Le Patient est détecté {ma_classe} avec une probabilité de
d'être dans cette classe
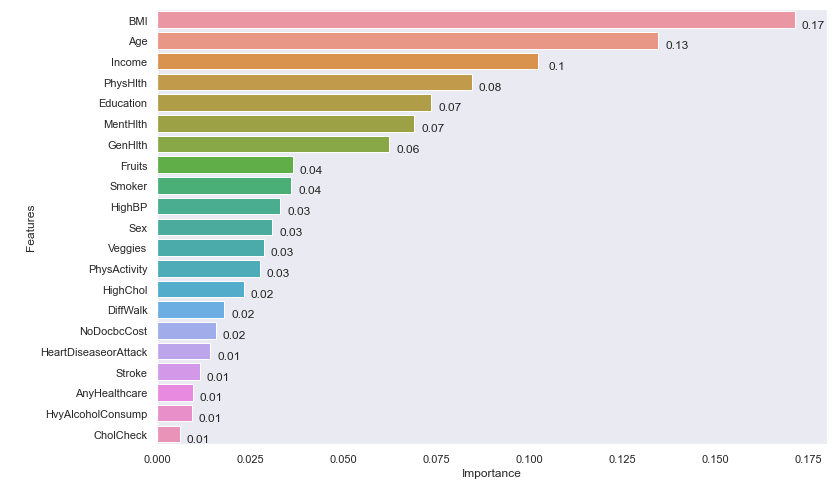
Importance des variables:
"""
)
OUTPUT_PREDANGER = (
STYLE
+ f"""
{round(results["No diabetes"],2)*100}%
{round(results["Prediabetes"],2)*100}%
{round(results["Diabetes"],2)*100}%
Le Patient est détecté {ma_classe} avec une probabilité de
d'être dans cette classe
Importance des variables:

"""
)
OUTPUT_DANGER = (
STYLE
+ f"""
{round(results["No diabetes"],2)*100}%
{round(results["Prediabetes"],2)*100}%
{round(results["Diabetes"],2)*100}%
Le Patient est détecté {ma_classe} avec une probabilité de
d'être dans cette classe
Importance des variables:

"""
)
if ma_classe=="No diabetes" and max_prob > 0.7 :
max_prob = OUTPUT_OK
elif (ma_classe=="No diabetes" and max_prob <= 0.7) or ma_classe=="Prediabetes" :
max_prob = OUTPUT_PREDANGER
else:
max_prob = OUTPUT_DANGER
return max_prob
# Get PDF File for user
def get_data_inputPDF(PDF_file):
path_file = PDF_file.name
with pdfplumber.open(path_file) as pdf:
num_page = 0
page = pdf.pages[num_page]
text = page.extract_text(x_tolerance=2, y_tolerance=0)
AnyHealthcare_pattern = re.compile('^(AnyHealthcare)\s+(\w{3})')
HighBP_pattern = re.compile('^(HighBP)\s+(\w{3})')
NoDocbcCost_pattern = re.compile('^(NoDocbcCost)\s+(\w{3})')
HighChol_pattern = re.compile('^(HighChol)\s+(\w{3})')
GenHlth_pattern = re.compile('^(GenHlth)\s+(\d+)')
CholCheck_pattern = re.compile('^(CholCheck)\s+(\w{3})')
MentHlth_pattern = re.compile('^(MentHlth)\s+(\d+)')
BMI_pattern = re.compile('^(BMI)\s+(\d+)')
PhysHlth_pattern = re.compile('^(PhysHlth)\s+(\d+)')
Smoker_pattern = re.compile('^(Smoker)\s+(\w{3})')
DiffWalk_pattern = re.compile('^(DiffWalk)\s+(\w{3})')
Stroke_pattern = re.compile('^(Stroke)\s+(\w{3})')
Sex_pattern = re.compile('^(Sex)\s+(\w+)')
HeartDiseaseorAttack_pattern = re.compile('^(HeartDiseaseorAttack)\s+(\w{3})')
Age_pattern = re.compile('^(Age)\s+(\d+)')
PhysActivity_pattern = re.compile('^(PhysActivity)\s+(\w{3})')
Education_pattern = re.compile('^(Education)\s+(\d+)')
Fruits_pattern = re.compile('^(Fruits)\s+(\w{3})')
Income_pattern = re.compile('^(Income)\s+(\d+)')
Veggies_pattern = re.compile('^(Veggies)\s+(\w{3})')
HvyAlcoholConsump_pattern = re.compile('^(HvyAlcoholConsump)\s+(\w{3})')
for line in text.split("\n"):
AnyHealthcare_ligne = AnyHealthcare_pattern.search(line)
HighBP_ligne = HighBP_pattern.search(line)
NoDocbcCost_ligne = NoDocbcCost_pattern.search(line)
HighChol_ligne = HighChol_pattern.search(line)
GenHlth_ligne = GenHlth_pattern.search(line)
CholCheck_ligne = CholCheck_pattern.search(line)
MentHlth_ligne = MentHlth_pattern.search(line)
BMI_ligne = BMI_pattern.search(line)
PhysHlth_ligne = PhysHlth_pattern.search(line)
Smoker_ligne = Smoker_pattern.search(line)
DiffWalk_ligne = DiffWalk_pattern.search(line)
Stroke_ligne = Stroke_pattern.search(line)
Sex_ligne = Sex_pattern.search(line)
HeartDiseaseorAttack_ligne = HeartDiseaseorAttack_pattern.search(line)
Age_ligne = Age_pattern.search(line)
PhysActivity_ligne = PhysActivity_pattern.search(line)
Education_ligne = Education_pattern.search(line)
Fruits_ligne = Fruits_pattern.search(line)
Income_ligne = Income_pattern.search(line)
Veggies_ligne = Veggies_pattern.search(line)
HvyAlcoholConsump_ligne = HvyAlcoholConsump_pattern.search(line)
if AnyHealthcare_ligne:
AnyHealthcare = AnyHealthcare_ligne.group(2)
if HvyAlcoholConsump_ligne:
HvyAlcoholConsump = HvyAlcoholConsump_ligne.group(2)
if HighBP_ligne:
HighBP = HighBP_ligne.group(2)
if NoDocbcCost_ligne:
NoDocbcCost = NoDocbcCost_ligne.group(2)
if HighChol_ligne:
HighChol= HighChol_ligne.group(2)
if GenHlth_ligne:
GenHlth = GenHlth_ligne.group(2)
if CholCheck_ligne:
CholCheck = CholCheck_ligne.group(2)
if MentHlth_ligne:
MentHlth = MentHlth_ligne.group(2)
if BMI_ligne:
BMI = BMI_ligne.group(2)
if PhysHlth_ligne:
PhysHlth = PhysHlth_ligne.group(2)
if Smoker_ligne:
Smoker = Smoker_ligne.group(2)
if DiffWalk_ligne:
DiffWalk = DiffWalk_ligne.group(2)
if Stroke_ligne:
Stroke = Stroke_ligne.group(2)
if Sex_ligne:
Sex = Sex_ligne.group(2)
if HeartDiseaseorAttack_ligne:
HeartDiseaseorAttack = HeartDiseaseorAttack_ligne.group(2)
if Age_ligne:
Age = Age_ligne.group(2)
if PhysActivity_ligne:
PhysActivity = PhysActivity_ligne.group(2)
if Education_ligne:
Education = Education_ligne.group(2)
if Fruits_ligne:
Fruits = Fruits_ligne.group(2)
if Income_ligne:
Income = Income_ligne.group(2)
if Veggies_ligne:
Veggies = Veggies_ligne.group(2)
tab = namedtuple('table','HighBP HighChol CholCheck BMI Smoker Stroke HeartDiseaseorAttack \
PhysActivity Fruits Veggies HvyAlcoholConsump AnyHealthcare NoDocbcCost GenHlth \
MentHlth PhysHlth DiffWalk \
Sex Age Education Income')
data_unpreprared = pd.DataFrame([tab(HighBP,HighChol, CholCheck, BMI, Smoker,Stroke,HeartDiseaseorAttack,
PhysActivity, Fruits, Veggies,HvyAlcoholConsump,AnyHealthcare, NoDocbcCost, GenHlth,
MentHlth, PhysHlth, DiffWalk,
Sex, Age, Education,Income)])
# Preprare data type
AnyHealthcare = 0 if AnyHealthcare=="Non" else 1
HvyAlcoholConsump = 0 if HvyAlcoholConsump=="Non" else 1
HighBP = 0 if HighBP=="Non" else 1
NoDocbcCost = 0 if NoDocbcCost=="Non" else 1
HighChol= 0 if HighChol=="Non" else 1
GenHlth = int(GenHlth)
CholCheck = 0 if CholCheck=="Non" else 1
MentHlth = int(MentHlth)
BMI = int(BMI)
PhysHlth = int(PhysHlth)
Smoker = 0 if Smoker=="Non" else 1
DiffWalk = 0 if DiffWalk=="Non" else 1
Stroke = 0 if Stroke=="Non" else 1
Sex = 0 if Sex=="Homme" else 1
HeartDiseaseorAttack = 0 if HeartDiseaseorAttack=="Non" else 1
Age = int(Age)
PhysActivity = 0 if PhysActivity=="Non" else 1
Education = int(Education)
Fruits = 0 if Fruits=="Non" else 1
Income = int(Income)
Veggies = 0 if Veggies=="Non" else 1
user_df = pd.DataFrame([tab(HighBP,HighChol, CholCheck, BMI, Smoker,Stroke,HeartDiseaseorAttack,
PhysActivity, Fruits, Veggies,HvyAlcoholConsump,AnyHealthcare, NoDocbcCost, GenHlth,
MentHlth, PhysHlth, DiffWalk,
Sex, Age, Education,Income)])
# #LOAD MODEL AND TRANSFORMERS
MODELS = load_models()
model = MODELS["model"]
transformers = loading_data()
num_col_trans = transformers["num_col_trans"]
cat_col_trans = transformers["cat_col_trans"]
# Data preparation before fitting to the model
user_df["Age2"] = user_df["Age"]**2
user_df_std = num_col_trans.transform(user_df)
user_df_std = pd.DataFrame(user_df_std ,columns=list(user_df.columns) )
user_df_encoded = cat_col_trans.transform(user_df_std)
user_encoded = np.array(user_df_encoded).reshape((1,-1))
#PREDICTION
target_names = ['No diabetes', 'Prediabetes', 'Diabetes']
pred_proba = model.predict_proba(user_encoded)[0]
results = {classe : pred_proba[i] for i, classe in enumerate(target_names)}
max_prob = 0
ma_classe = ""
for k,v in results.items():
if v > max_prob:
max_prob= v
ma_classe= k
STYLE = """
"""
OUTPUT_OK = (
STYLE
+ f"""
{round(results["No diabetes"],2)*100}%
{round(results["Prediabetes"],2)*100}%
{round(results["Diabetes"],2)*100}%
Le Patient est détecté {ma_classe} avec une probabilité de
d'être dans cette classe
Importance des variables:

"""
)
OUTPUT_PREDANGER = (
STYLE
+ f"""
{round(results["No diabetes"],2)*100}%
{round(results["Prediabetes"],2)*100}%
{round(results["Diabetes"],2)*100}%
Le Patient est détecté {ma_classe} avec une probabilité de
d'être dans cette classe
Importance des variables:

"""
)
OUTPUT_DANGER = (
STYLE
+ f"""
{round(results["No diabetes"],2)*100}%
{round(results["Prediabetes"],2)*100}%
{round(results["Diabetes"],2)*100}%
Le Patient est détecté {ma_classe} avec une probabilité de
d'être dans cette classe
Importance des variables:

"""
)
if ma_classe=="No diabetes" and max_prob > 0.7 :
max_prob = OUTPUT_OK
elif (ma_classe=="No diabetes" and max_prob <= 0.7) or ma_classe=="Prediabetes" :
max_prob = OUTPUT_PREDANGER
else:
max_prob = OUTPUT_DANGER
return data_unpreprared,max_prob
def Prediction_VGG16(image):
#Prepare image
IMG_SIZE = 224
image = img_to_array(image)
image = image*1.0/255.0
img_prepared = image.reshape((-1,IMG_SIZE,IMG_SIZE,3))
#Load model vgg6 package
path = "model_vgg16.h5"
my_model =load_model(path)
#Prediction
classes = ["Brain Tumor","Healthy"]
prediction = my_model.predict(img_prepared)[0]
prediction = prediction.tolist()
results = {k:v for k,v in zip(classes,prediction)}
max_prob = 0
ma_classe = ""
for k,v in results.items():
if v > max_prob:
max_prob= v
ma_classe= k
STYLE = """
"""
OUTPUT_OK = (
STYLE
+ f"""
{round(results["Brain Tumor"],2)*100}%
{round(results["Healthy"],2)*100}%
Le Patient est détecté {ma_classe} avec une probabilité de
d'être dans cette classe
"""
)
OUTPUT_PREDANGER = (
STYLE
+ f"""
{round(results["Brain Tumor"],2)*100}%
{round(results["Healthy"],2)*100}%
Le Patient est détecté {ma_classe} avec une probabilité de
d'être dans cette classe
"""
)
OUTPUT_DANGER = (
STYLE
+ f"""
{round(results["Brain Tumor"],2)*100}%
{round(results["Healthy"],2)*100}%
Le Patient est détecté {ma_classe} avec une probabilité de
d'être dans cette classe
"""
)
if ma_classe=="Healthy" and max_prob > 0.7 :
max_prob = OUTPUT_OK
elif ma_classe=="Healthy" and max_prob <= 0.7:
max_prob = OUTPUT_PREDANGER
else:
max_prob = OUTPUT_DANGER
return max_prob







