hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
d03bf34c90873d2cfcf9455c44c24f68ef0b1a1c | 4,003 | ipynb | Jupyter Notebook | mod09.ipynb | theabc50111/machine_learning_tibame | af4926537220e23d974e1299030dda573e7eb429 | [
"MIT"
] | null | null | null | mod09.ipynb | theabc50111/machine_learning_tibame | af4926537220e23d974e1299030dda573e7eb429 | [
"MIT"
] | null | null | null | mod09.ipynb | theabc50111/machine_learning_tibame | af4926537220e23d974e1299030dda573e7eb429 | [
"MIT"
] | null | null | null | 27.993007 | 104 | 0.590057 | [
[
[
"# Decision Tree \nref: http://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeClassifier.html",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.datasets import load_breast_cancer\nfrom sklearn.tree import DecisionTreeClassifier\n\n\n# Load data\ncancer = load_breast_cancer()\nprint(cancer.feature_names)\nprint(cancer.target_names)\nx = cancer.data \ny = cancer.target\n\nx_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, random_state=1)\n\nmodel = DecisionTreeClassifier(min_samples_leaf=3)\nmodel.fit(x_train, y_train)\n\ny_pred = model.predict(x_test)\n\n\naccuracy = accuracy_score(y_test, y_pred)\nnum_correct_samples = accuracy_score(y_test, y_pred, normalize=False)\n\n\nprint(f'number of correct sample: {num_correct_samples}')\nprint(f'accuracy: {accuracy}')\n",
"['mean radius' 'mean texture' 'mean perimeter' 'mean area'\n 'mean smoothness' 'mean compactness' 'mean concavity'\n 'mean concave points' 'mean symmetry' 'mean fractal dimension'\n 'radius error' 'texture error' 'perimeter error' 'area error'\n 'smoothness error' 'compactness error' 'concavity error'\n 'concave points error' 'symmetry error' 'fractal dimension error'\n 'worst radius' 'worst texture' 'worst perimeter' 'worst area'\n 'worst smoothness' 'worst compactness' 'worst concavity'\n 'worst concave points' 'worst symmetry' 'worst fractal dimension']\n['malignant' 'benign']\nnumber of correct sample: 109\naccuracy: 0.956140350877193\n"
]
],
[
[
"# Random Forest \nref: http://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\n\n# Load data\ncancer = load_breast_cancer()\nx = cancer.data \ny = cancer.target\n\nx_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)\n\n\nmodel = RandomForestClassifier(max_depth=6, n_estimators=10)\nmodel.fit(x_train, y_train)\n\ny_pred = model.predict(x_test)\n\n\naccuracy = accuracy_score(y_test, y_pred)\nnum_correct_samples = accuracy_score(y_test, y_pred, normalize=False)\n\nprint('number of correct sample: {}'.format(num_correct_samples))\nprint('accuracy: {}'.format(accuracy))",
"number of correct sample: 112\naccuracy: 0.9824561403508771\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d03bf834d9efc2da07119e36347e0259835c4b2d | 616,042 | ipynb | Jupyter Notebook | ml_training/dataset_generation/masks/segmentation_mask_metal.ipynb | viswambhar-yasa/LaDECO | 0172270a86c71e8c32913005ec07fd63293af0f7 | [
"MIT"
] | null | null | null | ml_training/dataset_generation/masks/segmentation_mask_metal.ipynb | viswambhar-yasa/LaDECO | 0172270a86c71e8c32913005ec07fd63293af0f7 | [
"MIT"
] | null | null | null | ml_training/dataset_generation/masks/segmentation_mask_metal.ipynb | viswambhar-yasa/LaDECO | 0172270a86c71e8c32913005ec07fd63293af0f7 | [
"MIT"
] | null | null | null | 308,021 | 616,041 | 0.96079 | [
[
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Mounted at /content/drive\n"
],
[
"cd /content/drive/MyDrive/ML-LaDECO/MLaDECO",
"/content/drive/MyDrive/ML-LaDECO/MLaDECO\n"
]
],
[
[
"### Importing libraries and methods from thermograms, ml_training and ultilites modules",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nprint('Project MLaDECO')\nprint('Author: Viswambhar Yasa')\nprint('Software version: 0.1')\nfrom sklearn.preprocessing import MinMaxScaler, StandardScaler\nimport tensorflow as tf\nfrom tensorflow.keras import models\nfrom thermograms.Utilities import Utilities\nfrom ml_training.dataset_generation.fourier_transformation import fourier_transformation\nfrom ml_training.dataset_generation.principal_componant_analysis import principal_componant_analysis\nfrom utilites.segmentation_colormap_anno import segmentation_colormap_anno\nfrom utilites.tolerance_maks_gen import tolerance_predicted_mask\nimport matplotlib.pyplot as plt",
"Project MLaDECO\nAuthor: Viswambhar Yasa\nSoftware version: 0.1\n"
]
],
[
[
"##### Importing dataset for training ",
"_____no_output_____"
]
],
[
[
"root_path = r'utilites/datasets'\ndata_file_name = r'metal_data.hdf5'\nthermal_class = Utilities()\nthermal_data,experiment_list=thermal_class.open_file(root_path, data_file_name,True)\nexperiment_name=r'2021-12-15-Materialstudie_Metallproben-ML3-laserbehandelte_Probe-1000W-10s'\nexperimental_data=thermal_data[experiment_name] ",
"Experiments in the file \n\n1 : 2021-12-15-Materialstudie_Metallproben-ML1-laserbehandelte_Probe-1000W-10s\n2 : 2021-12-15-Materialstudie_Metallproben-ML2-laserbehandelte_Probe-1000W-10s\n3 : 2021-12-15-Materialstudie_Metallproben-ML3-laserbehandelte_Probe-1000W-10s\n\n\nA total of 3 experiments are loaded in file \n\n"
]
],
[
[
"##### Checking the shape and file format of the thermographic experiment dataset",
"_____no_output_____"
]
],
[
[
"experimental_data",
"_____no_output_____"
]
],
[
[
"##### Identifying the reflection phase index",
"_____no_output_____"
]
],
[
[
"input_data, reflection_st_index, reflection_end_index = fourier_transformation(experimental_data,\n scaling_type='normalization', index=1)",
"reflection_start_index: 295 reflection_end_index: 798\nThe size of filtered data: (256, 256, 503)\n"
],
[
"from PIL import Image",
"_____no_output_____"
]
],
[
[
"##### Performing data normalization to improve the learning ability of machine learning model by scaling down the data between a smaller range",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import MinMaxScaler, StandardScaler \nfrom sklearn.model_selection import train_test_split\nexp_data=np.array(experimental_data)\nstandardizing = StandardScaler()\nstd_output_data = standardizing.fit_transform(\n exp_data.reshape(exp_data.shape[0], -1)).reshape(exp_data.shape)",
"_____no_output_____"
],
[
"normalizing = MinMaxScaler(feature_range=(0, 1))\nnrm_output_data = normalizing.fit_transform(\n exp_data.reshape(exp_data.shape[0], -1)).reshape(exp_data.shape)",
"_____no_output_____"
]
],
[
[
"##### Plotting thermograms after gaussian normalization and min max scaling operation ",
"_____no_output_____"
]
],
[
[
"from mpl_toolkits.axes_grid1 import make_axes_locatable\nfig = plt.figure(figsize=(10,5))\nax1 = fig.add_subplot(121)\nim1 = ax1.imshow(std_output_data[:,:,400].astype(np.float32), cmap='RdYlBu_r', interpolation='None')\nax1.set_title('Gaussian distribution scaling')\n\ndivider = make_axes_locatable(ax1)\ncax = divider.append_axes('right', size='5%', pad=0.05)\nfig.colorbar(im1, cax=cax, orientation='vertical')\nax1.axis('off')\nax2 = fig.add_subplot(122)\nim2 = ax2.imshow(nrm_output_data[:,:,400].astype(np.float32), cmap='RdYlBu_r', interpolation='None')\nax2.set_title('Min-Max Normalization')\nax2.axis('off')\ndivider = make_axes_locatable(ax2)\ncax = divider.append_axes('right', size='5%', pad=0.05)\nfig.colorbar(im2, cax=cax, orientation='vertical')\nplt.savefig(r\"Documents/temp/metal_scaling.png\",dpi=600,bbox_inches='tight',transparent=True)",
"_____no_output_____"
],
[
"plt.imshow(std_output_data[:,:,400].astype(np.float32),cmap='RdYlBu_r') \nplt.colorbar()",
"_____no_output_____"
],
[
"#experiment_name='2021-05-11 - Variantenvergleich - VarioTherm Halogenlampe - Winkel 30°'\nexperimental_data=np.array(thermal_data[experiment_name])",
"_____no_output_____"
],
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"def grayscale_image(data):\n \"\"\"\n Creates a gray scale image dataset from the input data \n\n Args:\n data (numpy array): Thermograms\n\n Returns:\n (numpy array): Gray scale images\n \"\"\"\n #print(data.shape)\n seq_data=np.zeros((data.shape))\n for i in range(data.shape[-1]):\n temp=np.expand_dims(data[:,:,i],axis=-1)\n #print(temp.shape)\n a_i=tf.keras.utils.array_to_img(temp).convert('L')\n #a_i=array_to_img(temp).convert('L')\n imgGray = tf.keras.utils.img_to_array(a_i)\n #print(imgGray.shape)\n seq_data[:,:,i]=np.squeeze(imgGray)\n return seq_data\nd=grayscale_image(experimental_data)",
"_____no_output_____"
],
[
"d.shape",
"_____no_output_____"
]
],
[
[
"#### Extracting information from gray scale image and saving it in png format",
"_____no_output_____"
]
],
[
[
"from PIL import Image\nfrom keras.preprocessing.image import array_to_img,img_to_array\ndata=experimental_data\n\nplt.figure()\nplt.imshow(d[:,:,500],cmap='RdYlBu_r')\n#plt.imshow(std_output_data[:,:,250].astype(np.float64),cmap='gray')\nplt.savefig(\"Documents/temp/metal_output.png\")\nplt.axis('off')",
"_____no_output_____"
],
[
"img=plt.imread('Documents/temp/metal_output.png')",
"_____no_output_____"
],
[
"img.shape",
"_____no_output_____"
]
],
[
[
"##### Performing principal companant analysis to features by filtering intensity ",
"_____no_output_____"
]
],
[
[
"EOFs=principal_componant_analysis(experimental_data)",
"_____no_output_____"
],
[
"img1 = Image.fromarray(EOFs[:,:,0].astype(np.int8))\n#img2 = Image.fromarray(EOFs[:,:,0].astype(np.float32))\nplt.imshow(np.squeeze(EOFs),cmap='YlOrRd_r')\nplt.colorbar()\nplt.savefig(\"Documents/temp/metal_PCA.png\",dpi=600,bbox_inches='tight',transparent=True)",
"_____no_output_____"
],
[
"mask=np.zeros(shape=(np.squeeze(EOFs).shape))\nmask[np.squeeze(EOFs) > 250]=1\nmask[np.squeeze(EOFs) < -450]=1\nplt.imsave('Documents/temp/metal_mask1.png',np.squeeze(mask),cmap='binary_r')",
"_____no_output_____"
],
[
"substrate=mask",
"_____no_output_____"
]
],
[
[
"##### Final mask of the dataset",
"_____no_output_____"
]
],
[
[
"plt.imshow(mask)",
"_____no_output_____"
],
[
"import cv2\nimg1 = cv2.imread('Documents/temp/metal_mask1.png',0)\nprint(img1.shape)",
"(256, 256)\n"
]
],
[
[
"##### converting the image format data to a numpy array format and scaling it between integer values based on number of features",
"_____no_output_____"
]
],
[
[
"img1[img1==255]=1\nplt.imshow(img1,cmap='binary_r')\nplt.colorbar()",
"_____no_output_____"
]
],
[
[
"##### Saving the segmentation mask",
"_____no_output_____"
]
],
[
[
"name='ml_training/dataset_generation/annots/'+experiment_name",
"_____no_output_____"
],
[
"np.save(name,img1)",
"_____no_output_____"
],
[
"ar=np.load(name+'.npy')",
"_____no_output_____"
],
[
"plt.imshow(ar,cmap='gray')\nplt.colorbar()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d03c240e0b13660f55bfb3d1ea065481ddd95689 | 59,637 | ipynb | Jupyter Notebook | 4-ml-clustering-and-retrieval/Week_6-assignment.ipynb | anilk991/Coursera-Machine-Learning-Specialization-UW | 9b43e0282b38f39c769f344ee42a30b2b4405a77 | [
"MIT"
] | 1 | 2020-10-21T12:57:23.000Z | 2020-10-21T12:57:23.000Z | 4-ml-clustering-and-retrieval/Week_6-assignment.ipynb | anilk991/Coursera-Machine-Learning-Specialization-UW | 9b43e0282b38f39c769f344ee42a30b2b4405a77 | [
"MIT"
] | null | null | null | 4-ml-clustering-and-retrieval/Week_6-assignment.ipynb | anilk991/Coursera-Machine-Learning-Specialization-UW | 9b43e0282b38f39c769f344ee42a30b2b4405a77 | [
"MIT"
] | null | null | null | 44.208302 | 645 | 0.557657 | [
[
[
"# Hierarchical Clustering",
"_____no_output_____"
],
[
"**Hierarchical clustering** refers to a class of clustering methods that seek to build a **hierarchy** of clusters, in which some clusters contain others. In this assignment, we will explore a top-down approach, recursively bipartitioning the data using k-means.",
"_____no_output_____"
],
[
"**Note to Amazon EC2 users**: To conserve memory, make sure to stop all the other notebooks before running this notebook.",
"_____no_output_____"
],
[
"## Import packages",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function # to conform python 2.x print to python 3.x\nimport turicreate\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport sys\nimport os\nimport time\nfrom scipy.sparse import csr_matrix\nfrom sklearn.cluster import KMeans\nfrom sklearn.metrics import pairwise_distances\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Load the Wikipedia dataset",
"_____no_output_____"
]
],
[
[
"wiki = turicreate.SFrame('people_wiki.sframe/')",
"_____no_output_____"
]
],
[
[
"As we did in previous assignments, let's extract the TF-IDF features:",
"_____no_output_____"
]
],
[
[
"wiki['tf_idf'] = turicreate.text_analytics.tf_idf(wiki['text'])",
"_____no_output_____"
]
],
[
[
"To run k-means on this dataset, we should convert the data matrix into a sparse matrix.",
"_____no_output_____"
]
],
[
[
"from em_utilities import sframe_to_scipy # converter\n\n# This will take about a minute or two.\nwiki = wiki.add_row_number()\ntf_idf, map_word_to_index = sframe_to_scipy(wiki, 'tf_idf')",
"_____no_output_____"
]
],
[
[
"To be consistent with the k-means assignment, let's normalize all vectors to have unit norm.",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import normalize\ntf_idf = normalize(tf_idf)",
"_____no_output_____"
]
],
[
[
"## Bipartition the Wikipedia dataset using k-means",
"_____no_output_____"
],
[
"Recall our workflow for clustering text data with k-means:\n\n1. Load the dataframe containing a dataset, such as the Wikipedia text dataset.\n2. Extract the data matrix from the dataframe.\n3. Run k-means on the data matrix with some value of k.\n4. Visualize the clustering results using the centroids, cluster assignments, and the original dataframe. We keep the original dataframe around because the data matrix does not keep auxiliary information (in the case of the text dataset, the title of each article).\n\nLet us modify the workflow to perform bipartitioning:\n\n1. Load the dataframe containing a dataset, such as the Wikipedia text dataset.\n2. Extract the data matrix from the dataframe.\n3. Run k-means on the data matrix with k=2.\n4. Divide the data matrix into two parts using the cluster assignments.\n5. Divide the dataframe into two parts, again using the cluster assignments. This step is necessary to allow for visualization.\n6. Visualize the bipartition of data.\n\nWe'd like to be able to repeat Steps 3-6 multiple times to produce a **hierarchy** of clusters such as the following:\n```\n (root)\n |\n +------------+-------------+\n | |\n Cluster Cluster\n +------+-----+ +------+-----+\n | | | |\n Cluster Cluster Cluster Cluster\n```\nEach **parent cluster** is bipartitioned to produce two **child clusters**. At the very top is the **root cluster**, which consists of the entire dataset.\n\nNow we write a wrapper function to bipartition a given cluster using k-means. There are three variables that together comprise the cluster:\n\n* `dataframe`: a subset of the original dataframe that correspond to member rows of the cluster\n* `matrix`: same set of rows, stored in sparse matrix format\n* `centroid`: the centroid of the cluster (not applicable for the root cluster)\n\nRather than passing around the three variables separately, we package them into a Python dictionary. The wrapper function takes a single dictionary (representing a parent cluster) and returns two dictionaries (representing the child clusters).",
"_____no_output_____"
]
],
[
[
"def bipartition(cluster, maxiter=400, num_runs=4, seed=None):\n '''cluster: should be a dictionary containing the following keys\n * dataframe: original dataframe\n * matrix: same data, in matrix format\n * centroid: centroid for this particular cluster'''\n \n data_matrix = cluster['matrix']\n dataframe = cluster['dataframe']\n \n # Run k-means on the data matrix with k=2. We use scikit-learn here to simplify workflow.\n kmeans_model = KMeans(n_clusters=2, max_iter=maxiter, n_init=num_runs, random_state=seed, n_jobs=1)\n kmeans_model.fit(data_matrix)\n centroids, cluster_assignment = kmeans_model.cluster_centers_, kmeans_model.labels_\n \n # Divide the data matrix into two parts using the cluster assignments.\n data_matrix_left_child, data_matrix_right_child = data_matrix[cluster_assignment==0], \\\n data_matrix[cluster_assignment==1]\n \n # Divide the dataframe into two parts, again using the cluster assignments.\n cluster_assignment_sa = turicreate.SArray(cluster_assignment) # minor format conversion\n dataframe_left_child, dataframe_right_child = dataframe[cluster_assignment_sa==0], \\\n dataframe[cluster_assignment_sa==1]\n \n \n # Package relevant variables for the child clusters\n cluster_left_child = {'matrix': data_matrix_left_child,\n 'dataframe': dataframe_left_child,\n 'centroid': centroids[0]}\n cluster_right_child = {'matrix': data_matrix_right_child,\n 'dataframe': dataframe_right_child,\n 'centroid': centroids[1]}\n \n return (cluster_left_child, cluster_right_child)",
"_____no_output_____"
]
],
[
[
"The following cell performs bipartitioning of the Wikipedia dataset. Allow 2+ minutes to finish.\n\nNote. For the purpose of the assignment, we set an explicit seed (`seed=1`) to produce identical outputs for every run. In pratical applications, you might want to use different random seeds for all runs.",
"_____no_output_____"
]
],
[
[
"%%time\nwiki_data = {'matrix': tf_idf, 'dataframe': wiki} # no 'centroid' for the root cluster\nleft_child, right_child = bipartition(wiki_data, maxiter=100, num_runs=1, seed=0)",
"/home/offbeat/Environments/turicreate/lib/python3.7/site-packages/sklearn/cluster/_kmeans.py:974: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 0.25.\n \" removed in 0.25.\", FutureWarning)\n"
]
],
[
[
"Let's examine the contents of one of the two clusters, which we call the `left_child`, referring to the tree visualization above.",
"_____no_output_____"
]
],
[
[
"left_child",
"_____no_output_____"
]
],
[
[
"And here is the content of the other cluster we named `right_child`.",
"_____no_output_____"
]
],
[
[
"right_child",
"_____no_output_____"
]
],
[
[
"## Visualize the bipartition",
"_____no_output_____"
],
[
"We provide you with a modified version of the visualization function from the k-means assignment. For each cluster, we print the top 5 words with highest TF-IDF weights in the centroid and display excerpts for the 8 nearest neighbors of the centroid.",
"_____no_output_____"
]
],
[
[
"def display_single_tf_idf_cluster(cluster, map_index_to_word):\n '''map_index_to_word: SFrame specifying the mapping betweeen words and column indices'''\n \n wiki_subset = cluster['dataframe']\n tf_idf_subset = cluster['matrix']\n centroid = cluster['centroid']\n \n # Print top 5 words with largest TF-IDF weights in the cluster\n idx = centroid.argsort()[::-1]\n for i in range(5):\n print('{0}:{1:.3f}'.format(map_index_to_word['category'], centroid[idx[i]])),\n print('')\n \n # Compute distances from the centroid to all data points in the cluster.\n distances = pairwise_distances(tf_idf_subset, [centroid], metric='euclidean').flatten()\n # compute nearest neighbors of the centroid within the cluster.\n nearest_neighbors = distances.argsort()\n # For 8 nearest neighbors, print the title as well as first 180 characters of text.\n # Wrap the text at 80-character mark.\n for i in range(8):\n text = ' '.join(wiki_subset[nearest_neighbors[i]]['text'].split(None, 25)[0:25])\n print('* {0:50s} {1:.5f}\\n {2:s}\\n {3:s}'.format(wiki_subset[nearest_neighbors[i]]['name'],\n distances[nearest_neighbors[i]], text[:90], text[90:180] if len(text) > 90 else ''))\n print('')",
"_____no_output_____"
]
],
[
[
"Let's visualize the two child clusters:",
"_____no_output_____"
]
],
[
[
"display_single_tf_idf_cluster(left_child, map_word_to_index)",
"113949:0.025\n113949:0.017\n113949:0.012\n113949:0.011\n113949:0.011\n\n* Anita Kunz 0.97401\n anita e kunz oc born 1956 is a canadianborn artist and illustratorkunz has lived in london\n new york and toronto contributing to magazines and working\n* Janet Jackson 0.97472\n janet damita jo jackson born may 16 1966 is an american singer songwriter and actress know\n n for a series of sonically innovative socially conscious and\n* Madonna (entertainer) 0.97475\n madonna louise ciccone tkoni born august 16 1958 is an american singer songwriter actress \n and businesswoman she achieved popularity by pushing the boundaries of lyrical\n* %C3%81ine Hyland 0.97536\n ine hyland ne donlon is emeritus professor of education and former vicepresident of univer\n sity college cork ireland she was born in 1942 in athboy co\n* Jane Fonda 0.97621\n jane fonda born lady jayne seymour fonda december 21 1937 is an american actress writer po\n litical activist former fashion model and fitness guru she is\n* Christine Robertson 0.97643\n christine mary robertson born 5 october 1948 is an australian politician and former austra\n lian labor party member of the new south wales legislative council serving\n* Pat Studdy-Clift 0.97643\n pat studdyclift is an australian author specialising in historical fiction and nonfictionb\n orn in 1925 she lived in gunnedah until she was sent to a boarding\n* Alexandra Potter 0.97646\n alexandra potter born 1970 is a british author of romantic comediesborn in bradford yorksh\n ire england and educated at liverpool university gaining an honors degree in\n\n"
],
[
"display_single_tf_idf_cluster(right_child, map_word_to_index)",
"113949:0.040\n113949:0.036\n113949:0.029\n113949:0.029\n113949:0.028\n\n* Todd Williams 0.95468\n todd michael williams born february 13 1971 in syracuse new york is a former major league \n baseball relief pitcher he attended east syracuseminoa high school\n* Gord Sherven 0.95622\n gordon r sherven born august 21 1963 in gravelbourg saskatchewan and raised in mankota sas\n katchewan is a retired canadian professional ice hockey forward who played\n* Justin Knoedler 0.95639\n justin joseph knoedler born july 17 1980 in springfield illinois is a former major league \n baseball catcherknoedler was originally drafted by the st louis cardinals\n* Chris Day 0.95648\n christopher nicholas chris day born 28 july 1975 is an english professional footballer who\n plays as a goalkeeper for stevenageday started his career at tottenham\n* Tony Smith (footballer, born 1957) 0.95653\n anthony tony smith born 20 february 1957 is a former footballer who played as a central de\n fender in the football league in the 1970s and\n* Ashley Prescott 0.95761\n ashley prescott born 11 september 1972 is a former australian rules footballer he played w\n ith the richmond and fremantle football clubs in the afl between\n* Leslie Lea 0.95802\n leslie lea born 5 october 1942 in manchester is an english former professional footballer \n he played as a midfielderlea began his professional career with blackpool\n* Tommy Anderson (footballer) 0.95818\n thomas cowan tommy anderson born 24 september 1934 in haddington is a scottish former prof\n essional footballer he played as a forward and was noted for\n\n"
]
],
[
[
"The right cluster consists of athletes and artists (singers and actors/actresses), whereas the left cluster consists of non-athletes and non-artists. So far, we have a single-level hierarchy consisting of two clusters, as follows:",
"_____no_output_____"
],
[
"```\n Wikipedia\n +\n |\n +--------------------------+--------------------+\n | |\n + +\n Non-athletes/artists Athletes/artists\n```",
"_____no_output_____"
],
[
"Is this hierarchy good enough? **When building a hierarchy of clusters, we must keep our particular application in mind.** For instance, we might want to build a **directory** for Wikipedia articles. A good directory would let you quickly narrow down your search to a small set of related articles. The categories of athletes and non-athletes are too general to facilitate efficient search. For this reason, we decide to build another level into our hierarchy of clusters with the goal of getting more specific cluster structure at the lower level. To that end, we subdivide both the `athletes/artists` and `non-athletes/artists` clusters.",
"_____no_output_____"
],
[
"## Perform recursive bipartitioning",
"_____no_output_____"
],
[
"### Cluster of athletes and artists",
"_____no_output_____"
],
[
"To help identify the clusters we've built so far, let's give them easy-to-read aliases:",
"_____no_output_____"
]
],
[
[
"non_athletes_artists = left_child\nathletes_artists = right_child",
"_____no_output_____"
]
],
[
[
"Using the bipartition function, we produce two child clusters of the athlete cluster:",
"_____no_output_____"
]
],
[
[
"# Bipartition the cluster of athletes and artists\nleft_child_athletes_artists, right_child_athletes_artists = bipartition(athletes_artists, maxiter=100, num_runs=6, seed=1)",
"/home/offbeat/Environments/turicreate/lib/python3.7/site-packages/sklearn/cluster/_kmeans.py:974: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 0.25.\n \" removed in 0.25.\", FutureWarning)\n"
]
],
[
[
"The left child cluster mainly consists of athletes:",
"_____no_output_____"
]
],
[
[
"display_single_tf_idf_cluster(left_child_athletes_artists, map_word_to_index)",
"113949:0.054\n113949:0.043\n113949:0.038\n113949:0.035\n113949:0.030\n\n* Tony Smith (footballer, born 1957) 0.94677\n anthony tony smith born 20 february 1957 is a former footballer who played as a central de\n fender in the football league in the 1970s and\n* Justin Knoedler 0.94746\n justin joseph knoedler born july 17 1980 in springfield illinois is a former major league \n baseball catcherknoedler was originally drafted by the st louis cardinals\n* Chris Day 0.94849\n christopher nicholas chris day born 28 july 1975 is an english professional footballer who\n plays as a goalkeeper for stevenageday started his career at tottenham\n* Todd Williams 0.94882\n todd michael williams born february 13 1971 in syracuse new york is a former major league \n baseball relief pitcher he attended east syracuseminoa high school\n* Todd Curley 0.95007\n todd curley born 14 january 1973 is a former australian rules footballer who played for co\n llingwood and the western bulldogs in the australian football league\n* Ashley Prescott 0.95015\n ashley prescott born 11 september 1972 is a former australian rules footballer he played w\n ith the richmond and fremantle football clubs in the afl between\n* Tommy Anderson (footballer) 0.95037\n thomas cowan tommy anderson born 24 september 1934 in haddington is a scottish former prof\n essional footballer he played as a forward and was noted for\n* Leslie Lea 0.95065\n leslie lea born 5 october 1942 in manchester is an english former professional footballer \n he played as a midfielderlea began his professional career with blackpool\n\n"
]
],
[
[
"On the other hand, the right child cluster consists mainly of artists (singers and actors/actresses):",
"_____no_output_____"
]
],
[
[
"display_single_tf_idf_cluster(right_child_athletes_artists, map_word_to_index)",
"113949:0.045\n113949:0.043\n113949:0.035\n113949:0.031\n113949:0.031\n\n* Alessandra Aguilar 0.93880\n alessandra aguilar born 1 july 1978 in lugo is a spanish longdistance runner who specialis\n es in marathon running she represented her country in the event\n* Heather Samuel 0.93999\n heather barbara samuel born 6 july 1970 is a retired sprinter from antigua and barbuda who\n specialized in the 100 and 200 metres in 1990\n* Viola Kibiwot 0.94037\n viola jelagat kibiwot born december 22 1983 in keiyo district is a runner from kenya who s\n pecialises in the 1500 metres kibiwot won her first\n* Ayelech Worku 0.94052\n ayelech worku born june 12 1979 is an ethiopian longdistance runner most known for winning\n two world championships bronze medals on the 5000 metres she\n* Krisztina Papp 0.94105\n krisztina papp born 17 december 1982 in eger is a hungarian long distance runner she is th\n e national indoor record holder over 5000 mpapp began\n* Petra Lammert 0.94230\n petra lammert born 3 march 1984 in freudenstadt badenwrttemberg is a former german shot pu\n tter and current bobsledder she was the 2009 european indoor champion\n* Morhad Amdouni 0.94231\n morhad amdouni born 21 january 1988 in portovecchio is a french middle and longdistance ru\n nner he was european junior champion in track and cross country\n* Brian Davis (golfer) 0.94378\n brian lester davis born 2 august 1974 is an english professional golferdavis was born in l\n ondon he turned professional in 1994 and became a member\n\n"
]
],
[
[
"Our hierarchy of clusters now looks like this:\n```\n Wikipedia\n +\n |\n +--------------------------+--------------------+\n | |\n + +\n Non-athletes/artists Athletes/artists\n +\n |\n +----------+----------+\n | |\n | |\n + |\n athletes artists\n```",
"_____no_output_____"
],
[
"Should we keep subdividing the clusters? If so, which cluster should we subdivide? To answer this question, we again think about our application. Since we organize our directory by topics, it would be nice to have topics that are about as coarse as each other. For instance, if one cluster is about baseball, we expect some other clusters about football, basketball, volleyball, and so forth. That is, **we would like to achieve similar level of granularity for all clusters.**\n\nBoth the athletes and artists node can be subdivided more, as each one can be divided into more descriptive professions (singer/actress/painter/director, or baseball/football/basketball, etc.). Let's explore subdividing the athletes cluster further to produce finer child clusters.",
"_____no_output_____"
],
[
"Let's give the clusters aliases as well:",
"_____no_output_____"
]
],
[
[
"athletes = left_child_athletes_artists\nartists = right_child_athletes_artists",
"_____no_output_____"
]
],
[
[
"### Cluster of athletes",
"_____no_output_____"
],
[
"In answering the following quiz question, take a look at the topics represented in the top documents (those closest to the centroid), as well as the list of words with highest TF-IDF weights.\n\nLet us bipartition the cluster of athletes.",
"_____no_output_____"
]
],
[
[
"left_child_athletes, right_child_athletes = bipartition(athletes, maxiter=100, num_runs=6, seed=1)",
"/home/offbeat/Environments/turicreate/lib/python3.7/site-packages/sklearn/cluster/_kmeans.py:974: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 0.25.\n \" removed in 0.25.\", FutureWarning)\n"
],
[
"display_single_tf_idf_cluster(left_child_athletes, map_word_to_index)\ndisplay_single_tf_idf_cluster(right_child_athletes, map_word_to_index)",
"113949:0.110\n113949:0.102\n113949:0.051\n113949:0.046\n113949:0.045\n\n* Steve Springer 0.89370\n steven michael springer born february 11 1961 is an american former professional baseball \n player who appeared in major league baseball as a third baseman and\n* Dave Ford 0.89622\n david alan ford born december 29 1956 is a former major league baseball pitcher for the ba\n ltimore orioles born in cleveland ohio ford attended lincolnwest\n* Todd Williams 0.89829\n todd michael williams born february 13 1971 in syracuse new york is a former major league \n baseball relief pitcher he attended east syracuseminoa high school\n* Justin Knoedler 0.90102\n justin joseph knoedler born july 17 1980 in springfield illinois is a former major league \n baseball catcherknoedler was originally drafted by the st louis cardinals\n* Kevin Nicholson (baseball) 0.90619\n kevin ronald nicholson born march 29 1976 is a canadian baseball shortstop he played part \n of the 2000 season for the san diego padres of\n* Joe Strong 0.90658\n joseph benjamin strong born september 9 1962 in fairfield california is a former major lea\n gue baseball pitcher who played for the florida marlins from 2000\n* James Baldwin (baseball) 0.90691\n james j baldwin jr born july 15 1971 is a former major league baseball pitcher he batted a\n nd threw righthanded in his 11season career he\n* James Garcia 0.90738\n james robert garcia born february 3 1980 is an american former professional baseball pitch\n er who played in the san francisco giants minor league system as\n\n113949:0.048\n113949:0.043\n113949:0.041\n113949:0.036\n113949:0.034\n\n* Todd Curley 0.94563\n todd curley born 14 january 1973 is a former australian rules footballer who played for co\n llingwood and the western bulldogs in the australian football league\n* Tony Smith (footballer, born 1957) 0.94590\n anthony tony smith born 20 february 1957 is a former footballer who played as a central de\n fender in the football league in the 1970s and\n* Chris Day 0.94605\n christopher nicholas chris day born 28 july 1975 is an english professional footballer who\n plays as a goalkeeper for stevenageday started his career at tottenham\n* Jason Roberts (footballer) 0.94617\n jason andre davis roberts mbe born 25 january 1978 is a former professional footballer and\n now a football punditborn in park royal london roberts was\n* Ashley Prescott 0.94618\n ashley prescott born 11 september 1972 is a former australian rules footballer he played w\n ith the richmond and fremantle football clubs in the afl between\n* David Hamilton (footballer) 0.94910\n david hamilton born 7 november 1960 is an english former professional association football\n player who played as a midfielder he won caps for the england\n* Richard Ambrose 0.94924\n richard ambrose born 10 june 1972 is a former australian rules footballer who played with \n the sydney swans in the australian football league afl he\n* Neil Grayson 0.94944\n neil grayson born 1 november 1964 in york is an english footballer who last played as a st\n riker for sutton towngraysons first club was local\n\n"
]
],
[
[
"**Quiz Question**. Which diagram best describes the hierarchy right after splitting the `athletes` cluster? Refer to the quiz form for the diagrams.",
"_____no_output_____"
],
[
"**Caution**. The granularity criteria is an imperfect heuristic and must be taken with a grain of salt. It takes a lot of manual intervention to obtain a good hierarchy of clusters.\n\n* **If a cluster is highly mixed, the top articles and words may not convey the full picture of the cluster.** Thus, we may be misled if we judge the purity of clusters solely by their top documents and words. \n* **Many interesting topics are hidden somewhere inside the clusters but do not appear in the visualization.** We may need to subdivide further to discover new topics. For instance, subdividing the `ice_hockey_football` cluster led to the appearance of runners and golfers.",
"_____no_output_____"
],
[
"### Cluster of non-athletes",
"_____no_output_____"
],
[
"Now let us subdivide the cluster of non-athletes.",
"_____no_output_____"
]
],
[
[
"%%time \n# Bipartition the cluster of non-athletes\nleft_child_non_athletes_artists, right_child_non_athletes_artists = bipartition(non_athletes_artists, maxiter=100, num_runs=3, seed=1)",
"/home/offbeat/Environments/turicreate/lib/python3.7/site-packages/sklearn/cluster/_kmeans.py:974: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 0.25.\n \" removed in 0.25.\", FutureWarning)\n"
],
[
"display_single_tf_idf_cluster(left_child_non_athletes_artists, map_word_to_index)",
"113949:0.016\n113949:0.013\n113949:0.013\n113949:0.012\n113949:0.012\n\n* Barry Sullivan (lawyer) 0.97227\n barry sullivan is a chicago lawyer and as of july 1 2009 the cooney conway chair in advoca\n cy at loyola university chicago school of law\n* Kayee Griffin 0.97444\n kayee frances griffin born 6 february 1950 is an australian politician and former australi\n an labor party member of the new south wales legislative council serving\n* Christine Robertson 0.97450\n christine mary robertson born 5 october 1948 is an australian politician and former austra\n lian labor party member of the new south wales legislative council serving\n* James A. Joseph 0.97464\n james a joseph born 1935 is an american former diplomatjoseph is professor of the practice\n of public policy studies at duke university and founder of\n* David Anderson (British Columbia politician) 0.97492\n david a anderson pc oc born august 16 1937 in victoria british columbia is a former canadi\n an cabinet minister educated at victoria college in victoria\n* Mary Ellen Coster Williams 0.97594\n mary ellen coster williams born april 3 1953 is a judge of the united states court of fede\n ral claims appointed to that court in 2003\n* Sven Erik Holmes 0.97600\n sven erik holmes is a former federal judge and currently the vice chairman legal risk and \n regulatory and chief legal officer for kpmg llp a\n* Andrew Fois 0.97652\n andrew fois is an attorney living and working in washington dc as of april 9 2012 he will \n be serving as the deputy attorney general\n\n"
],
[
"display_single_tf_idf_cluster(right_child_non_athletes_artists, map_word_to_index)",
"113949:0.039\n113949:0.030\n113949:0.023\n113949:0.021\n113949:0.015\n\n* Madonna (entertainer) 0.96092\n madonna louise ciccone tkoni born august 16 1958 is an american singer songwriter actress \n and businesswoman she achieved popularity by pushing the boundaries of lyrical\n* Janet Jackson 0.96153\n janet damita jo jackson born may 16 1966 is an american singer songwriter and actress know\n n for a series of sonically innovative socially conscious and\n* Cher 0.96540\n cher r born cherilyn sarkisian may 20 1946 is an american singer actress and television ho\n st described as embodying female autonomy in a maledominated industry\n* Laura Smith 0.96600\n laura smith is a canadian folk singersongwriter she is best known for her 1995 single shad\n e of your love one of the years biggest hits\n* Natashia Williams 0.96677\n natashia williamsblach born august 2 1978 is an american actress and former wonderbra camp\n aign model who is perhaps best known for her role as shane\n* Anita Kunz 0.96716\n anita e kunz oc born 1956 is a canadianborn artist and illustratorkunz has lived in london\n new york and toronto contributing to magazines and working\n* Maggie Smith 0.96747\n dame margaret natalie maggie smith ch dbe born 28 december 1934 is an english actress she \n made her stage debut in 1952 and has had\n* Lizzie West 0.96752\n lizzie west born in brooklyn ny on july 21 1973 is a singersongwriter her music can be des\n cribed as a blend of many genres including\n\n"
]
],
[
[
"The clusters are not as clear, but the left cluster has a tendency to show important female figures, and the right one to show politicians and government officials.\n\nLet's divide them further.",
"_____no_output_____"
]
],
[
[
"female_figures = left_child_non_athletes_artists\npoliticians_etc = right_child_non_athletes_artists",
"_____no_output_____"
],
[
"politicians_etc = left_child_non_athletes_artists\nfemale_figures = right_child_non_athletes_artists",
"_____no_output_____"
]
],
[
[
"**Quiz Question**. Let us bipartition the clusters `female_figures` and `politicians`. Which diagram best describes the resulting hierarchy of clusters for the non-athletes? Refer to the quiz for the diagrams.\n\n**Note**. Use `maxiter=100, num_runs=6, seed=1` for consistency of output.",
"_____no_output_____"
]
],
[
[
"left_female_figures, right_female_figures = bipartition(female_figures, maxiter=100, num_runs=6, seed=1)\nleft_politicians_etc, right_politicians_etc = bipartition(politicians_etc, maxiter=100, num_runs=6, seed=1)",
"/home/offbeat/Environments/turicreate/lib/python3.7/site-packages/sklearn/cluster/_kmeans.py:974: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 0.25.\n \" removed in 0.25.\", FutureWarning)\n/home/offbeat/Environments/turicreate/lib/python3.7/site-packages/sklearn/cluster/_kmeans.py:974: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 0.25.\n \" removed in 0.25.\", FutureWarning)\n"
],
[
"display_single_tf_idf_cluster(left_female_figures, map_word_to_index)\ndisplay_single_tf_idf_cluster(right_female_figures, map_word_to_index)",
"113949:0.039\n113949:0.036\n113949:0.033\n113949:0.028\n113949:0.026\n\n* Kayee Griffin 0.95170\n kayee frances griffin born 6 february 1950 is an australian politician and former australi\n an labor party member of the new south wales legislative council serving\n* Marcelle Mersereau 0.95417\n marcelle mersereau born february 14 1942 in pointeverte new brunswick is a canadian politi\n cian a civil servant for most of her career she also served\n* Lucienne Robillard 0.95453\n lucienne robillard pc born june 16 1945 is a canadian politician and a member of the liber\n al party of canada she sat in the house\n* Maureen Lyster 0.95590\n maureen anne lyster born 10 september 1943 is an australian politician she was an australi\n an labor party member of the victorian legislative assembly from 1985\n* Liz Cunningham 0.95690\n elizabeth anne liz cunningham is an australian politician she was an independent member of\n the legislative assembly of queensland from 1995 to 2015 representing the\n* Carol Skelton 0.95780\n carol skelton pc born december 12 1945 in biggar saskatchewan is a canadian politician she\n is a member of the security intelligence review committee which\n* Stephen Harper 0.95816\n stephen joseph harper pc mp born april 30 1959 is a canadian politician who is the 22nd an\n d current prime minister of canada and the\n* Doug Lewis 0.95875\n douglas grinslade doug lewis pc qc born april 17 1938 is a former canadian politician a ch\n artered accountant and lawyer by training lewis entered the\n\n113949:0.018\n113949:0.015\n113949:0.013\n113949:0.013\n113949:0.010\n\n* Lawrence W. Green 0.97495\n lawrence w green is best known by health education researchers as the originator of the pr\n ecede model and codeveloper of the precedeproceed model which has\n* James A. Joseph 0.97506\n james a joseph born 1935 is an american former diplomatjoseph is professor of the practice\n of public policy studies at duke university and founder of\n* Timothy Luke 0.97584\n timothy w luke is university distinguished professor of political science in the college o\n f liberal arts and human sciences as well as program chair of\n* Archie Brown 0.97628\n archibald haworth brown cmg fba commonly known as archie brown born 10 may 1938 is a briti\n sh political scientist and historian in 2005 he became\n* Jerry L. Martin 0.97687\n jerry l martin is chairman emeritus of the american council of trustees and alumni he serv\n ed as president of acta from its founding in 1995\n* Ren%C3%A9e Fox 0.97713\n rene c fox a summa cum laude graduate of smith college in 1949 earned her phd in sociology\n in 1954 from radcliffe college harvard university\n* Robert Bates (political scientist) 0.97732\n robert hinrichs bates born 1942 is an american political scientist he is eaton professor o\n f the science of government in the departments of government and\n* Ferdinand K. Levy 0.97737\n ferdinand k levy was a famous management scientist with several important contributions to\n system analysis he was a professor at georgia tech from 1972 until\n\n"
],
[
"display_single_tf_idf_cluster(left_politicians_etc, map_word_to_index)\ndisplay_single_tf_idf_cluster(right_politicians_etc, map_word_to_index)",
"113949:0.027\n113949:0.023\n113949:0.017\n113949:0.016\n113949:0.015\n\n* Julian Knowles 0.96904\n julian knowles is an australian composer and performer specialising in new and emerging te\n chnologies his creative work spans the fields of composition for theatre dance\n* Peter Combe 0.97080\n peter combe born 20 october 1948 is an australian childrens entertainer and musicianmusica\n l genre childrens musiche has had 22 releases including seven gold albums two\n* Craig Pruess 0.97121\n craig pruess born 1950 is an american composer musician arranger and gold platinum record \n producer who has been living in britain since 1973 his career\n* Ceiri Torjussen 0.97169\n ceiri torjussen born 1976 is a composer who has contributed music to dozens of film and te\n levision productions in the ushis music was described by\n* Brenton Broadstock 0.97192\n brenton broadstock ao born 1952 is an australian composerbroadstock was born in melbourne \n he studied history politics and music at monash university and later composition\n* Michael Peter Smith 0.97318\n michael peter smith born september 7 1941 is a chicagobased singersongwriter rolling stone\n magazine once called him the greatest songwriter in the english language he\n* Marc Hoffman 0.97356\n marc hoffman born april 16 1961 is a composer of concert music and music for film pianist \n vocalist recording artist and music educator hoffman grew\n* Tom Bancroft 0.97378\n tom bancroft born 1967 london is a british jazz drummer and composer he began drumming age\n d seven and started off playing jazz with his father\n\n113949:0.124\n113949:0.092\n113949:0.015\n113949:0.015\n113949:0.014\n\n* Janet Jackson 0.93374\n janet damita jo jackson born may 16 1966 is an american singer songwriter and actress know\n n for a series of sonically innovative socially conscious and\n* Barbara Hershey 0.93507\n barbara hershey born barbara lynn herzstein february 5 1948 once known as barbara seagull \n is an american actress in a career spanning nearly 50 years\n* Lauren Royal 0.93717\n lauren royal born march 3 circa 1965 is a book writer from california royal has written bo\n th historic and novelistic booksa selfproclaimed angels baseball fan\n* Alexandra Potter 0.93802\n alexandra potter born 1970 is a british author of romantic comediesborn in bradford yorksh\n ire england and educated at liverpool university gaining an honors degree in\n* Cher 0.93804\n cher r born cherilyn sarkisian may 20 1946 is an american singer actress and television ho\n st described as embodying female autonomy in a maledominated industry\n* Madonna (entertainer) 0.93806\n madonna louise ciccone tkoni born august 16 1958 is an american singer songwriter actress \n and businesswoman she achieved popularity by pushing the boundaries of lyrical\n* Jane Fonda 0.93836\n jane fonda born lady jayne seymour fonda december 21 1937 is an american actress writer po\n litical activist former fashion model and fitness guru she is\n* Ellina Graypel 0.93927\n ellina graypel born july 19 1972 is an awardwinning russian singersongwriter she was born \n near the volga river in the heart of russia she spent\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d03c2dbc1679223f5e044616172deb392b044fa9 | 14,246 | ipynb | Jupyter Notebook | MATH/17_joint_probability_conditional_probability.ipynb | CATERINA-SEUL/Data-Science-School | 6189b15a70274d77aa0430efab2d2db81cfe4408 | [
"MIT"
] | null | null | null | MATH/17_joint_probability_conditional_probability.ipynb | CATERINA-SEUL/Data-Science-School | 6189b15a70274d77aa0430efab2d2db81cfe4408 | [
"MIT"
] | null | null | null | MATH/17_joint_probability_conditional_probability.ipynb | CATERINA-SEUL/Data-Science-School | 6189b15a70274d77aa0430efab2d2db81cfe4408 | [
"MIT"
] | null | null | null | 21.984568 | 94 | 0.336586 | [
[
[
"### 콜모고로프의 공리\n\n 1) 모든 사건에 대해 확률은 실수이고 0 또는 양수이다.\n \n - $P(A) \\geq 0$\n \n \n 2) 표본공간(전체집합) 이라는 사건(부분집합)에대 대한 확률은 1이다.\n \n - $P(\\Omega) = 1$\n \n \n 3) 공통원소가 없는 두 사건의 합집합의 확률은 사건별 확률의 합이다\n \n - $A\\cap B = \\emptyset \\rightarrow P(A\\cup B) = P(A) + P(B)$\n \n \n \n-----\n\n### 확률 성질 요약\n\n 1) 공집합의 확률\n \n - $P(0) = \\emptyset$\n \n \n 2) 여집합의 확률\n \n - $P(A\\complement) = 1-P(A)$\n \n \n 3) 포함 배제의 원리\n \n - $P(A\\cup B) = P(A) + P(B) - P(A\\cap B) $\n \n \n 4) 전체확률의 법칙\n \n - $P(A) = \\sum_i(A,C_i)$\n \n \n----\n\n### 확률 분포\n\n - 표본의 개수가 유한한 경우 : 단순사건에 대한 정보만 전달하면 됨 \n \n - 확률질량함수 : $P({a})$\n \n - 표본의 개수가 무한한 경우 \n \n - 구간 : 표본공간이 실수의 집합이면 사건은 시작점과 끝점이라는 두 숫자로 표현\n \n $A = {a < x \\leq b}$\n \n 1) 누적분포함수 (cdf == $F(x)$)\n \n - $F(b) = F(a) + P(a,b)$\n $\\rightarrow P(a,b) = F(b) - F(a)$ \n \n 2) 확률밀도함수 (pdf) : $F(x)$ 를 미분하여, 도함수를 구해서 기울기 출력\n \n - $p(x) = \\frac{dF(x)}{dx}$\n \n - 누적분포함수와 확률밀도함수의 관계 $\\rightarrow$ 적분\n - $F(x) = \\int^x_{-\\infty}p(u)du$\n \n ###### 확률 밀도 함수의 특징 \n \n 1) 누적분포함수의 기울기가 음수가 될 수 없기 때문에 확률밀도함수는 0보다 크거나 같다.\n \n - $p(x) \\geq 0$\n \n 2) $-\\infty$부터 $\\infty$까지 적분하면 표본공간$(-\\infty , \\infty)$의 확률이 되므로 값은 1이다.\n \n - $\\int^\\infty_{-\\infty} p(u)du = 1$\n \n----",
"_____no_output_____"
],
[
"### 결합확률과 조건부 확률\n\n - 결합확률 (joint probability)\n \n - $P(A\\cap B) or P(A,B)$\n \n \n - 주변확률 (marginal probability)\n \n - $P(A) , P(B)$\n \n \n - 조건부 확률 (100% 참) \n \n - $P(A|B) = new P(A) if P(B) = 1$\n \n $ = \\frac{P(A,B)}{P(B)} = \\frac{P(Anew)}{P(\\Omega new)}$\n \n \n - 독립 (independent)\n - 사건A와 사건B의 결합확률의 값이 $P(A,B) = P(A)P(B)$ 관계가 성립하면서 두 사건 $A$와 $B$는 서로 독립이라고 정의한다.\n \n - $P(A) = P(A|B)$\n \n----\n\n#### 원인과 결과, 근거와 추론, 가정과 조건부 결론\n\n - 조건부확률 $P(A|B)$에서 사건 (주장/명제) B, A \n \n 1) 가정과 그 가정에 따른 조건부 결론\n 2) 원인과 결과\n 3) 근거와 추론\n \n $\\rightarrow P(A,B) = P(A|B)P(B)$ \n \n : A,B가 모두 발생할 확률은 B라는 사건이 발생할 확률과 그 사건이 발생한 경우 다시 A가 발생할 경우의 곱\n \n \n---\n\n#### 사슬법칙\n\n - $P(X_1,...,X_N) = P(X_1)\\prod^N_{i=2}P(N_i|X_1,...,X_{i-1})$\n \n----\n#### 확률변수 (random variable)\n\n - 확률적인 숫자를 출력하는 변수 ex) 랜덤박스",
"_____no_output_____"
],
[
"----\n#### JointProbabilityDistribution(variables, cardinality, values)\n\n - variables : 확률변수의 이름, 문자열의리스트, 정의하려는 확률변수가 하나인 경우에도 리스트로 넣어야 한다.\n \n - cardinality : 각 확률변수의 표본 혹은 배타적 사건의 수의 리스트\n - values : 확률 변수의 모든 표본(조합)에 대한 (결합) 확률값의 리스트\n ",
"_____no_output_____"
]
],
[
[
"from pgmpy.factors.discrete import JointProbabilityDistribution as JPD",
"_____no_output_____"
],
[
"px = JPD(['X'], [2], np.array([12, 8]) / 20)\nprint(px)",
"+------+--------+\n| X | P(X) |\n+======+========+\n| X(0) | 0.6000 |\n+------+--------+\n| X(1) | 0.4000 |\n+------+--------+\n"
],
[
"py = JPD(['Y'], [2], np.array([10, 10])/20)\nprint(py)",
"+------+--------+\n| Y | P(Y) |\n+======+========+\n| Y(0) | 0.5000 |\n+------+--------+\n| Y(1) | 0.5000 |\n+------+--------+\n"
],
[
"pxy = JPD(['X', 'Y'], [2, 2], np.array([3, 9, 7, 1])/20)\nprint(pxy)",
"+------+------+----------+\n| X | Y | P(X,Y) |\n+======+======+==========+\n| X(0) | Y(0) | 0.1500 |\n+------+------+----------+\n| X(0) | Y(1) | 0.4500 |\n+------+------+----------+\n| X(1) | Y(0) | 0.3500 |\n+------+------+----------+\n| X(1) | Y(1) | 0.0500 |\n+------+------+----------+\n"
],
[
"pxy2 = JPD(['X', 'Y'], [2, 2], np.array([6, 6, 4, 4, ])/20)\nprint(pxy2)",
"+------+------+----------+\n| X | Y | P(X,Y) |\n+======+======+==========+\n| X(0) | Y(0) | 0.3000 |\n+------+------+----------+\n| X(0) | Y(1) | 0.3000 |\n+------+------+----------+\n| X(1) | Y(0) | 0.2000 |\n+------+------+----------+\n| X(1) | Y(1) | 0.2000 |\n+------+------+----------+\n"
]
],
[
[
"----\n#### marginal_distribution() \n\n - 인수로 받은 확률변수에 대한 주변확률분포를 구함\n - $X$의 주변확률을 구해라\n \n----\n\n - 결합확률로 부터 주변확률 $P(A),P(A^\\complement)$ 를 계산",
"_____no_output_____"
]
],
[
[
"pmx = pxy.marginal_distribution(['X'], inplace=False)\nprint(pmx)",
"+------+--------+\n| X | P(X) |\n+======+========+\n| X(0) | 0.6000 |\n+------+--------+\n| X(1) | 0.4000 |\n+------+--------+\n"
]
],
[
[
"#### marginalize()\n\n - 인수로 받은 확률변수를 주변화(marginalize)하여 나머지 확률 변수에 대한 주변확률분포를 구한다.\n \n - $X$를 구하기 위해서 $Y$를 없애라! (== 배타적인 사건으로 써라)\n----\n\n - 결합확률로부터 $P(A) , P(A^\\complement)$ 를 계산",
"_____no_output_____"
]
],
[
[
"pmx = pxy.marginalize(['Y'], inplace=False)\nprint(pmx)",
"+------+--------+\n| X | P(X) |\n+======+========+\n| X(0) | 0.6000 |\n+------+--------+\n| X(1) | 0.4000 |\n+------+--------+\n"
]
],
[
[
"- 결합확률로부터 $P(B),P(B^\\complement)$를 계산",
"_____no_output_____"
]
],
[
[
"py = pxy.marginal_distribution(['Y'], inplace=False)\nprint(pmy)",
"+------+--------+\n| Y | P(Y) |\n+======+========+\n| Y(0) | 0.5000 |\n+------+--------+\n| Y(1) | 0.5000 |\n+------+--------+\n"
],
[
"py = pxy.marginalize(['X'], inplace=False)\nprint(py)",
"+------+--------+\n| Y | P(Y) |\n+======+========+\n| Y(0) | 0.5000 |\n+------+--------+\n| Y(1) | 0.5000 |\n+------+--------+\n"
]
],
[
[
"#### conditional_distribution()\n\n - 어떤 확률변수가 어떤 사건이 되는 조건에 대해 조건부확률값을 계산\n \n---\n\n - 결합확률로부터 조건부확률 $P(B|A),P(B^\\complement|A)$ 를 계산",
"_____no_output_____"
]
],
[
[
"py_on_x0 = pxy.conditional_distribution([('X', 0)], inplace=False)\nprint(py_on_x0)",
"+------+--------+\n| Y | P(Y) |\n+======+========+\n| Y(0) | 0.2500 |\n+------+--------+\n| Y(1) | 0.7500 |\n+------+--------+\n"
]
],
[
[
" - 결합확률로부터 조건부확률 $P(B|A^\\complement),P(B^\\complement|A^\\complement)$ 를 계산",
"_____no_output_____"
]
],
[
[
"py_on_x1 = pxy.conditional_distribution([('X', 1)], inplace=False)\nprint(py_on_x1)",
"+------+--------+\n| Y | P(Y) |\n+======+========+\n| Y(0) | 0.8750 |\n+------+--------+\n| Y(1) | 0.1250 |\n+------+--------+\n"
]
],
[
[
" - 결합확률로부터 조건부확률 $P(A|B),P(A^\\complement|B)$ 를 계산",
"_____no_output_____"
]
],
[
[
"px_on_y0 = pxy.conditional_distribution([('Y', 0)], inplace=False)\nprint(px_on_y0)",
"+------+--------+\n| X | P(X) |\n+======+========+\n| X(0) | 0.3000 |\n+------+--------+\n| X(1) | 0.7000 |\n+------+--------+\n"
],
[
"px_on_y1 = pxy.conditional_distribution([('Y', 1)], inplace=False)\nprint(px_on_y1)",
"+------+--------+\n| X | P(X) |\n+======+========+\n| X(0) | 0.9000 |\n+------+--------+\n| X(1) | 0.1000 |\n+------+--------+\n"
]
],
[
[
"#### check_independence()\n\n - 두 확률변수 간의 독립 확인 가능.\n \n * 독립 : $P(A,B) = P(A)P(B)$\n - 독립인 경우에는 결합확률을 다 구할 필요가 없다 .",
"_____no_output_____"
]
],
[
[
"pxy.check_independence(['X'], ['Y'])",
"_____no_output_____"
]
],
[
[
" - JointProbabilityDistribution 객체끼리 곱하면 두 분포가 독립이라는 가정하에 결합확률을 구함",
"_____no_output_____"
]
],
[
[
"# 독립이 아님\n\nprint(px*py)\nprint(pxy)",
"+------+------+----------+\n| X | Y | P(X,Y) |\n+======+======+==========+\n| X(0) | Y(0) | 0.3000 |\n+------+------+----------+\n| X(0) | Y(1) | 0.3000 |\n+------+------+----------+\n| X(1) | Y(0) | 0.2000 |\n+------+------+----------+\n| X(1) | Y(1) | 0.2000 |\n+------+------+----------+\n+------+------+----------+\n| X | Y | P(X,Y) |\n+======+======+==========+\n| X(0) | Y(0) | 0.1500 |\n+------+------+----------+\n| X(0) | Y(1) | 0.4500 |\n+------+------+----------+\n| X(1) | Y(0) | 0.3500 |\n+------+------+----------+\n| X(1) | Y(1) | 0.0500 |\n+------+------+----------+\n"
],
[
"pxy2.check_independence(['X'], ['Y'])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d03c35cf6989d2685730090a59be6c9a397e8f58 | 25,422 | ipynb | Jupyter Notebook | python/intro-to-python.ipynb | 0xaio/talks | 262b83b7df51c29166a486304ddd4f6730d7bc82 | [
"MIT"
] | null | null | null | python/intro-to-python.ipynb | 0xaio/talks | 262b83b7df51c29166a486304ddd4f6730d7bc82 | [
"MIT"
] | null | null | null | python/intro-to-python.ipynb | 0xaio/talks | 262b83b7df51c29166a486304ddd4f6730d7bc82 | [
"MIT"
] | null | null | null | 23.670391 | 573 | 0.464519 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d03c3600b5821218ea30309ec674f6e542856f8d | 398,942 | ipynb | Jupyter Notebook | Misc/.ipynb_checkpoints/preprocess-checkpoint.ipynb | dattasiddhartha/DataX-NeuralDecisionMaking | 68b0d79954d308c28febe07844674d9c034877d4 | [
"MIT"
] | null | null | null | Misc/.ipynb_checkpoints/preprocess-checkpoint.ipynb | dattasiddhartha/DataX-NeuralDecisionMaking | 68b0d79954d308c28febe07844674d9c034877d4 | [
"MIT"
] | null | null | null | Misc/.ipynb_checkpoints/preprocess-checkpoint.ipynb | dattasiddhartha/DataX-NeuralDecisionMaking | 68b0d79954d308c28febe07844674d9c034877d4 | [
"MIT"
] | null | null | null | 819.182752 | 135,554 | 0.940736 | [
[
[
"## This notebook preprocesses subject 8 for Question 1: Can we predict if the subject will select Gamble or Safebet *before* the button press time?",
"_____no_output_____"
],
[
"## Behavior data",
"_____no_output_____"
]
],
[
[
"## Explore behavior data using pandas\n\nimport pandas as pd\n\nbeh_dir = '../data/decision-making/data/data_behav'\n\n# os.listdir(beh_dir)",
"_____no_output_____"
],
[
"# S08\nbeh8_df = pd.read_csv(os.path.join(beh_dir,'gamble.data.s08.csv'))",
"_____no_output_____"
]
],
[
[
"### Choice.class will be our outcome variable",
"_____no_output_____"
]
],
[
[
"beh8_df.groupby('choice.class').nunique()",
"_____no_output_____"
]
],
[
[
"### Great, we have 100 trials per choice: Gamble vs Safebet.",
"_____no_output_____"
]
],
[
[
"# This will be the outcome variable: beh8_df['choice.class']",
"_____no_output_____"
],
[
"y8 = beh8_df['choice.class'].values",
"_____no_output_____"
]
],
[
[
"Save y-data",
"_____no_output_____"
]
],
[
[
"mkdir ../data/decision-making/data/data_preproc",
"_____no_output_____"
],
[
"np.save('../data/decision-making/data/data_preproc/y8',y8)",
"_____no_output_____"
],
[
"ls ../data/decision-making/data/data_preproc",
"y8.npy\r\n"
]
],
[
[
"## Neural data",
"_____no_output_____"
]
],
[
[
"sfreq = 1000\n\nneur_dir = '../data/decision-making/data/data_ephys'\n\n# os.listdir(neur_dir)\n\nfrom scipy.io import loadmat\n\nneur8 = loadmat(os.path.join(neur_dir, 's08_ofc_hg_events.mat'))",
"_____no_output_____"
],
[
"neur8['buttonpress_events_hg'].shape",
"_____no_output_____"
],
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\n\n# first electrode\nplt.plot(neur8['buttonpress_events_hg'][:,:,0].T)\nplt.axvline(1000, color='k')\npass",
"_____no_output_____"
],
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\n\n# second electrode\nplt.plot(neur8['buttonpress_events_hg'][:,:,1].T)\nplt.axvline(1000, color='k')\npass",
"_____no_output_____"
],
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\n\n# 10th electrode\nplt.plot(neur8['buttonpress_events_hg'][:,:,-1].T)\nplt.axvline(1000, color='k')\npass",
"_____no_output_____"
]
],
[
[
"### Convert format of data to work for \"decoding over time\"",
"_____no_output_____"
],
[
"For decoding over time the data X is the epochs data of shape n_epochs x n_channels x n_times. As the last dimension of X is the time an estimator will be fit on every time instant.",
"_____no_output_____"
]
],
[
[
"neur8['buttonpress_events_hg'].shape",
"_____no_output_____"
]
],
[
[
"### Notice that current shape is n_epochs (200) x n_times (3000) x n_channels (10)",
"_____no_output_____"
]
],
[
[
"X8 = np.swapaxes(neur8['buttonpress_events_hg'],1,2)",
"_____no_output_____"
],
[
"X8.shape",
"_____no_output_____"
]
],
[
[
"Hooray, now it's n_epochs x n_channels x n_times.",
"_____no_output_____"
],
[
"## Save out X8",
"_____no_output_____"
]
],
[
[
"np.save('../data/decision-making/data/data_preproc/X8',X8)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d03c51eda626561ffc58a667a04c4db42a7cf6b4 | 682,314 | ipynb | Jupyter Notebook | Untitled.ipynb | huaifeng1993/JinNanConpetition2 | e6b77282e8c9d03a956401ff620a563836da4505 | [
"MIT"
] | null | null | null | Untitled.ipynb | huaifeng1993/JinNanConpetition2 | e6b77282e8c9d03a956401ff620a563836da4505 | [
"MIT"
] | null | null | null | Untitled.ipynb | huaifeng1993/JinNanConpetition2 | e6b77282e8c9d03a956401ff620a563836da4505 | [
"MIT"
] | null | null | null | 37.642833 | 145 | 0.37735 | [
[
[
"import pandas as pd\nimport os\nimport json\nimport numpy as np\nfrom skimage import io",
"_____no_output_____"
],
[
"images='datasets/jinnan/images/'\nann='datasets/jinnan/annotations/train_no_poly.json'",
"_____no_output_____"
],
[
"label_path=ann",
"_____no_output_____"
],
[
"print(label_path)",
"datasets/jinnan/annotations/train_no_poly.json\n"
],
[
"f = open(label_path,encoding='utf-8')\ndata = json.load(f)",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"data.keys()",
"_____no_output_____"
],
[
"test_data={}\ntest_data['info']=data['info']\ntest_data['licenses']=data['licenses']\ntest_data['categories']=data['categories']\ntest_data['images']=[]",
"_____no_output_____"
],
[
"images_dir='datasets/jinnan/test/'",
"_____no_output_____"
],
[
"images_path=os.listdir(images_dir)",
"_____no_output_____"
],
[
"images_path.sort(key = lambda x: int(x[:-4]))\nimages_path",
"_____no_output_____"
],
[
"images=[]\nfor x in range(len(images_path)):\n image={}\n img=io.imread(os.path.join(images_dir,images_path[x]))\n image['file_name']=images_path[x]\n image['id']=x\n image['height']=img.shape[0]\n image['width']=img.shape[1]\n images.append(image)\nimages",
"_____no_output_____"
],
[
"test_data['images']=images",
"_____no_output_____"
],
[
"with open(\"./test.json\",'w',encoding='utf-8') as json_file:\n json.dump(test_data,json_file,ensure_ascii=False)",
"_____no_output_____"
],
[
"labes=[1,2]\nscores=[0.1,0.2]\nretc=[[1,2,3,5],[2,4,6,7]]",
"_____no_output_____"
],
[
"[ {\"xmin\":x[2][0],\"xmax\":x[2][2],\"ymin\":x[2][1],\"ymax\":x[2][3],\"label\":x[0],\"confidence\":x[1]} for x in zip(labes,scores,retc)]",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03c5c65a4cd10fbce11bf7cf9498dd201d1b5d7 | 4,047 | ipynb | Jupyter Notebook | eopf-notebooks/eopf_product_data_structure/EOPF_S2_MSI_L1AB.ipynb | CSC-DPR/notebooks | 6e1034ca5402a7abb11f428bd68ba03cf47a5e9f | [
"Apache-2.0"
] | null | null | null | eopf-notebooks/eopf_product_data_structure/EOPF_S2_MSI_L1AB.ipynb | CSC-DPR/notebooks | 6e1034ca5402a7abb11f428bd68ba03cf47a5e9f | [
"Apache-2.0"
] | null | null | null | eopf-notebooks/eopf_product_data_structure/EOPF_S2_MSI_L1AB.ipynb | CSC-DPR/notebooks | 6e1034ca5402a7abb11f428bd68ba03cf47a5e9f | [
"Apache-2.0"
] | null | null | null | 25.613924 | 135 | 0.473684 | [
[
[
"# EOPF S2 MSI L1 A/B Products Data Structure Proposal\n",
"_____no_output_____"
]
],
[
[
"from IPython.display import IFrame\nfrom utils import display\nfrom EOProductDataStructure import EOProductBuilder, EOVariableBuilder, EOGroupBuilder\nimport yaml",
"_____no_output_____"
]
],
[
[
"# Display function",
"_____no_output_____"
]
],
[
[
"def set_variables(yaml_group, eopf_group):\n variables = yaml_group['variables']\n for var in variables:\n variable = EOVariableBuilder(var, default_attrs=True)\n \n try:\n variable.dtype = variables[var].split('->')[1]\n except:\n variable.dtype = \"string\"\n pass\n variable.dims = [d.split('->')[0] for d in variables[var].replace(' ','').replace('F(','').replace(')','').split(',')]\n eopf_group.variables.append(variable)\n for d in variable.dims:\n if d not in eopf_group.dims:\n eopf_group.dims.append(d)",
"_____no_output_____"
]
],
[
[
"# 1. Read S1 MSI Product",
"_____no_output_____"
]
],
[
[
"path_product=\"data/s2_msi_l1ab.yaml\"\n\nproduct = None\n\nwith open(path_product, \"r\") as stream:\n try:\n product = yaml.safe_load(stream)['product']\n except yaml.YAMLError as exc:\n print(exc)\n \ns2_msi = EOProductBuilder(\"S2_MSI_L1AB__\", coords=EOGroupBuilder('coords'))\n\nfor key, values in product.items():\n \n if key == \"attributes\":\n s2_msi.attrs = values \n else:\n group = EOGroupBuilder(key)\n if \"groups\" in values:\n groups = values['groups']\n for ygp in groups:\n eopf_group = EOGroupBuilder(ygp)\n set_variables(values['groups'][ygp], eopf_group)\n group.groups.append(eopf_group)\n else:\n set_variables(values,group)\n \n s2_msi.groups.append(group)\n \ndisplay(s2_msi.compute()) ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d03c6a5bb8fc27faac61439c19fd1408a67e8ca8 | 5,594 | ipynb | Jupyter Notebook | demo/demo.ipynb | joshnn/SSD.Pytorch | df40a1fb85e04c555540ac930efdaaa769669dbe | [
"MIT"
] | 55 | 2020-01-21T09:16:09.000Z | 2022-03-29T13:31:55.000Z | demo/demo.ipynb | joshnn/SSD.Pytorch | df40a1fb85e04c555540ac930efdaaa769669dbe | [
"MIT"
] | 16 | 2020-02-03T16:33:40.000Z | 2022-02-28T14:19:08.000Z | demo/demo.ipynb | joshnn/SSD.Pytorch | df40a1fb85e04c555540ac930efdaaa769669dbe | [
"MIT"
] | 20 | 2020-02-26T11:21:17.000Z | 2021-11-10T22:07:31.000Z | 28.395939 | 112 | 0.562567 | [
[
[
"# Object Detection with SSD\n### Here we demostrate detection on example images using SSD with PyTorch",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\nmodule_path = os.path.abspath(os.path.join('..'))\nif module_path not in sys.path:\n sys.path.append(module_path)\n\nimport torch\nimport torch.nn as nn\nimport torch.backends.cudnn as cudnn\nfrom torch.autograd import Variable\nimport numpy as np\nimport cv2\nif torch.cuda.is_available():\n torch.set_default_tensor_type('torch.cuda.FloatTensor')\n\nfrom ssd import build_ssd",
"_____no_output_____"
]
],
[
[
"## Build SSD300 in Test Phase\n1. Build the architecture, specifyingsize of the input image (300),\n and number of object classes to score (21 for VOC dataset)\n2. Next we load pretrained weights on the VOC0712 trainval dataset ",
"_____no_output_____"
]
],
[
[
"net = build_ssd('test', 300, 21) # initialize SSD\nnet.load_weights('../weights/ssd300_VOC_28000.pth')",
"_____no_output_____"
]
],
[
[
"## Load Image \n### Here we just load a sample image from the VOC07 dataset ",
"_____no_output_____"
]
],
[
[
"# image = cv2.imread('./data/example.jpg', cv2.IMREAD_COLOR) # uncomment if dataset not downloaded\n%matplotlib inline\nfrom matplotlib import pyplot as plt\nfrom data import VOCDetection, VOC_ROOT, VOCAnnotationTransform\n# here we specify year (07 or 12) and dataset ('test', 'val', 'train') \ntestset = VOCDetection(VOC_ROOT, [('2007', 'val')], None, VOCAnnotationTransform())\nimg_id = 60\nimage = testset.pull_image(img_id)\nrgb_image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)\n# View the sampled input image before transform\nplt.figure(figsize=(10,10))\nplt.imshow(rgb_image)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Pre-process the input. \n#### Using the torchvision package, we can create a Compose of multiple built-in transorm ops to apply \nFor SSD, at test time we use a custom BaseTransform callable to\nresize our image to 300x300, subtract the dataset's mean rgb values, \nand swap the color channels for input to SSD300.",
"_____no_output_____"
]
],
[
[
"x = cv2.resize(image, (300, 300)).astype(np.float32)\nx -= (104.0, 117.0, 123.0)\nx = x.astype(np.float32)\nx = x[:, :, ::-1].copy()\nplt.imshow(x)\nx = torch.from_numpy(x).permute(2, 0, 1)",
"_____no_output_____"
]
],
[
[
"## SSD Forward Pass\n### Now just wrap the image in a Variable so it is recognized by PyTorch autograd",
"_____no_output_____"
]
],
[
[
"xx = Variable(x.unsqueeze(0)) # wrap tensor in Variable\nif torch.cuda.is_available():\n xx = xx.cuda()\ny = net(xx)",
"_____no_output_____"
]
],
[
[
"## Parse the Detections and View Results\nFilter outputs with confidence scores lower than a threshold \nHere we choose 60% ",
"_____no_output_____"
]
],
[
[
"from data import VOC_CLASSES as labels\ntop_k=10\n\nplt.figure(figsize=(10,10))\ncolors = plt.cm.hsv(np.linspace(0, 1, 21)).tolist()\nplt.imshow(rgb_image) # plot the image for matplotlib\ncurrentAxis = plt.gca()\n\ndetections = y.data\n# scale each detection back up to the image\nscale = torch.Tensor(rgb_image.shape[1::-1]).repeat(2)\nfor i in range(detections.size(1)):\n j = 0\n while detections[0,i,j,0] >= 0.6:\n score = detections[0,i,j,0]\n label_name = labels[i-1]\n display_txt = '%s: %.2f'%(label_name, score)\n pt = (detections[0,i,j,1:]*scale).cpu().numpy()\n coords = (pt[0], pt[1]), pt[2]-pt[0]+1, pt[3]-pt[1]+1\n color = colors[i]\n currentAxis.add_patch(plt.Rectangle(*coords, fill=False, edgecolor=color, linewidth=2))\n currentAxis.text(pt[0], pt[1], display_txt, bbox={'facecolor':color, 'alpha':0.5})\n j+=1",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d03c73a47555a001ab7c686ce3ed66b6180b5182 | 6,340 | ipynb | Jupyter Notebook | src/Python/create_edge_list_fulldoc_v3.ipynb | fikramulla/LCP_v1 | f2cd786a31d32722aa8fda31bc4b460943d0ba0d | [
"CC-BY-4.0"
] | null | null | null | src/Python/create_edge_list_fulldoc_v3.ipynb | fikramulla/LCP_v1 | f2cd786a31d32722aa8fda31bc4b460943d0ba0d | [
"CC-BY-4.0"
] | null | null | null | src/Python/create_edge_list_fulldoc_v3.ipynb | fikramulla/LCP_v1 | f2cd786a31d32722aa8fda31bc4b460943d0ba0d | [
"CC-BY-4.0"
] | null | null | null | 6,340 | 6,340 | 0.721451 | [
[
[
"#march 22, 2022\n#cont. from R script nlp4edge_doc1_allpages_autoplotsave_v1.R\n#load csv - full doc sdglist_doc1.csv",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom itertools import combinations",
"_____no_output_____"
],
[
"df = pd.read_csv('/content/sdglist_doc9.csv')\nmy_list = df['SDG'].tolist()",
"_____no_output_____"
],
[
"sample_list = df['SDG']\nlist_combinations = list()\nfor n in range(len(sample_list) + 1):\n list_combinations += list(combinations(my_list, 2))\nlabel_list = list_combinations",
"_____no_output_____"
],
[
"#remove duplicates (or can select the first n)\nres = []\nfor i in label_list:\n if i not in res:\n res.append(i)\n\nlen(res) #should be 120",
"_____no_output_____"
],
[
"edge = pd.DataFrame(res) #great it works, now need to add the sum of the \"weight\" column, and i have my edge list!!!\nedge.columns = ['from', 'to']",
"_____no_output_____"
],
[
"#it looks like it retained the order, so try the same on weight column\n#trying weight column\n\nsample_list = df['weight']\nlist_combinations = list()\nfor n in range(len(sample_list) + 1):\n list_combinations += list(combinations(sample_list, 2))\ncount_list = list_combinations\n\n#can not remove duplicates the same way\n#selecting first 120 items in list since it retained the order\ncount_list = count_list[0:120]\ncount = pd.DataFrame(count_list)",
"_____no_output_____"
],
[
"count[\"sum\"] = count.iloc[:, 0] + count.iloc[:, 1]\ncount[\"inertia\"] = (count.iloc[:, 0] * count.iloc[:, 1]) / (count.iloc[:, 0] + count.iloc[:, 1])\ncount[\"inertia\"] = count[\"inertia\"].fillna(0)",
"_____no_output_____"
],
[
"edge[\"sum\"] = count[\"sum\"]\nedge[\"inertia\"] = count[\"inertia\"]",
"_____no_output_____"
],
[
"#pd.set_option('display.max_rows', None)\n#edge #if i want to view all ",
"_____no_output_____"
],
[
"edge.to_csv('my_edge_docX.csv', index=False)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03c79ce57ca23dc043a667703329151d4803bee | 668,683 | ipynb | Jupyter Notebook | exercises/ex1-DAR/teched2020-INT260_Data_Attribute_Recommendation.ipynb | SAP-samples/teched2020-INT260 | 82471f62547f7dfc08c1142ddabf1f8f5a03d693 | [
"Apache-2.0"
] | 11 | 2020-11-18T12:26:15.000Z | 2021-07-28T09:21:50.000Z | exercises/ex1-DAR/teched2020-INT260_Data_Attribute_Recommendation.ipynb | SAP-samples/teched2020-INT260 | 82471f62547f7dfc08c1142ddabf1f8f5a03d693 | [
"Apache-2.0"
] | 1 | 2020-11-16T15:04:16.000Z | 2020-11-17T16:10:20.000Z | exercises/ex1-DAR/teched2020-INT260_Data_Attribute_Recommendation.ipynb | SAP-samples/teched2020-INT260 | 82471f62547f7dfc08c1142ddabf1f8f5a03d693 | [
"Apache-2.0"
] | 5 | 2020-12-14T13:44:57.000Z | 2022-03-22T12:47:44.000Z | 466.631542 | 157,004 | 0.937995 | [
[
[
"# Data Attribute Recommendation - TechED 2020 INT260\n\nGetting started with the Python SDK for the Data Attribute Recommendation service.",
"_____no_output_____"
],
[
"## Business Scenario\n\nWe will consider a business scenario involving product master data. The creation and maintenance of this product master data requires the careful manual selection of the correct categories for a given product from a pre-defined hierarchy of product categories.\n\nIn this workshop, we will explore how to automate this tedious manual task with the Data Attribute Recommendation service.",
"_____no_output_____"
],
[
"<video controls src=\"videos/dar_prediction_material_table.mp4\"/>\n\n",
"_____no_output_____"
],
[
"This workshop will cover:\n \n* Data Upload\n* Model Training and Deployment\n* Inference Requests\n \nWe will work through a basic example of how to achieve these tasks using the [Python SDK for Data Attribute Recommendation](https://github.com/SAP/data-attribute-recommendation-python-sdk).\n\n\n",
"_____no_output_____"
],
[
"*Note: if you are doing several runs of this notebook on a trial account, you may see errors stating 'The resource can no longer be used. Usage limit has been reached'. It can be beneficial to [clean up the service instance](#Cleaning-up-a-service-instance) to free up limited trial resources acquired by an earlier run of the notebook. [Some limits](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/c03b561eea1744c9b9892b416037b99a.html) cannot be reset this way.*",
"_____no_output_____"
],
[
"## Table of Contents\n\n* [Exercise 01.1](#Exercise-01.1) - Installing the SDK and preparing the service key\n * [Creating a service instance and key on BTP Trial](#Creating-a-service-instance-and-key)\n * [Installing the SDK](#Installing-the-SDK)\n * [Loading the service key into your Jupyter Notebook](#Loading-the-service-key-into-your-Jupyter-Notebook)\n* [Exercise 01.2](#Exercise-01.2) - Uploading the data\n* [Exercise 01.3](#Exercise-01.3) - Training the model\n* [Exercise 01.4](#Exercise-01.4) - Deploying the Model and predicting labels\n* [Resources](#Resources) - Additional reading\n* [Cleaning up a service instance](#Cleaning-up-a-service-instance) - Clean up all resources on the service instance\n* [Optional Exercises](#Optional-Exercises) - Optional exercises",
"_____no_output_____"
],
[
"## Requirements\n\n\nSee the [README in the Github repository for this workshop](https://github.com/SAP-samples/teched2020-INT260/blob/master/exercises/ex1-DAR/README.md).",
"_____no_output_____"
],
[
"# Exercise 01.1\n\n*Back to [table of contents](#Table-of-Contents)*\n\nIn exercise 01.1, we will install the SDK and prepare the service key.",
"_____no_output_____"
],
[
"## Creating a service instance and key on BTP Trial",
"_____no_output_____"
],
[
"Please log in to your trial account: https://cockpit.eu10.hana.ondemand.com/trial/\n\nIn the your global account screen, go to the \"Boosters\" tab:\n\n\n\n*Boosters are only available on the Trial landscape. If you are using a production environment, please follow this tutorial to manually [create a service instance and a service key](https://developers.sap.com/tutorials/cp-aibus-dar-service-instance.html)*.",
"_____no_output_____"
],
[
"In the Boosters tab, enter \"Data Attribute Recommendation\" into the search box. Then, select the\nservice tile from the search results: \n \n",
"_____no_output_____"
],
[
"The resulting screen shows details of the booster pack. Here, click the \"Start\" button and wait a few seconds.\n\n",
"_____no_output_____"
],
[
"Once the booster is finished, click the \"go to Service Key\" link to obtain your service key.\n\n",
"_____no_output_____"
],
[
"Finally, download the key and save it to disk.\n\n",
"_____no_output_____"
],
[
"## Installing the SDK",
"_____no_output_____"
],
[
"The Data Attribute Recommendation SDK is available from the Python package repository. It can be installed with the standard `pip` tool:",
"_____no_output_____"
]
],
[
[
"! pip install data-attribute-recommendation-sdk",
"_____no_output_____"
]
],
[
[
"*Note: If you are not using a Jupyter notebook, but instead a regular Python development environment, we recommend using a Python virtual environment to set up your development environment. Please see [the dedicated tutorial to learn how to install the SDK inside a Python virtual environment](https://developers.sap.com/tutorials/cp-aibus-dar-sdk-setup.html).*",
"_____no_output_____"
],
[
"## Loading the service key into your Jupyter Notebook",
"_____no_output_____"
],
[
"Once you downloaded the service key from the Cockpit, upload it to your notebook environment. The service key must be uploaded to same directory where the `teched2020-INT260_Data_Attribute_Recommendation.ipynb` is stored.\n\nWe first navigate to the file browser in Jupyter. On the top of your Jupyter notebook, right-click on the Jupyter logo and open in a new tab.\n\n",
"_____no_output_____"
],
[
"**In the file browser, navigate to the directory where the `teched2020-INT260_Data_Attribute_Recommendation.ipynb` notebook file is stored. The service key must reside next to this file.**\n\nIn the Jupyter file browser, click the **Upload** button (1). In the file selection dialog that opens, select the `defaultKey_*.json` file you downloaded previously from the SAP Cloud Platform Cockpit. Rename the file to `key.json`. \n\nConfirm the upload by clicking on the second **Upload** button (2).\n\n",
"_____no_output_____"
],
[
"The service key contains your credentials to access the service. Please treat this as carefully as you would treat any password. We keep the service key as a separate file outside this notebook to avoid leaking the secret credentials.\n\nThe service key is a JSON file. We will load this file once and use the credentials throughout this workshop. ",
"_____no_output_____"
]
],
[
[
"# First, set up logging so we can see the actions performed by the SDK behind the scenes\nimport logging\nimport sys\n\nlogging.basicConfig(level=logging.INFO, stream=sys.stdout)\n\nfrom pprint import pprint # for nicer output formatting",
"_____no_output_____"
],
[
"import json\nimport os\n\nif not os.path.exists(\"key.json\"):\n msg = \"key.json is not found. Please follow instructions above to create a service key of\"\n msg += \" Data Attribute Recommendation. Then, upload it into the same directory where\"\n msg += \" this notebook is saved.\"\n print(msg)\n raise ValueError(msg)\n\nwith open(\"key.json\") as file_handle:\n key = file_handle.read()\n SERVICE_KEY = json.loads(key)",
"_____no_output_____"
]
],
[
[
"## Summary Exercise 01.1\n\nIn exercise 01.1, we have covered the following topics:\n\n* How to install the Python SDK for Data Attribute Recommendation\n* How to obtain a service key for the Data Attribute Recommendation service",
"_____no_output_____"
],
[
"# Exercise 01.2\n\n*Back to [table of contents](#Table-of-Contents)*\n*To perform this exercise, you need to execute the code in all previous exercises.*\n\nIn exercise 01.2, we will upload our demo dataset to the service.",
"_____no_output_____"
],
[
"## The Dataset",
"_____no_output_____"
],
[
"### Obtaining the Data",
"_____no_output_____"
],
[
"The dataset we use in this workshop is a CSV file containing product master data. The original data was released by BestBuy, a retail company, under an [open license](https://github.com/SAP-samples/data-attribute-recommendation-postman-tutorial-sample#data-and-license). This makes it ideal for first experiments with the Data Attribute Recommendation service.",
"_____no_output_____"
],
[
"The dataset can be downloaded directly from Github using the following command:",
"_____no_output_____"
]
],
[
[
"! wget -O bestBuy.csv \"https://raw.githubusercontent.com/SAP-samples/data-attribute-recommendation-postman-tutorial-sample/master/Tutorial_Example_Dataset.csv\"\n# If you receive a \"command not found\" error (i.e. on Windows), try curl instead of wget:\n# ! curl -o bestBuy.csv \"https://raw.githubusercontent.com/SAP-samples/data-attribute-recommendation-postman-tutorial-sample/master/Tutorial_Example_Dataset.csv\"",
"_____no_output_____"
]
],
[
[
"Let's inspect the data:",
"_____no_output_____"
]
],
[
[
"# if you are experiencing an import error here, run the following in a new cell:\n# ! pip install pandas\nimport pandas as pd\n\ndf = pd.read_csv(\"bestBuy.csv\")\ndf.head(5)",
"_____no_output_____"
],
[
"print()\nprint(f\"Data has {df.shape[0]} rows and {df.shape[1]} columns.\")",
"_____no_output_____"
]
],
[
[
"The CSV contains the several products. For each product, the description, the manufacturer and the price are given. Additionally, three levels of the products hierarchy are given.\n\nThe first product, a set of AAA batteries, is located in the following place in the product hierarchy:\n\n```\nlevel1_category: Connected Home & Housewares\n |\nlevel2_category: Housewares\n |\nlevel3_category: Household Batteries\n```",
"_____no_output_____"
],
[
"We will use the Data Attribute Recommendation service to predict the categories for a given product based on its **description**, **manufacturer** and **price**.",
"_____no_output_____"
],
[
"### Creating the DatasetSchema",
"_____no_output_____"
],
[
"We first have to describe the shape of our data by creating a DatasetSchema. This schema informs the service about the individual column types found in the CSV. We also describe which are the target columns used for training. These columns will be later predicted. In our case, these are the three category columns.\n\nThe service currently supports three column types: **text**, **category** and **number**. For prediction, only **category** is currently supported.\n\nA DatasetSchema for the BestBuy dataset looks as follows:\n\n```json\n{\n \"features\": [\n {\"label\": \"manufacturer\", \"type\": \"CATEGORY\"},\n {\"label\": \"description\", \"type\": \"TEXT\"},\n {\"label\": \"price\", \"type\": \"NUMBER\"}\n ],\n \"labels\": [\n {\"label\": \"level1_category\", \"type\": \"CATEGORY\"},\n {\"label\": \"level2_category\", \"type\": \"CATEGORY\"},\n {\"label\": \"level3_category\", \"type\": \"CATEGORY\"}\n ],\n \"name\": \"bestbuy-category-prediction\",\n}\n```\n\nWe will now upload this DatasetSchema to the Data Attribute Recommendation service. The SDK provides the\n[`DataManagerClient.create_dataset_schema()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.create_dataset_schema) method for this purpose.\n",
"_____no_output_____"
]
],
[
[
"from sap.aibus.dar.client.data_manager_client import DataManagerClient\n\ndataset_schema = {\n \"features\": [\n {\"label\": \"manufacturer\", \"type\": \"CATEGORY\"},\n {\"label\": \"description\", \"type\": \"TEXT\"},\n {\"label\": \"price\", \"type\": \"NUMBER\"}\n ],\n \"labels\": [\n {\"label\": \"level1_category\", \"type\": \"CATEGORY\"},\n {\"label\": \"level2_category\", \"type\": \"CATEGORY\"},\n {\"label\": \"level3_category\", \"type\": \"CATEGORY\"}\n ],\n \"name\": \"bestbuy-category-prediction\",\n}\n\n\ndata_manager = DataManagerClient.construct_from_service_key(SERVICE_KEY)\nresponse = data_manager.create_dataset_schema(dataset_schema)\ndataset_schema_id = response[\"id\"]\n\nprint()\nprint(\"DatasetSchema created:\")\n\npprint(response)\n\nprint()\nprint(f\"DatasetSchema ID: {dataset_schema_id}\")",
"_____no_output_____"
]
],
[
[
"The API responds with the newly created DatasetSchema resource. The service assigned an ID to the schema. We save this ID in a variable, as we will need it when we upload the data.",
"_____no_output_____"
],
[
"### Uploading the Data to the service",
"_____no_output_____"
],
[
"The [`DataManagerClient`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient) class is also responsible for uploading data to the service. This data must fit to an existing DatasetSchema. After uploading the data, the service will validate the Dataset against the DataSetSchema in a background process. The data must be a CSV file which can optionally be `gzip` compressed.\n\nWe will now upload our `bestBuy.csv` file, using the DatasetSchema which we created earlier.\n\nData upload is a two-step process. We first create the Dataset using [`DataManagerClient.create_dataset()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.create_dataset). Then we can upload data to the Dataset using the [`DataManagerClient.upload_data_to_dataset()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.upload_data_to_dataset) method.",
"_____no_output_____"
]
],
[
[
"dataset_resource = data_manager.create_dataset(\"my-bestbuy-dataset\", dataset_schema_id)\ndataset_id = dataset_resource[\"id\"]\n\nprint()\nprint(\"Dataset created:\")\n\npprint(dataset_resource)\n\nprint()\nprint(f\"Dataset ID: {dataset_id}\")",
"_____no_output_____"
],
[
"# Compress file first for a faster upload\n! gzip -9 -c bestBuy.csv > bestBuy.csv.gz",
"_____no_output_____"
]
],
[
[
"Note that the data upload can take a few minutes. Please do not restart the process while the cell is still running.",
"_____no_output_____"
]
],
[
[
"# Open in binary mode.\nwith open('bestBuy.csv.gz', 'rb') as file_handle:\n dataset_resource = data_manager.upload_data_to_dataset(dataset_id, file_handle)\n\nprint()\nprint(\"Dataset after data upload:\")\nprint()\npprint(dataset_resource)",
"_____no_output_____"
]
],
[
[
"Note that the Dataset status changed from `NO_DATA` to `VALIDATING`.\n\nDataset validation is a background process. The status will eventually change from `VALIDATING` to `SUCCEEDED`.\nThe SDK provides the [`DataManagerClient.wait_for_dataset_validation()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.wait_for_dataset_validation) method to poll for the Dataset validation.",
"_____no_output_____"
]
],
[
[
"dataset_resource = data_manager.wait_for_dataset_validation(dataset_id)\n\nprint()\nprint(\"Dataset after validation has finished:\")\n\nprint()\npprint(dataset_resource)",
"_____no_output_____"
]
],
[
[
"If the status is `FAILED` instead of `SUCCEEDED`, then the `validationMessage` will contain details about the validation failure.",
"_____no_output_____"
],
[
"To better understand the Dataset lifecycle, refer to the [corresponding document on help.sap.com](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/a9b7429687a04e769dbc7955c6c44265.html).",
"_____no_output_____"
],
[
"## Summary Exercise 01.2\n\nIn exercise 01.2, we have covered the following topics:\n\n* How to create a DatasetSchema\n* How to upload a Dataset to the service\n\nYou can find optional exercises related to exercise 01.2 [below](#Optional-Exercises-for-01.2).",
"_____no_output_____"
],
[
"# Exercise 01.3\n\n*Back to [table of contents](#Table-of-Contents)*\n\n*To perform this exercise, you need to execute the code in all previous exercises.*\n\nIn exercise 01.3, we will train the model.",
"_____no_output_____"
],
[
"## Training the Model",
"_____no_output_____"
],
[
"The Dataset is now uploaded and has been validated successfully by the service.\n\nTo train a machine learning model, we first need to select the correct model template.",
"_____no_output_____"
],
[
"### Selecting the right ModelTemplate\n\nThe Data Attribute Recommendation service currently supports two different ModelTemplates:\n\n| ID | Name | Description |\n|--------------------------------------|---------------------------|---------------------------------------------------------------------------|\n| d7810207-ca31-4d4d-9b5a-841a644fd81f | **Hierarchical template** | Recommended for the prediction of multiple classes that form a hierarchy. |\n| 223abe0f-3b52-446f-9273-f3ca39619d2c | **Generic template** | Generic neural network for multi-label, multi-class classification. |\n| 188df8b2-795a-48c1-8297-37f37b25ea00 | **AutoML template** | Finds the [best traditional machine learning model out of several traditional algorithms](https://blogs.sap.com/2021/04/28/how-does-automl-works-in-data-attribute-recommendation/). Single label only. |\n\nWe are building a model to predict product hierarchies. The **Hierarchical Template** is correct for this scenario. In this template, the first label in the DatasetSchema is considered the top-level category. Each subsequent label is considered to be further down in the hierarchy. \n\nComing back to our example DatasetSchema:\n\n```json\n{\n \"labels\": [\n {\"label\": \"level1_category\", \"type\": \"CATEGORY\"},\n {\"label\": \"level2_category\", \"type\": \"CATEGORY\"},\n {\"label\": \"level3_category\", \"type\": \"CATEGORY\"}\n ]\n}\n```\n\nThe first defined label is `level1_category`, which is given more weight during training than `level3_category`.\n\nRefer to the [official documentation on ModelTemplates](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/1e76e8c636974a06967552c05d40e066.html) to learn more. Additional model templates may be added over time, so check back regularly.",
"_____no_output_____"
],
[
"## Starting the training",
"_____no_output_____"
],
[
"When working with models, we use the [`ModelManagerClient`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient) class.\n\nTo start the training, we need the IDs of the dataset and the desired model template. We also have to provide a name for the model.\nThe [`ModelManagerClient.create_job()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.create_job) method launches the training Job.\n\n*Only one model of a given name can exist. If you receive a message stating 'The model name specified is already in use', you either have to remove the job and its associated model first or you have to change the `model_name` variable name below. You can also [clean up the entire service instance](#Cleaning-up-a-service-instance).*",
"_____no_output_____"
]
],
[
[
"from sap.aibus.dar.client.model_manager_client import ModelManagerClient\nfrom sap.aibus.dar.client.exceptions import DARHTTPException\n\nmodel_manager = ModelManagerClient.construct_from_service_key(SERVICE_KEY)\n\nmodel_template_id = \"d7810207-ca31-4d4d-9b5a-841a644fd81f\" # hierarchical template\nmodel_name = \"bestbuy-hierarchy-model\"\n\njob_resource = model_manager.create_job(model_name, dataset_id, model_template_id)\njob_id = job_resource['id']\n\nprint()\nprint(\"Job resource:\")\nprint()\n\npprint(job_resource)\n\nprint()\nprint(f\"ID of submitted Job: {job_id}\")",
"_____no_output_____"
]
],
[
[
"The job is now running in the background. Similar to the DatasetValidation, we have to poll the job until it succeeds.\nThe SDK provides the [`ModelManagerClient.wait_for_job()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.wait_for_job) method:",
"_____no_output_____"
]
],
[
[
"job_resource = model_manager.wait_for_job(job_id)\n\nprint()\n\nprint(\"Job resource after training is finished:\")\n\npprint(job_resource)",
"_____no_output_____"
]
],
[
[
"To better understand the Training Job lifecycle, see the [corresponding document on help.sap.com](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/0fc40aa077ce4c708c1e5bfc875aa3be.html).",
"_____no_output_____"
],
[
"## Intermission\n\nThe model training will take between 5 and 10 minutes.\n\nIn the meantime, we can explore the available [resources](#Resources) for both the service and the SDK.",
"_____no_output_____"
],
[
"## Inspecting the Model\n\nOnce the training job is finished successfully, we can inspect the model using [`ModelManagerClient.read_model_by_name()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.read_model_by_name).\n",
"_____no_output_____"
]
],
[
[
"model_resource = model_manager.read_model_by_name(model_name)\n\nprint()\npprint(model_resource)",
"_____no_output_____"
]
],
[
[
"In the model resource, the `validationResult` key provides information about model performance. You can also use these metrics to compare performance of different [ModelTemplates](#Selecting-the-right-ModelTemplate) or different datasets.",
"_____no_output_____"
],
[
"## Summary Exercise 01.3\n\nIn exercise 01.3, we have covered the following topics:\n\n* How to select the appropriate ModelTemplate\n* How to train a Model from a previously uploaded Dataset\n\nYou can find optional exercises related to exercise 01.3 [below](#Optional-Exercises-for-01.3).",
"_____no_output_____"
],
[
"# Exercise 01.4\n\n*Back to [table of contents](#Table-of-Contents)*\n\n*To perform this exercise, you need to execute the code in all previous exercises.*\n\nIn exercise 01.4, we will deploy the model and predict labels for some unlabeled data.",
"_____no_output_____"
],
[
"## Deploying the Model",
"_____no_output_____"
],
[
"The training job has finished and the model is ready to be deployed. By deploying the model, we create a server process in the background on the Data Attribute Recommendation service which will serve inference requests.\n\nIn the SDK, the [`ModelManagerClient.create_deployment()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#module-sap.aibus.dar.client.model_manager_client) method lets us create a Deployment.\n",
"_____no_output_____"
]
],
[
[
"deployment_resource = model_manager.create_deployment(model_name)\ndeployment_id = deployment_resource[\"id\"]\n\nprint()\nprint(\"Deployment resource:\")\nprint()\n\npprint(deployment_resource)\n\nprint(f\"Deployment ID: {deployment_id}\")",
"_____no_output_____"
]
],
[
[
"*Note: if you are using a trial account and you see errors such as 'The resource can no longer be used. Usage limit has been reached', consider [cleaning up the service instance](#Cleaning-up-a-service-instance) to free up limited trial resources.*",
"_____no_output_____"
],
[
"Similar to the data upload and the training job, model deployment is an asynchronous process. We have to poll the API until the Deployment is in status `SUCCEEDED`. The SDK provides the [`ModelManagerClient.wait_for_deployment()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.wait_for_deployment) for this purposes.",
"_____no_output_____"
]
],
[
[
"deployment_resource = model_manager.wait_for_deployment(deployment_id)\n\nprint()\nprint(\"Finished deployment resource:\")\nprint()\n\npprint(deployment_resource)",
"_____no_output_____"
]
],
[
[
"Once the Deployment is in status `SUCCEEDED`, we can run inference requests.",
"_____no_output_____"
],
[
"To better understand the Deployment lifecycle, see the [corresponding document on help.sap.com](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/f473b5b19a3b469e94c40eb27623b4f0.html).",
"_____no_output_____"
],
[
"*For trial users: the deployment will be stopped after 8 hours. You can restart it by deleting the deployment and creating a new one for your model. The [`ModelManagerClient.ensure_deployment_exists()`](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/c03b561eea1744c9b9892b416037b99a.html) method will delete and re-create automatically. Then, you need to poll until the deployment is succeeded using [`ModelManagerClient.wait_for_deployment()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.wait_for_deployment) as above.*",
"_____no_output_____"
],
[
"## Executing Inference requests",
"_____no_output_____"
],
[
"With a single inference request, we can send up to 50 objects to the service to predict the labels. The data send to the service must match the `features` section of the DatasetSchema created earlier. The `labels` defined inside of the DatasetSchema will be predicted for each object and returned as a response to the request.\n\nIn the SDK, the [`InferenceClient.create_inference_request()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.inference_client.InferenceClient.create_inference_request) method handles submission of inference requests.",
"_____no_output_____"
]
],
[
[
"from sap.aibus.dar.client.inference_client import InferenceClient\n\ninference = InferenceClient.construct_from_service_key(SERVICE_KEY)\n\nobjects_to_be_classified = [\n {\n \"features\": [\n {\"name\": \"manufacturer\", \"value\": \"Energizer\"},\n {\"name\": \"description\", \"value\": \"Alkaline batteries; 1.5V\"},\n {\"name\": \"price\", \"value\": \"5.99\"},\n ],\n },\n]\n\ninference_response = inference.create_inference_request(model_name, objects_to_be_classified)\n\nprint()\nprint(\"Inference request processed. Response:\")\nprint()\npprint(inference_response)",
"_____no_output_____"
]
],
[
[
"*Note: For trial accounts, you only have a limited number of objects which you can classify.*",
"_____no_output_____"
],
[
"You can also try to come up with your own example:",
"_____no_output_____"
]
],
[
[
"\n\nmy_own_items = [\n {\n \"features\": [\n {\"name\": \"manufacturer\", \"value\": \"EDIT THIS\"},\n {\"name\": \"description\", \"value\": \"EDIT THIS\"},\n {\"name\": \"price\", \"value\": \"0.00\"},\n ],\n },\n]\n\ninference_response = inference.create_inference_request(model_name, my_own_items)\n\nprint()\nprint(\"Inference request processed. Response:\")\nprint()\npprint(inference_response)",
"_____no_output_____"
]
],
[
[
"You can also classify multiple objects at once. For each object, the `top_n` parameter determines how many predictions are returned.",
"_____no_output_____"
]
],
[
[
"objects_to_be_classified = [\n {\n \"objectId\": \"optional-identifier-1\",\n \"features\": [\n {\"name\": \"manufacturer\", \"value\": \"Energizer\"},\n {\"name\": \"description\", \"value\": \"Alkaline batteries; 1.5V\"},\n {\"name\": \"price\", \"value\": \"5.99\"},\n ],\n },\n {\n \"objectId\": \"optional-identifier-2\",\n \"features\": [\n {\"name\": \"manufacturer\", \"value\": \"Eidos\"},\n {\"name\": \"description\", \"value\": \"Unravel a grim conspiracy at the brink of Revolution\"},\n {\"name\": \"price\", \"value\": \"19.99\"},\n ],\n },\n {\n \"objectId\": \"optional-identifier-3\",\n \"features\": [\n {\"name\": \"manufacturer\", \"value\": \"Cadac\"},\n {\"name\": \"description\", \"value\": \"CADAC Grill Plate for Safari Chef Grills: 12\\\"\"\n + \"cooking surface; designed for use with Safari Chef grills;\"\n + \"105 sq. in. cooking surface; PTFE nonstick coating;\"\n + \" 2 grill surfaces\"\n },\n {\"name\": \"price\", \"value\": \"39.99\"},\n ],\n }\n]\n\n\ninference_response = inference.create_inference_request(model_name, objects_to_be_classified, top_n=3)\n\nprint()\nprint(\"Inference request processed. Response:\")\nprint()\npprint(inference_response)",
"_____no_output_____"
]
],
[
[
"We can see that the service now returns the `n-best` predictions for each label as indicated by the `top_n` parameter.\n\nIn some cases, the predicted category has the special value `nan`. In the `bestBuy.csv` data set, not all records have the full set of three categories. Some records only have a top-level category. The model learns this fact from the data and will occasionally suggest that a record should not have a category.",
"_____no_output_____"
]
],
[
[
"# Inspect all video games with just a top-level category entry\nvideo_games = df[df['level1_category'] == 'Video Games']\nvideo_games.loc[df['level2_category'].isna() & df['level3_category'].isna()].head(5)",
"_____no_output_____"
]
],
[
[
"To learn how to execute inference calls without the SDK just using the underlying RESTful API, see [Inference without the SDK](#Inference-without-the-SDK).",
"_____no_output_____"
],
[
"## Summary Exercise 01.4\n\nIn exercise 01.4, we have covered the following topics:\n\n* How to deploy a previously trained model\n* How to execute inference requests against a deployed model\n\nYou can find optional exercises related to exercise 01.4 [below](#Optional-Exercises-for-01.4).",
"_____no_output_____"
],
[
"# Wrapping up\n\nIn this workshop, we looked into the following topics:\n\n* Installation of the Python SDK for Data Attribute Recommendation\n* Modelling data with a DatasetSchema\n* Uploading data into a Dataset\n* Training a model\n* Predicting labels for unlabelled data\n\nUsing these tools, we are able to solve the problem of missing Master Data attributes starting from just a CSV file containing training data.\n\nFeel free to revisit the workshop materials at any time. The [resources](#Resources) section below contains additional reading.\n\nIf you would like to explore the additional capabilities of the SDK, visit the [optional exercises](#Optional-Exercises) below.",
"_____no_output_____"
],
[
"## Cleanup",
"_____no_output_____"
],
[
"During the course of the workshop, we have created several resources on the Data Attribute Recommendation Service:\n\n* DatasetSchema\n* Dataset\n* Job\n* Model\n* Deployment\n\nThe SDK provides several methods to delete these resources. Note that there are dependencies between objects: you cannot delete a Dataset without deleting the Model beforehand.\n\nYou will need to set `CLEANUP_SESSION = True` below to execute the cleanup.",
"_____no_output_____"
]
],
[
[
"# Clean up all resources created earlier\n\nCLEANUP_SESSION = False\n\ndef cleanup_session():\n model_manager.delete_deployment_by_id(deployment_id) # this can take a few seconds\n model_manager.delete_model_by_name(model_name)\n model_manager.delete_job_by_id(job_id)\n\n data_manager.delete_dataset_by_id(dataset_id)\n data_manager.delete_dataset_schema_by_id(dataset_schema_id)\n print(\"DONE cleaning up!\")\n\nif CLEANUP_SESSION:\n print(\"Cleaning up resources generated in this session.\")\n cleanup_session()\nelse:\n print(\"Not cleaning up. Set 'CLEANUP_SESSION = True' above and run again!\")",
"_____no_output_____"
]
],
[
[
"## Resources\n\n*Back to [table of contents](#Table-of-Contents)*\n\n### SDK Resources\n\n* [SDK source code on Github](https://github.com/SAP/data-attribute-recommendation-python-sdk)\n* [SDK documentation](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/)\n* [How to obtain support](https://github.com/SAP/data-attribute-recommendation-python-sdk/blob/master/README.md#how-to-obtain-support)\n* [Tutorials: Classify Data Records with the SDK for Data Attribute Recommendation](https://developers.sap.com/group.cp-aibus-data-attribute-sdk.html)\n\n### Data Attribute Recommendation\n\n* [SAP Help Portal](https://help.sap.com/viewer/product/Data_Attribute_Recommendation/SHIP/en-US)\n* [API Reference](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/b45cf9b24fd042d082c16191aa938c8d.html)\n* [Tutorials using Postman - interact with the service RESTful API directly](https://developers.sap.com/mission.cp-aibus-data-attribute.html)\n* [Trial Account Limits](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/c03b561eea1744c9b9892b416037b99a.html)\n* [Metering and Pricing](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/1e093326a2764c298759fcb92c5b0500.html)\n",
"_____no_output_____"
],
[
"## Addendum",
"_____no_output_____"
],
[
"### Inference without the SDK\n\n*Back to [table of contents](#Table-of-Contents)*",
"_____no_output_____"
],
[
"The Data Attribute Service exposes a RESTful API. The SDK we use in this workshop uses this API to interact with the DAR service.\n\nFor custom integration, you can implement your own client for the API. The tutorial \"[Use Machine Learning to Classify Data Records]\" is a great way to explore the Data Attribute Recommendation API with the Postman REST client. Beyond the tutorial, the [API Reference] is a comprehensive documentation of the RESTful interface.\n\n[Use Machine Learning to Classify Data Records]: https://developers.sap.com/mission.cp-aibus-data-attribute.html\n\n[API Reference]: https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/b45cf9b24fd042d082c16191aa938c8d.html\n\nTo demonstrate the underlying API, the next example uses the `curl` command line tool to perform an inference request against the Inference API.\n\nThe example uses the `jq` command to extract the credentials from the service. The authentication token is retrieved from the `uaa_url` and then used for the inference request.",
"_____no_output_____"
]
],
[
[
"# If the following example gives you errors that the jq or curl commands cannot be found,\n# you may be able to install them from conda by uncommenting one of the lines below:\n#%conda install -q jq\n#%conda install -q curl",
"_____no_output_____"
],
[
"%%bash -s \"$model_name\" # Pass the python model_name variable as the first argument to shell script\n\nmodel_name=$1\n\necho \"Model: $model_name\"\n\nkey=$(cat key.json)\nurl=$(echo $key | jq -r .url)\nuaa_url=$(echo $key | jq -r .uaa.url)\nclientid=$(echo $key | jq -r .uaa.clientid)\nclientsecret=$(echo $key | jq -r .uaa.clientsecret)\n\necho \"Service URL: $url\"\n\ntoken_url=${uaa_url}/oauth/token?grant_type=client_credentials\n\necho \"Obtaining token with clientid $clientid from $token_url\"\n\nbearer_token=$(curl \\\n --silent --show-error \\\n --user $clientid:$clientsecret \\\n $token_url \\\n | jq -r .access_token\n )\n\ninference_url=${url}/inference/api/v3/models/${model_name}/versions/1\n\necho \"Running inference request against endpoint $inference_url\"\n\necho \"\"\n\n# We pass the token in the Authorization header.\n# The payload for the inference request is passed as\n# the body of the POST request below.\n# The output of the curl command is piped through `jq`\n# for pretty-printing\ncurl \\\n --silent --show-error \\\n --header \"Authorization: Bearer ${bearer_token}\" \\\n --header \"Content-Type: application/json\" \\\n -XPOST \\\n ${inference_url} \\\n -d '{\n \"objects\": [\n {\n \"features\": [\n {\n \"name\": \"manufacturer\",\n \"value\": \"Energizer\"\n },\n {\n \"name\": \"description\",\n \"value\": \"Alkaline batteries; 1.5V\"\n },\n {\n \"name\": \"price\",\n \"value\": \"5.99\"\n }\n ]\n }\n ]\n }' | jq",
"_____no_output_____"
]
],
[
[
"### Cleaning up a service instance\n\n*Back to [table of contents](#Table-of-Contents)*\n\nTo clean all data on the service instance, you can run the following snippet. The code is self-contained and does not require you to execute any of the cells above. However, you will need to have the `key.json` containing a service key in place.\n\nYou will need to set `CLEANUP_EVERYTHING = True` below to execute the cleanup.\n\n**NOTE: This will delete all data on the service instance!**",
"_____no_output_____"
]
],
[
[
"CLEANUP_EVERYTHING = False\n\ndef cleanup_everything():\n import logging\n import sys\n\n logging.basicConfig(level=logging.INFO, stream=sys.stdout)\n\n import json\n import os\n\n if not os.path.exists(\"key.json\"):\n msg = \"key.json is not found. Please follow instructions above to create a service key of\"\n msg += \" Data Attribute Recommendation. Then, upload it into the same directory where\"\n msg += \" this notebook is saved.\"\n print(msg)\n raise ValueError(msg)\n\n with open(\"key.json\") as file_handle:\n key = file_handle.read()\n SERVICE_KEY = json.loads(key)\n\n from sap.aibus.dar.client.model_manager_client import ModelManagerClient\n\n model_manager = ModelManagerClient.construct_from_service_key(SERVICE_KEY)\n\n for deployment in model_manager.read_deployment_collection()[\"deployments\"]:\n model_manager.delete_deployment_by_id(deployment[\"id\"])\n\n for model in model_manager.read_model_collection()[\"models\"]:\n model_manager.delete_model_by_name(model[\"name\"])\n\n for job in model_manager.read_job_collection()[\"jobs\"]:\n model_manager.delete_job_by_id(job[\"id\"])\n\n from sap.aibus.dar.client.data_manager_client import DataManagerClient\n\n data_manager = DataManagerClient.construct_from_service_key(SERVICE_KEY)\n\n for dataset in data_manager.read_dataset_collection()[\"datasets\"]:\n data_manager.delete_dataset_by_id(dataset[\"id\"])\n\n for dataset_schema in data_manager.read_dataset_schema_collection()[\"datasetSchemas\"]:\n data_manager.delete_dataset_schema_by_id(dataset_schema[\"id\"])\n \n print(\"Cleanup done!\")\n\nif CLEANUP_EVERYTHING:\n print(\"Cleaning up all resources in this service instance.\")\n cleanup_everything()\nelse:\n print(\"Not cleaning up. Set 'CLEANUP_EVERYTHING = True' above and run again.\")",
"_____no_output_____"
]
],
[
[
"### Optional Exercises\n\n*Back to [table of contents](#Table-of-Contents)*\n\nTo work with the optional exercises, create a new cell in the Jupyter notebook by clicking the `+` button in the menu above or by using the `b` shortcut on your keyboard. You can then enter your code in the new cell and execute it.\n\n#### Optional Exercises for 01.2\n\n##### DatasetSchemas\n\nUse the [`DataManagerClient.read_dataset_schema_by_id()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.read_dataset_schema_by_id) and the [`DataManagerClient.read_dataset_schema_collection()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.read_dataset_schema_collection) methods to list the newly created and all DatasetSchemas, respectively.\n\n##### Datasets\n\nUse the [`DataManagerClient.read_dataset_by_id()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.read_dataset_by_id) and the [`DataManagerClient.read_dataset_collection()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.read_dataset_collection) methods to inspect the newly created dataset.\n\nInstead of using two separate methods to upload data and wait for validation to finish, you can also use [`DataManagerClient.upload_data_and_validate()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.upload_data_and_validate).\n\n#### Optional Exercises for 01.3\n\n##### ModelTemplates\n\nUse the [`ModelManagerClient.read_model_template_collection()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.read_model_template_collection) to list all existing model templates.\n\n##### Jobs\n\nUse [`ModelManagerClient.read_job_by_id()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.read_job_by_id) and [`ModelManagerClient.read_job_collection()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.read_job_collection) to inspect the job we just created.\n\nThe entire process of uploading the data and starting the training is also available as a single method call in [`ModelCreator.create()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.workflow.model.ModelCreator.create).\n\n#### Optional Exercises for 01.4\n\n##### Deployments\n\nUse [`ModelManagerClient.read_deployment_by_id()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.read_deployment_by_id) and [`ModelManagerClient.read_deployment_collection()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.read_deployment_collection) to inspect the Deployment.\n\nUse the [`ModelManagerclient.lookup_deployment_id_by_model_name()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.lookup_deployment_id_by_model_name) method to find the deployment ID for a given model name.\n\n##### Inference\n\nUse the [`InferenceClient.do_bulk_inference()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.inference_client.InferenceClient.do_bulk_inference) method to process more than fifty objects at a time. Note how the data format returned changes.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d03c84070a82ed2e56844b7fdf264a6e312b997d | 12,119 | ipynb | Jupyter Notebook | project_1/Rensselaer ML Data Acrchitecture Project 1 (Classification with Tensorflow).ipynb | jmullini/rensselaer_ml_data_architecture | 90da0846f7b809630b2e9a5e7ed9b3f74c240d84 | [
"MIT",
"Unlicense"
] | 2 | 2021-02-24T22:52:29.000Z | 2021-02-24T22:53:20.000Z | project_1/Rensselaer ML Data Acrchitecture Project 1 (Classification with Tensorflow).ipynb | jmullini/rensselaer_ml_data_architecture | 90da0846f7b809630b2e9a5e7ed9b3f74c240d84 | [
"MIT",
"Unlicense"
] | null | null | null | project_1/Rensselaer ML Data Acrchitecture Project 1 (Classification with Tensorflow).ipynb | jmullini/rensselaer_ml_data_architecture | 90da0846f7b809630b2e9a5e7ed9b3f74c240d84 | [
"MIT",
"Unlicense"
] | null | null | null | 44.229927 | 258 | 0.46654 | [
[
[
"# Turning off future warnings\nfrom warnings import simplefilter\nsimplefilter(action='ignore', category=FutureWarning)\n\n# General data handling/io\nimport datetime\nimport pandas as pd\nimport numpy as np\nimport pickle\n\nfrom sklearn.model_selection import train_test_split\nfrom tensorflow.keras import Sequential\nfrom tensorflow.keras.layers import Dense\nimport tensorflow as tf\n",
"_____no_output_____"
],
[
"def base_data():\n \n return pd.read_csv('processed_data.csv')",
"_____no_output_____"
],
[
"def select_features(df):\n \n features = [f for f in df.columns if f != 'serious_delinquencies_in_past_2_years']\n \n X = df[features]\n \n X = X.astype('float32')\n \n Y = df['serious_delinquencies_in_past_2_years'].values\n \n return X, Y",
"_____no_output_____"
],
[
"class tfCallback(tf.keras.callbacks.Callback):\n \n def on_epoch_end(self, epochs, logs={}) :\n threshold = 0.84\n if(logs.get('acc') is not None and logs.get('acc') >= threshold) :\n print('\\nReached {}% accuracy so cancelling further training'.format(str(threshold)))\n self.model.stop_training = True",
"_____no_output_____"
],
[
"def build_network():\n \n ## Making a classifier with tensorflow\n \n # 500 epochs yielded about 84%\n \n # Load data\n df = base_data()\n\n # Split out X, Y\n X, Y = select_features(df)\n \n X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.33)\n \n #print(X_train.shape, X_test.shape, y_train.shape, y_test.shape)\n \n n_features = X_train.shape[1]\n \n callbacks = tfCallback()\n \n model = tf.keras.models.Sequential([\n \n tf.keras.layers.Dense(10, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,)),\n tf.keras.layers.Dense(8, activation='relu', kernel_initializer='he_normal'),\n tf.keras.layers.Dense(1, activation='sigmoid')\n\n ])\n \n # compile the model\n model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])\n \n model.fit(X_train, y_train, epochs=500, callbacks=[callbacks]) ",
"_____no_output_____"
],
[
"## Results way beyond what you would get in the real world\nbuild_network()",
"WARNING:tensorflow:From /home/james/.local/lib/python3.6/site-packages/tensorflow/python/ops/init_ops.py:1251: calling VarianceScaling.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.\nInstructions for updating:\nCall initializer instance with the dtype argument instead of passing it to the constructor\nWARNING:tensorflow:From /home/james/.local/lib/python3.6/site-packages/tensorflow/python/ops/nn_impl.py:180: add_dispatch_support.<locals>.wrapper (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\nEpoch 1/500\n29688/29688 [==============================] - 1s 26us/sample - loss: 0.5110 - acc: 0.8120\nEpoch 2/500\n29688/29688 [==============================] - 1s 27us/sample - loss: 0.4031 - acc: 0.8345\nEpoch 3/500\n29688/29688 [==============================] - 1s 27us/sample - loss: 0.3991 - acc: 0.8361\nEpoch 4/500\n29688/29688 [==============================] - 1s 25us/sample - loss: 0.3968 - acc: 0.8353\nEpoch 5/500\n29688/29688 [==============================] - 1s 28us/sample - loss: 0.3965 - acc: 0.8351\nEpoch 6/500\n29688/29688 [==============================] - 1s 28us/sample - loss: 0.3962 - acc: 0.8367\nEpoch 7/500\n29688/29688 [==============================] - 1s 28us/sample - loss: 0.3960 - acc: 0.8354\nEpoch 8/500\n29688/29688 [==============================] - 1s 25us/sample - loss: 0.3954 - acc: 0.8374\nEpoch 9/500\n29688/29688 [==============================] - 1s 27us/sample - loss: 0.3958 - acc: 0.8356\nEpoch 10/500\n29688/29688 [==============================] - 1s 28us/sample - loss: 0.3951 - acc: 0.8361\nEpoch 11/500\n29688/29688 [==============================] - 1s 30us/sample - loss: 0.3949 - acc: 0.8377\nEpoch 12/500\n29688/29688 [==============================] - 1s 28us/sample - loss: 0.3949 - acc: 0.8370\nEpoch 13/500\n29688/29688 [==============================] - 1s 29us/sample - loss: 0.3949 - acc: 0.8364\nEpoch 14/500\n29688/29688 [==============================] - 1s 24us/sample - loss: 0.3944 - acc: 0.8370\nEpoch 15/500\n29688/29688 [==============================] - 1s 27us/sample - loss: 0.3941 - acc: 0.8367\nEpoch 16/500\n29688/29688 [==============================] - 1s 30us/sample - loss: 0.3941 - acc: 0.8376\nEpoch 17/500\n29688/29688 [==============================] - 1s 24us/sample - loss: 0.3939 - acc: 0.8378\nEpoch 18/500\n29688/29688 [==============================] - 1s 24us/sample - loss: 0.3938 - acc: 0.8373\nEpoch 19/500\n29688/29688 [==============================] - 1s 31us/sample - loss: 0.3937 - acc: 0.8396\nEpoch 20/500\n29688/29688 [==============================] - 1s 24us/sample - loss: 0.3935 - acc: 0.8371\nEpoch 21/500\n29688/29688 [==============================] - 1s 28us/sample - loss: 0.3934 - acc: 0.8370\nEpoch 22/500\n29688/29688 [==============================] - 1s 27us/sample - loss: 0.3936 - acc: 0.8372\nEpoch 23/500\n29688/29688 [==============================] - 1s 31us/sample - loss: 0.3934 - acc: 0.8387\nEpoch 24/500\n29688/29688 [==============================] - 1s 30us/sample - loss: 0.3928 - acc: 0.8368\nEpoch 25/500\n29688/29688 [==============================] - 1s 27us/sample - loss: 0.3920 - acc: 0.8377\nEpoch 26/500\n29688/29688 [==============================] - 1s 24us/sample - loss: 0.3921 - acc: 0.8388\nEpoch 27/500\n29688/29688 [==============================] - 1s 27us/sample - loss: 0.3923 - acc: 0.8380\nEpoch 28/500\n29688/29688 [==============================] - 1s 36us/sample - loss: 0.3920 - acc: 0.8394\nEpoch 29/500\n29688/29688 [==============================] - 1s 48us/sample - loss: 0.3917 - acc: 0.8399\nEpoch 30/500\n29688/29688 [==============================] - 1s 39us/sample - loss: 0.3915 - acc: 0.8392\nEpoch 31/500\n29688/29688 [==============================] - 1s 34us/sample - loss: 0.3910 - acc: 0.8386\nEpoch 32/500\n29688/29688 [==============================] - 1s 36us/sample - loss: 0.3913 - acc: 0.8391\nEpoch 33/500\n29688/29688 [==============================] - 1s 40us/sample - loss: 0.3909 - acc: 0.8397\nEpoch 34/500\n29688/29688 [==============================] - 1s 34us/sample - loss: 0.3911 - acc: 0.8394\nEpoch 35/500\n29688/29688 [==============================] - 1s 32us/sample - loss: 0.3911 - acc: 0.8387\nEpoch 36/500\n29688/29688 [==============================] - 1s 33us/sample - loss: 0.3907 - acc: 0.83940s - loss: 0.3963 \nEpoch 37/500\n29688/29688 [==============================] - 1s 37us/sample - loss: 0.3910 - acc: 0.8387\nEpoch 38/500\n29688/29688 [==============================] - 1s 38us/sample - loss: 0.3906 - acc: 0.8388\nEpoch 39/500\n29688/29688 [==============================] - 1s 43us/sample - loss: 0.3907 - acc: 0.8390\nEpoch 40/500\n29688/29688 [==============================] - 1s 39us/sample - loss: 0.3906 - acc: 0.8386\nEpoch 41/500\n29688/29688 [==============================] - 1s 50us/sample - loss: 0.3905 - acc: 0.8388\nEpoch 42/500\n29688/29688 [==============================] - 1s 34us/sample - loss: 0.3905 - acc: 0.8394\nEpoch 43/500\n29688/29688 [==============================] - 1s 34us/sample - loss: 0.3902 - acc: 0.8383\nEpoch 44/500\n29688/29688 [==============================] - 1s 34us/sample - loss: 0.3903 - acc: 0.8393\nEpoch 45/500\n29688/29688 [==============================] - 1s 39us/sample - loss: 0.3900 - acc: 0.8399\nEpoch 46/500\n29688/29688 [==============================] - 1s 32us/sample - loss: 0.3905 - acc: 0.8392\nEpoch 47/500\n29688/29688 [==============================] - 1s 35us/sample - loss: 0.3904 - acc: 0.8395\nEpoch 48/500\n29688/29688 [==============================] - 1s 32us/sample - loss: 0.3903 - acc: 0.8391\nEpoch 49/500\n29688/29688 [==============================] - 1s 34us/sample - loss: 0.3903 - acc: 0.8386\nEpoch 50/500\n29688/29688 [==============================] - 1s 31us/sample - loss: 0.3901 - acc: 0.8393\nEpoch 51/500\n29688/29688 [==============================] - 1s 34us/sample - loss: 0.3899 - acc: 0.8385\nEpoch 52/500\n29688/29688 [==============================] - 1s 32us/sample - loss: 0.3902 - acc: 0.8391\nEpoch 53/500\n29688/29688 [==============================] - 1s 29us/sample - loss: 0.3899 - acc: 0.8380\nEpoch 54/500\n29688/29688 [==============================] - 1s 33us/sample - loss: 0.3899 - acc: 0.8389\nEpoch 55/500\n29688/29688 [==============================] - 1s 34us/sample - loss: 0.3900 - acc: 0.8396\nEpoch 56/500\n29688/29688 [==============================] - 1s 32us/sample - loss: 0.3901 - acc: 0.8387\nEpoch 57/500\n29688/29688 [==============================] - 1s 32us/sample - loss: 0.3901 - acc: 0.8390\nEpoch 58/500\n29184/29688 [============================>.] - ETA: 0s - loss: 0.3892 - acc: 0.8403\nReached 0.84% accuracy so cancelling further training\n29688/29688 [==============================] - 1s 30us/sample - loss: 0.3897 - acc: 0.8400\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03c845c90af7907b8e4d2c62b8fcb01e5ec97e7 | 25,248 | ipynb | Jupyter Notebook | ES_espectro_vegetacion.ipynb | hectornieto/Curso-WUE | 8c1d36077c768daefe3ea13b50e0bca0a69f107d | [
"CC0-1.0"
] | 1 | 2022-01-28T13:05:17.000Z | 2022-01-28T13:05:17.000Z | ES_espectro_vegetacion.ipynb | hectornieto/Curso-WUE | 8c1d36077c768daefe3ea13b50e0bca0a69f107d | [
"CC0-1.0"
] | null | null | null | ES_espectro_vegetacion.ipynb | hectornieto/Curso-WUE | 8c1d36077c768daefe3ea13b50e0bca0a69f107d | [
"CC0-1.0"
] | null | null | null | 70.133333 | 729 | 0.709799 | [
[
[
"# Resumen\nEste cuaderno digital interactivo tiene como objetivo demostrar las relaciones entre las propiedades fisico-químicas de la vegetación y el espectro solar.\n\nPara ello haremos uso de modelos de simulación, en particular de modelos de transferencia radiativa tanto a nivel de hoja individual como a nivel de dosel vegetal.",
"_____no_output_____"
],
[
"# Instrucciones\nLee con detenimiento todo el texto, y sigue sus instrucciones.\n\nUna vez leida cada sección de texto ejecuta la celda de código siguiente (marcada como `In []`) presionando el icono de `Run`/`Ejecutar` o presionando en el teclado ALT + ENTER. Aparecerá una interfaz gráfica con la que poder realizar las tareas asignadas.\n\nComo ejemplo ejectuta la siguiente celda para importar todas las librerías necesarias para el correcto funcionamiento del cuaderno. Una vez ejecutada debería aparecer un mensaje de agradecimiento.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nfrom ipywidgets import interactive, fixed\nfrom IPython.display import display\nfrom functions import prosail_and_spectra as fn",
"_____no_output_____"
]
],
[
[
"# Espectro de una hoja\nLas propiedades espectrales de una hoja (tanto su transmisividad, su reflectividad y su absortividad) dependen de su concentración de pigmentos, de su contenido de agua, su peso específico y la estructura interna de sus tejidos. \n\nVamos a usar el modelo ProspectD, el cual es una simplificación de la realidad en la que simula el espectro mediante la concentración de clorofilas (`Cab`), carotenoides (`Car`), antocianinos (`Ant`), así como el peso de agua y por unidad de supeficie (`Cw`) y el peso del resto de la materia seca (`Cm`) que engloba las celulosas, ligninas (responsables principales de la biomasa foliar) y otros componentes proteicos. También incluye un parámetro semi-empírico que representa otros pigmentos responsables del color de las hojas senescentes y enfermas. Además con el fin de simular hojas con distintas estructuras celulares incluye un último parámetro (`Nf`) que emula las distitas capas y tejidos celulares de la hoja.\n\n con idénticas propiedades espectrales\")\n\n> Si quieres saber más sobre el modelo ProspectD pincha en esta [publicación](./lecturas_adicionales/ProspectD_model.pdf).\n>\n> Si quieres más detalles sobre el cálculo y el código del modelo pincha [aquí](https://github.com/hectornieto/pypro4sail/blob/b111891e0a2c01b8b3fa5ff41790687d31297e5f/pypro4sail/prospect.py#L46).\n\nEjecuta la siguiente célula y verás un espectro típico de la hoja. El gráfico muestra tanto la reflectividad (en el eje y) como la transmisividad (en el eje secundario y, con valores invertidos) y la absortividad (como el espacio entre las dos curvas de reflectividad y transmisividad) $\\rho + \\tau + \\alpha = 1$.\n\nPresta atención a cómo y en qué regiones cambia el espectro según el parámetro que modifiques.\n* Haz variar la clorofila. \n* Haz variar el contenido de agua\n* Haz variar la materia seca\n* Haz variar los pigmentos marrones desde un valor de 0 (hoja sana) a valores mayores (hoja enferma o seca)",
"_____no_output_____"
]
],
[
[
"w_rho_leaf = interactive(fn.update_prospect_spectrum, N_leaf=fn.w_nleaf, Cab=fn.w_cab, \n Car=fn.w_car, Ant=fn.w_ant, Cbrown=fn.w_cbrown, Cw=fn.w_cw, Cm=fn.w_cm)\ndisplay(w_rho_leaf)",
"_____no_output_____"
]
],
[
[
"Observa lo siguente:\n* La concentración de clorofila `Cab` afecta principalmente a la región del visible (RGB) y del *red egde* (R-E), con más absorción en la región del rojo y del azul y más reflexión en el verde. Es por ello que la mayoría de las hojas presentan color verde.\n* El contenido de agua `Cw` afecta principalmente a la absorción en el infrarrojo de onda corta (SWIR), con máximos de absorción en trono a los 1460 y 2100 nm.\n* La materia seca `Cm` afecta principalmente a la absorción en el infrarrojo cercano (NIR).\n* Otros pigmentos afectan en menor medida al espectro visible. Por ejemplo los antocianos `Ant` que suelen aparecer durante la senescencia desplazan el pico de reflexión del verde hacia el rojo, sobre todo cuando a su vez decrece la concentración de clorofila.\n* El parámetro `N` afecta a la relación entre reflectividad y transmisividad. Cuantas más *capas* tenga una hoja más fenómenos de dispersión múltiple habrá y reflejará más.\n> Puedes ver este fenómeno también en las ventanas con doble o triple cristal usadas como aislante, por ejemplo de los escaparates comerciales. A no ser que uno se sitúen justo de frente y cerca del escaparate, éste parece más un espejo que una ventana.",
"_____no_output_____"
],
[
"# Espectro del suelo\nEl espectro del dosel o de la supeficie vegetal no sólo depende del espectro y las propiedades de las hojas, sino que también de la propia estructura del dosel así como del suelo. En particular en doseles abiertos o poco densos, como en las primeras fases fenológicas, el comportamiento espectral del suelo puede influir de manera muy importante en la señal espectral que capten los sensores de teledetección.\n\nEl espectro del suelo depende de varios factores, como son su composición mineralógica, materia orgánica, su textura y densidad así como su humedad superficial. \n\nEjectuta la siguiente celda y mira los distintas características espectrales de distintos tipos de suelo.",
"_____no_output_____"
]
],
[
[
"w_rho_soil = interactive(fn.update_soil_spectrum, soil_name=fn.w_soil)\ndisplay(w_rho_soil)",
"_____no_output_____"
]
],
[
[
"Observa lo diferente que puede ser un espectro de suelo en comparación con el de una hoja. Esto es clave a la hora de clasificar tipos de coberturas mediante teledetección así como cuantificar el vigor/densidad vegetal del cultivo.\n\nObserva que suelos más salinos (`aridisol.salorthid`) o gipsicos (`aridisol.gypsiorthd`), tienen una mayor reflectividad, sobre todo en el visible (RGB). Es decir, son más blancos que otros suelos.",
"_____no_output_____"
],
[
"# Espectro del dosel\nFinalmente, integrando la firma espectral de una hoja y del suelo subyacente podemos obtener el espectro de un dosel vegetal. \n\nEl espectro de la superficie vegetal además depende de la estructura del dosel, principalmente de la cantidad de hojas por unidad de superficie (definido como el Índice de Área Foliar) y de cómo estas hojas se orientan con respecto a la vertical. Además, dado que se produce una interacción de la luz incidente y reflejada entre el volumen de hojas y el suelo, la posición del sol y del sensor influyen en la señal espectral que obtengamos.\n\nPara esta parte cobinaremos el modelo de transferencia ProspectD para simular el espectro de una hoja con otro modelo de trasnferencia a nivel de dosel (4SAIL). Este último modelo considera la superficie vegetal como una capa horizontal y verticalmente homogéna, por lo que se recomienda cautela en su aplicación en doseles arbóreos heterogéneos.\n\n\n\n> Si quieres saber más sobre el modelo 4SAIL pincha en esta [publicación](./lecturas_adicionales/4SAIL_model.pdf)\n> \n> Si quieres más detalles sobre el cálculo y el código del modelo pincha [aquí](https://github.com/hectornieto/pypro4sail/blob/b111891e0a2c01b8b3fa5ff41790687d31297e5f/pypro4sail/four_sail.py#L245)\n\nEjecuta la siguente celda y mira cómo los [espectros de hoja](#Espectro-de-una-hoja) y [suelo](#Espectro-del-suelo) que se han generado previamente se integran para obtener un espectro de la superficie vegetal.\n\n> Puedes modificar los espectros de hoja y suelo, y esta gráfica se actualizará automáticamente.",
"_____no_output_____"
]
],
[
[
"w_rho_canopy = interactive(fn.update_4sail_spectrum,\n lai=fn.w_lai, hotspot=fn.w_hotspot, leaf_angle=fn.w_leaf_angle, \n sza=fn.w_sza, vza=fn.w_vza, psi=fn.w_psi, skyl=fn.w_skyl, \n leaf_spectrum=fixed(w_rho_leaf), soil_spectrum=fixed(w_rho_soil))\ndisplay(w_rho_canopy)",
"_____no_output_____"
]
],
[
[
"Recuerda en la [práctica sobre la radiación neta](./ES_radiacion_neta.ipynb) que una superficie vegetal tiene ciertas propiedades anisotrópicas, lo que quiere decir que reflejará de manera distinta según la geometria de iluminación y de observación. \n\nMira cómo cambia el espectro variando los valores del ángulo de observación cenital (VZA), ańgulo cenital del sol (SZA) y el ángulo azimutal relativo (PSI) entre el sol y el observador.\n\nHaz variar el LAI, y ponlo en cero (sin vegetación). Comprueba que el espectro que sale es directamente el espectro del suelo. Ahora incrementa ligeramente el LAI, verás como el espectro va cambiando, disminuyendo la reflectividad en el rojo y azul (debido a la clorofila de la hoja), y aumentando la reflectividad en el *red-edge* y el NIR.\n\nRecuerda también de la [práctica sobre la radiación neta](./ES_radiacion_neta.ipynb) el efecto que también tiene la disposición angular de las hojas. Con una observación al nadir (VZA=0) haz variar el ángulo típico de la hoja (`Leaf Angle`) desde un valor predominantemente horizontal (0º) a un ángulo predominantemente vertical (90º) ",
"_____no_output_____"
],
[
"# Sensibilidad de los parámetros\nEn esta tarea podrás ver el comportamiento espectral de la vegetación según varían los parámetros fisico-químicos de la vegetación así como su sensibilidad a las condiciones de observación e iluminación.\n\nPara ello vamos a realizar un análisis de sensibilidad variando un sólo parámetro a la vez, mientras que el resto de los parámetros permanecerán constantes. Puedes variar los valores individuales para el resto de los parámetros individuales (también se actualizarán de las gráficas anteriores). A continuación selecciona qué parámetro quieres analizar y el rango de valores máximo y mínimo que quieras que tenga.",
"_____no_output_____"
]
],
[
[
"w_sensitivity = interactive(fn.prosail_sensitivity,\n N_leaf=fn.w_nleaf, Cab=fn.w_cab, Car=fn.w_car, Ant=fn.w_ant, Cbrown=fn.w_cbrown, \n Cw=fn.w_cw, Cm=fn.w_cm, lai=fn.w_lai, hotspot=fn.w_hotspot, leaf_angle=fn.w_leaf_angle, \n sza=fn.w_sza, vza=fn.w_vza, psi=fn.w_psi, skyl=fn.w_skyl, \n soil_name=fn.w_soil, var=fn.w_param, value_range=fn.w_range)\ndisplay(w_sensitivity)",
"_____no_output_____"
]
],
[
[
"Empieza con al sensiblidad del espectro a la concentración de clorofila. Verás que la zona donde sobre todo hay variaciones es en el verde y el rojo. Observa también que en el *red-edge*, la zona de transición entre el rojo y el NIR, se produce un \"desplazamiento\" de la señal, este fenómento es clave y es la razón por la que los nuevos sensores (Sentinel, nuevas cámaras UAV) incluyen esta región para ayudar en la estimación de la clorofila y por tanto en la actividad fotosintética.\n\nEvalúa la sensibilidad al espectro de otros pigmentos (`Car` o `Ant`). Verás que la respuesta espectral a estos otros pigmentos es menor, lo que implica que resulta más dificil estimarlos a partir de teledetección. En cambio la variación espectral con los pigmentos marrones es bastante fuerte, como recordatorio estos pigmentos representan las variaciones cromáticas que se producen en hojas enfermas y muertas.\n\n> Esto implica que es relativamente posible detectar y cuantificar problemas sanitarios en la vegetación.\n\nMira ahora la sensibilidad del LAI cuando su rango es pequeño (p.ej. de 0 a 2). Verás que el espectro cambia significativamente según incrementa el LAI. Ahora mira la sensibilidad cuando el LAI recorre valores mas altos (p.ej. de 2 a 4), verás que la variación en el espectro es mucho menor. Se suele decir que a valores altos de LAI el espectro tiende a \"saturarse\" por lo que la señal se hace menos sensible.\n\n> Es más fácil estimar el LAI con menor margen de error en cultivos con poca densidad foliar o fases fenológicas tempranas, que en cultivos o vegetación muy densa.\n\nAhora mantén el valor fijo de LAI en un valor alto (p.ej 3) y haz variar el ángulo de observación cenital entre 0º (nadir) y una obsrvación oblicua (p.ej 35º). Verás que a pesar de haber un LAI alto, y que a priori hemos visto que ya es menos sensible, hay mayores variaciones espectrales al variar la geometría de observación.\n\n> Gracias a la anisotropía de la vegetación, las variaciones espectrales con respecto a la geometría de observación e iluminación pueden ayudar a resolver el LAI en condiciones de alta densidad.\n\nAhora mira el peso específico de la hoja, o la cantidad de materia seca (`Cm`). Verás que según el peso específico de la hora se producen variaciones importantes en el NIR y SWIR.\n\n> La biomasa foliar puede calcularse a partir del producto entre el `LAI` y `Cm`, por lo que es viable estimar la biomasa foliar de un cultivo. Esta informaición puede ser útil por ejemplo para estimar el rendimiento final de algunos cultivos, como pueden ser los cereales.\n\nEl parámetro `hotspot` es un parámetro semi-empírico relacionado con el tamaño relativo de la hoja con respecto a la altura del dosel. Afecta a cómo las hojas ensombrecen otras hojas dentro del dosel, por lo que su efecto más fuerte se observará cuando el observador (sensor) está justo en la misma posición que el sol. Para ello valores similares para VZA y SZA, y el ángulo azimutal relativo PSI en 0º. Ahora haz variar el hotstpot. Al poner el observador en la zona iluminada de la vegetación, el tamaño relativo de las hojas juega un papel importante, ya que cuanto más grandes sean estas el volumen de copa directamente iluminado será mayor.\n\n",
"_____no_output_____"
],
[
"# La señal de un sensor\nHasta ahora hemos visto el comportamiento espectral detallado de la vegetación. Sin embargo los sensores a bordo de los satélites, aeroplanos y drones no miden todo el espectro en continuo, si no que muestrean tal espectro en torno a unas bandas específicas, estratégicamente seleccionadas con el fin de intentar capturar los aspectos biofísicos más relvantes.\n\nSe denomina función de respuesta espectral a la forma en que un sensor específico integra el espectro con el fin de proporcionar la información en sus distintas bandas. Cada sensor, con cada una de sus bandas, tiene una función de respuesta espectral propia.\n\nEn esta tarea veremos las respuestas espectrales de los sensores que utilizaremos más comunmente, Landsat, Sentinel-2 y Sentinel-3. También veremos el comportamiento espectral de una cámara típica que se usa con drones.\n\nPartimos de las simulaciones generadas anteriormente. Selecciona el sensor que quieras simular para ver como cada uno de los sensores \"verían\" esos mismos espectros.",
"_____no_output_____"
]
],
[
[
"w_rho_sensor = interactive(fn.sensor_sensitivity,\n sensor=fn.w_sensor, spectra=fixed(w_sensitivity))\ndisplay(w_rho_sensor)",
"_____no_output_____"
]
],
[
[
"Realiza de nuevo un análisis de sensibilidad para la clorofila y compara la respuesta espectral que daría Landsat, Sentinel-2 y una camára UAV",
"_____no_output_____"
],
[
"# Derivación de parámetros de la vegetación\nHasta ahora hemos visto cómo el espectro de la superficie varía con respecto a los distintos parámetros biofísicos.\n\nSin embargo, nuestro objetivo final es el contrario, es decir, a partir de un espectro, o de unas determinadas bandas espectrales estimar una o varias variables biofísicas que nos son de interés. En particular para el objetivo del cálculo de la evapotranspiración y la eficiencia en el uso en el agua, nos puede interesar estimar el índice de área foliar y/o la fracción de radiación PAR absorbida, así como las clorofilas u otros pigmentos.\n\nUna de los métodos típicos es desarrollar relaciones empíricas entre las bandas (o entre índices de vegetación) y datos muestreados en el campo. Esto puede darnos la respuesta más fiable para nuestra parcela de estudio, pero como has podido ver anteriormente la señal espectral depende de otros muchos factores, que pueden provocar que esa relación calibrada con unos cuantos muestreos locales no sea extrapolable o aplicable a otros cultivos o regiones.\n\nOtra alternativa es desarrollar bases de datos sintéticas a partir de simulaciones. Es lo que vamos a realizar en esta tarea.\n\nVamos a ejecutar 5000 simulaciones haciendo variar los valores de los parámetros aleatoriamente según un rango de valores que puedan ser esperado en nuestras zonas de estudio. \n\nPor ejemplo si trabajas con cultivos perennes puede que te interesa mantener un rango de LAI con valores mínimos sensiblemente superiores a cero, mientras si trabajas con cultivos anuales, el valor 0 es necesario para reflejar el desarrollo del cultivo desde su plantación, emergencia y madurez. \n\nYa que hay una gran cantidad de parámetros y es muy probable que desconozcamos el rango plausible en la mayoría de los cultivos, no te preocupes, deja los valores por defecto y céntrate en los parámetros en los que tengas más confianza.\n\nPuedes también elegir uno o varios tipos de suelo, en función de la edafología de tu lugar. \n\n> Incluso podrías subir un espectro de suelo típico de tu zona a la carpeta [./input/soil_spectral_library](./input/soil_spectral_library). Tan sólo asegúrate que el archivo de texto tenga dos columnas, la primera con las longitudes de onda de 400 a 2500 y la segunda columna con la reflectividad correspondiente. Para actualizar la librería espectral de suelos, tendrías que ejecutar también la [primera celda](#Instrucciones).\n\nFinalmente selecciona el sensor para el que quieras genera la señal.\n\nCuando tengas tu configurado tu entorno de simulación, pincha en el botón `Generar Simulaciones`. El sistema tardará un rato pero al cabo de unos minutos te retornará una serie de gráficos.\n\n> Es posible que recibas un mensaje de aviso, no te preocupes, en principio todo debería funcionar con normalidad.",
"_____no_output_____"
]
],
[
[
"w_rho_sensor = interactive(fn.build_random_simulations, {\"manual\": True, \"manual_name\": \"Generar simulaciones\"},\n n_sim=fixed(5000), n_leaf_range=fn.w_range_nleaf,\n cab_range=fn.w_range_cab, car_range=fn.w_range_car,\n ant_range=fn.w_range_ant, cbrown_range=fn.w_range_cbrown, \n cw_range=fn.w_range_cw, cm_range=fn.w_range_cm,\n lai_range=fn.w_range_lai, hotspot_range=fn.w_range_hotspot, \n leaf_angle_range=fn.w_range_leaf_angle, \n sza=fn.w_sza, vza=fn.w_vza, psi=fn.w_psi, \n skyl=fn.w_skyl, soil_names=fn.w_soils, sensor=fn.w_sensor)\ndisplay(w_rho_sensor)",
"_____no_output_____"
]
],
[
[
"El gráfico muestra 4 ejemplos de relaciones entre 3 índices de vegetación típicos y 4 variables biofísicas.\n\n* NDVI: Normalized Difference Vegetation Index. Es el índicie de vegetación más utilizado. Generalmente se relaciona con el LAI, la biomasa foliar y/o la fracción de radiación interceptada/absorbida\n$$NDVI = \\frac{\\rho_{NIR} - \\rho_{R}}{\\rho_{NIR} + \\rho_{R}}$$\n\n* NDRE: Normalized Difference Red-Edge. Es un índicie de vegetación que usa la región del red edge, por lo que no puede calcularse para cualquier sensor. Generalmente se relaciona con la clorofila.\n$$NDRE = \\frac{\\rho_{NIR} - \\rho_{R-E}}{\\rho_{NIR} + \\rho_{R-E}}$$\n\n* NDWI: Normalized Difference Water Index. Es un índicie de vegetación que usa la región del SWIR, por lo que no puede calcularse para cualquier sensor. Generalmente se relaciona con el contenido de agua de la vegetación.\n$$NDWI = \\frac{\\rho_{NIR} - \\rho_{SWIR}}{\\rho_{NIR} + \\rho_{SWIR}}$$\n\nLas simulaciones se han guardado en un archivo prosail_simulations.csv en la carpeta [./output](./output/prosail_simulations.csv). Descargate este archivo y calcula distintos índices de vegetación e intenta desarrollar relaciones y modelos estadísticos entre las bandas o índices de vegetación y los parámetros biofísicos. Para ello puedes usar cualquier software con el que estés habituado a trabajar (Excel, R, SPSS, ...).\n\nPuedes realizar tantas simulaciones como consideres necesarias, por ejemplo variando el sensor o modificando los rangos plausibles para cubrir distintos tipos funcionales de vegetación. Tan sólo ten en cuenta que cada vez que se genere una simulación el archivo csv se sobreesecribirá. **Por lo que descárcatelo o haz una copia en tu carpeta virtual antes de volver a ejectura las nuevas simulaciones**.",
"_____no_output_____"
],
[
"# Conclusiones\nEn esta práctica hemos visto cómo el espectro de la vegetación responde a las variables biofísicas de la superfice.\n\n* El LAI es probablemente la variable que influya más en la respuesta espectral de la vegetación.\n* La concentración de clorofila en la hoja influye sobre todo en la región del visible y del *red-edge*.\n* El contenido de agua y el peso específico de la hora influyen sobre todo a partir del NIR.\n* La geometría de observación e iluminación, así como la respuesta espectral del suelo, influyen también en la señal. Esto hace que sea difícil aplicar una relación universal a la hora de estimar un parámetro biofísico.\n* Los modelos de transferencia radiativa pueden ayudar a estimar estos parámetros. Si bien idealmente es necesario disponer de datos de campo para realizar tareas de validación y/o calibración estadística.\n* Los sensores muestrean una parte del espectro en torno a bandas espectrales específicas. Por tanto una relación empírica desarrollada para un sensor específico puede que no sea aplicable o válida para otro sensor.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d03c8c2bd9d8909095f004441896929a09e278a6 | 633,189 | ipynb | Jupyter Notebook | notebooks/experiments/Sound Demo 3 - Multi-label classifier pretrained on audioset.ipynb | fronovics/AI_playground | ac302c0694fa2182af343c257b28a033bc4cf5b9 | [
"Apache-2.0"
] | null | null | null | notebooks/experiments/Sound Demo 3 - Multi-label classifier pretrained on audioset.ipynb | fronovics/AI_playground | ac302c0694fa2182af343c257b28a033bc4cf5b9 | [
"Apache-2.0"
] | null | null | null | notebooks/experiments/Sound Demo 3 - Multi-label classifier pretrained on audioset.ipynb | fronovics/AI_playground | ac302c0694fa2182af343c257b28a033bc4cf5b9 | [
"Apache-2.0"
] | null | null | null | 196.948367 | 254,352 | 0.881929 | [
[
[
"# Detecting sound sources in YouTube videos",
"_____no_output_____"
],
[
"## First load all dependencies and set work and data paths",
"_____no_output_____"
]
],
[
[
"# set plotting parameters\n%matplotlib inline\nimport matplotlib.pyplot as plt\n# change notebook settings for wider screen\nfrom IPython.core.display import display, HTML\ndisplay(HTML(\"<style>.container { width:100% !important; }</style>\"))\n\n# For embedding YouTube videos in Ipython Notebook\nfrom IPython.display import YouTubeVideo\n# and setting the time of the video in seconds\nfrom datetime import timedelta\n\nimport numpy as np\nimport os\nimport sys\nimport urllib.request\nimport pandas as pd\n\nsys.path.append(os.path.join('src', 'audioset_demos')) \n\nfrom __future__ import print_function\n\n# signal processing library\nfrom scipy import signal\nfrom scipy.io import wavfile\nimport wave\nimport six\nimport tensorflow as tf\nimport h5py\n\n# Audio IO and fast plotting\nimport pyaudio\nfrom pyqtgraph.Qt import QtCore, QtGui\nimport pyqtgraph as pg\n\n# Multiprocessing and threading\nimport multiprocessing\n\n\n# Dependencies for creating deep VGGish embeddings\nfrom src.audioset_demos import vggish_input\nimport vggish_input\nimport vggish_params\nimport vggish_postprocess\nimport vggish_slim\npca_params = 'vggish_pca_params.npz'\nmodel_checkpoint = 'vggish_model.ckpt'\n\n# Our YouTube video downloader based on youtube-dl module\nfrom src.audioset_demos import download_youtube_wav as dl_yt\n\n# third-party sounds processing and visualization library\nimport librosa\nimport librosa.display\n\n# Set user\nusr = 'maxvo'\n\nMAXINT16 = np.iinfo(np.int16).max\nprint(MAXINT16)\n\nFOCUS_CLASSES_ID = [0, 137, 62, 63, 500, 37] \n#FOCUS_CLASSES_ID = [0, 137, 37, 40, 62, 63, 203, 208, 359, 412, 500]\nclass_labels = pd.read_csv(os.path.join('src', 'audioset_demos', 'class_labels_indices.csv'))\nCLASS_NAMES = class_labels.loc[:, 'display_name'].tolist()\n\nFOCUS_CLASS_NAME_FRAME = class_labels.loc[FOCUS_CLASSES_ID, 'display_name']\n\nFOCUS_CLASS_NAME = FOCUS_CLASS_NAME_FRAME.tolist()\n\nprint(\"Chosen classes for experiments:\")\nprint(FOCUS_CLASS_NAME_FRAME)\n\n\n# Set current working directory\nsrc_dir = os.getcwd()\n\n# Set raw wav-file data directories for placing downloaded audio\nraw_dir = os.path.join(src_dir, 'data' ,'audioset_demos', 'raw')\nshort_raw_dir = os.path.join(src_dir, 'data', 'audioset_demos', 'short_raw')\nif not os.path.exists(short_raw_dir):\n os.makedirs(short_raw_dir)\nif not os.path.exists(raw_dir):\n os.makedirs(raw_dir)\n \naudioset_data_path = os.path.join('data', 'audioset_demos', 'audioset', 'packed_features')",
"_____no_output_____"
]
],
[
[
"## Download model parameters and PCA embedding",
"_____no_output_____"
]
],
[
[
"if not os.path.isfile(os.path.join('src', 'audioset_demos', 'vggish_model.ckpt')):\n urllib.request.urlretrieve(\n \"https://storage.googleapis.com/audioset/vggish_model.ckpt\",\n filename=os.path.join('src', 'audioset_demos', 'vggish_model.ckpt')\n )\n\nif not os.path.isfile(os.path.join('src', 'audioset_demos', 'vggish_pca_params.npz')):\n urllib.request.urlretrieve(\n \"https://storage.googleapis.com/audioset/vggish_pca_params.npz\",\n filename=os.path.join('src', 'audioset_demos', 'vggish_pca_params.npz')\n )\n \nif not os.path.isfile(os.path.join('data', 'audioset_demos', 'features.tar.gz')):\n urllib.request.urlretrieve(\n \"https://storage.googleapis.com/eu_audioset/youtube_corpus/v1/features/features.tar.gz\",\n filename=os.path.join('data', 'audioset_demos', 'features.tar.gz')\n )\n\n",
"_____no_output_____"
],
[
"import gzip\nimport shutil\nwith gzip.open(os.path.join('data', 'audioset_demos', 'features.tar.gz'), 'rb') as f_in:\n with open('packed_features', 'wb') as f_out:\n shutil.copyfileobj(f_in, f_out)",
"_____no_output_____"
],
[
"def save_data(hdf5_path, x, video_id_list, y=None):\n with h5py.File(hdf5_path, 'w') as hf:\n hf.create_dataset('x', data=x)\n hf.create_dataset('y', data=y)\n hf.create_dataset('video_id_list', data=video_id_list, dtype='S11')\n\ndef load_data(hdf5_path):\n with h5py.File(hdf5_path, 'r') as hf:\n x = hf['x'][:]\n if hf['y'] is not None:\n y = hf['y'][:]\n else:\n y = hf['y']\n video_id_list = hf['video_id_list'][:].tolist()\n\n return x, y, video_id_list\n\ndef time_str_to_sec(time_str='00:00:00'):\n time_str_list = time_str.split(':')\n seconds = int(\n timedelta(\n hours=int(time_str_list[0]),\n minutes=int(time_str_list[1]),\n seconds=int(time_str_list[2])\n ).total_seconds()\n )\n return seconds\n\n\nclass miniRecorder:\n \n def __init__(self, seconds=4, sampling_rate=16000):\n self.FORMAT = pyaudio.paInt16 #paFloat32 #paInt16\n self.CHANNELS = 1 # Must be Mono \n self.RATE = sampling_rate # sampling rate (Hz), 22050 was used for this application\n self.FRAMESIZE = 4200 # buffer size, number of data points to read at a time\n self.RECORD_SECONDS = seconds + 1 # how long should the recording (approx) be\n self.NOFRAMES = int((self.RATE * self.RECORD_SECONDS) / self.FRAMESIZE) # number of frames needed\n \n def record(self):\n # instantiate pyaudio\n p = pyaudio.PyAudio()\n # open stream\n stream = p.open(format=self.FORMAT,\n channels=self.CHANNELS,\n rate=self.RATE,\n input=True,\n frames_per_buffer=self.FRAMESIZE)\n # discard the first part of the recording\n discard = stream.read(self.FRAMESIZE)\n print('Recording...')\n data = stream.read(self.NOFRAMES * self.FRAMESIZE)\n decoded = np.frombuffer(data, dtype=np.int16) #np.float32)\n print('Finished...')\n stream.stop_stream()\n stream.close()\n p.terminate()\n \n # Remove first second to avoid \"click\" sound from starting recording\n self.sound_clip = decoded[self.RATE:]\n\nclass Worker(QtCore.QRunnable):\n '''\n Worker thread\n\n Inherits from QRunnable to handler worker thread setup, signals and wrap-up.\n\n :param callback: The function callback to run on this worker thread. Supplied args and \n kwargs will be passed through to the runner.\n :type callback: function\n :param args: Arguments to pass to the callback function\n :param kwargs: Keywords to pass to the callback function\n\n '''\n\n def __init__(self, fn, *args, **kwargs):\n super(Worker, self).__init__()\n # Store constructor arguments (re-used for processing)\n self.fn = fn\n self.args = args\n self.kwargs = kwargs\n\n @QtCore.pyqtSlot()\n def run(self):\n '''\n Initialise the runner function with passed args, kwargs.\n '''\n self.fn(*self.args, **self.kwargs)\n\n \nclass AudioFile:\n def __init__(self, file, chunk):\n \"\"\" Init audio stream \"\"\"\n self.chunk = chunk\n self.data = ''\n self.wf = wave.open(file, 'rb')\n self.p = pyaudio.PyAudio()\n self.stream = self.p.open(\n format = self.p.get_format_from_width(self.wf.getsampwidth()),\n channels = self.wf.getnchannels(),\n rate = self.wf.getframerate(),\n output = True\n )\n\n def play(self):\n \"\"\" Play entire file \"\"\"\n self.data = self.wf.readframes(self.chunk)\n while self.data:\n self.stream.write(self.data)\n self.data = self.wf.readframes(self.chunk)\n self.close()\n\n def close(self):\n \"\"\" Graceful shutdown \"\"\" \n self.stream.close()\n self.p.terminate()\n \n def read(self, chunk, exception_on_overflow=False):\n return self.data\n \n\n \nclass App(QtGui.QMainWindow):\n def __init__(self,\n predictor,\n n_top_classes=10,\n plot_classes=FOCUS_CLASSES_ID,\n parent=None):\n super(App, self).__init__(parent)\n \n ### Predictor model ###\n self.predictor = predictor\n self.n_classes = predictor.n_classes\n self.n_top_classes = n_top_classes\n self.plot_classes = plot_classes\n self.n_plot_classes = len(self.plot_classes)\n \n ### Start/stop control variable\n self.continue_recording = False\n self._timerId = None\n\n ### Settings ###\n self.rate = 16000 # sampling rate\n self.chunk = 1000 # reading chunk sizes, \n #self.rate = 22050 # sampling rate\n #self.chunk = 2450 # reading chunk sizes, make it a divisor of sampling rate\n #self.rate = 44100 # sampling rate\n #self.chunk = 882 # reading chunk sizes, make it a divisor of sampling rate\n self.nperseg = 400 # samples pr segment for spectrogram, scipy default is 256\n # self.nperseg = 490 # samples pr segment for spectrogram, scipy default is 256\n self.noverlap = 0 # overlap between spectrogram windows, scipt default is nperseg // 8 \n self.tape_length = 20 # length of running tape\n self.plot_length = 10 * self.rate\n self.samples_passed = 0\n self.pred_length = 10\n self.pred_samples = self.rate * self.pred_length\n self.start_tape() # initialize the tape\n self.eps = np.finfo(float).eps\n # Interval between predictions in number of samples\n self.pred_intv = (self.tape_length // 4) * self.rate\n self.pred_step = 10 * self.chunk\n self.full_tape = False\n #### Create Gui Elements ###########\n self.mainbox = QtGui.QWidget()\n self.setCentralWidget(self.mainbox)\n self.mainbox.setLayout(QtGui.QVBoxLayout())\n self.canvas = pg.GraphicsLayoutWidget()\n self.mainbox.layout().addWidget(self.canvas)\n self.label = QtGui.QLabel()\n self.mainbox.layout().addWidget(self.label)\n \n # Thread pool for prediction worker coordination\n self.threadpool = QtCore.QThreadPool()\n # self.threadpool_plot = QtCore.QThreadPool()\n print(\"Multithreading with maximum %d threads\" % self.threadpool.maxThreadCount())\n \n # Play, record and predict button in toolbar\n '''\n self.playTimer = QtCore.QTimer()\n self.playTimer.setInterval(500)\n self.playTimer.timeout.connect(self.playTick)\n self.toolbar = self.addToolBar(\"Play\")\n self.playScansAction = QtGui.QAction(QtGui.QIcon(\"control_play_blue.png\"), \"play scans\", self)\n self.playScansAction.triggered.connect(self.playScansPressed)\n self.playScansAction.setCheckable(True)\n self.toolbar.addAction(self.playScansAction)\n '''\n \n # Buttons and user input\n btn_brow_1 = QtGui.QPushButton('Start/Stop Recording', self)\n btn_brow_1.setGeometry(300, 15, 250, 25)\n #btn_brow_4.clicked.connect(support.main(fname_points, self.fname_stl_indir, self.fname_stl_outdir))\n # Action: Start or stop recording\n btn_brow_1.clicked.connect(lambda: self.press_record())\n \n btn_brow_2 = QtGui.QPushButton('Predict', self)\n btn_brow_2.setGeometry(20, 15, 250, 25)\n # Action: predict on present tape roll\n btn_brow_2.clicked.connect(\n lambda: self.start_predictions(\n sound_clip=self.tape,\n full_tape=False\n )\n )\n \n self.le1 = QtGui.QLineEdit(self)\n self.le1.setGeometry(600, 15, 250, 21)\n self.yt_video_id = str(self.le1.text())\n \n self.statusBar().showMessage(\"Ready\")\n\n # self.toolbar = self.addToolBar('Exit')\n # self.toolbar.addAction(exitAction)\n\n self.setGeometry(300, 300, 1400, 1200)\n self.setWindowTitle('Live Audio Event Detector')\n # self.show()\n \n # line plot\n self.plot = self.canvas.addPlot()\n self.p1 = self.plot.plot(pen='r')\n self.plot.setXRange(0, self.plot_length)\n self.plot.setYRange(-0.5, 0.5)\n self.plot.hideAxis('left')\n self.plot.hideAxis('bottom')\n self.canvas.nextRow()\n \n # spectrogram\n self.view = self.canvas.addViewBox()\n self.view.setAspectLocked(False)\n self.view.setRange(QtCore.QRectF(0,0, self.spec.shape[1], 100))\n # image plot\n self.img = pg.ImageItem() #(border='w')\n self.view.addItem(self.img)\n \n # bipolar colormap\n pos = np.array([0., 1., 0.5, 0.25, 0.75])\n color = np.array([[0,255,255,255], [255,255,0,255], [0,0,0,255], (0, 0, 255, 255), (255, 0, 0, 255)], dtype=np.ubyte)\n cmap = pg.ColorMap(pos, color)\n lut = cmap.getLookupTable(0.0, 1.0, 256)\n self.img.setLookupTable(lut)\n self.img.setLevels([-15, -5])\n \n self.canvas.nextRow()\n \n # create bar chart\n #self.view2 = self.canvas.addViewBox()\n # dummy data\n #self.x = np.arange(self.n_top_classes)\n #self.y1 = np.linspace(0, self.n_classes, num=self.n_top_classes)\n #self.bg1 = pg.BarGraphItem(x=self.x, height=self.y1, width=0.6, brush='r')\n #self.view2.addItem(self.bg1)\n \n # Prediction line plot\n self.plot2 = self.canvas.addPlot()\n self.plot2.addLegend()\n self.plot_list = [None]*self.n_plot_classes\n for i in range(self.n_plot_classes):\n self.plot_list[i] = self.plot2.plot(\n pen=pg.intColor(i),\n name=CLASS_NAMES[self.plot_classes[i]]\n )\n \n self.plot2.setXRange(0, self.plot_length)\n self.plot2.setYRange(0.0, 1.0)\n self.plot2.hideAxis('left')\n self.plot2.hideAxis('bottom')\n\n # self.canvas.nextRow()\n #### Start #####################\n \n # self.p = pyaudio.PyAudio()\n # self.start_stream()\n # self._update()\n\n def playScansPressed(self):\n if self.playScansAction.isChecked():\n self.playTimer.start()\n else:\n self.playTimer.stop()\n\n def playTick(self):\n self._update()\n\n \n def start_stream(self):\n if not self.yt_video_id:\n self.stream = self.p.open(\n format=pyaudio.paFloat32, \n channels=1, \n rate=self.rate,\n input=True,\n frames_per_buffer=self.chunk\n )\n else: \n self.stream = AudioFile(self.yt_video_id, self.chunk)\n self.stream.play()\n\n \n def close_stream(self):\n self.stream.stop_stream()\n self.stream.close()\n self.p.terminate()\n # self.exit_pool()\n \n def read_stream(self):\n self.raw = self.stream.read(self.chunk, exception_on_overflow=False)\n data = np.frombuffer(self.raw, dtype=np.float32)\n return self.raw, data\n \n def start_tape(self):\n self.tape = np.zeros(self.tape_length * self.rate)\n # empty spectrogram tape\n self.f, self.t, self.Sxx = signal.spectrogram(\n self.tape[-self.plot_length:],\n self.rate, \n nperseg=self.nperseg,\n noverlap=self.noverlap,\n detrend=False,\n return_onesided=True,\n mode='magnitude'\n )\n self.spec = np.zeros(self.Sxx.shape)\n self.pred = np.zeros((self.n_plot_classes, self.plot_length))\n \n def tape_add(self):\n if self.continue_recording:\n raw, audio = self.read_stream()\n self.tape[:-self.chunk] = self.tape[self.chunk:]\n self.tape[-self.chunk:] = audio\n self.samples_passed += self.chunk\n \n # spectrogram on whole tape\n # self.f, self.t, self.Sxx = signal.spectrogram(self.tape, self.rate)\n # self.spec = self.Sxx\n \n # spectrogram on last added part of tape\n self.f, self.t, self.Sxx = signal.spectrogram(self.tape[-self.chunk:], \n self.rate, \n nperseg=self.nperseg,\n noverlap=self.noverlap)\n spec_chunk = self.Sxx.shape[1]\n self.spec[:, :-spec_chunk] = self.spec[:, spec_chunk:]\n # Extend spectrogram after converting to dB scale\n self.spec[:, -spec_chunk:] = np.log10(abs(self.Sxx) + self.eps)\n \n self.pred[:, :-self.chunk] = self.pred[:, self.chunk:]\n '''\n if (self.samples_passed % self.pred_intv) == 0:\n sound_clip = self.tape # (MAXINT16 * self.tape).astype('int16') / 32768.0\n if self.full_tape:\n # predictions on full tape\n pred_chunk = self.predictor.predict(\n sound_clip=sound_clip[-self.pred_intv:],\n sample_rate=self.rate\n )[0][self.plot_classes]\n self.pred[:, -self.pred_intv:] = np.asarray(\n (self.pred_intv) * [pred_chunk]).transpose()\n\n else:\n # prediction, on some snip of the last part of the signal\n # 1 s seems to be the shortest time frame with reliable predictions\n self.start_predictions(sound_clip)\n '''\n \n def start_predictions(self, sound_clip=None, full_tape=False):\n #self.samples_passed_at_predict = self.samples_passed\n if sound_clip is None:\n sound_clip = self.tape\n \n if full_tape:\n worker = Worker(self.provide_prediction, *(), **{\n \"sound_clip\": sound_clip,\n \"pred_start\": -self.pred_samples,\n \"pred_stop\": None,\n \"pred_step\": self.pred_samples\n }\n )\n self.threadpool.start(worker)\n else:\n for chunk in range(0, self.pred_intv, self.pred_step):\n pred_start = - self.pred_intv - self.pred_samples + chunk\n pred_stop = - self.pred_intv + chunk\n worker = Worker(self.provide_prediction, *(), **{\n \"sound_clip\": sound_clip,\n \"pred_start\": pred_start,\n \"pred_stop\": pred_stop,\n \"pred_step\": self.pred_step\n }\n )\n self.threadpool.start(worker)\n \n def provide_prediction(self, sound_clip, pred_start, pred_stop, pred_step):\n #samples_passed_since_predict = self.samples_passed - self.samples_passed_at_predict\n #pred_stop -= samples_passed_since_predict\n pred_chunk = self.predictor.predict(\n sound_clip=sound_clip[pred_start:pred_stop],\n sample_rate=self.rate\n )[0][self.plot_classes]\n #samples_passed_since_predict = self.samples_passed - self.samples_passed_at_predict - samples_passed_since_predict\n #pred_stop -= samples_passed_since_predict\n if pred_stop is not None:\n pred_stop_step = pred_stop - pred_step\n else:\n pred_stop_step = None\n self.pred[:, pred_stop_step:pred_stop] = np.asarray(\n (pred_step) * [pred_chunk]\n ).transpose()\n\n\n def exit_pool(self):\n \"\"\"\n Exit all QRunnables and delete QThreadPool\n\n \"\"\"\n\n # When trying to quit, the application takes a long time to stop\n self.threadpool.globalInstance().waitForDone()\n self.threadpool.deleteLater()\n\n sys.exit(0)\n \n def press_record(self):\n self.yt_video_id = str(self.le1.text())\n # Switch between continue recording or stopping it \n # Start or avoid starting recording dependent on last press\n if self.continue_recording:\n self.continue_recording = False\n #if self._timerId is not None:\n # self.killTimer(self._timerId)\n self.close_stream()\n else:\n self.continue_recording = True\n self.p = pyaudio.PyAudio()\n self.start_stream()\n self._update()\n \n def _update(self):\n try:\n if self.continue_recording:\n self.tape_add()\n\n # self.img.setImage(self.spec.T)\n #kwargs = {\n # \"image\": self.spec.T,\n # \"autoLevels\": False,\n #\n #worker = Worker(self.img.setImage, *(), **kwargs)\n #self.threadpool_plot.start(worker)\n\n self.img.setImage(self.spec.T, autoLevels=False)\n\n #worker = Worker(\n # self.p1.setData,\n # *(),\n # **{'y': self.tape[-self.plot_length:]}\n #)\n #self.threadpool_plot.start(worker)\n\n self.p1.setData(self.tape[-self.plot_length:])\n\n #pred_var = np.var(self.pred, axis=-1)\n #pred_mean = np.mean(self.pred, axis=-1)\n\n #class_cand = np.where( (pred_mean > 0.001)*(pred_var > 0.01) )\n\n # n_classes_incl = min(self.n_top_classes, class_cand[0].shape[0]) \n\n # print(n_classes_incl)\n\n for i in range(self.n_plot_classes):\n #worker = Worker(\n # self.plot_list[i].setData,\n # *(),\n # **{'y': self.pred[i,:]}\n #)\n #self.threadpool_plot.start(worker)\n\n self.plot_list[i].setData(self.pred[i,:]) # self.plot_classes[i],:])\n\n #self.bg1.setOpts(\n # height=self.y1\n #)\n #self.bg1.setOpts(\n # height=np.sort(\n # self.pred[:, -1]\n # )[-1:-(self.n_top_classes+1):-1]\n #)\n #print(np.max(self.tape), np.min(self.tape))\n # self.label.setText('Class: {0:0.3f}'.format(self.pred[-1]))\n\n QtCore.QTimer.singleShot(1, self._update)\n \n except KeyboardInterrupt:\n self.close_stream()",
"_____no_output_____"
],
[
"from AudioSetClassifier import AudioSetClassifier\n\n # model_type='decision_level_single_attention',\n # balance_type='balance_in_batch',\n # at_iteration=50000\n\n#ASC = AudioSetClassifier(\n# model_type='decision_level_max_pooling', #single_attention',\n# balance_type='balance_in_batch',\n# iters=50000\n#)\n\nASC = AudioSetClassifier()",
"Using TensorFlow backend.\n"
],
[
"app=0 #This is the solution\napp = QtGui.QApplication(sys.argv)\nMainApp = App(predictor=ASC)\nMainApp.show()\nsys.exit(app.exec_())",
"_____no_output_____"
],
[
"minirec = miniRecorder(seconds=10, sampling_rate=16000)\nminirec.record()",
"Recording...\nFinished...\n"
],
[
"minirec_pred = ASC.predict(sound_clip=minirec.sound_clip / 32768.0, sample_rate=16000)\n\nprint(minirec_pred[:,[0, 37, 62, 63]])",
"[[ 8.65663052e-01 1.39752665e-05 1.58889816e-04 1.18576922e-03]]\n"
],
[
"max_prob_classes = np.argsort(minirec_pred, axis=-1)[:, ::-1]\nmax_prob = np.sort(minirec_pred, axis=-1)[:,::-1]\n\nprint(max_prob.shape)\n\nexample = pd.DataFrame(class_labels['display_name'][max_prob_classes[0,:10]])\nexample.loc[:, 'prob'] = pd.Series(max_prob[0, :10], index=example.index)\nprint(example)\nexample.plot.bar(x='display_name', y='prob', rot=90)\nplt.show()\nprint()\n ",
"(1, 527)\n display_name prob\n0 Speech 0.865663\n506 Inside, small room 0.050520\n1 Male speech, man speaking 0.047573\n5 Narration, monologue 0.047426\n46 Snort 0.043561\n482 Ping 0.025956\n354 Door 0.017503\n458 Arrow 0.016863\n438 Chop 0.014797\n387 Writing 0.014618\n"
]
],
[
[
"## Parameters for how to plot audio",
"_____no_output_____"
]
],
[
[
"# Sample rate\n# this has to be at least twice of max frequency which we've entered\n# but you can play around with different sample rates and see how this\n# affects the results;\n# since we generated this audio, the sample rate is the bitrate\nsample_rate = vggish_params.SAMPLE_RATE\n\n# size of audio FFT window relative to sample_rate\nn_window = 1024\n# overlap between adjacent FFT windows\nn_overlap = 360\n# number of mel frequency bands to generate\nn_mels = 64\n\n# max duration of short video clips\nduration = 10\n\n# note frequencies https://pages.mtu.edu/~suits/notefreqs.html\nfreq1 = 512.\nfreq2 = 1024.\n# fmin and fmax for librosa filters in Hz - used for visualization purposes only\nfmax = max(freq1, freq2)*8 + 1000. \nfmin = 0.\n\n# stylistic change to the notebook\nfontsize = 14\n\nplt.rcParams['font.family'] = 'serif'\nplt.rcParams['font.serif'] = 'Ubuntu'\nplt.rcParams['font.monospace'] = 'Ubuntu Mono'\nplt.rcParams['font.size'] = fontsize\nplt.rcParams['axes.labelsize'] = fontsize\nplt.rcParams['axes.labelweight'] = 'bold'\nplt.rcParams['axes.titlesize'] = fontsize\nplt.rcParams['xtick.labelsize'] = fontsize\nplt.rcParams['ytick.labelsize'] = fontsize\nplt.rcParams['legend.fontsize'] = fontsize\nplt.rcParams['figure.titlesize'] = fontsize",
"_____no_output_____"
]
],
[
[
"## Choosing video IDs and start times before download",
"_____no_output_____"
]
],
[
[
"video_ids = [\n 'BaW_jenozKc',\n 'E6sS2d-NeTE',\n 'xV0eTva6SKQ',\n '2Szah76TMgo',\n 'g38kRk6YAA0',\n 'OkkkPAE9KvE',\n 'N1zUp9aPFG4'\n]\n\nvideo_start_time_str = [\n '00:00:00',\n '00:00:10',\n '00:00:05',\n '00:00:02',\n '00:03:10',\n '00:00:10',\n '00:00:06'\n]\nvideo_start_time = list(map(time_str_to_sec, video_start_time_str))",
"_____no_output_____"
]
],
[
[
"## Download, save and cut video audio",
"_____no_output_____"
]
],
[
[
"video_titles = []\nmaxv = np.iinfo(np.int16).max\n\nfor i, vid in enumerate(video_ids): \n # Download and store video under data/raw/\n video_title = dl_yt.download_youtube_wav(\n video_id=vid,\n raw_dir=raw_dir,\n short_raw_dir=short_raw_dir,\n start_sec=video_start_time[i],\n duration=duration,\n sample_rate=sample_rate\n )\n video_titles += [video_title]\n \n print()\n '''\n audio_path = os.path.join(raw_dir, vid) + '.wav'\n short_audio_path = os.path.join(short_raw_dir, vid) + '.wav'\n \n # Load and downsample audio to 16000\n # audio is a 1D time series of the sound\n # can also use (audio, fs) = soundfile.read(audio_path)\n (audio, fs) = librosa.load(\n audio_path,\n sr = sample_rate,\n offset = video_start_time[i],\n duration = duration\n )\n \n # Store downsampled 10sec clip under data/short_raw/\n wavfile.write(\n filename=short_audio_path,\n rate=sample_rate,\n data=(audio * maxv).astype(np.int16)\n )\n '''",
"Done downloading, now converting ...\nyoutube-dl test video ''_ä↭𝕐 \n\tshort clip saved in:\n\t\t data/short_raw/BaW_jenozKc.wav\nyoutube-dl test video ''_ä↭𝕐 \n\tfull clip saved in:\n\t\t data/raw/BaW_jenozKc.wav\n\nDone downloading, now converting ...\nBrandweer - Prio 1 Binnenbrand De Grevelingen Grevelingen Den Bosch - 634TS 651AL \n\tshort clip saved in:\n\t\t data/short_raw/E6sS2d-NeTE.wav\nBrandweer - Prio 1 Binnenbrand De Grevelingen Grevelingen Den Bosch - 634TS 651AL \n\tfull clip saved in:\n\t\t data/raw/E6sS2d-NeTE.wav\n\nDone downloading, now converting ...\nLED Lights + 100W Siren \n\tshort clip saved in:\n\t\t data/short_raw/xV0eTva6SKQ.wav\nLED Lights + 100W Siren \n\tfull clip saved in:\n\t\t data/raw/xV0eTva6SKQ.wav\n\nDone downloading, now converting ...\nStart \n\tshort clip saved in:\n\t\t data/short_raw/2Szah76TMgo.wav\nStart \n\tfull clip saved in:\n\t\t data/raw/2Szah76TMgo.wav\n\nDone downloading, now converting ...\nSplitting a Playing Card in ULTRA SLOW MOTION - Smarter Every Day 194 \n\tshort clip saved in:\n\t\t data/short_raw/g38kRk6YAA0.wav\nSplitting a Playing Card in ULTRA SLOW MOTION - Smarter Every Day 194 \n\tfull clip saved in:\n\t\t data/raw/g38kRk6YAA0.wav\n\nDone downloading, now converting ...\nCapuchin Monkeys _ Wild Caribbean _ BBC Earth \n\tshort clip saved in:\n\t\t data/short_raw/OkkkPAE9KvE.wav\nCapuchin Monkeys _ Wild Caribbean _ BBC Earth \n\tfull clip saved in:\n\t\t data/raw/OkkkPAE9KvE.wav\n\nDone downloading, now converting ...\nZoo Animal Sounds! \n\tshort clip saved in:\n\t\t data/short_raw/N1zUp9aPFG4.wav\nZoo Animal Sounds! \n\tfull clip saved in:\n\t\t data/raw/N1zUp9aPFG4.wav\n\n"
],
[
"# Usage example for pyaudio\ni = 6\na = AudioFile(\n os.path.join(short_raw_dir, video_ids[i]) + '.wav',\n chunk = 1000\n)\na.play()\na.close() ",
"_____no_output_____"
]
],
[
[
"## Retrieve VGGish PCA embeddings ",
"_____no_output_____"
]
],
[
[
"video_vggish_emb = []\n\n# Restore VGGish model trained on YouTube8M dataset\n# Retrieve PCA-embeddings of bottleneck features\nwith tf.Graph().as_default(), tf.Session() as sess:\n # Define the model in inference mode, load the checkpoint, and\n # locate input and output tensors.\n vggish_slim.define_vggish_slim(training=False)\n vggish_slim.load_vggish_slim_checkpoint(sess, model_checkpoint)\n features_tensor = sess.graph.get_tensor_by_name(\n vggish_params.INPUT_TENSOR_NAME)\n embedding_tensor = sess.graph.get_tensor_by_name(\n vggish_params.OUTPUT_TENSOR_NAME)\n\n for i, vid in enumerate(video_ids): \n audio_path = os.path.join(short_raw_dir, vid) + '.wav'\n\n examples_batch = vggish_input.wavfile_to_examples(audio_path)\n\n print(examples_batch.shape)\n\n # Prepare a postprocessor to munge the model embeddings.\n pproc = vggish_postprocess.Postprocessor(pca_params)\n\n # Run inference and postprocessing.\n [embedding_batch] = sess.run([embedding_tensor],\n feed_dict={features_tensor: examples_batch})\n print(embedding_batch.shape)\n postprocessed_batch = pproc.postprocess(embedding_batch)\n print(postprocessed_batch.shape)\n \n video_vggish_emb.extend([postprocessed_batch])\n\nprint(len(video_vggish_emb))",
"INFO:tensorflow:Restoring parameters from vggish_model.ckpt\n(10, 96, 64)\n(10, 128)\n(10, 128)\n(10, 96, 64)\n(10, 128)\n(10, 128)\n(10, 96, 64)\n(10, 128)\n(10, 128)\n(10, 96, 64)\n(10, 128)\n(10, 128)\n(10, 96, 64)\n(10, 128)\n(10, 128)\n(10, 96, 64)\n(10, 128)\n(10, 128)\n(10, 96, 64)\n(10, 128)\n(10, 128)\n7\n"
]
],
[
[
"## Plot audio, transformations and embeddings\n\n### Function for visualising audio",
"_____no_output_____"
]
],
[
[
"def plot_audio(audio, emb):\n audio_sec = audio.shape[0]/sample_rate\n # Make a new figure\n plt.figure(figsize=(18, 16), dpi= 60, facecolor='w', edgecolor='k')\n plt.subplot(511)\n # Display the spectrogram on a mel scale\n librosa.display.waveplot(audio, int(sample_rate), max_sr = int(sample_rate))\n plt.title('Raw audio waveform @ %d Hz' % sample_rate, fontsize = fontsize) \n plt.xlabel(\"Time (s)\")\n plt.ylabel(\"Amplitude\")\n\n # Define filters and windows\n melW =librosa.filters.mel(sr=sample_rate, n_fft=n_window, n_mels=n_mels, fmin=fmin, fmax=fmax)\n ham_win = np.hamming(n_window)\n \n # Compute fft to spectrogram\n [f, t, x] = signal.spectral.spectrogram(\n x=audio,\n window=ham_win,\n nperseg=n_window,\n noverlap=n_overlap,\n detrend=False,\n return_onesided=True,\n mode='magnitude')\n \n # Apply filters and log transformation\n x_filtered = np.dot(x.T, melW.T)\n x_logmel = np.log(x_filtered + 1e-8)\n x_logmel = x_logmel.astype(np.float32)\n\n # Display frequency power spectrogram\n plt.subplot(512)\n x_coords = np.linspace(0, audio_sec, x.shape[0])\n librosa.display.specshow(\n x.T,\n sr=sample_rate,\n x_axis='time',\n y_axis='hz',\n x_coords=x_coords\n )\n plt.xlabel(\"Time (s)\")\n plt.title(\"FFT spectrogram (dB)\", fontsize = fontsize)\n # optional colorbar plot\n plt.colorbar(format='%+02.0f dB')\n\n # Display log-mel freq. power spectrogram\n plt.subplot(513)\n x_coords = np.linspace(0, audio_sec, x_logmel.shape[0])\n librosa.display.specshow(\n x_logmel.T,\n sr=sample_rate,\n x_axis='time',\n y_axis='mel',\n x_coords=x_coords\n )\n plt.xlabel(\"Time (s)\")\n plt.title(\"Mel power spectrogram used in DCASE 2017 (dB)\", fontsize = fontsize)\n # optional colorbar plot\n plt.colorbar(format='%+02.0f dB')\n \n # Display embeddings\n plt.subplot(514)\n x_coords = np.linspace(0, audio_sec, emb.shape[0])\n librosa.display.specshow(\n emb.T,\n sr=sample_rate,\n x_axis='time',\n y_axis=None,\n x_coords=x_coords\n )\n plt.xlabel(\"Time (s)\")\n plt.colorbar()\n \n plt.subplot(515)\n plt.scatter(\n x=emb[:, 0],\n y=emb[:, 1],\n )\n plt.xlabel(\"PC_1\")\n plt.ylabel(\"PC_2\")\n\n # Make the figure layout compact\n plt.tight_layout()\n \n plt.show()",
"_____no_output_____"
]
],
[
[
"### Visualise all clips of audio chosen",
"_____no_output_____"
]
],
[
[
"for i, vid in enumerate(video_ids): \n print(\"\\nAnalyzing audio from video with title:\\n\", video_titles[i])\n audio_path = os.path.join(short_raw_dir, vid) + '.wav'\n \n # audio is a 1D time series of the sound\n # can also use (audio, fs) = soundfile.read(audio_path)\n (audio, fs) = librosa.load(\n audio_path,\n sr = sample_rate,\n )\n \n plot_audio(audio, video_vggish_emb[i])\n \n start=int(\n timedelta(\n hours=0,\n minutes=0,\n seconds=video_start_time[i]\n ).total_seconds()\n )\n\n YouTubeVideo(vid, start=start, autoplay=0, theme=\"light\", color=\"red\")\n print()",
"_____no_output_____"
]
],
[
[
"### Visualise one clip of audio and embed YouTube video for comparison",
"_____no_output_____"
]
],
[
[
"i = 4\nvid = video_ids[i]\n\naudio_path = os.path.join(raw_dir, vid) + '.wav'\n \n# audio is a 1D time series of the sound\n# can also use (audio, fs) = soundfile.read(audio_path)\n(audio, fs) = librosa.load(\n audio_path,\n sr = sample_rate,\n offset = video_start_time[i],\n duration = duration\n)\n\nplot_audio(audio, video_vggish_emb[i])\n\nstart=int(\n timedelta(\n hours=0,\n minutes=0,\n seconds=video_start_time[i]\n ).total_seconds()\n)\n\nYouTubeVideo(\n vid,\n start=start,\n end=start+duration,\n autoplay=0,\n theme=\"light\",\n color=\"red\"\n)\n\n# Plot emb with scatter\n# Check first couple of PCs, \n# for both train and test data, to see if the test is lacking variance",
"/usr/local/anaconda3/envs/audioset_tensorflow/lib/python3.6/site-packages/librosa/filters.py:284: UserWarning: Empty filters detected in mel frequency basis. Some channels will produce empty responses. Try increasing your sampling rate (and fmax) or reducing n_mels.\n warnings.warn('Empty filters detected in mel frequency basis. '\n/usr/local/anaconda3/envs/audioset_tensorflow/lib/python3.6/site-packages/matplotlib/font_manager.py:1328: UserWarning: findfont: Font family ['serif'] not found. Falling back to DejaVu Sans\n (prop.get_family(), self.defaultFamily[fontext]))\n"
]
],
[
[
"## Evaluate trained audio detection model",
"_____no_output_____"
]
],
[
[
"import audio_event_detection_model as AEDM\nimport utilities\nfrom sklearn import metrics\n\nmodel = AEDM.CRNN_audio_event_detector()\n\n",
"_____no_output_____"
]
],
[
[
"### Evaluating model on audio downloaded",
"_____no_output_____"
]
],
[
[
"(x_user_inp, y_user_inp) = utilities.transform_data(\n np.array(video_vggish_emb)\n)\npredictions = model.predict(\n x=x_user_inp\n)",
"_____no_output_____"
]
],
[
[
"### Evaluating model on training data",
"_____no_output_____"
]
],
[
[
"(x_tr, y_tr, vid_tr) = load_data(os.path.join(audioset_data_path, 'bal_train.h5'))\n(x_tr, y_tr) = utilities.transform_data(x_tr, y_tr)\n\npred_tr = model.predict(x=x_tr)",
"\n\nLoading top model with weights from:\nmodels/md_50000_iters.h5\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_8 (InputLayer) (None, 10, 128) 0 \n__________________________________________________________________________________________________\ndense_21 (Dense) (None, 10, 1024) 132096 input_8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_13 (BatchNo (None, 10, 1024) 4096 dense_21[0][0] \n__________________________________________________________________________________________________\nactivation_13 (Activation) (None, 10, 1024) 0 batch_normalization_13[0][0] \n__________________________________________________________________________________________________\ndropout_13 (Dropout) (None, 10, 1024) 0 activation_13[0][0] \n__________________________________________________________________________________________________\ndense_22 (Dense) (None, 10, 1024) 1049600 dropout_13[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_14 (BatchNo (None, 10, 1024) 4096 dense_22[0][0] \n__________________________________________________________________________________________________\nactivation_14 (Activation) (None, 10, 1024) 0 batch_normalization_14[0][0] \n__________________________________________________________________________________________________\ndropout_14 (Dropout) (None, 10, 1024) 0 activation_14[0][0] \n__________________________________________________________________________________________________\ndense_23 (Dense) (None, 10, 1024) 1049600 dropout_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_15 (BatchNo (None, 10, 1024) 4096 dense_23[0][0] \n__________________________________________________________________________________________________\nactivation_15 (Activation) (None, 10, 1024) 0 batch_normalization_15[0][0] \n__________________________________________________________________________________________________\ndropout_15 (Dropout) (None, 10, 1024) 0 activation_15[0][0] \n__________________________________________________________________________________________________\ndense_24 (Dense) (None, 10, 527) 540175 dropout_15[0][0] \n__________________________________________________________________________________________________\ndense_25 (Dense) (None, 10, 527) 540175 dropout_15[0][0] \n__________________________________________________________________________________________________\nlambda_5 (Lambda) (None, 527) 0 dense_24[0][0] \n dense_25[0][0] \n==================================================================================================\nTotal params: 3,323,934\nTrainable params: 3,317,790\nNon-trainable params: 6,144\n__________________________________________________________________________________________________\n"
],
[
"print(pred_tr.max())\n\nprint(metrics.accuracy_score(y_tr, (pred_tr > 0.5).astype(np.float32)))\n\nprint(metrics.roc_auc_score(y_tr, pred_tr))\nprint(np.mean(metrics.roc_auc_score(y_tr, pred_tr, average=None)))\n\nstats = utilities.calculate_stats(pred_tr, y_tr)\nmAUC = np.mean([stat['auc'] for stat in stats])\n\n",
"0.999973\n0.130054151625\n0.984930222934\n0.984930222934\n"
],
[
"max_prob_classes = np.argsort(predictions, axis=-1)[:, ::-1]\nmax_prob = np.sort(predictions, axis=-1)[:,::-1]",
"_____no_output_____"
],
[
"print(mAUC)\n\nprint(max_prob.max())\n\nprint(max_prob[:,:10])\n\nprint(predictions.shape)\n\nprint(max_prob_classes[:,:10])",
"0.984930222934\n0.87231\n[[ 0.85056537 0.84223366 0.27688268 0.23452662 0.12581593 0.05143182\n 0.02919197 0.02805768 0.02699612 0.01704121]\n [ 0.32978365 0.1321748 0.10872712 0.10825769 0.07952435 0.07356845\n 0.04511245 0.03998784 0.03936461 0.03507403]\n [ 0.87230974 0.86156976 0.81838012 0.65405416 0.33036518 0.19758119\n 0.15447959 0.08347502 0.06029229 0.05457394]\n [ 0.18422596 0.1432181 0.11541138 0.10077612 0.06737918 0.06132444\n 0.05907288 0.05696394 0.04590562 0.03907897]\n [ 0.73532987 0.50799 0.30799454 0.16915816 0.08796553 0.07880456\n 0.06258963 0.06124302 0.0446381 0.04250672]\n [ 0.76051241 0.3201108 0.24144822 0.24041927 0.21041867 0.16869307\n 0.16662784 0.10298688 0.09610511 0.08055314]\n [ 0.67560714 0.18969995 0.17008433 0.08974643 0.07025716 0.06953395\n 0.0580983 0.05612611 0.05410512 0.04845256]]\n(7, 527)\n[[ 7 0 5 518 525 1 137 407 506 4]\n [137 396 35 322 0 398 325 318 300 308]\n [322 396 323 324 325 300 310 316 74 73]\n [ 72 435 426 87 137 0 89 434 88 301]\n [ 0 137 442 470 443 365 438 364 440 418]\n [ 0 514 72 112 111 113 127 510 126 98]\n [ 0 300 110 72 307 109 348 108 135 343]]\n"
],
[
"from numpy import genfromtxt\nimport pandas as pd\n\nclass_labels = pd.read_csv('class_labels_indices.csv')",
"_____no_output_____"
],
[
"print(class_labels['display_name'][max_prob_classes[5,:10]])",
"0 Speech\n514 Environmental noise\n72 Animal\n112 Bird vocalization, bird call, bird song\n111 Bird\n113 Chirp, tweet\n127 Cricket\n510 Outside, rural or natural\n126 Insect\n98 Fowl\nName: display_name, dtype: object\n"
],
[
"for i, vid in enumerate(video_ids[0]):\n print(video_titles[i])\n print()\n \n example = pd.DataFrame(class_labels['display_name'][max_prob_classes[i,:10]])\n example.loc[:, 'prob'] = pd.Series(max_prob[i, :10], index=example.index)\n print(example)\n example.plot.bar(x='display_name', y='prob', rot=90)\n plt.show()\n print()",
"_____no_output_____"
]
],
[
[
"## Investigating model predictions on downloaded audio clips",
"_____no_output_____"
]
],
[
[
"i = 0\nvid = video_ids[i]\n\nprint(video_titles[i])\nprint()\n\nYouTubeVideo(\n vid,\n start=start,\n end=start+duration,\n autoplay=0,\n theme=\"light\",\n color=\"red\"\n)",
"youtube-dl test video ''_ä↭𝕐\n\n"
],
[
"example = pd.DataFrame(class_labels['display_name'][max_prob_classes[i,:10]])\nexample.loc[:, 'prob'] = pd.Series(max_prob[i, :10], index=example.index)\nprint(example)\nexample.plot.bar(x='display_name', y='prob', rot=90)\nplt.show()\nprint()",
" display_name prob\n7 Speech synthesizer 0.850565\n0 Speech 0.842234\n5 Narration, monologue 0.276883\n518 Sidetone 0.234527\n525 Radio 0.125816\n1 Male speech, man speaking 0.051432\n137 Music 0.029192\n407 Tick 0.028058\n506 Inside, small room 0.026996\n4 Conversation 0.017041\n"
],
[
"#eval_metrics = model.evaluate(x=x_tr, y=y_tr)",
"_____no_output_____"
],
[
"#for i, metric_name in enumerate(model.metrics_names): \n# print(\"{}: {:1.4f}\".format(metric_name, eval_metrics[i]))",
"_____no_output_____"
],
[
"#qtapp = App(model)\n\nfrom AudioSetClassifier import AudioSetClassifier\n\nimport time\n\nASC = AudioSetClassifier()",
"\nLoading VGGish base model:\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_6 (InputLayer) (None, 96, 64, 1) 0 \n_________________________________________________________________\nconv1 (Conv2D) (None, 96, 64, 64) 640 \n_________________________________________________________________\npool1 (MaxPooling2D) (None, 48, 32, 64) 0 \n_________________________________________________________________\nconv2 (Conv2D) (None, 48, 32, 128) 73856 \n_________________________________________________________________\npool2 (MaxPooling2D) (None, 24, 16, 128) 0 \n_________________________________________________________________\nconv3_1 (Conv2D) (None, 24, 16, 256) 295168 \n_________________________________________________________________\nconv3_2 (Conv2D) (None, 24, 16, 256) 590080 \n_________________________________________________________________\npool3 (MaxPooling2D) (None, 12, 8, 256) 0 \n_________________________________________________________________\nconv4_1 (Conv2D) (None, 12, 8, 512) 1180160 \n_________________________________________________________________\nconv4_2 (Conv2D) (None, 12, 8, 512) 2359808 \n_________________________________________________________________\npool4 (MaxPooling2D) (None, 6, 4, 512) 0 \n_________________________________________________________________\nflatten (Flatten) (None, 12288) 0 \n_________________________________________________________________\nfc1_1 (Dense) (None, 4096) 50335744 \n_________________________________________________________________\nfc1_2 (Dense) (None, 4096) 16781312 \n_________________________________________________________________\nfc2 (Dense) (None, 128) 524416 \n=================================================================\nTotal params: 72,141,184\nTrainable params: 72,141,184\nNon-trainable params: 0\n_________________________________________________________________\n\n\nLoading top model with weights from:\nmodels/md_50000_iters.h5\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_7 (InputLayer) (None, 10, 128) 0 \n__________________________________________________________________________________________________\ndense_16 (Dense) (None, 10, 1024) 132096 input_7[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_10 (BatchNo (None, 10, 1024) 4096 dense_16[0][0] \n__________________________________________________________________________________________________\nactivation_10 (Activation) (None, 10, 1024) 0 batch_normalization_10[0][0] \n__________________________________________________________________________________________________\ndropout_10 (Dropout) (None, 10, 1024) 0 activation_10[0][0] \n__________________________________________________________________________________________________\ndense_17 (Dense) (None, 10, 1024) 1049600 dropout_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_11 (BatchNo (None, 10, 1024) 4096 dense_17[0][0] \n__________________________________________________________________________________________________\nactivation_11 (Activation) (None, 10, 1024) 0 batch_normalization_11[0][0] \n__________________________________________________________________________________________________\ndropout_11 (Dropout) (None, 10, 1024) 0 activation_11[0][0] \n__________________________________________________________________________________________________\ndense_18 (Dense) (None, 10, 1024) 1049600 dropout_11[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_12 (BatchNo (None, 10, 1024) 4096 dense_18[0][0] \n__________________________________________________________________________________________________\nactivation_12 (Activation) (None, 10, 1024) 0 batch_normalization_12[0][0] \n__________________________________________________________________________________________________\ndropout_12 (Dropout) (None, 10, 1024) 0 activation_12[0][0] \n__________________________________________________________________________________________________\ndense_19 (Dense) (None, 10, 527) 540175 dropout_12[0][0] \n__________________________________________________________________________________________________\ndense_20 (Dense) (None, 10, 527) 540175 dropout_12[0][0] \n__________________________________________________________________________________________________\nlambda_4 (Lambda) (None, 527) 0 dense_19[0][0] \n dense_20[0][0] \n==================================================================================================\nTotal params: 3,323,934\nTrainable params: 3,317,790\nNon-trainable params: 6,144\n__________________________________________________________________________________________________\n"
],
[
"sound_clip = os.path.join(short_raw_dir, video_ids[1]) + '.wav'\n\n\n\nt0 = time.time()\ntest_pred = ASC.predict(sound_clip=sound_clip)\nt1 = time.time()\n\nprint('Time spent on 1 forward pass prediction:', t1-t0)",
"Time spent on 1 forward pass prediction: 0.42424607276916504\n"
],
[
"print(test_pred.shape)",
"(1, 527)\n"
],
[
"for i, vid in enumerate(video_ids):\n print(video_titles[i])\n print()\n sound_clip = os.path.join(short_raw_dir, vid) + '.wav'\n predictions = ASC.predict(sound_clip=sound_clip)\n max_prob_classes = np.argsort(predictions, axis=-1)[:, ::-1]\n max_prob = np.sort(predictions, axis=-1)[:,::-1]\n \n print(max_prob.shape)\n \n example = pd.DataFrame(class_labels['display_name'][max_prob_classes[0,:10]])\n example.loc[:, 'prob'] = pd.Series(max_prob[0, :10], index=example.index)\n print(example)\n example.plot.bar(x='display_name', y='prob', rot=90)\n plt.show()\n print()",
"youtube-dl test video ''_ä↭𝕐\n\n(1, 527)\n display_name prob\n7 Speech synthesizer 0.850565\n0 Speech 0.842234\n5 Narration, monologue 0.276883\n518 Sidetone 0.234527\n525 Radio 0.125816\n1 Male speech, man speaking 0.051432\n137 Music 0.029192\n407 Tick 0.028058\n506 Inside, small room 0.026996\n4 Conversation 0.017041\n"
],
[
"import sys\n\napp=0 #This is the solution\napp = QtGui.QApplication(sys.argv)\nMainApp = App(predictor=ASC)\nMainApp.show()\nsys.exit(app.exec_())",
"_____no_output_____"
],
[
"#from PyQt4 import QtGui, QtCore\n\nclass SimpleWindow(QtGui.QWidget):\n def __init__(self, parent=None):\n QtGui.QWidget.__init__(self, parent)\n\n self.setGeometry(300, 300, 200, 80)\n self.setWindowTitle('Hello World')\n\n quit = QtGui.QPushButton('Close', self)\n quit.setGeometry(10, 10, 60, 35)\n\n self.connect(quit, QtCore.SIGNAL('clicked()'),\n self, QtCore.SLOT('close()'))\n\nif __name__ == '__main__':\n app = QtCore.QCoreApplication.instance()\n if app is None:\n app = QtGui.QApplication([])\n\n sw = SimpleWindow()\n sw.show()\n\n try:\n from IPython.lib.guisupport import start_event_loop_qt4\n start_event_loop_qt4(app)\n except ImportError:\n app.exec_()",
"_____no_output_____"
]
],
[
[
"1. Understand attention\n2. Understand filters\n3. Understand Multi-label, hierachical, knowledge graphs\n4. Understand class imbalance \n5. CCA on VGGish vs. ResNet audioset emb. to check if there's a linear connection. \n6. Train linear layer to convert VGGish emb. to ResNet-50 emb. ",
"_____no_output_____"
],
[
"Plot in GUI:\n1. Exclude all non-active classes\n2. Draw class names on curves going up\n3. Remove histogram\n4. Make faster",
"_____no_output_____"
]
],
[
[
"video_vggish_emb = []\ntest_wav_path = os.path.join(src_dir, 'data', 'wav_file')\nwav_files = os.listdir(test_wav_path)\nexample_names = []\n\n# Restore VGGish model trained on YouTube8M dataset\n# Retrieve PCA-embeddings of bottleneck features\nwith tf.Graph().as_default(), tf.Session() as sess:\n # Define the model in inference mode, load the checkpoint, and\n # locate input and output tensors.\n vggish_slim.define_vggish_slim(training=False)\n vggish_slim.load_vggish_slim_checkpoint(sess, model_checkpoint)\n features_tensor = sess.graph.get_tensor_by_name(\n vggish_params.INPUT_TENSOR_NAME)\n embedding_tensor = sess.graph.get_tensor_by_name(\n vggish_params.OUTPUT_TENSOR_NAME)\n # Prepare a postprocessor to munge the model embeddings.\n pproc = vggish_postprocess.Postprocessor(pca_params)\n \n for i, vid in enumerate(wav_files): \n audio_path = os.path.join(test_wav_path, vid)\n print(vid)\n\n examples_batch = vggish_input.wavfile_to_examples(audio_path)\n\n print(examples_batch.shape)\n\n # Run inference and postprocessing.\n [embedding_batch] = sess.run([embedding_tensor],\n feed_dict={features_tensor: examples_batch})\n print(embedding_batch.shape)\n postprocessed_batch = pproc.postprocess(embedding_batch)\n batch_shape = postprocessed_batch.shape\n print(batch_shape)\n \n if batch_shape[0] > 10:\n postprocessed_batch = postprocessed_batch[:10]\n elif batch_shape[0] < 10:\n zero_pad = np.zeros((10, 128))\n zero_pad[:batch_shape[0]] = postprocessed_batch \n postprocessed_batch = zero_pad\n \n print(postprocessed_batch.shape)\n \n if postprocessed_batch.shape[0] == 10:\n video_vggish_emb.extend([postprocessed_batch])\n example_names.extend([vid])\n\nprint(len(video_vggish_emb))",
"_____no_output_____"
],
[
"import audio_event_detection_model as AEDM\nimport utilities\nmodel = AEDM.CRNN_audio_event_detector()",
"_____no_output_____"
],
[
"(x_user_inp, y_user_inp) = utilities.transform_data(\n np.array(video_vggish_emb)\n)\npredictions_AEDM = model.predict(\n x=x_user_inp\n)\n",
"_____no_output_____"
],
[
"predictions_ASC = np.zeros([len(wav_files), 527])\n\nfor i, vid in enumerate(wav_files): \n audio_path = os.path.join(test_wav_path, vid)\n predictions_ASC[i] = ASC.predict(sound_clip=audio_path)",
"_____no_output_____"
],
[
"qkong_res = '''2018Q1Q10Q17Q12Q59Q512440Q-5889Q.fea_lab ['Speech'] [0.8013877]\n12_4_train ambience.fea_lab ['Vehicle', 'Rail transport', 'Train', 'Railroad car, train wagon'] [0.38702238, 0.6618184, 0.7742054, 0.5886036]\n19_3_forest winter.fea_lab ['Animal'] [0.16109303]\n2018Q1Q10Q17Q58Q49Q512348Q-5732Q.fea_lab ['Speech'] [0.78335935]\n15_1_whistle.fea_lab ['Whistling'] [0.34013063] ['music']\n2018Q1Q10Q13Q52Q8Q512440Q-5889Q.fea_lab ['Speech'] [0.7389336]\n09_2_my guitar.fea_lab ['Music', 'Musical instrument', 'Plucked string instrument', 'Guitar'] [0.84308875, 0.48860216, 0.43791085, 0.47915566]\n2018Q1Q10Q13Q29Q46Q512440Q-5889Q.fea_lab ['Vehicle'] [0.18344605]\n05_2_DFA.fea_lab ['Music', 'Musical instrument', 'Plucked string instrument', 'Guitar'] [0.93665695, 0.57123834, 0.53891456, 0.63112855]\n'''\n\nq_kong_res = {\n '2018Q1Q10Q17Q12Q59Q512440Q-5889Q.wav': (['Speech'], [0.8013877]),\n '12_4_train ambience.wav': (['Vehicle', 'Rail transport', 'Train', 'Railroad car, train wagon'], [0.38702238, 0.6618184, 0.7742054, 0.5886036]),\n '19_3_forest winter.wav': (['Animal'], [0.16109303]),\n '2018Q1Q10Q17Q58Q49Q512348Q-5732Q.wav': (['Speech'], [0.78335935]),\n '15_1_whistle.wav': (['Whistling'], [0.34013063], ['music']),\n '2018Q1Q10Q13Q52Q8Q512440Q-5889Q.wav': (['Speech'], [0.7389336]),\n '09_2_my guitar.wav': (['Music', 'Musical instrument', 'Plucked string instrument', 'Guitar'], [0.84308875, 0.48860216, 0.43791085, 0.47915566]),\n '2018Q1Q10Q13Q29Q46Q512440Q-5889Q.wav': (['Vehicle'], [0.18344605]),\n '05_2_DFA.wav': (['Music', 'Musical instrument', 'Plucked string instrument', 'Guitar'], [0.93665695, 0.57123834, 0.53891456, 0.63112855])\n}\n\n\n#test_examples_res = qkong_res.split('\\n')\n#print(test_examples_res)\n#rint()\n#split_fun = lambda x: x.split(' [')\n#test_examples_res = list(map(split_fun, test_examples_res))#\n\n# print(test_examples_res)",
"_____no_output_____"
],
[
"max_prob_classes_AEDM = np.argsort(predictions_AEDM, axis=-1)[:, ::-1]\nmax_prob_AEDM = np.sort(predictions_AEDM, axis=-1)[:,::-1]\n\nmax_prob_classes_ASC = np.argsort(predictions_ASC, axis=-1)[:, ::-1]\nmax_prob_ASC = np.sort(predictions_ASC, axis=-1)[:,::-1]\n\nfor i in range(len(wav_files)):\n print(wav_files[i])\n print(max_prob_classes_AEDM[i,:10])\n print(max_prob_AEDM[i,:10])\n print()\n print(max_prob_classes_ASC[i,:10])\n print(max_prob_ASC[i,:10])\n print()\n print()",
"_____no_output_____"
]
],
[
[
"2018Q1Q10Q17Q12Q59Q512440Q-5889Q.wav\n2018Q1Q10Q13Q52Q8Q512440Q-5889Q.wav\n2018Q1Q10Q13Q29Q46Q512440Q-5889Q.wav\n2018Q1Q10Q17Q58Q49Q512348Q-5732Q.wav",
"_____no_output_____"
]
],
[
[
"for i, vid in enumerate(example_names):\n print(vid)\n print()\n example = pd.DataFrame(class_labels['display_name'][max_prob_classes_AEDM[i,:10]])\n example.loc[:, 'top_10_AEDM_pred'] = pd.Series(max_prob_AEDM[i, :10], index=example.index)\n example.loc[:, 'index_ASC'] = pd.Series(max_prob_classes_ASC[i,:10], index=example.index)\n example.loc[:, 'display_name_ASC'] = pd.Series(\n class_labels['display_name'][max_prob_classes_ASC[i,:10]],\n index=example.index_ASC\n )\n example.loc[:, 'top_10_ASC_pred'] = pd.Series(max_prob_ASC[i, :10], index=example.index)\n print(example)\n example.plot.bar(x='display_name', y=['top_10_AEDM_pred', 'top_10_ASC_pred'] , rot=90)\n plt.show()\n print()\n \n ex_lab = q_kong_res[vid][0]\n ex_pred = q_kong_res[vid][1]\n \n example = pd.DataFrame(class_labels[class_labels['display_name'].isin(ex_lab)])\n example.loc[:, 'AEDM_pred'] = pd.Series(\n predictions_AEDM[i, example.index.tolist()],\n index=example.index\n )\n example.loc[:, 'ASC_pred'] = pd.Series(\n predictions_ASC[i, example.index.tolist()],\n index=example.index\n )\n example.loc[:, 'qkong_pred'] = pd.Series(\n ex_pred,\n index=example.index\n )\n print(example)\n print()\n \n example.plot.bar(x='display_name', y=['AEDM_pred', 'ASC_pred', 'qkong_pred'], rot=90)\n plt.show()",
"_____no_output_____"
]
],
[
[
"## Audio set data collection pipeline\n\n### Download, cut and convert the audio of listed urls",
"_____no_output_____"
]
],
[
[
"colnames = '# YTID, start_seconds, end_seconds, positive_labels'.split(', ')\nprint(colnames)\nbal_train_csv = pd.read_csv('balanced_train_segments.csv', sep=', ', header=2) #usecols=colnames)\nbal_train_csv.rename(columns={colnames[0]: colnames[0][-4:]}, inplace=True)",
"_____no_output_____"
],
[
"print(bal_train_csv.columns.values)\nprint(bal_train_csv.loc[:10, colnames[3]])\nprint(bal_train_csv.YTID.tolist()[:10])\n\nbal_train_csv['pos_lab_list'] = bal_train_csv.positive_labels.apply(lambda x: x[1:-1].split(','))\ncolnames.extend('pos_lab_list')\n\nprint('Pos_lab_list')\nprint(bal_train_csv.loc[:10, 'pos_lab_list'])\n\nsample_rate = 16000\n\naudioset_short_raw_dir = os.path.join(src_dir, 'data', 'audioset_short_raw')\nif not os.path.exists(audioset_short_raw_dir):\n os.makedirs(audioset_short_raw_dir)\naudioset_raw_dir = os.path.join(src_dir, 'data', 'audioset_raw')\nif not os.path.exists(audioset_raw_dir):\n os.makedirs(audioset_raw_dir)\naudioset_embed_path = os.path.join(src_dir, 'data', 'audioset_embed')\nif not os.path.exists(audioset_embed_path):\n os.makedirs(audioset_embed_path)\n\naudioset_video_titles = []\naudioset_video_ids = bal_train_csv.YTID.tolist()\naudioset_video_ids_bin = bal_train_csv.YTID.astype('|S11').tolist()\nvideo_start_time = bal_train_csv.start_seconds.tolist()\nvideo_end_time = bal_train_csv.end_seconds.tolist()\n\n# Provide class dictionary for conversion from mid to either index [0] or display_name [1] \nclass_dict = class_labels.set_index('mid').T.to_dict('list')\n\nprint(class_dict['/m/09x0r'])\n\nprint(\n list(\n map(\n lambda x: class_dict[x][0],\n bal_train_csv.loc[0, 'pos_lab_list']\n )\n )\n)\n\nbal_train_csv['pos_lab_ind_list'] = bal_train_csv.pos_lab_list.apply(\n lambda x: [class_dict[y][0] for y in x]\n)\n\nclass_vec = np.zeros([1, 527])\nclass_vec[:, bal_train_csv.loc[0, 'pos_lab_ind_list']] = 1\nprint(class_vec)\n\nprint(bal_train_csv.dtypes)\n#print(bal_train_csv.loc[:10, colnames[4]])",
"_____no_output_____"
],
[
"video_ids_incl = []\nvideo_ids_incl_bin = []\nvideo_ids_excl = []\nvggish_embeds = []\nlabels = []",
"_____no_output_____"
],
[
"print(video_ids_incl)",
"_____no_output_____"
],
[
"video_ids_incl = video_ids_incl[:-1]\nprint(video_ids_incl)\nvideo_ids_checked = video_ids_incl + video_ids_excl\nvideo_ids = [vid for vid in audioset_video_ids if vid not in video_ids_checked]",
"_____no_output_____"
],
[
"for i, vid in enumerate(video_ids):\n print('{}.'.format(i))\n # Download and store video under data/audioset_short_raw/\n if (vid + '.wav') not in os.listdir(audioset_short_raw_dir):\n video_title = dl_yt.download_youtube_wav(\n video_id=vid,\n raw_dir=None,\n short_raw_dir=audioset_short_raw_dir,\n start_sec=video_start_time[i],\n duration=video_end_time[i]-video_start_time[i],\n sample_rate=sample_rate\n )\n audioset_video_titles += [video_title]\n wav_available = video_title is not None\n else:\n print(vid, 'already downloaded, so we skip this download.')\n wav_available = True\n \n if wav_available:\n video_ids_incl += [vid]\n video_ids_incl_bin += [audioset_video_ids_bin[i]]\n vggish_embeds.extend(\n ASC.embed(\n os.path.join(\n audioset_short_raw_dir,\n vid\n ) + '.wav'\n )\n )\n class_vec = np.zeros([1, 527])\n class_vec[:, bal_train_csv.loc[i, 'pos_lab_ind_list']] = 1\n labels.extend(class_vec)\n else:\n video_ids_excl += [vid]\n print()\n \n\n\n\n",
"_____no_output_____"
],
[
"jobs = []\nfor i, vid in enumerate(video_ids):\n # Download and store video under data/audioset_short_raw/\n if (vid + '.wav') not in os.listdir(audioset_short_raw_dir):\n args = (\n vid,\n None,\n audioset_short_raw_dir,\n video_start_time[i],\n video_end_time[i]-video_start_time[i],\n sample_rate\n )\n process = multiprocessing.Process(\n target=dl_yt.download_youtube_wav,\n args=args\n )\n jobs.append(process)\n\n# Start the processes (i.e. calculate the random number lists)\nfor j in jobs:\n j.start()\n\n# Ensure all of the processes have finished\nfor j in jobs:\n j.join()",
"_____no_output_____"
],
[
"save_data(\n hdf5_path=os.path.join(audioset_embed_path, 'bal_train.h5'),\n x=np.array(vggish_embeds),\n video_id_list=np.array(video_ids_incl_bin),\n y=np.array(labels)\n)",
"_____no_output_____"
],
[
"x, y, vid_list = load_data(os.path.join(audioset_embed_path, 'bal_train.h5'))",
"_____no_output_____"
],
[
"print(vid_list)",
"_____no_output_____"
],
[
"x_train, y_train, video_id_train = load_data(os.path.join(audioset_embed_path, 'bal_train.h5'))",
"_____no_output_____"
],
[
"print(video_id_train)",
"_____no_output_____"
],
[
"x_train, y_train, video_id_train = load_data(\n os.path.join(\n 'data',\n 'audioset',\n 'packed_features',\n 'bal_train.h5'\n )\n)\n\nprint(video_id_train[:100])",
"_____no_output_____"
],
[
"from retrieve_audioset import retrieve_embeddings\n\nretrieve_embeddings(\n data_path=os.path.join('data', 'audioset')\n)",
"\nLoading VGGish base model:\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03ca146e8cf5d4741fbbc01d01e56e36f6005d0 | 58,757 | ipynb | Jupyter Notebook | gradient_descent_tutorial/gradient_descent_tutorial.ipynb | fimoziq/tutorials | f47f1b59bf3c9e9f79d530c6fc8ca36c0d9ea93b | [
"MIT"
] | 670 | 2020-07-23T11:33:36.000Z | 2022-03-31T16:38:11.000Z | gradient_descent_tutorial/gradient_descent_tutorial.ipynb | terragord7/tutorials | a5c3f1fed6c5c4d23f59a41c024f7499055c8d81 | [
"MIT"
] | 3 | 2021-01-03T16:36:39.000Z | 2022-02-17T06:05:43.000Z | gradient_descent_tutorial/gradient_descent_tutorial.ipynb | terragord7/tutorials | a5c3f1fed6c5c4d23f59a41c024f7499055c8d81 | [
"MIT"
] | 281 | 2020-07-23T06:37:28.000Z | 2022-03-30T07:33:48.000Z | 133.842825 | 30,146 | 0.850111 | [
[
[
"<a href=\"https://colab.research.google.com/github/towardsai/tutorials/blob/master/gradient_descent_tutorial/gradient_descent_tutorial.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Gradient Descent Algorithm Implementation\r\n\r\n* Tutorial: https://towardsai.net/p/data-science/gradient-descent-algorithm-for-machine-learning-python-tutorial-ml-9ded189ec556\r\n* Github: https://github.com/towardsai/tutorials/tree/master/gradient_descent_tutorial",
"_____no_output_____"
]
],
[
[
"#Download the dataset\r\n!wget https://raw.githubusercontent.com/towardsai/tutorials/master/gradient_descent_tutorial/data.txt",
"--2020-12-21 02:45:22-- https://raw.githubusercontent.com/towardsai/tutorials/master/gradient_descent_tutorial/data.txt\nResolving raw.githubusercontent.com (raw.githubusercontent.com)... 151.101.0.133, 151.101.64.133, 151.101.128.133, ...\nConnecting to raw.githubusercontent.com (raw.githubusercontent.com)|151.101.0.133|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 1359 (1.3K) [text/plain]\nSaving to: ‘data.txt’\n\ndata.txt 100%[===================>] 1.33K --.-KB/s in 0s \n\n2020-12-21 02:45:22 (62.5 MB/s) - ‘data.txt’ saved [1359/1359]\n\n"
],
[
"import pandas as pd\r\nimport numpy as np\r\nimport matplotlib.pyplot as plt \r\n%matplotlib inline",
"_____no_output_____"
],
[
"column_names = ['Population', 'Profit']\r\ndf = pd.read_csv('data.txt', header=None, names=column_names) \r\ndf.head()",
"_____no_output_____"
],
[
"df.insert(0, 'Theta0', 1)\r\ncols = df.shape[1]\r\nX = df.iloc[:,0:cols-1] \r\nY = df.iloc[:,cols-1:cols]\r\n \r\ntheta = np.matrix(np.array([0]*X.shape[1])) \r\nX = np.matrix(X.values) \r\nY = np.matrix(Y.values)",
"_____no_output_____"
],
[
"def calculate_RSS(X, y, theta): \r\n inner = np.power(((X * theta.T) - y), 2)\r\n return np.sum(inner) / (2 * len(X))",
"_____no_output_____"
],
[
"def gradientDescent(X, Y, theta, alpha, iters): \r\n t = np.matrix(np.zeros(theta.shape))\r\n parameters = int(theta.ravel().shape[1])\r\n cost = np.zeros(iters)\r\n \r\n for i in range(iters):\r\n error = (X * theta.T) - Y\r\n\r\n for j in range(parameters):\r\n term = np.multiply(error, X[:,j])\r\n t[0,j] = theta[0,j] - ((alpha / len(X)) * np.sum(term))\r\n\r\n theta = t\r\n cost[i] = calculate_RSS(X, Y, theta)\r\n\r\n return theta, cost",
"_____no_output_____"
],
[
"df.plot(kind='scatter', x='Population', y='Profit', figsize=(12,8))",
"_____no_output_____"
]
],
[
[
"**Error before applying Gradient Descent**",
"_____no_output_____"
]
],
[
[
"error = calculate_RSS(X, Y, theta)\r\nerror",
"_____no_output_____"
]
],
[
[
"**Apply Gradient Descent**",
"_____no_output_____"
]
],
[
[
"g, cost = gradientDescent(X, Y, theta, 0.01, 1000) \r\ng",
"_____no_output_____"
]
],
[
[
"**Error after Applying Gradient Descent**",
"_____no_output_____"
]
],
[
[
"error = calculate_RSS(X, Y, g)\r\nerror",
"_____no_output_____"
],
[
"x = np.linspace(df.Population.min(), df.Population.max(), 100) \r\nf = g[0, 0] + (g[0, 1] * x)\r\nfig, ax = plt.subplots(figsize=(12,8)) \r\nax.plot(x, f, 'r', label='Prediction') \r\nax.scatter(df.Population, df.Profit, label='Traning Data') \r\nax.legend(loc=2) \r\nax.set_xlabel('Population') \r\nax.set_ylabel('Profit') \r\nax.set_title('Predicted Profit vs. Population Size')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d03ca4077f01366b5f884ebc61c4abfc173c7c0c | 16,469 | ipynb | Jupyter Notebook | examples/Train_ppo_cnn+eval_contact-(pretrained).ipynb | pleslabay/CarRacing-mod | e416d7f5d6dc49731e64d85094256c30c5f7d4b3 | [
"MIT"
] | null | null | null | examples/Train_ppo_cnn+eval_contact-(pretrained).ipynb | pleslabay/CarRacing-mod | e416d7f5d6dc49731e64d85094256c30c5f7d4b3 | [
"MIT"
] | null | null | null | examples/Train_ppo_cnn+eval_contact-(pretrained).ipynb | pleslabay/CarRacing-mod | e416d7f5d6dc49731e64d85094256c30c5f7d4b3 | [
"MIT"
] | 1 | 2020-12-29T23:03:44.000Z | 2020-12-29T23:03:44.000Z | 31.978641 | 178 | 0.543385 | [
[
[
"# Filter tensorflow version warnings\nimport os\n# https://stackoverflow.com/questions/40426502/is-there-a-way-to-suppress-the-messages-tensorflow-prints/40426709\nos.environ['TF_CPP_MIN_LOG_LEVEL'] = '3' # or any {'0', '1', '2'}\nimport warnings\n# https://stackoverflow.com/questions/15777951/how-to-suppress-pandas-future-warning\nwarnings.simplefilter(action='ignore', category=FutureWarning)\nwarnings.simplefilter(action='ignore', category=Warning)\nimport tensorflow as tf\ntf.get_logger().setLevel('INFO')\ntf.autograph.set_verbosity(0)\nimport logging\ntf.get_logger().setLevel(logging.ERROR)",
"_____no_output_____"
],
[
"import gym\nfrom stable_baselines.common.policies import CnnPolicy #, MlpPolicy, CnnLstmPolicy\nfrom stable_baselines.common.vec_env import DummyVecEnv \nfrom stable_baselines import PPO2\n\nfrom stable_baselines.common.evaluation import evaluate_policy as test\nfrom stable_baselines.common.callbacks import EvalCallback, StopTrainingOnRewardThreshold\n",
"_____no_output_____"
]
],
[
[
"## if you wish to set which cores to use\naffinity_mask = {4, 5, 7} \n#affinity_mask = {6, 7, 9} \n#affinity_mask = {0, 1, 3} \n#affinity_mask = {2, 3, 5} \naffinity_mask = {0, 2, 4, 6} \n\npid = 0\nos.sched_setaffinity(pid, affinity_mask) \nprint(\"CPU affinity mask is modified to %s for process id 0\" % affinity_mask) \n",
"_____no_output_____"
],
[
"## DEFAULT 'CarRacing-v3' environment values\n\n# continuos action = (steering_angle, throttle, brake)\nACT = [[0, 0, 0], [-0.4, 0, 0], [0.4, 0, 0], [0, 0.6, 0], [0, 0, 0.8]]\n# discrete actions: center_steering and no gas/brake, steer left, steer right, accel, brake \n# --> actually a good choice, because car_dynamics softens the action's diff for gas and steering\n\n##REWARDS \n# reward given each step: step taken, distance to centerline, normalized speed [0-1], normalized steer angle [0-1]\n# reward given on new tile touched: %proportional of advance, %advance/steps_taken\n# reward given at episode end: all tiles touched (track finished), patience or off-raod exceeded, out of bounds, max_steps exceeded\n# reward for obstacles: obstacle hit (each step), obstacle collided (episode end)\nGYM_REWARD = [ -0.1, 0.0, 0.0, 0.0, 10.0, 0.0, 0, -0, -100, -0, -0, -0 ]\nSTD_REWARD = [ -0.1, 0.0, 0.0, 0.0, 1.0, 0.0, 100, -20, -100, -50, -0, -0 ]\nCONT_REWARD =[-0.11, 0.1, 0.0, 0.0, 1.0, 0.0, 100, -20, -100, -50, -5, -100 ]\n# see docu for RETURN computation details\n\n## DEFAULT Environment Parameters (not related to RL Algorithm!)\ngame_color = 1 # State (frame) color option: 0 = RGB, 1 = Grayscale, 2 = Green only\nindicators = True # show or not bottom Info Panel\nframes_per_state = 4 # stacked (rolling history) Frames on each state [1-inf], latest observation always on first Frame\nskip_frames = 3 # number of consecutive Frames to skip between history saves [0-4]\ndiscre = ACT # Action discretization function, format [[steer0, throtle0, brake0], [steer1, ...], ...]. None for continuous\n\nuse_track = 1 # number of times to use the same Track, [1-100]. More than 20 high risk of overfitting!!\nepisodes_per_track = 1 # number of evenly distributed starting points on each track [1-20]. Every time you call reset(), the env automatically starts at the next point\ntr_complexity = 12 # generated Track geometric Complexity, [6-20]\ntr_width = 45 # relative Track Width, [30-50]\npatience = 2.0 # max time in secs without Progress, [0.5-20]\noff_track = 1.0 # max time in secs Driving on Grass, [0.0-5]\nf_reward = CONT_REWARD # Reward Funtion coefficients, refer to Docu for details\n\nnum_obstacles = 5 # Obstacle objects placed on track [0-10]\nend_on_contact = False # Stop Episode on contact with obstacle, not recommended for starting-phase of training\nobst_location = 0 # array pre-setting obstacle Location, in %track. Negative value means tracks's left-hand side. 0 for random location\noily_patch = False # use all obstacles as Low-friction road (oily patch)\nverbose = 2 \n",
"_____no_output_____"
]
],
[
[
"## Choose one agent, see Docu for description\n#agent='CarRacing-v0'\n#agent='CarRacing-v1'\nagent='CarRacing-v3'\n\n# Stop training when the model reaches the reward threshold\ncallback_on_best = StopTrainingOnRewardThreshold(reward_threshold = 170, verbose=1)\n\nseed = 2000",
"_____no_output_____"
],
[
"## SIMULATION param \n## Changing these makes world models incompatible!!\ngame_color = 2\nindicators = True\nfpst = 4\nskip = 3\nactions = [[0, 0, 0], [-0.4, 0, 0], [0.4, 0, 0], [0, 0.6, 0], [0, 0, 0.8]] #this is ACT\n\nobst_loc = [6, -12, 25, -50, 75, -37, 62, -87, 95, -29] #track percentage, negative for obstacle to the left-hand side\n",
"_____no_output_____"
],
[
"## Loading drive_pretained model\n\nimport pickle\nroot = 'ppo_cnn_gym-mod_'\nfile = root+'c{:d}_f{:d}_s{:d}_{}_a{:d}'.format(game_color,fpst,skip,indicators,len(actions))\n\nmodel = PPO2.load(file)",
"_____no_output_____"
],
[
"## This model param\nuse = 6 # number of times to use same track [1,100]\nept = 10 # different starting points on same track [1,20]\npatience = 1.0\ntrack_complexity = 12\n#REWARD2 = [-0.05, 0.1, 0.0, 0.0, 2.0, 0.0, 100, -20, -100, -50, -5, -100]\n\nif agent=='CarRacing-v3': \n env = gym.make(agent, seed=seed, \n game_color=game_color,\n indicators=indicators,\n frames_per_state=fpst,\n skip_frames=skip, \n# discre=actions, #passing custom actions\n use_track = use, \n episodes_per_track = ept, \n tr_complexity = track_complexity, \n tr_width = 45,\n patience = patience,\n off_track = patience,\n end_on_contact = True, #learning to avoid obstacles the-hard-way\n oily_patch = False,\n num_obstacles = 5, #some obstacles\n obst_location = obst_loc, #passing fixed obstacle location\n# f_reward = REWARD2, #passing a custom reward function\n verbose = 2 ) \nelse: \n env = gym.make(agent)\n\nenv = DummyVecEnv([lambda: env])",
"_____no_output_____"
],
[
"## Training on obstacles\nmodel.set_env(env)\nbatch_size = 256\nupdates = 700",
"_____no_output_____"
],
[
"model.learn(total_timesteps = updates*batch_size, log_interval=1) #, callback=eval_callback)",
"_____no_output_____"
],
[
"#Save last updated model\n\nfile = root+'c{:d}_f{:d}_s{:d}_{}_a{:d}__u{:d}_e{:d}_p{}_bs{:d}'.format(\n game_color,fpst,skip,indicators,len(actions),use,ept,patience,batch_size)\n\nmodel.save(file, cloudpickle=True)\nparam_list=model.get_parameter_list()\n",
"_____no_output_____"
],
[
"env.close()",
"_____no_output_____"
],
[
"## This model param #2\nuse = 6 # number of times to use same track [1,100]\nept = 10 # different starting points on same track [1,20]\npatience = 1.0\ntrack_complexity = 12\n#REWARD2 = [-0.05, 0.1, 0.0, 0.0, 2.0, 0.0, 100, -20, -100, -50, -5, -100]\nseed = 25000\n\nif agent=='CarRacing-v3': \n env2 = gym.make(agent, seed=seed, \n game_color=game_color,\n indicators=indicators,\n frames_per_state=fpst,\n skip_frames=skip, \n# discre=actions, #passing custom actions\n use_track = use, \n episodes_per_track = ept, \n tr_complexity = track_complexity, \n tr_width = 45,\n patience = patience,\n off_track = patience,\n end_on_contact = False, # CHANGED \n oily_patch = False,\n num_obstacles = 5, #some obstacles\n obst_location = 0, #using random obstacle location\n# f_reward = REWARD2, #passing a custom reward function\n verbose = 3 ) \nelse: \n env2 = gym.make(agent)\n\nenv2 = DummyVecEnv([lambda: env2])",
"_____no_output_____"
],
[
"## Training on obstacles\nmodel.set_env(env2)\n#batch_size = 384\nupdates = 1500",
"_____no_output_____"
],
[
"## Separate evaluation env\ntest_freq = 100 #policy updates until evaluation\ntest_episodes_per_track = 5 #number of starting points on test_track\neval_log = './evals/'\n\nenv_test = gym.make(agent, seed=int(3.14*seed), \n game_color=game_color,\n indicators=indicators,\n frames_per_state=fpst,\n skip_frames=skip, \n# discre=actions, #passing custom actions\n use_track = 1, #change test track after 1 ept round\n episodes_per_track = test_episodes_per_track, \n tr_complexity = 12, #test on a medium complexity track\n tr_width = 45,\n patience = 2.0,\n off_track = 2.0,\n end_on_contact = False,\n oily_patch = False,\n num_obstacles = 5,\n obst_location = obst_loc) #passing fixed obstacle location\n\nenv_test = DummyVecEnv([lambda: env_test])\n\neval_callback = EvalCallback(env_test, callback_on_new_best=callback_on_best, #None,\n n_eval_episodes=test_episodes_per_track*3, eval_freq=test_freq*batch_size,\n best_model_save_path=eval_log, log_path=eval_log, deterministic=True, \n render = False)\n",
"_____no_output_____"
],
[
"model.learn(total_timesteps = updates*batch_size, log_interval=1, callback=eval_callback)",
"_____no_output_____"
],
[
"#Save last updated model\n\n#file = root+'c{:d}_f{:d}_s{:d}_{}_a{:d}__u{:d}_e{:d}_p{}_bs{:d}'.format(\n# game_color,fpst,skip,indicators,len(actions),use,ept,patience,batch_size)\n\nmodel.save(file+'_II', cloudpickle=True)\nparam_list=model.get_parameter_list()\n",
"_____no_output_____"
],
[
"env2.close()\nenv_test.close()",
"_____no_output_____"
],
[
"## Enjoy last trained policy\n\nif agent=='CarRacing-v3': #create an independent test environment, almost everything in std/random definition\n env3 = gym.make(agent, seed=None, \n game_color=game_color,\n indicators = True,\n frames_per_state=fpst,\n skip_frames=skip, \n# discre=actions,\n use_track = 2, \n episodes_per_track = 1, \n patience = 5.0,\n off_track = 3.0 )\nelse:\n env3 = gym.make(agent)\n\nenv3 = DummyVecEnv([lambda: env3])\nobs = env3.reset()\nprint(obs.shape) \n\ndone = False\npasos = 0\n_states=None\n\nwhile not done: # and pasos<1500:\n action, _states = model.predict(obs, deterministic=True)\n obs, reward, done, info = env3.step(action)\n env3.render()\n pasos+=1\n \nenv3.close()\nprint()\nprint(reward, done, pasos) #, info)",
"_____no_output_____"
],
[
"## Enjoy best eval_policy\n\nobs = env3.reset()\nprint(obs.shape) \n\n## Load bestmodel from eval\n#if not isinstance(model_test, PPO2):\nmodel_test = PPO2.load(eval_log+'best_model', env3)\n\ndone = False\npasos = 0\n_states=None\n\nwhile not done: # and pasos<1500:\n action, _states = model_test.predict(obs, deterministic=True)\n obs, reward, done, info = env3.step(action)\n env3.render()\n pasos+=1\n \nenv3.close()\nprint()\nprint(reward, done, pasos)\nprint(action, _states)",
"_____no_output_____"
],
[
"model_test.save(file+'_evalbest', cloudpickle=True)",
"_____no_output_____"
],
[
"env2.close()",
"_____no_output_____"
],
[
"env3.close()",
"_____no_output_____"
],
[
"env_test.close()",
"_____no_output_____"
],
[
"print(action, _states)",
"_____no_output_____"
],
[
"obs.shape",
"_____no_output_____"
]
]
] | [
"code",
"raw",
"code"
] | [
[
"code",
"code"
],
[
"raw",
"raw"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03ca46602c35df1734327c8168ab66ada124849 | 1,631 | ipynb | Jupyter Notebook | examples/just_ship_it_example.ipynb | gstaff/just-ship-it | 7f2bc0867b92f7253b17e78b62760d5ccfeb46bf | [
"Apache-2.0"
] | null | null | null | examples/just_ship_it_example.ipynb | gstaff/just-ship-it | 7f2bc0867b92f7253b17e78b62760d5ccfeb46bf | [
"Apache-2.0"
] | null | null | null | examples/just_ship_it_example.ipynb | gstaff/just-ship-it | 7f2bc0867b92f7253b17e78b62760d5ccfeb46bf | [
"Apache-2.0"
] | null | null | null | 24.343284 | 146 | 0.450644 | [
[
[
"!pip install just-ship-it\n!pip install flask==0.12.2 # Newer versions of Flask have an issue running in\n # notebooks, see https://stackoverflow.com/questions/51180917/python-flask-unsupportedoperation-not-writable",
"_____no_output_____"
],
[
"# just_ship_it_example.py\ndef greet(name):\n \"\"\"Greets you by name!\"\"\"\n return 'Hello {name}!'.format(name=name)\n\ndef hello():\n return {'hello': 'world'}",
"_____no_output_____"
],
[
"if __name__ == '__main__':\n from just_ship_it import ship_it\n ship_it()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d03ca8c52283613f5443c3baf85d653ef41361c7 | 69,048 | ipynb | Jupyter Notebook | API_Chi_Busi_Licences.ipynb | oimartin/Real_Tech_Influence | 2c598ce889162a6562a7b5d0fbb7647f5ea55e3e | [
"CNRI-Python"
] | null | null | null | API_Chi_Busi_Licences.ipynb | oimartin/Real_Tech_Influence | 2c598ce889162a6562a7b5d0fbb7647f5ea55e3e | [
"CNRI-Python"
] | null | null | null | API_Chi_Busi_Licences.ipynb | oimartin/Real_Tech_Influence | 2c598ce889162a6562a7b5d0fbb7647f5ea55e3e | [
"CNRI-Python"
] | null | null | null | 40.640377 | 149 | 0.408542 | [
[
[
"## Connect to Chicago Data Portal API - Business Licenses Data",
"_____no_output_____"
]
],
[
[
"#Import dependencies\nimport pandas as pd\nimport requests\nimport json\n\n# Google developer API key\nfrom config2 import API_chi_key",
"_____no_output_____"
],
[
"# Build API URL\n# API calls = 8000 (based on zipcode and issued search results)\n# Filters: 'application type' Issued\ntarget_URL = f\"https://data.cityofchicago.org/resource/xqx5-8hwx.json?$$app_token={API_chi_key}&$limit=8000&application_type=ISSUE&zip_code=\"",
"_____no_output_____"
],
[
"# Create list of zipcodes we are examining based\n# on three different businesses of interest\nzipcodes = [\"60610\",\"60607\",\"60606\",\"60661\",\n \"60614\",\"60622\",\"60647\",\"60654\"]",
"_____no_output_____"
],
[
"# Create a request to get json data on business licences\nresponses = []\nfor zipcode in zipcodes:\n license_response = requests.get(target_URL + zipcode).json()\n responses.append(license_response)",
"_____no_output_____"
],
[
"len(responses)",
"_____no_output_____"
],
[
"# Create sepearte variables for the 8 responses for zipcodes\n# Data loaded in nested gropus based on zipcodes, so\n# needed to make them separate\nzip_60610 = responses[0]\nzip_60607 = responses[1]\nzip_60606 = responses[2]\nzip_60661 = responses[3]\nzip_60614 = responses[4]\nzip_60622 = responses[5]\nzip_60647 = responses[6]\nzip_60654 = responses[7]",
"_____no_output_____"
],
[
"# Read zipcode_responses_busi.json files into pd DF\nzip_60610_data = pd.DataFrame(zip_60610)",
"_____no_output_____"
],
[
"# Create list of the json object variables\n# excluding zip_60610 bc that will start as a DF\nzip_data = [zip_60607, zip_60606, zip_60661, zip_60614,\n zip_60622, zip_60647, zip_60654]",
"_____no_output_____"
],
[
"# Create a new DF to save compiled business data into\nall_7_zipcodes = zip_60610_data",
"_____no_output_____"
],
[
"# Append json objects to all_7_zipcode DF\n# Print length of all_7_zipcode to check adding correctly\nfor zipcodes_df in zip_data:\n all_7_zipcodes = all_7_zipcodes.append(zipcodes_df)",
"C:\\Users\\oimar\\Miniconda3\\lib\\site-packages\\pandas\\core\\indexing.py:1472: FutureWarning: \nPassing list-likes to .loc or [] with any missing label will raise\nKeyError in the future, you can use .reindex() as an alternative.\n\nSee the documentation here:\nhttps://pandas.pydata.org/pandas-docs/stable/indexing.html#deprecate-loc-reindex-listlike\n return self._getitem_tuple(key)\n"
],
[
"len(all_7_zipcodes)",
"_____no_output_____"
],
[
"# Get list of headers of all_7_zipcodes\nlist(all_7_zipcodes)",
"_____no_output_____"
],
[
"# Select certain columns to show \ncore_info_busi_licences = all_7_zipcodes[['legal_name', 'doing_business_as_name',\n 'zip_code', 'license_description', \n 'business_activity', 'application_type', \n 'license_start_date', 'latitude', 'longitude']]",
"_____no_output_____"
],
[
"# Get an idea of the number of null values in each column\ncore_info_busi_licences.isna().sum()",
"_____no_output_____"
],
[
"# Add sepearate column for just the start year\n# Will use later when selecting year businesess were created\ncore_info_busi_licences['start_year'] = core_info_busi_licences['license_start_date']",
"C:\\Users\\oimar\\Miniconda3\\lib\\site-packages\\ipykernel_launcher.py:3: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n This is separate from the ipykernel package so we can avoid doing imports until\n"
],
[
"# Edit 'start_year' to just include year from date information\ncore_info_busi_licences['start_year'] = core_info_busi_licences['start_year'].str[0:4]",
"C:\\Users\\oimar\\Miniconda3\\lib\\site-packages\\ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n"
],
[
"# Explore what kinds of businesses are missing \"latitude\" and \"longitude\"\n# Also, the 'business_activity' licenses have null values (limited Business Licences?)\ncore_info_busi_licences[core_info_busi_licences.isnull().any(axis=1)]",
"_____no_output_____"
],
[
"# Get rid of NaN values in 'latitude' and 'license_start_date'\ncore_info_busi_licences.dropna(subset=['latitude'], inplace=True)\ncore_info_busi_licences.dropna(subset=['license_start_date'], inplace=True)",
"C:\\Users\\oimar\\Miniconda3\\lib\\site-packages\\ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \nC:\\Users\\oimar\\Miniconda3\\lib\\site-packages\\ipykernel_launcher.py:3: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n This is separate from the ipykernel package so we can avoid doing imports until\n"
],
[
"core_info_busi_licences['application_type'].unique()",
"_____no_output_____"
],
[
"# Cast 'start_year' column as an integer\ncore_info_busi_licences['start_year'] = core_info_busi_licences['start_year'].astype('int64')",
"C:\\Users\\oimar\\Miniconda3\\lib\\site-packages\\ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n"
],
[
"# Confirm that NaN values for 'latitude' and 'license_start_date' \n# were dropped\ncore_info_busi_licences.isna().sum()",
"_____no_output_____"
],
[
"# Record number of businesses licenses pulled \nlen(core_info_busi_licences)",
"_____no_output_____"
]
],
[
[
"## Connect to sqlite database",
"_____no_output_____"
]
],
[
[
"# Python SQL toolkit and Object Relational Mapper\nimport sqlalchemy\nfrom sqlalchemy.ext.automap import automap_base\nfrom sqlalchemy import create_engine\nfrom config2 import mysql_password",
"_____no_output_____"
],
[
"# Declare a Base using `automap_base()`\nBase = automap_base()",
"_____no_output_____"
],
[
"# Create engine using the `demographics.sqlite` database file\n# engine = create_engine(\"sqlite://\", echo=False)\n\nengine = create_engine(f'mysql://root:coolcat1015@localhost:3306/real_tech_db')",
"_____no_output_____"
],
[
"# Copy 'core_info_busi_licenses' db to MySql database\ncore_info_busi_licences.to_sql('business_licenses', \n con=engine, \n if_exists='replace',\n index_label=True)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d03cac6a8ba1abd88bb936af9bca459991ce7d42 | 75,957 | ipynb | Jupyter Notebook | tutorials/W3D1_BayesianDecisions/W3D1_Tutorial1.ipynb | bgalbraith/course-content | 3db3bbba0fee7af1def2a67e34be073c43434f4a | [
"CC-BY-4.0",
"BSD-3-Clause"
] | null | null | null | tutorials/W3D1_BayesianDecisions/W3D1_Tutorial1.ipynb | bgalbraith/course-content | 3db3bbba0fee7af1def2a67e34be073c43434f4a | [
"CC-BY-4.0",
"BSD-3-Clause"
] | 1 | 2021-06-16T05:41:08.000Z | 2021-06-16T05:41:08.000Z | tutorials/W3D1_BayesianDecisions/W3D1_Tutorial1.ipynb | bgalbraith/course-content | 3db3bbba0fee7af1def2a67e34be073c43434f4a | [
"CC-BY-4.0",
"BSD-3-Clause"
] | null | null | null | 46.68531 | 925 | 0.587951 | [
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/tutorials/W3D1_BayesianDecisions/W3D1_Tutorial1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Tutorial 1: Bayes with a binary hidden state\n**Week 3, Day 1: Bayesian Decisions**\n\n**By Neuromatch Academy**\n\n__Content creators:__ [insert your name here]\n\n__Content reviewers:__ ",
"_____no_output_____"
],
[
"# Tutorial Objectives\nThis is the first in a series of two core tutorials on Bayesian statistics. In these tutorials, we will explore the fundemental concepts of the Bayesian approach from two perspectives. This tutorial will work through an example of Bayesian inference and decision making using a binary hidden state. The second main tutorial extends these concepts to a continuous hidden state. In the next days, each of these basic ideas will be extended--first through time as we consider what happens when we infere a hidden state using multiple observations and when the hidden state changes across time. In the third day, we will introduce the notion of how to use inference and decisions to select actions for optimal control. For this tutorial, you will be introduced to our binary state fishing problem!\n\nThis notebook will introduce the fundamental building blocks for Bayesian statistics: \n\n1. How do we use probability distributions to represent hidden states?\n2. How does marginalization work and how can we use it?\n3. How do we combine new information with our prior knowledge?\n4. How do we combine the possible loss (or gain) for making a decision with our probabilitic knowledge?\n",
"_____no_output_____"
]
],
[
[
"#@title Video 1: Introduction to Bayesian Statistics\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id='JiEIn9QsrFg', width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"## Setup \nPlease execute the cells below to initialize the notebook environment.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom matplotlib import patches\nfrom matplotlib import transforms\nfrom matplotlib import gridspec\nfrom scipy.optimize import fsolve\n\nfrom collections import namedtuple",
"_____no_output_____"
],
[
"#@title Figure Settings\nimport ipywidgets as widgets # interactive display\nfrom ipywidgets import GridspecLayout\nfrom IPython.display import clear_output\n%config InlineBackend.figure_format = 'retina'\nplt.style.use(\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/nma.mplstyle\")\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
],
[
"# @title Plotting Functions\n\ndef plot_joint_probs(P, ):\n assert np.all(P >= 0), \"probabilities should be >= 0\"\n # normalize if not\n P = P / np.sum(P)\n marginal_y = np.sum(P,axis=1)\n marginal_x = np.sum(P,axis=0)\n\n # definitions for the axes\n left, width = 0.1, 0.65\n bottom, height = 0.1, 0.65\n spacing = 0.005\n\n # start with a square Figure\n fig = plt.figure(figsize=(5, 5))\n\n joint_prob = [left, bottom, width, height]\n rect_histx = [left, bottom + height + spacing, width, 0.2]\n rect_histy = [left + width + spacing, bottom, 0.2, height]\n\n rect_x_cmap = plt.cm.Blues\n rect_y_cmap = plt.cm.Reds\n\n # Show joint probs and marginals\n ax = fig.add_axes(joint_prob)\n ax_x = fig.add_axes(rect_histx, sharex=ax)\n ax_y = fig.add_axes(rect_histy, sharey=ax)\n\n # Show joint probs and marginals\n ax.matshow(P,vmin=0., vmax=1., cmap='Greys')\n ax_x.bar(0, marginal_x[0], facecolor=rect_x_cmap(marginal_x[0]))\n ax_x.bar(1, marginal_x[1], facecolor=rect_x_cmap(marginal_x[1]))\n ax_y.barh(0, marginal_y[0], facecolor=rect_y_cmap(marginal_y[0]))\n ax_y.barh(1, marginal_y[1], facecolor=rect_y_cmap(marginal_y[1]))\n # set limits\n ax_x.set_ylim([0,1])\n ax_y.set_xlim([0,1])\n\n # show values\n ind = np.arange(2)\n x,y = np.meshgrid(ind,ind)\n for i,j in zip(x.flatten(), y.flatten()):\n c = f\"{P[i,j]:.2f}\"\n ax.text(j,i, c, va='center', ha='center', color='black')\n for i in ind:\n v = marginal_x[i]\n c = f\"{v:.2f}\"\n ax_x.text(i, v +0.1, c, va='center', ha='center', color='black')\n v = marginal_y[i]\n c = f\"{v:.2f}\"\n ax_y.text(v+0.2, i, c, va='center', ha='center', color='black')\n\n # set up labels\n ax.xaxis.tick_bottom()\n ax.yaxis.tick_left()\n ax.set_xticks([0,1])\n ax.set_yticks([0,1])\n ax.set_xticklabels(['Silver','Gold'])\n ax.set_yticklabels(['Small', 'Large'])\n ax.set_xlabel('color')\n ax.set_ylabel('size')\n ax_x.axis('off')\n ax_y.axis('off')\n return fig\n# test\n# P = np.random.rand(2,2)\n# P = np.asarray([[0.9, 0.8], [0.4, 0.1]])\n# P = P / np.sum(P)\n# fig = plot_joint_probs(P)\n# plt.show(fig)\n# plt.close(fig)\n\n\n# fig = plot_prior_likelihood(0.5, 0.3)\n# plt.show(fig)\n# plt.close(fig)\n\n\ndef plot_prior_likelihood_posterior(prior, likelihood, posterior):\n\n # definitions for the axes\n left, width = 0.05, 0.3\n bottom, height = 0.05, 0.9\n padding = 0.1\n small_width = 0.1\n left_space = left + small_width + padding\n added_space = padding + width\n\n fig = plt.figure(figsize=(10, 4))\n\n rect_prior = [left, bottom, small_width, height]\n rect_likelihood = [left_space , bottom , width, height]\n rect_posterior = [left_space + added_space, bottom , width, height]\n\n ax_prior = fig.add_axes(rect_prior)\n ax_likelihood = fig.add_axes(rect_likelihood, sharey=ax_prior)\n ax_posterior = fig.add_axes(rect_posterior, sharey = ax_prior)\n\n rect_colormap = plt.cm.Blues\n\n # Show posterior probs and marginals\n ax_prior.barh(0, prior[0], facecolor = rect_colormap(prior[0, 0]))\n ax_prior.barh(1, prior[1], facecolor = rect_colormap(prior[1, 0]))\n ax_likelihood.matshow(likelihood, vmin=0., vmax=1., cmap='Reds')\n ax_posterior.matshow(posterior, vmin=0., vmax=1., cmap='Greens')\n\n\n # Probabilities plot details\n ax_prior.set(xlim = [1, 0], yticks = [0, 1], yticklabels = ['left', 'right'],\n ylabel = 'state (s)', title = \"Prior p(s)\")\n ax_prior.axis('off')\n\n # Likelihood plot details\n ax_likelihood.set(xticks = [0, 1], xticklabels = ['fish', 'no fish'],\n yticks = [0, 1], yticklabels = ['left', 'right'],\n ylabel = 'state (s)', xlabel = 'measurement (m)',\n title = 'Likelihood p(m (right) | s)')\n ax_likelihood.xaxis.set_ticks_position('bottom')\n ax_likelihood.spines['left'].set_visible(False)\n ax_likelihood.spines['bottom'].set_visible(False)\n\n # Posterior plot details\n\n ax_posterior.set(xticks = [0, 1], xticklabels = ['fish', 'no fish'],\n yticks = [0, 1], yticklabels = ['left', 'right'],\n ylabel = 'state (s)', xlabel = 'measurement (m)',\n title = 'Posterior p(s | m)')\n ax_posterior.xaxis.set_ticks_position('bottom')\n ax_posterior.spines['left'].set_visible(False)\n ax_posterior.spines['bottom'].set_visible(False)\n\n\n # show values\n ind = np.arange(2)\n x,y = np.meshgrid(ind,ind)\n for i,j in zip(x.flatten(), y.flatten()):\n c = f\"{posterior[i,j]:.2f}\"\n ax_posterior.text(j,i, c, va='center', ha='center', color='black')\n for i,j in zip(x.flatten(), y.flatten()):\n c = f\"{likelihood[i,j]:.2f}\"\n ax_likelihood.text(j,i, c, va='center', ha='center', color='black')\n for i in ind:\n v = prior[i, 0]\n c = f\"{v:.2f}\"\n ax_prior.text(v+0.2, i, c, va='center', ha='center', color='black')\n\n\ndef plot_prior_likelihood(ps, p_a_s1, p_a_s0, measurement):\n likelihood = np.asarray([[p_a_s1, 1-p_a_s1],[p_a_s0, 1-p_a_s0]])\n assert 0.0 <= ps <= 1.0\n prior = np.asarray([ps, 1 - ps])\n if measurement:\n posterior = likelihood[:, 0] * prior\n else:\n posterior = (likelihood[:, 1] * prior).reshape(-1)\n posterior /= np.sum(posterior)\n\n # definitions for the axes\n left, width = 0.05, 0.3\n bottom, height = 0.05, 0.9\n padding = 0.1\n small_width = 0.22\n left_space = left + small_width + padding\n small_padding = 0.05\n\n fig = plt.figure(figsize=(10, 4))\n\n rect_prior = [left, bottom, small_width, height]\n rect_likelihood = [left_space , bottom , width, height]\n rect_posterior = [left_space + width + small_padding, bottom , small_width, height]\n\n ax_prior = fig.add_axes(rect_prior)\n ax_likelihood = fig.add_axes(rect_likelihood, sharey=ax_prior)\n ax_posterior = fig.add_axes(rect_posterior, sharey=ax_prior)\n\n prior_colormap = plt.cm.Blues\n posterior_colormap = plt.cm.Greens\n\n # Show posterior probs and marginals\n ax_prior.barh(0, prior[0], facecolor = prior_colormap(prior[0]))\n ax_prior.barh(1, prior[1], facecolor = prior_colormap(prior[1]))\n ax_likelihood.matshow(likelihood, vmin=0., vmax=1., cmap='Reds')\n # ax_posterior.matshow(posterior, vmin=0., vmax=1., cmap='')\n ax_posterior.barh(0, posterior[0], facecolor = posterior_colormap(posterior[0]))\n ax_posterior.barh(1, posterior[1], facecolor = posterior_colormap(posterior[1]))\n\n # Probabilities plot details\n ax_prior.set(xlim = [1, 0], yticks = [0, 1], yticklabels = ['left', 'right'],\n ylabel = 'state (s)', title = \"Prior p(s)\")\n ax_prior.axis('off')\n\n # Likelihood plot details\n ax_likelihood.set(xticks = [0, 1], xticklabels = ['fish', 'no fish'],\n yticks = [0, 1], yticklabels = ['left', 'right'],\n ylabel = 'state (s)', xlabel = 'measurement (m)',\n title = 'Likelihood p(m | s)')\n ax_likelihood.xaxis.set_ticks_position('bottom')\n ax_likelihood.spines['left'].set_visible(False)\n ax_likelihood.spines['bottom'].set_visible(False)\n\n # Posterior plot details\n ax_posterior.set(xlim = [0, 1], yticks = [0, 1], yticklabels = ['left', 'right'],\n ylabel = 'state (s)', title = \"Posterior p(s | m)\")\n ax_posterior.axis('off')\n # ax_posterior.set(xticks = [0, 1], xticklabels = ['fish', 'no fish'],\n # yticks = [0, 1], yticklabels = ['left', 'right'],\n # ylabel = 'state (s)', xlabel = 'measurement (m)',\n # title = 'Posterior p(s | m)')\n # ax_posterior.xaxis.set_ticks_position('bottom')\n # ax_posterior.spines['left'].set_visible(False)\n # ax_posterior.spines['bottom'].set_visible(False)\n\n\n # show values\n ind = np.arange(2)\n x,y = np.meshgrid(ind,ind)\n # for i,j in zip(x.flatten(), y.flatten()):\n # c = f\"{posterior[i,j]:.2f}\"\n # ax_posterior.text(j,i, c, va='center', ha='center', color='black')\n for i in ind:\n v = posterior[i]\n c = f\"{v:.2f}\"\n ax_posterior.text(v+0.2, i, c, va='center', ha='center', color='black')\n for i,j in zip(x.flatten(), y.flatten()):\n c = f\"{likelihood[i,j]:.2f}\"\n ax_likelihood.text(j,i, c, va='center', ha='center', color='black')\n for i in ind:\n v = prior[i]\n c = f\"{v:.2f}\"\n ax_prior.text(v+0.2, i, c, va='center', ha='center', color='black')\n return fig\n\n# fig = plot_prior_likelihood(0.5, 0.3)\n# plt.show(fig)\n# plt.close(fig)\n\n\nfrom matplotlib import colors\ndef plot_utility(ps):\n prior = np.asarray([ps, 1 - ps])\n\n utility = np.array([[2, -3], [-2, 1]])\n\n expected = prior @ utility\n\n # definitions for the axes\n left, width = 0.05, 0.16\n bottom, height = 0.05, 0.9\n padding = 0.04\n small_width = 0.1\n left_space = left + small_width + padding\n added_space = padding + width\n\n fig = plt.figure(figsize=(17, 3))\n\n rect_prior = [left, bottom, small_width, height]\n rect_utility = [left + added_space , bottom , width, height]\n rect_expected = [left + 2* added_space, bottom , width, height]\n\n ax_prior = fig.add_axes(rect_prior)\n ax_utility = fig.add_axes(rect_utility, sharey=ax_prior)\n ax_expected = fig.add_axes(rect_expected)\n\n rect_colormap = plt.cm.Blues\n\n # Data of plots\n ax_prior.barh(0, prior[0], facecolor = rect_colormap(prior[0]))\n ax_prior.barh(1, prior[1], facecolor = rect_colormap(prior[1]))\n ax_utility.matshow(utility, cmap='cool')\n norm = colors.Normalize(vmin=-3, vmax=3)\n ax_expected.bar(0, expected[0], facecolor = rect_colormap(norm(expected[0])))\n ax_expected.bar(1, expected[1], facecolor = rect_colormap(norm(expected[1])))\n\n # Probabilities plot details\n ax_prior.set(xlim = [1, 0], yticks = [0, 1], yticklabels = ['left', 'right'],\n ylabel = 'state (s)', title = \"Probability of state\")\n ax_prior.axis('off')\n\n # Utility plot details\n ax_utility.set(xticks = [0, 1], xticklabels = ['left', 'right'],\n yticks = [0, 1], yticklabels = ['left', 'right'],\n ylabel = 'state (s)', xlabel = 'action (a)',\n title = 'Utility')\n ax_utility.xaxis.set_ticks_position('bottom')\n ax_utility.spines['left'].set_visible(False)\n ax_utility.spines['bottom'].set_visible(False)\n\n # Expected utility plot details\n ax_expected.set(title = 'Expected utility', ylim = [-3, 3],\n xticks = [0, 1], xticklabels = ['left', 'right'],\n xlabel = 'action (a)',\n yticks = [])\n ax_expected.xaxis.set_ticks_position('bottom')\n ax_expected.spines['left'].set_visible(False)\n ax_expected.spines['bottom'].set_visible(False)\n\n # show values\n ind = np.arange(2)\n x,y = np.meshgrid(ind,ind)\n\n for i,j in zip(x.flatten(), y.flatten()):\n c = f\"{utility[i,j]:.2f}\"\n ax_utility.text(j,i, c, va='center', ha='center', color='black')\n for i in ind:\n v = prior[i]\n c = f\"{v:.2f}\"\n ax_prior.text(v+0.2, i, c, va='center', ha='center', color='black')\n for i in ind:\n v = expected[i]\n c = f\"{v:.2f}\"\n ax_expected.text(i, 2.5, c, va='center', ha='center', color='black')\n\n return fig\n\n\ndef plot_prior_likelihood_utility(ps, p_a_s1, p_a_s0,measurement):\n assert 0.0 <= ps <= 1.0\n assert 0.0 <= p_a_s1 <= 1.0\n assert 0.0 <= p_a_s0 <= 1.0\n prior = np.asarray([ps, 1 - ps])\n likelihood = np.asarray([[p_a_s1, 1-p_a_s1],[p_a_s0, 1-p_a_s0]])\n utility = np.array([[2.0, -3.0], [-2.0, 1.0]])\n # expected = np.zeros_like(utility)\n\n if measurement:\n posterior = likelihood[:, 0] * prior\n else:\n posterior = (likelihood[:, 1] * prior).reshape(-1)\n posterior /= np.sum(posterior)\n # expected[:, 0] = utility[:, 0] * posterior\n # expected[:, 1] = utility[:, 1] * posterior\n expected = posterior @ utility\n\n # definitions for the axes\n left, width = 0.05, 0.15\n bottom, height = 0.05, 0.9\n padding = 0.05\n small_width = 0.1\n large_padding = 0.07\n left_space = left + small_width + large_padding\n\n fig = plt.figure(figsize=(17, 4))\n\n rect_prior = [left, bottom+0.05, small_width, height-0.1]\n rect_likelihood = [left_space, bottom , width, height]\n rect_posterior = [left_space + padding + width - 0.02, bottom+0.05 , small_width, height-0.1]\n rect_utility = [left_space + padding + width + padding + small_width, bottom , width, height]\n rect_expected = [left_space + padding + width + padding + small_width + padding + width, bottom+0.05 , width, height-0.1]\n\n ax_likelihood = fig.add_axes(rect_likelihood)\n ax_prior = fig.add_axes(rect_prior, sharey=ax_likelihood)\n ax_posterior = fig.add_axes(rect_posterior, sharey=ax_likelihood)\n ax_utility = fig.add_axes(rect_utility, sharey=ax_posterior)\n ax_expected = fig.add_axes(rect_expected)\n\n\n prior_colormap = plt.cm.Blues\n posterior_colormap = plt.cm.Greens\n expected_colormap = plt.cm.Wistia\n\n # Show posterior probs and marginals\n ax_prior.barh(0, prior[0], facecolor = prior_colormap(prior[0]))\n ax_prior.barh(1, prior[1], facecolor = prior_colormap(prior[1]))\n ax_likelihood.matshow(likelihood, vmin=0., vmax=1., cmap='Reds')\n ax_posterior.barh(0, posterior[0], facecolor = posterior_colormap(posterior[0]))\n ax_posterior.barh(1, posterior[1], facecolor = posterior_colormap(posterior[1]))\n ax_utility.matshow(utility, vmin=0., vmax=1., cmap='cool')\n # ax_expected.matshow(expected, vmin=0., vmax=1., cmap='Wistia')\n ax_expected.bar(0, expected[0], facecolor = expected_colormap(expected[0]))\n ax_expected.bar(1, expected[1], facecolor = expected_colormap(expected[1]))\n\n # Probabilities plot details\n ax_prior.set(xlim = [1, 0], yticks = [0, 1], yticklabels = ['left', 'right'],\n ylabel = 'state (s)', title = \"Prior p(s)\")\n ax_prior.axis('off')\n\n # Likelihood plot details\n ax_likelihood.set(xticks = [0, 1], xticklabels = ['fish', 'no fish'],\n yticks = [0, 1], yticklabels = ['left', 'right'],\n ylabel = 'state (s)', xlabel = 'measurement (m)',\n title = 'Likelihood p(m | s)')\n ax_likelihood.xaxis.set_ticks_position('bottom')\n ax_likelihood.spines['left'].set_visible(False)\n ax_likelihood.spines['bottom'].set_visible(False)\n\n # Posterior plot details\n ax_posterior.set(xlim = [0, 1], yticks = [0, 1], yticklabels = ['left', 'right'],\n ylabel = 'state (s)', title = \"Posterior p(s | m)\")\n ax_posterior.axis('off')\n\n # Utility plot details\n ax_utility.set(xticks = [0, 1], xticklabels = ['left', 'right'],\n xlabel = 'action (a)',\n title = 'Utility')\n ax_utility.xaxis.set_ticks_position('bottom')\n ax_utility.spines['left'].set_visible(False)\n ax_utility.spines['bottom'].set_visible(False)\n\n # Expected Utility plot details\n ax_expected.set(ylim = [-2, 2], xticks = [0, 1], xticklabels = ['left', 'right'],\n xlabel = 'action (a)', title = 'Expected utility', yticks=[])\n # ax_expected.axis('off')\n ax_expected.spines['left'].set_visible(False)\n # ax_expected.set(xticks = [0, 1], xticklabels = ['left', 'right'],\n # xlabel = 'action (a)',\n # title = 'Expected utility')\n # ax_expected.xaxis.set_ticks_position('bottom')\n # ax_expected.spines['left'].set_visible(False)\n # ax_expected.spines['bottom'].set_visible(False)\n\n # show values\n ind = np.arange(2)\n x,y = np.meshgrid(ind,ind)\n for i in ind:\n v = posterior[i]\n c = f\"{v:.2f}\"\n ax_posterior.text(v+0.2, i, c, va='center', ha='center', color='black')\n for i,j in zip(x.flatten(), y.flatten()):\n c = f\"{likelihood[i,j]:.2f}\"\n ax_likelihood.text(j,i, c, va='center', ha='center', color='black')\n for i,j in zip(x.flatten(), y.flatten()):\n c = f\"{utility[i,j]:.2f}\"\n ax_utility.text(j,i, c, va='center', ha='center', color='black')\n # for i,j in zip(x.flatten(), y.flatten()):\n # c = f\"{expected[i,j]:.2f}\"\n # ax_expected.text(j,i, c, va='center', ha='center', color='black')\n for i in ind:\n v = prior[i]\n c = f\"{v:.2f}\"\n ax_prior.text(v+0.2, i, c, va='center', ha='center', color='black')\n for i in ind:\n v = expected[i]\n c = f\"{v:.2f}\"\n ax_expected.text(i, v, c, va='center', ha='center', color='black')\n\n # # show values\n # ind = np.arange(2)\n # x,y = np.meshgrid(ind,ind)\n # for i,j in zip(x.flatten(), y.flatten()):\n # c = f\"{P[i,j]:.2f}\"\n # ax.text(j,i, c, va='center', ha='center', color='white')\n # for i in ind:\n # v = marginal_x[i]\n # c = f\"{v:.2f}\"\n # ax_x.text(i, v +0.2, c, va='center', ha='center', color='black')\n # v = marginal_y[i]\n # c = f\"{v:.2f}\"\n # ax_y.text(v+0.2, i, c, va='center', ha='center', color='black')\n\n return fig",
"_____no_output_____"
],
[
"# @title Helper Functions\n\ndef compute_marginal(px, py, cor):\n # calculate 2x2 joint probabilities given marginals p(x=1), p(y=1) and correlation\n p11 = px*py + cor*np.sqrt(px*py*(1-px)*(1-py))\n p01 = px - p11\n p10 = py - p11\n p00 = 1.0 - p11 - p01 - p10\n return np.asarray([[p00, p01], [p10, p11]])\n# test\n# print(compute_marginal(0.4, 0.6, -0.8))\n\n\ndef compute_cor_range(px,py):\n # Calculate the allowed range of correlation values given marginals p(x=1) and p(y=1)\n def p11(corr):\n return px*py + corr*np.sqrt(px*py*(1-px)*(1-py))\n def p01(corr):\n return px - p11(corr)\n def p10(corr):\n return py - p11(corr)\n def p00(corr):\n return 1.0 - p11(corr) - p01(corr) - p10(corr)\n Cmax = min(fsolve(p01, 0.0), fsolve(p10, 0.0))\n Cmin = max(fsolve(p11, 0.0), fsolve(p00, 0.0))\n return Cmin, Cmax",
"_____no_output_____"
]
],
[
[
"---\n# Section 1: Gone Fishin'\n",
"_____no_output_____"
]
],
[
[
"#@title Video 2: Gone Fishin'\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id='McALsTzb494', width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"You were just introduced to the **binary hidden state problem** we are going to explore. You need to decide which side to fish on. We know fish like to school together. On different days the school of fish is either on the left or right side, but we don’t know what the case is today. We will represent our knowledge probabilistically, asking how to make a decision (where to decide the fish are or where to fish) and what to expect in terms of gains or losses. In the next two sections we will consider just the probability of where the fish might be and what you gain or lose by choosing where to fish.\n\nRemember, you can either think of your self as a scientist conducting an experiment or as a brain trying to make a decision. The Bayesian approach is the same!\n",
"_____no_output_____"
],
[
"---\n# Section 2: Deciding where to fish \n\n",
"_____no_output_____"
]
],
[
[
"#@title Video 3: Utility\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id='xvIVZrqF_5s', width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"You know the probability that the school of fish is on the left side of the dock today, $P(s = left)$. You also know the probability that it is on the right, $P(s = right)$, because these two probabilities must add up to 1. You need to decide where to fish. It may seem obvious - you could just fish on the side where the probability of the fish being is higher! Unfortunately, decisions and actions are always a little more complicated. Deciding to fish may be influenced by more than just the probability of the school of fish being there as we saw by the potential issues of submarines and sunburn. \n\nWe quantify these factors numerically using **utility**, which describes the consequences of your actions: how much value you gain (or if negative, lose) given the state of the world ($s$) and the action you take ($a$). In our example, our utility can be summarized as:\n\n| Utility: U(s,a) | a = left | a = right |\n| ----------------- |----------|----------|\n| s = Left | 2 | -3 |\n| s = right | -2 | 1 |\n\nTo use utility to choose an action, we calculate the **expected utility** of that action by weighing these utilities with the probability of that state occuring. This allows us to choose actions by taking probabilities of events into account: we don't care if the outcome of an action-state pair is a loss if the probability of that state is very low. We can formalize this as:\n\n$$\\text{Expected utility of action a} = \\sum_{s}U(s,a)P(s) $$\n\nIn other words, the expected utility of an action a is the sum over possible states of the utility of that action and state times the probability of that state.\n",
"_____no_output_____"
],
[
"## Interactive Demo 2: Exploring the decision\n\nLet's start to get a sense of how all this works. \n\nTake a look at the interactive demo below. You can change the probability that the school of fish is on the left side ($p(s = left)$ using the slider. You will see the utility matrix and the corresponding expected utility of each action.\n\nFirst, make sure you understand how the expected utility of each action is being computed from the probabilities and the utility values. In the initial state: the probability of the fish being on the left is 0.9 and on the right is 0.1. The expected utility of the action of fishing on the left is then $U(s = left,a = left)p(s = left) + U(s = right,a = left)p(s = right) = 2(0.9) + -2(0.1) = 1.6$.\n\nFor each of these scenarios, think and discuss first. Then use the demo to try out each and see if your action would have been correct (that is, if the expected value of that action is the highest).\n\n\n1. You just arrived at the dock for the first time and have no sense of where the fish might be. So you guess that the probability of the school being on the left side is 0.5 (so the probability on the right side is also 0.5). Which side would you choose to fish on given our utility values?\n2. You think that the probability of the school being on the left side is very low (0.1) and correspondingly high on the right side (0.9). Which side would you choose to fish on given our utility values?\n3. What would you choose if the probability of the school being on the left side is slightly lower than on the right side (0. 4 vs 0.6)?",
"_____no_output_____"
]
],
[
[
"# @markdown Execute this cell to use the widget\nps_widget = widgets.FloatSlider(0.9, description='p(s = left)', min=0.0, max=1.0, step=0.01)\n\n@widgets.interact(\n ps = ps_widget,\n)\ndef make_utility_plot(ps):\n fig = plot_utility(ps)\n plt.show(fig)\n plt.close(fig)\n return None",
"_____no_output_____"
],
[
"# to_remove explanation\n\n# 1) With equal probabilities, the expected utility is higher on the left side,\n# since that is the side without submarines, so you would choose to fish there.\n\n# 2) If the probability that the fish is on the right side is high, you would\n# choose to fish there. The high probability of fish being on the right far outweights\n# the slightly higher utilities from fishing on the left (as you are unlikely to gain these)\n\n# 3) If the probability that the fish is on the right side is just slightly higher\n#. than on the left, you would choose the left side as the expected utility is still\n#. higher on the left. Note that in this situation, you are not simply choosing the\n#. side with the higher probability - the utility really matters here for the decision",
"_____no_output_____"
]
],
[
[
"In this section, you have seen that both the utility of various state and action pairs and our knowledge of the probability of each state affects your decision. Importantly, we want our knowledge of the probability of each state to be as accurate as possible! \n\nSo how do we know these probabilities? We may have prior knowledge from years of fishing at the same dock. Over those years, we may have learned that the fish are more likely to be on the left side for example. We want to make sure this knowledge is as accurate as possible though. To do this, we want to collect more data, or take some more measurements! For the next few sections, we will focus on making our knowledge of the probability as accurate as possible, before coming back to using utility to make decisions.",
"_____no_output_____"
],
[
"---\n# Section 3: Likelihood of the fish being on either side\n \n",
"_____no_output_____"
]
],
[
[
"#@title Video 4: Likelihood\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id='l4m0JzMWGio', width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"First, we'll think about what it means to take a measurement (also often called an observation or just data) and what it tells you about what the hidden state may be. Specifically, we'll be looking at the **likelihood**, which is the probability of your measurement ($m$) given the hidden state ($s$): $P(m | s)$. Remember that in this case, the hidden state is which side of the dock the school of fish is on.\n\nWe will watch someone fish (for let's say 10 minutes) and our measurement is whether they catch a fish or not. We know something about what catching a fish means for the likelihood of the fish being on one side or the other. ",
"_____no_output_____"
],
[
"## Think! 3: Guessing the location of the fish\n\nLet's say we go to different dock from the one in the video. Here, there are different probabilities of catching fish given the state of the world. In this case, if they fish on the side of the dock where the fish are, they have a 70% chance of catching a fish. Otherwise, they catch a fish with only 20% probability. \n\nThe fisherperson is fishing on the left side. \n\n1) Figure out each of the following:\n- probability of catching a fish given that the school of fish is on the left side, $P(m = catch\\text{ } fish | s = left )$\n- probability of not catching a fish given that the school of fish is on the left side, $P(m = no \\text{ } fish | s = left)$\n- probability of catching a fish given that the school of fish is on the right side, $P(m = catch \\text{ } fish | s = right)$\n- probability of not catching a fish given that the school of fish is on the right side, $P(m = no \\text{ } fish | s = right)$\n\n2) If the fisherperson catches a fish, which side would you guess the school is on? Why?\n\n3) If the fisherperson does not catch a fish, which side would you guess the school is on? Why?\n",
"_____no_output_____"
]
],
[
[
"#to_remove explanation\n\n# 1) The fisherperson is on the left side so:\n# - P(m = catch fish | s = left) = 0.7 because they have a 70% chance of catching\n# a fish when on the same side as the school\n# - P(m = no fish | s = left) = 0.3 because the probability of catching a fish\n# and not catching a fish for a given state must add up to 1 as these\n# are the only options: 1 - 0.7 = 0.3\n# - P(m = catch fish | s = right) = 0.2\n# - P(m = no fish | s = right) = 0.8\n\n# 2) If the fisherperson catches a fish, you would guess the school of fish is on the\n# left side. This is because the probability of catching a fish given that the\n# school is on the left side (0.7) is higher than the probability given that\n# the school is on the right side (0.2).\n\n# 3) If the fisherperson does not catch a fish, you would guess the school of fish is on the\n# right side. This is because the probability of not catching a fish given that the\n# school is on the right side (0.8) is higher than the probability given that\n# the school is on the right side (0.3).",
"_____no_output_____"
]
],
[
[
"In the prior exercise, you guessed where the school of fish was based on the measurement you took (watching someone fish). You did this by choosing the state (side of school) that maximized the probability of the measurement. In other words, you estimated the state by maximizing the likelihood (had the highest probability of measurement given state $P(m|s$)). This is called maximum likelihood estimation (MLE) and you've encountered it before during this course, in W1D3!\n\nWhat if you had been going to this river for years and you knew that the fish were almost always on the left side? This would probably affect how you make your estimate - you would rely less on the single new measurement and more on your prior knowledge. This is the idea behind Bayesian inference, as we will see later in this tutorial!",
"_____no_output_____"
],
[
"---\n# Section 4: Correlation and marginalization\n",
"_____no_output_____"
]
],
[
[
"#@title Video 5: Correlation and marginalization\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id='vsDjtWi-BVo', width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"In this section, we are going to take a step back for a bit and think more generally about the amount of information shared between two random variables. We want to know how much information you gain when you observe one variable (take a measurement) if you know something about another. We will see that the fundamental concept is the same if we think about two attributes, for example the size and color of the fish, or the prior information and the likelihood.",
"_____no_output_____"
],
[
"## Math Exercise 4: Computing marginal likelihoods\n\nTo understand the information between two variables, let's first consider the size and color of the fish.\n\n| P(X, Y) | Y = silver | Y = gold |\n| ----------------- |----------|----------|\n| X = small | 0.4 | 0.2 |\n| X = large | 0.1 | 0.3 |\n\nThe table above shows us the **joint probabilities**: the probability of both specific attributes occuring together. For example, the probability of a fish being small and silver ($P(X = small, Y = silver$) is 0.4.\n\nWe want to know what the probability of a fish being small regardless of color. Since the fish are either silver or gold, this would be the probability of a fish being small and silver plus the probability of a fish being small and gold. This is an example of marginalizing, or averaging out, the variable we are not interested in across the rows or columns.. In math speak: $P(X = small) = \\sum_y{P(X = small, Y)}$. This gives us a **marginal probability**, a probability of a variable outcome (in this case size), regardless of the other variables (in this case color).\n\nPlease complete the following math problems to further practice thinking through probabilities:\n\n1. Calculate the probability of a fish being silver.\n2. Calculate the probability of a fish being small, large, silver, or gold.\n3. Calculate the probability of a fish being small OR gold. (Hint: $P(A\\ \\textrm{or}\\ B) = P(A) + P(B) - P(A\\ \\textrm{and}\\ B)$)\n\n\n",
"_____no_output_____"
]
],
[
[
"# to_remove explanation\n\n# 1) The probability of a fish being silver is the joint probability of it being\n#. small and silver plus the joint probability of it being large and silver:\n#\n#. P(Y = silver) = P(X = small, Y = silver) + P(X = large, Y = silver)\n#. = 0.4 + 0.1\n#. = 0.5\n\n\n# 2) This is all the possibilities as in this scenario, our fish can only be small\n#. or large, silver or gold. So the probability is 1 - the fish has to be at\n#. least one of these.\n\n\n#. 3) First we compute the marginal probabilities\n#. P(X = small) = P(X = small, Y = silver) + P(X = small, Y = gold) = 0.6\n#. P(Y = gold) = P(X = small, Y = gold) + P(X = large, Y = gold) = 0.5\n#. We already know the joint probability: P(X = small, Y = gold) = 0.2\n#. We can now use the given formula:\n#. P( X = small or Y = gold) = P(X = small) + P(Y = gold) - P(X = small, Y = gold)\n#. = 0.6 + 0.5 - 0.2\n#. = 0.9",
"_____no_output_____"
]
],
[
[
"## Think! 4: Covarying probability distributions\n\nThe relationship between the marginal probabilities and the joint probabilities is determined by the correlation between the two random variables - a normalized measure of how much the variables covary. We can also think of this as gaining some information about one of the variables when we observe a measurement from the other. We will think about this more formally in Tutorial 2. \n\nHere, we want to think about how the correlation between size and color of these fish changes how much information we gain about one attribute based on the other. See Bonus Section 1 for the formula for correlation.\n\nUse the widget below and answer the following questions:\n\n1. When the correlation is zero, $\\rho = 0$, what does the distribution of size tell you about color?\n2. Set $\\rho$ to something small. As you change the probability of golden fish, what happens to the ratio of size probabilities? Set $\\rho$ larger (can be negative). Can you explain the pattern of changes in the probabilities of size as you change the probability of golden fish?\n3. Set the probability of golden fish and of large fish to around 65%. As the correlation goes towards 1, how often will you see silver large fish?\n4. What is increasing the (absolute) correlation telling you about how likely you are to see one of the properties if you see a fish with the other?\n",
"_____no_output_____"
]
],
[
[
"# @markdown Execute this cell to enable the widget\nstyle = {'description_width': 'initial'}\ngs = GridspecLayout(2,2)\n\ncor_widget = widgets.FloatSlider(0.0, description='ρ', min=-1, max=1, step=0.01)\npx_widget = widgets.FloatSlider(0.5, description='p(color=golden)', min=0.01, max=0.99, step=0.01, style=style)\npy_widget = widgets.FloatSlider(0.5, description='p(size=large)', min=0.01, max=0.99, step=0.01, style=style)\ngs[0,0] = cor_widget\ngs[0,1] = px_widget\ngs[1,0] = py_widget\n\n\n@widgets.interact(\n px=px_widget,\n py=py_widget,\n cor=cor_widget,\n)\ndef make_corr_plot(px, py, cor):\n Cmin, Cmax = compute_cor_range(px, py) #allow correlation values\n cor_widget.min, cor_widget.max = Cmin+0.01, Cmax-0.01\n if cor_widget.value > Cmax:\n cor_widget.value = Cmax\n if cor_widget.value < Cmin:\n cor_widget.value = Cmin\n cor = cor_widget.value\n P = compute_marginal(px,py,cor)\n # print(P)\n fig = plot_joint_probs(P)\n plt.show(fig)\n plt.close(fig)\n return None\n\n# gs[1,1] = make_corr_plot()",
"_____no_output_____"
],
[
"# to_remove explanation\n\n#' 1. When the correlation is zero, the two properties are completely independent.\n#' This means you don't gain any information about one variable from observing the other.\n#' Importantly, the marginal distribution of one variable is therefore independent of the other.\n\n#' 2. The correlation controls the distribution of probability across the joint probabilty table.\n#' The higher the correlation, the more the probabilities are restricted by the fact that both rows\n#' and columns need to sum to one! While the marginal probabilities show the relative weighting, the\n#' absolute probabilities for one quality will become more dependent on the other as the correlation\n#' goes to 1 or -1.\n\n#' 3. The correlation will control how much probabilty mass is located on the diagonals. As the\n#' correlation goes to 1 (or -1), the probability of seeing the one of the two pairings has to goes\n#' towards zero!\n\n#' 4. If we think about what information we gain by observing one quality, the intution from (3.) tells\n#' us that we know more (have more information) about the other quality as a function of the correlation.",
"_____no_output_____"
]
],
[
[
"We have just seen how two random variables can be more or less independent. The more correlated, the less independent, and the more shared information. We also learned that we can marginalize to determine the marginal likelihood of a hidden state or to find the marginal probability distribution of two random variables. We are going to now complete our journey towards being fully Bayesian!",
"_____no_output_____"
],
[
"---\n# Section 5: Bayes' Rule and the Posterior",
"_____no_output_____"
],
[
"Marginalization is going to be used to combine our prior knowlege, which we call the **prior**, and our new information from a measurement, the **likelihood**. Only in this case, the information we gain about the hidden state we are interested in, where the fish are, is based on the relationship between the probabilities of the measurement and our prior. \n\nWe can now calculate the full posterior distribution for the hidden state ($s$) using Bayes' Rule. As we've seen, the posterior is proportional the the prior times the likelihood. This means that the posterior probability of the hidden state ($s$) given a measurement ($m$) is proportional to the likelihood of the measurement given the state times the prior probability of that state (the marginal likelihood):\n\n$$ P(s | m) \\propto P(m | s) P(s) $$\n\nWe say proportional to instead of equal because we need to normalize to produce a full probability distribution:\n\n$$ P(s | m) = \\frac{P(m | s) P(s)}{P(m)} $$\n\nNormalizing by this $P(m)$ means that our posterior is a complete probability distribution that sums or integrates to 1 appropriately. We now can use this new, complete probability distribution for any future inference or decisions we like! In fact, as we will see tomorrow, we can use it as a new prior! Finally, we often call this probability distribution our beliefs over the hidden states, to emphasize that it is our subjective knowlege about the hidden state.\n\nFor many complicated cases, like those we might be using to model behavioral or brain inferences, the normalization term can be intractable or extremely complex to calculate. We can be careful to choose probability distributions were we can analytically calculate the posterior probability or numerical approximation is reliable. Better yet, we sometimes don't need to bother with this normalization! The normalization term, $P(m)$, is the probability of the measurement. This does not depend on state so is essentially a constant we can often ignore. We can compare the unnormalized posterior distribution values for different states because how they relate to each other is unchanged when divided by the same constant. We will see how to do this to compare evidence for different hypotheses tomorrow. (It's also used to compare the likelihood of models fit using maximum likelihood estimation, as you did in W1D5.)\n\nIn this relatively simple example, we can compute the marginal probability $P(m)$ easily by using:\n$$P(m) = \\sum_s P(m | s) P(s)$$\nWe can then normalize so that we deal with the full posterior distribution.\n",
"_____no_output_____"
],
[
"## Math Exercise 5: Calculating a posterior probability\n\nOur prior is $p(s = left) = 0.3$ and $p(s = right) = 0.7$. In the video, we learned that the chance of catching a fish given they fish on the same side as the school was 50%. Otherwise, it was 10%. We observe a person fishing on the left side. Our likelihood is: \n\n\n| Likelihood: p(m \\| s) | m = catch fish | m = no fish |\n| ----------------- |----------|----------|\n| s = left | 0.5 | 0.5 |\n| s = right | 0.1 | 0.9 |\n\n\nCalculate the posterior probability (on paper) that:\n\n1. The school is on the left if the fisherperson catches a fish: $p(s = left | m = catch fish)$ (hint: normalize by compute $p(m = catch fish)$)\n2. The school is on the right if the fisherperson does not catch a fish: $p(s = right | m = no fish)$",
"_____no_output_____"
]
],
[
[
"# to_remove explanation\n\n# 1. Using Bayes rule, we know that P(s = left | m = catch fish) = P(m = catch fish | s = left)P(s = left) / P(m = catch fish)\n#. Let's first compute P(m = catch fish):\n#. P(m = catch fish) = P(m = catch fish | s = left)P(s = left) + P(m = catch fish | s = right)P(s = right)\n# = 0.5 * 0.3 + .1*.7\n# = 0.22\n#. Now we can plug in all parts of Bayes rule:\n# P(s = left | m = catch fish) = P(m = catch fish | s = left)P(s = left) / P(m = catch fish)\n# = 0.5*0.3/0.22\n# = 0.68\n\n# 2. Using Bayes rule, we know that P(s = right | m = no fish) = P(m = no fish | s = right)P(s = right) / P(m = no fish)\n#. Let's first compute P(m = no fish):\n#. P(m = no fish) = P(m = no fish | s = left)P(s = left) + P(m = no fish | s = right)P(s = right)\n# = 0.5 * 0.3 + .9*.7\n# = 0.78\n#. Now we can plug in all parts of Bayes rule:\n# P(s = right | m = no fish) = P(m = no fish | s = right)P(s = right) / P(m = no fish)\n# = 0.9*0.7/0.78\n# = 0.81",
"_____no_output_____"
]
],
[
[
"## Coding Exercise 5: Computing Posteriors\n\nLet's implement our above math to be able to compute posteriors for different priors and likelihood.s\n\nAs before, our prior is $p(s = left) = 0.3$ and $p(s = right) = 0.7$. In the video, we learned that the chance of catching a fish given they fish on the same side as the school was 50%. Otherwise, it was 10%. We observe a person fishing on the left side. Our likelihood is: \n\n\n| Likelihood: p(m \\| s) | m = catch fish | m = no fish |\n| ----------------- |----------|----------|\n| s = left | 0.5 | 0.5 |\n| s = right | 0.1 | 0.9 |\n\n\nWe want our full posterior to take the same 2 by 2 form. Make sure the outputs match your math answers!\n\n",
"_____no_output_____"
]
],
[
[
"def compute_posterior(likelihood, prior):\n \"\"\" Use Bayes' Rule to compute posterior from likelihood and prior\n\n Args:\n likelihood (ndarray): i x j array with likelihood probabilities where i is\n number of state options, j is number of measurement options\n prior (ndarray): i x 1 array with prior probability of each state\n\n Returns:\n ndarray: i x j array with posterior probabilities where i is\n number of state options, j is number of measurement options\n\n \"\"\"\n\n #################################################\n ## TODO for students ##\n # Fill out function and remove\n raise NotImplementedError(\"Student exercise: implement compute_posterior\")\n #################################################\n\n # Compute unnormalized posterior (likelihood times prior)\n posterior = ... # first row is s = left, second row is s = right\n\n # Compute p(m)\n p_m = np.sum(posterior, axis = 0)\n\n # Normalize posterior (divide elements by p_m)\n posterior /= ...\n\n return posterior\n\n# Make prior\nprior = np.array([0.3, 0.7]).reshape((2, 1)) # first row is s = left, second row is s = right\n\n# Make likelihood\nlikelihood = np.array([[0.5, 0.5], [0.1, 0.9]]) # first row is s = left, second row is s = right\n\n# Compute posterior\nposterior = compute_posterior(likelihood, prior)\n\n# Visualize\nwith plt.xkcd():\n plot_prior_likelihood_posterior(prior, likelihood, posterior)",
"_____no_output_____"
],
[
"# to_remove solution\n\ndef compute_posterior(likelihood, prior):\n \"\"\" Use Bayes' Rule to compute posterior from likelihood and prior\n\n Args:\n likelihood (ndarray): i x j array with likelihood probabilities where i is\n number of state options, j is number of measurement options\n prior (ndarray): i x 1 array with prior probability of each state\n\n Returns:\n ndarray: i x j array with posterior probabilities where i is\n number of state options, j is number of measurement options\n\n \"\"\"\n\n # Compute unnormalized posterior (likelihood times prior)\n posterior = likelihood * prior # first row is s = left, second row is s = right\n\n # Compute p(m)\n p_m = np.sum(posterior, axis = 0)\n\n # Normalize posterior (divide elements by p_m)\n posterior /= p_m\n\n return posterior\n\n# Make prior\nprior = np.array([0.3, 0.7]).reshape((2, 1)) # first row is s = left, second row is s = right\n\n# Make likelihood\nlikelihood = np.array([[0.5, 0.5], [0.1, 0.9]]) # first row is s = left, second row is s = right\n\n# Compute posterior\nposterior = compute_posterior(likelihood, prior)\n\n# Visualize\nwith plt.xkcd():\n plot_prior_likelihood_posterior(prior, likelihood, posterior)",
"_____no_output_____"
]
],
[
[
"## Interactive Demo 5: What affects the posterior?\n\nNow that we can understand the implementation of *Bayes rule*, let's vary the parameters of the prior and likelihood to see how changing the prior and likelihood affect the posterior. \n\nIn the demo below, you can change the prior by playing with the slider for $p( s = left)$. You can also change the likelihood by changing the probability of catching a fish given that the school is on the left and the probability of catching a fish given that the school is on the right. The fisherperson you are observing is fishing on the left.\n \n\n1. Keeping the likelihood constant, when does the prior have the strongest influence over the posterior? Meaning, when does the posterior look most like the prior no matter whether a fish was caught or not?\n2. Keeping the likelihood constant, when does the prior exert the weakest influence? Meaning, when does the posterior look least like the prior and depend most on whether a fish was caught or not?\n3. Set the prior probability of the state = left to 0.6 and play with the likelihood. When does the likelihood exert the most influence over the posterior?",
"_____no_output_____"
]
],
[
[
"# @markdown Execute this cell to enable the widget\nstyle = {'description_width': 'initial'}\nps_widget = widgets.FloatSlider(0.3, description='p(s = left)',\n min=0.01, max=0.99, step=0.01)\np_a_s1_widget = widgets.FloatSlider(0.5, description='p(fish | s = left)',\n min=0.01, max=0.99, step=0.01, style=style)\np_a_s0_widget = widgets.FloatSlider(0.1, description='p(fish | s = right)',\n min=0.01, max=0.99, step=0.01, style=style)\nobserved_widget = widgets.Checkbox(value=False, description='Observed fish (m)',\n disabled=False, indent=False, layout={'width': 'max-content'})\n\n@widgets.interact(\n ps=ps_widget,\n p_a_s1=p_a_s1_widget,\n p_a_s0=p_a_s0_widget,\n m_right=observed_widget\n)\ndef make_prior_likelihood_plot(ps,p_a_s1,p_a_s0,m_right):\n fig = plot_prior_likelihood(ps,p_a_s1,p_a_s0,m_right)\n plt.show(fig)\n plt.close(fig)\n return None",
"_____no_output_____"
],
[
"# to_remove explanation\n\n# 1). The prior exerts a strong influence over the posterior when it is very informative: when\n#. the probability of the school being on one side or the other. If the prior that the fish are\n#. on the left side is very high (like 0.9), the posterior probability of the state being left is\n#. high regardless of the measurement.\n\n# 2). The prior does not exert a strong influence when it is not informative: when the probabilities\n#. of the school being on the left vs right are similar (both are 0.5 for example). In this case,\n#. the posterior is more driven by the collected data (the measurement) and more closely resembles\n#. the likelihood.\n\n\n#. 3) Similarly to the prior, the likelihood exerts the most influence when it is informative: when catching\n#. a fish tells you a lot of information about which state is likely. For example, if the probability of the\n#. fisherperson catching a fish if he is fishing on the right side and the school is on the left is 0\n#. (p fish | s = left) = 0 and the probability of catching a fish if the school is on the right is 1, the\n#. prior does not affect the posterior at all. The measurement tells you the hidden state completely.",
"_____no_output_____"
]
],
[
[
"# Section 6: Making Bayesian fishing decisions\n\nWe will explore how to consider the expected utility of an action based on our belief (the posterior distribution) about where we think the fish are. Now we have all the components of a Bayesian decision: our prior information, the likelihood given a measurement, the posterior distribution (belief) and our utility (the gains and losses). This allows us to consider the relationship between the true value of the hidden state, $s$, and what we *expect* to get if we take action, $a$, based on our belief!\n\nLet's use the following widget to think about the relationship between these probability distributions and utility function.",
"_____no_output_____"
],
[
"## Think! 6: What is more important, the probabilities or the utilities?\n\nWe are now going to put everything we've learned together to gain some intuitions for how each of the elements that goes into a Bayesian decision comes together. Remember, the common assumption in neuroscience, psychology, economics, ecology, etc. is that we (humans and animals) are tying to maximize our expected utility.\n\n1. Can you find a situation where the expected utility is the same for both actions?\n2. What is more important for determining the expected utility: the prior or a new measurement (the likelihood)?\n3. Why is this a normative model?\n4. Can you think of ways in which this model would need to be extended to describe human or animal behavior?",
"_____no_output_____"
]
],
[
[
"# @markdown Execute this cell to enable the widget\nstyle = {'description_width': 'initial'}\nps_widget = widgets.FloatSlider(0.3, description='p(s)',\n min=0.01, max=0.99, step=0.01)\np_a_s1_widget = widgets.FloatSlider(0.5, description='p(fish | s = left)',\n min=0.01, max=0.99, step=0.01, style=style)\np_a_s0_widget = widgets.FloatSlider(0.1, description='p(fish | s = right)',\n min=0.01, max=0.99, step=0.01, style=style)\nobserved_widget = widgets.Checkbox(value=False, description='Observed fish (m)',\n disabled=False, indent=False, layout={'width': 'max-content'})\n\n@widgets.interact(\n ps=ps_widget,\n p_a_s1=p_a_s1_widget,\n p_a_s0=p_a_s0_widget,\n m_right=observed_widget\n)\ndef make_prior_likelihood_utility_plot(ps, p_a_s1, p_a_s0,m_right):\n fig = plot_prior_likelihood_utility(ps, p_a_s1, p_a_s0,m_right)\n plt.show(fig)\n plt.close(fig)\n return None",
"_____no_output_____"
],
[
"# to_remove explanation\n\n#' 1. There are actually many (infinite) combinations that can produce the same\n#. expected utility for both actions: but the posterior probabilities will always\n# have to balance out the differences in the utility function. So, what is\n# important is that for a given utility function, there will be some 'point\n# of indifference'\n\n#' 2. What matters is the relative information: if the prior is close to 50/50,\n# then the likelihood has more infuence, if the likelihood is 50/50 given a\n# measurement (the measurement is uninformative), the prior is more important.\n# But the critical insite from Bayes Rule and the Bayesian approach is that what\n# matters is the relative information you gain from a measurement, and that\n# you can use all of this information for your decision.\n\n#' 3. The model gives us a very precise way to think about how we *should* combine\n# information and how we *should* act, GIVEN some assumption about our goals.\n# In this case, if we assume we are trying to maximize expected utility--we can\n# state what an animal or subject should do.\n\n#' 4. There are lots of possible extensions. Humans may not always try to maximize\n# utility; humans and animals might not be able to calculate or represent probabiltiy\n# distributions exactly; The utility function might be more complicated; etc.",
"_____no_output_____"
]
],
[
[
"---\n# Summary\n\nIn this tutorial, you learned about combining prior information with new measurements to update your knowledge using Bayes Rulem, in the context of a fishing problem. \n\nSpecifically, we covered:\n\n* That the likelihood is the probability of the measurement given some hidden state\n\n* That how the prior and likelihood interact to create the posterior, the probability of the hidden state given a measurement, depends on how they covary\n\n* That utility is the gain from each action and state pair, and the expected utility for an action is the sum of the utility for all state pairs, weighted by the probability of that state happening. You can then choose the action with highest expected utility.\n",
"_____no_output_____"
],
[
"---\n# Bonus",
"_____no_output_____"
],
[
"## Bonus Section 1: Correlation Formula\nTo understand the way we calculate the correlation, we need to review the definition of covariance and correlation.\n\nCovariance:\n\n$$\ncov(X,Y) = \\sigma_{XY} = E[(X - \\mu_{x})(Y - \\mu_{y})] = E[X]E[Y] - \\mu_{x}\\mu_{y}\n$$\n\nCorrelation:\n\n$$\n\\rho_{XY} = \\frac{cov(Y,Y)}{\\sqrt{V(X)V(Y)}} = \\frac{\\sigma_{XY}}{\\sigma_{X}\\sigma_{Y}}\n$$",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d03cad60d7e1ac44bca5747b29eb15869099357a | 122,659 | ipynb | Jupyter Notebook | Surrey/sourcecode/20210204_Colab_D_Surrey_ASL_MobileNet.ipynb | leonlha/e2e-3m | 6f3f8a7aee8e74388e7e09d3fdf0df6bbf72526c | [
"MIT"
] | 8 | 2020-08-05T16:19:31.000Z | 2021-06-23T05:56:12.000Z | Surrey/sourcecode/20210204_Colab_D_Surrey_ASL_MobileNet.ipynb | leonlha/e2e-3m | 6f3f8a7aee8e74388e7e09d3fdf0df6bbf72526c | [
"MIT"
] | null | null | null | Surrey/sourcecode/20210204_Colab_D_Surrey_ASL_MobileNet.ipynb | leonlha/e2e-3m | 6f3f8a7aee8e74388e7e09d3fdf0df6bbf72526c | [
"MIT"
] | 3 | 2020-08-05T22:36:28.000Z | 2021-06-02T09:50:49.000Z | 46.549905 | 253 | 0.492569 | [
[
[
"ls -l| tail -10",
"total 4\ndrwxr-xr-x 1 root root 4096 Feb 4 15:26 \u001b[0m\u001b[01;34msample_data\u001b[0m/\n"
],
[
"#G4\nfrom google.colab import drive\ndrive.mount('/content/gdrive')",
"Mounted at /content/gdrive\n"
],
[
"cp gdrive/My\\ Drive/fingerspelling5.tar.bz2 fingerspelling5.tar.bz2",
"_____no_output_____"
],
[
"# rm -r surrey/\n%rm -r dataset5/",
"_____no_output_____"
],
[
"# rm fingerspelling5.tar.bz2",
"_____no_output_____"
],
[
"# cd /media/datastorage/Phong/",
"_____no_output_____"
],
[
"!tar xjf fingerspelling5.tar.bz2",
"_____no_output_____"
],
[
"cd dataset5",
"/content/dataset5\n"
],
[
"mkdir surrey",
"_____no_output_____"
],
[
"mkdir surrey/D",
"_____no_output_____"
],
[
"mv dataset5/* surrey/D/",
"_____no_output_____"
],
[
"cd surrey",
"/content/surrey\n"
],
[
"cd ..",
"/content\n"
],
[
"#remove depth files\nimport glob\nimport os\nimport shutil\n\n# get parts of image's path\ndef get_image_parts(image_path):\n \"\"\"Given a full path to an image, return its parts.\"\"\"\n parts = image_path.split(os.path.sep)\n #print(parts)\n filename = parts[2]\n filename_no_ext = filename.split('.')[0]\n classname = parts[1]\n train_or_test = parts[0]\n\n return train_or_test, classname, filename_no_ext, filename\n\n\n#del_folders = ['A','B','C','D','E'] \nmove_folders_1 = ['A','B','C','E'] \nmove_folders_2 = ['D']\n\n# look for all images in sub-folders\nfor folder in move_folders_1:\n class_folders = glob.glob(os.path.join(folder, '*'))\n \n for iid_class in class_folders:\n #move depth files\n class_files = glob.glob(os.path.join(iid_class, 'depth*.png'))\n \n print('copying %d files' %(len(class_files)))\n for idx in range(len(class_files)): \n src = class_files[idx]\n if \"0001\" not in src: \n train_or_test, classname, _, filename = get_image_parts(src)\n\n dst = os.path.join('train_depth', classname, train_or_test+'_'+ filename)\n\n # image directory\n img_directory = os.path.join('train_depth', classname)\n\n # create folder if not existed\n if not os.path.exists(img_directory):\n os.makedirs(img_directory)\n\n #copying\n shutil.copy(src, dst)\n else:\n print('ignor: %s' %src)\n \n #move color files \n for iid_class in class_folders:\n #move depth files\n class_files = glob.glob(os.path.join(iid_class, 'color*.png'))\n \n print('copying %d files' %(len(class_files)))\n for idx in range(len(class_files)): \n src = class_files[idx]\n train_or_test, classname, _, filename = get_image_parts(src)\n\n dst = os.path.join('train_color', classname, train_or_test+'_'+ filename)\n \n # image directory\n img_directory = os.path.join('train_color', classname)\n\n # create folder if not existed\n if not os.path.exists(img_directory):\n os.makedirs(img_directory)\n \n #copying\n shutil.copy(src, dst)\n \n# look for all images in sub-folders\nfor folder in move_folders_2:\n class_folders = glob.glob(os.path.join(folder, '*'))\n \n for iid_class in class_folders:\n #move depth files\n class_files = glob.glob(os.path.join(iid_class, 'depth*.png'))\n \n print('copying %d files' %(len(class_files)))\n for idx in range(len(class_files)): \n src = class_files[idx]\n if \"0001\" not in src: \n train_or_test, classname, _, filename = get_image_parts(src)\n\n dst = os.path.join('test_depth', classname, train_or_test+'_'+ filename)\n\n # image directory\n img_directory = os.path.join('test_depth', classname)\n\n # create folder if not existed\n if not os.path.exists(img_directory):\n os.makedirs(img_directory)\n\n #copying\n shutil.copy(src, dst)\n else:\n print('ignor: %s' %src)\n \n #move color files \n for iid_class in class_folders:\n #move depth files\n class_files = glob.glob(os.path.join(iid_class, 'color*.png'))\n \n print('copying %d files' %(len(class_files)))\n for idx in range(len(class_files)): \n src = class_files[idx]\n train_or_test, classname, _, filename = get_image_parts(src)\n\n dst = os.path.join('test_color', classname, train_or_test+'_'+ filename)\n \n # image directory\n img_directory = os.path.join('test_color', classname)\n\n # create folder if not existed\n if not os.path.exists(img_directory):\n os.makedirs(img_directory)\n \n #copying\n shutil.copy(src, dst) ",
"copying 516 files\nignor: A/b/depth_1_0001.png\ncopying 524 files\nignor: A/e/depth_4_0001.png\ncopying 536 files\nignor: A/d/depth_3_0001.png\ncopying 522 files\nignor: A/x/depth_23_0001.png\ncopying 518 files\nignor: A/k/depth_10_0001.png\ncopying 572 files\nignor: A/p/depth_15_0001.png\ncopying 512 files\nignor: A/o/depth_14_0001.png\ncopying 516 files\nignor: A/l/depth_11_0001.png\ncopying 528 files\nignor: A/r/depth_17_0001.png\ncopying 525 files\nignor: A/w/depth_22_0001.png\ncopying 524 files\nignor: A/t/depth_19_0001.png\ncopying 528 files\nignor: A/a/depth_0_0001.png\ncopying 507 files\nignor: A/v/depth_21_0001.png\ncopying 524 files\nignor: A/u/depth_20_0001.png\ncopying 519 files\nignor: A/f/depth_5_0001.png\ncopying 471 files\ncopying 516 files\nignor: A/q/depth_16_0001.png\ncopying 515 files\nignor: A/i/depth_8_0001.png\ncopying 557 files\nignor: A/c/depth_2_0001.png\ncopying 530 files\nignor: A/n/depth_13_0001.png\ncopying 523 files\nignor: A/h/depth_7_0001.png\ncopying 533 files\nignor: A/y/depth_24_0001.png\ncopying 527 files\nignor: A/m/depth_12_0001.png\ncopying 528 files\nignor: A/g/depth_6_0001.png\ncopying 515 files\ncopying 523 files\ncopying 535 files\ncopying 521 files\ncopying 517 files\ncopying 571 files\ncopying 511 files\ncopying 515 files\ncopying 527 files\ncopying 524 files\ncopying 523 files\ncopying 527 files\ncopying 506 files\ncopying 523 files\ncopying 518 files\ncopying 470 files\ncopying 515 files\ncopying 514 files\ncopying 556 files\ncopying 529 files\ncopying 522 files\ncopying 532 files\ncopying 526 files\ncopying 527 files\ncopying 577 files\nignor: B/b/depth_1_0001.png\ncopying 565 files\nignor: B/e/depth_4_0001.png\ncopying 552 files\nignor: B/d/depth_3_0001.png\ncopying 622 files\nignor: B/x/depth_23_0001.png\ncopying 777 files\nignor: B/k/depth_10_0001.png\ncopying 642 files\nignor: B/p/depth_15_0001.png\ncopying 529 files\nignor: B/o/depth_14_0001.png\ncopying 577 files\nignor: B/l/depth_11_0001.png\ncopying 550 files\nignor: B/r/depth_17_0001.png\ncopying 648 files\nignor: B/w/depth_22_0001.png\ncopying 514 files\nignor: B/t/depth_19_0001.png\ncopying 536 files\nignor: B/a/depth_0_0001.png\ncopying 628 files\nignor: B/v/depth_21_0001.png\ncopying 542 files\nignor: B/u/depth_20_0001.png\ncopying 530 files\nignor: B/f/depth_5_0001.png\ncopying 749 files\nignor: B/s/depth_18_0001.png\ncopying 540 files\nignor: B/q/depth_16_0001.png\ncopying 543 files\nignor: B/i/depth_8_0001.png\ncopying 549 files\nignor: B/c/depth_2_0001.png\ncopying 544 files\nignor: B/n/depth_13_0001.png\ncopying 545 files\nignor: B/h/depth_7_0001.png\ncopying 537 files\nignor: B/y/depth_24_0001.png\ncopying 582 files\nignor: B/m/depth_12_0001.png\ncopying 544 files\nignor: B/g/depth_6_0001.png\ncopying 576 files\ncopying 564 files\ncopying 551 files\ncopying 621 files\ncopying 776 files\ncopying 641 files\ncopying 528 files\ncopying 576 files\ncopying 549 files\ncopying 647 files\ncopying 513 files\ncopying 535 files\ncopying 627 files\ncopying 541 files\ncopying 529 files\ncopying 748 files\ncopying 539 files\ncopying 542 files\ncopying 548 files\ncopying 543 files\ncopying 544 files\ncopying 536 files\ncopying 581 files\ncopying 543 files\ncopying 533 files\nignor: C/b/depth_1_0001.png\ncopying 529 files\nignor: C/e/depth_4_0001.png\ncopying 526 files\nignor: C/d/depth_3_0001.png\ncopying 531 files\nignor: C/x/depth_23_0001.png\ncopying 539 files\nignor: C/k/depth_10_0001.png\ncopying 534 files\nignor: C/p/depth_15_0001.png\ncopying 548 files\nignor: C/o/depth_14_0001.png\ncopying 586 files\nignor: C/l/depth_11_0001.png\ncopying 530 files\nignor: C/r/depth_17_0001.png\ncopying 890 files\nignor: C/w/depth_22_0001.png\ncopying 529 files\nignor: C/t/depth_19_0001.png\ncopying 524 files\nignor: C/a/depth_0_0001.png\ncopying 536 files\nignor: C/v/depth_21_0001.png\ncopying 530 files\nignor: C/u/depth_20_0001.png\ncopying 518 files\nignor: C/f/depth_5_0001.png\ncopying 507 files\nignor: C/s/depth_18_0001.png\ncopying 541 files\nignor: C/q/depth_16_0001.png\ncopying 530 files\nignor: C/i/depth_8_0001.png\ncopying 743 files\nignor: C/c/depth_2_0001.png\ncopying 564 files\nignor: C/n/depth_13_0001.png\ncopying 536 files\nignor: C/h/depth_7_0001.png\ncopying 543 files\nignor: C/y/depth_24_0001.png\ncopying 538 files\nignor: C/m/depth_12_0001.png\ncopying 532 files\nignor: C/g/depth_6_0001.png\ncopying 532 files\ncopying 528 files\ncopying 525 files\ncopying 530 files\ncopying 538 files\ncopying 533 files\ncopying 547 files\ncopying 585 files\ncopying 529 files\ncopying 889 files\ncopying 528 files\ncopying 523 files\ncopying 535 files\ncopying 529 files\ncopying 517 files\ncopying 506 files\ncopying 540 files\ncopying 529 files\ncopying 742 files\ncopying 563 files\ncopying 535 files\ncopying 542 files\ncopying 537 files\ncopying 531 files\ncopying 535 files\nignor: E/b/depth_1_0001.png\ncopying 540 files\nignor: E/e/depth_4_0001.png\ncopying 545 files\nignor: E/d/depth_3_0001.png\ncopying 523 files\nignor: E/x/depth_23_0001.png\ncopying 570 files\nignor: E/k/depth_10_0001.png\ncopying 528 files\nignor: E/p/depth_15_0001.png\ncopying 533 files\nignor: E/o/depth_14_0001.png\ncopying 515 files\nignor: E/l/depth_11_0001.png\ncopying 563 files\nignor: E/r/depth_17_0001.png\ncopying 514 files\nignor: E/w/depth_22_0001.png\ncopying 532 files\nignor: E/t/depth_19_0001.png\ncopying 546 files\nignor: E/a/depth_0_0001.png\ncopying 528 files\nignor: E/v/depth_21_0001.png\ncopying 530 files\nignor: E/u/depth_20_0001.png\ncopying 528 files\nignor: E/f/depth_5_0001.png\ncopying 527 files\nignor: E/s/depth_18_0001.png\ncopying 530 files\nignor: E/q/depth_16_0001.png\ncopying 526 files\nignor: E/i/depth_8_0001.png\ncopying 541 files\nignor: E/c/depth_2_0001.png\ncopying 531 files\nignor: E/n/depth_13_0001.png\ncopying 535 files\nignor: E/h/depth_7_0001.png\ncopying 522 files\nignor: E/y/depth_24_0001.png\ncopying 526 files\nignor: E/m/depth_12_0001.png\ncopying 538 files\nignor: E/g/depth_6_0001.png\ncopying 534 files\ncopying 539 files\ncopying 544 files\ncopying 522 files\ncopying 569 files\ncopying 527 files\ncopying 532 files\ncopying 514 files\ncopying 562 files\ncopying 513 files\ncopying 531 files\ncopying 545 files\ncopying 527 files\ncopying 529 files\ncopying 527 files\ncopying 526 files\ncopying 529 files\ncopying 525 files\ncopying 540 files\ncopying 530 files\ncopying 534 files\ncopying 521 files\ncopying 525 files\ncopying 537 files\ncopying 572 files\nignor: D/b/depth_1_0001.png\ncopying 528 files\nignor: D/e/depth_4_0001.png\ncopying 526 files\nignor: D/d/depth_3_0001.png\ncopying 538 files\nignor: D/x/depth_23_0001.png\ncopying 540 files\nignor: D/k/depth_10_0001.png\ncopying 532 files\nignor: D/p/depth_15_0001.png\ncopying 539 files\nignor: D/o/depth_14_0001.png\ncopying 565 files\nignor: D/l/depth_11_0001.png\ncopying 774 files\nignor: D/r/depth_17_0001.png\ncopying 536 files\nignor: D/w/depth_22_0001.png\ncopying 530 files\nignor: D/t/depth_19_0001.png\ncopying 547 files\nignor: D/a/depth_0_0001.png\ncopying 544 files\nignor: D/v/depth_21_0001.png\ncopying 532 files\nignor: D/u/depth_20_0001.png\ncopying 525 files\nignor: D/f/depth_5_0001.png\ncopying 535 files\nignor: D/s/depth_18_0001.png\ncopying 550 files\nignor: D/q/depth_16_0001.png\ncopying 522 files\nignor: D/i/depth_8_0001.png\ncopying 531 files\nignor: D/c/depth_2_0001.png\ncopying 530 files\nignor: D/n/depth_13_0001.png\ncopying 562 files\nignor: D/h/depth_7_0001.png\ncopying 536 files\nignor: D/y/depth_24_0001.png\ncopying 542 files\nignor: D/m/depth_12_0001.png\ncopying 542 files\nignor: D/g/depth_6_0001.png\ncopying 571 files\ncopying 527 files\ncopying 525 files\ncopying 537 files\ncopying 539 files\ncopying 531 files\ncopying 538 files\ncopying 564 files\ncopying 773 files\ncopying 535 files\ncopying 529 files\ncopying 546 files\ncopying 543 files\ncopying 531 files\ncopying 524 files\ncopying 534 files\ncopying 549 files\ncopying 521 files\ncopying 530 files\ncopying 529 files\ncopying 561 files\ncopying 535 files\ncopying 541 files\ncopying 541 files\n"
],
[
"# #/content\n%cd ..",
"/media/datastorage/Phong\n"
],
[
"ls -l",
"_____no_output_____"
],
[
"mkdir surrey/E/checkpoints",
"_____no_output_____"
],
[
"cd surrey/",
"_____no_output_____"
],
[
"#MUL 1 - Inception - ST\n\nfrom keras.applications import MobileNet\n# from keras.applications import InceptionV3\n# from keras.applications import Xception\n# from keras.applications.inception_resnet_v2 import InceptionResNetV2\n# from tensorflow.keras.applications import EfficientNetB0\n\nfrom keras.models import Model\nfrom keras.layers import concatenate\nfrom keras.layers import Dense, GlobalAveragePooling2D, Input, Embedding, SimpleRNN, LSTM, Flatten, GRU, Reshape\n\n# from keras.applications.inception_v3 import preprocess_input\n# from tensorflow.keras.applications.efficientnet import preprocess_input\nfrom keras.applications.mobilenet import preprocess_input\n\nfrom keras.layers import GaussianNoise\n\ndef get_adv_model():\n# f1_base = EfficientNetB0(include_top=False, weights='imagenet', \n# input_shape=(299, 299, 3), \n# pooling='avg')\n# f1_x = f1_base.output\n\n\n f1_base = MobileNet(weights='imagenet', include_top=False, input_shape=(224,224,3)) \n f1_x = f1_base.output\n f1_x = GlobalAveragePooling2D()(f1_x) \n\n# f1_x = f1_base.layers[-151].output #layer 5\n\n# f1_x = GlobalAveragePooling2D()(f1_x)\n# f1_x = Flatten()(f1_x)\n\n# f1_x = Reshape([1,1280])(f1_x) \n# f1_x = SimpleRNN(2048, \n# return_sequences=False, \n# # dropout=0.8 \n# input_shape=[1,1280])(f1_x)\n \n #Regularization with noise\n f1_x = GaussianNoise(0.1)(f1_x)\n\n f1_x = Dense(1024, activation='relu')(f1_x)\n f1_x = Dense(24, activation='softmax')(f1_x)\n model_1 = Model(inputs=[f1_base.input],outputs=[f1_x])\n model_1.summary()\n \n return model_1",
"_____no_output_____"
],
[
"from keras.callbacks import Callback\nimport pickle\nimport sys\n\n#Stop training on val_acc\nclass EarlyStoppingByAccVal(Callback):\n def __init__(self, monitor='val_acc', value=0.00001, verbose=0):\n super(Callback, self).__init__()\n self.monitor = monitor\n self.value = value\n self.verbose = verbose\n\n def on_epoch_end(self, epoch, logs={}):\n current = logs.get(self.monitor)\n if current is None:\n warnings.warn(\"Early stopping requires %s available!\" % self.monitor, RuntimeWarning)\n\n if current >= self.value:\n if self.verbose > 0:\n print(\"Epoch %05d: early stopping\" % epoch)\n self.model.stop_training = True\n\n#Save large model using pickle formate instead of h5 \nclass SaveCheckPoint(Callback):\n def __init__(self, model, dest_folder):\n super(Callback, self).__init__()\n self.model = model\n self.dest_folder = dest_folder\n \n #initiate\n self.best_val_acc = 0\n self.best_val_loss = sys.maxsize #get max value\n \n def on_epoch_end(self, epoch, logs={}):\n val_acc = logs['val_acc']\n val_loss = logs['val_loss']\n\n if val_acc > self.best_val_acc:\n self.best_val_acc = val_acc\n \n # Save weights in pickle format instead of h5\n print('\\nSaving val_acc %f at %s' %(self.best_val_acc, self.dest_folder))\n weigh= self.model.get_weights()\n\n #now, use pickle to save your model weights, instead of .h5\n #for heavy model architectures, .h5 file is unsupported.\n fpkl= open(self.dest_folder, 'wb') #Python 3\n pickle.dump(weigh, fpkl, protocol= pickle.HIGHEST_PROTOCOL)\n fpkl.close()\n \n# model.save('tmp.h5')\n elif val_acc == self.best_val_acc:\n if val_loss < self.best_val_loss:\n self.best_val_loss=val_loss\n \n # Save weights in pickle format instead of h5\n print('\\nSaving val_acc %f at %s' %(self.best_val_acc, self.dest_folder))\n weigh= self.model.get_weights()\n\n #now, use pickle to save your model weights, instead of .h5\n #for heavy model architectures, .h5 file is unsupported.\n fpkl= open(self.dest_folder, 'wb') #Python 3\n pickle.dump(weigh, fpkl, protocol= pickle.HIGHEST_PROTOCOL)\n fpkl.close() ",
"_____no_output_____"
],
[
"# Training\nimport tensorflow as tf\nfrom keras.preprocessing.image import ImageDataGenerator\nfrom keras.callbacks import TensorBoard, ModelCheckpoint, EarlyStopping, CSVLogger, ReduceLROnPlateau\nfrom keras.optimizers import Adam\n\nimport time, os\nfrom math import ceil\n\ntrain_datagen = ImageDataGenerator(\n# rescale = 1./255,\n rotation_range=30,\n width_shift_range=0.3,\n height_shift_range=0.3,\n shear_range=0.3,\n zoom_range=0.3,\n# horizontal_flip=True,\n# vertical_flip=True,##\n# brightness_range=[0.5, 1.5],##\n channel_shift_range=10,##\n fill_mode='nearest',\n # preprocessing_function=get_cutout_v2(),\n preprocessing_function=preprocess_input,\n)\n\ntest_datagen = ImageDataGenerator(\n# rescale = 1./255\n preprocessing_function=preprocess_input\n)\n\nNUM_GPU = 1\nbatch_size = 64\n\ntrain_set = train_datagen.flow_from_directory('surrey/D/train_color/',\n target_size = (224, 224),\n batch_size = batch_size,\n class_mode = 'categorical',\n shuffle=True,\n seed=7,\n# subset=\"training\"\n )\n\nvalid_set = test_datagen.flow_from_directory('surrey/D/test_color/',\n target_size = (224, 224),\n batch_size = batch_size,\n class_mode = 'categorical',\n shuffle=False,\n seed=7,\n# subset=\"validation\"\n )\n\nmodel_txt = 'st'\n# Helper: Save the model.\nsavedfilename = os.path.join('surrey', 'D', 'checkpoints', 'Surrey_MobileNet_D_tmp.hdf5')\n\ncheckpointer = ModelCheckpoint(savedfilename,\n monitor='val_accuracy', verbose=1, \n save_best_only=True, mode='max',save_weights_only=True)########\n\n# Helper: TensorBoard\ntb = TensorBoard(log_dir=os.path.join('svhn_output', 'logs', model_txt))\n\n# Helper: Save results.\ntimestamp = time.time()\ncsv_logger = CSVLogger(os.path.join('svhn_output', 'logs', model_txt + '-' + 'training-' + \\\n str(timestamp) + '.log'))\n\nearlystopping = EarlyStoppingByAccVal(monitor='val_accuracy', value=0.9900, verbose=1)\n\nepochs = 40##!!!\nlr = 1e-3\ndecay = lr/epochs\noptimizer = Adam(lr=lr, decay=decay)\n\n# train on multiple-gpus\n\n# Create a MirroredStrategy.\nstrategy = tf.distribute.MirroredStrategy()\nprint(\"Number of GPUs: {}\".format(strategy.num_replicas_in_sync))\n\n# Open a strategy scope.\nwith strategy.scope():\n # Everything that creates variables should be under the strategy scope.\n # In general this is only model construction & `compile()`.\n model_mul = get_adv_model()\n model_mul.compile(optimizer=optimizer,loss='categorical_crossentropy',metrics=['accuracy'])\n\nstep_size_train=ceil(train_set.n/train_set.batch_size)\nstep_size_valid=ceil(valid_set.n/valid_set.batch_size)\n# step_size_test=ceil(testing_set.n//testing_set.batch_size)\n\n# result = model_mul.fit_generator(\n# generator = train_set, \n# steps_per_epoch = step_size_train,\n# validation_data = valid_set,\n# validation_steps = step_size_valid,\n# shuffle=True,\n# epochs=epochs,\n# callbacks=[checkpointer],\n# # callbacks=[csv_logger, checkpointer, earlystopping],\n# # callbacks=[tb, csv_logger, checkpointer, earlystopping], \n# verbose=1) ",
"Found 52620 images belonging to 24 classes.\nFound 13154 images belonging to 24 classes.\nINFO:tensorflow:Using MirroredStrategy with devices ('/job:localhost/replica:0/task:0/device:GPU:0',)\nNumber of GPUs: 1\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nModel: \"model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 224, 224, 3)] 0 \n_________________________________________________________________\nconv1 (Conv2D) (None, 112, 112, 32) 864 \n_________________________________________________________________\nconv1_bn (BatchNormalization (None, 112, 112, 32) 128 \n_________________________________________________________________\nconv1_relu (ReLU) (None, 112, 112, 32) 0 \n_________________________________________________________________\nconv_dw_1 (DepthwiseConv2D) (None, 112, 112, 32) 288 \n_________________________________________________________________\nconv_dw_1_bn (BatchNormaliza (None, 112, 112, 32) 128 \n_________________________________________________________________\nconv_dw_1_relu (ReLU) (None, 112, 112, 32) 0 \n_________________________________________________________________\nconv_pw_1 (Conv2D) (None, 112, 112, 64) 2048 \n_________________________________________________________________\nconv_pw_1_bn (BatchNormaliza (None, 112, 112, 64) 256 \n_________________________________________________________________\nconv_pw_1_relu (ReLU) (None, 112, 112, 64) 0 \n_________________________________________________________________\nconv_pad_2 (ZeroPadding2D) (None, 113, 113, 64) 0 \n_________________________________________________________________\nconv_dw_2 (DepthwiseConv2D) (None, 56, 56, 64) 576 \n_________________________________________________________________\nconv_dw_2_bn (BatchNormaliza (None, 56, 56, 64) 256 \n_________________________________________________________________\nconv_dw_2_relu (ReLU) (None, 56, 56, 64) 0 \n_________________________________________________________________\nconv_pw_2 (Conv2D) (None, 56, 56, 128) 8192 \n_________________________________________________________________\nconv_pw_2_bn (BatchNormaliza (None, 56, 56, 128) 512 \n_________________________________________________________________\nconv_pw_2_relu (ReLU) (None, 56, 56, 128) 0 \n_________________________________________________________________\nconv_dw_3 (DepthwiseConv2D) (None, 56, 56, 128) 1152 \n_________________________________________________________________\nconv_dw_3_bn (BatchNormaliza (None, 56, 56, 128) 512 \n_________________________________________________________________\nconv_dw_3_relu (ReLU) (None, 56, 56, 128) 0 \n_________________________________________________________________\nconv_pw_3 (Conv2D) (None, 56, 56, 128) 16384 \n_________________________________________________________________\nconv_pw_3_bn (BatchNormaliza (None, 56, 56, 128) 512 \n_________________________________________________________________\nconv_pw_3_relu (ReLU) (None, 56, 56, 128) 0 \n_________________________________________________________________\nconv_pad_4 (ZeroPadding2D) (None, 57, 57, 128) 0 \n_________________________________________________________________\nconv_dw_4 (DepthwiseConv2D) (None, 28, 28, 128) 1152 \n_________________________________________________________________\nconv_dw_4_bn (BatchNormaliza (None, 28, 28, 128) 512 \n_________________________________________________________________\nconv_dw_4_relu (ReLU) (None, 28, 28, 128) 0 \n_________________________________________________________________\nconv_pw_4 (Conv2D) (None, 28, 28, 256) 32768 \n_________________________________________________________________\nconv_pw_4_bn (BatchNormaliza (None, 28, 28, 256) 1024 \n_________________________________________________________________\nconv_pw_4_relu (ReLU) (None, 28, 28, 256) 0 \n_________________________________________________________________\nconv_dw_5 (DepthwiseConv2D) (None, 28, 28, 256) 2304 \n_________________________________________________________________\nconv_dw_5_bn (BatchNormaliza (None, 28, 28, 256) 1024 \n_________________________________________________________________\nconv_dw_5_relu (ReLU) (None, 28, 28, 256) 0 \n_________________________________________________________________\nconv_pw_5 (Conv2D) (None, 28, 28, 256) 65536 \n_________________________________________________________________\nconv_pw_5_bn (BatchNormaliza (None, 28, 28, 256) 1024 \n_________________________________________________________________\nconv_pw_5_relu (ReLU) (None, 28, 28, 256) 0 \n_________________________________________________________________\nconv_pad_6 (ZeroPadding2D) (None, 29, 29, 256) 0 \n_________________________________________________________________\nconv_dw_6 (DepthwiseConv2D) (None, 14, 14, 256) 2304 \n_________________________________________________________________\nconv_dw_6_bn (BatchNormaliza (None, 14, 14, 256) 1024 \n_________________________________________________________________\nconv_dw_6_relu (ReLU) (None, 14, 14, 256) 0 \n_________________________________________________________________\nconv_pw_6 (Conv2D) (None, 14, 14, 512) 131072 \n_________________________________________________________________\nconv_pw_6_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_pw_6_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_dw_7 (DepthwiseConv2D) (None, 14, 14, 512) 4608 \n_________________________________________________________________\nconv_dw_7_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_dw_7_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_pw_7 (Conv2D) (None, 14, 14, 512) 262144 \n_________________________________________________________________\nconv_pw_7_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_pw_7_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_dw_8 (DepthwiseConv2D) (None, 14, 14, 512) 4608 \n_________________________________________________________________\nconv_dw_8_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_dw_8_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_pw_8 (Conv2D) (None, 14, 14, 512) 262144 \n_________________________________________________________________\nconv_pw_8_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_pw_8_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_dw_9 (DepthwiseConv2D) (None, 14, 14, 512) 4608 \n_________________________________________________________________\nconv_dw_9_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_dw_9_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_pw_9 (Conv2D) (None, 14, 14, 512) 262144 \n_________________________________________________________________\nconv_pw_9_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_pw_9_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_dw_10 (DepthwiseConv2D) (None, 14, 14, 512) 4608 \n_________________________________________________________________\nconv_dw_10_bn (BatchNormaliz (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_dw_10_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_pw_10 (Conv2D) (None, 14, 14, 512) 262144 \n_________________________________________________________________\nconv_pw_10_bn (BatchNormaliz (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_pw_10_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_dw_11 (DepthwiseConv2D) (None, 14, 14, 512) 4608 \n_________________________________________________________________\nconv_dw_11_bn (BatchNormaliz (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_dw_11_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_pw_11 (Conv2D) (None, 14, 14, 512) 262144 \n_________________________________________________________________\nconv_pw_11_bn (BatchNormaliz (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_pw_11_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_pad_12 (ZeroPadding2D) (None, 15, 15, 512) 0 \n_________________________________________________________________\nconv_dw_12 (DepthwiseConv2D) (None, 7, 7, 512) 4608 \n_________________________________________________________________\nconv_dw_12_bn (BatchNormaliz (None, 7, 7, 512) 2048 \n_________________________________________________________________\nconv_dw_12_relu (ReLU) (None, 7, 7, 512) 0 \n_________________________________________________________________\nconv_pw_12 (Conv2D) (None, 7, 7, 1024) 524288 \n_________________________________________________________________\nconv_pw_12_bn (BatchNormaliz (None, 7, 7, 1024) 4096 \n_________________________________________________________________\nconv_pw_12_relu (ReLU) (None, 7, 7, 1024) 0 \n_________________________________________________________________\nconv_dw_13 (DepthwiseConv2D) (None, 7, 7, 1024) 9216 \n_________________________________________________________________\nconv_dw_13_bn (BatchNormaliz (None, 7, 7, 1024) 4096 \n_________________________________________________________________\nconv_dw_13_relu (ReLU) (None, 7, 7, 1024) 0 \n_________________________________________________________________\nconv_pw_13 (Conv2D) (None, 7, 7, 1024) 1048576 \n_________________________________________________________________\nconv_pw_13_bn (BatchNormaliz (None, 7, 7, 1024) 4096 \n_________________________________________________________________\nconv_pw_13_relu (ReLU) (None, 7, 7, 1024) 0 \n_________________________________________________________________\nglobal_average_pooling2d (Gl (None, 1024) 0 \n_________________________________________________________________\ngaussian_noise (GaussianNois (None, 1024) 0 \n_________________________________________________________________\ndense (Dense) (None, 1024) 1049600 \n_________________________________________________________________\ndense_1 (Dense) (None, 24) 24600 \n=================================================================\nTotal params: 4,303,064\nTrainable params: 4,281,176\nNon-trainable params: 21,888\n_________________________________________________________________\n"
],
[
"# Training\nimport tensorflow as tf\nfrom keras.preprocessing.image import ImageDataGenerator\nfrom keras.callbacks import TensorBoard, ModelCheckpoint, EarlyStopping, CSVLogger, ReduceLROnPlateau\nfrom keras.optimizers import Adam\n\nimport time, os\nfrom math import ceil\n\ntrain_datagen = ImageDataGenerator(\n# rescale = 1./255,\n rotation_range=30,\n width_shift_range=0.3,\n height_shift_range=0.3,\n shear_range=0.3,\n zoom_range=0.3,\n# horizontal_flip=True,\n# vertical_flip=True,##\n# brightness_range=[0.5, 1.5],##\n channel_shift_range=10,##\n fill_mode='nearest',\n # preprocessing_function=get_cutout_v2(),\n preprocessing_function=preprocess_input,\n)\n\ntest_datagen = ImageDataGenerator(\n# rescale = 1./255\n preprocessing_function=preprocess_input\n)\n\nNUM_GPU = 1\nbatch_size = 64\n\ntrain_set = train_datagen.flow_from_directory('surrey/D/train_color/',\n target_size = (224, 224),\n batch_size = batch_size,\n class_mode = 'categorical',\n shuffle=True,\n seed=7,\n# subset=\"training\"\n )\n\nvalid_set = test_datagen.flow_from_directory('surrey/D/test_color/',\n target_size = (224, 224),\n batch_size = batch_size,\n class_mode = 'categorical',\n shuffle=False,\n seed=7,\n# subset=\"validation\"\n )\n\nmodel_txt = 'st'\n# Helper: Save the model.\nsavedfilename = os.path.join('gdrive', 'My Drive', 'Surrey_ASL', '5_Surrey_MobileNet_D.hdf5')\n\ncheckpointer = ModelCheckpoint(savedfilename,\n monitor='val_accuracy', verbose=1, \n save_best_only=True, mode='max',save_weights_only=True)########\n\n# Helper: TensorBoard\ntb = TensorBoard(log_dir=os.path.join('svhn_output', 'logs', model_txt))\n\n# Helper: Save results.\ntimestamp = time.time()\ncsv_logger = CSVLogger(os.path.join('svhn_output', 'logs', model_txt + '-' + 'training-' + \\\n str(timestamp) + '.log'))\n\nearlystopping = EarlyStoppingByAccVal(monitor='val_accuracy', value=0.9900, verbose=1)\n\nepochs = 40##!!!\nlr = 1e-3\ndecay = lr/epochs\noptimizer = Adam(lr=lr, decay=decay)\n\n# train on multiple-gpus\n\n# Create a MirroredStrategy.\nstrategy = tf.distribute.MirroredStrategy()\nprint(\"Number of GPUs: {}\".format(strategy.num_replicas_in_sync))\n\n# Open a strategy scope.\nwith strategy.scope():\n # Everything that creates variables should be under the strategy scope.\n # In general this is only model construction & `compile()`.\n model_mul = get_adv_model()\n model_mul.compile(optimizer=optimizer,loss='categorical_crossentropy',metrics=['accuracy'])\n\nstep_size_train=ceil(train_set.n/train_set.batch_size)\nstep_size_valid=ceil(valid_set.n/valid_set.batch_size)\n# step_size_test=ceil(testing_set.n//testing_set.batch_size)\n\nresult = model_mul.fit_generator(\n generator = train_set, \n steps_per_epoch = step_size_train,\n validation_data = valid_set,\n validation_steps = step_size_valid,\n shuffle=True,\n epochs=epochs,\n callbacks=[checkpointer],\n# callbacks=[csv_logger, checkpointer, earlystopping],\n# callbacks=[tb, csv_logger, checkpointer, earlystopping], \n verbose=1) ",
"Found 52620 images belonging to 24 classes.\nFound 13154 images belonging to 24 classes.\nINFO:tensorflow:Using MirroredStrategy with devices ('/job:localhost/replica:0/task:0/device:GPU:0',)\nNumber of GPUs: 1\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nDownloading data from https://storage.googleapis.com/tensorflow/keras-applications/mobilenet/mobilenet_1_0_224_tf_no_top.h5\n17227776/17225924 [==============================] - 0s 0us/step\nModel: \"model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 224, 224, 3)] 0 \n_________________________________________________________________\nconv1 (Conv2D) (None, 112, 112, 32) 864 \n_________________________________________________________________\nconv1_bn (BatchNormalization (None, 112, 112, 32) 128 \n_________________________________________________________________\nconv1_relu (ReLU) (None, 112, 112, 32) 0 \n_________________________________________________________________\nconv_dw_1 (DepthwiseConv2D) (None, 112, 112, 32) 288 \n_________________________________________________________________\nconv_dw_1_bn (BatchNormaliza (None, 112, 112, 32) 128 \n_________________________________________________________________\nconv_dw_1_relu (ReLU) (None, 112, 112, 32) 0 \n_________________________________________________________________\nconv_pw_1 (Conv2D) (None, 112, 112, 64) 2048 \n_________________________________________________________________\nconv_pw_1_bn (BatchNormaliza (None, 112, 112, 64) 256 \n_________________________________________________________________\nconv_pw_1_relu (ReLU) (None, 112, 112, 64) 0 \n_________________________________________________________________\nconv_pad_2 (ZeroPadding2D) (None, 113, 113, 64) 0 \n_________________________________________________________________\nconv_dw_2 (DepthwiseConv2D) (None, 56, 56, 64) 576 \n_________________________________________________________________\nconv_dw_2_bn (BatchNormaliza (None, 56, 56, 64) 256 \n_________________________________________________________________\nconv_dw_2_relu (ReLU) (None, 56, 56, 64) 0 \n_________________________________________________________________\nconv_pw_2 (Conv2D) (None, 56, 56, 128) 8192 \n_________________________________________________________________\nconv_pw_2_bn (BatchNormaliza (None, 56, 56, 128) 512 \n_________________________________________________________________\nconv_pw_2_relu (ReLU) (None, 56, 56, 128) 0 \n_________________________________________________________________\nconv_dw_3 (DepthwiseConv2D) (None, 56, 56, 128) 1152 \n_________________________________________________________________\nconv_dw_3_bn (BatchNormaliza (None, 56, 56, 128) 512 \n_________________________________________________________________\nconv_dw_3_relu (ReLU) (None, 56, 56, 128) 0 \n_________________________________________________________________\nconv_pw_3 (Conv2D) (None, 56, 56, 128) 16384 \n_________________________________________________________________\nconv_pw_3_bn (BatchNormaliza (None, 56, 56, 128) 512 \n_________________________________________________________________\nconv_pw_3_relu (ReLU) (None, 56, 56, 128) 0 \n_________________________________________________________________\nconv_pad_4 (ZeroPadding2D) (None, 57, 57, 128) 0 \n_________________________________________________________________\nconv_dw_4 (DepthwiseConv2D) (None, 28, 28, 128) 1152 \n_________________________________________________________________\nconv_dw_4_bn (BatchNormaliza (None, 28, 28, 128) 512 \n_________________________________________________________________\nconv_dw_4_relu (ReLU) (None, 28, 28, 128) 0 \n_________________________________________________________________\nconv_pw_4 (Conv2D) (None, 28, 28, 256) 32768 \n_________________________________________________________________\nconv_pw_4_bn (BatchNormaliza (None, 28, 28, 256) 1024 \n_________________________________________________________________\nconv_pw_4_relu (ReLU) (None, 28, 28, 256) 0 \n_________________________________________________________________\nconv_dw_5 (DepthwiseConv2D) (None, 28, 28, 256) 2304 \n_________________________________________________________________\nconv_dw_5_bn (BatchNormaliza (None, 28, 28, 256) 1024 \n_________________________________________________________________\nconv_dw_5_relu (ReLU) (None, 28, 28, 256) 0 \n_________________________________________________________________\nconv_pw_5 (Conv2D) (None, 28, 28, 256) 65536 \n_________________________________________________________________\nconv_pw_5_bn (BatchNormaliza (None, 28, 28, 256) 1024 \n_________________________________________________________________\nconv_pw_5_relu (ReLU) (None, 28, 28, 256) 0 \n_________________________________________________________________\nconv_pad_6 (ZeroPadding2D) (None, 29, 29, 256) 0 \n_________________________________________________________________\nconv_dw_6 (DepthwiseConv2D) (None, 14, 14, 256) 2304 \n_________________________________________________________________\nconv_dw_6_bn (BatchNormaliza (None, 14, 14, 256) 1024 \n_________________________________________________________________\nconv_dw_6_relu (ReLU) (None, 14, 14, 256) 0 \n_________________________________________________________________\nconv_pw_6 (Conv2D) (None, 14, 14, 512) 131072 \n_________________________________________________________________\nconv_pw_6_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_pw_6_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_dw_7 (DepthwiseConv2D) (None, 14, 14, 512) 4608 \n_________________________________________________________________\nconv_dw_7_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_dw_7_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_pw_7 (Conv2D) (None, 14, 14, 512) 262144 \n_________________________________________________________________\nconv_pw_7_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_pw_7_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_dw_8 (DepthwiseConv2D) (None, 14, 14, 512) 4608 \n_________________________________________________________________\nconv_dw_8_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_dw_8_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_pw_8 (Conv2D) (None, 14, 14, 512) 262144 \n_________________________________________________________________\nconv_pw_8_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_pw_8_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_dw_9 (DepthwiseConv2D) (None, 14, 14, 512) 4608 \n_________________________________________________________________\nconv_dw_9_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_dw_9_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_pw_9 (Conv2D) (None, 14, 14, 512) 262144 \n_________________________________________________________________\nconv_pw_9_bn (BatchNormaliza (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_pw_9_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_dw_10 (DepthwiseConv2D) (None, 14, 14, 512) 4608 \n_________________________________________________________________\nconv_dw_10_bn (BatchNormaliz (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_dw_10_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_pw_10 (Conv2D) (None, 14, 14, 512) 262144 \n_________________________________________________________________\nconv_pw_10_bn (BatchNormaliz (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_pw_10_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_dw_11 (DepthwiseConv2D) (None, 14, 14, 512) 4608 \n_________________________________________________________________\nconv_dw_11_bn (BatchNormaliz (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_dw_11_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_pw_11 (Conv2D) (None, 14, 14, 512) 262144 \n_________________________________________________________________\nconv_pw_11_bn (BatchNormaliz (None, 14, 14, 512) 2048 \n_________________________________________________________________\nconv_pw_11_relu (ReLU) (None, 14, 14, 512) 0 \n_________________________________________________________________\nconv_pad_12 (ZeroPadding2D) (None, 15, 15, 512) 0 \n_________________________________________________________________\nconv_dw_12 (DepthwiseConv2D) (None, 7, 7, 512) 4608 \n_________________________________________________________________\nconv_dw_12_bn (BatchNormaliz (None, 7, 7, 512) 2048 \n_________________________________________________________________\nconv_dw_12_relu (ReLU) (None, 7, 7, 512) 0 \n_________________________________________________________________\nconv_pw_12 (Conv2D) (None, 7, 7, 1024) 524288 \n_________________________________________________________________\nconv_pw_12_bn (BatchNormaliz (None, 7, 7, 1024) 4096 \n_________________________________________________________________\nconv_pw_12_relu (ReLU) (None, 7, 7, 1024) 0 \n_________________________________________________________________\nconv_dw_13 (DepthwiseConv2D) (None, 7, 7, 1024) 9216 \n_________________________________________________________________\nconv_dw_13_bn (BatchNormaliz (None, 7, 7, 1024) 4096 \n_________________________________________________________________\nconv_dw_13_relu (ReLU) (None, 7, 7, 1024) 0 \n_________________________________________________________________\nconv_pw_13 (Conv2D) (None, 7, 7, 1024) 1048576 \n_________________________________________________________________\nconv_pw_13_bn (BatchNormaliz (None, 7, 7, 1024) 4096 \n_________________________________________________________________\nconv_pw_13_relu (ReLU) (None, 7, 7, 1024) 0 \n_________________________________________________________________\nglobal_average_pooling2d (Gl (None, 1024) 0 \n_________________________________________________________________\ngaussian_noise (GaussianNois (None, 1024) 0 \n_________________________________________________________________\ndense (Dense) (None, 1024) 1049600 \n_________________________________________________________________\ndense_1 (Dense) (None, 24) 24600 \n=================================================================\nTotal params: 4,303,064\nTrainable params: 4,281,176\nNon-trainable params: 21,888\n_________________________________________________________________\n"
],
[
"ls -l",
"_____no_output_____"
],
[
"# Open a strategy scope.\nwith strategy.scope():\n model_mul.load_weights(os.path.join('gdrive', 'My Drive', 'Surrey_ASL', '5_Surrey_MobileNet_D.hdf5'))",
"_____no_output_____"
],
[
"model_mul.evaluate(valid_set)",
"206/206 [==============================] - 34s 117ms/step - loss: 0.3835 - accuracy: 0.9023\n"
],
[
"\n# Helper: Save the model.\nsavedfilename = os.path.join('gdrive', 'My Drive', 'Surrey_ASL', '5_Surrey_MobileNet_D_L2.hdf5')\n\ncheckpointer = ModelCheckpoint(savedfilename,\n monitor='val_accuracy', verbose=1, \n save_best_only=True, mode='max',save_weights_only=True)########\n\nepochs = 15##!!!\nlr = 1e-4\ndecay = lr/epochs\noptimizer = Adam(lr=lr, decay=decay)\n\n# Open a strategy scope.\nwith strategy.scope():\n model_mul.compile(optimizer=optimizer,loss='categorical_crossentropy',metrics=['accuracy'])\n\nresult = model_mul.fit_generator(\n generator = train_set, \n steps_per_epoch = step_size_train,\n validation_data = valid_set,\n validation_steps = step_size_valid,\n shuffle=True,\n epochs=epochs,\n callbacks=[checkpointer],\n# callbacks=[csv_logger, checkpointer, earlystopping],\n# callbacks=[tb, csv_logger, checkpointer, earlystopping], \n verbose=1) ",
"/usr/local/lib/python3.6/dist-packages/tensorflow/python/keras/engine/training.py:1844: UserWarning: `Model.fit_generator` is deprecated and will be removed in a future version. Please use `Model.fit`, which supports generators.\n warnings.warn('`Model.fit_generator` is deprecated and '\n"
],
[
"# Open a strategy scope.\nwith strategy.scope():\n model_mul.load_weights(os.path.join('gdrive', 'My Drive', 'Surrey_ASL', '5_Surrey_MobileNet_D_L2.hdf5'))",
"_____no_output_____"
],
[
"model_mul.evaluate(valid_set)",
"206/206 [==============================] - 30s 116ms/step - loss: 0.4206 - accuracy: 0.9067\n"
],
[
"# Helper: Save the model.\nsavedfilename = os.path.join('gdrive', 'My Drive', 'Surrey_ASL', '5_Surrey_MobileNet_D_L3.hdf5')\n\ncheckpointer = ModelCheckpoint(savedfilename,\n monitor='val_accuracy', verbose=1, \n save_best_only=True, mode='max',save_weights_only=True)########\n\nepochs = 15##!!!\nlr = 1e-5\ndecay = lr/epochs\noptimizer = Adam(lr=lr, decay=decay)\n\n# Open a strategy scope.\nwith strategy.scope():\n model_mul.compile(optimizer=optimizer,loss='categorical_crossentropy',metrics=['accuracy'])\n\nresult = model_mul.fit_generator(\n generator = train_set, \n steps_per_epoch = step_size_train,\n validation_data = valid_set,\n validation_steps = step_size_valid,\n shuffle=True,\n epochs=epochs,\n callbacks=[checkpointer],\n# callbacks=[csv_logger, checkpointer, earlystopping],\n# callbacks=[tb, csv_logger, checkpointer, earlystopping], \n verbose=1) ",
"/usr/local/lib/python3.6/dist-packages/tensorflow/python/keras/engine/training.py:1844: UserWarning: `Model.fit_generator` is deprecated and will be removed in a future version. Please use `Model.fit`, which supports generators.\n warnings.warn('`Model.fit_generator` is deprecated and '\n"
],
[
"# Open a strategy scope.\nwith strategy.scope():\n model_mul.load_weights(os.path.join('gdrive', 'My Drive', 'Surrey_ASL', '5_Surrey_MobileNet_D_L3.hdf5'))",
"_____no_output_____"
],
[
"model_mul.evaluate(valid_set)",
"206/206 [==============================] - 30s 118ms/step - loss: 0.4298 - accuracy: 0.9053\n"
],
[
"import numpy as np\nfrom keras.preprocessing.image import ImageDataGenerator\nimport time, os\nfrom math import ceil\n\n# PREDICT ON OFFICIAL TEST\ntrain_datagen = ImageDataGenerator(\n# rescale = 1./255,\n rotation_range=30,\n width_shift_range=0.3,\n height_shift_range=0.3,\n shear_range=0.3,\n zoom_range=0.3,\n# horizontal_flip=True,\n# vertical_flip=True,##\n# brightness_range=[0.5, 1.5],##\n channel_shift_range=10,##\n fill_mode='nearest',\n preprocessing_function=preprocess_input,\n)\n\ntest_datagen1 = ImageDataGenerator(\n# rescale = 1./255,\n preprocessing_function=preprocess_input\n)\n\nbatch_size = 64\n\ntrain_set = train_datagen.flow_from_directory('surrey/D/train_color/',\n target_size = (224, 224),\n batch_size = batch_size,\n class_mode = 'categorical',\n shuffle=True,\n seed=7,\n# subset=\"training\"\n )\n\ntest_set1 = test_datagen1.flow_from_directory('surrey/D/test_color/',\n target_size = (224, 224),\n batch_size = batch_size,\n class_mode = 'categorical',\n shuffle=False,\n seed=7,\n# subset=\"validation\"\n )\n\n# if NUM_GPU != 1:\npredict1=model_mul.predict_generator(test_set1, steps = ceil(test_set1.n/test_set1.batch_size),verbose=1)\n# else:\n# predict1=model.predict_generator(test_set1, steps = ceil(test_set1.n/test_set1.batch_size),verbose=1)\n \npredicted_class_indices=np.argmax(predict1,axis=1)\nlabels = (train_set.class_indices)\nlabels = dict((v,k) for k,v in labels.items())\npredictions1 = [labels[k] for k in predicted_class_indices]\n\nimport pandas as pd\n\nfilenames=test_set1.filenames\nresults=pd.DataFrame({\"file_name\":filenames,\n \"predicted1\":predictions1,\n })\nresults.to_csv('Surrey_MobileNet_D_L3_0902.csv')\nresults.head()",
"Found 52620 images belonging to 24 classes.\nFound 13154 images belonging to 24 classes.\n"
],
[
"np.save(os.path.join('gdrive', 'My Drive', 'Surrey_ASL', 'npy', '5Colab_Surrey_MobileNet_D_L2_0902.hdf5'), predict1)",
"_____no_output_____"
],
[
"np.save(os.path.join('gdrive', 'My Drive', 'Surrey_ASL', 'npy', '5Colab_Surrey_MobileNet_D_L3_0902.hdf5'), predict1)",
"_____no_output_____"
],
[
"from sklearn.metrics import classification_report, confusion_matrix\nimport numpy as np\n\ntest_datagen = ImageDataGenerator(\n preprocessing_function=preprocess_input)\n\ntesting_set = test_datagen.flow_from_directory('surrey/D/test_color/',\n target_size = (224, 224),\n batch_size = 32,\n class_mode = 'categorical',\n seed=7,\n shuffle=False\n# subset=\"validation\"\n )\n\ny_pred = model_mul.predict_generator(testing_set)\ny_pred = np.argmax(y_pred, axis=1)\n\ny_true = testing_set.classes\n\nprint(confusion_matrix(y_true, y_pred))\n\n# print(model.evaluate_generator(testing_set,\n# steps = testing_set.n//testing_set.batch_size))",
"Found 13154 images belonging to 24 classes.\n"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03cbafa397d3c1c9ca6ec1690688b41422b2f0a | 936,023 | ipynb | Jupyter Notebook | Model backlog/Train/64-melanoma-5fold-seresnet18-radam.ipynb | dimitreOliveira/melanoma-classification | e366a645b872f035a34fd2df5ee96fa8a1615ce1 | [
"MIT"
] | 10 | 2020-08-19T02:54:32.000Z | 2021-11-14T16:04:08.000Z | Model backlog/Train/64-melanoma-5fold-seresnet18-radam.ipynb | dimitreOliveira/melanoma-classification | e366a645b872f035a34fd2df5ee96fa8a1615ce1 | [
"MIT"
] | null | null | null | Model backlog/Train/64-melanoma-5fold-seresnet18-radam.ipynb | dimitreOliveira/melanoma-classification | e366a645b872f035a34fd2df5ee96fa8a1615ce1 | [
"MIT"
] | 5 | 2020-09-16T14:04:36.000Z | 2021-03-05T12:44:44.000Z | 206.03632 | 250,328 | 0.847916 | [
[
[
"## Dependencies",
"_____no_output_____"
]
],
[
[
"# !pip install --quiet efficientnet\n!pip install --quiet image-classifiers",
"_____no_output_____"
],
[
"import warnings, json, re, glob, math\nfrom scripts_step_lr_schedulers import *\nfrom melanoma_utility_scripts import *\nfrom kaggle_datasets import KaggleDatasets\nfrom sklearn.model_selection import KFold\nimport tensorflow.keras.layers as L\nimport tensorflow.keras.backend as K\nfrom tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint\nfrom tensorflow.keras import optimizers, layers, metrics, losses, Model\n# import efficientnet.tfkeras as efn\nfrom classification_models.tfkeras import Classifiers\nimport tensorflow_addons as tfa\n\nSEED = 0\nseed_everything(SEED)\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
]
],
[
[
"## TPU configuration",
"_____no_output_____"
]
],
[
[
"strategy, tpu = set_up_strategy()\nprint(\"REPLICAS: \", strategy.num_replicas_in_sync)\nAUTO = tf.data.experimental.AUTOTUNE",
"REPLICAS: 1\n"
]
],
[
[
"# Model parameters",
"_____no_output_____"
]
],
[
[
"config = {\n \"HEIGHT\": 256,\n \"WIDTH\": 256,\n \"CHANNELS\": 3,\n \"BATCH_SIZE\": 64,\n \"EPOCHS\": 25,\n \"LEARNING_RATE\": 3e-4,\n \"ES_PATIENCE\": 10,\n \"N_FOLDS\": 5,\n \"N_USED_FOLDS\": 5,\n \"TTA_STEPS\": 25,\n \"BASE_MODEL\": 'seresnet18',\n \"BASE_MODEL_WEIGHTS\": 'imagenet',\n \"DATASET_PATH\": 'melanoma-256x256'\n}\n\nwith open('config.json', 'w') as json_file:\n json.dump(json.loads(json.dumps(config)), json_file)\n \nconfig",
"_____no_output_____"
]
],
[
[
"# Load data",
"_____no_output_____"
]
],
[
[
"database_base_path = '/kaggle/input/siim-isic-melanoma-classification/'\nk_fold = pd.read_csv(database_base_path + 'train.csv')\ntest = pd.read_csv(database_base_path + 'test.csv')\n\nprint('Train samples: %d' % len(k_fold))\ndisplay(k_fold.head())\nprint(f'Test samples: {len(test)}')\ndisplay(test.head())\n\nGCS_PATH = KaggleDatasets().get_gcs_path(config['DATASET_PATH'])\nTRAINING_FILENAMES = tf.io.gfile.glob(GCS_PATH + '/train*.tfrec')\nTEST_FILENAMES = tf.io.gfile.glob(GCS_PATH + '/test*.tfrec')",
"Train samples: 33126\n"
]
],
[
[
"# Augmentations",
"_____no_output_____"
]
],
[
[
"def data_augment(image, label):\n p_spatial = tf.random.uniform([1], minval=0, maxval=1, dtype='float32')\n p_spatial2 = tf.random.uniform([1], minval=0, maxval=1, dtype='float32')\n p_rotate = tf.random.uniform([1], minval=0, maxval=1, dtype='float32')\n p_crop = tf.random.uniform([1], minval=0, maxval=1, dtype='float32')\n p_pixel = tf.random.uniform([1], minval=0, maxval=1, dtype='float32')\n \n ### Spatial-level transforms\n if p_spatial >= .2: # flips\n image['input_image'] = tf.image.random_flip_left_right(image['input_image'])\n image['input_image'] = tf.image.random_flip_up_down(image['input_image'])\n if p_spatial >= .7:\n image['input_image'] = tf.image.transpose(image['input_image'])\n \n if p_rotate >= .8: # rotate 270º\n image['input_image'] = tf.image.rot90(image['input_image'], k=3)\n elif p_rotate >= .6: # rotate 180º\n image['input_image'] = tf.image.rot90(image['input_image'], k=2)\n elif p_rotate >= .4: # rotate 90º\n image['input_image'] = tf.image.rot90(image['input_image'], k=1)\n \n if p_spatial2 >= .6:\n if p_spatial2 >= .9:\n image['input_image'] = transform_rotation(image['input_image'], config['HEIGHT'], 180.)\n elif p_spatial2 >= .8:\n image['input_image'] = transform_zoom(image['input_image'], config['HEIGHT'], 8., 8.)\n elif p_spatial2 >= .7:\n image['input_image'] = transform_shift(image['input_image'], config['HEIGHT'], 8., 8.)\n else:\n image['input_image'] = transform_shear(image['input_image'], config['HEIGHT'], 2.)\n \n if p_crop >= .6: # crops\n if p_crop >= .8:\n image['input_image'] = tf.image.random_crop(image['input_image'], size=[int(config['HEIGHT']*.8), int(config['WIDTH']*.8), config['CHANNELS']])\n elif p_crop >= .7:\n image['input_image'] = tf.image.random_crop(image['input_image'], size=[int(config['HEIGHT']*.9), int(config['WIDTH']*.9), config['CHANNELS']])\n else:\n image['input_image'] = tf.image.central_crop(image['input_image'], central_fraction=.8)\n image['input_image'] = tf.image.resize(image['input_image'], size=[config['HEIGHT'], config['WIDTH']])\n\n if p_pixel >= .6: # Pixel-level transforms\n if p_pixel >= .9:\n image['input_image'] = tf.image.random_hue(image['input_image'], 0.01)\n elif p_pixel >= .8:\n image['input_image'] = tf.image.random_saturation(image['input_image'], 0.7, 1.3)\n elif p_pixel >= .7:\n image['input_image'] = tf.image.random_contrast(image['input_image'], 0.8, 1.2)\n else:\n image['input_image'] = tf.image.random_brightness(image['input_image'], 0.1)\n\n return image, label",
"_____no_output_____"
]
],
[
[
"## Auxiliary functions",
"_____no_output_____"
]
],
[
[
"# Datasets utility functions\ndef read_labeled_tfrecord(example, height=config['HEIGHT'], width=config['WIDTH'], channels=config['CHANNELS']):\n example = tf.io.parse_single_example(example, LABELED_TFREC_FORMAT)\n image = decode_image(example['image'], height, width, channels)\n label = tf.cast(example['target'], tf.float32)\n # meta features\n data = {}\n data['patient_id'] = tf.cast(example['patient_id'], tf.int32)\n data['sex'] = tf.cast(example['sex'], tf.int32)\n data['age_approx'] = tf.cast(example['age_approx'], tf.int32)\n data['anatom_site_general_challenge'] = tf.cast(tf.one_hot(example['anatom_site_general_challenge'], 7), tf.int32)\n \n return {'input_image': image, 'input_meta': data}, label # returns a dataset of (image, data, label)\n\ndef read_labeled_tfrecord_eval(example, height=config['HEIGHT'], width=config['WIDTH'], channels=config['CHANNELS']):\n example = tf.io.parse_single_example(example, LABELED_TFREC_FORMAT)\n image = decode_image(example['image'], height, width, channels)\n label = tf.cast(example['target'], tf.float32)\n image_name = example['image_name']\n # meta features\n data = {}\n data['patient_id'] = tf.cast(example['patient_id'], tf.int32)\n data['sex'] = tf.cast(example['sex'], tf.int32)\n data['age_approx'] = tf.cast(example['age_approx'], tf.int32)\n data['anatom_site_general_challenge'] = tf.cast(tf.one_hot(example['anatom_site_general_challenge'], 7), tf.int32)\n \n return {'input_image': image, 'input_meta': data}, label, image_name # returns a dataset of (image, data, label, image_name)\n\ndef load_dataset(filenames, ordered=False, buffer_size=-1):\n ignore_order = tf.data.Options()\n if not ordered:\n ignore_order.experimental_deterministic = False # disable order, increase speed\n\n dataset = tf.data.TFRecordDataset(filenames, num_parallel_reads=buffer_size) # automatically interleaves reads from multiple files\n dataset = dataset.with_options(ignore_order) # uses data as soon as it streams in, rather than in its original order\n dataset = dataset.map(read_labeled_tfrecord, num_parallel_calls=buffer_size)\n \n return dataset # returns a dataset of (image, data, label)\n\ndef load_dataset_eval(filenames, buffer_size=-1):\n dataset = tf.data.TFRecordDataset(filenames, num_parallel_reads=buffer_size) # automatically interleaves reads from multiple files\n dataset = dataset.map(read_labeled_tfrecord_eval, num_parallel_calls=buffer_size)\n \n return dataset # returns a dataset of (image, data, label, image_name)\n\ndef get_training_dataset(filenames, batch_size, buffer_size=-1):\n dataset = load_dataset(filenames, ordered=False, buffer_size=buffer_size)\n dataset = dataset.map(data_augment, num_parallel_calls=AUTO)\n dataset = dataset.repeat() # the training dataset must repeat for several epochs\n dataset = dataset.shuffle(2048)\n dataset = dataset.batch(batch_size, drop_remainder=True) # slighly faster with fixed tensor sizes\n dataset = dataset.prefetch(buffer_size) # prefetch next batch while training (autotune prefetch buffer size)\n return dataset\n\ndef get_validation_dataset(filenames, ordered=True, repeated=False, batch_size=32, buffer_size=-1):\n dataset = load_dataset(filenames, ordered=ordered, buffer_size=buffer_size)\n if repeated:\n dataset = dataset.repeat()\n dataset = dataset.shuffle(2048)\n dataset = dataset.batch(batch_size, drop_remainder=repeated)\n dataset = dataset.prefetch(buffer_size)\n return dataset\n\ndef get_eval_dataset(filenames, batch_size=32, buffer_size=-1):\n dataset = load_dataset_eval(filenames, buffer_size=buffer_size)\n dataset = dataset.batch(batch_size, drop_remainder=False)\n dataset = dataset.prefetch(buffer_size)\n return dataset\n\n# Test function\ndef read_unlabeled_tfrecord(example, height=config['HEIGHT'], width=config['WIDTH'], channels=config['CHANNELS']):\n example = tf.io.parse_single_example(example, UNLABELED_TFREC_FORMAT)\n image = decode_image(example['image'], height, width, channels)\n image_name = example['image_name']\n # meta features\n data = {}\n data['patient_id'] = tf.cast(example['patient_id'], tf.int32)\n data['sex'] = tf.cast(example['sex'], tf.int32)\n data['age_approx'] = tf.cast(example['age_approx'], tf.int32)\n data['anatom_site_general_challenge'] = tf.cast(tf.one_hot(example['anatom_site_general_challenge'], 7), tf.int32)\n \n return {'input_image': image, 'input_tabular': data}, image_name # returns a dataset of (image, data, image_name)\n\ndef load_dataset_test(filenames, buffer_size=-1):\n dataset = tf.data.TFRecordDataset(filenames, num_parallel_reads=buffer_size) # automatically interleaves reads from multiple files\n dataset = dataset.map(read_unlabeled_tfrecord, num_parallel_calls=buffer_size)\n # returns a dataset of (image, data, label, image_name) pairs if labeled=True or (image, data, image_name) pairs if labeled=False\n return dataset\n\ndef get_test_dataset(filenames, batch_size=32, buffer_size=-1, tta=False):\n dataset = load_dataset_test(filenames, buffer_size=buffer_size)\n if tta:\n dataset = dataset.map(data_augment, num_parallel_calls=AUTO)\n dataset = dataset.batch(batch_size, drop_remainder=False)\n dataset = dataset.prefetch(buffer_size)\n return dataset\n\n# Advanced augmentations\ndef transform_rotation(image, height, rotation):\n # input image - is one image of size [dim,dim,3] not a batch of [b,dim,dim,3]\n # output - image randomly rotated\n DIM = height\n XDIM = DIM%2 #fix for size 331\n \n rotation = rotation * tf.random.normal([1],dtype='float32')\n # CONVERT DEGREES TO RADIANS\n rotation = math.pi * rotation / 180.\n \n # ROTATION MATRIX\n c1 = tf.math.cos(rotation)\n s1 = tf.math.sin(rotation)\n one = tf.constant([1],dtype='float32')\n zero = tf.constant([0],dtype='float32')\n rotation_matrix = tf.reshape( tf.concat([c1,s1,zero, -s1,c1,zero, zero,zero,one],axis=0),[3,3] )\n\n # LIST DESTINATION PIXEL INDICES\n x = tf.repeat( tf.range(DIM//2,-DIM//2,-1), DIM )\n y = tf.tile( tf.range(-DIM//2,DIM//2),[DIM] )\n z = tf.ones([DIM*DIM],dtype='int32')\n idx = tf.stack( [x,y,z] )\n \n # ROTATE DESTINATION PIXELS ONTO ORIGIN PIXELS\n idx2 = K.dot(rotation_matrix,tf.cast(idx,dtype='float32'))\n idx2 = K.cast(idx2,dtype='int32')\n idx2 = K.clip(idx2,-DIM//2+XDIM+1,DIM//2)\n \n # FIND ORIGIN PIXEL VALUES \n idx3 = tf.stack( [DIM//2-idx2[0,], DIM//2-1+idx2[1,]] )\n d = tf.gather_nd(image, tf.transpose(idx3))\n \n return tf.reshape(d,[DIM,DIM,3])\n\ndef transform_shear(image, height, shear):\n # input image - is one image of size [dim,dim,3] not a batch of [b,dim,dim,3]\n # output - image randomly sheared\n DIM = height\n XDIM = DIM%2 #fix for size 331\n \n shear = shear * tf.random.normal([1],dtype='float32')\n shear = math.pi * shear / 180.\n \n # SHEAR MATRIX\n one = tf.constant([1],dtype='float32')\n zero = tf.constant([0],dtype='float32')\n c2 = tf.math.cos(shear)\n s2 = tf.math.sin(shear)\n shear_matrix = tf.reshape( tf.concat([one,s2,zero, zero,c2,zero, zero,zero,one],axis=0),[3,3] ) \n\n # LIST DESTINATION PIXEL INDICES\n x = tf.repeat( tf.range(DIM//2,-DIM//2,-1), DIM )\n y = tf.tile( tf.range(-DIM//2,DIM//2),[DIM] )\n z = tf.ones([DIM*DIM],dtype='int32')\n idx = tf.stack( [x,y,z] )\n \n # ROTATE DESTINATION PIXELS ONTO ORIGIN PIXELS\n idx2 = K.dot(shear_matrix,tf.cast(idx,dtype='float32'))\n idx2 = K.cast(idx2,dtype='int32')\n idx2 = K.clip(idx2,-DIM//2+XDIM+1,DIM//2)\n \n # FIND ORIGIN PIXEL VALUES \n idx3 = tf.stack( [DIM//2-idx2[0,], DIM//2-1+idx2[1,]] )\n d = tf.gather_nd(image, tf.transpose(idx3))\n \n return tf.reshape(d,[DIM,DIM,3])\n\ndef transform_shift(image, height, h_shift, w_shift):\n # input image - is one image of size [dim,dim,3] not a batch of [b,dim,dim,3]\n # output - image randomly shifted\n DIM = height\n XDIM = DIM%2 #fix for size 331\n \n height_shift = h_shift * tf.random.normal([1],dtype='float32') \n width_shift = w_shift * tf.random.normal([1],dtype='float32') \n one = tf.constant([1],dtype='float32')\n zero = tf.constant([0],dtype='float32')\n \n # SHIFT MATRIX\n shift_matrix = tf.reshape( tf.concat([one,zero,height_shift, zero,one,width_shift, zero,zero,one],axis=0),[3,3] )\n\n # LIST DESTINATION PIXEL INDICES\n x = tf.repeat( tf.range(DIM//2,-DIM//2,-1), DIM )\n y = tf.tile( tf.range(-DIM//2,DIM//2),[DIM] )\n z = tf.ones([DIM*DIM],dtype='int32')\n idx = tf.stack( [x,y,z] )\n \n # ROTATE DESTINATION PIXELS ONTO ORIGIN PIXELS\n idx2 = K.dot(shift_matrix,tf.cast(idx,dtype='float32'))\n idx2 = K.cast(idx2,dtype='int32')\n idx2 = K.clip(idx2,-DIM//2+XDIM+1,DIM//2)\n \n # FIND ORIGIN PIXEL VALUES \n idx3 = tf.stack( [DIM//2-idx2[0,], DIM//2-1+idx2[1,]] )\n d = tf.gather_nd(image, tf.transpose(idx3))\n \n return tf.reshape(d,[DIM,DIM,3])\n\ndef transform_zoom(image, height, h_zoom, w_zoom):\n # input image - is one image of size [dim,dim,3] not a batch of [b,dim,dim,3]\n # output - image randomly zoomed\n DIM = height\n XDIM = DIM%2 #fix for size 331\n \n height_zoom = 1.0 + tf.random.normal([1],dtype='float32')/h_zoom\n width_zoom = 1.0 + tf.random.normal([1],dtype='float32')/w_zoom\n one = tf.constant([1],dtype='float32')\n zero = tf.constant([0],dtype='float32')\n \n # ZOOM MATRIX\n zoom_matrix = tf.reshape( tf.concat([one/height_zoom,zero,zero, zero,one/width_zoom,zero, zero,zero,one],axis=0),[3,3] )\n\n # LIST DESTINATION PIXEL INDICES\n x = tf.repeat( tf.range(DIM//2,-DIM//2,-1), DIM )\n y = tf.tile( tf.range(-DIM//2,DIM//2),[DIM] )\n z = tf.ones([DIM*DIM],dtype='int32')\n idx = tf.stack( [x,y,z] )\n \n # ROTATE DESTINATION PIXELS ONTO ORIGIN PIXELS\n idx2 = K.dot(zoom_matrix,tf.cast(idx,dtype='float32'))\n idx2 = K.cast(idx2,dtype='int32')\n idx2 = K.clip(idx2,-DIM//2+XDIM+1,DIM//2)\n \n # FIND ORIGIN PIXEL VALUES \n idx3 = tf.stack( [DIM//2-idx2[0,], DIM//2-1+idx2[1,]] )\n d = tf.gather_nd(image, tf.transpose(idx3))\n \n return tf.reshape(d,[DIM,DIM,3])",
"_____no_output_____"
]
],
[
[
"## Learning rate scheduler",
"_____no_output_____"
]
],
[
[
"lr_min = 1e-6\n# lr_start = 0\nlr_max = config['LEARNING_RATE']\nsteps_per_epoch = 24844 // config['BATCH_SIZE']\ntotal_steps = config['EPOCHS'] * steps_per_epoch\nwarmup_steps = steps_per_epoch * 5\n# hold_max_steps = 0\n# step_decay = .8\n# step_size = steps_per_epoch * 1\n\n# rng = [i for i in range(0, total_steps, 32)]\n# y = [step_schedule_with_warmup(tf.cast(x, tf.float32), step_size=step_size, \n# warmup_steps=warmup_steps, hold_max_steps=hold_max_steps, \n# lr_start=lr_start, lr_max=lr_max, step_decay=step_decay) for x in rng]\n\n# sns.set(style=\"whitegrid\")\n# fig, ax = plt.subplots(figsize=(20, 6))\n# plt.plot(rng, y)\n# print(\"Learning rate schedule: {:.3g} to {:.3g} to {:.3g}\".format(y[0], max(y), y[-1]))",
"_____no_output_____"
]
],
[
[
"# Model",
"_____no_output_____"
]
],
[
[
"# Initial bias\npos = len(k_fold[k_fold['target'] == 1])\nneg = len(k_fold[k_fold['target'] == 0])\ninitial_bias = np.log([pos/neg])\nprint('Bias')\nprint(pos)\nprint(neg)\nprint(initial_bias)\n\n# class weights\ntotal = len(k_fold)\nweight_for_0 = (1 / neg)*(total)/2.0 \nweight_for_1 = (1 / pos)*(total)/2.0\n\nclass_weight = {0: weight_for_0, 1: weight_for_1}\nprint('Class weight')\nprint(class_weight)",
"Bias\n584\n32542\n[-4.02038586]\nClass weight\n{0: 0.5089730194825148, 1: 28.36130136986301}\n"
],
[
"def model_fn(input_shape):\n input_image = L.Input(shape=input_shape, name='input_image')\n BaseModel, preprocess_input = Classifiers.get(config['BASE_MODEL'])\n base_model = BaseModel(input_shape=input_shape, \n weights=config['BASE_MODEL_WEIGHTS'], \n include_top=False)\n \n x = base_model(input_image)\n x = L.GlobalAveragePooling2D()(x)\n \n output = L.Dense(1, activation='sigmoid', name='output', \n bias_initializer=tf.keras.initializers.Constant(initial_bias))(x)\n \n model = Model(inputs=input_image, outputs=output)\n \n return model",
"_____no_output_____"
]
],
[
[
"# Training",
"_____no_output_____"
]
],
[
[
"# Evaluation\neval_dataset = get_eval_dataset(TRAINING_FILENAMES, batch_size=config['BATCH_SIZE'], buffer_size=AUTO)\nimage_names = next(iter(eval_dataset.unbatch().map(lambda data, label, image_name: image_name).batch(count_data_items(TRAINING_FILENAMES)))).numpy().astype('U')\nimage_data = eval_dataset.map(lambda data, label, image_name: data)\n# Resample dataframe\nk_fold = k_fold[k_fold['image_name'].isin(image_names)]\n\n# Test\nNUM_TEST_IMAGES = len(test)\ntest_preds = np.zeros((NUM_TEST_IMAGES, 1))\ntest_preds_last = np.zeros((NUM_TEST_IMAGES, 1))\n\ntest_dataset = get_test_dataset(TEST_FILENAMES, batch_size=config['BATCH_SIZE'], buffer_size=AUTO, tta=True)\nimage_names_test = next(iter(test_dataset.unbatch().map(lambda data, image_name: image_name).batch(NUM_TEST_IMAGES))).numpy().astype('U')\ntest_image_data = test_dataset.map(lambda data, image_name: data)\n\n\nhistory_list = []\nk_fold_best = k_fold.copy()\nkfold = KFold(config['N_FOLDS'], shuffle=True, random_state=SEED)\n\nfor n_fold, (trn_idx, val_idx) in enumerate(kfold.split(TRAINING_FILENAMES)):\n if n_fold < config['N_USED_FOLDS']:\n n_fold +=1\n print('\\nFOLD: %d' % (n_fold))\n# tf.tpu.experimental.initialize_tpu_system(tpu)\n K.clear_session()\n\n ### Data\n train_filenames = np.array(TRAINING_FILENAMES)[trn_idx]\n valid_filenames = np.array(TRAINING_FILENAMES)[val_idx]\n steps_per_epoch = count_data_items(train_filenames) // config['BATCH_SIZE']\n\n # Train model\n model_path = f'model_fold_{n_fold}.h5'\n\n es = EarlyStopping(monitor='val_auc', mode='max', patience=config['ES_PATIENCE'], \n restore_best_weights=False, verbose=1)\n checkpoint = ModelCheckpoint(model_path, monitor='val_auc', mode='max', \n save_best_only=True, save_weights_only=True)\n\n with strategy.scope():\n model = model_fn((config['HEIGHT'], config['WIDTH'], config['CHANNELS']))\n\n optimizer = tfa.optimizers.RectifiedAdam(lr=lr_max,\n total_steps=total_steps,\n warmup_proportion=(warmup_steps / total_steps),\n min_lr=lr_min)\n model.compile(optimizer, loss=losses.BinaryCrossentropy(label_smoothing=0.05), \n metrics=[metrics.AUC()])\n\n history = model.fit(get_training_dataset(train_filenames, batch_size=config['BATCH_SIZE'], buffer_size=AUTO),\n validation_data=get_validation_dataset(valid_filenames, ordered=True, repeated=False, \n batch_size=config['BATCH_SIZE'], buffer_size=AUTO),\n epochs=config['EPOCHS'], \n steps_per_epoch=steps_per_epoch,\n callbacks=[checkpoint, es],\n class_weight=class_weight, \n verbose=2).history\n\n # save last epoch weights\n model.save_weights('last_' + model_path)\n\n history_list.append(history)\n\n # Get validation IDs\n valid_dataset = get_eval_dataset(valid_filenames, batch_size=config['BATCH_SIZE'], buffer_size=AUTO)\n valid_image_names = next(iter(valid_dataset.unbatch().map(lambda data, label, image_name: image_name).batch(count_data_items(valid_filenames)))).numpy().astype('U')\n k_fold[f'fold_{n_fold}'] = k_fold.apply(lambda x: 'validation' if x['image_name'] in valid_image_names else 'train', axis=1)\n k_fold_best[f'fold_{n_fold}'] = k_fold_best.apply(lambda x: 'validation' if x['image_name'] in valid_image_names else 'train', axis=1)\n \n ##### Last model #####\n print('Last model evaluation...')\n preds = model.predict(image_data)\n name_preds_eval = dict(zip(image_names, preds.reshape(len(preds))))\n k_fold[f'pred_fold_{n_fold}'] = k_fold.apply(lambda x: name_preds_eval[x['image_name']], axis=1)\n\n print(f'Last model inference (TTA {config[\"TTA_STEPS\"]} steps)...')\n for step in range(config['TTA_STEPS']):\n test_preds_last += model.predict(test_image_data)\n\n ##### Best model #####\n print('Best model evaluation...')\n model.load_weights(model_path)\n preds = model.predict(image_data)\n name_preds_eval = dict(zip(image_names, preds.reshape(len(preds))))\n k_fold_best[f'pred_fold_{n_fold}'] = k_fold_best.apply(lambda x: name_preds_eval[x['image_name']], axis=1)\n\n print(f'Best model inference (TTA {config[\"TTA_STEPS\"]} steps)...')\n for step in range(config['TTA_STEPS']):\n test_preds += model.predict(test_image_data)\n\n\n# normalize preds\ntest_preds /= (config['N_USED_FOLDS'] * config['TTA_STEPS'])\ntest_preds_last /= (config['N_USED_FOLDS'] * config['TTA_STEPS'])\n\nname_preds = dict(zip(image_names_test, test_preds.reshape(NUM_TEST_IMAGES)))\nname_preds_last = dict(zip(image_names_test, test_preds_last.reshape(NUM_TEST_IMAGES)))\n\ntest['target'] = test.apply(lambda x: name_preds[x['image_name']], axis=1)\ntest['target_last'] = test.apply(lambda x: name_preds_last[x['image_name']], axis=1)",
"\nFOLD: 1\nDownloading data from https://github.com/qubvel/classification_models/releases/download/0.0.1/seresnet18_imagenet_1000_no_top.h5\n45359104/45351256 [==============================] - 4s 0us/step\nEpoch 1/25\n408/408 - 147s - loss: 0.7536 - auc: 0.7313 - val_loss: 0.1771 - val_auc: 0.5180\nEpoch 2/25\n408/408 - 144s - loss: 0.5032 - auc: 0.8513 - val_loss: 0.1900 - val_auc: 0.7332\nEpoch 3/25\n408/408 - 145s - loss: 0.5233 - auc: 0.8501 - val_loss: 0.3951 - val_auc: 0.8278\nEpoch 4/25\n408/408 - 146s - loss: 0.5291 - auc: 0.8503 - val_loss: 0.5892 - val_auc: 0.8244\nEpoch 5/25\n408/408 - 146s - loss: 0.5110 - auc: 0.8528 - val_loss: 0.3490 - val_auc: 0.8255\nEpoch 6/25\n408/408 - 139s - loss: 0.4682 - auc: 0.8814 - val_loss: 0.6515 - val_auc: 0.8535\nEpoch 7/25\n408/408 - 142s - loss: 0.4610 - auc: 0.8793 - val_loss: 0.3841 - val_auc: 0.8673\nEpoch 8/25\n408/408 - 141s - loss: 0.4458 - auc: 0.8894 - val_loss: 0.3039 - val_auc: 0.8412\nEpoch 9/25\n408/408 - 141s - loss: 0.4535 - auc: 0.8867 - val_loss: 0.5310 - val_auc: 0.8819\nEpoch 10/25\n408/408 - 142s - loss: 0.4385 - auc: 0.8951 - val_loss: 0.6995 - val_auc: 0.8433\nEpoch 11/25\n408/408 - 141s - loss: 0.4285 - auc: 0.8992 - val_loss: 0.3153 - val_auc: 0.8760\nEpoch 12/25\n408/408 - 139s - loss: 0.4083 - auc: 0.9143 - val_loss: 0.5465 - val_auc: 0.8659\nEpoch 13/25\n408/408 - 142s - loss: 0.4043 - auc: 0.9146 - val_loss: 0.2805 - val_auc: 0.8740\nEpoch 14/25\n408/408 - 141s - loss: 0.3886 - auc: 0.9253 - val_loss: 0.4161 - val_auc: 0.8632\nEpoch 15/25\n408/408 - 143s - loss: 0.3700 - auc: 0.9338 - val_loss: 0.4457 - val_auc: 0.8666\nEpoch 16/25\n408/408 - 141s - loss: 0.3674 - auc: 0.9356 - val_loss: 0.3412 - val_auc: 0.8839\nEpoch 17/25\n408/408 - 140s - loss: 0.3375 - auc: 0.9480 - val_loss: 0.3924 - val_auc: 0.8703\nEpoch 18/25\n408/408 - 142s - loss: 0.3237 - auc: 0.9544 - val_loss: 0.2758 - val_auc: 0.8552\nEpoch 19/25\n408/408 - 143s - loss: 0.3311 - auc: 0.9535 - val_loss: 0.3325 - val_auc: 0.8610\nEpoch 20/25\n408/408 - 140s - loss: 0.2902 - auc: 0.9680 - val_loss: 0.2184 - val_auc: 0.8552\nEpoch 21/25\n408/408 - 145s - loss: 0.2846 - auc: 0.9694 - val_loss: 0.2595 - val_auc: 0.8624\nEpoch 22/25\n408/408 - 143s - loss: 0.2552 - auc: 0.9785 - val_loss: 0.2535 - val_auc: 0.8693\nEpoch 23/25\n408/408 - 145s - loss: 0.2528 - auc: 0.9791 - val_loss: 0.2828 - val_auc: 0.8782\nEpoch 24/25\n408/408 - 143s - loss: 0.2380 - auc: 0.9839 - val_loss: 0.2587 - val_auc: 0.8695\nEpoch 25/25\n408/408 - 140s - loss: 0.2331 - auc: 0.9850 - val_loss: 0.2575 - val_auc: 0.8700\nLast model evaluation...\nLast model inference (TTA 25 steps)...\nBest model evaluation...\nBest model inference (TTA 25 steps)...\n\nFOLD: 2\nEpoch 1/25\n408/408 - 145s - loss: 1.4535 - auc: 0.6980 - val_loss: 0.1884 - val_auc: 0.4463\nEpoch 2/25\n408/408 - 143s - loss: 0.5506 - auc: 0.8422 - val_loss: 0.1974 - val_auc: 0.7122\nEpoch 3/25\n408/408 - 143s - loss: 0.5113 - auc: 0.8634 - val_loss: 0.2947 - val_auc: 0.8525\nEpoch 4/25\n408/408 - 144s - loss: 0.5339 - auc: 0.8479 - val_loss: 0.6366 - val_auc: 0.7912\nEpoch 5/25\n408/408 - 139s - loss: 0.5006 - auc: 0.8572 - val_loss: 0.7557 - val_auc: 0.8531\nEpoch 6/25\n408/408 - 140s - loss: 0.4760 - auc: 0.8725 - val_loss: 0.2821 - val_auc: 0.8140\nEpoch 7/25\n408/408 - 146s - loss: 0.4729 - auc: 0.8781 - val_loss: 0.4087 - val_auc: 0.8631\nEpoch 8/25\n408/408 - 139s - loss: 0.4344 - auc: 0.9005 - val_loss: 0.3831 - val_auc: 0.8518\nEpoch 9/25\n408/408 - 138s - loss: 0.4148 - auc: 0.9081 - val_loss: 0.3328 - val_auc: 0.8447\nEpoch 10/25\n408/408 - 139s - loss: 0.4234 - auc: 0.9058 - val_loss: 0.2685 - val_auc: 0.8798\nEpoch 11/25\n408/408 - 142s - loss: 0.4121 - auc: 0.9129 - val_loss: 0.3698 - val_auc: 0.8625\nEpoch 12/25\n408/408 - 139s - loss: 0.4006 - auc: 0.9194 - val_loss: 0.3287 - val_auc: 0.8915\nEpoch 13/25\n408/408 - 139s - loss: 0.3891 - auc: 0.9254 - val_loss: 0.6094 - val_auc: 0.8477\nEpoch 14/25\n408/408 - 141s - loss: 0.3815 - auc: 0.9255 - val_loss: 0.3927 - val_auc: 0.8242\nEpoch 15/25\n408/408 - 139s - loss: 0.3694 - auc: 0.9361 - val_loss: 0.3596 - val_auc: 0.8811\nEpoch 16/25\n408/408 - 139s - loss: 0.3507 - auc: 0.9424 - val_loss: 0.3919 - val_auc: 0.8972\nEpoch 17/25\n408/408 - 139s - loss: 0.3349 - auc: 0.9512 - val_loss: 0.3917 - val_auc: 0.8796\nEpoch 18/25\n408/408 - 138s - loss: 0.3244 - auc: 0.9568 - val_loss: 0.4386 - val_auc: 0.8936\nEpoch 19/25\n408/408 - 140s - loss: 0.3032 - auc: 0.9635 - val_loss: 0.2599 - val_auc: 0.8978\nEpoch 20/25\n408/408 - 139s - loss: 0.2978 - auc: 0.9653 - val_loss: 0.2680 - val_auc: 0.8872\nEpoch 21/25\n408/408 - 141s - loss: 0.2585 - auc: 0.9775 - val_loss: 0.3013 - val_auc: 0.8902\nEpoch 22/25\n408/408 - 139s - loss: 0.2553 - auc: 0.9792 - val_loss: 0.2838 - val_auc: 0.9049\nEpoch 23/25\n408/408 - 141s - loss: 0.2520 - auc: 0.9792 - val_loss: 0.2890 - val_auc: 0.9038\nEpoch 24/25\n408/408 - 140s - loss: 0.2374 - auc: 0.9841 - val_loss: 0.2680 - val_auc: 0.9005\nEpoch 25/25\n408/408 - 139s - loss: 0.2374 - auc: 0.9843 - val_loss: 0.2697 - val_auc: 0.9005\nLast model evaluation...\nLast model inference (TTA 25 steps)...\nBest model evaluation...\nBest model inference (TTA 25 steps)...\n\nFOLD: 3\nEpoch 1/25\n408/408 - 149s - loss: 1.1720 - auc: 0.7271 - val_loss: 0.1744 - val_auc: 0.6360\nEpoch 2/25\n408/408 - 149s - loss: 0.5507 - auc: 0.8441 - val_loss: 0.1939 - val_auc: 0.6844\nEpoch 3/25\n408/408 - 148s - loss: 0.5058 - auc: 0.8589 - val_loss: 0.3885 - val_auc: 0.8607\nEpoch 4/25\n408/408 - 150s - loss: 0.5285 - auc: 0.8505 - val_loss: 0.4079 - val_auc: 0.8370\nEpoch 5/25\n408/408 - 149s - loss: 0.4942 - auc: 0.8637 - val_loss: 0.5155 - val_auc: 0.8279\nEpoch 6/25\n408/408 - 150s - loss: 0.4860 - auc: 0.8649 - val_loss: 0.5217 - val_auc: 0.8420\nEpoch 7/25\n408/408 - 145s - loss: 0.4461 - auc: 0.8891 - val_loss: 0.5477 - val_auc: 0.7847\nEpoch 8/25\n408/408 - 148s - loss: 0.4492 - auc: 0.8907 - val_loss: 0.4068 - val_auc: 0.8478\nEpoch 9/25\n408/408 - 145s - loss: 0.4383 - auc: 0.8924 - val_loss: 0.3346 - val_auc: 0.8607\nEpoch 10/25\n408/408 - 148s - loss: 0.4140 - auc: 0.9097 - val_loss: 0.4308 - val_auc: 0.8583\nEpoch 11/25\n408/408 - 143s - loss: 0.4326 - auc: 0.9001 - val_loss: 0.3784 - val_auc: 0.8515\nEpoch 12/25\n408/408 - 145s - loss: 0.4062 - auc: 0.9162 - val_loss: 0.2820 - val_auc: 0.8529\nEpoch 13/25\n408/408 - 152s - loss: 0.3969 - auc: 0.9224 - val_loss: 0.5535 - val_auc: 0.8801\nEpoch 14/25\n408/408 - 147s - loss: 0.3705 - auc: 0.9303 - val_loss: 0.3018 - val_auc: 0.8591\nEpoch 15/25\n408/408 - 150s - loss: 0.3705 - auc: 0.9347 - val_loss: 0.4215 - val_auc: 0.8524\nEpoch 16/25\n408/408 - 143s - loss: 0.3468 - auc: 0.9420 - val_loss: 0.3081 - val_auc: 0.8652\nEpoch 17/25\n408/408 - 150s - loss: 0.3313 - auc: 0.9511 - val_loss: 0.2978 - val_auc: 0.8725\nEpoch 18/25\n408/408 - 146s - loss: 0.3187 - auc: 0.9567 - val_loss: 0.2542 - val_auc: 0.8754\nEpoch 19/25\n408/408 - 150s - loss: 0.2957 - auc: 0.9648 - val_loss: 0.2455 - val_auc: 0.8672\nEpoch 20/25\n408/408 - 145s - loss: 0.2855 - auc: 0.9697 - val_loss: 0.2919 - val_auc: 0.8630\nEpoch 21/25\n408/408 - 148s - loss: 0.2806 - auc: 0.9714 - val_loss: 0.2798 - val_auc: 0.8683\nEpoch 22/25\n408/408 - 145s - loss: 0.2517 - auc: 0.9785 - val_loss: 0.3469 - val_auc: 0.8822\nEpoch 23/25\n408/408 - 148s - loss: 0.2355 - auc: 0.9841 - val_loss: 0.3256 - val_auc: 0.8883\nEpoch 24/25\n408/408 - 147s - loss: 0.2316 - auc: 0.9851 - val_loss: 0.2705 - val_auc: 0.8842\nEpoch 25/25\n408/408 - 143s - loss: 0.2232 - auc: 0.9873 - val_loss: 0.2706 - val_auc: 0.8848\nLast model evaluation...\nLast model inference (TTA 25 steps)...\nBest model evaluation...\nBest model inference (TTA 25 steps)...\n\nFOLD: 4\nEpoch 1/25\n408/408 - 150s - loss: 1.3047 - auc: 0.7255 - val_loss: 0.2129 - val_auc: 0.3756\nEpoch 2/25\n408/408 - 149s - loss: 0.5269 - auc: 0.8519 - val_loss: 0.2027 - val_auc: 0.6503\nEpoch 3/25\n408/408 - 148s - loss: 0.5088 - auc: 0.8602 - val_loss: 0.3177 - val_auc: 0.8657\nEpoch 4/25\n408/408 - 146s - loss: 0.4965 - auc: 0.8637 - val_loss: 0.3331 - val_auc: 0.8419\nEpoch 5/25\n408/408 - 148s - loss: 0.5099 - auc: 0.8562 - val_loss: 0.3309 - val_auc: 0.8765\nEpoch 6/25\n408/408 - 140s - loss: 0.4817 - auc: 0.8662 - val_loss: 1.0217 - val_auc: 0.8450\nEpoch 7/25\n408/408 - 143s - loss: 0.4676 - auc: 0.8770 - val_loss: 0.3531 - val_auc: 0.8796\nEpoch 8/25\n408/408 - 141s - loss: 0.4693 - auc: 0.8751 - val_loss: 0.5131 - val_auc: 0.9037\nEpoch 9/25\n408/408 - 142s - loss: 0.4275 - auc: 0.8990 - val_loss: 0.3979 - val_auc: 0.8893\nEpoch 10/25\n408/408 - 142s - loss: 0.4374 - auc: 0.8914 - val_loss: 0.8738 - val_auc: 0.8437\nEpoch 11/25\n408/408 - 142s - loss: 0.4223 - auc: 0.9046 - val_loss: 0.3543 - val_auc: 0.8808\nEpoch 12/25\n408/408 - 142s - loss: 0.4182 - auc: 0.9067 - val_loss: 0.3024 - val_auc: 0.8862\nEpoch 13/25\n408/408 - 144s - loss: 0.4118 - auc: 0.9144 - val_loss: 1.0908 - val_auc: 0.8471\nEpoch 14/25\n408/408 - 143s - loss: 0.4006 - auc: 0.9190 - val_loss: 0.4312 - val_auc: 0.8784\nEpoch 15/25\n408/408 - 142s - loss: 0.3657 - auc: 0.9344 - val_loss: 0.2889 - val_auc: 0.8589\nEpoch 16/25\n408/408 - 142s - loss: 0.3656 - auc: 0.9353 - val_loss: 0.4130 - val_auc: 0.8836\nEpoch 17/25\n408/408 - 142s - loss: 0.3435 - auc: 0.9453 - val_loss: 0.3969 - val_auc: 0.8719\nEpoch 18/25\n408/408 - 142s - loss: 0.3292 - auc: 0.9503 - val_loss: 0.5234 - val_auc: 0.8915\nEpoch 00018: early stopping\nLast model evaluation...\nLast model inference (TTA 25 steps)...\nBest model evaluation...\nBest model inference (TTA 25 steps)...\n\nFOLD: 5\nEpoch 1/25\n408/408 - 138s - loss: 1.1062 - auc: 0.7312 - val_loss: 0.1750 - val_auc: 0.4583\nEpoch 2/25\n408/408 - 138s - loss: 0.5368 - auc: 0.8487 - val_loss: 0.2057 - val_auc: 0.5969\nEpoch 3/25\n408/408 - 141s - loss: 0.5105 - auc: 0.8574 - val_loss: 0.5717 - val_auc: 0.8518\nEpoch 4/25\n408/408 - 140s - loss: 0.5071 - auc: 0.8597 - val_loss: 0.4976 - val_auc: 0.8366\nEpoch 5/25\n408/408 - 146s - loss: 0.5105 - auc: 0.8546 - val_loss: 0.3420 - val_auc: 0.8437\nEpoch 6/25\n408/408 - 140s - loss: 0.4721 - auc: 0.8772 - val_loss: 0.3763 - val_auc: 0.8288\nEpoch 7/25\n408/408 - 141s - loss: 0.4629 - auc: 0.8822 - val_loss: 0.4596 - val_auc: 0.8585\nEpoch 8/25\n408/408 - 143s - loss: 0.4467 - auc: 0.8933 - val_loss: 0.5264 - val_auc: 0.8700\nEpoch 9/25\n408/408 - 142s - loss: 0.4328 - auc: 0.8978 - val_loss: 0.4177 - val_auc: 0.8681\nEpoch 10/25\n408/408 - 142s - loss: 0.4295 - auc: 0.9040 - val_loss: 0.2681 - val_auc: 0.8794\nEpoch 11/25\n408/408 - 143s - loss: 0.4001 - auc: 0.9158 - val_loss: 0.3823 - val_auc: 0.8509\nEpoch 12/25\n408/408 - 141s - loss: 0.4058 - auc: 0.9154 - val_loss: 0.3573 - val_auc: 0.8603\nEpoch 13/25\n408/408 - 141s - loss: 0.3936 - auc: 0.9226 - val_loss: 0.4060 - val_auc: 0.8714\nEpoch 14/25\n408/408 - 142s - loss: 0.3788 - auc: 0.9292 - val_loss: 0.5670 - val_auc: 0.8686\nEpoch 15/25\n408/408 - 141s - loss: 0.3713 - auc: 0.9336 - val_loss: 0.5811 - val_auc: 0.8503\nEpoch 16/25\n408/408 - 143s - loss: 0.3567 - auc: 0.9418 - val_loss: 0.2765 - val_auc: 0.8795\nEpoch 17/25\n408/408 - 142s - loss: 0.3378 - auc: 0.9480 - val_loss: 0.5983 - val_auc: 0.8812\nEpoch 18/25\n408/408 - 141s - loss: 0.3140 - auc: 0.9601 - val_loss: 0.2750 - val_auc: 0.8484\nEpoch 19/25\n408/408 - 142s - loss: 0.3175 - auc: 0.9590 - val_loss: 0.3299 - val_auc: 0.8727\nEpoch 20/25\n408/408 - 141s - loss: 0.2859 - auc: 0.9698 - val_loss: 0.3239 - val_auc: 0.8892\nEpoch 21/25\n408/408 - 143s - loss: 0.2695 - auc: 0.9745 - val_loss: 0.2528 - val_auc: 0.8865\nEpoch 22/25\n408/408 - 144s - loss: 0.2526 - auc: 0.9795 - val_loss: 0.2476 - val_auc: 0.8705\nEpoch 23/25\n408/408 - 141s - loss: 0.2444 - auc: 0.9823 - val_loss: 0.2859 - val_auc: 0.8829\nEpoch 24/25\n408/408 - 143s - loss: 0.2326 - auc: 0.9857 - val_loss: 0.2601 - val_auc: 0.8804\nEpoch 25/25\n408/408 - 140s - loss: 0.2301 - auc: 0.9853 - val_loss: 0.2612 - val_auc: 0.8816\nLast model evaluation...\nLast model inference (TTA 25 steps)...\nBest model evaluation...\nBest model inference (TTA 25 steps)...\n"
]
],
[
[
"## Model loss graph",
"_____no_output_____"
]
],
[
[
"for n_fold in range(config['N_USED_FOLDS']):\n print(f'Fold: {n_fold + 1}')\n plot_metrics(history_list[n_fold])",
"Fold: 1\n"
]
],
[
[
"## Model loss graph aggregated",
"_____no_output_____"
]
],
[
[
"plot_metrics_agg(history_list, config['N_USED_FOLDS'])",
"_____no_output_____"
]
],
[
[
"# Model evaluation (best)",
"_____no_output_____"
]
],
[
[
"display(evaluate_model(k_fold_best, config['N_USED_FOLDS']).style.applymap(color_map))\ndisplay(evaluate_model_Subset(k_fold_best, config['N_USED_FOLDS']).style.applymap(color_map))",
"_____no_output_____"
]
],
[
[
"# Model evaluation (last)",
"_____no_output_____"
]
],
[
[
"display(evaluate_model(k_fold, config['N_USED_FOLDS']).style.applymap(color_map))\ndisplay(evaluate_model_Subset(k_fold, config['N_USED_FOLDS']).style.applymap(color_map))",
"_____no_output_____"
]
],
[
[
"# Confusion matrix",
"_____no_output_____"
]
],
[
[
"for n_fold in range(config['N_USED_FOLDS']):\n n_fold += 1\n pred_col = f'pred_fold_{n_fold}' \n train_set = k_fold_best[k_fold_best[f'fold_{n_fold}'] == 'train']\n valid_set = k_fold_best[k_fold_best[f'fold_{n_fold}'] == 'validation'] \n print(f'Fold: {n_fold}')\n plot_confusion_matrix(train_set['target'], np.round(train_set[pred_col]),\n valid_set['target'], np.round(valid_set[pred_col]))",
"Fold: 1\n"
]
],
[
[
"# Visualize predictions",
"_____no_output_____"
]
],
[
[
"k_fold['pred'] = 0\nfor n_fold in range(config['N_USED_FOLDS']):\n k_fold['pred'] += k_fold[f'pred_fold_{n_fold+1}'] / config['N_FOLDS']\n\nprint('Label/prediction distribution')\nprint(f\"Train positive labels: {len(k_fold[k_fold['target'] > .5])}\")\nprint(f\"Train positive predictions: {len(k_fold[k_fold['pred'] > .5])}\")\nprint(f\"Train positive correct predictions: {len(k_fold[(k_fold['target'] > .5) & (k_fold['pred'] > .5)])}\")\n \nprint('Top 10 samples')\ndisplay(k_fold[['image_name', 'sex', 'age_approx','anatom_site_general_challenge', 'diagnosis',\n 'target', 'pred'] + [c for c in k_fold.columns if (c.startswith('pred_fold'))]].head(10))\n\nprint('Top 10 positive samples')\ndisplay(k_fold[['image_name', 'sex', 'age_approx','anatom_site_general_challenge', 'diagnosis',\n 'target', 'pred'] + [c for c in k_fold.columns if (c.startswith('pred_fold'))]].query('target == 1').head(10))\n\nprint('Top 10 predicted positive samples')\ndisplay(k_fold[['image_name', 'sex', 'age_approx','anatom_site_general_challenge', 'diagnosis',\n 'target', 'pred'] + [c for c in k_fold.columns if (c.startswith('pred_fold'))]].query('pred > .5').head(10))",
"Label/prediction distribution\nTrain positive labels: 581\nTrain positive predictions: 2647\nTrain positive correct predictions: 578\nTop 10 samples\n"
]
],
[
[
"# Visualize test predictions",
"_____no_output_____"
]
],
[
[
"print(f\"Test predictions {len(test[test['target'] > .5])}|{len(test[test['target'] <= .5])}\")\nprint(f\"Test predictions (last) {len(test[test['target_last'] > .5])}|{len(test[test['target_last'] <= .5])}\")\n\nprint('Top 10 samples')\ndisplay(test[['image_name', 'sex', 'age_approx','anatom_site_general_challenge', 'target', 'target_last'] + \n [c for c in test.columns if (c.startswith('pred_fold'))]].head(10))\n\nprint('Top 10 positive samples')\ndisplay(test[['image_name', 'sex', 'age_approx','anatom_site_general_challenge', 'target', 'target_last'] + \n [c for c in test.columns if (c.startswith('pred_fold'))]].query('target > .5').head(10))\n\nprint('Top 10 positive samples (last)')\ndisplay(test[['image_name', 'sex', 'age_approx','anatom_site_general_challenge', 'target', 'target_last'] + \n [c for c in test.columns if (c.startswith('pred_fold'))]].query('target_last > .5').head(10))",
"Test predictions 1506|9476\nTest predictions (last) 1172|9810\nTop 10 samples\n"
]
],
[
[
"# Test set predictions",
"_____no_output_____"
]
],
[
[
"submission = pd.read_csv(database_base_path + 'sample_submission.csv')\nsubmission['target'] = test['target']\nsubmission['target_last'] = test['target_last']\nsubmission['target_blend'] = (test['target'] * .5) + (test['target_last'] * .5)\ndisplay(submission.head(10))\ndisplay(submission.describe())\n\n### BEST ###\nsubmission[['image_name', 'target']].to_csv('submission.csv', index=False)\n\n### LAST ###\nsubmission_last = submission[['image_name', 'target_last']]\nsubmission_last.columns = ['image_name', 'target']\nsubmission_last.to_csv('submission_last.csv', index=False)\n\n### BLEND ###\nsubmission_blend = submission[['image_name', 'target_blend']]\nsubmission_blend.columns = ['image_name', 'target']\nsubmission_blend.to_csv('submission_blend.csv', index=False)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d03ccb5c1a0a3ab1e162e79a2e87700e745408ed | 15,701 | ipynb | Jupyter Notebook | docs/tutorials/cta_data_analysis.ipynb | Jaleleddine/gammapy | de9195df40fa5bbf8840cda4e7cd5e8cc5eaadbb | [
"BSD-3-Clause"
] | null | null | null | docs/tutorials/cta_data_analysis.ipynb | Jaleleddine/gammapy | de9195df40fa5bbf8840cda4e7cd5e8cc5eaadbb | [
"BSD-3-Clause"
] | null | null | null | docs/tutorials/cta_data_analysis.ipynb | Jaleleddine/gammapy | de9195df40fa5bbf8840cda4e7cd5e8cc5eaadbb | [
"BSD-3-Clause"
] | null | null | null | 26.34396 | 247 | 0.568817 | [
[
[
"# CTA data analysis with Gammapy\n\n## Introduction\n\n**This notebook shows an example how to make a sky image and spectrum for simulated CTA data with Gammapy.**\n\nThe dataset we will use is three observation runs on the Galactic center. This is a tiny (and thus quick to process and play with and learn) subset of the simulated CTA dataset that was produced for the first data challenge in August 2017.\n",
"_____no_output_____"
],
[
"## Setup\n\nAs usual, we'll start with some setup ...",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"!gammapy info --no-envvar --no-system",
"_____no_output_____"
],
[
"import numpy as np\nimport astropy.units as u\nfrom astropy.coordinates import SkyCoord\nfrom astropy.convolution import Gaussian2DKernel\nfrom regions import CircleSkyRegion\nfrom gammapy.modeling import Fit\nfrom gammapy.data import DataStore\nfrom gammapy.datasets import (\n Datasets,\n FluxPointsDataset,\n SpectrumDataset,\n MapDataset,\n)\nfrom gammapy.modeling.models import (\n PowerLawSpectralModel,\n SkyModel,\n GaussianSpatialModel,\n)\nfrom gammapy.maps import MapAxis, WcsNDMap, WcsGeom, RegionGeom\nfrom gammapy.makers import (\n MapDatasetMaker,\n SafeMaskMaker,\n SpectrumDatasetMaker,\n ReflectedRegionsBackgroundMaker,\n)\nfrom gammapy.estimators import TSMapEstimator, FluxPointsEstimator\nfrom gammapy.estimators.utils import find_peaks\nfrom gammapy.visualization import plot_spectrum_datasets_off_regions",
"_____no_output_____"
],
[
"# Configure the logger, so that the spectral analysis\n# isn't so chatty about what it's doing.\nimport logging\n\nlogging.basicConfig()\nlog = logging.getLogger(\"gammapy.spectrum\")\nlog.setLevel(logging.ERROR)",
"_____no_output_____"
]
],
[
[
"## Select observations\n\nA Gammapy analysis usually starts by creating a `~gammapy.data.DataStore` and selecting observations.\n\nThis is shown in detail in the other notebook, here we just pick three observations near the galactic center.",
"_____no_output_____"
]
],
[
[
"data_store = DataStore.from_dir(\"$GAMMAPY_DATA/cta-1dc/index/gps\")",
"_____no_output_____"
],
[
"# Just as a reminder: this is how to select observations\n# from astropy.coordinates import SkyCoord\n# table = data_store.obs_table\n# pos_obs = SkyCoord(table['GLON_PNT'], table['GLAT_PNT'], frame='galactic', unit='deg')\n# pos_target = SkyCoord(0, 0, frame='galactic', unit='deg')\n# offset = pos_target.separation(pos_obs).deg\n# mask = (1 < offset) & (offset < 2)\n# table = table[mask]\n# table.show_in_browser(jsviewer=True)",
"_____no_output_____"
],
[
"obs_id = [110380, 111140, 111159]\nobservations = data_store.get_observations(obs_id)",
"_____no_output_____"
],
[
"obs_cols = [\"OBS_ID\", \"GLON_PNT\", \"GLAT_PNT\", \"LIVETIME\"]\ndata_store.obs_table.select_obs_id(obs_id)[obs_cols]",
"_____no_output_____"
]
],
[
[
"## Make sky images\n\n### Define map geometry\n\nSelect the target position and define an ON region for the spectral analysis",
"_____no_output_____"
]
],
[
[
"axis = MapAxis.from_edges(\n np.logspace(-1.0, 1.0, 10), unit=\"TeV\", name=\"energy\", interp=\"log\"\n)\ngeom = WcsGeom.create(\n skydir=(0, 0), npix=(500, 400), binsz=0.02, frame=\"galactic\", axes=[axis]\n)\ngeom",
"_____no_output_____"
]
],
[
[
"### Compute images\n\nExclusion mask currently unused. Remove here or move to later in the tutorial?",
"_____no_output_____"
]
],
[
[
"target_position = SkyCoord(0, 0, unit=\"deg\", frame=\"galactic\")\non_radius = 0.2 * u.deg\non_region = CircleSkyRegion(center=target_position, radius=on_radius)",
"_____no_output_____"
],
[
"exclusion_mask = geom.to_image().region_mask([on_region], inside=False)\nexclusion_mask = WcsNDMap(geom.to_image(), exclusion_mask)\nexclusion_mask.plot();",
"_____no_output_____"
],
[
"%%time\nstacked = MapDataset.create(geom=geom)\nstacked.edisp = None\nmaker = MapDatasetMaker(selection=[\"counts\", \"background\", \"exposure\", \"psf\"])\nmaker_safe_mask = SafeMaskMaker(methods=[\"offset-max\"], offset_max=2.5 * u.deg)\n\nfor obs in observations:\n cutout = stacked.cutout(obs.pointing_radec, width=\"5 deg\")\n dataset = maker.run(cutout, obs)\n dataset = maker_safe_mask.run(dataset, obs)\n stacked.stack(dataset)",
"_____no_output_____"
],
[
"# The maps are cubes, with an energy axis.\n# Let's also make some images:\ndataset_image = stacked.to_image()",
"_____no_output_____"
]
],
[
[
"### Show images\n\nLet's have a quick look at the images we computed ...",
"_____no_output_____"
]
],
[
[
"dataset_image.counts.smooth(2).plot(vmax=5);",
"_____no_output_____"
],
[
"dataset_image.background.plot(vmax=5);",
"_____no_output_____"
],
[
"dataset_image.excess.smooth(3).plot(vmax=2);",
"_____no_output_____"
]
],
[
[
"## Source Detection\n\nUse the class `~gammapy.estimators.TSMapEstimator` and function `gammapy.estimators.utils.find_peaks` to detect sources on the images. We search for 0.1 deg sigma gaussian sources in the dataset.",
"_____no_output_____"
]
],
[
[
"spatial_model = GaussianSpatialModel(sigma=\"0.05 deg\")\nspectral_model = PowerLawSpectralModel(index=2)\nmodel = SkyModel(spatial_model=spatial_model, spectral_model=spectral_model)",
"_____no_output_____"
],
[
"ts_image_estimator = TSMapEstimator(\n model,\n kernel_width=\"0.5 deg\",\n selection_optional=[],\n downsampling_factor=2,\n sum_over_energy_groups=False,\n energy_edges=[0.1, 10] * u.TeV,\n)",
"_____no_output_____"
],
[
"%%time\nimages_ts = ts_image_estimator.run(stacked)",
"_____no_output_____"
],
[
"sources = find_peaks(\n images_ts[\"sqrt_ts\"],\n threshold=5,\n min_distance=\"0.2 deg\",\n)\nsources",
"_____no_output_____"
],
[
"source_pos = SkyCoord(sources[\"ra\"], sources[\"dec\"])\nsource_pos",
"_____no_output_____"
],
[
"# Plot sources on top of significance sky image\nimages_ts[\"sqrt_ts\"].plot(add_cbar=True)\n\nplt.gca().scatter(\n source_pos.ra.deg,\n source_pos.dec.deg,\n transform=plt.gca().get_transform(\"icrs\"),\n color=\"none\",\n edgecolor=\"white\",\n marker=\"o\",\n s=200,\n lw=1.5,\n);",
"_____no_output_____"
]
],
[
[
"## Spatial analysis\n\nSee other notebooks for how to run a 3D cube or 2D image based analysis.",
"_____no_output_____"
],
[
"## Spectrum\n\nWe'll run a spectral analysis using the classical reflected regions background estimation method,\nand using the on-off (often called WSTAT) likelihood function.",
"_____no_output_____"
]
],
[
[
"energy_axis = MapAxis.from_energy_bounds(0.1, 40, 40, unit=\"TeV\", name=\"energy\")\nenergy_axis_true = MapAxis.from_energy_bounds(\n 0.05, 100, 200, unit=\"TeV\", name=\"energy_true\"\n)\n\ngeom = RegionGeom.create(region=on_region, axes=[energy_axis])\ndataset_empty = SpectrumDataset.create(\n geom=geom, energy_axis_true=energy_axis_true\n)",
"_____no_output_____"
],
[
"dataset_maker = SpectrumDatasetMaker(\n containment_correction=False, selection=[\"counts\", \"exposure\", \"edisp\"]\n)\nbkg_maker = ReflectedRegionsBackgroundMaker(exclusion_mask=exclusion_mask)\nsafe_mask_masker = SafeMaskMaker(methods=[\"aeff-max\"], aeff_percent=10)",
"_____no_output_____"
],
[
"%%time\ndatasets = Datasets()\n\nfor observation in observations:\n dataset = dataset_maker.run(\n dataset_empty.copy(name=f\"obs-{observation.obs_id}\"), observation\n )\n dataset_on_off = bkg_maker.run(dataset, observation)\n dataset_on_off = safe_mask_masker.run(dataset_on_off, observation)\n datasets.append(dataset_on_off)",
"_____no_output_____"
],
[
"plt.figure(figsize=(8, 8))\n_, ax, _ = dataset_image.counts.smooth(\"0.03 deg\").plot(vmax=8)\n\non_region.to_pixel(ax.wcs).plot(ax=ax, edgecolor=\"white\")\nplot_spectrum_datasets_off_regions(datasets, ax=ax)",
"_____no_output_____"
]
],
[
[
"### Model fit\n\nThe next step is to fit a spectral model, using all data (i.e. a \"global\" fit, using all energies).",
"_____no_output_____"
]
],
[
[
"%%time\nspectral_model = PowerLawSpectralModel(\n index=2, amplitude=1e-11 * u.Unit(\"cm-2 s-1 TeV-1\"), reference=1 * u.TeV\n)\n\nmodel = SkyModel(spectral_model=spectral_model, name=\"source-gc\")\n\ndatasets.models = model\n\nfit = Fit(datasets)\nresult = fit.run()\nprint(result)",
"_____no_output_____"
]
],
[
[
"### Spectral points\n\nFinally, let's compute spectral points. The method used is to first choose an energy binning, and then to do a 1-dim likelihood fit / profile to compute the flux and flux error.",
"_____no_output_____"
]
],
[
[
"# Flux points are computed on stacked observation\nstacked_dataset = datasets.stack_reduce(name=\"stacked\")\n\nprint(stacked_dataset)",
"_____no_output_____"
],
[
"energy_edges = MapAxis.from_energy_bounds(\"1 TeV\", \"30 TeV\", nbin=5).edges\n\nstacked_dataset.models = model\n\nfpe = FluxPointsEstimator(energy_edges=energy_edges, source=\"source-gc\")\nflux_points = fpe.run(datasets=[stacked_dataset])\nflux_points.table_formatted",
"_____no_output_____"
]
],
[
[
"### Plot\n\nLet's plot the spectral model and points. You could do it directly, but for convenience we bundle the model and the flux points in a `FluxPointDataset`:",
"_____no_output_____"
]
],
[
[
"flux_points_dataset = FluxPointsDataset(data=flux_points, models=model)",
"_____no_output_____"
],
[
"flux_points_dataset.plot_fit();",
"_____no_output_____"
]
],
[
[
"## Exercises\n\n* Re-run the analysis above, varying some analysis parameters, e.g.\n * Select a few other observations\n * Change the energy band for the map\n * Change the spectral model for the fit\n * Change the energy binning for the spectral points\n* Change the target. Make a sky image and spectrum for your favourite source.\n * If you don't know any, the Crab nebula is the \"hello world!\" analysis of gamma-ray astronomy.",
"_____no_output_____"
]
],
[
[
"# print('hello world')\n# SkyCoord.from_name('crab')",
"_____no_output_____"
]
],
[
[
"## What next?\n\n* This notebook showed an example of a first CTA analysis with Gammapy, using simulated 1DC data.\n* Let us know if you have any question or issues!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d03cce44c2080daeb8c89e6a969ae44fdca6a480 | 198,211 | ipynb | Jupyter Notebook | courses/08_Plotly_Bokeh/Fire_Australia19.ipynb | visiont3lab/data-visualization | dc1a49b61359bdee5d8a437e20378016ac7f9399 | [
"BSD-3-Clause"
] | 1 | 2021-02-24T17:40:19.000Z | 2021-02-24T17:40:19.000Z | courses/08_Plotly_Bokeh/Fire_Australia19.ipynb | visiont3lab/data-visualization | dc1a49b61359bdee5d8a437e20378016ac7f9399 | [
"BSD-3-Clause"
] | 3 | 2021-06-08T21:06:05.000Z | 2022-01-13T02:23:24.000Z | courses/08_Plotly_Bokeh/Fire_Australia19.ipynb | visiont3lab/data-visualization | dc1a49b61359bdee5d8a437e20378016ac7f9399 | [
"BSD-3-Clause"
] | 18 | 2020-04-01T06:52:36.000Z | 2021-02-24T17:47:26.000Z | 198,211 | 198,211 | 0.883811 | [
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport folium\nfrom folium.plugins import MarkerCluster\n%matplotlib inline\n\naustralia=pd.read_csv(\"https://frenzy86.s3.eu-west-2.amazonaws.com/fav/australia_cleaned.csv\")\naustralia.head()",
"_____no_output_____"
],
[
"plt.figure(figsize=(18,12))\n\nplt.hist(australia[\"confidence\"],label=\"Sicurezza Incendi\",color=\"red\");\nplt.xlabel(\"Livello di sicurezza degli incendi\")\nplt.ylabel(\"Numero di incendi\")\nplt.title(\"Grafico numero incendi e Livello di sicurezza\")\nplt.legend(loc=2);",
"_____no_output_____"
],
[
"plt.figure(figsize=(18,12))\nplt.scatter(australia[\"confidence\"],australia [\"brightness\"], label =\"Sicurezza Incendi\", color=\"orange\");\n\nplt.ylabel(\"Luminosità a 21 Kelvin\")\nplt.xlabel('Livello di sicurezza degli incendi')\nplt.title(\"Grafico Livello di sicurezza incendi e la luminosità 21 Kelvin\")\nplt.legend(loc=2);",
"_____no_output_____"
],
[
"plt.figure(figsize=(18,12))\nplt.scatter(australia[\"confidence\"],australia [\"bright_t31\"], label =\"Sicurezza Incendi\", color=\"yellow\");\n\n\nplt.ylabel(\"Luminosità a 31 Kelvin\")\nplt.xlabel('Livello di sicurezza degli incendi')\nplt.title(\"Grafico Livello di sicurezza incendi e la luminosità 31 Kelvin\")\nplt.legend(loc=2);",
"_____no_output_____"
],
[
"pd.crosstab(australia[\"sat_Terra\"], australia[\"time_N\"]).plot(kind=\"bar\",figsize=(20,10));\n\nplt.title(\"Rapporto tra gli incendi raccolti dal satellite terrestre in notturni e diurni\")\nplt.ylabel(\"N° di incendi riconosciuti dai satelliti\")\nplt.xlabel(\"Tipo di fuoco, notturno o diurno\");",
"_____no_output_____"
],
[
"australia_1 =australia.copy()\naustralia_1.head()",
"_____no_output_____"
],
[
"data=australia_1[(australia_1[\"confidence\"]>= 70)]\ndata.head()",
"_____no_output_____"
],
[
"data.shape",
"_____no_output_____"
],
[
"#Creare lista longitudine e latitudine\nlat=data[\"latitude\"].values.tolist()\nlong=data[\"longitude\"].values.tolist()",
"_____no_output_____"
],
[
"#Mappa Australia\nmap1=folium.Map([-25.274398,133.775136],zoom_start=4)",
"_____no_output_____"
],
[
"#Creare un cluster di mappa\naustralia_cluster = MarkerCluster()",
"_____no_output_____"
],
[
"for latV,longV in zip(lat,long):\n folium.Marker(location=[latV,longV]).add_to(australia_cluster)",
"_____no_output_____"
],
[
"#Aggiungere il cluster alla mappa che vogliamo stampare\naustralia_cluster.add_to(map1);",
"_____no_output_____"
],
[
"map1",
"_____no_output_____"
],
[
"localizacion=australia_1[(australia_1[\"frp\"]>= 2500)]\nlocalizacion.head()",
"_____no_output_____"
],
[
"\nmap_2 = folium.Map([-25.274398,133.775136],zoom_start=4.5,tiles='Stamen Terrain')",
"_____no_output_____"
],
[
"lat_2 = localizacion[\"latitude\"].values.tolist()\nlong_2 = localizacion[\"longitude\"].values.tolist()",
"_____no_output_____"
],
[
"australia_cluster_2 = MarkerCluster().add_to(map_2)",
"_____no_output_____"
],
[
"for lat_2,long_2 in zip(lat_2,long_2):\n folium.Marker([lat_2,long_2]).add_to(australia_cluster_2)",
"_____no_output_____"
],
[
"map_2",
"_____no_output_____"
]
],
[
[
"\nVuoi conoscere gli incendi divampati dopo il 15 settembre 2019?",
"_____no_output_____"
]
],
[
[
"mes = australia_1[(australia_1[\"acq_date\"]>= \"2019-09-15\")]\nmes.head()",
"_____no_output_____"
],
[
"mes.describe()",
"_____no_output_____"
],
[
"map_sett = folium.Map([-25.274398,133.775136], zoom_start=4)",
"_____no_output_____"
],
[
"lat_3 = mes[\"latitude\"].values.tolist()\nlong_3 = mes[\"longitude\"].values.tolist()",
"_____no_output_____"
],
[
"australia_cluster_3 = MarkerCluster().add_to(map_sett)",
"_____no_output_____"
],
[
"for lat_3,long_3 in zip(lat_3,long_3):\n folium.Marker([lat_3,long_3]).add_to(australia_cluster_3)",
"_____no_output_____"
],
[
"map_sett",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"#Play with Folium",
"_____no_output_____"
]
],
[
[
"44.4807035,11.3712528",
"_____no_output_____"
],
[
"import folium\nm1 = folium.Map(location=[44.48, 11.37], tiles='openstreetmap', zoom_start=18)\nm1.save('map1.html')",
"_____no_output_____"
],
[
"m1",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"m3.save(\"filename.png\")",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03cf614b2390f0ad9980c9e711c0fced0ac0732 | 704,263 | ipynb | Jupyter Notebook | Combineinator_Library.ipynb | combineinator/combine-inator-acikhack2021 | 526bd54208d85fbe7faad05d2116e91c5bfaaffa | [
"MIT"
] | null | null | null | Combineinator_Library.ipynb | combineinator/combine-inator-acikhack2021 | 526bd54208d85fbe7faad05d2116e91c5bfaaffa | [
"MIT"
] | null | null | null | Combineinator_Library.ipynb | combineinator/combine-inator-acikhack2021 | 526bd54208d85fbe7faad05d2116e91c5bfaaffa | [
"MIT"
] | 1 | 2021-12-16T19:02:32.000Z | 2021-12-16T19:02:32.000Z | 111.894344 | 202,154 | 0.770283 | [
[
[
"<a href=\"https://colab.research.google.com/github/combineinator/combine-inator-acikhack2021/blob/main/Combineinator_Library.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Drive already mounted at /content/drive; to attempt to forcibly remount, call drive.mount(\"/content/drive\", force_remount=True).\n"
]
],
[
[
"## CombineInator (parent class)",
"_____no_output_____"
]
],
[
[
"class CombineInator:\n \n def __init__(self):\n self.source = \"\"\n\n\n def translate_model(self, source):\n\n if source == \"en\":\n\n tokenizer_trs = AutoTokenizer.from_pretrained(\"Helsinki-NLP/opus-mt-en-trk\")\n model_trs = AutoModelForSeq2SeqLM.from_pretrained(\"Helsinki-NLP/opus-mt-en-trk\")\n pipe_trs = \"translation_en_to_trk\"\n\n elif source == \"tr\":\n\n tokenizer_trs = AutoTokenizer.from_pretrained(\"Helsinki-NLP/opus-mt-tr-en\")\n model_trs = AutoModelForSeq2SeqLM.from_pretrained(\"Helsinki-NLP/opus-mt-tr-en\")\n pipe_trs = \"translation_tr_to_en\"\n \n\n return model_trs, tokenizer_trs, pipe_trs \n\n \n def translate(self, pipe, model, tokenizer, response):\n \n translator = pipeline(pipe, model=model, tokenizer=tokenizer)\n \n # elde edilen cümleleri hedeflnen dile çevirme:\n trans = translator(response)[0][\"translation_text\"]\n\n return trans\n \n ",
"_____no_output_____"
]
],
[
[
"## WikiWebScraper (child)",
"_____no_output_____"
]
],
[
[
"import requests\nimport re\nfrom bs4 import BeautifulSoup\nfrom tqdm import tqdm\nfrom os.path import exists, basename, splitext",
"_____no_output_____"
],
[
"class WikiWebScraper(CombineInator):\n \n def __init__(self):\n \n self.__HEADERS_PARAM = {\n \"User-Agent\": \"Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/88.0.4324.104 Safari/537.36\"}\n \n \n def category_scraping_interface(self, CATEGORY_QUERY, LIMIT, SAVE_PATH, PAGE_PER_SAVE, REMOVE_NUMBERS, JUST_TITLE_ANALYSIS, TEXT_INTO_SENTENCES_PARAM):\n \n \"\"\"\n \n Kategorik verilerin ayıklanma işlemleri bu fonksiyonda yönetilir.\n \n :param CATEGORY_QUERY: Ayıklanacak kategori sorgusu.\n :type CATEGORY_QUERY: str\n\n :param SAVE_PATH: Ayıklanan verinin kaydedileceği yol.\n :type SAVE_PATH: str\n\n :param LIMIT: Ayıklanması istenen veri limiti. Verilmediği taktirde tüm verileri çeker.\n :type LIMIT: int\n\n :param PAGE_PER_SAVE: Belirlenen aralıkla ayıklanan kategorik verinin kaydedilmesini sağlar.\n :type PAGE_PER_SAVE: int\n\n :param TEXT_INTO_SENTENCES_PARAM: Ayıklanan verilerin cümleler halinde mi, yoksa metin halinde mi kaydedileceğini belirler.\n :type TEXT_INTO_SENTENCES_PARAM: bool\n\n :param REMOVE_NUMBERS: Ayıklanan verilerden rakamların silinip silinmemesini belirler.\n :type REMOVE_NUMBERS: bool\n\n :param JUST_TITLE_ANALYSIS: Sadece sayfaların başlık bilgilerinin toplanmasını sağlar.\n :type JUST_TITLE_ANALYSIS: bool\n \n \"\"\"\n \n sub_list = []\n page_list = []\n text_list = []\n \n page_list, sub_list = self.first_variable(CATEGORY_QUERY, (LIMIT - len(text_list)))\n fv = True\n \n if page_list and sub_list is not None: \n with tqdm(total=LIMIT, desc=\"Sayfa taranıyor.\") as pbar:\n while len(page_list) < LIMIT:\n \n if fv is True:\n pbar.update(len(page_list))\n fv = False \n \n temp_soup = \"\"\n \n if len(sub_list) == 0:\n break\n \n temp_soup = self.sub_scraper(sub_list[0]) \n \n if (temp_soup == False):\n break \n \n del sub_list[0] \n \n sub_list = sub_list + self.sub_category_scraper(temp_soup)\n \n temp_page_scraper = self.page_scraper(temp_soup, (LIMIT - len(page_list))) \n \n if temp_page_scraper is not None:\n for i in temp_page_scraper:\n if i not in page_list:\n page_list.append(i)\n pbar.update(1) \n \n if len(sub_list) == 0:\n sub_list = sub_list + self.sub_category_scraper(temp_soup)\n \n temp_range = 0\n loop_counter = 0\n \n if JUST_TITLE_ANALYSIS is False: \n for i in range(PAGE_PER_SAVE, len(page_list)+PAGE_PER_SAVE, PAGE_PER_SAVE):\n \n if loop_counter == (len(page_list) // PAGE_PER_SAVE):\n PATH = SAVE_PATH + \"/\" + CATEGORY_QUERY + \"_\" + str(temp_range) + \" - \" + str(len(page_list)) + \".txt\"\n temp_text_list = self.text_into_sentences(self.text_scraper(page_list[temp_range:i], (len(page_list) % PAGE_PER_SAVE)), REMOVE_NUMBERS,TEXT_INTO_SENTENCES_PARAM)\n else:\n PATH = SAVE_PATH + \"/\" + CATEGORY_QUERY + \"_\" + str(temp_range) + \" - \" + str(i) + \".txt\"\n temp_text_list = self.text_into_sentences(self.text_scraper(page_list[temp_range:i], PAGE_PER_SAVE), REMOVE_NUMBERS, TEXT_INTO_SENTENCES_PARAM)\n \n text_list += temp_text_list\n \n self.save_to_csv(PATH, temp_text_list) \n temp_range = i\n loop_counter += 1\n \n print(\"\\n\\n\"+str(len(page_list)) + \" adet sayfa bulundu ve içerisinden \" + str(len(text_list)) + \" satır farklı metin ayrıştırıldı.\")\n return text_list\n \n else:\n PATH = SAVE_PATH + \"/\" + CATEGORY_QUERY + \"_\" + str(len(page_list)) + \"_page_links\" + \".txt\"\n self.save_to_csv(PATH, page_list, JUST_TITLE_ANALYSIS)\n print(\"\\n\\n\"+str(len(page_list)) + \" adet sayfa bulundu ve sayfaların adresleri \\\"\" + PATH + \"\\\" konumunda kaydedildi.\")\n return page_list\n \n else:\n print(\"Aranan kategori bulunamadı.\")\n \n \n def categorical_scraper(self, CATEGORY_QUERY, save_path, LIMIT=-1, page_per_save=10000, text_into_sentences_param=True, remove_numbers=False, just_title_analysis=False):\n \n \"\"\"\n Wikipedia üzerinden kategorik olarak veri çekmek için kullanılır.\n \n :param CATEGORY_QUERY: Ayıklanacak kategori sorgusu.\n :type CATEGORY_QUERY: str\n\n :param save_path: Ayıklanan verinin kaydedileceği yol.\n :type save_path: str\n\n :param LIMIT: Ayıklanması istenen veri limiti. Verilmediği taktirde tüm verileri çeker.\n :type LIMIT: int\n\n :param page_per_save: Belirlenen aralıkla ayıklanan kategorik verinin kaydedilmesini sağlar.\n :type page_per_save: int\n\n :param text_into_sentences_param: Ayıklanan verilerin cümleler halinde mi, yoksa metin halinde mi kaydedileceğini belirler.\n :type text_into_sentences_param: bool\n\n :param remove_numbers: Ayıklanan verilerden rakamların silinip silinmemesini belirler.\n :type remove_numbers: bool\n\n :param just_title_analysis: Sadece sayfaların başlık bilgilerinin toplanmasını sağlar.\n :type just_title_analysis: bool\n \n \"\"\"\n\n if LIMIT == -1:\n LIMIT = 9999999\n \n CATEGORY_QUERY = CATEGORY_QUERY.replace(\" \",\"_\")\n \n return_list = self.category_scraping_interface(CATEGORY_QUERY, LIMIT, save_path, page_per_save, remove_numbers, just_title_analysis, text_into_sentences_param)\n \n if return_list is None:\n return []\n else:\n return return_list\n \n \n \n def text_scraper_from_pagelist(self, page_list_path, save_path, page_per_save=10000, remove_numbers=False, text_into_sentences_param=True, RANGE=None):\n \n \"\"\"\n \n Wikipedia üzerinden kategorik olarak veri çekmek için kullanılır.\n \n :param page_list_path: Toplanan sayfaların başlık bilgilerinin çıkartılmasını sağlar\n :type page_list_path: str\n \n :param save_path: Ayıklanan verinin kaydedileceği yol.\n :type save_path: str\n \n :param page_per_save: Belirlenen aralıkla ayıklanan kategorik verinin kaydedilmesini sağlar.\n :type page_per_save: int\n \n :param text_into_sentences_param: Ayıklanan verilerin cümleler halinde mi, yoksa metin halinde mi kaydedileceğini belirler.\n :type text_into_sentences_param: bool\n \n :param remove_numbers: Ayıklanan verilerden rakamların silinip silinmemesini belirler.\n :type remove_numbers: bool\n \n :param RANGE: Ayıklnacak verilerin aralığını belirler. \"RANGE = [500,1000]\" şeklinde kullanılır. Verilmediği zaman tüm veri ayıklanır.\n :type RANGE: list\n \n \"\"\"\n \n page_list = []\n text_list = []\n \n with open(page_list_path, 'r') as f:\n page_list = [line.strip() for line in f]\n \n if RANGE is not None:\n page_list = page_list[RANGE[0]:RANGE[1]]\n \n temp_range = 0\n loop_counter = 0\n \n for i in range(page_per_save, len(page_list)+page_per_save, page_per_save):\n \n if loop_counter == (len(page_list) // page_per_save):\n PATH = save_path + \"/\" + \"scraped_page\" + \"_\" + str(temp_range) + \" - \" + str(len(page_list)) + \".txt\"\n temp_text_list = self.text_into_sentences(self.text_scraper(page_list[temp_range:i], (len(page_list) % page_per_save), True), remove_numbers, text_into_sentences_param)\n else:\n PATH = save_path + \"/\" + \"scraped_page\" + \"_\" + str(temp_range) + \" - \" + str(i) + \".txt\"\n temp_text_list = self.text_into_sentences(self.text_scraper(page_list[temp_range:i], page_per_save, True), remove_numbers, text_into_sentences_param)\n \n text_list += temp_text_list\n \n save_to_csv(PATH, temp_text_list)\n temp_range = i\n loop_counter += 1\n \n print(\"\\n\\\"\" + page_list_path + \"\\\" konumundaki \" + str(len(page_list)) + \" adet sayfa içerisinden \" + str(len(text_list)) + \" satır metin ayrıştırıldı.\")\n \n return text_list\n \n \n \n def page_scraper(self, page_soup, LIMIT):\n \n \"\"\"\n \n Gönderilen wikipedia SOUP objesinin içerisindeki kategorik içerik sayfaları döndürür.\n\n :param page_soup: Wikipedia kategori sayfasının SOUP objesidir.\n \n :param LIMIT: Ayıklanacaj sayfa limitini belirler.\n :type LIMIT: int\n \n \"\"\"\n \n page_list = []\n \n try:\n pages = page_soup.find(\"div\", attrs={\"id\": \"mw-pages\"}).find_all(\"a\")\n for page in pages[1:]:\n if len(page_list) == LIMIT:\n break\n else:\n page_list.append([page.text, page[\"href\"]])\n return page_list\n \n except:\n pass\n \n \n def sub_category_scraper(self, sub_soup):\n \n \"\"\"\n \n Gönderilen wikipedia SOUP objesinin içerisindeki alt kategorileri döndürür.\n\n :param sub_soup: Alt kategori sayfasının SOUP objesidir.\n \n \"\"\"\n \n sub_list = []\n \n try:\n sub_categories = sub_soup.find_all(\"div\", attrs={\"class\": \"CategoryTreeItem\"})\n for sub in sub_categories[1:]:\n sub_list.append([sub.a.text, sub.a[\"href\"]])\n \n return sub_list\n \n except:\n print(\"Aranan kategori için yeterli sayfa bulunamadı.\")\n \n \n \n def sub_scraper(self, sub):\n \n \"\"\"\n \n Fonksiyona gelen wikipedia kategori/alt kategorisinin SOUP objesini döndürür.\n\n :param sub: Alt kategori sayfasının linkini içerir.\n \n \"\"\"\n \n try:\n req = requests.get(\"https://tr.wikipedia.org\" + str(sub[1]), headers=self.__HEADERS_PARAM)\n soup = BeautifulSoup(req.content, \"lxml\")\n return soup\n \n except:\n print(\"\\nAlt kategori kalmadı\")\n return False\n \n \n def text_scraper(self, page_list, LIMIT, IS_FROM_TXT=False):\n \n \"\"\"\n \n Önceden ayıklanmış sayfa listesini içerisindeki sayfaları ayıklayarak içerisindeki metin listesini döndürür.\n\n :param page_list: Sayfa listesini içerir.\n\n :parama LIMIT: Ayıklanacaj sayfa limitini belirler.\n :type LIMIT: int\n\n :param IS_FROM_TXT: Ayıklanacak sayfanın listeleden mi olup olmadığını kontrol eder.\n \n \"\"\"\n \n text_list = []\n \n with tqdm(total=LIMIT, desc=\"Sayfa Ayrıştırılıyor\") as pbar:\n for page in page_list:\n \n if len(text_list) == LIMIT:\n break\n if IS_FROM_TXT is False:\n req = requests.get(\"https://tr.wikipedia.org\" + str(page[1]), headers=self.__HEADERS_PARAM)\n else:\n req = requests.get(\"https://tr.wikipedia.org\" + str(page), headers=self.__HEADERS_PARAM)\n soup = BeautifulSoup(req.content, \"lxml\")\n page_text = soup.find_all(\"p\")\n temp_text = \"\"\n \n for i in page_text[1:]:\n temp_text = temp_text + i.text\n \n text_list.append(temp_text)\n pbar.update(1)\n \n return text_list\n \n \n \n def first_variable(self, CATEGORY_QUERY, LIMIT):\n \n \"\"\"\n \n Sorguda verilen kategorinin doğruluğunu kontrol eder ve eğer sorgu doğru ise ilk değerleri ayıklar.\n\n :param CATEGORY_QUERY: Ayıklanacak kategori sorgusu.\n :type CATEGORY_QUERY: str\n\n :param LIMIT: Ayıklanması istenen veri limiti. Verilmediği taktirde tüm verileri çeker.\n :type LIMIT: int\n \n \"\"\"\n \n first_req = requests.get(\"https://tr.wikipedia.org/wiki/Kategori:\" + CATEGORY_QUERY, headers=self.__HEADERS_PARAM)\n first_soup = BeautifulSoup(first_req.content, \"lxml\")\n page_list = self.page_scraper(first_soup, LIMIT)\n sub_list = self.sub_category_scraper(first_soup)\n \n return page_list, sub_list\n \n \n \n def text_into_sentences(self, texts, remove_numbers, text_into_sentences_param):\n \n \"\"\"\n \n Metin verilerini cümlelerine ayıklar.\n\n :param texts: Düzlenecek metin verisi.\n\n :param remove_numbers: Sayıların temizlenip temizlenmeyeceğini kontrol eder.\n\n :param text_into_sentences_param: Metinlerin cümlelere çevrilip çevrilmeyeceğini kontrol eder.\n \n \"\"\"\n \n flatlist = [] \n sent_list = []\n \n texts = self.sentence_cleaning(texts, remove_numbers)\n \n if text_into_sentences_param is True:\n for line in texts:\n temp_line = re.split(r'(?<![IVX0-9]\\S)(?<!\\w\\.\\w.)(?<![A-Z][a-z]\\.)(?<=\\.|\\?)\\s', line)\n for i in temp_line:\n if len(i.split(\" \")) > 3:\n sent_list.append(i)\n else:\n sent_list = texts\n \n flatlist = list(dict.fromkeys(self.flat(sent_list, flatlist)))\n \n return flatlist\n \n \n \n def flat(self, sl,fl): \n \n \"\"\"\n \n Metinler, cümlelerine ayırıldıktan sonra listenin düzlenmesine yarar.\n\n :param sl: Yollanan listle.\n\n :param fl: Düzlemem liste.\n\n \n \n \"\"\"\n \n for e in sl: \n if type(e) == list: \n flat(e,fl) \n elif len(e.split(\" \"))>3:\n fl.append(e) \n return fl\n \n \n \n def sentence_cleaning(self, sentences, remove_numbers):\n \n \"\"\"\n \n Ayıklanan wikipedia verilerinin temizlenmesi bu fonksiyonda gerçekleşir.\n\n :param sentences: Temizlenmek için gelen veri seti.\n \n :param remove_numbers: Sayıların temizlenip temizlenmeyeceğini kontrol eder.\n \n \"\"\"\n \n return_list = []\n \n if remove_numbers is False:\n removing_func = '[^[a-zA-ZğüışöçĞÜIİŞÖÇ0-9.,!:;`?%&\\-\\'\" ]'\n else:\n removing_func = '[^[a-zA-ZğüışöçĞÜIİŞÖÇ.,!:;`?%&\\-\\'\" ]'\n \n for input_text in sentences:\n try: \n input_text = re.sub(r'(\\[.*?\\])', '', input_text)\n input_text = re.sub(r'(\\(.*?\\))', '', input_text)\n input_text = re.sub(r'(\\{.*?\\})', '', input_text)\n input_text = re.sub(removing_func, '', input_text)\n input_text = re.sub(\"(=+(\\s|.)*)\", \"\", input_text)\n input_text = re.sub(\"(\\s{2,})\", \"\", input_text)\n input_text = input_text.replace(\"''\", \"\")\n input_text = input_text.replace(\"\\n\", \"\") \n return_list.append(input_text)\n except:\n pass\n \n return return_list\n \n \n def save_to_csv(self, PATH, data, is_just_title_analysis=False):\n \n \"\"\"\n \n Verilerin 'csv' formatında kaydedilmesini bu fonksiyonda gerçekleşir.\n\n :param PATH: Kaydedilecek yol.\n\n :param data: Kaydedilecek veri.\n\n :param is_just_title_analysis: Sadece analiz yapılıp yapılmadığını kontrol eder.\n \n \"\"\"\n \n if is_just_title_analysis is False:\n with open(PATH, \"w\") as output:\n for i in data:\n output.write(i+\"\\n\")\n \n else:\n temp_data = []\n for i in data:\n temp_data.append(i[1])\n with open(PATH, \"w\") as output:\n for i in temp_data:\n output.write(i+\"\\n\")",
"_____no_output_____"
]
],
[
[
"### Örnek kullanım",
"_____no_output_____"
]
],
[
[
"library = WikiWebScraper()",
"_____no_output_____"
],
[
"PATH = \"/content/\"",
"_____no_output_____"
],
[
"library.categorical_scraper(\"savaş\", PATH, 20, text_into_sentences_param=False)",
"Sayfa taranıyor.: 100%|██████████| 20/20 [00:00<00:00, 52.99it/s]\nSayfa Ayrıştırılıyor: 100%|██████████| 20/20 [00:04<00:00, 4.68it/s]"
]
],
[
[
"## speechModule (child)",
"_____no_output_____"
]
],
[
[
"!pip install transformers\n!pip install simpletransformers",
"Collecting transformers\n Downloading transformers-4.9.2-py3-none-any.whl (2.6 MB)\n\u001b[K |████████████████████████████████| 2.6 MB 4.3 MB/s \n\u001b[?25hRequirement already satisfied: tqdm>=4.27 in /usr/local/lib/python3.7/dist-packages (from transformers) (4.62.0)\nCollecting tokenizers<0.11,>=0.10.1\n Downloading tokenizers-0.10.3-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (3.3 MB)\n\u001b[K |████████████████████████████████| 3.3 MB 35.3 MB/s \n\u001b[?25hRequirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from transformers) (2.23.0)\nRequirement already satisfied: numpy>=1.17 in /usr/local/lib/python3.7/dist-packages (from transformers) (1.19.5)\nRequirement already satisfied: packaging in /usr/local/lib/python3.7/dist-packages (from transformers) (21.0)\nCollecting pyyaml>=5.1\n Downloading PyYAML-5.4.1-cp37-cp37m-manylinux1_x86_64.whl (636 kB)\n\u001b[K |████████████████████████████████| 636 kB 45.9 MB/s \n\u001b[?25hRequirement already satisfied: regex!=2019.12.17 in /usr/local/lib/python3.7/dist-packages (from transformers) (2019.12.20)\nCollecting sacremoses\n Downloading sacremoses-0.0.45-py3-none-any.whl (895 kB)\n\u001b[K |████████████████████████████████| 895 kB 49.0 MB/s \n\u001b[?25hRequirement already satisfied: importlib-metadata in /usr/local/lib/python3.7/dist-packages (from transformers) (4.6.4)\nRequirement already satisfied: filelock in /usr/local/lib/python3.7/dist-packages (from transformers) (3.0.12)\nCollecting huggingface-hub==0.0.12\n Downloading huggingface_hub-0.0.12-py3-none-any.whl (37 kB)\nRequirement already satisfied: typing-extensions in /usr/local/lib/python3.7/dist-packages (from huggingface-hub==0.0.12->transformers) (3.7.4.3)\nRequirement already satisfied: pyparsing>=2.0.2 in /usr/local/lib/python3.7/dist-packages (from packaging->transformers) (2.4.7)\nRequirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.7/dist-packages (from importlib-metadata->transformers) (3.5.0)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (2021.5.30)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (1.24.3)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (3.0.4)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (2.10)\nRequirement already satisfied: click in /usr/local/lib/python3.7/dist-packages (from sacremoses->transformers) (7.1.2)\nRequirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from sacremoses->transformers) (1.15.0)\nRequirement already satisfied: joblib in /usr/local/lib/python3.7/dist-packages (from sacremoses->transformers) (1.0.1)\nInstalling collected packages: tokenizers, sacremoses, pyyaml, huggingface-hub, transformers\n Attempting uninstall: pyyaml\n Found existing installation: PyYAML 3.13\n Uninstalling PyYAML-3.13:\n Successfully uninstalled PyYAML-3.13\nSuccessfully installed huggingface-hub-0.0.12 pyyaml-5.4.1 sacremoses-0.0.45 tokenizers-0.10.3 transformers-4.9.2\nCollecting simpletransformers\n Downloading simpletransformers-0.61.13-py3-none-any.whl (221 kB)\n\u001b[K |████████████████████████████████| 221 kB 4.1 MB/s \n\u001b[?25hRequirement already satisfied: transformers>=4.2.0 in /usr/local/lib/python3.7/dist-packages (from simpletransformers) (4.9.2)\nCollecting wandb>=0.10.32\n Downloading wandb-0.12.0-py2.py3-none-any.whl (1.6 MB)\n\u001b[K |████████████████████████████████| 1.6 MB 46.0 MB/s \n\u001b[?25hCollecting streamlit\n Downloading streamlit-0.87.0-py2.py3-none-any.whl (8.0 MB)\n\u001b[K |████████████████████████████████| 8.0 MB 32.8 MB/s \n\u001b[?25hCollecting tensorboardx\n Downloading tensorboardX-2.4-py2.py3-none-any.whl (124 kB)\n\u001b[K |████████████████████████████████| 124 kB 46.1 MB/s \n\u001b[?25hRequirement already satisfied: pandas in /usr/local/lib/python3.7/dist-packages (from simpletransformers) (1.1.5)\nRequirement already satisfied: regex in /usr/local/lib/python3.7/dist-packages (from simpletransformers) (2019.12.20)\nRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from simpletransformers) (1.19.5)\nRequirement already satisfied: tokenizers in /usr/local/lib/python3.7/dist-packages (from simpletransformers) (0.10.3)\nCollecting sentencepiece\n Downloading sentencepiece-0.1.96-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (1.2 MB)\n\u001b[K |████████████████████████████████| 1.2 MB 38.6 MB/s \n\u001b[?25hRequirement already satisfied: scipy in /usr/local/lib/python3.7/dist-packages (from simpletransformers) (1.4.1)\nRequirement already satisfied: scikit-learn in /usr/local/lib/python3.7/dist-packages (from simpletransformers) (0.22.2.post1)\nRequirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from simpletransformers) (2.23.0)\nCollecting datasets\n Downloading datasets-1.11.0-py3-none-any.whl (264 kB)\n\u001b[K |████████████████████████████████| 264 kB 50.9 MB/s \n\u001b[?25hRequirement already satisfied: tqdm>=4.47.0 in /usr/local/lib/python3.7/dist-packages (from simpletransformers) (4.62.0)\nCollecting seqeval\n Downloading seqeval-1.2.2.tar.gz (43 kB)\n\u001b[K |████████████████████████████████| 43 kB 2.0 MB/s \n\u001b[?25hRequirement already satisfied: importlib-metadata in /usr/local/lib/python3.7/dist-packages (from transformers>=4.2.0->simpletransformers) (4.6.4)\nRequirement already satisfied: huggingface-hub==0.0.12 in /usr/local/lib/python3.7/dist-packages (from transformers>=4.2.0->simpletransformers) (0.0.12)\nRequirement already satisfied: sacremoses in /usr/local/lib/python3.7/dist-packages (from transformers>=4.2.0->simpletransformers) (0.0.45)\nRequirement already satisfied: filelock in /usr/local/lib/python3.7/dist-packages (from transformers>=4.2.0->simpletransformers) (3.0.12)\nRequirement already satisfied: pyyaml>=5.1 in /usr/local/lib/python3.7/dist-packages (from transformers>=4.2.0->simpletransformers) (5.4.1)\nRequirement already satisfied: packaging in /usr/local/lib/python3.7/dist-packages (from transformers>=4.2.0->simpletransformers) (21.0)\nRequirement already satisfied: typing-extensions in /usr/local/lib/python3.7/dist-packages (from huggingface-hub==0.0.12->transformers>=4.2.0->simpletransformers) (3.7.4.3)\nRequirement already satisfied: pyparsing>=2.0.2 in /usr/local/lib/python3.7/dist-packages (from packaging->transformers>=4.2.0->simpletransformers) (2.4.7)\nCollecting GitPython>=1.0.0\n Downloading GitPython-3.1.18-py3-none-any.whl (170 kB)\n\u001b[K |████████████████████████████████| 170 kB 46.1 MB/s \n\u001b[?25hRequirement already satisfied: python-dateutil>=2.6.1 in /usr/local/lib/python3.7/dist-packages (from wandb>=0.10.32->simpletransformers) (2.8.2)\nRequirement already satisfied: promise<3,>=2.0 in /usr/local/lib/python3.7/dist-packages (from wandb>=0.10.32->simpletransformers) (2.3)\nRequirement already satisfied: protobuf>=3.12.0 in /usr/local/lib/python3.7/dist-packages (from wandb>=0.10.32->simpletransformers) (3.17.3)\nCollecting subprocess32>=3.5.3\n Downloading subprocess32-3.5.4.tar.gz (97 kB)\n\u001b[K |████████████████████████████████| 97 kB 5.6 MB/s \n\u001b[?25hRequirement already satisfied: Click!=8.0.0,>=7.0 in /usr/local/lib/python3.7/dist-packages (from wandb>=0.10.32->simpletransformers) (7.1.2)\nCollecting docker-pycreds>=0.4.0\n Downloading docker_pycreds-0.4.0-py2.py3-none-any.whl (9.0 kB)\nRequirement already satisfied: psutil>=5.0.0 in /usr/local/lib/python3.7/dist-packages (from wandb>=0.10.32->simpletransformers) (5.4.8)\nCollecting sentry-sdk>=1.0.0\n Downloading sentry_sdk-1.3.1-py2.py3-none-any.whl (133 kB)\n\u001b[K |████████████████████████████████| 133 kB 48.6 MB/s \n\u001b[?25hCollecting shortuuid>=0.5.0\n Downloading shortuuid-1.0.1-py3-none-any.whl (7.5 kB)\nCollecting pathtools\n Downloading pathtools-0.1.2.tar.gz (11 kB)\nCollecting configparser>=3.8.1\n Downloading configparser-5.0.2-py3-none-any.whl (19 kB)\nRequirement already satisfied: six>=1.13.0 in /usr/local/lib/python3.7/dist-packages (from wandb>=0.10.32->simpletransformers) (1.15.0)\nCollecting gitdb<5,>=4.0.1\n Downloading gitdb-4.0.7-py3-none-any.whl (63 kB)\n\u001b[K |████████████████████████████████| 63 kB 1.5 MB/s \n\u001b[?25hCollecting smmap<5,>=3.0.1\n Downloading smmap-4.0.0-py2.py3-none-any.whl (24 kB)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->simpletransformers) (2021.5.30)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->simpletransformers) (3.0.4)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->simpletransformers) (2.10)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests->simpletransformers) (1.24.3)\nRequirement already satisfied: pyarrow!=4.0.0,>=1.0.0 in /usr/local/lib/python3.7/dist-packages (from datasets->simpletransformers) (3.0.0)\nCollecting fsspec>=2021.05.0\n Downloading fsspec-2021.7.0-py3-none-any.whl (118 kB)\n\u001b[K |████████████████████████████████| 118 kB 49.5 MB/s \n\u001b[?25hRequirement already satisfied: multiprocess in /usr/local/lib/python3.7/dist-packages (from datasets->simpletransformers) (0.70.12.2)\nRequirement already satisfied: dill in /usr/local/lib/python3.7/dist-packages (from datasets->simpletransformers) (0.3.4)\nCollecting xxhash\n Downloading xxhash-2.0.2-cp37-cp37m-manylinux2010_x86_64.whl (243 kB)\n\u001b[K |████████████████████████████████| 243 kB 53.6 MB/s \n\u001b[?25hRequirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.7/dist-packages (from importlib-metadata->transformers>=4.2.0->simpletransformers) (3.5.0)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.7/dist-packages (from pandas->simpletransformers) (2018.9)\nRequirement already satisfied: joblib in /usr/local/lib/python3.7/dist-packages (from sacremoses->transformers>=4.2.0->simpletransformers) (1.0.1)\nCollecting watchdog\n Downloading watchdog-2.1.5-py3-none-manylinux2014_x86_64.whl (75 kB)\n\u001b[K |████████████████████████████████| 75 kB 3.5 MB/s \n\u001b[?25hCollecting blinker\n Downloading blinker-1.4.tar.gz (111 kB)\n\u001b[K |████████████████████████████████| 111 kB 48.1 MB/s \n\u001b[?25hRequirement already satisfied: tzlocal in /usr/local/lib/python3.7/dist-packages (from streamlit->simpletransformers) (1.5.1)\nRequirement already satisfied: altair>=3.2.0 in /usr/local/lib/python3.7/dist-packages (from streamlit->simpletransformers) (4.1.0)\nRequirement already satisfied: astor in /usr/local/lib/python3.7/dist-packages (from streamlit->simpletransformers) (0.8.1)\nRequirement already satisfied: attrs in /usr/local/lib/python3.7/dist-packages (from streamlit->simpletransformers) (21.2.0)\nRequirement already satisfied: tornado>=5.0 in /usr/local/lib/python3.7/dist-packages (from streamlit->simpletransformers) (5.1.1)\nCollecting base58\n Downloading base58-2.1.0-py3-none-any.whl (5.6 kB)\nCollecting validators\n Downloading validators-0.18.2-py3-none-any.whl (19 kB)\nRequirement already satisfied: toml in /usr/local/lib/python3.7/dist-packages (from streamlit->simpletransformers) (0.10.2)\nRequirement already satisfied: pillow>=6.2.0 in /usr/local/lib/python3.7/dist-packages (from streamlit->simpletransformers) (7.1.2)\nRequirement already satisfied: cachetools>=4.0 in /usr/local/lib/python3.7/dist-packages (from streamlit->simpletransformers) (4.2.2)\nCollecting pydeck>=0.1.dev5\n Downloading pydeck-0.6.2-py2.py3-none-any.whl (4.2 MB)\n\u001b[K |████████████████████████████████| 4.2 MB 36.2 MB/s \n\u001b[?25hRequirement already satisfied: jinja2 in /usr/local/lib/python3.7/dist-packages (from altair>=3.2.0->streamlit->simpletransformers) (2.11.3)\nRequirement already satisfied: jsonschema in /usr/local/lib/python3.7/dist-packages (from altair>=3.2.0->streamlit->simpletransformers) (2.6.0)\nRequirement already satisfied: toolz in /usr/local/lib/python3.7/dist-packages (from altair>=3.2.0->streamlit->simpletransformers) (0.11.1)\nRequirement already satisfied: entrypoints in /usr/local/lib/python3.7/dist-packages (from altair>=3.2.0->streamlit->simpletransformers) (0.3)\nRequirement already satisfied: ipywidgets>=7.0.0 in /usr/local/lib/python3.7/dist-packages (from pydeck>=0.1.dev5->streamlit->simpletransformers) (7.6.3)\nCollecting ipykernel>=5.1.2\n Downloading ipykernel-6.2.0-py3-none-any.whl (122 kB)\n\u001b[K |████████████████████████████████| 122 kB 43.5 MB/s \n\u001b[?25hRequirement already satisfied: traitlets>=4.3.2 in /usr/local/lib/python3.7/dist-packages (from pydeck>=0.1.dev5->streamlit->simpletransformers) (5.0.5)\nRequirement already satisfied: jupyter-client<8.0 in /usr/local/lib/python3.7/dist-packages (from ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (5.3.5)\nRequirement already satisfied: matplotlib-inline<0.2.0,>=0.1.0 in /usr/local/lib/python3.7/dist-packages (from ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.1.2)\nCollecting ipython<8.0,>=7.23.1\n Downloading ipython-7.26.0-py3-none-any.whl (786 kB)\n\u001b[K |████████████████████████████████| 786 kB 42.3 MB/s \n\u001b[?25hRequirement already satisfied: argcomplete>=1.12.3 in /usr/local/lib/python3.7/dist-packages (from ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (1.12.3)\nRequirement already satisfied: debugpy<2.0,>=1.0.0 in /usr/local/lib/python3.7/dist-packages (from ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (1.0.0)\nRequirement already satisfied: pickleshare in /usr/local/lib/python3.7/dist-packages (from ipython<8.0,>=7.23.1->ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.7.5)\nRequirement already satisfied: backcall in /usr/local/lib/python3.7/dist-packages (from ipython<8.0,>=7.23.1->ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.2.0)\nRequirement already satisfied: jedi>=0.16 in /usr/local/lib/python3.7/dist-packages (from ipython<8.0,>=7.23.1->ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.18.0)\nCollecting prompt-toolkit!=3.0.0,!=3.0.1,<3.1.0,>=2.0.0\n Downloading prompt_toolkit-3.0.20-py3-none-any.whl (370 kB)\n\u001b[K |████████████████████████████████| 370 kB 51.2 MB/s \n\u001b[?25hRequirement already satisfied: decorator in /usr/local/lib/python3.7/dist-packages (from ipython<8.0,>=7.23.1->ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (4.4.2)\nRequirement already satisfied: pexpect>4.3 in /usr/local/lib/python3.7/dist-packages (from ipython<8.0,>=7.23.1->ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (4.8.0)\nRequirement already satisfied: setuptools>=18.5 in /usr/local/lib/python3.7/dist-packages (from ipython<8.0,>=7.23.1->ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (57.4.0)\nRequirement already satisfied: pygments in /usr/local/lib/python3.7/dist-packages (from ipython<8.0,>=7.23.1->ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (2.6.1)\nRequirement already satisfied: widgetsnbextension~=3.5.0 in /usr/local/lib/python3.7/dist-packages (from ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (3.5.1)\nRequirement already satisfied: nbformat>=4.2.0 in /usr/local/lib/python3.7/dist-packages (from ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (5.1.3)\nRequirement already satisfied: jupyterlab-widgets>=1.0.0 in /usr/local/lib/python3.7/dist-packages (from ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (1.0.0)\nRequirement already satisfied: parso<0.9.0,>=0.8.0 in /usr/local/lib/python3.7/dist-packages (from jedi>=0.16->ipython<8.0,>=7.23.1->ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.8.2)\nRequirement already satisfied: MarkupSafe>=0.23 in /usr/local/lib/python3.7/dist-packages (from jinja2->altair>=3.2.0->streamlit->simpletransformers) (2.0.1)\nRequirement already satisfied: pyzmq>=13 in /usr/local/lib/python3.7/dist-packages (from jupyter-client<8.0->ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (22.2.1)\nRequirement already satisfied: jupyter-core>=4.6.0 in /usr/local/lib/python3.7/dist-packages (from jupyter-client<8.0->ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (4.7.1)\nRequirement already satisfied: ipython-genutils in /usr/local/lib/python3.7/dist-packages (from nbformat>=4.2.0->ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.2.0)\nRequirement already satisfied: ptyprocess>=0.5 in /usr/local/lib/python3.7/dist-packages (from pexpect>4.3->ipython<8.0,>=7.23.1->ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.7.0)\nRequirement already satisfied: wcwidth in /usr/local/lib/python3.7/dist-packages (from prompt-toolkit!=3.0.0,!=3.0.1,<3.1.0,>=2.0.0->ipython<8.0,>=7.23.1->ipykernel>=5.1.2->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.2.5)\nRequirement already satisfied: notebook>=4.4.1 in /usr/local/lib/python3.7/dist-packages (from widgetsnbextension~=3.5.0->ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (5.3.1)\nRequirement already satisfied: Send2Trash in /usr/local/lib/python3.7/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (1.8.0)\nRequirement already satisfied: nbconvert in /usr/local/lib/python3.7/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (5.6.1)\nRequirement already satisfied: terminado>=0.8.1 in /usr/local/lib/python3.7/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.11.0)\nRequirement already satisfied: mistune<2,>=0.8.1 in /usr/local/lib/python3.7/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.8.4)\nRequirement already satisfied: pandocfilters>=1.4.1 in /usr/local/lib/python3.7/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (1.4.3)\nRequirement already satisfied: bleach in /usr/local/lib/python3.7/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (4.0.0)\nRequirement already satisfied: testpath in /usr/local/lib/python3.7/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.5.0)\nRequirement already satisfied: defusedxml in /usr/local/lib/python3.7/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.7.1)\nRequirement already satisfied: webencodings in /usr/local/lib/python3.7/dist-packages (from bleach->nbconvert->notebook>=4.4.1->widgetsnbextension~=3.5.0->ipywidgets>=7.0.0->pydeck>=0.1.dev5->streamlit->simpletransformers) (0.5.1)\nBuilding wheels for collected packages: subprocess32, pathtools, seqeval, blinker\n Building wheel for subprocess32 (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for subprocess32: filename=subprocess32-3.5.4-py3-none-any.whl size=6502 sha256=edafa257c8c538c743626ca954d9ad2162bcd285b9e8b603a4e42e40bc0915b0\n Stored in directory: /root/.cache/pip/wheels/50/ca/fa/8fca8d246e64f19488d07567547ddec8eb084e8c0d7a59226a\n Building wheel for pathtools (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for pathtools: filename=pathtools-0.1.2-py3-none-any.whl size=8807 sha256=bb14f38637a9266577970b220d800cc0219eae7839138c878e142d6c1ed029aa\n Stored in directory: /root/.cache/pip/wheels/3e/31/09/fa59cef12cdcfecc627b3d24273699f390e71828921b2cbba2\n Building wheel for seqeval (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for seqeval: filename=seqeval-1.2.2-py3-none-any.whl size=16181 sha256=27fd50b7f536964cd2074bf74947dc6edf8ff89dd1f9ed4c9b42e359af39de58\n Stored in directory: /root/.cache/pip/wheels/05/96/ee/7cac4e74f3b19e3158dce26a20a1c86b3533c43ec72a549fd7\n Building wheel for blinker (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for blinker: filename=blinker-1.4-py3-none-any.whl size=13478 sha256=6b027cce8df1741f240444ffb96aeb86155f3aa0dc970cf152c5641a7fd0ae23\n Stored in directory: /root/.cache/pip/wheels/22/f5/18/df711b66eb25b21325c132757d4314db9ac5e8dabeaf196eab\nSuccessfully built subprocess32 pathtools seqeval blinker\nInstalling collected packages: prompt-toolkit, ipython, ipykernel, smmap, gitdb, xxhash, watchdog, validators, subprocess32, shortuuid, sentry-sdk, pydeck, pathtools, GitPython, fsspec, docker-pycreds, configparser, blinker, base58, wandb, tensorboardx, streamlit, seqeval, sentencepiece, datasets, simpletransformers\n Attempting uninstall: prompt-toolkit\n Found existing installation: prompt-toolkit 1.0.18\n Uninstalling prompt-toolkit-1.0.18:\n Successfully uninstalled prompt-toolkit-1.0.18\n Attempting uninstall: ipython\n Found existing installation: ipython 5.5.0\n Uninstalling ipython-5.5.0:\n Successfully uninstalled ipython-5.5.0\n Attempting uninstall: ipykernel\n Found existing installation: ipykernel 4.10.1\n Uninstalling ipykernel-4.10.1:\n Successfully uninstalled ipykernel-4.10.1\n\u001b[31mERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.\njupyter-console 5.2.0 requires prompt-toolkit<2.0.0,>=1.0.0, but you have prompt-toolkit 3.0.20 which is incompatible.\ngoogle-colab 1.0.0 requires ipykernel~=4.10, but you have ipykernel 6.2.0 which is incompatible.\ngoogle-colab 1.0.0 requires ipython~=5.5.0, but you have ipython 7.26.0 which is incompatible.\u001b[0m\nSuccessfully installed GitPython-3.1.18 base58-2.1.0 blinker-1.4 configparser-5.0.2 datasets-1.11.0 docker-pycreds-0.4.0 fsspec-2021.7.0 gitdb-4.0.7 ipykernel-6.2.0 ipython-7.26.0 pathtools-0.1.2 prompt-toolkit-3.0.20 pydeck-0.6.2 sentencepiece-0.1.96 sentry-sdk-1.3.1 seqeval-1.2.2 shortuuid-1.0.1 simpletransformers-0.61.13 smmap-4.0.0 streamlit-0.87.0 subprocess32-3.5.4 tensorboardx-2.4 validators-0.18.2 wandb-0.12.0 watchdog-2.1.5 xxhash-2.0.2\n"
],
[
"from os import path\nfrom IPython.display import Audio\nfrom transformers import pipeline, AutoTokenizer, AutoModelForSeq2SeqLM, Wav2Vec2Processor, Wav2Vec2ForCTC\nimport librosa\nimport torch",
"_____no_output_____"
],
[
"class speechModule(CombineInator):\n\n def __init__(self):\n self.SAMPLING_RATE = 16_000\n self.git_repo_url = 'https://github.com/CorentinJ/Real-Time-Voice-Cloning.git'\n self.project_name = splitext(basename(self.git_repo_url))[0]\n \n def get_repo(self):\n\n \"\"\"\n\n Metinin sese çevrilmesi sırasında kullanılacak ses klonlama kütüphanesini çeker.\n\n \"\"\"\n\n if not exists(self.project_name):\n # clone and install\n !git clone -q --recursive {self.git_repo_url}\n # install dependencies\n !cd {self.project_name} && pip install -q -r requirements.txt\n !pip install -q gdown\n !apt-get install -qq libportaudio2\n !pip install -q https://github.com/tugstugi/dl-colab-notebooks/archive/colab_utils.zip\n # download pretrained model\n !cd {self.project_name} && wget https://github.com/blue-fish/Real-Time-Voice-Cloning/releases/download/v1.0/pretrained.zip && unzip -o pretrained.zip\n from sys import path as syspath\n syspath.append(self.project_name)\n \n def wav2vec_model(self, source):\n\n \"\"\"\n\n Sesin metne çevrilmesi sırasında kullanılacak ilgili dile göre wav2vec modelini belirler.\n\n :param source: ses dosyası dili (\"tr\" / \"en\")\n :type source: str\n\n \"\"\"\n\n processor = None\n model = None\n\n if source == \"en\":\n processor = Wav2Vec2Processor.from_pretrained(\"facebook/wav2vec2-large-960h\")\n model = Wav2Vec2ForCTC.from_pretrained(\"facebook/wav2vec2-large-960h\")\n \n elif source ==\"tr\":\n processor = Wav2Vec2Processor.from_pretrained(\"m3hrdadfi/wav2vec2-large-xlsr-turkish\")\n model = Wav2Vec2ForCTC.from_pretrained(\"m3hrdadfi/wav2vec2-large-xlsr-turkish\") \n\n return model, processor\n\n \n def speech2text(self, audio_file, model, processor, language):\n\n \"\"\"\n\n Girdi olarak verilen sesi metne çevirir.\n\n :param audio_file: ses dosyasının yer aldığı dizin\n type audio_file: str\n \n :param model: sesin metne çevrilmesi esnasında kullanılacak huggingface kütüphanesinden çekilen model\n\n :param processor: sesin metne çevrilmesi esnasında kullanılacak huggingface kütüphanesinden çekilen istemci\n\n :param language: girdi olarak verilen ses doyasının dili (\"tr\" / \"en\")\n :type language: str\n\n \"\"\"\n\n #load any audio file of your choice\n speech, rate = librosa.load(audio_file, sr=self.SAMPLING_RATE)\n input_values = processor(speech, sampling_rate=self.SAMPLING_RATE, return_tensors = 'pt').input_values\n\n #Store logits (non-normalized predictions)\n logits = model(input_values).logits\n\n #Store predicted id's\n predicted_ids = torch.argmax(logits, dim =-1)\n\n #decode the audio to generate text\n response = processor.decode(predicted_ids[0]).lower()\n\n if language == \"en\":\n response = \">>tur<< \" + response\n\n return response\n \n \n def text2speech(self, audio, translation):\n\n \"\"\"\n\n Metini sese çevirir.\n\n :param audio: klonlanacak ses doyasının yer aldığı dizin\n :type audio: str\n\n :param translation: çevirisi yapılmış metin\n :type translation: str\n\n \"\"\"\n from numpy import pad as pad\n from synthesizer.inference import Synthesizer\n from encoder import inference as encoder\n from vocoder import inference as vocoder\n from pathlib import Path\n\n encoder.load_model(self.project_name / Path(\"encoder/saved_models/pretrained.pt\"))\n synthesizer = Synthesizer(self.project_name / Path(\"synthesizer/saved_models/pretrained/pretrained.pt\"))\n vocoder.load_model(self.project_name / Path(\"vocoder/saved_models/pretrained/pretrained.pt\"))\n\n embedding = encoder.embed_utterance(encoder.preprocess_wav(audio, self.SAMPLING_RATE))\n specs = synthesizer.synthesize_spectrograms([translation], [embedding])\n generated_wav = vocoder.infer_waveform(specs[0])\n generated_wav = pad(generated_wav, (0, self.SAMPLING_RATE), mode=\"constant\")\n\n return Audio(generated_wav, rate=self.SAMPLING_RATE, autoplay=True)\n\n \n\n\n \n def speech2text2trans2speech(self, filename:str, source_lang:str, output_type:str = \"text\"):\n \n \"\"\"\n\n Aldığı ses dosyasını text'e dönüştürüp, hedeflenen dile çeviren ve çevirdiği metni ses olarak\n döndüren fonksiyon.\n \n :param filename: Ses dosyasının adı\n :type filename: str\n\n :param lang: Ses dosyası dili (\"en\"/\"tr\") \n :type lang: str \n \n \"\"\"\n\n output_types = [\"text\", \"speech\"]\n source_languages = [\"en\", \"tr\"]\n\n if source_lang not in source_languages:\n print(\"Kaynak dil olarak yalnızca 'en' ve 'tr' parametreleri kullanılabilir.\")\n return None\n \n if output_type not in output_types:\n print(\"Çıkış türü için yalnızca 'text' ve 'speech' parametreleri desteklenmektedir.\")\n return None\n\n if source_lang == \"en\" and output_type==\"speech\":\n print(\"Üzgünüz, text2speech modülümüzde Türkçe dil desteği bulunmamaktadır.\\n\")\n return None\n\n model_trs, tokenizer_trs, pipe_trs = CombineInator.translate_model(self, source_lang)\n\n model_s2t, processor_s2t = self.wav2vec_model(source_lang)\n \n input_text = self.speech2text(filename, model_s2t, processor_s2t, source_lang)\n print(input_text)\n translation = CombineInator.translate(self, pipe_trs, model_trs, tokenizer_trs, input_text)\n \n if output_type == \"text\":\n return translation\n \n else:\n print(\"\\n\" + translation + \"\\n\")\n return self.text2speech(filename, translation)",
"_____no_output_____"
]
],
[
[
"### Örnek kullanım",
"_____no_output_____"
]
],
[
[
"filename = \"_path_to_wav_file\" # ses dosyası pathi verilmelidir",
"_____no_output_____"
],
[
"speechM = speechModule()",
"_____no_output_____"
],
[
"speechM.get_repo()",
"\u001b[K |████████████████████████████████| 80 kB 3.5 MB/s \n\u001b[K |████████████████████████████████| 676 kB 26.6 MB/s \n\u001b[K |████████████████████████████████| 10.3 MB 30.7 MB/s \n\u001b[K |████████████████████████████████| 14.5 MB 72 kB/s \n\u001b[K |████████████████████████████████| 241 kB 47.5 MB/s \n\u001b[K |████████████████████████████████| 8.3 MB 46.6 MB/s \n\u001b[K |████████████████████████████████| 66 kB 4.3 MB/s \n\u001b[K |████████████████████████████████| 1.1 MB 35.2 MB/s \n\u001b[K |████████████████████████████████| 52 kB 1.2 MB/s \n\u001b[K |████████████████████████████████| 59.9 MB 37 kB/s \n\u001b[K |████████████████████████████████| 317 kB 48.4 MB/s \n\u001b[?25h Building wheel for umap-learn (setup.py) ... \u001b[?25l\u001b[?25hdone\n Building wheel for pynndescent (setup.py) ... \u001b[?25l\u001b[?25hdone\n Building wheel for visdom (setup.py) ... \u001b[?25l\u001b[?25hdone\n Building wheel for webrtcvad (setup.py) ... \u001b[?25l\u001b[?25hdone\n Building wheel for torchfile (setup.py) ... \u001b[?25l\u001b[?25hdone\n\u001b[31mERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.\ngoogle-colab 1.0.0 requires ipykernel~=4.10, but you have ipykernel 6.2.0 which is incompatible.\ngoogle-colab 1.0.0 requires ipython~=5.5.0, but you have ipython 7.26.0 which is incompatible.\ndatascience 0.10.6 requires folium==0.2.1, but you have folium 0.8.3 which is incompatible.\nalbumentations 0.1.12 requires imgaug<0.2.7,>=0.2.5, but you have imgaug 0.2.9 which is incompatible.\u001b[0m\nSelecting previously unselected package libportaudio2:amd64.\n(Reading database ... 148489 files and directories currently installed.)\nPreparing to unpack .../libportaudio2_19.6.0-1_amd64.deb ...\nUnpacking libportaudio2:amd64 (19.6.0-1) ...\nSetting up libportaudio2:amd64 (19.6.0-1) ...\nProcessing triggers for libc-bin (2.27-3ubuntu1.2) ...\n/sbin/ldconfig.real: /usr/local/lib/python3.7/dist-packages/ideep4py/lib/libmkldnn.so.0 is not a symbolic link\n\n\u001b[K \\ 3.0 kB 3.1 MB/s\n\u001b[?25h Building wheel for Deep-Learning-Colab-Notebook-Utils (setup.py) ... \u001b[?25l\u001b[?25hdone\n--2021-08-26 18:25:38-- https://github.com/blue-fish/Real-Time-Voice-Cloning/releases/download/v1.0/pretrained.zip\nResolving github.com (github.com)... 52.69.186.44\nConnecting to github.com (github.com)|52.69.186.44|:443... connected.\nHTTP request sent, awaiting response... 302 Found\nLocation: https://github-releases.githubusercontent.com/274049886/9764da00-6919-11eb-8444-e8027183f04f?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=AKIAIWNJYAX4CSVEH53A%2F20210826%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Date=20210826T182538Z&X-Amz-Expires=300&X-Amz-Signature=20c4159d481be7c38c865b9d7c7db0b526a7a04e300c368cffb19e99a07ad177&X-Amz-SignedHeaders=host&actor_id=0&key_id=0&repo_id=274049886&response-content-disposition=attachment%3B%20filename%3Dpretrained.zip&response-content-type=application%2Foctet-stream [following]\n--2021-08-26 18:25:38-- https://github-releases.githubusercontent.com/274049886/9764da00-6919-11eb-8444-e8027183f04f?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=AKIAIWNJYAX4CSVEH53A%2F20210826%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Date=20210826T182538Z&X-Amz-Expires=300&X-Amz-Signature=20c4159d481be7c38c865b9d7c7db0b526a7a04e300c368cffb19e99a07ad177&X-Amz-SignedHeaders=host&actor_id=0&key_id=0&repo_id=274049886&response-content-disposition=attachment%3B%20filename%3Dpretrained.zip&response-content-type=application%2Foctet-stream\nResolving github-releases.githubusercontent.com (github-releases.githubusercontent.com)... 185.199.110.154, 185.199.108.154, 185.199.109.154, ...\nConnecting to github-releases.githubusercontent.com (github-releases.githubusercontent.com)|185.199.110.154|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 397171814 (379M) [application/octet-stream]\nSaving to: ‘pretrained.zip’\n\npretrained.zip 100%[===================>] 378.77M 29.1MB/s in 13s \n\n2021-08-26 18:25:52 (29.3 MB/s) - ‘pretrained.zip’ saved [397171814/397171814]\n\nArchive: pretrained.zip\n creating: encoder/saved_models/\n inflating: encoder/saved_models/pretrained.pt \n creating: synthesizer/saved_models/\n creating: synthesizer/saved_models/pretrained/\n inflating: synthesizer/saved_models/pretrained/pretrained.pt \n creating: vocoder/saved_models/\n creating: vocoder/saved_models/pretrained/\n inflating: vocoder/saved_models/pretrained/pretrained.pt \n"
],
[
"speechM.speech2text2trans2speech(filename, \"tr\", \"speech\")",
"_____no_output_____"
]
],
[
[
"## Lxmert (child)",
"_____no_output_____"
]
],
[
[
"!git clone https://github.com/hila-chefer/Transformer-MM-Explainability\n\nimport os\nos.chdir(f'./Transformer-MM-Explainability')\n\n!pip install -r requirements.txt",
"_____no_output_____"
],
[
"%cd Transformer-MM-Explainability",
"_____no_output_____"
],
[
"from lxmert.lxmert.src.modeling_frcnn import GeneralizedRCNN\nimport lxmert.lxmert.src.vqa_utils as utils\nfrom lxmert.lxmert.src.processing_image import Preprocess\nfrom transformers import LxmertTokenizer\nfrom lxmert.lxmert.src.huggingface_lxmert import LxmertForQuestionAnswering\nfrom lxmert.lxmert.src.lxmert_lrp import LxmertForQuestionAnswering as LxmertForQuestionAnsweringLRP\nfrom tqdm import tqdm\nfrom lxmert.lxmert.src.ExplanationGenerator import GeneratorOurs, GeneratorBaselines, GeneratorOursAblationNoAggregation\nimport random\nimport numpy as np\nimport cv2\nimport torch\nimport matplotlib.pyplot as plt\nfrom PIL import Image\nimport torchvision.transforms as transforms\nfrom captum.attr import visualization\nimport requests",
"_____no_output_____"
],
[
"class Lxmert(CombineInator):\n\n def __init__(self):\n self.OBJ_URL = \"https://raw.githubusercontent.com/airsplay/py-bottom-up-attention/master/demo/data/genome/1600-400-20/objects_vocab.txt\"\n self.ATTR_URL = \"https://raw.githubusercontent.com/airsplay/py-bottom-up-attention/master/demo/data/genome/1600-400-20/attributes_vocab.txt\"\n self.VQA_URL = \"https://raw.githubusercontent.com/airsplay/lxmert/master/data/vqa/trainval_label2ans.json\"\n self.model_lrp = self.ModelUsage()\n self.lrp = GeneratorOurs(self.model_lrp)\n self.baselines = GeneratorBaselines(self.model_lrp)\n self.vqa_answers = utils.get_data(self.VQA_URL)\n \n class ModelUsage:\n \n \"\"\"\n\n Model kullanımı için sınıf yapısı\n\n \"\"\"\n \n def __init__(self, use_lrp=True):\n self.VQA_URL = \"https://raw.githubusercontent.com/airsplay/lxmert/master/data/vqa/trainval_label2ans.json\"\n\n self.vqa_answers = utils.get_data(self.VQA_URL)\n\n # load models and model components\n self.frcnn_cfg = utils.Config.from_pretrained(\"unc-nlp/frcnn-vg-finetuned\")\n self.frcnn_cfg.MODEL.DEVICE = \"cuda\"\n\n self.frcnn = GeneralizedRCNN.from_pretrained(\"unc-nlp/frcnn-vg-finetuned\", config=self.frcnn_cfg)\n\n self.image_preprocess = Preprocess(self.frcnn_cfg)\n\n self.lxmert_tokenizer = LxmertTokenizer.from_pretrained(\"unc-nlp/lxmert-base-uncased\")\n\n if use_lrp:\n self.lxmert_vqa = LxmertForQuestionAnsweringLRP.from_pretrained(\"unc-nlp/lxmert-vqa-uncased\").to(\"cuda\")\n else:\n self.lxmert_vqa = LxmertForQuestionAnswering.from_pretrained(\"unc-nlp/lxmert-vqa-uncased\").to(\"cuda\")\n\n self.lxmert_vqa.eval()\n self.model = self.lxmert_vqa\n\n # self.vqa_dataset = vqa_data.VQADataset(splits=\"valid\")\n\n def forward(self, item):\n \n PATH, question = item\n \n self.image_file_path = PATH\n\n # run frcnn\n images, sizes, scales_yx = self.image_preprocess(PATH)\n output_dict = self.frcnn(\n images,\n sizes,\n scales_yx=scales_yx,\n padding=\"max_detections\",\n max_detections= self.frcnn_cfg.max_detections,\n return_tensors=\"pt\"\n )\n inputs = self.lxmert_tokenizer(\n question,\n truncation=True,\n return_token_type_ids=True,\n return_attention_mask=True,\n add_special_tokens=True,\n return_tensors=\"pt\"\n )\n self.question_tokens = self.lxmert_tokenizer.convert_ids_to_tokens(inputs.input_ids.flatten())\n self.text_len = len(self.question_tokens)\n # Very important that the boxes are normalized\n normalized_boxes = output_dict.get(\"normalized_boxes\")\n features = output_dict.get(\"roi_features\")\n self.image_boxes_len = features.shape[1]\n self.bboxes = output_dict.get(\"boxes\")\n self.output = self.lxmert_vqa(\n input_ids=inputs.input_ids.to(\"cuda\"),\n attention_mask=inputs.attention_mask.to(\"cuda\"),\n visual_feats=features.to(\"cuda\"),\n visual_pos=normalized_boxes.to(\"cuda\"),\n token_type_ids=inputs.token_type_ids.to(\"cuda\"),\n return_dict=True,\n output_attentions=False,\n )\n return self.output\n \n def ceviri(self, text: str, lang_src='tr'):\n \n \"\"\"\n \n Aldığı metni istenilen dile çeviren fonksiyon.\n\n :param text: Orjinal metin\n :type text: str\n\n :param lang_src: Metin dosyasının dili (kaynak dili)\n :type lang_src: str\n \n :param lang_tgt: Çevrilecek dil (hedef dil)\n :param lang_tgt: str\n\n :return: translated text\n\n \"\"\"\n\n if lang_src == \"en\":\n text = \">>tur<< \" + text\n\n model, tokenizer, pipeline = CombineInator.translate_model(self, lang_src)\n return (CombineInator.translate(self, pipeline, model, tokenizer, text))\n \n \n def save_image_vis(self, image_file_path, bbox_scores):\n \"\"\"\n Resim üzerinde sorunun cevabını çizer ve kaydeder.\n\n :param image_file_path: imgenin yer aldığı dizin\n :type image_file_path: str\n\n :param bbox_scores: tespit edilen nesnelerin skorlarını içeren tensor\n :type bbox_scores: tensor\n\n \"\"\"\n\n\n _, top_bboxes_indices = bbox_scores.topk(k=1, dim=-1)\n img = cv2.imread(image_file_path)\n mask = torch.zeros(img.shape[0], img.shape[1])\n for index in range(len(bbox_scores)):\n [x, y, w, h] = self.model_lrp.bboxes[0][index]\n curr_score_tensor = mask[int(y):int(h), int(x):int(w)]\n new_score_tensor = torch.ones_like(curr_score_tensor)*bbox_scores[index].item()\n mask[int(y):int(h), int(x):int(w)] = torch.max(new_score_tensor,mask[int(y):int(h), int(x):int(w)])\n mask = (mask - mask.min()) / (mask.max() - mask.min())\n mask = mask.unsqueeze_(-1)\n mask = mask.expand(img.shape)\n img = img * mask.cpu().data.numpy()\n cv2.imwrite('lxmert/lxmert/experiments/paper/new.jpg', img)\n \n \n \n def get_image_and_question(self, img_path:str, soru:str):\n \n \"\"\"\n\n Input olarak verilen imge ve soruyu döndürür.\n\n :param img_path: Soru sorulacak imgenin path bilgisi\n :type img_path: str\n\n :param soru: Resim özelinde modele sorulacak olan Türkçe soru \n :type soru: str\n\n :return: image_scores, text_scores\n\n \"\"\"\n \n ing_soru = self.ceviri(soru, \"tr\")\n R_t_t, R_t_i = self.lrp.generate_ours((img_path, ing_soru), use_lrp=False, normalize_self_attention=True, method_name=\"ours\")\n \n return R_t_i[0], R_t_t[0] \n\n \n def resim_uzerinden_soru_cevap(self, PATH:str, turkce_soru:str):\n \"\"\"\n Verilen girdi imgesi üzerinden yine veriler sorular ile sorgulama yapılabilmesini\n sağlar. \n\n PATH: imgenin path bilgisi\n turkce_soru: Resimde cevabı aranacak soru\n \"\"\"\n\n #Eğer sorgulanacak resim local'de yok ve internet üzerinden bir resim ise:\n if PATH.startswith(\"http\"):\n im = Image.open(requests.get(PATH, stream=True).raw)\n im.save('lxmert/lxmert/experiments/paper/online_image.jpg', 'JPEG')\n PATH = 'lxmert/lxmert/experiments/paper/online_image.jpg'\n\n\n image_scores, text_scores = self.get_image_and_question(PATH, turkce_soru)\n self.save_image_vis(PATH, image_scores)\n orig_image = Image.open(self.model_lrp.image_file_path)\n\n fig, axs = plt.subplots(ncols=2, figsize=(20, 5))\n axs[0].imshow(orig_image);\n axs[0].axis('off');\n axs[0].set_title('original');\n\n masked_image = Image.open('lxmert/lxmert/experiments/paper/new.jpg')\n axs[1].imshow(masked_image);\n axs[1].axis('off');\n axs[1].set_title('masked');\n\n text_scores = (text_scores - text_scores.min()) / (text_scores.max() - text_scores.min())\n vis_data_records = [visualization.VisualizationDataRecord(text_scores,0,0,0,0,0,self.model_lrp.question_tokens,1)]\n visualization.visualize_text(vis_data_records)\n cevap = self.ceviri(self.vqa_answers[self.model_lrp.output.question_answering_score.argmax()], lang_src='en')\n print(\"ANSWER:\", cevap)\n ",
"_____no_output_____"
]
],
[
[
"### Örnek kullanım",
"_____no_output_____"
]
],
[
[
"lxmert = Lxmert()\n\nPATH = '_path_to_jpg_' # jpg dosyası pathi verilmelidir\nturkce_soru = 'Resimde neler var'\n\nlxmert.resim_uzerinden_soru_cevap(PATH, turkce_soru)",
"loading configuration file cache\nloading weights file https://cdn.huggingface.co/unc-nlp/frcnn-vg-finetuned/pytorch_model.bin from cache at /root/.cache/torch/transformers/57f6df6abe353be2773f2700159c65615babf39ab5b48114d2b49267672ae10f.77b59256a4cf8343ae0f923246a81489fc8d82f98d082edc2d2037c977c0d9d0\n"
]
],
[
[
"## Web Arayüz",
"_____no_output_____"
]
],
[
[
"!pip install flask-ngrok",
"Collecting flask-ngrok\n Downloading flask_ngrok-0.0.25-py3-none-any.whl (3.1 kB)\nRequirement already satisfied: Flask>=0.8 in /usr/local/lib/python3.7/dist-packages (from flask-ngrok) (1.1.4)\nRequirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from flask-ngrok) (2.23.0)\nRequirement already satisfied: Jinja2<3.0,>=2.10.1 in /usr/local/lib/python3.7/dist-packages (from Flask>=0.8->flask-ngrok) (2.11.3)\nRequirement already satisfied: click<8.0,>=5.1 in /usr/local/lib/python3.7/dist-packages (from Flask>=0.8->flask-ngrok) (7.1.2)\nRequirement already satisfied: itsdangerous<2.0,>=0.24 in /usr/local/lib/python3.7/dist-packages (from Flask>=0.8->flask-ngrok) (1.1.0)\nRequirement already satisfied: Werkzeug<2.0,>=0.15 in /usr/local/lib/python3.7/dist-packages (from Flask>=0.8->flask-ngrok) (1.0.1)\nRequirement already satisfied: MarkupSafe>=0.23 in /usr/local/lib/python3.7/dist-packages (from Jinja2<3.0,>=2.10.1->Flask>=0.8->flask-ngrok) (2.0.1)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests->flask-ngrok) (1.24.3)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->flask-ngrok) (3.0.4)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->flask-ngrok) (2021.5.30)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->flask-ngrok) (2.10)\nInstalling collected packages: flask-ngrok\nSuccessfully installed flask-ngrok-0.0.25\n"
],
[
"from flask import Flask, redirect, url_for, render_template, request, flash\nfrom flask_ngrok import run_with_ngrok",
"_____no_output_____"
],
[
"# Burada web_dependencies klasörü içerisinde bulunan klasörlerin pathi verilmelidir.\ntemplate_folder = '_path_to_templates_folder_'\nstatic_folder = '_path_to_static_folder_'",
"_____no_output_____"
],
[
"app = Flask(__name__, template_folder=template_folder, static_folder=static_folder)\nrun_with_ngrok(app) # Start ngrok when app is run\n\n@app.route(\"/\", methods=['GET', 'POST'])\ndef home():\n if request.method == 'POST':\n konu = request.form[\"topic\"]\n library.categorical_scraper(konu, PATH, 20, text_into_sentences_param=False)\n \n return render_template(\"index.html\")\n\nif __name__ == \"__main__\":\n\n\t#app.debug = True\n\tapp.run()",
" * Serving Flask app \"__main__\" (lazy loading)\n * Environment: production\n\u001b[31m WARNING: This is a development server. Do not use it in a production deployment.\u001b[0m\n\u001b[2m Use a production WSGI server instead.\u001b[0m\n * Debug mode: off\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d03d130bcba804c0aaea8089e71ab7079a53e28c | 53,561 | ipynb | Jupyter Notebook | nb/dhod_centrals_gs.ipynb | gitter-badger/DHOD | f2f084fea6c299f95d15cbea5ec94d404bc946b5 | [
"MIT"
] | 6 | 2020-03-26T17:16:22.000Z | 2021-08-19T21:39:16.000Z | nb/dhod_centrals_gs.ipynb | gitter-badger/DHOD | f2f084fea6c299f95d15cbea5ec94d404bc946b5 | [
"MIT"
] | 23 | 2019-11-12T23:49:31.000Z | 2021-08-06T16:53:35.000Z | nb/dhod_centrals_gs.ipynb | gitter-badger/DHOD | f2f084fea6c299f95d15cbea5ec94d404bc946b5 | [
"MIT"
] | 1 | 2019-12-02T00:52:37.000Z | 2019-12-02T00:52:37.000Z | 85.6976 | 13,852 | 0.830007 | [
[
[
"import numpy as np \n\nimport tensorflow.compat.v1 as tf\ntf.disable_v2_behavior()\n\nimport tensorflow_probability as tfp\n# -- plotting\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nmpl.rcParams['text.usetex'] = True\nmpl.rcParams['font.family'] = 'serif'\nmpl.rcParams['axes.linewidth'] = 1.5\nmpl.rcParams['axes.xmargin'] = 1\nmpl.rcParams['xtick.labelsize'] = 'x-large'\nmpl.rcParams['xtick.major.size'] = 5\nmpl.rcParams['xtick.major.width'] = 1.5\nmpl.rcParams['ytick.labelsize'] = 'x-large'\nmpl.rcParams['ytick.major.size'] = 5\nmpl.rcParams['ytick.major.width'] = 1.5\nmpl.rcParams['legend.frameon'] = False",
"WARNING:tensorflow:From /usr/lib/python3.7/site-packages/tensorflow_core/python/compat/v2_compat.py:65: disable_resource_variables (from tensorflow.python.ops.variable_scope) is deprecated and will be removed in a future version.\nInstructions for updating:\nnon-resource variables are not supported in the long term\n"
],
[
"#import tensorflow as tf \n\"\"\"\nGumbel Softmax functions borrowed from http://blog.evjang.com/2016/11/tutorial-categorical-variational.html\n\"\"\"\n\ndef sample_gumbel(shape, eps=1e-7): \n \"\"\"Sample from Gumbel(0, 1)\"\"\"\n U = tf.random_uniform(shape,minval=0,maxval=1)\n return -tf.log(-tf.log(U + eps) + eps)\n\ndef gumbel_softmax_sample(logits, temperature): \n \"\"\" Draw a sample from the Gumbel-Softmax distribution\"\"\"\n y = logits + sample_gumbel(tf.shape(logits))\n return tf.nn.softmax( y / temperature)\n\ndef gumbel_softmax(logits, temperature, hard=False):\n \"\"\"Sample from the Gumbel-Softmax distribution and optionally discretize.\n Args:\n logits: [batch_size, n_class] unnormalized log-probs\n temperature: non-negative scalar\n hard: if True, take argmax, but differentiate w.r.t. soft sample y\n Returns:\n [batch_size,..., n_class] sample from the Gumbel-Softmax distribution.\n If hard=True, then the returned sample will be one-hot, otherwise it will\n be a probabilitiy distribution that sums to 1 across classes\n \"\"\"\n y = gumbel_softmax_sample(logits, temperature)\n if hard:\n k = tf.shape(logits)[-1]\n #y_hard = tf.cast(tf.one_hot(tf.argmax(y,1),k), y.dtype)\n y_hard = tf.cast(tf.equal(y,tf.reduce_max(y,-1,keep_dims=True)),y.dtype)\n y = tf.stop_gradient(y_hard - y) + y\n return y",
"_____no_output_____"
],
[
"d = gumbel_softmax(np.array([np.log(0.5), np.log(0.5)] ), 0.5, hard=True)",
"WARNING:tensorflow:From <ipython-input-2-abc763f8b025>:31: calling reduce_max_v1 (from tensorflow.python.ops.math_ops) with keep_dims is deprecated and will be removed in a future version.\nInstructions for updating:\nkeep_dims is deprecated, use keepdims instead\n"
],
[
"tfp.__version__",
"_____no_output_____"
],
[
"sess = tf.Session()",
"_____no_output_____"
],
[
"_Mmin = tf.get_variable(name='mass', initializer=13.2, dtype=tf.float32)",
"_____no_output_____"
],
[
"Mhalo = tf.convert_to_tensor(np.random.uniform(11., 14., 1000), dtype=tf.float32)\nsiglogm = tf.convert_to_tensor(0.2, dtype=tf.float32)\ntemperature = 0.5\n\ndef Ncen(Mmin): \n # mean occupation of centrals\n return tf.clip_by_value(0.5 * (1+tf.math.erf((Mhalo - Mmin)/siglogm)),1e-4,1-1e-4)\n\ndef hod(Mmin): \n p = Ncen(Mmin)\n samp = gumbel_softmax(tf.stack([tf.log(p), tf.log(1.-p)],axis=1), temperature, hard=True)\n return samp[...,0]\n\n\ndef numden(Mmin): \n return tf.reduce_sum(hod(Mmin))",
"_____no_output_____"
],
[
"ncen,mh,nh = sess.run([Ncen(12.5), Mhalo, hod(12.5)] )\nplt.scatter(mh, (ncen))\nplt.xlim(11, 13.5)",
"_____no_output_____"
],
[
"plt.scatter(mh, (ncen), c='k')\nplt.scatter(mh, nh)\nplt.xlim(11., 13.5)",
"_____no_output_____"
],
[
"Mmin_true = 12.5",
"_____no_output_____"
],
[
"loss = (numden(Mmin_true) - numden(_Mmin))**2",
"_____no_output_____"
],
[
"opt = tf.train.AdamOptimizer(learning_rate=0.01)",
"_____no_output_____"
],
[
"opt_op = opt.minimize(loss)",
"_____no_output_____"
],
[
"sess.run(tf.global_variables_initializer())",
"_____no_output_____"
],
[
"losses=[]\nmasses=[]\nfor i in range(200):\n _,l,m = sess.run([opt_op, loss, _Mmin])\n losses.append(l)\n masses.append(m)",
"_____no_output_____"
],
[
"losses",
"_____no_output_____"
],
[
"%pylab inline",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"plot(losses)",
"_____no_output_____"
],
[
"plot(masses)\naxhline(Mmin_true, color='r', label='True Mmin')\nxlim(0,200)\nxlabel('Number of iterations')\nylabel('Mmin')\nlegend()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03d19ee0c1a59993e2de39f82989a6efba3ec30 | 17,657 | ipynb | Jupyter Notebook | Data Modeling with Postgres/test.ipynb | yazeedalrubyli/DEND | bac308fd6f24c093a416ef1b6cc9d2e556344962 | [
"Apache-2.0"
] | null | null | null | Data Modeling with Postgres/test.ipynb | yazeedalrubyli/DEND | bac308fd6f24c093a416ef1b6cc9d2e556344962 | [
"Apache-2.0"
] | null | null | null | Data Modeling with Postgres/test.ipynb | yazeedalrubyli/DEND | bac308fd6f24c093a416ef1b6cc9d2e556344962 | [
"Apache-2.0"
] | null | null | null | 32.637708 | 312 | 0.415246 | [
[
[
"%load_ext sql",
"_____no_output_____"
],
[
"%sql postgresql://student:student@127.0.0.1/sparkifydb",
"_____no_output_____"
],
[
"%sql SELECT * FROM songplays LIMIT 5;",
" * postgresql://student:***@127.0.0.1/sparkifydb\n5 rows affected.\n"
],
[
"%sql SELECT * FROM songplays WHERE artist_id is not null;",
" * postgresql://student:***@127.0.0.1/sparkifydb\n1 rows affected.\n"
],
[
"%sql SELECT * FROM users LIMIT 5;",
" * postgresql://student:***@127.0.0.1/sparkifydb\n5 rows affected.\n"
],
[
"%sql SELECT * FROM songs LIMIT 5;",
" * postgresql://student:***@127.0.0.1/sparkifydb\n5 rows affected.\n"
],
[
"%sql SELECT * FROM artists LIMIT 5;",
" * postgresql://student:***@127.0.0.1/sparkifydb\n5 rows affected.\n"
],
[
"%sql SELECT * FROM time LIMIT 5;",
" * postgresql://student:***@127.0.0.1/sparkifydb\n5 rows affected.\n"
]
],
[
[
"## REMEMBER: Restart this notebook to close connection to `sparkifydb`\nEach time you run the cells above, remember to restart this notebook to close the connection to your database. Otherwise, you won't be able to run your code in `create_tables.py`, `etl.py`, or `etl.ipynb` files since you can't make multiple connections to the same database (in this case, sparkifydb).",
"_____no_output_____"
]
]
] | [
"code",
"markdown"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d03d1c3460fa7fa90b15bfd0d72935e7bfd35c73 | 231,075 | ipynb | Jupyter Notebook | climate_starter.ipynb | gracesco/HuefnerSQLAlchemyChallenge | 40895346dfccf4a657904b02cd92ea4dc7217447 | [
"ADSL"
] | null | null | null | climate_starter.ipynb | gracesco/HuefnerSQLAlchemyChallenge | 40895346dfccf4a657904b02cd92ea4dc7217447 | [
"ADSL"
] | null | null | null | climate_starter.ipynb | gracesco/HuefnerSQLAlchemyChallenge | 40895346dfccf4a657904b02cd92ea4dc7217447 | [
"ADSL"
] | null | null | null | 641.875 | 97,555 | 0.815272 | [
[
[
"%matplotlib inline\nfrom matplotlib import style\nstyle.use('fivethirtyeight')\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"import datetime as dt",
"_____no_output_____"
]
],
[
[
"# Reflect Tables into SQLAlchemy ORM",
"_____no_output_____"
]
],
[
[
"# Python SQL toolkit and Object Relational Mapper\nimport sqlalchemy\nfrom sqlalchemy.ext.automap import automap_base\nfrom sqlalchemy.orm import Session\nfrom sqlalchemy import create_engine, func",
"_____no_output_____"
],
[
"engine = create_engine(\"sqlite:///Resources/hawaii.sqlite\")",
"_____no_output_____"
],
[
"# reflect an existing database into a new model\nBase = automap_base()\n# reflect the tables\nBase.prepare(engine, reflect=True)",
"_____no_output_____"
],
[
"# We can view all of the classes that automap found\nBase.classes.keys()",
"_____no_output_____"
],
[
"# Save references to each table\nMeasurements = Base.classes.measurement\nStations = Base.classes.station",
"_____no_output_____"
],
[
"# Create our session (link) from Python to the DB\nsession = Session(engine)\n",
"_____no_output_____"
]
],
[
[
"# Exploratory Climate Analysis",
"_____no_output_____"
]
],
[
[
"# Design a query to retrieve the last 12 months of precipitation data and plot the results\nCY_precipitation = session.query(Measurements.date).filter(Measurements.date >= \"2016-08-23\").order_by(Measurements.date).all()\n# # Calculate the date 1 year ago from the last data point in the database\nLY_precipitation = session.query(Measurements.date).filter(Measurements.date).order_by(Measurements.date.desc()).first()\nlast_date = dt.date(2017,8,23) - dt.timedelta(days=365)\n\nlast_date",
"_____no_output_____"
],
[
"# # # Perform a query to retrieve the data and precipitation scores\nlast_year = session.query(Measurements.prcp, Measurements.date).order_by(Measurements.date.desc())\n# # # Save the query results as a Pandas DataFrame and set the index to the date column, Sort the dataframe by date\ndate = []\nprecipitation = []\nlast_year_df = last_year.filter(Measurements.date >=\"2016-08-23\")\nfor precip in last_year_df:\n date.append(precip.date)\n precipitation.append(precip.prcp)\nLY_df = pd.DataFrame({\n \"Date\": date,\n \"Precipitation\": precipitation\n})\nLY_df.set_index(\"Date\", inplace=True)\nLY_df = LY_df.sort_index(ascending=True)\n\n# # Use Pandas Plotting with Matplotlib to plot the data\nLY_graph = LY_df.plot(figsize = (20,10), rot=90, title= \"Hawaii Precipitation Data 8/23/16-8/23/17\")\nLY_graph.set_ylabel(\"Precipitation (in)\")\nplt.savefig(\"Images/PrecipitationAugtoAug.png\")\n\n\n",
"_____no_output_____"
],
[
"# Use Pandas to calcualte the summary statistics for the precipitation data\nLY_df.describe()",
"_____no_output_____"
],
[
"# Design a query to show how many stations are available in this dataset?\nstation_count = session.query(Measurements.station, func.count(Measurements.station)).\\\n group_by(Measurements.station).all()\n\nprint(\"There are \" + str(len(station_count)) + \" stations in this dataset.\")",
"There are 9 stations in this dataset.\n"
],
[
"# What are the most active stations? (i.e. what stations have the most rows)?\n# List the stations and the counts in descending order.\nactive_stations = session.query(Measurements.station, func.count(Measurements.station)).\\\n group_by(Measurements.station).\\\n order_by(func.count(Measurements.station).desc()).all()\nactive_stations",
"_____no_output_____"
],
[
"# Using the station id from the previous query, calculate the lowest temperature recorded, \n# highest temperature recorded, and average temperature of the most active station?\nmost_active = session.query(Measurements.station).group_by(Measurements.station).order_by(func.count(Measurements.station).desc()).first()\n\nmost_active_info = session.query(Measurements.station, func.min(Measurements.tobs), func.max(Measurements.tobs), func.avg(Measurements.tobs)).\\\n filter(Measurements.station == most_active[0]).all()\n\nprint(\"The lowest temperature at station \" + most_active_info[0][0] + \" is \" + str(most_active_info[0][1]) + \" degrees.\")\nprint(\"The highest temperature at station \" + most_active_info[0][0] + \" is \" + str(most_active_info[0][2]) + \" degrees.\")\nprint(\"The average temperature at station \" + most_active_info[0][0] + \" is \" + str(round(most_active_info[0][3], 2)) + \" degrees.\")",
"The lowest temperature at station USC00519281 is 54.0 degrees.\nThe highest temperature at station USC00519281 is 85.0 degrees.\nThe average temperature at station USC00519281 is 71.66 degrees.\n"
],
[
"# Choose the station with the highest number of temperature observations.\n# Query the last 12 months of temperature observation data for this station and plot the results as a histogram\ntobs_query = session.query(Measurements.station).group_by(Measurements.station).order_by(func.count(Measurements.id).desc()).first()\ntobs = session.query(Measurements.station, Measurements.tobs).filter(Measurements.station == tobs_query[0]).filter(Measurements.date > last_date).all()\n\nstation_df = pd.DataFrame(tobs, columns = ['station', 'tobs'])\nstation_df.hist(column='tobs', bins=12)\nplt.ylabel('Frequency')\nplt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03d250285f6c81f734e9c35084ecd3c0b6d92c5 | 15,324 | ipynb | Jupyter Notebook | Credit card fraud .ipynb | Boutayna98/Credit-Card-Fraud-Detection | f7bccc6c91057b83822422298f015a52fed2df24 | [
"Unlicense"
] | null | null | null | Credit card fraud .ipynb | Boutayna98/Credit-Card-Fraud-Detection | f7bccc6c91057b83822422298f015a52fed2df24 | [
"Unlicense"
] | null | null | null | Credit card fraud .ipynb | Boutayna98/Credit-Card-Fraud-Detection | f7bccc6c91057b83822422298f015a52fed2df24 | [
"Unlicense"
] | null | null | null | 29.077799 | 379 | 0.448512 | [
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn.model_selection import train_test_split, GridSearchCV\nfrom sklearn import metrics\nfrom sklearn.impute import MissingIndicator, SimpleImputer\nfrom sklearn.preprocessing import PolynomialFeatures, KBinsDiscretizer, FunctionTransformer\nfrom sklearn.preprocessing import StandardScaler, MinMaxScaler, MaxAbsScaler\nfrom sklearn.preprocessing import LabelEncoder, OneHotEncoder, LabelBinarizer, OrdinalEncoder\nfrom sklearn.ensemble import BaggingClassifier, BaggingRegressor,RandomForestClassifier,RandomForestRegressor\nfrom sklearn.ensemble import GradientBoostingClassifier,GradientBoostingRegressor, AdaBoostClassifier, AdaBoostRegressor \nfrom sklearn.svm import LinearSVC, LinearSVR, SVC, SVR\nfrom sklearn.metrics import f1_score\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.metrics import confusion_matrix",
"_____no_output_____"
]
],
[
[
"# Import dataset",
"_____no_output_____"
]
],
[
[
"bd=pd.read_csv('creditcard.csv')",
"_____no_output_____"
],
[
"bd.head()",
"_____no_output_____"
]
],
[
[
"# Exploring dataset",
"_____no_output_____"
]
],
[
[
"bd.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 284807 entries, 0 to 284806\nData columns (total 31 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Time 284807 non-null float64\n 1 V1 284807 non-null float64\n 2 V2 284807 non-null float64\n 3 V3 284807 non-null float64\n 4 V4 284807 non-null float64\n 5 V5 284807 non-null float64\n 6 V6 284807 non-null float64\n 7 V7 284807 non-null float64\n 8 V8 284807 non-null float64\n 9 V9 284807 non-null float64\n 10 V10 284807 non-null float64\n 11 V11 284807 non-null float64\n 12 V12 284807 non-null float64\n 13 V13 284807 non-null float64\n 14 V14 284807 non-null float64\n 15 V15 284807 non-null float64\n 16 V16 284807 non-null float64\n 17 V17 284807 non-null float64\n 18 V18 284807 non-null float64\n 19 V19 284807 non-null float64\n 20 V20 284807 non-null float64\n 21 V21 284807 non-null float64\n 22 V22 284807 non-null float64\n 23 V23 284807 non-null float64\n 24 V24 284807 non-null float64\n 25 V25 284807 non-null float64\n 26 V26 284807 non-null float64\n 27 V27 284807 non-null float64\n 28 V28 284807 non-null float64\n 29 Amount 284807 non-null float64\n 30 Class 284807 non-null int64 \ndtypes: float64(30), int64(1)\nmemory usage: 67.4 MB\n"
]
],
[
[
"# Pre processing",
"_____no_output_____"
]
],
[
[
"sc = StandardScaler()\namount = bd['Amount'].values\nbd['Amount'] = sc.fit_transform(amount.reshape(-1, 1))",
"_____no_output_____"
],
[
"bd.drop(['Time'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"bd.shape",
"_____no_output_____"
],
[
"bd.drop_duplicates(inplace=True)",
"_____no_output_____"
],
[
"bd.shape",
"_____no_output_____"
]
],
[
[
"# Modelling",
"_____no_output_____"
]
],
[
[
"X = bd.drop('Class', axis = 1).values\ny = bd['Class'].values",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.25, random_state = 1)",
"_____no_output_____"
],
[
"from sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor, export_graphviz, export",
"C:\\Users\\PC\\Nouveau dossier\\lib\\site-packages\\sklearn\\utils\\deprecation.py:143: FutureWarning: The sklearn.tree.export module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.tree. Anything that cannot be imported from sklearn.tree is now part of the private API.\n warnings.warn(message, FutureWarning)\n"
],
[
"DT = DecisionTreeClassifier(max_depth = 4, criterion = 'entropy')\nDT.fit(X_train, y_train)\ndt_yhat = DT.predict(X_test)",
"_____no_output_____"
],
[
"print('Accuracy score of the Decision Tree model is {}'.format(accuracy_score(y_test,dt_yhat)))",
"Accuracy score of the Decision Tree model is 0.9991583957281328\n"
],
[
"print('F1 score of the Decision Tree model is {}'.format(f1_score(y_test,dt_yhat)))",
"F1 score of the Decision Tree model is 0.7521367521367521\n"
],
[
"confusion_matrix(y_test, dt_yhat, labels = [0, 1])",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03d2b1bd3e1c50012c083a146803f23cdfcd7fe | 18,947 | ipynb | Jupyter Notebook | notebook/proc-cabocha.ipynb | samlet/stack | 47db17fd4fdab264032f224dca31a4bb1d19b754 | [
"Apache-2.0"
] | 3 | 2020-01-11T13:55:38.000Z | 2020-08-25T22:34:15.000Z | notebook/proc-cabocha.ipynb | samlet/stack | 47db17fd4fdab264032f224dca31a4bb1d19b754 | [
"Apache-2.0"
] | null | null | null | notebook/proc-cabocha.ipynb | samlet/stack | 47db17fd4fdab264032f224dca31a4bb1d19b754 | [
"Apache-2.0"
] | 1 | 2021-01-01T05:21:44.000Z | 2021-01-01T05:21:44.000Z | 26.388579 | 111 | 0.413575 | [
[
[
"import CaboCha\n\n# c = CaboCha.Parser(\"\");\nc = CaboCha.Parser()\n\n# sentence = \"太郎はこの本を二郎を見た女性に渡した。\"\nsentence = \"太郎は花子が読んでいる本を次郎に渡した。\"\nprint(c.parseToString(sentence))\n\ntree = c.parse(sentence)\n\nprint(tree.toString(CaboCha.FORMAT_TREE))\nprint(tree.toString(CaboCha.FORMAT_LATTICE))",
" 太郎は---------D\n 花子が-D |\n 読んでいる-D |\n 本を---D\n 次郎に-D\n 渡した。\nEOS\n\n 太郎は---------D\n 花子が-D |\n 読んでいる-D |\n 本を---D\n 次郎に-D\n 渡した。\nEOS\n\n* 0 5D 0/1 -0.742128\n太郎\t名詞,固有名詞,人名,名,*,*,太郎,タロウ,タロー\nは\t助詞,係助詞,*,*,*,*,は,ハ,ワ\n* 1 2D 0/1 1.700175\n花子\t名詞,固有名詞,人名,名,*,*,花子,ハナコ,ハナコ\nが\t助詞,格助詞,一般,*,*,*,が,ガ,ガ\n* 2 3D 0/2 1.825021\n読ん\t動詞,自立,*,*,五段・マ行,連用タ接続,読む,ヨン,ヨン\nで\t助詞,接続助詞,*,*,*,*,で,デ,デ\nいる\t動詞,非自立,*,*,一段,基本形,いる,イル,イル\n* 3 5D 0/1 -0.742128\n本\t名詞,一般,*,*,*,*,本,ホン,ホン\nを\t助詞,格助詞,一般,*,*,*,を,ヲ,ヲ\n* 4 5D 1/2 -0.742128\n次\t名詞,一般,*,*,*,*,次,ツギ,ツギ\n郎\t名詞,一般,*,*,*,*,郎,ロウ,ロー\nに\t助詞,格助詞,一般,*,*,*,に,ニ,ニ\n* 5 -1D 0/1 0.000000\n渡し\t動詞,自立,*,*,五段・サ行,連用形,渡す,ワタシ,ワタシ\nた\t助動詞,*,*,*,特殊・タ,基本形,た,タ,タ\n。\t記号,句点,*,*,*,*,。,。,。\nEOS\n\n"
],
[
"print(tree.toString(CaboCha.FORMAT_XML))",
"<sentence>\n <chunk id=\"0\" link=\"5\" rel=\"D\" score=\"-0.742128\" head=\"0\" func=\"1\">\n <tok id=\"0\" feature=\"名詞,固有名詞,人名,名,*,*,太郎,タロウ,タロー\">太郎</tok>\n <tok id=\"1\" feature=\"助詞,係助詞,*,*,*,*,は,ハ,ワ\">は</tok>\n </chunk>\n <chunk id=\"1\" link=\"2\" rel=\"D\" score=\"1.700175\" head=\"2\" func=\"3\">\n <tok id=\"2\" feature=\"名詞,固有名詞,人名,名,*,*,花子,ハナコ,ハナコ\">花子</tok>\n <tok id=\"3\" feature=\"助詞,格助詞,一般,*,*,*,が,ガ,ガ\">が</tok>\n </chunk>\n <chunk id=\"2\" link=\"3\" rel=\"D\" score=\"1.825021\" head=\"4\" func=\"6\">\n <tok id=\"4\" feature=\"動詞,自立,*,*,五段・マ行,連用タ接続,読む,ヨン,ヨン\">読ん</tok>\n <tok id=\"5\" feature=\"助詞,接続助詞,*,*,*,*,で,デ,デ\">で</tok>\n <tok id=\"6\" feature=\"動詞,非自立,*,*,一段,基本形,いる,イル,イル\">いる</tok>\n </chunk>\n <chunk id=\"3\" link=\"5\" rel=\"D\" score=\"-0.742128\" head=\"7\" func=\"8\">\n <tok id=\"7\" feature=\"名詞,一般,*,*,*,*,本,ホン,ホン\">本</tok>\n <tok id=\"8\" feature=\"助詞,格助詞,一般,*,*,*,を,ヲ,ヲ\">を</tok>\n </chunk>\n <chunk id=\"4\" link=\"5\" rel=\"D\" score=\"-0.742128\" head=\"10\" func=\"11\">\n <tok id=\"9\" feature=\"名詞,一般,*,*,*,*,次,ツギ,ツギ\">次</tok>\n <tok id=\"10\" feature=\"名詞,一般,*,*,*,*,郎,ロウ,ロー\">郎</tok>\n <tok id=\"11\" feature=\"助詞,格助詞,一般,*,*,*,に,ニ,ニ\">に</tok>\n </chunk>\n <chunk id=\"5\" link=\"-1\" rel=\"D\" score=\"0.000000\" head=\"12\" func=\"13\">\n <tok id=\"12\" feature=\"動詞,自立,*,*,五段・サ行,連用形,渡す,ワタシ,ワタシ\">渡し</tok>\n <tok id=\"13\" feature=\"助動詞,*,*,*,特殊・タ,基本形,た,タ,タ\">た</tok>\n <tok id=\"14\" feature=\"記号,句点,*,*,*,*,。,。,。\">。</tok>\n </chunk>\n</sentence>\n\n"
],
[
"print(tree.toString(CaboCha.FORMAT_TREE_LATTICE))",
" 太郎は---------D\n 花子が-D |\n 読んでいる-D |\n 本を---D\n 次郎に-D\n 渡した。\nEOS\n* 0 5D 0/1 -0.742128\n太郎\t名詞,固有名詞,人名,名,*,*,太郎,タロウ,タロー\nは\t助詞,係助詞,*,*,*,*,は,ハ,ワ\n* 1 2D 0/1 1.700175\n花子\t名詞,固有名詞,人名,名,*,*,花子,ハナコ,ハナコ\nが\t助詞,格助詞,一般,*,*,*,が,ガ,ガ\n* 2 3D 0/2 1.825021\n読ん\t動詞,自立,*,*,五段・マ行,連用タ接続,読む,ヨン,ヨン\nで\t助詞,接続助詞,*,*,*,*,で,デ,デ\nいる\t動詞,非自立,*,*,一段,基本形,いる,イル,イル\n* 3 5D 0/1 -0.742128\n本\t名詞,一般,*,*,*,*,本,ホン,ホン\nを\t助詞,格助詞,一般,*,*,*,を,ヲ,ヲ\n* 4 5D 1/2 -0.742128\n次\t名詞,一般,*,*,*,*,次,ツギ,ツギ\n郎\t名詞,一般,*,*,*,*,郎,ロウ,ロー\nに\t助詞,格助詞,一般,*,*,*,に,ニ,ニ\n* 5 -1D 0/1 0.000000\n渡し\t動詞,自立,*,*,五段・サ行,連用形,渡す,ワタシ,ワタシ\nた\t助動詞,*,*,*,特殊・タ,基本形,た,タ,タ\n。\t記号,句点,*,*,*,*,。,。,。\nEOS\n\n"
],
[
"from cabocha.analyzer import CaboChaAnalyzer\nanalyzer = CaboChaAnalyzer()\n# seq=\"日本語の形態素解析はすごい\"\n# seq=\"太郎はこの本を二郎を見た女性に渡した。\"\n# seq=\"あなたたち自身がこれをやらないといけない\"\nseq=\"太郎は花子が読んでいる本を次郎に渡した\"\ntree = analyzer.parse(seq)\nfor chunk in tree:\n print(str(chunk)+str(chunk.id)+\",\"+str(chunk.link))\n for token in chunk:\n print(\"\\t\"+str(token.surface)+\" ☞ \"+token.pos)",
"Chunk(\"太郎は\")0,5\n\t太郎 ☞ 名詞\n\tは ☞ 助詞\nChunk(\"花子が\")1,2\n\t花子 ☞ 名詞\n\tが ☞ 助詞\nChunk(\"読んでいる\")2,3\n\t読ん ☞ 動詞\n\tで ☞ 助詞\n\tいる ☞ 動詞\nChunk(\"本を\")3,5\n\t本 ☞ 名詞\n\tを ☞ 助詞\nChunk(\"次郎に\")4,5\n\t次 ☞ 名詞\n\t郎 ☞ 名詞\n\tに ☞ 助詞\nChunk(\"渡した\")5,-1\n\t渡し ☞ 動詞\n\tた ☞ 助動詞\n"
],
[
"from utils import dump\n# 遵循chunk的依赖\nchunks = tree.chunks\nstart_chunk = chunks[0]\nprint(start_chunk.next_link)\ntoken=start_chunk.tokens[0]\nprint(token, type(token))\ndump(token.dict())",
"Chunk(\"渡した\")\nToken(\"太郎\") <class 'cabocha.analyzer.Token'>\n{\n \"id\": 0,\n \"additional_info\": null,\n \"feature\": \"名詞,固有名詞,人名,名,*,*,太郎,タロウ,タロー\",\n \"feature_list\": [\n \"名詞\",\n \"固有名詞\",\n \"人名\",\n \"名\",\n \"*\",\n \"*\",\n \"太郎\",\n \"タロウ\",\n \"タロー\"\n ],\n \"feature_list_size\": 9,\n \"ne\": null,\n \"normalized_surface\": \"太郎\",\n \"surface\": \"太郎\",\n \"pos\": \"名詞\",\n \"pos1\": \"固有名詞\",\n \"pos2\": \"人名\",\n \"pos3\": \"名\",\n \"ctype\": \"*\",\n \"cform\": \"*\",\n \"genkei\": \"太郎\",\n \"yomi\": \"タロウ\"\n}\n"
],
[
"seq=\"太郎は花子が読んでいる本を次郎に渡した\"\ntree = analyzer.parse(seq)\nchunks = tree.chunks\n\n# 关注chunk的来源\nend_chunk = chunks[-1]\nprint(end_chunk.prev_links)\nprint(end_chunk.prev_links[0].prev_links)\n# print(end_chunk.prev_links[0].prev_links[0].prev_links)",
"[Chunk(\"太郎は\"), Chunk(\"本を\"), Chunk(\"次郎に\")]\n[]\n"
],
[
"seq=\"太郎は花子が読んでいる本を次郎に渡した\"\ntree = analyzer.parse(seq)\nchunks = tree.chunks\nchunk = chunks[0]\nprint(chunk)\ndump(chunk.dict())",
"Chunk(\"太郎は\")\n{\n \"id\": 0,\n \"additional_info\": null,\n \"feature_list\": [\n \"SCASE:は\",\n \"SHS:太郎\",\n \"SHP0:名詞\",\n \"SHP1:固有名詞\",\n \"SHP2:人名\",\n \"SHP3:名\",\n \"SFS:は\",\n \"SFP0:助詞\",\n \"SFP1:係助詞\",\n \"SLS:太郎\",\n \"SLP0:名詞\",\n \"SLP1:固有名詞\",\n \"SLP2:人名\",\n \"SLP3:名\",\n \"SRS:は\",\n \"SRP0:助詞\",\n \"SRP1:係助詞\",\n \"LF:は\",\n \"RL:太郎\",\n \"RH:太郎\",\n \"RF:は\",\n \"SBOS:1\",\n \"GCASE:は\",\n \"A:は\"\n ],\n \"feature_list_size\": 24,\n \"func_pos\": 1,\n \"head_pos\": 0,\n \"link\": 5,\n \"score\": -0.742127537727356,\n \"token_pos\": 0,\n \"token_size\": 2,\n \"tokens\": [\n {\n \"id\": 0,\n \"additional_info\": null,\n \"feature\": \"名詞,固有名詞,人名,名,*,*,太郎,タロウ,タロー\",\n \"feature_list\": [\n \"名詞\",\n \"固有名詞\",\n \"人名\",\n \"名\",\n \"*\",\n \"*\",\n \"太郎\",\n \"タロウ\",\n \"タロー\"\n ],\n \"feature_list_size\": 9,\n \"ne\": null,\n \"normalized_surface\": \"太郎\",\n \"surface\": \"太郎\",\n \"pos\": \"名詞\",\n \"pos1\": \"固有名詞\",\n \"pos2\": \"人名\",\n \"pos3\": \"名\",\n \"ctype\": \"*\",\n \"cform\": \"*\",\n \"genkei\": \"太郎\",\n \"yomi\": \"タロウ\"\n },\n {\n \"id\": 1,\n \"additional_info\": null,\n \"feature\": \"助詞,係助詞,*,*,*,*,は,ハ,ワ\",\n \"feature_list\": [\n \"助詞\",\n \"係助詞\",\n \"*\",\n \"*\",\n \"*\",\n \"*\",\n \"は\",\n \"ハ\",\n \"ワ\"\n ],\n \"feature_list_size\": 9,\n \"ne\": null,\n \"normalized_surface\": \"は\",\n \"surface\": \"は\",\n \"pos\": \"助詞\",\n \"pos1\": \"係助詞\",\n \"pos2\": \"*\",\n \"pos3\": \"*\",\n \"ctype\": \"*\",\n \"cform\": \"*\",\n \"genkei\": \"は\",\n \"yomi\": \"ハ\"\n }\n ],\n \"next_link_id\": 5,\n \"prev_link_ids\": []\n}\n"
],
[
"# 检索适合特定条件的块或令牌\ntree.find(function=lambda chunk: any(token.pos == \"名詞\" for token in chunk))",
"_____no_output_____"
],
[
"tree.find(function=lambda chunk: any(token.pos == \"形容詞\" for token in chunk))",
"_____no_output_____"
],
[
"# 太郎把这本书递给了看见次郎的女人。\n# Taro handed this book to the woman who saw Jiro.\nsentence=\"太郎はこの本を二郎を見た女性に渡した。\"\ntree = analyzer.parse(sentence)\ntree.find(function=lambda chunk: any(token.pos == \"名詞\" for token in chunk))",
"_____no_output_____"
],
[
"chunks=tree.find(function=lambda chunk: any(token.pos == \"名詞\" for token in chunk))\nend_chunk = chunks[-1]\nprint(end_chunk)\nprint(end_chunk.prev_links)\nprint(end_chunk.prev_links[0].prev_links)",
"Chunk(\"女性に\")\n[Chunk(\"見た\")]\n[Chunk(\"本を\"), Chunk(\"二郎を\")]\n"
],
[
"print(end_chunk)\nprint(end_chunk.next_link)\nprint(chunks[0])\nprint(chunks[0].next_link)\nprint(chunks[1])\nprint(chunks[1].next_link)",
"Chunk(\"女性に\")\nChunk(\"渡した。\")\nChunk(\"太郎は\")\nChunk(\"渡した。\")\nChunk(\"本を\")\nChunk(\"見た\")\n"
],
[
"c = CaboCha.Parser()\ntree = c.parse(sentence)\nprint(tree.toString(CaboCha.FORMAT_TREE))",
" 太郎は-----------D\n この-D |\n 本を---D |\n 二郎を-D |\n 見た-D |\n 女性に-D\n 渡した。\nEOS\n\n"
],
[
"sentence=\"太郎はこの本を二郎を見た女性に渡した。\"\ntree = analyzer.parse(sentence)\nfor chunk in tree.chunks:\n print(chunk, [token.feature for token in chunk])\ntree.find(function=lambda chunk: any(token.pos == \"名詞\" for token in chunk))",
"Chunk(\"太郎は\") ['名詞,固有名詞,人名,名,*,*,太郎,タロウ,タロー', '助詞,係助詞,*,*,*,*,は,ハ,ワ']\nChunk(\"この\") ['連体詞,*,*,*,*,*,この,コノ,コノ']\nChunk(\"本を\") ['名詞,一般,*,*,*,*,本,ホン,ホン', '助詞,格助詞,一般,*,*,*,を,ヲ,ヲ']\nChunk(\"二郎を\") ['名詞,数,*,*,*,*,二,ニ,ニ', '名詞,一般,*,*,*,*,郎,ロウ,ロー', '助詞,格助詞,一般,*,*,*,を,ヲ,ヲ']\nChunk(\"見た\") ['動詞,自立,*,*,一段,連用形,見る,ミ,ミ', '助動詞,*,*,*,特殊・タ,基本形,た,タ,タ']\nChunk(\"女性に\") ['名詞,一般,*,*,*,*,女性,ジョセイ,ジョセイ', '助詞,格助詞,一般,*,*,*,に,ニ,ニ']\nChunk(\"渡した。\") ['動詞,自立,*,*,五段・サ行,連用形,渡す,ワタシ,ワタシ', '助動詞,*,*,*,特殊・タ,基本形,た,タ,タ', '記号,句点,*,*,*,*,。,。,。']\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03d2f433b707ead9e1847e434f838bfa91ea754 | 102,884 | ipynb | Jupyter Notebook | notebooks/Data_EDA.ipynb | Prudhvi-Kumar-Kakani/Data-Science-CRISP-DM--Covid-19 | 908cb345c635cf8cfa265e98faa1d796b0836fd7 | [
"FTL"
] | 1 | 2021-12-01T22:01:31.000Z | 2021-12-01T22:01:31.000Z | notebooks/Data_EDA.ipynb | Prudhvi-Kumar-Kakani/Data-Science-CRISP-DM--Covid-19 | 908cb345c635cf8cfa265e98faa1d796b0836fd7 | [
"FTL"
] | null | null | null | notebooks/Data_EDA.ipynb | Prudhvi-Kumar-Kakani/Data-Science-CRISP-DM--Covid-19 | 908cb345c635cf8cfa265e98faa1d796b0836fd7 | [
"FTL"
] | null | null | null | 31.511179 | 23,915 | 0.387631 | [
[
[
"# Exploratory data analysis",
"_____no_output_____"
]
],
[
[
"# import libraries\nimport numpy as np\nimport pandas as pd\n\nimport plotly.graph_objects as go\nimport matplotlib.pyplot as plt\npd.set_option('display.max_rows', 500)\n%matplotlib inline",
"_____no_output_____"
],
[
"# load the processed data\nmain_df = pd.read_csv('../data/processed/COVID_small_table_confirmed.csv', sep=';')\nmain_df.head()",
"_____no_output_____"
],
[
"main_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 205 entries, 0 to 204\nData columns (total 6 columns):\ndate 205 non-null object\nGermany 205 non-null int64\nIndia 205 non-null int64\nUS 205 non-null int64\nBrazil 205 non-null int64\nUnited Kingdom 205 non-null int64\ndtypes: int64(5), object(1)\nmemory usage: 9.7+ KB\n"
],
[
"# GEt countries list\ncountry_list = main_df.columns[1:]\ncountry_list",
"_____no_output_____"
],
[
"fig = go.Figure()\n\n# define plot for individual trace\nfor country in country_list:\n fig.add_trace(go.Scatter(x = main_df.date,\n y = main_df[country],\n mode = 'markers+lines',\n name = country))\n\n# overall layout properties\nfig.update_layout( width = 1200,\n height = 800,\n title = 'Confirmed cases for specific countries',\n xaxis_title = 'Date',\n yaxis_title = 'No. of confirmed cases')\n\nfig.update_yaxes(type='log')",
"_____no_output_____"
],
[
"# Dash board visualization\n\nimport dash\nimport dash_core_components as dcc\nimport dash_html_components as html\n\napp = dash.Dash()\napp.layout = html.Div([\n \n \n \n \n dcc.Graph(figure=fig, id='main_window_slope')\n])",
"_____no_output_____"
],
[
"app.run_server(debug=True, use_reloader=False) # Turn off reloader if inside Jupyter",
"Running on http://127.0.0.1:8050/\nRunning on http://127.0.0.1:8050/\nDebugger PIN: 839-733-624\nDebugger PIN: 839-733-624\n * Serving Flask app \"__main__\" (lazy loading)\n * Environment: production\n WARNING: Do not use the development server in a production environment.\n Use a production WSGI server instead.\n * Debug mode: on\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03d38b149d92a75dfe16d1971311573e3e49ec0 | 1,588 | ipynb | Jupyter Notebook | Untitled5.ipynb | Aleena1970/AJ1997 | 3a762fb41720560ddc919b9160cd72e5e109851d | [
"MIT"
] | null | null | null | Untitled5.ipynb | Aleena1970/AJ1997 | 3a762fb41720560ddc919b9160cd72e5e109851d | [
"MIT"
] | null | null | null | Untitled5.ipynb | Aleena1970/AJ1997 | 3a762fb41720560ddc919b9160cd72e5e109851d | [
"MIT"
] | null | null | null | 21.459459 | 48 | 0.491184 | [
[
[
"x=int(input(\"Enter Salary\"))\nif x <=250000 :\n tax = 0\nif x <= 500000 and x > 250000 :\n tax = (x-250000)*5/100\nif x <= 750000 and x > 500000 :\n tax = (x-500000)*10/100+12500\nif x <= 1000000 and x > 750000 :\n tax = (x-750000)*15/100+37500\nif x <= 1250000 and x > 1000000 :\n tax = (x-1000000)*20/100+75000\nif x <= 1500000 and x > 1250000 :\n tax = (x-1250000)*25/100+125000\nif x > 1500000 :\n tax = (x-1500000)*30/100+187500\nprint(tax)",
"Enter Salary 350000\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d03d63fc3063c07feab549fb688836520cca5446 | 162,616 | ipynb | Jupyter Notebook | Music Recommendation.ipynb | sasikamaleshvadlani/Music-Recommendation-based-on-Facial-Emotions | 1aa2e6fee93d945fa0ef8e1d0ea52051a6d85358 | [
"MIT"
] | null | null | null | Music Recommendation.ipynb | sasikamaleshvadlani/Music-Recommendation-based-on-Facial-Emotions | 1aa2e6fee93d945fa0ef8e1d0ea52051a6d85358 | [
"MIT"
] | null | null | null | Music Recommendation.ipynb | sasikamaleshvadlani/Music-Recommendation-based-on-Facial-Emotions | 1aa2e6fee93d945fa0ef8e1d0ea52051a6d85358 | [
"MIT"
] | 1 | 2021-09-04T20:12:16.000Z | 2021-09-04T20:12:16.000Z | 495.780488 | 149,044 | 0.92446 | [
[
[
"#import libraries\nimport cv2\nimport matplotlib.pyplot as plt\nimport numpy as np\nfrom tensorflow.keras.models import model_from_json\n\nimport pickle\nimport tkinter as tk\nfrom tkinter import filedialog\nfrom tkinter import PhotoImage\nfrom pygame import mixer\nimport matplotlib.pyplot as plt\nimport random\nimport os",
"_____no_output_____"
],
[
"#Taking picture from webcam and detecting emotion\n#load model \nmodel = model_from_json(open(\"model.json\", \"r\").read()) \n#load weights\nmodel.load_weights('model.h5')\nemotion_dict = {0: \"Angry\", 1: \"Disgusted\", 2: \"Fearful\", 3: \"Happy\", 4: \"Neutral\", 5: \"Sad\", 6: \"Surprised\"}\ncap=cv2.VideoCapture(0)\nret, frame =cap.read()\n\nfacecasc = cv2.CascadeClassifier('haarcascade_frontalface_alt2.xml')\ngray = cv2.cvtColor(frame, cv2.COLOR_BGR2GRAY)\nfaces = facecasc.detectMultiScale(gray,scaleFactor=1.3, minNeighbors=5)\n\nfor (x, y, w, h) in faces:\n cv2.rectangle(frame, (x, y-50), (x+w, y+h+10), (255, 0, 0), 2)\n roi_gray = gray[y:y + h, x:x + w]\n cropped_img = np.expand_dims(np.expand_dims(cv2.resize(roi_gray, (48, 48)), -1), 0)\n prediction = model.predict(cropped_img)\n maxindex = int(np.argmax(prediction))\n cv2.putText(frame, emotion_dict[maxindex], (x+20, y-60), cv2.FONT_HERSHEY_SIMPLEX, 1, (255, 255, 255), 2, cv2.LINE_AA)\n dominantemotion=emotion_dict[maxindex]\n#cv2.imshow('Video', cv2.resize(frame,(1600,960),interpolation = cv2.INTER_CUBIC))\nplt.imshow(frame)\ncap.release()\ncv2.destroyAllWindows()",
"_____no_output_____"
],
[
"#Playing music from detected emotion\nclass Playe(tk.Frame):\n def __init__(self, dominantemotion , master=None):\n super().__init__(master)\n self.master = master\n self.pack()\n mixer.init()\n self.dominantemotion=dominantemotion\n print(self.dominantemotion)\n \n self.playlist=[]\n self.current = 0\n self.paused = True\n self.played = False\n\n\n self.create_frames()\n self.track_widgets()\n self.control_widgets()\n self.tracklist_widgets()\n self.retrieve_songs()\n\n def create_frames(self):\n self.track = tk.LabelFrame(self, text='Song Track',\n font=(\"times new roman\",15,\"bold\"),\n bg=\"grey\",fg=\"white\",bd=5,relief=tk.GROOVE)\n self.track.config(width=410,height=300)\n self.track.grid(row=0, column=0, padx=10)\n\n self.tracklist = tk.LabelFrame(self, text=f'PlayList - {str(len(self.playlist))}',\n font=(\"times new roman\",15,\"bold\"),\n bg=\"grey\",fg=\"white\",bd=5,relief=tk.GROOVE)\n self.tracklist.config(width=190,height=400)\n self.tracklist.grid(row=0, column=1, rowspan=3, pady=5)\n\n self.controls = tk.LabelFrame(self,\n font=(\"times new roman\",15,\"bold\"),\n bg=\"white\",fg=\"white\",bd=2,relief=tk.GROOVE)\n self.controls.config(width=410,height=80)\n self.controls.grid(row=2, column=0, pady=5, padx=10)\n\n def track_widgets(self):\n self.canvas = tk.Label(self.track, image=img)\n self.canvas.configure(width=400, height=240)\n self.canvas.grid(row=0,column=0)\n\n self.songtrack = tk.Label(self.track, font=(\"times new roman\",16,\"bold\"),\n bg=\"white\",fg=\"dark blue\")\n self.songtrack['text'] = 'VFSTR MP3 Player'\n self.songtrack.config(width=30, height=1)\n self.songtrack.grid(row=1,column=0,padx=10)\n\n def control_widgets(self):\n\n self.prev = tk.Button(self.controls, image=prev)\n self.prev['command'] = self.prev_song\n self.prev.grid(row=0, column=1)\n\n self.pause = tk.Button(self.controls, image=pause)\n self.pause['command'] = self.pause_song\n self.pause.grid(row=0, column=2)\n\n self.next = tk.Button(self.controls, image=next_)\n self.next['command'] = self.next_song\n self.next.grid(row=0, column=3)\n\n self.volume = tk.DoubleVar(self)\n self.slider = tk.Scale(self.controls, from_ = 0, to = 10, orient = tk.HORIZONTAL)\n self.slider['variable'] = self.volume\n self.slider.set(8)\n mixer.music.set_volume(0.8)\n self.slider['command'] = self.change_volume\n self.slider.grid(row=0, column=4, padx=5)\n\n\n def tracklist_widgets(self):\n self.scrollbar = tk.Scrollbar(self.tracklist, orient=tk.VERTICAL)\n self.scrollbar.grid(row=0,column=1, rowspan=5, sticky='ns')\n\n self.list = tk.Listbox(self.tracklist, selectmode=tk.SINGLE,\n yscrollcommand=self.scrollbar.set, selectbackground='sky blue')\n self.enumerate_songs()\n self.list.config(height=22)\n self.list.bind('<Double-1>', self.play_song)\n\n self.scrollbar.config(command=self.list.yview)\n self.list.grid(row=0, column=0, rowspan=5)\n\n def retrieve_songs(self):\n self.songlist = []\n if(self.dominantemotion=='angry'):\n directory = r'C:\\Users\\USER\\Downloads\\Code\\Angry'\n elif(self.dominantemotion=='Surprised'):\n directory = r'C:\\Users\\USER\\Downloads\\Code\\Sad'\n elif(self.dominantemotion=='Happy'):\n directory = r'C:\\Users\\USER\\Downloads\\Code\\Happy'\n elif(self.dominantemotion=='Neutral'):\n directory = r'C:\\Users\\USER\\Downloads\\Code\\Neutral'\n elif(self.dominantemotion=='fear'):\n directory = r'C:\\Users\\USER\\Downloads\\Code\\Fear'\n\n #filedialog.askdirectory()\n for root_, dirs, files in os.walk(directory):\n for file in files:\n if os.path.splitext(file)[1] == '.mp3':\n path = (root_ + '/' + file).replace('\\\\','/')\n self.songlist.append(path)\n self.playlist = self.songlist\n random.shuffle(self.playlist)\n self.tracklist['text'] = f'PlayList - {str(len(self.playlist))}'\n self.list.delete(0, tk.END)\n self.enumerate_songs()\n self.play_song()\n def enumerate_songs(self):\n for index, song in enumerate(self.playlist):\n self.list.insert(index, os.path.basename(song))\n\n\n def play_song(self, event=None):\n if event is not None:\n self.current = self.list.curselection()[0]\n for i in range(len(self.playlist)):\n self.list.itemconfigure(i, bg=\"white\")\n\n print(self.playlist[self.current])\n mixer.music.load(self.playlist[self.current])\n self.songtrack['anchor'] = 'w'\n self.songtrack['text'] = os.path.basename(self.playlist[self.current])\n\n self.pause['image'] = play\n self.paused = False\n self.played = True\n self.list.activate(self.current)\n self.list.itemconfigure(self.current, bg='sky blue')\n\n mixer.music.play()\n\n def pause_song(self):\n if not self.paused:\n self.paused = True\n mixer.music.pause()\n self.pause['image'] = pause\n else:\n if self.played == False:\n self.play_song()\n self.paused = False\n mixer.music.unpause()\n self.pause['image'] = play\n\n def prev_song(self):\n if self.current > 0:\n self.current -= 1\n self.list.itemconfigure(self.current + 1, bg='white')\n else:\n self.current = 0\n self.play_song()\n\n def next_song(self):\n if self.current < len(self.playlist) - 1:\n self.list.itemconfigure(self.current, bg='white')\n self.current += 1\n else:\n self.current = 0\n self.list.itemconfigure(len(self.playlist) - 1, bg='white')\n self.play_song()\n\n\n\n def change_volume(self, event=None):\n self.v = self.volume.get()\n mixer.music.set_volume(self.v / 10)\n \n\nroot = tk.Tk()\nroot.geometry('600x400')\nroot.wm_title('VFSTR Music Recommendation System')\n\nimg = PhotoImage(master=root,file=r'C:\\Users\\USER\\Downloads\\Code\\images\\music.gif')\nnext_ = PhotoImage(master=root,file = r'C:\\Users\\USER\\Downloads\\Code\\images\\next.gif')\nprev = PhotoImage(master=root,file=r'C:\\Users\\USER\\Downloads\\Code\\images\\previous.gif')\nplay = PhotoImage(master=root,file=r'C:\\Users\\USER\\Downloads\\Code\\images\\play.gif')\npause = PhotoImage(master=root,file=r'C:\\Users\\USER\\Downloads\\Code\\images\\pause.gif')\n\napp = Playe(dominantemotion, master=root)\napp.mainloop() \n",
"Happy\nC:/Users/USER/Downloads/Code/Happy/Bhale Manchi Roju Jarigina Katha Telugu Old Classics Ghantasala YouTube.mp3\n"
],
[
"pwd",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d03d64dd7121c821b0e283eb7a8fb8085727f12c | 11,953 | ipynb | Jupyter Notebook | 21octubre.ipynb | humbertoguell/daa2020_1 | 14a628002c86917f9954ab157bc653c7f4e281ac | [
"MIT"
] | null | null | null | 21octubre.ipynb | humbertoguell/daa2020_1 | 14a628002c86917f9954ab157bc653c7f4e281ac | [
"MIT"
] | null | null | null | 21octubre.ipynb | humbertoguell/daa2020_1 | 14a628002c86917f9954ab157bc653c7f4e281ac | [
"MIT"
] | null | null | null | 26.041394 | 351 | 0.381912 | [
[
[
"<a href=\"https://colab.research.google.com/github/humbertoguell/daa2020_1/blob/master/21octubre.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"2+3+\nfor r in range(n):\n sumaRenglon=0\n sumaRenglon=0\n sumaRenglon=0\n for c in range(n):\n sumaRenglon +=a2d.get_item(r,c)\n total += a2d.get_item(r,c)",
"_____no_output_____"
],
[
"def ejemplo1( n ):\n c = n + 1\n d = c * n\n e = n * n\n total = c + e - d\n print(f\"total={ total }\")\nejemplo1( 99999 )",
"_____no_output_____"
],
[
"def ejemplo2( n ):\n contador = 0\n for i in range( n ) :\n for j in range( n ) :\n contador += 1\n return contador\n\nejemplo2( 100 )",
"_____no_output_____"
],
[
"def ejemplo3( n ): # n=4\n x = n * 2 # x = 8\n y = 0 # y = 0\n for m in range( 100 ): #3\n y = x - n # y = 4\n return y\n\nejemplo3(1000000000)",
"_____no_output_____"
],
[
"def ejemplo4( n ):\n x = 3 * 3.1416 + n\n y = x + 3 * 3 - n\n z = x + y\n return z\n\nejemplo4(9)",
"_____no_output_____"
],
[
"def ejemplo5( x ):\n n = 10\n for j in range( 0 , x , 1 ):\n n = j + n\n return n\n\nejemplo5(1000000)",
"_____no_output_____"
],
[
"from time import time\n\ndef ejemplo6( n ):\n start_time = time()\n data=[[[1 for x in range(n)] for x in range(n)] \n for x in range(n)]\n suma = 0\n for d in range(n):\n for r in range(n):\n for c in range(n):\n suma += data[d][r][c]\n elapsed_time = time() - start_time\n print(\"Tiempo transcurrido: %0.10f segundos.\" % elapsed_time)\n return suma\nejemplo6( 500 )",
"_____no_output_____"
],
[
"def ejemplo7( n ):\n count = 0\n for i in range( n ) :\n for j in range( 25 ) :\n for k in range( n ):\n count += 1\n return count",
"_____no_output_____"
],
[
"def ejemplo7_2( n ):\n count = 1\n for i in range( n ) :\n for j in range( 25 ) :\n for k in range( n ):\n count += 1\n for k in range( n ):\n count += 1\n return count # 1 + 25n^2 +25n^2\n\nejemplo7_2(3)",
"_____no_output_____"
],
[
"def ejemplo8( numeros ): # numeros es una lista (arreglo en c)\n total = 0\n for index in range(len(numeros)):\n total = numeros[index]\n return total\nejemplo8(numeros)",
"_____no_output_____"
],
[
"def ejemplo9( n ):\n contador = 0\n basura = 0\n for i in range( n ) :\n contador += 1\n for j in range( n ) :\n contador += 1\n basura = basura + contador\n return contador\n\n\nprint(ejemplo9( 5 ))\n#3+2n",
"_____no_output_____"
],
[
"def ejemplo10( n ):\n count = 0\n for i in range( n ) :\n for j in range( i+1 ) :\n count += 1\n return count",
"_____no_output_____"
],
[
"def ejemplo10( n ):\n count = 0\n for i in range( n ) :\n for j in range( i ) :\n count += 1\n return count\n\nprint(ejemplo10(5))\n\"\"\"\n\nn= 3\n000\nn00 <-- aqui empieza el for interno\nnn0 <--- aqui termina el for interno\nnnn\nn = 4\n0000\nn000 <-- aqui empieza el for interno\nnn00\nnnn0 <--- aqui termina el for interno\nnnnn\n\nn =5\n00000\nn0000 <-- aqui empieza el for interno\nnn000\nnnn00\nnnnn0 <--- aqui termina el for interno\nnnnnn\n\"\"\"\n",
"10\n"
],
[
"def ejemplo11( n ):\n count = 0\n i = n\n while i > 1 :\n count += 1\n i = i // 2 \n return count\nprint(ejemplo11(16))\n# T(n) = 2 + (2 Log 2 n)",
"_____no_output_____"
],
[
"def ejemplo12( n ):\n contador = 0\n for x in range(n):\n contador += ejemplo11(x)\n return contador\ndef ejemplo12_bis( n=5 ):\n contador = 0\n contador = contador + ejemplo11(0) # 0\n contador = contador + ejemplo11(1) # 0\n contador = contador + ejemplo11(2) # 1\n contador = contador + ejemplo11(3) # 1\n contador = contador + ejemplo11(4) # 2\n return contador\nejemplo12_bis( 5 ) ",
"_____no_output_____"
],
[
"def ejemplo13( x ):\n bandera = x\n contador = 0\n while( bandera >= 10):\n print(f\" x = { bandera } \")\n bandera /= 10\n contador = contador + 1\n print(contador)\n# T(x) = log10 x +1\nejemplo13( 1000 )",
"_____no_output_____"
],
[
"def ejemplo14( n ):\n y = n\n z = n\n contador = 0\n while y >= 3: #3\n y /= 3 # 1\n contador += 1 # cont =3\n \n while z >= 3: #27\n z /= 3\n contador += 1\n return contador",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03d77874c26e082e852500eed27cb133dba6ee9 | 9,633 | ipynb | Jupyter Notebook | ML Algorithms/Naive Bayes Classifiers/Naive Bayes Classifiers.ipynb | jrderek/Data-science-master-resources | 95adab02dccbf5fbe6333389324a1f8d032d3165 | [
"MIT"
] | 14 | 2020-09-17T17:04:04.000Z | 2021-08-19T05:08:49.000Z | ML Algorithms/Naive Bayes Classifiers/Naive Bayes Classifiers.ipynb | jrderek/Data-science-master-resources | 95adab02dccbf5fbe6333389324a1f8d032d3165 | [
"MIT"
] | 85 | 2020-10-01T16:53:21.000Z | 2021-07-08T17:44:17.000Z | ML Algorithms/Naive Bayes Classifiers/Naive Bayes Classifiers.ipynb | jrderek/Data-science-master-resources | 95adab02dccbf5fbe6333389324a1f8d032d3165 | [
"MIT"
] | 5 | 2020-09-18T08:53:01.000Z | 2021-08-19T05:12:52.000Z | 23.100719 | 340 | 0.47358 | [
[
[
"### Naive Bayes Classifiers ",
"_____no_output_____"
],
[
"#### Author : Sanjoy Biswas \n#### Topic : NaiveNaive Bayes Classifiers : Spam Ham Email Datection \n#### Email : sanjoy.eee32@gmail.com ",
"_____no_output_____"
],
[
"It is a classification technique based on Bayes’ Theorem with an assumption of independence among predictors. In simple terms, a Naive Bayes classifier assumes that the presence of a particular feature in a class is unrelated to the presence of any other feature.\n\nFor example, a fruit may be considered to be an apple if it is red, round, and about 3 inches in diameter. Even if these features depend on each other or upon the existence of the other features, all of these properties independently contribute to the probability that this fruit is an apple and that is why it is known as ‘Naive’.\n\nNaive Bayes model is easy to build and particularly useful for very large data sets. Along with simplicity, Naive Bayes is known to outperform even highly sophisticated classification methods.\n\nBayes theorem provides a way of calculating posterior probability P(c|x) from P(c), P(x) and P(x|c). Look at the equation below:",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"Above,\n\nP(c|x) is the posterior probability of class (c, target) given predictor (x, attributes).<br>\n!P(c) is the prior probability of class.<br>\nP(x|c) is the likelihood which is the probability of predictor given class.<br>\nP(x) is the prior probability of predictor.\n",
"_____no_output_____"
],
[
"### Import Libraries",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"### Import Dataset",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(r'F:\\ML Algorithms By Me\\Naive Bayes Classifiers\\emails.csv')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.isnull().sum()",
"_____no_output_____"
]
],
[
[
"### Separate Dependent & Independent Value",
"_____no_output_____"
]
],
[
[
"x = df.text.values\ny = df.spam.values",
"_____no_output_____"
]
],
[
[
"### Split Train and Test Dataset",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"xtrain,xtest,ytrain,ytest = train_test_split(x,y,test_size=0.3)",
"_____no_output_____"
]
],
[
[
"### Data Preprocessing",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import CountVectorizer",
"_____no_output_____"
],
[
"cv = CountVectorizer()",
"_____no_output_____"
],
[
"x_train = cv.fit_transform(xtrain)",
"_____no_output_____"
],
[
"x_train.toarray()",
"_____no_output_____"
]
],
[
[
"### Apply Naive Bayes Classifiers Algorithm",
"_____no_output_____"
]
],
[
[
"from sklearn.naive_bayes import MultinomialNB",
"_____no_output_____"
],
[
"model = MultinomialNB()",
"_____no_output_____"
],
[
"model.fit(x_train,ytrain)",
"_____no_output_____"
],
[
"x_test = cv.fit_transform(xtest)",
"_____no_output_____"
],
[
"x_test.toarray()",
"_____no_output_____"
],
[
"model.score(x_train,ytrain)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03d88bb6832bb43b5bc5c849db2f9c1feb8617c | 772,180 | ipynb | Jupyter Notebook | notebooks/03-framing-models.ipynb | sflippl/patches | c19889e676e231af44669a01c61854e9e5791227 | [
"MIT"
] | null | null | null | notebooks/03-framing-models.ipynb | sflippl/patches | c19889e676e231af44669a01c61854e9e5791227 | [
"MIT"
] | null | null | null | notebooks/03-framing-models.ipynb | sflippl/patches | c19889e676e231af44669a01c61854e9e5791227 | [
"MIT"
] | null | null | null | 580.586466 | 283,656 | 0.943553 | [
[
[
"# Framing models",
"_____no_output_____"
]
],
[
[
"import lettertask\nimport patches\nimport torch\nimport torch.nn as nn\nimport torch.optim as optim\nimport numpy as np\nfrom tqdm import tqdm\nimport lazytools_sflippl as lazytools\nimport plotnine as gg\nimport pandas as pd",
"_____no_output_____"
],
[
"cbm = lettertask.data.CompositionalBinaryModel(\n width=[5, 5],\n change_probability=[0.05, 0.5],\n samples=10000,\n seed=1001\n)",
"_____no_output_____"
],
[
"cts = patches.data.Contrastive1DTimeSeries(cbm.to_array(), seed=202)",
"_____no_output_____"
]
],
[
[
"## Base-reconstructive model",
"_____no_output_____"
]
],
[
[
"class BaRec(nn.Module):\n def __init__(self, latent_features, input_features=None, timesteps=None,\n data=None, bias=True):\n super().__init__()\n if data:\n input_features = input_features or data.n_vars\n timesteps = timesteps or data.n_timesteps\n elif input_features is None or timesteps is None:\n raise ValueError('You must either provide data or both input '\n 'features and timesteps.')\n self.latent_features = latent_features\n self.input_features = input_features\n self.timesteps = timesteps\n self.encoder = nn.Linear(input_features, latent_features, bias=bias)\n self.predictor = nn.Linear(latent_features, timesteps, bias=bias)\n self.decoder = nn.Conv1d(latent_features, input_features, 1, bias=bias)\n\n def forward(self, x):\n code = self.encoder(x['current_values'])\n prediction = self.predictor(code)\n decoded = self.decoder(prediction).transpose(1, 2)\n return decoded",
"_____no_output_____"
],
[
"barec = BaRec(1, data=cts)\noptimizer = optim.Adam(barec.parameters())\ncriterion = nn.MSELoss()",
"_____no_output_____"
],
[
"data = cts[0]\nprediction = barec(data)\nprint(data['future_values'].shape)\nprint(prediction.shape)",
"torch.Size([1, 5, 10])\ntorch.Size([1, 5, 10])\n"
],
[
"ideal = np.array([[1,0],[0,1]], dtype=np.float32).repeat(5,1)/np.sqrt(5)\nideal",
"_____no_output_____"
],
[
"barec = BaRec(1, data=cts, bias=False)\noptimizer = optim.Adam(barec.parameters())\ncriterion = nn.MSELoss()\nloss_traj = []\nangles = []\nrunning_loss = 0\nfor epoch in tqdm(range(10)):\n for i, data in enumerate(cts):\n if i<len(cts):\n optimizer.zero_grad()\n prediction = barec(data)\n loss = criterion(prediction, data['future_values'])\n loss.backward()\n optimizer.step()\n running_loss += loss\n if i % 50 == 49:\n loss_traj.append(running_loss.detach.numpy()/50)\n running_loss = 0\n est = next(barec.parameters()).detach().numpy()\n angles.append(np.matmul(ideal, est.T)/np.sqrt(np.matmul(est, est.T)))",
"100%|██████████| 10/10 [03:09<00:00, 19.00s/it]\n"
],
[
"(gg.ggplot(\n lazytools.array_to_dataframe(\n np.array(\n [l.detach().numpy() for l in loss_traj]\n )\n ),\n gg.aes(x='dim0', y='array')\n) +\n gg.geom_smooth(method='loess'))",
"_____no_output_____"
],
[
"(gg.ggplot(\n lazytools.array_to_dataframe(\n np.concatenate(angles, axis=1)\n ),\n gg.aes(x='dim1', y='array', color='dim0', group='dim0')\n) + \n gg.geom_line())",
"_____no_output_____"
],
[
"np.save('angles.npy', np.concatenate(angles, axis=1))",
"_____no_output_____"
]
],
[
[
"## 1-part latent predictive model",
"_____no_output_____"
]
],
[
[
"class LaPred1P(nn.Module):\n def __init__(self, latent_features, input_features=None, timesteps=None,\n data=None, bias=True):\n super().__init__()\n if data:\n input_features = input_features or data.n_vars\n timesteps = timesteps or data.n_timesteps\n elif input_features is None or timesteps is None:\n raise ValueError('You must either provide data or both input '\n 'features and timesteps.')\n self.latent_features = latent_features\n self.input_features = input_features\n self.timesteps = timesteps\n self.encoder = nn.Linear(input_features, latent_features, bias=bias)\n self.predictor = nn.Linear(latent_features, timesteps*latent_features, bias=bias)\n\n def forward(self, x):\n code = self.encoder(x['input'])\n prediction = self.predictor(code).\\\n reshape(self.timesteps, self.latent_features)\n return prediction",
"_____no_output_____"
],
[
"hmm = patches.data.HiddenMarkovModel(cbm.to_array(), cbm.latent_array()[:, [0]])",
"_____no_output_____"
],
[
"lapred1p = LaPred1P(1, data=hmm, bias=False)",
"_____no_output_____"
],
[
"lapred1p(hmm[0])",
"_____no_output_____"
],
[
"hmm[0]['future_latent_values']",
"_____no_output_____"
],
[
"lapred1p = LaPred1P(1, data=hmm, bias=False)\noptimizer = optim.Adam(lapred1p.parameters())\ncriterion = nn.MSELoss()\nrunning_loss = 0\nloss_traj = []\nangles = []\nfor epoch in tqdm(range(10)):\n for i, data in enumerate(hmm):\n if i<len(hmm):\n if i % 10 == 0:\n est = list(lapred1p.parameters())[0].detach().numpy()\n angles.append(np.matmul(ideal, est.T)/np.sqrt(np.matmul(est, est.T)))\n optimizer.zero_grad()\n prediction = lapred1p(data)\n loss = criterion(prediction, data['future_latent_values'])\n loss.backward()\n optimizer.step()\n running_loss += loss\n if i % 50 == 49:\n loss_traj.append(running_loss.detach().numpy()/50)\n running_loss = 0",
"100%|██████████| 10/10 [01:06<00:00, 6.54s/it]\n"
],
[
"list(lapred1p.parameters())",
"_____no_output_____"
],
[
"(gg.ggplot(\n lazytools.array_to_dataframe(\n np.array(loss_traj)\n ),\n gg.aes(x='dim0', y='array')\n) +\n gg.geom_smooth(method='loess'))",
"_____no_output_____"
],
[
"np.concatenate(angles, axis=1).shape",
"_____no_output_____"
],
[
"(gg.ggplot(\n lazytools.array_to_dataframe(\n np.concatenate(angles, axis=1)\n ),\n gg.aes(x='dim1', y='array', color='dim0', group='dim0')\n) + \n gg.geom_line())",
"_____no_output_____"
]
],
[
[
"## 2-part latent predictive model",
"_____no_output_____"
]
],
[
[
"class LaPred2P(nn.Module):\n def __init__(self, latent_features, input_features=None, timesteps=None,\n data=None, bias=True):\n super().__init__()\n if data:\n input_features = input_features or data.n_vars\n timesteps = timesteps or data.n_timesteps\n elif input_features is None or timesteps is None:\n raise ValueError('You must either provide data or both input '\n 'features and timesteps.')\n self.latent_features = latent_features\n self.input_features = input_features\n self.timesteps = timesteps\n self.encoder = nn.Linear(input_features, latent_features, bias=bias)\n self.predictor = nn.Linear(latent_features, timesteps*latent_features, bias=bias)\n\n def forward(self, x):\n code = self.encoder(x['input'])\n prediction = self.predictor(x['latent_values']).\\\n reshape(self.timesteps, self.latent_features)\n return {\n 'latent_values': code,\n 'latent_prediction': prediction\n }",
"_____no_output_____"
],
[
"lapred2p = LaPred2P(1, data=hmm, bias=False)\noptimizer = optim.Adam(lapred2p.parameters())\ncriterion = nn.MSELoss()\nloss_traj = []\nangles = []\nrunning_loss = 0\nfor epoch in tqdm(range(10)):\n for i, data in enumerate(hmm):\n if i<len(hmm):\n if i % 10 == 0:\n est = list(lapred2p.parameters())[0].detach().numpy()\n angles.append(np.matmul(ideal, est.T)/np.sqrt(np.matmul(est, est.T)))\n optimizer.zero_grad()\n prediction = lapred2p(data)\n loss = criterion(prediction['latent_prediction'], data['future_latent_values']) + \\\n criterion(prediction['latent_values'], data['latent_values'])\n loss.backward()\n optimizer.step()\n running_loss += loss\n if i % 50 == 49:\n loss_traj.append(running_loss.detach().numpy()/50)\n running_loss = 0",
"100%|██████████| 10/10 [01:10<00:00, 7.08s/it]\n"
],
[
"list(lapred2p.parameters())[0]",
"_____no_output_____"
],
[
"(gg.ggplot(\n lazytools.array_to_dataframe(\n np.array(loss_traj)\n ),\n gg.aes(x='dim0', y='array')\n) +\n gg.geom_smooth(method='loess'))",
"_____no_output_____"
],
[
"(gg.ggplot(\n lazytools.array_to_dataframe(\n np.concatenate(angles, axis=1)\n ),\n gg.aes(x='dim1', y='array', color='dim0', group='dim0')\n) + \n gg.geom_line())",
"_____no_output_____"
]
],
[
[
"## Contrastive predictive model",
"_____no_output_____"
]
],
[
[
"cts = patches.data.Contrastive1DTimeSeries(data=cbm.to_array())\nce = patches.networks.LinearScaffold(latent_features=1, data=cts)\ncriterion = patches.losses.ContrastiveLoss(loss=nn.MSELoss())\noptimizer = optim.Adam(ce.parameters())\nangles = []\nloss_traj = []\nrunning_loss = 0\nfor epoch in tqdm(range(10)):\n for i, data in enumerate(cts):\n if i<len(cts):\n if i % 10 == 0:\n est = list(ce.parameters())[0].detach().numpy()\n angles.append(np.matmul(ideal, est.T)/np.sqrt(np.matmul(est, est.T)))\n optimizer.zero_grad()\n code = ce(data)\n loss = criterion(code)\n loss.backward()\n optimizer.step()\n running_loss += loss\n if i % 50 == 49:\n loss_traj.append(running_loss.detach().numpy()/50)\n running_loss = 0",
"100%|██████████| 10/10 [03:19<00:00, 19.86s/it]\n"
],
[
"(gg.ggplot(\n lazytools.array_to_dataframe(\n np.array(loss_traj)\n ),\n gg.aes(x='dim0', y='array')\n) +\n gg.geom_smooth(method='loess'))",
"_____no_output_____"
],
[
"(gg.ggplot(\n lazytools.array_to_dataframe(\n np.concatenate(angles, axis=1)\n ),\n gg.aes(x='dim1', y='array', color='dim0', group='dim0')\n) + \n gg.geom_line())",
"_____no_output_____"
],
[
"list(ce.parameters())",
"_____no_output_____"
]
],
[
[
"## Sampling bias",
"_____no_output_____"
]
],
[
[
"def moving_average(array):\n \"\"\"Moving average over axis 0.\"\"\"\n cumsum = array.cumsum(axis=0)\n length = cumsum.shape[0]\n rng = np.arange(1, length+1)\n if cumsum.ndim>1:\n rng = rng.reshape(length, 1).repeat(cumsum.shape[1], 1)\n return cumsum/rng",
"_____no_output_____"
],
[
"exposure = moving_average(np.abs(cbm.to_array()))",
"_____no_output_____"
],
[
"(gg.ggplot(lazytools.array_to_dataframe(exposure), gg.aes(x='dim0', group='dim1', y='array')) + \n gg.geom_line(alpha=0.2) + \n gg.scale_x_log10())",
"/home/sflippl/.local/lib/python3.7/site-packages/plotnine/scales/scale.py:549: RuntimeWarning: divide by zero encountered in log10\n return self.trans.transform(x)\n"
],
[
"coherence = moving_average(1-2*(cbm.latent_array()[:-1,:]!=cbm.latent_array()[1:,:]))",
"_____no_output_____"
],
[
"lazytools.array_to_dataframe(coherence)['dim1'].astype(str)",
"_____no_output_____"
],
[
"(gg.ggplot(lazytools.array_to_dataframe(coherence),\n gg.aes(x='dim0', color='dim1', y='array', group='dim1')) + \n gg.geom_line() + \n gg.scale_x_log10())",
"/home/sflippl/.local/lib/python3.7/site-packages/plotnine/scales/scale.py:549: RuntimeWarning: divide by zero encountered in log10\n return self.trans.transform(x)\n"
],
[
"coherence = moving_average(1-2*(cbm.latent_array()[:-2,:]!=cbm.latent_array()[2:,:]))\n(gg.ggplot(lazytools.array_to_dataframe(coherence),\n gg.aes(x='dim0', color='dim1', y='array', group='dim1')) + \n gg.geom_line() + \n gg.scale_x_log10())",
"/home/sflippl/.local/lib/python3.7/site-packages/plotnine/scales/scale.py:549: RuntimeWarning: divide by zero encountered in log10\n return self.trans.transform(x)\n"
],
[
"dfs = []\nfor t in tqdm(range(1, 10)):\n for pos_1 in range(10):\n for pos_2 in range(10):\n pos_subset = (cbm.to_array()[:-t,pos_1]!=0) & (cbm.to_array()[t:,pos_2]!=0)\n tmp_coherence = moving_average(\n 1-2*(cbm.to_array()[:-t][pos_subset,pos_1]!=cbm.to_array()[t:][pos_subset,pos_2])\n )\n tmp_df = lazytools.array_to_dataframe(tmp_coherence)\n tmp_df['pos_1'] = np.array(pos_1)\n tmp_df['pos_2'] = np.array(pos_2)\n tmp_df['t'] = np.array(t)\n tmp_df['n'] = len(tmp_df)\n dfs.append(tmp_df)",
"100%|██████████| 9/9 [01:00<00:00, 6.70s/it]\n"
],
[
"df = pd.concat(dfs)\ndf['dim0'] = (df['dim0']+1)/df['n']\ndf['coherent'] = (df['pos_1'] <= 4) & (df['pos_2']<= 4)\ndf['group'] = df['pos_1'].astype(str)+df['pos_2'].astype(str)",
"_____no_output_____"
],
[
"(gg.ggplot(df, gg.aes(x='dim0', y='array', group='group', color='coherent')) + \n gg.geom_line(alpha=0.2) + \n gg.facet_wrap('t') + \n gg.scale_x_log10())",
"_____no_output_____"
],
[
"(gg.ggplot(df[(df['dim0']==1)], gg.aes(x='array', fill='coherent')) + \n gg.geom_histogram(position='identity', alpha=.8) + \n gg.facet_wrap('t'))",
"/home/sflippl/.local/lib/python3.7/site-packages/plotnine/stats/stat_bin.py:93: UserWarning: 'stat_bin()' using 'bins = 24'. Pick better value with 'binwidth'.\n warn(msg.format(params['bins']))\n"
],
[
"help(gg.labs)",
"Help on class labs in module plotnine.labels:\n\nclass labs(builtins.object)\n | labs(*args, **kwargs)\n | \n | General class for all label adding classes\n | \n | Parameters\n | ----------\n | args : dict\n | Aesthetics to be renamed\n | kwargs : dict\n | Aesthetics to be renamed\n | \n | Methods defined here:\n | \n | __init__(self, *args, **kwargs)\n | Initialize self. See help(type(self)) for accurate signature.\n | \n | __radd__(self, gg, inplace=False)\n | \n | ----------------------------------------------------------------------\n | Data descriptors defined here:\n | \n | __dict__\n | dictionary for instance variables (if defined)\n | \n | __weakref__\n | list of weak references to the object (if defined)\n | \n | ----------------------------------------------------------------------\n | Data and other attributes defined here:\n | \n | labels = {}\n\n"
],
[
"str(cbm)",
"_____no_output_____"
],
[
"cbm.width",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03d89fb8101858754444e4ff48533e4b6a5b09d | 4,148 | ipynb | Jupyter Notebook | numbaTemplate.ipynb | kbrezinski/Project-Template | 048fb77e611231feee2dc28f17624cf7f96453a9 | [
"BSD-3-Clause"
] | null | null | null | numbaTemplate.ipynb | kbrezinski/Project-Template | 048fb77e611231feee2dc28f17624cf7f96453a9 | [
"BSD-3-Clause"
] | null | null | null | numbaTemplate.ipynb | kbrezinski/Project-Template | 048fb77e611231feee2dc28f17624cf7f96453a9 | [
"BSD-3-Clause"
] | null | null | null | 20.844221 | 80 | 0.496143 | [
[
[
"%matplotlib inline\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"from numba import jit, njit\nimport random",
"_____no_output_____"
],
[
"def monte_carlo_pi(n):\n acc = 0\n for i in range(n):\n if (random.random() ** 2 + random.random() ** 2) < 1.0:\n acc += 1\n return 4.0 * acc / n",
"_____no_output_____"
],
[
"%timeit -n 10 monte_carlo_pi(10000)",
"10.2 ms ± 52.3 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)\n"
],
[
"## use njit or nopython=True\nmonte_carlo_pi_jit = njit()(monte_carlo_pi)",
"_____no_output_____"
],
[
"%timeit -n 10 monte_carlo_pi_jit(10000)",
"98.7 µs ± 8.93 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)\n"
],
[
"## Need to remove reflection using Python lists\nimport numpy as np",
"_____no_output_____"
],
[
"@njit(nogil=True)\ndef scalar_computation(input_list):\n output_list = np.zeros_like(input_list)\n for ii, item in enumerate(input_list):\n if item % 2 == 0:\n output_list[ii] = 1\n else:\n output_list[ii] = 0\n return output_list\n\n#%time scalar_computation(list(range(100)))",
"_____no_output_____"
],
[
"@vectorize\ndef scalar_computation_vec(item): \n if item % 2 == 0:\n return 1\n else:\n return 0\n\n#%timeit -n 10 scalar_computation(list(range(100)))",
"_____no_output_____"
],
[
"%%timeit\nfrom concurrent.futures import ThreadPoolExecutor\n\nwith ThreadPoolExecutor(6) as ex:\n ex.map(scalar_computation_vec, list(range(10000)))",
"463 ms ± 105 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"%%timeit\nfrom concurrent.futures import ThreadPoolExecutor\n\nwith ThreadPoolExecutor(6) as ex:\n ex.map(scalar_computation, list(range(10000)))",
"502 ms ± 87.7 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03d916c13e20ae5a74624990817df7a443fc936 | 19,038 | ipynb | Jupyter Notebook | notebooks/train_test.ipynb | guillermoiglesiashernandez/StyleGAN | 6fb4a1d04c57f1f907cf35d9a3f08cef39fe7e7d | [
"MIT"
] | null | null | null | notebooks/train_test.ipynb | guillermoiglesiashernandez/StyleGAN | 6fb4a1d04c57f1f907cf35d9a3f08cef39fe7e7d | [
"MIT"
] | null | null | null | notebooks/train_test.ipynb | guillermoiglesiashernandez/StyleGAN | 6fb4a1d04c57f1f907cf35d9a3f08cef39fe7e7d | [
"MIT"
] | null | null | null | 19,038 | 19,038 | 0.725917 | [
[
[
"import os\n\nos.chdir('/content/')\n!git clone https://github.com/guillermoiglesiashernandez/StyleGAN\nos.chdir('/content/StyleGAN/')",
"fatal: destination path 'StyleGAN' already exists and is not an empty directory.\n"
],
[
"!pip3 install tensorflow_addons",
"Requirement already satisfied: tensorflow_addons in /usr/local/lib/python3.7/dist-packages (0.15.0)\nRequirement already satisfied: typeguard>=2.7 in /usr/local/lib/python3.7/dist-packages (from tensorflow_addons) (2.7.1)\n"
],
[
"import tensorflow as tf\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom tensorflow import keras\nfrom train.train import train\nfrom train.style_gan import StyleGAN\nfrom train.plot_images import plot_images\nfrom data_loader.data_download import download_data",
"_____no_output_____"
],
[
"style_gan = StyleGAN(start_res=4, target_res=16)",
"_____no_output_____"
],
[
"url = \"https://drive.google.com/uc?id=1O7m1010EJjLE5QxLZiM9Fpjs7Oj6e684\"\noutput_dir = \"celeba_gan\"\n\ndata = download_data(url, output_dir)",
"Found 202599 files belonging to 1 classes.\n"
],
[
"train(style_gan, data, start_res=4, target_res=16, steps_per_epoch=1, display_images=False)\n\n\"\"\"\n## Results\nWe can now run some inference using pre-trained 64x64 checkpoints. In general, the image\nfidelity increases with the resolution. You can try to train this StyleGAN to resolutions\nabove 128x128 with the CelebA HQ dataset.\n\"\"\"\n\nurl = \"https://github.com/soon-yau/stylegan_keras/releases/download/keras_example_v1.0/stylegan_128x128.ckpt.zip\"\n\nweights_path = keras.utils.get_file(\n \"stylegan_128x128.ckpt.zip\",\n url,\n extract=True,\n cache_dir=os.path.abspath(\".\"),\n cache_subdir=\"pretrained\",\n)\n\nstyle_gan.grow_model(128)\nstyle_gan.load_weights(os.path.join(\"pretrained/stylegan_128x128.ckpt\"))\n\ntf.random.set_seed(196)\nbatch_size = 2\nz = tf.random.normal((batch_size, style_gan.z_dim))\nw = style_gan.mapping(z)\nnoise = style_gan.generate_noise(batch_size=batch_size)\nimages = style_gan({\"style_code\": w, \"noise\": noise, \"alpha\": 1.0})\nplot_images(images, 5)\n\n\"\"\"\n## Style Mixing\nWe can also mix styles from two images to create a new image.\n\"\"\"\n\nalpha = 0.4\nw_mix = np.expand_dims(alpha * w[0] + (1 - alpha) * w[1], 0)\nnoise_a = [np.expand_dims(n[0], 0) for n in noise]\nmix_images = style_gan({\"style_code\": w_mix, \"noise\": noise_a})\nimage_row = np.hstack([images[0], images[1], mix_images[0]])\nplt.figure(figsize=(9, 3))\nplt.imshow(image_row)\nplt.axis(\"off\")",
"\nModel resolution:4x4\nSTABLE\n1/1 [==============================] - 10s 10s/step - d_loss: 4.7932 - g_loss: -0.6010\n\nModel resolution:8x8\nTRANSITION\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03dae4bf598dbfb9a9f27cdc37787f07805d120 | 273,996 | ipynb | Jupyter Notebook | IBM-statistics/2-Visualizing_Data.ipynb | brndnaxr/brndnaxr-micro-projects | 47bd3e7a627e1b1c0059133309e481c506a7c670 | [
"MIT"
] | null | null | null | IBM-statistics/2-Visualizing_Data.ipynb | brndnaxr/brndnaxr-micro-projects | 47bd3e7a627e1b1c0059133309e481c506a7c670 | [
"MIT"
] | null | null | null | IBM-statistics/2-Visualizing_Data.ipynb | brndnaxr/brndnaxr-micro-projects | 47bd3e7a627e1b1c0059133309e481c506a7c670 | [
"MIT"
] | null | null | null | 202.360414 | 68,636 | 0.898112 | [
[
[
"<center>\n <img src=\"https://gitlab.com/ibm/skills-network/courses/placeholder101/-/raw/master/labs/module%201/images/IDSNlogo.png\" width=\"300\" alt=\"cognitiveclass.ai logo\" />\n</center>\n",
"_____no_output_____"
],
[
"# **Data Visualization**\n",
"_____no_output_____"
],
[
"Estimated time needed: **30** minutes\n",
"_____no_output_____"
],
[
"In this lab, you will learn how to visualize and interpret data\n",
"_____no_output_____"
],
[
"## Objectives\n",
"_____no_output_____"
],
[
"* Import Libraries\n* Lab Exercises\n * Identifying duplicates\n * Plotting Scatterplots\n * Plotting Boxplots\n",
"_____no_output_____"
],
[
"***\n",
"_____no_output_____"
],
[
"## Import Libraries\n",
"_____no_output_____"
],
[
"All Libraries required for this lab are listed below. The libraries pre-installed on Skills Network Labs are commented. If you run this notebook in a different environment, e.g. your desktop, you may need to uncomment and install certain libraries.\n",
"_____no_output_____"
]
],
[
[
"# !pip install pandas\n# !pip install numpy\n# !pip install matplotlib\n# !pip install seaborn",
"_____no_output_____"
]
],
[
[
"Import the libraries we need for the lab\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt ",
"_____no_output_____"
]
],
[
[
"Read in the csv file from the url using the request library\n",
"_____no_output_____"
]
],
[
[
"ratings_url = 'https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-ST0151EN-SkillsNetwork/labs/teachingratings.csv'\nratings_df = pd.read_csv(ratings_url)",
"_____no_output_____"
]
],
[
[
"## Lab Exercises\n",
"_____no_output_____"
],
[
"### Identify all duplicate cases using prof. Using all observations, find the average and standard deviation for age. Repeat the analysis by first filtering the data set to include one observation for each instructor with a total number of observations restricted to 94.\n",
"_____no_output_____"
],
[
"Identify all duplicate cases using prof variable - find the unique values of the prof variables\n",
"_____no_output_____"
]
],
[
[
"ratings_df.prof.unique()",
"_____no_output_____"
]
],
[
[
"Print out the number of unique values in the prof variable\n",
"_____no_output_____"
]
],
[
[
"ratings_df.prof.nunique()",
"_____no_output_____"
]
],
[
[
"Using all observations, Find the average and standard deviation for age\n",
"_____no_output_____"
]
],
[
[
"ratings_df['age'].mean()",
"_____no_output_____"
],
[
"ratings_df['age'].std()",
"_____no_output_____"
]
],
[
[
"Repeat the analysis by first filtering the data set to include one observation for each instructor with a total number of observations restricted to 94.\n\n> first we drop duplicates using prof as a subset and assign it a new dataframe name called no_duplicates_ratings_df\n",
"_____no_output_____"
]
],
[
[
"no_duplicates_ratings_df = ratings_df.drop_duplicates(subset =['prof'])\nno_duplicates_ratings_df.head()",
"_____no_output_____"
]
],
[
[
"> Use the new dataset to get the mean of age\n",
"_____no_output_____"
]
],
[
[
"no_duplicates_ratings_df['age'].mean()",
"_____no_output_____"
],
[
"no_duplicates_ratings_df['age'].std()",
"_____no_output_____"
]
],
[
[
"### Using a bar chart, demonstrate if instructors teaching lower-division courses receive higher average teaching evaluations.\n",
"_____no_output_____"
]
],
[
[
"ratings_df.head()",
"_____no_output_____"
]
],
[
[
"Find the average teaching evaluation in both groups of upper and lower-division\n",
"_____no_output_____"
]
],
[
[
"division_eval = ratings_df.groupby('division')[['eval']].mean().reset_index()",
"_____no_output_____"
]
],
[
[
"Plot the barplot using the seaborn library\n",
"_____no_output_____"
]
],
[
[
"sns.set(style=\"whitegrid\")\nax = sns.barplot(x=\"division\", y=\"eval\", data=division_eval)",
"_____no_output_____"
]
],
[
[
"### Plot the relationship between age and teaching evaluation scores.\n",
"_____no_output_____"
],
[
"Create a scatterplot with the scatterplot function in the seaborn library\n",
"_____no_output_____"
]
],
[
[
"ax = sns.scatterplot(x='age', y='eval', data=ratings_df)",
"_____no_output_____"
]
],
[
[
"### Using gender-differentiated scatter plots, plot the relationship between age and teaching evaluation scores.\n",
"_____no_output_____"
],
[
"Create a scatterplot with the scatterplot function in the seaborn library this time add the <code>hue</code> argument\n",
"_____no_output_____"
]
],
[
[
"ax = sns.scatterplot(x='age', y='eval', hue='gender',\n data=ratings_df)",
"_____no_output_____"
]
],
[
[
"### Create a box plot for beauty scores differentiated by credits.\n",
"_____no_output_____"
],
[
"We use the <code>boxplot()</code> function from the seaborn library\n",
"_____no_output_____"
]
],
[
[
"ax = sns.boxplot(x='credits', y='beauty', data=ratings_df)",
"_____no_output_____"
]
],
[
[
"### What is the number of courses taught by gender?\n",
"_____no_output_____"
],
[
"We use the <code>catplot()</code> function from the seaborn library\n",
"_____no_output_____"
]
],
[
[
"sns.catplot(x='gender', kind='count', data=ratings_df)",
"_____no_output_____"
]
],
[
[
"### Create a group histogram of taught by gender and tenure\n",
"_____no_output_____"
],
[
"We will add the <code>hue = Tenure</code> argument\n",
"_____no_output_____"
]
],
[
[
"sns.catplot(x='gender', hue = 'tenure', kind='count', data=ratings_df)",
"_____no_output_____"
]
],
[
[
"### Add division as another factor to the above histogram\n",
"_____no_output_____"
],
[
"We add another argument named <code>row</code> and use the division variable as the row\n",
"_____no_output_____"
]
],
[
[
"sns.catplot(x='gender', hue = 'tenure', row = 'division',\n kind='count', data=ratings_df,\n height = 3, aspect = 2)",
"_____no_output_____"
]
],
[
[
"### Create a scatterplot of age and evaluation scores, differentiated by gender and tenure\n",
"_____no_output_____"
],
[
"Use the <code>relplot()</code> function for complex scatter plots\n",
"_____no_output_____"
]
],
[
[
"sns.relplot(x=\"age\", y=\"eval\", hue=\"gender\",\n row=\"tenure\",\n data=ratings_df, height = 3, aspect = 2)",
"_____no_output_____"
]
],
[
[
"### Create a distribution plot of teaching evaluation scores\n",
"_____no_output_____"
],
[
"We use the <code>distplot()</code> function from the seaborn library, set <code>kde = false</code> because we don'e need the curve\n",
"_____no_output_____"
]
],
[
[
"ax = sns.distplot(ratings_df['eval'], kde = False)",
"D:\\anaconda3\\lib\\site-packages\\seaborn\\distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n"
]
],
[
[
"### Create a distribution plot of teaching evaluation score with gender as a factor\n",
"_____no_output_____"
]
],
[
[
"## use the distplot function from the seaborn library\nsns.distplot(ratings_df[ratings_df['gender'] == 'female']['eval'], color='green', kde=False) \nsns.distplot(ratings_df[ratings_df['gender'] == 'male']['eval'], color=\"orange\", kde=False) \nplt.show()",
"_____no_output_____"
]
],
[
[
"### Create a box plot - age of the instructor by gender\n",
"_____no_output_____"
]
],
[
[
"ax = sns.boxplot(x=\"gender\", y=\"age\", data=ratings_df)",
"_____no_output_____"
]
],
[
[
"### Compare age along with tenure and gender\n",
"_____no_output_____"
]
],
[
[
"ax = sns.boxplot(x=\"tenure\", y=\"age\", hue=\"gender\",\n data=ratings_df)",
"_____no_output_____"
]
],
[
[
"## Practice Questions\n",
"_____no_output_____"
],
[
"### Question 1: Create a distribution plot of beauty scores with Native English speaker as a factor\n\n* Make the color of the native English speakers plot - orange and non - native English speakers - blue\n",
"_____no_output_____"
]
],
[
[
"## insert code\n",
"_____no_output_____"
]
],
[
[
"Double-click **here** for the solution.\n\n<!-- The answer is below:\nsns.distplot(ratings_df[ratings_df['native'] == 'yes']['beauty'], color=\"orange\", kde=False) \nsns.distplot(ratings_df[ratings_df['native'] == 'no']['beauty'], color=\"blue\", kde=False) \nplt.show()\n-->\n",
"_____no_output_____"
],
[
"### Question 2: Create a Horizontal box plot of the age of the instructors by visible minority\n",
"_____no_output_____"
]
],
[
[
"## insert code\n",
"_____no_output_____"
]
],
[
[
"Double-click **here** for a hint.\n\n<!-- The hint is below:\nRemember that the positions of the argument determine whether it will be vertical or horizontal\n-->\n",
"_____no_output_____"
],
[
"Double-click **here** for the solution.\n\n<!-- The answer is below:\nax = sns.boxplot(x=\"age\", y=\"minority\", data=ratings_df)\n-->\n",
"_____no_output_____"
],
[
"### Question 3: Create a group histogram of tenure by minority and add the gender factor\n",
"_____no_output_____"
]
],
[
[
"## insert code",
"_____no_output_____"
]
],
[
[
"Double-click **here** for the solution.\n\n<!-- The answer is below:\nsns.catplot(x='tenure', hue = 'minority', row = 'gender',\n kind='count', data=ratings_df,\n height = 3, aspect = 2)\n-->\n",
"_____no_output_____"
],
[
"### Question 4: Create a boxplot of the age variable\n",
"_____no_output_____"
]
],
[
[
"## insert code",
"_____no_output_____"
]
],
[
[
"Double-click **here** for the solution.\n\n<!-- The answer is below:\n## you only habve to specify the y-variable\nax = sns.boxplot(y=\"age\", data=ratings_df)\n-->\n",
"_____no_output_____"
],
[
"## Authors\n",
"_____no_output_____"
],
[
"[Aije Egwaikhide](https://www.linkedin.com/in/aije-egwaikhide/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkST0151ENSkillsNetwork20531532-2021-01-01) is a Data Scientist at IBM who holds a degree in Economics and Statistics from the University of Manitoba and a Post-grad in Business Analytics from St. Lawrence College, Kingston. She is a current employee of IBM where she started as a Junior Data Scientist at the Global Business Services (GBS) in 2018. Her main role was making meaning out of data for their Oil and Gas clients through basic statistics and advanced Machine Learning algorithms. The highlight of her time in GBS was creating a customized end-to-end Machine learning and Statistics solution on optimizing operations in the Oil and Gas wells. She moved to the Cognitive Systems Group as a Senior Data Scientist where she will be providing the team with actionable insights using Data Science techniques and further improve processes through building machine learning solutions. She recently joined the IBM Developer Skills Network group where she brings her real-world experience to the courses she creates.\n",
"_____no_output_____"
],
[
"## Change Log\n",
"_____no_output_____"
],
[
"| Date (YYYY-MM-DD) | Version | Changed By | Change Description |\n| ----------------- | ------- | --------------- | -------------------------------------- |\n| 2020-08-14 | 0.1 | Aije Egwaikhide | Created the initial version of the lab |\n",
"_____no_output_____"
],
[
"Copyright © 2020 IBM Corporation. This notebook and its source code are released under the terms of the [MIT License](https://cognitiveclass.ai/mit-license/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkST0151ENSkillsNetwork20531532-2021-01-01).\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d03daf5e7b0ebd5f6be6a10ae4b924adcb4473bb | 2,559 | ipynb | Jupyter Notebook | notebooks/run_demo.ipynb | yarden-livnat/ipyregulus | 971ab02cd3676b9ea8c712fd3940d42d974c445d | [
"BSD-3-Clause"
] | 1 | 2018-09-06T17:07:41.000Z | 2018-09-06T17:07:41.000Z | notebooks/run_demo.ipynb | yarden-livnat/ipyregulus | 971ab02cd3676b9ea8c712fd3940d42d974c445d | [
"BSD-3-Clause"
] | 3 | 2021-03-10T09:24:25.000Z | 2022-01-22T10:49:25.000Z | notebooks/run_demo.ipynb | yarden-livnat/ipyregulus | 971ab02cd3676b9ea8c712fd3940d42d974c445d | [
"BSD-3-Clause"
] | 2 | 2018-08-30T19:11:05.000Z | 2020-01-07T16:29:01.000Z | 22.252174 | 124 | 0.528722 | [
[
[
"import cyit",
"_____no_output_____"
],
[
"import demo",
"importing Jupyter notebook from demo.ipynb\n"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
d03dc47ae3565754701771556d3b5a3740f5b87d | 33,861 | ipynb | Jupyter Notebook | docs/docsource/theory.ipynb | kcbhamu/kaldo | 5926ac03f796f82841012ba1dd4ead8eee03a6c7 | [
"BSD-3-Clause"
] | 49 | 2020-07-04T21:50:46.000Z | 2022-03-17T21:05:12.000Z | docs/docsource/theory.ipynb | kaituohuo/kaldo | 537bceea2c3206711a8899d68e1dbd23fb0c38b6 | [
"BSD-3-Clause"
] | 27 | 2020-07-24T06:22:26.000Z | 2022-03-23T02:17:50.000Z | docs/docsource/theory.ipynb | kaituohuo/kaldo | 537bceea2c3206711a8899d68e1dbd23fb0c38b6 | [
"BSD-3-Clause"
] | 13 | 2020-08-20T15:07:52.000Z | 2021-11-07T19:18:59.000Z | 67.318091 | 765 | 0.621305 | [
[
[
"<!--  -->\n\n## Introduction\n\nUnderstanding heat transport in semiconductors and insulators is of fundamental importance because of its technological impact in electronics and renewable energy harvesting and conversion.\nAnharmonic Lattice Dynamics provides a powerful framework for the description of heat transport at the nanoscale. One of the advantages of this method is that it naturally includes quantum effects due to atoms vibrations, which are needed to compute thermal properties of semiconductors widely use in nanotechnology, like Silicon and Carbon, even at room temperature.\nWhile heat transport in amorphous and crystalline semiconductors has a different microscopic origin, a unified approach to simulate both crystals and glasses has been devised.\nHere we introduce a unified workflow, which implements both the Boltzmann Transport equation (BTE) and the Quasi Harmonic Green-Kubo (QHGK) methods. We discuss how the theory can be optimized to exploit modern parallel architectures, and how it is implemented in kALDo: a versatile and scalable open-source software to compute phonon transport in solids.\n\n## Theory\n\nIn semiconductors, electronic and vibrational dynamics often occur over different time scales, and can thus be decoupled using the Born Oppenheimer approximation. Under this assumption, the potential $\\phi$ of a system made of $N_{atoms}$ atoms, is a function of all the $x_{i\\alpha}$ atomic positions, where $i$ and $\\alpha$ refer to the atomic and Cartesian indices, respectively. Near thermal equilibrium, the potential energy can be Taylor expanded in the atomic displacements, $\\mathbf{u}=\\mathbf x-\\mathbf{x}_{\\rm equilibrium}$,\n\n$$\n \\phi(\\{x_{i\\alpha}\\})=\\phi_0 +\n\\sum_{i\\alpha}\\phi^{\\prime}_{i\\alpha }u_{i\\alpha}\n+\\frac{1}{2}\n\\sum_{i\\alpha i'\\alpha'}\n\\phi^{\\prime\\prime}_{i\\alpha i'\\alpha '}u_{i\\alpha} u_{i'\\alpha'}+\n$$\n\n$$\n+\n\\frac{1}{3!}\\sum_{i\\alpha i'\\alpha 'i''\\alpha ''}\n\\phi^{\\prime\\prime\\prime}_{i\\alpha i'\\alpha 'i''\\alpha ''} u_{i\\alpha }u_{i'\\alpha '} u_{i''\\alpha ''} + \\dots, \n$$\n\nwhere\n\n$$\n \\phi^{\\prime\\prime}_{i\\alpha i'\\alpha '}=\\frac{\\partial^{2} \\phi}{\\partial u_{i\\alpha } \\partial u_{i'\\alpha '} },\\qquad\n\\phi^{\\prime\\prime\\prime}_{i\\alpha i'\\alpha 'i''\\alpha ''}=\\frac{\\partial^{3} \\phi}{\\partial u_{i\\alpha } \\partial u_{i'\\alpha '} \\partial u_{i''\\alpha ''}}, \n$$\n\nare the second and third order interatomic force constants (IFC). The term $\\phi_0$ can be discarded, and the forces $F = - \\phi^{\\prime}$ are zero at equilibrium.\n\n\nThe IFCs can be evaluated by finite difference, which consists in calculating the difference between the forces acting on the system when one of the atoms is displaced by a small finite shift along a Cartesian direction. The second and third order IFCs need respectively, $2N_{atoms}$, and $4N_{atoms}^2$ forces calculations. In crystals, this amount can be reduced exploiting the spatial symmetries of the system, or adopting a compressed sensing approach. In the framework of DFT, it is also possible and often convenient to compute IFCs using perturbation theory.\n\nThe dynamical matrix is the second order IFC rescaled by the masses, $D_{i\\alpha i'\\alpha}=\\phi^{\\prime\\prime}_{i\\alpha i'\\alpha'}/\\sqrt{m_im_{i'}}$. It is diagonal in the phonons basis\n\n$$\n \\sum_{i'\\alpha'} D_{i\\alpha i'\\alpha'}\\eta_{i'\\alpha'\\mu} =\\eta_{i\\alpha\\mu} \\omega_\\mu^2 \n$$\n\nand $\\omega_\\mu/(2\\pi)$ are the frequencies of the normal modes of the system.\n\nFor crystals, where there is long range order due to the periodicity, the dimensionality of the problem can be reduced. The Fourier transfom maps the large direct space onto a compact volume in the reciprocal space: the Brillouin zone. More precisely we adopt a supercell approach, where we calculate the dynamical matrix on $N_{\\rm replicas}$ replicas of a unit cell of $N_{\\rm unit}$ atoms, at positions $\\mathbf R_l$, and calculate\n\n$$\nD_{i \\alpha k i' \\alpha'}=\\sum_l \\chi_{kl} D_{i \\alpha l i' \\alpha'},\\quad \\chi_{kl} = \\mathrm{e}^{-i \\mathbf{q_k}\\cdot \\mathbf{R}_{l} },\n$$\n\nwhere $\\mathbf q_k$ is a grid of size $N_k$ indexed by $k$ and the eigenvalue equation becomes\n\n$$\n\\sum_{i'\\alpha'} D_{i \\alpha k i' \\alpha'} \\eta_{i' \\alpha'k s}=\\omega_{k m}^{2} \\eta_{i \\alpha k s }.\n$$\n\nwhich now depends on the quasi-momentum index, $k$, and the phonons mode $s$.\n\n\n### Boltzman Transport Equation\n\nAt finite temperature $T$, the Bose Einstein statistic is the quantum distribution for atomic vibrations\n\n$$\nn_{\\mu}=n(\\omega_{\\mu})=\\frac{1}{e^{\\frac{\\hbar\\omega_{\\mu}}{k_B T}}-1}\n$$\n\nwhere $k_B$ is the Boltzmann constant and we use $\\mu =(k,s)$.\n\nWe consider a small temperature gradient applied along the $\\alpha$-axis of a crystalline material. If the phonons population depends on the position only through the temperature, $\\frac{\\partial n_{\\mu\\alpha}}{\\partial x_\\alpha} = \\frac{\\partial n_{\\mu\\alpha}}{\\partial T}\\nabla_\\alpha T$, we can Taylor expand it\n\n$$\n\\tilde n_{\\mu\\alpha} \\simeq n_\\mu + \\lambda_{\\mu\\alpha} \\frac{\\partial n_\\mu}{\\partial x_\\alpha} \\simeq n_\\mu + \\psi_{\\mu\\alpha}\\nabla_\\alpha T\n$$\n\nwith $\\psi_{\\mu\\alpha}=\\lambda_{\\mu\\alpha} \\frac{\\partial n_\\mu}{\\partial T}$, where $\\lambda_{\\mu\\alpha}$ is the phonons mean free path.\nBeing quantum quasi-particles, phonons have a well-defined group velocity, which, for the acoustic modes in the long wavelength limit, corresponds to the speed of sound in the material,\n\n$$\nv_{ ks\\alpha}=\\frac{\\partial \\omega_{k s}}{\\partial {q_{k\\alpha}}} = \\frac{1}{2\\omega_{ks}}\\sum_{i\\beta l i'\\beta'}\ni R_{l \\alpha} D_{i\\beta li'\\beta'}\\chi_{kl}\n\\eta_{ks i\\beta}\\eta_{ksi'\\beta}\n$$\n\nand the last equality is obtained by applying the derivative with respect to $\\mathbf{q}_k$ directly to tbe eigenvectors Equation\n\n\nThe heat current per mode is written in terms of the phonon energy $\\hbar \\omega$, velocity $v$, and out-of-equilibrium phonons population, $\\tilde n$:\n\n$$\nj_{\\mu\\alpha'} =\\sum_\\alpha \\hbar \\omega_\\mu v_{\\mu\\alpha'} (\\tilde n_{\\mu\\alpha} - n_{\\mu})\\simeq- \\sum_\\alpha c_\\mu v_{\\mu\\alpha'} \\mathbf{\\lambda}_{\\mu\\alpha} \\nabla_\\alpha T .\n$$\n\nAs we deal with extended systems, we can assume heat transport in the diffusive regime, and we can use Fourier's law\n\n$$\nJ_{\\alpha}=-\\sum_{\\alpha'}\\kappa_{\\alpha\\alpha'} \\nabla_{\\alpha'} T,\n$$\n\nwhere the heat current is the sum of the contribution from each phonon mode: $J_\\alpha = 1/(N_k V)\\sum_\\mu j_{\\mu\\alpha}$.\nThe thermal conductivity then results:\n\n$$\n\\kappa_{\\alpha \\alpha'}=\\frac{1}{ V N_k} \\sum_{\\mu} c_\\mu v_{\\mu\\alpha} \\lambda_{\\mu\\alpha'},\n$$\nwhere we defined the heat capacity per mode\n$$\nc_\\mu=\\hbar \\omega_\\mu \\frac{\\partial n_\\mu}{\\partial T},\n$$\n\nwhich is connected to total heat capacity through $C = \\sum_\\mu c_\\mu /NV$.\n\nWe can now introduce the BTE, which combines the kinetic theory of gases with collective phonons vibrations:\n$$\n{\\mathbf{v}}_{\\mu} \\cdot {\\boldsymbol{\\nabla}} T \\frac{\\partial n_{\\mu}}{\\partial T}=\\left.\\frac{\\partial n_{\\mu}}{\\partial t}\\right|_{\\text {scatt}},\n$$\nwhere the scattering term, in the linearized form is\n\n$$\n\\left.\\frac{\\partial n_{\\mu}}{\\partial t}\\right|_{\\text {scatt}}=\n$$\n\n$$\n\\frac{\\nabla_\\alpha T}{\\omega_\\mu N_k}\\sum_{\\mu^{\\prime} \\mu^{\\prime \\prime}}^{+} \\Gamma_{\\mu \\mu^{\\prime} \\mu^{\\prime \\prime}}^{+}\n\\left(\\omega_\\mu\\mathbf{\\psi}_{\\mu\\alpha}\n+\\omega_{\\mu^{\\prime}}\\mathbf{\\psi}_{\\mu^{\\prime}\\alpha}\n-\\omega_{\\mu^{\\prime \\prime}} \\mathbf{\\psi}_{\\mu^{\\prime \\prime}\\alpha}\\right)\n+\n$$\n\n$$\n+\\frac{\\nabla_\\alpha T}{\\omega_\\mu N_k}\\sum_{\\mu^{\\prime} \\mu^{\\prime \\prime}}^{-} \\frac{1}{2} \\Gamma_{\\mu \\mu^{\\prime} \\mu^{\\prime \\prime}}^{-}\n\\left(\\omega_\\mu\\mathbf{\\psi}_{\\mu\\alpha}\n-\\omega_{\\mu^{\\prime}} \\mathbf{\\psi}_{\\mu^{\\prime}\\alpha}\n-\\omega_{\\mu^{\\prime \\prime}} \\mathbf{\\psi}_{\\mu^{\\prime \\prime}\\alpha}\\right) .\n$$\n\n$\\Gamma^{+}_{\\mu\\mu'\\mu''}$ and $\\Gamma^{-}_{\\mu\\mu'\\mu''}$ are the scattering rates for three-phonon scattering processes, and they correspond to the events of phonons annihilation $\\mu, \\mu'\\rightarrow\\mu''$ and phonons creation $\\mu \\rightarrow\\mu',\\mu''\n\n$$\n\\Gamma_{\\mu \\mu^{\\prime} \\mu^{\\prime \\prime}}^{\\pm} =\\frac{\\hbar \\pi}{8} \\frac{g_{\\mu\\mu'\\mu''}^{\\pm}}{\\omega_{\\mu} \\omega_{\\mu'} \\omega_{\\mu''}}\\left|\\phi_{\\mu \\mu^{\\prime} \\mu^{\\prime \\prime}}^{\\pm}\\right|^{2},\n$$\n\nand the projection of the potentials on the phonon modes are given by\n\n$$\n\\phi^\\pm\n_{ksk's'k'' s''}=\n\\sum_{il'i'l''i''}\n\\frac{\n\\phi_{il'i'l''i''}}\n{\\sqrt{m_{i}m_{i'}m_{i''}}}\n\\eta_{i ks}\\eta^{\\pm}_{i'k' s'}\n\\eta^*_{i''k''s''}\\chi^\\pm_{k'l'}\\chi^*_{k''l''}\n$$\n\nwith $\\eta^+=\\eta$, $\\chi^+=\\chi$ and $\\eta^-=\\eta^*$, $\\chi^-=\\chi^*$.\nThe phase space volume $g^\\pm_{\\mu\\mu^\\prime\\mu^{\\prime\\prime}}$ in the previous equation are defined as\n\n$$\ng^+_{\\mu\\mu^\\prime\\mu^{\\prime\\prime}} = (n_{\\mu'}-n_{\\mu''})\n\\delta^+_{\\mu\\mu^\\prime\\mu^{\\prime\\prime}}\n$$\n\n$$\ng^-_{\\mu\\mu^\\prime\\mu^{\\prime\\prime}} = (1 + n_{\\mu'}+n_{\\mu''})\n\\delta^-_{\\mu\\mu^\\prime\\mu^{\\prime\\prime}},\n$$\n\nand include the $\\delta$ for the conservation of the energy and momentum in three-phonons scattering processes,\n\n$$\n\\delta_{ks k's' k''s''}^{\\pm}=\n\\delta_{\\mathbf q_{k}\\pm\\mathbf q_{k'}-\\mathbf q_{k''}, \\mathbf Q}\n\\delta\\left(\\omega_{ks}\\pm\\omega_{k's'}-\\omega_{k''s''}\\right),\n$$\n\nwith $Q$ the lattice vectors. Finally, the normalized phase-space per mode $g_\\mu=\\frac{1}{N}\\sum_{\\mu'\\mu''}g_{\\mu\\mu'\\mu''}$, provides useful information about the weight of a specific mode in the anharmonic scattering processes.\n\nIn order to calculate the conductivity, we express the mean free path in terms of the 3-phonon scattering rates\n$$\nv_{\\mu\\alpha} = \\tilde \\Gamma_{\\mu\\mu' }\\lambda_\\mu = (\\delta_{\\mu\\mu'}\\Gamma^0_\\mu + \\Gamma^{1}_{\\mu\\mu'})\\lambda_{\\mu\\alpha},\n$$\n\nwhere we introduced\n\n$$\n\\Gamma^{0}_\\mu=\\sum_{\\mu'\\mu''}(\\Gamma^+_{\\mu\\mu'\\mu''} + \\Gamma^-_{\\mu\\mu'\\mu''} ),\n$$\n\nand\n\n$$\n\\Gamma^{1}_{\\mu\\mu'}=\n\\frac{\\omega_{\\mu'}}{\\omega_\\mu}\n\\sum_{\\mu''}(\\Gamma^+_{\\mu\\mu'\\mu''}\n-\\Gamma^+_{\\mu\\mu''\\mu'}\n-\\Gamma^-_{\\mu\\mu'\\mu''}\n-\\Gamma^-_{\\mu\\mu''\\mu'}\n).\n$$\n\nIn RTA, the off-diagonal terms are ignored, $\\Gamma^{1}_{\\mu\\mu'}=0$, and the conductivity is\n\n$$\n\\kappa_{\\alpha\\alpha'} =\\frac{1}{N_kV} \\sum_{\\mu}c_\\mu v_{\\mu\\alpha}{\\lambda_{\\mu\\alpha'}}\n=\\frac{1}{N_kV} \\sum_\\mu c_\\mu v_{\\mu\\alpha} {\\tau_\\mu}{v_{\\mu\\alpha'}},\n$$\n\nwhere $\\tau_\\mu=1/2\\Gamma_{\\mu}^0$ corresponds the phonons lifetime calculated using the Fermi Golden Rule.\n\nIt has been shown that, to correctly capture the physics of phonon transport, especially in highly conductive materials, the off diagonal terms of the scattering rates cannot be disregarded.\nMore generally, the mean free path is calculated inverting the scattering tensor\n\n$$\n\\lambda_{\\mu\\alpha} = \\sum_{\\mu'}(\\tilde \\Gamma_{\\mu\\mu' })^{-1}v_{\\mu'\\alpha}.\n$$\n\n$$\n\\kappa_{\\alpha\\alpha'} =\\frac{1}{N_kV} \\sum_{\\mu\\mu'} c_\\mu v_{\\mu\\alpha}(\\tilde \\Gamma_{\\mu\\mu' })^{-1}v_{\\mu'\\alpha'}.\n$$\n\nThis inversion operation is computationally expensive; however, when the off-diagonal elements of the scattering rate matrix are much smaller than the diagonal, we can rewrite the mean free path obtained from the BTE as a series:\n\n$$\n\\lambda_{\\mu\\alpha} = \\sum_{\\mu'}\\left(\\delta_{\\mu\\mu'} + \\frac{1}{\\Gamma^0_\\mu}\\Gamma^{1}_{\\mu\\mu'}\\right)^{-1}\\frac{1}{\\Gamma^0_{\\mu'}}v_{\\mu'\\alpha} =\n$$\n\n$$\n=\n\\sum_{\\mu'}\n\\left[\n\\sum^{\\infty}_{n=0}\\left(- \\frac{1}{\\Gamma^0_\\mu}\\Gamma^{1}_{\\mu\\mu'}\\right)^n\n\\right]\\frac{1}{\\Gamma^0_{\\mu'}}v_{\\mu'\\alpha} ,\n$$\n\nwhere in the last step we used the identity $\\sum_0 q^n = (1 - q)^{-1}$, true when $|q|=|\\Gamma^1/\\Gamma^0|<1$.\nThis equation can then be written in an iterative form\n\n$$\n\\lambda^0_{\\mu\\alpha} = \\frac{1}{\\Gamma^0_\\mu}v_\\mu\n\\qquad\n\\lambda^{n+1}_{\\mu\\alpha} = - \\frac{1}{\\Gamma^0_\\mu}\\sum_{\\mu'}\\Gamma^{1}_{\\mu\\mu'} \\lambda^{n}_{\\mu'\\alpha}.\n$$\n\nHence, the inversion in of the scattering tensor is obtained by a recursive expression. Once the mean free path is calculated, the conductivity is straightforwardly computed.\n\n\n### Quasi-Harmonic Green Kubo\n\nIn non-crystalline solids with no long range order, such as glasses, alloys, nano-crystalline, and partially disordered systems, the phonon picture is formally not well-defined. While vibrational modes are still the heat carriers, their mean-free-paths may be so short that the quasi-particle picture of heat carriers breaks down and the BTE is no longer applicable.\nIn glasses heat transport is dominated by a diffusive processes in which delocalized modes with similar frequency transfer energy from one to another.\nWhereas this mechanism is intrinsically distinct from the underlying hypothesis of the BTE approach, the two transport pictures have been recently reconciled in a unified theory, in which the thermal conductivity is written as:\n\n$$\n\\kappa_{\\alpha \\alpha'}=\\frac{1}{V} \\sum_{\\mu \\mu'} c_{\\mu \\mu'} v_{\\mu \\mu' \\alpha} v_{\\mu \\mu' \\alpha'} \\tau_{\\mu \\mu'}.\n$$\n\nThis expression is analogous to the RTA one, where modal heat capacity, phonon group velocity and lifetimes are replaced by\nthe generalized heat capacity,\n\n$$\nc_{\\mu \\mu'}=\\frac{\\hbar \\omega_{\\mu} \\omega_{\\mu'}}{T} \\frac{n_{\\mu}-n_{\\mu'}}{\\omega_{\\mu}-\\omega_{\\mu'}},\n$$\n\nthe generalized velocities,\n\n$$\nv_{\\mu\\mu'\\alpha}=\\frac{1}{2\\sqrt{\\omega_\\mu\\omega_{\\mu'}}}\n\\sum_{ii'\\beta'\\beta''}(x_{i\\alpha}-x_{i'\\alpha })D_{i\\beta i'\\beta'}\\eta_{\\mu i\\beta}\\eta_{\\mu'i'\\beta'},\n$$\n\nand the generalized lifetime $\\tau_{\\mu\\mu'}$.\nThe latter is expressed as a Lorentzian, which weighs diffusive processes between phonons with nearly-resonant frequencies:\n\n$$\n\\tau_{\\mu\\mu'} =\n \\frac{\\gamma_{\\mu}+\\gamma_{\\mu'}}{\\left(\\omega_{\\mu}-\\omega_{\\mu'}\\right)^{2}+\\left(\\gamma_{\\mu}+\\gamma_{\\mu'}\\right)^{2}}\n$$\n\nwhere $\\gamma_\\mu$ is the line width of mode $\\mu$ that can be computed using Fermi Golden rule.\nThese equations have been derived from the Green-Kubo theory of linear response applied to thermal conductivity, by taking a quasi-harmonic approximation of the heat current, from which this approach is named quasi-harmonic Green-Kubo (QHGK).\nIt has been proven that for crystalline materials QHGK is formally equivalent to the BTE in the relaxation time approximation, and that its classical limit reproduces correctly molecular dynamics simulations for amorphous silicon up to relatively high temperature (600 K).\nFinally, we provide a microscopic definition of the mode diffusivity,\n\n$$\nD_{\\mu} =\\frac{1}{N_k V} \\sum_{\\mu'}v_{\\mu\\mu'} \\tau_{\\mu\\mu'}v_{\\mu\\mu'},\n$$\n\nwhich conveniently provide a measure of the temperature-independent contribution of each mode to thermal transport.\n\n## Benchmarks applications\nThe workflow for ALD calculations is illustrated below\n\n<img src=\"_resources/timeline.png\" width=\"650\">\n\nHere, we present two example simulations of both a periodic and an amorphous structure.\n\n### *Ab initio* silicon diamond\n\nIn this example we calculate the second order IFC using Density Functional Perturbation Theory as implemented in the Quantum-Espresso package. The phonon lifetimes and thermal conductivity calculations are performed using a (19, 19, 19) q-point grid.\n\nThe kALDo minimal input file, looks like\n\n```python\n# IFCs object creation using ase.build.bulk\nfc = ForceConstants(atoms=bulk('Si', 'diamond', a=2.699),\n supercell=(5, 5, 5)))\n\n# input is the ASE input for QE\nfc.second.calculate(calculator=Espresso(**input))\nfc.third.calculate(calculator=Espresso(**input))\n\n# Phonons object creation\nphonons = Phonons(force_constants=fc,\n kpts=[19, 19, 19],\n temperature=300)\n\n\n# Conductivity calculations\ncond = Conductivity(phonons=phonons))\n\nprint('Thermal conductivity matrix, in (W/m/K):')\nprint(cond.conductivity(method='inverse').sum(axis=0))\n ```\n\nWe performed the simulation using the local density approximation for the exchange and correlation functional and a Bachelet-Hamann-Schluter norm-conserving pseudoptential. Kohn-Sham orbitals are represented on a plane-waves basis set with a cutoff of 20 Ry and (8, 8, 8) k-points mesh. The minimized lattice parameter is 5.398A. The third-order IFC is calculated using finite difference displacement on (5, 5, 5) replicas of the irreducible fcc-unit cell, including up to the 5th nearest neighbor.\nWe obtained the following thermal properties\n\n<img src=\"_resources/si-diamond-observables.png\" width=\"650\">\n\nThe silicon diamond modes analysis is shown above. Quantum (red) and classical (blue) results are compared. a) Normalized density of states, b) Normalized phase-space per mode $g$, c) lifetime per mode $\\tau$, d) mean free path $\\lambda$, and e) cumulative conductivity $\\kappa_{cum}$.\n\n### Amorphous silicon\n\nHere we study a-Si generated by LAMMPS molecular dynamics simulations of quenching from the melt a 4096 atom crystal silicon structure, with 1989 Tersoff interatomic potential and the QHGK method. The minimal input file looks like the following\n\n```python\n# IFCs object creation\nfc = ForceConstants.from_folder(atoms,\n folder='./input_data'))\n\n# Phonons object creation\nphon = Phonons(force_constants=fc,\n temperature=300)\n\n# Conductivity calculations\ncond = Conductivity(phonons=phonons))\n\nprint('Thermal conductivity matrix, in (W/m/K):')\nprint(cond.conductivity(method='qhgk').sum(axis=0))\n ```\n\nIn a simliar treatment to the silicon crystal, a full battery of modal analysis can be calculated with both quantum and classical statistics on the amorphous systems re- turning the phonon DoS as well as the associated lifetimes, generalized diffusivities, normalized phase space and cumulative conductivity\n\n<img src=\"_resources/amorphous.png\" width=\"650\">\n\nClassical and quantum properties for 4096 atom amorphous silicon system are shown above. a) density of states, b) lifetimes, c) diffusivities, and e) cumulative thermal conductivity. In spite of the increased quantum lifetimes, a decrease of 0.17W/m/K is seen in the quantum conductivity. The difference in conductivity is primarily a result of the overestimation of classical high frequency heat capacities.\n\n\n### References\n\n[1]: B. J. Alder, D. M. Gass, and T. E. Wainwright, “Studies in Molecular Dynamics. VIII. The Transport Coefficients for a Hard-Sphere Fluid,” Journal Chemical Physics 53, 3813–3826 (1970).\n\n[2]: A. J. C. Ladd, B. Moran, and W. G. Hoover, “Lattice thermal conductivity: A comparison of molecular dynamics and anharmonic lattice dynamics,” Physical Review B 34, 5058–5064 (1986).\n\n[3]: A. Marcolongo, P. Umari, and S. Baroni, “Microscopic theory and quantum simulation of atomic heat transport,” Nature Physics 12, 80–84 (2015).\n\n[4]: R. Peierls, “Zur kinetischen Theorie der Wärmeleitung in Kristallen,” Annalen der Physik 395, 1055–1101 (1929).\n\n[5]: J. M. Ziman, Electrons and Phonons: The Theory of Transport Phenomena in Solids, International series of monographs on physics (OUP Oxford, 2001).\n\n[6]: A. J. H. McGaughey, A. Jain, and H.-Y. Kim, “Phonon properties and thermal conductivity from first principles, lattice dynamics, and the Boltzmann transport equation,” Journal of Applied Physics 125, 011101–20 (2019).\n\n[7]: M. Omini and A. Sparavigna, “Beyond the isotropic-model approximation in the theory of thermal conductivity,” Physical Review B 53, 9064–9073 (1996).\n\n[8]: A. Ward, D. A. Broido, D. A. Stewart, and G. Deinzer, “Ab initio theory of the lattice thermal conductivity in diamond,” Physical Review B 80, 125203 (2009).\n\n[9]: L. Chaput, A. Togo, I. Tanaka, and G. Hug, “Phonon-phonon interactions in transition metals,” Physical Review B 84, 094302 (2011).\n\n[10]: W. Li, J. Carrete, N. A. Katcho, and N. Mingo, “ShengBTE: A solver of the Boltzmann transport equation for phonons,” Computer Physics Communications 185, 1747–1758 (2014).\n\n[11]: G. Fugallo, M. Lazzeri, L. Paulatto, and F. M. B, “Ab initio variational approach for evaluating lattice thermal conductivity,” Physical Review B 88, 045430 (2013).\n\n[12]: A. Cepellotti and N. Marzari, “Thermal Transport in Crystals as a Kinetic Theory of Relaxons,” Physical Review X 6, 041013–14 (2016).\n\n[13]: A. Chernatynskiy and S. R. Phillpot, “Phonon Transport Simulator (PhonTS),” Computer Physics Communications 192, 196–204 (2015).\n\n[14]: A. Togo, L. Chaput, and I. Tanaka, “Distributions of phonon lifetimes in brillouin zones,” Physical Review B 91, 094306 (2015).\n\n[15]: J. Carrete, B. Vermeersch, A. Katre, A. van Roekeghem, T. Wang, G. K. H. Madsen, and N. Mingo, “almaBTE : A solver of the space–time dependent Boltzmann transport equation for phonons in structured materials,” Computer Physics Communications 220, 351–362 (2017).\n\n[16]: T. Tadano, Y. Gohda, and S. Tsuneyuki, “Anharmonic force constants extracted from first-principles molecular dynamics: applications to heat transfer simulations,” Journal of Physics: Condensed Matter 26, 225402–13 (2014).\n\n[17]: D. A. Broido, M. Malorny, G. Birner, N. Mingo, and D. A. Stewart, “Intrinsic lattice thermal conductivity of semiconductors from first principles,” Applied Physics Letters 91, 231922 (2007).\n\n[18]: L. Lindsay, A. Katre, A. Cepellotti, and N. Mingo, “Perspective on ab initio phonon thermal transport,” Journal Applied Physics 126, 050902–21 (2019).\n\n[19]: L. Lindsay, D. A. Broido, and T. L. Reinecke, “First-Principles Determination of Ultrahigh Thermal Conductivity of Boron Arsenide: A Competitor for Diamond?” Physical Review Letters 111, 025901–5 (2013).\n\n[20]: G. Fugallo, A. Cepellotti, L. Paulatto, M. Lazzeri, N. Marzari, and F. Mauri, “Thermal Conductivity of Graphene and Graphite: Collective Ex- citations and Mean Free Paths,” Nano Letters 14, 6109–6114 (2014).\n\n[21]: A. Cepellotti, G. Fugallo, L. Paulatto, M. Lazzeri, F. Mauri, and N. Marzari, “Phonon hydrodynamics in two-dimensional materials,” Na- ture Communications 6, 6400 (2015).\n\n[22]: A. Jain and A. J. H. Mcgaughey, “Strongly anisotropic in-plane thermal transport in single-layer black phosphorene,” Scientific Reports 5, 8501–5 (2015).\n\n[23]: M. Zeraati, S. M. Vaez Allaei, I. Abdolhosseini Sarsari, M. Pourfath, and D. Donadio, “Highly anisotropic thermal conductivity of arsenene: An ab initio study,” Physical Review B 93, 085424 (2016).\n\n[24]: B. Ouyang, S. Chen, Y. Jing, T. Wei, S. Xiong, and D. Donadio, “Enhanced thermoelectric performance of two dimensional MS2 (M=Mo,W) through phase engineering,” Journal of Materiomics 4, 329–337 (2018).\n\n[25]: S. Chen, A. Sood, E. Pop, K. E. Goodson, and D. Donadio, “Strongly tunable anisotropic thermal transport in MoS 2by strain and lithium inter- calation: first-principles calculations,” 2D Materials 6, 025033–10 (2019). 26A. Sood, F. Xiong, S. Chen, R. Cheaito, F. Lian, M. Asheghi, Y. Cui, D. Donadio, K. E. Goodson, and E. Pop, “Quasi-Ballistic Thermal Transport Across MoS 2Thin Films,” Nano Letters 19, 2434–2442 (2019).\n\n[27]: C. Ott, F. Reiter, M. Baumgartner, M. Pielmeier, A. Vogel, P. Walke, S. Burger, M. Ehrenreich, G. Kieslich, D. Daisenberger, J. Armstrong, U. K. Thakur, P. Kumar, S. Chen, D. Donadio, L. S. Walter, R. T. Weitz, K. Shankar, and T. Nilges, “Flexible and Ultrasoft Inorganic 1D Semiconductor and Heterostructure Systems Based on SnIP,” Advanced Functional Materials 271, 1900233 (2019).\n\n[28]: P. B. Allen and J. L. Feldman, “Thermal conductivity of disordered harmonic solids,” Physical Review B 48, 12581–12588 (1993).\n\n[29]: L. Isaeva, G. Barbalinardo, D. Donadio, and S. Baroni, “Modeling heat transport in crystals and glasses from a unified lattice-dynamical approach,”Nature Communications 10, 3853 (2019).\n\n[30]: M. Simoncelli, N. Marzari, and F. Mauri, “Unified theory of thermal transport in crystals and glasses,” Nature Physics 15, 809–813 (2019).\n\n[31]: F. Eriksson, E. Fransson, and P. Erhart, “The Hiphive Package for the Extraction of High-Order Force Constants by Machine Learning,” Advanced Theory and Simulations 2, 1800184–11 (2019).\n\n[32]: S. Baroni, S. de Gironcoli, A. Dal Corso, and P. Giannozzi, “Phonons and related crystal properties from density-functional perturbation theory,” Rev Mod Phys 73, 515–562 (2001).\n\n[33]: L. Paulatto, F. Mauri, and M. Lazzeri, “Anharmonic properties from a generalized third-order ab initioapproach: Theory and applications to graphite and graphene,” Phys. Rev. B 87, 214303–18 (2013).\n\n[34]: G. P. Srivastava, “The Physics of Phonons, ,” Adam Hilger, Bristol 1990. (1990).\n\n[35]: M. S. Green, “Markoff random processes and the statistical mechanics of time-dependent phenomena.” Journal Chemical Physics 20, 1281–1295 (1952).\n\n[36]: M. Green, “Markoff random processes and the statistical mechanics of time-dependent phenomena. ii. irreversible processes in fluids,” Journal Chemical Physics 22, 398–413 (1954).\n\n[37]: R. Kubo, “Statistical-Mechanical Theory of Irreversible Processes. I. General Theory and Simple Applications to Magnetic and Conduction Prob lems,” Journal of the Physical Society of Japan 12, 570–586 (1957).\n\n[38]: R. Kubo, M. Yokota, and S. Nakajima, “Statistical-Mechanical Theory of Irreversible Processes. II. Response to Thermal Disturbance,” Journal of the Physical Society of Japan 12, 1203–1211 (1957).\n\n[39]: Y. He, I. Savic ́, D. Donadio, and G. Galli, “Lattice thermal conductivity of semiconducting bulk materials: atomistic simulations,” Physical Chemistry Chemical Physics 14, 16209–14 (2012).\n\n[40]: A. H. Larsen, J. J. Mortensen, J. Blomqvist, I. E. Castelli, R. Christensen, M. Dułak, J. Friis, M. N. Groves, B. Hammer, C. Hargus, E. D. Hermes, P. C. Jennings, P. B. Jensen, J. Kermode, J. R. Kitchin, E. L. Kolsbjerg, J. Kubal, K. Kaasbjerg, S. Lysgaard, J. B. Maronsson, T. Maxson, T. Olsen, L. Pastewka, A. Peterson, C. Rostgaard, J. Schiøtz, O. Schütt, M. Strange, K. S. Thygesen, T. Vegge, L. Vilhelmsen, M. Walter, Z. Zeng, and K. W. Jacobsen, “The atomic simulation environment—a python library for working with atoms,” Journal of Physics: Condensed Matter 29, 273002 (2017).\n\n[41]: B. Aradi, B. Hourahine, and T. Frauenheim, “Dftb+, a sparse matrix-based implementation of the dftb method,” J Phys Chem A 111, 5678–5684 (2007).\n\n[42]: D. G A Smith and J. Gray, “opt_einsum - A Python package for optimizing contraction order for einsum-like expressions,” Journal of Open Source Software 3, 753–3 (2018).\n\n[43]: P. Giannozzi, O. Andreussi, T. Brumme, O. Bunau, M. B. Nardelli, M. Calandra, R. Car, C. Cavazzoni, D. Ceresoli, M. Cococcioni, N. Colonna, I. Carnimeo, A. D. Corso, S. de Gironcoli, P. Delugas, R. A. D. Jr, A. Ferretti, A. Floris, G. Fratesi, G. Fugallo, R. Gebauer, U. Gerstmann, F. Giustino, T. Gorni, J. Jia, M. Kawamura, H.-Y. Ko, A. Kokalj, E. Küçükbenli, M. Lazzeri, M. Marsili, N. Marzari, F. Mauri, N. L. Nguyen, H.-V. Nguyen, A. O. de-la Roza, L. Paulatto, S. Poncé, D. Rocca, R. Sabatini, B. Santra, M. Schlipf, A. P. Seitsonen, A. Smogunov, I. Timrov, T. Thonhauser, P. Umari, N. Vast, X. Wu, and S. Baroni, “Advanced capabilities for materials modelling with quantum espresso,” Journal of Physics: Condensed Matter 29, 465901 (2017).\n\n[44]: G. B. Bachelet, D. R. Hamann, and M. Schluter, “Pseudopotentials That Work - From H to Pu,” Physical Review B 26, 4199–4228 (1982).\n\n[45]: R. Kremer, K. Graf, M. Cardona, G. Devyatykh, A. Gusev, A. Gibin, A. In- yushkin, A. Taldenkov, and H. Pohl, “Thermal conductivity of isotopically enriched Si-28: revisited,” Solid State Communications 131, 499–503 (2004).\n\n[46]: J. Tersoff, “Modeling solid-state chemistry: Interatomic potentials for multicomponent systems,” Physical Review B 39, 5566–5568 (1989).\n\n[41]: A. Krylov, T. L. Windus, T. Barnes, E. Marin-Rimoldi, J. A. Nash, B. Pritchard, D. G. Smith, D. Altarawy, P. Saxe, C. Clementi, T. D. Crawford, R. J. Harrison, S. Jha, V. S. Pande, and T. Head-Gordon, “Perspective: Computational chemistry software and its advancement as illustrated through three grand challenge cases for molecular science,” Journal of Chemical Physics 149, 180901 (2018).\n\n[42]: N.Wilkins-Diehrand, T.D.Crawford,“NSF’s Inaugural Software Institutes: The Science Gateways Community Institute and the Molecular Sciences Software Institute.” Computing in Science and Engineering 20 (2018).\n\n[43]: W. Li, N. Mingo, L. Lindsay, D. A. Broido, D. A. Stewart, and N. A. Katcho, “Thermal conductivity of diamond nanowires from first princi- ples,” Physical Review B 85, 195436 (2012).",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
d03dcee14be99d4c73406c2b1fa614b9871d0171 | 312,526 | ipynb | Jupyter Notebook | analysis/apa/apa_max_isoform_genesis_doubledope_kl_vae_strong_margin_pos_2_lower_fitness.ipynb | johli/genesis | 5424c1888d4330e505ad87412e7f1cc5dd828888 | [
"MIT"
] | 12 | 2020-02-02T14:29:15.000Z | 2021-09-12T08:05:43.000Z | analysis/apa/apa_max_isoform_genesis_doubledope_kl_vae_strong_margin_pos_2_lower_fitness.ipynb | johli/genesis | 5424c1888d4330e505ad87412e7f1cc5dd828888 | [
"MIT"
] | 1 | 2022-01-04T08:04:00.000Z | 2022-01-10T08:49:04.000Z | analysis/apa/apa_max_isoform_genesis_doubledope_kl_vae_strong_margin_pos_2_lower_fitness.ipynb | johli/genesis | 5424c1888d4330e505ad87412e7f1cc5dd828888 | [
"MIT"
] | 3 | 2020-03-10T22:24:05.000Z | 2021-05-05T13:23:01.000Z | 351.152809 | 25,724 | 0.920602 | [
[
[
"import keras\nfrom keras.models import Sequential, Model, load_model\n\nfrom keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda\nfrom keras.layers import Conv2D, MaxPooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, BatchNormalization, LocallyConnected2D, Permute\nfrom keras.layers import Concatenate, Reshape, Softmax, Conv2DTranspose, Embedding, Multiply\nfrom keras.callbacks import ModelCheckpoint, EarlyStopping, Callback\nfrom keras import regularizers\nfrom keras import backend as K\nimport keras.losses\n\nimport tensorflow as tf\n#tf.compat.v1.enable_eager_execution()\n\nfrom tensorflow.python.framework import ops\n\nimport isolearn.keras as iso\n\nimport numpy as np\n\nimport tensorflow as tf\nimport logging\nlogging.getLogger('tensorflow').setLevel(logging.ERROR)\n\nimport pandas as pd\n\nimport os\nimport pickle\nimport numpy as np\n\nimport random\n\nimport scipy.sparse as sp\nimport scipy.io as spio\n\nimport matplotlib.pyplot as plt\n\nimport isolearn.io as isoio\nimport isolearn.keras as isol\n\nfrom genesis.visualization import *\nfrom genesis.generator import *\nfrom genesis.predictor import *\nfrom genesis.optimizer import *\n\nfrom definitions.generator.aparent_deconv_conv_generator_concat_trainmode import load_generator_network\nfrom definitions.predictor.aparent import load_saved_predictor\n\nimport sklearn\nfrom sklearn.decomposition import PCA\nfrom sklearn.manifold import TSNE\n\nfrom scipy.stats import pearsonr\n\nimport seaborn as sns\n\nfrom matplotlib import colors\n\nfrom scipy.stats import norm\n\nfrom genesis.vae import *\n\ndef set_seed(seed_value) :\n # 1. Set the `PYTHONHASHSEED` environment variable at a fixed value\n os.environ['PYTHONHASHSEED']=str(seed_value)\n\n # 2. Set the `python` built-in pseudo-random generator at a fixed value\n random.seed(seed_value)\n\n # 3. Set the `numpy` pseudo-random generator at a fixed value\n np.random.seed(seed_value)\n\n # 4. Set the `tensorflow` pseudo-random generator at a fixed value\n tf.set_random_seed(seed_value)\n\n # 5. Configure a new global `tensorflow` session\n session_conf = tf.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)\n sess = tf.Session(graph=tf.get_default_graph(), config=session_conf)\n K.set_session(sess)\n\ndef load_data(data_name, valid_set_size=0.05, test_set_size=0.05, batch_size=32) :\n \n #Load cached dataframe\n cached_dict = pickle.load(open(data_name, 'rb'))\n plasmid_df = cached_dict['plasmid_df']\n plasmid_cuts = cached_dict['plasmid_cuts']\n\n #print(\"len(plasmid_df) = \" + str(len(plasmid_df)) + \" (loaded)\")\n\n #Generate training and test set indexes\n plasmid_index = np.arange(len(plasmid_df), dtype=np.int)\n\n plasmid_train_index = plasmid_index[:-int(len(plasmid_df) * (valid_set_size + test_set_size))]\n plasmid_valid_index = plasmid_index[plasmid_train_index.shape[0]:-int(len(plasmid_df) * test_set_size)]\n plasmid_test_index = plasmid_index[plasmid_train_index.shape[0] + plasmid_valid_index.shape[0]:]\n\n #print('Training set size = ' + str(plasmid_train_index.shape[0]))\n #print('Validation set size = ' + str(plasmid_valid_index.shape[0]))\n #print('Test set size = ' + str(plasmid_test_index.shape[0]))\n\n\n data_gens = {\n gen_id : iso.DataGenerator(\n idx,\n {'df' : plasmid_df},\n batch_size=batch_size,\n inputs = [\n {\n 'id' : 'seq',\n 'source_type' : 'dataframe',\n 'source' : 'df',\n 'extractor' : lambda row, index: row['padded_seq'][180 + 40: 180 + 40 + 81] + \"G\" * (128-81),\n 'encoder' : iso.OneHotEncoder(seq_length=128),\n 'dim' : (1, 128, 4),\n 'sparsify' : False\n }\n ],\n outputs = [\n {\n 'id' : 'dummy_output',\n 'source_type' : 'zeros',\n 'dim' : (1,),\n 'sparsify' : False\n }\n ],\n randomizers = [],\n shuffle = True if gen_id == 'train' else False\n ) for gen_id, idx in [('all', plasmid_index), ('train', plasmid_train_index), ('valid', plasmid_valid_index), ('test', plasmid_test_index)]\n }\n\n x_train = np.concatenate([data_gens['train'][i][0][0] for i in range(len(data_gens['train']))], axis=0)\n x_test = np.concatenate([data_gens['test'][i][0][0] for i in range(len(data_gens['test']))], axis=0)\n \n return x_train, x_test\n",
"Using TensorFlow backend.\n"
],
[
"#Specfiy problem-specific parameters\n\nexperiment_suffix = '_strong_vae_very_high_kl_epoch_35_margin_pos_2_lower_fitness'\n\nvae_model_prefix = \"vae/saved_models/vae_apa_max_isoform_doubledope_strong_cano_pas_len_128_50_epochs_very_high_kl\"\nvae_model_suffix = \"_epoch_35\"#\"\"#\n\n#VAE model path\nsaved_vae_encoder_model_path = vae_model_prefix + \"_encoder\" + vae_model_suffix + \".h5\"\nsaved_vae_decoder_model_path = vae_model_prefix + \"_decoder\" + vae_model_suffix + \".h5\"\n\n#Padding for the VAE\nvae_upstream_padding = ''\nvae_downstream_padding = 'G' * 47\n\n#VAE sequence template\nvae_sequence_template = 'ATCCANNNNNNNNNNNNNNNNNNNNNNNNNAATAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCAGCC' + 'G' * (128 - 81)\n\n#VAE latent dim\nvae_latent_dim = 100\n\n#Oracle predictor model path\nsaved_predictor_model_path = '../../../aparent/saved_models/aparent_plasmid_iso_cut_distalpas_all_libs_no_sampleweights_sgd.h5'\n\n#Subtring indices for VAE\nvae_pwm_start = 40\nvae_pwm_end = 121\n\n#VAE parameter collection\nvae_params = [\n saved_vae_encoder_model_path,\n saved_vae_decoder_model_path,\n vae_upstream_padding,\n vae_downstream_padding,\n vae_latent_dim,\n vae_pwm_start,\n vae_pwm_end\n]\n\n#Load data set\nvae_data_path = \"vae/apa_doubledope_cached_set_strong_short_cano_pas.pickle\"\n\n_, x_test = load_data(vae_data_path, valid_set_size=0.005, test_set_size=0.095)\n",
"_____no_output_____"
],
[
"#Evaluate ELBO distribution on test set\n\n#Load VAE models\nvae_encoder_model = load_model(saved_vae_encoder_model_path, custom_objects={'st_sampled_softmax':st_sampled_softmax, 'st_hardmax_softmax':st_hardmax_softmax, 'min_pred':min_pred, 'InstanceNormalization':InstanceNormalization})\nvae_decoder_model = load_model(saved_vae_decoder_model_path, custom_objects={'st_sampled_softmax':st_sampled_softmax, 'st_hardmax_softmax':st_hardmax_softmax, 'min_pred':min_pred, 'InstanceNormalization':InstanceNormalization})\n\n#Compute multi-sample ELBO on test set\nlog_mean_p_vae_test, mean_log_p_vae_test, log_p_vae_test = evaluate_elbo(vae_encoder_model, vae_decoder_model, x_test, n_samples=128)\n\nprint(\"mean log(likelihood) = \" + str(mean_log_p_vae_test))\n\n#Log Likelihood Plot\nplot_min_val = None\nplot_max_val = None\n\nf = plt.figure(figsize=(6, 4))\n\nlog_p_vae_test_hist, log_p_vae_test_edges = np.histogram(log_mean_p_vae_test, bins=50, density=True)\nbin_width_test = log_p_vae_test_edges[1] - log_p_vae_test_edges[0]\n\nplt.bar(log_p_vae_test_edges[1:] - bin_width_test/2., log_p_vae_test_hist, width=bin_width_test, linewidth=2, edgecolor='black', color='orange')\n\nplt.xticks(fontsize=14)\nplt.yticks(fontsize=14)\n\nif plot_min_val is not None and plot_max_val is not None :\n plt.xlim(plot_min_val, plot_max_val)\n\nplt.xlabel(\"VAE Log Likelihood\", fontsize=14)\nplt.ylabel(\"Data Density\", fontsize=14)\n\nplt.axvline(x=mean_log_p_vae_test, linewidth=2, color='red', linestyle=\"--\")\n\nplt.tight_layout()\nplt.show()\n",
"mean log(likelihood) = -38.70989728805291\n"
],
[
"#Evaluate ELBO distribution on test set (training-level no. of samples)\n\n#Load VAE models\nvae_encoder_model = load_model(saved_vae_encoder_model_path, custom_objects={'st_sampled_softmax':st_sampled_softmax, 'st_hardmax_softmax':st_hardmax_softmax, 'min_pred':min_pred, 'InstanceNormalization':InstanceNormalization})\nvae_decoder_model = load_model(saved_vae_decoder_model_path, custom_objects={'st_sampled_softmax':st_sampled_softmax, 'st_hardmax_softmax':st_hardmax_softmax, 'min_pred':min_pred, 'InstanceNormalization':InstanceNormalization})\n\n#Compute multi-sample ELBO on test set\nlog_mean_p_vae_test, mean_log_p_vae_test, log_p_vae_test = evaluate_elbo(vae_encoder_model, vae_decoder_model, x_test, n_samples=32)\n\nprint(\"mean log(likelihood) = \" + str(mean_log_p_vae_test))\n\n#Log Likelihood Plot\nplot_min_val = None\nplot_max_val = None\n\nf = plt.figure(figsize=(6, 4))\n\nlog_p_vae_test_hist, log_p_vae_test_edges = np.histogram(log_mean_p_vae_test, bins=50, density=True)\nbin_width_test = log_p_vae_test_edges[1] - log_p_vae_test_edges[0]\n\nplt.bar(log_p_vae_test_edges[1:] - bin_width_test/2., log_p_vae_test_hist, width=bin_width_test, linewidth=2, edgecolor='black', color='orange')\n\nplt.xticks(fontsize=14)\nplt.yticks(fontsize=14)\n\nif plot_min_val is not None and plot_max_val is not None :\n plt.xlim(plot_min_val, plot_max_val)\n\nplt.xlabel(\"VAE Log Likelihood\", fontsize=14)\nplt.ylabel(\"Data Density\", fontsize=14)\n\nplt.axvline(x=mean_log_p_vae_test, linewidth=2, color='red', linestyle=\"--\")\n\nplt.tight_layout()\nplt.show()\n",
"mean log(likelihood) = -38.80713871551151\n"
],
[
"\n#Define target isoform loss function\ndef get_isoform_loss(target_isos, fitness_weight=2., batch_size=32, n_samples=1, n_z_samples=1, mini_batch_size=1, seq_length=205, vae_loss_mode='bound', vae_divergence_weight=1., ref_vae_log_p=-10, vae_log_p_margin=1, decoded_pwm_epsilon=10**-6, pwm_start=0, pwm_end=70, pwm_target_bits=1.8, vae_pwm_start=0, entropy_weight=0.0, entropy_loss_mode='margin', similarity_weight=0.0, similarity_margin=0.5) :\n \n target_iso = np.zeros((len(target_isos), 1))\n for i, t_iso in enumerate(target_isos) :\n target_iso[i, 0] = t_iso\n \n masked_entropy_mse = get_target_entropy_sme_masked(pwm_start=pwm_start, pwm_end=pwm_end, target_bits=pwm_target_bits)\n if entropy_loss_mode == 'margin' :\n masked_entropy_mse = get_margin_entropy_ame_masked(pwm_start=pwm_start, pwm_end=pwm_end, min_bits=pwm_target_bits)\n \n pwm_sample_entropy_func = get_pwm_margin_sample_entropy_masked(pwm_start=pwm_start, pwm_end=pwm_end, margin=similarity_margin, shift_1_nt=True)\n \n def loss_func(loss_tensors) :\n _, _, _, sequence_class, pwm_logits_1, pwm_logits_2, pwm_1, pwm_2, sampled_pwm_1, sampled_pwm_2, mask, sampled_mask, iso_pred, cut_pred, iso_score_pred, cut_score_pred, vae_pwm_1, vae_sampled_pwm_1, z_mean_1, z_log_var_1, z_1, decoded_pwm_1 = loss_tensors\n \n #Create target isoform with sample axis\n iso_targets = K.constant(target_iso)\n iso_true = K.gather(iso_targets, sequence_class[:, 0])\n iso_true = K.tile(K.expand_dims(iso_true, axis=-1), (1, K.shape(sampled_pwm_1)[1], 1))\n\n #Re-create iso_pred from cut_pred\n #iso_pred = K.expand_dims(K.sum(cut_pred[..., 76:76+35], axis=-1), axis=-1)\n \n #Specify costs\n iso_loss = fitness_weight * K.mean(symmetric_sigmoid_kl_divergence(iso_true, iso_pred), axis=1)\n \n #Construct VAE sequence inputs\n decoded_pwm_1 = K.clip(decoded_pwm_1, decoded_pwm_epsilon, 1. - decoded_pwm_epsilon)\n \n log_p_x_given_z_1 = K.sum(K.sum(vae_sampled_pwm_1[:, :, :, pwm_start-vae_pwm_start:pwm_end-vae_pwm_start, ...] * K.log(K.stop_gradient(decoded_pwm_1[:, :, :, pwm_start-vae_pwm_start:pwm_end-vae_pwm_start, ...])) / K.log(K.constant(10.)), axis=(-1, -2)), axis=-1)\n \n log_p_std_normal_1 = K.sum(normal_log_prob(z_1, 0., 1.) / K.log(K.constant(10.)), axis=-1)\n log_p_importance_1 = K.sum(normal_log_prob(z_1, z_mean_1, K.sqrt(K.exp(z_log_var_1))) / K.log(K.constant(10.)), axis=-1)\n \n log_p_vae_1 = log_p_x_given_z_1 + log_p_std_normal_1 - log_p_importance_1\n log_p_vae_div_n_1 = log_p_vae_1 - K.log(K.constant(n_z_samples, dtype='float32')) / K.log(K.constant(10.))\n\n #Calculate mean ELBO across samples (log-sum-exp trick)\n max_log_p_vae_1 = K.max(log_p_vae_div_n_1, axis=-1)\n\n log_mean_p_vae_1 = max_log_p_vae_1 + K.log(K.sum(10**(log_p_vae_div_n_1 - K.expand_dims(max_log_p_vae_1, axis=-1)), axis=-1)) / K.log(K.constant(10.))\n \n #Specify VAE divergence loss function\n vae_divergence_loss = 0.\n \n if vae_loss_mode == 'bound' :\n vae_divergence_loss = vae_divergence_weight * K.mean(K.switch(log_mean_p_vae_1 < ref_vae_log_p - vae_log_p_margin, -log_mean_p_vae_1 + (ref_vae_log_p - vae_log_p_margin), K.zeros_like(log_mean_p_vae_1)), axis=1)\n \n elif vae_loss_mode == 'penalty' :\n vae_divergence_loss = vae_divergence_weight * K.mean(-log_mean_p_vae_1, axis=1)\n \n elif vae_loss_mode == 'target' :\n vae_divergence_loss = vae_divergence_weight * K.mean((log_mean_p_vae_1 - (ref_vae_log_p - vae_log_p_margin))**2, axis=1)\n \n elif 'mini_batch_' in vae_loss_mode :\n mini_batch_log_mean_p_vae_1 = K.permute_dimensions(K.reshape(log_mean_p_vae_1, (int(batch_size / mini_batch_size), mini_batch_size, n_samples)), (0, 2, 1))\n mini_batch_mean_log_p_vae_1 = K.mean(mini_batch_log_mean_p_vae_1, axis=-1)\n tiled_mini_batch_mean_log_p_vae_1 = K.tile(mini_batch_mean_log_p_vae_1, (mini_batch_size, 1))\n \n if vae_loss_mode == 'mini_batch_bound' :\n vae_divergence_loss = vae_divergence_weight * K.mean(K.switch(tiled_mini_batch_mean_log_p_vae_1 < ref_vae_log_p - vae_log_p_margin, -tiled_mini_batch_mean_log_p_vae_1 + (ref_vae_log_p - vae_log_p_margin), K.zeros_like(tiled_mini_batch_mean_log_p_vae_1)), axis=1)\n elif vae_loss_mode == 'mini_batch_target' :\n vae_divergence_loss = vae_divergence_weight * K.mean((tiled_mini_batch_mean_log_p_vae_1 - (ref_vae_log_p - vae_log_p_margin))**2, axis=1)\n \n entropy_loss = entropy_weight * masked_entropy_mse(pwm_1, mask)\n entropy_loss += similarity_weight * K.mean(pwm_sample_entropy_func(sampled_pwm_1, sampled_pwm_2, sampled_mask), axis=1)\n \n #Compute total loss\n total_loss = iso_loss + entropy_loss + vae_divergence_loss\n \n return total_loss\n \n return loss_func\n\nclass EpochVariableCallback(Callback):\n def __init__(self, my_variable, my_func):\n self.my_variable = my_variable \n self.my_func = my_func\n def on_epoch_end(self, epoch, logs={}):\n K.set_value(self.my_variable, self.my_func(K.get_value(self.my_variable), epoch))\n\n#Function for running GENESIS\ndef run_genesis(sequence_templates, loss_func, library_contexts, model_path, batch_size=32, n_samples=1, n_z_samples=1, vae_params=None, n_epochs=10, steps_per_epoch=100) :\n \n #Build Generator Network\n _, generator = build_generator(batch_size, len(sequence_templates[0]), load_generator_network, n_classes=len(sequence_templates), n_samples=n_samples, sequence_templates=sequence_templates, batch_normalize_pwm=False)\n\n #Build Predictor Network and hook it on the generator PWM output tensor\n _, sample_predictor = build_predictor(generator, load_saved_predictor(model_path, library_contexts=library_contexts), batch_size, n_samples=n_samples, eval_mode='sample')\n\n #Build VAE model\n vae_tensors = []\n if vae_params is not None :\n encoder_model_path, decoder_model_path, vae_upstream_padding, vae_downstream_padding, vae_latent_dim, vae_pwm_start, vae_pwm_end = vae_params\n vae_tensors = build_vae(generator, encoder_model_path, decoder_model_path, batch_size=batch_size, seq_length=len(sequence_templates[0]), n_samples=n_samples, n_z_samples=n_z_samples, vae_latent_dim=vae_latent_dim, vae_upstream_padding=vae_upstream_padding, vae_downstream_padding=vae_downstream_padding, vae_pwm_start=vae_pwm_start, vae_pwm_end=vae_pwm_end)\n\n #Build Loss Model (In: Generator seed, Out: Loss function)\n _, loss_model = build_loss_model(sample_predictor, loss_func, extra_loss_tensors=vae_tensors)\n \n #Specify Optimizer to use\n opt = keras.optimizers.Adam(lr=0.001, beta_1=0.9, beta_2=0.999)\n\n #Compile Loss Model (Minimize self)\n loss_model.compile(loss=lambda true, pred: pred, optimizer=opt)\n\n #Fit Loss Model\n train_history = loss_model.fit(\n [], np.ones((1, 1)),\n epochs=n_epochs,\n steps_per_epoch=steps_per_epoch\n )\n\n return generator, sample_predictor, train_history\n",
"_____no_output_____"
],
[
"#Maximize isoform proportion\n\nsequence_templates = [\n 'CTTCCGATCTCTCGCTCTTTCTATGGCATTCATTACTCGCATCCANNNNNNNNNNNNNNNNNNNNNNNNNAATAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCAGCCAATTAAGCCTGTCGTCGTGGGTGTCGAAAATGAAATAAAACAAGTCAATTGCGTAGTTTATTCAGACGTACCCCGTGGACCTAC'\n]\n\nlibrary_contexts = [\n 'doubledope'\n]\n\nmargin_similarities = [\n 0.5\n]\n",
"_____no_output_____"
],
[
"#Generate new random seed\nprint(np.random.randint(low=0, high=1000000))",
"821745\n"
],
[
"#Train APA Cleavage GENESIS Network\n\nprint(\"Training GENESIS\")\n\n#Number of PWMs to generate per objective\nbatch_size = 64\nmini_batch_size = 8\n#Number of One-hot sequences to sample from the PWM at each grad step\nn_samples = 1\n#Number of VAE latent vector samples at each grad step\nn_z_samples = 32#128#32\n#Number of epochs per objective to optimize\nn_epochs = 50#10#5#25\n#Number of steps (grad updates) per epoch\nsteps_per_epoch = 50\n\nseed = 104590\n\nfor class_i in range(len(sequence_templates)) :\n lib_name = library_contexts[class_i].split(\"_\")[0]\n print(\"Library context = \" + str(lib_name))\n \n K.clear_session()\n \n set_seed(seed)\n \n loss = get_isoform_loss(\n [1.0],\n fitness_weight=0.1,#0.5,\n batch_size=batch_size,\n n_samples=n_samples,\n n_z_samples=n_z_samples,\n mini_batch_size=mini_batch_size,\n seq_length=len(sequence_templates[0]),\n vae_loss_mode='mini_batch_bound',#'target',\n vae_divergence_weight=40.0 * 1./71.,#5.0 * 1./71.,#0.5 * 1./71.,\n ref_vae_log_p=-38.807,\n vae_log_p_margin=2.0,\n #decoded_pwm_epsilon=0.05,\n pwm_start=vae_pwm_start + 5,\n pwm_end=vae_pwm_start + 5 + 71,\n vae_pwm_start=vae_pwm_start,\n pwm_target_bits=1.8,\n entropy_weight=0.5,#0.01,\n entropy_loss_mode='margin',\n similarity_weight=5.0,#0.5,#5.0,\n similarity_margin=margin_similarities[class_i]\n )\n\n genesis_generator, genesis_predictor, train_history = run_genesis([sequence_templates[class_i]], loss, [library_contexts[class_i]], saved_predictor_model_path, batch_size, n_samples, n_z_samples, vae_params, n_epochs, steps_per_epoch)\n\n genesis_generator.get_layer('lambda_rand_sequence_class').function = lambda inp: inp\n genesis_generator.get_layer('lambda_rand_input_1').function = lambda inp: inp\n genesis_generator.get_layer('lambda_rand_input_2').function = lambda inp: inp\n\n genesis_predictor.get_layer('lambda_rand_sequence_class').function = lambda inp: inp\n genesis_predictor.get_layer('lambda_rand_input_1').function = lambda inp: inp\n genesis_predictor.get_layer('lambda_rand_input_2').function = lambda inp: inp\n\n # Save model and weights\n save_dir = 'saved_models'\n\n if not os.path.isdir(save_dir):\n os.makedirs(save_dir)\n \n \n model_name = 'genesis_apa_max_isoform_' + str(lib_name) + experiment_suffix + '_vae_kl_generator.h5'\n model_path = os.path.join(save_dir, model_name)\n genesis_generator.save(model_path)\n print('Saved trained model at %s ' % model_path)\n\n model_name = 'genesis_apa_max_isoform_' + str(lib_name) + experiment_suffix + '_vae_kl_predictor.h5'\n model_path = os.path.join(save_dir, model_name)\n genesis_predictor.save(model_path)\n print('Saved trained model at %s ' % model_path)\n",
"Training GENESIS\nLibrary context = doubledope\nFalse\n"
],
[
"#Load GENESIS models and predict sample sequences\n\nlib_name = library_contexts[0].split(\"_\")[0]\nbatch_size = 64\n\nmodel_names = [\n 'genesis_apa_max_isoform_' + str(lib_name) + experiment_suffix + '_vae_kl',\n]\n\nsequence_templates = [\n 'CTTCCGATCTCTCGCTCTTTCTATGGCATTCATTACTCGCATCCANNNNNNNNNNNNNNNNNNNNNNNNNAATAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCAGCCAATTAAGCCTGTCGTCGTGGGTGTCGAAAATGAAATAAAACAAGTCAATTGCGTAGTTTATTCAGACGTACCCCGTGGACCTAC'\n]\n\nfor class_i in range(len(sequence_templates)-1, 0-1, -1) :\n save_dir = os.path.join(os.getcwd(), 'saved_models')\n model_name = model_names[class_i] + '_predictor.h5'\n model_path = os.path.join(save_dir, model_name)\n\n predictor = load_model(model_path, custom_objects={'st_sampled_softmax': st_sampled_softmax, 'st_hardmax_softmax': st_hardmax_softmax})\n \n n = batch_size\n\n sequence_class = np.array([0] * n).reshape(-1, 1) #np.random.uniform(-6, 6, (n, 1)) #\n\n noise_1 = np.random.uniform(-1, 1, (n, 100))\n noise_2 = np.random.uniform(-1, 1, (n, 100))\n\n pred_outputs = predictor.predict([sequence_class, noise_1, noise_2], batch_size=batch_size)\n\n _, _, _, optimized_pwm, _, _, _, _, _, iso_pred, cut_pred, _, _ = pred_outputs\n \n #Plot one PWM sequence logo per optimized objective (Experiment 'Punish A-runs')\n\n for pwm_index in range(10) :\n\n sequence_template = sequence_templates[class_i]\n\n pwm = np.expand_dims(optimized_pwm[pwm_index, :, :, 0], axis=0)\n cut = np.expand_dims(cut_pred[pwm_index, 0, :], axis=0)\n iso = np.expand_dims(np.sum(cut[:, 80: 115], axis=-1), axis=-1)\n\n plot_seqprop_logo(pwm, iso, cut, annotate_peaks='max', sequence_template=sequence_template, figsize=(12, 1.5), width_ratios=[1, 8], logo_height=0.8, usage_unit='fraction', plot_start=70-50, plot_end=76+50, save_figs=False, fig_name='genesis_apa_max_isoform_' + str(lib_name) + experiment_suffix + \"_pwm_index_\" + str(pwm_index), fig_dpi=150)\n",
"/home/ubuntu/anaconda3/envs/tensorflow_p36/lib/python3.6/site-packages/keras/engine/saving.py:292: UserWarning: No training configuration found in save file: the model was *not* compiled. Compile it manually.\n warnings.warn('No training configuration found in save file: '\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03ddac6621f96e3102f790155c56bcbb20f1781 | 9,171 | ipynb | Jupyter Notebook | Chapter03/Chapter 3 - Applying a 1D CNN to text.ipynb | ShubhInfotech-Bhilai/PythonDeep-Learning | 2183de8a1c8a050a07e2cb8e8dab01bacbd237f2 | [
"MIT"
] | 143 | 2017-10-27T22:39:27.000Z | 2022-02-10T19:05:07.000Z | Chapter03/Chapter 3 - Applying a 1D CNN to text.ipynb | ShubhInfotech-Bhilai/PythonDeep-Learning | 2183de8a1c8a050a07e2cb8e8dab01bacbd237f2 | [
"MIT"
] | 7 | 2017-12-31T18:26:59.000Z | 2020-12-01T19:48:08.000Z | Chapter03/Chapter 3 - Applying a 1D CNN to text.ipynb | ShubhInfotech-Bhilai/PythonDeep-Learning | 2183de8a1c8a050a07e2cb8e8dab01bacbd237f2 | [
"MIT"
] | 110 | 2017-10-27T05:33:48.000Z | 2022-01-21T13:54:43.000Z | 37.129555 | 881 | 0.505506 | [
[
[
"import numpy as np\n\nfrom keras.preprocessing import sequence\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Dropout, Activation\nfrom keras.layers import Embedding\nfrom keras.layers import Conv1D, GlobalMaxPooling1D\nfrom keras.callbacks import EarlyStopping\nfrom keras.datasets import imdb",
"Using TensorFlow backend.\n"
],
[
"n_words = 1000\n(X_train, y_train), (X_test, y_test) = imdb.load_data(num_words=n_words)\nprint('Train seq: {}'.format(len(X_train)))\nprint('Test seq: {}'.format(len(X_train)))",
"Train seq: 25000\nTest seq: 25000\n"
],
[
"print('Train example: \\n{}'.format(X_train[0]))\nprint('\\nTest example: \\n{}'.format(X_test[0]))\n\n# Note: the data is already preprocessed (words are mapped to vectors)",
"Train example: \n[1, 14, 22, 16, 43, 530, 973, 2, 2, 65, 458, 2, 66, 2, 4, 173, 36, 256, 5, 25, 100, 43, 838, 112, 50, 670, 2, 9, 35, 480, 284, 5, 150, 4, 172, 112, 167, 2, 336, 385, 39, 4, 172, 2, 2, 17, 546, 38, 13, 447, 4, 192, 50, 16, 6, 147, 2, 19, 14, 22, 4, 2, 2, 469, 4, 22, 71, 87, 12, 16, 43, 530, 38, 76, 15, 13, 2, 4, 22, 17, 515, 17, 12, 16, 626, 18, 2, 5, 62, 386, 12, 8, 316, 8, 106, 5, 4, 2, 2, 16, 480, 66, 2, 33, 4, 130, 12, 16, 38, 619, 5, 25, 124, 51, 36, 135, 48, 25, 2, 33, 6, 22, 12, 215, 28, 77, 52, 5, 14, 407, 16, 82, 2, 8, 4, 107, 117, 2, 15, 256, 4, 2, 7, 2, 5, 723, 36, 71, 43, 530, 476, 26, 400, 317, 46, 7, 4, 2, 2, 13, 104, 88, 4, 381, 15, 297, 98, 32, 2, 56, 26, 141, 6, 194, 2, 18, 4, 226, 22, 21, 134, 476, 26, 480, 5, 144, 30, 2, 18, 51, 36, 28, 224, 92, 25, 104, 4, 226, 65, 16, 38, 2, 88, 12, 16, 283, 5, 16, 2, 113, 103, 32, 15, 16, 2, 19, 178, 32]\n\nTest example: \n[1, 89, 27, 2, 2, 17, 199, 132, 5, 2, 16, 2, 24, 8, 760, 4, 2, 7, 4, 22, 2, 2, 16, 2, 17, 2, 7, 2, 2, 9, 4, 2, 8, 14, 991, 13, 877, 38, 19, 27, 239, 13, 100, 235, 61, 483, 2, 4, 7, 4, 20, 131, 2, 72, 8, 14, 251, 27, 2, 7, 308, 16, 735, 2, 17, 29, 144, 28, 77, 2, 18, 12]\n"
],
[
"# Pad sequences with max_len\nmax_len = 200\nX_train = sequence.pad_sequences(X_train, maxlen=max_len)\nX_test = sequence.pad_sequences(X_test, maxlen=max_len)",
"_____no_output_____"
],
[
"# Define network architecture and compile\nmodel = Sequential()\nmodel.add(Embedding(n_words, 50, input_length=max_len))\nmodel.add(Dropout(0.5))\nmodel.add(Conv1D(128, 3, padding='valid', activation='relu', strides=1,))\nmodel.add(GlobalMaxPooling1D())\nmodel.add(Dense(250, activation='relu'))\nmodel.add(Dropout(0.5))\nmodel.add(Dense(1, activation='sigmoid'))\n\nmodel.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding_1 (Embedding) (None, 200, 50) 50000 \n_________________________________________________________________\ndropout_1 (Dropout) (None, 200, 50) 0 \n_________________________________________________________________\nconv1d_1 (Conv1D) (None, 198, 128) 19328 \n_________________________________________________________________\nglobal_max_pooling1d_1 (Glob (None, 128) 0 \n_________________________________________________________________\ndense_1 (Dense) (None, 250) 32250 \n_________________________________________________________________\ndropout_2 (Dropout) (None, 250) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 1) 251 \n=================================================================\nTotal params: 101,829\nTrainable params: 101,829\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"callbacks = [EarlyStopping(monitor='val_acc', patience=3)]",
"_____no_output_____"
],
[
"batch_size = 64\nn_epochs = 100\n\nmodel.fit(X_train, y_train, batch_size=batch_size, epochs=n_epochs, validation_split=0.2, callbacks=callbacks)",
"Train on 20000 samples, validate on 5000 samples\nEpoch 1/100\n20000/20000 [==============================] - 4s - loss: 0.5609 - acc: 0.6845 - val_loss: 0.4027 - val_acc: 0.8196\nEpoch 2/100\n20000/20000 [==============================] - 2s - loss: 0.3884 - acc: 0.8278 - val_loss: 0.3528 - val_acc: 0.8488\nEpoch 3/100\n20000/20000 [==============================] - 2s - loss: 0.3499 - acc: 0.8472 - val_loss: 0.3503 - val_acc: 0.8430\nEpoch 4/100\n20000/20000 [==============================] - 2s - loss: 0.3275 - acc: 0.8617 - val_loss: 0.3173 - val_acc: 0.8648\nEpoch 5/100\n20000/20000 [==============================] - 2s - loss: 0.3144 - acc: 0.8659 - val_loss: 0.3168 - val_acc: 0.8606\nEpoch 6/100\n20000/20000 [==============================] - 2s - loss: 0.2990 - acc: 0.8728 - val_loss: 0.3103 - val_acc: 0.8672\nEpoch 7/100\n20000/20000 [==============================] - 2s - loss: 0.2967 - acc: 0.8756 - val_loss: 0.3084 - val_acc: 0.8688\nEpoch 8/100\n20000/20000 [==============================] - 2s - loss: 0.2856 - acc: 0.8792 - val_loss: 0.3053 - val_acc: 0.8706\nEpoch 9/100\n20000/20000 [==============================] - 2s - loss: 0.2783 - acc: 0.8835 - val_loss: 0.3063 - val_acc: 0.8708\nEpoch 10/100\n20000/20000 [==============================] - 2s - loss: 0.2726 - acc: 0.8854 - val_loss: 0.3032 - val_acc: 0.8722\nEpoch 11/100\n20000/20000 [==============================] - 2s - loss: 0.2667 - acc: 0.8894 - val_loss: 0.3015 - val_acc: 0.8706\nEpoch 12/100\n20000/20000 [==============================] - 2s - loss: 0.2569 - acc: 0.8945 - val_loss: 0.3064 - val_acc: 0.8684\nEpoch 13/100\n20000/20000 [==============================] - 2s - loss: 0.2581 - acc: 0.8931 - val_loss: 0.3051 - val_acc: 0.8682\nEpoch 14/100\n20000/20000 [==============================] - 2s - loss: 0.2489 - acc: 0.8979 - val_loss: 0.3031 - val_acc: 0.8698\n"
],
[
"print('\\nAccuracy on test set: {}'.format(model.evaluate(X_test, y_test)[1]))\n\n# Accuracy on test set: 0.873",
"24448/25000 [============================>.] - ETA: 0s\nAccuracy on test set: 0.873\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03df2048182d03cd6f8dffcfe09e67809a47657 | 34,615 | ipynb | Jupyter Notebook | without_outliers.ipynb | arjunfzk/iitbhu_enigma_ml_hackathon | 6a6ef5f37430b1087b5fb62a725f3b405293c9c3 | [
"MIT"
] | null | null | null | without_outliers.ipynb | arjunfzk/iitbhu_enigma_ml_hackathon | 6a6ef5f37430b1087b5fb62a725f3b405293c9c3 | [
"MIT"
] | null | null | null | without_outliers.ipynb | arjunfzk/iitbhu_enigma_ml_hackathon | 6a6ef5f37430b1087b5fb62a725f3b405293c9c3 | [
"MIT"
] | null | null | null | 30.205061 | 95 | 0.385729 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib \nfrom sklearn.preprocessing import LabelEncoder\nimport seaborn as sns",
"_____no_output_____"
],
[
"train=pd.read_csv('train.csv')\ntest=pd.read_csv('test.csv')\ntrain.head()",
"_____no_output_____"
],
[
"train.isnull().sum()",
"_____no_output_____"
],
[
"train.describe()",
"_____no_output_____"
],
[
"train['Tag'].nunique()",
"_____no_output_____"
],
[
"tag_le=LabelEncoder()\ntrain['Tag']=tag_le.fit_transform(train['Tag'])\ntest['Tag']=tag_le.fit_transform(test['Tag'])",
"_____no_output_____"
],
[
"train['Username'].nunique()",
"_____no_output_____"
],
[
"train['Reputation'].nunique()",
"_____no_output_____"
],
[
"def remove_outlier(df_in, col_name, train):\n q1 = train[col_name].quantile(0.25)\n q3 = train[col_name].quantile(0.75)\n iqr = q3-q1\n fence_low = q1-1.5*iqr\n fence_high = q3+1.5*iqr\n df_out = df_in.loc[(df_in[col_name] > fence_low) & (df_in[col_name] < fence_high)]\n return df_out\n\ntrain1=train.copy()\ntrain1=remove_outlier(train1, 'Answers', train)\ntrain1=remove_outlier(train1, 'Views', train)\ntrain1=remove_outlier(train1, 'Reputation', train)",
"_____no_output_____"
],
[
"train.head(10)",
"_____no_output_____"
],
[
"train.describe()",
"_____no_output_____"
],
[
"train1['conversion']=train1['Views'] * train1['Reputation']",
"_____no_output_____"
],
[
"test.describe()",
"_____no_output_____"
],
[
"test1=test.copy()\ntest1=remove_outlier(test1, 'Answers', test)\ntest1=remove_outlier(test1, 'Views', test)\ntest1=remove_outlier(test1, 'Reputation', test)\ntest1['conversion']=test1['Reputation'] * test1['Views']\nID=test1['ID']\ntest1=test1.drop(['ID'], axis=1)",
"_____no_output_____"
],
[
"test1.describe()",
"_____no_output_____"
],
[
"ax = sns.boxplot(x=train1['Answers'])",
"_____no_output_____"
],
[
"ax = sns.boxplot(x=train1['Reputation'])",
"_____no_output_____"
],
[
"ax = sns.boxplot(x=train1['Views'])",
"_____no_output_____"
],
[
"from sklearn.preprocessing import MinMaxScaler\nX=train1.drop(['ID', 'Upvotes'], axis=1).values\ny=train1['Upvotes'].values.reshape(-1,1)",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test= train_test_split(X, y, test_size=0.2, random_state=0)",
"_____no_output_____"
],
[
"import xgboost as xgb\nxgbc=xgb.XGBRegressor(eval_metric='rmse')\nxgbc.fit(X,y)",
"_____no_output_____"
],
[
"'''from sklearn.model_selection import KFold\nfrom sklearn.metrics import mean_squared_error\ncv=[]\nkfold = KFold(10)\nfor train_index, test_index in kfold.split(train1):\n X_train, X_test = X[train_index], X[test_index]\n y_train, y_test = y[train_index], y[test_index]\n xgbc.fit(X_train,y_train)\n cv.append(np.sqrt(mean_squared_error(y_test, xgbc.predict(X_test))))\nprint(sum(cv)/ len(cv))'''",
"_____no_output_____"
],
[
"xgb.plot_importance(xgbc)",
"_____no_output_____"
],
[
"#from sklearn.metrics import mean_squared_error\n#print(np.sqrt(mean_squared_error(y, xgbc.predict(X))))",
"_____no_output_____"
],
[
"Submission = pd.DataFrame({ 'ID': ID,\n 'Upvotes':xgbc.predict(test1.values) })\nSubmission.to_csv(\"without_outliers.csv\", index=False)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03dfb4a6b8fe65cc8475ed1f5eca61fcf3fd230 | 22,532 | ipynb | Jupyter Notebook | examples/sql_connector/sql-airbnb.ipynb | thomassonobe/more-data | b3d4a8e32f385a69749c8139915e3638fcced37b | [
"BSD-3-Clause"
] | null | null | null | examples/sql_connector/sql-airbnb.ipynb | thomassonobe/more-data | b3d4a8e32f385a69749c8139915e3638fcced37b | [
"BSD-3-Clause"
] | null | null | null | examples/sql_connector/sql-airbnb.ipynb | thomassonobe/more-data | b3d4a8e32f385a69749c8139915e3638fcced37b | [
"BSD-3-Clause"
] | null | null | null | 38.516239 | 154 | 0.3827 | [
[
[
"import os\nimport sys\n\nfrom moredata.enricher import EnricherBuilder, Enricher\nfrom moredata.enricher.sql_connector import SqlConnector\nfrom moredata.models.data import JsonData\nfrom moredata.parser import parse_document\nfrom moredata.utils import read_json_from_file, Converter, write_json_generator_to_json\nfrom moredata.datasets import get_path",
"_____no_output_____"
],
[
"import pandas as pd\n\ndf = pd.read_csv(get_path(\"airbnb-berlin-main\"))\ndf = df.loc[(~df['latitude'].isna()) & (~df['longitude'].isna())]\ndf.to_json('./data/airbnb-berlin.json', orient='records')",
"_____no_output_____"
],
[
"from sqlalchemy import create_engine\nURL = \"mysql+pymysql://root:root@localhost:3306/moredata\"\nengine = create_engine(URL, echo=False)\n\nextra = pd.read_csv(get_path(\"airbnb-berlin-extra\"))\nextra.to_sql('extra', con=engine, if_exists='replace')",
"_____no_output_____"
],
[
"pd.read_sql('SELECT id, amenities, accommodates, beds, bedrooms FROM extra', engine)",
"_____no_output_____"
],
[
"data = JsonData(data_file='./data/airbnb-berlin.json', parser=parse_document)\n\nsql_enricher = Enricher(connector=SqlConnector(connection_url=URL, table_name=\"extra\", column='id', result_attr=\"extra\", dict_keys=[\"id\"]))\n\ndata_enriched = sql_enricher.enrich(data)\n\nwrite_json_generator_to_json(\"./data/airbnb-berlin-enriched\", data_enriched, 100000) ",
"_____no_output_____"
],
[
"data_enriched = pd.read_json(\"./data/airbnb-berlin-enriched-0.json\", orient='records')",
"_____no_output_____"
],
[
"type(data_enriched.extra[0])",
"_____no_output_____"
],
[
"pd.concat([data_enriched, pd.json_normalize(data_enriched['extra'], max_level=0).drop(['index', 'id'], axis=1)], axis=1)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03e1350b8830c0ff9a16794a852ff1e4b4830df | 373,913 | ipynb | Jupyter Notebook | Jupyter/BMRS20_mos_our_cuts.ipynb | bsafdi/BlankSkyfor3p5 | 0b9d37bd9811b1ed77ea71c0fa3d15a16de365b4 | [
"MIT"
] | null | null | null | Jupyter/BMRS20_mos_our_cuts.ipynb | bsafdi/BlankSkyfor3p5 | 0b9d37bd9811b1ed77ea71c0fa3d15a16de365b4 | [
"MIT"
] | null | null | null | Jupyter/BMRS20_mos_our_cuts.ipynb | bsafdi/BlankSkyfor3p5 | 0b9d37bd9811b1ed77ea71c0fa3d15a16de365b4 | [
"MIT"
] | null | null | null | 295.116811 | 54,208 | 0.916154 | [
[
[
"# An analysis of the dataset presented in [this technical comment](https://arxiv.org/abs/2004.06601), but with our quality cuts applied\n\nAs a response to our paper [Dessert et al. _Science_ 2020](https://science.sciencemag.org/content/367/6485/1465) (DRS20), we received [a technical comment](https://arxiv.org/abs/2004.06601) (BRMS). BRMS performed a simplified version of our analysis in a partially overlapping dataset using 17 Ms of MOS observations spanning 20$^\\circ$ to 35$^\\circ$ from the Galactic Center. They assumed a single power-law background with additional lines at 3.1, 3.3, 3.7, and 3.9 keV, and claim a 4$\\sigma$ detection of a line at 3.48 keV using an energy window of 3-4 keV. However, it is important to note that the BRMS analysis do not apply any (stated) quality cuts to their dataset. On the other hand, as detailed in DRS20, we selected low-background or blank-sky observations, so the data is much cleaner.\n\nIn our formal response to the technical comment, we repeat this analysis on the 8.5 Ms of the BRMS dataset that passes the quality cuts. In this notebook, we show this data and analysis in detail. Many of the details will follow the procedure used in the notebook `DRS20_mos_stacked`. For a pedagogical introduction to the analysis here, we refer to that notebook.\n\nIf you use the data in this example in a publication, please cite Dessert et al. _Science_ 2020.\n\n**Please direct any questions to bsafdi@umich.edu.**",
"_____no_output_____"
]
],
[
[
"# Import required modules\n\n%matplotlib inline\n%load_ext autoreload\n%autoreload 2\n \nimport sys,os\nimport numpy as np\n\nfrom scipy.stats import chi2 as chi2_scipy\nfrom scipy.optimize import dual_annealing\nfrom scipy.optimize import minimize\n\nimport matplotlib.pyplot as plt\nfrom matplotlib import rc\nfrom matplotlib import rcParams\n\nrc('font', **{'family': 'serif', 'serif': ['Computer Modern']})\nrcParams['text.usetex'] = True\nrcParams['text.latex.unicode'] = True",
"_____no_output_____"
]
],
[
[
"**NB**: In this notebook, we minimize with `scipy` so that it is easy to run for the interested reader. For scientific analysis, we recommend [Minuit](https://iminuit.readthedocs.io/en/latest/) as a minimizer. In our paper, we used Minuit.",
"_____no_output_____"
],
[
"# Define signal line energy\n\nBy default we will look for an anomalous line at 3.48 keV, as defined by the EUXL parameter below, denoting the energy of the unidentified X-ray line. Lines at different energies can be searched for by changing this parameter accordingly (for example to 3.55 keV as in the previous notebook). We start with 3.48 keV as this is the fiducial line energy in BMRS. We note that 3.48 keV is the energy where the weakest limit is obtained, although on the clean data we will not find any evidence for a feature there.",
"_____no_output_____"
]
],
[
[
"EUXL = 3.48 # [keV]",
"_____no_output_____"
]
],
[
[
"**NB:** changing EUXL will of course vary the results below, and values in the surrounding discussion will not necessarily be reflective.\n\n# Load in the data and models\n\nFirst we will load in the data products that we will use in the analysis. These include the stacked MOS data, associated energy bins, and uncertainties. \n\nWe will use data from two regions of interest (ROI):\n- **Signal Region (SR)**: 20-35 degrees from the Galactic Center, this was the fiducial ROI in BRMS (DRS20 instead used 5-45);\n- **Background Region (BR)**: 60-90 degrees from the Galactic Center, a useful region for studying background as it contains less dark matter.\n\nWe also load the appropriately averaged D-factors for these two regions (ROIs) for our fiducial NFW profile, along with the respective exposure times.",
"_____no_output_____"
]
],
[
[
"## Signal Region (20-35 degrees)\ndata = np.load(\"../data/data_mos_boyarsky_ROI_our_cuts.npy\") # [cts/s/keV]\ndata_yerrs = np.load(\"../data/data_yerrs_mos_boyarsky_ROI_our_cuts.npy\") # [cts/s/keV]\nQPB = np.load(\"../data/QPB_mos_boyarsky_ROI_our_cuts.npy\") # [cts/s/keV]\n\n# Exposure time\nExp = 8.49e6 # [s]\n\n# D-factor averaged over the signal ROI\nD_signal = 4.4e28 # [keV/cm^2]\n\n\n## Background Region (60-90 degrees)\n\n# Data and associated errors\ndata_bkg = np.load(\"../data/data_mos_bkg_ROI.npy\") # [cts/s/keV]\ndata_yerrs_bkg = np.load(\"../data/data_yerrs_mos_bkg_ROI.npy\") # [cts/s/keV]\n\n# Exposure time\nExp_bkg = 67.64e6 # [s]\n\n# D-factor averaged over the background ROI\nD_bkg = 1.91e28 # [keV/cm^2]\n\n## Energy binning appropriate for both the signal and background\nEnergies=np.load(\"../data/mos_energies.npy\") # [keV]",
"_____no_output_____"
]
],
[
[
"## Load in the Models\n\nNext we use the models that will be used in fitting the above data.\n\nThere are a sequence of models corresponding to physical line fluxes at the energies specified by `Es_line`. That is, `mod_UXL` gives the detectors counts as a function of energy after forward modeling a physical line at EUXL keV with a flux of 1 cts/cm$^2$/s/sr.",
"_____no_output_____"
]
],
[
[
"# Load the forward-modeled lines and energies\nmods = np.load(\"../data/mos_mods.npy\")\nEs_line = np.load(\"../data/mos_mods_line_energies.npy\")\n\n# Load the detector response\ndet_res = np.load(\"../data/mos_det_res.npy\")\n\narg_UXL = np.argmin((Es_line-EUXL)**2)\nmod_UXL = mods[arg_UXL]\n\nprint \"The energy of our \"+str(EUXL)+\" keV line example will be: \"+str(Es_line[arg_UXL])+\" keV\"\n\n# How to go from flux to sin^2(2\\theta)\ndef return_sin_theta_lim(E_line,flux,D_factor):\n \"\"\"\n D_factor [keV/cm^2]\n flux [cts/cm^2/s/sr]\n E_line [keV] (dark matter mass is twice this value)\n returns: associated sin^2(2theta)\n \"\"\"\n DMmass = 2.*E_line\n res = (4.*np.pi*DMmass/D_factor)/1.361e-22*(1/DMmass)**5*flux\n return res",
"The energy of our 3.48 keV line example will be: 3.4824707846410687 keV\n"
]
],
[
[
"# Visualize the data\n\nData in the signal region, where the dashed vertical line denotes the location of a putative signal line. Note in particular the flux is similar to that in Fig. 2 of DRS20, indicating that the included observations are low-background. ",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(10,8))\nplt.errorbar(Energies,data,yerr=data_yerrs,xerr=(Energies[1]-Energies[0])/2.,\n color=\"black\",label=\"data\",marker=\"o\", fmt='none',capsize=4)\nplt.axvline(EUXL,color=\"black\",linestyle=\"dashed\")\nplt.xlim(EUXL-0.25,EUXL+0.25)\nplt.ylim(7.9e-2,0.1)\nplt.xticks(fontsize=22)\nplt.yticks(fontsize=22)\nplt.xlabel(r\"$E$ [keV]\",fontsize=22)\nplt.ylabel(r\"SR Flux [cts/s/keV]\",fontsize=22)\nplt.show()",
"/sw/lsa/centos7/python-anaconda2/2019.03/lib/python2.7/site-packages/matplotlib/font_manager.py:1331: UserWarning: findfont: Font family [u'serif'] not found. Falling back to DejaVu Sans\n (prop.get_family(), self.defaultFamily[fontext]))\n"
]
],
[
[
"# Statistical analysis\n\nNow, let's perform a rigorous statistical analysis, using profile likelihood. As we operate in the large counts limit for the stacked data, we can perform a simple $\\chi^2$ analysis rather than a full joint likelihood analysis as used by default in Dessert et al. 2020.",
"_____no_output_____"
]
],
[
[
"## Define the functions we will use\n\nclass chi2:\n \"\"\" A set offunctions for calculation the chisq associated with different hypotheses\n \"\"\"\n def __init__(self,ens,dat,err,null_mod,sig_template):\n self._ens = ens\n self._dat = dat\n self._err = err\n self._null_mod = null_mod\n self._sig_template = sig_template\n self._A_sig = 0.0\n \n def chi2(self,x):\n null_mod = self._null_mod(self._ens,x[1:])\n sig_mod = self._sig_template*x[0]\n return np.sum((self._dat - null_mod - sig_mod)**2/self._err**2)\n \n def chi2_null(self,x):\n null_mod = self._null_mod(self._ens,x)\n return np.sum((self._dat - null_mod)**2/self._err**2)\n \n def chi2_fixed_signal(self,x):\n null_mod = self._null_mod(self._ens,x)\n sig_mod = self._sig_template*self._A_sig\n return np.sum((self._dat - null_mod - sig_mod)**2/self._err**2) \n \n def fix_signal_strength(self,A_sig):\n self._A_sig = A_sig",
"_____no_output_____"
]
],
[
[
"## Fit within $E_{\\rm UXL} \\pm 0.25$ keV\n\nFirst, we will fit the models from $[E_{\\rm UXL}-0.25,\\,E_{\\rm UXL}+0.25]$ keV. Later in this notebook, we broaden this range to 3.0 to 4.0 keV. For the default $E_{\\rm UXL} = 3.48$ keV, this corresponds to $3.23~{\\rm keV} < E < 3.73~{\\rm keV}$.\n\nTo begin with then, let's reduce the dataset to this restricted range.",
"_____no_output_____"
]
],
[
[
"whs_reduced = np.where((Energies >= EUXL-0.25) & (Energies <= EUXL+0.25))[0]\nEnergies_reduced = Energies[whs_reduced]\ndata_reduced = data[whs_reduced]\ndata_yerrs_reduced = data_yerrs[whs_reduced]\ndata_bkg_reduced = data_bkg[whs_reduced]\ndata_yerrs_bkg_reduced = data_yerrs_bkg[whs_reduced]\nmod_UXL_reduced = mod_UXL[whs_reduced]",
"_____no_output_____"
]
],
[
[
"Let's fit this data with the background only hypothesis and consider the quality of fit.",
"_____no_output_____"
],
[
"## A polynomial background model\n\nHere we model the continuum background as a quadratic. In addition, we add degrees of freedom associated with the possible background lines at 3.3 keV and 3.7 keV. ",
"_____no_output_____"
]
],
[
[
"arg_3p3 = np.argmin((Es_line-3.32)**2)\nmod_3p3 = mods[arg_3p3]\n\narg_3p7 = np.argmin((Es_line-3.68)**2)\nmod_3p7 = mods[arg_3p7]\n\ndef mod_poly_two_lines(ens,x):\n \"An extended background model to include two additional lines\"\n A, B, C, S1, S2 = x\n\n return A+B*ens + C*ens**2 + S1*mod_3p3[whs_reduced] + S2*mod_3p7[whs_reduced]\n\nchi2_instance = chi2(Energies_reduced,data_reduced,data_yerrs_reduced,mod_poly_two_lines,mod_UXL_reduced)\n\nmn_null_line = minimize(chi2_instance.chi2_null,np.array([0.282,-0.098, 0.011,0.1,0.1]),method='Nelder-Mead')\n\nmn_line = minimize(chi2_instance.chi2,np.array([1.e-2,mn_null_line.x[0],mn_null_line.x[1],mn_null_line.x[2],mn_null_line.x[3],mn_null_line.x[4]]),method='Nelder-Mead',options={'fatol':1e-10,'xatol':1e-10,'adaptive':True})\n\n\nprint \"The Delta chi^2 between signal and null model is:\", mn_null_line.fun - mn_line.fun\nprint \"The chi^2/DOF of the null-model fit is:\", mn_null_line.fun/(len(whs_reduced)-5.)\n\nprint \"Expected 68% containment for the chi^2/DOF:\", np.array(chi2_scipy.interval(0.68,len(whs_reduced)-5.))/float(len(whs_reduced)-5.)\nprint \"Expected 99% containment for the chi^2/DOF:\", np.array(chi2_scipy.interval(0.99,len(whs_reduced)-5.))/float(len(whs_reduced)-5.)",
"The Delta chi^2 between signal and null model is: 0.18286814612878288\nThe chi^2/DOF of the null-model fit is: 0.9413439893438815\nExpected 68% containment for the chi^2/DOF: [0.85614219 1.14370943]\nExpected 99% containment for the chi^2/DOF: [0.66578577 1.41312157]\n"
]
],
[
[
"The null model is a good fit to the data, and the best-fit signal strength is still consistent with zero at 1$\\sigma$.\n\nNext we plot the best fit signal and background model, in particular we see the model is almost identical in the two cases, emphasizing the lack of preference for a new emission line at 3.48 keV in this dataset.",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(10,8))\nplt.errorbar(Energies,data,yerr=data_yerrs,xerr=(Energies[1]-Energies[0])/2.,\n color=\"black\",label=\"data\",marker=\"o\", fmt='none',capsize=4)\nplt.plot(Energies_reduced,mod_poly_two_lines(Energies_reduced,mn_null_line.x),'k-',label =r\"Null model\")\nplt.plot(Energies_reduced,mod_poly_two_lines(Energies_reduced,mn_line.x[1:])+mn_line.x[0]*mod_UXL_reduced,\n 'r-',label =r\"Signal model\")\nplt.axvline(EUXL,color=\"black\",linestyle=\"dashed\")\nplt.xlim(EUXL-0.25,EUXL+0.25)\nplt.ylim(0.08,0.1)\nplt.xticks(fontsize=22)\nplt.yticks(fontsize=22)\nplt.xlabel(r\"$E$ [keV]\",fontsize=22)\nplt.ylabel(r\"SR Flux [cts/s/keV]\",fontsize=22)\nplt.legend(fontsize=22)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Finally let's compute the associated limit via profile likelihood.",
"_____no_output_____"
]
],
[
[
"A_sig_array = np.linspace(mn_line.x[0],0.05,100)\nchi2_sig_array = np.zeros(len(A_sig_array))\nbf = mn_line.x[1:]\nfor i in range(len(A_sig_array)):\n chi2_instance.fix_signal_strength(A_sig_array[i])\n mn_profile = minimize(chi2_instance.chi2_fixed_signal,bf,method='Nelder-Mead',\n options={'fatol':1e-10,'xatol':1e-10,'adaptive':True})\n bf = mn_profile.x\n chi2_sig_array[i] = mn_profile.fun\n\namin = np.argmin((chi2_sig_array-chi2_sig_array[0] - 2.71)**2)\nlimit_signal_strength = A_sig_array[amin]\n\nprint \"The 95% upper limit on the signal flux is\", limit_signal_strength, \"cts/cm^2/s/sr\"\nprint \"This corresponds to a limit on sin^2(2theta) of\", return_sin_theta_lim(EUXL,limit_signal_strength,D_signal)",
"The 95% upper limit on the signal flux is 0.02664201119758925 cts/cm^2/s/sr\nThis corresponds to a limit on sin^2(2theta) of 2.382479159553265e-11\n"
]
],
[
[
"## Power law background model",
"_____no_output_____"
],
[
"Now let's try a power law for the continuum background model (along with the two lines) as done in BMRS. Given that the stacked data is the sum of power laws, we would not expect the stacked data to be a power law itself, although in our relatively clean dataset we find it to be a reasonable description. ",
"_____no_output_____"
]
],
[
[
"def mod_power_two_lines(ens,x):\n \"An extended background model to include two additional lines\"\n A, n, S1, S2 = x\n\n return A*ens**n + S1*mod_3p3[whs_reduced] + S2*mod_3p7[whs_reduced]\n\nchi2_instance = chi2(Energies_reduced,data_reduced,data_yerrs_reduced,mod_power_two_lines,mod_UXL_reduced)\n\nmn_null_line = minimize(chi2_instance.chi2_null,np.array([0.18244131, -0.58714693, 0.02237754, 0.01157593]),method='Nelder-Mead')\n\nmn_line = minimize(chi2_instance.chi2,np.array([1.e-2,mn_null_line.x[0],mn_null_line.x[1],mn_null_line.x[2],mn_null_line.x[3]]),method='Nelder-Mead',options={'fatol':1e-10,'xatol':1e-10,'adaptive':True})\n\n\nprint \"The Delta chi^2 between signal and null model is:\", mn_null_line.fun - mn_line.fun\nprint \"The chi^2/DOF of the null-model fit is:\", mn_null_line.fun/(len(whs_reduced)-4.)",
"The Delta chi^2 between signal and null model is: 0.00043135939542082724\nThe chi^2/DOF of the null-model fit is: 0.9426290489941959\n"
],
[
"fig = plt.figure(figsize=(10,8))\nplt.errorbar(Energies,data,yerr=data_yerrs,xerr=(Energies[1]-Energies[0])/2.,\n color=\"black\",label=\"data\",marker=\"o\", fmt='none',capsize=4)\nplt.plot(Energies_reduced,mod_power_two_lines(Energies_reduced,mn_null_line.x),'k-',label =r\"Null model\")\nplt.plot(Energies_reduced,mod_power_two_lines(Energies_reduced,mn_line.x[1:])+mn_line.x[0]*mod_UXL_reduced,\n 'r-',label =r\"Signal model\")\nplt.axvline(EUXL,color=\"black\",linestyle=\"dashed\")\nplt.xlim(EUXL-0.25,EUXL+0.25)\nplt.ylim(0.08,0.1)\nplt.xticks(fontsize=22)\nplt.yticks(fontsize=22)\nplt.xlabel(r\"$E$ [keV]\",fontsize=22)\nplt.ylabel(r\"SR Flux [cts/s/keV]\",fontsize=22)\nplt.legend(fontsize=22)\nplt.show()",
"_____no_output_____"
],
[
"A_sig_array = np.linspace(mn_line.x[0],0.05,100)\nchi2_sig_array = np.zeros(len(A_sig_array))\nbf = mn_line.x[1:]\nfor i in range(len(A_sig_array)):\n chi2_instance.fix_signal_strength(A_sig_array[i])\n mn_profile = minimize(chi2_instance.chi2_fixed_signal,bf,method='Nelder-Mead',\n options={'fatol':1e-10,'xatol':1e-10,'adaptive':True})\n bf = mn_profile.x\n chi2_sig_array[i] = mn_profile.fun\n\namin = np.argmin((chi2_sig_array-chi2_sig_array[0] - 2.71)**2)\nlimit_signal_strength = A_sig_array[amin]\nprint \"The 95% upper limit on the signal flux is\", limit_signal_strength, \"cts/cm^2/s/sr\"\nprint \"This corresponds to a limit on sin^2(2theta) of\", return_sin_theta_lim(EUXL,limit_signal_strength,D_signal)",
"The 95% upper limit on the signal flux is 0.020575238409308062 cts/cm^2/s/sr\nThis corresponds to a limit on sin^2(2theta) of 1.8399540616307525e-11\n"
]
],
[
[
"The power law continuum background does not substantively change the results: we still find no evidence for a line. Note this is the same procedure as in BMRS's test color-coded red in their Fig. 1 and Tab. 1. In that analysis, they find marginal 1.3$\\sigma$ evidence for a line, although on our cleaner dataset we found no evidence. \n\n**NB:** As an aside, BMRS also perform an analysis, color-coded green in their Fig. 1 and Tab. 1, in which they fix the 3.3 keV and 3.7 keV emission lines to their best fit fluxes in the fit. They claim that DRS20, in our Supplementary Material Sec 2.7, also fixed the fluxes of these lines. This statement is incorrect.",
"_____no_output_____"
],
[
"# Departing from the narrow window\n\nWe now fit the same dataset over the 3-4 keV range. \n\nOur procedure is as follows. Firstly, we update the dataset. Then we will define a new background model incorporating these additional lines. Finally we repeat our default $\\chi^2$ fit procedure. Note that we continue to use a power law continuum background model here. As such, the following analysis is a repetition of the BMRS magenta color-coded analysis on this reduced and low-background dataset. In that magenta analysis, they claim a 4.0$\\sigma$ detection of a line at 3.48 keV. Let us see what we obtain on when we include only the observations passing our quality cuts.",
"_____no_output_____"
]
],
[
[
"whs_reduced = np.where((Energies >= 3.0) & (Energies <= 4.0))[0]\nEnergies_reduced = Energies[whs_reduced]\ndata_reduced = data[whs_reduced]\ndata_yerrs_reduced = data_yerrs[whs_reduced]\ndata_bkg_reduced = data_bkg[whs_reduced]\ndata_yerrs_bkg_reduced = data_yerrs_bkg[whs_reduced]\nmod_UXL_reduced = mod_UXL[whs_reduced]\n\narg_3p1 = np.argmin((Es_line-3.12)**2) \nmod_3p1 = mods[arg_3p1]\n\narg_3p9 = np.argmin((Es_line-3.90)**2)\nmod_3p9 = mods[arg_3p9]\n\narg_3p7 = np.argmin((Es_line-3.68)**2)\nmod_3p7 = mods[arg_3p7]\n\narg_3p3 = np.argmin((Es_line-3.32)**2) \nmod_3p3 = mods[arg_3p3]\n\n\ndef mod_power_four_lines(ens,x):\n A, n,S1, S2, S3, S4 = x\n return A*ens**n + S1*mod_3p3[whs_reduced] + S2*mod_3p7[whs_reduced]+ S3*mod_3p1[whs_reduced] + S4*mod_3p9[whs_reduced]\n\nchi2_instance = chi2(Energies_reduced,data_reduced,data_yerrs_reduced,mod_power_four_lines,mod_UXL_reduced)\n\n\nx0 = np.array([0.18088868 ,-0.58201284 , 0.02472505 , 0.01364361 , 0.08959867,\n 0.03220519])\nbounds = np.array([[1e-6,5],[-3,0],[0,0.5],[0,0.5],[0,0.5],[0,0.5]])\nmn_null = dual_annealing(chi2_instance.chi2_null,x0=x0,bounds=bounds,local_search_options={\"method\": \"Nelder-Mead\"},seed=1234,maxiter=500)\n\nboundss = np.array([[-0.5,0.5],[1e-6,5],[-3,0],[0,0.1],[0,0.1],[0,0.1],[0,0.2]])\nx0s=np.array([1.e-2,mn_null.x[0],mn_null.x[1],mn_null.x[2],mn_null.x[3],mn_null.x[4],mn_null.x[5]])\nmn = dual_annealing(chi2_instance.chi2,x0=x0s,bounds=boundss,local_search_options={\"method\": \"Nelder-Mead\"},seed=1234,maxiter=500)\n\nprint \"Best fit background parameters:\", mn_null.x\nprint \"Best fit signal+background parameters:\", mn.x\n\nprint \"The Delta chi^2 between signal and null model is:\", mn_null.fun - mn.fun\nprint \"The chi^2/DOF of the null-model fit is:\", mn_null.fun/(len(whs_reduced)-6.)\n\nprint \"NB: the best-fit signal strength in this case is\", mn.x[0], \"cts/cm$^2$/s/sr\"",
"Best fit background parameters: [ 0.1807753 -0.58187775 0.02547398 0.01436228 0.09052193 0.03304785]\nBest fit signal+background parameters: [ 0.0015216 0.18110231 -0.58372105 0.02608541 0.0154532 0.0911047\n 0.03434406]\nThe Delta chi^2 between signal and null model is: 0.03379302143113705\nThe chi^2/DOF of the null-model fit is: 0.9791145496670692\nNB: the best-fit signal strength in this case is 0.0015216044839330481 cts/cm$^2$/s/sr\n"
]
],
[
[
"We find no evidence for a 3.5 keV line when we expand the energy window. Although the best-fit signal strength is positive, the $\\Delta \\chi^2 \\sim 0.03$, which is entirely negligable significance. \n\nLet's have a look at the best fit signal and background models in this case. There are subtle difference between the two, but no excess is appearing at 3.48 keV.\n\nAdditionally, we are defining a fixed signal to plot overtop the data for illustration. The default signal parameters here corresponds to a 2$\\sigma$ downward fluctuationn in the signal reported in [Cappelluti et. al. ApJ 2018](https://iopscience.iop.org/article/10.3847/1538-4357/aaaa68/meta) from observations of the Chandra Deep Fields. Note that even taking the conservative downward flucutation, it is not a good fit to the data. This plot appears in our response to BMRS.",
"_____no_output_____"
]
],
[
[
"flux_ill = 4.8e-11 / return_sin_theta_lim(EUXL,1.,D_signal)\nprint \"Flux [cts/cm^2/s/sr] and sin^(2theta) for illustration: \", flux_ill, return_sin_theta_lim(EUXL,flux_ill,D_signal)\n\nchi2_instance.fix_signal_strength(flux_ill)\n\nmn_f = dual_annealing(chi2_instance.chi2_fixed_signal,x0=x0,bounds=bounds,local_search_options={\"method\": \"Nelder-Mead\"},seed=1234,maxiter=500)\nprint \"Delta chi^2 between fixed signal and null:\", mn_null.fun-mn_f.fun",
"Flux [cts/cm^2/s/sr] and sin^(2theta) for illustration: 0.0536758750798 4.8e-11\nDelta chi^2 between fixed signal and null: -40.04152650916589\n"
],
[
"def avg_data(data,n):\n return np.mean(data.reshape(-1, n), axis=1)\n\nfig = plt.figure(figsize=(10,8))\n\nplt.errorbar(avg_data(Energies,6),avg_data(data,6),yerr=np.sqrt(6*avg_data(data_yerrs**2,6))/6.,xerr=6*(Energies[1]-Energies[0])/2.,\n color=\"black\",marker=\"o\", fmt='none',capsize=4) \n\nplt.plot(Energies_reduced,mod_power_four_lines(Energies_reduced,mn_null.x),\n 'k-',label =r\"Null P.L. model\")\nplt.plot(Energies_reduced,mod_power_four_lines(Energies_reduced,mn.x[1:])+mn.x[0]*mod_UXL_reduced,\n 'r-',label =r\"Best fit signal model\")\nplt.plot(Energies_reduced,mod_power_four_lines(Energies_reduced,mn_f.x)+chi2_instance._A_sig*mod_UXL_reduced,\n 'r--',label =r\"$\\sin^2(2\\theta) = 4.8 \\times 10^{-11}$\")\n\nplt.xlim(3,4)\nplt.ylim(0.08,0.1)\n\nplt.xticks(fontsize=22)\nplt.yticks(fontsize=22)\nplt.xlabel(r\"$E$ [keV]\",fontsize=22)\nplt.ylabel(r\"Flux [cts/s/keV]\",fontsize=22)\nplt.legend(fontsize=22)\nplt.show()",
"_____no_output_____"
]
],
[
[
"**NB:** In the plot above we averaged the data solely for presentation purposes, no averaging was performed in the analysis.\n\nFinally, we compute the limit in this case using the by now familiar procedure.",
"_____no_output_____"
]
],
[
[
"A_sig_array = np.linspace(mn.x[0],0.05,100)\nchi2_sig_array = np.zeros(len(A_sig_array))\nbf = mn.x[1:]\nfor i in range(len(A_sig_array)):\n chi2_instance.fix_signal_strength(A_sig_array[i])\n mn_profile = minimize(chi2_instance.chi2_fixed_signal,bf,method='Nelder-Mead')\n bf = mn_profile.x\n chi2_sig_array[i] = mn_profile.fun\n\namin = np.argmin((chi2_sig_array-chi2_sig_array[0] - 2.71)**2)\nlimit_signal_strength = A_sig_array[amin]\nprint \"The 95% upper limit on the signal flux is\", limit_signal_strength, \"cts/cm^2/s/sr\"\nprint \"This corresponds to a limit on sin^2(2theta) of\", return_sin_theta_lim(EUXL,limit_signal_strength,D_signal)",
"The 95% upper limit on the signal flux is 0.015232665842012591 cts/cm^2/s/sr\nThis corresponds to a limit on sin^2(2theta) of 1.362191038952712e-11\n"
]
],
[
[
"## Now with a polynomial background",
"_____no_output_____"
],
[
"Here we repeat the earlier analysis but with a polynomial background model, as used in the stacked analysis in DRS20 Supplementary Material Sec. 2.9.",
"_____no_output_____"
]
],
[
[
"whs_reduced = np.where((Energies >= 3.0) & (Energies <= 4.0))[0]\nEnergies_reduced = Energies[whs_reduced]\ndata_reduced = data[whs_reduced]\ndata_yerrs_reduced = data_yerrs[whs_reduced]\ndata_bkg_reduced = data_bkg[whs_reduced]\ndata_yerrs_bkg_reduced = data_yerrs_bkg[whs_reduced]\nmod_UXL_reduced = mod_UXL[whs_reduced]\n\narg_3p1 = np.argmin((Es_line-3.12)**2) #3.12 #should really be 3.128\nmod_3p1 = mods[arg_3p1]\n\narg_3p9 = np.argmin((Es_line-3.90)**2)\nmod_3p9 = mods[arg_3p9]\n\narg_3p7 = np.argmin((Es_line-3.68)**2)\nmod_3p7 = mods[arg_3p7]\n\narg_3p3 = np.argmin((Es_line-3.32)**2) \nmod_3p3 = mods[arg_3p3]\n\ndef mod_poly_four_lines(ens,x):\n A, B, C,S1, S2, S3, S4 = x\n return A+B*ens + C*ens**2 + S1*mod_3p3[whs_reduced] + S2*mod_3p7[whs_reduced]+ S3*mod_3p1[whs_reduced] + S4*mod_3p9[whs_reduced]\n\nchi2_instance = chi2(Energies_reduced,data_reduced,data_yerrs_reduced,mod_poly_four_lines,mod_UXL_reduced)\n\n\nx0 = np.array([ 0.2015824 , -0.05098609 , 0.0052141 , 0.02854594 , 0.01742288,\n 0.08976637 , 0.029351 ])\nbounds = np.array([[-1,1],[-0.5,0.5],[-0.1,0.1],[0,0.2],[0,0.2],[0,0.2],[0,0.2]])\nmn_null = dual_annealing(chi2_instance.chi2_null,x0=x0,bounds=bounds,local_search_options={\"method\": \"Nelder-Mead\"},seed=1234,maxiter=3000)\n\nboundss = np.array([[-0.5,0.5],[-1,1],[-0.5,0.5],[-0.1,0.1],[0,0.2],[0,0.2],[0,0.2],[0,0.2]])\nx0s=np.array([1.e-2,mn_null.x[0],mn_null.x[1],mn_null.x[2],mn_null.x[3],mn_null.x[4],mn_null.x[5],mn_null.x[6]])\nmn = dual_annealing(chi2_instance.chi2,x0=x0s,bounds=boundss,local_search_options={\"method\": \"Nelder-Mead\"},seed=1234,maxiter=3000)\n\nprint \"Best fit background parameters:\", mn_null.x\nprint \"Best fit signal+background parameters:\", mn.x\n\nprint \"The Delta chi^2 between signal and null model is:\", mn_null.fun - mn.fun\nprint \"The chi^2/DOF of the null-model fit is:\", mn_null.fun/(len(whs_reduced)-7.)\n\nprint \"NB: the best-fit signal strength in this case is:\", mn.x[0], \"cts/cm$^2$/s/sr\"",
"Best fit background parameters: [ 0.20091551 -0.05052423 0.00514508 0.02652786 0.01548994 0.08761685\n 0.02739853]\nBest fit signal+background parameters: [ 0.00607216 0.2010101 -0.05069341 0.00517223 0.02940035 0.01881783\n 0.09116903 0.03089509]\nThe Delta chi^2 between signal and null model is: 0.5487446049041864\nThe chi^2/DOF of the null-model fit is: 0.980518891711762\nNB: the best-fit signal strength in this case is: 0.00607216339159919 cts/cm$^2$/s/sr\n"
],
[
"fig = plt.figure(figsize=(10,8))\n\nplt.errorbar(avg_data(Energies,6),avg_data(data,6),yerr=np.sqrt(6*avg_data(data_yerrs**2,6))/6.,xerr=6*(Energies[1]-Energies[0])/2.,\n color=\"black\",marker=\"o\", fmt='none',capsize=4) \n\nplt.plot(Energies_reduced,mod_poly_four_lines(Energies_reduced,mn_null.x),\n 'k-',label =r\"Null P.L. model\")\nplt.plot(Energies_reduced,mod_poly_four_lines(Energies_reduced,mn.x[1:])+mn.x[0]*mod_UXL_reduced,\n 'r-',label =r\"Best fit signal model\")\n\nplt.xlim(3,4)\nplt.ylim(0.08,0.1)\n\nplt.xticks(fontsize=22)\nplt.yticks(fontsize=22)\nplt.xlabel(r\"$E$ [keV]\",fontsize=22)\nplt.ylabel(r\"Flux [cts/s/keV]\",fontsize=22)\nplt.legend(fontsize=22)\nplt.show()",
"_____no_output_____"
],
[
"A_sig_array = np.linspace(mn.x[0],0.05,100)\nchi2_sig_array = np.zeros(len(A_sig_array))\nbf = mn.x[1:]\nfor i in range(len(A_sig_array)):\n chi2_instance.fix_signal_strength(A_sig_array[i])\n mn_profile = minimize(chi2_instance.chi2_fixed_signal,bf,method='Nelder-Mead',\n options={'fatol':1e-10,'xatol':1e-10,'adaptive':True})\n bf = mn_profile.x\n chi2_sig_array[i] = mn_profile.fun\n\namin = np.argmin((chi2_sig_array-chi2_sig_array[0] - 2.71)**2)\nlimit_signal_strength = A_sig_array[amin]\nprint \"The 95% upper limit on the signal flux is\", limit_signal_strength, \"cts/cm^2/s/sr\"\nprint \"This corresponds to a limit on sin^2(2theta) of\", return_sin_theta_lim(EUXL,limit_signal_strength,D_signal)",
"The 95% upper limit on the signal flux is 0.02781422393515111 cts/cm^2/s/sr\nThis corresponds to a limit on sin^2(2theta) of 2.487305045147695e-11\n"
]
],
[
[
"This change to the background continuum model does not change any conclusions. The 3.5 keV line is in tension with these limits.",
"_____no_output_____"
],
[
"## Subtract the background data\n\nNow, we subtract off the data taken far away from the Galactic Center. We use a folded powerlaw for the background continuum under the assumption that the residual flux in the signal region should be astrophysical.",
"_____no_output_____"
]
],
[
[
"# A folded powerlaw function\ndef folded_PL(A,n):\n mod_F = np.matmul(det_res,A*Energies**n) \n return mod_F\n\ndef mod_folded_power_four_lines(ens,x):\n A, n,S1, S2, S3, S4 = x\n return folded_PL(A,n)[whs_reduced] + S1*mod_3p3[whs_reduced] + S2*mod_3p7[whs_reduced]+ S3*mod_3p1[whs_reduced] + S4*mod_3p9[whs_reduced]\n\n\nchi2_instance = chi2(Energies_reduced,data_reduced- data_bkg[whs_reduced],np.sqrt(data_yerrs_reduced**2+data_yerrs_bkg_reduced**2),mod_folded_power_four_lines,mod_UXL_reduced)\n\n\nx0 = np.array([1.80533176e-02, -5.18514882e-01, 9.80776897e-03, 1.45353856e-04, 6.39560515e-02, 1.84053386e-02])\nbounds = np.array([[0.0,0.1],[-2,0],[0,0.1],[0,0.2],[0,0.2],[0,0.2]])\nmn_null = dual_annealing(chi2_instance.chi2_null,x0=x0,bounds=bounds,local_search_options={\"method\": \"Nelder-Mead\"},seed=1234,maxiter=1000)\n\nboundss = np.array([[-0.5,0.5],[0.0,0.1],[-2,0],[0,0.1],[0,0.2],[0,0.2],[0,0.2]])\nx0s=np.array([1.e-2,mn_null.x[0],mn_null.x[1],mn_null.x[2],mn_null.x[3],mn_null.x[4],mn_null.x[5]])\nmn = dual_annealing(chi2_instance.chi2,x0=x0s,bounds=boundss,local_search_options={\"method\": \"Nelder-Mead\"},seed=1234,maxiter=1000)\n\nprint \"Best fit background parameters:\", mn_null.x\nprint \"Best fit signal+background parameters:\", mn.x\n\nprint \"The Delta chi^2 between signal and null model is:\", mn_null.fun - mn.fun\nprint \"The chi^2/DOF of the null-model fit is:\", mn_null.fun/(len(whs_reduced)-6.)\n\nprint \"NB: the best-fit signal strength in this case is:\", mn.x[0], \"cts/cm$^2$/s/sr\"",
"Best fit background parameters: [ 1.80533176e-02 -5.18652456e-01 9.90575404e-03 1.02427156e-04\n 6.39560515e-02 1.86563894e-02]\nBest fit signal+background parameters: [ 1.44916562e-03 1.60687313e-02 -4.29522829e-01 1.13028621e-02\n 3.83633951e-05 6.65749498e-02 1.75828890e-02]\nThe Delta chi^2 between signal and null model is: 0.10125021484452645\nThe chi^2/DOF of the null-model fit is: 1.0259200982344832\nNB: the best-fit signal strength in this case is: 0.0014491656186800237 cts/cm$^2$/s/sr\n"
],
[
"fig = plt.figure(figsize=(10,6))\n\nplt.errorbar(avg_data(Energies,6),avg_data(data-data_bkg,6),yerr=np.sqrt(6*avg_data(data_yerrs**2+data_yerrs_bkg**2,6))/6.,xerr=6*(Energies[1]-Energies[0])/2.,\n color=\"black\",marker=\"o\", fmt='none',capsize=4) #label=\"data\"\n\nplt.plot(Energies_reduced,mod_folded_power_four_lines(Energies_reduced,mn_null.x),\n 'k-',label =r\"Null model\")\nplt.plot(Energies_reduced,mod_folded_power_four_lines(Energies_reduced,mn.x[1:])+mn.x[0]*mod_UXL_reduced,\n 'r-',label =r\"Best fit signal model\")\n\nplt.xlim(3,4)\nplt.ylim(0.006,0.015)\nplt.xticks(fontsize=22)\nplt.yticks(fontsize=22)\nplt.xlabel(r\"$E$ [keV]\",fontsize=22)\nplt.ylabel(r\"SR Flux [cts/s/keV]\",fontsize=22)\nplt.legend(fontsize=22)\nplt.show()",
"_____no_output_____"
],
[
"A_sig_array = np.linspace(mn.x[0],0.05,100)\nchi2_sig_array = np.zeros(len(A_sig_array))\nbf = mn.x[1:]\nfor i in range(len(A_sig_array)):\n chi2_instance.fix_signal_strength(A_sig_array[i])\n mn_profile = minimize(chi2_instance.chi2_fixed_signal,bf,method='Nelder-Mead')\n bf = mn_profile.x\n chi2_sig_array[i] = mn_profile.fun\n\namin = np.argmin((chi2_sig_array-chi2_sig_array[0] - 2.71)**2)\nlimit_signal_strength = A_sig_array[amin]\n\nprint \"The 95% upper limit on the signal flux is\", limit_signal_strength, \"cts/cm^2/s/sr\"\nprint \"This corresponds to a limit on sin^2(2theta) of\", return_sin_theta_lim(EUXL,limit_signal_strength,D_signal-D_bkg)",
"The 95% upper limit on the signal flux is 0.01567112720512729 cts/cm^2/s/sr\nThis corresponds to a limit on sin^2(2theta) of 2.476370769990894e-11\n"
]
],
[
[
"In this version of the analysis, too, we see no evidence for a 3.5 keV line and obtain comparable limits as in the stacked analyses in the previous sections.\n\n# Include the Quiescent Particle Background (QPB)\n\nNow we will do a joint likelihood including the QPB data. The QPB data is complicated because the data is correlated from observation to observation. Thus, summing the data leads to correlated uncertainties. To account for this, we will estimate the uncertainties on the QPB data in a data-driven way by fixing the normalization of the $\\chi^2$ function such that the powerlaw gives the expected $\\chi^2/{\\rm DOF}$. We note that this is just an approximation, which is not necessary within the context of the full joint likelihood framework.",
"_____no_output_____"
]
],
[
[
"# We are going to fix a powerlaw to the QPB data and then renormalize the chi^2 function\ndef PL(A,n,ens):\n return A*ens**n\n\ndef chi2_QPB_UN(x):\n A,n = x\n mod = PL(A,n,Energies_reduced)\n return np.sum((mod-QPB[whs_reduced])**2)\n\nmn_QPB = minimize(chi2_QPB_UN,[0.084,-0.20],method=\"Nelder-Mead\")\nbf_QPB=mn_QPB.x\n\nchi2_not_reduced = chi2_QPB_UN(bf_QPB)\n\n# The function below has the expected normalization\nchi2_QPB = lambda x: chi2_QPB_UN(x)/chi2_not_reduced*((len(QPB[whs_reduced])-2.))\n\n\nfig = plt.figure(figsize=(10,8))\nplt.scatter(Energies_reduced,QPB[whs_reduced],marker=\"o\",color=\"black\")\nplt.plot(Energies_reduced,PL(bf_QPB[0],bf_QPB[1],Energies_reduced),'r-',label=\"best-fit P.L.\")\n\nplt.xlim(3,4)\nplt.ylim(0.04,0.065)\nplt.xlabel(r\"$E$ [keV]\",fontsize=22)\nplt.ylabel(r\"QPB [cts/s/keV]\",fontsize=22)\nplt.legend(fontsize=22)\nplt.xticks(fontsize=22)\nplt.yticks(fontsize=22)\nplt.show()",
"_____no_output_____"
],
[
"def mod_2power_four_lines(ens,x):\n AQPB, nQPB,A, n,S1, S2, S3, S4 = x\n return PL(AQPB,nQPB,ens)+ folded_PL(A,n)[whs_reduced] + S1*mod_3p3[whs_reduced] + S2*mod_3p7[whs_reduced]+ S3*mod_3p1[whs_reduced] + S4*mod_3p9[whs_reduced]\n\nchi2_instance = chi2(Energies_reduced,data_reduced,data_yerrs_reduced,mod_2power_four_lines,mod_UXL_reduced)\n\n\nx0 = np.array([0.07377512 ,-0.28001362 , 0.15844243, -1.07912658 , 0.02877547,\n 0.01134023 , 0.08755627 , 0.03134949])\nbounds = np.array([[0.75*bf_QPB[0],1.25*bf_QPB[0]],[-1,0],[0.0001,2.0],[-3,0],[0,0.1],[0,0.1],[0,0.1],[0,0.1]])\n\n# Below is the joint likelihood for the null model\ndef joint_chi2(x):\n return chi2_QPB(x[:2])+chi2_instance.chi2_null(x)\n\nmn_null = dual_annealing(joint_chi2,x0=x0,bounds=bounds,local_search_options={\"method\": \"Nelder-Mead\"},seed=1234,maxiter=1000)\n\n# Below is the joint likelihood for the signal model\ndef joint_chi2_sig(x):\n return chi2_QPB(x[1:3])+chi2_instance.chi2(x)\nboundss = np.array([[-0.5,0.5],[0.5*bf_QPB[0],2*bf_QPB[0]],[-1,0],[0.0001,2.0],[-3,0],[0,0.1],[0,0.1],[0,0.1],[0,0.1]])\nx0s=np.array([1.e-2,mn_null.x[0],mn_null.x[1],mn_null.x[2],mn_null.x[3],mn_null.x[4],mn_null.x[5],mn_null.x[6],mn_null.x[7]])\nmn = dual_annealing(joint_chi2_sig,x0=x0s,bounds=boundss,local_search_options={\"method\": \"Nelder-Mead\"},seed=1234,maxiter=1000)\n\nprint \"The Delta chi^2 between signal and null model is:\", mn_null.fun - mn.fun\nprint \"NB: the best-fit signal strength in this case is:\", mn.x[0], \"cts/cm$^2$/s/sr\"",
"The Delta chi^2 between signal and null model is: 0.45016371056283333\nNB: the best-fit signal strength in this case is: 0.005545658335402866 cts/cm$^2$/s/sr\n"
],
[
"fig = plt.figure(figsize=(10,8))\n\nplt.errorbar(avg_data(Energies,6),avg_data(data,6),yerr=np.sqrt(6*avg_data(data_yerrs**2,6))/6.,xerr=6*(Energies[1]-Energies[0])/2.,\n color=\"black\",marker=\"o\", fmt='none',capsize=4) #label=\"data\"\n\nplt.plot(Energies_reduced,mod_2power_four_lines(Energies_reduced,mn.x[1:])+mn.x[0]*mod_UXL_reduced,\n 'r-',label =r\"Best fit signal model\")\nx0 = np.array([bf_QPB[0],bf_QPB[1], 0.064218, -0.4306988 , 0.02542355 , 0.01451921 , 0.09027154, 0.03331636])\nplt.plot(Energies_reduced,mod_2power_four_lines(Energies_reduced,mn_null.x),\n 'k-',label =r\"Null P.L. model\")\n\nplt.xlim(3,4)\n\nplt.ylim(0.08,0.1)\n\nplt.xticks(fontsize=22)\nplt.yticks(fontsize=22)\nplt.xlabel(r\"$E$ [keV]\",fontsize=22)\nplt.ylabel(r\"Flux [cts/s/keV]\",fontsize=22)\nplt.legend(fontsize=22)\nplt.show()",
"_____no_output_____"
],
[
"A_sig_array = np.linspace(mn.x[0],0.05,100)\nchi2_sig_array = np.zeros(len(A_sig_array))\nbf = mn.x[1:]\nfor i in range(len(A_sig_array)):\n chi2_instance.fix_signal_strength(A_sig_array[i])\n mn_profile = minimize(chi2_instance.chi2_fixed_signal,bf,method='Nelder-Mead')\n bf = mn_profile.x\n chi2_sig_array[i] = mn_profile.fun\n\namin = np.argmin((chi2_sig_array-chi2_sig_array[0] - 2.71)**2)\nlimit_signal_strength = A_sig_array[amin]\n\nprint \"The 95% upper limit on the signal flux is\", limit_signal_strength, \"cts/cm^2/s/sr\"\nprint \"This corresponds to a limit on sin^2(2theta) of\", return_sin_theta_lim(EUXL,limit_signal_strength,D_signal)",
"The 95% upper limit on the signal flux is 0.019016670961038363 cts/cm^2/s/sr\nThis corresponds to a limit on sin^2(2theta) of 1.700578155032655e-11\n"
]
],
[
[
"Finally, including the QPB in our analysis does not significantly change the results.\n\n# Summary\n\nTo summarize, we see no evidence of a 3.5 keV line in any of our analysis variations here. We obtain the following limits on $\\sin^2(2\\theta)$ for $E_{\\rm UXL} = 3.48$ keV:\n\n* Quadratic background fit within $E_{\\rm UXL} \\pm 0.25$ keV: $2.35 \\times 10^{-11}$\n\n* Power law background fit within $E_{\\rm UXL} \\pm 0.25$ keV: $1.82 \\times 10^{-11}$\n\n* Power law background fit from 3 to 4 keV: $1.34 \\times 10^{-11}$\n\n* Quadratic background fit from 3 to 4 keV: $2.45 \\times 10^{-11}$\n\n* Power law background fit on background-subtracted data from 3 to 4 keV: $1.87 \\times 10^{-11}$\n\n* Power law background fit with joint (X-ray + QPB) likelihood from 3 to 4 keV: $1.68 \\times 10^{-11}$\n\nAlthough these limits are much weaker than our fiducial limit presented in DRS20, they still strongly constrain the 3.5 keV line.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d03e1da35524f987332691e4a3aab3dcb2477ae6 | 58,157 | ipynb | Jupyter Notebook | hassio/HA-cloud-SQL usage.ipynb | WillKoehrsen/HASS-data-science | f4191054c9e437b47a908f054275e330fe4747c3 | [
"MIT"
] | 3 | 2018-01-09T09:53:06.000Z | 2021-07-26T07:05:53.000Z | hassio/HA-cloud-SQL usage.ipynb | WillKoehrsen/HASS-data-science | f4191054c9e437b47a908f054275e330fe4747c3 | [
"MIT"
] | null | null | null | hassio/HA-cloud-SQL usage.ipynb | WillKoehrsen/HASS-data-science | f4191054c9e437b47a908f054275e330fe4747c3 | [
"MIT"
] | 4 | 2018-01-26T03:23:13.000Z | 2018-08-15T13:16:31.000Z | 106.710092 | 1,582 | 0.650257 | [
[
[
"import pandas as pd\nfrom sqlalchemy import create_engine, text",
"_____no_output_____"
],
[
"settings = {\n 'user': 'hass',\n 'pass': 'password',\n 'host': '192.100.123.100',\n 'db': 'ha_db'\n}",
"_____no_output_____"
],
[
"url = 'postgresql+psycopg2://{user}:{pass}@{host}:5432/{db}'.format(**settings)\nurl",
"_____no_output_____"
],
[
"engine = create_engine(url, client_encoding='utf8')",
"_____no_output_____"
],
[
"sql = \"SELECT entity_id, COUNT(*) FROM states \\\nGROUP BY entity_id ORDER by 2 DESC\"\nprint('Performing query...')\ndf = pd.read_sql(sql, con=engine)",
"Performing query...\n"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df['entity_id'].unique()",
"_____no_output_____"
],
[
"%%time\ns = text(\"SELECT * FROM states\")\nallquery = engine.execute(s)\n\n# get rows from query into a pandas dataframe\nallqueryDF = pd.DataFrame(allquery.fetchall())",
"CPU times: user 354 ms, sys: 63.7 ms, total: 417 ms\nWall time: 3 s\n"
],
[
"allqueryDF.head()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03e2a208d518b02bcd7ec3255da41173c68bf2c | 793,359 | ipynb | Jupyter Notebook | frameworks/docker/itorch/tutorials/2_supervised_learning.ipynb | zhhengcs/sunny-side-up | 0ddefdd1adf1c7ea3f48b669ffc763c0678481a3 | [
"Apache-2.0"
] | 581 | 2015-06-24T15:04:18.000Z | 2022-02-19T23:22:01.000Z | frameworks/docker/itorch/tutorials/2_supervised_learning.ipynb | zhhengcs/sunny-side-up | 0ddefdd1adf1c7ea3f48b669ffc763c0678481a3 | [
"Apache-2.0"
] | 32 | 2015-06-22T21:29:11.000Z | 2020-06-26T22:16:39.000Z | frameworks/docker/itorch/tutorials/2_supervised_learning.ipynb | zhhengcs/sunny-side-up | 0ddefdd1adf1c7ea3f48b669ffc763c0678481a3 | [
"Apache-2.0"
] | 168 | 2015-06-02T18:19:25.000Z | 2020-12-28T19:25:07.000Z | 302.808779 | 203,510 | 0.915669 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d03e2c74fccf26951a8022478a73552aefe9bfe8 | 7,229 | ipynb | Jupyter Notebook | notebooks/multi_convnet_sentiment_classifier.ipynb | ewbolme/DLTFpT | 4841554b686bdf74f18c1ce3f2204513c0c9dccc | [
"MIT"
] | null | null | null | notebooks/multi_convnet_sentiment_classifier.ipynb | ewbolme/DLTFpT | 4841554b686bdf74f18c1ce3f2204513c0c9dccc | [
"MIT"
] | null | null | null | notebooks/multi_convnet_sentiment_classifier.ipynb | ewbolme/DLTFpT | 4841554b686bdf74f18c1ce3f2204513c0c9dccc | [
"MIT"
] | null | null | null | 24.756849 | 147 | 0.555679 | [
[
[
"# Multi-ConvNet Sentiment Classifier",
"_____no_output_____"
],
[
"In this notebook, we concatenate the outputs of *multiple, parallel convolutional layers* to classify IMDB movie reviews by their sentiment.",
"_____no_output_____"
],
[
"#### Load dependencies",
"_____no_output_____"
]
],
[
[
"import tensorflow\nfrom tensorflow.keras.datasets import imdb\nfrom tensorflow.keras.preprocessing.sequence import pad_sequences\nfrom tensorflow.keras.models import Model # new!\nfrom tensorflow.keras.layers import Input, concatenate # new! \nfrom tensorflow.keras.layers import Dense, Dropout, Embedding, SpatialDropout1D, Conv1D, GlobalMaxPooling1D\nfrom tensorflow.keras.callbacks import ModelCheckpoint\nimport os\nfrom sklearn.metrics import roc_auc_score \nimport matplotlib.pyplot as plt ",
"_____no_output_____"
]
],
[
[
"#### Set hyperparameters",
"_____no_output_____"
]
],
[
[
"# output directory name:\noutput_dir = 'model_output/multiconv'\n\n# training:\nepochs = 4\nbatch_size = 128\n\n# vector-space embedding: \nn_dim = 64\nn_unique_words = 5000 \nmax_review_length = 400\npad_type = trunc_type = 'pre'\ndrop_embed = 0.2 \n\n# convolutional layer architecture:\nn_conv_1 = n_conv_2 = n_conv_3 = 256 \nk_conv_1 = 3\nk_conv_2 = 2\nk_conv_3 = 4\n\n# dense layer architecture: \nn_dense = 256\ndropout = 0.2",
"_____no_output_____"
]
],
[
[
"#### Load data",
"_____no_output_____"
]
],
[
[
"(x_train, y_train), (x_valid, y_valid) = imdb.load_data(num_words=n_unique_words) ",
"_____no_output_____"
]
],
[
[
"#### Preprocess data",
"_____no_output_____"
]
],
[
[
"x_train = pad_sequences(x_train, maxlen=max_review_length, padding=pad_type, truncating=trunc_type, value=0)\nx_valid = pad_sequences(x_valid, maxlen=max_review_length, padding=pad_type, truncating=trunc_type, value=0)",
"_____no_output_____"
]
],
[
[
"#### Design neural network architecture",
"_____no_output_____"
]
],
[
[
"input_layer = Input(shape=(max_review_length,), \n dtype='int16', name='input') \n\n# embedding: \nembedding_layer = Embedding(n_unique_words, n_dim, \n name='embedding')(input_layer)\ndrop_embed_layer = SpatialDropout1D(drop_embed, \n name='drop_embed')(embedding_layer)\n\n# three parallel convolutional streams: \nconv_1 = Conv1D(n_conv_1, k_conv_1, \n activation='relu', name='conv_1')(drop_embed_layer)\nmaxp_1 = GlobalMaxPooling1D(name='maxp_1')(conv_1)\n\nconv_2 = Conv1D(n_conv_2, k_conv_2, \n activation='relu', name='conv_2')(drop_embed_layer)\nmaxp_2 = GlobalMaxPooling1D(name='maxp_2')(conv_2)\n\nconv_3 = Conv1D(n_conv_3, k_conv_3, \n activation='relu', name='conv_3')(drop_embed_layer)\nmaxp_3 = GlobalMaxPooling1D(name='maxp_3')(conv_3)\n\n# concatenate the activations from the three streams: \nconcat = concatenate([maxp_1, maxp_2, maxp_3])\n\n# dense hidden layers: \ndense_layer = Dense(n_dense, \n activation='relu', name='dense')(concat)\ndrop_dense_layer = Dropout(dropout, name='drop_dense')(dense_layer)\ndense_2 = Dense(int(n_dense/4), \n activation='relu', name='dense_2')(drop_dense_layer)\ndropout_2 = Dropout(dropout, name='drop_dense_2')(dense_2)\n\n# sigmoid output layer: \npredictions = Dense(1, activation='sigmoid', name='output')(dropout_2)\n\n# create model: \nmodel = Model(input_layer, predictions)",
"_____no_output_____"
],
[
"model.summary() ",
"_____no_output_____"
]
],
[
[
"#### Configure model",
"_____no_output_____"
]
],
[
[
"model.compile(loss='binary_crossentropy', optimizer='nadam', metrics=['accuracy'])",
"_____no_output_____"
],
[
"modelcheckpoint = ModelCheckpoint(filepath=output_dir+\"/weights.{epoch:02d}.hdf5\")\nif not os.path.exists(output_dir):\n os.makedirs(output_dir)",
"_____no_output_____"
]
],
[
[
"#### Train!",
"_____no_output_____"
]
],
[
[
"model.fit(x_train, y_train, batch_size=batch_size, epochs=epochs, verbose=1, validation_data=(x_valid, y_valid), callbacks=[modelcheckpoint])",
"_____no_output_____"
]
],
[
[
"#### Evaluate",
"_____no_output_____"
]
],
[
[
"model.load_weights(output_dir+\"/weights.02.hdf5\") ",
"_____no_output_____"
],
[
"y_hat = model.predict(x_valid)",
"_____no_output_____"
],
[
"plt.hist(y_hat)\n_ = plt.axvline(x=0.5, color='orange')",
"_____no_output_____"
],
[
"\"{:0.2f}\".format(roc_auc_score(y_valid, y_hat)*100.0)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d03e31beef0142d74434646b47c3041442cb3ebd | 33,114 | ipynb | Jupyter Notebook | documents/Intermediate/FactoryPlanning1&2/factory_planning_1.ipynb | biancaitian/gurobi-official-examples | 175e1d0aa88dffde644fac188c48c7d3c185d007 | [
"Apache-2.0"
] | 4 | 2021-08-01T11:50:14.000Z | 2022-03-13T01:49:24.000Z | documents/Intermediate/FactoryPlanning1&2/factory_planning_1.ipynb | biancaitian/gurobi-official-examples | 175e1d0aa88dffde644fac188c48c7d3c185d007 | [
"Apache-2.0"
] | null | null | null | documents/Intermediate/FactoryPlanning1&2/factory_planning_1.ipynb | biancaitian/gurobi-official-examples | 175e1d0aa88dffde644fac188c48c7d3c185d007 | [
"Apache-2.0"
] | 4 | 2021-04-14T06:58:00.000Z | 2022-03-15T10:37:40.000Z | 37.374718 | 680 | 0.477653 | [
[
[
"# 工厂规划\n\n等级:中级\n\n## 目的和先决条件\n\n\n此模型和Factory Planning II都是生产计划问题的示例。在生产计划问题中,必须选择要生产哪些产品,要生产多少产品以及要使用哪些资源,以在满足一系列限制的同时最大化利润或最小化成本。这些问题在广泛的制造环境中都很常见。\n\n### What You Will Learn\n\n在此特定示例中,我们将建模并解决生产组合问题:在每个阶段中,我们可以制造一系列产品。每种产品在不同的机器上生产需要不同的时间,并产生不同的利润。目的是创建最佳的多周期生产计划,以使利润最大化。由于维护,某些机器在特定时期内不可用。由于市场限制,每个产品每个月的销售量都有上限,并且存储容量也受到限制。\n\nIn Factory Planning II, we’ll add more complexity to this example; the month in which each machine is down for maintenance will be chosen as a part of the optimized plan.\n\nMore information on this type of model can be found in example # 3 of the fifth edition of Modeling Building in Mathematical Programming by H. P. Williams on pages 255-256 and 300-302.\n\nThis modeling example is at the intermediate level, where we assume that you know Python and are familiar with the Gurobi Python API. In addition, you should have some knowledge about building mathematical optimization models.\n\n**Note:** You can download the repository containing this and other examples by clicking [here](https://github.com/Gurobi/modeling-examples/archive/master.zip). In order to run this Jupyter Notebook properly, you must have a Gurobi license. If you do not have one, you can request an [evaluation license](https://www.gurobi.com/downloads/request-an-evaluation-license/?utm_source=Github&utm_medium=website_JupyterME&utm_campaign=CommercialDataScience) as a *commercial user*, or download a [free license](https://www.gurobi.com/academia/academic-program-and-licenses/?utm_source=Github&utm_medium=website_JupyterME&utm_campaign=AcademicDataScience) as an *academic user*.\n\n---\n## Problem Description\n\nA factory makes seven products (Prod 1 to Prod 7) using a range of machines including:\n\n- Four grinders\n- Two vertical drills\n- Three horizontal drills\n- One borer\n- One planer\n\nEach product has a defined profit contribution per unit sold (defined as the sales price per unit minus the cost of raw materials). In addition, the manufacturing of each product requires a certain amount of time on each machine (in hours). The contribution and manufacturing time value are shown below. A dash indicates that the manufacturing process for the given product does not require that machine.\n\n| <i></i> | PROD1 | PROD2 | PROD3 | PROD4 | PROD5 | PROD6 | PROD7 |\n| --- | --- | --- | --- | --- | --- | --- | --- |\n| Profit | 10 | 6 | 8 | 4 | 11 | 9 | 3 |\n| Grinding | 0.5 | 0.7 | - | - | 0.3 | 0.2 | 0.5 |\n| Vertical Drilling | 0.1 | 0.2 | - | 0.3 | - | 0.6 | - |\n| Horizontal Drilling | 0.2 | - | 0.8 | - | - | - | 0.6 |\n| Boring | 0.05 | 0.03 | - | 0.07 | 0.1 | - | 0.08 |\n| Planning | - | - | 0.01 | - | 0.05 | - | 0.05 |\n\nIn each of the six months covered by this model, one or more of the machines is scheduled to be down for maintenance and as a result will not be available to use for production that month. The maintenance schedule is as follows:\n\n| Month | Machine |\n| --- | --- |\n| January | One grinder |\n| February | Two horizontal drills |\n| March | One borer |\n| April | One vertical drill |\n| May | One grinder and one vertical drill |\n| June | One horizontal drill |\n\nThere are limitations on how many of each product can be sold in a given month. These limits are shown below:\n\n| Month | PROD1 | PROD2 | PROD3 | PROD4 | PROD5 | PROD6 | PROD7 |\n| --- | --- | --- | --- | --- | --- | --- | --- |\n| January | 500 | 1000 | 300 | 300 | 800 | 200 | 100 |\n| February | 600 | 500 | 200 | 0 | 400 | 300 | 150 |\n| March | 300 | 600 | 0 | 0 | 500 | 400 | 100 |\n| April | 200 | 300 | 400 | 500 | 200 | 0 | 100 |\n| May | 0 | 100 | 500 | 100 | 1000 | 300 | 0 |\n| June | 500 | 500 | 100 | 300 | 1100 | 500 | 60 |\n\nUp to 100 units of each product may be stored in inventory at a cost of $\\$0.50$ per unit per month. At the start of January, there is no product inventory. However, by the end of June, there should be 50 units of each product in inventory.\n\nThe factory produces products six days a week using two eight-hour shifts per day. It may be assumed that each month consists of 24 working days. Also, for the purposes of this model, there are no production sequencing issues that need to be taken into account.\n\nWhat should the production plan look like? Also, is it possible to recommend any price increases and determine the value of acquiring any new machines?\n\nThis problem is based on a larger model built for the Cornish engineering company of Holman Brothers.\n\n---\n## Model Formulation\n\n### Sets and Indices\n\n$t \\in \\text{Months}=\\{\\text{Jan},\\text{Feb},\\text{Mar},\\text{Apr},\\text{May},\\text{Jun}\\}$: Set of months.\n\n$p \\in \\text{Products}=\\{1,2,\\dots,7\\}$: Set of products.\n\n$m \\in \\text{Machines}=\\{\\text{Grinder},\\text{VertDrill},\\text{horiDrill},\\text{Borer},\\text{Planer}\\}$: Set of machines.\n\n### Parameters\n\n$\\text{hours_per_month} \\in \\mathbb{R}^+$: Time (in hours/month) available at any machine on a monthly basis. It results from multiplying the number of working days (24 days) by the number of shifts per day (2) by the duration of a shift (8 hours).\n\n$\\text{max_inventory} \\in \\mathbb{N}$: Maximum number of units of a single product type that can be stored in inventory at any given month.\n\n$\\text{holding_cost} \\in \\mathbb{R}^+$: Monthly cost (in USD/unit/month) of keeping in inventory a unit of any product type.\n\n$\\text{store_target} \\in \\mathbb{N}$: Number of units of each product type to keep in inventory at the end of the planning horizon.\n\n$\\text{profit}_p \\in \\mathbb{R}^+$: Profit (in USD/unit) of product $p$.\n\n$\\text{installed}_m \\in \\mathbb{N}$: Number of machines of type $m$ installed in the factory.\n\n$\\text{down}_{t,m} \\in \\mathbb{N}$: Number of machines of type $m$ scheduled for maintenance at month $t$.\n\n$\\text{time_req}_{m,p} \\in \\mathbb{R}^+$: Time (in hours/unit) needed on machine $m$ to manufacture one unit of product $p$.\n\n$\\text{max_sales}_{t,p} \\in \\mathbb{N}$: Maximum number of units of product $p$ that can be sold at month $t$.\n\n\n### Decision Variables\n\n$\\text{make}_{t,p} \\in \\mathbb{R}^+$: Number of units of product $p$ to manufacture at month $t$.\n\n$\\text{store}_{t,p} \\in [0, \\text{max_inventory}] \\subset \\mathbb{R}^+$: Number of units of product $p$ to store at month $t$.\n\n$\\text{sell}_{t,p} \\in [0, \\text{max_sales}_{t,p}] \\subset \\mathbb{R}^+$: Number of units of product $p$ to sell at month $t$.\n\n**Assumption:** We can produce fractional units.\n\n### Objective Function\n\n- **Profit:** Maximize the total profit (in USD) of the planning horizon.\n\n\\begin{equation}\n\\text{Maximize} \\quad Z = \\sum_{t \\in \\text{Months}}\\sum_{p \\in \\text{Products}}\n(\\text{profit}_p*\\text{make}_{t,p} - \\text{holding_cost}*\\text{store}_{t,p})\n\\tag{0}\n\\end{equation}\n\n### Constraints\n\n- **Initial Balance:** For each product $p$, the number of units produced should be equal to the number of units sold plus the number stored (in units of product).\n\n\\begin{equation}\n\\text{make}_{\\text{Jan},p} = \\text{sell}_{\\text{Jan},p} + \\text{store}_{\\text{Jan},p} \\quad \\forall p \\in \\text{Products}\n\\tag{1}\n\\end{equation}\n\n- **Balance:** For each product $p$, the number of units produced in month $t$ and the ones previously stored should be equal to the number of units sold and stored in that month (in units of product).\n\n\\begin{equation}\n\\text{store}_{t-1,p} + \\text{make}_{t,p} = \\text{sell}_{t,p} + \\text{store}_{t,p} \\quad \\forall (t,p) \\in \\text{Months} \\setminus \\{\\text{Jan}\\} \\times \\text{Products}\n\\tag{2}\n\\end{equation}\n\n- **Inventory Target:** The number of units of product $p$ kept in inventory at the end of the planning horizon should hit the target (in units of product).\n\n\\begin{equation}\n\\text{store}_{\\text{Jun},p} = \\text{store_target} \\quad \\forall p \\in \\text{Products}\n\\tag{3}\n\\end{equation}\n- **Machine Capacity:** Total time used to manufacture any product at machine type $m$ cannot exceed its monthly capacity (in hours).\n\n\\begin{equation}\n\\sum_{p \\in \\text{Products}}\\text{time_req}_{m,p}*\\text{make}_{t,p} \\leq \\text{hours_per_month}*(\\text{installed}_m - \\text{down}_{t,m}) \\quad \\forall (t,m) \\in \\text{Months} \\times \\text{Machines}\n\\tag{4}\n\\end{equation}\n\n---\n## Python Implementation\n\nWe import the Gurobi Python Module and other Python libraries.",
"_____no_output_____"
]
],
[
[
"import gurobipy as gp\nimport numpy as np\nimport pandas as pd\nfrom gurobipy import GRB\n\n# tested with Python 3.7.0 & Gurobi 9.0",
"_____no_output_____"
]
],
[
[
"### Input Data\nWe define all the input data of the model.",
"_____no_output_____"
]
],
[
[
"# Parameters\n\nproducts = [\"Prod1\", \"Prod2\", \"Prod3\", \"Prod4\", \"Prod5\", \"Prod6\", \"Prod7\"]\nmachines = [\"grinder\", \"vertDrill\", \"horiDrill\", \"borer\", \"planer\"]\nmonths = [\"Jan\", \"Feb\", \"Mar\", \"Apr\", \"May\", \"Jun\"]\n\nprofit = {\"Prod1\":10, \"Prod2\":6, \"Prod3\":8, \"Prod4\":4, \"Prod5\":11, \"Prod6\":9, \"Prod7\":3}\n\ntime_req = {\n \"grinder\": { \"Prod1\": 0.5, \"Prod2\": 0.7, \"Prod5\": 0.3,\n \"Prod6\": 0.2, \"Prod7\": 0.5 },\n \"vertDrill\": { \"Prod1\": 0.1, \"Prod2\": 0.2, \"Prod4\": 0.3,\n \"Prod6\": 0.6 },\n \"horiDrill\": { \"Prod1\": 0.2, \"Prod3\": 0.8, \"Prod7\": 0.6 },\n \"borer\": { \"Prod1\": 0.05,\"Prod2\": 0.03,\"Prod4\": 0.07,\n \"Prod5\": 0.1, \"Prod7\": 0.08 },\n \"planer\": { \"Prod3\": 0.01,\"Prod5\": 0.05,\"Prod7\": 0.05 }\n}\n\n\n# number of machines down\ndown = {(\"Jan\",\"grinder\"): 1, (\"Feb\", \"horiDrill\"): 2, (\"Mar\", \"borer\"): 1,\n (\"Apr\", \"vertDrill\"): 1, (\"May\", \"grinder\"): 1, (\"May\", \"vertDrill\"): 1,\n (\"Jun\", \"planer\"): 1, (\"Jun\", \"horiDrill\"): 1}\n\n# number of each machine available\ninstalled = {\"grinder\":4, \"vertDrill\":2, \"horiDrill\":3, \"borer\":1, \"planer\":1} \n\n# market limitation of sells\nmax_sales = {\n (\"Jan\", \"Prod1\") : 500,\n (\"Jan\", \"Prod2\") : 1000,\n (\"Jan\", \"Prod3\") : 300,\n (\"Jan\", \"Prod4\") : 300,\n (\"Jan\", \"Prod5\") : 800,\n (\"Jan\", \"Prod6\") : 200,\n (\"Jan\", \"Prod7\") : 100,\n (\"Feb\", \"Prod1\") : 600,\n (\"Feb\", \"Prod2\") : 500,\n (\"Feb\", \"Prod3\") : 200,\n (\"Feb\", \"Prod4\") : 0,\n (\"Feb\", \"Prod5\") : 400,\n (\"Feb\", \"Prod6\") : 300,\n (\"Feb\", \"Prod7\") : 150,\n (\"Mar\", \"Prod1\") : 300,\n (\"Mar\", \"Prod2\") : 600,\n (\"Mar\", \"Prod3\") : 0,\n (\"Mar\", \"Prod4\") : 0,\n (\"Mar\", \"Prod5\") : 500,\n (\"Mar\", \"Prod6\") : 400,\n (\"Mar\", \"Prod7\") : 100,\n (\"Apr\", \"Prod1\") : 200,\n (\"Apr\", \"Prod2\") : 300,\n (\"Apr\", \"Prod3\") : 400,\n (\"Apr\", \"Prod4\") : 500,\n (\"Apr\", \"Prod5\") : 200,\n (\"Apr\", \"Prod6\") : 0,\n (\"Apr\", \"Prod7\") : 100,\n (\"May\", \"Prod1\") : 0,\n (\"May\", \"Prod2\") : 100,\n (\"May\", \"Prod3\") : 500,\n (\"May\", \"Prod4\") : 100,\n (\"May\", \"Prod5\") : 1000,\n (\"May\", \"Prod6\") : 300,\n (\"May\", \"Prod7\") : 0,\n (\"Jun\", \"Prod1\") : 500,\n (\"Jun\", \"Prod2\") : 500,\n (\"Jun\", \"Prod3\") : 100,\n (\"Jun\", \"Prod4\") : 300,\n (\"Jun\", \"Prod5\") : 1100,\n (\"Jun\", \"Prod6\") : 500,\n (\"Jun\", \"Prod7\") : 60,\n}\n\nholding_cost = 0.5\nmax_inventory = 100\nstore_target = 50\nhours_per_month = 2*8*24",
"_____no_output_____"
]
],
[
[
"## Model Deployment\nWe create a model and the variables. For each product (seven kinds of products) and each time period (month), we will create variables for the amount of which products get manufactured, held, and sold. In each month, there is an upper limit on the amount of each product that can be sold. This is due to market limitations.",
"_____no_output_____"
]
],
[
[
"factory = gp.Model('Factory Planning I')\n\nmake = factory.addVars(months, products, name=\"Make\") # quantity manufactured\nstore = factory.addVars(months, products, ub=max_inventory, name=\"Store\") # quantity stored\nsell = factory.addVars(months, products, ub=max_sales, name=\"Sell\") # quantity sold",
"Using license file c:\\gurobi\\gurobi.lic\nSet parameter TokenServer to value SANTOS-SURFACE-\n"
]
],
[
[
"Next, we insert the constraints. The balance constraints ensure that the amount of product that is in storage in the prior month plus the amount that gets manufactured equals the amount that is sold and held for each product in the current month. This ensures that all products in the model are manufactured in some month. The initial storage is empty.",
"_____no_output_____"
]
],
[
[
"#1. Initial Balance\nBalance0 = factory.addConstrs((make[months[0], product] == sell[months[0], product] \n + store[months[0], product] for product in products), name=\"Initial_Balance\")\n \n#2. Balance\nBalance = factory.addConstrs((store[months[months.index(month) -1], product] + \n make[month, product] == sell[month, product] + store[month, product] \n for product in products for month in months \n if month != months[0]), name=\"Balance\")",
"_____no_output_____"
]
],
[
[
"The Inventory Target constraints force that at the end of the last month the storage contains the specified amount of each product.",
"_____no_output_____"
]
],
[
[
"#3. Inventory Target\nTargetInv = factory.addConstrs((store[months[-1], product] == store_target for product in products), name=\"End_Balance\")",
"_____no_output_____"
]
],
[
[
"The capacity constraints ensure that, for each month, the time all products require on a certain kind of machine is less than or equal to the available hours for that type of machine in that month multiplied by the number of available machines in that period. Each product requires some machine hours on different machines. Each machine is down in one or more months due to maintenance, so the number and type of available machines varies per month. There can be multiple machines per machine type.",
"_____no_output_____"
]
],
[
[
"#4. Machine Capacity\n\nMachineCap = factory.addConstrs((gp.quicksum(time_req[machine][product] * make[month, product]\n for product in time_req[machine])\n <= hours_per_month * (installed[machine] - down.get((month, machine), 0))\n for machine in machines for month in months),\n name = \"Capacity\")",
"_____no_output_____"
]
],
[
[
"The objective is to maximize the profit of the company, which consists of\nthe profit for each product minus the cost for storing the unsold products. This can be stated as:",
"_____no_output_____"
]
],
[
[
"#0. Objective Function\nobj = gp.quicksum(profit[product] * sell[month, product] - holding_cost * store[month, product] \n for month in months for product in products)\n\nfactory.setObjective(obj, GRB.MAXIMIZE)",
"_____no_output_____"
]
],
[
[
"Next, we start the optimization and Gurobi finds the optimal solution.",
"_____no_output_____"
]
],
[
[
"factory.optimize()",
"Gurobi Optimizer version 9.0.0 build v9.0.0rc2 (win64)\nOptimize a model with 79 rows, 126 columns and 288 nonzeros\nModel fingerprint: 0xead11e9d\nCoefficient statistics:\n Matrix range [1e-02, 1e+00]\n Objective range [5e-01, 1e+01]\n Bounds range [6e+01, 1e+03]\n RHS range [5e+01, 2e+03]\nPresolve removed 74 rows and 110 columns\nPresolve time: 0.01s\nPresolved: 5 rows, 16 columns, 21 nonzeros\n\nIteration Objective Primal Inf. Dual Inf. Time\n 0 1.2466500e+05 3.640000e+02 0.000000e+00 0s\n 2 9.3715179e+04 0.000000e+00 0.000000e+00 0s\n\nSolved in 2 iterations and 0.01 seconds\nOptimal objective 9.371517857e+04\n"
]
],
[
[
"---\n## Analysis\n\nThe result of the optimization model shows that the maximum profit we can achieve is $\\$93,715.18$.\nLet's see the solution that achieves that optimal result.\n\n### Production Plan\n\nThis plan determines the amount of each product to make at each period of the planning horizon. For example, in February we make 700 units of product Prod1.",
"_____no_output_____"
]
],
[
[
"rows = months.copy()\ncolumns = products.copy()\nmake_plan = pd.DataFrame(columns=columns, index=rows, data=0.0)\n\nfor month, product in make.keys():\n if (abs(make[month, product].x) > 1e-6):\n make_plan.loc[month, product] = np.round(make[month, product].x, 1)\nmake_plan",
"_____no_output_____"
]
],
[
[
"### Sales Plan\n\nThis plan defines the amount of each product to sell at each period of the planning horizon. For example, in February we sell 600 units of product Prod1.",
"_____no_output_____"
]
],
[
[
"rows = months.copy()\ncolumns = products.copy()\nsell_plan = pd.DataFrame(columns=columns, index=rows, data=0.0)\n\nfor month, product in sell.keys():\n if (abs(sell[month, product].x) > 1e-6):\n sell_plan.loc[month, product] = np.round(sell[month, product].x, 1)\nsell_plan",
"_____no_output_____"
]
],
[
[
"### Inventory Plan\n\nThis plan reflects the amount of product in inventory at the end of each period of the planning horizon. For example, at the end of February we have 100 units of Prod1 in inventory.",
"_____no_output_____"
]
],
[
[
"rows = months.copy()\ncolumns = products.copy()\nstore_plan = pd.DataFrame(columns=columns, index=rows, data=0.0)\n\nfor month, product in store.keys():\n if (abs(store[month, product].x) > 1e-6):\n store_plan.loc[month, product] = np.round(store[month, product].x, 1)\nstore_plan",
"_____no_output_____"
]
],
[
[
"**Note:** If you want to write your solution to a file, rather than print it to the terminal, you can use the model.write() command. An example implementation is:\n\n`factory.write(\"factory-planning-1-output.sol\")`\n\n---\n## References\n\nH. Paul Williams, Model Building in Mathematical Programming, fifth edition.\n\nCopyright © 2020 Gurobi Optimization, LLC",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d03e31fa6a88b7cddcfe13d5bba576402477baf7 | 2,072 | ipynb | Jupyter Notebook | 02_25_20_working_with_smile_strings.ipynb | pmont056/62k-crystals | 1384089210fedd87282bf06b5c949cce22098880 | [
"MIT"
] | 1 | 2020-02-25T23:10:12.000Z | 2020-02-25T23:10:12.000Z | 02_25_20_working_with_smile_strings.ipynb | pmont056/61k-crystals | 1384089210fedd87282bf06b5c949cce22098880 | [
"MIT"
] | 15 | 2020-02-26T00:02:51.000Z | 2020-05-09T21:58:38.000Z | 02_25_20_working_with_smile_strings.ipynb | pmont056/61k-crystals | 1384089210fedd87282bf06b5c949cce22098880 | [
"MIT"
] | null | null | null | 20.116505 | 113 | 0.508687 | [
[
[
"# This is the first cell of code, importing the necessary packages.\n# Should this raise an error on your system, you might still need to install these packages, see above. \n\nimport os\nfrom copy import deepcopy\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\npd.set_option('display.width',5000)\npd.set_option('display.max_columns',200)",
"_____no_output_____"
],
[
"df_5k = pd.read_json(\"C:/Users/koolk/Downloads/m1507656/df_5k.json\", orient='split') \nprint(df_5k.shape)",
"(5239, 29)\n"
],
[
"df_5k = df_5k.reset_index()",
"_____no_output_____"
],
[
"df_5k.canonical_smiles[3]",
"_____no_output_____"
],
[
"smile_map = {'C': 2,\n 'O': 3,\n '2': 4}\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d03e40ca5cec51339fc4f58f224c77e30a48c769 | 70,575 | ipynb | Jupyter Notebook | PyCitySchools_starter.ipynb | pratixashah/Pandas_City_School | 2e753a69b5c983ae70febdb97028279d01f37d8d | [
"MIT"
] | null | null | null | PyCitySchools_starter.ipynb | pratixashah/Pandas_City_School | 2e753a69b5c983ae70febdb97028279d01f37d8d | [
"MIT"
] | null | null | null | PyCitySchools_starter.ipynb | pratixashah/Pandas_City_School | 2e753a69b5c983ae70febdb97028279d01f37d8d | [
"MIT"
] | null | null | null | 36.662338 | 202 | 0.399504 | [
[
[
"### Note\n* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.",
"_____no_output_____"
]
],
[
[
"# Dependencies and Setup\nimport pandas as pd\n\n# File to Load (Remember to Change These)\nschool_data_to_load = \"Resources/schools_complete.csv\"\nstudent_data_to_load = \"Resources/students_complete.csv\"\n\n# Read School and Student Data File and store into Pandas DataFrames\nschool_data = pd.read_csv(school_data_to_load)\nstudent_data = pd.read_csv(student_data_to_load)\n\n# Combine the data into a single dataset. \nschool_data_complete = pd.merge(student_data, school_data, how=\"left\", on=[\"school_name\", \"school_name\"])\nschool_data_complete",
"_____no_output_____"
]
],
[
[
"## District Summary\n\n* Calculate the total number of schools\n\n* Calculate the total number of students\n\n* Calculate the total budget\n\n* Calculate the average math score \n\n* Calculate the average reading score\n\n* Calculate the percentage of students with a passing math score (70 or greater)\n\n* Calculate the percentage of students with a passing reading score (70 or greater)\n\n* Calculate the percentage of students who passed math **and** reading (% Overall Passing)\n\n* Create a dataframe to hold the above results\n\n* Optional: give the displayed data cleaner formatting",
"_____no_output_____"
]
],
[
[
"total_school = len(school_data_complete[\"school_name\"].unique())\ntotal_student = sum(school_data_complete[\"size\"].unique())\n\nstudent_pass_math = school_data_complete.loc[school_data_complete[\"math_score\"] >= 70]\nstudent_pass_reading = school_data_complete.loc[school_data_complete[\"reading_score\"] >= 70]\nstudent_pass_math_and_reading = school_data_complete.loc[(school_data_complete[\"math_score\"] >= 70) & (school_data_complete[\"reading_score\"] >= 70)]\n\nstudent_pass_math_percentage = student_pass_math[\"Student ID\"].count()/total_student\nstudent_pass_reading_percentage = student_pass_reading[\"Student ID\"].count()/total_student\nstudent_pass_math_and_reading_percentage = student_pass_math_and_reading[\"Student ID\"].count()/total_student\n\ndistrict_summary_df = pd.DataFrame({\n \"Total Schools\": [total_school],\n \"Total Students\": [f\"{total_student:,}\"],\n \"Total Budget\": [f\"${sum(school_data_complete['budget'].unique()):,}\"],\n \"Average Math Score\":[f\"${school_data_complete['math_score'].mean():.2f}\"],\n \"Average Reading Score\":[f\"${school_data_complete['reading_score'].mean():.2f}\"],\n \"% Passing Math\":[f\"{student_pass_math_percentage:.2%}\"],\n \"% Passing Reading\":[f\"{student_pass_reading_percentage:.2%}\"],\n \"% Overall Passing\":[f\"{student_pass_math_and_reading_percentage:.2%}\"]\n})\ndistrict_summary_df",
"_____no_output_____"
]
],
[
[
"## School Summary",
"_____no_output_____"
],
[
"* Create an overview table that summarizes key metrics about each school, including:\n * School Name\n * School Type\n * Total Students\n * Total School Budget\n * Per Student Budget\n * Average Math Score\n * Average Reading Score\n * % Passing Math\n * % Passing Reading\n * % Overall Passing (The percentage of students that passed math **and** reading.)\n \n* Create a dataframe to hold the above results",
"_____no_output_____"
]
],
[
[
"group_by_school_data = school_data_complete.groupby([\"school_name\",\"type\"])\n\ngroup_by_school_pass_math = student_pass_math.groupby([\"school_name\",\"type\"])\ngroup_by_school_pass_reading = student_pass_reading.groupby([\"school_name\",\"type\"])\ngroup_by_school_pass_math_and_reading = student_pass_math_and_reading.groupby([\"school_name\",\"type\"])\n\nschool_summary_df = pd.DataFrame({\n \"Total Students\": group_by_school_data[\"school_name\"].count(),\n \"Total School Budget\": group_by_school_data['budget'].mean(),\n \"Per Student Budget\": group_by_school_data[\"budget\"].mean()/group_by_school_data[\"school_name\"].count(),\n \"Average Math Score\": group_by_school_data[\"math_score\"].mean(),\n \"Average Reading Score\": group_by_school_data[\"reading_score\"].mean(),\n \"% Passing Math\": group_by_school_pass_math[\"school_name\"].count()/group_by_school_data[\"school_name\"].count(),\n \"% Passing Reading\": group_by_school_pass_reading[\"school_name\"].count()/group_by_school_data[\"school_name\"].count(),\n \"% Overall Passing\": group_by_school_pass_math_and_reading[\"school_name\"].count()/group_by_school_data[\"school_name\"].count()\n})\n\nschool_summary = school_summary_df.copy()\n\nschool_summary_df[\"Total School Budget\"] = school_summary_df[\"Total School Budget\"].map(\"${:,}\".format)\nschool_summary_df[\"Per Student Budget\"] = school_summary_df[\"Per Student Budget\"].map(\"${:,.0f}\".format)\nschool_summary_df[\"Average Math Score\"] = school_summary_df[\"Average Math Score\"].map(\"{:,.2f}\".format)\nschool_summary_df[\"Average Reading Score\"] = school_summary_df[\"Average Reading Score\"].map(\"{:,.2f}\".format)\nschool_summary_df[\"% Passing Math\"] = school_summary_df[\"% Passing Math\"].map(\"{:.2%}\".format)\nschool_summary_df[\"% Passing Reading\"] = school_summary_df[\"% Passing Reading\"].map(\"{:,.2%}\".format)\nschool_summary_df[\"% Overall Passing\"] = school_summary_df[\"% Overall Passing\"].map(\"{:,.2%}\".format)\n\nschool_summary_df\n",
"_____no_output_____"
]
],
[
[
"## Top Performing Schools (By % Overall Passing)",
"_____no_output_____"
],
[
"* Sort and display the top five performing schools by % overall passing.",
"_____no_output_____"
]
],
[
[
"group_by_school_data_sorted = school_summary_df.sort_values(\"% Overall Passing\", ascending=False)\ngroup_by_school_data_sorted.head(5)",
"_____no_output_____"
]
],
[
[
"## Bottom Performing Schools (By % Overall Passing)",
"_____no_output_____"
],
[
"* Sort and display the five worst-performing schools by % overall passing.",
"_____no_output_____"
]
],
[
[
"group_by_school_data_sorted = school_summary_df.sort_values(\"% Overall Passing\")\ngroup_by_school_data_sorted.head(5)",
"_____no_output_____"
]
],
[
[
"## Math Scores by Grade",
"_____no_output_____"
],
[
"* Create a table that lists the average Reading Score for students of each grade level (9th, 10th, 11th, 12th) at each school.\n\n * Create a pandas series for each grade. Hint: use a conditional statement.\n \n * Group each series by school\n \n * Combine the series into a dataframe\n \n * Optional: give the displayed data cleaner formatting",
"_____no_output_____"
]
],
[
[
"# math_score_by_grade = school_data_complete.groupby([\"school_name\",\"grade\"])\n# math_score_by_grade[\"math_score\"].mean()\n\nmath_score_by_grade_9 = school_data_complete.loc[school_data_complete[\"grade\"] == \"9th\"]\nmath_score_by_grade_9_school = math_score_by_grade_9.groupby(\"school_name\")\n\nmath_score_by_grade_10 = school_data_complete.loc[school_data_complete[\"grade\"] == \"10th\"]\nmath_score_by_grade_10_school = math_score_by_grade_10.groupby(\"school_name\")\n\nmath_score_by_grade_11 = school_data_complete.loc[school_data_complete[\"grade\"] == \"11th\"]\nmath_score_by_grade_11_school = math_score_by_grade_11.groupby(\"school_name\")\n\nmath_score_by_grade_12 = school_data_complete.loc[school_data_complete[\"grade\"] == \"12th\"]\nmath_score_by_grade_12_school = math_score_by_grade_12.groupby(\"school_name\")\n\nmath_score_by_grade_df = pd.DataFrame({\n \"9th\": math_score_by_grade_9_school[\"math_score\"].mean().map(\"{:.2f}\".format),\n \"10th\":math_score_by_grade_10_school[\"math_score\"].mean().map(\"{:.2f}\".format),\n \"11th\": math_score_by_grade_11_school[\"math_score\"].mean().map(\"{:.2f}\".format),\n \"12th\":math_score_by_grade_12_school[\"math_score\"].mean().map(\"{:.2f}\".format)\n})\nmath_score_by_grade_df",
"_____no_output_____"
]
],
[
[
"## Reading Score by Grade ",
"_____no_output_____"
],
[
"* Perform the same operations as above for reading scores",
"_____no_output_____"
]
],
[
[
"# reading_score_by_grade = school_data_complete.groupby([\"school_name\",\"grade\"])\n# reading_score_by_grade[\"reading_score\"].mean()\n\nreading_score_by_grade_9 = school_data_complete.loc[school_data_complete[\"grade\"] == \"9th\"]\nreading_score_by_grade_9_school = reading_score_by_grade_9.groupby(\"school_name\")\n\nreading_score_by_grade_10 = school_data_complete.loc[school_data_complete[\"grade\"] == \"10th\"]\nreading_score_by_grade_10_school = reading_score_by_grade_10.groupby(\"school_name\")\n\nreading_score_by_grade_11 = school_data_complete.loc[school_data_complete[\"grade\"] == \"11th\"]\nreading_score_by_grade_11_school = reading_score_by_grade_11.groupby(\"school_name\")\n\nreading_score_by_grade_12 = school_data_complete.loc[school_data_complete[\"grade\"] == \"12th\"]\nreading_score_by_grade_12_school = reading_score_by_grade_12.groupby(\"school_name\")\n\nreading_score_by_grade_df = pd.DataFrame({\n \"9th\": reading_score_by_grade_9_school[\"reading_score\"].mean().map(\"{:.2f}\".format),\n \"10th\":reading_score_by_grade_10_school[\"reading_score\"].mean().map(\"{:.2f}\".format),\n \"11th\":reading_score_by_grade_11_school[\"reading_score\"].mean().map(\"{:.2f}\".format),\n \"12th\":reading_score_by_grade_12_school[\"reading_score\"].mean().map(\"{:.2f}\".format)\n})\nreading_score_by_grade_df",
"_____no_output_____"
]
],
[
[
"## Scores by School Spending",
"_____no_output_____"
],
[
"* Create a table that breaks down school performances based on average Spending Ranges (Per Student). Use 4 reasonable bins to group school spending. Include in the table each of the following:\n * Average Math Score\n * Average Reading Score\n * % Passing Math\n * % Passing Reading\n * Overall Passing Rate (Average of the above two)",
"_____no_output_____"
]
],
[
[
"score_by_spending = school_summary.copy()\n\nbins=[0,585,630,645,680]\nlabels=[\"<$585\",\"$585-630\",\"$630-645\",\"$645-680\"]\n\nscore_by_spending[\"Spending Ranges (Per Student)\"] = pd.cut(score_by_spending[\"Per Student Budget\"],bins,labels=labels, include_lowest=True)\nscore_by_spending\nscore_by_spending_group = score_by_spending.groupby([\"Spending Ranges (Per Student)\"])\n\nscore_by_spending_df = pd.DataFrame({\n \"Average Math Score\":map(\"{:.2f}\".format,score_by_spending_group[\"Average Math Score\"].mean()),\n \"Average Reading Score\":map(\"{:.2f}\".format,score_by_spending_group[\"Average Reading Score\"].mean()),\n \"% Passing Math\": map(\"{:.2%}\".format,score_by_spending_group[\"% Passing Math\"].mean()),\n \"% Passing Reading\": map(\"{:.2%}\".format,score_by_spending_group[\"% Passing Reading\"].mean()),\n \"% Overall Passing\":map(\"{:.2%}\".format, score_by_spending_group[\"% Overall Passing\"].mean())\n})\nscore_by_spending_df\n",
"_____no_output_____"
]
],
[
[
"## Scores by School Size",
"_____no_output_____"
],
[
"* Perform the same operations as above, based on school size.",
"_____no_output_____"
]
],
[
[
"score_by_school_size = school_summary.copy()\n\nbins=[0,1000,2000,5000]\nlabels = [\"Small (<1000)\",\"Medium (1000-2000)\",\"Large (2000-5000)\"]\n\nscore_by_school_size[\"School Type\"] = pd.cut(score_by_school_size[\"Total Students\"],bins,labels=labels)\nscore_by_school_size_group = score_by_school_size.groupby([\"School Type\"])\n\nscore_by_school_size_df = pd.DataFrame({\n \"Average Math Score\":map(\"{:.2f}\".format,score_by_school_size_group[\"Average Math Score\"].mean()),\n \"Average Reading Score\":map(\"{:.2f}\".format,score_by_school_size_group[\"Average Reading Score\"].mean()),\n \"% Passing Math\": map(\"{:.2%}\".format,score_by_school_size_group[\"% Passing Math\"].mean()),\n \"% Passing Reading\": map(\"{:.2%}\".format,score_by_school_size_group[\"% Passing Reading\"].mean()),\n \"% Overall Passing\":map(\"{:.2%}\".format, score_by_school_size_group[\"% Overall Passing\"].mean())\n}) \nscore_by_school_size_df",
"_____no_output_____"
]
],
[
[
"## Scores by School Type",
"_____no_output_____"
],
[
"* Perform the same operations as above, based on school type",
"_____no_output_____"
]
],
[
[
"score_by_school_type = school_summary.copy()\nscore_by_school_type_group = score_by_school_type.groupby([\"type\"])\n\nscore_by_school_type_df = pd.DataFrame({\n \"Average Math Score\":map(\"{:.2f}\".format,score_by_school_type_group[\"Average Math Score\"].mean()),\n \"Average Reading Score\":map(\"{:.2f}\".format,score_by_school_type_group[\"Average Reading Score\"].mean()),\n \"% Passing Math\": map(\"{:.2%}\".format,score_by_school_type_group[\"% Passing Math\"].mean()),\n \"% Passing Reading\": map(\"{:.2%}\".format,score_by_school_type_group[\"% Passing Reading\"].mean()),\n \"% Overall Passing\":map(\"{:.2%}\".format, score_by_school_type_group[\"% Overall Passing\"].mean())\n}) \nscore_by_school_type_df",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d03e4500f7330523baacbafa0ff1507b309e14d8 | 264,650 | ipynb | Jupyter Notebook | L2_computer_vision.ipynb | schwickert/auto | 32e86f8bda3d657ac8f9d2f01cb126429301d2a3 | [
"MIT"
] | null | null | null | L2_computer_vision.ipynb | schwickert/auto | 32e86f8bda3d657ac8f9d2f01cb126429301d2a3 | [
"MIT"
] | null | null | null | L2_computer_vision.ipynb | schwickert/auto | 32e86f8bda3d657ac8f9d2f01cb126429301d2a3 | [
"MIT"
] | null | null | null | 749.716714 | 114,760 | 0.953225 | [
[
[
"# Lesson 2: Computer Vision Fundamentals\n## Submission, Markus Schwickert, 2018-02-22\n\n### Photos",
"_____no_output_____"
]
],
[
[
"#importing some useful packages\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimport numpy as np\nimport cv2\nimport os\n%matplotlib inline",
"_____no_output_____"
],
[
"#reading in an image\nk1=0 # select here which of the images in the directory you want to process (0-5)\ntest_images=os.listdir(\"test_images/\")\nprint ('test_images/'+test_images[k1])\nimage = mpimg.imread('test_images/'+test_images[k1])\n\n#printing out some stats and plotting\nprint('This image is:', type(image), 'with dimensions:', image.shape)\nplt.imshow(image) # if you wanted to show a single color channel image called 'gray', for example, call as plt.imshow(gray, cmap='gray')\n",
"test_images/solidYellowCurve.jpg\nThis image is: <class 'numpy.ndarray'> with dimensions: (540, 960, 3)\n"
],
[
"# Grab the x and y size and make a copy of the image\nysize = image.shape[0]\nxsize = image.shape[1]",
"_____no_output_____"
],
[
"# Note: always make a copy rather than simply using \"=\"\ncolor_select = np.copy(image)\n\n# Define our color selection criteria\n# Note: if you run this code, you'll find these are not sensible values!!\n# But you'll get a chance to play with them soon in a quiz\nred_threshold = 180\ngreen_threshold = 180\nblue_threshold = 100\nrgb_threshold = [red_threshold, green_threshold, blue_threshold]\n\n# Identify pixels below the threshold\nthresholds = (image[:,:,0] < rgb_threshold[0]) \\\n | (image[:,:,1] < rgb_threshold[1]) \\\n | (image[:,:,2] < rgb_threshold[2])\ncolor_select[thresholds] = [0,0,0]\n\n# Display the image \nplt.imshow(color_select)\nplt.show()",
"_____no_output_____"
],
[
"gray = cv2.cvtColor(color_select, cv2.COLOR_RGB2GRAY) #grayscale conversion\nplt.imshow(gray, cmap='gray')",
"_____no_output_____"
],
[
"# Define a polygon region of interest \n# Keep in mind the origin (x=0, y=0) is in the upper left in image processing\n\nleft_bottom = [0, ysize]\nright_bottom = [xsize, ysize]\nfp1 = [450, 320]\nfp2 = [490, 320]\n\nmask = np.zeros_like(gray) \nignore_mask_color = 255 \n\n# This time we are defining a four sided polygon to mask\nvertices = np.array([[left_bottom, fp1, fp2, right_bottom]], dtype=np.int32)\ncv2.fillPoly(mask, vertices, ignore_mask_color)\ngrayROI = cv2.bitwise_and(gray, mask)\n\n# Display the image\nplt.imshow(grayROI, cmap='gray')",
"_____no_output_____"
],
[
"# Canny edge detection\n\n# Define a kernel size for Gaussian smoothing / blurring\n# Note: this step is optional as cv2.Canny() applies a 5x5 Gaussian internally\nkernel_size = 5\nblur_gray = cv2.GaussianBlur(grayROI,(kernel_size, kernel_size), 0)\n\n# Define parameters for Canny and run it\nlow_threshold = 50\nhigh_threshold = 150\nedges = cv2.Canny(blur_gray, low_threshold, high_threshold)\n\n# Display the image\nplt.imshow(edges, cmap='Greys_r')",
"_____no_output_____"
],
[
"# Hough Transformation\n\n# Define the Hough transform parameters\n# Make a blank the same size as our image to draw on\nrho = 1\ntheta = np.pi/180\nthreshold = 1\nmin_line_length = 16\nmax_line_gap = 20\nline_image = np.copy(image)*0 #creating a blank to draw lines on\n\n# Run Hough on edge detected image\nlines = cv2.HoughLinesP(edges, rho, theta, threshold, np.array([]),\n min_line_length, max_line_gap)\n\n# Iterate over the output \"lines\" and draw lines on the blank\nfor line in lines:\n for x1,y1,x2,y2 in line:\n cv2.line(line_image,(x1,y1),(x2,y2),(255,0,0),10)\n\n# Create a \"color\" binary image to combine with line image\n#color_edges = np.dstack((edges, edges, edges)) \n\n# Draw the lines on the edge image\nprint (test_images[k1])\ncombo = cv2.addWeighted(image, 0.8, line_image, 1, 0) \nplt.imshow(combo)",
"solidYellowCurve.jpg\n"
],
[
"mpimg.imsave('MS_images/'+test_images[k1], combo)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03e4eb701724afbb0ea7c59082468a22cb3cbc1 | 82,277 | ipynb | Jupyter Notebook | sankey.ipynb | numalariamodeling/chicago_sentinel_surveillance | 1f4f25348ba97cfe481875d41e6da4c91edd1584 | [
"Apache-2.0"
] | 2 | 2022-03-17T17:41:44.000Z | 2022-03-31T22:49:51.000Z | sankey.ipynb | numalariamodeling/chicago_sentinel_surveillance | 1f4f25348ba97cfe481875d41e6da4c91edd1584 | [
"Apache-2.0"
] | null | null | null | sankey.ipynb | numalariamodeling/chicago_sentinel_surveillance | 1f4f25348ba97cfe481875d41e6da4c91edd1584 | [
"Apache-2.0"
] | null | null | null | 213.15285 | 68,072 | 0.89285 | [
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport datetime as dt\nimport seaborn as sns\nimport numpy as np\nimport matplotlib.dates as mdates\nimport datetime\n#sns.set(color_codes=True)\nimport matplotlib as mpl\nmpl.rcParams['pdf.fonttype'] = 42\nimport statistics as st\nsns.set_style('whitegrid', {'axes.linewidth' : 0.5})\nfrom statsmodels.distributions.empirical_distribution import ECDF\nimport scipy\nimport gc\n\nfrom helpers import *",
"_____no_output_____"
],
[
"today_str = dt.datetime.today().strftime('%y%m%d')",
"_____no_output_____"
],
[
"def curve(startx, starty, endx, endy):\n x1 = np.linspace(0,(endx-startx),100)\n x2 = x1+startx\n x = x1/(endx-startx)\n y = (endy-starty)*(6*x**5-15*x**4+10*x**3)+starty\n y = (endy-starty)*(-20*x**7+70*x**6-84*x**5+35*x**4)+starty\n return x2, y",
"_____no_output_____"
],
[
"curative = pd.read_csv('~/Box/covid_CDPH/2021.07.06 Master Set Data Only_Deidentified.csv', encoding= 'unicode_escape')\ncurative['patient_symptom_date'] = pd.to_datetime(curative['patient_symptom_date'], errors='coerce')\ncurative['collection_time'] = pd.to_datetime(curative['collection_time'], errors='coerce')\ncurative['days'] = (pd.to_datetime(curative['collection_time'], utc=True) - pd.to_datetime(curative['patient_symptom_date'], utc=True)).dt.days",
"C:\\Users\\Ibis Grad\\anaconda3\\lib\\site-packages\\IPython\\core\\interactiveshell.py:3063: DtypeWarning: Columns (0,4,10,11,13,15,16,18,19,20,21,22,23,24,25,30,31) have mixed types.Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
],
[
"idph = pd.read_csv('~/Box/covid_IDPH/sentinel_surveillance/210706_SS_epic.csv', encoding= 'unicode_escape')\nidph['test_date'] = pd.to_datetime(idph['test_date'])\nidph['test_time'] = pd.to_datetime(idph['test_time'])\nidph['date_symptoms_start'] = pd.to_datetime(idph['date_symptoms_start'])\nidph['days'] = (idph['test_date'] - idph['date_symptoms_start']).dt.days\nss_cond = (idph['days'] <= 4) & (idph['days'] >= 0)\npos_cond = (idph['result'] == 'DETECTED') | (idph['result'] == 'POSITIVE') | (idph['result'] == 'Detected')\nchi_cond = (idph['test_site_city'] == 'CHICAGO')",
"_____no_output_____"
],
[
"zips = pd.read_csv('./data/Chicago_ZIP_codes.txt', header=None)[0].values",
"_____no_output_____"
],
[
"idph['chicago'] = idph['pat_zip_code'].apply(lambda x: zip_in_zips(x, zips))",
"_____no_output_____"
],
[
"curative['chicago'] = curative['patient_city'] == 'Chicago'",
"_____no_output_____"
],
[
"curative_time_frame_cond = (curative['collection_time'] >= pd.to_datetime('9-27-20')) & (curative['collection_time'] <= pd.to_datetime('6-13-21'))\ncurative_ss = (curative['days'] >= 0) & (curative['days'] <= 4)\ncurative_symptom = curative['patient_is_symptomatic']\n\nidph_time_frame_cond = (idph['test_date'] >= pd.to_datetime('9-27-20')) & (idph['test_date'] <= pd.to_datetime('6-13-21'))\nidph_ss = (idph['days'] >= 0) & (idph['days'] <= 4)\nidph_symptom = idph['symptomatic_per_cdc'] == 'Yes'\nidph_chicago_site = (idph['test_site'] == 'IDPH COMMUNITY TESTING AUBURN GRESHAM') | (idph['test_site'] == 'IDPH AUBURN GRESHAM COMMUNITY TESTING') | (idph['test_site'] == 'IDPH HARWOOD HEIGHTS COMMUNITY TESTING')\n\nidph_count = np.sum(idph_time_frame_cond & idph_ss & idph['chicago'] & idph_chicago_site)\ncurative_count = np.sum(curative_time_frame_cond & curative_ss & curative['chicago'])\n\npos_cond_curative = curative['test_result'] == 'POSITIVE'\ncurative['positive'] = pos_cond_curative\nchi_idph = (idph['test_site_city'] == 'Chicago') | (idph['test_site_city'] == 'CHICAGO')\npos_cond_idph = (idph['result'] == 'DETECTED') | (idph['result'] == 'POSITIVE') | (idph['result'] == 'Detected')\nidph['positive'] = pos_cond_idph\n\nprint(idph_count)\nprint(curative_count)",
"7478\n6474\n"
],
[
"print('Tests collected at sentinel sites in study period: ')\nsentinel_sites_total = len(curative[curative_time_frame_cond]) + len(idph[idph_time_frame_cond & idph_chicago_site])\nprint(sentinel_sites_total)\n\nprint('with Chicago residence: ')\nchicago_residents = len(curative[curative_time_frame_cond & curative['chicago']]) + \\\nlen(idph[idph_time_frame_cond & idph_chicago_site & idph['chicago']])\nprint(chicago_residents)\n\nprint('with valid symptom date: ')\nwith_symptom_date = len(curative[curative_time_frame_cond & curative['chicago']].dropna(subset=['days'])) + \\\nlen(idph[idph_time_frame_cond & idph_chicago_site & idph['chicago']].dropna(subset=['days']))\nprint(with_symptom_date)\n\nprint('symptom date 4 or fewer days before test: ')\ntot_ss = len(curative[curative_time_frame_cond & curative['chicago'] & curative_ss].dropna(subset=['days'])) + \\\nlen(idph[idph_time_frame_cond & idph_chicago_site & idph['chicago'] & idph_ss].dropna(subset=['days']))\nprint(tot_ss)\n\nprint('and positive: ')\ntot_sc = len(curative[curative_time_frame_cond & curative['chicago'] & curative_ss & pos_cond_curative].dropna(subset=['days'])) + \\\nlen(idph[idph_time_frame_cond & idph_chicago_site & idph['chicago'] & idph_ss & pos_cond_idph].dropna(subset=['days']))\nprint(tot_sc)",
"Tests collected at sentinel sites in study period: \n324872\nwith Chicago residence: \n274934\nwith valid symptom date: \n21406\nsymptom date 4 or fewer days before test: \n13952\nand positive: \n3607\n"
],
[
"h = 10\nw = 8\n\nfig = plt.figure(figsize=(w, h))\n\nfigh = h-0\nfigw = w-0\n\nax = fig.add_axes([0,0,figw/w,figh/h])\n\nstop_location = np.arange(0,5,1)\nline_width = 0.05\n#ax.set_xlim([-0.05,1.05])\nh_padding = 0.15\nv_padding = 0.2\nline_width = 0.2\nline_height = 4.5\nmidpoint = (v_padding + line_height)/2\n\ntot_height = sentinel_sites_total\n\nax.fill_between([stop_location[0], stop_location[0]+line_width], \n [midpoint+line_height/2]*2, \n [midpoint-line_height/2]*2, \n color='gold', zorder=15)\n\n#ax.text(x=stop_location[0]+line_width/1.75, \n# y=midpoint, s=\"specimens collected at sentinel sites in study period n = \" + \"{:,}\".format(sentinel_sites_total), \n# ha='center', va='center', \n# rotation=90, zorder=16, color='k', fontsize=14)\n\nsplits = [chicago_residents, with_symptom_date, tot_ss, tot_sc]\nd = tot_height\nsplits_array = np.array(splits)/d\nd_t = 1\nd_ts = d\nd_top = midpoint+line_height/2\nd_bot = midpoint-line_height/2\nd_x = stop_location[0]\n# midpoint = figh/2\n\ninclude_color_array = ['gold']*(len(splits)-1) + ['blue']\nexclude_color_array = ['crimson']*(len(splits)-1) + ['blue']\n\nfor s, l_l, s1, include_color, exclude_color in zip(splits_array, \n stop_location[1:], \n splits, \n include_color_array, \n exclude_color_array):\n \n t_line = line_height*d_t + v_padding\n \n ax.fill_between([l_l, l_l+line_width], \n [midpoint+t_line/2]*2, \n [midpoint+t_line/2-line_height*s]*2, \n color=include_color, zorder=13)\n \n ax.fill_between([l_l, l_l+line_width], \n [midpoint-t_line/2]*2, \n [midpoint-t_line/2+line_height*(d_t-s)]*2, \n color=exclude_color)\n \n \n a1 = curve(d_x+line_width, d_bot, \n l_l, midpoint-t_line/2)\n a2 = curve(d_x+line_width, d_bot+line_height*(d_t-s), \n l_l, midpoint-t_line/2+line_height*(d_t-s))\n \n ax.fill_between(a1[0], a1[1], a2[1], color=exclude_color, alpha=0.25, linewidth=0)\n \n ax.text((d_x+l_l+line_width)/2, \n midpoint+t_line/2-line_height*(s)/2, \n \"n = \"+\"{:,}\".format(s1), \n ha='center', va='center', \n rotation=0, fontsize=14)\n \n ax.text((d_x+l_l+line_width)/2, \n midpoint-t_line/2+line_height*(d_t-s)/2, \n \"n = \"+\"{:,}\".format(d_ts - s1), \n ha='center', va='center', \n rotation=0, fontsize=14)\n \n a1 = curve(d_x+line_width, d_top, \n l_l, midpoint+t_line/2)\n a2 = curve(d_x+line_width, d_bot+line_height*(d_t-s), \n l_l, midpoint+t_line/2-line_height*s)\n \n ax.fill_between(a1[0], a1[1], a2[1], color=include_color, alpha=0.25, linewidth=0)\n \n d_t = s\n d_ts = s1\n d_top = midpoint+t_line/2\n d_bot = midpoint+t_line/2-line_height*s\n d_x = l_l\n midpoint = midpoint+t_line/2-line_height*s/2\n \nax.text(x=stop_location[1]+line_width+0.05, y=0.35, s='not Chicago resident', \n ha='left', va='center', fontsize=14)\nax.text(x=stop_location[2]+line_width+0.05, y=2.5, s='no valid date of symptom onset', \n ha='left', va='center', fontsize=14)\nax.text(x=stop_location[3]+line_width+0.05, y=4.5, s='symptom onset > 4 days\\nbefore specimen collection', \n ha='left', va='top', fontsize=14)\nax.text(x=stop_location[4]+line_width+0.05, y=5.02, s=\" positive test → sentinel case\", \n ha='left', va='top', fontsize=14, weight='bold')\nax.text(x=stop_location[4]+line_width+0.05, y=4.75, s=\" negative or inconclusive test\", \n ha='left', va='top', fontsize=14)\nax.text(x=stop_location[0]-0.1, \n y=2.5, s=\"specimens collected at\\ntesting sites in study period\\nn = \" + \"{:,}\".format(sentinel_sites_total), \n ha='right', va='center', \n rotation=0, zorder=16, color='k', fontsize=14)\n\nax.fill_between(x=[2.95, 4 + line_width+0.05], y1=4.55, y2=5.075, \n color='black', alpha=0.1, edgecolor='black', linewidth=0, linestyle='dashed', zorder=0)\n\nax.text(x=3.6, y=5.11, s=\"sentinel samples\", \n ha='center', va='bottom', fontsize=14, weight='bold')\n\nax.grid(False)\nax.axis('off')\nfig.savefig('sankey_diagram_' + today_str + '.png', dpi=200, bbox_inches='tight')\nfig.savefig('sankey_diagram_' + today_str + '.pdf', bbox_inches='tight')",
"meta NOT subset; don't know how to subset; dropped\nmeta NOT subset; don't know how to subset; dropped\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03e6b917bbc77dbffe8e4576d71c0ef9c437dd6 | 244,753 | ipynb | Jupyter Notebook | presentations/How To - Estimate Pi.ipynb | johnmathews/quant1 | 77030f5de646ca21e951d81d823f265bef87445e | [
"CC-BY-4.0"
] | 2 | 2018-11-20T15:11:11.000Z | 2021-02-14T18:55:15.000Z | presentations/How To - Estimate Pi.ipynb | johnmathews/quant1 | 77030f5de646ca21e951d81d823f265bef87445e | [
"CC-BY-4.0"
] | null | null | null | presentations/How To - Estimate Pi.ipynb | johnmathews/quant1 | 77030f5de646ca21e951d81d823f265bef87445e | [
"CC-BY-4.0"
] | 2 | 2017-07-13T12:39:13.000Z | 2018-04-11T06:11:21.000Z | 1,133.115741 | 198,262 | 0.94349 | [
[
[
"#Estimating $\\pi$ by Sampling Points\nBy Evgenia \"Jenny\" Nitishinskaya and Delaney Granizo-Mackenzie\n\nNotebook released under the Creative Commons Attribution 4.0 License.\n\n---\n\nA stochastic way to estimate the value of $\\pi$ is to sample points from a square area. Some of the points will fall within the area of a circle as defined by $x^2 + y^2 = 1$, we count what percentage all points fall within this area, which allows us to estimate the area of the circle and therefore $\\pi$.",
"_____no_output_____"
]
],
[
[
"# Import libraries\nimport math\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"in_circle = 0\noutside_circle = 0\n\nn = 10 ** 4\n\n# Draw many random points\nX = np.random.rand(n)\nY = np.random.rand(n)\n\nfor i in range(n):\n \n if X[i]**2 + Y[i]**2 > 1:\n outside_circle += 1\n else:\n in_circle += 1\n\narea_of_quarter_circle = float(in_circle)/(in_circle + outside_circle)\npi_estimate = area_of_circle = area_of_quarter_circle * 4",
"_____no_output_____"
],
[
"pi_estimate",
"_____no_output_____"
]
],
[
[
"We can visualize the process to see how it works.",
"_____no_output_____"
]
],
[
[
"# Plot a circle for reference\ncircle1=plt.Circle((0,0),1,color='r', fill=False, lw=2)\nfig = plt.gcf()\nfig.gca().add_artist(circle1)\n# Set the axis limits so the circle doesn't look skewed\nplt.xlim((0, 1.8))\nplt.ylim((0, 1.2))\nplt.scatter(X, Y)",
"_____no_output_____"
]
],
[
[
"Finally, let's see how our estimate gets better as we increase $n$. We'll do this by computing the estimate for $\\pi$ at each step and plotting that estimate to see how it converges.",
"_____no_output_____"
]
],
[
[
"in_circle = 0\noutside_circle = 0\n\nn = 10 ** 3\n\n# Draw many random points\nX = np.random.rand(n)\nY = np.random.rand(n)\n\n# Make a new array\npi = np.ndarray(n)\n\nfor i in range(n):\n \n if X[i]**2 + Y[i]**2 > 1:\n outside_circle += 1\n else:\n in_circle += 1\n\n area_of_quarter_circle = float(in_circle)/(in_circle + outside_circle)\n pi_estimate = area_of_circle = area_of_quarter_circle * 4\n \n pi[i] = pi_estimate\n \nplt.plot(range(n), pi)\nplt.xlabel('n')\nplt.ylabel('pi estimate')\nplt.plot(range(n), [math.pi] * n)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d03e6f34ddfc792600c4a62ec11091474066444e | 34,468 | ipynb | Jupyter Notebook | wp/notebooks/active learning/binary/.ipynb_checkpoints/active_learning-checkpoint.ipynb | ExLeonem/master-thesis-code | 559ad55f15c99772358384146bd30dd517b1dfe8 | [
"MIT"
] | null | null | null | wp/notebooks/active learning/binary/.ipynb_checkpoints/active_learning-checkpoint.ipynb | ExLeonem/master-thesis-code | 559ad55f15c99772358384146bd30dd517b1dfe8 | [
"MIT"
] | null | null | null | wp/notebooks/active learning/binary/.ipynb_checkpoints/active_learning-checkpoint.ipynb | ExLeonem/master-thesis-code | 559ad55f15c99772358384146bd30dd517b1dfe8 | [
"MIT"
] | null | null | null | 31.797048 | 143 | 0.495416 | [
[
[
"import os, importlib, sys\n\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom sklearn.model_selection import train_test_split\n\n%load_ext autoreload ",
"_____no_output_____"
],
[
"# Common paths\nBASE_PATH = os.path.join(os.getcwd(), \"..\", \"..\")\nMODULE_PATH = os.path.join(BASE_PATH, \"modules\")\nDS_PATH = os.path.join(BASE_PATH, \"datasets\")",
"_____no_output_____"
],
[
"sys.path.append(MODULE_PATH)\n\nimport mp.MomentPropagation as mp\nimportlib.reload(mp)\n\nimport data.mnist as mnist_loader\nimportlib.reload(mnist_loader)",
"_____no_output_____"
],
[
"import tensorflow as tf\nfrom tensorflow import keras\nimport tensorflow.keras\nfrom tensorflow.keras import Sequential\nfrom tensorflow.keras.layers import Conv2D, MaxPool2D, Dropout, Flatten, Dense, Softmax",
"_____no_output_____"
],
[
"gpus = tf.config.experimental.list_physical_devices(\"GPU\")\ncpus = tf.config.experimental.list_physical_devices(\"CPU\")\n\nif gpus:\n try:\n for gpu in gpus:\n tf.config.experimental.set_memory_growth(gpu, True)\n \n logical_gpus = tf.config.experimental.list_logical_devices(\"GPU\")\n print(len(gpus), \"Physical GPU's, \", len(logical_gpus), \"Logical GPU's\")\n \n except RuntimeError as e:\n print(e)\n \nelif cpus:\n try:\n logical_cpus = tf.config.experimental.list_logical_devices(\"CPU\")\n print(len(cpus), \"Physical CPU,\", len(logical_cpus), \"Logical CPU\")\n \n except RuntimeError as e:\n print(e)\n \n \ntfk = tf.keras",
"1 Physical GPU's, 1 Logical GPU's\n"
],
[
"input_shape = (28, 28, 1)\nmodel = Sequential([\n Conv2D(128, 4,activation=\"relu\", input_shape=input_shape),\n MaxPool2D(),\n Dropout(.2),\n Conv2D(64, 3, activation=\"relu\"),\n MaxPool2D(),\n Dropout(.2),\n Flatten(),\n Dense(512, activation=\"relu\"),\n Dense(256, activation=\"relu\"),\n Dense(128, activation=\"relu\"),\n Dense(1, activation=\"sigmoid\")\n])\n\nmodel.build(input_shape)\nmodel.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d (Conv2D) (None, 25, 25, 128) 2176 \n_________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 12, 12, 128) 0 \n_________________________________________________________________\ndropout (Dropout) (None, 12, 12, 128) 0 \n_________________________________________________________________\nconv2d_1 (Conv2D) (None, 10, 10, 64) 73792 \n_________________________________________________________________\nmax_pooling2d_1 (MaxPooling2 (None, 5, 5, 64) 0 \n_________________________________________________________________\ndropout_1 (Dropout) (None, 5, 5, 64) 0 \n_________________________________________________________________\nflatten (Flatten) (None, 1600) 0 \n_________________________________________________________________\ndense (Dense) (None, 512) 819712 \n_________________________________________________________________\ndense_1 (Dense) (None, 256) 131328 \n_________________________________________________________________\ndense_2 (Dense) (None, 128) 32896 \n_________________________________________________________________\ndense_3 (Dense) (None, 1) 129 \n=================================================================\nTotal params: 1,060,033\nTrainable params: 1,060,033\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"%autoreload 2\nimport active_learning as active\nimportlib.reload(active)",
"_____no_output_____"
],
[
"import mp.MomentPropagation as mp\nimportlib.reload(mp)\n\nmp = mp.MP()\nmp_model = mp.create_MP_Model(model=model, use_mp=True, verbose=True)",
"_____no_output_____"
],
[
"import data.mnist as mnist_loader\nimportlib.reload(mnist_loader)\n\n# Load Data\nmnist_path = os.path.join(DS_PATH, \"mnist\")\ninputs, targets = mnist_loader.load(mnist_path)\n\n# Select only first and second class\nselector = (targets==0) | (targets==1)\nnew_inputs = inputs[selector].astype(\"float32\")/255.0\nnew_targets = targets[selector]\n\n# Create splits\nx_train, x_test, y_train, y_test = train_test_split(new_inputs, new_targets)\nx_test, x_val, y_test, y_val = train_test_split(x_test, y_test)",
"_____no_output_____"
],
[
"new_inputs.shape",
"_____no_output_____"
],
[
"new_inputs[:, None, ...].shape",
"_____no_output_____"
],
[
"sample_input = np.random.randn(10, 28, 28, 1)\nsample_input.shape",
"_____no_output_____"
],
[
"# pred_mp,var_mp = mp_model(sample_input)",
"_____no_output_____"
],
[
"mp_model",
"_____no_output_____"
],
[
"%autoreload 2\nimport bayesian \nfrom bayesian import McDropout, MomentPropagation\n\nmp_m = MomentPropagation(mp_model)",
"_____no_output_____"
],
[
"prediction = mp_m.predict(sample_input)",
"_____no_output_____"
],
[
"prediction",
"_____no_output_____"
],
[
"variance = mp_m.variance(prediction)\nvariance",
"_____no_output_____"
],
[
"exepctation = mp_m.expectation(prediction)\nexepctation",
"_____no_output_____"
],
[
"%autoreload 2\n\nfrom acl import ActiveLearning\nfrom active_learning import TrainConfig, Config, Metrics, aggregates_per_key\n\nimport bayesian \nfrom bayesian import McDropout, MomentPropagation\n\n\ntrain_config = TrainConfig(\n batch_size=2,\n epochs=1\n)\n\nacq_config = Config(\n name=\"std_mean\",\n pseudo=True\n)\n\nmodel_name = \"mp\"\nacq_name = \"max_entropy\"\ndp_model = McDropout(model)\nmp_m = MomentPropagation(mp_model)\n\n\nactive_learning = ActiveLearning(\n dp_model, \n np.expand_dims(new_inputs, axis=-1), labels=new_targets, \n train_config=train_config,\n acq_name=acq_name\n)\n\nhistory = active_learning.start(step_size=40)\n\n\n# Save history\nMETRICS_PATH = os.path.join(BASE_PATH, \"metrics\")\nmetrics = Metrics(METRICS_PATH, keys=[\"iteration\", \"train_time\", \"query_time\", \"loss\"])\nmetrics.write()",
"100%|██████████| 28/28 [03:59<00:00, 8.57s/it]\n"
],
[
"# Compare mc dropout, moment propagation (max_entropy, bald)",
"_____no_output_____"
],
[
"history",
"_____no_output_____"
],
[
"%autoreload 2\n\n",
"_____no_output_____"
],
[
"\n\nmetrics.write(\"test\", history)",
"_____no_output_____"
],
[
"read_metrics = metrics.read(\"test\")\nread_metrics[:5]",
"_____no_output_____"
],
[
"pd.DataFrame(read_metrics)[[\"iteration\", \"loss\"]]",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03e8f718d54d75d97fa7dc6e30da10f53ad8014 | 19,672 | ipynb | Jupyter Notebook | ch5/training_recommender_systems_on_standalone_dataset.ipynb | Vega95/Deep-Learning-with-fastai-Cookbook | e692fee0e7d8de184cb57deb222123c94483acd7 | [
"MIT"
] | 38 | 2020-12-29T05:01:09.000Z | 2022-03-17T06:06:01.000Z | ch5/training_recommender_systems_on_standalone_dataset.ipynb | itsshaikaslam/Deep-Learning-with-fastai-Cookbook | e692fee0e7d8de184cb57deb222123c94483acd7 | [
"MIT"
] | 1 | 2021-08-12T06:55:50.000Z | 2021-08-23T11:59:00.000Z | ch5/training_recommender_systems_on_standalone_dataset.ipynb | itsshaikaslam/Deep-Learning-with-fastai-Cookbook | e692fee0e7d8de184cb57deb222123c94483acd7 | [
"MIT"
] | 22 | 2021-02-17T13:53:55.000Z | 2022-03-31T03:54:58.000Z | 24.437267 | 118 | 0.444947 | [
[
[
"# Training a recommender system on a standalone dataset\nTrain a recommender system with fastai using a standalone dataset\n\n- This notebook ingests the Amazon reviews dataset (https://www.kaggle.com/saurav9786/amazon-product-reviews)\n\n",
"_____no_output_____"
]
],
[
[
"# imports for notebook boilerplate\n!pip install -Uqq fastbook\nimport fastbook\nfrom fastbook import *\nfrom fastai.collab import *",
"_____no_output_____"
],
[
"# set up the notebook for fast.ai\nfastbook.setup_book()",
"_____no_output_____"
],
[
"modifier = 'apr13'",
"_____no_output_____"
]
],
[
[
"# Ingest the dataset\n- define the path object\n- define a dataframe to contain the dataset",
"_____no_output_____"
]
],
[
[
"# ingest the standalone dataset\n# this step assumes you have completed the steps in \"Getting Ready\"\n# in section \"Training a recommender system on a standalone dataset\" of Chapter 5\npath = URLs.path('amazon_reviews')",
"_____no_output_____"
],
[
"# examine the directory structure\npath.ls()",
"_____no_output_____"
],
[
"# ingest the dataset into a Pandas dataframe\ndf = pd.read_csv(path/'ratings_Electronics.csv',header = None)\n# add the column names described in https://www.kaggle.com/saurav9786/amazon-product-reviews\ndf.columns = ['userID','productID','rating','timestamp']",
"_____no_output_____"
]
],
[
[
"# Examine the dataset",
"_____no_output_____"
]
],
[
[
"# examine the first few records in the dataframe\ndf.head()",
"_____no_output_____"
],
[
"# get the number of records in the dataset\ndf.shape",
"_____no_output_____"
],
[
"# get the count of unique values in each column of the dataset\ndf.nunique()",
"_____no_output_____"
],
[
"# count the number of missing values in each column of the dataset\ndf.isnull().sum()",
"_____no_output_____"
],
[
"df['rating'].nunique()",
"_____no_output_____"
],
[
"%%time\n# defined a CollabDataLoaders object\ndls=CollabDataLoaders.from_df(df,bs= 64)",
"CPU times: user 36.7 s, sys: 3.86 s, total: 40.6 s\nWall time: 39.2 s\n"
],
[
"dls.show_batch()",
"_____no_output_____"
]
],
[
[
"# Define and train the model",
"_____no_output_____"
]
],
[
[
"%%time\n# define the model\nlearn=collab_learner(dls,y_range= [ 0 , 5.0 ] )",
"CPU times: user 8.66 s, sys: 671 ms, total: 9.33 s\nWall time: 5.65 s\n"
],
[
"%%time\n# train the model\nlearn.fit_one_cycle( 1 )",
"_____no_output_____"
]
],
[
[
"# Exercise the trained model\n- define a dataframe containing test data\n- apply the trained model to the dataframe",
"_____no_output_____"
]
],
[
[
"# set values for test dataframe\nscoring_columns = ['userID','productID']\ntest_df = pd.DataFrame(columns=scoring_columns)\ntest_df.at[0,'userID'] = 'A2NYK9KWFMJV4Y'\ntest_df.at[0,'productID'] = 'B008ABOJKS'\ntest_df.at[1,'userID'] = 'A29ZTEO6EKSRDV'\ntest_df.at[1,'productID'] = 'B006202R44'\ntest_df.head()",
"_____no_output_____"
],
[
"dl = learn.dls.test_dl(test_df)\nlearn.get_preds(dl=dl)",
"_____no_output_____"
],
[
"learn.summary()",
"_____no_output_____"
],
[
"# save the model - first save the current path\nkeep_path = learn.path",
"_____no_output_____"
],
[
"learn.path",
"_____no_output_____"
],
[
"learn.path = Path('/notebooks/temp')",
"_____no_output_____"
],
[
"learn.model_dir",
"_____no_output_____"
],
[
"learn.save('recomm_'+modifier)",
"_____no_output_____"
],
[
"learn.path = keep_path",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03e9667b896579ca3bb7ec6cfa70237f813a49f | 125,197 | ipynb | Jupyter Notebook | 04-Linear_Regresion_Python/NumpyIntro-Answers.ipynb | Chilefase/MIT-1.001 | c07e88aed0636894750eda93f37e8911666c7173 | [
"MIT"
] | null | null | null | 04-Linear_Regresion_Python/NumpyIntro-Answers.ipynb | Chilefase/MIT-1.001 | c07e88aed0636894750eda93f37e8911666c7173 | [
"MIT"
] | null | null | null | 04-Linear_Regresion_Python/NumpyIntro-Answers.ipynb | Chilefase/MIT-1.001 | c07e88aed0636894750eda93f37e8911666c7173 | [
"MIT"
] | null | null | null | 149.044048 | 22,356 | 0.895005 | [
[
[
"## Good review of numpy https://www.youtube.com/watch?v=GB9ByFAIAH4 ",
"_____no_output_____"
],
[
"## Numpy library - Remember to do pip install numpy\n### Numpy provides support for math and logical operations on arrays \n#### https://www.tutorialspoint.com/numpy/index.htm\n### It supports many more data types than python \n#### https://www.tutorialspoint.com/numpy/numpy_data_types.htm \n### Only a single data type is allowed in any particular array",
"_____no_output_____"
]
],
[
[
"a = np.array([1,2,3,4])\nprint(id(a))\nprint(type(a))\nb = np.array(a)\nprint(f'b = {id(b)}')\na = a + 1\na",
"140321654298944\n<class 'numpy.ndarray'>\nb = 140321654357568\n"
]
],
[
[
"# <img src='numpyArray.png' width ='400'>",
"_____no_output_____"
]
],
[
[
"# arange vs linspace - both generate a numpy array of numbers\nimport numpy as np\nnp.linspace(0,10,5) # specifies No. of values with 0 and 10 being first and last\nnp.arange(0, 10, 5) # specifies step size=5 starting at 0 up to but NOT including last",
"_____no_output_____"
],
[
"x = np.linspace(0,10,11) # generate 10 numbers \nx = x + 1 # operates on all elements of the array\ntype(x)",
"_____no_output_____"
],
[
"# generate points and use function to transform them\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nx = np.arange(0,10,0.1)\ny = np.sin(x)\nplt.plot(x,y)",
"_____no_output_____"
],
[
"\nimport numpy as np\nimport matplotlib.pyplot as plt\na = np.random.choice(np.linspace(0,10,10),100)\nplt.hist(a,bins=np.arange(0,11,1))\nnp.linspace(0,10,11)",
"_____no_output_____"
],
[
"plt.hist(a,bins=np.arange(0,11,1),density=True)",
"_____no_output_____"
],
[
"# Use Bins 1/2 wide - What does plot meean? \nplt.hist(a,bins=np.arange(0,11,0.5),density=True)",
"_____no_output_____"
],
[
"# Data as sampling from an unseen population\n# Choose at random from 1 through 10 \nimport numpy as np\nimport matplotlib.pyplot as plt\na = np.random.choice(np.arange(0,10),100)\na = np.random.random(100)*10.0\na\n\n",
"_____no_output_____"
]
],
[
[
"# Normal Distribution \n\n$\n\\text{the normal distribution is given by} \\\\\n$\n$$\nf(z)=\\frac{1}{\\sqrt{2 \\pi}}e^{-\\frac{(z)^2}{2}} \n$$\n$\n\\text{This can be rewritten in term of the mean and variance} \\\\\n$\n$$\nf(x)=\\frac{1}{\\sigma \\sqrt{2 \\pi}}e^{-\\frac{(x- \\mu)^2}{2 \\sigma^2}}\n$$\nThe random variable $X$ described by the PDF is a normal variable that follows a normal distribution with mean $\\mu$ and variance $\\sigma^2$.\n\n$\n\\text{Normal distribution notation is} \\\\\n$\n$$\nX \\sim N(\\mu,\\sigma^2) \\\\\n$$\n\nThe total area under the PDF curve equals 1.",
"_____no_output_____"
]
],
[
[
"# Normal Data \n\na = np.random.normal(10,2,10)\n\n\nplt.hist(a,bins=np.arange(5,16,1),density=True)\nplt.scatter(np.arange(5,15,1),a)\nplt.plot(a)",
"_____no_output_____"
],
[
"plt.hist(a,bins=np.arange(5,16,0.1), density=True)",
"_____no_output_____"
],
[
"plt.hist(a,bins=np.arange(5,16,1))",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\na = np.random.normal(0,2,200)\nplt.hist(a, bins=np.arange(-5,5,1))\n\n",
"_____no_output_____"
]
],
[
[
"## Mean and Variance\n\n$$\n\\mu = \\frac{\\sum(x)}{N}\n$$\n$$\n\\sigma^{2} =\\sum{\\frac{(x - \\mu)^{2}}{N} }\n$$\n",
"_____no_output_____"
]
],
[
[
"# IN CLASS - Generate a Population and calculate its mean and variance\nimport matplotlib.pyplot as plt\nNpoints = 10\np = np.random.normal(0,10,Npoints*100)\n\ndef myMean(sample):\n N = len(sample)\n total = 0\n for x in sample:\n total = total + x\n return x/N\npmean = myMean(p)\nprint(f'mean= {pmean}')\n\ndef myVar(sample,mean):\n tsample = sample - mean\n var = sum(tsample * tsample)/len(sample)\n return var\n \npvar = myVar(p, pmean)\nprint(f'Variance = {pvar}')\nprint(f'realVar = ')",
"mean= 0.000683255410669019\nVariance = 102.89071952384164\nrealVar = \n"
],
[
"import numpy as np\nimport scipy as scipy\nimport matplotlib.pyplot as plt\nfrom scipy.stats import norm\n\nplt.style.use('ggplot')\nfig, ax = plt.subplots()\nx= np.arange(34,40,0.01)\ny = np.random.normal(x)\nlines = ax.plot(x, norm.pdf(x,loc=37,scale=1))\nax.set_ylim(0,0.45) # range\n\nax.set_xlabel('x',fontsize=20) # set x label\nax.set_ylabel('pdf(x)',fontsize=20,rotation=90) # set y label\nax.xaxis.set_label_coords(0.55, -0.05) # x label coordinate\nax.yaxis.set_label_coords(-0.1, 0.5) # y label coordinate\npx=np.arange(36,37,0.1)\nplt.fill_between(px,norm.pdf(px,loc=37,scale=1),color='r',alpha=0.5)\nplt.show()\n\n\n\n\n\n",
"_____no_output_____"
],
[
"a = np.random.normal(10,1,20)\na",
"_____no_output_____"
]
],
[
[
"## Calculate the mean and subtract the mean from each data value\n$$ ",
"_____no_output_____"
]
],
[
[
"from matplotlib import collections as matcoll\nNpoints = 20\nx = np.arange(0,Npoints)\ny = np.random.normal(loc=10, scale=2, size=Npoints )\nlines = []\nfor i in range(Npoints):\n pair=[(x[i],0), (x[i], y[i])]\n lines.append(pair)\nlinecoll = matcoll.LineCollection(lines)\nfig, ax = plt.subplots()\nax.add_collection(linecoll)\n\nplt.scatter(x,y, marker='o', color='blue')\nplt.xticks(x)\nplt.ylim(0,40)\nplt.show()\nylim=(0,10)\n",
"_____no_output_____"
]
],
[
[
"### Numpy 2D Arrays\n",
"_____no_output_____"
]
],
[
[
"## Multi-Dimensional Arrays\n<img src='multiArray.png' width = 500>",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"# Numpy 2_D Arrays\na = [0,1,2]\nb = [3,4,5]\nc = [6,7,8]\nz = [a, \n b, \n c]\n",
"_____no_output_____"
],
[
"a = np.arange(0,9)\nz = a.reshape(3,3)\nz\n",
"_____no_output_____"
],
[
"z[2,2]",
"_____no_output_____"
],
[
"z[0:3:2,0:3:2]",
"_____no_output_____"
],
[
"## Exercise - Produce a 10x10 checkerboard of 1s and 0s",
"_____no_output_____"
],
[
"\nimport numpy as np\nimport seaborn as sns\nfrom matplotlib.colors import ListedColormap as lc\nZ = np.zeros((8,8),dtype=int)\nZ[1::2,::2] = 1\nZ[::2,1::2] = 1\nprint(Z)\nsns.heatmap(Z, annot=True,linewidths=5,cbar=False)\nimport seaborn as sns\nsns.heatmap(Z, annot=True,linewidths=5,cbar=False)\n\n\n",
"[[0 1 0 1 0 1 0 1]\n [1 0 1 0 1 0 1 0]\n [0 1 0 1 0 1 0 1]\n [1 0 1 0 1 0 1 0]\n [0 1 0 1 0 1 0 1]\n [1 0 1 0 1 0 1 0]\n [0 1 0 1 0 1 0 1]\n [1 0 1 0 1 0 1 0]]\n"
],
[
"# IN CLASS - use the above formula to plot the normal distribution over x = -4 to 4\n# takee mean = 0, and sigma = 1\nimport numpy as np\nimport matplotlib.pyplot as plt\nx = np.linspace(-4,4,100)\ny = (np.exp(-(x*x)/2))/np.sqrt(2*np.pi)\nplt.plot(x,y)",
"_____no_output_____"
],
[
"import scipy.integrate as integrate\nresult = integrate.quad(lambda x: (np.exp(-(x*x)))/np.sqrt(2*np.pi) , -5, 5)\nresult",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03eb8d47badcef985fc5cd5b792fd3ca0cafad8 | 10,205 | ipynb | Jupyter Notebook | Machine Learning With Python/colab/final/11 - Lesson - Natural Language Processing.ipynb | shreejitverma/Data-Scientist | 03c06936e957f93182bb18362b01383e5775ffb1 | [
"MIT"
] | 2 | 2022-03-12T04:53:03.000Z | 2022-03-27T12:39:21.000Z | Machine Learning With Python/colab/final/11 - Lesson - Natural Language Processing.ipynb | shreejitverma/Data-Scientist | 03c06936e957f93182bb18362b01383e5775ffb1 | [
"MIT"
] | null | null | null | Machine Learning With Python/colab/final/11 - Lesson - Natural Language Processing.ipynb | shreejitverma/Data-Scientist | 03c06936e957f93182bb18362b01383e5775ffb1 | [
"MIT"
] | 2 | 2022-03-12T04:52:21.000Z | 2022-03-27T12:45:32.000Z | 24.709443 | 236 | 0.508084 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d03ec6c888b8d978ae240c5867e8ee7b5ac38599 | 781,087 | ipynb | Jupyter Notebook | Session_2_Practical_Data_Science.ipynb | MattFinney/practical_data_science_in_python | cd0e0221307875c78a34f064c18c4b6bf6e4842d | [
"MIT"
] | 8 | 2020-11-20T19:02:18.000Z | 2021-11-17T01:51:30.000Z | Session_2_Practical_Data_Science.ipynb | MattFinney/practical_data_science_in_python | cd0e0221307875c78a34f064c18c4b6bf6e4842d | [
"MIT"
] | null | null | null | Session_2_Practical_Data_Science.ipynb | MattFinney/practical_data_science_in_python | cd0e0221307875c78a34f064c18c4b6bf6e4842d | [
"MIT"
] | 3 | 2021-02-06T18:38:13.000Z | 2021-11-17T01:44:20.000Z | 883.582579 | 482,553 | 0.94674 | [
[
[
"<a href=\"https://colab.research.google.com/github/MattFinney/practical_data_science_in_python/blob/main/Session_2_Practical_Data_Science.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/><a>\n\n# Practical Data Science in Python\n## Unsupervised Learning: Classifying Spotify Tracks by Genre with $k$-Means Clustering\nAuthors: Matthew Finney, Paulina Toro Isaza\n",
"_____no_output_____"
],
[
"#### Run this First! (Function Definitions)",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set_palette('Set1')\nfrom sklearn.decomposition import PCA\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.cluster import KMeans\nfrom IPython.display import Audio, Image, clear_output\nrs = 123\nnp.random.seed(rs)\n\ndef pca_plot(df, classes=None):\n\n # Scale data for PCA\n scaled_df = StandardScaler().fit_transform(df)\n \n # Fit the PCA and extract the first two components\n pca_results = PCA().fit_transform(scaled_df)\n pca1_scores = pca_results[:,0]\n pca2_scores = pca_results[:,1]\n \n # Sort the legend labels\n if classes is None:\n hue_order = None\n n_classes = 0\n elif str(classes[0]).isnumeric():\n classes = ['Cluster {}'.format(x) for x in classes]\n hue_order = sorted(np.unique(classes))\n n_classes = np.max(np.unique(classes).shape)\n else:\n hue_order = sorted(np.unique(classes))\n n_classes = np.max(np.unique(classes).shape)\n\n # Plot the first two principal components\n plt.figure(figsize=(8.5,8.5))\n plt.grid()\n sns.scatterplot(pca1_scores, pca2_scores, s=50, hue=classes,\n hue_order=hue_order, palette='Set1')\n plt.xlabel(\"Principal Component {}\".format(1))\n plt.ylabel(\"Principal Component {}\".format(2))\n plt.title('Principal Component Plot')\n plt.show()\n\ndef tracklist_player(track_list, df, header=\"Track Player\"):\n action = ''\n\n for track in track_list:\n print('{}\\nTrack Name: {}\\nArtist Name(s): {}'.format(header, df.loc[track,'name'],df.loc[track,'artist']))\n try:\n display(Image(df.loc[track,'cover_url'], format='jpeg', height=150))\n except:\n print('No cover art available')\n \n try:\n display(Audio(df.loc[track,'preview_url']+'.mp3', autoplay=True))\n except:\n print('No audio preview available')\n \n print('Press <Enter> for the next track or q then <Enter> to quit: ')\n action = input()\n clear_output()\n\n if action=='q':\n break\n\n print('No more clusters. Goodbye!')\n\n\ndef play_cluster_tracks(track_df, cluster_column=\"best_cluster\"):\n for cluster in sorted(track_df[cluster_column].unique()):\n\n # Get the tracks in the cluster, and shuffle them for variety\n tracks_list = track_df[track_df[cluster_column] == cluster].index.values\n np.random.shuffle(tracks_list)\n\n # Instantiate a tracklist player\n tracklist_player(tracks_list, df=track_df, header='{}'.format(cluster))\n\n# Load Track DataFrame\npath = 'https://raw.githubusercontent.com/MattFinney/practical_data_science_in_python/main/spotify_track_data.csv'\ntracks_df = pd.read_csv(path)\n\n# Columns from the track dataframe which are relevant for our analysis\naudio_feature_cols = ['danceability', 'energy', 'key', 'loudness', 'mode', \n 'speechiness', 'acousticness', 'instrumentalness',\n 'liveness', 'valence', 'tempo', 'duration_ms',\n 'time_signature']\n\n# Show the first five rows of our dataframe\ntracks_df.head()",
"/usr/local/lib/python3.6/dist-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
]
],
[
[
"## Recap from Session 1",
"_____no_output_____"
],
[
"In our earlier session, we started working with a dataset of Spotify tracks. We explored the variables in the dataset, and determined that audio features - like danceability, accousticness, and tempo - vary across the songs in our dataset and might help us to thoughtfully group the tracks into different playlists. We then used Principal Component Analysis (PCA), a dimensionality reduction technique, to visualize the variation in songs.\n\nWe'll pick up where we left off, with the PCA plot from last time. If you're just joining us for Session 2, don't fret! Attending Session 1 is NOT a prerequisite to learn and have fun in Session 2 today!",
"_____no_output_____"
]
],
[
[
"# Plot the principal component analysis results\npca_plot(tracks_df[audio_feature_cols])",
"_____no_output_____"
]
],
[
[
"## Today: Classification using $k$-Means Clustering",
"_____no_output_____"
],
[
"Our Principal Component Analysis in the first session helped us to visualize the variation of track audio features in just two dimensions. Looking at the scatterplot of the first two principal components above, we can see that there are a few different groups of tracks. But how do we mathematically separate the tracks into these meaningful groups?\n\nOne way to separate the tracks into meaningful groups based on similar audio features is to use clustering. Clustering is a machine learning technique that is very powerful for identifying patterns in unlabeled data where the ground truth is not known.",
"_____no_output_____"
],
[
"### What is $k$-Means Clustering?",
"_____no_output_____"
],
[
"$k$-Means Clustering is one of the most popular clustering algorithms. The algorithm assigns each data point to a cluster using four main steps.",
"_____no_output_____"
],
[
"**Step 1: Initialize the Clusters**\\\nBased on the user's desired number of clusters $k$, the algorithm randomly chooses a centroid for each cluster. In this example, we choose a $k=3$, therefore the algorithm randomly picks 3 centroids.\n\n",
"_____no_output_____"
],
[
"**Step 2: Assign Each Data Point**\\\nThe algorithm assigns each point to the closest centroid to get $k$ initial clusters.\n\n",
"_____no_output_____"
],
[
"**Step 3: Recompute the Cluster Centers**\\\nFor every cluster, the algorithm recomputes the centroid by taking the average of all points in the cluster. The changes in centroids are shown below by arrows.\n\n",
"_____no_output_____"
],
[
"**Step 4: Reassign the Points**\\\nSince the centroids change, the algorithm then re-assigns the points to the closest centroid. The image below shows the new clusters after re-assignment.\n\n",
"_____no_output_____"
],
[
"\nThe algorithm repeats the calculation of centroids and assignment of points until points stop changing clusters. When clustering large datasets, you stop the algorithm before reaching convergence, using other criteria instead.\n\n*Note: Some content in this section was [adapted](https://creativecommons.org/licenses/by/4.0/) from Google's free [Clustering in Machine Learning](https://developers.google.com/machine-learning/clustering) course. The course is a great resource if you want to explore clustering in more detail!*",
"_____no_output_____"
],
[
"### Cluster the Spotify Tracks using their Audio Features",
"_____no_output_____"
],
[
"Now, we will use the `sklearn.cluster.KMeans` Python library to apply the $k$-means algorithm to our `tracks_df` data. Based on our visual inspection of the PCA plot, let's start with a guess k=3 to get 3 clusters.",
"_____no_output_____"
]
],
[
[
"initial_k = ____\n\n# Scale the data, so that the units of features don't impact feature importance\nscaled_df = StandardScaler().fit_transform(tracks_df[audio_feature_cols])\n\n# Cluster the data using the k means algorithm\ninitial_cluster_results = ______(n_clusters=initial_k, n_init=25, random_state=rs).fit(scaled_df)",
"_____no_output_____"
]
],
[
[
"Now, let's print the cluster results. Notice that we're given a number (0 or 1) for each observation in our data set. This number is the id of the cluster assigned to each track.",
"_____no_output_____"
]
],
[
[
"# Print the cluster results\nprint(initial_cluster_results._______)",
"_____no_output_____"
]
],
[
[
"And let's save the cluster results in our `tracks_df` dataframe as a column named `initial_cluster` so we can access them later.",
"_____no_output_____"
]
],
[
[
"# Save the cluster labels in our dataframe\ntracks_df[______________] = ['Cluster ' + str(i) for i in __________.______]",
"_____no_output_____"
]
],
[
[
"Let's plot the PCA plot and color each observation based on the assigned cluster to visualize our $k$-means results.",
"_____no_output_____"
]
],
[
[
"# Show a PCA plot of the clusters\npca_plot(tracks_df[audio_feature_cols], classes=tracks_df['initial_cluster'])",
"_____no_output_____"
]
],
[
[
"Does it look like our $k$-means algorithm correctly separated the tracks into clusters? Does each color map to a distinct group of points?",
"_____no_output_____"
],
[
"### How do our clusters of songs differ?",
"_____no_output_____"
],
[
"One way we can evaluate our clusters is by looking how the distribution of each data feature varies by cluster. In our case, let's check to see if tracks in the different clusters tend to have different values of energy, loudness, or speechiness.",
"_____no_output_____"
]
],
[
[
"# Plot the distribution of audio features by cluster\ng = sns.pairplot(tracks_df, hue=\"initial_cluster\",\n vars=['danceability', 'energy', 'loudness', 'speechiness', 'tempo'],\n hue_order=sorted(tracks_df.initial_cluster.unique()), palette='Set1')\ng.fig.suptitle('Distribution of Audio Features by Cluster', y=1.05)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Experiment with different values of $k$",
"_____no_output_____"
],
[
"Use the slider to select different values of $k$, then run the cell below to see how the choice of the number of clusters affects our results.",
"_____no_output_____"
]
],
[
[
"trial_k = 10 #@param {type:\"slider\", min:1, max:10, step:1}\n\n# Cluster the data using the k means algorithm\ntrial_cluster_results = KMeans(n_clusters=trial_k, n_init=25, random_state=rs).fit(scaled_df)\n\n# Save the cluster labels in our dataframe\ntracks_df['trial_cluster'] = ['Cluster ' + str(i) for i in trial_cluster_results.labels_]\n\n# Show a PCA plot of the clusters\npca_plot(tracks_df[audio_feature_cols], classes=tracks_df['trial_cluster'])\n\n# Plot the distribution of audio features by cluster\ng = sns.pairplot(tracks_df, hue=\"trial_cluster\",\n vars=['danceability', 'energy', 'loudness', 'speechiness', 'tempo'],\n hue_order=sorted(tracks_df.trial_cluster.unique()), palette='Set1')\ng.fig.suptitle('Distribution of Audio Features by Cluster', y=1.05)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Which value of $k$ works best for our data?",
"_____no_output_____"
],
[
"You may have noticed that the $k$-means algorithm requires you to choose $k$ and decide the number of clusters before you run the algorithm. But how do we know which value of $k$ is the best fit for our data? \n\nOne approach is to track the total distance from points to their cluster centroid as we increase the number of clusters, $k$. Usually, the total distance decreases as we increase $k$, but we reach a value of $k$ where increasing $k$ only marginally decreases the total distance. An elbow plot helps us to find that value of $k$; it's the value of $k$ where the slope of the line in the elbow plot crosses the threshold of slope $=-1$. When you plot distance vs $k$, this point often looks like an \"elbow\".\n\nLet's build an elbow plot to select the value of $k$ that will give us the highest quality clusters that best explain the variation in our data.",
"_____no_output_____"
]
],
[
[
"# Calculate the Total Distance for each value of k between 1 and 10\nscores = []\nk_list = np.arange(____,____)\n\nfor i in k_list:\n fit_k = _____(n_clusters=i, n_init=5, random_state=rs).fit(scaled_df)\n scores.append(fit_k.inertia_)\n\n# Plot this in an elbow plot\nplt.figure(figsize=(11,8.5))\nsns.lineplot(______, ______)\nplt.xlabel('Number of clusters $k$')\nplt.ylabel('Total Point to Centroid Distance')\nplt.grid()\nplt.title('The Elbow Method showing the optimal $k$')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Do you see the \"elbow\"? At what value of $k$ does it occur?",
"_____no_output_____"
],
[
"### Evaluate the results of our clustering algorithm for the best $k$",
"_____no_output_____"
],
[
"Use the slider below to choose the \"best\" $k$ that you determined from looking at the elbow plot. Evaluate the results in the PCA plot. Does this look like a good value of $k$ to separate the data into meaningful clusters?",
"_____no_output_____"
]
],
[
[
"best_k = 1 #@param {type:\"slider\", min:1, max:10, step:1}\n\n# Cluster the data using the k means algorithm\nbest_cluster_results = KMeans(n_clusters=best_k, n_init=25, random_state=rs).fit(scaled_df)\n\n# Save the cluster labels in our dataframe\ntracks_df['best_cluster'] = ['Cluster ' + str(i) for i in best_cluster_results.labels_]\n\n# Show a PCA plot of the clusters\npca_plot(tracks_df[audio_feature_cols], classes=tracks_df['best_cluster'])",
"_____no_output_____"
]
],
[
[
"## How did we do?",
"_____no_output_____"
],
[
"In addition to the mathematical ways to validate the selection of the best $k$ parameter for our model and the quality of our resulting clusters, there's another very important way to evaluate our results: listening to the tracks!\n\nLet's listen to the tracks in each cluster! What do you notice about the attributes that tracks in each cluster have in common? What do you notice about how the clusters are different? What makes each cluster unique?",
"_____no_output_____"
]
],
[
[
"play_cluster_tracks(tracks_df, cluster_column='best_cluster')",
"Cluster 0\nTrack Name: Needy Bees\nArtist Name(s): Nick Hakim\n"
]
],
[
[
"## Wrap Up and Next Session",
"_____no_output_____"
],
[
"That's a wrap! Now that you've learned some practical skills in data science, please join us tomorrow afternoon for the third and final session in our series, where we'll talk about how to continue your studies and/or pursue a career in Data Science!\n\n**Making Your Next Professional Play in Data Science**\\\nFriday, October 2 | 11:30am - 12:45pm PT\\\n[https://sched.co/dtqZ](https://sched.co/dtqZ)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d03ecdda609e9f761e8a248b789206682e362435 | 364,463 | ipynb | Jupyter Notebook | topic1_introduction_of_Python/Part1_Python_Introduction.ipynb | AbnerCui/Python_Lectures | 2004ed7901de9731c0715e25d4b63064369ea6ad | [
"MIT"
] | 2 | 2020-09-04T03:45:44.000Z | 2020-09-16T05:49:58.000Z | topic1_introduction_of_Python/Part1_Python_Introduction.ipynb | AbnerCui/Python_Lectures | 2004ed7901de9731c0715e25d4b63064369ea6ad | [
"MIT"
] | null | null | null | topic1_introduction_of_Python/Part1_Python_Introduction.ipynb | AbnerCui/Python_Lectures | 2004ed7901de9731c0715e25d4b63064369ea6ad | [
"MIT"
] | 7 | 2020-07-19T02:00:05.000Z | 2021-05-29T07:41:48.000Z | 130.913434 | 61,256 | 0.86969 | [
[
[
"# Python是什么?",
"_____no_output_____"
],
[
"\n### Python是一种高级的多用途编程语言,广泛用于各种非技术和技术领域。Python是一种具备动态语义、面向对象的解释型高级编程语言。它的高级内建数据结构和动态类型及动态绑定相结合,使其在快速应用开发上极具吸引力,也适合于作为脚本或者“粘合剂”语言,将现有组件连接起来。Python简单、易学的语法强调可读性,因此可以降低程序维护成本。Python支持模块和软件包,鼓励模块化的代码重用。",
"_____no_output_____"
]
],
[
[
"print('hellow world')",
"hellow world\n"
]
],
[
[
"## Python简史\n### 1989,为了度过圣诞假期,Guido开始编写Python语言编译器。Python这个名字来自Guido的喜爱的电视连续剧《蒙蒂蟒蛇的飞行马戏团》。他希望新的语言Python能够满足他在C和Shell之间创建全功能、易学、可扩展的语言的愿景。\n### 1989年由荷兰人Guido van Rossum于1989年发明,第一个公开发行版发行于1991年\n### Granddaddy of Python web frameworks, Zope 1 was released in 1999\n### Python 1.0 - January 1994 增加了 lambda, map, filter and reduce.\n### Python 2.0 - October 16, 2000,加入了内存回收机制,构成了现在Python语言框架的基础\n### Python 2.4 - November 30, 2004, 同年目前最流行的WEB框架Django 诞生\n### Python 2.5 - September 19, 2006\n### Python 2.6 - October 1, 2008\n### Python 2.7 - July 3, 2010\n### Python 3.0 - December 3, 2008\n### Python 3.1 - June 27, 2009\n### Python 3.2 - February 20, 2011\n### Python 3.3 - September 29, 2012\n### Python 3.4 - March 16, 2014\n### Python 3.5 - September 13, 2015\n### Python 3.6 - December 23, 2016\n### Python 3.7 - June 15, 2018\n## Python的主要运用领域有:\n### 云计算:云计算最热的语言,典型的应用OpenStack\n### WEB开发:许多优秀的WEB框架,许多大型网站是Python开发、YouTube、Dropbox、Douban……典型的Web框架包括Django\n### 科学计算和人工智能:典型的图书馆NumPy、SciPy、Matplotlib、Enided图书馆、熊猫\n### 系统操作和维护:操作和维护人员的基本语言\n### 金融:定量交易、金融分析,在金融工程领域,Python不仅使用最多,而且使用最多,其重要性逐年增加。",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"## Python在一些公司的运用有:\n### 谷歌:谷歌应用程序引擎,代码。谷歌。com、Google.、Google爬虫、Google广告和其他项目正在广泛使用Python。\n### CIA:美国中情局网站是用Python开发的\n### NASA:美国航天局广泛使用Python进行数据分析和计算\n### YouTube:世界上最大的视频网站YouTube是用Python开发的。\n### Dropbox:美国最大的在线云存储网站,全部用Python实现,每天处理10亿的文件上传和下载。\n### Instagram:美国最大的照片共享社交网站,每天有3000多万张照片被共享,所有这些都是用Python开发的\n### Facebook:大量的基本库是通过Python实现的\n### Red.:世界上最流行的Linux发行版中的Yum包管理工具是用Python开发的\n### Douban:几乎所有公司的业务都是通过Python开发的。\n### 知识:中国最大的Q&A社区,通过Python开发(国外Quora)\n### 除此之外,还有搜狐、金山、腾讯、盛大、网易、百度、阿里、淘宝、土豆、新浪、果壳等公司正在使用Python来完成各种任务。",
"_____no_output_____"
],
[
"## Python有如下特征:\n### 1. 开放源码:Python和大部分可用的支持库及工具都是开源的,通常使用相当灵活和开放的许可证。\n### 2. 多重范型:Python支持不同的编程和实现范型,例如面向对象和命令式/函数式或者过程式编程。\n### 3. 多用途:Python可以用用于快速、交互式代码开发,也可以用于构建大型应用程序;它可以用于低级系统操作,也可以承担高级分析任务。\n### 4. 跨平台:Python可用于大部分重要的操作系统,如Windows、Linux和Mac OS;它用于构建桌面应用和Web应用。\n### 5. 运行速度慢:这里是指与C和C++相比。",
"_____no_output_____"
],
[
"## Python 常用标准库",
"_____no_output_____"
],
[
"### math模块为浮点运算提供了对底层C函数库的访问: ",
"_____no_output_____"
]
],
[
[
"import math\nprint(math.pi)\nprint(math.log(1024, 2))",
"3.141592653589793\n10.0\n"
]
],
[
[
"### random提供了生成随机数的工具。",
"_____no_output_____"
]
],
[
[
"import random\nprint(random.choice(['apple', 'pear', 'banana']))\nprint(random.random())",
"apple\n0.0034954793658343863\n"
]
],
[
[
"### datetime模块为日期和时间处理同时提供了简单和复杂的方法。",
"_____no_output_____"
]
],
[
[
"from datetime import date\nnow = date.today()\nbirthday = date(1999, 8, 20)\nage = now - birthday\nprint(age.days)",
"7625\n"
]
],
[
[
"### Numpy是高性能科学计算和数据分析的基础包。\n### Pandas 纳入了大量库和一些标准的数据模型,提供了高效地操作大型数据集所需的工具。\n### Statismodels是一个Python模块,它提供对许多不同统计模型估计的类和函数,并且可以进行统计测试和统计数据的探索。\n### matplotlib一个绘制数据图的库。对于数据科学家或分析师非常有用。\n### 更多https://docs.python.org/zh-cn/3/library/",
"_____no_output_____"
],
[
"# 基础架构工具",
"_____no_output_____"
],
[
"## Anaconda安装",
"_____no_output_____"
],
[
"https://www.anaconda.com/products/individual",
"_____no_output_____"
],
[
"## Spyder使用",
"_____no_output_____"
],
[
"## GitHub创建与使用",
"_____no_output_____"
],
[
"### GitHub 是一个面向开源及私有软件项目的托管平台,因为只支持 Git 作为唯一的版本库格式进行托管,故名 GitHub。 GitHub 于 2008 年 4 月 10 日正式上线,除了 Git 代码仓库托管及基本的 Web 管理界面以外,还提供了订阅、讨论组、文本渲染、在线文件编辑器、协作图谱(报表)、代码片段分享(Gist)等功能。目前,其注册用户已经超过350万,托管版本数量也是非常之多,其中不乏知名开源项目 Ruby on Rails、jQuery、python 等。GitHub 去年为漏洞支付了 16.6 万美元赏金。 2018年6月,GitHub被微软以75亿美元的价格收购。https://github.com/",
"_____no_output_____"
],
[
"# Python基础语法",
"_____no_output_____"
]
],
[
[
"print (\"Hello, Python!\")",
"Hello, Python!\n"
]
],
[
[
"## 行和缩进",
"_____no_output_____"
],
[
"### python 最具特色的就是用缩进来写模块。\n### 缩进的空白数量是可变的,但是所有代码块语句必须包含相同的缩进空白数量,这个必须严格执行。\n### 以下实例缩进为四个空格:",
"_____no_output_____"
]
],
[
[
"if 1>2:\n print (\"True\")\nelse:\n print (\"False\")",
"False\n"
],
[
"if True:\n print (\"Answer\")\n print (\"True\")\nelse:\n print (\"Answer\")\n # 没有严格缩进,在执行时会报错\n print (\"False\")",
"_____no_output_____"
]
],
[
[
"## 多行语句",
"_____no_output_____"
],
[
"### Python语句中一般以新行作为语句的结束符。\n### 但是我们可以使用斜杠( \\)将一行的语句分为多行显示,如下所示:",
"_____no_output_____"
]
],
[
[
"total = 1 + \\\n 2 + \\\n 3\nprint(total)",
"6\n"
]
],
[
[
"### 语句中包含 [], {} 或 () 括号就不需要使用多行连接符。如下实例:",
"_____no_output_____"
]
],
[
[
"days = ['Monday', 'Tuesday', 'Wednesday',\n 'Thursday', 'Friday']\nprint(days)",
"['Monday', 'Tuesday', 'Wednesday', 'Thursday', 'Friday']\n"
]
],
[
[
"## Python 引号",
"_____no_output_____"
],
[
"### Python 可以使用引号( ' )、双引号( \" )、三引号( ''' 或 \"\"\" ) 来表示字符串,引号的开始与结束必须是相同类型的。\n### 其中三引号可以由多行组成,编写多行文本的快捷语法,常用于文档字符串,在文件的特定地点,被当做注释。",
"_____no_output_____"
]
],
[
[
"word = 'word'\nsentence = \"这是一个句子。\"\nparagraph = \"\"\"这是一个段落。\n包含了多个语句\"\"\"\nprint(word)\nprint(sentence)\nprint(paragraph)",
"word\n这是一个句子。\n这是一个段落。\n包含了多个语句\n"
]
],
[
[
"## Python注释",
"_____no_output_____"
],
[
"### python中单行注释采用 # 开头。",
"_____no_output_____"
]
],
[
[
"# 第一个注释\nprint (\"Hello, Python!\") # 第二个注释",
"Hello, Python!\n"
]
],
[
[
"# Python变量类型",
"_____no_output_____"
],
[
"## 标准数据类型",
"_____no_output_____"
],
[
"### 在内存中存储的数据可以有多种类型。\n### 例如,一个人的年龄可以用数字来存储,他的名字可以用字符来存储。\n### Python 定义了一些标准类型,用于存储各种类型的数据。\n### Python有五个标准的数据类型:\n### Numbers(数字)\n### String(字符串)\n### List(列表)\n### Tuple(元组)\n### Dictionary(字典)",
"_____no_output_____"
],
[
"## Python数字",
"_____no_output_____"
],
[
"### Python支持三种不同的数字类型:\n### int(有符号整型)\n### float(浮点型)\n### complex(复数)",
"_____no_output_____"
]
],
[
[
"int1 = 1\nfloat2 = 2.0\ncomplex3 = 1+2j\nprint(type(int1),type(float2),type(complex3))",
"<class 'int'> <class 'float'> <class 'complex'>\n"
]
],
[
[
"## Python字符串",
"_____no_output_____"
],
[
"### 字符串或串(String)是由数字、字母、下划线组成的一串字符。",
"_____no_output_____"
]
],
[
[
"st = '123asd_'\nst1 = st[0:3]\nprint(st)\nprint(st1)",
"123asd_\n123\n"
]
],
[
[
"## Python列表",
"_____no_output_____"
],
[
"### List(列表) 是 Python 中使用最频繁的数据类型。\n### 列表可以完成大多数集合类的数据结构实现。它支持字符,数字,字符串甚至可以包含列表(即嵌套)。\n### 列表用 [ ] 标识,是 python 最通用的复合数据类型。",
"_____no_output_____"
]
],
[
[
"list1 = [ 'runoob', 786 , 2.23, 'john', 70.2 ]\ntinylist = [123, 'john']\n \nprint (list1) # 输出完整列表\nprint (list1[0]) # 输出列表的第一个元素\nprint (list1[1:3]) # 输出第二个至第三个元素 \nprint (list1[2:]) # 输出从第三个开始至列表末尾的所有元素\nprint (tinylist * 2) # 输出列表两次\nprint (list1 + tinylist) # 打印组合的列表\n\nlist1[0] = 0\nprint(list1)",
"['runoob', 786, 2.23, 'john', 70.2]\nrunoob\n[786, 2.23]\n[2.23, 'john', 70.2]\n[123, 'john', 123, 'john']\n['runoob', 786, 2.23, 'john', 70.2, 123, 'john']\n[0, 786, 2.23, 'john', 70.2]\n"
]
],
[
[
"## Python元组",
"_____no_output_____"
],
[
"### 元组是另一个数据类型,类似于 List(列表)。\n### 元组用 () 标识。内部元素用逗号隔开。但是元组不能二次赋值,相当于只读列表。",
"_____no_output_____"
]
],
[
[
"tuple1 = ( 'runoob', 786 , 2.23, 'john', 70.2 )\ntinytuple = (123, 'john')\nprint(tuple1[0])\nprint(tuple1+tinytuple)",
"runoob\n('runoob', 786, 2.23, 'john', 70.2, 123, 'john')\n"
]
],
[
[
"## Python 字典",
"_____no_output_____"
],
[
"### 字典(dictionary)是除列表以外python之中最灵活的内置数据结构类型。列表是有序的对象集合,字典是无序的对象集合。\n### 两者之间的区别在于:字典当中的元素是通过键来存取的,而不是通过偏移存取。\n### 字典用\"{ }\"标识。字典由索引(key)和它对应的值value组成。",
"_____no_output_____"
]
],
[
[
"dict1 = {}\ndict1['one'] = \"This is one\"\n \ntinydict = {'name': 'john','code':6734, 'dept': 'sales'}\n\nprint(tinydict['name'])\nprint(dict1)\nprint(tinydict.keys())\nprint(tinydict.values())",
"john\n{'one': 'This is one'}\ndict_keys(['name', 'code', 'dept'])\ndict_values(['john', 6734, 'sales'])\n"
]
],
[
[
"# Python运算符",
"_____no_output_____"
],
[
"## Python算术运算符",
"_____no_output_____"
]
],
[
[
"a = 21\nb = 10\nc = 0\n \nc = a + b\nprint (\"c 的值为:\", c)\n \nc = a - b\nprint (\"c 的值为:\", c)\n \nc = a * b\nprint (\"c 的值为:\", c) \n \nc = a / b\nprint (\"c 的值为:\", c) \n \nc = a % b #取余数\nprint (\"c 的值为:\", c)\n \n# 修改变量 a 、b 、c\na = 2\nb = 3\nc = a**b \nprint (\"c 的值为:\", c)\n \na = 10\nb = 5\nc = a//b #取整\nprint (\"c 的值为:\", c)",
"c 的值为: 31\nc 的值为: 11\nc 的值为: 210\nc 的值为: 2.1\nc 的值为: 1\nc 的值为: 8\nc 的值为: 2\n"
]
],
[
[
"## Python比较运算符",
"_____no_output_____"
]
],
[
[
"a = 21\nb = 10\nc = 0\n \nif a == b :\n print (\"a 等于 b\")\nelse:\n print (\"a 不等于 b\")\n\nif a != b :\n print (\"a 不等于 b\")\nelse:\n print (\"a 等于 b\")\n\nif a < b :\n print (\"a 小于 b\")\nelse:\n print (\"a 大于等于 b\")\n\nif a > b :\n print (\"a 大于 b\")\nelse:\n print (\"a 小于等于 b\")\n\n# 修改变量 a 和 b 的值\na = 5\nb = 20\nif a <= b :\n print (\"a 小于等于 b\")\nelse:\n print (\"a 大于 b\")\n\nif b >= a :\n print (\"b 大于等于 a\")\nelse:\n print (\"b 小于 a\")",
"a 不等于 b\na 不等于 b\na 大于等于 b\na 大于 b\na 小于等于 b\nb 大于等于 a\n"
]
],
[
[
"## Python逻辑运算符",
"_____no_output_____"
]
],
[
[
"a = True\nb = False\n \nif a and b :\n print (\"变量 a 和 b 都为 true\")\nelse:\n print (\"变量 a 和 b 有一个不为 true\")\n\nif a or b :\n print (\"变量 a 和 b 都为 true,或其中一个变量为 true\")\nelse:\n print (\"变量 a 和 b 都不为 true\")\n\nif not( a and b ):\n print (\"变量 a 和 b 都为 false,或其中一个变量为 false\")\nelse:\n print (\"变量 a 和 b 都为 true\")",
"变量 a 和 b 有一个不为 true\n变量 a 和 b 都为 true,或其中一个变量为 true\n变量 a 和 b 都为 false,或其中一个变量为 false\n"
]
],
[
[
"## Python赋值运算符",
"_____no_output_____"
]
],
[
[
"a = 21\nb = 10\nc = 0\n \nc = a + b\nprint (\"c 的值为:\", c)\n \nc += a\nprint (\"c 的值为:\", c)\n \nc *= a\nprint (\"c 的值为:\", c) \n \nc /= a \nprint (\"c 的值为:\", c) \n \nc = 2\nc %= a\nprint (\"c 的值为:\", c)\n \nc **= a\nprint (\"c 的值为:\", c)\n \nc //= a\nprint (\"c 的值为:\", c)",
"c 的值为: 31\nc 的值为: 52\nc 的值为: 1092\nc 的值为: 52.0\nc 的值为: 2\nc 的值为: 2097152\nc 的值为: 99864\n"
]
],
[
[
"# Python 条件语句",
"_____no_output_____"
],
[
"### Python条件语句是通过一条或多条语句的执行结果(True或者False)来决定执行的代码块。",
"_____no_output_____"
]
],
[
[
"flag = False\nname = 'luren'\nif name == 'python': # 判断变量是否为 python \n flag = True # 条件成立时设置标志为真\n print ('welcome boss') # 并输出欢迎信息\nelse:\n print (name) # 条件不成立时输出变量名称",
"luren\n"
],
[
"num = 5 \nif num == 3: # 判断num的值\n print ('boss') \nelif num == 2:\n print ('user')\nelif num == 1:\n print ('worker')\nelif num < 0: # 值小于零时输出\n print ('error')\nelse:\n print ('roadman') # 条件均不成立时输出",
"roadman\n"
],
[
"num = 9\nif num >= 0 and num <= 10: # 判断值是否在0~10之间\n print ('hello')\n\nnum = 10\nif num < 0 or num > 10: # 判断值是否在小于0或大于10\n print ('hello')\nelse:\n print ('undefine')\n\nnum = 8\n# 判断值是否在0~5或者10~15之间\nif (num >= 0 and num <= 5) or (num >= 10 and num <= 15): \n print ('hello')\nelse:\n print ('undefine')",
"hello\nundefine\nundefine\n"
]
],
[
[
"# Python循环语句",
"_____no_output_____"
],
[
"## Python 提供了 for 循环和 while 循环",
"_____no_output_____"
],
[
"## Python While 循环语句",
"_____no_output_____"
],
[
"### Python 编程中 while 语句用于循环执行程序,即在某条件下,循环执行某段程序,以处理需要重复处理的相同任务。",
"_____no_output_____"
]
],
[
[
"count = 0\nwhile (count < 9):\n print ('The count is:', count)\n count = count + 1\n\nprint (\"Good bye!\")",
"The count is: 0\nThe count is: 1\nThe count is: 2\nThe count is: 3\nThe count is: 4\nThe count is: 5\nThe count is: 6\nThe count is: 7\nThe count is: 8\nGood bye!\n"
],
[
"count = 0\nwhile count < 5:\n print (count, \" is less than 5\")\n count = count + 1\nelse:\n print (count, \" is not less than 5\")",
"0 is less than 5\n1 is less than 5\n2 is less than 5\n3 is less than 5\n4 is less than 5\n5 is not less than 5\n"
]
],
[
[
"## Python for 循环语句",
"_____no_output_____"
]
],
[
[
"fruits = ['banana', 'apple', 'mango']\nfor index in range(len(fruits)):\n print ('当前水果 :', fruits[index])\nprint (\"Good bye!\")",
"当前水果 : banana\n当前水果 : apple\n当前水果 : mango\nGood bye!\n"
]
],
[
[
"## Python 循环嵌套",
"_____no_output_____"
]
],
[
[
"num=[];\ni=2\nfor i in range(2,100):\n j=2\n for j in range(2,i):\n if(i%j==0):\n break\n else:\n num.append(i)\nprint(num)\nprint (\"Good bye!\")",
"[2, 3, 5, 7, 11, 13, 17, 19, 23, 29, 31, 37, 41, 43, 47, 53, 59, 61, 67, 71, 73, 79, 83, 89, 97]\nGood bye!\n"
]
],
[
[
"## Python break 语句",
"_____no_output_____"
]
],
[
[
"for letter in 'Python': \n if letter == 'h':\n break\n print ('当前字母 :', letter)",
"当前字母 : P\n当前字母 : y\n当前字母 : t\n"
]
],
[
[
"## Python continue 语句",
"_____no_output_____"
],
[
"### Python continue 语句跳出本次循环,而break跳出整个循环。",
"_____no_output_____"
]
],
[
[
"for letter in 'Python': \n if letter == 'h':\n continue\n print ('当前字母 :', letter)",
"当前字母 : P\n当前字母 : y\n当前字母 : t\n当前字母 : o\n当前字母 : n\n"
]
],
[
[
"## Python pass 语句",
"_____no_output_____"
],
[
"### Python pass 是空语句,是为了保持程序结构的完整性。\n### pass 不做任何事情,一般用做占位语句。",
"_____no_output_____"
]
],
[
[
"# 输出 Python 的每个字母\nfor letter in 'Python':\n if letter == 'h':\n pass\n print ('这是 pass 块')\n print ('当前字母 :', letter)\n\nprint (\"Good bye!\")",
"当前字母 : P\n当前字母 : y\n当前字母 : t\n这是 pass 块\n当前字母 : h\n当前字母 : o\n当前字母 : n\nGood bye!\n"
]
],
[
[
"# Python应用实例(链家二手房数据分析)",
"_____no_output_____"
],
[
"## 一、根据上海的部分二手房信息,从多角度进行观察和分析房价与哪些因素有关以及房屋不同状况所占比例\n## 二、先对数据进行预处理、构造预测房价的模型、并输入参数对房价进行预测\n备注:数据来源CSDN下载。上海链家二手房.csv.因文件读入问题,改名为sh.csv",
"_____no_output_____"
],
[
"## 一、导入数据 对数据进行一些简单的预处理",
"_____no_output_____"
]
],
[
[
"#导入需要用到的包\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nfrom IPython.display import display\nsns.set_style({'font.sans-serif':['simhei','Arial']})\n%matplotlib inline",
"_____no_output_____"
],
[
"shanghai=pd.read_csv('sh.csv')# 将已有数据导进来\nshanghai.head(n=1)#显示第一行数据 查看数据是否导入成功",
"_____no_output_____"
]
],
[
[
"### 每项数据类型均为object 不方便处理,需要对一些项删除单位转换为int或者float类型\n### 有些列冗余 像house_img需要删除\n### 有些列 如何house_desc包含多种信息 需要逐个提出来单独处理",
"_____no_output_____"
]
],
[
[
"shanghai.describe()",
"_____no_output_____"
],
[
"# 检查缺失值情况\nshanghai.info()\n#np.isnan(shanghai).any()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 73316 entries, 0 to 73315\nData columns (total 12 columns):\nhouse_title 73314 non-null object\nhouse_img 70428 non-null object\ns_cate_href 73316 non-null object\nhouse_desc 73316 non-null object\nzone_href 73316 non-null object\ndistrict 73316 non-null object\nhouse_detail 73316 non-null object\nhouse_price 73316 non-null int64\nhouse_href 73316 non-null object\ns_cate 73316 non-null object\nsingel_price 73316 non-null object\nhouse_time 70515 non-null object\ndtypes: int64(1), object(11)\nmemory usage: 6.7+ MB\n"
],
[
"shanghai.dropna(inplace=True)\n#数据处理 删除带有NAN项的行",
"_____no_output_____"
],
[
"df=shanghai.copy()\nhouse_desc=df['house_desc']\nhouse_desc[0]",
"_____no_output_____"
]
],
[
[
"### house_desc 中带有 室厅的信息 房子面积 楼层 朝向信息 需要分别提出来当一列 下面进行提取",
"_____no_output_____"
]
],
[
[
"df['layout']=df['house_desc'].map(lambda x:x.split('|')[0])\ndf['area']=df['house_desc'].map(lambda x:x.split('|')[1])\ndf['temp']=df['house_desc'].map(lambda x:x.split('|')[2])\n#df['Dirextion']=df['house_desc'].map(lambda x:x.split('|')[3])\ndf['floor']=df['temp'].map(lambda x:x.split('/')[0])\ndf.head(n=1)",
"_____no_output_____"
]
],
[
[
"### 一些列中带有单位 不利于后期处理 去掉单位 并把数据类型转换为float或int",
"_____no_output_____"
]
],
[
[
"df['area']=df['area'].apply(lambda x:x.rstrip('平'))\ndf['singel_price']=df['singel_price'].apply(lambda x:x.rstrip('元/平'))\ndf['singel_price']=df['singel_price'].apply(lambda x:x.lstrip('单价'))\ndf['district']=df['district'].apply(lambda x:x.rstrip('二手房'))\ndf['house_time']=df['house_time'].apply(lambda x:str(x))\ndf['house_time']=df['house_time'].apply(lambda x:x.rstrip('年建'))\ndf.head(n=1)",
"_____no_output_____"
]
],
[
[
"### 删除一些不需要用到的列 以及 house_desc、temp",
"_____no_output_____"
]
],
[
[
"del df['house_img']\ndel df['s_cate_href']\ndel df['house_desc']\ndel df['zone_href']\ndel df['house_href']\ndel df['temp']",
"_____no_output_____"
]
],
[
[
"### 根据房子总价和房子面积 计算房子每平方米的价格\n### 从house_title 描述房子信息中提取关键词。若带有 交通便利、地铁则认为其交通方便,否则交通不便",
"_____no_output_____"
]
],
[
[
"df.head(n=1)\ndf['singel_price']=df['singel_price'].apply(lambda x:float(x))\ndf['area']=df['area'].apply(lambda x:float(x))\ndf.head(n=1)\ndf.head(n=1)\ndf['house_title']=df['house_title'].apply(lambda x:str(x))\ndf['trafic']=df['house_title'].apply(lambda x:'交通便利' if x.find(\"交通便利\")>=0 or x.find(\"地铁\")>=0 else \"交通不便\" )\ndf.head(n=1)",
"_____no_output_____"
]
],
[
[
"## 二、根据各列信息 用可视化的形式展现 房价与不同因素如地区、房子面积、所在楼层等之间的关系",
"_____no_output_____"
]
],
[
[
"df_house_count = df.groupby('district')['house_price'].count().sort_values(ascending=False).to_frame().reset_index()\ndf_house_mean = df.groupby('district')['singel_price'].mean().sort_values(ascending=False).to_frame().reset_index()\n\nf, [ax1,ax2,ax3] = plt.subplots(3,1,figsize=(20,15))\nsns.barplot(x='district', y='singel_price', palette=\"Reds_d\", data=df_house_mean, ax=ax1)\nax1.set_title('上海各大区二手房每平米单价对比',fontsize=15)\nax1.set_xlabel('区域')\nax1.set_ylabel('每平米单价')\nsns.countplot(df['district'], ax=ax2)\nsns.boxplot(x='district', y='house_price', data=df, ax=ax3)\nax3.set_title('上海各大区二手房房屋总价',fontsize=15)\nax3.set_xlabel('区域')\nax3.set_ylabel('房屋总价')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 上面三幅图显示了 房子单价、总数量、总价与地区之间的关系。\n#### 由上面第一幅图可以看到房子单价与地区有关,其中黄浦以及静安地区房价最高。这与地区的发展水平、交通便利程度以及离市中心远近程度有关\n#### 由上面第二幅图可以直接看出不同地区的二手房数量,其中浦东最多\n#### 由上面第三幅图可以看出上海二手房房价基本在一千万上下,很少有高于两千万的",
"_____no_output_____"
]
],
[
[
"f, [ax1,ax2] = plt.subplots(1, 2, figsize=(15, 5))\n# 二手房的面积分布\nsns.distplot(df['area'], bins=20, ax=ax1, color='r')\nsns.kdeplot(df['area'], shade=True, ax=ax1)\n# 二手房面积和价位的关系\nsns.regplot(x='area', y='house_price', data=df, ax=ax2)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 由从左到右第一幅图可以看出 基本二手房面积在60-200平方米之间,其中一百平方米左右的占比更大\n### 由第二幅看出,二手房总结与二手房面积基本成正比,和我们的常识吻合",
"_____no_output_____"
]
],
[
[
"areas=[len(df[df.area<100]),len(df[(df.area>100)&(df.area<200)]),len(df[df.area>200])]\nlabels=['area<100' , '100<area<200','area>200']\nplt.pie(areas,labels= labels,autopct='%0f%%',shadow=True)\nplt.show()\n# 绘制饼图",
"_____no_output_____"
]
],
[
[
"### 将面积划分为三个档次,面积大于200、面积小与100、面积在一百到两百之间 三者的占比情况可以发现 百分之六十九的房子面积在一百平方米一下,高于一百大于200的只有百分之二十五而面积大于两百的只有百分之四",
"_____no_output_____"
]
],
[
[
"df.loc[df['area']>1000]\n# 查看size>1000的样本 发现只有一个是大于1000 ",
"_____no_output_____"
],
[
"f, ax1= plt.subplots(figsize=(20,20))\nsns.countplot(y='layout', data=df, ax=ax1)\nax1.set_title('房屋户型',fontsize=15)\nax1.set_xlabel('数量')\nax1.set_ylabel('户型')\nf, ax2= plt.subplots(figsize=(20,20))\nsns.barplot(y='layout', x='house_price', data=df, ax=ax2)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 上述两幅图显示了 不同户型的数量和价格\n#### 由第一幅图看出2室1厅最多 2室2厅 3室2厅也较多 是主流的户型选择 \n#### 由第二幅看出 室和厅的数量增加随之价格也增加,但是室和厅之间的比例要适合",
"_____no_output_____"
]
],
[
[
"a1=0\na2=0\nfor x in df['trafic']:\n \n if x=='交通便利':\n a1=a1+1\n else:\n a2=a2+1\nsizes=[a1,a2]\nlabels=['交通便利' , '交通不便']\nplt.pie(sizes,labels= labels,autopct='%0f%%',shadow=True)\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### 上述图显示了上海二手房交通不便利情况。其中百分之六十一为交通不便,百分之三十八为交通不便。由于交通便利情况仅仅是根据对房屋的描述情况提取出来的,实际上 交通便利的占比会更高些",
"_____no_output_____"
]
],
[
[
"f, [ax1,ax2] = plt.subplots(1, 2, figsize=(20, 10))\nsns.countplot(df['trafic'], ax=ax1)\nax1.set_title('交通是否便利数量对比',fontsize=15)\nax1.set_xlabel('交通是否便利')\nax1.set_ylabel('数量')\nsns.barplot(x='trafic', y='house_price', data=df, ax=ax2)\nax2.set_title('交通是否便利房价对比',fontsize=15)\nax2.set_xlabel('交通是否便利')\nax2.set_ylabel('总价')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 左边那幅图显示了交通便利以及不便的二手房数量,这与我们刚才的饼图信息一致\n### 右边那幅图显示了交通便利与否与房价的关系。交通便利的房子价格更高",
"_____no_output_____"
]
],
[
[
"f, ax1= plt.subplots(figsize=(20,5))\nsns.countplot(x='floor', data=df, ax=ax1)\nax1.set_title('楼层',fontsize=15)\nax1.set_xlabel('楼层数')\nax1.set_ylabel('数量')\nf, ax2 = plt.subplots(figsize=(20, 5))\nsns.barplot(x='floor', y='house_price', data=df, ax=ax2)\nax2.set_title('楼层',fontsize=15)\nax2.set_xlabel('楼层数')\nax2.set_ylabel('总价')\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### 楼层(地区、高区、中区、地下几层)与数量、房价的关系。高区、中区、低区居多",
"_____no_output_____"
],
[
"## 三、根据已有数据建立简单的上海二手房房间预测模型",
"_____no_output_____"
],
[
"### 对数据再次进行简单的预处理 把户型这列拆成室和厅",
"_____no_output_____"
]
],
[
[
"df[['室','厅']] = df['layout'].str.extract(r'(\\d+)室(\\d+)厅')\ndf['室'] = df['室'].astype(float)\ndf['厅'] = df['厅'].astype(float)\ndel df['layout']\ndf.head()",
"_____no_output_____"
],
[
"df.dropna(inplace=True)\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 67753 entries, 0 to 73304\nData columns (total 12 columns):\nhouse_title 67753 non-null object\ndistrict 67753 non-null object\nhouse_detail 67753 non-null object\nhouse_price 67753 non-null int64\ns_cate 67753 non-null object\nsingel_price 67753 non-null float64\nhouse_time 67753 non-null object\narea 67753 non-null float64\nfloor 67753 non-null object\ntrafic 67753 non-null object\n室 67753 non-null float64\n厅 67753 non-null float64\ndtypes: float64(4), int64(1), object(7)\nmemory usage: 6.7+ MB\n"
],
[
"df.columns",
"_____no_output_____"
]
],
[
[
"### 删除不需要用到的信息如房子的基本信息描述 ",
"_____no_output_____"
]
],
[
[
"del df['house_title']\ndel df['house_detail']\ndel df['s_cate']",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression\nlinear = LinearRegression()\narea=df['area']\nprice=df['house_price']\narea = np.array(area).reshape(-1,1) # 这里需要注意新版的sklearn需要将数据转换为矩阵才能进行计算\nprice = np.array(price).reshape(-1,1)\n# 训练模型\nmodel = linear.fit(area,price)\n# 打印截距和回归系数\n\nprint(model.intercept_, model.coef_)",
"[-61.47368693] [[6.41045431]]\n"
],
[
"linear_p = model.predict(area)\nplt.figure(figsize=(12,6))\nplt.scatter(area,price)\nplt.plot(area,linear_p,'red')\nplt.xlabel(\"area\")\nplt.ylabel(\"price\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### 上面用线性回归模型对房价进行简单的预测 红色的代表预测房价,而蓝色点代表真实值。可以看出在面积小于1000时真实值紧密分布在预测值两旁",
"_____no_output_____"
],
[
" # 注意!",
"_____no_output_____"
],
[
"## 当是用Jupyter Notebook编程时,第一步请检查Notebook是否可读性可写",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"## 如果显示read-only,请打开终端(CMD),输入sudo chmod -R 777 filename,给文件夹授权,之后重新打开Jupyter Notebook方可保存文件。",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d03ed716e90bfe9ba35c81a959273d17024e4fb8 | 2,353 | ipynb | Jupyter Notebook | 20200907/Answers/Electrical_Cars.ipynb | SNStatComp/CBSAcademyBD | ae82e9f79ec4bd58f5446a40154ad1fe3c25b602 | [
"CC-BY-4.0"
] | null | null | null | 20200907/Answers/Electrical_Cars.ipynb | SNStatComp/CBSAcademyBD | ae82e9f79ec4bd58f5446a40154ad1fe3c25b602 | [
"CC-BY-4.0"
] | null | null | null | 20200907/Answers/Electrical_Cars.ipynb | SNStatComp/CBSAcademyBD | ae82e9f79ec4bd58f5446a40154ad1fe3c25b602 | [
"CC-BY-4.0"
] | null | null | null | 26.738636 | 108 | 0.549086 | [
[
[
"## Exercise: electrical cars",
"_____no_output_____"
]
],
[
[
"# Imports:\nimport requests\nfrom bs4 import BeautifulSoup\nimport re\nimport time\nheaders = {'user-agent': 'scrapingCourseBot'}",
"_____no_output_____"
],
[
"# Retrieve the number of electrical Renaults built between 2010 and 2014 and\n# print the tag containing the number of occasions:\n\npars = {'bmin': 2010, 'bmax': 2014}\nr2 = requests.get('https://www.gaspedaal.nl/renault/elektrisch?', params=pars, headers=headers)\n\n# The number of occasions is in the HTML as:\n# <h1 class=\"listing__header__total_found\">xxxxx occasions gevonden</h1>\n#match = re.search(r'<h1 class=\"listing__header__total_found\">.*</h1>', r2.text)\n# or alternatively:\nmatch = re.search(r'[\\d\\.]* occasions gevonden', r2.text)\nif match:\n print(match.group())\nelse:\n print('not found')\n",
"_____no_output_____"
],
[
"### Retrieve the number of electrical Renaults for every year between 2010 and 2019:\nfor year in range(2010, 2020):\n pars = {'bmin': year, 'bmax': year}\n r3 = requests.get('https://www.gaspedaal.nl/renault/elektrisch?', params=pars, headers=headers)\n match = re.search(r'[\\d\\.]* occasions gevonden', r3.text)\n if match:\n print(str(year) + \": \" + match.group())\n else:\n print('not found')\n time.sleep(1)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d03ee57156154b28760a60d7de797eb7748e2f9d | 49,851 | ipynb | Jupyter Notebook | Concise_Chit_Chat.ipynb | huan/python-concise-chit-chat | c919d857611826764f080f1cabc92f1b3327b4f0 | [
"Apache-2.0"
] | 1 | 2018-11-20T12:14:38.000Z | 2018-11-20T12:14:38.000Z | Concise_Chit_Chat.ipynb | huan/python-concise-chit-chat | c919d857611826764f080f1cabc92f1b3327b4f0 | [
"Apache-2.0"
] | 3 | 2018-10-08T18:35:55.000Z | 2018-11-20T07:59:56.000Z | Concise_Chit_Chat.ipynb | zixia/concise-chit-chat | c919d857611826764f080f1cabc92f1b3327b4f0 | [
"Apache-2.0"
] | 1 | 2018-11-20T12:14:40.000Z | 2018-11-20T12:14:40.000Z | 46.76454 | 1,924 | 0.513009 | [
[
[
"<a href=\"https://colab.research.google.com/github/huan/concise-chit-chat/blob/master/Concise_Chit_Chat.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Concise Chit Chat\n\nGitHub Repository: <https://github.com/huan/concise-chit-chat>\n\n## Code TODO:\n\n1. create a DataLoader class for dataset preprocess. (Use tf.data.Dataset inside?)\n1. Create a PyPI package for easy load cornell movie curpos dataset(?)\n1. Use PyPI module `embeddings` to load `GLOVES`, or use tfhub to load `GLOVES`?\n1. How to do a `clip_norm`(or set `clip_value`) in Keras with Eager mode but without `tf.contrib`?\n1. Better name for variables & functions\n1. Code clean\n1. Encapsulate all layers to Model Class: \n 1. ChitChatEncoder\n 1. ChitChatDecoder\n 1. ChitChatModel\n1. Re-style to follow the book\n1. ...?\n\n## Book Todo\n\n1. Outlines\n1. What's seq2seq\n1. What's word embedding\n1. \n1. Split code into snips\n1. Write for snips\n1. Content cleaning and optimizing\n1. ...?\n\n## Other\n\n1. `keras.callbacks.TensorBoard` instead of `tf.contrib.summary`?\n - `model.fit(callbacks=[TensorBoard(...)])`\n1. download url? - http://old.pep.com.cn/gzsx/jszx_1/czsxtbjxzy/qrzptgjzxjc/dzkb/dscl/",
"_____no_output_____"
],
[
"### config.py",
"_____no_output_____"
]
],
[
[
"'''doc'''\n\n# GO for start of the sentence\n# DONE for end of the sentence\nGO = '\\b'\nDONE = '\\a'\n\n# max words per sentence\nMAX_LEN = 20\n",
"_____no_output_____"
]
],
[
[
"### data_loader.py",
"_____no_output_____"
]
],
[
[
"'''\ndata loader\n'''\nimport gzip\nimport re\nfrom typing import (\n # Any,\n List,\n Tuple,\n)\n\nimport tensorflow as tf\nimport numpy as np\n\n# from .config import (\n# GO,\n# DONE,\n# MAX_LEN,\n# )\n\nDATASET_URL = 'https://github.com/huan/concise-chit-chat/releases/download/v0.0.1/dataset.txt.gz'\nDATASET_FILE_NAME = 'concise-chit-chat-dataset.txt.gz'\n\n\nclass DataLoader():\n '''data loader'''\n\n def __init__(self) -> None:\n print('DataLoader', 'downloading dataset from:', DATASET_URL)\n dataset_file = tf.keras.utils.get_file(\n DATASET_FILE_NAME,\n origin=DATASET_URL,\n )\n print('DataLoader', 'loading dataset from:', dataset_file)\n\n # dataset_file = './data/dataset.txt.gz'\n\n # with open(path, encoding='iso-8859-1') as f:\n with gzip.open(dataset_file, 'rt') as f:\n self.raw_text = f.read().lower()\n\n self.queries, self.responses \\\n = self.__parse_raw_text(self.raw_text)\n self.size = len(self.queries)\n\n def get_batch(\n self,\n batch_size=32,\n ) -> Tuple[List[List[str]], List[List[str]]]:\n '''get batch'''\n # print('corpus_list', self.corpus)\n batch_indices = np.random.choice(\n len(self.queries),\n size=batch_size,\n )\n batch_queries = self.queries[batch_indices]\n batch_responses = self.responses[batch_indices]\n\n return batch_queries, batch_responses\n\n def __parse_raw_text(\n self,\n raw_text: str\n ) -> Tuple[List[List[str]], List[List[str]]]:\n '''doc'''\n query_list = []\n response_list = []\n\n for line in raw_text.strip('\\n').split('\\n'):\n query, response = line.split('\\t')\n query, response = self.preprocess(query), self.preprocess(response)\n query_list.append('{} {} {}'.format(GO, query, DONE))\n response_list.append('{} {} {}'.format(GO, response, DONE))\n\n return np.array(query_list), np.array(response_list)\n\n def preprocess(self, text: str) -> str:\n '''doc'''\n new_text = text\n\n new_text = re.sub('[^a-zA-Z0-9 .,?!]', ' ', new_text)\n new_text = re.sub(' +', ' ', new_text)\n new_text = re.sub(\n '([\\w]+)([,;.?!#&-\\'\\\"-]+)([\\w]+)?',\n r'\\1 \\2 \\3',\n new_text,\n )\n if len(new_text.split()) > MAX_LEN:\n new_text = (' ').join(new_text.split()[:MAX_LEN])\n match = re.search('[.?!]', new_text)\n if match is not None:\n idx = match.start()\n new_text = new_text[:idx+1]\n\n new_text = new_text.strip().lower()\n\n return new_text\n",
"_____no_output_____"
]
],
[
[
"### vocabulary.py",
"_____no_output_____"
]
],
[
[
"'''doc'''\nimport re\nfrom typing import (\n List,\n)\n\nimport tensorflow as tf\n\n# from .config import (\n# DONE,\n# GO,\n# MAX_LEN,\n# )\n\n\nclass Vocabulary:\n '''voc'''\n def __init__(self, text: str) -> None:\n self.tokenizer = tf.keras.preprocessing.text.Tokenizer(filters='')\n self.tokenizer.fit_on_texts(\n [GO, DONE] + re.split(\n r'[\\s\\t\\n]',\n text,\n )\n )\n # additional 1 for the index 0\n self.size = 1 + len(self.tokenizer.word_index.keys())\n\n def texts_to_padded_sequences(\n self,\n text_list: List[List[str]]\n ) -> tf.Tensor:\n '''doc'''\n sequence_list = self.tokenizer.texts_to_sequences(text_list)\n padded_sequences = tf.keras.preprocessing.sequence.pad_sequences(\n sequence_list,\n maxlen=MAX_LEN,\n padding='post',\n truncating='post',\n )\n\n return padded_sequences\n\n def padded_sequences_to_texts(self, sequence: List[int]) -> str:\n return 'tbw'\n",
"_____no_output_____"
]
],
[
[
"### model.py",
"_____no_output_____"
]
],
[
[
"'''doc'''\nimport tensorflow as tf\nimport numpy as np\nfrom typing import (\n List,\n)\n\n# from .vocabulary import Vocabulary\n# from .config import (\n# DONE,\n# GO,\n# MAX_LENGTH,\n# )\n\nEMBEDDING_DIM = 300\nLATENT_UNIT_NUM = 500\n\n\nclass ChitEncoder(tf.keras.Model):\n '''encoder'''\n def __init__(\n self,\n ) -> None:\n super().__init__()\n\n self.lstm_encoder = tf.keras.layers.CuDNNLSTM(\n units=LATENT_UNIT_NUM,\n return_state=True,\n )\n\n def call(\n self,\n inputs: tf.Tensor, # shape: [batch_size, max_len, embedding_dim]\n training=None,\n mask=None,\n ) -> tf.Tensor:\n _, *state = self.lstm_encoder(inputs)\n return state # shape: ([latent_unit_num], [latent_unit_num])\n\n\nclass ChatDecoder(tf.keras.Model):\n '''decoder'''\n def __init__(\n self,\n voc_size: int,\n ) -> None:\n super().__init__()\n\n self.lstm_decoder = tf.keras.layers.CuDNNLSTM(\n units=LATENT_UNIT_NUM,\n return_sequences=True,\n return_state=True,\n )\n\n self.dense = tf.keras.layers.Dense(\n units=voc_size,\n )\n\n self.time_distributed_dense = tf.keras.layers.TimeDistributed(\n self.dense\n )\n\n self.initial_state = None\n\n def set_state(self, state=None):\n '''doc'''\n # import pdb; pdb.set_trace()\n self.initial_state = state\n\n def call(\n self,\n inputs: tf.Tensor, # shape: [batch_size, None, embedding_dim]\n training=False,\n mask=None,\n ) -> tf.Tensor:\n '''chat decoder call'''\n\n # batch_size = tf.shape(inputs)[0]\n # max_len = tf.shape(inputs)[0]\n\n # outputs = tf.zeros(shape=(\n # batch_size, # batch_size\n # max_len, # max time step\n # LATENT_UNIT_NUM, # dimention of hidden state\n # ))\n\n # import pdb; pdb.set_trace()\n outputs, *states = self.lstm_decoder(inputs, initial_state=self.initial_state)\n self.initial_state = states\n\n outputs = self.time_distributed_dense(outputs)\n return outputs\n\n\nclass ChitChat(tf.keras.Model):\n '''doc'''\n def __init__(\n self,\n vocabulary: Vocabulary,\n ) -> None:\n super().__init__()\n\n self.word_index = vocabulary.tokenizer.word_index\n self.index_word = vocabulary.tokenizer.index_word\n self.voc_size = vocabulary.size\n\n # [batch_size, max_len] -> [batch_size, max_len, voc_size]\n self.embedding = tf.keras.layers.Embedding(\n input_dim=self.voc_size,\n output_dim=EMBEDDING_DIM,\n mask_zero=True,\n )\n\n self.encoder = ChitEncoder()\n # shape: [batch_size, state]\n\n self.decoder = ChatDecoder(self.voc_size)\n # shape: [batch_size, max_len, voc_size]\n\n def call(\n self,\n inputs: List[List[int]], # shape: [batch_size, max_len]\n teacher_forcing_targets: List[List[int]]=None, # shape: [batch_size, max_len]\n training=None,\n mask=None,\n ) -> tf.Tensor: # shape: [batch_size, max_len, embedding_dim]\n '''call'''\n batch_size = tf.shape(inputs)[0]\n\n inputs_embedding = self.embedding(tf.convert_to_tensor(inputs))\n state = self.encoder(inputs_embedding)\n\n self.decoder.set_state(state)\n\n if training:\n teacher_forcing_targets = tf.convert_to_tensor(teacher_forcing_targets)\n teacher_forcing_embeddings = self.embedding(teacher_forcing_targets)\n\n # outputs[:, 0, :].assign([self.__go_embedding()] * batch_size)\n batch_go_embedding = tf.ones([batch_size, 1, 1]) * [self.__go_embedding()]\n batch_go_one_hot = tf.ones([batch_size, 1, 1]) * [tf.one_hot(self.word_index[GO], self.voc_size)]\n\n outputs = batch_go_one_hot\n output = self.decoder(batch_go_embedding)\n\n for t in range(1, MAX_LEN):\n outputs = tf.concat([outputs, output], 1)\n if training:\n target = teacher_forcing_embeddings[:, t, :]\n decoder_input = tf.expand_dims(target, axis=1)\n else:\n decoder_input = self.__indice_to_embedding(tf.argmax(output))\n\n output = self.decoder(decoder_input)\n\n return outputs\n\n def predict(self, inputs: List[int], temperature=1.) -> List[int]:\n '''doc'''\n\n outputs = self([inputs])\n outputs = tf.squeeze(outputs)\n\n word_list = []\n for t in range(1, MAX_LEN):\n output = outputs[t]\n\n indice = self.__logit_to_indice(output, temperature=temperature)\n\n word = self.index_word[indice]\n\n if indice == self.word_index[DONE]:\n break\n\n word_list.append(word)\n\n return ' '.join(word_list)\n\n def __go_embedding(self) -> tf.Tensor:\n return self.embedding(\n tf.convert_to_tensor(self.word_index[GO]))\n\n def __logit_to_indice(\n self,\n inputs,\n temperature=1.,\n ) -> int:\n '''\n [vocabulary_size]\n convert one hot encoding to indice with temperature\n '''\n inputs = tf.squeeze(inputs)\n prob = tf.nn.softmax(inputs / temperature).numpy()\n indice = np.random.choice(self.voc_size, p=prob)\n return indice\n\n def __indice_to_embedding(self, indice: int) -> tf.Tensor:\n tensor = tf.convert_to_tensor([[indice]])\n return self.embedding(tensor)\n",
"_____no_output_____"
]
],
[
[
"### Train",
"_____no_output_____"
],
[
"### Tensor Board\n\n[Quick guide to run TensorBoard in Google Colab](https://www.dlology.com/blog/quick-guide-to-run-tensorboard-in-google-colab/)\n\n`tensorboard` vs `tensorboard/` ?",
"_____no_output_____"
]
],
[
[
"\nLOG_DIR = '/content/data/tensorboard/'\nget_ipython().system_raw(\n 'tensorboard --logdir {} --host 0.0.0.0 --port 6006 &'\n .format(LOG_DIR)\n)\n# Install\n! npm install -g localtunnel\n\n# Tunnel port 6006 (TensorBoard assumed running)\nget_ipython().system_raw('lt --port 6006 >> url.txt 2>&1 &')\n\n# Get url\n! cat url.txt",
"\u001b[K\u001b[?25h/tools/node/bin/lt -> /tools/node/lib/node_modules/localtunnel/bin/client\n\u001b[K\u001b[?25h+ localtunnel@1.9.1\nadded 54 packages from 31 contributors in 2.667s\nyour url is: https://tame-walrus-3.localtunnel.me\n"
],
[
"'''train'''\nimport tensorflow as tf\n\n# from chit_chat import (\n# ChitChat,\n# DataLoader,\n# Vocabulary,\n# )\n\ntf.enable_eager_execution()\n\ndata_loader = DataLoader()\nvocabulary = Vocabulary(data_loader.raw_text)\nchitchat = ChitChat(vocabulary=vocabulary)\n\n\ndef loss(model, x, y) -> tf.Tensor:\n '''doc'''\n weights = tf.cast(\n tf.not_equal(y, 0),\n tf.float32,\n )\n\n prediction = model(\n inputs=x,\n teacher_forcing_targets=y,\n training=True,\n )\n\n # implment the following contrib function in a loop ?\n # https://stackoverflow.com/a/41135778/1123955\n # https://stackoverflow.com/q/48025004/1123955\n return tf.contrib.seq2seq.sequence_loss(\n prediction,\n tf.convert_to_tensor(y),\n weights,\n )\n\n\ndef grad(model, inputs, targets):\n '''doc'''\n with tf.GradientTape() as tape:\n loss_value = loss(model, inputs, targets)\n\n return tape.gradient(loss_value, model.variables)\n\n\ndef train() -> int:\n '''doc'''\n learning_rate = 1e-3\n num_batches = 8000\n batch_size = 128\n\n print('Dataset size: {}, Vocabulary size: {}'.format(\n data_loader.size,\n vocabulary.size,\n ))\n\n optimizer = tf.train.AdamOptimizer(learning_rate=learning_rate)\n\n root = tf.train.Checkpoint(\n optimizer=optimizer,\n model=chitchat,\n optimizer_step=tf.train.get_or_create_global_step(),\n )\n\n root.restore(tf.train.latest_checkpoint('./data/save'))\n print('checkpoint restored.')\n\n writer = tf.contrib.summary.create_file_writer('./data/tensorboard')\n writer.set_as_default()\n\n global_step = tf.train.get_or_create_global_step()\n\n for batch_index in range(num_batches):\n global_step.assign_add(1)\n\n queries, responses = data_loader.get_batch(batch_size)\n\n encoder_inputs = vocabulary.texts_to_padded_sequences(queries)\n decoder_outputs = vocabulary.texts_to_padded_sequences(responses)\n\n grads = grad(chitchat, encoder_inputs, decoder_outputs)\n\n optimizer.apply_gradients(\n grads_and_vars=zip(grads, chitchat.variables)\n )\n\n if batch_index % 10 == 0:\n print(\"batch %d: loss %f\" % (batch_index, loss(\n chitchat, encoder_inputs, decoder_outputs).numpy()))\n root.save('./data/save/model.ckpt')\n print('checkpoint saved.')\n\n with tf.contrib.summary.record_summaries_every_n_global_steps(1):\n # your model code goes here\n tf.contrib.summary.scalar('loss', loss(\n chitchat, encoder_inputs, decoder_outputs).numpy())\n # print('summary had been written.')\n\n return 0\n\n\ndef main() -> int:\n '''doc'''\n return train()\n\n\nmain()\n",
"DataLoader downloading dataset from: https://github.com/huan/concise-chit-chat/releases/download/v0.0.1/dataset.txt.gz\nDownloading data from https://github.com/huan/concise-chit-chat/releases/download/v0.0.1/dataset.txt.gz\n2310144/2304624 [==============================] - 2s 1us/step\nDataLoader loading dataset from: /root/.keras/datasets/concise-chit-chat-dataset.txt.gz\nDataset size: 158015, Vocabulary size: 5001\ncheckpoint restored.\nbatch 0: loss 8.378550\ncheckpoint saved.\nbatch 10: loss 5.363611\ncheckpoint saved.\nbatch 20: loss 5.347839\ncheckpoint saved.\nbatch 30: loss 5.355069\ncheckpoint saved.\nbatch 40: loss 5.263589\ncheckpoint saved.\nbatch 50: loss 5.107776\ncheckpoint saved.\nbatch 60: loss 5.089061\ncheckpoint saved.\nbatch 70: loss 4.997322\ncheckpoint saved.\nbatch 80: loss 4.978524\ncheckpoint saved.\nbatch 90: loss 4.913832\ncheckpoint saved.\nbatch 100: loss 4.860683\ncheckpoint saved.\nbatch 110: loss 4.823889\ncheckpoint saved.\nbatch 120: loss 4.892643\ncheckpoint saved.\nbatch 130: loss 4.658375\ncheckpoint saved.\nbatch 140: loss 4.795345\ncheckpoint saved.\nbatch 150: loss 4.598568\ncheckpoint saved.\nbatch 160: loss 4.728464\ncheckpoint saved.\nbatch 170: loss 4.666811\ncheckpoint saved.\nbatch 180: loss 4.597885\ncheckpoint saved.\nbatch 190: loss 4.595983\ncheckpoint saved.\nbatch 200: loss 4.565698\ncheckpoint saved.\nbatch 210: loss 4.488758\ncheckpoint saved.\nbatch 220: loss 4.618034\ncheckpoint saved.\nbatch 230: loss 4.423510\ncheckpoint saved.\nbatch 240: loss 4.498684\ncheckpoint saved.\nbatch 250: loss 4.484734\ncheckpoint saved.\nbatch 260: loss 4.350574\ncheckpoint saved.\nbatch 270: loss 4.410893\ncheckpoint saved.\nbatch 280: loss 4.302926\ncheckpoint saved.\nbatch 290: loss 4.379134\ncheckpoint saved.\nbatch 300: loss 4.427905\ncheckpoint saved.\nbatch 310: loss 4.361248\ncheckpoint saved.\nbatch 320: loss 4.323555\ncheckpoint saved.\nbatch 330: loss 4.307600\ncheckpoint saved.\nbatch 340: loss 4.380899\ncheckpoint saved.\nbatch 350: loss 4.399303\ncheckpoint saved.\nbatch 360: loss 4.326337\ncheckpoint saved.\nbatch 370: loss 4.202069\ncheckpoint saved.\nbatch 380: loss 4.299171\ncheckpoint saved.\nbatch 390: loss 4.236771\ncheckpoint saved.\n"
],
[
"#! rm -fvr data/tensorboard\n# ! pwd\n# ! rm -frv data/save\n# ! rm -fr /content/data/tensorboard\n# ! kill 2823\n# ! kill -9 2823\n# ! ps axf | grep lt\n",
"removed 'data/save/model.ckpt-8.data-00000-of-00001'\nremoved 'data/save/checkpoint'\nremoved 'data/save/model.ckpt-11.data-00000-of-00001'\nremoved 'data/save/model.ckpt-8.index'\nremoved 'data/save/model.ckpt-1.data-00000-of-00001'\nremoved 'data/save/model.ckpt-6.index'\nremoved 'data/save/model.ckpt-4.index'\nremoved 'data/save/model.ckpt-4.data-00000-of-00001'\nremoved 'data/save/model.ckpt-12.index'\nremoved 'data/save/model.ckpt-9.data-00000-of-00001'\nremoved 'data/save/model.ckpt-3.data-00000-of-00001'\nremoved 'data/save/model.ckpt-9.index'\nremoved 'data/save/model.ckpt-7.index'\nremoved 'data/save/model.ckpt-10.data-00000-of-00001'\nremoved 'data/save/model.ckpt-2.index'\nremoved 'data/save/model.ckpt-3.index'\nremoved 'data/save/model.ckpt-6.data-00000-of-00001'\nremoved 'data/save/model.ckpt-11.index'\nremoved 'data/save/model.ckpt-7.data-00000-of-00001'\nremoved 'data/save/model.ckpt-2.data-00000-of-00001'\nremoved 'data/save/model.ckpt-1.index'\nremoved 'data/save/model.ckpt-5.index'\nremoved 'data/save/model.ckpt-10.index'\nremoved 'data/save/model.ckpt-12.data-00000-of-00001'\nremoved 'data/save/model.ckpt-5.data-00000-of-00001'\nremoved directory 'data/save'\n"
],
[
"! cat url.txt",
"your url is: https://bright-fox-51.localtunnel.me\n"
]
],
[
[
"### chat.py",
"_____no_output_____"
]
],
[
[
"'''train'''\n# import tensorflow as tf\n\n# from chit_chat import (\n# ChitChat,\n# DataLoader,\n# Vocabulary,\n# DONE,\n# GO,\n# )\n\n# tf.enable_eager_execution()\n\n\ndef main() -> int:\n '''chat main'''\n data_loader = DataLoader()\n vocabulary = Vocabulary(data_loader.raw_text)\n\n print('Dataset size: {}, Vocabulary size: {}'.format(\n data_loader.size,\n vocabulary.size,\n ))\n\n chitchat = ChitChat(vocabulary)\n checkpoint = tf.train.Checkpoint(model=chitchat)\n checkpoint.restore(tf.train.latest_checkpoint('./data/save'))\n print('checkpoint restored.')\n\n return cli(chitchat, vocabulary=vocabulary, data_loader=data_loader)\n\n\ndef cli(chitchat: ChitChat, data_loader: DataLoader, vocabulary: Vocabulary):\n '''command line interface'''\n index_word = vocabulary.tokenizer.index_word\n word_index = vocabulary.tokenizer.word_index\n query = ''\n while True:\n try:\n # Get input sentence\n query = input('> ').lower()\n # Check if it is quit case\n if query == 'q' or query == 'quit':\n break\n # Normalize sentence\n query = data_loader.preprocess(query)\n query = '{} {} {}'.format(GO, query, DONE)\n # Evaluate sentence\n query_sequence = vocabulary.texts_to_padded_sequences([query])[0]\n\n response_sequence = chitchat.predict(query_sequence, 1)\n\n # Format and print response sentence\n response_word_list = [\n index_word[indice]\n for indice in response_sequence\n if indice != 0 and indice != word_index[DONE]\n ]\n\n print('Bot:', ' '.join(response_word_list))\n\n except KeyError:\n print(\"Error: Encountered unknown word.\")\n\n\nmain()\n",
"_____no_output_____"
],
[
"! cat /proc/cpuinfo",
"processor\t: 0\nvendor_id\t: GenuineIntel\ncpu family\t: 6\nmodel\t\t: 63\nmodel name\t: Intel(R) Xeon(R) CPU @ 2.30GHz\nstepping\t: 0\nmicrocode\t: 0x1\ncpu MHz\t\t: 2299.998\ncache size\t: 46080 KB\nphysical id\t: 0\nsiblings\t: 2\ncore id\t\t: 0\ncpu cores\t: 1\napicid\t\t: 0\ninitial apicid\t: 0\nfpu\t\t: yes\nfpu_exception\t: yes\ncpuid level\t: 13\nwp\t\t: yes\nflags\t\t: fpu vme de pse tsc msr pae mce cx8 apic sep mtrr pge mca cmov pat pse36 clflush mmx fxsr sse sse2 ss ht syscall nx pdpe1gb rdtscp lm constant_tsc rep_good nopl xtopology nonstop_tsc cpuid pni pclmulqdq ssse3 fma cx16 sse4_1 sse4_2 x2apic movbe popcnt aes xsave avx f16c rdrand hypervisor lahf_lm abm pti fsgsbase tsc_adjust bmi1 avx2 smep bmi2 erms xsaveopt arch_capabilities\nbugs\t\t: cpu_meltdown spectre_v1 spectre_v2 spec_store_bypass l1tf\nbogomips\t: 4599.99\nclflush size\t: 64\ncache_alignment\t: 64\naddress sizes\t: 46 bits physical, 48 bits virtual\npower management:\n\nprocessor\t: 1\nvendor_id\t: GenuineIntel\ncpu family\t: 6\nmodel\t\t: 63\nmodel name\t: Intel(R) Xeon(R) CPU @ 2.30GHz\nstepping\t: 0\nmicrocode\t: 0x1\ncpu MHz\t\t: 2299.998\ncache size\t: 46080 KB\nphysical id\t: 0\nsiblings\t: 2\ncore id\t\t: 0\ncpu cores\t: 1\napicid\t\t: 1\ninitial apicid\t: 1\nfpu\t\t: yes\nfpu_exception\t: yes\ncpuid level\t: 13\nwp\t\t: yes\nflags\t\t: fpu vme de pse tsc msr pae mce cx8 apic sep mtrr pge mca cmov pat pse36 clflush mmx fxsr sse sse2 ss ht syscall nx pdpe1gb rdtscp lm constant_tsc rep_good nopl xtopology nonstop_tsc cpuid pni pclmulqdq ssse3 fma cx16 sse4_1 sse4_2 x2apic movbe popcnt aes xsave avx f16c rdrand hypervisor lahf_lm abm pti fsgsbase tsc_adjust bmi1 avx2 smep bmi2 erms xsaveopt arch_capabilities\nbugs\t\t: cpu_meltdown spectre_v1 spectre_v2 spec_store_bypass l1tf\nbogomips\t: 4599.99\nclflush size\t: 64\ncache_alignment\t: 64\naddress sizes\t: 46 bits physical, 48 bits virtual\npower management:\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d03efc220407211499745a0d86f912bd8327f124 | 54,542 | ipynb | Jupyter Notebook | CaracteristicasPorEquipo.ipynb | Howl24/Lineup-Prediction | 9b890a52a444010a6a07de807426305fae5a4058 | [
"MIT"
] | null | null | null | CaracteristicasPorEquipo.ipynb | Howl24/Lineup-Prediction | 9b890a52a444010a6a07de807426305fae5a4058 | [
"MIT"
] | null | null | null | CaracteristicasPorEquipo.ipynb | Howl24/Lineup-Prediction | 9b890a52a444010a6a07de807426305fae5a4058 | [
"MIT"
] | null | null | null | 52.953398 | 18,004 | 0.567141 | [
[
[
"# Scraping de Características Generales",
"_____no_output_____"
],
[
"Se obtienen características generales del equipo de la página whoscored.\nLas estadísticas son de la temporada más reciente.\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"from selenium import webdriver \nfrom selenium.webdriver.common.by import By \nfrom selenium.webdriver.support.ui import WebDriverWait \nfrom selenium.webdriver.support import expected_conditions as EC \nfrom selenium.common.exceptions import TimeoutException\n\nfrom selenium.webdriver.support.ui import Select\n",
"_____no_output_____"
],
[
"option = webdriver.ChromeOptions()\noption.add_argument(\" — incognito\")",
"_____no_output_____"
],
[
"browser = webdriver.Chrome(executable_path='./chromedriver',\n chrome_options=option)",
"_____no_output_____"
],
[
"team_links = [\n 'https://es.whoscored.com/Teams/326/Archive/Rusia-Russia',\n 'https://es.whoscored.com/Teams/967/Archive/Uruguay-Uruguay',\n 'https://es.whoscored.com/Teams/340/Archive/Portugal-Portugal',\n 'https://es.whoscored.com/Teams/338/Archive/Espa%C3%B1a-Spain',\n 'https://es.whoscored.com/Teams/1293/Archive/Ir%C3%A1n-Iran',\n 'https://es.whoscored.com/Teams/341/Archive/Francia-France',\n 'https://es.whoscored.com/Teams/328/Archive/Australia-Australia',\n 'https://es.whoscored.com/Teams/416/Archive/Per%C3%BA-Peru',\n 'https://es.whoscored.com/Teams/425/Archive/Dinamarca-Denmark',\n 'https://es.whoscored.com/Teams/346/Archive/Argentina-Argentina',\n 'https://es.whoscored.com/Teams/770/Archive/Islandia-Iceland',\n 'https://es.whoscored.com/Teams/337/Archive/Croacia-Croatia',\n 'https://es.whoscored.com/Teams/977/Archive/Nigeria-Nigeria',\n 'https://es.whoscored.com/Teams/409/Archive/Brasil-Brazil',\n 'https://es.whoscored.com/Teams/423/Archive/Suiza-Switzerland',\n 'https://es.whoscored.com/Teams/970/Archive/Costa-Rica-Costa-Rica',\n 'https://es.whoscored.com/Teams/336/Archive/Alemania-Germany',\n 'https://es.whoscored.com/Teams/972/Archive/M%C3%A9xico-Mexico',\n 'https://es.whoscored.com/Teams/344/Archive/Suecia-Sweden',\n 'https://es.whoscored.com/Teams/1159/Archive/Corea-Del-sur-South-Korea',\n 'https://es.whoscored.com/Teams/339/Archive/B%C3%A9lgica-Belgium',\n 'https://es.whoscored.com/Teams/959/Archive/T%C3%BAnez-Tunisia',\n 'https://es.whoscored.com/Teams/345/Archive/Inglaterra-England',\n 'https://es.whoscored.com/Teams/342/Archive/Polonia-Poland',\n 'https://es.whoscored.com/Teams/957/Archive/Senegal-Senegal',\n 'https://es.whoscored.com/Teams/408/Archive/Colombia-Colombia',\n 'https://es.whoscored.com/Teams/986/Archive/Japan-Japan'\n]",
"_____no_output_____"
],
[
"def wait_browser(browser, load_xpath, timeout=20):\n try:\n WebDriverWait(browser, timeout).until(\n EC.visibility_of_element_located(\n (By.XPATH, load_xpath)))\n except TimeoutException:\n print(\"Timed out waiting for page to load\")\n browser.quit()",
"_____no_output_____"
],
[
"import pandas as pd\n\ndata = []\n\nfor team_link in team_links:\n team_data = {}\n \n browser.get(team_link)\n wait_browser(browser, '//a[@class=\"team-link\"]') \n team_data['Equipo'] = browser.find_element_by_xpath('//a[@class=\"team-link\"]').text\n sidebox = browser.find_element_by_xpath('//div[@class=\"team-profile-side-box\"]')\n team_data['rating'] = sidebox.find_element_by_xpath('//div[@class=\"rating\"]').text \n \n \n stats_container = browser.find_element_by_xpath('//div[@class=\"stats-container\"]')\n labels = stats_container.find_elements_by_tag_name('dt')\n values = stats_container.find_elements_by_tag_name('dd')\n for l, v in zip(labels, values):\n team_data[l.text] = v.text\n \n data.append(team_data)\n \ndf = pd.DataFrame(data)\ndf.head()",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.to_csv(\"características_equipos.csv\", index=False)",
"_____no_output_____"
]
],
[
[
"# Procesamiento de datos de equipos\n",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport pandas as pd",
"_____no_output_____"
],
[
"df = pd.read_csv(\"características_equipos.csv\")",
"_____no_output_____"
],
[
"df['rating'] = df['rating'].apply(lambda x: x.replace(',', '.')).astype(float)",
"_____no_output_____"
],
[
"x = range(len(df['rating']))",
"_____no_output_____"
],
[
"df = df.sort_values('rating')",
"_____no_output_____"
],
[
"plt.barh(x, df['rating'])\nplt.yticks(x, df['Equipo'])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03f1325eacd252f9f80709a100aef116d17c81d | 18,436 | ipynb | Jupyter Notebook | heuristic_LWR.ipynb | rubensolozabal/Job-shop-Scheduling-using-Policy-Gradients | 355c617a6cd80bc7cedca2934a5b94c7d523b0bb | [
"MIT"
] | null | null | null | heuristic_LWR.ipynb | rubensolozabal/Job-shop-Scheduling-using-Policy-Gradients | 355c617a6cd80bc7cedca2934a5b94c7d523b0bb | [
"MIT"
] | null | null | null | heuristic_LWR.ipynb | rubensolozabal/Job-shop-Scheduling-using-Policy-Gradients | 355c617a6cd80bc7cedca2934a5b94c7d523b0bb | [
"MIT"
] | null | null | null | 85.748837 | 12,200 | 0.785203 | [
[
[
"from IPython.core.debugger import set_trace\n\nimport numpy as np\n\nimport import_ipynb\nfrom environment import *\n",
"importing Jupyter notebook from environment.ipynb\nimporting Jupyter notebook from config.ipynb\n"
],
[
"def heuristicLWR(num_jobs, num_mc, machines, durations):\n \n \n machines_ = np.array(machines)\n tmp = np.zeros((num_jobs,num_mc+1), dtype=int)\n tmp[:,:-1] = machines_\n machines_ = tmp\n \n \n durations_ = np.array(durations)\n tmp = np.zeros((num_jobs,num_mc+1), dtype=int)\n tmp[:,:-1] = durations_\n durations_ = tmp\n \n indices = np.zeros([num_jobs], dtype=int)\n \n # Internal variables\n previousTaskReadyTime = np.zeros([num_jobs], dtype=int)\n machineReadyTime = np.zeros([num_mc], dtype=int)\n\n placements = [[] for _ in range(num_mc)]\n \n\n # While...\n while(not np.array_equal(indices, np.ones([num_jobs], dtype=int)*num_mc)):\n \n machines_Idx = machines_[range(num_jobs),indices]\n durations_Idx = durations_[range(num_jobs),indices]\n\n \n # 1: Check previous Task and machine availability\n mask = np.zeros([num_jobs], dtype=bool)\n \n for j in range(num_jobs):\n \n if previousTaskReadyTime[j] == 0 and machineReadyTime[machines_Idx[j]] == 0 and indices[j]<num_mc:\n mask[j] = True\n\n\n \n # 2: Competition SPT\n \n for m in range(num_mc):\n \n job = None\n remaining = 99999\n \n for j in range(num_jobs):\n \n tmp = np.sum(durations_[j][indices[j]:])\n \n if machines_Idx[j] == m and tmp < remaining and mask[j]:\n job = j\n remaining = tmp\n \n \n if job != None:\n \n placements[m].append([job, indices[job]])\n\n previousTaskReadyTime[job] += durations_Idx[job]\n machineReadyTime[m] += durations_Idx[job]\n\n indices[job] += 1\n\n \n \n # time +1\n \n previousTaskReadyTime = np.maximum(previousTaskReadyTime - 1 , np.zeros([num_jobs], dtype=int)) \n machineReadyTime = np.maximum(machineReadyTime - 1 , np.zeros([num_mc], dtype=int)) \n \n \n return placements\n",
"_____no_output_____"
],
[
"if __name__ == \"__main__\":\n \n \n # Import environment\n config = Config()\n config.machine_profile = \"xsmall_default\"\n config.job_profile = \"xsmall_default\"\n config.reconfigure()\n \n # Configure environment\n env = Environment(config)\n env.clear()\n \n \n # Read problem instance\n filename = \"datasets/inference/dataset_xsmall.data\"\n\n with open(filename, \"r\") as file:\n NB_JOBS, NB_MACHINES = [int(v) for v in file.readline().split()]\n JOBS = [[int(v) for v in file.readline().split()] for i in range(NB_JOBS)]\n\n #-----------------------------------------------------------------------------\n # Prepare the data for modeling\n #-----------------------------------------------------------------------------\n\n # Build list of machines. MACHINES[j][s] = id of the machine for the operation s of the job j\n machines = [[JOBS[j][2 * s] for s in range(NB_MACHINES)] for j in range(NB_JOBS)]\n\n\n # Build list of durations. DURATION[j][s] = duration of the operation s of the job j\n durations = [[JOBS[j][2 * s + 1] for s in range(NB_MACHINES)] for j in range(NB_JOBS)]\n\n\n placements = heuristicLWR(NB_JOBS, NB_MACHINES, machines, durations)\n \n\n env.step(machines, durations, placements)\n \n print(\"Makespan: \", env.makespan)\n \n env.plot(save=False)\n ",
"Makespan: 41\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d03f14f65a8a87a0ae6889743249f914b0f073e0 | 40,365 | ipynb | Jupyter Notebook | TEST/Database/Database - Project 3.ipynb | LoriWard/COVID-Trends-Website | 6b4a4dd30a38ff1be7549df9e49bff766c1c2659 | [
"MIT"
] | null | null | null | TEST/Database/Database - Project 3.ipynb | LoriWard/COVID-Trends-Website | 6b4a4dd30a38ff1be7549df9e49bff766c1c2659 | [
"MIT"
] | null | null | null | TEST/Database/Database - Project 3.ipynb | LoriWard/COVID-Trends-Website | 6b4a4dd30a38ff1be7549df9e49bff766c1c2659 | [
"MIT"
] | null | null | null | 33.977273 | 217 | 0.352731 | [
[
[
"# Database Load",
"_____no_output_____"
]
],
[
[
"# Dependencies and Setup\nimport json\nimport os\nimport pandas as pd\nimport urllib.request\nimport requests\n# from config import db_pwd, db_user\nfrom config import URI\nfrom sqlalchemy import create_engine",
"_____no_output_____"
]
],
[
[
"## Load Data into DataFrame",
"_____no_output_____"
]
],
[
[
"csv_file = \"data/Merged_data.csv\"\ngoogle_data_df = pd.read_csv(csv_file)\ngoogle_data_df.head()",
"_____no_output_____"
],
[
"google_df = google_data_df[[\"states\",\"dates\",\"SMA_retail_recreation\", \"SMA_grocery_pharmacy\", \"SMA_parks\",\n \"SMA_transit\", \"SMA_workplaces\", \"SMA_residential\", \"case_count\", \"new_case_count\", \"revenue_all\", \"revenue_ss60\", \"deaths\"]]\ngoogle_df.head()",
"_____no_output_____"
],
[
"google_df = google_df.rename(columns = {\"SMA_retail_recreation\":'sma_retail_recreation',\n \"SMA_grocery_pharmacy\":'sma_grocery_pharmacy', \"SMA_parks\":'sma_parks',\"SMA_transit\":'sma_transit',\"SMA_workplaces\":'sma_workplaces',\"SMA_residential\":'sma_residential'})\ngoogle_df",
"_____no_output_____"
],
[
"# Lisa Connection to the database\nrds_connection_string = f\"{db_user}:{{db_pwd}}@localhost:5432/mobility_db\"\nengine = create_engine(f'postgresql://{rds_connection_string}')",
"_____no_output_____"
],
[
"# Stojancho Connection to the database\n# rds_connection_string = f\"postgres:{pcode}@localhost:5432/mobility2_db\"\nengine = create_engine(f'{URI}')",
"_____no_output_____"
]
],
[
[
"### Check for tables",
"_____no_output_____"
]
],
[
[
"engine.table_names()",
"_____no_output_____"
]
],
[
[
"### Use pandas to load csv converted DataFrame into database",
"_____no_output_____"
]
],
[
[
"google_df.to_sql(name='merged_data', con=engine, if_exists='append', index=False)",
"_____no_output_____"
]
],
[
[
"### Confirm data has been added by querying the tables",
"_____no_output_____"
]
],
[
[
"pd.read_sql_query('select * from merged_data', con=engine).head(10)",
"_____no_output_____"
],
[
"pd.read_sql_query('select sma_workplaces from merged_data', con=engine).head(10)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d03f225417383657bdaeb101d41412d29802b0c1 | 404,222 | ipynb | Jupyter Notebook | community/awards/teach_me_qiskit_2018/quantum_machine_learning/1_K_Means/Quantum K-Means Algorithm.ipynb | Chibikuri/qiskit-tutorials | 15c121b95249de17e311c869fbc455210b2fcf5e | [
"Apache-2.0"
] | 2 | 2017-11-09T16:33:14.000Z | 2018-02-26T00:42:17.000Z | community/awards/teach_me_qiskit_2018/quantum_machine_learning/1_K_Means/Quantum K-Means Algorithm.ipynb | Chibikuri/qiskit-tutorials | 15c121b95249de17e311c869fbc455210b2fcf5e | [
"Apache-2.0"
] | 1 | 2020-05-08T20:25:11.000Z | 2020-05-08T20:25:11.000Z | community/awards/teach_me_qiskit_2018/quantum_machine_learning/1_K_Means/Quantum K-Means Algorithm.ipynb | Chibikuri/qiskit-tutorials | 15c121b95249de17e311c869fbc455210b2fcf5e | [
"Apache-2.0"
] | 2 | 2019-03-24T21:00:25.000Z | 2019-03-24T21:57:10.000Z | 325.460548 | 9,352 | 0.937962 | [
[
[
"<img src=\"../../../../images/qiskit-heading.gif\" alt=\"Note: In order for images to show up in this jupyter notebook you need to select File => Trusted Notebook\" width=\"500 px\" align=\"left\">",
"_____no_output_____"
],
[
"\n# _*Quantum K-Means algorithm*_ \n\nThe latest version of this notebook is available on https://github.com/qiskit/qiskit-tutorial.\n\n***\n### Contributors \nShan Jin, Xi He, Xiaokai Hou, Li Sun, Dingding Wen, Shaojun Wu and Xiaoting Wang$^{1}$\n\n1. Institute of Fundamental and Frontier Sciences, University of Electronic Science and Technology of China,Chengdu, China,610051\n***",
"_____no_output_____"
],
[
"## Introduction\nClustering algorithm is a typical unsupervised learning algorithm, which is mainly used to automatically classify similar samples into one category.In the clustering algorithm, according to the similarity between the samples, the samples are divided into different categories. For different similarity calculation methods, different clustering results will be obtained. The commonly used similarity calculation method is the Euclidean distance method.\nWhat we want to show is the quantum K-Means algorithm. The K-Means algorithm is a distance-based clustering algorithm that uses distance as an evaluation index for similarity, that is, the closer the distance between two objects is, the greater the similarity. The algorithm considers the cluster to be composed of objects that are close together, so the compact and independent cluster is the ultimate target.\n\n\n#### Experiment design\nThe implementation of the quantum K-Means algorithm mainly uses the swap test to compare the distances among the input data points. Select K points randomly from N data points as centroids, measure the distance from each point to each centroid, and assign it to the nearest centroid- class, recalculate centroids of each class that has been obtained, and iterate 2 to 3 steps until the new centroid is equal to or less than the specified threshold, and the algorithm ends. In our example, we selected 6 data points, 2 centroids, and used the swap test circuit to calculate the distance. Finally, we obtained two clusters of data points.\n$|0\\rangle$ is an auxiliary qubit, through left $H$ gate, it will be changed to $\\frac{1}{\\sqrt{2}}(|0\\rangle + |1\\rangle)$. Then under the control of $|1\\rangle$, the circuit will swap two vectors $|x\\rangle$ and $|y\\rangle$. Finally, we get the result at the right end of the circuit:\n$$|0_{anc}\\rangle |x\\rangle |y\\rangle \\rightarrow \\frac{1}{2}|0_{anc}\\rangle(|xy\\rangle + |yx\\rangle) + \\frac{1}{2}|1_{anc}\\rangle(|xy\\rangle - |yx\\rangle)$$\nIf we measure auxiliary qubit alone, then the probability of final state in the ground state $|1\\rangle$ is:\n$$P(|1_{anc}\\rangle) = \\frac{1}{2} - \\frac{1}{2}|\\langle x | y \\rangle|^2$$\nIf we measure auxiliary qubit alone, then the probability of final state in the ground state $|1\\rangle$ is:\n$$Euclidean \\ distance = \\sqrt{(2 - 2|\\langle x | y \\rangle|)}$$\nSo, we can see that the probability of measuring $|1\\rangle$ has positive correlation with the Euclidean distance.\nThe schematic diagram of quantum K-Means is as the follow picture.[[1]](#cite) \n<img src=\"../images/k_means_circuit.png\">\nTo make our algorithm can be run using qiskit, we design a more detailed circuit to achieve our algorithm. \n|\n#### Quantum K-Means circuit\n<img src=\"../images/k_means.png\">",
"_____no_output_____"
],
[
"## Data points\n<table border=\"1\">\n<tr>\n<td>point num</td>\n<td>theta</td>\n<td>phi</td>\n<td>lam</td>\n<td>x</td>\n<td>y</td>\n</tr>\n<tr>\n<td>1</td>\n<td>0.01</td>\n<td>pi</td>\n<td>pi</td>\n<td>0.710633</td>\n<td>0.703562</td>\n</tr>\n<tr>\n<td>2</td>\n<td>0.02</td>\n<td>pi</td>\n<td>pi</td>\n<td>0.714142</td>\n<td>0.7</td>\n</tr>\n<tr>\n<td>3</td>\n<td>0.03</td>\n<td>pi</td>\n<td>pi</td>\n<td>0.717633</td>\n<td>0.696421</td>\n</tr>\n<tr>\n<td>4</td>\n<td>0.04</td>\n<td>pi</td>\n<td>pi</td>\n<td>0.721107</td>\n<td>0.692824</td>\n</tr>\n<tr>\n<td>5</td>\n<td>0.05</td>\n<td>pi</td>\n<td>pi</td>\n<td>0.724562</td>\n<td>0.68921</td>\n</tr>\n<tr>\n<td>6</td>\n<td>1.31</td>\n<td>pi</td>\n<td>pi</td>\n<td>0.886811</td>\n<td>0.462132</td>\n</tr>\n<tr>\n<td>7</td>\n<td>1.32</td>\n<td>pi</td>\n<td>pi</td>\n<td>0.889111</td>\n<td>0.457692</td>\n</tr>\n<tr>\n<td>8</td>\n<td>1.33</td>\n<td>pi</td>\n<td>pi</td>\n<td>0.891388</td>\n<td>0.453241</td>\n</tr>\n<tr>\n<td>9</td>\n<td>1.34</td>\n<td>pi</td>\n<td>pi</td>\n<td>0.893643</td>\n<td>0.448779</td>\n</tr>\n<tr>\n<td>10</td>\n<td>1.35</td>\n<td>pi</td>\n<td>pi</td>\n<td>0.895876</td>\n<td>0.444305</td>\n</tr>\n",
"_____no_output_____"
],
[
"## Quantum K-Means algorithm program",
"_____no_output_____"
]
],
[
[
"# import math lib\nfrom math import pi\n\n# import Qiskit\nfrom qiskit import Aer, IBMQ, execute\nfrom qiskit import QuantumCircuit, ClassicalRegister, QuantumRegister\n\n# import basic plot tools\nfrom qiskit.tools.visualization import plot_histogram",
"_____no_output_____"
],
[
"# To use local qasm simulator\nbackend = Aer.get_backend('qasm_simulator')",
"_____no_output_____"
]
],
[
[
"In this section, we first judge the version of Python and import the packages of qiskit, math to implement the following code. We show our algorithm on the ibm_qasm_simulator, if you need to run it on the real quantum conputer, please remove the \"#\" in frint of \"import Qconfig\".",
"_____no_output_____"
]
],
[
[
"theta_list = [0.01, 0.02, 0.03, 0.04, 0.05, 1.31, 1.32, 1.33, 1.34, 1.35]",
"_____no_output_____"
]
],
[
[
"Here we define the number pi in the math lib, because we need to use u3 gate. And we also define a list about the parameter theta which we need to use in the u3 gate. As the same above, if you want to implement on the real quantum comnputer, please remove the symbol \"#\" and configure your local Qconfig.py file. ",
"_____no_output_____"
]
],
[
[
"# create Quantum Register called \"qr\" with 5 qubits\nqr = QuantumRegister(5, name=\"qr\")\n# create Classical Register called \"cr\" with 5 bits\ncr = ClassicalRegister(5, name=\"cr\")\n \n# Creating Quantum Circuit called \"qc\" involving your Quantum Register \"qr\"\n# and your Classical Register \"cr\"\nqc = QuantumCircuit(qr, cr, name=\"k_means\")\n \n#Define a loop to compute the distance between each pair of points\nfor i in range(9):\n for j in range(1,10-i):\n # Set the parament theta about different point\n theta_1 = theta_list[i]\n theta_2 = theta_list[i+j]\n #Achieve the quantum circuit via qiskit\n qc.h(qr[2])\n qc.h(qr[1])\n qc.h(qr[4])\n qc.u3(theta_1, pi, pi, qr[1])\n qc.u3(theta_2, pi, pi, qr[4])\n qc.cswap(qr[2], qr[1], qr[4])\n qc.h(qr[2])\n \n qc.measure(qr[2], cr[2])\n qc.reset(qr)\n \n job = execute(qc, backend=backend, shots=1024)\n result = job.result() \n print(result)\n print('theta_1:' + str(theta_1))\n print('theta_2:' + str(theta_2))\n# print( result.get_data(qc))\n plot_histogram(result.get_counts())",
"COMPLETED\ntheta_1:0.01\ntheta_2:0.02\n"
]
],
[
[
"Here we achieve the function k_means() and the test main function to run the program. Considering the qubits controlling direction of ibmqx4, we takes the quantum register 1, 2, 4 as our working register, if you want to run this program on other computer, please redesign the circuit structure to ensure your program can be run accurately. ",
"_____no_output_____"
],
[
"## Result analysis\nIn this program, we take the quantum register 1, 2, 4 as our operated register (considering the condition when using ibmqx4.) We take the quantum register 1, 4 storing the input information about data points, and the quantum register 2 as controlling register to decide whether to use the swap operator. To estimate the distance of any pair of data points, we use a loop to implement the K-Means Circuit. In the end, we measure the controlling register to judge the distance between two data points. The probability when we get 1 means that the distance between two data points.",
"_____no_output_____"
],
[
"## Reference\n<cite>[1].Quantum algorithms for supervised and unsupervised machine learning(*see open access: [ arXiv:1307.0411v2](https://arxiv.org/abs/1307.0411)*)</cite><a id='cite'></a>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d03f24d523f90ebb9d973522c733f1bf324ad7ee | 3,477 | ipynb | Jupyter Notebook | python/basicml/basicml.ipynb | imjoseangel/100DaysOfCode | bff90569033e2b02a56e893bd45727125962aeb3 | [
"MIT"
] | 1 | 2022-03-30T12:59:44.000Z | 2022-03-30T12:59:44.000Z | python/basicml/basicml.ipynb | imjoseangel/100DaysOfCode | bff90569033e2b02a56e893bd45727125962aeb3 | [
"MIT"
] | null | null | null | python/basicml/basicml.ipynb | imjoseangel/100DaysOfCode | bff90569033e2b02a56e893bd45727125962aeb3 | [
"MIT"
] | 3 | 2019-08-13T11:33:36.000Z | 2022-03-08T22:00:09.000Z | 3,477 | 3,477 | 0.69025 | [
[
[
"#! /usr/bin/env python\n# -*- coding: utf-8 -*-\nfrom __future__ import (division, absolute_import, print_function,\n unicode_literals)\nfrom sklearn import tree",
"_____no_output_____"
],
[
"# features = [[155, \"rough\"], [180, \"rough\"], [135, \"smooth\"],\n# [110, \"smooth\"], etc] # Input to classifier\nfeatures = [[155, 0], [180, 0], [135, 1], [110, 1], [300, 0], [320, 0],\n [350, 1], [380, 1]] # scikit-learn requires real-valued features",
"_____no_output_____"
],
[
"# labels = [\"orange\", \"orange\", \"apple\", \"apple\", \"melon\", \"melon\",\n# \"watermelon\", \"watermelon\"] # output values\nlabels = ['🍊', '🍊', '🍎', '🍎', '🍈', '🍈', '🍉', '🍉']",
"_____no_output_____"
],
[
"# Training classifier\nclassifier = tree.DecisionTreeClassifier() # using decision tree classifier\nclassifier = classifier.fit(features, labels) # Find patterns in data",
"_____no_output_____"
],
[
"# Making predictions\nresult = classifier.predict([[120, 1]])",
"_____no_output_____"
],
[
"# Output is apple\nprint(result[0])",
"🍎\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03f31d5f5cd5f92c37616657b32f371ae5b2ead | 7,254 | ipynb | Jupyter Notebook | experiments/BatchBALD/AMN_ens_random_original_breakout_larger_batch_size.ipynb | Tony-Cheng/Active-Reinforcement-Learning | 50bb65106ae1f957d8cb6cb5706ce1285519e6b4 | [
"MIT"
] | null | null | null | experiments/BatchBALD/AMN_ens_random_original_breakout_larger_batch_size.ipynb | Tony-Cheng/Active-Reinforcement-Learning | 50bb65106ae1f957d8cb6cb5706ce1285519e6b4 | [
"MIT"
] | null | null | null | experiments/BatchBALD/AMN_ens_random_original_breakout_larger_batch_size.ipynb | Tony-Cheng/Active-Reinforcement-Learning | 50bb65106ae1f957d8cb6cb5706ce1285519e6b4 | [
"MIT"
] | null | null | null | 34.216981 | 359 | 0.610973 | [
[
[
"from tqdm.notebook import tqdm\nimport math\nimport gym\nimport torch\nimport torch.optim as optim \nfrom torch.utils.tensorboard import SummaryWriter\nfrom collections import deque\n\nfrom active_rl.networks.dqn_atari import ENS_DQN\nfrom active_rl.utils.memory import LabelledReplayMemory\nfrom active_rl.utils.optimization import AMN_optimization_ensemble\nfrom active_rl.environments.atari_wrappers import make_atari, wrap_deepmind\nfrom active_rl.utils.atari_utils import fp, ActionSelector, evaluate\nfrom active_rl.utils.acquisition_functions import ens_random",
"_____no_output_____"
],
[
"env_name = 'Breakout'\nenv_raw = make_atari('{}NoFrameskip-v4'.format(env_name))\nenv = wrap_deepmind(env_raw, frame_stack=False, episode_life=False, clip_rewards=True)\nc,h,w = c,h,w = fp(env.reset()).shape\nn_actions = env.action_space.n",
"_____no_output_____"
],
[
"BATCH_SIZE = 64\nLR = 0.0000625\nGAMMA = 0.99\nEPS = 0.05\nNUM_STEPS = 10000000\nNOT_LABELLED_CAPACITY = 1000\nLABELLED_CAPACITY = 100000\nINITIAL_STEPS=NOT_LABELLED_CAPACITY\nTRAINING_PER_LABEL = 20.\nPERCENTAGE = 0.1\nTRAINING_ITER = int(TRAINING_PER_LABEL * NOT_LABELLED_CAPACITY * PERCENTAGE)\n\nNAME = f\"AMN_ens_random_breakout_original_label_percentage_{PERCENTAGE}_batch_size_{PERCENTAGE * NOT_LABELLED_CAPACITY}\"",
"_____no_output_____"
],
[
"device = 'cuda:0'\n# AMN_net = MC_DQN(n_actions).to(device)\nAMN_net = ENS_DQN(n_actions).to(device)\nexpert_net = torch.load(\"models/dqn_expert_breakout_model\", map_location=device)\nAMN_net.apply(AMN_net.init_weights)\nexpert_net.eval()\n# optimizer = optim.Adam(AMN_net.parameters(), lr=LR, eps=1.5e-4)\noptimizer = optim.Adam(AMN_net.parameters(), lr=LR, eps=1.5e-4)",
"/home/tony/anaconda3/envs/rl/lib/python3.8/site-packages/torch/serialization.py:658: SourceChangeWarning: source code of class 'torch.nn.modules.conv.Conv2d' has changed. you can retrieve the original source code by accessing the object's source attribute or set `torch.nn.Module.dump_patches = True` and use the patch tool to revert the changes.\n warnings.warn(msg, SourceChangeWarning)\n/home/tony/anaconda3/envs/rl/lib/python3.8/site-packages/torch/serialization.py:658: SourceChangeWarning: source code of class 'torch.nn.modules.linear.Linear' has changed. you can retrieve the original source code by accessing the object's source attribute or set `torch.nn.Module.dump_patches = True` and use the patch tool to revert the changes.\n warnings.warn(msg, SourceChangeWarning)\n"
],
[
"memory = LabelledReplayMemory(NOT_LABELLED_CAPACITY, LABELLED_CAPACITY, [5,h,w], n_actions, ens_random, AMN_net, device=device)\naction_selector = ActionSelector(EPS, EPS, AMN_net, 1, n_actions, device)",
"_____no_output_____"
],
[
"steps_done = 0\nwriter = SummaryWriter(f'runs/{NAME}')",
"_____no_output_____"
],
[
"q = deque(maxlen=5)\ndone=True\neps = 0\nepisode_len = 0\nnum_labels = 0",
"_____no_output_____"
],
[
"progressive = tqdm(range(NUM_STEPS), total=NUM_STEPS, ncols=400, leave=False, unit='b')\nfor step in progressive:\n if done:\n env.reset()\n sum_reward = 0\n episode_len = 0\n img, _, _, _ = env.step(1) # BREAKOUT specific !!!\n for i in range(10): # no-op\n n_frame, _, _, _ = env.step(0)\n n_frame = fp(n_frame)\n q.append(n_frame)\n \n # Select and perform an action\n state = torch.cat(list(q))[1:].unsqueeze(0)\n action, eps = action_selector.select_action(state)\n n_frame, reward, done, info = env.step(action)\n n_frame = fp(n_frame)\n\n # 5 frame as memory\n q.append(n_frame)\n memory.push(torch.cat(list(q)).unsqueeze(0), action, reward, done) # here the n_frame means next frame from the previous time step\n episode_len += 1\n\n # Perform one step of the optimization (on the target network)\n if step % NOT_LABELLED_CAPACITY == 0 and step > 0:\n num_labels += memory.label_sample(percentage=PERCENTAGE,batch_size=BATCH_SIZE);\n loss = 0\n for _ in range(TRAINING_ITER):\n loss += AMN_optimization_ensemble(AMN_net, expert_net, optimizer, memory, batch_size=BATCH_SIZE, \n device=device)\n loss /= TRAINING_ITER\n writer.add_scalar('Performance/loss', loss, num_labels)\n \n if step % 10000 == 0 and step > 0:\n evaluated_reward = evaluate(step, AMN_net, device, env_raw, n_actions, eps=0.05, num_episode=15)\n writer.add_scalar('Performance/reward_vs_label', evaluated_reward, num_labels)\n writer.add_scalar('Performance/reward_vs_step', evaluated_reward, step)\n \n evaluated_reward_expert = evaluate(step, expert_net, device, env_raw, n_actions, eps=0.05, num_episode=15)\n writer.add_scalar('Performance/reward_expert_vs_step', evaluated_reward_expert, step)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03f3ec93fec40f8d51ad5c096b76367028a0522 | 594,733 | ipynb | Jupyter Notebook | week_13/seminar.ipynb | ishalyminov/shad_speech | e1345d2de929e150b2683190b127a837fbcb34f3 | [
"MIT"
] | null | null | null | week_13/seminar.ipynb | ishalyminov/shad_speech | e1345d2de929e150b2683190b127a837fbcb34f3 | [
"MIT"
] | null | null | null | week_13/seminar.ipynb | ishalyminov/shad_speech | e1345d2de929e150b2683190b127a837fbcb34f3 | [
"MIT"
] | null | null | null | 991.221667 | 567,448 | 0.877365 | [
[
[
"# Seminar for Lecture 13 \"VAE Vocoder\"\n\n\nIn the lectures, we studied various approaches to creating vocoders. The problem of sound generation is solved by deep generative models. We've discussed autoregressive models that can be reduced to **MAF**. We've considered the reverse analogue of MAF – **IAF**. We've seen how **normalizing flows** can help us directly optimize likelihood without using autoregression. And alse we've considered a vocoder built with the **GAN** paradigm.\n\nAt this seminar we will try to apply another popular generative model: the **variational autoencoder (VAE)**. We will try to build an encoder-decoder architecture with **MAF** as encoder and **IAF** as decoder. We will train this network by maximizing ELBO with a couple of additional losses (in vocoders, you can't do without it yet 🤷♂️).\n\n⚠️ In this seminar we call **\"MAF\"** not the generative model discussed on lecture, but network which architecture is like MAF's one and accepting audio as input. So we won't model data distribution with our **\"MAF\"**.",
"_____no_output_____"
]
],
[
[
"# ! pip install torch==1.7.1+cu101 torchvision==0.8.2+cu101 torchaudio==0.7.2 -f https://download.pytorch.org/whl/torch_stable.html\n# ! pip install numpy==1.17.5 matplotlib==3.3.3 tqdm==4.54.0",
"_____no_output_____"
],
[
"import torch\nfrom torch import nn\nfrom torch.nn import functional as F\nfrom typing import Union\nfrom math import log, pi, sqrt\nfrom IPython.display import display, Audio\nimport numpy as np\nimport librosa\n\nimport os\nos.environ[\"CUDA_DEVICE_ORDER\"] = \"PCI_BUS_ID\"\nos.environ[\"CUDA_VISIBLE_DEVICES\"] = \"0\"\n\ndevice = torch.device(\"cpu\")\nif False and torch.cuda.is_available():\n print('GPU found! 🎉')\n device = torch.device(\"cuda\")",
"_____no_output_____"
]
],
[
[
"Introduce auxiliary modules:\n1. causal convolution – simple convolution with `kernel_size` and `dilation` hyper-parameters, but working in causal way (does not look in the future)\n2. residual block – main building component of WaveNet architecture\n\nYes, WaveNet is everywhere. We can build MAF and IAF with any architecture, but WaveNet declared oneself as simple yet powerfull architecture. We will use WaveNet with conditioning on mel spectrograms, because we are building a vocoder.",
"_____no_output_____"
]
],
[
[
"class CausalConv(nn.Module):\n def __init__(self, in_channels, out_channels, kernel_size, dilation=1):\n super(CausalConv, self).__init__()\n\n self.padding = dilation * (kernel_size - 1)\n self.conv = nn.Conv1d(\n in_channels,\n out_channels,\n kernel_size,\n padding=self.padding,\n dilation=dilation)\n self.conv = nn.utils.weight_norm(self.conv)\n nn.init.kaiming_normal_(self.conv.weight)\n\n def forward(self, x):\n x = self.conv(x)\n x = x[:, :, :-self.padding]\n return x\n\n\nclass ResBlock(nn.Module):\n def __init__(self, in_channels, out_channels, skip_channels, kernel_size, dilation, cin_channels):\n super(ResBlock, self).__init__()\n self.cin_channels = cin_channels\n\n self.filter_conv = CausalConv(in_channels, out_channels, kernel_size, dilation)\n self.gate_conv = CausalConv(in_channels, out_channels, kernel_size, dilation)\n self.res_conv = nn.Conv1d(out_channels, in_channels, kernel_size=1)\n self.skip_conv = nn.Conv1d(out_channels, skip_channels, kernel_size=1)\n self.res_conv = nn.utils.weight_norm(self.res_conv)\n self.skip_conv = nn.utils.weight_norm(self.skip_conv)\n nn.init.kaiming_normal_(self.res_conv.weight)\n nn.init.kaiming_normal_(self.skip_conv.weight)\n\n self.filter_conv_c = nn.Conv1d(cin_channels, out_channels, kernel_size=1)\n self.gate_conv_c = nn.Conv1d(cin_channels, out_channels, kernel_size=1)\n self.filter_conv_c = nn.utils.weight_norm(self.filter_conv_c)\n self.gate_conv_c = nn.utils.weight_norm(self.gate_conv_c)\n nn.init.kaiming_normal_(self.filter_conv_c.weight)\n nn.init.kaiming_normal_(self.gate_conv_c.weight)\n\n def forward(self, x, c=None):\n h_filter = self.filter_conv(x)\n h_gate = self.gate_conv(x)\n h_filter += self.filter_conv_c(c)\n h_gate += self.gate_conv_c(c)\n out = torch.tanh(h_filter) * torch.sigmoid(h_gate)\n res = self.res_conv(out)\n skip = self.skip_conv(out)\n return (x + res) * sqrt(0.5), skip",
"_____no_output_____"
]
],
[
[
"For WaveNet it doesn't matter what it is used for: MAF or IAF - it all depends on our interpretation of the input and output variables.\n\nBelow is the WaveNet architecture that you are already familiar with from the last seminar. But this time, you will need to implement not inference but forward pass - and it's very simple 😉.",
"_____no_output_____"
]
],
[
[
"class WaveNet(nn.Module):\n def __init__(self, params):\n super(WaveNet, self). __init__()\n\n self.front_conv = nn.Sequential(\n CausalConv(1, params.residual_channels, params.front_kernel_size),\n nn.ReLU())\n\n self.res_blocks = nn.ModuleList()\n for b in range(params.num_blocks):\n for n in range(params.num_layers):\n self.res_blocks.append(ResBlock(\n in_channels=params.residual_channels,\n out_channels=params.gate_channels,\n skip_channels=params.skip_channels,\n kernel_size=params.kernel_size,\n dilation=2 ** n,\n cin_channels=params.mel_channels))\n\n self.final_conv = nn.Sequential(\n nn.ReLU(),\n nn.Conv1d(params.skip_channels, params.skip_channels, kernel_size=1),\n nn.ReLU(),\n nn.Conv1d(params.skip_channels, params.out_channels, kernel_size=1))\n\n def forward(self, x, c):\n # x: input tensor with signal or noise [B, 1, T]\n # c: local conditioning [B, C_mel, T]\n out = 0\n ################################################################################\n x = self.front_conv(x)\n for b in range(len(self.res_blocks)):\n x, x_skip = self.res_blocks[b](x, c)\n out = out + x_skip\n\n out = self.final_conv(out)\n ################################################################################\n return out",
"_____no_output_____"
],
[
"# check that works and gives expected output size\n# full correctness we will check later, when the whole network will be assembled\n\nclass Params:\n mel_channels: int = 80\n num_blocks: int = 4\n num_layers: int = 6\n out_channels: int = 3\n front_kernel_size: int = 2\n residual_channels: int = 64\n gate_channels: int = 64\n skip_channels: int = 128\n kernel_size: int = 2\n\nnet = WaveNet(Params()).to(device).eval()\nwith torch.no_grad():\n z = torch.FloatTensor(5, 1, 4096).normal_().to(device)\n c = torch.FloatTensor(5, 80, 4096).zero_().to(device)\n assert list(net(z, c).size()) == [5, 3, 4096]",
"_____no_output_____"
]
],
[
[
"Excellent 👍! Now we are ready to get started on more complex and interesting things.\n\nDo you remember our talks about vocoders built on IAF (Parallel WaveNet or ClariNet Vocoder)? We casually said that IAF we use not just one WaveNet (predicting mu and sigma), but a stack of WaveNets. Actually, let's implement this stack, but first, a few formulas that will help you.\n\nConsider transformations of random variable $z^{(0)} \\sim \\mathcal{N}(0, I)$: \n$$z^{(0)} \\rightarrow z^{(1)} \\rightarrow \\dots \\rightarrow z^{(n)}.$$\n\nEach transformation has the form: \n$$ z^{(k)} = f^{(k)}(z^{(k-1)}) = z{(k-1)} \\cdot \\sigma^{(k)} + \\mu^{(k)},$$ \nwhere $\\mu^{(k)}_t = \\mu(z_{<t}^{(k-1)}; \\theta_k)$ and $\\sigma^{(k)}_t = \\sigma(z_{<t}^{(k-1)}; \\theta_k)$ – are shifting and scaling variables modeled by a Gaussan WaveNet. \n\nIt is easy to deduce that the whole transformation $f^{(k)} \\circ \\dots \\circ f^{(2)} \\circ f^{(1)}$ can be represented as $f^{(\\mathrm{total})}(z) = z \\cdot \\sigma^{(\\mathrm{total})} + \\mu^{(\\mathrm{total})}$, where\n$$\\sigma^{(\\mathrm{total})} = \\prod_{k=1}^n \\sigma^{(k)}, ~ ~ ~ \\mu^{(\\mathrm{total})} = \\sum_{k=1}^n \\mu^{(k)} \\prod_{j > k}^n \\sigma^{(j)} $$\n\n$\\mu^{(\\mathrm{total})}$ and $\\sigma^{(\\mathrm{total})}$ we will need in the future for $p(\\hat x | z) estimation$.\n\nYou need to **implement** `forward` method of `WaveNetFlows` model.\n\n📝 Notes: \n1. WaveNet outputs tensor `output` of size `[B, 2, T]`, where `output[:, 0, :]` is $\\mu$ and `output[:, 1, :]` is $\\log \\sigma$. We model logarithms of $\\sigma$ insead of $\\sigma$ for stable gradients. \n2. As we model $\\mu(z_{<t}^{(k-1)}; \\theta_k)$ and $\\sigma(z_{<t}^{(k-1)}; \\theta_k)$ – their output we have length `T - 1`. To keep constant length `T` of modelled noise variable we need to pad it on the left side (with zero).\n3. $\\mu^{(\\mathrm{total})}$ and $\\sigma^{(\\mathrm{total})}$ wil have length `T - 1`, because we do not pad distribution parameters.",
"_____no_output_____"
]
],
[
[
"class WaveNetFlows(nn.Module):\n def __init__(self, params):\n super(WaveNetFlows, self).__init__()\n self.device = params.device\n\n self.iafs = nn.ModuleList()\n for i in range(params.num_flows):\n self.iafs.append(WaveNet(params))\n\n def forward(self, z, c):\n # z: random sample from standart distribution [B, 1, T]\n # c: local conditioning for WaveNet [B, C_mel, T]\n mu_tot, logs_tot = 0., 0.\n ################################################################################\n mus, log_sigmas = [], []\n for iaf in self.iafs:\n out_i = iaf(z, c)\n mu = out_i[:, 0, :-1].unsqueeze(1)\n mu_padded = torch.cat([torch.zeros((*(z.shape[:-1]), 1), dtype=torch.float32).to(device), mu], axis=-1)\n mus.append(mu)\n log_sigma = out_i[:, 1, :-1].unsqueeze(1)\n log_sigma_padded = torch.cat([torch.zeros((*(z.shape[:-1]), 1), dtype=torch.float32).to(device), log_sigma], axis=-1)\n log_sigmas.append(log_sigma)\n z = torch.exp(log_sigma_padded) * z + mu_padded\n logs_tot = torch.sum(torch.stack(log_sigmas, axis=0), axis=0)\n\n for i in range(len(self.iafs) - 1):\n mu_tot += mus[i] * torch.exp(torch.sum(torch.stack(log_sigmas[i + 1:], axis=0), axis=0))\n mu_tot += mus[-1]\n ################################################################################\n return z, mu_tot, logs_tot",
"_____no_output_____"
],
[
"class Params:\n num_flows: int = 4\n mel_channels: int = 80\n num_blocks: int = 1\n num_layers: int = 5\n out_channels: int = 2\n front_kernel_size: int = 2\n residual_channels: int = 64\n gate_channels: int = 64\n skip_channels: int = 64\n kernel_size: int = 3\n device = device\n \nnet = WaveNetFlows(Params()).to(device)\n\nwith torch.no_grad():\n z = torch.FloatTensor(3, 1, 4096).normal_().to(device)\n c = torch.FloatTensor(3, 80, 4096).zero_().to(device)\n z_hat, mu, log_sigma = net(z, c)\n assert list(z_hat.size()) == [3, 1, 4096] # same length as input\n assert list(mu.size()) == [3, 1, 4096 - 1] # shorter by one sample\n assert list(log_sigma.size()) == [3, 1, 4096 - 1] # shorted by one sample",
"_____no_output_____"
]
],
[
[
"If you are not familiar with VAE framework, please try to figure it out. For example, please familiarize with this [blog post](https://wiseodd.github.io/techblog/2016/12/10/variational-autoencoder/).\n\n\nIn short, VAE – is just \"modification\" of AutoEncoder, which consists of encoder and decoder. VAE allows you to sample from data distribution $p(x)$ as $p(x|z)$ via its decoder, where $p(z)$ is simple and known, e.g. $\\mathcal{N}(0, I)$. The interesting part is that $p(x | z)$ cannot be optimized with Maximum Likelihood Estimation, because $p(x | z)$ is not tractable. \n\nBut we can maximize Evidence Lower Bound (ELBO) which has a form:\n\n$$\\max_{\\phi, \\theta} \\mathbb{E}_{q_{\\phi}(z | x)} \\log p_{\\theta}(x | z) - \\mathbb{D}_{KL}(q_{\\phi}(z | x) || p(z))$$\n\nwhere $p_{\\theta}(x | z)$ is VAE decoder and $q_{\\phi}(z | x)$ is VAE encoder. For more details please read mentioned blog post or any other materials on this theme.\n\nIn our case $q_{\\phi}(z | x)$ is represented by MAF WaveNet, and $p_{\\theta}(x | z)$ – by IAF build with WaveNet stack. To be more precise our decoder $p_{\\theta}(x | z)$ is parametrised by the **one-step-ahead prediction** from an IAF.\n\n🧑💻 **let's practice..**\n\nWe will start from easy part: generation (or sampling). \n\n**Implement** `generate` method, which accepts mel spectrogram as conditioning tensor. Inside this method random tensor from standart distribution N(0, I) is sampled. This tensor than transformed to tensor from audio distribution via `decoder`. In the cell bellow you will see code for loading pretrained model and mel spectrogram. Listen to result – it should sound passable, but MOS 5.0 is not expected. 😄",
"_____no_output_____"
]
],
[
[
"class WaveNetVAE(nn.Module):\n def __init__(self, encoder_params, decoder_params):\n super(WaveNetVAE, self).__init__()\n assert encoder_params.device == decoder_params.device\n self.device = encoder_params.device\n self.mse_loss = torch.nn.MSELoss()\n\n self.encoder = WaveNet(encoder_params)\n self.decoder = WaveNetFlows(decoder_params)\n self.log_eps = nn.Parameter(torch.zeros(1))\n\n self.upsample_conv = nn.ModuleList()\n for s in [16, 16]:\n conv = nn.ConvTranspose2d(1, 1, (3, 2 * s), padding=(1, s // 2), stride=(1, s))\n conv = nn.utils.weight_norm(conv)\n nn.init.kaiming_normal_(conv.weight)\n self.upsample_conv.append(conv)\n self.upsample_conv.append(nn.LeakyReLU(0.4))\n\n def forward(self, x, c):\n # x: audio signal [B, 1, T]\n # c: mel spectrogram [B, 1, T / HOP_SIZE]\n loss_rec = 0\n loss_kl = 0\n loss_frame_rec = 0\n loss_frame_prior = 0\n ################################################################################\n c_up = self.upsample(c)\n mu_log_sigma = self.encoder(x, c_up)\n mu = mu_log_sigma[:, 0, :].unsqueeze(1)\n log_sigma = mu_log_sigma[:, 1, :].unsqueeze(1)\n mu_whitened = (x - mu) / torch.exp(log_sigma)\n eps = torch.randn_like(log_sigma).to(self.device)\n z = mu_whitened + torch.exp(log_sigma / 2) * eps\n x_rec, mu_tot, log_sigma_tot = self.decoder(z, c_up)\n x_prior = self.generate(c)\n x_rec_stft = librosa.stft(x_rec.view(-1).detach().numpy())\n x_stft = librosa.stft(x.view(-1).detach().numpy())\n x_prior_stft = librosa.stft(x_prior.view(-1).detach().numpy())\n\n loss_frame_rec = self.mse_loss(torch.FloatTensor(x_stft), torch.FloatTensor(x_rec_stft))\n loss_frame_prior = self.mse_loss(torch.FloatTensor(x_stft), torch.FloatTensor(x_prior_stft))\n loss_kl = torch.sum(-self.log_eps + (1 / 2.0) * (torch.exp(self.log_eps) ** 2.0 - 1.0 + mu_whitened ** 2.0))\n loss_rec = torch.sum(torch.normal(mu_tot, torch.exp(log_sigma_tot)))\n ################################################################################\n alpha = 1e-9 # for annealing during training\n return loss_rec + alpha * loss_kl + loss_frame_rec + loss_frame_prior\n\n def generate(self, c):\n # c: mel spectrogram [B, 80, L] where L - number of mel frames\n # outputs: audio [B, 1, L * HOP_SIZE]\n ################################################################################\n c_up = self.upsample(c)\n frames_number = c_up.shape[-1]\n z = torch.randn(c_up.shape[0], 1, frames_number).to(self.device)\n x_sample, _, _ = self.decoder(z, c_up)\n ################################################################################\n return x_sample\n\n def upsample(self, c):\n c = c.unsqueeze(1) # [B, 1, C, L]\n for f in self.upsample_conv:\n c = f(c)\n c = c.squeeze(1) # [B, C, T], where T = L * HOP_SIZE\n return c",
"_____no_output_____"
],
[
"# saved checkpoint model has following architecture parameters\n\nclass ParamsMAF:\n mel_channels: int = 80\n num_blocks: int = 2\n num_layers: int = 10\n out_channels: int = 2\n front_kernel_size: int = 32\n residual_channels: int = 128\n gate_channels: int = 256\n skip_channels: int = 128\n kernel_size: int = 2\n device: str = device\n\n\nclass ParamsIAF:\n num_flows: int = 6\n mel_channels: int = 80\n num_blocks: int = 1\n num_layers: int = 10\n out_channels: int = 2\n front_kernel_size: int = 32\n residual_channels: int = 64\n gate_channels: int = 128\n skip_channels: int = 64\n kernel_size: int = 3\n device: str = device\n \n# load checkpoint\nckpt_path = 'data/checkpoint.pth'\nnet = WaveNetVAE(ParamsMAF(), ParamsIAF()).eval().to(device)\nckpt = torch.load(ckpt_path, map_location='cpu')\nnet.load_state_dict(ckpt['state_dict'])\n\n# load original audio and it's mel\nx = torch.load('data/x.pth').to(device)\nc = torch.load('data/c.pth').to(device)\n\n# generate audio from \nwith torch.no_grad():\n x_prior = net.generate(c.unsqueeze(0)).squeeze()\n\ndisplay(Audio(x_prior.cpu(), rate=22050))",
"_____no_output_____"
]
],
[
[
"If it sounds plausible **5 points** 🥉 are already yours 🎉! And here the most interesting and difficult part comes: loss function implementation. The `forward` method will return the loss. But lets talk more precisly about our architecture and how it was trained.\n\nThe encoder of our model $q_{\\phi}(z|x)$ is parametrerized by a Gaussian autoregressive WaveNet, which maps the audio $x$ into the sample length latent representation $z$. Specifically, the Gaussian WaveNet (if we talk about **real MAF**) models $x_t$ given the previous samples $x_{<t}$ with $x_t ∼ \\mathcal{N}(\\mu(x_{<t}; \\phi), \\sigma(x_{<t}; \\phi))$, where the mean $\\mu(x_{<t}; \\phi)$ and log-scale $\\log \\sigma(x_{<t}; \\phi)$ are predicted by WaveNet, respectively.\n\nOur **encoder** posterior is constructed as\n\n$$q_{\\phi}(z | x) = \\prod_{t} q_{\\phi}(z_t | x_{\\leq t})$$\n\nwhere\n\n$$q_{\\phi}(z_t | x_{\\leq t}) = \\mathcal{N}(\\frac{x_t - \\mu(x_{<t}; \\phi)}{\\sigma(x_{<t}; \\phi)}, \\varepsilon)$$\n\nWe apply the mean $\\mu(x_{<t}; \\phi)$ and scale $\\sigma(x_{<t})$ for \"whitening\" the posterior distribution. Also we introduce a trainable scalar $\\varepsilon > 0$ to decouple the global variation, which will make optimization process easier.\n\nSubstitution of our model formulas in $\\mathbb{D}_{KL}$ formula gives:\n\n$$\\mathbb{D}_{KL}(q_{\\phi}(z | x) || p(z)) = \\sum_t \\log\\frac{1}{\\varepsilon} + \\frac{1}{2}(\\varepsilon^2 - 1 + (\\frac{x_t - \\mu(x_{<t})}{\\sigma(x_{<t})})^2)$$\n\n**Implement** calculation of `loss_kld` in `forward` method as KL divergence.\n\n---\n\nThe other term in ELBO formula can be interpreted as reconstruction loss. It can be evaluated by sampling from $p_{\\theta}(x | z)$, where $z$ is from $q_{\\phi}(z | \\hat x)$, $\\hat x$ is our ground truth audio. But sampling is not differential operation! 🤔 We can apply reparametrization trick!\n\n**Implement** calculation of `loss_rec` in `forward` method as recontruction loss – which is just log likelihood of ground truth sample $x$ in predicted by IAF distribution $p_{\\theta}(x | \\hat z)$ where $\\hat z \\sim q_{\\phi}(z | \\hat x)$.\n\n--- \n\nVocoders without MLE are still not able to train without auxilary losses. We studied many of them, but STFT-loss is our favourite!\n\n**Implement** calculation of `loss_frame_rec` which stands for MSE loss in STFT domain between original audio and its reconstruction.\n\n--- \n\nWe can go even further and calculate STFT loss with random sample from $p_\\theta(x | z)$. Conditioning on mel spectrogram allows us to do so.\n\n**Implement** calculation of `loss_frame_prior` which stands for MSE loss in STFT domain between original audio and sample from prior.",
"_____no_output_____"
]
],
[
[
"net = WaveNetVAE(ParamsMAF(), ParamsIAF()).to(device).train()\n\nx = x[:64 * 256]\nc = c[:, :64]\n\nnet.zero_grad()\nloss = net.forward(x.unsqueeze(0).unsqueeze(0), c.unsqueeze(0))\nloss.backward()\nprint(f\"Initial loss: {loss.item():.2f}\")\n\nckpt = torch.load(ckpt_path, map_location='cpu')\nnet.load_state_dict(ckpt['state_dict'])\n\nnet.zero_grad()\nloss = net.forward(x.unsqueeze(0).unsqueeze(0), c.unsqueeze(0))\nloss.backward()\nprint(f\"Optimized loss: {loss.item():.2f}\")",
"Initial loss: 6629.40\nOptimized loss: 6.45\n"
]
],
[
[
"If you correctly implemented losses and the backward pass works smoothly, **8 more points**🥈 are yours 🎉!",
"_____no_output_____"
],
[
"For **2 additional points** 🥇 please write a short essay (in russian) about your thoughts on vocoders. Try to avoid obvious statements as \"vocoder is very important part of TTS pipeline\". We are interested in insights you've got from studying vocoders. \n\n`YOUR TEXT HERE`",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d03f40554a01fe6b181cea57c2577b3208acb47b | 7,476 | ipynb | Jupyter Notebook | Jupyter_Notebooks/ibm_db-closeSync.ipynb | ibmdb/jupyter-node-ibm_db | 588bf32955237aa2829aa796a9ce10865a5dfa99 | [
"Apache-2.0"
] | null | null | null | Jupyter_Notebooks/ibm_db-closeSync.ipynb | ibmdb/jupyter-node-ibm_db | 588bf32955237aa2829aa796a9ce10865a5dfa99 | [
"Apache-2.0"
] | 1 | 2019-06-12T10:23:03.000Z | 2019-06-12T10:23:03.000Z | Jupyter_Notebooks/ibm_db-closeSync.ipynb | ibmdb/jupyter-node-ibm_db | 588bf32955237aa2829aa796a9ce10865a5dfa99 | [
"Apache-2.0"
] | 2 | 2019-11-03T17:23:38.000Z | 2021-12-28T11:00:46.000Z | 32.789474 | 154 | 0.422151 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d03f4b5d4672d2c97c8399179e31e9d306beab77 | 5,601 | ipynb | Jupyter Notebook | src/examples/tutorial/ascent_intro/notebooks/08_ascent_trigger_examples.ipynb | goodbadwolf/ascent | 70662ebc6fd550d2d13349cb750022d9ce3b29a6 | [
"BSD-3-Clause"
] | 97 | 2017-10-23T23:59:46.000Z | 2022-03-02T22:21:00.000Z | src/examples/tutorial/ascent_intro/notebooks/08_ascent_trigger_examples.ipynb | goodbadwolf/ascent | 70662ebc6fd550d2d13349cb750022d9ce3b29a6 | [
"BSD-3-Clause"
] | 475 | 2017-09-12T22:46:37.000Z | 2022-03-18T19:19:04.000Z | src/examples/tutorial/ascent_intro/notebooks/08_ascent_trigger_examples.ipynb | goodbadwolf/ascent | 70662ebc6fd550d2d13349cb750022d9ce3b29a6 | [
"BSD-3-Clause"
] | 39 | 2017-09-12T20:18:29.000Z | 2022-01-20T00:22:55.000Z | 29.324607 | 286 | 0.578647 | [
[
[
"# Trigger Examples\nTriggers allow the user to specify a set of actions that are triggered by the result of a boolean expression.\nThey provide flexibility to adapt what analysis and visualization actions are taken in situ. Triggers leverage Ascent's Query and Expression infrastructure. See Ascent's [Triggers](https://ascent.readthedocs.io/en/latest/Actions/Triggers.html) docs for deeper details on Triggers.",
"_____no_output_____"
]
],
[
[
"# cleanup any old results\n!./cleanup.sh\n\n# ascent + conduit imports\nimport conduit\nimport conduit.blueprint\nimport ascent\n\nimport numpy as np\n\n# helpers we use to create tutorial data\nfrom ascent_tutorial_jupyter_utils import img_display_width\nfrom ascent_tutorial_jupyter_utils import tutorial_gyre_example\n\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Trigger Example 1\n### Using triggers to render when conditions occur",
"_____no_output_____"
]
],
[
[
"# Use triggers to render when conditions occur\na = ascent.Ascent()\na.open()\n\n# setup actions\nactions = conduit.Node()\n\n# declare a question to ask \nadd_queries = actions.append()\nadd_queries[\"action\"] = \"add_queries\"\n\n# add our entropy query (q1)\nqueries = add_queries[\"queries\"] \nqueries[\"q1/params/expression\"] = \"entropy(histogram(field('gyre'), num_bins=128))\"\nqueries[\"q1/params/name\"] = \"entropy\"\n\n# declare triggers \nadd_triggers = actions.append()\nadd_triggers[\"action\"] = \"add_triggers\"\ntriggers = add_triggers[\"triggers\"] \n\n# add a simple trigger (t1_ that fires at cycle 500\ntriggers[\"t1/params/condition\"] = \"cycle() == 500\"\ntriggers[\"t1/params/actions_file\"] = \"cycle_trigger_actions.yaml\"\n\n# add trigger (t2) that fires when the change in entroy exceeds 0.5\n\n# the history function allows you to access query results of previous\n# cycles. relative_index indicates how far back in history to look.\n\n# Looking at the plot of gyre entropy in the previous notebook, we see a jump\n# in entropy at cycle 200, so we expect the trigger to fire at cycle 200\ntriggers[\"t2/params/condition\"] = \"entropy - history(entropy, relative_index = 1) > 0.5\"\ntriggers[\"t2/params/actions_file\"] = \"entropy_trigger_actions.yaml\"\n\n# view our full actions tree\nprint(actions.to_yaml())\n\n# gyre time varying params\nnsteps = 10\ntime = 0.0\ndelta_time = 0.5\n\nfor step in range(nsteps):\n # call helper that generates a double gyre time varying example mesh.\n # gyre ref :https://shaddenlab.berkeley.edu/uploads/LCS-tutorial/examples.html\n mesh = tutorial_gyre_example(time)\n \n # update the example cycle\n cycle = 100 + step * 100\n mesh[\"state/cycle\"] = cycle\n print(\"time: {} cycle: {}\".format(time,cycle))\n \n # publish mesh to ascent\n a.publish(mesh)\n \n # execute the actions\n a.execute(actions)\n \n # update time\n time = time + delta_time\n\n# retrieve the info node that contains the trigger and query results\ninfo = conduit.Node()\na.info(info)\n\n# close ascent\na.close()",
"_____no_output_____"
],
[
"# we expect our cycle trigger to render only at cycle 500\n! ls cycle_trigger*.png",
"_____no_output_____"
],
[
"# show the result image from the cycle trigger\nascent.jupyter.AscentImageSequenceViewer([\"cycle_trigger_out_500.png\"]).show()",
"_____no_output_____"
],
[
"# we expect our entropy trigger to render only at cycle 200\n! ls entropy_trigger*.png",
"_____no_output_____"
],
[
"# show the result image from the entropy trigger\nascent.jupyter.AscentImageSequenceViewer([\"entropy_trigger_out_200.png\"]).show()",
"_____no_output_____"
],
[
"print(info[\"expressions\"].to_yaml())",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03f4ee121b35f7818dd6f7ebe06dcc6cec9aa84 | 110,192 | ipynb | Jupyter Notebook | basic-matplotlib-plotting.ipynb | jaspajjr/pydata-visualisation | cb1d804c9ba522ecddcf55f92ebbe96363d24bb3 | [
"MIT"
] | 1 | 2018-05-17T08:49:50.000Z | 2018-05-17T08:49:50.000Z | basic-matplotlib-plotting.ipynb | jaspajjr/pydata-visualisation | cb1d804c9ba522ecddcf55f92ebbe96363d24bb3 | [
"MIT"
] | 5 | 2020-03-24T15:46:10.000Z | 2021-12-13T19:47:54.000Z | basic-matplotlib-plotting.ipynb | jaspajjr/pydata-visualisation | cb1d804c9ba522ecddcf55f92ebbe96363d24bb3 | [
"MIT"
] | 1 | 2018-10-17T17:48:32.000Z | 2018-10-17T17:48:32.000Z | 216.913386 | 19,400 | 0.914368 | [
[
[
"## These notebooks can be found at https://github.com/jaspajjr/pydata-visualisation if you want to follow along",
"_____no_output_____"
],
[
"https://matplotlib.org/users/intro.html\n\nMatplotlib is a library for making 2D plots of arrays in Python.\n\n* Has it's origins in emulating MATLAB, it can also be used in a Pythonic, object oriented way. \n\n\n* Easy stuff should be easy, difficult stuff should be possible",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\n\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"Everything in matplotlib is organized in a hierarchy. At the top of the hierarchy is the matplotlib “state-machine environment” which is provided by the matplotlib.pyplot module. At this level, simple functions are used to add plot elements (lines, images, text, etc.) to the current axes in the current figure.\n\nPyplot’s state-machine environment behaves similarly to MATLAB and should be most familiar to users with MATLAB experience.\nThe next level down in the hierarchy is the first level of the object-oriented interface, in which pyplot is used only for a few functions such as figure creation, and the user explicitly creates and keeps track of the figure and axes objects. At this level, the user uses pyplot to create figures, and through those figures, one or more axes objects can be created. These axes objects are then used for most plotting actions.",
"_____no_output_____"
],
[
"## Scatter Plot",
"_____no_output_____"
],
[
"To start with let's do a really basic scatter plot:",
"_____no_output_____"
]
],
[
[
"plt.plot([0, 1, 2, 3, 4, 5], [0, 2, 4, 6, 8, 10])",
"_____no_output_____"
],
[
"x = [0, 1, 2, 3, 4, 5]\ny = [0, 2, 4, 6, 8, 10]\nplt.plot(x, y)",
"_____no_output_____"
]
],
[
[
"What if we don't want a line?",
"_____no_output_____"
]
],
[
[
"plt.plot([0, 1, 2, 3, 4, 5],\n [0, 2, 5, 7, 8, 10],\n marker='o',\n linestyle='')\nplt.xlabel('The X Axis')\nplt.ylabel('The Y Axis')\n\nplt.show();",
"_____no_output_____"
]
],
[
[
"#### Simple example from matplotlib\n\nhttps://matplotlib.org/tutorials/intermediate/tight_layout_guide.html#sphx-glr-tutorials-intermediate-tight-layout-guide-py",
"_____no_output_____"
]
],
[
[
"def example_plot(ax, fontsize=12):\n ax.plot([1, 2])\n\n ax.locator_params(nbins=5)\n ax.set_xlabel('x-label', fontsize=fontsize)\n ax.set_ylabel('y-label', fontsize=fontsize)\n ax.set_title('Title', fontsize=fontsize)\n\nfig, ax = plt.subplots()\nexample_plot(ax, fontsize=24)",
"_____no_output_____"
],
[
"fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(nrows=2, ncols=2)\n# fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(nrows=2, ncols=2, sharex=True, sharey=True)\nax1.plot([0, 1, 2, 3, 4, 5],\n [0, 2, 5, 7, 8, 10])\nax2.plot([0, 1, 2, 3, 4, 5],\n [0, 2, 4, 9, 16, 25])\nax3.plot([0, 1, 2, 3, 4, 5],\n [0, 13, 18, 21, 23, 25])\nax4.plot([0, 1, 2, 3, 4, 5],\n [0, 1, 2, 3, 4, 5])\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"## Date Plotting",
"_____no_output_____"
]
],
[
[
"import pandas_datareader as pdr\n\ndf = pdr.get_data_fred('GS10')\ndf = df.reset_index()\n\nprint(df.info())\n\ndf.head()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 101 entries, 0 to 100\nData columns (total 2 columns):\nDATE 101 non-null datetime64[ns]\nGS10 101 non-null float64\ndtypes: datetime64[ns](1), float64(1)\nmemory usage: 1.7 KB\nNone\n"
],
[
"fig = plt.figure(figsize=(12, 8))\nax = fig.add_subplot(111)\n\nax.plot_date(df['DATE'], df['GS10'])",
"_____no_output_____"
]
],
[
[
"## Bar Plot",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(12, 8))\nax = fig.add_subplot(111)\n\nx_data = [0, 1, 2, 3, 4]\nvalues = [20, 35, 30, 35, 27]\nax.bar(x_data, values)\n\nax.set_xticks(x_data)\nax.set_xticklabels(('A', 'B', 'C', 'D', 'E'))\n;",
"_____no_output_____"
]
],
[
[
"## Matplotlib basics\n\nhttp://pbpython.com/effective-matplotlib.html",
"_____no_output_____"
],
[
"### Behind the scenes\n\n* matplotlib.backend_bases.FigureCanvas is the area onto which the figure is drawn \n\n* matplotlib.backend_bases.Renderer is the object which knows how to draw on the FigureCanvas \n\n* matplotlib.artist.Artist is the object that knows how to use a renderer to paint onto the canvas \n\n\nThe typical user will spend 95% of their time working with the Artists.\n\n\nhttps://matplotlib.org/tutorials/intermediate/artists.html#sphx-glr-tutorials-intermediate-artists-py",
"_____no_output_____"
]
],
[
[
"fig, (ax1, ax2) = plt.subplots(\n nrows=1,\n ncols=2,\n sharey=True,\n figsize=(12, 8))\n\nfig.suptitle(\"Main Title\", fontsize=14, fontweight='bold');\n\nx_data = [0, 1, 2, 3, 4]\nvalues = [20, 35, 30, 35, 27]\n\nax1.barh(x_data, values);\nax1.set_xlim([0, 55])\n#ax1.set(xlabel='Unit of measurement', ylabel='Groups')\nax1.set(title='Foo', xlabel='Unit of measurement')\nax1.grid()\n\n\nax2.barh(x_data, [y / np.sum(values) for y in values], color='r');\nax2.set_title('Transformed', fontweight='light')\nax2.axvline(x=.1, color='k', linestyle='--')\nax2.set(xlabel='Unit of measurement') # Worth noticing this\nax2.set_axis_off();\n\nfig.savefig('example_plot.png', dpi=80, bbox_inches=\"tight\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d03f5d21220efb7a7cebc477ad400ba2345767b5 | 4,423 | ipynb | Jupyter Notebook | exercises/exercise-explore-your-data.ipynb | bobrokerson/kaggle | 96c13e85476e2fe0fdb2af74075f82510db90573 | [
"MIT"
] | null | null | null | exercises/exercise-explore-your-data.ipynb | bobrokerson/kaggle | 96c13e85476e2fe0fdb2af74075f82510db90573 | [
"MIT"
] | null | null | null | exercises/exercise-explore-your-data.ipynb | bobrokerson/kaggle | 96c13e85476e2fe0fdb2af74075f82510db90573 | [
"MIT"
] | null | null | null | 4,423 | 4,423 | 0.742935 | [
[
[
"**[Machine Learning Course Home Page](https://www.kaggle.com/learn/machine-learning)**\n\n---\n",
"_____no_output_____"
],
[
"This exercise will test your ability to read a data file and understand statistics about the data.\n\nIn later exercises, you will apply techniques to filter the data, build a machine learning model, and iteratively improve your model.\n\nThe course examples use data from Melbourne. To ensure you can apply these techniques on your own, you will have to apply them to a new dataset (with house prices from Iowa).\n\nThe exercises use a \"notebook\" coding environment. In case you are unfamiliar with notebooks, we have a [90-second intro video](https://www.youtube.com/watch?v=4C2qMnaIKL4).\n\n# Exercises\n\nRun the following cell to set up code-checking, which will verify your work as you go.",
"_____no_output_____"
]
],
[
[
"# Set up code checking\nfrom learntools.core import binder\nbinder.bind(globals())\nfrom learntools.machine_learning.ex2 import *\nprint(\"Setup Complete\")",
"_____no_output_____"
]
],
[
[
"## Step 1: Loading Data\nRead the Iowa data file into a Pandas DataFrame called `home_data`.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\n# Path of the file to read\niowa_file_path ='../input/home-data-for-ml-course/train.csv'\n\n# Fill in the line below to read the file into a variable home_data\nhome_data = pd.read_csv(iowa_file_path)\n\n\n\n\n# Call line below with no argument to check that you've loaded the data correctly\nstep_1.check()",
"_____no_output_____"
]
],
[
[
"## Step 2: Review The Data\nUse the command you learned to view summary statistics of the data. Then fill in variables to answer the following questions",
"_____no_output_____"
]
],
[
[
"# Print summary statistics in next line\n#print(home_data.info())\nhome_data[['YearBuilt','YrSold']].describe()",
"_____no_output_____"
],
[
"# What is the average lot size (rounded to nearest integer)?\navg_lot_size_1 = home_data.LotArea.mean()\navg_lot_size = round(avg_lot_size_1)\nprint(avg_lot_size)\n# As of today, how old is the newest home (current year - the date in which it was built)\nimport matplotlib.pyplot as plt\n\nhome_data['YrSold'].value_counts().plot(kind='bar');\nplt.xlabel(\"Year Sold\", labelpad=14)\nplt.ylabel(\"Count of Houses\", labelpad=14)\nplt.title(\"Count of Houses Sold by Year\", y=1.02);\n\nnewest_home_age = 2022-home_data['YearBuilt'].max()\nprint(newest_home_age)\n\n# Checks your answers\nstep_2.check()",
"_____no_output_____"
]
],
[
[
"---\n**[Machine Learning Course Home Page](https://www.kaggle.com/learn/machine-learning)**\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d03f71389900c19317a13091b61f9f5d036690b0 | 6,120 | ipynb | Jupyter Notebook | Day-2/Activities/04-Par_CruddyDB/Unsolved/.ipynb_checkpoints/Par_CruddyDB-checkpoint.ipynb | racheltrindle/sqlalchemy-challenge | 0315f13ab66b61589969fd06306e11a90e6c88bb | [
"ADSL"
] | null | null | null | Day-2/Activities/04-Par_CruddyDB/Unsolved/.ipynb_checkpoints/Par_CruddyDB-checkpoint.ipynb | racheltrindle/sqlalchemy-challenge | 0315f13ab66b61589969fd06306e11a90e6c88bb | [
"ADSL"
] | null | null | null | Day-2/Activities/04-Par_CruddyDB/Unsolved/.ipynb_checkpoints/Par_CruddyDB-checkpoint.ipynb | racheltrindle/sqlalchemy-challenge | 0315f13ab66b61589969fd06306e11a90e6c88bb | [
"ADSL"
] | null | null | null | 23.90625 | 112 | 0.572876 | [
[
[
"* Within a Python file, create new SQLAlchemy class called `Garbage` that holds the following values...\n* `__tablename__`: Should be \"garbage_collection\"\n* `id`: The primary key for the table that is an integer and automatically increments\n* `item`: A string that describes what kind of item was collected\n* `weight`: A double that explains how heavy the item is\n* `collector`: A string that lets users know which garbage man collected the item\n* Create a connection and a session before adding a few items into the SQLite database crafted.\n* Update the values within at least two of the rows added to the table.\n* Delete the row with the lowest weight from the table.\n* Print out all of the data within the database.\n",
"_____no_output_____"
],
[
"# Import SQL Alchemy\nfrom sqlalchemy import create_engine\n\n# Import and establish Base for which classes will be constructed \nfrom sqlalchemy.ext.declarative import declarative_base\nBase = declarative_base()\n\n# Import modules to declare columns and column data types\nfrom sqlalchemy import Column, Integer, String, Float",
"_____no_output_____"
],
[
"class Garbage(base):\n id=Column(Integer)\n item=Column(String)\n weight=Column(Integer)\n collecter=Column(String)\n\n# YOUR CODE HERE",
"_____no_output_____"
],
[
"# Create a connection to a SQLite database\n# YOUR CODE HERE",
"_____no_output_____"
],
[
"# Create the garbage_collection table within the database\nBase.metadata.create_all(engine)",
"_____no_output_____"
],
[
"# To push the objects made and query the server we use a Session object\nfrom sqlalchemy.orm import Session\nsession = Session(bind=engine)",
"_____no_output_____"
],
[
"# Create some instances of the Garbage class\n# YOUR CODE HERE",
"_____no_output_____"
],
[
"# Add these objects to the session\n# YOUR CODE HERE",
"_____no_output_____"
],
[
"# Update two rows of data\n# YOUR CODE HERE",
"_____no_output_____"
],
[
"# Delete the row with the lowest weight\n# YOUR CODE HERE",
"_____no_output_____"
],
[
"# Collect all of the items and print their information\n# YOUR CODE HERE",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03f8724d659eadb9e0311ddeaf8a0f0e2cef165 | 140,125 | ipynb | Jupyter Notebook | container_files/tutorials/Procedures and Functions Tutorial.ipynb | mldbai/mldb | 0554aa390a563a6294ecc841f8026a88139c3041 | [
"Apache-2.0"
] | 665 | 2015-12-09T17:00:14.000Z | 2022-03-25T07:46:46.000Z | container_files/tutorials/Procedures and Functions Tutorial.ipynb | mldbai/mldb | 0554aa390a563a6294ecc841f8026a88139c3041 | [
"Apache-2.0"
] | 797 | 2015-12-09T19:48:19.000Z | 2022-03-07T02:19:47.000Z | container_files/tutorials/Procedures and Functions Tutorial.ipynb | mldbai/mldb | 0554aa390a563a6294ecc841f8026a88139c3041 | [
"Apache-2.0"
] | 103 | 2015-12-25T04:39:29.000Z | 2022-02-03T02:55:22.000Z | 163.69743 | 108,070 | 0.842326 | [
[
[
"# Procedures and Functions Tutorial\n\nMLDB is the Machine Learning Database, and all machine learning operations are done via Procedures and Functions. Training a model happens via Procedures, and applying a model happens via Functions.",
"_____no_output_____"
],
[
"The notebook cells below use `pymldb`'s `Connection` class to make [REST API](../../../../doc/#builtin/WorkingWithRest.md.html) calls. You can check out the [Using `pymldb` Tutorial](../../../../doc/nblink.html#_tutorials/Using pymldb Tutorial) for more details.",
"_____no_output_____"
]
],
[
[
"from pymldb import Connection\nmldb = Connection(\"http://localhost\")",
"_____no_output_____"
]
],
[
[
"## Loading a Dataset \n\nThe classic [Iris Flower Dataset](http://en.wikipedia.org/wiki/Iris_flower_data_set) isn't very big but it's well-known and easy to reason about so it's a good example dataset to use for machine learning examples.\n\nWe can import it directly from a remote URL:\n",
"_____no_output_____"
]
],
[
[
"mldb.put('/v1/procedures/import_iris', {\n \"type\": \"import.text\",\n \"params\": {\n \"dataFileUrl\": \"file://mldb/mldb_test_data/iris.data\",\n \"headers\": [ \"sepal length\", \"sepal width\", \"petal length\", \"petal width\", \"class\" ],\n \"outputDataset\": \"iris\",\n \"runOnCreation\": True\n }\n})",
"_____no_output_____"
]
],
[
[
"## A quick look at the data\n\nWe can use the [Query API](../../../../doc/#builtin/sql/QueryAPI.md.html) to get the data into a Pandas DataFrame to take a quick look at it.",
"_____no_output_____"
]
],
[
[
"df = mldb.query(\"select * from iris\")\ndf.head()",
"_____no_output_____"
],
[
"%matplotlib inline\nimport seaborn as sns, pandas as pd\n\nsns.pairplot(df, hue=\"class\", size=2.5)",
"_____no_output_____"
]
],
[
[
"## Unsupervised Machine Learning with a `kmeans.train` Procedure\n\nWe will create and run a [Procedure](../../../../doc/#builtin/procedures/Procedures.md.html) of type [`kmeans.train`](../../../../doc/#builtin/procedures/KmeansProcedure.md.html). This will train an unsupervised K-Means model and use it to assign each row in the input to a cluster, in the output dataset.",
"_____no_output_____"
]
],
[
[
"mldb.put('/v1/procedures/iris_train_kmeans', {\n 'type' : 'kmeans.train',\n 'params' : {\n 'trainingData' : 'select * EXCLUDING(class) from iris',\n 'outputDataset' : 'iris_clusters',\n 'numClusters' : 3,\n 'metric': 'euclidean',\n \"runOnCreation\": True\n }\n})",
"_____no_output_____"
]
],
[
[
"Now we can look at the output dataset and compare the clusters the model learned with the three types of flower in the dataset.",
"_____no_output_____"
]
],
[
[
"mldb.query(\"\"\"\n select pivot(class, num) as *\n from (\n select cluster, class, count(*) as num\n from merge(iris_clusters, iris)\n group by cluster, class\n )\n group by cluster\n\"\"\")",
"_____no_output_____"
]
],
[
[
"As you can see, the K-means algorithm doesn't do a great job of clustering this data (as is mentioned in the Wikipedia article!).",
"_____no_output_____"
],
[
"## Supervised Machine Learning with `classifier.train` and `.test` Procedures\n\nWe will now create and run a [Procedure](../../../../doc/#builtin/procedures/Procedures.md.html) of type [`classifier.train`](../../../../doc/#builtin/procedures/Classifier.md.html). The configuration below will use 20% of the data to train a decision tree to classify rows into the three classes of Iris. The output of this procedure is a [Function](../../../../doc/#builtin/functions/Functions.md.html), which we will be able to call from REST or SQL.",
"_____no_output_____"
]
],
[
[
"mldb.put('/v1/procedures/iris_train_classifier', {\n 'type' : 'classifier.train',\n 'params' : {\n 'trainingData' : \"\"\"\n select \n {* EXCLUDING(class)} as features, \n class as label \n from iris \n where rowHash() % 5 = 0\n \"\"\",\n \"algorithm\": \"dt\",\n \"modelFileUrl\": \"file://models/iris.cls\",\n \"mode\": \"categorical\",\n \"functionName\": \"iris_classify\",\n \"runOnCreation\": True\n }\n})",
"_____no_output_____"
]
],
[
[
"We can now test the classifier we just trained on the subset of the data we didn't use for training. To do so we use a procedure of type [`classifier.test`](../../../../doc/#builtin/procedures/Accuracy.md.html).",
"_____no_output_____"
]
],
[
[
"rez = mldb.put('/v1/procedures/iris_test_classifier', {\n 'type' : 'classifier.test',\n 'params' : {\n 'testingData' : \"\"\"\n select \n iris_classify({\n features: {* EXCLUDING(class)}\n }) as score,\n class as label \n from iris \n where rowHash() % 5 != 0\n \"\"\",\n \"mode\": \"categorical\",\n \"runOnCreation\": True\n }\n})\n\nrunResults = rez.json()[\"status\"][\"firstRun\"][\"status\"]\nprint rez",
"<Response [201]>\n"
]
],
[
[
"The procedure returns a confusion matrix, which you can compare with the one that resulted from the K-means procedure.",
"_____no_output_____"
]
],
[
[
"pd.DataFrame(runResults[\"confusionMatrix\"])\\\n .pivot_table(index=\"actual\", columns=\"predicted\", fill_value=0)",
"_____no_output_____"
]
],
[
[
"As you can see, the decision tree does a much better job of classifying the data than the K-means model, using 20% of the examples as training data.\n\nThe procedure also returns standard classification statistics on how the classifier performed on the test set. Below are performance statistics for each label:",
"_____no_output_____"
]
],
[
[
"pd.DataFrame.from_dict(runResults[\"labelStatistics\"]).transpose()",
"_____no_output_____"
]
],
[
[
"They are also available, averaged over all labels:",
"_____no_output_____"
]
],
[
[
"pd.DataFrame.from_dict({\"weightedStatistics\": runResults[\"weightedStatistics\"]})",
"_____no_output_____"
]
],
[
[
"### Scoring new examples\n\nWe can call the Function REST API endpoint to classify a never-before-seen set of measurements like this:",
"_____no_output_____"
]
],
[
[
"mldb.get('/v1/functions/iris_classify/application', input={\n \"features\":{\n \"petal length\": 1,\n \"petal width\": 2,\n \"sepal length\": 3,\n \"sepal width\": 4\n }\n})",
"_____no_output_____"
]
],
[
[
"## Where to next?\n\nCheck out the other [Tutorials and Demos](../../../../doc/#builtin/Demos.md.html).\n\nYou can also take a look at the [`classifier.experiment`](../../../../doc/#builtin/procedures/ExperimentProcedure.md.html) procedure type that can be used to train and test a classifier in a single call.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d03f9047c6c6a514a0e061efcb811b2ddc2535e5 | 20,809 | ipynb | Jupyter Notebook | A2/A2-probability-information-theory.ipynb | Colmine/NLP_Stuff | 0e91f2b3184076de3ccb68922db6f37b388d4547 | [
"AFL-3.0"
] | null | null | null | A2/A2-probability-information-theory.ipynb | Colmine/NLP_Stuff | 0e91f2b3184076de3ccb68922db6f37b388d4547 | [
"AFL-3.0"
] | null | null | null | A2/A2-probability-information-theory.ipynb | Colmine/NLP_Stuff | 0e91f2b3184076de3ccb68922db6f37b388d4547 | [
"AFL-3.0"
] | null | null | null | 23.781714 | 239 | 0.46033 | [
[
[
"# Assignment 2 - Elementary Probability and Information Theory \n# Boise State University NLP - Dr. Kennington\n\n### Instructions and Hints:\n\n* This notebook loads some data into a `pandas` dataframe, then does a small amount of preprocessing. Make sure your data can load by stepping through all of the cells up until question 1. \n* Most of the questions require you to write some code. In many cases, you will write some kind of probability function like we did in class using the data. \n* Some of the questions only require you to write answers, so be sure to change the cell type to markdown or raw text\n* Don't worry about normalizing the text this time (e.g., lowercase, etc.). Just focus on probabilies. \n* Most questions can be answered in a single cell, but you can make as many additional cells as you need. \n* Follow the instructions on the corresponding assignment Trello card for submitting your assignment. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd \n\ndata = pd.read_csv('pnp-train.txt',delimiter='\\t',encoding='latin-1', # utf8 encoding didn't work for this\n names=['type','name']) # supply the column names for the dataframe\n\n# this next line creates a new column with the lower-cased first word\ndata['first_word'] = data['name'].map(lambda x: x.lower().split()[0])",
"_____no_output_____"
],
[
"data[:10]",
"_____no_output_____"
],
[
"data.describe()",
"_____no_output_____"
]
],
[
[
"## 1. Write a probability function/distribution $P(T)$ over the types. \n\nHints:\n\n* The Counter library might be useful: `from collections import Counter`\n* Write a function `def P(T='')` that returns the probability of the specific value for T\n* You can access the types from the dataframe by calling `data['type']`",
"_____no_output_____"
]
],
[
[
"from collections import Counter\n\ndef P(T=''):\n global counts\n global data\n counts = Counter(data['type'])\n return counts[T] / len(data['type'])\ncounts",
"_____no_output_____"
]
],
[
[
"## 2. What is `P(T='movie')` ?",
"_____no_output_____"
]
],
[
[
"P(T='movie')",
"_____no_output_____"
]
],
[
[
"## 3. Show that your probability distribution sums to one.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nround(np.sum([P(T=x) for x in set(data['type'])]), 4)",
"_____no_output_____"
]
],
[
[
"## 4. Write a joint distribution using the type and the first word of the name\n\nHints:\n\n* The function is $P2(T,W_1)$\n* You will need to count up types AND the first words, for example: ('person','bill)\n* Using the [itertools.product](https://docs.python.org/2/library/itertools.html#itertools.product) function was useful for me here",
"_____no_output_____"
]
],
[
[
"def P2(T='', W1=''):\n global count\n count = data[['type', 'first_word']]\n return len(count.loc[(count['type'] == T) & (count['first_word'] == W1)]) / len(count)",
"_____no_output_____"
]
],
[
[
"## 5. What is P2(T='person', W1='bill')? What about P2(T='movie',W1='the')?",
"_____no_output_____"
]
],
[
[
"P2(T='person', W1='bill')",
"_____no_output_____"
],
[
"P2(T='movie', W1='the')",
"_____no_output_____"
]
],
[
[
"## 6. Show that your probability distribution P(T,W1) sums to one.",
"_____no_output_____"
]
],
[
[
"types = Counter(data['type'])\nwords = Counter(data['first_word'])\nretVal = 0\nfor x in types:\n for y in words:\n retVal = retVal + P2(T=x,W1=y)\nprint(round(retVal,4))",
"1.0\n"
]
],
[
[
"## 7. Make a new function Q(T) from marginalizing over P(T,W1) and make sure that Q(T) sums to one.\n\nHints:\n\n* Your Q function will call P(T,W1)\n* Your check for the sum to one should be the same answer as Question 3, only it calls Q instead of P.",
"_____no_output_____"
]
],
[
[
"def Q(T=''):\n words = Counter(data['first_word'])\n retVal = 0\n for x in words:\n retVal = retVal + P2(T,W1=x)\n return retVal",
"_____no_output_____"
],
[
"Q('movie')",
"_____no_output_____"
],
[
"round(np.sum([Q(T=x) for x in set(data['type'])]), 4)",
"_____no_output_____"
]
],
[
[
"## 8. What is the KL Divergence of your Q function and your P function for Question 1?\n\n* Even if you know the answer, you still need to write code that computes it.",
"_____no_output_____"
],
[
"I wasn't quite sure how to properly do this question so it's kind of just half implemented. Although I do know that it should be 0.0",
"_____no_output_____"
]
],
[
[
"import math\n(P('drug') * math.log(P('drug') / Q('drug')) + P('movie') * math.log(P('movie') / Q('movie')))",
"_____no_output_____"
]
],
[
[
"## 9. Convert from P(T,W1) to P(W1|T) \n\nHints:\n\n* Just write a comment cell, no code this time. \n* Note that $P(T,W1) = P(W1,T)$",
"_____no_output_____"
],
[
"Given that P(T,W1) = P(W1,T) then we can infer that P(W1|T) = (P(W1,T)/P(T))",
"_____no_output_____"
],
[
"(try to use markdown math formating, answer in this cell)",
"_____no_output_____"
],
[
"## 10. Write a function `Pwt` (that calls the functions you already have) to compute $P(W_1|T)$.\n\n* This will be something like the multiplication rule, but you may need to change something",
"_____no_output_____"
]
],
[
[
"def Pwt(W1='',T=''):\n return P2(T=T,W1=W1)/P(T=T)",
"_____no_output_____"
]
],
[
[
"## 11. What is P(W1='the'|T='movie')?",
"_____no_output_____"
]
],
[
[
"Pwt(W1='the',T='movie')",
"_____no_output_____"
]
],
[
[
"## 12. Use Baye's rule to convert from P(W1|T) to P(T|W1). Write a function Ptw to reflect this. \n\nHints:\n\n* Call your other functions.\n* You may need to write a function for P(W1) and you may need a new counter for `data['first_word']`",
"_____no_output_____"
]
],
[
[
"def Pw(W1=''):\n words = Counter(data['first_word'])\n return words[W1] / len(data['first_word'])\n\n\ndef Ptw(T='',W1=''):\n return (Pwt(W1=W1,T=T)*P(T=T))/Pw(W1=W1)",
"_____no_output_____"
]
],
[
[
"## 13 \n### What is P(T='movie'|W1='the')? \n### What about P(T='person'|W1='the')?\n### What about P(T='drug'|W1='the')?\n### What about P(T='place'|W1='the')\n### What about P(T='company'|W1='the')",
"_____no_output_____"
]
],
[
[
"Ptw(T='movie',W1='the')",
"_____no_output_____"
],
[
"Ptw(T='person',W1='the')",
"_____no_output_____"
],
[
"Ptw(T='drug',W1='the')",
"_____no_output_____"
],
[
"Ptw(T='place',W1='the')",
"_____no_output_____"
],
[
"Ptw(T='company',W1='the')",
"_____no_output_____"
]
],
[
[
"## 14 Given this, if the word 'the' is found in a name, what is the most likely type?",
"_____no_output_____"
]
],
[
[
"Pwt('the', 'movie')",
"_____no_output_____"
]
],
[
[
"## 15. Is Ptw(T='movie'|W1='the') the same as Pwt(W1='the'|T='movie') the same? Why or why not?",
"_____no_output_____"
]
],
[
[
"Ptw(T='movie',W1='the')",
"_____no_output_____"
],
[
"Pwt(W1='the', T='movie')",
"_____no_output_____"
]
],
[
[
"They are not the same because it's basically the probability of getting a movie type and then getting the word 'the' after, whereas the other way is getting the word 'the' and then getting a movie type after.",
"_____no_output_____"
],
[
"## 16. Do you think modeling Ptw(T|W1) would be better with a continuous function like a Gaussian? Why or why not?\n\n- Answer in a markdown cell\n",
"_____no_output_____"
],
[
"No, I don't think modeling the Ptw(T|W1) would be better with a continuous function like a Gaussian function. This is \nbecause the set of data that we're given is finite and is likely not completely randomly distributed and likely has bias somewhere in it. Because of this fact using a continuous function might not yield the results we're looking for.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d03f92443d5d302df99a96fc0a2f8863b432335d | 108,533 | ipynb | Jupyter Notebook | module3-model-interpretation/LS_DS_243_BONUS_Feature_Selection.ipynb | standroidbeta/DS-Unit-2-Sprint-4-Practicing-Understanding | a43a5bb5404beed9a0127127b157e677d4eb281a | [
"MIT"
] | null | null | null | module3-model-interpretation/LS_DS_243_BONUS_Feature_Selection.ipynb | standroidbeta/DS-Unit-2-Sprint-4-Practicing-Understanding | a43a5bb5404beed9a0127127b157e677d4eb281a | [
"MIT"
] | null | null | null | module3-model-interpretation/LS_DS_243_BONUS_Feature_Selection.ipynb | standroidbeta/DS-Unit-2-Sprint-4-Practicing-Understanding | a43a5bb5404beed9a0127127b157e677d4eb281a | [
"MIT"
] | null | null | null | 40.771225 | 17,084 | 0.555029 | [
[
[
"_Lambda School Data Science - Model Validation_\n\n\n# Feature Selection\n\nObjectives:\n* Feature importance\n* Feature selection ",
"_____no_output_____"
],
[
"## Yesterday we saw that...\n\n## Less isn't always more (but sometimes it is)\n\n## More isn't always better (but sometimes it is)\n\n\n",
"_____no_output_____"
],
[
"Saavas, Ando [Feature Selection (4 parts)](https://blog.datadive.net/selecting-good-features-part-i-univariate-selection/)\n\n>There are in general two reasons why feature selection is used:\n1. Reducing the number of features, to reduce overfitting and improve the generalization of models.\n2. To gain a better understanding of the features and their relationship to the response variables.\n\n>These two goals are often at odds with each other and thus require different approaches: depending on the data at hand a feature selection method that is good for goal (1) isn’t necessarily good for goal (2) and vice versa. What seems to happen often though is that people use their favourite method (or whatever is most conveniently accessible from their tool of choice) indiscriminately, especially methods more suitable for (1) for achieving (2).",
"_____no_output_____"
],
[
"While they are not always mutually exclusive, here's a little bit about what's going on with these two goals",
"_____no_output_____"
],
[
"### Goal 1: Reducing Features, Reducing Overfitting, Improving Generalization of Models\n\nThis is when you're actually trying to engineer a packaged, machine learning pipeline that is streamlined and highly generalizable to novel data as more is collected, and you don't really care \"how\" it works as long as it does work. \n\nApproaches that are good at this tend to fail at Goal 2 because they handle multicollinearity by (sometime randomly) choosing/indicating just one of a group of strongly correlated features. This is good to reduce redundancy, but bad if you want to interpret the data.",
"_____no_output_____"
],
[
"### Goal 2: Gaining a Better Understanding of the Features and their Relationships\n\nThis is when you want a good, interpretable model or you're doing data science more for analysis than engineering. Company asks you \"How do we increase X?\" and you can tell them all the factors that correlate to it and their predictive power.\n\nApproaches that are good at this tend to fail at Goal 1 because, well, they *don't* handle the multicollinearity problem. If three features are all strongly correlated to each other as well as the output, they will all have high scores. But including all three features in a model is redundant.",
"_____no_output_____"
],
[
"### Each part in Saavas's Blog series describes an increasingly complex (and computationally costly) set of methods for feature selection and interpretation.\n\nThe ultimate comparison is completed using an adaptation of a dataset called Friedman's 1 regression dataset from Friedman, Jerome H.'s '[Multivariate Adaptive Regression Splines](http://www.stat.ucla.edu/~cocteau/stat204/readings/mars.pdf).\n>The data is generated according to formula $y=10sin(πX_1X_2)+20(X_3–0.5)^2+10X_4+5X_5+ϵ$, where the $X_1$ to $X_5$ are drawn from uniform distribution and ϵ is the standard normal deviate N(0,1). Additionally, the original dataset had five noise variables $X_6,…,X_{10}$, independent of the response variable. We will increase the number of variables further and add four variables $X_{11},…,X_{14}$ each of which are very strongly correlated with $X_1,…,X_4$, respectively, generated by $f(x)=x+N(0,0.01)$. This yields a correlation coefficient of more than 0.999 between the variables. This will illustrate how different feature ranking methods deal with correlations in the data.\n\n**Okay, that's a lot--here's what you need to know:**\n1. $X_1$ and $X_2$ have the same non-linear relationship to $Y$ -- though together they do have a not-quite-linear relationship to $Y$ (with sinusoidal noise--but the range of the values doesn't let it get negative)\n2. $X_3$ has a quadratic relationship with $Y$\n3. $X_4$ and $X_5$ have linear relationships to $Y$, with $X_4$ being weighted twice as heavily as $X_5$\n4. $X_6$ through $X_{10}$ are random and have NO relationship to $Y$\n5. $X_{11}$ through $X_{14}$ correlate strongly to $X_1$ through $X_4$ respectively (and thus have the same respective relationships with $Y$)\n\n\nThis will help us see the difference between the models in selecting features and interpreting features\n* how well they deal with multicollinearity (#5)\n* how well they identify noise (#4)\n* how well they identify different kinds of relationships\n* how well they identify/interpret predictive power of individual variables.",
"_____no_output_____"
]
],
[
[
"# import\nimport numpy as np\n\n# Create the dataset\n# from https://blog.datadive.net/selecting-good-features-part-iv-stability-selection-rfe-and-everything-side-by-side/\n\nnp.random.seed(42)\n\nsize = 1500 # I increased the size from what's given in the link\nXs = np.random.uniform(0, 1, (size, 14)) \n# Changed variable name to Xs to use X later\n \n#\"Friedamn #1” regression problem\nY = (10 * np.sin(np.pi*Xs[:,0]*Xs[:,1]) + 20*(Xs[:,2] - .5)**2 +\n 10*Xs[:,3] + 5*Xs[:,4] + np.random.normal(0,1))\n#Add 4 additional correlated variables (correlated with X1-X4)\nXs[:,10:] = Xs[:,:4] + np.random.normal(0, .025, (size,4))\n \nnames = [\"X%s\" % i for i in range(1,15)]",
"_____no_output_____"
],
[
"# Putting it into pandas--because... I like pandas. And usually you'll be\n# working with dataframes not arrays (you'll care what the column titles are)\nimport pandas as pd\n\nfriedmanX = pd.DataFrame(data=Xs, columns=names)\nfriedmanY = pd.Series(data=Y, name='Y')\n\nfriedman = friedmanX.join(friedmanY)\n\nfriedman.head()",
"_____no_output_____"
]
],
[
[
"We want to be able to look at classification problems too, so let's bin the Y values to create a categorical feature from the Y values. It should have *roughly* similar relationships to the X features as Y does.",
"_____no_output_____"
]
],
[
[
"# First, let's take a look at what Y looks like\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nsns.distplot(friedmanY);",
"_____no_output_____"
]
],
[
[
"That's pretty normal, let's make two binary categories--one balanced, one unbalanced, to see the difference.\n* balanced binary variable will be split evenly in half\n* unbalanced binary variable will indicate whether $Y <5$.",
"_____no_output_____"
]
],
[
[
"friedman['Y_bal'] = friedman['Y'].apply(lambda y: 1 if (y < friedman.Y.median()) else 0)\nfriedman['Y_un'] = friedman['Y'].apply(lambda y: 1 if (y < 5) else 0)\n\nprint(friedman.Y_bal.value_counts(), '\\n\\n', friedman.Y_un.value_counts())",
"1 750\n0 750\nName: Y_bal, dtype: int64 \n\n 0 1461\n1 39\nName: Y_un, dtype: int64\n"
],
[
"friedman.head()",
"_____no_output_____"
],
[
"# Finally, let's put it all into our usual X and y's\n# (I already have the X dataframe as friedmanX, but I'm working backward to\n# follow a usual flow)\n\nX = friedman.drop(columns=['Y', 'Y_bal', 'Y_un'])\n\ny = friedman.Y\n\ny_bal = friedman.Y_bal\n\ny_un = friedman.Y_un",
"_____no_output_____"
]
],
[
[
"#### Alright! Let's get to it! Remember, with each part, we are increasing complexity of the analysis and thereby increasing the computational costs and runtime.",
"_____no_output_____"
],
[
"So even before univariate selection--which compares each feature to the output feature one by one--there is a [VarianceThreshold](https://scikit-learn.org/stable/modules/generated/sklearn.feature_selection.VarianceThreshold.html#sklearn.feature_selection.VarianceThreshold) object in sklearn.feature_selection. It defaults to getting rid of any features that are the same across all samples. Great for cleaning data in that respect. \n\nThe `threshold` parameter defaults to `0` to show the above behavior. if you change it, make sure you have good reason. Use with caution.",
"_____no_output_____"
],
[
"## Part 1: univariate selection\n* Best for goal 2 - getting \"a better understanding of the data, its structure and characteristics\"\n* unable to remove redundancy (for example selecting only the best feature among a subset of strongly correlated features)\n* Super fast - can be used for baseline models or just after baseline\n\n[sci-kit's univariariate feature selection objects and techniques](https://scikit-learn.org/stable/modules/feature_selection.html#univariate-feature-selection)",
"_____no_output_____"
],
[
"#### Y (continuous output)\n\noptions (they do what they sound like they do)\n* SelectKBest\n* SelectPercentile\n\nboth take the same parameter options for `score_func`\n* `f_regression`: scores by correlation coefficient, f value, p value--basically automates what you can do by looking at a correlation matrix except without the ability to recognize collinearity\n* `mutual_info_regression`: can capture non-linear correlations, but doesn't handle noise well\n\nLet's take a look at mutual information (MI)",
"_____no_output_____"
]
],
[
[
"import sklearn.feature_selection as fe\n\nMIR = fe.SelectKBest(fe.mutual_info_regression, k='all').fit(X, y)\n\nMIR_scores = pd.Series(data=MIR.scores_, name='MI_Reg_Scores', index=names)\n\nMIR_scores",
"_____no_output_____"
]
],
[
[
"#### Y_bal (balanced binary output)\n\noptions\n* SelectKBest\n* SelectPercentile\n\nthese options will cut out features with error rates above a certain tolerance level, define in parameter -`alpha`\n* SelectFpr (false positive rate--false positives predicted/total negatives in dataset)\n* SelectFdr (false discovery rate--false positives predicted/total positives predicted)\n* ~~SelectFwe (family-wise error--for multinomial classification tasks)~~\n\nall have the same optons for parameter `score_func`\n* `chi2`\n* `f_classif`\n* `mutual_info_classif`",
"_____no_output_____"
]
],
[
[
"MIC_b = fe.SelectFpr(fe.mutual_info_classif).fit(X, y_bal)\n\nMIC_b_scores = pd.Series(data=MIC_b.scores_, \n name='MIC_Bal_Scores', index=names)\n\nMIC_b_scores",
"_____no_output_____"
]
],
[
[
"#### Y_un (unbalanced binary output)",
"_____no_output_____"
]
],
[
[
"MIC_u = fe.SelectFpr(fe.mutual_info_classif).fit(X, y_un)\n\nMIC_u_scores = pd.Series(data=MIC_u.scores_, \n name='MIC_Unbal_Scores', index=names)\n\nMIC_u_scores",
"_____no_output_____"
]
],
[
[
"## Part 2: linear models and regularization\n* L1 Regularization (Lasso for regression) is best for goal 1: \"produces sparse solutions and as such is very useful selecting a strong subset of features for improving model performance\" (forces coefficients to zero, telling you which you could remove--but doesn't handle multicollinearity)\n* L2 Regularization (Ridge for regression) is best for goal 2: \"can be used for data interpretation due to its stability and the fact that useful features tend to have non-zero coefficients\n* Also fast\n\n[sci-kit's L1 feature selection](https://scikit-learn.org/stable/modules/feature_selection.html#l1-based-feature-selection) (can easily be switched to L2 using the parameter `penalty='l2'` for categorical targets or using `Ridge` instead of Lasso for continuous targets)\n\nWe won't do this here, because\n1. You know regression\n2. The same principles apply as shown in Part 3 below with `SelectFromModel`\n3. There's way cooler stuff coming up",
"_____no_output_____"
],
[
"## Part 3: random forests\n* Best for goal 1, not 2 because:\n * strong features can end up with low scores \n * biased towards variables with many categories\n* \"require very little feature engineering and parameter tuning\"\n* Takes a little more time depending on your dataset - but a popular technique\n\n[sci-kit's implementation of tree-based feature selection](https://scikit-learn.org/stable/modules/feature_selection.html#tree-based-feature-selection)",
"_____no_output_____"
],
[
"#### Y",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestRegressor as RFR\n\n# Fitting a random forest regression\nrfr = RFR().fit(X, y)\n\n# Creating scores from feature_importances_ ranking (some randomness here)\nrfr_scores = pd.Series(data=rfr.feature_importances_, name='RFR', index=names)\n\nrfr_scores",
"/home/seek/anaconda3/lib/python3.7/site-packages/sklearn/ensemble/forest.py:246: FutureWarning: The default value of n_estimators will change from 10 in version 0.20 to 100 in 0.22.\n \"10 in version 0.20 to 100 in 0.22.\", FutureWarning)\n"
]
],
[
[
"#### Y_bal",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier as RFC\n\n# Fitting a Random Forest Classifier\nrfc_b = RFC().fit(X, y_bal)\n\n# Creating scores from feature_importances_ ranking (some randomness here)\nrfc_b_scores = pd.Series(data=rfc_b.feature_importances_, name='RFC_bal', \n index=names)\n\nrfc_b_scores",
"/home/seek/anaconda3/lib/python3.7/site-packages/sklearn/ensemble/forest.py:246: FutureWarning: The default value of n_estimators will change from 10 in version 0.20 to 100 in 0.22.\n \"10 in version 0.20 to 100 in 0.22.\", FutureWarning)\n"
]
],
[
[
"#### Y_un",
"_____no_output_____"
]
],
[
[
"# Fitting a Random Forest Classifier\nrfc_u = RFC().fit(X, y_un)\n\n# Creating scores from feature_importances_ ranking (some randomness here)\nrfc_u_scores = pd.Series(data=rfc_u.feature_importances_, \n name='RFC_unbal', index=names)\n\nrfc_u_scores",
"/home/seek/anaconda3/lib/python3.7/site-packages/sklearn/ensemble/forest.py:246: FutureWarning: The default value of n_estimators will change from 10 in version 0.20 to 100 in 0.22.\n \"10 in version 0.20 to 100 in 0.22.\", FutureWarning)\n"
]
],
[
[
"### SelectFromModel \nis a meta-transformer that can be used along with any estimator that has a `coef_` or `feature_importances_` attribute after fitting. The features are considered unimportant and removed, if the corresponding `coef_` or `feature_importances_` values are below the provided `threshold` parameter. Apart from specifying the `threshold` numerically, there are built-in heuristics for finding a `threshold` using a string argument. Available heuristics are `'mean'`, `'median'` and float multiples of these like `'0.1*mean'`.",
"_____no_output_____"
]
],
[
[
"# Random forest regression transformation of X (elimination of least important\n# features)\nrfr_transform = fe.SelectFromModel(rfr, prefit=True)\n\nX_rfr = rfr_transform.transform(X)\n\n\n# Random forest classifier transformation of X_bal (elimination of least important\n# features)\nrfc_b_transform = fe.SelectFromModel(rfc_b, prefit=True)\n\nX_rfc_b = rfc_b_transform.transform(X)\n\n\n# Random forest classifier transformation of X_un (elimination of least important\n# features)\nrfc_u_transform = fe.SelectFromModel(rfc_u, prefit=True)\n\nX_rfc_u = rfc_u_transform.transform(X)",
"_____no_output_____"
],
[
"RF_comparisons = pd.DataFrame(data=np.array([rfr_transform.get_support(),\n rfc_b_transform.get_support(),\n rfc_u_transform.get_support()]).T,\n columns=['RF_Regressor', 'RF_balanced_classifier',\n 'RF_unbalanced_classifier'],\n index=names)\n\nRF_comparisons",
"_____no_output_____"
]
],
[
[
"## Part 4: stability selection, RFE, and everything side by side\n* These methods take longer since they are *wrapper methods* and build multiple ML models before giving results. \"They both build on top of other (model based) selection methods such as regression or SVM, building models on different subsets of data and extracting the ranking from the aggregates.\"\n* Stability selection is good for both goal 1 and 2: \"among the top performing methods for many different datasets and settings\"\n * For categorical targets\n * ~~[RandomizedLogisticRegression](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.RandomizedLogisticRegression.html)~~ (Deprecated) use [RandomizedLogisticRegression](https://thuijskens.github.io/stability-selection/docs/randomized_lasso.html#stability_selection.randomized_lasso.RandomizedLogisticRegression)\n \n * [ExtraTreesClassifier](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.ExtraTreesClassifier.html#sklearn.ensemble.ExtraTreesClassifier)\n \n * For continuous targets\n * ~~[RandomizedLasso](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.RandomizedLasso.html)~~ (Deprecated) use [RandomizedLasso](https://thuijskens.github.io/stability-selection/docs/randomized_lasso.html#stability_selection.randomized_lasso.RandomizedLogisticRegression) \n * [ExtraTreesRegressor](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.ExtraTreesRegressor.html#sklearn.ensemble.ExtraTreesRegressor)\n \n Welcome to open-source, folks! [Here](https://github.com/scikit-learn/scikit-learn/issues/8995) is the original discussion to deprecate `RandomizedLogisticRegression` and `RandomizedLasso`. [Here](https://github.com/scikit-learn/scikit-learn/issues/9657) is a failed attempt to resurrect it. It looks like it'll be gone for good soon. So we shouldn't get dependent on it.\n\n The alternatives from the deprecated scikit objects come from an official scikit-learn-contrib module called [stability_selection](https://github.com/scikit-learn-contrib/stability-selection). They also have a `StabilitySelection` object that acts similarly scikit's `SelectFromModel`.\n\n* recursive feature elimination (RFE) is best for goal 1\n * [sci-kit's RFE and RFECV (RFE with built-in cross-validation)](https://scikit-learn.org/stable/modules/feature_selection.html#recursive-feature-elimination)",
"_____no_output_____"
]
],
[
[
"!pip install git+https://github.com/scikit-learn-contrib/stability-selection.git",
"Collecting git+https://github.com/scikit-learn-contrib/stability-selection.git\n Cloning https://github.com/scikit-learn-contrib/stability-selection.git to /tmp/pip-req-build-axvzrzpv\nRequirement already satisfied: nose>=1.1.2 in /home/seek/anaconda3/lib/python3.7/site-packages (from stability-selection==0.0.1) (1.3.7)\nRequirement already satisfied: scikit-learn>=0.19 in /home/seek/anaconda3/lib/python3.7/site-packages (from stability-selection==0.0.1) (0.20.3)\nRequirement already satisfied: matplotlib>=2.0.0 in /home/seek/anaconda3/lib/python3.7/site-packages (from stability-selection==0.0.1) (3.0.3)\nRequirement already satisfied: numpy>=1.8.0 in /home/seek/anaconda3/lib/python3.7/site-packages (from stability-selection==0.0.1) (1.16.2)\nRequirement already satisfied: scipy>=0.13.3 in /home/seek/anaconda3/lib/python3.7/site-packages (from scikit-learn>=0.19->stability-selection==0.0.1) (1.2.1)\nRequirement already satisfied: cycler>=0.10 in /home/seek/anaconda3/lib/python3.7/site-packages (from matplotlib>=2.0.0->stability-selection==0.0.1) (0.10.0)\nRequirement already satisfied: kiwisolver>=1.0.1 in /home/seek/anaconda3/lib/python3.7/site-packages (from matplotlib>=2.0.0->stability-selection==0.0.1) (1.0.1)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /home/seek/anaconda3/lib/python3.7/site-packages (from matplotlib>=2.0.0->stability-selection==0.0.1) (2.3.1)\nRequirement already satisfied: python-dateutil>=2.1 in /home/seek/anaconda3/lib/python3.7/site-packages (from matplotlib>=2.0.0->stability-selection==0.0.1) (2.8.0)\nRequirement already satisfied: six in /home/seek/anaconda3/lib/python3.7/site-packages (from cycler>=0.10->matplotlib>=2.0.0->stability-selection==0.0.1) (1.12.0)\nRequirement already satisfied: setuptools in /home/seek/anaconda3/lib/python3.7/site-packages (from kiwisolver>=1.0.1->matplotlib>=2.0.0->stability-selection==0.0.1) (40.8.0)\nBuilding wheels for collected packages: stability-selection\n Building wheel for stability-selection (setup.py) ... \u001b[?25ldone\n\u001b[?25h Stored in directory: /tmp/pip-ephem-wheel-cache-woii323n/wheels/58/be/39/79880712b91ffa56e341ff10586a1956527813437ddd759473\nSuccessfully built stability-selection\nInstalling collected packages: stability-selection\nSuccessfully installed stability-selection-0.0.1\n"
]
],
[
[
"Okay, I tried this package... it seems to have some problems... hopefully a good implementation of stability selection for Lasso and Logistic Regression will be created soon! In the meantime, scikit's RandomLasso and RandomLogisticRegression have not been removed, so you can fiddle some! Just alter the commented out code!\n* import from scikit instead of stability-selection\n* use scikit's `SelectFromModel` as shown above!\n\nTa Da!",
"_____no_output_____"
],
[
"#### Y",
"_____no_output_____"
]
],
[
[
"'''from stability_selection import (RandomizedLogisticRegression,\n RandomizedLasso, StabilitySelection,\n plot_stability_path)\n\n# Stability selection using randomized lasso method\nrl = RandomizedLasso(max_iter=2000)\nrl_selector = StabilitySelection(base_estimator=rl, lambda_name='alpha',\n n_jobs=2)\nrl_selector.fit(X, y);\n'''",
"_____no_output_____"
],
[
"from sklearn.ensemble import ExtraTreesRegressor as ETR\n\n# Stability selection using randomized decision trees\netr = ETR(n_estimators=50).fit(X, y)\n\n# Creating scores from feature_importances_ ranking (some randomness here)\netr_scores = pd.Series(data=etr.feature_importances_, \n name='ETR', index=names)\n\netr_scores",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression\n\n# Recursive feature elimination with cross validaiton using linear regression \n# as the model\nlr = LinearRegression()\n# rank all features, i.e continue the elimination until the last one\nrfe = fe.RFECV(lr)\nrfe.fit(X, y)\n\nrfe_score = pd.Series(data=(-1*rfe.ranking_), name='RFE', index=names)\n\nrfe_score",
"/home/seek/anaconda3/lib/python3.7/site-packages/sklearn/model_selection/_split.py:2053: FutureWarning: You should specify a value for 'cv' instead of relying on the default value. The default value will change from 3 to 5 in version 0.22.\n warnings.warn(CV_WARNING, FutureWarning)\n"
]
],
[
[
"#### Y_bal",
"_____no_output_____"
]
],
[
[
"# stability selection using randomized logistic regression\n'''rlr_b = RandomizedLogisticRegression()\nrlr_b_selector = StabilitySelection(base_estimator=rlr_b, lambda_name='C',\n n_jobs=2)\nrlr_b_selector.fit(X, y_bal);'''",
"_____no_output_____"
],
[
"from sklearn.ensemble import ExtraTreesClassifier as ETC\n\n# Stability selection using randomized decision trees\netc_b = ETC(n_estimators=50).fit(X, y_bal)\n\n# Creating scores from feature_importances_ ranking (some randomness here)\netc_b_scores = pd.Series(data=etc_b.feature_importances_, \n name='ETC_bal', index=names)\n\netc_b_scores",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression\n\n# Recursive feature elimination with cross validaiton using logistic regression \n# as the model\nlogr_b = LogisticRegression(solver='lbfgs')\n# rank all features, i.e continue the elimination until the last one\nrfe_b = fe.RFECV(logr_b)\nrfe_b.fit(X, y_bal)\n\nrfe_b_score = pd.Series(data=(-1*rfe_b.ranking_), name='RFE_bal', index=names)\n\nrfe_b_score",
"/home/seek/anaconda3/lib/python3.7/site-packages/sklearn/model_selection/_split.py:2053: FutureWarning: You should specify a value for 'cv' instead of relying on the default value. The default value will change from 3 to 5 in version 0.22.\n warnings.warn(CV_WARNING, FutureWarning)\n"
]
],
[
[
"#### Y_un",
"_____no_output_____"
]
],
[
[
"# stability selection uisng randomized logistic regression\n'''rlr_u = RandomizedLogisticRegression(max_iter=2000)\nrlr_u_selector = StabilitySelection(base_estimator=rlr_u, lambda_name='C')\n\nrlr_u_selector.fit(X, y_un);'''",
"_____no_output_____"
],
[
"# Stability selection using randomized decision trees\netc_u = ETC(n_estimators=50).fit(X, y_un)\n\n# Creating scores from feature_importances_ ranking (some randomness here)\netc_u_scores = pd.Series(data=etc_u.feature_importances_, \n name='ETC_unbal', index=names)\n\netc_u_scores",
"_____no_output_____"
],
[
"# Recursive feature elimination with cross validaiton using logistic regression \n# as the model\nlogr_u = LogisticRegression(solver='lbfgs')\n# rank all features, i.e continue the elimination until the last one\nrfe_u = fe.RFECV(logr_u)\nrfe_u.fit(X, y_un)\n\nrfe_u_score = pd.Series(data=(-1*rfe_u.ranking_), name='RFE_unbal', index=names)\n\nrfe_u_score",
"/home/seek/anaconda3/lib/python3.7/site-packages/sklearn/model_selection/_split.py:2053: FutureWarning: You should specify a value for 'cv' instead of relying on the default value. The default value will change from 3 to 5 in version 0.22.\n warnings.warn(CV_WARNING, FutureWarning)\n"
],
[
"'''RL_comparisons = pd.DataFrame(data=np.array([rl_selector.get_support(),\n rlr_b_selector.get_support(),\n rlr_u_selector.get_support()]).T,\n columns=['RandomLasso', 'RandomLog_bal',\n 'RandomLog_unbal'],\n index=names)\n\nRL_comparisons'''",
"_____no_output_____"
],
[
"comparisons = pd.concat([MIR_scores, MIC_b_scores, MIC_u_scores, rfr_scores,\n rfc_b_scores, rfc_u_scores, etr_scores, etc_b_scores,\n etc_u_scores, rfe_score, rfe_b_score, rfe_u_score], \n axis=1)\n\ncomparisons",
"_____no_output_____"
],
[
"from sklearn.preprocessing import MinMaxScaler\n\nscaler = MinMaxScaler()\nscaled_df = scaler.fit_transform(comparisons)\nscaled_comparisons = pd.DataFrame(scaled_df, columns=comparisons.columns,\n index=names)\n\nscaled_comparisons",
"/home/seek/anaconda3/lib/python3.7/site-packages/sklearn/preprocessing/data.py:334: DataConversionWarning: Data with input dtype int64, float64 were all converted to float64 by MinMaxScaler.\n return self.partial_fit(X, y)\n"
]
],
[
[
"### What do you notice from the diagram below?",
"_____no_output_____"
]
],
[
[
"sns.heatmap(scaled_comparisons);",
"_____no_output_____"
]
],
[
[
"Head, Tim [Cross Validation Gone Wrong](https://betatim.github.io/posts/cross-validation-gone-wrong/)\n\n>Choosing your input features is just one of the many choices you have to make when building your machine-learning application. Remember to make all decisions during the cross validation, otherwise you are in for a rude awakening when your model is confronted with unseen data for the first time.",
"_____no_output_____"
],
[
"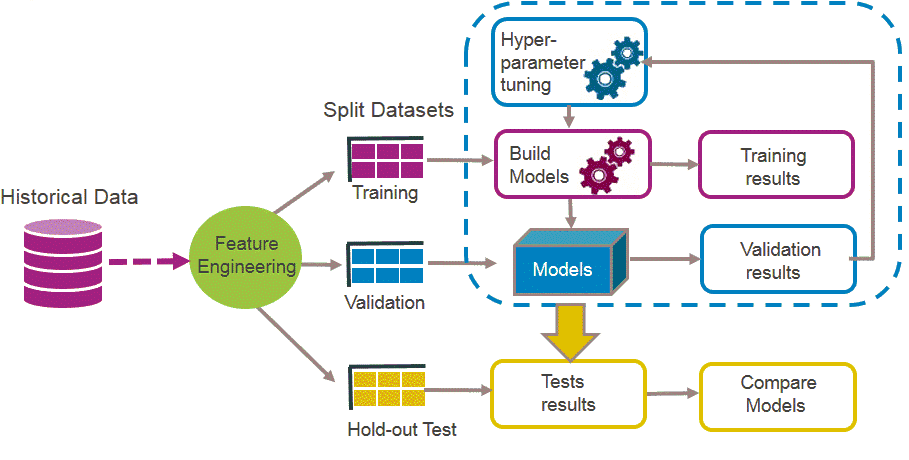\n\n\\[Image by Angarita, Natalia for [Capgemini](https://www.capgemini.com/2016/05/machine-learning-has-transformed-many-aspects-of-our-everyday-life/)\\]",
"_____no_output_____"
],
[
"**I don't fully support this diagram for a few reasons.**\n\n* I would replace \"Feature Engineering\" with \"Data Cleaning\"\n* Feature engineering can be done alongside either data cleaning or training your model--it can be done before *or* after splitting your data. (But it will need to be part of the final pipeline.)\n* Any feature standardization happens **after** the split\n* And you can use cross validation instead of an independent validation set\n\nHowever **feature selection (Goal 1) is part of choosing and training a model and should happen *after* splitting**. Feature selection belongs safely **inside the dotted line**.\n\n\"But doesn't it make sense to make your decisions based on all the information?\"\n\nNO! Mr. Head has a point!\n\n## The number of features you end up using *is* a hyperparameter. Don't cross the dotted line while hyperparameter tuning!!! Work on goal 1 AFTER splitting.\n\nI know you want to see how your model is performing... \"just real quick\"... but don't do it!\n\n...\n\nDon't!\n\n*(Kaggle does the initial train-test split for you. It doesn't even let you **see** the target values for the test data. How you like dem apples?)*\n\n\n\nWhat you **can** do is create multiple \"final\" models by hyperparameter tuning different types of models (all inside the dotted line!), then use the final hold-out test to see which does best.\n\n**All this is said with the caveat that you have a large enough dataset to support three way validation or a test set plus cross-validation**\n\n## On the flip side, feature *interpretation* (Goal 2) can be done with all the data, before splitting, since you are looking to get a full understanding underlying the relationships in the dataset.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d03f947cd342ebe79961959f3cb35f0578c85f2e | 15,613 | ipynb | Jupyter Notebook | _notebooks/2022-04-07-scraping2.ipynb | christopherGuan/sample-ds-blog | 52859b8801125ee8a5a330557e1248937952f43f | [
"Apache-2.0"
] | null | null | null | _notebooks/2022-04-07-scraping2.ipynb | christopherGuan/sample-ds-blog | 52859b8801125ee8a5a330557e1248937952f43f | [
"Apache-2.0"
] | null | null | null | _notebooks/2022-04-07-scraping2.ipynb | christopherGuan/sample-ds-blog | 52859b8801125ee8a5a330557e1248937952f43f | [
"Apache-2.0"
] | null | null | null | 30.978175 | 505 | 0.596746 | [
[
[
"# Scraping Amazon Reviews using Scrapy in Python Part 2\n> Are you looking for a method of scraping Amazon reviews and do not know where to begin with? In that case, you may find this blog very useful in scraping Amazon reviews.\n- toc: true \n- badges: true\n- comments: true\n- author: Zeyu Guan\n- categories: [spaCy, Python, Machine Learning, Data Mining, NLP, RandomForest]\n- annotations: true\n- image: https://www.freecodecamp.org/news/content/images/2020/09/wall-5.jpeg\n- hide: false",
"_____no_output_____"
],
[
"## Required Packages\n[wordcloud](https://github.com/amueller/word_cloud), \n[geopandas](https://geopandas.org/en/stable/getting_started/install.html), \n[nbformat](https://pypi.org/project/nbformat/), \n[seaborn](https://seaborn.pydata.org/installing.html), \n[scikit-learn](https://scikit-learn.org/stable/install.html)",
"_____no_output_____"
],
[
"## Now let's get started!\nFirst thing first, you need to load all the necessary libraries:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom matplotlib import pyplot as plt\nimport numpy as np\nfrom wordcloud import WordCloud\nfrom wordcloud import STOPWORDS\nimport re\nimport plotly.graph_objects as go\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"# Data Cleaning\n\nFollowing the previous blog, the raw data we scraped from Amazon look like below. \n\n\nEven thought that looks relatively clean, but there are still some inperfections such as star1 and star2 need to be combined, date need to be splited, and etc. The whole process could be found from my [github notebooks](https://github.com/christopherGuan/sample-ds-blog).\n\nBelow is data after cleaning. It contains 6 columns and more than 500 rows. \n\n",
"_____no_output_____"
],
[
"\n# EDA\nBelow are the questions I curioused about, and the result generated by doing data analysis.\n\n- Which rating (1-5) got the most and least?\n\n\n- Which country are they targeting?\n\n\n- Which month people prefer to give a higher rating?\n\n\n- Which month people leave commons the most?\n\n\n\n- What are the useful words that people mentioned in the reviews?\n\n",
"_____no_output_____"
],
[
"# Sentiment Analysis (Method 1)\n\n## What is sentiment analysis?\n\nEssentially, sentiment analysis or sentiment classification fall under the broad category of text classification tasks in which you are given a phrase or a list of phrases and your classifier is expected to determine whether the sentiment behind that phrase is positive, negative, or neutral. To keep the problem as a binary classification problem, the third attribute is sometimes ignored. Recent tasks have taken into account sentiments such as \"somewhat positive\" and \"somewhat negative.\" \n\nIn this specific case, we catogrize 4 and 5 stars to the positive group and 1 & 2 stars to the negative gorup. \n\n\n\nBelow are the most frequently words in reviews from positive group and negative group respectively. \n\nPositive review\n\n\nNegative review\n",
"_____no_output_____"
],
[
"## Build up the first model\n\nNow we can build up a easy model that, as input, it will accept reviews. It will then predict whether the review will be positive or negative.\n\nBecause this is a classification task, we will train a simple logistic regression model.\n\n\n- **Clean Data**\nFirst, we create a new function to remove all punctuations from the data for later use.\n",
"_____no_output_____"
]
],
[
[
"def remove_punctuation(text):\n final = \"\".join(u for u in text if u not in (\"?\", \".\", \";\", \":\", \"!\",'\"'))\n return final",
"_____no_output_____"
]
],
[
[
"\n- **Split the Dataframe**\n\nNow, we split 80% of the dataset for training and 20% for testing. Meanwhile, each dataset should contain only two variables, one is to indicate positive or negative and another one is the reviews.\n\n\n",
"_____no_output_____"
]
],
[
[
"df['random_number'] = np.random.randn(len(index))\ntrain = df[df['random_number'] <= 0.8]\ntest = df[df['random_number'] > 0.8]",
"_____no_output_____"
]
],
[
[
"\n- **Create a bag of words**\n\nHere I would like to introduce a new package.\n\n[Scikit-learn](https://scikit-learn.org/stable/install.html) is an open source machine learning library that supports supervised and unsupervised learning. It also provides various tools for model fitting, data preprocessing, model selection, model evaluation, and many other utilities.\n\nIn this example, we are going to use [sklearn.feature_extraction.text.CountVectorizer](https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.CountVectorizer.html?highlight=countvectorizer#sklearn.feature_extraction.text.CountVectorizer) to convert a collection of text documents to a matrix of token counts.\n\nThe reason why we need to convert the text into a bag-of-words model is because the logistic regression algorithm cannot understand text.\n",
"_____no_output_____"
]
],
[
[
"train_matrix = vectorizer.fit_transform(train['title'])\ntest_matrix = vectorizer.transform(test['title'])",
"_____no_output_____"
]
],
[
[
"- **Import Logistic Regression**",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nlr = LogisticRegression()",
"_____no_output_____"
]
],
[
[
"- **Split target and independent variables**",
"_____no_output_____"
]
],
[
[
"X_train = train_matrix\nX_test = test_matrix\ny_train = train['sentiment']\ny_test = test['sentiment']",
"_____no_output_____"
]
],
[
[
"- **Fit model on data**",
"_____no_output_____"
]
],
[
[
"lr.fit(X_train,y_train)",
"_____no_output_____"
]
],
[
[
"- **Make predictionsa**",
"_____no_output_____"
]
],
[
[
"predictions = lr.predict(X_test)",
"_____no_output_____"
]
],
[
[
"The output will be either 1 or -1. As we assumed before, 1 presents the model predict the review is a positive review and vice versa.",
"_____no_output_____"
],
[
"## Testing\n\nNow, we can test the accuracy of our model!",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import confusion_matrix, classification_report\nnew = np.asarray(y_test)\nconfusion_matrix(predictions,y_test)\n\nprint(classification_report(predictions,y_test))",
"_____no_output_____"
]
],
[
[
"\n\nThe accuracy is as high as 89%!",
"_____no_output_____"
],
[
"# Sentiment Analysis (Method 2)\n\nIn this process, you will learn how to build your own sentiment analysis classifier using Python and understand the basics of NLP (natural language processing). First, let's try to use a quick and dirty method to utilize the [Naive Bayes classifier](https://www.datacamp.com/community/tutorials/simplifying-sentiment-analysis-python) to predict the sentiments of Amazon product review. \n\nBased on the application's requirements, we should first put each review in a txt file and catogorize them as negative or positive review in different folder. ",
"_____no_output_____"
]
],
[
[
"#Find all negative review\nneg = df[df[\"sentiment\"] == -1].review\n\n#Reset the index\nneg.index = range(len(neg.index))\n\n## Write each DataFrame to separate txt\nfor i in range(len(neg)): \n data = neg[i]\n with open(str(i) + \".txt\",\"w\") as file:\n file.write(data + \"\\n\")",
"_____no_output_____"
]
],
[
[
"Next, we sort the order of the official data and remove all the content. In other words, we only keep the file name. ",
"_____no_output_____"
]
],
[
[
"import os\nimport pandas as pd\n\n#Get file names\nfile_names = os.listdir('/Users/zeyu/nltk_data/corpora/movie_reviews/neg')\n\n#Convert pandas\nneg_df = pd.DataFrame (file_names, columns = ['file_name'])\n\n#split to sort\nneg_df[['number','id']] = neg_df.file_name.apply(\n lambda x: pd.Series(str(x).split(\"_\")))\n\n#change the number to be the index\nneg_df_index = neg_df.set_index('number')\nneg_org = neg_df_index.sort_index(ascending=True)\n\n#del neg[\"id\"]\nneg_org.reset_index(inplace=True)\n\nneg_org = neg_org.drop([0], axis=0).reset_index(drop=True)\nneg_names = neg_org['file_name']\n\nfor file_name in neg_names:\n t = open(f'/Users/zeyu/nltk_data/corpora/movie_reviews/neg/{file_name}', 'w')\n t.write(\"\")\n t.close()",
"_____no_output_____"
]
],
[
[
"Next, we insert the content of amazon review to the official files with their original file names.",
"_____no_output_____"
]
],
[
[
"#Get file names\nfile_names = os.listdir('/Users/zeyu/Desktop/DS/neg')\n\n#Convert pandas\npos_df = pd.DataFrame (file_names, columns = ['file_name'])\n\npos_names = pos_df['file_name']\n\nfor index, file_name in enumerate(pos_names):\n try: \n\n t = open(f'/Users/zeyu/Desktop/DS/neg/{file_name}', 'r')\n # t.write(\"\")\n t_val = ascii(t.read())\n t.close()\n \n writefname = pos_names_org[index]\n t = open(f'/Users/zeyu/nltk_data/corpora/movie_reviews/neg/{writefname}', 'w')\n t.write(t_val)\n t.close()\n except:\n print(f'{index} Reading/writing Error')",
"_____no_output_____"
]
],
[
[
"Eventually, we can just run these few lines to predict the sentiments of Amazon product review.",
"_____no_output_____"
]
],
[
[
"import nltk\nfrom nltk.corpus import movie_reviews\nimport random\n\ndocuments = [(list(movie_reviews.words(fileid)), category)\n for category in movie_reviews.categories()\n for fileid in movie_reviews.fileids(category)]\n\n#All words, not unique. \nrandom.shuffle(documents)\n",
"_____no_output_____"
],
[
"#Change to lower case. Count word appears.\nall_words = nltk.FreqDist(w.lower() for w in movie_reviews.words())\n\n\n#Only show first 2000.\nword_features = list(all_words)[:2000]\n\n\ndef document_features(document):\n document_words = set(document)\n features = {}\n for word in word_features:\n features['contains({})'.format(word)] = (word in document_words)\n return features",
"_____no_output_____"
],
[
"#Calculate the accuracy of the given.\n\nfeaturesets = [(document_features(d), c) for (d,c) in documents]\ntrain_set, test_set = featuresets[100:], featuresets[:100]\nclassifier = nltk.NaiveBayesClassifier.train(train_set)\n\nprint(nltk.classify.accuracy(classifier, test_set))",
"_____no_output_____"
],
[
"classifier.show_most_informative_features(5)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d03fa4b1cd8d8b9fb81c510b4e50e33d872208d5 | 4,102 | ipynb | Jupyter Notebook | run/monitor-flir-service.ipynb | johnnewto/FLIR-pubsub | 71eec3a5bf8fe5959cf997f7f12d9cfe0f58d8fc | [
"Apache-2.0"
] | null | null | null | run/monitor-flir-service.ipynb | johnnewto/FLIR-pubsub | 71eec3a5bf8fe5959cf997f7f12d9cfe0f58d8fc | [
"Apache-2.0"
] | 6 | 2021-05-20T12:36:50.000Z | 2022-02-26T06:25:11.000Z | run/monitor-flir-service.ipynb | johnnewto/FLIR-pubsub | 71eec3a5bf8fe5959cf997f7f12d9cfe0f58d8fc | [
"Apache-2.0"
] | 1 | 2020-03-24T02:07:21.000Z | 2020-03-24T02:07:21.000Z | 25.798742 | 183 | 0.568016 | [
[
[
"### Install and monitor the FLIR camera service\nInstall",
"_____no_output_____"
]
],
[
[
"! sudo cp flir-server.service /etc/systemd/system/flir-server.service",
"_____no_output_____"
]
],
[
[
"Start the service",
"_____no_output_____"
]
],
[
[
"! sudo systemctl start flir-server.service",
"_____no_output_____"
]
],
[
[
"Stop the service",
"_____no_output_____"
]
],
[
[
"! sudo systemctl stop flir-server.service",
"\u001b[0;1;31mWarning:\u001b[0m The unit file, source configuration file or drop-ins of flir-server.service changed on disk. Run 'systemctl daemon-reload' to reload units.\r\n"
]
],
[
[
"Enable it so that it starts on boot",
"_____no_output_____"
]
],
[
[
"! sudo systemctl enable flir-server.service # enable at boot",
"_____no_output_____"
]
],
[
[
"Disable it so that it does not start on boot",
"_____no_output_____"
]
],
[
[
"! sudo systemctl enable flir-server.service # enable at boot",
"_____no_output_____"
]
],
[
[
"To show status of the service ",
"_____no_output_____"
]
],
[
[
"! sudo systemctl status flir-server.service\n",
"\u001b[0;1;32m●\u001b[0m flir-server.service - FLIR-camera server service\u001b[m\n Loaded: loaded (/etc/systemd/system/flir-server.service; enabled; vendor pres\u001b[m\n Active: \u001b[0;1;32mactive (running)\u001b[0m since Tue 2020-03-24 07:53:04 NZDT; 20min ago\u001b[m\n Main PID: 765 (python)\u001b[m\n Tasks: 17 (limit: 4915)\u001b[m\n CGroup: /system.slice/flir-server.service\u001b[m\n └─765 /home/rov/.virtualenvs/flir/bin/python -u run/flir-server.py\u001b[m\n\u001b[m\nMar 24 07:53:06 rov-UP python[765]: 19444715 - executing: \"Gain.SetValue(6)\"\u001b[m\nMar 24 07:53:06 rov-UP python[765]: 19444715 - executing: \"BlackLevelSelector.Se\u001b[m\nMar 24 07:53:06 rov-UP python[765]: 19444715 - executing: \"BlackLevel.SetValue(0\u001b[m\nMar 24 07:53:07 rov-UP python[765]: 19444715 - executing: \"GammaEnable.SetValue(\u001b[m\nMar 24 07:53:07 rov-UP python[765]: Starting : FrontLeft\u001b[m\nMar 24 07:53:17 rov-UP python[765]: Stopping FrontLeft due to inactivity.\u001b[m\nMar 24 07:54:12 rov-UP python[765]: Starting : FrontLeft\u001b[m\nMar 24 07:54:26 rov-UP python[765]: Stopping FrontLeft due to inactivity.\u001b[m\nMar 24 07:54:33 rov-UP python[765]: Starting : FrontLeft\u001b[m\nMar 24 07:54:48 rov-UP python[765]: Stopping FrontLeft due to inactivity.\u001b[m\n\u001b[K\u001b[?1l\u001b>1-18/18 (END)\u001b[m\u001b[K\u0007"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d03fa74764df0377c4c01df5dd598ac61905798c | 255,859 | ipynb | Jupyter Notebook | yolodemo.ipynb | 240897/ResearchProjectT2 | 22b6efb6586a82f4cc6cb84e0929e64503288894 | [
"Unlicense"
] | null | null | null | yolodemo.ipynb | 240897/ResearchProjectT2 | 22b6efb6586a82f4cc6cb84e0929e64503288894 | [
"Unlicense"
] | null | null | null | yolodemo.ipynb | 240897/ResearchProjectT2 | 22b6efb6586a82f4cc6cb84e0929e64503288894 | [
"Unlicense"
] | null | null | null | 471.195212 | 175,038 | 0.937821 | [
[
[
"!pip install opencv-python",
"Requirement already satisfied: opencv-python in /usr/local/lib/python3.7/dist-packages (4.1.2.30)\nRequirement already satisfied: numpy>=1.14.5 in /usr/local/lib/python3.7/dist-packages (from opencv-python) (1.19.5)\n"
],
[
"import cv2\nimport matplotlib.pyplot as plt\nimport numpy as np\n",
"_____no_output_____"
],
[
"image= cv2.imread(\"/content/team-image2.jpg\")",
"_____no_output_____"
],
[
"type(image)",
"_____no_output_____"
],
[
"image.shape",
"_____no_output_____"
],
[
"image.size",
"_____no_output_____"
],
[
"plt.imshow(image)",
"_____no_output_____"
],
[
"new_image= cv2.cvtColor(image, cv2.COLOR_BGR2RGB)",
"_____no_output_____"
],
[
"plt.imshow(new_image)",
"_____no_output_____"
],
[
"r,g,b=cv2.split(new_image)",
"_____no_output_____"
],
[
"yolo = cv2.dnn.readNet(\"/content/yolov3-tiny.weights\",\"/content/yolov2-tiny.cfg\")",
"_____no_output_____"
],
[
"classes=[]\n\nwith open(\"/content/coco.names\", 'r') as f:\n classes=f.read().splitlines()",
"_____no_output_____"
],
[
"len(classes)",
"_____no_output_____"
],
[
"image= cv2.imread(\"/content/team-image2.jpg\")",
"_____no_output_____"
],
[
"blob=cv2.dnn.blobFromImage(image, 1/225,(100,100),(0,0,0),swapRB=True,crop=False)",
"_____no_output_____"
],
[
"blob.shape",
"_____no_output_____"
],
[
"yolo.setInput(blob)",
"_____no_output_____"
],
[
"output_layes=yolo.getUnconnectedOutLayersNames()\nlayer_oupt=yolo.forward(output_layes)",
"_____no_output_____"
],
[
"boxes=[]\nconfidences=[]\nclass_ids=[]\n\nfor output in layer_oupt:\n for detection in output:\n score=detection[5:]\n class_id=np.argmax(score)\n confidence=score[class_ids]\n if confidence > 0.7:\n center_x= int(detection[0]*width)\n center_y= int(detection[0]*height)\n w= int(detection[0]*width)\n h= int(detection[0]*height)\n\n x=int(center_x-w/2)\n y=int(center_y-h/2)\n\n\n boxes.append([x,y,w,h])\n confidences.append(float(confidence))\n class_ids.append(class_id)\n",
"/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:10: DeprecationWarning: The truth value of an empty array is ambiguous. Returning False, but in future this will result in an error. Use `array.size > 0` to check that an array is not empty.\n # Remove the CWD from sys.path while we load stuff.\n"
],
[
"len(boxes)",
"_____no_output_____"
],
[
"indexes= cv2.dnn.NMSBoxes(boxes,confidences,0.5,0.4)",
"_____no_output_____"
],
[
"font=cv2.FONT_HERSHEY_PLAIN\ncolors=np.random.uniform(0,225,size=(len(boxes),3))",
"_____no_output_____"
],
[
"len(indexes)",
"_____no_output_____"
],
[
"for i in indexes.flatten():\n x,y,w,h=boxes=[i]\n label=str(classes[class_ids[i]])\n confi=str(round(confidences[i],2))\n color=colors[i]\n\n cv2.rectangle(image,(x,y),(x+w,y+h),color,2)\n cv2.putText(image,label +\"\"+confi,(x+y+20),font,2,(225,225,225),2)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03fb9aad7b0fcf16ab7768133c1e5714bc16dad | 125,807 | ipynb | Jupyter Notebook | motif_miner_comparison/motif_miner_performance.ipynb | andrewguy/CCARL | 0afda67bcc58be2b6b6bf426cccaab04453c0590 | [
"MIT"
] | 2 | 2020-05-13T12:50:44.000Z | 2020-07-27T08:32:42.000Z | motif_miner_comparison/motif_miner_performance.ipynb | andrewguy/CCARL | 0afda67bcc58be2b6b6bf426cccaab04453c0590 | [
"MIT"
] | 1 | 2020-04-30T15:33:45.000Z | 2021-11-30T01:53:18.000Z | motif_miner_comparison/motif_miner_performance.ipynb | andrewguy/CCARL | 0afda67bcc58be2b6b6bf426cccaab04453c0590 | [
"MIT"
] | 2 | 2020-12-05T00:25:43.000Z | 2022-02-10T13:58:35.000Z | 170.009459 | 47,964 | 0.878671 | [
[
[
"### Analysis of motifs using Motif Miner (RINGS tool that employs alpha frequent subtree mining)",
"_____no_output_____"
]
],
[
[
"csv_files = [\"ABA_14361_100ug_v5.0_DATA.csv\",\n \"ConA_13799-10ug_V5.0_DATA.csv\",\n 'PNA_14030_10ug_v5.0_DATA.csv',\n \"RCAI_10ug_14110_v5.0_DATA.csv\",\n \"PHA-E-10ug_13853_V5.0_DATA.csv\",\n \"PHA-L-10ug_13856_V5.0_DATA.csv\",\n \"LCA_10ug_13934_v5.0_DATA.csv\",\n \"SNA_10ug_13631_v5.0_DATA.csv\",\n \"MAL-I_10ug_13883_v5.0_DATA.csv\",\n \"MAL_II_10ug_13886_v5.0_DATA.csv\",\n \"GSL-I-B4_10ug_13920_v5.0_DATA.csv\",\n \"jacalin-1ug_14301_v5.0_DATA.csv\",\n 'WGA_14057_1ug_v5.0_DATA.csv',\n \"UEAI_100ug_13806_v5.0_DATA.csv\",\n \"SBA_14042_10ug_v5.0_DATA.csv\",\n \"DBA_100ug_13897_v5.0_DATA.csv\",\n \"PSA_14040_10ug_v5.0_DATA.csv\",\n \"HA_PuertoRico_8_34_13829_v5_DATA.csv\",\n 'H3N8-HA_16686_v5.1_DATA.csv',\n \"Human-DC-Sign-tetramer_15320_v5.0_DATA.csv\"]\n\ncsv_file_normal_names = [\n r\"\\textit{Agaricus bisporus} agglutinin (ABA)\",\n r\"Concanavalin A (Con A)\",\n r'Peanut agglutinin (PNA)',\n r\"\\textit{Ricinus communis} agglutinin I (RCA I/RCA\\textsubscript{120})\",\n r\"\\textit{Phaseolus vulgaris} erythroagglutinin (PHA-E)\",\n r\"\\textit{Phaseolus vulgaris} leucoagglutinin (PHA-L)\",\n r\"\\textit{Lens culinaris} agglutinin (LCA)\",\n r\"\\textit{Sambucus nigra} agglutinin (SNA)\",\n r\"\\textit{Maackia amurensis} lectin I (MAL-I)\",\n r\"\\textit{Maackia amurensis} lectin II (MAL-II)\",\n r\"\\textit{Griffonia simplicifolia} Lectin I isolectin B\\textsubscript{4} (GSL I-B\\textsubscript{4})\",\n r\"Jacalin\",\n r'Wheat germ agglutinin (WGA)',\n r\"\\textit{Ulex europaeus} agglutinin I (UEA I)\",\n r\"Soybean agglutinin (SBA)\",\n r\"\\textit{Dolichos biflorus} agglutinin (DBA)\",\n r\"\\textit{Pisum sativum} agglutinin (PSA)\",\n r\"Influenza hemagglutinin (HA) (A/Puerto Rico/8/34) (H1N1)\",\n r'Influenza HA (A/harbor seal/Massachusetts/1/2011) (H3N8)',\n r\"Human DC-SIGN tetramer\"]",
"_____no_output_____"
],
[
"import sys\nimport os\nimport pandas as pd\nimport numpy as np\nfrom scipy import interp\n\nsys.path.append('..')\n\nfrom ccarl.glycan_parsers.conversions import kcf_to_digraph, cfg_to_kcf\nfrom ccarl.glycan_plotting import draw_glycan_diagram\nfrom ccarl.glycan_graph_methods import generate_digraph_from_glycan_string\nfrom ccarl.glycan_features import generate_features_from_subtrees\nimport ccarl.glycan_plotting\n\nfrom sklearn.linear_model import LogisticRegressionCV, LogisticRegression\nfrom sklearn.metrics import matthews_corrcoef, make_scorer, roc_curve, auc\n\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"from collections import defaultdict\n\naucs = defaultdict(list)\nys = defaultdict(list)\nprobs = defaultdict(list)\nmotifs = defaultdict(list)\n\n\nfor fold in [1,2,3,4,5]:\n print(f\"Running fold {fold}...\")\n for csv_file in csv_files:\n alpha = 0.8\n minsup = 0.2\n input_file = f'./temp_{csv_file}'\n training_data = pd.read_csv(f\"../Data/CV_Folds/fold_{fold}/training_set_{csv_file}\")\n test_data = pd.read_csv(f\"../Data/CV_Folds/fold_{fold}/test_set_{csv_file}\")\n pos_glycan_set = training_data['glycan'][training_data.binding == 1].values\n kcf_string = '\\n'.join([cfg_to_kcf(x) for x in pos_glycan_set])\n with open(input_file, 'w') as f:\n f.write(kcf_string)\n min_sup = int(len(pos_glycan_set) * minsup)\n subtrees = os.popen(f\"ruby Miner_cmd.rb {min_sup} {alpha} {input_file}\").read()\n subtree_graphs = [kcf_to_digraph(x) for x in subtrees.split(\"///\")[0:-1]]\n motifs[csv_file].append(subtree_graphs)\n\n os.remove(input_file)\n\n binding_class = training_data.binding.values\n glycan_graphs = [generate_digraph_from_glycan_string(x, parse_linker=True,\n format='CFG')\n for x in training_data.glycan]\n\n glycan_graphs_test = [generate_digraph_from_glycan_string(x, parse_linker=True,\n format='CFG')\n for x in test_data.glycan]\n\n features = [generate_features_from_subtrees(subtree_graphs, glycan) for \n glycan in glycan_graphs]\n\n features_test = [generate_features_from_subtrees(subtree_graphs, glycan) for \n glycan in glycan_graphs_test]\n\n logistic_clf = LogisticRegression(penalty='l2', C=100, solver='lbfgs',\n class_weight='balanced', max_iter=1000)\n\n X = features\n y = binding_class\n logistic_clf.fit(X, y)\n\n y_test = test_data.binding.values\n X_test = features_test\n fpr, tpr, _ = roc_curve(y_test, logistic_clf.predict_proba(X_test)[:,1], drop_intermediate=False)\n aucs[csv_file].append(auc(fpr, tpr))\n ys[csv_file].append(y_test)\n probs[csv_file].append(logistic_clf.predict_proba(X_test)[:,1])",
"Running fold 1...\nRunning fold 2...\nRunning fold 3...\nRunning fold 4...\nRunning fold 5...\n"
],
[
"# Assess the number of subtrees generated for each CV round.\n\nsubtree_lengths = defaultdict(list)\n\nfor fold in [1,2,3,4,5]:\n print(f\"Running fold {fold}...\")\n for csv_file in csv_files:\n alpha = 0.8\n minsup = 0.2\n input_file = f'./temp_{csv_file}'\n training_data = pd.read_csv(f\"../Data/CV_Folds/fold_{fold}/training_set_{csv_file}\")\n test_data = pd.read_csv(f\"../Data/CV_Folds/fold_{fold}/test_set_{csv_file}\")\n pos_glycan_set = training_data['glycan'][training_data.binding == 1].values\n kcf_string = '\\n'.join([cfg_to_kcf(x) for x in pos_glycan_set])\n with open(input_file, 'w') as f:\n f.write(kcf_string)\n min_sup = int(len(pos_glycan_set) * minsup)\n subtrees = os.popen(f\"ruby Miner_cmd.rb {min_sup} {alpha} {input_file}\").read()\n subtree_graphs = [kcf_to_digraph(x) for x in subtrees.split(\"///\")[0:-1]]\n subtree_lengths[csv_file].append(len(subtree_graphs))\n os.remove(input_file)",
"Running fold 1...\nRunning fold 2...\nRunning fold 3...\nRunning fold 4...\nRunning fold 5...\n"
],
[
"subtree_lengths = [y for x in subtree_lengths.values() for y in x]\nprint(np.mean(subtree_lengths))\nprint(np.max(subtree_lengths))\nprint(np.min(subtree_lengths))",
"12.59\n24\n7\n"
],
[
"def plot_multiple_roc(data):\n '''Plot multiple ROC curves.\n \n Prints out key AUC values (mean, median etc).\n \n Args:\n data (list): A list containing [y, probs] for each model, where:\n y: True class labels\n probs: Predicted probabilities\n Returns:\n Figure, Axes, Figure, Axes\n '''\n mean_fpr = np.linspace(0, 1, 100)\n\n fig, axes = plt.subplots(figsize=(4, 4))\n\n ax = axes\n ax.set_title('')\n #ax.legend(loc=\"lower right\")\n ax.set_xlabel('False Positive Rate')\n ax.set_ylabel('True Positive Rate')\n ax.set_aspect('equal', adjustable='box')\n auc_values = []\n tpr_list = []\n\n for y, probs in data:\n #data_point = data[csv_file]\n #y = data_point[7] # test binding\n #X = data_point[8] # test features\n #logistic_clf = data_point[0] # model\n fpr, tpr, _ = roc_curve(y, probs, drop_intermediate=False)\n tpr_list.append(interp(mean_fpr, fpr, tpr))\n auc_values.append(auc(fpr, tpr))\n ax.plot(fpr, tpr, color='blue', alpha=0.1, label=f'ROC curve (area = {auc(fpr, tpr): 2.3f})')\n ax.plot([0,1], [0,1], linestyle='--', color='grey', linewidth=0.8, dashes=(5, 10))\n\n mean_tpr = np.mean(tpr_list, axis=0)\n median_tpr = np.median(tpr_list, axis=0)\n upper_tpr = np.percentile(tpr_list, 75, axis=0)\n lower_tpr = np.percentile(tpr_list, 25, axis=0)\n ax.plot(mean_fpr, median_tpr, color='black')\n\n ax.fill_between(mean_fpr, lower_tpr, upper_tpr, color='grey', alpha=.5,\n label=r'$\\pm$ 1 std. dev.')\n fig.savefig(\"Motif_Miner_CV_ROC_plot_all_curves.svg\")\n\n fig2, ax2 = plt.subplots(figsize=(4, 4))\n ax2.hist(auc_values, range=[0.5,1], bins=10, rwidth=0.9, color=(0, 114/255, 178/255))\n ax2.set_xlabel(\"AUC value\")\n ax2.set_ylabel(\"Counts\")\n fig2.savefig(\"Motif_Miner_CV_AUC_histogram.svg\")\n\n print(f\"Mean AUC value: {np.mean(auc_values): 1.3f}\")\n\n print(f\"Median AUC value: {np.median(auc_values): 1.3f}\")\n print(f\"IQR of AUC values: {np.percentile(auc_values, 25): 1.3f} - {np.percentile(auc_values, 75): 1.3f}\")\n return fig, axes, fig2, ax2, auc_values\n",
"_____no_output_____"
],
[
"# Plot ROC curves for all test sets\nroc_data = [[y, prob] for y_fold, prob_fold in zip(ys.values(), probs.values()) for y, prob in zip(y_fold, prob_fold)]\n_, _, _, _, auc_values = plot_multiple_roc(roc_data)",
"Mean AUC value: 0.866\nMedian AUC value: 0.874\nIQR of AUC values: 0.825 - 0.940\n"
],
[
"auc_values_ccarl = [0.950268817204301,\n 0.9586693548387097,\n 0.9559811827956988,\n 0.8686155913978494,\n 0.9351222826086956,\n 0.989010989010989,\n 0.9912587412587414,\n 0.9090909090909092,\n 0.9762626262626264,\n 0.9883597883597884,\n 0.9065533980582524,\n 0.9417475728155339,\n 0.8268608414239482,\n 0.964349376114082,\n 0.9322638146167558,\n 0.9178037686809616,\n 0.96361273554256,\n 0.9362139917695472,\n 0.9958847736625515,\n 0.9526748971193415,\n 0.952300785634119,\n 0.9315375982042648,\n 0.9705387205387206,\n 0.9865319865319865,\n 0.9849773242630385,\n 0.9862385321100917,\n 0.9862385321100918,\n 0.9606481481481481,\n 0.662037037037037,\n 0.7796296296296297,\n 0.9068627450980392,\n 0.915032679738562,\n 0.9820261437908496,\n 0.9893790849673203,\n 0.9882988298829882,\n 0.9814814814814815,\n 1.0,\n 0.8439153439153441,\n 0.9859813084112149,\n 0.9953271028037383,\n 0.8393308080808081,\n 0.8273358585858586,\n 0.7954545454545453,\n 0.807070707070707,\n 0.8966329966329966,\n 0.8380952380952381,\n 0.6201058201058202,\n 0.7179894179894181,\n 0.6778846153846154,\n 0.75,\n 0.9356060606060607,\n 0.8619528619528619,\n 0.8787878787878789,\n 0.9040816326530613,\n 0.7551020408163266,\n 0.9428694158075602,\n 0.9226804123711341,\n 0.8711340206185567,\n 0.7840909090909091,\n 0.8877840909090909,\n 0.903225806451613,\n 0.8705594120049,\n 0.9091465904450796,\n 0.8816455696202531,\n 0.8521097046413502,\n 0.8964521452145213,\n 0.9294554455445544,\n 0.8271452145214522,\n 0.8027272727272727,\n 0.8395454545454546,\n 0.8729967948717949,\n 0.9306891025641025,\n 0.9550970873786407,\n 0.7934686672550749,\n 0.8243601059135041,\n 0.8142100617828772,\n 0.9179611650485436,\n 0.8315533980582525,\n 0.7266990291262136,\n 0.9038834951456312,\n 0.9208916083916084,\n 0.7875,\n 0.9341346153846154,\n 0.9019230769230768,\n 0.9086538461538461,\n 0.9929245283018868,\n 0.9115566037735848,\n 0.9952830188679246,\n 0.9658018867924528,\n 0.7169811320754716,\n 0.935981308411215,\n 0.9405660377358491,\n 0.9905660377358491,\n 0.9937106918238994,\n 0.9302935010482181,\n 0.7564814814814815,\n 0.9375,\n 0.8449074074074074,\n 0.8668981481481483,\n 0.7978971962616823]",
"_____no_output_____"
],
[
"auc_value_means = [np.mean(auc_values[x*5:x*5+5]) for x in range(int(len(auc_values) / 5))]\nauc_value_means_ccarl = [np.mean(auc_values_ccarl[x*5:x*5+5]) for x in range(int(len(auc_values_ccarl) / 5))]",
"_____no_output_____"
],
[
"auc_value_mean_glymmr = np.array([0.6067939 , 0.76044574, 0.66786624, 0.69578298, 0.81659623,\n 0.80536403, 0.77231548, 0.96195032, 0.70013384, 0.60017685,\n 0.77336818, 0.78193305, 0.66269668, 0.70333122, 0.54247748,\n 0.63003707, 0.79619231, 0.85141509, 0.9245296 , 0.63366329])\n\nauc_value_mean_glymmr_best = np.array([0.77559242, 0.87452658, 0.75091636, 0.7511371 , 0.87450697,\n 0.82895628, 0.81083123, 0.96317065, 0.75810185, 0.82680149,\n 0.84747054, 0.8039597 , 0.69651882, 0.73431593, 0.582194 ,\n 0.67407767, 0.83049825, 0.88891509, 0.9345188 , 0.72702016])\n\nauc_value_motiffinder = [0.9047619047619048, 0.9365601503759399, 0.6165413533834586, 0.9089068825910931,\n 0.4962962962962963, 0.6358816964285713, 0.8321078431372548, 0.8196576151121606, 0.8725400457665904,\n 0.830220713073005, 0.875, 0.7256367663344407, 0.8169291338582677, 0.9506818181818182, 0.7751351351351351,\n 0.9362947658402204, 0.6938461538461539, 0.6428571428571428, 0.7168021680216802, 0.5381136950904392] #Note, only from a single test-train split.",
"_____no_output_____"
],
[
"import seaborn as sns\nsns.set(style=\"ticks\")\nplot_data = np.array([auc_value_mean_glymmr, auc_value_mean_glymmr_best, auc_value_motiffinder, auc_value_means, auc_value_means_ccarl]).T\nax = sns.violinplot(data=plot_data, cut=2, inner='quartile')\nsns.swarmplot(data=plot_data, color='black')\nax.set_ylim([0.5, 1.05])\nax.set_xticklabels([\"GLYMMR\\n(mean)\", \"GLYMMR\\n(best)\", \"MotifFinder\", \"Glycan\\nMiner Tool\", \"CCARL\"])\n#ax.grid('off')\nax.set_ylabel(\"AUC\")\nax.figure.savefig('method_comparison_violin_plot.svg')",
"_____no_output_____"
],
[
"auc_value_means_ccarl",
"_____no_output_____"
],
[
"print(\"CCARL Performance\")\nprint(f\"Median AUC value: {np.median(auc_value_means_ccarl): 1.3f}\")\nprint(f\"IQR of AUC values: {np.percentile(auc_value_means_ccarl, 25): 1.3f} - {np.percentile(auc_value_means_ccarl, 75): 1.3f}\")",
"CCARL Performance\nMedian AUC value: 0.887\nIQR of AUC values: 0.865 - 0.954\n"
],
[
"print(\"Glycan Miner Tool Performance\")\nprint(f\"Median AUC value: {np.median(auc_value_means): 1.3f}\")\nprint(f\"IQR of AUC values: {np.percentile(auc_value_means, 25): 1.3f} - {np.percentile(auc_value_means, 75): 1.3f}\")",
"Glycan Miner Tool Performance\nMedian AUC value: 0.862\nIQR of AUC values: 0.845 - 0.898\n"
],
[
"print(\"Glycan Miner Tool Performance\")\nprint(f\"Median AUC value: {np.median(auc_value_mean_glymmr_best): 1.3f}\")\nprint(f\"IQR of AUC values: {np.percentile(auc_value_mean_glymmr_best, 25): 1.3f} - {np.percentile(auc_value_mean_glymmr_best, 75): 1.3f}\")",
"Glycan Miner Tool Performance\nMedian AUC value: 0.807\nIQR of AUC values: 0.747 - 0.854\n"
],
[
"print(\"Glycan Miner Tool Performance\")\nprint(f\"Median AUC value: {np.median(auc_value_mean_glymmr): 1.3f}\")\nprint(f\"IQR of AUC values: {np.percentile(auc_value_mean_glymmr, 25): 1.3f} - {np.percentile(auc_value_mean_glymmr, 75): 1.3f}\")",
"Glycan Miner Tool Performance\nMedian AUC value: 0.732\nIQR of AUC values: 0.655 - 0.798\n"
],
[
"from matplotlib.backends.backend_pdf import PdfPages\nsns.reset_orig()\nimport networkx as nx\n\nfor csv_file in csv_files:\n with PdfPages(f\"./motif_miner_motifs/glycan_motif_miner_motifs_{csv_file}.pdf\") as pdf:\n for motif in motifs[csv_file][0]:\n fig, ax = plt.subplots()\n ccarl.glycan_plotting.draw_glycan_diagram(motif, ax)\n pdf.savefig(fig)\n plt.close(fig)",
"_____no_output_____"
],
[
"\nglymmr_mean_stdev = np.array([0.15108904, 0.08300011, 0.11558078, 0.05259819, 0.061275 ,\n 0.09541182, 0.09239553, 0.05114523, 0.05406571, 0.16180131,\n 0.10345311, 0.06080207, 0.0479003 , 0.09898648, 0.06137992,\n 0.09813596, 0.07010635, 0.14010784, 0.05924527, 0.13165457])\n\nglymmr_best_stdev = np.array([0.08808868, 0.04784959, 0.13252895, 0.03163248, 0.04401516,\n 0.08942411, 0.08344247, 0.05714308, 0.05716086, 0.05640053,\n 0.08649275, 0.05007289, 0.05452531, 0.05697662, 0.0490626 ,\n 0.1264917 , 0.04994508, 0.1030053 , 0.03359648, 0.12479809])\n\nauc_value_std_ccarl = [np.std(auc_values_ccarl[x*5:x*5+5]) for x in range(int(len(auc_values_ccarl) / 5))]\n \nprint(r\"Lectin & GLYMMR(mean) & GLYMMR(best) & Glycan Miner Tool & MotifFinder & CCARL \\\\ \\hline\")\nfor i, csv_file, name in zip(list(range(len(csv_files))), csv_files, csv_file_normal_names):\n print(f\"{name} & {auc_value_mean_glymmr[i]:0.3f} ({glymmr_mean_stdev[i]:0.3f}) & {auc_value_mean_glymmr_best[i]:0.3f} ({glymmr_best_stdev[i]:0.3f}) \\\n& {np.mean(aucs[csv_file]):0.3f} ({np.std(aucs[csv_file]):0.3f}) & {auc_value_motiffinder[i]:0.3f} & {auc_value_means_ccarl[i]:0.3f} ({auc_value_std_ccarl[i]:0.3f}) \\\\\\\\\") ",
"Lectin & GLYMMR(mean) & GLYMMR(best) & Glycan Miner Tool & MotifFinder & CCARL \\\\ \\hline\n\\textit{Agaricus bisporus} agglutinin (ABA) & 0.607 (0.151) & 0.776 (0.088) & 0.888 (0.067) & 0.905 & 0.934 (0.034) \\\\\nConcanavalin A (Con A) & 0.760 (0.083) & 0.875 (0.048) & 0.951 (0.042) & 0.937 & 0.971 (0.031) \\\\\nPeanut agglutinin (PNA) & 0.668 (0.116) & 0.751 (0.133) & 0.894 (0.041) & 0.617 & 0.914 (0.048) \\\\\n\\textit{Ricinus communis} agglutinin I (RCA I/RCA\\textsubscript{120}) & 0.696 (0.053) & 0.751 (0.032) & 0.848 (0.034) & 0.909 & 0.953 (0.026) \\\\\n\\textit{Phaseolus vulgaris} erythroagglutinin (PHA-E) & 0.817 (0.061) & 0.875 (0.044) & 0.910 (0.016) & 0.496 & 0.965 (0.021) \\\\\n\\textit{Phaseolus vulgaris} leucoagglutinin (PHA-L) & 0.805 (0.095) & 0.829 (0.089) & 0.858 (0.110) & 0.636 & 0.875 (0.132) \\\\\n\\textit{Lens culinaris} agglutinin (LCA) & 0.772 (0.092) & 0.811 (0.083) & 0.908 (0.083) & 0.832 & 0.956 (0.037) \\\\\n\\textit{Sambucus nigra} agglutinin (SNA) & 0.962 (0.051) & 0.963 (0.057) & 0.962 (0.050) & 0.820 & 0.961 (0.059) \\\\\n\\textit{Maackia amurensis} lectin I (MAL-I) & 0.700 (0.054) & 0.758 (0.057) & 0.868 (0.050) & 0.873 & 0.833 (0.035) \\\\\n\\textit{Maackia amurensis} lectin II (MAL-II) & 0.600 (0.162) & 0.827 (0.056) & 0.850 (0.091) & 0.830 & 0.721 (0.073) \\\\\n\\textit{Griffonia simplicifolia} Lectin I isolectin B\\textsubscript{4} (GSL I-B\\textsubscript{4}) & 0.773 (0.103) & 0.847 (0.086) & 0.875 (0.066) & 0.875 & 0.867 (0.061) \\\\\nJacalin & 0.782 (0.061) & 0.804 (0.050) & 0.848 (0.026) & 0.726 & 0.882 (0.055) \\\\\nWheat germ agglutinin (WGA) & 0.663 (0.048) & 0.697 (0.055) & 0.831 (0.034) & 0.817 & 0.883 (0.021) \\\\\n\\textit{Ulex europaeus} agglutinin I (UEA I) & 0.703 (0.099) & 0.734 (0.057) & 0.866 (0.023) & 0.951 & 0.859 (0.047) \\\\\nSoybean agglutinin (SBA) & 0.542 (0.061) & 0.582 (0.049) & 0.781 (0.046) & 0.775 & 0.875 (0.061) \\\\\n\\textit{Dolichos biflorus} agglutinin (DBA) & 0.630 (0.098) & 0.674 (0.126) & 0.722 (0.083) & 0.936 & 0.839 (0.069) \\\\\n\\textit{Pisum sativum} agglutinin (PSA) & 0.796 (0.070) & 0.830 (0.050) & 0.858 (0.064) & 0.694 & 0.891 (0.053) \\\\\nInfluenza hemagglutinin (HA) (A/Puerto Rico/8/34) (H1N1) & 0.851 (0.140) & 0.889 (0.103) & 0.838 (0.144) & 0.643 & 0.917 (0.104) \\\\\nInfluenza HA (A/harbor seal/Massachusetts/1/2011) (H3N8) & 0.925 (0.059) & 0.935 (0.034) & 0.947 (0.021) & 0.717 & 0.958 (0.028) \\\\\nHuman DC-SIGN tetramer & 0.634 (0.132) & 0.727 (0.125) & 0.823 (0.130) & 0.538 & 0.841 (0.062) \\\\\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03fc058d825f851b6ab42a3d7ea9a4145d153be | 7,757 | ipynb | Jupyter Notebook | src/analyze.ipynb | hcrrch/SimpleHTR-learn | ba31112bf30d88559a1eae0ce7ea62f349ac8e2f | [
"MIT"
] | null | null | null | src/analyze.ipynb | hcrrch/SimpleHTR-learn | ba31112bf30d88559a1eae0ce7ea62f349ac8e2f | [
"MIT"
] | null | null | null | src/analyze.ipynb | hcrrch/SimpleHTR-learn | ba31112bf30d88559a1eae0ce7ea62f349ac8e2f | [
"MIT"
] | null | null | null | 31.921811 | 141 | 0.585407 | [
[
[
"from __future__ import division\nfrom __future__ import print_function",
"_____no_output_____"
],
[
"import sys\nimport math\nimport pickle\nimport copy\nimport numpy as np\nimport cv2\nimport matplotlib.pyplot as plt\nfrom DataLoader import Batch\nfrom Model import Model, DecoderType\nfrom SamplePreprocessor import preprocess",
"_____no_output_____"
],
[
"# constants like filepaths\nclass Constants:\n\t\"filenames and paths to data\"\n\tfnCharList = '../model/charList.txt'\n\tfnAnalyze = '../data/analyze.png'\n\tfnPixelRelevance = '../data/pixelRelevance.npy'\n\tfnTranslationInvariance = '../data/translationInvariance.npy'\n\tfnTranslationInvarianceTexts = '../data/translationInvarianceTexts.pickle'\n\tgtText = 'are'\n\tdistribution = 'histogram' # 'histogram' or 'uniform'",
"_____no_output_____"
],
[
"def odds(val):\n\treturn val / (1 - val)",
"_____no_output_____"
],
[
"def weightOfEvidence(origProb, margProb):\n\treturn math.log2(odds(origProb)) - math.log2(odds(margProb))\n\t\n\t",
"_____no_output_____"
],
[
"def analyzePixelRelevance():\n\t\"simplified implementation of paper: Zintgraf et al - Visualizing Deep Neural Network Decisions: Prediction Difference Analysis\"\n\t\n\t# setup model\n\tmodel = Model(open(Constants.fnCharList).read(), DecoderType.BestPath, mustRestore=True)\n\t\n\t# read image and specify ground-truth text\n\timg = cv2.imread(Constants.fnAnalyze, cv2.IMREAD_GRAYSCALE)\n\t(w, h) = img.shape\n\tassert Model.imgSize[1] == w\n\t\n\t# compute probability of gt text in original image\n\tbatch = Batch([Constants.gtText], [preprocess(img, Model.imgSize)])\n\t(_, probs) = model.inferBatch(batch, calcProbability=True, probabilityOfGT=True)\n\torigProb = probs[0]\n\t\n\tgrayValues = [0, 63, 127, 191, 255]\n\tif Constants.distribution == 'histogram':\n\t\tbins = [0, 31, 95, 159, 223, 255]\n\t\t(hist, _) = np.histogram(img, bins=bins)\n\t\tpixelProb = hist / sum(hist)\n\telif Constants.distribution == 'uniform':\n\t\tpixelProb = [1.0 / len(grayValues) for _ in grayValues]\n\telse:\n\t\traise Exception('unknown value for Constants.distribution')\n\t\n\t# iterate over all pixels in image\n\tpixelRelevance = np.zeros(img.shape, np.float32)\n\tfor x in range(w):\n\t\tfor y in range(h):\n\t\t\t\n\t\t\t# try a subset of possible grayvalues of pixel (x,y)\n\t\t\timgsMarginalized = []\n\t\t\tfor g in grayValues:\n\t\t\t\timgChanged = copy.deepcopy(img)\n\t\t\t\timgChanged[x, y] = g\n\t\t\t\timgsMarginalized.append(preprocess(imgChanged, Model.imgSize))\n\n\t\t\t# put them all into one batch\n\t\t\tbatch = Batch([Constants.gtText]*len(imgsMarginalized), imgsMarginalized)\n\t\t\t\n\t\t\t# compute probabilities\n\t\t\t(_, probs) = model.inferBatch(batch, calcProbability=True, probabilityOfGT=True)\n\t\t\t\n\t\t\t# marginalize over pixel value (assume uniform distribution)\n\t\t\tmargProb = sum([probs[i] * pixelProb[i] for i in range(len(grayValues))])\n\t\t\t\n\t\t\tpixelRelevance[x, y] = weightOfEvidence(origProb, margProb)\n\t\t\t\n\t\t\tprint(x, y, pixelRelevance[x, y], origProb, margProb)\n\t\t\t\n\tnp.save(Constants.fnPixelRelevance, pixelRelevance)",
"_____no_output_____"
],
[
"def analyzeTranslationInvariance():\n\t# setup model\n\tmodel = Model(open(Constants.fnCharList).read(), DecoderType.BestPath, mustRestore=True)\n\t\n\t# read image and specify ground-truth text\n\timg = cv2.imread(Constants.fnAnalyze, cv2.IMREAD_GRAYSCALE)\n\t(w, h) = img.shape\n\tassert Model.imgSize[1] == w\n\t\n\timgList = []\n\tfor dy in range(Model.imgSize[0]-h+1):\n\t\ttargetImg = np.ones((Model.imgSize[1], Model.imgSize[0])) * 255\n\t\ttargetImg[:,dy:h+dy] = img\n\t\timgList.append(preprocess(targetImg, Model.imgSize))\n\t\n\t# put images and gt texts into batch\n\tbatch = Batch([Constants.gtText]*len(imgList), imgList)\n\t\n\t# compute probabilities\n\t(texts, probs) = model.inferBatch(batch, calcProbability=True, probabilityOfGT=True)\n\t\n\t# save results to file\n\tf = open(Constants.fnTranslationInvarianceTexts, 'wb')\n\tpickle.dump(texts, f)\n\tf.close()\n\tnp.save(Constants.fnTranslationInvariance, probs)",
"_____no_output_____"
],
[
"def showResults():\n\t# 1. pixel relevance\n\tpixelRelevance = np.load(Constants.fnPixelRelevance)\n\tplt.figure('Pixel relevance')\n\t\n\tplt.imshow(pixelRelevance, cmap=plt.cm.jet, vmin=-0.25, vmax=0.25)\n\tplt.colorbar()\n\t\n\timg = cv2.imread(Constants.fnAnalyze, cv2.IMREAD_GRAYSCALE)\n\tplt.imshow(img, cmap=plt.cm.gray, alpha=.4)\n\t\n\n\t# 2. translation invariance\n\tprobs = np.load(Constants.fnTranslationInvariance)\n\tf = open(Constants.fnTranslationInvarianceTexts, 'rb')\n\ttexts = pickle.load(f)\n\ttexts = ['%d:'%i + texts[i] for i in range(len(texts))]\n\tf.close()\n\t\n\tplt.figure('Translation invariance')\n\t\n\tplt.plot(probs, 'o-')\n\tplt.xticks(np.arange(len(texts)), texts, rotation='vertical')\n\tplt.xlabel('horizontal translation and best path')\n\tplt.ylabel('text probability of \"%s\"'%Constants.gtText)\n\t\n\t# show both plots\n\tplt.show()",
"_____no_output_____"
],
[
"if __name__ == '__main__':\n\tif len(sys.argv)>1:\n\t\tif sys.argv[1]=='--relevance':\n\t\t\tprint('Analyze pixel relevance')\n\t\t\tanalyzePixelRelevance()\n\t\telif sys.argv[1]=='--invariance':\n\t\t\tprint('Analyze translation invariance')\n\t\t\tanalyzeTranslationInvariance()\n\telse:\n\t\tprint('Show results')\n\t\tshowResults()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03fc0bd39689f87a8eead2b6fc3ae56fe099590 | 3,065 | ipynb | Jupyter Notebook | pretrained-model/prepare-stt/benchmark-google-speech-malay-dataset.ipynb | ishine/malaya-speech | fd34afc7107af1656dff4b3201fa51dda54fde18 | [
"MIT"
] | 111 | 2020-08-31T04:58:54.000Z | 2022-03-29T15:44:18.000Z | pretrained-model/prepare-stt/benchmark-google-speech-malay-dataset.ipynb | ishine/malaya-speech | fd34afc7107af1656dff4b3201fa51dda54fde18 | [
"MIT"
] | 14 | 2020-12-16T07:27:22.000Z | 2022-03-15T17:39:01.000Z | pretrained-model/prepare-stt/benchmark-google-speech-malay-dataset.ipynb | ishine/malaya-speech | fd34afc7107af1656dff4b3201fa51dda54fde18 | [
"MIT"
] | 29 | 2021-02-09T08:57:15.000Z | 2022-03-12T14:09:19.000Z | 23.945313 | 277 | 0.448287 | [
[
[
"import json\n\nwith open('bahasa-asr-test.json') as fopen:\n data = json.load(fopen)\n \nX, Y = data['X'], data['Y']",
"_____no_output_____"
],
[
"import malaya_speech\nimport speech_recognition as sr\n\nr = sr.Recognizer()",
"_____no_output_____"
],
[
"import unicodedata\nimport re\nimport itertools\n\nvocabs = [\" \", \"a\", \"e\", \"n\", \"i\", \"t\", \"o\", \"u\", \"s\", \"k\", \"r\", \"l\", \"h\", \"d\", \"m\", \"g\", \"y\", \"b\", \"p\", \"w\", \"c\", \"f\", \"j\", \"v\", \"z\", \"0\", \"1\", \"x\", \"2\", \"q\", \"5\", \"3\", \"4\", \"6\", \"9\", \"8\", \"7\"]\n\ndef preprocessing_text(string):\n \n string = unicodedata.normalize('NFC', string.lower())\n string = ''.join([c if c in vocabs else ' ' for c in string])\n string = re.sub(r'[ ]+', ' ', string).strip()\n string = (\n ''.join(''.join(s)[:2] for _, s in itertools.groupby(string))\n )\n return string",
"_____no_output_____"
],
[
"from tqdm import tqdm\n\nwer, cer = [], []\nfor i in tqdm(range(len(X))):\n try:\n with sr.AudioFile(X[i]) as source:\n a = r.record(source)\n\n text = r.recognize_google(a, language = 'ms')\n text = preprocessing_text(text)\n \n wer.append(malaya_speech.metrics.calculate_wer(Y[i], text))\n cer.append(malaya_speech.metrics.calculate_cer(Y[i], text))\n except Exception as e:\n print(e)",
"100%|██████████| 780/780 [05:33<00:00, 2.34it/s]\n"
],
[
"import numpy as np\n\nnp.mean(wer), np.mean(cer)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d03fc747669193175910cd8a8fcfa8045c5ab8cf | 56,960 | ipynb | Jupyter Notebook | Consignas TP_RLC.ipynb | EmanuelDri/CalculadoraRLC | b295b79f291185f5af6b2ce64781ce1ed77357b9 | [
"MIT"
] | null | null | null | Consignas TP_RLC.ipynb | EmanuelDri/CalculadoraRLC | b295b79f291185f5af6b2ce64781ce1ed77357b9 | [
"MIT"
] | null | null | null | Consignas TP_RLC.ipynb | EmanuelDri/CalculadoraRLC | b295b79f291185f5af6b2ce64781ce1ed77357b9 | [
"MIT"
] | null | null | null | 128.577878 | 27,658 | 0.854178 | [
[
[
"# Descripción del TP",
"_____no_output_____"
],
[
"## Introducción\nEn este práctico se pide desarrollar un programa que calcule parámetros de interés para un circuito RLC serie. \nTomando como datos de entrada:\n* εmax\n* la frecuencia de la fuente en Hz\n* los valores de R, L y C: \n\nEl programa debe determinar: \n* Imax \n* la diferencia de potencial de cada componente \n* la potencia media\n* el factor Q\n* el factor de potencia (cosφ)\n* la impedancia compleja Z\n* La frecuencia de resonancia del sistema.\n* Cuál de las tres impedancias domina el comportamiento del circuito\n\nTambién debe representar gráficamente:\n * El diagrama de fasores de tensiones del circuito RLC (`función quiver`).\n * Las tensiones VL, VC, VR y ε superpuestas en el dominio del tiempo.\n * Las ecuaciones de la FEM y la corriente con los valores correspondientes a los datos de entrada.",
"_____no_output_____"
],
[
"## Requerimientos de la implementación\n1. El desarrollo debe realizarse empleando el entorno Google Collaboratory (BTW donde estás leyendo ahora), el cual está basado en el lenguaje de programación Python.\n2. Crear un nuevo notebook para este práctico. Cuando se entregue el práctico el notebook debe estar con visibilidad pública. \n * Durante el desarrollo debe compartirse con sbrulh@gmail.com, contediego13@gmail.com y emanueldri@gmail.com, con permiso de visualización y generar comentarios *(no edición)*\n * Para crear un notebook nuevo:\n>Archivo → Bloc de notas nuevo \n3. Cada cambio significativo debe guardarse fijando una revisión del archivo `Archivo → Guardar y fijar revisión`. \n4. Cada bloque de código debe de estar documentado mediante comentarios que expliquen cómo funciona. No es necesario comentar cada sentencia de programación, pero sí es necesario entender qué es lo que hacen.\n5. La implementación de cada funcionalidad del programa debe estar implementada en un bloque de código diferente.\n6. Cada bloque de código debe ir acompañado por un título introductorio a su funcionalidad (introduciendo un bloque de texto antes).\n7. El ingreso de datos para la ejecución del programa debe hacerse mediante un [formulario interactivo](https://colab.research.google.com/notebooks/forms.ipynb)\n8. Incluir un esquema del circuito, se puede dibujar en https://www.circuit-diagram.org/editor/#",
"_____no_output_____"
],
[
"#Tutorial\n",
"_____no_output_____"
],
[
"##Google Colaboratory (Colab) y Jupyter Notebook\nEl espacio en el que vamos a trabajar se llama Google Colaboratory, esta es una plataforma de cómputo en la nube. La forma que dispone para interactuar con ella es [Jupiter Notebook](https://jupyter.org/), que permite de manera interactiva escribir texto y ejecutar código en Python.",
"_____no_output_____"
],
[
"## Usando Markdown\nMarkdown son un conjunto de instrucciones mediante las cuales le indicamos a Jupyter Notebook qué formato darle al texto. ***Doble click en cualquier parte del texto de este notebook ver cómo está escrito usando markdown***.",
"_____no_output_____"
],
[
"## Trabajando en equipo\nPara trabajar colaborativamente, hacer click en el botón `Compartir` del panel superior.",
"_____no_output_____"
],
[
"# Programación en Python",
"_____no_output_____"
],
[
"##Importar librerías",
"_____no_output_____"
]
],
[
[
"#hola, soy un comentario\nimport numpy #importamos la librería de funciones matemáticas\nfrom matplotlib import pyplot #importamos la librería de gráficos cartesianos",
"_____no_output_____"
]
],
[
[
"## Trabajar con variables\nPara instanciar o asignarle valor a una variable usamos el signo \"=\"\nLas variables persisten en todo el notebook, se pueden usar en cualquier bloque de código una vez que son instanciadas",
"_____no_output_____"
]
],
[
[
"a=7 #creamos la variable \"a\" y le asignamos el valor 7\n#se puede destinar un bloque de código para ",
"_____no_output_____"
]
],
[
[
"En el bloque de abajo usamos la variable \"a\" de arriba",
"_____no_output_____"
]
],
[
[
"a+5 #realizamos una operación con la variable \"a\", debajo aparece el resultado de la operación",
"_____no_output_____"
]
],
[
[
"##Ingresar datos usando formularios\nLos formularios permiten capturar datos de usuario para usarlos posteriormente en la ejecución de código. \n* Para insertar un formulario se debe presionar el botón más acciones de celda (**⋮**) a la derecha de la celda de código. *No se debe usar `Insertar→Agregar campo de formulario` porque actualmente no funciona esta opción*. \n* Ejemplos del uso de formularios se pueden encontrar [aquí](https://colab.research.google.com/notebooks/forms.ipynb#scrollTo=_7gRpQLXQSID). \n* Desde (**⋮**) se puede ocultar el código asociado a un formulario.\n\n",
"_____no_output_____"
]
],
[
[
"#@title Ejemplos de formulario\nvalor_formulario_numerico = 5 #@param {type:\"number\"}\nvalor_formulario_slider = 16 #@param {type:\"slider\", min:0, max:100, step:1}\n",
"_____no_output_____"
]
],
[
[
"##Funciones matemáticas\n* Las funciones aritméticas se encuentran incluidas en Python mediante [operadores](https://www.aprendeprogramando.es/cursos-online/python/operadores-aritmeticos/operadores-aritmeticos)\n* Funciones adicionales como las trigonométricas se invocan desde la librería numpy. Una vez importada, la podemos usar mediante la sintaxis:\n>`numpy.nombre de la función`\n*[Tutorial de funciones de la librería numpy](https://www.interactivechaos.com/manual/tutorial-de-numpy/funciones-universales-trigonometricas)",
"_____no_output_____"
]
],
[
[
"#Ejemplos de uso de funciones matemáticas\n#La función print muestra en pantalla un mensaje suministrado por el programador,\n#el contenido de una variable o alguna operación más compleja con variables\n\nprint(f'Valor del formulario numerico: {valor_formulario_numerico}')\nprint(f'Valor del formulario numerico al cuadrado: {valor_formulario_numerico**2}')\nprint(f'Raiz cuadrada del formulario numerico al cuadrado: {numpy.sqrt(valor_formulario_numerico)}')\nprint(f'Seno del formulario slider al cuadrado: {numpy.sin(valor_formulario_slider)}')",
"Valor del formulario numerico: 5\nValor del formulario numerico al cuadrado: 25\nRaiz cuadrada del formulario numerico al cuadrado: 2.23606797749979\nSeno del formulario slider al cuadrado: -0.2879033166650653\n"
]
],
[
[
"##Generando gráficas\n",
"_____no_output_____"
]
],
[
[
"pi=numpy.pi #Obtenemos el valor de π desde numpy y lo guardamos en la variable pi\nt = [i for i in numpy.arange(0,2*pi,pi/1000)] #generamos un vector de 2000 elementos,\n#con valores entre 0 y 2π en pasos de π/1000\ncorriente=numpy.sin(t); #calculamos el seno de los valores de \"t\"\ntension=numpy.sin(t+numpy.ones(len(t))*pi/2); #calculamos el seno de los valores\n#de \"t\" y le sumamos un ángulo de fase de π/2\npyplot.plot(t,corriente,t,tension, '-') #generamos un gráfico de tensión y corriente\n#en función de t grafic\npyplot.xlabel(\"Eje X\") #damos nombre al eje X\npyplot.ylabel(\"Eje Y\") #damos nombre al eje Y\npyplot.title(\"Onda seno\") #agregamos el título al gráfico\npyplot.legend([\"Corriente\",\"Tensión\"])\npyplot.figure(dpi=100) #ajustamos la resolución de la figura generada\npyplot.show() #mostramos el gráfico cuando ya lo tenemos listo",
"_____no_output_____"
]
],
[
[
"### Mostrar vectores\nPara mostrar vectores usamos la funcíon quiver: \n `(origen, coordenadas X de las puntas, coordenadas Y de las flechas, color=colores de las flechas)`",
"_____no_output_____"
]
],
[
[
"V = numpy.array([[1,1],[-2,2],[4,-7]])\norigin = [0,0,0], [0,0,0] # origin point\npyplot.grid()\npyplot.quiver([0,0,0], [0,0,0], V[:,0], V[:,1], color=['r','b','g'], scale=23)\n\npyplot.show()",
"_____no_output_____"
]
],
[
[
"##Mostrar fórmulas\nPara mostrar fórmulas podemos acudir a pyplot `(se podría usar otra librería u otra función si se quisiera)` \nLas fórmulas introducidas de esta forma se escriben en LaTex\n> Un buen editor de fórmulas de LaTex: https://www.latex4technics.com/ \n\nPara generar la fórmula usamos pyplot.text\n* `pyplot.text(pos X, pos Y, r 'formula en latex', fontsize=tamaño de fuente)`\n> * En el ejemplo %i indica que se debe reemplazar este símbolo por un valor entero o el contenido de una variable a la derecha del texto \n> * Cambiando %i por %f indicaría que se debe interpretar el número externo al texto como valor con coma\n* `pyplot.axis('off')` indica que no se debe mostrar nada más que el texto\n* Finalmente, `pyplot.show()` muestra la ecuación\n\n",
"_____no_output_____"
]
],
[
[
"#add text\npyplot.text(1, 1,r'$\\alpha > \\beta > %i$'% valor_formulario_slider,fontsize=40)\npyplot.axis('off')\npyplot.show() #or savefig",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d03fe9102e78e343652005a047d2857942a16cf3 | 1,652 | ipynb | Jupyter Notebook | Problem_3.ipynb | Nickamaes/Numerical-Methods-58011 | 3e3d0ed7c2045aadf32e36433ddc00d2a083f3b9 | [
"Apache-2.0"
] | null | null | null | Problem_3.ipynb | Nickamaes/Numerical-Methods-58011 | 3e3d0ed7c2045aadf32e36433ddc00d2a083f3b9 | [
"Apache-2.0"
] | null | null | null | Problem_3.ipynb | Nickamaes/Numerical-Methods-58011 | 3e3d0ed7c2045aadf32e36433ddc00d2a083f3b9 | [
"Apache-2.0"
] | null | null | null | 25.415385 | 239 | 0.459443 | [
[
[
"<a href=\"https://colab.research.google.com/github/Nickamaes/Numerical-Methods-58011/blob/main/Problem_3.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import math\ndef f(x):\n return(math.exp(x)) #Trigo function\na = -1\nb = 1\nn = 10\nh = (b-a)/n #Width of Trapezoid\nS = h * (f(a)+f(b)) #Value of summation\nfor i in range(1,n):\n S += f(a+i*h)\nIntegral = S*h\nprint('Integral = %0.4f' %Integral)",
"Integral = 2.1731\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
d03fea012f2f6d690de1a0cf1982dbd72b2ae14d | 638 | ipynb | Jupyter Notebook | notebooks/book1/20/fig_20_24.ipynb | karm-patel/pyprobml | af8230a0bc0d01bb0f779582d87e5856d25e6211 | [
"MIT"
] | null | null | null | notebooks/book1/20/fig_20_24.ipynb | karm-patel/pyprobml | af8230a0bc0d01bb0f779582d87e5856d25e6211 | [
"MIT"
] | 1 | 2022-03-27T04:59:50.000Z | 2022-03-27T04:59:50.000Z | notebooks/book1/20/fig_20_24.ipynb | karm-patel/pyprobml | af8230a0bc0d01bb0f779582d87e5856d25e6211 | [
"MIT"
] | 2 | 2022-03-26T11:52:36.000Z | 2022-03-27T05:17:48.000Z | 39.875 | 467 | 0.714734 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d03fee3e32ae43c7a80fb0895f7083cdda85bae1 | 100,656 | ipynb | Jupyter Notebook | train_algo_relocation/analysis/get_solved_new_instances-24h-relocation-14151617-expensive_relocation-185k.ipynb | hsinyic/tusp-archive | 21ce50038bb7b9a4c099a3926a9eae108323304e | [
"MIT"
] | 2 | 2020-11-10T05:51:06.000Z | 2022-03-27T14:01:10.000Z | train_algo_relocation/analysis/get_solved_new_instances-24h-relocation-14151617-expensive_relocation-185k.ipynb | hsinyic/tusp-archive | 21ce50038bb7b9a4c099a3926a9eae108323304e | [
"MIT"
] | null | null | null | train_algo_relocation/analysis/get_solved_new_instances-24h-relocation-14151617-expensive_relocation-185k.ipynb | hsinyic/tusp-archive | 21ce50038bb7b9a4c099a3926a9eae108323304e | [
"MIT"
] | 1 | 2020-11-10T05:51:09.000Z | 2020-11-10T05:51:09.000Z | 42.668928 | 11,612 | 0.498192 | [
[
[
"import json\nimport random\nimport numpy as np\nimport tensorflow as tf\nfrom collections import deque\nfrom keras.models import Sequential\nfrom keras.optimizers import RMSprop\nfrom keras.layers import Dense, Flatten\nfrom keras.layers.convolutional import Conv2D\nfrom keras import backend as K\nimport datetime\nimport itertools\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport scipy as sp\nimport time\nimport math\n\nfrom matplotlib.colors import LinearSegmentedColormap\nimport colorsys\nimport numpy as np\n \nfrom data_retrieval_relocation_3ksol_reloc import INSTANCEProvider\nfrom kbh_yard_b2b_relocation import KBH_Env #This is the environment of the shunting yard\nfrom dqn_kbh_colfax_relocation_test_agent import DQNAgent",
"_____no_output_____"
],
[
"# this function returns random colors for visualisation of learning.\ndef rand_cmap(nlabels, type='soft', first_color_black=True, last_color_black=False):\n # Generate soft pastel colors, by limiting the RGB spectrum\n if type == 'soft':\n low = 0.6\n high = 0.95\n randRGBcolors = [(np.random.uniform(low=low, high=high),\n np.random.uniform(low=low, high=high),\n np.random.uniform(low=low, high=high)) for i in range(nlabels)]\n\n if first_color_black:\n randRGBcolors[0] = [0, 0, 0]\n\n if last_color_black:\n randRGBcolors[-1] = [0, 0, 0]\n random_colormap = LinearSegmentedColormap.from_list('new_map', randRGBcolors, N=nlabels)\n return random_colormap\n",
"_____no_output_____"
],
[
"#1525445230 is the 185k expensive relocation model.",
"_____no_output_____"
],
[
"for model_nr in ['1525445230']:\n #which model to load.\n test_case = model_nr\n \n #LOAD THE INSTANCE PROVIDER\n ig = INSTANCEProvider()\n instances = ig.instances\n # Create environment KBH\n yrd = KBH_Env()\n\n # Create the DQNAgent with the CNN approximation of the Q-function and its experience replay and training functions.\n # load the trained model.\n agent = DQNAgent(yrd, True, test_case)\n\n # set epsilon to 0 to act just greedy\n agent.epsilon = 0\n\n #new_cmap = rand_cmap(200, type='soft', first_color_black=True, last_color_black=False, verbose=True)\n \n visualization = False\n \n n = len(instances)\n \n # result vectors\n original_lengths = []\n terminated_at_step = []\n success = []\n relocations = []\n print_count = 0\n \n \n # train types different tracks? \n type_step_track = []\n \n \n for instance in instances:\n nr_relocations = 0\n if print_count % 100 == 0:\n print(print_count)\n print_count = print_count + 1\n #Initialize problem\n event_list = ig.get_instance(instance)\n \n steps, t, total_t, score= len(event_list), 0, 0, 0\n\n state = yrd.reset(event_list) # Get first observation based on the first train arrival.\n history = np.reshape(state, (\n 1, yrd.shape[0], yrd.shape[1], yrd.shape[2])) # reshape state into tensor, which we call history.\n\n done, busy_relocating = False, False\n \n if visualization:\n #visualize learning\n new_cmap = rand_cmap(200, type='soft', first_color_black=True, last_color_black=False)\n\n if visualization == True:\n plt.imshow(np.float32(history[0][0]), cmap=new_cmap, interpolation='nearest')\n plt.show()\n\n while not done:\n action = agent.get_action(history) # RL choose action based on observation\n\n if visualization == True:\n print(agent.model.predict(history))\n print(action+1)\n# # RL take action and get next observation and reward\n# # note the +1 at action\n\n # save for arrival activities the parking location\n event_list_temp = event_list.reset_index(drop=True).copy()\n if event_list_temp.event_type[0]=='arrival':\n train_type = event_list_temp.composition[0]\n type_step_track.append({'type': train_type, 'action': action+1, 'step':t, 'instance_id': instance})\n\n\n # based on that action now let environment go to new state\n event = event_list.iloc[0]\n # check if after this we are done... \n done_ = True if len(event_list) == 1 else False # then there is no next event\n# if done_:\n# print(\"Reached the end of a problem!\")\n if busy_relocating:\n # here we do not drop an event from the event list.\n coming_arrivals = event_list.loc[event_list['event_type'] == 'arrival'].reset_index(drop=True)\n coming_departures = event_list.loc[event_list['event_type'] == 'departure'].reset_index(drop=True)\n\n next_state, reward, done = yrd.reloc_destination_step(event, event_list, action+1, coming_arrivals, coming_departures, done_)\n nr_relocations += 1\n busy_relocating = False\n else: \n # These operations below are expensive: maybe just use indexing.\n event_list.drop(event_list.index[:1], inplace=True)\n coming_arrivals = event_list.loc[event_list['event_type'] == 'arrival'].reset_index(drop=True)\n coming_departures = event_list.loc[event_list['event_type'] == 'departure'].reset_index(drop=True)\n\n # do step\n next_state, reward, done = yrd.step(action+1, coming_arrivals, coming_departures, event, event_list, done_)\n\n busy_relocating = True if reward == -0.5 else False\n\n history_ = np.float32(np.reshape(next_state, (1, yrd.shape[0], yrd.shape[1], yrd.shape[2])))\n \n score += reward # log direct reward of action\n\n if visualization == True: \n #show action\n plt.imshow(np.float32(history_[0][0]), cmap=new_cmap, interpolation='nearest')\n plt.show()\n time.sleep(0.05)\n if reward == -1:\n time.sleep(1)\n print(reward)\n\n if done: # based on what the environment returns.\n #print('ended at step' , t+1)\n #print('original length', steps)\n original_lengths.append(steps)\n terminated_at_step.append(t+1)\n relocations.append(nr_relocations)\n if int(np.unique(history_)[0]) == 1: #then we are in win state\n success.append(1)\n else: \n success.append(0)\n break;\n\n history = history_ # next state now becomes the current state.\n t += 1 # next step in this episode\n \n #save data needed for Entropy calculations.\n df_type_step_track = pd.DataFrame.from_records(type_step_track)\n df_type_step_track['strtype'] = df_type_step_track.apply(lambda row: str(row.type), axis = 1)\n df_type_step_track.strtype = df_type_step_track.strtype.astype('category')\n filename = 'data_'+model_nr+'_relocation_arrival_actions.csv'\n df_type_step_track.to_csv(filename)\n\n# analysis_runs = pd.DataFrame(\n# {'instance_id': instances,\n# 'original_length': original_lengths,\n# 'terminated_at_step': terminated_at_step\n# })\n \n# analysis_runs['solved'] = analysis_runs.apply(lambda row: 1 if row.original_length == row.terminated_at_step else 0, axis =1 )\n# analysis_runs['tried'] = analysis_runs.apply(lambda row: 1 if row.terminated_at_step != -1 else 0, axis =1)\n# analysis_runs['percentage'] = analysis_runs.apply(lambda row: row.solved/755, axis=1)\n \n# analysis_runs.to_csv('best_model_solved_instances.csv')\n# print('Model: ', model_nr)\n# summary = analysis_runs.groupby('original_length', as_index=False)[['solved', 'tried', 'percentage']].sum()\n# print(summary)\n \n# #print hist\n# %matplotlib inline \n# #%%\n# # analyse the parking actions per step and train type\n# df_type_step_track = pd.DataFrame.from_records(type_step_track)\n# bins = [1,2,3,4,5,6,7,8,9,10]\n# plt.hist(df_type_step_track.action, bins, align='left')\n \n# #prepare for save\n# df_type_step_track['strtype'] = df_type_step_track.apply(lambda row: str(row.type), axis = 1)\n# df_type_step_track.strtype = df_type_step_track.strtype.astype('category')\n# filename = 'data_'+model_nr+'_paper.csv'\n# df_type_step_track.to_csv(filename)\n\n\n\n",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d_5 (Conv2D) (None, 1, 33, 32) 16928 \n_________________________________________________________________\nconv2d_6 (Conv2D) (None, 1, 33, 64) 8256 \n_________________________________________________________________\nflatten_3 (Flatten) (None, 2112) 0 \n_________________________________________________________________\ndense_5 (Dense) (None, 256) 540928 \n_________________________________________________________________\ndense_6 (Dense) (None, 9) 2313 \n=================================================================\nTotal params: 568,425\nTrainable params: 568,425\nNon-trainable params: 0\n_________________________________________________________________\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d_7 (Conv2D) (None, 1, 33, 32) 16928 \n_________________________________________________________________\nconv2d_8 (Conv2D) (None, 1, 33, 64) 8256 \n_________________________________________________________________\nflatten_4 (Flatten) (None, 2112) 0 \n_________________________________________________________________\ndense_7 (Dense) (None, 256) 540928 \n_________________________________________________________________\ndense_8 (Dense) (None, 9) 2313 \n=================================================================\nTotal params: 568,425\nTrainable params: 568,425\nNon-trainable params: 0\n_________________________________________________________________\ntarget_model_updated\n1526895996\nmodel loaded\n0\n100\n200\n300\n400\n500\n600\n700\n800\n900\n1000\n1100\n1200\n1300\n1400\n1500\n1600\n1700\n1800\n1900\n2000\n2100\n2200\n2300\n2400\n2500\n2600\n2700\n2800\n2900\n3000\n"
],
[
"analysis_runs = pd.DataFrame(\n{'instance_id': instances,\n 'original_length': original_lengths,\n 'terminated_at_step': terminated_at_step,\n 'success': success,\n 'nr_relocations': relocations\n})\n",
"_____no_output_____"
],
[
"analysis_runs.sort_values('terminated_at_step')",
"_____no_output_____"
],
[
"print(analysis_runs.loc[analysis_runs.success == 0].instance_id.to_string(index=False))",
"4020\n4022\n4033\n4083\n4106\n4109\n4112\n4172\n4200\n4201\n4237\n4245\n4257\n4261\n4263\n4267\n4280\n4313\n4320\n4332\n4341\n4345\n4355\n4364\n4365\n4370\n4374\n4382\n4468\n4486\n4487\n4499\n4510\n4530\n4538\n4544\n4556\n4569\n4609\n4628\n4667\n4681\n4690\n4699\n4710\n4712\n4743\n4790\n4795\n4803\n4824\n4839\n4846\n4847\n4853\n4857\n4858\n4860\n4862\n4863\n4884\n4890\n4894\n4901\n4904\n4908\n4927\n4928\n4936\n4962\n4966\n4969\n4974\n4989\n4991\n5006\n5008\n5011\n5012\n5014\n5031\n5036\n5037\n5039\n5041\n5048\n5056\n5063\n5086\n5087\n5118\n5127\n5167\n5175\n5178\n5181\n5194\n5202\n5221\n5223\n5239\n5241\n5242\n5243\n5246\n5249\n5262\n5265\n5273\n5274\n5275\n5294\n5304\n5305\n5307\n5308\n5312\n5313\n5333\n5338\n5359\n5385\n5388\n5390\n5399\n5402\n5409\n5411\n5412\n5422\n5440\n5442\n5452\n5453\n5463\n5465\n5473\n5483\n5488\n5493\n5494\n5503\n5507\n5511\n5513\n5524\n5535\n5545\n5548\n5561\n5572\n5590\n5595\n5596\n5597\n5603\n5610\n5613\n5618\n5622\n5623\n5626\n5636\n5656\n5668\n5673\n5676\n5685\n5688\n5691\n5694\n5709\n5715\n5719\n5724\n5733\n5735\n5742\n5743\n5746\n5747\n5748\n5749\n5751\n5756\n5768\n5782\n5790\n5794\n5814\n5823\n5829\n5831\n5843\n5849\n5850\n5851\n5857\n5860\n5871\n5872\n5893\n5899\n5909\n5914\n5928\n5932\n5944\n5970\n5973\n5988\n5996\n6001\n6002\n6005\n6006\n6016\n6018\n6029\n6035\n6051\n6053\n6061\n6066\n6068\n6079\n6082\n6088\n6104\n6122\n6129\n6131\n6136\n6144\n6149\n6160\n6162\n6167\n6169\n6180\n6181\n6183\n6184\n6193\n6195\n6198\n6208\n6219\n6220\n6229\n6233\n6240\n6255\n6256\n6260\n6266\n6275\n6278\n6286\n6303\n6307\n6310\n6311\n6319\n6320\n6333\n6335\n6336\n6340\n6347\n6356\n6359\n6361\n6369\n6374\n6375\n6376\n6380\n6384\n6387\n6388\n6396\n6400\n6402\n6407\n6419\n6420\n6421\n6431\n6433\n6437\n6446\n6450\n6462\n6464\n6465\n6470\n6472\n6482\n6489\n6490\n6491\n6495\n6502\n6506\n6513\n6519\n6523\n6524\n6533\n6536\n6538\n6540\n6542\n6557\n6560\n6564\n6569\n6574\n6584\n6585\n6588\n6593\n6594\n6595\n6596\n6598\n6624\n6625\n6628\n6638\n6640\n6646\n6649\n6657\n6659\n6660\n6662\n6673\n6676\n6683\n6698\n6700\n6702\n6708\n6713\n6715\n6730\n6731\n6734\n6737\n6744\n6745\n6752\n6753\n6757\n6768\n6769\n6775\n6783\n6784\n6793\n6796\n6798\n6806\n6808\n6809\n6817\n6820\n6829\n6840\n6851\n6854\n6856\n6859\n6872\n6879\n6881\n6886\n6887\n6894\n6895\n6901\n6912\n6922\n6935\n6936\n6947\n6960\n6961\n6964\n6965\n6970\n6979\n6981\n6984\n6985\n6989\n6991\n6996\n6997\n7001\n7002\n7011\n7015\n7017\n7018\n7019\n"
],
[
"analysis_runs.loc[analysis_runs.success == 1].copy().groupby('nr_relocations')[['instance_id']].count()",
"_____no_output_____"
],
[
"summary = analysis_runs.groupby('original_length', as_index=False)[['success']].sum()\nprint(summary)\n",
" original_length success\n0 37 708\n1 41 659\n2 43 643\n3 46 602\n"
],
[
"summary = analysis_runs.groupby('original_length', as_index=False)[['success']].mean()\nprint(summary)\n",
" original_length success\n0 37 0.937748\n1 41 0.872848\n2 43 0.851656\n3 46 0.797351\n"
],
[
"max_reloc = max(analysis_runs.nr_relocations)\nprint(max_reloc)\nplt.hist(analysis_runs.nr_relocations, bins=range(0,max_reloc+2), align='left')\n",
"5\n"
],
[
"import seaborn as sns\nsns.set(style=\"darkgrid\")\n\ng = sns.FacetGrid(analysis_runs, col=\"original_length\", margin_titles=True)\nbins = range(0,max_reloc+2)\ng.map(plt.hist, \"nr_relocations\", color=\"steelblue\", bins=bins, lw=0, align='left')\n\nprint(analysis_runs.loc[analysis_runs.success == 1].groupby('original_length', as_index=False)[['nr_relocations']].mean())",
" original_length nr_relocations\n0 37 0.936441\n1 41 0.817906\n2 43 0.839813\n3 46 0.923588\n"
]
],
[
[
"# CODE HAS BEEN RUN UNTILL HERE.\n.\n\n.\n\n.\n\n.\n\n.\n\n.\n\n.\n\n.\n\nv\n\n\n\n\n",
"_____no_output_____"
],
[
"# analysis of mistakes\n\n",
"_____no_output_____"
]
],
[
[
"analysis_runs.loc[analysis_runs.success == 0].sort_values('terminated_at_step')",
"_____no_output_____"
],
[
"#plt.hist(analysis_runs.loc[analysis_runs.success == 0].terminated_at_step, bins=8)\n\nlen(analysis_runs.loc[analysis_runs.success == 0])\n\nanalysis_runs['instance_size'] = analysis_runs.apply(lambda row: str(row.original_length).replace('37', '14').replace('41', '15').replace('43', '16').replace('46','17'), axis=1)\n\nimport seaborn as sns\nsns.set(style=\"darkgrid\")\n\nbins = [0,5,10,15,20,25,30,35,40,45,50]\ng = sns.FacetGrid(analysis_runs.loc[analysis_runs.success == 0], col=\"instance_size\", margin_titles=True)\ng.set(ylim=(0, 100), xlim=(0,50))\n\ng.map(plt.hist, \"terminated_at_step\", color=\"steelblue\", bins=bins, lw=0)\nsns.plt.savefig('185k_failures.eps')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
d03ff007d79767fb25651a9fe35e0b24a2c6c1d1 | 157,095 | ipynb | Jupyter Notebook | VAE_cell_cycle.ipynb | dauparas/tensorflow_examples | abed941d75e0d4be0a9cd6aefbd344acd88decdc | [
"MIT"
] | 4 | 2019-06-19T18:31:59.000Z | 2020-10-19T19:19:00.000Z | VAE_cell_cycle.ipynb | dauparas/tensorflow_examples | abed941d75e0d4be0a9cd6aefbd344acd88decdc | [
"MIT"
] | null | null | null | VAE_cell_cycle.ipynb | dauparas/tensorflow_examples | abed941d75e0d4be0a9cd6aefbd344acd88decdc | [
"MIT"
] | null | null | null | 208.072848 | 39,106 | 0.876883 | [
[
[
"<a href=\"https://colab.research.google.com/github/dauparas/tensorflow_examples/blob/master/VAE_cell_cycle.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"https://github.com/PMBio/scLVM/blob/master/tutorials/tcell_demo.ipynb",
"_____no_output_____"
],
[
"Variational Autoencoder Model (VAE) with latent subspaces based on:\nhttps://arxiv.org/pdf/1812.06190.pdf",
"_____no_output_____"
]
],
[
[
"#Step 1: import dependencies\nfrom tensorflow.keras import layers\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport tensorflow as tf\nfrom keras import regularizers\nimport time\nfrom __future__ import division\n\nimport tensorflow as tf\nimport tensorflow_probability as tfp\ntfd = tfp.distributions\n\n%matplotlib inline\nplt.style.use('dark_background')\n\nimport pandas as pd",
"Using TensorFlow backend.\n"
],
[
"import os\nfrom matplotlib import cm\nimport h5py\nimport scipy as SP\nimport pylab as PL",
"_____no_output_____"
],
[
"data = os.path.join('data_Tcells_normCounts.h5f')\nf = h5py.File(data,'r')\nY = f['LogNcountsMmus'][:] # gene expression matrix\ntech_noise = f['LogVar_techMmus'][:] # technical noise\ngenes_het_bool=f['genes_heterogen'][:] # index of heterogeneous genes\ngeneID = f['gene_names'][:] # gene names\ncellcyclegenes_filter = SP.unique(f['cellcyclegenes_filter'][:].ravel() -1) # idx of cell cycle genes from GO\ncellcyclegenes_filterCB = f['ccCBall_gene_indices'][:].ravel() -1 # idx of cell cycle genes from cycle base ...",
"_____no_output_____"
],
[
"# filter cell cycle genes\nidx_cell_cycle = SP.union1d(cellcyclegenes_filter,cellcyclegenes_filterCB)\n# determine non-zero counts\nidx_nonzero = SP.nonzero((Y.mean(0)**2)>0)[0]\nidx_cell_cycle_noise_filtered = SP.intersect1d(idx_cell_cycle,idx_nonzero)\n# subset gene expression matrix\nYcc = Y[:,idx_cell_cycle_noise_filtered]",
"_____no_output_____"
],
[
"plt = PL.subplot(1,1,1);\nPL.imshow(Ycc,cmap=cm.RdBu,vmin=-3,vmax=+3,interpolation='None');\n#PL.colorbar();\nplt.set_xticks([]);\nplt.set_yticks([]);\nPL.xlabel('genes');\nPL.ylabel('cells');",
"_____no_output_____"
],
[
"X = np.delete(Y, idx_cell_cycle_noise_filtered, axis=1)\nX = Y #base case\nU = Y[:,idx_cell_cycle_noise_filtered]\n\nmean = np.mean(X, axis=0)\nvariance = np.var(X, axis=0)\n\nindx_small_mean = np.argwhere(mean < 0.00001)\nX = np.delete(X, indx_small_mean, axis=1)\n\nmean = np.mean(X, axis=0)\nvariance = np.var(X, axis=0)",
"_____no_output_____"
],
[
"fano = variance/mean\n\nprint(fano.shape)",
"(30233,)\n"
],
[
"indx_small_fano = np.argwhere(fano < 1.0)",
"_____no_output_____"
],
[
"X = np.delete(X, indx_small_fano, axis=1)\n\nmean = np.mean(X, axis=0)\nvariance = np.var(X, axis=0)\n\nfano = variance/mean",
"_____no_output_____"
],
[
"print(fano.shape)",
"(8892,)\n"
],
[
"#Reconstruction loss\ndef x_given_z(z, output_size):\n with tf.variable_scope('M/x_given_w_z'):\n act = tf.nn.leaky_relu\n \n h = z\n h = tf.layers.dense(h, 8, act)\n h = tf.layers.dense(h, 16, act)\n h = tf.layers.dense(h, 32, act)\n h = tf.layers.dense(h, 64, act)\n h = tf.layers.dense(h, 128, act)\n h = tf.layers.dense(h, 256, act)\n loc = tf.layers.dense(h, output_size)\n #log_variance = tf.layers.dense(x, latent_size)\n #scale = tf.nn.softplus(log_variance)\n scale = 0.01*tf.ones(tf.shape(loc))\n return tfd.MultivariateNormalDiag(loc, scale)\n\n#KL term for z\ndef z_given_x(x, latent_size): #+\n with tf.variable_scope('M/z_given_x'):\n act = tf.nn.leaky_relu\n h = x\n \n h = tf.layers.dense(h, 256, act)\n h = tf.layers.dense(h, 128, act)\n h = tf.layers.dense(h, 64, act)\n h = tf.layers.dense(h, 32, act)\n h = tf.layers.dense(h, 16, act)\n h = tf.layers.dense(h, 8, act)\n \n loc = tf.layers.dense(h,latent_size)\n log_variance = tf.layers.dense(h, latent_size)\n scale = tf.nn.softplus(log_variance)\n# scale = 0.01*tf.ones(tf.shape(loc))\n return tfd.MultivariateNormalDiag(loc, scale)\n\ndef z_given(latent_size):\n with tf.variable_scope('M/z_given'):\n loc = tf.zeros(latent_size)\n scale = 0.01*tf.ones(tf.shape(loc))\n return tfd.MultivariateNormalDiag(loc, scale)",
"_____no_output_____"
],
[
"#Connect encoder and decoder and define the loss function\ntf.reset_default_graph()\n\nx_in = tf.placeholder(tf.float32, shape=[None, X.shape[1]], name='x_in')\nx_out = tf.placeholder(tf.float32, shape=[None, X.shape[1]], name='x_out')\n\nz_latent_size = 2\n\nbeta = 0.000001\n\n#KL_z\nzI = z_given(z_latent_size)\nzIx = z_given_x(x_in, z_latent_size)\nzIx_sample = zIx.sample()\nzIx_mean = zIx.mean()\n#kl_z = tf.reduce_mean(zIx.log_prob(zIx_sample)- zI.log_prob(zIx_sample))\nkl_z = tf.reduce_mean(tfd.kl_divergence(zIx, zI)) #analytical\n\n#Reconstruction\nxIz = x_given_z(zIx_sample, X.shape[1])\nrec_out = xIz.mean()\nrec_loss = tf.losses.mean_squared_error(x_out, rec_out)\n\nloss = rec_loss + beta*kl_z\noptimizer = tf.train.AdamOptimizer(0.001).minimize(loss)",
"_____no_output_____"
],
[
"#Helper function\ndef batch_generator(features, x, u, batch_size):\n \"\"\"Function to create python generator to shuffle and split features into batches along the first dimension.\"\"\"\n idx = np.arange(features.shape[0])\n np.random.shuffle(idx)\n for start_idx in range(0, features.shape[0], batch_size):\n end_idx = min(start_idx + batch_size, features.shape[0])\n part = idx[start_idx:end_idx]\n yield features[part,:], x[part,:] , u[part, :]",
"_____no_output_____"
],
[
"n_epochs = 5000\nbatch_size = X.shape[0]\nstart = time.time()\nwith tf.Session() as sess:\n sess.run(tf.global_variables_initializer())\n \n \n for i in range(n_epochs):\n gen = batch_generator(X, X, U, batch_size) #create batch generator\n rec_loss_ = 0\n kl_z_ = 0\n \n for j in range(np.int(X.shape[0]/batch_size)):\n x_in_batch, x_out_batch, u_batch = gen.__next__()\n \n \n _, rec_loss__, kl_z__= sess.run([optimizer, rec_loss, kl_z], feed_dict={x_in: x_in_batch, x_out: x_out_batch})\n rec_loss_ += rec_loss__\n kl_z_ += kl_z__\n \n if (i+1)% 50 == 0 or i == 0:\n zIx_mean_, rec_out_= sess.run([zIx_mean, rec_out], feed_dict ={x_in:X, x_out:X})\n end = time.time()\n print('epoch: {0}, rec_loss: {1:.3f}, kl_z: {2:.2f}'.format((i+1), rec_loss_/(1+np.int(X.shape[0]/batch_size)), kl_z_/(1+np.int(X.shape[0]/batch_size))))\n start = time.time()",
"epoch: 1, rec_loss: 0.488, kl_z: 5038.03\nepoch: 50, rec_loss: 0.256, kl_z: 1169.19\nepoch: 100, rec_loss: 0.254, kl_z: 1150.47\nepoch: 150, rec_loss: 0.256, kl_z: 1138.06\nepoch: 200, rec_loss: 0.253, kl_z: 891.83\nepoch: 250, rec_loss: 0.250, kl_z: 493.30\nepoch: 300, rec_loss: 0.248, kl_z: 730.26\nepoch: 350, rec_loss: 0.243, kl_z: 559.81\nepoch: 400, rec_loss: 0.238, kl_z: 564.55\nepoch: 450, rec_loss: 0.234, kl_z: 710.93\nepoch: 500, rec_loss: 0.228, kl_z: 784.23\nepoch: 550, rec_loss: 0.222, kl_z: 896.33\nepoch: 600, rec_loss: 0.217, kl_z: 1257.52\nepoch: 650, rec_loss: 0.208, kl_z: 1527.80\nepoch: 700, rec_loss: 0.195, kl_z: 1799.51\nepoch: 750, rec_loss: 0.185, kl_z: 2238.79\nepoch: 800, rec_loss: 0.169, kl_z: 2253.14\nepoch: 850, rec_loss: 0.163, kl_z: 2524.81\nepoch: 900, rec_loss: 0.153, kl_z: 2371.30\nepoch: 950, rec_loss: 0.138, kl_z: 2567.25\nepoch: 1000, rec_loss: 0.129, kl_z: 2703.91\nepoch: 1050, rec_loss: 0.119, kl_z: 2730.01\nepoch: 1100, rec_loss: 0.110, kl_z: 2854.48\nepoch: 1150, rec_loss: 0.104, kl_z: 2838.82\nepoch: 1200, rec_loss: 0.099, kl_z: 2848.78\nepoch: 1250, rec_loss: 0.092, kl_z: 2902.38\nepoch: 1300, rec_loss: 0.094, kl_z: 2743.52\nepoch: 1350, rec_loss: 0.086, kl_z: 2913.92\nepoch: 1400, rec_loss: 0.079, kl_z: 2738.15\nepoch: 1450, rec_loss: 0.075, kl_z: 2695.68\nepoch: 1500, rec_loss: 0.072, kl_z: 2631.96\nepoch: 1550, rec_loss: 0.066, kl_z: 2619.38\nepoch: 1600, rec_loss: 0.276, kl_z: 5195.00\nepoch: 1650, rec_loss: 0.247, kl_z: 2940.30\nepoch: 1700, rec_loss: 0.241, kl_z: 1825.31\nepoch: 1750, rec_loss: 0.236, kl_z: 1415.02\nepoch: 1800, rec_loss: 0.230, kl_z: 1225.71\nepoch: 1850, rec_loss: 0.226, kl_z: 1231.15\nepoch: 1900, rec_loss: 0.220, kl_z: 1264.15\nepoch: 1950, rec_loss: 0.212, kl_z: 1333.83\nepoch: 2000, rec_loss: 0.205, kl_z: 1449.89\nepoch: 2050, rec_loss: 0.198, kl_z: 1722.17\nepoch: 2100, rec_loss: 0.192, kl_z: 1952.83\nepoch: 2150, rec_loss: 0.186, kl_z: 2188.21\nepoch: 2200, rec_loss: 0.180, kl_z: 2312.91\nepoch: 2250, rec_loss: 0.175, kl_z: 2320.98\nepoch: 2300, rec_loss: 0.167, kl_z: 2494.97\nepoch: 2350, rec_loss: 0.162, kl_z: 2534.96\nepoch: 2400, rec_loss: 0.159, kl_z: 2485.46\nepoch: 2450, rec_loss: 0.152, kl_z: 2470.98\nepoch: 2500, rec_loss: 0.150, kl_z: 2546.58\nepoch: 2550, rec_loss: 0.143, kl_z: 2635.48\nepoch: 2600, rec_loss: 0.137, kl_z: 2575.49\nepoch: 2650, rec_loss: 0.132, kl_z: 2556.10\nepoch: 2700, rec_loss: 0.128, kl_z: 2502.45\nepoch: 2750, rec_loss: 0.126, kl_z: 2708.18\nepoch: 2800, rec_loss: 0.120, kl_z: 2577.95\nepoch: 2850, rec_loss: 0.113, kl_z: 2625.05\nepoch: 2900, rec_loss: 0.113, kl_z: 2510.52\nepoch: 2950, rec_loss: 0.107, kl_z: 2573.50\nepoch: 3000, rec_loss: 0.105, kl_z: 2498.15\nepoch: 3050, rec_loss: 0.103, kl_z: 2485.70\nepoch: 3100, rec_loss: 0.101, kl_z: 2390.64\nepoch: 3150, rec_loss: 0.101, kl_z: 2433.73\nepoch: 3200, rec_loss: 0.097, kl_z: 2279.95\nepoch: 3250, rec_loss: 0.090, kl_z: 2303.65\nepoch: 3300, rec_loss: 0.088, kl_z: 2240.98\nepoch: 3350, rec_loss: 0.087, kl_z: 2256.19\nepoch: 3400, rec_loss: 0.085, kl_z: 2204.10\nepoch: 3450, rec_loss: 0.082, kl_z: 2199.10\nepoch: 3500, rec_loss: 0.082, kl_z: 2147.36\nepoch: 3550, rec_loss: 0.080, kl_z: 2091.81\nepoch: 3600, rec_loss: 0.078, kl_z: 2094.17\nepoch: 3650, rec_loss: 0.075, kl_z: 2095.87\nepoch: 3700, rec_loss: 0.076, kl_z: 2082.63\nepoch: 3750, rec_loss: 0.074, kl_z: 2055.43\nepoch: 3800, rec_loss: 0.070, kl_z: 2018.21\nepoch: 3850, rec_loss: 0.070, kl_z: 2024.04\nepoch: 3900, rec_loss: 0.067, kl_z: 1994.23\nepoch: 3950, rec_loss: 0.071, kl_z: 1973.09\nepoch: 4000, rec_loss: 0.066, kl_z: 1976.65\nepoch: 4050, rec_loss: 0.062, kl_z: 1942.60\nepoch: 4100, rec_loss: 0.061, kl_z: 1930.08\nepoch: 4150, rec_loss: 0.058, kl_z: 1919.03\nepoch: 4200, rec_loss: 0.060, kl_z: 1893.53\nepoch: 4250, rec_loss: 0.056, kl_z: 1897.14\nepoch: 4300, rec_loss: 0.060, kl_z: 1889.32\nepoch: 4350, rec_loss: 0.057, kl_z: 1847.00\nepoch: 4400, rec_loss: 0.056, kl_z: 1810.34\nepoch: 4450, rec_loss: 0.055, kl_z: 1853.19\nepoch: 4500, rec_loss: 0.056, kl_z: 1782.88\nepoch: 4550, rec_loss: 0.051, kl_z: 1817.94\nepoch: 4600, rec_loss: 0.050, kl_z: 1781.25\nepoch: 4650, rec_loss: 0.056, kl_z: 1756.28\nepoch: 4700, rec_loss: 0.052, kl_z: 1764.26\nepoch: 4750, rec_loss: 0.049, kl_z: 1793.30\nepoch: 4800, rec_loss: 0.047, kl_z: 1734.55\nepoch: 4850, rec_loss: 0.045, kl_z: 1763.27\nepoch: 4900, rec_loss: 0.043, kl_z: 1716.00\nepoch: 4950, rec_loss: 0.045, kl_z: 1727.20\nepoch: 5000, rec_loss: 0.042, kl_z: 1735.31\n"
],
[
"from sklearn.decomposition import TruncatedSVD\nsvd = TruncatedSVD(n_components=2, n_iter=7, random_state=42)\nsvd.fit(U.T) \n\n\nprint(svd.explained_variance_ratio_) \n\nprint(svd.explained_variance_ratio_.sum()) \n\nprint(svd.singular_values_) \n\nU_ = svd.components_\n\nU_ = U_.T",
"[0.55983905 0.04389005]\n0.6037290921647739\n[482.32958745 71.86901165]\n"
],
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"fig, axs = plt.subplots(1, 2, figsize=(14,5))\naxs[0].scatter(zIx_mean_[:,0],zIx_mean_[:,1], c=U_[:,0], cmap='viridis', s=5.0);\naxs[0].set_xlabel('z1')\naxs[0].set_ylabel('z2')\n\nfig.suptitle('X1')\nplt.show()",
"_____no_output_____"
],
[
"fig, axs = plt.subplots(1, 2, figsize=(14,5))\naxs[0].scatter(wIxy_mean_[:,0],wIxy_mean_[:,1], c=U_[:,1], cmap='viridis', s=5.0);\naxs[0].set_xlabel('w1')\naxs[0].set_ylabel('w2')\n\naxs[1].scatter(zIx_mean_[:,0],zIx_mean_[:,1], c=U_[:,1], cmap='viridis', s=5.0);\naxs[1].set_xlabel('z1')\naxs[1].set_ylabel('z2')\n\nfig.suptitle('X1')\nplt.show()",
"_____no_output_____"
],
[
"error = np.abs(X-rec_out_)",
"_____no_output_____"
],
[
"plt.plot(np.reshape(error, -1), '*', markersize=0.1);",
"_____no_output_____"
],
[
"plt.hist(np.reshape(error, -1), bins=50);",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d03ff7f81c3f38dbb98e697788556feba84b8870 | 1,072 | ipynb | Jupyter Notebook | code-snippet/rust/notebook/example-evcxr.ipynb | zhoujiagen/ml_hacks | 6e68e5227c472562f1c6330fba726a92bf4a7499 | [
"MIT"
] | null | null | null | code-snippet/rust/notebook/example-evcxr.ipynb | zhoujiagen/ml_hacks | 6e68e5227c472562f1c6330fba726a92bf4a7499 | [
"MIT"
] | null | null | null | code-snippet/rust/notebook/example-evcxr.ipynb | zhoujiagen/ml_hacks | 6e68e5227c472562f1c6330fba726a92bf4a7499 | [
"MIT"
] | null | null | null | 18.169492 | 81 | 0.526119 | [
[
[
"- [Evcxr](https://github.com/google/evcxr): An evaluation context for Rust.",
"_____no_output_____"
]
],
[
[
"println!(\"Hello Rust!\");",
"Hello Rust!\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
d03ffda932d8b916979d249d5432108a82066456 | 57,534 | ipynb | Jupyter Notebook | 6_sagemaker-deployment/Tutorials/Boston Housing - XGBoost (Hyperparameter Tuning) - Low Level.ipynb | JasmineMou/udacity_DL | 5e33a2aae8a062948cc17e147ec25de334f0ed41 | [
"MIT"
] | null | null | null | 6_sagemaker-deployment/Tutorials/Boston Housing - XGBoost (Hyperparameter Tuning) - Low Level.ipynb | JasmineMou/udacity_DL | 5e33a2aae8a062948cc17e147ec25de334f0ed41 | [
"MIT"
] | null | null | null | 6_sagemaker-deployment/Tutorials/Boston Housing - XGBoost (Hyperparameter Tuning) - Low Level.ipynb | JasmineMou/udacity_DL | 5e33a2aae8a062948cc17e147ec25de334f0ed41 | [
"MIT"
] | null | null | null | 61.798067 | 14,856 | 0.682866 | [
[
[
"# Predicting Boston Housing Prices\n\n## Using XGBoost in SageMaker (Hyperparameter Tuning)\n\n_Deep Learning Nanodegree Program | Deployment_\n\n---\n\nAs an introduction to using SageMaker's Low Level API for hyperparameter tuning, we will look again at the [Boston Housing Dataset](https://www.cs.toronto.edu/~delve/data/boston/bostonDetail.html) to predict the median value of a home in the area of Boston Mass.\n\nThe documentation reference for the API used in this notebook is the [SageMaker Developer's Guide](https://docs.aws.amazon.com/sagemaker/latest/dg/)\n\n## General Outline\n\nTypically, when using a notebook instance with SageMaker, you will proceed through the following steps. Of course, not every step will need to be done with each project. Also, there is quite a lot of room for variation in many of the steps, as you will see throughout these lessons.\n\n1. Download or otherwise retrieve the data.\n2. Process / Prepare the data.\n3. Upload the processed data to S3.\n4. Train a chosen model.\n5. Test the trained model (typically using a batch transform job).\n6. Deploy the trained model.\n7. Use the deployed model.\n\nIn this notebook we will only be covering steps 1 through 5 as we are only interested in creating a tuned model and testing its performance.",
"_____no_output_____"
]
],
[
[
"# Make sure that we use SageMaker 1.x\n!pip install sagemaker==1.72.0",
"Collecting sagemaker==1.72.0\n Downloading sagemaker-1.72.0.tar.gz (297 kB)\n\u001b[K |████████████████████████████████| 297 kB 16.6 MB/s eta 0:00:01\n\u001b[?25hRequirement already satisfied: boto3>=1.14.12 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (1.17.55)\nRequirement already satisfied: numpy>=1.9.0 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (1.19.5)\nRequirement already satisfied: protobuf>=3.1 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (3.15.2)\nRequirement already satisfied: scipy>=0.19.0 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (1.5.3)\nRequirement already satisfied: protobuf3-to-dict>=0.1.5 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (0.1.5)\nCollecting smdebug-rulesconfig==0.1.4\n Downloading smdebug_rulesconfig-0.1.4-py2.py3-none-any.whl (10 kB)\nRequirement already satisfied: importlib-metadata>=1.4.0 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (3.7.0)\nRequirement already satisfied: packaging>=20.0 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (20.9)\nRequirement already satisfied: jmespath<1.0.0,>=0.7.1 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from boto3>=1.14.12->sagemaker==1.72.0) (0.10.0)\nRequirement already satisfied: botocore<1.21.0,>=1.20.55 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from boto3>=1.14.12->sagemaker==1.72.0) (1.20.55)\nRequirement already satisfied: s3transfer<0.5.0,>=0.4.0 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from boto3>=1.14.12->sagemaker==1.72.0) (0.4.1)\nRequirement already satisfied: python-dateutil<3.0.0,>=2.1 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from botocore<1.21.0,>=1.20.55->boto3>=1.14.12->sagemaker==1.72.0) (2.8.1)\nRequirement already satisfied: urllib3<1.27,>=1.25.4 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from botocore<1.21.0,>=1.20.55->boto3>=1.14.12->sagemaker==1.72.0) (1.26.4)\nRequirement already satisfied: zipp>=0.5 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from importlib-metadata>=1.4.0->sagemaker==1.72.0) (3.4.0)\nRequirement already satisfied: typing-extensions>=3.6.4 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from importlib-metadata>=1.4.0->sagemaker==1.72.0) (3.7.4.3)\nRequirement already satisfied: pyparsing>=2.0.2 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from packaging>=20.0->sagemaker==1.72.0) (2.4.7)\nRequirement already satisfied: six>=1.9 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from protobuf>=3.1->sagemaker==1.72.0) (1.15.0)\nBuilding wheels for collected packages: sagemaker\n Building wheel for sagemaker (setup.py) ... \u001b[?25ldone\n\u001b[?25h Created wheel for sagemaker: filename=sagemaker-1.72.0-py2.py3-none-any.whl size=386358 sha256=918ca6ca3fd8db5bdf10a9a6e2b41c5779c041e02424f8e8ff5b4fb7d7129a62\n Stored in directory: /home/ec2-user/.cache/pip/wheels/c3/58/70/85faf4437568bfaa4c419937569ba1fe54d44c5db42406bbd7\nSuccessfully built sagemaker\nInstalling collected packages: smdebug-rulesconfig, sagemaker\n Attempting uninstall: smdebug-rulesconfig\n Found existing installation: smdebug-rulesconfig 1.0.1\n Uninstalling smdebug-rulesconfig-1.0.1:\n Successfully uninstalled smdebug-rulesconfig-1.0.1\n Attempting uninstall: sagemaker\n Found existing installation: sagemaker 2.38.0\n Uninstalling sagemaker-2.38.0:\n Successfully uninstalled sagemaker-2.38.0\nSuccessfully installed sagemaker-1.72.0 smdebug-rulesconfig-0.1.4\n"
]
],
[
[
"## Step 0: Setting up the notebook\n\nWe begin by setting up all of the necessary bits required to run our notebook. To start that means loading all of the Python modules we will need.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport os\n\nimport time\nfrom time import gmtime, strftime\n\nimport numpy as np\nimport pandas as pd\n\nimport matplotlib.pyplot as plt\n\nfrom sklearn.datasets import load_boston\nimport sklearn.model_selection",
"_____no_output_____"
]
],
[
[
"In addition to the modules above, we need to import the various bits of SageMaker that we will be using. ",
"_____no_output_____"
]
],
[
[
"import sagemaker\nfrom sagemaker import get_execution_role\nfrom sagemaker.amazon.amazon_estimator import get_image_uri\n\n# This is an object that represents the SageMaker session that we are currently operating in. This\n# object contains some useful information that we will need to access later such as our region.\nsession = sagemaker.Session()\n\n# This is an object that represents the IAM role that we are currently assigned. When we construct\n# and launch the training job later we will need to tell it what IAM role it should have. Since our\n# use case is relatively simple we will simply assign the training job the role we currently have.\nrole = get_execution_role()",
"_____no_output_____"
]
],
[
[
"## Step 1: Downloading the data\n\nFortunately, this dataset can be retrieved using sklearn and so this step is relatively straightforward.",
"_____no_output_____"
]
],
[
[
"boston = load_boston()",
"_____no_output_____"
]
],
[
[
"## Step 2: Preparing and splitting the data\n\nGiven that this is clean tabular data, we don't need to do any processing. However, we do need to split the rows in the dataset up into train, test and validation sets.",
"_____no_output_____"
]
],
[
[
"# First we package up the input data and the target variable (the median value) as pandas dataframes. This\n# will make saving the data to a file a little easier later on.\n\nX_bos_pd = pd.DataFrame(boston.data, columns=boston.feature_names)\nY_bos_pd = pd.DataFrame(boston.target)\n\n# We split the dataset into 2/3 training and 1/3 testing sets.\nX_train, X_test, Y_train, Y_test = sklearn.model_selection.train_test_split(X_bos_pd, Y_bos_pd, test_size=0.33)\n\n# Then we split the training set further into 2/3 training and 1/3 validation sets.\nX_train, X_val, Y_train, Y_val = sklearn.model_selection.train_test_split(X_train, Y_train, test_size=0.33)",
"_____no_output_____"
]
],
[
[
"## Step 3: Uploading the data files to S3\n\nWhen a training job is constructed using SageMaker, a container is executed which performs the training operation. This container is given access to data that is stored in S3. This means that we need to upload the data we want to use for training to S3. In addition, when we perform a batch transform job, SageMaker expects the input data to be stored on S3. We can use the SageMaker API to do this and hide some of the details.\n\n### Save the data locally\n\nFirst we need to create the test, train and validation csv files which we will then upload to S3.",
"_____no_output_____"
],
[
"**My Comment**:\n\nTo solve the \"no space left\" issue: remove `-sagemaker-deployment/cache/sentiment_analysis`. ",
"_____no_output_____"
]
],
[
[
"# This is our local data directory. We need to make sure that it exists.\ndata_dir = '../data/boston'\nif not os.path.exists(data_dir):\n os.makedirs(data_dir)",
"_____no_output_____"
],
[
"# We use pandas to save our test, train and validation data to csv files. Note that we make sure not to include header\n# information or an index as this is required by the built in algorithms provided by Amazon. Also, for the train and\n# validation data, it is assumed that the first entry in each row is the target variable.\n\nX_test.to_csv(os.path.join(data_dir, 'test.csv'), header=False, index=False)\n\npd.concat([Y_val, X_val], axis=1).to_csv(os.path.join(data_dir, 'validation.csv'), header=False, index=False)\npd.concat([Y_train, X_train], axis=1).to_csv(os.path.join(data_dir, 'train.csv'), header=False, index=False)",
"_____no_output_____"
]
],
[
[
"### Upload to S3\n\nSince we are currently running inside of a SageMaker session, we can use the object which represents this session to upload our data to the 'default' S3 bucket. Note that it is good practice to provide a custom prefix (essentially an S3 folder) to make sure that you don't accidentally interfere with data uploaded from some other notebook or project.",
"_____no_output_____"
]
],
[
[
"prefix = 'boston-xgboost-tuning-LL'\n\ntest_location = session.upload_data(os.path.join(data_dir, 'test.csv'), key_prefix=prefix)\nval_location = session.upload_data(os.path.join(data_dir, 'validation.csv'), key_prefix=prefix)\ntrain_location = session.upload_data(os.path.join(data_dir, 'train.csv'), key_prefix=prefix)",
"_____no_output_____"
]
],
[
[
"## Step 4: Train and construct the XGBoost model\n\nNow that we have the training and validation data uploaded to S3, we can construct our XGBoost model and train it. Unlike in the previous notebooks, instead of training a single model, we will use SageMakers hyperparameter tuning functionality to train multiple models and use the one that performs the best on the validation set.\n\n### Set up the training job\n\nFirst, we will set up a training job for our model. This is very similar to the way in which we constructed the training job in previous notebooks. Essentially this describes the *base* training job from which SageMaker will create refinements by changing some hyperparameters during the hyperparameter tuning job.",
"_____no_output_____"
]
],
[
[
"# We will need to know the name of the container that we want to use for training. SageMaker provides\n# a nice utility method to construct this for us.\ncontainer = get_image_uri(session.boto_region_name, 'xgboost')\n\n# We now specify the parameters we wish to use for our training job\ntraining_params = {}\n\n# We need to specify the permissions that this training job will have. For our purposes we can use\n# the same permissions that our current SageMaker session has.\ntraining_params['RoleArn'] = role\n\n# Here we describe the algorithm we wish to use. The most important part is the container which\n# contains the training code.\ntraining_params['AlgorithmSpecification'] = {\n \"TrainingImage\": container,\n \"TrainingInputMode\": \"File\"\n}\n\n# We also need to say where we would like the resulting model artifacts stored.\ntraining_params['OutputDataConfig'] = {\n \"S3OutputPath\": \"s3://\" + session.default_bucket() + \"/\" + prefix + \"/output\"\n}\n\n# We also need to set some parameters for the training job itself. Namely we need to describe what sort of\n# compute instance we wish to use along with a stopping condition to handle the case that there is\n# some sort of error and the training script doesn't terminate.\ntraining_params['ResourceConfig'] = {\n \"InstanceCount\": 1,\n \"InstanceType\": \"ml.m4.xlarge\",\n \"VolumeSizeInGB\": 5\n}\n \ntraining_params['StoppingCondition'] = {\n \"MaxRuntimeInSeconds\": 86400\n}\n\n# Next we set the algorithm specific hyperparameters. In this case, since we are setting up\n# a training job which will serve as the base training job for the eventual hyperparameter\n# tuning job, we only specify the _static_ hyperparameters. That is, the hyperparameters that\n# we do _not_ want SageMaker to change.\ntraining_params['StaticHyperParameters'] = {\n \"gamma\": \"4\",\n \"subsample\": \"0.8\",\n \"objective\": \"reg:linear\",\n \"early_stopping_rounds\": \"10\",\n \"num_round\": \"200\"\n}\n\n# Now we need to tell SageMaker where the data should be retrieved from.\ntraining_params['InputDataConfig'] = [\n {\n \"ChannelName\": \"train\",\n \"DataSource\": {\n \"S3DataSource\": {\n \"S3DataType\": \"S3Prefix\",\n \"S3Uri\": train_location,\n \"S3DataDistributionType\": \"FullyReplicated\"\n }\n },\n \"ContentType\": \"csv\",\n \"CompressionType\": \"None\"\n },\n {\n \"ChannelName\": \"validation\",\n \"DataSource\": {\n \"S3DataSource\": {\n \"S3DataType\": \"S3Prefix\",\n \"S3Uri\": val_location,\n \"S3DataDistributionType\": \"FullyReplicated\"\n }\n },\n \"ContentType\": \"csv\",\n \"CompressionType\": \"None\"\n }\n]",
"'get_image_uri' method will be deprecated in favor of 'ImageURIProvider' class in SageMaker Python SDK v2.\nThere is a more up to date SageMaker XGBoost image. To use the newer image, please set 'repo_version'='1.0-1'. For example:\n\tget_image_uri(region, 'xgboost', '1.0-1').\n"
]
],
[
[
"### Set up the tuning job\n\nNow that the *base* training job has been set up, we can describe the tuning job that we would like SageMaker to perform. In particular, like in the high level notebook, we will specify which hyperparameters we wish SageMaker to change and what range of values they may take on.\n\nIn addition, we specify the *number* of models to construct (`max_jobs`) and the number of those that can be trained in parallel (`max_parallel_jobs`). In the cell below we have chosen to train `20` models, of which we ask that SageMaker train `3` at a time in parallel. Note that this results in a total of `20` training jobs being executed which can take some time, in this case almost a half hour. With more complicated models this can take even longer so be aware!",
"_____no_output_____"
]
],
[
[
"# We need to construct a dictionary which specifies the tuning job we want SageMaker to perform\ntuning_job_config = {\n # First we specify which hyperparameters we want SageMaker to be able to vary,\n # and we specify the type and range of the hyperparameters.\n \"ParameterRanges\": {\n \"CategoricalParameterRanges\": [],\n \"ContinuousParameterRanges\": [\n {\n \"MaxValue\": \"0.5\",\n \"MinValue\": \"0.05\",\n \"Name\": \"eta\"\n },\n ],\n \"IntegerParameterRanges\": [\n {\n \"MaxValue\": \"12\",\n \"MinValue\": \"3\",\n \"Name\": \"max_depth\"\n },\n {\n \"MaxValue\": \"8\",\n \"MinValue\": \"2\",\n \"Name\": \"min_child_weight\"\n }\n ]},\n # We also need to specify how many models should be fit and how many can be fit in parallel\n \"ResourceLimits\": {\n \"MaxNumberOfTrainingJobs\": 20,\n \"MaxParallelTrainingJobs\": 3\n },\n # Here we specify how SageMaker should update the hyperparameters as new models are fit\n \"Strategy\": \"Bayesian\",\n # And lastly we need to specify how we'd like to determine which models are better or worse\n \"HyperParameterTuningJobObjective\": {\n \"MetricName\": \"validation:rmse\",\n \"Type\": \"Minimize\"\n }\n }",
"_____no_output_____"
]
],
[
[
"### Execute the tuning job\n\nNow that we've built the data structures that describe the tuning job we want SageMaker to execute, it is time to actually start the job.",
"_____no_output_____"
]
],
[
[
"# First we need to choose a name for the job. This is useful for if we want to recall information about our\n# tuning job at a later date. Note that SageMaker requires a tuning job name and that the name needs to\n# be unique, which we accomplish by appending the current timestamp.\ntuning_job_name = \"tuning-job\" + strftime(\"%Y-%m-%d-%H-%M-%S\", gmtime())\n\n# And now we ask SageMaker to create (and execute) the training job\nsession.sagemaker_client.create_hyper_parameter_tuning_job(HyperParameterTuningJobName = tuning_job_name,\n HyperParameterTuningJobConfig = tuning_job_config,\n TrainingJobDefinition = training_params)",
"_____no_output_____"
]
],
[
[
"The tuning job has now been created by SageMaker and is currently running. Since we need the output of the tuning job, we may wish to wait until it has finished. We can do so by asking SageMaker to output the logs generated by the tuning job and continue doing so until the job terminates.",
"_____no_output_____"
]
],
[
[
"session.wait_for_tuning_job(tuning_job_name)",
"............................................................................................................................................................................................................................................................................................................................................................!\n"
]
],
[
[
"### Build the model\n\nNow that the tuning job has finished, SageMaker has fit a number of models, the results of which are stored in a data structure which we can access using the name of the tuning job.",
"_____no_output_____"
]
],
[
[
"tuning_job_info = session.sagemaker_client.describe_hyper_parameter_tuning_job(HyperParameterTuningJobName=tuning_job_name)",
"_____no_output_____"
]
],
[
[
"Among the pieces of information included in the `tuning_job_info` object is the name of the training job which performed best out of all of the models that SageMaker fit to our data. Using this training job name we can get access to the resulting model artifacts, from which we can construct a model.",
"_____no_output_____"
]
],
[
[
"# We begin by asking SageMaker to describe for us the results of the best training job. The data\n# structure returned contains a lot more information than we currently need, try checking it out\n# yourself in more detail.\nbest_training_job_name = tuning_job_info['BestTrainingJob']['TrainingJobName']\ntraining_job_info = session.sagemaker_client.describe_training_job(TrainingJobName=best_training_job_name)\n\nmodel_artifacts = training_job_info['ModelArtifacts']['S3ModelArtifacts']",
"_____no_output_____"
],
[
"# Just like when we created a training job, the model name must be unique\nmodel_name = best_training_job_name + \"-model\"\n\n# We also need to tell SageMaker which container should be used for inference and where it should\n# retrieve the model artifacts from. In our case, the xgboost container that we used for training\n# can also be used for inference.\nprimary_container = {\n \"Image\": container,\n \"ModelDataUrl\": model_artifacts\n}\n\n# And lastly we construct the SageMaker model\nmodel_info = session.sagemaker_client.create_model(\n ModelName = model_name,\n ExecutionRoleArn = role,\n PrimaryContainer = primary_container)",
"_____no_output_____"
]
],
[
[
"## Step 5: Testing the model\n\nNow that we have fit our model to the training data, using the validation data to avoid overfitting, we can test our model. To do this we will make use of SageMaker's Batch Transform functionality. In other words, we need to set up and execute a batch transform job, similar to the way that we constructed the training job earlier.\n\n### Set up the batch transform job\n\nJust like when we were training our model, we first need to provide some information in the form of a data structure that describes the batch transform job which we wish to execute.\n\nWe will only be using some of the options available here but to see some of the additional options please see the SageMaker documentation for [creating a batch transform job](https://docs.aws.amazon.com/sagemaker/latest/dg/API_CreateTransformJob.html).",
"_____no_output_____"
]
],
[
[
"# Just like in each of the previous steps, we need to make sure to name our job and the name should be unique.\ntransform_job_name = 'boston-xgboost-batch-transform-' + strftime(\"%Y-%m-%d-%H-%M-%S\", gmtime())\n\n# Now we construct the data structure which will describe the batch transform job.\ntransform_request = \\\n{\n \"TransformJobName\": transform_job_name,\n \n # This is the name of the model that we created earlier.\n \"ModelName\": model_name,\n \n # This describes how many compute instances should be used at once. If you happen to be doing a very large\n # batch transform job it may be worth running multiple compute instances at once.\n \"MaxConcurrentTransforms\": 1,\n \n # This says how big each individual request sent to the model should be, at most. One of the things that\n # SageMaker does in the background is to split our data up into chunks so that each chunks stays under\n # this size limit.\n \"MaxPayloadInMB\": 6,\n \n # Sometimes we may want to send only a single sample to our endpoint at a time, however in this case each of\n # the chunks that we send should contain multiple samples of our input data.\n \"BatchStrategy\": \"MultiRecord\",\n \n # This next object describes where the output data should be stored. Some of the more advanced options which\n # we don't cover here also describe how SageMaker should collect output from various batches.\n \"TransformOutput\": {\n \"S3OutputPath\": \"s3://{}/{}/batch-bransform/\".format(session.default_bucket(),prefix)\n },\n \n # Here we describe our input data. Of course, we need to tell SageMaker where on S3 our input data is stored, in\n # addition we need to detail the characteristics of our input data. In particular, since SageMaker may need to\n # split our data up into chunks, it needs to know how the individual samples in our data file appear. In our\n # case each line is its own sample and so we set the split type to 'line'. We also need to tell SageMaker what\n # type of data is being sent, in this case csv, so that it can properly serialize the data.\n \"TransformInput\": {\n \"ContentType\": \"text/csv\",\n \"SplitType\": \"Line\",\n \"DataSource\": {\n \"S3DataSource\": {\n \"S3DataType\": \"S3Prefix\",\n \"S3Uri\": test_location,\n }\n }\n },\n \n # And lastly we tell SageMaker what sort of compute instance we would like it to use.\n \"TransformResources\": {\n \"InstanceType\": \"ml.m4.xlarge\",\n \"InstanceCount\": 1\n }\n}",
"_____no_output_____"
]
],
[
[
"### Execute the batch transform job\n\nNow that we have created the request data structure, it is time to as SageMaker to set up and run our batch transform job. Just like in the previous steps, SageMaker performs these tasks in the background so that if we want to wait for the transform job to terminate (and ensure the job is progressing) we can ask SageMaker to wait of the transform job to complete.",
"_____no_output_____"
]
],
[
[
"transform_response = session.sagemaker_client.create_transform_job(**transform_request)",
"_____no_output_____"
],
[
"transform_desc = session.wait_for_transform_job(transform_job_name)",
"............................................................!\n"
]
],
[
[
"### Analyze the results\n\nNow that the transform job has completed, the results are stored on S3 as we requested. Since we'd like to do a bit of analysis in the notebook we can use some notebook magic to copy the resulting output from S3 and save it locally.",
"_____no_output_____"
]
],
[
[
"transform_output = \"s3://{}/{}/batch-bransform/\".format(session.default_bucket(),prefix)",
"_____no_output_____"
],
[
"!aws s3 cp --recursive $transform_output $data_dir",
"download: s3://sagemaker-us-east-1-888201120197/boston-xgboost-tuning-LL/batch-bransform/test.csv.out to ../data/boston/test.csv.out\r\n"
]
],
[
[
"To see how well our model works we can create a simple scatter plot between the predicted and actual values. If the model was completely accurate the resulting scatter plot would look like the line $x=y$. As we can see, our model seems to have done okay but there is room for improvement.",
"_____no_output_____"
]
],
[
[
"Y_pred = pd.read_csv(os.path.join(data_dir, 'test.csv.out'), header=None)",
"_____no_output_____"
],
[
"plt.scatter(Y_test, Y_pred)\nplt.xlabel(\"Median Price\")\nplt.ylabel(\"Predicted Price\")\nplt.title(\"Median Price vs Predicted Price\")",
"_____no_output_____"
]
],
[
[
"## Optional: Clean up\n\nThe default notebook instance on SageMaker doesn't have a lot of excess disk space available. As you continue to complete and execute notebooks you will eventually fill up this disk space, leading to errors which can be difficult to diagnose. Once you are completely finished using a notebook it is a good idea to remove the files that you created along the way. Of course, you can do this from the terminal or from the notebook hub if you would like. The cell below contains some commands to clean up the created files from within the notebook.",
"_____no_output_____"
]
],
[
[
"# First we will remove all of the files contained in the data_dir directory\n!rm $data_dir/*\n\n# And then we delete the directory itself\n!rmdir $data_dir",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d04001d0fa14ff603cfd0c957fcbc36eb2dacf0f | 24,184 | ipynb | Jupyter Notebook | Exploratory-data-analysis-with-Julia.ipynb | schwartx/data-science-with-julia | 8823b7825bde9292b6ecd78e2c01d98334c4740d | [
"MIT"
] | null | null | null | Exploratory-data-analysis-with-Julia.ipynb | schwartx/data-science-with-julia | 8823b7825bde9292b6ecd78e2c01d98334c4740d | [
"MIT"
] | null | null | null | Exploratory-data-analysis-with-Julia.ipynb | schwartx/data-science-with-julia | 8823b7825bde9292b6ecd78e2c01d98334c4740d | [
"MIT"
] | null | null | null | 38.943639 | 2,459 | 0.468285 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d040081d1fbd3625663aee562e72ed6ad47b7d9f | 7,086 | ipynb | Jupyter Notebook | Python Programming Basic Assignment/Assignment_18.ipynb | kpsanjeet/Python-Programming-Basic-Assignment | 75b9304095cc79cf399bed35eab5bba88195e108 | [
"MIT"
] | null | null | null | Python Programming Basic Assignment/Assignment_18.ipynb | kpsanjeet/Python-Programming-Basic-Assignment | 75b9304095cc79cf399bed35eab5bba88195e108 | [
"MIT"
] | null | null | null | Python Programming Basic Assignment/Assignment_18.ipynb | kpsanjeet/Python-Programming-Basic-Assignment | 75b9304095cc79cf399bed35eab5bba88195e108 | [
"MIT"
] | null | null | null | 21.736196 | 184 | 0.470929 | [
[
[
"Question 1:",
"_____no_output_____"
]
],
[
[
"Create a function that takes a list of non-negative integers and strings and return a new list\nwithout the strings.\nExamples\nfilter_list([1, 2, \"a\", \"b\"]) ➞ [1, 2]\nfilter_list([1, \"a\", \"b\", 0, 15]) ➞ [1, 0, 15]\nfilter_list([1, 2, \"aasf\", \"1\", \"123\", 123]) ➞ [1, 2, 123]",
"_____no_output_____"
]
],
[
[
"def filter_list(lst):\n return list(filter(lambda x: type(x) == int,lst))\n\nprint(filter_list([1, 2, \"a\", \"b\"]))\nprint(filter_list([1, \"a\", \"b\", 0, 15]))\nprint(filter_list([1, 2, \"aasf\", \"1\", \"123\", 123]))",
"[1, 2]\n[1, 0, 15]\n[1, 2, 123]\n"
]
],
[
[
"Question 2:",
"_____no_output_____"
]
],
[
[
"The \"Reverser\" takes a string as input and returns that string in reverse order, with the\nopposite case.\nExamples\nreverse(\"Hello World\") ➞ \"DLROw OLLEh\"\nreverse(\"ReVeRsE\") ➞ \"eSrEvEr\"\nreverse(\"Radar\") ➞ \"RADAr\"",
"_____no_output_____"
]
],
[
[
"Answer :",
"_____no_output_____"
]
],
[
[
"def reverse(string):\n return string[::-1].swapcase()\n\nprint(reverse(\"Hello World\"))\nprint(reverse(\"ReVeRsE\"))\nprint(reverse(\"Radar\"))",
"DLROw OLLEh\neSrEvEr\nRADAr\n"
]
],
[
[
"Question 3:",
"_____no_output_____"
]
],
[
[
"You can assign variables from lists like this:\nlst = [1, 2, 3, 4, 5, 6]\nfirst = lst[0]\nmiddle = lst[1:-1]\nlast = lst[-1]\nprint(first) ➞ outputs 1\nprint(middle) ➞ outputs [2, 3, 4, 5]\nprint(last) ➞ outputs 6\nWith Python 3, you can assign variables from lists in a much more succinct way. Create\nvariables first, middle and last from the given list using destructuring assignment\n(check the Resources tab for some examples), where:\nfirst ➞ 1\nmiddle ➞ [2, 3, 4, 5]\nlast ➞ 6\nYour task is to unpack the list writeyourcodehere into three variables, being first, middle, and last, with middle being everything in between the first and last element. Then\nprint all three variables.",
"_____no_output_____"
]
],
[
[
"Answer :",
"_____no_output_____"
]
],
[
[
"def unpack_list(lst):\n first = lst[0]\n middle = lst[1:-1]\n last = lst[-1]\n return first,middle,last\n\n\nlst = [1, 2, 3, 4, 5, 6]\n\nfirst,middle,last = unpack_list(lst)\n\nprint(first)\nprint(middle)\nprint(last)",
"1\n[2, 3, 4, 5]\n6\n"
]
],
[
[
"Question 4:",
"_____no_output_____"
]
],
[
[
"Write a function that calculates the factorial of a number recursively. Examples\nfactorial(5) ➞ 120\nfactorial(3) ➞ 6\nfactorial(1) ➞ 1\nfactorial(0) ➞ 1",
"_____no_output_____"
]
],
[
[
"def factorial(num):\n if num<0:\n return f\"{num} is negative number.\"\n if num == 0 or num == 1:\n return 1\n else: \n return num*factorial(num-1)\n \n \n \nprint(factorial(5))\nprint(factorial(3))\nprint(factorial(1))\nprint(factorial(0))",
"120\n6\n1\n1\n"
]
],
[
[
"Question 5:",
"_____no_output_____"
]
],
[
[
"Write a function that moves all elements of one type to the end of the list.\nExamples\nmove_to_end([1, 3, 2, 4, 4, 1], 1) ➞ [3, 2, 4, 4, 1, 1]\n# Move all the 1s to the end of the array.\nmove_to_end([7, 8, 9, 1, 2, 3, 4], 9) ➞ [7, 8, 1, 2, 3, 4, 9]\nmove_to_end([\"a\", \"a\", \"a\", \"b\"], \"a\") ➞ [\"b\", \"a\", \"a\", \"a\"]",
"_____no_output_____"
]
],
[
[
"Answer :",
"_____no_output_____"
]
],
[
[
"def move_to_end(lst,x):\n lst.remove(x)\n lst.append(x)\n return lst\n\nprint(move_to_end([1, 3, 2, 4, 4, 1], 1))\nprint(move_to_end([7, 8, 9, 1, 2, 3, 4], 9))\nprint(move_to_end([\"a\", \"a\", \"a\", \"b\"], \"a\"))",
"[3, 2, 4, 4, 1, 1]\n[7, 8, 1, 2, 3, 4, 9]\n['a', 'a', 'b', 'a']\n"
]
]
] | [
"markdown",
"raw",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"raw",
"code",
"markdown",
"raw",
"markdown",
"code"
] | [
[
"markdown"
],
[
"raw"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
]
] |
d0401ab1be6cb9fa3cf8274146db23e1089571d7 | 22,965 | ipynb | Jupyter Notebook | Chapter04/weight_distribution.ipynb | jayanthvarma134/Computational-neuroscience | 55660999d4d56047fa370fdb906069ae194ffe0b | [
"MIT"
] | 3 | 2021-06-07T14:13:01.000Z | 2021-12-19T14:14:12.000Z | Chapter04/weight_distribution.ipynb | jayanthvarma134/Computational-neuroscience | 55660999d4d56047fa370fdb906069ae194ffe0b | [
"MIT"
] | 10 | 2021-04-24T15:55:44.000Z | 2021-06-11T18:56:43.000Z | Chapter04/weight_distribution.ipynb | jayanthvarma134/Computational-neuroscience | 55660999d4d56047fa370fdb906069ae194ffe0b | [
"MIT"
] | 5 | 2021-04-21T12:38:36.000Z | 2021-05-04T04:42:06.000Z | 234.336735 | 20,342 | 0.909123 | [
[
[
"# Weight distribution of Hebbian synapses in rate model\n\n%matplotlib inline\nfrom scipy.stats import norm\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nnn = 500 # number of nodes\nnpat = 1000 # number of patterns\n\n# Random pattern; firing rates are exponential distributed\n\nar = 40 # average firing rate of pattern\nrPre = -ar*np.log(np.random.rand(nn,npat)) # exponential distr. pre rates\nrPost = -ar*np.log(np.random.rand(1,npat)) # exponential distr. post rate\n\n# Weight matrix\n\nw = np.dot((rPost-ar),np.transpose(rPre-ar)) # Hebbian covariance rule\nw = w/np.sqrt(npat) # standard scaling to keep variance constant\nw = np.transpose(w)\nw = w/nn\n\n# Histogram plotting and fitting a gaussian\n\nmean,std=norm.fit(w)\nfig, ax = plt.subplots()\nax.hist(w, bins=21, density=\"true\")\nax.set_title(\"Normalized histograms of weight values\")\nax.set_xlabel(\"Synaptic Weight\")\nax.set_ylabel(\"Probability\")\nxmin, xmax = plt.xlim()\nx = np.linspace(xmin, xmax, 100)\ny = norm.pdf(x, mean, std)\nplt.plot(x, y)\n\nplt.show()\n\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
d0401b4d157d970c8c116e47bdb6338b65efd395 | 4,685 | ipynb | Jupyter Notebook | examples/ext_mod_fapar.ipynb | akuhnregnier/wildfires | 4d31cbdd4a1303ecebc391a35c73b8f07d8fe400 | [
"MIT"
] | 1 | 2021-01-30T15:38:32.000Z | 2021-01-30T15:38:32.000Z | examples/ext_mod_fapar.ipynb | akuhnregnier/wildfires | 4d31cbdd4a1303ecebc391a35c73b8f07d8fe400 | [
"MIT"
] | null | null | null | examples/ext_mod_fapar.ipynb | akuhnregnier/wildfires | 4d31cbdd4a1303ecebc391a35c73b8f07d8fe400 | [
"MIT"
] | null | null | null | 25.741758 | 98 | 0.504376 | [
[
[
"import warnings\nfrom datetime import datetime\n\nimport cartopy.crs as ccrs\nimport iris\nimport matplotlib.pyplot as plt\nimport numpy as np\nfrom tqdm import tqdm\n\nfrom wildfires.analysis import cube_plotting\nfrom wildfires.data import (\n Ext_MOD15A2H_fPAR,\n MOD15A2H_LAI_fPAR,\n dataset_times,\n dummy_lat_lon_cube,\n)\nfrom wildfires.logging_config import enable_logging\nfrom wildfires.utils import ensure_datetime, get_land_mask, match_shape\n\nwarnings.filterwarnings(\"ignore\", \"Collapsing a non-contiguous coordinate.*\")\n\nenable_logging(mode=\"jupyter\")",
"_____no_output_____"
],
[
"def mask_water(cube):\n assert isinstance(cube.data, np.ndarray)\n if not hasattr(cube.data, \"mask\"):\n cube.data = np.ma.MaskedArray(\n cube.data, mask=np.zeros_like(cube.data, dtype=np.bool_)\n )\n cube.data.mask |= ~match_shape(get_land_mask(), cube.shape)\n return cube",
"_____no_output_____"
],
[
"ext_mod = Ext_MOD15A2H_fPAR()\nmod = MOD15A2H_LAI_fPAR()",
"_____no_output_____"
],
[
"min_time, max_time, time_df = dataset_times((ext_mod, mod))\n# Discard anything but the year and month.\nmin_time = datetime(min_time.year, min_time.month, 1)\nmax_time = datetime(max_time.year, max_time.month, 1)\ntime_df",
"_____no_output_____"
],
[
"fapar_name = \"Fraction of Absorbed Photosynthetically Active Radiation\"",
"_____no_output_____"
],
[
"mod_fapar = mod.get_monthly_data(start=min_time, end=max_time).extract_cube(\n iris.Constraint(fapar_name)\n)\next_mod_fapar = ext_mod.get_monthly_data(start=min_time, end=max_time).extract_cube(\n iris.Constraint(fapar_name)\n)",
"_____no_output_____"
],
[
"mod_fapar.units = \"1\"",
"_____no_output_____"
],
[
"ext_mod_fapar",
"_____no_output_____"
],
[
"mod_fapar",
"_____no_output_____"
],
[
"fig_kwargs = dict(figsize=(17, 4), dpi=200)",
"_____no_output_____"
],
[
"for i in tqdm(range(mod_fapar.shape[0])):\n fig, axes = plt.subplots(\n 1, 3, subplot_kw=dict(projection=ccrs.Robinson()), **fig_kwargs\n )\n for ax, cube, title in zip(\n axes,\n (\n mod_fapar[i],\n ext_mod_fapar[i],\n dummy_lat_lon_cube(ext_mod_fapar[i].data - mod_fapar[i].data),\n ),\n (\"MOD\", \"Ext MOD\", \"Ext MOD - MOD\"),\n ):\n ax.set_title(title)\n kwargs = {}\n if \"-\" in title:\n kwargs = {\n **kwargs,\n **dict(\n cmap=\"RdBu_r\",\n cmap_midpoint=0,\n cmap_symmetric=True,\n vmin_vmax_percentiles=(2, 98),\n ),\n }\n\n cube_plotting(\n mask_water(cube),\n ax=ax,\n title=f\"{ensure_datetime(mod_fapar[i].coord('time').cell(0).point):%Y-%m}\",\n **kwargs,\n )",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0404a0872061dc8d9178b115cfa0b78d7a47650 | 30,385 | ipynb | Jupyter Notebook | eda/hyper-parameter_tuning/random_forest-Level0.ipynb | jimthompson5802/model-stacking-framework | 486461340c41072627a8c6c69ef6902297d2ada7 | [
"MIT"
] | 3 | 2018-06-10T23:45:52.000Z | 2019-08-04T08:08:05.000Z | eda/hyper-parameter_tuning/random_forest-Level0.ipynb | jimthompson5802/model-stacking-framework | 486461340c41072627a8c6c69ef6902297d2ada7 | [
"MIT"
] | 25 | 2018-06-09T11:08:00.000Z | 2019-02-06T22:47:26.000Z | eda/hyper-parameter_tuning/random_forest-Level0.ipynb | jimthompson5802/model-stacking-framework | 486461340c41072627a8c6c69ef6902297d2ada7 | [
"MIT"
] | 1 | 2018-06-10T23:45:53.000Z | 2018-06-10T23:45:53.000Z | 53.494718 | 119 | 0.710614 | [
[
[
"# required to get access to model_stacking package\nimport os\nos.chdir('../..')",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport os.path\nfrom framework.model_stacking import getConfigParameters\nimport pickle",
"retrieving configuration file: config.yml from current working directory\n"
],
[
"CONFIG = getConfigParameters()\nROOT_DIR = CONFIG['ROOT_DIR']\nDATA_DIR = CONFIG['DATA_DIR']\nID_VAR = CONFIG['ID_VAR']\nTARGET_VAR = CONFIG['TARGET_VAR']",
"_____no_output_____"
],
[
"from sklearn.ensemble import RandomForestRegressor as ThisModel\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.model_selection import RandomizedSearchCV\nfrom sklearn.metrics import mean_squared_error, make_scorer\nfrom scipy.stats import randint",
"_____no_output_____"
],
[
"FEATURE_SET = 'KFS01'\nMODEL_ALGO = 'randomforest_level0_'+FEATURE_SET+'_hyp.pkl'",
"_____no_output_____"
],
[
"PARAM_GRID = dict(this_model__n_estimators=randint(100,800),\n this_model__min_samples_split=randint(2,8),\n this_model__max_depth=randint(2,15))\n\nN_ITER = 30",
"_____no_output_____"
]
],
[
[
"## Get Training Data",
"_____no_output_____"
]
],
[
[
"# get training data\ntrain_df = pd.read_csv(os.path.join(ROOT_DIR,DATA_DIR,FEATURE_SET,'train.csv.gz'))",
"_____no_output_____"
],
[
"X_train = train_df.drop(ID_VAR + [TARGET_VAR],axis=1)\ny_train = train_df.loc[:,TARGET_VAR]",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"y_train.shape",
"_____no_output_____"
],
[
"y_train[:10]",
"_____no_output_____"
]
],
[
[
"## Setup pipeline for hyper-parameter tuning",
"_____no_output_____"
]
],
[
[
"# set up pipeline\npipe = Pipeline([('this_model',ThisModel(n_jobs=-1))])",
"_____no_output_____"
]
],
[
[
"this_scorer = make_scorer(lambda y, y_hat: np.sqrt(mean_squared_error(y,y_hat)),greater_is_better=False)",
"_____no_output_____"
]
],
[
[
"def kag_rmsle(y,y_hat):\n return np.sqrt(mean_squared_error(y,y_hat))\n\nthis_scorer = make_scorer(kag_rmsle, greater_is_better=False)",
"_____no_output_____"
],
[
"\ngrid_search = RandomizedSearchCV(pipe, \n param_distributions=PARAM_GRID,\n scoring=this_scorer,cv=5,\n n_iter=N_ITER,\n verbose=2,\n n_jobs=1,\n refit=False)",
"_____no_output_____"
],
[
"grid_search.fit(X_train,y_train)",
"Fitting 5 folds for each of 30 candidates, totalling 150 fits\n[CV] this_model__max_depth=2, this_model__min_samples_split=4, this_model__n_estimators=202 \n[CV] this_model__max_depth=2, this_model__min_samples_split=4, this_model__n_estimators=202, total= 4.7s\n[CV] this_model__max_depth=2, this_model__min_samples_split=4, this_model__n_estimators=202 \n"
],
[
"grid_search.best_params_",
"_____no_output_____"
],
[
"grid_search.best_score_",
"_____no_output_____"
],
[
"df = pd.DataFrame(grid_search.cv_results_).sort_values('rank_test_score')\ndf",
"_____no_output_____"
],
[
"hyper_parameters = dict(FeatureSet=FEATURE_SET,cv_run=df)\nwith open(os.path.join(CONFIG['ROOT_DIR'],'eda','hyper-parameter_tuning',MODEL_ALGO),'wb') as f:\n pickle.dump(hyper_parameters,f)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"raw",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"raw"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0404f8f54d165988c459f3794d424d13fb3edc6 | 48,779 | ipynb | Jupyter Notebook | notebooks/Profiling_parallel_code.ipynb | Eandreas1857/dsgrn_acdc | cfbccbd6cc27ffa4b0bd570ffb4f206b2ca9705c | [
"MIT"
] | null | null | null | notebooks/Profiling_parallel_code.ipynb | Eandreas1857/dsgrn_acdc | cfbccbd6cc27ffa4b0bd570ffb4f206b2ca9705c | [
"MIT"
] | null | null | null | notebooks/Profiling_parallel_code.ipynb | Eandreas1857/dsgrn_acdc | cfbccbd6cc27ffa4b0bd570ffb4f206b2ca9705c | [
"MIT"
] | null | null | null | 64.352243 | 223 | 0.523442 | [
[
[
"import DSGRN\nimport cProfile\nimport sys\nsys.setrecursionlimit(10**9)\nsys.path.insert(0,'/home/elizabeth/Desktop/GIT/dsgrn_acdc/src')\n\nimport PhenotypeGraphviz\nimport PhenotypeGraphFun\nimport CondensationGraph_iter",
"_____no_output_____"
],
[
"database = Database(\"/home/elizabeth/Desktop/ACDC/ACDC_Fullconn.db\") \nnetwork = Network(\"/home/elizabeth/Desktop/ACDC/ACDC_Fullconn\") ",
"_____no_output_____"
],
[
"parameter_graph = ParameterGraph(network)\nprint(parameter_graph.size())",
"2560000\n"
],
[
"AP35 = {\"Hb\":[0,2], \"Gt\":2, \"Kr\":0, \"Kni\":0}\nAP37 = {\"Hb\":2, \"Gt\":[0,2], \"Kr\":0, \"Kni\":0}\nAP40 = {\"Hb\":2, \"Gt\":0, \"Kr\":[0,2], \"Kni\":0} \nAP45 = {\"Hb\":[0,2], \"Gt\":0, \"Kr\":2, \"Kni\":0} \nAP47 = {\"Hb\":[0,2], \"Gt\":0, \"Kr\":2, \"Kni\":0} \nAP51 = {\"Hb\":0, \"Gt\":0, \"Kr\":2, \"Kni\":[0,2]} \nAP57 = {\"Hb\":0, \"Gt\":0, \"Kr\":[0,2], \"Kni\":2} \nAP61 = {\"Hb\":0, \"Gt\":0, \"Kr\":[0,2], \"Kni\":2}\nAP63 = {\"Hb\":0, \"Gt\":[0,2], \"Kr\":0, \"Kni\":2} \nAP67 = {\"Hb\":0, \"Gt\":2, \"Kr\":0, \"Kni\":[0,2]}",
"_____no_output_____"
],
[
"E = [[AP37], [AP40]]\nparamslist = get_paramslist_optimized(database, E, '=')",
"_____no_output_____"
]
],
[
[
"We can see in the next two cells that the parallelized code took off 16 minutes of computation time here. I haven't run on anything larger, so I dont know how it scales yet. There is another example past this as well.",
"_____no_output_____"
]
],
[
[
"cProfile.runctx('get_phenotype_graph_optimized(database, paramslist)', globals(), {'database':database,'paramslist':paramslist})",
" 1076681 function calls in 2674.774 seconds\n\n Ordered by: standard name\n\n ncalls tottime percall cumtime percall filename:lineno(function)\n 1 0.072 0.072 2674.774 2674.774 <string>:1(<module>)\n 1 2652.066 2652.066 2674.702 2674.702 PhenotypeGraphFun.py:121(get_phenotype_graph_optimized)\n 215335 5.489 0.000 5.489 0.000 PhenotypeGraphFun.py:143(<listcomp>)\n 215335 0.272 0.000 0.272 0.000 PhenotypeGraphFun.py:145(<listcomp>)\n 1 0.000 0.000 2674.774 2674.774 {built-in method builtins.exec}\n 215335 0.090 0.000 0.090 0.000 {built-in method builtins.len}\n 1 0.000 0.000 0.000 0.000 {built-in method from_iterable}\n 1 0.024 0.024 0.024 0.024 {built-in method fromkeys}\n 1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}\n 215335 3.431 0.000 3.431 0.000 {method 'intersection' of 'set' objects}\n 215335 13.330 0.000 13.330 0.000 {method 'pop' of 'list' objects}\n\n\n"
],
[
"cProfile.runctx('get_phenotype_graph_parallel(database, paramslist, num_processes)', globals(), {'database':database,'paramslist':paramslist, 'num_processes':8})",
" 3243 function calls (3213 primitive calls) in 1706.558 seconds\n\n Ordered by: standard name\n\n ncalls tottime percall cumtime percall filename:lineno(function)\n 31/26 0.000 0.000 0.001 0.000 <frozen importlib._bootstrap>:1009(_handle_fromlist)\n 8 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:103(release)\n 8 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:143(__init__)\n 8 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:147(__enter__)\n 8 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:151(__exit__)\n 8 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:157(_get_module_lock)\n 8 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:176(cb)\n 10/4 0.000 0.000 0.004 0.001 <frozen importlib._bootstrap>:211(_call_with_frames_removed)\n 106 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:222(_verbose_message)\n 7 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:307(__init__)\n 7 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:311(__enter__)\n 7 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:318(__exit__)\n 28 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:321(<genexpr>)\n 6 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:35(_new_module)\n 8 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:369(__init__)\n 13 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:403(cached)\n 35 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:416(parent)\n 7 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:424(has_location)\n 7 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:504(_init_module_attrs)\n 7 0.000 0.000 0.003 0.000 <frozen importlib._bootstrap>:576(module_from_spec)\n 8 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:58(__init__)\n 7/4 0.000 0.000 0.005 0.001 <frozen importlib._bootstrap>:663(_load_unlocked)\n 8 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:719(find_spec)\n 8 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:78(acquire)\n 8 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:792(find_spec)\n 25 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:855(__enter__)\n 25 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:859(__exit__)\n 1 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:873(_find_spec_legacy)\n 8 0.000 0.000 0.001 0.000 <frozen importlib._bootstrap>:882(_find_spec)\n 8/4 0.000 0.000 0.005 0.001 <frozen importlib._bootstrap>:948(_find_and_load_unlocked)\n 8/4 0.000 0.000 0.005 0.001 <frozen importlib._bootstrap>:978(_find_and_load)\n 1 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:1029(__init__)\n 1 0.000 0.000 0.003 0.003 <frozen importlib._bootstrap_external>:1040(create_module)\n 1 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:1048(exec_module)\n 21 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:1203(_path_importer_cache)\n 8 0.000 0.000 0.001 0.000 <frozen importlib._bootstrap_external>:1240(_get_spec)\n 8 0.000 0.000 0.001 0.000 <frozen importlib._bootstrap_external>:1272(find_spec)\n 7 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:1351(_get_spec)\n 19 0.000 0.000 0.001 0.000 <frozen importlib._bootstrap_external>:1356(find_spec)\n 1 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:1404(_fill_cache)\n 12 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:271(cache_from_source)\n 7 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:369(_get_cached)\n 19 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:40(_relax_case)\n 6 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:401(_check_name_wrapper)\n 6 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:438(_classify_pyc)\n 6 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:471(_validate_timestamp_pyc)\n 18 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:51(_r_long)\n 6 0.000 0.000 0.001 0.000 <frozen importlib._bootstrap_external>:523(_compile_bytecode)\n 97 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:56(_path_join)\n 7 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:574(spec_from_file_location)\n 97 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:58(<listcomp>)\n 12 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:62(_path_split)\n 6 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:719(create_module)\n 6/4 0.000 0.000 0.004 0.001 <frozen importlib._bootstrap_external>:722(exec_module)\n 32 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:74(_path_stat)\n 6 0.000 0.000 0.001 0.000 <frozen importlib._bootstrap_external>:793(get_code)\n 7 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:84(_path_is_mode_type)\n 6 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:884(__init__)\n 6 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:909(get_filename)\n 6 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:914(get_data)\n 7 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:93(_path_isfile)\n 6 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap_external>:951(path_stats)\n 1 0.091 0.091 1706.557 1706.557 <string>:1(<module>)\n 1 0.052 0.052 1706.467 1706.467 PhenotypeGraphFun.py:252(get_phenotype_graph_parallel)\n 8 0.000 0.000 0.000 0.000 __init__.py:212(_acquireLock)\n 8 0.000 0.000 0.000 0.000 __init__.py:221(_releaseLock)\n 11 0.000 0.000 0.000 0.000 _weakrefset.py:81(add)\n 1 0.000 0.000 0.000 0.000 connection.py:10(<module>)\n 1 0.000 0.000 0.000 0.000 connection.py:114(_ConnectionBase)\n 4 0.000 0.000 0.000 0.000 connection.py:117(__init__)\n 1 0.000 0.000 0.000 0.000 connection.py:348(Connection)\n 1 0.000 0.000 0.000 0.000 connection.py:422(Listener)\n 2 0.000 0.000 0.000 0.000 connection.py:506(Pipe)\n 1 0.000 0.000 0.000 0.000 connection.py:569(SocketListener)\n 1 0.000 0.000 0.000 0.000 connection.py:754(ConnectionWrapper)\n 1 0.000 0.000 0.000 0.000 connection.py:776(XmlListener)\n 2 0.000 0.000 0.004 0.002 context.py:109(SimpleQueue)\n 1 0.000 0.000 0.027 0.027 context.py:114(Pool)\n 6 0.000 0.000 0.000 0.000 context.py:186(get_context)\n 4 0.000 0.000 0.000 0.000 context.py:196(get_start_method)\n 1 0.000 0.000 0.000 0.000 context.py:232(get_context)\n 8 0.000 0.000 0.020 0.002 context.py:274(_Popen)\n 4 0.000 0.000 0.001 0.000 context.py:64(Lock)\n 32 0.000 0.000 0.002 0.000 iostream.py:197(schedule)\n 16 0.000 0.000 0.006 0.000 iostream.py:337(flush)\n 32 0.000 0.000 0.000 0.000 iostream.py:93(_event_pipe)\n 1 0.000 0.000 0.000 0.000 pool.py:10(<module>)\n 1 0.000 0.000 0.000 0.000 pool.py:146(Pool)\n 8 0.000 0.000 0.000 0.000 pool.py:152(Process)\n 1 0.000 0.000 0.026 0.026 pool.py:155(__init__)\n 1 0.000 0.000 0.021 0.021 pool.py:227(_repopulate_pool)\n 1 0.000 0.000 0.004 0.004 pool.py:250(_setup_queues)\n 1 0.000 0.000 0.001 0.001 pool.py:367(map_async)\n 1 0.000 0.000 0.001 0.001 pool.py:375(_map_async)\n 1 0.000 0.000 0.000 0.000 pool.py:53(RemoteTraceback)\n 1 0.000 0.000 0.000 0.000 pool.py:59(ExceptionWithTraceback)\n 1 0.000 0.000 0.000 0.000 pool.py:629(ApplyResult)\n 1 0.000 0.000 0.000 0.000 pool.py:631(__init__)\n 1 0.000 0.000 0.000 0.000 pool.py:639(ready)\n 1 0.000 0.000 1706.387 1706.387 pool.py:647(wait)\n 1 0.000 0.000 1706.387 1706.387 pool.py:650(get)\n 1 0.000 0.000 0.000 0.000 pool.py:674(MapResult)\n 1 0.001 0.001 0.001 0.001 pool.py:676(__init__)\n 1 0.000 0.000 0.000 0.000 pool.py:715(IMapIterator)\n 1 0.000 0.000 0.000 0.000 pool.py:76(MaybeEncodingError)\n 1 0.000 0.000 0.000 0.000 pool.py:779(IMapUnorderedIterator)\n 1 0.000 0.000 0.000 0.000 pool.py:793(ThreadPool)\n 1 0.000 0.000 0.000 0.000 popen_fork.py:1(<module>)\n 1 0.000 0.000 0.000 0.000 popen_fork.py:13(Popen)\n 8 0.000 0.000 0.019 0.002 popen_fork.py:16(__init__)\n 28 0.000 0.000 0.000 0.000 popen_fork.py:25(poll)\n 8 0.000 0.000 0.013 0.002 popen_fork.py:67(_launch)\n 8 0.000 0.000 0.020 0.003 process.py:101(start)\n 8 0.000 0.000 0.000 0.000 process.py:180(name)\n 8 0.000 0.000 0.000 0.000 process.py:184(name)\n 8 0.000 0.000 0.000 0.000 process.py:196(daemon)\n 4 0.000 0.000 0.000 0.000 process.py:36(current_process)\n 8 0.000 0.000 0.000 0.000 process.py:53(_cleanup)\n 8 0.000 0.000 0.000 0.000 process.py:72(__init__)\n 16 0.000 0.000 0.000 0.000 process.py:85(<genexpr>)\n 8 0.000 0.000 0.000 0.000 process.py:90(_check_closed)\n 1 0.000 0.000 0.003 0.003 queues.py:10(<module>)\n 1 0.000 0.000 0.000 0.000 queues.py:286(JoinableQueue)\n 1 0.000 0.000 0.000 0.000 queues.py:328(SimpleQueue)\n 2 0.000 0.000 0.001 0.000 queues.py:330(__init__)\n 1 0.000 0.000 0.000 0.000 queues.py:34(Queue)\n 32 0.000 0.000 0.000 0.000 random.py:224(_randbelow)\n 32 0.000 0.000 0.000 0.000 random.py:256(choice)\n 1 0.000 0.000 0.000 0.000 random.py:88(__init__)\n 1 0.000 0.000 0.000 0.000 random.py:97(seed)\n 1 0.000 0.000 0.000 0.000 reduction.py:43(register)\n 1 0.000 0.000 0.000 0.000 six.py:184(find_module)\n 32 0.001 0.000 0.001 0.000 socket.py:342(send)\n 1 0.000 0.000 0.000 0.000 synchronize.py:11(<module>)\n 4 0.000 0.000 0.000 0.000 synchronize.py:114(_make_name)\n 1 0.000 0.000 0.000 0.000 synchronize.py:123(Semaphore)\n 1 0.000 0.000 0.000 0.000 synchronize.py:142(BoundedSemaphore)\n 1 0.000 0.000 0.000 0.000 synchronize.py:159(Lock)\n 4 0.000 0.000 0.000 0.000 synchronize.py:161(__init__)\n 1 0.000 0.000 0.000 0.000 synchronize.py:184(RLock)\n 1 0.000 0.000 0.000 0.000 synchronize.py:210(Condition)\n 1 0.000 0.000 0.000 0.000 synchronize.py:321(Event)\n 1 0.000 0.000 0.000 0.000 synchronize.py:360(Barrier)\n 1 0.000 0.000 0.000 0.000 synchronize.py:46(SemLock)\n 4 0.000 0.000 0.000 0.000 synchronize.py:50(__init__)\n 4 0.000 0.000 0.000 0.000 synchronize.py:90(_make_methods)\n 4 0.000 0.000 0.000 0.000 tempfile.py:142(rng)\n 4 0.000 0.000 0.000 0.000 tempfile.py:153(__next__)\n 4 0.000 0.000 0.000 0.000 tempfile.py:156(<listcomp>)\n 48 0.000 0.000 0.000 0.000 threading.py:1050(_wait_for_tstate_lock)\n 48 0.000 0.000 0.000 0.000 threading.py:1092(is_alive)\n 3 0.000 0.000 0.000 0.000 threading.py:1116(daemon)\n 3 0.000 0.000 0.000 0.000 threading.py:1131(daemon)\n 3 0.000 0.000 0.000 0.000 threading.py:1225(current_thread)\n 20 0.000 0.000 0.000 0.000 threading.py:216(__init__)\n 20 0.000 0.000 0.000 0.000 threading.py:240(__enter__)\n 20 0.000 0.000 0.000 0.000 threading.py:243(__exit__)\n 19 0.000 0.000 0.000 0.000 threading.py:249(_release_save)\n 19 0.000 0.000 0.000 0.000 threading.py:252(_acquire_restore)\n 19 0.000 0.000 0.000 0.000 threading.py:255(_is_owned)\n 19 0.000 0.000 1706.391 89.810 threading.py:264(wait)\n 20 0.000 0.000 0.000 0.000 threading.py:499(__init__)\n 55 0.000 0.000 0.000 0.000 threading.py:507(is_set)\n 20 0.000 0.000 1706.391 85.320 threading.py:534(wait)\n 3 0.000 0.000 0.000 0.000 threading.py:728(_newname)\n 3 0.000 0.000 0.000 0.000 threading.py:763(__init__)\n 3 0.000 0.000 0.001 0.000 threading.py:834(start)\n 1 0.000 0.000 0.000 0.000 util.py:10(<module>)\n 4 0.000 0.000 0.000 0.000 util.py:148(register_after_fork)\n 1 0.000 0.000 0.000 0.000 util.py:159(Finalize)\n 9 0.000 0.000 0.000 0.000 util.py:163(__init__)\n 1 0.000 0.000 0.000 0.000 util.py:345(ForkAwareThreadLock)\n 1 0.000 0.000 0.000 0.000 util.py:362(ForkAwareLocal)\n 8 0.000 0.000 0.006 0.001 util.py:410(_flush_std_streams)\n 12 0.000 0.000 0.000 0.000 util.py:48(debug)\n 1 0.000 0.000 0.000 0.000 weakref.py:102(__init__)\n 4 0.000 0.000 0.000 0.000 weakref.py:165(__setitem__)\n 1 0.000 0.000 0.000 0.000 weakref.py:290(update)\n 4 0.000 0.000 0.000 0.000 weakref.py:336(__new__)\n 4 0.000 0.000 0.000 0.000 weakref.py:341(__init__)\n 4 0.000 0.000 0.000 0.000 {built-in method __new__ of type object at 0x565487df0240}\n 6 0.000 0.000 0.000 0.000 {built-in method _imp._fix_co_filename}\n 41 0.000 0.000 0.000 0.000 {built-in method _imp.acquire_lock}\n 1 0.003 0.003 0.003 0.003 {built-in method _imp.create_dynamic}\n 1 0.000 0.000 0.000 0.000 {built-in method _imp.exec_dynamic}\n 2 0.000 0.000 0.000 0.000 {built-in method _imp.is_builtin}\n 8 0.000 0.000 0.000 0.000 {built-in method _imp.is_frozen}\n 16 0.000 0.000 0.000 0.000 {built-in method _imp.lock_held}\n 41 0.000 0.000 0.000 0.000 {built-in method _imp.release_lock}\n 55 0.000 0.000 0.000 0.000 {built-in method _thread.allocate_lock}\n 19 0.000 0.000 0.000 0.000 {built-in method _thread.get_ident}\n 3 0.000 0.000 0.000 0.000 {built-in method _thread.start_new_thread}\n 1 0.000 0.000 0.000 0.000 {built-in method atexit.register}\n 30 0.000 0.000 0.000 0.000 {built-in method builtins.__build_class__}\n 2 0.000 0.000 0.001 0.000 {built-in method builtins.__import__}\n 7 0.000 0.000 0.000 0.000 {built-in method builtins.any}\n 1 0.000 0.000 0.000 0.000 {built-in method builtins.divmod}\n 7/1 0.000 0.000 1706.558 1706.558 {built-in method builtins.exec}\n 46 0.000 0.000 0.000 0.000 {built-in method builtins.getattr}\n 80 0.000 0.000 0.000 0.000 {built-in method builtins.hasattr}\n 4 0.000 0.000 0.000 0.000 {built-in method builtins.id}\n 50 0.000 0.000 0.000 0.000 {built-in method builtins.isinstance}\n 58 0.000 0.000 0.000 0.000 {built-in method builtins.len}\n 26 0.000 0.000 0.000 0.000 {built-in method builtins.next}\n 6 0.000 0.000 0.000 0.000 {built-in method builtins.setattr}\n 18 0.000 0.000 0.000 0.000 {built-in method from_bytes}\n 1 0.000 0.000 0.000 0.000 {built-in method from_iterable}\n 6 0.001 0.000 0.001 0.000 {built-in method marshal.loads}\n 8 0.000 0.000 0.000 0.000 {built-in method posix.close}\n 8 0.012 0.001 0.012 0.001 {built-in method posix.fork}\n 19 0.000 0.000 0.000 0.000 {built-in method posix.fspath}\n 1 0.000 0.000 0.000 0.000 {built-in method posix.getcwd}\n 29 0.000 0.000 0.000 0.000 {built-in method posix.getpid}\n 1 0.000 0.000 0.000 0.000 {built-in method posix.listdir}\n 10 0.000 0.000 0.000 0.000 {built-in method posix.pipe}\n 32 0.000 0.000 0.000 0.000 {built-in method posix.stat}\n 1 0.000 0.000 0.000 0.000 {built-in method posix.sysconf}\n 28 0.000 0.000 0.000 0.000 {built-in method posix.waitpid}\n 1 0.000 0.000 0.000 0.000 {function Random.seed at 0x7f6a716928c0}\n 20 0.000 0.000 0.000 0.000 {method '__enter__' of '_thread.lock' objects}\n 20 0.000 0.000 0.000 0.000 {method '__exit__' of '_thread.lock' objects}\n 8 0.000 0.000 0.000 0.000 {method 'acquire' of '_thread.RLock' objects}\n 124 1706.391 13.761 1706.391 13.761 {method 'acquire' of '_thread.lock' objects}\n 19 0.000 0.000 0.000 0.000 {method 'add' of 'set' objects}\n 51 0.000 0.000 0.000 0.000 {method 'append' of 'collections.deque' objects}\n 8 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}\n 32 0.000 0.000 0.000 0.000 {method 'bit_length' of 'int' objects}\n 8 0.000 0.000 0.000 0.000 {method 'copy' of 'dict' objects}\n 1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}\n 8 0.000 0.000 0.000 0.000 {method 'endswith' of 'str' objects}\n 3 0.000 0.000 0.000 0.000 {method 'format' of 'str' objects}\n 24 0.000 0.000 0.000 0.000 {method 'get' of 'dict' objects}\n 48 0.000 0.000 0.000 0.000 {method 'getrandbits' of '_random.Random' objects}\n 121 0.000 0.000 0.000 0.000 {method 'join' of 'str' objects}\n 1 0.000 0.000 0.000 0.000 {method 'put' of '_queue.SimpleQueue' objects}\n 6 0.000 0.000 0.000 0.000 {method 'read' of '_io.FileIO' objects}\n 8 0.000 0.000 0.000 0.000 {method 'release' of '_thread.RLock' objects}\n 19 0.000 0.000 0.000 0.000 {method 'release' of '_thread.lock' objects}\n 8 0.000 0.000 0.000 0.000 {method 'replace' of 'str' objects}\n 92 0.000 0.000 0.000 0.000 {method 'rpartition' of 'str' objects}\n 206 0.000 0.000 0.000 0.000 {method 'rstrip' of 'str' objects}\n 2 0.000 0.000 0.000 0.000 {method 'setter' of 'property' objects}\n 2 0.000 0.000 0.000 0.000 {method 'startswith' of 'str' objects}\n\n\n"
],
[
"database = Database(\"/home/elizabeth/Desktop/ACDC/ACDC_FullconnE.db\") \nnetwork = Network(\"/home/elizabeth/Desktop/ACDC/ACDC_FullconnE\")\nparameter_graph = ParameterGraph(network)\nprint(parameter_graph.size())",
"38416\n"
],
[
"AP35 = {\"Hb\":[0,2], \"Gt\":2, \"Kr\":0, \"Kni\":0}\nAP37 = {\"Hb\":2, \"Gt\":[0,1], \"Kr\":0, \"Kni\":0}\nAP40 = {\"Hb\":2, \"Gt\":1, \"Kr\":[0,1], \"Kni\":0} #edit\nAP45 = {\"Hb\":[0,1], \"Gt\":1, \"Kr\":2, \"Kni\":0} #edit\nAP47 = {\"Hb\":[0,1], \"Gt\":0, \"Kr\":2, \"Kni\":0} \nAP51 = {\"Hb\":1, \"Gt\":0, \"Kr\":2, \"Kni\":[0,1]} #edit\nAP57 = {\"Hb\":1, \"Gt\":0, \"Kr\":[0,1], \"Kni\":2} #edit\nAP61 = {\"Hb\":0, \"Gt\":0, \"Kr\":[0,1], \"Kni\":2}\nAP63 = {\"Hb\":0, \"Gt\":[0,1], \"Kr\":1, \"Kni\":2} #edit\nAP67 = {\"Hb\":0, \"Gt\":2, \"Kr\":1, \"Kni\":[0,1]} #edit",
"_____no_output_____"
],
[
"E = [[AP37], [AP40], [AP45], [AP47], [AP51], [AP57], [AP61], [AP63], [AP67]]\nparamslist = get_paramslist(database, E, '<')",
"_____no_output_____"
]
],
[
[
"This one is smaller than the first example, but I feel shows promising results in speeding up computation time. ",
"_____no_output_____"
]
],
[
[
"cProfile.runctx('get_phenotype_graph_optimized(database, paramslist)', globals(), {'database':database,'paramslist':paramslist})",
" 61026 function calls in 1.541 seconds\n\n Ordered by: standard name\n\n ncalls tottime percall cumtime percall filename:lineno(function)\n 1 0.004 0.004 1.541 1.541 <string>:1(<module>)\n 1 1.480 1.480 1.537 1.537 PhenotypeGraphFun.py:121(get_phenotype_graph_optimized)\n 12204 0.019 0.000 0.019 0.000 PhenotypeGraphFun.py:143(<listcomp>)\n 12204 0.006 0.000 0.006 0.000 PhenotypeGraphFun.py:145(<listcomp>)\n 1 0.000 0.000 1.541 1.541 {built-in method builtins.exec}\n 12204 0.001 0.000 0.001 0.000 {built-in method builtins.len}\n 1 0.000 0.000 0.000 0.000 {built-in method from_iterable}\n 1 0.001 0.001 0.001 0.001 {built-in method fromkeys}\n 1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}\n 12204 0.018 0.000 0.018 0.000 {method 'intersection' of 'set' objects}\n 12204 0.013 0.000 0.013 0.000 {method 'pop' of 'list' objects}\n\n\n"
],
[
"cProfile.runctx('get_phenotype_graph_parallel(database, paramslist, num_processes)', globals(), {'database':database,'paramslist':paramslist, 'num_processes':8})",
" 1871 function calls in 0.325 seconds\n\n Ordered by: standard name\n\n ncalls tottime percall cumtime percall filename:lineno(function)\n 15 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:1009(_handle_fromlist)\n 15 0.000 0.000 0.000 0.000 <frozen importlib._bootstrap>:416(parent)\n 1 0.004 0.004 0.325 0.325 <string>:1(<module>)\n 1 0.003 0.003 0.321 0.321 PhenotypeGraphFun.py:252(get_phenotype_graph_parallel)\n 8 0.000 0.000 0.000 0.000 __init__.py:212(_acquireLock)\n 8 0.000 0.000 0.000 0.000 __init__.py:221(_releaseLock)\n 11 0.000 0.000 0.000 0.000 _weakrefset.py:81(add)\n 4 0.000 0.000 0.000 0.000 connection.py:117(__init__)\n 2 0.000 0.000 0.000 0.000 connection.py:506(Pipe)\n 2 0.000 0.000 0.000 0.000 context.py:109(SimpleQueue)\n 1 0.000 0.000 0.028 0.028 context.py:114(Pool)\n 6 0.000 0.000 0.000 0.000 context.py:186(get_context)\n 4 0.000 0.000 0.000 0.000 context.py:196(get_start_method)\n 1 0.000 0.000 0.000 0.000 context.py:232(get_context)\n 8 0.000 0.000 0.024 0.003 context.py:274(_Popen)\n 4 0.000 0.000 0.000 0.000 context.py:64(Lock)\n 32 0.000 0.000 0.001 0.000 iostream.py:197(schedule)\n 16 0.000 0.000 0.005 0.000 iostream.py:337(flush)\n 32 0.000 0.000 0.000 0.000 iostream.py:93(_event_pipe)\n 8 0.000 0.000 0.001 0.000 pool.py:152(Process)\n 1 0.000 0.000 0.028 0.028 pool.py:155(__init__)\n 1 0.000 0.000 0.027 0.027 pool.py:227(_repopulate_pool)\n 1 0.000 0.000 0.000 0.000 pool.py:250(_setup_queues)\n 1 0.000 0.000 0.000 0.000 pool.py:367(map_async)\n 1 0.000 0.000 0.000 0.000 pool.py:375(_map_async)\n 1 0.000 0.000 0.000 0.000 pool.py:631(__init__)\n 1 0.000 0.000 0.000 0.000 pool.py:639(ready)\n 1 0.000 0.000 0.290 0.290 pool.py:647(wait)\n 1 0.000 0.000 0.290 0.290 pool.py:650(get)\n 1 0.000 0.000 0.000 0.000 pool.py:676(__init__)\n 8 0.000 0.000 0.024 0.003 popen_fork.py:16(__init__)\n 220 0.000 0.000 0.001 0.000 popen_fork.py:25(poll)\n 8 0.000 0.000 0.018 0.002 popen_fork.py:67(_launch)\n 8 0.000 0.000 0.026 0.003 process.py:101(start)\n 8 0.000 0.000 0.000 0.000 process.py:180(name)\n 8 0.000 0.000 0.000 0.000 process.py:184(name)\n 8 0.000 0.000 0.000 0.000 process.py:196(daemon)\n 4 0.000 0.000 0.000 0.000 process.py:36(current_process)\n 8 0.000 0.000 0.002 0.000 process.py:53(_cleanup)\n 8 0.000 0.000 0.000 0.000 process.py:72(__init__)\n 16 0.000 0.000 0.000 0.000 process.py:85(<genexpr>)\n 8 0.000 0.000 0.000 0.000 process.py:90(_check_closed)\n 2 0.000 0.000 0.000 0.000 queues.py:330(__init__)\n 32 0.000 0.000 0.000 0.000 random.py:224(_randbelow)\n 32 0.000 0.000 0.000 0.000 random.py:256(choice)\n 32 0.000 0.000 0.000 0.000 socket.py:342(send)\n 4 0.000 0.000 0.000 0.000 synchronize.py:114(_make_name)\n 4 0.000 0.000 0.000 0.000 synchronize.py:161(__init__)\n 4 0.000 0.000 0.000 0.000 synchronize.py:50(__init__)\n 4 0.000 0.000 0.000 0.000 synchronize.py:90(_make_methods)\n 4 0.000 0.000 0.000 0.000 tempfile.py:142(rng)\n 4 0.000 0.000 0.000 0.000 tempfile.py:153(__next__)\n 4 0.000 0.000 0.000 0.000 tempfile.py:156(<listcomp>)\n 48 0.000 0.000 0.000 0.000 threading.py:1050(_wait_for_tstate_lock)\n 48 0.000 0.000 0.000 0.000 threading.py:1092(is_alive)\n 3 0.000 0.000 0.000 0.000 threading.py:1116(daemon)\n 3 0.000 0.000 0.000 0.000 threading.py:1131(daemon)\n 3 0.000 0.000 0.000 0.000 threading.py:1225(current_thread)\n 20 0.000 0.000 0.000 0.000 threading.py:216(__init__)\n 20 0.000 0.000 0.000 0.000 threading.py:240(__enter__)\n 20 0.000 0.000 0.000 0.000 threading.py:243(__exit__)\n 19 0.000 0.000 0.000 0.000 threading.py:249(_release_save)\n 19 0.000 0.000 0.000 0.000 threading.py:252(_acquire_restore)\n 19 0.000 0.000 0.000 0.000 threading.py:255(_is_owned)\n 19 0.000 0.000 0.294 0.015 threading.py:264(wait)\n 20 0.000 0.000 0.000 0.000 threading.py:499(__init__)\n 55 0.000 0.000 0.000 0.000 threading.py:507(is_set)\n 20 0.000 0.000 0.294 0.015 threading.py:534(wait)\n 3 0.000 0.000 0.000 0.000 threading.py:728(_newname)\n 3 0.000 0.000 0.000 0.000 threading.py:763(__init__)\n 3 0.000 0.000 0.000 0.000 threading.py:834(start)\n 4 0.000 0.000 0.000 0.000 util.py:148(register_after_fork)\n 9 0.000 0.000 0.000 0.000 util.py:163(__init__)\n 8 0.000 0.000 0.005 0.001 util.py:410(_flush_std_streams)\n 12 0.000 0.000 0.000 0.000 util.py:48(debug)\n 4 0.000 0.000 0.000 0.000 weakref.py:165(__setitem__)\n 4 0.000 0.000 0.000 0.000 weakref.py:336(__new__)\n 4 0.000 0.000 0.000 0.000 weakref.py:341(__init__)\n 4 0.000 0.000 0.000 0.000 {built-in method __new__ of type object at 0x565487df0240}\n 16 0.000 0.000 0.000 0.000 {built-in method _imp.lock_held}\n 39 0.000 0.000 0.000 0.000 {built-in method _thread.allocate_lock}\n 3 0.000 0.000 0.000 0.000 {built-in method _thread.get_ident}\n 3 0.000 0.000 0.000 0.000 {built-in method _thread.start_new_thread}\n 1 0.000 0.000 0.000 0.000 {built-in method builtins.divmod}\n 1 0.000 0.000 0.325 0.325 {built-in method builtins.exec}\n 4 0.000 0.000 0.000 0.000 {built-in method builtins.getattr}\n 16 0.000 0.000 0.000 0.000 {built-in method builtins.hasattr}\n 4 0.000 0.000 0.000 0.000 {built-in method builtins.id}\n 9 0.000 0.000 0.000 0.000 {built-in method builtins.isinstance}\n 37 0.000 0.000 0.000 0.000 {built-in method builtins.len}\n 26 0.000 0.000 0.000 0.000 {built-in method builtins.next}\n 1 0.000 0.000 0.000 0.000 {built-in method from_iterable}\n 8 0.000 0.000 0.000 0.000 {built-in method posix.close}\n 8 0.017 0.002 0.018 0.002 {built-in method posix.fork}\n 29 0.000 0.000 0.000 0.000 {built-in method posix.getpid}\n 10 0.000 0.000 0.000 0.000 {built-in method posix.pipe}\n 220 0.001 0.000 0.001 0.000 {built-in method posix.waitpid}\n 20 0.000 0.000 0.000 0.000 {method '__enter__' of '_thread.lock' objects}\n 20 0.000 0.000 0.000 0.000 {method '__exit__' of '_thread.lock' objects}\n 8 0.000 0.000 0.000 0.000 {method 'acquire' of '_thread.RLock' objects}\n 124 0.294 0.002 0.294 0.002 {method 'acquire' of '_thread.lock' objects}\n 19 0.000 0.000 0.000 0.000 {method 'add' of 'set' objects}\n 51 0.000 0.000 0.000 0.000 {method 'append' of 'collections.deque' objects}\n 8 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}\n 32 0.000 0.000 0.000 0.000 {method 'bit_length' of 'int' objects}\n 8 0.000 0.000 0.000 0.000 {method 'copy' of 'dict' objects}\n 1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}\n 8 0.000 0.000 0.000 0.000 {method 'get' of 'dict' objects}\n 53 0.000 0.000 0.000 0.000 {method 'getrandbits' of '_random.Random' objects}\n 12 0.000 0.000 0.000 0.000 {method 'join' of 'str' objects}\n 1 0.000 0.000 0.000 0.000 {method 'put' of '_queue.SimpleQueue' objects}\n 8 0.000 0.000 0.000 0.000 {method 'release' of '_thread.RLock' objects}\n 19 0.000 0.000 0.000 0.000 {method 'release' of '_thread.lock' objects}\n 8 0.000 0.000 0.000 0.000 {method 'replace' of 'str' objects}\n 15 0.000 0.000 0.000 0.000 {method 'rpartition' of 'str' objects}\n\n\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d040555e21b256d6512941d475e9eda9e1f39a84 | 456,883 | ipynb | Jupyter Notebook | Notebooks/Scala/04 Using Delta Lake in Azure Synapse.ipynb | buswrecker/Synapse | dc94db93659ccd9c478b22b406154e97aec8255e | [
"MIT"
] | 256 | 2020-04-15T22:55:22.000Z | 2022-03-28T19:40:28.000Z | Notebooks/Scala/04 Using Delta Lake in Azure Synapse.ipynb | buswrecker/Synapse | dc94db93659ccd9c478b22b406154e97aec8255e | [
"MIT"
] | 27 | 2020-04-16T21:58:29.000Z | 2022-03-28T18:51:28.000Z | Notebooks/Scala/04 Using Delta Lake in Azure Synapse.ipynb | isabella232/Synapse-1 | 52d7e3b952a183e5dce13a1b51b8139f43ac114f | [
"MIT"
] | 241 | 2020-04-16T21:39:21.000Z | 2022-03-26T19:24:18.000Z | 31.67739 | 6,049 | 0.281556 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d04059d9950c86e6bc50e191bf5033b99e3c777f | 16,396 | ipynb | Jupyter Notebook | DeepRL_For_HPE/FC_RNN_Evaluater.ipynb | muratcancicek/Deep_RL_For_Head_Pose_Est | b3436a61a44d20d8bcfd1341792e0533e3ff9fc2 | [
"Apache-2.0"
] | null | null | null | DeepRL_For_HPE/FC_RNN_Evaluater.ipynb | muratcancicek/Deep_RL_For_Head_Pose_Est | b3436a61a44d20d8bcfd1341792e0533e3ff9fc2 | [
"Apache-2.0"
] | null | null | null | DeepRL_For_HPE/FC_RNN_Evaluater.ipynb | muratcancicek/Deep_RL_For_Head_Pose_Est | b3436a61a44d20d8bcfd1341792e0533e3ff9fc2 | [
"Apache-2.0"
] | null | null | null | 44.313514 | 261 | 0.593193 | [
[
[
"from FC_RNN_Evaluater.FC_RNN_Evaluater import *\nfrom FC_RNN_Evaluater.Stateful_FC_RNN_Configuration import *\nfrom FC_RNN_Evaluater.runFC_RNN_Experiment import *",
"/home/mcicek/anaconda3/lib/python3.6/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
],
[
"from keras import Model\nfrom keras.layers import TimeDistributed, LSTM, Dense, Dropout, Flatten, Input",
"_____no_output_____"
],
[
"def getFinalModel(timesteps = timesteps, lstm_nodes = lstm_nodes, lstm_dropout = lstm_dropout, \n lstm_recurrent_dropout = lstm_recurrent_dropout, num_outputs = num_outputs, \n lr = learning_rate, include_vgg_top = include_vgg_top, use_vgg16 = use_vgg16):\n if use_vgg16:\n modelID = 'VGG16' \n inp = (224, 224, 3)\n modelPackage = vgg16\n margins = (8, 8, 48, 48)\n Target_Frame_Shape = (240, 320, 3)\n cnn_model = vgg16.VGG16(weights='imagenet', input_shape = inp, include_top=include_vgg_top) \n\n def preprocess_input(imagePath): return preprocess_input_for_model(imagePath, Target_Frame_Shape, margins, modelPackage)\n \n if include_vgg_top:\n modelID = modelID + '_inc_top'\n cnn_model.layers.pop()\n cnn_model.outputs = [cnn_model.layers[-1].output]\n cnn_model.output_layers = [cnn_model.layers[-1]] \n cnn_model.layers[-1].outbound_nodes = []\n x = cnn_model.layers[-1].output\n x = Dense(1024, activation='relu', name='predictions')(x)\n model1 = Model(input=cnn_model.input,output=x)\n\n cnn_model.summary()\n rnn = Sequential()\n rnn.add(TimeDistributed(model1, batch_input_shape=(train_batch_size, timesteps, inp[0], inp[1], inp[2]), name = 'tdCNN')) \n\n if not include_vgg_top:\n rnn.add(TimeDistributed(Flatten()))\n \n \"\"\"\n cnn_model.pop()\n rnn.add(TimeDistributed(Dropout(0.25), name = 'dropout025_conv'))\n rnn.add(TimeDistributed(Dense(1024), name = 'fc1024')) # , activation='relu', activation='relu', kernel_regularizer=regularizers.l2(0.001)\n rnn.add(TimeDistributed(Dropout(0.25), name = 'dropout025'))\n \"\"\"\n rnn.add(TimeDistributed(Dense(num_outputs), name = 'fc3'))\n\n rnn.add(LSTM(lstm_nodes, dropout=lstm_dropout, recurrent_dropout=lstm_recurrent_dropout, stateful=True))#, activation='relu'\n \n \n #model = Model(inputs=cnn_model.input, outputs=rnn(TimeDistributed(cnn_model.output)))\n \n modelID = modelID + '_seqLen%d' % timesteps\n modelID = modelID + '_stateful'\n modelID = modelID + '_lstm%d' % lstm_nodes\n rnn.add(Dense(num_outputs))\n \n modelID = modelID + '_output%d' % num_outputs\n\n modelID = modelID + '_BatchSize%d' % train_batch_size\n modelID = modelID + '_inEpochs%d' % in_epochs\n modelID = modelID + '_outEpochs%d' % out_epochs\n \n for layer in rnn.layers[:1]: \n layer.trainable = False\n adam = Adam(lr=lr)\n modelID = modelID + '_AdamOpt_lr-%f' % lr\n rnn.compile(optimizer=adam, loss='mean_absolute_error') #'mean_squared_error', metrics=['mae'])#\n modelID = modelID + '_%s' % now()[:-7].replace(' ', '_').replace(':', '-')\n return cnn_model, rnn, modelID, preprocess_input",
"_____no_output_____"
],
[
"vgg_model, full_model, modelID, preprocess_input = getFinalModel(timesteps = timesteps, lstm_nodes = lstm_nodes, lstm_dropout = lstm_dropout, lstm_recurrent_dropout = lstm_recurrent_dropout, \n num_outputs = num_outputs, lr = learning_rate, include_vgg_top = include_vgg_top)",
"/home/mcicek/anaconda3/lib/python3.6/site-packages/ipykernel_launcher.py:22: UserWarning: Update your `Model` call to the Keras 2 API: `Model(inputs=Tensor(\"in..., outputs=Tensor(\"pr...)`\n"
],
[
"full_model = trainCNN_LSTM(full_model, modelID, out_epochs, trainingSubjects, timesteps, output_begin, num_outputs, \n batch_size = train_batch_size, in_epochs = in_epochs, stateful = STATEFUL, preprocess_input = preprocess_input)",
"All frames and annotations from 1 datasets have been read by 2019-01-29 00:45:23.596839\n1. set (Dataset 9) being trained for epoch 1 by 2019-01-29 00:45:32.509373!\nEpoch 1/1\n882/882 [==============================] - 27s 31ms/step - loss: 0.3977\nEpoch 1 completed!\n"
],
[
"def unscaleEstimations(test_labels, predictions, scalers, output_begin, num_outputs):\n \"\"\"* label_rescaling_factor * label_rescaling_factor\n \"\"\"\n sclrs = [scalers[0][output_begin:output_begin+num_outputs], scalers[1][output_begin:output_begin+num_outputs]]\n test_labels = unscaleAnnoByScalers(test_labels, sclrs)\n predictions = unscaleAnnoByScalers(predictions, sclrs)\n return test_labels, predictions",
"_____no_output_____"
],
[
"def evaluateSubject(full_model, subject, test_gen, test_labels, timesteps, output_begin, num_outputs, angles, batch_size, stateful = False, record = False):\n if num_outputs == 1: angles = ['Yaw']\n printLog('For the Subject %d (%s):' % (subject, BIWI_Subject_IDs[subject]), record = record)\n predictions = full_model.predict_generator(test_gen, steps = int(len(test_labels)/batch_size), verbose = 1)\n #kerasEval = full_model.evaluate_generator(test_gen) \n test_labels, predictions = unscaleEstimations(test_labels, predictions, BIWI_Lebel_Scalers, output_begin, num_outputs)\n full_model.reset_states()\n outputs = []\n for i in range(num_outputs):\n if stateful:\n start_index = (test_labels.shape[0] % batch_size) if batch_size > 1 else 0\n matrix = numpy.concatenate((test_labels[start_index:, i:i+1], predictions[:, i:i+1]), axis=1)\n differences = (test_labels[start_index:, i:i+1] - predictions[:, i:i+1])\n else:\n print(test_labels[:, i:i+1].shape, predictions[:, i:i+1].shape)\n matrix = numpy.concatenate((test_labels[:, i:i+1], predictions[:, i:i+1]), axis=1)\n differences = (test_labels[:, i:i+1] - predictions[:, i:i+1])\n absolute_mean_error = np.abs(differences).mean()\n printLog(\"\\tThe absolute mean error on %s angle estimation: %.2f Degree\" % (angles[i], absolute_mean_error), record = record)\n outputs.append((matrix, absolute_mean_error))\n return full_model, outputs\n",
"_____no_output_____"
],
[
"def evaluateCNN_LSTM(full_model, label_rescaling_factor, testSubjects, timesteps, output_begin, \n num_outputs, batch_size, angles, stateful = False, record = False, preprocess_input = None):\n if num_outputs == 1: angles = ['Yaw']\n test_generators, test_labelSets = getTestBiwiForImageModel(testSubjects, timesteps, False, output_begin, num_outputs, \n batch_size = batch_size, stateful = stateful, record = record, preprocess_input = preprocess_input)\n results = []\n for subject, test_gen, test_labels in zip(testSubjects, test_generators, test_labelSets):\n full_model, outputs = evaluateSubject(full_model, subject, test_gen, test_labels, timesteps, output_begin, num_outputs, angles, batch_size = batch_size, stateful = stateful, record = record)\n results.append((subject, outputs))\n means = evaluateAverage(results, angles, num_outputs, record = record)\n return full_model, means, results \n",
"_____no_output_____"
],
[
" full_model, means, results = evaluateCNN_LSTM(full_model, label_rescaling_factor = label_rescaling_factor, \n testSubjects = testSubjects, timesteps = timesteps, output_begin = output_begin, \n num_outputs = num_outputs, batch_size = test_batch_size, angles = angles, stateful = STATEFUL, preprocess_input = preprocess_input)",
"All frames and annotations from 1 datasets have been read by 2019-01-29 00:52:13.451180\nFor the Subject 9 (M03):\n882/882 [==============================] - 12s 14ms/step\n\tThe absolute mean error on Pitch angle estimation: 20.35 Degree\n\tThe absolute mean error on Yaw angle estimation: 29.29 Degree\n\tThe absolute mean error on Roll angle estimation: 31.73 Degree\n"
],
[
"test_generators, test_labelSets = getTestBiwiForImageModel(testSubjects, timesteps, False, output_begin, num_outputs, \n batch_size = 1, stateful = True, preprocess_input = preprocess_input)",
"All frames and annotations from 1 datasets have been read by 2019-01-28 23:01:54.311180\n"
],
[
"test_gen, test_labels = test_generators[0], test_labelSets[0]",
"_____no_output_____"
],
[
"test_labels",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d04068ae263c2eb907b2a0cd014804536e4e07b1 | 4,270 | ipynb | Jupyter Notebook | Data/Car_Data/connectTesting.ipynb | CSCI4850/s22-team6-project | 8e290f3725fdea79ed62777dd14ecc9f7d670648 | [
"MIT"
] | null | null | null | Data/Car_Data/connectTesting.ipynb | CSCI4850/s22-team6-project | 8e290f3725fdea79ed62777dd14ecc9f7d670648 | [
"MIT"
] | null | null | null | Data/Car_Data/connectTesting.ipynb | CSCI4850/s22-team6-project | 8e290f3725fdea79ed62777dd14ecc9f7d670648 | [
"MIT"
] | 1 | 2022-03-14T15:31:40.000Z | 2022-03-14T15:31:40.000Z | 35.289256 | 983 | 0.559485 | [
[
[
"print('Setting UP')\nimport os\nos.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'\nimport socketio\nimport eventlet\nimport numpy as np\nfrom flask import Flask\nfrom tensorflow.keras.models import load_model\nimport base64\nfrom io import BytesIO\nfrom PIL import Image\nimport cv2\n \n#### FOR REAL TIME COMMUNICATION BETWEEN CLIENT AND SERVER\nsio = socketio.Server()\n#### FLASK IS A MICRO WEB FRAMEWORK WRITTEN IN PYTHON\napp = Flask(__name__) # '__main__'\n \nmaxSpeed = 10\n \n \ndef preProcess(img):\n img = img[60:135,:,:]\n img = cv2.cvtColor(img, cv2.COLOR_RGB2YUV)\n img = cv2.GaussianBlur(img, (3, 3), 0)\n img = cv2.resize(img, (200, 66))\n img = img/255\n return img\n \n \n@sio.on('telemetry')\ndef telemetry(sid, data):\n speed = float(data['speed'])\n image = Image.open(BytesIO(base64.b64decode(data['image'])))\n image = np.asarray(image)\n image = preProcess(image)\n image = np.array([image])\n steering = float(model.predict(image))\n throttle = 1.0 - speed / maxSpeed\n print(f'{steering}, {throttle}, {speed}')\n sendControl(steering, throttle)\n \n \n@sio.on('connect')\ndef connect(sid, environ):\n print('Connected')\n sendControl(0, 0)\n \n \ndef sendControl(steering, throttle):\n sio.emit('steer', data={\n 'steering_angle': steering.__str__(),\n 'throttle': throttle.__str__()\n })\n \n \nif __name__ == '__main__':\n model = load_model('model.h5')\n app = socketio.Middleware(sio, app)\n ### LISTEN TO PORT 4567\n eventlet.wsgi.server(eventlet.listen(('', 4567)), app)",
"Setting UP\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d040739f9700be3b163b766b0befedcedda6000c | 22,437 | ipynb | Jupyter Notebook | python/python3.ipynb | shiyeli/jupyter_notebook | eba4c070385708f8589bdc2dac4b785a70a9f460 | [
"MIT"
] | null | null | null | python/python3.ipynb | shiyeli/jupyter_notebook | eba4c070385708f8589bdc2dac4b785a70a9f460 | [
"MIT"
] | null | null | null | python/python3.ipynb | shiyeli/jupyter_notebook | eba4c070385708f8589bdc2dac4b785a70a9f460 | [
"MIT"
] | null | null | null | 36.423701 | 1,504 | 0.571511 | [
[
[
"import csv\nfrom io import StringIO\nwith open('/Users/yetongxue/Downloads/网站管理.csv')\nreader = csv.reader(f, delimiter=',')\nfor index,row in enumerate(reader):\n if index == 0:\n continue\n print(row)",
"_____no_output_____"
],
[
"import tldextract\nresult = tldextract.extract('http://*.sometime')\nresult",
"_____no_output_____"
],
[
"from queue import PriorityQueue\nimport numpy as np\nq = PriorityQueue(5)\narr = np.random.randint(1, 20, size=5)\narr",
"_____no_output_____"
],
[
"a = [[1,2], [3,4]]\na.reverse()\na",
"_____no_output_____"
],
[
"bin(int('2000000000000000004008020', 16))",
"_____no_output_____"
],
[
"len('10000000000000000000000000000000000000000000000000000000000000000000000100000000001000000000100000')",
"_____no_output_____"
],
[
"bin(int('00000000000004008020', 16))",
"_____no_output_____"
],
[
"len('100000000001000000000100000')",
"_____no_output_____"
],
[
"root = {}\nnode = root\nfor char in 'qwerasdf':\n node = node.setdefault(char, {})\nnode['#'] = '#'\nroot",
"_____no_output_____"
],
[
"import datetime\nt = 1531732030883 * 0.001\ndatetime.datetime.fromtimestamp(1531732030883 * 0.001)\ntoday ",
"_____no_output_____"
],
[
"import collections\ndic = collections.OrderedDict()\ndic[1] = 1\ndic[2] = 2\ndic",
"_____no_output_____"
],
[
"dic.popitem(last=False)",
"_____no_output_____"
],
[
"import datetime\ntoday = datetime.datetime.now().date()\nupdate_time = datetime.datetime.strptime('2020-08-07 23:20:10', '%Y-%m-%d %H:%M:%S').date()\nif today - update_time > datetime.timedelta(days=7):\n print(1)\n(today - update_time).days \n\n\n'1'.strip().startswith('=')",
"1\n"
],
[
"def check_row(row):\n data = []\n for i in row:\n i = i.strip()\n while i.startswith('='):\n i = i[1:]\n data.append(i)\n return data\n\ncheck_row(['12', ' ==12', '1=1'])",
"_____no_output_____"
],
[
"import re\nres = re.match('^=+\\s.', '=ab')\nres",
"_____no_output_____"
],
[
"cc_ai = {\n 'normal': 'JS',\n 'low': 'JS',\n 'middle': 'JS',\n 'high': 'JSPAGE'\n }\ncc_ai",
"_____no_output_____"
],
[
"import datetime\nstart = datetime.datetime.now()\nend = datetime.datetime.strptime('2020-09-24T07:14:04.252394', '%Y-%m-%dT%H:%M:%S.%f')\ndatetime.datetime.strptime(str(start.date()), '%Y-%m-%d') - datetime.timedelta(days=7)\nisinstance(datetime.datetime.now(), datetime.datetime)",
"_____no_output_____"
],
[
"''.join('\\t\\ra-_sdf\\n a sdf '.split())",
"_____no_output_____"
],
[
"SITE_MODULE = [\n 'app_ipv6', 'site_shield', 'subdomain_limit', 'custom_page',\n 'app_extra', 'service_charge', 'app_ipv6_defend', 'privacy_shield',\n 'app_ipv6_custom', 'app_subdomain_limit', 'app_ip_limit', 'app_bandwidth',\n 'app_subuser', 'batch', 'service_charge', 'cn2_line', 'site_shield_monitor',\n 'app_httpdns_1k', 'app_domain_limit'\n ]\nSITE_MODULE",
"_____no_output_____"
],
[
"''.join('a, \\rd'.replace(',', ',').split())",
"_____no_output_____"
],
[
"from docxtpl import DocxTemplate\n\npath = '/Users/yetongxue/Downloads/test1.docx'\nsave_path = '/Users/yetongxue/Downloads/test1_save.docx'\ntemplate = DocxTemplate(path)\ndata = {'sites': [{'domain':'asdf'},{'domain':'asdf2323'}]}\ntemplate.render(context=data)\ntemplate.save(save_path)",
"_____no_output_____"
],
[
"import json\njson.dumps([{'chargeType': 'plan', 'number': 1, 'expireTime': '2021-04-23', 'chargeId': '604d75cc261b3b0011a1a496'}, {'chargeType': 'additionPackage', 'number': 1, 'expireTime': '2021-04-23', 'chargeId': '604d75cd261b3b0011a1a498'}])\n ",
"_____no_output_____"
],
[
"import commonds\nstatus, output = commands.getstatusoutput('date')\n# 2016年 06月 30日 星期四 19:26:21 CST\nstatus, output",
"_____no_output_____"
],
[
"import os\nres = os.popen('dig -x 2001:4d0:9700:903:198:9:3:30').read()\nif 'intra.jiasule.com' in res:\n print('ok')\nres",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |