repo_name
stringlengths 9
75
| topic
stringclasses 30
values | issue_number
int64 1
203k
| title
stringlengths 1
976
| body
stringlengths 0
254k
| state
stringclasses 2
values | created_at
stringlengths 20
20
| updated_at
stringlengths 20
20
| url
stringlengths 38
105
| labels
sequencelengths 0
9
| user_login
stringlengths 1
39
| comments_count
int64 0
452
|
|---|---|---|---|---|---|---|---|---|---|---|---|
ydataai/ydata-profiling | jupyter | 741 | pandas profilling too dlow: make multi processing | Have a stable of 10000 rows, 4000 columns,
panda profile is too slow...
Can you make it in multi-processing ?
(ie column stats can be done in multi-processing easily).
thanks | closed | 2021-03-30T06:11:08Z | 2021-09-27T22:45:44Z | https://github.com/ydataai/ydata-profiling/issues/741 | [
"performance 🚀"
] | arita37 | 1 |
CorentinJ/Real-Time-Voice-Cloning | tensorflow | 695 | No module named pathlib | > matteo@MBP-di-matteo Real-Time-Voice-Cloning-master % python demo_cli.py
> Traceback (most recent call last):
> File "demo_cli.py", line 2, in <module>
> from utils.argutils import print_args
> File "/Users/matteo/Real-Time-Voice-Cloning-master/utils/argutils.py", line 22
> def print_args(args: argparse.Namespace, parser=None):
> ^
> SyntaxError: invalid syntax
> matteo@MBP-di-matteo Real-Time-Voice-Cloning-master % python demo_toolbox.py
> Traceback (most recent call last):
> File "demo_toolbox.py", line 1, in <module>
> from pathlib import Path
> ImportError: No module named pathlib
> matteo@MBP-di-matteo Real-Time-Voice-Cloning-master % sudo python demo_toolbox.py
> Password:
> Traceback (most recent call last):
> File "demo_toolbox.py", line 1, in <module>
> from pathlib import Path
> ImportError: No module named pathlib
What should I do? | closed | 2021-03-07T10:40:27Z | 2021-03-08T21:24:20Z | https://github.com/CorentinJ/Real-Time-Voice-Cloning/issues/695 | [] | matteopuppis | 3 |
axnsan12/drf-yasg | django | 129 | Is it possible to set example value for fields | Hello,
I am looking to set *example* attribute of model property but cannot find example in documentation or source code.
Basically, I would like swagger.yaml to create attribute [here](https://github.com/axnsan12/drf-yasg/blob/aca0c4713e0163fb0deea8ea397368084a7c83e5/tests/reference.yaml#L1485-L1489), like this:
```yaml
properties:
title:
description: title model help_text
type: string
maxLength: 255
minLength: 1
example: My title
```
Then result documentation would include "My title" instead of "string" like in screenshot:

Is that possible? Would appreciate direction where to look. | closed | 2018-05-21T20:21:19Z | 2024-06-20T00:35:36Z | https://github.com/axnsan12/drf-yasg/issues/129 | [] | bmihelac | 11 |
huggingface/transformers | deep-learning | 36,145 | Problems with Training ModernBERT | ### System Info
- `transformers` version: 4.48.3
- Platform: Linux-6.8.0-52-generic-x86_64-with-glibc2.35
- Python version: 3.12.9
- Huggingface_hub version: 0.28.1
- Safetensors version: 0.5.2
- Accelerate version: 1.3.0
- Accelerate config: not found
- PyTorch version (GPU?): 2.6.0+cu126 (True)
- Tensorflow version (GPU?): not installed (NA)
- Flax version (CPU?/GPU?/TPU?): not installed (NA)
- Jax version: not installed
- JaxLib version: not installed
- Using distributed or parallel set-up in script?: Parallel (I'm not sure; I'm using a single GPU on a single machine)
- Using GPU in script?: Yes
- GPU type: NVIDIA GeForce RTX 2060
I have also tried installing `python3.12-dev` in response to the following initial error message (included with the code snippet later)
```python
/usr/include/python3.12/pyconfig.h:3:12: fatal error: x86_64-linux-gnu/python3.12/pyconfig.h: No such file or directory
# include <x86_64-linux-gnu/python3.12/pyconfig.h>
^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
compilation terminated.
```
but the error persists.
### Who can help?
@ArthurZucker
### Information
- [ ] The official example scripts
- [x] My own modified scripts
### Tasks
- [ ] An officially supported task in the `examples` folder (such as GLUE/SQuAD, ...)
- [x] My own task or dataset (give details below)
### Reproduction
I am essentially replicating the code from the following:
- https://github.com/di37/ner-electrical-engineering-finetuning/blob/main/notebooks/01_data_tokenization.ipynb
- https://github.com/di37/ner-electrical-engineering-finetuning/blob/main/notebooks/02_finetuning.ipynb
- See also: https://blog.cubed.run/automating-electrical-engineering-text-analysis-with-named-entity-recognition-ner-part-1-babd2df422d8
The following is the code for setting up the training process:
```python
import os
from transformers import BertTokenizerFast
from datasets import load_dataset
# from utilities import MODEL_ID, DATASET_ID, OUTPUT_DATASET_PATH
DATASET_ID = "disham993/ElectricalNER"
MODEL_ID = "answerdotai/ModernBERT-large"
LOGS = "logs"
OUTPUT_DATASET_PATH = os.path.join(
"data", "tokenized_electrical_ner_modernbert"
) # "data"
OUTPUT_DIR = "models"
MODEL_PATH = os.path.join(OUTPUT_DIR, MODEL_ID)
OUTPUT_MODEL = os.path.join(OUTPUT_DIR, f"electrical-ner-{MODEL_ID.split('/')[-1]}")
EVAL_STRATEGY = "epoch"
LEARNING_RATE = 1e-5
PER_DEVICE_TRAIN_BATCH_SIZE = 64
PER_DEVICE_EVAL_BATCH_SIZE = 64
NUM_TRAIN_EPOCHS = 5
WEIGHT_DECAY = 0.01
LOCAL_MODELS = {
"google-bert/bert-base-uncased": "electrical-ner-bert-base-uncased",
"distilbert/distilbert-base-uncased": "electrical-ner-distilbert-base-uncased",
"google-bert/bert-large-uncased": "electrical-ner-bert-large-uncased",
"answerdotai/ModernBERT-base": "electrical-ner-ModernBERT-base",
"answerdotai/ModernBERT-large": "electrical-ner-ModernBERT-large",
}
ONLINE_MODELS = {
"google-bert/bert-base-uncased": "disham993/electrical-ner-bert-base",
"distilbert/distilbert-base-uncased": "disham993/electrical-ner-distilbert-base",
"google-bert/bert-large-uncased": "disham993/electrical-ner-bert-large",
"answerdotai/ModernBERT-base": "disham993/electrical-ner-ModernBERT-base",
"answerdotai/ModernBERT-large": "disham993/electrical-ner-ModernBERT-large",
}
electrical_ner_dataset = load_dataset(DATASET_ID, trust_remote_code=True)
print(electrical_ner_dataset)
from datasets import DatasetDict
shrunk_train = electrical_ner_dataset['train'].select(range(10))
shrunk_valid = electrical_ner_dataset['validation'].select(range(5))
shrunk_test = electrical_ner_dataset['test'].select(range(5))
electrical_ner_dataset = DatasetDict({
'train': shrunk_train,
'validation': shrunk_valid,
'test': shrunk_test
})
electrical_ner_dataset.shape
tokenizer = BertTokenizerFast.from_pretrained(MODEL_ID)
def tokenize_and_align_labels(examples, label_all_tokens=True):
"""
Function to tokenize and align labels with respect to the tokens. This function is specifically designed for
Named Entity Recognition (NER) tasks where alignment of the labels is necessary after tokenization.
Parameters:
examples (dict): A dictionary containing the tokens and the corresponding NER tags.
- "tokens": list of words in a sentence.
- "ner_tags": list of corresponding entity tags for each word.
label_all_tokens (bool): A flag to indicate whether all tokens should have labels.
If False, only the first token of a word will have a label,
the other tokens (subwords) corresponding to the same word will be assigned -100.
Returns:
tokenized_inputs (dict): A dictionary containing the tokenized inputs and the corresponding labels aligned with the tokens.
"""
tokenized_inputs = tokenizer(examples["tokens"], truncation=True, is_split_into_words=True)
labels = []
for i, label in enumerate(examples["ner_tags"]):
word_ids = tokenized_inputs.word_ids(batch_index=i)
# word_ids() => Return a list mapping the tokens
# to their actual word in the initial sentence.
# It Returns a list indicating the word corresponding to each token.
previous_word_idx = None
label_ids = []
# Special tokens like `<s>` and `<\s>` are originally mapped to None
# We need to set the label to -100 so they are automatically ignored in the loss function.
for word_idx in word_ids:
if word_idx is None:
# set –100 as the label for these special tokens
label_ids.append(-100)
# For the other tokens in a word, we set the label to either the current label or -100, depending on
# the label_all_tokens flag.
elif word_idx != previous_word_idx:
# if current word_idx is != prev then its the most regular case
# and add the corresponding token
label_ids.append(label[word_idx])
else:
# to take care of sub-words which have the same word_idx
# set -100 as well for them, but only if label_all_tokens == False
label_ids.append(label[word_idx] if label_all_tokens else -100)
# mask the subword representations after the first subword
previous_word_idx = word_idx
labels.append(label_ids)
tokenized_inputs["labels"] = labels
return tokenized_inputs
tokenized_datasets = electrical_ner_dataset.map(tokenize_and_align_labels, batched=True)
tokenized_electrical_ner_dataset = tokenized_datasets
import os
import numpy as np
from transformers import AutoTokenizer
from transformers import DataCollatorForTokenClassification
from transformers import AutoModelForTokenClassification
from datasets import load_from_disk
from transformers import TrainingArguments, Trainer
import evaluate
import json
import pandas as pd
label_list= tokenized_electrical_ner_dataset["train"].features["ner_tags"].feature.names
num_labels = len(label_list)
print(f"Labels: {label_list}")
print(f"Number of labels: {num_labels}")
model = AutoModelForTokenClassification.from_pretrained(MODEL_ID, num_labels=num_labels)
tokenizer = AutoTokenizer.from_pretrained(MODEL_ID)
args = TrainingArguments(
output_dir=MODEL_PATH,
eval_strategy=EVAL_STRATEGY,
learning_rate=LEARNING_RATE,
per_device_train_batch_size=1,
per_device_eval_batch_size=1,
num_train_epochs=NUM_TRAIN_EPOCHS,
weight_decay=WEIGHT_DECAY,
push_to_hub=False
)
data_collator = DataCollatorForTokenClassification(tokenizer)
def compute_metrics(eval_preds):
"""
Function to compute the evaluation metrics for Named Entity Recognition (NER) tasks.
The function computes precision, recall, F1 score and accuracy.
Parameters:
eval_preds (tuple): A tuple containing the predicted logits and the true labels.
Returns:
A dictionary containing the precision, recall, F1 score and accuracy.
"""
pred_logits, labels = eval_preds
pred_logits = np.argmax(pred_logits, axis=2)
# the logits and the probabilities are in the same order,
# so we don’t need to apply the softmax
# We remove all the values where the label is -100
predictions = [
[label_list[eval_preds] for (eval_preds, l) in zip(prediction, label) if l != -100]
for prediction, label in zip(pred_logits, labels)
]
true_labels = [
[label_list[l] for (eval_preds, l) in zip(prediction, label) if l != -100]
for prediction, label in zip(pred_logits, labels)
]
metric = evaluate.load("seqeval")
results = metric.compute(predictions=predictions, references=true_labels)
return {
"precision": results["overall_precision"],
"recall": results["overall_recall"],
"f1": results["overall_f1"],
"accuracy": results["overall_accuracy"],
}
trainer = Trainer(
model,
args,
train_dataset=tokenized_electrical_ner_dataset["train"],
eval_dataset=tokenized_electrical_ner_dataset["validation"],
data_collator=data_collator,
tokenizer=tokenizer,
compute_metrics=compute_metrics,
)
trainer.train()
```
```python
The tokenizer class you load from this checkpoint is not the same type as the class this function is called from. It may result in unexpected tokenization.
The tokenizer class you load from this checkpoint is 'PreTrainedTokenizerFast'.
The class this function is called from is 'BertTokenizerFast'.
Map: 100%|█████████████████████████████| 10/10 [00:00<00:00, 1220.41 examples/s]
Map: 100%|████████████████████████████████| 5/5 [00:00<00:00, 818.40 examples/s]
Map: 100%|███████████████████████████████| 5/5 [00:00<00:00, 1246.82 examples/s]
Some weights of ModernBertForTokenClassification were not initialized from the model checkpoint at answerdotai/ModernBERT-large and are newly initialized: ['classifier.bias', 'classifier.weight']
You should probably TRAIN this model on a down-stream task to be able to use it for predictions and inference.
/home/hyunjong/Documents/Development/Python/trouver_personal_playground/playgrounds/ml_model_training_playground/modernbert_training_error.py:183: FutureWarning: `tokenizer` is deprecated and will be removed in version 5.0.0 for `Trainer.__init__`. Use `processing_class` instead.
trainer = Trainer(
0%| | 0/50 [00:00<?, ?it/s]In file included from /usr/include/python3.12/Python.h:12:0,
from /tmp/tmpjbmobkir/main.c:5:
/usr/include/python3.12/pyconfig.h:3:12: fatal error: x86_64-linux-gnu/python3.12/pyconfig.h: No such file or directory
# include <x86_64-linux-gnu/python3.12/pyconfig.h>
^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
compilation terminated.
Traceback (most recent call last):
File "/home/hyunjong/Documents/Development/Python/trouver_personal_playground/playgrounds/ml_model_training_playground/modernbert_training_error.py", line 193, in <module>
trainer.train()
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/transformers/trainer.py", line 2171, in train
return inner_training_loop(
^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/transformers/trainer.py", line 2531, in _inner_training_loop
tr_loss_step = self.training_step(model, inputs, num_items_in_batch)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/transformers/trainer.py", line 3675, in training_step
loss = self.compute_loss(model, inputs, num_items_in_batch=num_items_in_batch)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/transformers/trainer.py", line 3731, in compute_loss
outputs = model(**inputs)
^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/nn/modules/module.py", line 1739, in _wrapped_call_impl
return self._call_impl(*args, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/nn/modules/module.py", line 1750, in _call_impl
return forward_call(*args, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/transformers/models/modernbert/modeling_modernbert.py", line 1349, in forward
outputs = self.model(
^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/nn/modules/module.py", line 1739, in _wrapped_call_impl
return self._call_impl(*args, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/nn/modules/module.py", line 1750, in _call_impl
return forward_call(*args, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/transformers/models/modernbert/modeling_modernbert.py", line 958, in forward
hidden_states = self.embeddings(input_ids=input_ids, inputs_embeds=inputs_embeds)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/nn/modules/module.py", line 1739, in _wrapped_call_impl
return self._call_impl(*args, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/nn/modules/module.py", line 1750, in _call_impl
return forward_call(*args, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/transformers/models/modernbert/modeling_modernbert.py", line 217, in forward
self.compiled_embeddings(input_ids)
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/eval_frame.py", line 574, in _fn
return fn(*args, **kwargs)
^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/convert_frame.py", line 1380, in __call__
return self._torchdynamo_orig_callable(
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/convert_frame.py", line 1164, in __call__
result = self._inner_convert(
^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/convert_frame.py", line 547, in __call__
return _compile(
^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/convert_frame.py", line 986, in _compile
guarded_code = compile_inner(code, one_graph, hooks, transform)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/convert_frame.py", line 715, in compile_inner
return _compile_inner(code, one_graph, hooks, transform)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_utils_internal.py", line 95, in wrapper_function
return function(*args, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/convert_frame.py", line 750, in _compile_inner
out_code = transform_code_object(code, transform)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/bytecode_transformation.py", line 1361, in transform_code_object
transformations(instructions, code_options)
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/convert_frame.py", line 231, in _fn
return fn(*args, **kwargs)
^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/convert_frame.py", line 662, in transform
tracer.run()
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/symbolic_convert.py", line 2868, in run
super().run()
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/symbolic_convert.py", line 1052, in run
while self.step():
^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/symbolic_convert.py", line 962, in step
self.dispatch_table[inst.opcode](self, inst)
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/symbolic_convert.py", line 3048, in RETURN_VALUE
self._return(inst)
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/symbolic_convert.py", line 3033, in _return
self.output.compile_subgraph(
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/output_graph.py", line 1101, in compile_subgraph
self.compile_and_call_fx_graph(
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/output_graph.py", line 1382, in compile_and_call_fx_graph
compiled_fn = self.call_user_compiler(gm)
^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/output_graph.py", line 1432, in call_user_compiler
return self._call_user_compiler(gm)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/output_graph.py", line 1483, in _call_user_compiler
raise BackendCompilerFailed(self.compiler_fn, e).with_traceback(
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/output_graph.py", line 1462, in _call_user_compiler
compiled_fn = compiler_fn(gm, self.example_inputs())
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/repro/after_dynamo.py", line 130, in __call__
compiled_gm = compiler_fn(gm, example_inputs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/__init__.py", line 2340, in __call__
return compile_fx(model_, inputs_, config_patches=self.config)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/compile_fx.py", line 1863, in compile_fx
return aot_autograd(
^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/backends/common.py", line 83, in __call__
cg = aot_module_simplified(gm, example_inputs, **self.kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_functorch/aot_autograd.py", line 1155, in aot_module_simplified
compiled_fn = dispatch_and_compile()
^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_functorch/aot_autograd.py", line 1131, in dispatch_and_compile
compiled_fn, _ = create_aot_dispatcher_function(
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_functorch/aot_autograd.py", line 580, in create_aot_dispatcher_function
return _create_aot_dispatcher_function(
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_functorch/aot_autograd.py", line 830, in _create_aot_dispatcher_function
compiled_fn, fw_metadata = compiler_fn(
^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_functorch/_aot_autograd/jit_compile_runtime_wrappers.py", line 678, in aot_dispatch_autograd
compiled_fw_func = aot_config.fw_compiler(fw_module, adjusted_flat_args)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_functorch/aot_autograd.py", line 489, in __call__
return self.compiler_fn(gm, example_inputs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/compile_fx.py", line 1741, in fw_compiler_base
return inner_compile(
^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/compile_fx.py", line 569, in compile_fx_inner
return wrap_compiler_debug(_compile_fx_inner, compiler_name="inductor")(
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_dynamo/repro/after_aot.py", line 102, in debug_wrapper
inner_compiled_fn = compiler_fn(gm, example_inputs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/compile_fx.py", line 685, in _compile_fx_inner
mb_compiled_graph = fx_codegen_and_compile(
^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/compile_fx.py", line 1129, in fx_codegen_and_compile
return scheme.codegen_and_compile(gm, example_inputs, inputs_to_check, graph_kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/compile_fx.py", line 1044, in codegen_and_compile
compiled_fn = graph.compile_to_module().call
^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/graph.py", line 2027, in compile_to_module
return self._compile_to_module()
^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/graph.py", line 2033, in _compile_to_module
self.codegen_with_cpp_wrapper() if self.cpp_wrapper else self.codegen()
^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/graph.py", line 1968, in codegen
self.scheduler.codegen()
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/scheduler.py", line 3477, in codegen
return self._codegen()
^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/scheduler.py", line 3554, in _codegen
self.get_backend(device).codegen_node(node)
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/codegen/cuda_combined_scheduling.py", line 80, in codegen_node
return self._triton_scheduling.codegen_node(node)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/codegen/simd.py", line 1219, in codegen_node
return self.codegen_node_schedule(
^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/codegen/simd.py", line 1263, in codegen_node_schedule
src_code = kernel.codegen_kernel()
^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/codegen/triton.py", line 3154, in codegen_kernel
**self.inductor_meta_common(),
^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/_inductor/codegen/triton.py", line 3013, in inductor_meta_common
"backend_hash": torch.utils._triton.triton_hash_with_backend(),
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/utils/_triton.py", line 111, in triton_hash_with_backend
backend = triton_backend()
^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/torch/utils/_triton.py", line 103, in triton_backend
target = driver.active.get_current_target()
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/triton/runtime/driver.py", line 23, in __getattr__
self._initialize_obj()
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/triton/runtime/driver.py", line 20, in _initialize_obj
self._obj = self._init_fn()
^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/triton/runtime/driver.py", line 9, in _create_driver
return actives[0]()
^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/triton/backends/nvidia/driver.py", line 450, in __init__
self.utils = CudaUtils() # TODO: make static
^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/triton/backends/nvidia/driver.py", line 80, in __init__
mod = compile_module_from_src(Path(os.path.join(dirname, "driver.c")).read_text(), "cuda_utils")
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/triton/backends/nvidia/driver.py", line 57, in compile_module_from_src
so = _build(name, src_path, tmpdir, library_dirs(), include_dir, libraries)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/triton/runtime/build.py", line 50, in _build
ret = subprocess.check_call(cc_cmd)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/usr/lib/python3.12/subprocess.py", line 415, in check_call
raise CalledProcessError(retcode, cmd)
torch._dynamo.exc.BackendCompilerFailed: backend='inductor' raised:
CalledProcessError: Command '['/home/hyunjong/anaconda3/bin/x86_64-conda-linux-gnu-cc', '/tmp/tmpjbmobkir/main.c', '-O3', '-shared', '-fPIC', '-Wno-psabi', '-o', '/tmp/tmpjbmobkir/cuda_utils.cpython-312-x86_64-linux-gnu.so', '-lcuda', '-L/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/triton/backends/nvidia/lib', '-L/lib/x86_64-linux-gnu', '-L/lib/i386-linux-gnu', '-I/home/hyunjong/Documents/Development/Python/trouver_py312_venv/lib/python3.12/site-packages/triton/backends/nvidia/include', '-I/tmp/tmpjbmobkir', '-I/usr/include/python3.12']' returned non-zero exit status 1.
Set TORCH_LOGS="+dynamo" and TORCHDYNAMO_VERBOSE=1 for more information
You can suppress this exception and fall back to eager by setting:
import torch._dynamo
torch._dynamo.config.suppress_errors = True
0%| | 0/50 [00:01<?, ?it/s]
```
### Expected behavior
For the training process to happen. | closed | 2025-02-12T04:04:50Z | 2025-02-14T04:21:22Z | https://github.com/huggingface/transformers/issues/36145 | [
"bug"
] | hyunjongkimmath | 4 |
psf/black | python | 4,507 | Re-condensation / Simplification Of Code After Line Length Reduction | Generally i've come to really love the black formatter, so first off thanks for that!
There is really only one thing I dont like about it which is that if the length of a line is reduced such that it could be put simply on one line, the nested format remains and leaves sometimes goofy looking artifacts.
I believe the black formatter would be better off it attempted to (perhaps via option) to first eliminate any line nesting or extra space then performed a format.
Example - a list comprehension with conditional logic removed
```
existing = list(
[bw.uuid for bw in session.query(Benchmark_Whitelist_DB).all()]
)
```
Could easily be replaced with:
```
existing = [bw.uuid for bw in session.query(Benchmark_Whitelist_DB).all()]
```
No doubt that is more readable, and I think this would reduce some of the opposition to regular use of black.
I think this case could be isolated for strings of a certain line length, between the tab and a colon?
I feel like someone has to have suggested this before?
| closed | 2024-11-08T03:06:46Z | 2024-11-12T04:38:37Z | https://github.com/psf/black/issues/4507 | [
"T: style"
] | SoundsSerious | 8 |
keras-team/autokeras | tensorflow | 1,209 | Set minimum epochs before early stopping | ### Feature Description
The ability to run a minimum amount of epochs before early stopping is activated. For example. Min_epochs=100, the model will run a minimum of 100 epochs before early stopping will be able to stop the model training.
### Code Example
<!---
Please provide a code example for using that feature
given the proposed feature is implemented.
-->
```classifier = ak.AutoModel(
inputs=input_node, outputs=output_node, max_trials=100, min_epochs=100, overwrite=True)
```
### Reason
<!---
It allows for more model control and the ability to run a minimum amount of epochs before stopping the model.
-->
### Solution
<!---
Add it as a parameter for the automodel or fit function.
-->
| closed | 2020-06-25T14:17:53Z | 2020-12-10T06:23:15Z | https://github.com/keras-team/autokeras/issues/1209 | [
"feature request",
"wontfix"
] | sword134 | 3 |
ansible/ansible | python | 84,381 | exctract list of ips from inventory without providing vault password | ### Summary
I want to extract the list of ips from ansible inventory,
I can do that as follows:
```
ansible-inventory --list --ask-vault-pass | jq -r '._meta.hostvars[].ansible_host'
```
but this requires giving vault pass which theoretically is not required.
please provide a way to extract ip list from inventory without querying the encrypted vault
A generalization of the requested feature is as follows:
provide a way to parse and extract information from inventory without querying the encrypted vault
### Issue Type
Feature Idea
### Component Name
cli
### Additional Information
It is useful in bash scripts that need ip list. this feature help one to not have a duplicate ip list for things which could not be done directly with ansible playbooks and needs scripting
### Code of Conduct
- [X] I agree to follow the Ansible Code of Conduct | open | 2024-11-24T04:48:24Z | 2024-12-09T15:48:18Z | https://github.com/ansible/ansible/issues/84381 | [
"feature"
] | zxsimba | 5 |
Miserlou/Zappa | flask | 1,735 | "package" create zip with files modified date set to Jan 1, 1980 (this prevents django collectstatic from working properly) | ## Context
Running `zappa package <stage> --output package.zip` will create a package.zip, however all of the files in the package have a modified date of January 1, 1980. This breaks django's collectstatic, which seems to depend on the modified date of the file changing.
## Expected Behavior
modified dates in the package zip should be the correct date
## Actual Behavior
modified dates in the package zip are fixed to January 1, 1980, preventing collectstatic from updating existing files that have changed.
## Your Environment
* Zappa version used: 0.47
* Operating System and Python version: ubuntu 16.04, python 3.6.5
## temporary work-around
Delete files from the s3 bucket before running collectstatic | open | 2018-12-21T15:53:46Z | 2018-12-21T16:14:56Z | https://github.com/Miserlou/Zappa/issues/1735 | [] | kylegibson | 1 |
ScottfreeLLC/AlphaPy | scikit-learn | 38 | Error in importing BalanceCascade from imblearn.ensemble | **Describe the bug**
ImportError: cannot import name 'BalanceCascade' from 'imblearn.ensemble' (/opt/conda/lib/python3.7/site-packages/imblearn/ensemble/__init__.py)
**To Reproduce**
Steps to reproduce the behavior:
1. Following instructions from here: https://alphapy.readthedocs.io/en/latest/tutorials/kaggle.html
2. Running step 2 (alphapy) throws the following error. Seems like there is no BalanceCascade in imblearn
**Expected behavior**
No error thrown.
**Screenshots**
If applicable, add screenshots to help explain your problem.

**Desktop (please complete the following information):**
- OS: Google Cloud Platform Virtual Machine running Linux
**Additional context**
Not sure what I'm doing wrong. Googled the imblearn package and they dont seem to have a BalanceCascade class in ensemble: https://github.com/scikit-learn-contrib/imbalanced-learn/blob/master/imblearn/ensemble/__init__.py
Any help is appreciated!
| closed | 2020-07-06T08:47:21Z | 2020-08-25T23:48:49Z | https://github.com/ScottfreeLLC/AlphaPy/issues/38 | [
"bug"
] | toko-stephen-leo | 4 |
indico/indico | flask | 6,538 | Fix faulty checking for empty string and extend to more formats | **Describe the bug**
The `check-format-strings` function should warn us when we remove `{}` from a translation string. Currently with empty braces this is not the case (see below). We also want to extend the feature so that we support `%(...)` notation.
**To Reproduce**
Steps to reproduce the behavior:
1. Go to a translation file
2. Change a `msgstr` that contains `{}` to not contain that `{}` anymore.
3. Run `indico i18n check-format-strings`
4. It should show that there were `No issues found!`
**Screenshots**

| closed | 2024-09-13T14:37:18Z | 2024-09-18T09:07:04Z | https://github.com/indico/indico/issues/6538 | [
"bug"
] | AjobK | 7 |
jmcnamara/XlsxWriter | pandas | 653 | XlsxWriter Roadmap | I write and maintain 4 libraries for writing Xlsx files in 4 different programming languages with more or less the same APIs:
* [Excel::Writer::XLSX][1] in Perl,
* [XlsxWriter][2] in Python,
* [Libxlsxwriter][3] in C and
* [rust_xlsxwriter][4] in Rust.
See also Note 1.
New features get added to the Perl version first, then the Python version and then the C version. As such a feature request has to be implemented 4 times, with tests and documentation. The Perl and Python versions are almost completely feature compatible. The C version is somewhat behind the others and the Rust version is a work in progress.
This document gives a broad overview of features that are planned to be implemented, in order.
1. Bugs. These have the highest priority. (Note 2)
2. ~~Add user defined types to the write() method in XlsxWriter.~~ #631 **Done**.
3. ~~Add hyperlinks to images in Excel::Writer::XLSX [Issue 161][5], fix it in the Python version, and add it to the C version.~~ **Done**
4. ~~Learn the phrygian dominant mode in all keys.~~ Close enough.
5. ~~Fix the issue where duplicate images aren't removed/merged.~~ #615 In Python, Perl and C versions. **Done**
6. ~~Add support for comments to the C version~~ https://github.com/jmcnamara/libxlsxwriter/issues/38. **Done**
7. ~~Add support for object positioning to the C version.~~ **Done**
8. ~~Add support for user defined chart data labels.~~ #343 This is the most frequently requested feature across all the libraries. **Done**
9. ~~Add header/footer image support to the C version.~~ **Done**
10. Learn the altered scale in all keys.
11. ~~Add conditional formatting to the C library.~~ **Done**
12. ~~Add support for new Excel dynamic functions.~~ **Done**
13. ~~Add autofilter filter conditions to the C library.~~ **Done**
14. ~~Drop Python 2 support.~~ #720
15. ~~Add table support to libxlsxwriter.~~ **Done**
16. Implement missing features in the C library.
17. Other frequently requested, and feasible, features, in all 3 versions.
**Update for 2023**: I will implement a simulated column autofit method in the Python library. The majority of any other effort will go into getting the Rust version of the library to feature compatibility with the Python version.
Notes:
1. I also wrote a version in Lua, and two other Perl versions (for older Excel file formats) that I no longer actively maintain. I wrote, and open sourced, the first version in January 2000.
2. Some avoidable bugs have lower priority.
[1]: https://github.com/jmcnamara/excel-writer-xlsx
[2]: https://github.com/jmcnamara/XlsxWriter
[3]: https://github.com/jmcnamara/libxlsxwriter
[4]: https://github.com/jmcnamara/rust_xlsxwriter
[5]: https://github.com/jmcnamara/excel-writer-xlsx/issues/161
| open | 2019-08-31T17:13:10Z | 2024-11-19T11:15:01Z | https://github.com/jmcnamara/XlsxWriter/issues/653 | [] | jmcnamara | 21 |
nschloe/tikzplotlib | matplotlib | 452 | Figure alignment in subfloats | Hi,
what is your workflow when generating plots to be used as subfloats?
(I'm not happy with groupplots because of the lack of configurability, the visual appeal and it being complicated to add/change subcaptions of the plots)
I've tried it with the code below. But i have several size issues. Is it possible so set the 'drawing area' to a fixed size or sth? And how to do this from tikzplotlib?
Thanks!
Python:
```
import matplotlib.pyplot as plt
import tikzplotlib
import numpy as np
x1=np.arange(0,10)*10e9
x2=np.arange(0,1000)
y1=np.random.randn(1,len(x1))[0]
y2=0.01*x2*np.random.randn(1,len(x2))[0]
KIT_green=(0/255,150/255,130/255)
KIT_blue=(70/255,100/255,170/255)
plt.figure()
plt.plot(x2,y2,label="second trace",color=KIT_green)
plt.xlabel(r"Time $t$ (in \si{\milli\second})")
plt.ylabel(r"Amplitude $S_{11}$ \\ (some measurement) \\ (and another meaningless line) (in \si{\volt})");
tikzplotlib.save("subfigs_left.tikz",extra_axis_parameters=["ylabel style={align=center}"],axis_width="5cm",axis_height="5cm")
plt.figure()
plt.plot(x1,y1,label="first trace",color=KIT_blue)
plt.xlabel(r"Time $t$ (in \si{\milli\second})")
plt.ylabel(r"Amplitude $S_{11}$, $S_{35}$ (in \si{\volt})");
tikzplotlib.save("subfigs_right.tikz",extra_axis_parameters=["ylabel style={align=center}"],axis_width="5cm",axis_height="5cm")
```
LaTeX:
```
\documentclass[11pt]{article}
\usepackage[utf8]{inputenc}
\usepackage{graphicx}
\usepackage{subfig}
\usepackage{siunitx}
\usepackage{tikz}
\usepackage{pgfplots}
\usepackage{tikzscale}
\begin{document}
\begin{figure}
\centering
\subfloat[Plot 1: this shows this]{\includegraphics[width=0.4\textwidth]{subfigs_left.tikz}}
\qquad
\subfloat[Plot 2: and this shows that. But this explanation is quite long. blablabla]{\includegraphics[width=0.4\textwidth]{subfigs_right.tikz}}
\caption{Two plots}
\label{fig:subfig}
\end{figure}
\end{document}
```

| open | 2020-12-04T22:52:00Z | 2020-12-04T22:52:00Z | https://github.com/nschloe/tikzplotlib/issues/452 | [] | youcann | 0 |
desec-io/desec-stack | rest-api | 655 | GUI: Domains not selectable using Firefox | From a user's report:
> One note, using firefox on OSX ( ffv 109.0 (64-bit) ) I was unable to select a domain to edit,
>
> I tested with chrome and safari and was able to select a domain to edit. | closed | 2023-01-26T12:53:01Z | 2024-10-07T16:59:39Z | https://github.com/desec-io/desec-stack/issues/655 | [
"bug",
"gui"
] | peterthomassen | 0 |
vitalik/django-ninja | pydantic | 828 | [BUG] ForeignKey field in modelSchema do not use the related alias_generator | **Describe the bug**
When using an alias_generator in the config of a `modelSchema` the id's returned for ForeignKey Fields do not use that generator
**Versions (please complete the following information):**
- Python version: [ 3.11.4]
- Django version: [4.1.5]
- Django-Ninja version: [0.20.0]
- Pydantic version: [1.10.4]
I have this `Dealer` model
```
class Dealer(AbstractDisableModel, AddressBase, ContactInfoBase):
...
distributor = models.ForeignKey(
Distributor, on_delete=models.DO_NOTHING, related_name="dealers"
)
...
```
Which adheres to this modelschema
```
class DealerSchema(ModelSchema):
...
class Config(CamelModelSchema.Config):
...
```
which uses this schema that converts properties to camelcase
```
class CamelModelSchema(Schema):
class Config:
alias_generator = to_camel
allow_population_by_field_name = True
```
All of this works for the fields directly attached to the instances, but foreignkey fields (that end in `_id`) don't seem to be converted. Would it be possible to have the generator adjust ALL fields?
E.g. this is the schema of the response of a dealer instance
<img width="242" alt="image" src="https://github.com/vitalik/django-ninja/assets/8971598/584a1d91-ef33-4312-b41e-e4c51493d7d3">
`distributor_id` is still not camelcased although the rest of the keys are. The `distributor_id` field is not sent through the alias_generator. Applying the same alias generator to the modelschema of the distributor also doesn't fix the issue.
I would guess this is a bug with the framework? Or potentially it is expected to behave like this... | open | 2023-08-18T09:27:14Z | 2024-03-21T19:29:39Z | https://github.com/vitalik/django-ninja/issues/828 | [] | stvdrsch | 3 |
roboflow/supervision | deep-learning | 1,411 | Increasing Video FPS running on CPU Using Threading | ### Search before asking
- [X] I have searched the Supervision [issues](https://github.com/roboflow/supervision/issues) and found no similar feature requests.
### Description
I want to increase FPS of a video running on my CPU system. I tested with few annotated and object tracking videos. When I am running the frames without passing through the model the fps is still low thus resulting lesser while passing them through YOLO or any model.
The code snippet I am using is
-----
<img width="410" alt="VideoSpeed1" src="https://github.com/user-attachments/assets/f36f708c-b9aa-477f-af1d-031a90d6fa01">
So, with the following method and running the normal frames I am getting something like the following :
<img width="469" alt="VideoSpeed2" src="https://github.com/user-attachments/assets/438503da-2746-4bc4-8bd4-be12099cef15">
With normal supervision's frame generator - fps is around 1-10 max
With threading its increasing to a greater value
### Use case
If we notice there is a significant change with threading. I was wondering if we could add a MainThread Class in the supervision utils in sv.VideoInfo or add a total new class so that frames running on CPU can have such fps. Let me know if we can handle such case. I can share the python file on drive if necesssary.
Thanks
### Additional
_No response_
### Are you willing to submit a PR?
- [X] Yes I'd like to help by submitting a PR! | open | 2024-07-27T21:45:43Z | 2024-10-19T01:27:38Z | https://github.com/roboflow/supervision/issues/1411 | [
"enhancement"
] | dsaha21 | 13 |
zappa/Zappa | django | 616 | [Migrated] KeyError: 'events' on running cli command `zappa unschedule STAGE` | Originally from: https://github.com/Miserlou/Zappa/issues/1577 by [monkut](https://github.com/monkut)
<!--- Provide a general summary of the issue in the Title above -->
## Context
In an attempt to reset scheduled events via zappa cli:
```
zappa unschedule prod
```
Seems that on unschedule 'events' key is expected from the defined `event_source` settings, but not required?
## Expected Behavior
<!--- Tell us what should happen -->
If unschedule is successful, the output should reflect that.
## Actual Behavior
<!--- Tell us what happens instead -->
```
zappa unschedule prod
Calling unschedule for stage prod..
Unscheduling..
Oh no! An error occurred! :(
==============
Traceback (most recent call last):
File "/project/.venv/lib/python3.6/site-packages/zappa/cli.py", line 2693, in handle
sys.exit(cli.handle())
File "/project/.venv/lib/python3.6/site-packages/zappa/cli.py", line 504, in handle
self.dispatch_command(self.command, stage)
File "/project/.venv/lib/python3.6/site-packages/zappa/cli.py", line 605, in dispatch_command
self.unschedule()
File "/project/.venv/lib/python3.6/site-packages/zappa/cli.py", line 1228, in unschedule
events=events,
File "/project/.venv/lib/python3.6/site-packages/zappa/core.py", line 2615, in unschedule_events
print("Removed event " + name + " (" + str(event_source['events']) + ").")
KeyError: 'events'
```
## Possible Fix
<!--- Not obligatory, but suggest a fix or reason for the bug -->
Could remove the attempt to output the missing key
(since it's not outputting now, i'm not sure what this is... but seems to work when I comment out this line)
## Steps to Reproduce
<!--- Provide a link to a live example, or an unambiguous set of steps to -->
<!--- reproduce this bug include code to reproduce, if relevant -->
Not sure if this is a problem with my setup or not.. but
1. create project with kinesis stream scheduled events
2. deploy
3. turn of scheduling with `zappa unschedule STAGE`
## Your Environment
<!--- Include as many relevant details about the environment you experienced the bug in -->
* Zappa version used: 0.46.2 (was also a problem in 0.46.1)
* Operating System and Python version: macOS (3.6.2)
* The output of `pip freeze`:
```
argcomplete==1.9.3
aws-requests-auth==0.4.1
awscli==1.15.55
base58==1.0.0
boto3==1.7.68
botocore==1.10.68
Cerberus==1.2
certifi==2018.4.16
cfn-flip==1.0.3
chardet==3.0.4
click==6.7
colorama==0.3.9
Django==2.0.7
django-extensions==2.0.7
django-logging-json==1.15
django-reversion==2.0.13
django-storages==1.6.6
djangorestframework==3.8.2
djangorestframework-csv==2.1.0
docutils==0.14
durationpy==0.5
elasticsearch==6.3.0
future==0.16.0
hjson==3.0.1
idna==2.7
jmespath==0.9.3
jsonlines==1.2.0
kappa==0.6.0
lambda-packages==0.20.0
placebo==0.8.1
psycopg2-binary==2.7.5
pyasn1==0.4.4
PyJWT==1.6.4
python-dateutil==2.6.1
python-slugify==1.2.4
pytz==2018.5
PyYAML==3.12
redis==2.10.6
requests==2.19.1
rsa==3.4.2
s3transfer==0.1.13
six==1.11.0
toml==0.9.4
tqdm==4.19.1
troposphere==2.3.1
unicodecsv==0.14.1
Unidecode==1.0.22
urllib3==1.23
Werkzeug==0.14.1
wsgi-request-logger==0.4.6
zappa==0.46.2
```
* Link to your project (optional):
* Your `zappa_settings.py`:
```
{
"prod": {
"aws_region": "region",
"django_settings": "project.settings",
"profile_name": "myprofile",
"project_name": "project",
"runtime": "python3.6",
"memory_size": 2048,
"timeout_seconds": 300,
"s3_bucket": "zappa-project-prod",
"vpc_config": {
"SubnetIds": [
"subnet-yyyy88",
"subnet-zzz99"
],
"SecurityGroupIds": [
"sg-119191"
]
}
"events": [
{
"function": "project.event_handler.x_data_handler",
"event_source": {
"arn": "arn:aws:kinesis...",
"starting_position": "LATEST",
"batch_size": 500,
"enabled": true
},
"name": "lumbergh_prod_track_data_handler"
},
{
"function": "project.track_event_handler.y_data_handler",
"event_source": {
"arn": "kinesis-stream..",
"starting_position": "LATEST",
"batch_size": 500,
"enabled": true
},
"name": "y_event_data_handler"
}
],
"aws_environment_variables": {
"GDAL_DATA": "/var/task/geolib/usr/local/share/gdal/",
"GDAL_LIBRARY_PATH": "/var/task/libgdal.so.1.18.2",
"GEOS_LIBRARY_PATH": "/var/task/libgeos_c.so.1"
},
"manage_roles": false,
"role_name": "xxx"
}
}
``` | closed | 2021-02-20T12:26:41Z | 2022-07-16T06:52:03Z | https://github.com/zappa/Zappa/issues/616 | [] | jneves | 1 |
hyperspy/hyperspy | data-visualization | 2,774 | BLO output writer not working for lazy signals | It seems that the BLO writer does not work for lazy signals. The error seems to be in the line containing `tofile`. This is problematic for converting larger 4DSTEM/NBED datasets to BLO. I'll try to fix this.
In addition, the writer is very particular about units and does not seem to accept angstroms. | closed | 2021-06-22T14:44:49Z | 2021-09-05T17:08:07Z | https://github.com/hyperspy/hyperspy/issues/2774 | [
"type: bug"
] | din14970 | 3 |
graphql-python/gql | graphql | 471 | Shopify error: Prefix "query" when sending a mutation | When I have my mutation written I get an error when the request is sent that there is an unexpected sting (\"query\") at the start of my code. when the response is sent i can see that there is a hardcoded prefix "query" at the start of the request. I am unsure what to do now as I dont have that hard coded in my mutation so any advice on what to do would be greatly appreciated. There might be something I am doing wrong but I am just unsure. Thanks. I have transport and a client already defined in my code so not sure if there is an error in that but its not relevant to my traceback error as well which is the same as log errors.
2024-02-28 10:05:26 INFO: >>> {"query": "mutation customerRequestDataErasure($customerId: ID!) {\n customerRequestDataErasure(customerId: $customerId) {\n customerId\n userErrors {\n field\n message\n code\n }\n }\n}", "variables": {"customerId": "global_id (removed for security purposes)"}}
2024-02-28 10:05:26 INFO: <<< {"errors":[{"message":"syntax error, unexpected STRING (\"query\") at [1, 2]","locations":[{"line":1,"column":2}]}]}
My mutation:
query = gql("""
mutation customerRequestDataErasure($customerId: ID!) {
customerRequestDataErasure(customerId: $customerId) {
customerId
userErrors {
field
message
code
}
}
}
""")
params = {"customerId": ""}
result = client.execute(query, variable_values=params)
| closed | 2024-02-28T00:17:31Z | 2024-03-06T01:14:02Z | https://github.com/graphql-python/gql/issues/471 | [
"type: question or discussion",
"status: needs more information"
] | mattroberts96 | 3 |
python-visualization/folium | data-visualization | 1,521 | How to add custom `script` content to Folium output? | #### Please add a code sample or a nbviewer link, copy-pastable if possible
```html
<!DOCTYPE html>
<html lang="en">
<head>
<title></title>
<meta charset="utf-8" />
<meta http-equiv="x-ua-compatible" content="IE=Edge" />
<meta name="viewport" content="width=device-width, initial-scale=1, shrink-to-fit=no" />
<!-- Add references to the Leaflet JS map control resources. -->
<link rel="stylesheet" href="https://unpkg.com/leaflet@1.3.1/dist/leaflet.css"
integrity="sha512-Rksm5RenBEKSKFjgI3a41vrjkw4EVPlJ3+OiI65vTjIdo9brlAacEuKOiQ5OFh7cOI1bkDwLqdLw3Zg0cRJAAQ=="
crossorigin="" />
<script src="https://unpkg.com/leaflet@1.3.1/dist/leaflet.js"
integrity="sha512-/Nsx9X4HebavoBvEBuyp3I7od5tA0UzAxs+j83KgC8PU0kgB4XiK4Lfe4y4cgBtaRJQEIFCW+oC506aPT2L1zw=="
crossorigin=""></script>
<script type='text/javascript'>
var map;
function GetMap() {
map = L.map('myMap').setView([25,0], 3);
//Create a tile layer that points to the Azure Maps tiles.
L.tileLayer('https://atlas.microsoft.com/map/tile?subscription-key={subscriptionKey}&api-version=2.0&tilesetId={tilesetId}&zoom={z}&x={x}&y={y}&tileSize=256&language={language}&view={view}', {
attribution: `© ${new Date().getFullYear()} TomTom, Microsoft`,
//Add your Azure Maps key to the map SDK. Get an Azure Maps key at https://azure.com/maps. NOTE: The primary key should be used as the key.
subscriptionKey: 'key_goes_here',
tilesetId: 'microsoft.base.road',
language: 'en-US',
view: 'Auto'
}).addTo(map);
}
</script>
</head>
<body onload="GetMap()">
<div id="myMap" style="position:relative;width:100%;height:1000px;"></div>
</body>
</html>
```
#### Problem description
How do I add the `<script>` shown above (the one with `var map` and `subscriptionKey`) to Folium output? I'd like to use Azure basemap and my own subscription key.
| closed | 2021-10-21T20:13:43Z | 2022-11-18T11:01:56Z | https://github.com/python-visualization/folium/issues/1521 | [] | SeaDude | 2 |
microsoft/qlib | deep-learning | 901 | loss_fn() takes 3 positional arguments but 4 were given | Hi author,
When i run examples/benchmarks/LSTM/workflow_config_lstm_Alpha158.yaml, I got errors below, can you give me some favor?
i do not use GPU and the version of qlib is 0.8.3
(venv) aurora@aurora-MS-7D22:/data/vcodes/qlib/workshop$ python ../qlib/workflow/cli.py workflow_config_lstm_Alpha158.yaml
[64471:MainThread](2022-01-27 09:12:13,773) INFO - qlib.Initialization - [config.py:405] - default_conf: client.
[64471:MainThread](2022-01-27 09:12:13,774) INFO - qlib.Initialization - [__init__.py:73] - qlib successfully initialized based on client settings.
[64471:MainThread](2022-01-27 09:12:13,774) INFO - qlib.Initialization - [__init__.py:75] - data_path={'__DEFAULT_FREQ': PosixPath('/home/aurora/.qlib/qlib_data/cn_data')}
[64471:MainThread](2022-01-27 09:12:13,774) INFO - qlib.workflow - [expm.py:320] - <mlflow.tracking.client.MlflowClient object at 0x7fea7a241c10>
[64471:MainThread](2022-01-27 09:12:13,783) INFO - qlib.workflow - [exp.py:257] - Experiment 1 starts running ...
[64471:MainThread](2022-01-27 09:12:13,848) INFO - qlib.workflow - [recorder.py:290] - Recorder 22a03d2d5a164b45a1beebd9cea24de3 starts running under Experiment 1 ...
Please install necessary libs for CatBoostModel.
Please install necessary libs for XGBModel, such as xgboost.
[64471:MainThread](2022-01-27 09:12:14,194) INFO - qlib.LSTM - [pytorch_lstm_ts.py:62] - LSTM pytorch version...
[64471:MainThread](2022-01-27 09:12:14,194) INFO - qlib.LSTM - [pytorch_lstm_ts.py:80] - LSTM parameters setting:
d_feat : 20
hidden_size : 64
num_layers : 2
dropout : 0.0
n_epochs : 200
lr : 0.001
metric : loss
batch_size : 800
early_stop : 10
optimizer : adam
loss_type : mse
device : cpu
n_jobs : 20
use_GPU : False
seed : None
/data/vcodes/qlib/venv/lib/python3.8/site-packages/qlib/utils/__init__.py:689: FutureWarning: MultiIndex.is_lexsorted is deprecated as a public function, users should use MultiIndex.is_monotonic_increasing instead.
if idx.is_monotonic_increasing and not (isinstance(idx, pd.MultiIndex) and not idx.is_lexsorted()):
[64471:MainThread](2022-01-27 09:12:47,336) INFO - qlib.timer - [log.py:113] - Time cost: 33.140s | Loading data Done
[64471:MainThread](2022-01-27 09:12:47,696) INFO - qlib.timer - [log.py:113] - Time cost: 0.016s | FilterCol Done
/data/vcodes/qlib/venv/lib/python3.8/site-packages/qlib/data/dataset/processor.py:288: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
df[self.cols] = X
/data/vcodes/qlib/venv/lib/python3.8/site-packages/qlib/data/dataset/processor.py:290: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
df.clip(-3, 3, inplace=True)
[64471:MainThread](2022-01-27 09:12:48,510) INFO - qlib.timer - [log.py:113] - Time cost: 0.813s | RobustZScoreNorm Done
/data/vcodes/qlib/venv/lib/python3.8/site-packages/qlib/data/dataset/processor.py:192: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
df.fillna({col: self.fill_value for col in cols}, inplace=True)
[64471:MainThread](2022-01-27 09:12:48,718) INFO - qlib.timer - [log.py:113] - Time cost: 0.208s | Fillna Done
[64471:MainThread](2022-01-27 09:12:48,788) INFO - qlib.timer - [log.py:113] - Time cost: 0.027s | DropnaLabel Done
/data/vcodes/qlib/venv/lib/python3.8/site-packages/qlib/data/dataset/processor.py:334: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
df[cols] = t
[64471:MainThread](2022-01-27 09:12:48,970) INFO - qlib.timer - [log.py:113] - Time cost: 0.182s | CSRankNorm Done
[64471:MainThread](2022-01-27 09:12:48,970) INFO - qlib.timer - [log.py:113] - Time cost: 1.634s | fit & process data Done
[64471:MainThread](2022-01-27 09:12:48,970) INFO - qlib.timer - [log.py:113] - Time cost: 34.774s | Init data Done
/data/vcodes/qlib/venv/lib/python3.8/site-packages/qlib/utils/__init__.py:689: FutureWarning: MultiIndex.is_lexsorted is deprecated as a public function, users should use MultiIndex.is_monotonic_increasing instead.
if idx.is_monotonic_increasing and not (isinstance(idx, pd.MultiIndex) and not idx.is_lexsorted()):
/data/vcodes/qlib/venv/lib/python3.8/site-packages/qlib/utils/__init__.py:689: FutureWarning: MultiIndex.is_lexsorted is deprecated as a public function, users should use MultiIndex.is_monotonic_increasing instead.
if idx.is_monotonic_increasing and not (isinstance(idx, pd.MultiIndex) and not idx.is_lexsorted()):
/data/vcodes/qlib/venv/lib/python3.8/site-packages/torch/utils/data/dataloader.py:478: UserWarning: This DataLoader will create 20 worker processes in total. Our suggested max number of worker in current system is 12, which is smaller than what this DataLoader is going to create. Please be aware that excessive worker creation might get DataLoader running slow or even freeze, lower the worker number to avoid potential slowness/freeze if necessary.
warnings.warn(_create_warning_msg(
[64471:MainThread](2022-01-27 09:12:50,313) INFO - qlib.LSTM - [pytorch_lstm_ts.py:250] - training...
[64471:MainThread](2022-01-27 09:12:50,313) INFO - qlib.LSTM - [pytorch_lstm_ts.py:254] - Epoch0:
[64471:MainThread](2022-01-27 09:12:50,313) INFO - qlib.LSTM - [pytorch_lstm_ts.py:255] - training...
[64471:MainThread](2022-01-27 09:12:51,220) INFO - qlib.timer - [log.py:113] - Time cost: 0.000s | waiting `async_log` Done
[64471:MainThread](2022-01-27 09:12:51,220) ERROR - qlib.workflow - [utils.py:38] - An exception has been raised[TypeError: loss_fn() takes 3 positional arguments but 4 were given].
File "../qlib/workflow/cli.py", line 67, in <module>
run()
File "../qlib/workflow/cli.py", line 63, in run
fire.Fire(workflow)
File "/data/vcodes/qlib/venv/lib/python3.8/site-packages/fire/core.py", line 141, in Fire
component_trace = _Fire(component, args, parsed_flag_args, context, name)
File "/data/vcodes/qlib/venv/lib/python3.8/site-packages/fire/core.py", line 466, in _Fire
component, remaining_args = _CallAndUpdateTrace(
File "/data/vcodes/qlib/venv/lib/python3.8/site-packages/fire/core.py", line 681, in _CallAndUpdateTrace
component = fn(*varargs, **kwargs)
File "../qlib/workflow/cli.py", line 57, in workflow
recorder = task_train(config.get("task"), experiment_name=experiment_name)
File "/data/vcodes/qlib/venv/lib/python3.8/site-packages/qlib/model/trainer.py", line 122, in task_train
_exe_task(task_config)
File "/data/vcodes/qlib/venv/lib/python3.8/site-packages/qlib/model/trainer.py", line 44, in _exe_task
auto_filter_kwargs(model.fit)(dataset, reweighter=reweighter)
File "/data/vcodes/qlib/venv/lib/python3.8/site-packages/qlib/utils/__init__.py", line 852, in _func
return func(*args, **new_kwargs)
File "/data/vcodes/qlib/venv/lib/python3.8/site-packages/qlib/contrib/model/pytorch_lstm_ts.py", line 256, in fit
self.train_epoch(train_loader)
File "/data/vcodes/qlib/venv/lib/python3.8/site-packages/qlib/contrib/model/pytorch_lstm_ts.py", line 172, in train_epoch
loss = self.loss_fn(pred, label, weight.to(self.device))
TypeError: loss_fn() takes 3 positional arguments but 4 were given
| closed | 2022-01-27T01:23:50Z | 2022-02-06T14:40:15Z | https://github.com/microsoft/qlib/issues/901 | [
"question"
] | aurora5161 | 4 |
onnx/onnx | machine-learning | 5,916 | Should numpy_helper.to_array() support segments? | # Ask a Question
### Question
Should numpy_helper.to_array() be expected to support segments?
### Further information
Currently in [numpy_helper](https://github.com/onnx/onnx/blob/48501e0ff59843a33f89e028cbcbf080edd0c1df/onnx/numpy_helper.py#L231-L232) there is:
```
def to_array(tensor: TensorProto, base_dir: str = "") -> np.ndarray: # noqa: PLR0911
"""Converts a tensor def object to a numpy array.
...
"""
if tensor.HasField("segment"):
raise ValueError("Currently not supporting loading segments.")
```
Issue https://github.com/onnx/onnx/issues/2984 hit this in the past and [comment](https://github.com/onnx/onnx/issues/2984#issuecomment-748358644) that says it was closed by https://github.com/onnx/onnx/pull/3136.
However it looks like to_array() still doesn't support segments. If I'm reading it correctly the "fix" for #2984 was to use [_load_proto](https://github.com/jcwchen/onnx/blob/main/onnx/backend/test/runner/__init__.py#L461) instead of to_array().
I'm currently hitting this error from to_array() trying to load resnet-preproc-v1-18/test_data_set_0/input_0.pb from [resnet-preproc-v1-18.tar.gz in the onnx model zoo](https://github.com/onnx/models/blob/main/validated/vision/classification/resnet/preproc/resnet-preproc-v1-18.tar.gz) using:
```
with open('resnet-preproc-v1-18/test_data_set_0/input_0.pb', 'rb') as pb:
proto.ParseFromString(pb.read())
arr = onnx.numpy_helper.to_array(proto)
```
So should numpy_helper.to_array() support segments?
* If yes, can the logic for load_segments copied/moved/made accessible to to_array()
* If no, should the to_array() error be updated to use _load_proto()?
### Relevant Area: <!--e.g., model usage, backend, best practices, converters, shape_inference, version_converter, training, test, operators, IR, ONNX Hub, data preprocessing, CI pipelines. -->
loading model zoo test data
### Is this issue related to a specific model?
It's not model specific, but I hit it attempting to load test data for resnet-preproc-v1-18.tar.gz from the onnx model zoo
### Notes
<!-- Any additional information, code snippets. -->
| closed | 2024-02-07T23:19:47Z | 2024-03-07T20:27:37Z | https://github.com/onnx/onnx/issues/5916 | [
"question"
] | cjvolzka | 1 |
voila-dashboards/voila | jupyter | 947 | Widgets not rendering with xeus-python kernel. | ## Description
<!--Describe the bug clearly and concisely. Include screenshots/gifs if possible-->
Widgets do not render in Voila with `xeus-python` kernel.
## Reproduce
<!--Describe step-by-step instructions to reproduce the behavior-->
1. Create new environnement:
```bash
mamba create -n testenv python=3.9 jupyterlab voila ipywidgets xeus-python -y
```
2. Create a new notebook with content:
```python
from ipywidgets import widgets
widgets.Button(description="Click Me!")
```
2. Select `XPython` as kernel then save notebook.
3. Start `Voila` with the newly created notebook.
4. See error :
```python
Traceback (most recent call last):
File "/home/********/miniconda3/lib/python3.9/site-packages/voila/handler.py", line 209, in _jinja_cell_generator
output_cell = await task
File "/home/********/miniconda3/lib/python3.9/site-packages/voila/execute.py", line 69, in execute_cell
result = await self.async_execute_cell(cell, cell_index, store_history)
File "/home/********/miniconda3/lib/python3.9/site-packages/nbclient/client.py", line 846, in async_execute_cell
exec_reply = await self.task_poll_for_reply
File "/home/********/miniconda3/lib/python3.9/site-packages/nbclient/client.py", line 632, in _async_poll_for_reply
await asyncio.wait_for(task_poll_output_msg, self.iopub_timeout)
File "/home/********/miniconda3/lib/python3.9/asyncio/tasks.py", line 481, in wait_for
return fut.result()
File "/home/********/miniconda3/lib/python3.9/site-packages/nbclient/client.py", line 665, in _async_poll_output_msg
self.process_message(msg, cell, cell_index)
File "/home/********/miniconda3/lib/python3.9/site-packages/nbclient/client.py", line 904, in process_message
display_id = content.get('transient', {}).get('display_id', None)
AttributeError: 'NoneType' object has no attribute 'get'
```
<!--Describe how you diagnosed the issue -->
## Expected behavior
A button with text "Click Me!" should be shown.
## Context
<!--Complete the following for context, and add any other relevant context-->
- Version:
```
ipykernel 6.3.1 py39hef51801_0 conda-forge
ipython 7.27.0 py39hef51801_0 conda-forge
ipywidgets 7.6.4 pyhd8ed1ab_0 conda-forge
jupyter_client 7.0.2 pyhd8ed1ab_0 conda-forge
jupyter_core 4.7.1 py39hf3d152e_0 conda-forge
jupyter_server 1.10.2 pyhd8ed1ab_0 conda-forge
jupyterlab 3.1.10 pyhd8ed1ab_0 conda-forge
jupyterlab_pygments 0.1.2 pyh9f0ad1d_0 conda-forge
jupyterlab_server 2.7.2 pyhd8ed1ab_0 conda-forge
jupyterlab_widgets 1.0.1 pyhd8ed1ab_0 conda-forge
nbclient 0.5.4 pyhd8ed1ab_0 conda-forge
nbconvert 6.1.0 py39hf3d152e_0 conda-forge
nbformat 5.1.3 pyhd8ed1ab_0 conda-forge
notebook 6.4.3 pyha770c72_0 conda-forge
python 3.9.7 h49503c6_0_cpython conda-forge
tornado 6.1 py39h3811e60_1 conda-forge
traitlets 5.1.0 pyhd8ed1ab_0 conda-forge
voila 0.2.11 pyhd8ed1ab_0 conda-forge
widgetsnbextension 3.5.1 py39hf3d152e_4 conda-forge
xeus 1.0.4 h7d0c39e_0 conda-forge
xeus-python 0.12.5 py39h1aaad98_2 conda-forge
```
- Operating System and version: Windows 10
- Browser and version: Chrome 92.0.4515.159
| closed | 2021-09-03T14:17:04Z | 2021-09-09T12:37:23Z | https://github.com/voila-dashboards/voila/issues/947 | [
"bug"
] | trungleduc | 4 |
Lightning-AI/pytorch-lightning | deep-learning | 19,624 | IterableDataset with CORRECT length causes validation loop to be skipped | ### Bug description
This is related to this issue:
https://github.com/Lightning-AI/pytorch-lightning/issues/10290
Whereby an IterableDataset with a length defined wont trigger a validation epoch, even if the defined length is correct so long as the following conditions met:
1. Accurate length of IterableDataset defined
2. Dataset accurately split between multiple workers with no overlap
3. Drop last = True for the dataloader
4. Dataset size does not evenly divide into the batches
In this instance multiple workers may be left with an incomplete batch right at the end of the training epoch. So the number of "dropped batches" exceeds 1. Then the dataloader will raise a StopIteration before the length is reached, causing the validation epoch to be skipped.
This is standard PyTorch behavior as the collation function is called per worker in an IterableDataset.
https://github.com/pytorch/pytorch/issues/33413
I am having this issue right now my current fix is artificially subtract from the length of my IterableDataset to account for this. Unfortunately I really would like the length to be defined, so can't set it to inf which was the hotfix in the previous thread.
The progress bar is useful for me to judge which partition I need to run certain jobs on plus I use the dataset length to sync up my cyclic learning rate with the number of steps in an epoch.
### What version are you seeing the problem on?
master
### How to reproduce the bug
```python
import torch as T
import numpy as np
from torch.utils.data import DataLoader, Dataset, IterableDataset, get_worker_info
from lightning import LightningModule, Trainer, LightningDataModule
nwrkrs = 4
drop = True
class Data(IterableDataset):
def __init__(self) -> None:
super().__init__()
self.data = np.random.rand(100, 10).astype(np.float32)
def __len__(self) -> int:
return len(self.data)
def __iter__(self):
worker_info = get_worker_info()
worker_id = 0 if worker_info is None else worker_info.id
num_workers = 1 if worker_info is None else worker_info.num_workers
worker_samples = np.array_split(self.data, num_workers)[worker_id]
for i in worker_samples:
yield i
class Model(LightningModule):
def __init__(self) -> None:
super().__init__()
self.layer = T.nn.Linear(10, 1)
self.did_validation = False
def forward(self, x: T.Tensor) -> T.Tensor:
return self.layer(x)
def training_step(self, batch):
return self(batch).mean()
def validation_step(self, batch):
self.did_validation = True
return self(batch).mean()
def configure_optimizers(self):
return T.optim.Adam(self.parameters())
model = Model()
trainer = Trainer(logger=False, max_epochs=2, num_sanity_val_steps=0)
train_loader = DataLoader(Data(), num_workers=nwrkrs, batch_size=32, drop_last=drop)
valid_loader = DataLoader(Data(), num_workers=nwrkrs, batch_size=32, drop_last=drop)
trainer.fit(model, train_loader, valid_loader)
print("Performed validation:", model.did_validation)
```
Setting up the code above and running it with the following settings gives these results:
```python
nwrkrs = 0, drop = True
```
`Performed validation: True`
```python
nwrkrs = 4, drop = False
```
`Performed validation: True`
```python
nwrkrs = 4, drop = True
```
`Performed validation: False`
cc @justusschock @awaelchli | open | 2024-03-13T09:14:56Z | 2025-03-03T10:33:07Z | https://github.com/Lightning-AI/pytorch-lightning/issues/19624 | [
"question",
"data handling",
"ver: 2.2.x"
] | mattcleigh | 10 |
ahmedfgad/GeneticAlgorithmPython | numpy | 150 | Problems with multithreading and generation step | Hi,
I realy appreciate your works on PyGAD!
I'm using it to make some chaotic learning with thousands of model, and a greedy fitness function. the parallelization is realy efficient in my case.
I have found some problems with multithreading using keras models.
To reproduce the problem, i use this regression sample : `https://pygad.readthedocs.io/en/latest/README_pygad_kerasga_ReadTheDocs.html#example-1-regression-example`
I only reduce the **num_generations** to 100.
Steps to reproduce :
I run a few times the sample,
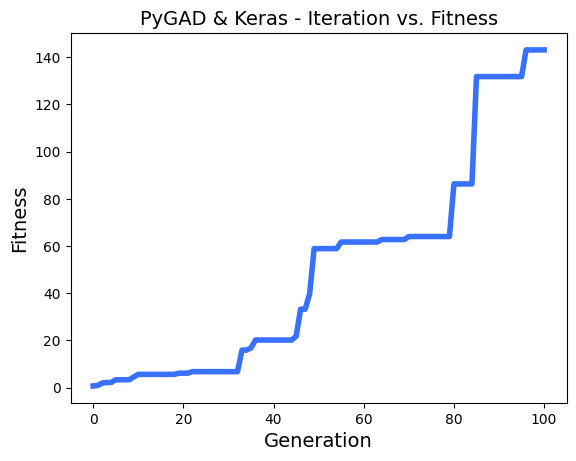
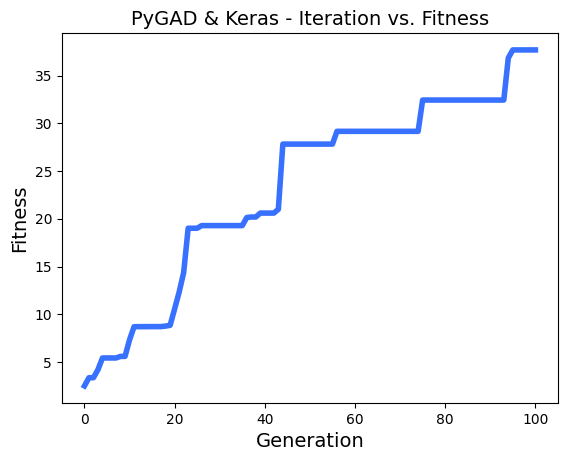
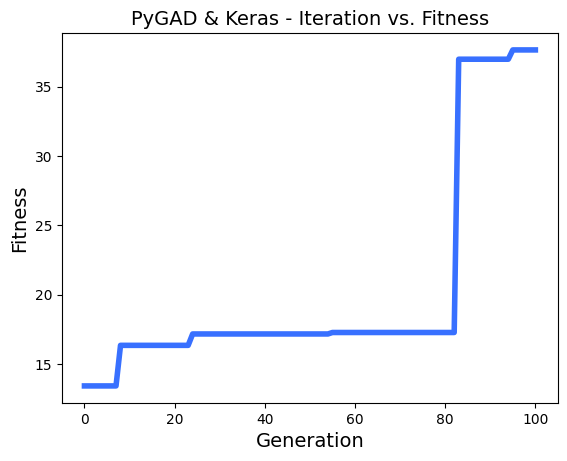
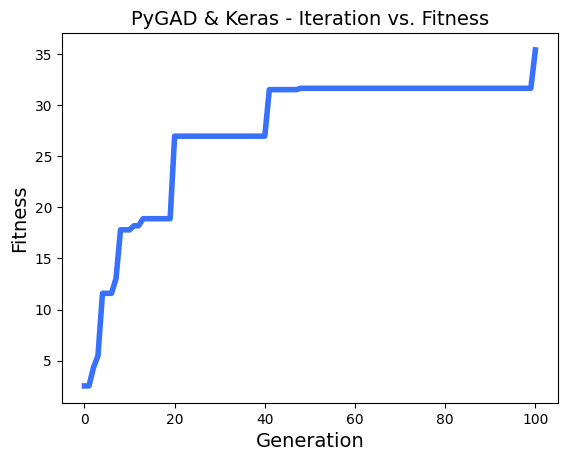
- then, i enable the parallel processing on 8 threads :

- then, run again a few times :




- sometimes, i see in logs a fitness lower than the n-1 generation, example :

- I printed all solutions used in each epoch, and i saw thats solutions are most of time the same, so the parallel_processing seems to break the generation of the next population in the most of cases.
Thanks!
EDIT :
In addition i tried to reproduce the same problem with this [classification problem sample ](https://pygad.readthedocs.io/en/latest/README_pygad_gann_ReadTheDocs.html#image-classification),
Adding the multiprocessing support cause the same problem.
| open | 2022-12-13T13:09:47Z | 2024-01-28T05:47:51Z | https://github.com/ahmedfgad/GeneticAlgorithmPython/issues/150 | [
"bug"
] | BenoitMiquey | 1 |
schemathesis/schemathesis | pytest | 2,072 | [BUG] config allow_x00=False doesn't work | ### Checklist
- [x] I checked the [FAQ section](https://schemathesis.readthedocs.io/en/stable/faq.html#frequently-asked-questions) of the documentation
- [x] I looked for similar issues in the [issue tracker](https://github.com/schemathesis/schemathesis/issues)
- [x] I am using the latest version of Schemathesis
### Describe the bug
I have a FastAPI backend. Schemathesis generate failing case `ValueError: A string literal cannot contain NUL (0x00) characters.`
`parameters = {'name_1': '0\x00'}, context = <sqlalchemy.dialects.postgresql.psycopg2.PGExecutionContext_psycopg2 object at 0x7d142f430cd0>`
Because schemathesis generate case with `\x00` even though I have explicitly used `generation_config=GenerationConfig(allow_x00=False)`
### To Reproduce
My schemathesis pytest file look like this:
```python
def application_provider():
return create_app()
schemathesis.experimental.OPEN_API_3_1.enable()
schema = schemathesis.from_asgi(
f"/{CONFIG['SERVICE_NAME']}/api/v1/openapi.json",
application_provider(),
generation_config=GenerationConfig(allow_x00=False),
)
schemathesis.fixups.install()
# run it manually with "pytest -m schemathesis"
@pytest.mark.schemathesis
@schema.parametrize(skip_deprecated_operations=True)
@settings(
deadline=2000,
verbosity=Verbosity.normal,
suppress_health_check=[HealthCheck.too_slow, HealthCheck.filter_too_much, HealthCheck.function_scoped_fixture],
)
@pytest.mark.parametrize("mocked_database_function", [DATA_SET], indirect=True)
def test_api(case, mocked_database_function, mock_keycloak_client):
headers = get_header_authentication(IDENTITY_ROOT_ADMIN_PROVIDER)
response = case.call_asgi(headers=headers, base_url=f"http://localhost:8090/{CONFIG['SERVICE_NAME']}")
case.validate_response(response, checks=DEFAULT_CHECKS)
```
The code of the endpoint that fail:
```python
class PostOrganizationRoleSchema(BaseModel):
name: str
async def post_create_organization_role(payload: PostOrganizationRoleSchema) -> OrganizationRoleSchema:
"""
Create a new OrganizationRole
"""
payload = payload.model_dump()
organization_role = (
OrganizationRoles.query.options(joinedload("permissions"))
.filter(OrganizationRoles.name == payload.get("name"))
.one_or_none()
) #fail here because name='\x00'
....
```
### Expected behavior
Schemathesis should not create case with special character `\x00`
### Environment
```
- OS: Debian
- Python version: 3.9
- Schemathesis version: 3.25.2
- Spec version: Open API 3.1.0
```
| closed | 2024-02-26T16:21:08Z | 2024-02-29T13:26:37Z | https://github.com/schemathesis/schemathesis/issues/2072 | [
"Priority: High",
"Type: Bug"
] | croumegous | 1 |
postmanlabs/httpbin | api | 274 | Plaintext IP endpoint | Plaintext IP endpoint would be really helpful especially in scripts.
```
$ curl http://httpbin.org/ip.txt --proxy $PROXY >> ips.txt
```
| closed | 2016-01-17T16:07:14Z | 2018-04-26T17:51:09Z | https://github.com/postmanlabs/httpbin/issues/274 | [] | nounder | 3 |
flasgger/flasgger | api | 428 | Failed to get parameters by POST method in “try it out” feature | ### problem
- I‘ve developed an API called ANSWER,in both GET and POST, and then tested by "try it out".
- However, POST api didn't passed the parameter, while GET api ran normal, and so did the POSTMAN
- I don't know what's the problem is
### environments
- Python 3.7
- flasgger 0.9.5
### others
- swagger ui, the parameter "query" shows nothing

- POSTMAN screenshot, "query" ran normal

| open | 2020-08-26T03:48:37Z | 2023-08-01T14:39:52Z | https://github.com/flasgger/flasgger/issues/428 | [] | wqw547243068 | 7 |
healthchecks/healthchecks | django | 991 | Slack legacy webhook integration | Hello!
Slack will eventually be deprecating the webhook integration. The new suggested way to send messages to Slack channels is by creating a custom Slack App in the user's Workspace, which uses a OAuth token (with appropriate scopes) instead of a webhook URL. Could this new method of sending Slack notifications please be incorporated into Healthchecks?
Thanks!
Jim | closed | 2024-04-13T18:53:17Z | 2024-04-14T15:46:53Z | https://github.com/healthchecks/healthchecks/issues/991 | [] | jbuitt | 2 |
Asabeneh/30-Days-Of-Python | pandas | 359 | Translation Spanish | Hi. Thank you for your tutorials.
I want to add Spanish translation to this repository.
Can I work with that? | open | 2023-02-20T18:34:10Z | 2023-08-17T18:16:04Z | https://github.com/Asabeneh/30-Days-Of-Python/issues/359 | [] | Misio942 | 3 |
keras-team/keras | python | 20,566 | Typo on the ReadME | The ReadME had a typo where time series functionality was spelt timeseries. I have submitted a PR to fix this issue,
| closed | 2024-11-30T06:05:49Z | 2024-11-30T11:12:01Z | https://github.com/keras-team/keras/issues/20566 | [] | orcanshul | 0 |
Neoteroi/BlackSheep | asyncio | 211 | Support for OpenAPI parameter serialization | ### Discussed in https://github.com/Neoteroi/BlackSheep/discussions/209
<div type='discussions-op-text'>
<sup>Originally posted by **adriangb** November 16, 2021</sup>
Does BlackSheep support for [OpenAPI parameter serialization](https://swagger.io/docs/specification/serialization/) (aside from the defaults)?</div> | closed | 2021-11-16T18:42:09Z | 2021-11-16T18:42:23Z | https://github.com/Neoteroi/BlackSheep/issues/211 | [] | RobertoPrevato | 1 |
zihangdai/xlnet | nlp | 188 | tpu_estimator.py won't work in tensorflow 1.14 | The XLNET's tpu_estimator.py code won't work in tensorflow 1.14. What's XLNET's plan to support newer tensorflow like 1.14 and beyond?
The XLNET's tpu_esimator.py is based on tf 1.13.1. However, the tpu_estimator design in tensorflow-estimator 1.14 (which is used by tf 1.14), has significant change from previous versions. This caused problem when running tensorflow 1.14 on XLNET.
For example, one error I got is
tpu_estimator.py, line 2224, in __init__
self._ctx = tpu_context._get_tpu_context(
AttributeError: 'module' object has no attribute '_get_tpu_context'
The reason the above code failed is that 'tpu_context' was moved from 'tensorflow.contrib.tpu.python.tpu' to 'tensorflow_estimator.python.estimator.tpu' in 1.14 | open | 2019-07-24T23:51:48Z | 2019-07-24T23:53:08Z | https://github.com/zihangdai/xlnet/issues/188 | [] | LiweiPeng | 0 |
tableau/server-client-python | rest-api | 740 | Updating workbook fields (such as name) is failing with "Payload is either malformed or incomplete" error | TSC: latest (v0.14.0)
Tableau server versions tested:
* exhibit the failure: 2019.1, 2019.3
* works correctly: 2020.4
Sample script:
```py
tableau_auth = TSC.TableauAuth(username, password, site_id=site_url)
tableau_server = TSC.Server(server_url, use_server_version=True)
tableau_server.add_http_options({'verify': False})
with tableau_server.auth.sign_in(tableau_auth):
workbook_item = tableau_server.workbooks.get_by_id(workbook_luid)
new_name = "{}_{}".format(workbook_item.name, "Test")
workbook_item.name = new_name
tableau_server.workbooks.update(workbook_item)
```
Result:
```
Traceback (most recent call last):
File "c:\dev\- Tests\workbook_actions.py", line 30, in <module>
main()
File "c:\dev\- Tests\workbook_actions.py", line 27, in main
tableau_server.workbooks.update(workbook_item)
File "C:\Python39\lib\site-packages\tableauserverclient\server\endpoint\endpoint.py", line 127, in wrapper
return func(self, *args, **kwargs)
File "C:\Python39\lib\site-packages\tableauserverclient\server\endpoint\workbooks_endpoint.py", line 107, in update
server_response = self.put_request(url, update_req)
File "C:\Python39\lib\site-packages\tableauserverclient\server\endpoint\endpoint.py", line 93, in put_request
return self._make_request(self.parent_srv.session.put, url,
File "C:\Python39\lib\site-packages\tableauserverclient\server\endpoint\endpoint.py", line 55, in _make_request
self._check_status(server_response)
File "C:\Python39\lib\site-packages\tableauserverclient\server\endpoint\endpoint.py", line 70, in _check_status
raise ServerResponseError.from_response(server_response.content, self.parent_srv.namespace)
tableauserverclient.server.endpoint.exceptions.ServerResponseError:
400000: Bad Request
Payload is either malformed or incomplete
``` | closed | 2020-11-19T19:08:15Z | 2021-02-19T18:47:12Z | https://github.com/tableau/server-client-python/issues/740 | [] | bcantoni | 2 |
scrapy/scrapy | web-scraping | 6,095 | (With 2-line reproducible sample) When getting inner html, it gives *all* html after it instead of inside it; bug happens for both `.xpath()` and `.css()` | <!--
Thanks for taking an interest in Scrapy!
If you have a question that starts with "How to...", please see the Scrapy Community page: https://scrapy.org/community/.
The GitHub issue tracker's purpose is to deal with bug reports and feature requests for the project itself.
Keep in mind that by filing an issue, you are expected to comply with Scrapy's Code of Conduct, including treating everyone with respect: https://github.com/scrapy/scrapy/blob/master/CODE_OF_CONDUCT.md
The following is a suggested template to structure your issue, you can find more guidelines at https://doc.scrapy.org/en/latest/contributing.html#reporting-bugs
-->
### Description
Hi thanks for the lib! However, When getting inner html, it gives *all* html after it instead of inside it.
### Steps to Reproduce
```
selector = Selector(text='<html><body><div><a>Apple</a><a>Orange</a></div><span>Banana</span></body></html>')
print(selector.xpath('//div/node()').get())
# other attempts
print(selector.xpath('//div/node()').extract())
print(selector.css('div').get())
print(selector.css('div').extract())
```
**Expected behavior:** [What you expect to happen]
Output sth like
```
<a>Apple</a><a>Orange</a>
```
**Actual behavior:** [What actually happens]
```
<a>Apple</a><a>Orange</a></div><span>Banana</span></body></html>
```
**Reproduces how often:** [What percentage of the time does it reproduce?]
100%
### Versions
2.11
### Additional context
Any additional information, configuration, data or output from commands that might be necessary to reproduce or understand the issue. Please try not to include screenshots of code or the command line, paste the contents as text instead. You can use [GitHub Flavored Markdown](https://help.github.com/en/articles/creating-and-highlighting-code-blocks) to make the text look better.
| closed | 2023-10-15T01:48:40Z | 2023-10-15T10:37:17Z | https://github.com/scrapy/scrapy/issues/6095 | [] | fzyzcjy | 2 |
jonaswinkler/paperless-ng | django | 1,186 | [FEATURE REQUEST] Document translation | Would be great if there was a feature where I could translate and OCRd document from one language to another. | open | 2021-07-23T11:26:13Z | 2021-08-13T11:34:24Z | https://github.com/jonaswinkler/paperless-ng/issues/1186 | [] | landcraft | 5 |
apify/crawlee-python | web-scraping | 315 | Document how to switch between HTTP clients | Describe how to use curl impersonate, how to wrap it so it can be used as alternative to httpx | closed | 2024-07-16T11:24:03Z | 2024-08-06T13:13:58Z | https://github.com/apify/crawlee-python/issues/315 | [
"documentation",
"t-tooling"
] | vdusek | 0 |
pyg-team/pytorch_geometric | deep-learning | 10,110 | Can we get a new release? | ### 😵 Describe the installation problem
Currently the last 2.6.1 release is not compatible with numpy 2 as some functions make use of `np.math` resulting in attribute errors. As far as I can tell these calls were removed in #9752 but a release has not yet been made.
Lack of a release is preventing other packages such as https://github.com/FAIR-Chem/fairchem/pull/1003 from supporting numpy 2.
### Environment
| open | 2025-03-12T00:15:51Z | 2025-03-17T22:14:57Z | https://github.com/pyg-team/pytorch_geometric/issues/10110 | [
"installation"
] | CompRhys | 1 |
RobertCraigie/prisma-client-py | pydantic | 969 | feat(prisma): upgrade to v5.15.0 | Hi,
Would it be possible to bump the version to v5.15.0 as there is the benefit of the below preview feature that will help organise folders.
```
generator client {
provider = "prisma-client-js"
previewFeatures = ["prismaSchemaFolder"]
}
``` | closed | 2024-06-06T18:56:27Z | 2024-08-04T17:59:59Z | https://github.com/RobertCraigie/prisma-client-py/issues/969 | [] | SuperP4rks | 2 |
dmlc/gluon-nlp | numpy | 1,406 | [nlp_data] Add BookCorpus | ## Description
The book corpus can now have a reliable, stable download link from https://the-eye.eu/public/AI/pile_preliminary_components/books1.tar.gz. Also, there are more links in https://the-eye.eu/public/AI/pile_preliminary_components/ that are worthwhile being included in `nlp_data`. We may try to download from their link and provide the corresponding license.
| open | 2020-10-26T15:18:36Z | 2020-11-17T20:53:57Z | https://github.com/dmlc/gluon-nlp/issues/1406 | [
"enhancement"
] | sxjscience | 9 |
stanford-oval/storm | nlp | 30 | The article outline does not match the one in storm_gen_outline.txt | This project is very good, I carefully read the project code, some of the ideas are very wonderful. But I still have some confusion when I read the code.
I noticed that the outline argument was not used in the forward function of the ConvToSection class. In this way, articles generated by the write_section object only correspond to the level 1 outline. The suboutline generated in the chapter content is inconsistent with the suboutline generated in storm_gen_outline.txt, which appears to serve only the function of retrieving fragments under the first level outline.
```python
class ConvToSection(dspy.Module):
"""Use the information collected from the information-seeking conversation to write a section."""
def __init__(self, engine: Union[dspy.dsp.LM, dspy.dsp.HFModel]):
super().__init__()
self.write_section = dspy.Predict(WriteSection)
self.engine = engine
def forward(self, topic: str, outline: str, section: str, searched_url_to_snippets: dict):
info = ''
for n, r in enumerate(searched_url_to_snippets.values()):
info += f'[{n + 1}]\n' + '\n'.join(r)
info += '\n\n'
info = limit_word_count_preserve_newline(info, 1500)
with dspy.settings.context(lm=self.engine):
section = clean_up_section(
self.write_section(topic=topic, info=info, section=section).output)
return dspy.Prediction(section=section)
```
| closed | 2024-04-24T09:42:41Z | 2024-04-25T06:42:40Z | https://github.com/stanford-oval/storm/issues/30 | [] | xuxiangwork | 3 |
horovod/horovod | pytorch | 3,569 | Multiple-GPU pytorch training can fail with some cluster setups on Databricks | **Environment:**
1. Framework: PyTorch
2. Framework version: 1.11.0
3. Horovod version: 0.24.3
4. MPI version: 4.1.2
5. CUDA version: 11.3
6. NCCL version: 2.10.3
7. Python version: 3.9.5
8. Spark / PySpark version: 3.3.0
9. Ray version: N/A
10. OS and version: Ubuntu 20.04
11. GCC version: 9.4.0
12. CMake version: 3.16.3
**Bug report:**
The [pytroch_spark.mnist.py](https://github.com/horovod/horovod/blob/master/examples/spark/pytorch/pytorch_spark_mnist.py) example fails with num_proc=4 on a cluster with one driver and one worker using AWS p3.8xlarge instances, which have 4 V100 GPUs.
Error seems to be related to NCCL GPU initialization.
```
[1,6]<stdout>:
[1,6]<stdout>:0601-182219-q4uu569j-10-68-186-195:1374:1448 [0] init.cc:497 NCCL WARN Duplicate GPU detected : rank 6 and rank 0 both on CUDA device 170
[1,6]<stdout>:0601-182219-q4uu569j-10-68-186-195:1374:1448 [0] NCCL INFO init.cc:867 -> 5
[1,6]<stdout>:0601-182219-q4uu569j-10-68-186-195:1374:1448 [0] NCCL INFO init.cc:903 -> 5
[1,6]<stdout>:0601-182219-q4uu569j-10-68-186-195:1374:1448 [0] NCCL INFO init.cc:916 -> 5
[1,6]<stdout>:
[1,6]<stdout>:0601-182219-q4uu569j-10-68-186-195:1374:1448 [0] init.cc:497 NCCL WARN Duplicate GPU detected : rank 6 and rank 0 both on CUDA device 170
[1,6]<stdout>:0601-182219-q4uu569j-10-68-186-195:1374:1448 [0] NCCL INFO init.cc:867 -> 5
[1,6]<stdout>:0601-182219-q4uu569j-10-68-186-195:1374:1448 [0] NCCL INFO init.cc:903 -> 5
[1,6]<stdout>:0601-182219-q4uu569j-10-68-186-195:1374:1448 [0] NCCL INFO init.cc:916 -> 5
[1,6]<stdout>:
[1,6]<stdout>:0601-182219-q4uu569j-10-68-186-195:1374:1448 [0] misc/argcheck.cc:30 NCCL WARN ncclGetAsyncError : comm argument is NULL
[1,6]<stdout>:0601-182219-q4uu569j-10-68-186-195:1374:1448 [0] NCCL INFO init.cc:1007 -> 4
[1,6]<stderr>:Traceback (most recent call last):
[1,6]<stderr>: File "/databricks/python/lib/python3.9/site-packages/horovod/torch/mpi_ops.py", line 946, in synchronize
[1,6]<stderr>: mpi_lib.horovod_torch_wait_and_clear(handle)
[1,6]<stderr>:RuntimeError: ncclCommInitRank failed: invalid usage
[1,6]<stderr>:
[1,6]<stderr>:During handling of the above exception, another exception occurred:
[1,6]<stderr>:
[1,6]<stderr>:Traceback (most recent call last):
[1,6]<stderr>: File "/usr/lib/python3.9/runpy.py", line 197, in _run_module_as_main
[1,6]<stderr>: return _run_code(code, main_globals, None,
[1,6]<stderr>: File "/usr/lib/python3.9/runpy.py", line 87, in _run_code
[1,6]<stderr>: exec(code, run_globals)
[1,6]<stderr>: File "/databricks/python/lib/python3.9/site-packages/horovod/spark/task/mpirun_exec_fn.py", line 52, in <module>
[1,6]<stderr>: main(codec.loads_base64(sys.argv[1]), codec.loads_base64(sys.argv[2]))
[1,6]<stderr>: File "/databricks/python/lib/python3.9/site-packages/horovod/spark/task/mpirun_exec_fn.py", line 45, in main
[1,6]<stderr>: task_exec(driver_addresses, settings, 'OMPI_COMM_WORLD_RANK', 'OMPI_COMM_WORLD_LOCAL_RANK')
[1,6]<stderr>: File "/databricks/python/lib/python3.9/site-packages/horovod/spark/task/__init__.py", line 61, in task_exec
[1,6]<stderr>: result = fn(*args, **kwargs)
[1,6]<stderr>: File "/databricks/python/lib/python3.9/site-packages/horovod/spark/torch/remote.py", line 410, in train
[1,6]<stderr>: 'train': _train(epoch)
[1,6]<stderr>: File "/databricks/python/lib/python3.9/site-packages/horovod/spark/torch/remote.py", line 354, in _train
[1,6]<stderr>: outputs, loss = train_minibatch(model, optimizer, transform_outputs,
[1,6]<stderr>: File "/databricks/python/lib/python3.9/site-packages/horovod/spark/torch/remote.py", line 445, in train_minibatch
[1,6]<stderr>: optimizer.step()
[1,6]<stderr>: File "/databricks/python/lib/python3.9/site-packages/torch/optim/optimizer.py", line 88, in wrapper
[1,6]<stderr>: return func(*args, **kwargs)
[1,6]<stderr>: File "/databricks/python/lib/python3.9/site-packages/horovod/torch/optimizer.py", line 333, in step
[1,6]<stderr>: self.synchronize()
[1,6]<stderr>: File "/databricks/python/lib/python3.9/site-packages/horovod/torch/optimizer.py", line 284, in synchronize
[1,6]<stderr>: output = synchronize(handle) if not callable(handle) else handle()
[1,6]<stderr>: File "/databricks/python/lib/python3.9/site-packages/horovod/torch/mpi_ops.py", line 951, in synchronize
[1,6]<stderr>: raise HorovodInternalError(e)
[1,6]<stderr>:horovod.common.exceptions.HorovodInternalError: ncclCommInitRank failed: invalid usage
--------------------------------------------------------------------------
Primary job terminated normally, but 1 process returned
a non-zero exit code. Per user-direction, the job has been aborted.
--------------------------------------------------------------------------
--------------------------------------------------------------------------
mpirun detected that one or more processes exited with non-zero status, thus causing
the job to be terminated. The first process to do so was:
Process name: [[9819,1],4]
Exit code: 1
--------------------------------------------------------------------------
Exception in thread Thread-19:
Traceback (most recent call last):
File "/usr/lib/python3.9/threading.py", line 954, in _bootstrap_inner
self.run()
File "/usr/lib/python3.9/threading.py", line 892, in run
self._target(*self._args, **self._kwargs)
File "/databricks/python/lib/python3.9/site-packages/horovod/spark/runner.py", line 141, in run_spark
result = procs.mapPartitionsWithIndex(mapper).collect()
File "/databricks/spark/python/pyspark/rdd.py", line 1217, in collect
sock_info = self.ctx._jvm.PythonRDD.collectAndServe(self._jrdd.rdd())
File "/databricks/spark/python/lib/py4j-0.10.9.1-src.zip/py4j/java_gateway.py", line 1304, in __call__
File "/databricks/spark/python/pyspark/sql/utils.py", line 196, in deco
return f(*a, **kw)
File "/databricks/spark/python/lib/py4j-0.10.9.1-src.zip/py4j/protocol.py", line 326, in get_return_value
py4j.protocol.Py4JJavaError: An error occurred while calling z:org.apache.spark.api.python.PythonRDD.collectAndServe.
: org.apache.spark.SparkException: Job 6 cancelled part of cancelled job group horovod.spark.run.0
at org.apache.spark.scheduler.DAGScheduler.failJobAndIndependentStages(DAGScheduler.scala:3203)
at org.apache.spark.scheduler.DAGScheduler.handleJobCancellation(DAGScheduler.scala:3077)
at org.apache.spark.scheduler.DAGScheduler.$anonfun$handleJobGroupCancelled$4(DAGScheduler.scala:1336)
at scala.runtime.java8.JFunction1$mcVI$sp.apply(JFunction1$mcVI$sp.java:23)
at scala.collection.mutable.HashSet.foreach(HashSet.scala:79)
at org.apache.spark.scheduler.DAGScheduler.handleJobGroupCancelled(DAGScheduler.scala:1335)
at org.apache.spark.scheduler.DAGSchedulerEventProcessLoop.doOnReceive(DAGScheduler.scala:3373)
at org.apache.spark.scheduler.DAGSchedulerEventProcessLoop.onReceive(DAGScheduler.scala:3352)
at org.apache.spark.scheduler.DAGSchedulerEventProcessLoop.onReceive(DAGScheduler.scala:3340)
at org.apache.spark.util.EventLoop$$anon$1.run(EventLoop.scala:49)
at org.apache.spark.scheduler.DAGScheduler.runJob(DAGScheduler.scala:1121)
at org.apache.spark.SparkContext.runJobInternal(SparkContext.scala:2659)
at org.apache.spark.rdd.RDD.$anonfun$collect$1(RDD.scala:1027)
at org.apache.spark.rdd.RDDOperationScope$.withScope(RDDOperationScope.scala:165)
at org.apache.spark.rdd.RDDOperationScope$.withScope(RDDOperationScope.scala:125)
at org.apache.spark.rdd.RDDOperationScope$.withScope(RDDOperationScope.scala:112)
at org.apache.spark.rdd.RDD.withScope(RDD.scala:411)
at org.apache.spark.rdd.RDD.collect(RDD.scala:1025)
at org.apache.spark.api.python.PythonRDD$.collectAndServe(PythonRDD.scala:282)
at org.apache.spark.api.python.PythonRDD.collectAndServe(PythonRDD.scala)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43)
at java.lang.reflect.Method.invoke(Method.java:498)
at py4j.reflection.MethodInvoker.invoke(MethodInvoker.java:244)
at py4j.reflection.ReflectionEngine.invoke(ReflectionEngine.java:380)
at py4j.Gateway.invoke(Gateway.java:306)
at py4j.commands.AbstractCommand.invokeMethod(AbstractCommand.java:132)
at py4j.commands.CallCommand.execute(CallCommand.java:79)
at py4j.ClientServerConnection.waitForCommands(ClientServerConnection.java:195)
at py4j.ClientServerConnection.run(ClientServerConnection.java:115)
at java.lang.Thread.run(Thread.java:748)
```
One workaround is to use the [Single Node clusters](https://docs.databricks.com/clusters/single-node.html), which works fine with num_proc=4 on the same AWS p3.8xlarge instance. | closed | 2022-06-07T17:01:19Z | 2022-09-16T02:02:57Z | https://github.com/horovod/horovod/issues/3569 | [
"wontfix"
] | wenfeiy-db | 1 |
plotly/dash | dash | 2,701 | [Feature Request] Making setProps available |
### Feature request
Make `setProps` function available in Dash so it's possible to update a prop in JavaScript and have it trigger a Dash callback.
### Use Cases:
__Dash AG Grid__
Dash Ag Grid has a limited number of props that trigger callbacks. While the Grid's full API is accessible clientside, there are many cases where the `setProps` function would be useful Here are just a few examples:
- Triggering a callback that includes the value of a cell when using the grid’s [context menu.](https://community.plotly.com/t/context-menu-in-dash-ag-grid/75674/14?u=annmariew)
- Knowing which [action updated `virtualRowData`](https://community.plotly.com/t/deleting-rows-in-dash-ag-grid/78700/6?u=annmariew)
- [Triggering a callback from a custom header component](https://community.plotly.com/t/ag-grid-invoke-callbacks-from-custom-header-component/78335/2?u=annmariew)
- Having a button in a cell update other Dash components on the page
__Custom Event Listeners__
- This is a popular request, and it’s available in [Dash Extensions]( https://www.dash-extensions.com/components/event_listener)
> The EventListener component makes it possible to listen to (arbitrary) JavaScript events. The intended usage of the EventListener component is when the event is not already available as a Dash prop. Say that you need to listen to double-click events, but the Dash component only exposes a (single) click property; or some data that you need is not propagated from the JavaScript layer. In these cases, the EventListener component makes it possible to achieve the desired behaviour without editing the component source code.
- [Setting an event listener on a component rather than the whole document](
https://community.plotly.com/t/how-to-get-trigger-from-keyboard/76960/7)
- [Updating a dcc.Store component clientside](
https://community.plotly.com/t/how-to-return-javascript-variable-from-clientside-callback-in-dcc-store-element/59601/3)
There are many more requests and work-arounds posted on the forum. Let me know if you would like more examples
| closed | 2023-11-25T21:55:11Z | 2024-03-11T16:37:00Z | https://github.com/plotly/dash/issues/2701 | [
"feature",
"P1"
] | AnnMarieW | 1 |
dask/dask | numpy | 11,186 | Most tests in `test_parquet.py` fail on s390x (big-endian) | **Describe the issue**: Most tests in `test_parquet.py` fail on s390x (a big-endian arch, which is rare these days):
```
FAILED ../dataframe/io/tests/test_parquet.py::test_local[pyarrow-pyarrow-False] - AssertionError: assert False
FAILED ../dataframe/io/tests/test_parquet.py::test_local[pyarrow-pyarrow-True] - AssertionError: assert False
FAILED ../dataframe/io/tests/test_parquet.py::test_read_glob[pyarrow-pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_calculate_divisions_false[pyarrow-pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_read_list[pyarrow-pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_columns_index[pyarrow-pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_columns_no_index[pyarrow-pyarrow] - AssertionError: DataFrame.iloc[:, 2] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_columns_index_with_multi_index[pyarrow] - AssertionError: DataFrame.iloc[:, 0] (column name="a") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_roundtrip_from_pandas[pyarrow-pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_roundtrip_nullable_dtypes - OSError: Unexpected end of stream: Decoded values 0 does not match expected 1
FAILED ../dataframe/io/tests/test_parquet.py::test_use_nullable_dtypes_with_types_mapper[pyarrow] - AssertionError: DataFrame.iloc[:, 0] (column name="a") NA mask are different
FAILED ../dataframe/io/tests/test_parquet.py::test_append[pyarrow-False] - AssertionError: DataFrame.iloc[:, 2] (column name="f") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_append[pyarrow-True] - AssertionError: DataFrame.iloc[:, 2] (column name="f") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_append_create[pyarrow] - AssertionError: DataFrame.iloc[:, 2] (column name="f") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_append_wo_index[pyarrow-False] - AssertionError: DataFrame.index are different
FAILED ../dataframe/io/tests/test_parquet.py::test_append_wo_index[pyarrow-True] - AssertionError: DataFrame.index are different
FAILED ../dataframe/io/tests/test_parquet.py::test_read_parquet_custom_columns[pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="f") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_timestamp_index[pyarrow-True] - AssertionError: DataFrame.iloc[:, 0] (column name="A") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_timestamp_index[pyarrow-False] - AssertionError: DataFrame.iloc[:, 0] (column name="A") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_pyarrow_schema_mismatch_explicit_schema_none - AssertionError: DataFrame.iloc[:, 1] (column name="y") NA mask are different
FAILED ../dataframe/io/tests/test_parquet.py::test_partition_on_string[aa] - AssertionError: assert {0.0491608291...24936683, ...} == {nan, nan, na...na...
FAILED ../dataframe/io/tests/test_parquet.py::test_partition_on_string[partition_on1] - AssertionError: assert {0.0319944601...08035897, ...} == {nan, nan, na...na...
FAILED ../dataframe/io/tests/test_parquet.py::test_to_parquet_lazy[pyarrow-threads] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_to_parquet_lazy[pyarrow-processes] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_writing_parquet_with_kwargs[pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_roundtrip_arrow[df4] - AssertionError: Attributes of DataFrame.iloc[:, 0] (column name="x") are di...
FAILED ../dataframe/io/tests/test_parquet.py::test_roundtrip_arrow[df11] - AssertionError: DataFrame.iloc[:, 0] (column name="x") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_roundtrip_arrow[df17] - AssertionError: DataFrame.iloc[:, 0] (column name="x") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_roundtrip_arrow[df18] - AssertionError: DataFrame.iloc[:, 0] (column name="-") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_roundtrip_arrow[df19] - AssertionError: DataFrame.iloc[:, 0] (column name=".") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_roundtrip_arrow[df20] - AssertionError: DataFrame.iloc[:, 0] (column name=" ") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_datasets_timeseries[pyarrow] - AssertionError: DataFrame.iloc[:, 2] (column name="x") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_read_glob_no_meta[pyarrow-pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_read_glob_yes_meta[pyarrow-pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_read_dir_nometa[pyarrow-pyarrow-True-True] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_read_dir_nometa[pyarrow-pyarrow-True-False] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_read_dir_nometa[pyarrow-pyarrow-False-True] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_read_dir_nometa[pyarrow-pyarrow-False-False] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_statistics_nometa[pyarrow-pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_timeseries_nulls_in_schema[pyarrow-infer] - OSError: Unexpected end of stream: Decoded values 0 does not match expected 1
FAILED ../dataframe/io/tests/test_parquet.py::test_timeseries_nulls_in_schema[pyarrow-None] - OSError: Unexpected end of stream: Decoded values 0 does not match expected 1
FAILED ../dataframe/io/tests/test_parquet.py::test_split_row_groups_int_aggregate_files[pyarrow-8] - AssertionError: DataFrame.iloc[:, 1] (column name="f") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_row_groups_int_aggregate_files[pyarrow-25] - AssertionError: DataFrame.iloc[:, 1] (column name="f") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_filter_nulls[pyarrow-True-filters1-<lambda>-2] - OSError: Unexpected end of stream: Decoded values 0 does not match expected 2
FAILED ../dataframe/io/tests/test_parquet.py::test_filter_nulls[pyarrow-False-filters1-<lambda>-2] - OSError: Unexpected end of stream: Decoded values 0 does not match expected 2
FAILED ../dataframe/io/tests/test_parquet.py::test_filter_isna[True] - assert 15 == 10
FAILED ../dataframe/io/tests/test_parquet.py::test_filter_isna[False] - assert 15 == 10
FAILED ../dataframe/io/tests/test_parquet.py::test_split_row_groups_filter[pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="f") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_files[4096-None-True] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_files[4096-None-False] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_files[4096-a-True] - AssertionError: DataFrame.iloc[:, 0] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_files[4096-a-False] - AssertionError: DataFrame.iloc[:, 0] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_files[1MiB-None-True] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_files[1MiB-None-False] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_files[1MiB-a-True] - AssertionError: DataFrame.iloc[:, 0] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_files[1MiB-a-False] - AssertionError: DataFrame.iloc[:, 0] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_aggregate_files[a] - AssertionError: DataFrame.iloc[:, 0] (column name="c") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_aggregate_files[b] - AssertionError: DataFrame.iloc[:, 0] (column name="c") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_blocksize[pyarrow-None-True] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_blocksize[pyarrow-None-False] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_blocksize[pyarrow-1024-True] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_blocksize[pyarrow-1024-False] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_blocksize[pyarrow-4096-True] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_blocksize[pyarrow-4096-False] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_blocksize[pyarrow-1MiB-True] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_split_adaptive_blocksize[pyarrow-1MiB-False] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_blocksize[pyarrow-default-True] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_blocksize[pyarrow-default-False] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_blocksize[pyarrow-512-True] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_blocksize[pyarrow-512-False] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_blocksize[pyarrow-1024-True] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_blocksize[pyarrow-1024-False] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_blocksize[pyarrow-1MiB-True] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_blocksize[pyarrow-1MiB-False] - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_roundtrip_pandas_blocksize[pyarrow-pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_pandas_metadata_nullable_pyarrow - AssertionError: DataFrame.iloc[:, 0] (column name="A") NA mask are different
FAILED ../dataframe/io/tests/test_parquet.py::test_partitioned_preserve_index[pyarrow-pyarrow] - AssertionError: DataFrame.iloc[:, 0] (column name="A") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_multi_partition_none_index_false[pyarrow] - AssertionError: DataFrame.iloc[:, 1] (column name="y") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_dir_filter[pyarrow] - AssertionError: DataFrame.iloc[:, 0] (column name="A") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_roundtrip_decimal_dtype - AssertionError: DataFrame.iloc[:, 1] (column name="col1") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_roundtrip_rename_columns[pyarrow] - AssertionError: DataFrame.iloc[:, 0] (column name="d") are different
FAILED ../dataframe/io/tests/test_parquet.py::test_dtype_backend[pyarrow-numpy_nullable] - OSError: Unexpected end of stream: Decoded values 0 does not match expected 1
FAILED ../dataframe/io/tests/test_parquet.py::test_dtype_backend[pyarrow-pyarrow] - OSError: Unexpected end of stream: Decoded values 0 does not match expected 1
FAILED ../dataframe/io/tests/test_parquet.py::test_read_parquet_lists_not_converting - AssertionError: DataFrame.iloc[:, 1] (column name="b") are different
```
**Minimal Complete Verifiable Example**:
Not a code bug, exactly. To reproduce, run the test suite on a big-endian arch, and ensure the `test_parquet.py` tests run. I am a Fedora packager working on the Fedora package; it seems we didn't notice this before because all of these tests were skipped before, I'm not sure why, but in the s390x build log from our last successful build, they all [showed up as SKIPPED](https://kojipkgs.fedoraproject.org//packages/python-dask/2024.2.1/3.fc41~bootstrap/data/logs/s390x/build.log).
The tests do seem to make quite a lot of assumptions that look LE-specific, so it's not exactly surprising that they fail, I guess. I don't know if the underyling code is BE-safe or not.
**Anything else we need to know?**:
**Environment**:
- Dask version: 2024.6.0
- Python version: 3.13
- Operating System: Fedora Rawhide
- Install method (conda, pip, source): happens while building distribution package
| closed | 2024-06-17T23:03:11Z | 2024-06-20T07:55:26Z | https://github.com/dask/dask/issues/11186 | [
"needs triage"
] | AdamWill | 4 |
JaidedAI/EasyOCR | machine-learning | 1,326 | How to limit VRam GPU usage | Is there anyway to limit the GPU VRam usage? | open | 2024-10-22T12:21:47Z | 2024-12-09T11:06:36Z | https://github.com/JaidedAI/EasyOCR/issues/1326 | [] | Pabloferex | 2 |
scikit-learn/scikit-learn | data-science | 30,430 | Example of binning of continous variables for chi2 | ### Describe the issue linked to the documentation
The [chi2](https://scikit-learn.org/stable/modules/generated/sklearn.feature_selection.chi2.html) doesn't work on continuous variables. This issue has numerous discussions, e.g. [here](https://stats.stackexchange.com/questions/369945/feature-selection-using-chi-squared-for-continuous-features).
The Matlab counterpart command, [fscchi2](https://www.mathworks.com/help/stats/fscchi2.html), solves this issue by automatically binning data. I believe that the example of chi2 feature selection with pre-binning may be beneficial.
### Suggest a potential alternative/fix
_No response_ | closed | 2024-12-08T08:05:54Z | 2025-01-06T11:02:06Z | https://github.com/scikit-learn/scikit-learn/issues/30430 | [
"Documentation"
] | bykhov | 4 |
Ehco1996/django-sspanel | django | 518 | 使用docker-compose up -命令出错 | **问题的描述**
因为视频下方没有附带docker的和docker-compose的安装链接,所以就用yum来装的docker和docker-compose
运行环境是CentOS7
[root@host django-sspanel]# docker --version
Docker version 1.13.1, build 7d71120/1.13.1
[root@host django-sspanel]# docker-compose --version
docker-compose version 1.18.0, build 8dd22a9
[root@host django-sspanel]# docker-compose up -d
ERROR: Couldn't find env file: /root/django-sspanel/.env
**项目的配置文件**
**如何复现**
**相关截图/log**
**其他信息**
| closed | 2021-05-04T06:38:13Z | 2021-05-04T07:00:44Z | https://github.com/Ehco1996/django-sspanel/issues/518 | [
"bug"
] | NoString | 2 |
koxudaxi/datamodel-code-generator | pydantic | 1,819 | References to other files don't work | **Describe the bug**
References to other files don't work.
I have a custom schema that uses file references like so:
```json
{
"$schema": "https://json-schema.org/draft/2020-12/schema",
"$id": "file:Foo.schema.json",
"title": "A schema file",
"description": "Schema for validating the serialization of a Foo object.",
"type": "object",
"properties": {
"bar": {
"$ref": "Bar.schema.json"
}
}
```
When calling `datamodel-codegen`:
```sh
$ datamodel-codegen --input /home/user/schemas/Foo.schema.json --output out --input-file-type jsonschema
```
Only the `out` folder is created with a `Bar.py` file.
**Expected behavior**
The `out` folder to have both Python files.
**Version:**
- OS: Ubuntu 22.04.3 LTS
- Python version: Python 3.10.12
- datamodel-code-generator version: `80266b4f6ea09bc7675466af459c7f6617f0c307` | open | 2024-01-25T17:40:01Z | 2024-06-17T12:31:27Z | https://github.com/koxudaxi/datamodel-code-generator/issues/1819 | [] | leonardopsantos | 1 |
RobertCraigie/prisma-client-py | pydantic | 202 | Support configuring the CLI binary path | ## Problem
<!-- A clear and concise description of what the problem is. Ex. I'm always frustrated when [...] -->
Unlike the engine binary paths, the CLI binary path cannot be dynamically set by the user, we should support this to improve the user experience on unsupported architectures.
## Suggested solution
<!-- A clear and concise description of what you want to happen. -->
Use an environment variable `PRISMA_CLI_BINARY` to represent the CLI binary path.
## Additional context
<!-- Add any other context or screenshots about the feature request here. -->
Related issue: #195
We should also look into loading dotenv files before running CLI commands so that binary paths can be configured in a `.env` file instead of having to be set by the user's shell but that should be a separate issue.
| closed | 2022-01-01T15:30:47Z | 2022-02-01T12:08:36Z | https://github.com/RobertCraigie/prisma-client-py/issues/202 | [
"kind/improvement"
] | RobertCraigie | 0 |
graphistry/pygraphistry | pandas | 456 | [BUG] Plugins docs nav missing packages and methods | **Describe the bug**
2
nav issues
- [ ] `graphistry.plugins.cugraph` is missing from the nav
- [ ] methods are not listed in the nav for `graphistry.plugins.cugraph` and `graphistry.plugins.igraph`
| open | 2023-04-03T22:12:56Z | 2023-04-03T22:29:53Z | https://github.com/graphistry/pygraphistry/issues/456 | [
"bug",
"p2",
"docs"
] | lmeyerov | 0 |
redis/redis-om-python | pydantic | 572 | Issue with validate_model using FieldInfo in pydantic v2 | **Environment:**
redis-om: 0.2.1
pydantic: 2.4.2
pydantic-core: 2.11.0
fastapi: 0.104.0
**Description:**
When using the check method in the custom RedisModel which internally calls the validate_model function from pydantic v1, the validation fails with pydantic v2 models having FieldInfo.
**Problematic Code:**
The issue seems to arise from the line:
`value = field.get_default()`
**Possible solution**
Changing the code to the following seems to fix that:
```
def validate_model( # noqa: C901 (ignore complexity)
model: Type[BaseModel], input_data: 'DictStrAny', cls: 'ModelOrDc' = None
) -> Tuple['DictStrAny', 'SetStr', Optional[ValidationError]]:
"""
validate data against a model.
"""
values = {}
errors = []
# input_data names, possibly alias
names_used = set()
# field names, never aliases
fields_set = set()
config = model.__config__
check_extra = config.extra is not Extra.ignore
cls_ = cls or model
for validator in model.__pre_root_validators__:
try:
input_data = validator(cls_, input_data)
except (ValueError, TypeError, AssertionError) as exc:
return {}, set(), ValidationError([ErrorWrapper(exc, loc=ROOT_KEY)], cls_)
for name, field in model.__fields__.items():
value = input_data.get(field.alias, _missing)
using_name = False
if value is _missing and config.allow_population_by_field_name and field.alt_alias:
value = input_data.get(field.name, _missing)
using_name = True
if value is _missing:
if field.required:
errors.append(ErrorWrapper(MissingError(), loc=field.alias))
continue
value = field.get_default()
**//////////////////////////BEGINNING OF ADDED CODE**
if value and hasattr(value,'default'):
if hasattr(value,'default_factory') and value.default_factory:
value = value.default_factory()
else:
value=value.default
if str(value) == 'PydanticUndefined':
value=None
**/////////////////////////////END OF ADDED CODE**
if not config.validate_all and not field.validate_always:
values[name] = value
continue
else:
fields_set.add(name)
if check_extra:
names_used.add(field.name if using_name else field.alias)
v_, errors_ = field.validate(value, values, loc=field.alias, cls=cls_)
if isinstance(errors_, ErrorWrapper):
errors.append(errors_)
elif isinstance(errors_, list):
errors.extend(errors_)
else:
values[name] = v_
if check_extra:
if isinstance(input_data, GetterDict):
extra = input_data.extra_keys() - names_used
else:
extra = input_data.keys() - names_used
if extra:
fields_set |= extra
if config.extra is Extra.allow:
for f in extra:
values[f] = input_data[f]
else:
for f in sorted(extra):
errors.append(ErrorWrapper(ExtraError(), loc=f))
for skip_on_failure, validator in model.__post_root_validators__:
if skip_on_failure and errors:
continue
try:
values = validator(cls_, values)
except (ValueError, TypeError, AssertionError) as exc:
errors.append(ErrorWrapper(exc, loc=ROOT_KEY))
if errors:
return values, fields_set, ValidationError(errors, cls_)
else:
return values, fields_set, None
```
| closed | 2023-10-22T18:09:52Z | 2024-05-02T14:34:20Z | https://github.com/redis/redis-om-python/issues/572 | [] | CodeToFreedom | 1 |
waditu/tushare | pandas | 1,075 | ggt_top10返回值rank部分记录为空 | ggt_top10返回的数据记录,深港通的部分记录rank值为空,沪港通的没有问题。
可测试:20190620、20190621的数据。
https://tushare.pro/register?reg=263568
2019-06-21 00:00:00 00700 腾讯控股 354.4 0.113 2 4 367672240 -88478680
2019-06-21 00:00:00 00002 中电控股 86.4 -4.2659 4 73271825 -26887325
2019-06-21 00:00:00 00763 中兴通讯 22 -2.0045 4 106951050 42836290
2019-06-21 00:00:00 00883 中国海洋石油 13.48 2.5875 4 78048160 71707880
2019-06-21 00:00:00 01109 华润置地 33.95 0.1475 4 49238400 45332000
2019-06-21 00:00:00 01776 广发证券 9.26 -1.2793 4 125526748 31402880
2019-06-21 00:00:00 01787 山东黄金 21 4.2184 4 137955625 72538775
2019-06-21 00:00:00 01918 融创中国 38.7 0.1294 4 65921350 -39657950
2019-06-21 00:00:00 02202 万科企业 29.15 -1.8519 4 85983260 65566630
2019-06-21 00:00:00 06066 中信建投证券 6.08 -1.1382 4 120004980 -70052450
| open | 2019-06-22T03:06:40Z | 2019-06-25T03:32:18Z | https://github.com/waditu/tushare/issues/1075 | [] | pennill | 1 |
youfou/wxpy | api | 310 | 控制台二维码输出和二维码图片输出两者不能同时进行 | site-packages/itchat/components/login.py中get_QR方法中控制的代码如下:
if enableCmdQR:
utils.print_cmd_qr(qrCode.text(1), enableCmdQR=enableCmdQR)
else:
with open(picDir, 'wb') as f:
f.write(qrStorage.getvalue())
utils.print_qr(picDir)
当enableCmdQR有值时就不能输出二维码图片啦,可根据需要自行修改 | open | 2018-06-16T07:36:14Z | 2018-06-16T07:39:47Z | https://github.com/youfou/wxpy/issues/310 | [] | dododogamila | 0 |
microsoft/RD-Agent | automation | 500 | QR 过期了 | 麻烦更新下,谢谢 | closed | 2024-12-01T15:08:46Z | 2024-12-03T06:08:46Z | https://github.com/microsoft/RD-Agent/issues/500 | [
"question"
] | hydenli | 1 |
matplotlib/matplotlib | data-visualization | 29,202 | [Bug]: `fontsize` in tables not working | ### Bug summary
Specifying `fontsize` kwarg in `matplotlib.pyplot.table` doesn't have any effect.
### Code for reproduction
```Python
import matplotlib.pyplot as plt
import numpy as np
x = np.linspace(0, 10, 100)
y = x + 1
tableData = [['a', 1], ['b', 1]]
fig, ax = plt.subplots()
ax.plot(x, y)
t = ax.table(
cellText=tableData,
loc='top',
cellLoc='center',
fontsize=30
)
plt.show()
```
### Actual outcome

### Expected outcome
A table with bigger font size.
### Additional information
This works:
```
import matplotlib.pyplot as plt
import numpy as np
x = np.linspace(0, 10, 100)
y = x + 1
tableData = [['a', 1], ['b', 1]]
fig, ax = plt.subplots()
ax.plot(x, y)
t = ax.table(
cellText=tableData,
loc='top',
cellLoc='center'
)
t.set_fontsize(30) # <----------------
plt.show()
```

### Operating system
Windows 11
### Matplotlib Version
3.8.2
### Matplotlib Backend
QtAgg
### Python version
3.12.0
### Jupyter version
_No response_
### Installation
pip | closed | 2024-11-28T09:57:58Z | 2024-12-16T14:55:17Z | https://github.com/matplotlib/matplotlib/issues/29202 | [
"Difficulty: Easy",
"Good first issue",
"topic: table"
] | acgc99 | 8 |
xuebinqin/U-2-Net | computer-vision | 204 | How to slim the u2net-human-seg.pth? | u2net-human-seg.pth is 168 MB which is too large. How to slim it? | open | 2021-05-18T07:06:18Z | 2021-05-20T07:23:31Z | https://github.com/xuebinqin/U-2-Net/issues/204 | [] | fastscannerapps | 3 |
Miserlou/Zappa | django | 1,344 | Event schedule for async task is not updated | ## Context
I'm using dynamodb triggers which are calling my Lambda function. I did a setup in zappa_settings using "events" list and deployed it. DynamoDB triggers were created successfully.
There are two probelms with it:
1. I tried to change batch_size attribute.
2. I have deleted configuration for one of the triggers
## Expected Behavior
1. DynamoDb trigger should be updated with new settings
2. DynamoDb trigger should be deleted if it don't exist in config anymore
## Actual Behavior
Trigger was not updated nor deleted later. script just said:
`dynamodb event schedule for func_name already exists - Nothing to do here.`
I have to remove it from aws console manually in order to get changes applied.
## Possible Fix
Triggers have to be recreated either all time or if config changes are detected
| open | 2018-01-09T22:34:14Z | 2018-02-23T22:10:58Z | https://github.com/Miserlou/Zappa/issues/1344 | [
"enhancement",
"non-bug",
"good-idea"
] | chekan-o | 1 |
approximatelabs/sketch | pandas | 5 | Valid OpenApi Key Not Accepted with df.sektch.apply | 
I copied your original Google Colb Example Sheet https://colab.research.google.com/gist/bluecoconut/410a979d94613ea2aaf29987cf0233bc/sketch-demo.ipynb#scrollTo=6xZgjwWypy91
https://colab.research.google.com/gist/bluecoconut/410a979d94613ea2aaf29987cf0233bc/sketch-demo.ipynb
`top_5_states = state_sales.sort_values(by='Price Each', ascending=False).head(5).copy()`
`top_5_states.sketch.apply("new column with full name of the states. just the top 5")`
added the lines from the screenshot, the error code is always
`Not sure what happened: {'error': {'message': "Incorrect API key provided: 'sk-ZCTf*****************************************cUZ'. You can find your API key at https://beta.openai.com/.", 'type': 'invalid_request_error', 'param': None, 'code': 'invalid_api_key'}}
Not sure what happened: {'error': {'message': "Incorrect API key provided: 'sk-ZCTf*****************************************cUZ'. You can find your API key at https://beta.openai.com/.", 'type': 'invalid_request_error', 'param': None, 'code': 'invalid_api_key'}}
Not sure what happened: {'error': {'message': "Incorrect API key provided: 'sk-ZCTf*****************************************cUZ'. You can find your API key at https://beta.openai.com/.", 'type': 'invalid_request_error', 'param': None, 'code': 'invalid_api_key'}}
Not sure what happened: {'error': {'message': "Incorrect API key provided: 'sk-ZCTf*****************************************cUZ'. You can find your API key at https://beta.openai.com/.", 'type': 'invalid_request_error', 'param': None, 'code': 'invalid_api_key'}}`
I copied the API key from
https://beta.openai.com/account/api-keys
Bug? or
What am I missing?
Thx | closed | 2023-01-21T13:54:48Z | 2023-01-21T14:29:04Z | https://github.com/approximatelabs/sketch/issues/5 | [] | franzenzenhofer | 2 |
geopandas/geopandas | pandas | 3,103 | DOC: Errors in website providing Geoplot examples | When looking through Geoplot examples in Geopandas I ran across multiple errors inside the online documentation.
#### Screenshot
I think the issue stems from here:

But it breaks the code on the whole page:

#### Problem description
[Link to page](https://geopandas.org/en/stable/gallery/plotting_with_geoplot.html)
Data is not loaded properly into Geopandas and therefore it can't be used for plotting.
#### Expected Output
The expected output is a properly rendered map.
| closed | 2023-12-05T15:15:29Z | 2024-06-08T07:53:57Z | https://github.com/geopandas/geopandas/issues/3103 | [
"documentation",
"downstream issue"
] | AlexMuresan | 7 |
Urinx/WeixinBot | api | 243 | 手动调用wechat.webwxsendmsg给好友发消息失败,求大神点播 | {"BaseResponse":{"Ret":-1,"ErrMsg":""},"MsgID":"","LocalID":""}发送失败返回信息 | closed | 2017-11-30T02:26:07Z | 2017-12-11T08:20:09Z | https://github.com/Urinx/WeixinBot/issues/243 | [] | kun910202 | 0 |
slackapi/python-slack-sdk | asyncio | 891 | Warn when chat.postMessage is called without `text` argument | It's a best practice to always provide a `text` argument when posting a message, even though the platform doesn't technically require it when `blocks` are provided. The `text` argument is used in places where `blocks` cannot be rendered such as: system push notifications, assistive technology such as screen readers, etc.
In order to help apps adhere to this best practice and give users a more accessible experience using Slack, we should add a warning when the `chat.postMessage` (or possibly related methods like `chat.update`) are called without a `text` argument. This warning will be proactive.
One day, the platform may also emit a warning in the response metadata. This SDK should already be set up to log response metadata warnings. When this happens, we should remove the proactive warning.
### Category (place an `x` in each of the `[ ]`)
- [x] **slack_sdk.web.WebClient** (Web API client)
- [ ] **slack_sdk.webhook.WebhookClient** (Incoming Webhook, response_url sender)
- [ ] **slack_sdk.models** (UI component builders)
- [ ] **slack_sdk.oauth** (OAuth Flow Utilities)
- [ ] **slack_sdk.rtm.RTMClient** (RTM client)
- [ ] **slack_sdk.signature** (Request Signature Verifier)
### Requirements
Please read the [Contributing guidelines](https://github.com/slackapi/python-slack-sdk/blob/main/.github/contributing.md) and [Code of Conduct](https://slackhq.github.io/code-of-conduct) before creating this issue or pull request. By submitting, you are agreeing to those rules.
| closed | 2020-12-07T05:55:32Z | 2021-01-08T04:19:24Z | https://github.com/slackapi/python-slack-sdk/issues/891 | [
"enhancement",
"web-client",
"good first issue"
] | aoberoi | 0 |
stitchfix/hamilton | numpy | 75 | Add ResultMixin implementations for Dask native types | **Is your feature request related to a problem? Please describe.**
We should implement useful implementations of:
```python
class ResultMixin(object):
"""Base class housing the static function.
Why a static function? That's because certain frameworks can only pickle a static function, not an entire
object.
"""
@staticmethod
@abc.abstractmethod
def build_result(**outputs: typing.Dict[str, typing.Any]) -> typing.Any:
"""This function builds the result given the computed values."""
pass
```
for use with Dask. E.g. returning a Dask native array, dataframe, bag, etc. Currently the default is to return a *pandas dataframe*.
See the `build_result` function in `DaskGraphAdapter` for a reference point on how it could be used.
**Describe the solution you'd like**
These should probably be placed in the `h_dask.py` module for now. Otherwise open to naming.
Alternatively, we could include more options in `DaskGraphAdapter`. Open to thinking what way is the most user friendly solution going forward.
**Additional context**
The addition of these ResultMixins should enable a user who is using Dask, to not have to implement their own version,
instead they can use the ones that come with Hamilton.
| closed | 2022-02-11T21:50:07Z | 2023-02-26T17:04:40Z | https://github.com/stitchfix/hamilton/issues/75 | [
"enhancement",
"good first issue",
"dask"
] | skrawcz | 1 |
python-restx/flask-restx | flask | 13 | Add support to config swagger-ui using app.config | I came across a problem that probably can only be solved by asking you maintainers of this package.
My need: Show `model` tab by default, instead of `example value`, in the `parameters` section in API method documentation.

The method that I tried: According [docs for `Swagger-UI` configuration](https://github.com/swagger-api/swagger-ui/blob/master/docs/usage/configuration.md#configuration), I can set `DEFAULT_MODEL_RENDERING` to `"model"` to make this happen.
Why didn't work: Since `flask-restplus` doesn't seems to do anything when I set `app.config.SWAGGER_UI_DEFAULT_MODEL_RENDERING` to `"model"`, I suppose allowing users to set custom configurations of `Swagger-UI` could be a feature added to `flask-restx`. | closed | 2020-01-16T13:26:34Z | 2020-01-17T10:37:20Z | https://github.com/python-restx/flask-restx/issues/13 | [
"enhancement"
] | tjysdsg | 3 |
NVlabs/neuralangelo | computer-vision | 192 | ModuleNotFoundError: No module named 'tinycudann' | Can someone help me take a look at the errors I have encountered?Here are the error codes I encountered.
Training with 1 GPUs.
Using random seed 0
Make folder logs/example_group/example_name
* checkpoint:
* save_epoch: 9999999999
* save_iter: 20000
* save_latest_iter: 9999999999
* save_period: 9999999999
* strict_resume: True
* cudnn:
* benchmark: True
* deterministic: False
* data:
* name: dummy
* num_images: None
* num_workers: 4
* preload: True
* readjust:
* center: [0.0, 0.0, 0.0]
* scale: 1.0
* root: datasets/lego_ds2
* train:
* batch_size: 2
* image_size: [802, 802]
* subset: None
* type: projects.neuralangelo.data
* use_multi_epoch_loader: True
* val:
* batch_size: 2
* image_size: [300, 300]
* max_viz_samples: 16
* subset: 4
* image_save_iter: 9999999999
* inference_args:
* local_rank: 0
* logdir: logs/example_group/example_name
* logging_iter: 9999999999999
* max_epoch: 9999999999
* max_iter: 500000
* metrics_epoch: None
* metrics_iter: None
* model:
* appear_embed:
* dim: 8
* enabled: False
* background:
* enabled: True
* encoding:
* levels: 10
* type: fourier
* encoding_view:
* levels: 3
* type: spherical
* mlp:
* activ: relu
* activ_density: softplus
* activ_density_params:
* activ_params:
* hidden_dim: 256
* hidden_dim_rgb: 128
* num_layers: 8
* num_layers_rgb: 2
* skip: [4]
* skip_rgb: []
* view_dep: True
* white: False
* object:
* rgb:
* encoding_view:
* levels: 3
* type: spherical
* mlp:
* activ: relu_
* activ_params:
* hidden_dim: 256
* num_layers: 4
* skip: []
* weight_norm: True
* mode: idr
* s_var:
* anneal_end: 0.1
* init_val: 3.0
* sdf:
* encoding:
* coarse2fine:
* enabled: True
* init_active_level: 4
* step: 5000
* hashgrid:
* dict_size: 22
* dim: 8
* max_logres: 11
* min_logres: 5
* range: [-2, 2]
* levels: 16
* type: hashgrid
* gradient:
* mode: numerical
* taps: 4
* mlp:
* activ: softplus
* activ_params:
* beta: 100
* geometric_init: True
* hidden_dim: 256
* inside_out: False
* num_layers: 1
* out_bias: 0.5
* skip: []
* weight_norm: True
* render:
* num_sample_hierarchy: 4
* num_samples:
* background: 32
* coarse: 64
* fine: 16
* rand_rays: 512
* stratified: True
* type: projects.neuralangelo.model
* nvtx_profile: False
* optim:
* fused_opt: False
* params:
* lr: 0.001
* weight_decay: 0.01
* sched:
* gamma: 10.0
* iteration_mode: True
* step_size: 9999999999
* two_steps: [300000, 400000]
* type: two_steps_with_warmup
* warm_up_end: 5000
* type: AdamW
* pretrained_weight: None
* source_filename: projects/neuralangelo/configs/custom/lego.yaml
* speed_benchmark: False
* test_data:
* name: dummy
* num_workers: 0
* test:
* batch_size: 1
* is_lmdb: False
* roots: None
* type: imaginaire.datasets.images
* timeout_period: 9999999
* trainer:
* amp_config:
* backoff_factor: 0.5
* enabled: False
* growth_factor: 2.0
* growth_interval: 2000
* init_scale: 65536.0
* ddp_config:
* find_unused_parameters: False
* static_graph: True
* depth_vis_scale: 0.5
* ema_config:
* beta: 0.9999
* enabled: False
* load_ema_checkpoint: False
* start_iteration: 0
* grad_accum_iter: 1
* image_to_tensorboard: False
* init:
* gain: None
* type: none
* loss_weight:
* curvature: 0.0005
* eikonal: 0.1
* render: 1.0
* type: projects.neuralangelo.trainer
* validation_iter: 5000
* wandb_image_iter: 10000
* wandb_scalar_iter: 100
cudnn benchmark: True
cudnn deterministic: False
Setup trainer.
Using random seed 0
Traceback (most recent call last):
File "train.py", line 104, in <module>
main()
File "train.py", line 79, in main
trainer = get_trainer(cfg, is_inference=False, seed=args.seed)
File "/home/intel/neuralangelo/imaginaire/trainers/utils/get_trainer.py", line 32, in get_trainer
trainer = trainer_lib.Trainer(cfg, is_inference=is_inference, seed=seed)
File "/home/intel/neuralangelo/projects/neuralangelo/trainer.py", line 26, in __init__
super().__init__(cfg, is_inference=is_inference, seed=seed)
File "/home/intel/neuralangelo/projects/nerf/trainers/base.py", line 28, in __init__
super().__init__(cfg, is_inference=is_inference, seed=seed)
File "/home/intel/neuralangelo/imaginaire/trainers/base.py", line 50, in __init__
self.model = self.setup_model(cfg, seed=seed)
File "/home/intel/neuralangelo/imaginaire/trainers/base.py", line 116, in setup_model
lib_model = importlib.import_module(cfg.model.type)
File "/home/intel/miniconda3/envs/neuralangelo/lib/python3.8/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1014, in _gcd_import
File "<frozen importlib._bootstrap>", line 991, in _find_and_load
File "<frozen importlib._bootstrap>", line 975, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 671, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 843, in exec_module
File "<frozen importlib._bootstrap>", line 219, in _call_with_frames_removed
File "/home/intel/neuralangelo/projects/neuralangelo/model.py", line 21, in <module>
from projects.neuralangelo.utils.modules import NeuralSDF, NeuralRGB, BackgroundNeRF
File "/home/intel/neuralangelo/projects/neuralangelo/utils/modules.py", line 16, in <module>
import tinycudann as tcnn
ModuleNotFoundError: No module named 'tinycudann'
[2024-03-25 20:11:48,544] torch.distributed.elastic.multiprocessing.api: [ERROR] failed (exitcode: 1) local_rank: 0 (pid: 37238) of binary: /home/intel/miniconda3/envs/neuralangelo/bin/python
Traceback (most recent call last):
File "/home/intel/miniconda3/envs/neuralangelo/bin/torchrun", line 10, in <module>
sys.exit(main())
File "/home/intel/miniconda3/envs/neuralangelo/lib/python3.8/site-packages/torch/distributed/elastic/multiprocessing/errors/__init__.py", line 346, in wrapper
return f(*args, **kwargs)
File "/home/intel/miniconda3/envs/neuralangelo/lib/python3.8/site-packages/torch/distributed/run.py", line 806, in main
run(args)
File "/home/intel/miniconda3/envs/neuralangelo/lib/python3.8/site-packages/torch/distributed/run.py", line 797, in run
elastic_launch(
File "/home/intel/miniconda3/envs/neuralangelo/lib/python3.8/site-packages/torch/distributed/launcher/api.py", line 134, in __call__
return launch_agent(self._config, self._entrypoint, list(args))
File "/home/intel/miniconda3/envs/neuralangelo/lib/python3.8/site-packages/torch/distributed/launcher/api.py", line 264, in launch_agent
raise ChildFailedError(
torch.distributed.elastic.multiprocessing.errors.ChildFailedError:
============================================================
train.py FAILED
------------------------------------------------------------
Failures:
<NO_OTHER_FAILURES>
------------------------------------------------------------
Root Cause (first observed failure):
[0]:
time : 2024-03-25_20:11:48
host : intel-MD72-HB1-00
rank : 0 (local_rank: 0)
exitcode : 1 (pid: 37238)
error_file: <N/A>
traceback : To enable traceback see: https://pytorch.org/docs/stable/elastic/errors.html
| open | 2024-03-25T14:41:20Z | 2024-03-25T14:41:20Z | https://github.com/NVlabs/neuralangelo/issues/192 | [] | kerrigenwan | 0 |
huggingface/datasets | pytorch | 6,532 | [Feature request] Indexing datasets by a customly-defined id field to enable random access dataset items via the id | ### Feature request
Some datasets may contain an id-like field, for example the `id` field in [wikimedia/wikipedia](https://huggingface.co/datasets/wikimedia/wikipedia) and the `_id` field in [BeIR/dbpedia-entity](https://huggingface.co/datasets/BeIR/dbpedia-entity). HF datasets support efficient random access via row, but not via this kinds of id fields. I wonder if it is possible to add support for indexing by a custom "id-like" field to enable random access via such ids. The ids may be numbers or strings.
### Motivation
In some cases, especially during inference/evaluation, I may want to find out the item that has a specified id, defined by the dataset itself.
For example, in a typical re-ranking setting in information retrieval, the user may want to re-rank the set of candidate documents of each query. The input is usually presented in a TREC-style run file, with the following format:
```
<qid> Q0 <docno> <rank> <score> <tag>
```
The re-ranking program should be able to fetch the queries and documents according to the `<qid>` and `<docno>`, which are the original id defined in the query/document datasets. To accomplish this, I have to iterate over the whole HF dataset to get the mapping from real ids to row ids every time I start the program, which is time-consuming. Thus I want HF dataset to provide options for users to index by a custom id column, not by row.
### Your contribution
I'm not an expert in this project and I'm afraid that I'm not able to make contributions on the code. | open | 2023-12-25T11:37:10Z | 2025-01-22T10:59:45Z | https://github.com/huggingface/datasets/issues/6532 | [
"enhancement"
] | Yu-Shi | 9 |
google/seq2seq | tensorflow | 215 | Anybody succeeded to run the image captioning task? | I am just wondering is it possible to use the current code to run the image captioning task. If so, can anybody give me some hint?
Thanks!
| open | 2017-05-10T12:25:39Z | 2017-05-14T18:03:53Z | https://github.com/google/seq2seq/issues/215 | [] | Jiakui | 2 |
JaidedAI/EasyOCR | pytorch | 331 | Window 10 Pillow Install error: The headers or library files could not be found for zlib | I am trying to install the library on a Windows 10 PC with pip 20.3.3 and I get the following error:
`The headers or library files could not be found for zlib,
a required dependency when compiling Pillow from source.`
[Maybe the used Pillow version is too old and an update to Pillow 8 would fix this?](https://github.com/python-pillow/Pillow/issues/4242)
Full Log:
```Installing collected packages: Pillow, easyocr
Attempting uninstall: Pillow
Found existing installation: pillow 8.0.1
Uninstalling pillow-8.0.1:
Successfully uninstalled pillow-8.0.1
Running setup.py install for Pillow ... error
ERROR: Command errored out with exit status 1:
command: 'c:\users\marc\pycharmprojects\test\venv\scripts\python.exe' -u -c 'import sys, setuptools, tokenize; sys.argv[0] = '"'"'C:\\Users\\Marc\\AppData\\Local\\Temp\\pip-install
-n0sv2674\\pillow_ea060bd110994f7cb64699bfd27baf90\\setup.py'"'"'; __file__='"'"'C:\\Users\\Marc\\AppData\\Local\\Temp\\pip-install-n0sv2674\\pillow_ea060bd110994f7cb64699bfd27baf90\\setu
p.py'"'"';f=getattr(tokenize, '"'"'open'"'"', open)(__file__);code=f.read().replace('"'"'\r\n'"'"', '"'"'\n'"'"');f.close();exec(compile(code, __file__, '"'"'exec'"'"'))' install --record
'C:\Users\Marc\AppData\Local\Temp\pip-record-7jimv80y\install-record.txt' --single-version-externally-managed --compile --install-headers 'c:\users\marc\pycharmprojects\test\venv\inclu
de\site\python3.9\Pillow'
cwd: C:\Users\Marc\AppData\Local\Temp\pip-install-n0sv2674\pillow_ea060bd110994f7cb64699bfd27baf90\
Complete output (176 lines):
C:\Users\Marc\AppData\Local\Temp\pip-install-n0sv2674\pillow_ea060bd110994f7cb64699bfd27baf90\setup.py:28: RuntimeWarning: Pillow does not yet support Python 3.9 and does not yet prov
ide prebuilt Windows binaries. We do not recommend building from source on Windows.
warnings.warn(
running install
running build
running build_py
creating build
creating build\lib.win-amd64-3.9
creating build\lib.win-amd64-3.9\PIL
copying src\PIL\BdfFontFile.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\BlpImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\BmpImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\BufrStubImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ContainerIO.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\CurImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\DcxImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\DdsImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\EpsImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ExifTags.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\features.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\FitsStubImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\FliImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\FontFile.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\FpxImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\FtexImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\GbrImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\GdImageFile.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\GifImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\GimpGradientFile.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\GimpPaletteFile.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\GribStubImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\Hdf5StubImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\IcnsImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\IcoImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\Image.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageChops.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageCms.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageColor.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageDraw.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageDraw2.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageEnhance.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageFile.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageFilter.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageFont.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageGrab.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageMath.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageMode.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageMorph.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageOps.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImagePalette.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImagePath.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageQt.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageSequence.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageShow.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageStat.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageTk.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageTransform.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImageWin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\ImtImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\IptcImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\Jpeg2KImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\JpegImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\JpegPresets.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\McIdasImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\MicImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\MpegImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\MpoImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\MspImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PaletteFile.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PalmImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PcdImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PcfFontFile.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PcxImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PdfImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PdfParser.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PixarImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PngImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PpmImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PsdImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PSDraw.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\PyAccess.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\SgiImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\SpiderImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\SunImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\TarIO.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\TgaImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\TiffImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\TiffTags.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\WalImageFile.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\WebPImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\WmfImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\XbmImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\XpmImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\XVThumbImagePlugin.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\_binary.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\_tkinter_finder.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\_util.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\_version.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\__init__.py -> build\lib.win-amd64-3.9\PIL
copying src\PIL\__main__.py -> build\lib.win-amd64-3.9\PIL
running egg_info
writing src\Pillow.egg-info\PKG-INFO
writing dependency_links to src\Pillow.egg-info\dependency_links.txt
writing top-level names to src\Pillow.egg-info\top_level.txt
reading manifest file 'src\Pillow.egg-info\SOURCES.txt'
reading manifest template 'MANIFEST.in'
warning: no files found matching '*.c'
warning: no files found matching '*.h'
warning: no files found matching '*.sh'
warning: no previously-included files found matching '.appveyor.yml'
warning: no previously-included files found matching '.coveragerc'
warning: no previously-included files found matching '.codecov.yml'
warning: no previously-included files found matching '.editorconfig'
warning: no previously-included files found matching '.readthedocs.yml'
warning: no previously-included files found matching 'azure-pipelines.yml'
warning: no previously-included files matching '.git*' found anywhere in distribution
warning: no previously-included files matching '*.pyc' found anywhere in distribution
warning: no previously-included files matching '*.so' found anywhere in distribution
no previously-included directories found matching '.azure-pipelines'
no previously-included directories found matching '.travis'
writing manifest file 'src\Pillow.egg-info\SOURCES.txt'
running build_ext
The headers or library files could not be found for zlib,
a required dependency when compiling Pillow from source.
Please see the install instructions at:
https://pillow.readthedocs.io/en/latest/installation.html
Traceback (most recent call last):
File "C:\Users\Marc\AppData\Local\Temp\pip-install-n0sv2674\pillow_ea060bd110994f7cb64699bfd27baf90\setup.py", line 852, in <module>
setup(
File "c:\users\marc\pycharmprojects\test\venv\lib\site-packages\setuptools\__init__.py", line 153, in setup
return distutils.core.setup(**attrs)
File "C:\Users\Marc\AppData\Local\Programs\Python\Python39\lib\distutils\core.py", line 148, in setup
dist.run_commands()
File "C:\Users\Marc\AppData\Local\Programs\Python\Python39\lib\distutils\dist.py", line 966, in run_commands
self.run_command(cmd)
File "C:\Users\Marc\AppData\Local\Programs\Python\Python39\lib\distutils\dist.py", line 985, in run_command
cmd_obj.run()
File "c:\users\marc\pycharmprojects\test\venv\lib\site-packages\setuptools\command\install.py", line 61, in run
return orig.install.run(self)
File "C:\Users\Marc\AppData\Local\Programs\Python\Python39\lib\distutils\command\install.py", line 546, in run
self.run_command('build')
File "C:\Users\Marc\AppData\Local\Programs\Python\Python39\lib\distutils\cmd.py", line 313, in run_command
self.distribution.run_command(command)
File "C:\Users\Marc\AppData\Local\Programs\Python\Python39\lib\distutils\dist.py", line 985, in run_command
cmd_obj.run()
File "C:\Users\Marc\AppData\Local\Programs\Python\Python39\lib\distutils\command\build.py", line 135, in run
self.run_command(cmd_name)
File "C:\Users\Marc\AppData\Local\Programs\Python\Python39\lib\distutils\cmd.py", line 313, in run_command
self.distribution.run_command(command)
File "C:\Users\Marc\AppData\Local\Programs\Python\Python39\lib\distutils\dist.py", line 985, in run_command
cmd_obj.run()
File "C:\Users\Marc\AppData\Local\Programs\Python\Python39\lib\distutils\command\build_ext.py", line 340, in run
self.build_extensions()
File "C:\Users\Marc\AppData\Local\Temp\pip-install-n0sv2674\pillow_ea060bd110994f7cb64699bfd27baf90\setup.py", line 687, in build_extensions
raise RequiredDependencyException(f)
__main__.RequiredDependencyException: zlib
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "<string>", line 1, in <module>
File "C:\Users\Marc\AppData\Local\Temp\pip-install-n0sv2674\pillow_ea060bd110994f7cb64699bfd27baf90\setup.py", line 903, in <module>
raise RequiredDependencyException(msg)
__main__.RequiredDependencyException:
The headers or library files could not be found for zlib,
a required dependency when compiling Pillow from source.
Please see the install instructions at:
https://pillow.readthedocs.io/en/latest/installation.html
----------------------------------------
Rolling back uninstall of pillow
Moving to c:\users\marc\pycharmprojects\test\venv\lib\site-packages\pil\
from C:\Users\Marc\PycharmProjects\test\venv\Lib\site-packages\~il
Moving to c:\users\marc\pycharmprojects\test\venv\lib\site-packages\pillow-8.0.1.dist-info\
from C:\Users\Marc\PycharmProjects\test\venv\Lib\site-packages\~illow-8.0.1.dist-info
ERROR: Command errored out with exit status 1: 'c:\users\marc\pycharmprojects\test\venv\scripts\python.exe' -u -c 'import sys, setuptools, tokenize; sys.argv[0] = '"'"'C:\\Users\\Marc\\
AppData\\Local\\Temp\\pip-install-n0sv2674\\pillow_ea060bd110994f7cb64699bfd27baf90\\setup.py'"'"'; __file__='"'"'C:\\Users\\Marc\\AppData\\Local\\Temp\\pip-install-n0sv2674\\pillow_ea060
bd110994f7cb64699bfd27baf90\\setup.py'"'"';f=getattr(tokenize, '"'"'open'"'"', open)(__file__);code=f.read().replace('"'"'\r\n'"'"', '"'"'\n'"'"');f.close();exec(compile(code, __file__, '
"'"'exec'"'"'))' install --record 'C:\Users\Marc\AppData\Local\Temp\pip-record-7jimv80y\install-record.txt' --single-version-externally-managed --compile --install-headers 'c:\users\marc\
pycharmprojects\test\venv\include\site\python3.9\Pillow' Check the logs for full command output.
``` | closed | 2020-12-15T20:15:55Z | 2022-05-14T10:59:09Z | https://github.com/JaidedAI/EasyOCR/issues/331 | [] | liebig | 4 |
graphql-python/graphene-sqlalchemy | sqlalchemy | 80 | Database ID | 1. Is there a way to get/map database id so that you can get this value in query?
2. Can we map only selected fields from the database table instead of all by default? | closed | 2017-10-08T23:59:42Z | 2023-02-25T00:49:15Z | https://github.com/graphql-python/graphene-sqlalchemy/issues/80 | [] | navinesh | 8 |
miguelgrinberg/Flask-SocketIO | flask | 1,978 | TypeError: <engineio.packet.Packet object at 0xabec08e0> is not a byte | **IMPORTANT**:
python 3.9.8
flask 2.0.2
Flask-SocketIO 5.3.3
python-engineio 4.4.0
python-socketio 5.8.0
eventlet 0.30.1
gunicorn 20.1.0
**Describe the bug**

**To Reproduce**
Steps to reproduce the behavior:
1. Go to '...'
2. Click on '....'
3. Scroll down to '....'
4. See error
**Expected behavior**
A clear and concise description of what you expected to happen.
**Logs**

**Additional context**
Add any other context about the problem here.
| closed | 2023-04-24T02:39:35Z | 2023-04-24T06:06:57Z | https://github.com/miguelgrinberg/Flask-SocketIO/issues/1978 | [] | wxhou | 1 |
FlareSolverr/FlareSolverr | api | 1,219 | Error solving the challenge, will always timeout. Ends with 500 Internal Server Error. | ### Have you checked our README?
- [X] I have checked the README
### Have you followed our Troubleshooting?
- [X] I have followed your Troubleshooting
### Is there already an issue for your problem?
- [X] I have checked older issues, open and closed
### Have you checked the discussions?
- [X] I have read the Discussions
### Environment
```markdown
- FlareSolverr version:3.3.19
- Last working FlareSolverr version:3.3.19
- Operating system: Docker Unraid
- Are you using Docker: Yes
- FlareSolverr User-Agent (see log traces or / endpoint):Mozilla/5.0 (X11; Linux x86_64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/120.0.0.0 Safari/537.36
- Are you using a VPN: No
- Are you using a Proxy: No
- Are you using Captcha Solver: No
- If using captcha solver, which one:
- URL to test this issue: https://www.ygg.re/
```
### Description
Using flaresolverr with Jacket on unraid docker, requests will always timeout. It was working fine yesterday and then stopped today. Seems to be the same issue that was present in early May of 2024, which was fixed with the ENV LANG. I didn't changed anything to my config since that fix.
### Logged Error Messages
```text
2024-06-20 16:40:00 INFO Incoming request => POST /v1 body: {'maxTimeout': 60000, 'cmd': 'request.get', 'url': 'https://www.ygg.re/engine/search?do=search&order=desc&sort=publish_date&category=all'}
version_main cannot be converted to an integer
2024-06-20 16:40:01 INFO Challenge detected. Title found: Just a moment...
2024-06-20 16:40:01 INFO Challenge detected. Title found: Just a moment...
2024-06-20 16:41:00 ERROR Error: Error solving the challenge. Timeout after 60.0 seconds.
2024-06-20 16:41:00 INFO Response in 60.843 s
2024-06-20 16:41:00 INFO 172.17.0.1 POST http://192.168.1.5:8191/v1 500 Internal Server Error
```
### Screenshots
_No response_ | closed | 2024-06-20T16:44:28Z | 2024-07-04T14:04:06Z | https://github.com/FlareSolverr/FlareSolverr/issues/1219 | [
"duplicate"
] | remy4409 | 35 |
explosion/spaCy | data-science | 13,500 | Batch processing does not speed up `en_core_web_trf` | ## How to reproduce the behaviour
```python
spacy.prefer_gpu()
nlp = spacy.load(
"en_core_web_trf",
disable=['tagger', 'ner', 'lemmatizer', 'textcat']
)
node = """Some really long string, 3000 characters"""
# simulating 96 pretty long docs
nodes = [node*25]*96
```
Then, run each of the below lines separately and time it:
```
# 1 minute 7.5 s
[list(doc.sents) for doc in nlp.pipe(nodes, batch_size=96)]
# 1 minute 7.3 s
[list(doc.sents) for doc in nlp.pipe(nodes, batch_size=32)]
# 1 m 8.2 s
[list(doc.sents) for doc in nlp.pipe(nodes, batch_size=1)]
```
Running the same thing with `en_core_web_lg` results in substantial gains due to batching. Largest batch size is roughly 1/4 of the runtime of batch_size=1.
## Your Environment
<!-- Include details of your environment. You can also type `python -m spacy info --markdown` and copy-paste the result here.-->
Using a single RTX A6000
python -m spacy info --markdown:
## Info about spaCy
- **spaCy version:** 3.7.4
- **Platform:** Linux-5.15.0-94-generic-x86_64-with-glibc2.35
- **Python version:** 3.10.12
- **Pipelines:** en_core_web_lg (3.7.1), en_core_web_trf (3.7.3), en_core_web_sm (3.7.1), de_core_news_sm (3.7.0)
## Expected Behavior
My understanding from the documentation and this [issue](https://github.com/explosion/spaCy/issues/4935) is that we should expect significant gains from batching, as observed with `en_core_web_lg`. However, using `en_core_web_trf` does not yield significant gains from batching.
I'm wondering if this is a bug, or if we should not expect improved performance due to batching for a Transformer-Parser pipeline. Thanks for this awesome package, and in advance for your help! | open | 2024-05-16T17:06:17Z | 2024-05-16T17:06:17Z | https://github.com/explosion/spaCy/issues/13500 | [] | njaramish | 0 |
scanapi/scanapi | rest-api | 89 | Add a changelog linter | ## Description
Configure a changelog linter. Maybe https://github.com/rcmachado/changelog
Related with: #88 | closed | 2019-12-20T20:04:08Z | 2020-01-09T14:21:02Z | https://github.com/scanapi/scanapi/issues/89 | [
"Documentation",
"Automation"
] | camilamaia | 0 |
flairNLP/flair | nlp | 3,308 | [Bug]: I have (in a fresh conda env) with pip install flair this | ### Describe the bug
---------------------------------------------------------------------------
RuntimeError Traceback (most recent call last)
Cell In[4], line 5
2 from flair.models import SequenceTagger
4 # load tagger
----> 5 tagger = SequenceTagger.load("flair/ner-spanish-large")
7 # make example sentence
8 sentence = Sentence("George Washington fue a Washington")
File [~/miniconda3/envs/flair/lib/python3.11/site-packages/flair/models/sequence_tagger_model.py:1035](https://file+.vscode-resource.vscode-cdn.net/home/jncc/Documents/Aprendizaje_Profundo/Repositorios/PRIVATE_REPOS/RNEC_fullstack/scripts/protonb/~/miniconda3/envs/flair/lib/python3.11/site-packages/flair/models/sequence_tagger_model.py:1035), in SequenceTagger.load(cls, model_path)
1031 @classmethod
1032 def load(cls, model_path: Union[str, Path, Dict[str, Any]]) -> "SequenceTagger":
1033 from typing import cast
-> 1035 return cast("SequenceTagger", super().load(model_path=model_path))
File [~/miniconda3/envs/flair/lib/python3.11/site-packages/flair/nn/model.py:559](https://file+.vscode-resource.vscode-cdn.net/home/jncc/Documents/Aprendizaje_Profundo/Repositorios/PRIVATE_REPOS/RNEC_fullstack/scripts/protonb/~/miniconda3/envs/flair/lib/python3.11/site-packages/flair/nn/model.py:559), in Classifier.load(cls, model_path)
555 @classmethod
556 def load(cls, model_path: Union[str, Path, Dict[str, Any]]) -> "Classifier":
557 from typing import cast
--> 559 return cast("Classifier", super().load(model_path=model_path))
File [~/miniconda3/envs/flair/lib/python3.11/site-packages/flair/nn/model.py:191](https://file+.vscode-resource.vscode-cdn.net/home/jncc/Documents/Aprendizaje_Profundo/Repositorios/PRIVATE_REPOS/RNEC_fullstack/scripts/protonb/~/miniconda3/envs/flair/lib/python3.11/site-packages/flair/nn/model.py:191), in Model.load(cls, model_path)
189 if not isinstance(model_path, dict):
190 model_file = cls._fetch_model(str(model_path))
--> 191 state = load_torch_state(model_file)
...
2042 self.__class__.__name__, "\n\t".join(error_msgs)))
2043 return _IncompatibleKeys(missing_keys, unexpected_keys)
RuntimeError: Error(s) in loading state_dict for XLMRobertaModel:
Unexpected key(s) in state_dict: "embeddings.position_ids".
### To Reproduce
```python
from flair.data import Sentence
from flair.models import SequenceTagger
# load tagger
tagger = SequenceTagger.load("flair/ner-spanish-large")
# make example sentence
sentence = Sentence("George Washington fue a Washington")
# predict NER tags
tagger.predict(sentence)
# print sentence
print(sentence)
```
### Expected behavior
The code show be working ...
### Logs and Stack traces
_No response_
### Screenshots
_No response_
### Additional Context
_No response_
### Environment
conda env | closed | 2023-09-01T19:10:33Z | 2023-09-01T19:18:47Z | https://github.com/flairNLP/flair/issues/3308 | [
"bug"
] | jocastrocUnal | 1 |
gradio-app/gradio | data-science | 10,517 | Add supported hook for rewriting user input before it reaches the chat interface (for anonymizing user input) | - [x] I have searched to see if a similar issue already exists.
**Is your feature request related to a problem? Please describe.**
Yes, we need to anonymize user input before it reaches the LLM for data privacy reasons. I'd also like the user to see how their input is anonymized to ease data-privacy concerns.
Here's what I envision what it might look like:
```python
def anonymize_input(user_input, history):
# rewrite user input here...
return user_input
with gr.Blocks() as demo:
chat_interface = gr.ChatInterface(
fn=dummy_chat,
type="messages",
on_user_input=anonymize_input,
)
```
**Additional context**
Here's what it currently looks like:
```python
from gliner import GLiNER # Ensure you have the GLiNER library installed
import gradio as gr
model = GLiNER.from_pretrained("urchade/gliner_medium-v2.1")
def dummy_chat(user_input: str, history):
return f"You said '{user_input}'"
class ChatInterfaceWithAnonymizeOverride(gr.ChatInterface):
def _append_message_to_history(self, message, history, role):
history = super()._append_message_to_history(message, history, role)
text = history[-1]["content"]
labels = ["Name", "Nationality", "City", "Birthdate", "Phone number", "Email", "Address"]
# Perform entity prediction
entities = model.predict_entities(text, labels, threshold=0.5)
# Replace detected entities in the text
for entity in entities:
if entity["label"] in labels: # For standard PII
placeholder = f"[{entity['label']}]"
text = text.replace(entity["text"], placeholder)
history[-1]["content"] = text
return history
with gr.Blocks() as demo:
chat_interface = ChatInterfaceWithAnonymizeOverride(
fn=dummy_chat,
type="messages",
)
if __name__ == "__main__":
demo.launch()
```
As you can see I'm overriding an internal function for this to work (good job on class design btw)

| closed | 2025-02-05T21:00:37Z | 2025-03-11T15:11:07Z | https://github.com/gradio-app/gradio/issues/10517 | [
"enhancement",
"needs designing"
] | brycepg | 3 |
xonsh/xonsh | data-science | 5,522 | Bug: `xthread` not found | ## Current Behavior
```console
$ xthread
xonsh: For full traceback set: $XONSH_SHOW_TRACEBACK = True
xonsh: subprocess mode: command not found: 'xthread'
xthread: command not found
```
<detail>
```console
<xonsh-code>:1:0 - xthread
<xonsh-code>:1:0 + ![xthread]
TRACE SUBPROC: (['xthread'],), captured=hiddenobject
xonsh: To log full traceback to a file set: $XONSH_TRACEBACK_LOGFILE = <filename>
Traceback (most recent call last):
File "/nix/store/w3jmcy5z80zamcbjp3x2wx101f3p8a0n-python3-3.11.9-env/lib/python3.11/site-packages/xonsh/procs/specs.py", line 474, in _run_binary
p = self.cls(cmd, bufsize=bufsize, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/nix/store/7hnr99nxrd2aw6lghybqdmkckq60j6l9-python3-3.11.9/lib/python3.11/subprocess.py", line 1026, in __init__
self._execute_child(args, executable, preexec_fn, close_fds,
File "/nix/store/7hnr99nxrd2aw6lghybqdmkckq60j6l9-python3-3.11.9/lib/python3.11/subprocess.py", line 1955, in _execute_child
raise child_exception_type(errno_num, err_msg, err_filename)
FileNotFoundError: [Errno 2] No such file or directory: 'xthread'
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/nix/store/w3jmcy5z80zamcbjp3x2wx101f3p8a0n-python3-3.11.9-env/lib/python3.11/site-packages/xonsh/procs/pipelines.py", line 167, in __init__
proc = spec.run(pipeline_group=pipeline_group)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/nix/store/w3jmcy5z80zamcbjp3x2wx101f3p8a0n-python3-3.11.9-env/lib/python3.11/site-packages/xonsh/procs/specs.py", line 456, in run
p = self._run_binary(kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^
File "/nix/store/w3jmcy5z80zamcbjp3x2wx101f3p8a0n-python3-3.11.9-env/lib/python3.11/site-packages/xonsh/procs/specs.py", line 493, in _run_binary
raise xt.XonshError(e) from ex
xonsh.tools.XonshError: xonsh: subprocess mode: command not found: 'xthread'
xthread: command not found
TRACE SUBPROC: (['/etc/profiles/per-user/midirhee12/bin/starship', 'prompt', '--status=1', '--jobs=0', '--cmd-duration=89'],), captured=stdout
TRACE SUBPROC: (['/etc/profiles/per-user/midirhee12/bin/starship', 'prompt', '--status=1', '--jobs=0', '--cmd-duration=89', '--right'],), captured=stdout
```
</detail>
## Expected Behavior
For `xthread` to be found and ran.
## For community
⬇️ **Please click the 👍 reaction instead of leaving a `+1` or 👍 comment**
| closed | 2024-06-20T16:10:02Z | 2024-06-20T22:34:53Z | https://github.com/xonsh/xonsh/issues/5522 | [] | midirhee12 | 3 |
plotly/dash-cytoscape | dash | 58 | [Feature request] ctxt menu | Please consider this addition:
https://github.com/cytoscape/cytoscape.js-cxtmenu
Enables a lot of interactivity (yet I got a feeling that it is challenging to implement)
Anyway, are there any plans about cytoscape addons in near future?Lots of precious functions there.
Thanks! | open | 2019-05-05T17:57:45Z | 2021-05-19T10:40:03Z | https://github.com/plotly/dash-cytoscape/issues/58 | [
"suggestion"
] | vetertann | 2 |
PaddlePaddle/PaddleHub | nlp | 2,155 | no module |
1)PaddleHub2.0.0和PaddlePaddle2.0.0版本
2)系统环境Windows
| closed | 2022-12-02T05:39:46Z | 2022-12-02T09:44:00Z | https://github.com/PaddlePaddle/PaddleHub/issues/2155 | [] | MaxokDavid | 0 |
predict-idlab/plotly-resampler | plotly | 18 | Roadmap | This issue is a request to the community to submit what their vision of plotly-resampler is, which features are still worth implementing?
Some features which I find worth pursuing:
* Summary dataset statistics of the respective view (initially in a table)
-> e.g. a Table with df.describe() for each series that is shown.
* Options to **annotate your data**
No idea how to implement this with plotly, would think that you would need to define your own OO structure to do so (which re-uses underlying annotations), but this would still imply a lot of logic and design decisions (e.g. loading annotations, saving annotations, ...)
* Other aggregation methods (e.g. using width of trace for uncertainty / downsampling rate)
just a gist, but using a plot-per-pixel ratio of 2 with LTTB seems a rather good IDEA to me.
Also, playing with the line-width seems a valid path to embark upon. | open | 2022-01-15T17:28:25Z | 2023-02-06T02:35:22Z | https://github.com/predict-idlab/plotly-resampler/issues/18 | [
"discussion"
] | jonasvdd | 20 |
seleniumbase/SeleniumBase | web-scraping | 2,562 | how to close or quit uc_driver ? | I couldn't find any documentation about closing uc_driver after it's finished running. sb.quit() does not exist

| closed | 2024-03-04T09:50:37Z | 2024-03-04T13:10:11Z | https://github.com/seleniumbase/SeleniumBase/issues/2562 | [
"question",
"UC Mode / CDP Mode"
] | zxczxcdev | 1 |
fastapi/fastapi | asyncio | 12,323 | Should the alias parameter in the Path method be removed? | ### Privileged issue
- [ ] I'm @tiangolo or he asked me directly to create an issue here.
### Issue Content
This is my MRE code:
```python
from __future__ import annotations
from typing import Annotated
import uvicorn
from fastapi import FastAPI, Path
app = FastAPI()
@app.get('/{item_id}')
async def async_root(item_id: Annotated[int, Path(title = 'Item ID', alias = 'xid')]):
return {'item_id': item_id}
if __name__ == '__main__':
uvicorn.run(app, host = '127.0.0.1', port = 18081)
```
and I got the swagger docs:
http://127.0.0.1:18081/docs#/default/async_root__item_id__get

Question:
1. Why is the content of the title field not displayed in the documentation?
2.

The generated curl request is incorrect; the placeholder {item_id} has not been replaced with the value 123 for xid
3. When I try to manually send a curl request to access it, I still get an error:
```python
curl http://127.0.0.1:18081/123
{"detail":[{"type":"missing","loc":["path","xid"],"msg":"Field required","input":null}]}
```
4. When I remove the alias field from the Path method, it works normally:
```python
@app.get('/{item_id}')
async def async_root(item_id: Annotated[int, Path(title = 'Item ID')]):
return {'item_id': item_id}
```

```python
curl http://127.0.0.1:18081/123
{"item_id":123}
```
the `alias` field description:

My Environment:
Python 3.12.4
FastAPI 0.115.0
macOS M2 Max 14.6.1
PyCharm 2024.2.3 (Professional Edition) | closed | 2024-09-30T03:44:47Z | 2024-09-30T12:35:34Z | https://github.com/fastapi/fastapi/issues/12323 | [] | bowenerchen | 1 |
kynan/nbstripout | jupyter | 115 | Nbdime error on Windows with nbstripout | Hey,
We got an issue reported in https://github.com/jupyterlab/jupyterlab-git/issues/471#issuecomment-574326817 when using nbstripout and nbdime on Windows.
I was able to reproduce that issue on the following system:
- Windows 10 x64
- Relevant packages
```
# Name Version Build Channel
jupyterlab-git 0.9.0 dev_0 <develop>
nbdime 1.1.0 pypi_0 pypi
nbformat 4.4.0 py_1 conda-forge
nbstripout 0.3.7 py_0 conda-forge
notebook 6.0.1 py37_0 conda-forge
python 3.7.6 h5b45d93_1 conda-forge
```
Digging into that issue, `nbdime` finds out the filter applied on the notebook by querying the `git config` command. Then it applies the obtained command on the file before processing with the diff operation.
On Windows, git uses a unix shell. But `nbdime` with that logic uses the native shell of the OS. And in the particular case of nbstripout, it is failing.
- Call from nbdime
```
subprocess.CalledProcessError: Command ''C:/Users/freud/Anaconda3/envs/jlab/python.exe' -m nbstripout' returned non-zero exit status 1.
```
- Call executed directly in Windows shell:
```
(jlab) ...\jupyterlab-git\examples>'C:/Users/freud/Anaconda3/envs/jlab/python.exe' -m nbstripout < demo.ipynb
''C:' is not recognized as valid command
```
The error raises due to the use of single quote in
https://github.com/kynan/nbstripout/blob/df6a925b34b067e5ff8a01329a78c86e7f2181a3/nbstripout/_nbstripout.py#L138
Using double quote `"` removes the error. | closed | 2020-01-25T15:50:54Z | 2021-04-25T17:04:46Z | https://github.com/kynan/nbstripout/issues/115 | [
"type:enhancement",
"resolution:fixed",
"platform:windows"
] | fcollonval | 2 |
grillazz/fastapi-sqlalchemy-asyncpg | sqlalchemy | 195 | switch project to uv | closed | 2025-03-01T13:45:35Z | 2025-03-08T10:03:22Z | https://github.com/grillazz/fastapi-sqlalchemy-asyncpg/issues/195 | [] | grillazz | 0 |
|
serpapi/google-search-results-python | web-scraping | 53 | requests.exceptions.SSLError: HTTPSConnectionPool(host='serpapi.com', port=443) | ````
from langchain import SerpAPIWrapper
from langchain.requests import RequestsWrapper
import requests
#verfy=False
search = SerpAPIWrapper(serpapi_api_key="api")
results = search.run("how to make a cake")`
``` | open | 2023-07-14T14:49:47Z | 2023-07-14T22:37:09Z | https://github.com/serpapi/google-search-results-python/issues/53 | [] | abhishekmm | 1 |
microsoft/unilm | nlp | 1,379 | BEiT3 pretrained parameters can't be found. Or the input_size does not match the model? | I am trying to fine-tuning the BEiT3ForVQAV2 model based on the pretrained parameters.
I downloaded the pretrained parameters from this link:
https://conversationhub.blob.core.windows.net/beit-share-public/beit3/pretraining/beit3_base_patch16_224.pth?sv=2021-10-04&st=2023-06-08T11%3A16%3A02Z&se=2033-06-09T11%3A16%3A00Z&sr=c&sp=r&sig=N4pfCVmSeq4L4tS8QbrFVsX6f6q844eft8xSuXdxU48%3D
According to the README, I train the model with these hyper parameters:
```
-- model beit3_base_patch16_480
-- input_size 480
-- finetune ./beit3_base_patch16_224.zip
```
Yes, I set `finetune` to `./beit3_base_patch16_224.zip` but not `./beit3_base_patch16_224.pth`. Because the downloaded file is a .zip file, and there is no .pth file after decompression.
I ran the code but got:
```
RuntimeError: "compute_indices_weights_cubic" not implemented for 'Half'
```
I think the reason is that the `input_size` did not match to the pretrained parameters. So I changed the value:
```
-- model beit3_base_patch16_224
-- input_size 224
-- finetune ./beit3_base_patch16_224.zip
```
There were no problems and the code ran successfully. But the model performed poorly with a vqa score of 0.67 (77.65 from the paper)
I'm sure I didn't make any changes to the code or the dataset, other than setting input_size to 224.
So I re-downloaded the pre-training parameters via this link:
https://conversationhub.blob.core.windows.net/beit-share-public/beit3/vqa/beit3_base_patch16_480_vqa.pth?sv=2021-10-04&st=2023-06-08T11%3A16%3A02Z&se=2033-06-09T11%3A16%3A00Z&sr=c&sp=r&sig=N4pfCVmSeq4L4tS8QbrFVsX6f6q844eft8xSuXdxU48%3D
and ran the code with:
```
-- model beit3_base_patch16_480
-- input_size 480
-- finetune ./beit3_base_patch16_480_vqa.zip
```
but this time:
RuntimeError: Error(s) in loading state_dict for BEiTForVisualQuestionAnswering:
size mismatch for beit3.encoder.embed_positions.A.weight: coping a param with shape torch.Size([199, 768]) from checkpoint, the shape in current model is torch.Size([903, 768])
I'm wondering if I downloaded the wrong pre-trained parameters file. | open | 2023-11-30T15:37:46Z | 2024-11-14T05:03:03Z | https://github.com/microsoft/unilm/issues/1379 | [] | gaoCleo | 2 |
Textualize/rich | python | 2,386 | [BUG] ModuleNotFoundError: No module named 'typing_extensions' for v12.5.0 | Version: v12.5.0, Python 3.9
import rich.logging
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "C:\Users\clientadmin\miniconda3\envs\bio2print3\lib\site-packages\rich\logging.py", line 13, in <module>
from .traceback import Traceback
File "C:\Users\clientadmin\miniconda3\envs\bio2print3\lib\site-packages\rich\traceback.py", line 26, in <module>
from .syntax import Syntax
File "C:\Users\clientadmin\miniconda3\envs\bio2print3\lib\site-packages\rich\syntax.py", line 52, in <module>
from typing_extensions import TypeAlias
ModuleNotFoundError: No module named 'typing_extensions' | closed | 2022-07-11T14:10:23Z | 2022-07-11T14:35:12Z | https://github.com/Textualize/rich/issues/2386 | [] | l-spiecker | 2 |
Johnserf-Seed/TikTokDownload | api | 87 | [BUG]长链接无法下载下页视频 | **描述出现的错误**
应该是大佬忘记改了
使用长链接时无法下载下页视频,短连接正常
**截图**

| closed | 2022-02-08T11:56:16Z | 2022-02-09T07:14:46Z | https://github.com/Johnserf-Seed/TikTokDownload/issues/87 | [
"故障(bug)",
"额外求助(help wanted)",
"无效(invalid)"
] | gaucnet | 2 |
ray-project/ray | machine-learning | 51,060 | [Ray Core] `ray.data.Dataset.repartition` not working consistently with doc/error message | ### What happened + What you expected to happen
```
ds = ds.repartition(num_blocks=x, target_num_rows_per_block=y)
```
Obviously it's conflicting to setting both. Calling this gives the error `ValueError: Either num_blocks or target_num_rows_per_block must be set, but not both.`, as expected.
But this doesn't work `ds = ds.repartition(target_num_rows_per_block=y)`. I got `TypeError: Dataset.repartition() missing 1 required positional argument: 'num_blocks'`
This doesn't seem to make sense to me. According to [doc](https://docs.ray.io/en/latest/data/api/doc/ray.data.Dataset.repartition.html), it should be happy with one of the 2 arguments given.
### Versions / Dependencies
ray 2.43.0
Python 3.11.11
Ubuntu 22.04.3 LTS
### Reproduction script
```
import ray
ds = ray.data.range(100)
ds.repartition(target_num_rows_per_block=10)
```
### Issue Severity
None | closed | 2025-03-04T10:12:28Z | 2025-03-10T22:08:28Z | https://github.com/ray-project/ray/issues/51060 | [
"bug",
"P1",
"triage",
"data"
] | marcmk6 | 3 |
horovod/horovod | deep-learning | 3,923 | fail to build horovod 0.28.0 from the source with gcc 12 due to gloo issue | **Environment:**
1. Framework: tensorflow 2.12.0, pytorch 2.0.1
2. Framework version:
3. Horovod version: 0.28.0
4. MPI version:
5. CUDA version: 12.1.1
6. NCCL version: 2.17.1
7. Python version: 3.11
8. Spark / PySpark version:
9. Ray version:
10. OS and version: ArchLinux
11. GCC version: 12.3.0
12. CMake version: 3.26.3
**Checklist:**
1. Did you search issues to find if somebody asked this question before?
2. If your question is about hang, did you read [this doc](https://github.com/horovod/horovod/blob/master/docs/running.rst)?
3. If your question is about docker, did you read [this doc](https://github.com/horovod/horovod/blob/master/docs/docker.rst)?
4. Did you check if you question is answered in the [troubleshooting guide](https://github.com/horovod/horovod/blob/master/docs/troubleshooting.rst)?
**Bug report:**
```bash
git clone https://github.com/horovod/horovod.git
cd horovod
git submodule update --init --recursive
# modify these environment variable as you need, see also https://github.com/horovod/horovod/blob/master/docs/install.rst
export HOROVOD_CUDA_HOME=/opt/cuda
export HOROVOD_CPU_OPERATIONS=GLOO
export HOROVOD_GPU=CUDA
export HOROVOD_GPU_ALLREDUCE=NCCL
export HOROVOD_GPU_BROADCAST=NCCL
export HOROVOD_WITH_GLOO=1
export HOROVOD_WITH_MPI=1
export HOROVOD_WITHOUT_MXNET=0
export HOROVOD_WITH_PYTORCH=1
export HOROVOD_WITH_TENSORFLOW=1
export HOROVOD_BUILD_CUDA_CC_LIST="60,61,62,70,72,75,80,86,89,90"
export CC=gcc-12
export CXX=g++-12
python setup.py build
```
error logs:
```text
/build/python-horovod/src/horovod-0.28.0/third_party/gloo/gloo/mpi/context.cc:43:3: note: in expansion of macro ‘GLOO_ENFORCE_EQ’
43 | GLOO_ENFORCE_EQ(rv, MPI_SUCCESS);
| ^~~~~~~~~~~~~~~
make[2]: *** [third_party/gloo/gloo/CMakeFiles/gloo.dir/build.make:300: third_party/gloo/gloo/CMakeFiles/gloo.dir/common/linux.cc.o] Error 1
make[2]: *** Waiting for unfinished jobs....
/build/python-horovod/src/horovod-0.28.0/third_party/gloo/gloo/transport/tcp/device.cc: In function ‘gloo::transport::tcp::attr gloo::transport::tcp::CreateDeviceAttr(const attr&)’:
/build/python-horovod/src/horovod-0.28.0/third_party/gloo/gloo/transport/tcp/device.cc:151:39: error: aggregate ‘std::array<char, 64> hostname’ has incomplete type and cannot be defined
151 | std::array<char, HOST_NAME_MAX> hostname;
| ^~~~~~~~
make[2]: *** [third_party/gloo/gloo/CMakeFiles/gloo.dir/build.make:524: third_party/gloo/gloo/CMakeFiles/gloo.dir/transport/tcp/device.cc.o] Error 1
make[2]: Leaving directory '/build/python-horovod/src/horovod-0.28.0/build/temp.linux-x86_64-cpython-311/RelWithDebInfo'
make[1]: *** [CMakeFiles/Makefile2:524: third_party/compatible17_gloo/gloo/CMakeFiles/compatible17_gloo.dir/all] Error 2
make[1]: *** Waiting for unfinished jobs....
```
According to gloo upstream issue https://github.com/facebookincubator/gloo/issues/332, this is fixed by https://github.com/facebookincubator/gloo/commit/4a5e339b764261d20fc409071dc7a8b8989aa195. We only need to update submodule `third_party/gloo` to at least this commit. I could confirm that this works by:
```bash
cd third_party/gloo
git pull https://github.com/facebookincubator/gloo.git
```
I update the submodule, and then I could build it.
| closed | 2023-05-12T15:15:13Z | 2023-05-24T16:52:41Z | https://github.com/horovod/horovod/issues/3923 | [
"bug"
] | hubutui | 3 |
graphql-python/graphene-sqlalchemy | sqlalchemy | 39 | Fragments doesn't seem to work | Graphene does not seem to resolve the other object Type using fragments in the query.
Here is my schema:
```python
class User(db.Model):
__tablename__ = 'tbl_users'
id = Column(String(40), primary_key=True)
username = Column(String(64), index=True, unique=True)
email = Column(String(64), index=True, unique=True)
claims = None
class User(SQLAlchemyObjectType):
class Meta:
interfaces = (relay.Node,)
model = UserModel
class UserMeta(SQLAlchemyObjectType):
class Meta:
interfaces = (relay.Node,)
model = UserModel
claims = json.JSONString()
class Queries(graphene.ObjectType):
me = graphene.Field(UserInterface)
def resolve_me(_, args, context, info):
query = User.get_query(context)
user = query.get(1)
user.claims = {'hello': 'world'}
return user
schema = graphene.Schema(
query=Queries,
types=[UserMeta, User]
)
```
And here is my query:
```gql
{
me {
...userData
...userMeta
}
}
fragment userMeta on UserMeta {
claims
}
fragment userData on User {
username
email
}
```
It will return:
```json
{
"data": {
"me": {
"createdAt": "2017-03-31T05:29:32+00:00",
"email": "email@nowhere.space"
}
}
}
```
However, if I move `claims` to the `User` object type, it works as expected.
Also, even if I only use the `UserMeta` fragment in the query, it still doesn't resolve it.
Update:
Im sorry, my way was wrong. | closed | 2017-04-03T06:07:49Z | 2023-02-26T00:53:17Z | https://github.com/graphql-python/graphene-sqlalchemy/issues/39 | [] | richmondwang | 1 |
deepfakes/faceswap | machine-learning | 787 | Microsoft SmartScrren detected a Malware in your exe file | **Describe the bug**
Microsoft Antivirus find a malware in your files. Virustotal find via heuristic a malware, too.
URL: https://www.virustotal.com/gui/file/066ad1b0c174372e3fdc0869a293e0bfb23b23e7cce2b114addc35225825505b/detection
**To Reproduce**
Steps to reproduce the behavior:
Open the installer on windows 10 with all updates (09.07.2019)
**Expected behavior**
No malware warning.
**Desktop (please complete the following information):**
Windows 10 1903 Build: 18362.175
**Smartphone (please complete the following information):**
Not tested on smartphones.
| closed | 2019-07-09T17:12:36Z | 2019-07-09T17:13:59Z | https://github.com/deepfakes/faceswap/issues/787 | [] | gittyhub2018 | 1 |
junyanz/pytorch-CycleGAN-and-pix2pix | pytorch | 1,517 | test came out cropped | Hello. I am a student who has just started studying deeplearning and there are many things I don't know.
I trained the pix2pix model with options `—model pix2pix --direction AtoB --save_epoch_freq 100 --n_epochs 400 --n_epochs_decay 400 --preprocess scale_width_and_crop --load_size 768 --crop_size 768`
and tried to test with options `—model pix2pix --direction AtoB --preprocess scale_width_and_crop --load_size 1024 --crop_size 512`
but the result came out cropped. (image size of test is 1488*837)
I want to know why this happens even if test and train have the same options.
and I also tried with scale_witdth option but I got this error:
`RuntimeError: Sizes of tensors must match except in dimension 1. Expected size 13 but got size 12 for tensor number 1 in the list.`
I want to know why these errors occur and how to solve! Thanks. | open | 2022-12-08T01:44:31Z | 2022-12-15T11:10:46Z | https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix/issues/1517 | [] | GyuRiiii | 1 |
AUTOMATIC1111/stable-diffusion-webui | pytorch | 16,901 | [Feature Request]: Random Num Generator Filename pattern | ### Is there an existing issue for this?
- [x] I have searched the existing issues and checked the recent builds/commits
### What would your feature do ?
A way to add [random number generator source](https://github.com/AUTOMATIC1111/stable-diffusion-webui/wiki/Custom-Images-Filename-Name-and-Subdirectory) (randn_source) to the filename. So it either adds GPU, CPU or NV to the image filename.
### Proposed workflow
1. Go to ....
2. Press ....
3. ...
### Additional information
_No response_ | open | 2025-03-19T01:34:46Z | 2025-03-19T01:35:38Z | https://github.com/AUTOMATIC1111/stable-diffusion-webui/issues/16901 | [
"enhancement"
] | JingJang | 0 |
autogluon/autogluon | computer-vision | 4,843 | [timeseries] reference to python 3.8 should be removed/updated | Autogluon no longer supports Python 3.8. The minimum version is now 3.9, so the reference to Python 3.8 below should be removed or updated:
https://github.com/autogluon/autogluon/blob/f1bd5f42b2da0099c8d7319f38f811127446d9af/timeseries/tests/unittests/utils/test_features.py#L50
cc: @shchur @canerturkmen | closed | 2025-01-27T10:09:59Z | 2025-01-28T14:56:17Z | https://github.com/autogluon/autogluon/issues/4843 | [
"module: timeseries"
] | celestinoxp | 1 |
junyanz/pytorch-CycleGAN-and-pix2pix | deep-learning | 1,229 | How to set options to train pix2pix with 128x128 data? | Of course I have searched and found these:
#406
#578
However, I didn't get any useful infomation, in #406, there is only one reply by @junyanz:
> You can resize your images to 200x200 by setting --display_winsize 200 during test time.
And @haoxiangchnchn said :
> Thank you for your reply, I have solved this problem and wish you a smooth job!
It's really weird, cause the reply said "during test time", what I want to do is "during traing time", and display_winsize just change display size, cannot change model or input data.
While I set crop_size=128, I got this error:
> ValueError: Expected more than 1 value per channel when training, got input size [1, 512, 1, 1]
So anyone can help me? Thx. | closed | 2021-01-21T08:41:28Z | 2022-04-17T01:22:25Z | https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix/issues/1229 | [] | fire717 | 6 |
cookiecutter/cookiecutter-django | django | 4,865 | ci.yml for running pytest is building docs image unnecessarily (run time 4:30m instead of 3:00m) | ## What happened?
With docker setup enabled ci.yml is building all images (including the docs) during the Github Actions workflow. But only the images related to the django service are needed to run pytest in the next step. This causes the CI workflow to last 4m 30s instead of just below 3m (incl. linter).
## What should've happened instead?
Only the images related to running the django service and therefore the pytest command should be build.
Please see my pull request for the minor required change: #4863 | closed | 2024-02-15T22:00:06Z | 2024-02-16T09:55:25Z | https://github.com/cookiecutter/cookiecutter-django/issues/4865 | [
"bug"
] | mtmpaulwulff | 0 |
explosion/spaCy | nlp | 13,547 | ImportError: cannot import name symnols | <!-- NOTE: For questions or install related issues, please open a Discussion instead. -->
## How to reproduce the behaviour
trying to import `Corpus` or `Example` as shown here: https://spacy.io/usage/training#custom-code-readers-batchers
```python
from spacy.training import Corpus
```
produces import error:
```txt
---------------------------------------------------------------------------
ImportError
Traceback (most recent call last)
Cell In[3], line 1
----> 1 from spacy.training import Corpus
File ~/dev/project/.venv/lib/python3.10/site-packages/spacy/__init__.py:13
10 # These are imported as part of the API
11 from thinc.api import Config, prefer_gpu, require_cpu, require_gpu # noqa: F401
---> 13 from . import pipeline # noqa: F401
14 from . import util
15 from .about import __version__ # noqa: F401
File ~/dev/project/.venv/lib/python3.10/site-packages/spacy/pipeline/__init__.py:1
----> 1 from .attributeruler import AttributeRuler
2 from .dep_parser import DependencyParser
3 from .edit_tree_lemmatizer import EditTreeLemmatizer
File ~/dev/project/.venv/lib/python3.10/site-packages/spacy/pipeline/attributeruler.py:8
6 from .. import util
7 from ..errors import Errors
----> 8 from ..language import Language
9 from ..matcher import Matcher
10 from ..scorer import Scorer
File ~/dev/project/.venv/lib/python3.10/site-packages/spacy/language.py:43
41 from .lang.tokenizer_exceptions import BASE_EXCEPTIONS, URL_MATCH
42 from .lookups import load_lookups
---> 43 from .pipe_analysis import analyze_pipes, print_pipe_analysis, validate_attrs
44 from .schemas import (
45 ConfigSchema,
46 ConfigSchemaInit,
(...)
49 validate_init_settings,
50 )
51 from .scorer import Scorer
File ~/dev/project/.venv/lib/python3.10/site-packages/spacy/pipe_analysis.py:6
3 from wasabi import msg
5 from .errors import Errors
----> 6 from .tokens import Doc, Span, Token
7 from .util import dot_to_dict
9 if TYPE_CHECKING:
10 # This lets us add type hints for mypy etc. without causing circular imports
File ~/dev/project/.venv/lib/python3.10/site-packages/spacy/tokens/__init__.py:1
----> 1 from ._serialize import DocBin
2 from .doc import Doc
3 from .morphanalysis import MorphAnalysis
File ~/dev/project/.venv/lib/python3.10/site-packages/spacy/tokens/_serialize.py:14
12 from ..errors import Errors
13 from ..util import SimpleFrozenList, ensure_path
---> 14 from ..vocab import Vocab
15 from ._dict_proxies import SpanGroups
16 from .doc import DOCBIN_ALL_ATTRS as ALL_ATTRS
File ~/dev/project/.venv/lib/python3.10/site-packages/spacy/vocab.pyx:1, in init spacy.vocab()
File ~/dev/project/.venv/lib/python3.10/site-packages/spacy/morphology.pyx:9, in init spacy.morphology()
ImportError: cannot import name symbols
```
I'm not sure why there are dots being used to navigate the submodules instead of the full module path.
i.e. `from . import pipeline` opposed to `from spacy import pipeline`
## Your Environment
<!-- Include details of your environment. You can also type `python -m spacy info --markdown` and copy-paste the result here.-->
- **spaCy version:** 3.7.5
- **Platform:** macOS-14.5-arm64-arm-64bit
- **Python version:** 3.10.13
- **Environment Information**: env created with pypoetry
| closed | 2024-06-25T12:38:48Z | 2024-07-26T00:02:37Z | https://github.com/explosion/spaCy/issues/13547 | [] | fschlz | 2 |
joeyespo/grip | flask | 173 | Display size mismatch | ## Issue:
- Display size is mismatched, images below for verification. It looks like the issue occurs when using
`#` or `##`, causing it to be left shifted compared to the rest of the document, which then removes the side lines and causes the overflow.
## Potential fix:
Add a css over ride for a right shift on headers, or wrap everything inside of a div to ensure it's consistent, and not overflowing to the left.
## Personal solution
So I reinstalled multiple times. It resulted in the error on 3.5.1 the first install, error on 2.7.11 on the second install, but then fixed itself on 3.5.1 on the third install. Not sure if it's a result of all the reinstalls, but end result is the bug disappears. Opening this issue in case others run into the same issue, though you can close if I'm incorrect.
## OS:
- OSX 10.11.3 El Capitan
## Browsers checked:
- Firefox (45.0.2)
- Chrome (49.0.2623.112 (64-bit))
## Python versions checked:
- 2.7.11
- 3.5.1
## Image verification - issue:
Python 2.7.11, Chrome example 1:

Python 2.7.11, Firefox example 1:

Python 2.7.11, Firefox example 2:

## Image verification - fixed
Python 3.5.1, Chrome: fixed after 3rd reinstall

Python 3.5.1, Firefox: fixed after 3rd reinstall

| closed | 2016-04-13T04:11:35Z | 2016-04-13T04:43:00Z | https://github.com/joeyespo/grip/issues/173 | [
"already-implemented"
] | JasonYao | 3 |
tatsu-lab/stanford_alpaca | deep-learning | 11 | We are thinking about why this small model can store enough world knowledge | Hi, we find your work in home page.
https://crfm.stanford.edu/2023/03/13/alpaca.html
This work inspires us how to adjust large language models in a good way.
Now, We are thinking about why this small model can store enough world knowledge.
Best. | closed | 2023-03-14T06:32:42Z | 2023-03-14T07:07:18Z | https://github.com/tatsu-lab/stanford_alpaca/issues/11 | [] | RedBlack888 | 1 |
pytest-dev/pytest-django | pytest | 1,019 | Test works on local machine but in Docker I get "no module named 'test' " | I have a pytest.ini and a test_something.py file in the same dir as my manage.py. If I run 'pytest' directly, it works. If I run the whole thing with docker-compose, get an interactive console on the docker container and run 'pytest' there, I get:
> root@06070aaebbbd:/code/coco# pytest
> Traceback (most recent call last):
> File "/usr/local/lib/python3.7/site-packages/pytest_django/plugin.py", line 179, in _handle_import_error
> yield
> File "/usr/local/lib/python3.7/site-packages/pytest_django/plugin.py", line 351, in pytest_load_initial_conftests
> dj_settings.DATABASES
> File "/usr/local/lib/python3.7/site-packages/django/conf/__init__.py", line 82, in __getattr__
> self._setup(name)
> File "/usr/local/lib/python3.7/site-packages/django/conf/__init__.py", line 69, in _setup
> self._wrapped = Settings(settings_module)
> File "/usr/local/lib/python3.7/site-packages/django/conf/__init__.py", line 170, in __init__
> mod = importlib.import_module(self.SETTINGS_MODULE)
> File "/usr/local/lib/python3.7/importlib/__init__.py", line 127, in import_module
> return _bootstrap._gcd_import(name[level:], package, level)
> File "<frozen importlib._bootstrap>", line 1006, in _gcd_import
> File "<frozen importlib._bootstrap>", line 983, in _find_and_load
> File "<frozen importlib._bootstrap>", line 953, in _find_and_load_unlocked
> File "<frozen importlib._bootstrap>", line 219, in _call_with_frames_removed
> File "<frozen importlib._bootstrap>", line 1006, in _gcd_import
> File "<frozen importlib._bootstrap>", line 983, in _find_and_load
> File "<frozen importlib._bootstrap>", line 965, in _find_and_load_unlocked
> ModuleNotFoundError: No module named 'test'
>
> During handling of the above exception, another exception occurred:
>
> Traceback (most recent call last):
> File "/usr/local/bin/pytest", line 8, in <module>
> sys.exit(console_main())
> File "/usr/local/lib/python3.7/site-packages/_pytest/config/__init__.py", line 187, in console_main
> code = main()
> File "/usr/local/lib/python3.7/site-packages/_pytest/config/__init__.py", line 145, in main
> config = _prepareconfig(args, plugins)
> File "/usr/local/lib/python3.7/site-packages/_pytest/config/__init__.py", line 325, in _prepareconfig
> pluginmanager=pluginmanager, args=args
> File "/usr/local/lib/python3.7/site-packages/pluggy/_hooks.py", line 265, in __call__
> return self._hookexec(self.name, self.get_hookimpls(), kwargs, firstresult)
> File "/usr/local/lib/python3.7/site-packages/pluggy/_manager.py", line 80, in _hookexec
> return self._inner_hookexec(hook_name, methods, kwargs, firstresult)
> File "/usr/local/lib/python3.7/site-packages/pluggy/_manager.py", line 80, in _hookexec
> return self._inner_hookexec(hook_name, methods, kwargs, firstresult)
> File "/usr/local/lib/python3.7/site-packages/pluggy/_callers.py", line 55, in _multicall
> gen.send(outcome)
> File "/usr/local/lib/python3.7/site-packages/_pytest/helpconfig.py", line 102, in pytest_cmdline_parse
> config: Config = outcome.get_result()
> File "/usr/local/lib/python3.7/site-packages/pluggy/_result.py", line 60, in get_result
> raise ex[1].with_traceback(ex[2])
> File "/usr/local/lib/python3.7/site-packages/pluggy/_callers.py", line 39, in _multicall
> res = hook_impl.function(*args)
> File "/usr/local/lib/python3.7/site-packages/_pytest/config/__init__.py", line 1016, in pytest_cmdline_parse
> self.parse(args)
> File "/usr/local/lib/python3.7/site-packages/_pytest/config/__init__.py", line 1304, in parse
> self._preparse(args, addopts=addopts)
> File "/usr/local/lib/python3.7/site-packages/_pytest/config/__init__.py", line 1207, in _preparse
> early_config=self, args=args, parser=self._parser
> File "/usr/local/lib/python3.7/site-packages/pluggy/_hooks.py", line 265, in __call__
> return self._hookexec(self.name, self.get_hookimpls(), kwargs, firstresult)
> File "/usr/local/lib/python3.7/site-packages/pluggy/_manager.py", line 80, in _hookexec
> return self._inner_hookexec(hook_name, methods, kwargs, firstresult)
> File "/usr/local/lib/python3.7/site-packages/pluggy/_callers.py", line 60, in _multicall
> return outcome.get_result()
> File "/usr/local/lib/python3.7/site-packages/pluggy/_result.py", line 60, in get_result
> raise ex[1].with_traceback(ex[2])
> File "/usr/local/lib/python3.7/site-packages/pluggy/_callers.py", line 39, in _multicall
> res = hook_impl.function(*args)
> File "/usr/local/lib/python3.7/site-packages/pytest_django/plugin.py", line 351, in pytest_load_initial_conftests
> dj_settings.DATABASES
> File "/usr/local/lib/python3.7/contextlib.py", line 130, in __exit__
> self.gen.throw(type, value, traceback)
> File "/usr/local/lib/python3.7/site-packages/pytest_django/plugin.py", line 183, in _handle_import_error
> raise ImportError(msg)
> ImportError: No module named 'test'
Any idea what might be the problem here? | open | 2022-07-07T15:29:14Z | 2023-05-02T11:24:07Z | https://github.com/pytest-dev/pytest-django/issues/1019 | [] | NiklasRz | 4 |
nvbn/thefuck | python | 957 | `&&` or `|` support | Feature request:
First off let me say I <3 thefuck. use it every day. props yo.
ok so feat req: One of my favorite uses of `fuck` is to push a new git branch. Would be nice to either `&&` or `|` stuff to the fuck so I could use it like this.

| open | 2019-09-24T04:48:08Z | 2019-09-24T14:05:37Z | https://github.com/nvbn/thefuck/issues/957 | [] | benmonro | 1 |
aleju/imgaug | deep-learning | 291 | [Documentation Question] Upsizing segmentation masks | I'm curious how a type uint segmentation mask is able to be resized larger. The only method that makes sense to me would be KNN, but the docs say, "Note that segmentation maps are handled internally as heatmaps (one per class) and as such can be resized using cubic interpolation."
But wouldn't cubic interpolation fill intermediate pixels with float values instead of only using the integer values present in the original mask? | closed | 2019-03-27T22:51:47Z | 2019-03-29T19:52:09Z | https://github.com/aleju/imgaug/issues/291 | [] | austinmw | 2 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.