---
license: mit
language:
- en
tags:
- medical
- vision
widget:
- src: "https://huggingface.co/flaviagiammarino/pubmed-clip-vit-base-patch32/resolve/main/scripts/input.jpeg"
candidate_labels: "Chest X-Ray, Brain MRI, Abdomen CT Scan"
example_title: "Abdomen CT Scan"
---
# Model Card for PubMedCLIP
PubMedCLIP is a fine-tuned version of [CLIP](https://huggingface.co/docs/transformers/model_doc/clip) for the medical domain.
## Model Description
PubMedCLIP was trained on the [Radiology Objects in COntext (ROCO)](https://github.com/razorx89/roco-dataset) dataset, a large-scale multimodal medical imaging dataset.
The ROCO dataset includes diverse imaging modalities (such as X-Ray, MRI, ultrasound, fluoroscopy, etc.) from various human body regions (such as head, spine, chest, abdomen, etc.)
captured from open-access [PubMed](https://pubmed.ncbi.nlm.nih.gov/) articles.
PubMedCLIP was trained for 50 epochs with a batch size of 64 using the Adam optimizer with a learning rate of 10−5.
The authors have released three different pre-trained models at this [link](https://1drv.ms/u/s!ApXgPqe9kykTgwD4Np3-f7ODAot8?e=zLVlJ2)
which use ResNet-50, ResNet-50x4 and ViT32 as image encoders. This repository includes only the ViT32 variant of the PubMedCLIP model.
- **Repository:** [PubMedCLIP Official GitHub Repository](https://github.com/sarahESL/PubMedCLIP)
- **Paper:** [Does CLIP Benefit Visual Question Answering in the Medical Domain as Much as it Does in the General Domain?](https://arxiv.org/abs/2112.13906)
## Usage
```python
import requests
from PIL import Image
import matplotlib.pyplot as plt
from transformers import CLIPProcessor, CLIPModel
model = CLIPModel.from_pretrained("flaviagiammarino/pubmed-clip-vit-base-patch32")
processor = CLIPProcessor.from_pretrained("flaviagiammarino/pubmed-clip-vit-base-patch32")
url = "https://huggingface.co/flaviagiammarino/pubmed-clip-vit-base-patch32/resolve/main/scripts/input.jpeg"
image = Image.open(requests.get(url, stream=True).raw)
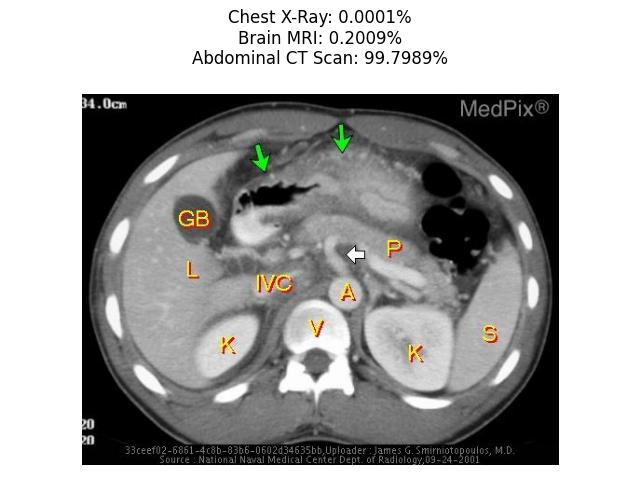
text = ["Chest X-Ray", "Brain MRI", "Abdominal CT Scan"]
inputs = processor(text=text, images=image, return_tensors="pt", padding=True)
probs = model(**inputs).logits_per_image.softmax(dim=1).squeeze()
plt.subplots()
plt.imshow(image)
plt.title("".join([x[0] + ": " + x[1] + "\n" for x in zip(text, [format(prob, ".4%") for prob in probs])]))
plt.axis("off")
plt.tight_layout()
plt.show()
```
## Additional Information
### Licensing Information
The authors have released the model code and pre-trained checkpoints under the [MIT License](https://github.com/sarahESL/PubMedCLIP/blob/main/LICENSE).
### Citation Information
```
@article{eslami2021does,
title={Does clip benefit visual question answering in the medical domain as much as it does in the general domain?},
author={Eslami, Sedigheh and de Melo, Gerard and Meinel, Christoph},
journal={arXiv preprint arXiv:2112.13906},
year={2021}
}
```