Spaces:
Running
title: Medirag
emoji: 🐨
colorFrom: yellow
colorTo: red
sdk: gradio
sdk_version: 5.5.0
app_file: app.py
pinned: false
license: mit

|
MediRAG |
MediRAG is a Retrieval-Augmented Generation (RAG) system designed to make drug-related information more accessible to patients. Many patients struggle with the patient information leaflets that accompany medications, which are often packed with crucial information in small print, leading to confusion about dosages, side effects, and other essential drug details. MedRAG aims to bridge this gap by providing a digital platform where users can easily ask questions and receive clear, understandable answers.
Features
- Bot and Website Development: A user-friendly interface where patients can input questions and receive responses derived from drug guides and patient leaflets.
- Input and Output Guardrails: Safeguards to ensure the reliability, appropriateness, and safety of the information provided, preventing potential misuse.
- DSPy Prompting: Utilizes Dynamic Scripting in Python (DSPy) to create reliable and adaptable prompts for the question-answering process.
- Retrieval-Augmented Generation (RAG) with Semantic Caching: Enhances data retrieval and response time by caching semantically similar queries, ensuring both speed and accuracy.
- Vector Database: Leverages a vector database for efficient retrieval of drug information, enabling quick and precise responses to user queries.
- Observability: Integrated monitoring tools to track system health, retrieval performance, and ensure consistent, high-quality user experiences.
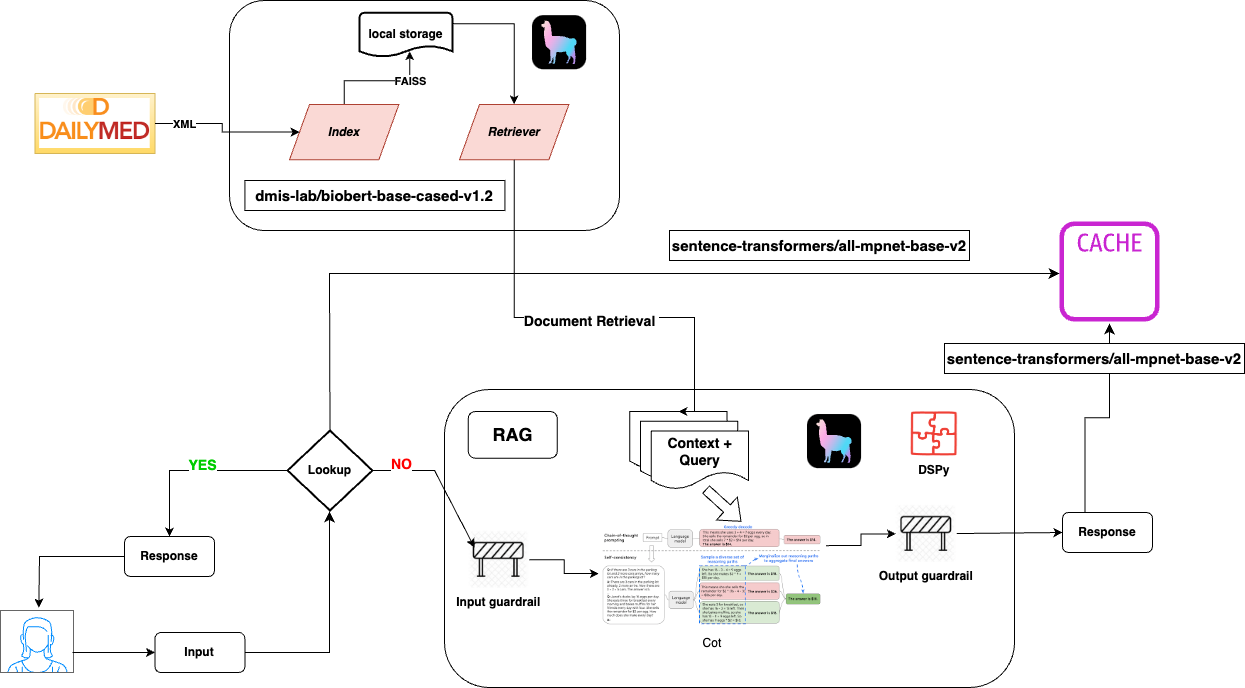
High-Level Architecture
Question-Answering Bot and Website:
- Users can interact with a bot on the website to ask drug-related questions.
- The bot retrieves information from drug guides and patient information leaflets to provide clear and concise answers.
Input and Output Guardrails:
- Implemented to filter inappropriate or potentially harmful queries.
- Ensures the bot's responses are accurate and aligned with medical guidelines.
DSPy Prompting:
- Uses DSPy to dynamically generate prompts that guide the retrieval process.
- Helps in crafting responses that are both contextually relevant and easy to understand.
LlamaIndex streaming workflows:
- Uses LlamaIndex to construct the streaming workflow.
- Helps in crafting responses that are both contextually relevant and easy to understand.
Retrieval-Augmented Generation (RAG) with Semantic Caching:
- Utilizes a RAG model to combine real-time retrieval with language generation.
- Semantic caching improves the response time by reusing answers to similar questions.
Vector Database:
- Employs a vector database for fast and effective retrieval of information.
- Enhances the bot's ability to search and retrieve relevant content from large datasets.
Getting Started
To get started with MedRAG:
- Clone the repository:
git clone https://github.com/alvinhenrick/medirag.git - Create
.envand insert your tokensHF_TOKEN=Your token OPENAI_API_KEY=Your token - Install the required dependencies:
cd medirag poetry install - Run the app:
poetry run python app.py
To-Do List
High Priority
- Implement comprehensive observability tools to monitor and log system performance effectively.
- Explore and implement semantic chunking to enhance retrieval performance and accuracy.
- Build an comprehensive LLM evaluation with respect to Q&A on Drug Label Data.
Medium Priority
- Experiment with different embeddings and other models to enhance retrieval performance and accuracy.
- Experiment with different embeddings and other models to improve the accuracy and relevance of bot responses.
- Index all five DailyMed datasets to ensure complete data coverage and retrieval capabilities.
- https://dailymed-data.nlm.nih.gov/public-release-files/dm_spl_release_human_rx_part1.zip
- https://dailymed-data.nlm.nih.gov/public-release-files/dm_spl_release_human_rx_part2.zip
- https://dailymed-data.nlm.nih.gov/public-release-files/dm_spl_release_human_rx_part3.zip
- https://dailymed-data.nlm.nih.gov/public-release-files/dm_spl_release_human_rx_part4.zip
- https://dailymed-data.nlm.nih.gov/public-release-files/dm_spl_release_human_rx_part5.zip
Low Priority
- Add an LLM agent to further enhance the system’s capabilities and improve interaction dynamics.
License
This project is licensed under the MIT License - see the LICENSE file for details.