Commit
•
35c4f9e
1
Parent(s):
d1760e8
all files added
Browse files- Kidney-Disease-Classifcation/.dvc/.gitignore +3 -0
- Kidney-Disease-Classifcation/.dvc/config +0 -0
- Kidney-Disease-Classifcation/.dvcignore +3 -0
- Kidney-Disease-Classifcation/.gitattributes +2 -0
- Kidney-Disease-Classifcation/.github/workflows/.gitkeep +0 -0
- Kidney-Disease-Classifcation/.gitignore +161 -0
- Kidney-Disease-Classifcation/.pre-commit-config.yaml +10 -0
- Kidney-Disease-Classifcation/README.md +70 -0
- Kidney-Disease-Classifcation/app.py +50 -0
- Kidney-Disease-Classifcation/config/config.yaml +19 -0
- Kidney-Disease-Classifcation/dvc.lock +112 -0
- Kidney-Disease-Classifcation/dvc.yaml +50 -0
- Kidney-Disease-Classifcation/main.py +62 -0
- Kidney-Disease-Classifcation/model/model.h5 +3 -0
- Kidney-Disease-Classifcation/params.yaml +6 -0
- Kidney-Disease-Classifcation/requirements.txt +18 -0
- Kidney-Disease-Classifcation/research/01_data_ingestion.ipynb +252 -0
- Kidney-Disease-Classifcation/research/02_prepare_base_model.ipynb +281 -0
- Kidney-Disease-Classifcation/research/03_model_training.ipynb +341 -0
- Kidney-Disease-Classifcation/research/04_model_evaluation_with_mlflow.ipynb +346 -0
- Kidney-Disease-Classifcation/research/trials.ipynb +91 -0
- Kidney-Disease-Classifcation/scores.json +4 -0
- Kidney-Disease-Classifcation/setup.py +27 -0
- Kidney-Disease-Classifcation/src/kidney_classification/__init__.py +18 -0
- Kidney-Disease-Classifcation/src/kidney_classification/components/__init__.py +0 -0
- Kidney-Disease-Classifcation/src/kidney_classification/components/data_ingestion.py +45 -0
- Kidney-Disease-Classifcation/src/kidney_classification/components/model_evaluation_mlflow.py +59 -0
- Kidney-Disease-Classifcation/src/kidney_classification/components/model_training.py +49 -0
- Kidney-Disease-Classifcation/src/kidney_classification/components/prepare_base_model.py +46 -0
- Kidney-Disease-Classifcation/src/kidney_classification/config/__init__.py +0 -0
- Kidney-Disease-Classifcation/src/kidney_classification/config/configuration.py +83 -0
- Kidney-Disease-Classifcation/src/kidney_classification/constants/__init__.py +4 -0
- Kidney-Disease-Classifcation/src/kidney_classification/entity/__init__.py +0 -0
- Kidney-Disease-Classifcation/src/kidney_classification/entity/config_entity.py +42 -0
- Kidney-Disease-Classifcation/src/kidney_classification/pipeline/__init__.py +0 -0
- Kidney-Disease-Classifcation/src/kidney_classification/pipeline/prediction.py +31 -0
- Kidney-Disease-Classifcation/src/kidney_classification/pipeline/stage_01_data_ingestion.py +31 -0
- Kidney-Disease-Classifcation/src/kidney_classification/pipeline/stage_02_prepare_base_model.py +30 -0
- Kidney-Disease-Classifcation/src/kidney_classification/pipeline/stage_03_model_training.py +32 -0
- Kidney-Disease-Classifcation/src/kidney_classification/pipeline/stage_04_model_evaluation_with_mlflow.py +32 -0
- Kidney-Disease-Classifcation/src/kidney_classification/utils/__init__.py +0 -0
- Kidney-Disease-Classifcation/src/kidney_classification/utils/common.py +79 -0
- Kidney-Disease-Classifcation/templates.py +44 -0
- Kidney-Disease-Classifcation/templates/dagshub-kidney_disease_classification.png +0 -0
- Kidney-Disease-Classifcation/templates/index.html +353 -0
- Kidney-Disease-Classifcation/templates/kidney_ctscan.png +0 -0
- Kidney-Disease-Classifcation/templates/mlflow-kidney_disease_classification.png +0 -0
Kidney-Disease-Classifcation/.dvc/.gitignore
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
/config.local
|
| 2 |
+
/tmp
|
| 3 |
+
/cache
|
Kidney-Disease-Classifcation/.dvc/config
ADDED
|
File without changes
|
Kidney-Disease-Classifcation/.dvcignore
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Add patterns of files dvc should ignore, which could improve
|
| 2 |
+
# the performance. Learn more at
|
| 3 |
+
# https://dvc.org/doc/user-guide/dvcignore
|
Kidney-Disease-Classifcation/.gitattributes
ADDED
|
@@ -0,0 +1,2 @@
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Auto detect text files and perform LF normalization
|
| 2 |
+
* text=auto
|
Kidney-Disease-Classifcation/.github/workflows/.gitkeep
ADDED
|
File without changes
|
Kidney-Disease-Classifcation/.gitignore
ADDED
|
@@ -0,0 +1,161 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Byte-compiled / optimized / DLL files
|
| 2 |
+
__pycache__/
|
| 3 |
+
*.py[cod]
|
| 4 |
+
*$py.class
|
| 5 |
+
|
| 6 |
+
# C extensions
|
| 7 |
+
*.so
|
| 8 |
+
|
| 9 |
+
# Distribution / packaging
|
| 10 |
+
.Python
|
| 11 |
+
build/
|
| 12 |
+
develop-eggs/
|
| 13 |
+
dist/
|
| 14 |
+
downloads/
|
| 15 |
+
eggs/
|
| 16 |
+
.eggs/
|
| 17 |
+
lib/
|
| 18 |
+
lib64/
|
| 19 |
+
parts/
|
| 20 |
+
sdist/
|
| 21 |
+
var/
|
| 22 |
+
wheels/
|
| 23 |
+
share/python-wheels/
|
| 24 |
+
*.egg-info/
|
| 25 |
+
.installed.cfg
|
| 26 |
+
*.egg
|
| 27 |
+
MANIFEST
|
| 28 |
+
|
| 29 |
+
# PyInstaller
|
| 30 |
+
# Usually these files are written by a python script from a template
|
| 31 |
+
# before PyInstaller builds the exe, so as to inject date/other infos into it.
|
| 32 |
+
*.manifest
|
| 33 |
+
*.spec
|
| 34 |
+
|
| 35 |
+
# Installer logs
|
| 36 |
+
pip-log.txt
|
| 37 |
+
pip-delete-this-directory.txt
|
| 38 |
+
|
| 39 |
+
# Unit test / coverage reports
|
| 40 |
+
htmlcov/
|
| 41 |
+
.tox/
|
| 42 |
+
.nox/
|
| 43 |
+
.coverage
|
| 44 |
+
.coverage.*
|
| 45 |
+
.cache
|
| 46 |
+
nosetests.xml
|
| 47 |
+
coverage.xml
|
| 48 |
+
*.cover
|
| 49 |
+
*.py,cover
|
| 50 |
+
.hypothesis/
|
| 51 |
+
.pytest_cache/
|
| 52 |
+
cover/
|
| 53 |
+
|
| 54 |
+
# Translations
|
| 55 |
+
*.mo
|
| 56 |
+
*.pot
|
| 57 |
+
|
| 58 |
+
# Django stuff:
|
| 59 |
+
*.log
|
| 60 |
+
local_settings.py
|
| 61 |
+
db.sqlite3
|
| 62 |
+
db.sqlite3-journal
|
| 63 |
+
|
| 64 |
+
# Flask stuff:
|
| 65 |
+
instance/
|
| 66 |
+
.webassets-cache
|
| 67 |
+
|
| 68 |
+
# Scrapy stuff:
|
| 69 |
+
.scrapy
|
| 70 |
+
|
| 71 |
+
# Sphinx documentation
|
| 72 |
+
docs/_build/
|
| 73 |
+
|
| 74 |
+
# PyBuilder
|
| 75 |
+
.pybuilder/
|
| 76 |
+
target/
|
| 77 |
+
|
| 78 |
+
# Jupyter Notebook
|
| 79 |
+
.ipynb_checkpoints
|
| 80 |
+
|
| 81 |
+
# IPython
|
| 82 |
+
profile_default/
|
| 83 |
+
ipython_config.py
|
| 84 |
+
|
| 85 |
+
# pyenv
|
| 86 |
+
# For a library or package, you might want to ignore these files since the code is
|
| 87 |
+
# intended to run in multiple environments; otherwise, check them in:
|
| 88 |
+
# .python-version
|
| 89 |
+
|
| 90 |
+
# pipenv
|
| 91 |
+
# According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
|
| 92 |
+
# However, in case of collaboration, if having platform-specific dependencies or dependencies
|
| 93 |
+
# having no cross-platform support, pipenv may install dependencies that don't work, or not
|
| 94 |
+
# install all needed dependencies.
|
| 95 |
+
#Pipfile.lock
|
| 96 |
+
|
| 97 |
+
# poetry
|
| 98 |
+
# Similar to Pipfile.lock, it is generally recommended to include poetry.lock in version control.
|
| 99 |
+
# This is especially recommended for binary packages to ensure reproducibility, and is more
|
| 100 |
+
# commonly ignored for libraries.
|
| 101 |
+
# https://python-poetry.org/docs/basic-usage/#commit-your-poetrylock-file-to-version-control
|
| 102 |
+
#poetry.lock
|
| 103 |
+
|
| 104 |
+
# PEP 582; used by e.g. github.com/David-OConnor/pyflow
|
| 105 |
+
__pypackages__/
|
| 106 |
+
|
| 107 |
+
# Celery stuff
|
| 108 |
+
celerybeat-schedule
|
| 109 |
+
celerybeat.pid
|
| 110 |
+
|
| 111 |
+
# SageMath parsed files
|
| 112 |
+
*.sage.py
|
| 113 |
+
|
| 114 |
+
# Environments
|
| 115 |
+
.env
|
| 116 |
+
.venv
|
| 117 |
+
env/
|
| 118 |
+
venv/
|
| 119 |
+
ENV/
|
| 120 |
+
env.bak/
|
| 121 |
+
venv.bak/
|
| 122 |
+
|
| 123 |
+
# Spyder project settings
|
| 124 |
+
.spyderproject
|
| 125 |
+
.spyproject
|
| 126 |
+
|
| 127 |
+
# Rope project settings
|
| 128 |
+
.ropeproject
|
| 129 |
+
|
| 130 |
+
# mkdocs documentation
|
| 131 |
+
/site
|
| 132 |
+
|
| 133 |
+
# mypy
|
| 134 |
+
.mypy_cache/
|
| 135 |
+
.dmypy.json
|
| 136 |
+
dmypy.json
|
| 137 |
+
|
| 138 |
+
# Pyre type checker
|
| 139 |
+
.pyre/
|
| 140 |
+
|
| 141 |
+
# pytype static type analyzer
|
| 142 |
+
.pytype/
|
| 143 |
+
|
| 144 |
+
# Cython debug symbols
|
| 145 |
+
cython_debug/
|
| 146 |
+
|
| 147 |
+
# Data Artifacts
|
| 148 |
+
artifacts/*
|
| 149 |
+
|
| 150 |
+
# PyCharm
|
| 151 |
+
# JetBrains specific template is maintainted in a separate JetBrains.gitignore that can
|
| 152 |
+
# be found at https://github.com/github/gitignore/blob/main/Global/JetBrains.gitignore
|
| 153 |
+
# and can be added to the global gitignore or merged into this file. For a more nuclear
|
| 154 |
+
# option (not recommended) you can uncomment the following to ignore the entire idea folder.
|
| 155 |
+
#.idea/
|
| 156 |
+
|
| 157 |
+
#mlflow runs
|
| 158 |
+
mlruns/
|
| 159 |
+
|
| 160 |
+
#input images
|
| 161 |
+
inputImage.jpg
|
Kidney-Disease-Classifcation/.pre-commit-config.yaml
ADDED
|
@@ -0,0 +1,10 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
repos:
|
| 2 |
+
- repo: https://github.com/pre-commit/pre-commit-hooks
|
| 3 |
+
rev: v4.5.0
|
| 4 |
+
hooks:
|
| 5 |
+
- id: trailing-whitespace
|
| 6 |
+
- id: end-of-file-fixer
|
| 7 |
+
- repo: https://github.com/psf/black
|
| 8 |
+
rev: 23.3.0
|
| 9 |
+
hooks:
|
| 10 |
+
- id: black
|
Kidney-Disease-Classifcation/README.md
ADDED
|
@@ -0,0 +1,70 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Kidney Tumor, Cyst, or Stone Classification
|
| 2 |
+

|
| 3 |
+
|
| 4 |
+
## Project Overview
|
| 5 |
+
The main goal of this project is to develop a reliable and efficient deep-learning model that can accurately classify kidney tumors and Stone from medical images.
|
| 6 |
+
|
| 7 |
+
## Introduction
|
| 8 |
+
Kidney Disease Classification is a project utilizing deep learning techniques to classify Kidney Tumor and Stone diseases from [medical images dataset](https://www.kaggle.com/datasets/nazmul0087/ct-kidney-dataset-normal-cyst-tumor-and-stone/). This project leverages the power of Deep Learning, Machine Learning Operations (MLOps) practices, Data Version Control (DVC). It integrates with DagsHub for collaboration and versioning.
|
| 9 |
+
|
| 10 |
+
## Dagshub Project Pipeline
|
| 11 |
+

|
| 12 |
+
|
| 13 |
+
## Mlflow Stats
|
| 14 |
+

|
| 15 |
+
|
| 16 |
+
## Importance of the Project
|
| 17 |
+
- **Enhancing Healthcare**: By providing accurate and quick disease classification, this project aims to improve patient care and diagnostic accuracy significantly.
|
| 18 |
+
- **Research and Development**: It serves as a tool for researchers to analyze medical images more effectively, paving the way for discoveries in the medical field.
|
| 19 |
+
- **Educational Value**: This project can be a learning platform for students and professionals interested in deep learning and medical image analysis.
|
| 20 |
+
|
| 21 |
+
## Technical Overview
|
| 22 |
+
- **Deep Learning Frameworks**: Utilizes popular frameworks like TensorFlow or PyTorch for building and training the classification models.
|
| 23 |
+
- **Data Version Control (DVC)**: Manages and versions large datasets and machine learning models, ensuring reproducibility and streamlined data pipelines.
|
| 24 |
+
- **Git Integration**: For source code management and version control, making the project easily maintainable and scalable.
|
| 25 |
+
- **MLOps Practices**: Incorporates best practices in machine learning operations to automate workflows, from data preparation to model deployment.
|
| 26 |
+
- **DagsHub Integration**: Facilitates collaboration, data and model versioning, experiment tracking, and more in a user-friendly platform.
|
| 27 |
+
|
| 28 |
+
## How to run?
|
| 29 |
+
### STEPS:
|
| 30 |
+
|
| 31 |
+
Clone the repository
|
| 32 |
+
|
| 33 |
+
```bash
|
| 34 |
+
https://github.com/krishnaik06/Kidney-Disease-Classification-Deep-Learning-Project
|
| 35 |
+
```
|
| 36 |
+
### STEP 01- Create a conda environment after opening the repository
|
| 37 |
+
|
| 38 |
+
```bash
|
| 39 |
+
conda create -n venv python=3.11 -y
|
| 40 |
+
```
|
| 41 |
+
|
| 42 |
+
```bash
|
| 43 |
+
conda activate venv
|
| 44 |
+
```
|
| 45 |
+
|
| 46 |
+
|
| 47 |
+
### STEP 02- install the requirements
|
| 48 |
+
```bash
|
| 49 |
+
pip install -r requirements.txt
|
| 50 |
+
```
|
| 51 |
+
|
| 52 |
+
```bash
|
| 53 |
+
# Finally run the following command
|
| 54 |
+
python app.py
|
| 55 |
+
```
|
| 56 |
+
|
| 57 |
+
Now,
|
| 58 |
+
```bash
|
| 59 |
+
open up your local host and port
|
| 60 |
+
```
|
| 61 |
+
|
| 62 |
+
## To Run the Pipeline
|
| 63 |
+
```bash
|
| 64 |
+
dvc repro
|
| 65 |
+
```
|
| 66 |
+
---
|
| 67 |
+
|
| 68 |
+
This project is still in development, and we welcome contributions of all kinds: from model development and data processing to documentation and bug fixes.
|
| 69 |
+
|
| 70 |
+
**Join me in this exciting journey to revolutionize the field of medical image classification with AI!**
|
Kidney-Disease-Classifcation/app.py
ADDED
|
@@ -0,0 +1,50 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import sys
|
| 2 |
+
|
| 3 |
+
sys.path.append("./src")
|
| 4 |
+
|
| 5 |
+
from kidney_classification.pipeline.prediction import PredictionPipeline
|
| 6 |
+
from kidney_classification.utils.common import decodeImage
|
| 7 |
+
from flask_cors import CORS, cross_origin
|
| 8 |
+
import os
|
| 9 |
+
from flask import Flask, request, jsonify, render_template
|
| 10 |
+
|
| 11 |
+
|
| 12 |
+
os.putenv("LANG", "en_US.UTF-8")
|
| 13 |
+
os.putenv("LC_ALL", "en_US.UTF-8")
|
| 14 |
+
|
| 15 |
+
app = Flask(__name__)
|
| 16 |
+
CORS(app)
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
class ClientApp:
|
| 20 |
+
def __init__(self):
|
| 21 |
+
self.filename = "inputImage.jpg"
|
| 22 |
+
self.classifier = PredictionPipeline(self.filename)
|
| 23 |
+
|
| 24 |
+
|
| 25 |
+
@app.route("/", methods=["GET"])
|
| 26 |
+
@cross_origin()
|
| 27 |
+
def home():
|
| 28 |
+
return render_template("index.html")
|
| 29 |
+
|
| 30 |
+
|
| 31 |
+
@app.route("/train", methods=["GET", "POST"])
|
| 32 |
+
@cross_origin()
|
| 33 |
+
def trainRoute():
|
| 34 |
+
os.system("dvc repro")
|
| 35 |
+
return "Training done successfully!"
|
| 36 |
+
|
| 37 |
+
|
| 38 |
+
@app.route("/predict", methods=["POST"])
|
| 39 |
+
@cross_origin()
|
| 40 |
+
def predictRoute():
|
| 41 |
+
image = request.json["image"]
|
| 42 |
+
decodeImage(image, clApp.filename)
|
| 43 |
+
result = clApp.classifier.predict()
|
| 44 |
+
return jsonify(result)
|
| 45 |
+
|
| 46 |
+
|
| 47 |
+
if __name__ == "__main__":
|
| 48 |
+
clApp = ClientApp()
|
| 49 |
+
|
| 50 |
+
app.run(host="0.0.0.0", port=8080) # for AWS
|
Kidney-Disease-Classifcation/config/config.yaml
ADDED
|
@@ -0,0 +1,19 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
artifacts_root: artifacts
|
| 2 |
+
|
| 3 |
+
|
| 4 |
+
data_ingestion:
|
| 5 |
+
root_dir: artifacts/data_ingestion
|
| 6 |
+
source_URL: https://drive.google.com/file/d/1BBxDX8JqV-WvNyOzYbqfrk3THFLBpEWF/view?usp=sharing
|
| 7 |
+
local_data_file: artifacts/data_ingestion/data.zip
|
| 8 |
+
unzip_dir: artifacts/data_ingestion
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
prepare_base_model:
|
| 12 |
+
root_dir: artifacts/prepare_base_model
|
| 13 |
+
base_model_path: artifacts/prepare_base_model/base_model.h5
|
| 14 |
+
updated_base_model_path: artifacts/prepare_base_model/base_model_updated.keras
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
training:
|
| 18 |
+
root_dir: artifacts/training
|
| 19 |
+
trained_model_path: artifacts/training/model.h5
|
Kidney-Disease-Classifcation/dvc.lock
ADDED
|
@@ -0,0 +1,112 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
schema: '2.0'
|
| 2 |
+
stages:
|
| 3 |
+
data_ingestion:
|
| 4 |
+
cmd: python src/kidney_classification/pipeline/stage_01_data_ingestion.py
|
| 5 |
+
deps:
|
| 6 |
+
- path: config/config.yaml
|
| 7 |
+
hash: md5
|
| 8 |
+
md5: 23d61aa500e4da63569da56d61ddb49e
|
| 9 |
+
size: 568
|
| 10 |
+
- path: src/kidney_classification/pipeline/stage_01_data_ingestion.py
|
| 11 |
+
hash: md5
|
| 12 |
+
md5: 0496157b33ff2c7182935a33c605f461
|
| 13 |
+
size: 955
|
| 14 |
+
outs:
|
| 15 |
+
- path: artifacts/data_ingestion/CT-KIDNEY-DATASET-Normal-Cyst-Tumor-Stone
|
| 16 |
+
hash: md5
|
| 17 |
+
md5: 47292b3e13804acbce0f2b4ce55edc57.dir
|
| 18 |
+
size: 887719156
|
| 19 |
+
nfiles: 5511
|
| 20 |
+
prepare_base_model:
|
| 21 |
+
cmd: python src/kidney_classification/pipeline/stage_02_prepare_base_model.py
|
| 22 |
+
deps:
|
| 23 |
+
- path: config/config.yaml
|
| 24 |
+
hash: md5
|
| 25 |
+
md5: 23d61aa500e4da63569da56d61ddb49e
|
| 26 |
+
size: 568
|
| 27 |
+
- path: src/kidney_classification/pipeline/stage_02_prepare_base_model.py
|
| 28 |
+
hash: md5
|
| 29 |
+
md5: 27f16408dee8b42a215b576052e54bb2
|
| 30 |
+
size: 950
|
| 31 |
+
params:
|
| 32 |
+
params.yaml:
|
| 33 |
+
CLASSES: 4
|
| 34 |
+
IMAGE_SIZE:
|
| 35 |
+
- 150
|
| 36 |
+
- 150
|
| 37 |
+
- 3
|
| 38 |
+
INCLUDE_TOP: false
|
| 39 |
+
WEIGHTS: imagenet
|
| 40 |
+
outs:
|
| 41 |
+
- path: artifacts/prepare_base_model
|
| 42 |
+
hash: md5
|
| 43 |
+
md5: 2721c46e50849883acd55e5d4e3dcefb.dir
|
| 44 |
+
size: 60010269
|
| 45 |
+
nfiles: 1
|
| 46 |
+
training:
|
| 47 |
+
cmd: python src/kidney_classification/pipeline/stage_03_model_training.py
|
| 48 |
+
deps:
|
| 49 |
+
- path: artifacts/data_ingestion/CT-KIDNEY-DATASET-Normal-Cyst-Tumor-Stone
|
| 50 |
+
hash: md5
|
| 51 |
+
md5: ec42dfce2ae993cf49f6d499a389c93e.dir
|
| 52 |
+
size: 1661580918
|
| 53 |
+
nfiles: 12446
|
| 54 |
+
- path: artifacts/prepare_base_model
|
| 55 |
+
hash: md5
|
| 56 |
+
md5: a4f718a24c253b4e539f7ba2dc9d3442.dir
|
| 57 |
+
size: 59997688
|
| 58 |
+
nfiles: 1
|
| 59 |
+
- path: config/config.yaml
|
| 60 |
+
hash: md5
|
| 61 |
+
md5: bc47b5f88a0220822ff7921144b69204
|
| 62 |
+
size: 565
|
| 63 |
+
- path: src/kidney_classification/pipeline/stage_03_model_training.py
|
| 64 |
+
hash: md5
|
| 65 |
+
md5: cb8342c5c2f23c4d395f299924161231
|
| 66 |
+
size: 941
|
| 67 |
+
params:
|
| 68 |
+
params.yaml:
|
| 69 |
+
BATCH_SIZE: 32
|
| 70 |
+
EPOCHS: 15
|
| 71 |
+
IMAGE_SIZE:
|
| 72 |
+
- 150
|
| 73 |
+
- 150
|
| 74 |
+
- 3
|
| 75 |
+
outs:
|
| 76 |
+
- path: artifacts/training/model.h5
|
| 77 |
+
hash: md5
|
| 78 |
+
md5: 17969099556c165c584938f53a3fc085
|
| 79 |
+
size: 62136448
|
| 80 |
+
evaluation:
|
| 81 |
+
cmd: python src/kidney_classification/pipeline/stage_04_model_evaluation_with_mlflow.py
|
| 82 |
+
deps:
|
| 83 |
+
- path: artifacts/data_ingestion/CT-KIDNEY-DATASET-Normal-Cyst-Tumor-Stone
|
| 84 |
+
hash: md5
|
| 85 |
+
md5: ec42dfce2ae993cf49f6d499a389c93e.dir
|
| 86 |
+
size: 1661580918
|
| 87 |
+
nfiles: 12446
|
| 88 |
+
- path: artifacts/training/model.h5
|
| 89 |
+
hash: md5
|
| 90 |
+
md5: 17969099556c165c584938f53a3fc085
|
| 91 |
+
size: 62136448
|
| 92 |
+
- path: config/config.yaml
|
| 93 |
+
hash: md5
|
| 94 |
+
md5: bc47b5f88a0220822ff7921144b69204
|
| 95 |
+
size: 565
|
| 96 |
+
- path: src/kidney_classification/pipeline/stage_04_model_evaluation_with_mlflow.py
|
| 97 |
+
hash: md5
|
| 98 |
+
md5: 7c91c2fdf529e4dcf7dec2684eb3d212
|
| 99 |
+
size: 892
|
| 100 |
+
params:
|
| 101 |
+
params.yaml:
|
| 102 |
+
BATCH_SIZE: 32
|
| 103 |
+
EPOCHS: 15
|
| 104 |
+
IMAGE_SIZE:
|
| 105 |
+
- 150
|
| 106 |
+
- 150
|
| 107 |
+
- 3
|
| 108 |
+
outs:
|
| 109 |
+
- path: scores.json
|
| 110 |
+
hash: md5
|
| 111 |
+
md5: e80b69c44a4cf771b208a549d9e5ae30
|
| 112 |
+
size: 58
|
Kidney-Disease-Classifcation/dvc.yaml
ADDED
|
@@ -0,0 +1,50 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
stages:
|
| 2 |
+
data_ingestion:
|
| 3 |
+
cmd: python src/kidney_classification/pipeline/stage_01_data_ingestion.py
|
| 4 |
+
deps:
|
| 5 |
+
- src/kidney_classification/pipeline/stage_01_data_ingestion.py
|
| 6 |
+
- config/config.yaml
|
| 7 |
+
outs:
|
| 8 |
+
- artifacts/data_ingestion/CT-KIDNEY-DATASET-Normal-Cyst-Tumor-Stone
|
| 9 |
+
|
| 10 |
+
prepare_base_model:
|
| 11 |
+
cmd: python src/kidney_classification/pipeline/stage_02_prepare_base_model.py
|
| 12 |
+
deps:
|
| 13 |
+
- src/kidney_classification/pipeline/stage_02_prepare_base_model.py
|
| 14 |
+
- config/config.yaml
|
| 15 |
+
params:
|
| 16 |
+
- IMAGE_SIZE
|
| 17 |
+
- INCLUDE_TOP
|
| 18 |
+
- CLASSES
|
| 19 |
+
- WEIGHTS
|
| 20 |
+
outs:
|
| 21 |
+
- artifacts/prepare_base_model
|
| 22 |
+
|
| 23 |
+
training:
|
| 24 |
+
cmd: python src/kidney_classification/pipeline/stage_03_model_training.py
|
| 25 |
+
deps:
|
| 26 |
+
- src/kidney_classification/pipeline/stage_03_model_training.py
|
| 27 |
+
- config/config.yaml
|
| 28 |
+
- artifacts/data_ingestion/CT-KIDNEY-DATASET-Normal-Cyst-Tumor-Stone
|
| 29 |
+
- artifacts/prepare_base_model
|
| 30 |
+
params:
|
| 31 |
+
- IMAGE_SIZE
|
| 32 |
+
- BATCH_SIZE
|
| 33 |
+
- EPOCHS
|
| 34 |
+
outs:
|
| 35 |
+
- artifacts/training/model.h5
|
| 36 |
+
|
| 37 |
+
evaluation:
|
| 38 |
+
cmd: python src/kidney_classification/pipeline/stage_04_model_evaluation_with_mlflow.py
|
| 39 |
+
deps:
|
| 40 |
+
- src/kidney_classification/pipeline/stage_04_model_evaluation_with_mlflow.py
|
| 41 |
+
- config/config.yaml
|
| 42 |
+
- artifacts/data_ingestion/CT-KIDNEY-DATASET-Normal-Cyst-Tumor-Stone
|
| 43 |
+
- artifacts/training/model.h5
|
| 44 |
+
params:
|
| 45 |
+
- IMAGE_SIZE
|
| 46 |
+
- BATCH_SIZE
|
| 47 |
+
- EPOCHS
|
| 48 |
+
metrics:
|
| 49 |
+
- scores.json:
|
| 50 |
+
cache: false
|
Kidney-Disease-Classifcation/main.py
ADDED
|
@@ -0,0 +1,62 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import sys
|
| 2 |
+
|
| 3 |
+
sys.path.append("./src")
|
| 4 |
+
from kidney_classification.pipeline.stage_04_model_evaluation_with_mlflow import (
|
| 5 |
+
EvaluationPipeline,
|
| 6 |
+
)
|
| 7 |
+
from kidney_classification.pipeline.stage_03_model_training import ModelTrainingPipeline
|
| 8 |
+
from kidney_classification.pipeline.stage_02_prepare_base_model import (
|
| 9 |
+
PrepareBaseModelTrainingPipeline,
|
| 10 |
+
)
|
| 11 |
+
from kidney_classification.pipeline.stage_01_data_ingestion import (
|
| 12 |
+
DataIngestionTrainingPipeline,
|
| 13 |
+
)
|
| 14 |
+
from kidney_classification import logger
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
STAGE_NAME = "Data Ingestion"
|
| 18 |
+
|
| 19 |
+
try:
|
| 20 |
+
logger.info(f"-------------Running stage: {STAGE_NAME}-------------")
|
| 21 |
+
pipeline = DataIngestionTrainingPipeline()
|
| 22 |
+
pipeline.main()
|
| 23 |
+
logger.info(f"-------------Stage: {STAGE_NAME} completed-------------")
|
| 24 |
+
except Exception as e:
|
| 25 |
+
logger.exception(e)
|
| 26 |
+
raise e
|
| 27 |
+
|
| 28 |
+
|
| 29 |
+
STAGE_NAME = "Prepare base model"
|
| 30 |
+
|
| 31 |
+
try:
|
| 32 |
+
logger.info(f"-------------Running stage: {STAGE_NAME}-------------")
|
| 33 |
+
pipeline = PrepareBaseModelTrainingPipeline()
|
| 34 |
+
pipeline.main()
|
| 35 |
+
logger.info(f"-------------Stage: {STAGE_NAME} completed-------------")
|
| 36 |
+
except Exception as e:
|
| 37 |
+
logger.exception(e)
|
| 38 |
+
raise e
|
| 39 |
+
|
| 40 |
+
|
| 41 |
+
STAGE_NAME = "Training Model"
|
| 42 |
+
|
| 43 |
+
try:
|
| 44 |
+
logger.info(f"-------------Running stage: {STAGE_NAME}-------------")
|
| 45 |
+
pipeline = ModelTrainingPipeline()
|
| 46 |
+
pipeline.main()
|
| 47 |
+
logger.info(f"-------------Stage: {STAGE_NAME} completed-------------")
|
| 48 |
+
except Exception as e:
|
| 49 |
+
logger.exception(e)
|
| 50 |
+
raise e
|
| 51 |
+
|
| 52 |
+
|
| 53 |
+
STAGE_NAME = "Evaluation Stage"
|
| 54 |
+
|
| 55 |
+
try:
|
| 56 |
+
logger.info(f"-------------Running stage: {STAGE_NAME}-------------")
|
| 57 |
+
pipeline = EvaluationPipeline()
|
| 58 |
+
pipeline.main()
|
| 59 |
+
logger.info(f"-------------Stage: {STAGE_NAME} completed-------------")
|
| 60 |
+
except Exception as e:
|
| 61 |
+
logger.exception(e)
|
| 62 |
+
raise e
|
Kidney-Disease-Classifcation/model/model.h5
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:909a0b1a2101b57412a20ecda3513ab35e757cbf565875ea9c7803fdc3e8b6ab
|
| 3 |
+
size 62136504
|
Kidney-Disease-Classifcation/params.yaml
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
IMAGE_SIZE: [150, 150, 3]
|
| 2 |
+
BATCH_SIZE: 32
|
| 3 |
+
INCLUDE_TOP: False
|
| 4 |
+
EPOCHS: 15
|
| 5 |
+
CLASSES: 4
|
| 6 |
+
WEIGHTS: imagenet
|
Kidney-Disease-Classifcation/requirements.txt
ADDED
|
@@ -0,0 +1,18 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
tensorflow==2.15.0.post1
|
| 2 |
+
pandas==2.1.4
|
| 3 |
+
dvc==3.35.0
|
| 4 |
+
mlflow==2.9.2
|
| 5 |
+
notebook==7.0.6
|
| 6 |
+
numpy==1.26.2
|
| 7 |
+
matplotlib==3.8.2
|
| 8 |
+
seaborn==0.13.0
|
| 9 |
+
python-box==7.1.1
|
| 10 |
+
PyYAML==6.0.1
|
| 11 |
+
tqdm==4.66.1
|
| 12 |
+
ensure==1.0.4
|
| 13 |
+
joblib==1.3.2
|
| 14 |
+
types-PyYAML==6.0.12.12
|
| 15 |
+
scipy==1.11.4
|
| 16 |
+
Flask==3.0.0
|
| 17 |
+
Flask-Cors==4.0.0
|
| 18 |
+
gdown==4.7.1
|
Kidney-Disease-Classifcation/research/01_data_ingestion.ipynb
ADDED
|
@@ -0,0 +1,252 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "code",
|
| 5 |
+
"execution_count": 2,
|
| 6 |
+
"metadata": {},
|
| 7 |
+
"outputs": [],
|
| 8 |
+
"source": [
|
| 9 |
+
"import os"
|
| 10 |
+
]
|
| 11 |
+
},
|
| 12 |
+
{
|
| 13 |
+
"cell_type": "code",
|
| 14 |
+
"execution_count": 29,
|
| 15 |
+
"metadata": {},
|
| 16 |
+
"outputs": [
|
| 17 |
+
{
|
| 18 |
+
"data": {
|
| 19 |
+
"text/plain": [
|
| 20 |
+
"'/home/shrey/Desktop/Kidney-Disease-Classifcation/src'"
|
| 21 |
+
]
|
| 22 |
+
},
|
| 23 |
+
"execution_count": 29,
|
| 24 |
+
"metadata": {},
|
| 25 |
+
"output_type": "execute_result"
|
| 26 |
+
}
|
| 27 |
+
],
|
| 28 |
+
"source": [
|
| 29 |
+
"%pwd"
|
| 30 |
+
]
|
| 31 |
+
},
|
| 32 |
+
{
|
| 33 |
+
"cell_type": "code",
|
| 34 |
+
"execution_count": 30,
|
| 35 |
+
"metadata": {},
|
| 36 |
+
"outputs": [],
|
| 37 |
+
"source": [
|
| 38 |
+
"os.chdir('../')"
|
| 39 |
+
]
|
| 40 |
+
},
|
| 41 |
+
{
|
| 42 |
+
"cell_type": "code",
|
| 43 |
+
"execution_count": 31,
|
| 44 |
+
"metadata": {},
|
| 45 |
+
"outputs": [
|
| 46 |
+
{
|
| 47 |
+
"data": {
|
| 48 |
+
"text/plain": [
|
| 49 |
+
"'/home/shrey/Desktop/Kidney-Disease-Classifcation'"
|
| 50 |
+
]
|
| 51 |
+
},
|
| 52 |
+
"execution_count": 31,
|
| 53 |
+
"metadata": {},
|
| 54 |
+
"output_type": "execute_result"
|
| 55 |
+
}
|
| 56 |
+
],
|
| 57 |
+
"source": [
|
| 58 |
+
"%pwd"
|
| 59 |
+
]
|
| 60 |
+
},
|
| 61 |
+
{
|
| 62 |
+
"cell_type": "code",
|
| 63 |
+
"execution_count": 32,
|
| 64 |
+
"metadata": {},
|
| 65 |
+
"outputs": [],
|
| 66 |
+
"source": [
|
| 67 |
+
"from dataclasses import dataclass\n",
|
| 68 |
+
"from pathlib import Path\n",
|
| 69 |
+
"\n",
|
| 70 |
+
"\n",
|
| 71 |
+
"@dataclass(frozen=True)\n",
|
| 72 |
+
"class DataIngestionConfig:\n",
|
| 73 |
+
" root_dir: Path\n",
|
| 74 |
+
" source_URL: str\n",
|
| 75 |
+
" local_data_file: Path\n",
|
| 76 |
+
" unzip_dir: Path"
|
| 77 |
+
]
|
| 78 |
+
},
|
| 79 |
+
{
|
| 80 |
+
"cell_type": "code",
|
| 81 |
+
"execution_count": 33,
|
| 82 |
+
"metadata": {},
|
| 83 |
+
"outputs": [],
|
| 84 |
+
"source": [
|
| 85 |
+
"from kidney_classification.constants import *\n",
|
| 86 |
+
"from kidney_classification.utils.common import read_yaml, create_directories"
|
| 87 |
+
]
|
| 88 |
+
},
|
| 89 |
+
{
|
| 90 |
+
"cell_type": "code",
|
| 91 |
+
"execution_count": 34,
|
| 92 |
+
"metadata": {},
|
| 93 |
+
"outputs": [],
|
| 94 |
+
"source": [
|
| 95 |
+
"class ConfigurationManager:\n",
|
| 96 |
+
" def __init__(\n",
|
| 97 |
+
" self,\n",
|
| 98 |
+
" config_filepath=CONFIG_FILE_PATH,\n",
|
| 99 |
+
" params_filepath=PARAMS_FILE_PATH):\n",
|
| 100 |
+
"\n",
|
| 101 |
+
" self.config = read_yaml(config_filepath)\n",
|
| 102 |
+
" self.params = read_yaml(params_filepath)\n",
|
| 103 |
+
"\n",
|
| 104 |
+
" create_directories([self.config.artifacts_root])\n",
|
| 105 |
+
"\n",
|
| 106 |
+
" def get_data_ingestion_config(self) -> DataIngestionConfig:\n",
|
| 107 |
+
" config = self.config.data_ingestion\n",
|
| 108 |
+
"\n",
|
| 109 |
+
" create_directories([config.root_dir])\n",
|
| 110 |
+
"\n",
|
| 111 |
+
" data_ingestion_config = DataIngestionConfig(\n",
|
| 112 |
+
" root_dir=config.root_dir,\n",
|
| 113 |
+
" source_URL=config.source_URL,\n",
|
| 114 |
+
" local_data_file=config.local_data_file,\n",
|
| 115 |
+
" unzip_dir=config.unzip_dir\n",
|
| 116 |
+
" )\n",
|
| 117 |
+
"\n",
|
| 118 |
+
" return data_ingestion_config"
|
| 119 |
+
]
|
| 120 |
+
},
|
| 121 |
+
{
|
| 122 |
+
"cell_type": "code",
|
| 123 |
+
"execution_count": 35,
|
| 124 |
+
"metadata": {},
|
| 125 |
+
"outputs": [],
|
| 126 |
+
"source": [
|
| 127 |
+
"import os\n",
|
| 128 |
+
"import zipfile\n",
|
| 129 |
+
"import gdown\n",
|
| 130 |
+
"from kidney_classification import logger"
|
| 131 |
+
]
|
| 132 |
+
},
|
| 133 |
+
{
|
| 134 |
+
"cell_type": "code",
|
| 135 |
+
"execution_count": 36,
|
| 136 |
+
"metadata": {},
|
| 137 |
+
"outputs": [],
|
| 138 |
+
"source": [
|
| 139 |
+
"class DataIngestion:\n",
|
| 140 |
+
" def __init__(self, config: DataIngestionConfig):\n",
|
| 141 |
+
" self.config = config\n",
|
| 142 |
+
"\n",
|
| 143 |
+
" def download_file(self) -> str:\n",
|
| 144 |
+
" '''\n",
|
| 145 |
+
" Fetch data from the url\n",
|
| 146 |
+
" '''\n",
|
| 147 |
+
"\n",
|
| 148 |
+
" try:\n",
|
| 149 |
+
" dataset_url = self.config.source_URL\n",
|
| 150 |
+
" zip_download_dir = self.config.local_data_file\n",
|
| 151 |
+
" os.makedirs(\"artifacts/data_ingestion\", exist_ok=True)\n",
|
| 152 |
+
" logger.info(\n",
|
| 153 |
+
" f\"Downloading data from {dataset_url} into file {zip_download_dir}\")\n",
|
| 154 |
+
"\n",
|
| 155 |
+
" file_id = dataset_url.split(\"/\")[-2]\n",
|
| 156 |
+
" prefix = 'https://drive.google.com/uc?/export=download&id='\n",
|
| 157 |
+
" gdown.download(prefix+file_id, zip_download_dir)\n",
|
| 158 |
+
"\n",
|
| 159 |
+
" logger.info(\n",
|
| 160 |
+
" f\"Downloaded data from {dataset_url} into file {zip_download_dir}\")\n",
|
| 161 |
+
"\n",
|
| 162 |
+
" except Exception as e:\n",
|
| 163 |
+
" raise e\n",
|
| 164 |
+
"\n",
|
| 165 |
+
" def extract_zip_file(self):\n",
|
| 166 |
+
" \"\"\"\n",
|
| 167 |
+
" zip_file_path: str\n",
|
| 168 |
+
" Extracts the zip file into the data directory\n",
|
| 169 |
+
" Function returns None\n",
|
| 170 |
+
" \"\"\"\n",
|
| 171 |
+
" unzip_path = self.config.unzip_dir\n",
|
| 172 |
+
" os.makedirs(unzip_path, exist_ok=True)\n",
|
| 173 |
+
" with zipfile.ZipFile(self.config.local_data_file, 'r') as zip_ref:\n",
|
| 174 |
+
" zip_ref.extractall(unzip_path)"
|
| 175 |
+
]
|
| 176 |
+
},
|
| 177 |
+
{
|
| 178 |
+
"cell_type": "code",
|
| 179 |
+
"execution_count": 37,
|
| 180 |
+
"metadata": {},
|
| 181 |
+
"outputs": [
|
| 182 |
+
{
|
| 183 |
+
"name": "stdout",
|
| 184 |
+
"output_type": "stream",
|
| 185 |
+
"text": [
|
| 186 |
+
"[2023-12-23 01:06:36,655: INFO: common yaml file: config/config.yaml loaded successfully]\n",
|
| 187 |
+
"[2023-12-23 01:06:36,656: INFO: common yaml file: params.yaml loaded successfully]\n",
|
| 188 |
+
"[2023-12-23 01:06:36,657: INFO: common created directory at: artifacts]\n",
|
| 189 |
+
"[2023-12-23 01:06:36,658: INFO: common created directory at: artifacts/data_ingestion]\n",
|
| 190 |
+
"[2023-12-23 01:06:36,659: INFO: 986331348 Downloading data from https://drive.google.com/file/d/1CGXriP_nlctsaWVbz0W3mFdPK05lQmHK/view?usp=sharing into file artifacts/data_ingestion/data.zip]\n"
|
| 191 |
+
]
|
| 192 |
+
},
|
| 193 |
+
{
|
| 194 |
+
"name": "stderr",
|
| 195 |
+
"output_type": "stream",
|
| 196 |
+
"text": [
|
| 197 |
+
"Downloading...\n",
|
| 198 |
+
"From (uriginal): https://drive.google.com/uc?/export=download&id=1CGXriP_nlctsaWVbz0W3mFdPK05lQmHK\n",
|
| 199 |
+
"From (redirected): https://drive.google.com/uc?/export=download&id=1CGXriP_nlctsaWVbz0W3mFdPK05lQmHK&confirm=t&uuid=3aeb0b9b-c680-4ec4-a1ab-78891e4f5025\n",
|
| 200 |
+
"To: /home/shrey/Desktop/Kidney-Disease-Classifcation/artifacts/data_ingestion/data.zip\n",
|
| 201 |
+
"100%|██████████| 1.63G/1.63G [02:31<00:00, 10.7MB/s]"
|
| 202 |
+
]
|
| 203 |
+
},
|
| 204 |
+
{
|
| 205 |
+
"name": "stdout",
|
| 206 |
+
"output_type": "stream",
|
| 207 |
+
"text": [
|
| 208 |
+
"[2023-12-23 01:09:10,818: INFO: 986331348 Downloaded data from https://drive.google.com/file/d/1CGXriP_nlctsaWVbz0W3mFdPK05lQmHK/view?usp=sharing into file artifacts/data_ingestion/data.zip]\n"
|
| 209 |
+
]
|
| 210 |
+
},
|
| 211 |
+
{
|
| 212 |
+
"name": "stderr",
|
| 213 |
+
"output_type": "stream",
|
| 214 |
+
"text": [
|
| 215 |
+
"\n"
|
| 216 |
+
]
|
| 217 |
+
}
|
| 218 |
+
],
|
| 219 |
+
"source": [
|
| 220 |
+
"try:\n",
|
| 221 |
+
" config = ConfigurationManager()\n",
|
| 222 |
+
" data_ingestion_config = config.get_data_ingestion_config()\n",
|
| 223 |
+
" data_ingestion = DataIngestion(config=data_ingestion_config)\n",
|
| 224 |
+
" data_ingestion.download_file()\n",
|
| 225 |
+
" data_ingestion.extract_zip_file()\n",
|
| 226 |
+
"except Exception as e:\n",
|
| 227 |
+
" raise e"
|
| 228 |
+
]
|
| 229 |
+
}
|
| 230 |
+
],
|
| 231 |
+
"metadata": {
|
| 232 |
+
"kernelspec": {
|
| 233 |
+
"display_name": "env",
|
| 234 |
+
"language": "python",
|
| 235 |
+
"name": "python3"
|
| 236 |
+
},
|
| 237 |
+
"language_info": {
|
| 238 |
+
"codemirror_mode": {
|
| 239 |
+
"name": "ipython",
|
| 240 |
+
"version": 3
|
| 241 |
+
},
|
| 242 |
+
"file_extension": ".py",
|
| 243 |
+
"mimetype": "text/x-python",
|
| 244 |
+
"name": "python",
|
| 245 |
+
"nbconvert_exporter": "python",
|
| 246 |
+
"pygments_lexer": "ipython3",
|
| 247 |
+
"version": "3.11.6"
|
| 248 |
+
}
|
| 249 |
+
},
|
| 250 |
+
"nbformat": 4,
|
| 251 |
+
"nbformat_minor": 2
|
| 252 |
+
}
|
Kidney-Disease-Classifcation/research/02_prepare_base_model.ipynb
ADDED
|
@@ -0,0 +1,281 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "code",
|
| 5 |
+
"execution_count": 13,
|
| 6 |
+
"metadata": {},
|
| 7 |
+
"outputs": [],
|
| 8 |
+
"source": [
|
| 9 |
+
"import os"
|
| 10 |
+
]
|
| 11 |
+
},
|
| 12 |
+
{
|
| 13 |
+
"cell_type": "code",
|
| 14 |
+
"execution_count": 14,
|
| 15 |
+
"metadata": {},
|
| 16 |
+
"outputs": [
|
| 17 |
+
{
|
| 18 |
+
"data": {
|
| 19 |
+
"text/plain": [
|
| 20 |
+
"'/home/shrey/Desktop/Kidney-Disease-Classifcation'"
|
| 21 |
+
]
|
| 22 |
+
},
|
| 23 |
+
"execution_count": 14,
|
| 24 |
+
"metadata": {},
|
| 25 |
+
"output_type": "execute_result"
|
| 26 |
+
}
|
| 27 |
+
],
|
| 28 |
+
"source": [
|
| 29 |
+
"%pwd"
|
| 30 |
+
]
|
| 31 |
+
},
|
| 32 |
+
{
|
| 33 |
+
"cell_type": "code",
|
| 34 |
+
"execution_count": 3,
|
| 35 |
+
"metadata": {},
|
| 36 |
+
"outputs": [],
|
| 37 |
+
"source": [
|
| 38 |
+
"os.chdir(\"../\")"
|
| 39 |
+
]
|
| 40 |
+
},
|
| 41 |
+
{
|
| 42 |
+
"cell_type": "code",
|
| 43 |
+
"execution_count": 4,
|
| 44 |
+
"metadata": {},
|
| 45 |
+
"outputs": [
|
| 46 |
+
{
|
| 47 |
+
"data": {
|
| 48 |
+
"text/plain": [
|
| 49 |
+
"'/home/shrey/Desktop/Kidney-Disease-Classifcation'"
|
| 50 |
+
]
|
| 51 |
+
},
|
| 52 |
+
"execution_count": 4,
|
| 53 |
+
"metadata": {},
|
| 54 |
+
"output_type": "execute_result"
|
| 55 |
+
}
|
| 56 |
+
],
|
| 57 |
+
"source": [
|
| 58 |
+
"%pwd"
|
| 59 |
+
]
|
| 60 |
+
},
|
| 61 |
+
{
|
| 62 |
+
"cell_type": "code",
|
| 63 |
+
"execution_count": 15,
|
| 64 |
+
"metadata": {},
|
| 65 |
+
"outputs": [],
|
| 66 |
+
"source": [
|
| 67 |
+
"os.chdir(\"src\")"
|
| 68 |
+
]
|
| 69 |
+
},
|
| 70 |
+
{
|
| 71 |
+
"cell_type": "code",
|
| 72 |
+
"execution_count": 16,
|
| 73 |
+
"metadata": {},
|
| 74 |
+
"outputs": [],
|
| 75 |
+
"source": [
|
| 76 |
+
"from dataclasses import dataclass\n",
|
| 77 |
+
"from pathlib import Path\n",
|
| 78 |
+
"\n",
|
| 79 |
+
"\n",
|
| 80 |
+
"@dataclass(frozen=True)\n",
|
| 81 |
+
"class PrepareBaseModelConfig:\n",
|
| 82 |
+
" root_dir: Path\n",
|
| 83 |
+
" base_model_path: Path\n",
|
| 84 |
+
" updated_base_model_path: Path\n",
|
| 85 |
+
" params_image_size: list\n",
|
| 86 |
+
" params_include_top: bool\n",
|
| 87 |
+
" params_weights: str\n",
|
| 88 |
+
" params_classes: int"
|
| 89 |
+
]
|
| 90 |
+
},
|
| 91 |
+
{
|
| 92 |
+
"cell_type": "code",
|
| 93 |
+
"execution_count": 17,
|
| 94 |
+
"metadata": {},
|
| 95 |
+
"outputs": [],
|
| 96 |
+
"source": [
|
| 97 |
+
"from kidney_classification.constants import *\n",
|
| 98 |
+
"from kidney_classification.utils.common import read_yaml, create_directories"
|
| 99 |
+
]
|
| 100 |
+
},
|
| 101 |
+
{
|
| 102 |
+
"cell_type": "code",
|
| 103 |
+
"execution_count": 18,
|
| 104 |
+
"metadata": {},
|
| 105 |
+
"outputs": [],
|
| 106 |
+
"source": [
|
| 107 |
+
"class ConfigurationManager:\n",
|
| 108 |
+
" def __init__(\n",
|
| 109 |
+
" self,\n",
|
| 110 |
+
" config_filepath = CONFIG_FILE_PATH,\n",
|
| 111 |
+
" params_filepath = PARAMS_FILE_PATH):\n",
|
| 112 |
+
"\n",
|
| 113 |
+
" self.config = read_yaml(config_filepath)\n",
|
| 114 |
+
" self.params = read_yaml(params_filepath)\n",
|
| 115 |
+
"\n",
|
| 116 |
+
" create_directories([self.config.artifacts_root])\n",
|
| 117 |
+
"\n",
|
| 118 |
+
" \n",
|
| 119 |
+
"\n",
|
| 120 |
+
" def get_prepare_base_model_config(self) -> PrepareBaseModelConfig:\n",
|
| 121 |
+
" config = self.config.prepare_base_model\n",
|
| 122 |
+
" \n",
|
| 123 |
+
" create_directories([config.root_dir])\n",
|
| 124 |
+
"\n",
|
| 125 |
+
" prepare_base_model_config = PrepareBaseModelConfig(\n",
|
| 126 |
+
" root_dir=Path(config.root_dir),\n",
|
| 127 |
+
" base_model_path=Path(config.base_model_path),\n",
|
| 128 |
+
" updated_base_model_path=Path(config.updated_base_model_path),\n",
|
| 129 |
+
" params_image_size=self.params.IMAGE_SIZE,\n",
|
| 130 |
+
" params_include_top=self.params.INCLUDE_TOP,\n",
|
| 131 |
+
" params_weights=self.params.WEIGHTS,\n",
|
| 132 |
+
" params_classes=self.params.CLASSES\n",
|
| 133 |
+
" )\n",
|
| 134 |
+
"\n",
|
| 135 |
+
" return prepare_base_model_config"
|
| 136 |
+
]
|
| 137 |
+
},
|
| 138 |
+
{
|
| 139 |
+
"cell_type": "code",
|
| 140 |
+
"execution_count": 19,
|
| 141 |
+
"metadata": {},
|
| 142 |
+
"outputs": [],
|
| 143 |
+
"source": [
|
| 144 |
+
"import tensorflow as tf\n",
|
| 145 |
+
"from tensorflow.keras.layers import Dense, Flatten, BatchNormalization, Dropout\n",
|
| 146 |
+
"from tensorflow.keras.models import Sequential"
|
| 147 |
+
]
|
| 148 |
+
},
|
| 149 |
+
{
|
| 150 |
+
"cell_type": "code",
|
| 151 |
+
"execution_count": 26,
|
| 152 |
+
"metadata": {},
|
| 153 |
+
"outputs": [],
|
| 154 |
+
"source": [
|
| 155 |
+
"class PrepareBaseModel:\n",
|
| 156 |
+
" @staticmethod\n",
|
| 157 |
+
" def prepare_full_model():\n",
|
| 158 |
+
" VGG_model = Sequential()\n",
|
| 159 |
+
"\n",
|
| 160 |
+
" pretrained_model= tf.keras.applications.VGG16(\n",
|
| 161 |
+
" include_top=False,\n",
|
| 162 |
+
" input_shape=(150, 150, 3),\n",
|
| 163 |
+
" pooling='max',\n",
|
| 164 |
+
" classes=4,\n",
|
| 165 |
+
" weights='imagenet'\n",
|
| 166 |
+
" )\n",
|
| 167 |
+
"\n",
|
| 168 |
+
" VGG_model.add(pretrained_model)\n",
|
| 169 |
+
" VGG_model.add(Flatten())\n",
|
| 170 |
+
" VGG_model.add(Dense(512, activation='relu'))\n",
|
| 171 |
+
" VGG_model.add(BatchNormalization())\n",
|
| 172 |
+
" VGG_model.add(Dropout(0.5))\n",
|
| 173 |
+
"\n",
|
| 174 |
+
" VGG_model.add(Dense(4, activation='softmax'))\n",
|
| 175 |
+
" pretrained_model.trainable = False\n",
|
| 176 |
+
"\n",
|
| 177 |
+
" VGG_model.compile(\n",
|
| 178 |
+
" optimizer='adam',\n",
|
| 179 |
+
" loss='sparse_categorical_crossentropy',\n",
|
| 180 |
+
" metrics=['accuracy']\n",
|
| 181 |
+
" )\n",
|
| 182 |
+
"\n",
|
| 183 |
+
" return VGG_model\n",
|
| 184 |
+
"\n",
|
| 185 |
+
" def update_base_model(self, config: PrepareBaseModelConfig):\n",
|
| 186 |
+
" full_model = self.prepare_full_model()\n",
|
| 187 |
+
"\n",
|
| 188 |
+
" full_model.summary()\n",
|
| 189 |
+
" full_model.save(config.updated_base_model_path)\n",
|
| 190 |
+
" \n",
|
| 191 |
+
" \n",
|
| 192 |
+
" @staticmethod\n",
|
| 193 |
+
" def save_model(path: Path, model: tf.keras.Model):\n",
|
| 194 |
+
" model.save(path)"
|
| 195 |
+
]
|
| 196 |
+
},
|
| 197 |
+
{
|
| 198 |
+
"cell_type": "code",
|
| 199 |
+
"execution_count": 22,
|
| 200 |
+
"metadata": {},
|
| 201 |
+
"outputs": [],
|
| 202 |
+
"source": [
|
| 203 |
+
"os.chdir(\"../\")"
|
| 204 |
+
]
|
| 205 |
+
},
|
| 206 |
+
{
|
| 207 |
+
"cell_type": "code",
|
| 208 |
+
"execution_count": 27,
|
| 209 |
+
"metadata": {},
|
| 210 |
+
"outputs": [
|
| 211 |
+
{
|
| 212 |
+
"name": "stdout",
|
| 213 |
+
"output_type": "stream",
|
| 214 |
+
"text": [
|
| 215 |
+
"[2024-01-03 15:58:32,485: INFO: common yaml file: config/config.yaml loaded successfully]\n",
|
| 216 |
+
"[2024-01-03 15:58:32,487: INFO: common yaml file: params.yaml loaded successfully]\n",
|
| 217 |
+
"Model: \"sequential_3\"\n",
|
| 218 |
+
"_________________________________________________________________\n",
|
| 219 |
+
" Layer (type) Output Shape Param # \n",
|
| 220 |
+
"=================================================================\n",
|
| 221 |
+
" vgg16 (Functional) (None, 512) 14714688 \n",
|
| 222 |
+
" \n",
|
| 223 |
+
" flatten_3 (Flatten) (None, 512) 0 \n",
|
| 224 |
+
" \n",
|
| 225 |
+
" dense_5 (Dense) (None, 512) 262656 \n",
|
| 226 |
+
" \n",
|
| 227 |
+
" batch_normalization_3 (Bat (None, 512) 2048 \n",
|
| 228 |
+
" chNormalization) \n",
|
| 229 |
+
" \n",
|
| 230 |
+
" dropout_3 (Dropout) (None, 512) 0 \n",
|
| 231 |
+
" \n",
|
| 232 |
+
" dense_6 (Dense) (None, 4) 2052 \n",
|
| 233 |
+
" \n",
|
| 234 |
+
"=================================================================\n",
|
| 235 |
+
"Total params: 14981444 (57.15 MB)\n",
|
| 236 |
+
"Trainable params: 265732 (1.01 MB)\n",
|
| 237 |
+
"Non-trainable params: 14715712 (56.14 MB)\n",
|
| 238 |
+
"_________________________________________________________________\n"
|
| 239 |
+
]
|
| 240 |
+
}
|
| 241 |
+
],
|
| 242 |
+
"source": [
|
| 243 |
+
"try:\n",
|
| 244 |
+
" config = ConfigurationManager()\n",
|
| 245 |
+
" prepare_base_model_config = config.get_prepare_base_model_config()\n",
|
| 246 |
+
" prepare_base_model = PrepareBaseModel()\n",
|
| 247 |
+
" prepare_base_model.update_base_model(prepare_base_model_config)\n",
|
| 248 |
+
"except Exception as e:\n",
|
| 249 |
+
" raise e"
|
| 250 |
+
]
|
| 251 |
+
},
|
| 252 |
+
{
|
| 253 |
+
"cell_type": "code",
|
| 254 |
+
"execution_count": null,
|
| 255 |
+
"metadata": {},
|
| 256 |
+
"outputs": [],
|
| 257 |
+
"source": []
|
| 258 |
+
}
|
| 259 |
+
],
|
| 260 |
+
"metadata": {
|
| 261 |
+
"kernelspec": {
|
| 262 |
+
"display_name": "env",
|
| 263 |
+
"language": "python",
|
| 264 |
+
"name": "python3"
|
| 265 |
+
},
|
| 266 |
+
"language_info": {
|
| 267 |
+
"codemirror_mode": {
|
| 268 |
+
"name": "ipython",
|
| 269 |
+
"version": 3
|
| 270 |
+
},
|
| 271 |
+
"file_extension": ".py",
|
| 272 |
+
"mimetype": "text/x-python",
|
| 273 |
+
"name": "python",
|
| 274 |
+
"nbconvert_exporter": "python",
|
| 275 |
+
"pygments_lexer": "ipython3",
|
| 276 |
+
"version": "3.11.6"
|
| 277 |
+
}
|
| 278 |
+
},
|
| 279 |
+
"nbformat": 4,
|
| 280 |
+
"nbformat_minor": 2
|
| 281 |
+
}
|
Kidney-Disease-Classifcation/research/03_model_training.ipynb
ADDED
|
@@ -0,0 +1,341 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "code",
|
| 5 |
+
"execution_count": 1,
|
| 6 |
+
"metadata": {},
|
| 7 |
+
"outputs": [],
|
| 8 |
+
"source": [
|
| 9 |
+
"import os"
|
| 10 |
+
]
|
| 11 |
+
},
|
| 12 |
+
{
|
| 13 |
+
"cell_type": "code",
|
| 14 |
+
"execution_count": 2,
|
| 15 |
+
"metadata": {},
|
| 16 |
+
"outputs": [
|
| 17 |
+
{
|
| 18 |
+
"data": {
|
| 19 |
+
"text/plain": [
|
| 20 |
+
"'/home/shrey/Desktop/Kidney-Disease-Classifcation/research'"
|
| 21 |
+
]
|
| 22 |
+
},
|
| 23 |
+
"execution_count": 2,
|
| 24 |
+
"metadata": {},
|
| 25 |
+
"output_type": "execute_result"
|
| 26 |
+
}
|
| 27 |
+
],
|
| 28 |
+
"source": [
|
| 29 |
+
"%pwd"
|
| 30 |
+
]
|
| 31 |
+
},
|
| 32 |
+
{
|
| 33 |
+
"cell_type": "code",
|
| 34 |
+
"execution_count": 3,
|
| 35 |
+
"metadata": {},
|
| 36 |
+
"outputs": [],
|
| 37 |
+
"source": [
|
| 38 |
+
"os.chdir(\"../\")"
|
| 39 |
+
]
|
| 40 |
+
},
|
| 41 |
+
{
|
| 42 |
+
"cell_type": "code",
|
| 43 |
+
"execution_count": 4,
|
| 44 |
+
"metadata": {},
|
| 45 |
+
"outputs": [
|
| 46 |
+
{
|
| 47 |
+
"data": {
|
| 48 |
+
"text/plain": [
|
| 49 |
+
"'/home/shrey/Desktop/Kidney-Disease-Classifcation'"
|
| 50 |
+
]
|
| 51 |
+
},
|
| 52 |
+
"execution_count": 4,
|
| 53 |
+
"metadata": {},
|
| 54 |
+
"output_type": "execute_result"
|
| 55 |
+
}
|
| 56 |
+
],
|
| 57 |
+
"source": [
|
| 58 |
+
"%pwd"
|
| 59 |
+
]
|
| 60 |
+
},
|
| 61 |
+
{
|
| 62 |
+
"cell_type": "code",
|
| 63 |
+
"execution_count": 5,
|
| 64 |
+
"metadata": {},
|
| 65 |
+
"outputs": [],
|
| 66 |
+
"source": [
|
| 67 |
+
"from dataclasses import dataclass\n",
|
| 68 |
+
"from pathlib import Path\n",
|
| 69 |
+
"\n",
|
| 70 |
+
"\n",
|
| 71 |
+
"@dataclass(frozen=True)\n",
|
| 72 |
+
"class TrainingConfig:\n",
|
| 73 |
+
" root_dir: Path\n",
|
| 74 |
+
" trained_model_path: Path\n",
|
| 75 |
+
" updated_base_model_path: Path\n",
|
| 76 |
+
" training_data: Path\n",
|
| 77 |
+
" params_epochs: int\n",
|
| 78 |
+
" params_batch_size: int\n",
|
| 79 |
+
" params_image_size: list"
|
| 80 |
+
]
|
| 81 |
+
},
|
| 82 |
+
{
|
| 83 |
+
"cell_type": "code",
|
| 84 |
+
"execution_count": 6,
|
| 85 |
+
"metadata": {},
|
| 86 |
+
"outputs": [],
|
| 87 |
+
"source": [
|
| 88 |
+
"os.chdir(\"./src\")"
|
| 89 |
+
]
|
| 90 |
+
},
|
| 91 |
+
{
|
| 92 |
+
"cell_type": "code",
|
| 93 |
+
"execution_count": 7,
|
| 94 |
+
"metadata": {},
|
| 95 |
+
"outputs": [
|
| 96 |
+
{
|
| 97 |
+
"name": "stderr",
|
| 98 |
+
"output_type": "stream",
|
| 99 |
+
"text": [
|
| 100 |
+
"2024-01-03 15:58:59.001906: I tensorflow/core/util/port.cc:113] oneDNN custom operations are on. You may see slightly different numerical results due to floating-point round-off errors from different computation orders. To turn them off, set the environment variable `TF_ENABLE_ONEDNN_OPTS=0`.\n",
|
| 101 |
+
"2024-01-03 15:58:59.003769: I external/local_tsl/tsl/cuda/cudart_stub.cc:31] Could not find cuda drivers on your machine, GPU will not be used.\n",
|
| 102 |
+
"2024-01-03 15:58:59.032703: E external/local_xla/xla/stream_executor/cuda/cuda_dnn.cc:9261] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered\n",
|
| 103 |
+
"2024-01-03 15:58:59.032753: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:607] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered\n",
|
| 104 |
+
"2024-01-03 15:58:59.033922: E external/local_xla/xla/stream_executor/cuda/cuda_blas.cc:1515] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered\n",
|
| 105 |
+
"2024-01-03 15:58:59.038705: I external/local_tsl/tsl/cuda/cudart_stub.cc:31] Could not find cuda drivers on your machine, GPU will not be used.\n",
|
| 106 |
+
"2024-01-03 15:58:59.039146: I tensorflow/core/platform/cpu_feature_guard.cc:182] This TensorFlow binary is optimized to use available CPU instructions in performance-critical operations.\n",
|
| 107 |
+
"To enable the following instructions: AVX2 AVX512F AVX512_VNNI FMA, in other operations, rebuild TensorFlow with the appropriate compiler flags.\n",
|
| 108 |
+
"2024-01-03 15:58:59.655806: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Could not find TensorRT\n"
|
| 109 |
+
]
|
| 110 |
+
}
|
| 111 |
+
],
|
| 112 |
+
"source": [
|
| 113 |
+
"from kidney_classification.constants import *\n",
|
| 114 |
+
"from kidney_classification.utils.common import read_yaml, create_directories\n",
|
| 115 |
+
"import tensorflow as tf"
|
| 116 |
+
]
|
| 117 |
+
},
|
| 118 |
+
{
|
| 119 |
+
"cell_type": "code",
|
| 120 |
+
"execution_count": 8,
|
| 121 |
+
"metadata": {},
|
| 122 |
+
"outputs": [],
|
| 123 |
+
"source": [
|
| 124 |
+
"class ConfigurationManager:\n",
|
| 125 |
+
" def __init__(\n",
|
| 126 |
+
" self,\n",
|
| 127 |
+
" config_filepath = CONFIG_FILE_PATH,\n",
|
| 128 |
+
" params_filepath = PARAMS_FILE_PATH):\n",
|
| 129 |
+
"\n",
|
| 130 |
+
" self.config = read_yaml(config_filepath)\n",
|
| 131 |
+
" self.params = read_yaml(params_filepath)\n",
|
| 132 |
+
"\n",
|
| 133 |
+
" create_directories([self.config.artifacts_root])\n",
|
| 134 |
+
"\n",
|
| 135 |
+
"\n",
|
| 136 |
+
" \n",
|
| 137 |
+
" def get_training_config(self) -> TrainingConfig:\n",
|
| 138 |
+
" training = self.config.training\n",
|
| 139 |
+
" prepare_base_model = self.config.prepare_base_model\n",
|
| 140 |
+
" params = self.params\n",
|
| 141 |
+
" training_data = os.path.join(self.config.data_ingestion.unzip_dir, \"CT-KIDNEY-DATASET-Normal-Cyst-Tumor-Stone\")\n",
|
| 142 |
+
" create_directories([\n",
|
| 143 |
+
" Path(training.root_dir)\n",
|
| 144 |
+
" ])\n",
|
| 145 |
+
"\n",
|
| 146 |
+
" training_config = TrainingConfig(\n",
|
| 147 |
+
" root_dir=Path(training.root_dir),\n",
|
| 148 |
+
" trained_model_path=Path(training.trained_model_path),\n",
|
| 149 |
+
" updated_base_model_path=Path(prepare_base_model.updated_base_model_path),\n",
|
| 150 |
+
" training_data=Path(training_data),\n",
|
| 151 |
+
" params_epochs=params.EPOCHS,\n",
|
| 152 |
+
" params_batch_size=params.BATCH_SIZE,\n",
|
| 153 |
+
" params_image_size=params.IMAGE_SIZE\n",
|
| 154 |
+
" )\n",
|
| 155 |
+
"\n",
|
| 156 |
+
" return training_config"
|
| 157 |
+
]
|
| 158 |
+
},
|
| 159 |
+
{
|
| 160 |
+
"cell_type": "code",
|
| 161 |
+
"execution_count": 9,
|
| 162 |
+
"metadata": {},
|
| 163 |
+
"outputs": [],
|
| 164 |
+
"source": [
|
| 165 |
+
"class Training:\n",
|
| 166 |
+
" def __init__(self, config: TrainingConfig):\n",
|
| 167 |
+
" self.config = config\n",
|
| 168 |
+
"\n",
|
| 169 |
+
" \n",
|
| 170 |
+
" def get_base_model(self):\n",
|
| 171 |
+
" self.model = tf.keras.models.load_model(\n",
|
| 172 |
+
" self.config.updated_base_model_path\n",
|
| 173 |
+
" )\n",
|
| 174 |
+
"\n",
|
| 175 |
+
" def train_valid_generator(self):\n",
|
| 176 |
+
" img_height, img_width = self.config.params_image_size[:-1]\n",
|
| 177 |
+
"\n",
|
| 178 |
+
" train = tf.keras.utils.image_dataset_from_directory(\n",
|
| 179 |
+
" self.config.training_data,\n",
|
| 180 |
+
" image_size=(img_height, img_width),\n",
|
| 181 |
+
" validation_split=0.1,\n",
|
| 182 |
+
" subset='training',\n",
|
| 183 |
+
" seed=123\n",
|
| 184 |
+
" )\n",
|
| 185 |
+
" \n",
|
| 186 |
+
" val = tf.keras.utils.image_dataset_from_directory(\n",
|
| 187 |
+
" self.config.training_data,\n",
|
| 188 |
+
" image_size=(img_height, img_width),\n",
|
| 189 |
+
" validation_split=0.2,\n",
|
| 190 |
+
" subset='validation',\n",
|
| 191 |
+
" seed=123\n",
|
| 192 |
+
" )\n",
|
| 193 |
+
" train = train.map(lambda x, y: (x / 255, y))\n",
|
| 194 |
+
" val = val.map(lambda x, y: (x / 255, y))\n",
|
| 195 |
+
" AUTOTUNE = tf.data.AUTOTUNE\n",
|
| 196 |
+
"\n",
|
| 197 |
+
" self.train_dataset = train.cache().prefetch(buffer_size=AUTOTUNE)\n",
|
| 198 |
+
" self.val_dataset = val.cache().prefetch(buffer_size=AUTOTUNE)\n",
|
| 199 |
+
"\n",
|
| 200 |
+
" \n",
|
| 201 |
+
" @staticmethod\n",
|
| 202 |
+
" def save_model(path: Path, model: tf.keras.Model):\n",
|
| 203 |
+
" model.save(path)\n",
|
| 204 |
+
"\n",
|
| 205 |
+
" \n",
|
| 206 |
+
" def define_and_train_model(self):\n",
|
| 207 |
+
"\n",
|
| 208 |
+
" self.model.fit(\n",
|
| 209 |
+
" self.train_dataset,\n",
|
| 210 |
+
" validation_data=self.val_dataset,\n",
|
| 211 |
+
" epochs=self.config.params_epochs,\n",
|
| 212 |
+
" )\n",
|
| 213 |
+
"\n",
|
| 214 |
+
" self.save_model(\n",
|
| 215 |
+
" path=self.config.trained_model_path,\n",
|
| 216 |
+
" model=self.model\n",
|
| 217 |
+
" )"
|
| 218 |
+
]
|
| 219 |
+
},
|
| 220 |
+
{
|
| 221 |
+
"cell_type": "code",
|
| 222 |
+
"execution_count": 10,
|
| 223 |
+
"metadata": {},
|
| 224 |
+
"outputs": [],
|
| 225 |
+
"source": [
|
| 226 |
+
"os.chdir(\"../\")"
|
| 227 |
+
]
|
| 228 |
+
},
|
| 229 |
+
{
|
| 230 |
+
"cell_type": "code",
|
| 231 |
+
"execution_count": 11,
|
| 232 |
+
"metadata": {},
|
| 233 |
+
"outputs": [
|
| 234 |
+
{
|
| 235 |
+
"name": "stdout",
|
| 236 |
+
"output_type": "stream",
|
| 237 |
+
"text": [
|
| 238 |
+
"[2024-01-03 15:59:11,299: INFO: common yaml file: config/config.yaml loaded successfully]\n",
|
| 239 |
+
"[2024-01-03 15:59:11,301: INFO: common yaml file: params.yaml loaded successfully]\n"
|
| 240 |
+
]
|
| 241 |
+
},
|
| 242 |
+
{
|
| 243 |
+
"name": "stderr",
|
| 244 |
+
"output_type": "stream",
|
| 245 |
+
"text": [
|
| 246 |
+
"2024-01-03 15:59:11.602362: E external/local_xla/xla/stream_executor/cuda/cuda_driver.cc:274] failed call to cuInit: CUDA_ERROR_NO_DEVICE: no CUDA-capable device is detected\n"
|
| 247 |
+
]
|
| 248 |
+
},
|
| 249 |
+
{
|
| 250 |
+
"name": "stdout",
|
| 251 |
+
"output_type": "stream",
|
| 252 |
+
"text": [
|
| 253 |
+
"[2024-01-03 15:59:12,070: WARNING: optimizer Skipping variable loading for optimizer 'Adam', because it has 13 variables whereas the saved optimizer has 1 variables. ]\n",
|
| 254 |
+
"Found 12446 files belonging to 4 classes.\n",
|
| 255 |
+
"Using 11202 files for training.\n",
|
| 256 |
+
"Found 12446 files belonging to 4 classes.\n",
|
| 257 |
+
"Using 2489 files for validation.\n",
|
| 258 |
+
"Epoch 1/15\n",
|
| 259 |
+
"351/351 [==============================] - 561s 2s/step - loss: 0.5099 - accuracy: 0.8103 - val_loss: 0.2366 - val_accuracy: 0.9152\n",
|
| 260 |
+
"Epoch 2/15\n",
|
| 261 |
+
"351/351 [==============================] - 566s 2s/step - loss: 0.2120 - accuracy: 0.9229 - val_loss: 0.2941 - val_accuracy: 0.8803\n",
|
| 262 |
+
"Epoch 3/15\n",
|
| 263 |
+
"351/351 [==============================] - 571s 2s/step - loss: 0.1573 - accuracy: 0.9466 - val_loss: 0.0994 - val_accuracy: 0.9667\n",
|
| 264 |
+
"Epoch 4/15\n",
|
| 265 |
+
"351/351 [==============================] - 547s 2s/step - loss: 0.1370 - accuracy: 0.9528 - val_loss: 0.0742 - val_accuracy: 0.9787\n",
|
| 266 |
+
"Epoch 5/15\n",
|
| 267 |
+
"351/351 [==============================] - 552s 2s/step - loss: 0.1217 - accuracy: 0.9565 - val_loss: 0.0564 - val_accuracy: 0.9835\n",
|
| 268 |
+
"Epoch 6/15\n",
|
| 269 |
+
"351/351 [==============================] - 550s 2s/step - loss: 0.1064 - accuracy: 0.9621 - val_loss: 0.0626 - val_accuracy: 0.9791\n",
|
| 270 |
+
"Epoch 7/15\n",
|
| 271 |
+
"351/351 [==============================] - 550s 2s/step - loss: 0.1028 - accuracy: 0.9622 - val_loss: 0.0845 - val_accuracy: 0.9658\n",
|
| 272 |
+
"Epoch 8/15\n",
|
| 273 |
+
"351/351 [==============================] - 551s 2s/step - loss: 0.0953 - accuracy: 0.9655 - val_loss: 0.0527 - val_accuracy: 0.9811\n",
|
| 274 |
+
"Epoch 9/15\n",
|
| 275 |
+
"351/351 [==============================] - 526s 2s/step - loss: 0.0900 - accuracy: 0.9692 - val_loss: 0.0698 - val_accuracy: 0.9731\n",
|
| 276 |
+
"Epoch 10/15\n",
|
| 277 |
+
"351/351 [==============================] - 526s 1s/step - loss: 0.0806 - accuracy: 0.9708 - val_loss: 0.0454 - val_accuracy: 0.9867\n",
|
| 278 |
+
"Epoch 11/15\n",
|
| 279 |
+
"351/351 [==============================] - 529s 2s/step - loss: 0.0756 - accuracy: 0.9723 - val_loss: 0.0404 - val_accuracy: 0.9871\n",
|
| 280 |
+
"Epoch 12/15\n",
|
| 281 |
+
"351/351 [==============================] - 533s 2s/step - loss: 0.0762 - accuracy: 0.9723 - val_loss: 0.0458 - val_accuracy: 0.9867\n",
|
| 282 |
+
"Epoch 13/15\n",
|
| 283 |
+
"351/351 [==============================] - 538s 2s/step - loss: 0.0651 - accuracy: 0.9760 - val_loss: 0.0331 - val_accuracy: 0.9912\n",
|
| 284 |
+
"Epoch 14/15\n",
|
| 285 |
+
"351/351 [==============================] - 547s 2s/step - loss: 0.0740 - accuracy: 0.9746 - val_loss: 0.0726 - val_accuracy: 0.9771\n",
|
| 286 |
+
"Epoch 15/15\n",
|
| 287 |
+
"351/351 [==============================] - 562s 2s/step - loss: 0.0742 - accuracy: 0.9744 - val_loss: 0.0441 - val_accuracy: 0.9859\n"
|
| 288 |
+
]
|
| 289 |
+
},
|
| 290 |
+
{
|
| 291 |
+
"name": "stderr",
|
| 292 |
+
"output_type": "stream",
|
| 293 |
+
"text": [
|
| 294 |
+
"/home/shrey/Desktop/Kidney-Disease-Classifcation/env/lib/python3.11/site-packages/keras/src/engine/training.py:3103: UserWarning: You are saving your model as an HDF5 file via `model.save()`. This file format is considered legacy. We recommend using instead the native Keras format, e.g. `model.save('my_model.keras')`.\n",
|
| 295 |
+
" saving_api.save_model(\n"
|
| 296 |
+
]
|
| 297 |
+
}
|
| 298 |
+
],
|
| 299 |
+
"source": [
|
| 300 |
+
"try:\n",
|
| 301 |
+
" config = ConfigurationManager()\n",
|
| 302 |
+
" training_config = config.get_training_config()\n",
|
| 303 |
+
" training = Training(config=training_config)\n",
|
| 304 |
+
" training.get_base_model()\n",
|
| 305 |
+
" training.train_valid_generator()\n",
|
| 306 |
+
" training.define_and_train_model()\n",
|
| 307 |
+
" \n",
|
| 308 |
+
"except Exception as e:\n",
|
| 309 |
+
" raise e"
|
| 310 |
+
]
|
| 311 |
+
},
|
| 312 |
+
{
|
| 313 |
+
"cell_type": "code",
|
| 314 |
+
"execution_count": null,
|
| 315 |
+
"metadata": {},
|
| 316 |
+
"outputs": [],
|
| 317 |
+
"source": []
|
| 318 |
+
}
|
| 319 |
+
],
|
| 320 |
+
"metadata": {
|
| 321 |
+
"kernelspec": {
|
| 322 |
+
"display_name": "env",
|
| 323 |
+
"language": "python",
|
| 324 |
+
"name": "python3"
|
| 325 |
+
},
|
| 326 |
+
"language_info": {
|
| 327 |
+
"codemirror_mode": {
|
| 328 |
+
"name": "ipython",
|
| 329 |
+
"version": 3
|
| 330 |
+
},
|
| 331 |
+
"file_extension": ".py",
|
| 332 |
+
"mimetype": "text/x-python",
|
| 333 |
+
"name": "python",
|
| 334 |
+
"nbconvert_exporter": "python",
|
| 335 |
+
"pygments_lexer": "ipython3",
|
| 336 |
+
"version": "3.11.6"
|
| 337 |
+
}
|
| 338 |
+
},
|
| 339 |
+
"nbformat": 4,
|
| 340 |
+
"nbformat_minor": 2
|
| 341 |
+
}
|
Kidney-Disease-Classifcation/research/04_model_evaluation_with_mlflow.ipynb
ADDED
|
@@ -0,0 +1,346 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "code",
|
| 5 |
+
"execution_count": 1,
|
| 6 |
+
"metadata": {},
|
| 7 |
+
"outputs": [],
|
| 8 |
+
"source": [
|
| 9 |
+
"import os"
|
| 10 |
+
]
|
| 11 |
+
},
|
| 12 |
+
{
|
| 13 |
+
"cell_type": "code",
|
| 14 |
+
"execution_count": 2,
|
| 15 |
+
"metadata": {},
|
| 16 |
+
"outputs": [
|
| 17 |
+
{
|
| 18 |
+
"data": {
|
| 19 |
+
"text/plain": [
|
| 20 |
+
"'/home/shrey/Desktop/Kidney-Disease-Classifcation/research'"
|
| 21 |
+
]
|
| 22 |
+
},
|
| 23 |
+
"execution_count": 2,
|
| 24 |
+
"metadata": {},
|
| 25 |
+
"output_type": "execute_result"
|
| 26 |
+
}
|
| 27 |
+
],
|
| 28 |
+
"source": [
|
| 29 |
+
"%pwd"
|
| 30 |
+
]
|
| 31 |
+
},
|
| 32 |
+
{
|
| 33 |
+
"cell_type": "code",
|
| 34 |
+
"execution_count": 3,
|
| 35 |
+
"metadata": {},
|
| 36 |
+
"outputs": [],
|
| 37 |
+
"source": [
|
| 38 |
+
"os.chdir('../')"
|
| 39 |
+
]
|
| 40 |
+
},
|
| 41 |
+
{
|
| 42 |
+
"cell_type": "code",
|
| 43 |
+
"execution_count": 4,
|
| 44 |
+
"metadata": {},
|
| 45 |
+
"outputs": [
|
| 46 |
+
{
|
| 47 |
+
"data": {
|
| 48 |
+
"text/plain": [
|
| 49 |
+
"'/home/shrey/Desktop/Kidney-Disease-Classifcation'"
|
| 50 |
+
]
|
| 51 |
+
},
|
| 52 |
+
"execution_count": 4,
|
| 53 |
+
"metadata": {},
|
| 54 |
+
"output_type": "execute_result"
|
| 55 |
+
}
|
| 56 |
+
],
|
| 57 |
+
"source": [
|
| 58 |
+
"%pwd"
|
| 59 |
+
]
|
| 60 |
+
},
|
| 61 |
+
{
|
| 62 |
+
"cell_type": "code",
|
| 63 |
+
"execution_count": 5,
|
| 64 |
+
"metadata": {},
|
| 65 |
+
"outputs": [],
|
| 66 |
+
"source": [
|
| 67 |
+
"os.environ[\"MLFLOW_TRACKING_URI\"]=\"https://dagshub.com/Shrey-patel-07/Kidney-Disease-Classifcation.mlflow\"\n",
|
| 68 |
+
"os.environ[\"MLFLOW_TRACKING_USERNAME\"]=\"Shrey-patel-07\"\n",
|
| 69 |
+
"os.environ[\"MLFLOW_TRACKING_PASSWORD\"]=\"6a425a20bb5b645c5efa1ca49ab28097a92a76ee\""
|
| 70 |
+
]
|
| 71 |
+
},
|
| 72 |
+
{
|
| 73 |
+
"cell_type": "code",
|
| 74 |
+
"execution_count": 6,
|
| 75 |
+
"metadata": {},
|
| 76 |
+
"outputs": [
|
| 77 |
+
{
|
| 78 |
+
"name": "stderr",
|
| 79 |
+
"output_type": "stream",
|
| 80 |
+
"text": [
|
| 81 |
+
"2024-01-04 12:24:18.257148: I tensorflow/core/util/port.cc:113] oneDNN custom operations are on. You may see slightly different numerical results due to floating-point round-off errors from different computation orders. To turn them off, set the environment variable `TF_ENABLE_ONEDNN_OPTS=0`.\n",
|
| 82 |
+
"2024-01-04 12:24:18.258826: I external/local_tsl/tsl/cuda/cudart_stub.cc:31] Could not find cuda drivers on your machine, GPU will not be used.\n",
|
| 83 |
+
"2024-01-04 12:24:18.285564: E external/local_xla/xla/stream_executor/cuda/cuda_dnn.cc:9261] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered\n",
|
| 84 |
+
"2024-01-04 12:24:18.285595: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:607] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered\n",
|
| 85 |
+
"2024-01-04 12:24:18.286297: E external/local_xla/xla/stream_executor/cuda/cuda_blas.cc:1515] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered\n",
|
| 86 |
+
"2024-01-04 12:24:18.290364: I external/local_tsl/tsl/cuda/cudart_stub.cc:31] Could not find cuda drivers on your machine, GPU will not be used.\n",
|
| 87 |
+
"2024-01-04 12:24:18.290802: I tensorflow/core/platform/cpu_feature_guard.cc:182] This TensorFlow binary is optimized to use available CPU instructions in performance-critical operations.\n",
|
| 88 |
+
"To enable the following instructions: AVX2 AVX512F AVX512_VNNI FMA, in other operations, rebuild TensorFlow with the appropriate compiler flags.\n",
|
| 89 |
+
"2024-01-04 12:24:18.864121: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Could not find TensorRT\n"
|
| 90 |
+
]
|
| 91 |
+
}
|
| 92 |
+
],
|
| 93 |
+
"source": [
|
| 94 |
+
"import tensorflow as tf"
|
| 95 |
+
]
|
| 96 |
+
},
|
| 97 |
+
{
|
| 98 |
+
"cell_type": "code",
|
| 99 |
+
"execution_count": 7,
|
| 100 |
+
"metadata": {},
|
| 101 |
+
"outputs": [
|
| 102 |
+
{
|
| 103 |
+
"name": "stderr",
|
| 104 |
+
"output_type": "stream",
|
| 105 |
+
"text": [
|
| 106 |
+
"2024-01-04 12:24:20.808055: E external/local_xla/xla/stream_executor/cuda/cuda_driver.cc:274] failed call to cuInit: CUDA_ERROR_NO_DEVICE: no CUDA-capable device is detected\n"
|
| 107 |
+
]
|
| 108 |
+
}
|
| 109 |
+
],
|
| 110 |
+
"source": [
|
| 111 |
+
"model = tf.keras.models.load_model('artifacts/training/model.h5')"
|
| 112 |
+
]
|
| 113 |
+
},
|
| 114 |
+
{
|
| 115 |
+
"cell_type": "code",
|
| 116 |
+
"execution_count": 8,
|
| 117 |
+
"metadata": {},
|
| 118 |
+
"outputs": [],
|
| 119 |
+
"source": [
|
| 120 |
+
"from dataclasses import dataclass\n",
|
| 121 |
+
"from pathlib import Path\n",
|
| 122 |
+
"\n",
|
| 123 |
+
"@dataclass(frozen=True)\n",
|
| 124 |
+
"class EvaluationConfig:\n",
|
| 125 |
+
" path_of_model: Path\n",
|
| 126 |
+
" training_data: Path\n",
|
| 127 |
+
" all_params: dict\n",
|
| 128 |
+
" mlflow_uri: str\n",
|
| 129 |
+
" params_image_size: list\n",
|
| 130 |
+
" params_batch_size: int"
|
| 131 |
+
]
|
| 132 |
+
},
|
| 133 |
+
{
|
| 134 |
+
"cell_type": "code",
|
| 135 |
+
"execution_count": 9,
|
| 136 |
+
"metadata": {},
|
| 137 |
+
"outputs": [],
|
| 138 |
+
"source": [
|
| 139 |
+
"os.chdir('src/')"
|
| 140 |
+
]
|
| 141 |
+
},
|
| 142 |
+
{
|
| 143 |
+
"cell_type": "code",
|
| 144 |
+
"execution_count": 10,
|
| 145 |
+
"metadata": {},
|
| 146 |
+
"outputs": [],
|
| 147 |
+
"source": [
|
| 148 |
+
"from kidney_classification.constants import *\n",
|
| 149 |
+
"from kidney_classification.utils.common import read_yaml, create_directories, save_json"
|
| 150 |
+
]
|
| 151 |
+
},
|
| 152 |
+
{
|
| 153 |
+
"cell_type": "code",
|
| 154 |
+
"execution_count": 11,
|
| 155 |
+
"metadata": {},
|
| 156 |
+
"outputs": [],
|
| 157 |
+
"source": [
|
| 158 |
+
"class ConfigurationManager:\n",
|
| 159 |
+
" def __init__(\n",
|
| 160 |
+
" self,\n",
|
| 161 |
+
" config_filepath=CONFIG_FILE_PATH,\n",
|
| 162 |
+
" param_filepath=PARAMS_FILE_PATH\n",
|
| 163 |
+
" ):\n",
|
| 164 |
+
" self.config = read_yaml(config_filepath)\n",
|
| 165 |
+
" self.params = read_yaml(param_filepath)\n",
|
| 166 |
+
" create_directories([self.config.artifacts_root])\n",
|
| 167 |
+
"\n",
|
| 168 |
+
" def get_evaluation_config(self) -> EvaluationConfig:\n",
|
| 169 |
+
" eval_config = EvaluationConfig(\n",
|
| 170 |
+
" path_of_model=\"artifacts/training/model.h5\",\n",
|
| 171 |
+
" training_data=\"artifacts/data_ingestion/CT-KIDNEY-DATASET-Normal-Cyst-Tumor-Stone\",\n",
|
| 172 |
+
" mlflow_uri=\"https://dagshub.com/Shrey-patel-07/Kidney-Disease-Classifcation.mlflow\",\n",
|
| 173 |
+
" all_params=self.params,\n",
|
| 174 |
+
" params_image_size=self.params.IMAGE_SIZE,\n",
|
| 175 |
+
" params_batch_size=self.params.BATCH_SIZE\n",
|
| 176 |
+
" )\n",
|
| 177 |
+
" return eval_config"
|
| 178 |
+
]
|
| 179 |
+
},
|
| 180 |
+
{
|
| 181 |
+
"cell_type": "code",
|
| 182 |
+
"execution_count": 12,
|
| 183 |
+
"metadata": {},
|
| 184 |
+
"outputs": [],
|
| 185 |
+
"source": [
|
| 186 |
+
"import mlflow\n",
|
| 187 |
+
"import mlflow.keras\n",
|
| 188 |
+
"from urllib.parse import urlparse"
|
| 189 |
+
]
|
| 190 |
+
},
|
| 191 |
+
{
|
| 192 |
+
"cell_type": "code",
|
| 193 |
+
"execution_count": 13,
|
| 194 |
+
"metadata": {},
|
| 195 |
+
"outputs": [],
|
| 196 |
+
"source": [
|
| 197 |
+
"class Evaluation:\n",
|
| 198 |
+
" def __init__(self, config: EvaluationConfig):\n",
|
| 199 |
+
" self.config = config\n",
|
| 200 |
+
" self.valid_generator = None # Initialize to None\n",
|
| 201 |
+
"\n",
|
| 202 |
+
" def _valid_generator(self):\n",
|
| 203 |
+
" img_height, img_width = self.config.params_image_size[:-1]\n",
|
| 204 |
+
"\n",
|
| 205 |
+
" self.valid_generator = tf.keras.utils.image_dataset_from_directory(\n",
|
| 206 |
+
" self.config.training_data,\n",
|
| 207 |
+
" image_size=(img_height, img_width),\n",
|
| 208 |
+
" validation_split=0.30,\n",
|
| 209 |
+
" subset='validation',\n",
|
| 210 |
+
" seed=123\n",
|
| 211 |
+
" )\n",
|
| 212 |
+
"\n",
|
| 213 |
+
" self.valid_generator = self.valid_generator.map(lambda x, y: (x / 255, y))\n",
|
| 214 |
+
" AUTOTUNE = tf.data.AUTOTUNE\n",
|
| 215 |
+
" self.valid_generator = self.valid_generator.cache().prefetch(buffer_size=AUTOTUNE)\n",
|
| 216 |
+
"\n",
|
| 217 |
+
"\n",
|
| 218 |
+
" @staticmethod\n",
|
| 219 |
+
" def load_model(path: Path) -> tf.keras.Model:\n",
|
| 220 |
+
" return tf.keras.models.load_model(path)\n",
|
| 221 |
+
" \n",
|
| 222 |
+
"\n",
|
| 223 |
+
" def evaluation(self):\n",
|
| 224 |
+
" self.model = self.load_model(self.config.path_of_model)\n",
|
| 225 |
+
" self._valid_generator()\n",
|
| 226 |
+
" self.score = self.model.evaluate(self.valid_generator)\n",
|
| 227 |
+
" self.save_score()\n",
|
| 228 |
+
"\n",
|
| 229 |
+
" def save_score(self):\n",
|
| 230 |
+
" scores = {\"loss\": self.score[0], \"accuracy\": self.score[1]}\n",
|
| 231 |
+
" save_json(path=Path(\"scores.json\"), data=scores)\n",
|
| 232 |
+
"\n",
|
| 233 |
+
" def log_into_mlflow(self):\n",
|
| 234 |
+
" mlflow.set_registry_uri(self.config.mlflow_uri)\n",
|
| 235 |
+
" tracking_url_type_store = urlparse(mlflow.get_tracking_uri()).scheme\n",
|
| 236 |
+
" \n",
|
| 237 |
+
" with mlflow.start_run():\n",
|
| 238 |
+
" mlflow.log_params(self.config.all_params)\n",
|
| 239 |
+
" mlflow.log_metrics(\n",
|
| 240 |
+
" {\"loss\": self.score[0], \"accuracy\": self.score[1]}\n",
|
| 241 |
+
" )\n",
|
| 242 |
+
" # Model registry does not work with file store\n",
|
| 243 |
+
" if tracking_url_type_store != \"file\":\n",
|
| 244 |
+
"\n",
|
| 245 |
+
" # Register the model\n",
|
| 246 |
+
" mlflow.keras.log_model(self.model, \"model\", registered_model_name=\"VGG16Model\")\n",
|
| 247 |
+
" else:\n",
|
| 248 |
+
" mlflow.keras.log_model(self.model, \"model\")"
|
| 249 |
+
]
|
| 250 |
+
},
|
| 251 |
+
{
|
| 252 |
+
"cell_type": "code",
|
| 253 |
+
"execution_count": 14,
|
| 254 |
+
"metadata": {},
|
| 255 |
+
"outputs": [],
|
| 256 |
+
"source": [
|
| 257 |
+
"os.chdir('../')"
|
| 258 |
+
]
|
| 259 |
+
},
|
| 260 |
+
{
|
| 261 |
+
"cell_type": "code",
|
| 262 |
+
"execution_count": 15,
|
| 263 |
+
"metadata": {},
|
| 264 |
+
"outputs": [
|
| 265 |
+
{
|
| 266 |
+
"name": "stdout",
|
| 267 |
+
"output_type": "stream",
|
| 268 |
+
"text": [
|
| 269 |
+
"[2024-01-04 12:24:51,908: INFO: common yaml file: config/config.yaml loaded successfully]\n",
|
| 270 |
+
"[2024-01-04 12:24:51,910: INFO: common yaml file: params.yaml loaded successfully]\n",
|
| 271 |
+
"Found 12446 files belonging to 1 classes.\n",
|
| 272 |
+
"Using 3733 files for validation.\n",
|
| 273 |
+
"117/117 [==============================] - 147s 1s/step - loss: 3.2269e-06 - accuracy: 1.0000\n",
|
| 274 |
+
"[2024-01-04 12:27:19,911: INFO: common json file saved at: scores.json]\n",
|
| 275 |
+
"[2024-01-04 12:27:19,912: INFO: common json file saved at: scores.json]\n"
|
| 276 |
+
]
|
| 277 |
+
},
|
| 278 |
+
{
|
| 279 |
+
"name": "stderr",
|
| 280 |
+
"output_type": "stream",
|
| 281 |
+
"text": [
|
| 282 |
+
"2024/01/04 12:27:22 WARNING mlflow.tensorflow: You are saving a TensorFlow Core model or Keras model without a signature. Inference with mlflow.pyfunc.spark_udf() will not work unless the model's pyfunc representation accepts pandas DataFrames as inference inputs.\n"
|
| 283 |
+
]
|
| 284 |
+
},
|
| 285 |
+
{
|
| 286 |
+
"name": "stdout",
|
| 287 |
+
"output_type": "stream",
|
| 288 |
+
"text": [
|
| 289 |
+
"INFO:tensorflow:Assets written to: /tmp/tmpdrlg6a4p/model/data/model/assets\n",
|
| 290 |
+
"[2024-01-04 12:27:23,234: INFO: builder_impl Assets written to: /tmp/tmpdrlg6a4p/model/data/model/assets]\n"
|
| 291 |
+
]
|
| 292 |
+
},
|
| 293 |
+
{
|
| 294 |
+
"name": "stderr",
|
| 295 |
+
"output_type": "stream",
|
| 296 |
+
"text": [
|
| 297 |
+
"/home/shrey/Desktop/Kidney-Disease-Classifcation/env/lib/python3.11/site-packages/_distutils_hack/__init__.py:33: UserWarning: Setuptools is replacing distutils.\n",
|
| 298 |
+
" warnings.warn(\"Setuptools is replacing distutils.\")\n",
|
| 299 |
+
"Registered model 'VGG16Model' already exists. Creating a new version of this model...\n",
|
| 300 |
+
"2024/01/04 12:27:47 INFO mlflow.store.model_registry.abstract_store: Waiting up to 300 seconds for model version to finish creation. Model name: VGG16Model, version 3\n",
|
| 301 |
+
"Created version '3' of model 'VGG16Model'.\n"
|
| 302 |
+
]
|
| 303 |
+
}
|
| 304 |
+
],
|
| 305 |
+
"source": [
|
| 306 |
+
"try:\n",
|
| 307 |
+
" config = ConfigurationManager()\n",
|
| 308 |
+
" eval_config = config.get_evaluation_config()\n",
|
| 309 |
+
" evaluation = Evaluation(eval_config)\n",
|
| 310 |
+
" evaluation.evaluation()\n",
|
| 311 |
+
" evaluation.log_into_mlflow()\n",
|
| 312 |
+
" \n",
|
| 313 |
+
"except Exception as e:\n",
|
| 314 |
+
" raise e"
|
| 315 |
+
]
|
| 316 |
+
},
|
| 317 |
+
{
|
| 318 |
+
"cell_type": "code",
|
| 319 |
+
"execution_count": null,
|
| 320 |
+
"metadata": {},
|
| 321 |
+
"outputs": [],
|
| 322 |
+
"source": []
|
| 323 |
+
}
|
| 324 |
+
],
|
| 325 |
+
"metadata": {
|
| 326 |
+
"kernelspec": {
|
| 327 |
+
"display_name": "env",
|
| 328 |
+
"language": "python",
|
| 329 |
+
"name": "python3"
|
| 330 |
+
},
|
| 331 |
+
"language_info": {
|
| 332 |
+
"codemirror_mode": {
|
| 333 |
+
"name": "ipython",
|
| 334 |
+
"version": 3
|
| 335 |
+
},
|
| 336 |
+
"file_extension": ".py",
|
| 337 |
+
"mimetype": "text/x-python",
|
| 338 |
+
"name": "python",
|
| 339 |
+
"nbconvert_exporter": "python",
|
| 340 |
+
"pygments_lexer": "ipython3",
|
| 341 |
+
"version": "3.11.6"
|
| 342 |
+
}
|
| 343 |
+
},
|
| 344 |
+
"nbformat": 4,
|
| 345 |
+
"nbformat_minor": 2
|
| 346 |
+
}
|
Kidney-Disease-Classifcation/research/trials.ipynb
ADDED
|
@@ -0,0 +1,91 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "code",
|
| 5 |
+
"execution_count": 1,
|
| 6 |
+
"metadata": {},
|
| 7 |
+
"outputs": [],
|
| 8 |
+
"source": [
|
| 9 |
+
"import gdown"
|
| 10 |
+
]
|
| 11 |
+
},
|
| 12 |
+
{
|
| 13 |
+
"cell_type": "code",
|
| 14 |
+
"execution_count": 5,
|
| 15 |
+
"metadata": {},
|
| 16 |
+
"outputs": [],
|
| 17 |
+
"source": [
|
| 18 |
+
"url = \"https://drive.google.com/file/d/1CGXriP_nlctsaWVbz0W3mFdPK05lQmHK/view?usp=sharing\""
|
| 19 |
+
]
|
| 20 |
+
},
|
| 21 |
+
{
|
| 22 |
+
"cell_type": "code",
|
| 23 |
+
"execution_count": 6,
|
| 24 |
+
"metadata": {},
|
| 25 |
+
"outputs": [],
|
| 26 |
+
"source": [
|
| 27 |
+
"file_id = url.split(\"/\")[-2]"
|
| 28 |
+
]
|
| 29 |
+
},
|
| 30 |
+
{
|
| 31 |
+
"cell_type": "code",
|
| 32 |
+
"execution_count": 7,
|
| 33 |
+
"metadata": {},
|
| 34 |
+
"outputs": [
|
| 35 |
+
{
|
| 36 |
+
"name": "stderr",
|
| 37 |
+
"output_type": "stream",
|
| 38 |
+
"text": [
|
| 39 |
+
"Downloading...\n",
|
| 40 |
+
"From (uriginal): https://drive.google.com/uc?export=download&id=1CGXriP_nlctsaWVbz0W3mFdPK05lQmHK\n",
|
| 41 |
+
"From (redirected): https://drive.google.com/uc?export=download&id=1CGXriP_nlctsaWVbz0W3mFdPK05lQmHK&confirm=t&uuid=17666f87-2ddb-4a4f-a8fb-421ff82803bd\n",
|
| 42 |
+
"To: /home/shrey/Desktop/Kidney-Disease-Classifcation/research/kidney-CT-scan.zip\n",
|
| 43 |
+
"100%|██████████| 1.63G/1.63G [07:37<00:00, 3.56MB/s]\n"
|
| 44 |
+
]
|
| 45 |
+
},
|
| 46 |
+
{
|
| 47 |
+
"data": {
|
| 48 |
+
"text/plain": [
|
| 49 |
+
"'kidney-CT-scan.zip'"
|
| 50 |
+
]
|
| 51 |
+
},
|
| 52 |
+
"execution_count": 7,
|
| 53 |
+
"metadata": {},
|
| 54 |
+
"output_type": "execute_result"
|
| 55 |
+
}
|
| 56 |
+
],
|
| 57 |
+
"source": [
|
| 58 |
+
"prefix = \"https://drive.google.com/uc?export=download&id=\"\n",
|
| 59 |
+
"gdown.download(prefix + file_id, \"kidney-CT-scan.zip\", quiet=False)"
|
| 60 |
+
]
|
| 61 |
+
},
|
| 62 |
+
{
|
| 63 |
+
"cell_type": "code",
|
| 64 |
+
"execution_count": null,
|
| 65 |
+
"metadata": {},
|
| 66 |
+
"outputs": [],
|
| 67 |
+
"source": []
|
| 68 |
+
}
|
| 69 |
+
],
|
| 70 |
+
"metadata": {
|
| 71 |
+
"kernelspec": {
|
| 72 |
+
"display_name": "env",
|
| 73 |
+
"language": "python",
|
| 74 |
+
"name": "python3"
|
| 75 |
+
},
|
| 76 |
+
"language_info": {
|
| 77 |
+
"codemirror_mode": {
|
| 78 |
+
"name": "ipython",
|
| 79 |
+
"version": 3
|
| 80 |
+
},
|
| 81 |
+
"file_extension": ".py",
|
| 82 |
+
"mimetype": "text/x-python",
|
| 83 |
+
"name": "python",
|
| 84 |
+
"nbconvert_exporter": "python",
|
| 85 |
+
"pygments_lexer": "ipython3",
|
| 86 |
+
"version": "3.11.6"
|
| 87 |
+
}
|
| 88 |
+
},
|
| 89 |
+
"nbformat": 4,
|
| 90 |
+
"nbformat_minor": 2
|
| 91 |
+
}
|
Kidney-Disease-Classifcation/scores.json
ADDED
|
@@ -0,0 +1,4 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"loss": 0.013196375221014023,
|
| 3 |
+
"accuracy": 0.996370255947113
|
| 4 |
+
}
|
Kidney-Disease-Classifcation/setup.py
ADDED
|
@@ -0,0 +1,27 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import setuptools
|
| 2 |
+
|
| 3 |
+
with open("README.md", "r") as fh:
|
| 4 |
+
long_description = fh.read()
|
| 5 |
+
|
| 6 |
+
__version__ = "0.0.1"
|
| 7 |
+
|
| 8 |
+
REPO_NAME = "Kidney-Disease-Classification"
|
| 9 |
+
AUTHOR_NAME = "Shrey-patel-07"
|
| 10 |
+
SRC_REPO = "kidney_classification"
|
| 11 |
+
AUTHOR_EMAIL = "shrey07patel@gmail.com"
|
| 12 |
+
|
| 13 |
+
setuptools.setup(
|
| 14 |
+
name=SRC_REPO,
|
| 15 |
+
version=__version__,
|
| 16 |
+
author=AUTHOR_NAME,
|
| 17 |
+
author_email=AUTHOR_EMAIL,
|
| 18 |
+
description="A small package for kidney disease classification",
|
| 19 |
+
long_description=long_description,
|
| 20 |
+
long_description_content_type="text/markdown",
|
| 21 |
+
url=f"https://github.com/{AUTHOR_NAME}/{REPO_NAME}",
|
| 22 |
+
project_urls={
|
| 23 |
+
"Bug Tracker": f"https://github.com/{AUTHOR_NAME}/{REPO_NAME}/issues",
|
| 24 |
+
},
|
| 25 |
+
package_dir={"": "src"},
|
| 26 |
+
packages=setuptools.find_packages(where="src"),
|
| 27 |
+
)
|
Kidney-Disease-Classifcation/src/kidney_classification/__init__.py
ADDED
|
@@ -0,0 +1,18 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import sys
|
| 3 |
+
import logging
|
| 4 |
+
|
| 5 |
+
logging_str = "[%(asctime)s: %(levelname)s: %(module)s %(message)s]"
|
| 6 |
+
|
| 7 |
+
__name__ = "kidney_classification"
|
| 8 |
+
log_dir = "logs"
|
| 9 |
+
log_filepath = os.path.join(log_dir, "running_logs.log")
|
| 10 |
+
os.makedirs(log_dir, exist_ok=True)
|
| 11 |
+
|
| 12 |
+
logging.basicConfig(
|
| 13 |
+
level=logging.INFO,
|
| 14 |
+
format=logging_str,
|
| 15 |
+
handlers=[logging.FileHandler(log_filepath), logging.StreamHandler(sys.stdout)],
|
| 16 |
+
)
|
| 17 |
+
|
| 18 |
+
logger = logging.getLogger(__name__)
|
Kidney-Disease-Classifcation/src/kidney_classification/components/__init__.py
ADDED
|
File without changes
|
Kidney-Disease-Classifcation/src/kidney_classification/components/data_ingestion.py
ADDED
|
@@ -0,0 +1,45 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import zipfile
|
| 3 |
+
import gdown
|
| 4 |
+
from kidney_classification import logger
|
| 5 |
+
from kidney_classification.entity.config_entity import DataIngestionConfig
|
| 6 |
+
|
| 7 |
+
|
| 8 |
+
class DataIngestion:
|
| 9 |
+
def __init__(self, config: DataIngestionConfig):
|
| 10 |
+
self.config = config
|
| 11 |
+
|
| 12 |
+
def download_file(self) -> str:
|
| 13 |
+
"""
|
| 14 |
+
Fetch data from the url
|
| 15 |
+
"""
|
| 16 |
+
|
| 17 |
+
try:
|
| 18 |
+
dataset_url = self.config.source_URL
|
| 19 |
+
zip_download_dir = self.config.local_data_file
|
| 20 |
+
os.makedirs("artifacts/data_ingestion", exist_ok=True)
|
| 21 |
+
logger.info(
|
| 22 |
+
f"Downloading data from {dataset_url} into file {zip_download_dir}"
|
| 23 |
+
)
|
| 24 |
+
|
| 25 |
+
file_id = dataset_url.split("/")[-2]
|
| 26 |
+
prefix = "https://drive.google.com/uc?/export=download&id="
|
| 27 |
+
gdown.download(prefix + file_id, zip_download_dir)
|
| 28 |
+
|
| 29 |
+
logger.info(
|
| 30 |
+
f"Downloaded data from {dataset_url} into file {zip_download_dir}"
|
| 31 |
+
)
|
| 32 |
+
|
| 33 |
+
except Exception as e:
|
| 34 |
+
raise e
|
| 35 |
+
|
| 36 |
+
def extract_zip_file(self):
|
| 37 |
+
"""
|
| 38 |
+
zip_file_path: str
|
| 39 |
+
Extracts the zip file into the data directory
|
| 40 |
+
Function returns None
|
| 41 |
+
"""
|
| 42 |
+
unzip_path = self.config.unzip_dir
|
| 43 |
+
os.makedirs(unzip_path, exist_ok=True)
|
| 44 |
+
with zipfile.ZipFile(self.config.local_data_file, "r") as zip_ref:
|
| 45 |
+
zip_ref.extractall(unzip_path)
|
Kidney-Disease-Classifcation/src/kidney_classification/components/model_evaluation_mlflow.py
ADDED
|
@@ -0,0 +1,59 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import tensorflow as tf
|
| 2 |
+
from urllib.parse import urlparse
|
| 3 |
+
import mlflow
|
| 4 |
+
import mlflow.keras
|
| 5 |
+
from pathlib import Path
|
| 6 |
+
from kidney_classification.utils.common import save_json
|
| 7 |
+
from kidney_classification.entity.config_entity import EvaluationConfig
|
| 8 |
+
|
| 9 |
+
|
| 10 |
+
class Evaluation:
|
| 11 |
+
def __init__(self, config: EvaluationConfig):
|
| 12 |
+
self.config = config
|
| 13 |
+
self.valid_generator = None # Initialize to None
|
| 14 |
+
|
| 15 |
+
def _valid_generator(self):
|
| 16 |
+
img_height, img_width = self.config.params_image_size[:-1]
|
| 17 |
+
|
| 18 |
+
self.valid_generator = tf.keras.utils.image_dataset_from_directory(
|
| 19 |
+
self.config.training_data,
|
| 20 |
+
image_size=(img_height, img_width),
|
| 21 |
+
validation_split=0.30,
|
| 22 |
+
subset="validation",
|
| 23 |
+
seed=123,
|
| 24 |
+
)
|
| 25 |
+
|
| 26 |
+
self.valid_generator = self.valid_generator.map(lambda x, y: (x / 255, y))
|
| 27 |
+
AUTOTUNE = tf.data.AUTOTUNE
|
| 28 |
+
self.valid_generator = self.valid_generator.cache().prefetch(
|
| 29 |
+
buffer_size=AUTOTUNE
|
| 30 |
+
)
|
| 31 |
+
|
| 32 |
+
@staticmethod
|
| 33 |
+
def load_model(path: Path) -> tf.keras.Model:
|
| 34 |
+
return tf.keras.models.load_model(path)
|
| 35 |
+
|
| 36 |
+
def evaluation(self):
|
| 37 |
+
self.model = self.load_model(self.config.path_of_model)
|
| 38 |
+
self._valid_generator()
|
| 39 |
+
self.score = self.model.evaluate(self.valid_generator)
|
| 40 |
+
self.save_score()
|
| 41 |
+
|
| 42 |
+
def save_score(self):
|
| 43 |
+
scores = {"loss": self.score[0], "accuracy": self.score[1]}
|
| 44 |
+
save_json(path=Path("scores.json"), data=scores)
|
| 45 |
+
|
| 46 |
+
def log_into_mlflow(self):
|
| 47 |
+
mlflow.set_registry_uri(self.config.mlflow_uri)
|
| 48 |
+
tracking_url_type_store = urlparse(mlflow.get_tracking_uri()).scheme
|
| 49 |
+
|
| 50 |
+
with mlflow.start_run():
|
| 51 |
+
mlflow.log_params(self.config.all_params)
|
| 52 |
+
mlflow.log_metrics({"loss": self.score[0], "accuracy": self.score[1]})
|
| 53 |
+
# Model registry does not work with file store
|
| 54 |
+
if tracking_url_type_store != "file":
|
| 55 |
+
mlflow.keras.log_model(
|
| 56 |
+
self.model, "model", registered_model_name="VGG16Model"
|
| 57 |
+
)
|
| 58 |
+
else:
|
| 59 |
+
mlflow.keras.log_model(self.model, "model")
|
Kidney-Disease-Classifcation/src/kidney_classification/components/model_training.py
ADDED
|
@@ -0,0 +1,49 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import tensorflow as tf
|
| 2 |
+
from pathlib import Path
|
| 3 |
+
from kidney_classification.entity.config_entity import TrainingConfig
|
| 4 |
+
|
| 5 |
+
|
| 6 |
+
class Training:
|
| 7 |
+
def __init__(self, config: TrainingConfig):
|
| 8 |
+
self.config = config
|
| 9 |
+
|
| 10 |
+
def get_base_model(self):
|
| 11 |
+
self.model = tf.keras.models.load_model(self.config.updated_base_model_path)
|
| 12 |
+
|
| 13 |
+
def train_valid_generator(self):
|
| 14 |
+
img_height, img_width = self.config.params_image_size[:-1]
|
| 15 |
+
|
| 16 |
+
train = tf.keras.utils.image_dataset_from_directory(
|
| 17 |
+
self.config.training_data,
|
| 18 |
+
image_size=(img_height, img_width),
|
| 19 |
+
validation_split=0.1,
|
| 20 |
+
subset="training",
|
| 21 |
+
seed=123,
|
| 22 |
+
)
|
| 23 |
+
|
| 24 |
+
val = tf.keras.utils.image_dataset_from_directory(
|
| 25 |
+
self.config.training_data,
|
| 26 |
+
image_size=(img_height, img_width),
|
| 27 |
+
validation_split=0.2,
|
| 28 |
+
subset="validation",
|
| 29 |
+
seed=123,
|
| 30 |
+
)
|
| 31 |
+
train = train.map(lambda x, y: (x / 255, y))
|
| 32 |
+
val = val.map(lambda x, y: (x / 255, y))
|
| 33 |
+
AUTOTUNE = tf.data.AUTOTUNE
|
| 34 |
+
|
| 35 |
+
self.train_dataset = train.cache().prefetch(buffer_size=AUTOTUNE)
|
| 36 |
+
self.val_dataset = val.cache().prefetch(buffer_size=AUTOTUNE)
|
| 37 |
+
|
| 38 |
+
@staticmethod
|
| 39 |
+
def save_model(path: Path, model: tf.keras.Model):
|
| 40 |
+
model.save(path)
|
| 41 |
+
|
| 42 |
+
def define_and_train_model(self):
|
| 43 |
+
self.model.fit(
|
| 44 |
+
self.train_dataset,
|
| 45 |
+
validation_data=self.val_dataset,
|
| 46 |
+
epochs=self.config.params_epochs,
|
| 47 |
+
)
|
| 48 |
+
|
| 49 |
+
self.save_model(path=self.config.trained_model_path, model=self.model)
|
Kidney-Disease-Classifcation/src/kidney_classification/components/prepare_base_model.py
ADDED
|
@@ -0,0 +1,46 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import tensorflow as tf
|
| 2 |
+
from pathlib import Path
|
| 3 |
+
from kidney_classification.entity.config_entity import PrepareBaseModelConfig
|
| 4 |
+
from tensorflow.keras.models import Sequential
|
| 5 |
+
from tensorflow.keras.layers import Dense, Flatten, BatchNormalization, Dropout
|
| 6 |
+
|
| 7 |
+
|
| 8 |
+
class PrepareBaseModel:
|
| 9 |
+
@staticmethod
|
| 10 |
+
def prepare_full_model():
|
| 11 |
+
VGG_model = Sequential()
|
| 12 |
+
|
| 13 |
+
pretrained_model = tf.keras.applications.VGG16(
|
| 14 |
+
include_top=False,
|
| 15 |
+
input_shape=(150, 150, 3),
|
| 16 |
+
pooling="max",
|
| 17 |
+
classes=4,
|
| 18 |
+
weights="imagenet",
|
| 19 |
+
)
|
| 20 |
+
|
| 21 |
+
VGG_model.add(pretrained_model)
|
| 22 |
+
VGG_model.add(Flatten())
|
| 23 |
+
VGG_model.add(Dense(512, activation="relu"))
|
| 24 |
+
VGG_model.add(BatchNormalization())
|
| 25 |
+
VGG_model.add(Dropout(0.5))
|
| 26 |
+
|
| 27 |
+
VGG_model.add(Dense(4, activation="softmax"))
|
| 28 |
+
pretrained_model.trainable = False
|
| 29 |
+
|
| 30 |
+
VGG_model.compile(
|
| 31 |
+
optimizer="adam",
|
| 32 |
+
loss="sparse_categorical_crossentropy",
|
| 33 |
+
metrics=["accuracy"],
|
| 34 |
+
)
|
| 35 |
+
|
| 36 |
+
return VGG_model
|
| 37 |
+
|
| 38 |
+
def update_base_model(self, config: PrepareBaseModelConfig):
|
| 39 |
+
full_model = self.prepare_full_model()
|
| 40 |
+
|
| 41 |
+
full_model.summary()
|
| 42 |
+
full_model.save(config.updated_base_model_path)
|
| 43 |
+
|
| 44 |
+
@staticmethod
|
| 45 |
+
def save_model(path: Path, model: tf.keras.Model):
|
| 46 |
+
model.save(path)
|
Kidney-Disease-Classifcation/src/kidney_classification/config/__init__.py
ADDED
|
File without changes
|
Kidney-Disease-Classifcation/src/kidney_classification/config/configuration.py
ADDED
|
@@ -0,0 +1,83 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
from kidney_classification.constants import *
|
| 3 |
+
from kidney_classification.utils.common import read_yaml, create_directories, save_json
|
| 4 |
+
from kidney_classification.entity.config_entity import (
|
| 5 |
+
DataIngestionConfig,
|
| 6 |
+
PrepareBaseModelConfig,
|
| 7 |
+
TrainingConfig,
|
| 8 |
+
EvaluationConfig,
|
| 9 |
+
)
|
| 10 |
+
|
| 11 |
+
|
| 12 |
+
class ConfigurationManager:
|
| 13 |
+
def __init__(
|
| 14 |
+
self, config_filepath=CONFIG_FILE_PATH, params_filepath=PARAMS_FILE_PATH
|
| 15 |
+
):
|
| 16 |
+
self.config = read_yaml(config_filepath)
|
| 17 |
+
self.params = read_yaml(params_filepath)
|
| 18 |
+
|
| 19 |
+
create_directories([self.config.artifacts_root])
|
| 20 |
+
|
| 21 |
+
def get_data_ingestion_config(self) -> DataIngestionConfig:
|
| 22 |
+
config = self.config.data_ingestion
|
| 23 |
+
|
| 24 |
+
create_directories([config.root_dir])
|
| 25 |
+
|
| 26 |
+
data_ingestion_config = DataIngestionConfig(
|
| 27 |
+
root_dir=config.root_dir,
|
| 28 |
+
source_URL=config.source_URL,
|
| 29 |
+
local_data_file=config.local_data_file,
|
| 30 |
+
unzip_dir=config.unzip_dir,
|
| 31 |
+
)
|
| 32 |
+
|
| 33 |
+
return data_ingestion_config
|
| 34 |
+
|
| 35 |
+
def get_prepare_base_model_config(self) -> PrepareBaseModelConfig:
|
| 36 |
+
config = self.config.prepare_base_model
|
| 37 |
+
|
| 38 |
+
create_directories([config.root_dir])
|
| 39 |
+
|
| 40 |
+
prepare_base_model_config = PrepareBaseModelConfig(
|
| 41 |
+
root_dir=Path(config.root_dir),
|
| 42 |
+
base_model_path=Path(config.base_model_path),
|
| 43 |
+
updated_base_model_path=Path(config.updated_base_model_path),
|
| 44 |
+
params_image_size=self.params.IMAGE_SIZE,
|
| 45 |
+
params_include_top=self.params.INCLUDE_TOP,
|
| 46 |
+
params_weights=self.params.WEIGHTS,
|
| 47 |
+
params_classes=self.params.CLASSES,
|
| 48 |
+
)
|
| 49 |
+
|
| 50 |
+
return prepare_base_model_config
|
| 51 |
+
|
| 52 |
+
def get_training_config(self) -> TrainingConfig:
|
| 53 |
+
training = self.config.training
|
| 54 |
+
prepare_base_model = self.config.prepare_base_model
|
| 55 |
+
params = self.params
|
| 56 |
+
training_data = os.path.join(
|
| 57 |
+
self.config.data_ingestion.unzip_dir,
|
| 58 |
+
"CT-KIDNEY-DATASET-Normal-Cyst-Tumor-Stone",
|
| 59 |
+
)
|
| 60 |
+
create_directories([Path(training.root_dir)])
|
| 61 |
+
|
| 62 |
+
training_config = TrainingConfig(
|
| 63 |
+
root_dir=Path(training.root_dir),
|
| 64 |
+
trained_model_path=Path(training.trained_model_path),
|
| 65 |
+
updated_base_model_path=Path(prepare_base_model.updated_base_model_path),
|
| 66 |
+
training_data=Path(training_data),
|
| 67 |
+
params_epochs=params.EPOCHS,
|
| 68 |
+
params_batch_size=params.BATCH_SIZE,
|
| 69 |
+
params_image_size=params.IMAGE_SIZE,
|
| 70 |
+
)
|
| 71 |
+
|
| 72 |
+
return training_config
|
| 73 |
+
|
| 74 |
+
def get_evaluation_config(self) -> EvaluationConfig:
|
| 75 |
+
eval_config = EvaluationConfig(
|
| 76 |
+
path_of_model="artifacts/training/model.h5",
|
| 77 |
+
training_data="artifacts/data_ingestion/CT-KIDNEY-DATASET-Normal-Cyst-Tumor-Stone",
|
| 78 |
+
mlflow_uri="https://dagshub.com/Shrey-patel-07/Kidney-Disease-Classifcation.mlflow",
|
| 79 |
+
all_params=self.params,
|
| 80 |
+
params_image_size=self.params.IMAGE_SIZE,
|
| 81 |
+
params_batch_size=self.params.BATCH_SIZE,
|
| 82 |
+
)
|
| 83 |
+
return eval_config
|
Kidney-Disease-Classifcation/src/kidney_classification/constants/__init__.py
ADDED
|
@@ -0,0 +1,4 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from pathlib import Path
|
| 2 |
+
|
| 3 |
+
CONFIG_FILE_PATH = Path("config/config.yaml")
|
| 4 |
+
PARAMS_FILE_PATH = Path("params.yaml")
|
Kidney-Disease-Classifcation/src/kidney_classification/entity/__init__.py
ADDED
|
File without changes
|
Kidney-Disease-Classifcation/src/kidney_classification/entity/config_entity.py
ADDED
|
@@ -0,0 +1,42 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from dataclasses import dataclass
|
| 2 |
+
from pathlib import Path
|
| 3 |
+
|
| 4 |
+
|
| 5 |
+
@dataclass(frozen=True)
|
| 6 |
+
class DataIngestionConfig:
|
| 7 |
+
root_dir: Path
|
| 8 |
+
source_URL: str
|
| 9 |
+
local_data_file: Path
|
| 10 |
+
unzip_dir: Path
|
| 11 |
+
|
| 12 |
+
|
| 13 |
+
@dataclass(frozen=True)
|
| 14 |
+
class PrepareBaseModelConfig:
|
| 15 |
+
root_dir: Path
|
| 16 |
+
base_model_path: Path
|
| 17 |
+
updated_base_model_path: Path
|
| 18 |
+
params_image_size: list
|
| 19 |
+
params_include_top: bool
|
| 20 |
+
params_weights: str
|
| 21 |
+
params_classes: int
|
| 22 |
+
|
| 23 |
+
|
| 24 |
+
@dataclass(frozen=True)
|
| 25 |
+
class TrainingConfig:
|
| 26 |
+
root_dir: Path
|
| 27 |
+
trained_model_path: Path
|
| 28 |
+
updated_base_model_path: Path
|
| 29 |
+
training_data: Path
|
| 30 |
+
params_epochs: int
|
| 31 |
+
params_batch_size: int
|
| 32 |
+
params_image_size: list
|
| 33 |
+
|
| 34 |
+
|
| 35 |
+
@dataclass(frozen=True)
|
| 36 |
+
class EvaluationConfig:
|
| 37 |
+
path_of_model: Path
|
| 38 |
+
training_data: Path
|
| 39 |
+
all_params: dict
|
| 40 |
+
mlflow_uri: str
|
| 41 |
+
params_image_size: list
|
| 42 |
+
params_batch_size: int
|
Kidney-Disease-Classifcation/src/kidney_classification/pipeline/__init__.py
ADDED
|
File without changes
|
Kidney-Disease-Classifcation/src/kidney_classification/pipeline/prediction.py
ADDED
|
@@ -0,0 +1,31 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import numpy as np
|
| 2 |
+
from tensorflow.keras.models import load_model
|
| 3 |
+
from tensorflow.keras.preprocessing import image
|
| 4 |
+
import os
|
| 5 |
+
|
| 6 |
+
|
| 7 |
+
class PredictionPipeline:
|
| 8 |
+
def __init__(self, filename):
|
| 9 |
+
self.filename = filename
|
| 10 |
+
|
| 11 |
+
def predict(self):
|
| 12 |
+
model = load_model(os.path.join("model", "model.h5"))
|
| 13 |
+
|
| 14 |
+
imagename = self.filename
|
| 15 |
+
test_image = image.load_img(imagename, target_size=(150, 150))
|
| 16 |
+
test_image = image.img_to_array(test_image)
|
| 17 |
+
test_image = np.expand_dims(test_image, axis=0)
|
| 18 |
+
result = np.argmax(model.predict(test_image), axis=1)
|
| 19 |
+
|
| 20 |
+
if result[0] == 0:
|
| 21 |
+
prediction = "Cyst"
|
| 22 |
+
return [{"image": prediction}]
|
| 23 |
+
elif result[0] == 1:
|
| 24 |
+
prediction = "Normal"
|
| 25 |
+
return [{"image": prediction}]
|
| 26 |
+
elif result[0] == 2:
|
| 27 |
+
prediction = "Stone"
|
| 28 |
+
return [{"image": prediction}]
|
| 29 |
+
else:
|
| 30 |
+
prediction = "Tumor"
|
| 31 |
+
return [{"image": prediction}]
|
Kidney-Disease-Classifcation/src/kidney_classification/pipeline/stage_01_data_ingestion.py
ADDED
|
@@ -0,0 +1,31 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import sys
|
| 2 |
+
|
| 3 |
+
sys.path.append("./src")
|
| 4 |
+
from kidney_classification.config.configuration import ConfigurationManager
|
| 5 |
+
from kidney_classification.components.data_ingestion import DataIngestion
|
| 6 |
+
from kidney_classification import logger
|
| 7 |
+
|
| 8 |
+
STAGE_NAME = "Data Ingestion"
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
class DataIngestionTrainingPipeline:
|
| 12 |
+
def __init__(self):
|
| 13 |
+
pass
|
| 14 |
+
|
| 15 |
+
def main(self):
|
| 16 |
+
config = ConfigurationManager()
|
| 17 |
+
data_ingestion_config = config.get_data_ingestion_config()
|
| 18 |
+
data_ingestion = DataIngestion(config=data_ingestion_config)
|
| 19 |
+
data_ingestion.download_file()
|
| 20 |
+
data_ingestion.extract_zip_file()
|
| 21 |
+
|
| 22 |
+
|
| 23 |
+
if __name__ == "__main__":
|
| 24 |
+
try:
|
| 25 |
+
logger.info(f"-------------Running stage: {STAGE_NAME}-------------")
|
| 26 |
+
pipeline = DataIngestionTrainingPipeline()
|
| 27 |
+
pipeline.main()
|
| 28 |
+
logger.info(f"-------------Stage: {STAGE_NAME} completed-------------")
|
| 29 |
+
except Exception as e:
|
| 30 |
+
logger.exception(e)
|
| 31 |
+
raise e
|
Kidney-Disease-Classifcation/src/kidney_classification/pipeline/stage_02_prepare_base_model.py
ADDED
|
@@ -0,0 +1,30 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import sys
|
| 2 |
+
|
| 3 |
+
sys.path.append("./src")
|
| 4 |
+
from kidney_classification.config.configuration import ConfigurationManager
|
| 5 |
+
from kidney_classification.components.prepare_base_model import PrepareBaseModel
|
| 6 |
+
from kidney_classification import logger
|
| 7 |
+
|
| 8 |
+
STAGE_NAME = "Prepare base model"
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
class PrepareBaseModelTrainingPipeline:
|
| 12 |
+
def __init__(self):
|
| 13 |
+
pass
|
| 14 |
+
|
| 15 |
+
def main(self):
|
| 16 |
+
config = ConfigurationManager()
|
| 17 |
+
prepare_base_model_config = config.get_prepare_base_model_config()
|
| 18 |
+
prepare_base_model = PrepareBaseModel()
|
| 19 |
+
prepare_base_model.update_base_model(prepare_base_model_config)
|
| 20 |
+
|
| 21 |
+
|
| 22 |
+
if __name__ == "__main__":
|
| 23 |
+
try:
|
| 24 |
+
logger.info(f"-------------Running stage: {STAGE_NAME}-------------")
|
| 25 |
+
pipeline = PrepareBaseModelTrainingPipeline()
|
| 26 |
+
pipeline.main()
|
| 27 |
+
logger.info(f"-------------Stage: {STAGE_NAME} completed-------------")
|
| 28 |
+
except Exception as e:
|
| 29 |
+
logger.exception(e)
|
| 30 |
+
raise e
|
Kidney-Disease-Classifcation/src/kidney_classification/pipeline/stage_03_model_training.py
ADDED
|
@@ -0,0 +1,32 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import sys
|
| 2 |
+
|
| 3 |
+
sys.path.append("./src")
|
| 4 |
+
from kidney_classification.config.configuration import ConfigurationManager
|
| 5 |
+
from kidney_classification.components.model_training import Training
|
| 6 |
+
from kidney_classification import logger
|
| 7 |
+
|
| 8 |
+
STAGE_NAME = "Training Model"
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
class ModelTrainingPipeline:
|
| 12 |
+
def __init__(self):
|
| 13 |
+
pass
|
| 14 |
+
|
| 15 |
+
def main(self):
|
| 16 |
+
config = ConfigurationManager()
|
| 17 |
+
training_config = config.get_training_config()
|
| 18 |
+
training = Training(config=training_config)
|
| 19 |
+
training.get_base_model()
|
| 20 |
+
training.train_valid_generator()
|
| 21 |
+
training.define_and_train_model()
|
| 22 |
+
|
| 23 |
+
|
| 24 |
+
if __name__ == "__main__":
|
| 25 |
+
try:
|
| 26 |
+
logger.info(f"-------------Running stage: {STAGE_NAME}-------------")
|
| 27 |
+
pipeline = ModelTrainingPipeline()
|
| 28 |
+
pipeline.main()
|
| 29 |
+
logger.info(f"-------------Stage: {STAGE_NAME} completed-------------")
|
| 30 |
+
except Exception as e:
|
| 31 |
+
logger.exception(e)
|
| 32 |
+
raise e
|
Kidney-Disease-Classifcation/src/kidney_classification/pipeline/stage_04_model_evaluation_with_mlflow.py
ADDED
|
@@ -0,0 +1,32 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import sys
|
| 2 |
+
|
| 3 |
+
sys.path.append("./src")
|
| 4 |
+
from kidney_classification.config.configuration import ConfigurationManager
|
| 5 |
+
from kidney_classification.components.model_evaluation_mlflow import Evaluation
|
| 6 |
+
from kidney_classification import logger
|
| 7 |
+
|
| 8 |
+
|
| 9 |
+
STAGE_NAME = "Evaluation Stage"
|
| 10 |
+
|
| 11 |
+
|
| 12 |
+
class EvaluationPipeline:
|
| 13 |
+
def __init__(self):
|
| 14 |
+
pass
|
| 15 |
+
|
| 16 |
+
def main(self):
|
| 17 |
+
config = ConfigurationManager()
|
| 18 |
+
eval_config = config.get_evaluation_config()
|
| 19 |
+
evaluation = Evaluation(eval_config)
|
| 20 |
+
evaluation.evaluation()
|
| 21 |
+
evaluation.log_into_mlflow()
|
| 22 |
+
|
| 23 |
+
|
| 24 |
+
if __name__ == "__main__":
|
| 25 |
+
try:
|
| 26 |
+
logger.info(f"-------------Running stage: {STAGE_NAME}-------------")
|
| 27 |
+
pipeline = EvaluationPipeline()
|
| 28 |
+
pipeline.main()
|
| 29 |
+
logger.info(f"-------------Stage: {STAGE_NAME} completed-------------")
|
| 30 |
+
except Exception as e:
|
| 31 |
+
logger.exception(e)
|
| 32 |
+
raise e
|
Kidney-Disease-Classifcation/src/kidney_classification/utils/__init__.py
ADDED
|
File without changes
|
Kidney-Disease-Classifcation/src/kidney_classification/utils/common.py
ADDED
|
@@ -0,0 +1,79 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import json
|
| 3 |
+
import yaml
|
| 4 |
+
import base64
|
| 5 |
+
import joblib
|
| 6 |
+
from typing import Any
|
| 7 |
+
from pathlib import Path
|
| 8 |
+
from box import ConfigBox
|
| 9 |
+
from ensure import ensure_annotations
|
| 10 |
+
from box.exceptions import BoxValueError
|
| 11 |
+
from kidney_classification import logger
|
| 12 |
+
|
| 13 |
+
|
| 14 |
+
@ensure_annotations
|
| 15 |
+
def read_yaml(path_to_yaml: Path) -> ConfigBox:
|
| 16 |
+
"""reads yaml file and returns
|
| 17 |
+
|
| 18 |
+
Args:
|
| 19 |
+
path_to_yaml (str): path like input
|
| 20 |
+
|
| 21 |
+
Raises:
|
| 22 |
+
ValueError: if yaml file is empty
|
| 23 |
+
e: empty file
|
| 24 |
+
|
| 25 |
+
Returns:
|
| 26 |
+
ConfigBox: ConfigBox type
|
| 27 |
+
"""
|
| 28 |
+
try:
|
| 29 |
+
with open(path_to_yaml) as yaml_file:
|
| 30 |
+
content = yaml.safe_load(yaml_file)
|
| 31 |
+
logger.info(f"yaml file: {path_to_yaml} loaded successfully")
|
| 32 |
+
return ConfigBox(content)
|
| 33 |
+
except BoxValueError:
|
| 34 |
+
raise ValueError("yaml file is empty")
|
| 35 |
+
except Exception as e:
|
| 36 |
+
raise e
|
| 37 |
+
|
| 38 |
+
|
| 39 |
+
@ensure_annotations
|
| 40 |
+
def create_directories(path_to_directories: list, verbose=True):
|
| 41 |
+
"""create list of directories
|
| 42 |
+
|
| 43 |
+
Args:
|
| 44 |
+
path_to_directories (list): list of path of directories
|
| 45 |
+
ignore_log (bool, optional): ignore if multiple dirs is to be created. Defaults to False.
|
| 46 |
+
"""
|
| 47 |
+
for path in path_to_directories:
|
| 48 |
+
if not os.path.exists(path):
|
| 49 |
+
os.makedirs(path)
|
| 50 |
+
if verbose:
|
| 51 |
+
logger.info(f"created directory at: {path}")
|
| 52 |
+
else:
|
| 53 |
+
continue
|
| 54 |
+
|
| 55 |
+
|
| 56 |
+
def decodeImage(imgstring, fileName):
|
| 57 |
+
imgdata = base64.b64decode(imgstring)
|
| 58 |
+
with open(fileName, "wb") as f:
|
| 59 |
+
f.write(imgdata)
|
| 60 |
+
f.close()
|
| 61 |
+
|
| 62 |
+
|
| 63 |
+
def encodeImageIntoBase64(croppedImagePath):
|
| 64 |
+
with open(croppedImagePath, "rb") as f:
|
| 65 |
+
return base64.b64encode(f.read())
|
| 66 |
+
|
| 67 |
+
|
| 68 |
+
@ensure_annotations
|
| 69 |
+
def save_json(path: Path, data: dict):
|
| 70 |
+
"""save json data
|
| 71 |
+
|
| 72 |
+
Args:
|
| 73 |
+
path (Path): path to json file
|
| 74 |
+
data (dict): data to be saved in json file
|
| 75 |
+
"""
|
| 76 |
+
with open(path, "w") as f:
|
| 77 |
+
json.dump(data, f, indent=4)
|
| 78 |
+
|
| 79 |
+
logger.info(f"json file saved at: {path}")
|
Kidney-Disease-Classifcation/templates.py
ADDED
|
@@ -0,0 +1,44 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
from pathlib import Path
|
| 3 |
+
import logging
|
| 4 |
+
|
| 5 |
+
# logging string
|
| 6 |
+
logging.basicConfig(level=logging.INFO, format="[%(asctime)s]: %(message)s:")
|
| 7 |
+
|
| 8 |
+
project_name = "kidney_classification"
|
| 9 |
+
|
| 10 |
+
list_of_files = [
|
| 11 |
+
".github/workflows/.gitkeep",
|
| 12 |
+
f"src/{project_name}/__init__.py",
|
| 13 |
+
f"src/{project_name}/components/__init__.py",
|
| 14 |
+
f"src/{project_name}/utils/__init__.py",
|
| 15 |
+
f"src/{project_name}/config/__init__.py",
|
| 16 |
+
f"src/{project_name}/config/configuration.py",
|
| 17 |
+
f"src/{project_name}/pipeline/__init__.py",
|
| 18 |
+
f"src/{project_name}/entity/__init__.py",
|
| 19 |
+
f"src/{project_name}/constants/__init__.py",
|
| 20 |
+
"config/config.yaml",
|
| 21 |
+
"dvc.yaml",
|
| 22 |
+
"params.yaml",
|
| 23 |
+
"requirements.txt",
|
| 24 |
+
"setup.py",
|
| 25 |
+
"research/trials.ipynb",
|
| 26 |
+
"templates/index.html",
|
| 27 |
+
]
|
| 28 |
+
|
| 29 |
+
|
| 30 |
+
for filepath in list_of_files:
|
| 31 |
+
filepath = Path(filepath)
|
| 32 |
+
filedir, filename = os.path.split(filepath)
|
| 33 |
+
|
| 34 |
+
if filedir != "":
|
| 35 |
+
os.makedirs(filedir, exist_ok=True)
|
| 36 |
+
logging.info(f"Creating directory; {filedir} for the file: {filename}")
|
| 37 |
+
|
| 38 |
+
if (not os.path.exists(filepath)) or (os.path.getsize(filepath) == 0):
|
| 39 |
+
with open(filepath, "w") as f:
|
| 40 |
+
pass
|
| 41 |
+
logging.info(f"Creating empty file: {filepath}")
|
| 42 |
+
|
| 43 |
+
else:
|
| 44 |
+
logging.info(f"{filename} is already exists")
|
Kidney-Disease-Classifcation/templates/dagshub-kidney_disease_classification.png
ADDED

|
Kidney-Disease-Classifcation/templates/index.html
ADDED
|
@@ -0,0 +1,353 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
<!DOCTYPE html>
|
| 2 |
+
<html lang="en">
|
| 3 |
+
|
| 4 |
+
<head>
|
| 5 |
+
<meta charset="UTF-8">
|
| 6 |
+
<meta name="viewport" content="width=device-width, initial-scale=1.0">
|
| 7 |
+
<meta http-equiv="X-UA-Compatible" content="ie=edge">
|
| 8 |
+
<title>Kidney Disease Classification</title>
|
| 9 |
+
<link rel="stylesheet" href="https://maxcdn.bootstrapcdn.com/bootstrap/4.0.0/css/bootstrap.min.css"
|
| 10 |
+
integrity="sha384-Gn5384xqQ1aoWXA+058RXPxPg6fy4IWvTNh0E263XmFcJlSAwiGgFAW/dAiS6JXm" crossorigin="anonymous">
|
| 11 |
+
<link rel="stylesheet" href="https://cdnjs.cloudflare.com/ajax/libs/font-awesome/5.15.4/css/all.min.css" />
|
| 12 |
+
<style>
|
| 13 |
+
body {
|
| 14 |
+
background-color: #eff2f9;
|
| 15 |
+
}
|
| 16 |
+
|
| 17 |
+
.iupload h3 {
|
| 18 |
+
color: #1b2d6b;
|
| 19 |
+
font-size: 30px;
|
| 20 |
+
font-weight: 700;
|
| 21 |
+
}
|
| 22 |
+
|
| 23 |
+
.img-part {
|
| 24 |
+
height: 300px;
|
| 25 |
+
width: 300px;
|
| 26 |
+
margin: 0px auto;
|
| 27 |
+
}
|
| 28 |
+
|
| 29 |
+
.image-part {
|
| 30 |
+
height: 300px;
|
| 31 |
+
width: 300px;
|
| 32 |
+
border: 1px solid #1b2d6b;
|
| 33 |
+
}
|
| 34 |
+
|
| 35 |
+
.image-part img {
|
| 36 |
+
position: absolute;
|
| 37 |
+
height: 300px;
|
| 38 |
+
width: 300px;
|
| 39 |
+
display: none;
|
| 40 |
+
padding: 5px;
|
| 41 |
+
}
|
| 42 |
+
|
| 43 |
+
.image-part #video {
|
| 44 |
+
display: block;
|
| 45 |
+
height: 300px;
|
| 46 |
+
width: 300px;
|
| 47 |
+
padding: 5px;
|
| 48 |
+
}
|
| 49 |
+
|
| 50 |
+
.res-part {
|
| 51 |
+
border: 1px solid #dedede;
|
| 52 |
+
margin-left: 20px;
|
| 53 |
+
height: 310px;
|
| 54 |
+
width: 100%;
|
| 55 |
+
padding: 5px;
|
| 56 |
+
margin: 0px auto;
|
| 57 |
+
overflow: auto;
|
| 58 |
+
}
|
| 59 |
+
|
| 60 |
+
.res-part2 {
|
| 61 |
+
border: 1px solid #dedede;
|
| 62 |
+
height: 310px;
|
| 63 |
+
width: 100%;
|
| 64 |
+
padding: 5px;
|
| 65 |
+
margin: 0px auto;
|
| 66 |
+
}
|
| 67 |
+
|
| 68 |
+
.resp-img {
|
| 69 |
+
height: 298px;
|
| 70 |
+
width: 233px;
|
| 71 |
+
margin: 0px auto;
|
| 72 |
+
}
|
| 73 |
+
|
| 74 |
+
.jsonRes {
|
| 75 |
+
margin-left: 30px;
|
| 76 |
+
}
|
| 77 |
+
|
| 78 |
+
#send {
|
| 79 |
+
cursor: pointer;
|
| 80 |
+
}
|
| 81 |
+
|
| 82 |
+
.btn-part {
|
| 83 |
+
width: 325px;
|
| 84 |
+
}
|
| 85 |
+
|
| 86 |
+
textarea,
|
| 87 |
+
select,
|
| 88 |
+
.form-control,
|
| 89 |
+
.custom-select,
|
| 90 |
+
button.btn,
|
| 91 |
+
.btn-primary,
|
| 92 |
+
input[type="text"],
|
| 93 |
+
input[type="url"],
|
| 94 |
+
.uneditable-input {
|
| 95 |
+
border: 1px solid #363e75;
|
| 96 |
+
outline: 0 !important;
|
| 97 |
+
border-radius: 0px;
|
| 98 |
+
box-shadow: none;
|
| 99 |
+
-webkit-box-shadow: none;
|
| 100 |
+
-moz-box-shadow: none;
|
| 101 |
+
}
|
| 102 |
+
|
| 103 |
+
textarea:focus,
|
| 104 |
+
select:focus,
|
| 105 |
+
.form-control:focus,
|
| 106 |
+
.btn:focus,
|
| 107 |
+
.btn-primary:focus,
|
| 108 |
+
.custom-select:focus,
|
| 109 |
+
input[type="text"]:focus,
|
| 110 |
+
.uneditable-input:focus {
|
| 111 |
+
border: 1px solid #007bff;
|
| 112 |
+
outline: 0 !important;
|
| 113 |
+
border-radius: 0px;
|
| 114 |
+
box-shadow: none;
|
| 115 |
+
-webkit-box-shadow: none;
|
| 116 |
+
-moz-box-shadow: none;
|
| 117 |
+
}
|
| 118 |
+
|
| 119 |
+
#loading {
|
| 120 |
+
position: fixed;
|
| 121 |
+
left: 0px;
|
| 122 |
+
top: 0px;
|
| 123 |
+
width: 100%;
|
| 124 |
+
height: 100%;
|
| 125 |
+
z-index: 9999999999;
|
| 126 |
+
overflow: hidden;
|
| 127 |
+
background: rgba(255, 255, 255, 0.7);
|
| 128 |
+
}
|
| 129 |
+
|
| 130 |
+
.loader {
|
| 131 |
+
border: 8px solid #f3f3f3;
|
| 132 |
+
border-top: 8px solid #363e75;
|
| 133 |
+
border-radius: 50%;
|
| 134 |
+
width: 60px;
|
| 135 |
+
height: 60px;
|
| 136 |
+
left: 50%;
|
| 137 |
+
margin-left: -4em;
|
| 138 |
+
display: block;
|
| 139 |
+
animation: spin 2s linear infinite;
|
| 140 |
+
}
|
| 141 |
+
|
| 142 |
+
.loader,
|
| 143 |
+
.loader:after {
|
| 144 |
+
display: block;
|
| 145 |
+
position: absolute;
|
| 146 |
+
top: 50%;
|
| 147 |
+
margin-top: -4.05em;
|
| 148 |
+
}
|
| 149 |
+
|
| 150 |
+
@keyframes spin {
|
| 151 |
+
0% {
|
| 152 |
+
transform: rotate(0deg);
|
| 153 |
+
}
|
| 154 |
+
|
| 155 |
+
100% {
|
| 156 |
+
transform: rotate(360deg);
|
| 157 |
+
}
|
| 158 |
+
}
|
| 159 |
+
|
| 160 |
+
.right-part {
|
| 161 |
+
border: 1px solid #dedede;
|
| 162 |
+
padding: 5px;
|
| 163 |
+
}
|
| 164 |
+
|
| 165 |
+
.socials {
|
| 166 |
+
position: fixed;
|
| 167 |
+
bottom: 20px;
|
| 168 |
+
left: 0;
|
| 169 |
+
width: 100%;
|
| 170 |
+
display: flex;
|
| 171 |
+
justify-content: center;
|
| 172 |
+
align-items: center;
|
| 173 |
+
}
|
| 174 |
+
|
| 175 |
+
.icons {
|
| 176 |
+
display: flex;
|
| 177 |
+
justify-content: center;
|
| 178 |
+
align-items: center;
|
| 179 |
+
}
|
| 180 |
+
|
| 181 |
+
.icons a {
|
| 182 |
+
margin: 0 10px;
|
| 183 |
+
color: #333;
|
| 184 |
+
font-size: 24px;
|
| 185 |
+
}
|
| 186 |
+
|
| 187 |
+
.credit {
|
| 188 |
+
position: absolute;
|
| 189 |
+
bottom: 0;
|
| 190 |
+
left: 10px;
|
| 191 |
+
color: #333;
|
| 192 |
+
}
|
| 193 |
+
</style>
|
| 194 |
+
</head>
|
| 195 |
+
|
| 196 |
+
<body>
|
| 197 |
+
<div class="main container">
|
| 198 |
+
<section class="iupload">
|
| 199 |
+
<h3 class="text-center py-4">Object Classification</h3>
|
| 200 |
+
<div class="row">
|
| 201 |
+
<div class="img-part col-md-6">
|
| 202 |
+
<div class="image-part">
|
| 203 |
+
<video autoplay id="video"
|
| 204 |
+
poster="https://img.freepik.com/free-vector/group-young-people-posing-photo_52683-18824.jpg?size=338&ext=jpg"></video>
|
| 205 |
+
<img src="" id="photo">
|
| 206 |
+
<canvas style="display:none;" id="canvas"></canvas>
|
| 207 |
+
</div>
|
| 208 |
+
<div class="btn-part">
|
| 209 |
+
<form id="upload-data pt-3" class="">
|
| 210 |
+
<div class="input-group mt-3 row">
|
| 211 |
+
<button type="button" class="btn btn-primary col-md-5 col-xs-5 ml-3 mr-4"
|
| 212 |
+
id="uload">Upload</button>
|
| 213 |
+
<button id="send" type="button"
|
| 214 |
+
class="btn btn-success col-md-5 col-xs-5">Predict</button>
|
| 215 |
+
</div>
|
| 216 |
+
|
| 217 |
+
<input type="hidden" class="form-control mr-2" id="url" placeholder="Enter REST Api url..."
|
| 218 |
+
value="../predict" />
|
| 219 |
+
<input name="upload" type="file" id="fileinput"
|
| 220 |
+
style="position:absolute;top:-500px;" /><br />
|
| 221 |
+
</form>
|
| 222 |
+
</div>
|
| 223 |
+
</div>
|
| 224 |
+
<div class="col-md-6 col-xs-12 right-part">
|
| 225 |
+
<h5 class="mb-2">
|
| 226 |
+
<center>Prediction Results</center>
|
| 227 |
+
</h5>
|
| 228 |
+
<div class="row">
|
| 229 |
+
<div class="res-part col-md-5 col-xs-12">
|
| 230 |
+
<div class="jsonRes"></div>
|
| 231 |
+
</div>
|
| 232 |
+
</div>
|
| 233 |
+
</div>
|
| 234 |
+
</div>
|
| 235 |
+
</section>
|
| 236 |
+
</div>
|
| 237 |
+
<div class="socials">
|
| 238 |
+
<div class="credit">Designed by Shrey Patel</div>
|
| 239 |
+
<div class="icons">
|
| 240 |
+
<a href="https://www.linkedin.com/in/shreypatel07" target="_blank">
|
| 241 |
+
<i class="fab fa-linkedin"></i>
|
| 242 |
+
</a>
|
| 243 |
+
<a href="https://github.com/Shrey-patel-07" target="_blank">
|
| 244 |
+
<i class="fab fa-github"></i>
|
| 245 |
+
</a>
|
| 246 |
+
<a href="https://x.com/Shrey_Patel_07" target="_blank">
|
| 247 |
+
<i class="fab fa-twitter"></i>
|
| 248 |
+
</a>
|
| 249 |
+
</div>
|
| 250 |
+
</div>
|
| 251 |
+
|
| 252 |
+
|
| 253 |
+
<div id="loading">
|
| 254 |
+
<div class="loader"></div>
|
| 255 |
+
</div>
|
| 256 |
+
<script src="https://ajax.googleapis.com/ajax/libs/jquery/3.4.1/jquery.min.js"></script>
|
| 257 |
+
<script src="https://cdnjs.cloudflare.com/ajax/libs/popper.js/1.12.9/umd/popper.min.js"
|
| 258 |
+
integrity="sha384-ApNbgh9B+Y1QKtv3Rn7W3mgPxhU9K/ScQsAP7hUibX39j7fakFPskvXusvfa0b4Q"
|
| 259 |
+
crossorigin="anonymous"></script>
|
| 260 |
+
<script src="https://maxcdn.bootstrapcdn.com/bootstrap/4.0.0/js/bootstrap.min.js"
|
| 261 |
+
integrity="sha384-JZR6Spejh4U02d8jOt6vLEHfe/JQGiRRSQQxSfFWpi1MquVdAyjUar5+76PVCmYl"
|
| 262 |
+
crossorigin="anonymous"></script>
|
| 263 |
+
|
| 264 |
+
<script>
|
| 265 |
+
var mybtn = document.getElementById('startbtn');
|
| 266 |
+
var myvideo = document.getElementById('video');
|
| 267 |
+
var mycanvas = document.getElementById('canvas');
|
| 268 |
+
var myphoto = document.getElementById('photo');
|
| 269 |
+
var base_data = "";
|
| 270 |
+
|
| 271 |
+
function sendRequest(base64Data) {
|
| 272 |
+
var type = "json";
|
| 273 |
+
if (base64Data != "" || base64Data != null) {
|
| 274 |
+
if (type == "imgtobase") {
|
| 275 |
+
$(".res-part").html("");
|
| 276 |
+
$(".res-part").html(base64Data);
|
| 277 |
+
}
|
| 278 |
+
else if (type == "basetoimg") {
|
| 279 |
+
var imageData = $("#imgstring").val();
|
| 280 |
+
$(".res-part").html("");
|
| 281 |
+
$(".res-part").append("<img src='data:image/jpeg;base64," + imageData + "' alt='' />");
|
| 282 |
+
}
|
| 283 |
+
else {
|
| 284 |
+
var url = $("#url").val();
|
| 285 |
+
$("#loading").show();
|
| 286 |
+
$.ajax({
|
| 287 |
+
url: url,
|
| 288 |
+
type: "post",
|
| 289 |
+
cache: false,
|
| 290 |
+
async: true,
|
| 291 |
+
crossDomain: true,
|
| 292 |
+
headers: {
|
| 293 |
+
'Content-Type': 'application/json',
|
| 294 |
+
'Access-Control-Allow-Origin': '*'
|
| 295 |
+
},
|
| 296 |
+
data: JSON.stringify({ image: base64Data }),
|
| 297 |
+
success: function (res) {
|
| 298 |
+
$(".res-part").html("");
|
| 299 |
+
$(".res-part2").html("");
|
| 300 |
+
try {
|
| 301 |
+
var imageData = res[1].image;
|
| 302 |
+
if (imageData.length > 100) {
|
| 303 |
+
if (imageData.length > 10) { $(".res-part2").append("<img class='resp-img' src='data:image/jpeg;base64," + imageData + "' alt='' />"); }
|
| 304 |
+
}
|
| 305 |
+
} catch (e) { }
|
| 306 |
+
$(".res-part").html("<pre>" + res[0].image + "</pre>");
|
| 307 |
+
$("#loading").hide();
|
| 308 |
+
}
|
| 309 |
+
});
|
| 310 |
+
}
|
| 311 |
+
}
|
| 312 |
+
}
|
| 313 |
+
|
| 314 |
+
$(document).ready(function () {
|
| 315 |
+
$("#loading").hide();
|
| 316 |
+
|
| 317 |
+
$('#send').click(function (evt) {
|
| 318 |
+
sendRequest(base_data);
|
| 319 |
+
});
|
| 320 |
+
|
| 321 |
+
$('#uload').click(function (evt) {
|
| 322 |
+
$('#fileinput').focus().trigger('click');
|
| 323 |
+
});
|
| 324 |
+
$("#fileinput").change(function () {
|
| 325 |
+
if (this.files && this.files[0]) {
|
| 326 |
+
var reader = new FileReader();
|
| 327 |
+
reader.onload = function (e) {
|
| 328 |
+
var url = e.target.result;
|
| 329 |
+
var img = new Image();
|
| 330 |
+
img.crossOrigin = 'Anonymous';
|
| 331 |
+
img.onload = function () {
|
| 332 |
+
var canvas = document.createElement('CANVAS');
|
| 333 |
+
var ctx = canvas.getContext('2d');
|
| 334 |
+
canvas.height = this.height;
|
| 335 |
+
canvas.width = this.width;
|
| 336 |
+
ctx.drawImage(this, 0, 0);
|
| 337 |
+
base_data = canvas.toDataURL('image/jpeg', 1.0).replace(/^data:image.+;base64,/, '');
|
| 338 |
+
canvas = null;
|
| 339 |
+
};
|
| 340 |
+
img.src = url;
|
| 341 |
+
$('#photo').attr('src', url);
|
| 342 |
+
$('#photo').show();
|
| 343 |
+
$('#video').hide();
|
| 344 |
+
}
|
| 345 |
+
reader.readAsDataURL(this.files[0]);
|
| 346 |
+
}
|
| 347 |
+
});
|
| 348 |
+
});
|
| 349 |
+
|
| 350 |
+
</script>
|
| 351 |
+
</body>
|
| 352 |
+
|
| 353 |
+
</html>
|
Kidney-Disease-Classifcation/templates/kidney_ctscan.png
ADDED

|
Kidney-Disease-Classifcation/templates/mlflow-kidney_disease_classification.png
ADDED
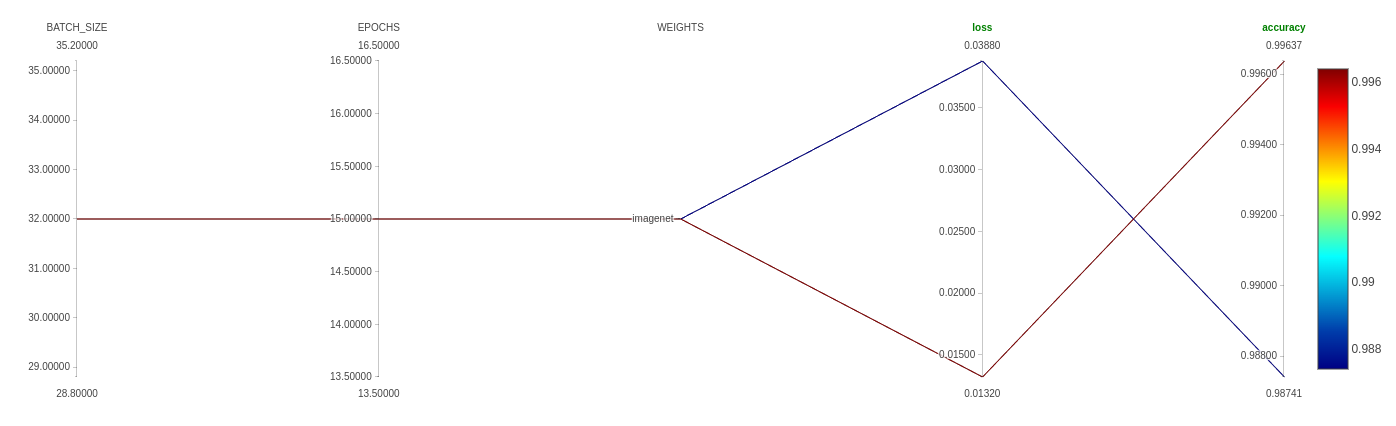
|