED TO A DIFFERENT CLUSTER':'CLUSTERING'
}
# Apply the function
mostly_used_techniques__ = process_list(mostly_used_techniques_, elements_to_remove, mapping_dict)
print(len(mostly_used_techniques__))
</code>
#### This script replaces specified substrings in a list of techniques with their corresponding values from a replacement dictionary, standardizing the terminology.
<code>
def replace_substrings(input_list, replacements):
processed_list = []
for item in input_list:
for old, new in replacements.items():
item = item.replace(old, new)
processed_list.append(item)
return processed_list
# Replacement dictionary
replacements = {
'ML': 'MACHINE LEARNING',
'DL': 'DEEP LEARNING',
'CNN': 'CONVOLUTIONAL NEURAL NETWORKS',
'NLP': 'NATURAL LANGUAGE PROCESSING',
'LLM': 'LARGE LANGUAGE MODEL',
'LSTM':'LONG-SHORT TERM MEMORY',
'RNN': 'RECURRENT NEURAL NETWORKS',
'UNET':'UNET CONVOLUTIONAL NEURAL NETWORKS',
'U-NET':'UNET CONVOLUTIONAL NEURAL NETWORKS',
'RESNET':'RESIDUAL NEURAL NETWORKS',
'CHAT BOT': 'CHATBOT',
'CHATBOTS': 'CHATBOT'
}
# Apply the function
processed_list = replace_substrings(mostly_used_techniques__, replacements)
processed_list
</code>
<code>
new_column_order = ['Title', 'Agency', 'Department', 'Department_code','Summary']
df_hhs_trim=df_hhs_trim[new_column_order]
df_hhs_trim.head(1)
</code>
<code>
new_column_order = ['Title', 'Agency', 'Department', 'Department_code', 'Techniques','Summary']
df_AI_inv_trim=df_AI_inv_trim[new_column_order]
df_AI_inv_trim.head(1)
</code>
<code>
def columns_to_dict(df, key_col, value_col):
result_dict = {}
for key, value in zip(df[key_col], df[value_col]):
if key in result_dict:
if value not in result_dict[key]:
result_dict[key].append(value)
else:
result_dict[key] = [value]
# Flatten lists with a single item
result_dict = {k: v[0] if len(v) == 1 else v for k, v in result_dict.items()}
return result_dict
# Convert the two columns into a dictionary
columns_to_dict(df_AI_inv_trim, 'Department', 'Department_code')
</code>
#### Adding AI techniques found in other dataset
<code>
processed_list=processed_list+['AI', 'CHATBOT', 'ML', 'NLP', 'dl', 'deep learning', 'CHATBOTS', 'machine-learning', 'cyberthreats', 'seq2seq', 'text summarization',
'associated topics', 'data-driven', 'decision making', 'Long Short-Term Memory', 'lstm', 'recurrent neural network', 'Chat Bot', 'topic modeling', 'entity recognition',
'Predictive Intelligence', 'decision-making', 'Zero-shot learning']
</code>
<code>
techniques_lower = [tech.lower() for tech in processed_list]
# Function to identify techniques in the summary
def identify_techniques(summary):
found_techniques = []
summary_lower = summary.lower()
for tech in techniques_lower:
if re.search(r'\b' + re.escape(tech) + r'\b', summary_lower):
if tech not in found_techniques:
found_techniques.append(tech)
return ', '.join(found_techniques)
# Apply the function to each row in the DataFrame
df_hhs_trim['Techniques'] = df_hhs_trim['Summary'].apply(identify_techniques)
# Display the DataFrame with the new column
# df_hhs_trim[['Summary', 'Techniques']]
</code>
<code>
new_column_order = ['Title', 'Agency', 'Department', 'Department_code', 'Techniques','Summary']
df_hhs_trim=df_hhs_trim[new_column_order]
</code>
<code>
df_hhs_trim.head(1)
</code>
<code>
df_AI_inv_trim[df_AI_inv_trim['Techniques'].isna()].head(1)
</code>
<code>
for index, row in df_AI_inv_trim.iterrows():
if pd.isna(row['Techniques']):
df_AI_inv_trim.at[index, 'Techniques'] = identify_techniques(row['Summary'])
</code>
<code>
df_AI_inv_trim[['Techniques', 'Summary']].iloc[41]
</code>
#### Combining the two datasets and filter the rows based on the departments of intrest
<code>
ai_use_cases=pd.concat([df_hhs_trim,df_AI_inv_trim], axis=0,ignore_index=True)
ai_use_cases.shape
ai_use_cases['Department_code']=ai_use_cases['Department_code'].str.upper()
relevant_agencies = ["HHS", "USDA", "ED", "DOE", "HUD", "SSA", "SBA", "VA"]
ai_use_cases_relevant=ai_use_cases[ai_use_cases['Department_code'].isin(relevant_agencies)]
</code>
#### Saving the dataset containing 430 use cases to a csv file
<code>
ai_use_cases_relevant.to_csv('ai_use_cases_relevant_430.csv')
</code>
<code>
def split_and_flatten(strings):
split_lists = [s.split(',') for s in strings]
flattened_list_ = list(itertools.chain.from_iterable(split_lists))
flattened_list_ = [item.strip().upper() for item in flattened_list_ if item != '' ]
return flattened_list_
techniques_relavant=ai_use_cases_relevant['Techniques'].tolist()
# techniques_relavant=list(set(techniques_relavant))
techniques_relavant=list(set(split_and_flatten(techniques_relavant)))
</code>
<code>
mostly_used_techniques_relavant = process_list(techniques_relavant, elements_to_remove, mapping_dict)
print(len(mostly_used_techniques_relavant))
</code>
#### AI techniques used in the 430 AI use cases
<code>
techniques_used = replace_substrings(mostly_used_techniques_relavant, replacements)
list(set(techniques_used))
</code>
#### Saving the AI techniques to text file
<code>
file_name = "AI_echniques_used.txt"
# Open the file in write mode and save the list
with open(file_name, "w") as file:
for item in techniques_used:
file.write(item + "\n")
</code>
<code>
ai_use_cases_relevant['Techniques']=ai_use_cases_relevant['Techniques'].apply(lambda x: x.upper())
</code>
#### clustering the similar use cases followed by preprocessing the summaries of the use cases
<code>
import pandas as pd
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.cluster import KMeans
import numpy as np
import nltk
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
# # Download necessary NLTK packages for tokenization and stopwords
# nltk.download('punkt')
# nltk.download('stopwords')
def preprocess_text(text):
tokens = word_tokenize(text)
tokens = [word.lower() for word in tokens]
tokens = [word for word in tokens if word.isalpha()]
stop_words = set(stopwords.words('english'))
tokens = [word for word in tokens if word not in stop_words]
preprocessed_text = ' '.join(tokens)
return preprocessed_text
# Apply preprocessing
ai_use_cases_relevant['Processed_Summary'] = ai_use_cases_relevant['Summary'].apply(preprocess_text)
# Vectorization
vectorizer = TfidfVectorizer()
tfidf_matrix = vectorizer.fit_transform(ai_use_cases_relevant['Processed_Summary'])
# Similarity Calculation
similarity_matrix = cosine_similarity(tfidf_matrix)
num_clusters = 20
km = KMeans(n_clusters=num_clusters)
km.fit(tfidf_matrix)
clusters = km.labels_
# Assign clusters back to the DataFrame
ai_use_cases_relevant['Cluster'] = clusters
</code>
#### Plotting the elbow plot
<code>
import matplotlib.pyplot as plt
# Function to calculate the distortions for different numbers of clusters
def calculate_distortions(data, max_clusters):
distortions = []
for i in range(1, max_clusters + 1):
km = KMeans(n_clusters=i, random_state=0)
km.fit(data)
distortions.append(km.inertia_)
return distortions
distortions = calculate_distortions(tfidf_matrix, num_clusters)
plt.figure(figsize=(10, 6))
plt.plot(range(1, num_clusters+1), distortions, marker='o')
plt.xlabel('Number of clusters')
plt.ylabel('Distortion')
plt.title('Elbow Method For Optimal k')
plt.show()
</code>
#### Print contents of first few clusters
<code>
def print_cluster_contents(cluster_id, num_samples=5):
print(f"\nCluster - {cluster_id}:")
samples = ai_use_cases_relevant[ai_use_cases_relevant['Cluster'] == cluster_id]['Summary'].sample(n=num_samples, random_state=1)
for i, sample in enumerate(samples, 1):
print(f"Sample {i}: {sample}")
for i in range(min(num_clusters, num_clusters)):
print_cluster_contents(i)
</code>
#### Concatinating the summaries in each cluster
<code>
clustered_texts = ai_use_cases_relevant.groupby('Cluster')['Summary'].apply(' '.join)
pd.set_option('display.max_colwidth', 2000)
print(clustered_texts)
</code>
#### Summary generation using the HugginFace's BART model
<code>
from transformers import BartForConditionalGeneration, BartTokenizer
def abstractive_summary(text, model, tokenizer, max_length=1024, num_beams=4):
inputs = tokenizer("summarize: " +text, max_length=max_length, return_tensors="pt", truncation=True)
summary_ids = model.generate(inputs["input_ids"], num_beams=num_beams, max_length=200, early_stopping=True)
summary = tokenizer.decode(summary_ids[0], skip_special_tokens=True)
return summary
# Load pre-trained BART model and tokenizer
model = BartForConditionalGeneration.from_pretrained('facebook/bart-large-cnn')
tokenizer = BartTokenizer.from_pretrained('facebook/bart-large-cnn')
# Apply abstractive summarization on each cluster
abstractive_summaries_bart = clustered_texts.apply(lambda x: abstractive_summary(x, model, tokenizer))
print(abstractive_summaries_bart)
</code>
#### Summary generation by the phi3 model using ollama api
<code>
import ollama
def gen_title_phi3(text: str):
response = ollama.chat(model='phi3', messages=[
{"role" : "system", "content" : "You are a Summary generator. Generate a summary from the provided text."},
{"role" : "user", "content" : "Convert the following text into a summary of the form 'Summary: <summary>':"},
{"role" : "user", "content" : text}
])
title = response['message']['content']
return title
abstractive_summaries_phi3 = clustered_texts.apply(lambda x: gen_title_phi3(x))
print(abstractive_summaries_phi3)
</code>
#### Summary generation by the llama3 model using ollama api
<code>
import ollama
def gen_summaries_llama3(text: str):
response = ollama.chat(model='llama3', messages=[
{"role" : "system", "content" : "You are a Summary generator. Generate a summary from the provided text."},
{"role" : "user", "content" : "Convert the following text into a summary of the form 'Summary: <summary>':"},
{"role" : "user", "content" : text}
])
title = response['message']['content']
return title
abstractive_summaries_llama3 = clustered_texts.apply(lambda x: gen_summaries_llama3(x))
print(abstractive_summaries_llama3)
</code>
#### Saving the clustered text, summaried by BART, phi3 and llama3 models to a csv file
<code>
df_summaries = pd.concat([clustered_texts, abstractive_summaries_bart, abstractive_summaries_phi3, abstractive_summaries_llama3], axis=1)
df_summaries.columns=['clustered_summaries','BART_summaries','phi3_summaries','llama3_summaries']
df_summaries.to_csv('summaries_of_usecases.csv')
</code>
#### Reading the dataset containing 430 use cases to a csv file for topic identification
<code>
df_topics=pd.read_csv('ai_use_cases_relevant_430.csv', index_col='Unnamed: 0')
df_topics.head()
</code>
#### Function to identify the topics using llama3 model using ollama API
<code>
import ollama
def gen_topic_llama3(text: str):
response = ollama.chat(model='llama3', messages=[
{"role" : "system", "content" : "You are a topic labeller, I want you to identiy the topic for given text from the list of topics given below?, note just give me the topic name and limit your answer to two tokens"},
{"role" : "user", "content" : """
Topics:
1. Accessibility: using AI for translation / interpretation, section 508 compliance, plain language, or other activities to increase accessibility of documents and interactions with the government
2. Policy-making and public engagement: use of AI in any stage of developing regulations or gathering input
3. Asset management: use of AI to manage both physical and digital assets
4. Hotlines and service desks: use of AI to triage, respond, and refer to calls, texts, emails
5. Service / benefits access: use of AI to support determining eligibility for services, streamlining applications, etc.
6. Program integrity: use of AI to detect potential fraud or other wrong-doing in use of public benefits and services
7. Case management: use of AI to document and summarize interactions, suggest and enable referrals
8. Service delivery: use of AI to provide direct services either to the public or to state/local/tribal/territorial governments
9. People operations: use of AI for purposes related to recruiting, retaining, and off-boarding employees
10. Internal operations: administrative use cases for AI, e.g. notetaking, virtual assistants
11. Other
Identify the topic the text belongs to '<topic_name>':"""},
{"role" : "user", "content" : text}
])
Topic = response['message']['content']
return Topic
</code>
<code>
gen_topic_llama3(df_topics['Summary'].tolist()[0])
</code>
<code>
# Applying function on the summary column to identify the topics using llama3 model
df_topics['topics']=df_topics['Summary'].apply(lambda x: gen_topic_llama3(x))
</code>
<code>
# saving the topics to the csv file
df_topics.to_csv('ai_use_case_topics.csv')
</code>
|
{
"filename": "use_cases_topics_s3_1.ipynb",
"repository": "HHS/acf-nlp-on-gfe-testing",
"query": "transformed_from_existing",
"size": 281376,
"sha": ""
}
|
# Book_Embeddings_LitQA2_keyphrase_3.ipynb
Repository: AdrianDimitrov1/Lab
<code>
import os
from openai import OpenAI
from pydantic import BaseModel, Field, conlist
import time
import numpy as np
from numpy.linalg import norm
from scipy.interpolate import UnivariateSpline
from matplotlib import pyplot as plt
import re
from IPython.display import clear_output
import sys
from rake_nltk import Rake
import nltk
client = OpenAI(api_key = os.environ["OPENAI_API_KEY"])
class gen_format(BaseModel):
Question: conlist(str, min_length=5, max_length=5) = Field(description="A list of five questions that could be answered by the given answer")
class rag_format(BaseModel):
Ranked_Relevant_Information: list[str] = Field(description="The ranked list of information relevant to the query, with no paraphrasing or changing of the text from the files.")
def preprocess_text(text):
# Replace decimals/commas in numbers with an underscore and replace hyphens with underscores, generally (except for negative numbers).
#It is only these cases that the sentence tokenizer in Rake doesn't seem to handle well
text = re.sub(r'(\d+)\.(\d+)', r'\1_\2', text)
text = re.sub(r'(\d+)\,(\d+)', r'\1\2', text)
# Pattern explanation:
# (?<!\s)-(?!\d) - matches hyphens not preceded by whitespace or followed by digit
# | - OR
# (?<=\s)-(?=\D) - matches hyphens preceded by whitespace and followed by non-digit
text = re.sub(r'(?<!\s)-(?!\d)|(?<=\s)-(?=\D)', '_', text)
return text
def embedding_answers(answer, ideal, custom_stopwords, english_words) -> str:
#tell Rake to leave logical comparatives alone
r = Rake(stopwords=custom_stopwords)
# Extraction given the text.
text1=preprocess_text(answer)
text2=preprocess_text(ideal)
r.extract_keywords_from_text(text1)
key_phrases1=r.get_ranked_phrases()
r.extract_keywords_from_text(text2)
key_phrases2=r.get_ranked_phrases()
print(key_phrases1)
print(key_phrases2)
result_1=[]
i=0
indicies=[]
for string_ideal in key_phrases2:
#check for "names" that need to be matched exactly
#checks that string_ideal is one word with at least one letter and that is not in english
if (not (" " in string_ideal)) and (any(char.isalpha() for char in string_ideal)) and (not (string_ideal in english_words)):
#if this word does exist in the answer...
if (string_ideal in text1.lower()):
#we have a match!
result_1.append(1)
else:
#if not, no match, therefore "incorrect"
result_1.append(0.7)
else:
max_cos=0
resp1 = client.embeddings.create(
input=string_ideal,
model="text-embedding-ada-002",
encoding_format= "float",
)
check=0
j=0
indicies.append(0)
for string_gen in key_phrases1:
if (string_ideal==string_gen and check==0):
max_cos=1
result_1.append(max_cos)
indicies[i]=j
check=1
j+=1
if (max_cos!=1):
j=0
for string_gen in key_phrases1:
resp2 = client.embeddings.create(
input=string_gen,
model="text-embedding-ada-002",
encoding_format= "float",
)
a=np.array(resp1.data[0].embedding)
b=np.array(resp2.data[0].embedding)
cos=np.dot(a,b)/(norm(a)*norm(b))
if (cos>max_cos):
max_cos=cos
indicies[i]=j
j+=1
result_1.append(max_cos)
i+=1
return np.array(result_1)
def embedding_questions(question, answer, question_model) -> str:
og_question = client.embeddings.create(
input=question,
model="text-embedding-ada-002",
encoding_format= "float",
)
gen_message="""
You are a question generating agent. Your task is to generate a list of questions for the given answer.
"""
gen_assistant = client.beta.assistants.create(
name="gen_test",
instructions=gen_message,
model=question_model,
temperature = 0.0,
top_p = 0.2,
response_format= {
"type": "json_schema",
"json_schema": {
"name": "answer",
"schema": gen_format.model_json_schema()
},
}
)
thread = client.beta.threads.create(
messages=[],
)
parsed = client.beta.threads.messages.create(
thread_id=thread.id,
content=answer,
role='user',
)
run = client.beta.threads.runs.create(
thread_id=thread.id,
assistant_id=gen_assistant.id,
# pass the latest system message as instructions
instructions=gen_message,
)
run = client.beta.threads.runs.retrieve(run.id, thread_id=thread.id)
while run.status!="completed":
run = client.beta.threads.runs.retrieve(run.id, thread_id=thread.id)
response_messages = client.beta.threads.messages.list(thread.id, order="asc")
for message in response_messages.data:
for content in message.content:
output=content.text.value
if output.startswith("{"):
data=json.loads(output)
generated=data["Question"]
client.beta.assistants.delete(assistant_id=gen_assistant.id)
result=[]
for gen_question in generated:
gen_question_vector = client.embeddings.create(
input=gen_question,
model="text-embedding-ada-002",
encoding_format= "float",
)
a=np.array(gen_question_vector.data[0].embedding)
b=np.array(og_question.data[0].embedding)
result.append(np.dot(a, b)/(norm(a)*norm(b)))
return result
def embedding_search(vector_store, question, answer, search_model) -> str:
rag_message="""You are a retrieval agent tasked with performing file searches to find information for the purpose of providing answers.
Find pieces of information that will be directly relevant for answering the query and rank these pieces of information from most relevant to least relevant.
You must quote the passages from the files directly. Do not paraphrase or change the text in any way.
Do not add anything else to the passage quotations, including sources and filenames.
If no information is relevant, you must return a single piece of information, where you state "No information found".
Ideally, these pieces of information will be sentences, phrases, data points or sets of data points, but you have limited flexiblility to include other pieces of information if you think they are appropriate.
You must use tool call (i.e., file search).
You know about the content of the code-base.
"""
rag_assistant = client.beta.assistants.create(
name="rag_test",
instructions=rag_message,
tools=[
{"type": "file_search",
"file_search":{
'max_num_results': 10,
"ranking_options": {
"ranker": "auto",
"score_threshold": 0.6
}
}
}
],
tool_resources={"file_search": {"vector_store_ids":[vector_store.id]}},
model=search_model,
temperature = 0,
top_p = 0.2,
response_format= {
"type": "json_schema",
"json_schema": {
"name": "answer",
"schema": rag_format.model_json_schema()
},
}
)
thread = client.beta.threads.create(
messages=[],
)
parsed = client.beta.threads.messages.create(
thread_id=thread.id,
content=question,
role='user',
)
run = client.beta.threads.runs.create(
thread_id=thread.id,
assistant_id=rag_assistant.id,
# pass the latest system message as instructions
instructions=rag_message,
)
run = client.beta.threads.runs.retrieve(run.id, thread_id=thread.id)
while run.status!="completed":
run = client.beta.threads.runs.retrieve(run.id, thread_id=thread.id)
response_messages = client.beta.threads.messages.list(thread.id, order="asc")
for message in response_messages.data:
for content in message.content:
output=content.text.value
if output.startswith("{"):
data=json.loads(output)
try:
information=data["Ranked_Relevant_Information"]
except:
information=data["Ranked Relevant Information"]
if ("information" in locals()):
#uncomment for hallucination guarding
"""run_steps = client.beta.threads.runs.steps.list(
thread_id=thread.id,
run_id=run.id
)
j=0
for step in run_steps.data:
#wait until the runs.steps.list has finished
while step.status!="completed":
run_steps = client.beta.threads.runs.steps.list(
thread_id=thread.id,
run_id=run.id
)
if (j!=0):
retrieved_step = client.beta.threads.runs.steps.retrieve(
thread_id=step.thread_id,
run_id=run.id,
step_id=step.id,
include=["step_details.tool_calls[*].file_search.results[*].content"]
)
#check for hallucinations and flag all "offending text" from the passed information list
information=hallucination_check(retrieved_step, information, question, 20)
j+=1
"""
pass
else:
information=["No information."]
answer_resp = client.embeddings.create(
input=answer,
model="text-embedding-ada-002",
encoding_format= "float",
)
results=[]
for i in range(len(information)):
info_resp = client.embeddings.create(
input=information[i],
model="text-embedding-ada-002",
encoding_format= "float",
)
a=np.array(info_resp.data[0].embedding)
b=np.array(answer_resp.data[0].embedding)
results.append(np.dot(a, b)/(norm(a)*norm(b)))
i=0
#add is binary True/False
add=1
my_diff=0
mean_score=[]
for score in results:
if (i!=0):
my_diff=np.absolute(score-store)
if (i==1):
ref_diff=my_diff
if (my_diff>=2*ref_diff):
add=0
if (add==1):
mean_score.append(score)
store=score
i+=1
return mean_score
def hallucination_check(retrieved_step, information, question, char_match):
raw_information=""
for result in retrieved_step.step_details.tool_calls[0].file_search.results:
raw_information=raw_information+result.content[0].text
#regex expression to remove all references from the text before "cleaning".
raw_information = re.sub(r"\(\d+\)|\[\d+\]|\(.*Fig.*\)|\(.*Table.*\)", "", raw_information)
file_information=''.join(ch for ch in raw_information if ch.isalnum())
for i in range(len(information)):
verify=information[i].split("...")
for split in verify:
filtered_information=''.join(ch for ch in split if ch.isalnum())
if not (filtered_information in file_information):
clear_output(wait=True)
print("AI is answering this question:\n"+question)
print("AI understood:\n"+split)
length=len(filtered_information)
if length < char_match:
char_match = length
index=file_information.find(filtered_information[:char_match])
if (index!=-1):
print("The file contained:\n"+file_information[index:index+length])
else:
index=file_information.find(filtered_information[length-char_match:])
if (index!=-1):
print("The file contained:\n"+file_information[index-length+char_match:index+char_match])
else:
print("The file contained:\n"+raw_information)
print("Waiting for user input...")
print("Correct the potential hallucination. If the AI is correct, type 'y'. If information given by the AI is not in the file segment, type 'n'.", end='', flush=True)
user=input()
print("User entered:", user)
if (user!='y'):
print("The file contained:\n"+raw_information)
print("Correct the potential hallucination. If the AI is correct, type 'y'. If no information from these chunks is relevant, type 'n'. Otherwise, input what the information should be.", end='', flush=True)
user=input()
if (user!='y'):
if(user=='n'):
split="Null information."
else:
split=user
information[i]=""
for split in verify:
information[i]=information[i]+"..."+split
information[i]=information[i][3:]
return information
</code>
<code>
import pandas as pd
nltk.download('stopwords')
nltk.download('words')
english_words = set(nltk.corpus.words.words())
custom_stopwords = set(nltk.corpus.stopwords.words('english')) - {"no", "not", "than", "more", "same", "before", "after", "now", "then", "above", "below", "over", "under", "like", "other", "such", "few", "most", "some", "between"} # Keep logical comparatives- important for RAG analysis
assistant_data = "/home/adrian/Documents/University Work/Part III Project/PaperQA2/LitQA2_Papers"
lit = pd.read_csv('../PaperQA2/LitQA2_edit.csv')
question=[]
for i in range(lit.shape[0]):
question.append(lit.loc[i, "question"])
#extract answer from these agent and ideal from LitQA2
with open("output_4o_mini_exact_extended.txt", 'r', encoding='utf-8') as file:
file_content = file.read()
spaces=0
tab=0
i=0
answer=[]
ideal=[]
for char in file_content:
if (spaces==2 and char != "\t" and tab==0):
answer[i]+=char
elif (char =="\t"):
spaces=0
ideal.append("")
tab+=1
elif (char != "\n" and tab==1):
ideal[i]+=char
elif (char == "\n" and tab==1):
i+=1
tab=0
if (char == " " and spaces<2 and tab==0):
spaces+=1
if (spaces==2):
answer.append("")
for i in range(len(answer)):
answer[i]=" ".join(word for word in answer[i].split() if ".pdf" not in word)
</code>
<code>
answer_store=[]
print("Embedding Answers")
for i in range(len(answer)):
print(i/len(answer)*100, end="")
print("\r", end="")
answer_store.append(embedding_answers(answer[i], ideal[i], custom_stopwords, english_words))
print(answer[i]+"\t"+ideal[i]+" "+str(answer_store[i])+"\n")
</code>
|
{
"filename": "Book_Embeddings_LitQA2_keyphrase_3.ipynb",
"repository": "AdrianDimitrov1/Lab",
"query": "transformed_from_existing",
"size": 91871,
"sha": ""
}
|
# 3.ipynb
Repository: Anika-Roy/Exploring-Unsupervised-Learning-Methods
# Hierarchical Clustering
Implement the required, using classes and methods.
- routines like hc.linkages(X, linkage type) (takes the data and provides linkage matrix)
- hc.dendogram(Z) (takes the linkage matrix and plots a dendogram).
<code>
from scipy.cluster.hierarchy import dendrogram, linkage
from matplotlib import pyplot as plt
import pandas as pd
</code>
<code>
# class for hierarchical clustering using scipy
class HierarchicalClustering:
def __init__(self, data, linkage_method='single'):
self.data = data
self.linkage_method = linkage_method
self.linkage_matrix = None
def linkages(self):
# self.linkage_matrix = linkage(self.data, self.linkage_method,optimal_ordering=True)
self.linkage_matrix = linkage(self.data, self.linkage_method)
return self.linkage_matrix
def plot_dendrogram(self, Z):
# create a figure
plt.figure(figsize=(25, 10))
# plot the dendrogram
dendrogram(Z)
# show the graph
plt.show()
</code>
### Hierarchical clustering on Customer Dataset
<code>
# load csv into pandas dataframe
df = pd.read_csv('SMAI-Dataset-hc-dataset/new_customers.csv')
df.head()
</code>
Rudimentary Feature Preprocessing
<code>
# drop the 'CustomerID' column
df.drop('CustomerID', axis=1, inplace=True)
# binary encode 'Genre'(Gender) column
# Replace 'Male' with 0 and 'Female' with 1 in the 'Genre' column
df['Genre'] = df['Genre'].replace({'Male': 0, 'Female': 1})
df.head()
</code>
<code>
df.info()
</code>
<code>
# convert df to numpy array
X = df.values
# perform hierarchical clustering using all features and linkage method as 'single'
hc = HierarchicalClustering(X, linkage_method='single')
linkage_matrix = hc.linkages()
# print(linkage_matrix)
hc.plot_dendrogram(linkage_matrix)
</code>
### Hierarchical Clustering on Gene Expression Dataset
<code>
# load csv into pandas dataframe
df = pd.read_csv('SMAI-Dataset-gene-expression/gene.csv')
df.head()
</code>
<code>
df.info()
</code>
<code>
# drop the 'ID_REF' column
df.drop('ID_REF', axis=1, inplace=True)
df.head()
</code>
<code>
# convert df to numpy array
X = df.values
# perform hierarchical clustering using all features and linkage method as 'single'
hc = HierarchicalClustering(X, linkage_method='single')
linkage_matrix = hc.linkages()
# print(linkage_matrix)
hc.plot_dendrogram(linkage_matrix)
</code>
|
{
"filename": "3.ipynb",
"repository": "Anika-Roy/Exploring-Unsupervised-Learning-Methods",
"query": "transformed_from_existing",
"size": 102901,
"sha": ""
}
|
# ENCODE_FCC_02_region_macs_annotate_chipseq_subset_SH_3.ipynb
Repository: ReddyLab/Proj
**Set environment**
<code>
source ../run_config_project.sh
show_env
</code>
**Set global variables**
<code>
ls -1 ${FD_RES}/region
</code>
<code>
ls -1 ${FD_RES}/region/summary
</code>
<code>
FP_REGION_LABEL_A=${FD_RES}/region/summary/metadata.label.astarr_macs.tsv
FP_REGION_LABEL_B=${FD_RES}/region/summary/metadata.label.chipseq_subset.tsv
</code>
<code>
ls ${FP_REGION_LABEL_A}
cat ${FP_REGION_LABEL_A}
</code>
<code>
ls ${FP_REGION_LABEL_B}
cat ${FP_REGION_LABEL_B}
</code>
## Preview
<code>
### Loop region A
while read FOLDER_REG_A FNAME_REG_A LABEL_REG_A; do
### Set input A
FD_INP_A=${FD_RES}/region/${FOLDER_REG_A}
FN_INP_A=${FNAME_REG_A}
FP_INP_A=${FD_INP_A}/${FN_INP_A}
FOLDER_A=${LABEL_REG_A}
### Loop region B
while read FOLDER_REG_B FNAME_REG_B LABEL_REG_B; do
### Set input B
FD_INP_B=${FD_RES}/region/${FOLDER_REG_B}
FN_INP_B=${FNAME_REG_B}
FP_INP_B=${FD_INP_B}/${FN_INP_B}
FOLDER_B=${FOLDER_REG_B}
### Set output
FD_OUT=${FD_RES}/region_annotation/${FOLDER_A}/${FOLDER_B}
FN_OUT=${LABEL_REG_A}.${LABEL_REG_B}.bed.gz
FP_OUT=${FD_OUT}/${FN_OUT}
### setup log file
FN_LOG=region.annotation.${LABEL_REG_A}.${LABEL_REG_B}.txt
FP_LOG=${FD_LOG}/${FN_LOG}
### Set script
FP_EXE=${FD_EXE}/run_bedtools_intersect.sh
### show progress
echo ==============================
echo "Output Label A:" ${LABEL_REG_A}
echo "Output Label B:" ${LABEL_REG_B}
echo
echo "Output FDiry: " ${FD_OUT}
echo "Output FName: " ${FN_OUT}
echo "Log FPath: " '${FD_LOG}/'${FN_LOG}
echo
done < <(cat ${FP_REGION_LABEL_B} | awk 'NR >=2 {print}')
done < <(cat ${FP_REGION_LABEL_A} | awk 'NR >=2 {print}')
</code>
## Execute
<code>
### Loop region A
while read FOLDER_REG_A FNAME_REG_A LABEL_REG_A; do
### Set input A
FD_INP_A=${FD_RES}/region/${FOLDER_REG_A}
FN_INP_A=${FNAME_REG_A}
FP_INP_A=${FD_INP_A}/${FN_INP_A}
FOLDER_A=${LABEL_REG_A}
### Loop region B
while read FOLDER_REG_B FNAME_REG_B LABEL_REG_B; do
### Set input B
FD_INP_B=${FD_RES}/region/${FOLDER_REG_B}
FN_INP_B=${FNAME_REG_B}
FP_INP_B=${FD_INP_B}/${FN_INP_B}
FOLDER_B=${FOLDER_REG_B}
### Set output
FD_OUT=${FD_RES}/region_annotation/${FOLDER_A}/${FOLDER_B}
FN_OUT=${LABEL_REG_A}.${LABEL_REG_B}.bed.gz
FP_OUT=${FD_OUT}/${FN_OUT}
### setup log file
FN_LOG=region.annotation.${LABEL_REG_A}.${LABEL_REG_B}.txt
FP_LOG=${FD_LOG}/${FN_LOG}
### Set script
FP_EXE=${FD_EXE}/run_bedtools_intersect.sh
### show progress
echo ==============================
echo "Output Label A:" ${LABEL_REG_A}
echo "Output Label B:" ${LABEL_REG_B}
echo
echo "Output FDiry: " ${FD_OUT}
echo "Output FName: " ${FN_OUT}
echo "Log FPath: " '${FD_LOG}/'${FN_LOG}
echo
### execute
mkdir -p ${FD_OUT}
sbatch -p ${NODE} \
--exclude=dl-01 \
--cpus-per-task 4 \
--mem 4G \
--output ${FP_LOG} \
${FP_EXE} ${FD_PRJ} ${FP_INP_A} ${FP_INP_B} ${FP_OUT}
echo
done < <(cat ${FP_REGION_LABEL_B} | awk 'NR >=2 {print}')
done < <(cat ${FP_REGION_LABEL_A} | awk 'NR >=2 {print}')
</code>
## Review
<code>
ls -1 /data/reddylab/Kuei/repo/Proj_ENCODE_FCC/results/region_annotation
</code>
<code>
FDIRY=${FD_RES}/region
ls ${FDIRY}/encode_chipseq_subset/*.bed.gz | wc -l
ls -1 ${FDIRY}/encode_chipseq_subset/*.bed.gz | xargs -n 1 basename
</code>
<code>
FDIRY=${FD_RES}/region_annotation/fcc_astarr_macs_input_overlap/encode_chipseq_subset
ls ${FDIRY}/*.bed.gz | wc -l
ls -1 ${FDIRY}/*.bed.gz | xargs -n 1 basename
</code>
<code>
FDIRY=${FD_RES}/region_annotation/fcc_astarr_macs_input_union/encode_chipseq_subset
ls ${FDIRY}/*.bed.gz | wc -l
ls -1 ${FDIRY}/*.bed.gz | xargs -n 1 basename
</code>
<code>
cat ${FD_LOG}/region.annotation.fcc_astarr_macs_input_overlap.encode_chipseq_ATF1_ENCFF627RSK.txt
</code>
<code>
cat ${FD_LOG}/region.annotation.fcc_astarr_macs_input_union.encode_chipseq_ATF1_ENCFF627RSK.txt
</code>
|
{
"filename": "ENCODE_FCC_02_region_macs_annotate_chipseq_subset_SH_3.ipynb",
"repository": "ReddyLab/Proj",
"query": "transformed_from_existing",
"size": 80473,
"sha": ""
}
|
# MLP_dataset-split_predict.ipynb
Repository: huangyiru123/mace
<code>
import numpy as np
from ase.io import read, write
from ase import units
from ase.md.langevin import Langevin
from ase.optimize import BFGS
from mace.calculators import MACECalculator
from matplotlib import pyplot as plt
from tqdm import tqdm
import os
# 1️⃣ 加载训练好的 MACE 模型
model_path = 'MACE_model.model' # 你的模型文件路径
# 检查模型文件是否存在
if not os.path.exists(model_path):
print(f"Model file {model_path} not found!")
else:
calculator = MACECalculator(
model_path=model_path, # 你的模型文件
device='cuda' # 如果没有 GPU,则改成 'cpu'
)
# 1️⃣ 加载分子结构数据(读取第一帧)
init_conf = read('test.extxyz', '0') # 读取第一帧
init_conf.set_calculator(calculator) # 设置计算器(此处calculator为MACE)
# 2️⃣ 设置 Langevin 动力学参数
dyn = Langevin(
init_conf, # 初始分子结构
2.0 * units.fs, # 时间步长
temperature_K=310, # 目标温度 310K
friction=5e-3 # 摩擦系数
)
# 3️⃣ 记录每 10 步的数据,并附加计算结果(如能量,力等)
def write_frame():
# 获取力
forces = dyn.atoms.get_forces()
# 获取势能和温度
energy = dyn.atoms.get_potential_energy()
temperature = dyn.atoms.get_temperature()
# 将能量、力、温度等附加到原子信息中(使用不同的键名避免重复)
dyn.atoms.info['energy_langevin'] = energy
dyn.atoms.arrays['forces_langevin'] = forces
dyn.atoms.info['temperature'] = temperature
# 保存到文件
dyn.atoms.write('test.xyz', append=True) # 追加方式写入数据
# 输出相关信息
print(f"Time: {dyn.get_time() / units.fs:.2f} fs, Energy: {energy:.3f} eV, Temperature: {temperature:.2f} K")
# 4️⃣ 将 write_frame 函数附加到 MD 动力学模拟中,每 10 步记录一次
dyn.attach(write_frame, interval=10) # 每 10 步存一次数据
# 5️⃣ 运行 1000 个时间步(你可以根据需求调整步数)
print("Running Molecular Dynamics Simulation...")
dyn.run(1000) # 运行 1000 步
print("Simulation completed. Results saved in test.xyz.")
# 6️⃣ 进行 MACE 力的预测
mace_calcs = MACECalculator(model_path=model_path, device='cuda')
# 7️⃣ 进行 MACE 力的预测
# 读取模拟过程中保存的轨迹
traj = read('test.xyz', ':')
forces_mace = []
energies_mace = []
for at in tqdm(traj):
at.calc = mace_calcs # 使用 MACE 计算器
forces = at.get_forces() # 获取预测的力
energies_mace.append(at.get_potential_energy()) # 获取势能
# 保存力到原子数组中(使用不同的键名避免重复)
at.arrays['forces_mace'] = forces
at.info['energy_mace'] = at.get_potential_energy() # 保存MACE能量
forces_mace.append(forces)
# 8️⃣ 绘制能量和力随时间变化的图
# 绘制 MACE 力和势能随时间的变化
plt.figure(figsize=(10, 6))
# 绘制能量随时间的变化
plt.subplot(2, 1, 1)
plt.plot(np.arange(len(traj)), energies_mace, label='MACE Energy')
plt.xlabel('Time (fs)')
plt.ylabel('Energy (eV/atom)')
plt.legend()
# 绘制力随时间的变化
plt.subplot(2, 1, 2)
force_magnitude = np.linalg.norm(np.array(forces_mace), axis=1)
plt.plot(np.arange(len(traj)), force_magnitude, label='MACE Force', color='r')
plt.xlabel('Time (fs)')
plt.ylabel('Force (eV/Å)')
plt.legend()
plt.tight_layout()
plt.show()
# 9️⃣ 进行结构优化:使用 BFGS 优化器进行结构优化,直到力趋近于零
# 使用 BFGS 优化器进行结构优化
opt = BFGS(traj[-1]) # 使用最后一帧(或选择其他帧进行优化)
opt.run(fmax=0.01) # 设置最大力为 0.01 eV/Å
# 10️⃣ 保存优化后的结果
write('test_optimized.xyz', traj[-1]) # 保存优化后的轨迹文件
print("Optimization completed. Optimized structure saved in test_optimized.xyz.")
</code>
|
{
"filename": "MLP_dataset-split_predict.ipynb",
"repository": "huangyiru123/mace",
"query": "transformed_from_existing",
"size": 189920,
"sha": ""
}
|
# image_processing_00-Index.ipynb
Repository: guiwitz/Python
# Image processing with Python
#### **Guillaume Witz**, Science IT Support, Bern University
## Table of Contents
### [1. Introduction](01-Introduction.ipynb)
### [2. Numpy refresh with images](02-Numpy_images.ipynb)
### [3. Importing images](03-Image_import.ipynb)
### [4. Basics: scaling, thresholding, filtering](04-Basics.ipynb)
### [4b. Basics with non-biology data](04b-Other_image_types.ipynb)
### [5. Binary operations](05-Binary_operations.ipynb)
### [6. Wrapping code into functions](06-Functions.ipynb)
### [7. Segmentation: active contours](07-Active_contours.ipynb)
### [8. Segmentation: pattern matching](08-Pattern_matching.ipynb)
### [9. Segmentation: Watershed algorithm](09-Watershed.ipynb)
### [10. Operations in 3D](10-3D_case.ipynb)
### [11. A complete pipeline](11-Complete_analysis.ipynb)
### [12. Image registration](12-Registration.ipynb)
### [13. Machine learning: Pixel classification](13-Pixel_classification.ipynb)
### [14.Machine learning: segmentation with deep learning](14-DeepLearning.ipynb)
|
{
"filename": "image_processing_00-Index.ipynb",
"repository": "guiwitz/Python",
"query": "transformed_from_existing",
"size": 2301,
"sha": ""
}
|
# Data_Analysis_Pipeline.ipynb
Repository: LewisLabUCSD/GlycoMSNetworking
1. Oxonium MS/MS filtrering
<code>
import pandas as pd
from pyteomics import mgf, pylab_aux
import pylab
import math
def filter_mgf(source, seed_mass=204.09, tol=0.01):
filtered_spectra=[]
spectra = mgf.read(source)
for i in range(len(spectra)):
spectrum = next(spectra)
if any([math.isclose(seed_mass, k, abs_tol=tol) for k in spectrum['m/z array']]):
filtered_spectra.append(spectrum)
return filtered_spectra
</code>
<code>
files = [] # list of MGF files
for file in files:
mgf.write(filter_mgf(source=file), output=file.replace('.mgf', '_filtered_001.mgf'))
</code>
2. Byonics search + METABOLOMICS-SNETS-V2 output Network walk
<code>
import pandas as pd
import numpy as np
import networkx as nx
import math
import json
</code>
<code>
node_data = pd.read_csv('', index_col='shared name',low_memory=False) # METABOLOMICS-SNETS-V2 output node information
node_data = node_data.loc[node_data['number of spectra']>=3]
edge_data = pd.read_csv('',low_memory=False) # METABOLOMICS-SNETS-V2 output output edge information
data = pd.read_excel('') # Byonics search data
data = data.loc[~data['Glycan Composition'].str.contains('Na')]
</code>
<code>
G = nx.DiGraph()
for index in edge_data.index:
if edge_data.loc[index]['node1'] != edge_data.loc[index]['node2'] \
and edge_data.loc[index]['node1'] in node_data.index \
and edge_data.loc[index]['node2'] in node_data.index:
node1_mass = node_data.loc[edge_data.loc[index]['node1']]['parent mass']
node2_mass = node_data.loc[edge_data.loc[index]['node2']]['parent mass']
mass_difference = edge_data.loc[index]['mass_difference']
if mass_difference < 0:
if node1_mass >= node2_mass:
node1=edge_data.loc[index]['node1']
node2=edge_data.loc[index]['node2']
else:
node1=edge_data.loc[index]['node2']
node2=edge_data.loc[index]['node1']
if mass_difference >= 0:
if node1_mass >= node2_mass:
node1=edge_data.loc[index]['node2']
node2=edge_data.loc[index]['node1']
else:
node1=edge_data.loc[index]['node1']
node2=edge_data.loc[index]['node2']
G.add_edge(node1, node2, mass_difference=mass_difference)
nx.set_node_attributes(G, {i[0]:{'parent mass':i[1], 'precursor mass':i[2], 'number of spectra':i[3], 'UniqueFileSources':i[4]} for i in zip(node_data.index,
node_data['parent mass'],
node_data['precursor mass'],
node_data['number of spectra'],
node_data['UniqueFileSources'])})
node_parent_mass_d = {i[1]['parent mass']:i[0]for i in list(G.nodes(data=True))}
count=0
for seed_mass, seed_pep, seed_glycan in zip(data['Theo. MH+ [Da]'], data['Annotated Sequence'], data['Glycan Composition']):
for k,v in node_parent_mass_d.items():
if math.isclose(seed_mass, k, abs_tol=1.10):# corrected
count+=1
try:
G.nodes[v]['glycoform'].append((seed_pep, seed_glycan))
#print( G.nodes[v], v, seed_pep, seed_glycan, node_parent_mass_d[k])
except:
G.nodes[v].update({'glycoform':[(seed_pep, seed_glycan)]})
print(nx.info(G))
</code>
<code>
def glycan_motifs(glycan):
motifs = {}
for a in glycan.split(')')[:-1]:
motifs[a.split('(')[0]]=int(a.split('(')[1])
return motifs
from itertools import combinations_with_replacement
def find_possible_glycoforms(seed_glycoform, mass_diff, n, mono_mass, tol=0.10):
new_glycans=[]
mono_mass_rev = {v:k for k,v in mono_mass.items()}
num = list(mono_mass.values()) + [-1*i for i in mono_mass.values()]
for m in range(1,n+1):
for masses in combinations_with_replacement(num,m):
if math.isclose(mass_diff, sum(masses), abs_tol=tol) and [abs(i) for i in masses].count(1.0)<=1:
foo=True
test = list(zip(masses, [mono_mass_rev[abs(j)] for j in masses]))
for index in range(len(test)):
if foo:
for j_index in range(len(test[index+1:])):
if sum((test[index][0], test[index+1+j_index][0]))==0:
foo=False
break
else:
break
if not foo:
pass
else:
bar = True
motifs = glycan_motifs(seed_glycoform)
motifs.update({'HYDRO':1.0})
for item, val in zip([mono_mass_rev[abs(j)] for j in masses], masses):
if val > 0:
try:
motifs[item]+=1
except:
motifs.update({item:1})
elif val < 0:
try:
motifs[item]-=1
except:
bar=False
break
if bar:
motifs.pop('HYDRO', None)
order = {'HexNAc':0, 'Hex': 1, 'Fuc': 2, 'NeuAc': 3, 'NeuGc': 4}
new_glycan = ''.join(['{}({})'.format(i,j) for i,j in sorted(motifs.items(), key=lambda x: order[x[0]]) if j!=0])
elif not bar:
new_glycan = ''
if '-' in new_glycan or len(new_glycan)==0:
break
new_glycans.append(new_glycan)
if mass_diff <= tol and mass_diff >= 0:
new_glycans.append(seed_glycoform)
return new_glycans
</code>
<code>
# go through nodes until len new_nodes == 0 new nodes added if brand new or mutated by addition
n=2
#mono_mass = {'HexNAc':203.080,'Hex':162.053,'Fuc':146.058,'NeuAc':291.095,'HYDRO':1.0}
mono_mass = {'HexNAc':203.080,'Hex':162.053,'Fuc':146.058,'NeuAc':291.095,'NeuGc':307.090,'HYDRO':1.0}
orig_nodes = [k for k,v in G.nodes(data=True) if 'glycoform' in v.keys()]
new_nodes = orig_nodes
edges_done = []
while len(new_nodes) != 0:
prev_nodes = new_nodes
new_nodes = list()
for node in prev_nodes:
for edge in nx.edges(G,node):
new_node = edge[1]
if new_node not in orig_nodes:
mass_diff = G.edges[edge]['mass_difference']
if (mass_diff < 0 and G.nodes(data=True)[node]['parent mass'] > G.nodes(data=True)[new_node]['parent mass']) or \
(mass_diff >= 0 and G.nodes(data=True)[node]['parent mass'] <= G.nodes(data=True)[new_node]['parent mass']):
if 'glycoform' in G.nodes[node].keys():
for seed_pep, seed_glycan in G.nodes[node]['glycoform']:
new_glycans = find_possible_glycoforms(seed_glycoform=seed_glycan, mass_diff=mass_diff, n=n, mono_mass=mono_mass)
if len(new_glycans) != 0:
#print(node, new_node, seed_pep, seed_glycan, new_glycans, mass_diff)
#print()
for new_glycan in new_glycans:
try:
if (seed_pep, new_glycan) not in G.nodes[new_node]['glycoform']:
G.nodes[new_node]['glycoform'].append((seed_pep, new_glycan))
new_nodes.append(new_node)
except:
G.nodes[new_node].update({'glycoform':[(seed_pep, new_glycan)]})
new_nodes.append(new_node)
# all nodes coming in
for inbound_node in [i[0] for i in G.edges() if i[1]==node]:
new_node = inbound_node
if 'glycoform' in G.nodes[new_node].keys():
if len(set([i[0] for i in G.nodes[new_node]['glycoform']]).intersection(set([i[0] for i in G.nodes[node]['glycoform']]))) > 0:
continue
for edge in [i for i in nx.edges(G,new_node) if i[1]==node]:
if new_node not in orig_nodes:
mass_diff = G.edges[edge]['mass_difference']
if (mass_diff < 0 and G.nodes(data=True)[new_node]['parent mass'] > G.nodes(data=True)[node]['parent mass']) or \
(mass_diff >= 0 and G.nodes(data=True)[new_node]['parent mass'] <= G.nodes(data=True)[node]['parent mass']):
if 'glycoform' in G.nodes[node].keys():
for seed_pep, seed_glycan in G.nodes[node]['glycoform']:
new_glycans = find_possible_glycoforms(seed_glycoform=seed_glycan, mass_diff=-1*(mass_diff), n=n, mono_mass=mono_mass)
if len(new_glycans) != 0:
#print(node, new_node, seed_pep, seed_glycan, new_glycans, mass_diff)
#print()
for new_glycan in new_glycans:
try:
if (seed_pep, new_glycan) not in G.nodes[new_node]['glycoform']:
G.nodes[new_node]['glycoform'].append((seed_pep, new_glycan))
new_nodes.append(new_node)
except:
G.nodes[new_node].update({'glycoform':[(seed_pep, new_glycan)]})
new_nodes.append(new_node)
</code>
<code>
nx.to_pandas_edgelist(G).to_csv('', index=False)
nodelist_data = pd.DataFrame.from_dict(G.nodes, orient='index')
seed_d = {}
for index in nodelist_data.loc[nodelist_data.notna()['glycoform']].index:
if index in orig_nodes:
seed_d[index]='Y'
else:
seed_d[index]='N'
nodelist_data['Byonics Seed'] = pd.Series(seed_d)
accession_map = dict(zip(data['Annotated Sequence'], data['Master Protein Accessions']))
accession_d = {}
for index in nodelist_data.loc[nodelist_data.notna()['glycoform']].index:
accession_d[index] = [accession_map[i[0]] for i in nodelist_data.loc[index]['glycoform']]
nodelist_data['Protein Accession'] = pd.Series(accession_d)
nodelist_data.to_csv('')
</code>
3. Glycan Fragmentation Network
<code>
import pandas as pd
import numpy as np
</code>
<code>
data = pd.read_csv('', index_col=0) # nodelist_data from previous section
data = data.dropna(subset=['glycoform'])
glycans = set([i[1] for j in data['glycoform'] for i in eval(j)])
glycans = (glycans - set([i for i in glycans if 'HexNAc' not in i]))
</code>
<code>
def glycan_motifs(glycan):
motifs = {}
for a in glycan.split(')')[:-1]:
motifs[a.split('(')[0]]=int(a.split('(')[1])
return motifs
def tree_builder(mod_motifs):
tree = {}
for k,v in mod_motifs.items():
for k2,v2 in mod_motifs.items():
bar=False
if k!=k2:
if len(v2)==len(v):
foo=[]
for mod in v:
if mod[0] in [i[0] for i in v2]:
foo.append(True)
else:
foo.append(False)
if sum(foo)==len(v2):
foo=[]
for mod in v:
if mod[1] == [i[1] for i in v2 if mod[0]==i[0]][0]:
foo.append(True)
elif mod[1] == str(int([i[1] for i in v2 if mod[0]==i[0]][0])-1):
foo.append(False)
else:
for x in range(len(v2)*2):
foo.append(True)
if sum(foo)==len(v2)-1:
if k2 in tree.keys():
tree[k2].append(k)
bar=True
else:
tree[k2]=[]
tree[k2].append(k)
bar=True
if len(v2)-len(v)==1:
foo=[]
for mod in v:
if mod[0] in [i[0] for i in v2]:
foo.append(True)
else:
foo.append(False)
if [i for i in v2 if i[0] not in [i[0] for i in v]][0][1]==str(1):
if sum(foo)==len(v):
foo=[]
for mod in v:
if mod[1] == [i[1] for i in v2 if mod[0]==i[0]][0]:
foo.append(True)
else:
foo.append(False)
if sum(foo)==len(v):
if k2 in tree.keys():
tree[k2].append(k)
bar=True
else:
tree[k2]=[]
tree[k2].append(k)
bar=True
return tree
</code>
<code>
mod_motifs = {}
for glycan in glycans:
mod_motifs[glycan]=[[k, str(v)] for k,v in glycan_motifs(glycan).items()]
tree = tree_builder(mod_motifs)
</code>
<code>
while len(glycans - set(tree.keys())) > 1:
new_glycans = set()
for glycan in (glycans - set(tree.keys())):
temp_new_glycans = []
for k in glycan_motifs(glycan).keys():
temp = glycan_motifs(glycan)
temp[k] = temp[k]-1
order = {'HexNAc':0, 'Hex': 1, 'Fuc': 2, 'NeuAc': 3, 'NeuGc': 4}
temp_new_glycans.append(''.join(['{}({})'.format(i,j) for i,j in sorted(temp.items(), key=lambda x: order[x[0]]) if j!=0]))
if any([temp_new_glycan in glycans for temp_new_glycan in temp_new_glycans]):
temp_new_glycans = [temp_new_glycan for temp_new_glycan in temp_new_glycans if temp_new_glycan in glycans]
new_glycans = new_glycans.union(set(temp_new_glycans))
new_glycans = new_glycans - set([''])
new_glycans = set([new_glycan for new_glycan in new_glycans if 'HexNAc' in new_glycan])
glycans = glycans.union(new_glycans)
mod_motifs = {}
for glycan in glycans:
mod_motifs[glycan]=[[k, str(v)] for k,v in glycan_motifs(glycan).items()]
tree = tree_builder(mod_motifs)
</code>
<code>
need_fuc_variant = []
for k in tree.keys():
if 'Fuc' in k:
foo=True
keys = [k]
while foo:
try:
need_fuc_variant+=[i for j in keys for i in tree[j]]
keys = [i for j in keys for i in tree[j]]
except:
foo=False
new_glycans=set()
for i in set([i for i in need_fuc_variant if 'Fuc' not in i]):
temp = glycan_motifs(i)
temp.update({'Fuc':1})
order = {'HexNAc':0, 'Hex': 1, 'Fuc': 2, 'NeuAc': 3, 'NeuGc': 4}
new_glycans.add(''.join(['{}({})'.format(l,m) for l,m in sorted(temp.items(), key=lambda x: order[x[0]]) if m!=0]))
#if 'Hex(' in i:
#new_glycans.add(i[:15]+'Fuc(1)'+i[15:])
#else:
#new_glycans.add(i[:9]+'Fuc(1)'+i[9:])
mod_motifs = {}
for glycan in (glycans.union(new_glycans)):
mod_motifs[glycan]=[[k, str(v)] for k,v in glycan_motifs(glycan).items()]
tree = tree_builder(mod_motifs)
</code>
<code>
source=[]
target=[]
for k,v in tree.items():
for i in range(len(v)):
source.append(k)
target.append(v[i])
cyto_df = pd.DataFrame(columns=['source','target'])
cyto_df['source']=source
cyto_df['target']=target
cyto_df.to_csv('', index=False) # glycan_fragmentation_tree for cytoscape
import pickle
with open('', 'wb') as fp:
pickle.dump(tree, fp) # glycan_fragmentation_tree for library builder
</code>
4. Spectral Library Builder and Precursor Library Builder
<code>
import pandas as pd
import numpy as np
from pyteomics import mass
# oxonium ions
ox_d = {}
ox_d[0] = ['[HexNAc-C2H6O3]', '[+126.0550]', 'OX', float('126.0550'), int('1'), float('126.0550')]
ox_d[1] = ['[HexNAc-CH6O3]', '[+138.0550]', 'OX', float('138.0550'), int('1'), float('138.0550')]
ox_d[2] = ['[HexNAc-C2H4O2]', '[+144.0656]', 'OX', float('144.0656'), int('1'), float('144.0656')]
ox_d[3] = ['[Hex-H2O]', '[+145.0495]', 'OX', float('145.0495'), int('1'), float('145.0495')]
ox_d[4] = ['[Hex]', '[+163.0601]', 'OX', float('163.0601'), int('1'), float('163.0601')]
ox_d[5] = ['[HexNAc-2H2O]', '[+168.0655]', 'OX', float('168.0655'), int('1'), float('168.0655')]
ox_d[6] = ['[HexNAc-H2O]', '[+186.0761]', 'OX', float('186.0761'), int('1'), float('186.0761')]
ox_d[7] = ['[HexNAc]', '[+204.0867]', 'OX', float('204.0867'), int('1'), float('204.0867')]
ox_d[8] = ['[HexHexNAc]', '[+366.1395]', 'OX', float('366.1395'), int('1'), float('366.1395')]
ox_d[9] = ['[Neu5Ac-H2O]', '[+274.092]', 'OX', float('274.092'), int('1'), float('274.092')]
ox_d[10] = ['[Neu5Ac]', '[+292.103]', 'OX', float('292.103'), int('1'), float('292.103')]
ox_df = pd.DataFrame(ox_d).T
ox_df.columns=['Annotation', 'FragmentSeq', 'FragmentType', 'TheoFragmentMass', 'z', 'FragmentMZ']
</code>
<code>
import pickle
with open('', 'rb') as input_file:
glycan_frag_network = pickle.load(input_file) # glycan_fragmentation_tree for library builder
</code>
<code>
def find_children(k, tree):
children = []
keys = [k]
foo=True
while foo:
new_children=[]
for j in keys:
try:
new_children+=[i for i in tree[j]]
except:
continue
if len(new_children)>0:
keys=new_children
children+=new_children
else:
foo=False
return set(children)
def glycan_motifs(glycan):
motifs = {}
for a in glycan.split(')')[:-1]:
motifs[a.split('(')[0]]=int(a.split('(')[1])
return motifs
</code>
<code>
data = pd.read_csv('', index_col=0) # nodelist_data from previous section
data = data.dropna(subset=['glycoform'])
data['glycoform'] = [eval(i) for i in data['glycoform']]
mol_network_data = pd.read_csv('', index_col='name',low_memory=False).loc[data.index] # METABOLOMICS-SNETS-V2 output node information
</code>
<code>
mono_mass = {'HexNAc':203.080,'Hex':162.053,'Fuc':146.058,'NeuAc':291.095,'NeuGc':307.090}
precursor_mzs = []
frag_mzs = []
rela_frag_ints = []
precursor_rts = []
frag_annotations = []
frag_types = []
frag_zs = []
theo_frag_masses = []
precursor_zs = []
peptide_seqs = []
precursor_annotations = []
decoys = []
for index in data.index:
for precursor_peptide, precursor_glycan in data.loc[index]['glycoform']:
precursor_rt = round(mol_network_data.loc[index]['RTMean']/60, 4)
precursor_motifs = glycan_motifs(precursor_glycan)
precursor_pep_mass = mass.calculate_mass(sequence=precursor_peptide.split('.')[1])
precursor_pep_glycan_mass = precursor_pep_mass + sum([mono_mass[k]*int(v) for k,v in precursor_motifs.items()])
#precursor_pep_glycan_mass = data.loc[index]['parent mass']
#precursor_pep_mass = data.loc[index]['parent mass'] - sum([mono_mass[k]*int(v) for k,v in precursor_motifs.items()])
for z in range(2,4+1):
precursor_charge = z
precursor_mz = (precursor_pep_glycan_mass+z)/z
for glycan_frag in find_children(k=precursor_glycan,tree=glycan_frag_network):
frag_motifs = glycan_motifs(glycan_frag)
for i in range(1,z):
frag_mz = (precursor_pep_mass+sum([mono_mass[k]*int(v) for k,v in frag_motifs.items()])+i)/i
frag_charge = i
theo_frag_mass = precursor_pep_mass+sum([mono_mass[k]*int(v) for k,v in frag_motifs.items()])+1
precursor_mzs.append(precursor_mz)
frag_mzs.append(frag_mz)
rela_frag_ints.append(0.5)
precursor_rts.append(precursor_rt)
frag_annotations.append(glycan_frag)
frag_types.append('Y')
frag_zs.append(frag_charge)
theo_frag_masses.append(theo_frag_mass)
precursor_zs.append(precursor_charge)
peptide_seqs.append(precursor_peptide)
precursor_annotations.append(precursor_glycan)
decoys.append(0)
#oxonium ions
for ox_ion in ox_df.index:
if ox_df.loc[ox_ion]['Annotation'] == '[Hex-H2O]' or ox_df.loc[ox_ion]['Annotation'] == '[Hex]':
try:
if precursor_motifs['HexNAc']==2 and precursor_motifs['Hex']>=4:
frag_mz = ox_df.loc[ox_ion]['FragmentMZ']
glycan_frag = ox_df.loc[ox_ion]['Annotation']
frag_charge = ox_df.loc[ox_ion]['z']
theo_frag_mass = ox_df.loc[ox_ion]['TheoFragmentMass']
#print(precursor_glycan, glycan_frag, frag_mz, frag_charge)
precursor_mzs.append(precursor_mz)
frag_mzs.append(frag_mz)
rela_frag_ints.append(0.5)
precursor_rts.append(precursor_rt)
frag_annotations.append(glycan_frag)
frag_types.append('OX')
frag_zs.append(frag_charge)
theo_frag_masses.append(theo_frag_mass)
precursor_zs.append(precursor_charge)
peptide_seqs.append(precursor_peptide)
precursor_annotations.append(precursor_glycan)
decoys.append(0)
except:
continue
if ox_df.loc[ox_ion]['Annotation'] == '[HexHexNAc]':
try:
if precursor_motifs['HexNAc']>2 and precursor_motifs['Hex']>3:
frag_mz = ox_df.loc[ox_ion]['FragmentMZ']
glycan_frag = ox_df.loc[ox_ion]['Annotation']
frag_charge = ox_df.loc[ox_ion]['z']
theo_frag_mass = ox_df.loc[ox_ion]['TheoFragmentMass']
#print(precursor_glycan, glycan_frag, frag_mz, frag_charge)
precursor_mzs.append(precursor_mz)
frag_mzs.append(frag_mz)
rela_frag_ints.append(0.5)
precursor_rts.append(precursor_rt)
frag_annotations.append(glycan_frag)
frag_types.append('OX')
frag_zs.append(frag_charge)
theo_frag_masses.append(theo_frag_mass)
precursor_zs.append(precursor_charge)
peptide_seqs.append(precursor_peptide)
precursor_annotations.append(precursor_glycan)
decoys.append(0)
except:
continue
if ox_df.loc[ox_ion]['Annotation'] == '[Neu5Ac-H2O]' or ox_df.loc[ox_ion]['Annotation'] == '[Neu5Ac]':
if 'NeuAc' in precursor_glycan or 'NeuGc' in precursor_glycan:
frag_mz = ox_df.loc[ox_ion]['FragmentMZ']
glycan_frag = ox_df.loc[ox_ion]['Annotation']
frag_charge = ox_df.loc[ox_ion]['z']
theo_frag_mass = ox_df.loc[ox_ion]['TheoFragmentMass']
#print(precursor_glycan, glycan_frag, frag_mz, frag_charge)
precursor_mzs.append(precursor_mz)
frag_mzs.append(frag_mz)
rela_frag_ints.append(0.5)
precursor_rts.append(precursor_rt)
frag_annotations.append(glycan_frag)
frag_types.append('OX')
frag_zs.append(frag_charge)
theo_frag_masses.append(theo_frag_mass)
precursor_zs.append(precursor_charge)
peptide_seqs.append(precursor_peptide)
precursor_annotations.append(precursor_glycan)
decoys.append(0)
elif ox_df.loc[ox_ion]['Annotation'] == '[HexNAc-C2H6O3]' or \
ox_df.loc[ox_ion]['Annotation'] == '[HexNAc-CH6O3]' or \
ox_df.loc[ox_ion]['Annotation'] == '[HexNAc-C2H4O2]' or \
ox_df.loc[ox_ion]['Annotation'] == '[HexNAc-2H2O]' or \
ox_df.loc[ox_ion]['Annotation'] == '[HexNAc-H2O]' or \
ox_df.loc[ox_ion]['Annotation'] == '[HexNAc]':
frag_mz = ox_df.loc[ox_ion]['FragmentMZ']
glycan_frag = ox_df.loc[ox_ion]['Annotation']
frag_charge = ox_df.loc[ox_ion]['z']
theo_frag_mass = ox_df.loc[ox_ion]['TheoFragmentMass']
#print(precursor_glycan, glycan_frag, frag_mz, frag_charge)
precursor_mzs.append(precursor_mz)
frag_mzs.append(frag_mz)
rela_frag_ints.append(0.5)
precursor_rts.append(precursor_rt)
frag_annotations.append(glycan_frag)
frag_types.append('OX')
frag_zs.append(frag_charge)
theo_frag_masses.append(theo_frag_mass)
precursor_zs.append(precursor_charge)
peptide_seqs.append(precursor_peptide)
precursor_annotations.append(precursor_glycan)
decoys.append(0)
</code>
<code>
spectral_lib = pd.DataFrame()
spectral_lib['PrecursorMz'] = precursor_mzs
spectral_lib['FragmentMz'] = frag_mzs
spectral_lib['RelativeFragmentIntensity'] = rela_frag_ints
spectral_lib['RetentionTime'] = precursor_rts
spectral_lib['Annotation'] = frag_annotations
spectral_lib['FragmentType'] = frag_types
spectral_lib['FragmentCharge'] = frag_zs
spectral_lib['TheoreticalFragmentMass'] = theo_frag_masses
spectral_lib['PrecursorCharge'] = precursor_zs
spectral_lib['PeptideSequence'] = peptide_seqs
spectral_lib['PrecursorAnnotation'] = precursor_annotations
spectral_lib['Decoy'] = decoys
spectral_lib.to_csv('', index=False,sep='\t')
</code>
<code>
precursor_lib = spectral_lib[['PeptideSequence','PrecursorAnnotation','PrecursorMz','PrecursorCharge','PrecursorMz','PrecursorCharge']]
precursor_lib.columns = ['Molecule List Name','Molecule Name','Precursor Mz','Precursor Charge','Product Mz','Product Charge']
precursor_lib = precursor_lib.drop_duplicates()
precursor_lib['Molecule List Name']=[i.split('.')[1] for i in precursor_lib['Molecule List Name']]
precursor_lib.insert(6,'Fragment Ion', 'precursor')
precursor_lib.to_csv('', index=False)
</code>
|
{
"filename": "Data_Analysis_Pipeline.ipynb",
"repository": "LewisLabUCSD/GlycoMSNetworking",
"query": "transformed_from_existing",
"size": 38022,
"sha": ""
}
|
# QnA_Platform_for_EdTech_google_palm.ipynb
Repository: Latisha-cpu/EduGenie
<code>
from langchain.llms import GooglePalm
api_key = 'AIzaSyCAMmIjOar_aiZfl-Ds-XgRj4-CD06B7ig' # get this free api key from https://makersuite.google.com/
llm = GooglePalm(google_api_key=api_key, temperature=0.1)
</code>
<code>
facts = llm("AI for biologists")
print(facts)
</code>
<code>
from langchain.chains import RetrievalQA
from langchain.embeddings import GooglePalmEmbeddings
from langchain.llms import GooglePalm
</code>
<code>
from langchain.document_loaders.csv_loader import CSVLoader
loader = CSVLoader(file_path='query.csv', source_column="prompt")
# Store the loaded data in the 'data' variable
data = loader.load()
</code>
<code>
from langchain.embeddings import HuggingFaceInstructEmbeddings
# Initialize instructor embeddings using the Hugging Face model
instructor_embeddings = HuggingFaceInstructEmbeddings(model_name="hkunlp/instructor-large")
e = instructor_embeddings.embed_query("What is your refund policy?")
</code>
<code>
len(e)
</code>
<code>
e[:5]
</code>
<code>
from langchain.vectorstores import FAISS
# Create a FAISS instance for vector database from 'data'
vectordb = FAISS.from_documents(documents=data,
embedding=instructor_embeddings)
# Create a retriever for querying the vector database
retriever = vectordb.as_retriever(score_threshold = 0.7)
</code>
<code>
rdocs = retriever.get_relevant_documents("how about job placement support?")
rdocs
</code>
<code>
from langchain.prompts import PromptTemplate
prompt_template = """Given the following context and a question, generate an answer based on this context only.
In the answer try to provide as much text as possible from "response" section in the source document context without making much changes.
If the answer is not found in the context, kindly state "I don't know." Don't try to make up an answer.
CONTEXT: {context}
QUESTION: {question}"""
PROMPT = PromptTemplate(
template=prompt_template, input_variables=["context", "question"]
)
chain_type_kwargs = {"prompt": PROMPT}
from langchain.chains import RetrievalQA
chain = RetrievalQA.from_chain_type(llm=llm,
chain_type="stuff",
retriever=retriever,
input_key="query",
return_source_documents=True,
chain_type_kwargs=chain_type_kwargs)
</code>
<code>
chain('Do you provide job assistance and also do you provide job gurantee?')
</code>
|
{
"filename": "QnA_Platform_for_EdTech_google_palm.ipynb",
"repository": "Latisha-cpu/EduGenie",
"query": "transformed_from_existing",
"size": 14628,
"sha": ""
}
|
# what_how_why_1.ipynb
Repository: ben-silke/biol3209
# Why is there a problem?
Answering all these is your justification for doing the work and, at the very least, should make you comfortable you may be doing something worthwhile.
Seems like there are two different problems:
1. Finding a set of genes/ orthologs from a metagenomic sample
2. finding the orthologs
### Why does the problem even exist?
What is metagenomics?
Why is metagenomics important?
What insights can we gain from a large sample of DNA.
We can learn the type of orgisms and the array of this difference from a sample. e.g. What bacteria live in the gut?
We can learn the functional abilities of the organisms within the enviroment. e.g. What processes are the bacteria in the gut responsible for? What proteins are preset? What can this tell us about the environment?
### Why hasn’t it already been solved?
#### BLAST
BLAST allows one to one comparison however this complexity of this is great and a batch run on a large sample is not so effective. Is it possible to narrow the possibilities and then feed them to blast? A blast wrapper of sorts which can be used to minimie the operations which the BLAST search must undertake.
In a metagenomic analysis; there is a large amount of data. This means that any system which looks at a one to one comparsion of sequences is looking at $O(n^2)$ complexity. Because of this; this compelxity either needs to decrease or the time cost of each operation at each comparsion needs to decrease to make this a viable option for analysis.
# Examples of current solutions
Questions to answer / points to consider:
What is the evidence the solutions are inadequate? (That evidence can be measurement, or a theoretical property.)
The prior work was likely developed against different technological inputs 2.
Much of the information available to us now may not have been known to the authors of the original work.
## JustOrthologs:
### algorthim.
Just orthologs first looks for all the CDS's within the sample which are the exact same length. Then JustOrthologs looks to compare the CDS's to each other through the dinucleotide ratio's between the different CDS's.
1. This has the potential that a large number of false negatives are found. What is the actual likelihood that the CDS regions are of the exact same length? Therefore the sample space is decreased rapidly and very significantly. Proteins may differ in size by 5-10 amin acids; there 15-30 bps. This means that this restriction may simply be too much.
2. The amino acid ratio may not take into account the bp content of different species. Some species may be more GC rich and therefore could be additionally excluded in this calculation.
3. A dinucleotide ratio does not acutally inform the protein content. The redundancy of the genetic code means it may be better to determine proteins based upon the actual amino acid produced by a codon.
4. The program does not appear to take into account the fact the direction of the sequence upon entry. In true metagenomic samples; the direction of the sequence is unknown. In other words; which sample is actually being looked at. This can affect the ratio. E.g. is TA a true TA or is it an AT from another direction?
4.1. This acutally may be considered because there are 16 dinucleotides which are used.
5. The program does not seem to take into account rRNA or tRNA genes.
5.1. I expect that these are regions which are CDS (ATG-...-TAG) but it does not seem to pick up. The alternative is that the program uses prior annotated sequenes. This also represents issues because then you have to manuall annotate the sequences.
## MetaGeneMark 2:
## wtdbg2-assembler
This program is a genome assembler. It takes the reads from the metagenomic sample and assembles them into a sequence. It uses a fuzzy de Bruijn graph to do this.
<code>
import itertools
perms = itertools.permutations('ATCG', 2)
perms = [f'{perm[0]}{perm[1]}' for perm in perms]
print(perms)
</code>
# What is the problem?
1. finding genes within a metagenomic sample
2. are any of the genes found likely to be orthologs/ (related/ the same)?
3. Should we do anything we these orthologs once they have been found.
What is the problem you will solve?
Try and express this as succinctly as possible. This statement may need to be updated as your knowledge of the domain increases 3. Critically, the statement must be something you can actually solve.
# Important Questions to answer
### What is a gene?
A gene is a region of DNA. Not all genes code for proteins. A gene may code for a protein or an piece of RNA.
### What is an ortholog? Why are orthologs important?
Othologs and paralogs are two categories of homologs. Homologs are genes which are similar due to some degree of shared ancestry.
Orthologs and paralogs constitute two major types of homologs:
1. Orthologs evolved from a common ancestor by speciation
1.1. Speciation is when two genes or organisms diverge due to external enviromental pressures. Speciation is the when two organisms begin to split and become different species.
2. Paralogs are related by duplication events
### What is a paralog? Why are paralogs important?
Paralogs are related by duplication events. The two genes arise as a consequence of genetic duplication.
Therefore; paralogs are found within the same species rather than in different species.
Is it possible to determine if the overall organim is of the same sequence or if they are of a different sequence?
### are the other defined types of relationships between genes?
Homology can be refined into different categories further. Some papers talk about [primary homologs](https://doi.org/10.1186/s12859-020-3384-2). The definition of what a primary homolog is; and what other categories of homologs are is often depedent on the method/ algorithm used to find the specific relationship/ region.
### What is a protein - and what is the relationship between protein and gene?
Some genes code for proteins. A protein is a functional element which serves a function within the cell. Some proteins can confer functionals/ abilities upon the organism. Proteins can vary in length; and the addition and deletion of some Amino acids may not directly comprimise the function of the protein. Some regions are redundant.
### How to we talk about related genes/ proteins?
/
### How do we find genes / proteins in a DNA sample.
Find CDS/ ORFs. (ATG - ... - TAG).
If there is a CDS? does that mean that there is a gene/ further: does that mean we have a protein?
#### Are there markers within proteins which might help us find them?
1. Types of Amino Acid; cysteine/ histidine residues; acidic/ basic/ other FGs.
2. Are there regions of a protein which might be similar.
3. length of a CDS.
# Inputs and Outputs?
Inputs:
1. large sampled collection of DNA from a metagenomic sample.
Outputs:
1. a list of genes found in the sample / a list of proteins found in the sample.
2. a list of orthologs found in the sample.
3. a list of paralogs found in the sample.
4. a list of other relationships found in the sample.
# How will you solve it?
/
Is the problem:
1. finding genes within a DNA.
1.1. a full sequence
1.2. a set of contigs
2. finding the orthologs of specific genes. If this is the case; it would potentially be pertinent to utilise GeneMarkS-2 as this is a solid solution for this problem.
### Why is it applicable to restrict the search to only bacteria/ prokaryotes?
The problem space is decreased by this restriction. In addition; the handling of introns and exons is unnecessary. The actual determination of these might be quite complex.
# Pan Genomes and Core Genomes
Pan genome is the full set of genes for a species. Bacteria of the same species within different environments will have different genes as is required for the environment.
Core genome is the set of shared genes within a species.
Looking at the shared set of genes in an environment allows the knowledge of the metabolic content of the environment to be determined.
|
{
"filename": "what_how_why_1.ipynb",
"repository": "ben-silke/biol3209",
"query": "transformed_from_existing",
"size": 11091,
"sha": ""
}
|
# GBS-pyrad-3_1.ipynb
Repository: sr320/nb-2016
# Example _de novo_ RADseq assembly using _pyRAD_
# Modification to start looking at Ostrea data
----------
Please direct questions about _pyRAD_ analyses to the google group thread ([link](https://groups.google.com/forum/#!forum/pyrad-users))
--------------
+ This tutorial is meant as a walkthrough for a single-end RADseq analyses. If you have not yet read the [__full tutorial__](http://www.dereneaton.com/software/pyrad), you should start there for a broader description of how _pyRAD_ works. If you are new to RADseq analyses, this tutorial will provide a simple overview of how to execute _pyRAD_, what the data files look like, and how to check that your analysis is working, and the expected output formats.
+ Each cell in this tutorial begins with the header (%%bash) indicating that the code should be executed in a command line shell, for example by copying and pasting the text into your terminal (but excluding the %%bash header).
-------------
Begin by executing the command below. This will download an example simulated RADseq data set and unarchive it into your current directory.
<code>
pwd
</code>
<code>
cd /Volumes/web/halfshell/working-directory
</code>
<code>
mkdir 16-05-17b
</code>
<code>
cd 16-05-17b
</code>
<code>
ls | head
</code>
------------
The params file lists on each line one parameter followed by a __##__ mark, after which any comments can be left. In the comments section there is a description of the parameter and in parentheses the step of the analysis affected by the parameter. Lines 1-12 are required, the remaining lines are optional. The params.txt file is further described in the general tutorial.
### evolving params file
<code>
%%bash
cat params.txt
</code>
#### To change parameters you can edit params.txt in any text editor. Here to automate things I use the script below.
--------------
__Let's take a look at what the raw data look like.__
Your input data will be in fastQ format, usually ending in .fq or .fastq. Your data could be split among multiple files, or all within a single file (de-multiplexing goes much faster if they happen to be split into multiple files). The file/s may be compressed with gzip so that they have a .gz ending, but they do not need to be. The location of these files should be entered on line 2 of the params file. Below are the first three reads in the example file.
## Sample Description
<img src="http://eagle.fish.washington.edu/cnidarian/skitch/Genotype_by_sequencing_November_2015_·_RobertsLab_project-olympia_oyster-genomic_Wiki_🔊_1CEB70ED.png" alt="Genotype_by_sequencing_November_2015_·_RobertsLab_project-olympia_oyster-genomic_Wiki_🔊_1CEB70ED.png"/>
<code>
mkdir fastq
</code>
<code>
!cp /Volumes/web/nightingales/O_lurida/20160223_gbs/*1.fq.gz fastq/
</code>
<code>
ls
</code>
<code>
!gunzip *.gz
</code>
<code>
ls | head
</code>
<code>
%%bash
less simRADs_R1.fastq | head -n 12 | cut -c 1-90
</code>
------------
Each read takes four lines. The first is the name of the read (its location on the plate). The second line contains the sequence data. The third line is a spacer. And the fourth line the quality scores for the base calls. In this case arbitrarily high since the data were simulated.
These are 100 bp single-end reads prepared as RADseq. The first six bases form the barcode and the next five bases (TGCAG) the restriction site overhang. All following bases make up the sequence data.
----------------
## Step 1: de-multiplexing ##
<code>
already done by bgi
</code>
<code>
pwd
</code>
### Step 2: quality filtering
<code>
%%bash
pyRAD -p params.txt -s 2
</code>
<code>
%%bash
ls edits/
</code>
The filtered data are written in fasta format (quality scores removed) into a new directory called edits/. Below I show a preview of the file which you can view most easily using the `less` command (I use `head` here to make it fit in the text window better).
<code>
%%bash
head -n 10 edits/1A0.edit | cut -c 1-80
</code>
### Step 3: clustering within-samples
Step 3 de-replicates and then clusters reads within each sample by the set clustering threshold and writes the clusters to new files in a directory called clust.xx
<code>
%%bash
pyRAD -p params.txt -s 3
</code>
Once again, I recommend you use the unix command 'less' to look at the clustS files. These contain each cluster separated by "//". For the first few clusters below you can see that there is one or two alleles in the cluster and one or a few reads that contained a (simulated) sequencing error.
<code>
%%bash
less clust.85/1A0.clustS.gz | head -n 26 | cut -c 1-80
</code>
---------------
The stats output tells you how many clusters were found, and their mean depth of coverage. It also tells you how many pass your minimum depth setting. You can use this information to decide if you wish to increase or decrease the mindepth before it is applied for making consensus base calls in steps 4 & 5.
<code>
%%bash
head -n 40 stats/s3.clusters.txt
</code>
### Steps 4 & 5: Call consensus sequences
#### Step 4 jointly infers the error-rate and heterozygosity across samples.
<code>
%%bash
pyRAD -p params.txt -s 4
</code>
<code>
%%bash
less stats/Pi_E_estimate.txt
</code>
#### Step 5 calls consensus sequences using the parameters inferred above, and filters for paralogs.
<code>
%%bash
pyRAD -p params.txt -s 5
</code>
#### The stats output for step 5
<code>
%%bash
less stats/s5.consens.txt
</code>
### Step 6: Cluster across samples
Step 6 clusters consensus sequences across samples. It will print its progress to the screen. This uses 6 threads by default. If you enter 0 for param 37 it will use all available processors.
<code>
%%bash
pyRAD -p params.txt -s 6
</code>
## Step 7: Assemble final data sets
The final step is to output data only for the loci that you want to have included in your data set. This filters once again for potential paralogs or highly repetitive regions, and includes options to minimize the amount of missing data in the output.
<code>
%%bash
pyRAD -p params.txt -s 7
</code>
### Final stats output
<code>
%%bash
less stats/c85m4p3.stats
</code>
---------------
## Output formats ##
We created 11 output files from our analysis. The standard two (.loci and .excluded_loci), as well as the 9 additional ones listed in the params file. These are all shown below.
<code>
%%bash
ls outfiles/
</code>
### Loci format
The ".loci" file contains each locus listed in a fasta-like format that also shows which sites are variable below each locus. Autapomorphies are listed as '-' and shared SNPs as '*'. This is a custom format that is human readable and also used as input to perform D-statistic tests in pyRAD. This is the easiest way to visualize your results. I recommend viewing the file with the command `less`. Below I use a head and cut to make it easy to view in this window.
<code>
%%bash
head -n 39 outfiles/c85m4p3.loci | cut -c 1-75
</code>
### PHY format
<code>
%%bash
head -n 50 outfiles/c85m4p3.phy | cut -c 1-85
</code>
### NEX format
<code>
%%bash
head -n 50 outfiles/c85m4p3.nex | cut -c 1-85
</code>
### Alleles format
<code>
%%bash
head -n 50 outfiles/c85m4p3.alleles| cut -c 1-85
</code>
### STRUCTURE (.str) format
<code>
%%bash
head -n 50 outfiles/c85m4p3.str | cut -c 1-20
</code>
### GENO (.geno) format (used in _Admixture_)
<code>
%%bash
head -n 40 outfiles/c85m4p3.geno
</code>
### SNPs format
<code>
%%bash
head -n 50 outfiles/c85m4p3.snps | cut -c 1-85
</code>
### UNLINKED_SNPs format
<code>
%%bash
head -n 50 outfiles/c85m4p3.unlinked_snps | cut -c 1-85
</code>
## OTHER FORMATS
You may also produce some more complicated formatting options that involve pooling individuals into groups or populations. This can be done for the "treemix" and "migrate" outputs, which are formatted for input into the programs _TreeMix_ and _migrate-n_, respectively. Grouping individuals into populations is done with the final lines of the params file as shown below, and similar to the assignment of individuals into clades for hierarchical clustering (see full tutorial).
Each line designates a group, and has three arguments that are separated by space or tab. The first is the group name, the second is the minimum number of individuals that must have data in that group for a locus to be included in the output, and the third is a list of the members of that group. Lists of taxa can include comma-separated names and wildcard selectors, like below. Example:
<code>
%%bash
## append group designations to the params file
echo "pop1 4 1A0,1B0,1C0,1D0 " >> params.txt
echo "pop2 4 2E0,2F0,2G0,2H0 " >> params.txt
echo "pop3 4 3* " >> params.txt
## view params file
cat params.txt
</code>
## Creating population output files
Now if we run _pyRAD_ with the 'm' (migrate) or 't' (treemix) output options, it will create their output files.
<code>
%%bash
pyRAD -p params.txt -s 7
</code>
## TREEMIX format
<code>
%%bash
less outfiles/c85m4p3.treemix.gz | head -n 30
</code>
## MIGRATE-n FORMAT
<code>
%%bash
head -n 40 outfiles/c85m4p3.migrate | cut -c 1-85
</code>
|
{
"filename": "GBS-pyrad-3_1.ipynb",
"repository": "sr320/nb-2016",
"query": "transformed_from_existing",
"size": 35147,
"sha": ""
}
|
# Report_1.ipynb
Repository: 27410/27410-group-assignment-group-3-progesterone-in-s-cerevisiae
# Progesterone production in *Saccharomyces cerevisiae*
## 1. Introduction
### 1.1 Literature review of the compound
Steroids are ring-structured lipophilic compounds serving various functions in cells. They are of huge pharmaceutical interest, as they are used to treat various diseases and manage both male and female fertility (Tong et al. 2009). Progesterone (**Figure 1**), a female sex hormone, is among the most valuable steroid drugs (Batth et al. 2020) . It is mainly used as a contraceptive, which has been on the market for decades (Howie 1985; Nath et al. 2010)
The market size for progesterone alone is currently estimated to be USD 800 million but is expected to reach a staggering USD 1569 million by 2027; In other words, the market size will nearly double within the next five years, due to the increasing progesterone demand (Market Data Forecast, 2022).
<p>
<img src="figures/progesterone.png" width="75%" />
</p>
**Figure 1.** The structure of progesterone $\textrm{C}_{21}\textrm{H}_{30}\textrm{O}_{2}$; a female sex steroid hormone.
The chemical synthesis of steroids was a highly competitive research field during the 20th century (Slater 2000). In 1940, Bachmann and Wilds produced the sex hormone equilenin as the first complex molecule to be chemically synthesized (Bachmann et al. 1940). In the following decades, many other steroids, including progesterone, were fully chemically synthesized with RB Woodward revolutionizing the field in 1952 (Woodward et al. 1952; Al Jasem 2014). He later won the Nobel prize for this work (Bartlett et al. 1965).
Despite the great efforts in the chemical synthesis of steroids, the structural complexity of steroids complicates their synthesis which often requires harsh conditions and contributes to heavy environmental pollution (Tong et al. 2009). Therefore, most steroid drugs are produced semi-synthetically, using a naturally abundant complex precursor as a starting point. Diosgenin, a steroidal sapogenin, is commonly used as a precursor and can be extracted from plants of the *Dioscorea* genus (Jesus et al. 2016; Al Jasem 2014; Dong et al. 2015). For example, progesterone can be produced from diosgenin via the so-called Marker synthesis (Al Jasem 2014). However, the protection and limited availability of these *Dioscorea* plants have caused increasing market prices of diosgenin.
Collectively, the obstacles in the synthesis of steroids pushed researchers to look towards alternative production methods, namely through natural biosynthesis using microbial cell factories (Tong et al. 2009). Mild reaction conditions, lower chemical pollution, and higher conversion rates are among the advantages of using microbial cell factories in steroid production as compared to chemical synthesis (Tong et al. 2009). To our knowledge, no progesterone-producing cell factory has been published. Therefore, we decided to design one.
Understanding the biosynthetic pathway of progesterone is crucial to design a cell factory since it aids in choosing a suitable type of cell host for the cell factory. The biosynthesis of all steroids starts in the production of the triterpenoid precursor, squalene, in the mevalonate pathway (**Figure 2**). Through several enzymatic steps, squalene can then be cyclized to establish the foundation of all steroids including progesterone (Buhaescu et al. 2007). With this knowledge, it seemed reasonable to choose a cell host already skilled at producing squalene as the starting point.

**Figure 2.** Progesterone biosynthesis.
### 1.2 Literature review of the cell factory
When designing a cell factory, the first decisive decision to make is which cell to pick as the chassis. The cell host must be culturable. To integrate a heterologous pathway, the cell must be genetically engineerable as well. Additionally, it is a huge advantage if the cell host is well-known in the industry, has status as GRAS (generally regarded as safe), and naturally produces the compound of interest or a suitable precursor. Sometimes, some enzymes required in the heterologous pathway depend on certain features, such as eukaryotic organelles, which is the case for progesterone production, precluding prokaryotes. After considering various cell hosts – including the mammalian CHO-cells, microalgae, and non-conventional yeasts – our final choice ended at baker’s yeast, *Saccharomyces cerevisiae*.
*S. cerevisiae* is the most studied eukaryote that lives up to all the above-mentioned requirements except that it does not produce progesterone naturally (Parapouli et al. 2020). It does, however, produce squalene, which is used to produce the steroid, ergosterol (Xu et al. 2020). Extensive research of *S. cerevisiae* recently enabled researchers to generate a strain producing a staggering 21 g squalene pr. L, placing *S. cerevisiae* at the forefront of squalene-producing cell factories (Paramasivan et al. 2022; Zhu et al. 2021). Overproduction of the squalene is a good starting point for the overproduction of steroids as well, as more precursors will be available.
Several issues challenge the production of steroids with *S. cerevisiae*. One issue is that many steroids are non-exportable and might cause toxic effects when accumulating in the cell. Using non-conventional yeasts, like *Yarrowia lipolytica* or *Pichia pastoris*, might aid in solving these issues. For example, *Y. lipolytica* also efficiently produces steroid precursors and is known for its ability to accumulate lipids and lipophilic compounds, and therefore maybe also steroids (Xu et al. 2020; Worland et al. 2020; Adrio 2017). *P. pastoris* holds an efficient secretion system, which has been suggested to allow extracellular steroid synthesis to overcome the toxic effect of steroid accumulation (Xu et al. 2020; Ahmad et al. 2014). The downside of these non-conventional strains is that they are much less studied compared to *S. cerevisiae*. As designing cell factories computationally heavily rely on representative and detailed models, we still decided to use *S. cerevisiae* as our model organism. Nonetheless, we believe that the findings for *S. cerevisiae* in this report will be applicable for other yeast strains.
## 2. Problem definition
Current production methods of steroid drugs rely on the extraction of precursors from plants combined with chemical synthesis, which causes a burden to the environment. Due to the increasing demand for steroid drugs, the development of efficient and sustainable production methods is a highly relevant and important topic.
**In this project, we aim to design a cell factory of *S. cerevisiae* that is optimized to produce the steroid drug, progesterone.**
First, we will analyze different genome-scale models (GSMs) of *S. cerevisiae* to use for our computer-aided analysis and design. We will identify and implement the necessary heterologous pathway into our model. This model will serve as the foundation to identify reaction targets for knockouts, up-regulation, and down-regulation, in order to improve the progesterone yield. Additionally, we perform a co-factor swapping analysis to test the effect on growth and progesterone productivity when swapping NAD(H) and NADP(H) in the identified reactions. To understand the effect on growth and progesterone productivity of our implemented alterations, we generate phenotypic phase plane plots, calculate maximum theoretical yields, and perform a dynamic flux balance analysis of a batch fermentation.
Lastly, we will assess 12 strains that we designed to conclude which of them that are most likely to be the best progesterone-producing cell factory.
## 3. Selection and assessment of existing GSM
Our choice of host organism, *S. cerevisiae*, is a very common and well-researched organism, thus multiple genome-scale metabolic models (GSMs) exist. GSMs can be used to computationally calculate predicted outcomes, which can then be verified experimentally. From BIGG and EMBL-EBI's BioModels we found four candidate GSMs: iFF708, iMM904, iND750, and yeast-GEM (version 8.6.2 newest on GitHub). iND750 is an improved version of the model iFF708, containing more genes, metabolites, and reactions (Duarte NC, Herrgård MJ, Palsson BØ. 2004).
Using `memote`, it is possible to assess different quality measures of the GSMs, including stochiometry, annotation, and reaction/metabolite statistics. Each model was individually tested using `memote` and the results of the `memote` runs can be seen in the `models/memote` folder. The results are summarised in the table below.
**Table 1** shows the `memote` results evaluating the four GSMs (iFF708, iMM904, iND750, and yeast8.6.2).
| Measure | iMM904 | yeast8.6.2 | iFF708 | iND750 |
| ---- | ---- | - | - | - |
| Total Metabolites | 1,226 | 2,744 | 796 | 1,059 |
| Total Reactions | 1,577 | 4,063 | 1,379 | 1.266 |
| Total Genes | 905 | 1,160 | 619 | 750 |
| Stochiometric Consistency | 100.0% | 0.0% | 0.0% | 100.0% |
| Mass Balance | 96.0% | 93.7% | 0.0% | 97.3% |
| Charge Balance | 98.5% | 98.2% | 100.0% | 100.0% |
| Metabolite Connectivity | 100.0% | 100.0% | 100.0% | 100.0% |
| Unbounded Flux In Default Medium | 76.2% | 58.8% | 71.7% | 83.0% |
| Metabolite Annotation | 80% | 68% | 25% | 80% |
| Reaction Annotation | 82% | 65% | 25% | 83% |
| Gene Annotation | 43% | 54% | 0% | 43% |
| **Total score** | 85% | 68% | 19% | 86% |
As seen in **Table 1**, yeast8.6.2 contains by far the most metabolites, reactions, and genes. The difference is largest for the number of reactions and metabolites, of which it has approximately two times the number, compared to the other GSMs. iMM904, IFF708, and iND750 have a more similar number of reactions, with iMM904 having the most metabolites annotated. For the measure of stoichiometric consistency, the best models are iMM904 and iND750, which both have a consistency score of 100%. At last, iND750 has the highest total score (= 86%), with iMM904 coming just second (= 85%), followed by yeast8.6.2 (= 68%), and iFF708 (= 19%).
We decided that the stochiometric consistency was a more important parameter than the number of reactions, as we expect the data generated using GSMs with a stochiometric consistency of 100% to be more reliable than using GSMs with a stochiometric consistency of 0%; In that way, we eliminated yeast8.6.2 and iFF708 as GSMs. This left us with either iMM904 or iND750 to choose from. Since the scores of these two GSMs are so close, we decided to go for iMM904, as it contains the most reactions, metabolites, and genes, which we expect will provide us with a better result that, again, will provide us with more reliable data when simulating using this model.
_[Notebook: GSM comparison](01_GSM_Comparison.ipynb)_
## 4. Computer-Aided Cell Factory Engineering
#### **Implementation and characterization of the cell factory**
**1. Implementation of heterologous pathway**
Yeast cells naturally produce the steroid ergosterol, which is produced in a long biosynthetic pathway from the precursor squalene (**Figure 3**). Progesterone can be produced from the intermediates zymosterol and 5-dehydroepisterol via a heterologous pathway (Jiang, Yi-qi, and Jian-ping Lin. 2022). Since the progesterone biosynthesis from 5-dehydroepisterol is not validated, we chose to implement the heterologous pathway starting from zymesterol as the precursor.
<!-- However, the biosynthesis of progesterone from 5-dehydroepisterol rely on an enzymatic reaction that to our knowledge is not validated. Therefore, the production of progesterone from zymosterol is the heterologous pathway we have implemented in our model (**Figure 3**). -->

**Figure 3.** Steroid biosynthesis. Natural ergosterol pathway is shown with a green box and the implemented progesterone heterologous pathway is shown with a blue box. The enzymes are represented by their gene name where endogenous yeast genes are represented in black and heterologous genes are represented in red. Arrows indicate the direction of reaction. Co-enzymes and co-substrates are shown in light grey.
We investigated other potential progesterone production pathways using the `pathway_prediction` algorithm from `cameo` (Cardoso, Joao GR, et al. 2018). In all the pathways suggested by `cameo`, zymosterol is converted into progesterone in six steps but with different paths.
Interestingly, `cameo` found another reaction (MNXR4011) between cholesterol and pregnenolone where only one NADP(H), instead of six in the manually curated pathway (CYP11A1), is needed. Therefore, this reaction was implemented instead.
<!-- All `cameo` pathways agree with the implemented heterologous pathway in the way that zymosterol is in four steps converted into cholesterol which is afterwards converted by two steps to progesterone. -->
_[Notebook: Heterologous pathway implementation](02_heterologous_pathway_implementation.ipynb)_
**2. Calculating the maximum theoretical yield and productivity on default and alternative carbon sources**
At an uptake rate of 10 mmol/(gDW\*h) glucose, the maximum theoretical growth rate for the strain is 0.288 /h (objective set at growth), the maximum theoretical productivity of progesterone was 0.167 mmol/(gDW\*h), and the maximum theoretical progesterone yield was 0.017 mmol progesterone/mmol glucose (objective set at progesterone production).
When both biomass and progesterone was set as the objective, the values changed; maximum possible growth rate was 0.119 /h, the maximum progesterone productivity was 0.156 mmol/(gDW\*h), and the maximum progesterone yield was 0.016 mmol progesterone/mmol glucose.
<!-- When the objective was changed, to account for both maximum growth and maximum production of progesterone, the values changed; Maximum possible growth rate was 0.119 /h, the maximum progesterone productivity was 0.156 mmol/(gDW\*h), and the maximum progesterone yield was 0.016 mmol progesterone/mmol glucose. -->
By increasing the availability of glucose in the medium, the maximum theoretical productivity of progesterone only slightly changed, whereas the maximum theoretical progesterone yield was drastically reduced.
As the yield is defined as product over substrate, the substrate concentration is increased but the productivity is the same.
Thus, the yield will be significantly lowered.
It can therefore can be concluded that solely increasing the glucose concentration is not a valid approach for increasing progesterone yield;
This makes sense, as there are two limiting exchanges in the medium (glucose and $\textrm{O}_{2}$), and it is possible that both need to be changed to increase the yield of progesterone.
Furthermore, using the alternative carbon sources, fructose and galactose, did not improve the yield.
<!-- It was also found that the alternative carbon sources, fructose and galactose, was utilized not better than the default media containing glucose. -->
_[Notebook: Maximum theoretical yield](03_maximum_theoretical_yield.ipynb)_
**3. Phenotypic phase plane analysis using `cameo` and `cobrapy`**
To fully elucidate how the cells production capabilities behave in relation to changes in the medium and objective, we perform a phenotypic phase plane analysis using `cameo` and `cobrapy`. Since our medium only restricts uptake of oxygen and glucose, these are the parameters we specifically look at. Before analysing the response to changes in these conditions, we must first understand the trade-off between progesterone production in our cell factory and its growth.
<p float="left">
<img src="figures/04_phenotypic_phase_plane_biomass_progesterone.jpg" width="50%" />
</p>
**Figure 4.** Phenotypic phase plane for progesterone flux (through the demand `DM_progesterone_c`) over biomass flux (cell growth).
**Figure 4** clearly depicts the trade-off between growth and production of progesterone. For the highest possible cell growth, it cannot prioritise the production of progesterone. Interestingly, the reverse isn't actually true. We see an almost constant plateau in progesterone production at a cell growth, $\mu\sim 0.1$. This seems to show that if we optimise for the production of progesterone and choose the maximum, the cell factory is still able to grow.
But which conditions create the highest productivity - and maybe more importantly the highest yield? By plotting the productivities and yield of progesterone and biomass in the ranges $-10 < \textrm{O}_2 < 0$ and $-20 < \textrm{Glc} < 0$, we explore the entire realistic space of values in search of the optimal.
<p float="left">
<img src="figures/04_phenotypic_phase_plane_progesterone_productivity.jpg" width="40%" />
<img src="figures/04_phenotypic_phase_plane_biomass_productivity.jpg" width="40%" />
</p>
**Figure 5(a, b).** Phenotypic phase plane for progesterone and biomass productivity/flux (where each is maximised) as a function of oxygen and glucose.
<br/>
<p float="left">
<img src="figures/04_phenotypic_phase_plane_progesterone_yield.jpg" width="40%" />
<img src="figures/04_phenotypic_phase_plane_biomass_yield.jpg" width="40%" />
</p>
**Figure 6(a, b).** Phenotypic phase plane for progesterone and biomass yield (where each is maximised) as a function of oxygen and glucose.
In **Figure 5a** and **5b**, we observe that an increased glucose in general increases the cell growth, but for progesterone productivity is quite quickly stagnates and remains constant as glucose increases; it seems to be much more dependent on $\textrm{O}_2$ flux. But while a higher oxygen flux increases productivity, higher levels of oxygen decrease productivity until it reaches 0 for very high levels of oxygen. This may be explained biologically by oxygen toxicity.
The difference in how glucose affects progesterone and biomass production can also be seen in the respective yield plots **Figure 6a** and **6b**. Progesterone yield decreases as glucose flux increases, while biomass yield approaches a constant.
For low values of glucose, varying the oxygen for either objective gives the same tendency: an optimal ridge with high yield. For the default oxygen level in our model `EX_o2_e = -2`, we find the ridge in the sectional plots **Figure 7a** and **7b**:
<p float="left">
<img src="figures/04_phenotypic_phase_plane_progesterone_yield_optimum.jpg" width="35%" />
<img src="figures/04_phenotypic_phase_plane_biomass_yield_optimum.jpg" width="35%" />
</p>
**Figure 7(a, b).** Phenotypic phase plane for progesterone and biomass yield (where each is maximised) as a function of glucose for oxygen flux at -2. At the maximum yield we find the conditions in **Table 2**, shown below.
*Table 2. Phenotypic phase plane analysis results.*
| Objective | Productivity | Yield | Glucose flux | Oxygen flux |
| - | - | - | - | - |
| Progesterone | 0.084 | 0.098 | -0.856 | -2.0 |
| Biomass | 0.070 | 0.082 | -0.856 | -2.0 |
Interestingly we observe the same optimal glucose flux for both.
The carbon yield in Cmole for progesterone is 0.344.
_[Notebook: Phenotypic phase plane analysis](04_phenotypic_phase_plane_analysis.ipynb)_
#### **Cell factory engineering strategies**
<!-- Cell factories can be engineered in various ways in order to make them stable and productive. -->
<!-- Using computer-aided cell factory design, we investigated different cell factory engineering strategies for improving the progesterone producing *S. cerevisiae* strain (iMM904_progesterone). -->
**1. Gene targets for knock-outs**
<!-- **1. Knocking out ERG5 and ERG6** -->
To increase the flux towards progesterone production, we searched for gene targets for knock-outs using `OptGene` and searching literature.
`OptGene` is an evolutionary programming based tool to find knockout targets (Patil, Kiran Raosaheb, et al. 2005). Unfortunately, we did not identify any knockout targets using `OptGene`.
<!-- The heterologous pathway for production of progesterone starts from zymosterol which naturally is an important precursor for ergosterol (**Figure 3**). -->
It is a common engineering strategy to knockout ERG5 and ERG6 (**Figure 3**) to improve the production of cholesterol and similar steroids that are produced from a precursor in the ergosterol pathway (Jiang, Yi-qi, and Jian-ping Lin. 2022; Xu, Shanhui, and Yanran Li. 2020).
<!-- Therefore, we investigated the effect of knocking out these genes in our model optimized for growth and progesterone productivity. -->
Surprisingly, knocking out ERG5 and ERG6 in our model had no effect on cell growth (µ = 0.119 /h) or progesterone productivity (0.156 mmol/(gDW*h)) when biomass and progesterone were set to be the objective.
This might be because the flux through ERG5 and ERG6 in this optimized model is already 0, which in principle simulates that they are knock-outed (for further details, see _[Notebook: Gene target analysis](05_gene_target_analysis.ipynb)_).
<!-- if knocking out ERG5 and ERG6 in our model improves the progesterone production. -->
<!-- Surprisingly, our simulation showed that knocking out ERG5 and ERG6 had no effect on cell growth (µ = 0.119 /h) or progesterone productivity (0.156 mmol/(gDW*h)) when biomass and progesterone were set to be the objective. -->
<!-- **Rune: Jeg tænker at det her bliver nødt til at blive skåret lidt ned. Måske man kunne undlade noget af varificeringen af knockouts? Så afsnit med HSD3B og 61 ændrede reactioner. Tænker måske heller ikke nødvendigvis at vi behøver at henvise til tablen her(?) Og så tror jeg også der skal cuttes lidt ned så vi måske går lidt hurtigere til konklusionerne (?) Måske kunne afsnit 1 og 2 kombineres i et (Knockout analysis). Det er lidt sjovt at have et afsnit med OptGene hvor der bare står at det ikke virker**
**Caro: har slettet noget af det, måske skal der slettes mere**
_[Notebook: Gene target analysis](05_gene_target_analysis.ipynb)_ -->
**2. Up- and downregulation targets using FSEOF**
Flux Scanning based on Enforced Objective Flux (FSEOF) analysis identifies gene targets for up- or down-regulation to increase the flux towards the compound of interest (Choi, Hyung Seok, et al. 2010).
By performing an FSEOF analysis, we identified 117 reactions with a large flux change when the flux was enforced towards progesterone.
<!-- is a tool for identification of gene amplification targets (Choi, Hyung Seok, et al. 2010). -->
<!-- The flux is increased for a compound of interest (enforced objective) at the same time of maximizing biomass formation flux. -->
<!-- The output is reactions of which the flux changes when increasing the flux towards the compound of interest making these reactions good targets for up- or downregulation. -->
<!-- We performed the analysis on our model (iMM904_progesterone) setting the enforced objective to be progesterone. -->
<!-- The flux of 117 reactions changed as a result of an increasing flux towards progesterone. -->
<!-- Reactions which are a part of the heterologous pathway were removed because they are not relevant targets: It is obvious that their increase in flux follows the same as the enforced increased progesterone flux. Also, reactions which flux was close to 0 were not of interest. -->
**Figure 8** shows 20 reactions (including DM_progesterone_c reference) with the highest relative flux change, which are therefore promising targets for up- or down-regulation.
The most up-regulated reactions are G3PD1ir, GLYCDy, G3PT, and DHAK with a flux increase of 4.58 mmol/(gDW\*h).
These reactions form a cycle where NAD(+) and NADPH are formed.
By increasing the flux through this cycle, the concentration of NAD(+) and NADPH are increased in the cell.
Since NAD(+) and NADPH are used to produce progesterone (**Figure 3**), it makes sense that a higher concentration of these co-factors results in increased flux towards progesterone as well.
<!-- it makes sense that when these co-factors are increased then the flux through progesterone is increased as well. -->

**Figure 8.** The 20 reactions (including DM_progesterone_c reference) with the highest relative flux change when increasing progesterone flux. The x-axis shows an increasing progesterone flux over 10 steps. The progesterone flux is increased with 0.015 mmol/(gDW\*h) per step.
In total, the progesterone flux is increased 0.135 mmol/(gDW\*h).
We investigated the influence of this reaction cycle (G3PD1ir, GLYCDy, G3PT, and DHAK) on the production of progesterone (**Figure 9**). Having the optimized reaction cycle in the model resulted in an 3.91% increase in maximum progesterone productivity when $\mu$ = 0.1187 compared to a model where no cycling happens.
Thereby, the model suggests that up-regulation of the reaction cycle will result in higher progesterone production.

**Figure 9.** Phase plane plot of progesterone productivity (mmol/(gDW\*h)) and growth rate (/h). The blue line reflects the model when it has the optimized reaction cycle (G3PD1ir, GLYCDy, G3PT, and DHAK). The orange line reflects the model when the reaction cycle is turned off.
_[Notebook: Gene target analysis](05_gene_target_analysis.ipynb)_
**3. Co-factor swap targets**
The balance of co-factors within a cell is important to obtain a high theoretical yield of a given product (King, Zachary A., and Adam M. Feist, 2014). In our implemented pathway, the cell uses four NADPH and one NAD(+) to produce progesterone.
Due to this extensive use of NADPH, we investigated if we could improve the co-factor balance by producing more NADPH on the cost of NADH.
Using the algorithm `CofactorSwapOptimization`, we identified 20 reactions where swapping the co-factor NAD(H) with NADP(H) could potentially increase progesterone productivity.
Of the 20 reactions, the GADP reaction from the glycolysis seemed to be the reaction with most potential to investigate further.
The GADP reaction produces NADH from the following reaction:
Glyceraldehyde-3-phosphate + NAD(+) + Pi <=> 3-Phospho-D-glyceroyl-phosphate + H(+) + NADH
We exchanged this reaction with a similar one producing NADPH instead as described in _[Notebook: co-factor swap](06_cp-factor_swap.ipynb)_ and tested how it would affect the theoretical maximum progesterone and biomass productivity.
The theoretical progesterone productivity did not increase, but the theoretical growth rate increased with 2%.
While this might not seem impressive, it is more informative to plot the phase plane of progesterone productivity versus biomass productivity for the old model and the model with NAD(H) swapped with NADP(H) for GAPD (**Figure 10**).
This plot reveals, that the co-factor swap allow for maximum progesterone production at much higher growth rates. For the initial model, the progesterone productivity decreases around a biomass productivity of 0.10. With the co-factor swap, the progesterone productivity decreases around a biomass productivity of 0.16. In other words, the biomass productivity increases by 60% when maximum progesterone productivity is priotized.
<!-- , where the progesterone productivity would have decreased from 0.167 to about 0.125 in the initial model. -->
<!-- _[Notebook: co-factor swap](06_cp-factor_swap.ipynb)_ -->

**Figure 10.** The phase plane of progesterone productivity (mmol/gDW*h) and growth rate (/h) of a model with and without NAD(H) swapped with NADP(H) in GAPD.
_[Notebook: Co-factor swap](06_co-factor_swap.ipynb)_
**4. Dynamic Flux Balance Analysis**
Calculating a single number for the progesterone yield and flux gives little information about what the final titres will be.
We performed Dynamic Flux Based Analysis (DFBA) to estimate the final titres of biomass, progesterone, and the precursor squalene.
<!-- To get a better idea of how the progesterone and biomass titres change over time, w -->
<!-- it can be insightful to mimic real conditions by simulating a simple batch fermentation. This is the purpose of Dynamic Flux Based Analysis (DFBA). Using DFBA, we estimated the titre of progesterone and also the precursor squalene in order to compare our estimates with experimental results from literature. -->
Since our model with pathway 1 and co-factor swapping implemented seemed to be one of the promising strains, we used it to simulate an aerobic batch fermentation with a constant $\textrm{O}_{2}$ level of 2 mmol/L and an initial glucose concentration of 10 mmol/L.
The simulation was visualized as seen in **Figure 11**. The batch fermentation ran for 5.1 hours before all the glucose got consumed. The final progesterone titre reached 0.212 mmol/L (0.067 g/L) and the squalene titre reached 0.214 mmol/L (0.088 g/L). In our simulation, there is a linear relationship between the initial glucose added and the final progesterone titre (See _[Notebook: DFBA](07_DFBA.ipynb)_). This does not seem fully realistic, but it is still likely that adding more glucose to a certain level would result in a higher titre.

**Figure 11.** Simulation of a batch fermentation with DFBA. Initial glucose concentration was 10 mmol/L and a constant O2 level of 2 mmol/L. The final progesterone titre reached 0.21 mmol/L.
_[Notebook: Dynamic Flux Balance Analysis](07_DFBA.ipynb)_
#### **Promising cell factory designs**
**1. Metabolic pathway visualisations using `Escher`**
The computed fluxes were visualized using the online version of escher.
Flux going through the central carbon metabolism (**Figure 12**) and the heterologous pathway (**Figure 13**) was visualized. The two color scales are identical.

**Figure 12.** The fluxes of the central carbon metabolism. Created using the online version of `escher` (https://escher.github.io/#/). *Red* represents the highest flux, *blue* is the lowest flux, and *purple* represents flux somewhere between the *red* and *blue* flux values. Lastly, *grey* represents no flux.
From **Figure 12**, it can be seen that one of the highest fluxes produces ethanol. This could be a result of overflow metabolism - also known as the Crabtree effect in yeast - where ethanol is produced in excess as cells utilize aerobic fermentation over respiration (Malina, Carl, et al. 2021).

**Figure 13.** Flux through the pathway producing progesterone. Created using the online version of escher (https://escher.github.io/#/). *Red* represents flux through the pathway, *grey* represents no flux.
The heterologous pathway producing progesterone can be seen in **Figure 13**; It appears that the pathway going through cholesta-8-en-3beta-ol is preferred over the pathway going through cholesta-7,24-dien-3beta-ol for the production of progesterone. However, this could be because the fluxes loaded are saved following one simulation, and if doing enough simulations, the flux would go through the other pathway.
**2. Strain assessment**
<!-- *Table 3. Optimized models results.*
| Model number | Max µ (/h) | Max progesterone yield (mmol/mmol) | Optimized µ (/h) | Optimized progesterone yield (mmol/mmol) | Progesterone yield at µ=0.18 (mmol/mmol) |
| - | - | - | - | - | - |
| Model 1 | 0.2879 | **0.0167** | 0.1187 | **0.0156** | 0.0104 |
| Model 2 | 0.2879 | **0.0167** | 0.1113 | **0.0156** | 0.01 |
| Model 3 | **0.2937** | **0.0167** | 0.1771 | 0.0155 | **0.0152** |
| Model 4 | **0.2937** | **0.0167** | 0.1771 | 0.0155 | **0.0152** |
| Model 5 | 0.2879 | 0.0143 | 0.1313 | 0.0135 | 0.0096 |
| Model 6 | 0.2879 | 0.0143 | 0.1237 | 0.0135 | 0.0092 |
| Model 7 | **0.2937** | 0.0143 | **0.1919** | 0.0133 | 0.0139 |
| Model 8 | **0.2937** | 0.0143 | **0.1919** | 0.0133 | 0.0139 |
| Model 9 | 0.2879 | **0.0167** | 0.1187 | **0.0156** | 0.0104 |
| Model 10 | 0.2879 | **0.0167** | 0.1113 | **0.0156** | 0.01 |
| Model 11 | **0.2937** | **0.0167** | 0.1771 | 0.0155 | **0.0152** |
| Model 12 | **0.2937** | **0.0167** | 0.1771 | 0.0155 | **0.0152** | -->
12 different cell factory designs were assessed under standard medium conditions; glucose uptake = 10 mmol/(gDW\*h) and $\text{O}_2$ uptake = 2 mmol/(gDW\*h) (**Table 3** and **Figure 14**).
The four best performing cell factory designs (model 3, 4, 11, and 12) perform equally well (**Figure 14**), however, due to a different number of modifications these models are scores differently leaving only model 3 in the top (**Table 3**).
Interestingly, all six models containing the co-factor swapping are in the top-7 of best scored models. Therefore, this modification seems to be important to get a high performing model. By upregulating the NAD(+)/NADPH cycling as an additionally modification to co-factor swapping (model 4, 8, and 12 compared to model 3, 7, and 11) there are no observable change in performance (**Figure 14**).
This suggests that the co-factor swapping takes over the role that NAD(+)/NADPH cycling has. Also, the upregulation of the NAD(+)/NADPH cycling only increase the performance slightly compared to models only with an implemented pathway (**Figure 14**), and taking the extra modification into account this upregulation does not even improve the score of the model (**Table 3**).
Models with the manually derived pathway (model 5-8) follow the same trends that for the other pathways, but with a lower maximum progesterone productivity.
**Table 3**. Quantitative strain assessment of 12 different cell factory designs.
|Model number |Features and modifications |Number of modifications |Max µ (/h) |Max progesterone yield (mmol/mmol) |Optimized µ (/h) |Optimized progesterone yield (mmol/mmol) |Progesterone yield at µ=0.18 (mmol/mmol) |Score |
| - | - | :-: | :-: | :-: | :-: | :-: | :-: | :-: |
| Model 3 | Pathway 1, co-swap | 8 | **0.2937** | **0.0167** | 0.1771 | 0.0155 | **0.0152** | **90.2%** |
| Model 4 | Pathway 1, up NAD/NADPH cycling, co-swap | 12 | **0.2937** | **0.0167** | 0.1771 | 0.0155 | **0.0152** | 86.0% |
| Model 11 | Combined pathway, co-swap | 12 | **0.2937** | **0.0167** | 0.1771 | 0.0155 | **0.0152** | 86.0% |
| Model 7 | Manuel pathway, co-swap | 8 | **0.2937** | 0.0143 | **0.1919** | 0.0133 | 0.0139 | 85.4% |
| Model 1 | Pathway 1 | **4** | 0.2879 | **0.0167** | 0.1113 | **0.0156** | 0.01 | 82.8% |
| Model 12 | Combined pathway, up NAD/NADPH cycling, co-swap | 16 | **0.2937** | **0.0167** | 0.1771 | 0.0155 | **0.0152** | 81.9% |
| Model 8 | Manuel pathway, up NAD/NADPH cycling, co-swap | 12 | **0.2937** | 0.0143 | **0.1919** | 0.0133 | 0.0139 | 81.2% |
| Model 2 | Pathway 1, up NAD/NADPH cycling | 8 | 0.2879 | **0.0167** | 0.1187 | **0.0156** | 0.0104 | 79.7% |
| Model 9 | Combined pathway | 8 | 0.2879 | **0.0167** | 0.1113 | **0.0156** | 0.01 | 78.6% |
| Model 5 | Manuel pathway | **4** | 0.2879 | 0.0143 | 0.1237 | 0.0135 | 0.0092 | 78.3% |
| Model 10 | Combined pathway, up NAD/NADPH cycling | 12 | 0.2879 | **0.0167** | 0.1187 | **0.0156** | 0.0104 | 75.5% |
| Model 6 | Manuel pathway, up NAD/NADPH cycling | 8 | 0.2879 | 0.0143 | 0.1313 | 0.0135 | 0.0096 | 75.3% |

**Figure 14.** Phenotypic phase plane of progesterone productivity (mmol/gDW\*h) and growth rate (/h) results of 12 different cell factory desings.
_[Notebook: Strain assessment](08_strain_assessment.ipynb)_
## 5. Discussion
We successfully designed and evaluated 12 progesterone-producing *S. cerevisiae* cell factories. Our four best performing strains reached low progesterone and squalene titres compared to what has been achieved experimentally in literature (Paramasivan et al. 2022), especially compared to the record of 21 g/L (Zhu et al. 2021). However, it is difficult to compare our result to the record, as this sky-high titer was achieved by more advanced compartmentalization engineering strategies followed by a two-staged fed-batch fermentation (Zhu et al. 2021). Simulating these strategies was unfortunately beyond the scope of this project.
The features, that are likely to have the biggest effect on progesterone productivity, are the choice of pathway and the co-factor balance of NADP(H) and NAD(H) (see **Figure 14**). Choosing pathway 1 increases the theoretical maximal progesterone productivity and improving the co-factor balance of NADP(H) and NAD(H) increases the growth rate when the maximum progesterone productivity is prioritized. All of the investigated modifications relate to the availability of NADP(H) in the cell, which in conclusion must be very important for growth and progesterone productivity.
Other than ensuring availability of NADP(H), we need optimal substrate levels in our growth medium - particularly oxygen and glucose - for the cell factory to perform well. With our phase plane analysis, it seems that we need higher levels of oxygen than that of glucose for optimal progesterone yield. The yield of model 1 in Cmole/Cmol is 0.344, indicating that around a third of the input carbons are used to produce our product. Theoretically, this could be increased with higher glucose and especially oxygen flux. We do have to be careful with that, though, as increasing the oxygen by a lot is probably unrealistic, owing to oxygen toxicity, etc.
The success of implementing this heterologous pathway and modifications in real life depends, first of all, on whether the enzymes we have found will work efficiently in yeast, as we assume they will in this model. Other assumptions used in these simulations do not necessarily represent reality. For example, we assume that only one substrate, glucose, is limiting growth and that the fluxes of all metabolites are constant in steady state. Also, the degree of detail about the cell in the model is limited. For example, we do not model the effect of the accumulation of progesterone and contingent intermediates, which might be toxic to the cell and inhibit growth (Csáky et al. 2020; Xu et al. 2020).
For the abovementioned reasons, the tools used in this report mainly aid in finding a suitable heterologous pathway and gene targets for knock-outs, knock-downs, and up-regulation. The calculated yields and titres might aid in assessing the theoretical impact of the implemented modifications, but the numbers themselves should not be regarded as conclusive. To get better estimates of productivity, yields and titres, it is possible to advance the model by for example including enzyme kinetics (Domenzain et al. 2021). What might be even more realistic is to start experimenting in the lab, starting by introducing the heterologous pathway and thereafter attempt to improve the growth and productivity by engineering the gene targets found in this report.
<!-- We successfully designed and evaluated 12 progesterone-producing *S. cerevisiae* cell factories where four of them turned out to be equally performing in our simulations. The features, that are likely to have the biggest effect on progesterone productivity, are the choice of pathway and the co-factor balance of NADP(H) and NAD(H) (see **Figure X**). Choosing pathway 1 seems to increase the theoretical maximal progesterone productivity from **XX** to **XX**. The main difference of this pathway from the others is that it requires six times less NADPH in the second last reaction. Changing the co-factor balance of NADP(H) and NAD(H) increases the growth rate when the maximum progesterone productivity is prioritized. Also, the FSEOF analysis revealed that the reactions with the biggest flux increase when optimizing for progesterone productivity are connected in a cycle where NAD and NADPH is produced. Thereby, the upregulation of these genes leads to a slight improvement in growth and progesterone productivity due to increased availability of NADPH and NAD. Notably, all these investigated modifications relate to the availability of NADP(H) in the cell, which in conclusion must be very important for growth and progesterone productivity. (188 words) **FIND ARTICLE ABOUT THE IMPORTANCE OF NADPH.**
- could have used OptKnock -->
<!-- We successfully designed and evaluated 12 progesterone-producing *S. cerevisiae* cell factories where four of them turned out to be equally performing in our simulations. The features, that are likely to have the biggest effect on progesterone productivity, are the choice of pathway and the co-factor balance of NADP(H) and NAD(H) (see **Figure X**). Choosing pathway 1 seems to increase the theoretical maximal progesterone productivity from **XX** to **XX**. The main difference of this pathway from the others is that it requires six times less NADPH in the second last reaction. Changing the co-factor balance of NADP(H) and NAD(H) increases the growth rate when the maximum progesterone productivity is prioritized. Also, the FSEOF analysis revealed that the reactions with the biggest flux increase when optimizing for progesterone productivity are connected in a cycle where NAD and NADPH is produced. Thereby, the upregulation of these genes leads to a slight improvement in growth and progesterone productivity due to increased availability of NADPH and NAD. Notably, all these investigated modifications relate to the availability of NADP(H) in the cell, which in conclusion must be very important for growth and progesterone productivity. **FIND ARTICLE ABOUT THE IMPORTANCE OF NADPH.** -->
<!-- Other than ensuring availability of NADP(H), we need optimal substrate levels in our growth medium - particularly oxygen and glucose - for the cell factory to perform well. With our phase plane analysis, it seems that we need higher levels of oxygen than that of glucose for optimal progesterone yield. The yield of model 1 in Cmole/Cmol is 0.344, indicating that around a third of the input carbons are used to produce our product. Theoretically, this could be increased with higher glucose and especially oxygen flux. We do have to be careful with that, though, as increasing the oxygen by a lot is probably unrealistic, owing to oxygen toxicity, etc. -->
<!-- The success of implementing this heterologous pathway and modifications in real life depends, first of all, on whether the enzymes we have found will work efficiently in yeast, as we assume they will in this model. Other assumptions used in these simulations do not necessarily represent reality. For example, we assume that only one substrate, glucose, is limiting growth and that the fluxes of all metabolites are constant in steady state. Also, the degree of details about the cell in the model is limited. For example, we do not model the effect of the accumulation of progesterone and contingent intermediates, which might be toxic to the cell and inhibit growth (**REF**). -->
<!-- For the abovementioned reasons, the tools used in this report mainly aid in finding a suitable heterologous pathway and gene targets for knock-outs, knock-downs, and up-regulation. The calculated yields and titres might aid in assessing the theoretical impact of the implemented modifications, but the numbers themselves should not be regarded as conclusive. To get better estimates of productivity, yields and titres, it is possible to advance the model by for example including enzyme kinetics or **XXX** data (**REF**). What might be even more realistic is to start experimenting in the lab, starting by introducing the heterologous pathway and thereafter attempt to improve the growth and productivity by engineering the gene targets found in this report **Er det her for flabet skrevet hehe**. -->
## 6. Conclusion
We computationally generated 12 progesterone-producing *S. cerevisiae* cell factories.
Using phenotypic simulations, our best-performing strains reached a progesterone productivity of 0.167 mmol/(gDW\*h). Additionally, the progesterone titre was estimated to 0.212 mmol/L in a batch fermentation simulation with 10 mmol/L glucose initially and a constant $\textrm{O}_{2}$ uptake of 2 mmol/(gDW\*h).
Our work indicates that it is possible to produce the heterologous steroid progesterone using *S. cerevisiae* as host and that the production can be optimized computationally, to find the best strategies for maximizing the yield obtained.
Such a production would be more sustainable than the current production and, thus, contribute to several of the UN sustainable development goals (SDG); These goals describe the steps necessary to ensure a sustainable future for all. Our work is related to SDGs 3 (by promoting health), 9 (by being innovative), and 12 (by ensuring responsible production), and if successful, will provide a better alternative for steroid production and contribute to a sustainable tomorrow.

**Figure 15.** Our project contributes to SDG3 - Good health and well-being (by promoting good health), SDG9 - Industry, innovation and infrastructure (by being innovative), and SDG12 - Responsible consumption and production (by ensuring sustainable production).
## References
Al Jasem, Yosef, et al. "Preparation of steroidal hormones with an emphasis on transformations of phytosterols and cholesterol-a review." Mediterranean Journal of Chemistry 3.2 (2014): 796-830.
Bachmann, W. E., Wayne Cole, and A. L. Wilds. "The total synthesis of the sex hormone equilenin and its stereoisomers." Journal of the American Chemical Society 62.4 (1940): 824-839.
Bartlett, Paul D., Frank Henry Westheimer, and G. Büchi. "Robert Burns Woodward, Nobel Prize in Chemistry for 1965." Science 150.3696 (1965): 585-587.
Batth, Rituraj, et al. "Biosynthesis and industrial production of androsteroids." Plants 9.9 (2020): 1144.
Buhaescu, Irina, and Hassane Izzedine. "Mevalonate pathway: a review of clinical and therapeutical implications." Clinical biochemistry 40.9-10 (2007): 575-584.
Cardoso, Joao GR, et al. "Cameo: a Python library for computer aided metabolic engineering and optimization of cell factories." ACS synthetic biology 7.4 (2018): 1163-1166.
Choi, Hyung Seok, et al. "In silico identification of gene amplification targets for improvement of lycopene production." Applied and environmental microbiology 76.10 (2010): 3097-3105.
Csáky, Zsófia, et al. "Squalene lipotoxicity in a lipid droplet‐less yeast mutant is linked to plasma membrane dysfunction." Yeast 37.1 (2020): 45-62.
Domenzain, Iván, et al. "Reconstruction of a catalogue of genome-scale metabolic models with enzymatic constraints using GECKO 2.0." BioRxiv (2021).
Dong, Jingzhou, et al. "Direct biotransformation of dioscin into diosgenin in rhizome of Dioscorea zingiberensis by Penicillium dioscin." Indian journal of microbiology 55.2 (2015): 200-206.
Duarte NC, Herrgård MJ, Palsson BØ. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 2004 Jul;14(7):1298-309. doi: 10.1101/gr.2250904. Epub 2004 Jun 14. PMID: 15197165; PMCID: PMC442145.
Howie, Peter W. "The progestogen-only pill." British journal of obstetrics and gynaecology 92.10 (1985): 1001-1002.
Jesus, Mafalda, et al. "Diosgenin: recent highlights on pharmacology and analytical methodology." Journal of analytical methods in chemistry 2016 (2016).
Jiang, Yi-qi, and Jian-ping Lin. "Recent progress in strategies for steroid production in yeasts." World Journal of Microbiology and Biotechnology 38.6 (2022): 1-14.
Jordá, Tania, and Sergi Puig. "Regulation of ergosterol biosynthesis in Saccharomyces cerevisiae." Genes 11.7 (2020): 795.
Malina, Carl, et al. "Adaptations in metabolism and protein translation give rise to the Crabtree effect in yeast." Proceedings of the National Academy of Sciences of the United States of America (2021): vol. 118,51.
Nath, Anita, and Regine Sitruk-Ware. "Progesterone vaginal ring for contraceptive use during lactation." Contraception 82.5 (2010): 428-434.
Paramasivan, Kalaivani, and Sarma Mutturi. "Recent advances in the microbial production of squalene." World Journal of Microbiology and Biotechnology 38.5 (2022): 1-21.
Parapouli, Maria, et al. "Saccharomyces cerevisiae and its industrial applications." AIMS microbiology 6.1 (2020): 1.
Patil, Kiran Raosaheb, et al. "Evolutionary programming as a platform for in silico metabolic engineering." BMC bioinformatics 6.1 (2005): 1-12.
Slater, Leo B. "Industry and academy: The synthesis of steroids." Historical studies in the physical and biological sciences 30.2 (2000): 443-480.
Tong, Wang-Yu, and Xiang Dong. "Microbial biotransformation: recent developments on steroid drugs." Recent patents on biotechnology 3.2 (2009): 141-153.
Woodward, R. B., et al. "The total synthesis of steroids1." Journal of the American Chemical Society 74.17 (1952): 4223-4251.
Xu, Shanhui, and Yanran Li. "Yeast as a promising heterologous host for steroid bioproduction." Journal of Industrial Microbiology & Biotechnology: Official Journal of the Society for Industrial Microbiology and Biotechnology 47.9-10 (2020): 829-843.
Zhu, Zhan-Tao, et al. "Metabolic compartmentalization in yeast mitochondria: Burden and solution for squalene overproduction." Metabolic engineering 68 (2021): 232-245.
|
{
"filename": "Report_1.ipynb",
"repository": "27410/27410-group-assignment-group-3-progesterone-in-s-cerevisiae",
"query": "transformed_from_existing",
"size": 58895,
"sha": ""
}
|
# Summarization.ipynb
Repository: SanchitSah12/Text-Summarization
<code>
f = open("text1.txt","r")
content = f.read()
print(content)
path_online_source = "summarized_from_online_source.txt"
path_model_source = "summary_from_model.txt"
text_path = "text1.txt"
</code>
<code>
import nltk
nltk.download('stopwords')
nltk.download('punkt')
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize, sent_tokenize
</code>
<code>
# Tokenizing the text
stopWords = set(stopwords.words("english"))
words = word_tokenize(content)
print(stopWords)
print()
print(words)
</code>
<code>
# Creating a frequency table to keep the
# score of each word
freqTable = dict()
for word in words:
word = word.lower()
if word in stopWords:
continue
if word in freqTable:
freqTable[word] += 1
else:
freqTable[word] = 1
</code>
<code>
print(freqTable)
</code>
<code>
# Creating a dictionary to keep the score of each sentence
sentences = sent_tokenize(content)
sentenceValue = dict()
for sentence in sentences:
for word, freq in freqTable.items():
if word in sentence.lower():
if sentence in sentenceValue:
sentenceValue[sentence] += freq
else:
sentenceValue[sentence] = freq
</code>
<code>
print(sentenceValue)
</code>
<code>
sumValues = 0
for sentence in sentenceValue:
sumValues += sentenceValue[sentence]
</code>
<code>
# Average value of a sentence from the original text
average = int(sumValues / len(sentenceValue))
# Storing sentences into our summary.
summary = ''
for sentence in sentences:
if (sentence in sentenceValue) and (sentenceValue[sentence] > (1.2 * average)):
summary += " " + sentence
print(summary)
</code>
<code>
len(summary)
</code>
<code>
import spacy
</code>
<code>
nlp = spacy.load("en_core_web_sm")
</code>
<code>
doc = nlp(summary)
</code>
<code>
print(doc)
</code>
<code>
summary_length = len(doc)
print(summary_length)
</code>
<code>
#reading summary obtained from online sources
with open (path_online_source, "r") as f:
data = f.read()
</code>
<code>
# summarized = nlp(data)
</code>
<code>
print(data)
</code>
<code>
f.close()
</code>
<code>
summarized = nlp(data)
</code>
<code>
print(summarized)
</code>
<code>
online_length = len(summarized)
print(online_length)
</code>
<code>
import numpy as np
import matplotlib.pyplot as plt
</code>
<code>
x =[online_length]
</code>
<code>
y = [summary_length]
</code>
<code>
X_axis = np.arange(1)
</code>
<code>
plt.bar(X_axis - 0.3, x, 0.1, label = 'summary_from_online_source')
plt.bar(X_axis + 0.3, y, 0.1, label = 'summary from_model')
plt.xticks(X_axis)
plt.xlabel("Models")
plt.ylabel("Length of Summary")
plt.title("Length of summary from models")
plt.legend()
plt.show()
</code>
<code>
import math
import string
import sys
</code>
<code>
# reading the text file
# This function will return a list of the lines of text in the file.
def read_file(filename):
try:
with open(filename, 'r') as f:
data = f.read()
return data
except IOError:
print("Error opening or reading input file: ", filename)
sys.exit()
</code>
<code>
# splitting the text lines into words
# translation table is a global variable mapping upper case to lower case and punctuation to spaces
translation_table = str.maketrans(string.punctuation+string.ascii_uppercase,
" "*len(string.punctuation)+string.ascii_lowercase)
</code>
<code>
# returns a list of the words in the file
def get_words_from_line_list(text):
text = text.translate(translation_table)
word_list = text.split()
return word_list
</code>
<code>
# counts frequency of each word returns a dictionary which maps the words to their frequency.
def count_frequency(word_list):
D = {}
for new_word in word_list:
if new_word in D:
D[new_word] = D[new_word] + 1
else:
D[new_word] = 1
return D
</code>
<code>
# returns dictionary of (word, frequency) pairs from the previous dictionary.
def word_frequencies_for_file(filename):
line_list = read_file(filename)
word_list = get_words_from_line_list(line_list)
freq_mapping = count_frequency(word_list)
print("File", filename, ":", )
print(len(line_list), "lines, ", )
print(len(word_list), "words, ", )
print(len(freq_mapping), "distinct words")
return freq_mapping
</code>
<code>
# returns the dot product of two documents
def dotProduct(D1, D2):
Sum = 0.0
for key in D1:
if key in D2:
Sum += (D1[key] * D2[key])
return Sum
</code>
<code>
# returns the angle in radians between document vectors
def vector_angle(D1, D2):
numerator = dotProduct(D1, D2)
denominator = math.sqrt(dotProduct(D1, D1)*dotProduct(D2, D2))
return math.acos(numerator / denominator)
</code>
<code>
def documentSimilarity(filename_1, filename_2):
sorted_word_list_1 = word_frequencies_for_file(filename_1)
sorted_word_list_2 = word_frequencies_for_file(filename_2)
distance = vector_angle(sorted_word_list_1, sorted_word_list_2)
print("The distance between the documents is: % 0.6f (radians)"% distance)
</code>
<code>
with open (path_model_source, "a") as f:
f.write(summary)
</code>
<code>
f.close()
</code>
|
{
"filename": "Summarization.ipynb",
"repository": "SanchitSah12/Text-Summarization",
"query": "transformed_from_existing",
"size": 59219,
"sha": ""
}
|
# final_model.ipynb
Repository: sunoo2468/capstone1
<code>
!pip install requests pandas sqlalchemy beautifulsoup4 nltk
</code>
<code>
pip install torch torchvision torchaudio
</code>
## 전체 코드: 뉴스 크롤링 → 키워드 추출 → 임베딩 → 대조학습 → 유망도 예측 → 추천
1. 데이터 수집
- 기능
- NewsAPI를 통해 최근 CNN 뉴스 30일치 수집
- NASDAQ 기업 리스트 로딩 및 필터링
- 주요 함수
- fetch_news_articles()
- load_nasdaq_tickers()
2. 전처리 및 키워드 분석
- 기능
- vader로 감성 분석
- spaCy로 ORG 엔터티 추출
- 산업 키워드 포함 여부 체크
- 주요 함수
- extract_industry_keywords(text)
- extract_positive_orgs(text)
3. 임베딩 및 대조학습 모델
- 기능
- GloVe 로딩 및 PCA 차원 축소
- 뉴스/산업/기업 벡터 생성
- 대조학습(Projector 모델) 정의 및 학습
- 주요 함수
- load_glove_embeddings(path)
- get_text_vector(text)
- generate_company_vector_from_ticker(ticker, glove_pca, projector_model)
- train_contrastive_model(samples, company_vectors)
4. 유망도 예측 모델
- 기능
- 산업 키워드 빈도/감성/연결성 기반 유망도 계산
- 산업 벡터 + 유망도 라벨로 MLP 회귀 모델 학습
- 이후 기업 벡터 입력으로 유망도 예측
- 주요 함수
- calculate_industry_score(df)
- train_potential_predictor(industry_vectors, industry_score)
- get_company_potential_score(ticker)
5. 응용 기능: 유사 기업 추천 및 스코어 조회
- 기능
- 유사 기업 추천 (코사인 유사도)
- 스코어 기반 순위화 가능
- 주요 함수
- recommend_similar_companies(ticker, company_vectors, top_k=3)
- get_company_potential_score(ticker)
6. 시각화 및 유망도 랭킹화
- 기능
- 산업 및 기업 벡터를 PCA(2D)로 시각화하여 클러스터 구조 확인
- 산업/기업 유망도 점수를 z-score로 정규화하여 상위 랭킹 도출
- 산업 vs 기업 관계를 하나의 시각 공간에 통합적으로 분석 가능
- 주요 함수 및 처리 흐름
- PCA(n_components=2) → 벡터 2D 축소
- plt.scatter + plt.text → 산업/기업별 plot 시각화
- scipy.stats.zscore → 유망도 점수 정규화
- DataFrame.sort_values("zscore") → 랭킹화 정렬 및 출력
# 1. 모듈 임포트 및 초기 설정
<code>
import os
import requests
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import nltk
import time
from datetime import datetime, timedelta
from bs4 import BeautifulSoup
from collections import Counter
from sklearn.decomposition import PCA
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
import torch
import torch.nn as nn
import spacy
from nltk.sentiment.vader import SentimentIntensityAnalyzer
nltk.download('vader_lexicon')
vader_analyzer = SentimentIntensityAnalyzer()
nlp = spacy.load("en_core_web_lg")
# 날짜 범위 설정
API_KEY = os.getenv('API_KEY')
today = datetime.today()
from_date = (today - timedelta(days=30)).strftime("%Y-%m-%d")
to_date = today.strftime("%Y-%m-%d")
# 산업키워드 정의
industry_keywords = [
"ai", "artificial intelligence", "machine learning", "deep learning", "neural network",
"automotive", "car", "vehicle", "electric vehicle", "ev", "self-driving", "autonomous vehicle",
"space", "nasa", "spacex", "rocket", "satellite", "aerospace",
"semiconductor", "chip", "microchip", "integrated circuit", "ic",
"robot", "robotics", "automation", "industrial automation", "drone", "drone delivery",
"cloud computing", "cloud", "big data", "data center", "database", "analytics",
"biotech", "biotechnology", "pharmaceutical", "vaccine", "healthcare", "gene editing", "crispr",
"renewable energy", "solar", "wind", "green energy", "hydrogen", "nuclear energy", "clean energy",
"fintech", "digital banking", "mobile payment", "blockchain", "crypto", "bitcoin", "ethereum",
"streaming", "netflix", "disney", "hbo", "youtube", "media",
"telecom", "5g", "6g", "broadband", "wireless", "satellite internet",
"manufacturing", "industrial", "machinery", "supply chain", "smart factory",
"construction", "infrastructure", "civil engineering", "smart city",
"logistics", "delivery", "shipping", "e-commerce logistics", "transportation",
"cybersecurity", "security", "data protection", "encryption",
"gaming", "video game", "esports", "game development",
"climate", "carbon", "net zero", "sustainability", "esg",
"edtech", "online education", "e-learning", "virtual classroom",
"retail", "fashion", "e-commerce", "online shopping",
"agriculture", "agritech", "smart farming"
]
</code>
# 2. GloVe 임베딩 로딩 및 PCA 차원 축소
<code>
def load_glove_embeddings(file_path):
embeddings = {}
with open(file_path, 'r', encoding='utf-8') as f:
for line in f:
values = line.split()
word = values[0]
vector = np.array(values[1:], dtype='float32')
embeddings[word] = vector
return embeddings
glove_path = "data/glove.6B.300d.txt"
glove_embeddings = load_glove_embeddings(glove_path)
words = list(glove_embeddings.keys())
vectors = np.stack([glove_embeddings[word] for word in words])
pca = PCA(n_components=64)
reduced_vectors = pca.fit_transform(vectors)
glove_pca = dict(zip(words, reduced_vectors))
</code>
# 3. NASDAQ 기업 로드 및 필터링
<code>
nasdaq_df = pd.read_csv("data/nasdaq_screener_1744184912302.csv")
nasdaq_df = nasdaq_df[(nasdaq_df["Market Cap"] > 0) & nasdaq_df["Sector"].notnull()]
nasdaq_df = nasdaq_df[~nasdaq_df["Name"].str.contains("Units|Rights|Warrant|Preferred|Depositary|Series", case=False)]
nasdaq_df["Symbol"] = nasdaq_df["Symbol"].astype(str).str.lower()
ticker_name_map = dict(zip(nasdaq_df["Symbol"], nasdaq_df["Name"].str.lower()))
name_to_ticker_map = {v: k for k, v in ticker_name_map.items()}
</code>
# 4. 뉴스 크롤링(수집) (CNN)
<code>
API_URL = "https://newsapi.org/v2/everything"
params = {
'q': 'nasdaq OR stock OR technology OR innovation',
'from': from_date,
'to': to_date,
'language': 'en',
'pageSize': 100,
'domains': 'cnn.com',
'apiKey': '4c1f9d63d25843a4917cce95a9b2186c',
'sortBy': 'publishedAt'
}
headers = {'User-Agent': 'Mozilla/5.0'}
news_list = []
page = 1
while page <= 5:
params['page'] = page
response = requests.get(API_URL, params=params, headers=headers)
if response.status_code != 200:
break
articles = response.json().get("articles", [])
for article in articles:
full_text = f"{article.get('title', '')} {article.get('description', '')} {article.get('content', '')}"
news_list.append({
"title": article.get("title", ""),
"description": article.get("description", ""),
"content": article.get("content", ""),
"url": article.get("url", ""),
"published_at": article.get("publishedAt", ""),
"full_text": full_text
})
if len(articles) < 100:
break
page += 1
time.sleep(0.5)
df = pd.DataFrame(news_list)
</code>
# 5. 키워드 추출
<code>
def extract_industry_keywords(text):
if not isinstance(text, str): return []
text = text.lower()
return [kw for kw in industry_keywords if kw in text]
def extract_positive_orgs(text, max_orgs=3):
if not isinstance(text, str): return []
score = vader_analyzer.polarity_scores(text)
if score["compound"] <= 0.2: return []
doc = nlp(text)
orgs = set(ent.text.strip().lower()
for ent in doc.ents if ent.label_ == "ORG")
matched = []
for org in orgs:
for name, ticker in name_to_ticker_map.items():
if name in org or org in name:
matched.append(ticker)
break
if len(matched) >= max_orgs:
break
return matched
df["positive_org_keywords"] = df["full_text"].apply(lambda x: extract_positive_orgs(x))
df["industry_keywords"] = df["full_text"].apply(lambda x: extract_industry_keywords(x))
</code>
### 5.1 결과확인
<code>
# 키워드 통계 출력
org_counts = Counter(sum(df["positive_org_keywords"].tolist(), []))
industry_counts = Counter(sum(df["industry_keywords"].tolist(), []))
print("\n 상위 기업 키워드:")
for keyword, count in org_counts.most_common(20):
print(f"{keyword}: {count}")
print("\n 상위 산업 키워드:")
for keyword, count in industry_counts.most_common(20):
print(f"{keyword}: {count}")
</code>
# 6. 임베딩 및 대조학습
<code>
# 기업 벡터 생성 함수들
def get_cik(ticker):
url = f"https://www.sec.gov/files/company_tickers.json"
headers = {'User-Agent': 'sj0juu@gmail.com'}
data = requests.get(url, headers=headers).json()
for k, v in data.items():
if v['ticker'].lower() == ticker.lower():
return str(v['cik_str']).zfill(10)
return None
def get_10k_filing_urls(cik):
url = f"https://data.sec.gov/submissions/CIK{cik}.json"
headers = {'User-Agent': 'sj0juu@gmail.com'}
data = requests.get(url, headers=headers).json()
urls = []
for i, filing in enumerate(data['filings']['recent']['form']):
if filing == "10-K":
accession = data['filings']['recent']['accessionNumber'][i].replace("-", "")
doc_url = f"https://www.sec.gov/Archives/edgar/data/{int(cik)}/{accession}/index.json"
doc_data = requests.get(doc_url, headers=headers).json()
for file in doc_data['directory']['item']:
if file['name'].endswith(".htm") or file['name'].endswith(".txt"):
urls.append(f"https://www.sec.gov/Archives/edgar/data/{int(cik)}/{accession}/{file['name']}")
break
return urls
def extract_text_from_url(url):
headers = {'User-Agent': 'sj0juu@gmail.com'}
res = requests.get(url, headers=headers)
try:
soup = BeautifulSoup(res.content, 'html.parser')
return soup.get_text(separator=' ')
except:
return None
def extract_top_keywords(text, top_k=10):
vectorizer = TfidfVectorizer(stop_words='english', max_features=1000)
tfidf_matrix = vectorizer.fit_transform([text])
scores = tfidf_matrix.toarray()[0]
features = vectorizer.get_feature_names_out()
top_indices = scores.argsort()[::-1][:top_k]
return [features[i] for i in top_indices]
def build_company_vector(keywords, glove_pca, projector_model):
vectors = [glove_pca[word] for word in keywords if word in glove_pca]
if vectors:
avg_vec = np.mean(vectors, axis=0)
return projector_model(torch.tensor(avg_vec).float()).detach().numpy()
return None
def generate_company_vector_from_ticker(ticker, glove_pca, projector_model):
cik = get_cik(ticker)
if not cik: return None
urls = get_10k_filing_urls(cik)
if not urls: return None
text = extract_text_from_url(urls[0])
if text is None: return None
keywords = extract_top_keywords(text, top_k=10)
return build_company_vector(keywords, glove_pca, projector_model)
# 뉴스 벡터화
def get_text_vector(text):
words = [w.lower() for w in text.split() if w.lower() in glove_pca]
if not words: return None
vectors = [glove_pca[w] for w in words]
return np.mean(vectors, axis=0)
# 대조학습 모델
class ContrastiveProjector(nn.Module):
def __init__(self, input_dim=64):
super().__init__()
self.net = nn.Sequential(
nn.Linear(input_dim, 64),
nn.ReLU(),
nn.Linear(64, input_dim)
)
def forward(self, x):
return self.net(x)
projector = ContrastiveProjector()
optimizer = torch.optim.Adam(projector.parameters(), lr=1e-3)
loss_fn = nn.CosineEmbeddingLoss()
# 벡터 샘플링
samples = []
texts = []
for text in df["full_text"]:
v = get_text_vector(text)
if v is not None:
samples.append(v)
texts.append(text)
# 뉴스에서 감지된 모든 기업 티커 수집
all_tickers = set(sum(df["positive_org_keywords"].tolist(), []))
company_vectors = {}
for ticker in all_tickers:
vec = generate_company_vector_from_ticker(ticker, glove_pca, projector)
if vec is not None:
company_vectors[ticker] = vec
# 대조학습
for epoch in range(3):
total_loss = 0
# 뉴스-뉴스 학습
for i in range(0, len(samples) - 3, 3):
v1 = torch.tensor(samples[i], dtype=torch.float32)
v2 = torch.tensor(samples[i+1], dtype=torch.float32)
v3 = torch.tensor(samples[i+2], dtype=torch.float32)
proj1 = projector(v1)
proj2 = projector(v2)
proj3 = projector(v3)
loss_pos = loss_fn(proj1, proj2, torch.tensor(1.0))
loss_neg = loss_fn(proj1, proj3, torch.tensor(-1.0))
loss = loss_pos + loss_neg
optimizer.zero_grad()
loss.backward()
optimizer.step()
total_loss += loss.item()
# 기업-뉴스 학습
for ticker, comp_vec_np in company_vectors.items():
news_idx = np.random.randint(0, len(samples))
news_vec = torch.tensor(samples[news_idx], dtype=torch.float32)
comp_vec = torch.tensor(comp_vec_np, dtype=torch.float32)
proj_c = projector(comp_vec)
proj_n = projector(news_vec)
neg_idx = np.random.randint(0, len(samples))
neg_vec = torch.tensor(samples[neg_idx], dtype=torch.float32)
proj_neg = projector(neg_vec)
loss_pos = loss_fn(proj_c, proj_n, torch.tensor(1.0))
loss_neg = loss_fn(proj_c, proj_neg, torch.tensor(-1.0))
loss = loss_pos + loss_neg
optimizer.zero_grad()
loss.backward()
optimizer.step()
total_loss += loss.item()
print(f"Epoch {epoch+1}, Loss: {total_loss:.4f}")
</code>
# 7. 산업 벡터 생성 및 예측
<code>
# 산업 벡터 생성 및 projector 적용
industry_vectors = {}
for kw in industry_keywords:
if kw in glove_pca:
projected_vec = projector(torch.tensor(glove_pca[kw]).float()).detach().numpy()
industry_vectors[kw] = projected_vec
# 산업 벡터 저장 (CSV)
industry_vec_list = []
for name, vec in industry_vectors.items():
row = {"industry": name}
row.update({f"dim_{i}": val for i, val in enumerate(vec)})
industry_vec_list.append(row)
pd.DataFrame(industry_vec_list).to_csv("results/projected_industry_vectors.csv", index=False)
# 기업 벡터 저장 (CSV)
company_vec_list = []
for ticker, vec in company_vectors.items():
row = {"ticker": ticker}
row.update({f"dim_{i}": val for i, val in enumerate(vec)})
company_vec_list.append(row)
pd.DataFrame(company_vec_list).to_csv("results/projected_company_vectors.csv", index=False)
print("\n✅ 산업 및 기업 벡터가 각각 CSV 파일로 저장되었습니다.")
</code>
# 8. 유망도 회귀 모델
<code>
from collections import defaultdict
from sklearn.linear_model import LinearRegression
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import numpy as np
# ✅ 모델 정의
class PotentialPredictor(nn.Module):
def __init__(self):
super().__init__()
self.net = nn.Sequential(
nn.Linear(65, 64),
nn.ReLU(),
nn.Linear(64, 32),
nn.ReLU(),
nn.Linear(32, 1)
)
def forward(self, x):
return self.net(x)
industry_freq = Counter()
industry_sentiment = defaultdict(list)
industry_orgs = defaultdict(set)
industry_freq = Counter()
industry_sentiment = defaultdict(list)
industry_orgs = defaultdict(set)
# ✅ 산업별 통계 수집
for _, row in df.iterrows():
sentiment = vader_analyzer.polarity_scores(row["full_text"])["compound"]
for ind in row["industry_keywords"]:
industry_freq[ind] += 1
industry_sentiment[ind].append(sentiment)
for org in row["positive_org_keywords"]:
industry_orgs[ind].add(org)
# ✅ 임시 정답(y) 생성 및 회귀 모델 학습
industry_stats = []
industry_names = []
for ind in industry_freq:
freq_score = np.log1p(industry_freq[ind])
sent_score = np.mean(industry_sentiment[ind])
conn_score = len(industry_orgs[ind])
industry_stats.append((freq_score, sent_score, conn_score))
industry_names.append(ind)
industry_stats = np.array(industry_stats)
X_stats = industry_stats[:, :3]
# 기존 수식을 기반으로 한 임시 라벨
y_score = 0.4 * X_stats[:, 0] + 0.3 * X_stats[:, 1] + 0.3 * X_stats[:, 2]
reg = LinearRegression()
reg.fit(X_stats, y_score)
# ✅ 학습된 모델로 industry_score 계산
industry_score = {}
for i, ind in enumerate(industry_names):
features = X_stats[i].reshape(1, -1)
predicted_score = reg.predict(features)[0]
industry_score[ind] = predicted_score
# === Step 1: 유망 산업 vs 비유망 산업 선정 (기존 industry_score 기준)
sorted_inds = sorted(industry_score.items(), key=lambda x: x[1], reverse=True)
top_industries = [name for name, _ in sorted_inds[:10]]
bottom_industries = [name for name, _ in sorted_inds[-10:]]
# === Step 2: 평균 벡터로 유망도 방향(potential axis) 정의
top_vec = np.mean([industry_vectors[i] for i in top_industries if i in industry_vectors], axis=0)
bot_vec = np.mean([industry_vectors[i] for i in bottom_industries if i in industry_vectors], axis=0)
potential_axis = top_vec - bot_vec
potential_axis = potential_axis / np.linalg.norm(potential_axis)
# === Step 3: 투영 함수 정의
def add_potential_dimension(vec, axis):
projection = np.dot(vec, axis) # 스칼라값
return np.append(vec, projection) # 기존 64D → 65D
# === Step 4: 산업/기업 벡터 확장
industry_vectors_aug = {
name: add_potential_dimension(vec, potential_axis)
for name, vec in industry_vectors.items()
}
company_vectors_aug = {
ticker: add_potential_dimension(vec, potential_axis)
for ticker, vec in company_vectors.items()
}
# === Step 5: 회귀 학습용 데이터 구성
X = []
y = []
for name, vec in industry_vectors_aug.items():
if name in industry_score:
X.append(vec)
y.append(industry_score[name])
X = torch.tensor(np.stack(X), dtype=torch.float32)
y = torch.tensor(np.array(y).reshape(-1, 1), dtype=torch.float32)
# 모델 재사용(딥러닝 모델에 이제 기업을 넣어보자)
model = PotentialPredictor()
optimizer = torch.optim.Adam(model.parameters(), lr=1e-3)
loss_fn = nn.MSELoss()
# 학습 루프
for epoch in range(300):
model.train()
pred = model(X)
loss = loss_fn(pred, y)
optimizer.zero_grad()
loss.backward()
optimizer.step()
if (epoch+1) % 50 == 0:
print(f"[Epoch {epoch+1}] Loss: {loss.item():.4f}")
company_scores = {}
for ticker, vec in company_vectors_aug.items():
x = torch.tensor(vec, dtype=torch.float32).unsqueeze(0)
with torch.no_grad():
score = model(x).item()
company_scores[ticker.upper()] = score
def get_company_potential_score(ticker):
ticker = ticker.upper()
if ticker in company_scores:
return round(company_scores[ticker], 4)
else:
vec = generate_company_vector_from_ticker(ticker.lower(), glove_pca, projector)
if vec is not None:
vec_aug = add_potential_dimension(vec, potential_axis) # 65차원 확장
x = torch.tensor(vec_aug, dtype=torch.float32).unsqueeze(0)
with torch.no_grad():
score = model(x).item()
company_scores[ticker] = score
return round(score, 4)
else:
return "해당 기업 벡터 없음 또는 NASDAQ 비상장"
</code>
### 8.1 (예시)결과
<code>
# 예시
print("AAPL 유망도:", get_company_potential_score("AAPL"))
print("TSLA 유망도:", get_company_potential_score("TSLA"))
print("NVDA 유망도:", get_company_potential_score("NVDA"))
</code>
<code>
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from sklearn.decomposition import PCA
#3D
#각 점은 65차원 기업 벡터를 3차원으로 축소한 것입니다.
#색상은 **잠재력 축에의 정렬 정도(= 유망도 점수)**를 나타냅니다.
#밝은 색 = 현재 산업 흐름과 가장 유사한 방향에 있는 기업
#흰색 텍스트는 유망도 상위 5개 기업입니다.
#축 해석
#x축 (PCA 1) 데이터에서 가장 큰 분산 방향 (가장 큰 정보량)
#y축 (PCA 2) 두 번째로 큰 분산 방향 (PCA1과 직교)
#z축 (PCA 3) 세 번째 주성분 방향 (PCA1,2와 직교)
#색상 (컬러바) ✔️ 잠재력 축 방향에의 투영값 = 유망도 점수
# 데이터 준비
tickers = list(company_vectors_aug.keys())
vecs = np.stack([company_vectors_aug[t] for t in tickers])
scores = [v[-1] for v in vecs] # 65번째 차원 = 유망도 축 투영값
# PCA로 3D 축소
pca = PCA(n_components=3)
vecs_3d = pca.fit_transform(vecs)
# 3D 시각화
fig = plt.figure(figsize=(12, 8))
ax = fig.add_subplot(111, projection='3d')
scatter = ax.scatter(vecs_3d[:, 0], vecs_3d[:, 1], vecs_3d[:, 2],
c=scores, cmap='plasma', s=60, alpha=0.9)
# 상위 유망 기업 라벨링
top_indices = np.argsort(scores)[-5:]
for i in top_indices:
ax.text(vecs_3d[i, 0], vecs_3d[i, 1], vecs_3d[i, 2], tickers[i],
fontsize=9, weight='bold', color='white')
# 축 및 색상바 설정
ax.set_title("3D 기업 벡터 시각화 (잠재력 축 강조)", fontsize=14)
ax.set_xlabel("PCA 1")
ax.set_ylabel("PCA 2")
ax.set_zlabel("PCA 3")
cbar = fig.colorbar(scatter, ax=ax, shrink=0.6)
cbar.set_label("유망도 점수 (잠재력 축 투영값)")
plt.tight_layout()
plt.show()
</code>
# 9. 시각화 (PCA)
### 9.1 산업 시각화
<code>
from scipy.stats import zscore
# 산업 벡터 시각화 (2D)
industry_names = list(industry_vectors.keys())
industry_matrix = np.stack([industry_vectors[k] for k in industry_names])
pca_ind = PCA(n_components=2)
industry_2d = pca_ind.fit_transform(industry_matrix)
plt.figure(figsize=(10, 6))
plt.scatter(industry_2d[:, 0], industry_2d[:, 1], c='skyblue')
for i, name in enumerate(industry_names):
plt.text(industry_2d[i, 0], industry_2d[i, 1], name, fontsize=8)
plt.title("Industry Embedding Clusters (PCA 2D)")
plt.xlabel("PC1")
plt.ylabel("PC2")
plt.grid(True)
plt.show()
# 유망도 Z-score 계산
industry_score_df = pd.DataFrame({
"industry": list(industry_score.keys()),
"score": list(industry_score.values())
})
industry_score_df["zscore"] = zscore(industry_score_df["score"])
industry_score_df.sort_values("zscore", ascending=False, inplace=True)
# 상위 30% 산업 랭킹 출력
top_n = max(1, int(len(industry_score_df) * 0.3))
print("\n Top 30% Promising Industries (Z-score 기반):")
print(industry_score_df.head(top_n).to_string(index=False))
</code>
### 9.2 기업 벡터 시각화 + 산업/기업 클러스터 통합 시각화
<code>
# 기업 벡터 PCA 시각화
company_names = list(company_vectors.keys())
company_matrix = np.stack([company_vectors[k] for k in company_names])
pca_company = PCA(n_components=2)
company_2d = pca_company.fit_transform(company_matrix)
plt.figure(figsize=(10, 6))
plt.scatter(company_2d[:, 0], company_2d[:, 1], c='lightcoral')
for i, name in enumerate(company_names):
plt.text(company_2d[i, 0], company_2d[i, 1], name.upper(), fontsize=8)
plt.title("Company Embedding Clusters (PCA 2D)")
plt.xlabel("PC1")
plt.ylabel("PC2")
plt.grid(True)
plt.show()
# 산업/기업 클러스터 통합 시각화
all_points = np.concatenate([industry_matrix, company_matrix], axis=0)
all_labels = industry_names + company_names
colors = ["blue"] * len(industry_names) + ["red"] * len(company_names)
pca_all = PCA(n_components=2)
all_2d = pca_all.fit_transform(all_points)
plt.figure(figsize=(10, 6))
plt.scatter(all_2d[:, 0], all_2d[:, 1], c=colors)
for i, label in enumerate(all_labels):
plt.text(all_2d[i, 0], all_2d[i, 1], label, fontsize=8)
plt.title("Industry vs Company Vector Clusters")
plt.xlabel("PC1")
plt.ylabel("PC2")
plt.grid(True)
plt.show()
</code>
### 9.3 기업 유망도 Z-score 기반 랭킹
<code>
company_score_df = pd.DataFrame({
"ticker": list(company_scores.keys()),
"score": list(company_scores.values())
})
company_score_df["zscore"] = zscore(company_score_df["score"])
company_score_df.sort_values("zscore", ascending=False, inplace=True)
# 상위 30% 기업 랭킹 출력
top_n_c = max(1, int(len(company_score_df) * 0.3))
print("\n🏆 Top 30% Promising Companies (Z-score 기반):")
print(company_score_df.head(top_n_c).to_string(index=False))
</code>
<code>
import matplotlib.pyplot as plt
import matplotlib.font_manager as fm
import platform
# 한글 폰트 설정 (OS별 분기)
if platform.system() == 'Darwin': # macOS
plt.rcParams['font.family'] = 'AppleGothic'
elif platform.system() == 'Windows':
plt.rcParams['font.family'] = 'Malgun Gothic'
else: # 리눅스 (colab 포함)
plt.rcParams['font.family'] = 'NanumGothic'
plt.rcParams['axes.unicode_minus'] = False # 마이너스(-) 깨짐 방지
</code>
<code>
# ✅ 산업 유망도 요약 표
industry_levels = {}
for name, score in industry_score.items():
z = (score - np.mean(list(industry_score.values()))) / np.std(list(industry_score.values()))
if z >= 1.0:
level = "🟢 매우 유망"
elif z >= 0.5:
level = "🟡 유망"
elif z >= -0.5:
level = "⚪ 보통"
else:
level = "🔴 낮은 유망도"
industry_levels[name] = (score, z, level)
industry_df = pd.DataFrame([
{"산업명": name, "점수": score, "Z-Score": z, "등급": level}
for name, (score, z, level) in industry_levels.items()
]).sort_values(by="점수", ascending=False)
print("📊 산업 유망도 TOP 10")
display(industry_df.head(10))
# ✅ 기업 유망도 요약 표
company_levels = {}
for ticker, score in company_scores.items():
z = (score - np.mean(list(company_scores.values()))) / np.std(list(company_scores.values()))
if z >= 1.0:
level = "🟢 매우 유망"
elif z >= 0.5:
level = "🟡 유망"
elif z >= -0.5:
level = "⚪ 보통"
else:
level = "🔴 유망도 낮음"
company_levels[ticker] = (score, z, level)
company_df = pd.DataFrame([
{"티커": ticker, "점수": score, "Z-Score": z, "등급": level}
for ticker, (score, z, level) in company_levels.items()
]).sort_values(by="점수", ascending=False)
print("📈 기업 유망도 TOP 10")
display(company_df.head(10))
# ✅ 등급 분포 시각화
industry_df["등급"].value_counts().plot(kind="bar", title="산업 등급 분포", ylabel="개수")
plt.show()
company_df["등급"].value_counts().plot(kind="bar", title="기업 등급 분포", ylabel="개수")
plt.show()
</code>
### 9.4 유사한 기업 3개 추천 (cosine similarity 기반)
<code>
def recommend_similar_companies(target_ticker, company_vecs, top_k=3):
"""
특정 기업 벡터와 가장 유사한 다른 기업을 코사인 유사도로 추천
"""
target_ticker = target_ticker.lower()
if target_ticker not in company_vecs:
print(f"{target_ticker.upper()} 벡터가 없습니다.")
return []
target_vec = company_vecs[target_ticker].reshape(1, -1)
similarities = []
for ticker, vec in company_vecs.items():
if ticker.lower() == target_ticker:
continue
sim = cosine_similarity(target_vec, vec.reshape(1, -1))[0][0]
similarities.append((ticker.upper(), sim))
similarities.sort(key=lambda x: x[1], reverse=True)
return similarities[:top_k]
</code>
<code>
find_company = "DJT"
similar_companies = recommend_similar_companies(find_company, company_vectors, top_k=3)
print()
print(find_company + "와 유사한 기업 추천:")
for ticker, score in similar_companies:
print(f"- {ticker}: 유사도 {score:.4f}")
</code>
# 전체코드
<code>
import os
import requests
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import nltk
import time
from datetime import datetime, timedelta
from bs4 import BeautifulSoup
from collections import Counter
from sklearn.decomposition import PCA
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
import torch
import torch.nn as nn
import spacy
from nltk.sentiment.vader import SentimentIntensityAnalyzer
nltk.download('vader_lexicon')
vader_analyzer = SentimentIntensityAnalyzer()
nlp = spacy.load("en_core_web_lg")
# 날짜 범위 설정
API_KEY = os.getenv('API_KEY')
today = datetime.today()
from_date = (today - timedelta(days=30)).strftime("%Y-%m-%d")
to_date = today.strftime("%Y-%m-%d")
# 산업키워드 정의
industry_keywords = [
"ai", "artificial intelligence", "machine learning", "deep learning", "neural network",
"automotive", "car", "vehicle", "electric vehicle", "ev", "self-driving", "autonomous vehicle",
"space", "nasa", "spacex", "rocket", "satellite", "aerospace",
"semiconductor", "chip", "microchip", "integrated circuit", "ic",
"robot", "robotics", "automation", "industrial automation", "drone", "drone delivery",
"cloud computing", "cloud", "big data", "data center", "database", "analytics",
"biotech", "biotechnology", "pharmaceutical", "vaccine", "healthcare", "gene editing", "crispr",
"renewable energy", "solar", "wind", "green energy", "hydrogen", "nuclear energy", "clean energy",
"fintech", "digital banking", "mobile payment", "blockchain", "crypto", "bitcoin", "ethereum",
"streaming", "netflix", "disney", "hbo", "youtube", "media",
"telecom", "5g", "6g", "broadband", "wireless", "satellite internet",
"manufacturing", "industrial", "machinery", "supply chain", "smart factory",
"construction", "infrastructure", "civil engineering", "smart city",
"logistics", "delivery", "shipping", "e-commerce logistics", "transportation",
"cybersecurity", "security", "data protection", "encryption",
"gaming", "video game", "esports", "game development",
"climate", "carbon", "net zero", "sustainability", "esg",
"edtech", "online education", "e-learning", "virtual classroom",
"retail", "fashion", "e-commerce", "online shopping",
"agriculture", "agritech", "smart farming"
]
def load_glove_embeddings(file_path):
embeddings = {}
with open(file_path, 'r', encoding='utf-8') as f:
for line in f:
values = line.split()
word = values[0]
vector = np.array(values[1:], dtype='float32')
embeddings[word] = vector
return embeddings
glove_path = "data/glove.6B.300d.txt"
glove_embeddings = load_glove_embeddings(glove_path)
words = list(glove_embeddings.keys())
vectors = np.stack([glove_embeddings[word] for word in words])
pca = PCA(n_components=64)
reduced_vectors = pca.fit_transform(vectors)
glove_pca = dict(zip(words, reduced_vectors))
nasdaq_df = pd.read_csv("data/nasdaq_screener_1744184912302.csv")
nasdaq_df = nasdaq_df[(nasdaq_df["Market Cap"] > 0) & nasdaq_df["Sector"].notnull()]
nasdaq_df = nasdaq_df[~nasdaq_df["Name"].str.contains("Units|Rights|Warrant|Preferred|Depositary|Series", case=False)]
nasdaq_df["Symbol"] = nasdaq_df["Symbol"].astype(str).str.lower()
ticker_name_map = dict(zip(nasdaq_df["Symbol"], nasdaq_df["Name"].str.lower()))
name_to_ticker_map = {v: k for k, v in ticker_name_map.items()}
API_URL = "https://newsapi.org/v2/everything"
params = {
'q': 'nasdaq OR stock OR technology OR innovation',
'from': from_date,
'to': to_date,
'language': 'en',
'pageSize': 100,
'domains': 'cnn.com',
'apiKey': '4c1f9d63d25843a4917cce95a9b2186c',
'sortBy': 'publishedAt'
}
headers = {'User-Agent': 'Mozilla/5.0'}
news_list = []
page = 1
while page <= 5:
params['page'] = page
response = requests.get(API_URL, params=params, headers=headers)
if response.status_code != 200:
break
articles = response.json().get("articles", [])
for article in articles:
full_text = f"{article.get('title', '')} {article.get('description', '')} {article.get('content', '')}"
news_list.append({
"title": article.get("title", ""),
"description": article.get("description", ""),
"content": article.get("content", ""),
"url": article.get("url", ""),
"published_at": article.get("publishedAt", ""),
"full_text": full_text
})
if len(articles) < 100:
break
page += 1
time.sleep(0.5)
df = pd.DataFrame(news_list)
def extract_industry_keywords(text):
if not isinstance(text, str): return []
text = text.lower()
return [kw for kw in industry_keywords if kw in text]
def extract_positive_orgs(text, max_orgs=3):
if not isinstance(text, str): return []
score = vader_analyzer.polarity_scores(text)
if score["compound"] <= 0: return []
doc = nlp(text)
orgs = set(ent.text.strip().lower() for ent in doc.ents if ent.label_ == "ORG")
matched = []
for org in orgs:
for name, ticker in name_to_ticker_map.items():
if name in org or org in name:
matched.append(ticker)
break
if len(matched) >= max_orgs:
break
return matched
df["positive_org_keywords"] = df["full_text"].apply(lambda x: extract_positive_orgs(x))
df["industry_keywords"] = df["full_text"].apply(lambda x: extract_industry_keywords(x))
# 키워드 통계 출력
org_counts = Counter(sum(df["positive_org_keywords"].tolist(), []))
industry_counts = Counter(sum(df["industry_keywords"].tolist(), []))
print("\n🏢 상위 기업 키워드:")
for keyword, count in org_counts.most_common(20):
print(f"{keyword}: {count}")
print("\n🏭 상위 산업 키워드:")
for keyword, count in industry_counts.most_common(20):
print(f"{keyword}: {count}")
# 기업 벡터 생성 함수들
def get_cik(ticker):
url = f"https://www.sec.gov/files/company_tickers.json"
headers = {'User-Agent': 'sj0juu@gmail.com'}
data = requests.get(url, headers=headers).json()
for k, v in data.items():
if v['ticker'].lower() == ticker.lower():
return str(v['cik_str']).zfill(10)
return None
def get_10k_filing_urls(cik):
url = f"https://data.sec.gov/submissions/CIK{cik}.json"
headers = {'User-Agent': 'sj0juu@gmail.com'}
data = requests.get(url, headers=headers).json()
urls = []
for i, filing in enumerate(data['filings']['recent']['form']):
if filing == "10-K":
accession = data['filings']['recent']['accessionNumber'][i].replace("-", "")
doc_url = f"https://www.sec.gov/Archives/edgar/data/{int(cik)}/{accession}/index.json"
doc_data = requests.get(doc_url, headers=headers).json()
for file in doc_data['directory']['item']:
if file['name'].endswith(".htm") or file['name'].endswith(".txt"):
urls.append(f"https://www.sec.gov/Archives/edgar/data/{int(cik)}/{accession}/{file['name']}")
break
return urls
def extract_text_from_url(url):
headers = {'User-Agent': 'sj0juu@gmail.com'}
res = requests.get(url, headers=headers)
try:
soup = BeautifulSoup(res.content, 'html.parser')
return soup.get_text(separator=' ')
except:
return None
def extract_top_keywords(text, top_k=10):
vectorizer = TfidfVectorizer(stop_words='english', max_features=1000)
tfidf_matrix = vectorizer.fit_transform([text])
scores = tfidf_matrix.toarray()[0]
features = vectorizer.get_feature_names_out()
top_indices = scores.argsort()[::-1][:top_k]
return [features[i] for i in top_indices]
def build_company_vector(keywords, glove_pca, projector_model):
vectors = [glove_pca[word] for word in keywords if word in glove_pca]
if vectors:
avg_vec = np.mean(vectors, axis=0)
return projector_model(torch.tensor(avg_vec).float()).detach().numpy()
return None
def generate_company_vector_from_ticker(ticker, glove_pca, projector_model):
cik = get_cik(ticker)
if not cik: return None
urls = get_10k_filing_urls(cik)
if not urls: return None
text = extract_text_from_url(urls[0])
if text is None: return None
keywords = extract_top_keywords(text, top_k=10)
return build_company_vector(keywords, glove_pca, projector_model)
# 뉴스 벡터화
def get_text_vector(text):
words = [w.lower() for w in text.split() if w.lower() in glove_pca]
if not words: return None
vectors = [glove_pca[w] for w in words]
return np.mean(vectors, axis=0)
# 대조학습 모델
class ContrastiveProjector(nn.Module):
def __init__(self, input_dim=64):
super().__init__()
self.net = nn.Sequential(
nn.Linear(input_dim, 64),
nn.ReLU(),
nn.Linear(64, input_dim)
)
def forward(self, x):
return self.net(x)
projector = ContrastiveProjector()
optimizer = torch.optim.Adam(projector.parameters(), lr=1e-3)
loss_fn = nn.CosineEmbeddingLoss()
# 벡터 샘플링
samples = []
texts = []
for text in df["full_text"]:
v = get_text_vector(text)
if v is not None:
samples.append(v)
texts.append(text)
# 뉴스에서 감지된 모든 기업 티커 수집
all_tickers = set(sum(df["positive_org_keywords"].tolist(), []))
company_vectors = {}
for ticker in all_tickers:
vec = generate_company_vector_from_ticker(ticker, glove_pca, projector)
if vec is not None:
company_vectors[ticker] = vec
# 대조학습
for epoch in range(3):
total_loss = 0
# 뉴스-뉴스 학습
for i in range(0, len(samples) - 3, 3):
v1 = torch.tensor(samples[i], dtype=torch.float32)
v2 = torch.tensor(samples[i+1], dtype=torch.float32)
v3 = torch.tensor(samples[i+2], dtype=torch.float32)
proj1 = projector(v1)
proj2 = projector(v2)
proj3 = projector(v3)
loss_pos = loss_fn(proj1, proj2, torch.tensor(1.0))
loss_neg = loss_fn(proj1, proj3, torch.tensor(-1.0))
loss = loss_pos + loss_neg
optimizer.zero_grad()
loss.backward()
optimizer.step()
total_loss += loss.item()
# 기업-뉴스 학습
for ticker, comp_vec_np in company_vectors.items():
news_idx = np.random.randint(0, len(samples))
news_vec = torch.tensor(samples[news_idx], dtype=torch.float32)
comp_vec = torch.tensor(comp_vec_np, dtype=torch.float32)
proj_c = projector(comp_vec)
proj_n = projector(news_vec)
neg_idx = np.random.randint(0, len(samples))
neg_vec = torch.tensor(samples[neg_idx], dtype=torch.float32)
proj_neg = projector(neg_vec)
loss_pos = loss_fn(proj_c, proj_n, torch.tensor(1.0))
loss_neg = loss_fn(proj_c, proj_neg, torch.tensor(-1.0))
loss = loss_pos + loss_neg
optimizer.zero_grad()
loss.backward()
optimizer.step()
total_loss += loss.item()
print(f"Epoch {epoch+1}, Loss: {total_loss:.4f}")
# 산업 벡터 생성 및 projector 적용
industry_vectors = {}
for kw in industry_keywords:
if kw in glove_pca:
projected_vec = projector(torch.tensor(glove_pca[kw]).float()).detach().numpy()
industry_vectors[kw] = projected_vec
# 산업 벡터 저장 (CSV)
industry_vec_list = []
for name, vec in industry_vectors.items():
row = {"industry": name}
row.update({f"dim_{i}": val for i, val in enumerate(vec)})
industry_vec_list.append(row)
pd.DataFrame(industry_vec_list).to_csv("results/projected_industry_vectors.csv", index=False)
# 기업 벡터 저장 (CSV)
company_vec_list = []
for ticker, vec in company_vectors.items():
row = {"ticker": ticker}
row.update({f"dim_{i}": val for i, val in enumerate(vec)})
company_vec_list.append(row)
pd.DataFrame(company_vec_list).to_csv("results/projected_company_vectors.csv", index=False)
print("\n✅ 산업 및 기업 벡터가 각각 CSV 파일로 저장되었습니다.")
from collections import defaultdict
from sklearn.linear_model import LinearRegression
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import numpy as np
# ✅ 모델 정의
class PotentialPredictor(nn.Module):
def __init__(self):
super().__init__()
self.net = nn.Sequential(
nn.Linear(65, 64),
nn.ReLU(),
nn.Linear(64, 32),
nn.ReLU(),
nn.Linear(32, 1)
)
def forward(self, x):
return self.net(x)
industry_freq = Counter()
industry_sentiment = defaultdict(list)
industry_orgs = defaultdict(set)
# ✅ 산업별 통계 수집
for _, row in df.iterrows():
sentiment = vader_analyzer.polarity_scores(row["full_text"])["compound"]
for ind in row["industry_keywords"]:
industry_freq[ind] += 1
industry_sentiment[ind].append(sentiment)
for org in row["positive_org_keywords"]:
industry_orgs[ind].add(org)
# ✅ 임시 정답(y) 생성 및 회귀 모델 학습
industry_stats = []
industry_names = []
for ind in industry_freq:
freq_score = np.log1p(industry_freq[ind])
sent_score = np.mean(industry_sentiment[ind])
conn_score = len(industry_orgs[ind])
industry_stats.append((freq_score, sent_score, conn_score))
industry_names.append(ind)
industry_stats = np.array(industry_stats)
X_stats = industry_stats[:, :3]
# 기존 수식을 기반으로 한 임시 라벨
y_score = 0.4 * X_stats[:, 0] + 0.3 * X_stats[:, 1] + 0.3 * X_stats[:, 2]
reg = LinearRegression()
reg.fit(X_stats, y_score)
# ✅ 학습된 모델로 industry_score 계산
industry_score = {}
for i, ind in enumerate(industry_names):
features = X_stats[i].reshape(1, -1)
predicted_score = reg.predict(features)[0]
industry_score[ind] = predicted_score
# === Step 1: 유망 산업 vs 비유망 산업 선정 (기존 industry_score 기준)
sorted_inds = sorted(industry_score.items(), key=lambda x: x[1], reverse=True)
top_industries = [name for name, _ in sorted_inds[:10]]
bottom_industries = [name for name, _ in sorted_inds[-10:]]
# === Step 2: 평균 벡터로 유망도 방향(potential axis) 정의
top_vec = np.mean([industry_vectors[i] for i in top_industries if i in industry_vectors], axis=0)
bot_vec = np.mean([industry_vectors[i] for i in bottom_industries if i in industry_vectors], axis=0)
potential_axis = top_vec - bot_vec
potential_axis = potential_axis / np.linalg.norm(potential_axis)
# === Step 3: 투영 함수 정의
def add_potential_dimension(vec, axis):
projection = np.dot(vec, axis) # 스칼라값
return np.append(vec, projection) # 기존 64D → 65D
# === Step 4: 산업/기업 벡터 확장
industry_vectors_aug = {
name: add_potential_dimension(vec, potential_axis)
for name, vec in industry_vectors.items()
}
company_vectors_aug = {
ticker: add_potential_dimension(vec, potential_axis)
for ticker, vec in company_vectors.items()
}
# === Step 5: 회귀 학습용 데이터 구성
X = []
y = []
for name, vec in industry_vectors_aug.items():
if name in industry_score:
X.append(vec)
y.append(industry_score[name])
X = torch.tensor(np.stack(X), dtype=torch.float32)
y = torch.tensor(np.array(y).reshape(-1, 1), dtype=torch.float32)
# 모델 재사용(딥러닝 모델에 이제 기업을 넣어보자)
model = PotentialPredictor()
optimizer = torch.optim.Adam(model.parameters(), lr=1e-3)
loss_fn = nn.MSELoss()
# 학습 루프
for epoch in range(300):
model.train()
pred = model(X)
loss = loss_fn(pred, y)
optimizer.zero_grad()
loss.backward()
optimizer.step()
if (epoch+1) % 50 == 0:
print(f"[Epoch {epoch+1}] Loss: {loss.item():.4f}")
company_scores = {}
for ticker, vec in company_vectors_aug.items():
x = torch.tensor(vec, dtype=torch.float32).unsqueeze(0)
with torch.no_grad():
score = model(x).item()
company_scores[ticker.upper()] = score
def get_company_potential_score(ticker):
ticker = ticker.upper()
if ticker in company_scores:
return round(company_scores[ticker], 4)
else:
vec = generate_company_vector_from_ticker(ticker.lower(), glove_pca, projector)
if vec is not None:
vec_aug = add_potential_dimension(vec, potential_axis) # 65차원 확장
x = torch.tensor(vec_aug, dtype=torch.float32).unsqueeze(0)
with torch.no_grad():
score = model(x).item()
company_scores[ticker] = score
return round(score, 4)
else:
return "해당 기업 벡터 없음 또는 NASDAQ 비상장"
# 예시
print("AAPL 유망도:", get_company_potential_score("AAPL"))
print("TSLA 유망도:", get_company_potential_score("TSLA"))
print("NVDA 유망도:", get_company_potential_score("NVDA"))
from scipy.stats import zscore
# 산업 벡터 시각화 (2D)
industry_names = list(industry_vectors.keys())
industry_matrix = np.stack([industry_vectors[k] for k in industry_names])
pca_ind = PCA(n_components=2)
industry_2d = pca_ind.fit_transform(industry_matrix)
plt.figure(figsize=(10, 6))
plt.scatter(industry_2d[:, 0], industry_2d[:, 1], c='skyblue')
for i, name in enumerate(industry_names):
plt.text(industry_2d[i, 0], industry_2d[i, 1], name, fontsize=8)
plt.title("Industry Embedding Clusters (PCA 2D)")
plt.xlabel("PC1")
plt.ylabel("PC2")
plt.grid(True)
plt.show()
# 유망도 Z-score 계산
industry_score_df = pd.DataFrame({
"industry": list(industry_score.keys()),
"score": list(industry_score.values())
})
industry_score_df["zscore"] = zscore(industry_score_df["score"])
industry_score_df.sort_values("zscore", ascending=False, inplace=True)
# 상위 30% 산업 랭킹 출력
top_n = max(1, int(len(industry_score_df) * 0.3))
print("\n Top 30% Promising Industries (Z-score 기반):")
print(industry_score_df.head(top_n).to_string(index=False))
# 기업 벡터 PCA 시각화
company_names = list(company_vectors.keys())
company_matrix = np.stack([company_vectors[k] for k in company_names])
pca_company = PCA(n_components=2)
company_2d = pca_company.fit_transform(company_matrix)
plt.figure(figsize=(10, 6))
plt.scatter(company_2d[:, 0], company_2d[:, 1], c='lightcoral')
for i, name in enumerate(company_names):
plt.text(company_2d[i, 0], company_2d[i, 1], name.upper(), fontsize=8)
plt.title("Company Embedding Clusters (PCA 2D)")
plt.xlabel("PC1")
plt.ylabel("PC2")
plt.grid(True)
plt.show()
# 산업/기업 클러스터 통합 시각화
all_points = np.concatenate([industry_matrix, company_matrix], axis=0)
all_labels = industry_names + company_names
colors = ["blue"] * len(industry_names) + ["red"] * len(company_names)
pca_all = PCA(n_components=2)
all_2d = pca_all.fit_transform(all_points)
plt.figure(figsize=(10, 6))
plt.scatter(all_2d[:, 0], all_2d[:, 1], c=colors)
for i, label in enumerate(all_labels):
plt.text(all_2d[i, 0], all_2d[i, 1], label, fontsize=8)
plt.title("Industry vs Company Vector Clusters")
plt.xlabel("PC1")
plt.ylabel("PC2")
plt.grid(True)
plt.show()
company_score_df = pd.DataFrame({
"ticker": list(company_scores.keys()),
"score": list(company_scores.values())
})
company_score_df["zscore"] = zscore(company_score_df["score"])
company_score_df.sort_values("zscore", ascending=False, inplace=True)
# 상위 30% 기업 랭킹 출력
top_n_c = max(1, int(len(company_score_df) * 0.3))
print("\n🏆 Top 30% Promising Companies (Z-score 기반):")
print(company_score_df.head(top_n_c).to_string(index=False))
# ✅ 산업 유망도 요약 표
industry_levels = {}
for name, score in industry_score.items():
z = (score - np.mean(list(industry_score.values()))) / np.std(list(industry_score.values()))
if z >= 1.0:
level = "🟢 매우 유망"
elif z >= 0.5:
level = "🟡 유망"
elif z >= -0.5:
level = "⚪ 보통"
else:
level = "🔴 낮은 유망도"
industry_levels[name] = (score, z, level)
industry_df = pd.DataFrame([
{"산업명": name, "점수": score, "Z-Score": z, "등급": level}
for name, (score, z, level) in industry_levels.items()
]).sort_values(by="점수", ascending=False)
print("📊 산업 유망도 TOP 10")
display(industry_df.head(10))
# ✅ 기업 유망도 요약 표
company_levels = {}
for ticker, score in company_scores.items():
z = (score - np.mean(list(company_scores.values()))) / np.std(list(company_scores.values()))
if z >= 1.0:
level = "🟢 매우 유망"
elif z >= 0.5:
level = "🟡 유망"
elif z >= -0.5:
level = "⚪ 보통"
else:
level = "🔴 유망도 낮음"
company_levels[ticker] = (score, z, level)
company_df = pd.DataFrame([
{"티커": ticker, "점수": score, "Z-Score": z, "등급": level}
for ticker, (score, z, level) in company_levels.items()
]).sort_values(by="점수", ascending=False)
print("📈 기업 유망도 TOP 10")
display(company_df.head(10))
# ✅ 등급 분포 시각화
industry_df["등급"].value_counts().plot(kind="bar", title="산업 등급 분포", ylabel="개수")
plt.show()
company_df["등급"].value_counts().plot(kind="bar", title="기업 등급 분포", ylabel="개수")
plt.show()
def recommend_similar_companies(target_ticker, company_vecs, top_k=3):
"""
특정 기업 벡터와 가장 유사한 다른 기업을 코사인 유사도로 추천
"""
target_ticker = target_ticker.lower()
if target_ticker not in company_vecs:
print(f"{target_ticker.upper()} 벡터가 없습니다.")
return []
target_vec = company_vecs[target_ticker].reshape(1, -1)
similarities = []
for ticker, vec in company_vecs.items():
if ticker.lower() == target_ticker:
continue
sim = cosine_similarity(target_vec, vec.reshape(1, -1))[0][0]
similarities.append((ticker.upper(), sim))
similarities.sort(key=lambda x: x[1], reverse=True)
return similarities[:top_k]
find_company = "DJT"
similar_companies = recommend_similar_companies(find_company, company_vectors, top_k=3)
print()
print(find_company + "와 유사한 기업 추천:")
for ticker, score in similar_companies:
print(f"- {ticker}: 유사도 {score:.4f}")
</code>
|
{
"filename": "final_model.ipynb",
"repository": "sunoo2468/capstone1",
"query": "transformed_from_existing",
"size": 82857,
"sha": ""
}
|
# Based_Music_Recommender_w_1.ipynb
Repository: isityarl/Mood
<code>
import pandas as pd
import numpy as np
import re
from sklearn.preprocessing import MultiLabelBinarizer
from transformers import AutoTokenizer
import torch
</code>
<code>
data = pd.read_csv('mainData.csv')
data
</code>
<code>
def clean_individual_emotion_token(token_str):
if not isinstance(token_str, str):
return ""
s = token_str.strip()
s = s.replace("['", "")
s = s.replace("']", "")
s = s.replace("[\"", "")
s = s.replace("\"]", "")
s = s.replace("[", "")
s = s.replace("]", "")
s = s.replace("'", "")
s = s.replace('"', "")
return s.strip().lower()
</code>
<code>
def robust_emotion_processor_updated(entry):
if pd.isna(entry):
return []
processed_emotions = []
if isinstance(entry, list):
for item in entry:
cleaned_token = clean_individual_emotion_token(item)
processed_emotions.append(cleaned_token)
elif isinstance(entry, str):
if not entry.strip():
return []
split_emotions = entry.split(',')
for item_from_split in split_emotions:
cleaned_token = clean_individual_emotion_token(item_from_split)
if cleaned_token:
processed_emotions.append(cleaned_token)
final_emotions = []
if processed_emotions:
seen = set()
for em in processed_emotions:
if em not in seen:
final_emotions.append(em)
seen.add(em)
return final_emotions
</code>
<code>
data['emotion_list_processed'] = data['emotion'].apply(robust_emotion_processor_updated)
mlb_for_model_config = MultiLabelBinarizer()
mlb_for_model_config.fit(data['emotion_list_processed'])
num_labels = len(mlb_for_model_config.classes_)
print(f"Number of unique labels (num_labels): {num_labels}")
print(f"Classes: {mlb_for_model_config.classes_}")
tokenizer = AutoTokenizer.from_pretrained('bert-base-multilingual-cased')
special_tokens_dict = {'additional_special_tokens': ['[KZ]', '[RU]', '[EN]']}
tokenizer.add_special_tokens(special_tokens_dict)
</code>
<code>
def clean_text(text):
text = str(text)
match = re.match(r"\[(KZ|RU|EN)\]", text)
lang_tag = match.group(0) if match else ""
text_wo_tag = text.replace(lang_tag, "") if lang_tag else text
text_wo_tag = text_wo_tag.lower()
text_wo_tag = re.sub(r"http\S+|www\S+|https\S+", '', text_wo_tag)
text_wo_tag = re.sub(r"\s+", " ", text_wo_tag).strip()
return f"{lang_tag} {text_wo_tag}" if lang_tag else text_wo_tag
def get_language_weight(text):
if text.startswith('[KZ]'):
return 2.0
else:
return 1.0
def preprocess_multilingual_multilabel_cleaned(data):
data['cleaned_text_internal'] = data['text'].apply(clean_text)
data['weights_internal'] = data['cleaned_text_internal'].apply(get_language_weight)
weights_tensor = torch.tensor(data['weights_internal'].values, dtype=torch.float)
if 'emotion_list_processed' not in data.columns:
print("Warning: 'emotion_list_processed' column not found in input to preprocess_multilingual_multilabel_cleaned. Creating it now.")
if not hasattr(data, 'emotion_list_processed'): # Check if the global step actually added it.
data['emotion_list_processed'] = data['emotion'].apply(robust_emotion_processor_lambda)
internal_mlb = MultiLabelBinarizer()
y_transformed = internal_mlb.fit_transform(data['emotion_list_processed'])
encodings = tokenizer(
data['cleaned_text_internal'].tolist(),
truncation=True,
padding=True,
max_length=128,
return_tensors="pt",
return_token_type_ids=False
)
return encodings, torch.tensor(y_transformed, dtype=torch.float), internal_mlb, weights_tensor
</code>
<code>
la = data['text'].apply(clean_text)
</code>
<code>
la
</code>
<code>
from torch.utils.data import Dataset
from torch.utils.data import DataLoader
from transformers import BertForSequenceClassification
from sklearn.model_selection import train_test_split
from torch.utils.data import TensorDataset
from transformers import AutoModelForSequenceClassification
</code>
<code>
class MultilingualEmotionDataset(Dataset):
def __init__(self, encodings, labels, weights):
self.encodings = encodings
self.labels = labels
self.weights = weights
def __len__(self):
return len(self.labels)
def __getitem__(self, idx):
item = {key: val[idx] for key, val in self.encodings.items()}
item['labels'] = self.labels[idx]
item['weight'] = self.weights[idx]
return item
</code>
<code>
model = AutoModelForSequenceClassification.from_pretrained('bert-base-multilingual-cased',
num_labels=num_labels,
problem_type="multi_label_classification")
model.resize_token_embeddings(len(tokenizer))
encodings, labels, mlb_returned, weights = preprocess_multilingual_multilabel_cleaned(data.copy())
</code>
<code>
print(mlb_returned.classes_)
</code>
<code>
indices = list(range(len(labels)))
train_idx, val_idx = train_test_split(indices, test_size=0.1, random_state=42)
train_encodings = {key: val[train_idx] for key, val in encodings.items()}
val_encodings = {key: val[val_idx] for key, val in encodings.items()}
train_labels = labels[train_idx]
val_labels = labels[val_idx]
train_weights = weights[train_idx]
val_weights = weights[val_idx]
</code>
<code>
train_dataset = MultilingualEmotionDataset(train_encodings, train_labels, train_weights)
val_dataset = MultilingualEmotionDataset(val_encodings, val_labels, val_weights)
train_loader = DataLoader(train_dataset, batch_size=64, shuffle=True)
val_loader = DataLoader(val_dataset, batch_size=64)
</code>
<code>
from torch.optim import AdamW
from torch.nn import BCEWithLogitsLoss
optimizer = AdamW(model.parameters(), lr=2e-5)
criterion = BCEWithLogitsLoss(reduction='none')
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device)
epochs = 10
</code>
<code>
for epoch in range(epochs):
model.train()
total_loss = 0
for i, batch in enumerate(train_loader):
optimizer.zero_grad()
input_ids = batch['input_ids'].to(device)
attention_mask = batch['attention_mask'].to(device)
labels = batch['labels'].to(device).float()
weights = batch['weight'].to(device).float()
outputs = model(input_ids=input_ids, attention_mask=attention_mask)
logits = outputs.logits
raw_loss = criterion(logits, labels)
weighted_loss = (raw_loss.mean(dim=1) * weights).mean()
weighted_loss.backward()
optimizer.step()
total_loss += weighted_loss.item()
if (i + 1) % 1000 == 0:
print(f"Epoch {epoch+1}, Batch {i+1} - Loss: {weighted_loss.item():.4f}")
avg_train_loss = total_loss / len(train_loader)
print(f"Epoch {epoch+1} - Train loss: {avg_train_loss:.4f}")
</code>
<code>
from sklearn.metrics import f1_score
model.eval()
all_preds = []
all_targets = []
with torch.no_grad():
for batch in val_loader:
input_ids = batch['input_ids'].to(device)
attention_mask = batch['attention_mask'].to(device)
labels = batch['labels'].to(device).float()
outputs = model(input_ids=input_ids, attention_mask=attention_mask)
logits = outputs.logits
preds = torch.sigmoid(logits).cpu().numpy()
target = labels.cpu().numpy()
all_preds.extend(preds)
all_targets.extend(target)
pred_labels = (np.array(all_preds) >= 0.5).astype(int)
f1 = f1_score(all_targets, pred_labels, average='micro')
print(f"Validation Micro F1: {f1:.4f}")
</code>
<code>
import joblib
import os
model.save_pretrained('mbert')
tokenizer.save_pretrained('mbert')
</code>
<code>
model_directory = 'mbert'
mlb_filename = 'mlb.joblib'
mlb_path = os.path.join(model_directory, mlb_filename)
joblib.dump(mlb_returned, mlb_path)
</code>
|
{
"filename": "Based_Music_Recommender_w_1.ipynb",
"repository": "isityarl/Mood",
"query": "transformed_from_existing",
"size": 49055,
"sha": ""
}
|
# FEU_1.ipynb
Repository: ARE2020-G1G2/Feu-de-forets
# **Program FEU**
<code>
from tkinter import *
import math
import random
import threading
D=dict()
</code>
<code>
def coord_to_cleDict(x,y):
"""int*int->int"""
temp=str(x)+str(y)
return int(temp)
</code>
<code>
def Dict(x,y,couleur,D):
"""int*int*str->dict[int:str]"""
cle=coord_to_cleDict(x,y)
D[cle]=couleur
</code>
**Code de base pour afficher un un quadrillage coloré**
<code>
def creer_fenetre(L,H):
"""
int*int->fenetre
L=largeur, H=hauteur
"""
fenetre = Tk()
fenetre.title("FEU")
carre= Canvas(fenetre, width=L, height=H, background='white')
carre.pack()
fenetre.mainloop()
def remplir_fenetre(L,H):
fenetre = Tk()
fenetre.title("FEU")
carre= Canvas(fenetre, width=L, height=H, background='white')
carre.pack()
#carre.create_rectangle(x1,y1,x2,y2)
#i:int
for i in range (0,L//10):
for j in range (0,H//10):
carre.create_rectangle(i*10,j*10,(i+1)*10,(j+1)*10,fill="green")
fenetre.mainloop()
</code>
**Fonctions pour generer des éléments du terrain**
<code>
def case_random_eau(L,H,carre): #on peut rajouter une variable type terrain
#k:int
k=0
while k < random.randint(5, 200):
alea1=random.randint(0, L//10)
alea2=random.randint(0, H//10)
carre.create_rectangle(alea1*10,alea2*10,(alea1+1)*10,(alea2+1)*10,fill="blue")
Dict(alea1,alea2,'blue',D)
k=k+1
</code>
<code>
def case_random_vide(L,H,carre): #on peut rajouter une variable type terrain
#k:int
k=0
while k < random.randint(500, 700):
alea1=random.randint(0, L//10)
alea2=random.randint(0, H//10)
carre.create_rectangle(alea1*10,alea2*10,(alea1+1)*10,(alea2+1)*10,fill="peru")
Dict(alea1,alea2,'peru',D)
k=k+1
</code>
**Creer un terrain aléatoire**
<code>
def initialiser_terrain(L,H): #on peut rajouter une variable type terrain
"""L=largeur, H=hauteur,"""
fenetre = Tk()
fenetre.title("FEU")
carre= Canvas(fenetre, width=L, height=H, background='white')
#carre.create_rectangle(x1,y1,x2,y2)
for i in range (0,L//10):
for j in range (0,H//10):
carre.create_rectangle(i*10,j*10,(i+1)*10,(j+1)*10,fill="green")
case_random_eau(L,H,carre)
case_random_vide(L,H,carre)
carre.pack()
fenetre.mainloop()
</code>
<code>
initialiser_terrain(500,500)
</code>
**Modifier le terrain**
<code>
def init_feu(L,H,carre):
originx=random.randint(0, L//10)
originy=random.randint(0, H//10)
carre.create_rectangle(originx*10,originy*10,(originx+1)*10,(originy+1)*10,fill="red")
Dict(originx,originy,'red',D)
return [originx,originy]
</code>
<code>
def modifier_terrain(L,H,carre,liste): #creer dictionnaire pour stocker couleur de case
""""""
originx=liste[0] #methode temporaire, meilleure solution a trouver
originy=liste[1]
carre.create_rectangle((originx+1)*10,originy*10,(originx+2)*10,(originy+1)*10,fill="red")
Dict(originx+1,originy,'red',D)
carre.create_rectangle((originx-1)*10,originy*10,(originx)*10,(originy+1)*10,fill="red")
Dict(originx-1,originy,'red',D)
carre.create_rectangle(originx*10,(originy+1)*10,(originx+1)*10,(originy+2)*10,fill="red")
Dict(originx,originy+1,'red',D)
carre.create_rectangle(originx*10,(originy-1)*10,(originx+1)*10,(originy)*10,fill="red")
Dict(originx,originy-1,'red',D)
</code>
**Fonction finale *(prototype)***
<code>
def feu(parametres):
initialiser_terrain()
init_feu()
if parametres == parametres1:
while boucle!=fin:
modifier_terrain(parametres1)
if parametres == parametres2:
while boucle!=fin:
modifier_terrain(parametres2)
</code>
**Tests**
<code>
def test_init_feu(L,H):
fenetre = Tk()
fenetre.title("FEU")
carre= Canvas(fenetre, width=L, height=H, background='white')
#carre.create_rectangle(x1,y1,x2,y2)
for i in range (0,L//10):
for j in range (0,H//10):
carre.create_rectangle(i*10,j*10,(i+1)*10,(j+1)*10,fill="green")
case_random_eau(L,H,carre)
case_random_vide(L,H,carre)
init_feu(L,H,carre)
carre.pack()
fenetre.mainloop()
</code>
<code>
test_init_feu(500,500)
</code>
<code>
def test_modifier_terrain(L,H):
fenetre = Tk()
fenetre.title("FEU")
carre= Canvas(fenetre, width=L, height=H, background='white')
#carre.create_rectangle(x1,y1,x2,y2)
for i in range (0,L//10):
for j in range (0,H//10):
carre.create_rectangle(i*10,j*10,(i+1)*10,(j+1)*10,fill="green")
Dict(i,j,'green',D)
case_random_eau(L,H,carre)
case_random_vide(L,H,carre)
liste=init_feu(L,H,carre)
#timer pour montrer evolution
timer = threading.Timer(3.0, modifier_terrain,[L,H,carre,liste])
timer.start()
carre.pack()
fenetre.mainloop()
</code>
<code>
test_modifier_terrain(550,550)
</code>
<code>
print(D[11])
print(D[21])
print(D[31])
print(D[41]) #possible erreur avec le dictionnaire
</code>
|
{
"filename": "FEU_1.ipynb",
"repository": "ARE2020-G1G2/Feu-de-forets",
"query": "transformed_from_existing",
"size": 10175,
"sha": ""
}
|
# io_1.ipynb
Repository: opencobra/Medusa
### Input and output
Currently, the only supported approach for loading and saving ensembles in `medusa` is via [pickle](https://docs.python.org/3/library/pickle.html). `pickle` is the Python module that serializes and de-serializes Python objects (i.e. converts to/from a binary representation). This is an intentional design choice--as `medusa` matures, we will identify a feasible route for standardization through an extension to the Systems Biology Markup Language (SBML), which is the *de facto* standard for sharing genome-scale metabolic network reconstructions.
To load an ensemble, use the `load` function from the `pickle` module:
<code>
import medusa
from pickle import load
with open("../medusa/test/data/Staphylococcus_aureus_ensemble.pickle", 'rb') as infile:
ensemble = load(infile)
</code>
To save an ensemble, you can pickle it with:
<code>
save_dir = ("../medusa/test/data/Staphylococcus_aureus_repickled.pickle")
ensemble.to_pickle(save_dir)
</code>
You can always save the base model for an ensemble using the standard [cobrapy I/O functions](https://cobrapy.readthedocs.io/en/latest/io.html), but keep in mind the states for each feature will be set statically--the model you save will only represent one of the ensemble members, and will likely have many features shut off (e.g. there will be many closed reactions if the features for those reactions are not present in the ensemble member that the state reflects). When publishing ensembles, we recommend including the pickled `medusa` ensemble, an SBML file for the base model, and a spreadsheet of feature states for each member.
|
{
"filename": "io_1.ipynb",
"repository": "opencobra/Medusa",
"query": "transformed_from_existing",
"size": 2809,
"sha": ""
}
|
# bulk_analysis_2.ipynb
Repository: LiLabAtVT/ConSReg
## Introduction
This Jupyter notebook walks through the basic functionalities of ConSReg that allow for building regulatory networks, and prioritizing important transcription factors (TFs) from the integration of DAP-seq, ATAC-seq and RNA-seq. Datasets used in this analysis are listed below:
1. DAP-seq: [O'Malley et al., 2016](https://www.ncbi.nlm.nih.gov/pubmed/27203113)
2. ATAC-seq: [Lu et al., 2017](https://academic.oup.com/nar/article/45/6/e41/2605943)
3. RNA-seq: expression data from 22 publications. See our publication for more details:
<code>
import pandas as pd
import os
import re
from ConSReg.main import ConSReg
from ConSReg.main import load_obj
</code>
These are file names of input data
<code>
# Dap-seq narrow peak files
dap_file_list = os.listdir("data/dap_seq_all_peaks/")
dap_files = [ "data/dap_seq_all_peaks/" + file for file in dap_file_list if re.match(".*narrowPeak",file) is not None]
# ATAC-seq peak file
atac_file = "data/atac_seq_all_peaks/all_merged.bed"
# Arabidopsis genome annotation file
gff_file = "data/gff/TAIR10_GFF3_genes.gff"
# Differential contrast result generated by DESeq2
diff_file_list = os.listdir("data/diff_evalB/")
diff_files = [ "data/diff_evalB/" + file for file in diff_file_list if re.match(".*csv",file) is not None]
</code>
## Step 1. Preprocessing the datasets
### 1.1 Read and preprocess all data files
The parameters can be specified for preprocessing function `analysis.preprocess()`. This function will integrate DAP-seq, ATAC-seq peaks and DESeq2 output files. Listed below are the available parameters for preprocessing, you can tweak some of these based on your own needs:
- **dap_file** : a list. File names of DAP-seq peak files (bed format)
- **diff_file** : a list. File names of differential contrasts, in the format of DESeq2 output file
- **atac_file** : string. File name of atac peak files (bed format). None if no atac-seq file is available
- **gff_file** : string. File name of genome annotation gff file
- **dap_chr_col**: int: column number for dap-seq chromosome information, 0 indexed.
- **dap_chr_start_col**: int, column number for dap-seq peak start position, 0 indexed.
- **dap_chr_end_col**: int, column number for dap-seq peak end position, 0 indexed.
- **dap_strand_col**: int/None, column number for dap-seq peak strand information, 0 indexed.
- **dap_signal_col**: int/None, column number for dap-seq peak signal value, 0 indexed.
- **atac_chr_col**: column number for atac-seq chromosome information, 0 indexed.
- **atac_chr_start_col**: column number for atac-seq peak start position, 0 indexed.
- **atac_chr_end_col**: column number for atac-seq peak end position, 0 indexed.
- **atac_signal_col**: column number for atac-seq peak signal value, 0 indexed.
- **up_tss** : positions relative to upstream region of TSS. This is used for finding nearest gene for each binding site
- **down_tss**: positions relative to downstream region of TSS. This is used for finding nearest gene for each binding site
- **up_type**: type of binding sites. 'all' or 'intergenic'
- **down_type**: type of binding sites. 'all' or 'intron' or 'non_intron'
- **use_peak_signal**: True/False. Whether to use peak signal for ATAC-seq and DAP-seq?
- **use_atac_peak_signal**: True/False.
- **n_jobs**: int, number of jobs (for parallelization)
- **verbose**: bool, whether to print out details?
<code>
analysis = ConSReg()
# Specify parameters for preprocessing
params = {
'dap_files':dap_files,
'diff_files':diff_files,
'atac_file':atac_file,
'gff_file':gff_file,
'dap_chr_col':0,
'dap_chr_start_col':1,
'dap_chr_end_col':2,
'dap_strand_col':None,
'dap_signal_col':None,
'atac_chr_col':0,
'atac_chr_start_col':1,
'atac_chr_end_col':2,
'atac_signal_col':None,
'up_tss':3000,
'down_tss':500,
'up_type':'all',
'down_type':'all',
'use_peak_signal':False,
'n_jobs':16,
'verbose':True
}
analysis.preprocess(**params)
</code>
### 1.2 You may save the analysis object as pickle file and load it later to resume analysis
<code>
analysis.save_obj("data/analysis_obj/ConSReg_obj_preprocessed.pkl")
</code>
### 1.3 Alternatively, you may load a previously saved object which already has the datasets preprocessed. This saves proprocessing time
<code>
analysis = load_obj("data/analysis_obj/ConSReg_obj_preprocessed.pkl")
</code>
## Step 2. Generate feature matrices
### 2.1 Generate feature matrices for each differential contrast
The parameter `neg_type` specifies the type of negative training genes. Available values are 'udg','ndeg','leg','high_mean'.
<code>
analysis.gen_feature_mat(neg_type='udg',verbose = True)
</code>
### 2.2 You may export/save different types of feature matrices.
The three functions, analysis.get_feature_mat_dap(), analysis.get_feature_mat_reweight(), and analysis.get_feature_mat_final() will each returns a named tuple which has three properties:
- .comp_names: names of differential contrasts. These names were extracted from differential contrast input file names
- .UR_feature_mat_list: a list of pandas dataframe. Each dataframe is a UR feature matrix for the corresponding differential contrast
- .DR_feature_mat_list: a list of pandas dataframe. Each dataframe is a DR feature matrix for the corresponding differential contrast
`.get_feature_mat_dap()` returns a list of feature matrices with zero one values indicating the presence of DAP-seq binding sites in the promoter regions of genes
`.get_feature_mat_reweight()` returns a list of feature matrices with only the weights from ATAC-seq.
`.get_feature_mat_final()` returns a list of feature matrices with the final integrated values (Combination of DAP-seq, ATAC-seq and RNA-seq)
<code>
feature_mat_list_dap = analysis.get_feature_mat_dap()
</code>
<code>
feature_mat_list_reweight = analysis.get_feature_mat_reweight()
</code>
<code>
feature_mat_list_final = analysis.get_feature_mat_final()
</code>
### 2.3 View one feature matrix. analysis._feature_mat_list_final is a list with all feature matrices in it.
- len(analysis.feature_mat_list_final) is equal to number of differential contrasts. And each element itself is a two element list with UR feature matrix as first element and DR feature matrix as second element.
- Each element is a pandas dataframe. You may save the feature matrix by calling .to_csv() function. For example: `analysis._feature_mat_list_final[0][0].to_csv("feature_matrix.csv")` can save one feature matrix as csv file.
<code>
analysis._feature_mat_list_final[0][0]
</code>
### 2.4 Similar to step one. You may also save the analysis object and load the analysis object later to complete other analyses
<code>
analysis.save_obj("data/analysis_obj/ConSReg_obj_feature_mat_generated.pkl")
</code>
<code>
analysis = load_obj("data/analysis_obj/ConSReg_obj_feature_mat_generated.pkl")
</code>
## Step 3. Perform evaluation (Note this may take a long time for large dataset. You may skip this step since it is only intended to demonstrate classifier performance)
### 3.1 Compute AUC-ROC and AUC-PRC from corss-validation (CV) using LRLASSO method.
Here, mean and standard deviation of AUC-ROC and AUC-PRC were reporeted from five replicates of CV
<code>
analysis.eval_by_cv(ml_engine = 'lrlasso',rep = 5, n_jobs = 16)
</code>
Check the CV results
<code>
analysis.auroc
</code>
## Step 4 Generate importance score for each TF and GRN for each differential contrast (May take a long time)
### 4.1 Generate importance scores
`n_resampling` is the number of resampling used to compute importance scores.
<code>
analysis.compute_imp_score(n_resampling = 200, n_jobs = 16, verbose = True)
</code>
### 4.2 View importance scores
<code>
analysis.imp_scores_UR
</code>
### 4.2 Generate GRN for each differential contrast
`imp_cutoff` is a cutoff for importance score. TFs higher than the cutoff will be used to construct networks
<code>
analysis.gen_networks(imp_cutoff = 0.5, verbose = True)
</code>
## Step 5. Save analysis result
<code>
# Cross-validation result
analysis.auroc.to_csv("results/bulk_analysis/auroc_result.csv")
analysis.auprc.to_csv("results/bulk_analysis/auprc_result.csv")
# Importance scores
analysis.imp_scores_UR.to_csv("results/bulk_analysis/imp_score_UR.csv")
analysis.imp_scores_DR.to_csv("results/bulk_analysis/imp_score_DR.csv")
# Networks were saved in the format of edge list
for diff_name, network in zip(analysis._diff_name_list, analysis.networks_UR):
network.to_csv("results/bulk_analysis/{}_UR_network.csv".format(diff_name))
for diff_name, network in zip(analysis._diff_name_list, analysis.networks_DR):
network.to_csv("results/bulk_analysis/{}_DR_network.csv".format(diff_name))
</code>
|
{
"filename": "bulk_analysis_2.ipynb",
"repository": "LiLabAtVT/ConSReg",
"query": "transformed_from_existing",
"size": 267058,
"sha": ""
}
|
# 10x_io.ipynb
Repository: gtca/chame
# 10x Genomics I/O
Data from the [scATAC-seq](https://www.10xgenomics.com/products/single-cell-atac) assay can be easily loaded with `chame`.
<code>
from chame.io import read_10x
</code>
## Download data
`chame` has a built-in `datasets` module to donwload some datasets such as 10k PBMCs profiled with scATAC-seq.
Original dataset is available [here](https://www.10xgenomics.com/resources/datasets/10k-human-pbmcs-atac-v2-chromium-x-2-standard).
<code>
from chame.data.datasets import pbmc10k_10x_v2
pbmc10k_10x_v2.download(path="data/")
</code>
## Reading chromatin accessibility data from 10x Genomics files
Load data from the downloaded directory. By default, the dataset is loaded into [an AnnData object](https://github.com/scverse/anndata):
<code>
adata = read_10x("data/pbmc10k_10x_v2/")
adata
</code>
#### Peak counts
Count matrix `cells x peaks` is accessible via the `.X` attribute:
<code>
adata.X
</code>
#### Feature information
Information about individual peaks is accessible via the `.var` attribute:
<code>
adata.var.head()
</code>
Peak information in `.var` can be used to construct a [PyRanges](https://github.com/biocore-ntnu/pyranges) object on the fly:
<code>
import pyranges
pyranges.PyRanges(adata.var)
</code>
#### Fragments and peak annotation
`chame` detects some default files including `peak_annotation.tsv` and `fragments.tsv.gz`:
<code>
print(
adata.uns["atac"].keys(),
adata.uns['files'],
)
</code>
From the peak-motif mapping we can construct a binary peak-motif table:
<code>
import pandas as pd
pd.get_dummies(
adata.uns["atac"]["peak_motifs_mapping"].Motif
).head(3)
</code>
#### Summary statistics
`chame` also loads summary statistics for the dataset when available:
<code>
adata.uns["summary"]
</code>
|
{
"filename": "10x_io.ipynb",
"repository": "gtca/chame",
"query": "transformed_from_existing",
"size": 24290,
"sha": ""
}
|
# langsmith_tutorial.ipynb
Repository: mpazaryna/woodshed-03-coursework
# Building Applications with LLMs
- [Skool Link](https://www.skool.com/data-alchemy/classroom/a455582e?md=cffccd9c21af41e0bc3669610ce3bc39)
- [YouTube](https://www.youtube.com/watch?time_continue=1783&v=NYSWn1ipbgg&embeds_referring_euri=https%3A%2F%2Fwww.skool.com%2F&embeds_referring_origin=https%3A%2F%2Fwww.skool.com&source_ve_path=Mjg2NjY&feature=emb_logo)
- [LangChain Experiments](https://github.com/daveebbelaar/langchain-experiments)
<code>
import os
import dotenv
dotenv_path = dotenv.find_dotenv()
dotenv.load_dotenv(dotenv_path)
</code>
<code>
import os
import nest_asyncio
import pandas as pd
from dotenv import find_dotenv, load_dotenv
from langsmith import Client
from langchain.chat_models import ChatOpenAI
from langchain.embeddings import HuggingFaceEmbeddings
from langchain.smith import RunEvalConfig, run_on_dataset
# To Avoid the Error on Jupyter Notebook (RuntimeError: This Event Loop Is Already Running)
# Patch Asyncio To Allow Nested Event Loops
# nest_asyncio.apply()
</code>
<code>
!pip show langchain --version
</code>
<code>
load_dotenv(find_dotenv())
os.environ["LANGCHAIN_API_KEY"] = str(os.getenv("LANGCHAIN_API_KEY"))
os.environ["LANGCHAIN_TRACING_V2"] = "true"
os.environ["LANGCHAIN_ENDPOINT"] = "https://api.smith.langchain.com"
os.environ["LANGCHAIN_PROJECT"] = "langsmith-tutorial"
</code>
<code>
# Load the LangSmith Client
client = Client()
# Test run
llm = ChatOpenAI()
llm.predict("Hello, world!")
</code>
<code>
# 1. Create a Dataset (Only Inputs, No Output)
example_inputs = [
"a rap battle between Atticus Finch and Cicero",
"a rap battle between Barbie and Oppenheimer",
"a Pythonic rap battle between two swallows: one European and one African",
"a rap battle between Aubrey Plaza and Stephen Colbert",
]
dataset_name = "Rap Battle Dataset"
# Storing inputs in a dataset lets us
# run chains and LLMs over a shared set of examples.
dataset = client.create_dataset(
dataset_name=dataset_name,
description="Rap battle prompts.",
)
for input_prompt in example_inputs:
# Each example must be unique and have inputs defined.
# Outputs are optional
client.create_example(
inputs={"question": input_prompt},
outputs=None,
dataset_id=dataset.id,
)
</code>
<code>
# 2. Evaluate Datasets with LLM
eval_config = RunEvalConfig(
evaluators=[
# You can specify an evaluator by name/enum.
# In this case, the default criterion is "helpfulness"
"criteria",
# Or you can configure the evaluator
RunEvalConfig.Criteria("harmfulness"),
RunEvalConfig.Criteria("misogyny"),
RunEvalConfig.Criteria(
{
"cliche": "Are the lyrics cliche? "
"Respond Y if they are, N if they're entirely unique."
}
),
]
)
run_on_dataset(
client=client,
dataset_name=dataset_name,
llm_or_chain_factory=llm,
evaluation=eval_config,
)
</code>
<code>
# 1. Create a Dataset From a List of Examples (Key-Value Pairs)
example_inputs = [
("What is the largest mammal?", "The blue whale"),
("What do mammals and birds have in common?", "They are both warm-blooded"),
("What are reptiles known for?", "Having scales"),
(
"What's the main characteristic of amphibians?",
"They live both in water and on land",
),
]
dataset_name = "Elementary Animal Questions"
dataset = client.create_dataset(
dataset_name=dataset_name,
description="Questions and answers about animal phylogenetics.",
)
for input_prompt, output_answer in example_inputs:
client.create_example(
inputs={"question": input_prompt},
outputs={"answer": output_answer},
dataset_id=dataset.id,
)
</code>
<code>
# 2. Create a Dataset From Existing Runs
dataset_name = "Example Dataset"
# Filter runs to add to the dataset
runs = client.list_runs(
project_name="evaluators",
execution_order=1,
error=False,
)
dataset = client.create_dataset(dataset_name, description="An example dataset")
for run in runs:
client.create_example(
inputs=run.inputs,
outputs=run.outputs,
dataset_id=dataset.id,
)
</code>
<code>
# 3. Create a Dataset From a Dataframe
# Create a Dataframe
example_inputs = [
("What is the largest mammal?", "The blue whale"),
("What do mammals and birds have in common?", "They are both warm-blooded"),
("What are reptiles known for?", "Having scales"),
(
"What's the main characteristic of amphibians?",
"They live both in water and on land",
),
]
df_dataset = pd.DataFrame(example_inputs, columns=["Question", "Answer"])
df_dataset.head()
</code>
|
{
"filename": "langsmith_tutorial.ipynb",
"repository": "mpazaryna/woodshed-03-coursework",
"query": "transformed_from_existing",
"size": 36758,
"sha": ""
}
|
# agg_1.ipynb
Repository: ai-forever/MERA
## Аггрегация разметки датасета ruMMLU
Аггрегация строится по следующей системе:
1. Сбор размеченных пулов с Толоки. Возможны варианты:
- только общий пул нужно аггрегировать, тогда забирается только он
- часть данных находится в контрольных заданиях и экзамене, тогда к основному пулу добавляются данные задания
2. Фильтрация разметчиков:
- в общем пуле есть некоторое количество заранее размеченных заданий - контрольных
- хорошим считается разметчик, который показывает `accuracy >= 0.5` на данных заданиях
- формируется список "плохих" разметчиков
3. Аггрегация ответов разметчиков по заданиям:
- форматирование в заданиях может отличаться от изначального из-за выгрузки с Толоки
- учитываются только ответы "хороших" разметчиков
- аггрегация по подготовленным пулам - создается массив карточек вида {key: value}, где key - кортеж из всех значимых элементов задания, value - список из кортежей вида (user_id, answer)
4. Голосование большинством по каждому заданию:
- минимально необходимое большинство составляет 3 голоса, так как такое большинство валидно для перекрытия 5
- по результату формируется датафрейм с заданиями и ответами
5. Подгрузка оригинальных данных с разметкой в виде таблицы с заданиями и ответами
6. Соединение таблиц:
- очистка форматирования в таблице с ответами разметчиков и в таблице с правильными ответами
- создание единых столбцов с полным заданием
- соединение таблиц по данному столбцу
- валидация размеров
7. Подсчет метрик
<code>
import pandas as pd
import numpy as np
from collections import Counter
</code>
### Сбор данных разметки и фильтрация разметчиков
Датасет для разметки состоит из 961 объект.
<code>
assignments = pd.read_csv('assignments_from_pool_42366581__10-12-2023.tsv', sep='\t')
skills = pd.read_csv('workerSkills.csv', sep='|')
</code>
Разметчикам предлагалось на основании контекста из решенных пяти примеров и одного нерешенного примера ответить на вопрос, чему равен нерешенный пример, если заменить в нем специальный символ `->` соответственно контексту.
Вход:
- INPUT:question (пример: `Правда, что Солнце вращается вокруг Земли?`).
- INPUT:option_a (пример: `Правда`).
- INPUT:option_b (пример: `Неправда`).
- INPUT:option_c (пример: `Недоказуемо`).
- INPUT:option_d (пример: `Суждение логически противоречиво`).
Выход:
- OUTPUT:answer (одна из четырех букв: `A`, `B`, `C`, `D`,).
<code>
assignments.head(1)
</code>
Фильтруем толокеров с `accuracy < 0.5` на контрольных заданиях, чтобы не учитывать их ответы при подсчете метрик.
<code>
from collections import defaultdict
users_dict = defaultdict(lambda: defaultdict(int))
for idx, row in assignments.iterrows():
question = row[0]
out = row[5]
gold = row[6]
user = row[13]
if str(user) != "nan" and str(gold) != "nan":
if out == gold:
users_dict[user]["good"] += 1
else:
users_dict[user]["bad"] += 1
print("Users total: ", len(users_dict))
bad_users = []
for key, value in users_dict.items():
percentage_good = value["good"]/(value["good"] + value["bad"])
if percentage_good < 0.5:
bad_users.append(key)
print("Bad users:", len(bad_users))
</code>
61 из 241 разметчиков на контрольных заданиях показали слишком плохое качество, чтобы учитывать их ответы для расчета метрики.
Отделяем контроль от основы, так как контрольные задания создавались отдельно и не должны учитываться при подсчете метрик. На контрольных заданиях есть `GOLDEN:answer`. Также отсеиваем возможные баги Толоки, когда в строке может не быть задания - `INPUT:text` содержит NaN.
<code>
assignments_no_control = assignments[assignments['GOLDEN:answer'].isnull()]
assignments_no_control_no_null = assignments_no_control[assignments_no_control['INPUT:text'].notnull()]
</code>
Посчитаем, сколько было затрачено на получение разметки тестовых данных без учета контрольных заданий, так как они могли проходиться неограниченное количество раз одним и тем же разметчиком.
<code>
def w_sum(df):
idx = df.index.values
vals = df.values
summ = idx * vals
return summ.sum()
d1 = assignments_no_control_no_null['ASSIGNMENT:reward'].value_counts(normalize=True)
d2 = assignments_no_control_no_null['ASSIGNMENT:reward'].value_counts()
print(f'взвешенная цена айтема в тесте: {round(w_sum(d1), 3)}')
print(f'потрачено на разметку теста: {round(w_sum(d2), 3)}')
print(f'{round(w_sum(d2), 3)} / {round(w_sum(d1), 3)}')
</code>
Выделим, сколько составила средняя часовая ставка для разметки тестовой части датасета. Это будет простое среднее из следующих величин: количество заданий, которое разметчк может сделать за час на основе данного задания, помноженное на цену задания.
<code>
def get_hour_pay(df):
try:
times = pd.to_datetime(df['ASSIGNMENT:submitted']) - pd.to_datetime(df['ASSIGNMENT:started'])
except Exception as e:
times = []
for i in range(len(assignments_no_control_no_null)):
try:
start = pd.to_datetime(assignments_no_control_no_null['ASSIGNMENT:started'].iloc[i])
except Exception as e:
start = pd.to_datetime(assignments_no_control_no_null['ASSIGNMENT:started'].apply(lambda x: x.split('T')[1]).iloc[i])
try:
end = pd.to_datetime(assignments_no_control_no_null['ASSIGNMENT:submitted'].iloc[i])
except Exception as e:
start = pd.to_datetime(assignments_no_control_no_null['ASSIGNMENT:submitted'].apply(lambda x: x.split('T')[1]).iloc[i])
delta = end - start
times.extend([delta])
times = pd.Series(times)
# times = pd.to_datetime(df['ASSIGNMENT:submitted'].apply(lambda x: x.split('T')[1])) - pd.to_datetime(df['ASSIGNMENT:started'].apply(lambda x: x.split('T')[1]))
sums = 3600 / times.apply(lambda x: x.seconds) * df['ASSIGNMENT:reward']
return sums.mean()
get_hour_pay(assignments_no_control_no_null)
</code>
### Сбор ответов разметчиков и голосование
Собираем ответы голосования большинством для каждого задания.
<code>
from collections import defaultdict
text_dict = defaultdict(list)
for task, op1, op2, op3, op4, user, out in zip(
assignments_no_control_no_null["INPUT:text"], assignments_no_control_no_null["INPUT:option_a"],
assignments_no_control_no_null["INPUT:option_b"], assignments_no_control_no_null["INPUT:option_c"],
assignments_no_control_no_null["INPUT:option_d"],
assignments_no_control_no_null["ASSIGNMENT:worker_id"], assignments_no_control_no_null["OUTPUT:answer"]
):
if user not in bad_users:
text_dict[(task, op1, op2, op3, op4)].append([
user,
{"out": out}
])
print(len(text_dict))
</code>
<code>
keys = list(text_dict.keys())
Counter([len(text_dict[keys[i]]) for i in range(len(keys))])
</code>
Есть 110 заданий, где перекрытие меньше 5. Для формирования итоговых лейблов нужно, чтобы было простое большинство разметчиков, проголосовавших за данную опцию. Если большинства нет, то оценка строится, исходя из оценки навыков разметчиков. В таком случае, финальный лейбл будет присвоен по голосу группы с наилучшими навыками. Если по навыкам будет равенство, то решаем по ответам топ-3 по навыкам разметчиков. Если и данный способ дает равенство, то используются оценки навыков разметчиков из EM-алгоритма (реализация GLAD).
<code>
preds_full = {}
user2skill = {k:v for k, v in zip(skills['worker_id'], skills['skill_value'])}
control_acc = assignments[assignments['GOLDEN:answer'].notna()]\
.groupby('ASSIGNMENT:worker_id')\
.apply(lambda x: (np.array(x['OUTPUT:answer']) == np.array(x['GOLDEN:answer'])).mean())
user2control = {k:v for k, v in zip(control_acc.index, control_acc.values)}
from crowdkit.aggregation.classification.glad import GLAD
full = assignments['INPUT:text'] + ' ' + assignments['INPUT:option_a'] + ' ' + assignments['INPUT:option_b'] + ' ' + assignments['INPUT:option_c'] + ' ' + assignments['INPUT:option_d']
id2task = dict(enumerate(full))
task2id = {k:v for v, k in id2task.items()}
id2user = dict(enumerate(assignments['ASSIGNMENT:worker_id']))
user2id = {k:v for v, k in id2user.items()}
codes = full.map(task2id)
res = pd.DataFrame({'task': codes, 'worker': assignments['ASSIGNMENT:worker_id'].map(user2id), 'label': assignments['OUTPUT:answer']})
model = GLAD(n_iter=10000, tol=1e-06, m_step_max_iter=1000, m_step_tol=1e-03)
model.fit(res)
user2alpha = dict(enumerate(model.alphas_))
tb = model.alphas_.copy()
tb.index = tb.index.map(id2user)
user2alpha = {k:v for k, v in zip(tb.index, tb.values)}
stats = {
'total_agreement': 0,
'majority': 0,
'skill_based': 0,
'major_based': 0,
'em_based': 0,
'rest': 0,
}
for i in range(len(keys)):
ans = text_dict[keys[i]]
lst = [[ans[j][0], ans[j][1]['out']] for j in range(len(ans))]
users, votes = list(zip(*lst))
cnt = pd.Series(Counter(votes)).sort_values(ascending=False)
# # total agreement
if len(cnt) == 1:
res = cnt.index[0]
stats['total_agreement'] += 1
# simple majority
elif cnt.iloc[0] > cnt.iloc[1]:
res = cnt.index[0]
stats['majority'] += 1
# (> 1 options) & (1 option == 2 option)
else:
# try overall skill based comparison
vals = list(map(lambda x: user2skill[x], users))
table = pd.DataFrame({'user': users, 'votes': votes, 'skill': vals})
agg = table.groupby('votes').agg(
sum_skill=pd.NamedAgg(column='skill', aggfunc='sum'),
sum_votes=pd.NamedAgg(column='user', aggfunc='count')
).sort_values(by=['sum_votes', 'sum_skill'], ascending=False)
# check there is a leader by skills
if agg['sum_skill'].iloc[0] > agg['sum_skill'].iloc[1]:
res = agg.index[0]
stats['skill_based'] += 1
else:
# top-3 answers by overall skills
vals = list(map(lambda x: user2skill[x], users))
table = pd.DataFrame({'user': users, 'votes': votes, 'skill': vals})
table = table.sort_values(by='skill', ascending=False)
if len(table) >= 3:
sub = table.iloc[:3]
else:
sub = table
agg = sub.groupby('votes').agg(
sum_skill=pd.NamedAgg(column='skill', aggfunc='sum'),
sum_votes=pd.NamedAgg(column='user', aggfunc='count')
).sort_values(by=['sum_votes', 'sum_skill'], ascending=False)
if agg['sum_skill'].iloc[0] != agg['sum_skill'].iloc[1]:
res = agg.index[0]
stats['major_based'] += 1
else:
vals = list(map(lambda x: user2alpha[x], users))
table = pd.DataFrame({'user': users, 'votes': votes, 'skill': vals})
agg = table.groupby('votes').agg(
sum_skill=pd.NamedAgg(column='skill', aggfunc='sum'),
sum_votes=pd.NamedAgg(column='user', aggfunc='count')
).sort_values(by=['sum_votes', 'sum_skill'], ascending=False)
# check there is a leader by skills
if agg['sum_skill'].iloc[0] != agg['sum_skill'].iloc[1]:
res = agg.index[0]
stats['em_based'] += 1
else:
res = agg.index[0]
stats['rest'] += 1
preds_full[keys[i]] = res
</code>
<code>
stats
</code>
<code>
preds_full_df = pd.concat([pd.DataFrame(preds_full.keys(), columns=['task', 'op1', 'op2', 'op3', 'op4']), pd.DataFrame(preds_full.values(), columns=['lb'])], axis=1).astype(str)
</code>
### Сопоставление разметки и ground truth
Забираем задания из датасета с правильными ответами.
<code>
res_df = pd.read_csv('general_wa.tsv', sep='\t')
</code>
<code>
res_df = res_df.rename({
'INPUT:text': 'task',
'INPUT:option_a': 'op1',
'INPUT:option_b': 'op2',
'INPUT:option_c': 'op3',
'INPUT:option_d': 'op4',
'GOLDEN:answer': 'lb',
}, axis=1).astype(str)
</code>
После скачивания с Толоки в текстах рушится форматирование, потому нельзя просто сделать join двух табличек. Нужно убрать все "лишнее" форматирование сразу из двух табличек, чтобы остались только тексты, пунктуация и пробелы.
<code>
def format_text(text):
text = (text.strip().replace('\n', ' ').replace('\t', ' ')
.replace('\r', ' ').replace(' ', ' ').replace(' ', ' ')
.replace(' ', ' '))
return text
res_df['task'] = res_df['task'].apply(format_text)
res_df['op1'] = res_df['op1'].apply(format_text)
res_df['op2'] = res_df['op2'].apply(format_text)
res_df['op3'] = res_df['op3'].apply(format_text)
res_df['op4'] = res_df['op4'].apply(format_text)
preds_full_df['task'] = preds_full_df['task'].apply(format_text)
preds_full_df['op1'] = preds_full_df['op1'].apply(format_text)
preds_full_df['op2'] = preds_full_df['op2'].apply(format_text)
preds_full_df['op3'] = preds_full_df['op3'].apply(format_text)
preds_full_df['op4'] = preds_full_df['op4'].apply(format_text)
res_df['full'] = res_df['task'] + ' ' + res_df['op1'] + ' ' + res_df['op2'] + ' ' + res_df['op3'] + ' ' + res_df['op4']
preds_full_df['full'] = preds_full_df['task'] + ' ' + preds_full_df['op1'] + ' ' + preds_full_df['op2'] + ' ' + preds_full_df['op3'] + ' ' + preds_full_df['op4']
res_df['full'] = res_df['full'].apply(format_text)
preds_full_df['full'] = preds_full_df['full'].apply(format_text)
</code>
Делаем left join, чтобы соединить голосование и правильные метки для одних и тех же заданий.
<code>
new = res_df.merge(preds_full_df.drop(['task', 'op1', 'op2', 'op3', 'op4'], axis=1), on='full', how='left')
</code>
<code>
new_valid = new[new['lb_y'].notna()].copy()
len(new_valid)
</code>
Ни одна строка не была утеряна.
<code>
new_valid.head(1)
</code>
### Подсчет метрики
Если в правом столбце меток осталось 961 непустых строк, значит, форматирование было подчищено корректно и ничего не потерялось
Попробуем посчитать разные метрики
<code>
(new_valid['lb_x'] == new_valid['lb_y']).mean()
</code>
<code>
d = new_valid.groupby('domain').apply(lambda x: (x['lb_x'] == x['lb_y']).mean().round(3))
d
</code>
`Accuracy = 0.844`
|
{
"filename": "agg_1.ipynb",
"repository": "ai-forever/MERA",
"query": "transformed_from_existing",
"size": 39444,
"sha": ""
}
|
# 12_nlp_dive_1.ipynb
Repository: immc-lab/fastbook-zh
<code>
#hide
! [ -e /content ] && pip install -Uqq fastbook
import fastbook
fastbook.setup_book()
</code>
<code>
#hide
from fastbook import *
</code>
# A Language Model from Scratch
一个来自Scratch的语言模型
We're now ready to go deep... deep into deep learning! You already learned how to train a basic neural network, but how do you go from there to creating state-of-the-art models? In this part of the book we're going to uncover all of the mysteries, starting with language models.
You saw in <<chapter_nlp>> how to fine-tune a pretrained language model to build a text classifier. In this chapter, we will explain to you what exactly is inside that model, and what an RNN is. First, let's gather some data that will allow us to quickly prototype our various models.
我们现在准备好深入...深入深度学习!您已经学习了如何训练基本的神经网络,但是您如何从那里开始创建最先进的模型?在本书的这一部分,我们将揭开所有的谜团,从语言模型开始。
您在<>中看到了如何微调预训练的语言模型以构建文本分类器。在本章中,我们将向您解释该模型内部到底是什么,以及RNN是什么。首先,让我们收集一些数据,使我们能够快速原型化我们的各种模型。
## The Data
数据
Whenever we start working on a new problem, we always first try to think of the simplest dataset we can that will allow us to try out methods quickly and easily, and interpret the results. When we started working on language modeling a few years ago we didn't find any datasets that would allow for quick prototyping, so we made one. We call it *Human Numbers*, and it simply contains the first 10,000 numbers written out in English.
每当我们开始处理一个新问题时,我们总是首先尝试想出我们能想到的最简单的数据集,这将使我们能够快速轻松地尝试我们的方法,并解释结果。当我们几年前开始研究语言建模时,我们没有找到任何可以快速原型化的数据集,所以我们做了一个。我们称之为人类数字,它只是包含用英语写出来的前10,000个数字。
> j: One of the most common practical mistakes I see even amongst highly experienced practitioners is failing to use appropriate datasets at appropriate times during the analysis process. In particular, most people tend to start with datasets that are too big and too complicated.
J:即使在经验丰富的从业者中,我也看到最常见的实际错误之一是在分析过程中未能在适当的时间使用适当的数据集。特别是,大多数人倾向于从太大太复杂的数据集开始。
We can download, extract, and take a look at our dataset in the usual way:
我们可以以通常的方式下载、提取和查看我们的数据集:
<code>
from fastai.text.all import *
path = untar_data(URLs.HUMAN_NUMBERS)
</code>
<code>
#hide
Path.BASE_PATH = path
</code>
<code>
path.ls()
</code>
Let's open those two files and see what's inside. At first we'll join all of the texts together and ignore the train/valid split given by the dataset (we'll come back to that later):
让我们打开这两个文件,看看里面有什么。首先,我们将所有文本连接在一起,忽略数据集给出的训练集和验证集。(我们稍后会回到这个问题):
<code>
lines = L()
with open(path/'train.txt') as f: lines += L(*f.readlines())
with open(path/'valid.txt') as f: lines += L(*f.readlines())
lines
</code>
We take all those lines and concatenate them in one big stream. To mark when we go from one number to the next, we use a `.` as a separator:
我们将所有这些线连接在一个大流中。当我们从一个数字到下一个数字时,我们使用.作为分隔符:
<code>
text = ' . '.join([l.strip() for l in lines])
text[:100]
</code>
We can tokenize this dataset by splitting on spaces:
我们可以使用空格划分此数据集:
<code>
tokens = text.split(' ')
tokens[:10]
</code>
To numericalize, we have to create a list of all the unique tokens (our *vocab*):
要进行数值化,我们必须创建包含所有唯一令牌(我们的词汇表)的列表:
<code>
vocab = L(*tokens).unique()
vocab
</code>
Then we can convert our tokens into numbers by looking up the index of each in the vocab:
然后我们可以通过在词汇表中查找每个词的索引将我们的标记转换为数字:
<code>
word2idx = {w:i for i,w in enumerate(vocab)}
nums = L(word2idx[i] for i in tokens)
nums
</code>
Now that we have a small dataset on which language modeling should be an easy task, we can build our first model.
现在我们有了一个小的数据集,语言模型化应该是一项简单的任务,我们可以构建我们的第一个模型。
## Our First Language Model from Scratch
我们从零开始的第一个语言模型
One simple way to turn this into a neural network would be to specify that we are going to predict each word based on the previous three words. We could create a list of every sequence of three words as our independent variables, and the next word after each sequence as the dependent variable.
We can do that with plain Python. Let's do it first with tokens just to confirm what it looks like:
将其转化为神经网络的一个简单方法是指定我们将根据前三个单词预测每个单词。我们可以创建一个列表,将三个单词的每个序列作为自变量,将每个序列后的下一个单词作为因变量。
我们可以用普通Python 编程语言编程语言来做到这一点。让我们首先使用令牌来确认它的外观:
<code>
L((tokens[i:i+3], tokens[i+3]) for i in range(0,len(tokens)-4,3))
</code>
Now we will do it with tensors of the numericalized values, which is what the model will actually use:
现在我们将使用数值的张量来完成,这是模型实际使用的:
<code>
seqs = L((tensor(nums[i:i+3]), nums[i+3]) for i in range(0,len(nums)-4,3))
seqs
</code>
We can batch those easily using the `DataLoader` class. For now we will split the sequences randomly:
我们可以使用DataLoader类轻松批处理它们。现在我们将随机拆分序列:
<code>
bs = 64
cut = int(len(seqs) * 0.8)
dls = DataLoaders.from_dsets(seqs[:cut], seqs[cut:], bs=64, shuffle=False)
</code>
We can now create a neural network architecture that takes three words as input, and returns a prediction of the probability of each possible next word in the vocab. We will use three standard linear layers, but with two tweaks.
The first tweak is that the first linear layer will use only the first word's embedding as activations, the second layer will use the second word's embedding plus the first layer's output activations, and the third layer will use the third word's embedding plus the second layer's output activations. The key effect of this is that every word is interpreted in the information context of any words preceding it.
The second tweak is that each of these three layers will use the same weight matrix. The way that one word impacts the activations from previous words should not change depending on the position of a word. In other words, activation values will change as data moves through the layers, but the layer weights themselves will not change from layer to layer. So, a layer does not learn one sequence position; it must learn to handle all positions.
Since layer weights do not change, you might think of the sequential layers as "the same layer" repeated. In fact, PyTorch makes this concrete; we can just create one layer, and use it multiple times.
我们现在可以创建一个神经网络结构,将三个单词作为输入,并返回词汇中每个可能的下一个单词的概率预测。我们将使用三个标准线性的层,但有两个调整。
第一个调整是第一个线性的层将只使用第一个单词的嵌入作为激活,第二层将使用第二个单词的嵌入加上第一层的输出激活,第三层将使用第三个单词的嵌入加上第二层的输出激活。这样做的关键效果是每个单词都在它之前的任何单词的信息上下文中被解释。
第二个调整是这三层中的每一层都将使用相同的权矩阵。一个单词影响前一个单词激活的方式不应根据单词的位置而改变。换句话说,激活值会随着数据在层中移动而改变,但层权重本身不会因层而改变。因此,一个层不会学习一个序列位置;它必须学会处理所有位置。
由于层权重不会改变,您可能会认为顺序层是重复的“同一层”。事实上,PyTorch使这变得具体;我们可以只创建一层,并多次使用它。
### Our Language Model in PyTorch
PyTorch中的语言模型
We can now create the language model module that we described earlier:
我们现在可以创建前面描述的语言模型模块:
<code>
class LMModel1(Module):
def __init__(self, vocab_sz, n_hidden):
self.i_h = nn.Embedding(vocab_sz, n_hidden)
self.h_h = nn.Linear(n_hidden, n_hidden)
self.h_o = nn.Linear(n_hidden,vocab_sz)
def forward(self, x):
h = F.relu(self.h_h(self.i_h(x[:,0])))
h = h + self.i_h(x[:,1])
h = F.relu(self.h_h(h))
h = h + self.i_h(x[:,2])
h = F.relu(self.h_h(h))
return self.h_o(h)
</code>
As you see, we have created three layers:
- The embedding layer (`i_h`, for *input* to *hidden*)
- The linear layer to create the activations for the next word (`h_h`, for *hidden* to *hidden*)
- A final linear layer to predict the fourth word (`h_o`, for *hidden* to *output*)
This might be easier to represent in pictorial form, so let's define a simple pictorial representation of basic neural networks. <<img_simple_nn>> shows how we're going to represent a neural net with one hidden layer.
如您所见,我们创建了三层:
嵌入层(i_h,用于输入隐藏)
线性的层创建下一个单词的激活(h_h,从隐藏到隐藏)
最后一个线性的层来预测第四个单词(h_o,隐藏到输出)
这可能更容易以图形形式表示,因此让我们定义基本神经网络的简单图形表示。<>显示了我们将如何表示具有一个隐含层的神经网络。
<img alt="Pictorial representation of simple neural network" width="400" src="images/att_00020.png" caption="Pictorial representation of a simple neural network" id="img_simple_nn">
Each shape represents activations: rectangle for input, circle for hidden (inner) layer activations, and triangle for output activations. We will use those shapes (summarized in <<img_shapes>>) in all the diagrams in this chapter.
每个形状代表激活:矩形用于输入,圆形用于隐藏(内部)层激活,三角形用于输出激活。我们将在本章的所有图表中使用这些形状(总结在<>中)。
<img alt="Shapes used in our pictorial representations" width="200" src="images/att_00021.png" id="img_shapes" caption="Shapes used in our pictorial representations">
An arrow represents the actual layer computation—i.e., the linear layer followed by the activation function. Using this notation, <<lm_rep>> shows what our simple language model looks like.
箭头表示实际的层计算——即线性的层,后跟激活函数。使用这种表示法,<>显示了我们的简单语言模型的样子。
<img alt="Representation of our basic language model" width="500" caption="Representation of our basic language model" id="lm_rep" src="images/att_00022.png">
To simplify things, we've removed the details of the layer computation from each arrow. We've also color-coded the arrows, such that all arrows with the same color have the same weight matrix. For instance, all the input layers use the same embedding matrix, so they all have the same color (green).
Let's try training this model and see how it goes:
为了简化,我们从每个箭头中删除了层计算的细节。我们还对箭头进行了颜色编码,使得所有具有相同颜色的箭头都具有相同的权矩阵。例如,所有输入层使用相同的嵌入矩阵,因此它们都具有相同的颜色(绿色)。
让我们试着训练这个模型,看看它是如何进行的:
<code>
learn = Learner(dls, LMModel1(len(vocab), 64), loss_func=F.cross_entropy,
metrics=accuracy)
learn.fit_one_cycle(4, 1e-3)
</code>
To see if this is any good, let's check what a very simple model would give us. In this case we could always predict the most common token, so let's find out which token is most often the target in our validation set:
为了看看这是否有任何好处,让我们检查一个非常简单的模型会给我们什么。在这种情况下,我们总是可以预测最常见的令牌,所以让我们找出哪个令牌是我们验证集中最常见的目标:
<code>
n,counts = 0,torch.zeros(len(vocab))
for x,y in dls.valid:
n += y.shape[0]
for i in range_of(vocab): counts[i] += (y==i).long().sum()
idx = torch.argmax(counts)
idx, vocab[idx.item()], counts[idx].item()/n
</code>
The most common token has the index 29, which corresponds to the token `thousand`. Always predicting this token would give us an accuracy of roughly 15\%, so we are faring way better!
最常见的令牌有索引29,对应于令牌千。总是预测这个令牌会给我们大约15%的准确率,所以我们做得更好!
> A: My first guess was that the separator would be the most common token, since there is one for every number. But looking at `tokens` reminded me that large numbers are written with many words, so on the way to 10,000 you write "thousand" a lot: five thousand, five thousand and one, five thousand and two, etc. Oops! Looking at your data is great for noticing subtle features and also embarrassingly obvious ones.
A:我的第一个猜测是分隔符将是最常见的令牌,因为每个数字都有一个。但是看着令牌提醒我,大的数字是用很多单词写的,所以在到达10,000的路上,你会写很多“千”: 5000、51000、52000,等等。哎呀!查看你的数据非常有助于注意微妙的特征和令人尴尬的明显特征。
This is a nice first baseline. Let's see how we can refactor it with a loop.
这是一个很好的第一个基线。让我们看看如何用循环重构它。
### Our First Recurrent Neural Network
我们的第一个循环神经网络
Looking at the code for our module, we could simplify it by replacing the duplicated code that calls the layers with a `for` loop. As well as making our code simpler, this will also have the benefit that we will be able to apply our module equally well to token sequences of different lengths—we won't be restricted to token lists of length three:
看看我们模块的代码,我们可以通过用for循环替换调用层的重复代码来简化它。除了使我们的代码更简单之外,这还有一个好处,那就是我们将能够同样好地将我们的模块应用于不同长度的令牌序列——我们不会局限于长度为3的令牌列表:
<code>
class LMModel2(Module):
def __init__(self, vocab_sz, n_hidden):
self.i_h = nn.Embedding(vocab_sz, n_hidden)
self.h_h = nn.Linear(n_hidden, n_hidden)
self.h_o = nn.Linear(n_hidden,vocab_sz)
def forward(self, x):
h = 0
for i in range(3):
h = h + self.i_h(x[:,i])
h = F.relu(self.h_h(h))
return self.h_o(h)
</code>
Let's check that we get the same results using this refactoring:
让我们检查使用此重构是否得到相同的结果:
<code>
learn = Learner(dls, LMModel2(len(vocab), 64), loss_func=F.cross_entropy,
metrics=accuracy)
learn.fit_one_cycle(4, 1e-3)
</code>
We can also refactor our pictorial representation in exactly the same way, as shown in <<basic_rnn>> (we're also removing the details of activation sizes here, and using the same arrow colors as in <<lm_rep>>).
我们还可以以完全相同的方式重构图形表示,如<>中所示(我们还在此处删除了激活大小的详细信息,并使用与<>中相同的箭头颜色)。
<img alt="Basic recurrent neural network" width="400" caption="Basic recurrent neural network" id="basic_rnn" src="images/att_00070.png">
You will see that there is a set of activations that are being updated each time through the loop, stored in the variable `h`—this is called the *hidden state*.
您将看到有一组激活每次都通过循环更新,存储在变量h中——这称为隐状态。
> Jargon: hidden state: The activations that are updated at each step of a recurrent neural network.
行话:隐状态:在循环神经网络的每一步更新的激活。
A neural network that is defined using a loop like this is called a *recurrent neural network* (RNN). It is important to realize that an RNN is not a complicated new architecture, but simply a refactoring of a multilayer neural network using a `for` loop.
> A: My true opinion: if they were called "looping neural networks," or LNNs, they would seem 50% less daunting!
使用这样的循环定义的神经网络称为循环神经网络(RNN)。重要的是要意识到RNN不是复杂的新架构,而只是使用for循环重构多层神经网络。
A:我的真实观点是:如果它们被称为“循环神经网络”或LNNs,它们看起来可怕的程度会减少50%!
Now that we know what an RNN is, let's try to make it a little bit better.
现在我们知道了什么是RNN,让我们试着把它做得更好一点。
## Improving the RNN
改进RNN
Looking at the code for our RNN, one thing that seems problematic is that we are initializing our hidden state to zero for every new input sequence. Why is that a problem? We made our sample sequences short so they would fit easily into batches. But if we order the samples correctly, those sample sequences will be read in order by the model, exposing the model to long stretches of the original sequence.
Another thing we can look at is having more signal: why only predict the fourth word when we could use the intermediate predictions to also predict the second and third words?
Let's see how we can implement those changes, starting with adding some state.
看看我们的RNN代码,有一件事似乎有问题,那就是我们正在为每个新的输入序列初始化我们的隐状态为零。为什么会有问题?我们缩短了样本序列,这样它们就可以很容易地成批。但是如果我们正确排序样本,这些样本序列将被模型按顺序读取,从而使模型暴露在原始序列的很长一段中。
我们可以考虑的另一件事是有更多的信号:当我们可以使用中间预测来预测第二个和第三个单词时,为什么只预测第四个单词?
让我们看看如何实现这些更改,从添加一些状态开始。
### Maintaining the State of an RNN
维护RNN的状态
Because we initialize the model's hidden state to zero for each new sample, we are throwing away all the information we have about the sentences we have seen so far, which means that our model doesn't actually know where we are up to in the overall counting sequence. This is easily fixed; we can simply move the initialization of the hidden state to `__init__`.
But this fix will create its own subtle, but important, problem. It effectively makes our neural network as deep as the entire number of tokens in our document. For instance, if there were 10,000 tokens in our dataset, we would be creating a 10,000-layer neural network.
To see why this is the case, consider the original pictorial representation of our recurrent neural network in <<lm_rep>>, before refactoring it with a `for` loop. You can see each layer corresponds with one token input. When we talk about the representation of a recurrent neural network before refactoring with the `for` loop, we call this the *unrolled representation*. It is often helpful to consider the unrolled representation when trying to understand an RNN.
The problem with a 10,000-layer neural network is that if and when you get to the 10,000th word of the dataset, you will still need to calculate the derivatives all the way back to the first layer. This is going to be very slow indeed, and very memory-intensive. It is unlikely that you'll be able to store even one mini-batch on your GPU.
The solution to this problem is to tell PyTorch that we do not want to back propagate the derivatives through the entire implicit neural network. Instead, we will just keep the last three layers of gradients. To remove all of the gradient history in PyTorch, we use the `detach` method.
Here is the new version of our RNN. It is now stateful, because it remembers its activations between different calls to `forward`, which represent its use for different samples in the batch:
因为我们为每个新样本初始化模型的隐状态为零,所以我们丢弃了迄今为止所看到的关于句子的所有信息,这意味着我们的模型实际上不知道我们在整个计数序列中的位置。这很容易修复;我们可以简单地将隐状态的初始化移动到__init__。
但是这个修复会产生一个微妙但重要的问题。它有效地使我们的神经网络与文档中的所有标记一样深。例如,如果我们的数据集中有10,000个标记,我们将创建一个10,000层的神经网络。
要了解为什么会出现这种情况,请考虑<>中循环神经网络的原始图形表示,然后再使用for循环对其进行重构。您可以看到每一层对应一个令牌输入。当我们在使用for循环重构之前讨论循环神经网络的表示时,我们称之为展开表示。在尝试理解RNN时考虑展开表示通常很有帮助。
10,000层神经网络的问题是,如果并且当您到达数据集的第10,000个单词时,您仍然需要计算导数直到回到第一层。这确实会非常慢,并且非常占用内存。您不太可能在GPU上存储哪怕一个迷你批次。
这个问题的解决方案是告诉机器学习库,我们不想通过整个隐式神经网络反向传播导数。相反,我们将只保留最后三层梯度。为了删除机器学习库中的所有梯度历史,我们使用分离方法。
这是我们RNN的新版本。它现在是有状态的,因为它会记住它在不同的转发调用之间的激活,这代表它对批处理中不同样本的使用:
<code>
class LMModel3(Module):
def __init__(self, vocab_sz, n_hidden):
self.i_h = nn.Embedding(vocab_sz, n_hidden)
self.h_h = nn.Linear(n_hidden, n_hidden)
self.h_o = nn.Linear(n_hidden,vocab_sz)
self.h = 0
def forward(self, x):
for i in range(3):
self.h = self.h + self.i_h(x[:,i])
self.h = F.relu(self.h_h(self.h))
out = self.h_o(self.h)
self.h = self.h.detach()
return out
def reset(self): self.h = 0
</code>
This model will have the same activations whatever sequence length we pick, because the hidden state will remember the last activation from the previous batch. The only thing that will be different is the gradients computed at each step: they will only be calculated on sequence length tokens in the past, instead of the whole stream. This approach is called *backpropagation through time* (BPTT).
无论我们选择什么序列长度,这个模型都将具有相同的激活,因为隐状态将记住前一批的最后一次激活。唯一不同的是在每一步计算的梯度:它们只会在过去的序列长度标记上计算,而不是整个流。这种方法称为通过时间的反向传播算法(BPTT)。
> jargon: Back propagation through time (BPTT): Treating a neural net with effectively one layer per time step (usually refactored using a loop) as one big model, and calculating gradients on it in the usual way. To avoid running out of memory and time, we usually use _truncated_ BPTT, which "detaches" the history of computation steps in the hidden state every few time steps.
行话:通过时间反向传播(BPTT):将每个时间步有效地一层(通常使用循环重构)的神经网络视为一个大模型,并以通常的方式计算其上的梯度。为了避免运行内存溢出和时间,我们通常使用截断的BPTT,它每隔几个时间步“分离”隐状态中计算步骤的历史。
To use `LMModel3`, we need to make sure the samples are going to be seen in a certain order. As we saw in <<chapter_nlp>>, if the first line of the first batch is our `dset[0]` then the second batch should have `dset[1]` as the first line, so that the model sees the text flowing.
`LMDataLoader` was doing this for us in <<chapter_nlp>>. This time we're going to do it ourselves.
To do this, we are going to rearrange our dataset. First we divide the samples into `m = len(dset) // bs` groups (this is the equivalent of splitting the whole concatenated dataset into, for example, 64 equally sized pieces, since we're using `bs=64` here). `m` is the length of each of these pieces. For instance, if we're using our whole dataset (although we'll actually split it into train versus valid in a moment), that will be:
要使用LMModel3,我们需要确保以一定的顺序看到样本。正如我们在<>中看到的,如果第一批的第一行是我们的dset[0],那么第二批应该有dset[1]作为第一行,以便模型看到文本流动。
LMDataLoader在<>中为我们做了这件事。这次我们要自己做。
为此,我们将重新排列我们的数据集。首先,我们将样本划分为m=len(dset)//bs组(这相当于将整个串联数据集拆分为64个相同大小的块,因为我们在这里使用bs=64)。m是这些片段中每个片段的长度。例如,如果我们使用我们的整个数据集(尽管我们实际上会将其拆分为训练与有效),那将是:
<code>
m = len(seqs)//bs
m,bs,len(seqs)
</code>
The first batch will be composed of the samples:
(0, m, 2*m, ..., (bs-1)*m)
the second batch of the samples:
(1, m+1, 2*m+1, ..., (bs-1)*m+1)
and so forth. This way, at each epoch, the model will see a chunk of contiguous text of size `3*m` (since each text is of size 3) on each line of the batch.
The following function does that reindexing:
第一个批次由以下的样本组成:
(0, m, 2*m, ..., (bs-1)*m)
第二个批次由一下的样本组成:
(1, m+1, 2*m+1, ..., (bs-1)*m+1)
第四层也是如此。用这种方法,在每一个批次,模型将在批处理的每一行上看到一块大小为3*m的连续文本(因为每个文本的大小都是3)。
以下函数执行此重新索引操作:
<code>
def group_chunks(ds, bs):
m = len(ds) // bs
new_ds = L()
for i in range(m): new_ds += L(ds[i + m*j] for j in range(bs))
return new_ds
</code>
Then we just pass `drop_last=True` when building our `DataLoaders` to drop the last batch that does not have a shape of `bs`. We also pass `shuffle=False` to make sure the texts are read in order:
然后,在构建DataLoader时,我们只需传递drop_last=True以删除最后一批没有bs形状的文本。我们还传递shuffle=False以确保按顺序读取文本:
<code>
cut = int(len(seqs) * 0.8)
dls = DataLoaders.from_dsets(
group_chunks(seqs[:cut], bs),
group_chunks(seqs[cut:], bs),
bs=bs, drop_last=True, shuffle=False)
</code>
The last thing we add is a little tweak of the training loop via a `Callback`. We will talk more about callbacks in <<chapter_accel_sgd>>; this one will call the `reset` method of our model at the beginning of each epoch and before each validation phase. Since we implemented that method to zero the hidden state of the model, this will make sure we start with a clean state before reading those continuous chunks of text. We can also start training a bit longer:
我们添加的最后一件事是通过回调对训练循环进行一点调整。我们将在<>中更多地讨论回调;这个将在每个epoch的开始和每个验证阶段之前调用模型的重置方法。由于我们实现了该方法以将模型的隐状态归零,这将确保我们在阅读那些连续的文本块之前从干净的状态开始。我们还可以开始更长时间的训练:
<code>
learn = Learner(dls, LMModel3(len(vocab), 64), loss_func=F.cross_entropy,
metrics=accuracy, cbs=ModelResetter)
learn.fit_one_cycle(10, 3e-3)
</code>
This is already better! The next step is to use more targets and compare them to the intermediate predictions.
这已经更好了!下一步是使用更多的目标,并将它们与中间预测进行比较。
### Creating More Signal
创造更多信号
Another problem with our current approach is that we only predict one output word for each three input words. That means that the amount of signal that we are feeding back to update weights with is not as large as it could be. It would be better if we predicted the next word after every single word, rather than every three words, as shown in <<stateful_rep>>.
我们当前方法的另一个问题是,我们每三个输入词只预测一个输出词。这意味着,我们反馈给更新权重的信号量没有可能的大。如果我们在每个单词之后预测下一个单词,而不是每三个单词预测一次,会更好,如<>所示。
<img alt="RNN predicting after every token" width="400" caption="RNN predicting after every token" id="stateful_rep" src="images/att_00024.png">
This is easy enough to add. We need to first change our data so that the dependent variable has each of the three next words after each of our three input words. Instead of `3`, we use an attribute, `sl` (for sequence length), and make it a bit bigger:
这很容易添加。我们需要首先更改我们的数据,以便因变量在我们的三个输入词之后都有三个接下来的词。我们使用属性sl(表示序列长度)而不是3,并使其大一点:
<code>
sl = 16
seqs = L((tensor(nums[i:i+sl]), tensor(nums[i+1:i+sl+1]))
for i in range(0,len(nums)-sl-1,sl))
cut = int(len(seqs) * 0.8)
dls = DataLoaders.from_dsets(group_chunks(seqs[:cut], bs),
group_chunks(seqs[cut:], bs),
bs=bs, drop_last=True, shuffle=False)
</code>
Looking at the first element of `seqs`, we can see that it contains two lists of the same size. The second list is the same as the first, but offset by one element:
查看seqs的第一个元素,我们可以看到它包含两个大小相同的列表。第二个列表与第一个相同,但偏移量为一个元素:
<code>
[L(vocab[o] for o in s) for s in seqs[0]]
</code>
Now we need to modify our model so that it outputs a prediction after every word, rather than just at the end of a three-word sequence:
现在我们需要修改我们的模型,以便它在每个单词之后输出预测,而不仅仅是在三个单词序列的末尾:
<code>
class LMModel4(Module):
def __init__(self, vocab_sz, n_hidden):
self.i_h = nn.Embedding(vocab_sz, n_hidden)
self.h_h = nn.Linear(n_hidden, n_hidden)
self.h_o = nn.Linear(n_hidden,vocab_sz)
self.h = 0
def forward(self, x):
outs = []
for i in range(sl):
self.h = self.h + self.i_h(x[:,i])
self.h = F.relu(self.h_h(self.h))
outs.append(self.h_o(self.h))
self.h = self.h.detach()
return torch.stack(outs, dim=1)
def reset(self): self.h = 0
</code>
This model will return outputs of shape `bs x sl x vocab_sz` (since we stacked on `dim=1`). Our targets are of shape `bs x sl`, so we need to flatten those before using them in `F.cross_entropy`:
此模型将返回形状bs x sl xvocab_sz的输出(因为我们堆叠在dim=1上)。我们的目标是形状bs x sl,因此我们需要在将它们用于F.cross_entropy之前将它们展平:
<code>
def loss_func(inp, targ):
return F.cross_entropy(inp.view(-1, len(vocab)), targ.view(-1))
</code>
We can now use this loss function to train the model:
现在我们可以使用损失函数来训练模型
<code>
learn = Learner(dls, LMModel4(len(vocab), 64), loss_func=loss_func,
metrics=accuracy, cbs=ModelResetter)
learn.fit_one_cycle(15, 3e-3)
</code>
We need to train for longer, since the task has changed a bit and is more complicated now. But we end up with a good result... At least, sometimes. If you run it a few times, you'll see that you can get quite different results on different runs. That's because effectively we have a very deep network here, which can result in very large or very small gradients. We'll see in the next part of this chapter how to deal with this.
Now, the obvious way to get a better model is to go deeper: we only have one linear layer between the hidden state and the output activations in our basic RNN, so maybe we'll get better results with more.
我们需要训练更长时间,因为任务已经发生了一些变化,现在变得更加复杂了。但是我们最终会得到一个好结果...至少有时是这样。如果你运行几次,你会发现你可以在不同的运行中得到完全不同的结果。这是因为实际上我们这里有一个非常深的网络,这可能会导致非常大或非常小的梯度。我们将在本章的下一部分看到如何处理这个问题。
现在,获得更好模型的明显方法是深入研究:我们在基本RNN中的隐状态和输出激活之间只有一个线性层,所以也许我们会得到更好的结果。
## Multilayer RNNs
多层RNN
In a multilayer RNN, we pass the activations from our recurrent neural network into a second recurrent neural network, like in <<stacked_rnn_rep>>.
在多层RNN中,我们将循环神经网络的激活传递到第二个循环神经网络,如<>中。
<img alt="2-layer RNN" width="550" caption="2-layer RNN" id="stacked_rnn_rep" src="images/att_00025.png">
The unrolled representation is shown in <<unrolled_stack_rep>> (similar to <<lm_rep>>).
展开的表示显示在<>中(类似于<>)。
<img alt="2-layer unrolled RNN" width="500" caption="Two-layer unrolled RNN" id="unrolled_stack_rep" src="images/att_00026.png">
Let's see how to implement this in practice.
现在我们来看看怎样在实际中实现。
### The Model
模型
We can save some time by using PyTorch's `RNN` class, which implements exactly what we created earlier, but also gives us the option to stack multiple RNNs, as we have discussed:
我们可以通过使用 PyTorch的RNN类来节省一些时间,它完全实现了我们之前创建的内容,但也为我们提供了堆叠多个RNN的选项,正如我们已经讨论过的:
<code>
class LMModel5(Module):
def __init__(self, vocab_sz, n_hidden, n_layers):
self.i_h = nn.Embedding(vocab_sz, n_hidden)
self.rnn = nn.RNN(n_hidden, n_hidden, n_layers, batch_first=True)
self.h_o = nn.Linear(n_hidden, vocab_sz)
self.h = torch.zeros(n_layers, bs, n_hidden)
def forward(self, x):
res,h = self.rnn(self.i_h(x), self.h)
self.h = h.detach()
return self.h_o(res)
def reset(self): self.h.zero_()
</code>
<code>
learn = Learner(dls, LMModel5(len(vocab), 64, 2),
loss_func=CrossEntropyLossFlat(),
metrics=accuracy, cbs=ModelResetter)
learn.fit_one_cycle(15, 3e-3)
</code>
Now that's disappointing... our previous single-layer RNN performed better. Why? The reason is that we have a deeper model, leading to exploding or vanishing activations.
现在这令人失望...我们之前的单层RNN表现更好。为什么?原因是我们有一个更深的模型,导致爆炸或消失激活。
### Exploding or Disappearing Activations
爆炸或消失激活
In practice, creating accurate models from this kind of RNN is difficult. We will get better results if we call `detach` less often, and have more layers—this gives our RNN a longer time horizon to learn from, and richer features to create. But it also means we have a deeper model to train. The key challenge in the development of deep learning has been figuring out how to train these kinds of models.
The reason this is challenging is because of what happens when you multiply by a matrix many times. Think about what happens when you multiply by a number many times. For example, if you multiply by 2, starting at 1, you get the sequence 1, 2, 4, 8,... after 32 steps you are already at 4,294,967,296. A similar issue happens if you multiply by 0.5: you get 0.5, 0.25, 0.125… and after 32 steps it's 0.00000000023. As you can see, multiplying by a number even slightly higher or lower than 1 results in an explosion or disappearance of our starting number, after just a few repeated multiplications.
Because matrix multiplication is just multiplying numbers and adding them up, exactly the same thing happens with repeated matrix multiplications. And that's all a deep neural network is —each extra layer is another matrix multiplication. This means that it is very easy for a deep neural network to end up with extremely large or extremely small numbers.
This is a problem, because the way computers store numbers (known as "floating point") means that they become less and less accurate the further away the numbers get from zero. The diagram in <<float_prec>>, from the excellent article ["What You Never Wanted to Know About Floating Point but Will Be Forced to Find Out"](http://www.volkerschatz.com/science/float.html), shows how the precision of floating-point numbers varies over the number line.
在实践中,从这种RNN创建准确的模型是很困难的。如果我们更少地调用分离,并拥有更多的层,我们将获得更好的结果——这给了我们的RNN更长的时间范围来学习,以及更丰富的特征来创建。但这也意味着我们有更深层次的模型要训练。深度学习发展的关键挑战一直是弄清楚如何训练这类模型。
这之所以具有挑战性,是因为当你多次乘以一个矩阵时会发生什么。想想当你多次乘以一个数字时会发生什么。例如,如果你乘以2,从1开始,你得到的序列是1,2,4,8,...32步后你已经是4,294,967,296了。如果你乘以0.5也会发生类似的问题:你得到0.5,0.25,0.125...32步后是0.00000000023。正如你所看到的,乘以一个比1稍高或稍低的数字会导致我们的起始数爆炸或消失,只需重复几次乘法。
因为矩阵乘法只是将数字相乘并相加,重复的矩阵乘法也会发生完全相同的事情。这就是深度神经网络的全部——每多一层都是另一个矩阵乘法。这意味着深度神经网络很容易得到非常大或非常小的数字。
这是一个问题,因为计算机存储数字的方式(称为“浮点”)意味着数字离零越远,它们就变得越来越不准确。<>中的图表来自优秀文章“你从未想过要知道的关于浮点但将被迫找出”,显示了浮点数的查准率/精确度如何在数线上变化。
<img alt="Precision of floating point numbers" width="1000" caption="Precision of floating-point numbers" id="float_prec" src="images/fltscale.svg">
This inaccuracy means that often the gradients calculated for updating the weights end up as zero or infinity for deep networks. This is commonly referred to as the *vanishing gradients* or *exploding gradients* problem. It means that in SGD, the weights are either not updated at all or jump to infinity. Either way, they won't improve with training.
Researchers have developed a number of ways to tackle this problem, which we will be discussing later in the book. One option is to change the definition of a layer in a way that makes it less likely to have exploding activations. We'll look at the details of how this is done in <<chapter_convolutions>>, when we discuss batch normalization, and <<chapter_resnet>>, when we discuss ResNets, although these details don't generally matter in practice (unless you are a researcher that is creating new approaches to solving this problem). Another strategy for dealing with this is by being careful about initialization, which is a topic we'll investigate in <<chapter_foundations>>.
For RNNs, there are two types of layers that are frequently used to avoid exploding activations: *gated recurrent units* (GRUs) and *long short-term memory* (LSTM) layers. Both of these are available in PyTorch, and are drop-in replacements for the RNN layer. We will only cover LSTMs in this book; there are plenty of good tutorials online explaining GRUs, which are a minor variant on the LSTM design.
这种不准确性意味着,对于深度网络,为更新权重而计算的梯度通常最终为零或无穷大。这通常被称为梯度消失或梯度爆炸问题。这意味着在SGD中,权重要么根本不更新,要么跳到无穷大。无论哪种方式,它们都不会通过训练得到改善。
研究人员已经开发了许多方法来解决这个问题,我们将在本书的后面讨论。一种选择是改变层的定义,使其不太可能发生爆炸式激活。当我们讨论批处理规范化时,我们将在<>和<>中查看如何做到这一点的细节,尽管这些细节在实践中通常并不重要(除非您是正在创建新方法来解决这个问题的研究人员)。另一种解决这个问题的策略是小心初始化,这是我们将在<>中研究的主题。
对于RNN,有两种类型的层经常用于避免爆炸激活:门控循环单元(GRU)和长时记忆(LSTM)层。这两种都可以在PyTorch中使用,并且是RNN层的直接替代品。我们将在本书中只介绍LSTM;网上有很多很好的教程解释GRU,这是LSTM设计的一个小变体。
## LSTM
LSTM is an architecture that was introduced back in 1997 by Jürgen Schmidhuber and Sepp Hochreiter. In this architecture, there are not one but two hidden states. In our base RNN, the hidden state is the output of the RNN at the previous time step. That hidden state is then responsible for two things:
- Having the right information for the output layer to predict the correct next token
- Retaining memory of everything that happened in the sentence
Consider, for example, the sentences "Henry has a dog and he likes his dog very much" and "Sophie has a dog and she likes her dog very much." It's very clear that the RNN needs to remember the name at the beginning of the sentence to be able to predict *he/she* or *his/her*.
In practice, RNNs are really bad at retaining memory of what happened much earlier in the sentence, which is the motivation to have another hidden state (called *cell state*) in the LSTM. The cell state will be responsible for keeping *long short-term memory*, while the hidden state will focus on the next token to predict. Let's take a closer look at how this is achieved and build an LSTM from scratch.
LSTM是一个由Jürgen Schmidhuber和Sepp Hochreiter于1997年引入的架构。在这个架构中,不是一个而是两个隐藏状态。在我们的基础RNN中,隐状态是RNN在前一个时间步的输出。然后,隐状态负责两件事:
为输出层提供正确的信息来预测正确的下一个令牌
保留对句子中发生的一切的记忆
例如,考虑句子“亨利有一只狗,他非常喜欢他的狗”和“索菲有一只狗,她非常喜欢她的狗”很明显,RNN需要记住句子开头的名字才能预测他/她或他/她。
在实践中,RNN真的不擅长保留对句子中更早发生的事情的记忆,这是LSTM中另一个隐状态(称为单元格状态)的动机。单元格状态将负责保持长时记忆,而隐状态将专注于下一个要预测的令牌。让我们仔细看看这是如何实现的,并从头开始构建LSTM。
### Building an LSTM from Scratch
从头开始构建LSTM
In order to build an LSTM, we first have to understand its architecture. <<lstm>> shows its inner structure.
<img src="images/LSTM.png" id="lstm" caption="Architecture of an LSTM" alt="A graph showing the inner architecture of an LSTM" width="700">
为了构建LSTM,我们首先必须了解它的架构。<>显示了它的内部结构。
In this picture, our input $x_{t}$ enters on the left with the previous hidden state ($h_{t-1}$) and cell state ($c_{t-1}$). The four orange boxes represent four layers (our neural nets) with the activation being either sigmoid ($\sigma$) or tanh. tanh is just a sigmoid function rescaled to the range -1 to 1. Its mathematical expression can be written like this:
$$\tanh(x) = \frac{e^{x} - e^{-x}}{e^{x}+e^{-x}} = 2 \sigma(2x) - 1$$
where $\sigma$ is the sigmoid function. The green circles are elementwise operations. What goes out on the right is the new hidden state ($h_{t}$) and new cell state ($c_{t}$), ready for our next input. The new hidden state is also used as output, which is why the arrow splits to go up.
Let's go over the four neural nets (called *gates*) one by one and explain the diagram—but before this, notice how very little the cell state (at the top) is changed. It doesn't even go directly through a neural net! This is exactly why it will carry on a longer-term state.
First, the arrows for input and old hidden state are joined together. In the RNN we wrote earlier in this chapter, we were adding them together. In the LSTM, we stack them in one big tensor. This means the dimension of our embeddings (which is the dimension of $x_{t}$) can be different than the dimension of our hidden state. If we call those `n_in` and `n_hid`, the arrow at the bottom is of size `n_in + n_hid`; thus all the neural nets (orange boxes) are linear layers with `n_in + n_hid` inputs and `n_hid` outputs.
The first gate (looking from left to right) is called the *forget gate*. Since it’s a linear layer followed by a sigmoid, its output will consist of scalars between 0 and 1. We multiply this result by the cell state to determine which information to keep and which to throw away: values closer to 0 are discarded and values closer to 1 are kept. This gives the LSTM the ability to forget things about its long-term state. For instance, when crossing a period or an `xxbos` token, we would expect to it to (have learned to) reset its cell state.
The second gate is called the *input gate*. It works with the third gate (which doesn't really have a name but is sometimes called the *cell gate*) to update the cell state. For instance, we may see a new gender pronoun, in which case we'll need to replace the information about gender that the forget gate removed. Similar to the forget gate, the input gate decides which elements of the cell state to update (values close to 1) or not (values close to 0). The third gate determines what those updated values are, in the range of –1 to 1 (thanks to the tanh function). The result is then added to the cell state.
The last gate is the *output gate*. It determines which information from the cell state to use to generate the output. The cell state goes through a tanh before being combined with the sigmoid output from the output gate, and the result is the new hidden state.
In terms of code, we can write the same steps like this:
在这张图中,我们的输入在左侧输入之前的隐藏状态( ℎ𝑡−1 )和单元格状态( 𝑐𝑡−1 )。四个橙色框代表四层(我们的神经网络),激活要么是sigmoid ( 𝜎 ) 要么是tanh。tanh只是一个重新缩放到-1到1范围内的sigmoid函数。它的数学表达式可以这样写:
$$\tanh(x) = \frac{e^{x} - e^{-x}}{e^{x}+e^{-x}} = 2 \sigma(2x) - 1$$
其中𝜎是sigmoid函数。绿色圆圈是元素操作。右侧输出的是新的隐藏状态 ( ℎ𝑡 ) 和新的单元格状态 ( 𝑐𝑡 ), 为我们的下一个输入做好准备。新的隐藏状态也用作输出,这就是箭头向上分裂的原因。
让我们一个接一个地检查四个神经网络(称为门)并解释图表——但在此之前,请注意细胞状态(在顶部)变化很小。它甚至没有直接通过神经网络!这正是为什么它会持续更长时间的状态。
首先,输入和旧隐藏状态的箭头连接在一起。在本章前面写的RNN中,我们将它们相加在一起。在LSTM中,我们将它们堆叠在一个大张量中。这意味着我们嵌入的维度(即的维度)可能与我们隐藏状态的维度不同。如果我们调用这些n_in和n_hid,底部的箭头大小为n_in+n_hid;因此所有神经网络(橙色框)都是线性层,具有n_in+n_hid输入和n_hid输出。
第一个门(从左到右)被称为遗忘门。由于它是一个线性层,后跟一个sigmoid,它的输出将由0到1之间的标量组成。我们将此结果乘以单元格状态以确定哪些信息要保留,哪些信息要丢弃:更接近0的值被丢弃,更接近1的值被保留。这使LSTM能够忘记有关其长期状态的事情。例如,当跨越一个句点或xxbos标记时,我们希望它(已经学会)重置其单元格状态。
第二个门称为输入门。它与第三个门(实际上没有名字,但有时称为单元格门)一起更新单元格状态。例如,我们可能会看到一个新的性别代词,在这种情况下,我们需要替换忘记门删除的有关性别的信息。与忘记门类似,输入门决定要更新单元格状态的哪些元素(值接近1)或不更新(值接近0)。第三个门确定这些更新的值是什么,范围在-1到1之间(感谢tanh函数)。然后将结果添加到单元格状态。
最后一个门是输出门。它决定使用来自单元状态的哪些信息来生成输出。单元状态在与输出门的sigmoid输出组合之前经过tanh,结果是新的隐藏状态。
在代码方面,我们可以编写如下相同的步骤:
<code>
class LSTMCell(Module):
def __init__(self, ni, nh):
self.forget_gate = nn.Linear(ni + nh, nh)
self.input_gate = nn.Linear(ni + nh, nh)
self.cell_gate = nn.Linear(ni + nh, nh)
self.output_gate = nn.Linear(ni + nh, nh)
def forward(self, input, state):
h,c = state
h = torch.cat([h, input], dim=1)
forget = torch.sigmoid(self.forget_gate(h))
c = c * forget
inp = torch.sigmoid(self.input_gate(h))
cell = torch.tanh(self.cell_gate(h))
c = c + inp * cell
out = torch.sigmoid(self.output_gate(h))
h = out * torch.tanh(c)
return h, (h,c)
</code>
In practice, we can then refactor the code. Also, in terms of performance, it's better to do one big matrix multiplication than four smaller ones (that's because we only launch the special fast kernel on the GPU once, and it gives the GPU more work to do in parallel). The stacking takes a bit of time (since we have to move one of the tensors around on the GPU to have it all in a contiguous array), so we use two separate layers for the input and the hidden state. The optimized and refactored code then looks like this:
在实践中,我们可以重构代码。此外,就性能而言,做一个大矩阵乘法比做四个小矩阵乘法更好(这是因为我们只在GPU上启动一次特殊的快速内核,它让GPU有更多的工作要并行完成)。堆叠需要一点时间(因为我们必须在GPU上移动一个张量以将其全部放在一个连续的数组中),所以我们使用两个单独的层来输入和隐状态。优化和重构的代码如下所示:
在实践中,我们可以重构代码。此外,就性能而言,做一个大矩阵乘法比做四个小矩阵乘法更好(这是因为我们只在GPU上启动一次特殊的快速内核,它让GPU有更多的工作要并行完成)。堆叠需要一点时间(因为我们必须在GPU上移动一个张量以将其全部放在一个连续的数组中),所以我们使用两个单独的层来输入和隐藏状态。优化和重构的代码如下所示: ...
<code>
class LSTMCell(Module):
def __init__(self, ni, nh):
self.ih = nn.Linear(ni,4*nh)
self.hh = nn.Linear(nh,4*nh)
def forward(self, input, state):
h,c = state
# One big multiplication for all the gates is better than 4 smaller ones
gates = (self.ih(input) + self.hh(h)).chunk(4, 1)
ingate,forgetgate,outgate = map(torch.sigmoid, gates[:3])
cellgate = gates[3].tanh()
c = (forgetgate*c) + (ingate*cellgate)
h = outgate * c.tanh()
return h, (h,c)
</code>
Here we use the PyTorch `chunk` method to split our tensor into four pieces. It works like this:
这里我们使用PyTorch的chunk方法将我们的张量分成四块。它是这样工作的:
<code>
t = torch.arange(0,10); t
</code>
<code>
t.chunk(2)
</code>
Let's now use this architecture to train a language model!
现在让我们使用这种架构来训练语言模型!
### Training a Language Model Using LSTMs
使用LSTMs训练一个语言模型
Here is the same network as `LMModel5`, using a two-layer LSTM. We can train it at a higher learning rate, for a shorter time, and get better accuracy:
这是与LMModel5相同的网络,使用两层LSTM。我们可以以更高的学习率、更短的时间训练它,并获得更好的准确率:
<code>
class LMModel6(Module):
def __init__(self, vocab_sz, n_hidden, n_layers):
self.i_h = nn.Embedding(vocab_sz, n_hidden)
self.rnn = nn.LSTM(n_hidden, n_hidden, n_layers, batch_first=True)
self.h_o = nn.Linear(n_hidden, vocab_sz)
self.h = [torch.zeros(n_layers, bs, n_hidden) for _ in range(2)]
def forward(self, x):
res,h = self.rnn(self.i_h(x), self.h)
self.h = [h_.detach() for h_ in h]
return self.h_o(res)
def reset(self):
for h in self.h: h.zero_()
</code>
<code>
learn = Learner(dls, LMModel6(len(vocab), 64, 2),
loss_func=CrossEntropyLossFlat(),
metrics=accuracy, cbs=ModelResetter)
learn.fit_one_cycle(15, 1e-2)
</code>
Now that's better than a multilayer RNN! We can still see there is a bit of overfitting, however, which is a sign that a bit of regularization might help.
这比多层RNN更好!然而,我们仍然可以看到有一点过拟合,这是一点正则化可能有所帮助的迹象。
## Regularizing an LSTM
正则化一个LSTM
Recurrent neural networks, in general, are hard to train, because of the problem of vanishing activations and gradients we saw before. Using LSTM (or GRU) cells makes training easier than with vanilla RNNs, but they are still very prone to overfitting. Data augmentation, while a possibility, is less often used for text data than for images because in most cases it requires another model to generate random augmentations (e.g., by translating the text into another language and then back into the original language). Overall, data augmentation for text data is currently not a well-explored space.
However, there are other regularization techniques we can use instead to reduce overfitting, which were thoroughly studied for use with LSTMs in the paper ["Regularizing and Optimizing LSTM Language Models"](https://arxiv.org/abs/1708.02182) by Stephen Merity, Nitish Shirish Keskar, and Richard Socher. This paper showed how effective use of *dropout*, *activation regularization*, and *temporal activation regularization* could allow an LSTM to beat state-of-the-art results that previously required much more complicated models. The authors called an LSTM using these techniques an *AWD-LSTM*. We'll look at each of these techniques in turn.
一般来说,循环神经网络很难训练,因为我们之前看到的激活和梯度消失的问题。使用LSTM(或GRU)细胞比使用普通RNN更容易训练,但它们仍然非常容易过拟合。数据增强虽然是一种可能性,但经常用于图像而不是文本,因为在大多数情况下,它需要另一个模型来生成随机增强(例如,通过将文本翻译成另一种语言,然后返回到原始语言)。总的来说,文本数据的数据目前不是一个很好探索的空间。
### Dropout
Dropout is a regularization technique that was introduced by Geoffrey Hinton et al. in [Improving neural networks by preventing co-adaptation of feature detectors](https://arxiv.org/abs/1207.0580). The basic idea is to randomly change some activations to zero at training time. This makes sure all neurons actively work toward the output, as seen in <<img_dropout>> (from "Dropout: A Simple Way to Prevent Neural Networks from Overfitting" by Nitish Srivastava et al.).
<img src="images/Dropout1.png" alt="A figure from the article showing how neurons go off with dropout" width="800" id="img_dropout" caption="Applying dropout in a neural network (courtesy of Nitish Srivastava et al.)">
Hinton used a nice metaphor when he explained, in an interview, the inspiration for dropout:
> : I went to my bank. The tellers kept changing and I asked one of them why. He said he didn’t know but they got moved around a lot. I figured it must be because it would require cooperation between employees to successfully defraud the bank. This made me realize that randomly removing a different subset of neurons on each example would prevent conspiracies and thus reduce overfitting.
In the same interview, he also explained that neuroscience provided additional inspiration:
> : We don't really know why neurons spike. One theory is that they want to be noisy so as to regularize, because we have many more parameters than we have data points. The idea of dropout is that if you have noisy activations, you can afford to use a much bigger model.
This explains the idea behind why dropout helps to generalize: first it helps the neurons to cooperate better together, then it makes the activations more noisy, thus making the model more robust.
We can see, however, that if we were to just zero those activations without doing anything else, our model would have problems training: if we go from the sum of five activations (that are all positive numbers since we apply a ReLU) to just two, this won't have the same scale. Therefore, if we apply dropout with a probability `p`, we rescale all activations by dividing them by `1-p` (on average `p` will be zeroed, so it leaves `1-p`), as shown in <<img_dropout1>>.
<img src="images/Dropout.png" alt="A figure from the article introducing dropout showing how a neuron is on/off" width="600" id="img_dropout1" caption="Why scale the activations when applying dropout (courtesy of Nitish Srivastava et al.)">
This is a full implementation of the dropout layer in PyTorch (although PyTorch's native layer is actually written in C, not Python):
<code>
class Dropout(Module):
def __init__(self, p): self.p = p
def forward(self, x):
if not self.training: return x
mask = x.new(*x.shape).bernoulli_(1-p)
return x * mask.div_(1-p)
</code>
The `bernoulli_` method is creating a tensor of random zeros (with probability `p`) and ones (with probability `1-p`), which is then multiplied with our input before dividing by `1-p`. Note the use of the `training` attribute, which is available in any PyTorch `nn.Module`, and tells us if we are doing training or inference.
> note: Do Your Own Experiments: In previous chapters of the book we'd be adding a code example for `bernoulli_` here, so you can see exactly how it works. But now that you know enough to do this yourself, we're going to be doing fewer and fewer examples for you, and instead expecting you to do your own experiments to see how things work. In this case, you'll see in the end-of-chapter questionnaire that we're asking you to experiment with `bernoulli_`—but don't wait for us to ask you to experiment to develop your understanding of the code we're studying; go ahead and do it anyway!
Using dropout before passing the output of our LSTM to the final layer will help reduce overfitting. Dropout is also used in many other models, including the default CNN head used in `fastai.vision`, and is available in `fastai.tabular` by passing the `ps` parameter (where each "p" is passed to each added `Dropout` layer), as we'll see in <<chapter_arch_details>>.
Dropout has different behavior in training and validation mode, which we specified using the `training` attribute in `Dropout`. Calling the `train` method on a `Module` sets `training` to `True` (both for the module you call the method on and for every module it recursively contains), and `eval` sets it to `False`. This is done automatically when calling the methods of `Learner`, but if you are not using that class, remember to switch from one to the other as needed.
### Activation Regularization and Temporal Activation Regularization
*Activation regularization* (AR) and *temporal activation regularization* (TAR) are two regularization methods very similar to weight decay, discussed in <<chapter_collab>>. When applying weight decay, we add a small penalty to the loss that aims at making the weights as small as possible. For activation regularization, it's the final activations produced by the LSTM that we will try to make as small as possible, instead of the weights.
To regularize the final activations, we have to store those somewhere, then add the means of the squares of them to the loss (along with a multiplier `alpha`, which is just like `wd` for weight decay):
``` python
loss += alpha * activations.pow(2).mean()
```
Temporal activation regularization is linked to the fact we are predicting tokens in a sentence. That means it's likely that the outputs of our LSTMs should somewhat make sense when we read them in order. TAR is there to encourage that behavior by adding a penalty to the loss to make the difference between two consecutive activations as small as possible: our activations tensor has a shape `bs x sl x n_hid`, and we read consecutive activations on the sequence length axis (the dimension in the middle). With this, TAR can be expressed as:
``` python
loss += beta * (activations[:,1:] - activations[:,:-1]).pow(2).mean()
```
`alpha` and `beta` are then two hyperparameters to tune. To make this work, we need our model with dropout to return three things: the proper output, the activations of the LSTM pre-dropout, and the activations of the LSTM post-dropout. AR is often applied on the dropped-out activations (to not penalize the activations we turned into zeros afterward) while TAR is applied on the non-dropped-out activations (because those zeros create big differences between two consecutive time steps). There is then a callback called `RNNRegularizer` that will apply this regularization for us.
### Training a Weight-Tied Regularized LSTM
We can combine dropout (applied before we go into our output layer) with AR and TAR to train our previous LSTM. We just need to return three things instead of one: the normal output of our LSTM, the dropped-out activations, and the activations from our LSTMs. The last two will be picked up by the callback `RNNRegularization` for the contributions it has to make to the loss.
Another useful trick we can add from [the AWD LSTM paper](https://arxiv.org/abs/1708.02182) is *weight tying*. In a language model, the input embeddings represent a mapping from English words to activations, and the output hidden layer represents a mapping from activations to English words. We might expect, intuitively, that these mappings could be the same. We can represent this in PyTorch by assigning the same weight matrix to each of these layers:
self.h_o.weight = self.i_h.weight
In `LMModel7`, we include these final tweaks:
<code>
class LMModel7(Module):
def __init__(self, vocab_sz, n_hidden, n_layers, p):
self.i_h = nn.Embedding(vocab_sz, n_hidden)
self.rnn = nn.LSTM(n_hidden, n_hidden, n_layers, batch_first=True)
self.drop = nn.Dropout(p)
self.h_o = nn.Linear(n_hidden, vocab_sz)
self.h_o.weight = self.i_h.weight
self.h = [torch.zeros(n_layers, bs, n_hidden) for _ in range(2)]
def forward(self, x):
raw,h = self.rnn(self.i_h(x), self.h)
out = self.drop(raw)
self.h = [h_.detach() for h_ in h]
return self.h_o(out),raw,out
def reset(self):
for h in self.h: h.zero_()
</code>
We can create a regularized `Learner` using the `RNNRegularizer` callback:
<code>
learn = Learner(dls, LMModel7(len(vocab), 64, 2, 0.5),
loss_func=CrossEntropyLossFlat(), metrics=accuracy,
cbs=[ModelResetter, RNNRegularizer(alpha=2, beta=1)])
</code>
A `TextLearner` automatically adds those two callbacks for us (with those values for `alpha` and `beta` as defaults), so we can simplify the preceding line to:
<code>
learn = TextLearner(dls, LMModel7(len(vocab), 64, 2, 0.4),
loss_func=CrossEntropyLossFlat(), metrics=accuracy)
</code>
We can then train the model, and add additional regularization by increasing the weight decay to `0.1`:
<code>
learn.fit_one_cycle(15, 1e-2, wd=0.1)
</code>
Now this is far better than our previous model!
## Conclusion
You have now seen everything that is inside the AWD-LSTM architecture we used in text classification in <<chapter_nlp>>. It uses dropout in a lot more places:
- Embedding dropout (inside the embedding layer, drops some random lines of embeddings)
- Input dropout (applied after the embedding layer)
- Weight dropout (applied to the weights of the LSTM at each training step)
- Hidden dropout (applied to the hidden state between two layers)
This makes it even more regularized. Since fine-tuning those five dropout values (including the dropout before the output layer) is complicated, we have determined good defaults and allow the magnitude of dropout to be tuned overall with the `drop_mult` parameter you saw in that chapter (which is multiplied by each dropout).
Another architecture that is very powerful, especially in "sequence-to-sequence" problems (that is, problems where the dependent variable is itself a variable-length sequence, such as language translation), is the Transformers architecture. You can find it in a bonus chapter on the [book's website](https://book.fast.ai/).
## Questionnaire
1. If the dataset for your project is so big and complicated that working with it takes a significant amount of time, what should you do?
1. Why do we concatenate the documents in our dataset before creating a language model?
1. To use a standard fully connected network to predict the fourth word given the previous three words, what two tweaks do we need to make to our model?
1. How can we share a weight matrix across multiple layers in PyTorch?
1. Write a module that predicts the third word given the previous two words of a sentence, without peeking.
1. What is a recurrent neural network?
1. What is "hidden state"?
1. What is the equivalent of hidden state in ` LMModel1`?
1. To maintain the state in an RNN, why is it important to pass the text to the model in order?
1. What is an "unrolled" representation of an RNN?
1. Why can maintaining the hidden state in an RNN lead to memory and performance problems? How do we fix this problem?
1. What is "BPTT"?
1. Write code to print out the first few batches of the validation set, including converting the token IDs back into English strings, as we showed for batches of IMDb data in <<chapter_nlp>>.
1. What does the `ModelResetter` callback do? Why do we need it?
1. What are the downsides of predicting just one output word for each three input words?
1. Why do we need a custom loss function for `LMModel4`?
1. Why is the training of `LMModel4` unstable?
1. In the unrolled representation, we can see that a recurrent neural network actually has many layers. So why do we need to stack RNNs to get better results?
1. Draw a representation of a stacked (multilayer) RNN.
1. Why should we get better results in an RNN if we call `detach` less often? Why might this not happen in practice with a simple RNN?
1. Why can a deep network result in very large or very small activations? Why does this matter?
1. In a computer's floating-point representation of numbers, which numbers are the most precise?
1. Why do vanishing gradients prevent training?
1. Why does it help to have two hidden states in the LSTM architecture? What is the purpose of each one?
1. What are these two states called in an LSTM?
1. What is tanh, and how is it related to sigmoid?
1. What is the purpose of this code in `LSTMCell`: `h = torch.cat([h, input], dim=1)`
1. What does `chunk` do in PyTorch?
1. Study the refactored version of `LSTMCell` carefully to ensure you understand how and why it does the same thing as the non-refactored version.
1. Why can we use a higher learning rate for `LMModel6`?
1. What are the three regularization techniques used in an AWD-LSTM model?
1. What is "dropout"?
1. Why do we scale the acitvations with dropout? Is this applied during training, inference, or both?
1. What is the purpose of this line from `Dropout`: `if not self.training: return x`
1. Experiment with `bernoulli_` to understand how it works.
1. How do you set your model in training mode in PyTorch? In evaluation mode?
1. Write the equation for activation regularization (in math or code, as you prefer). How is it different from weight decay?
1. Write the equation for temporal activation regularization (in math or code, as you prefer). Why wouldn't we use this for computer vision problems?
1. What is "weight tying" in a language model?
### Further Research
1. In ` LMModel2`, why can `forward` start with `h=0`? Why don't we need to say `h=torch.zeros(...)`?
1. Write the code for an LSTM from scratch (you may refer to <<lstm>>).
1. Search the internet for the GRU architecture and implement it from scratch, and try training a model. See if you can get results similar to those we saw in this chapter. Compare your results to the results of PyTorch's built in `GRU` module.
1. Take a look at the source code for AWD-LSTM in fastai, and try to map each of the lines of code to the concepts shown in this chapter.
|
{
"filename": "12_nlp_dive_1.ipynb",
"repository": "immc-lab/fastbook-zh",
"query": "transformed_from_existing",
"size": 133161,
"sha": ""
}
|
# Untitled-Copy1.ipynb
Repository: Sshanu/QALearn
<code>
import os
</code>
<code>
os.system("pdfminer/tools/pdf2txt.py " + "docs/pdfs/02.pdf" + " > " + "02" + ".txt")
</code>
<code>
os.system("pdfminer/tools/pdf2txt.py " + "docs/pdfs/03.pdf" + " > " + "03" + ".txt")
os.system("pdfminer/tools/pdf2txt.py " + "docs/pdfs/04.pdf" + " > " + "04" + ".txt")
os.system("pdfminer/tools/pdf2txt.py " + "docs/pdfs/05.pdf" + " > " + "05" + ".txt")
</code>
<code>
pdfs = ['content.pdf', '01.pdf', '02.pdf', '03.pdf', '04.pdf', '05.pdf']
</code>
<code>
from PyPDF2 import PdfFileMerger
</code>
<code>
merger = PdfFileMerger()
for pdf in pdfs:
merger.append(open("docs/pdfs/" + pdf, 'rb'))
with open('result.pdf', 'wb') as fout:
merger.write(fout)
</code>
<code>
from file2id import file2id
from sim2id import sim2id
import pickle as pkl
</code>
<code>
index_list, sections, flag = file2id("docs/NCERT-Class-12-Biology.txt")
</code>
<code>
f = open("qalearn/media/data/NCERT-Class-12-Biology", "wb")
pkl.dump([index_list, sections], f)
f.close()
</code>
<code>
sections[-1]
</code>
<code>
import re
</code>
<code>
def breakdown(text, max_char):
regex = "\.\s"
match = re.search(regex, text[max_char-100: max_char+500])
print(match)
return text[:match.start()+1+max_char], text[max_char + match.start()+1:]
</code>
<code>
a, b = breakdown(sections[-1], 2000)
</code>
<code>
a
</code>
<code>
b
</code>
|
{
"filename": "Untitled-Copy1.ipynb",
"repository": "Sshanu/QALearn",
"query": "transformed_from_existing",
"size": 25216,
"sha": ""
}
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.