markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
Self-Driving Car Engineer Nanodegree Project: **Finding Lane Lines on the Road** ***In this project, you will use the tools you learned about in the lesson to identify lane lines on the road. You can develop your pipeline on a series of individual images, and later apply the result to a video stream (really just a series of images). Check out the video clip "raw-lines-example.mp4" (also contained in this repository) to see what the output should look like after using the helper functions below. Once you have a result that looks roughly like "raw-lines-example.mp4", you'll need to get creative and try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video "P1_example.mp4". Ultimately, you would like to draw just one line for the left side of the lane, and one for the right.In addition to implementing code, there is a brief writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) that can be used to guide the writing process. Completing both the code in the Ipython notebook and the writeup template will cover all of the [rubric points](https://review.udacity.com/!/rubrics/322/view) for this project.---Let's have a look at our first image called 'test_images/solidWhiteRight.jpg'. Run the 2 cells below (hit Shift-Enter or the "play" button above) to display the image.**Note: If, at any point, you encounter frozen display windows or other confounding issues, you can always start again with a clean slate by going to the "Kernel" menu above and selecting "Restart & Clear Output".**--- **The tools you have are color selection, region of interest selection, grayscaling, Gaussian smoothing, Canny Edge Detection and Hough Tranform line detection. You are also free to explore and try other techniques that were not presented in the lesson. Your goal is piece together a pipeline to detect the line segments in the image, then average/extrapolate them and draw them onto the image for display (as below). Once you have a working pipeline, try it out on the video stream below.**--- Your output should look something like this (above) after detecting line segments using the helper functions below Your goal is to connect/average/extrapolate line segments to get output like this **Run the cell below to import some packages. If you get an `import error` for a package you've already installed, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.** Import Packages | #importing some useful packages
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import numpy as np
import cv2
%matplotlib inline | _____no_output_____ | MIT | P1.ipynb | MohamedHeshamMustafa/CarND-LaneLines-P1 |
Read in an Image | #reading in an image
image = mpimg.imread('test_images/solidWhiteRight.jpg')
#printing out some stats and plotting
print('This image is:', type(image), 'with dimensions:', image.shape)
plt.imshow(image) # if you wanted to show a single color channel image called 'gray', for example, call as plt.imshow(gray, cmap='gray') | This image is: <class 'numpy.ndarray'> with dimensions: (540, 960, 3)
| MIT | P1.ipynb | MohamedHeshamMustafa/CarND-LaneLines-P1 |
Ideas for Lane Detection Pipeline **Some OpenCV functions (beyond those introduced in the lesson) that might be useful for this project are:**`cv2.inRange()` for color selection `cv2.fillPoly()` for regions selection `cv2.line()` to draw lines on an image given endpoints `cv2.addWeighted()` to coadd / overlay two images`cv2.cvtColor()` to grayscale or change color`cv2.imwrite()` to output images to file `cv2.bitwise_and()` to apply a mask to an image**Check out the OpenCV documentation to learn about these and discover even more awesome functionality!** Helper Functions Below are some helper functions to help get you started. They should look familiar from the lesson! | import math
def grayscale(img):
"""Applies the Grayscale transform
This will return an image with only one color channel
but NOTE: to see the returned image as grayscale
(assuming your grayscaled image is called 'gray')
you should call plt.imshow(gray, cmap='gray')"""
return cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)
# Or use BGR2GRAY if you read an image with cv2.imread()
# return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
def canny(img, low_threshold, high_threshold):
"""Applies the Canny transform"""
return cv2.Canny(img, low_threshold, high_threshold)
def gaussian_blur(img, kernel_size):
"""Applies a Gaussian Noise kernel"""
return cv2.GaussianBlur(img, (kernel_size, kernel_size), 0)
def region_of_interest(img, vertices):
"""
Applies an image mask.
Only keeps the region of the image defined by the polygon
formed from `vertices`. The rest of the image is set to black.
`vertices` should be a numpy array of integer points.
"""
#defining a blank mask to start with
mask = np.zeros_like(img)
#defining a 3 channel or 1 channel color to fill the mask with depending on the input image
if len(img.shape) > 2:
channel_count = img.shape[2] # i.e. 3 or 4 depending on your image
ignore_mask_color = (255,) * channel_count
else:
ignore_mask_color = 255
#filling pixels inside the polygon defined by "vertices" with the fill color
cv2.fillPoly(mask, vertices, ignore_mask_color)
#returning the image only where mask pixels are nonzero
masked_image = cv2.bitwise_and(img, mask)
return masked_image
def draw_lines(img, lines, color=[255, 0, 0], thickness=8):
"""
NOTE: this is the function you might want to use as a starting point once you want to
average/extrapolate the line segments you detect to map out the full
extent of the lane (going from the result shown in raw-lines-example.mp4
to that shown in P1_example.mp4).
Think about things like separating line segments by their
slope ((y2-y1)/(x2-x1)) to decide which segments are part of the left
line vs. the right line. Then, you can average the position of each of
the lines and extrapolate to the top and bottom of the lane.
This function draws `lines` with `color` and `thickness`.
Lines are drawn on the image inplace (mutates the image).
If you want to make the lines semi-transparent, think about combining
this function with the weighted_img() function below
"""
negative_slopes = []
positive_slopes = []
negetive_intercepts = []
positive_intercepts = []
left_line_x = []
left_line_y = []
right_line_x = []
right_line_y = []
y_max = img.shape[0]
y_min = img.shape[0]
#Drawing Lines
for line in lines:
for x1,y1,x2,y2 in line:
current_slope = (y2-y1)/(x2-x1)
if current_slope < 0.0 and current_slope > -math.inf:
negative_slopes.append(current_slope) # left line
left_line_x.append(x1)
left_line_x.append(x2)
left_line_y.append(y1)
left_line_y.append(y2)
negetive_intercepts.append(y1 -current_slope*x1)
if current_slope > 0.0 and current_slope < math.inf:
positive_slopes.append(current_slope) # right line
right_line_x.append(x1)
right_line_x.append(x2)
right_line_y.append(y1)
right_line_y.append(y2)
positive_intercepts.append(y1 - current_slope*x1)
y_min = min(y_min, y1, y2)
y_min += 20 # add small threshold
if len(positive_slopes) > 0 and len(right_line_x) > 0 and len(right_line_y) > 0:
ave_positive_slope = sum(positive_slopes) / len(positive_slopes)
ave_right_line_x = sum(right_line_x) / len(right_line_x)
ave_right_line_y = sum(right_line_y ) / len(right_line_y)
intercept = sum(positive_intercepts) / len(positive_intercepts)
x_min=int((y_min-intercept)/ave_positive_slope)
x_max = int((y_max - intercept)/ ave_positive_slope)
cv2.line(img, (x_min, y_min), (x_max, y_max), color, thickness)
if len(negative_slopes) > 0 and len(left_line_x) > 0 and len(left_line_y) > 0:
ave_negative_slope = sum(negative_slopes) / len(negative_slopes)
ave_left_line_x = sum(left_line_x) / len(left_line_x)
ave_left_line_y = sum(left_line_y ) / len(left_line_y)
intercept = sum(negetive_intercepts) / len(negetive_intercepts)
x_min = int((y_min-intercept)/ave_negative_slope)
x_max = int((y_max - intercept)/ ave_negative_slope)
cv2.line(img, (x_min, y_min), (x_max, y_max), color, thickness)
def hough_lines(img, rho, theta, threshold, min_line_len, max_line_gap):
"""
`img` should be the output of a Canny transform.
Returns an image with hough lines drawn.
"""
lines = cv2.HoughLinesP(img, rho, theta, threshold, np.array([]), minLineLength=min_line_len, maxLineGap=max_line_gap)
line_img = np.zeros((img.shape[0], img.shape[1], 3), dtype=np.uint8)
draw_lines(line_img, lines)
return line_img
# Python 3 has support for cool math symbols.
def weighted_img(img, initial_img, α=0.8, β=1., γ=0.):
"""
`img` is the output of the hough_lines(), An image with lines drawn on it.
Should be a blank image (all black) with lines drawn on it.
`initial_img` should be the image before any processing.
The result image is computed as follows:
initial_img * α + img * β + γ
NOTE: initial_img and img must be the same shape!
"""
return cv2.addWeighted(initial_img, α, img, β, γ) | _____no_output_____ | MIT | P1.ipynb | MohamedHeshamMustafa/CarND-LaneLines-P1 |
Test ImagesBuild your pipeline to work on the images in the directory "test_images" **You should make sure your pipeline works well on these images before you try the videos.** | import os
os.listdir("test_images/") | _____no_output_____ | MIT | P1.ipynb | MohamedHeshamMustafa/CarND-LaneLines-P1 |
Build a Lane Finding Pipeline Build the pipeline and run your solution on all test_images. Make copies into the `test_images_output` directory, and you can use the images in your writeup report.Try tuning the various parameters, especially the low and high Canny thresholds as well as the Hough lines parameters. | # TODO: Build your pipeline that will draw lane lines on the test_images
# then save them to the test_images_output directory.
##1) We Have To read our Image in a grey scale fromat
Input_Image = mpimg.imread('test_images/solidWhiteCurve.jpg')
Input_Grey_Img = grayscale(Input_Image)
plt.imshow(Input_Grey_Img, cmap='gray')
plt.title('Image in Grey Scale Format')
##2) Apply Canny Detection with a low threshold 1 : 3 to high threshold
## we do further smoothing before applying canny algorithm
Kernel_size = 3 #always put an odd number (3, 5, 7, ..)
img_Smoothed = gaussian_blur(Input_Grey_Img, Kernel_size)
High_threshold = 150
Low_threshold = 75
imga_fter_Canny = canny(img_Smoothed, Low_threshold, High_threshold)
plt.imshow(imga_fter_Canny, cmap='gray')
plt.title('Image after Applying Canny')
##3) Determine Region of interest to detect Lane lines in Image
## Set Verticies Parameter to determine regoin of interest first
#Vertices : Left_bottom, Right_bottom, Apex (Area of interest)
vertices = np.array([[(0,image.shape[0]),(470, 320), (500, 320), (image.shape[1],image.shape[0])]], dtype=np.int32)
Masked_Image = region_of_interest(imga_fter_Canny, vertices)
plt.imshow(Masked_Image,cmap='gray')
plt.title('Massked Image')
##4)using hough transfrom to find lines
# Define the Hough transform parameters
# Make a blank the same size as our image to draw on
rho = 2
theta = np.pi/180
threshold = 15
min_line_length = 40
max_line_gap = 20
lines = hough_lines(Masked_Image, rho, theta, threshold, min_line_length, max_line_gap)
plt.imshow(lines,cmap='gray')
plt.title('lines Image')
##5) Draw Lines on the real Image
Final_out = weighted_img(lines, Input_Image, α=0.8, β=1., γ=0.)
plt.imshow(Final_out)
plt.title('Final Image with lane detected') | _____no_output_____ | MIT | P1.ipynb | MohamedHeshamMustafa/CarND-LaneLines-P1 |
Test on VideosYou know what's cooler than drawing lanes over images? Drawing lanes over video!We can test our solution on two provided videos:`solidWhiteRight.mp4``solidYellowLeft.mp4`**Note: if you get an import error when you run the next cell, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.****If you get an error that looks like this:**```NeedDownloadError: Need ffmpeg exe. You can download it by calling: imageio.plugins.ffmpeg.download()```**Follow the instructions in the error message and check out [this forum post](https://discussions.udacity.com/t/project-error-of-test-on-videos/274082) for more troubleshooting tips across operating systems.** | # Import everything needed to edit/save/watch video clips
from moviepy.editor import VideoFileClip
from IPython.display import HTML
def process_image(image):
# NOTE: The output you return should be a color image (3 channel) for processing video below
# TODO: put your pipeline here,
# you should return the final output (image where lines are drawn on lanes)
##1) We Have To read our Image in a grey scale fromat
Input_Grey_Img = grayscale(image)
##2) Apply Canny Detection with a low threshold 1 : 3 to high threshold
## we do further smoothing before applying canny algorithm
Kernel_size = 3 #always put an odd number (3, 5, 7, ..)
img_Smoothed = gaussian_blur(Input_Grey_Img, Kernel_size)
High_threshold = 150
Low_threshold = 50
imga_fter_Canny = canny(img_Smoothed, Low_threshold, High_threshold)
##3) Determine Region of interest to detect Lane lines in Image
## Set Verticies Parameter to determine regoin of interest first
#Vertices : Left_bottom, Right_bottom, Apex (Area of interest)
vertices = np.array([[(0,image.shape[0]),
(470, 320),
(500, 320),
(image.shape[1],
image.shape[0])]],
dtype=np.int32)
Masked_Image = region_of_interest(imga_fter_Canny, vertices)
##4)using hough transfrom to find lines
# Define the Hough transform parameters
# Make a blank the same size as our image to draw on
rho = 2
theta = np.pi/180
threshold = 55
min_line_length = 100
max_line_gap = 150
lines = hough_lines(Masked_Image, rho, theta, threshold, min_line_length, max_line_gap)
##5)Draw Lines on the real Image
result = weighted_img(lines, image, α=0.8, β=1., γ=0.)
return result | _____no_output_____ | MIT | P1.ipynb | MohamedHeshamMustafa/CarND-LaneLines-P1 |
Let's try the one with the solid white lane on the right first ... | white_output = 'test_videos_output/solidWhiteRight.mp4'
## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video
## To do so add .subclip(start_second,end_second) to the end of the line below
## Where start_second and end_second are integer values representing the start and end of the subclip
## You may also uncomment the following line for a subclip of the first 5 seconds
##clip1 = VideoFileClip("test_videos/solidWhiteRight.mp4").subclip(0,5)
clip1 = VideoFileClip("test_videos/solidWhiteRight.mp4")
white_clip = clip1.fl_image(process_image) #NOTE: this function expects color images!!
%time white_clip.write_videofile(white_output, audio=False) | t: 10%|▉ | 21/221 [00:00<00:00, 209.97it/s, now=None] | MIT | P1.ipynb | MohamedHeshamMustafa/CarND-LaneLines-P1 |
Play the video inline, or if you prefer find the video in your filesystem (should be in the same directory) and play it in your video player of choice. | HTML("""
<video width="960" height="540" controls>
<source src="{0}">
</video>
""".format(white_output)) | _____no_output_____ | MIT | P1.ipynb | MohamedHeshamMustafa/CarND-LaneLines-P1 |
Improve the draw_lines() function**At this point, if you were successful with making the pipeline and tuning parameters, you probably have the Hough line segments drawn onto the road, but what about identifying the full extent of the lane and marking it clearly as in the example video (P1_example.mp4)? Think about defining a line to run the full length of the visible lane based on the line segments you identified with the Hough Transform. As mentioned previously, try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video "P1_example.mp4".****Go back and modify your draw_lines function accordingly and try re-running your pipeline. The new output should draw a single, solid line over the left lane line and a single, solid line over the right lane line. The lines should start from the bottom of the image and extend out to the top of the region of interest.** Now for the one with the solid yellow lane on the left. This one's more tricky! | yellow_output = 'test_videos_output/solidYellowLeft.mp4'
## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video
## To do so add .subclip(start_second,end_second) to the end of the line below
## Where start_second and end_second are integer values representing the start and end of the subclip
## You may also uncomment the following line for a subclip of the first 5 seconds
##clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4').subclip(0,5)
clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4')
yellow_clip = clip2.fl_image(process_image)
%time yellow_clip.write_videofile(yellow_output, audio=False)
HTML("""
<video width="960" height="540" controls>
<source src="{0}">
</video>
""".format(yellow_output)) | _____no_output_____ | MIT | P1.ipynb | MohamedHeshamMustafa/CarND-LaneLines-P1 |
Writeup and SubmissionIf you're satisfied with your video outputs, it's time to make the report writeup in a pdf or markdown file. Once you have this Ipython notebook ready along with the writeup, it's time to submit for review! Here is a [link](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) to the writeup template file. Optional ChallengeTry your lane finding pipeline on the video below. Does it still work? Can you figure out a way to make it more robust? If you're up for the challenge, modify your pipeline so it works with this video and submit it along with the rest of your project! | def process_image1(image):
# NOTE: The output you return should be a color image (3 channel) for processing video below
# TODO: put your pipeline here,
# you should return the final output (image where lines are drawn on lanes)
##1) We Have To read our Image in a grey scale fromat
Input_Grey_Img = grayscale(image)
##2) Apply Canny Detection with a low threshold 1 : 3 to high threshold
## we do further smoothing before applying canny algorithm
Kernel_size = 3 #always put an odd number (3, 5, 7, ..)
img_Smoothed = gaussian_blur(Input_Grey_Img, Kernel_size)
High_threshold = 150
Low_threshold = 50
imga_fter_Canny = canny(img_Smoothed, Low_threshold, High_threshold)
##3) Determine Region of interest to detect Lane lines in Image
## Set Verticies Parameter to determine regoin of interest first
vertices = np.array([[(226, 680),
(614,436),
(714,436),
(1093,634)]])
Masked_Image = region_of_interest(imga_fter_Canny, vertices)
##4)using hough transfrom to find lines
# Define the Hough transform parameters
# Make a blank the same size as our image to draw on
rho = 2
theta = np.pi/180
threshold = 55
min_line_length = 100
max_line_gap = 150
lines = hough_lines(Masked_Image, rho, theta, threshold, min_line_length, max_line_gap)
##5)Draw Lines on the real Image
result = weighted_img(lines, image, α=0.8, β=1., γ=0.)
return result
challenge_output = 'test_videos_output/challenge.mp4'
## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video
## To do so add .subclip(start_second,end_second) to the end of the line below
## Where start_second and end_second are integer values representing the start and end of the subclip
## You may also uncomment the following line for a subclip of the first 5 seconds
##clip3 = VideoFileClip('test_videos/challenge.mp4').subclip(0,5)
clip3 = VideoFileClip('test_videos/challenge.mp4')
challenge_clip = clip3.fl_image(process_image1)
%time challenge_clip.write_videofile(challenge_output, audio=False)
HTML("""
<video width="960" height="540" controls>
<source src="{0}">
</video>
""".format(challenge_output)) | _____no_output_____ | MIT | P1.ipynb | MohamedHeshamMustafa/CarND-LaneLines-P1 |
Bayes Classifier | import util
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import multivariate_normal as mvn
%matplotlib inline
def clamp_sample(x):
x = np.minimum(x, 1)
x = np.maximum(x, 0)
return x
class BayesClassifier:
def fit(self, X, Y):
# assume classes are numbered 0...K-1
self.K = len(set(Y))
self.gaussians = []
self.p_y = np.zeros(self.K)
for k in range(self.K):
Xk = X[Y == k]
self.p_y[k] = len(Xk)
mean = Xk.mean(axis=0) # describe gaussian
cov = np.cov(Xk.T) # describe gaussian
g = {'m': mean, 'c': cov}
self.gaussians.append(g)
# normalize p(y)
self.p_y /= self.p_y.sum()
def sample_given_y(self, y):
g = self.gaussians[y]
return clamp_sample( mvn.rvs(mean=g['m'], cov=g['c']) )
def sample(self):
y = np.random.choice(self.K, p=self.p_y)
return clamp_sample( self.sample_given_y(y) )
X, Y = util.get_mnist()
clf = BayesClassifier()
clf.fit(X, Y)
for k in range(clf.K):
# show one sample for each class
# also show the mean image learned from Gaussian Bayes Classifier
sample = clf.sample_given_y(k).reshape(28, 28)
mean = clf.gaussians[k]['m'].reshape(28, 28)
plt.subplot(1,2,1)
plt.imshow(sample, cmap='gray')
plt.title("Sample")
plt.subplot(1,2,2)
plt.imshow(mean, cmap='gray')
plt.title("Mean")
plt.show() | _____no_output_____ | MIT | Week2/Bayes Classifier.ipynb | yumengdong/GANs |
Bayes Classifier with Gaussian Mixture Models | from sklearn.mixture import BayesianGaussianMixture
class BayesClassifier:
def fit(self, X, Y):
# assume classes are numbered 0...K-1
self.K = len(set(Y))
self.gaussians = []
self.p_y = np.zeros(self.K)
for k in range(self.K):
print("Fitting gmm", k)
Xk = X[Y == k]
self.p_y[k] = len(Xk)
gmm = BayesianGaussianMixture(10) # number of clusters
gmm.fit(Xk)
self.gaussians.append(gmm)
# normalize p(y)
self.p_y /= self.p_y.sum()
def sample_given_y(self, y):
gmm = self.gaussians[y]
sample = gmm.sample()
# note: sample returns a tuple containing 2 things:
# 1) the sample
# 2) which cluster it came from
# we'll use (2) to obtain the means so we can plot
# them like we did in the previous script
# we cheat by looking at "non-public" params in
# the sklearn source code
mean = gmm.means_[sample[1]]
return clamp_sample( sample[0].reshape(28, 28) ), mean.reshape(28, 28)
def sample(self):
y = np.random.choice(self.K, p=self.p_y)
return clamp_sample( self.sample_given_y(y) )
clf = BayesClassifier()
clf.fit(X, Y)
for k in range(clf.K):
# show one sample for each class
# also show the mean image learned
sample, mean = clf.sample_given_y(k)
plt.subplot(1,2,1)
plt.imshow(sample, cmap='gray')
plt.title("Sample")
plt.subplot(1,2,2)
plt.imshow(mean, cmap='gray')
plt.title("Mean")
plt.show()
# generate a random sample
sample, mean = clf.sample()
plt.subplot(1,2,1)
plt.imshow(sample, cmap='gray')
plt.title("Random Sample from Random Class")
plt.subplot(1,2,2)
plt.imshow(mean, cmap='gray')
plt.title("Corresponding Cluster Mean")
plt.show() | _____no_output_____ | MIT | Week2/Bayes Classifier.ipynb | yumengdong/GANs |
Neural Network and Autoencoder | import tensorflow as tf
class Autoencoder:
def __init__(self, D, M):
# represents a batch of training data
self.X = tf.placeholder(tf.float32, shape=(None, D))
# input -> hidden
self.W = tf.Variable(tf.random_normal(shape=(D, M)) * np.sqrt(2.0 / M))
self.b = tf.Variable(np.zeros(M).astype(np.float32))
# hidden -> output
self.V = tf.Variable(tf.random_normal(shape=(M, D)) * np.sqrt(2.0 / D))
self.c = tf.Variable(np.zeros(D).astype(np.float32))
# construct the reconstruction
self.Z = tf.nn.relu(tf.matmul(self.X, self.W) + self.b)
logits = tf.matmul(self.Z, self.V) + self.c
self.X_hat = tf.nn.sigmoid(logits)
# compute the cost
self.cost = tf.reduce_sum(
tf.nn.sigmoid_cross_entropy_with_logits(
labels=self.X,
logits=logits
)
)
# make the trainer
self.train_op = tf.train.RMSPropOptimizer(learning_rate=0.001).minimize(self.cost)
# set up session and variables for later
self.init_op = tf.global_variables_initializer()
self.sess = tf.InteractiveSession()
self.sess.run(self.init_op)
def fit(self, X, epochs=30, batch_sz=64):
costs = []
n_batches = len(X) // batch_sz
print("n_batches:", n_batches)
for i in range(epochs):
if i % 5 == 0:
print("epoch:", i)
np.random.shuffle(X)
for j in range(n_batches):
batch = X[j*batch_sz:(j+1)*batch_sz]
_, c, = self.sess.run((self.train_op, self.cost), feed_dict={self.X: batch})
c /= batch_sz # just debugging
costs.append(c)
if (j % 100 == 0) and (i % 5 == 0):
print("iter: %d, cost: %.3f" % (j, c))
plt.plot(costs)
plt.show()
def predict(self, X):
return self.sess.run(self.X_hat, feed_dict={self.X: X})
model = Autoencoder(784, 300)
model.fit(X)
done = False
while not done:
i = np.random.choice(len(X))
x = X[i]
im = model.predict([x]).reshape(28, 28)
plt.subplot(1,2,1)
plt.imshow(x.reshape(28, 28), cmap='gray')
plt.title("Original")
plt.subplot(1,2,2)
plt.imshow(im, cmap='gray')
plt.title("Reconstruction")
plt.show()
ans = input("Generate another?")
if ans and ans[0] in ('n' or 'N'):
done = True | _____no_output_____ | MIT | Week2/Bayes Classifier.ipynb | yumengdong/GANs |
Our data exists as vectors in matrixes Linear algeabra helps us manipulate data to eventually find the smallest sum squared errors of our data which will give us our beta value for our regression model | import numpy as np
# create array to be transformed into vectors
x1 = np.array([1,2,1])
x2 = np.array([4,1,5])
x3 = np.array([6,8,6])
print("Array 1:", x1, sep="\n")
print("Array 2:", x2, sep="\n")
print("Array 3:", x3, sep="\n") | Array 1:
[1 2 1]
Array 2:
[4 1 5]
Array 3:
[6 8 6]
| MIT | In-Class Projects/Project 8 - Working with OLS.ipynb | zacharyejohnson/ECON411 |
Next, transform these arrays into row vectors using matrix(). | x1 = np.matrix(x1)
x2 = np.matrix(x2)
x3 = np.matrix(x3) | _____no_output_____ | MIT | In-Class Projects/Project 8 - Working with OLS.ipynb | zacharyejohnson/ECON411 |
use np.concatenate() to combine the rows | X = np.concatenate((x1, x2, x3), axis = 0)
X | _____no_output_____ | MIT | In-Class Projects/Project 8 - Working with OLS.ipynb | zacharyejohnson/ECON411 |
X.getI method gets inverse of matrix | X_inverse = X.getI()
X_inverse = np.round(X_inverse, 2)
X_inverse | _____no_output_____ | MIT | In-Class Projects/Project 8 - Working with OLS.ipynb | zacharyejohnson/ECON411 |
Regression function - Pulling necessary dataWe now know the necessary operations for inverting matrices and minimizing squared residuals. We can import real data and begin to analyze how variables influence one another. To start, we will use the Fraser economic freedom data. | import pandas as pd
import statsmodels.api as sm
import numpy as np
data = pd.read_csv('fraserDataWithRGDPPC.csv',
index_col = [0,1],
parse_dates = True)
data
years = np.array(sorted(list(set(data.index.get_level_values("Year")))))
years = pd.date_range(years[0], years[-2], freq = "AS")
countries = sorted(list(set(data.index.get_level_values("ISO_Code"))))
index_names = list(data.index.names)
multi_index = pd.MultiIndex.from_product([countries, years[:-1]], names = data.index.names)
data = data.reindex(multi_index)
data["RGDP Per Capita Lag"] = data.groupby("ISO_Code")["RGDP Per Capita"].shift()
data
data.dropna(axis = 0).loc['GBR'] | _____no_output_____ | MIT | In-Class Projects/Project 8 - Working with OLS.ipynb | zacharyejohnson/ECON411 |
Running Regression Model: | y_vars = ['RGDP Per Capita']
x_vars = [
'Size of Government', 'Legal System & Property Rights', 'Sound Money',
'Freedom to trade internationally', 'Regulation'
]
reg_vars = y_vars + x_vars
reg_data = data[reg_vars].dropna()
reg_data.corr().round(2)
reg_data.describe().round(2)
y = reg_data[y_vars]
x = reg_data[x_vars]
x['Constant'] = 1
results = sm.OLS(y, x).fit()
results.summary()
predictor = results.predict()
reg_data[y_vars[0] + " Predictor"] = predictor
reg_data.loc["GBR", [y_vars[0], y_vars[0] + " Predictor"]].plot() | _____no_output_____ | MIT | In-Class Projects/Project 8 - Working with OLS.ipynb | zacharyejohnson/ECON411 |
OLS Statistics We have calculated beta values for each independent variable, meaning that we estimated the average effect of a change in each independent variable upon the dependent variable. While this is useful, we have not yet measured the statistical significance of these estimations; neither have we determined the explanatory power of our particular regression.Our regression has estimated predicted values for our dependent variable given the values of the independent variables for each observation. Together, these estimations for an array of predicted values that we will refer to as $y ̂ $. We will refer to individual predicted values as ($y_i$) ̂. We will also refer to the mean value of observations of our dependent variable as $y ̅ $ and individual observed values of our dependent variable as $y_i$. These values will be use to estimate the sum of squares due to regression ($SSR$), sum of squared errors ($SSE$), and the total sum of squares ($SST$). By comparing the estimated $y$ values, the observed $y$ values, and the mean of $y$, we will estimate the standard error for each coefficient and other values that estimate convey the significance of the estimation.We define these values as follows:$SSR = \sum_{i=0}^{n} (y ̂ _{i} - y ̅ )^2$$SSE = \sum_{i=0}^{n} (y_{i} - y ̂ _{i})^2$$SST = \sum_{i=0}^{n} (y_{i} - y ̅ _{i})^2$It happens that the sum of the squared distances between the estimated values and mean of observed values and the squared distances between the observed and estimated values add up to the sum of the squared distances between the observed values and the mean of observed values. We indicate this as:$SST = SSR + SSE$The script below will estimate these statistics. It calls the sum_square_stats method from the which is passed in the calculate_regression_stats method. | y_name = y_vars[0]
y_hat = reg_data[y_name + " Predictor"]
y_mean = reg_data[y_name].mean()
y = reg_data[y_name]
y_hat, y_mean, y
reg_data["Residuals"] = y_hat.sub(y_mean)
reg_data["Squared Residuals"] = reg_data["Residuals"].pow(2)
reg_data["Squared Errors"] = (y.sub(y_hat)) ** 2
reg_data["Squared Totals"] = (y.sub(y_mean)) ** 2
SSR = reg_data["Squared Residuals"].sum()
SSE = reg_data["Squared Errors"].sum()
SST = reg_data["Squared Totals"].sum()
SSR, SSE, SST
n = results.nobs
k = len(results.params)
estimator_variance = SSE / (n-k)
n, k, estimator_variance
cov_matrix = results.cov_params()
cov_matrix | _____no_output_____ | MIT | In-Class Projects/Project 8 - Working with OLS.ipynb | zacharyejohnson/ECON411 |
Calculate t-stats | parameters = {}
for x_var in cov_matrix.keys():
parameters[x_var] = {}
parameters[x_var]["Beta"] = results.params[x_var]
parameters[x_var]["Standard Error"] = cov_matrix.loc[x_var, x_var]**(1 / 2)
parameters[x_var]["t_stats"] = parameters[x_var]["Beta"] / parameters[
x_var]["Standard Error"]
pd.DataFrame(parameters).T
r2 = SSR / SST
r2
results.summary() | _____no_output_____ | MIT | In-Class Projects/Project 8 - Working with OLS.ipynb | zacharyejohnson/ECON411 |
Plot Residuals | import matplotlib.pyplot as plt
plt.rcParams.update({"font.size": 26})
fig, ax = plt.subplots(figsize=(12, 8))
reg_data[["Residuals"]].plot.hist(bins=100, ax=ax)
plt.xticks(rotation=60) | _____no_output_____ | MIT | In-Class Projects/Project 8 - Working with OLS.ipynb | zacharyejohnson/ECON411 |
slightly skewed left. Need to log the data in order to normally distrbute it Regression using rates | reg_data = data
reg_data["RGDP Per Capita"] = data.groupby("ISO_Code")["RGDP Per Capita"].pct_change()
reg_data["RGDP Per Capita Lag"] = reg_data["RGDP Per Capita"].shift()
reg_data = reg_data.replace([np.inf, -np.inf], np.nan).dropna(axis = 0, how = "any")
reg_data.loc["USA"]
reg_data.corr().round(2)
y_var = ["RGDP Per Capita"]
x_vars = ["Size of Government",
"Legal System & Property Rights",
"Sound Money",
"Freedom to trade internationally",
"Regulation",
"RGDP Per Capita Lag"]
y = reg_data[y_var]
X = reg_data[x_vars]
x["Constant"] = 1
results = sm.OLS(y, X).fit()
reg_data["Predictor"] = results.predict()
results.summary()
reg_data["Residuals"] = results.resid
fig, ax = plt.subplots(figsize = (12,8))
reg_data[["Residuals"]].plot.hist(bins = 100, ax = ax)
betaEstimates = results.params
tStats = results.tvalues
pValues = results.pvalues
stdErrors = results.bse
resultsDict = {"Beta Estimates" : betaEstimates,
"t-stats":tStats,
"p-values":pValues,
"Standard Errors":stdErrors}
resultsDF = pd.DataFrame(resultsDict)
resultsDF.round(3)
fig, ax = plt.subplots(figsize = (14,10))
reg_data.plot.scatter(x = y_var[0],
y = "Predictor",
s = 30, ax = ax)
plt.xticks(rotation=90)
plt.show()
plt.close()
fig, ax = plt.subplots(figsize = (14,10))
reg_data.plot.scatter(x = y_var[0],
y = "Residuals",
s = 30, ax = ax)
ax.axhline(0, ls = "--", color = "k")
plt.xticks(rotation=90)
plt.show()
plt.close()
| _____no_output_____ | MIT | In-Class Projects/Project 8 - Working with OLS.ipynb | zacharyejohnson/ECON411 |
DatafaucetDatafaucet is a productivity framework for ETL, ML application. Simplifying some of the common activities which are typical in Data pipeline such as project scaffolding, data ingesting, start schema generation, forecasting etc. | import datafaucet as dfc | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Loading and Saving Data | dfc.project.load()
query = """
SELECT
p.payment_date,
p.amount,
p.rental_id,
p.staff_id,
c.*
FROM payment p
INNER JOIN customer c
ON p.customer_id = c.customer_id;
"""
df = dfc.load(query, 'pagila') | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Select cols | df.cols.find('id').columns
df.cols.find(by_type='string').columns
df.cols.find(by_func=lambda x: x.startswith('st')).columns
df.cols.find('^st').columns | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Collect data, oriented by rows or cols | df.cols.find(by_type='numeric').rows.collect(3)
df.cols.find(by_type='string').collect(3)
df.cols.find('name', 'date').data.collect(3) | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Get just one row or column | df.cols.find('active', 'amount', 'name').one()
df.cols.find('active', 'amount', 'name').rows.one() | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Grid view | df.cols.find('amount', 'id', 'name').data.grid(5) | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Data Exploration | df.cols.find('amount', 'id', 'name').data.facets() | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Rename columns | df.cols.find(by_type='timestamp').rename('new_', '***').columns
# to do
# df.cols.rename(transform=['unidecode', 'alnum', 'alpha', 'num', 'lower', 'trim', 'squeeze', 'slice', tr("abc", "_", mode='')'])
# df.cols.rename(transform=['unidecode', 'alnum', 'lower', 'trim("_")', 'squeeze("_")'])
# as a dictionary
mapping = {
'staff_id': 'foo',
'first_name': 'bar',
'email': 'qux',
'active':'active'
}
# or as a list of 2-tuples
mapping = [
('staff_id','foo'),
('first_name','bar'),
'active'
]
dict(zip(df.columns, df.cols.rename('new_', '***', mapping).columns)) | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Drop multiple columns | df.cols.find('id').drop().rows.collect(3) | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Apply to multiple columns | from pyspark.sql import functions as F
(df
.cols.find(by_type='string').lower()
.cols.get('email').split('@')
.cols.get('email').expand(2)
.cols.find('name', 'email')
.rows.collect(3)
) | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Aggregations | from datafaucet.spark import aggregations as A
df.cols.find('amount', '^st.*id', 'first_name').agg(A.all).cols.collect(10) | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
group by a set of columns | df.cols.find('amount').groupby('staff_id', 'store_id').agg(A.all).cols.collect(4) | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Aggregate specific metrics | # by function
df.cols.get('amount', 'active').groupby('customer_id').agg({'count':F.count, 'sum': F.sum}).rows.collect(10)
# or by alias
df.cols.get('amount', 'active').groupby('customer_id').agg('count','sum').rows.collect(10)
# or a mix of the two
df.cols.get('amount', 'active').groupby('customer_id').agg('count',{'sum': F.sum}).rows.collect(10) | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Featurize specific metrics in a single row | (df
.cols.get('amount', 'active')
.groupby('customer_id', 'store_id')
.featurize({'count':A.count, 'sum':A.sum, 'avg':A.avg})
.rows.collect(10)
)
# todo:
# different features per different column | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Plot dataset statistics | df.data.summary()
from bokeh.io import output_notebook
output_notebook()
from bokeh.plotting import figure, show, output_file
p = figure(plot_width=400, plot_height=400)
p.hbar(y=[1, 2, 3], height=0.5, left=0,
right=[1.2, 2.5, 3.7], color="navy")
show(p)
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(style="whitegrid")
# Initialize the matplotlib figure
f, ax = plt.subplots(figsize=(6, 6))
# Load the example car crash dataset
crashes = sns.load_dataset("car_crashes").sort_values("total", ascending=False)[:10]
# Plot the total crashes
sns.set_color_codes("pastel")
sns.barplot(x="total", y="abbrev", data=crashes,
label="Total", color="b")
# Plot the crashes where alcohol was involved
sns.set_color_codes("muted")
sns.barplot(x="alcohol", y="abbrev", data=crashes,
label="Alcohol-involved", color="b")
# Add a legend and informative axis label
ax.legend(ncol=2, loc="lower right", frameon=True)
ax.set(xlim=(0, 24), ylabel="",
xlabel="Automobile collisions per billion miles")
sns.despine(left=True, bottom=True)
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(style="white", palette="muted", color_codes=True)
# Generate a random univariate dataset
rs = np.random.RandomState(10)
d = rs.normal(size=100)
# Plot a simple histogram with binsize determined automatically
sns.distplot(d, hist=True, kde=True, rug=True, color="b");
import seaborn as sns
sns.set(style="ticks")
df = sns.load_dataset("iris")
sns.pairplot(df, hue="species")
from IPython.display import HTML
HTML('''
<!-- Bootstrap CSS -->
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/css/bootstrap.min.css" crossorigin="anonymous">
<div class="container-fluid">
<div class="jumbotron">
<h1 class="display-4">Hello, world!</h1>
<p class="lead">This is a simple hero unit, a simple jumbotron-style component for calling extra attention to featured content or information.</p>
<hr class="my-4">
<p>It uses utility classes for typography and spacing to space content out within the larger container.</p>
<a class="btn btn-primary btn-lg" href="#" role="button">Learn more</a>
</div>
<button type="button" class="btn btn-secondary" data-toggle="tooltip" data-placement="top" title="Tooltip on top">
Tooltip on top
</button>
<button type="button" class="btn btn-secondary" data-toggle="tooltip" data-placement="right" title="Tooltip on right">
Tooltip on right
</button>
<button type="button" class="btn btn-secondary" data-toggle="tooltip" data-placement="bottom" title="Tooltip on bottom">
Tooltip on bottom
</button>
<button type="button" class="btn btn-secondary" data-toggle="tooltip" data-placement="left" title="Tooltip on left">
Tooltip on left
</button>
<table class="table">
<thead>
<tr>
<th scope="col">#</th>
<th scope="col">First</th>
<th scope="col">Last</th>
<th scope="col">Handle</th>
</tr>
</thead>
<tbody>
<tr>
<th scope="row">1</th>
<td>Mark</td>
<td>Otto</td>
<td>@mdo</td>
</tr>
<tr>
<th scope="row">2</th>
<td>Jacob</td>
<td>Thornton</td>
<td>@fat</td>
</tr>
<tr>
<th scope="row">3</th>
<td>Larry</td>
<td>the Bird</td>
<td>@twitter</td>
</tr>
</tbody>
</table>
<span class="badge badge-primary">Primary</span>
<span class="badge badge-secondary">Secondary</span>
<span class="badge badge-success">Success</span>
<span class="badge badge-danger">Danger</span>
<span class="badge badge-warning">Warning</span>
<span class="badge badge-info">Info</span>
<span class="badge badge-light">Light</span>
<span class="badge badge-dark">Dark</span>
<table class="table table-sm" style="text-align:left">
<thead>
<tr>
<th scope="col">#</th>
<th scope="col">First</th>
<th scope="col">Last</th>
<th scope="col">Handle</th>
<th scope="col">bar</th>
</tr>
</thead>
<tbody>
<tr>
<th scope="row">1</th>
<td>Mark</td>
<td>Otto</td>
<td>@mdo</td>
<td class="text-left"><span class="badge badge-primary" style="width: 75%">Primary</span></td>
</tr>
<tr>
<th scope="row">2</th>
<td>Jacob</td>
<td>Thornton</td>
<td>@fat</td>
<td class="text-left"><span class="badge badge-secondary" style="width: 25%">Primary</span></td>
</tr>
<tr>
<th scope="row">3</th>
<td colspan="2">Larry the Bird</td>
<td>@twitter</td>
<td class="text-left"><span class="badge badge-warning" style="width: 55%">Primary</span></td>
</div>
</tr>
</tbody>
</table>
</div>''')
tbl = '''
<table class="table table-sm">
<thead>
<tr>
<th scope="col">#</th>
<th scope="col">First</th>
<th scope="col">Last</th>
<th scope="col">Handle</th>
<th scope="col">bar</th>
</tr>
</thead>
<tbody>
<tr>
<th scope="row">1</th>
<td>Mark</td>
<td>Otto</td>
<td>@mdo</td>
<td class="text-left"><span class="badge badge-primary" style="width: 75%">75%</span></td>
</tr>
<tr>
<th scope="row">2</th>
<td>Jacob</td>
<td>Thornton</td>
<td>@fat</td>
<td class="text-left"><span class="badge badge-secondary" style="width: 25%" title="Tooltip on top">25%</span></td>
</tr>
<tr>
<th scope="row">3</th>
<td colspan="2">Larry the Bird</td>
<td>@twitter</td>
<td class="text-left"><span class="badge badge-warning" style="width: 0%">0%</span></td>
</tr>
</tbody>
</table>
'''
drp = '''
<div class="dropdown">
<button class="btn btn-secondary dropdown-toggle" type="button" id="dropdownMenuButton" data-toggle="dropdown" aria-haspopup="true" aria-expanded="false">
Dropdown button
</button>
<div class="dropdown-menu" aria-labelledby="dropdownMenuButton">
<a class="dropdown-item" href="#">Action</a>
<a class="dropdown-item" href="#">Another action</a>
<a class="dropdown-item" href="#">Something else here</a>
</div>
</div>'''
tabs = f'''
<nav>
<div class="nav nav-tabs" id="nav-tab" role="tablist">
<a class="nav-item nav-link active" id="nav-home-tab" data-toggle="tab" href="#nav-home" role="tab" aria-controls="nav-home" aria-selected="true">Home</a>
<a class="nav-item nav-link" id="nav-profile-tab" data-toggle="tab" href="#nav-profile" role="tab" aria-controls="nav-profile" aria-selected="false">Profile</a>
<a class="nav-item nav-link" id="nav-contact-tab" data-toggle="tab" href="#nav-contact" role="tab" aria-controls="nav-contact" aria-selected="false">Contact</a>
</div>
</nav>
<div class="tab-content" id="nav-tabContent">
<div class="tab-pane fade show active" id="nav-home" role="tabpanel" aria-labelledby="nav-home-tab">..jjj.</div>
<div class="tab-pane fade" id="nav-profile" role="tabpanel" aria-labelledby="nav-profile-tab">..kkk.</div>
<div class="tab-pane fade" id="nav-contact" role="tabpanel" aria-labelledby="nav-contact-tab">{tbl}</div>
</div>
'''
from IPython.display import HTML
HTML(f'''
<!-- Bootstrap CSS -->
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/css/bootstrap.min.css" crossorigin="anonymous">
<div class="container-fluid">
<div class="row">
<div class="col">
{drp}
</div>
<div class="col">
{tabs}
</div>
<div class="col">
{tbl}
</div>
</div>
</div>
<script src="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/js/bootstrap.bundle.min.js" crossorigin="anonymous" >
''')
from IPython.display import HTML
HTML(f'''
<!-- Bootstrap CSS -->
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/css/bootstrap.min.css" crossorigin="anonymous">
<script src="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/js/bootstrap.bundle.min.js" crossorigin="anonymous" >
''')
d =df.cols.find('id', 'name').sample(10)
d.columns
tbl_head = '''
<thead>
<tr>
'''
tbl_head += '\n'.join([' <th scope="col">'+str(x)+'</th>' for x in d.columns])
tbl_head +='''
</tr>
</thead>
'''
print(tbl_head)
tbl_body = '''
<tbody>
<tr>
<th scope="row">1</th>
<td>Mark</td>
<td>Otto</td>
<td>@mdo</td>
<td class="text-left"><span class="badge badge-primary" style="width: 75%">75%</span></td>
</tr>
<tr>
<th scope="row">2</th>
<td>Jacob</td>
<td>Thornton</td>
<td>@fat</td>
<td class="text-left"><span class="badge badge-secondary" style="width: 25%" title="Tooltip on top">25%</span></td>
</tr>
<tr>
<th scope="row">3</th>
<td colspan="2">Larry the Bird</td>
<td>@twitter</td>
<td class="text-left"><span class="badge badge-warning" style="width: 0%">0%</span></td>
</tr>
</tbody>
</table>
'''
HTML(f'''
<!-- Bootstrap CSS -->
<div class="container-fluid">
<div class="row">
<div class="col">
<table class="table table-sm">
{tbl_head}
{tbl_body}
</table>
</div>
</div>
</div>
''')
# .rows.sample()
# .cols.select('name', 'id', 'amount')\
# .cols.apply(F.lower, 'name')\
# .cols.apply(F.floor, 'amount', output_prefix='_')\
# .cols.drop('^amount$')\
# .cols.rename()
# .cols.unicode()
.grid()
df = df.cols.select('name')
df = df.rows.overwrite([('Nhập mật', 'khẩu')])
df.columns
# .rows.overwrite(['Nhập mật', 'khẩu'])\
# .cols.apply(F.lower)\
# .grid()
# #withColumn('pippo', F.lower(F.col('first_name'))).grid()
import pandas as pd
df = pd.DataFrame({'lab':['A', 'B', 'C'], 'val':[10, 30, 20]})
df.plot.bar(x='lab', y='val', rot=0); | _____no_output_____ | MIT | examples/tutorial/patched.ipynb | natbusa/datalabframework |
Indexer for SantaScript score queryedit https://www.elastic.co/guide/en/elasticsearch/reference/current/query-dsl-script-score-query.htmlvector-functionsELASTICSEARCHで分散表現を使った類似文書検索 https://yag-ays.github.io/project/elasticsearch-similarity-search/Image Search for ICDAR WML 2019 https://github.com/taniokah/icdar-wml-2019/blob/master/Image%20Search%20for%20ICDAR%20WML%202019.ipynb | # Crawling Santa images.
!pip install icrawler
!rm -rf google_images/*
!rm -rf bing_images/*
!rm -rf baidu_images/*
from icrawler.builtin import BaiduImageCrawler, BingImageCrawler, GoogleImageCrawler
crawler = GoogleImageCrawler(storage={"root_dir": "google_images"}, downloader_threads=4)
crawler.crawl(keyword="Santa", offset=0, max_num=1000)
#bing_crawler = BingImageCrawler(storage={'root_dir': 'bing_images'}, downloader_threads=4)
#bing_crawler.crawl(keyword='Santa', filters=None, offset=0, max_num=1000)
#baidu_crawler = BaiduImageCrawler(storage={'root_dir': 'baidu_images'})
#baidu_crawler.crawl(keyword='Santa', offset=0, max_num=1000)
!wget https://artifacts.elastic.co/downloads/elasticsearch/elasticsearch-7.5.1-linux-x86_64.tar.gz -q
!tar -xzf elasticsearch-7.5.1-linux-x86_64.tar.gz
!chown -R daemon:daemon elasticsearch-7.5.1/
#!elasticsearch-7.5.1/bin/elasticsearch
import os
from subprocess import Popen, PIPE, STDOUT
es_server = Popen(['elasticsearch-7.5.1/bin/elasticsearch'],
stdout=PIPE, stderr=STDOUT,
preexec_fn=lambda: os.setuid(1) # as daemon
)
!ps aux | grep elastic
!sleep 30
!curl -X GET "localhost:9200/"
!pip install elasticsearch
from datetime import datetime
from elasticsearch import Elasticsearch
es = Elasticsearch(timeout=60)
doc = {
'author': 'Santa Claus',
'text': 'Where is Santa Claus?',
'timestamp': datetime.now(),
}
res = es.index(index="test-index", doc_type='tweet', id=1, body=doc)
print(res['result'])
res = es.get(index="test-index", doc_type='tweet', id=1)
print(res['_source'])
es.indices.refresh(index="test-index")
res = es.search(index="test-index", body={"query": {"match_all": {}}})
print("Got %d Hits:" % res['hits']['total']['value'])
for hit in res['hits']['hits']:
print("%(timestamp)s %(author)s: %(text)s" % hit["_source"])
# Load libraries
from keras.applications.vgg16 import VGG16, preprocess_input, decode_predictions
from keras.preprocessing import image
from PIL import Image
import matplotlib.pyplot as plt
import numpy as np
import sys
model = VGG16(weights='imagenet')
def predict(filename, featuresize, scale=1.0):
img = image.load_img(filename, target_size=(224, 224))
return predictimg(img, featuresize, scale=1.0)
def predictpart(filename, featuresize, scale=1.0, size=1):
im = Image.open(filename)
width, height = im.size
im = im.resize((width * size, height * size))
im_list = np.asarray(im)
# partition
out_img = []
if size > 1:
v_split = size
h_split = size
[out_img.extend(np.hsplit(h_img, h_split)) for h_img in np.vsplit(im_list, v_split)]
else:
out_img.append(im_list)
reslist = []
for offset in range(size * size):
img = Image.fromarray(out_img[offset])
reslist.append(predictimg(img, featuresize, scale))
return reslist
def predictimg(img, featuresize, scale=1.0):
width, height = img.size
img = img.resize((int(width * scale), int(height * scale)))
img = img.resize((224, 224))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
preds = model.predict(preprocess_input(x))
results = decode_predictions(preds, top=featuresize)[0]
return results
def showimg(filename, title, i, scale=1.0, col=2, row=5):
im = Image.open(filename)
width, height = im.size
im = im.resize((int(width * scale), int(height * scale)))
im = im.resize((width, height))
im_list = np.asarray(im)
plt.subplot(col, row, i)
plt.title(title)
plt.axis("off")
plt.imshow(im_list)
def showpartimg(filename, title, i, size, scale=1.0, col=2, row=5):
im = Image.open(filename)
width, height = im.size
im = im.resize((int(width * scale), int(height * scale)))
#im = im.resize((width, height))
im = im.resize((width * size, height * size))
im_list = np.asarray(im)
# partition
out_img = []
if size > 1:
v_split = size
h_split = size
[out_img.extend(np.hsplit(h_img, h_split)) for h_img in np.vsplit(im_list, v_split)]
else:
out_img.append(im_list)
# draw image
for offset in range(size * size):
im_list = out_img[offset]
pos = i + offset
print(str(col) + ' ' + str(row) + ' ' + str(pos))
plt.subplot(col, row, pos)
plt.title(title)
plt.axis("off")
plt.imshow(im_list)
out_img[offset] = Image.fromarray(im_list)
return out_img
# Predict an image
scale = 1.0
filename = "google_images/000046.jpg"
plt.figure(figsize=(20, 10))
#showimg(filename, "query", i+1, scale)
imgs = showpartimg(filename, "query", 1, 1, scale)
plt.show()
for img in imgs:
reslist = predictpart(filename, 10, scale)
for results in reslist:
for result in results:
print(result)
print()
def createindex(indexname):
if es.indices.exists(index=indexname):
es.indices.delete(index=indexname)
es.indices.create(index=indexname, body={
"settings": {
"index.mapping.total_fields.limit": 10000,
}
})
mapping = {
"image": {
"properties": {
"f": {
"type": "text"
},
's': {
"type": "sparse_vector"
}
}
}
}
es.indices.put_mapping(index=indexname, doc_type='image', body=mapping, include_type_name=True)
wnidmap = {}
def loadimages(directory):
imagefiles = []
for file in os.listdir(directory):
if file.rfind('.jpg') < 0:
continue
filepath = os.path.join(directory, file)
imagefiles.append(filepath)
return imagefiles
def indexfiles(indexname, directory, featuresize=10, docsize=1000):
imagefiles = loadimages(directory)
for i in range(len(imagefiles)):
if i >= docsize:
return
filename = imagefiles[i]
indexfile(indexname, filename, i, featuresize)
sys.stdout.write("\r%d" % (i + 1))
sys.stdout.flush()
es.indices.refresh(index=indexname)
def indexfile(indexname, filename, i, featuresize):
global wnidmap
rounddown = 16
doc = {'f': filename, 's':{}}
results = predict(filename, featuresize)
#print(len(results))
synset = doc['s']
for result in results:
score = float(str(result[2]))
wnid = result[0]
id = 0
if wnid in wnidmap.keys():
id = wnidmap[wnid]
else:
id = len(wnidmap)
wnidmap[wnid] = id
synset[str(id)] = score
#print(doc)
#count = es.count(index=indexname, doc_type='image')['count']
count = i
res = es.index(index=indexname, doc_type='image', id=count, body=doc)
createindex("santa-search")
directory = "google_images/"
indexfiles("santa-search", directory, 100, 1000)
#directory = "bing_images/"
#indexfiles("santa-search", directory, 100, 1000)
#directory = "baidu_images/"
#indexfiles("santa-search", directory, 100, 1000)
res = es.search(index="santa-search", request_timeout=60, body={"query": {"match_all": {}}})
print("Got " + str(res['hits']['total']) + " Hits:" )
for hit in res['hits']['hits']:
print(hit["_source"])
#print("%(timestamp)s %(author)s: %(text)s" % hit["_source"])
def searchimg(indexname, filename, num=10, topk=10, scoretype='dot', scale=1.0, partition=1):
plt.figure(figsize=(20, 10))
imgs = showpartimg(filename, "query", 1, partition, scale)
plt.show()
reslist = []
for img in imgs:
results = predictimg(img, num, scale)
for result in results:
print(result)
print()
res = search(indexname, results, num, topk, scoretype)
reslist.append(res)
return reslist
def search(indexname, synsets, num, topk, scoretype='dot', disp=True):
if scoretype == 'vcos':
inline = {}
for synset in synsets:
score = synset[2]
if score <= 0.0:
continue
wnid = synset[0]
if wnid not in wnidmap.keys():
continue
id = wnidmap[wnid]
inline[str(id)] = float(score)
if inline == {}:
print("Got " + str(0) + " Hits:")
return
#print('wnidmap = ' + str(wnidmap))
#print('inline = ' + str(inline))
b = {
"size": topk,
"query": {
"script_score": {
"query": {"match_all": {}},
"script": {
"source": "cosineSimilaritySparse(params.s, doc['s']) + 0.01",
"params": {
's': {}
}
}
}
}}
b['query']['script_score']['script']['params']['s'] = inline
res = es.search(index=indexname, body=b)
#print(str(b))
if disp==True:
print("Got " + str(res['hits']['total']['value']) + " Hits:")
topres = res['hits']['hits'][0:topk]
for hit in topres:
print(str(hit["_id"]) + " " + str(hit["_source"]["f"]) + " " + str(hit["_score"]))
plt.figure(figsize=(20, 10))
for i in range(len(topres)):
hit = topres[i]
row = 5
col = int(topk / 5)
if i >= 25:
break
showimg(hit["_source"]["f"], hit["_id"], i+1, col, row)
plt.show()
return res
filename = "google_images/000001.jpg"
_ = searchimg('santa-search', filename, 10, 10, 'vcos', 1.0, 1)
| _____no_output_____ | MIT | Indexer_for_Santa.ipynb | taniokah/where-is-santa- |
===================================================================Determine the observable time of the Canopus on the Vernal and Autumnal equinox among -2000 B.C.E. ~ 0 B.C. | %matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from astropy.visualization import astropy_mpl_style
plt.style.use(astropy_mpl_style)
import astropy.units as u
from astropy.time import Time
from astropy.coordinates import SkyCoord, EarthLocation, AltAz, ICRS | _____no_output_____ | BSD-2-Clause | multi_epoch-max-duration-Autumnal.ipynb | Niu-LIU/Canopus |
The observing period is the whole year of -2000 B.C.E. ~ 0 B.C.To represent the epoch before the common era, I use the Julian date. | We can see that if we transformate the dates into UTC, they don't exactly respond to March 21 or September 23.
This is normal since UTC is used only after 1960-01-01.
In my opinion, this won't affect our results. | _____no_output_____ | BSD-2-Clause | multi_epoch-max-duration-Autumnal.ipynb | Niu-LIU/Canopus |
I calculate the altitude and azimuth of Sun and Canopus among 4:00~8:00 in autumnal equinox and 16:00~20:00 in vernal equinox for every year. | def observable_duration(obs_time):
"""
"""
# Assume we have an observer in Tai Mountain.
taishan = EarthLocation(lat=36.2*u.deg, lon=117.1*u.deg, height=1500*u.m)
utcoffset = +8 * u.hour # Daylight Time
midnight = obs_time - utcoffset
# Position of the Canopus with the proper motion correction at the beginning of the year.
# This effect is very small.
dt_jyear = obs_time.jyear - 2000.0
ra = 95.98787790 * u.deg + 19.93 * u.mas * dt_jyear
dec = -52.69571787 * u.deg + 23.24 * u.mas * dt_jyear
hip30438 = SkyCoord(ra=ra, dec=dec, frame="icrs")
delta_midnight = np.arange(0, 24, 1./30) * u.hour # Interval of 2 minutes
obser_time = midnight + delta_midnight
local_frame = AltAz(obstime=obser_time,
location=taishan)
hip30438altazs = hip30438.transform_to(local_frame)
# position of Sun
from astropy.coordinates import get_sun
sunaltazs = get_sun(obser_time).transform_to(local_frame)
mask = (sunaltazs.alt < -0*u.deg) & (hip30438altazs.alt > 0)
observable_time = delta_midnight[mask]
# observable_time
if len(observable_time):
beg_time = observable_time.min().to('hr').value
end_time = observable_time.max().to('hr').value
else:
beg_time, end_time = 0, 0
return beg_time, end_time
year_arr = np.arange(0, 2000, 1)
# Number of days for every year
date_nb = np.ones_like(year_arr)
date_nb = np.where(year_arr % 4 == 0, 366, 365)
date_nb = np.where((year_arr % 100 == 0) & (
year_arr % 400 != 0), 365, date_nb)
total_date_nb = np.zeros_like(year_arr)
for i in range(year_arr.size):
total_date_nb[i] = np.sum(date_nb[:i+1])
# Autumnal equinox of every year
obs_time_aut = Time("0000-09-23 00:00:00") - total_date_nb * u.day
# Calculate the observable time of everyday
beg_time = np.zeros_like(obs_time_aut)
end_time = np.zeros_like(obs_time_aut)
obs_dur = np.zeros_like(obs_time_aut) # Observable duration
for i, obs_timei in enumerate(obs_time_aut):
# we calculate the 30 days before and after the equinox
delta_date = np.arange(-5, 5, 1) * u.day
obs_time0 = obs_timei + delta_date
beg_time_aut = np.zeros_like(obs_time0)
end_time_aut = np.zeros_like(obs_time0)
for j, obs_time0j in enumerate(obs_time0):
# Vernal equninox
beg_time_aut[j], end_time_aut[j] = observable_duration(obs_time0j)
obs_dur_aut = end_time_aut - beg_time_aut
obs_dur[i] = np.max(obs_dur_aut)
beg_time[i] = beg_time_aut[obs_dur_aut == obs_dur[i]][0]
end_time[i] = end_time_aut[obs_dur_aut == obs_dur[i]][0] | WARNING: ErfaWarning: ERFA function "dtf2d" yielded 1 of "dubious year (Note 6)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "utctai" yielded 1 of "dubious year (Note 3)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "taiutc" yielded 2000 of "dubious year (Note 4)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "taiutc" yielded 10 of "dubious year (Note 4)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "taiutc" yielded 1 of "dubious year (Note 4)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "taiutc" yielded 720 of "dubious year (Note 4)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "utctai" yielded 720 of "dubious year (Note 3)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "epv00" yielded 720 of "warning: date outsidethe range 1900-2100 AD" [astropy._erfa.core]
WARNING: Tried to get polar motions for times before IERS data is valid. Defaulting to polar motion from the 50-yr mean for those. This may affect precision at the 10s of arcsec level [astropy.coordinates.builtin_frames.utils]
WARNING: ErfaWarning: ERFA function "apio13" yielded 720 of "dubious year (Note 2)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "utctai" yielded 720 of "dubious year (Note 3)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "taiutc" yielded 720 of "dubious year (Note 4)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "epv00" yielded 720 of "warning: date outsidethe range 1900-2100 AD" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "utcut1" yielded 720 of "dubious year (Note 3)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "utctai" yielded 1 of "dubious year (Note 3)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "taiutc" yielded 1 of "dubious year (Note 4)" [astropy._erfa.core]
WARNING: ErfaWarning: ERFA function "taiutc" yielded 10 of "dubious year (Note 4)" [astropy._erfa.core]
| BSD-2-Clause | multi_epoch-max-duration-Autumnal.ipynb | Niu-LIU/Canopus |
I assume that the Canopus can be observed by the local observer only when the observable duration in one day is longer than 10 minitues.With such an assumption, I determine the observable period of the Canopus. | # Save data
np.save("multi_epoch-max-duration-Autumnal-output", [obs_time_aut.jyear, obs_dur])
# For Autumnal equinox
# mask = (obs_dur >= 1./6)
mask = (obs_dur >= 1.0/60)
observable_date = obs_time_aut[mask]
fig, ax = plt.subplots(figsize=(12, 8))
ax.plot(observable_date.jyear, obs_dur[mask],
"r.", ms=3, label="Autumnal")
# ax.fill_between(obs_time.jyear, 0, 24,
# (obs_dur1 >= 1./6) & (obs_dur2 >= 1./6), color="0.8", zorder=0)
ax.set_xlabel("Year", fontsize=15)
ax.set_xlim([-2000, 0])
ax.set_xticks(np.arange(-2000, 1, 100))
ax.set_ylim([0, 2.0])
ax.set_ylabel("Time (hour)", fontsize=15)
ax.set_title("Observable duration of Canopus among $-2000$ B.C.E and 0")
ax.legend(fontsize=15)
fig.tight_layout()
plt.savefig("multi_epoch-max-duration-Autumnal.eps", dpi=100)
plt.savefig("multi_epoch-max-duration-Autumnal.png", dpi=100) | _____no_output_____ | BSD-2-Clause | multi_epoch-max-duration-Autumnal.ipynb | Niu-LIU/Canopus |
Build a Pipeline> A tutorial on building pipelines to orchestrate your ML workflowA Kubeflow pipeline is a portable and scalable definition of a machine learning(ML) workflow. Each step in your ML workflow, such as preparing data ortraining a model, is an instance of a pipeline component. This documentprovides an overview of pipeline concepts and best practices, and instructionsdescribing how to build an ML pipeline. Before you begin1. Run the following command to install the Kubeflow Pipelines SDK. If you run this command in a Jupyter notebook, restart the kernel after installing the SDK. | !pip install kfp --upgrade | _____no_output_____ | CC-BY-4.0 | content/en/docs/components/pipelines/sdk/build-pipeline.ipynb | droctothorpe/website |
2. Import the `kfp` and `kfp.components` packages. | import kfp
import kfp.components as comp | _____no_output_____ | CC-BY-4.0 | content/en/docs/components/pipelines/sdk/build-pipeline.ipynb | droctothorpe/website |
Understanding pipelinesA Kubeflow pipeline is a portable and scalable definition of an ML workflow,based on containers. A pipeline is composed of a set of input parameters and alist of the steps in this workflow. Each step in a pipeline is an instance of acomponent, which is represented as an instance of [`ContainerOp`][container-op].You can use pipelines to:* Orchestrate repeatable ML workflows.* Accelerate experimentation by running a workflow with different sets of hyperparameters. Understanding pipeline componentsA pipeline component is a containerized application that performs one step in apipeline's workflow. Pipeline components are defined in[component specifications][component-spec], which define the following:* The component's interface, its inputs and outputs.* The component's implementation, the container image and the command to execute.* The component's metadata, such as the name and description of the component.You can build components by [defining a component specification for acontainerized application][component-dev], or you can [use the KubeflowPipelines SDK to generate a component specification for a Pythonfunction][python-function-component]. You can also [reuse prebuilt componentsin your pipeline][prebuilt-components]. Understanding the pipeline graphEach step in your pipeline's workflow is an instance of a component. Whenyou define your pipeline, you specify the source of each step's inputs. Stepinputs can be set from the pipeline's input arguments, constants, or stepinputs can depend on the outputs of other steps in this pipeline. KubeflowPipelines uses these dependencies to define your pipeline's workflow asa graph.For example, consider a pipeline with the following steps: ingest data,generate statistics, preprocess data, and train a model. The followingdescribes the data dependencies between each step.* **Ingest data**: This step loads data from an external source which is specified using a pipeline argument, and it outputs a dataset. Since this step does not depend on the output of any other steps, this step can run first.* **Generate statistics**: This step uses the ingested dataset to generate and output a set of statistics. Since this step depends on the dataset produced by the ingest data step, it must run after the ingest data step.* **Preprocess data**: This step preprocesses the ingested dataset and transforms the data into a preprocessed dataset. Since this step depends on the dataset produced by the ingest data step, it must run after the ingest data step.* **Train a model**: This step trains a model using the preprocessed dataset, the generated statistics, and pipeline parameters, such as the learning rate. Since this step depends on the preprocessed data and the generated statistics, it must run after both the preprocess data and generate statistics steps are complete.Since the generate statistics and preprocess data steps both depend on theingested data, the generate statistics and preprocess data steps can run inparallel. All other steps are executed once their data dependencies areavailable. Designing your pipelineWhen designing your pipeline, think about how to split your ML workflow intopipeline components. The process of splitting an ML workflow into pipelinecomponents is similar to the process of splitting a monolithic script intotestable functions. The following rules can help you define the componentsthat you need to build your pipeline.* Components should have a single responsibility. Having a single responsibility makes it easier to test and reuse a component. For example, if you have a component that loads data you can reuse that for similar tasks that load data. If you have a component that loads and transforms a dataset, the component can be less useful since you can use it only when you need to load and transform that dataset. * Reuse components when possible. Kubeflow Pipelines provides [components for common pipeline tasks and for access to cloud services][prebuilt-components].* Consider what you need to know to debug your pipeline and research the lineage of the models that your pipeline produces. Kubeflow Pipelines stores the inputs and outputs of each pipeline step. By interrogating the artifacts produced by a pipeline run, you can better understand the variations in model quality between runs or track down bugs in your workflow.In general, you should design your components with composability in mind. Pipelines are composed of component instances, also called steps. Steps candefine their inputs as depending on the output of another step. Thedependencies between steps define the pipeline workflow graph. Building pipeline componentsKubeflow pipeline components are containerized applications that perform astep in your ML workflow. Here are the ways that you can define pipelinecomponents:* If you have a containerized application that you want to use as a pipeline component, create a component specification to define this container image as a pipeline component. This option provides the flexibility to include code written in any language in your pipeline, so long as you can package the application as a container image. Learn more about [building pipeline components][component-dev].* If your component code can be expressed as a Python function, [evaluate if your component can be built as a Python function-based component][python-function-component]. The Kubeflow Pipelines SDK makes it easier to build lightweight Python function-based components by saving you the effort of creating a component specification.Whenever possible, [reuse prebuilt components][prebuilt-components] to saveyourself the effort of building custom components.The example in this guide demonstrates how to build a pipeline that uses aPython function-based component and reuses a prebuilt component. Understanding how data is passed between componentsWhen Kubeflow Pipelines runs a component, a container image is started in aKubernetes Pod and your component’s inputs are passed in as command-linearguments. When your component has finished, the component's outputs arereturned as files.In your component's specification, you define the components inputs and outputsand how the inputs and output paths are passed to your program as command-linearguments. You can pass small inputs, such as short strings or numbers, to yourcomponent by value. Large inputs, such as datasets, must be passed to yourcomponent as file paths. Outputs are written to the paths that KubeflowPipelines provides.Python function-based components make it easier to build pipeline componentsby building the component specification for you. Python function-basedcomponents also handle the complexity of passing inputs into your componentand passing your function’s outputs back to your pipeline.Learn more about how [Python function-based components handle inputs andoutputs][python-function-component-data-passing]. Getting started building a pipelineThe following sections demonstrate how to get started building a Kubeflowpipeline by walking through the process of converting a Python script intoa pipeline. Design your pipelineThe following steps walk through some of the design decisions you may facewhen designing a pipeline.1. Evaluate the process. In the following example, a Python function downloads a zipped tar file (`.tar.gz`) that contains several CSV files, from a public website. The function extracts the CSV files and then merges them into a single file.[container-op]: https://kubeflow-pipelines.readthedocs.io/en/latest/source/kfp.dsl.htmlkfp.dsl.ContainerOp[component-spec]: https://www.kubeflow.org/docs/components/pipelines/reference/component-spec/[python-function-component]: https://www.kubeflow.org/docs/components/pipelines/sdk/python-function-components/[component-dev]: https://www.kubeflow.org/docs/components/pipelines/sdk/component-development/[python-function-component-data-passing]: https://www.kubeflow.org/docs/components/pipelines/sdk/python-function-components/understanding-how-data-is-passed-between-components[prebuilt-components]: https://www.kubeflow.org/docs/examples/shared-resources/ | import glob
import pandas as pd
import tarfile
import urllib.request
def download_and_merge_csv(url: str, output_csv: str):
with urllib.request.urlopen(url) as res:
tarfile.open(fileobj=res, mode="r|gz").extractall('data')
df = pd.concat(
[pd.read_csv(csv_file, header=None)
for csv_file in glob.glob('data/*.csv')])
df.to_csv(output_csv, index=False, header=False) | _____no_output_____ | CC-BY-4.0 | content/en/docs/components/pipelines/sdk/build-pipeline.ipynb | droctothorpe/website |
2. Run the following Python command to test the function. | download_and_merge_csv(
url='https://storage.googleapis.com/ml-pipeline-playground/iris-csv-files.tar.gz',
output_csv='merged_data.csv') | _____no_output_____ | CC-BY-4.0 | content/en/docs/components/pipelines/sdk/build-pipeline.ipynb | droctothorpe/website |
3. Run the following to print the first few rows of the merged CSV file. | !head merged_data.csv | _____no_output_____ | CC-BY-4.0 | content/en/docs/components/pipelines/sdk/build-pipeline.ipynb | droctothorpe/website |
4. Design your pipeline. For example, consider the following pipeline designs. * Implement the pipeline using a single step. In this case, the pipeline contains one component that works similarly to the example function. This is a straightforward function, and implementing a single-step pipeline is a reasonable approach in this case. The down side of this approach is that the zipped tar file would not be an artifact of your pipeline runs. Not having this artifact available could make it harder to debug this component in production. * Implement this as a two-step pipeline. The first step downloads a file from a website. The second step extracts the CSV files from a zipped tar file and merges them into a single file. This approach has a few benefits: * You can reuse the [Web Download component][web-download-component] to implement the first step. * Each step has a single responsibility, which makes the components easier to reuse. * The zipped tar file is an artifact of the first pipeline step. This means that you can examine this artifact when debugging pipelines that use this component. This example implements a two-step pipeline. Build your pipeline components 1. Build your pipeline components. This example modifies the initial script to extract the contents of a zipped tar file, merge the CSV files that were contained in the zipped tar file, and return the merged CSV file. This example builds a Python function-based component. You can also package your component's code as a Docker container image and define the component using a ComponentSpec. In this case, the following modifications were required to the original function. * The file download logic was removed. The path to the zipped tar file is passed as an argument to this function. * The import statements were moved inside of the function. Python function-based components require standalone Python functions. This means that any required import statements must be defined within the function, and any helper functions must be defined within the function. Learn more about [building Python function-based components][python-function-components]. * The function's arguments are decorated with the [`kfp.components.InputPath`][input-path] and the [`kfp.components.OutputPath`][output-path] annotations. These annotations let Kubeflow Pipelines know to provide the path to the zipped tar file and to create a path where your function stores the merged CSV file. The following example shows the updated `merge_csv` function.[web-download-component]: https://github.com/kubeflow/pipelines/blob/master/components/web/Download/component.yaml[python-function-components]: https://www.kubeflow.org/docs/components/pipelines/sdk/python-function-components/[input-path]: https://kubeflow-pipelines.readthedocs.io/en/latest/source/kfp.components.html?highlight=inputpathkfp.components.InputPath[output-path]: https://kubeflow-pipelines.readthedocs.io/en/latest/source/kfp.components.html?highlight=outputpathkfp.components.OutputPath | def merge_csv(file_path: comp.InputPath('Tarball'),
output_csv: comp.OutputPath('CSV')):
import glob
import pandas as pd
import tarfile
tarfile.open(name=file_path, mode="r|gz").extractall('data')
df = pd.concat(
[pd.read_csv(csv_file, header=None)
for csv_file in glob.glob('data/*.csv')])
df.to_csv(output_csv, index=False, header=False) | _____no_output_____ | CC-BY-4.0 | content/en/docs/components/pipelines/sdk/build-pipeline.ipynb | droctothorpe/website |
2. Use [`kfp.components.create_component_from_func`][create_component_from_func] to return a factory function that you can use to create pipeline steps. This example also specifies the base container image to run this function in, the path to save the component specification to, and a list of PyPI packages that need to be installed in the container at runtime.[create_component_from_func]: (https://kubeflow-pipelines.readthedocs.io/en/latest/source/kfp.components.htmlkfp.components.create_component_from_func[container-op]: https://kubeflow-pipelines.readthedocs.io/en/stable/source/kfp.dsl.htmlkfp.dsl.ContainerOp | create_step_merge_csv = kfp.components.create_component_from_func(
func=merge_csv,
output_component_file='component.yaml', # This is optional. It saves the component spec for future use.
base_image='python:3.7',
packages_to_install=['pandas==1.1.4']) | _____no_output_____ | CC-BY-4.0 | content/en/docs/components/pipelines/sdk/build-pipeline.ipynb | droctothorpe/website |
Build your pipeline1. Use [`kfp.components.load_component_from_url`][load_component_from_url] to load the component specification YAML for any components that you are reusing in this pipeline.[load_component_from_url]: https://kubeflow-pipelines.readthedocs.io/en/latest/source/kfp.components.html?highlight=load_component_from_urlkfp.components.load_component_from_url | web_downloader_op = kfp.components.load_component_from_url(
'https://raw.githubusercontent.com/kubeflow/pipelines/master/components/contrib/web/Download/component.yaml') | _____no_output_____ | CC-BY-4.0 | content/en/docs/components/pipelines/sdk/build-pipeline.ipynb | droctothorpe/website |
2. Define your pipeline as a Python function. Your pipeline function's arguments define your pipeline's parameters. Use pipeline parameters to experiment with different hyperparameters, such as the learning rate used to train a model, or pass run-level inputs, such as the path to an input file, into a pipeline run. Use the factory functions created by `kfp.components.create_component_from_func` and `kfp.components.load_component_from_url` to create your pipeline's tasks. The inputs to the component factory functions can be pipeline parameters, the outputs of other tasks, or a constant value. In this case, the `web_downloader_task` task uses the `url` pipeline parameter, and the `merge_csv_task` uses the `data` output of the `web_downloader_task`. | # Define a pipeline and create a task from a component:
def my_pipeline(url):
web_downloader_task = web_downloader_op(url=url)
merge_csv_task = create_step_merge_csv(file=web_downloader_task.outputs['data'])
# The outputs of the merge_csv_task can be referenced using the
# merge_csv_task.outputs dictionary: merge_csv_task.outputs['output_csv'] | _____no_output_____ | CC-BY-4.0 | content/en/docs/components/pipelines/sdk/build-pipeline.ipynb | droctothorpe/website |
Compile and run your pipelineAfter defining the pipeline in Python as described in the preceding section, use one of the following options to compile the pipeline and submit it to the Kubeflow Pipelines service. Option 1: Compile and then upload in UI1. Run the following to compile your pipeline and save it as `pipeline.yaml`. | kfp.compiler.Compiler().compile(
pipeline_func=my_pipeline,
package_path='pipeline.yaml') | _____no_output_____ | CC-BY-4.0 | content/en/docs/components/pipelines/sdk/build-pipeline.ipynb | droctothorpe/website |
2. Upload and run your `pipeline.yaml` using the Kubeflow Pipelines user interface.See the guide to [getting started with the UI][quickstart].[quickstart]: https://www.kubeflow.org/docs/components/pipelines/overview/quickstart Option 2: run the pipeline using Kubeflow Pipelines SDK client1. Create an instance of the [`kfp.Client` class][kfp-client] following steps in [connecting to Kubeflow Pipelines using the SDK client][connect-api].[kfp-client]: https://kubeflow-pipelines.readthedocs.io/en/latest/source/kfp.client.htmlkfp.Client[connect-api]: https://www.kubeflow.org/docs/components/pipelines/sdk/connect-api | client = kfp.Client() # change arguments accordingly | _____no_output_____ | CC-BY-4.0 | content/en/docs/components/pipelines/sdk/build-pipeline.ipynb | droctothorpe/website |
2. Run the pipeline using the `kfp.Client` instance: | client.create_run_from_pipeline_func(
my_pipeline,
arguments={
'url': 'https://storage.googleapis.com/ml-pipeline-playground/iris-csv-files.tar.gz'
}) | _____no_output_____ | CC-BY-4.0 | content/en/docs/components/pipelines/sdk/build-pipeline.ipynb | droctothorpe/website |
 https://www.kaggle.com/danofer/sarcasmContextThis dataset contains 1.3 million Sarcastic comments from the Internet commentary website Reddit. The dataset was generated by scraping comments from Reddit (not by me :)) containing the \s ( sarcasm) tag. This tag is often used by Redditors to indicate that their comment is in jest and not meant to be taken seriously, and is generally a reliable indicator of sarcastic comment content.ContentData has balanced and imbalanced (i.e true distribution) versions. (True ratio is about 1:100). Thecorpus has 1.3 million sarcastic statements, along with what they responded to as well as many non-sarcastic comments from the same source.Labelled comments are in the train-balanced-sarcasm.csv file.AcknowledgementsThe data was gathered by: Mikhail Khodak and Nikunj Saunshi and Kiran Vodrahalli for their article "A Large Self-Annotated Corpus for Sarcasm". The data is hosted here.Citation:@unpublished{SARC, authors={Mikhail Khodak and Nikunj Saunshi and Kiran Vodrahalli}, title={A Large Self-Annotated Corpus for Sarcasm}, url={https://arxiv.org/abs/1704.05579}, year=2017}Annotation of files in the original dataset: readme.txt.InspirationPredicting sarcasm and relevant NLP features (e.g. subjective determinant, racism, conditionals, sentiment heavy words, "Internet Slang" and specific phrases). Sarcasm vs SentimentUnusual linguistic features such as caps, italics, or elongated words. e.g., "Yeahhh, I'm sure THAT is the right answer".Topics that people tend to react to sarcastically | import os
# Install java
! apt-get update -qq
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ["PATH"] = os.environ["JAVA_HOME"] + "/bin:" + os.environ["PATH"]
! java -version
# Install pyspark
! pip install --ignore-installed pyspark==2.4.4
# Install Spark NLP
! pip install --ignore-installed spark-nlp
import sys
import time
import sparknlp
from pyspark.sql import SparkSession
packages = [
'JohnSnowLabs:spark-nlp: 2.5.5'
]
spark = SparkSession \
.builder \
.appName("ML SQL session") \
.config('spark.jars.packages', ','.join(packages)) \
.config('spark.executor.instances','2') \
.config("spark.executor.memory", "2g") \
.config("spark.driver.memory","16g") \
.getOrCreate()
print("Spark NLP version: ", sparknlp.version())
print("Apache Spark version: ", spark.version)
! wget -N https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/resources/en/sarcasm/train-balanced-sarcasm.csv -P /tmp
from pyspark.sql import SQLContext
sql = SQLContext(spark)
trainBalancedSarcasmDF = spark.read.option("header", True).option("inferSchema", True).csv("/tmp/train-balanced-sarcasm.csv")
trainBalancedSarcasmDF.printSchema()
# Let's create a temp view (table) for our SQL queries
trainBalancedSarcasmDF.createOrReplaceTempView('data')
sql.sql('SELECT COUNT(*) FROM data').collect()
sql.sql('select * from data limit 20').show()
sql.sql('select label,count(*) as cnt from data group by label order by cnt desc').show()
sql.sql('select count(*) from data where comment is null').collect()
df = sql.sql('select label,concat(parent_comment,"\n",comment) as comment from data where comment is not null and parent_comment is not null limit 100000')
print(type(df))
df.printSchema()
df.show()
from sparknlp.annotator import *
from sparknlp.common import *
from sparknlp.base import *
from pyspark.ml import Pipeline
document_assembler = DocumentAssembler() \
.setInputCol("comment") \
.setOutputCol("document")
sentence_detector = SentenceDetector() \
.setInputCols(["document"]) \
.setOutputCol("sentence") \
.setUseAbbreviations(True)
tokenizer = Tokenizer() \
.setInputCols(["sentence"]) \
.setOutputCol("token")
stemmer = Stemmer() \
.setInputCols(["token"]) \
.setOutputCol("stem")
normalizer = Normalizer() \
.setInputCols(["stem"]) \
.setOutputCol("normalized")
finisher = Finisher() \
.setInputCols(["normalized"]) \
.setOutputCols(["ntokens"]) \
.setOutputAsArray(True) \
.setCleanAnnotations(True)
nlp_pipeline = Pipeline(stages=[document_assembler, sentence_detector, tokenizer, stemmer, normalizer, finisher])
nlp_model = nlp_pipeline.fit(df)
processed = nlp_model.transform(df).persist()
processed.count()
processed.show()
train, test = processed.randomSplit(weights=[0.7, 0.3], seed=123)
print(train.count())
print(test.count())
from pyspark.ml import feature as spark_ft
stopWords = spark_ft.StopWordsRemover.loadDefaultStopWords('english')
sw_remover = spark_ft.StopWordsRemover(inputCol='ntokens', outputCol='clean_tokens', stopWords=stopWords)
tf = spark_ft.CountVectorizer(vocabSize=500, inputCol='clean_tokens', outputCol='tf')
idf = spark_ft.IDF(minDocFreq=5, inputCol='tf', outputCol='idf')
feature_pipeline = Pipeline(stages=[sw_remover, tf, idf])
feature_model = feature_pipeline.fit(train)
train_featurized = feature_model.transform(train).persist()
train_featurized.count()
train_featurized.show()
train_featurized.groupBy("label").count().show()
train_featurized.printSchema()
from pyspark.ml import classification as spark_cls
rf = spark_cls. RandomForestClassifier(labelCol="label", featuresCol="idf", numTrees=100)
model = rf.fit(train_featurized)
test_featurized = feature_model.transform(test)
preds = model.transform(test_featurized)
preds.show()
pred_df = preds.select('comment', 'label', 'prediction').toPandas()
pred_df.head()
import pandas as pd
from sklearn import metrics as skmetrics
pd.DataFrame(
data=skmetrics.confusion_matrix(pred_df['label'], pred_df['prediction']),
columns=['pred ' + l for l in ['0','1']],
index=['true ' + l for l in ['0','1']]
)
print(skmetrics.classification_report(pred_df['label'], pred_df['prediction'],
target_names=['0','1']))
spark.stop() | _____no_output_____ | Apache-2.0 | tutorials/old_generation_notebooks/colab/6- Sarcasm Classifiers (TF-IDF).ipynb | fcivardi/spark-nlp-workshop |
import re
from collections import defaultdict
from tqdm import tnrange, tqdm_notebook
import random
from tqdm.auto import tqdm
import os
from sklearn.model_selection import train_test_split
import numpy as np
from matplotlib import pyplot as plt
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch.utils.data.dataset import Dataset
from torch.nn.utils.rnn import pad_sequence
from nltk.stem.snowball import SnowballStemmer
def make_reproducible(seed, make_cuda_reproducible):
random.seed(seed)
os.environ['PYTHONHASHSEED'] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
if make_cuda_reproducible:
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
SEED = 2341
make_reproducible(SEED, False)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
print(device)
def indices_from_sentence(words):
cur_id = 0
result = []
for word in words:
result.append((cur_id, len(word)))
cur_id += len(word)
return result
print(indices_from_sentence(['word1', 'a', ',', 'word2']))
print(indices_from_sentence(re.split('(\W)', 'Барак Обама принимает в Белом доме своего французского коллегу Николя Саркози.')))
test_words = re.split('(\W)', 'Скотланд-Ярд{ORG} вызвал на допрос Руперта{PERSON} Мердока{PERSON}')
test_words_clean = re.split('(\W)', 'Скотланд-Ярд вызвал на допрос Руперта Мердока')
print(test_words)
print(indices_from_sentence(test_words_clean))
def extract_tags(words):
i = 0
res_tags = []
res_source = []
cur_id = 0
while i < len(words):
if words[i] == '{':
res_tags.append((cur_id - len(words[i - 1]), len(words[i - 1]), words[i + 1]))
i += 2
else:
res_source.append(words[i])
cur_id += len(words[i])
i += 1
return res_tags, res_source
extract_tags(test_words)
def combine_datasets():
with open('train_nes.txt', 'r') as train_nes, \
open('train_sentences.txt', 'r') as train_sentences, \
open('train_sentences_enhanced.txt', 'r') as train_sentences_enhanced, \
open('combined_sentences.txt', 'w') as combined_sentences, \
open('combined_nes.txt', 'w') as combined_nes:
combined_nes.write(train_nes.read())
combined_sentences.write(train_sentences.read())
for line in train_sentences_enhanced:
words = re.split('(\W)', line)
res_tags, res_source = extract_tags(words)
res_tags_flatten = []
for tag in res_tags:
res_tags_flatten.append(str(tag[0]))
res_tags_flatten.append(str(tag[1]))
res_tags_flatten.append(tag[2])
res_tags_flatten.append('EOL')
combined_nes.write(' '.join(res_tags_flatten) + '\n')
combined_sentences.write(''.join(res_source))
combine_datasets()
def read_training_data():
with open('train_nes.txt', 'r') as combined_nes, open('train_sentences.txt', 'r') as combined_sentences:
X, y = [], []
for line in combined_sentences:
X.append(re.split('(\W)', line))
for i, line in enumerate(combined_nes):
words = line.split()[:-1]
tags_in_line = []
i = 0
while i < len(words):
tags_in_line.append((int(words[i]), int(words[i + 1]), words[i + 2]))
i += 3
y.append(tags_in_line)
return X, y
X, y = read_training_data()
print(X[0])
print(y[0])
print(X[-1])
print(y[-1])
stemmer = SnowballStemmer("russian")
def preprocess(word):
return stemmer.stem(word.lower())
def build_vocab(data):
vocab = defaultdict(lambda: 0)
for sent in data:
for word in sent:
stemmed = preprocess(word)
if stemmed not in vocab:
vocab[stemmed] = len(vocab) + 1
return vocab
VOCAB = build_vocab(X)
PAD_VALUE = len(VOCAB) + 1
print(len(VOCAB))
def get_positions(sent):
pos = []
idx = 0
for word in sent:
cur_l = len(word)
pos.append((idx, cur_l))
idx += cur_l
return pos
def pad_dataset(dataset, vocab):
num_dataset = [torch.tensor([vocab[preprocess(word)] for word in sent]) for sent in dataset]
return pad_sequence(num_dataset, batch_first=True, padding_value=PAD_VALUE)
X_padded = pad_dataset(X, VOCAB)
def pos_dataset(dataset):
return [get_positions(sent) for sent in dataset]
X_pos = pos_dataset(X)
def pair_X_Y(X_padded, X_pos, Y):
dataset = []
tag_to_int = {
'NONE': 0,
'PERSON': 1,
'ORG': 2
}
for sent, pos, tags in zip(X_padded, X_pos, Y):
y = []
pos_i = 0
tag_i = 0
for word in sent:
if pos_i < len(pos) and tag_i < len(tags) and pos[pos_i][0] == tags[tag_i][0]:
y.append(tag_to_int[tags[tag_i][2]])
tag_i += 1
else:
y.append(tag_to_int['NONE'])
pos_i += 1
dataset.append([sent.numpy(), y])
return np.array(dataset)
pairs_dataset = pair_X_Y(X_padded, X_pos, y)
print(pairs_dataset.shape)
TRAIN_X_Y, VAL_X_Y = train_test_split(pairs_dataset, test_size=0.1, random_state=SEED)
class Model(nn.Module):
def __init__(self, embedding_dim, hidden_dim, vocab_size):
super(Model, self).__init__()
self.emb = nn.Embedding(vocab_size, embedding_dim)
self.lstm = nn.LSTM(embedding_dim, hidden_dim, num_layers=2, bidirectional=False, batch_first=True)
self.fc2 = nn.Linear(hidden_dim, 3)
def forward(self, batch):
emb = self.emb(batch)
out, _ = self.lstm(emb)
tag_hidden = self.fc2(out)
tag_probs = F.log_softmax(tag_hidden, dim=-1)
return tag_probs
def train(model, train, val, epoch_cnt, batch_size):
train_loader = torch.utils.data.DataLoader(train, batch_size=batch_size, shuffle=True)
val_loader = torch.utils.data.DataLoader(val, batch_size=batch_size)
loss_function = nn.NLLLoss()
optimizer = optim.Adam(model.parameters(), lr=5e-4)
train_loss_values = []
val_loss_values = []
for epoch in tnrange(epoch_cnt, desc='Epoch'):
for batch_data in train_loader:
x, y = batch_data[:, 0].to(device), batch_data[:, 1].to(device)
optimizer.zero_grad()
output = model(x.long())
output = output.view(-1, 3)
y = y.reshape(-1)
loss = loss_function(output, y.long())
train_loss_values.append(loss)
loss.backward()
nn.utils.clip_grad_norm_(model.parameters(), 5)
optimizer.step()
with torch.no_grad():
loss_values = []
for batch_data in val_loader:
x, y = batch_data[:, 0].to(device), batch_data[:, 1].to(device)
output = model(x.long())
output = output.view(-1, 3)
y = y.reshape(-1)
loss = loss_function(output, y.long())
loss_values.append(loss.item())
val_loss_values.append(np.mean(np.array(loss_values)))
return train_loss_values, val_loss_values
embed = 128
hidden_dim = 256
vocab_size = len(VOCAB) + 1
epoch_cnt = 290
batch_size = 512
model = Model(embed, hidden_dim, vocab_size)
model = model.float()
model = model.to(device)
train_loss_values, val_loss_values =\
train(model, TRAIN_X_Y, VAL_X_Y, epoch_cnt, batch_size)
plt.plot(train_loss_values, label='train')
plt.plot(np.arange(0, len(train_loss_values), len(train_loss_values) / epoch_cnt), val_loss_values, label='validation')
plt.legend()
plt.title("Loss values")
plt.show()
def read_test():
test_filename = "test.txt"
lines = []
with open(test_filename, 'r') as test_file:
for line in test_file:
lines.append(re.split('(\W)', line))
return lines
TEST = read_test()
print(TEST[0])
def produce_test_results():
test_padded = pad_dataset(TEST, VOCAB)
test_pos = pos_dataset(TEST)
with torch.no_grad():
test_loader = torch.utils.data.DataLoader(test_padded, batch_size=batch_size)
ans = None
for batch_data in test_loader:
x = batch_data.to(device)
output = model(x.long())
_, ansx = output.max(dim=-1)
ansx = ansx.cpu().numpy()
if ans is None:
ans = ansx
else:
ans = np.append(ans, ansx, axis=0)
out_filename = "out.txt"
int_to_tag = {1:"PERSON" , 2:"ORG"}
with open(out_filename, "w") as out:
for sent, pos, tags in zip(test_padded, test_pos, ans):
for i in range(len(pos)):
if tags[i] in int_to_tag:
out.write("%d %d %s " % (pos[i][0], pos[i][1], int_to_tag[tags[i]]))
out.write("EOL\n")
produce_test_results()
| _____no_output_____ | MIT | HW_03_LSTM.ipynb | RamSaw/NLP |
|
Fashion MNIST con terminación tempranaUsando el modelo del ejercicio anterior, en este notebooks aprenderás a crear tu callback y terminar tempranamente el entrenamiento de tu modelo. Ejercicio 1 - importar tensorflowprimero que nada, importa las bibliotecas que consideres necesarias | %tensorflow_version 2.x
import tensorflow as tf | _____no_output_____ | MIT | ejercicios/D1_E2_callbacks_SOLUCION.ipynb | lcmencia/penguin-tf-workshop |
Ejercicio 2 - crear el callbackEscribe un callback que resulte en la terminación temprana del entrenamiento cuando el modelo llegue a más de 80% de precisión. Imprime un mensaje en la consola explicando el motivo de la terminación temprana y el número de *epoch* al usuario. | class CallbackPenguin(tf.keras.callbacks.Callback):
def on_epoch_end(self, epoch, logs={}):
if logs.get('accuracy') > 0.85:
print('\nEl modelo ha llegado a 85% de precisión, terminando entrenamiento en el epoch', epoch + 1)
self.model.stop_training = True | _____no_output_____ | MIT | ejercicios/D1_E2_callbacks_SOLUCION.ipynb | lcmencia/penguin-tf-workshop |
Ejercicio 3 - cargar el *dataset*Carga el *dataset* de Fashion MNIST y normaliza las imágenes del dataset (recuerda que se deben normalizar tanto las imágenes del *training set* y las del *testing set*) | (train_imgs, train_labels), (test_imgs, test_labels) = tf.keras.datasets.fashion_mnist.load_data()
train_imgs = train_imgs/255.0
test_imgs = test_imgs/255.0 | _____no_output_____ | MIT | ejercicios/D1_E2_callbacks_SOLUCION.ipynb | lcmencia/penguin-tf-workshop |
Ejercicio 4 - crear el modeloRecrea el modelo del ejercicio anterior, y compila el modelo. | # crear el modelo
model = tf.keras.models.Sequential([tf.keras.layers.Flatten(input_shape=(28, 28)),
tf.keras.layers.Dense(100, activation=tf.nn.relu),
tf.keras.layers.Dense(10, activation=tf.nn.softmax)])
# compilar el modelo
model.compile(optimizer='sgd', loss='sparse_categorical_crossentropy', metrics=['accuracy']) | _____no_output_____ | MIT | ejercicios/D1_E2_callbacks_SOLUCION.ipynb | lcmencia/penguin-tf-workshop |
Ejercicio 4 - entrenar el modeloEntrena el modelo usando el comando `fit` y el callback que escribiste en el ejercicio 2. | callback_penguin = CallbackPenguin()
model.fit(train_imgs, train_labels, epochs=50, callbacks=[callback_penguin]) | _____no_output_____ | MIT | ejercicios/D1_E2_callbacks_SOLUCION.ipynb | lcmencia/penguin-tf-workshop |
Visit the NASA mars news site | # Visit the Mars news site
url = 'https://redplanetscience.com/'
browser.visit(url)
# Optional delay for loading the page
browser.is_element_present_by_css('div.list_text', wait_time=1)
# Convert the browser html to a soup object
html = browser.html
news_soup = soup(html, 'html.parser')
slide_elem = news_soup.select_one('div.list_text')
print(news_soup.prettify())
slide_elem = news_soup.body.find('div', class_="content_title")
#display the current title content
news_title = slide_elem.find('div', class_="content_title").get_text()
news_title
# Use the parent element to find the first a tag and save it as `news_title`
news_title
news_p = slide_elem.find('div', class_="article_teaser_body").get_text()
news_p
# Use the parent element to find the paragraph text
news_p | _____no_output_____ | ADSL | .ipynb_checkpoints/Mission_to_Mars-checkpoint.ipynb | danelle1126/web-scraping-challenge |
JPL Space Images Featured Image | # Visit URL
url = 'https://spaceimages-mars.com'
browser.visit(url)
# Find and click the full image button
full_image_link = browser.find_by_tag('button')[1]
full_image_link.click()
# Parse the resulting html with soup
html = browser.html
img_soup = soup(html, 'html.parser')
print(img_soup.prettify())
img_url_rel = img_soup.find('img',class_='fancybox-image').get('src')
img_url_rel
# find the relative image url
img_url_rel
img_url = f'https://spaceimages-mars.com/{img_url_rel}'
img_url
# Use the base url to create an absolute url
img_url | _____no_output_____ | ADSL | .ipynb_checkpoints/Mission_to_Mars-checkpoint.ipynb | danelle1126/web-scraping-challenge |
Mars Facts | url = 'https://galaxyfacts-mars.com'
browser.visit(url)
html = browser.html
facts_soup = soup(html, 'html.parser')
html = browser.html
facts_soup = soup(html, 'html.parser')
tables = pd.read_html(url)
tables
df = tables[0]
df.head()
# Use `pd.read_html` to pull the data from the Mars-Earth Comparison section
# hint use index 0 to find the table
df.head()
df.columns = ['Description','Mars','Earth']
df = df.iloc[1:]
df.set_index('Description',inplace=True)
df
df
df.to_html()
df.to_html() | _____no_output_____ | ADSL | .ipynb_checkpoints/Mission_to_Mars-checkpoint.ipynb | danelle1126/web-scraping-challenge |
Hemispheres | url = 'https://marshemispheres.com/'
browser.visit(url)
html = browser.html
hems_soup = soup(html, 'html.parser')
print(hems_soup.prettify())
# Create a list to hold the images and titles.
hemisphere_image_urls = []
# Get a list of all of the hemispheres
links = browser.find_by_css('a.product-item img')
# Next, loop through those links, click the link, find the sample anchor, return the href
for i in range(len(links)):
hemisphereInfo = {}
# We have to find the elements on each loop to avoid a stale element exception
browser.find_by_css('a.product-item img')[i].click()
# Next, we find the Sample image anchor tag and extract the href
sample = browser.links.find_by_text('Sample').first
hemisphereInfo['img_url'] = sample['href']
# Get Hemisphere title
titleA = browser.find_by_css('h2.title').text
hemisphereInfo['title'] = titleA.rpartition(' Enhanced')[0]
# Append hemisphere object to list
hemisphere_image_urls.append(hemisphereInfo)
# Finally, we navigate backwards
browser.back()
hemisphere_image_urls
hemisphere_image_urls
browser.quit() | _____no_output_____ | ADSL | .ipynb_checkpoints/Mission_to_Mars-checkpoint.ipynb | danelle1126/web-scraping-challenge |
Object Detection with TRTorch (SSD) --- OverviewIn PyTorch 1.0, TorchScript was introduced as a method to separate your PyTorch model from Python, make it portable and optimizable.TRTorch is a compiler that uses TensorRT (NVIDIA's Deep Learning Optimization SDK and Runtime) to optimize TorchScript code. It compiles standard TorchScript modules into ones that internally run with TensorRT optimizations.TensorRT can take models from any major framework and specifically tune them to perform better on specific target hardware in the NVIDIA family, and TRTorch enables us to continue to remain in the PyTorch ecosystem whilst doing so. This allows us to leverage the great features in PyTorch, including module composability, its flexible tensor implementation, data loaders and more. TRTorch is available to use with both PyTorch and LibTorch. To get more background information on this, we suggest the **lenet-getting-started** notebook as a primer for getting started with TRTorch. Learning objectivesThis notebook demonstrates the steps for compiling a TorchScript module with TRTorch on a pretrained SSD network, and running it to test the speedup obtained. Contents1. [Requirements](1)2. [SSD Overview](2)3. [Creating TorchScript modules](3)4. [Compiling with TRTorch](4)5. [Running Inference](5)6. [Measuring Speedup](6)7. [Conclusion](7) --- 1. RequirementsFollow the steps in `notebooks/README` to prepare a Docker container, within which you can run this demo notebook.In addition to that, run the following cell to obtain additional libraries specific to this demo. | # Known working versions
!pip install numpy==1.21.2 scipy==1.5.2 Pillow==6.2.0 scikit-image==0.17.2 matplotlib==3.3.0 | _____no_output_____ | BSD-3-Clause | notebooks/ssd-object-detection-demo.ipynb | p1x31/TRTorch |
--- 2. SSD Single Shot MultiBox Detector model for object detection_ | _- | -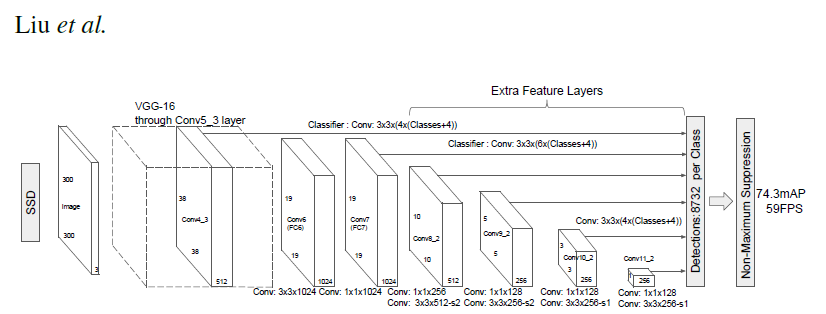 |  PyTorch has a model repository called the PyTorch Hub, which is a source for high quality implementations of common models. We can get our SSD model pretrained on [COCO](https://cocodataset.org/home) from there. Model DescriptionThis SSD300 model is based on the[SSD: Single Shot MultiBox Detector](https://arxiv.org/abs/1512.02325) paper, whichdescribes SSD as “a method for detecting objects in images using a single deep neural network".The input size is fixed to 300x300.The main difference between this model and the one described in the paper is in the backbone.Specifically, the VGG model is obsolete and is replaced by the ResNet-50 model.From the[Speed/accuracy trade-offs for modern convolutional object detectors](https://arxiv.org/abs/1611.10012)paper, the following enhancements were made to the backbone:* The conv5_x, avgpool, fc and softmax layers were removed from the original classification model.* All strides in conv4_x are set to 1x1.The backbone is followed by 5 additional convolutional layers.In addition to the convolutional layers, we attached 6 detection heads:* The first detection head is attached to the last conv4_x layer.* The other five detection heads are attached to the corresponding 5 additional layers.Detector heads are similar to the ones referenced in the paper, however,they are enhanced by additional BatchNorm layers after each convolution.More information about this SSD model is available at Nvidia's "DeepLearningExamples" Github [here](https://github.com/NVIDIA/DeepLearningExamples/tree/master/PyTorch/Detection/SSD). | import torch
torch.hub._validate_not_a_forked_repo=lambda a,b,c: True
# List of available models in PyTorch Hub from Nvidia/DeepLearningExamples
torch.hub.list('NVIDIA/DeepLearningExamples:torchhub')
# load SSD model pretrained on COCO from Torch Hub
precision = 'fp32'
ssd300 = torch.hub.load('NVIDIA/DeepLearningExamples:torchhub', 'nvidia_ssd', model_math=precision); | Using cache found in /root/.cache/torch/hub/NVIDIA_DeepLearningExamples_torchhub
Downloading checkpoint from https://api.ngc.nvidia.com/v2/models/nvidia/ssd_pyt_ckpt_amp/versions/20.06.0/files/nvidia_ssdpyt_amp_200703.pt
| BSD-3-Clause | notebooks/ssd-object-detection-demo.ipynb | p1x31/TRTorch |
Setting `precision="fp16"` will load a checkpoint trained with mixed precision into architecture enabling execution on Tensor Cores. Handling mixed precision data requires the Apex library. Sample Inference We can now run inference on the model. This is demonstrated below using sample images from the COCO 2017 Validation set. | # Sample images from the COCO validation set
uris = [
'http://images.cocodataset.org/val2017/000000397133.jpg',
'http://images.cocodataset.org/val2017/000000037777.jpg',
'http://images.cocodataset.org/val2017/000000252219.jpg'
]
# For convenient and comprehensive formatting of input and output of the model, load a set of utility methods.
utils = torch.hub.load('NVIDIA/DeepLearningExamples:torchhub', 'nvidia_ssd_processing_utils')
# Format images to comply with the network input
inputs = [utils.prepare_input(uri) for uri in uris]
tensor = utils.prepare_tensor(inputs, False)
# The model was trained on COCO dataset, which we need to access in order to
# translate class IDs into object names.
classes_to_labels = utils.get_coco_object_dictionary()
# Next, we run object detection
model = ssd300.eval().to("cuda")
detections_batch = model(tensor)
# By default, raw output from SSD network per input image contains 8732 boxes with
# localization and class probability distribution.
# Let’s filter this output to only get reasonable detections (confidence>40%) in a more comprehensive format.
results_per_input = utils.decode_results(detections_batch)
best_results_per_input = [utils.pick_best(results, 0.40) for results in results_per_input] | /opt/conda/lib/python3.8/site-packages/torch/nn/functional.py:718: UserWarning: Named tensors and all their associated APIs are an experimental feature and subject to change. Please do not use them for anything important until they are released as stable. (Triggered internally at ../c10/core/TensorImpl.h:1153.)
return torch.max_pool2d(input, kernel_size, stride, padding, dilation, ceil_mode)
| BSD-3-Clause | notebooks/ssd-object-detection-demo.ipynb | p1x31/TRTorch |
Visualize results | from matplotlib import pyplot as plt
import matplotlib.patches as patches
# The utility plots the images and predicted bounding boxes (with confidence scores).
def plot_results(best_results):
for image_idx in range(len(best_results)):
fig, ax = plt.subplots(1)
# Show original, denormalized image...
image = inputs[image_idx] / 2 + 0.5
ax.imshow(image)
# ...with detections
bboxes, classes, confidences = best_results[image_idx]
for idx in range(len(bboxes)):
left, bot, right, top = bboxes[idx]
x, y, w, h = [val * 300 for val in [left, bot, right - left, top - bot]]
rect = patches.Rectangle((x, y), w, h, linewidth=1, edgecolor='r', facecolor='none')
ax.add_patch(rect)
ax.text(x, y, "{} {:.0f}%".format(classes_to_labels[classes[idx] - 1], confidences[idx]*100), bbox=dict(facecolor='white', alpha=0.5))
plt.show()
# Visualize results without TRTorch/TensorRT
plot_results(best_results_per_input) | _____no_output_____ | BSD-3-Clause | notebooks/ssd-object-detection-demo.ipynb | p1x31/TRTorch |
Benchmark utility | import time
import numpy as np
import torch.backends.cudnn as cudnn
cudnn.benchmark = True
# Helper function to benchmark the model
def benchmark(model, input_shape=(1024, 1, 32, 32), dtype='fp32', nwarmup=50, nruns=1000):
input_data = torch.randn(input_shape)
input_data = input_data.to("cuda")
if dtype=='fp16':
input_data = input_data.half()
print("Warm up ...")
with torch.no_grad():
for _ in range(nwarmup):
features = model(input_data)
torch.cuda.synchronize()
print("Start timing ...")
timings = []
with torch.no_grad():
for i in range(1, nruns+1):
start_time = time.time()
pred_loc, pred_label = model(input_data)
torch.cuda.synchronize()
end_time = time.time()
timings.append(end_time - start_time)
if i%10==0:
print('Iteration %d/%d, avg batch time %.2f ms'%(i, nruns, np.mean(timings)*1000))
print("Input shape:", input_data.size())
print("Output location prediction size:", pred_loc.size())
print("Output label prediction size:", pred_label.size())
print('Average batch time: %.2f ms'%(np.mean(timings)*1000))
| _____no_output_____ | BSD-3-Clause | notebooks/ssd-object-detection-demo.ipynb | p1x31/TRTorch |
We check how well the model performs **before** we use TRTorch/TensorRT | # Model benchmark without TRTorch/TensorRT
model = ssd300.eval().to("cuda")
benchmark(model, input_shape=(128, 3, 300, 300), nruns=100) | Warm up ...
Start timing ...
Iteration 10/100, avg batch time 382.30 ms
Iteration 20/100, avg batch time 382.72 ms
Iteration 30/100, avg batch time 382.63 ms
Iteration 40/100, avg batch time 382.83 ms
Iteration 50/100, avg batch time 382.90 ms
Iteration 60/100, avg batch time 382.86 ms
Iteration 70/100, avg batch time 382.88 ms
Iteration 80/100, avg batch time 382.86 ms
Iteration 90/100, avg batch time 382.95 ms
Iteration 100/100, avg batch time 382.97 ms
Input shape: torch.Size([128, 3, 300, 300])
Output location prediction size: torch.Size([128, 4, 8732])
Output label prediction size: torch.Size([128, 81, 8732])
Average batch time: 382.97 ms
| BSD-3-Clause | notebooks/ssd-object-detection-demo.ipynb | p1x31/TRTorch |
--- 3. Creating TorchScript modules To compile with TRTorch, the model must first be in **TorchScript**. TorchScript is a programming language included in PyTorch which removes the Python dependency normal PyTorch models have. This conversion is done via a JIT compiler which given a PyTorch Module will generate an equivalent TorchScript Module. There are two paths that can be used to generate TorchScript: **Tracing** and **Scripting**. - Tracing follows execution of PyTorch generating ops in TorchScript corresponding to what it sees. - Scripting does an analysis of the Python code and generates TorchScript, this allows the resulting graph to include control flow which tracing cannot do. Tracing however due to its simplicity is more likely to compile successfully with TRTorch (though both systems are supported). | model = ssd300.eval().to("cuda")
traced_model = torch.jit.trace(model, [torch.randn((1,3,300,300)).to("cuda")]) | _____no_output_____ | BSD-3-Clause | notebooks/ssd-object-detection-demo.ipynb | p1x31/TRTorch |
If required, we can also save this model and use it independently of Python. | # This is just an example, and not required for the purposes of this demo
torch.jit.save(traced_model, "ssd_300_traced.jit.pt")
# Obtain the average time taken by a batch of input with Torchscript compiled modules
benchmark(traced_model, input_shape=(128, 3, 300, 300), nruns=100) | Warm up ...
Start timing ...
Iteration 10/100, avg batch time 382.67 ms
Iteration 20/100, avg batch time 382.54 ms
Iteration 30/100, avg batch time 382.73 ms
Iteration 40/100, avg batch time 382.53 ms
Iteration 50/100, avg batch time 382.56 ms
Iteration 60/100, avg batch time 382.50 ms
Iteration 70/100, avg batch time 382.54 ms
Iteration 80/100, avg batch time 382.54 ms
Iteration 90/100, avg batch time 382.57 ms
Iteration 100/100, avg batch time 382.62 ms
Input shape: torch.Size([128, 3, 300, 300])
Output location prediction size: torch.Size([128, 4, 8732])
Output label prediction size: torch.Size([128, 81, 8732])
Average batch time: 382.62 ms
| BSD-3-Clause | notebooks/ssd-object-detection-demo.ipynb | p1x31/TRTorch |
--- 4. Compiling with TRTorchTorchScript modules behave just like normal PyTorch modules and are intercompatible. From TorchScript we can now compile a TensorRT based module. This module will still be implemented in TorchScript but all the computation will be done in TensorRT. | import trtorch
# The compiled module will have precision as specified by "op_precision".
# Here, it will have FP16 precision.
trt_model = trtorch.compile(traced_model, {
"inputs": [trtorch.Input((3, 3, 300, 300))],
"enabled_precisions": {torch.float, torch.half}, # Run with FP16
"workspace_size": 1 << 20
}) | _____no_output_____ | BSD-3-Clause | notebooks/ssd-object-detection-demo.ipynb | p1x31/TRTorch |
--- 5. Running Inference Next, we run object detection | # using a TRTorch module is exactly the same as how we usually do inference in PyTorch i.e. model(inputs)
detections_batch = trt_model(tensor.to(torch.half)) # convert the input to half precision
# By default, raw output from SSD network per input image contains 8732 boxes with
# localization and class probability distribution.
# Let’s filter this output to only get reasonable detections (confidence>40%) in a more comprehensive format.
results_per_input = utils.decode_results(detections_batch)
best_results_per_input_trt = [utils.pick_best(results, 0.40) for results in results_per_input] | _____no_output_____ | BSD-3-Clause | notebooks/ssd-object-detection-demo.ipynb | p1x31/TRTorch |
Now, let's visualize our predictions! | # Visualize results with TRTorch/TensorRT
plot_results(best_results_per_input_trt) | _____no_output_____ | BSD-3-Clause | notebooks/ssd-object-detection-demo.ipynb | p1x31/TRTorch |
We get similar results as before! --- 6. Measuring SpeedupWe can run the benchmark function again to see the speedup gained! Compare this result with the same batch-size of input in the case without TRTorch/TensorRT above. | batch_size = 128
# Recompiling with batch_size we use for evaluating performance
trt_model = trtorch.compile(traced_model, {
"inputs": [trtorch.Input((batch_size, 3, 300, 300))],
"enabled_precisions": {torch.float, torch.half}, # Run with FP16
"workspace_size": 1 << 20
})
benchmark(trt_model, input_shape=(batch_size, 3, 300, 300), nruns=100, dtype="fp16") | Warm up ...
Start timing ...
Iteration 10/100, avg batch time 72.90 ms
Iteration 20/100, avg batch time 72.95 ms
Iteration 30/100, avg batch time 72.92 ms
Iteration 40/100, avg batch time 72.94 ms
Iteration 50/100, avg batch time 72.99 ms
Iteration 60/100, avg batch time 73.01 ms
Iteration 70/100, avg batch time 73.04 ms
Iteration 80/100, avg batch time 73.04 ms
Iteration 90/100, avg batch time 73.04 ms
Iteration 100/100, avg batch time 73.06 ms
Input shape: torch.Size([128, 3, 300, 300])
Output location prediction size: torch.Size([128, 4, 8732])
Output label prediction size: torch.Size([128, 81, 8732])
Average batch time: 73.06 ms
| BSD-3-Clause | notebooks/ssd-object-detection-demo.ipynb | p1x31/TRTorch |
3. Markov Models Example ProblemsWe will now look at a model that examines our state of healthiness vs. being sick. Keep in mind that this is very much like something you could do in real life. If you wanted to model a certain situation or environment, we could take some data that we have gathered, build a maximum likelihood model on it, and do things like study the properties that emerge from the model, or make predictions from the model, or generate the next most likely state. Let's say we have 2 states: **sick** and **healthy**. We know that we spend most of our time in a healthy state, so the probability of transitioning from healthy to sick is very low:$$p(sick \; | \; healthy) = 0.005$$Hence, the probability of going from healthy to healthy is:$$p(healthy \; | \; healthy) = 0.995$$Now, on the other hand the probability of going from sick to sick is also very high. This is because if you just got sick yesterday then you are very likely to be sick tomorrow.$$p(sick \; | \; sick) = 0.8$$However, the probability of transitioning from sick to healthy should be higher than the reverse, because you probably won't stay sick for as long as you would stay healthy:$$p(healthy \; | \; sick) = 0.02$$We have now fully defined our state transition matrix, and we can now do some calculations. 1.1 Example Calculations 1.1.1 What is the probability of being healthy for 10 days in a row, given that we already start out as healthy? Well that is:$$p(healthy \; 10 \; days \; in \; a \; row \; | \; healthy \; at \; t=0) = 0.995^9 = 95.6 \%$$How about the probability of being healthy for 100 days in a row? $$p(healthy \; 100 \; days \; in \; a \; row \; | \; healthy \; at \; t=0) = 0.995^{99} = 60.9 \%$$ 2. Expected Number of Continuously Sick DaysWe can now look at the expected number of days that you would remain in the same state (e.g. how many days would you expect to stay sick given the model?). This is a bit more difficult than the last problem, but completely doable, only involving the mathematics of infinite sums.First, we can look at the probability of being in state $i$, and going to state $i$ in the next state. That is just $A(i,i)$:$$p \big(s(t)=i \; | \; s(t-1)=i \big) = A(i, i)$$Now, what is the probability distribution that we actually want to calculate? How about we calculate the probability that we stay in state $i$ for $n$ transitions, at which point we move to another state:$$p \big(s(t) \;!=i \; | \; s(t-1)=i \big) = 1 - A(i, i)$$So, the joint probability that we are trying to model is:$$p\big(s(1)=i, s(2)=i,...,s(n)=i, s(n+1) \;!= i\big) = A(i,i)^{n-1}\big(1-A(i,i)\big)$$In english this means that we are multiplying the transition probability of staying in the same state, $A(i,i)$, times the number of times we stayed in the same state, $n$, (note it is $n-1$ because we are given that we start in that state, hence there is no transition associated with it) times $1 - A(i,i)$, the probability of transitioning from that state. This leaves us with an expected value for $n$ of:$$E(n) = \sum np(n) = \sum_{n=1..\infty} nA(i,i)^{n-1}(1-A(i,i))$$Note, in the above equation $p(n)$ is the probability that we will see state $i$ $n-1$ times after starting from $i$ and then see a state that is not $i$. Also, we know that the expected value of $n$ should be the sum of all possible values of $n$ times $p(n)$. 2.1 Expected $n$So, we can now expand this function and calculate the two sums separately. $$E(n) = \sum_{n=1..\infty}nA(i,i)^{n-1}(1 - A(i,i)) = \sum nA(i, i)^{n-1} - \sum nA(i,i)^n$$**First Sum**With our first sum, we can say that:$$S = \sum na(i, i)^{n-1}$$$$S = 1 + 2a + 3a^2 + 4a^3+ ...$$And we can then multiply that sum, $S$, by $a$, to get:$$aS = a + 2a^2 + 3a^3 + 4a^4+...$$And then we can subtract $aS$ from $S$:$$S - aS = S'= 1 + a + a^2 + a^3+...$$This $S'$ is another infinite sum, but it is one that is much easier to solve! $$S'= 1 + a + a^2 + a^3+...$$And then $aS'$ is:$$aS' = a + a^2 + a^3+ + a^4 + ...$$Which, when we then do $S' - aS'$, we end up with:$$S' - aS' = 1$$$$S' = \frac{1}{1 - a}$$And if we then substitute that value in for $S'$ above:$$S - aS = S'= 1 + a + a^2 + a^3+... = \frac{1}{1 - a}$$$$S - aS = \frac{1}{1 - a}$$$$S = \frac{1}{(1 - a)^2}$$**Second Sum**We can now look at our second sum:$$S = \sum na(i,i)^n$$$$S = 1a + 2a^2 + 3a^3 +...$$$$Sa = 1a^2 + 2a^3 +...$$$$S - aS = S' = a + a^2 + a^3 + ...$$$$aS' = a^2 + a^3 + a^4 +...$$$$S' - aS' = a$$$$S' = \frac{a}{1 - a}$$And we can plug back in $S'$ to get:$$S - aS = \frac{a}{1 - a}$$$$S = \frac{a}{(1 - a)^2}$$**Combine** We can now combine these two sums as follows:$$E(n) = \frac{1}{(1 - a)^2} - \frac{a}{(1-a)^2}$$$$E(n) = \frac{1}{1-a}$$**Calculate Number of Sick Days**So, how do we calculate the correct number of sick days? That is just:$$\frac{1}{1 - 0.8} = 5$$ 3. SEO and Bounce Rate Optimization We are now going to look at SEO and Bounch Rate Optimization. This is a problem that every developer and website owner can relate to. You have a website and obviously you would like to increase traffic, increase conversions, and avoid a high bounce rate (which could lead to google assigning your page a low ranking). What would a good way of modeling this data be? Without even looking at any code we can look at some examples of things that we want to know, and how they relate to markov models. 3.1 ArrivalFirst and foremost, how do people arrive on your page? Is it your home page? Your landing page? Well, this is just the very first page of what is hopefully a sequence of pages. So, the markov analogy here is that this is just the initial state distribution or $\pi$. So, once we have our markov model, the $\pi$ vector will tell us which of our pages a user is most likely to start on. 3.2 Sequences of Pages What about sequences of pages? Well, if you think people are getting to your landing page, hitting the buy button, checking out, and then closing the browser window, you can test the validity of that assumption by calculating the probability of that sequence. Of course, the probability of any sequence is probability going to be much less than 1. This is because for a longer sequence, we have more multiplication, and hence smaller final numbers. We do have two alternatives however:> * 1) You can compare the probability of two different sequences. So, are people going through the entire checkout process? Or is it more probable that they are just bouncing? * 2) Another option is to just find the transition probabilities themselves. These are conditional probabilities instead of joint probabilities. You want to know, once they have made it to the landing page, what is the probability of hitting buy. Then, once they have hit buy, what is the probability of them completing the checkout. 3.3 Bounce RateThis is hard to measure, unless you are google and hence have analytics on nearly every page on the web. This is because once a user has left your site, you can no longer run code on their computer or track what they are doing. However, let's pretend that we can determine this information. Once we have done this, we can measure which page has the highest bounce rate. At this point we can manually analyze that page and ask our marketing people "what is different about this page that people don't find it useful/want to leave?" We can then address that problem, and the hopefully later analysis shows that the fixed page no longer has a high bounce right. In the markov model, we can just represents this as the null state. 3.4 DataSo, the data we are going to be working with has two columns: `last_page_id` and `next_page_id`. This can be interpreted as the current page and the next page. The site has 10 pages with the id's 0-9. We can represent start pages by making the current page -1, and the next page the actual page. We can represent the end of the page with two different codes, `B`(bounce) or `C` (close). In the case of bounce, the user saw the page and then immediately bounced. In the case of close, the user saw the page stayed and potentially saw some useful information, and then closed the window. So, you can imagine that our engineer may use time as a factor in determining if it is a bounce or a close. | import numpy as np
import pandas as pd
"""Goal here is to store start page and end page, and the count how many times that happens. After that
we are going to turn it into a probability distribution. We can divide all transitions that start with specific
start state, by row_sum"""
transitions = {} # getting all specific transitions from start pg to end pg, tallying up # of times each occurs
row_sums = {} # start date as key -> getting number of times each starting pg occurs
# Collect our counts
for line in open('../../../data/site/site_data.csv'):
s, e = line.rstrip().split(',') # get start and end page
transitions[(s, e)] = transitions.get((s, e), 0.) + 1
row_sums[s] = row_sums.get(s, 0.) + 1
# Normalize the counts so they become real probability distributions
for k, v in transitions.items():
s, e = k
transitions[k] = v / row_sums[s]
# Calculate initial state distribution
print('Initial state distribution')
for k, v in transitions.items():
s, e = k
if s == '-1': # this means it is the start of the sequence.
print (e, v)
# Which page has the highest bounce rate?
for k, v in transitions.items():
s, e = k
if e == 'B':
print(f'Bounce rate for {s}: {v}') | Initial state distribution
8 0.10152591025834719
2 0.09507982071813466
5 0.09779926474291183
9 0.10384247368686106
0 0.10298635241980159
6 0.09800070504104345
7 0.09971294757516241
1 0.10348995316513068
4 0.10243239159993957
3 0.09513018079266758
Bounce rate for 1: 0.125939617991374
Bounce rate for 2: 0.12649551345962112
Bounce rate for 8: 0.12529550827423167
Bounce rate for 6: 0.1208153180975911
Bounce rate for 7: 0.12371650388179314
Bounce rate for 3: 0.12743384922616077
Bounce rate for 4: 0.1255756067205974
Bounce rate for 5: 0.12369559684398065
Bounce rate for 0: 0.1279673590504451
Bounce rate for 9: 0.13176232104396302
| MIT | Machine_Learning/05-Hidden_Markov_Models-03-Markov-Models-Example-Problems-and-Applications.ipynb | NathanielDake/NathanielDake.github.io |
We can see that page with `id` 9 has the highest value in the initial state distribution, so we are most likely to start on that page. We can then see that the page with highest bounce rate is also at page `id` 9. 4. Build a 2nd-order language model and generate phrasesSo, we are now going to work with non first order markov chains for a little bit. In this example we are going to try and create a language model. So we are going to first train a model on some data to determine the distribution of a word given the previous two words. We can then use this model to generate new phrases. Note that another step of this model would be to calculate the probability of a phrase.So the data that we are going to look at is just a collection of Robert Frost Poems. It is just a text file with all of the poems concatenated together. So, the first thing we are going to want to do is tokenize each sentence, and remove punctuation. It will look similar to this:```def remove_punctuation(s): return s.translate(None, string.punctuation) tokens = [t for t in remove_puncuation(line.rstrip().lower()).split()]```Once we have tokenized each line, we want to perform various counts in addition to the second order model counts. We need to measure the initial distribution of words, or stated another way the distribution of the first word of a sentence. We also want to know the distribution of the second word of a sentence. Both of these do not have two previous words, so they are not second order. We could technically include them in the second order measurement by using `None` in place of the previous words, but we won't do that here. We also want to keep track of how to end the sentence (end of sentence distribution, will look similar to (w(t-2), w(t-1) -> END)), so we will include a special token for that too. When we do this counting, what we first want to do is create an array of all possibilities. So, for example if we had two sentences:```I love dogsI love cats```Then we could have a dictionary where the key was `(I, love)` and the value was an array `[dogs, cats]`. If "I love" was also a stand alone sentence, then the value would be `[dogs, cats, END]`. The function below can help us with this, since we first need to check if there is any value for the key, create an array if not, otherwise just append to the array. ```def add2dict(d, k, v): if k not in d: d[k] = [] else: d[k].append(v)```One we have collected all of these arrays of possible next words, we need to turn them into **probability distributions**. For example, the array `[cat, cat, dog]` would become the dictionary `{"cat": 2/3, "dog": 1/3}`. Here is a function that can do this:```def list2pdict(ts): d = {} n = len(ts) for t in ts: d[t] = d.get(t, 0.) + 1 for t, c in d.items(): d[t] = c / n return d```Next, we will need a function that can sample from this dictionary. To do this we will need to generate a random number between 0 and 1, and then use the distribution of the words to sample a word given a random number. Here is a function that can do that:```def sample_word(d): p0 = np.random.random() cumulative = 0 for t, p in d.items(): cumulative += p if p0 < cumulative: return t assert(False) should never get here```Because all of our distributions are structured as dictionaries, we can use the same function for all of them. | import numpy as np
import string
"""3 dicts. 1st store pdist for the start of a phrase, then a second word dict which stores the distributions
for the 2nd word of a sentence, and then we are going to have a dict for all second order transitions"""
initial = {}
second_word = {}
transitions = {}
def remove_punctuation(s):
return s.translate(str.maketrans('', '', string.punctuation))
def add2dict(d, k, v):
"""Parameters: Dictionary, Key, Value"""
if k not in d:
d[k] = []
d[k].append(v)
# Loop through file of poems
for line in open('../../../data/poems/robert_frost.txt'):
tokens = remove_punctuation(line.rstrip().lower()).split() # Get all tokens for specific line we are looping over
T = len(tokens) # Length of sequence
for i in range(T): # Loop through every token in sequence
t = tokens[i]
if i == 0: # We are looking at first word
initial[t] = initial.get(t, 0.) + 1
else:
t_1 = tokens[i - 1]
if i == T - 1: # Looking at last word
add2dict(transitions, (t_1, t), 'END')
if i == 1: # second word of sentence, hence only 1 previous word
add2dict(second_word, t_1, t)
else:
t_2 = tokens[i - 2] # Get second previous word
add2dict(transitions, (t_2, t_1), t) # add previous and 2nd previous word as key, and current word as val
# Normalize the distributions
initial_total = sum(initial.values())
for t, c in initial.items():
initial[t] = c / initial_total
# Take our list and turn it into a dictionary of probabilities
def list2pdict(ts):
d = {}
n = len(ts) # get total number of values
for t in ts: # look at each token
d[t] = d.get(t, 0.) + 1
for t, c in d.items(): # go through dictionary, divide frequency by sum
d[t] = c / n
return d
for t_1, ts in second_word.items():
second_word[t_1] = list2pdict(ts)
for k, ts in transitions.items():
transitions[k] = list2pdict(ts)
def sample_word(d):
p0 = np.random.random() # Generate random number from 0 to 1
cumulative = 0 # cumulative count for all probabilities seen so far
for t, p in d.items():
cumulative += p
if p0 < cumulative:
return t
assert(False) # should never hit this
"""Function to generate a poem"""
def generate():
for i in range(4):
sentence = []
# initial word
w0 = sample_word(initial)
sentence.append(w0)
# sample second word
w1 = sample_word(second_word[w0])
sentence.append(w1)
# second-order transitions until END -> enter infinite loop
while True:
w2 = sample_word(transitions[(w0, w1)]) # sample next word given previous two words
if w2 == 'END':
break
sentence.append(w2)
w0 = w1
w1 = w2
print(' '.join(sentence))
generate() | another from the childrens house of makebelieve
they dont go with the dead race of the lettered
i never heard of clara robinson
where he can eat off a barrel from the sense of our having been together
| MIT | Machine_Learning/05-Hidden_Markov_Models-03-Markov-Models-Example-Problems-and-Applications.ipynb | NathanielDake/NathanielDake.github.io |
Quantization of Signals*This jupyter notebook is part of a [collection of notebooks](../index.ipynb) on various topics of Digital Signal Processing. Please direct questions and suggestions to [Sascha.Spors@uni-rostock.de](mailto:Sascha.Spors@uni-rostock.de).* Spectral Shaping of the Quantization NoiseThe quantized signal $x_Q[k]$ can be expressed by the continuous amplitude signal $x[k]$ and the quantization error $e[k]$ as\begin{equation}x_Q[k] = \mathcal{Q} \{ x[k] \} = x[k] + e[k]\end{equation}According to the [introduced model](linear_uniform_quantization_error.ipynbModel-for-the-Quantization-Error), the quantization noise can be modeled as uniformly distributed white noise. Hence, the noise is distributed over the entire frequency range. The basic concept of [noise shaping](https://en.wikipedia.org/wiki/Noise_shaping) is a feedback of the quantization error to the input of the quantizer. This way the spectral characteristics of the quantization noise can be modified, i.e. spectrally shaped. Introducing a generic filter $h[k]$ into the feedback loop yields the following structureThe quantized signal can be deduced from the block diagram above as\begin{equation}x_Q[k] = \mathcal{Q} \{ x[k] - e[k] * h[k] \} = x[k] + e[k] - e[k] * h[k]\end{equation}where the additive noise model from above has been introduced and it has been assumed that the impulse response $h[k]$ is normalized such that the magnitude of $e[k] * h[k]$ is below the quantization step $Q$. The overall quantization error is then\begin{equation}e_H[k] = x_Q[k] - x[k] = e[k] * (\delta[k] - h[k])\end{equation}The power spectral density (PSD) of the quantization error with noise shaping is calculated to\begin{equation}\Phi_{e_H e_H}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) = \Phi_{ee}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) \cdot \left| 1 - H(\mathrm{e}^{\,\mathrm{j}\,\Omega}) \right|^2\end{equation}Hence the PSD $\Phi_{ee}(\mathrm{e}^{\,\mathrm{j}\,\Omega})$ of the quantizer without noise shaping is weighted by $| 1 - H(\mathrm{e}^{\,\mathrm{j}\,\Omega}) |^2$. Noise shaping allows a spectral modification of the quantization error. The desired shaping depends on the application scenario. For some applications, high-frequency noise is less disturbing as low-frequency noise. Example - First-Order Noise ShapingIf the feedback of the error signal is delayed by one sample we get with $h[k] = \delta[k-1]$\begin{equation}\Phi_{e_H e_H}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) = \Phi_{ee}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) \cdot \left| 1 - \mathrm{e}^{\,-\mathrm{j}\,\Omega} \right|^2\end{equation}For linear uniform quantization $\Phi_{ee}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) = \sigma_e^2$ is constant. Hence, the spectral shaping constitutes a high-pass characteristic of first order. The following simulation evaluates the noise shaping quantizer of first order. | %matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import scipy.signal as sig
w = 8 # wordlength of the quantized signal
xmin = -1 # minimum of input signal
N = 32768 # number of samples
def uniform_midtread_quantizer_w_ns(x, Q):
# limiter
x = np.copy(x)
idx = np.where(x <= -1)
x[idx] = -1
idx = np.where(x > 1 - Q)
x[idx] = 1 - Q
# linear uniform quantization with noise shaping
xQ = Q * np.floor(x/Q + 1/2)
e = xQ - x
xQ = xQ - np.concatenate(([0], e[0:-1]))
return xQ[1:]
# quantization step
Q = 1/(2**(w-1))
# compute input signal
np.random.seed(5)
x = np.random.uniform(size=N, low=xmin, high=(-xmin-Q))
# quantize signal
xQ = uniform_midtread_quantizer_w_ns(x, Q)
e = xQ - x[1:]
# estimate PSD of error signal
nf, Pee = sig.welch(e, nperseg=64)
# estimate SNR
SNR = 10*np.log10((np.var(x)/np.var(e)))
print('SNR = {:2.1f} dB'.format(SNR))
plt.figure(figsize=(10,5))
Om = nf*2*np.pi
plt.plot(Om, Pee*6/Q**2, label='estimated PSD')
plt.plot(Om, np.abs(1 - np.exp(-1j*Om))**2, label='theoretic PSD')
plt.plot(Om, np.ones(Om.shape), label='PSD w/o noise shaping')
plt.title('PSD of quantization error')
plt.xlabel(r'$\Omega$')
plt.ylabel(r'$\hat{\Phi}_{e_H e_H}(e^{j \Omega}) / \sigma_e^2$')
plt.axis([0, np.pi, 0, 4.5]);
plt.legend(loc='upper left')
plt.grid() | SNR = 45.2 dB
| MIT | quantization/noise_shaping.ipynb | davidjustin1974/digital-signal-processing-lecture |
Neural networks with PyTorchDeep learning networks tend to be massive with dozens or hundreds of layers, that's where the term "deep" comes from. You can build one of these deep networks using only weight matrices as we did in the previous notebook, but in general it's very cumbersome and difficult to implement. PyTorch has a nice module `nn` that provides a nice way to efficiently build large neural networks. | # Import necessary packages
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import numpy as np
import torch
import helper
import matplotlib.pyplot as plt | _____no_output_____ | MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
Now we're going to build a larger network that can solve a (formerly) difficult problem, identifying text in an image. Here we'll use the MNIST dataset which consists of greyscale handwritten digits. Each image is 28x28 pixels, you can see a sample belowOur goal is to build a neural network that can take one of these images and predict the digit in the image.First up, we need to get our dataset. This is provided through the `torchvision` package. The code below will download the MNIST dataset, then create training and test datasets for us. Don't worry too much about the details here, you'll learn more about this later. | ### Run this cell
from torchvision import datasets, transforms
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5,), (0.5,)),
])
# Download and load the training data
trainset = datasets.MNIST('~/.pytorch/MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True) | _____no_output_____ | MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
We have the training data loaded into `trainloader` and we make that an iterator with `iter(trainloader)`. Later, we'll use this to loop through the dataset for training, like```pythonfor image, label in trainloader: do things with images and labels```You'll notice I created the `trainloader` with a batch size of 64, and `shuffle=True`. The batch size is the number of images we get in one iteration from the data loader and pass through our network, often called a *batch*. And `shuffle=True` tells it to shuffle the dataset every time we start going through the data loader again. But here I'm just grabbing the first batch so we can check out the data. We can see below that `images` is just a tensor with size `(64, 1, 28, 28)`. So, 64 images per batch, 1 color channel, and 28x28 images. | dataiter = iter(trainloader)
images, labels = dataiter.next()
print(type(images))
print(images.shape)
print(labels.shape) | <class 'torch.Tensor'>
torch.Size([64, 1, 28, 28])
torch.Size([64])
| MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
This is what one of the images looks like. | plt.imshow(images[1].numpy().squeeze(), cmap='Greys_r'); | _____no_output_____ | MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
First, let's try to build a simple network for this dataset using weight matrices and matrix multiplications. Then, we'll see how to do it using PyTorch's `nn` module which provides a much more convenient and powerful method for defining network architectures.The networks you've seen so far are called *fully-connected* or *dense* networks. Each unit in one layer is connected to each unit in the next layer. In fully-connected networks, the input to each layer must be a one-dimensional vector (which can be stacked into a 2D tensor as a batch of multiple examples). However, our images are 28x28 2D tensors, so we need to convert them into 1D vectors. Thinking about sizes, we need to convert the batch of images with shape `(64, 1, 28, 28)` to a have a shape of `(64, 784)`, 784 is 28 times 28. This is typically called *flattening*, we flattened the 2D images into 1D vectors.Previously you built a network with one output unit. Here we need 10 output units, one for each digit. We want our network to predict the digit shown in an image, so what we'll do is calculate probabilities that the image is of any one digit or class. This ends up being a discrete probability distribution over the classes (digits) that tells us the most likely class for the image. That means we need 10 output units for the 10 classes (digits). We'll see how to convert the network output into a probability distribution next.> **Exercise:** Flatten the batch of images `images`. Then build a multi-layer network with 784 input units, 256 hidden units, and 10 output units using random tensors for the weights and biases. For now, use a sigmoid activation for the hidden layer. Leave the output layer without an activation, we'll add one that gives us a probability distribution next. | ## Your solution
images_flat = images.view(64, 784)
def act(x):
return 1/(1+torch.exp(-x))
torch.manual_seed(42)
n_input = 784
n_hidden = 256
n_output = 10
W1 = torch.randn((n_input, n_hidden))
W2 = torch.randn((n_hidden, n_output))
B1 = torch.randn((1, 1))
B2 = torch.randn((1, 1))
def network(features):
return act(torch.mm(act(torch.mm(features, W1) + B1) ,W2) + B2)
out = network(images_flat)
out.shape
#out = # output of your network, should have shape (64,10) | _____no_output_____ | MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
Now we have 10 outputs for our network. We want to pass in an image to our network and get out a probability distribution over the classes that tells us the likely class(es) the image belongs to. Something that looks like this:Here we see that the probability for each class is roughly the same. This is representing an untrained network, it hasn't seen any data yet so it just returns a uniform distribution with equal probabilities for each class.To calculate this probability distribution, we often use the [**softmax** function](https://en.wikipedia.org/wiki/Softmax_function). Mathematically this looks like$$\Large \sigma(x_i) = \cfrac{e^{x_i}}{\sum_k^K{e^{x_k}}}$$What this does is squish each input $x_i$ between 0 and 1 and normalizes the values to give you a proper probability distribution where the probabilites sum up to one.> **Exercise:** Implement a function `softmax` that performs the softmax calculation and returns probability distributions for each example in the batch. Note that you'll need to pay attention to the shapes when doing this. If you have a tensor `a` with shape `(64, 10)` and a tensor `b` with shape `(64,)`, doing `a/b` will give you an error because PyTorch will try to do the division across the columns (called broadcasting) but you'll get a size mismatch. The way to think about this is for each of the 64 examples, you only want to divide by one value, the sum in the denominator. So you need `b` to have a shape of `(64, 1)`. This way PyTorch will divide the 10 values in each row of `a` by the one value in each row of `b`. Pay attention to how you take the sum as well. You'll need to define the `dim` keyword in `torch.sum`. Setting `dim=0` takes the sum across the rows while `dim=1` takes the sum across the columns. | def softmax(x):
return torch.exp(x)/torch.sum(torch.exp(x), dim=1).reshape(64, 1)
## TODO: Implement the softmax function here
# Here, out should be the output of the network in the previous excercise with shape (64,10)
probabilities = softmax(out)
# Does it have the right shape? Should be (64, 10)
print(probabilities.shape)
# Does it sum to 1?
print(probabilities.sum(dim=1)) | torch.Size([64, 10])
tensor([1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,
1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,
1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,
1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,
1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,
1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,
1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,
1.0000])
| MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
Building networks with PyTorchPyTorch provides a module `nn` that makes building networks much simpler. Here I'll show you how to build the same one as above with 784 inputs, 256 hidden units, 10 output units and a softmax output. | from torch import nn
class Network(nn.Module):
def __init__(self):
super().__init__()
# Inputs to hidden layer linear transformation
self.hidden = nn.Linear(784, 256)
# Output layer, 10 units - one for each digit
self.output = nn.Linear(256, 10)
# Define sigmoid activation and softmax output
self.sigmoid = nn.Sigmoid()
self.softmax = nn.Softmax(dim=1)
def forward(self, x):
# Pass the input tensor through each of our operations
x = self.hidden(x)
x = self.sigmoid(x)
x = self.output(x)
x = self.softmax(x)
return x | _____no_output_____ | MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
Let's go through this bit by bit.```pythonclass Network(nn.Module):```Here we're inheriting from `nn.Module`. Combined with `super().__init__()` this creates a class that tracks the architecture and provides a lot of useful methods and attributes. It is mandatory to inherit from `nn.Module` when you're creating a class for your network. The name of the class itself can be anything.```pythonself.hidden = nn.Linear(784, 256)```This line creates a module for a linear transformation, $x\mathbf{W} + b$, with 784 inputs and 256 outputs and assigns it to `self.hidden`. The module automatically creates the weight and bias tensors which we'll use in the `forward` method. You can access the weight and bias tensors once the network (`net`) is created with `net.hidden.weight` and `net.hidden.bias`.```pythonself.output = nn.Linear(256, 10)```Similarly, this creates another linear transformation with 256 inputs and 10 outputs.```pythonself.sigmoid = nn.Sigmoid()self.softmax = nn.Softmax(dim=1)```Here I defined operations for the sigmoid activation and softmax output. Setting `dim=1` in `nn.Softmax(dim=1)` calculates softmax across the columns.```pythondef forward(self, x):```PyTorch networks created with `nn.Module` must have a `forward` method defined. It takes in a tensor `x` and passes it through the operations you defined in the `__init__` method.```pythonx = self.hidden(x)x = self.sigmoid(x)x = self.output(x)x = self.softmax(x)```Here the input tensor `x` is passed through each operation and reassigned to `x`. We can see that the input tensor goes through the hidden layer, then a sigmoid function, then the output layer, and finally the softmax function. It doesn't matter what you name the variables here, as long as the inputs and outputs of the operations match the network architecture you want to build. The order in which you define things in the `__init__` method doesn't matter, but you'll need to sequence the operations correctly in the `forward` method.Now we can create a `Network` object. | # Create the network and look at it's text representation
model = Network()
model | _____no_output_____ | MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
You can define the network somewhat more concisely and clearly using the `torch.nn.functional` module. This is the most common way you'll see networks defined as many operations are simple element-wise functions. We normally import this module as `F`, `import torch.nn.functional as F`. | import torch.nn.functional as F
class Network(nn.Module):
def __init__(self):
super().__init__()
# Inputs to hidden layer linear transformation
self.hidden = nn.Linear(784, 256)
# Output layer, 10 units - one for each digit
self.output = nn.Linear(256, 10)
def forward(self, x):
# Hidden layer with sigmoid activation
x = F.sigmoid(self.hidden(x))
# Output layer with softmax activation
x = F.softmax(self.output(x), dim=1)
return x | _____no_output_____ | MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
Activation functionsSo far we've only been looking at the sigmoid activation function, but in general any function can be used as an activation function. The only requirement is that for a network to approximate a non-linear function, the activation functions must be non-linear. Here are a few more examples of common activation functions: Tanh (hyperbolic tangent), and ReLU (rectified linear unit).In practice, the ReLU function is used almost exclusively as the activation function for hidden layers. Your Turn to Build a Network> **Exercise:** Create a network with 784 input units, a hidden layer with 128 units and a ReLU activation, then a hidden layer with 64 units and a ReLU activation, and finally an output layer with a softmax activation as shown above. You can use a ReLU activation with the `nn.ReLU` module or `F.relu` function.It's good practice to name your layers by their type of network, for instance 'fc' to represent a fully-connected layer. As you code your solution, use `fc1`, `fc2`, and `fc3` as your layer names. | ## Your solution here
class FirstNet(nn.Module):
def __init__(self):
super().__init__()
self.fc1 = nn.Linear(784, 128)
self.fc2 = nn.Linear(128, 64)
self.fc3 = nn.Linear(64, 10)
def forward(self, x):
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = F.softmax(self.fc3(x), dim=1)
return x
model = FirstNet()
| _____no_output_____ | MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
Initializing weights and biasesThe weights and such are automatically initialized for you, but it's possible to customize how they are initialized. The weights and biases are tensors attached to the layer you defined, you can get them with `model.fc1.weight` for instance. | print(model.fc1.weight)
print(model.fc1.bias) | Parameter containing:
tensor([[-0.0324, 0.0352, -0.0258, ..., 0.0276, -0.0145, -0.0265],
[ 0.0058, 0.0206, -0.0277, ..., 0.0027, -0.0107, -0.0135],
[-0.0175, -0.0071, 0.0020, ..., -0.0303, 0.0014, -0.0020],
...,
[-0.0153, -0.0351, -0.0131, ..., -0.0250, 0.0067, -0.0284],
[ 0.0173, -0.0294, 0.0263, ..., -0.0160, 0.0226, -0.0053],
[ 0.0124, -0.0154, 0.0274, ..., 0.0156, 0.0218, -0.0344]],
requires_grad=True)
Parameter containing:
tensor([ 0.0177, -0.0303, -0.0252, -0.0185, -0.0159, 0.0209, -0.0233, 0.0078,
-0.0006, 0.0265, 0.0153, 0.0204, -0.0302, -0.0021, -0.0076, -0.0075,
0.0357, 0.0261, -0.0172, 0.0036, 0.0261, -0.0217, 0.0093, -0.0073,
0.0035, 0.0165, 0.0037, -0.0039, 0.0139, -0.0182, 0.0091, -0.0335,
0.0334, 0.0294, 0.0281, 0.0304, -0.0251, -0.0110, 0.0209, 0.0265,
0.0242, -0.0241, 0.0032, -0.0322, 0.0065, -0.0212, -0.0006, 0.0007,
0.0006, 0.0322, -0.0046, -0.0328, 0.0060, 0.0189, -0.0153, 0.0214,
-0.0122, 0.0064, 0.0167, 0.0233, 0.0340, 0.0207, -0.0257, 0.0185,
-0.0009, -0.0320, 0.0239, -0.0226, 0.0093, 0.0098, -0.0124, 0.0063,
-0.0062, 0.0022, -0.0144, 0.0011, 0.0053, 0.0161, -0.0220, 0.0323,
-0.0197, 0.0168, 0.0340, 0.0330, -0.0035, 0.0030, -0.0253, 0.0044,
-0.0017, -0.0034, -0.0259, 0.0183, -0.0257, 0.0126, 0.0293, -0.0269,
0.0178, 0.0098, 0.0264, 0.0125, 0.0160, 0.0170, 0.0312, 0.0148,
0.0223, -0.0148, -0.0042, 0.0080, -0.0098, 0.0243, -0.0257, 0.0050,
0.0167, -0.0194, 0.0257, -0.0018, 0.0061, -0.0322, -0.0059, -0.0114,
0.0315, -0.0073, 0.0253, -0.0096, 0.0028, 0.0145, 0.0022, -0.0301],
requires_grad=True)
| MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
For custom initialization, we want to modify these tensors in place. These are actually autograd *Variables*, so we need to get back the actual tensors with `model.fc1.weight.data`. Once we have the tensors, we can fill them with zeros (for biases) or random normal values. | # Set biases to all zeros
model.fc1.bias.data.fill_(0)
# sample from random normal with standard dev = 0.01
model.fc1.weight.data.normal_(std=0.01) | _____no_output_____ | MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
Forward passNow that we have a network, let's see what happens when we pass in an image. | # Grab some data
dataiter = iter(trainloader)
images, labels = dataiter.next()
# Resize images into a 1D vector, new shape is (batch size, color channels, image pixels)
images.resize_(64, 1, 784)
# or images.resize_(images.shape[0], 1, 784) to automatically get batch size
# Forward pass through the network
img_idx = 0
ps = model.forward(images[img_idx,:])
img = images[img_idx]
helper.view_classify(img.view(1, 28, 28), ps) | _____no_output_____ | MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
As you can see above, our network has basically no idea what this digit is. It's because we haven't trained it yet, all the weights are random! Using `nn.Sequential`PyTorch provides a convenient way to build networks like this where a tensor is passed sequentially through operations, `nn.Sequential` ([documentation](https://pytorch.org/docs/master/nn.htmltorch.nn.Sequential)). Using this to build the equivalent network: | # Hyperparameters for our network
input_size = 784
hidden_sizes = [128, 64]
output_size = 10
# Build a feed-forward network
model = nn.Sequential(nn.Linear(input_size, hidden_sizes[0]),
nn.ReLU(),
nn.Linear(hidden_sizes[0], hidden_sizes[1]),
nn.ReLU(),
nn.Linear(hidden_sizes[1], output_size),
nn.Softmax(dim=1))
print(model)
# Forward pass through the network and display output
images, labels = next(iter(trainloader))
images.resize_(images.shape[0], 1, 784)
ps = model.forward(images[0,:])
helper.view_classify(images[0].view(1, 28, 28), ps) | Sequential(
(0): Linear(in_features=784, out_features=128, bias=True)
(1): ReLU()
(2): Linear(in_features=128, out_features=64, bias=True)
(3): ReLU()
(4): Linear(in_features=64, out_features=10, bias=True)
(5): Softmax()
)
| MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
Here our model is the same as before: 784 input units, a hidden layer with 128 units, ReLU activation, 64 unit hidden layer, another ReLU, then the output layer with 10 units, and the softmax output.The operations are available by passing in the appropriate index. For example, if you want to get first Linear operation and look at the weights, you'd use `model[0]`. | print(model[0])
model[0].weight | Linear(in_features=784, out_features=128, bias=True)
| MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
You can also pass in an `OrderedDict` to name the individual layers and operations, instead of using incremental integers. Note that dictionary keys must be unique, so _each operation must have a different name_. | from collections import OrderedDict
model = nn.Sequential(OrderedDict([
('fc1', nn.Linear(input_size, hidden_sizes[0])),
('relu1', nn.ReLU()),
('fc2', nn.Linear(hidden_sizes[0], hidden_sizes[1])),
('relu2', nn.ReLU()),
('output', nn.Linear(hidden_sizes[1], output_size)),
('softmax', nn.Softmax(dim=1))]))
model | _____no_output_____ | MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
Now you can access layers either by integer or the name | print(model[0])
print(model.fc1) | Linear(in_features=784, out_features=128, bias=True)
Linear(in_features=784, out_features=128, bias=True)
| MIT | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | jr7/deep-learning-v2-pytorch |
------------ First A.I. activity ------------ 1. IBOVESPA volume prediction -> Importing libraries that are going to be used in the code | import pandas as pd
import numpy as np
import matplotlib.pyplot as plt | _____no_output_____ | MIT | drafts/exercises/ibovespa.ipynb | ItamarRocha/introduction-to-AI |
-> Importing the datasets | dataset = pd.read_csv("datasets/ibovespa.csv",delimiter = ";") | _____no_output_____ | MIT | drafts/exercises/ibovespa.ipynb | ItamarRocha/introduction-to-AI |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.