
This article revisits the metaphor of the genome as a computer program, a concept first proposed publicly by the author in 1995. Drawing on historical discussions in computational biology, including previously unpublished exchanges from the bionet.genome.chromosome newsgroup, we explore how the genome functions not merely as a passive database of genes but as an active, logic-driven computational system. The genome executes massively parallel processes—driven by environmental inputs, chemical conditions, and internal state—using a computational architecture fundamentally different from conventional computing. From early visual metaphors in Mendelian genetics to contemporary logic circuits in synthetic biology, this paper traces the historical development of computational models that express genomic logic, while critically examining both the utility and limitations of the program metaphor. We conclude that the genome represents a unique computational paradigm that could inform the development of novel computing architectures and artificial intelligence systems.
Target Audience: This article is written for researchers and enthusiasts in computational biology, synthetic biology, artificial intelligence, and related fields. While some background in biology or computer science is helpful, we provide explanations and analogies to make the concepts accessible to interdisciplinary audiences.
Biological processes have often been described through metaphor: the cell as a factory, DNA as a blueprint, and most provocatively—the genome as a computer program. Unlike static descriptions, this metaphor opens the door to seeing life itself as computation: a dynamic process with inputs, logic conditions, iterative loops, subroutines, and termination conditions.
In 1995, the author explored this idea in an essay published in The X Advisor, proposing that gene regulation could be modeled as a logic program. That same year, in discussions on the bionet.genome.chromosome newsgroup, computational biologists including Robert Robbins of Johns Hopkins University developed this metaphor further, exploring profound differences between genomic and conventional computation. This article revisits and expands that vision through both historical analysis and modern advances in biology and AI.
As we will explore, the genome-as-program metaphor provides valuable insights but also requires us to stretch conventional computational thinking into new paradigms—ones that might ultimately inform the future of computing itself.
The visualization of biological logic began with Gregor Mendel in the 19th century. Though his work predates formal computational thinking, Mendel's charts—showing ratios of inherited traits—used symbolic logic to track biological outcomes. Later, chromosome theory and operon models introduced control diagrams that represented genetic regulatory mechanisms.
The Punnett square, named after British geneticist Reginald Punnett (1875-1967), represents one of the earliest systematic approaches to modeling genetic inheritance as a computational process. Punnett, a collaborator of William Bateson (1861-1926) who coined the term "genetics" and was a key figure in establishing genetics as a scientific discipline, developed this visualization method to predict the outcomes of genetic crosses. The square format provides a systematic way to compute all possible combinations of parental alleles, making it one of the first "genetic algorithms" in computational biology.
The Punnett square in Figure 1 demonstrates a monohybrid cross between two heterozygous parents (Aa × Aa). Each cell in the 2×2 grid represents a possible genotype outcome, with the probability of each outcome determined by the rules of Mendelian inheritance. This systematic enumeration of possibilities mirrors the truth table approach used in digital logic design, where all possible input combinations are explicitly listed to determine output states.
The computational logic underlying the Punnett square can be expressed through Boolean operations. Consider a simple genetic system where allele A is dominant and allele a is recessive. The phenotypic expression follows these logical rules:
Dominance Logic (OR operation):
Phenotype = A OR A = Dominant trait
This follows the logical rule: if either allele is A, the dominant phenotype is expressed.
Recessive Logic (AND operation):
Phenotype = a AND a = Recessive trait
This follows the logical rule: only if both alleles are a is the recessive phenotype expressed.
The Punnett square can be extended to more complex genetic systems. For example, a dihybrid cross (AaBb × AaBb) creates a 4×4 grid with 16 possible combinations, demonstrating how genetic complexity scales exponentially with the number of genes involved. This combinatorial explosion is a fundamental characteristic of genetic computation that distinguishes it from simple linear processes.
The logical structure of Mendelian inheritance can be formalized using truth tables, similar to those used in digital circuit design:
Truth Table for Dominant/Recessive Inheritance:
| Allele 1 | Allele 2 | Genotype | Phenotype | Logic |
|---|---|---|---|---|
| A | A | AA | Dominant | 1 OR 1 = 1 |
| A | a | Aa | Dominant | 1 OR 0 = 1 |
| a | A | aA | Dominant | 0 OR 1 = 1 |
| a | a | aa | Recessive | 0 AND 0 = 0 |
This truth table approach reveals that genetic inheritance operates through fundamental logical operations: OR for dominance (presence of dominant allele) and AND for recessiveness (absence of dominant alleles). These same logical operations form the basis of digital computation, establishing a direct parallel between genetic and computational logic.
The Punnett square method demonstrates several key principles of genetic computation: (1) systematic enumeration of possibilities, (2) probabilistic outcomes based on combinatorial rules, (3) hierarchical organization of genetic information, and (4) the ability to predict complex outcomes from simple rules. These principles would later be formalized in computational genetics and serve as the foundation for modern genetic algorithms and evolutionary computation.

The transition from Mendelian genetics to molecular biology in the mid-20th century marked a crucial evolution in computational thinking about biological systems. This period saw the emergence of sophisticated models that explicitly treated genetic regulation as a computational process, moving beyond simple inheritance patterns to complex regulatory networks.
In the 1960s, François Jacob and Jacques Monod's lac operon model introduced a logic gate–like system for regulating gene expression, paving the way for computational thinking in molecular biology. This revolutionary model showed how gene expression could be controlled through what resembled conditional logic, establishing the foundation for understanding genetic regulation as a computational process.
Jacob and Monod's work on the lac operon in Escherichia coli revealed a sophisticated regulatory system that operates through logical principles. The operon consists of three structural genes (lacZ, lacY, lacA) that are coordinately regulated by a single promoter and operator region. The system responds to two environmental inputs: the presence of lactose (the substrate) and the absence of glucose (the preferred energy source).
The computational logic of the lac operon can be expressed as a Boolean function:
Lac Operon Logic:
Expression = (Lactose present) AND (Glucose absent)
This logical function determines whether the operon is transcribed and the enzymes are produced.
The regulatory mechanism involves two key proteins: the lac repressor (encoded by lacI) and the catabolite activator protein (CAP). The lac repressor acts as a NOT gate—it binds to the operator and prevents transcription unless lactose is present. CAP acts as an AND gate—it enhances transcription only when glucose is absent. Together, these regulatory proteins implement a complex logical circuit that integrates multiple environmental signals.
The lac operon model demonstrated several key principles of biological computation: (1) the use of regulatory proteins as logic gates, (2) the integration of multiple inputs through logical operations, (3) the ability to respond to environmental conditions through conditional logic, and (4) the coordination of multiple genes through shared regulatory elements. These principles would later be formalized in computational models of gene regulatory networks and serve as the foundation for synthetic biology.
Jacob and Monod's work earned them the Nobel Prize in Physiology or Medicine in 1965, recognizing the profound implications of their discovery for understanding how genetic information is processed and regulated. Their model established the conceptual framework for viewing genetic regulation as a computational process, influencing generations of researchers in molecular biology and computational biology.

In April 1995, during the early days of the internet and computational biology, a significant exchange on the bionet.genome.chromosome newsgroup explored the genome-as-program metaphor in depth. This discussion occurred at a pivotal moment when the Human Genome Project was gaining momentum and computational approaches to biology were emerging as a new paradigm. The author initiated this discussion by asking whether "an organism's genome can be regarded as a computer program" and whether its structure could be represented as "a flowchart with genes as objects connected by logical terms."
Robert Robbins of Johns Hopkins University responded with a comprehensive analysis that both supported and complicated the metaphor. While acknowledging the digital nature of the genetic code, Robbins highlighted that the genome functions more like "a mass storage device" with properties not shared by electronic counterparts, and that genomic programs operate with unprecedented levels of parallelism—"in excess of 10^18 parallel processes" in the human body. These discussions represented one of the earliest sophisticated analyses of the computational nature of genomic function and laid the groundwork for modern computational biology approaches.
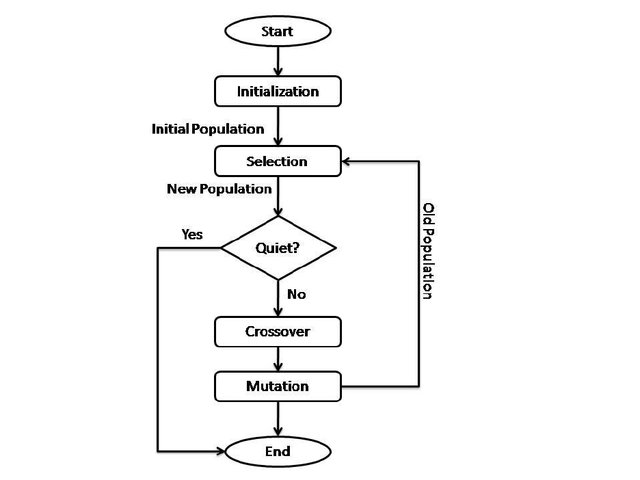
In 1995, the author's speculative essay proposed treating gene expression as an executing program with logical flow. To demonstrate this concept, the author created one of the first computational flowcharts representing gene regulation—a diagram of the lac operon's β-galactosidase expression system that explicitly modeled genetic regulation using programming logic constructs (see Figure 1).

This original flowchart depicted the lac operon as a decision tree with conditional branches, feedback loops, and termination conditions—showing how the presence or absence of lactose and glucose created logical pathways leading to different outcomes for β-galactosidase production. The diagram used programming-style logic gates (decision diamonds for yes/no conditions, process rectangles for actions) to represent biological regulatory mechanisms, making explicit the parallel between genetic circuits and computer logic circuits.
The article was featured on a bioinformatics resource list curated by Professor Inge Jonassen at the University of Bergen, where it appeared alongside foundational references like PubMed, In Silico Biology, and DNA Computers.
The use of flowcharts to represent biological processes has become increasingly sophisticated in modern computational biology. Contemporary flowcharts often integrate multiple data types, computational algorithms, and biological processes into unified visual representations. These modern flowcharts serve as computational roadmaps, guiding researchers through complex analytical pipelines and decision-making processes.
Modern biological flowcharts typically include several key elements: (1) data input nodes representing experimental or computational data sources, (2) processing nodes showing analytical algorithms or computational methods, (3) decision points representing conditional logic based on statistical thresholds or biological criteria, (4) output nodes displaying results or predictions, and (5) feedback loops showing iterative refinement processes. This structure mirrors the computational architecture of modern bioinformatics pipelines.
The flowchart in Figure 3.1 demonstrates a fascinating example of how biological metaphors have been adopted in computer science. This figure, from a network security paper (Al-Haija et al., 2014), shows a genetic algorithm flowchart that uses biological terminology—"thrive," "extinct," "mutate"—to describe computational processes for intrusion detection. This illustrates the profound influence of biological thinking on computational approaches, even in domains far removed from biology itself.
The use of biological metaphors in this network security application is particularly revealing. The algorithm treats potential security threats as a "population" that can "thrive" (successful attacks), "go extinct" (failed attacks), or "mutate" (evolve new attack strategies). This demonstrates how the genome-as-program metaphor has influenced computational thinking across multiple disciplines, creating a shared language between biological and computational systems.
This example shows that the computational principles underlying biological systems—population dynamics, selection pressure, adaptation, and evolution—have become fundamental tools in computer science. The fact that network security researchers chose biological terminology to describe their algorithms underscores the intuitive appeal and explanatory power of biological metaphors in computational contexts.
Since then, influential graphical systems have emerged for representing genomic data and processes: Martin Krzywinski's Circos (2009), Höhna's probabilistic phylogenetic networks (2014), Koutrouli's network visualizations (2020), and O'Donoghue's reviews (2018). These systems have grappled with the challenge of representing the multi-dimensional and massively parallel nature of genomic processes.
Martin Krzywinski's Circos visualization system represents a breakthrough in genomic data representation, using circular layouts to display complex multi-dimensional relationships between genomic regions. This innovative approach addresses the fundamental challenge of representing massive amounts of genomic data in an intuitive format, allowing researchers to identify patterns and relationships that would be impossible to see in linear representations. The circular layout enables the display of multiple data types simultaneously, making it an essential tool for modern comparative genomics and evolutionary studies. The Circos plot shows how different chromosomes (represented as segments around the circle) are connected by syntenic links (curved ribbons), revealing evolutionary relationships and structural variations that provide insights into genome evolution and organization.

Höhna et al.'s probabilistic phylogenetic networks represent a significant advancement in phylogenetic analysis, incorporating uncertainty and probabilistic relationships into evolutionary tree representations. This sophisticated approach acknowledges that biological processes are inherently stochastic and that our understanding of evolutionary relationships contains uncertainty. The model demonstrates how modern computational approaches can handle the inherent uncertainty in biological data, using probabilistic frameworks to represent evolutionary relationships rather than deterministic trees. This probabilistic approach has become essential for modern evolutionary biology and demonstrates how computational thinking has evolved to handle biological complexity, providing more realistic and nuanced representations of evolutionary processes.

Koutrouli et al.'s biological network visualization demonstrates how modern computational biology uses graph theory to model complex biological systems. This sophisticated network representation shows genes as nodes and their interactions as edges, revealing the intricate web of regulatory relationships that govern cellular processes. This network-based approach represents a fundamental shift from linear, sequential thinking to systems-level understanding of biological complexity. The graph structure allows researchers to identify hubs, modules, and emergent properties that would be invisible in traditional linear representations, acknowledging that biological systems are inherently networked and that understanding requires analysis of the entire system rather than individual components.

O'Donoghue et al.'s multi-dimensional biomedical data visualization represents a crucial advancement in handling the massive datasets generated by modern genomics. The heatmap format allows researchers to visualize complex multi-dimensional data in an intuitive color-coded format, where each cell represents the expression level of a gene under specific conditions. This approach enables the identification of expression patterns, clustering of genes with similar expression profiles, and the discovery of regulatory relationships across multiple conditions. The visualization demonstrates how computational methods can transform raw numerical data into meaningful biological insights, revealing patterns that would be impossible to detect through manual analysis. This approach has become essential for modern genomics, transcriptomics, and systems biology, enabling researchers to handle the complexity and scale of contemporary biological datasets.

Before we can understand genomic "programs," we must first understand the unique storage medium they operate on. As Robbins noted in 1995, the genome functions like a specialized mass storage device with properties unlike any electronic counterpart:
Unlike computer hard drives that store files at specific locations (like "sector 1, track 2"), the genome uses a smarter system called associative addressing. Think of it like a library where you find books by their content rather than their shelf position. As Robbins described it, "All addressing is associative, with multiple read heads scanning the device in parallel, looking for specific START LOADING HERE signals." This means the genome doesn't use absolute positions but rather characteristic patterns recognized by cellular machinery.
The genome resembles "a mass-storage device based on a linked-list architecture, rather than a physical platter." Information is encountered sequentially as cellular machinery moves along the DNA strand, with "pointers" in the form of regulatory sequences directing the machinery to relevant sections.
With diploid organisms possessing two sets of chromosomes, the genome exhibits built-in redundancy. However, as G. Dellaire noted in the 1995 discussions, mechanisms like imprinting and allelic silencing create a situation where "you only actually have one 'program' running" from certain loci, raising questions about "gene dosage" without clear parallels in conventional computing.
Dellaire also highlighted that "the actual structure of genome and not just the linear sequence may 'encode' sets of instructions for the 'reading and accessing' of this genetic code." This insight presaged modern understanding of epigenetics, chromatin structure, and the "histone code" as additional layers of information storage and processing.
Despite the differences in storage medium, the genome operates with recognizable computational logic structures:
The genome employs structures analogous to:
Bootloader: zygotic genome activation initiates development
Conditional logic: expression dependent on chemical signals
Loops: circadian cycles, metabolism, cell cycles
Subroutines: growth, repair, reproduction
Shutdown: apoptosis and programmed cell death
These resemble constructs such as IF-THEN, WHILE, SWITCH-CASE, and HALT in conventional computation.
At the molecular level, chemical reactions function as the basic operational units of genomic computation. These reactions operate through principles that can be understood as computational processes, though they differ fundamentally from digital computation in their analog, probabilistic nature.
Enzyme-Substrate Interactions as Logic Gates: Enzymes function as molecular logic gates, where the presence of specific substrates triggers catalytic reactions. These interactions follow Michaelis-Menten kinetics, creating sigmoidal response curves that resemble threshold logic functions. The enzyme's specificity for its substrate acts as a recognition mechanism, similar to how a logic gate responds only to specific input combinations.
Concentration Thresholds as Decision Points: Biological systems use concentration gradients and threshold mechanisms to make decisions. For example, the lac operon's response to lactose depends on the concentration of allolactose exceeding a critical threshold. These thresholds create binary-like decision points in otherwise continuous systems, enabling discrete logic-like behavior from analog chemical processes.
Feedback Loops as Iterative Processing: Biochemical feedback mechanisms implement iterative computational processes. Positive feedback creates amplification cascades (similar to computational scaling), while negative feedback provides stability and regulation. These loops can create oscillatory behavior, bistable switches, and other complex dynamics that resemble computational algorithms for pattern generation and control.
Signal Amplification as Computational Scaling: Biological systems use cascading reactions to amplify weak signals, similar to how computational systems use amplifiers and buffers. The phosphorylation cascade in signal transduction pathways, for example, can amplify a single extracellular signal into thousands of intracellular responses, demonstrating how biological systems achieve computational scaling through chemical mechanisms.
Stochastic Processes as Probabilistic Computation: Unlike deterministic digital computation, biological reactions are inherently stochastic. This probabilistic nature creates computational properties not found in conventional computing, including noise tolerance, adaptive responses, and emergent behaviors that arise from the statistical properties of molecular interactions.
Perhaps the most profound difference between genomic and conventional computation lies in the scale and nature of parallelism involved.
As Robbins calculated in 1995, "The expression of the human genome involves the simultaneous expression and (potential) interaction of something probably in excess of 10^18 parallel processes." This number derives from approximately 10^13 cells in the human body, each running 10^5-10^6 processes in parallel, with potential interactions between any processes in any cells.
This scale of parallelism is fundamentally different from any human-engineered computing system. To put this in perspective, the world's most powerful supercomputers operate with approximately 10^6-10^7 processing cores, while the human body operates with 10^18 parallel processes. This represents a difference of 11-12 orders of magnitude, making biological computation the most massively parallel system known to exist.
The implications of this scale are profound. Each cell in the human body is simultaneously executing thousands of biochemical reactions, processing environmental signals, maintaining homeostasis, and coordinating with neighboring cells. These processes are not merely concurrent but truly parallel, with each reaction occurring independently and simultaneously. The coordination between these processes emerges from the physical and chemical properties of the system rather than from centralized control mechanisms.
This massive parallelism enables biological systems to achieve computational capabilities that are impossible with sequential or even moderately parallel systems. For example, the immune system can simultaneously monitor for thousands of different pathogens, the nervous system can process multiple sensory inputs in real-time, and the metabolic system can maintain homeostasis across multiple organ systems simultaneously. These capabilities arise not from sophisticated algorithms but from the sheer scale of parallel processing available in biological systems.
Unlike computer "parallel processing" that often involves time-sharing a smaller number of processors, genomic parallelism involves true simultaneous execution: "each single cell has millions of programs executing in a truly parallel (i.e., independent execution, no time sharing) mode."
This distinction between true parallelism and time-sharing is crucial for understanding biological computation. In conventional computing, "parallel" systems typically use time-sharing, where a limited number of processors rapidly switch between different tasks, creating the illusion of simultaneous execution. Even modern multi-core processors use sophisticated scheduling algorithms to manage task allocation and context switching.
In contrast, biological systems achieve true parallelism through physical separation and chemical independence. Each molecule in a cell can react independently and simultaneously with other molecules, without requiring any scheduling or coordination mechanism. This independence arises from the fundamental properties of chemical reactions—each reaction occurs based on local conditions and molecular interactions, not on system-wide scheduling decisions.
This true parallelism has profound implications for system design and behavior. In time-shared systems, bottlenecks can occur when multiple processes compete for limited resources. In biological systems, such bottlenecks are rare because each process operates independently with its own local resources. This independence also means that biological systems are inherently fault-tolerant—the failure of one process does not necessarily affect others, and the system can continue operating even with significant component failures.
The absence of centralized control in biological systems is both a strength and a challenge. On one hand, it eliminates single points of failure and enables robust, adaptive behavior. On the other hand, it makes biological systems difficult to understand and predict, as their behavior emerges from the collective interactions of countless independent processes rather than from explicit algorithms or control structures.
Development begins with a specialized "bootloader" sequence that activates the zygotic genome after fertilization. This process transitions from maternal to zygotic control, initiates cascades of gene expression in precise sequence, establishes the initial conditions for all subsequent development, and creates a developmental trajectory with remarkable robustness.
The zygotic genome activation (ZGA) represents one of the most critical computational events in development. During early development, the embryo relies on maternal RNA and proteins deposited in the egg, but at a specific developmental stage, the zygotic genome "boots up" and begins transcribing its own genes. This transition is analogous to a computer bootloader that initializes the operating system, establishing the basic computational environment for all subsequent operations.
The bootloader process involves several computational elements that mirror those found in computer systems. First, there is a precise timing mechanism that determines when ZGA occurs—this timing is critical and must be coordinated with other developmental events. Second, there is a hierarchical activation sequence, where certain genes (often called "pioneer" genes) must be activated first to establish the conditions for subsequent gene expression. Third, there are feedback mechanisms that ensure the bootloader process is robust and can recover from errors or perturbations.
This bootloader analogy extends beyond the initial activation. Throughout development, there are multiple "reboot" events where cells transition between different developmental states. For example, during cellular differentiation, cells undergo transcriptional reprogramming that resembles a system reboot, where the cell's computational state is reset and a new program begins executing. These transitions are often triggered by specific signals or environmental conditions, similar to how computer systems can be configured to boot different operating systems based on user input or system state.
The robustness of the developmental bootloader is remarkable. Despite variations in environmental conditions, genetic background, and random molecular noise, development proceeds with remarkable consistency. This robustness suggests that the bootloader process has evolved sophisticated error-checking and recovery mechanisms, similar to those found in reliable computer systems. The ability to maintain developmental integrity despite perturbations is essential for the survival and reproduction of organisms, making the bootloader one of the most critical computational systems in biology.
This unprecedented parallelism enables emergent properties not found in sequential computing: robust error correction through redundant processes, self-organization without central control, pattern formation through reaction-diffusion dynamics, and adaptation to changing conditions without explicit programming.
Robust Error Correction Through Redundancy: Biological systems achieve remarkable reliability through massive redundancy rather than through precise error-free operation. Each cell contains multiple copies of critical genes, and many cellular processes have backup mechanisms that can compensate for failures. This redundancy is made possible by the massive parallelism of biological systems—if one process fails, others can take over without affecting overall system function. This approach to error correction is fundamentally different from conventional computing, where reliability is typically achieved through precise design and error detection rather than through redundancy.
Self-Organization Without Central Control: The massive parallelism of biological systems enables self-organization, where complex patterns and behaviors emerge from the collective interactions of many simple components. This self-organization occurs without any central controller or coordinator—each component follows simple local rules, and the overall system behavior emerges from their collective interactions. Examples include the formation of cellular patterns during development, the synchronization of circadian rhythms across multiple cells, and the coordination of immune responses across the body. This emergent behavior is a direct consequence of the massive parallelism and local interactions that characterize biological systems.
Pattern Formation Through Reaction-Diffusion Dynamics: The parallel nature of biological systems enables complex pattern formation through reaction-diffusion mechanisms. These patterns emerge from the interaction between chemical reactions (which create and destroy molecules) and diffusion (which spreads molecules through space). The classic example is Alan Turing's model of animal coat patterns, where simple chemical reactions occurring in parallel across a developing embryo create complex spatial patterns. These patterns emerge spontaneously from the parallel execution of simple chemical rules, demonstrating how massive parallelism can create complex, organized structures without explicit programming.
Adaptation Without Explicit Programming: Biological systems can adapt to changing conditions without any explicit programming for those conditions. This adaptation occurs through the parallel operation of many different processes, each responding to local conditions. When environmental conditions change, some processes may be enhanced while others are suppressed, leading to an overall adaptation of the system. This adaptive behavior emerges from the collective response of many parallel processes rather than from explicit algorithms for adaptation. The ability to adapt to novel conditions without explicit programming is one of the most remarkable properties of biological systems and is a direct consequence of their massive parallelism.
Collective Intelligence Through Distributed Processing: The massive parallelism of biological systems enables forms of collective intelligence that are impossible in sequential systems. For example, the immune system can simultaneously monitor for thousands of different pathogens, learn from encounters with new pathogens, and mount appropriate responses. This collective intelligence emerges from the parallel operation of many different cell types, each contributing specialized knowledge and capabilities to the overall system. The intelligence of the system as a whole exceeds the capabilities of any individual component, demonstrating how massive parallelism can create emergent computational capabilities.
One of Robbins' most profound insights was that genomic programs execute on virtual machines defined by other genomic programs.
"Genome programs execute on a virtual machine that is defined by some of the genomic programs that are executing. Thus, in trying to understand the genome, we are trying to reverse engineer binaries for an unknown CPU, in fact for a virtual CPU whose properties are encoded in the binaries we are trying to reverse engineer."
This insight reveals one of the most profound challenges in understanding biological computation. Unlike conventional computing, where the hardware (CPU, memory, etc.) is designed independently of the software that runs on it, in biological systems the "hardware" and "software" are co-evolved and mutually dependent. The cellular machinery that interprets the genome (the virtual machine) is itself encoded in the genome, creating a circular dependency that makes biological systems fundamentally different from engineered computing systems.
This self-defining nature has several important implications. First, it means that biological systems are inherently self-modifying—the programs can change the machine that executes them. This capability enables biological systems to adapt and evolve in ways that are impossible for conventional computers. For example, during development, cells can change their transcriptional machinery, modify their chromatin structure, and alter their metabolic networks, effectively reprogramming the virtual machine on which they run.
Second, this self-defining nature creates a fundamental challenge for reverse engineering. In conventional computing, we can understand a program by understanding the hardware it runs on. In biological systems, we must simultaneously understand both the program (the genome) and the machine (the cellular machinery), even though each depends on the other. This circular dependency makes biological systems much more difficult to understand and model than conventional computing systems.
Third, this self-defining nature enables biological systems to achieve levels of integration and optimization that are impossible in conventional computing. Because the hardware and software co-evolved, they are perfectly matched to each other, enabling biological systems to achieve remarkable efficiency and robustness. This integration also means that biological systems can adapt to new challenges by modifying both their programs and their execution environment simultaneously.
Unlike the deterministic operations of conventional computers, "genomic op codes are probabilistic, rather than deterministic. That is, when control hits a particular op code, there is a certain probability that a certain action will occur."
Think of it like rolling dice instead of flipping a light switch. Every biochemical reaction, every gene expression event, and every cellular process has an inherent element of randomness. This randomness is not a defect but a fundamental feature that enables unique capabilities.
The probabilistic nature arises from molecular chaos—molecules bouncing around randomly, transcription factors binding and unbinding, and constantly changing cellular conditions. This creates uncertainty about when and how biological operations will occur.
This probabilistic nature has profound implications. Biological systems must be robust to noise and uncertainty, and they can exploit randomness to achieve behaviors impossible in deterministic systems. For example, probabilistic gene expression enables cells to explore different states and adapt to changing conditions.
However, this also creates challenges for prediction. Unlike computers where the same inputs always produce the same outputs, biological systems can produce different outcomes even under identical conditions. This makes them harder to model but also more robust and adaptable.
This self-modifying, probabilistic system bears more resemblance to modern AI architectures than to conventional computing: Like neural networks, it operates with weighted probabilities; like reinforcement learning systems, it optimizes toward outcomes; like agent-based systems, it balances multiple objectives; unlike current AI, it developed through natural selection rather than design.
Neural Network Parallels: Biological systems operate through networks of interacting components that process information in parallel, similar to artificial neural networks. In both cases, the behavior of the system emerges from the collective activity of many simple processing units. However, biological networks are more sophisticated than artificial neural networks in several ways. They can modify their own structure and connectivity, they operate with multiple types of signals (chemical, electrical, mechanical), and they can change their computational properties based on context and experience.
Reinforcement Learning Analogies: Biological systems learn through trial and error, optimizing their behavior based on feedback from the environment. This learning process resembles reinforcement learning, where an agent learns to maximize rewards by exploring different actions and observing their consequences. However, biological reinforcement learning is more sophisticated than artificial versions, as it can modify not only its behavior but also its own learning mechanisms and objectives. This meta-learning capability enables biological systems to adapt their learning strategies to different environments and challenges.
Multi-Objective Optimization: Biological systems must balance multiple competing objectives simultaneously, such as growth, reproduction, survival, and energy efficiency. This multi-objective optimization is similar to the challenges faced by AI agents in complex environments. However, biological systems have evolved sophisticated mechanisms for balancing these objectives, including hierarchical control systems, priority-based decision making, and adaptive trade-offs that change based on environmental conditions.
Emergent Intelligence: The intelligence of biological systems emerges from the collective behavior of many simple components, rather than from a centralized control system. This emergent intelligence is similar to the behavior of swarm intelligence systems and multi-agent AI systems. However, biological systems achieve levels of coordination and cooperation that far exceed current artificial multi-agent systems, demonstrating how evolution can discover sophisticated solutions to complex coordination problems.
Adaptive Architecture: Unlike artificial AI systems, which have fixed architectures designed by humans, biological systems can modify their own computational architecture in response to experience and environmental conditions. This adaptive architecture enables biological systems to optimize their computational capabilities for specific tasks and environments, creating specialized processing systems that are perfectly suited to their particular challenges.
Different organisms demonstrate different "programming paradigms" at the genomic level:
Program: Infect → Reproduce → Die
Trigger: Contact with host cell
Computational simplicity: Limited conditionals, linear execution
Optimization: Maximum efficiency in minimal code
Viruses represent the most minimal form of biological computation, with genomes that are optimized for maximum efficiency in minimal code. The viral "program" is essentially a bootloader that hijacks the host cell's computational machinery to reproduce itself. This minimalism makes viruses excellent models for understanding the fundamental principles of biological computation, as they demonstrate how complex behaviors can emerge from simple, linear programs.
The viral life cycle follows a simple linear sequence: attachment to a host cell, entry into the cell, replication of viral components, assembly of new virus particles, and release from the cell. This linear execution is similar to a simple computer program with minimal branching and no complex control structures. However, even this simple program must handle multiple contingencies, such as different types of host cells, varying environmental conditions, and host immune responses.
The computational efficiency of viruses is remarkable. Some viruses can encode their entire program in fewer than 10,000 nucleotides, yet they can successfully infect, replicate, and spread through host populations. This efficiency is achieved through several strategies: overlapping genes that encode multiple proteins, regulatory sequences that serve multiple functions, and the exploitation of host cell machinery for most computational tasks. This minimalism demonstrates how biological systems can achieve complex outcomes through the efficient use of limited computational resources.
However, this minimalism also creates vulnerabilities. Viruses have limited ability to adapt to changing conditions, and they are highly dependent on their host cells for most computational functions. This dependence makes viruses excellent models for understanding the trade-offs between computational efficiency and robustness, as well as the relationship between program complexity and adaptability.
Program: Eat → Grow → Divide
Loop structure: WHILE food_present DO grow
Event triggers: Mitosis on threshold conditions
State-based logic: Different metabolic states based on environmental conditions
Unicellular organisms represent a more sophisticated form of biological computation, with programs that must balance multiple objectives while operating autonomously in complex environments. Unlike viruses, which are essentially parasites that hijack host machinery, unicellular organisms must implement their own computational infrastructure while also performing the basic functions of life: metabolism, growth, reproduction, and response to environmental changes.
The computational architecture of unicellular organisms is based on state machines that can transition between different metabolic states based on environmental conditions. For example, bacteria can switch between aerobic and anaerobic metabolism, between different carbon sources, and between growth and survival modes. These state transitions are triggered by environmental signals and are implemented through complex regulatory networks that integrate multiple inputs to make decisions about cellular behavior.
The cell cycle represents a fundamental computational loop that drives cellular behavior. This loop includes phases for growth, DNA replication, and cell division, with checkpoints that ensure each phase is completed correctly before proceeding to the next. These checkpoints implement error detection and correction mechanisms that are essential for maintaining genomic integrity. The cell cycle demonstrates how biological systems can implement complex control structures using simple molecular mechanisms.
Unicellular organisms also demonstrate sophisticated signal processing capabilities. They can detect and respond to multiple environmental signals simultaneously, integrating information about nutrient availability, temperature, pH, and the presence of other organisms. This signal integration enables cells to make complex decisions about their behavior, such as whether to grow, divide, form spores, or enter a dormant state. These decision-making processes resemble the control systems used in autonomous robots and other artificial agents.
The computational capabilities of unicellular organisms are particularly impressive given their simplicity. A single bacterial cell can implement complex behaviors such as chemotaxis (movement toward or away from chemicals), quorum sensing (communication with other cells), and biofilm formation (cooperative behavior with other cells). These capabilities demonstrate how biological systems can achieve sophisticated computational outcomes through the coordinated action of simple molecular components.
Subroutines: Cellular differentiation, immune responses
Conditional branches: Hormone levels, cell signaling
Coordinated processes: Development, aging, reproduction
Distributed computation: Different cells executing different aspects of the overall program
Multicellular organisms represent the most complex form of biological computation, with programs that must coordinate the behavior of thousands to trillions of cells while maintaining the integrity and functionality of the entire organism. This coordination requires sophisticated communication systems, hierarchical control structures, and distributed decision-making mechanisms that far exceed the complexity of any artificial distributed system.
The computational architecture of multicellular organisms is based on cellular differentiation, where different cells execute different programs while sharing the same genome. This differentiation is controlled by complex regulatory networks that integrate multiple signals to determine cellular fate. The process of differentiation resembles the creation of specialized subroutines in a computer program, where different components perform different functions while working together to achieve overall system goals.
Communication between cells is essential for coordinating the behavior of multicellular organisms. This communication occurs through multiple mechanisms, including direct cell-to-cell contact, secreted signaling molecules, and electrical signals in the nervous system. These communication systems enable cells to share information about their state, coordinate their activities, and respond collectively to environmental changes. The complexity of these communication networks rivals that of modern computer networks, with multiple protocols, routing mechanisms, and error correction systems.
The immune system represents one of the most sophisticated computational systems in multicellular organisms. It must simultaneously monitor for thousands of different pathogens, learn from encounters with new pathogens, and mount appropriate responses while avoiding attacks on the organism's own cells. This system operates through distributed algorithms that involve multiple cell types, each contributing specialized knowledge and capabilities to the overall immune response. The immune system demonstrates how biological systems can achieve collective intelligence through the coordinated action of many simple components.
Development represents another remarkable computational achievement of multicellular organisms. Starting from a single cell, development creates complex three-dimensional structures with precise spatial organization and functional specialization. This process involves the coordinated action of thousands of genes across millions of cells, with precise timing and spatial control. The computational complexity of development is staggering, involving the simultaneous execution of thousands of parallel processes with complex interdependencies and feedback loops.
The computational capabilities of multicellular organisms are particularly impressive given the challenges they face. They must maintain homeostasis across multiple organ systems, respond to changing environmental conditions, and coordinate complex behaviors such as movement, feeding, and reproduction. These capabilities demonstrate how biological systems can achieve sophisticated computational outcomes through the coordinated action of many simple components, creating emergent properties that exceed the capabilities of any individual component.
The evolution from the author's original 1995 β-galactosidase flowchart to today's sophisticated Mermaid-based visualizations represents not just a technological advancement, but a fundamental transformation in how we create and share biological knowledge. This transformation exemplifies the democratization of computational biology through the convergence of human insight, AI assistance, and modern visualization tools.
In 1995, creating the original β-galactosidase flowchart (Figure 3) was an arduous, month-long process that required:
This process, while thorough, was limited by the tools available and the manual nature of knowledge synthesis. The author, drawing on an education in mathematics and philosophy at Bedford College, London in the 1970s, and working as a web developer and journalist in the 1990s, spent countless hours transforming biological concepts into computational visualizations for a monthly column in The X Advisor, a computer industry trade publication.
Today, the same process that took a month in 1995 can be accomplished in hours or days, thanks to the revolutionary combination of:

2025 Mermaid-Based β-Galactosidase Analysis - Using modern tools and AI assistance, we can now create far more sophisticated and detailed visualizations:
This comparison reveals a profound transformation in scientific practice:
1995 Characteristics:
2025 Capabilities:
The Remarkable Achievement: What once required a month of dedicated work by a trained biologist can now be accomplished in days, with far greater detail and sophistication. Yet this transformation was only possible through the convergence of human biological understanding (rooted in solid educational foundations), innovative visualization tools (Mermaid), and AI assistance (LLMs).
This evolution represents more than just technological progress—it represents the democratization of computational biology. In 1995, creating biological flowcharts required specialized knowledge, significant time investment, and access to academic communities. Today, the combination of educational background, AI assistance, and modern tools enables rapid creation of sophisticated biological visualizations.
The author's journey from manually creating single flowcharts to generating hundreds of detailed biological process diagrams exemplifies how AI can amplify human expertise rather than replace it. The mathematical and philosophical training from Bedford College, combined with decades of experience in journalism and web development, provided the analytical framework necessary to guide AI systems in creating meaningful visualizations. Now at 72 and retired, the author continues the amateur science tradition with vastly improved tools.
Rarely Used for Biological Applications: While Mermaid has been implemented in numerous documentation platforms since its 2014 release, its application to biological process modeling—particularly the systematic extraction of .mmd files from scientific literature by humans and AI working together—represents a novel and innovative use case. This approach transforms static biological knowledge into dynamic, visual computational models.
This work represents a genuine innovation in biological visualization and computational thinking. By systematically applying the Programming Framework methodology to biological processes, we have created:
This innovation bridges the gap between computational thinking and biological understanding, creating new possibilities for research, education, and synthetic biology applications. The transformation from 1995 to 2025 demonstrates how the combination of solid educational foundations, innovative thinking, and modern AI tools can enable individual researchers to make significant contributions to scientific understanding.
The exchange between Welz and Robison in 1995 highlighted a fundamental challenge that persists today: how to visually represent massively parallel processes using tools designed for sequential thinking. The author's β-galactosidase flowchart exemplified both the promise and the problems of this approach.
As Robison noted: "Flowcharts are inherently linear beasts, ill-suited for parallel processes, especially biological ones with many non-linearly combined inputs." Traditional flowcharts suggest a sequence of operations that misrepresents the simultaneous nature of genomic processes.
Contemporary approaches to representing genomic computation have attempted to address these limitations through network diagrams showing interaction rather than sequence, heat maps representing multiple states simultaneously, multi-dimensional representations capturing regulatory relationships, and dynamic simulations rather than static diagrams. However, even these advanced visualization systems struggle with the fundamental challenge identified in 1995: representing true parallelism in comprehensible visual formats.

The visualization challenges raised by Robison's critique of the β-galactosidase flowchart continue to influence how we think about representing biological systems. Modern synthetic biology, systems biology, and computational biology all grapple with the same fundamental tension between the need for clear, understandable representations and the reality of massively parallel, probabilistic biological processes.
While the genome-as-program metaphor provides valuable insights, it is important to acknowledge its limitations and consider alternative perspectives. Several criticisms and challenges have been raised regarding this approach.
A fundamental challenge to the metaphor is the absence of a programmer. Unlike human-written software:
The genome evolved through natural selection; there is no separate "specification" from "implementation"; the "debugging" process (evolution) occurs across generations; the line between program and programmer blurs as the genome modifies itself.
In conventional computing, hardware and software are distinct. In genomic systems: the genome is both the program and the machine that interprets itself; the distinction between "data" and "process" blurs; physical structure and information content are inseparable.
Unlike most computer programs: no central processing unit coordinates execution; no master clock synchronizes operations; no operating system manages resources; control emerges from distributed interactions.
Several alternative metaphors have been proposed for understanding biological systems:
Network Metaphor: Some researchers prefer to view biological systems as complex networks rather than programs, emphasizing the interconnected nature of biological components and the emergent properties that arise from network dynamics.
Ecosystem Metaphor: Others argue that biological systems are better understood as ecosystems, where multiple agents interact in complex ways, creating dynamic equilibria and co-evolutionary processes.
Information Processing Metaphor: An alternative approach focuses on information processing and communication rather than computation, emphasizing how biological systems encode, transmit, and process information.
These alternative perspectives highlight different aspects of biological complexity and may be more appropriate for certain types of analysis. The genome-as-program metaphor should be viewed as one useful framework among many, rather than a complete description of biological reality.
The genome-as-program metaphor has profound implications for both synthetic biology and artificial intelligence.
Viewing the genome as a program enables engineered cells to be written, debugged, and optimized. Synthetic biology gains logic tools to regulate traits, behaviors, and lifecycles. The β-galactosidase flowchart represents an early conceptual bridge toward this engineering approach, demonstrating how biological regulatory circuits can be understood and potentially redesigned using computational logic.
The genomic computational paradigm offers lessons for AI design: massive parallelism with simple components; probabilistic operations with emergent determinism; self-modifying code and execution environment; integration of digital and analog processing.
The Genome Logic Modeling Project (GLMP) aims to formalize the metaphor of the genome as a computer program. It models organisms as logic-executing agents, with internal subroutines and external triggers. GLMP frames biology as structured, conditional, recursive, and state-driven.
Goals and Objectives: The GLMP seeks to create a unified framework for understanding biological systems through computational logic, develop tools for modeling genetic circuits, and establish a collaborative platform for interdisciplinary research. The project aims to bridge the gap between theoretical computational biology and practical applications in synthetic biology and AI.
Expected Outcomes: The GLMP will produce computational models of genetic circuits, visualization tools for genomic logic, educational materials for teaching computational biology, and a community platform for researchers to share insights and collaborate on genomic modeling projects.
This article represents a foundational publication for this project, which will explore topics including: Life as a Running Logic Program; Bootloaders of Life: Zygotic Genome Activation; Subroutines in Biology: Modular Design; Shutdown Protocols: Senescence and Apoptosis; Synthetic Biology Through Logic Gates; Agent-Based Models of Organism Logic.
Concrete Examples of GLMP Research:
The GLMP is designed as an open, collaborative platform that invites researchers, computational biologists, AI specialists, and interested parties from all disciplines to participate in this endeavor. The project recognizes that understanding the genome as a computational system requires diverse perspectives and expertise, from molecular biologists who understand the biochemical details to computer scientists who can formalize computational models.
We encourage contributions in several key areas: (1) Specific Gene Circuit Analysis—detailed computational models of individual genetic circuits, similar to the β-galactosidase example but for other genes and processes; (2) Cross-Species Comparisons—how different organisms implement similar computational functions; (3) Computational Tool Development—software and visualization tools for representing genomic logic; and (4) Integration with Modern AI—connections between genomic computation and contemporary artificial intelligence systems.
The recent announcement of DeepMind's Cell project, led by Demis Hassabis, represents a significant validation of the genome-as-program metaphor and demonstrates how this perspective is gaining traction in the AI community. Like the GLMP, DeepMind's Cell project aims to model cellular processes as computational systems, beginning with the yeast cell as a model organism.
This convergence of approaches is particularly significant because it shows that the computational perspective on biology is not merely a metaphor but a practical framework for understanding and modeling biological systems. The fact that one of the world's leading AI research organizations is pursuing this approach validates the fundamental insights that motivated the GLMP.
The GLMP can complement and extend DeepMind's work by providing a broader theoretical framework and encouraging community participation. While DeepMind focuses on building comprehensive cell models, the GLMP can serve as a platform for researchers to contribute specific computational analyses of genetic circuits, regulatory networks, and cellular processes. This collaborative approach can accelerate progress in both understanding biological computation and developing new computational paradigms.
We invite researchers and enthusiasts to contribute to the GLMP in several ways:
For Molecular Biologists: Share your knowledge of specific genetic circuits and regulatory mechanisms. Help us understand how your research area can be represented as computational logic. Contribute examples of gene regulation that could be modeled as flowcharts or logic circuits.
For Computer Scientists: Develop computational models of genetic processes. Create visualization tools for representing genomic logic. Design algorithms inspired by biological computation. Help formalize the computational languages needed to describe genomic processes.
For AI Researchers: Explore connections between genomic computation and artificial intelligence. Investigate how biological learning and adaptation mechanisms can inform AI design. Develop AI systems that can analyze and model genomic logic.
For Educators: Help develop educational materials that use computational metaphors to teach biology. Create interactive simulations of genetic processes. Bridge the gap between computer science and biology education.
For Enthusiasts: Participate in discussions, share ideas, and help build the GLMP community. Contribute to documentation, visualization, and communication efforts. Help make complex biological concepts accessible to broader audiences.
The GLMP represents an opportunity to fundamentally change how we understand and interact with biological systems. By treating the genome as a computational system, we can develop new tools for understanding life, new approaches to synthetic biology, and new paradigms for computing itself. The time is right for this perspective, as evidenced by the convergence of approaches from multiple research communities.
This metaphor opens several promising research avenues:
Develop specialized notation for genomic computation; create simulation environments based on genomic logic; bridge between biological description and computational models. The insights from early flowcharts like Figure 1 suggest the need for new visual languages that can better represent parallel, probabilistic biological processes.
Design computing systems inspired by genomic parallelism; explore probabilistic processing at massive scale; develop self-modifying execution environments. The scale of parallelism identified by Robbins—exceeding 10^18 processes—suggests computational architectures fundamentally different from current designs.
Teach genomic function using computational metaphors; develop interactive simulations of genomic processes; bridge disciplinary gaps between computer science and biology. The historical progression from simple flowcharts to modern network visualizations illustrates the ongoing challenge of making complex biological computation comprehensible.
The choice of yeast (Saccharomyces cerevisiae) as a model organism for both DeepMind's Cell project and potential GLMP analyses is particularly apt. Yeast represents an ideal intermediate complexity system—more sophisticated than bacteria but simpler than multicellular organisms—making it perfect for developing computational models of cellular processes.
Yeast cells offer several advantages for computational analysis: (1) Well-characterized genome—extensive genetic and biochemical data available; (2) Modular processes—clear separation of cellular functions that can be modeled as computational modules; (3) Experimental tractability—easy to manipulate and observe; and (4) Evolutionary conservation—many processes conserved in higher organisms.
Specific yeast processes that could be modeled as computational systems include: (1) Cell cycle regulation—a complex state machine with checkpoints and feedback loops; (2) Metabolic networks—dynamic systems responding to nutrient availability; (3) Stress response pathways—adaptive systems that modify cellular behavior based on environmental conditions; and (4) Mating type switching—a sophisticated genetic program that controls cellular identity and behavior.
The GLMP community can contribute to this effort by developing computational models of specific yeast processes, creating visualization tools for yeast genetic circuits, and comparing yeast computational logic with that of other organisms. This work can serve as a foundation for understanding more complex cellular systems and provide valuable insights for both basic biology and synthetic biology applications.
Associative Addressing: A memory system where data is found by content rather than location (like finding a book by its subject rather than shelf position).
Probabilistic Op Codes: Computational operations that have a probability of occurring rather than being deterministic (like rolling dice instead of flipping a light switch).
Massive Parallelism: The simultaneous execution of billions of processes, as opposed to sequential processing where operations happen one after another.
Virtual Machine: A computational environment that is defined by the programs it runs, creating a circular dependency between hardware and software.
Zygotic Genome Activation: The "bootloader" process where an embryo transitions from using maternal RNA to transcribing its own genes.
Summary of Key Findings:
The genome is not a static archive but a living program in execution—one that operates on computational principles fundamentally different from those of conventional computers. Each organism runs a massively parallel set of probabilistic processes driven by chemistry, inheritance, and context.
The β-galactosidase flowchart of 1995, while limited in its linear representation, marked an important step in recognizing the computational nature of genetic regulation. The critiques it received—particularly regarding the challenge of representing parallel processes—highlighted fundamental issues that continue to shape how we visualize and understand biological computation today.
As Robert Robbins presciently noted in 1995, "It would be really interesting to think about the computational properties that might emerge in a system with probabilistic op codes and with as much parallelism as biological computers." Nearly three decades later, this observation points toward a rich frontier of research at the intersection of computation and biology.
Implications and Future Directions: By understanding the genome as a unique computational paradigm, we gain insights not only into how life functions but also into new possibilities for computing itself. The Genome Logic Modeling Project (GLMP) provides a framework for advancing this understanding through collaborative research. The genome-as-program metaphor invites us to reimagine biology not only as a science of what life is, but how it computes. The tension between linear representations and parallel realities, first exemplified in early flowcharts, continues to drive innovation in both biological understanding and computational design.