# Description
A pre-trained model for the endoscopic tool segmentation task.
# Model Overview
This model is trained using a flexible unet structure with an efficient-b2 [1] as the backbone and a UNet architecture [2] as the decoder. Datasets use private samples from [Activ Surgical](https://www.activsurgical.com/).
The [pytorch model](https://drive.google.com/file/d/14r6WmzaZrgaWLGu0O9vSAzdeIGVFQ3cs/view?usp=sharing) and [torchscript model](https://drive.google.com/file/d/1i-e5xXHtmvmqitwUP8Q3JqvnmN3mlrEm/view?usp=sharing) are shared in google drive. Details can be found in large_files.yml file. Modify the "bundle_root" parameter specified in configs/train.json and configs/inference.json to reflect where models are downloaded. Expected directory path to place downloaded models is "models/" under "bundle_root".
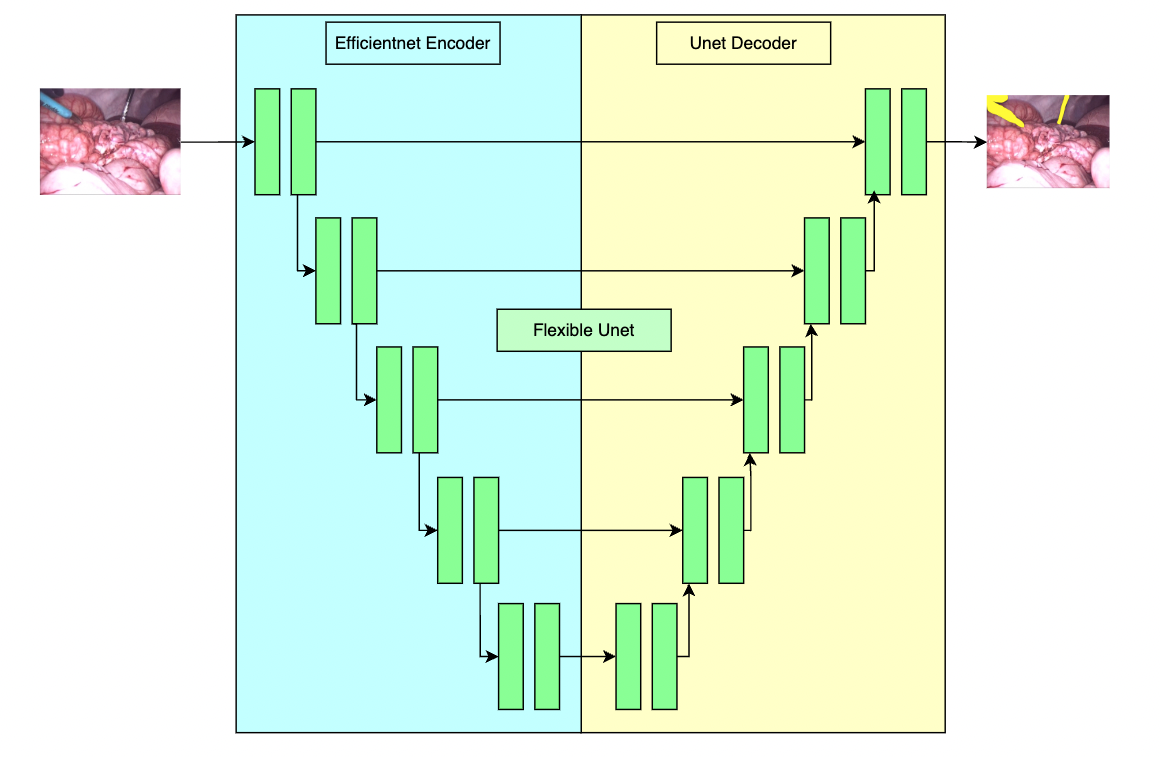
## Data
Datasets used in this work were provided by [Activ Surgical](https://www.activsurgical.com/).
Since datasets are private, existing public datasets like [EndoVis 2017](https://endovissub2017-roboticinstrumentsegmentation.grand-challenge.org/Data/) can be used to train a similar model.
When using EndoVis or any other dataset, it should be divided into "train", "valid" and "test" folders. Samples in each folder would better be images and converted to jpg format. Otherwise, "images", "labels", "val_images" and "val_labels" parameters in "configs/train.json" and "datalist" in "configs/inference.json" should be modified to fit given dataset. After that, "dataset_dir" parameter in "configs/train.json" and "configs/inference.json" should be changed to root folder which contains previous "train", "valid" and "test" folders.
Please notice that loading data operation in this bundle is adaptive. If images and labels are not in the same format, it may lead to a mismatching problem. For example, if images are in jpg format and labels are in npy format, PIL and Numpy readers will be used separately to load images and labels. Since these two readers have their own way to parse file's shape, loaded labels will be transpose of the correct ones and incur a missmatching problem.
## Training configuration
The training was performed with an at least 12GB-memory GPU.
Actual Model Input: 736 x 480 x 3
## Input and output formats
Input: 3 channel video frames
Output: 2 channels: Label 1: tools; Label 0: everything else
## Scores
This model achieves the following IoU score on the test dataset (our own split from the first batch data):
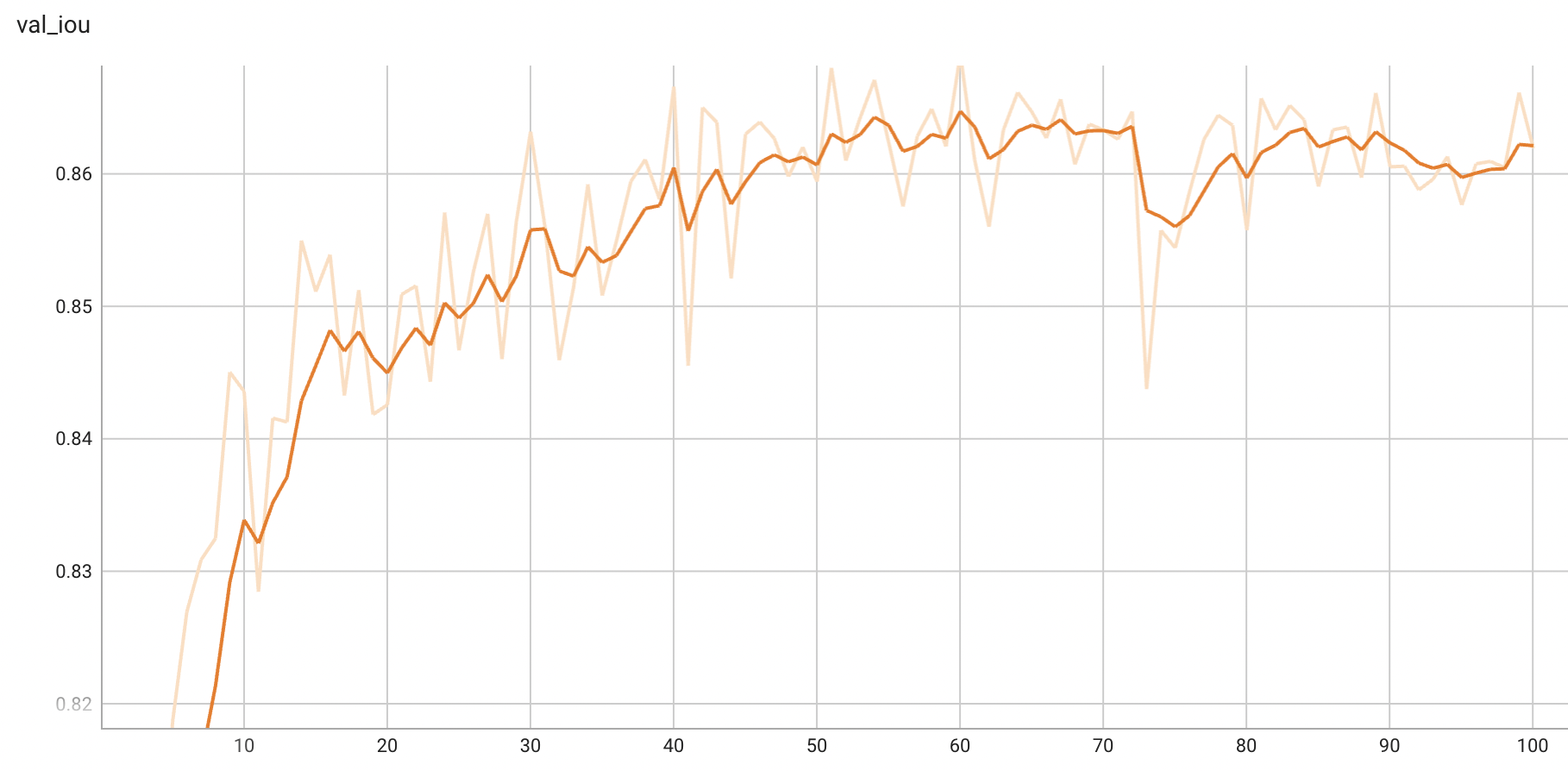
Mean IoU = 0.87
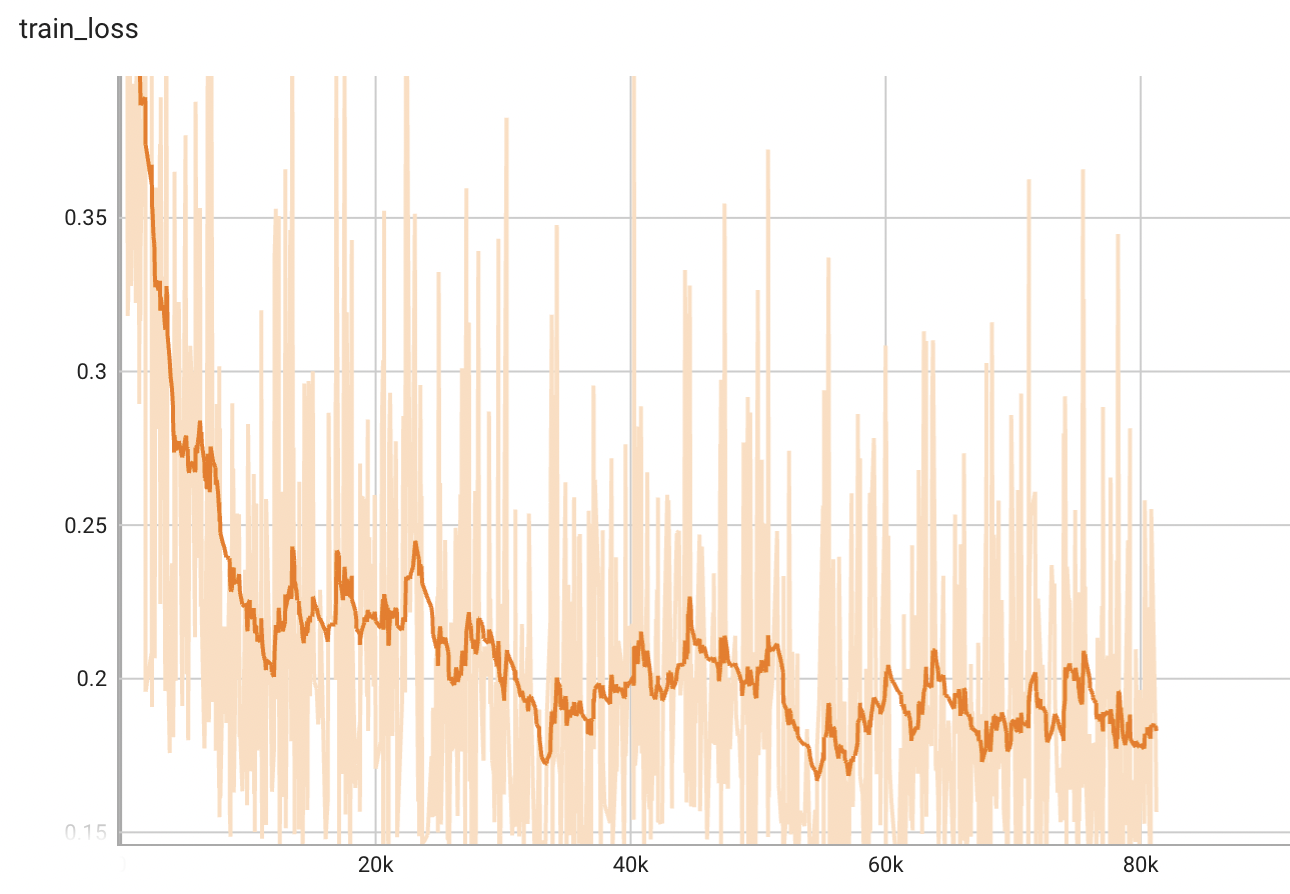
## Training Performance
A graph showing the training loss over 100 epochs.
## Validation Performance
A graph showing the validation mean IoU over 100 epochs.
## commands example
Execute training:
```
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
```
Override the `train` config to execute evaluation with the trained model:
```
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
```
Execute inference:
```
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
```
Export checkpoint to TorchScript file:
```
python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.json
```
# References
[1] Tan, M. and Le, Q. V. Efficientnet: Rethinking model scaling for convolutional neural networks. ICML, 2019a. https://arxiv.org/pdf/1905.11946.pdf
[2] O. Ronneberger, P. Fischer, and T. Brox. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention, pages 234–241. Springer, 2015. https://arxiv.org/pdf/1505.04597.pdf
# License
Copyright (c) MONAI Consortium
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.