---
license: cc-by-4.0
---
# TreeSatAI-Time-Series
****
This dataset was introduced in the [ECCV24 paper](https://arxiv.org/pdf/2404.08351) Omnisat.
Ahlswede et al. (https://essd.copernicus.org/articles/15/681/2023/) introduced the TreeSatAI Benchmark Archive, a new dataset for tree species classification in Central Europe based on multi-sensor data from aerial,
Sentinel-1 and Sentinel-2. The dataset contains labels of 20 European tree species (*i.e.*, 15 tree genera) derived from forest administration data of the federal state of Lower Saxony, Germany.
The authors propose models and guidelines for the application of the latest machine learning techniques for the task of tree species classification with multi-label data.
Finally, they provide various benchmark experiments showcasing the information which can be derived from the different sensors including artificial neural networks and tree-based machine learning methods.
The hereby proposed dataset is an extension of the existing dataset TreeSatAI by Ahlswede et al.
While the original dataset only grants access to a single Sentinel-1 & -2 image for each patch, this new dataset compiles all available Sentinel-1 & -2 data spanning a year.
This integration of temporal information assists in distinguishing between different tree species.
Notably, we aligned the year of the Sentinel Time Series with that of the aerial patch if it was 2017 or later.
For preceding years, considering minimal changes in the forest and the need for sufficient temporal context, we specifically chose the year 2017.
****
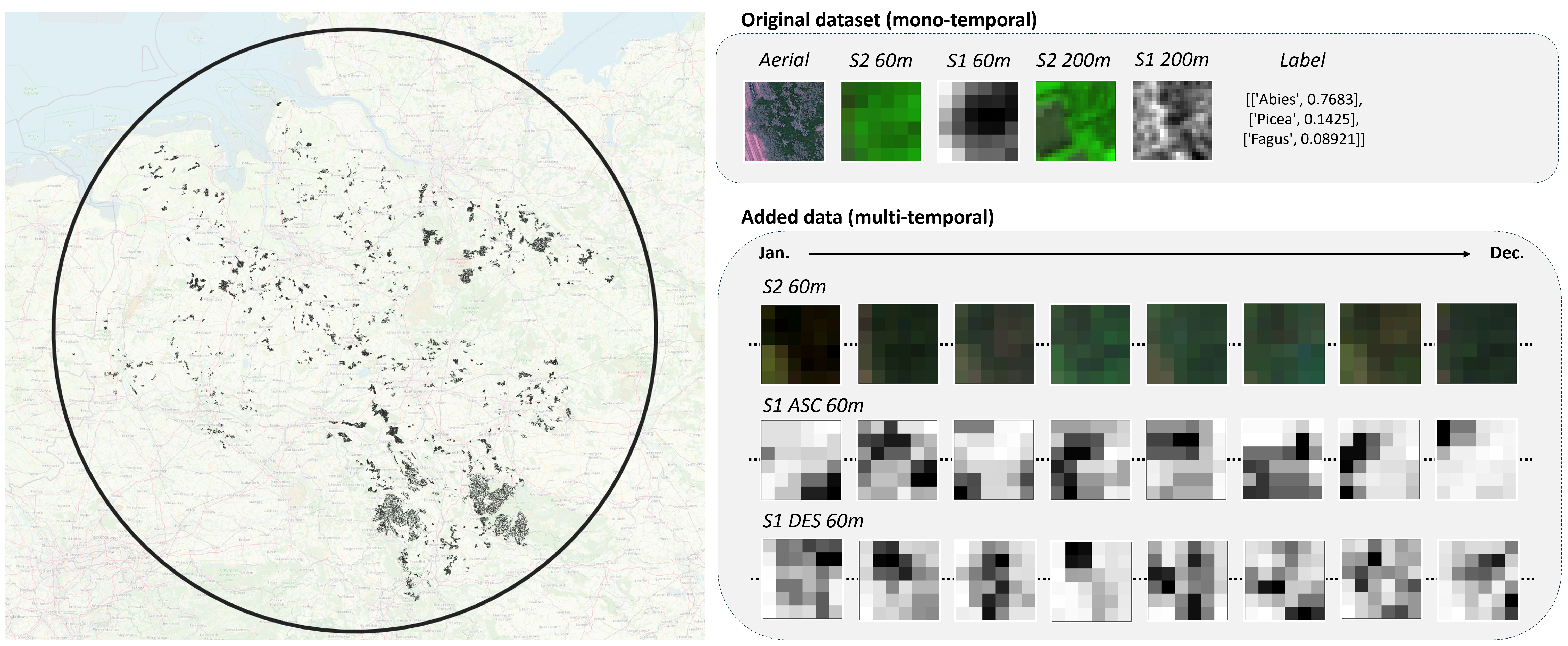 ****
The dataset covers 50 381 patches of 60mx60m located in Germany.
****
The dataset covers 50 381 patches of 60mx60m located in Germany.
The following zip files are available :
📦 **aerial** (from the original dataset): aerial acquisitions at 0.2m spatial resolution with RGB and Infrared bands.
📦 **sentinel** (from the original dataset): the single acquisition of Sentinel-1 & -2 covering the patch extent (60m) or a wider area (200m)
📦 **sentinel-ts**: the yearly time series of Sentinel-1 & -2.
📦 **labels** (from the original dataset): patchwise labels of present tree species and proprotion.
📦 **geojson** (from the original dataset): vector file providing geographical location of the patches.
📦 **split** (from the original dataset): train, val and tests patches split.
****
The **Sentinel Time Series** are provided for each patch in HDF format (.h5) with several datasets :
sen-1-asc-data : Sentinel-1 ascending orbit backscattering coefficient data (Tx2x6x6) | Channels: VV, VH
sen-1-asc-products : Sentinel-1 ascending orbit product names (T)
sen-1-des-data: Sentinel-1 descending orbit backscattering coefficient data (Tx2x6x6) | Channels: VV, VH
sen-1-des-data : Sentinel-1 ascending orbit product names (T)
sen-2-data : Sentinel-2 Level-2 BOA reflectances (Tx10x6x6) | Channels: B02,B03,B04,B05,B06,B07,B08,B8A,B11,B12
sen-2-masks : Sentinel-2 cloud cover masks (Tx2x6x6) | Channels: snow probability, cloud probability
sen-2-products : Sentinel-2 product names (T)
Sentinel product names follow the official naming convention from the European Space Agency.
To access the Sentinel Time Series data in python you can use :
```
import h5py
with h5py.File(path/to/file.h5, 'r') as h5file:
sen_1_asc_data = h5file['sen-1-asc-data'][:]
sen_1_asc_products = h5file['sen-1-asc-products'][:]
sen_1_des_data = h5file['sen-1-des-data'][:]
sen_1_des_products = h5file['sen-1-des-products'][:]
sen_2_data = h5file['sen-2-data'][:]
sen_2_products = h5file['sen-2-products'][:]
sen_2_masks = h5file['sen-2-masks'][:]
```
****
### Licence
This dataset is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
### Contact
If you have any questions, issues or feedback, you can contact us at: ai-challenge@ign.fr
### Citation
```
@article{astruc2024omnisat,
title={OmniSat: Self-Supervised Modality Fusion for Earth Observation},
author={Astruc, Guillaume and Gonthier, Nicolas and Mallet, Clement and Landrieu, Loic},
journal={ECCV},
year={2024}
}
```  ****
The dataset covers 50 381 patches of 60mx60m located in Germany.
****
The dataset covers 50 381 patches of 60mx60m located in Germany.